hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ecb308205a93bb7d267dfb3607532159f20e15a0 | 2,855 | ipynb | Jupyter Notebook | 0_mean_median_mode.ipynb | ayushsubedi/10daysofstatistics | a9719781d91b4bee975125155290df4d752bf748 | [
"MIT"
] | null | null | null | 0_mean_median_mode.ipynb | ayushsubedi/10daysofstatistics | a9719781d91b4bee975125155290df4d752bf748 | [
"MIT"
] | null | null | null | 0_mean_median_mode.ipynb | ayushsubedi/10daysofstatistics | a9719781d91b4bee975125155290df4d752bf748 | [
"MIT"
] | 1 | 2021-03-20T07:42:39.000Z | 2021-03-20T07:42:39.000Z | 17.955975 | 78 | 0.440981 | [
[
[
"## Brute Force",
"_____no_output_____"
]
],
[
[
"# Enter your code here. Read input from STDIN. Print output to STDOUT\nN = int(input())\nraw_X = input()\nX = []\nfor x in raw_X.split(\" \"):\n X.append(int(x))\n\n# mean\nsum_ = 0\nfor x in X:\n sum_ = sum_ + x\nprint ('%.1f'%(sum_/N))\n\n# median\nX.sort()\nif len(X)%2==0:\n median = (X[(len(X)//2)-1]+X[len(X)//2])/2\nelse:\n median = X[len(X)//2]\n\nprint ('%.1f'%median)\n\n# mode\nmode_list = []\nfor i in X:\n mode_list.append(X.count(i))\n \nmode = X[mode_list.index(max(mode_list))]\nprint (mode)\n \n",
" 10\n 64630 11735 14216 99233 14470 4978 73429 38120 51135 67060\n"
],
[
"test1 = [x for x in range(101)]",
"_____no_output_____"
]
],
[
[
"## Difference between plus equals and equals plus",
"_____no_output_____"
]
],
[
[
"sum_ = 0\nfor x in test1:\n sum_ += x\nprint (sum_)",
"5050\n"
],
[
"sum_ = 0\nfor x in test1:\n sum_ =+ x\nprint (sum_)",
"100\n"
]
],
[
[
"## Better solutions",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
ecb30ad8829d513d28de451ad36de76ebe1b2cbd | 6,233 | ipynb | Jupyter Notebook | 01-basics/09-backpropagation.ipynb | shuhua-wang/pytorch-tutorials | 83665071dfcac247a0de547fe792f2f818544c87 | [
"MIT"
] | null | null | null | 01-basics/09-backpropagation.ipynb | shuhua-wang/pytorch-tutorials | 83665071dfcac247a0de547fe792f2f818544c87 | [
"MIT"
] | null | null | null | 01-basics/09-backpropagation.ipynb | shuhua-wang/pytorch-tutorials | 83665071dfcac247a0de547fe792f2f818544c87 | [
"MIT"
] | null | null | null | 32.128866 | 332 | 0.535376 | [
[
[
"### 09. Backpropagation",
"_____no_output_____"
],
[
"#### Table of Contents\n\n- [1. Chian rule](#heading09-1)\n- [2. Computational graph and local gradients](#heading09-2)\n- [3. Forward and backward pass](#heading09-3)\n- [4. Backpropagation with pytorch code](#heading09-4)",
"_____no_output_____"
],
[
"<a id=\"heading09-1\"></a>\n\n#### 1. Chian rule\n\n<img src=\"./images/chain_rule.png\" alt=\"chain_rule\" width=\"400\"/>\n\n**Chian rule** is a formula to compute the derivative of a composite function. If a variable $z$ depends on the variable $y$, $z=f(y)$, where the $y$ itself depends on the variable $x$, $y=f(x)$. The derivative of $z$ with respect to $x$ can be calculated by:\n\n$\\frac{\\partial z}{\\partial x}=\\frac{\\partial z}{\\partial y} \\frac{\\partial y}{\\partial x}$\n\n<a id=\"heading09-2\"></a>\n\n#### 2. Computational graph and local gradients\n\n<img src=\"./images/computational_graph.png\" alt=\"computational_graph\" width=\"400\"/>\n\n**A computational graph** is defined as a directed graph where the nodes correspond to mathematical operations. Computational graphs are a way of expressing and evaluating a mathematical expression.\n\nIf we assume that the $z$ is a function of variables $x$ and $y$, $z=f(x,y)=xy$. The $Loss$ is a function of $z$, $Loss=f(z)$. Assume that we have already known the derivative of $Loss$ with respect to $z$ is equal to $\\frac{\\partial Loss}{\\partial z}$, how can we calculate the derivative of $Loss$ with respect to $x$?\n\nFirst, we can obtain the **local gradients** (the derivative of $z$ with respect to $x$ or $y$):\n\n$\\frac{\\partial z}{\\partial x}=\\frac{\\partial xy}{\\partial x}=y$, $\\frac{\\partial z}{\\partial y}=\\frac{\\partial xy}{\\partial y}=x$\n\nThen, based on the **chian rule**, the derivative of $Loss$ with respect to $x$ is given by:\n\n$\\frac{\\partial Loss}{\\partial x}=\\frac{\\partial Loss}{\\partial z} \\frac{\\partial z}{\\partial x}=y\\frac{\\partial Loss}{\\partial z}$\n\n<a id=\"heading09-3\"></a>\n\n#### 3. Forward and backward pass\n\nThe backpropagation consists of 3 steps:\n - (1) forward pass: compute loss\n - (2) compute local gradients\n - (3) backward pass: compute _dLoss/dWeights_ using the chain rule\n\n<img src=\"./images/forward_backward_pass.png\" alt=\"forward_backward_pass\" width=\"400\"/>\n\n**Linear regression:**\n - The linear regression is defines as: $y=wx$\n - The prediction value is represented as: $\\hat{y}$\n - Error between predicted and real value is: $s=\\hat{y}-y$\n - Loss function is defined as squared error: $Loss=(\\hat{y}-y)^{2}$\n - Given an example as: $(x=1, y=2)$, the initial weight: $w=1$\n\n**Forward pass:**\n - $\\hat{y}=wx=1 \\times 1=1$\n - $s=\\hat{y}-y=1-2=-1$\n - $Loss=(\\hat{y}-y)^{2}=(-1)^{2}=1$\n\n**Local gradients:**\n - $\\frac{\\partial Loss}{\\partial s}=\\frac{\\partial s^{2}}{\\partial s}=2s$\n - $\\frac{\\partial s}{\\partial \\hat{y}}=\\frac{\\partial (\\hat{y}-y)}{\\partial \\hat{y}}=1$\n - $\\frac{\\partial \\hat{y}}{\\partial w}=\\frac{\\partial wx}{\\partial w}=x$\n\n**Backward pass:** using the chian rule to compute _dLoss/dWeights_\n - $\\frac{\\partial Loss}{\\partial w}=\\frac{\\partial Loss}{\\partial s} \\cdot \\frac{\\partial s}{\\partial \\hat{y}} \\cdot \\frac{\\partial \\hat{y}}{\\partial w}=2sx=2 \\times (-1) \\times 1=-2$\n \n <a id=\"heading09-4\"></a>\n\n#### 4. Backpropagation with pytorch code",
"_____no_output_____"
]
],
[
[
"import torch",
"_____no_output_____"
],
[
"if torch.cuda.is_available():\n device = torch.device(\"cuda\")\nelse:\n device = torch.device(\"cpu\")\n\n# define x, y and initial weights\nx = torch.tensor(1.0, device=device)\ny = torch.tensor(2.0, device=device)\nw = torch.tensor(1.0, device=device, requires_grad=True)\nprint(x)\nprint(y)\nprint(w)",
"tensor(1., device='cuda:0')\ntensor(2., device='cuda:0')\ntensor(1., device='cuda:0', requires_grad=True)\n"
],
[
"# forward pass and compute the loss\ny_hat = w * x\nloss = (y_hat - y)**2\nprint(loss)",
"tensor(1., device='cuda:0', grad_fn=<PowBackward0>)\n"
],
[
"# backward pass\nloss.backward()\nprint(w.grad)\n\n# update weights\n# next forward and backward",
"tensor(-2., device='cuda:0')\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecb3159e05d4e15b764d5df7c03df73d66a2fd6e | 20,074 | ipynb | Jupyter Notebook | Notebooks/connor_notebooks/toll_scrape/toll_parser_illinois.ipynb | standroidbeta/data-science | 8f18133e676ac2ea334fadea8894ec4453f286cc | [
"MIT"
] | null | null | null | Notebooks/connor_notebooks/toll_scrape/toll_parser_illinois.ipynb | standroidbeta/data-science | 8f18133e676ac2ea334fadea8894ec4453f286cc | [
"MIT"
] | 1 | 2021-06-02T00:23:07.000Z | 2021-06-02T00:23:07.000Z | Notebooks/connor_notebooks/toll_scrape/toll_parser_illinois.ipynb | Labs17-RVNav/rvnav-ds | d357dc78f2efc51d65b0fd5f378e6fd4d1916ca4 | [
"MIT"
] | 2 | 2019-08-21T00:11:51.000Z | 2019-09-17T20:27:37.000Z | 30.415152 | 220 | 0.342084 | [
[
[
"import pandas as pd\nfrom bs4 import BeautifulSoup\nimport requests",
"_____no_output_____"
],
[
"website_url = requests.get('https://www.illinoistollway.com/tolling-information#2019Rates').text",
"_____no_output_____"
],
[
"soup = BeautifulSoup(website_url, 'html.parser')",
"_____no_output_____"
],
[
"my_table = soup.find('div', {'class':'page-content'})",
"_____no_output_____"
],
[
"smaller_table = my_table.find('div', {'class':'columns-1'})",
"_____no_output_____"
],
[
"table_table = smaller_table.find('div', {'class':'portlet-boundary portlet-boundary_56_ portlet-static portlet-static-end portlet-borderless portlet-journal-content ', 'id': 'p_p_id_56_INSTANCE_bltNHBO1zL1X_'})",
"_____no_output_____"
],
[
"highway_lst = []\nhighway_lst.append(table_table.h3.text)",
"_____no_output_____"
],
[
"table_body = table_table.tbody",
"_____no_output_____"
],
[
"tolling_location = table_body.find_all('td', {'align':'left'})",
"_____no_output_____"
],
[
"tolling_location_list = []\nfor i in tolling_location:\n tolling_location_list.append(i.text)",
"_____no_output_____"
]
],
[
[
"### Jane Addams Toll Locations",
"_____no_output_____"
]
],
[
[
"tolling_location_list",
"_____no_output_____"
],
[
"all_times = table_body.find_all('td', {'align':'center'})",
"_____no_output_____"
],
[
"# HTML tags containing Jane Addams table\nall_times",
"_____no_output_____"
],
[
"catch_all = []\nfor i in all_times:\n catch_all.append(i.text)",
"_____no_output_____"
],
[
"catch_all",
"_____no_output_____"
]
],
[
[
"### Create DataFrame (Jane Addams)",
"_____no_output_____"
]
],
[
[
"df_jane = pd.DataFrame(columns=['id', 'Autos: I-Pass All Times', 'Autos: I-Pass Cash', 'Small Trucks Daytime',\n 'Medium Trucks Daytime', 'Large Trucks Daytime', 'Small Trucks Overnight', \n 'Medium Trucks Overnight', 'Large Trucks Overnight'])",
"_____no_output_____"
],
[
"def divide_chunks(l, n): \n \n # looping till length l \n for i in range(0, len(l), n): \n yield l[i:i + n] ",
"_____no_output_____"
],
[
"n = 9\n\nx = list(divide_chunks(catch_all, n))",
"_____no_output_____"
],
[
"df_jane = pd.DataFrame(x, columns=['id', 'Autos: I-Pass All Times', 'Autos: I-Pass Cash', 'Small Trucks Daytime',\n 'Medium Trucks Daytime', 'Large Trucks Daytime', 'Small Trucks Overnight', \n 'Medium Trucks Overnight', 'Large Trucks Overnight'])",
"_____no_output_____"
],
[
"df_jane.head()",
"_____no_output_____"
],
[
"df_jane['Location'] = tolling_location_list",
"_____no_output_____"
],
[
"df_jane.head()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb31f0117fc329940007a29e6c7546bb1586f2a | 53,172 | ipynb | Jupyter Notebook | nbs/03_xtras.ipynb | abhinavm24/fastcore | b8598c3be540168e4ba58195b0f6eaedf7fa27e1 | [
"Apache-2.0"
] | null | null | null | nbs/03_xtras.ipynb | abhinavm24/fastcore | b8598c3be540168e4ba58195b0f6eaedf7fa27e1 | [
"Apache-2.0"
] | null | null | null | nbs/03_xtras.ipynb | abhinavm24/fastcore | b8598c3be540168e4ba58195b0f6eaedf7fa27e1 | [
"Apache-2.0"
] | null | null | null | 26.244817 | 257 | 0.51892 | [
[
[
"#default_exp xtras",
"_____no_output_____"
],
[
"#export\nfrom fastcore.imports import *\nfrom fastcore.foundation import *\nfrom fastcore.basics import *\nfrom functools import wraps\n\nimport mimetypes,pickle,random,json,subprocess,shlex,bz2,gzip,zipfile,tarfile\nimport imghdr,struct,distutils.util,tempfile,time,string,collections\nfrom contextlib import contextmanager,ExitStack\nfrom pdb import set_trace\nfrom datetime import datetime, timezone\nfrom timeit import default_timer",
"_____no_output_____"
],
[
"from fastcore.test import *\nfrom nbdev.showdoc import *\nfrom fastcore.nb_imports import *\nfrom time import sleep",
"_____no_output_____"
]
],
[
[
"# Utility functions\n\n> Utility functions used in the fastai library",
"_____no_output_____"
],
[
"## Collections",
"_____no_output_____"
]
],
[
[
"#export\ndef dict2obj(d):\n \"Convert (possibly nested) dicts (or lists of dicts) to `AttrDict`\"\n if isinstance(d, (L,list)): return L(d).map(dict2obj)\n if not isinstance(d, dict): return d\n return AttrDict(**{k:dict2obj(v) for k,v in d.items()})",
"_____no_output_____"
]
],
[
[
"This is a convenience to give you \"dotted\" access to (possibly nested) dictionaries, e.g:",
"_____no_output_____"
]
],
[
[
"d1 = dict(a=1, b=dict(c=2,d=3))\nd2 = dict2obj(d1)\ntest_eq(d2.b.c, 2)\ntest_eq(d2.b['c'], 2)",
"_____no_output_____"
]
],
[
[
"It can also be used on lists of dicts.",
"_____no_output_____"
]
],
[
[
"_list_of_dicts = [d1, d1]\nds = dict2obj(_list_of_dicts)\ntest_eq(ds[0].b.c, 2)",
"_____no_output_____"
],
[
"#export\ndef obj2dict(d):\n \"Convert (possibly nested) AttrDicts (or lists of AttrDicts) to `dict`\"\n if isinstance(d, (L,list)): return list(L(d).map(obj2dict))\n if not isinstance(d, dict): return d\n return dict(**{k:obj2dict(v) for k,v in d.items()})",
"_____no_output_____"
]
],
[
[
"`obj2dict` can be used to reverse what is done by `dict2obj`:",
"_____no_output_____"
]
],
[
[
"test_eq(obj2dict(d2), d1)\ntest_eq(obj2dict(ds), _list_of_dicts) ",
"_____no_output_____"
],
[
"#export\ndef _repr_dict(d, lvl):\n if isinstance(d,dict):\n its = [f\"{k}: {_repr_dict(v,lvl+1)}\" for k,v in d.items()]\n elif isinstance(d,(list,L)): its = [_repr_dict(o,lvl+1) for o in d]\n else: return str(d)\n return '\\n' + '\\n'.join([\" \"*(lvl*2) + \"- \" + o for o in its])",
"_____no_output_____"
],
[
"#export\ndef repr_dict(d):\n \"Print nested dicts and lists, such as returned by `dict2obj`\"\n return _repr_dict(d,0).strip()",
"_____no_output_____"
],
[
"print(repr_dict(d2))",
"- a: 1\n- b: \n - c: 2\n - d: 3\n"
]
],
[
[
"`repr_dict` is used to display `AttrDict` both with `repr` and in Jupyter Notebooks:",
"_____no_output_____"
]
],
[
[
"#export\n@patch\ndef __repr__(self:AttrDict): return repr_dict(self)\n\nAttrDict._repr_markdown_ = AttrDict.__repr__",
"_____no_output_____"
],
[
"print(repr(d2))",
"- a: 1\n- b: \n - c: 2\n - d: 3\n"
],
[
"d2",
"_____no_output_____"
],
[
"#export\ndef is_listy(x):\n \"`isinstance(x, (tuple,list,L,slice,Generator))`\"\n return isinstance(x, (tuple,list,L,slice,Generator))",
"_____no_output_____"
],
[
"assert is_listy((1,))\nassert is_listy([1])\nassert is_listy(L([1]))\nassert is_listy(slice(2))\nassert not is_listy(array([1]))",
"_____no_output_____"
],
[
"#export\ndef shufflish(x, pct=0.04):\n \"Randomly relocate items of `x` up to `pct` of `len(x)` from their starting location\"\n n = len(x)\n return L(x[i] for i in sorted(range_of(x), key=lambda o: o+n*(1+random.random()*pct)))",
"_____no_output_____"
],
[
"#export\ndef mapped(f, it):\n \"map `f` over `it`, unless it's not listy, in which case return `f(it)`\"\n return L(it).map(f) if is_listy(it) else f(it)",
"_____no_output_____"
],
[
"def _f(x,a=1): return x-a\n\ntest_eq(mapped(_f,1),0)\ntest_eq(mapped(_f,[1,2]),[0,1])\ntest_eq(mapped(_f,(1,)),(0,))",
"_____no_output_____"
]
],
[
[
"## Reindexing Collections",
"_____no_output_____"
]
],
[
[
"#export\n#hide\nclass IterLen:\n \"Base class to add iteration to anything supporting `__len__` and `__getitem__`\"\n def __iter__(self): return (self[i] for i in range_of(self))",
"_____no_output_____"
],
[
"#export\n@docs\nclass ReindexCollection(GetAttr, IterLen):\n \"Reindexes collection `coll` with indices `idxs` and optional LRU cache of size `cache`\"\n _default='coll'\n def __init__(self, coll, idxs=None, cache=None, tfm=noop):\n if idxs is None: idxs = L.range(coll)\n store_attr()\n if cache is not None: self._get = functools.lru_cache(maxsize=cache)(self._get)\n\n def _get(self, i): return self.tfm(self.coll[i])\n def __getitem__(self, i): return self._get(self.idxs[i])\n def __len__(self): return len(self.coll)\n def reindex(self, idxs): self.idxs = idxs\n def shuffle(self): random.shuffle(self.idxs)\n def cache_clear(self): self._get.cache_clear()\n def __getstate__(self): return {'coll': self.coll, 'idxs': self.idxs, 'cache': self.cache, 'tfm': self.tfm}\n def __setstate__(self, s): self.coll,self.idxs,self.cache,self.tfm = s['coll'],s['idxs'],s['cache'],s['tfm']\n\n _docs = dict(reindex=\"Replace `self.idxs` with idxs\",\n shuffle=\"Randomly shuffle indices\",\n cache_clear=\"Clear LRU cache\")",
"_____no_output_____"
],
[
"show_doc(ReindexCollection, title_level=4)",
"_____no_output_____"
]
],
[
[
"This is useful when constructing batches or organizing data in a particular manner (i.e. for deep learning). This class is primarly used in organizing data for language models in fastai.",
"_____no_output_____"
],
[
"You can supply a custom index upon instantiation with the `idxs` argument, or you can call the `reindex` method to supply a new index for your collection.\n\nHere is how you can reindex a list such that the elements are reversed:",
"_____no_output_____"
]
],
[
[
"rc=ReindexCollection(['a', 'b', 'c', 'd', 'e'], idxs=[4,3,2,1,0])\nlist(rc)",
"_____no_output_____"
]
],
[
[
"Alternatively, you can use the `reindex` method:",
"_____no_output_____"
]
],
[
[
"show_doc(ReindexCollection.reindex, title_level=6)",
"_____no_output_____"
],
[
"rc=ReindexCollection(['a', 'b', 'c', 'd', 'e'])\nrc.reindex([4,3,2,1,0])\nlist(rc)",
"_____no_output_____"
]
],
[
[
"You can optionally specify a LRU cache, which uses [functools.lru_cache](https://docs.python.org/3/library/functools.html#functools.lru_cache) upon instantiation:",
"_____no_output_____"
]
],
[
[
"sz = 50\nt = ReindexCollection(L.range(sz), cache=2)\n\n#trigger a cache hit by indexing into the same element multiple times\nt[0], t[0]\nt._get.cache_info()",
"_____no_output_____"
]
],
[
[
"You can optionally clear the LRU cache by calling the `cache_clear` method:",
"_____no_output_____"
]
],
[
[
"show_doc(ReindexCollection.cache_clear, title_level=5)",
"_____no_output_____"
],
[
"sz = 50\nt = ReindexCollection(L.range(sz), cache=2)\n\n#trigger a cache hit by indexing into the same element multiple times\nt[0], t[0]\nt.cache_clear()\nt._get.cache_info()",
"_____no_output_____"
],
[
"show_doc(ReindexCollection.shuffle, title_level=5)",
"_____no_output_____"
]
],
[
[
"Note that an ordered index is automatically constructed for the data structure even if one is not supplied.",
"_____no_output_____"
]
],
[
[
"rc=ReindexCollection(['a', 'b', 'c', 'd', 'e', 'f', 'g', 'h'])\nrc.shuffle()\nlist(rc)",
"_____no_output_____"
],
[
"sz = 50\nt = ReindexCollection(L.range(sz), cache=2)\ntest_eq(list(t), range(sz))\ntest_eq(t[sz-1], sz-1)\ntest_eq(t._get.cache_info().hits, 1)\nt.shuffle()\ntest_eq(t._get.cache_info().hits, 1)\ntest_ne(list(t), range(sz))\ntest_eq(set(t), set(range(sz)))\nt.cache_clear()\ntest_eq(t._get.cache_info().hits, 0)\ntest_eq(t.count(0), 1)",
"_____no_output_____"
],
[
"#hide\n#Test ReindexCollection pickles\nt1 = pickle.loads(pickle.dumps(t))\ntest_eq(list(t), list(t1))",
"_____no_output_____"
]
],
[
[
"## File Functions",
"_____no_output_____"
],
[
"Utilities (other than extensions to Pathlib.Path) for dealing with IO.",
"_____no_output_____"
]
],
[
[
"# export\n@contextmanager\ndef maybe_open(f, mode='r', **kwargs):\n \"Context manager: open `f` if it is a path (and close on exit)\"\n if isinstance(f, (str,os.PathLike)):\n with open(f, mode, **kwargs) as f: yield f\n else: yield f",
"_____no_output_____"
]
],
[
[
"This is useful for functions where you want to accept a path *or* file. `maybe_open` will not close your file handle if you pass one in.",
"_____no_output_____"
]
],
[
[
"def _f(fn):\n with maybe_open(fn) as f: return f.encoding\n\nfname = '00_test.ipynb'\nsys_encoding = 'cp1252' if sys.platform == 'win32' else 'UTF-8'\ntest_eq(_f(fname), sys_encoding)\nwith open(fname) as fh: test_eq(_f(fh), sys_encoding)",
"_____no_output_____"
]
],
[
[
"For example, we can use this to reimplement [`imghdr.what`](https://docs.python.org/3/library/imghdr.html#imghdr.what) from the Python standard library, which is [written in Python 3.9](https://github.com/python/cpython/blob/3.9/Lib/imghdr.py#L11) as:",
"_____no_output_____"
]
],
[
[
"def what(file, h=None):\n f = None\n try:\n if h is None:\n if isinstance(file, (str,os.PathLike)):\n f = open(file, 'rb')\n h = f.read(32)\n else:\n location = file.tell()\n h = file.read(32)\n file.seek(location)\n for tf in imghdr.tests:\n res = tf(h, f)\n if res: return res\n finally:\n if f: f.close()\n return None",
"_____no_output_____"
]
],
[
[
"Here's an example of the use of this function:",
"_____no_output_____"
]
],
[
[
"fname = 'images/puppy.jpg'\nwhat(fname)",
"_____no_output_____"
]
],
[
[
"With `maybe_open`, `Self`, and `L.map_first`, we can rewrite this in a much more concise and (in our opinion) clear way:",
"_____no_output_____"
]
],
[
[
"def what(file, h=None):\n if h is None:\n with maybe_open(file, 'rb') as f: h = f.peek(32)\n return L(imghdr.tests).map_first(Self(h,file))",
"_____no_output_____"
]
],
[
[
"...and we can check that it still works:",
"_____no_output_____"
]
],
[
[
"test_eq(what(fname), 'jpeg')",
"_____no_output_____"
]
],
[
[
"...along with the version passing a file handle:",
"_____no_output_____"
]
],
[
[
"with open(fname,'rb') as f: test_eq(what(f), 'jpeg')",
"_____no_output_____"
]
],
[
[
"...along with the `h` parameter version:",
"_____no_output_____"
]
],
[
[
"with open(fname,'rb') as f: test_eq(what(None, h=f.read(32)), 'jpeg')",
"_____no_output_____"
],
[
"def _jpg_size(f):\n size,ftype = 2,0\n while not 0xc0 <= ftype <= 0xcf:\n f.seek(size, 1)\n byte = f.read(1)\n while ord(byte) == 0xff: byte = f.read(1)\n ftype = ord(byte)\n size = struct.unpack('>H', f.read(2))[0] - 2\n f.seek(1, 1) # `precision'\n h,w = struct.unpack('>HH', f.read(4))\n return w,h\n\ndef _gif_size(f): return struct.unpack('<HH', head[6:10])\n\ndef _png_size(f):\n assert struct.unpack('>i', head[4:8])[0]==0x0d0a1a0a\n return struct.unpack('>ii', head[16:24])",
"_____no_output_____"
],
[
"#export\ndef image_size(fn):\n \"Tuple of (w,h) for png, gif, or jpg; `None` otherwise\"\n d = dict(png=_png_size, gif=_gif_size, jpeg=_jpg_size)\n with maybe_open(fn, 'rb') as f: return d[imghdr.what(f)](f)",
"_____no_output_____"
],
[
"test_eq(image_size(fname), (1200,803))",
"_____no_output_____"
],
[
"#export\ndef bunzip(fn):\n \"bunzip `fn`, raising exception if output already exists\"\n fn = Path(fn)\n assert fn.exists(), f\"{fn} doesn't exist\"\n out_fn = fn.with_suffix('')\n assert not out_fn.exists(), f\"{out_fn} already exists\"\n with bz2.BZ2File(fn, 'rb') as src, out_fn.open('wb') as dst:\n for d in iter(lambda: src.read(1024*1024), b''): dst.write(d)",
"_____no_output_____"
],
[
"f = Path('files/test.txt')\nif f.exists(): f.unlink()\nbunzip('files/test.txt.bz2')\nt = f.open().readlines()\ntest_eq(len(t),1)\ntest_eq(t[0], 'test\\n')\nf.unlink()",
"_____no_output_____"
],
[
"#export\ndef join_path_file(file, path, ext=''):\n \"Return `path/file` if file is a string or a `Path`, file otherwise\"\n if not isinstance(file, (str, Path)): return file\n path.mkdir(parents=True, exist_ok=True)\n return path/f'{file}{ext}'",
"_____no_output_____"
],
[
"path = Path.cwd()/'_tmp'/'tst'\nf = join_path_file('tst.txt', path)\nassert path.exists()\ntest_eq(f, path/'tst.txt')\nwith open(f, 'w') as f_: assert join_path_file(f_, path) == f_\nshutil.rmtree(Path.cwd()/'_tmp')",
"_____no_output_____"
],
[
"#export\ndef loads(s, cls=None, object_hook=None, parse_float=None,\n parse_int=None, parse_constant=None, object_pairs_hook=None, **kw):\n \"Same as `json.loads`, but handles `None`\"\n if not s: return {}\n return json.loads(s, cls=cls, object_hook=object_hook, parse_float=parse_float,\n parse_int=parse_int, parse_constant=parse_constant, object_pairs_hook=object_pairs_hook, **kw)",
"_____no_output_____"
],
[
"#export\ndef loads_multi(s:str):\n \"Generator of >=0 decoded json dicts, possibly with non-json ignored text at start and end\"\n _dec = json.JSONDecoder()\n while s.find('{')>=0:\n s = s[s.find('{'):]\n obj,pos = _dec.raw_decode(s)\n if not pos: raise ValueError(f'no JSON object found at {pos}')\n yield obj\n s = s[pos:]",
"_____no_output_____"
],
[
"tst = \"\"\"\n# ignored\n{ \"a\":1 }\nhello\n{\n\"b\":2\n}\n\"\"\"\n\ntest_eq(list(loads_multi(tst)), [{'a': 1}, {'b': 2}])",
"_____no_output_____"
],
[
"#export\ndef untar_dir(file, dest):\n with tempfile.TemporaryDirectory(dir='.') as d:\n d = Path(d)\n with tarfile.open(mode='r:gz', fileobj=file) as t: t.extractall(d)\n next(d.iterdir()).rename(dest)",
"_____no_output_____"
],
[
"#export\ndef repo_details(url):\n \"Tuple of `owner,name` from ssh or https git repo `url`\"\n res = remove_suffix(url.strip(), '.git')\n res = res.split(':')[-1]\n return res.split('/')[-2:]",
"_____no_output_____"
],
[
"test_eq(repo_details('https://github.com/fastai/fastai.git'), ['fastai', 'fastai'])\ntest_eq(repo_details('[email protected]:fastai/nbdev.git\\n'), ['fastai', 'nbdev'])",
"_____no_output_____"
],
[
"#export\ndef run(cmd, *rest, ignore_ex=False, as_bytes=False, stderr=False):\n \"Pass `cmd` (splitting with `shlex` if string) to `subprocess.run`; return `stdout`; raise `IOError` if fails\"\n if rest: cmd = (cmd,)+rest\n elif isinstance(cmd,str): cmd = shlex.split(cmd)\n res = subprocess.run(cmd, stdout=subprocess.PIPE, stderr=subprocess.PIPE)\n stdout = res.stdout\n if stderr and res.stderr: stdout += b' ;; ' + res.stderr\n if not as_bytes: stdout = stdout.decode().strip()\n if ignore_ex: return (res.returncode, stdout)\n if res.returncode: raise IOError(stdout)\n return stdout",
"_____no_output_____"
]
],
[
[
"You can pass a string (which will be split based on standard shell rules), a list, or pass args directly:",
"_____no_output_____"
]
],
[
[
"if sys.platform == 'win32':\n assert 'ipynb' in run('cmd /c dir /p')\n assert 'ipynb' in run(['cmd', '/c', 'dir', '/p'])\n assert 'ipynb' in run('cmd', '/c', 'dir', '/p')\nelse:\n assert 'ipynb' in run('ls -ls')\n assert 'ipynb' in run(['ls', '-l'])\n assert 'ipynb' in run('ls', '-l')",
"_____no_output_____"
]
],
[
[
"Some commands fail in non-error situations, like `grep`. Use `ignore_ex` in those cases, which will return a tuple of stdout and returncode:",
"_____no_output_____"
]
],
[
[
"if sys.platform == 'win32':\n test_eq(run('cmd /c findstr asdfds 00_test.ipynb', ignore_ex=True)[0], 1)\nelse:\n test_eq(run('grep asdfds 00_test.ipynb', ignore_ex=True)[0], 1)",
"_____no_output_____"
]
],
[
[
"`run` automatically decodes returned bytes to a `str`. Use `as_bytes` to skip that:",
"_____no_output_____"
]
],
[
[
"if sys.platform == 'win32':\n # why I ignore as_types, because every time nbdev_clean_nbs will update \\n to \\nn\n test_eq(run('cmd /c echo hi'), 'hi')\nelse:\n test_eq(run('echo hi', as_bytes=True), b'hi\\n')",
"_____no_output_____"
],
[
"#export\ndef open_file(fn, mode='r', **kwargs):\n \"Open a file, with optional compression if gz or bz2 suffix\"\n if isinstance(fn, io.IOBase): return fn\n fn = Path(fn)\n if fn.suffix=='.bz2': return bz2.BZ2File(fn, mode, **kwargs)\n elif fn.suffix=='.gz' : return gzip.GzipFile(fn, mode, **kwargs)\n elif fn.suffix=='.zip': return zipfile.ZipFile(fn, mode, **kwargs)\n else: return open(fn,mode, **kwargs)",
"_____no_output_____"
],
[
"#export\ndef save_pickle(fn, o):\n \"Save a pickle file, to a file name or opened file\"\n with open_file(fn, 'wb') as f: pickle.dump(o, f)",
"_____no_output_____"
],
[
"#export\ndef load_pickle(fn):\n \"Load a pickle file from a file name or opened file\"\n with open_file(fn, 'rb') as f: return pickle.load(f)",
"_____no_output_____"
],
[
"for suf in '.pkl','.bz2','.gz':\n # delete=False is added for Windows. https://stackoverflow.com/questions/23212435/permission-denied-to-write-to-my-temporary-file\n with tempfile.NamedTemporaryFile(suffix=suf, delete=False) as f:\n fn = Path(f.name)\n save_pickle(fn, 't')\n t = load_pickle(fn)\n f.close()\n test_eq(t,'t')",
"_____no_output_____"
]
],
[
[
"## Extensions to Pathlib.Path",
"_____no_output_____"
],
[
"The following methods are added to the standard python libary [Pathlib.Path](https://docs.python.org/3/library/pathlib.html#basic-use).",
"_____no_output_____"
]
],
[
[
"#export\n@patch\ndef readlines(self:Path, hint=-1, encoding='utf8'):\n \"Read the content of `self`\"\n with self.open(encoding=encoding) as f: return f.readlines(hint)",
"_____no_output_____"
],
[
"#export\n@patch\ndef read_json(self:Path, encoding=None, errors=None):\n \"Same as `read_text` followed by `loads`\"\n return loads(self.read_text(encoding=encoding, errors=errors))",
"_____no_output_____"
],
[
"#export\n@patch\ndef mk_write(self:Path, data, encoding=None, errors=None, mode=511):\n \"Make all parent dirs of `self`, and write `data`\"\n self.parent.mkdir(exist_ok=True, parents=True, mode=mode)\n self.write_text(data, encoding=encoding, errors=errors)",
"_____no_output_____"
],
[
"#export\n@patch\ndef ls(self:Path, n_max=None, file_type=None, file_exts=None):\n \"Contents of path as a list\"\n extns=L(file_exts)\n if file_type: extns += L(k for k,v in mimetypes.types_map.items() if v.startswith(file_type+'/'))\n has_extns = len(extns)==0\n res = (o for o in self.iterdir() if has_extns or o.suffix in extns)\n if n_max is not None: res = itertools.islice(res, n_max)\n return L(res)",
"_____no_output_____"
]
],
[
[
"We add an `ls()` method to `pathlib.Path` which is simply defined as `list(Path.iterdir())`, mainly for convenience in REPL environments such as notebooks.",
"_____no_output_____"
]
],
[
[
"path = Path()\nt = path.ls()\nassert len(t)>0\nt1 = path.ls(10)\ntest_eq(len(t1), 10)\nt2 = path.ls(file_exts='.ipynb')\nassert len(t)>len(t2)\nt[0]",
"_____no_output_____"
]
],
[
[
"You can also pass an optional `file_type` MIME prefix and/or a list of file extensions.",
"_____no_output_____"
]
],
[
[
"lib_path = (path/'../fastcore')\ntxt_files=lib_path.ls(file_type='text')\nassert len(txt_files) > 0 and txt_files[0].suffix=='.py'\nipy_files=path.ls(file_exts=['.ipynb'])\nassert len(ipy_files) > 0 and ipy_files[0].suffix=='.ipynb'\ntxt_files[0],ipy_files[0]",
"_____no_output_____"
],
[
"#hide\npath = Path()\npkl = pickle.dumps(path)\np2 = pickle.loads(pkl)\ntest_eq(path.ls()[0], p2.ls()[0])",
"_____no_output_____"
],
[
"#export\n@patch\ndef __repr__(self:Path):\n b = getattr(Path, 'BASE_PATH', None)\n if b:\n try: self = self.relative_to(b)\n except: pass\n return f\"Path({self.as_posix()!r})\"",
"_____no_output_____"
]
],
[
[
"fastai also updates the `repr` of `Path` such that, if `Path.BASE_PATH` is defined, all paths are printed relative to that path (as long as they are contained in `Path.BASE_PATH`:",
"_____no_output_____"
]
],
[
[
"t = ipy_files[0].absolute()\ntry:\n Path.BASE_PATH = t.parent.parent\n test_eq(repr(t), f\"Path('nbs/{t.name}')\")\nfinally: Path.BASE_PATH = None",
"_____no_output_____"
]
],
[
[
"## Other Helpers",
"_____no_output_____"
]
],
[
[
"#export\ndef truncstr(s:str, maxlen:int, suf:str='…', space='')->str:\n \"Truncate `s` to length `maxlen`, adding suffix `suf` if truncated\"\n return s[:maxlen-len(suf)]+suf if len(s)+len(space)>maxlen else s+space",
"_____no_output_____"
],
[
"w = 'abacadabra'\ntest_eq(truncstr(w, 10), w)\ntest_eq(truncstr(w, 5), 'abac…')\ntest_eq(truncstr(w, 5, suf=''), 'abaca')\ntest_eq(truncstr(w, 11, space='_'), w+\"_\")\ntest_eq(truncstr(w, 10, space='_'), w[:-1]+'…')\ntest_eq(truncstr(w, 5, suf='!!'), 'aba!!')",
"_____no_output_____"
],
[
"#export\nspark_chars = '▁▂▃▅▆▇'",
"_____no_output_____"
],
[
"#export\ndef _ceil(x, lim=None): return x if (not lim or x <= lim) else lim\n\ndef _sparkchar(x, mn, mx, incr, empty_zero):\n if x is None or (empty_zero and not x): return ' '\n if incr == 0: return spark_chars[0]\n res = int((_ceil(x,mx)-mn)/incr-0.5)\n return spark_chars[res]",
"_____no_output_____"
],
[
"#export\ndef sparkline(data, mn=None, mx=None, empty_zero=False):\n \"Sparkline for `data`, with `None`s (and zero, if `empty_zero`) shown as empty column\"\n valid = [o for o in data if o is not None]\n if not valid: return ' '\n mn,mx,n = ifnone(mn,min(valid)),ifnone(mx,max(valid)),len(spark_chars)\n res = [_sparkchar(x=o, mn=mn, mx=mx, incr=(mx-mn)/n, empty_zero=empty_zero) for o in data]\n return ''.join(res)",
"_____no_output_____"
],
[
"data = [9,6,None,1,4,0,8,15,10]\nprint(f'without \"empty_zero\": {sparkline(data, empty_zero=False)}')\nprint(f' with \"empty_zero\": {sparkline(data, empty_zero=True )}')",
"without \"empty_zero\": ▅▂ ▁▂▁▃▇▅\n with \"empty_zero\": ▅▂ ▁▂ ▃▇▅\n"
]
],
[
[
"You can set a maximum and minimum for the y-axis of the sparkline with the arguments `mn` and `mx` respectively:",
"_____no_output_____"
]
],
[
[
"sparkline([1,2,3,400], mn=0, mx=3)",
"_____no_output_____"
],
[
"#export\ndef autostart(g):\n \"Decorator that automatically starts a generator\"\n @functools.wraps(g)\n def f():\n r = g()\n next(r)\n return r\n return f",
"_____no_output_____"
],
[
"#export\nclass EventTimer:\n \"An event timer with history of `store` items of time `span`\"\n def __init__(self, store=5, span=60):\n self.hist,self.span,self.last = collections.deque(maxlen=store),span,default_timer()\n self._reset()\n\n def _reset(self): self.start,self.events = self.last,0\n\n def add(self, n=1):\n \"Record `n` events\"\n if self.duration>self.span:\n self.hist.append(self.freq)\n self._reset()\n self.events +=n\n self.last = default_timer()\n\n @property\n def duration(self): return default_timer()-self.start\n @property\n def freq(self): return self.events/self.duration",
"_____no_output_____"
],
[
"show_doc(EventTimer, title_level=4)",
"_____no_output_____"
]
],
[
[
"Add events with `add`, and get number of `events` and their frequency (`freq`).",
"_____no_output_____"
]
],
[
[
"# Random wait function for testing\ndef _randwait(): yield from (sleep(random.random()/200) for _ in range(100))\n\nc = EventTimer(store=5, span=0.03)\nfor o in _randwait(): c.add(1)\nprint(f'Num Events: {c.events}, Freq/sec: {c.freq:.01f}')\nprint('Most recent: ', sparkline(c.hist), *L(c.hist).map('{:.01f}'))",
"Num Events: 8, Freq/sec: 423.0\nMost recent: ▂▂▁▁▇ 318.5 319.0 266.9 275.6 427.7\n"
],
[
"#export\n_fmt = string.Formatter()",
"_____no_output_____"
],
[
"#export\ndef stringfmt_names(s:str)->list:\n \"Unique brace-delimited names in `s`\"\n return uniqueify(o[1] for o in _fmt.parse(s) if o[1])",
"_____no_output_____"
],
[
"s = '/pulls/{pull_number}/reviews/{review_id}'\ntest_eq(stringfmt_names(s), ['pull_number','review_id'])",
"_____no_output_____"
],
[
"#export\nclass PartialFormatter(string.Formatter):\n \"A `string.Formatter` that doesn't error on missing fields, and tracks missing fields and unused args\"\n def __init__(self):\n self.missing = set()\n super().__init__()\n\n def get_field(self, nm, args, kwargs):\n try: return super().get_field(nm, args, kwargs)\n except KeyError:\n self.missing.add(nm)\n return '{'+nm+'}',nm\n\n def check_unused_args(self, used, args, kwargs):\n self.xtra = filter_keys(kwargs, lambda o: o not in used)",
"_____no_output_____"
],
[
"show_doc(PartialFormatter, title_level=4)",
"_____no_output_____"
],
[
"#export\ndef partial_format(s:str, **kwargs):\n \"string format `s`, ignoring missing field errors, returning missing and extra fields\"\n fmt = PartialFormatter()\n res = fmt.format(s, **kwargs)\n return res,list(fmt.missing),fmt.xtra",
"_____no_output_____"
]
],
[
[
"The result is a tuple of `(formatted_string,missing_fields,extra_fields)`, e.g:",
"_____no_output_____"
]
],
[
[
"res,missing,xtra = partial_format(s, pull_number=1, foo=2)\ntest_eq(res, '/pulls/1/reviews/{review_id}')\ntest_eq(missing, ['review_id'])\ntest_eq(xtra, {'foo':2})",
"_____no_output_____"
],
[
"#export\ndef utc2local(dt:datetime)->datetime:\n \"Convert `dt` from UTC to local time\"\n return dt.replace(tzinfo=timezone.utc).astimezone(tz=None)",
"_____no_output_____"
],
[
"dt = datetime(2000,1,1,12)\nprint(f'{dt} UTC is {utc2local(dt)} local time')",
"2000-01-01 12:00:00 UTC is 2000-01-01 12:00:00+00:00 local time\n"
],
[
"#export\ndef local2utc(dt:datetime)->datetime:\n \"Convert `dt` from local to UTC time\"\n return dt.replace(tzinfo=None).astimezone(tz=timezone.utc)",
"_____no_output_____"
],
[
"print(f'{dt} local is {local2utc(dt)} UTC time')",
"2000-01-01 12:00:00 local is 2000-01-01 12:00:00+00:00 UTC time\n"
],
[
"#export\ndef trace(f):\n \"Add `set_trace` to an existing function `f`\"\n if getattr(f, '_traced', False): return f\n def _inner(*args,**kwargs):\n set_trace()\n return f(*args,**kwargs)\n _inner._traced = True\n return _inner",
"_____no_output_____"
]
],
[
[
"You can add a breakpoint to an existing function, e.g:\n\n```python\nPath.cwd = trace(Path.cwd)\nPath.cwd()\n```\n\nNow, when the function is called it will drop you into the debugger. Note, you must issue the `s` command when you begin to step into the function that is being traced.",
"_____no_output_____"
]
],
[
[
"#export\ndef round_multiple(x, mult, round_down=False):\n \"Round `x` to nearest multiple of `mult`\"\n def _f(x_): return (int if round_down else round)(x_/mult)*mult\n res = L(x).map(_f)\n return res if is_listy(x) else res[0]",
"_____no_output_____"
],
[
"test_eq(round_multiple(63,32), 64)\ntest_eq(round_multiple(50,32), 64)\ntest_eq(round_multiple(40,32), 32)\ntest_eq(round_multiple( 0,32), 0)\ntest_eq(round_multiple(63,32, round_down=True), 32)\ntest_eq(round_multiple((63,40),32), (64,32))",
"_____no_output_____"
],
[
"#export\n@contextmanager\ndef modified_env(*delete, **replace):\n \"Context manager temporarily modifying `os.environ` by deleting `delete` and replacing `replace`\"\n prev = dict(os.environ)\n try:\n os.environ.update(replace)\n for k in delete: os.environ.pop(k, None)\n yield\n finally:\n os.environ.clear()\n os.environ.update(prev)",
"_____no_output_____"
],
[
"# USER isn't in Cloud Linux Environments\nenv_test = 'USERNAME' if sys.platform == \"win32\" else 'SHELL'\noldusr = os.environ[env_test]\n\nreplace_param = {env_test: 'a'}\nwith modified_env('PATH', **replace_param):\n test_eq(os.environ[env_test], 'a')\n assert 'PATH' not in os.environ\n\nassert 'PATH' in os.environ\ntest_eq(os.environ[env_test], oldusr)",
"_____no_output_____"
],
[
"#export\nclass ContextManagers(GetAttr):\n \"Wrapper for `contextlib.ExitStack` which enters a collection of context managers\"\n def __init__(self, mgrs): self.default,self.stack = L(mgrs),ExitStack()\n def __enter__(self): self.default.map(self.stack.enter_context)\n def __exit__(self, *args, **kwargs): self.stack.__exit__(*args, **kwargs)",
"_____no_output_____"
],
[
"show_doc(ContextManagers, title_level=4)",
"_____no_output_____"
],
[
"#export\ndef str2bool(s):\n \"Case-insensitive convert string `s` too a bool (`y`,`yes`,`t`,`true`,`on`,`1`->`True`)\"\n if not isinstance(s,str): return bool(s)\n return bool(distutils.util.strtobool(s)) if s else False",
"_____no_output_____"
],
[
"for o in \"y YES t True on 1\".split(): assert str2bool(o)\nfor o in \"n no FALSE off 0\".split(): assert not str2bool(o)\nfor o in 0,None,'',False: assert not str2bool(o)\nfor o in 1,True: assert str2bool(o)",
"_____no_output_____"
],
[
"#export\ndef _is_instance(f, gs):\n tst = [g if type(g) in [type, 'function'] else g.__class__ for g in gs]\n for g in tst:\n if isinstance(f, g) or f==g: return True\n return False\n\ndef _is_first(f, gs):\n for o in L(getattr(f, 'run_after', None)):\n if _is_instance(o, gs): return False\n for g in gs:\n if _is_instance(f, L(getattr(g, 'run_before', None))): return False\n return True",
"_____no_output_____"
],
[
"#export\ndef sort_by_run(fs):\n end = L(fs).attrgot('toward_end')\n inp,res = L(fs)[~end] + L(fs)[end], L()\n while len(inp):\n for i,o in enumerate(inp):\n if _is_first(o, inp):\n res.append(inp.pop(i))\n break\n else: raise Exception(\"Impossible to sort\")\n return res",
"_____no_output_____"
]
],
[
[
"# Export -",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.export import notebook2script\nnotebook2script()",
"Converted 00_test.ipynb.\nConverted 01_basics.ipynb.\nConverted 02_foundation.ipynb.\nConverted 03_xtras.ipynb.\nConverted 03a_parallel.ipynb.\nConverted 03b_net.ipynb.\nConverted 04_dispatch.ipynb.\nConverted 05_transform.ipynb.\nConverted 07_meta.ipynb.\nConverted 08_script.ipynb.\nConverted index.ipynb.\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb327c3951d236b2b49c55287edfa4439844b15 | 15,000 | ipynb | Jupyter Notebook | docs/Performances of Dimension.ipynb | mocquin/physipy | a44805dbf4e68544c987e07564dd4a8d50be8b4c | [
"MIT"
] | 5 | 2021-01-23T11:23:07.000Z | 2022-02-28T15:38:58.000Z | docs/Performances of Dimension.ipynb | mocquin/physipy | a44805dbf4e68544c987e07564dd4a8d50be8b4c | [
"MIT"
] | null | null | null | docs/Performances of Dimension.ipynb | mocquin/physipy | a44805dbf4e68544c987e07564dd4a8d50be8b4c | [
"MIT"
] | 2 | 2020-11-07T20:08:08.000Z | 2021-06-09T02:58:04.000Z | 29.296875 | 150 | 0.4998 | [
[
[
"# Dimension performance",
"_____no_output_____"
]
],
[
[
"from physipy import Dimension",
"_____no_output_____"
],
[
"d = Dimension(\"L\")",
"_____no_output_____"
],
[
"%prun -D prun d*d",
" \n*** Profile stats marshalled to file 'prun'. \n"
],
[
"!snakeviz prun",
"snakeviz web server started on 127.0.0.1:8080; enter Ctrl-C to exit\nhttp://127.0.0.1:8080/snakeviz/%2FUsers%2Fmocquin%2FDocuments%2FCLE%2FOptique%2FPython%2FJUPYTER%2FMYLIB10%2FMODULES%2Fphysipy%2Fdocs%2Fprun\n"
]
],
[
[
"We need operations on array-like objects.\nThe solutions are :\n - a dict\n - list\n - numpy array\n - ordered dict\n - counter\nAmong these solutions",
"_____no_output_____"
],
[
"Most important operators : \n - equality check, to check if the dimensions are equal (for `Dimension.__eq__`)\n - addition of values key-wise, when computing the product of 2 dimension (for `Dimension.__mul__`)\n - substration of values key-wise, when computing the division of 2 dimensions (for `Dimension.__truediv__`)\n - multiplication of all values, when computing the exp of a dimension by a scalar (for `Dimension.__pow__`)\n ",
"_____no_output_____"
],
[
"We can rely on the operators, but the actual implementation matters. Exemple for ",
"_____no_output_____"
]
],
[
[
"import operator as op\noperators = {\n \"op.eq\":(\"binary\", op.eq), \n \"op.add\":(\"binary\", op.add),\n \"op.sub\":(\"binary\", op.sub),\n \"op.mul\":(\"binary\", op.mul),\n}",
"_____no_output_____"
],
[
"import time\nclass Timer():\n def __enter__(self):\n self.start = time.time()\n return self\n def __exit__(self, *args):\n self.end = time.time()\n self.secs = self.end - self.start\n self.msecs = self.secs * 1000 # millisecs",
"_____no_output_____"
],
[
"class Implem():\n def __init__(self, name, creator):\n self.name = name\n self.creator = creator\n def __call__(self, *args, **kwargs):\n return self.creator(*args, **kwargs)\n\n \n \nimplemetations = [DimAsDict, DimAsArray, DimAsList]\n \ndef bench_dimension_base_data(ns=[3, 4, 5, 6, 7, 8, 10, 15, 20, 50, 100, 1000, 10000]):\n # 4 operations to time\n # for various number of dimensions \n # for all implemetations\n # need to store the result of each test\n res = []\n for implem in implemetations:\n for opmeta in operators: \n for n in ns:\n obj = implem(n)\n if opmeta[0] == \"binary\":\n op = opmeta[1]\n with Timer() as t:\n resop = op(obj, obj)\n res_dict = {\n \"implem\":implem.name,\n \"n\":n,\n \"result\":resop,\n \"time\":t.msecs,\n }\n res.append(res_dict)\n \n \n ",
"_____no_output_____"
],
[
"import physipy\nfrom physipy import m, Dimension",
"_____no_output_____"
],
[
"d = Dimension(\"L\")\n%timeit d**2",
"4.14 µs ± 40.3 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)\n"
],
[
"d = Dimension(\"L\")\n%timeit d**2",
"4.1 µs ± 16.3 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)\n"
],
[
" ",
"_____no_output_____"
],
[
"import numpy as np\n\nclass DimAsListArray():\n \"\"\"\n Benefit the speed of array when computing mul/div, and speed of list equality for keys\n \"\"\"\n \n def __init__(self, values=np.zeros(3), KEYS=BASEKEYS):\n self.dims_keys = KEYS\n self.dim_values = values\n \n def __mul__(self, other):\n return DimAsListArray(self.dim_values+other.dim_values)\n ",
"_____no_output_____"
],
[
"import numpy as np\nimport collections\n\"\"\"Goal : return True if 2 vectors of numbers are equal\nInputs :\n - vectors are assured to be the same size\n - vector values can be int, float, np.numbers, fractions\n - the order of the numbers matters (like with dict comparison or ordered dict)\n\"\"\"\n \nas_dictl = {\"A\":0, \"B\":0, \"C\":0}\nas_dictr = {\"A\":0, \"B\":0, \"C\":0}\nas_listl = [0, 0, 0]\nas_listr = [0, 0, 0]\nas_arryl = np.array([0, 0, 0])\nas_arryr = np.array([0, 0, 0])\nas_odictl = collections.OrderedDict( {\"A\":0, \"B\":0, \"C\":0})\nas_odictr = collections.OrderedDict( {\"A\":0, \"B\":0, \"C\":0})\nas_counterl = collections.Counter(\"AAABBBCCC\")\nas_counterr = collections.Counter(\"AAABBBCCC\")",
"_____no_output_____"
],
[
"%timeit as_listl == as_listr\n%timeit as_dictl == as_dictr\n%timeit as_counterl == as_counterr\n%timeit as_odictl == as_odictr\n%timeit as_arryl.tolist() == as_arryr.tolist()\n%timeit list(as_odictl.values()) == list(as_odictr.values())\n%timeit np.array_equal(as_arryl, as_arryr)\n%timeit np.all(as_arryl == as_arryr)\n",
"47.2 ns ± 2.02 ns per loop (mean ± std. dev. of 7 runs, 10000000 loops each)\n77.4 ns ± 1.95 ns per loop (mean ± std. dev. of 7 runs, 10000000 loops each)\n79.2 ns ± 0.916 ns per loop (mean ± std. dev. of 7 runs, 10000000 loops each)\n86.9 ns ± 1.27 ns per loop (mean ± std. dev. of 7 runs, 10000000 loops each)\n324 ns ± 16.2 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n799 ns ± 14.4 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n5.22 µs ± 572 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)\n5.35 µs ± 409 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)\n"
],
[
"a = np.arange(500)\nb = np.arange(500)\n\n%timeit np.all(a == b)\n%timeit a.tolist() == b.tolist()\n",
"5 µs ± 123 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)\n17.7 µs ± 124 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)\n"
],
[
"import numpy as np\nimport collections\nfrom operator import add\n\n\nas_dictl = {\"A\":0, \"B\":0, \"C\":0}\nas_dictr = {\"A\":0, \"B\":0, \"C\":0}\nas_listl = [0, 0, 0]\nas_listr = [0, 0, 0]\nas_arryl = np.array([0, 0, 0])\nas_arryr = np.array([0, 0, 0])\nas_odictl = collections.OrderedDict( {\"A\":0, \"B\":0, \"C\":0})\nas_odictr = collections.OrderedDict( {\"A\":0, \"B\":0, \"C\":0})\n\n%timeit [l+r for l,r in zip(as_listl, as_listr)]\n%timeit {k:as_dictl[k]+as_dictr[k] for k in (as_dictl.keys() & as_dictr.keys())}\n#%timeit as_odictl == as_odictr\n#%timeit as_arryl.tolist() == as_arryr.tolist()\n#%timeit list(as_odictl.values()) == list(as_odictr.values())\n#%timeit np.array_equal(as_arryl, as_arryr)\n%timeit as_arryl + as_arryr\n%timeit list(map(add, as_listl, as_listr))",
"616 ns ± 27.4 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n1.35 µs ± 264 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n600 ns ± 45.5 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n624 ns ± 61 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n"
],
[
"import numpy as np\nimport collections\nfrom operator import mul\n\nas_dictl = {\"A\":0, \"B\":0, \"C\":0}\nas_dictr = {\"A\":0, \"B\":0, \"C\":0}\nas_listl = [0, 0, 0]\nas_listr = [0, 0, 0]\nas_arryl = np.array([0, 0, 0])\nas_arryr = np.array([0, 0, 0])\nas_odictl = collections.OrderedDict( {\"A\":0, \"B\":0, \"C\":0})\nas_odictr = collections.OrderedDict( {\"A\":0, \"B\":0, \"C\":0})\n\n%timeit [l*r for l,r in zip(as_listl, as_listr)]\n%timeit {k:as_dictl[k]*as_dictr[k] for k in (as_dictl.keys() & as_dictr.keys())}\n#%timeit as_odictl == as_odictr\n#%timeit as_arryl.tolist() == as_arryr.tolist()\n#%timeit list(as_odictl.values()) == list(as_odictr.values())\n#%timeit np.array_equal(as_arryl, as_arryr)\n%timeit as_arryl * as_arryr\n%timeit list(map(mul, as_listl, as_listr))",
"685 ns ± 145 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n1.15 µs ± 87.4 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n544 ns ± 50.9 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n574 ns ± 36.5 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n"
],
[
"import numpy as np\nimport collections\nfrom operator import pow\n\nas_dictl = {\"A\":1, \"B\":1, \"C\":1}\nas_dictr = 2\nas_listl = [1, 1, 1]\nas_listr = 2\nas_arryl = np.array([1, 1, 1])\nas_arryr = 2\nas_odictl = collections.OrderedDict( {\"A\":1, \"B\":1, \"C\":1})\nas_odictr = 2\n\n%timeit [l**as_dictr for l in as_listl]\n%timeit {k:as_dictl[k]**as_dictr for k in as_dictl.keys()}\n%timeit as_arryl ** as_arryr\n%timeit list(map(lambda x:x**2, as_listl))",
"980 ns ± 12.9 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n1.19 µs ± 12.1 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n733 ns ± 11.5 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n1.27 µs ± 16.1 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb33801dbe11286e6df486b6c702ac072cc5344 | 20,826 | ipynb | Jupyter Notebook | KCP.ipynb | leanluig/jup_old | 1dd47469c89c884131b21f11d457d0a18d3e90f2 | [
"Apache-2.0"
] | null | null | null | KCP.ipynb | leanluig/jup_old | 1dd47469c89c884131b21f11d457d0a18d3e90f2 | [
"Apache-2.0"
] | null | null | null | KCP.ipynb | leanluig/jup_old | 1dd47469c89c884131b21f11d457d0a18d3e90f2 | [
"Apache-2.0"
] | null | null | null | 36.15625 | 128 | 0.507731 | [
[
[
"%matplotlib inline\n\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport numpy as np\n\nsns.set(style=\"darkgrid\")\n\n# CSV einlesen (Achtung keine '/' in den Spaltenüberschriften wegen späterem Namen)\n# geht davon aus, dass keine Fehler in der Datei sind und alle Felder richtig gefüllt\n# zum Überprüfen dien Notebooks LPS_first bzw. ARC_first nutzen\n\n#Team='ARC'\nTeam='LPS'\n\nif Team == 'ARC':\n df = pd.read_csv('input/20190521_Board_ARC_Input_für_J_Datum_rckwrts.csv', sep=';', \n header=4, usecols=[0,1,4,5,6,7], parse_dates=[3,4,5], encoding='iso8859_15')\n df.columns = ['art', 'titel', 'plan', 'next', 'input', 'out']\nelse:\n df = pd.read_csv('input/20190410LPS_CFD.csv', sep=';', usecols=[9,11,18], parse_dates=[1,2], encoding='iso8859_15')\n df.columns = ['art', 'input', 'out']\n#end it TEAM\n \nfeiertage = ['2018-10-03', # Tag der Einheit\n '2018-11-01', # Allerheiligen\n '2018-12-24', # Heiligabend\n '2018-12-25', # Weihnachten\n '2018-12-26', # Weihnachten\n '2018-12-31', # Silvester\n '2019-01-01', # Neujahr\n '2019-02-28', # Wieverfastelovend\n '2019-03-04', # Rosenmontag\n '2019-04-19', # Karfreitag\n '2019-04-22', # Ostermontag\n '2019-05-01', # 1. Mai\n '2019-05-30', # Christi Himmelfahrt\n '2019-06-10', # Pfingst-Montag\n '2019-06-20' # Fronleichnam\n ]\n\n# astype('datetime64[D]') notwendig\ndauer=np.busday_count(df.input.values.astype('datetime64[D]'), \n df.out.values.astype('datetime64[D]'), \n holidays=feiertage)+1\n\ndf['dauer'] = dauer\n\nvon=(min(df.input))\nvon=von.strftime('%d.%m.%Y')\n\nbis=(max(df.out))\nbis=bis.strftime('%d.%m.%Y')\n\ndef calc_8090_percentile(ldf, art, von, bis):\n perc80 = np.percentile(ldf.dauer,80,interpolation='lower') # lower ist richtig\n perc90 = np.percentile(ldf.dauer,90,interpolation='lower')\n return (perc80, perc90)\n#end 8090_percentile\n \n\ndef plot_leadtime(ldf, art, von, bis):\n perc80 = np.percentile(ldf.dauer,80,interpolation='lower')\n perc90 = np.percentile(ldf.dauer,90,interpolation='lower')\n \n # gca stands for 'get current axis'\n ax = plt.gca()\n #hist zählt direkt die Elemente\n ldf.plot(kind='hist', y='dauer',bins=max(dauer),rwidth=1, color='black', ax=ax, label='')\n \n text80 = '80% fertig in ' + str(perc80) + ' Tagen'\n # vertical dotted line originating at mean value\n plt.axvline(perc80+.2, linestyle='-.', linewidth=1, color='blue', label=text80)\n\n text90 = '90% fertig in ' + str(perc90) + ' Tagen'\n # vertical dotted line originating at mean value\n plt.axvline(perc90+.2, linestyle='--', linewidth=1, color='red', label=text90)\n\n # Beschriftung\n plt.suptitle(art)\n \n title = von +' bis ' + bis + ': ' + str(len(ldf)) + ' Zettel'\n plt.title(title, fontsize=12)\n plt.xlabel('Dauer in Tagen')\n plt.ylabel('Anzahl')\n plt.legend(loc=0)\n \n # yticks ändern, so dass der Mindestabstand 1 ist\n ymin, ymax = plt.ylim()\n ax.yaxis.set_major_locator(plt.MultipleLocator(int(ymax/10+1))) \n \n # x-Achse ab 0\n xmin, xmax = plt.xlim()\n plt.xlim(0, xmax)\n \n \n #plt.show()\n filename='out/'+ Team + '_' + art +'.png'\n plt.savefig(filename, dpi=300)\n plt.close()\n#enddef plotte()\n\nvon=(min(df.input))\nvon=von.strftime('%d.%m.%Y')\n\nbis=(max(df.out))\nbis=bis.strftime('%d.%m.%Y')\n\n# muss kein DataFrame sein, ist meine komplizierte, aber laufende Lösung\narten = pd.DataFrame(columns=['Anzahl', 'Art'])\narten = arten.astype( dtype={'Anzahl': int, 'Art' : str})\n\n\nfor gew_art in df.art.drop_duplicates():\n gewaehlt = df.art == gew_art\n# gewaehlt = df.input > '2018-12-31'\n plot_leadtime(df[gewaehlt], gew_art, von, bis)\n laenge = len(df[gewaehlt])\n print gew_art, laenge\n arten.loc[len(arten)] = [int(laenge), gew_art]\n#endfor gew_art in ....\nprint arten\n\nplot_leadtime(df, 'Alle', von, bis)",
"Linux Plattform weiterentwickeln 144\nSecurity 26\nBerechtigung verwalten 37\nLinux patchen updaten 48\nÜbergabe alter Themen 22\nLinux System zur Verfügung stellen 41\nVMWare SAN LUN 5\nITR 18\nVMWare verwalten 39\nRhel7 Mig 22\nIncident 6\nAm Board 19\n Anzahl Art\n0 144 Linux Plattform weiterentwickeln\n1 26 Security\n2 37 Berechtigung verwalten\n3 48 Linux patchen updaten\n4 22 Übergabe alter Themen\n5 41 Linux System zur Verfügung stellen\n6 5 VMWare SAN LUN\n7 18 ITR\n8 39 VMWare verwalten\n9 22 Rhel7 Mig\n10 6 Incident\n11 19 Am Board\n"
],
[
"def plot_turbulence(ldf, art, von, bis):\n \n #alle Arbeitstage von ... bis\n bdate = pd.bdate_range(von, bis, holidays=feiertage, freq='C')\n Tage = bdate.strftime('%d.%m.%y')\n\n # erste Spalte des neuen Dataframes tur\n series = pd.DataFrame(bdate, columns=['days'])\n series['Tage'] = Tage\n \n series['lfd_no'] = np.arange(1, len(series)+1)\n #print series.lfd\n\n # number of pull-actions (System liquidity)\n i = 0\n sum = 0\n col = np.zeros(len(series), int) \n for day in series.days:\n sum = len(df[df.out == day])\n sum += len(df[df.input == day])\n col[i] = sum\n i += 1\n #endfor day in series.days\n series['Pulls'] = col\n \n sum_pulls = np.sum(col)\n \n first = np.gradient(series.Pulls)\n series['First_Deviation'] = first\n # 2nd deviation = Volatility\n \n vol = np.gradient(first)\n series['Volatility'] = vol\n \n third = np.gradient(vol)\n \n # 2nd 2nd deviation = turbulence\n fourth = np.gradient(third)\n series['Volatility_of_Volatility'] = fourth\n \n from scipy import stats \n series['Fourth_Moment'] = stats.moment(col, moment = 4)\n \n # Turbulence as in Swiftkanban\n frequency = np.zeros(max(col)+1, int) \n for i in range (0, max(col)+1):\n frequency[i] = len (col[col == i])\n # print i, frequency[i]\n \n wahrscheinlichkeit = np.zeros(len(series), dtype = float) \n\n i = 0\n for p in series.Pulls:\n wahrscheinlichkeit[i] = frequency[p] \n i += 1\n \n real_turb = np.power(col - np.mean(col), 4) * wahrscheinlichkeit\n print real_turb \n \n series['Turbulence'] = real_turb \n #/ sum_pulls\n\n #gewaehltes y\n gew_y = 8\n print (gew_y)\n \n #ax=tur.plot(kind='bar', stacked=True, linewidth=.1 , x='Tage', y=[2,3])\n ax=series.plot(kind='line' , x='lfd_no', y=[gew_y])\n \n #nur jeden xstep-ten Wert plotten auf x-achse\n #xstep = 5\n #xindex = np.arange(0, len(series), xstep, int)\n \n #einigeTage=Tage[xindex]\n #ax.xaxis.set_major_locator(plt.MultipleLocator(xstep)) \n #ax.set_xticklabels(labels=einigeTage)\n \n # integers for y-Axis\n from matplotlib.ticker import MaxNLocator\n ax.yaxis.set_major_locator(MaxNLocator(integer=True))\n \n filename='turbulence/'+ Team + '_' + art + '_' + str(gew_y) +'.png'\n #filename='turbulence/'+ Team + '_' + art + '.png'\n\n #ax.autoscale_view()\n plt.setp(plt.gca().get_xticklabels(), rotation=45, horizontalalignment='right', size = 5) \n \n # Beschriftung\n #plt.suptitle('Turbulenz von Team ' + Team)\n #Subtitle = 'Pulls'\n \n #title = von +' bis ' + bis + ': ' + str(len(ldf)) + ' Zettel'\n #title = str(len(ldf)) + ' Zettel'\n title = 'Auswertung von Team ' + Team\n \n plt.title(title)\n #plt.ylabel('Anzahl')\n plt.xlabel('')\n \n plt.legend(loc=0) \n \n # noch Testen\n # x-Achse sinnvoll verschieben!! Jeweils manuel anzupassen!!!\n xmin, xmax = plt.xlim()\n \n #plt.xlim(xmin+4, xmax-22) \n \n #plt.ylim(-12,12)\n \n plt.savefig(filename, dpi=300)\n plt.close()\n\n#end plot_turbulence\n\nplot_turbulence(df, 'Turbulence', '2018-08-21', '2019-05-04')",
"[1.86457821e+02 1.71392768e+03 1.71392768e+03 2.23774985e+03\n 2.66608107e+01 9.99756567e-06 1.01022556e+03 5.43234703e+02\n 1.58114027e+03 5.43234703e+02 1.71392768e+03 2.66608107e+01\n 2.01860704e+01 2.66608107e+01 3.83797280e+03 1.01167871e+04\n 1.32923580e+04 2.44113285e+03 3.87360649e+04 9.99756567e-06\n 4.15586017e+03 1.86457821e+02 1.58114027e+03 1.01022556e+03\n 2.66608107e+01 9.99756567e-06 5.43234703e+02 2.66608107e+01\n 1.86457821e+02 1.01022556e+03 2.66608107e+01 1.71392768e+03\n 2.23774985e+03 1.71392768e+03 2.01860704e+01 1.86457821e+02\n 5.43234703e+02 5.43234703e+02 1.71392768e+03 2.01860704e+01\n 9.99756567e-06 1.58114027e+03 1.58114027e+03 3.83797280e+03\n 2.66608107e+01 2.01860704e+01 9.99756567e-06 2.01860704e+01\n 2.66608107e+01 1.01022556e+03 2.64259880e+03 5.43234703e+02\n 3.83797280e+03 2.01860704e+01 2.66608107e+01 3.83797280e+03\n 5.43234703e+02 1.01022556e+03 1.01022556e+03 1.86457821e+02\n 1.01022556e+03 2.66608107e+01 2.01860704e+01 1.86457821e+02\n 9.99756567e-06 2.66608107e+01 9.99756567e-06 1.71392768e+03\n 1.71392768e+03 5.43234703e+02 1.01022556e+03 2.01860704e+01\n 2.66608107e+01 5.43234703e+02 1.71392768e+03 9.99756567e-06\n 1.86457821e+02 2.01860704e+01 5.43234703e+02 9.99756567e-06\n 1.86457821e+02 9.99756567e-06 9.99756567e-06 5.43234703e+02\n 5.43234703e+02 1.71392768e+03 1.71392768e+03 1.71392768e+03\n 2.23774985e+03 2.23774985e+03 2.66608107e+01 5.43234703e+02\n 2.23774985e+03 5.43234703e+02 5.43234703e+02 1.86457821e+02\n 1.71392768e+03 2.23774985e+03 2.01860704e+01 1.58114027e+03\n 2.01860704e+01 1.58114027e+03 2.01860704e+01 2.01860704e+01\n 2.01860704e+01 1.71392768e+03 9.99756567e-06 2.01860704e+01\n 5.43234703e+02 2.66608107e+01 2.23774985e+03 5.43234703e+02\n 2.66608107e+01 2.01860704e+01 2.23774985e+03 1.01022556e+03\n 1.71392768e+03 5.43234703e+02 1.71392768e+03 2.66608107e+01\n 2.66608107e+01 1.71392768e+03 1.86457821e+02 9.99756567e-06\n 1.01022556e+03 2.66608107e+01 5.43234703e+02 2.66608107e+01\n 5.43234703e+02 5.43234703e+02 5.43234703e+02 2.66608107e+01\n 2.66608107e+01 5.43234703e+02 1.71392768e+03 3.83797280e+03\n 2.23774985e+03 5.43234703e+02 1.71392768e+03 5.43234703e+02\n 5.43234703e+02 2.66608107e+01 1.86457821e+02 2.66608107e+01\n 5.43234703e+02 2.66608107e+01 2.66608107e+01 2.66608107e+01\n 1.01022556e+03 9.99756567e-06 2.66608107e+01 2.01860704e+01\n 5.43234703e+02 2.64259880e+03 2.01860704e+01 5.43234703e+02\n 2.66608107e+01 1.01022556e+03 1.32923580e+04 3.83797280e+03\n 5.43234703e+02 5.43234703e+02 2.66608107e+01 2.66608107e+01\n 1.71392768e+03 1.71392768e+03 5.43234703e+02 5.43234703e+02\n 5.43234703e+02 1.71392768e+03 5.43234703e+02 5.43234703e+02]\n8\n"
],
[
"def plot_cfd(ldf, art, von, bis):\n \n #alle Arbeitstage von ... bis\n bdate = pd.bdate_range(von, bis, holidays=feiertage, freq='C')\n Tage = bdate.strftime('%d.%m.%y')\n\n # erste Spalte des neuen Dataframes cfd\n cfd = pd.DataFrame(bdate, columns=['days'])\n cfd['Tage'] = Tage\n\n i = 0\n sum = 0\n col = np.zeros(len(cfd), int) \n for day in cfd.days:\n sum += len(df[df.out == day])\n col[i] = sum\n i += 1\n #endfor day in cfd.days\n cfd['fertig'] = col\n\n i = 0\n sum = 0\n col = np.zeros(len(cfd), int) \n for day in cfd.days:\n sum += len(df[df.input == day])\n# print str(day), len(df[df.input == day]), len(df[df.out == day])\n sum -= len(df[df.out == day])\n col[i] = sum\n if (col[i] < 0): \n # print \"neg\", str(day), str(col[i])\n col[i] = 0\n \n i += 1\n #endfor day in cfd.days\n \n cfd['Umsetzung'] = col\n \n ax=cfd.plot(kind='bar', stacked=True, linewidth=.1 , x='Tage', y=[2,3])\n \n #nur jeden xstep-ten Wert plotten\n xstep = 10\n xindex = np.arange(2, len(cfd), xstep, int)\n \n einigeTage=Tage[xindex]\n ax.xaxis.set_major_locator(plt.MultipleLocator(xstep)) \n ax.set_xticklabels(labels=einigeTage)\n \n filename='out/'+ Team + '_' + art +'.png'\n\n #ax.autoscale_view()\n plt.setp(plt.gca().get_xticklabels(), rotation=45, horizontalalignment='right', size = 5) \n \n # Beschriftung\n plt.suptitle('CFD von Team ' + Team)\n \n title = von +' bis ' + bis + ': ' + str(len(ldf)) + ' Zettel'\n plt.title(title)\n plt.ylabel('Anzahl')\n plt.xlabel('')\n \n plt.legend(loc=0)\n \n # noch Testen\n # x-Achse sinnvoll verschieben!! Jeweils manuel anzupassen!!!\n xmin, xmax = plt.xlim()\n plt.xlim(xmin+4, xmax-22) \n \n plt.savefig(filename, dpi=300)\n plt.close()\n\n#end plot_cfd\n\nplot_cfd(df, 'CFD', von, bis)\n\n",
"_____no_output_____"
],
[
"def plot_art(ldf, von, bis):\n ",
"_____no_output_____"
],
[
"# Breite des Intervals für den \"gleitenden Durchschnitt\" in Tagen\n#breite = \n\n#alle Arbeitstage von ... bis\ndata = pd.bdate_range(von, bis, holidays=feiertage, freq='C')\n\n# erste Spalte des neuen Dataframes perc\nperc = pd.DataFrame(data, columns=['days'])",
"_____no_output_____"
],
[
"i = 0\np80 = np.zeros(len(perc), int) \nfor day in perc.days:\n gewaehlt = df.input > day\n if (len (df.dauer[gewaehlt]) != 0):\n p80[i] = np.percentile(df.dauer[gewaehlt],90,interpolation='lower')\n i = i + 1\n#endfor day in perc.days\n\nperc['Alle'] = p80\n\nfor gew_art in df.art.drop_duplicates():\n i = 0\n p80 = np.zeros(len(perc), int) \n gewaehlt = df.art == gew_art\n for day in perc.days:\n gewaehlt &= df.input > day\n if (len (df.dauer[gewaehlt]) != 0):\n p80[i] = np.percentile(df.dauer[gewaehlt],90,interpolation='lower')\n i = i + 1\n #endfor day in perc.days\n # perc[gew_art] = p80\n#endfor gew_art in ...\nperc.tail()",
"_____no_output_____"
],
[
"\nperc.plot(x='days')",
"_____no_output_____"
],
[
"perc.plot(x='days')\nfilename='out/'+ 'perc_over_time' +'.png'\nplt.savefig(filename, dpi=300)\nplt.close()",
"_____no_output_____"
],
[
"fig1, ax1 = plt.subplots()\nax1.pie(arten.Anzahl, autopct='%1.1f%%', labels=arten.Art,\n startangle=90)\nax1.axis('equal') # Equal aspect ratio ensures that pie is drawn as a circle.\n\nplt.show()\n",
"_____no_output_____"
],
[
"arten = arten.sort_values(by='Anzahl', ascending=False)\n\nfig, ax = plt.subplots(figsize=(16, 8), subplot_kw=dict(aspect=\"equal\"))\n\n# Absolutwerte und Prozente\ndef func(pct, allvals):\n absolute = int(pct/100.*np.sum(allvals)+.5)\n return \"{:.0f}%\\n({:d})\".format(pct, absolute)\n\n\nwedges, texts, autotexts = ax.pie(arten.Anzahl, autopct=lambda pct: func(pct, arten.Anzahl),\n textprops=dict(color=\"w\"), counterclock=False, startangle = 150)\n\nax.legend(wedges, arten.Art,\n title=\"Arten der Arbeit\",\n loc=\"center left\",\n bbox_to_anchor=(1, 0, 0.5, 1))\n\nplt.setp(autotexts, size=10, weight=\"bold\")\n\nax.set_title(Team + \": Verteilung nach Arten der Arbeit\")\n\n#plt.show()\nart='Arten_der_Arbeit'\nfilename='out/'+ Team + '_' + art +'.png'\nplt.savefig(filename, dpi=300)\nplt.close()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb3389cc32d3302fad529179c66d83f116da650 | 42,281 | ipynb | Jupyter Notebook | 02-Sep-2018/Part2.ipynb | ankushdata/New_Batch | 870808e04b1131c084ce51bb5097407022800e8e | [
"MIT"
] | null | null | null | 02-Sep-2018/Part2.ipynb | ankushdata/New_Batch | 870808e04b1131c084ce51bb5097407022800e8e | [
"MIT"
] | null | null | null | 02-Sep-2018/Part2.ipynb | ankushdata/New_Batch | 870808e04b1131c084ce51bb5097407022800e8e | [
"MIT"
] | 24 | 2019-03-13T04:04:12.000Z | 2021-04-21T14:59:12.000Z | 28.224967 | 141 | 0.330858 | [
[
[
" Objective of this sheet!\n\n* Handling missing value data using fillna as 0?\n* Filling NAN with different values for different columns?\n* Using ffill and bfill to copy the before and after value within a column\n* Replacing the NAN values vertically\n* Droping the NA values on the basis of the conditionality\n* When we want to replace the outliers(or other values) with NAN.\n* Replacing different values (column wise) not just the NAN ones, this time dictionary will come in rescue. \n* Giving numerical value to the categorical data for the problem solving purpose.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# another way of importing the file\ndf = pd.read_csv('weather.csv')",
"_____no_output_____"
],
[
"#1 we are going to put some other values instead of NA\nnew_df = df.fillna(0)\nnew_df",
"_____no_output_____"
],
[
"#2 I am using dictionary to fill NA with the different values in specific columns\nnew_df = df.fillna({'temperature':0,\n 'windspeed': 99,\n 'event': 'no event'})\nnew_df",
"_____no_output_____"
],
[
"#3 to carry forward previous day value for a NA value, we use ffill\nnew_df = df.fillna(method = 'ffill')\nnew_df",
"_____no_output_____"
],
[
"# to copy the next value in the row, we use bfill\nnew_df = df.fillna(method = 'bfill')\nnew_df",
"_____no_output_____"
],
[
"# 4 adding axis to the equation. It will copy vertically\nnew_df = df.fillna(method = 'bfill', axis = 'columns')\nnew_df",
"_____no_output_____"
],
[
"#5 dropping the NA values\nnew_df = df.dropna()\nnew_df",
"_____no_output_____"
],
[
"# droping those rows where all the rows have na\nnew_df = df.dropna(how = 'all')\nnew_df",
"_____no_output_____"
],
[
"# Importing another file for the coming events\ndf = pd.read_excel('weather_nan.xlsx')\ndf",
"_____no_output_____"
],
[
"#6 When I want to replace my values with NaN\nnew_df = df.replace(-99999, np.NAN)\nnew_df",
"_____no_output_____"
],
[
"#7 replace by using the dictionary, not just for the NAN but also could be handy for other values replacement\n\nnew_df = df.replace({\n 'temperature': -99999,\n 'windspeed' : -88888,\n 'event' : '0'\n },np.NaN)\nnew_df",
"_____no_output_____"
],
[
"#8 Again replacing the catg. or other string values to numerical.\ndf = pd.DataFrame({\n 'score' : ['exceptional','average','good','poor','average','exceptional'],\n 'student': ['rob','maya','parthiv','tom','julain','erica']\n })\ndf",
"_____no_output_____"
],
[
"# now I want to replace some scores into numbers\nnew_df = df.replace(['poor','average','good','exceptional'], [1,2,3,4])\nnew_df",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n*Questions and Answers.*\n</div>",
"_____no_output_____"
],
[
"\n#### Q1 - Please try the ffill and bfill for last element and first element of any column?\n",
"_____no_output_____"
],
[
"#### Q2 - Please write few lines on what is interpolate, also how can it be of great use during modeling?\n",
"_____no_output_____"
],
[
"#### Q3 - Please create(by using dict.) a dataset of your own choice and also keep some NAN in it, please demonstrate the bfill, ffil\n",
"_____no_output_____"
],
[
"#### Q4 - Please use the column mean(average) in place of the missing values (NAN). Please carry this on the dataset created by you?\n",
"_____no_output_____"
],
[
"#### Q5 - How can we change the date format given in the second sheet that we uploaded. It has to be aligned?",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecb33b51169113a29ad01764fd8d2a68bfff0b20 | 42,162 | ipynb | Jupyter Notebook | 05_gpt2_lm.ipynb | sofiedwardsv/Composer-Cassification | a64b87feb206c374a6680c9aa94a21017003194d | [
"MIT"
] | 2 | 2020-09-20T10:02:36.000Z | 2020-09-23T08:08:03.000Z | 05_gpt2_lm.ipynb | sofiedwardsv/Composer-Cassification | a64b87feb206c374a6680c9aa94a21017003194d | [
"MIT"
] | null | null | null | 05_gpt2_lm.ipynb | sofiedwardsv/Composer-Cassification | a64b87feb206c374a6680c9aa94a21017003194d | [
"MIT"
] | 1 | 2020-12-22T17:56:18.000Z | 2020-12-22T17:56:18.000Z | 129.331288 | 17,612 | 0.889806 | [
[
[
"# Training a GPT-2 Language Model",
"_____no_output_____"
],
[
"In this notebook we train a GPT-2 language model on the IMSLP and/or target data. This code can be used to train two different language models: (a) one that is trained on target data, and (b) one that is trained on IMSLP data and finetuned on target data. For (a), you can stop at the end of the section entitled \"Train Language Model\".",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"from pathlib import Path\nimport json\nfrom train_utils import plotLosses",
"_____no_output_____"
],
[
"bpe_path = Path('/home/tjtsai/.fastai/data/bscore_lm/bpe_data')\nbpe_path.mkdir(exist_ok=True, parents=True)",
"_____no_output_____"
]
],
[
[
"### Train Language Model",
"_____no_output_____"
]
],
[
[
"data_type = 'target' # 'target' or 'imslp'",
"_____no_output_____"
],
[
"lm_train_file = bpe_path/f'bpe_lm_{data_type}_train.txt'\nlm_valid_file = bpe_path/f'bpe_lm_{data_type}_valid.txt'\ntok_path = bpe_path/f'tokenizer_{data_type}'\noutput_model_path = bpe_path/f'models/gpt2_train-{data_type}_lm'",
"_____no_output_____"
],
[
"# changes from defaults:\n# vocab_size: 50257 -> 30000\n# n_positions: 1024 -> 514\n# n_ctx: 1024 -> 514\n# n_layer: 12 -> 6\nconfig = {\n \"architectures\": [\n \"GPT2LMHeadModel\"\n ],\n \"vocab_size\": 30000,\n \"n_positions\": 514,\n \"n_ctx\": 514,\n \"n_embd\": 768,\n \"n_layer\": 6,\n \"n_head\": 12,\n \"resid_pdrop\": 0.1,\n \"embd_pdrop\": 0.1,\n \"attn_pdrop\": 0.1,\n \"layer_norm_epsilon\": 1e-5,\n \"initializer_range\": 0.02,\n \"summary_type\": \"cls_index\",\n \"summary_use_proj\": True,\n \"summary_activation\": None,\n \"summary_proj_to_labels\": True,\n \"summary_first_dropout\": 0.1\n }",
"_____no_output_____"
],
[
"with open(f\"{tok_path}/config.json\", 'w') as fp:\n json.dump(config, fp)",
"_____no_output_____"
],
[
"cmd = f\"\"\"\npython ./run_language_modeling.py\n--train_data_file {lm_train_file}\n--output_dir {output_model_path}\n--model_type gpt2\n--eval_data_file {lm_valid_file}\n--line_by_line\n--config_name {tok_path}\n--tokenizer_name {tok_path}\n--do_train\n--do_eval\n--evaluate_during_training\n--per_gpu_train_batch_size 16\n--per_gpu_eval_batch_size 16\n--learning_rate 1e-4\n--num_train_epochs 12\n--logging_steps 7180\n--save_steps 7180\n--seed 42\n--overwrite_output_dir\n\"\"\".replace(\"\\n\", \" \")\n#--save_total_limit 2\n#--should_continue\n# target data: batch size 16, 204 steps per epoch, 12 epochs\n# imslp data: batch size 16, 7180 steps per epoch, ? epochs",
"_____no_output_____"
],
[
"!echo {cmd} > train_lm.sh",
"_____no_output_____"
],
[
"# you may need to run this in a bash shell with the appropriate virtual environment\n#!./train_lm.sh",
"_____no_output_____"
],
[
"plotLosses(output_model_path/'eval_results.txt')",
"_____no_output_____"
]
],
[
[
"### Finetune Language Model",
"_____no_output_____"
],
[
"This section only applies for the LM trained on IMSLP data.",
"_____no_output_____"
]
],
[
[
"finetuned_models_path = bpe_path/'models/gpt2_train-imslp_finetune-target_lm'\nlm_train_file = bpe_path/'bpe_lm_target_train.txt'\nlm_valid_file = bpe_path/'bpe_lm_target_valid.txt'",
"_____no_output_____"
],
[
"cmd = f\"\"\"\npython ./run_language_modeling.py\n--train_data_file {lm_train_file}\n--output_dir {finetuned_models_path}\n--model_type gpt2\n--eval_data_file {lm_valid_file}\n--line_by_line\n--model_name_or_path {output_model_path}\n--tokenizer_name {output_model_path}\n--do_train\n--do_eval\n--evaluate_during_training\n--per_gpu_train_batch_size 16\n--per_gpu_eval_batch_size 16\n--learning_rate 5e-5\n--num_train_epochs 12\n--logging_steps 204\n--save_steps 204\n--seed 42\n--overwrite_output_dir\n\"\"\".replace(\"\\n\", \" \")\n#--save_total_limit 2\n#--should_continue",
"_____no_output_____"
],
[
"!echo {cmd} > train_lm.sh",
"_____no_output_____"
],
[
"# you may need to run this in a bash shell with different virtual environment\n#!./train_roberta.sh",
"_____no_output_____"
],
[
"plotLosses(finetuned_models_path/'eval_results.txt')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecb345680d34186d7f2d88eb63378502feb65c17 | 10,482 | ipynb | Jupyter Notebook | __site/generated/notebooks/ISL-lab-5.ipynb | ven-k/MLJTutorials | 42151c8a96ad701aeaf763d53c8b7c6689eb6e8d | [
"MIT"
] | null | null | null | __site/generated/notebooks/ISL-lab-5.ipynb | ven-k/MLJTutorials | 42151c8a96ad701aeaf763d53c8b7c6689eb6e8d | [
"MIT"
] | null | null | null | __site/generated/notebooks/ISL-lab-5.ipynb | ven-k/MLJTutorials | 42151c8a96ad701aeaf763d53c8b7c6689eb6e8d | [
"MIT"
] | null | null | null | 24.897862 | 255 | 0.541118 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecb3466a236513f8d9d6d8925c1233bf301bda74 | 25,458 | ipynb | Jupyter Notebook | 100days/day 17 - perceptron.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
] | 13 | 2021-03-11T00:25:22.000Z | 2022-03-19T00:19:23.000Z | 100days/day 17 - perceptron.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
] | 160 | 2021-04-26T19:04:15.000Z | 2022-03-26T20:18:37.000Z | 100days/day 17 - perceptron.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
] | 12 | 2021-04-26T19:43:01.000Z | 2022-01-31T08:36:29.000Z | 53.259414 | 8,188 | 0.507306 | [
[
[
"import numpy as np\nfrom bokeh.plotting import figure, show, output_notebook",
"_____no_output_____"
]
],
[
[
"## data",
"_____no_output_____"
]
],
[
[
"X = np.array([[0, 1, 1], [1, 0, 1], [1, 1, 1], [-1, 1, 1], [1, -1, 1]])\nY = np.array([1, 1, 1, 0, 0])\nW = np.zeros(3)",
"_____no_output_____"
]
],
[
[
"## algorithm",
"_____no_output_____"
]
],
[
[
"def perceptron(x, w):\n return (x @ w >= 0).astype(int)",
"_____no_output_____"
],
[
"def train(x, y, w):\n for i in range(len(x)):\n # evaluate perceptron\n h = perceptron(x[i, :], w)\n \n # misclassification\n if h != y[i]:\n # positive sample\n if y[i] == 1: \n w += x[i, :]\n # negative sample\n else: \n w -= x[i, :]\n \n # evaluate\n return perceptron(x, w)",
"_____no_output_____"
]
],
[
[
"## training",
"_____no_output_____"
]
],
[
[
"print('y=', Y)\nfor _ in range(5):\n h = train(X, Y, W)\n print('w=', W, 'acc=', np.mean(h == Y))",
"y= [1 1 1 0 0]\nw= [ 0. 0. -2.] acc= 0.4\nw= [ 1. 1. -2.] acc= 0.6\nw= [ 2. 1. -2.] acc= 0.8\nw= [ 2. 2. -1.] acc= 1.0\nw= [ 2. 2. -1.] acc= 1.0\n"
]
],
[
[
"## plot",
"_____no_output_____"
]
],
[
[
"output_notebook()\n\ncolor = list(map({0: 'red', 1: 'green'}.__getitem__, Y))\nx0, y0 = -1.5, (-1.5 * -W[0] - W[2]) / W[1]\nx1, y1 = 1.5, (1.5 * -W[0] - W[2]) / W[1]\n\nplot = figure()\nplot.circle(x=X[:, 0], y=X[:, 1], color=color, size=10)\nplot.line(x=[x0, x1], y=[y0, y1])\nshow(plot)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb34c3a6113e99484d0adab12ab8285625ebf13 | 330,744 | ipynb | Jupyter Notebook | calcificationTestImageProcessing_LBP2.ipynb | emyesme/CalcificationDetection | b84ab4968b4cf4a37b5c2e85f70b9ec4e63ec5ad | [
"MIT"
] | null | null | null | calcificationTestImageProcessing_LBP2.ipynb | emyesme/CalcificationDetection | b84ab4968b4cf4a37b5c2e85f70b9ec4e63ec5ad | [
"MIT"
] | null | null | null | calcificationTestImageProcessing_LBP2.ipynb | emyesme/CalcificationDetection | b84ab4968b4cf4a37b5c2e85f70b9ec4e63ec5ad | [
"MIT"
] | null | null | null | 253.638037 | 285,769 | 0.902441 | [
[
[
"<a href=\"https://colab.research.google.com/github/emyesme/CalcificationDetection/blob/main/calcificationTestImageProcessing_LBP2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"#Libraries and Data Setup",
"_____no_output_____"
],
[
"## Import libraries",
"_____no_output_____"
]
],
[
[
"# just once to install opencv\n!pip install opencv-python\n!pip install matplotlib\n!pip install numpy\n#!pip install google-colab\n\n#!pip install PyWavelets\n#!pip install image_dehazer\n!pip install -U scikit-image\n#!pip install fastprogress\nfrom fastprogress import master_bar, progress_bar\n\n# import opencv\nimport cv2\n# import numpy\nimport numpy as np\nimport math\nfrom skimage import feature\nimport itertools\n#import show special for google colab\nfrom google.colab.patches import cv2_imshow\n#import plt for display\nimport matplotlib.pyplot as plt",
"Requirement already satisfied: opencv-python in /usr/local/lib/python3.7/dist-packages (4.1.2.30)\nRequirement already satisfied: numpy>=1.14.5 in /usr/local/lib/python3.7/dist-packages (from opencv-python) (1.21.6)\nRequirement already satisfied: matplotlib in /usr/local/lib/python3.7/dist-packages (3.2.2)\nRequirement already satisfied: numpy>=1.11 in /usr/local/lib/python3.7/dist-packages (from matplotlib) (1.21.6)\nRequirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib) (2.8.2)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib) (1.4.2)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib) (3.0.8)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/dist-packages (from matplotlib) (0.11.0)\nRequirement already satisfied: typing-extensions in /usr/local/lib/python3.7/dist-packages (from kiwisolver>=1.0.1->matplotlib) (4.2.0)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.7/dist-packages (from python-dateutil>=2.1->matplotlib) (1.15.0)\nRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (1.21.6)\nRequirement already satisfied: scikit-image in /usr/local/lib/python3.7/dist-packages (0.18.3)\nCollecting scikit-image\n Downloading scikit_image-0.19.2-cp37-cp37m-manylinux_2_12_x86_64.manylinux2010_x86_64.whl (13.5 MB)\n\u001b[K |████████████████████████████████| 13.5 MB 14.1 MB/s \n\u001b[?25hRequirement already satisfied: networkx>=2.2 in /usr/local/lib/python3.7/dist-packages (from scikit-image) (2.6.3)\nRequirement already satisfied: PyWavelets>=1.1.1 in /usr/local/lib/python3.7/dist-packages (from scikit-image) (1.3.0)\nRequirement already satisfied: numpy>=1.17.0 in /usr/local/lib/python3.7/dist-packages (from scikit-image) (1.21.6)\nRequirement already satisfied: tifffile>=2019.7.26 in /usr/local/lib/python3.7/dist-packages (from scikit-image) (2021.11.2)\nRequirement already satisfied: imageio>=2.4.1 in /usr/local/lib/python3.7/dist-packages (from scikit-image) (2.4.1)\nRequirement already satisfied: pillow!=7.1.0,!=7.1.1,!=8.3.0,>=6.1.0 in /usr/local/lib/python3.7/dist-packages (from scikit-image) (7.1.2)\nRequirement already satisfied: scipy>=1.4.1 in /usr/local/lib/python3.7/dist-packages (from scikit-image) (1.4.1)\nRequirement already satisfied: packaging>=20.0 in /usr/local/lib/python3.7/dist-packages (from scikit-image) (21.3)\nRequirement already satisfied: pyparsing!=3.0.5,>=2.0.2 in /usr/local/lib/python3.7/dist-packages (from packaging>=20.0->scikit-image) (3.0.8)\nInstalling collected packages: scikit-image\n Attempting uninstall: scikit-image\n Found existing installation: scikit-image 0.18.3\n Uninstalling scikit-image-0.18.3:\n Successfully uninstalled scikit-image-0.18.3\n\u001b[31mERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.\nalbumentations 0.1.12 requires imgaug<0.2.7,>=0.2.5, but you have imgaug 0.2.9 which is incompatible.\u001b[0m\nSuccessfully installed scikit-image-0.19.2\n"
]
],
[
[
"## Drive Setup",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/drive') # This will prompt for authorization.",
"Mounted at /content/drive\n"
]
],
[
[
"We put a shortcut in our drive to the image processing folder for this.",
"_____no_output_____"
]
],
[
[
"import os \nDATA_DIR = os.path.join('/content',\n 'drive',\n 'MyDrive',\n 'Image Processing and Analysis 2022',\n 'projects',\n 'Calcification Detection',\n 'dataset')",
"_____no_output_____"
]
],
[
[
"## Data Setup",
"_____no_output_____"
]
],
[
[
"#go into de directory of the images\n\n# this have 3 outputs root directory, the folders in the path and the files in the path.\n# we ignore _ the two first because we are not interested in those\n_, _, images = next(os.walk(os.path.join(DATA_DIR,'images')))\n_, _, breastMasks = next(os.walk(os.path.join(DATA_DIR,'masks')))\n_, _, groundTruths = next(os.walk(os.path.join(DATA_DIR, 'groundtruths')))\n\nimages.sort()\nbreastMasks.sort()\ngroundTruths.sort()\n\n# read numbers of normal images\nnormals = []\nwith open(os.path.join(DATA_DIR,'normals.txt')) as f:\n for line in f:\n normals.append(line[:-1])",
"_____no_output_____"
]
],
[
[
"## Google Colab Like a Pro",
"_____no_output_____"
]
],
[
[
"# https://medium.com/@robertbracco1/configuring-google-colab-like-a-pro-d61c253f7573#a642\n%%javascript\nfunction ClickConnect(){\nconsole.log(\"Working\");\ndocument.querySelector(\"colab-toolbar-button#connect\").click()\n}setInterval(ClickConnect,60000)",
"_____no_output_____"
]
],
[
[
"# Preprocessing",
"_____no_output_____"
],
[
"## DeHazing Using Dark Channel Prior and Guided Filter",
"_____no_output_____"
],
[
"Dehazing method proposed by the professor (also used in his paper)\n\nTaken from here:\nhttps://github.com/He-Zhang/image_dehaze\nInfo on readme.md of the repo",
"_____no_output_____"
],
[
"### Dark Channel",
"_____no_output_____"
],
[
"Here goes the theory behind this function ",
"_____no_output_____"
]
],
[
[
"# Here goes inputs --> output types\ndef DarkChannel(im,sz):\n b,g,r = cv2.split(im)\n dc = cv2.min(cv2.min(r,g),b);\n kernel = cv2.getStructuringElement(cv2.MORPH_RECT,(sz,sz))\n dark = cv2.erode(dc,kernel)\n return dark",
"_____no_output_____"
]
],
[
[
"### AtmLight",
"_____no_output_____"
]
],
[
[
"# Possibly change to grayscale would be nice\ndef AtmLight(im,dark):\n [h,w] = im.shape[:2]\n imsz = h*w\n numpx = int(max(math.floor(imsz/1000),1))\n darkvec = dark.reshape(imsz);\n imvec = im.reshape(imsz,3);\n\n indices = darkvec.argsort();\n indices = indices[imsz-numpx::]\n\n atmsum = np.zeros([1,3])\n for ind in range(1,numpx):\n atmsum = atmsum + imvec[indices[ind]]\n\n A = atmsum / numpx;\n return A",
"_____no_output_____"
]
],
[
[
"### TransmissionEstimate",
"_____no_output_____"
]
],
[
[
"def TransmissionEstimate(im,A,sz):\n omega = 0.95;# the closer to 1 the stronger the darkenning\n im3 = np.empty(im.shape,im.dtype);\n\n for ind in range(0,3):\n im3[:,:,ind] = im[:,:,ind]/A[0,ind]\n\n transmission = 1 - omega*DarkChannel(im3,sz);\n return transmission",
"_____no_output_____"
]
],
[
[
"### GuidedFilter",
"_____no_output_____"
]
],
[
[
"def Guidedfilter(im,p,r,eps):\n mean_I = cv2.boxFilter(im,cv2.CV_64F,(r,r));\n mean_p = cv2.boxFilter(p, cv2.CV_64F,(r,r));\n mean_Ip = cv2.boxFilter(im*p,cv2.CV_64F,(r,r));\n cov_Ip = mean_Ip - mean_I*mean_p;\n\n mean_II = cv2.boxFilter(im*im,cv2.CV_64F,(r,r));\n var_I = mean_II - mean_I*mean_I;\n\n a = cov_Ip/(var_I + eps);\n b = mean_p - a*mean_I;\n\n mean_a = cv2.boxFilter(a,cv2.CV_64F,(r,r));\n mean_b = cv2.boxFilter(b,cv2.CV_64F,(r,r));\n\n q = mean_a*im + mean_b;\n return q;",
"_____no_output_____"
]
],
[
[
"### TransmissionRefine",
"_____no_output_____"
]
],
[
[
"def TransmissionRefine(im,et):\n gray = cv2.cvtColor(im,cv2.COLOR_BGR2GRAY);\n gray = np.float64(gray)/255;\n r = 60;\n eps = 0.0001;\n t = Guidedfilter(gray,et,r,eps);\n\n return t;",
"_____no_output_____"
]
],
[
[
"### Recover",
"_____no_output_____"
]
],
[
[
"def Recover(im,t,A,tx = 0.1):\n res = np.empty(im.shape,im.dtype);\n t = cv2.max(t,tx);\n\n for ind in range(0,3):\n res[:,:,ind] = (im[:,:,ind]-A[0,ind])/t + A[0,ind]\n return res",
"_____no_output_____"
]
],
[
[
"### deHazingDarkChannelPriorPy",
"_____no_output_____"
]
],
[
[
"def deHazingDarkChannelPriorPy(matrix, mask):\n\n I = matrix.astype(np.float64)/255\n \n dark = DarkChannel(I,15)\n A = AtmLight(I,dark)\n te = TransmissionEstimate(I,A,15)\n t = TransmissionRefine(matrix,te)\n J = Recover(I,t,A,0.1)\n preprocessed = J\n return preprocessed\n\n# image = cv2.imread(DATA_DIR+\"/images/53582422_3f0db31711fc9795_MG_R_ML_ANON.tif\")\n# dark, t, matrix, J = deHazingDarkChannelPriorPy(image, image)",
"_____no_output_____"
]
],
[
[
"## Image Dilation",
"_____no_output_____"
]
],
[
[
"def imgDilation(matrix):\n kernel = np.ones((3,3), np.uint8)\n img_dilation = cv2.dilate(matrix, kernel, iterations=3)\n return img_dilation",
"_____no_output_____"
]
],
[
[
"## CLAHE",
"_____no_output_____"
]
],
[
[
"def imgCLAHE(matrix):\n matrix = matrix.astype(np.uint16)\n #gray = cv2.cvtColor(matrix, cv2.COLOR_RGB2GRAY)\n \n clahe = cv2.createCLAHE(clipLimit=2.0, tileGridSize=(8,8))\n cl1 = clahe.apply(matrix)\n return cl1",
"_____no_output_____"
]
],
[
[
"## De-noising",
"_____no_output_____"
]
],
[
[
"def gaussianBlur(matrix):\n img_blurred = cv2.GaussianBlur(matrix, (5,5), 0)\n return img_blurred",
"_____no_output_____"
]
],
[
[
"## Other Tried Methods",
"_____no_output_____"
],
[
"**CLAHE (adaptive histogram equalization)**\n * CLAHE + dehazing = bad results, black image \n * points less visible with CLAHE\n\n**Linear stretching**\n * still missing linear streching\n * The code goes on forever (high computational cost)\n\n",
"_____no_output_____"
],
[
"# Candidate Extraction",
"_____no_output_____"
],
[
"##Hessian-Matrix-Based Analysis",
"_____no_output_____"
],
[
"Hessian-matrix-based analysis or difference of gaussians (DoH) blob detection from skimage:\n\nhttps://scikit-image.org/docs/stable/api/skimage.feature.html?highlight=local%20binary%20pattern#skimage.feature.blob_doh",
"_____no_output_____"
]
],
[
[
"def candidateExtraction(matrix, mask):\n\n from skimage import feature\n \n # returns x,y,sigma of the blob\n blobs = feature.blob_doh(matrix,\n min_sigma=1,\n max_sigma=30,\n num_sigma=10,\n # The absolute lower bound for scale space maxima.\n # Local maxima smaller than threshold are ignored.\n # Reduce this to detect blobs with lower intensities.\n # If threshold_rel is also specified, whichever threshold is larger will be used.\n # If None, threshold_rel is used instead.\n threshold=0.005,\n # lower more sensible, more false positives bad also tinier calcifications detected\n overlap=0.5,\n log_scale=False,\n threshold_rel=None\n )\n # taken from the documentation\n # ...The downside is that this method can’t be used for detecting blobs of radius less than 3px\n # due to the box filters used in the approximation of Hessian Determinant.\n result = blobs\n return result",
"_____no_output_____"
]
],
[
[
"## Difference of Gaussians",
"_____no_output_____"
],
[
"Taken from:\n\nhttps://scikit-image.org/docs/stable/api/skimage.feature.html?highlight=local%20binary%20pattern#skimage.feature.blob_dog",
"_____no_output_____"
]
],
[
[
"from math import sqrt\ndef candidateExtractionDoG(matrix, mask):\n from skimage import data, feature\n\n blobs = feature.blob_dog(matrix,\n min_sigma=0.005,\n max_sigma=50,\n threshold=0.04)\n\n blobs[:, 2] = blobs[:, 2] * sqrt(2) # https://scikit-image.org/docs/stable/auto_examples/features_detection/plot_blob.html\n\n result = blobs\n return result",
"_____no_output_____"
]
],
[
[
"# Feature Extraction",
"_____no_output_____"
]
],
[
[
"# Get Ground Truth for each patch\ndef patchGroundTruth(candidate, groundTruth):\n\n left = int((candidate[0] - candidate[2]) if ((candidate[0] - candidate[2]) > 0) else 0)\n right = int((candidate[0] + candidate[2]) if ((candidate[0] + candidate[2]) < groundTruth.shape[0]) else groundTruth.shape[0])\n top = int((candidate[1] - candidate[2]) if ((candidate[1] - candidate[2]) > 0) else 0)\n bottom = int((candidate[1] + candidate[2]) if ((candidate[1] + candidate[2]) < groundTruth.shape[1]) else groundTruth.shape[1])\n\n truePatch = groundTruth[left : right,\n top : bottom]\n sum = np.sum(truePatch)\n if sum > 0:\n return str(1)\n else:\n return str(0)",
"_____no_output_____"
]
],
[
[
"Documentation and example on LBP:\n\n\n\n* https://scikit-image.org/docs/dev/api/skimage.feature.html#skimage.feature.local_binary_pattern\n* https://scikit-image.org/docs/dev/auto_examples/features_detection/plot_local_binary_pattern.html\n\nReference Paper:\n\n\n\n* Sadad T, Munir A, Saba T, Hussain A, Fuzzy C-Means and Region Growing based Classification of Tumor from Mammograms using Hybrid Texture Features, Journal of Computational Science (2018), https://doi.org/10.1016/j.jocs.2018.09.015\n\nInput of LBP:\n\nGrayscale image, radius (pixel distance from center point), points (number of surrounding pixels to be taken at the defined radius, usually 8*radius), and method.\n\nOutput of LBP:\n\nLBP grayscale image for textural classification\n\n\n\n",
"_____no_output_____"
],
[
"Video tutorial on how to use GLCM:\n\n* https://www.youtube.com/watch?v=5x-CIHRmMNY\n\nDocumentation and example on GLCM:\n\n* https://scikit-image.org/docs/dev/auto_examples/features_detection/plot_glcm.html\n\nPaper:\n\n* https://ijcrr.com/uploads/3454_pdf.pdf\n\nGLCM Properties documentation:\n\n* https://scikit-image.org/docs/dev/api/skimage.feature.html#skimage.feature.graycoprops\n\nGLCM Gray co-matrix documentation:\n\n* https://stackoverflow.com/questions/54512617/creating-gray-level-co-occurrence-matrix-from-16-bit-image\n(why can't we have 16 bit type for the coocurrence matrix computation)\n\n* https://www.sciencedirect.com/topics/engineering/cooccurrence-matrix (simple explanation GLCM)",
"_____no_output_____"
],
[
"Output of the GLCM command: \n\nthe gray-level co-occurrence histogram. The value P[i,j,d,theta]\nis the number of times that gray-level j occurs at a distance d\nand at an angle theta from gray-level i.\nIf normed is False, the output is of type uint32, otherwise it is float64.\nThe dimensions are: levels x levels x number of distances x number of angles.",
"_____no_output_____"
]
],
[
[
"# Using GLCM with the given candidates\ndef featuresExtraction(matrix, candidates, features, mask, groundTruth, image, folder):\n \n from skimage.feature import local_binary_pattern\n\n # No candidates, no extraction needed\n if (len(candidates) == 0):\n return []\n # angles\n angles = [0, np.pi/4, np.pi/2, 3*np.pi/4]\n\n flag = True\n for index, candidate in enumerate(progress_bar(candidates)):\n\n distances = [1] # probably we need bigger\n\n # combination of distances and angles as couples of values\n distancesAngles = list(itertools.product(distances, angles))\n\n # to use them as coordinates they have to be integers\n \n candidate = candidate.astype(np.int64)\n\n # changed for LBP+GLCM\n \n if candidate[2] == 0:\n candidate[2] = 1\n\n\n # candidates are y,x and sigma\n left = int((candidate[0] - candidate[2]) if ((candidate[0] - candidate[2]) > 0) else 0)\n right = int((candidate[0] + candidate[2]) if ((candidate[0] + candidate[2]) < matrix.shape[0]) else matrix.shape[0])\n top = int((candidate[1] - candidate[2]) if ((candidate[1] - candidate[2]) > 0) else 0)\n bottom = int((candidate[1] + candidate[2]) if ((candidate[1] + candidate[2]) < matrix.shape[1]) else matrix.shape[1])\n\n patchCandidate = matrix[left: right,\n top : bottom]\n\n \n # LBP received grayscale\n\n radius= 1\n points= 8 * radius\n\n patchCandidate = cv2.cvtColor(patchCandidate, cv2.COLOR_BGR2GRAY)\n patchCandidate = local_binary_pattern(patchCandidate, points, radius, method='default')\n\n # graycomatrix, glcm, receive unsigned integer type\n patchCandidate = patchCandidate.astype(np.uint8)\n\n \n dictFeatures = {}\n dictFeatures = {'name': 'patch_' + str(index) + '_' + str(image.split(\".\")[0]),\n 'label': patchGroundTruth(candidate, groundTruth)}\n\n for distanceAngle in distancesAngles:\n distance = distanceAngle[0]\n angle = distanceAngle[1]\n \n # get the degree to use it as name for the column\n name = str(angle*(180.0/np.pi))\n\n # input image, distance in pixels, angles\n glcm = feature.graycomatrix(patchCandidate, [ distance ], [ angle ])\n\n # properties\n dictFeatures['LBP' + 'contrast'+ str(distance) + name] = feature.graycoprops(glcm, 'contrast')[0][0]\n dictFeatures['LBP' +'dissimilarity' + str(distance) + name] = feature.graycoprops(glcm, 'dissimilarity')[0][0]\n dictFeatures['LBP' +'homogeneity' + str(distance) + name] = feature.graycoprops(glcm, 'homogeneity')[0][0]\n dictFeatures['LBP' +'energy' + str(distance) + name] = feature.graycoprops(glcm, 'energy')[0][0]\n dictFeatures['LBP' +'correlation' + str(distance) + name] = feature.graycoprops(glcm, 'correlation')[0][0]\n dictFeatures['LBP' +'ASM' + str(distance) + name] = feature.graycoprops(glcm, 'ASM')[0][0]\n\n # add to the dataframe the features for this patch\n features = features.append(dictFeatures, ignore_index=True)\n\n # save in the csv\n flag = writeFeatures(features, flag, folder, image, \"LBPfeatures\")\n\n return features",
"_____no_output_____"
],
[
"def writeFeatures(features, flag, folder, image, name):\n if(flag):\n features.to_csv(os.path.join('/content',\n 'drive',\n 'MyDrive',\n 'Results',\n folder,\n str(image.split('.')[0]) + '_'+ name + '.csv'),\n mode='a',\n index=False)\n flag = False\n else:\n features.to_csv(os.path.join('/content',\n 'drive',\n 'MyDrive',\n 'Results',\n folder,\n str(image.split('.')[0]) + '_'+ name + '.csv'),\n mode='a',\n header=False,\n index=False)\n return flag",
"_____no_output_____"
]
],
[
[
"# Connected Components",
"_____no_output_____"
]
],
[
[
"# function to get connected components of the ground truth binary image\ndef componentsStatsGroundTruth(matrix):\n # getting the info of the components in the ground truth\n # second value is connectivity 4 or 8\n connectedComponentsGroundTruth = cv2.connectedComponentsWithStats(matrix, 8, cv2.CV_32S)\n\n # Get the results\n # The first cell is the number of labels\n num_labels = connectedComponentsGroundTruth[0]\n # The second cell is the label matrix\n labels = connectedComponentsGroundTruth[1]\n # The third cell is the stat matrix\n stats = connectedComponentsGroundTruth[2]\n # The fourth cell is the centroid matrix\n centroids = connectedComponentsGroundTruth[3]\n\n return num_labels, labels, stats, centroids",
"_____no_output_____"
]
],
[
[
"# Show Images",
"_____no_output_____"
]
],
[
[
"from matplotlib.patches import Circle\nimport matplotlib.patches as mpatches\n\n# function to draw the grid to display\ndef display_grid(figure, axis, img, imgGroundTruth, preprocessed, candidates, features):\n # draw in the axis the img\n axis[0][0].imshow(img)\n # switch off the axis of the plot\n axis[0][0].axis('off')\n # set a title for the plot\n axis[0][0].set_title('Image')\n\n axis[0][1].imshow(imgGroundTruth, cmap='gray')\n axis[0][1].axis('off')\n axis[0][1].set_title('Ground Truth')\n\n axis[0][2].imshow(imgMask)\n axis[0][2].axis('off')\n axis[0][2].set_title('Breast Mask')\n\n axis[1][0].imshow(preprocessed, cmap='gray')\n axis[1][0].axis('off')\n axis[1][0].set_title('Preprocessed')\n\n # draw candidates as circles\n axis[1][1].imshow(preprocessed, cmap='gray')\n axis[1][1].axis('off')\n axis[1][1].set_title('Candidates')\n\n # Now, loop through coord arrays, and create a circle at each x,y pair\n for y,x,sigma in candidates:\n\n blob = Circle((x,y), sigma*5, color='blue', fill=False)\n axis[1][1].add_patch(blob)\n\n rect=mpatches.Rectangle((x,y),sigma,sigma, \n fill=False,\n color=\"red\",\n linewidth=2)\n axis[1][1].add_patch(rect)\n\n axis[1][2].imshow(imgGroundTruth, cmap='gray')\n axis[1][2].axis('off')\n axis[1][2].set_title('compare with ground truth and candidates')\n\n # Now, loop through coord arrays, and create a circle at each x,y pair\n for y,x,sigma in candidates:\n blob = Circle((x,y), sigma, color='blue', fill=False)\n axis[1][2].add_patch(blob)\n \n return figure, axis",
"_____no_output_____"
]
],
[
[
"# Main",
"_____no_output_____"
]
],
[
[
"import copy\nimport pandas as pd\n\n#go through the image files \nfor image, breastMask, groundTruth in zip(progress_bar(images), breastMasks, groundTruths):\n\n # to save the features generated with the glcm\n features = pd.DataFrame(dtype=np.float64)\n\n # 20588020, 7717, 5328, 3787, 5725, 3859, 6934, 50995872\n digits = '6934'\n\n if ((digits in image) and (digits in breastMask) and ('mask' in breastMask)):\n #if ('mask' in breastMask):\n #if image not in already:\n #upload images\n img = cv2.imread(os.path.join(DATA_DIR,'images',image))\n imgMask = cv2.imread(os.path.join(DATA_DIR, 'masks', breastMask))\n imgGroundTruth = cv2.imread(os.path.join(DATA_DIR, 'groundtruths', image), cv2.IMREAD_GRAYSCALE)\n imgCopy = copy.deepcopy(img)\n\n imgCopy = cv2.cvtColor(imgCopy, cv2.COLOR_RGB2GRAY)\n preprocessed = imgCLAHE(imgCopy)\n preprocessed = cv2.cvtColor(preprocessed, cv2.COLOR_GRAY2RGB)\n preprocessed = deHazingDarkChannelPriorPy(preprocessed, imgMask)\n \n \n preprocessedBlurDil = imgDilation(preprocessed)\n\n\n # candidate extraction #\n copyPreprocessed = copy.deepcopy(preprocessed)\n copyPreprocessedBlurDil = copy.deepcopy(preprocessedBlurDil)\n\n # we have to change np.float64 to np.float32 for the grayscale conversion\n copyPreprocessedBlurDil = copyPreprocessedBlurDil.astype(np.float32)\n copyPreprocessedBlurDil = cv2.cvtColor(copyPreprocessedBlurDil, cv2.COLOR_BGR2GRAY)\n\n candidates = candidateExtractionDoG(copyPreprocessedBlurDil, imgMask)\n\n # feature extraction #\n\n copyPreprocessed = copyPreprocessed.astype(np.float32)\n \n\n# copyPreprocessed = cv2.cvtColor(copyPreprocessed, cv2.COLOR_BGR2GRAY)\n\n features = featuresExtraction(copyPreprocessed, candidates, features, imgMask, imgGroundTruth, image, \"TestLBP\")\n \n # ML must be applied for the classification of the features extracted\n\n ################ ERASE MEMORY\n # import gc\n # del features\n # del preprocessed\n # del candidates\n # # del blobs\n # del copyPreprocessed\n # del imgCopy\n # del img\n # del imgMask\n # del imgGroundTruth\n # gc.collect()\n ############################\n\n # end image processing part #\n\n # display related #\n\n # matrix of plots and size of the figure\n figure, axis = plt.subplots(2, 3, figsize=(15,15))\n display_grid(figure, axis, img, imgGroundTruth, preprocessed, candidates, features)\n plt.subplots_adjust(wspace=0, hspace=0)\n\n # display figure with image\n plt.show() \n print(len(candidates))",
"_____no_output_____"
]
],
[
[
"## CONCLUSIONS FOR PREPROCESSING",
"_____no_output_____"
],
[
"* still missing quantum noise supression\n\n* details in the phd defense file\n\n* Observations from the results:\n * fiber intersections may also appear as bright spots (false positives)\n\n* THINGS WE NOTICE BETWEEN BOTH DEHAZING METHODS\n * Better suppression of fatty tissue (noise) and greater enhancement of brightness of desired feature (microcalcifications)\n * sometimes for the other dehazing method black patches become present in the fatty tissue\n * this did not happen in the dehazing with dark channel prior (and guided filter)\n * sharper\n * enhances the contrast and details\n\n* Observations from the results:\n * images with pectoral muscule cause false positives",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecb367952ce5847b6aae9627c7fa2e6e8cd7c34e | 150,412 | ipynb | Jupyter Notebook | docs/source/examples/Catch that asteroid!.ipynb | wumpus/poliastro | 6ef314f3b80528018ce489fd51d26db106daac91 | [
"MIT"
] | null | null | null | docs/source/examples/Catch that asteroid!.ipynb | wumpus/poliastro | 6ef314f3b80528018ce489fd51d26db106daac91 | [
"MIT"
] | null | null | null | docs/source/examples/Catch that asteroid!.ipynb | wumpus/poliastro | 6ef314f3b80528018ce489fd51d26db106daac91 | [
"MIT"
] | null | null | null | 268.114082 | 66,956 | 0.928457 | [
[
[
"# Catch that asteroid!",
"_____no_output_____"
],
[
"First, we need to increase the timeout time to allow the download of data occur properly",
"_____no_output_____"
]
],
[
[
"from astropy.utils.data import conf\nconf.dataurl",
"_____no_output_____"
],
[
"conf.remote_timeout ",
"_____no_output_____"
],
[
"conf.remote_timeout = 10000",
"_____no_output_____"
]
],
[
[
"Then, we do the rest of the imports.",
"_____no_output_____"
]
],
[
[
"from astropy import units as u\nfrom astropy.time import Time, TimeDelta\nfrom astropy.coordinates import solar_system_ephemeris\nsolar_system_ephemeris.set(\"jpl\")\n\nfrom poliastro.bodies import Sun, Earth, Moon\nfrom poliastro.ephem import Ephem\nfrom poliastro.frames import Planes\nfrom poliastro.twobody import Orbit\nfrom poliastro.plotting import StaticOrbitPlotter\nfrom poliastro.plotting.misc import plot_solar_system\nfrom poliastro.util import time_range\n\nEPOCH = Time(\"2017-09-01 12:05:50\", scale=\"tdb\")\nC_FLORENCE = \"#000\"\nC_MOON = \"#999\"",
"_____no_output_____"
],
[
"Earth.plot(EPOCH);",
"_____no_output_____"
]
],
[
[
"Our first option to retrieve the orbit of the Florence asteroid is to use `Orbit.from_sbdb`, which gives us the osculating elements at a certain epoch:",
"_____no_output_____"
]
],
[
[
"florence_osc = Orbit.from_sbdb(\"Florence\")\nflorence_osc",
"_____no_output_____"
]
],
[
[
"However, the epoch of the result is not close to the time of the close approach we are studying:",
"_____no_output_____"
]
],
[
[
"florence_osc.epoch.iso",
"_____no_output_____"
]
],
[
[
"Therefore, if we `propagate` this orbit to `EPOCH`, the results will be a bit different from the reality. Therefore, we need to find some other means.\n\nLet's use the `Ephem.from_horizons` method as an alternative, sampling over a period of 6 months:",
"_____no_output_____"
]
],
[
[
"from poliastro.ephem import Ephem",
"_____no_output_____"
],
[
"epochs = time_range(\n EPOCH - TimeDelta(3 * 30 * u.day), end=EPOCH + TimeDelta(3 * 30 * u.day)\n)",
"_____no_output_____"
],
[
"florence = Ephem.from_horizons(\"Florence\", epochs, plane=Planes.EARTH_ECLIPTIC)\nflorence",
"_____no_output_____"
],
[
"florence.plane",
"_____no_output_____"
]
],
[
[
"And now, let's compute the distance between Florence and the Earth at that epoch:",
"_____no_output_____"
]
],
[
[
"earth = Ephem.from_body(Earth, epochs, plane=Planes.EARTH_ECLIPTIC)\nearth",
"_____no_output_____"
],
[
"from poliastro.util import norm",
"_____no_output_____"
],
[
"min_distance = norm(florence.rv(EPOCH)[0] - earth.rv(EPOCH)[0]) - Earth.R\nmin_distance.to(u.km)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-success\">This value is consistent with what ESA says! $7\\,060\\,160$ km</div>",
"_____no_output_____"
]
],
[
[
"abs((min_distance - 7060160 * u.km) / (7060160 * u.km)).decompose()",
"_____no_output_____"
],
[
"from IPython.display import HTML\n\nHTML(\n\"\"\"<blockquote class=\"twitter-tweet\" data-lang=\"en\"><p lang=\"es\" dir=\"ltr\">La <a href=\"https://twitter.com/esa_es\">@esa_es</a> ha preparado un resumen del asteroide <a href=\"https://twitter.com/hashtag/Florence?src=hash\">#Florence</a> 😍 <a href=\"https://t.co/Sk1lb7Kz0j\">pic.twitter.com/Sk1lb7Kz0j</a></p>— AeroPython (@AeroPython) <a href=\"https://twitter.com/AeroPython/status/903197147914543105\">August 31, 2017</a></blockquote>\n<script src=\"//platform.twitter.com/widgets.js\" charset=\"utf-8\"></script>\"\"\"\n)",
"_____no_output_____"
]
],
[
[
"And now we can plot!",
"_____no_output_____"
]
],
[
[
"frame = plot_solar_system(outer=False, epoch=EPOCH)\nframe.plot_ephem(florence, EPOCH, label=\"Florence\", color=C_FLORENCE);",
"_____no_output_____"
]
],
[
[
"Finally, we are going to visualize the orbit of Florence with respect to the Earth. For that, we set a narrower time range, and specify that we want to retrieve the ephemerides with respect to our planet:",
"_____no_output_____"
]
],
[
[
"epochs = time_range(EPOCH - TimeDelta(5 * u.day), end=EPOCH + TimeDelta(5 * u.day))",
"_____no_output_____"
],
[
"florence_e = Ephem.from_horizons(\"Florence\", epochs, attractor=Earth)\nflorence_e",
"_____no_output_____"
]
],
[
[
"We now retrieve the ephemerides of the Moon, which are given directly in GCRS:",
"_____no_output_____"
]
],
[
[
"moon = Ephem.from_body(Moon, epochs, attractor=Earth)\nmoon",
"_____no_output_____"
],
[
"from poliastro.plotting.static import StaticOrbitPlotter\n\nplotter = StaticOrbitPlotter()\nplotter.set_attractor(Earth)\nplotter.set_body_frame(Moon)\nplotter.plot_ephem(moon, EPOCH, label=Moon, color=C_MOON);",
"_____no_output_____"
]
],
[
[
"And now, the glorious final plot:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nframe = StaticOrbitPlotter()\n\nframe.set_attractor(Earth)\nframe.set_orbit_frame(Orbit.from_ephem(Earth, florence_e, EPOCH))\n\nframe.plot_ephem(florence_e, EPOCH, label=\"Florence\", color=C_FLORENCE)\nframe.plot_ephem(moon, EPOCH, label=Moon, color=C_MOON);",
"_____no_output_____"
]
],
[
[
"<div style=\"text-align: center; font-size: 3em;\"><em>Per Python ad astra!</em></div>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb3703c5ef9cd0612df78101a3c9f8e1d8594c4 | 16,076 | ipynb | Jupyter Notebook | _notebooks/2022-01-23-Optimization-Review-of-Linear-Algebra-and-Geometry.ipynb | v-poghosyan/blog | 9852c4ac08feed5c2b3407022a2500d19b767a21 | [
"Apache-2.0"
] | null | null | null | _notebooks/2022-01-23-Optimization-Review-of-Linear-Algebra-and-Geometry.ipynb | v-poghosyan/blog | 9852c4ac08feed5c2b3407022a2500d19b767a21 | [
"Apache-2.0"
] | null | null | null | _notebooks/2022-01-23-Optimization-Review-of-Linear-Algebra-and-Geometry.ipynb | v-poghosyan/blog | 9852c4ac08feed5c2b3407022a2500d19b767a21 | [
"Apache-2.0"
] | null | null | null | 60.664151 | 849 | 0.615389 | [
[
[
"# Optimization - Review of Linear Algebra and Geometry\n\n> Preliminary mathematical concepts in the study of optimization - Eigenpairs, Fundamental Subspaces, Symmetry, Spectral Decomposition, Convexity, etc.\n\n- hide: false\n- toc: true\n- badges: true\n- comments: true\n- categories: ['Optimization','Applied Mathematics','Proofs']",
"_____no_output_____"
],
[
"# Introduction\n\nThe study of optimization can be summed up as the attempt to find those parameter(s) that optimize some objective function, if such exist. The objective function can be almost anything — cost, profit, nodes in a wireless network, distance to a destination, similarity to a target image, etc. If the objective function describes cost we may wish to minimize it. If, on the other hand, it describes profit then a natural goal would be to maximize it. \n\nThe problems of minimization and maximization, summed up as *optimization* in one word, are the same problem up to a reflection with respect to the domain of the parameter(s). \n\nFormally, let the objective function be $f: \\mathbb{R^n} \\to \\mathbb{R}$, and let it have minimizer $x^* \\in \\mathbb{R^n}$. Then, by definition of minimizer, $f(x^*) \\leq f(x) \\ \\ \\forall x \\in \\mathbb{R^n}$. It follows that $-f(x^*) \\geq -f(x) \\ \\ \\forall x \\in \\mathbb{R^n}$, so $x^*$ is the maximizer for $-f$.\n\n## Model of a Convex Optimization Problem\n\nThis series of posts will cover the ways in which we can solve an optimization problem of the form\n\n$\n\\textrm{minimize}: f(x)\n\\\\\n\\textrm{subject to}: x \\in \\mathcal{X}\n$\n\nwhere the *objective function* $f$ is a *convex function*, and the *constraint set* $\\mathcal{X}$ is a *convex set*. Importantly, we will *not* cover the ways in which we can model a real-world problem as a convex optimization problem of the above form.\n\n## Why Convex Optimization?\n\nFirst, let's define the *size* of an optimization problem as the dimensionality of the parameter $x$ added to the number of the problem constraints.\n\nConvex optimization problems are a class of *easy* optimization problems — problems whose time and/or space complexity grows slowly with respect to problem size.\n\nThese problems are general enough to capture many scenarios of interest, even some that do not fall strictly into the convex category, but specific enough to be solvable through generic algorithms and numerical methods.\n\n",
"_____no_output_____"
],
[
"# Review of Linear Algebra and Geometry\n\nWe start our exploration of convex optimization with a refresher on convexity and the linear algebra that's in common use in the subject. \n\n## Convexity\n\nSet convexity is defined as follows:\n\n> Definition: A set $C \\subseteq \\mathbb{R^d}$ is **convex** if, for all points $x_1,x_2 \\in C$ and any $\\theta \\in [0,1]$, the point $\\theta x_1 + (1-\\theta) x_2$ (i.e. the parametrized line segment between $x_1$ and $x_2$) is also in $C$.\n<br>\n\n\n### Some Operations that Preserve Convexity\n\nShifting, scaling, and rotation (i.e. *affine* transformations) preserve convexity. Let the matrix $A$ define such a transformation, and $b$ be a shift vector. Then $C' = \\{Ax + b \\ | \\ x \\in C \\}$ is convex provided that $C$ was convex.\n\nAn *intersection* of convex sets is also convex. That is, $C' = \\{ x \\ | \\ x \\in C_1 \\cap x \\in C_2 \\}$ is convex provided that $C_1$ and $C_2$ were convex to begin with. The proof follows directly from the definition of intersection...\n\nHowever, *unions* of convex sets need not be convex...\n\n## Examples of Convex Sets\n\nThe following are some common convex sets we will come across in practice.\n\n### Convex Hull of $n$ Points\n\nA *convex combination* of points $x_1, ..., x_n$ is a point of the form $x = \\theta_1 x_1 + ... + \\theta_n x_n$ where $\\sum_{i = 1}^{n} \\theta_i = 1$ and $\\theta_i \\geq 0 \\ \\ \\forall i$.\n\nLet $x_1,x_2,...,x_n$ be $n$ points in space. Their *convex hull* is the set of all points which can be written as some convex combination of them. Equivalently, by varying the $\\theta_i$'s we generate the convex hull as the set of all convex combinations of these points.\n\nThe convex hull can be visualized as the closed polygon formed when a rubber band is stretched around the $n$ points. The convex hull of two points is the line segment joining them. That of three points is the polygon (complete with its inner region). In general, for $n$ points, the concept generalizes to an $n$-dimensional polygon.\n\nFormally, the convex hull is the set $\\{ \\theta_1 x_1 + ... + \\theta_n x_n \\ | \\ \\theta_1 + ... + \\theta_n = 1 \\ \\ \\textrm{and} \\ \\ \\theta_i \\geq 0 \\ \\ \\forall i \\}$\n\n\n\n> Note: A handy interactive visualization, along with an efficient algorithm that generates a convex hull of $n$ points on a 2D plane can be found in the following [blog post](https://www.jgibson.id.au/articles/convex-hull/) by Joel Gibson.\n<br>\n\n### Convex Hull of a Set\n\nThe convex hull of a set can be similarly defined as all the convex combinations of the elements in the set. However, since the set may contain infinite elements, there's a more helpful, equivalent definition...\n\nLet $C$ be a non-convex set. The convex hull of $C$ is the intersection of all convex supersets of $C$. That is, it's the intersection of all convex sets containing $C$. The result of such an intersection will be the smallest convex superset of $C$. \n\nIn fact, this minimal convex superset is unique {% fn 1 %} and can therefore be taken as yet another, equivalent, definition for the convex hull of a set.\n\nVisualizing the convex hull of a non-convex set is similar to visualizing that of $n$ points — simply imagine the shape enclosed by a rubber band stretched around the non-convex set.\n\n### Affine Combination of $n$ Points\n\nAn *affine combination* of points $x_1,...,x_n$ is a point of the form $x = \\theta_1 x_1 + ... + \\theta_n x_n$ with $\\sum_{i=1}^{n}\\theta_i = 1$ but where the $\\theta_i$'s need not be non-negative. \n\nFor two points, the set of all affine combinations is the *line* that passes through them, whereas for three points it's the *plane*. in general, it is the plane in $(n-1)$-dimensions passing through the $n$ points.\n\n### Linear Combinations - Hyperplanes and Halfspaces\n\nA *linear combination* of $n$ vectors is all vectors of the form $x = \\theta_1 x_1 + ... + \\theta_n x_n$ with the $\\theta_i$'s totally unrestricted. \n\nThe set of all linear combinations of $n$ vectors (i.e. points) is called their *span*. Formally, it is the set $\\{ \\theta_1 x_1 + ... + \\theta_n x_n \\ \\ | \\ \\ \\forall \\theta_1,...,\\theta_n \\}$.\n\nThe span of a single vector is the line passing through it. For two vectors the span is the plane passing through them and, in general, the span of $n$ vectors is a plane in $n$-dimensions that contains these vectors.\n\n\n#### Hyperplanes\n\nFor fixed weights $\\theta_i = a_i \\ \\ \\forall i$, a *hyperplane* is the set of all points $x \\in \\mathbb{R^n}$ whose linear combination equals a fixed constant $b \\in \\mathbb{R}$.\n\nFormally, a hyperplane is the set $\\{ x \\ \\ | \\ \\ a_1 x_1 + ... a_n x_n = b\\} = \\{ x \\ \\ | \\ \\ a^T x = b\\}$ \n\nThere's a geometric interpretation of the parameters $a \\in \\mathbb{R^n}$ and $b \\in \\mathbb{R}$. Since the dot-product between perpendicular vectors is $0$, $\\{ x \\ \\ | \\ \\ a^T x = 0\\}$ is simply the set of all vectors perpendicular to $a$ (whose tail, as with all vectors in linear algebra, is considered to be fixed at the origin), making $a$ the *normal vector* to the hyperplane passing through the origin. To allow for parallel hyperplanes that are translated from the origin, the *offset* $b \\in \\mathbb{R}$ is introduced in the generalization $\\{ x \\ \\ | \\ \\ a^T x = b \\}$. This is now the set of all vectors whose dot-product with $a$ is constant. These vectors are not quite perpendicular to $a$, but they form a parallel hyperplane that's been shifted from the origin by a distance of $\\frac{|b|}{\\|a\\|_2}$.\n\nSince the sum $a_1 x_1 + ... a_n x_n = b$ is fixed, the last coordinate, which we'll call $x_k$ for some $k \\in [1,...,n]$, is fixed by the choice of the other $n-1$ coordinates. Therefore, a hyperplane in $\\mathbb{R^n}$ spans $n-1$ dimensions instead of $n$.\n<br>",
"_____no_output_____"
],
[
"#### Halfspaces\n\nA *halfspace* is either of the two sub-spaces a hyperplane partitions the whole space into. Since the dot-product between vectors which are roughly in the same direction is positive, and vice versa, the two halfspaces associated to a hyperplane $\\{ x \\ \\ | \\ \\ a^T x = b\\}$ are $\\{ x \\ \\ | \\ \\ a^T x \\geq b\\}$ and $\\{ x \\ \\ | \\ \\ a^T x \\leq b\\} $.",
"_____no_output_____"
],
[
"### Conic Combinations of $n$ Points\n\nA *conic combination* of $x_1,...x_n$ is a point $x = \\sum_{i=1}^{n} \\theta_i x_i$ where $\\theta_i \\geq 0 \\ \\ \\forall i$. Note that the absence of the restriction that $\\sum_{i=1}^{n} \\theta_i = 1$ is what distinguishes a conic combination from a convex combination. \n\n**A visual example:**\n\n and (1,1)\")",
"_____no_output_____"
],
[
"### Ellipses\n\nRecall from Euclidean geometry that ellipses are conic sections. In general we define ellipses in $n$-dimensions as the [sub-level sets](https://en.wikipedia.org/wiki/Level_set) of [quadratic forms](https://en.wikipedia.org/wiki/Quadratic_form). That is $\\{ x \\ \\ | \\ \\ (x-c)^T M (x-c) \\leq 1 \\}$ where $M \\succeq 0$ defines the stretch along each principal axis, and $c \\in \\mathbb{R^n}$ is the center. \n\nAn equivalent definition of an ellipse using the L2-norm is $\\{ x \\ \\ | \\ \\ \\|Ax - b\\|_2 \\leq 1 \\}$. That is, for a given $A$ and $b$ in the L2-norm definition, we can find an $M$ and $c$ in the sub-level set definition and vice versa. \n\n> Note: More generally, the ellipse is $\\{ x \\ \\ | \\ \\ (x-c)^T M (x-c) \\leq r \\}$. However, since the scaling factor $r$ is positive, it can simply be absorbed into $Q$ without affecting $Q$'s positive semidefiniteness.\n<br>\n\nTo quickly convince ourselves in the equivalence of these definitions, we take the simple case where $b = 0$.\n\n$$\n\\begin{aligned}\n \\|Ax\\|_2 &= ((Ax)^T(Ax))^{1/2} \\\\\n &= (x^TA^TAx)^{1/2} \\\\\n &= (x^TU D U^Tx)^{1/2} \\\\\n &= x^TU D^{1/2} U^Tx \\\\\n \\end{aligned}\n$$\n\nWhere the third equality is by the [spectral decomposition](https://en.wikipedia.org/wiki/Eigendecomposition_of_a_matrix#Real_symmetric_matrices) of the real symmetric matrix $A^TA$, in which $D = diag(\\lambda_1,...,\\lambda_n)$ is the diagnonal matrix of eigenvalues and the columns of $U$ are the corresponding eigenvectors. Taking $M= UD^{1/2}U^T$, where $D^{1/2}$ is simply $D^{1/2} = diag(\\sqrt\\lambda_1,...,\\sqrt\\lambda_n)$, we have the equivalent sub-level set definition of the ellipse. ",
"_____no_output_____"
],
[
"### Norm Balls\n\nRelated to ellipses are *Euclidean balls*, which are *norm balls* for the choice of the L2-norm. A Euclidean ball has the form $\\{ x \\ \\ | \\ \\ \\|x\\|_2 \\leq r \\}$, and is clearly convex as it's a generalizations of the sphere in $n$-dimensions. \n\nBut also, a Euclidean ball is the special ellipse for the choice of $M = rI$, and $c = 0$. \n\nIn general, norm balls $\\{ x \\ \\ | \\ \\ \\|x\\|_p \\leq r\\}$ where $\\|x\\|_p = (x_1^p + ... + x_n^p)^{1/p}$ are convex for any choice of $p \\geq 1$.",
"_____no_output_____"
],
[
"### Polyhedra\n\nWhere a halfspace is a set with one linear inequality constraint, a *polyhedron* is a set with many, but finite, such linear inequality constraints. These constraints can be packed into a matrix $A \\in \\mathbb{R^{m \\times n}}$ by vector $b \\in \\mathbb{R^m}$ multiplication form, making the polyhedron into the set $\\{x \\ \\ | \\ \\ Ax \\leq b\\}$.\n\nSince polyhedra are simply intersections of halfspaces and hyperplanes, and the latter are both convex, polyhedra are also convex sets.",
"_____no_output_____"
],
[
"### The Set of All Positive Semidefinite Matrices\n\nThe set of all PSD matrices $\\{ Q \\ \\ | \\ \\ x^TQx \\geq 0 \\ \\ \\forall x \\in \\mathbb{R^m}\\}$ is convex. We can, of course, use the definition of convexity to show this. But, a more elucidative approach would be the following remark. \n\nNote that $Q \\mapsto x^TQx$ is a [linear functional](https://en.wikipedia.org/wiki/Linear_form) that maps the space of all PSD matrices to its field of scalars. This is analogous to how $a \\mapsto x^Ta$ is a linear functional so, just as $\\{ a \\ \\ | \\ \\ x^Ta \\geq 0 \\}$ is a halfspace in the space of vectors, $H_x = \\{ Q \\ \\ | \\ \\ x^TQx \\geq 0 \\}$ for a given choice of $x \\in \\mathbb{R^m}$ is a halfspace in the space of PSD matrices. Halfspaces, as we already know, are convex and $\\{ Q \\ \\ | \\ \\ x^TQx \\geq 0 \\ \\ \\forall x \\in \\mathbb{R^m}\\}$ is nothing but an intersection of halfspaces for each choice of $x$. That is, $\\{ Q \\ \\ | \\ \\ x^TQx \\geq 0 \\ \\ \\forall x \\in \\mathbb{R^m}\\} = \\bigcap_x H_x$, concluding the proof of its convexity. \n",
"_____no_output_____"
],
[
"{{ '**Proof of uniqueness of the minimal, convex superset:** \nSuppose $C_1$ and $C_2$ are both minimal, convex supersets of $C$. Any convex set $D$ that contains $C$ must clearly contain the minimal, convex superset. Hence, $C_1 \\subseteq C_2$ and $C_2 \\subseteq C_1$, which implies $C_1 = C_2$.' | fndetail: 1 }}\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecb38489135b75fd6ec092cf1cda818c56badc8c | 64,704 | ipynb | Jupyter Notebook | tests/.ipynb_checkpoints/functional_testing-checkpoint.ipynb | geek-yang/DLACs | 0998e2ec0cb68b246da2ceea4e5cd54797d01a54 | [
"Apache-2.0"
] | 5 | 2020-03-21T14:37:40.000Z | 2022-03-28T11:47:13.000Z | tests/.ipynb_checkpoints/functional_testing-checkpoint.ipynb | geek-yang/DLACs | 0998e2ec0cb68b246da2ceea4e5cd54797d01a54 | [
"Apache-2.0"
] | 8 | 2022-01-20T16:05:11.000Z | 2022-02-13T18:19:44.000Z | tests/.ipynb_checkpoints/functional_testing-checkpoint.ipynb | geek-yang/DLACs | 0998e2ec0cb68b246da2ceea4e5cd54797d01a54 | [
"Apache-2.0"
] | 2 | 2021-01-29T03:25:05.000Z | 2021-03-22T12:15:15.000Z | 45.694915 | 276 | 0.423529 | [
[
[
"# Copyright Netherlands eScience Center <br>\n** Function : Predict the Spatial Sea Ice Concentration with BayesConvLSTM at weekly time scale** <br>\n** Author : Yang Liu ** <br>\n** First Built : 2020.03.02 ** <br>\n** Last Update : 2020.03.06 ** <br>\n** Library : Pytorth, Numpy, NetCDF4, os, iris, cartopy, dlacs, matplotlib **<br>\nDescription : This notebook serves to predict the Arctic sea ice using deep learning. The Bayesian Convolutional Long Short Time Memory neural network is used to deal with this spatial-temporal sequence problem. We use Pytorch as the deep learning framework. <br>\n<br>\n** Here we predict sea ice concentration with one extra relevant field from either ocean or atmosphere to test the predictor.** <br>\n\nReturn Values : Time series and figures <br>\n\nThe regionalization adopted here follows that of the MASIE (Multisensor Analyzed Sea Ice Extent) product available from the National Snow and Ice Data Center:<br>\nhttps://nsidc.org/data/masie/browse_regions<br>\nIt is given by paper J.Walsh et. al., 2019. Benchmark seasonal prediction skill estimates based on regional indices.<br>",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport sys\nimport warnings\nimport numbers\n\n# for data loading\nimport os\nfrom netCDF4 import Dataset\n# for pre-processing and machine learning\nimport numpy as np\nimport sklearn\n#import scipy\nimport torch\nimport torch.nn.functional\n\n#sys.path.append(os.path.join('C:','Users','nosta','ML4Climate','Scripts','DLACs'))\n#sys.path.append(\"C:\\\\Users\\\\nosta\\\\ML4Climate\\\\Scripts\\\\DLACs\")\nsys.path.append(\"../\")\nimport dlacs\nimport dlacs.BayesConvLSTM\nimport dlacs.preprocess\nimport dlacs.function\n\n# for visualization\nimport dlacs.visual\nimport matplotlib\nimport matplotlib.pyplot as plt\nfrom matplotlib.pyplot import cm\nimport iris # also helps with regriding\nimport cartopy\nimport cartopy.crs as ccrs\n\n# ignore all the DeprecationWarnings by pytorch\nif not sys.warnoptions:\n warnings.simplefilter(\"ignore\")",
"_____no_output_____"
]
],
[
[
"The testing device is Dell Inspirion 5680 with Intel Core i7-8700 x64 CPU and Nvidia GTX 1060 6GB GPU.<br>\nHere is a benchmark about cpu v.s. gtx 1060 <br>\nhttps://www.analyticsindiamag.com/deep-learning-tensorflow-benchmark-intel-i5-4210u-vs-geforce-nvidia-1060-6gb/",
"_____no_output_____"
]
],
[
[
"# constants\nconstant = {'g' : 9.80616, # gravititional acceleration [m / s2]\n 'R' : 6371009, # radius of the earth [m]\n 'cp': 1004.64, # heat capacity of air [J/(Kg*K)]\n 'Lv': 2500000, # Latent heat of vaporization [J/Kg]\n 'R_dry' : 286.9, # gas constant of dry air [J/(kg*K)]\n 'R_vap' : 461.5, # gas constant for water vapour [J/(kg*K)]\n 'rho' : 1026, # sea water density [kg/m3]\n }\n\n################################################################################# \n######### datapath ########\n#################################################################################\n# please specify data path\ndatapath_ERAI = '/home/ESLT0068/WorkFlow/Core_Database_DeepLearn/ERA-Interim'\n#datapath_ERAI = 'H:\\\\Creator_Zone\\\\Core_Database_DeepLearn\\\\ERA-Interim'\ndatapath_ORAS4 = '/home/ESLT0068/WorkFlow/Core_Database_DeepLearn/ORAS4'\n#datapath_ORAS4 = 'H:\\\\Creator_Zone\\\\Core_Database_DeepLearn\\\\ORAS4'\ndatapath_ORAS4_mask = '/home/ESLT0068/WorkFlow/Core_Database_DeepLearn/ORAS4'\n#datapath_ORAS4_mask = 'H:\\\\Creator_Zone\\\\Core_Database_DeepLearn\\\\ORAS4'\n#datapath_PIOMASS = '/home/ESLT0068/WorkFlow/Core_Database_AMET_OMET_reanalysis/PIOMASS'\n#datapath_PIOMASS = 'H:\\\\Creator_Zone\\\\Core_Database_AMET_OMET_reanalysis\\\\PIOMASS'\n#datapath_clim_index = '/home/ESLT0068/WorkFlow/Core_Database_AMET_OMET_reanalysis/Climate_index'\n#datapath_clim_index = 'F:\\\\PhD_essential\\\\Core_Database_AMET_OMET_reanalysis\\\\Climate_index'\noutput_path = '/home/ESLT0068/NLeSC/Computation_Modeling/ML4Climate/PredictArctic/BayesMaps'\n#output_path = 'C:\\\\Users\\\\nosta\\\\ML4Climate\\\\PredictArctic\\\\BayesMaps'",
"_____no_output_____"
],
[
"if __name__==\"__main__\":\n print ('*********************** get the key to the datasets *************************')\n # weekly variables on ERAI grid\n dataset_ERAI_fields_sic = Dataset(os.path.join(datapath_ERAI,\n 'sic_weekly_erai_1979_2017.nc'))\n# dataset_ERAI_fields_slp = Dataset(os.path.join(datapath_ERAI,\n# 'slp_weekly_erai_1979_2017.nc'))\n# dataset_ERAI_fields_t2m = Dataset(os.path.join(datapath_ERAI,\n# 't2m_weekly_erai_1979_2017.nc'))\n# dataset_ERAI_fields_z500 = Dataset(os.path.join(datapath_ERAI,\n# 'z500_weekly_erai_1979_2017.nc'))\n# dataset_ERAI_fields_z850 = Dataset(os.path.join(datapath_ERAI,\n# 'z850_weekly_erai_1979_2017.nc'))\n# dataset_ERAI_fields_uv10m = Dataset(os.path.join(datapath_ERAI,\n# 'uv10m_weekly_erai_1979_2017.nc'))\n# dataset_ERAI_fields_rad = Dataset(os.path.join(datapath_ERAI,\n# 'rad_flux_weekly_erai_1979_2017.nc'))\n #dataset_PIOMASS_siv = Dataset(os.path.join(datapath_PIOMASS,\n # 'siv_monthly_PIOMASS_1979_2017.nc'))\n # OHC interpolated on ERA-Interim grid\n dataset_ORAS4_OHC = Dataset(os.path.join(datapath_ORAS4,\n 'ohc_monthly_oras2erai_1978_2017.nc'))\n# dataset_index = Dataset(os.path.join(datapath_clim_index,\n# 'index_climate_monthly_regress_1950_2017.nc'))\n #dataset_ERAI_fields_flux = Dataset(os.path.join(datapath_ERAI_fields,\n # 'surface_erai_monthly_regress_1979_2017_radiation.nc'))\n # mask\n dataset_ORAS4_mask = Dataset(os.path.join(datapath_ORAS4_mask, 'mesh_mask.nc'))\n print ('*********************** extract variables *************************')\n #################################################################################\n ######### data gallery #########\n #################################################################################\n # we use time series from 1979 to 2016 (468 months in total)\n # training data: 1979 - 2013\n # validation: 2014 - 2016\n # variables list:\n # SIC (ERA-Interim) / SIV (PIOMASS) / SST (ERA-Interim) / ST (ERA-Interim) / OHC (ORAS4) / AO-NAO-AMO-NINO3.4 (NOAA)\n # integrals from spatial fields cover the area from 20N - 90N (4D fields [year, month, lat, lon])\n # *************************************************************************************** #\n # SIC (ERA-Interim) - benckmark\n SIC_ERAI = dataset_ERAI_fields_sic.variables['sic'][:-1,:,:,:] # 4D fields [year, week, lat, lon]\n year_ERAI = dataset_ERAI_fields_sic.variables['year'][:-1]\n week_ERAI = dataset_ERAI_fields_sic.variables['week'][:]\n latitude_ERAI = dataset_ERAI_fields_sic.variables['latitude'][:]\n longitude_ERAI = dataset_ERAI_fields_sic.variables['longitude'][:]\n # T2M (ERA-Interim)\n# T2M_ERAI = dataset_ERAI_fields_t2m.variables['t2m'][:-1,:,:,:] # 4D fields [year, week, lat, lon]\n# year_ERAI_t2m = dataset_ERAI_fields_t2m.variables['year'][:-1]\n# week_ERAI_t2m = dataset_ERAI_fields_t2m.variables['week'][:]\n# latitude_ERAI_t2m = dataset_ERAI_fields_t2m.variables['latitude'][:]\n# longitude_ERAI_t2m = dataset_ERAI_fields_t2m.variables['longitude'][:]\n # SLP (ERA-Interim)\n# SLP_ERAI = dataset_ERAI_fields_slp.variables['slp'][:-1,:,:,:] # 4D fields [year, week, lat, lon]\n# year_ERAI_slp = dataset_ERAI_fields_slp.variables['year'][:-1]\n# week_ERAI_slp = dataset_ERAI_fields_slp.variables['week'][:]\n# latitude_ERAI_slp = dataset_ERAI_fields_slp.variables['latitude'][:]\n# longitude_ERAI_slp = dataset_ERAI_fields_slp.variables['longitude'][:]\n # Z500 (ERA-Interim)\n# Z500_ERAI = dataset_ERAI_fields_z500.variables['z'][:-1,:,:,:] # 4D fields [year, week, lat, lon]\n# year_ERAI_z500 = dataset_ERAI_fields_z500.variables['year'][:-1]\n# week_ERAI_z500 = dataset_ERAI_fields_z500.variables['week'][:]\n# latitude_ERAI_z500 = dataset_ERAI_fields_z500.variables['latitude'][:]\n# longitude_ERAI_z500 = dataset_ERAI_fields_z500.variables['longitude'][:]\n # Z850 (ERA-Interim)\n# Z850_ERAI = dataset_ERAI_fields_z850.variables['z'][:-1,:,:,:] # 4D fields [year, week, lat, lon]\n# year_ERAI_z850 = dataset_ERAI_fields_z850.variables['year'][:-1]\n# week_ERAI_z850 = dataset_ERAI_fields_z850.variables['week'][:]\n# latitude_ERAI_z850 = dataset_ERAI_fields_z850.variables['latitude'][:]\n# longitude_ERAI_z850 = dataset_ERAI_fields_z850.variables['longitude'][:]\n # UV10M (ERA-Interim)\n# U10M_ERAI = dataset_ERAI_fields_uv10m.variables['u10m'][:-1,:,:,:] # 4D fields [year, week, lat, lon]\n# V10M_ERAI = dataset_ERAI_fields_uv10m.variables['v10m'][:-1,:,:,:] # 4D fields [year, week, lat, lon]\n# year_ERAI_uv10m = dataset_ERAI_fields_uv10m.variables['year'][:-1]\n# week_ERAI_uv10m = dataset_ERAI_fields_uv10m.variables['week'][:]\n# latitude_ERAI_uv10m = dataset_ERAI_fields_uv10m.variables['latitude'][:]\n# longitude_ERAI_uv10m = dataset_ERAI_fields_uv10m.variables['longitude'][:]\n # SFlux (ERA-Interim)\n# SFlux_ERAI = dataset_ERAI_fields_rad.variables['SFlux'][:-1,:,:,:] # 4D fields [year, week, lat, lon]\n# year_ERAI_SFlux = dataset_ERAI_fields_rad.variables['year'][:-1]\n# week_ERAI_SFlux = dataset_ERAI_fields_rad.variables['week'][:]\n# latitude_ERAI_SFlux = dataset_ERAI_fields_rad.variables['latitude'][:]\n# longitude_ERAI_SFlux = dataset_ERAI_fields_rad.variables['longitude'][:]\n #SIV (PIOMASS)\n #SIV_PIOMASS = dataset_PIOMASS_siv.variables['SIV'][:-12]\n #year_SIV = dataset_PIOMASS_siv.variables['year'][:-1]\n # OHC (ORAS4)\n # from 1978 - 2017 (for interpolation) / from 90 N upto 40 N\n OHC_300_ORAS4 = dataset_ORAS4_OHC.variables['OHC'][:-1,:,:67,:]/1000 # unit Peta Joule\n latitude_ORAS4 = dataset_ORAS4_OHC.variables['latitude'][:]\n longitude_ORAS4 = dataset_ORAS4_OHC.variables['longitude'][:]\n mask_OHC = np.ma.getmask(OHC_300_ORAS4[0,0,:,:])\n # AO-NAO-AMO-NINO3.4 (NOAA)\n# AO = dataset_index.variables['AO'][348:-1] # from 1979 - 2017\n# NAO = dataset_index.variables['NAO'][348:-1]\n# NINO = dataset_index.variables['NINO'][348:-1]\n# AMO = dataset_index.variables['AMO'][348:-1]\n# PDO = dataset_index.variables['PDO'][348:-1]",
"*********************** get the key to the datasets *************************\n*********************** extract variables *************************\n"
],
[
" #################################################################################\n ########### global land-sea mask ###########\n #################################################################################\n sea_ice_mask_global = np.ones((len(latitude_ERAI),len(longitude_ERAI)),dtype=float)\n sea_ice_mask_global[SIC_ERAI[0,0,:,:]==-1] = 0\n #################################################################################\n ########### regionalization sea mask ###########\n #################################################################################\n print ('*********************** create mask *************************')\n # W:-156 E:-124 N:80 S:67\n mask_Beaufort = np.zeros((len(latitude_ERAI),len(longitude_ERAI)),dtype=int)\n # W:-180 E:-156 N:80 S:66\n mask_Chukchi = np.zeros((len(latitude_ERAI),len(longitude_ERAI)),dtype=int)\n # W:146 E:180 N:80 S:67\n mask_EastSiberian = np.zeros((len(latitude_ERAI),len(longitude_ERAI)),dtype=int)\n # W:100 E:146 N:80 S:67\n mask_Laptev = np.zeros((len(latitude_ERAI),len(longitude_ERAI)),dtype=int)\n # W:60 E:100 N:80 S:67\n mask_Kara = np.zeros((len(latitude_ERAI),len(longitude_ERAI)),dtype=int)\n # W:18 E:60 N:80 S:64\n mask_Barents = np.zeros((len(latitude_ERAI),len(longitude_ERAI)),dtype=int)\n # W:-44 E:18 N:80 S:55\n mask_Greenland = np.zeros((len(latitude_ERAI),len(longitude_ERAI)),dtype=int)\n # W:-180 E:180 N:90 S:80\n mask_CenArctic = np.zeros((len(latitude_ERAI),len(longitude_ERAI)),dtype=int)\n print ('*********************** calc mask *************************')\n mask_Beaufort[13:31,32:76] = 1\n\n mask_Chukchi[13:32,0:32] = 1\n mask_Chukchi[13:32,-1] = 1\n\n mask_EastSiberian[13:31,434:479] = 1\n\n mask_Laptev[13:31,374:434] = 1\n\n mask_Kara[13:31,320:374] = 1\n\n mask_Barents[13:36,264:320] = 1\n\n mask_Greenland[13:47,179:264] = 1\n mask_Greenland[26:47,240:264] = 0\n\n mask_CenArctic[:13,:] = 1\n print ('*********************** packing *************************')\n mask_dict = {'Beaufort': mask_Beaufort[:,:],\n 'Chukchi': mask_Chukchi[:,:],\n 'EastSiberian': mask_EastSiberian[:,:],\n 'Laptev': mask_Laptev[:,:],\n 'Kara': mask_Kara[:,:],\n 'Barents': mask_Barents[:,:],\n 'Greenland': mask_Greenland[:,:],\n 'CenArctic': mask_CenArctic[:,:]}\n seas_namelist = ['Beaufort','Chukchi','EastSiberian','Laptev',\n 'Kara', 'Barents', 'Greenland','CenArctic']\n #################################################################################\n ######## temporal interpolation matrix ########\n #################################################################################\n # interpolate from monthly to weekly\n # original monthly data will be taken as the last week of the month\n OHC_300_ORAS4_weekly_series = np.zeros(SIC_ERAI.reshape(len(year_ERAI)*48,len(latitude_ERAI),len(longitude_ERAI)).shape,\n dtype=float)\n OHC_300_ORAS4_series= dlacs.preprocess.operator.unfold(OHC_300_ORAS4)\n # calculate the difference between two months\n OHC_300_ORAS4_deviation_series = (OHC_300_ORAS4_series[1:,:,:] - OHC_300_ORAS4_series[:-1,:,:]) / 4\n for i in np.arange(4):\n OHC_300_ORAS4_weekly_series[3-i::4,:,:] = OHC_300_ORAS4_series[12:,:,:] - i * OHC_300_ORAS4_deviation_series[11:,:,:]\n\n print ('****************** calculate extent from spatial fields *******************')\n # size of the grid box\n dx = 2 * np.pi * constant['R'] * np.cos(2 * np.pi * latitude_ERAI /\n 360) / len(longitude_ERAI)\n dy = np.pi * constant['R'] / 480\n # calculate the sea ice area\n SIC_ERAI_area = np.zeros(SIC_ERAI.shape, dtype=float)\n# SFlux_ERAI_area = np.zeros(SFlux_ERAI.shape, dtype=float)\n for i in np.arange(len(latitude_ERAI[:])):\n # change the unit to terawatt\n SIC_ERAI_area[:,:,i,:] = SIC_ERAI[:,:,i,:]* dx[i] * dy / 1E+6 # unit km2\n# SFlux_ERAI_area[:,:,i,:] = SFlux_ERAI[:,:,i,:]* dx[i] * dy / 1E+12 # unit TeraWatt\n SIC_ERAI_area[SIC_ERAI_area<0] = 0 # switch the mask from -1 to 0\n print ('================ reshape input data into time series =================')\n SIC_ERAI_area_series = dlacs.preprocess.operator.unfold(SIC_ERAI_area)\n# T2M_ERAI_series = dlacs.preprocess.operator.unfold(T2M_ERAI)\n# SLP_ERAI_series = dlacs.preprocess.operator.unfold(SLP_ERAI)\n# Z500_ERAI_series = dlacs.preprocess.operator.unfold(Z500_ERAI)\n# Z850_ERAI_series = dlacs.preprocess.operator.unfold(Z850_ERAI)\n# U10M_ERAI_series = dlacs.preprocess.operator.unfold(U10M_ERAI)\n# V10M_ERAI_series = dlacs.preprocess.operator.unfold(V10M_ERAI)\n# SFlux_ERAI_area_series = dlacs.preprocess.operator.unfold(SFlux_ERAI_area)\n print ('****************** choose the fields from target region *******************')\n # select land-sea mask\n sea_ice_mask_barents = sea_ice_mask_global[12:36,264:320]\n print ('****************** choose the fields from target region *******************')\n # select the area between greenland and ice land for instance 60-70 N / 44-18 W\n sic_exp = SIC_ERAI_area_series[:,12:36,264:320]\n# t2m_exp = T2M_ERAI_series[:,12:36,264:320]\n# slp_exp = SLP_ERAI_series[:,12:36,264:320]\n# z500_exp = Z500_ERAI_series[:,12:36,264:320]\n# z850_exp = Z850_ERAI_series[:,12:36,264:320]\n# u10m_exp = U10M_ERAI_series[:,12:36,264:320]\n# v10m_exp = V10M_ERAI_series[:,12:36,264:320]\n# sflux_exp = SFlux_ERAI_area_series[:,12:36,264:320]\n ohc_exp = OHC_300_ORAS4_weekly_series[:,12:36,264:320]\n print(sic_exp.shape)\n# print(t2m_exp.shape)\n# print(slp_exp.shape)\n# print(z500_exp.shape)\n# print(u10m_exp.shape)\n# print(v10m_exp.shape)\n# print(sflux_exp.shape)\n print(ohc_exp.shape)\n print(latitude_ERAI[12:36])\n print(longitude_ERAI[264:320])\n print(latitude_ORAS4[12:36])\n print(longitude_ORAS4[264:320])\n #print(latitude_ERAI[26:40])\n #print(longitude_ERAI[180:216])\n #print(sic_exp[:])\n print ('******************* pre-processing *********************')\n print ('========================= normalize data ===========================')\n sic_exp_norm = dlacs.preprocess.operator.normalize(sic_exp)\n# t2m_exp_norm = deepclim.preprocess.operator.normalize(t2m_exp)\n# slp_exp_norm = deepclim.preprocess.operator.normalize(slp_exp)\n# z500_exp_norm = deepclim.preprocess.operator.normalize(z500_exp)\n# z850_exp_norm = deepclim.preprocess.operator.normalize(z850_exp)\n# u10m_exp_norm = deepclim.preprocess.operator.normalize(u10m_exp)\n# v10m_exp_norm = deepclim.preprocess.operator.normalize(v10m_exp)\n# sflux_exp_norm = deepclim.preprocess.operator.normalize(sflux_exp)\n ohc_exp_norm = dlacs.preprocess.operator.normalize(ohc_exp)\n print('================ save the normalizing factor =================')\n sic_max = np.amax(sic_exp)\n sic_min = np.amin(sic_exp)\n print(sic_max,\"km2\")\n print(sic_min,\"km2\")\n print ('==================== A series of time (index) ====================')\n _, yy, xx = sic_exp_norm.shape # get the lat lon dimension\n year = np.arange(1979,2017,1)\n year_cycle = np.repeat(year,48)\n month_cycle = np.repeat(np.arange(1,13,1),4)\n month_cycle = np.tile(month_cycle,len(year)+1) # one extra repeat for lead time dependent prediction\n month_cycle.astype(float)\n month_2D = np.repeat(month_cycle[:,np.newaxis],yy,1)\n month_exp = np.repeat(month_2D[:,:,np.newaxis],xx,2)\n print ('=================== artificial data for evaluation ====================')\n # calculate climatology of SIC\n# seansonal_cycle_SIC = np.zeros(48,dtype=float)\n# for i in np.arange(48):\n# seansonal_cycle_SIC[i] = np.mean(SIC_ERAI_sum_norm[i::48],axis=0)\n # weight for loss\n# weight_month = np.array([0,1,1,\n# 1,0,0,\n# 1,1,1,\n# 0,0,0])\n #weight_loss = np.repeat(weight_month,4)\n #weight_loss = np.tile(weight_loss,len(year))",
"*********************** create mask *************************\n*********************** calc mask *************************\n*********************** packing *************************\n****************** calculate extent from spatial fields *******************\n================ reshape input data into time series =================\n****************** choose the fields from target region *******************\n****************** choose the fields from target region *******************\n(1824, 24, 56)\n(1824, 24, 56)\n[80.5 79.75 79. 78.25 77.5 76.75 76. 75.25 74.5 73.75 73. 72.25\n 71.5 70.75 70. 69.25 68.5 67.75 67. 66.25 65.5 64.75 64. 63.25]\n[18. 18.75 19.5 20.25 21. 21.75 22.5 23.25 24. 24.75 25.5 26.25\n 27. 27.75 28.5 29.25 30. 30.75 31.5 32.25 33. 33.75 34.5 35.25\n 36. 36.75 37.5 38.25 39. 39.75 40.5 41.25 42. 42.75 43.5 44.25\n 45. 45.75 46.5 47.25 48. 48.75 49.5 50.25 51. 51.75 52.5 53.25\n 54. 54.75 55.5 56.25 57. 57.75 58.5 59.25]\n[80.5 79.75 79. 78.25 77.5 76.75 76. 75.25 74.5 73.75 73. 72.25\n 71.5 70.75 70. 69.25 68.5 67.75 67. 66.25 65.5 64.75 64. 63.25]\n[18. 18.75 19.5 20.25 21. 21.75 22.5 23.25 24. 24.75 25.5 26.25\n 27. 27.75 28.5 29.25 30. 30.75 31.5 32.25 33. 33.75 34.5 35.25\n 36. 36.75 37.5 38.25 39. 39.75 40.5 41.25 42. 42.75 43.5 44.25\n 45. 45.75 46.5 47.25 48. 48.75 49.5 50.25 51. 51.75 52.5 53.25\n 54. 54.75 55.5 56.25 57. 57.75 58.5 59.25]\n******************* pre-processing *********************\n========================= normalize data ===========================\n================ save the normalizing factor =================\n1565.2049481856002 km2\n0.0 km2\n==================== A series of time (index) ====================\n=================== artificial data for evaluation ====================\n"
]
],
[
[
"# Procedure for LSTM <br>\n** We use Pytorth to implement LSTM neural network with time series of climate data. ** <br>",
"_____no_output_____"
]
],
[
[
" print ('******************* parameter for check *********************')\n choice_exp_norm = ohc_exp_norm\n print ('******************* create basic dimensions for tensor and network *********************')\n # specifications of neural network\n input_channels = 3\n hidden_channels = [3, 2, 1] # number of channels & hidden layers, the channels of last layer is the channels of output, too\n #hidden_channels = [3, 3, 3, 3, 2]\n #hidden_channels = [2]\n kernel_size = 3\n # here we input a sequence and predict the next step only\n #step = 1 # how many steps to predict ahead\n #effective_step = [0] # step to output\n batch_size = 1\n #num_layers = 1\n learning_rate = 0.01\n num_epochs = 150#0\n print ('******************* cross validation and testing data *********************')\n # take 10% data as cross-validation data\n cross_valid_year = 4\n # take 10% years as testing data\n test_year = 4\n # minibatch\n #iterations = 3 # training data divided into 3 sets\n print ('******************* check the environment *********************')\n print (\"Pytorch version {}\".format(torch.__version__))\n # check if CUDA is available\n use_cuda = torch.cuda.is_available()\n print(\"Is CUDA available? {}\".format(use_cuda))\n # CUDA settings torch.__version__ must > 0.4\n # !!! This is important for the model!!! The first option is gpu\n device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\") \n print ('******************* run BayesConvLSTM *********************')\n print ('The model is designed to make many to one prediction.')\n print ('A series of multi-chanel variables will be input to the model.')\n print ('The model learns by verifying the output at each timestep.')\n # check the sequence length\n sequence_len, height, width = sic_exp_norm.shape\n # initialize our model\n model = dlacs.BayesConvLSTM.BayesConvLSTM(input_channels, hidden_channels, kernel_size).to(device)\n # use Evidence Lower Bound (ELBO) to quantify the loss\n ELBO = dlacs.function.ELBO(height*width)\n # for classification, target must be integers (label)\n #ELBO = dlacs.function.ELBO(height*width,loss_function=torch.nn.KLDivLoss())\n #ELBO = dlacs.function.ELBO(height*width,loss_function=torch.CrossEntropyLoss())\n #ELBO = dlacs.function.ELBO(height*width,loss_function=torch.NLLLoss(reduction='mean'))\n # penalty for kl\n penalty_kl = sequence_len\n # stochastic gradient descent\n #optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate, momentum=0.9)\n # Adam optimizer\n optimiser = torch.optim.Adam(model.parameters(), lr=learning_rate)\n print(model)\n print(ELBO)\n print(optimiser)",
"******************* parameter for check *********************\n******************* create basic dimensions for tensor and network *********************\n******************* cross validation and testing data *********************\n******************* check the environment *********************\nPytorch version 1.1.0\nIs CUDA available? False\n******************* run BayesConvLSTM *********************\nThe model is designed to make many to one prediction.\nA series of multi-chanel variables will be input to the model.\nThe model learns by verifying the output at each timestep.\nBayesConvLSTM(\n (cell0): BayesConvLSTMCell()\n (cell1): BayesConvLSTMCell()\n (cell2): BayesConvLSTMCell()\n)\nELBO(\n (loss_function): MSELoss()\n)\nAdam (\nParameter Group 0\n amsgrad: False\n betas: (0.9, 0.999)\n eps: 1e-08\n lr: 0.01\n weight_decay: 0\n)\n"
],
[
" for name, param in model.named_parameters():\n if param.requires_grad:\n print (name)\n print (param.data)\n print (param.size())\n print (\"=========================\")",
"cell0.Wxi_mu\ntensor([[[[-1.0558e-01, -1.8014e-01, 3.3321e-02],\n [ 2.3716e-02, -1.2316e-01, 1.5326e-01],\n [-1.4931e-01, -1.2037e-01, -2.9395e-03]],\n\n [[-1.6941e-01, 1.2226e-01, -6.5187e-02],\n [-1.0834e-01, -1.4857e-01, -1.3921e-04],\n [-1.3112e-01, 1.2654e-01, -6.7747e-02]],\n\n [[ 1.7094e-01, -4.0709e-02, 1.6852e-01],\n [ 6.5585e-03, -1.6063e-01, 1.8219e-01],\n [ 1.2504e-01, 9.6949e-02, -7.3622e-02]]],\n\n\n [[[ 1.3706e-01, -1.2122e-01, 6.7758e-02],\n [ 8.9354e-02, 1.5358e-01, 1.2304e-01],\n [-6.3727e-02, 7.3968e-02, -1.8771e-01]],\n\n [[ 1.2527e-01, -1.1710e-02, -7.6872e-02],\n [ 3.5652e-03, 1.4838e-01, -5.1766e-02],\n [ 1.2488e-02, 1.5517e-02, -1.2831e-02]],\n\n [[ 1.7406e-01, 1.8051e-01, 4.7447e-02],\n [ 9.8560e-02, -1.8229e-03, -1.1217e-01],\n [ 5.5913e-03, 4.4141e-03, -1.0908e-01]]],\n\n\n [[[ 4.8881e-02, -5.0010e-02, 1.7787e-01],\n [ 7.9141e-02, 6.8945e-02, -1.5756e-01],\n [ 1.5758e-01, -1.7167e-01, -9.1566e-02]],\n\n [[ 9.8564e-02, 1.4169e-01, -4.1926e-02],\n [-1.2288e-03, -1.0169e-01, 1.8602e-01],\n [ 1.7055e-01, -6.6630e-02, -1.7616e-02]],\n\n [[ 1.0433e-01, -6.5166e-02, -7.2140e-02],\n [ 1.2388e-01, -1.6532e-01, -1.3893e-01],\n [-7.9466e-02, 2.5198e-02, -1.2264e-01]]]])\ntorch.Size([3, 3, 3, 3])\n=========================\ncell0.Whi_mu\ntensor([[[[ 0.1505, 0.1594, 0.1507],\n [-0.1064, -0.1529, 0.0767],\n [-0.0612, -0.0013, -0.0842]],\n\n [[-0.1894, -0.1463, 0.0028],\n [ 0.0290, 0.0451, -0.0291],\n [ 0.1440, -0.0578, -0.1537]],\n\n [[-0.1109, -0.0751, 0.0480],\n [ 0.1751, -0.0272, -0.1116],\n [ 0.0330, -0.1047, 0.0708]]],\n\n\n [[[-0.0114, 0.1160, 0.1861],\n [-0.0486, -0.0762, 0.0408],\n [-0.1535, 0.1055, -0.1539]],\n\n [[ 0.1408, 0.0064, 0.1595],\n [ 0.1489, -0.0719, -0.1464],\n [ 0.1878, 0.0596, -0.1849]],\n\n [[-0.0016, 0.1308, -0.0777],\n [ 0.0976, 0.1232, 0.0970],\n [-0.0749, 0.0472, 0.1033]]],\n\n\n [[[-0.0918, -0.0488, -0.1228],\n [ 0.1324, -0.0902, 0.0612],\n [-0.0488, 0.1842, -0.1201]],\n\n [[ 0.0284, -0.0162, -0.0321],\n [ 0.1476, -0.1896, 0.0177],\n [ 0.0410, -0.1147, -0.1073]],\n\n [[ 0.0245, 0.0552, 0.0043],\n [-0.0861, 0.0531, -0.0379],\n [-0.0255, -0.0181, 0.1451]]]])\ntorch.Size([3, 3, 3, 3])\n=========================\ncell0.Wxf_mu\ntensor([[[[ 0.1483, -0.1208, 0.0284],\n [ 0.0200, -0.0550, -0.1284],\n [ 0.1364, 0.1188, -0.0278]],\n\n [[ 0.0073, 0.0267, 0.1735],\n [-0.0877, 0.1521, 0.1651],\n [ 0.1610, 0.1607, 0.0480]],\n\n [[-0.0760, 0.0976, 0.1390],\n [ 0.1454, 0.1334, -0.0760],\n [ 0.1114, 0.1230, -0.0989]]],\n\n\n [[[ 0.0878, -0.1020, -0.0035],\n [ 0.1706, -0.0822, 0.0816],\n [-0.0940, -0.0863, 0.0037]],\n\n [[ 0.0222, -0.1566, -0.1362],\n [-0.0844, -0.1022, 0.0365],\n [-0.1225, 0.1563, 0.0662]],\n\n [[ 0.0304, -0.1415, -0.0393],\n [-0.1836, -0.0835, -0.0122],\n [-0.0296, -0.0322, 0.1411]]],\n\n\n [[[ 0.0464, -0.1525, 0.1017],\n [-0.0647, 0.0233, -0.1618],\n [ 0.1650, 0.0671, 0.1499]],\n\n [[-0.0495, -0.1173, -0.1195],\n [-0.1611, 0.0425, -0.1830],\n [ 0.1597, 0.0825, -0.0109]],\n\n [[ 0.0979, 0.0147, 0.0652],\n [ 0.1144, 0.0251, 0.0099],\n [-0.0434, 0.0506, -0.1574]]]])\ntorch.Size([3, 3, 3, 3])\n=========================\ncell0.Whf_mu\ntensor([[[[-0.0477, 0.1021, 0.1133],\n [ 0.1019, -0.1736, -0.1368],\n [ 0.1167, -0.1414, 0.0797]],\n\n [[ 0.0577, 0.1485, -0.0892],\n [-0.0392, -0.0002, -0.0093],\n [ 0.0752, 0.0591, -0.1756]],\n\n [[-0.1076, 0.0940, -0.0927],\n [-0.0192, -0.0187, -0.0515],\n [-0.0295, 0.1501, 0.0820]]],\n\n\n [[[ 0.0443, 0.0992, -0.0708],\n [ 0.1801, 0.0410, -0.1901],\n [ 0.1426, 0.1340, 0.1905]],\n\n [[-0.1884, 0.0514, 0.1214],\n [ 0.1535, -0.1456, 0.0904],\n [ 0.1441, -0.1416, 0.0159]],\n\n [[ 0.0998, -0.0896, 0.0262],\n [ 0.1321, 0.1180, 0.1093],\n [ 0.1409, 0.0826, -0.1799]]],\n\n\n [[[ 0.0978, 0.1783, 0.0761],\n [-0.0497, 0.1332, 0.1554],\n [-0.0412, 0.0712, 0.1806]],\n\n [[-0.1848, 0.0492, 0.1601],\n [ 0.1624, 0.0586, -0.1298],\n [ 0.0999, 0.0259, 0.0918]],\n\n [[ 0.1636, -0.0670, -0.0600],\n [-0.0835, -0.1121, -0.0713],\n [-0.1210, 0.0723, -0.0130]]]])\ntorch.Size([3, 3, 3, 3])\n=========================\ncell0.Wxc_mu\ntensor([[[[-0.0057, -0.0202, -0.0485],\n [-0.0967, 0.1473, -0.0356],\n [-0.0162, -0.0720, -0.1881]],\n\n [[-0.0987, -0.0084, 0.1601],\n [ 0.1424, -0.1698, -0.0228],\n [-0.1683, 0.1419, -0.0611]],\n\n [[-0.1773, -0.1575, 0.1331],\n [ 0.1831, 0.1064, 0.1431],\n [-0.0737, -0.0624, -0.0597]]],\n\n\n [[[-0.1334, 0.0719, 0.1027],\n [ 0.1069, -0.1807, -0.1687],\n [-0.1189, -0.1354, -0.1711]],\n\n [[ 0.1653, -0.0884, 0.0597],\n [-0.1120, -0.1139, 0.1678],\n [ 0.1285, -0.0140, 0.0315]],\n\n [[ 0.0852, -0.0090, 0.0941],\n [-0.1844, -0.1168, 0.1344],\n [-0.0234, 0.0558, -0.0977]]],\n\n\n [[[-0.0627, -0.1281, 0.1190],\n [-0.1080, -0.1252, 0.1044],\n [-0.0424, 0.0172, 0.0418]],\n\n [[-0.0770, 0.0221, -0.0586],\n [-0.0362, -0.1121, -0.0629],\n [ 0.1090, -0.0433, -0.1642]],\n\n [[-0.0198, -0.1160, 0.0252],\n [-0.0019, 0.1053, 0.0765],\n [-0.0321, -0.1601, 0.0124]]]])\ntorch.Size([3, 3, 3, 3])\n=========================\ncell0.Whc_mu\ntensor([[[[ 0.0359, 0.1472, 0.1581],\n [-0.1290, -0.0004, 0.0275],\n [-0.0004, 0.0909, -0.1282]],\n\n [[ 0.0801, 0.0296, -0.1655],\n [ 0.1732, 0.0651, -0.0822],\n [-0.0551, -0.1902, 0.1069]],\n\n [[-0.1731, 0.1697, -0.1525],\n [-0.0669, -0.0647, -0.1256],\n [ 0.0472, -0.1045, -0.0320]]],\n\n\n [[[-0.1885, -0.1612, 0.1851],\n [ 0.1781, -0.1923, 0.0423],\n [ 0.1899, 0.1653, -0.0224]],\n\n [[-0.0145, 0.0156, -0.0249],\n [-0.1112, -0.0055, -0.0352],\n [ 0.0536, 0.0455, 0.1236]],\n\n [[-0.0340, -0.1064, -0.0361],\n [-0.0933, 0.0180, 0.0059],\n [ 0.0303, -0.1235, 0.0433]]],\n\n\n [[[ 0.0081, 0.1448, 0.1891],\n [ 0.1265, 0.1450, -0.1162],\n [-0.0990, -0.1284, 0.1863]],\n\n [[-0.0763, -0.1483, -0.0290],\n [ 0.1244, 0.0433, -0.1442],\n [-0.0900, -0.1422, 0.1674]],\n\n [[-0.0737, 0.1878, -0.1239],\n [-0.0063, -0.0742, -0.1087],\n [ 0.0585, 0.1563, -0.0684]]]])\ntorch.Size([3, 3, 3, 3])\n=========================\ncell0.Wxo_mu\ntensor([[[[-0.0006, -0.1884, -0.0080],\n [-0.1371, 0.0886, 0.1734],\n [ 0.0219, -0.1607, 0.1174]],\n\n [[-0.1835, 0.1636, 0.0334],\n [ 0.0926, 0.1284, 0.1311],\n [ 0.1581, 0.1168, -0.1597]],\n\n [[ 0.1251, 0.0225, -0.1540],\n [-0.0835, -0.1881, -0.1867],\n [-0.1786, -0.1777, -0.0792]]],\n\n\n [[[-0.0390, 0.1583, -0.1335],\n [-0.0334, -0.0339, -0.1427],\n [-0.0163, 0.0857, -0.1614]],\n\n [[ 0.0259, 0.0045, 0.0927],\n [-0.1736, -0.0424, 0.1609],\n [ 0.1830, 0.0239, -0.0536]],\n\n [[ 0.1002, -0.0555, -0.1863],\n [ 0.0137, 0.0472, 0.1629],\n [ 0.1072, -0.0688, -0.1815]]],\n\n\n [[[ 0.1561, 0.1189, -0.1054],\n [-0.0654, 0.0633, 0.0789],\n [-0.0077, -0.0585, 0.1569]],\n\n [[ 0.0418, 0.0187, -0.1913],\n [ 0.1730, 0.0185, 0.0857],\n [-0.1578, -0.0213, 0.1000]],\n\n [[-0.1295, -0.0706, -0.0222],\n [ 0.0388, 0.0221, -0.0255],\n [ 0.1079, 0.0573, -0.0127]]]])\ntorch.Size([3, 3, 3, 3])\n=========================\ncell0.Who_mu\ntensor([[[[-0.1418, 0.1296, 0.1494],\n [-0.1297, 0.0966, -0.1601],\n [ 0.0589, -0.1354, 0.1033]],\n\n [[-0.1342, 0.0837, -0.0151],\n [-0.0918, -0.0557, 0.1827],\n [ 0.1472, 0.1182, 0.0730]],\n\n [[-0.0610, 0.1449, -0.1682],\n [ 0.0557, 0.1713, -0.1626],\n [-0.1836, -0.1490, 0.0024]]],\n\n\n [[[ 0.1771, -0.0781, -0.1357],\n [-0.0530, 0.1234, 0.0002],\n [ 0.0061, 0.0391, 0.1904]],\n\n [[ 0.0308, 0.0320, 0.0450],\n [ 0.1424, -0.1130, -0.1656],\n [ 0.0455, 0.1137, 0.1095]],\n\n [[-0.1099, 0.0360, -0.0398],\n [-0.0466, 0.1522, 0.0596],\n [-0.1342, 0.1874, 0.1896]]],\n\n\n [[[-0.1889, -0.0500, -0.0976],\n [-0.0876, -0.0597, -0.1398],\n [-0.0772, -0.1107, -0.0526]],\n\n [[ 0.0040, -0.0116, -0.1339],\n [ 0.1082, 0.0162, 0.0092],\n [-0.0347, -0.0388, -0.0249]],\n\n [[-0.0752, 0.1624, -0.0845],\n [ 0.1130, 0.0603, -0.0400],\n [-0.0488, 0.1633, 0.1463]]]])\ntorch.Size([3, 3, 3, 3])\n=========================\ncell0.Wxi_bias\ntensor([0.0449, 0.1731, 0.0882])\ntorch.Size([3])\n=========================\ncell0.Wxf_bias\ntensor([ 0.1077, 0.0077, -0.0217])\ntorch.Size([3])\n=========================\ncell0.Wxc_bias\ntensor([ 0.0084, -0.1140, 0.0357])\ntorch.Size([3])\n=========================\ncell0.Wxo_bias\ntensor([ 0.1414, -0.0581, -0.1559])\ntorch.Size([3])\n=========================\ncell0.Wxi_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell0.Whi_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell0.Wxf_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell0.Whf_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell0.Wxc_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell0.Whc_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell0.Wxo_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell0.Who_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell1.Wxi_mu\ntensor([[[[-0.1404, 0.0992, 0.0712],\n [ 0.1500, -0.0049, -0.1585],\n [ 0.0450, 0.0732, 0.1892]],\n\n [[ 0.0362, -0.1619, 0.1274],\n [ 0.1129, -0.0810, 0.1015],\n [-0.0978, 0.0430, -0.0125]],\n\n [[-0.1353, -0.0158, 0.1046],\n [-0.1306, -0.0575, -0.0956],\n [ 0.1300, -0.1472, -0.1036]]],\n\n\n [[[-0.0360, -0.1872, 0.0382],\n [-0.1581, -0.1454, -0.0130],\n [ 0.0750, -0.0926, 0.0594]],\n\n [[ 0.0781, -0.1598, -0.1888],\n [ 0.1771, -0.0096, -0.0051],\n [-0.0063, 0.1794, 0.1344]],\n\n [[-0.1107, 0.1619, -0.1914],\n [-0.1832, 0.1638, -0.0908],\n [ 0.0348, 0.1907, 0.0779]]]])\ntorch.Size([2, 3, 3, 3])\n=========================\ncell1.Whi_mu\ntensor([[[[-0.0060, 0.1114, 0.1306],\n [ 0.1781, 0.1436, 0.1708],\n [-0.1407, 0.1659, 0.0237]],\n\n [[-0.0003, 0.1765, -0.1130],\n [ 0.0117, 0.1632, 0.0438],\n [-0.1570, -0.1841, 0.0727]]],\n\n\n [[[ 0.0648, -0.0630, 0.0190],\n [-0.0354, -0.1580, -0.0695],\n [-0.0177, 0.1496, 0.0449]],\n\n [[ 0.1066, 0.1308, 0.1854],\n [ 0.0280, -0.0853, 0.0243],\n [-0.1214, -0.1760, -0.1087]]]])\ntorch.Size([2, 2, 3, 3])\n=========================\ncell1.Wxf_mu\ntensor([[[[ 1.8089e-01, 4.6241e-02, -1.5846e-02],\n [-5.0193e-02, -1.3801e-01, -2.7986e-03],\n [-1.3488e-01, -1.6418e-01, -4.1823e-02]],\n\n [[-1.7122e-01, -1.0898e-01, 7.3998e-02],\n [ 1.0235e-01, 1.4572e-02, 1.7708e-01],\n [ 1.8915e-01, -1.5504e-01, 1.1076e-01]],\n\n [[ 1.3813e-01, -1.3368e-02, 3.3334e-02],\n [-3.3073e-02, 7.7808e-02, -1.1030e-01],\n [-3.9036e-02, -5.6320e-02, 2.0027e-02]]],\n\n\n [[[ 1.5672e-01, -9.8987e-02, 4.5164e-02],\n [ 1.1657e-01, 6.4120e-05, -1.3413e-01],\n [-3.0016e-02, 8.5811e-04, -1.7133e-01]],\n\n [[ 4.8487e-02, -1.0534e-01, -1.4458e-01],\n [ 9.2448e-02, -9.0670e-03, 6.7324e-02],\n [-1.4408e-01, 1.4100e-01, -3.7544e-02]],\n\n [[-1.0125e-01, 8.9484e-02, -1.2644e-01],\n [ 6.6773e-02, 1.3655e-01, -6.9855e-03],\n [-7.1238e-02, -2.5733e-03, 1.5021e-01]]]])\ntorch.Size([2, 3, 3, 3])\n=========================\ncell1.Whf_mu\ntensor([[[[-0.0731, -0.0869, -0.0946],\n [-0.0206, 0.1421, -0.1010],\n [-0.1215, -0.1730, -0.0944]],\n\n [[-0.0532, 0.1439, 0.0485],\n [-0.0335, 0.1732, 0.1497],\n [-0.0009, -0.1924, 0.0016]]],\n\n\n [[[ 0.0568, -0.0796, 0.0265],\n [-0.0356, 0.0204, -0.1383],\n [-0.1770, -0.0752, -0.1090]],\n\n [[ 0.1912, -0.0911, 0.0309],\n [-0.1404, 0.1690, -0.0901],\n [-0.1060, -0.0982, -0.0329]]]])\ntorch.Size([2, 2, 3, 3])\n=========================\ncell1.Wxc_mu\ntensor([[[[-0.0222, 0.1284, 0.1002],\n [ 0.1631, -0.1533, 0.0350],\n [ 0.1828, -0.1165, 0.0352]],\n\n [[-0.0629, 0.1134, -0.0560],\n [-0.0066, 0.0927, 0.1167],\n [-0.1769, 0.0404, -0.1750]],\n\n [[-0.1779, 0.0923, -0.0512],\n [-0.1366, 0.0809, 0.1696],\n [-0.1831, -0.0611, 0.0865]]],\n\n\n [[[ 0.0611, -0.0180, -0.1381],\n [-0.1713, -0.0810, -0.0440],\n [-0.1204, 0.1552, 0.1176]],\n\n [[-0.1674, 0.0036, 0.1364],\n [ 0.1514, 0.1678, 0.1255],\n [-0.1083, -0.1702, 0.1447]],\n\n [[ 0.1181, 0.0820, -0.1202],\n [ 0.0926, -0.1522, 0.1392],\n [-0.0532, -0.0579, 0.0461]]]])\ntorch.Size([2, 3, 3, 3])\n=========================\ncell1.Whc_mu\ntensor([[[[-0.1240, 0.1753, 0.0938],\n [-0.1022, 0.1047, -0.1400],\n [-0.1333, -0.1533, 0.0638]],\n\n [[ 0.1651, -0.0441, 0.0830],\n [-0.1517, -0.0811, 0.0503],\n [-0.1181, -0.0214, 0.1019]]],\n\n\n [[[ 0.0463, -0.1071, 0.0453],\n [-0.0066, -0.1868, -0.1811],\n [-0.1088, 0.0688, 0.0407]],\n\n [[-0.0922, -0.1232, -0.1846],\n [ 0.0690, -0.1158, 0.1553],\n [-0.1189, 0.0874, -0.0758]]]])\ntorch.Size([2, 2, 3, 3])\n=========================\ncell1.Wxo_mu\ntensor([[[[ 0.1527, -0.1640, 0.0508],\n [ 0.0962, -0.0682, -0.0372],\n [-0.1589, 0.1528, 0.0121]],\n\n [[ 0.1067, 0.0870, -0.0544],\n [-0.0982, 0.0931, 0.0710],\n [-0.0285, -0.1295, -0.0802]],\n\n [[-0.0122, 0.0565, -0.1265],\n [ 0.0528, -0.0288, 0.0336],\n [ 0.0388, 0.1410, -0.0516]]],\n\n\n [[[-0.0034, 0.1200, -0.0667],\n [-0.1649, -0.1221, -0.0091],\n [ 0.0953, 0.0078, -0.0263]],\n\n [[-0.0107, -0.0407, 0.0003],\n [ 0.0730, 0.0168, 0.0869],\n [ 0.0928, -0.1398, -0.1093]],\n\n [[ 0.1082, 0.1917, 0.0315],\n [ 0.1872, 0.1334, 0.0763],\n [ 0.1679, 0.0947, 0.0454]]]])\ntorch.Size([2, 3, 3, 3])\n=========================\ncell1.Who_mu\ntensor([[[[-0.0509, 0.0025, -0.1129],\n [ 0.0715, 0.0297, 0.0817],\n [-0.0365, 0.0852, -0.1337]],\n\n [[ 0.1213, -0.0770, 0.0929],\n [-0.0727, -0.1370, -0.0354],\n [-0.1425, 0.1710, -0.1473]]],\n\n\n [[[ 0.1704, 0.1538, 0.1436],\n [-0.0588, 0.1103, -0.1793],\n [-0.0882, -0.0427, -0.1215]],\n\n [[ 0.0544, -0.1143, 0.0111],\n [-0.1670, 0.0416, 0.0137],\n [-0.0586, -0.0527, -0.0813]]]])\ntorch.Size([2, 2, 3, 3])\n=========================\ncell1.Wxi_bias\ntensor([0.1286, 0.0473])\ntorch.Size([2])\n=========================\ncell1.Wxf_bias\ntensor([-0.0822, -0.1662])\ntorch.Size([2])\n=========================\ncell1.Wxc_bias\ntensor([ 0.1198, -0.1193])\ntorch.Size([2])\n=========================\ncell1.Wxo_bias\ntensor([-0.0934, -0.1523])\ntorch.Size([2])\n=========================\ncell1.Wxi_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell1.Whi_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell1.Wxf_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell1.Whf_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell1.Wxc_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell1.Whc_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell1.Wxo_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell1.Who_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell2.Wxi_mu\ntensor([[[[-0.1606, 0.0787, 0.1289],\n [-0.0449, 0.0370, -0.2151],\n [ 0.2200, 0.0090, -0.2147]],\n\n [[ 0.0652, -0.0208, 0.1788],\n [-0.1789, -0.0495, 0.1428],\n [ 0.2312, 0.2153, -0.1653]]]])\ntorch.Size([1, 2, 3, 3])\n=========================\ncell2.Whi_mu\ntensor([[[[ 0.0746, -0.1864, -0.0186],\n [-0.1168, 0.0553, -0.0861],\n [-0.1042, -0.1182, 0.1272]]]])\ntorch.Size([1, 1, 3, 3])\n=========================\ncell2.Wxf_mu\ntensor([[[[-0.2300, 0.2292, 0.2332],\n [ 0.0055, -0.1534, 0.1039],\n [ 0.0683, -0.1853, 0.1626]],\n\n [[ 0.0172, 0.1180, 0.0007],\n [-0.0547, -0.2348, 0.2096],\n [-0.0505, 0.0312, 0.1056]]]])\ntorch.Size([1, 2, 3, 3])\n=========================\ncell2.Whf_mu\ntensor([[[[-0.1685, 0.0699, -0.1502],\n [-0.0669, -0.0573, -0.1454],\n [ 0.1328, 0.0575, -0.0792]]]])\ntorch.Size([1, 1, 3, 3])\n=========================\ncell2.Wxc_mu\ntensor([[[[ 0.0426, 0.1938, 0.1131],\n [ 0.1673, -0.1285, 0.1556],\n [ 0.0333, -0.0428, -0.0106]],\n\n [[ 0.1117, 0.0892, -0.0455],\n [-0.0538, -0.1840, -0.2090],\n [ 0.2158, 0.1657, -0.1914]]]])\ntorch.Size([1, 2, 3, 3])\n=========================\ncell2.Whc_mu\ntensor([[[[-0.1140, 0.1616, -0.0362],\n [ 0.2270, -0.2153, -0.0376],\n [-0.0136, -0.0810, -0.2291]]]])\ntorch.Size([1, 1, 3, 3])\n=========================\ncell2.Wxo_mu\ntensor([[[[-0.0806, 0.1536, 0.0378],\n [-0.1392, -0.0544, -0.0027],\n [ 0.0450, 0.1036, 0.0378]],\n\n [[-0.1259, -0.1299, -0.0302],\n [ 0.0685, 0.1393, -0.0400],\n [-0.1050, -0.1544, 0.2353]]]])\ntorch.Size([1, 2, 3, 3])\n=========================\ncell2.Who_mu\ntensor([[[[ 0.0191, 0.0763, 0.0068],\n [ 0.1463, 0.2203, 0.1838],\n [-0.1304, 0.0951, 0.0110]]]])\ntorch.Size([1, 1, 3, 3])\n=========================\ncell2.Wxi_bias\ntensor([0.1835])\ntorch.Size([1])\n=========================\ncell2.Wxf_bias\ntensor([-0.2281])\ntorch.Size([1])\n=========================\ncell2.Wxc_bias\ntensor([-0.0220])\ntorch.Size([1])\n=========================\ncell2.Wxo_bias\ntensor([-0.0880])\ntorch.Size([1])\n=========================\ncell2.Wxi_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell2.Whi_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell2.Wxf_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell2.Whf_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell2.Wxc_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell2.Whc_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell2.Wxo_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\ncell2.Who_log_alpha\ntensor([[-3.]])\ntorch.Size([1, 1])\n=========================\n"
],
[
" print('##############################################################')\n print('############# preview model parameters matrix ###############')\n print('##############################################################')\n print('Number of parameter matrices: ', len(list(model.parameters())))\n for i in range(len(list(model.parameters()))):\n print(list(model.parameters())[i].size())",
"_____no_output_____"
],
[
" %%time\n print('##############################################################')\n print('################## start training loop #####################')\n print('##############################################################')\n # track training loss\n hist = np.zeros(num_epochs)\n # loop of epoch\n for t in range(num_epochs):\n # Clear stored gradient\n model.zero_grad()\n # loop of timestep\n for timestep in range(sequence_len - cross_valid_year*12*4 - test_year*12*4):\n # hidden state re-initialized inside the model when timestep=0\n #################################################################################\n ######## create input tensor with multi-input dimension ########\n #################################################################################\n # create variables\n x_input = np.stack((sic_exp_norm[timestep,:,:],\n choice_exp_norm[timestep,:,:],\n month_exp[timestep,:,:])) #vstack,hstack,dstack\n x_var = torch.autograd.Variable(torch.Tensor(x_input).view(-1,input_channels,height,width)).to(device)\n #################################################################################\n ######## create training tensor with multi-input dimension ########\n #################################################################################\n y_train_stack = sic_exp_norm[timestep+1,:,:] #vstack,hstack,dstack\n y_var = torch.autograd.Variable(torch.Tensor(y_train_stack).view(-1,hidden_channels[-1],height,width)).to(device)\n ################################################################################# \n # Forward pass\n y_pred, kl_loss, _ = model(x_var, timestep)\n # choose training data\n y_target = y_var\n # torch.nn.functional.mse_loss(y_pred, y_train) can work with (scalar,vector) & (vector,vector)\n # Please Make Sure y_pred & y_train have the same dimension\n # accumulate loss\n #print (timestep)\n if timestep == 0:\n loss = ELBO(y_pred, y_target, kl_loss,\n 1 / (len(hidden_channels) * 8 * penalty_kl * kernel_size**2)) # weight of KL due to 8 gates in each layer\n else:\n loss += ELBO(y_pred, y_target, kl_loss,\n 1 / (len(hidden_channels) * 8 * penalty_kl * kernel_size**2)) \n #print(y_pred.shape)\n #print(y_train.shape)\n # print loss at certain iteration\n if t % 5 == 0:\n print(\"Epoch \", t, \"MSE: \", loss.item())\n # gradient check\n # Gradcheck requires double precision numbers to run\n #res = torch.autograd.gradcheck(loss_fn, (y_pred.double(), y_train.double()), eps=1e-6, raise_exception=True)\n #print(res)\n hist[t] = loss.item()\n\n # Zero out gradient, else they will accumulate between epochs\n optimiser.zero_grad()\n \n # Backward pass\n loss.backward()\n\n # Update parametersdd\n optimiser.step()\n \n # save the model\n # (recommended) save the model parameters only\n torch.save(model.state_dict(), os.path.join(output_path,'bayesconvlstm.pkl'))\n # save the entire model\n # torch.save(model, os.path.join(output_path,'bayesconvlstm.pkl'))",
"##############################################################\n################## start training loop #####################\n##############################################################\nEpoch 0 MSE: 89783.515625\nEpoch 5 MSE: 59760.1484375\nEpoch 10 MSE: 39905.62890625\nEpoch 15 MSE: 33778.2265625\nEpoch 20 MSE: 29385.525390625\nEpoch 25 MSE: 26195.1796875\nEpoch 30 MSE: 19809.05859375\nEpoch 35 MSE: 18534.54296875\nEpoch 40 MSE: 16601.708984375\nEpoch 45 MSE: 15840.9091796875\nEpoch 50 MSE: 15043.3095703125\nEpoch 55 MSE: 14217.677734375\nEpoch 60 MSE: 13625.8056640625\nEpoch 65 MSE: 13082.3955078125\nEpoch 70 MSE: 12588.7265625\n"
],
[
" print (\"******************* Loss with time **********************\")\n fig00 = plt.figure()\n try:\n plt.plot(hist, label=\"Training loss\")\n plt.legend()\n plt.show()\n fig00.savefig(os.path.join(output_path,'SIC_ERAI_LSTM_pred_error.png'),dpi=200)\n except:\n print('Model is reloaded instead of trained!')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecb3b02a38a8d19281c941b7bb753abbe8a555dc | 2,844 | ipynb | Jupyter Notebook | empty_gitenberg_repos.ipynb | rdhyee/nypl50 | 3538083725bc78cce5e3e2e271290dc74ea50507 | [
"Apache-2.0"
] | null | null | null | empty_gitenberg_repos.ipynb | rdhyee/nypl50 | 3538083725bc78cce5e3e2e271290dc74ea50507 | [
"Apache-2.0"
] | null | null | null | empty_gitenberg_repos.ipynb | rdhyee/nypl50 | 3538083725bc78cce5e3e2e271290dc74ea50507 | [
"Apache-2.0"
] | null | null | null | 22.393701 | 120 | 0.526371 | [
[
[
"# Goal\n\nrespond to [empty repos · Issue #103 · gitenberg-dev/gitberg](https://github.com/gitenberg-dev/gitberg/issues/103)",
"_____no_output_____"
]
],
[
[
"from github3 import (login, GitHub)\nfrom github_settings import (username, password, token)\nfrom itertools import islice\n\n#gh = login(username, password=password)\ngh = login(token=token)\n\nfrom github3 import (login, GitHub)\nfrom github_settings import (username, password, token)\nfrom itertools import islice\n\n#gh = login(username, password=password)\ngh = login(token=token)\n\n\ndef repo_is_empty(repo_name, repo_owner='GITenberg', branch='master'):\n try:\n repo = gh.repository(repo_owner, repo_name)\n repo_branch = repo.branch(branch)\n # if there no branch\n if repo_branch is None:\n return True\n # if there are no files in tree\n tree = repo.tree(repo_branch.commit.sha)\n return len(tree.tree) == 0 \n except Exception as e:\n return e\n",
"_____no_output_____"
],
[
"repo_is_empty('United-States-Declaration-of-Independence_1')",
"_____no_output_____"
],
[
"# no master branch at all\nrepo = gh.repository('GITenberg', 'United-States-Declaration-of-Independence_1')\nrepo.branch('master') is None",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecb3b04ed3a19d583fe200d569190e3f5dbda622 | 28,649 | ipynb | Jupyter Notebook | TLCS_98_0/.ipynb_checkpoints/coach0-checkpoint.ipynb | chantal000/Deep-QLearning-Agent-for-Traffic-Signal-Control | 49a7082ca05f9add218f10cacc3fabffb5beb996 | [
"MIT"
] | null | null | null | TLCS_98_0/.ipynb_checkpoints/coach0-checkpoint.ipynb | chantal000/Deep-QLearning-Agent-for-Traffic-Signal-Control | 49a7082ca05f9add218f10cacc3fabffb5beb996 | [
"MIT"
] | null | null | null | TLCS_98_0/.ipynb_checkpoints/coach0-checkpoint.ipynb | chantal000/Deep-QLearning-Agent-for-Traffic-Signal-Control | 49a7082ca05f9add218f10cacc3fabffb5beb996 | [
"MIT"
] | null | null | null | 53.952919 | 486 | 0.679081 | [
[
[
"# Getting Started Guide",
"_____no_output_____"
],
[
"## Table of Contents\n- [Using Coach from the Command Line](#Using-Coach-from-the-Command-Line)\n- [Using Coach as a Library](#Using-Coach-as-a-Library)\n - [Preset based - using `CoachInterface`](#Preset-based---using-CoachInterface)\n - [Training a preset](#Training-a-preset)\n - [Running each training or inference iteration manually](#Running-each-training-or-inference-iteration-manually)\n - [Non-preset - using `GraphManager` directly](#Non-preset---using-GraphManager-directly)\n - [Training an agent with a custom Gym environment](#Training-an-agent-with-a-custom-Gym-environment)\n - [Advanced functionality - proprietary exploration policy, checkpoint evaluation](#Advanced-functionality---proprietary-exploration-policy,-checkpoint-evaluation)",
"_____no_output_____"
],
[
"## Using Coach from the Command Line",
"_____no_output_____"
],
[
"When running Coach from the command line, we use a Preset module to define the experiment parameters.\nAs its name implies, a preset is a predefined set of parameters to run some agent on some environment.\nCoach has many predefined presets that follow the algorithms definitions in the published papers, and allows training some of the existing algorithms with essentially no coding at all. This presets can easily be run from the command line. For example:\n\n`coach -p CartPole_DQN`\n\nYou can find all the predefined presets under the `presets` directory, or by listing them using the following command:\n\n`coach -l`\n\nCoach can also be used with an externally defined preset by passing the absolute path to the module and the name of the graph manager object which is defined in the preset: \n\n`coach -p /home/my_user/my_agent_dir/my_preset.py:graph_manager`\n\nSome presets are generic for multiple environment levels, and therefore require defining the specific level through the command line:\n\n`coach -p Atari_DQN -lvl breakout`\n\nThere are plenty of other command line arguments you can use in order to customize the experiment. A full documentation of the available arguments can be found using the following command:\n\n`coach -h`",
"_____no_output_____"
],
[
"## Using Coach as a Library",
"_____no_output_____"
],
[
"Alternatively, Coach can be used a library directly from python. As described above, Coach uses the presets mechanism to define the experiments. A preset is essentially a python module which instantiates a `GraphManager` object. The graph manager is a container that holds the agents and the environments, and has some additional parameters for running the experiment, such as visualization parameters. The graph manager acts as the scheduler which orchestrates the experiment.\n\n**Note: Each one of the examples in this section is independent, so notebook kernels need to be restarted before running it. Make sure you run the next cell before running any of the examples.**",
"_____no_output_____"
]
],
[
[
"# Adding module path to sys path if not there, so rl_coach submodules can be imported\nimport os\nimport sys\nimport tensorflow as tf\nmodule_path = os.path.abspath(os.path.join('..'))\nresources_path = os.path.abspath(os.path.join('Resources'))\nif module_path not in sys.path:\n sys.path.append(module_path)\nif resources_path not in sys.path:\n sys.path.append(resources_path)\n \nfrom rl_coach.coach import CoachInterface",
"C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:516: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\nC:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:517: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\nC:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:518: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\nC:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:519: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\nC:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:520: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\nC:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:525: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\nC:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:541: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\nC:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:542: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\nC:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:543: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\nC:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:544: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\nC:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:545: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\nC:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:550: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\n"
]
],
[
[
"### Preset based - using `CoachInterface`\n\nThe basic method to run Coach directly from python is through a `CoachInterface` object, which uses the same arguments as the command line invocation but allowes for more flexibility and additional control of the training/inference process.\n\nLet's start with some examples.",
"_____no_output_____"
],
[
"#### Training a preset\nIn this example, we'll create a very simple graph containing a Clipped PPO agent running with the CartPole-v0 Gym environment. `CoachInterface` has a few useful parameters such as `custom_parameter` that enables overriding preset settings, and other optional parameters enabling control over the training process. We'll override the preset's schedule parameters, train with a single rollout worker, and save checkpoints every 10 seconds:",
"_____no_output_____"
]
],
[
[
"coach = CoachInterface(preset='/Users/Chantal/anaconda/envs/coach/Lib/site-packages/rl_coach/presets/CartPole_ClippedPPO.py',\n # The optional custom_parameter enables overriding preset settings\n custom_parameter='heatup_steps=EnvironmentSteps(5);improve_steps=TrainingSteps(3)',\n # Other optional parameters enable easy access to advanced functionalities\n num_workers=1, checkpoint_save_secs=10)",
"\u001b]2;\u0007\n\u001b[30;46mCreating graph - name: BasicRLGraphManager\u001b[0m\n\u001b[30;46mCreating agent - name: agent\u001b[0m\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\general_network.py:71: The name tf.variable_scope is deprecated. Please use tf.compat.v1.variable_scope instead.\n\nWARNING:tensorflow:\nThe TensorFlow contrib module will not be included in TensorFlow 2.0.\nFor more information, please see:\n * https://github.com/tensorflow/community/blob/master/rfcs/20180907-contrib-sunset.md\n * https://github.com/tensorflow/addons\n * https://github.com/tensorflow/io (for I/O related ops)\nIf you depend on functionality not listed there, please file an issue.\n\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\architecture.py:102: The name tf.train.get_or_create_global_step is deprecated. Please use tf.compat.v1.train.get_or_create_global_step instead.\n\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\general_network.py:240: The name tf.GraphKeys is deprecated. Please use tf.compat.v1.GraphKeys instead.\n\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\general_network.py:241: The name tf.placeholder is deprecated. Please use tf.compat.v1.placeholder instead.\n\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\general_network.py:242: The name tf.assign is deprecated. Please use tf.compat.v1.assign instead.\n\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\layers.py:182: dense (from tensorflow.python.layers.core) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse keras.layers.dense instead.\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorflow\\contrib\\layers\\python\\layers\\layers.py:1634: flatten (from tensorflow.python.layers.core) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse keras.layers.flatten instead.\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\heads\\v_head.py:38: The name tf.losses.mean_squared_error is deprecated. Please use tf.compat.v1.losses.mean_squared_error instead.\n\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\general_network.py:313: The name tf.get_variable is deprecated. Please use tf.compat.v1.get_variable instead.\n\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\heads\\head.py:156: The name tf.losses.add_loss is deprecated. Please use tf.compat.v1.losses.add_loss instead.\n\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\heads\\ppo_head.py:113: Categorical.__init__ (from tensorflow.python.ops.distributions.categorical) is deprecated and will be removed after 2019-01-01.\nInstructions for updating:\nThe TensorFlow Distributions library has moved to TensorFlow Probability (https://github.com/tensorflow/probability). You should update all references to use `tfp.distributions` instead of `tf.distributions`.\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorflow\\python\\ops\\distributions\\categorical.py:242: Distribution.__init__ (from tensorflow.python.ops.distributions.distribution) is deprecated and will be removed after 2019-01-01.\nInstructions for updating:\nThe TensorFlow Distributions library has moved to TensorFlow Probability (https://github.com/tensorflow/probability). You should update all references to use `tfp.distributions` instead of `tf.distributions`.\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\heads\\ppo_head.py:66: kl_divergence (from tensorflow.python.ops.distributions.kullback_leibler) is deprecated and will be removed after 2019-01-01.\nInstructions for updating:\nThe TensorFlow Distributions library has moved to TensorFlow Probability (https://github.com/tensorflow/probability). You should update all references to use `tfp.distributions` instead of `tf.distributions`.\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\general_network.py:352: The name tf.losses.get_losses is deprecated. Please use tf.compat.v1.losses.get_losses instead.\n\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\general_network.py:391: The name tf.train.AdamOptimizer is deprecated. Please use tf.compat.v1.train.AdamOptimizer instead.\n\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\tensorflow\\python\\ops\\math_grad.py:1250: add_dispatch_support.<locals>.wrapper (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\graph_managers\\graph_manager.py:277: The name tf.train.write_graph is deprecated. Please use tf.io.write_graph instead.\n\nWARNING:tensorflow:From C:\\Users\\Chantal\\anaconda\\envs\\coach\\lib\\site-packages\\rl_coach\\architectures\\tensorflow_components\\savers.py:46: The name tf.train.Saver is deprecated. Please use tf.compat.v1.train.Saver instead.\n\n"
],
[
"coach.run()",
"_____no_output_____"
]
],
[
[
"#### Running each training or inference iteration manually",
"_____no_output_____"
],
[
"The graph manager (which was instantiated in the preset) can be accessed from the `CoachInterface` object. The graph manager simplifies the scheduling process by encapsulating the calls to each of the training phases. Sometimes, it can be beneficial to have a more fine grained control over the scheduling process. This can be easily done by calling the individual phase functions directly:",
"_____no_output_____"
]
],
[
[
"from rl_coach.environments.gym_environment import GymEnvironment, GymVectorEnvironment\nfrom rl_coach.base_parameters import VisualizationParameters\nfrom rl_coach.core_types import EnvironmentSteps\n\ntf.reset_default_graph()\ncoach = CoachInterface(preset='CartPole_ClippedPPO')\n\n# registering an iteration signal before starting to run\ncoach.graph_manager.log_signal('iteration', -1)\n\ncoach.graph_manager.heatup(EnvironmentSteps(100))\n\n# training\nfor it in range(10):\n # logging the iteration signal during training\n coach.graph_manager.log_signal('iteration', it)\n # using the graph manager to train and act a given number of steps\n coach.graph_manager.train_and_act(EnvironmentSteps(100))\n # reading signals during training\n training_reward = coach.graph_manager.get_signal_value('Training Reward')",
"_____no_output_____"
]
],
[
[
"Sometimes we may want to track the agent's decisions, log or maybe even modify them.\nWe can access the agent itself through the `CoachInterface` as follows. \n\nNote that we also need an instance of the environment to do so. In this case we use instantiate a `GymEnvironment` object with the CartPole `GymVectorEnvironment`:",
"_____no_output_____"
]
],
[
[
"# inference\nenv_params = GymVectorEnvironment(level='CartPole-v0')\nenv = GymEnvironment(**env_params.__dict__, visualization_parameters=VisualizationParameters())\n\nresponse = env.reset_internal_state()\nfor _ in range(10):\n action_info = coach.graph_manager.get_agent().choose_action(response.next_state)\n print(\"State:{}, Action:{}\".format(response.next_state,action_info.action))\n response = env.step(action_info.action)\n print(\"Reward:{}\".format(response.reward))",
"_____no_output_____"
]
],
[
[
"### Non-preset - using `GraphManager` directly",
"_____no_output_____"
],
[
"It is also possible to invoke coach directly in the python code without defining a preset (which is necessary for `CoachInterface`) by using the `GraphManager` object directly. Using Coach this way won't allow you access functionalities such as multi-threading, but it might be convenient if you don't want to define a preset file.",
"_____no_output_____"
],
[
"#### Training an agent with a custom Gym environment\n\nHere we show an example of how to use the `GraphManager` to train an agent on a custom Gym environment.\n\nWe first construct a `GymEnvironmentParameters` object describing the environment parameters. For Gym environments with vector observations, we can use the more specific `GymVectorEnvironment` object. \n\nThe path to the custom environment is defined in the `level` parameter and it can be the absolute path to its class (e.g. `'/home/user/my_environment_dir/my_environment_module.py:MyEnvironmentClass'`) or the relative path to the module as in this example. In any case, we can use the custom gym environment without registering it.\n\nCustom parameters for the environment's `__init__` function can be passed as `additional_simulator_parameters`.",
"_____no_output_____"
]
],
[
[
"from rl_coach.agents.clipped_ppo_agent import ClippedPPOAgentParameters\nfrom rl_coach.environments.gym_environment import GymVectorEnvironment\nfrom rl_coach.graph_managers.basic_rl_graph_manager import BasicRLGraphManager\nfrom rl_coach.graph_managers.graph_manager import SimpleSchedule\nfrom rl_coach.architectures.embedder_parameters import InputEmbedderParameters\n\n# Resetting tensorflow graph as the network has changed.\ntf.reset_default_graph()\n\n# define the environment parameters\nbit_length = 10\nenv_params = GymVectorEnvironment(level='rl_coach.environments.toy_problems.bit_flip:BitFlip')\nenv_params.additional_simulator_parameters = {'bit_length': bit_length, 'mean_zero': True}\n\n# Clipped PPO\nagent_params = ClippedPPOAgentParameters()\nagent_params.network_wrappers['main'].input_embedders_parameters = {\n 'state': InputEmbedderParameters(scheme=[]),\n 'desired_goal': InputEmbedderParameters(scheme=[])\n}\n\ngraph_manager = BasicRLGraphManager(\n agent_params=agent_params,\n env_params=env_params,\n schedule_params=SimpleSchedule()\n)",
"_____no_output_____"
],
[
"graph_manager.improve()",
"_____no_output_____"
]
],
[
[
"#### Advanced functionality - proprietary exploration policy, checkpoint evaluation",
"_____no_output_____"
],
[
"Agent modules, such as exploration policy, memory and neural network topology can be replaced with proprietary ones. In this example we'll show how to replace the default exploration policy of the DQN agent with a different one that is defined under the Resources folder. We'll also show how to change the default checkpoint save settings, and how to load a checkpoint for evaluation.",
"_____no_output_____"
],
[
"We'll start with the standard definitions of a DQN agent solving the CartPole environment (taken from the Cartpole_DQN preset)",
"_____no_output_____"
]
],
[
[
"from rl_coach.agents.dqn_agent import DQNAgentParameters\nfrom rl_coach.base_parameters import VisualizationParameters, TaskParameters\nfrom rl_coach.core_types import TrainingSteps, EnvironmentEpisodes, EnvironmentSteps\nfrom rl_coach.environments.gym_environment import GymVectorEnvironment\nfrom rl_coach.graph_managers.basic_rl_graph_manager import BasicRLGraphManager\nfrom rl_coach.graph_managers.graph_manager import ScheduleParameters\nfrom rl_coach.memories.memory import MemoryGranularity\n\n\n####################\n# Graph Scheduling #\n####################\n\n# Resetting tensorflow graph as the network has changed.\ntf.reset_default_graph()\n\nschedule_params = ScheduleParameters()\nschedule_params.improve_steps = TrainingSteps(4000)\nschedule_params.steps_between_evaluation_periods = EnvironmentEpisodes(10)\nschedule_params.evaluation_steps = EnvironmentEpisodes(1)\nschedule_params.heatup_steps = EnvironmentSteps(1000)\n\n#########\n# Agent #\n#########\nagent_params = DQNAgentParameters()\n\n# DQN params\nagent_params.algorithm.num_steps_between_copying_online_weights_to_target = EnvironmentSteps(100)\nagent_params.algorithm.discount = 0.99\nagent_params.algorithm.num_consecutive_playing_steps = EnvironmentSteps(1)\n\n# NN configuration\nagent_params.network_wrappers['main'].learning_rate = 0.00025\nagent_params.network_wrappers['main'].replace_mse_with_huber_loss = False\n\n# ER size\nagent_params.memory.max_size = (MemoryGranularity.Transitions, 40000)\n\n################\n# Environment #\n################\nenv_params = GymVectorEnvironment(level='CartPole-v0')",
"_____no_output_____"
]
],
[
[
"Next, we'll override the exploration policy with our own policy defined in `Resources/exploration.py`.\nWe'll also define the checkpoint save directory and interval in seconds.\n\nMake sure the first cell at the top of this notebook is run before the following one, such that module_path and resources_path are adding to sys path.",
"_____no_output_____"
]
],
[
[
"from exploration import MyExplorationParameters\n\n# Overriding the default DQN Agent exploration policy with my exploration policy\nagent_params.exploration = MyExplorationParameters()\n\n# Creating a graph manager to train a DQN agent to solve CartPole\ngraph_manager = BasicRLGraphManager(agent_params=agent_params, env_params=env_params,\n schedule_params=schedule_params, vis_params=VisualizationParameters())\n\n# Resources path was defined at the top of this notebook\nmy_checkpoint_dir = resources_path + '/checkpoints'\n\n# Checkpoints will be stored every 5 seconds to the given directory\ntask_parameters1 = TaskParameters()\ntask_parameters1.checkpoint_save_dir = my_checkpoint_dir\ntask_parameters1.checkpoint_save_secs = 5\n\ngraph_manager.create_graph(task_parameters1)\ngraph_manager.improve()\n",
"_____no_output_____"
]
],
[
[
"Last, we'll load the latest checkpoint from the checkpoint directory, and evaluate it.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport shutil\n\n# Clearing the previous graph before creating the new one to avoid name conflicts\ntf.reset_default_graph()\n\n# Updating the graph manager's task parameters to restore the latest stored checkpoint from the checkpoints directory\ntask_parameters2 = TaskParameters()\ntask_parameters2.checkpoint_restore_path = my_checkpoint_dir\n\ngraph_manager.create_graph(task_parameters2)\ngraph_manager.evaluate(EnvironmentSteps(5))\n\n# Clearning up\nshutil.rmtree(my_checkpoint_dir)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb3c202633e94eca5673c44f4dc65ec7cf248cc | 36,049 | ipynb | Jupyter Notebook | HMM warmup (optional).ipynb | DeepanshKhurana/udacityproject-hmm-tagger-nlp | 5426617123f2eb246840e3b7ccc7cbba59ace54c | [
"MIT"
] | null | null | null | HMM warmup (optional).ipynb | DeepanshKhurana/udacityproject-hmm-tagger-nlp | 5426617123f2eb246840e3b7ccc7cbba59ace54c | [
"MIT"
] | null | null | null | HMM warmup (optional).ipynb | DeepanshKhurana/udacityproject-hmm-tagger-nlp | 5426617123f2eb246840e3b7ccc7cbba59ace54c | [
"MIT"
] | null | null | null | 74.481405 | 13,044 | 0.74837 | [
[
[
"# Intro to Hidden Markov Models (optional)\n---\n### Introduction\n\nIn this notebook, you'll use the [Pomegranate](http://pomegranate.readthedocs.io/en/latest/index.html) library to build a simple Hidden Markov Model and explore the Pomegranate API.\n\n<div class=\"alert alert-block alert-info\">\n**Note:** You are not required to complete this notebook and it will not be submitted with your project, but it is designed to quickly introduce the relevant parts of the Pomegranate library that you will need to complete the part of speech tagger.\n</div>\n\nThe notebook already contains some code to get you started. You only need to add some new functionality in the areas indicated; you will not need to modify the included code beyond what is requested. Sections that begin with **'IMPLEMENTATION'** in the header indicate that you need to fill in code in the block that follows. Instructions will be provided for each section, and the specifics of the implementation are marked in the code block with a 'TODO' statement. Please be sure to read the instructions carefully!\n\n<div class=\"alert alert-block alert-info\">\n**Note:** Code and Markdown cells can be executed using the `Shift + Enter` keyboard shortcut. Markdown cells can be edited by double-clicking the cell to enter edit mode.\n</div>\n<hr>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-warning\">\n**Note:** Make sure you have selected a **Python 3** kernel in Workspaces or the hmm-tagger conda environment if you are running the Jupyter server on your own machine.\n</div>",
"_____no_output_____"
]
],
[
[
"# Jupyter \"magic methods\" -- only need to be run once per kernel restart\n%load_ext autoreload\n%aimport helpers\n%autoreload 1",
"_____no_output_____"
],
[
"# import python modules -- this cell needs to be run again if you make changes to any of the files\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nfrom helpers import show_model\nfrom pomegranate import State, HiddenMarkovModel, DiscreteDistribution",
"_____no_output_____"
]
],
[
[
"## Build a Simple HMM\n---\nYou will start by building a simple HMM network based on an example from the textbook [Artificial Intelligence: A Modern Approach](http://aima.cs.berkeley.edu/).\n\n> You are the security guard stationed at a secret under-ground installation. Each day, you try to guess whether it’s raining today, but your only access to the outside world occurs each morning when you see the director coming in with, or without, an umbrella.\n\nA simplified diagram of the required network topology is shown below.\n\n\n\n### Describing the Network\n\n<div class=\"alert alert-block alert-warning\">\n$\\lambda = (A, B)$ specifies a Hidden Markov Model in terms of an emission probability distribution $A$ and a state transition probability distribution $B$.\n</div>\n\nHMM networks are parameterized by two distributions: the emission probabilties giving the conditional probability of observing evidence values for each hidden state, and the transition probabilities giving the conditional probability of moving between states during the sequence. Additionally, you can specify an initial distribution describing the probability of a sequence starting in each state.\n\n<div class=\"alert alert-block alert-warning\">\nAt each time $t$, $X_t$ represents the hidden state, and $Y_t$ represents an observation at that time.\n</div>\n\nIn this problem, $t$ corresponds to each day of the week and the hidden state represent the weather outside (whether it is Rainy or Sunny) and observations record whether the security guard sees the director carrying an umbrella or not.\n\nFor example, during some particular week the guard may observe an umbrella ['yes', 'no', 'yes', 'no', 'yes'] on Monday-Friday, while the weather outside is ['Rainy', 'Sunny', 'Sunny', 'Sunny', 'Rainy']. In that case, $t=Wednesday$, $Y_{Wednesday}=yes$, and $X_{Wednesday}=Sunny$. (It might be surprising that the guard would observe an umbrella on a sunny day, but it is possible under this type of model.)\n\n### Initializing an HMM Network with Pomegranate\nThe Pomegranate library supports [two initialization methods](http://pomegranate.readthedocs.io/en/latest/HiddenMarkovModel.html#initialization). You can either explicitly provide the three distributions, or you can build the network line-by-line. We'll use the line-by-line method for the example network, but you're free to use either method for the part of speech tagger.",
"_____no_output_____"
]
],
[
[
"# create the HMM model\nmodel = HiddenMarkovModel(name=\"Example Model\")",
"_____no_output_____"
]
],
[
[
"### **IMPLEMENTATION**: Add the Hidden States\nWhen the HMM model is specified line-by-line, the object starts as an empty container. The first step is to name each state and attach an emission distribution.\n\n#### Observation Emission Probabilities: $P(Y_t | X_t)$\nWe need to assume that we have some prior knowledge (possibly from a data set) about the director's behavior to estimate the emission probabilities for each hidden state. In real problems you can often estimate the emission probabilities empirically, which is what we'll do for the part of speech tagger. Our imaginary data will produce the conditional probability table below. (Note that the rows sum to 1.0)\n\n| | $yes$ | $no$ |\n| --- | --- | --- |\n| $Sunny$ | 0.10 | 0.90 |\n| $Rainy$ | 0.80 | 0.20 |",
"_____no_output_____"
]
],
[
[
"# create the HMM model\nmodel = HiddenMarkovModel(name=\"Example Model\")\n\n# emission probability distributions, P(umbrella | weather)\nsunny_emissions = DiscreteDistribution({\"yes\": 0.1, \"no\": 0.9})\nsunny_state = State(sunny_emissions, name=\"Sunny\")\n\n# TODO: create a discrete distribution for the rainy emissions from the probability table\n# above & use that distribution to create a state named Rainy\nrainy_emissions = DiscreteDistribution({\"yes\": 0.8, \"no\": 0.2})\nrainy_state = State(rainy_emissions, name=\"Rainy\")\n\n# add the states to the model\nmodel.add_states(sunny_state, rainy_state)\n\nassert rainy_emissions.probability(\"yes\") == 0.8, \"The director brings his umbrella with probability 0.8 on rainy days\"\nprint(\"Looks good so far!\")",
"Looks good so far!\n"
]
],
[
[
"### **IMPLEMENTATION:** Adding Transitions\nOnce the states are added to the model, we can build up the desired topology of individual state transitions.\n\n#### Initial Probability $P(X_0)$:\nWe will assume that we don't know anything useful about the likelihood of a sequence starting in either state. If the sequences start each week on Monday and end each week on Friday (so each week is a new sequence), then this assumption means that it's equally likely that the weather on a Monday may be Rainy or Sunny. We can assign equal probability to each starting state by setting $P(X_0=Rainy) = 0.5$ and $P(X_0=Sunny)=0.5$:\n\n| $Sunny$ | $Rainy$ |\n| --- | ---\n| 0.5 | 0.5 |\n\n#### State transition probabilities $P(X_{t} | X_{t-1})$\nFinally, we will assume for this example that we can estimate transition probabilities from something like historical weather data for the area. In real problems you can often use the structure of the problem (like a language grammar) to impose restrictions on the transition probabilities, then re-estimate the parameters with the same training data used to estimate the emission probabilities. Under this assumption, we get the conditional probability table below. (Note that the rows sum to 1.0)\n\n| | $Sunny$ | $Rainy$ |\n| --- | --- | --- |\n|$Sunny$| 0.80 | 0.20 |\n|$Rainy$| 0.40 | 0.60 |",
"_____no_output_____"
]
],
[
[
"# create edges for each possible state transition in the model\n# equal probability of a sequence starting on either a rainy or sunny day\nmodel.add_transition(model.start, sunny_state, 0.5)\nmodel.add_transition(model.start, rainy_state, 0.5)\n\n# add sunny day transitions (we already know estimates of these probabilities\n# from the problem statement)\nmodel.add_transition(sunny_state, sunny_state, 0.8) # 80% sunny->sunny\nmodel.add_transition(sunny_state, rainy_state, 0.2) # 20% sunny->rainy\n\n# TODO: add rainy day transitions using the probabilities specified in the transition table\nmodel.add_transition(rainy_state, sunny_state, 0.4) # 40% rainy->sunny\nmodel.add_transition(rainy_state, rainy_state, 0.6) # 60% rainy->rainy\n\n# finally, call the .bake() method to finalize the model\nmodel.bake()\n\nassert model.edge_count() == 6, \"There should be two edges from model.start, two from Rainy, and two from Sunny\"\nassert model.node_count() == 4, \"The states should include model.start, model.end, Rainy, and Sunny\"\nprint(\"Great! You've finished the model.\")",
"Great! You've finished the model.\n"
]
],
[
[
"## Visualize the Network\n---\nWe have provided a helper function called `show_model()` that generates a PNG image from a Pomegranate HMM network. You can specify an optional filename to save the file to disk. Setting the \"show_ends\" argument True will add the model start & end states that are included in every Pomegranate network.",
"_____no_output_____"
]
],
[
[
"show_model(model, figsize=(5, 5), filename=\"example.png\", overwrite=True, show_ends=False)",
"_____no_output_____"
]
],
[
[
"### Checking the Model\nThe states of the model can be accessed using array syntax on the `HMM.states` attribute, and the transition matrix can be accessed by calling `HMM.dense_transition_matrix()`. Element $(i, j)$ encodes the probability of transitioning from state $i$ to state $j$. For example, with the default column order specified, element $(2, 1)$ gives the probability of transitioning from \"Rainy\" to \"Sunny\", which we specified as 0.4.\n\nRun the next cell to inspect the full state transition matrix, then read the . ",
"_____no_output_____"
]
],
[
[
"column_order = [\"Example Model-start\", \"Sunny\", \"Rainy\", \"Example Model-end\"] # Override the Pomegranate default order\ncolumn_names = [s.name for s in model.states]\norder_index = [column_names.index(c) for c in column_order]\n\n# re-order the rows/columns to match the specified column order\ntransitions = model.dense_transition_matrix()[:, order_index][order_index, :]\nprint(\"The state transition matrix, P(Xt|Xt-1):\\n\")\nprint(transitions)\nprint(\"\\nThe transition probability from Rainy to Sunny is {:.0f}%\".format(100 * transitions[2, 1]))",
"The state transition matrix, P(Xt|Xt-1):\n\n[[ 0. 0.5 0.5 0. ]\n [ 0. 0.8 0.2 0. ]\n [ 0. 0.4 0.6 0. ]\n [ 0. 0. 0. 0. ]]\n\nThe transition probability from Rainy to Sunny is 40%\n"
]
],
[
[
"## Inference in Hidden Markov Models\n---\nBefore moving on, we'll use this simple network to quickly go over the Pomegranate API to perform the three most common HMM tasks:\n\n<div class=\"alert alert-block alert-info\">\n**Likelihood Evaluation**<br>\nGiven a model $\\lambda=(A,B)$ and a set of observations $Y$, determine $P(Y|\\lambda)$, the likelihood of observing that sequence from the model\n</div>\n\nWe can use the weather prediction model to evaluate the likelihood of the sequence [yes, yes, yes, yes, yes] (or any other state sequence). The likelihood is often used in problems like machine translation to weight interpretations in conjunction with a statistical language model.\n\n<div class=\"alert alert-block alert-info\">\n**Hidden State Decoding**<br>\nGiven a model $\\lambda=(A,B)$ and a set of observations $Y$, determine $Q$, the most likely sequence of hidden states in the model to produce the observations\n</div>\n\nWe can use the weather prediction model to determine the most likely sequence of Rainy/Sunny states for a known observation sequence, like [yes, no] -> [Rainy, Sunny]. We will use decoding in the part of speech tagger to determine the tag for each word of a sentence. The decoding can be further split into \"smoothing\" when we want to calculate past states, \"filtering\" when we want to calculate the current state, or \"prediction\" if we want to calculate future states. \n\n<div class=\"alert alert-block alert-info\">\n**Parameter Learning**<br>\nGiven a model topography (set of states and connections) and a set of observations $Y$, learn the transition probabilities $A$ and emission probabilities $B$ of the model, $\\lambda=(A,B)$\n</div>\n\nWe don't need to learn the model parameters for the weather problem or POS tagging, but it is supported by Pomegranate.\n\n### IMPLEMENTATION: Calculate Sequence Likelihood\n\nCalculating the likelihood of an observation sequence from an HMM network is performed with the [forward algorithm](https://en.wikipedia.org/wiki/Forward_algorithm). Pomegranate provides the the `HMM.forward()` method to calculate the full matrix showing the likelihood of aligning each observation to each state in the HMM, and the `HMM.log_probability()` method to calculate the cumulative likelihood over all possible hidden state paths that the specified model generated the observation sequence.\n\nFill in the code in the next section with a sample observation sequence and then use the `forward()` and `log_probability()` methods to evaluate the sequence.",
"_____no_output_____"
]
],
[
[
"# TODO: input a sequence of 'yes'/'no' values in the list below for testing\nobservations = ['yes', 'no', 'yes']\n\nassert len(observations) > 0, \"You need to choose a sequence of 'yes'/'no' observations to test\"\n\n# TODO: use model.forward() to calculate the forward matrix of the observed sequence,\n# and then use np.exp() to convert from log-likelihood to likelihood\nforward_matrix = np.exp(model.forward(observations))\n\n# TODO: use model.log_probability() to calculate the all-paths likelihood of the\n# observed sequence and then use np.exp() to convert log-likelihood to likelihood\nprobability_percentage = np.exp(model.log_probability(observations))\n\n# Display the forward probabilities\nprint(\" \" + \"\".join(s.name.center(len(s.name)+6) for s in model.states))\nfor i in range(len(observations) + 1):\n print(\" <start> \" if i==0 else observations[i - 1].center(9), end=\"\")\n print(\"\".join(\"{:.0f}%\".format(100 * forward_matrix[i, j]).center(len(s.name) + 6)\n for j, s in enumerate(model.states)))\n\nprint(\"\\nThe likelihood over all possible paths \" + \\\n \"of this model producing the sequence {} is {:.2f}%\\n\\n\"\n .format(observations, 100 * probability_percentage))",
" Rainy Sunny Example Model-start Example Model-end \n <start> 0% 0% 100% 0% \n yes 40% 5% 0% 0% \n no 5% 18% 0% 0% \n yes 5% 2% 0% 0% \n\nThe likelihood over all possible paths of this model producing the sequence ['yes', 'no', 'yes'] is 6.92%\n\n\n"
]
],
[
[
"### IMPLEMENTATION: Decoding the Most Likely Hidden State Sequence\n\nThe [Viterbi algorithm](https://en.wikipedia.org/wiki/Viterbi_algorithm) calculates the single path with the highest likelihood to produce a specific observation sequence. Pomegranate provides the `HMM.viterbi()` method to calculate both the hidden state sequence and the corresponding likelihood of the viterbi path.\n\nThis is called \"decoding\" because we use the observation sequence to decode the corresponding hidden state sequence. In the part of speech tagging problem, the hidden states map to parts of speech and the observations map to sentences. Given a sentence, Viterbi decoding finds the most likely sequence of part of speech tags corresponding to the sentence.\n\nFill in the code in the next section with the same sample observation sequence you used above, and then use the `model.viterbi()` method to calculate the likelihood and most likely state sequence. Compare the Viterbi likelihood against the forward algorithm likelihood for the observation sequence.",
"_____no_output_____"
]
],
[
[
"# TODO: input a sequence of 'yes'/'no' values in the list below for testing\nobservations = ['yes', 'no', 'yes']\n\n# TODO: use model.viterbi to find the sequence likelihood & the most likely path\nviterbi_likelihood, viterbi_path = model.viterbi(observations)\n\nprint(\"The most likely weather sequence to have generated \" + \\\n \"these observations is {} at {:.2f}%.\"\n .format([s[1].name for s in viterbi_path[1:]], np.exp(viterbi_likelihood)*100)\n)",
"The most likely weather sequence to have generated these observations is ['Rainy', 'Sunny', 'Rainy'] at 2.30%.\n"
]
],
[
[
"### Forward likelihood vs Viterbi likelihood\nRun the cells below to see the likelihood of each sequence of observations with length 3, and compare with the viterbi path.",
"_____no_output_____"
]
],
[
[
"from itertools import product\n\nobservations = ['no', 'no', 'yes']\n\np = {'Sunny': {'Sunny': np.log(.8), 'Rainy': np.log(.2)}, 'Rainy': {'Sunny': np.log(.4), 'Rainy': np.log(.6)}}\ne = {'Sunny': {'yes': np.log(.1), 'no': np.log(.9)}, 'Rainy':{'yes':np.log(.8), 'no':np.log(.2)}}\no = observations\nk = []\nvprob = np.exp(model.viterbi(o)[0])\nprint(\"The likelihood of observing {} if the weather sequence is...\".format(o))\nfor s in product(*[['Sunny', 'Rainy']]*3):\n k.append(np.exp(np.log(.5)+e[s[0]][o[0]] + p[s[0]][s[1]] + e[s[1]][o[1]] + p[s[1]][s[2]] + e[s[2]][o[2]]))\n print(\"\\t{} is {:.2f}% {}\".format(s, 100 * k[-1], \" <-- Viterbi path\" if k[-1] == vprob else \"\"))\nprint(\"\\nThe total likelihood of observing {} over all possible paths is {:.2f}%\".format(o, 100*sum(k)))",
"The likelihood of observing ['no', 'no', 'yes'] if the weather sequence is...\n\t('Sunny', 'Sunny', 'Sunny') is 2.59% \n\t('Sunny', 'Sunny', 'Rainy') is 5.18% <-- Viterbi path\n\t('Sunny', 'Rainy', 'Sunny') is 0.07% \n\t('Sunny', 'Rainy', 'Rainy') is 0.86% \n\t('Rainy', 'Sunny', 'Sunny') is 0.29% \n\t('Rainy', 'Sunny', 'Rainy') is 0.58% \n\t('Rainy', 'Rainy', 'Sunny') is 0.05% \n\t('Rainy', 'Rainy', 'Rainy') is 0.58% \n\nThe total likelihood of observing ['no', 'no', 'yes'] over all possible paths is 10.20%\n"
]
],
[
[
"### Congratulations!\nYou've now finished the HMM warmup. You should have all the tools you need to complete the part of speech tagger project.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb3c3ce49858fe567907d344e43163feae13dcb | 51,429 | ipynb | Jupyter Notebook | NAFLD 2020.ipynb | xDB-9/NAFLD-2020-RACIPE | f8efedb76b064295c096a6c5581c199b8208650f | [
"Apache-2.0"
] | null | null | null | NAFLD 2020.ipynb | xDB-9/NAFLD-2020-RACIPE | f8efedb76b064295c096a6c5581c199b8208650f | [
"Apache-2.0"
] | null | null | null | NAFLD 2020.ipynb | xDB-9/NAFLD-2020-RACIPE | f8efedb76b064295c096a6c5581c199b8208650f | [
"Apache-2.0"
] | null | null | null | 87.762799 | 32,250 | 0.729899 | [
[
[
"import numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"filepath= '~/Desktop/IISc/Tools/RACIPE-1.0/NAFLD 2020.csv'\ndf= pd.read_csv(filepath)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.dtypes",
"_____no_output_____"
],
[
"print(df.isnull().any()) ",
"_____no_output_____"
],
[
"unique_train=pd.DataFrame([(col,df[col].nunique()) for col in df.columns],\n columns=['Columns', 'Unique categories'])\nunique_train=unique_train[1:]\nfig, ax = plt.subplots(2, 1, sharex=True, sharey=True)\nax[0].bar(unique_train.Columns, unique_train['Unique categories'])\nplt.xticks(rotation=90)\n\nsns.heatmap(df.isnull(),yticklabels=False,cbar=False,cmap='viridis')",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"arr1=[]\narr2=[]\narr3=[]\narr4=[]\narr5=[]\narr6=[]\nfor j in range(1,100):\n if df.iloc[j,0]==1:\n for i in range(1,25):\n arr1.append(df.iloc[j,i])\n elif df.iloc[j,0]==2:\n for i in range(1,25):\n arr2.append(df.iloc[j,i])\n elif df.iloc[j,0]==3:\n for i in range(1,25):\n arr3.append(df.iloc[j,i])\n elif df.iloc[j,0]==4:\n for i in range(1,25):\n arr4.append(df.iloc[j,i])\n elif df.iloc[j,0]==5:\n for i in range(1,25):\n arr5.append(df.iloc[j,i])\n elif df.iloc[j,0]==6:\n for i in range(1,25):\n arr6.append(df.iloc[j,i])",
"_____no_output_____"
],
[
"s1=pd.Series(arr1)\ns2=pd.Series(arr2)\ns3=pd.Series(arr3)\ns4=pd.Series(arr4)\ns5=pd.Series(arr5)\ns6=pd.Series(arr6)",
"_____no_output_____"
],
[
"s1.plot.kde()\ns2.plot.kde()\ns3.plot.kde()\ns4.plot.kde()\nplt.legend(labels=['PPARG','HNF1A','SREBF1','HNF4A'])\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb3d36bc74b9dafc22a8867def43812ee76b4c3 | 30,585 | ipynb | Jupyter Notebook | Zipf's Law.ipynb | sktxdev/MSRA | 05a3605d2056b7d2d21db3264074e6434f675b6a | [
"MIT"
] | 1 | 2018-11-14T14:44:14.000Z | 2018-11-14T14:44:14.000Z | Zipf's Law.ipynb | sktxdev/MSRA | 05a3605d2056b7d2d21db3264074e6434f675b6a | [
"MIT"
] | null | null | null | Zipf's Law.ipynb | sktxdev/MSRA | 05a3605d2056b7d2d21db3264074e6434f675b6a | [
"MIT"
] | 1 | 2019-06-29T04:11:09.000Z | 2019-06-29T04:11:09.000Z | 104.743151 | 20,298 | 0.778584 | [
[
[
"<h3>Zipf's Law</h3>\n\nZipf's law (/ˈzɪf/) is an empirical law formulated using mathematical statistics that refers to the fact that many types of data studied in the physical and social sciences can be approximated with a Zipfian distribution, one of a family of related discrete power law probability distributions. Zipf distribution is related to the zeta distribution, but is not identical.\n\nFor example, Zipf's law states that given some corpus of natural language utterances, the frequency of any word is inversely proportional to its rank in the frequency table. Thus the most frequent word will occur approximately twice as often as the second most frequent word, three times as often as the third most frequent word, etc.: the rank-frequency distribution is an inverse relation. For example, in the Brown Corpus of American English text, the word the is the most frequently occurring word, and by itself accounts for nearly 7% of all word occurrences (69,971 out of slightly over 1 million). True to Zipf's Law, the second-place word of accounts for slightly over 3.5% of words (36,411 occurrences), followed by and (28,852). Only 135 vocabulary items are needed to account for half the Brown Corpus.\n\nReference: https://en.wikipedia.org/wiki/Zipf%27s_law\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport re\nimport seaborn as sns\n\n",
"_____no_output_____"
],
[
"df = pd.read_csv('data/train.csv', sep='|')\ndf = df[['Phrase']]\ndf['clean_text'] = df['Phrase'].apply(lambda x: re.sub('[^A-Za-z\\']', ' ', x.lower()))\ndf.head()",
"_____no_output_____"
],
[
"word_list = ' '.join(df.clean_text.values).split(' ')\nwords = pd.DataFrame(word_list, columns=['word'])\nword_counts = words.word.value_counts().reset_index()\nword_counts.columns = ['word', 'n']\nword_counts['word_rank'] = word_counts.n.rank(ascending=False)\n",
"_____no_output_____"
],
[
"%matplotlib inline\n\nf, ax = plt.subplots(figsize=(7, 7))\nax.set(xscale=\"log\", yscale=\"log\")\nsns.regplot(\"n\", \"word_rank\", word_counts, ax=ax, scatter_kws={\"s\": 100})",
"_____no_output_____"
]
],
[
[
"<h4>Summary</h4>\nThe frequency chart shows that spaces, the, a, of, and etc., \n(i.e., stopwords make up the highest percentage of words)\nHow low to set the weed out point?\n ",
"_____no_output_____"
]
],
[
[
"print(word_counts)",
" word n word_rank\n0 175845 1.0\n1 the 51633 2.0\n2 a 36415 3.0\n3 of 32702 4.0\n4 and 32177 5.0\n5 to 22761 6.0\n6 's 16971 7.0\n7 in 13997 8.0\n8 is 13476 9.0\n9 that 12338 10.0\n10 it 11734 11.0\n11 as 8651 12.0\n12 with 7750 13.0\n13 for 7553 14.0\n14 its 7051 15.0\n15 film 6733 16.0\n16 an 6502 17.0\n17 movie 6241 18.0\n18 this 5677 19.0\n19 but 5126 20.0\n20 be 5053 21.0\n21 on 4893 22.0\n22 you 4855 23.0\n23 by 3990 24.0\n24 n't 3970 25.0\n25 more 3895 26.0\n26 his 3827 27.0\n27 one 3784 28.0\n28 about 3682 29.0\n29 not 3668 30.0\n... ... ... ...\n15108 turturro 2 15070.0\n15109 audrey 2 15070.0\n15110 execrable 2 15070.0\n15111 oliveira 2 15070.0\n15112 ruh 2 15070.0\n15113 ryosuke 2 15070.0\n15114 recovers 1 15126.5\n15115 underventilated 1 15126.5\n15116 roland 1 15126.5\n15117 implied 1 15126.5\n15118 retrospective 1 15126.5\n15119 anciently 1 15126.5\n15120 piles 1 15126.5\n15121 y 1 15126.5\n15122 upends 1 15126.5\n15123 tu 1 15126.5\n15124 prechewed 1 15126.5\n15125 lifted 1 15126.5\n15126 foreshadowing 1 15126.5\n15127 aggrieved 1 15126.5\n15128 luis 1 15126.5\n15129 casings 1 15126.5\n15130 omitted 1 15126.5\n15131 unsaid 1 15126.5\n15132 joshua 1 15126.5\n15133 petter 1 15126.5\n15134 overstylized 1 15126.5\n15135 credulity 1 15126.5\n15136 harmlessly 1 15126.5\n15137 marinated 1 15126.5\n\n[15138 rows x 3 columns]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb3d64fbe8985093fa0500cbd62f5ff624c76a1 | 13,263 | ipynb | Jupyter Notebook | battery-state-estimation/experiments/dataset_a/soc/lstm_soc_percentage_all_sufficient_types.ipynb | abhignya2110/battery-state-estimation | a0584000f2f19e7004054e822904eb98e0333780 | [
"Apache-2.0"
] | 1 | 2022-01-17T06:26:13.000Z | 2022-01-17T06:26:13.000Z | battery-state-estimation/experiments/dataset_a/soc/lstm_soc_percentage_all_sufficient_types.ipynb | abhignya2110/battery-state-estimation | a0584000f2f19e7004054e822904eb98e0333780 | [
"Apache-2.0"
] | null | null | null | battery-state-estimation/experiments/dataset_a/soc/lstm_soc_percentage_all_sufficient_types.ipynb | abhignya2110/battery-state-estimation | a0584000f2f19e7004054e822904eb98e0333780 | [
"Apache-2.0"
] | null | null | null | 26.473054 | 135 | 0.525371 | [
[
[
"# Main notebook for battery state estimation",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport scipy.io\nimport math\nimport os\nimport ntpath\nimport sys\nimport logging\nimport time\nimport sys\n\nfrom importlib import reload\nimport plotly.graph_objects as go\n\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.keras import layers\n\nfrom keras.models import Sequential\nfrom keras.layers.core import Dense, Dropout, Activation\nfrom keras.optimizers import SGD, Adam\nfrom keras.utils import np_utils\nfrom keras.layers import LSTM, Embedding, RepeatVector, TimeDistributed, Masking\nfrom keras.callbacks import EarlyStopping, ModelCheckpoint, LambdaCallback\n\n\nIS_COLAB = False\n\nif IS_COLAB:\n from google.colab import drive\n drive.mount('/content/drive')\n data_path = \"/content/drive/My Drive/battery-state-estimation/battery-state-estimation/\"\nelse:c\n data_path = \"../../../\"\n\nsys.path.append(data_path)\nfrom data_processing.dataset_a import DatasetA, CycleCols\nfrom data_processing.model_data_handler import ModelDataHandler",
"_____no_output_____"
]
],
[
[
"### Config logging",
"_____no_output_____"
]
],
[
[
"reload(logging)\nlogging.basicConfig(format='%(asctime)s [%(levelname)s]: %(message)s', level=logging.DEBUG, datefmt='%Y/%m/%d %H:%M:%S')",
"_____no_output_____"
]
],
[
[
"# Load Data",
"_____no_output_____"
],
[
"### Initial the data object\n\nLoad the cycle and capacity data to memory based on the specified chunk size",
"_____no_output_____"
]
],
[
[
"dataset = DatasetA(\n test_types=[],\n chunk_size=1000000,\n lines=[37, 40],\n charge_line=37,\n discharge_line=40,\n base_path=data_path\n)",
"_____no_output_____"
]
],
[
[
"### Determine the training and testing name\n\nPrepare the training and testing data for model data handler to load the model input and output data.",
"_____no_output_____"
]
],
[
[
"train_data_test_names = [\n '000-DM-3.0-4019-S', \n '001-DM-3.0-4019-S', \n '002-DM-3.0-4019-S', \n\n '006-EE-2.85-0820-S', \n '007-EE-2.85-0820-S',\n '042-EE-2.85-0820-S',\n\n '009-DM-3.0-4019-H', \n '010-DM-3.0-4019-H',\n\n '013-DM-3.0-4019-P', \n '014-DM-3.0-4019-P',\n '015-DM-3.0-4019-P', \n '016-DM-3.0-4019-P', \n\n '018-DP-2.00-1320-S', \n '019-DP-2.00-1320-S', \n '036-DP-2.00-1720-S',\n '037-DP-2.00-1720-S', \n '038-DP-2.00-2420-S',\n\n '043-EE-2.85-0820-H',\n \n #'040-DM-4.00-2320-S', \n #'045-BE-2.75-2019-S'\n]\n\ntest_data_test_names = [\n '003-DM-3.0-4019-S', \n '008-EE-2.85-0820-S', \n '011-DM-3.0-4019-H', \n '017-DM-3.0-4019-P', \n '039-DP-2.00-2420-S',\n '044-EE-2.85-0820-H',\n \n #'041-DM-4.00-2320-S',\n]\n\ndataset.prepare_data(train_data_test_names, test_data_test_names)",
"_____no_output_____"
]
],
[
[
"### Initial the model data handler\n\nModel data handler will be used to get the model input and output data for further training purpose.",
"_____no_output_____"
]
],
[
[
"mdh = ModelDataHandler(dataset, [\n CycleCols.VOLTAGE,\n CycleCols.CURRENT,\n CycleCols.TEMPERATURE\n])",
"_____no_output_____"
]
],
[
[
"# Model training",
"_____no_output_____"
]
],
[
[
"train_x, train_y, test_x, test_y = mdh.get_discharge_whole_cycle(soh = False, output_capacity = False)\n",
"_____no_output_____"
],
[
"train_y = mdh.keep_only_capacity(train_y, is_multiple_output = True)\ntest_y = mdh.keep_only_capacity(test_y, is_multiple_output = True)",
"_____no_output_____"
],
[
"EXPERIMENT = \"lstm_soc_percentage_all_sufficient_types\"\n\nexperiment_name = time.strftime(\"%Y-%m-%d-%H-%M-%S\") + '_' + EXPERIMENT\nprint(experiment_name)\n\nos.environ[\"CUDA_VISIBLE_DEVICES\"] = \"3\"\n\n# Model definition\n\nopt = tf.keras.optimizers.Adam(lr=0.00003)\n\nmodel = Sequential()\nmodel.add(LSTM(256, activation='selu',\n return_sequences=True,\n input_shape=(train_x.shape[1], train_x.shape[2])))\nmodel.add(LSTM(256, activation='selu', return_sequences=True))\nmodel.add(LSTM(128, activation='selu', return_sequences=True))\nmodel.add(Dense(64, activation='selu'))\nmodel.add(Dense(1, activation='linear'))\nmodel.summary()\n\nmodel.compile(optimizer=opt, loss='huber', metrics=['mse', 'mae', 'mape', tf.keras.metrics.RootMeanSquaredError(name='rmse')])\n\nes = EarlyStopping(monitor='val_loss', patience=50)\nmc = ModelCheckpoint(data_path + 'results/trained_model/%s_best.h5' % experiment_name, \n save_best_only=True, \n monitor='val_loss')",
"_____no_output_____"
],
[
"history = model.fit(train_x, train_y, \n epochs=1000, \n batch_size=32, \n verbose=2,\n validation_split=0.2,\n callbacks = [es, mc]\n )",
"_____no_output_____"
],
[
"model.save(data_path + 'results/trained_model/%s.h5' % experiment_name)\n\nhist_df = pd.DataFrame(history.history)\nhist_csv_file = data_path + 'results/trained_model/%s_history.csv' % experiment_name\nwith open(hist_csv_file, mode='w') as f:\n hist_df.to_csv(f)",
"_____no_output_____"
]
],
[
[
"### Testing",
"_____no_output_____"
]
],
[
[
"results = model.evaluate(test_x, test_y)\nprint(results)",
"_____no_output_____"
]
],
[
[
"# Data Visualization",
"_____no_output_____"
]
],
[
[
"# fig = go.Figure()\n# fig.add_trace(go.Scatter(y=history.history['loss'],\n# mode='lines', name='train'))\n# fig.add_trace(go.Scatter(y=history.history['val_loss'],\n# mode='lines', name='validation'))\n# fig.update_layout(title='Loss trend',\n# xaxis_title='epoch',\n# yaxis_title='loss')\n# fig.show()",
"_____no_output_____"
],
[
"# train_predictions = model.predict(train_x)",
"_____no_output_____"
],
[
"# cycle_num = 0\n# steps_num = 8000\n# step_index = np.arange(cycle_num*steps_num, (cycle_num+1)*steps_num)\n\n# fig = go.Figure()\n# fig.add_trace(go.Scatter(x=step_index, y=train_predictions.flatten()[cycle_num*steps_num:(cycle_num+1)*steps_num],\n# mode='lines', name='SoC predicted'))\n# fig.add_trace(go.Scatter(x=step_index, y=train_y.flatten()[cycle_num*steps_num:(cycle_num+1)*steps_num],\n# mode='lines', name='SoC actual'))\n# fig.update_layout(title='Results on training',\n# xaxis_title='Cycle',\n# yaxis_title='SoC percentage')\n# fig.show()",
"_____no_output_____"
],
[
"# test_predictions = model.predict(test_x)",
"_____no_output_____"
],
[
"# cycle_num = 0\n# steps_num = 1000\n# step_index = np.arange(cycle_num*steps_num, (cycle_num+1)*steps_num)\n\n# fig = go.Figure()\n# fig.add_trace(go.Scatter(x=step_index, y=test_predictions.flatten()[cycle_num*steps_num:(cycle_num+1)*steps_num],\n# mode='lines', name='SoC predicted'))\n# fig.add_trace(go.Scatter(x=step_index, y=test_y.flatten()[cycle_num*steps_num:(cycle_num+1)*steps_num],\n# mode='lines', name='SoC actual'))\n# fig.update_layout(title='Results on testing',\n# xaxis_title='Cycle',\n# yaxis_title='SoC percentage')\n# fig.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecb4141066fedd150a7411e4bee0671ee0a26237 | 135,764 | ipynb | Jupyter Notebook | header_footer/biosignalsnotebooks_environment/categories/Train_and_Classify/.ipynb_checkpoints/classification_game_volume_3-checkpoint.ipynb | biosignalsnotebooks/biosignalsnotebooks | 72b1f053320747683bb9ff123ca180cb1bd47f6a | [
"MIT"
] | 7 | 2018-11-07T14:40:13.000Z | 2019-11-03T20:38:52.000Z | biosignalsnotebooks_notebooks/Categories/Train_and_Classify/classification_game_volume_3.ipynb | Boris69bg/biosignalsnotebooks | ed183aeb8161ff8a829a5444e956cb0b368ec51b | [
"MIT"
] | null | null | null | biosignalsnotebooks_notebooks/Categories/Train_and_Classify/classification_game_volume_3.ipynb | Boris69bg/biosignalsnotebooks | ed183aeb8161ff8a829a5444e956cb0b368ec51b | [
"MIT"
] | 1 | 2019-06-02T07:50:41.000Z | 2019-06-02T07:50:41.000Z | 57.575912 | 16,641 | 0.571889 | [
[
[
"<link rel=\"stylesheet\" href=\"../../styles/theme_style.css\">\n<!--link rel=\"stylesheet\" href=\"../../styles/header_style.css\"-->\n<link rel=\"stylesheet\" href=\"https://cdnjs.cloudflare.com/ajax/libs/font-awesome/4.7.0/css/font-awesome.min.css\">\n\n<table width=\"100%\">\n <tr>\n <td id=\"image_td\" width=\"15%\" class=\"header_image_color_7\"><div id=\"image_img\"\n class=\"header_image_7\"></div></td>\n <td class=\"header_text\"> Rock, Paper or Scissor Game - Train and Classify [Volume 3] </td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"<div id=\"flex-container\">\n <div id=\"diff_level\" class=\"flex-item\">\n <strong>Difficulty Level:</strong> <span class=\"fa fa-star checked\"></span>\n <span class=\"fa fa-star checked\"></span>\n <span class=\"fa fa-star checked\"></span>\n <span class=\"fa fa-star\"></span>\n <span class=\"fa fa-star\"></span>\n </div>\n <div id=\"tag\" class=\"flex-item-tag\">\n <span id=\"tag_list\">\n <table id=\"tag_list_table\">\n <tr>\n <td class=\"shield_left\">Tags</td>\n <td class=\"shield_right\" id=\"tags\">train_and_classify☁machine-learning☁features☁train☁nearest-neighbour</td>\n </tr>\n </table>\n </span>\n <!-- [OR] Visit https://img.shields.io in order to create a tag badge-->\n </div>\n</div>",
"_____no_output_____"
],
[
"<span class=\"color4\"><strong>Previous Notebooks that are part of \"Rock, Paper or Scissor Game - Train and Classify\" module</strong></span>\n<ul>\n <li><a href=\"classification_game_volume_1.ipynb\"><strong>Rock, Paper or Scissor Game - Train and Classify [Volume 1] | Experimental Setup <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></strong></a></li>\n <li><a href=\"classification_game_volume_2.ipynb\"><strong>Rock, Paper or Scissor Game - Train and Classify [Volume 2] | Feature Extraction <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></strong></a></li>\n</ul>\n\n<span class=\"color7\"><strong>Following Notebooks that are part of \"Rock, Paper or Scissor Game - Train and Classify\" module</strong></span>\n<ul>\n <li><a href=\"../Evaluate/classification_game_volume_4.ipynb\"><strong>Rock, Paper or Scissor Game - Train and Classify [Volume 4] | Performance Evaluation <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></strong></a></li>\n</ul> \n\n<table width=\"100%\">\n <tr>\n <td style=\"text-align:left;font-size:12pt;border-top:dotted 2px #62C3EE\">\n <span class=\"color1\">☌</span> After the previous two volumes of the <span class=\"color4\"><strong>Jupyter Notebook</strong></span> dedicated to our \"Classification Game\", we are reaching a decisive stage: Training of Classifier.\n <br>\n Currently, as demonstrated in the previous <a href=\"classification_game_volume_2.ipynb\">volume <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></a>, all the training data (examples and respective features) are ready to be applied to a classification algorithm.\n <br>\n The choice of classification algorithm resulted in the selection of <span class=\"color13\"><strong>k-Nearest Neighbour</strong></span> classifier.\n <br>\n On current <span class=\"color4\"><strong>Jupyter Notebook</strong></span> it will be described relevant steps to achieve our goal of training a <span class=\"color13\"><strong>k-Nearest Neighbour</strong></span> classifier.\n </td>\n </tr>\n</table>\n<hr>",
"_____no_output_____"
],
[
"<p style=\"font-size:20pt;color:#62C3EE;padding-bottom:5pt\">Starting Point (Setup)</p>\n<strong>List of Available Classes:</strong>\n<br>\n<ol start=\"0\">\n <li><span class=\"color1\"><strong>\"No Action\"</strong></span> [When the hand is relaxed]</li>\n <li><span class=\"color4\"><strong>\"Paper\"</strong></span> [All fingers are extended]</li>\n <li><span class=\"color7\"><strong>\"Rock\"</strong></span> [All fingers are flexed]</li>\n <li><span class=\"color13\"><strong>\"Scissor\"</strong></span> [Forefinger and middle finger are extended and the remaining ones are flexed]</li>\n</ol>\n<table align=\"center\">\n <tr>\n <td height=\"200px\">\n <img src=\"../../images/train_and_classify/classification_game_paper.png\" style=\"display:block;height:100%\">\n </td>\n <td height=\"200px\">\n <img src=\"../../images/train_and_classify/classification_game_stone.png\" style=\"display:block;height:100%\">\n </td>\n <td height=\"200px\">\n <img src=\"../../images/train_and_classify/classification_game_scissor.png\" style=\"display:block;height:100%\">\n </td>\n </tr>\n <tr>\n <td style=\"text-align:center\">\n <strong>Paper</strong>\n </td>\n <td style=\"text-align:center\">\n <strong>Rock</strong>\n </td>\n <td style=\"text-align:center\">\n <strong>Scissor</strong>\n </td>\n </tr>\n</table>\n\n<strong>Acquired Data:</strong>\n<br>\n<ul>\n <li>Electromyography (EMG) | 2 muscles | Adductor pollicis and Flexor digitorum superficialis</li>\n <li>Accelerometer (ACC) | 1 axis | Sensor parallel to the thumb nail (Axis perpendicular)</li>\n</ul>",
"_____no_output_____"
],
[
"<p style=\"font-size:20pt;color:#62C3EE;padding-bottom:5pt\">Protocol/Feature Extraction</p>\n<strong>Extracted Features</strong>\n<ul>\n <li><span style=\"color:#E84D0E\"><strong>[From] EMG signal</strong></span></li>\n <ul>\n <li>Standard Deviation ☆</li>\n <li>Maximum sampled value ☝</li>\n <li><a href=\"https://en.wikipedia.org/wiki/Zero-crossing_rate\">Zero-Crossing Rate</a> ☌</li>\n <li>Standard Deviation of the absolute signal ☇</li>\n </ul>\n <li><span style=\"color:#FDC400\"><strong>[From] ACC signal</strong></span></li>\n <ul>\n <li>Average Value ☉</li>\n <li>Standard Deviation ☆</li>\n <li>Maximum sampled value ☝</li>\n <li><a href=\"https://en.wikipedia.org/wiki/Zero-crossing_rate\">Zero-Crossing Rate</a> ☌</li>\n <li><a href=\"https://en.wikipedia.org/wiki/Slope\">Slope of the regression curve</a> ☍</li>\n </ul>\n</ul>\n\n<strong>Formal definition of parameters</strong>\n<br>\n☝ | Maximum Sample Value of a set of elements is equal to the last element of the sorted set\n\n☉ | $\\mu = \\frac{1}{N}\\sum_{i=1}^N (sample_i)$\n\n☆ | $\\sigma = \\sqrt{\\frac{1}{N}\\sum_{i=1}^N(sample_i - \\mu_{signal})^2}$\n\n☌ | $zcr = \\frac{1}{N - 1}\\sum_{i=1}^{N-1}bin(i)$ \n\n☇ | $\\sigma_{abs} = \\sqrt{\\frac{1}{N}\\sum_{i=1}^N(|sample_i| - \\mu_{signal_{abs}})^2}$\n\n☍ | $m = \\frac{\\Delta signal}{\\Delta t}$\n\n... being $N$ the number of acquired samples (that are part of the signal), $sample_i$ the value of the sample number $i$, $signal_{abs}$ the absolute signal, $\\Delta signal$ is the difference between the y coordinate of two points of the regression curve and $\\Delta t$ the difference between the x (time) coordinate of the same two points of the regression curve.\n\n... and \n\n$bin(i)$ a binary function defined as:\n\n$bin(i) = \\begin{cases} 1, & \\mbox{if } signal_i \\times signal_{i-1} \\leq 0 \\\\ 0, & \\mbox{if } signal_i \\times signal_{i-1}>0 \\end{cases}$\n<hr>",
"_____no_output_____"
],
[
"<p style=\"font-size:20pt;color:#62C3EE;padding-bottom:5pt\">Feature Selection</p>\n<strong>Intro</strong>\n<br>With <span class=\"color7\"><strong>Feature Selection</strong></span> we will start to use the resources contained inside an extremely useful <span class=\"color1\"><strong>Python</strong></span> package: <a href=\"https://scikit-learn.org/stable/index.html\">scikit-learn <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></a>\n\nLike described before, <span class=\"color7\"><strong>Feature Selection</strong></span> is intended to remove redundant or meaningless parameters which would increase the complexity of the classifier and not always translate into an improved performance. Without this step, the risk of overfitting to the training examples increases, making the classifier less able to categorize a new testing example.\n\nThere are different approaches to feature selection such as <span class=\"color4\"><strong>filter methods</strong></span> or <span class=\"color1\"><strong>wrapper methods</strong></span>.\n\nIn the first method (<span class=\"color4\"><strong>filter methods</strong></span>), a ranking will be attributed to the features, using the <strong>Pearson correlation coefficient</strong> to evaluate the impact that the feature under analysis has on the target class of the training example, or the <strong>Mutual Information parameter</strong> which defines whether two variables convey shared information. \n\nThe least relevant features will be excluded and the classifier will be trained later (for a deeper explanation, please, visit the article of Girish Chandrashekar and Ferat Sahin at <a href=\"https://www.sciencedirect.com/science/article/pii/S0045790613003066\"><strong>ScienceDirect <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></strong></a>).\n\nThe second methodology (<span class=\"color1\"><strong>wrapper methods</strong></span>) is characterised by the fact that the selection phase includes a classification algorithm, and features will be excluded or selected according to the quality of the trained classifier.\n\nThere are also a third major methodology applicable on <span class=\"color7\"><strong>Feature Selection</strong></span>, including the so called <span class=\"color1\"><strong>embedded methods</strong></span>. Essentially this methods are a combination of <span class=\"color4\"><strong>filter</strong></span> and <span class=\"color1\"><strong>wrapper</strong></span>, being characterised by the simultaneous execution of <span class=\"color7\"><strong>Feature Selection</strong></span> and <span class=\"color13\"><strong>Training</strong></span> stages.\n\nOne of the most intuitive <span class=\"color7\"><strong>Feature Selection</strong></span> methods is <span class=\"color1\"><strong>Recursive Feature Elimination</strong></span>, which will be used in the current <span class=\"color4\"><strong>Jupyter Notebook</strong></span>.\n\nEssentially the steps of this method consists in:\n<ol>\n <li>Original set of training examples is segmented into multiple ($K$) subsets of training examples and test examples</li>\n For each one of the $K$ subsets of training/test examples:\n <ol>\n <li>The training examples are used for training a \"virtual\" classifier (for example a <a href=\"https://en.wikipedia.org/wiki/Support-vector_machine\"><strong>Support Vector Machine <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></strong></a>)</li>\n <li>The test examples are given as inputs of the trained classifier and the \"virtual\" classifier quality is estimated</li>\n </ol>\n <li>At this point we can estimate the average quality of the $K$ \"virtual\" classifiers and know the weight of each feature on the training stage</li>\n <li>The feature with a smaller weight is excluded</li>\n <li>Repetition of steps <strong>1</strong>, <strong>2</strong> and <strong>3</strong> until only remains one feature</li>\n <li>Finally, when the \"feature elimination\" procedure ends, the set of features that provide a \"virtual\" classifier with the best average quality (step <strong>2</strong>) define the relevant features to be used during our final training stage</li>\n</ol> ",
"_____no_output_____"
],
[
"<p style=\"font-size:20pt;color:#62C3EE;padding-bottom:5pt\">k-Nearest Neighbour Classifier</p>\n<strong>Brief Intro</strong>\n<br>\nFollowing a \"Cartesian Logic\" each training example is formed by a set of features (in our case we have 20 training examples and each training example is composed by 8 features). Each feature can be viewed as a dimension, so, the training example would be reduced to a 8th dimensional point on the <a href=\"https://en.wikipedia.org/wiki/Cartesian_coordinate_system\"><strong>Cartesian Coordinate System <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></strong></a>.\n<br>\nThus, the training stage of a <span class=\"color4\"><strong>Jupyter Notebook</strong></span> classifier is really simple, consisting in filling the Cartesian Coordinate System with all the training examples/training points.\n<br>\nFor the standard <strong>Nearest Neighbour</strong>, when a test example is given as input of the classifier, the returned result/class will be the class of the training example nearest to our new test example.\n<br>\nOn the improved <span class=\"color13\"><strong>k-Nearest Neighbour</strong></span> classifier will be selected the $k$ nearest training points of test example. By a voting mechanism the returned class will be the one that has more training examples inside the $k$ set.\n<br>\nThe distance between training points can be estimated through the <a href=\"https://en.wikipedia.org/wiki/Norm_(mathematics)#Euclidean_norm\"><strong>Euclidean Norm <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></strong></a>:\n<br>\n<div style=\"text-align:center\">$||xy|| = \\sqrt{\\sum_{i=1}^N (x_{dim\\,i} - y_{dim\\,i})^2}$</div>\n... being $||xy||$ the Euclidean distance between two $N$ dimensional points, $x_{dim\\,i}$ is the value of coordinate $dim\\,i$ of point $x$ while $y_{dim\\,i}$ is the value of coordinate $dim\\,i$ of point $y$. \n<img src=\"../../images/train_and_classify/nn_concepts.gif\" width=\"50%\">",
"_____no_output_____"
],
[
"<p class=\"steps\">0 - Import of the needed packages for a correct execution of the current <span class=\"color4\">Jupyter Notebook</span></p>",
"_____no_output_____"
]
],
[
[
"# Python package that contains functions specialized on \"Machine Learning\" tasks.\nfrom sklearn.preprocessing import normalize\nfrom sklearn.neighbors import KNeighborsClassifier\n\n# biosignalsnotebooks own package that supports some functionalities used on the Jupyter Notebooks.\nimport biosignalsnotebooks as bsnb\n\n# Package containing a diversified set of function for statistical processing and also provide support to array operations.\nfrom numpy import array",
"_____no_output_____"
]
],
[
[
"<span class=\"color13\" style=\"font-size:30px\">⚠</span> This step was done internally !!! For now don't be worried about that. \n<p class=\"steps\">1 - Loading of the dictionary created on <a href=\"classification_game_volume_2.ipynb\">Volume 2 of \"Classification Game\" Jupyter Notebook <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></a></p>\nThis dictionary is formed by two levels:\n<ul>\n <li><span class=\"color1\"><strong>Level 1 | Class Number ($class_i$)</strong></span></li>\n [4 available keys - 1/training class]\n <li><span class=\"color7\"><strong>Level 2 | Trial Number ($trial_j$)</strong></span></li>\n [5 available keys - 1/trial]\n</ul>\n\nA list containing the extracted features values for a training example belonging to <span class=\"color1\"><strong>$class_i$</strong></span> and collected at <span class=\"color7\"><strong>$trial_j$</strong></span> is stored on the <strong>Level 2</strong> of our dictionary",
"_____no_output_____"
]
],
[
[
"# Package dedicated to the manipulation of json files.\nfrom json import loads\n\n# Specification of filename and relative path.\nrelative_path = \"../../signal_samples/classification_game/features\"\nfilename = \"classification_game_features.json\"\n\n# Load of data inside file storing it inside a Python dictionary.\nwith open(relative_path + \"/\" + filename) as file:\n features_dict = loads(file.read())",
"_____no_output_____"
],
[
"from sty import fg, rs\nprint(fg(98,195,238) + \"\\033[1mDict Keys\\033[0m\" + fg.rs + \" define the class number\")\nprint(fg(232,77,14) + \"\\033[1mDict Sub-Keys\\033[0m\" + fg.rs + \" define the trial number\\n\")\nprint(features_dict)",
"\u001b[38;2;98;195;238m\u001b[1mDict Keys\u001b[0m\u001b[39m define the class number\n\u001b[38;2;232;77;14m\u001b[1mDict Sub-Keys\u001b[0m\u001b[39m define the trial number\n\n{'0': {'1': [0.002128164580188196, 0.00732421875, 0.3148858143023837, 0.0013299640761190862, 0.00525897736063944, 0.0177154541015625, 0.14585764294049008, 0.0032250390995378314, 0.5878418, 0.004769659606303164, 0.6044, 0.0, 1.4062325168397938e-06], '2': [0.002029433963100043, 0.0075531005859375, 0.3459899981478051, 0.0012865379359157589, 0.00426341220793342, 0.0205078125, 0.24356362289312836, 0.0032271742477853944, 0.5960790740740741, 0.005347679929104084, 0.6175999999999999, 0.0, 1.7843743867526657e-06], '3': [0.004812456585924175, 0.01629638671875, 0.1500312565117733, 0.0027146265743094667, 0.002620978804585002, 0.01263427734375, 0.17816211710773078, 0.0022142130046097727, 0.9737463333333332, 0.008456826821502778, 1.0055999999999998, 0.0, 4.284292720672414e-07], '4': [0.003288393293733703, 0.0120849609375, 0.2182839094577996, 0.001892522517093399, 0.006623739508638536, 0.024169921875, 0.21266888927882086, 0.004546908635468114, 0.4644140350877193, 0.00591195751107905, 0.5671999999999999, 0.0, 5.204352203726982e-07], '5': [0.003974582803167046, 0.01190185546875, 0.18929431376180775, 0.0021560792451752937, 0.015274954840938857, 0.0413360595703125, 0.13502500463048714, 0.008656094595054242, 0.2837205925925926, 0.011517276154508253, 0.3273999999999999, 0.0, 2.2944983716296495e-06]}, '1': {'1': [0.01745991778366743, 0.13330078125, 0.1666919230186392, 0.012498507929444395, 0.008794508626081544, 0.0683441162109375, 0.2518563418699803, 0.006557225017506427, 0.6551165151515151, 0.1461447530029049, 1.5952000000000002, 0.00030307622367025305, -6.71839817994486e-06], '2': [0.01576872398048997, 0.11004638671875, 0.17624797260767705, 0.011874382612913703, 0.007355703497563003, 0.063262939453125, 0.2759055685709137, 0.005353108416775609, 0.664985945945946, 0.22564751043706982, 2.3568, 0.0007208506037123806, 3.0184467432070763e-05], '3': [0.016834862817464734, 0.10711669921875, 0.12385397566261043, 0.010210459675732673, 0.006991805896638525, 0.05291748046875, 0.23737289548258042, 0.005118249131484526, 0.7038323999999999, 0.09012218944433163, 1.2955999999999999, 0.0, 1.7932120498114455e-06], '4': [0.01624700006560064, 0.08184814453125, 0.13230391296718721, 0.010096274630523346, 0.006410319455413151, 0.03900146484375, 0.24232321459905246, 0.004563770456971738, 0.694470701754386, 0.07905537027731246, 1.1703999999999999, 0.0, 1.2355520070555934e-05], '5': [0.020006202433146783, 0.11279296875, 0.1737049068216789, 0.014510097131530179, 0.00870274334484326, 0.0604248046875, 0.2576508804923919, 0.006020127976422262, 0.7650033504273503, 0.18888218438624177, 2.2648, 0.00034193879295606086, -7.484701743981861e-07]}, '2': {'1': [0.03701667312138605, 0.48175048828125, 0.13714182735628042, 0.028527836988579556, 0.00962890534505662, 0.0972747802734375, 0.2526581366011894, 0.007592204871521871, -0.05535329729729731, 0.389147481076091, 1.12, 0.0016219138583528565, -0.00017734880190042993], '2': [0.05605906972012585, 0.728759765625, 0.15079500769362283, 0.04570180889395357, 0.022246936792071167, 0.2032470703125, 0.28363822875705247, 0.0181818341541995, -0.1621129230769231, 0.4012124197110927, 0.7678, 0.0020516327577363653, -0.00018692108787173123], '3': [0.04336534865689463, 0.39495849609375, 0.2081066853834006, 0.03180753348005212, 0.019210994245302326, 0.104644775390625, 0.2675908054044569, 0.013625052459912818, -0.08241263157894738, 0.33346391541823117, 1.7416, 0.0029829794700824705, -0.00014185053469919516], '4': [0.06487298554636435, 0.8785400390625, 0.19967561722832944, 0.05166313713613995, 0.013633174787322466, 0.128814697265625, 0.30762299513425845, 0.009650441229459253, -0.13143430630630631, 0.3800528622757343, 1.1568, 0.002342764462065237, -0.0001757656867122758], '5': [0.04605573833347097, 0.56634521484375, 0.21383160179872848, 0.03490825767448176, 0.022935690711269243, 0.1263427734375, 0.3056287796557606, 0.01751656809433346, -0.037762759689922494, 0.25814329874621555, 1.0832000000000002, 0.0021708792060784617, -0.00010479693645550052]}, '3': {'1': [0.06666059775795927, 0.36566162109375, 0.11758009432428038, 0.03798634625145099, 0.09589783278118581, 0.936767578125, 0.31566108310294355, 0.0724352171785137, -0.28372952845528454, 0.2879444621409897, 0.52, 0.0017889087656529517, -0.00010216214518629092], '2': [0.028400519040188962, 0.36090087890625, 0.11831082236279707, 0.01845200002389393, 0.06906328567307285, 0.3870391845703125, 0.34416139511027527, 0.050492947208441975, -0.2559360683760684, 0.28748525778615397, 0.5438000000000001, 0.001538724568302274, -0.0001163346319399635], '3': [0.026872172099736805, 0.25762939453125, 0.19161771709795206, 0.021019732851432105, 0.09220916129668498, 0.75457763671875, 0.33910144467375775, 0.07045219784614146, -0.3003303492063492, 0.3241576808540921, 0.696, 0.0014287982219399905, -0.00011106434030924326], '4': [0.03224804078389381, 0.36859130859375, 0.14737561976406224, 0.023877465471875494, 0.08250524718428286, 1.1531982421875, 0.32894511882373056, 0.06375121644479873, -0.27145336752136756, 0.29059513319336294, 1.4832, 0.0018806633612583347, -0.00011592650334266791], '5': [0.024408102937141216, 0.19610595703125, 0.1247592235886798, 0.015037408860519162, 0.06386941034280033, 0.44805908203125, 0.3390131871388354, 0.046133417352076656, -0.3375715851851852, 0.30169159148351743, 0.6961999999999999, 0.0010371906949177656, -9.453002551262363e-05]}}\n"
]
],
[
[
"<p class=\"steps\">2 - Restructuring of \"features_dict\" to a compatible format of <a href=\"https://scikit-learn.org/stable/index.html\">scikit-learn <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></a> package</p>\nfeatures_dict must be converted to a list, containing inside it a number of sub-lists equal to the number of training examples (in our case 20). In its turn, each sub-list is formed by a number of entries equal to the number of extracted features (13 for our original formulation of the problem).",
"_____no_output_____"
]
],
[
[
"# Initialisation of a list containing our training data and another list containing the labels of each training example.\nfeatures_list = []\nclass_training_examples = []\n\n# Access each feature list inside dictionary.\nlist_classes = features_dict.keys()\nfor class_i in list_classes:\n list_trials = features_dict[class_i].keys()\n for trial in list_trials:\n # Storage of the class label.\n class_training_examples += [int(class_i)]\n features_list += [features_dict[class_i][trial]]",
"_____no_output_____"
],
[
"print(fg(232,77,14) + \"\\033[1m[Number of list entries;Number of sub-list entries]:\\033[0m\" + fg.rs + \" [\" + str(len(features_list)) + \"; \" + str(len(features_list[0])) + \"]\" + u'\\u2713')\nprint(fg(253,196,0) + \"\\033[1mClass of each training example:\\033[0m\" + fg.rs)\nprint(class_training_examples)\nprint(fg(98,195,238) + \"\\033[1mFeatures List:\\033[0m\" + fg.rs)\nprint(features_list)",
"\u001b[38;2;232;77;14m\u001b[1m[Number of list entries;Number of sub-list entries]:\u001b[0m\u001b[39m [20; 13]✓\n\u001b[38;2;253;196;0m\u001b[1mClass of each training example:\u001b[0m\u001b[39m\n[0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3]\n\u001b[38;2;98;195;238m\u001b[1mFeatures List:\u001b[0m\u001b[39m\n[[0.002128164580188196, 0.00732421875, 0.3148858143023837, 0.0013299640761190862, 0.00525897736063944, 0.0177154541015625, 0.14585764294049008, 0.0032250390995378314, 0.5878418, 0.004769659606303164, 0.6044, 0.0, 1.4062325168397938e-06], [0.002029433963100043, 0.0075531005859375, 0.3459899981478051, 0.0012865379359157589, 0.00426341220793342, 0.0205078125, 0.24356362289312836, 0.0032271742477853944, 0.5960790740740741, 0.005347679929104084, 0.6175999999999999, 0.0, 1.7843743867526657e-06], [0.004812456585924175, 0.01629638671875, 0.1500312565117733, 0.0027146265743094667, 0.002620978804585002, 0.01263427734375, 0.17816211710773078, 0.0022142130046097727, 0.9737463333333332, 0.008456826821502778, 1.0055999999999998, 0.0, 4.284292720672414e-07], [0.003288393293733703, 0.0120849609375, 0.2182839094577996, 0.001892522517093399, 0.006623739508638536, 0.024169921875, 0.21266888927882086, 0.004546908635468114, 0.4644140350877193, 0.00591195751107905, 0.5671999999999999, 0.0, 5.204352203726982e-07], [0.003974582803167046, 0.01190185546875, 0.18929431376180775, 0.0021560792451752937, 0.015274954840938857, 0.0413360595703125, 0.13502500463048714, 0.008656094595054242, 0.2837205925925926, 0.011517276154508253, 0.3273999999999999, 0.0, 2.2944983716296495e-06], [0.01745991778366743, 0.13330078125, 0.1666919230186392, 0.012498507929444395, 0.008794508626081544, 0.0683441162109375, 0.2518563418699803, 0.006557225017506427, 0.6551165151515151, 0.1461447530029049, 1.5952000000000002, 0.00030307622367025305, -6.71839817994486e-06], [0.01576872398048997, 0.11004638671875, 0.17624797260767705, 0.011874382612913703, 0.007355703497563003, 0.063262939453125, 0.2759055685709137, 0.005353108416775609, 0.664985945945946, 0.22564751043706982, 2.3568, 0.0007208506037123806, 3.0184467432070763e-05], [0.016834862817464734, 0.10711669921875, 0.12385397566261043, 0.010210459675732673, 0.006991805896638525, 0.05291748046875, 0.23737289548258042, 0.005118249131484526, 0.7038323999999999, 0.09012218944433163, 1.2955999999999999, 0.0, 1.7932120498114455e-06], [0.01624700006560064, 0.08184814453125, 0.13230391296718721, 0.010096274630523346, 0.006410319455413151, 0.03900146484375, 0.24232321459905246, 0.004563770456971738, 0.694470701754386, 0.07905537027731246, 1.1703999999999999, 0.0, 1.2355520070555934e-05], [0.020006202433146783, 0.11279296875, 0.1737049068216789, 0.014510097131530179, 0.00870274334484326, 0.0604248046875, 0.2576508804923919, 0.006020127976422262, 0.7650033504273503, 0.18888218438624177, 2.2648, 0.00034193879295606086, -7.484701743981861e-07], [0.03701667312138605, 0.48175048828125, 0.13714182735628042, 0.028527836988579556, 0.00962890534505662, 0.0972747802734375, 0.2526581366011894, 0.007592204871521871, -0.05535329729729731, 0.389147481076091, 1.12, 0.0016219138583528565, -0.00017734880190042993], [0.05605906972012585, 0.728759765625, 0.15079500769362283, 0.04570180889395357, 0.022246936792071167, 0.2032470703125, 0.28363822875705247, 0.0181818341541995, -0.1621129230769231, 0.4012124197110927, 0.7678, 0.0020516327577363653, -0.00018692108787173123], [0.04336534865689463, 0.39495849609375, 0.2081066853834006, 0.03180753348005212, 0.019210994245302326, 0.104644775390625, 0.2675908054044569, 0.013625052459912818, -0.08241263157894738, 0.33346391541823117, 1.7416, 0.0029829794700824705, -0.00014185053469919516], [0.06487298554636435, 0.8785400390625, 0.19967561722832944, 0.05166313713613995, 0.013633174787322466, 0.128814697265625, 0.30762299513425845, 0.009650441229459253, -0.13143430630630631, 0.3800528622757343, 1.1568, 0.002342764462065237, -0.0001757656867122758], [0.04605573833347097, 0.56634521484375, 0.21383160179872848, 0.03490825767448176, 0.022935690711269243, 0.1263427734375, 0.3056287796557606, 0.01751656809433346, -0.037762759689922494, 0.25814329874621555, 1.0832000000000002, 0.0021708792060784617, -0.00010479693645550052], [0.06666059775795927, 0.36566162109375, 0.11758009432428038, 0.03798634625145099, 0.09589783278118581, 0.936767578125, 0.31566108310294355, 0.0724352171785137, -0.28372952845528454, 0.2879444621409897, 0.52, 0.0017889087656529517, -0.00010216214518629092], [0.028400519040188962, 0.36090087890625, 0.11831082236279707, 0.01845200002389393, 0.06906328567307285, 0.3870391845703125, 0.34416139511027527, 0.050492947208441975, -0.2559360683760684, 0.28748525778615397, 0.5438000000000001, 0.001538724568302274, -0.0001163346319399635], [0.026872172099736805, 0.25762939453125, 0.19161771709795206, 0.021019732851432105, 0.09220916129668498, 0.75457763671875, 0.33910144467375775, 0.07045219784614146, -0.3003303492063492, 0.3241576808540921, 0.696, 0.0014287982219399905, -0.00011106434030924326], [0.03224804078389381, 0.36859130859375, 0.14737561976406224, 0.023877465471875494, 0.08250524718428286, 1.1531982421875, 0.32894511882373056, 0.06375121644479873, -0.27145336752136756, 0.29059513319336294, 1.4832, 0.0018806633612583347, -0.00011592650334266791], [0.024408102937141216, 0.19610595703125, 0.1247592235886798, 0.015037408860519162, 0.06386941034280033, 0.44805908203125, 0.3390131871388354, 0.046133417352076656, -0.3375715851851852, 0.30169159148351743, 0.6961999999999999, 0.0010371906949177656, -9.453002551262363e-05]]\n"
]
],
[
[
"<p class=\"steps\">2.1 - Normalisation of the features values, ensuring that the training stage is not affected by scale factors</p>",
"_____no_output_____"
]
],
[
[
"features_list = normalize(features_list, axis=0, norm=\"max\") \n# axis=0 specifies that each feature is normalised independently from the others \n# and norm=\"max\" defines that the normalization reference value will be the feature maximum value.",
"_____no_output_____"
],
[
"print(features_list)\n\n# Store features_list inside a .json file.\nfrom json import dump\nfilename = \"classification_game_features_final.json\"\n\n# Generation of .json file in our previously mentioned \"relative_path\".\n# [Generation of new file]\nfeatures_list_json = list(map(list, features_list))\nwith open(relative_path + \"/\" + filename, 'w') as file:\n dump({\"features_list_final\": features_list_json, \"class_labels\": list(class_training_examples)}, file)",
"[[ 0.03192537 0.00833681 0.91010092 0.025743 0.05483938 0.01536202\n 0.42380594 0.04452308 0.6036909 0.01188812 0.25644942 0.\n 0.04658795]\n [ 0.03044428 0.00859733 1. 0.02490244 0.04445786 0.01778342\n 0.70770175 0.04455256 0.61215026 0.0133288 0.26205024 0.\n 0.05911565]\n [ 0.07219342 0.0185494 0.43362888 0.05254475 0.02733095 0.01095586\n 0.51767025 0.03056818 1. 0.02107818 0.42668024 0.\n 0.0141937 ]\n [ 0.04933039 0.01375573 0.63089659 0.03663197 0.06907079 0.02095903\n 0.6179336 0.06277207 0.47693534 0.01473523 0.24066531 0.\n 0.01724182]\n [ 0.05962417 0.01354731 0.54710921 0.04173342 0.15928363 0.03584471\n 0.39233048 0.11950119 0.29137013 0.02870618 0.13891718 0.\n 0.07601586]\n [ 0.26192261 0.15172989 0.48178249 0.24192313 0.09170706 0.05926485\n 0.73179719 0.09052537 0.67277944 0.3642578 0.67684997 0.10160185\n -0.22257799]\n [ 0.23655239 0.12526053 0.50940193 0.22984246 0.07670354 0.05485869\n 0.80167495 0.07390201 0.68291497 0.56241407 1. 0.24165456\n 1. ]\n [ 0.25254593 0.1219258 0.35796982 0.1976353 0.0729089 0.04588758\n 0.68971389 0.07065968 0.72280878 0.22462463 0.54972845 0.\n 0.05940844]\n [ 0.24372719 0.09316382 0.38239231 0.19542512 0.0668453 0.03382026\n 0.70409761 0.06300486 0.71319468 0.19704118 0.49660557 0.\n 0.40933371]\n [ 0.30012036 0.12838683 0.50205182 0.28085978 0.09075016 0.05239759\n 0.74863388 0.08311051 0.78562899 0.47077851 0.96096402 0.11462995\n -0.02479653]\n [ 0.55530065 0.54835348 0.39637512 0.55218941 0.10040796 0.08435218\n 0.7341269 0.10481372 -0.05684571 0.9699288 0.47522064 0.54372277\n -5.87549879]\n [ 0.8409626 0.8295123 0.43583632 0.88461157 0.23198581 0.17624643\n 0.82414307 0.25100821 -0.16648373 1. 0.32578072 0.68777971\n -6.19262501]\n [ 0.65053945 0.44956232 0.6014818 0.61567174 0.20032772 0.09074309\n 0.77751546 0.18809984 -0.0846346 0.83114056 0.73896809 1.\n -4.69945461]\n [ 0.97318338 1. 0.57711384 1. 0.14216353 0.11170213\n 0.89383353 0.13322858 -0.13497797 0.94726096 0.49083503 0.78537733\n -5.82305078]\n [ 0.69089897 0.6446436 0.61802828 0.67568986 0.23916798 0.10955859\n 0.88803911 0.24182392 -0.0387809 0.64340805 0.45960625 0.72775533\n -3.4718829 ]\n [ 1. 0.41621509 0.33983669 0.73526983 1. 0.81232137\n 0.91718911 1. -0.2913793 0.71768581 0.22063815 0.59970536\n -3.38459327]\n [ 0.42604657 0.41079617 0.34194868 0.35715988 0.72017567 0.33562242\n 1. 0.69707732 -0.26283649 0.71654127 0.23073659 0.51583478\n -3.8541224 ]\n [ 0.40311928 0.29324719 0.55382444 0.40686133 0.9615354 0.65433471\n 0.98529774 0.97262355 -0.30842771 0.80794528 0.29531568 0.47898359\n -3.67951963]\n [ 0.48376465 0.41954981 0.42595341 0.46217607 0.86034527 1.\n 0.95578738 0.88011355 -0.27877216 0.72429247 0.6293279 0.63046474\n -3.84060125]\n [ 0.36615488 0.22321801 0.36058621 0.29106651 0.66601516 0.38853604\n 0.9850413 0.6368921 -0.34667302 0.75194978 0.29540054 0.34770293\n -3.13174402]]\n"
]
],
[
[
"Each training array has the following structure/content:\n<br>\n\\[$\\sigma_{emg\\,flexor}$, $max_{emg\\,flexor}$, $zcr_{emg\\,flexor}$, $\\sigma_{emg\\,flexor}^{abs}$, $\\sigma_{emg\\,adductor}$, $max_{emg\\,adductor}$, $zcr_{emg\\,adductor}$, $\\sigma_{emg\\,adductor}^{abs}$, $\\mu_{acc\\,z}$, $\\sigma_{acc\\,z}$, $max_{acc\\,z}$, $zcr_{acc\\,z}$, $m_{acc\\,z}$\\] \n\nSelecting a good set of features is a really important stage for training an effective classification system. For now we will simply select a set of features without explaining the real reason for choosing them.\n\nIn order to understand the relevance of selecting a valuable set of features (and how this choice can affect the performance of our classifier), our last volume of \"Classification Game\" (<a href=\"../Evaluate/classification_game_volume_4.ipynb\"><strong>Rock, Paper or Scissor Game - Train and Classify [Volume 4] | Performance Evaluation <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></strong></a>) can be a useful resource to go deeper into this question !\n\n<span class=\"color1\"><strong>Set of Features A</strong></span>\n<ul>\n <li>$\\sigma_{emg\\,flexor}$</li>\n <li>$zcr_{emg\\,flexor}$</li>\n <li>$\\sigma_{emg\\,flexor}^{abs}$</li>\n <li>$\\sigma_{emg\\,adductor}$</li>\n <li>$\\sigma_{emg\\,adductor}^{abs}$</li>\n <li>$\\sigma_{acc\\,z}$</li>\n <li>$max_{acc\\,z}$</li>\n <li>$m_{acc\\,z}$</li>\n</ul>",
"_____no_output_____"
],
[
"<p class=\"steps\">3 - Removal of meaningless features from our \"features_list\" list</p>",
"_____no_output_____"
],
[
"\\[$\\sigma_{emg\\,flexor}$, $max_{emg\\,flexor}$, $zcr_{emg\\,flexor}$, $\\sigma_{emg\\,flexor}^{abs}$, $\\sigma_{emg\\,adductor}$, $max_{emg\\,adductor}$, $zcr_{emg\\,adductor}$, $\\sigma_{emg\\,adductor}^{abs}$, $\\mu_{acc\\,z}$, $\\sigma_{acc\\,z}$, $max_{acc\\,z}$, $zcr_{acc\\,z}$, $m_{acc\\,z}$\\] \n\n= \\[True, False, True, True, True, False, False, True, False, True, True, False, True\\] <span class=\"color1\">(List of entries that contain relevant features are flagged with \"True\")</span>",
"_____no_output_____"
]
],
[
[
"# Access each training example and exclude meaningless entries.\n# Entries that we want to keep are marked with \"True\" flag.\nacception_labels = [True, False, True, True, True, False, False, True, \n False, True, True, False, True]\ntraining_examples = []\nfor example_nbr in range(0, len(features_list)):\n training_examples += [list(array(features_list[example_nbr])[array(acception_labels)])]",
"_____no_output_____"
]
],
[
[
"<span class=\"color7\">Checkpoint !!!</span>\nCurrently all the information needed for training our classifier is stored on the following variables:\n<ul>\n <li><strong>training_examples</strong> (list where each entry is a sublist representative of a training example, containing the respective feature values for <span class=\"color1\">set A</span>)</li>\n <li><strong>class_training_examples</strong> (list where each entry contains the class label linked to each training example)</li>\n</ul>\n<p class=\"steps\">4 - Creation of a \"k-Nearest Neighbour\" <a href=\"https://scikit-learn.org/stable/index.html\">scikit-learn <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></a> objects</p>\nWe use the predefined $k$ (number of neighbours) which is 5.",
"_____no_output_____"
]
],
[
[
"# k-Nearest Neighbour object initialisation.\nknn_classifier = KNeighborsClassifier()",
"_____no_output_____"
]
],
[
[
"<p class=\"steps\">5 - Begin the training stage of classifier (fitting model to data)</p>",
"_____no_output_____"
]
],
[
[
"knn_classifier.fit(training_examples, class_training_examples)",
"_____no_output_____"
]
],
[
[
"The following interactive plot ensures a deep understanding about the class separation provided by each pair of dimensions/features.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom numpy import array\n\nfrom bokeh.layouts import layout\nfrom bokeh.models import CustomJS, Slider, Select, ColumnDataSource, WidgetBox\nfrom bokeh.plotting import figure, show\n\ntools = 'pan'\nfeatures_identifiers = [\"std_emg_flexor\", \"zcr_emg_flexor\", \"std_abs_emg_flexor\", \"std_emg_adductor\", \"std_abs_emg_adductor\", \"std_acc_z\", \"max_acc_z\", \"m_acc_z\"]\n\ndef slider():\n dict_features = {}\n for feature_nbr in range(0, len(training_examples[0])):\n values_feature = array(training_examples)[:, feature_nbr]\n \n # Fill of dict.\n for class_of_example in range(0, len(class_training_examples)):\n current_keys = list(dict_features.keys())\n if class_training_examples[class_of_example] not in current_keys:\n dict_features[class_training_examples[class_of_example]] = {}\n \n current_sub_keys = list(dict_features[class_training_examples[class_of_example]].keys())\n if features_identifiers[feature_nbr] not in current_sub_keys:\n dict_features[class_training_examples[class_of_example]][features_identifiers[feature_nbr]] = []\n \n dict_features[class_training_examples[class_of_example]][features_identifiers[feature_nbr]] += [values_feature[class_of_example]]\n \n # Add of two additional keys that will store the data currently being ploted.\n if feature_nbr == 0:\n if \"x\" not in current_sub_keys:\n dict_features[class_training_examples[class_of_example]][\"x\"] = []\n dict_features[class_training_examples[class_of_example]][\"x\"] += [values_feature[class_of_example]]\n elif feature_nbr == 1:\n if \"y\" not in current_sub_keys:\n dict_features[class_training_examples[class_of_example]][\"y\"] = []\n dict_features[class_training_examples[class_of_example]][\"y\"] += [values_feature[class_of_example]]\n \n source_class_0 = ColumnDataSource(data=dict_features[0])\n source_class_1 = ColumnDataSource(data=dict_features[1])\n source_class_2 = ColumnDataSource(data=dict_features[2])\n source_class_3 = ColumnDataSource(data=dict_features[3])\n\n plot = figure(x_range=(-1.5, 1.5), y_range=(-1.5, 1.5), tools='', toolbar_location=None, title=\"Pairing Classification Dimensions\")\n bsnb.opensignals_style([plot])\n \n # Define different colours for points of each class.\n # [Class 0]\n plot.circle('x', 'y', source=source_class_0, line_width=3, line_alpha=0.6, color=\"red\")\n # [Class 1]\n plot.circle('x', 'y', source=source_class_1, line_width=3, line_alpha=0.6, color=\"green\")\n # [Class 2]\n plot.circle('x', 'y', source=source_class_2, line_width=3, line_alpha=0.6, color=\"orange\")\n # [Class 3]\n plot.circle('x', 'y', source=source_class_3, line_width=3, line_alpha=0.6, color=\"blue\")\n\n callback = CustomJS(args=dict(source=[source_class_0, source_class_1, source_class_2, source_class_3]), code=\"\"\"\n // Each class has an independent data structure.\n var data_0 = source[0].data;\n var data_1 = source[1].data;\n var data_2 = source[2].data;\n var data_3 = source[3].data;\n \n // Selected values in the interface.\n var feature_identifier_x = x_feature.value;\n var feature_identifier_y = y_feature.value;\n console.log(\"x_feature: \" + feature_identifier_x);\n console.log(\"y_feature: \" + feature_identifier_y);\n \n // Update of values.\n var x_0 = data_0[\"x\"];\n var y_0 = data_0[\"y\"];\n for (var i = 0; i < x_0.length; i++) {\n x_0[i] = data_0[feature_identifier_x][i];\n y_0[i] = data_0[feature_identifier_y][i];\n }\n \n var x_1 = data_1[\"x\"];\n var y_1 = data_1[\"y\"];\n for (var i = 0; i < x_1.length; i++) {\n x_1[i] = data_1[feature_identifier_x][i];\n y_1[i] = data_1[feature_identifier_y][i];\n }\n \n var x_2 = data_2[\"x\"];\n var y_2 = data_2[\"y\"];\n for (var i = 0; i < x_2.length; i++) {\n x_2[i] = data_2[feature_identifier_x][i];\n y_2[i] = data_2[feature_identifier_y][i];\n }\n \n var x_3 = data_3[\"x\"];\n var y_3 = data_3[\"y\"];\n for (var i = 0; i < x_3.length; i++) {\n x_3[i] = data_3[feature_identifier_x][i];\n y_3[i] = data_3[feature_identifier_y][i];\n }\n \n // Communicate update.\n source[0].change.emit();\n source[1].change.emit();\n source[2].change.emit();\n source[3].change.emit();\n \"\"\")\n\n x_feature_select = Select(title=\"Select the Feature of Axis x:\", value=\"std_emg_flexor\", options=features_identifiers, callback=callback)\n callback.args[\"x_feature\"] = x_feature_select\n \n y_feature_select = Select(title=\"Select the Feature of Axis y:\", value=\"zcr_emg_flexor\", options=features_identifiers, callback=callback)\n callback.args[\"y_feature\"] = y_feature_select\n\n widgets = WidgetBox(x_feature_select, y_feature_select)\n return [widgets, plot]\n\nl = layout([slider(),], sizing_mode='scale_width')\n\nshow(l)",
"_____no_output_____"
]
],
[
[
"<p class=\"steps\">6 - For classifying a new \"test\" example (with unknown class) it will only be necessary to give an input to the classifier, i.e., a list with the features values of the \"test\" example</p>",
"_____no_output_____"
]
],
[
[
"# A list with 8 arbitrary entries.\ntest_examples_features = [0.65, 0.51, 0.70, 0.10, 0.20, 0.17, 0.23, 0.88]\n\n# Classification.\nprint(\"Returned Class: \")\nprint(knn_classifier.predict([test_examples_features]))\n\n# Probability of Accuracy.\nprint(\"Probability of each class:\")\nprint(knn_classifier.predict_proba([test_examples_features]))",
"Returned Class: \n[1]\nProbability of each class:\n[[0.4 0.6 0. 0. ]]\n"
]
],
[
[
"There is a clear doubt between class \"0\" (\"No Action\") and class \"1\" (\"Paper\"), with 40 % and 60 % of accuracy probability, respectively. ",
"_____no_output_____"
],
[
"With the steps described on the current volume of \"Classification Game\", our classifier is trained and ready to receive new examples and classify them immediately.\n\nThere is only one remaining task, that will be briefly explained on the <a href=\"../Evaluate/classification_game_volume_4.ipynb\">final volume <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></a>, consisting in the objective evaluation of the classifier quality.\n\n<strong><span class=\"color7\">We hope that you have enjoyed this guide. </span><span class=\"color2\">biosignalsnotebooks</span><span class=\"color4\"> is an environment in continuous expansion, so don't stop your journey and learn more with the remaining <a href=\"../MainFiles/biosignalsnotebooks.ipynb\">Notebooks <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></a></span></strong> !",
"_____no_output_____"
],
[
"<span class=\"color6\">**Auxiliary Code Segment (should not be replicated by\nthe user)**</span>",
"_____no_output_____"
]
],
[
[
"from biosignalsnotebooks.__notebook_support__ import css_style_apply\ncss_style_apply()",
".................... CSS Style Applied to Jupyter Notebook .........................\n"
],
[
"%%html\n<script>\n // AUTORUN ALL CELLS ON NOTEBOOK-LOAD!\n require(\n ['base/js/namespace', 'jquery'],\n function(jupyter, $) {\n $(jupyter.events).on(\"kernel_ready.Kernel\", function () {\n console.log(\"Auto-running all cells-below...\");\n jupyter.actions.call('jupyter-notebook:run-all-cells-below');\n jupyter.actions.call('jupyter-notebook:save-notebook');\n });\n }\n );\n</script>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
ecb416302d680f9dea6966ba2668bef7cdfbaa6d | 30,686 | ipynb | Jupyter Notebook | homework_5.ipynb | LivLilli/WikipediaHyperlinkGraph_Analysis | 5745e581c86019b16e5da45fe55d301c59fc2f26 | [
"MIT"
] | null | null | null | homework_5.ipynb | LivLilli/WikipediaHyperlinkGraph_Analysis | 5745e581c86019b16e5da45fe55d301c59fc2f26 | [
"MIT"
] | null | null | null | homework_5.ipynb | LivLilli/WikipediaHyperlinkGraph_Analysis | 5745e581c86019b16e5da45fe55d301c59fc2f26 | [
"MIT"
] | 1 | 2018-12-23T20:59:08.000Z | 2018-12-23T20:59:08.000Z | 33.06681 | 1,547 | 0.562341 | [
[
[
"<div>\n <h1 style=\"margin-top: 50px; font-size: 33px; text-align: center\"> Homework 5 - Visit the Wikipedia hyperlinks graph! </h1>\n <br>\n <div style=\"font-weight:200; font-size: 20px; padding-bottom: 15px; width: 100%; text-align: center;\">\n <right>Maria Luisa Croci, Livia Lilli, Pavan Kumar Alikana</right>\n <br>\n </div>\n <hr>\n</div>",
"_____no_output_____"
],
[
"# RQ1",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport json\nimport pickle\nfrom tqdm import tqdm\n\nfrom collections import defaultdict\nfrom heapq import *\nimport numpy as np\nimport collections\nimport networkx as nx",
"_____no_output_____"
]
],
[
[
"For our first requests, we can use 2 different approaches:\n\n* We can start from the file building a dictionary that describe our graph; we do it because we will need this dictionary for the request 2;\n\n* Or, better, we can use an easy command <b>nx.info</b>, to get all the informations we need.\n\nSo let's see!\n",
"_____no_output_____"
],
[
"## Approach 1",
"_____no_output_____"
],
[
"Let's start downloading <a href=\"https://drive.google.com/file/d/1ghPJ4g6XMCUDFQ2JPqAVveLyytG8gBfL/view\">Wikicat hyperlink graph</a> . \n\nIt is a reduced version of the one we can find on SNAP. \n\nEvery row is an <b>edge</b>. The two elements of each row are the <b>nodes</b>, in particular the first is the <b> source</b>, the second represents the <b>destination</b>.\n\nSo, our first goal is open and read the file with python, and split each line with the new line charachter.\nThen we take all the <i>source nodes</i> for each row, and we put them as keys into our <b>graph</b> dictionary. The values will be all the correspondent destination nodes.\n\nBut we have not done! Infact our scope is to analyze the graph, in particular discovering the following informations:\n\n* If it is direct or not;\n\n* The number of nodes;\n\n* The number of edges;\n\n* The average node degree. Is the graph dense?\n\nTo do this we want that our dictionary has as keys <u>all the nodes</u>, sources and destinations, and for now we have just the first ones. So we add as new keys all the nodes that are just destinations, putting as values empty lists.\n\n\nNow we have the dictionary with all the nodes of our graph as keys, and as values there are all the eventual destinations!",
"_____no_output_____"
]
],
[
[
"F = open('wiki-topcats-reduced.txt','r') \nall_rows = F.read().split('\\n')\n\ngraph = {}\nfor row in all_rows:\n row = row.split(\"\\t\")\n if row[0] not in graph:\n try:\n graph[row[0]] = [row[1]]\n except:\n pass\n else:\n graph[row[0]].append(row[1])\n \n ",
"_____no_output_____"
],
[
"lista = []\nfor l in graph.values():\n lista+= l\n ",
"_____no_output_____"
],
[
"for node in lista:\n if node not in graph:\n graph[node] = []\n else:\n pass",
"_____no_output_____"
]
],
[
[
"So, what can we say?\n\n* We are in a case of <b>page ranking</b>. So for definition we have nodes representing sources and destinations, with edges with a particular direction. In other words, our graph has a set of edges which are <i>ordered pairs</i> of nodes, and for the graph theory we have a <b>directed graph</b>.\n\n\n* The number of nodes is <u>461193</u>. It's just the number of keys of our dictionary.\n\n\n* The number of edges is <u>2645247</u> and it's computed looking at all the lenghts of the <b>adjacency list</b>.\n\n\n* In graph theory, the <b>degree</b> (or <i>valency</i>) of a vertex of a graph is the number of edges incident to the vertex. We need the <b>average node degree</b>, so we compute the ratio between number of edges and number of nodes. It results <u>5.735661642739591</u>.",
"_____no_output_____"
],
[
"#### Number of nodes",
"_____no_output_____"
]
],
[
[
"V = list(graph.keys())\nn_nodes = len(V)\nn_nodes",
"_____no_output_____"
]
],
[
[
"#### Number of edges",
"_____no_output_____"
]
],
[
[
"n_edges = 0\nfor l in graph.values():\n n_edges += len(l)\nn_edges ",
"_____no_output_____"
]
],
[
[
"#### Avarage node degree",
"_____no_output_____"
]
],
[
[
"avg_degree = n_edges/n_nodes\navg_degree",
"_____no_output_____"
]
],
[
[
"## Approach 2",
"_____no_output_____"
],
[
"Although, since we need the average in and out degree because our graph is directed, we could use an easy command nx.info as follow, in order to obtain the basic informations.\n\nFirst import the graph from the reduced file of the list of edges indicating with nx.DiGraph that what we want is an oriented graph.",
"_____no_output_____"
]
],
[
[
"graph = nx.read_edgelist(\"wiki-topcats-reduced.txt\", delimiter=\"\\t\", create_using=nx.DiGraph())\nprint(nx.info(graph))",
"Name: \nType: DiGraph\nNumber of nodes: 461193\nNumber of edges: 2645247\nAverage in degree: 5.7357\nAverage out degree: 5.7357\n"
]
],
[
[
"**Is the graph dense?**\n\nWith the following formula $D=\\frac{E}{N(N-1)}$ we obtain a value that could go from 0 up to 1. It measure the probability that any pairs of vertex is connected, so technically if the density is close to 1 the number of edges is close to the maximal number of edges, viceversa if the density is close to 0 we have a graph with only few edges (called sparse graph).\n\n",
"_____no_output_____"
]
],
[
[
"nx.density(graph)",
"_____no_output_____"
]
],
[
[
"As we could expect, according to the number of nodes and edges that we already know, the density is very low, so it means that our graph is sparse.",
"_____no_output_____"
],
[
"# RQ2",
"_____no_output_____"
],
[
"Let's start creating a dictionary called <b>categories</b> where for every category taken from <i>wiki-topcats-categories.txt</i> file, we have the list of all its articles. But attention! We must take into account all the categories that has a number of articles greater than <b>3500</b>. So we filter our dictionary considering the categories with more that 3500 articles. Another, we take just the articles that are the result of the intersection between the set of articles of the category and the articles of our <b>graph</b>; in other words, we don't consider those nodes that are in our graph but not in our categories!\n\n\nWe create also a dictionary called <b>inv_dic</b> that shows for every node (article), a set of all the categories associated. \n",
"_____no_output_____"
]
],
[
[
"C = open('wiki-topcats-categories.txt','r') ",
"_____no_output_____"
],
[
"categories = {}\nfor line in C.readlines():\n l = line.split(' ')\n cat = l[0].replace(\"Category:\",\"\").replace(\";\", \"\")\n art = l[1:]\n art[-1] = art[-1].replace(\"\\n\",\"\")\n if len(art) >= 3500:\n categories[cat]= set(art).intersection(set(V))\n\n",
"_____no_output_____"
],
[
"all_set = categories.values()\nall_nodes = []\nfor s in all_set:\n all_nodes += s\ninv_dic = {}\nfor node in all_nodes:\n for cat in categories:\n if node in categories[cat] and node not in inv_dic:\n inv_dic[node] = [cat]\n elif node in categories[cat] and node in inv_dic and cat not in inv_dic[node]:\n inv_dic[node].append(cat)\n else:\n pass\n",
"_____no_output_____"
]
],
[
[
"## Block Ranking ",
"_____no_output_____"
],
[
"Our scope now is, to take in input a category $C_0 = \\{article_1, article_2, \\dots \\}$. Then we want to rank all of the nodes according to the following criterion:\n\nObtain a <b>block-ranking</b>, where the blocks are represented by the categories.\nThe first category of the rank, $C_0$, always corresponds to the input category. The order of the remaining categories is given by: $$distance(C_0, C_1) = median(ShortestPath(C_0, C_i))$$",
"_____no_output_____"
],
[
"How we do that? At first we create the functions we need.\n\nOur input category is 'Year_of_birth_unknown' for convention because the one with the smaller number of nodes.\n\n* The first function we write is the <b> ShortestPath</b> which takes in input a node (of the input category) and our graph. It computes the distances, using the visit in amplitude of the graph. For this we apply the <b><i>BFS</i></b> algorithm, that consists in searching graph data structures. It starts at the <i>tree root</i> (or some arbitrary node of a graph called <i>search key</i>), and at first it explores all of the neighbor nodes at the present depth prior, moving on to the nodes at the next depth level. The gif below shows this concept.\n\nSo the SorthestPath function creates a dictionary where the keys are the nodes (including the input node) and their value are the distances from the node of the input category. \n\nThe distance from the node of the input category to itself is written as zero. The others are started as -1, and then eventually incremented.\n\n\n* Now it's the turn of <b>createDistancesDict</b> function, which take 4 elements as input: the input category, the graph, the <i>categories</i> dictionary and finally the <i>inv_dic</i>. In easy words, it applies the ShortestPath function to every node of the input cateogory creating a dictionary where each key is one of these nodes, and the value is a dictionary where for every other node of the graph there is the distance from the starting node of C0.\n\n\n* Now we create the <b>dictDistanceCi</b> dictionary, where we wanna show for each category a list of all the distances of the correspondent nodes from the nodes of the input category. Of course we don't need the distances among the nodes of the input cateogory, so we don't consider them.\n\n\n* A the end of our process, we compute for each category (taken from the precedent dictionary) the <b>median</b> of the correspondent distances. Then we add in an Ordered Dictionary called <b>rank</b> each category with its value of median. So we obtain our <b>BLOCK RANKING</b>.\n\n\n",
"_____no_output_____"
],
[
"<img src=\"https://upload.wikimedia.org/wikipedia/commons/5/5d/Breadth-First-Search-Algorithm.gif\">",
"_____no_output_____"
]
],
[
[
"input_category = input()\n",
"Year_of_birth_unknown\n"
],
[
"def ShortestPath(c0, graph):\n queue = []\n queue.append(c0)\n \n distanceDict = dict()\n for node in graph:\n distanceDict[node] = -1\n distanceDict[c0] = 0\n\n while queue:\n vertex = queue.pop(0)\n for i in graph[vertex]:\n if distanceDict[i] == -1:\n queue.append(i)\n distanceDict[i] = distanceDict[vertex] + 1\n return distanceDict\n ",
"_____no_output_____"
],
[
"def calculateMedian(lista):\n lung = len(lista)\n #ordinata = sorted(lista)\n ordinata = lista\n if(lung % 2 != 0):\n return ordinata[lung/2]\n else:\n return (ordinata[lung/2]) + (ordinata[lung/2 + 1]) /2 ",
"_____no_output_____"
],
[
"from collections import OrderedDict\nimport pickle\n\ndef createDistancesDict(c0, graph, dizionarioCatNodi, listNode):\n \n #listNode è un dizionario <articolo, [categorie]>\n \n #Prendo come categoria 0 la lista di nodi della categoria 0\n Category0 = dizionarioCatNodi[c0]\n \n #Dizionario dove per ogni chiave(articolo in C0) c'è un dizionatio (nodo, distanza) con la distanza verso tutti gli altri nodi \n dictDistances = dict()\n \n for node in tqdm(Category0):\n try:\n dictDistances[node] = ShortestPath(node, graph)\n except Exception as e: print(e)\n \n with open(\"distance_dict.p\", 'wb') as handle:\n pickle.dump(dictDistances, handle, protocol=pickle.HIGHEST_PROTOCOL)",
"_____no_output_____"
],
[
"createDistancesDict(input_category, graph, categories, inv_dic)",
"100%|██████████| 2536/2536 [2:39:26<00:00, 2.93s/it] \n"
],
[
"with open(\"distance_dict.p\", 'rb') as handle:\n dist_dict = pickle.load(handle)",
"_____no_output_____"
],
[
"dictDistanceCi = dict()\n#inizializzo le distanze da C0 per ogni categoria ad una lista vuota\nfor cat in categories:\n dictDistanceCi[cat] = []",
"_____no_output_____"
],
[
"#for every cat the distances of its nodes from nodes of C0\nfor node in dist_dict:\n for node2 in dist_dict[node]:\n for cat in inv_dic[node2]:\n if cat != inv_dic[node]:\n dictDistanceCi[cat].append(dist_dict[node][node2])\n\nwith open(\"dictDistanceCi.p\", 'ab') as handle:\n pickle.dump(dictDistanceCi, handle, protocol=pickle.HIGHEST_PROTOCOL)",
"_____no_output_____"
],
[
"with open(\"dictDistanceCi.p\", 'rb') as handle:\n dictDistanceCi = pickle.load(handle)",
"_____no_output_____"
],
[
"rank = OrderedDict()\nfor cat in tqdm(dictDistanceCi):\n distance = np.median(dictDistanceCi[cat])\n rank[cat] = distance\n\nrank['Year_of_birth_unknown'] = 0",
"_____no_output_____"
],
[
"block_rank = {}\nfor tupla in rank:\n block_rank[tupla[0]] = tupla[1]",
"_____no_output_____"
],
[
"for el in block_rank:\n if block_rank[el] == -1.0:\n block_rank[el] = 10000.0\nblock_rank = sorted(block_rank.items(), key=lambda x: x[1])",
"_____no_output_____"
],
[
"block_rank",
"_____no_output_____"
]
],
[
[
"Obtained the Ordered Dictionary <b>rank</b> we notice that there are some categories with median equal to -1. This means that these categories are not reachable from the input category and so the values of distance among their nodes and the input category ones didn't change its initial values -1 associated during the inizialization in the BFS. For this reason we give to them a big value, for example 10000, so that in the block rank they will stay at the end.",
"_____no_output_____"
],
[
"## Sorting nodes of category",
"_____no_output_____"
],
[
"Once we obtain the <i>block ranking vector</i>, we want to sort the nodes in each category. The way we should sort them is the following...",
"_____no_output_____"
],
[
" We have to compute the subgraph induced by $C_0$. Then, for each node, we compute the sum of the weigths of the <b>in-edges</b>. The nodes will be ordered by this score.\n The following image explains how to do it for each step.\n \n In other words, we have to consider a category, and for that category we must compute for each node the number of in-edges, but considering just those that have the source of the same category! For example, in the first image, the B node of the category \"0\" has got 2 in-edges, but only one is from a node of the same category.",
"_____no_output_____"
],
[
"<img src=\"https://raw.githubusercontent.com/CriMenghini/ADM-2018/master/Homework_5/imgs/algorithm.PNG\">",
"_____no_output_____"
],
[
"For this scope we have created a function called <b>in_edges</b> that implements our idea of sorting, given as input a category. \n\nSo we apply this function for each category saving the correspondent dictionary on a file <i>pickle</i>, naming it as <i>\"cat_i.p\"</i> where i is the index of the i-category. To control the correspondence index-category, we create a dictionary where for each category we have its index; we call it <b>indexing</b>. ",
"_____no_output_____"
],
[
"What does our <i>in_edge()</i> function do exactly? \n\nWell, we can see that for a node <i>n1</i> of the choosen category, it starts a contator and for every node <i>n2</i> of our graph checks two important things:\n\n* if there is an edge from <i>n2</i> to <i>n1</i>;\n\n* if <i>n2</i>, the source node, is in the same category of <i>n1</i>;\n\nIf these 2 points are respected, then it increments the contator of <i>n1</i>. \n\nAt the end, it saves in a dictionary each node n1 and its contator, or in other words, the number of its in-edges.\nBut it's not finished! We want to sort the nodes in the dictionary in base of their values, so we just do it. Now the output is ready!\n\n\nWe have reported as examples some of our dictionaries saved on pickle. In particular you can see that for \"the category 7\" (that in our block ranking correponds to the <b>Category0</b>).",
"_____no_output_____"
]
],
[
[
"all_cat = list(categories.keys())",
"_____no_output_____"
],
[
"def in_edges(cat, graph):\n n_cat = categories[cat]\n d = {}\n for n1 in tqdm(n_cat):\n count = 0\n for n2 in graph:\n if n1 in graph[n2] and n2 in n_cat:\n count += 1\n d[n1] = count\n d = sorted(d.items(), key=lambda x: x[1])\n return d\n ",
"_____no_output_____"
],
[
"for i in range(len(all_cat)):\n dd = in_edges(all_cat[i], graph)\n \n #pickle.dump(dd, open( \"cat\"+str(i)+\".p\", \"wb\" ) )\n with open(\"cat\"+str(i)+\".p\", 'wb') as handle:\n pickle.dump(dd, handle, protocol=pickle.HIGHEST_PROTOCOL)",
"_____no_output_____"
],
[
"indexing = {}\nfor i in range(len(all_cat)):\n indexing[all_cat[i]] = i\n ",
"_____no_output_____"
],
[
"indexing",
"_____no_output_____"
]
],
[
[
"Here there is the indexing dictionary, that occurs to us to find the in_edge dictionary of a particular category, starting from its index.",
"_____no_output_____"
],
[
"Here, as promised, we have the dictionary for the category 0 of our block ranking, or in other words, the category7 of our indexing.\n\nFor convention we print just a portion of it, in particular a part where we can see the moment where the score changes.",
"_____no_output_____"
]
],
[
[
"with open(\"cat\"+str(7)+\".p\", 'rb') as handle:\n dd7 = pickle.load(handle)",
"_____no_output_____"
],
[
"print(dd7[1600:1700])\n",
"[('484342', 0), ('62178', 0), ('1149543', 0), ('1766275', 0), ('1550129', 0), ('56442', 0), ('568716', 0), ('456768', 0), ('1408474', 0), ('62175', 0), ('1147610', 0), ('1340876', 0), ('1489713', 0), ('246778', 0), ('603141', 0), ('666878', 0), ('1638138', 0), ('1046336', 0), ('752751', 0), ('174850', 0), ('358851', 0), ('1656546', 1), ('43293', 1), ('1190380', 1), ('1203249', 1), ('1203361', 1), ('165957', 1), ('1341803', 1), ('215630', 1), ('171716', 1), ('175433', 1), ('1656576', 1), ('175333', 1), ('1766243', 1), ('1656762', 1), ('359312', 1), ('159886', 1), ('1469343', 1), ('167751', 1), ('174834', 1), ('953683', 1), ('1479797', 1), ('173382', 1), ('888207', 1), ('417673', 1), ('1203360', 1), ('1656431', 1), ('159764', 1), ('185804', 1), ('196154', 1), ('166402', 1), ('173549', 1), ('1144878', 1), ('171403', 1), ('957992', 1), ('34425', 1), ('413030', 1), ('1766072', 1), ('166521', 1), ('1144436', 1), ('1341563', 1), ('171426', 1), ('1656575', 1), ('171971', 1), ('62855', 1), ('1142120', 1), ('1345938', 1), ('1143085', 1), ('1517069', 1), ('180086', 1), ('1143248', 1), ('1343089', 1), ('1203022', 1), ('184948', 1), ('1765825', 1), ('1421753', 1), ('1122637', 1), ('159737', 1), ('159860', 1), ('159732', 1), ('834580', 1), ('167775', 1), ('1342972', 1), ('748772', 1), ('1341499', 1), ('129328', 1), ('417685', 1), ('666879', 1), ('129329', 1), ('1344834', 1), ('194766', 1), ('175328', 1), ('1340892', 1), ('1404830', 1), ('168161', 1), ('1341564', 1), ('1143113', 1), ('171793', 1), ('62620', 1), ('1779627', 1)]\n"
]
],
[
[
"<img src=\"http://scalar.usc.edu/works/querying-social-media-with-nodexl/media/Network_theoryarticlenetworkonWikipedia1point5deg.jpg\" height=\"200\" width=\"400\">",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
ecb42aa5cf0afc8a138f7542f37d031413e7bc07 | 71,114 | ipynb | Jupyter Notebook | workbooks/2_Cleaning.ipynb | kokorikii/lyric_nlp | 114af3f99bc6cc8430da9f84f1a2b5107a859d59 | [
"CC0-1.0"
] | null | null | null | workbooks/2_Cleaning.ipynb | kokorikii/lyric_nlp | 114af3f99bc6cc8430da9f84f1a2b5107a859d59 | [
"CC0-1.0"
] | null | null | null | workbooks/2_Cleaning.ipynb | kokorikii/lyric_nlp | 114af3f99bc6cc8430da9f84f1a2b5107a859d59 | [
"CC0-1.0"
] | null | null | null | 85.064593 | 19,592 | 0.781956 | [
[
[
"# Data Cleaning",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport re\nfrom better_profanity import profanity\n\n\nfrom nltk.tokenize import sent_tokenize, word_tokenize, RegexpTokenizer\nfrom nltk.corpus import stopwords\nfrom nltk.stem import WordNetLemmatizer\n\npd.set_option('display.max_rows', 500)",
"_____no_output_____"
],
[
"lyric_df = pd.read_csv('../data/full_lyrics_1941_2014.csv')",
"_____no_output_____"
],
[
"lyric_df.rename(columns = {'Unnamed: 0': 'song_rank'}, inplace = True)\n\nlyric_df['song_rank'] = lyric_df['song_rank'] + 1",
"_____no_output_____"
],
[
"lyric_df.shape",
"_____no_output_____"
],
[
"print(f\"{round(lyric_df['lyrics'].isnull().sum() / len(lyric_df) * 100, 2)}% of lyric values are null\")",
"5.79% of lyric values are null\n"
],
[
"# dropping null values as these are only ~5% of the total song dataset\nlyric_df = lyric_df.dropna()",
"_____no_output_____"
],
[
"lyric_df.groupby('year')['year'].count().plot();",
"_____no_output_____"
],
[
"# lowercase all words in the lyrics column\nlyric_df['lyrics'] = lyric_df['lyrics'].str.lower()",
"_____no_output_____"
],
[
"# adding column indicating the decade that the song made the billboard top 100\n\nlyric_df['decade'] = (lyric_df['year']//10)*10 # integer divididing by 10 drops the year, multiplying by 10 again rounds the year to the nearest decade",
"_____no_output_____"
],
[
"lyric_df.head()",
"_____no_output_____"
]
],
[
[
"## Removing Annotation and API debris",
"_____no_output_____"
]
],
[
[
"lyric_df = lyric_df.replace(\"\\[.*?\\]\", \"\", regex = True)",
"_____no_output_____"
],
[
"lyric_df = lyric_df.replace(\"youembedshare\", \"\", regex = True) \nlyric_df = lyric_df.replace(\"embedshare\", \"\", regex = True)\nlyric_df = lyric_df.replace(\"yeahembedshare\", \"\", regex = True) \nlyric_df = lyric_df.replace(\"urlcopyembedcopy\", \"\", regex = True)",
"_____no_output_____"
]
],
[
[
"## Word/Character Counts & API Weirdness",
"_____no_output_____"
],
[
"The inclusion of a word count column has surfaced some irregularities with the Genius Lyric API. There are ~500 songs with excessive word counts (many greater than 100,000 words). These appear to be the text of screenplays, novels and poems that are completely unrelated to the song. As an example, the lyrics for the 1977 Star Wars theme (an instrumental, so I would have expected to not generate results) actually returned the entire text of a 1920 classic French novel 'The Guermantes Way'.\n<p>\n It's unclear why this is happening. I had initially thought this may be placeholder text for instrumental songs, but this does not appear to be the case as songs with lyrics are also impacted. The incorrect lyrics also look to repeat, with some showing up on as many as five different songs. \n<p>\n In order to correct for this I've removed all songs which returned lyrics with a word count greater than 1,100. This dropped the overall count from 6,304 to 5,366. While not ideal, this still leaves enough data to perform analysis and modeling. ",
"_____no_output_____"
]
],
[
[
"# adding word count and unique word count for each song\n\nlyric_df['word_count']=[len(x.split()) for x in lyric_df['lyrics'].tolist()]",
"_____no_output_____"
],
[
"lyric_df['characters'] = lyric_df['lyrics'].apply(len)\nlyric_df['word_length'] = round(lyric_df['characters'] / (lyric_df['word_count']+1), 1)",
"_____no_output_____"
],
[
"lyric_df.drop_duplicates(subset = ['lyrics'], keep = False, inplace = True)",
"_____no_output_____"
],
[
"lyric_df.sort_values(by = 'word_count', ascending = False).head()",
"_____no_output_____"
],
[
"lyric_df = lyric_df[lyric_df['word_count'] < 1100]",
"_____no_output_____"
],
[
"lyric_df.shape",
"_____no_output_____"
],
[
"lyric_df.sort_values(by = 'word_count', ascending = False).head()",
"_____no_output_____"
]
],
[
[
"## Lemmatize and Remove Punctuation",
"_____no_output_____"
]
],
[
[
"# Function to clean and separate titles using regex, and then Stem/Lem the cleaned text and filter stop words. Courtesy Breakfast Hour lesson. Thanks Katie!\ndef cleaner_rev(review):\n # Set token & instantiate Lem/Stem\n lemmatizer = WordNetLemmatizer()\n my_tokenizer = RegexpTokenizer(\"[\\w']+|\\$[\\d\\.]+\") \n \n # Tokenize words\n words = my_tokenizer.tokenize(review.lower())\n # What about stop words??\n stop_word_list = stopwords.words('english')\n no_stops = [i for i in words if i not in stop_word_list]\n # Lem/Stem\n rev_lem = [lemmatizer.lemmatize(i) for i in no_stops]\n # Put words back together\n return ' '.join(rev_lem)",
"_____no_output_____"
],
[
"# adding column of the cleaned lyrics\nlyric_df['clean_lyrics'] = lyric_df['lyrics'].map(cleaner_rev)",
"/var/folders/x1/nngn1kws60393982xbp6dwyc0000gn/T/ipykernel_32782/54294245.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n lyric_df['clean_lyrics'] = lyric_df['lyrics'].map(cleaner_rev)\n"
]
],
[
[
"## Adding Profanity Check",
"_____no_output_____"
]
],
[
[
"lyric_df['profanity'] = lyric_df['clean_lyrics'].apply(lambda x: profanity.contains_profanity(x))",
"/var/folders/x1/nngn1kws60393982xbp6dwyc0000gn/T/ipykernel_32782/1443096844.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n lyric_df['profanity'] = lyric_df['clean_lyrics'].apply(lambda x: profanity.contains_profanity(x))\n"
],
[
"lyric_df['suggestive'] = np.where(lyric_df['profanity'] == True, 1, 0)",
"/var/folders/x1/nngn1kws60393982xbp6dwyc0000gn/T/ipykernel_32782/1006202724.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n lyric_df['suggestive'] = np.where(lyric_df['profanity'] == True, 1, 0)\n"
],
[
"lyric_df.groupby('year')['year'].count().plot();",
"_____no_output_____"
],
[
"lyric_df.groupby('decade')['decade'].count().plot(kind = 'bar');",
"_____no_output_____"
],
[
"lyric_df.to_csv('../data/lyrics_clean.csv')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecb42ac93e1cc80b47dc827c3b33a839ebd126d7 | 3,950 | ipynb | Jupyter Notebook | onnx/examples/make_model.ipynb | chenbohua3/onnx | c940fa3fea84948e46603cab2f86467291443beb | [
"Apache-2.0"
] | 1 | 2022-03-04T03:29:37.000Z | 2022-03-04T03:29:37.000Z | onnx/examples/make_model.ipynb | chenbohua3/onnx | c940fa3fea84948e46603cab2f86467291443beb | [
"Apache-2.0"
] | null | null | null | onnx/examples/make_model.ipynb | chenbohua3/onnx | c940fa3fea84948e46603cab2f86467291443beb | [
"Apache-2.0"
] | 1 | 2022-03-27T19:17:02.000Z | 2022-03-27T19:17:02.000Z | 25.483871 | 83 | 0.427089 | [
[
[
"import onnx\nfrom onnx import helper\nfrom onnx import AttributeProto, TensorProto, GraphProto\n\n\n# The protobuf definition can be found here:\n# https://github.com/onnx/onnx/blob/main/onnx/onnx.proto\n\n\n# Create one input (ValueInfoProto)\nX = helper.make_tensor_value_info('X', TensorProto.FLOAT, [1, 2])\n\n# Create second input (ValueInfoProto)\nPads = helper.make_tensor_value_info('Pads', TensorProto.INT64, [4])\n\n# Create one output (ValueInfoProto)\nY = helper.make_tensor_value_info('Y', TensorProto.FLOAT, [1, 4])\n\n# Create a node (NodeProto)\nnode_def = helper.make_node(\n 'Pad', # node name\n ['X', 'Pads'], # inputs\n ['Y'], # outputs\n mode='constant', # Attributes\n)\n\n# Create the graph (GraphProto)\ngraph_def = helper.make_graph(\n [node_def],\n \"test-model\",\n [X, Pads],\n [Y],\n [helper.make_tensor('Pads', TensorProto.INT64, [4,], [0, 0, 1, 1,])],\n)\n\n# Create the model (ModelProto)\nmodel_def = helper.make_model(graph_def,\n producer_name='onnx-example')\n\nprint('The producer_name in model: {}\\n'.format(model_def.producer_name))\nprint('The graph in model:\\n{}'.format(model_def.graph))\nonnx.checker.check_model(model_def)\nprint('The model is checked!')",
"The producer_name in model: onnx-example\n\nThe graph in model:\nnode {\n input: \"X\"\n input: \"Pads\"\n output: \"Y\"\n op_type: \"Pad\"\n attribute {\n name: \"mode\"\n s: \"constant\"\n type: STRING\n }\n}\nname: \"test-model\"\ninitializer {\n dims: 4\n data_type: 7\n int64_data: 0\n int64_data: 0\n int64_data: 1\n int64_data: 1\n name: \"Pads\"\n}\ninput {\n name: \"X\"\n type {\n tensor_type {\n elem_type: 1\n shape {\n dim {\n dim_value: 1\n }\n dim {\n dim_value: 2\n }\n }\n }\n }\n}\ninput {\n name: \"Pads\"\n type {\n tensor_type {\n elem_type: 7\n shape {\n dim {\n dim_value: 4\n }\n }\n }\n }\n}\noutput {\n name: \"Y\"\n type {\n tensor_type {\n elem_type: 1\n shape {\n dim {\n dim_value: 1\n }\n dim {\n dim_value: 4\n }\n }\n }\n }\n}\n\nThe model is checked!\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
ecb4312103fbb0bd1b0f0b3ace62d251a9710482 | 59,683 | ipynb | Jupyter Notebook | Q1 Notebook.ipynb | RajanPatel97/EE4-68-Pattern-Recognition-CW1 | b2454d050574f487a26cac806ef6c49c95a37f8a | [
"MIT"
] | null | null | null | Q1 Notebook.ipynb | RajanPatel97/EE4-68-Pattern-Recognition-CW1 | b2454d050574f487a26cac806ef6c49c95a37f8a | [
"MIT"
] | null | null | null | Q1 Notebook.ipynb | RajanPatel97/EE4-68-Pattern-Recognition-CW1 | b2454d050574f487a26cac806ef6c49c95a37f8a | [
"MIT"
] | null | null | null | 54.956722 | 14,505 | 0.660289 | [
[
[
"#Load dependencies\nimport pandas as pd\nimport numpy as np\nfrom matplotlib import*\nimport matplotlib.pyplot as plt\nfrom matplotlib.cm import register_cmap\nfrom scipy import stats\nfrom sklearn.decomposition import PCA as sklearnPCA\nimport seaborn",
"_____no_output_____"
],
[
"import scipy.io as sio\nfaces = sio.loadmat('face.mat')\nx = faces['X'] #image dataset\nl = faces['l'] #image ids",
"_____no_output_____"
],
[
"x,l",
"_____no_output_____"
],
[
"mean_vec = np.mean(x, axis=0)\ncov_mat = (x - mean_vec).T.dot((x - mean_vec)) / (x.shape[0]-1)\nprint('Covariance matrix \\n%s' %cov_mat)",
"Covariance matrix \n[[2806.55046192 1490.76930592 1887.07935024 ... 585.03080896\n 796.14844962 729.26134053]\n [1490.76930592 1931.84058855 1195.12587348 ... 270.38959416\n 362.35489357 533.36781523]\n [1887.07935024 1195.12587348 2282.04871118 ... 383.76760282\n 789.34487668 355.62480341]\n ...\n [ 585.03080896 270.38959416 383.76760282 ... 1245.59399732\n 540.61168245 519.11243291]\n [ 796.14844962 362.35489357 789.34487668 ... 540.61168245\n 2099.50525478 1024.47584755]\n [ 729.26134053 533.36781523 355.62480341 ... 519.11243291\n 1024.47584755 1354.14630827]]\n"
],
[
"mean_vec",
"_____no_output_____"
],
[
"print('NumPy covariance matrix: \\n%s' %np.cov(x.T))",
"NumPy covariance matrix: \n[[2806.55046192 1490.76930592 1887.07935024 ... 585.03080896\n 796.14844962 729.26134053]\n [1490.76930592 1931.84058855 1195.12587348 ... 270.38959416\n 362.35489357 533.36781523]\n [1887.07935024 1195.12587348 2282.04871118 ... 383.76760282\n 789.34487668 355.62480341]\n ...\n [ 585.03080896 270.38959416 383.76760282 ... 1245.59399732\n 540.61168245 519.11243291]\n [ 796.14844962 362.35489357 789.34487668 ... 540.61168245\n 2099.50525478 1024.47584755]\n [ 729.26134053 533.36781523 355.62480341 ... 519.11243291\n 1024.47584755 1354.14630827]]\n"
],
[
"#Perform eigendecomposition on covariance matrix\ncov_mat = np.cov(x.T)\neig_vals, eig_vecs = np.linalg.eig(cov_mat)\nprint('Eigenvectors \\n%s' %eig_vecs)\nprint('\\nEigenvalues \\n%s' %eig_vals)",
"Eigenvectors \n[[ 0.06602667 0.01104711 0.04436251 ... 0.00631561 0.03435145\n -0.02028382]\n [ 0.03181648 -0.00148726 0.04757846 ... 0.04218133 0.00346875\n -0.01201677]\n [ 0.0608956 -0.0087206 -0.00051175 ... 0.1509385 -0.00894678\n -0.00702069]\n ...\n [ 0.02779266 -0.0001546 0.04325758 ... 0.00973427 -0.07504817\n 0.00278474]\n [ 0.04399205 0.02754092 0.00523021 ... -0.00779072 -0.00617477\n 0.02583282]\n [ 0.03155445 0.03014632 0.04273412 ... 0.03711877 -0.02471916\n -0.01075868]]\n\nEigenvalues \n[3.11022442e+05 1.05982034e+05 9.39576251e+04 4.82405102e+04\n 4.45400049e+04 2.82798368e+04 2.56788052e+04 2.14673998e+04\n 1.68948815e+04 1.45153236e+04 1.30131318e+04 1.12523586e+04\n 1.04400262e+04 9.43327641e+03 8.72040191e+03 7.73985483e+03\n 7.18384586e+03 6.84208732e+03 6.58537080e+03 6.21656822e+03\n 5.61879002e+03 5.23735386e+03 5.13109587e+03 4.67997367e+03\n 4.48396374e+03 4.42970357e+03 4.34614492e+03 3.81170206e+03\n 3.55777587e+03 3.51545309e+03 3.44634577e+03 3.19349512e+03\n 3.13855786e+03 3.06337710e+03 2.85819975e+03 2.76889946e+03\n 2.67454953e+03 2.55637486e+03 2.52690849e+03 2.43987390e+03\n 2.41889090e+03 2.20195430e+03 2.15134132e+03 2.12175114e+03\n 2.06610292e+03 2.00483148e+03 1.95998389e+03 1.93154898e+03\n 1.86944822e+03 1.81320369e+03 1.76887432e+03 1.71364971e+03\n 1.66183253e+03 1.58316968e+03 1.52967747e+03 1.51278163e+03\n 1.48054124e+03 1.43220763e+03 1.39114015e+03 1.37952551e+03\n 1.36124255e+03 1.34202223e+03 1.31753363e+03 1.26246723e+03\n 1.24414656e+03 1.19587782e+03 1.17266947e+03 1.15404958e+03\n 1.10818986e+03 1.09054486e+03 1.11073862e+03 1.04363200e+03\n 1.05172566e+03 1.02829600e+03 9.99653464e+02 9.92740373e+02\n 9.51223719e+02 9.44796955e+02 9.28543784e+02 9.09353380e+02\n 9.04628309e+02 8.88343463e+02 8.64921145e+02 8.75193807e+02\n 8.41879602e+02 8.27852733e+02 8.09234232e+02 7.80732720e+02\n 7.58077383e+02 7.70677237e+02 7.34451910e+02 7.44617415e+02\n 7.40175238e+02 7.15935409e+02 6.56928414e+02 6.50481399e+02\n 7.05039973e+02 6.99689661e+02 6.69238529e+02 6.88798234e+02\n 6.39838928e+02 6.42719226e+02 6.21576598e+02 6.09889583e+02\n 5.99277272e+02 5.91487649e+02 5.75174255e+02 5.68418139e+02\n 5.66871414e+02 5.54983400e+02 5.49808774e+02 5.42355320e+02\n 5.38830212e+02 5.36397752e+02 5.24611736e+02 5.18706769e+02\n 5.12051035e+02 5.14544303e+02 5.04181783e+02 4.93884997e+02\n 4.90174682e+02 4.86988294e+02 4.83186023e+02 4.75811155e+02\n 4.68346417e+02 4.60925926e+02 4.58456967e+02 4.53610566e+02\n 4.46799724e+02 4.38341166e+02 4.35501730e+02 4.34109820e+02\n 4.23597536e+02 4.21777626e+02 4.12168298e+02 4.09495552e+02\n 4.04807054e+02 4.00211238e+02 3.93002078e+02 3.92242961e+02\n 3.82610672e+02 3.79872946e+02 3.68613851e+02 3.73116506e+02\n 3.74869444e+02 3.65399219e+02 3.55638131e+02 3.56748315e+02\n 3.03112259e+02 3.60088284e+02 3.07609320e+02 3.29873902e+02\n 3.39414664e+02 3.48627282e+02 3.41551136e+02 3.49451557e+02\n 3.21471900e+02 3.31677209e+02 3.15972709e+02 3.23351245e+02\n 3.06573443e+02 3.11923060e+02 3.13247771e+02 3.23976241e+02\n 2.99286878e+02 2.96965147e+02 2.95336183e+02 2.89598325e+02\n 2.85174720e+02 2.86054548e+02 2.82916300e+02 2.78441159e+02\n 2.73736784e+02 2.71308498e+02 2.72298495e+02 2.67427223e+02\n 2.64816049e+02 2.61293678e+02 2.58786994e+02 2.47860735e+02\n 2.56047162e+02 2.51428690e+02 2.53256037e+02 2.54752306e+02\n 2.46113788e+02 2.39372040e+02 2.43171050e+02 2.44106160e+02\n 2.37566360e+02 2.01441897e+02 2.02446438e+02 2.04776015e+02\n 2.26767866e+02 2.32045770e+02 2.23831739e+02 2.07174459e+02\n 2.21370052e+02 2.34079944e+02 2.16619566e+02 2.11926407e+02\n 2.09220193e+02 2.30059714e+02 2.19562816e+02 2.10082776e+02\n 2.10387756e+02 2.15249705e+02 2.19032508e+02 2.00305382e+02\n 1.98807175e+02 1.92374960e+02 1.94485920e+02 1.96266085e+02\n 1.90160384e+02 1.88330010e+02 1.86597959e+02 1.52449676e+02\n 1.52986791e+02 1.85411071e+02 1.83603645e+02 1.81563184e+02\n 1.79777271e+02 1.78119482e+02 1.75544043e+02 1.54962197e+02\n 1.74598943e+02 1.72837992e+02 1.59180899e+02 1.56320585e+02\n 1.55504587e+02 1.63201780e+02 1.62144577e+02 1.67867119e+02\n 1.69434888e+02 1.65994134e+02 1.84690886e+02 1.56724087e+02\n 1.59989119e+02 1.72323405e+02 1.68601275e+02 1.76276630e+02\n 1.62472546e+02 1.51891388e+02 1.48691491e+02 1.22195065e+02\n 1.22871716e+02 1.47014245e+02 1.24322089e+02 1.25749757e+02\n 1.29697541e+02 1.27133264e+02 1.45493283e+02 1.44986848e+02\n 1.44292492e+02 1.42256315e+02 1.41641482e+02 1.31600091e+02\n 1.26643234e+02 1.33671790e+02 1.31903805e+02 1.36983612e+02\n 1.24647608e+02 1.34822207e+02 1.38027125e+02 1.39914240e+02\n 1.39575155e+02 1.38980146e+02 1.35298814e+02 1.35848084e+02\n 1.00931105e+02 1.01440682e+02 1.02508673e+02 1.21741562e+02\n 1.20762124e+02 1.17820914e+02 1.19794107e+02 1.16068847e+02\n 1.14701482e+02 1.13670669e+02 1.19346754e+02 1.18922705e+02\n 1.12606113e+02 1.04493239e+02 1.18484111e+02 1.11630612e+02\n 1.09283568e+02 1.08520187e+02 1.11248178e+02 1.05537487e+02\n 1.07816835e+02 1.06912712e+02 1.05324001e+02 1.15101922e+02\n 1.06350044e+02 1.06055780e+02 1.10247980e+02 1.03543526e+01\n 1.25743149e+01 1.17383761e+01 1.00077021e+02 9.90040769e+01\n 9.59412803e+01 9.85126894e+01 9.72468170e+01 9.80371291e+01\n 9.51959695e+01 8.44581577e+01 8.55403777e+01 9.39775853e+01\n 9.29270478e+01 9.24829378e+01 8.65652566e+01 9.01268525e+01\n 9.36586847e+01 8.82808147e+01 8.94029842e+01 9.12148307e+01\n 9.15876164e+01 8.88286430e+01 8.46813434e+01 8.79510920e+01\n 9.41803363e+01 8.71009304e+01 1.22062872e+01 1.30721421e+01\n 1.34931362e+01 1.37604091e+01 1.41562037e+01 1.44548404e+01\n 1.46086555e+01 1.48681883e+01 1.51388095e+01 1.55021211e+01\n 1.57271830e+01 1.62894797e+01 1.64344641e+01 8.35444749e+01\n 8.29916145e+01 8.24633479e+01 8.17628003e+01 1.67448043e+01\n 8.10907928e+01 7.98658710e+01 7.98135760e+01 7.91295341e+01\n 7.86031738e+01 7.81932175e+01 7.33847246e+01 7.53072980e+01\n 7.43446300e+01 7.46997836e+01 7.60536552e+01 7.66151019e+01\n 7.68887801e+01 7.70280240e+01 1.69400013e+01 1.72050893e+01\n 1.73556265e+01 1.76450577e+01 1.80134317e+01 1.83718779e+01\n 1.84467191e+01 1.87843925e+01 1.90056498e+01 1.93758818e+01\n 1.97724634e+01 1.90799362e+01 1.99768643e+01 2.02123301e+01\n 2.06904662e+01 2.08875690e+01 2.10210661e+01 7.31625425e+01\n 7.24018746e+01 7.15062685e+01 7.20050754e+01 7.22414289e+01\n 6.96459840e+01 7.06778549e+01 7.04597777e+01 6.93504577e+01\n 6.80171304e+01 6.83561400e+01 6.73845643e+01 6.69131279e+01\n 6.63381185e+01 6.55733695e+01 6.51916109e+01 6.42377282e+01\n 6.45061579e+01 6.29510056e+01 6.26577325e+01 6.21413124e+01\n 5.77608855e+01 5.90022786e+01 5.87749854e+01 5.64350995e+01\n 5.68790986e+01 6.61862626e+01 5.73858422e+01 6.34594384e+01\n 5.70665642e+01 6.09795278e+01 5.97933878e+01 6.13829684e+01\n 6.04373379e+01 5.98927928e+01 2.13392008e+01 2.15831861e+01\n 2.20551738e+01 2.18964897e+01 5.41144730e+01 5.46755418e+01\n 5.60865800e+01 5.54287965e+01 5.55408218e+01 5.59498938e+01\n 2.22567934e+01 2.26746303e+01 2.29583724e+01 2.28374093e+01\n 2.35622404e+01 5.04181042e+01 4.94808572e+01 5.10896626e+01\n 5.35675201e+01 5.22630878e+01 4.90740785e+01 4.91632877e+01\n 5.24282310e+01 5.33867670e+01 5.15721085e+01 5.21523110e+01\n 5.49634867e+01 4.85577277e+01 5.00601229e+01 4.80520195e+01\n 4.77270889e+01 4.66771911e+01 4.71939671e+01 4.71585789e+01\n 2.37702931e+01 4.60990430e+01 4.63069715e+01 4.56472986e+01\n 2.34608772e+01 4.51335513e+01 4.37721170e+01 4.50660269e+01\n 2.40979879e+01 2.45667453e+01 2.50769093e+01 2.47452634e+01\n 2.56087858e+01 2.57961967e+01 2.87150104e+01 2.41596804e+01\n 2.64779936e+01 2.69663470e+01 2.79126012e+01 2.82543107e+01\n 2.75511772e+01 2.85512199e+01 4.33043505e+01 2.67228329e+01\n 4.43722280e+01 4.45697224e+01 2.61744629e+01 2.77094511e+01\n 4.39769910e+01 4.30362904e+01 4.21120801e+01 2.46741720e+01\n 4.24576317e+01 4.15214308e+01 4.04782648e+01 2.63560163e+01\n 3.97531940e+01 4.07791286e+01 2.72824718e+01 3.85982431e+01\n 3.79464080e+01 4.26724518e+01 3.05161236e+01 3.02040851e+01\n 3.08330063e+01 3.72897244e+01 3.11150466e+01 3.20346606e+01\n 2.96706821e+01 2.97929140e+01 3.45214712e+01 3.29304226e+01\n 3.42716181e+01 3.31589099e+01 2.93303350e+01 3.17407682e+01\n 3.58077619e+01 3.63345801e+01 3.61765807e+01 2.60364958e+01\n 4.10130933e+01 3.49803143e+01 3.55278773e+01 3.16093525e+01\n 2.90935830e+01 4.12593503e+01 3.98722879e+01 3.75628122e+01\n 3.71384954e+01 3.92840097e+01 3.39628558e+01 3.15239245e+01\n 3.36048020e+01 3.89103416e+01 3.27782027e+01 3.64777688e+01\n 3.36181642e+01 3.48729709e+01 3.89992278e+01 3.90335786e+01]\n"
],
[
"# Visually confirm that the list is correctly sorted by decreasing eigenvalues\neig_pairs = [(np.abs(eig_vals[i]), eig_vecs[:,i]) for i in range(len(eig_vals))]\nprint('Eigenvalues in descending order:')\nfor i in eig_pairs:\n print(i[0])",
"Eigenvalues in descending order:\n311022.4420849844\n105982.03353361382\n93957.62505013801\n48240.510209135056\n44540.00490112329\n28279.836809758977\n25678.805248032084\n21467.399846539174\n16894.881478801017\n14515.323622661926\n13013.13182960087\n11252.358606274463\n10440.02621979263\n9433.276408770507\n8720.401909281629\n7739.854830825744\n7183.845863089899\n6842.087317192625\n6585.370804791674\n6216.568219030426\n5618.790020050505\n5237.353861294975\n5131.095872870628\n4679.973665946103\n4483.963735341708\n4429.703573415428\n4346.144918060908\n3811.702055672763\n3557.7758697754457\n3515.453088781213\n3446.345766410721\n3193.495118063144\n3138.557862504642\n3063.3771021689386\n2858.199746994838\n2768.89945567036\n2674.5495281215694\n2556.374861075687\n2526.908494401603\n2439.873904964791\n2418.890895682103\n2201.9543025474986\n2151.3413153450174\n2121.751139201031\n2066.1029168634536\n2004.8314845741309\n1959.983886001329\n1931.5489803190724\n1869.4482217858101\n1813.2036868897644\n1768.8743234713374\n1713.6497123060776\n1661.8325298828374\n1583.1696780801135\n1529.677469050175\n1512.7816295325815\n1480.541239014482\n1432.2076317447013\n1391.1401529447178\n1379.5255051093907\n1361.2425527031523\n1342.0222281438753\n1317.5336346661836\n1262.4672278341322\n1244.1465562291826\n1195.8778185841327\n1172.669469778237\n1154.0495792358056\n1108.1898590746164\n1090.5448597419881\n1110.738619740711\n1043.6319984331392\n1051.7256572298122\n1028.2959968957373\n999.6534644934983\n992.7403731793862\n951.223718783496\n944.7969551692623\n928.543783722767\n909.3533804130085\n904.6283088024742\n888.3434625471021\n864.9211446649406\n875.1938067556389\n841.879602007456\n827.8527331115025\n809.2342319065043\n780.732719616406\n758.0773830055925\n770.6772369055755\n734.4519100899873\n744.6174147881574\n740.1752383472858\n715.9354091123731\n656.9284140819905\n650.481398792497\n705.0399726915158\n699.6896612598927\n669.2385291795309\n688.7982341183695\n639.8389277912969\n642.7192257210561\n621.5765975108885\n609.8895829070166\n599.2772718758473\n591.4876491953141\n575.1742553625694\n568.4181385188098\n566.8714142117092\n554.9834001418958\n549.8087743487538\n542.3553198078141\n538.8302115917961\n536.3977521917859\n524.6117356509207\n518.7067686148162\n512.0510351953983\n514.5443031674581\n504.18178291067085\n493.8849971008431\n490.1746817048139\n486.98829392636816\n483.1860233009167\n475.8111549893896\n468.346417116772\n460.92592620514876\n458.4569674245789\n453.6105659874256\n446.7997241599576\n438.34116635168533\n435.5017304193622\n434.1098200273594\n423.5975355580008\n421.77762578594565\n412.1682984575998\n409.49555173189157\n404.8070540728779\n400.2112376352482\n393.00207758611\n392.24296093199894\n382.6106723725907\n379.87294624592107\n368.61385070581804\n373.1165063432442\n374.8694439681696\n365.3992189043871\n355.6381311640695\n356.7483149622654\n303.1122592143829\n360.0882837205311\n307.6093201643627\n329.8739019262263\n339.41466394767644\n348.62728193667556\n341.551136456638\n349.45155697819865\n321.47189986712675\n331.67720875998447\n315.9727087211659\n323.35124451219247\n306.57344252840767\n311.9230603261564\n313.24777134787223\n323.9762408887289\n299.2868775884438\n296.96514730239807\n295.33618337196805\n289.5983251207867\n285.1747203747023\n286.0545475846372\n282.91630005248214\n278.44115898488445\n273.73678364084725\n271.3084980363303\n272.2984954146691\n267.42722346910716\n264.81604903328724\n261.2936780889148\n258.7869943742711\n247.86073467579428\n256.0471624323299\n251.42868962893434\n253.2560369369451\n254.7523059802875\n246.1137879657186\n239.3720397628648\n243.1710495816511\n244.10615960493934\n237.56635957597842\n201.44189669885452\n202.44643786958935\n204.77601462134479\n226.76786602563942\n232.045769540486\n223.83173948934882\n207.17445891188112\n221.3700519219688\n234.07994388810883\n216.6195661583438\n211.92640739885317\n209.2201934318225\n230.05971355883344\n219.56281590447247\n210.0827759724295\n210.3877558292852\n215.24970451846244\n219.03250796977284\n200.3053817282664\n198.80717453247001\n192.37496017832228\n194.4859200291724\n196.26608490518674\n190.16038404034106\n188.33001030942287\n186.59795894234918\n152.44967583425563\n152.9867907450378\n185.41107129325275\n183.603644971385\n181.56318396868807\n179.77727087824462\n178.11948152654705\n175.54404300463253\n154.9621974491339\n174.59894283991397\n172.83799187551722\n159.18089898460383\n156.32058506387676\n155.50458673359728\n163.20177969533094\n162.1445769390631\n167.86711924270836\n169.43488836179708\n165.99413412547773\n184.6908860510799\n156.72408651712854\n159.98911945909066\n172.32340472392718\n168.60127541151775\n176.27663014326959\n162.47254565567073\n151.89138795490024\n148.69149138710765\n122.19506483156069\n122.87171552419184\n147.01424483782517\n124.32208852612376\n125.74975672142394\n129.69754081127186\n127.13326395803749\n145.49328342089967\n144.98684759726675\n144.29249199066982\n142.2563150812547\n141.6414822334281\n131.60009092075467\n126.64323444104535\n133.67178999585258\n131.90380480995867\n136.98361186666304\n124.64760820470353\n134.8222070651401\n138.0271250778343\n139.9142402262684\n139.57515523744846\n138.98014592612316\n135.29881425540492\n135.84808384137096\n100.93110512484068\n101.4406816983162\n102.50867287300088\n121.74156240612972\n120.76212432790678\n117.82091357092945\n119.79410736892713\n116.06884713454814\n114.70148209311365\n113.67066941293243\n119.3467542303102\n118.9227046069753\n112.6061134553927\n104.49323944919497\n118.48411059176185\n111.6306121870291\n109.28356802159284\n108.52018720437377\n111.24817753011297\n105.53748719611046\n107.81683467043189\n106.91271218867328\n105.32400118322202\n115.10192228175929\n106.35004398623855\n106.05578003222696\n110.24798036868545\n10.354352569043755\n12.574314874978565\n11.738376115693057\n100.07702123009844\n99.00407687385622\n95.94128029657749\n98.51268936171795\n97.24681696210352\n98.03712907675741\n95.19596950978199\n84.45815768572865\n85.54037773464415\n93.9775852621647\n92.92704781518445\n92.48293781151592\n86.56525661427264\n90.1268524538254\n93.65868469191301\n88.28081465712344\n89.40298415729639\n91.2148306653318\n91.58761640307368\n88.82864295527875\n84.68134335392314\n87.95109201001695\n94.18033634019824\n87.10093041576144\n12.206287237769732\n13.072142110660923\n13.493136150243828\n13.760409133842565\n14.156203661238127\n14.454840380075817\n14.60865552653458\n14.868188333530458\n15.13880945851226\n15.502121099544963\n15.727182986273679\n16.289479709321135\n16.43446411811313\n83.54447487858593\n82.99161453897716\n82.46334791198936\n81.76280028923522\n16.744804289243888\n81.09079281908762\n79.86587104107647\n79.8135759603323\n79.12953412383224\n78.60317376103626\n78.19321749652845\n73.38472457158032\n75.30729797907048\n74.34462995350079\n74.69978357307694\n76.05365524996395\n76.61510192668065\n76.88878011804033\n77.02802404134489\n16.940001320066465\n17.205089304509553\n17.35562651495091\n17.645057685741875\n18.013431685353694\n18.37187785015319\n18.446719104837896\n18.784392468269015\n19.00564984466105\n19.375881845527374\n19.77246344826039\n19.079936206745348\n19.9768643472044\n20.212330136904992\n20.69046618161945\n20.887568967011454\n21.021066148168835\n73.16254254013482\n72.4018745955013\n71.50626851243285\n72.00507535054888\n72.24142891012984\n69.64598404563236\n70.6778549478173\n70.45977772045376\n69.35045772607114\n68.01713044113674\n68.35613998441167\n67.38456426138742\n66.9131278657809\n66.33811847543956\n65.57336949014672\n65.19161091240386\n64.23772819546446\n64.50615787396956\n62.95100562828669\n62.65773252304973\n62.141312445815004\n57.76088550459144\n59.002278552844366\n58.774985429716516\n56.43509947538791\n56.87909864535524\n66.18626264296901\n57.38584217385791\n63.45943842070624\n57.06656415451938\n60.97952784521\n59.79338781198762\n61.38296842141059\n60.437337943642305\n59.8927928244259\n21.339200836529322\n21.58318608320171\n22.055173753461734\n21.896489716123586\n54.11447297049053\n54.67554177371473\n56.08657997192318\n55.42879647526362\n55.54082183606163\n55.94989377062544\n22.25679342850529\n22.674630306741356\n22.958372361349134\n22.83740930330869\n23.562240429098022\n50.418104157415506\n49.48085718728814\n51.08966261469677\n53.56752012964555\n52.26308777202658\n49.074078549465305\n49.163287664054\n52.428231003959866\n53.386766995197505\n51.57210848119675\n52.15231095887517\n54.96348665126723\n48.55772765450421\n50.06012289251242\n48.052019490955225\n47.72708892416213\n46.677191089745676\n47.19396709455897\n47.15857889968895\n23.770293142874735\n46.09904302937909\n46.30697151249598\n45.64729860125598\n23.46087719188255\n45.13355128838178\n43.77211697576519\n45.06602685972443\n24.097987881560176\n24.56674530492258\n25.0769092641604\n24.745263429647455\n25.608785797657088\n25.796196747106034\n28.71501038382733\n24.159680362279193\n26.477993551484012\n26.96634701934903\n27.912601222382524\n28.25431071885446\n27.551177228808047\n28.551219947303114\n43.30435052381595\n26.722832924083377\n44.37222803344144\n44.56972244275259\n26.17446289255599\n27.70945106590651\n43.97699096139477\n43.03629041960201\n42.1120801127691\n24.67417203385585\n42.457631720006596\n41.52143081226566\n40.47826476879794\n26.356016348133213\n39.753194048814514\n40.779128625832364\n27.28247182978233\n38.598243118027455\n37.94640798378368\n42.672451846409714\n30.51612363540293\n30.204085124046593\n30.83300626959807\n37.28972443118738\n31.11504656418226\n32.03466061802931\n29.670682140375426\n29.792913992509053\n34.52147116761381\n32.93042258301972\n34.271618066476506\n33.158909876613265\n29.3303349556431\n31.740768224259618\n35.80776192560801\n36.3345801461499\n36.17658072152051\n26.03649577779809\n41.01309325728534\n34.98031427254047\n35.52787729789352\n31.609352456877247\n29.093582972565724\n41.259350267191564\n39.872287872380866\n37.562812227824736\n37.13849537412439\n39.28400970688531\n33.96285575230838\n31.523924497793914\n33.604801950772995\n38.910341556001605\n32.7782026988712\n36.477768784450156\n33.61816419268564\n34.872970914998376\n38.999227761181366\n39.03357857654357\n"
],
[
"pca = sklearnPCA(n_components=2)\npca.fit_transform(x)\npca.explained_variance_ratio_",
"_____no_output_____"
],
[
"#Explained variance\npca = sklearnPCA().fit(x)\nplt.plot(np.cumsum(pca.explained_variance_ratio_))\nplt.xlabel('number of components')\nplt.ylabel('cumulative explained variance')\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb4373dfddb49019e792dff3adeb4e24feec636 | 12,823 | ipynb | Jupyter Notebook | colab-export-image-classifier.ipynb | butchland/build-your-own-image-classifier | 977245efdb3238ef257ad5ea6252e16baed454e3 | [
"MIT"
] | null | null | null | colab-export-image-classifier.ipynb | butchland/build-your-own-image-classifier | 977245efdb3238ef257ad5ea6252e16baed454e3 | [
"MIT"
] | null | null | null | colab-export-image-classifier.ipynb | butchland/build-your-own-image-classifier | 977245efdb3238ef257ad5ea6252e16baed454e3 | [
"MIT"
] | null | null | null | 35.918768 | 371 | 0.555096 | [
[
[
"<a href=\"https://colab.research.google.com/github/butchland/build-your-own-image-classifier/blob/master/colab-export-image-classifier.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Export your Image Classifier to Github\n\n## Instructions\n\n1. In the **Specify Project Name and Github Credentials** section below, fill out the project name first _(this is the name of the project you used in the previous notebook. If you didn't change the name of the default project in the previous notebook, you shouldn't have to change the default project name here either so just leave the project name as is)_.\n\n Also enter your `Github ID` and the name of the `Github repo` you created from the `build-your-own-image-classifier-template` project as discussed in the article. \n\n You will also need to provide your `real name` and an `email address` as this will be used to configure your git credentials (this will show up as the name and email of the author who made what change to your repo) \n \n _(Your github password will be asked to be entered separately later after connecting and starting the command to start running (Cmd/Ctrl+F9 or the menu Runtime/Run all) in the section **Enter Github Password**)_\n\n This notebook assumes that you have already built and exported your image classifier (i.e. the _`export.pkl`_ file already been saved to your Google Drive under the _`/My Drive/build-your-own-image-classifier/models/pets`_ directory or its equivalent.\n\n If the exported image classifier does not exist, this will trigger an error. Please make sure to run the previous notebook ([Build your Image Classifier](https://colab.research.google.com/github/butchland/build-your-own-image-classifier/blob/master/colab-build-image-classifier.ipynb)) before running this one. \n\n1. Click on the `Connect` button on the top right area of the page. This will change into a checkmark with the RAM and Disk health bars once the connection is complete.\n1. Press `Cmd/Ctrl+F9` or Click on the menu `Runtime/Run all`\n1. Click on the link to `accounts.google.com` that appears and login in to your Google Account if neccessary or select the Google Account to use for your Google Drive. (This will open a new tab)\n1. Authorize `Google Drive File Stream` to access your Google Drive.\n\n1. Copy the generated authentication token and paste it on the input box that appears.\n\n1. Once the text 'Please enter your Password ...' is displayed at the **Enter Github Password** section near the bottom of the notebook, enter it in the password box and press enter.\n\n1. If you entered your Github ID, repo or password incorrectly an error should appear, otherwise, the text 'DONE! DONE! DONE!' should printed at the end of the notebook, You can click on the menu `Runtime/Factory reset runtime` and click `Yes` on the dialog box to end your session.\n\nYour exported image classifier(`export.pkl`) will now be visible in the list of file in your Github repo after you refresh your github repo page.\n\nIf no `export.pkl` appears on the list, then something might have gone awry with the process. You can rerun the previous step by closing the notebook and reopen it from the link in the article.",
"_____no_output_____"
],
[
"## What is going on?\n\nThis section explains the code behind this notebook\n\n_(Click on SHOW CODE to display the code)_",
"_____no_output_____"
],
[
"### Connect to your Google Drive\n\nWe'll need to connect to your Google Drive in order to retrieve your exported image classifier.",
"_____no_output_____"
]
],
[
[
"#@title {display-mode: \"form\"}\nfrom google.colab import drive\ndrive.mount('/content/drive')",
"_____no_output_____"
]
],
[
[
"### Specify Project Name and Github Credentials\n\nFill out the `project name` -- the project name should be the same one used as the project name used in the previous notebook. Your `github_id` and `github_repo` should contain the information you previously used to create your Github ID and repo. \n",
"_____no_output_____"
]
],
[
[
"#@title Enter your project name {display-mode: \"form\"}\nproject = \"pets\" #@param {type: \"string\"}\ngithub_id = \"\" #@param {type: \"string\"}\ngithub_repo = \"\" #@param {type: \"string\"}\nuser_email = \"\" #@param {type: \"string\"}\nreal_name = \"\" #@param {type: \"string\"}",
"_____no_output_____"
]
],
[
[
"Check that the github ID, repo, email and name have been filled out",
"_____no_output_____"
]
],
[
[
"#@title {display-mode: \"form\"}\nif github_id == \"\" or github_repo == \"\" or user_email == \"\" or real_name == \"\":\n print(\"Rerun your notebook by pressing Cmd/Ctrl-F9 or menu Runtime/Run all\")\n raise RuntimeError(\"Please enter your Github ID and Repo as well as your user email and name\")",
"_____no_output_____"
]
],
[
[
"### Install Python Packages and Git Extensions\n\nInstall all the python packages as well as git extensions to enable exporting your image classifier. ",
"_____no_output_____"
]
],
[
[
"#@title {display-mode: \"form\"} \n!pip install -Uqq fastai --upgrade\n!curl -s https://packagecloud.io/install/repositories/github/git-lfs/script.deb.sh | bash\n!apt-get -qq install git-lfs\n!git lfs install",
"_____no_output_____"
]
],
[
[
"### Copy your Image Classifier from Google Drive",
"_____no_output_____"
]
],
[
[
"#@title {display-mode: \"form\"} \nfrom fastai.vision.all import *\nfrom fastai.vision.widgets import *\nfrom ipywidgets import widgets\nfile_name = f'export.pkl'\nfolder_path = f'build-your-own-image-classifier/models/{project}' \nif not (Path('/content/drive/My Drive')/folder_path/file_name).is_file():\n raise RuntimeError(f'Exported image classifier does not exist: at My Drive/f{folder_path}/{file_name}')",
"_____no_output_____"
],
[
"#@title {display-mode: \"form\"} \n!cp /content/drive/My\\ Drive/{folder_path}/{file_name} /content/.",
"_____no_output_____"
],
[
"#@title {display-mode: \"form\"} \npath = Path(f'/content')\nPath.BASE_PATH = path\nif not (path/file_name).is_file():\n raise RuntimeError(\"Could not find export.pkl -- Please run notebook to build your classifier first!\")\n",
"_____no_output_____"
]
],
[
[
"### Enter Github Password \nPlease enter your github password as requested",
"_____no_output_____"
]
],
[
[
"#@title {display-mode: \"form\"}\nprint('Please enter your password.')\nimport getpass\ngithub_password = getpass.getpass()",
"_____no_output_____"
]
],
[
[
"Configure git and \"push\" exported image classifier to Github",
"_____no_output_____"
]
],
[
[
"#@title {display-mode: \"form\"}\n!git config --global user.name \"{real_name}\"\n!git config --global user.email \"{user_email}\"\n!git clone -q https://github.com/{github_id}/{github_repo}.git",
"_____no_output_____"
],
[
"#@title {display-mode: \"form\"}\nif not (Path('/content')/github_repo).is_dir():\n print('You might have entered the wrong github credentials')\n raise RuntimeError(f'Could not download your github repo https://github.com/{github_id}/{github_project}.git')\n%cd /content/{github_repo}\n!cp /content/export.pkl .\n!git add export.pkl\n!git commit -m \"Add exported image classifier\"",
"_____no_output_____"
],
[
"#@title {display-mode: \"form\"}\n!git config credential.helper store\n!echo \"https://{github_id}:{github_password}@github.com\" > /root/.git-credentials\n!git push\n!rm -f /root/.git-credentials\n",
"_____no_output_____"
],
[
"#@title {display-mode: \"form\"}\nprint(\"DONE! DONE! DONE!\")\nprint(\"Make sure to end your session (Click on menu Runtime/Factory reset runtime and click 'Yes' on the dialog box to end your session)\")\nprint(\"before closing this notebook.\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecb4417994fa7d9f29eac2b0da9eb5ca43e825f3 | 43,229 | ipynb | Jupyter Notebook | Python/EnergyMinimization/UnitTests.ipynb | SouslovLab/ActiveElastocapillarity | 5882abf0b15461d8dc5b54887ec14f0bba84650f | [
"MIT"
] | null | null | null | Python/EnergyMinimization/UnitTests.ipynb | SouslovLab/ActiveElastocapillarity | 5882abf0b15461d8dc5b54887ec14f0bba84650f | [
"MIT"
] | null | null | null | Python/EnergyMinimization/UnitTests.ipynb | SouslovLab/ActiveElastocapillarity | 5882abf0b15461d8dc5b54887ec14f0bba84650f | [
"MIT"
] | null | null | null | 26.900436 | 472 | 0.535705 | [
[
[
"import meshio\nimport pygmsh\nimport pygalmesh\nimport numpy as np\nimport copy\nfrom mshr import *\nfrom dolfin import *\nfrom collections import Counter\nimport matplotlib\nimport matplotlib.pyplot as plt\nimport os\nimport json\nimport shutil\nimport scipy.optimize as opt\nfrom EnergyMinimization import *\nfrom AnalysisFunctions import *\nimport numba\nimport timeit\nfrom timeit import default_timer as timer",
"_____no_output_____"
],
[
"# root folder for data\nDataFolder=os.getcwd()+'/Data/Scratch'\n# Folder for the run data\"",
"_____no_output_____"
]
],
[
[
"# Testing the Mesh Generation",
"_____no_output_____"
],
[
"Make a mesh with pygmsh, with some dummy values to play with:",
"_____no_output_____"
]
],
[
[
"with pygmsh.occ.Geometry() as geom:\n geom.characteristic_length_max = 0.1\n ellipsoid = geom.add_ball([0.0, 0.0, 0.0], 1)\n InputMesh = geom.generate_mesh()",
"/home/jackbinysh/miniconda3/lib/python3.8/site-packages/numpy/ctypeslib.py:521: RuntimeWarning: A builtin ctypes object gave a PEP3118 format string that does not match its itemsize, so a best-guess will be made of the data type. Newer versions of python may behave correctly.\n return array(obj, copy=False)\n"
],
[
"interiorbonds,edgebonds,boundarytris, bidxTotidx, tetras= MakeMeshData3D(InputMesh)\nbonds=np.concatenate((interiorbonds,edgebonds))\norientedboundarytris=OrientTriangles(InputMesh.points,boundarytris,np.array([0,0,0]))",
"_____no_output_____"
],
[
"cells=[ (\"line\", bonds ), (\"triangle\",boundarytris ), (\"tetra\",tetras)]\nisbond= np.ones(len(bonds))\nisedgebond= np.concatenate( ( np.zeros(len(interiorbonds)),np.ones(len(edgebonds)) ) )\nCellDataDict={'isedgebond':[isedgebond,np.zeros(len(boundarytris)),np.zeros(len(tetras))]\n ,'isbond':[isbond,np.zeros(len(boundarytris)),np.zeros(len(tetras))]}\n\nOutputMesh=meshio.Mesh(InputMesh.points, cells, {},CellDataDict)\nOutputMesh.write(DataFolder+\"InitialMesh.vtk\",binary=True) ",
"_____no_output_____"
]
],
[
[
"## Testing the boundary triangle finding",
"_____no_output_____"
],
[
"when pygmsh generates a sphere, it gives the tetrahedrons and boundary triangles. We can use this to check our bondary finding is working. First, make the ball. Now, compare the lists of boundary triangles that we find to those that pygmsh finds. We need to sort the pygmsh ones, as the vertices dont always appear in ascending order",
"_____no_output_____"
]
],
[
[
"np.array_equal(boundarytris,np.sort(InputMesh.cells[1].data,axis=1))",
"_____no_output_____"
],
[
"boundarytris.shape",
"_____no_output_____"
],
[
"InputMesh.cells[1].data.shape",
"_____no_output_____"
]
],
[
[
"This seems a good verification that our boundary list is correct",
"_____no_output_____"
],
[
"## Check total surface area",
"_____no_output_____"
],
[
"For a sphere",
"_____no_output_____"
]
],
[
[
"vTotalArea3D(InputMesh.points,boundarytris)",
"_____no_output_____"
],
[
"4*np.pi",
"_____no_output_____"
]
],
[
[
"## Check total volume",
"_____no_output_____"
],
[
"Ive written two functions which do this, lets check both",
"_____no_output_____"
]
],
[
[
"Volume3D(InputMesh.points,orientedboundarytris,bidxTotidx).sum()",
"_____no_output_____"
],
[
"Volume3D_tetras(InputMesh.points,tetras).sum()",
"_____no_output_____"
],
[
"(4/3)*np.pi",
"_____no_output_____"
]
],
[
[
"# Physical Tests",
"_____no_output_____"
],
[
"## Checking the Bending Modulus Energy",
"_____no_output_____"
],
[
" As implemented, the bending modulus approximates the continuum limit $F= \\frac{\\kappa_c}{2}\\int dA(C_1+C_2-C_0)^2 + k_g \\int dA C_1C_2$ for a closed surface, where $C_1$ etc. are the principal curvatures. According to Boal and Rao 1992, the energy of a sphere without a spontaneous curvature is $\\frac{4\\pi k_{rig}}{\\sqrt{3}}$, where $k_{rig}$ is the microscopic modulus. This seems to be true for a triangulation by equilateral triangles. Lets check this:",
"_____no_output_____"
]
],
[
[
"theta_0=0\nkbend=1\nenergies= BendingEnergy(InputMesh.points,orientedboundarytris,bidxTotidx,kbend)\nenergies.sum()",
"_____no_output_____"
],
[
"(4/np.sqrt(3))*np.pi",
"_____no_output_____"
]
],
[
[
"## Checking the Spring Energy",
"_____no_output_____"
],
[
"A basic test: lets just make 2 springs (to check the vectorization), and confirm their behaviour. We are supposed to be implementing:\n\n$V(r,r_0) = k_{\\mathrm{neo}}\\left( \\frac{1-\\alpha}{2}\\left(\\frac{1}{\\lambda}+2\\lambda^2\\right)+\\frac{\\alpha}{2}\\left(\\frac{1}{\\lambda^2}+2\\lambda\\right) \\right)$, where $V=r/r_0$, and $k_{neo}=\\frac{r_0^2 k_{hook}}{3}$. Some tests:\n\n$V(1)= \\frac{1}{2} k_{hook} r_0^2=\\frac{3}{2} k_{neo}$, indepdent of the mat non.",
"_____no_output_____"
]
],
[
[
"MatNon=1\nkhook=1\n#rest lengths\nr0_ij=np.array([1,2])\nSpringEnergy=NeoHookean3D(r0_ij,r0_ij,khook,MatNon)\nprint(SpringEnergy) ",
"_____no_output_____"
],
[
"np.log(0.5)",
"_____no_output_____"
]
],
[
[
"Lets do the same with our shifted energy:",
"_____no_output_____"
]
],
[
[
"MatNon=1\nkhook=1\n#rest lengths\nr0_ij=np.array([1,2])\nSpringEnergy=NeoHookeanShifted(r0_ij,r0_ij,khook,MatNon)\nprint(SpringEnergy)",
"_____no_output_____"
]
],
[
[
"Plotting both energies:",
"_____no_output_____"
]
],
[
[
"lam=np.arange(0.1, 10, 0.01);\nMatNon=0.7\nkhook=2\n\nEnergy=NeoHookean3D(lam,1,khook,MatNon)\nplt.plot(lam,Energy)\n\nkneo_ij = (1**2)*khook/3 \nEnergy=NeoHookeanShifted(lam,1,khook,MatNon)+1.5*kneo_ij\nplt.plot(lam,Energy)",
"_____no_output_____"
]
],
[
[
"Plotting the energy: Pure Neohookean on a loglogplot. Expectation: Minimum at $(0,log(0.5)$=$(0,-0.693)$ Asmptotes to grad -1 and grad 2 in either limit.\n\nPure MR: same minimum, but the opposite grad behaviours",
"_____no_output_____"
]
],
[
[
"lam=np.arange(0.1, 10, 0.01);\nMatNon=0\nkhook=1\nEnergy=NeoHookean3D(lam,1,khook,MatNon)\nplt.plot(np.log(lam),np.log(Energy))\n\nMatNon=1\nkhook=1\nEnergy=NeoHookean3D(lam,1,khook,MatNon)\nplt.plot(np.log(lam),np.log(Energy))",
"_____no_output_____"
],
[
"lam=10**10\nMatNon=0\nkhook=1\nprint(( np.log(NeoHookean3D(lam,1,khook,MatNon))-np.log(NeoHookean3D(1,1,khook,MatNon)) )/(np.log(lam)))\nlam=10**(-10)\nMatNon=0\nkhook=1\nprint(( np.log(NeoHookean3D(lam,1,khook,MatNon))-np.log(NeoHookean3D(1,1,khook,MatNon)) )/(np.log(lam)))\n",
"_____no_output_____"
],
[
"lam=10**10\nMatNon=1\nkhook=1\nprint(( np.log(NeoHookean3D(lam,1,khook,MatNon))-np.log(NeoHookean3D(1,1,khook,MatNon)) )/(np.log(lam)))\nlam=10**(-10)\nMatNon=1\nkhook=1\nprint(( np.log(NeoHookean3D(lam,1,khook,MatNon))-np.log(NeoHookean3D(1,1,khook,MatNon)) )/(np.log(lam)))\n",
"_____no_output_____"
],
[
"matplotlib.rcParams.update({'font.size': 20})\nlam=np.arange(0.5,2 , 0.01);\nMatNon=0\nkhook=1\nEnergy=NeoHookean3D(lam,1,khook,MatNon)\nplt.plot((lam),(Energy),linewidth=4.0)\n\nMatNon=1\nkhook=1\nEnergy=NeoHookean3D(lam,1,khook,MatNon)\nplt.plot((lam),(Energy),linewidth=4.0)\n\nplt.xlabel('$\\lambda$')\nplt.ylabel('F')\nplt.legend(['NH: $\\\\alpha/\\mu=0$','MR: $\\\\alpha/\\\\mu=1$'],prop={'size':20})\nplt.savefig(\"Energies.png\", bbox_inches='tight',dpi=400)",
"_____no_output_____"
]
],
[
[
"# Checking that the Numba versions of functions match the regular ones",
"_____no_output_____"
],
[
"Lets generate a problem similar to our own:",
"_____no_output_____"
]
],
[
[
"# Target mesh size:\ntarget_a = 0.2\n# continuum bending modulus:\nkc=0.5\n# continuum shear modulus:\nmu=1\n# Energetic penalty for volume change\nB=100000\n# The Material Nonlinearity parameter, between 0 and 1\nMatNon=0.99\n# the spring prestress values \ng0=1\n\n# The microscopic values\nkbend=kc/target_a\nkhook = mu\ntheta0=0.2",
"_____no_output_____"
],
[
"with pygmsh.occ.Geometry() as geom:\n geom.characteristic_length_max = target_a\n ellipsoid = geom.add_ball([0.0, 0.0, 0.0], 1)\n #ellipsoid = geom.add_ellipsoid([0.0, 0.0, 0.0], [0.95, 0.95, 1.05])\n InputMesh = geom.generate_mesh()",
"_____no_output_____"
],
[
"interiorbonds,edgebonds,boundarytris, bidxTotidx, tetras= MakeMeshData3D(InputMesh)\nbonds=np.concatenate((interiorbonds,edgebonds))\norientedboundarytris=OrientTriangles(InputMesh.points,boundarytris,np.array([0,0,0]))\nboundarytris=orientedboundarytris",
"_____no_output_____"
],
[
"# make the preferred rest lengths of the interior springs\ninteriorpairs=InputMesh.points[interiorbonds]\ninteriorvecs = np.subtract(interiorpairs[:,0,:],interiorpairs[:,1,:])\nInteriorBondRestLengths=np.linalg.norm(interiorvecs,axis=1)\n\n# make the preferred rest lengths of the edge springs. Initially have the at g0=1, but then\n#update them in the loop\nedgepairs=InputMesh.points[edgebonds]\nedgevecs = np.subtract(edgepairs[:,0,:],edgepairs[:,1,:])\nInitialEdgeBondRestLengths=np.linalg.norm(edgevecs,axis=1)\n\n# The volume constraint is simply that the target volume should be the initial volume\nTargetVolumes=Volume3D_tetras(InputMesh.points,tetras)\n\nP =InputMesh.points\n\n # the important bit! Giving it the prestress\nEdgeBondRestLengths= g0*InitialEdgeBondRestLengths\nr0_ij=np.concatenate((InteriorBondRestLengths,EdgeBondRestLengths)) \n ",
"_____no_output_____"
]
],
[
[
"To test numerical equality, we can use numpy's testing module:\nhttps://numpy.org/doc/stable/reference/generated/numpy.testing.assert_allclose.html#numpy.testing.assert_allclose\n",
"_____no_output_____"
]
],
[
[
"x=Volume3D_tetras(P,tetras)\nNumbax=NumbaVolume3D_tetras_2(P,tetras)\nnp.testing.assert_allclose(x,Numbax)",
"_____no_output_____"
],
[
"x=BendingEnergy(P,orientedboundarytris,bidxTotidx,kbend)\nNumbax=NumbaBendingEnergy_2(P,orientedboundarytris,bidxTotidx,kbend)\nnp.testing.assert_allclose(x,Numbax)",
"_____no_output_____"
],
[
"x=energy3D(P,bonds,orientedboundarytris,bidxTotidx,tetras,r0_ij,khook,kbend,theta0,B,MatNon,TargetVolumes)\nNumbax=Numbaenergy3D(P,bonds,orientedboundarytris,bidxTotidx,tetras,r0_ij,khook,kbend,theta0,B,MatNon,TargetVolumes)\nnp.testing.assert_allclose(x,Numbax)",
"_____no_output_____"
]
],
[
[
"## Checking Timings",
"_____no_output_____"
],
[
"## volume",
"_____no_output_____"
]
],
[
[
"start = timer()\nfor i in range(0,5000):\n x=Volume3D_tetras(P,tetras)\nend = timer()\nprint(end-start)",
"_____no_output_____"
],
[
"# directly sum the triple product over all tetrahedra\n@jit(nopython=True)\ndef NumbaVolume3D_tetras_2(P,tetras):\n \n Tot=np.zeros(len(tetras))\n for i in range(len(tetras)):\n \n P0= P[tetras[i,0]]\n P1= P[tetras[i,1]] \n P2= P[tetras[i,2]] \n P3= P[tetras[i,3]] \n \n t0x=P1[0]-P0[0]\n t0y=P1[1]-P0[1]\n t0z=P1[2]-P0[2]\n \n t1x=P2[0]-P0[0]\n t1y=P2[1]-P0[1]\n t1z=P2[2]-P0[2]\n \n t2x=P3[0]-P0[0]\n t2y=P3[1]-P0[1]\n t2z=P3[2]-P0[2]\n \n \n t0ct1x = t0y*t1z- t0z*t1y\n t0ct1y = t0z*t1x- t0x*t1z\n t0ct1z = t0x*t1y- t0y*t1x\n \n t2dott0ct1=t2x*t0ct1x+t2y*t0ct1y+t2z*t0ct1z\n \n Tot[i]=np.abs(t2dott0ct1/6)\n \n return Tot",
"_____no_output_____"
],
[
"x=NumbaVolume3D_tetras_2(P,tetras)\nstart = timer()\nfor i in range(0,5000):\n x=NumbaVolume3D_tetras_2(P,tetras)\nend = timer()\nprint(end-start)",
"_____no_output_____"
]
],
[
[
"## Bending",
"_____no_output_____"
]
],
[
[
"start = timer()\nfor i in range(0,5000):\n x=BendingEnergy(P,orientedboundarytris,bidxTotidx,kbend)\nend = timer()\nprint(end-start)",
"_____no_output_____"
],
[
"x=NumbaBendingEnergy_2(P,orientedboundarytris,bidxTotidx,kbend)\nstart = timer()\nfor i in range(0,5000):\n x=NumbaBendingEnergy_2(P,orientedboundarytris,bidxTotidx,kbend)\nend = timer()\nprint(end-start)",
"_____no_output_____"
]
],
[
[
"## Spring",
"_____no_output_____"
]
],
[
[
"start = timer()\nfor i in range(0,5000):\n x=NeoHookean3D(r0_ij,r0_ij,khook,MatNon).sum() \nend = timer()\nprint(end-start)",
"_____no_output_____"
],
[
"x=NumbaNeoHookean3D(r0_ij,r0_ij,khook,MatNon).sum() \nstart = timer()\nfor i in range(0,5000):\n x=NumbaNeoHookean3D(r0_ij,r0_ij,khook,MatNon).sum() \nend = timer()\nprint(end-start)",
"_____no_output_____"
]
],
[
[
"## Making Spring rests",
"_____no_output_____"
]
],
[
[
"start = timer()\nfor i in range(0,5000):\n # We convert it to a matrix here.\n P_ij = P.reshape((-1, 3))\n # from the bond list, work out what the current bond lengths are:\n AB=P_ij[bonds]\n t1 = np.subtract(AB[:,0,:],AB[:,1,:])\n r_ij=np.linalg.norm(t1,axis=1)\nend = timer()\nprint(end-start)\n ",
"_____no_output_____"
],
[
"# We convert it to a matrix here.\nP_ij = P.reshape((-1, 3))\nr_ij=NumbaMakeBondLengths(P_ij,bonds)\nstart = timer()\nfor i in range(0,5000):\n # We convert it to a matrix here.\n P_ij = P.reshape((-1, 3))\n r_ij=NumbaMakeBondLengths(P_ij,bonds)\nend = timer()\nprint(end-start)",
"_____no_output_____"
],
[
"start = timer()\nfor i in range(0,5000):\n # We convert it to a matrix here.\n P_ij = P.reshape((-1, 3))\n # from the bond list, work out what the current bond lengths are:\n AB=P_ij[bonds]\n t1 = np.subtract(AB[:,0,:],AB[:,1,:])\n r_ij=np.linalg.norm(t1,axis=1)\nend = timer()\nprint(end-start)",
"_____no_output_____"
]
],
[
[
"## Totals",
"_____no_output_____"
]
],
[
[
"start = timer()\nfor i in range(0,5000):\n x=energy3D(P,bonds,orientedboundarytris,bidxTotidx,tetras,r0_ij,khook,kbend,theta0,B,MatNon,TargetVolumes)\nend = timer()\nprint(end-start)",
"_____no_output_____"
],
[
"Numbax=Numbaenergy3D(P,bonds,orientedboundarytris,bidxTotidx,tetras,r0_ij,khook,kbend,theta0,B,MatNon,TargetVolumes)\nstart = timer()\nfor i in range(0,5000):\n Numbax=Numbaenergy3D(P,bonds,orientedboundarytris,bidxTotidx,tetras,r0_ij,khook,kbend,theta0,B,MatNon,TargetVolumes)\nend = timer()\nprint(end-start)",
"_____no_output_____"
],
[
"## Energy Minimization",
"_____no_output_____"
],
[
"start = timer()\nopt.minimize(Numbaenergy3D, P.ravel()\n ,options={'gtol':1e-01,'disp': True} \n ,args=(bonds\n ,orientedboundarytris\n ,bidxTotidx\n ,tetras\n ,r0_ij\n ,khook\n ,kbend\n ,theta0\n ,B\n ,MatNon\n ,TargetVolumes)\n ).x.reshape((-1, 3))\nend = timer()\nprint(end-start)",
"_____no_output_____"
],
[
"start = timer()\nopt.minimize(energy3D, P.ravel()\n ,options={'gtol':1e-01,'disp': True} \n ,args=(bonds\n ,orientedboundarytris\n ,bidxTotidx\n ,tetras\n ,r0_ij\n ,khook\n ,kbend\n ,theta0\n ,B\n ,MatNon\n ,TargetVolumes)\n ).x.reshape((-1, 3))\nend = timer()\nprint(end-start)",
"_____no_output_____"
],
[
"x=NumbaNeoHookean3D(r0_ij,r0_ij,khook,MatNon).sum() \nstart = timer()\nfor i in range(0,5000):\n x=NumbaNeoHookean3D(r0_ij,r0_ij,khook,MatNon).sum() \nend = timer()\nprint(end-start)",
"_____no_output_____"
]
],
[
[
"# Testing the ellipse distance function",
"_____no_output_____"
]
],
[
[
"import scipy.optimize as opt\nimport numpy as np\nimport matplotlib\nimport matplotlib.pyplot as plt\nfrom numba import jit",
"_____no_output_____"
],
[
"def newton(f,Df,x0,epsilon,max_iter):\n '''Approximate solution of f(x)=0 by Newton's method.\n\n Parameters\n ----------\n f : function\n Function for which we are searching for a solution f(x)=0.\n Df : function\n Derivative of f(x).\n x0 : number\n Initial guess for a solution f(x)=0.\n epsilon : number\n Stopping criteria is abs(f(x)) < epsilon.\n max_iter : integer\n Maximum number of iterations of Newton's method.\n\n Returns\n -------\n xn : number\n Implement Newton's method: compute the linear approximation\n of f(x) at xn and find x intercept by the formula\n x = xn - f(xn)/Df(xn)\n Continue until abs(f(xn)) < epsilon and return xn.\n If Df(xn) == 0, return None. If the number of iterations\n exceeds max_iter, then return None.\n\n Examples\n --------\n >>> f = lambda x: x**2 - x - 1\n >>> Df = lambda x: 2*x - 1\n >>> newton(f,Df,1,1e-8,10)\n Found solution after 5 iterations.\n 1.618033988749989\n '''\n xn = x0\n for n in range(0,max_iter):\n fxn = f(xn)\n if abs(fxn) < epsilon:\n print('Found solution after',n,'iterations.')\n return xn\n Dfxn = Df(xn)\n if Dfxn == 0:\n print('Zero derivative. No solution found.')\n return None\n xn = xn - fxn/Dfxn\n print('Exceeded maximum iterations. No solution found.')\n return None",
"_____no_output_____"
],
[
"alpha=1\nbeta=1\nR=1\nr0=np.array([0,0.5])\nf=lambda theta: (alpha**2-beta**2)*R*np.sin(theta)*np.cos(theta)- alpha*r0[0]*np.sin(theta)+beta*r0[1]*np.cos(theta)\nDf = lambda theta: (alpha**2-beta**2)*R*(np.cos(theta)**2-np.sin(theta)**2)- alpha*r0[0]*np.cos(theta)-beta*r0[1]*np.sin(theta)\nx0=3\nepsilon=0.01\nmax_iter=5\nnewton(f,Df,x0,epsilon,max_iter)\n",
"_____no_output_____"
],
[
"@jit(nopython=True)\ndef f(theta,r0,R,alpha,beta):\n return (alpha**2-beta**2)*R*np.sin(theta)*np.cos(theta)- alpha*r0[0]*np.sin(theta)+beta*r0[1]*np.cos(theta)\n@jit(nopython=True)\ndef Df(theta,r0,R,alpha,beta):\n return (alpha**2-beta**2)*R*(np.cos(theta)**2-np.sin(theta)**2)- alpha*r0[0]*np.cos(theta)-beta*r0[1]*np.sin(theta)\n\n@jit(nopython=True)\ndef DistanceToEllipse(r0,R,alpha,beta):\n \n # Initial guess\n theta0=np.arctan2((alpha*r0[1]),(beta*r0[0]))\n\n # run newtons method\n max_iter=5\n theta = theta0\n for n in range(0,max_iter):\n fxn = f(theta,r0,R,alpha,beta)\n Dfxn = Df(theta,r0,R,alpha,beta)\n theta = theta - fxn/Dfxn\n \n thetafinal=theta \n \n xellipse=R*alpha*np.cos(thetafinal)\n yellipse=R*beta*np.sin(thetafinal)\n \n deltax= r0[0]-xellipse\n deltay= r0[1]-yellipse\n \n return (thetafinal,xellipse,yellipse,np.sqrt(deltax**2+deltay**2))",
"_____no_output_____"
],
[
"start = timer()\nfor i in range(0,5000):\n x=DistanceToEllipse(r0,R,alpha,beta)\nend = timer()\nprint(end-start)",
"_____no_output_____"
]
],
[
[
"try out some different r0 values below. It seems to work okay!",
"_____no_output_____"
]
],
[
[
"R=1\nalpha=1.3\nbeta=1.5\ntheta = np.linspace(0.0, 2.0 * np.pi, 100)\nx = R*alpha*np.cos(theta)\ny = R*beta*np.sin(theta)\nplt.plot(x,y)\nplt.plot(0,0,'go')\n\nr0=np.array([0.6,0.4])\nplt.plot(r0[0],r0[1],'ro')\n\n(thetafinal, Ellipsex,Ellipsey, distance)=DistanceToEllipse(r0,R,alpha,beta)\n\nplt.plot(Ellipsex,Ellipsey,'bo')\n\n# draw a ray normal to the ellipse point:\nvx=-beta*np.cos(thetafinal)\nvy=-alpha*np.sin(thetafinal)\n\nplt.plot([Ellipsex,Ellipsex+vx],[Ellipsey,Ellipsey+vy])\n\nplt.axes().set_aspect('equal')\n\nprint(distance)\n",
"_____no_output_____"
]
],
[
[
"# Testing the ellipsoid fitting functions",
"_____no_output_____"
]
],
[
[
"DataFolder",
"_____no_output_____"
],
[
"# Make the Mesh\nwith pygmsh.occ.Geometry() as geom:\n geom.characteristic_length_max = 0.1\n #ellipsoid = geom.add_ball([0.0, 0.0, 0.0], 1)\n ellipsoid = geom.add_ellipsoid([0.0, 0.0, 0.0], [0.95, 0.95, 1.0556])\n InputMesh = geom.generate_mesh()\n \nInputMesh.write(DataFolder+\"/\"+\"InitialMesh.vtk\",binary=True) \n\ninteriorbonds,edgebonds,boundarytris, bidxTotidx, tetras= MakeMeshData3D(InputMesh)\nbonds=np.concatenate((interiorbonds,edgebonds))\norientedboundarytris=OrientTriangles(InputMesh.points,boundarytris,np.array([0,0,0]))\n\n# Get the points on the boundary:\nBoundaryPoints= np.unique(edgebonds.ravel())",
"_____no_output_____"
],
[
"P=InputMesh.points[BoundaryPoints]\nxx=P[:,0]\nyy=P[:,1]\nzz=P[:,2]\n\ncenter,axes,ec,inve,vec = ls_ellipsoid(xx,yy,zz)",
"_____no_output_____"
],
[
"print(center)",
"_____no_output_____"
],
[
"print(axes)\n",
"_____no_output_____"
],
[
"print(ec)",
"_____no_output_____"
],
[
"print(inve)",
"_____no_output_____"
],
[
"print(vec)",
"_____no_output_____"
]
],
[
[
"This seems to work pretty well!",
"_____no_output_____"
],
[
"# Testing the Analytic Prediction Functions:",
"_____no_output_____"
],
[
"To test these functions are working fine, I will compare a few values to the mathematica code, with some randomly chosen parameter values:\n\n",
"_____no_output_____"
]
],
[
[
"FTot",
"_____no_output_____"
]
],
[
[
"Expectation:\n\nalpha0=0.5\nkappa0=0.3\ngamma0=0.2\n\nFTot(1,alpha0,gamma0=0.2,kappa0)= 16.3363\nFTot(1.2,alpha0,gamma0=0.2,kappa0)= 16.8611\nFTot(0.2,alpha0,gamma0=0.2,kappa0)= 205.077\n\nalpha0=0.5\nkappa0=0.3\ngamma0=-0.2\n\nFTot(1,alpha0,gamma0=0.2,kappa0)= 11.3097\nFTot(1.2,alpha0,gamma0=0.2,kappa0)= 11.7705\nFTot(0.2,alpha0,gamma0=0.2,kappa0)= 192.198\n",
"_____no_output_____"
]
],
[
[
"lam=np.array([1.0001,1.2,0.2])\nalpha0=0.5\nkappa0=0.3\ngamma0=0.2\nFTot(lam,alpha0,gamma0,kappa0)",
"_____no_output_____"
],
[
"alpha0=0.5\nkappa0=0.3\ngamma0=-0.2\nFTot(lam,alpha0,gamma0,kappa0)",
"_____no_output_____"
]
],
[
[
"Conclusion: This function seems to be working okay",
"_____no_output_____"
]
],
[
[
"FindGlobalMinimum",
"_____no_output_____"
]
],
[
[
"Expectation:\n\nalpha0=0.6\nkappa0=0.4\ngamma0=-1\nFindGlobalMinimum(alpha0,gamma0,kappa0,0.1,5)-> lam =1 , F =3.76991\n\n\nalpha0=0.6\nkappa0=0.4\ngamma0=-4.0\nFindGlobalMinimum(alpha0,gamma0,kappa0,0.1,5)-> lam =1.55262 , F =-34.1081\n\nalpha0=0.1\nkappa0=0.4\ngamma0=-4.0\nFindGlobalMinimum(alpha0,gamma0,kappa0,0.1,5)-> lam =0.691294 , F =-34.0731\n\n",
"_____no_output_____"
]
],
[
[
"alpha0=0.6\nkappa0=0.4\ngamma0=-1\nFindGlobalMinimum(alpha0,gamma0,kappa0,0.1,5)",
"/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in power\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in arcsin\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:177: RuntimeWarning: invalid value encountered in arctanh\n Fbend=(2/3)*np.pi*kappa*(7+(2/lam**3)+3*lam**3*np.arctanh(np.lib.scimath.sqrt(1-lam**3))/np.lib.scimath.sqrt(1-lam**3))\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in power\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in arcsin\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:177: RuntimeWarning: invalid value encountered in arctanh\n Fbend=(2/3)*np.pi*kappa*(7+(2/lam**3)+3*lam**3*np.arctanh(np.lib.scimath.sqrt(1-lam**3))/np.lib.scimath.sqrt(1-lam**3))\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in power\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in arcsin\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:177: RuntimeWarning: invalid value encountered in arctanh\n Fbend=(2/3)*np.pi*kappa*(7+(2/lam**3)+3*lam**3*np.arctanh(np.lib.scimath.sqrt(1-lam**3))/np.lib.scimath.sqrt(1-lam**3))\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in power\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in arcsin\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:177: RuntimeWarning: invalid value encountered in arctanh\n Fbend=(2/3)*np.pi*kappa*(7+(2/lam**3)+3*lam**3*np.arctanh(np.lib.scimath.sqrt(1-lam**3))/np.lib.scimath.sqrt(1-lam**3))\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in power\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in arcsin\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:177: RuntimeWarning: invalid value encountered in arctanh\n Fbend=(2/3)*np.pi*kappa*(7+(2/lam**3)+3*lam**3*np.arctanh(np.lib.scimath.sqrt(1-lam**3))/np.lib.scimath.sqrt(1-lam**3))\n"
],
[
"alpha0=0.6\nkappa0=0.4\ngamma0=-4.0\nFindGlobalMinimum(alpha0,gamma0,kappa0,0.1,5)",
"_____no_output_____"
],
[
"alpha0=0.1\nkappa0=0.4\ngamma0=-4.0\nFindGlobalMinimum(alpha0,gamma0,kappa0,0.1,5)",
"/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in power\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:175: RuntimeWarning: invalid value encountered in arcsin\n Area = (2*np.pi/lam)*( 1+(lam**(3/2)/e)*np.arcsin(e) )\n/home/jackbinysh/Code/ActiveElastocapillarity/Python/EnergyMinimization/AnalysisFunctions.py:177: RuntimeWarning: invalid value encountered in arctanh\n Fbend=(2/3)*np.pi*kappa*(7+(2/lam**3)+3*lam**3*np.arctanh(np.lib.scimath.sqrt(1-lam**3))/np.lib.scimath.sqrt(1-lam**3))\n"
]
],
[
[
"Conclusion: The minimizer also appears to work okay",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"raw",
"code",
"markdown",
"code",
"raw",
"code",
"markdown"
] | [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"raw"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"raw"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecb44f6b67acb65276a9a90e693e1b02816c0447 | 68,209 | ipynb | Jupyter Notebook | pandas/pandas1.ipynb | mengwangk/Machine-learning-tutorials | 75c172f5b136cd17fd31479d26e626665fe49f18 | [
"MIT"
] | 1 | 2021-08-07T12:58:46.000Z | 2021-08-07T12:58:46.000Z | pandas/pandas1.ipynb | mengwangk/Machine-learning-tutorials | 75c172f5b136cd17fd31479d26e626665fe49f18 | [
"MIT"
] | null | null | null | pandas/pandas1.ipynb | mengwangk/Machine-learning-tutorials | 75c172f5b136cd17fd31479d26e626665fe49f18 | [
"MIT"
] | 1 | 2021-08-07T12:58:46.000Z | 2021-08-07T12:58:46.000Z | 68,209 | 68,209 | 0.784852 | [
[
[
"import pandas as pd\ndf = pd.read_csv('../input/renewable/renewable_power_plants_PL.csv')\ndf.head(5)",
"_____no_output_____"
],
[
"df.tail(5)",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1041 entries, 0 to 1040\nData columns (total 9 columns):\ndistrict 1041 non-null object\nenergy_source_level_1 1041 non-null object\nenergy_source_level_2 1041 non-null object\nenergy_source_level_3 323 non-null object\ntechnology 718 non-null object\nelectrical_capacity 1041 non-null float64\nnumber_of_installations 1041 non-null int64\ndata_source 1041 non-null object\nas_of_year 1041 non-null int64\ndtypes: float64(1), int64(2), object(6)\nmemory usage: 73.3+ KB\n"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df.rename(columns={\n 'energy_source_level_1': 'energy_1', \n 'energy_source_level_2': 'energy_2',\n 'energy_source_level_3': 'energy_3'\n }, inplace=True)\n\ndf.columns",
"_____no_output_____"
],
[
"df.isnull().sum()",
"_____no_output_____"
],
[
"df.dropna()\ndf.dropna(axis=1)\ndf.head(5)",
"_____no_output_____"
],
[
"capacity = df['electrical_capacity']\ncapacity.head()",
"_____no_output_____"
],
[
"capacity.mean()",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
],
[
"df['energy_3'].describe()",
"_____no_output_____"
],
[
"df.corr()",
"_____no_output_____"
],
[
"df[df['electrical_capacity'] >= 50].head(5)",
"_____no_output_____"
],
[
"df[(df['energy_2'] == 'solar') | (df['energy_3'] == 'Sewage and landfill gas')].head()",
"_____no_output_____"
],
[
"df[df['technology'].isin(['Hydro'])].head()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nplt.rcParams.update({'font.size': 12, 'figure.figsize': (5, 5)})",
"_____no_output_____"
],
[
"df.plot(kind='scatter', x='number_of_installations', y='electrical_capacity', title='Number of installations vs Electrical capacity');",
"_____no_output_____"
],
[
"df['electrical_capacity'].plot(kind='hist', title='Electrical Capacity');",
"_____no_output_____"
],
[
"df['electrical_capacity'].plot(kind=\"box\");",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb45632a4bce2d68fb17b416dd4306ab50a95e1 | 375,109 | ipynb | Jupyter Notebook | sequence-labeling/sequence-labeling-training.ipynb | luist18/feup-pln | c08a810549a5f994d6779ac610ee24cd9e766a7b | [
"MIT"
] | null | null | null | sequence-labeling/sequence-labeling-training.ipynb | luist18/feup-pln | c08a810549a5f994d6779ac610ee24cd9e766a7b | [
"MIT"
] | null | null | null | sequence-labeling/sequence-labeling-training.ipynb | luist18/feup-pln | c08a810549a5f994d6779ac610ee24cd9e766a7b | [
"MIT"
] | null | null | null | 49.265695 | 34,240 | 0.433933 | [
[
[
"Notebook prepared by Henrique Lopes Cardoso ([email protected]), based on [Named Entity Recognition and Classification with Scikit-Learn](https://www.kdnuggets.com/2018/10/named-entity-recognition-classification-scikit-learn.html) by Susan Li.\n\n# SEQUENCE LABELING TRAINING",
"_____no_output_____"
],
[
"## Training a NER model\n\nTo train a model on NER, we need to rely on an annotated dataset. For the purpose of this notebook, we'll use the [Annotated Corpus for Named Entity Recognition](https://www.kaggle.com/abhinavwalia95/entity-annotated-corpus), which has been annotated with POS and named entities, using BIO encoding.\n\nThis is what the dataset (a CSV file) looks like:\n\n",
"_____no_output_____"
],
[
"Let's load the data and have a look on it.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\n# loading NER data\ndf = pd.read_csv('ner_dataset.csv', encoding = \"ISO-8859-1\")\n\ndf.head()",
"_____no_output_____"
]
],
[
[
"There are several rows for which the first column (*Sentence #*) is *NaN*. This is because only the first token in each sentence is signaled with the sentence number.\n\nHow many *NaN*s are there?",
"_____no_output_____"
]
],
[
[
"# your code here\ndf['Sentence #'].isna().sum()",
"_____no_output_____"
]
],
[
[
"We could choose to get rid of that first column. However, for reasons that will later become evident, we'll fill down the sentence number to all tokens in each sentence.",
"_____no_output_____"
]
],
[
[
"df = df.fillna(method='ffill')\ndf[:25]",
"_____no_output_____"
]
],
[
[
"Let's see now how many sentences we've got:",
"_____no_output_____"
]
],
[
[
"df['Sentence #'].nunique()",
"_____no_output_____"
]
],
[
[
"How many words, POS and BIO tags are there?",
"_____no_output_____"
]
],
[
[
"# your code here\nprint(df['POS'].unique())\nprint(df['Tag'].unique())",
"['NNS' 'IN' 'VBP' 'VBN' 'NNP' 'TO' 'VB' 'DT' 'NN' 'CC' 'JJ' '.' 'VBD' 'WP'\n '``' 'CD' 'PRP' 'VBZ' 'POS' 'VBG' 'RB' ',' 'WRB' 'PRP$' 'MD' 'WDT' 'JJR'\n ':' 'JJS' 'WP$' 'RP' 'PDT' 'NNPS' 'EX' 'RBS' 'LRB' 'RRB' '$' 'RBR' ';'\n 'UH' 'FW']\n['O' 'B-geo' 'B-gpe' 'B-per' 'I-geo' 'B-org' 'I-org' 'B-tim' 'B-art'\n 'I-art' 'I-per' 'I-gpe' 'I-tim' 'B-nat' 'B-eve' 'I-eve' 'I-nat']\n"
]
],
[
[
"Let's check the distribution of POS tags:",
"_____no_output_____"
]
],
[
[
"df.groupby('POS').size().reset_index(name='counts')",
"_____no_output_____"
]
],
[
[
"What is the distribution of named entity BIO tags?",
"_____no_output_____"
]
],
[
[
"# your code here\n# distribution of tags with seaborn\nplt.xticks(rotation=90)\nsns.countplot(x='Tag', data=df)\nplt.show()\n\n# tag without \"O\"\nplt.xticks(rotation=90)\nsns.countplot(x='Tag', data=df[df['Tag'] != 'O'])\nplt.show()",
"_____no_output_____"
]
],
[
[
"As expected, we have a very unbalanced dataset in terms of NER tags. Which is the most prevalent named entity type? What types tend to be composed of a single word?",
"_____no_output_____"
],
[
"### Can we do it with \"traditional\" classifiers?",
"_____no_output_____"
],
[
"First we generate our dataset for training the NER model. Adding the POS tag to the word seems to be a good idea -- hopefully the word's POS is helpful to determine whether the word corresponds to a named entity. We can get rid of the sentence number, though, as it does not seem to add anything useful for the task.",
"_____no_output_____"
]
],
[
[
"X = df[['Word', 'POS']]",
"_____no_output_____"
]
],
[
[
"Let's also collect the BIO labels for the words:",
"_____no_output_____"
]
],
[
[
"y = df['Tag'].values",
"_____no_output_____"
]
],
[
[
"Let's check the shape of our feature matrix:",
"_____no_output_____"
]
],
[
[
"print(X.shape)\nprint(y.shape)",
"(1048575, 2)\n(1048575,)\n"
]
],
[
[
"We need to transform each data entry into a 1-hot vector, for which we can use [DictVectorizer](https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.DictVectorizer.html). This will create a table with as many columns as the number of unique words and POS tags we have in our dataset -- the features we'll use to represent each token of the text.",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction import DictVectorizer\n\nv = DictVectorizer(sparse=True) # sparse=False will use much more memory, and take much longer to train...\nv.fit(X.to_dict('records'))",
"_____no_output_____"
]
],
[
[
"Now we split the dataset into training and test sets (we should be more careful by obtaining a test set that starts at the beginning of a sentence, but let's disregard this for now):",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.10, random_state=0, stratify=y)",
"_____no_output_____"
]
],
[
[
"If the full data set is too big to fit into memory, we'll need to use mini-batches of data to train the model with an [out-of-core learning](https://scikit-learn.org/0.15/modules/scaling_strategies.html) algorithm.\n\nTry training a traditional classifier, such as MultinomialNB. How well does it perform?",
"_____no_output_____"
]
],
[
[
"# your code here\n",
"_____no_output_____"
]
],
[
[
"The model seems to have a good accuracy, but that's too misleading -- the dataset is very unbalanced, with many *O* (outside) labels. If we look at macro average f1-score, we observe unsatisfactory results.\n\nTry out other classifiers, namely those that support [out-of-core](https://scikit-learn.org/0.15/modules/scaling_strategies.html) learning, such as:\n- Perceptron\n- SGDClassifier\n- PassiveAggressiveClassifier",
"_____no_output_____"
]
],
[
[
"# your code here\n",
"_____no_output_____"
]
],
[
[
"Turning to classifiers tailored for sequence labeling tasks!",
"_____no_output_____"
],
[
"### Conditional Random Fields\n\nConditional Random Fields is one of the most well known algorithms for dealing with sequential data, and is very useful for sequence labeling tasks.\nWe will make use of [sklearn-crfsuite](https://sklearn-crfsuite.readthedocs.io/en/latest/), which implements a CRF classifier.",
"_____no_output_____"
],
[
"#### Splitting into sentences\n\nLet's first reconstruct the sentences with their POS and NER tags:",
"_____no_output_____"
]
],
[
[
"class SentenceGetter(object):\n \n def __init__(self, data):\n self.n_sent = 1\n self.data = data\n self.empty = False\n agg_func = lambda s: [(w, p, t) for w, p, t in zip(s['Word'].values.tolist(), \n s['POS'].values.tolist(), \n s['Tag'].values.tolist())]\n self.grouped = self.data.groupby('Sentence #').apply(agg_func)\n self.sentences = [s for s in self.grouped]\n \n def get_next(self):\n try: \n s = self.grouped['Sentence: {}'.format(self.n_sent)]\n self.n_sent += 1\n return s \n except:\n return None\n\nsentences = SentenceGetter(df).sentences",
"_____no_output_____"
]
],
[
[
"Let's see what we've got, by printing the sequences of words, POS and NER labels for a few sentences:",
"_____no_output_____"
]
],
[
[
"def pprint_sentence(sent):\n for token in sent:\n print(token[0], end =\" \")\n print()\n for token in sent:\n print(token[1], end =\" \")\n print()\n for token in sent:\n print(token[2], end =\" \")\n print('\\n')\n return\n\npprint_sentence(sentences[0])\npprint_sentence(sentences[1])\npprint_sentence(sentences[2])",
"Thousands of demonstrators have marched through London to protest the war in Iraq and demand the withdrawal of British troops from that country . \nNNS IN NNS VBP VBN IN NNP TO VB DT NN IN NNP CC VB DT NN IN JJ NNS IN DT NN . \nO O O O O O B-geo O O O O O B-geo O O O O O B-gpe O O O O O \n\nIranian officials say they expect to get access to sealed sensitive parts of the plant Wednesday , after an IAEA surveillance system begins functioning . \nJJ NNS VBP PRP VBP TO VB NN TO JJ JJ NNS IN DT NN NNP , IN DT NNP NN NN VBZ VBG . \nB-gpe O O O O O O O O O O O O O O B-tim O O O B-org O O O O O \n\nHelicopter gunships Saturday pounded militant hideouts in the Orakzai tribal region , where many Taliban militants are believed to have fled to avoid an earlier military offensive in nearby South Waziristan . \nNN NNS NNP VBD JJ NNS IN DT NNP JJ NN , WRB JJ NNP NNS VBP VBN TO VB VBN TO VB DT JJR JJ NN IN JJ NNP NNP . \nO O B-tim O O O O O B-geo O O O O O B-org O O O O O O O O O O O O O O B-geo I-geo O \n\n"
]
],
[
[
"#### Feature extraction\n\nNext, we extract more features (word parts, simplified POS tags, lower/title/upper flags, features of nearby words) and convert them to sklearn-crfsuite format -- each sentence should be converted to a list of dicts. (The following code was taken from the [sklearn-crfsuite tutorial](https://sklearn-crfsuite.readthedocs.io/en/latest/tutorial.html).)",
"_____no_output_____"
]
],
[
[
"def word2features(sent, i):\n word = sent[i][0]\n postag = sent[i][1]\n \n features = {\n 'bias': 1.0, \n 'word.lower()': word.lower(), \n 'word[-3:]': word[-3:],\n 'word[-2:]': word[-2:],\n 'word.isupper()': word.isupper(),\n 'word.istitle()': word.istitle(),\n 'word.isdigit()': word.isdigit(),\n 'postag': postag,\n 'postag[:2]': postag[:2],\n }\n if i > 0:\n word1 = sent[i-1][0]\n postag1 = sent[i-1][1]\n features.update({\n '-1:word.lower()': word1.lower(),\n '-1:word.istitle()': word1.istitle(),\n '-1:word.isupper()': word1.isupper(),\n '-1:postag': postag1,\n '-1:postag[:2]': postag1[:2],\n })\n else:\n features['BOS'] = True\n if i < len(sent)-1:\n word1 = sent[i+1][0]\n postag1 = sent[i+1][1]\n features.update({\n '+1:word.lower()': word1.lower(),\n '+1:word.istitle()': word1.istitle(),\n '+1:word.isupper()': word1.isupper(),\n '+1:postag': postag1,\n '+1:postag[:2]': postag1[:2],\n })\n else:\n features['EOS'] = True\n\n return features\n\ndef sent2features(sent):\n return [word2features(sent, i) for i in range(len(sent))]\n\ndef sent2labels(sent):\n return [label for token, postag, label in sent]\n\ndef sent2tokens(sent):\n return [token for token, postag, label in sent]",
"_____no_output_____"
],
[
"X = [sent2features(s) for s in sentences]\ny = [sent2labels(s) for s in sentences]",
"_____no_output_____"
]
],
[
[
"Let's have a look at the features we get for a specific token:",
"_____no_output_____"
]
],
[
[
"pprint_sentence(sentences[0])\nX[0][6]",
"Thousands of demonstrators have marched through London to protest the war in Iraq and demand the withdrawal of British troops from that country . \nNNS IN NNS VBP VBN IN NNP TO VB DT NN IN NNP CC VB DT NN IN JJ NNS IN DT NN . \nO O O O O O B-geo O O O O O B-geo O O O O O B-gpe O O O O O \n\n"
]
],
[
[
"We're good to go: let's split the data into training and test sets so that we can employ the CRF model.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.15, random_state=0)",
"_____no_output_____"
]
],
[
[
"#### Training a CRF model\n\nWe will make use of [sklearn-crfsuite](https://sklearn-crfsuite.readthedocs.io/en/latest/), which implements a CRF classifier.",
"_____no_output_____"
]
],
[
[
"import sklearn_crfsuite\n\ncrf = sklearn_crfsuite.CRF(algorithm='lbfgs', c1=0.1, c2=0.1, max_iterations=100, all_possible_transitions=True)\ncrf.fit(X_train, y_train)",
"C:\\Users\\hlc\\AppData\\Local\\Packages\\PythonSoftwareFoundation.Python.3.8_qbz5n2kfra8p0\\LocalCache\\local-packages\\Python38\\site-packages\\sklearn\\base.py:209: FutureWarning: From version 0.24, get_params will raise an AttributeError if a parameter cannot be retrieved as an instance attribute. Previously it would return None.\n warnings.warn('From version 0.24, get_params will raise an '\n"
],
[
"from sklearn_crfsuite import metrics\n\ny_pred = crf.predict(X_test)\nprint(metrics.flat_classification_report(y_test, y_pred))",
"C:\\Users\\hlc\\AppData\\Local\\Packages\\PythonSoftwareFoundation.Python.3.8_qbz5n2kfra8p0\\LocalCache\\local-packages\\Python38\\site-packages\\sklearn\\utils\\validation.py:67: FutureWarning: Pass labels=None as keyword args. From version 0.25 passing these as positional arguments will result in an error\n warnings.warn(\"Pass {} as keyword args. From version 0.25 \"\n"
]
],
[
[
"What did the CRF classifier learn? Look at the output of the following code and interpret it.",
"_____no_output_____"
]
],
[
[
"from collections import Counter\n\ndef print_transitions(trans_features):\n for (label_from, label_to), weight in trans_features:\n print(\"%-6s -> %-7s %0.6f\" % (label_from, label_to, weight))\n\nprint(\"Top likely transitions:\")\nprint_transitions(Counter(crf.transition_features_).most_common(20))\n\nprint(\"\\nTop unlikely transitions:\")\nprint_transitions(Counter(crf.transition_features_).most_common()[-20:])",
"Top likely transitions:\nI-art -> I-art 7.391358\nB-art -> I-art 7.278202\nB-nat -> I-nat 6.479038\nB-eve -> I-eve 5.839108\nI-eve -> I-eve 5.758097\nI-tim -> I-tim 4.742140\nI-gpe -> I-gpe 4.730158\nB-gpe -> I-gpe 4.557278\nB-tim -> I-tim 4.334957\nI-org -> I-org 4.290097\nB-geo -> I-geo 4.277619\nB-org -> I-org 4.241753\nI-nat -> I-nat 3.921685\nB-per -> I-per 3.738044\nO -> O 3.633986\nI-geo -> I-geo 3.570474\nI-per -> I-per 3.148476\nI-geo -> B-art 1.880411\nO -> B-per 1.824350\nB-org -> B-art 1.528514\n\nTop unlikely transitions:\nI-org -> B-org -4.203153\nI-per -> I-org -4.227562\nB-org -> I-geo -4.255245\nI-org -> I-geo -4.298919\nB-geo -> B-geo -4.499268\nB-per -> I-org -4.704401\nB-geo -> I-per -4.778787\nB-tim -> B-tim -4.786172\nI-org -> I-per -4.928529\nB-geo -> I-org -5.109382\nB-org -> I-per -5.115562\nB-gpe -> I-geo -5.174884\nB-gpe -> I-org -5.675065\nI-per -> B-per -5.787749\nB-gpe -> B-gpe -5.939528\nO -> I-per -6.344294\nO -> I-tim -6.991131\nO -> I-org -7.414431\nO -> I-geo -7.802651\nB-per -> B-per -10.779160\n"
]
],
[
[
"Checking state features:",
"_____no_output_____"
]
],
[
[
"def print_state_features(state_features):\n for (attr, label), weight in state_features:\n print(\"%0.6f %-8s %s\" % (weight, label, attr))\n\nprint(\"Top positive:\")\nprint_state_features(Counter(crf.state_features_).most_common(20))\n\nprint(\"\\nTop negative:\")\nprint_state_features(Counter(crf.state_features_).most_common()[-20:])",
"Top positive:\n7.845206 O word.lower():last\n7.786359 O word.lower():month\n7.580241 B-per word.lower():vice\n7.133286 B-org word.lower():philippine\n7.017612 B-tim word.lower():multi-candidate\n6.509159 B-gpe word.lower():afghan\n6.494892 B-gpe word.lower():nepal\n6.474656 B-tim word.lower():2000\n6.408125 B-gpe word.lower():niger\n6.340409 B-gpe word.lower():german\n6.220968 B-per word.lower():obama\n6.020412 B-tim word.lower():february\n5.957609 B-tim word.lower():january\n5.838123 B-org word.lower():al-qaida\n5.836919 B-org word.lower():mid-march\n5.777816 B-geo word.lower():mid-march\n5.750286 O word.lower():chairman\n5.706728 B-nat word.lower():katrina\n5.662584 O BOS\n5.659179 I-gpe +1:word.lower():mayor\n\nTop negative:\n-3.604030 O postag:NNP\n-3.650103 O word[-3:]:1st\n-3.667834 O word.lower():one-fourth\n-3.669180 O +1:word.lower():ms.\n-3.681832 I-org word.lower():secretary\n-3.710592 O +1:word.lower():next\n-3.737056 O word.lower():westerners\n-3.751114 O word.lower():re-establishment\n-3.773720 O word.lower():three-year\n-3.774647 O +1:word.lower():months\n-3.897074 O +1:word.lower():last\n-3.977821 O +1:word.lower():year\n-3.991292 O +1:word.lower():years\n-4.032212 B-gpe word.lower():european\n-4.378263 O word.lower():morning\n-4.411121 O word.lower():multi-party\n-4.608025 O word.lower():afternoon\n-4.608862 B-geo word[-3:]:The\n-4.992223 O word[-2:]:0s\n-5.020282 O word.lower():summer\n"
]
],
[
[
"Using [ELI5](https://eli5.readthedocs.io/en/latest/index.html) we can visualize the CRF model weights (for state transitions and feature importance):",
"_____no_output_____"
]
],
[
[
"import eli5\n\neli5.show_weights(crf, top=10)",
"C:\\Users\\hlc\\AppData\\Local\\Packages\\PythonSoftwareFoundation.Python.3.8_qbz5n2kfra8p0\\LocalCache\\local-packages\\Python38\\site-packages\\sklearn\\base.py:209: FutureWarning: From version 0.24, get_params will raise an AttributeError if a parameter cannot be retrieved as an instance attribute. Previously it would return None.\n warnings.warn('From version 0.24, get_params will raise an '\n"
],
[
"eli5.show_weights(crf, top=10, targets=['O', 'B-org', 'I-per'])",
"_____no_output_____"
],
[
"eli5.show_weights(crf, top=10, feature_re='^word\\.is', horizontal_layout=False, show=['targets'])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecb45ca2a4014585d6cde8a24abf4dafae58ab4c | 11,384 | ipynb | Jupyter Notebook | tutorials/tutorial05.ipynb | xbresson/CE9010_2019 | 9436d8122d6dc0511907c229979768f30faf82bd | [
"MIT"
] | 10 | 2019-01-16T04:38:07.000Z | 2021-04-30T03:05:43.000Z | tutorials/tutorial05.ipynb | xbresson/CE9010_2019 | 9436d8122d6dc0511907c229979768f30faf82bd | [
"MIT"
] | null | null | null | tutorials/tutorial05.ipynb | xbresson/CE9010_2019 | 9436d8122d6dc0511907c229979768f30faf82bd | [
"MIT"
] | 4 | 2019-02-23T08:34:50.000Z | 2020-12-03T07:07:07.000Z | 25.931663 | 151 | 0.495432 | [
[
[
"## CE9010: Introduction to Data Analysis\n## Semester 2 2018/19\n## Xavier Bresson\n<hr>\n\n## Tutorial 5: Supervised classification - improving capacity learning\n## Objectives\n### $\\bullet$ Code linear and higher-order logistic regression models\n### $\\bullet$ Explore results\n<hr>",
"_____no_output_____"
]
],
[
[
"# Import libraries\n\n# math library\nimport numpy as np\n\n# visualization library\n%matplotlib inline\nfrom IPython.display import set_matplotlib_formats\nset_matplotlib_formats('png2x','pdf')\nimport matplotlib.pyplot as plt\n\n# machine learning library\nfrom sklearn.linear_model import LogisticRegression\n\n# 3d visualization\nfrom mpl_toolkits.mplot3d import axes3d\n\n# computational time\nimport time\n",
"_____no_output_____"
]
],
[
[
"## 1.1 Load dataset #1\n<hr>\nThe data features for each data $i$ are $x_i=(x_{i(1)},x_{i(2)})$. <br>\nThe data label/target, $y_i$, indicates two classes with value 0 or 1.\n\nPlot the data points.<br>\nHint: You may use matplotlib function `scatter(x,y)`.",
"_____no_output_____"
]
],
[
[
"# import data with numpy\ndata = np.loadtxt('data/two_circles.txt', delimiter=',')\n\n# number of training data\nn = data.shape[0] \nprint('Number of training data=',n)\n\n# print\nprint(data[:10,:])\nprint(data.shape)\nprint(data.dtype)\n\n# plot\nx1 = data[:,0] # feature 1\nx2 = data[:,1] # feature 2\nidx_class0 = (data[:,2]==0) # index of class0\nidx_class1 = (data[:,2]==1) # index of class1\n\nplt.figure(1,figsize=(6,6))\nplt.#YOUR CODE HERE\nplt.#YOUR CODE HERE\nplt.title('Training data')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 1.2 Linear logistic regression/classification task.\n<hr>\n\nThe logistic regression/classification predictive function is defined as:\n\n$$\n\\begin{aligned}\np_w(x) &= \\sigma(X w)\n\\end{aligned}\n$$\n\nIn the case of **linear** prediction, we have:\n\n<br>\n$$\nX = \n\\left[ \n\\begin{array}{cccc}\n1 & x_{1(1)} & x_{1(2)} \\\\ \n1 & x_{2(1)} & x_{2(2)} \\\\ \n\\vdots\\\\\n1 & x_{n(1)} & x_{n(2)} \n\\end{array} \n\\right]\n\\quad\n\\textrm{ and }\n\\quad\nw = \n\\left[ \n\\begin{array}{cccc}\nw_0 \\\\ \nw_1 \\\\ \nw_2\n\\end{array} \n\\right]\n\\quad\n\\Rightarrow \n\\quad\np_w(x) = \\sigma(X w) =\n\\left[ \n\\begin{array}{cccc}\n\\sigma(w_0 + w_1 x_{1(1)} + w_2 x_{1(2)}) \\\\ \n\\sigma(w_0 + w_1 x_{2(1)} + w_2 x_{2(2)}) \\\\ \n\\vdots\\\\\n\\sigma(w_0 + w_1 x_{n(1)} + w_2 x_{n(2)})\n\\end{array} \n\\right]\n$$\n\nImplement the linear logistic regression function with gradient descent or scikit-learn. Visualize the boundary decision.<br>\n\nCheck your code correctness: The loss value should be around 0.693. <br>",
"_____no_output_____"
]
],
[
[
"#YOUR CODE HERE",
"_____no_output_____"
],
[
"# compute values p(x) for multiple data points x\nx1_min, x1_max = X[:,1].min(), X[:,1].max() # min and max of grade 1\nx2_min, x2_max = X[:,2].min(), X[:,2].max() # min and max of grade 2\nxx1, xx2 = np.meshgrid(np.linspace(x1_min, x1_max), np.linspace(x2_min, x2_max)) # create meshgrid\nX2 = np.ones([np.prod(xx1.shape),3]) \nX2[:,1] = xx1.reshape(-1)\nX2[:,2] = xx2.reshape(-1)\np = f_pred(X2,w)\np = p.reshape(xx1.shape)\n\n\n# plot\nplt.figure(4,figsize=(6,6))\nplt.scatter(x1[idx_class0], x2[idx_class0], s=60, c='r', marker='+', label='Class0') \nplt.scatter(x1[idx_class1], x2[idx_class1], s=30, c='b', marker='o', label='Class1')\nplt.contour(xx1, xx2, p, [0.5], linewidths=2, colors='k') \nplt.legend()\nplt.title('Decision boundary (linear)')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"## 1.3 Quadratic logistic regression/classification task.\n<hr>\n\nThe logistic regression/classification predictive function is defined as:\n\n$$\n\\begin{aligned}\np_w(x) &= \\sigma(X w)\n\\end{aligned}\n$$\n\nIn the case of **quadratic** prediction, we have:\n\n<br>\n$$\nX = \n\\left[ \n\\begin{array}{cccccc}\n1 & x_{1(1)} & x_{1(2)} & x_{1(1)}^2 & x_{1(2)}^2 & x_{1(1)}x_{1(2)} \\\\ \n1 & x_{2(1)} & x_{2(2)} & x_{2(1)}^2 & x_{2(2)}^2 & x_{2(1)}x_{2(2)}\\\\ \n\\vdots\\\\\n1 & x_{n(1)} & x_{n(2)} & x_{n(1)}^2 & x_{n(2)}^2 & x_{n(1)}x_{n(2)}\n\\end{array} \n\\right]\n\\quad\n\\textrm{ and }\n\\quad\nw = \n\\left[ \n\\begin{array}{cccc}\nw_0 \\\\ \nw_1 \\\\ \nw_2\\\\ \nw_3\\\\ \nw_4\\\\ \nw_5\n\\end{array} \n\\right]\n\\quad\n$$\n\nImplement the quadratic logistic regression function with gradient descent or scikit-learn. Visualize the boundary decision.<br>\n\nCheck your code correctness: The loss value should be around 0.011. <br>",
"_____no_output_____"
]
],
[
[
"#YOUR CODE HERE",
"_____no_output_____"
]
],
[
[
"## 2.1 Load dataset #2\n<hr>\nThe data features for each data $i$ are $x_i=(x_{i(1)},x_{i(2)})$. <br>\nThe data label/target, $y_i$, indicates two classes with value 0 or 1.\n\nPlot the data points.<br>\nHint: You may use matplotlib function `scatter(x,y)`.",
"_____no_output_____"
]
],
[
[
"# import data with numpy\ndata = np.loadtxt('data/two_moons.txt', delimiter=',')\n\n# number of training data\nn = data.shape[0] \nprint('Number of training data=',n)\n\n# print\nprint(data[:10,:])\nprint(data.shape)\nprint(data.dtype)\n\n# plot\nx1 = data[:,0] # feature 1\nx2 = data[:,1] # feature 2\nidx_class0 = (data[:,2]==0) # index of class0\nidx_class1 = (data[:,2]==1) # index of class1\n\nplt.figure(1,figsize=(6,6))\nplt.#YOUR CODE HERE\nplt.#YOUR CODE HERE\nplt.title('Training data')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 2.2 Linear logistic regression/classification task.\n<hr>\n\n\nImplement the linear logistic regression function with gradient descent or scikit-learn. Visualize the boundary decision.<br>\n\nCheck your code correctness: The loss value should be around 0.255. <br>",
"_____no_output_____"
]
],
[
[
"#YOUR CODE HERE",
"_____no_output_____"
]
],
[
[
"## 2.3 Quadratic logistic regression/classification task.\n<hr>\n\n\nImplement the quadratic logistic regression function with gradient descent or scikit-learn. Visualize the boundary decision.<br>\n\nCheck your code correctness: The loss value should be around 0.255. <br>",
"_____no_output_____"
]
],
[
[
"#YOUR CODE HERE",
"_____no_output_____"
]
],
[
[
"## 2.4 Cubic logistic regression/classification task.\n<hr>\n\nThe logistic regression/classification predictive function is defined as:\n\n$$\n\\begin{aligned}\np_w(x) &= \\sigma(X w)\n\\end{aligned}\n$$\n\nIn the case of **cubic** prediction, we have:\n\n<br>\n$$\nX = \n\\left[ \n\\begin{array}{cccccccc}\n1 & x_{1(1)} & x_{1(2)} & x_{1(1)}^2 & x_{1(2)}^2 & x_{1(1)}x_{1(2)} & x_{1(2)}^3 & x_{1(2)}^3 & x_{1(1)}^2x_{1(2)} & x_{1(1)}x_{1(2)}^2 \\\\ \n1 & x_{2(1)} & x_{2(2)} & x_{2(1)}^2 & x_{2(2)}^2 & x_{2(1)}x_{2(2)} & x_{2(2)}^3 & x_{2(2)}^3 & x_{2(1)}^2x_{2(2)} & x_{2(1)}x_{2(2)}^2\\\\ \n\\vdots\\\\\n1 & x_{n(1)} & x_{n(2)} & x_{n(1)}^2 & x_{n(2)}^2 & x_{n(1)}x_{n(2)} & x_{n(2)}^3 & x_{n(2)}^3 & x_{n(1)}^2x_{n(2)} & x_{n(1)}x_{n(2)}^2\n\\end{array} \n\\right]\n\\quad\n\\textrm{ and }\n\\quad\nw = \n\\left[ \n\\begin{array}{cccc}\nw_0 \\\\ \nw_1 \\\\ \nw_2\\\\ \nw_3\\\\ \nw_4\\\\ \nw_5\\\\ \nw_6\\\\ \nw_7\\\\ \nw_8\\\\ \nw_9\n\\end{array} \n\\right]\n\\quad\n$$\n\nImplement the cubic logistic regression function with gradient descent or scikit-learn. Visualize the boundary decision.<br>\n\nCheck your code correctness: The loss value should be around 0.043. <br>",
"_____no_output_____"
]
],
[
[
"#YOUR CODE HERE",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb47e1ffd56c4a7a960b47150484055a80fbbb2 | 4,582 | ipynb | Jupyter Notebook | 00-Syllabus.ipynb | bcku/Spring-2018 | cddf0bfc52743523e734a29e286e4d8fb6de5195 | [
"Apache-2.0"
] | null | null | null | 00-Syllabus.ipynb | bcku/Spring-2018 | cddf0bfc52743523e734a29e286e4d8fb6de5195 | [
"Apache-2.0"
] | null | null | null | 00-Syllabus.ipynb | bcku/Spring-2018 | cddf0bfc52743523e734a29e286e4d8fb6de5195 | [
"Apache-2.0"
] | null | null | null | 28.81761 | 155 | 0.542994 | [
[
[
"# PSTAT 134/234 - Statistical Data Science\n\n---\n\n## Instructor: Sang-Yun Oh\n\n- Lectures: MW 11 am - 12:15 pm\n\n- Office: South Hall 5514\n\n- Office hours: Tuesday 4-6 pm\n\n\n## Teaching Assistant: Sergio Rodriguez \n\n- Sections: F 9 - 9:50 am / 12 - 12:50 pm\n\n- Office: South Hall 6432-W\n\n- Office hours: Thursday 1-3 pm\n",
"_____no_output_____"
],
[
"# Course Information \n\n---\n\n## Grading\n\n* Attendance in lectures and sections are required (20%) \n Total of five will be dropped. No exceptions\n\n* Individual in-class midterm (20%)\n\n* Individual assignments (30%)\n\n* Group final project & presentations (30%)\n\n\n## Textbooks\n\n- [Python Data Science Handbook](https://jakevdp.github.io/PythonDataScienceHandbook/) by Jake Vanderplas\n\n- [R for Data Science](http://r4ds.had.co.nz/) by Hadley Wickham and Garrett Grolemund\n\n- Other resources as necessary\n\n\n## Learn by doing\n\n- Critical statistical thinking is crucial\n\n- Significant programming is required\n\n- Many software tools will be new \n e.g., R, Python, command line tools, etc\n\n- Proactive attitude is a must! \n e.g., asking questions, discussing, experimenting, RTM (read-the-manual)\n\n- Diverse backgrounds mean you will have different strengths! \n Help each other, and assess your own areas of improvement\n\n- You don't have to be an expert at everything\n\n- But you have to be willing to dig deeper on your own",
"_____no_output_____"
],
[
"# Course outline\n\n---\n\n* **Week 1** (4/2-4/6): Data and uncertainty \n - Computing: Jupyter notebook and Python primer\n - Reading: [Chapter 1 (skim)-2](https://jakevdp.github.io/PythonDataScienceHandbook/index.html#1.-IPython:-Beyond-Normal-Python) in Vanderplas\n \n* **Week 2** (4/9-4/13): Data scraping, transformation, and wrangling\n - Computing: Shell commands and Pandas\n - Reading: [Chapter 3](https://jakevdp.github.io/PythonDataScienceHandbook/index.html#3.-Data-Manipulation-with-Pandas) in Vanderplas \n [The Unix Shell](http://swcarpentry.github.io/shell-novice/) by Software Carpentry \n \n* **Week 3-4** (4/16-4/27): Visualization and exploratory analysis\n - Computing: Matplotlib and Scikit-learn\n - Reading: [Chapter 4 (skim) - 5](https://jakevdp.github.io/PythonDataScienceHandbook/index.html#4.-Visualization-with-Matplotlib)\n\n* **In-class midterm** (4/30)\n\n* **Week 5-6** (5/2-5/11): Finance data module\n\n* **Week 7-8** (5/14-5/24): Health data module\n \n* **Week 9** (5/28-6/1): Text data module\n\n* **Week 10** (6/4-6/8): Final project presentations\n\n* **Final Projects** (6/14): Final project presentations\n",
"_____no_output_____"
],
[
"# Computational Environment\n\n---\n\n## Github\n\n* [Github Student Account](https://education.github.com/pack)\n\n\n## Jupyterhub\n\n* [Course Jupyter Hub](https://jupyterhub.lsit.ucsb.edu)\n\n* PSTAT 134/234 coursework only\n\n* Your work can be inspected by teaching staff at any time\n\n* Sign the [privacy policy](https://goo.gl/forms/pwa0FKNy6F0ZT8U32)",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecb4869e08de10887b0c9704a2ed36491b6f5747 | 3,675 | ipynb | Jupyter Notebook | notebooks/test_model.ipynb | Yossarian0916/audio_source_separator | 52c95d8b3a2da3ed059301adcd12886dd6b1eed5 | [
"Apache-2.0"
] | 2 | 2020-10-14T20:45:34.000Z | 2021-04-12T09:39:03.000Z | notebooks/test_model.ipynb | Yossarian0916/audio_source_separator | 52c95d8b3a2da3ed059301adcd12886dd6b1eed5 | [
"Apache-2.0"
] | 3 | 2020-11-13T18:40:16.000Z | 2022-02-10T01:29:08.000Z | notebooks/test_model.ipynb | Yossarian0916/audio_source_separator | 52c95d8b3a2da3ed059301adcd12886dd6b1eed5 | [
"Apache-2.0"
] | null | null | null | 26.438849 | 113 | 0.567619 | [
[
[
"import os\nimport sys\n\n\nmodule_path = os.path.abspath(os.path.join(os.getcwd(), os.pardir)) \nif module_path not in sys.path: \n sys.path.append(module_path)",
"_____no_output_____"
],
[
"import numpy as np\nimport tensorflow as tf\nimport IPython.display as ipd\n\nfrom utils.helper import wav_to_spectrogram_clips, rebuild_audio_from_spectro_clips\nfrom utils.dataset import create_samples\nfrom models.conv_denoising_unet import ConvDenoisingUnet\nfrom training.plot import plot_curve, plot_learning_curves",
"_____no_output_____"
],
[
"samples = create_samples('Dev')\ntrain_sample = samples[0]\n\nx_train = wav_to_spectrogram_clips(train_sample['mix'])\ny_train = dict()\ny_train['vocals'] = wav_to_spectrogram_clips(train_sample['vocals'])\ny_train['bass'] = wav_to_spectrogram_clips(train_sample['bass'])\ny_train['drums'] = wav_to_spectrogram_clips(train_sample['drums'])\ny_train['other'] = wav_to_spectrogram_clips(train_sample['other'])",
"_____no_output_____"
],
[
"# separator model\nseparator = ConvDenoisingUnet(1025, 100, (3, 3))\nmodel = separator.get_model()\nmodel.summary()\n\n\n# BEGIN TRAINING\nmodel.compile(optimizer=tf.keras.optimizers.Adam(lr=0.001),\n loss={'vocals': tf.keras.losses.MeanSquaredError(),\n 'bass': tf.keras.losses.MeanSquaredError(),\n 'drums': tf.keras.losses.MeanSquaredError(),\n 'other': tf.keras.losses.MeanSquaredError()})\n\nhistory = model.fit(x_train, y_train,\n batch_size=1,\n epochs=50,\n verbose=2)",
"_____no_output_____"
],
[
"pred = model.predict(wav_to_spectrogram_clips(train_sample['mix']))\npred_vocal = np.squeeze(pred[0], axis=-1)\nprint(pred_vocal.shape)",
"_____no_output_____"
],
[
"separated_vocals = rebuild_audio_from_spectro_clips(pred_vocal)\nipd.Audio(separated_vocals, rate=44100)",
"_____no_output_____"
],
[
"reconstructed_vocal = rebuild_audio_from_spectro_clips(wav_to_spectrogram_clips(train_sample['vocals']))\nipd.Audio(train_sample['vocals'], rate=44100)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb48d09e408ca3170ce36a143fe7d2b9315ef72 | 56,721 | ipynb | Jupyter Notebook | homework/HW02_numbers_solutions.ipynb | mtldatascience/sta-663-2019 | ce565879c3c22618db8d28e72daf08023b915a94 | [
"BSD-3-Clause"
] | 68 | 2019-01-09T21:53:55.000Z | 2022-02-16T17:14:22.000Z | homework/HW02_numbers_solutions.ipynb | mtldatascience/sta-663-2019 | ce565879c3c22618db8d28e72daf08023b915a94 | [
"BSD-3-Clause"
] | null | null | null | homework/HW02_numbers_solutions.ipynb | mtldatascience/sta-663-2019 | ce565879c3c22618db8d28e72daf08023b915a94 | [
"BSD-3-Clause"
] | 62 | 2019-01-09T21:43:48.000Z | 2021-11-15T04:26:25.000Z | 86.596947 | 39,016 | 0.818462 | [
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"# Homework 02: Working with numbers",
"_____no_output_____"
],
[
"**1**. (10 points) \n\nNormalize the $3 \\times 4$ diagonal matrix with diagonal (1, ,2, 3) so all rows have mean 0 and standard deviation 1. The matrix has 0 everywhere not on the diagonal.\n\n<font color=red>This is straightforward test of matrix construction, marginalization and broadcasting</font>",
"_____no_output_____"
]
],
[
[
"x = np.c_[np.diag([1,2,3]), np.zeros(3)]",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"x_scaled = (x - x.mean(axis=1)[:, None])/x.std(axis=1)[:, None]\nx_scaled",
"_____no_output_____"
],
[
"np.around(x_scaled.mean(axis=1))",
"_____no_output_____"
],
[
"x_scaled.std(axis=1)",
"_____no_output_____"
]
],
[
[
"**2**. (10 points) \n\nA fixed point of a funciton is a value that remains the same when the funciton is applied to it, that is $f(x) = x$. Write a function that finds the fixed poitn of another function $f$ given an intiial value $x_0$. For example, if\n\n$$f(x) \\rightarrow \\sqrt{x}$$\n\nand \n\n$x_0$ is any positive real number, then the function should return 1 since\n\n$$\\sqrt{1} = 1$$\n\nNot all funcitons have a fixed point - if it taakes over 1,000 iterations, the fucntion shold return None.\n\n- Use the function signature `fixed_point(f, x0, max_iter=1000)`. \n- Test with `fixed_point(np.sqrt, 10)`.\n\n<font color=red>This tests custom function writing, use of looping and checking a floating point condition, and the use of a higher order function.</font>",
"_____no_output_____"
]
],
[
[
"def fixed_point(f, x0, max_iter=1000):\n \"\"\"Fixed point iteration of f.\"\"\"\n \n tol = 1e-12\n x = x0\n i = 0\n while np.abs(x - f(x)) > tol:\n x = f(x)\n i += 1\n if i > max_iter:\n return None\n return x",
"_____no_output_____"
],
[
"fixed_point(np.sqrt, 10)",
"_____no_output_____"
]
],
[
[
"**3**. (10 points) \n\nUse `np.fromfunction` to construc the following matrix\n\n```python\narray([[5, 0, 0, 0, 5],\n [0, 4, 0, 4, 0],\n [0, 0, 3, 0, 0],\n [0, 2, 0, 2, 0],\n [1, 0, 0, 0, 1]])\n```\n\n<font color=red>This tests understanding of how `fromfunction` works and the use of element-wise selection with `np.where`. You need to see that we need to pull out the diagonals and anti-diagonals and write an expression to do so.</font>",
"_____no_output_____"
]
],
[
[
"j = np.repeat([np.arange(5)], 5, axis=0)\ni = j.T",
"_____no_output_____"
],
[
"i",
"_____no_output_____"
],
[
"j",
"_____no_output_____"
],
[
"np.fromfunction(lambda i, j: np.where((i==j) | (i==4-j), 5-i, 0), (5,5), dtype='int')",
"_____no_output_____"
]
],
[
[
"**4**. (15 points)\n\nSimulate $n$ coin toss experiments, in which you toss a coin $k$ times for each experiment. Find the maximum run length of heads (e.g. the sequence `T,T,H,H,H,T,H,H` has a maximum run length of 3 heads in each experiment. What is the most common maximum run length?\n\nLet $n$ = 10,000 and $k=100$.\n\n<font color=red>This tests if you can construct a simple simulation. The counting of runs was covered in the lecture notes using a finite state machine or regular expressions.</font>",
"_____no_output_____"
]
],
[
[
"def alt_runs(seq):\n \"\"\"Count using a finite state machine.\n \n A regular expression solution is also ok - see lecture notes for implementtation.\n \"\"\"\n\n current = seq[0]\n max_run = 1\n n = 1\n\n for s in seq[1:]:\n if s != current:\n n += 1\n else:\n max_run = max(max_run, n)\n n = 1 \n current = s\n max_run = max(max_run, n)\n return max_run",
"_____no_output_____"
],
[
"n = 10000\nk = 100\nseqs = np.random.choice(['H', 'T'], (n, k))\nmax_runs = [alt_runs(seq) for seq in seqs]",
"_____no_output_____"
],
[
"d = {}\nfor run in max_runs:\n d[run] = d.get(run, 0) + 1",
"_____no_output_____"
],
[
"sorted(d.items(), key=lambda x: x[1], reverse=True)[0]",
"_____no_output_____"
]
],
[
[
"**5**. (15 points)\n\nWikipedia gives this algorithm for finding prime numbers\n\nTo find all the prime numbers less than or equal to a given integer n by Eratosthenes' method:\n\n- Create a list of consecutive integers from 2 through n: (2, 3, 4, ..., n).\n- Initially, let p equal 2, the smallest prime number.\n- Enumerate the multiples of p by counting to n from 2p in increments of p, and mark them in the list (these will be 2p, 3p, 4p, ...; the p itself should not be marked).\n- Find the first number greater than p in the list that is not marked. If there was no such number, stop. Otherwise, let p now equal this new number (which is the next prime), and repeat from step 3.\n- When the algorithm terminates, the numbers remaining not marked in the list are all the primes below n.\n\nFind all primes less than 1,000 using this method.\n\n- You may use `numpy` and do not have to follow the algorithm exactly if you can achieve the same results.\n\n<font color=red>Test ability to construct a slightly more complex algorithm.</font>",
"_____no_output_____"
]
],
[
[
"def sieve(n):\n \"\"\"Sieve of Erastothenes.\"\"\"\n \n xs = np.arange(n+1)\n keep = np.ones(len(xs)).astype('bool')\n keep[:2] = 0\n p = 2\n while True:\n idx = p + p\n while idx <= n:\n keep[idx] = 0\n idx += p\n p = xs[p+1]\n while keep[p] == 0:\n p += 1\n if p == n:\n return xs[keep]",
"_____no_output_____"
],
[
"sieve(1000)",
"_____no_output_____"
]
],
[
[
"**6**. (40 points)\n\nWrite code to generate a plot similar to those shown below using the explanation for generation of 1D Cellular Automata found [here](http://mathworld.wolfram.com/ElementaryCellularAutomaton.html). You should only need to use standard Python, `numpy` and `matplotllib`.\n\n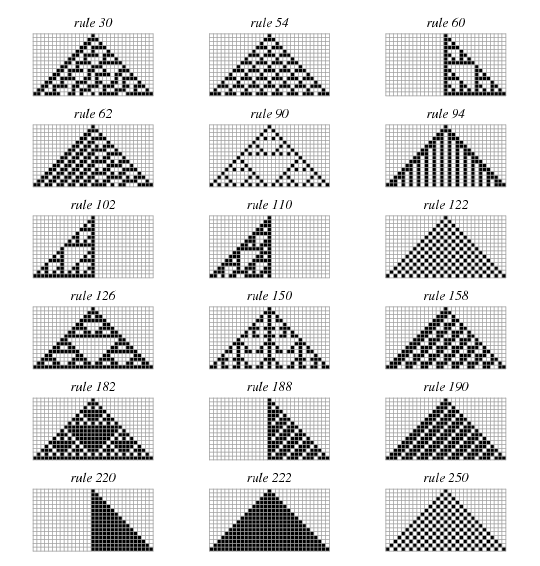\n\n\n\nThe input to the function making the plots should be a simple list of rules\n\n```python\nrules = [30, 54, 60, 62, 90, 94, 102, 110, 122, 126, \n 150, 158, 182, 188, 190, 220, 222, 250]\nmake_plots(rules, niter)\n```\n\nYou may, of course, write other helper functions to keep your code modular.\n\n<font color=red>Tests ability to break down a complex algorithm into simpler steps.\n\n- Convert integer rule into a map of next states\n- Use map to construct next state vector from current state vector by neighbor counting\n- Practice low level graphics construction (provided)\n</font>",
"_____no_output_____"
]
],
[
[
"def make_map(rule):\n \"\"\"Convert an integer into a rule mapping nbr states to new state.\"\"\"\n \n bits = map(int, list(bin(rule)[2:].zfill(8)))\n return dict(zip(range(7, -1, -1), bits))",
"_____no_output_____"
]
],
[
[
"Note: You can convert an integer into a binary representation using one of the methods below.",
"_____no_output_____"
]
],
[
[
"rule = 7\nprint(bin(rule)[2:].zfill(8))\nprint(format(7, '08b'))",
"00000111\n00000111\n"
],
[
"def make_ca(rule, init, niters):\n \"\"\"Run a 1d CA from init state for niters for given rule.\"\"\"\n \n mapper = make_map(rule)\n grid = np.zeros((niters, len(init)), 'int')\n grid[0] = init\n old = np.r_[init[-1:], init, init[0:1]]\n for i in range(1, niters):\n nbrs = zip(old[0:], old[1:], old[2:])\n cells = (int(''.join(map(str, nbr)), base=2) for nbr in nbrs)\n new = np.array([mapper[cell] for cell in cells])\n grid[i] = new\n old = np.r_[new[-1:], new, new[0:1]]\n return grid",
"_____no_output_____"
],
[
"%matplotlib inline",
"_____no_output_____"
],
[
"from matplotlib.ticker import NullFormatter, IndexLocator\nimport matplotlib.pyplot as plt\n\ndef plot_grid(rule, grid, ax=None):\n \"\"\"Plot a single grid.\"\"\"\n \n if ax is None:\n ax = plt.subplot(111)\n with plt.style.context('seaborn-white'):\n ax.grid(True, which='major', color='grey', linewidth=0.5)\n ax.imshow(grid, interpolation='none', cmap='Greys', aspect=1, alpha=0.8)\n ax.xaxis.set_major_locator(IndexLocator(1, 0))\n ax.yaxis.set_major_locator(IndexLocator(1, 0))\n ax.xaxis.set_major_formatter( NullFormatter() )\n ax.yaxis.set_major_formatter( NullFormatter() )\n ax.set_title('Rule %d' % rule)",
"_____no_output_____"
],
[
"def make_plots(rules, niter):\n \"\"\"Plot array of grids.\"\"\"\n \n nrows, ncols = rules.shape\n fig, axes = plt.subplots(nrows, ncols, figsize=(ncols*3, nrows*2))\n for i in range(nrows):\n for j in range(ncols):\n grid = make_ca(rules[i, j], init, niter)\n plot_grid(rules[i, j], grid, ax=axes[i,j])\n plt.tight_layout()",
"_____no_output_____"
],
[
"niter = 15\nwidth = niter*2+1\ninit = np.zeros(width, 'int')\ninit[width//2] = 1\nrules = np.array([30, 54, 60, 62, 90, 94, 102, 110, 122, 126, \n 150, 158, 182, 188, 190, 220, 222, 250]).reshape((-1, 3))\n\nncols = width\nmake_plots(rules, niter)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb49345c8118bcfa888218d61adbc2844d24d64 | 113,515 | ipynb | Jupyter Notebook | Code/Assignment-4/Inferential_Depressed.ipynb | Upward-Spiral-Science/spect-team | b5876fd76fc1da376b5d1fc6fd9337f620df142c | [
"Apache-2.0"
] | null | null | null | Code/Assignment-4/Inferential_Depressed.ipynb | Upward-Spiral-Science/spect-team | b5876fd76fc1da376b5d1fc6fd9337f620df142c | [
"Apache-2.0"
] | 3 | 2016-02-11T21:18:53.000Z | 2016-04-27T03:50:34.000Z | Code/Assignment-4/Inferential_Depressed.ipynb | Upward-Spiral-Science/spect-team | b5876fd76fc1da376b5d1fc6fd9337f620df142c | [
"Apache-2.0"
] | null | null | null | 267.094118 | 67,394 | 0.907422 | [
[
[
"import pandas as pd\nimport numpy as np\n\ndf_feats = pd.read_csv('reduced_data.csv')\ndf_labels = pd.read_csv('disorders.csv')['Depressed']\n\ndf = pd.concat([df_feats, df_labels], axis=1)\ndf_depr = df.loc[df['Depressed'] == 1].drop(['Depressed'], axis=1, inplace=False)\ndf_not_depr = df.loc[df['Depressed'] == 0].drop(['Depressed'], axis=1, inplace=False)",
"_____no_output_____"
],
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\n\n# Have a look at the distribution of Depressed and Not Depressed features\ndf_depr.plot(kind='hist', alpha=0.5, legend=None, title='Depressed')\ndf_not_depr.plot(kind='hist', alpha=0.5, legend=None, title='Not Depressed')",
"_____no_output_____"
],
[
"depr = df_depr.get_values().T\nnot_depr = df_not_depr.get_values().T",
"_____no_output_____"
]
],
[
[
"### State assumptions about your data\n\nLet X<sub>D</sub> denote features of depressed people, X<sub>ND</sub> denote features of people that are not depressed. Let μ<sub>D</sub> be the mean of X<sub>D</sub> and μ<sub>ND</sub> be the mean of X<sub>ND</sub>.\n\nAssume the real X<sub>D</sub> and X<sub>ND</sub> are both from a normal distribution.",
"_____no_output_____"
],
[
"### Formally define statistical test\n\nThe null and alternative hypotheses are:\n\n- H<sub>0</sub>: μ<sub>D</sub> = μ<sub>ND</sub>\n- H<sub>A</sub>: μ<sub>D</sub> != μ<sub>ND</sub>",
"_____no_output_____"
],
[
"### Provide algorithm for implementing test\n\nWe use Kolmogorov-Smirnov Goodness-of-Fit Test, which is a two-sided test for the null hypothesis that 2 independent samples are drawn from the same continuous distribution.\n\nThe original 754 features was reduced to 27 features, and on each of the 27 features, K-S test is applied below.",
"_____no_output_____"
]
],
[
[
"from scipy.stats import pearsonr\nfrom scipy.stats import chisquare\nfrom scipy.stats import ks_2samp\nfrom scipy.stats import anderson_ksamp\n\npearsonr_test = lambda x: pearsonr(x[0], x[1])[1]\nchi_test = lambda x: chisquare(x[0], x[1])[1]\nks_test = lambda x: ks_2samp(x[0], x[1])[1]\nanderson_ksamp_test = lambda x: anderson_ksamp(x)[2]",
"_____no_output_____"
]
],
[
[
"#### Note: Generate random samples and plot power vs n on null set based on Greg's code.",
"_____no_output_____"
],
[
"### Random Sampling Setup",
"_____no_output_____"
]
],
[
[
"import itertools\n\nnp.random.seed(123456789) # for reproducibility, set random seed\nalpha = 0.05 \nr = 20 # number of rois\nN = 100 # number of samples at each iteration\n\n# define number of subjects per class\nS = np.array((4, 6, 8, 10, 14, 18, 20, 26, 30, 40,\n 50, 60, 70, 80, 100, 120, 150, 200, 250,\n 300, 400, 500, 750, 1000, 1500, 2000,\n 3000, 5000))",
"_____no_output_____"
]
],
[
[
"### Sample data from null",
"_____no_output_____"
]
],
[
[
"pow_null = np.array((), dtype=np.dtype('float64'))\n\n# compute this statistic for various sizes of datasets\nfor s in S:\n s0 = s/2\n s1 = s - s0\n\n # compute this many times for each operating point to get average\n pval = np.array((), dtype=np.dtype('float64')) \n for _ in itertools.repeat(None,N):\n g0 = 1 * (np.random.rand(r, r, s0) > 0.5) # (null), 0.52 (classes)\n g1 = 1 * (np.random.rand(r, r, s1) > 0.5) # (null), 0.48 (classes)\n\n # compute feature of data\n pbar0 = 1.0*np.sum(g0, axis=(0,1))/(r**2 * s0)\n pbar1 = 1.0*np.sum(g1, axis=(0,1))/(r**2 * s1)\n\n # compute K-S test on feature\n pval = np.append(pval, ks_2samp(pbar0, pbar1)[1])\n \n # record average p value at operating point\n pow_null = np.append(pow_null, np.sum(1.0*(pval < alpha))/N)",
"_____no_output_____"
]
],
[
[
"### Sample data from alternate",
"_____no_output_____"
]
],
[
[
"pow_alt = np.array((), dtype=np.dtype('float64'))\n\n# compute this statistic for various sizes of datasets\nfor s in S:\n s0 = s/2\n s1 = s - s0\n\n # compute this many times for each operating point to get average\n pval = np.array((), dtype=np.dtype('float64')) \n for _ in itertools.repeat(None,N):\n g0 = 1 * (np.random.rand(r, r, s0) > 0.52) # (null), 0.52 (classes)\n g1 = 1 * (np.random.rand(r, r, s1) > 0.48) # (null), 0.48 (classes)\n\n # compute feature of data\n pbar0 = 1.0*np.sum(g0, axis=(0,1))/(r**2 * s0)\n pbar1 = 1.0*np.sum(g1, axis=(0,1))/(r**2 * s0)\n\n # compute K-S test on feature\n pval = np.append(pval, ks_2samp(pbar0, pbar1)[1])\n \n # record average p value at operating point\n pow_alt = np.append(pow_alt, np.sum(1.0*(pval < alpha))/N)",
"_____no_output_____"
]
],
[
[
"### Plot power vs n on null set",
"_____no_output_____"
]
],
[
[
"plt.scatter(S, pow_null, hold=True, label='null')\nplt.scatter(S, pow_alt, color='green', hold=True, label='alt')\nplt.xscale('log')\nplt.xlabel('number of samples')\nplt.ylabel('power')\nplt.title('Strength of depression classification under null model')\nplt.axhline(alpha, color='red', linestyle='--', label='alpha')\nplt.legend(loc=5)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Compute p-value on your real data\n",
"_____no_output_____"
]
],
[
[
"p_vals = list()\nfor a, b in zip(depr, not_depr):\n p_vals.append(round(ks_2samp(a, b)[1], 5))\nprint p_vals",
"[0.0, 3e-05, 0.99957, 0.04338, 0.08634, 0.70096, 0.18738, 0.0, 0.99991, 0.02431, 0.0, 0.0, 0.42373, 0.0, 0.0, 0.0003, 0.06074, 0.0, 0.0, 0.00034, 0.0, 0.07464, 0.28323, 0.0441, 0.23293, 0.60365, 0.00489]\n"
]
],
[
[
"### Sample from real data and plot power",
"_____no_output_____"
]
],
[
[
"from skbio.stats.power import subsample_power\n\n# Computer power of a sub sample set\ndef compute_sub_power(test, samples):\n pwr, counts = subsample_power(test=test,\n samples=samples,\n max_counts=1205,\n min_counts=100,\n counts_interval=100,\n draw_mode=\"ind\",\n alpha_pwr=0.05)\n return pwr, counts",
"_____no_output_____"
],
[
"from mpl_toolkits.axes_grid1 import Grid\n\nplt.close('all')\nfig = plt.figure()\nfig.set_size_inches(18, 9)\n\ngrid = Grid(fig, rect=111, nrows_ncols=(4, 7),\n axes_pad=0.25, label_mode='L',\n )\n \ndef plot_power(i, ax):\n a, b = depr[i], not_depr[i]\n samples = [np.array(a), np.array(b)]\n pwr, counts = compute_sub_power(ks_test, samples) \n ax.plot(counts, pwr.mean(0))\n \nfor i, ax in enumerate(grid):\n if i < 27:\n plot_power(i, ax)\n title = 'p = ' + str(p_vals[i])\n ax.set_title(title)\n\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"### Explain the degree to which you believe the result and why\n\nMost (17 out of 27) of the p-values is less than the significance level (0.05), for these we can reject the corresponding null hypotheses. \n\nThe third feature (whose p-value is almost 1.0) is race_id. This makes sense since people of various race are similarly likely to be depressed. The fifth to twenty-seventh features are all sparse-reconstructed and reduced features, it is hard to explain these results intuitively. \n\nSome of the power curves plotted using real data suspiciously decrease with the increase of number of samples. We know that there exists a lot of zeros in several column and some negative values across the entire feature matrix, since these features are reconstructed, this might be one of the reasons why the power curves act weirdly. \n\nAlso our original data is possibly noisy. We can see that the 10 power curves corresponding to the 10 large p-values (> 0.05) look like power curves of the null distribution, thereby the power curve and the p-value agree with each other on the real data.",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
ecb4b556b45c77381679b0b4b9f0d69d12829f0c | 6,588 | ipynb | Jupyter Notebook | backup/vrnn.ipynb | yizhouzhao/GenMotion | 67e73d06155d888eabda1187aa6ed3bbd796a814 | [
"MIT"
] | 32 | 2021-11-15T07:20:19.000Z | 2022-03-15T11:54:19.000Z | backup/vrnn.ipynb | yizhouzhao/GenMotion | 67e73d06155d888eabda1187aa6ed3bbd796a814 | [
"MIT"
] | null | null | null | backup/vrnn.ipynb | yizhouzhao/GenMotion | 67e73d06155d888eabda1187aa6ed3bbd796a814 | [
"MIT"
] | 3 | 2021-12-05T22:04:27.000Z | 2022-03-05T16:30:57.000Z | 23.528571 | 372 | 0.535671 | [
[
[
"# Tutorial: Human Motion Generative Model Using Variational Autoencoder\n\nVariational recurrent neural network (VRNN) extracts human motion features we use an autoencoder, and to represent the extracted features as a probability density function in a latent space we use a variational autoencoder. Motion generator is modeled as a map from a latent variable sampled in the latent space to a motion capture data.\n\n<div>\n<img src=\"../../../images/vrnn.png\" width=\"500\"/>\n</div>\n\n[original image link](http://www.ijmo.org/vol8/616-GV015.pdf)\n",
"_____no_output_____"
]
],
[
[
"import os\nos.chdir(\"../../../../genmotion/\")\nos.listdir(os.getcwd())",
"_____no_output_____"
],
[
"import torch",
"_____no_output_____"
],
[
"from algorithm.vrnn.params import HDM05Params\n\nopt = HDM05Params().get_params()\nprint(\"opt:\",opt)",
"opt: {'exp_mode': 'train', 'model_name': 'vrnn', 'learning_rate': 0.0001, 'input_dim': 62, 'output_dim': 62, 'position_loss_weight': 0.1, 'rotation_loss_weight': 1.0, 'model_save_path': 'e:\\\\researches\\\\GenMotion\\\\genmotion\\\\pretrained_models\\\\vrnn4hdm05', 'frame_interval': 10, 'input_motion_length': 50, 'hidden_dim': 128, 'n_layers': 1, 'z_dim': 32}\n"
],
[
"from dataset.hdm05.hdm05_data_utils import HDM05Dataset\n\ndata_path = \"E:/researches/GenMotion/dataset/HDM05/HDM_01-01_amc\"\ndataset = HDM05Dataset(data_path, opt)",
" 6%|▋ | 1/16 [00:00<00:02, 6.17it/s]"
],
[
"from algorithm.vrnn.models import VRNN\n\nmodel = VRNN(opt)",
"_____no_output_____"
],
[
"# set up model device\ndevice = torch.device(\"cuda\") if torch.cuda.is_available() else torch.device(\"cpu\")",
"_____no_output_____"
],
[
"# define the path your want to save the model\n# save_path = os.path.join(genmotion.__path__, \"../pretrained\")\nsave_path = os.path.join(os.getcwd(), \"pretrained\") \nprint(save_path) \nopt[\"model_save_path\"] = save_path ",
"_____no_output_____"
],
[
"from algorithm.vrnn.trainer import HDM05Trainer\n\ntrainer = HDM05Trainer(dataset, model, opt, device)\n\nprint(\"training dataset size: \", len(trainer.train_dataset))\nprint(\"evaluation dataset size: \", len(trainer.test_dataset)) ",
"_____no_output_____"
],
[
"trainer.train(epochs=1)",
"_____no_output_____"
],
[
"import genmotion\nsave_path = os.path.join(genmotion.__path__[0], \"pretrained/best.pth\")\n# save_path = os.path.join(os.getcwd(), \"/../pretrained\")\nprint(save_path)",
"e:\\researches\\genmotion\\genmotion\\pretrained/best.pth\n"
],
[
"# set up model device\ndevice = torch.device(\"cuda\") if torch.cuda.is_available() else torch.device(\"cpu\")",
"_____no_output_____"
],
[
"from algorithm.vrnn.sampler import HDM05Sampler\nsampler = HDM05Sampler(save_path, opt, device)",
"_____no_output_____"
],
[
"input_motion = dataset[0]",
"_____no_output_____"
],
[
"sampled_motion = sampler.sample(input_motion)",
"_____no_output_____"
],
[
"torch.FloatTensor(sampled_motion).shape",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb4c534424365277e59b946f74abc0e7df32507 | 8,135 | ipynb | Jupyter Notebook | 01 - Intro.ipynb | rhodeskl/ml_class | a4071751c69cfc3ed1ae44ac29daf4da29bd07d6 | [
"Unlicense"
] | null | null | null | 01 - Intro.ipynb | rhodeskl/ml_class | a4071751c69cfc3ed1ae44ac29daf4da29bd07d6 | [
"Unlicense"
] | 1 | 2018-05-09T22:27:31.000Z | 2018-05-11T17:59:40.000Z | 01 - Intro.ipynb | rhodeskl/ml_class | a4071751c69cfc3ed1ae44ac29daf4da29bd07d6 | [
"Unlicense"
] | null | null | null | 31.777344 | 224 | 0.583774 | [
[
[
"# Machine Learning Demystified\n\n\n## Agenda\n\n> 3 Hour Hands-On Workshop\n\n1. [Introduction (Python and Jupyter Basics)](01%20-%20Intro.ipynb)\n2. [Demistifying ML Terms](02%20-%20Demistifying%20ML%20Terms.ipynb)\n2. [Regression and Classification](03%20-%20Regression%20or%20Classification.ipynb)\n2. [Classification and Unsupervised Learning Examples (Clustering)](04%20-%20Classification%20and%20Unsupervised%20Learning%20Examples.ipynb)\n2. Bio Break\n2. [Preparing Data (Data Science!)](05 - Preparing Data.ipynb)\n2. [Regression Examples (Linear Regression and Neural Network)](06 - Regression Examples.ipynb)\n2. Bio Break\n2. [Where Do You Go From Here?](07%20-%20From%20Here.ipynb)\n\n## Get Started\nVisit **https://github.com/atomantic/ml_class** and follow the setup instructions\n\n## Follow Along!\nThis section focuses on getting comfortable with the Juypter Notebook, reading Python source code, and executing Python statements.",
"_____no_output_____"
]
],
[
[
"# In Jupyter you can execute commandline programs by prefixing with a '!'\n# hit ctrl+enter (or shift+enter) to execute\n!python --version",
"_____no_output_____"
],
[
"# Import the common packages for exploring Machine Learning\nimport numpy as np # <-- common convention for short names of packages...\nimport pandas as pd\nimport sklearn\nimport matplotlib\nimport matplotlib.pyplot as plt\n\n# Always good to check versions - because DOCS differ!\nprint('NumPy Version',np.__version__)\nprint('Pandas Version',pd.__version__)\nprint('Scikit Learn Version',sklearn.__version__)\nprint('MatplotLib Version',matplotlib.__version__)",
"_____no_output_____"
]
],
[
[
"\nNumPy is a library for the Python programming language, adding support for large, multi-dimensional arrays and matrices, along with a large collection of high-level mathematical functions to operate on these arrays.\n- [docs](https://docs.scipy.org/doc/)\n- n-dimensional array object\n- random numbers\n- complex array navigation: https://docs.scipy.org/doc/numpy-1.13.0/reference/arrays.indexing.html",
"_____no_output_____"
]
],
[
[
"# Create a simple NumPy array\na = np.array([[1,2],\n [3,4],\n [5,6],\n [7,8],\n [9,10],\n [11,12]])\n\nprint(\"full array:\", a)\n\n# Numpy uses interesting syntax for slicing data\n# Zero-indexed!\nprint(\"\\nfirst row:\", a[0])\n\n# query segments from an array: array[start:stop:step]\nb = np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])\n# note that stop is non-inclusive\n# step defaults to 1\nprint(\"\\n1st to 4th index (non-inclusive of last)\", b[1:4])\nprint(\"\\nevery 2 items from 0-4 (non-inclusive of last)\", b[0:4:2])\n\n# let's play with the first array:\n#print(\"\\nfirst column values from all rows:\", a[:,0])\n#print(\"\\nsecond column, second row:\", a[1,1])\n#print(\"\\nmore complex value pulling:\", a[2:4,0])\n\n# Your Turn\n#print(\"\\nPlayground:\", a[2,:])",
"_____no_output_____"
]
],
[
[
"\nPandas is a Python library that provides powerful data structures for data analysis, time series,and statistics. \n- [docs](https://pandas.pydata.org/pandas-docs/stable/)\n- powerful data analysis and manipulation\n- makes data into something like a spreadsheet",
"_____no_output_____"
]
],
[
[
"# Lets create a DataFrame with Pandas that has more advanced utility functions built in\n# Load the previously created NumPy array as an input argument known aka function parameter\ndf = pd.DataFrame(a)\n# with column names for ease of use\ndf.columns = ['Feature 1','Feature 2']\n\n# ** note: Jupyter will 'pretty print' the LAST object you reference without a print()\n# But you have to use print('') to show any others before it\n\nprint(df) # <--- this gets printed\ndf.values # <--- but this DOESN'T get printed\ndf # <--- but this does (last direct item)",
"_____no_output_____"
]
],
[
[
"\nmatplotlib is a plotting library for the Python programming language and its numerical mathematics extension NumPy.\n- [docs](https://matplotlib.org/contents.html)\n- powerful data visualization\n- interactive with iPython/Jupyter Notebooks",
"_____no_output_____"
]
],
[
[
"# multiple plots can be created and shown by giving the plots a figure number\nplt.figure(1)\n# generate some random data (10K numbers between 0-1)\nx = np.random.rand(10000)\n# create a histogram, placing the values in x into 100 buckets\nplt.hist(x, 100)\n# render it\nplt.show()",
"_____no_output_____"
],
[
"# Use the 'magic' % have iPython load matplotlib in interactive mode\n%matplotlib notebook",
"_____no_output_____"
],
[
"# interactive scatterplot\nN = 50\nx = np.random.rand(N)\ny = np.random.rand(N)\ncolors = np.random.rand(N)\narea = np.pi * (15 * np.random.rand(N))**2 # 0 to 15 point radii\n\nplt.figure(1)\nplt.scatter(x, y, s=area, c=colors, alpha=0.5)\nplt.show()",
"_____no_output_____"
],
[
"# Use the 'magic' % to see what variables are in memory\n%who\n%whos",
"_____no_output_____"
]
],
[
[
"See [Magic Commands Docs](http://ipython.readthedocs.io/en/stable/interactive/magics.html)",
"_____no_output_____"
],
[
"\nScikit-learn is a machine learning library for the Python programming langauge. It is simple and provides efficient tools for data mining and data analysis.\n- [docs](http://scikit-learn.org/stable/documentation.html)\n- complete machine learning toolkit\n- clustering tools\n- neural networks\n- experimental data\n\n### ...We'll Get to This\n\n## But First\n\nLet's continue to [Demistifying ML Terms](02%20-%20Demistifying%20ML%20Terms.ipynb)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ecb4cb46ffa8e1dbdc3729b6e23badfd8d7aaef1 | 7,380 | ipynb | Jupyter Notebook | seminario1/10. Graficos.ipynb | jmarinma/Julia-seminarios | 114ae06bfd9c3a889c87dc289cc41620553c9c7b | [
"MIT"
] | null | null | null | seminario1/10. Graficos.ipynb | jmarinma/Julia-seminarios | 114ae06bfd9c3a889c87dc289cc41620553c9c7b | [
"MIT"
] | 1 | 2021-03-11T11:53:43.000Z | 2021-03-11T11:53:43.000Z | seminario1/10. Graficos.ipynb | jmarinma/Julia-seminarios | 114ae06bfd9c3a889c87dc289cc41620553c9c7b | [
"MIT"
] | 1 | 2021-03-08T16:44:44.000Z | 2021-03-08T16:44:44.000Z | 23.503185 | 334 | 0.549322 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecb4d8fc08b056d5643847e1b32f6890b1f35b7e | 37,249 | ipynb | Jupyter Notebook | notebook/procs-ofbiz-party-note.ipynb | samlet/stack | 47db17fd4fdab264032f224dca31a4bb1d19b754 | [
"Apache-2.0"
] | 3 | 2020-01-11T13:55:38.000Z | 2020-08-25T22:34:15.000Z | notebook/procs-ofbiz-party-note.ipynb | samlet/stack | 47db17fd4fdab264032f224dca31a4bb1d19b754 | [
"Apache-2.0"
] | null | null | null | notebook/procs-ofbiz-party-note.ipynb | samlet/stack | 47db17fd4fdab264032f224dca31a4bb1d19b754 | [
"Apache-2.0"
] | 1 | 2021-01-01T05:21:44.000Z | 2021-01-01T05:21:44.000Z | 33.287757 | 1,687 | 0.398856 | [
[
[
"from sagas.ofbiz.services import OfService as s, oc, track\nok, r=track(lambda a: s().testScv(defaultValue=5.5, message=\"hello world\"))\nprint(ok, r) ",
"✔ testScv default 21 2019-02-10 22:13:54.522 ➷ 20 ms\nTrue {'responseMessage': 'success', 'resp': 'service done'}\n"
],
[
"s('meta').createPartyNote",
"_____no_output_____"
],
[
"# noteId='DemoNote', 这个参数是用于引用已经存在的note-data\nok,r=track(lambda a: s().createPartyNote(partyId='DemoCustomer', \n noteName='Demo Note',\n note='This is demo note to test createPartyNote service'))\nif ok:\n note_id=r['noteId']\n print(note_id)",
"✔ createPartyNote default 21 2019-02-10 22:33:33.424 ➷ 18 ms\n✔ createNote default 21 2019-02-10 22:33:33.424 ➷ 2 ms\n10004\n"
],
[
"from sagas.ofbiz.entities import OfEntity as e\ne('meta').PartyNote",
"_____no_output_____"
],
[
"e('df').queryPartyNote(partyId='DemoCustomer')",
"_____no_output_____"
],
[
"e('meta').NoteData",
"_____no_output_____"
],
[
"e('df').listNoteData()",
"_____no_output_____"
],
[
"from requests import put, get\njson_data=get('https://jsonplaceholder.typicode.com/posts?_start={startIndex}&_limit={limit}'\n .format(startIndex=0, limit=100)).json()",
"_____no_output_____"
],
[
"print(len(json_data))\n# print(json_data)\nfor r in json_data:\n# print(r['id'], r['title'], r['body'])\n ok,r=s().createPartyNote(partyId='DemoCustomer', \n noteName=r['title'],\n note=r['body'])\n if not ok:\n print(r)\n break",
"100\n"
],
[
"from sagas.ofbiz.entities import OfEntity as e\ne('df').listNoteData(_offset=5,_limit=3)",
"_____no_output_____"
],
[
"e().listNoteData(_offset=105,_limit=3)",
"_____no_output_____"
],
[
"import datetime\nprint(\"todo \"+str(datetime.datetime.now()))",
"todo 2019-02-10 23:14:14.914151\n"
],
[
"from sagas.ofbiz.entities import OfEntity as e, oc\nimport json\n\nrs=e().listNoteData(_offset=5,_limit=3)\njson_rs=oc.j.ValueHelper.valueListToJson(rs)\nfor r in json.loads(json_rs):\n print(r['noteName'])",
"sunt aut facere repellat provident occaecati excepturi optio reprehenderit\nqui est esse\nea molestias quasi exercitationem repellat qui ipsa sit aut\n"
],
[
"from sagas.ofbiz.entities import OfEntity as e\ne('json').listNoteData(_offset=5,_limit=3)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb4e3e4e27622154c1e0f2f65802cea5f32f58e | 8,730 | ipynb | Jupyter Notebook | airbnb_sample_test.ipynb | JamesBarciz/Datascience | d856a72c8053e499999bb304d742c6a36af8113a | [
"MIT"
] | null | null | null | airbnb_sample_test.ipynb | JamesBarciz/Datascience | d856a72c8053e499999bb304d742c6a36af8113a | [
"MIT"
] | null | null | null | airbnb_sample_test.ipynb | JamesBarciz/Datascience | d856a72c8053e499999bb304d742c6a36af8113a | [
"MIT"
] | 1 | 2020-03-03T01:02:24.000Z | 2020-03-03T01:02:24.000Z | 28.436482 | 93 | 0.466552 | [
[
[
"import requests\nimport json\nimport pandas as pd\nfrom pandas.io.json import json_normalize\nimport joblib",
"_____no_output_____"
],
[
"# Testing with the original parameters",
"_____no_output_____"
],
[
"url = 'https://airbnb-berlin-price-predict.herokuapp.com/'\ndata = {\n \"accommodates\": 2,\n \"bedrooms\": 1,\n \"cleaning_fee\": 30.50,\n \"extra_people\": 3,\n \"guests_included\": 3,\n \"minimum_nights\": 3,\n \"neighbourhood_group_cleansed_Mitte\": 0,\n \"neighbourhood_group_cleansed_Pankow\": 1,\n \"neighbourhood_group_cleansed_Tempelhof_Schoneberg\": 0,\n \"neighbourhood_group_cleansed_Friedrichshain_Kreuzberg\": 0,\n \"neighbourhood_group_cleansed_Neukolln\": 0,\n \"neighbourhood_group_cleansed_Charlottenburg_Wilm\": 0,\n \"neighbourhood_group_cleansed_Treptow_Kopenick\": 0,\n \"neighbourhood_group_cleansed_Steglitz_Zehlendorf\": 0,\n \"neighbourhood_group_cleansed_Reinickendorf\": 0,\n \"neighbourhood_group_cleansed_Lichtenberg\": 0,\n \"neighbourhood_group_cleansed_Marzahn_Hellersdorf\": 0,\n \"neighbourhood_group_cleansed_Spandau\": 0, \n \"property_type_Guesthouse\": 0,\n \"property_type_Apartment\": 0,\n \"property_type_Condominium\": 0,\n \"property_type_Loft\": 0,\n \"property_type_House\": 0,\n \"property_type_Serviced_apartment\": 0,\n \"property_type_Townhouse\": 0,\n \"property_type_Other\": 1,\n \"property_type_Bed_and_breakfast\": 0,\n \"property_type_Guest_suite\": 0,\n \"property_type_Hostel\": 0,\n \"room_type_Entire_home/apt\": 0,\n \"room_type_Private_room\": 1,\n \"room_type_Shared_room\": 0,\n \"bed_type_Real_Bed\": 0,\n \"bed_type_Sofa_Other\": 1,\n \"instant_bookable_f\": 1,\n \"instant_bookable_t\": 0,\n \"cancellation_policy_strict\": 1,\n \"cancellation_policy_flexible\": 0,\n \"cancellation_policy_moderate\": 0\n}",
"_____no_output_____"
],
[
"feature_data = json.dumps(feature_data)",
"_____no_output_____"
],
[
"# A response of 200 means everything went alright\n\npost_request = requests.post(url, feature_data)\nprint(post_request)",
"_____no_output_____"
],
[
"print(post_request.json())",
"_____no_output_____"
],
[
"with open('features.json', 'r') as f:\n features_dict = json.load(f)",
"_____no_output_____"
],
[
"type(features_dict)",
"_____no_output_____"
],
[
"json_normalize(features_dict)",
"_____no_output_____"
],
[
"model = joblib.load(open('berlin_model.gz', 'rb'))",
"_____no_output_____"
],
[
"model.predict(json_normalize(features_dict))[0]",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb4e5fb8b250a403d5cff6b63fdbeb6b263ed03 | 35,723 | ipynb | Jupyter Notebook | Starter_Code/credit_risk_ensemble.ipynb | amilamev/Classification_Homework | 7fd228fbdd2bdbf74d3cd558f6d2e511e2dee1a1 | [
"ADSL"
] | null | null | null | Starter_Code/credit_risk_ensemble.ipynb | amilamev/Classification_Homework | 7fd228fbdd2bdbf74d3cd558f6d2e511e2dee1a1 | [
"ADSL"
] | null | null | null | Starter_Code/credit_risk_ensemble.ipynb | amilamev/Classification_Homework | 7fd228fbdd2bdbf74d3cd558f6d2e511e2dee1a1 | [
"ADSL"
] | null | null | null | 31.753778 | 275 | 0.402206 | [
[
[
"# Ensemble Learning\n\n## Initial Imports",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nfrom pathlib import Path\nfrom collections import Counter",
"_____no_output_____"
],
[
"from sklearn.metrics import balanced_accuracy_score\nfrom sklearn.metrics import confusion_matrix\nfrom imblearn.metrics import classification_report_imbalanced",
"_____no_output_____"
]
],
[
[
"## Read the CSV and Perform Basic Data Cleaning",
"_____no_output_____"
]
],
[
[
"# Load the data\nfile_path = Path('Resources/LoanStats_2019Q1.csv')\ndf = pd.read_csv(file_path)\n\n# Preview the data\ndf.head()",
"_____no_output_____"
]
],
[
[
"## Split the Data into Training and Testing",
"_____no_output_____"
]
],
[
[
"# Create our features\nX = pd.get_dummies(df.drop(\"loan_status\", axis=1))\n\n\n# Create our target\ny = df.loc[:,'loan_status']",
"_____no_output_____"
],
[
"X.describe()",
"_____no_output_____"
],
[
"# Check the balance of our target values\n# YOUR CODE HERE\n\ny.value_counts()",
"_____no_output_____"
],
[
"# Split the X and y into X_train, X_test, y_train, y_test\n# YOUR CODE HERE\n\nfrom sklearn.model_selection import train_test_split\n\nX_train , X_test, y_train, y_test = train_test_split(X, y, random_state=1 , stratify = y)",
"_____no_output_____"
]
],
[
[
"## Data Pre-Processing\n\nScale the training and testing data using the `StandardScaler` from `sklearn`. Remember that when scaling the data, you only scale the features data (`X_train` and `X_testing`).",
"_____no_output_____"
]
],
[
[
"# Create the StandardScaler instance\n# YOUR CODE HERE\n\nfrom sklearn.preprocessing import StandardScaler\n\n\nscaler = StandardScaler()\nscaler\n",
"_____no_output_____"
],
[
"# Fit the Standard Scaler with the training data\n# When fitting scaling functions, only train on the training dataset\n# YOUR CODE HERE\nX_scaler = scaler.fit(X_train)\nX_scaler\n",
"_____no_output_____"
],
[
"# Scale the training and testing data\n# YOUR CODE HERE\nX_train_scaled = X_scaler.transform(X_train)\nX_test_scaled = X_scaler.transform(X_test)\nX_train_scaled.shape",
"_____no_output_____"
]
],
[
[
"## Ensemble Learners\n\nIn this section, you will compare two ensemble algorithms to determine which algorithm results in the best performance. You will train a Balanced Random Forest Classifier and an Easy Ensemble classifier . For each algorithm, be sure to complete the folliowing steps:\n\n1. Train the model using the training data. \n2. Calculate the balanced accuracy score from sklearn.metrics.\n3. Display the confusion matrix from sklearn.metrics.\n4. Generate a classication report using the `imbalanced_classification_report` from imbalanced-learn.\n5. For the Balanced Random Forest Classifier only, print the feature importance sorted in descending order (most important feature to least important) along with the feature score\n\nNote: Use a random state of 1 for each algorithm to ensure consistency between tests",
"_____no_output_____"
],
[
"### Balanced Random Forest Classifier",
"_____no_output_____"
]
],
[
[
"# Resample the training data with the BalancedRandomForestClassifier\n# YOUR CODE HERE\nfrom imblearn.ensemble import BalancedRandomForestClassifier\nrf_model = BalancedRandomForestClassifier(n_estimators=100, random_state=1)\nrf_model.fit(X_train, y_train)\n",
"_____no_output_____"
],
[
"# Calculated the balanced accuracy score\n# YOUR CODE HERE\n\ny_pred = rf_model.predict(X_test)\nbalanced_accuracy_score(y_test, y_pred)\n",
"_____no_output_____"
],
[
"# Display the confusion matrix\n# YOUR CODE HERE\n\nconfusion_matrix(y_test, y_pred)\n",
"_____no_output_____"
],
[
"# Print the imbalanced classification report\n# YOUR CODE HERE\n\nprint(classification_report_imbalanced(y_test, y_pred))\n",
" pre rec spe f1 geo iba sup\n\n high_risk 0.04 0.67 0.91 0.07 0.78 0.59 87\n low_risk 1.00 0.91 0.67 0.95 0.78 0.62 17118\n\navg / total 0.99 0.91 0.67 0.95 0.78 0.62 17205\n\n"
],
[
"# List the features sorted in descending order by feature importance\n# YOUR CODE HERE\nimportances = pd.DataFrame(rf_model.feature_importances_, index = X_train.columns, columns = ['Importance']).sort_values('Importance', ascending = False)\nimportances.head(10)",
"_____no_output_____"
]
],
[
[
"### Easy Ensemble Classifier",
"_____no_output_____"
]
],
[
[
"# Train the Classifier\n# YOUR CODE HERE\nfrom imblearn.ensemble import EasyEnsembleClassifier\n\n\nee_model = EasyEnsembleClassifier(n_estimators=100, random_state=1)\nee_model.fit(X_train, y_train)",
"_____no_output_____"
],
[
"# Calculated the balanced accuracy score\n# YOUR CODE HERE\n\ny_pred = ee_model.predict(X_test)\nbalanced_accuracy_score(y_test, y_pred)",
"_____no_output_____"
],
[
"# Display the confusion matrix\n\n\nconfusion_matrix(y_test, y_pred)\n",
"_____no_output_____"
],
[
"# Print the imbalanced classification report\n\nprint(classification_report_imbalanced(y_test, y_pred))\n",
" pre rec spe f1 geo iba sup\n\n high_risk 0.07 0.91 0.94 0.14 0.93 0.85 87\n low_risk 1.00 0.94 0.91 0.97 0.93 0.86 17118\n\navg / total 0.99 0.94 0.91 0.97 0.93 0.86 17205\n\n"
]
],
[
[
"### Final Questions\n\n1. Which model had the best balanced accuracy score?\n\n The Easy Ensemble Classifer model had the best score at .9254.\n\n2. Which model had the best recall score?\n\n The Easy Ensemble Classifer model had the best recall score at .94.\n \n3. Which model had the best geometric mean score?\n\n The Easy Ensemble Classifer model had the best geo score at .93.\n\n4. What are the top three features?\n\n The top three features are total_rec_prncp, total_rec_int, and total_pymnt_inv at importance values of 0.073767, 0.063903, and 0.060733.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecb4e78e44a9de38c08e7c5f434a1085653c3471 | 333,827 | ipynb | Jupyter Notebook | Lanl2.ipynb | tylerdn7/project-445 | 6e46141f1d01a0342518593aa0806fa6ebdae3b0 | [
"Apache-2.0"
] | null | null | null | Lanl2.ipynb | tylerdn7/project-445 | 6e46141f1d01a0342518593aa0806fa6ebdae3b0 | [
"Apache-2.0"
] | null | null | null | Lanl2.ipynb | tylerdn7/project-445 | 6e46141f1d01a0342518593aa0806fa6ebdae3b0 | [
"Apache-2.0"
] | null | null | null | 74.514955 | 25,798 | 0.623431 | [
[
[
"<a href=\"https://colab.research.google.com/github/tylerdn7/project-445/blob/master/Lanl2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!pip install kaggle\n\n!pip install numpy==1.15.0\n\n!pip install catboost",
"Requirement already satisfied: kaggle in /usr/local/lib/python3.6/dist-packages (1.5.3)\nRequirement already satisfied: urllib3<1.25,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from kaggle) (1.22)\nRequirement already satisfied: six>=1.10 in /usr/local/lib/python3.6/dist-packages (from kaggle) (1.11.0)\nRequirement already satisfied: certifi in /usr/local/lib/python3.6/dist-packages (from kaggle) (2019.3.9)\nRequirement already satisfied: python-dateutil in /usr/local/lib/python3.6/dist-packages (from kaggle) (2.5.3)\nRequirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from kaggle) (2.18.4)\nRequirement already satisfied: tqdm in /usr/local/lib/python3.6/dist-packages (from kaggle) (4.28.1)\nRequirement already satisfied: python-slugify in /usr/local/lib/python3.6/dist-packages (from kaggle) (3.0.2)\nRequirement already satisfied: idna<2.7,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->kaggle) (2.6)\nRequirement already satisfied: chardet<3.1.0,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->kaggle) (3.0.4)\nRequirement already satisfied: text-unidecode==1.2 in /usr/local/lib/python3.6/dist-packages (from python-slugify->kaggle) (1.2)\nRequirement already satisfied: numpy==1.15.0 in /usr/local/lib/python3.6/dist-packages (1.15.0)\nRequirement already satisfied: catboost in /usr/local/lib/python3.6/dist-packages (0.14.2)\nRequirement already satisfied: numpy>=1.11.1 in /usr/local/lib/python3.6/dist-packages (from catboost) (1.15.0)\nRequirement already satisfied: pandas>=0.19.1 in /usr/local/lib/python3.6/dist-packages (from catboost) (0.24.2)\nRequirement already satisfied: enum34 in /usr/local/lib/python3.6/dist-packages (from catboost) (1.1.6)\nRequirement already satisfied: graphviz in /usr/local/lib/python3.6/dist-packages (from catboost) (0.10.1)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from catboost) (1.11.0)\nRequirement already satisfied: pytz>=2011k in /usr/local/lib/python3.6/dist-packages (from pandas>=0.19.1->catboost) (2018.9)\nRequirement already satisfied: python-dateutil>=2.5.0 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.19.1->catboost) (2.5.3)\n"
],
[
"import pandas as pd\n\nimport numpy as np\n\nimport seaborn as sns\n\nimport io\n\nimport os\n\nimport xgboost as xgb\n\nimport seaborn as sns\n\nfrom sklearn.model_selection import train_test_split\n\nfrom sklearn.metrics import mean_absolute_error\n\nfrom sklearn.linear_model import SGDRegressor\n\nfrom sklearn.ensemble import RandomForestRegressor\n\nfrom catboost import CatBoostRegressor, Pool\n\nfrom sklearn.preprocessing import StandardScaler\n\nfrom sklearn.model_selection import GridSearchCV\n\nfrom sklearn.model_selection import train_test_split\n\nfrom sklearn.svm import NuSVR, SVR\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"from google.colab import files\n\nuploaded = files.upload()\n\nfor fn in uploaded.keys():\n print('User uploaded file \"{name}\" with length {length} bytes'.format(\n name=fn, length=len(uploaded[fn])))\n \n!mkdir -p ~/.kaggle/ && mv kaggle.json ~/.kaggle/ && chmod 600 ~/.kaggle/kaggle.json",
"_____no_output_____"
],
[
"!kaggle competitions download -c LANL-Earthquake-Prediction",
"sample_submission.csv: Skipping, found more recently modified local copy (use --force to force download)\ntest.zip: Skipping, found more recently modified local copy (use --force to force download)\ntrain.csv.zip: Skipping, found more recently modified local copy (use --force to force download)\n"
],
[
"!unzip train.csv.zip",
"Archive: train.csv.zip\n inflating: train.csv \n"
],
[
"\nseg_id = pd.read_csv('sample_submission.csv')\nseg_id.head()",
"_____no_output_____"
],
[
"!unzip test.zip",
"Archive: test.zip\n inflating: seg_430e66.csv \n inflating: seg_d1a281.csv \n inflating: seg_05a1b0.csv \n inflating: seg_f8dd7e.csv \n inflating: seg_b9bdd7.csv \n inflating: seg_24c1c9.csv \n inflating: seg_c5abaa.csv \n inflating: seg_6262c4.csv \n inflating: seg_734a88.csv \n inflating: seg_94a133.csv \n inflating: seg_d0c280.csv \n inflating: seg_d36737.csv \n inflating: seg_f80e44.csv \n inflating: seg_07c815.csv \n inflating: seg_7c9433.csv \n inflating: seg_211486.csv \n inflating: seg_78ded2.csv \n inflating: seg_f11f77.csv \n inflating: seg_b3883e.csv \n inflating: seg_3db0a8.csv \n inflating: seg_81f798.csv \n inflating: seg_0a45a1.csv \n inflating: seg_dc188b.csv \n inflating: seg_4a9e8d.csv \n inflating: seg_32fc4e.csv \n inflating: seg_7b2994.csv \n inflating: seg_7fd6b7.csv \n inflating: seg_4ce234.csv \n inflating: seg_6fc8b3.csv \n inflating: seg_50a667.csv \n inflating: seg_69a230.csv \n inflating: seg_2642d0.csv \n inflating: seg_5e70a7.csv \n inflating: seg_592807.csv \n inflating: seg_140bc5.csv \n inflating: seg_d5dbc1.csv \n inflating: seg_49336f.csv \n inflating: seg_bdf84b.csv \n inflating: seg_786ff6.csv \n inflating: seg_d14524.csv \n inflating: seg_b782c7.csv \n inflating: seg_d35274.csv \n inflating: seg_ced992.csv \n inflating: seg_24458d.csv \n inflating: seg_d098df.csv \n inflating: seg_51f0a2.csv \n inflating: seg_77c546.csv \n inflating: seg_fdd50e.csv \n inflating: seg_153d6a.csv \n inflating: seg_5cde88.csv \n inflating: seg_b91011.csv \n inflating: seg_460436.csv \n inflating: seg_27de37.csv \n inflating: seg_8ae847.csv \n inflating: seg_b52710.csv \n inflating: seg_063865.csv \n inflating: seg_abaad2.csv \n inflating: seg_87a67a.csv \n inflating: seg_47d374.csv \n inflating: seg_d40bb2.csv \n inflating: seg_d19980.csv \n inflating: seg_a1edc1.csv \n inflating: seg_bbf805.csv \n inflating: seg_e8a4f4.csv \n inflating: seg_8472f3.csv \n inflating: seg_8866f0.csv \n inflating: seg_79c47a.csv \n inflating: seg_500c80.csv \n inflating: seg_4d7c56.csv \n inflating: seg_75a878.csv \n inflating: seg_9a1a4f.csv \n inflating: seg_e4f403.csv \n inflating: seg_42c4c9.csv \n inflating: seg_d702b2.csv \n inflating: seg_9e61da.csv \n inflating: seg_db6d5f.csv \n inflating: seg_593c34.csv \n inflating: seg_2f8f6d.csv \n inflating: seg_04fd93.csv \n inflating: seg_3cba49.csv \n inflating: seg_88a81d.csv \n inflating: seg_1562cb.csv \n inflating: seg_89f975.csv \n inflating: seg_db694a.csv \n inflating: seg_17adc0.csv \n inflating: seg_59818b.csv \n inflating: seg_60b696.csv \n inflating: seg_f003ca.csv \n inflating: seg_e051bc.csv \n inflating: seg_d0598e.csv \n inflating: seg_5ad847.csv \n inflating: seg_be62ef.csv \n inflating: seg_9a1c76.csv \n inflating: seg_1010ad.csv \n inflating: seg_3452b2.csv \n inflating: seg_37f3fb.csv \n inflating: seg_7848f8.csv \n inflating: seg_ce472b.csv \n inflating: seg_355717.csv \n inflating: seg_eea20e.csv \n inflating: seg_baa745.csv \n inflating: seg_28fc32.csv \n inflating: seg_97e4a9.csv \n inflating: seg_a1a511.csv \n inflating: seg_c0260d.csv \n inflating: seg_c70fde.csv \n inflating: seg_eb7f91.csv \n inflating: seg_fb11ba.csv \n inflating: seg_35269b.csv \n inflating: seg_919c5c.csv \n inflating: seg_91eaeb.csv \n inflating: seg_9ab405.csv \n inflating: seg_37bf85.csv \n inflating: seg_136695.csv \n inflating: seg_35ba8f.csv \n inflating: seg_7f9b3a.csv \n inflating: seg_45e4ed.csv \n inflating: seg_702e03.csv \n inflating: seg_bf7224.csv \n inflating: seg_5f7fd9.csv \n inflating: seg_d85e2e.csv \n inflating: seg_e41fc9.csv \n inflating: seg_23b123.csv \n inflating: seg_660fef.csv \n inflating: seg_572172.csv \n inflating: seg_6d4fa6.csv \n inflating: seg_6f60b2.csv \n inflating: seg_4f48b4.csv \n inflating: seg_f86c41.csv \n inflating: seg_d516e3.csv \n inflating: seg_cbcce9.csv \n inflating: seg_34a8f7.csv \n inflating: seg_238242.csv \n inflating: seg_cc096e.csv \n inflating: seg_5bc0b0.csv \n inflating: seg_d27812.csv \n inflating: seg_b8141c.csv \n inflating: seg_e14149.csv \n inflating: seg_f6bae8.csv \n inflating: seg_e90c0d.csv \n inflating: seg_b917b5.csv \n inflating: seg_b72413.csv \n inflating: seg_195eda.csv \n inflating: seg_43597f.csv \n inflating: seg_324447.csv \n inflating: seg_06b8c9.csv \n inflating: seg_8310ea.csv \n inflating: seg_91fc29.csv \n inflating: seg_0e7cc5.csv \n inflating: seg_f165f6.csv \n inflating: seg_1a791c.csv \n inflating: seg_6ae32d.csv \n inflating: seg_d07ce8.csv \n inflating: seg_fa0ac7.csv \n inflating: seg_c671f7.csv \n inflating: seg_f2a20d.csv \n inflating: seg_da8b88.csv \n inflating: seg_bf1a72.csv \n inflating: seg_208ba5.csv \n inflating: seg_be19e2.csv \n inflating: seg_0cdcc8.csv \n inflating: seg_c0c0ed.csv \n inflating: seg_8be76c.csv \n inflating: seg_8f1127.csv \n inflating: seg_23323e.csv \n inflating: seg_35b753.csv \n inflating: seg_3d06cc.csv \n inflating: seg_986976.csv \n inflating: seg_222c5f.csv \n inflating: seg_e4f203.csv \n inflating: seg_a4208a.csv \n inflating: seg_521a88.csv \n inflating: seg_8c6d4e.csv \n inflating: seg_3bf9ac.csv \n inflating: seg_1cb942.csv \n inflating: seg_98be13.csv \n inflating: seg_a41c2c.csv \n inflating: seg_1afa29.csv \n inflating: seg_8f5589.csv \n inflating: seg_4d2671.csv \n inflating: seg_86d847.csv \n inflating: seg_7b0936.csv \n inflating: seg_185ad6.csv \n inflating: seg_ff0f1b.csv \n inflating: seg_5e8ef4.csv \n inflating: seg_8785e2.csv \n inflating: seg_f69c38.csv \n inflating: seg_b8bb87.csv \n inflating: seg_179d90.csv \n inflating: seg_81bebd.csv \n inflating: seg_76c2fa.csv \n inflating: seg_72858d.csv \n inflating: seg_d41daa.csv \n inflating: seg_e2b8b1.csv \n inflating: seg_83ef67.csv \n inflating: seg_5765b6.csv \n inflating: seg_57dd68.csv \n inflating: seg_8fc754.csv \n inflating: seg_bcf2d9.csv \n inflating: seg_ee1cfc.csv \n inflating: seg_304df5.csv \n inflating: seg_2e88dd.csv \n inflating: seg_b1fea8.csv \n inflating: seg_4b4ffb.csv \n inflating: seg_9a43ef.csv \n inflating: seg_abda04.csv \n inflating: seg_41be18.csv \n inflating: seg_2db7dc.csv \n inflating: seg_42648c.csv \n inflating: seg_20e9ad.csv \n inflating: seg_ea4d3a.csv \n inflating: seg_517345.csv \n inflating: seg_efd9ec.csv \n inflating: seg_6a45a0.csv \n inflating: seg_e5b510.csv \n inflating: seg_9ae1a1.csv \n inflating: seg_151d92.csv \n inflating: seg_ebb79f.csv \n inflating: seg_4e9646.csv \n inflating: seg_b88baa.csv \n inflating: seg_3865bc.csv \n inflating: seg_01c775.csv \n inflating: seg_4ea3fa.csv \n inflating: seg_326eb7.csv \n inflating: seg_90e904.csv \n inflating: seg_c243a6.csv \n inflating: seg_64daae.csv \n inflating: seg_f579ba.csv \n inflating: seg_9bee43.csv \n inflating: seg_20cbac.csv \n inflating: seg_6e7f73.csv \n inflating: seg_3d1f2e.csv \n inflating: seg_77fe77.csv \n inflating: seg_3bc9ed.csv \n inflating: seg_cc7e39.csv \n inflating: seg_c550b1.csv \n inflating: seg_8da61a.csv \n inflating: seg_bb6fcb.csv \n inflating: seg_dc6e94.csv \n inflating: seg_7d2535.csv \n inflating: seg_3b4414.csv \n inflating: seg_eb32bc.csv \n inflating: seg_60dacd.csv \n inflating: seg_304b87.csv \n inflating: seg_8d135f.csv \n inflating: seg_d062a6.csv \n inflating: seg_f741af.csv \n inflating: seg_d2551a.csv \n inflating: seg_1e61d6.csv \n inflating: seg_739643.csv \n inflating: seg_5924f5.csv \n inflating: seg_2883ec.csv \n inflating: seg_507065.csv \n inflating: seg_753190.csv \n inflating: seg_37608b.csv \n inflating: seg_65f01e.csv \n inflating: seg_37669c.csv \n inflating: seg_4f76a2.csv \n inflating: seg_566efe.csv \n inflating: seg_c5fc3b.csv \n inflating: seg_741fc1.csv \n inflating: seg_29022f.csv \n inflating: seg_911066.csv \n inflating: seg_b284b2.csv \n inflating: seg_2cadc0.csv \n inflating: seg_2ee95c.csv \n inflating: seg_158764.csv \n inflating: seg_d44589.csv \n inflating: seg_f5d2dd.csv \n inflating: seg_1a671a.csv \n inflating: seg_c267a5.csv \n inflating: seg_004cd2.csv \n inflating: seg_1969c8.csv \n inflating: seg_9c8bc8.csv \n inflating: seg_4c8db6.csv \n inflating: seg_8d4435.csv \n inflating: seg_ab2a78.csv \n inflating: seg_cf74e8.csv \n inflating: seg_b50239.csv \n inflating: seg_8f25b0.csv \n inflating: seg_a975df.csv \n inflating: seg_e3cf1a.csv \n inflating: seg_2c762c.csv \n inflating: seg_efc5fb.csv \n inflating: seg_d1a8b3.csv \n inflating: seg_392019.csv \n inflating: seg_83f928.csv \n inflating: seg_421da1.csv \n inflating: seg_ab70bd.csv \n inflating: seg_bd2b0c.csv \n inflating: seg_00cc91.csv \n inflating: seg_7d2e57.csv \n inflating: seg_ca407b.csv \n inflating: seg_ee7224.csv \n inflating: seg_7a521c.csv \n inflating: seg_d1b91e.csv \n inflating: seg_bbfd4e.csv \n inflating: seg_f679c6.csv \n inflating: seg_dcfb4b.csv \n inflating: seg_b57e76.csv \n inflating: seg_d4c763.csv \n inflating: seg_69dbad.csv \n inflating: seg_d36342.csv \n inflating: seg_850d95.csv \n inflating: seg_7f3ab0.csv \n inflating: seg_0c89ce.csv \n inflating: seg_9e8ca4.csv \n inflating: seg_c41d1d.csv \n inflating: seg_c5064e.csv \n inflating: seg_086a61.csv \n inflating: seg_4cb9c6.csv \n inflating: seg_7ed2dd.csv \n inflating: seg_7b3017.csv \n inflating: seg_a6e99c.csv \n inflating: seg_caa919.csv \n inflating: seg_614b50.csv \n inflating: seg_1c849d.csv \n inflating: seg_121446.csv \n inflating: seg_89791c.csv \n inflating: seg_7e3b3e.csv \n inflating: seg_6eedcc.csv \n inflating: seg_2eddf6.csv \n inflating: seg_efb639.csv \n inflating: seg_325790.csv \n inflating: seg_b64c9c.csv \n inflating: seg_a5f4dd.csv \n inflating: seg_d9190c.csv \n inflating: seg_b568c6.csv \n inflating: seg_a704ee.csv \n inflating: seg_dca2c2.csv \n inflating: seg_9e962b.csv \n inflating: seg_fdff11.csv \n inflating: seg_e8a2c4.csv \n inflating: seg_89885a.csv \n inflating: seg_77ea14.csv \n inflating: seg_fa34c7.csv \n inflating: seg_643d1f.csv \n inflating: seg_e00465.csv \n inflating: seg_e66697.csv \n inflating: seg_98c544.csv \n inflating: seg_812a50.csv \n inflating: seg_ed07b0.csv \n inflating: seg_56ebc7.csv \n inflating: seg_55b566.csv \n inflating: seg_4eef28.csv \n inflating: seg_634f05.csv \n inflating: seg_2ed79b.csv \n inflating: seg_b5aee9.csv \n inflating: seg_d5df41.csv \n inflating: seg_c52ae6.csv \n inflating: seg_3f3983.csv \n inflating: seg_8c1cca.csv \n inflating: seg_ed7f69.csv \n inflating: seg_3bb90a.csv \n inflating: seg_c01d9a.csv \n inflating: seg_14c371.csv \n inflating: seg_4ba3d1.csv \n inflating: seg_673c60.csv \n inflating: seg_fdc1d4.csv \n inflating: seg_6167f0.csv \n inflating: seg_170684.csv \n inflating: seg_e53a52.csv \n inflating: seg_7efdd5.csv \n inflating: seg_83f89e.csv \n inflating: seg_49322d.csv \n inflating: seg_ac45ae.csv \n inflating: seg_b42170.csv \n inflating: seg_7d718e.csv \n inflating: seg_f11620.csv \n inflating: seg_ad759c.csv \n inflating: seg_72c307.csv \n inflating: seg_0be6ee.csv \n inflating: seg_45a8a9.csv \n inflating: seg_854d91.csv \n inflating: seg_696621.csv \n inflating: seg_b31961.csv \n inflating: seg_e481c7.csv \n inflating: seg_46b84d.csv \n inflating: seg_75cd6a.csv \n inflating: seg_8cf573.csv \n inflating: seg_05e0d7.csv \n inflating: seg_13468a.csv \n inflating: seg_6c909f.csv \n inflating: seg_e9a09f.csv \n inflating: seg_3f2086.csv \n inflating: seg_a8e6df.csv \n inflating: seg_bce305.csv \n inflating: seg_2d3828.csv \n inflating: seg_d9750c.csv \n inflating: seg_92b095.csv \n inflating: seg_e20d76.csv \n inflating: seg_adf94a.csv \n inflating: seg_7fac26.csv \n inflating: seg_f4ad4b.csv \n inflating: seg_27a282.csv \n inflating: seg_2a0ace.csv \n inflating: seg_19ec5c.csv \n inflating: seg_72aba8.csv \n inflating: seg_fc8fd6.csv \n inflating: seg_16c3a7.csv \n inflating: seg_272a47.csv \n inflating: seg_c565bd.csv \n inflating: seg_5e7abf.csv \n inflating: seg_284223.csv \n inflating: seg_830cff.csv \n inflating: seg_ea9d37.csv \n inflating: seg_b00f05.csv \n inflating: seg_30fd84.csv \n inflating: seg_74b3bd.csv \n inflating: seg_311b5e.csv \n inflating: seg_151368.csv \n inflating: seg_7ef97b.csv \n inflating: seg_0d13a7.csv \n inflating: seg_dcee28.csv \n inflating: seg_c42952.csv \n inflating: seg_74235f.csv \n inflating: seg_b7b098.csv \n inflating: seg_de9e32.csv \n inflating: seg_cd352f.csv \n inflating: seg_d42c77.csv \n inflating: seg_c01f63.csv \n inflating: seg_0cb81b.csv \n inflating: seg_dd780d.csv \n inflating: seg_5009d9.csv \n inflating: seg_423ebe.csv \n inflating: seg_48feba.csv \n inflating: seg_9a6025.csv \n inflating: seg_fbd66b.csv \n inflating: seg_e13bd8.csv \n inflating: seg_d1ca05.csv \n inflating: seg_d1fbb8.csv \n inflating: seg_651808.csv \n inflating: seg_08b136.csv \n inflating: seg_073696.csv \n inflating: seg_83f476.csv \n inflating: seg_64be91.csv \n inflating: seg_2abd33.csv \n inflating: seg_1b08a4.csv \n inflating: seg_f639cd.csv \n inflating: seg_514b85.csv \n inflating: seg_7f786b.csv \n inflating: seg_9c090f.csv \n inflating: seg_457234.csv \n inflating: seg_08fd5b.csv \n inflating: seg_ec73d5.csv \n inflating: seg_5e1bba.csv \n inflating: seg_5ef47e.csv \n inflating: seg_d9bb68.csv \n inflating: seg_8f3dfe.csv \n inflating: seg_c7a3bc.csv \n inflating: seg_1a0eca.csv \n inflating: seg_98c44d.csv \n inflating: seg_490d6e.csv \n inflating: seg_c5dada.csv \n inflating: seg_193404.csv \n inflating: seg_4fce5b.csv \n inflating: seg_1cf1b5.csv \n inflating: seg_27b7c8.csv \n inflating: seg_d0c8e2.csv \n inflating: seg_f8db0a.csv \n inflating: seg_dcd9df.csv \n inflating: seg_6153ca.csv \n inflating: seg_8f3247.csv \n inflating: seg_afca81.csv \n inflating: seg_91f664.csv \n inflating: seg_988e69.csv \n inflating: seg_40b3aa.csv \n inflating: seg_373a04.csv \n inflating: seg_58e9f9.csv \n inflating: seg_dab0f9.csv \n inflating: seg_a39ebc.csv \n inflating: seg_82c138.csv \n inflating: seg_92be9f.csv \n inflating: seg_8c1760.csv \n inflating: seg_4e3bd7.csv \n inflating: seg_740837.csv \n inflating: seg_64dcf0.csv \n inflating: seg_ae32a6.csv \n inflating: seg_23c947.csv \n inflating: seg_a133bc.csv \n inflating: seg_a80f6f.csv \n inflating: seg_d21066.csv \n inflating: seg_a437cb.csv \n inflating: seg_ad82ca.csv \n inflating: seg_c20a27.csv \n inflating: seg_f073e2.csv \n inflating: seg_4d5a4a.csv \n inflating: seg_7c2beb.csv \n inflating: seg_ae3d92.csv \n inflating: seg_7aeca4.csv \n inflating: seg_9e3837.csv \n inflating: seg_c12d7d.csv \n inflating: seg_1ebe6b.csv \n inflating: seg_d5b100.csv \n inflating: seg_9a812d.csv \n inflating: seg_eb0b19.csv \n inflating: seg_07f37c.csv \n inflating: seg_3319b5.csv \n inflating: seg_2f9581.csv \n inflating: seg_fbce13.csv \n inflating: seg_f3063a.csv \n inflating: seg_d89369.csv \n inflating: seg_324537.csv \n inflating: seg_d9dab4.csv \n inflating: seg_b63de1.csv \n inflating: seg_0f8961.csv \n inflating: seg_dbf3f2.csv \n inflating: seg_e067ab.csv \n inflating: seg_6b9511.csv \n inflating: seg_f61b0e.csv \n inflating: seg_7189ca.csv \n inflating: seg_e86ed1.csv \n inflating: seg_b6cb2b.csv \n inflating: seg_f865be.csv \n inflating: seg_c654e7.csv \n inflating: seg_176542.csv \n inflating: seg_1c929e.csv \n inflating: seg_fa4df0.csv \n inflating: seg_666056.csv \n inflating: seg_abc5fd.csv \n inflating: seg_f21a22.csv \n inflating: seg_5bdb47.csv \n inflating: seg_b23489.csv \n inflating: seg_e2c82c.csv \n inflating: seg_5bd9ae.csv \n inflating: seg_0d434c.csv \n inflating: seg_1bf7b3.csv \n inflating: seg_b36c8a.csv \n inflating: seg_287fef.csv \n inflating: seg_425462.csv \n inflating: seg_ef74dc.csv \n inflating: seg_9c635d.csv \n inflating: seg_d7f015.csv \n inflating: seg_9c8162.csv \n inflating: seg_148698.csv \n inflating: seg_1fc7ab.csv \n inflating: seg_ef27c8.csv \n inflating: seg_a12acb.csv \n inflating: seg_d7ab50.csv \n inflating: seg_ce5832.csv \n inflating: seg_6fc3ef.csv \n inflating: seg_bd5022.csv \n inflating: seg_18d307.csv \n inflating: seg_c179f8.csv \n inflating: seg_46a42b.csv \n inflating: seg_5ec0e3.csv \n inflating: seg_2a8351.csv \n inflating: seg_555f63.csv \n inflating: seg_b3cf21.csv \n inflating: seg_0b9ad8.csv \n inflating: seg_8c1e7d.csv \n inflating: seg_c9f80a.csv \n inflating: seg_e49750.csv \n inflating: seg_0d762e.csv \n inflating: seg_2d5544.csv \n inflating: seg_075340.csv \n inflating: seg_30da11.csv \n inflating: seg_0e9bb3.csv \n inflating: seg_2f1664.csv \n inflating: seg_650de2.csv \n inflating: seg_616c8b.csv \n inflating: seg_04cceb.csv \n inflating: seg_3edf51.csv \n inflating: seg_00e5f7.csv \n inflating: seg_59e1db.csv \n inflating: seg_39300d.csv \n inflating: seg_38001b.csv \n inflating: seg_35dd45.csv \n inflating: seg_168d1c.csv \n inflating: seg_ddbadc.csv \n inflating: seg_e6b236.csv \n inflating: seg_b9365c.csv \n inflating: seg_da80c4.csv \n inflating: seg_8cd159.csv \n inflating: seg_743775.csv \n inflating: seg_23fe27.csv \n inflating: seg_9940e8.csv \n inflating: seg_0165c6.csv \n inflating: seg_4eb127.csv \n inflating: seg_3b95dc.csv \n inflating: seg_ee87e2.csv \n inflating: seg_2b2cb3.csv \n inflating: seg_b9d8a0.csv \n inflating: seg_e3d751.csv \n inflating: seg_c183e4.csv \n inflating: seg_b89e47.csv \n inflating: seg_635770.csv \n inflating: seg_932828.csv \n inflating: seg_ea098b.csv \n inflating: seg_e65e69.csv \n inflating: seg_a1e5b0.csv \n inflating: seg_bcfad6.csv \n inflating: seg_d3f6b0.csv \n inflating: seg_35a2d7.csv \n inflating: seg_15bf25.csv \n inflating: seg_fbff25.csv \n inflating: seg_6fd5bb.csv \n inflating: seg_3bfcf6.csv \n inflating: seg_75e669.csv \n inflating: seg_a82229.csv \n inflating: seg_d795ab.csv \n inflating: seg_ee4ae5.csv \n inflating: seg_77bf79.csv \n inflating: seg_b4f7a8.csv \n inflating: seg_cc2e9b.csv \n inflating: seg_cf5764.csv \n inflating: seg_73b0f7.csv \n inflating: seg_b35174.csv \n inflating: seg_eabb2c.csv \n inflating: seg_138807.csv \n inflating: seg_77e7bf.csv \n inflating: seg_46dd7c.csv \n inflating: seg_dbbb44.csv \n inflating: seg_96cd5d.csv \n inflating: seg_8a7201.csv \n inflating: seg_03d386.csv \n inflating: seg_7bb771.csv \n inflating: seg_f21bf4.csv \n inflating: seg_58e74b.csv \n inflating: seg_d36681.csv \n inflating: seg_dd4594.csv \n inflating: seg_b735a5.csv \n inflating: seg_25e76d.csv \n inflating: seg_9dcae1.csv \n inflating: seg_9a81a3.csv \n inflating: seg_2d7cc4.csv \n inflating: seg_ddd206.csv \n inflating: seg_77db7b.csv \n inflating: seg_aeeb31.csv \n inflating: seg_dc5e63.csv \n inflating: seg_d1cb37.csv \n inflating: seg_8dfc7e.csv \n inflating: seg_691a72.csv \n inflating: seg_ae42b3.csv \n inflating: seg_9bab11.csv \n inflating: seg_57908c.csv \n inflating: seg_d35e50.csv \n inflating: seg_e52f8c.csv \n inflating: seg_af1650.csv \n inflating: seg_342ce6.csv \n inflating: seg_306756.csv \n inflating: seg_6a26de.csv \n inflating: seg_c5325c.csv \n inflating: seg_ef779d.csv \n inflating: seg_725772.csv \n inflating: seg_36b77f.csv \n inflating: seg_aed434.csv \n inflating: seg_1eeb8f.csv \n inflating: seg_63c9a3.csv \n inflating: seg_0870ab.csv \n inflating: seg_813aab.csv \n inflating: seg_de3d5e.csv \n inflating: seg_df0df7.csv \n inflating: seg_2383c2.csv \n inflating: seg_0e4833.csv \n inflating: seg_f6c928.csv \n inflating: seg_f01368.csv \n inflating: seg_4c3a2b.csv \n inflating: seg_755d50.csv \n inflating: seg_ceb942.csv \n inflating: seg_c20c06.csv \n inflating: seg_154361.csv \n inflating: seg_8e7fca.csv \n inflating: seg_ada42c.csv \n inflating: seg_cc4199.csv \n inflating: seg_cff594.csv \n inflating: seg_38ee24.csv \n inflating: seg_7428e7.csv \n inflating: seg_b69455.csv \n inflating: seg_e5deec.csv \n inflating: seg_34a2b8.csv \n inflating: seg_ef47c4.csv \n inflating: seg_ebd77b.csv \n inflating: seg_e76e6e.csv \n inflating: seg_d2ad9a.csv \n inflating: seg_2018c8.csv \n inflating: seg_24e071.csv \n inflating: seg_f6659e.csv \n inflating: seg_b691c5.csv \n inflating: seg_c5256d.csv \n inflating: seg_e8385f.csv \n inflating: seg_428d66.csv \n inflating: seg_74537f.csv \n inflating: seg_ac5c61.csv \n inflating: seg_ca9e39.csv \n inflating: seg_c11828.csv \n inflating: seg_a56c04.csv \n inflating: seg_35b009.csv \n inflating: seg_eacccf.csv \n inflating: seg_d247da.csv \n inflating: seg_87a616.csv \n inflating: seg_482745.csv \n inflating: seg_df352f.csv \n inflating: seg_141f34.csv \n inflating: seg_8244bf.csv \n inflating: seg_24c078.csv \n inflating: seg_0d6d68.csv \n inflating: seg_db4185.csv \n inflating: seg_29d4c6.csv \n inflating: seg_6b536d.csv \n inflating: seg_2cac57.csv \n inflating: seg_56e3e7.csv \n inflating: seg_1a8f2c.csv \n inflating: seg_94c48c.csv \n inflating: seg_98ebaa.csv \n inflating: seg_bc1d67.csv \n inflating: seg_2c5e9f.csv \n inflating: seg_7ddc30.csv \n inflating: seg_16566c.csv \n inflating: seg_c509ff.csv \n inflating: seg_76f3f2.csv \n inflating: seg_71348a.csv \n inflating: seg_eba22d.csv \n inflating: seg_287e60.csv \n inflating: seg_6ab188.csv \n inflating: seg_65fbac.csv \n inflating: seg_377134.csv \n inflating: seg_7cb417.csv \n inflating: seg_8599ec.csv \n inflating: seg_827c07.csv \n inflating: seg_259dd3.csv \n inflating: seg_ea9fa4.csv \n inflating: seg_ffbd6a.csv \n inflating: seg_607559.csv \n inflating: seg_4a6096.csv \n inflating: seg_d508db.csv \n inflating: seg_bdae02.csv \n inflating: seg_e7ac12.csv \n inflating: seg_5a947e.csv \n inflating: seg_d2a533.csv \n inflating: seg_8e6a3f.csv \n inflating: seg_3ce740.csv \n inflating: seg_71c9e9.csv \n inflating: seg_78d817.csv \n inflating: seg_2dc6ff.csv \n inflating: seg_fe86f5.csv \n inflating: seg_c5fc87.csv \n inflating: seg_709b6a.csv \n inflating: seg_1a8f0d.csv \n inflating: seg_3878d0.csv \n inflating: seg_11860f.csv \n inflating: seg_e9df42.csv \n inflating: seg_de4e1d.csv \n inflating: seg_264c7d.csv \n inflating: seg_86720f.csv \n inflating: seg_4c65e6.csv \n inflating: seg_b54024.csv \n inflating: seg_bf1430.csv \n inflating: seg_b36650.csv \n inflating: seg_94d150.csv \n inflating: seg_3d19fc.csv \n inflating: seg_6fb17f.csv \n inflating: seg_3b7175.csv \n inflating: seg_e71dda.csv \n inflating: seg_d62e9c.csv \n inflating: seg_0f74a3.csv \n inflating: seg_d7a805.csv \n inflating: seg_7d65c7.csv \n inflating: seg_76b76f.csv \n inflating: seg_2fb672.csv \n inflating: seg_257b8a.csv \n inflating: seg_96fa35.csv \n inflating: seg_3d581c.csv \n inflating: seg_8b798b.csv \n inflating: seg_cedf3e.csv \n inflating: seg_e24f69.csv \n inflating: seg_0c247b.csv \n inflating: seg_dd8805.csv \n inflating: seg_511d25.csv \n inflating: seg_27db4d.csv \n inflating: seg_cc7a19.csv \n inflating: seg_1da819.csv \n inflating: seg_b2b29d.csv \n inflating: seg_a39fa8.csv \n inflating: seg_7630c5.csv \n inflating: seg_4db886.csv \n inflating: seg_5a67b5.csv \n inflating: seg_e405fe.csv \n inflating: seg_86892f.csv \n inflating: seg_f48db9.csv \n inflating: seg_1db443.csv \n inflating: seg_8c6e34.csv \n inflating: seg_29a073.csv \n inflating: seg_6e12ee.csv \n inflating: seg_11647f.csv \n inflating: seg_7fa6ec.csv \n inflating: seg_9d1dbe.csv \n inflating: seg_edecaa.csv \n inflating: seg_b4c040.csv \n inflating: seg_0b48c4.csv \n inflating: seg_591954.csv \n inflating: seg_0445d7.csv \n inflating: seg_d398df.csv \n inflating: seg_d9b162.csv \n inflating: seg_cbfb19.csv \n inflating: seg_0fd3ff.csv \n inflating: seg_2a0afc.csv \n inflating: seg_588f61.csv \n inflating: seg_a173d8.csv \n inflating: seg_2bf9dd.csv \n inflating: seg_389906.csv \n inflating: seg_f46cc1.csv \n inflating: seg_3b95d2.csv \n inflating: seg_7d994f.csv \n inflating: seg_3065ba.csv \n inflating: seg_0bc877.csv \n inflating: seg_5467c8.csv \n inflating: seg_2700bd.csv \n inflating: seg_dd0b11.csv \n inflating: seg_77f9e3.csv \n inflating: seg_ebec3e.csv \n inflating: seg_14d1d3.csv \n inflating: seg_31d763.csv \n inflating: seg_0e8b79.csv \n inflating: seg_440106.csv \n inflating: seg_853302.csv \n inflating: seg_774ad2.csv \n inflating: seg_974049.csv \n inflating: seg_899bf0.csv \n inflating: seg_e40348.csv \n inflating: seg_f0ce4a.csv \n inflating: seg_4d53c2.csv \n inflating: seg_8e5704.csv \n inflating: seg_41a267.csv \n inflating: seg_63c518.csv \n inflating: seg_0a1bd0.csv \n inflating: seg_293dc2.csv \n inflating: seg_a436bc.csv \n inflating: seg_d9d0e4.csv \n inflating: seg_01ecb0.csv \n inflating: seg_327ed4.csv \n inflating: seg_c2b8f7.csv \n inflating: seg_3cbccf.csv \n inflating: seg_34abd2.csv \n inflating: seg_a8bb2f.csv \n inflating: seg_74ce61.csv \n inflating: seg_695380.csv \n inflating: seg_e30ef5.csv \n inflating: seg_00c35b.csv \n inflating: seg_d16534.csv \n inflating: seg_06893f.csv \n inflating: seg_ec4645.csv \n inflating: seg_ca8389.csv \n inflating: seg_98c0b6.csv \n inflating: seg_3128e6.csv \n inflating: seg_8fb29e.csv \n inflating: seg_266c6e.csv \n inflating: seg_74f330.csv \n inflating: seg_d693b0.csv \n inflating: seg_a8ddf7.csv \n inflating: seg_920132.csv \n inflating: seg_bc437b.csv \n inflating: seg_1815fe.csv \n inflating: seg_81c432.csv \n inflating: seg_7ebdc3.csv \n inflating: seg_d962cc.csv \n inflating: seg_db3446.csv \n inflating: seg_2f4f14.csv \n inflating: seg_a35e7c.csv \n inflating: seg_5639b3.csv \n inflating: seg_730ebe.csv \n inflating: seg_476700.csv \n inflating: seg_a96b47.csv \n inflating: seg_eca2eb.csv \n inflating: seg_90ec68.csv \n inflating: seg_d661ce.csv \n inflating: seg_a0a423.csv \n inflating: seg_419b2a.csv \n inflating: seg_e1a9e0.csv \n inflating: seg_97d9ed.csv \n inflating: seg_a6c5cc.csv \n inflating: seg_15c9f9.csv \n inflating: seg_18d1d0.csv \n inflating: seg_7949da.csv \n inflating: seg_585872.csv \n inflating: seg_7c88d1.csv \n inflating: seg_e4c15a.csv \n inflating: seg_59cfdb.csv \n inflating: seg_ecaef0.csv \n inflating: seg_f34877.csv \n inflating: seg_931244.csv \n inflating: seg_f9eea3.csv \n inflating: seg_814b1f.csv \n inflating: seg_75654d.csv \n inflating: seg_3c8545.csv \n inflating: seg_84a65d.csv \n inflating: seg_84b9d9.csv \n inflating: seg_7c6c31.csv \n inflating: seg_5e635a.csv \n inflating: seg_f2750c.csv \n inflating: seg_330579.csv \n inflating: seg_bf58ce.csv \n inflating: seg_fd19b8.csv \n inflating: seg_335170.csv \n inflating: seg_cb806c.csv \n inflating: seg_826208.csv \n inflating: seg_4cb0d7.csv \n inflating: seg_5495ca.csv \n inflating: seg_4e627b.csv \n inflating: seg_0c1d3a.csv \n inflating: seg_a45455.csv \n inflating: seg_85762d.csv \n inflating: seg_7c0376.csv \n inflating: seg_07f8df.csv \n inflating: seg_4e0c4a.csv \n inflating: seg_913ffc.csv \n inflating: seg_6dd0fc.csv \n inflating: seg_1b78af.csv \n inflating: seg_6681ee.csv \n inflating: seg_65b962.csv \n inflating: seg_845a2a.csv \n inflating: seg_a86231.csv \n inflating: seg_3af649.csv \n inflating: seg_eee89d.csv \n inflating: seg_3fdae0.csv \n inflating: seg_750e7e.csv \n inflating: seg_95f45e.csv \n inflating: seg_b291c0.csv \n inflating: seg_a91c03.csv \n inflating: seg_23f823.csv \n inflating: seg_1d596d.csv \n inflating: seg_b253ee.csv \n inflating: seg_fd8add.csv \n inflating: seg_e5c033.csv \n inflating: seg_cfed24.csv \n inflating: seg_0ba585.csv \n inflating: seg_946d9a.csv \n inflating: seg_5965d8.csv \n inflating: seg_b0d251.csv \n inflating: seg_f0db55.csv \n inflating: seg_aa1adf.csv \n inflating: seg_fd9ac1.csv \n inflating: seg_3f80ef.csv \n inflating: seg_23efb0.csv \n inflating: seg_45e062.csv \n inflating: seg_13c2f0.csv \n inflating: seg_c6f9c2.csv \n inflating: seg_76f1af.csv \n inflating: seg_204dc6.csv \n inflating: seg_492dc6.csv \n inflating: seg_6b2ce0.csv \n inflating: seg_9df32d.csv \n inflating: seg_266d4e.csv \n inflating: seg_66ed4d.csv \n inflating: seg_725e84.csv \n inflating: seg_63582a.csv \n inflating: seg_5254ce.csv \n inflating: seg_903aba.csv \n inflating: seg_25aa9f.csv \n inflating: seg_952faa.csv \n inflating: seg_661629.csv \n inflating: seg_727fc7.csv \n inflating: seg_f455bc.csv \n inflating: seg_ab4bca.csv \n inflating: seg_0bb5fd.csv \n inflating: seg_a5fbcc.csv \n inflating: seg_aa78e0.csv \n inflating: seg_337b8c.csv \n inflating: seg_e5f9ec.csv \n inflating: seg_629822.csv \n inflating: seg_2c3203.csv \n inflating: seg_13e25f.csv \n inflating: seg_d2f588.csv \n inflating: seg_eebbc2.csv \n inflating: seg_e3e762.csv \n inflating: seg_ff1a62.csv \n inflating: seg_3aca44.csv \n inflating: seg_832a4d.csv \n inflating: seg_c80857.csv \n inflating: seg_32f72d.csv \n inflating: seg_d71352.csv \n inflating: seg_48a80c.csv \n inflating: seg_d668c0.csv \n inflating: seg_4280d9.csv \n inflating: seg_35ee83.csv \n inflating: seg_2e2372.csv \n inflating: seg_72b68c.csv \n inflating: seg_902074.csv \n inflating: seg_bb180c.csv \n inflating: seg_136285.csv \n inflating: seg_72ebda.csv \n inflating: seg_1e617a.csv \n inflating: seg_d78e7d.csv \n inflating: seg_30661b.csv \n inflating: seg_2396a2.csv \n inflating: seg_c73df6.csv \n inflating: seg_41aadc.csv \n inflating: seg_ecdca4.csv \n inflating: seg_2465ce.csv \n inflating: seg_3d9aa0.csv \n inflating: seg_c15d21.csv \n inflating: seg_5147e1.csv \n inflating: seg_ddddf0.csv \n inflating: seg_161f5f.csv \n inflating: seg_9a9041.csv \n inflating: seg_c9201c.csv \n inflating: seg_117ff4.csv \n inflating: seg_f96d0e.csv \n inflating: seg_1db8e8.csv \n inflating: seg_ae3388.csv \n inflating: seg_70edf5.csv \n inflating: seg_1827c5.csv \n inflating: seg_dff078.csv \n inflating: seg_20d9de.csv \n inflating: seg_0c3943.csv \n inflating: seg_90a0be.csv \n inflating: seg_8328f5.csv \n inflating: seg_4ae712.csv \n inflating: seg_395e0e.csv \n inflating: seg_59d484.csv \n inflating: seg_597bd6.csv \n inflating: seg_659ff8.csv \n inflating: seg_420d4e.csv \n inflating: seg_f229bf.csv \n inflating: seg_cd9f9b.csv \n inflating: seg_b947ae.csv \n inflating: seg_d78093.csv \n inflating: seg_94d573.csv \n inflating: seg_d63175.csv \n inflating: seg_45b051.csv \n inflating: seg_db90c0.csv \n inflating: seg_d3992e.csv \n inflating: seg_1ece48.csv \n inflating: seg_2dccec.csv \n inflating: seg_b13fb9.csv \n inflating: seg_ea096b.csv \n inflating: seg_c05a7d.csv \n inflating: seg_58ea74.csv \n inflating: seg_afd665.csv \n inflating: seg_a6cd86.csv \n inflating: seg_ff7478.csv \n inflating: seg_5165a4.csv \n inflating: seg_4c12b2.csv \n inflating: seg_c747ad.csv \n inflating: seg_397983.csv \n inflating: seg_2916e3.csv \n inflating: seg_7211f1.csv \n inflating: seg_a0646a.csv \n inflating: seg_14c054.csv \n inflating: seg_708349.csv \n inflating: seg_82303e.csv \n inflating: seg_3ec967.csv \n inflating: seg_bb5e15.csv \n inflating: seg_ea78c0.csv \n inflating: seg_383b6b.csv \n inflating: seg_f1a771.csv \n inflating: seg_316f86.csv \n inflating: seg_2f5695.csv \n inflating: seg_4f83a3.csv \n inflating: seg_055127.csv \n inflating: seg_462c1d.csv \n inflating: seg_a7b454.csv \n inflating: seg_6df48e.csv \n inflating: seg_6d36a1.csv \n inflating: seg_cdf88e.csv \n inflating: seg_49d84b.csv \n inflating: seg_6074f9.csv \n inflating: seg_c90be5.csv \n inflating: seg_c10a58.csv \n inflating: seg_20a990.csv \n inflating: seg_d014a3.csv \n inflating: seg_1fa983.csv \n inflating: seg_d30a9d.csv \n inflating: seg_8d6a7d.csv \n inflating: seg_e50f16.csv \n inflating: seg_18f633.csv \n inflating: seg_533613.csv \n inflating: seg_546864.csv \n inflating: seg_e6e24e.csv \n inflating: seg_e0c329.csv \n inflating: seg_84c7fb.csv \n inflating: seg_d11ca8.csv \n inflating: seg_0a42ba.csv \n inflating: seg_8baeab.csv \n inflating: seg_bf1294.csv \n inflating: seg_5c57a8.csv \n inflating: seg_f97274.csv \n inflating: seg_d3d8fc.csv \n inflating: seg_8456d3.csv \n inflating: seg_a009e6.csv \n inflating: seg_afadfb.csv \n inflating: seg_0dfe8c.csv \n inflating: seg_f1f5b9.csv \n inflating: seg_6a05e7.csv \n inflating: seg_3661de.csv \n inflating: seg_4435bd.csv \n inflating: seg_71b975.csv \n inflating: seg_71f571.csv \n inflating: seg_f5a6ab.csv \n inflating: seg_7a9228.csv \n inflating: seg_e86e44.csv \n inflating: seg_2bf5be.csv \n inflating: seg_0b082e.csv \n inflating: seg_d24095.csv \n inflating: seg_6b4e04.csv \n inflating: seg_38ad16.csv \n inflating: seg_86cfe9.csv \n inflating: seg_ba0485.csv \n inflating: seg_64f8dc.csv \n inflating: seg_301c1e.csv \n inflating: seg_9fc7c2.csv \n inflating: seg_2b4959.csv \n inflating: seg_6e98aa.csv \n inflating: seg_1e0523.csv \n inflating: seg_5559d9.csv \n inflating: seg_fc407c.csv \n inflating: seg_4743ab.csv \n inflating: seg_fb8af5.csv \n inflating: seg_a31fe3.csv \n inflating: seg_f2cb5e.csv \n inflating: seg_75c34b.csv \n inflating: seg_8c0114.csv \n inflating: seg_1f1e60.csv \n inflating: seg_496dad.csv \n inflating: seg_ca6d70.csv \n inflating: seg_289d99.csv \n inflating: seg_4d8f0f.csv \n inflating: seg_ad949a.csv \n inflating: seg_986b6e.csv \n inflating: seg_e80726.csv \n inflating: seg_818382.csv \n inflating: seg_032822.csv \n inflating: seg_74b264.csv \n inflating: seg_d31583.csv \n inflating: seg_93f935.csv \n inflating: seg_b86ce1.csv \n inflating: seg_2e50fb.csv \n inflating: seg_e10d3b.csv \n inflating: seg_7012af.csv \n inflating: seg_ec0e7b.csv \n inflating: seg_226e60.csv \n inflating: seg_31d810.csv \n inflating: seg_8a41bf.csv \n inflating: seg_e96140.csv \n inflating: seg_468fdb.csv \n inflating: seg_a674d2.csv \n inflating: seg_97ad82.csv \n inflating: seg_5a1adb.csv \n inflating: seg_7cbb45.csv \n inflating: seg_419531.csv \n inflating: seg_f639ec.csv \n inflating: seg_89e207.csv \n inflating: seg_db3fbe.csv \n inflating: seg_91607c.csv \n inflating: seg_78fc4b.csv \n inflating: seg_b80366.csv \n inflating: seg_a50d28.csv \n inflating: seg_9d2528.csv \n inflating: seg_a55540.csv \n inflating: seg_fb9491.csv \n inflating: seg_6f3d90.csv \n inflating: seg_0d0250.csv \n inflating: seg_a00985.csv \n inflating: seg_6dac5d.csv \n inflating: seg_92bd37.csv \n inflating: seg_c3f672.csv \n inflating: seg_266805.csv \n inflating: seg_f0cedb.csv \n inflating: seg_2d402d.csv \n inflating: seg_d91492.csv \n inflating: seg_b8745c.csv \n inflating: seg_70a60e.csv \n inflating: seg_4b0839.csv \n inflating: seg_5e020f.csv \n inflating: seg_3e87b4.csv \n inflating: seg_29e799.csv \n inflating: seg_14c89d.csv \n inflating: seg_f7de40.csv \n inflating: seg_2d92f0.csv \n inflating: seg_68da94.csv \n inflating: seg_b26368.csv \n inflating: seg_ccdc28.csv \n inflating: seg_e9272e.csv \n inflating: seg_a9b957.csv \n inflating: seg_14ad27.csv \n inflating: seg_7df3dd.csv \n inflating: seg_478327.csv \n inflating: seg_b0a794.csv \n inflating: seg_b8b72f.csv \n inflating: seg_8ebdea.csv \n inflating: seg_e256bd.csv \n inflating: seg_eb7595.csv \n inflating: seg_53caf1.csv \n inflating: seg_c09a41.csv \n inflating: seg_b9361a.csv \n inflating: seg_166f70.csv \n inflating: seg_5ed47e.csv \n inflating: seg_943de0.csv \n inflating: seg_c3507f.csv \n inflating: seg_4401d8.csv \n inflating: seg_c6c29f.csv \n inflating: seg_e8d02f.csv \n inflating: seg_0fc67a.csv \n inflating: seg_73858d.csv \n inflating: seg_2f9336.csv \n inflating: seg_093a0a.csv \n inflating: seg_a99be9.csv \n inflating: seg_b03de5.csv \n inflating: seg_e9e65b.csv \n inflating: seg_86e874.csv \n inflating: seg_d1a2b9.csv \n inflating: seg_a99799.csv \n inflating: seg_fde86d.csv \n inflating: seg_4743fa.csv \n inflating: seg_c62a06.csv \n inflating: seg_502fab.csv \n inflating: seg_ef9105.csv \n inflating: seg_003339.csv \n inflating: seg_be65f2.csv \n inflating: seg_b21273.csv \n inflating: seg_67aa76.csv \n inflating: seg_115617.csv \n inflating: seg_9d452d.csv \n inflating: seg_a9089b.csv \n inflating: seg_d56d32.csv \n inflating: seg_077b7e.csv \n inflating: seg_df8385.csv \n inflating: seg_a03e6a.csv \n inflating: seg_c01a57.csv \n inflating: seg_90c258.csv \n inflating: seg_1cd352.csv \n inflating: seg_dc1cf0.csv \n inflating: seg_b9ec4b.csv \n inflating: seg_bd7109.csv \n inflating: seg_3fc7ca.csv \n inflating: seg_f59974.csv \n inflating: seg_cc4932.csv \n inflating: seg_60277f.csv \n inflating: seg_14f5f0.csv \n inflating: seg_a58a6e.csv \n inflating: seg_1c904f.csv \n inflating: seg_214492.csv \n inflating: seg_ba5e7e.csv \n inflating: seg_8f4654.csv \n inflating: seg_421a96.csv \n inflating: seg_e0cfd4.csv \n inflating: seg_8be2da.csv \n inflating: seg_cf0e43.csv \n inflating: seg_0df469.csv \n inflating: seg_00be11.csv \n inflating: seg_ba9dbf.csv \n inflating: seg_1baf33.csv \n inflating: seg_be30ce.csv \n inflating: seg_c666dd.csv \n inflating: seg_622e9d.csv \n inflating: seg_de8180.csv \n inflating: seg_8bc20c.csv \n inflating: seg_114b79.csv \n inflating: seg_c9e533.csv \n inflating: seg_f93ce5.csv \n inflating: seg_19b9dd.csv \n inflating: seg_15adff.csv \n inflating: seg_8515fd.csv \n inflating: seg_434c96.csv \n inflating: seg_e5fbc1.csv \n inflating: seg_43c6f4.csv \n inflating: seg_724df9.csv \n inflating: seg_81c1a6.csv \n inflating: seg_8db6f0.csv \n inflating: seg_259d65.csv \n inflating: seg_075f77.csv \n inflating: seg_ac9de5.csv \n inflating: seg_4c4995.csv \n inflating: seg_bd6f5f.csv \n inflating: seg_9d1b26.csv \n inflating: seg_6f2222.csv \n inflating: seg_232559.csv \n inflating: seg_5c8ae5.csv \n inflating: seg_d2cb02.csv \n inflating: seg_6edcb8.csv \n inflating: seg_0488a1.csv \n inflating: seg_42e7e8.csv \n inflating: seg_40f810.csv \n inflating: seg_130e17.csv \n inflating: seg_6696d0.csv \n inflating: seg_c46ce6.csv \n inflating: seg_3631ec.csv \n inflating: seg_d1045e.csv \n inflating: seg_2c69e4.csv \n inflating: seg_1d044c.csv \n inflating: seg_d32f8a.csv \n inflating: seg_9c3d6a.csv \n inflating: seg_0339ca.csv \n inflating: seg_1ac9e2.csv \n inflating: seg_868c85.csv \n inflating: seg_a86a05.csv \n inflating: seg_77df38.csv \n inflating: seg_b89218.csv \n inflating: seg_fe70c0.csv \n inflating: seg_d8014d.csv \n inflating: seg_fbddb9.csv \n inflating: seg_a8e71a.csv \n inflating: seg_59cd7d.csv \n inflating: seg_369570.csv \n inflating: seg_12ac20.csv \n inflating: seg_970e86.csv \n inflating: seg_df45b0.csv \n inflating: seg_69a53e.csv \n inflating: seg_9c3ef8.csv \n inflating: seg_aa0180.csv \n inflating: seg_18f853.csv \n inflating: seg_12ee6e.csv \n inflating: seg_eb692c.csv \n inflating: seg_62c582.csv \n inflating: seg_e8ac98.csv \n inflating: seg_d9a2bb.csv \n inflating: seg_91cff2.csv \n inflating: seg_758ca4.csv \n inflating: seg_004ee5.csv \n inflating: seg_476bdb.csv \n inflating: seg_e05de2.csv \n inflating: seg_fad815.csv \n inflating: seg_91f512.csv \n inflating: seg_739856.csv \n inflating: seg_7480f4.csv \n inflating: seg_08f441.csv \n inflating: seg_b4402d.csv \n inflating: seg_5d92af.csv \n inflating: seg_091c9d.csv \n inflating: seg_ae9761.csv \n inflating: seg_947700.csv \n inflating: seg_060ce4.csv \n inflating: seg_4848e3.csv \n inflating: seg_85e9c6.csv \n inflating: seg_2642d4.csv \n inflating: seg_36e32d.csv \n inflating: seg_1144bc.csv \n inflating: seg_ab9654.csv \n inflating: seg_919a6a.csv \n inflating: seg_739fb0.csv \n inflating: seg_1c4c5b.csv \n inflating: seg_54af0d.csv \n inflating: seg_ad3a44.csv \n inflating: seg_09ed4e.csv \n inflating: seg_01a8dc.csv \n inflating: seg_3d1cb3.csv \n inflating: seg_55a0c3.csv \n inflating: seg_a84c13.csv \n inflating: seg_e30e44.csv \n inflating: seg_586c0f.csv \n inflating: seg_8d030a.csv \n inflating: seg_6e1ee7.csv \n inflating: seg_e266f4.csv \n inflating: seg_3c4106.csv \n inflating: seg_aee9bf.csv \n inflating: seg_3b30fd.csv \n inflating: seg_bf2d1f.csv \n inflating: seg_e3dafa.csv \n inflating: seg_9e7dff.csv \n inflating: seg_e15cb9.csv \n inflating: seg_7f5b6f.csv \n inflating: seg_e217fc.csv \n inflating: seg_0cca14.csv \n inflating: seg_f931de.csv \n inflating: seg_89d8a0.csv \n inflating: seg_e47d2b.csv \n inflating: seg_ee8f87.csv \n inflating: seg_c365de.csv \n inflating: seg_51865e.csv \n inflating: seg_2c6011.csv \n inflating: seg_a5271e.csv \n inflating: seg_9caf50.csv \n inflating: seg_a7d0eb.csv \n inflating: seg_14c435.csv \n inflating: seg_e1daa0.csv \n inflating: seg_73565e.csv \n inflating: seg_55a9a4.csv \n inflating: seg_cd0691.csv \n inflating: seg_f88d4a.csv \n inflating: seg_8bf229.csv \n inflating: seg_e742dc.csv \n inflating: seg_117278.csv \n inflating: seg_004314.csv \n inflating: seg_c26fb4.csv \n inflating: seg_e1f081.csv \n inflating: seg_c24946.csv \n inflating: seg_76d16c.csv \n inflating: seg_29d497.csv \n inflating: seg_f70b66.csv \n inflating: seg_687a5c.csv \n inflating: seg_1201e8.csv \n inflating: seg_4ae37c.csv \n inflating: seg_c6f533.csv \n inflating: seg_8f6284.csv \n inflating: seg_f056d8.csv \n inflating: seg_016cdb.csv \n inflating: seg_484901.csv \n inflating: seg_4dfa03.csv \n inflating: seg_9485c0.csv \n inflating: seg_fe2aac.csv \n inflating: seg_54490e.csv \n inflating: seg_878d2c.csv \n inflating: seg_2977cd.csv \n inflating: seg_f5406b.csv \n inflating: seg_065588.csv \n inflating: seg_c65135.csv \n inflating: seg_6b3a9d.csv \n inflating: seg_7f93b2.csv \n inflating: seg_d56ae2.csv \n inflating: seg_9872f9.csv \n inflating: seg_30207d.csv \n inflating: seg_061a33.csv \n inflating: seg_bff05e.csv \n inflating: seg_f99b64.csv \n inflating: seg_31d7bf.csv \n inflating: seg_42ff14.csv \n inflating: seg_c60dca.csv \n inflating: seg_d1eee8.csv \n inflating: seg_81a8ea.csv \n inflating: seg_4a91a9.csv \n inflating: seg_596ae3.csv \n inflating: seg_b25dde.csv \n inflating: seg_c44bbc.csv \n inflating: seg_0ebfd9.csv \n inflating: seg_f6e1ca.csv \n inflating: seg_f3c578.csv \n inflating: seg_5255c0.csv \n inflating: seg_f383fb.csv \n inflating: seg_b1b95e.csv \n inflating: seg_be0dfa.csv \n inflating: seg_5887a8.csv \n inflating: seg_1dec9c.csv \n inflating: seg_dc23c0.csv \n inflating: seg_a1b3cc.csv \n inflating: seg_8dd2ab.csv \n inflating: seg_27f532.csv \n inflating: seg_00648a.csv \n inflating: seg_a06228.csv \n inflating: seg_a802fa.csv \n inflating: seg_b6c10d.csv \n inflating: seg_8f592b.csv \n inflating: seg_6f17de.csv \n inflating: seg_cacd0a.csv \n inflating: seg_a7561c.csv \n inflating: seg_6de4fd.csv \n inflating: seg_331f2b.csv \n inflating: seg_b2b331.csv \n inflating: seg_97a161.csv \n inflating: seg_0e9987.csv \n inflating: seg_6e572a.csv \n inflating: seg_9757ac.csv \n inflating: seg_4acc4b.csv \n inflating: seg_bb2d77.csv \n inflating: seg_7ce9cb.csv \n inflating: seg_be0cdc.csv \n inflating: seg_a3d4d6.csv \n inflating: seg_512b3e.csv \n inflating: seg_d6b198.csv \n inflating: seg_07fc6d.csv \n inflating: seg_715ff6.csv \n inflating: seg_c81fec.csv \n inflating: seg_59eb97.csv \n inflating: seg_e98217.csv \n inflating: seg_85fb0a.csv \n inflating: seg_c25855.csv \n inflating: seg_a3ab46.csv \n inflating: seg_b2e045.csv \n inflating: seg_66820a.csv \n inflating: seg_c7a401.csv \n inflating: seg_070bb0.csv \n inflating: seg_496d07.csv \n inflating: seg_d9352b.csv \n inflating: seg_76106f.csv \n inflating: seg_268956.csv \n inflating: seg_5c0929.csv \n inflating: seg_8f4781.csv \n inflating: seg_83bef8.csv \n inflating: seg_acd715.csv \n inflating: seg_50b048.csv \n inflating: seg_05f126.csv \n inflating: seg_4e2c66.csv \n inflating: seg_5311d1.csv \n inflating: seg_6d4109.csv \n inflating: seg_f52cdc.csv \n inflating: seg_049957.csv \n inflating: seg_8dfd7a.csv \n inflating: seg_49fdf0.csv \n inflating: seg_561cf4.csv \n inflating: seg_d39131.csv \n inflating: seg_a4e745.csv \n inflating: seg_397dc9.csv \n inflating: seg_790108.csv \n inflating: seg_7020c5.csv \n inflating: seg_b95010.csv \n inflating: seg_e6e1df.csv \n inflating: seg_63c13d.csv \n inflating: seg_ed3e4c.csv \n inflating: seg_686edc.csv \n inflating: seg_5d94b2.csv \n inflating: seg_02042f.csv \n inflating: seg_46012e.csv \n inflating: seg_53e645.csv \n inflating: seg_0aa0fb.csv \n inflating: seg_b108c9.csv \n inflating: seg_a88097.csv \n inflating: seg_a53e4c.csv \n inflating: seg_edda79.csv \n inflating: seg_06bbc5.csv \n inflating: seg_b687fc.csv \n inflating: seg_979a8f.csv \n inflating: seg_2f86cd.csv \n inflating: seg_ad3cf0.csv \n inflating: seg_90bf60.csv \n inflating: seg_9e25e9.csv \n inflating: seg_a55a17.csv \n inflating: seg_753ba6.csv \n inflating: seg_fea3e2.csv \n inflating: seg_06d7ba.csv \n inflating: seg_de225c.csv \n inflating: seg_2ba79a.csv \n inflating: seg_20cc25.csv \n inflating: seg_cc2c46.csv \n inflating: seg_57ea44.csv \n inflating: seg_82316b.csv \n inflating: seg_b0bf65.csv \n inflating: seg_8bec6f.csv \n inflating: seg_06d4a2.csv \n inflating: seg_fa6383.csv \n inflating: seg_9f4fd4.csv \n inflating: seg_ae9626.csv \n inflating: seg_5975f4.csv \n inflating: seg_bf4ec0.csv \n inflating: seg_c607eb.csv \n inflating: seg_9c2843.csv \n inflating: seg_a93a88.csv \n inflating: seg_c186dc.csv \n inflating: seg_bd00c4.csv \n inflating: seg_5b7325.csv \n inflating: seg_e3ca24.csv \n inflating: seg_8777bd.csv \n inflating: seg_162fc4.csv \n inflating: seg_2dcd3a.csv \n inflating: seg_983778.csv \n inflating: seg_8d6578.csv \n inflating: seg_464839.csv \n inflating: seg_580b65.csv \n inflating: seg_b52add.csv \n inflating: seg_4db997.csv \n inflating: seg_f0574b.csv \n inflating: seg_447972.csv \n inflating: seg_105cd9.csv \n inflating: seg_d26f7d.csv \n inflating: seg_a420d4.csv \n inflating: seg_759fb1.csv \n inflating: seg_53a5fd.csv \n inflating: seg_500108.csv \n inflating: seg_71238c.csv \n inflating: seg_652cc2.csv \n inflating: seg_6a116b.csv \n inflating: seg_aa9b8e.csv \n inflating: seg_2be983.csv \n inflating: seg_fbf17a.csv \n inflating: seg_715e2c.csv \n inflating: seg_a99161.csv \n inflating: seg_7e9eb7.csv \n inflating: seg_9122aa.csv \n inflating: seg_58d52d.csv \n inflating: seg_b77bf5.csv \n inflating: seg_5e668a.csv \n inflating: seg_2aa101.csv \n inflating: seg_dc73b8.csv \n inflating: seg_934087.csv \n inflating: seg_07cd72.csv \n inflating: seg_29c906.csv \n inflating: seg_d59e4e.csv \n inflating: seg_0b3b94.csv \n inflating: seg_e877a5.csv \n inflating: seg_beda44.csv \n inflating: seg_50e270.csv \n inflating: seg_cc667b.csv \n inflating: seg_04b87a.csv \n inflating: seg_517425.csv \n inflating: seg_8e7194.csv \n inflating: seg_524b39.csv \n inflating: seg_9b7ef8.csv \n inflating: seg_ecf28d.csv \n inflating: seg_b853f8.csv \n inflating: seg_311499.csv \n inflating: seg_8dd3ba.csv \n inflating: seg_6422d1.csv \n inflating: seg_51f6fd.csv \n inflating: seg_1a8e06.csv \n inflating: seg_b5804a.csv \n inflating: seg_5a0479.csv \n inflating: seg_66b7c3.csv \n inflating: seg_3ba485.csv \n inflating: seg_7cd6b1.csv \n inflating: seg_735cdd.csv \n inflating: seg_c747b4.csv \n inflating: seg_32f6df.csv \n inflating: seg_123dd4.csv \n inflating: seg_5f46a5.csv \n inflating: seg_61219c.csv \n inflating: seg_6be48e.csv \n inflating: seg_fe5945.csv \n inflating: seg_bcf6af.csv \n inflating: seg_acf2f4.csv \n inflating: seg_75e93c.csv \n inflating: seg_a5934a.csv \n inflating: seg_0b54ac.csv \n inflating: seg_26a2a0.csv \n inflating: seg_2e917f.csv \n inflating: seg_aaa2ae.csv \n inflating: seg_aae025.csv \n inflating: seg_b87cf5.csv \n inflating: seg_0c4e31.csv \n inflating: seg_9a92db.csv \n inflating: seg_93211c.csv \n inflating: seg_b0cf32.csv \n inflating: seg_a9f80e.csv \n inflating: seg_7c2c48.csv \n inflating: seg_80b9b4.csv \n inflating: seg_04ae38.csv \n inflating: seg_5a242e.csv \n inflating: seg_d60d18.csv \n inflating: seg_4ea81c.csv \n inflating: seg_c94941.csv \n inflating: seg_3761d0.csv \n inflating: seg_bbbc30.csv \n inflating: seg_fdba93.csv \n inflating: seg_82ce27.csv \n inflating: seg_b51b7c.csv \n inflating: seg_dbc615.csv \n inflating: seg_80622d.csv \n inflating: seg_ce7537.csv \n inflating: seg_dcb51d.csv \n inflating: seg_0d04b9.csv \n inflating: seg_e59dca.csv \n inflating: seg_a1acb2.csv \n inflating: seg_d6dad6.csv \n inflating: seg_0125d9.csv \n inflating: seg_bff392.csv \n inflating: seg_f60872.csv \n inflating: seg_a2e2c8.csv \n inflating: seg_bb6904.csv \n inflating: seg_170a0c.csv \n inflating: seg_827a21.csv \n inflating: seg_e24496.csv \n inflating: seg_c22bd4.csv \n inflating: seg_e81b50.csv \n inflating: seg_e27c0a.csv \n inflating: seg_fd6340.csv \n inflating: seg_6c34c2.csv \n inflating: seg_006e4a.csv \n inflating: seg_f2b0c5.csv \n inflating: seg_545fda.csv \n inflating: seg_70be4d.csv \n inflating: seg_d3f967.csv \n inflating: seg_2d427f.csv \n inflating: seg_21eebb.csv \n inflating: seg_bee792.csv \n inflating: seg_e9aa44.csv \n inflating: seg_d9ce67.csv \n inflating: seg_bd4d30.csv \n inflating: seg_355d19.csv \n inflating: seg_04a696.csv \n inflating: seg_46c6d3.csv \n inflating: seg_456149.csv \n inflating: seg_5445bc.csv \n inflating: seg_bfe0b6.csv \n inflating: seg_f1b916.csv \n inflating: seg_0ba210.csv \n inflating: seg_4d0041.csv \n inflating: seg_10f3a6.csv \n inflating: seg_17d1ff.csv \n inflating: seg_d27193.csv \n inflating: seg_211299.csv \n inflating: seg_c27591.csv \n inflating: seg_477c83.csv \n inflating: seg_5b8c39.csv \n inflating: seg_9d3013.csv \n inflating: seg_d1abaa.csv \n inflating: seg_0775a7.csv \n inflating: seg_b402ac.csv \n inflating: seg_fa488e.csv \n inflating: seg_718b53.csv \n inflating: seg_92092a.csv \n inflating: seg_b47c19.csv \n inflating: seg_36147d.csv \n inflating: seg_b7a8a1.csv \n inflating: seg_6b9d02.csv \n inflating: seg_ba4ea3.csv \n inflating: seg_f71db8.csv \n inflating: seg_1f5240.csv \n inflating: seg_6d01a3.csv \n inflating: seg_679dd2.csv \n inflating: seg_259c63.csv \n inflating: seg_9825e0.csv \n inflating: seg_c47bdc.csv \n inflating: seg_961d02.csv \n inflating: seg_d7967e.csv \n inflating: seg_a6eed3.csv \n inflating: seg_1f7cf8.csv \n inflating: seg_af703b.csv \n inflating: seg_73ab4a.csv \n inflating: seg_fcca66.csv \n inflating: seg_703cea.csv \n inflating: seg_263deb.csv \n inflating: seg_5759c9.csv \n inflating: seg_b9de03.csv \n inflating: seg_0df75c.csv \n inflating: seg_f6d51b.csv \n inflating: seg_8c341c.csv \n inflating: seg_aee7c0.csv \n inflating: seg_74d8cc.csv \n inflating: seg_1084af.csv \n inflating: seg_ec973c.csv \n inflating: seg_b91ae2.csv \n inflating: seg_e7d3e7.csv \n inflating: seg_f734c8.csv \n inflating: seg_63c983.csv \n inflating: seg_5f352e.csv \n inflating: seg_78852d.csv \n inflating: seg_b695b3.csv \n inflating: seg_6ac82a.csv \n inflating: seg_0c12cc.csv \n inflating: seg_7862ed.csv \n inflating: seg_c18d5a.csv \n inflating: seg_56e6f1.csv \n inflating: seg_96db5e.csv \n inflating: seg_f96d5a.csv \n inflating: seg_739679.csv \n inflating: seg_f0d278.csv \n inflating: seg_c571ef.csv \n inflating: seg_318135.csv \n inflating: seg_0b4b24.csv \n inflating: seg_bd40f2.csv \n inflating: seg_f6b70c.csv \n inflating: seg_804fb5.csv \n inflating: seg_8cdf83.csv \n inflating: seg_4ab7bf.csv \n inflating: seg_5597ed.csv \n inflating: seg_f0181d.csv \n inflating: seg_eb2e11.csv \n inflating: seg_92310f.csv \n inflating: seg_9d6ff0.csv \n inflating: seg_7540f3.csv \n inflating: seg_20b8d0.csv \n inflating: seg_95357d.csv \n inflating: seg_69787a.csv \n inflating: seg_c11a4f.csv \n inflating: seg_18096a.csv \n inflating: seg_c714d0.csv \n inflating: seg_bc2186.csv \n inflating: seg_c1fe9a.csv \n inflating: seg_87f6c8.csv \n inflating: seg_a87e79.csv \n inflating: seg_b8fd73.csv \n inflating: seg_fa9f00.csv \n inflating: seg_c047b7.csv \n inflating: seg_c21d60.csv \n inflating: seg_da21ee.csv \n inflating: seg_89eed6.csv \n inflating: seg_29475f.csv \n inflating: seg_98a819.csv \n inflating: seg_4c067a.csv \n inflating: seg_530a02.csv \n inflating: seg_241266.csv \n inflating: seg_9f57c4.csv \n inflating: seg_2cdbe3.csv \n inflating: seg_ab3f14.csv \n inflating: seg_f7d792.csv \n inflating: seg_71936f.csv \n inflating: seg_758374.csv \n inflating: seg_f02d32.csv \n inflating: seg_75438e.csv \n inflating: seg_a543e5.csv \n inflating: seg_bc07c8.csv \n inflating: seg_180481.csv \n inflating: seg_18fb65.csv \n inflating: seg_b1a35c.csv \n inflating: seg_274a1a.csv \n inflating: seg_1ae362.csv \n inflating: seg_48b545.csv \n inflating: seg_91bf8c.csv \n inflating: seg_aa508a.csv \n inflating: seg_66e45a.csv \n inflating: seg_6951e0.csv \n inflating: seg_683ddf.csv \n inflating: seg_2367fb.csv \n inflating: seg_84fc12.csv \n inflating: seg_f650cc.csv \n inflating: seg_a6e801.csv \n inflating: seg_2a0dc0.csv \n inflating: seg_25ab3f.csv \n inflating: seg_a18246.csv \n inflating: seg_4587bb.csv \n inflating: seg_7bec10.csv \n inflating: seg_0f565c.csv \n inflating: seg_c60f1d.csv \n inflating: seg_f2f5a3.csv \n inflating: seg_c08d36.csv \n inflating: seg_feb312.csv \n inflating: seg_2eccb9.csv \n inflating: seg_bd31bf.csv \n inflating: seg_e34052.csv \n inflating: seg_a6959c.csv \n inflating: seg_24f338.csv \n inflating: seg_6eb146.csv \n inflating: seg_e29670.csv \n inflating: seg_ca67ee.csv \n inflating: seg_17e596.csv \n inflating: seg_eb47a9.csv \n inflating: seg_d90c5f.csv \n inflating: seg_c0e1b9.csv \n inflating: seg_21faa9.csv \n inflating: seg_00184e.csv \n inflating: seg_885f53.csv \n inflating: seg_72f40a.csv \n inflating: seg_a5d37f.csv \n inflating: seg_0fca83.csv \n inflating: seg_280863.csv \n inflating: seg_a1d8da.csv \n inflating: seg_9ad925.csv \n inflating: seg_ce9f59.csv \n inflating: seg_6c292b.csv \n inflating: seg_2005a7.csv \n inflating: seg_b52dac.csv \n inflating: seg_5abfdd.csv \n inflating: seg_0b07c7.csv \n inflating: seg_9626a4.csv \n inflating: seg_373ba6.csv \n inflating: seg_f9e4f2.csv \n inflating: seg_9802c3.csv \n inflating: seg_cc4950.csv \n inflating: seg_5d45db.csv \n inflating: seg_a2c108.csv \n inflating: seg_610379.csv \n inflating: seg_7bf143.csv \n inflating: seg_613702.csv \n inflating: seg_c0d7da.csv \n inflating: seg_41f45e.csv \n inflating: seg_f19f8e.csv \n inflating: seg_158691.csv \n inflating: seg_eaa0ec.csv \n inflating: seg_b32d1d.csv \n inflating: seg_e48c23.csv \n inflating: seg_0b76f5.csv \n inflating: seg_2dcd84.csv \n inflating: seg_961ddf.csv \n inflating: seg_760d70.csv \n inflating: seg_e6bd3f.csv \n inflating: seg_655cae.csv \n inflating: seg_14c8ce.csv \n inflating: seg_d5d3a1.csv \n inflating: seg_b78883.csv \n inflating: seg_f43730.csv \n inflating: seg_7d17eb.csv \n inflating: seg_dc4698.csv \n inflating: seg_3340b9.csv \n inflating: seg_970b35.csv \n inflating: seg_e54932.csv \n inflating: seg_eccb17.csv \n inflating: seg_0b9ba3.csv \n inflating: seg_bb92b1.csv \n inflating: seg_0042cc.csv \n inflating: seg_907c52.csv \n inflating: seg_4d18f4.csv \n inflating: seg_5c1a4d.csv \n inflating: seg_70b375.csv \n inflating: seg_3d6e79.csv \n inflating: seg_3f5be6.csv \n inflating: seg_31a6cd.csv \n inflating: seg_455b16.csv \n inflating: seg_9f042e.csv \n inflating: seg_43383f.csv \n inflating: seg_f6e89f.csv \n inflating: seg_24157a.csv \n inflating: seg_a6bf91.csv \n inflating: seg_217eed.csv \n inflating: seg_53bdf5.csv \n inflating: seg_705463.csv \n inflating: seg_aec276.csv \n inflating: seg_30b043.csv \n inflating: seg_529c99.csv \n inflating: seg_1e0b82.csv \n inflating: seg_a63573.csv \n inflating: seg_570a20.csv \n inflating: seg_9ee0aa.csv \n inflating: seg_939f5c.csv \n inflating: seg_3adb1a.csv \n inflating: seg_0968f1.csv \n inflating: seg_a35c82.csv \n inflating: seg_2bcbde.csv \n inflating: seg_fa796b.csv \n inflating: seg_e5750a.csv \n inflating: seg_5407b0.csv \n inflating: seg_746d1d.csv \n inflating: seg_5e0902.csv \n inflating: seg_210388.csv \n inflating: seg_1c4e72.csv \n inflating: seg_83cb2c.csv \n inflating: seg_abb06a.csv \n inflating: seg_cecd29.csv \n inflating: seg_ee4479.csv \n inflating: seg_67330f.csv \n inflating: seg_be4612.csv \n inflating: seg_71b4e0.csv \n inflating: seg_b8725b.csv \n inflating: seg_6c8a45.csv \n inflating: seg_aa98cc.csv \n inflating: seg_1d980f.csv \n inflating: seg_e1f045.csv \n inflating: seg_8773cf.csv \n inflating: seg_2313d1.csv \n inflating: seg_c901c0.csv \n inflating: seg_00a37e.csv \n inflating: seg_4a2525.csv \n inflating: seg_4dbbd1.csv \n inflating: seg_a247ac.csv \n inflating: seg_633651.csv \n inflating: seg_071067.csv \n inflating: seg_9a7c46.csv \n inflating: seg_d740b2.csv \n inflating: seg_88b289.csv \n inflating: seg_6e5a38.csv \n inflating: seg_658bf4.csv \n inflating: seg_85c1c0.csv \n inflating: seg_2efd5c.csv \n inflating: seg_17799c.csv \n inflating: seg_f6828d.csv \n inflating: seg_8d3a9d.csv \n inflating: seg_3bd97e.csv \n inflating: seg_d0f803.csv \n inflating: seg_692e7a.csv \n inflating: seg_74ffde.csv \n inflating: seg_59058a.csv \n inflating: seg_83dc2e.csv \n inflating: seg_63130f.csv \n inflating: seg_19515c.csv \n inflating: seg_c17817.csv \n inflating: seg_1f3ede.csv \n inflating: seg_6ed49a.csv \n inflating: seg_2a6343.csv \n inflating: seg_8aeb99.csv \n inflating: seg_2e9a47.csv \n inflating: seg_586726.csv \n inflating: seg_310230.csv \n inflating: seg_ba3e74.csv \n inflating: seg_eb1d6e.csv \n inflating: seg_398a25.csv \n inflating: seg_b95a77.csv \n inflating: seg_c07b7c.csv \n inflating: seg_80fb86.csv \n inflating: seg_22e509.csv \n inflating: seg_32ad0f.csv \n inflating: seg_0012b5.csv \n inflating: seg_ade769.csv \n inflating: seg_3b2013.csv \n inflating: seg_1e572b.csv \n inflating: seg_f43ab6.csv \n inflating: seg_1ef708.csv \n inflating: seg_52c139.csv \n inflating: seg_7cdfe5.csv \n inflating: seg_0eb333.csv \n inflating: seg_54b0ee.csv \n inflating: seg_26cba3.csv \n inflating: seg_670bde.csv \n inflating: seg_65c4e1.csv \n inflating: seg_53fa13.csv \n inflating: seg_523945.csv \n inflating: seg_9c00d7.csv \n inflating: seg_836aac.csv \n inflating: seg_411225.csv \n inflating: seg_8a6b0e.csv \n inflating: seg_49b0cb.csv \n inflating: seg_c2a0c8.csv \n inflating: seg_957ece.csv \n inflating: seg_9fe8b9.csv \n inflating: seg_dcbe02.csv \n inflating: seg_1bd38e.csv \n inflating: seg_47a48f.csv \n inflating: seg_cf9a49.csv \n inflating: seg_5d333e.csv \n inflating: seg_4f5931.csv \n inflating: seg_99f76c.csv \n inflating: seg_6a4ad1.csv \n inflating: seg_115a92.csv \n inflating: seg_00030f.csv \n inflating: seg_fedbd1.csv \n inflating: seg_c3f8d7.csv \n inflating: seg_376908.csv \n inflating: seg_d4ea17.csv \n inflating: seg_f342a3.csv \n inflating: seg_f6cfd4.csv \n inflating: seg_99ccbd.csv \n inflating: seg_c5dee4.csv \n inflating: seg_6cfb76.csv \n inflating: seg_8a3306.csv \n inflating: seg_db606c.csv \n inflating: seg_8200d2.csv \n inflating: seg_f6f523.csv \n inflating: seg_3e00cd.csv \n inflating: seg_950048.csv \n inflating: seg_fb1f5c.csv \n inflating: seg_946d71.csv \n inflating: seg_bca500.csv \n inflating: seg_3e55d5.csv \n inflating: seg_2e64af.csv \n inflating: seg_c42490.csv \n inflating: seg_295b1c.csv \n inflating: seg_8b39c4.csv \n inflating: seg_3d6aac.csv \n inflating: seg_8826f4.csv \n inflating: seg_6f650f.csv \n inflating: seg_1a0e94.csv \n inflating: seg_0b32f7.csv \n inflating: seg_7a5243.csv \n inflating: seg_552b8e.csv \n inflating: seg_c627fc.csv \n inflating: seg_2ece02.csv \n inflating: seg_ebe36c.csv \n inflating: seg_b6254c.csv \n inflating: seg_74dda9.csv \n inflating: seg_b3d886.csv \n inflating: seg_a39f3b.csv \n inflating: seg_77dbe7.csv \n inflating: seg_ab644e.csv \n inflating: seg_99f677.csv \n inflating: seg_4ca6de.csv \n inflating: seg_30501b.csv \n inflating: seg_5cfdc4.csv \n inflating: seg_2b372b.csv \n inflating: seg_0144cb.csv \n inflating: seg_25c8dc.csv \n inflating: seg_39a886.csv \n inflating: seg_750244.csv \n inflating: seg_9d57a1.csv \n inflating: seg_beb650.csv \n inflating: seg_5c9077.csv \n inflating: seg_247262.csv \n inflating: seg_a2fd8b.csv \n inflating: seg_0620e6.csv \n inflating: seg_76f76d.csv \n inflating: seg_e86f77.csv \n inflating: seg_9a74d8.csv \n inflating: seg_1eae76.csv \n inflating: seg_aba501.csv \n inflating: seg_6de935.csv \n inflating: seg_ff2f2d.csv \n inflating: seg_fbe3c2.csv \n inflating: seg_112b81.csv \n inflating: seg_4eeaef.csv \n inflating: seg_fde767.csv \n inflating: seg_848695.csv \n inflating: seg_9d79d4.csv \n inflating: seg_750c93.csv \n inflating: seg_8353f5.csv \n inflating: seg_79e301.csv \n inflating: seg_1e677e.csv \n inflating: seg_e7d1f8.csv \n inflating: seg_9a7d1d.csv \n inflating: seg_0c8502.csv \n inflating: seg_f24292.csv \n inflating: seg_2a2f1e.csv \n inflating: seg_284923.csv \n inflating: seg_96dab2.csv \n inflating: seg_00f3b9.csv \n inflating: seg_70e891.csv \n inflating: seg_cb0e21.csv \n inflating: seg_da7c3b.csv \n inflating: seg_b197e2.csv \n inflating: seg_ca2d1b.csv \n inflating: seg_c58ca5.csv \n inflating: seg_c35940.csv \n inflating: seg_74d58a.csv \n inflating: seg_0dcc40.csv \n inflating: seg_c7a579.csv \n inflating: seg_fb76ca.csv \n inflating: seg_c9481f.csv \n inflating: seg_05a4ad.csv \n inflating: seg_0dc38f.csv \n inflating: seg_f95fd3.csv \n inflating: seg_8509db.csv \n inflating: seg_b66b87.csv \n inflating: seg_5f24d3.csv \n inflating: seg_1b2298.csv \n inflating: seg_b9619c.csv \n inflating: seg_bb171f.csv \n inflating: seg_b7eeb6.csv \n inflating: seg_122ba9.csv \n inflating: seg_6b0107.csv \n inflating: seg_f6c0cb.csv \n inflating: seg_518b1e.csv \n inflating: seg_64bcb9.csv \n inflating: seg_53d818.csv \n inflating: seg_ceab44.csv \n inflating: seg_6f9a98.csv \n inflating: seg_d21eb6.csv \n inflating: seg_2597af.csv \n inflating: seg_35c587.csv \n inflating: seg_9ad261.csv \n inflating: seg_339f80.csv \n inflating: seg_de3237.csv \n inflating: seg_407b2b.csv \n inflating: seg_a25c46.csv \n inflating: seg_c522f8.csv \n inflating: seg_cacd58.csv \n inflating: seg_ba3c47.csv \n inflating: seg_ecf81b.csv \n inflating: seg_441c6d.csv \n inflating: seg_abeca6.csv \n inflating: seg_fb4c26.csv \n inflating: seg_4d0c45.csv \n inflating: seg_9f12c6.csv \n inflating: seg_53a557.csv \n inflating: seg_ead30e.csv \n inflating: seg_bbb01e.csv \n inflating: seg_ce72e2.csv \n inflating: seg_4d0008.csv \n inflating: seg_762188.csv \n inflating: seg_967ae4.csv \n inflating: seg_82456a.csv \n inflating: seg_b02e31.csv \n inflating: seg_447b8b.csv \n inflating: seg_32e763.csv \n inflating: seg_60df2a.csv \n inflating: seg_263fb3.csv \n inflating: seg_b378bc.csv \n inflating: seg_ea8270.csv \n inflating: seg_e6c973.csv \n inflating: seg_8658b3.csv \n inflating: seg_e2209b.csv \n inflating: seg_468bc9.csv \n inflating: seg_85e90e.csv \n inflating: seg_72859b.csv \n inflating: seg_fcb7d0.csv \n inflating: seg_b686e5.csv \n inflating: seg_007a37.csv \n inflating: seg_f83d28.csv \n inflating: seg_8fb0d0.csv \n inflating: seg_aabc52.csv \n inflating: seg_fb74f0.csv \n inflating: seg_63d651.csv \n inflating: seg_03f380.csv \n inflating: seg_12b9ba.csv \n inflating: seg_cec2f2.csv \n inflating: seg_8324a4.csv \n inflating: seg_514543.csv \n inflating: seg_8a8375.csv \n inflating: seg_b6bdaa.csv \n inflating: seg_df8e0a.csv \n inflating: seg_b08e9d.csv \n inflating: seg_e85f55.csv \n inflating: seg_c8459f.csv \n inflating: seg_8d5113.csv \n inflating: seg_1f9aed.csv \n inflating: seg_e25aca.csv \n inflating: seg_efbf3e.csv \n inflating: seg_05bef4.csv \n inflating: seg_0c74cf.csv \n inflating: seg_db3a95.csv \n inflating: seg_2f8e19.csv \n inflating: seg_0e1370.csv \n inflating: seg_e64dfa.csv \n inflating: seg_383f2e.csv \n inflating: seg_b44c2e.csv \n inflating: seg_94ecad.csv \n inflating: seg_5e541f.csv \n inflating: seg_9c6715.csv \n inflating: seg_941759.csv \n inflating: seg_61f504.csv \n inflating: seg_c23429.csv \n inflating: seg_68a484.csv \n inflating: seg_cd2b34.csv \n inflating: seg_d29a4b.csv \n inflating: seg_af959f.csv \n inflating: seg_09f0ff.csv \n inflating: seg_e09301.csv \n inflating: seg_2fe382.csv \n inflating: seg_c146a8.csv \n inflating: seg_a2d47d.csv \n inflating: seg_aa7a4b.csv \n inflating: seg_9f22a0.csv \n inflating: seg_d328bb.csv \n inflating: seg_225320.csv \n inflating: seg_966f3d.csv \n inflating: seg_ff4236.csv \n inflating: seg_ab001b.csv \n inflating: seg_d0f262.csv \n inflating: seg_4b8044.csv \n inflating: seg_a5dea6.csv \n inflating: seg_1fe311.csv \n inflating: seg_94aa0a.csv \n inflating: seg_09dd59.csv \n inflating: seg_4ddddc.csv \n inflating: seg_e8ce6a.csv \n inflating: seg_e8ad6b.csv \n inflating: seg_4adeda.csv \n inflating: seg_34ef79.csv \n inflating: seg_3c3ddf.csv \n inflating: seg_62a403.csv \n inflating: seg_643f86.csv \n inflating: seg_709d08.csv \n inflating: seg_b20487.csv \n inflating: seg_90ef18.csv \n inflating: seg_c106ca.csv \n inflating: seg_5cd98b.csv \n inflating: seg_e1468f.csv \n inflating: seg_915b3e.csv \n inflating: seg_b0f9cd.csv \n inflating: seg_a2986f.csv \n inflating: seg_b4cb44.csv \n inflating: seg_c5c455.csv \n inflating: seg_7a9f2b.csv \n inflating: seg_ab15ad.csv \n inflating: seg_be811d.csv \n inflating: seg_f22163.csv \n inflating: seg_0a0fbb.csv \n inflating: seg_e3005e.csv \n inflating: seg_bce23c.csv \n inflating: seg_23bdf9.csv \n inflating: seg_ce00bb.csv \n inflating: seg_47fed6.csv \n inflating: seg_3788c5.csv \n inflating: seg_29c772.csv \n inflating: seg_75211e.csv \n inflating: seg_0c9aa8.csv \n inflating: seg_0981f3.csv \n inflating: seg_b80b89.csv \n inflating: seg_8ce632.csv \n inflating: seg_c5f986.csv \n inflating: seg_807901.csv \n inflating: seg_914a62.csv \n inflating: seg_d6b386.csv \n inflating: seg_288001.csv \n inflating: seg_d47aba.csv \n inflating: seg_4ad674.csv \n inflating: seg_bcc8f1.csv \n inflating: seg_75dc3e.csv \n inflating: seg_578e84.csv \n inflating: seg_e0f296.csv \n inflating: seg_d753f8.csv \n inflating: seg_ce5520.csv \n inflating: seg_660fe1.csv \n inflating: seg_9fd9b8.csv \n inflating: seg_4ad5a2.csv \n inflating: seg_2dfb91.csv \n inflating: seg_8ab3a7.csv \n inflating: seg_53498a.csv \n inflating: seg_a11dd5.csv \n inflating: seg_91596c.csv \n inflating: seg_6e12ae.csv \n inflating: seg_a16a1b.csv \n inflating: seg_ca88e3.csv \n inflating: seg_19e1ff.csv \n inflating: seg_ee2781.csv \n inflating: seg_dd699c.csv \n inflating: seg_27ff17.csv \n inflating: seg_25cca7.csv \n inflating: seg_038879.csv \n inflating: seg_ff79d9.csv \n inflating: seg_a88dde.csv \n inflating: seg_9bd388.csv \n inflating: seg_33c30d.csv \n inflating: seg_563059.csv \n inflating: seg_78db0a.csv \n inflating: seg_d07c62.csv \n inflating: seg_876904.csv \n inflating: seg_279725.csv \n inflating: seg_922990.csv \n inflating: seg_0536c9.csv \n inflating: seg_fcd32e.csv \n inflating: seg_836ef0.csv \n inflating: seg_b853c1.csv \n inflating: seg_26049e.csv \n inflating: seg_890181.csv \n inflating: seg_d356ab.csv \n inflating: seg_b9ad7f.csv \n inflating: seg_2099f4.csv \n inflating: seg_eefd4a.csv \n inflating: seg_9b0439.csv \n inflating: seg_a47cce.csv \n inflating: seg_0d833c.csv \n inflating: seg_a8685e.csv \n inflating: seg_4abc29.csv \n inflating: seg_c7b424.csv \n inflating: seg_121cba.csv \n inflating: seg_dcf242.csv \n inflating: seg_f7050a.csv \n inflating: seg_3506d6.csv \n inflating: seg_7eb108.csv \n inflating: seg_5cfba9.csv \n inflating: seg_ef660a.csv \n inflating: seg_b80358.csv \n inflating: seg_19b08e.csv \n inflating: seg_343571.csv \n inflating: seg_d4dec8.csv \n inflating: seg_0879a8.csv \n inflating: seg_ea05c1.csv \n inflating: seg_afa4a1.csv \n inflating: seg_75eb90.csv \n inflating: seg_5b392b.csv \n inflating: seg_5bfbf0.csv \n inflating: seg_cdadb5.csv \n inflating: seg_3151ff.csv \n inflating: seg_03d63e.csv \n inflating: seg_ddc800.csv \n inflating: seg_a489e1.csv \n inflating: seg_41be7d.csv \n inflating: seg_c703be.csv \n inflating: seg_2f60db.csv \n inflating: seg_004f1f.csv \n inflating: seg_cea185.csv \n inflating: seg_2a4551.csv \n inflating: seg_e72f10.csv \n inflating: seg_ba83da.csv \n inflating: seg_902bc1.csv \n inflating: seg_6dfab7.csv \n inflating: seg_b7da0a.csv \n inflating: seg_420bcc.csv \n inflating: seg_3ae4d9.csv \n inflating: seg_67599b.csv \n inflating: seg_5eb380.csv \n inflating: seg_a61e1c.csv \n inflating: seg_ba98dc.csv \n inflating: seg_76e914.csv \n inflating: seg_26edaa.csv \n inflating: seg_e37b8e.csv \n inflating: seg_cd699e.csv \n inflating: seg_95644e.csv \n inflating: seg_f6f683.csv \n inflating: seg_447cb3.csv \n inflating: seg_0e1fbe.csv \n inflating: seg_7c10c1.csv \n inflating: seg_cf1371.csv \n inflating: seg_f6abc5.csv \n inflating: seg_e6307f.csv \n inflating: seg_996c37.csv \n inflating: seg_3f3689.csv \n inflating: seg_92af10.csv \n inflating: seg_8fd465.csv \n inflating: seg_82d7b6.csv \n inflating: seg_24ba8d.csv \n inflating: seg_3d059a.csv \n inflating: seg_0e3739.csv \n inflating: seg_74fcfc.csv \n inflating: seg_6d6fad.csv \n inflating: seg_df99d4.csv \n inflating: seg_a69246.csv \n inflating: seg_5f0a92.csv \n inflating: seg_d68930.csv \n inflating: seg_d5f344.csv \n inflating: seg_c5c6ea.csv \n inflating: seg_f77ee5.csv \n inflating: seg_29acb7.csv \n inflating: seg_5c530f.csv \n inflating: seg_e3b1f1.csv \n inflating: seg_5090fa.csv \n inflating: seg_ba2c48.csv \n inflating: seg_a68007.csv \n inflating: seg_c87c24.csv \n inflating: seg_d2db6b.csv \n inflating: seg_55d50b.csv \n inflating: seg_8d4965.csv \n inflating: seg_509d55.csv \n inflating: seg_45838b.csv \n inflating: seg_25b38a.csv \n inflating: seg_90b174.csv \n inflating: seg_4729a7.csv \n inflating: seg_4a9e6b.csv \n inflating: seg_ed7741.csv \n inflating: seg_590fa5.csv \n inflating: seg_490092.csv \n inflating: seg_c472cf.csv \n inflating: seg_3a22ac.csv \n inflating: seg_6d35cd.csv \n inflating: seg_cf646e.csv \n inflating: seg_7d88e3.csv \n inflating: seg_f57ca1.csv \n inflating: seg_b41d3c.csv \n inflating: seg_7a09ec.csv \n inflating: seg_64be5d.csv \n inflating: seg_70fb30.csv \n inflating: seg_3b7724.csv \n inflating: seg_61b50d.csv \n inflating: seg_9dea8d.csv \n inflating: seg_0e3d44.csv \n inflating: seg_570d69.csv \n inflating: seg_ac68bb.csv \n inflating: seg_9b44d2.csv \n inflating: seg_e8b2b3.csv \n inflating: seg_414d0f.csv \n inflating: seg_54d0f3.csv \n inflating: seg_8681aa.csv \n inflating: seg_f5e682.csv \n inflating: seg_ea0091.csv \n inflating: seg_dc7e0d.csv \n inflating: seg_78addc.csv \n inflating: seg_3234ca.csv \n inflating: seg_025e78.csv \n inflating: seg_f5a4ee.csv \n inflating: seg_70991d.csv \n inflating: seg_9cb464.csv \n inflating: seg_32c904.csv \n inflating: seg_c21723.csv \n inflating: seg_268249.csv \n inflating: seg_529be4.csv \n inflating: seg_87232d.csv \n inflating: seg_68102c.csv \n inflating: seg_fd374e.csv \n inflating: seg_c0ea9f.csv \n inflating: seg_0e3ed2.csv \n inflating: seg_d146a6.csv \n inflating: seg_d6b546.csv \n inflating: seg_738be6.csv \n inflating: seg_137f8a.csv \n inflating: seg_e7ced6.csv \n inflating: seg_3fb24b.csv \n inflating: seg_c3836b.csv \n inflating: seg_a9a820.csv \n inflating: seg_92a5f1.csv \n inflating: seg_5fe414.csv \n inflating: seg_c6b514.csv \n inflating: seg_d6ea73.csv \n inflating: seg_bd629b.csv \n inflating: seg_f2ae5e.csv \n inflating: seg_cf25d2.csv \n inflating: seg_16a270.csv \n inflating: seg_d37b05.csv \n inflating: seg_486973.csv \n inflating: seg_59de0b.csv \n inflating: seg_89b090.csv \n inflating: seg_6c309f.csv \n inflating: seg_cd43b4.csv \n inflating: seg_581fac.csv \n inflating: seg_98f624.csv \n inflating: seg_9a8a1f.csv \n inflating: seg_e92526.csv \n inflating: seg_4185b3.csv \n inflating: seg_812962.csv \n inflating: seg_5f355e.csv \n inflating: seg_f322b1.csv \n inflating: seg_a0979e.csv \n inflating: seg_0dae4b.csv \n inflating: seg_0a97c4.csv \n inflating: seg_af6204.csv \n inflating: seg_fb00b3.csv \n inflating: seg_9ddc99.csv \n inflating: seg_655ad6.csv \n inflating: seg_63c8e4.csv \n inflating: seg_e8bcde.csv \n inflating: seg_b13220.csv \n inflating: seg_e5e238.csv \n inflating: seg_b5a447.csv \n inflating: seg_8a8220.csv \n inflating: seg_a80123.csv \n inflating: seg_10c09d.csv \n inflating: seg_016913.csv \n inflating: seg_361f5a.csv \n inflating: seg_ca44a8.csv \n inflating: seg_cd2b53.csv \n inflating: seg_95b321.csv \n inflating: seg_d320f5.csv \n inflating: seg_fe73b1.csv \n inflating: seg_d3bb14.csv \n inflating: seg_9f6315.csv \n inflating: seg_75ffc9.csv \n inflating: seg_9e8323.csv \n inflating: seg_48a4b3.csv \n inflating: seg_2774c4.csv \n inflating: seg_d32f71.csv \n inflating: seg_857304.csv \n inflating: seg_4fa87d.csv \n inflating: seg_010eab.csv \n inflating: seg_41ab7e.csv \n inflating: seg_184222.csv \n inflating: seg_864fcf.csv \n inflating: seg_96642e.csv \n inflating: seg_9aa6e2.csv \n inflating: seg_cf3825.csv \n inflating: seg_ada81a.csv \n inflating: seg_80211c.csv \n inflating: seg_17067e.csv \n inflating: seg_de98fa.csv \n inflating: seg_7b5f90.csv \n inflating: seg_304581.csv \n inflating: seg_d83890.csv \n inflating: seg_5470ca.csv \n inflating: seg_037461.csv \n inflating: seg_30ad2a.csv \n inflating: seg_31631c.csv \n inflating: seg_55239a.csv \n inflating: seg_3f99c8.csv \n inflating: seg_046c28.csv \n inflating: seg_51b68c.csv \n inflating: seg_6968c8.csv \n inflating: seg_1c401b.csv \n inflating: seg_e0ce38.csv \n inflating: seg_8fb828.csv \n inflating: seg_c9831a.csv \n inflating: seg_3cfb73.csv \n inflating: seg_0cf651.csv \n inflating: seg_b51380.csv \n inflating: seg_ba941e.csv \n inflating: seg_57b6c0.csv \n inflating: seg_944c98.csv \n inflating: seg_8bde47.csv \n inflating: seg_af2472.csv \n inflating: seg_10a595.csv \n inflating: seg_9b7f93.csv \n inflating: seg_7cef95.csv \n inflating: seg_1d11e5.csv \n inflating: seg_f48363.csv \n inflating: seg_293dbf.csv \n inflating: seg_a68ffb.csv \n inflating: seg_31b808.csv \n inflating: seg_dad4f2.csv \n inflating: seg_65ef95.csv \n inflating: seg_e60719.csv \n inflating: seg_31a51f.csv \n inflating: seg_945bc8.csv \n inflating: seg_9d7490.csv \n inflating: seg_05f9d6.csv \n inflating: seg_bf4bf2.csv \n inflating: seg_60ab20.csv \n inflating: seg_827804.csv \n inflating: seg_816b7a.csv \n inflating: seg_146926.csv \n inflating: seg_766e42.csv \n inflating: seg_218049.csv \n inflating: seg_f30f0c.csv \n inflating: seg_8dcf3c.csv \n inflating: seg_2e71dd.csv \n inflating: seg_32d747.csv \n inflating: seg_e197b8.csv \n inflating: seg_d7f53a.csv \n inflating: seg_32f3a9.csv \n inflating: seg_3a8a76.csv \n inflating: seg_017314.csv \n inflating: seg_929ded.csv \n inflating: seg_1f3d44.csv \n inflating: seg_f6a217.csv \n inflating: seg_9d68cf.csv \n inflating: seg_d36055.csv \n inflating: seg_03d680.csv \n inflating: seg_f66db1.csv \n inflating: seg_2ea616.csv \n inflating: seg_655780.csv \n inflating: seg_dceeca.csv \n inflating: seg_461ab5.csv \n inflating: seg_62331a.csv \n inflating: seg_6da1ff.csv \n inflating: seg_93d864.csv \n inflating: seg_4b4c91.csv \n inflating: seg_495573.csv \n inflating: seg_a31d6e.csv \n inflating: seg_a14212.csv \n inflating: seg_f7290f.csv \n inflating: seg_9f7542.csv \n inflating: seg_9ca72e.csv \n inflating: seg_05b66a.csv \n inflating: seg_f618a8.csv \n inflating: seg_161aeb.csv \n inflating: seg_922914.csv \n inflating: seg_44d3ca.csv \n inflating: seg_ef644a.csv \n inflating: seg_4fd191.csv \n inflating: seg_ed52ac.csv \n inflating: seg_04ee93.csv \n inflating: seg_7fd3a3.csv \n inflating: seg_3a1600.csv \n inflating: seg_f67ce2.csv \n inflating: seg_4a719c.csv \n inflating: seg_268625.csv \n inflating: seg_ef24bd.csv \n inflating: seg_6ac67a.csv \n inflating: seg_a05c22.csv \n inflating: seg_53103b.csv \n inflating: seg_adf986.csv \n inflating: seg_c588c1.csv \n inflating: seg_3c64fb.csv \n inflating: seg_d86eb6.csv \n inflating: seg_762be4.csv \n inflating: seg_31c0f4.csv \n inflating: seg_620ee4.csv \n inflating: seg_c4af54.csv \n inflating: seg_0d663a.csv \n inflating: seg_39ad50.csv \n inflating: seg_f30b2f.csv \n inflating: seg_32906d.csv \n inflating: seg_a7e49b.csv \n inflating: seg_d88da7.csv \n inflating: seg_79dc90.csv \n inflating: seg_750d20.csv \n inflating: seg_229eda.csv \n inflating: seg_2b6d52.csv \n inflating: seg_c81e10.csv \n inflating: seg_3a80c5.csv \n inflating: seg_ba1d68.csv \n inflating: seg_87e4e8.csv \n inflating: seg_fb9b2e.csv \n inflating: seg_45e7ad.csv \n inflating: seg_c23641.csv \n inflating: seg_0d540e.csv \n inflating: seg_bfdd14.csv \n inflating: seg_abb03a.csv \n inflating: seg_103b32.csv \n inflating: seg_cee7d0.csv \n inflating: seg_ed7dee.csv \n inflating: seg_ad591c.csv \n inflating: seg_31064b.csv \n inflating: seg_079e34.csv \n inflating: seg_e40d64.csv \n inflating: seg_52636c.csv \n inflating: seg_ce46ba.csv \n inflating: seg_461b63.csv \n inflating: seg_31ddc5.csv \n inflating: seg_f4d644.csv \n inflating: seg_fc57d4.csv \n inflating: seg_3706be.csv \n inflating: seg_a4ad7c.csv \n inflating: seg_7120ac.csv \n inflating: seg_4b953a.csv \n inflating: seg_1abcaf.csv \n inflating: seg_280e3b.csv \n inflating: seg_ffe7cc.csv \n inflating: seg_a49ccc.csv \n inflating: seg_634180.csv \n inflating: seg_1b1ad8.csv \n inflating: seg_4c18e2.csv \n inflating: seg_b58542.csv \n inflating: seg_c6f0a2.csv \n inflating: seg_8344ee.csv \n"
],
[
"file_names = []\n\nfor ids in range(seg_id.shape[0]):\n file_names.append(seg_id['seg_id'][ids] + '.csv')\n \nfile_names[0:5]",
"_____no_output_____"
],
[
"len(file_names)",
"_____no_output_____"
],
[
"y_trains = pd.read_csv(file_names[0])\ny_trains.head()",
"_____no_output_____"
],
[
"y_trains.describe()",
"_____no_output_____"
],
[
"signal = pd.read_csv('train.csv', nrows=6000000, dtype={'acoustic_data': np.int16, 'time_to_failure': np.float64})",
"_____no_output_____"
],
[
"signal.head()",
"_____no_output_____"
],
[
"\n#Data Types of given signal\n\nsignal.dtypes",
"_____no_output_____"
],
[
"# Statistical Analysis of data\n\nsignal.describe()",
"_____no_output_____"
],
[
"# Training Dataset for Acoustic data\n\nplt.plot(signal['acoustic_data'].values)\nplt.show()",
"_____no_output_____"
],
[
"# time to failure training data plot\n\nplt.plot(signal['time_to_failure'].values)\nplt.show()",
"_____no_output_____"
],
[
"# testing dataset plot\n\nplt.plot(y_trains['acoustic_data'].values)\nplt.show()",
"_____no_output_____"
],
[
"# Distribution of acoustic data\n\nsns.distplot(signal.acoustic_data.values, color=\"Red\", kde=False)\nplt.show()",
"_____no_output_____"
],
[
"# Distribution of small part of acoustic data\n\nsns.distplot(signal.acoustic_data.values[0:50000], color=\"Green\", bins=100, kde=False)\nplt.show()",
"_____no_output_____"
],
[
"# Distribution of small part of acoustic data\n\nsns.distplot(y_trains.acoustic_data.values[0:50000], color=\"Blue\", bins=100, kde=False)\nplt.show()",
"_____no_output_____"
],
[
"# Plotting first 50000 values of time_to_failure\n\nplt.plot(signal['time_to_failure'].values[0:50000])\nplt.show()",
"_____no_output_____"
],
[
"# Plotting in-between 5000 values of time_to_failure\n\nplt.plot(signal['time_to_failure'].values[5950000:6500000])\nplt.show()",
"_____no_output_____"
],
[
"# Acoustic-data vs time-to-failure\n\nfig, ax1 = plt.subplots(figsize=(8,5))\nplt.title('acoustic_data vs time_to_failure')\nplt.plot(signal['acoustic_data'].values[::100], color='b')\nax1.set_ylabel('acoustic data', color='b')\nplt.legend(['acoustic data'], loc=(0.01, 0.9))\nax2 = ax1.twinx()\nplt.plot(signal['time_to_failure'].values[::100], color='r')\nax2.set_ylabel('time to failure', color='r')\nplt.legend(['time to failure'], loc=(0.01, 0.8))\nplt.show()",
"_____no_output_____"
],
[
"signalplot = pd.read_csv('train.csv', nrows=6000000, dtype={'acoustic_data': np.int16, 'time_to_failure': np.float64})\nsignalplot.head()\nsignalplot.info()\nsignalplot.describe()\nsignalplot.columns",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 6000000 entries, 0 to 5999999\nData columns (total 2 columns):\nacoustic_data int16\ntime_to_failure float64\ndtypes: float64(1), int16(1)\nmemory usage: 57.2 MB\n"
],
[
"sns.pairplot(signalplot)",
"_____no_output_____"
],
[
"# correlation between acoustic_data and time_to_failure\nsignalplot.corr()",
"_____no_output_____"
],
[
"rows = 150000\nsegments = int(np.floor(signal.shape[0] / rows))\nsegments\ntrain_X = pd.DataFrame(index=range(segments), dtype=np.float64)\ntrain_y = pd.DataFrame(index=range(segments), dtype=np.float64, columns=['time_to_failure'])",
"_____no_output_____"
],
[
"train_X.to_csv('statistical_features.csv', header=True, index=False) ",
"_____no_output_____"
],
[
"train_y.to_csv('output.csv', header=True, index=False) ",
"_____no_output_____"
],
[
"def gen_features(X):\n strain = []\n strain.append(X.mean())\n strain.append(X.std())\n strain.append(X.min())\n strain.append(X.max())\n strain.append(X.kurtosis())\n strain.append(X.skew())\n strain.append(np.quantile(X,0.01))\n strain.append(np.quantile(X,0.05))\n strain.append(np.quantile(X,0.95))\n strain.append(np.quantile(X,0.99))\n strain.append(np.abs(X).max())\n strain.append(np.abs(X).mean())\n strain.append(np.abs(X).std())\n return pd.Series(strain)",
"_____no_output_____"
],
[
"train = pd.read_csv('train.csv', iterator=True, chunksize=150_000, dtype={'acoustic_data': np.int16, 'time_to_failure': np.float64})\n\nX_train = pd.DataFrame()\ny_train = pd.Series()\nfor df in train:\n ch = gen_features(df['acoustic_data'])\n X_train = X_train.append(ch, ignore_index=True)\n y_train = y_train.append(pd.Series(df['time_to_failure'].values[-1]))",
"_____no_output_____"
],
[
"X_train.describe()",
"_____no_output_____"
],
[
"# catboost algorithm\n\ntrain_pool = Pool(X_train, y_train)\nm = CatBoostRegressor(iterations=10000, loss_function='MAE', boosting_type='Ordered')\nm.fit(X_train, y_train, silent=True)\nm.best_score_",
"_____no_output_____"
],
[
"# support vector machine algortihm\n\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.svm import NuSVR, SVR\n\n\nscaler = StandardScaler()\nscaler.fit(X_train)\nX_train_scaled = scaler.transform(X_train)\n\nparameters = [{'gamma': [0.001, 0.005, 0.01, 0.02, 0.05, 0.1],\n 'C': [0.1, 0.2, 0.25, 0.5, 1, 1.5, 2]}]\n #'nu': [0.75, 0.8, 0.85, 0.9, 0.95, 0.97]}]\n\nclf = GridSearchCV(SVR(kernel='rbf', tol=0.01), parameters, cv=5, scoring='neg_mean_absolute_error')\nclf.fit(X_train_scaled, y_train.values.flatten())\ny_pred1 = clf.predict(X_train_scaled)\n\nMAE_SVR = mean_absolute_error(y_train,y_pred1)",
"_____no_output_____"
],
[
"print (MAE_SVR)",
"2.094214896368042\n"
],
[
"",
"_____no_output_____"
],
[
"grid_hyperparameter = [{'n_estimators' : [10,20,30],'max_depth':[5,10]}]\n\nclf = GridSearchCV(RandomForestRegressor(max_features='sqrt',min_samples_leaf=4,min_samples_split=3), grid_hyperparameter, cv=2)\nclf.fit(X_train,y_train)\n\nclf_nr = clf.best_estimator_.get_params()['n_estimators']\nclf_depthr = clf.best_estimator_.get_params()['max_depth']\n\nprint(clf_nr,clf_depthr)\n",
"20 5\n"
],
[
"clf_RF = RandomForestRegressor(max_features='sqrt',min_samples_leaf=4,min_samples_split=3,n_estimators=clf_nr,max_depth = clf_depthr)\nclf_RF.fit(X_train_scaled,y_train.values.flatten())\n\ny_pred = clf_RF.predict(X_train_scaled)\nMAE_RF = mean_absolute_error(y_train,y_pred)",
"_____no_output_____"
],
[
"print(MAE_RF)",
"2.087707631770648\n"
],
[
"from prettytable import PrettyTable\n \nTable = PrettyTable()\n\nTable.field_names = [\"Model\",\"MAE\"]\n\n\nTable.add_row([\"CatboostRegressor\", m.best_score_])\nTable.add_row([\"Random Forest\",MAE_RF])\nTable.add_row([\"SVR\",MAE_SVR])\n\n\nprint(Table)",
"+-------------------+----------------------------------------+\n| Model | MAE |\n+-------------------+----------------------------------------+\n| CatboostRegressor | {'learn': {'MAE': 1.7836623258199857}} |\n| Random Forest | 2.087707631770648 |\n| SVR | 2.094214896368042 |\n+-------------------+----------------------------------------+\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb50524a3f80b7768b65c8dff07e975f313e6f8 | 174,694 | ipynb | Jupyter Notebook | pymaceuticals_starter.ipynb | skaur2/Matplotlib-challenge | 4a92fcddb28d104ff312d124ea06b54279d86123 | [
"ADSL"
] | null | null | null | pymaceuticals_starter.ipynb | skaur2/Matplotlib-challenge | 4a92fcddb28d104ff312d124ea06b54279d86123 | [
"ADSL"
] | null | null | null | pymaceuticals_starter.ipynb | skaur2/Matplotlib-challenge | 4a92fcddb28d104ff312d124ea06b54279d86123 | [
"ADSL"
] | null | null | null | 103.003538 | 19,716 | 0.796244 | [
[
[
"## Observations and Insights ",
"_____no_output_____"
]
],
[
[
"# Dependencies and Setup\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport scipy.stats as st\nimport numpy as np\n\n# Study data files\nmouse_metadata_path = \"data/Mouse_metadata.csv\"\nstudy_results_path = \"data/Study_results.csv\"\n\n# Read the mouse data and the study results\nmouse_metadata = pd.read_csv(mouse_metadata_path)\nstudy_results = pd.read_csv(study_results_path)\n\n# Combine the data into a single dataset\nmerged_df = pd.merge(mouse_metadata, study_results, on=\"Mouse ID\",how=\"left\")\n\n# Display the data table for preview\nmerged_df.head()",
"_____no_output_____"
],
[
"# Checking the number of mice.\nmouse_count = merged_df[\"Mouse ID\"].count()\nmouse_count\n",
"_____no_output_____"
],
[
"# Getting the duplicate mice by ID number that shows up for Mouse ID and Timepoint. \nduplicate_rows = merged_df[merged_df.duplicated(['Mouse ID', 'Timepoint'])]\nduplicate_rows\n",
"_____no_output_____"
],
[
"# Optional: Get all the data for the duplicate mouse ID. \nall_duplicate_rows = merged_df[merged_df.duplicated(['Mouse ID',])]\nall_duplicate_rows\n",
"_____no_output_____"
],
[
"# Create a clean DataFrame by dropping the duplicate mouse by its ID.\nclean_df = merged_df.drop_duplicates(\"Mouse ID\")\nclean_df",
"_____no_output_____"
],
[
"# Checking the number of mice in the clean DataFrame.\n",
"_____no_output_____"
]
],
[
[
"## Summary Statistics",
"_____no_output_____"
]
],
[
[
"# Generate a summary statistics table of mean, median, variance, standard deviation, and SEM of the tumor volume for each regimen\n\n# Use groupby and summary statistical methods to calculate the following properties of each drug regimen: \n# mean, median, variance, standard deviation, and SEM of the tumor volume. \n# Assemble the resulting series into a single summary dataframe.\n\nnewthing = merged_df.groupby('Drug Regimen')\nfirst = newthing.agg(['mean','median','var','std','sem'])[\"Tumor Volume (mm3)\"]\nfirst\n",
"_____no_output_____"
],
[
"# Generate a summary statistics table of mean, median, variance, standard deviation, and SEM of the tumor volume for each regimen\n\n# Using the aggregation method, produce the same summary statistics in a single line\n\nmeans = merged_df.groupby('Drug Regimen').mean()['Tumor Volume (mm3)']\nmedians = merged_df.groupby('Drug Regimen').median()['Tumor Volume (mm3)']\nvariances = merged_df.groupby('Drug Regimen').var()['Tumor Volume (mm3)']\nstandards = merged_df.groupby('Drug Regimen').std()['Tumor Volume (mm3)']\nsems = merged_df.groupby('Drug Regimen').sem()['Tumor Volume (mm3)']\n\nnewtable = pd.DataFrame(means)\nnewtable2 = newtable.rename(columns={\"Tumor Volume (mm3)\": \"Mean\"})\n\nnewtable2[\"Median\"] = medians\nnewtable2[\"Variance\"] = variances\nnewtable2[\"std\"] = standards\nnewtable2[\"sem\"] = sems\n\nnewtable2",
"_____no_output_____"
]
],
[
[
"## Bar and Pie Charts",
"_____no_output_____"
]
],
[
[
"# Generate a bar plot showing the total number of measurements taken on each drug regimen using pandas.\n\ndatapts2 = merged_df.groupby('Drug Regimen').count()['Tumor Volume (mm3)']\nforpanbar = pd.DataFrame(datapts2)\n#newtry = forpanbar.reset_index()\n#newtry\n\nalso = forpanbar.plot.bar(legend=False,rot=50)\nalso\nplt.ylabel(\"Number of Data Points\")\nplt.title(\"Points Per Drug Treatment\")\nplt.savefig('barplot1')\n",
"_____no_output_____"
],
[
"forpanbar.head()",
"_____no_output_____"
],
[
"# Generate a bar plot showing the total number of measurements taken on each drug regimen using pyplot.\n\nx_axis = np.arange(len(datapts2))\n\ntick_locations = [x for x in x_axis]\n#for x in x_axis:\n#tick_locations.append(x)\n\nplt.figure(figsize=(5,3))\n#plt.bar(x_axis, rain_df[\"Inches\"], color='r', alpha=0.5, align=\"center\")\n#plt.xticks(tick_locations, rain_df[\"State\"], rotation=\"vertical\")\n\nnewtry = forpanbar.reset_index()\nnewtry\n\nplt.bar(x_axis, forpanbar['Tumor Volume (mm3)'], alpha=0.75, align=\"center\")\nplt.xticks(tick_locations, newtry['Drug Regimen'],rotation=\"vertical\")\n\nplt.xlim(-0.75, len(datapts2)-.25)\nplt.ylim(0, 250)\n\nplt.title(\"Data Points Per Drug Treatment Regimen\")\nplt.xlabel(\"Drug Regimen\")\nplt.ylabel(\"Number of Data Points\")\n\nplt.savefig('barplot2')\nplt.show()",
"_____no_output_____"
],
[
"# Generate a pie plot showing the distribution of female versus male mice using pandas\ngender_data = clean_df[\"Sex\"].value_counts()\nplt.title(\"Female vs. Male Mice\")\ngender_data.plot.pie(autopct= \"%1.1f%%\")\nplt.show()",
"_____no_output_____"
],
[
"# Generate a pie plot showing the distribution of female versus male mice using pyplot\nlabels = ['Female', 'Male']\nsizes = [49.7999197, 50.200803]\nplot = gender_data.plot.pie(y='Total Count', autopct=\"%1.1f%%\")\nplt.title('Male vs Female Mouse Population')\nplt.ylabel('Sex')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"## Quartiles, Outliers and Boxplots",
"_____no_output_____"
]
],
[
[
"# Calculate the final tumor volume of each mouse across four of the treatment regimens: \n",
"_____no_output_____"
],
[
"# Capomulin\nmerged_df.head() \n\nsorted_df = merged_df.sort_values([\"Drug Regimen\", \"Mouse ID\", \"Timepoint\"], ascending=True)\nlast_df = sorted_df.loc[sorted_df[\"Timepoint\"] == 45]\nlast_df.head().reset_index()\n\ncapo_df = last_df[last_df[\"Drug Regimen\"].isin([\"Capomulin\"])]\ncapo_df.head().reset_index()\n\ncapo_obj = capo_df.sort_values([\"Tumor Volume (mm3)\"], ascending=True).reset_index()\ncapo_obj = capo_obj[\"Tumor Volume (mm3)\"]\ncapo_obj\n\nquartiles = capo_obj.quantile([.25,.5,.75])\nlowerq = quartiles[0.25]\nupperq = quartiles[0.75]\niqr = upperq - lowerq\n\nprint(f\"The lower quartile of temperatures is: {lowerq}\")\nprint(f\"The upper quartile of temperatures is: {upperq}\")\nprint(f\"The interquartile range of temperatures is: {iqr}\")\nprint(f\"The median of temperatures is: {quartiles[0.5]}\")\n\nlower_bound = lowerq - (1.5*iqr)\nupper_bound = upperq + (1.5*iqr)\nprint(f\"Values below {lower_bound} could be outliers.\")\nprint(f\"Values above {upper_bound} could be outliers.\")\n\nfig1, ax1 = plt.subplots()\nax1.set_title(\"Final Tumor Volume in Capomulin Regimen\")\nax1.set_ylabel(\"Final Tumor Volume (mm3)\")\nax1.boxplot(capo_obj)\nplt.show()\n",
"The lower quartile of temperatures is: 32.37735684\nThe upper quartile of temperatures is: 40.1592203\nThe interquartile range of temperatures is: 7.781863460000004\nThe median of temperatures is: 37.31184577\nValues below 20.70456164999999 could be outliers.\nValues above 51.83201549 could be outliers.\n"
],
[
"#Ramicane\nram_df = last_df[last_df[\"Drug Regimen\"].isin([\"Ramicane\"])]\nram_df.head().reset_index()\n\nram_obj = ram_df.sort_values([\"Tumor Volume (mm3)\"], ascending=True).reset_index()\nram_obj = ram_obj[\"Tumor Volume (mm3)\"]\nram_obj\n\nquartiles = capo_obj.quantile([.25,.5,.75])\nlowerq = quartiles[0.25]\nupperq = quartiles[0.75]\niqr = upperq - lowerq\n\nprint(f\"The lower quartile of temperatures is: {lowerq}\")\nprint(f\"The upper quartile of temperatures is: {upperq}\")\nprint(f\"The interquartile range of temperatures is: {iqr}\")\nprint(f\"The median of temperatures is: {quartiles[0.5]}\")\n\nlower_bound = lowerq - (1.5*iqr)\nupper_bound = upperq + (1.5*iqr)\nprint(f\"Values below {lower_bound} could be outliers.\")\nprint(f\"Values above {upper_bound} could be outliers.\")\n\nfig1, ax1 = plt.subplots()\nax1.set_title(\"Final Tumor Volume in Ramicane Regimen\")\nax1.set_ylabel(\"Final Tumor Volume (mm3)\")\nax1.boxplot(capo_obj)\nplt.show()\n",
"The lower quartile of temperatures is: 32.37735684\nThe upper quartile of temperatures is: 40.1592203\nThe interquartile range of temperatures is: 7.781863460000004\nThe median of temperatures is: 37.31184577\nValues below 20.70456164999999 could be outliers.\nValues above 51.83201549 could be outliers.\n"
],
[
"#Infubinol\ninfu_df = last_df[last_df[\"Drug Regimen\"].isin([\"Infubinol\"])]\ninfu_df.head().reset_index()\n\ninfu_obj = infu_df.sort_values([\"Tumor Volume (mm3)\"], ascending=True).reset_index()\ninfu_obj = infu_obj[\"Tumor Volume (mm3)\"]\ninfu_obj\n\nquartiles = capo_obj.quantile([.25,.5,.75])\nlowerq = quartiles[0.25]\nupperq = quartiles[0.75]\niqr = upperq - lowerq\n\nprint(f\"The lower quartile of temperatures is: {lowerq}\")\nprint(f\"The upper quartile of temperatures is: {upperq}\")\nprint(f\"The interquartile range of temperatures is: {iqr}\")\nprint(f\"The median of temperatures is: {quartiles[0.5]}\")\n\nlower_bound = lowerq - (1.5*iqr)\nupper_bound = upperq + (1.5*iqr)\nprint(f\"Values below {lower_bound} could be outliers.\")\nprint(f\"Values above {upper_bound} could be outliers.\")\n\nfig1, ax1 = plt.subplots()\nax1.set_title(\"Final Tumor Volume in Infubinol Regimen\")\nax1.set_ylabel(\"Final Tumor Volume (mm3)\")\nax1.boxplot(capo_obj)\nplt.show()",
"The lower quartile of temperatures is: 32.37735684\nThe upper quartile of temperatures is: 40.1592203\nThe interquartile range of temperatures is: 7.781863460000004\nThe median of temperatures is: 37.31184577\nValues below 20.70456164999999 could be outliers.\nValues above 51.83201549 could be outliers.\n"
],
[
"#Ceftamin\nceft_df = last_df[last_df[\"Drug Regimen\"].isin([\"Ceftamin\"])]\nceft_df.head().reset_index()\n\nceft_obj = ceft_df.sort_values([\"Tumor Volume (mm3)\"], ascending=True).reset_index()\nceft_obj = ceft_obj[\"Tumor Volume (mm3)\"]\nceft_obj\n\nquartiles = capo_obj.quantile([.25,.5,.75])\nlowerq = quartiles[0.25]\nupperq = quartiles[0.75]\niqr = upperq - lowerq\n\nprint(f\"The lower quartile of temperatures is: {lowerq}\")\nprint(f\"The upper quartile of temperatures is: {upperq}\")\nprint(f\"The interquartile range of temperatures is: {iqr}\")\nprint(f\"The median of temperatures is: {quartiles[0.5]}\")\n\nlower_bound = lowerq - (1.5*iqr)\nupper_bound = upperq + (1.5*iqr)\nprint(f\"Values below {lower_bound} could be outliers.\")\nprint(f\"Values above {upper_bound} could be outliers.\")\n\nfig1, ax1 = plt.subplots()\nax1.set_title(\"Final Tumor Volume in Infubinol Regimen\")\nax1.set_ylabel(\"Final Tumor Volume (mm3)\")\nax1.boxplot(capo_obj)\nplt.show()",
"The lower quartile of temperatures is: 32.37735684\nThe upper quartile of temperatures is: 40.1592203\nThe interquartile range of temperatures is: 7.781863460000004\nThe median of temperatures is: 37.31184577\nValues below 20.70456164999999 could be outliers.\nValues above 51.83201549 could be outliers.\n"
]
],
[
[
"## Line and Scatter Plots",
"_____no_output_____"
]
],
[
[
"# Generate a line plot of tumor volume vs. time point for a mouse treated with Capomulin\ncapomulin_df = merged_df.loc[merged_df[\"Drug Regimen\"] == \"Capomulin\"]\ncapomulin_df = capomulin_df.reset_index()\ncapomulin_df.head()\n\ncapo_mouse = capomulin_df.loc[capomulin_df[\"Mouse ID\"] == \"s185\"]\ncapo_mouse\n\ncapo_mouse = capo_mouse.loc[:, [\"Timepoint\", \"Tumor Volume (mm3)\"]]\n\ncapo_mouse = capo_mouse.reset_index(drop=True)\ncapo_mouse.set_index(\"Timepoint\").plot(figsize=(10,8), linewidth=2.5, color=\"orange\")",
"_____no_output_____"
],
[
"# Generate a scatter plot of average tumor volume vs. mouse weight for the Capomulin regimen\ncapomulin_df.head() \n\nweight_df = capomulin_df.loc[:, [\"Mouse ID\", \"Weight (g)\", \"Tumor Volume (mm3)\"]]\nweight_df.head()\n\navg_capo = pd.DataFrame(weight_df.groupby([\"Mouse ID\", \"Weight (g)\"])[\"Tumor Volume (mm3)\"].mean()).reset_index()\navg_capo.head()\n\navg_capo = avg_capo.rename(columns={\"Tumor Volume (mm3)\": \"Average Volume\"})\navg_capo.head()\n\navg_capo.plot(kind=\"scatter\", x=\"Weight (g)\", y=\"Average Volume\", grid=False, figsize=(4,4), title=\"Weight vs. Average Tumor Volume\")\nplt.show()",
"_____no_output_____"
],
[
"plt.clf()\nplt.cla()\nplt.close()",
"_____no_output_____"
]
],
[
[
"## Correlation and Regression",
"_____no_output_____"
]
],
[
[
"# Calculate the correlation coefficient and linear regression model \n# for mouse weight and average tumor volume for the Capomulin regimen\nmouse_weight = avg_capo.iloc[:,0]\navg_tumor_volume = avg_capo.iloc[:,1]\n\n# We then compute the Pearson correlation coefficient between \"Mouse Weight\" and \"Average Tumor Volume\"\ncorrelation = st.pearsonr(mouse_weight,avg_tumor_volume)\nprint(f\"The correlation between both factors is {round(correlation[0],2)}\")\n\n# import linregress\nfrom scipy.stats import linregress\n\n# Add the lineear regression equation and line to the scatter plot\nx_values = avg_capo[\"Weight (g)\"]\ny_values = avg_capo[\"Average Volume\"]\n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nregress_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.scatter(x_values, y_values)\nplt.plot(x_values,regress_values,\"r-\")\nplt.annotate(line_eq,(6,10),fontsize=15,color=\"red\")\nplt.xlabel(\"Mouse Weight\")\nplt.ylabel(\"Average Tumor Volume\")\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb50a3a0d9a2436eb18e402250de41cf87d0d93 | 27,594 | ipynb | Jupyter Notebook | Untitled.ipynb | aparecidovieira/keras_segmentation | 9e80cb502378af6021dbfa565877adebbb4b674f | [
"Net-SNMP",
"Xnet"
] | 5 | 2021-04-17T07:13:35.000Z | 2022-03-22T18:06:42.000Z | Untitled.ipynb | aparecidovieira/keras_segmentation | 9e80cb502378af6021dbfa565877adebbb4b674f | [
"Net-SNMP",
"Xnet"
] | null | null | null | Untitled.ipynb | aparecidovieira/keras_segmentation | 9e80cb502378af6021dbfa565877adebbb4b674f | [
"Net-SNMP",
"Xnet"
] | 2 | 2021-04-17T07:13:33.000Z | 2021-07-15T15:51:05.000Z | 33.086331 | 160 | 0.525477 | [
[
[
"import os\nos.environ[\"CUDA_VISIBLE_DEVICES\"] = \"-1\"\nimport numpy as np\nimport keras\nfrom keras.models import *\nfrom keras.layers import *\nfrom tensorflow.python.keras import losses\nfrom keras.applications.vgg16 import VGG16\nfrom keras.preprocessing.image import ImageDataGenerator\nfrom keras.optimizers import *\nfrom keras.callbacks import ModelCheckpoint\nimport cv2\nimport glob\nimport matplotlib.pyplot as plt\nimport itertools\nfrom util import losses, custom_data_generator, metrics \nfrom keras.utils import multi_gpu_model\nfrom models import model_loader\nimport datetime\n\n# import tensorflow as tf\n# from keras.backend.tensorflow_backend import set_session\n# config = tf.ConfigProto()\n# config.gpu_options.per_process_gpu_memory_fraction = 0.8\n# set_session(tf.Session(config=config))\n\n\nbatch_size =30\nis_train = False\nmodel_name = 'lanenet'\nimage_width,image_height = 256,256\nchanneles = 3\ncheckpoint_name = \"checkpoint_lanenet\"\none_hot_label= False\ndata_aug = False\n\n \n\n\n\n",
"_____no_output_____"
],
[
"checkpoint_dir = \"./checkpoints/%s/\"%(checkpoint_name)\nif not os.path.exists(checkpoint_dir):\n os.makedirs(checkpoint_dir)\n\n\n\n\n# train_inputs_path = \"/home/beemap/Documents/noor-workspace/Semantic-Segmentation-Suite/airbus/train/\"\n# train_masks_path = \"/home/beemap/Documents/noor-workspace/Semantic-Segmentation-Suite/airbus/train_label/\"\n# val_inputs_path = \"/home/beemap/Documents/noor-workspace/Semantic-Segmentation-Suite/airbus/val/\"\n# val_masks_path = \"/home/beemap/Documents/noor-workspace/Semantic-Segmentation-Suite/airbus/val_label/\"\n\n\n\n# train_inputs_path = \"/media/HDD_4T/noor-workspace/airbus-dataset/train/\"\n# train_masks_path = \"/media/HDD_4T/noor-workspace/airbus-dataset/train_label/\"\n# val_inputs_path = \"/media/HDD_4T/noor-workspace/airbus-dataset/val/\"\n# val_masks_path = \"/media/HDD_4T/noor-workspace/airbus-dataset/val_label/\"\n\npath = '../LANE_20_All'\n# train_inputs_path = path + \"/train/\"\n# train_masks_path = path +\"/train_labels/\"\nval_inputs_path = path +\"/images/\"\nval_masks_path = path +\"/val/\"\n\n \n#train_samples = glob.glob(train_inputs_path + \"*.png\")\n# train_samples = [s for s in train_samples if \"seoul\" in s] + \\\n# [s for s in train_samples if \"suwon\" in s] + \\\n# [s for s in train_samples if \"daegu\" in s]\n \nval_samples = glob.glob(val_inputs_path + \"*\")\n# val_samples = [s for s in val_samples if \"seoul\" in s] + \\\n# [s for s in val_samples if \"suwon\" in s] + \\\n# [s for s in val_samples if \"daegu\" in s]\n\n\n\n#print(\"\\n\\nTraining samples = %s\"%(len(train_samples)))\nprint(\"Validation samples = %s\\n\\n\"%(len(val_samples)))\n#train_generator = custom_data_generator.image_generator(train_samples,train_inputs_path,train_masks_path, batch_size,one_hot_label,data_aug)\n#val_generator = custom_data_generator.image_generator(val_samples,val_inputs_path,val_masks_path,batch_size,one_hot_label)\n \n",
"_____no_output_____"
],
[
"print(len(val_samples))",
"_____no_output_____"
],
[
"img = cv2.imread(val_samples[0], -1)\n#img = cv2.resize(img, (256, 256))\nimg.shape\n#plt.imshow(img)",
"_____no_output_____"
],
[
"new_dir = './predictions_lanes_20_NEW_2/'\nif not os.path.isdir(new_dir):\n os.makedirs(new_dir)\n \n",
"_____no_output_____"
],
[
"if is_train: \n \n print(\"Training .....\")\n\nelse:\n print(\"Loading Model .... \",checkpoint_dir)\n json_file = open(checkpoint_dir+model_name+\".json\", 'r')\n loaded_model_json = json_file.read()\n json_file.close()\n model = model_from_json(loaded_model_json)\n model.load_weights(\"./checkpoints/%s/%s_weights_200.h5\"%(checkpoint_name,model_name))\n #asd\n# print(checkpoint_path)\n# files = glob.glob('/home/beemap/Documents/noor-workspace/Semantic-Segmentation-Suite/airbus/train/*.png')\n# accu = []\n# BG_IU, BD_IU, BG_P, BD_P = metrics.calculate_IoU_Per_Epoch(model,val_inputs_path,val_masks_path,checkpoint_dir,1,False)\n# for i in range(50):\n# gen = next(train_generator)\n# gen = next(train_generator)",
"_____no_output_____"
],
[
"from processing import abs_sobel_thresh\nfrom processing import mag_threshold\nfrom processing import *\nfrom util import custom_data_generator as data_util\nfrom models.common import lanenet_wavelet\n\n",
"_____no_output_____"
],
[
"def binary_pipeline(img, mask):\n \n ind = (mask == 0)\n img[ind] = 0\n img_copy = cv.GaussianBlur(img, (3, 3), 0)\n #img_copy = np.copy(img)\n \n # color channels\n s_binary = hls_select(img_copy, sthresh=(140, 255), lthresh=(120, 255))\n #red_binary = red_select(img_copy, thresh=(200,255))\n \n # Sobel x\n x_binary = abs_sobel_thresh(img_copy,thresh=(25, 200))\n y_binary = abs_sobel_thresh(img_copy,thresh=(25, 200), orient='y')\n xy = cv.bitwise_and(x_binary, y_binary)\n \n #magnitude & direction\n mag_binary = mag_threshold(img_copy, sobel_kernel=3, thresh=(30,100))\n dir_binary = dir_threshold(img_copy, sobel_kernel=3, thresh=(0.8, 1.2))\n \n # Stack each channel\n gradient = np.zeros_like(s_binary)\n gradient[((x_binary == 1) & (y_binary == 1)) | ((mag_binary == 1) & (dir_binary == 1))] = 1\n final_binary = cv.bitwise_or(s_binary, gradient)\n \n return final_binary",
"_____no_output_____"
],
[
"from keras.callbacks import TensorBoard,Callback\nfrom PIL import Image\nimport shutil\nimport matplotlib.pyplot as plt\n\nn = 29\nm = 20\n#_, ax = plt.subplots(m, 4, figsize=(20, 50))\n\n# if is_train: \n \n# print(\"Training .....\")\n\n# else:\n# print(\"Loading Model .... \",checkpoint_dir)\n# json_file = open(checkpoint_dir+model_name+\".json\", 'r')\n# loaded_model_json = json_file.read()\n# json_file.close()\n# model = model_from_json(loaded_model_json)\n# model.load_weights(\"./checkpoints/%s/%s_weights_40.h5\"%(checkpoint_name,model_name))\n #asd\n# print(checkpoint_path)\n# files = glob.glob('/home/beemap/Documents/noor-workspace/Semantic-Segmentation-Suite/airbus/train/*.png')\n# accu = []\n# BG_IU, BD_IU, BG_P, BD_P = metrics.calculate_IoU_Per_Epoch(model,val_inputs_path,val_masks_path,checkpoint_dir,1,False)\n# for i in range(50):\n# gen = next(train_generator)\n# gen = next(train_generator)\ni = 0\nfor image in val_samples[:]:\n #gen = next(val_generator)\n #x = gen[0]\n #y = gen[1]\n img = cv2.imread(image, -1)[:, :, :3]\n #img = cv2.resize(img, (256, 256))\n r_img = img\n img = cv2.resize(img, (256, 256))\n #print(img.shape)\n img = np.float32(img)/255.0\n name = os.path.basename(image)\n maskPath = val_masks_path + name#[3:]\n# if not os.path.isfile(maskPath):\n# continue\n# gt = cv2.imread(maskPath, -1)\n #gt = cv2.resize(gt, (256, 256))\n #print(img.shape)\n input_image_gray = data_util.get_image(image,do_aug=[],gray=True, change=False)\n input_image_gray = cv2.resize(input_image_gray, (256, 256))\n \n w1, w2, w3, w4 = lanenet_wavelet(input_image_gray)\n w1 = np.expand_dims(w1, axis=0)\n w2 = np.expand_dims(w2, axis=0)\n w3 = np.expand_dims(w3, axis=0)\n w4 = np.expand_dims(w4, axis=0)\n mask = model.predict([np.expand_dims(img, axis=0), w1, w2, w3, w4], batch_size=None, verbose=0, steps=None)\n #print(mask.shape)\n mask = np.round(mask[0, :, :, 0]).astype(int)\n #print(mask.shape)\n seg = np.zeros((256, 256, 3))\n #print((mask.shape))\n #for c in range(3):\n seg[:, :, 0] += ((mask[:, :] == 1) * (255)).astype('uint8')\n seg[:, :, 1] += ((mask[:, :] == 1) * ( 255)).astype('uint8')\n seg[:, :, 2] += ((mask[:, :] == 1) * ( 255)).astype('uint8')\n\n #mask = model.predict(img, batch_size=None, verbose=0, steps=1)\n# filename = new_dir + name[:-4] + '_pred' + '.png'\n# gt_name = val_masks_path + name[3:]\n# gt_dst = new_dir + name[:-4] + '_gt' + '.png'\n #gt_img = cv2.imread(gt_name, -1)\n\n #print(filename)\n #cv2.imwrite(filename, seg)\n #cv2.imwrite(gt_dst, gt_img)\n name = os.path.basename(image)\n dest = new_dir + name\n result = binary_pipeline(img, mask)\n #if np.any(mask == 1):\n #shutil.copy(gt_name, gt_dst)\n# img = cv2.resize(img, (512, 512))\n# print(mask.shape)\n# mask = cv2.resize(mask, (512, 512))\n# result = cv2.resize(result, (512, 512))\n #new_res = np.concatenate((r_img, seg),axis=1)\n seg = cv2.resize(seg, (512, 512))\n \n cv2.imwrite(dest, seg)\n# ax[i, 0].imshow(img)\n# ax[i, 1].imshow(result)\n# ax[i, 2].imshow(seg)\n# ax[i, 3].imshow(r_img)\n\n# #plt.imshow(abs_sobel_thresh(image, thresh=(20,110)), cmap='gray');\n# #result = binary_pipeline(image)\n i+=1\n #if i >= m:\n #break\n #shutil.copy(image, dest)\n #print(mask.shape)\n# # thr = 0.5 \n# # mask[mask >= thr] = 1\n# # mask[mask < 1] = 0\n# gt = np.reshape(np.argmax(y, axis=-1), (10,256 , 256))\n# pr = np.reshape(np.argmax(mask, axis=-1), (10,256 , 256))\n\n# overlap = np.array(gt,dtype=bool)*np.array(pr,dtype=bool)\n# union = np.array(gt,dtype=bool) + np.array(pr,dtype=bool)\n# IOU = overlap.sum()/float(union.sum())\n# accu.append(IOU)\n# print(IOU)\n\n# f, axarr = plt.subplots(batch_size,3,figsize=(20,50))\n# for j in range(batch_size):\n# axarr[j,0].imshow(x[j,:,:,:])\n# axarr[j,1].imshow(y[j,:,:,0])\n# axarr[j,2].imshow(np.round(mask[j,:,:,0]))\n# axarr[1,0].imshow(x[1,:,:,:])\n# axarr[1,1].imshow(y[1,:,:,0])\n# axarr[1,2].imshow(mask[1,:,:,0])\n# axarr[2,0].imshow(x[2,:,:,:])\n# axarr[2,1].imshow(y[2,:,:,0])\n# axarr[2,2].imshow(mask[2,:,:,0])\n\n\n# overlap = np.array(y[:,:,:,0],dtype=bool)*np.array(np.round(mask[:,:,:,0]),dtype=bool)\n# union = np.array(y[:,:,:,0],dtype=bool) + np.array(np.round(mask[:,:,:,0]),dtype=bool)\n# IOU = overlap.sum()/float(union.sum())\n# print(IOU)\n",
"_____no_output_____"
],
[
"mask",
"_____no_output_____"
],
[
"ind = (mask==1)\n",
"_____no_output_____"
],
[
"gen = next(val_generator)\nx = gen[0]\ny = gen[1]\nf, axarr = plt.subplots(batch_size,3,figsize=(20,50))\nfor j in range(batch_size):\n axarr[j,0].imshow(x[j,:,:,:])\n axarr[j,1].imshow(y[j,:,:,0])\n axarr[j,2].imshow(y[j,:,:,0])",
"_____no_output_____"
],
[
"np.unique(y[j,:,:,0])",
"_____no_output_____"
],
[
"import metrics\nBG_IU, BD_IU, BG_P, BD_P = calculate_IoU_Per_Epoch(model,val_inputs_path,val_masks_path,\"./checkpoints/%s/\"%(checkpoint_name),11,one_hot_label=False)",
"_____no_output_____"
],
[
"BG_IU, BD_IU, BG_P, BD_P",
"_____no_output_____"
],
[
"plt.imshow(y[14,:,:,0])",
"_____no_output_____"
],
[
"import numpy as np\nimport cv2\nfrom util import custom_data_generator as data_util\nimport os\nimport glob\n\n\n\ndef calculate_IoU_Per_Epoch(model,val_inputs_path,val_masks_path,checkpoint_path,epoch_number,one_hot_label=False):\n \n val_samples = [os.path.basename(x) for x in glob.glob(val_inputs_path + \"*.png\")]\n# val_samples = [s for s in val_samples if \"seoul\" in s] + \\\n# [s for s in val_samples if \"suwon\" in s] + \\\n# [s for s in val_samples if \"daegu\" in s]\n val_samples_mini = val_samples[0:15] #+ val_samples[2000:2015]\n\n \n save_path = \"%s/epoch_%s/\"%(checkpoint_path,epoch_number)\n if not os.path.exists(save_path):\n os.makedirs(save_path)\n IU_BG_TP,IU_BG_FN,IU_BG_FP,IU_BG_TN,IU_BD_TP,IU_BD_FN,IU_BD_FP,IU_BD_TN=[],[],[],[],[],[],[],[]\n for j in range(len(val_samples_mini)):\n img = val_samples_mini[j]\n# print(img)\n \n gt = data_util.get_mask(val_masks_path+img,one_hot_label=one_hot_label,do_aug=[])[:,:,0].astype(int) \n input_image = data_util.get_image(val_inputs_path+img,do_aug=[])\n pred = model.predict(np.expand_dims(input_image, axis=0), batch_size=None, verbose=0, steps=None)\n \n \n if one_hot_label:\n pred = np.reshape(pred, (256 , 256, 2))\n pred = np.argmax(pred, axis=-1).astype(int) \n else:\n pred = np.round(pred[0,:,:,0]).astype(int) #(pred *255).astype(int)\n \n classes = np.array([0, 1])\n \n for ii in classes:\n \n TP, FN, FP, TN = IoU(pred, gt, ii)\n \n if ii == 0:\n IU_BG_TP.append(TP)\n IU_BG_FN.append(FN)\n IU_BG_FP.append(FP)\n IU_BG_TN.append(TN)\n\n elif ii == 1:\n print(TP, FN, FP, TN,ii)\n IU_BD_TP.append(TP)\n IU_BD_FN.append(FN)\n IU_BD_FP.append(FP)\n IU_BD_TN.append(TN)\n gt_pred = np.concatenate((data_util.changelabels(gt,'1d2rgb'),data_util.changelabels(pred,'1d2rgb')),axis=1) \n cv2.imwrite(save_path+img,np.concatenate((input_image*255,gt_pred),axis=1))\n# break\n\n print(IU_BD_TP,IU_BD_FN,IU_BD_FP)\n BG_IU = 100 * divided_IoU(IU_BG_TP, IU_BG_FN, IU_BG_FP)\n BD_IU = 100 * divided_IoU(IU_BD_TP, IU_BD_FN, IU_BD_FP)\n BG_P = 100 * divided_PixelAcc(IU_BG_TP, IU_BG_FN)\n BD_P = 100 * divided_PixelAcc(IU_BD_TP, IU_BD_FN)\n \n return BG_IU, BD_IU, BG_P, BD_P \n \n\n \n\ndef IoU(pred, valid, cl):\n tp = np.count_nonzero(np.logical_and(pred == cl, valid == cl))\n fn = np.count_nonzero(np.logical_and(pred != cl, valid == cl))\n fp = np.count_nonzero(np.logical_and(pred == cl, valid != cl))\n tn = np.count_nonzero(np.logical_and(pred != cl, valid != cl))\n return tp, fn, fp, tn\n\n\ndef divided_IoU(tp, fn, fp):\n try:\n return float(sum(tp)) / (sum(tp) + sum(fn) + sum(fp))\n except ZeroDivisionError:\n return 0\n\n\ndef divided_PixelAcc(tp, fn):\n try:\n return float(sum(tp)) / (sum(tp) + sum(fn))\n except ZeroDivisionError:\n return 0\n",
"_____no_output_____"
],
[
"\nBG_IU, BD_IU, BG_P, BD_P = calculate_IoU_Per_Epoch(model,val_inputs_path,val_masks_path,\"./checkpoints/%s/\"%(checkpoint_name),11,one_hot_label=False)\n \nprint(\"\\nBackground IOU = %02f\"%BG_IU)\nprint(\"Main-Class IOU = %02f\"%BD_IU)\nprint(\"Mean IOU = %02f\"%((BG_IU + BD_IU)/2))\nprint(\"Background P-Accuracy = %02f\"%BG_P)\nprint(\"Main-Class P-Accuracy = %02f\\n\"%BD_P)",
"_____no_output_____"
],
[
"# f, axarr = plt.subplots(1,3,figsize=(10,50))\n\nj = 3\n# axarr[j,0].imshow(x[j,:,:,:])\n# plt.imshow()\nfrom matplotlib.pyplot import figure\n\nfigure(figsize=(10,10))\nplt.imshow(np.concatenate((y[j,:,:,0],np.round(mask[j,:,:,0])),axis=-1))",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"# val_generator = custom_data_generator.image_generator(val_samples,val_inputs_path,val_masks_path, batch_size,False)\n\ngen = next(val_generator)\ngen = next(val_generator)\nx = gen[0]\ny = gen[1]\nmask = model.predict(x, batch_size=None, verbose=0, steps=None)\nf, axarr = plt.subplots(batch_size,3,figsize=(20,50))\nfor j in range(10):\n axarr[j,0].imshow(x[j,:,:,:])\n axarr[j,1].imshow(y[j,:,:,0])\n axarr[j,2].imshow(mask[j,:,:,0])\n",
"_____no_output_____"
],
[
"overlap = np.array(gt,dtype=bool)*np.array(pr,dtype=bool)\nunion = np.array(gt,dtype=bool) + np.array(pr,dtype=bool)\nIOU = overlap.sum()/float(union.sum())\naccu.append(IOU)\nprint(IOU)",
"_____no_output_____"
],
[
"from util import custom_data_generator as data_util\nimg = '18_44950_104646.png'\ngt = data_util.get_mask(val_masks_path+img)[:,:,0]\ninput_image = data_util.get_image(val_inputs_path+img)\ninput_image = np.expand_dims(input_image, axis=0)\nprint(gt.dtype)\n# mask = model.predict(input_image, batch_size=None, verbose=0, steps=None)\nplt.imshow(gt[:,:])",
"_____no_output_____"
],
[
"plt.imshow(mask[1])",
"_____no_output_____"
],
[
"def compute_class_weights(labels_dir, label_values):\n '''\n Arguments:\n labels_dir(list): Directory where the image segmentation labels are\n num_classes(int): the number of classes of pixels in all images\n\n Returns:\n class_weights(list): a list of class weights where each index represents each class label and the element is the class weight for that label.\n\n '''\n image_files = [os.path.join(labels_dir, file) for file in os.listdir(labels_dir) if file.endswith('.png')]\n num_classes = len(label_values)\n class_pixels = np.zeros(num_classes)\n total_pixels = 0.0\n for n in range(len(image_files)):\n image = imread(image_files[n], mode=\"RGB\")\n for index, colour in enumerate(label_values):\n class_map = np.all(np.equal(image, colour), axis = -1)\n class_map = class_map.astype(np.float32)\n class_pixels[index] += np.sum(class_map)\n\n print(\"\\rProcessing image: \" + str(n) + \" / \" + str(len(image_files)), end=\"\")\n sys.stdout.flush()\n total_pixels = float(np.sum(class_pixels))\n index_to_delete = np.argwhere(class_pixels==0.0)\n class_pixels = np.delete(class_pixels, index_to_delete)\n class_weights = total_pixels / class_pixels\n class_weights = class_weights / np.sum(class_weights)\n return class_weights",
"_____no_output_____"
],
[
"from scipy.misc import imread\nimport sys\nres=compute_class_weights(\"/media/HDD_4T/Documents/cesar-workspace/lashan/train_labels/\",[(0,0,0),(0,255,0)])",
"_____no_output_____"
],
[
"print(\"\\nBackground IOU = %02f\"%BG_IU)\nprint(\"Main-Class IOU = %02f\"%BD_IU)\nprint(\"Mean IOU = %02f\"%((BG_IU + BD_IU)/2))\nprint(\"Background P-Accuracy = %02f\"%BG_P)\nprint(\"Main-Class P-Accuracy = %02f\\n\"%BD_P)",
"_____no_output_____"
],
[
"pred = np.argmax(img, axis=-1)\npred = np.reshape(pred, (256 , 256))\nplt.imshow(pred)",
"_____no_output_____"
],
[
"# BG_IU, BD_IU, BG_P, BD_P = metrics.calculate_IoU_Per_Epoch(model,val_inputs_path,val_masks_path,checkpoint_dir,1,False)\noverlap = np.array(y[:,:,:,0],dtype=bool)*np.array(np.round(mask[:,:,:,0]),dtype=bool)\nunion = np.array(y[:,:,:,0],dtype=bool) + np.array(np.round(mask[:,:,:,0]),dtype=bool)\nIOU = overlap.sum()/float(union.sum())\nprint(IOU)",
"_____no_output_____"
],
[
"plt.imshow(gt)",
"_____no_output_____"
],
[
"# f, axarr = plt.subplots(batch_size,2,figsize=(30,50))\nfor j in range(40):\n scipy.misc.toimage(np.concatenate((y[j,:,:,0],mask[j,:,:,0]),axis=1), cmin=0.0, cmax=1).save(\"./pred/%s.png\"%j)\n# cv2.imwrite(\"./pred/%s.png\"%j,np.concatenate((y[j,:,:,0].astype(int),mask[j,:,:,0].astype(int)),axis=1))\n# axarr[j,0].imshow(x[j,:,:,:])\n# axarr[j,0].imshow(y[j,:,:,0])\n# axarr[j,1].imshow(mask[j,:,:,0])\nplt.imshow(np.concatenate((y[1,:,:,0].astype(int),mask[1,:,:,0].astype(int)),axis=1))",
"_____no_output_____"
],
[
"plt.imshow(mask[1,:,:,0])",
"_____no_output_____"
],
[
"import scipy.misc\nscipy.misc.toimage(mask[0,:,:,0], cmin=0.0, cmax=1).save('outfile.jpg')",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 10))\nf, axarr = plt.subplots(3,3,figsize=(50,50))\naxarr[0,0].imshow(x[0,:,:,:])\naxarr[0,1].imshow(y[0,:,:,0])\naxarr[0,2].imshow(mask[0,:,:,0])\naxarr[1,0].imshow(x[1,:,:,:])\naxarr[1,1].imshow(y[1,:,:,0])\naxarr[1,2].imshow(mask[1,:,:,0])\naxarr[2,0].imshow(x[2,:,:,:])\naxarr[2,1].imshow(y[2,:,:,0])\naxarr[2,2].imshow(mask[2,:,:,0])\n\n",
"_____no_output_____"
],
[
"plt.imshow(np.round(mask[1,:,:,0]))",
"_____no_output_____"
],
[
"mask[0,:,:,0]",
"_____no_output_____"
],
[
"sat = x[0,:,:,:]\npred = mask[0,:,:,0]",
"_____no_output_____"
],
[
"markers = cv2.watershed(sat,pred)\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb50c38f9ea64a7c636fb9ebb575bb3dbe68fd9 | 3,810 | ipynb | Jupyter Notebook | 10_handlers.ipynb | tightai/tightai | 3a440ad780f5c7ff84a54ad9dc6b342ce3b420b6 | [
"Apache-2.0"
] | 1 | 2020-09-13T08:10:59.000Z | 2020-09-13T08:10:59.000Z | 10_handlers.ipynb | tightai/tightai | 3a440ad780f5c7ff84a54ad9dc6b342ce3b420b6 | [
"Apache-2.0"
] | 1 | 2022-02-26T08:32:58.000Z | 2022-02-26T08:32:58.000Z | 10_handlers.ipynb | tightai/tightai | 3a440ad780f5c7ff84a54ad9dc6b342ce3b420b6 | [
"Apache-2.0"
] | null | null | null | 34.636364 | 202 | 0.497638 | [
[
[
"# default_exp handlers",
"_____no_output_____"
],
[
"#export\nimport base64\nimport os\nfrom tightai.conf import USER_HOME, TIGHTAI_LOCAL_DIRECTORY, TIGHTAI_LOCAL_CREDENTIALS\n\n\nclass CredentialHandler:\n def to_environ(self, username=None, token=None):\n if username is None or token is None:\n raise Exception(\"Username and Token are required.\")\n os.environ['TIGHTAI_USER'] = username\n os.environ['TIGHTAI_TOKEN'] = token\n return\n \n def get_username(self):\n username, _ = self.from_file()\n return username\n\n def get_encoded_token(self):\n username, token = self.from_file()\n username_token = \"{username}:{token}\".format(username=username, token=token)\n encoded = base64.b64encode(username_token.encode())\n return encoded.decode()\n\n\n def from_file(self):\n if not TIGHTAI_LOCAL_CREDENTIALS.exists():\n msg = \"Tight.ai credentials not found.\\n\\nSign up:\\nhttps://tight.ai/signup.\\n\\nLogin:\\n$ python -c \\\"from tightai.auth import login; login();\\\"\\nor\\n$ tight login\\n\"\n print(msg)\n raise Exception(msg)\n with open(TIGHTAI_LOCAL_CREDENTIALS) as inf:\n username = None\n token = None\n for line in inf:\n line = line.split('=')\n line[0] = line[0].strip()\n if line[0] == 'TIGHTAI_USER':\n username = line[1].strip().replace(\"'\", \"\").replace('\"', '')\n elif line[0] == 'TIGHTAI_TOKEN':\n token = line[1].strip().replace(\"'\", \"\").replace('\"', '')\n self.to_environ(username=username, token=token)\n return username, token\n return\n\n def remove(self):\n if TIGHTAI_LOCAL_CREDENTIALS.exists():\n TIGHTAI_LOCAL_CREDENTIALS.unlink()\n print(\"Successfully logged out.\")\n return \n print(\"Not logged in. Do you need to run tightai.login()?\")\n return\n\n def to_file(self, username=None, token=None):\n if username is None or token is None:\n raise Exception(\"Username and Token are required.\")\n if TIGHTAI_LOCAL_CREDENTIALS.exists():\n TIGHTAI_LOCAL_CREDENTIALS.parent.mkdir(parents=True, exist_ok=True)\n with open(TIGHTAI_LOCAL_CREDENTIALS, 'w') as cred_txt:\n docstring = f\"[apikey]\\nTIGHTAI_USER='{username}'\\nTIGHTAI_TOKEN='{token}'\"\n cred_txt.writelines(docstring)\n return True\n return None\n\n\ncredentials = CredentialHandler()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
ecb513d728beeb9c6faaf03e80e8c0b34c9df163 | 14,671 | ipynb | Jupyter Notebook | Lesson01_Variables/lesson01.ipynb | WomensCodingCircle/CodingCirclePython | 703aff89ffa0a3933baa02881f325d62087eb0af | [
"MIT"
] | 4 | 2017-02-09T20:05:04.000Z | 2018-12-06T13:13:35.000Z | Lesson01_Variables/lesson01.ipynb | WomensCodingCircle/CodingCirclePython | 703aff89ffa0a3933baa02881f325d62087eb0af | [
"MIT"
] | null | null | null | Lesson01_Variables/lesson01.ipynb | WomensCodingCircle/CodingCirclePython | 703aff89ffa0a3933baa02881f325d62087eb0af | [
"MIT"
] | 12 | 2015-12-07T17:22:01.000Z | 2021-12-29T02:50:15.000Z | 23.933116 | 452 | 0.560085 | [
[
[
"# Variables, expressions, and statements",
"_____no_output_____"
],
[
"## Values and Types\nValues are basic things a program works with. Values come in several different types:\n* A `string` is any value between quotes ('' single or double \"\") e.g. 'Hello Coding Circle', \"I am very smart\", \"342\"\n* An `integer` is any whole number, positive or negative e.g. 145, -3, 5\n* A `float` is any number with a decimal point e.g. 3.14, -2.5 \n\nTo tell what type a value is, use the built-in function `type()`",
"_____no_output_____"
]
],
[
[
"type('I am amazing!')",
"_____no_output_____"
],
[
"type(145)",
"_____no_output_____"
],
[
"type(2.5)",
"_____no_output_____"
]
],
[
[
"To print a value to the screen, we use the function `print()`\n\ne.g. `print(1)`",
"_____no_output_____"
]
],
[
[
"print(\"Hello World\")",
"_____no_output_____"
]
],
[
[
"Jupyter notebooks will always print the value of the last line so you don't have to. You can suppress this with a semicolon ';'",
"_____no_output_____"
]
],
[
[
"\"Hello World\"",
"_____no_output_____"
],
[
"\"Hello World\";",
"_____no_output_____"
]
],
[
[
"### TRY IT\nPredict and then print the type of 'Orca'",
"_____no_output_____"
],
[
"## Variables\nA variable is a name that you give a value. You can then use this name anywhere you would use the value that the name refers to. \n\nIt has some rules.\n* It must only contain letters, numbers and/or the underscore character. \n* However, it cannot start with a number.\n* It can start with an underscore but this usually means something special so stick to letters for now. \n\nTo assign a value to a variable, you use the assignment operator, which is '`=`' e.g., `my_name = 'Charlotte'`",
"_____no_output_____"
]
],
[
[
"WHALE = 'Orca'\nnumber_of_whales = 10\nweight_of_1_whale = 5003.2",
"_____no_output_____"
]
],
[
[
"Notice that when you ran that, nothing printed out. To print a variable, you use the same statement you would use to print the value. e.g. `print(WHALE)`",
"_____no_output_____"
]
],
[
[
"print(number_of_whales)",
"_____no_output_____"
]
],
[
[
"### TRY IT\nAssign the name of a sea creature to the variable `sea_creature`. Then print the value.",
"_____no_output_____"
],
[
"*Reccomendation* \nName your variables with descriptive names. Naming a variable 'a' is easy to type but won't help you figure out what it is doing when you come back to your code six months later.",
"_____no_output_____"
],
[
"## Operators and operands\nOperators are special symbols that represent computations that the computer performs. We have already learned one operator: the assignment operator '='.\n\nOperands are the values the operator is applied to.\n\nBasic math operators\n* \\+ addition\n* \\- subtraction\n* \\* multiplication\n* / division\n* \\*\\* power (exponentiation)\n\nTo use these operators, put a value or variable on either side of them. You can even assign the new value to a variable or print it out. They work with both integers or floats. ",
"_____no_output_____"
]
],
[
[
"1 + 2",
"_____no_output_____"
],
[
"fish = 15\nfish_left = fish - 3\nprint(fish_left)",
"_____no_output_____"
],
[
"print(3 * 2.1)",
"_____no_output_____"
],
[
"number_of_whales ** 2",
"_____no_output_____"
],
[
"print(5 / 2)",
"_____no_output_____"
]
],
[
[
"Hint: You can use a variable and assign it to the same variable name in the same statement.",
"_____no_output_____"
]
],
[
[
"number_of_whales = 8\nnumber_of_whales = number_of_whales + 2 \nprint(number_of_whales)",
"_____no_output_____"
]
],
[
[
"### TRY IT\nFind the result of 6^18.",
"_____no_output_____"
],
[
"## Order of operations\n\nYou can combine many operators in a single python statement. The way python evaluates it is the same way you were taught to in elementary school. PEMDAS for Please Excuse My Dear Aunt Sally. Or 1. Parentheses, 2. Exponents, 3. Multiplication, 4. Division, 5. Addition, 6. Subtraction. Left to right, with that precedence. It is good practice to always include parentheses to make your intention clear, even if order of operations is on your side.",
"_____no_output_____"
]
],
[
[
"2 * 3 + 4 / 2",
"_____no_output_____"
],
[
"(2 * (3 + 4)) / 2",
"_____no_output_____"
]
],
[
[
"## Modulus operator\n\nThe modulus operator is not one you were taught in school. It returns the remainder of integer division. It is useful in a few specific cases, but you could go months without using it.",
"_____no_output_____"
]
],
[
[
"5 % 2",
"_____no_output_____"
]
],
[
[
"### TRY IT\nFind if 12342872 is divisible by 3",
"_____no_output_____"
],
[
"## String operations\nThe `+` operator also works on strings. It is the concatenation operator, meaning it joins the two strings together.",
"_____no_output_____"
]
],
[
[
"print('Hello ' + 'Coding Circle')",
"_____no_output_____"
],
[
"print(\"The \" + WHALE + \" lives in the sea.\")",
"_____no_output_____"
]
],
[
[
"Hint: Be careful with spaces",
"_____no_output_____"
]
],
[
[
"print(\"My name is\" + \"Charlotte\")",
"_____no_output_____"
]
],
[
[
"### TRY IT\nPrint out Good morning to the sea creature you stored in variable named `sea_creature` earlier.",
"_____no_output_____"
],
[
"## Asking the user for input\n\nTo get an input for the user we use the built-in function `input()` and assign it to a variable.\n\nNOTE: The result is always a string.\n\n**WARNING** if you leave an input box without ever putting input in, jupyter won't be able to run any code. Ex. you run a cell with `input` and then re-run that cell before submitting input. To fix this hang the stop button in the menu.",
"_____no_output_____"
]
],
[
[
"my_name = input()\nprint(my_name)",
"_____no_output_____"
]
],
[
[
"You can pass a string to the `input()` function to prompt the user for what you are looking for.\n\ne.g. `input('How are you feeling?')`\n\nHint, add a new line character `\"\\n\"` to the end of the prompt to make the user enter it on a new line.",
"_____no_output_____"
]
],
[
[
"favorite_ocean_animal = input(\"What is your favorite sea creature?\\n\")\nprint(\"The \" + favorite_ocean_animal + \" is so cool!\")",
"_____no_output_____"
]
],
[
[
"If you want the user to enter a number, you will have to convert the string. Here are the conversion commands.\n\n* To convert a variable to an integer, use `int` -- e.g., `int(variable_name)`\n* To convert a variable to a float, use `float` -- e.g., `float(variable_name)`\n* To convert a variable to a string, use `str` -- e.g., `str(variable_name)`",
"_____no_output_____"
]
],
[
[
"number_of_fish = input(\"How many fish do you want?\\n\")\nnumber_of_fish_int = int(number_of_fish)\nprint(number_of_fish_int * 1.05)",
"_____no_output_____"
]
],
[
[
"### TRY IT\nPrompt the user for their favorite whale and store the value in a variable called `favorite_whale`.",
"_____no_output_____"
],
[
"## Comments\nComments let you explain your progam to someone who is reading your code. Do you know who that person is? It is almost always you in six months. Don't screw over future you. Comment your code.\n\nTo make a comment: you use the `#` symbol. You can put a comment on its own line or at the end of a statement.",
"_____no_output_____"
]
],
[
[
"# Calculate the price of fish that a user wants\nnumber_of_fish = input(\"How many fish do you want?\\n\") # Ask user for quantity of fish\nnumber_of_fish_int = int(number_of_fish) # raw_input returns string, so convert to integer\nprint(number_of_fish_int * 1.05) # multiply by price of fish",
"_____no_output_____"
]
],
[
[
"### TRY IT\nWrite a comment. ",
"_____no_output_____"
],
[
"# Project: Milestones\nWe are going to create an application that prompts the user for their (or their child's) birth year and will calculate and tell them the years of their milestones: Drive a car at 16, Drink alcohol at 21, and Run for president at 35.\n\n1. Ask the user for the birth year and store in a variable called `birth_year`.\n2. Convert `birth_year` to an int and store in variable called `birth_year`.\n3. Add 16 to the `birth_year` and store in a variable called `drive_car_year`.\n4. Add 21 to the `birth_year` and store in a variable called `alcohol_year`.\n5. Add 35 to the `birth_year` and store in a variable called `president_year`.\n6. Print out the message \"You can drive a car in: `drive_car_year`\" and similar messages for the other years. Hint: you will need to use string concatenation, you will also have to cast the integer years to strings using the `str()` method.",
"_____no_output_____"
]
],
[
[
"\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
ecb51f1a16e7dcecfa515bc902d769b4fadfb99b | 56,983 | ipynb | Jupyter Notebook | lecture_notebooks/L02 Variables and Expressions.ipynb | zrtamg/intro_python | cc7a246c2a0bbe22020c664ea86fce7360ce2f68 | [
"MIT"
] | 4 | 2022-01-04T21:58:01.000Z | 2022-01-06T22:51:34.000Z | lecture_notebooks/L02 Variables and Expressions.ipynb | zrtamg/intro_python | cc7a246c2a0bbe22020c664ea86fce7360ce2f68 | [
"MIT"
] | null | null | null | lecture_notebooks/L02 Variables and Expressions.ipynb | zrtamg/intro_python | cc7a246c2a0bbe22020c664ea86fce7360ce2f68 | [
"MIT"
] | 13 | 2022-01-04T22:02:42.000Z | 2022-03-31T23:41:53.000Z | 22.103569 | 483 | 0.522103 | [
[
[
"# Lecture 2 - Variables and Expressions (https://bit.ly/intro_python_02)\n\nToday:\n* Variables\n* Assignment is not equals\n* Copying References\n* Expressions\n* Statements\n* Variable Names\n* Operators\n* Abbreviated assignment\n* Logical operators\n* Order of operations\n",
"_____no_output_____"
],
[
"# Variables",
"_____no_output_____"
]
],
[
[
"# As in algebra, we use variables to 'refer' to things\n\nx = 1 # x is a variable refering to the int 1\n\ny = \"foo\" # y is a variable refering to the string \"foo\"",
"_____no_output_____"
]
],
[
[
"Think of x as \"referring to 1\". That is x is really a reference to the object representing 1. \n * x -----> 1\n * y ------> \"foo\"\n\nUnder the hood, in general, there is a piece of memory (bits) representing x \nthat stores the location of where the value 1 is actually stored in memory. \n\nSometimes, in some languages, x or y stores the content directly, e.g. x is actually \nmemory representing 1. However in Python variables are references.",
"_____no_output_____"
],
[
"# Assignment is not equals",
"_____no_output_____"
]
],
[
[
"# The consequence of this \"refers to\" notion is that \n# equals sign in Python means \"assignment\" not \"equals\"\n\nx = 1 # x refers to 1\n\n# Because = means assignment, we can update assignments, without it\n# being meaningless in a math sense\n\nx = \"foo\" # x refers to \"foo\"\n\n#What's the value of x?\nprint(x)",
"foo\n"
]
],
[
[
"# Copying references",
"_____no_output_____"
]
],
[
[
"# We can also copy variables\n\nx = 5\n\ny = x # Now x and y refer to the same thing (5)\n\nprint(y)",
"5\n"
],
[
"# But note, reassignment and copying can be co-mingled:\n\nx = 5\n\ny = x # Now x and y refer to the same thing (5)\n\nx = 10 # But now we change what x refers to, this doesn't change y!\n\nprint(x) \nprint(y)",
"10\n5\n"
]
],
[
[
"# Variable Names\n\nVariables don't have to be single letters (in fact, that's a terrible idea), but can be **any combination of alpha-numeric characters and the '_' symbol, providing they don't start with a number and aren't already a reserved keyword**. Remember this!",
"_____no_output_____"
]
],
[
[
"# Variable names\n\nfoo = 1 # Legit variable name\n\nbar2 = 2 # Also legit\n\n_boo = 3 # Legit\n\nsuper_helpful_variable_name = 4 # Legit\n\nsomePeopleLikeCamelCase = 5 # Fine variable name\n\nprint(super_helpful_variable_name)",
"4\n"
],
[
"# Not okay\n\n3l33t = 4 # Not okay\n\nnot^okay = 5 # Not okay\n\n# Etc.",
"_____no_output_____"
]
],
[
[
"Reserved key words, see Chapter 2 of the open textbook:\n\nhttp://openbookproject.net/thinkcs/python/english3e/variables_expressions_statements.html",
"_____no_output_____"
],
[
"# Expressions",
"_____no_output_____"
]
],
[
[
"# Now we have basic types and variables, we can do stuff with them\n\nx = 1\n\ny = 2\n\nx + y - 2 # This is an expression, the intepretter \n# has to \"evaluate it\" to figure out the value by \n# combining variables (x, y), \n# values (2) and operators (+, -)",
"_____no_output_____"
],
[
"# When we're calling a function (we'll learn about functions soon) (here the \"type\" function), \n# we're also evaluating an expression\n\ntype(3.14)",
"_____no_output_____"
]
],
[
[
"**Definition:** An expression is a combination of values, variables, operators, and calls to functions. Expressions need to be evaluated and result in a value (could be number, a string, an object, a list, etc.) \n",
"_____no_output_____"
],
[
"# Challenge 1",
"_____no_output_____"
]
],
[
[
"# Write an expression that uses x and y and evaluates to 3\nx = 1\ny = 2",
"_____no_output_____"
]
],
[
[
"# Statements",
"_____no_output_____"
]
],
[
[
"# Statements are instructions to the intepreter \n\nx = 1 # This is a statement, you're telling Python, make x refer to the value 1\n\nif x > 0: # Conditionals (like if (and others we'll see soon, \n # are also \"statements\"))\n print(\"boo\")",
"boo\n"
]
],
[
[
"* A statement is everything that can make up a line of Python code.\n\n* A statement does something.\n\nNote, therefore, that expressions are statements as well.\n\n* All expressions are statements.\n* Not all statements are expressions.\n\n\nNote: the definition of statement is a little contested (you may see slightly different versions in textbooks, but I like this one)",
"_____no_output_____"
],
[
"# Operators",
"_____no_output_____"
]
],
[
[
"# Operators perform basic operations on objects\n\n# Let's start with arithmetic operators\n\nx = 12\ny = 5\n\nx + y # + is the addition operator, duh",
"_____no_output_____"
],
[
"x * y # multiplication",
"_____no_output_____"
],
[
"x ** y # exponentiation ",
"_____no_output_____"
],
[
"x / y # What's the value of this one ?",
"_____no_output_____"
],
[
"x // y # And this one?",
"_____no_output_____"
],
[
"-x # Yup, we have negation",
"_____no_output_____"
]
],
[
[
"Note: \n * Negation is an example of a unary operator (it takes one operand)\n\n * The other operators are binary (they take two operands)",
"_____no_output_____"
]
],
[
[
"5 % 2 # The modulus operator, it returns the \"remainder\"",
"_____no_output_____"
],
[
"# It's a good way to determine if a number is divisible by another,\n# because if it is, the expression value will be 0\n4 % 2",
"_____no_output_____"
]
],
[
[
"# Challenge 2",
"_____no_output_____"
]
],
[
[
"x = 5\ny = 2\n# Write a statement that divides x by y, forgetting the remainder, storing the result in a variable z",
"_____no_output_____"
]
],
[
[
"# Operator Overloading",
"_____no_output_____"
]
],
[
[
"5 + 10 + 12 # We can use \"+\" to add together strings of numbers",
"_____no_output_____"
],
[
"# Some arithmetic operators are also \"overloaded\" to work with strings\n# (This is an example of \"polymorphism\", that we'll meet again much later)\n\n\"This\" + \"is\" + \"contrived\" # The '+' concatenates strings",
"_____no_output_____"
],
[
"# Note, this doesn't work because Python doesn't know how to add a string and\n# a number together\n\n\"this\" + 5",
"_____no_output_____"
]
],
[
[
"# Abbreviated Assignment\n\nYou will see this a lot:",
"_____no_output_____"
]
],
[
[
"x = 1\n\n# Instead of writing\n\nx = x + 5\n\n# you can use the shorthand:\n\nx += 5 # This means \"add 5 to value x refers to\"\n\nx",
"_____no_output_____"
]
],
[
[
"This works for all the obvious math operators:",
"_____no_output_____"
]
],
[
[
"\n\nx *= 2 # multiply x by 2, aka x = x * 2\n\nprint(x)",
"22\n"
],
[
"x -= 3 # subtract three, aka x = x - 3\n\nprint(x)",
"19\n"
],
[
"x /= 2 # divide by 2 \n\nprint(x)",
"9.5\n"
],
[
"x //= 2 # divide by 2 and forget fraction\n\nprint(x)",
"4.0\n"
],
[
"x %= 2 # Take x mod 2 and store the result in x, i.e. x = x % 2\n\nprint(x)",
"0.0\n"
]
],
[
[
"# Challenge 3",
"_____no_output_____"
]
],
[
[
"x = 7 \ny = 2\n# Use abbreviated assignment to subtract y from x, storing the result as x",
"_____no_output_____"
]
],
[
[
"# Boolean Type\n\nPython has a special boolean (binary) type, which can either be 'True' or 'False'. \n\nThis is essential for logical expressions.\n",
"_____no_output_____"
]
],
[
[
"type(True)",
"_____no_output_____"
],
[
"type(False)",
"_____no_output_____"
]
],
[
[
"# Logical Operators\n\n Booleans are used for making decisions in your program.\n \n To do this we use logical operators, which do, err, logic and evaluate to booleans.",
"_____no_output_____"
]
],
[
[
"x = 5\ny = 10\n\nx > y # The greater than operator compares two things",
"_____no_output_____"
],
[
"# To see the relationship with Booleans\nx = 5\ny = 10\n\n\ntype(x > y)",
"_____no_output_____"
],
[
"# There are a bunch of these\n\nx >= y # Is x greater than or equals to y?",
"_____no_output_____"
],
[
"x < y # Is x less than y?",
"_____no_output_____"
],
[
"x <= y # Less than or equals?",
"_____no_output_____"
],
[
"# What about this one?\n\nx == y",
"_____no_output_____"
]
],
[
[
"* As we discussed earlier, in Python (and many languages) '=' is the assignment operator\n* Logical equals is '=='. Some people find this weird, but you get over it.\n",
"_____no_output_____"
]
],
[
[
"# Python also has the not logical equals operator !=\n\nx != y # Read this as \"does x not equal y?\"",
"_____no_output_____"
],
[
"# We can compose logical statements into complex expressions \n# using logical 'and' and 'or'\n\nx = 5\ny = 10\nz = 7\n\nx >= y or z > x # Says: True if x is greater than or equal to y or z is greater than x",
"_____no_output_____"
],
[
"# Similarly\n\ny > x and y > z # Says: True if y is greater than x AND y is greater than z",
"_____no_output_____"
],
[
"# There is also the unary negation operator: not\n\nnot True",
"_____no_output_____"
],
[
"# Use it to switch True to False and vice versa:\n\ny = 0\nx = 1\n\nnot y > x",
"_____no_output_____"
]
],
[
[
"Logical comparisons also work with strings",
"_____no_output_____"
]
],
[
[
"# String comparison\n\n\"cats\" > \"dogs\" # ?",
"_____no_output_____"
],
[
"# Suggested exercise: play with >, <, >=, <=, not, and, or, () \n# and see if they do what you expect\n\n## Also definitely read text book on this for more thorough treatment",
"_____no_output_____"
]
],
[
[
"# Challenge 4",
"_____no_output_____"
]
],
[
[
"x = int(input(\"Enter a number\"))\ny = int(input(\"Enter a second number\"))\n# Write a logical expression that is True if and only if x is greater than y or x is divisible by y",
"_____no_output_____"
]
],
[
[
"# Order of operations\n\nThis is a boring topic, but if you don't understand it, you'll write lots of bugs",
"_____no_output_____"
]
],
[
[
"# Just like in math, it is important to know the order of operands\n# in Python\n\nx = 2\ny = 3\n\nx * y + x # Yup, this works just like math",
"_____no_output_____"
],
[
"x * (y + x) # If you want to force the addition \n# before the multiplication you can use brackets, like in math\n\n# Technically, the brackets have highest precedence of any operator",
"_____no_output_____"
],
[
"# Exponents have higher precedence than division/multiplication\n\n2 * 2**2 # The exponent happens first",
"_____no_output_____"
],
[
"# Arithmetic operators have precidence over logical operators\n\nx * y > x + y # This could also be written (x * y) > (x + y)",
"_____no_output_____"
],
[
"# Sometimes it is helpful to use brackets, because it's hard to remember\n# the order of operations, consider this..\n\nnot 3 * 5 > 10 and 2 > 1",
"_____no_output_____"
],
[
"# Which I think is much clearer as..\n\n(not 3 * 5 > 10) and (2 > 1) # The brackets are just there for clarity",
"_____no_output_____"
],
[
"# Or maybe even.. \n\n(not (3 * 5 > 10)) or (2 > 1)",
"_____no_output_____"
]
],
[
[
"# Challenge 5",
"_____no_output_____"
]
],
[
[
"# Change the expression below so that the 'not' is applied after the 'or' \n(not (3 * 5 > 10)) or (2 > 1)",
"_____no_output_____"
]
],
[
[
"# Reading\n\nOpenbook:\n\n* Read Chapter 2 on expressions, variables, statements and operators:\n * http://openbookproject.net/thinkcs/python/english3e/variables_expressions_statements.html\n\n \n# Homework\n\n* Go to Canvas and complete the second lecture quiz, which involves completing each challenge problem\n* See \"Reading 2\" in Zybooks\n* Assignment 1 is now available, it is due a week Friday at 11:59pm.\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb541fe85c47c5dc48cbe08e93882f6dd256795 | 566,171 | ipynb | Jupyter Notebook | radio_model.ipynb | M-T3K/RadiographyClassifier | 0734a850941994162406ebe3b4f3192ed8ad7b22 | [
"MIT"
] | null | null | null | radio_model.ipynb | M-T3K/RadiographyClassifier | 0734a850941994162406ebe3b4f3192ed8ad7b22 | [
"MIT"
] | null | null | null | radio_model.ipynb | M-T3K/RadiographyClassifier | 0734a850941994162406ebe3b4f3192ed8ad7b22 | [
"MIT"
] | null | null | null | 1,031.276867 | 245,606 | 0.946484 | [
[
[
"# Respiratory Condition Radiography Model\n\nIncludes Covid-19, Viral Pneumonia, and Metastasis (Lung Opacity)",
"_____no_output_____"
]
],
[
[
"import os\nimport imageio\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport random\nimport shutil\n\nimport scipy.ndimage as ndi\nfrom sklearn.metrics import classification_report, confusion_matrix\nimport tensorflow.keras.backend as tfback\nfrom keras.regularizers import l1, l2\nfrom keras.utils.vis_utils import plot_model\nfrom keras.models import Model, Sequential\nfrom keras.layers import Input, Dense, Flatten, Dropout, BatchNormalization\nfrom keras.layers import Conv2D, MaxPooling2D, SeparableConv2D\nfrom tensorflow.keras.optimizers import SGD, Adam\nfrom keras.preprocessing.image import ImageDataGenerator\nfrom keras.callbacks import ModelCheckpoint, ReduceLROnPlateau, EarlyStopping\nimport tensorflow as tf\n\n%matplotlib inline\n#Kaggle notebook input directory\ninput_path = 'Dataset/COVID_Radiography_Dataset/'",
"_____no_output_____"
]
],
[
[
"## Dataset",
"_____no_output_____"
]
],
[
[
"#Ploting some samples\n\nfig, ax = plt.subplots(1, 4, figsize=(15, 7))\nax=ax.ravel()\nplt.tight_layout()\n\nfor i,_dir in enumerate(['COVID/', 'NORMAL/', 'Viral Pneumonia/', 'Lung_Opacity/']):\n im_file = os.listdir(input_path+_dir)[0]\n full_path = input_path+_dir+im_file\n ax[i].imshow(plt.imread(full_path), cmap='gray')\n ax[i].set_title('Condition: {}'.format(_dir[:-1]))",
"_____no_output_____"
]
],
[
[
"Checking how many samples we have for each condition and total files:",
"_____no_output_____"
]
],
[
[
"n_covid = len(os.listdir(input_path + 'COVID/'))\nn_normal = len(os.listdir(input_path + 'NORMAL/'))\nn_pneumonia = len(os.listdir(input_path + 'Viral Pneumonia'))\nn_metastasis = len(os.listdir(input_path + 'Lung_Opacity')) # Lung_Opacity\nprint(f'COVID: {n_covid} images, Normal: {n_normal} images, Pneumonia: {n_pneumonia} images, Metastasis: {n_metastasis} \\nTotal images: {(n_covid+n_normal+n_pneumonia+n_metastasis)}.')",
"COVID: 3616 images, Normal: 10192 images, Pneumonia: 1345 images, Metastasis: 6012 \nTotal images: 21165.\n"
],
[
"# Data visualization for pixel distribution\n\n\nfig, ax = plt.subplots(2, 4, figsize=(15, 7))\nax=ax.ravel()\nplt.subplots_adjust(hspace=0.4)\n\nfor i,_dir in enumerate(['NORMAL/', 'COVID/', 'Viral Pneumonia/', 'Lung_Opacity/']):\n im_file = os.listdir(input_path+_dir)[0]\n full_path = input_path+_dir+im_file\n im=imageio.imread(full_path)\n hist=ndi.histogram(im, min=0, max=255, bins=256)\n cdf = hist.cumsum() / hist.sum()\n ax[i].plot(hist)\n ax[i].set_title(f'Histogram {_dir[:-1]}')\n ax[i].grid()\n ax[i+3].plot(cdf, 'g')\n ax[i+3].set_title(f'Cummulative distribution {_dir[:-1]}')\n ax[i+3].grid()",
"_____no_output_____"
],
[
"# Creating folders for train and test samples\n\nif os.path.isdir('training'):\n shutil.rmtree('training')\nif os.path.isdir('training/train'):\n shutil.rmtree('training/train')\nif os.path.isdir('training/test/'):\n shutil.rmtree('training/test')\n\nos.mkdir('training/')\n# os.mkdir('training/train')\nos.mkdir('training/test')\n\nshutil.copytree(input_path, 'training/train')\n",
"_____no_output_____"
],
[
"# Checking how many files we have in the newest folders\n\n# Removing excelsheets \nif os.path.isfile('training/train/COVID.metadata.xlsx'):\n os.remove('training/train/COVID.metadata.xlsx')\nif os.path.isfile('training/train/README.md.txt'):\n os.remove('training/train/README.md.txt')\nif os.path.isfile('training/train/Viral Pneumonia.metadata.xlsx'):\n os.remove('training/train/Viral Pneumonia.metadata.xlsx')\nif os.path.isfile('training/train/NORMAL.metadata.xlsx'):\n os.remove('training/train/NORMAL.metadata.xlsx')\nif os.path.isfile('training/train/Lung_Opacity.metadata.xlsx'):\n os.remove('training/train/Lung_Opacity.metadata.xlsx')\n\ncnt_files=0\nos.listdir('training/train')\nfor file in {'COVID', 'NORMAL', 'Viral Pneumonia', 'Lung_Opacity'}:\n for files in os.listdir(os.path.join('training/train/',file)):\n cnt_files += 1\nprint(cnt_files)",
"21165\n"
],
[
"# Performing data train/test split\n\nsplit_percentage = 0.3 # 30% Testing, 70% training\n\nsrc = 'training/train'\ndst = 'training/test'\n\nif os.path.isdir('training/test/COVID'):\n shutil.rmtree('training/test/COVID')\nif os.path.isdir('training/test/Viral Pneumonia'):\n shutil.rmtree('training/test/Viral Pneumonia')\nif os.path.isdir('training/test/NORMAL'):\n shutil.rmtree('training/test/NORMAL')\nif os.path.isdir('training/test/Lung_Opacity'):\n shutil.rmtree('training/test/Lung_Opacity')\n\nos.mkdir('training/test/COVID')\nos.mkdir('training/test/Viral Pneumonia')\nos.mkdir('training/test/NORMAL')\nos.mkdir('training/test/Lung_Opacity')\n\nsplit_length = int(cnt_files * split_percentage)\ntotal_files_copied = 0\nfor folders in os.listdir(src):\n dir = os.path.join(src, folders)\n cut_length = len([name for name in os.listdir(dir) if os.path.isfile(os.path.join(dir, name))]) * split_percentage\n count=0\n for files in os.listdir(dir):\n shutil.move(os.path.join(src, folders, files), os.path.join(dst, folders, files))\n if count >= cut_length:\n break\n total_files_copied += 1\n count += 1\n\nprint(f\"Expected Files: {cnt_files * split_percentage} \\nCopied Files: {total_files_copied} ({total_files_copied / cnt_files * 100})\")",
"Expected Files: 6349.5 \nCopied Files: 6351 (30.007087172218284)\n"
]
],
[
[
"## Relevant Variables",
"_____no_output_____"
]
],
[
[
"# Defining some constant variables to use in the preprocessor\ntrain_path = 'training/train'\ntest_path = 'training/test'\nIMAGE_SIZE = (250, 250) # 150x150 (height, width) pixels\nNUM_CLASSES = 4 #COVID, NORMAL, PNEUMONIA, LungOpacity (Potential Metastasis)\nBATCH_SIZE = 32 # try reducing batch size or freeze more layers if your GPU runs out of memory\nNUM_EPOCHS = 20 # idem for epochs",
"_____no_output_____"
]
],
[
[
"## Model",
"_____no_output_____"
]
],
[
[
"#Train datagen here is a preprocessor\n\nvalidation_percentage = 0.2\n\ntrain_ds = tf.keras.utils.image_dataset_from_directory(\n 'training/train',\n validation_split=validation_percentage,\n subset=\"training\",\n seed=42,\n image_size=IMAGE_SIZE,\n batch_size=BATCH_SIZE\n )\n\nval_ds = tf.keras.utils.image_dataset_from_directory(\n 'training/train',\n validation_split=validation_percentage,\n subset=\"validation\",\n seed=42,\n image_size=IMAGE_SIZE,\n batch_size=BATCH_SIZE\n )\n\nclass_names = train_ds.class_names\nprint(class_names)\n\nimport matplotlib.pyplot as plt\n\nplt.figure(figsize=(10, 10))\nfor images, labels in train_ds.take(1):\n for i in range(9):\n ax = plt.subplot(3, 3, i + 1)\n plt.imshow(images[i].numpy().astype(\"uint8\"))\n plt.title(class_names[labels[i]])\n plt.axis(\"off\")\n\n\nnormalization_layer = tf.keras.layers.Rescaling(1./255)\nnormalized_ds = train_ds.map(lambda x, y: (normalization_layer(x), y))\nimage_batch, labels_batch = next(iter(normalized_ds))\nfirst_image = image_batch[0]\n# Notice the pixel values are now in `[0,1]`.\nprint(np.min(first_image), np.max(first_image))\n\nAUTOTUNE = tf.data.AUTOTUNE\n\ntrain_ds = train_ds.cache().prefetch(buffer_size=AUTOTUNE)\nval_ds = val_ds.cache().prefetch(buffer_size=AUTOTUNE)\n\nmodel = tf.keras.Sequential([\n tf.keras.layers.Rescaling(1./255),\n tf.keras.layers.Conv2D(32, 3, activation='relu', padding='same'),\n tf.keras.layers.MaxPooling2D(2, strides=2),\n tf.keras.layers.Conv2D(64, 3, activation='relu', padding='same'),\n tf.keras.layers.MaxPooling2D(2, strides=2),\n tf.keras.layers.Conv2D(32, 3, activation='relu'),\n tf.keras.layers.BatchNormalization(),\n tf.keras.layers.Dropout(0.6),\n tf.keras.layers.Conv2D(128, 5, activation='relu', padding='same'),\n tf.keras.layers.MaxPooling2D(2,strides=2),\n tf.keras.layers.Dropout(0.3),\n tf.keras.layers.Conv2D(256, 3, activation='relu', padding='same'),\n tf.keras.layers.MaxPooling2D(2,strides=2),\n tf.keras.layers.Dropout(0.3),\n tf.keras.layers.Conv2D(256, 5, activation='relu', padding='same'),\n tf.keras.layers.MaxPooling2D(2,strides=2),\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(64, activation='relu'),\n tf.keras.layers.Dense(32, activation='relu'),\n tf.keras.layers.Dense(NUM_CLASSES, activation='softmax')\n])\n\nmodel.compile(\n optimizer='adam',\n loss=tf.losses.SparseCategoricalCrossentropy(from_logits=True),\n metrics=['accuracy'])\n\nresult = model.fit(\n train_ds,\n validation_data=val_ds,\n epochs=3\n)",
"Found 14810 files belonging to 4 classes.\nUsing 11848 files for training.\nFound 14810 files belonging to 4 classes.\nUsing 2962 files for validation.\n['COVID', 'Lung_Opacity', 'Normal', 'Viral Pneumonia']\n0.008331215 0.9589022\nEpoch 1/3\n371/371 [==============================] - 1394s 4s/step - loss: 0.7529 - accuracy: 0.6974 - val_loss: 0.8668 - val_accuracy: 0.7600\nEpoch 2/3\n371/371 [==============================] - 1349s 4s/step - loss: 0.4872 - accuracy: 0.8139 - val_loss: 0.5788 - val_accuracy: 0.7890\nEpoch 3/3\n371/371 [==============================] - 1358s 4s/step - loss: 0.3993 - accuracy: 0.8486 - val_loss: 0.4109 - val_accuracy: 0.8406\n"
],
[
"def plot_accs(result, epochs):\n acc = result.history['accuracy']\n loss = result.history['loss']\n val_acc = result.history['val_accuracy']\n val_loss = result.history['val_loss']\n plt.figure(figsize=(15, 5))\n plt.subplot(121)\n plt.plot(range(1,epochs), acc[1:], label='Train_acc')\n plt.plot(range(1,epochs), val_acc[1:], label='Test_acc')\n plt.title('Accuracy over ' + str(epochs) + ' Epochs', size=15)\n plt.legend()\n plt.grid(True)\n plt.subplot(122)\n plt.plot(range(1,epochs), loss[1:], label='Train_loss')\n plt.plot(range(1,epochs), val_loss[1:], label='Test_loss')\n plt.title('Loss over ' + str(epochs) + ' Epochs', size=15)\n plt.legend()\n plt.grid(True)\n plt.show()\n\nmodel.summary()\n# The best accuracy\nmax(result.history['accuracy'])\n\nplot_accs(result, 3)\nplot_model(model, to_file='new_classifier.png', show_shapes=True, show_layer_names=True)",
"Model: \"sequential_14\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n rescaling_16 (Rescaling) (None, 250, 250, 3) 0 \n \n conv2d_65 (Conv2D) (None, 250, 250, 32) 896 \n \n max_pooling2d_59 (MaxPoolin (None, 125, 125, 32) 0 \n g2D) \n \n conv2d_66 (Conv2D) (None, 125, 125, 64) 18496 \n \n max_pooling2d_60 (MaxPoolin (None, 62, 62, 64) 0 \n g2D) \n \n conv2d_67 (Conv2D) (None, 60, 60, 32) 18464 \n \n batch_normalization_10 (Bat (None, 60, 60, 32) 128 \n chNormalization) \n \n dropout_32 (Dropout) (None, 60, 60, 32) 0 \n \n conv2d_68 (Conv2D) (None, 60, 60, 128) 102528 \n \n max_pooling2d_61 (MaxPoolin (None, 30, 30, 128) 0 \n g2D) \n \n dropout_33 (Dropout) (None, 30, 30, 128) 0 \n \n conv2d_69 (Conv2D) (None, 30, 30, 256) 295168 \n \n max_pooling2d_62 (MaxPoolin (None, 15, 15, 256) 0 \n g2D) \n \n dropout_34 (Dropout) (None, 15, 15, 256) 0 \n \n conv2d_70 (Conv2D) (None, 15, 15, 256) 1638656 \n \n max_pooling2d_63 (MaxPoolin (None, 7, 7, 256) 0 \n g2D) \n \n flatten_14 (Flatten) (None, 12544) 0 \n \n dense_38 (Dense) (None, 64) 802880 \n \n dense_39 (Dense) (None, 32) 2080 \n \n dense_40 (Dense) (None, 4) 132 \n \n=================================================================\nTotal params: 2,879,428\nTrainable params: 2,879,364\nNon-trainable params: 64\n_________________________________________________________________\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecb556e622908f7d3856836d6c261a97f3a836c1 | 963,216 | ipynb | Jupyter Notebook | notebooks/ptCLM_multi.ipynb | wwieder/CTSM_py | 228f118df7fe369cd237bce398b5aacea21bc4d2 | [
"Apache-2.0"
] | 2 | 2020-01-31T22:01:41.000Z | 2021-04-13T18:23:13.000Z | notebooks/ptCLM_multi.ipynb | wwieder/CTSM_py | 228f118df7fe369cd237bce398b5aacea21bc4d2 | [
"Apache-2.0"
] | null | null | null | notebooks/ptCLM_multi.ipynb | wwieder/CTSM_py | 228f118df7fe369cd237bce398b5aacea21bc4d2 | [
"Apache-2.0"
] | 1 | 2020-12-22T08:47:09.000Z | 2020-12-22T08:47:09.000Z | 233.337209 | 171,816 | 0.835054 | [
[
[
"## ptCLM_multi\n#### Plots simulated soil temperature and water state for multiple sites\n- Will Wieder\n- Created Oct 2020",
"_____no_output_____"
]
],
[
[
"import xarray as xr\nimport cf_units as cf\nimport numpy as np\nimport pandas as pd\nfrom ctsm_py import utils\nfrom scipy import signal,stats\n\n# some resources for plotting\nimport matplotlib.pyplot as plt\nimport matplotlib.colors as colors\nimport matplotlib.lines as mlines\nimport matplotlib.dates as mdates\n\n# supress Runtime warnings that let you know when code isn't too efficiently\nimport warnings\nwarnings.simplefilter(\"ignore\", category=RuntimeWarning)\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Point to files\n* All sims use newPHS parameterization and low SLA (0.01 m2/gC)\n* FF and DM also have sandier sols and higher leaf CN (32 vs 24).",
"_____no_output_____"
]
],
[
[
"# Niwot LTER simulations\nyears = range(2015,2019)\nnmon = 12\nrollHour = -12 \n\nlongSite = ['fell_field','dry_meadow','moist_meadow','snowbed']\nveg = ['ff','dm','mm','sb']\nsite = [veg[v]+'_newPHS_lowSLA' for v in range(len(veg)) ]\nsite[0] = site[0]+'_SAND_cn32'\nsite[1] = site[1]+'_SAND_cn32'\nprint(site)\ncase = ['clm50bgc_NWT_'+site[v] for v in range(len(veg)) ]\n\nOBSdir = '/glade/p/cgd/tss/people/wwieder/inputdata/single_point/datmdata_NWT_Tvan/'\nOBSdir = [OBSdir +longSite[v]+'/' for v in range(len(veg)) ]\nOUTdir = OBSdir\n\n# Points to simulation files\nCLMdir = ['/glade/scratch/wwieder/archive/'+case[v]+'/lnd/hist/' \n for v in range(len(veg)) ]\nCLMfile = [[]] *len(veg)\nCLMmon = [[]] *len(veg)\nfor v in range(len(veg)):\n # each h1 file\n CLMfile[v] = [CLMdir[v] + case[v] +'.clm2.h1.'+str(years[i]) +'-01-01-00000.nc' \n for i in range(len(years)) ] \n # single month of data for soil C pools \n CLMmon[v] = [CLMdir[v] + case[v] +'.clm2.h0.'+str(years[0])+'-01.nc'] \n\nCLMfile[0][0]",
"['ff_newPHS_lowSLA_SAND_cn32', 'dm_newPHS_lowSLA_SAND_cn32', 'mm_newPHS_lowSLA', 'sb_newPHS_lowSLA']\n"
]
],
[
[
"### Read in dataset \n- combine along new dimension `vegdim`\n- get rid of extra `lndgrid` dimension\n- remove CLM time shift",
"_____no_output_____"
]
],
[
[
"dsCLM = [xr.open_mfdataset(CLMfile[v], decode_times=True, combine='by_coords') for v in range(len(veg)) ]\nvegdim = xr.DataArray(longSite, dims='veg', name='veg') \ndsCLM = xr.concat(dsCLM, dim=vegdim)\ndsCLM = dsCLM.isel(lndgrid=0)\ndsCLM = dsCLM.shift(time=-1)\nprint('---- read in data ----')\ndsCLM",
"---- read in data ----\n"
],
[
"#dsCLM.levgrnd",
"_____no_output_____"
],
[
"# create new variables to subset data\ndsCLM['year'] = dsCLM['time.year']\ndsCLM['month'] = dsCLM['time.month']\ndsCLM['season'] = dsCLM['time.season']\n\n#Can't groupby hour & minutes, so combine them here\ndsCLM['HourMin'] = np.round(dsCLM['time.hour'] + dsCLM['time.minute']/60,1)\ndsCLM['MonDay'] = np.round(dsCLM['time.month'] + dsCLM['time.day']/100,2)\n\n# total preciptiation\ndsCLM['ppt'] = dsCLM.RAIN + dsCLM.SNOW\ndsCLM['ppt'].attrs['units'] = dsCLM.RAIN.attrs['units']\ndsCLM['ppt'].attrs['long_name'] = 'RAIN + SNOW'",
"_____no_output_____"
]
],
[
[
"## Read in observatons ",
"_____no_output_____"
]
],
[
[
"nwtOBS = '/glade/p/cgd/tss/people/wwieder/inputdata/single_point/datmdata_NWT_Tvan/NWT_lter_obs_downloads/'\ndsNET = pd.read_table(nwtOBS+'sensor_network_soil_data_30_min.txt')#.to_xarray() # Saddle sensors\ndsTVan = pd.read_table(nwtOBS+'tvan_soil_data_30_min.txt') # Tvan soil sensors\ndsSNO = nwtOBS+'saddle_grid_snow_depth_data_biweekly.txt' # Saddle snow \ndsNPP = nwtOBS+'saddle_grid_productivity_data.txt' # Saddle productivity\ndsNET.date = pd.to_datetime(dsNET.date)\ndsTVan.date = pd.to_datetime(dsTVan.date)\n#dsTVan.insert(0, 'veg_com', 'ff')\ndsSOI = pd.concat([dsNET,dsTVan]) ",
"_____no_output_____"
],
[
"dsSOIgroup = dsSOI.groupby(['date','veg_com'])\ndsSOIgroup",
"_____no_output_____"
],
[
"dsSOIdaily = dsSOIgroup.aggregate(np.mean)\ndsSOIdailySTD = dsSOIgroup.aggregate(np.std)\n#dsSOIdaily\n#dsSOIdaily.groupby(['veg_com'])['soiltemp_upper_avg'].plot(x='date',by='veg_com',legend=True);\n#dsTVandaily['soiltemp_upper_avg'].plot();\n#dsNETdaily['soiltemp_upper_avg']['mean']",
"_____no_output_____"
]
],
[
[
"### Convert Saddle network data to xarray",
"_____no_output_____"
]
],
[
[
"dsSOI = dsSOIdaily.to_xarray()\ndsSOIstd = dsSOIdailySTD.to_xarray()\nVeg = [\"FF\", \"DM\", \"MM\",'WM','SB']\nfullVeg = [\"fell_field\", \"dry_meadow\", \"moist_meadow\",'wet_meadow','snowbed']\ndsSOI = dsSOI.reindex({'veg_com': Veg})\ndsSOIstd = dsSOIstd.reindex({'veg_com': Veg})\ndsCLM = dsCLM.reindex({'veg': fullVeg})\n# Quick look at data\ndsSOI.where(dsSOI['date.year']==2018).soilmoisture_lower_avg.plot(hue='veg_com');\nplt.title('Observed soil moisture', loc='left', fontsize='large', fontweight='bold');",
"_____no_output_____"
],
[
"dsCLM.where(dsCLM['year']>2017).H2OSOI.isel(levsoi=2).plot.line(hue='veg');\nplt.title('Simulated soil moisture', loc='left', fontsize='large', fontweight='bold');",
"/glade/u/home/wwieder/miniconda3/envs/python-tutorial/lib/python3.7/site-packages/IPython/core/pylabtools.py:132: UserWarning: Creating legend with loc=\"best\" can be slow with large amounts of data.\n fig.canvas.print_figure(bytes_io, **kw)\n"
]
],
[
[
"--------------------------\n## comparison plots\n--------------------------",
"_____no_output_____"
]
],
[
[
"annPPT = dsCLM['ppt'].groupby(dsCLM['year']).mean()\nannPPT = annPPT * 3600 * 24 * 365 / 10 #convert mm/s to cm/y\n#print(annPPT)\n\nannGPP = dsCLM['GPP'].groupby(dsCLM['year']).mean()\nannGPP = annGPP * 3600 * 24 * 365 #convert gC/m2/s to annual\nprint(annGPP)\nannGPP.isel(veg=range(len(veg))).plot.line(x='year');\nplt.title('Simulated GPP', loc='left', fontsize='large', fontweight='bold');",
"<xarray.DataArray 'GPP' (veg: 5, year: 4)>\ndask.array<mul, shape=(5, 4), dtype=float32, chunksize=(2, 1)>\nCoordinates:\n * veg (veg) object 'fell_field' 'dry_meadow' ... 'wet_meadow' 'snowbed'\n * year (year) int64 2015 2016 2017 2018\n"
],
[
"plotVars = ['ppt','SNOW_DEPTH','ELAI','TSOI','H2OSOI','BTRAN2']\nplt.figure(figsize=[18,8])\n\nplt.subplot(231)\nannPPT.plot.line(x='year', hue=\"veg\",add_legend=True)\nplt.subplot(232)\ndsCLM[plotVars[1]].plot.line(x='time', hue=\"veg\",add_legend=False)\nplt.subplot(233)\ndsCLM[plotVars[2]].plot.line(x='time', hue=\"veg\",add_legend=False);\nplt.subplot(234)\ndsCLM[plotVars[3]].isel(levgrnd=2).plot.line(x='time', hue=\"veg\",add_legend=False);\nplt.subplot(235)\ndsCLM[plotVars[4]].isel(levsoi=2).plot.line(x='time', hue=\"veg\",add_legend=False);\nplt.subplot(236)\ndsCLM[plotVars[5]].plot.line(x='time', hue=\"veg\",add_legend=False);\n",
"_____no_output_____"
],
[
"dsCLM[\"levgrnd\"].isel(levgrnd=slice(0,20)).values",
"_____no_output_____"
]
],
[
[
"### Plots of depth resolved temperature and moisture",
"_____no_output_____"
]
],
[
[
"simple = dsCLM.TSOI.isel(levgrnd=(slice(0,6))).plot(x=\"time\",yincrease=False, robust=True, col='veg', col_wrap=5,cmap='bwr'); ",
"_____no_output_____"
],
[
"################### Function to truncate color map ###################\ndef truncate_colormap(cmapIn='jet', minval=0.0, maxval=1.0, n=100):\n '''truncate_colormap(cmapIn='jet', minval=0.0, maxval=1.0, n=100)''' \n cmapIn = plt.get_cmap(cmapIn)\n\n new_cmap = colors.LinearSegmentedColormap.from_list(\n 'trunc({n},{a:.2f},{b:.2f})'.format(n=cmapIn.name, a=minval, b=maxval),\n cmapIn(np.linspace(minval, maxval, n)))\n\n arr = np.linspace(0, 50, 100).reshape((10, 10))\n #fig, ax = plt.subplots(ncols=2)\n #ax[0].imshow(arr, interpolation='nearest', cmap=cmapIn)\n #ax[1].imshow(arr, interpolation='nearest', cmap=new_cmap)\n #plt.show()\n\n return new_cmap\n\ncmap_mod = truncate_colormap(cmapIn='gist_earth_r',minval=.2, maxval=0.95) # calls function to truncate colormap\n\ntemp = dsCLM['H2OSOI'].copy(deep=True)\ntemp = temp.assign_coords({\"levsoi\": dsCLM[\"levgrnd\"].isel(levgrnd=slice(0,20)).values})\nsimple = temp.isel(levsoi=(slice(0,6))).plot(\n x=\"time\",yincrease=False, robust=True, col='veg', col_wrap=5,cmap=cmap_mod);",
"_____no_output_____"
],
[
"nyear = len(years) \nnlev = 1\nnveg = 1\nfor i in range(nyear):\n x = dsCLM['time.dayofyear'].where(dsCLM['year']==years[i])\n y = dsCLM.H2OSOI.isel(levsoi=nlev,veg=nveg).where(dsCLM['year']==years[i])\n plt.plot(x, y, '-')\ndepth = str(np.round(dsCLM[\"levgrnd\"].isel(levgrnd=nlev).values * 100,0)) \nplt.legend(years, frameon=False, loc='upper right');\nplt.title(longSite[nveg]+ \" \" +depth + \" cm\", loc='left', fontsize='x-large', fontweight='bold')\nplt.ylabel('volumetric soil water (mm3/mm3)');",
"_____no_output_____"
],
[
"nyear = len(years) \nnveg =2\nfor i in range(nyear):\n x = dsCLM['time.dayofyear'].where(dsCLM['year']==years[i])\n y = dsCLM.QRUNOFF.isel(veg=nveg).where(dsCLM['year']==years[i])\n y = y.cumsum(dim='time')*30*60 #convert mm/s flux to mm water\n plt.plot(x, y, '-')\nplt.legend(years, frameon=False, loc='lower right');\nplt.title(longSite[nveg]+ \" runnoff\", loc='left', fontsize='x-large', fontweight='bold')\nplt.ylabel('Cumulative sum runoff (mm)');",
"_____no_output_____"
],
[
"x = dsCLM.SNOW_DEPTH.groupby(dsCLM['year']).max()\ny = dsCLM.QRUNOFF.groupby(dsCLM['year']).sum() * 24*3600\nplt.plot(x, y, 'o', color='b');\nplt.xlabel('Snow Depth, annual max (m)');\nplt.ylabel('RUNOFF (mm/y)');\ndsCLM.SNOW_DEPTH.attrs",
"_____no_output_____"
]
],
[
[
"### plot soil moisture and temperature for single year",
"_____no_output_____"
]
],
[
[
"nyear = len(years) \nclmVARS = ['H2OSOI','TSOI']\nobsVARSu = ['soilmoisture_upper_avg','soiltemp_upper_avg']\nobsVARSl = ['soilmoisture_lower_avg','soiltemp_lower_avg']\n\nfor v in range(len(clmVARS)):\n fig, axs = plt.subplots(2, len(Veg), \n sharey='row', sharex=True,\n gridspec_kw={'wspace': 0, 'hspace': 0.12},\n figsize=(15,6)) \n for i in range(len(Veg)):\n for j in range(2):\n if i == 0: \n nlev = 2 #10 cm for fell field\n plotYear = 2018\n else: \n nlev = 1 # 5 cm for sensor network\n plotYear = 2018\n if j == 1: nlev = 4\n\n # Srelect data to plot \n x = dsCLM['time.dayofyear'].where(dsCLM['year']==plotYear)\n x2 = dsSOI['date.dayofyear'].where(dsSOI['date.year']==plotYear)\n\n if v == 0:\n y = dsCLM[clmVARS[v]].isel(levsoi=nlev,veg=i).where(dsCLM['year']==plotYear).groupby(x).mean() * 100\n else:\n y = dsCLM[clmVARS[v]].isel(levgrnd=nlev,veg=i).where(dsCLM['year']==plotYear).groupby(x).mean() -273.15\n \n if j == 0: \n y2 = dsSOI[obsVARSu[v]].isel(veg_com=i).where(dsSOI['date.year']==plotYear)\n ySTD = dsSOIstd[obsVARSu[v]].isel(veg_com=i).where(dsSOI['date.year']==plotYear)\n if j == 1: \n y2 = dsSOI[obsVARSl[v]].isel(veg_com=i).where(dsSOI['date.year']==plotYear)\n ySTD = dsSOIstd[obsVARSl[v]].isel(veg_com=i).where(dsSOI['date.year']==plotYear)\n x = x.groupby(x).mean()\n \n axs[j,i].plot(x2, y2, '-', color='k')\n axs[j,i].plot(x, y, '-',color='tab:red')\n axs[j,i].fill_between(x2, y2-ySTD, y2+ySTD, alpha=0.2,color='k')\n axs[j,i].yaxis.set_ticks_position('both')# Ticks on all 4 sides \n axs[j,i].xaxis.set_ticks_position('both') \n\n\n # control titles & axis labels\n # label colums of data with variables name\n depth = str(np.round(dsCLM[\"levgrnd\"].isel(levgrnd=nlev).values * 100,0))\n if j == 0:\n axs[j,i].set_title(depth + \" cm \"+fullVeg[i], \n loc='left', fontsize='large', fontweight='bold')\n if i == 0 :\n axs[j,i].legend(('Obs','CLM5'), frameon=False,fontsize='large')\n if j == 1:\n axs[j,i].set_title(depth + \" cm \", loc='left', fontsize='large', fontweight='bold')\n if i == 0:\n if v == 0:\n axs[j,i].set_ylabel('Vol. Soil Water (mm3/mm3)');\n else:\n axs[j,i].set_ylabel('Soil Temperature (C)');",
"_____no_output_____"
],
[
"#dsSOI.soiltemp_lower_avg.isel(veg_com=0).plot()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecb560911678d0939fad9b06e47ec9f79c4835e4 | 151,566 | ipynb | Jupyter Notebook | datasets/cil-gdpcir/indicators.ipynb | RadiantMLHub/PlanetaryComputerExamples | cd7f7f2f19a369d51f8fe991cb7103e560c74e22 | [
"MIT"
] | null | null | null | datasets/cil-gdpcir/indicators.ipynb | RadiantMLHub/PlanetaryComputerExamples | cd7f7f2f19a369d51f8fe991cb7103e560c74e22 | [
"MIT"
] | 1 | 2022-03-29T21:08:22.000Z | 2022-03-30T22:24:44.000Z | datasets/cil-gdpcir/indicators.ipynb | RadiantMLHub/PlanetaryComputerExamples | cd7f7f2f19a369d51f8fe991cb7103e560c74e22 | [
"MIT"
] | 1 | 2022-03-24T17:23:17.000Z | 2022-03-24T17:23:17.000Z | 62.37284 | 24,424 | 0.58556 | [
[
[
"## Computing climate indicators with xclim\n\nThe Climate Impact Lab Downscaled Projections for Climate Impacts Research (CIL-GDPCR) collections contain bias corrected and downscaled 1/4° CMIP6 projections for temperature and precipitation.\n\nSee the project homepage for more information: [github.com/ClimateImpactLab/downscaleCMIP6](https://github.com/ClimateImpactLab/downscaleCMIP6).\n\nThis tutorial covers constructing a time series across the CMIP:historical and ScenarioMIP:ssp126 experiments, and computing transformations using the [xclim](https://xclim.readthedocs.io/) package. Additional tutorials are available at [github.com/microsoft/PlanetaryComputerExamples](https://github.com/microsoft/PlanetaryComputerExamples/blob/main/datasets/cil-gdpcir).",
"_____no_output_____"
]
],
[
[
"# required to locate and authenticate with the stac collection\nimport planetary_computer\nimport pystac_client\nimport pystac\n\n# required to load a zarr array using xarray\nimport xarray as xr\n\n# climate indicators with xclim\nimport xclim.indicators\n\n# optional imports used in this notebook\nfrom dask.diagnostics import ProgressBar",
"_____no_output_____"
]
],
[
[
"### Building a joint historical and projection time series\n\nLet's work with the FGOALS-g3 historical and ssp1-2.6 simulations. We'll use the Planetary Computer's STAC API to search for the items we want, which contain all the information necessary to load the data with xarray.\n\nThe FGOALS-g3 data are available under the `cil-gdpcir-cc0` collection (which you can check in the `cmip6:institution_id` summary of the collection).",
"_____no_output_____"
]
],
[
[
"catalog = pystac_client.Client.open(\n \"https://planetarycomputer-staging.microsoft.com/api/stac/v1\"\n)\n\ncollection_cc0 = pystac.read_file(\n \"https://planetarycomputer-staging.microsoft.com/api/stac/v1/collections/cil-gdpcir-cc0\" # noqa\n)\n\nitems = catalog.search(\n collections=[\"cil-gdpcir-cc0\"],\n query={\n \"cmip6:source_id\": {\"eq\": \"FGOALS-g3\"},\n \"cmip6:experiment_id\": {\"in\": [\"historical\", \"ssp126\"]},\n },\n).get_all_items()",
"_____no_output_____"
],
[
"[item.id for item in items]",
"_____no_output_____"
]
],
[
[
"Retrieve object URLs by authenticating with Planetary Computer",
"_____no_output_____"
]
],
[
[
"# use the planetary computer API to sign the asset\nsigned_items = planetary_computer.sign(items)\n\n# select this variable ID for all models in the collection\nvariable_id = \"tasmin\"\n\n# get the API key and other important keyword arguments\nopen_kwargs = signed_items[0].assets[variable_id].extra_fields[\"xarray:open_kwargs\"]",
"_____no_output_____"
]
],
[
[
"### Reading a single variable",
"_____no_output_____"
]
],
[
[
"ds = xr.open_mfdataset(\n [item.assets[variable_id].href for item in signed_items],\n combine=\"by_coords\",\n combine_attrs=\"drop_conflicts\",\n parallel=True,\n **open_kwargs,\n)\n\nds",
"_____no_output_____"
]
],
[
[
"Let's take a look at the variable `tasmin`. Note the summary provided by the dask preview. This array is 213 GB in total, in 180 MB chunks. The data is chunked such that each year and 90 degrees of latitude and longitude form a chunk.\n\nTo read in the full time series for a single point, you'd need to work through 180.45 MB/chunk * 151 annual chunks = 27 GB of data. This doesn't all need to be held in memory, but it gives a sense of what the operation might look like in terms of download & compute time.",
"_____no_output_____"
]
],
[
[
"ds.tasmin",
"_____no_output_____"
]
],
[
[
"### Applying a climate indicator from xclim",
"_____no_output_____"
],
[
"The [`xclim`](https://xclim.readthedocs.io) package provides a large number of useful [indicators](https://xclim.readthedocs.io/en/stable/indicators.html) for analyzing climate data. Here, we'll use the Atmospheric Indicator: [Frost Days (`xclim.indicators.atmos.frost_days`)](https://xclim.readthedocs.io/en/stable/indicators_api.html#xclim.indicators.atmos.frost_days):",
"_____no_output_____"
]
],
[
[
"frost_days = xclim.indicators.atmos.frost_days(ds=ds)\nfrost_days",
"_____no_output_____"
]
],
[
[
"Here, the state data requirement has been reduced significantly - but careful - this is the size required by the final product *once computed*. But this is a scheduled [dask](https://docs.xarray.dev/en/latest/user-guide/dask.html) operation, and because of dask's [Lazy Evaluation](https://tutorial.dask.org/01x_lazy.html), we haven't done any work yet. Dask is waiting for us to require operations, e.g. by calling `.compute()`, `.persist()`, or because of blocking opreations like writing to disk or plotting. Until we do one of those, we haven't actually read any data yet!\n\n### Loading a subset of the data\n\nLet's subset the data and call `.compute()` so we can work with it in locally (in the notebook).\n\nI'll pick Oslo, Norway, as our oft-frosty location to inspect, and extract one year a decade to plot as a time series. Ideally, we'd look at all of the years and compute a statistic based on a moving multi-decadal window, but this is just an example ;) See [Scale with Dask](https://planetarycomputer.microsoft.com/docs/quickstarts/scale-with-dask/) if you'd like to run this example on a larger amount of data.\n\nThanks to [Wikipedia](https://en.wikipedia.org/wiki/Oslo) for the geographic info!",
"_____no_output_____"
]
],
[
[
"with ProgressBar():\n oslo_frost_days_summary = (\n frost_days.sel(lat=59.913889, lon=10.752222, method=\"nearest\").sel(\n time=frost_days.time.dt.year.isin(range(1950, 2101, 10))\n )\n ).compute()",
"[########################################] | 100% Completed | 13.7s\n"
],
[
"oslo_frost_days_summary",
"_____no_output_____"
],
[
"oslo_frost_days_summary.plot();",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecb563a01a7d8e3d08f1db187187a5cbe3821370 | 114,912 | ipynb | Jupyter Notebook | project1/task1_5_proj1.ipynb | jutanke/bit_projects | 0aeff78d2007b1b8b908dc5128d93b36aaccc0ff | [
"MIT"
] | null | null | null | project1/task1_5_proj1.ipynb | jutanke/bit_projects | 0aeff78d2007b1b8b908dc5128d93b36aaccc0ff | [
"MIT"
] | null | null | null | project1/task1_5_proj1.ipynb | jutanke/bit_projects | 0aeff78d2007b1b8b908dc5128d93b36aaccc0ff | [
"MIT"
] | 1 | 2018-12-05T13:01:18.000Z | 2018-12-05T13:01:18.000Z | 700.682927 | 101,032 | 0.941024 | [
[
[
"#!/usr/bin/env python 3\n#task 1.5: plotting the data without the outliers\n__author__ = \"Akhilesh Vyas\"\n__email__ = \"[email protected]\"\n\nimport numpy as np\nimport scipy.misc as msc\nimport scipy.ndimage as img\nimport matplotlib.pyplot as plt\nimport math",
"_____no_output_____"
],
[
"def foreground2BinImg(f):\n d = img.filters.gaussian_filter(f, sigma=0.50, mode='reflect') - img.filters.gaussian_filter(f, sigma=1.00, mode='reflect')\n d = np.abs(d)\n m = d.max()\n d[d< 0.1*m] = 0\n d[d>=0.1*m] = 1\n return img.morphology.binary_closing(d)\n\nimgName = 'lightning-3'\nf = msc.imread(imgName+'.png', flatten=True).astype(np.float)\ng = foreground2BinImg(f)\n\nplt.imshow(f)\nplt.show()\n\nplt.imshow(g)\nplt.show()\n",
"/Users/vyas/Library/Python/3.6/lib/python/site-packages/ipykernel_launcher.py:10: DeprecationWarning: `imread` is deprecated!\n`imread` is deprecated in SciPy 1.0.0, and will be removed in 1.2.0.\nUse ``imageio.imread`` instead.\n # Remove the CWD from sys.path while we load stuff.\n"
],
[
"#scaling Factor\n\n#GetMatrix\ndef getMatrix(i,j,m,n):\n X = g[i:j,m:n]\n return X\n\nH, W = g.shape\n#print (h,w)\n\nL = int(np.log2(W))\nN_Boxes = []\n\nfor l in range(1, L-1):\n n_boxes = 0\n f = int(math.pow(2, l))\n d = int(H/f)\n for s in range(f):\n i = s*d\n j = (s+1)*d\n for s in range(f):\n m = s*d\n n = (s+1)*d\n #print (i, j, m, n)\n X = getMatrix(i, j, m, n)\n if True in X[:,:]:\n n_boxes = n_boxes + 1 \n N_Boxes.append(n_boxes)\n \n \nN_Boxes_Log= np.log2(np.asarray(N_Boxes))\nscale_log = np.asarray([i for i in range(1, L-2)])\nprint (list(zip(scale_log, N_Boxes_Log)))\n ",
"[(1, 2.0), (2, 3.8073549220576042), (3, 5.4918530963296748), (4, 7.1996723448363644), (5, 8.6582114827517955), (6, 10.055282435501189)]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecb56ac9dd98f18c8b818ab3ed590ee8ab6ced16 | 12,618 | ipynb | Jupyter Notebook | talleres_inov_docente/1-05-aprendizaje_supervisado_clasificacion.ipynb | jfcaballero/Tutorial-sobre-scikit-learn-abreviado | 1e2aa1f9132c277162135a5463068801edab8d15 | [
"CC0-1.0"
] | 4 | 2019-02-20T14:36:39.000Z | 2019-02-21T22:55:57.000Z | talleres_inov_docente/1-05-aprendizaje_supervisado_clasificacion.ipynb | jfcaballero/Tutorial-sobre-scikit-learn-abreviado | 1e2aa1f9132c277162135a5463068801edab8d15 | [
"CC0-1.0"
] | null | null | null | talleres_inov_docente/1-05-aprendizaje_supervisado_clasificacion.ipynb | jfcaballero/Tutorial-sobre-scikit-learn-abreviado | 1e2aa1f9132c277162135a5463068801edab8d15 | [
"CC0-1.0"
] | null | null | null | 26.34238 | 372 | 0.577588 | [
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"# Aprendizaje supervisado parte 1 -- Clasificación",
"_____no_output_____"
],
[
"Para visualizar como funcionan los algoritmos de aprendizaje automático, es mejor considerar datos de una o dos dimensiones, esto es datasets con solo una o dos características. Aunque, en la práctica los datasets tienen muchas más características, es difícil representar datos de alta dimensionalidad en pantallas 2D.\n\nVamos a ilustrar ejemplos muy simples antes de comenzar con datasets del mundo real.",
"_____no_output_____"
],
[
"\nPrimero, vamos a inspeccionar un problema de clasificación binaria con dos dimensiones. Utilizaremos los datos sintéticos que nos proporciona la función ``make_blobs``.",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import make_blobs\n\nX, y = make_blobs(centers=2, random_state=0)\n\nprint('X ~ n_samples x n_features:', X.shape)\nprint('y ~ n_samples:', y.shape)\n\nprint('\\n5 primeros ejemplos:\\n', X[:5, :])\nprint('\\n5 primeras etiquetas:', y[:5])",
"_____no_output_____"
]
],
[
[
"Como los datos son bidimensionales, podemos representar cada punto en un sistema de coordenadas (ejes x e y).",
"_____no_output_____"
]
],
[
[
"plt.scatter(X[y == 0, 0], X[y == 0, 1], \n c='blue', s=40, label='0')\nplt.scatter(X[y == 1, 0], X[y == 1, 1], \n c='red', s=40, label='1', marker='s')\n\nplt.xlabel(u'primera característica')\nplt.ylabel(u'segunda característica')\nplt.legend(loc='upper right');",
"_____no_output_____"
]
],
[
[
"La clasificación es una tarea supervisada y, ya que estamos interesados en su rendimiento en datos no utilizados para entrenar, vamos a dividir los datos en dos partes:\n\n1. un conjunto de entrenamiento que el algoritmo de aprendizaje utiliza para ajustar los parámetros del modelo\n2. un conjunto de test para evaluar la capacidad de generalización del modelo\n\nLa función ``train_test_split`` del paquete ``model_selection`` hace justo esto por nosotros - la usaremos para generar una partición con un 75%//25% en entrenamiento y test, respectivamente.\n\n<img src=\"figures/train_test_split_matrix.svg\" width=\"100%\">\n",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(X, y,\n test_size=0.25,\n random_state=1234,\n stratify=y)",
"_____no_output_____"
]
],
[
[
"### El API de un estimador de scikit-learn\n<img src=\"figures/supervised_workflow.svg\" width=\"100%\">\n",
"_____no_output_____"
],
[
"Cualquier algoritmo de scikit-learn se maneja a través de una interfaz denominada ''Estimator'' (una de las ventajas de scikit-learn es que todos los modelos y algoritmos tienen una interfaz consistente). Por ejemplo, importamos la clase correspondiente al algoritmo de regresión logística:",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression",
"_____no_output_____"
]
],
[
[
"Ahora, instanciamos el estimador:",
"_____no_output_____"
]
],
[
[
"classifier = LogisticRegression()",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"y_train.shape",
"_____no_output_____"
]
],
[
[
"Para construir el modelo a partir de nuestros datos, esto es, aprender a clasificar nuevos puntos, llamamos a la función ``fit`` pasándole los datos de entrenamiento, y las etiquetas correspondientes (la salida deseada para los datos de entrenamiento):",
"_____no_output_____"
]
],
[
[
"classifier.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"Algunos métodos de los estimadores se devuelven a sí mismos por defecto. Esto es, después de ejecutar el código anterior, verás los parámetros por defecto de esta instancia particular de `LogisticRegression`. Otra forma de obtener los parámetros de inicialización de un estimador es usar `classifier.get_params()`, que devuelve un diccionario de parámetros.",
"_____no_output_____"
],
[
"Podemos aplicar el modelo a datos no utilizados anteriormente para predecir la respuesta estimada mediante el método ``predict``:",
"_____no_output_____"
]
],
[
[
"prediction = classifier.predict(X_test)",
"_____no_output_____"
]
],
[
[
"Podemos comparar el resultado con las etiquetas reales:",
"_____no_output_____"
]
],
[
[
"print(prediction)\nprint(y_test)",
"_____no_output_____"
]
],
[
[
"Podemos evaluar nuestro modelo cuantitativamente utilizando la proporción de patrones correctos. A esto se le llama **accuracy**:",
"_____no_output_____"
]
],
[
[
"np.mean(prediction == y_test)",
"_____no_output_____"
]
],
[
[
"Existe una función útil, ``score``, que incluyen todos los clasificadores de scikit-learn para obtener la medida de rendimiento a partir de los datos de test:\n ",
"_____no_output_____"
]
],
[
[
"classifier.score(X_test, y_test)",
"_____no_output_____"
]
],
[
[
"A veces es útil comparar el rendimiento en generalización (en el conjunto de test) con el rendimiento en entrenamiento:",
"_____no_output_____"
]
],
[
[
"classifier.score(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"LogisticRegression es un modelo lineal, lo que significa que creará una frontera de decisión que es lineal en el espacio de entrada. En 2D, esto quiere decir que generará una línea recta para separar los puntos azules de los rojos:",
"_____no_output_____"
]
],
[
[
"from figures import plot_2d_separator\n\nplt.scatter(X[y == 0, 0], X[y == 0, 1], \n c='blue', s=40, label='0')\nplt.scatter(X[y == 1, 0], X[y == 1, 1], \n c='red', s=40, label='1', marker='s')\n\nplt.xlabel(u\"primera característica\")\nplt.ylabel(u\"segunda característica\")\nplot_2d_separator(classifier, X)\nplt.legend(loc='upper right');",
"_____no_output_____"
]
],
[
[
"**Parámetros estimados**: todos los parámetros estimados del modelo son atributos del objeto estimador cuyo nombre termina en guión bajo. Para la regresión logística, serían los coeficientes y la coordenada en el origen de la línea:",
"_____no_output_____"
]
],
[
[
"print(classifier.coef_)\nprint(classifier.intercept_)",
"_____no_output_____"
]
],
[
[
"Otro clasificador: K Nearest Neighbors\n------------------------------------------------\nOtro clasificador popular y fácil de entender es el *k Nearest Neighbors (kNN)*. Implementa una de las estrategias más simples de aprendizaje (de hecho, en realidad no aprende): dado un nuevo ejemplo desconocido, buscar en la base de datos de referencia (entrenamiento) aquellos ejemplos que tengan características más parecidas y asignarle la clase predominante.\n\nLa interfaz es exactamente la misma que para ``LogisticRegression``.",
"_____no_output_____"
]
],
[
[
"from sklearn.neighbors import KNeighborsClassifier",
"_____no_output_____"
]
],
[
[
"Ahora vamos a modificar un parámetro de KNeighborsClassifier para que solo se examine el vecino más cercano:",
"_____no_output_____"
]
],
[
[
"knn = KNeighborsClassifier(n_neighbors=20)",
"_____no_output_____"
]
],
[
[
"Ajustamos el modelo con nuestros datos de entrenamiento.",
"_____no_output_____"
]
],
[
[
"knn.fit(X_train, y_train)",
"_____no_output_____"
],
[
"plt.scatter(X[y == 0, 0], X[y == 0, 1], \n c='blue', s=40, label='0')\nplt.scatter(X[y == 1, 0], X[y == 1, 1], \n c='red', s=40, label='1', marker='s')\n\nplt.xlabel(u\"primera característica\")\nplt.ylabel(u\"segunda característica\")\nplot_2d_separator(knn, X)\nplt.legend(loc='upper right');",
"_____no_output_____"
],
[
"knn.score(X_test, y_test)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-success\">\n <b>EJERCICIO</b>:\n <ul>\n <li>\n Aplicar KNeighborsClassifier al dataset ``iris``. Prueba con distintos valores para el parámetro ``n_neighbors`` y observa como cambian las puntuaciones de entrenamiento y test.\n </li>\n </ul>\n</div>",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecb57748c8d2289716a8938a64058993d5fea789 | 233,145 | ipynb | Jupyter Notebook | docs/_static/notebooks/telescopes_tutorial_5.ipynb | bshapiroalbert/PsrSigSim | 74bb40814295fb6ef84aa932a0de2f684162b8c4 | [
"MIT"
] | 1 | 2021-09-06T09:03:38.000Z | 2021-09-06T09:03:38.000Z | docs/_static/notebooks/telescopes_tutorial_5.ipynb | bshapiroalbert/PsrSigSim | 74bb40814295fb6ef84aa932a0de2f684162b8c4 | [
"MIT"
] | 1 | 2020-12-21T18:02:57.000Z | 2020-12-21T22:07:17.000Z | docs/_static/notebooks/telescopes_tutorial_5.ipynb | bshapiroalbert/PsrSigSim | 74bb40814295fb6ef84aa932a0de2f684162b8c4 | [
"MIT"
] | null | null | null | 399.90566 | 71,224 | 0.936181 | [
[
[
"# Telescopes: Tutorial 5\n\nThis notebook will build on the previous tutorials, showing more features of the `PsrSigSim`. Details will be given for new features, while other features have been discussed in the previous tutorial notebook. This notebook shows the details of different telescopes currently included in the `PsrSigSim`, how to call them, and how to define a user `telescope` for a simulated observation.\n\nWe again simulate precision pulsar timing data with high signal-to-noise pulse profiles in order to clearly show the input pulse profile in the final simulated data product. We note that the use of different telescopes will result in different signal strengths, as would be expected. \n\nThis example will follow previous notebook in defining all necessary classes except for `telescope`.",
"_____no_output_____"
]
],
[
[
"# import some useful packages\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n# import the pulsar signal simulator\nimport psrsigsim as pss",
"_____no_output_____"
]
],
[
[
"## The Folded Signal\n\nHere we will use the same `Signal` definitions that have been used in the previous tutorials. We will again simulate a 20-minute-long observation total, with subintegrations of 1 minute. The other simulation parameters will be 64 frequency channels each 12.5 MHz wide (for 800 MHz bandwidth).\n\nWe will simulate a real pulsar, J1713+0747, as we have a premade profile for this pulsar. The period, dm, and other relavent pulsar parameters come from the NANOGrav 11-yr data release. ",
"_____no_output_____"
]
],
[
[
"# Define our signal variables.\nf0 = 1500 # center observing frequecy in MHz\nbw = 800.0 # observation MHz\nNf = 64 # number of frequency channels\n# We define the pulse period early here so we can similarly define the frequency\nperiod = 0.00457 # pulsar period in seconds for J1713+0747\nf_samp = (1.0/period)*2048*10**-6 # sample rate of data in MHz (here 2048 samples across the pulse period)\nsublen = 60.0 # subintegration length in seconds, or rate to dump data at\n# Now we define our signal\nsignal_1713_GBT = pss.signal.FilterBankSignal(fcent = f0, bandwidth = bw, Nsubband=Nf, sample_rate = f_samp,\n sublen = sublen, fold = True) # fold is set to `True`",
"Warning: specified sample rate 0.4481400437636761 MHz < Nyquist frequency 1600.0 MHz\n"
]
],
[
[
"## The Pulsar and Profiles\n\nNow we will load the pulse profile as in Tutorial 3 and initialize a single `Pulsar` object. ",
"_____no_output_____"
]
],
[
[
"# First we load the data array\npath = 'psrsigsim/data/J1713+0747_profile.npy'\nJ1713_dataprof = np.load(path)\n\n# Now we define the data profile\nJ1713_prof = pss.pulsar.DataProfile(J1713_dataprof)",
"_____no_output_____"
],
[
"# Define the values needed for the puslar\nSmean = 0.009 # The mean flux of the pulsar, J1713+0747 at 1400 MHz from the ATNF pulsar catatlog, here 0.009 Jy\npsr_name = \"J1713+0747\" # The name of our simulated pulsar\n\n# Now we define the pulsar with the scaled J1713+0747 profiles\npulsar_J1713 = pss.pulsar.Pulsar(period, Smean, profiles=J1713_prof, name = psr_name)",
"_____no_output_____"
],
[
"# define the observation length\nobslen = 60.0*20 # seconds, 20 minutes in total",
"_____no_output_____"
]
],
[
[
"## The ISM\n\nHere we define the `ISM` class used to disperse the simulated pulses.",
"_____no_output_____"
]
],
[
[
"# Define the dispersion measure\ndm = 15.921200 # pc cm^-3\n# And define the ISM object, note that this class takes no initial arguements\nism_sim = pss.ism.ISM()",
"_____no_output_____"
]
],
[
[
"## Defining Telescopes\n\nHere we will show how to use the two predefined telescopes, Green Bank and Arecibo, and the systems accociated with them. We will also show how to define a `telescope` from scratch, so that any current or future telescopes and systems can be simulated.",
"_____no_output_____"
],
[
"### Predefined Telescopes\n\nWe start off by showing the two predefined telescopes.",
"_____no_output_____"
]
],
[
[
"# Define the Green Bank Telescope\ntscope_GBT = pss.telescope.telescope.GBT()\n\n# Define the Arecibo Telescope\ntscope_AO = pss.telescope.telescope.Arecibo()",
"_____no_output_____"
]
],
[
[
"Each telescope is made up of one or more `systems` consisting of a `Reciever` and a `Backend`. For the predefined telescopes, the systems for the `GBT` are the L-band-GUPPI system or the 800 MHz-GUPPI system. For `Arecibo` these are the 430 MHz-PUPPI system or the L-band-PUPPI system. One can check to see what these systems and their parameters are as we show below.",
"_____no_output_____"
]
],
[
[
"# Information about the GBT systems\nprint(tscope_GBT.systems)\n# We can also find out information about a receiver that has been defined here\nrcvr_LGUP = tscope_GBT.systems['Lband_GUPPI'][0]\nprint(rcvr_LGUP.bandwidth, rcvr_LGUP.fcent, rcvr_LGUP.name)",
"{'820_GUPPI': (Receiver(820), Backend(GUPPI)), 'Lband_GUPPI': (Receiver(Lband), Backend(GUPPI)), '800_GASP': (Receiver(800), Backend(GASP)), 'Lband_GASP': (Receiver(Lband), Backend(GASP))}\n800.0 MHz 1400.0 MHz Lband\n"
]
],
[
[
"### Defining a new system\n\nOne can also add a new system to one of these existing telescopes, similarly to what will be done when define a new telescope from scratch. Here we will add the 350 MHz receiver with the GUPPI backend to the Green Bank Telescope.\n\nFirst we define a new `Receiver` and `Backend` object. The `Receiver` object needs a center frequency of the receiver in MHz, a bandwidth in MHz to be centered on that center frequency, and a name. The `Backend` object needs only a name and a sampling rate in MHz. This sampling rate should be the maximum sampling rate of the backend, as it will allow lower sampling rates, but not higher sampling rates.",
"_____no_output_____"
]
],
[
[
"# First we define a new receiver\nrcvr_350 = pss.telescope.receiver.Receiver(fcent=350, bandwidth=100, name=\"350\")\n# And then we want to use the GUPPI backend\nguppi = pss.telescope.backend.Backend(samprate=3.125, name=\"GUPPI\")",
"_____no_output_____"
],
[
"# Now we add the new system. This needs just the receiver, backend, and a name\ntscope_GBT.add_system(name=\"350_GUPPI\", receiver=rcvr_350, backend=guppi)\n# And now we check that it has been added\nprint(tscope_GBT.systems[\"350_GUPPI\"])",
"(Receiver(350), Backend(GUPPI))\n"
]
],
[
[
"### Defining a new telescope\n\nWe can also define a new telescope from scratch. In addition to needing the `Receiver` and `Backend` objects to define at least one system, the `telescope` also needs the aperture size in meters, the total area in meters^2, the system temperature in kelvin, and a name. Here we will define a small 3-meter aperture circular radio telescope that you might find at a University or somebody's backyard.",
"_____no_output_____"
]
],
[
[
"# We first need to define the telescope parameters\naperture = 3.0 # meters\narea = (0.5*aperture)**2*np.pi # meters^2\nTsys = 250.0 # kelvin, note this is not a realistic system temperature for a backyard telescope\nname = \"Backyard_Telescope\"",
"_____no_output_____"
],
[
"# Now we can define the telescope\ntscope_bkyd = pss.telescope.Telescope(aperture, area=area, Tsys=Tsys, name=name)",
"_____no_output_____"
]
],
[
[
"Now similarly to defining a new system before, we must add a system to our new telescope by defining a receiver and a backend. Since this just represents a little telescope, the system won't be comparable to the previously defined telescope.",
"_____no_output_____"
]
],
[
[
"rcvr_bkyd = pss.telescope.receiver.Receiver(fcent=1400, bandwidth=20, name=\"Lband\")\n\nbackend_bkyd = pss.telescope.backend.Backend(samprate=0.25, name=\"Laptop\") # Note this is not a realistic sampling rate",
"_____no_output_____"
],
[
"# Add the system to our telecope\ntscope_bkyd.add_system(name=\"bkyd\", receiver=rcvr_bkyd, backend=backend_bkyd)\n# And now we check that it has been added\nprint(tscope_bkyd.systems)",
"{'bkyd': (Receiver(Lband), Backend(Laptop))}\n"
]
],
[
[
"## Observing with different telescopes\n\nNow that we have three different telescopes, we can observe our simulated pulsar with all three and compare the sensitivity of each telescope for the same initial `Signal` and `Pulsar`. Since the radiometer noise from the telescope is added directly to the signal though, we will need to define two additional `Signals` and create pulses for them before we can observe them with different telescopes.",
"_____no_output_____"
]
],
[
[
"# We define three new, similar, signals, one for each telescope\nsignal_1713_AO = pss.signal.FilterBankSignal(fcent = f0, bandwidth = bw, Nsubband=Nf, sample_rate = f_samp,\n sublen = sublen, fold = True)\n# Our backyard telescope will need slightly different parameters to be comparable to the other signals\nf0_bkyd = 1400.0 # center frequency of our backyard telescope\nbw_bkyd = 20.0 # Bandwidth of our backyard telescope\nNf_bkyd = 1 # only process one frequency channel 20 MHz wide for our backyard telescope\nsignal_1713_bkyd = pss.signal.FilterBankSignal(fcent = f0_bkyd, bandwidth = bw_bkyd, Nsubband=Nf_bkyd, \\\n sample_rate = f_samp, sublen = sublen, fold = True)",
"Warning: specified sample rate 0.4481400437636761 MHz < Nyquist frequency 1600.0 MHz\nWarning: specified sample rate 0.4481400437636761 MHz < Nyquist frequency 40.0 MHz\n"
],
[
"# Now we make pulses for all three signals\npulsar_J1713.make_pulses(signal_1713_GBT, tobs = obslen)\npulsar_J1713.make_pulses(signal_1713_AO, tobs = obslen)\npulsar_J1713.make_pulses(signal_1713_bkyd, tobs = obslen)\n# And disperse them\nism_sim.disperse(signal_1713_GBT, dm)\nism_sim.disperse(signal_1713_AO, dm)\nism_sim.disperse(signal_1713_bkyd, dm)",
"100% dispersed in 0.001 seconds."
],
[
"# And now we observe with each telescope, note the only change is the system name. First the GBT\ntscope_GBT.observe(signal_1713_GBT, pulsar_J1713, system=\"Lband_GUPPI\", noise=True)\n# Then Arecibo\ntscope_AO.observe(signal_1713_AO, pulsar_J1713, system=\"Lband_PUPPI\", noise=True)\n# And finally our little backyard telescope\ntscope_bkyd.observe(signal_1713_bkyd, pulsar_J1713, system=\"bkyd\", noise=True)",
"WARNING: AstropyDeprecationWarning: The truth value of a Quantity is ambiguous. In the future this will raise a ValueError. [astropy.units.quantity]\n"
]
],
[
[
"Now we can look at the simulated data and compare the sensitivity of the different telescopes. We first plot the observation from the GBT, then Arecibo, and then our newly defined backyard telescope.",
"_____no_output_____"
]
],
[
[
"# We first plot the first two pulses in frequency-time space to show the undispersed pulses\ntime = np.linspace(0, obslen, len(signal_1713_GBT.data[0,:]))\n\n# Since we know there are 2048 bins per pulse period, we can index the appropriate amount\nplt.plot(time[:4096], signal_1713_GBT.data[0,:4096], label = signal_1713_GBT.dat_freq[0])\nplt.plot(time[:4096], signal_1713_GBT.data[-1,:4096], label = signal_1713_GBT.dat_freq[-1])\nplt.ylabel(\"Intensity\")\nplt.xlabel(\"Time [s]\")\nplt.legend(loc = 'best')\nplt.title(\"L-band GBT Simulation\")\nplt.show()\nplt.close()\n\n# And the 2-D plot\nplt.imshow(signal_1713_GBT.data[:,:4096], aspect = 'auto', interpolation='nearest', origin = 'lower', \\\n extent = [min(time[:4096]), max(time[:4096]), signal_1713_GBT.dat_freq[0].value, signal_1713_GBT.dat_freq[-1].value])\nplt.ylabel(\"Frequency [MHz]\")\nplt.xlabel(\"Time [s]\")\nplt.colorbar(label = \"Intensity\")\nplt.show()\nplt.close()",
"_____no_output_____"
],
[
"# Since we know there are 2048 bins per pulse period, we can index the appropriate amount\nplt.plot(time[:4096], signal_1713_AO.data[0,:4096], label = signal_1713_AO.dat_freq[0])\nplt.plot(time[:4096], signal_1713_AO.data[-1,:4096], label = signal_1713_AO.dat_freq[-1])\nplt.ylabel(\"Intensity\")\nplt.xlabel(\"Time [s]\")\nplt.legend(loc = 'best')\nplt.title(\"L-band AO Simulation\")\nplt.show()\nplt.close()\n\n# And the 2-D plot\nplt.imshow(signal_1713_AO.data[:,:4096], aspect = 'auto', interpolation='nearest', origin = 'lower', \\\n extent = [min(time[:4096]), max(time[:4096]), signal_1713_AO.dat_freq[0].value, signal_1713_AO.dat_freq[-1].value])\nplt.ylabel(\"Frequency [MHz]\")\nplt.xlabel(\"Time [s]\")\nplt.colorbar(label = \"Intensity\")\nplt.show()\nplt.close()",
"_____no_output_____"
],
[
"# Since we know there are 2048 bins per pulse period, we can index the appropriate amount\nplt.plot(time[:4096], signal_1713_bkyd.data[0,:4096], label = \"1400.0 MHz\")\nplt.ylabel(\"Intensity\")\nplt.xlabel(\"Time [s]\")\nplt.legend(loc = 'best')\nplt.title(\"L-band Backyard Telescope Simulation\")\nplt.show()\nplt.close()",
"_____no_output_____"
]
],
[
[
"We can see that, as expected, the Arecibo telescope is more sensitive than the GBT when observing over the same timescale. We can also see that even though the simulated pulsar here is easily visible with these large telescopes, our backyard telescope is not able to see the pulsar over the same amount of time, since the output is pure noise. The `PsrSigSim` can be used to determine the approximate sensitivity of an observation of a simulated pulsar with any given telescope that can be defined.",
"_____no_output_____"
],
[
"### Note about randomly generated pulses and noise\n\n`PsrSigSim` uses `numpy.random` under the hood in order to generate the radio pulses and various types of noise. If a user desires or requires that this randomly generated data is reproducible we recommend using a call the seed generator native to `Numpy` before calling the function that produces the random noise/pulses. Newer versions of `Numpy` are moving toward slightly different [functionality/syntax](https://numpy.org/doc/stable/reference/random/index.html), but is essentially used in the same way. \n```\nnumpy.random.seed(1776)\npulsar_1.make_pulses(signal_1, tobs=obslen)\n\n```",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ecb579c55ea36de2a65631fbbec962d8201384da | 63,595 | ipynb | Jupyter Notebook | Notebooks/Continous Recording.ipynb | uwasystemhealth/PyDAQHAT | 3cdef8c0b9870b6bd0ac7ff3d7e55ca00b6c8e1b | [
"MIT"
] | null | null | null | Notebooks/Continous Recording.ipynb | uwasystemhealth/PyDAQHAT | 3cdef8c0b9870b6bd0ac7ff3d7e55ca00b6c8e1b | [
"MIT"
] | 3 | 2021-11-26T07:05:36.000Z | 2022-03-11T07:09:16.000Z | Notebooks/Continous Recording.ipynb | uwasystemhealth/PyDAQHAT | 3cdef8c0b9870b6bd0ac7ff3d7e55ca00b6c8e1b | [
"MIT"
] | null | null | null | 238.183521 | 56,822 | 0.921032 | [
[
[
"This notebook performs a continous recording using a start and stop button",
"_____no_output_____"
],
[
"## Import required modules",
"_____no_output_____"
]
],
[
[
"%matplotlib widget\nimport pydaqhat as py\nimport matplotlib.pyplot as plt\nimport matplotlib.ticker as ticker\nimport soundfile as sf\nfrom mutagen.flac import FLAC\nfrom ipywidgets import widgets",
"_____no_output_____"
]
],
[
[
"## Continous recording\nUse start/stop buttons to control recording",
"_____no_output_____"
]
],
[
[
"channels = [0] # Channels to use\niepe_enable = False # Enable/disable IEPE\nsensitivity = 1000 # Sensitivity in mV/unit\nsample_rate = 20480 # Number of samples per second\nbuffer_size = 10000 # Number of samples to keep in buffer before overwriting \nunit = (\"Voltage\", \"V\") # Unit of measurement. Format is (unit_name, unit_unit)",
"_____no_output_____"
],
[
"start = False\ndata = []\nhat = None\n\ndef start_click(i):\n global start\n \n if(start == False):\n main()\n start = True\n\ndef stop_click(i):\n global data\n global start\n \n if(start == True):\n data = stop_record()\n start = False\n \ndef stop_record():\n # Dump entire contents of buffer\n data = hat.a_in_scan_read(-1,0).data\n \n hat.a_in_scan_stop()\n hat.a_in_scan_cleanup()\n \n print(\"Scan has finished with length {}\".format(len(data)))\n \n return data\n\ndef continous_record():\n return py.continous_scan_start(\n channels=channels, \n iepe_enable=iepe_enable, \n sensitivity=sensitivity, \n sample_rate=sample_rate, \n buffer_size=buffer_size\n )\n\ndef main():\n global hat\n hat = continous_record()\n\nstart_button = widgets.Button(description = \"Start\")\nstart_button.on_click(start_click)\nstop_button = widgets.Button(description = \"Stop\")\nstop_button.on_click(stop_click)\ndisplay(start_button, stop_button)\n",
"_____no_output_____"
]
],
[
[
"## Save Recording to FLAC",
"_____no_output_____"
]
],
[
[
"filename = \"data/continous_recording_0.flac\"\n\nhat = py.get_hat()\nactual_sample_rate = round(hat.a_in_scan_actual_rate(sample_rate))\nsf.write(filename, data, actual_sample_rate)\n\nfile = FLAC(filename)\nfile[\"Title\"] = \"Finite Recording\"\nfile[\"Channels Used\"] = str(channels)\nfile[\"IEPE Enable\"] = str(iepe_enable)\nfile[\"Sensitivity\"] = str(sensitivity)\nfile[\"Sample Rate\"] = str(actual_sample_rate )\nfile[\"Unit\"] = \"{} ({})\".format(unit[0], unit[1])\nfile.save()\n\nprint(\"Recording saved to {}\".format(filename))",
"Recording saved to data/continous_recording_0.flac\n"
]
],
[
[
"## Visualise ",
"_____no_output_____"
]
],
[
[
"fig = plt.figure()\n\nax = fig.add_subplot(111)\nax.set_ylabel(\"Voltage (V)\")\nax.set_xlabel(\"Sample\")\nax.set_title(\"Continous Recording\")\nax.plot(range(len(data)), data)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb5942a4282dc83ce35165a8788aa2c690cb267 | 59,127 | ipynb | Jupyter Notebook | Demo.ipynb | redondoself/Mybiotools | ebdaabbd13846458bf64281ff96836eb4b68ae73 | [
"Apache-2.0"
] | 5 | 2016-05-12T14:33:34.000Z | 2019-06-24T13:39:44.000Z | Demo.ipynb | redondoself/Mybiotools | ebdaabbd13846458bf64281ff96836eb4b68ae73 | [
"Apache-2.0"
] | 1 | 2021-04-15T21:09:15.000Z | 2021-04-15T21:09:15.000Z | Demo.ipynb | redondoself/Mybiotools | ebdaabbd13846458bf64281ff96836eb4b68ae73 | [
"Apache-2.0"
] | 1 | 2020-03-24T11:44:05.000Z | 2020-03-24T11:44:05.000Z | 33.348562 | 2,602 | 0.378626 | [
[
[
"# Mybiotools -- A biotools collection for bench scientists\n\n## Python scripts to make bench work easier\n\n### Author: Mingzhang Yang",
"_____no_output_____"
],
[
"This notebook shows how you can take advantage of Mybiotools to make your life easier",
"_____no_output_____"
]
],
[
[
"#import Mybiotools package\nimport Mybiotools as mb",
"_____no_output_____"
],
[
"p53 = mb.Gene('TP53')",
"Please look up in the table below and set your gene of interest.\n Gene ID Gene description \\\n0 7157 tumor protein p53 \n1 24842 tumor protein p53 \n2 30590 tumor protein p53 \n3 7158 tumor protein p53 binding protein 1 \n4 403869 tumor protein p53 \n5 397276 tumor protein p53 \n6 281542 tumor protein p53 \n7 493847 tumor protein p53 \n8 100062044 tumor protein p53 \n9 100009292 tumor protein p53 \n10 100682525 tumor protein p53 \n11 100049321 tumor protein p53 \n12 716170 tumor protein p53 \n13 100379269 tumor protein p53 \n14 455214 tumor protein p53 \n15 443421 tumor protein p53 \n16 431679 tumor protein p53 \n17 102402069 tumor protein p53 \n18 100583326 tumor protein p53 \n19 100435218 tumor protein p53 \n\n Species \n0 Homo sapiens (human) \n1 Rattus norvegicus (Norway rat) \n2 Danio rerio (zebrafish) \n3 Homo sapiens (human) \n4 Canis lupus familiaris (dog) \n5 Sus scrofa (pig) \n6 Bos taurus (cattle) \n7 Felis catus (domestic cat) \n8 Equus caballus (horse) \n9 Oryctolagus cuniculus (rabbit) \n10 Cricetulus griseus (Chinese hamster) \n11 Oryzias latipes (Japanese medaka) \n12 Macaca mulatta (Rhesus monkey) \n13 Cavia porcellus (domestic guinea pig) \n14 Pan troglodytes (chimpanzee) \n15 Ovis aries (sheep) \n16 Xenopus tropicalis (tropical clawed frog) \n17 Bubalus bubalis (water buffalo) \n18 Nomascus leucogenys (northern white-cheeked gi... \n19 Pongo abelii (Sumatran orangutan) \nPlease select the index number(starts from 0) of the gene of your interest: 0\n\n\nYour gene object tumor protein p53 from Homo sapiens (human) has been created successfully.\n"
],
[
"p53.name",
"_____no_output_____"
],
[
"p53.Species",
"_____no_output_____"
],
[
"p53.mRNA",
"_____no_output_____"
],
[
"p53.default_mRNA",
"_____no_output_____"
],
[
"p53.get_Gene_ID()",
"7157\n"
],
[
"p53.get_mRNA()",
" NM_id name \\\n0 NM_000546.5 cellular tumor antigen p53 isoform a \n1 NM_001126112.2 cellular tumor antigen p53 isoform a \n2 NM_001126113.2 cellular tumor antigen p53 isoform c \n3 NM_001126114.2 cellular tumor antigen p53 isoform b \n4 NM_001126115.1 cellular tumor antigen p53 isoform d \n5 NM_001126116.1 cellular tumor antigen p53 isoform e \n6 NM_001126117.1 cellular tumor antigen p53 isoform f \n7 NM_001126118.1 cellular tumor antigen p53 isoform g \n8 NM_001276695.1 cellular tumor antigen p53 isoform h \n9 NM_001276696.1 cellular tumor antigen p53 isoform i \n10 NM_001276697.1 cellular tumor antigen p53 isoform j \n11 NM_001276698.1 cellular tumor antigen p53 isoform k \n12 NM_001276699.1 cellular tumor antigen p53 isoform l \n13 NM_001276760.1 cellular tumor antigen p53 isoform g \n14 NM_001276761.1 cellular tumor antigen p53 isoform g \n\n description \n0 This variant (1) can initiate translation from... \n1 This variant (2) uses an alternate splice site... \n2 This variant (4) contains an additional exon i... \n3 This variant (3) contains an additional exon i... \n4 This variant (5) uses an alternate promoter an... \n5 This variant (6) uses an alternate promoter an... \n6 This variant (7) uses an alternate promoter an... \n7 This variant (8, also known as p53I2) differs ... \n8 This variant (4) contains an additional exon i... \n9 This variant (3) contains an additional exon i... \n10 This variant (5) uses an alternate promoter an... \n11 This variant (6) uses an alternate promoter an... \n12 This variant (7) uses an alternate promoter an... \n13 This variant (1) can initiate translation from... \n14 This variant (2) uses an alternate splice site... \n"
],
[
"p53.mRNA",
"_____no_output_____"
],
[
"p53.mRNA[1]",
"_____no_output_____"
],
[
"p53.mRNA[1].NM_id",
"_____no_output_____"
],
[
"p53.mRNA[1].name",
"_____no_output_____"
],
[
"p53.mRNA[1].description",
"_____no_output_____"
],
[
"p53.default_mRNA",
"_____no_output_____"
],
[
"p53.get_mRNA_seq()",
"_____no_output_____"
],
[
"p53.default_mRNA_seq",
"_____no_output_____"
],
[
"p53.cds_seq",
"_____no_output_____"
],
[
"len(p53.default_mRNA_seq)",
"_____no_output_____"
],
[
"mb.help_info()",
"\n *clean_seq(input_seq)*: remove white space or numbers in the input gene or protein sequence.\n *reverse_complementory(input_seq)*: get the reversed complementory sequence of the input DNA sequence.\n *GC_content(input_seq)*: calculate the GC content of the input sequence.\n \n --help or -h: get help info of Mybiotools\n \n"
],
[
"mb.uniprot_search('TP53')",
"_____no_output_____"
],
[
"mb.list_files('.')",
"_____no_output_____"
],
[
"mb.list_files('.', '.pickle')",
"_____no_output_____"
],
[
"mb.list_files('.', Type=['.pickle', '.py'])",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"enzymes = pd.read_pickle('Restriction_Enzyme_list.pickle')",
"_____no_output_____"
],
[
"enzymes",
"_____no_output_____"
],
[
"enzymes.describe()",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"aa = pd.read_pickle('./AA_table.pickle')",
"_____no_output_____"
],
[
"aa",
"_____no_output_____"
],
[
"aa[['abrr', 'MW']]",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb594814d8d60b4f50c696e96366f1cb25c425b | 511,209 | ipynb | Jupyter Notebook | Model backlog/EfficientNet/EfficientNetB5/225 - EfficientNetB5-Reg-Img224 0,5data TTA 10.ipynb | ThinkBricks/APTOS2019BlindnessDetection | e524fd69f83a1252710076c78b6a5236849cd885 | [
"MIT"
] | 23 | 2019-09-08T17:19:16.000Z | 2022-02-02T16:20:09.000Z | Model backlog/EfficientNet/EfficientNetB5/225 - EfficientNetB5-Reg-Img224 0,5data TTA 10.ipynb | ThinkBricks/APTOS2019BlindnessDetection | e524fd69f83a1252710076c78b6a5236849cd885 | [
"MIT"
] | 1 | 2020-03-10T18:42:12.000Z | 2020-09-18T22:02:38.000Z | Model backlog/EfficientNet/EfficientNetB5/225 - EfficientNetB5-Reg-Img224 0,5data TTA 10.ipynb | ThinkBricks/APTOS2019BlindnessDetection | e524fd69f83a1252710076c78b6a5236849cd885 | [
"MIT"
] | 16 | 2019-09-21T12:29:59.000Z | 2022-03-21T00:42:26.000Z | 135.347895 | 81,804 | 0.74474 | [
[
[
"## Dependencies",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nimport cv2\nimport shutil\nimport random\nimport warnings\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport multiprocessing as mp\nimport matplotlib.pyplot as plt\nfrom tensorflow import set_random_seed\nfrom sklearn.utils import class_weight\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import confusion_matrix, cohen_kappa_score\nfrom keras import backend as K\nfrom keras.models import Model\nfrom keras.utils import to_categorical\nfrom keras import optimizers, applications\nfrom keras.preprocessing.image import ImageDataGenerator\nfrom keras.layers import Dense, Dropout, GlobalAveragePooling2D, Input\nfrom keras.callbacks import EarlyStopping, ReduceLROnPlateau, Callback, LearningRateScheduler\n\ndef seed_everything(seed=0):\n random.seed(seed)\n os.environ['PYTHONHASHSEED'] = str(seed)\n np.random.seed(seed)\n set_random_seed(0)\n\nseed = 0\nseed_everything(seed)\n%matplotlib inline\nsns.set(style=\"whitegrid\")\nwarnings.filterwarnings(\"ignore\")\nsys.path.append(os.path.abspath('../input/efficientnet/efficientnet-master/efficientnet-master/'))\nfrom efficientnet import *",
"/opt/conda/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:516: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\n/opt/conda/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:517: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\n/opt/conda/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:518: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\n/opt/conda/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:519: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\n/opt/conda/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:520: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\n/opt/conda/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:525: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\n/opt/conda/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:541: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\n/opt/conda/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:542: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\n/opt/conda/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:543: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\n/opt/conda/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:544: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\n/opt/conda/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:545: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\n/opt/conda/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:550: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\nUsing TensorFlow backend.\n"
]
],
[
[
"## Load data",
"_____no_output_____"
]
],
[
[
"hold_out_set = pd.read_csv('../input/aptos-split-oldnew/hold-out_5.csv')\nX_train = hold_out_set[hold_out_set['set'] == 'train']\nX_val = hold_out_set[hold_out_set['set'] == 'validation']\ntest = pd.read_csv('../input/aptos2019-blindness-detection/test.csv')\n\ntest[\"id_code\"] = test[\"id_code\"].apply(lambda x: x + \".png\")\n\nprint('Number of train samples: ', X_train.shape[0])\nprint('Number of validation samples: ', X_val.shape[0])\nprint('Number of test samples: ', test.shape[0])\ndisplay(X_train.head())",
"Number of train samples: 17599\nNumber of validation samples: 1831\nNumber of test samples: 1928\n"
]
],
[
[
"# Model parameters",
"_____no_output_____"
]
],
[
[
"# Model parameters\nFACTOR = 4\nBATCH_SIZE = 8 * FACTOR\nEPOCHS = 20\nWARMUP_EPOCHS = 5\nLEARNING_RATE = 1e-4 * FACTOR\nWARMUP_LEARNING_RATE = 1e-3 * FACTOR\nHEIGHT = 224\nWIDTH = 224\nCHANNELS = 3\nTTA_STEPS = 10\nES_PATIENCE = 5\nRLROP_PATIENCE = 3\nDECAY_DROP = 0.5\nLR_WARMUP_EPOCHS_1st = 2\nLR_WARMUP_EPOCHS_2nd = 5\nSTEP_SIZE = len(X_train) // BATCH_SIZE\nTOTAL_STEPS_1st = WARMUP_EPOCHS * STEP_SIZE\nTOTAL_STEPS_2nd = EPOCHS * STEP_SIZE\nWARMUP_STEPS_1st = LR_WARMUP_EPOCHS_1st * STEP_SIZE\nWARMUP_STEPS_2nd = LR_WARMUP_EPOCHS_2nd * STEP_SIZE",
"_____no_output_____"
]
],
[
[
"# Pre-procecess images",
"_____no_output_____"
]
],
[
[
"old_data_base_path = '../input/diabetic-retinopathy-resized/resized_train/resized_train/'\nnew_data_base_path = '../input/aptos2019-blindness-detection/train_images/'\ntest_base_path = '../input/aptos2019-blindness-detection/test_images/'\ntrain_dest_path = 'base_dir/train_images/'\nvalidation_dest_path = 'base_dir/validation_images/'\ntest_dest_path = 'base_dir/test_images/'\n\n# Making sure directories don't exist\nif os.path.exists(train_dest_path):\n shutil.rmtree(train_dest_path)\nif os.path.exists(validation_dest_path):\n shutil.rmtree(validation_dest_path)\nif os.path.exists(test_dest_path):\n shutil.rmtree(test_dest_path)\n \n# Creating train, validation and test directories\nos.makedirs(train_dest_path)\nos.makedirs(validation_dest_path)\nos.makedirs(test_dest_path)\n\ndef crop_image(img, tol=7):\n if img.ndim ==2:\n mask = img>tol\n return img[np.ix_(mask.any(1),mask.any(0))]\n elif img.ndim==3:\n gray_img = cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)\n mask = gray_img>tol\n check_shape = img[:,:,0][np.ix_(mask.any(1),mask.any(0))].shape[0]\n if (check_shape == 0): # image is too dark so that we crop out everything,\n return img # return original image\n else:\n img1=img[:,:,0][np.ix_(mask.any(1),mask.any(0))]\n img2=img[:,:,1][np.ix_(mask.any(1),mask.any(0))]\n img3=img[:,:,2][np.ix_(mask.any(1),mask.any(0))]\n img = np.stack([img1,img2,img3],axis=-1)\n \n return img\n\ndef circle_crop(img):\n img = crop_image(img)\n\n height, width, depth = img.shape\n largest_side = np.max((height, width))\n img = cv2.resize(img, (largest_side, largest_side))\n\n height, width, depth = img.shape\n\n x = width//2\n y = height//2\n r = np.amin((x, y))\n\n circle_img = np.zeros((height, width), np.uint8)\n cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)\n img = cv2.bitwise_and(img, img, mask=circle_img)\n img = crop_image(img)\n\n return img\n \ndef preprocess_image(image_id, base_path, save_path, HEIGHT=HEIGHT, WIDTH=WIDTH, sigmaX=10):\n image = cv2.imread(base_path + image_id)\n image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)\n image = circle_crop(image)\n image = cv2.resize(image, (HEIGHT, WIDTH))\n# image = cv2.addWeighted(image, 4, cv2.GaussianBlur(image, (0,0), sigmaX), -4 , 128)\n cv2.imwrite(save_path + image_id, image)\n \ndef preprocess_data(df, HEIGHT=HEIGHT, WIDTH=WIDTH, sigmaX=10):\n df = df.reset_index()\n for i in range(df.shape[0]):\n item = df.iloc[i]\n image_id = item['id_code']\n item_set = item['set']\n item_data = item['data']\n if item_set == 'train':\n if item_data == 'new':\n preprocess_image(image_id, new_data_base_path, train_dest_path)\n if item_data == 'old':\n preprocess_image(image_id, old_data_base_path, train_dest_path)\n if item_set == 'validation':\n if item_data == 'new':\n preprocess_image(image_id, new_data_base_path, validation_dest_path)\n if item_data == 'old':\n preprocess_image(image_id, old_data_base_path, validation_dest_path)\n \ndef preprocess_test(df, base_path=test_base_path, save_path=test_dest_path, HEIGHT=HEIGHT, WIDTH=WIDTH, sigmaX=10):\n df = df.reset_index()\n for i in range(df.shape[0]):\n image_id = df.iloc[i]['id_code']\n preprocess_image(image_id, base_path, save_path)\n\nn_cpu = mp.cpu_count()\ntrain_n_cnt = X_train.shape[0] // n_cpu\nval_n_cnt = X_val.shape[0] // n_cpu\ntest_n_cnt = test.shape[0] // n_cpu\n\n# Pre-procecss old data train set\npool = mp.Pool(n_cpu)\ndfs = [X_train.iloc[train_n_cnt*i:train_n_cnt*(i+1)] for i in range(n_cpu)]\ndfs[-1] = X_train.iloc[train_n_cnt*(n_cpu-1):]\nres = pool.map(preprocess_data, [x_df for x_df in dfs])\npool.close()\n\n# Pre-procecss validation set\npool = mp.Pool(n_cpu)\ndfs = [X_val.iloc[val_n_cnt*i:val_n_cnt*(i+1)] for i in range(n_cpu)]\ndfs[-1] = X_val.iloc[val_n_cnt*(n_cpu-1):] \nres = pool.map(preprocess_data, [x_df for x_df in dfs])\npool.close()\n\n# Pre-procecss test set\npool = mp.Pool(n_cpu)\ndfs = [test.iloc[test_n_cnt*i:test_n_cnt*(i+1)] for i in range(n_cpu)]\ndfs[-1] = test.iloc[test_n_cnt*(n_cpu-1):] \nres = pool.map(preprocess_test, [x_df for x_df in dfs])\npool.close()",
"_____no_output_____"
]
],
[
[
"# Data generator",
"_____no_output_____"
]
],
[
[
"datagen=ImageDataGenerator(rescale=1./255, \n rotation_range=360,\n horizontal_flip=True,\n vertical_flip=True)\n\ntrain_generator=datagen.flow_from_dataframe(\n dataframe=X_train,\n directory=train_dest_path,\n x_col=\"id_code\",\n y_col=\"diagnosis\",\n class_mode=\"raw\",\n batch_size=BATCH_SIZE,\n target_size=(HEIGHT, WIDTH),\n seed=seed)\n\nvalid_generator=datagen.flow_from_dataframe(\n dataframe=X_val,\n directory=validation_dest_path,\n x_col=\"id_code\",\n y_col=\"diagnosis\",\n class_mode=\"raw\",\n batch_size=BATCH_SIZE,\n target_size=(HEIGHT, WIDTH),\n seed=seed)\n\ntest_generator=datagen.flow_from_dataframe( \n dataframe=test,\n directory=test_dest_path,\n x_col=\"id_code\",\n batch_size=1,\n class_mode=None,\n shuffle=False,\n target_size=(HEIGHT, WIDTH),\n seed=seed)",
"Found 17599 validated image filenames.\nFound 1831 validated image filenames.\nFound 1928 validated image filenames.\n"
],
[
"def cosine_decay_with_warmup(global_step,\n learning_rate_base,\n total_steps,\n warmup_learning_rate=0.0,\n warmup_steps=0,\n hold_base_rate_steps=0):\n \"\"\"\n Cosine decay schedule with warm up period.\n In this schedule, the learning rate grows linearly from warmup_learning_rate\n to learning_rate_base for warmup_steps, then transitions to a cosine decay\n schedule.\n :param global_step {int}: global step.\n :param learning_rate_base {float}: base learning rate.\n :param total_steps {int}: total number of training steps.\n :param warmup_learning_rate {float}: initial learning rate for warm up. (default: {0.0}).\n :param warmup_steps {int}: number of warmup steps. (default: {0}).\n :param hold_base_rate_steps {int}: Optional number of steps to hold base learning rate before decaying. (default: {0}).\n :param global_step {int}: global step.\n :Returns : a float representing learning rate.\n :Raises ValueError: if warmup_learning_rate is larger than learning_rate_base, or if warmup_steps is larger than total_steps.\n \"\"\"\n\n if total_steps < warmup_steps:\n raise ValueError('total_steps must be larger or equal to warmup_steps.')\n learning_rate = 0.5 * learning_rate_base * (1 + np.cos(\n np.pi *\n (global_step - warmup_steps - hold_base_rate_steps\n ) / float(total_steps - warmup_steps - hold_base_rate_steps)))\n if hold_base_rate_steps > 0:\n learning_rate = np.where(global_step > warmup_steps + hold_base_rate_steps,\n learning_rate, learning_rate_base)\n if warmup_steps > 0:\n if learning_rate_base < warmup_learning_rate:\n raise ValueError('learning_rate_base must be larger or equal to warmup_learning_rate.')\n slope = (learning_rate_base - warmup_learning_rate) / warmup_steps\n warmup_rate = slope * global_step + warmup_learning_rate\n learning_rate = np.where(global_step < warmup_steps, warmup_rate,\n learning_rate)\n return np.where(global_step > total_steps, 0.0, learning_rate)\n\n\nclass WarmUpCosineDecayScheduler(Callback):\n \"\"\"Cosine decay with warmup learning rate scheduler\"\"\"\n\n def __init__(self,\n learning_rate_base,\n total_steps,\n global_step_init=0,\n warmup_learning_rate=0.0,\n warmup_steps=0,\n hold_base_rate_steps=0,\n verbose=0):\n \"\"\"\n Constructor for cosine decay with warmup learning rate scheduler.\n :param learning_rate_base {float}: base learning rate.\n :param total_steps {int}: total number of training steps.\n :param global_step_init {int}: initial global step, e.g. from previous checkpoint.\n :param warmup_learning_rate {float}: initial learning rate for warm up. (default: {0.0}).\n :param warmup_steps {int}: number of warmup steps. (default: {0}).\n :param hold_base_rate_steps {int}: Optional number of steps to hold base learning rate before decaying. (default: {0}).\n :param verbose {int}: quiet, 1: update messages. (default: {0}).\n \"\"\"\n\n super(WarmUpCosineDecayScheduler, self).__init__()\n self.learning_rate_base = learning_rate_base\n self.total_steps = total_steps\n self.global_step = global_step_init\n self.warmup_learning_rate = warmup_learning_rate\n self.warmup_steps = warmup_steps\n self.hold_base_rate_steps = hold_base_rate_steps\n self.verbose = verbose\n self.learning_rates = []\n\n def on_batch_end(self, batch, logs=None):\n self.global_step = self.global_step + 1\n lr = K.get_value(self.model.optimizer.lr)\n self.learning_rates.append(lr)\n\n def on_batch_begin(self, batch, logs=None):\n lr = cosine_decay_with_warmup(global_step=self.global_step,\n learning_rate_base=self.learning_rate_base,\n total_steps=self.total_steps,\n warmup_learning_rate=self.warmup_learning_rate,\n warmup_steps=self.warmup_steps,\n hold_base_rate_steps=self.hold_base_rate_steps)\n K.set_value(self.model.optimizer.lr, lr)\n if self.verbose > 0:\n print('\\nBatch %02d: setting learning rate to %s.' % (self.global_step + 1, lr))",
"_____no_output_____"
]
],
[
[
"# Model",
"_____no_output_____"
]
],
[
[
"def create_model(input_shape):\n input_tensor = Input(shape=input_shape)\n base_model = EfficientNetB5(weights=None, \n include_top=False,\n input_tensor=input_tensor)\n base_model.load_weights('../input/efficientnet-keras-weights-b0b5/efficientnet-b5_imagenet_1000_notop.h5')\n\n x = GlobalAveragePooling2D()(base_model.output)\n final_output = Dense(1, activation='linear', name='final_output')(x)\n model = Model(input_tensor, final_output)\n \n return model",
"_____no_output_____"
]
],
[
[
"# Train top layers",
"_____no_output_____"
]
],
[
[
"model = create_model(input_shape=(HEIGHT, WIDTH, CHANNELS))\n\nfor layer in model.layers:\n layer.trainable = False\n\nfor i in range(-2, 0):\n model.layers[i].trainable = True\n\ncosine_lr_1st = WarmUpCosineDecayScheduler(learning_rate_base=WARMUP_LEARNING_RATE,\n total_steps=TOTAL_STEPS_1st,\n warmup_learning_rate=0.0,\n warmup_steps=WARMUP_STEPS_1st,\n hold_base_rate_steps=(2 * STEP_SIZE))\n\nmetric_list = [\"accuracy\"]\ncallback_list = [cosine_lr_1st]\noptimizer = optimizers.Adam(lr=WARMUP_LEARNING_RATE)\nmodel.compile(optimizer=optimizer, loss='mean_squared_error', metrics=metric_list)\nmodel.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) (None, 224, 224, 3) 0 \n__________________________________________________________________________________________________\nconv2d_1 (Conv2D) (None, 112, 112, 48) 1296 input_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_1 (BatchNor (None, 112, 112, 48) 192 conv2d_1[0][0] \n__________________________________________________________________________________________________\nswish_1 (Swish) (None, 112, 112, 48) 0 batch_normalization_1[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_1 (DepthwiseCo (None, 112, 112, 48) 432 swish_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_2 (BatchNor (None, 112, 112, 48) 192 depthwise_conv2d_1[0][0] \n__________________________________________________________________________________________________\nswish_2 (Swish) (None, 112, 112, 48) 0 batch_normalization_2[0][0] \n__________________________________________________________________________________________________\nlambda_1 (Lambda) (None, 1, 1, 48) 0 swish_2[0][0] \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 1, 1, 12) 588 lambda_1[0][0] \n__________________________________________________________________________________________________\nswish_3 (Swish) (None, 1, 1, 12) 0 conv2d_2[0][0] \n__________________________________________________________________________________________________\nconv2d_3 (Conv2D) (None, 1, 1, 48) 624 swish_3[0][0] \n__________________________________________________________________________________________________\nactivation_1 (Activation) (None, 1, 1, 48) 0 conv2d_3[0][0] \n__________________________________________________________________________________________________\nmultiply_1 (Multiply) (None, 112, 112, 48) 0 activation_1[0][0] \n swish_2[0][0] \n__________________________________________________________________________________________________\nconv2d_4 (Conv2D) (None, 112, 112, 24) 1152 multiply_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_3 (BatchNor (None, 112, 112, 24) 96 conv2d_4[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_2 (DepthwiseCo (None, 112, 112, 24) 216 batch_normalization_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_4 (BatchNor (None, 112, 112, 24) 96 depthwise_conv2d_2[0][0] \n__________________________________________________________________________________________________\nswish_4 (Swish) (None, 112, 112, 24) 0 batch_normalization_4[0][0] \n__________________________________________________________________________________________________\nlambda_2 (Lambda) (None, 1, 1, 24) 0 swish_4[0][0] \n__________________________________________________________________________________________________\nconv2d_5 (Conv2D) (None, 1, 1, 6) 150 lambda_2[0][0] \n__________________________________________________________________________________________________\nswish_5 (Swish) (None, 1, 1, 6) 0 conv2d_5[0][0] \n__________________________________________________________________________________________________\nconv2d_6 (Conv2D) (None, 1, 1, 24) 168 swish_5[0][0] \n__________________________________________________________________________________________________\nactivation_2 (Activation) (None, 1, 1, 24) 0 conv2d_6[0][0] \n__________________________________________________________________________________________________\nmultiply_2 (Multiply) (None, 112, 112, 24) 0 activation_2[0][0] \n swish_4[0][0] \n__________________________________________________________________________________________________\nconv2d_7 (Conv2D) (None, 112, 112, 24) 576 multiply_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_5 (BatchNor (None, 112, 112, 24) 96 conv2d_7[0][0] \n__________________________________________________________________________________________________\ndrop_connect_1 (DropConnect) (None, 112, 112, 24) 0 batch_normalization_5[0][0] \n__________________________________________________________________________________________________\nadd_1 (Add) (None, 112, 112, 24) 0 drop_connect_1[0][0] \n batch_normalization_3[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_3 (DepthwiseCo (None, 112, 112, 24) 216 add_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_6 (BatchNor (None, 112, 112, 24) 96 depthwise_conv2d_3[0][0] \n__________________________________________________________________________________________________\nswish_6 (Swish) (None, 112, 112, 24) 0 batch_normalization_6[0][0] \n__________________________________________________________________________________________________\nlambda_3 (Lambda) (None, 1, 1, 24) 0 swish_6[0][0] \n__________________________________________________________________________________________________\nconv2d_8 (Conv2D) (None, 1, 1, 6) 150 lambda_3[0][0] \n__________________________________________________________________________________________________\nswish_7 (Swish) (None, 1, 1, 6) 0 conv2d_8[0][0] \n__________________________________________________________________________________________________\nconv2d_9 (Conv2D) (None, 1, 1, 24) 168 swish_7[0][0] \n__________________________________________________________________________________________________\nactivation_3 (Activation) (None, 1, 1, 24) 0 conv2d_9[0][0] \n__________________________________________________________________________________________________\nmultiply_3 (Multiply) (None, 112, 112, 24) 0 activation_3[0][0] \n swish_6[0][0] \n__________________________________________________________________________________________________\nconv2d_10 (Conv2D) (None, 112, 112, 24) 576 multiply_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_7 (BatchNor (None, 112, 112, 24) 96 conv2d_10[0][0] \n__________________________________________________________________________________________________\ndrop_connect_2 (DropConnect) (None, 112, 112, 24) 0 batch_normalization_7[0][0] \n__________________________________________________________________________________________________\nadd_2 (Add) (None, 112, 112, 24) 0 drop_connect_2[0][0] \n add_1[0][0] \n__________________________________________________________________________________________________\nconv2d_11 (Conv2D) (None, 112, 112, 144 3456 add_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_8 (BatchNor (None, 112, 112, 144 576 conv2d_11[0][0] \n__________________________________________________________________________________________________\nswish_8 (Swish) (None, 112, 112, 144 0 batch_normalization_8[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_4 (DepthwiseCo (None, 56, 56, 144) 1296 swish_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_9 (BatchNor (None, 56, 56, 144) 576 depthwise_conv2d_4[0][0] \n__________________________________________________________________________________________________\nswish_9 (Swish) (None, 56, 56, 144) 0 batch_normalization_9[0][0] \n__________________________________________________________________________________________________\nlambda_4 (Lambda) (None, 1, 1, 144) 0 swish_9[0][0] \n__________________________________________________________________________________________________\nconv2d_12 (Conv2D) (None, 1, 1, 6) 870 lambda_4[0][0] \n__________________________________________________________________________________________________\nswish_10 (Swish) (None, 1, 1, 6) 0 conv2d_12[0][0] \n__________________________________________________________________________________________________\nconv2d_13 (Conv2D) (None, 1, 1, 144) 1008 swish_10[0][0] \n__________________________________________________________________________________________________\nactivation_4 (Activation) (None, 1, 1, 144) 0 conv2d_13[0][0] \n__________________________________________________________________________________________________\nmultiply_4 (Multiply) (None, 56, 56, 144) 0 activation_4[0][0] \n swish_9[0][0] \n__________________________________________________________________________________________________\nconv2d_14 (Conv2D) (None, 56, 56, 40) 5760 multiply_4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_10 (BatchNo (None, 56, 56, 40) 160 conv2d_14[0][0] \n__________________________________________________________________________________________________\nconv2d_15 (Conv2D) (None, 56, 56, 240) 9600 batch_normalization_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_11 (BatchNo (None, 56, 56, 240) 960 conv2d_15[0][0] \n__________________________________________________________________________________________________\nswish_11 (Swish) (None, 56, 56, 240) 0 batch_normalization_11[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_5 (DepthwiseCo (None, 56, 56, 240) 2160 swish_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_12 (BatchNo (None, 56, 56, 240) 960 depthwise_conv2d_5[0][0] \n__________________________________________________________________________________________________\nswish_12 (Swish) (None, 56, 56, 240) 0 batch_normalization_12[0][0] \n__________________________________________________________________________________________________\nlambda_5 (Lambda) (None, 1, 1, 240) 0 swish_12[0][0] \n__________________________________________________________________________________________________\nconv2d_16 (Conv2D) (None, 1, 1, 10) 2410 lambda_5[0][0] \n__________________________________________________________________________________________________\nswish_13 (Swish) (None, 1, 1, 10) 0 conv2d_16[0][0] \n__________________________________________________________________________________________________\nconv2d_17 (Conv2D) (None, 1, 1, 240) 2640 swish_13[0][0] \n__________________________________________________________________________________________________\nactivation_5 (Activation) (None, 1, 1, 240) 0 conv2d_17[0][0] \n__________________________________________________________________________________________________\nmultiply_5 (Multiply) (None, 56, 56, 240) 0 activation_5[0][0] \n swish_12[0][0] \n__________________________________________________________________________________________________\nconv2d_18 (Conv2D) (None, 56, 56, 40) 9600 multiply_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_13 (BatchNo (None, 56, 56, 40) 160 conv2d_18[0][0] \n__________________________________________________________________________________________________\ndrop_connect_3 (DropConnect) (None, 56, 56, 40) 0 batch_normalization_13[0][0] \n__________________________________________________________________________________________________\nadd_3 (Add) (None, 56, 56, 40) 0 drop_connect_3[0][0] \n batch_normalization_10[0][0] \n__________________________________________________________________________________________________\nconv2d_19 (Conv2D) (None, 56, 56, 240) 9600 add_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_14 (BatchNo (None, 56, 56, 240) 960 conv2d_19[0][0] \n__________________________________________________________________________________________________\nswish_14 (Swish) (None, 56, 56, 240) 0 batch_normalization_14[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_6 (DepthwiseCo (None, 56, 56, 240) 2160 swish_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_15 (BatchNo (None, 56, 56, 240) 960 depthwise_conv2d_6[0][0] \n__________________________________________________________________________________________________\nswish_15 (Swish) (None, 56, 56, 240) 0 batch_normalization_15[0][0] \n__________________________________________________________________________________________________\nlambda_6 (Lambda) (None, 1, 1, 240) 0 swish_15[0][0] \n__________________________________________________________________________________________________\nconv2d_20 (Conv2D) (None, 1, 1, 10) 2410 lambda_6[0][0] \n__________________________________________________________________________________________________\nswish_16 (Swish) (None, 1, 1, 10) 0 conv2d_20[0][0] \n__________________________________________________________________________________________________\nconv2d_21 (Conv2D) (None, 1, 1, 240) 2640 swish_16[0][0] \n__________________________________________________________________________________________________\nactivation_6 (Activation) (None, 1, 1, 240) 0 conv2d_21[0][0] \n__________________________________________________________________________________________________\nmultiply_6 (Multiply) (None, 56, 56, 240) 0 activation_6[0][0] \n swish_15[0][0] \n__________________________________________________________________________________________________\nconv2d_22 (Conv2D) (None, 56, 56, 40) 9600 multiply_6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_16 (BatchNo (None, 56, 56, 40) 160 conv2d_22[0][0] \n__________________________________________________________________________________________________\ndrop_connect_4 (DropConnect) (None, 56, 56, 40) 0 batch_normalization_16[0][0] \n__________________________________________________________________________________________________\nadd_4 (Add) (None, 56, 56, 40) 0 drop_connect_4[0][0] \n add_3[0][0] \n__________________________________________________________________________________________________\nconv2d_23 (Conv2D) (None, 56, 56, 240) 9600 add_4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_17 (BatchNo (None, 56, 56, 240) 960 conv2d_23[0][0] \n__________________________________________________________________________________________________\nswish_17 (Swish) (None, 56, 56, 240) 0 batch_normalization_17[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_7 (DepthwiseCo (None, 56, 56, 240) 2160 swish_17[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_18 (BatchNo (None, 56, 56, 240) 960 depthwise_conv2d_7[0][0] \n__________________________________________________________________________________________________\nswish_18 (Swish) (None, 56, 56, 240) 0 batch_normalization_18[0][0] \n__________________________________________________________________________________________________\nlambda_7 (Lambda) (None, 1, 1, 240) 0 swish_18[0][0] \n__________________________________________________________________________________________________\nconv2d_24 (Conv2D) (None, 1, 1, 10) 2410 lambda_7[0][0] \n__________________________________________________________________________________________________\nswish_19 (Swish) (None, 1, 1, 10) 0 conv2d_24[0][0] \n__________________________________________________________________________________________________\nconv2d_25 (Conv2D) (None, 1, 1, 240) 2640 swish_19[0][0] \n__________________________________________________________________________________________________\nactivation_7 (Activation) (None, 1, 1, 240) 0 conv2d_25[0][0] \n__________________________________________________________________________________________________\nmultiply_7 (Multiply) (None, 56, 56, 240) 0 activation_7[0][0] \n swish_18[0][0] \n__________________________________________________________________________________________________\nconv2d_26 (Conv2D) (None, 56, 56, 40) 9600 multiply_7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_19 (BatchNo (None, 56, 56, 40) 160 conv2d_26[0][0] \n__________________________________________________________________________________________________\ndrop_connect_5 (DropConnect) (None, 56, 56, 40) 0 batch_normalization_19[0][0] \n__________________________________________________________________________________________________\nadd_5 (Add) (None, 56, 56, 40) 0 drop_connect_5[0][0] \n add_4[0][0] \n__________________________________________________________________________________________________\nconv2d_27 (Conv2D) (None, 56, 56, 240) 9600 add_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_20 (BatchNo (None, 56, 56, 240) 960 conv2d_27[0][0] \n__________________________________________________________________________________________________\nswish_20 (Swish) (None, 56, 56, 240) 0 batch_normalization_20[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_8 (DepthwiseCo (None, 56, 56, 240) 2160 swish_20[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_21 (BatchNo (None, 56, 56, 240) 960 depthwise_conv2d_8[0][0] \n__________________________________________________________________________________________________\nswish_21 (Swish) (None, 56, 56, 240) 0 batch_normalization_21[0][0] \n__________________________________________________________________________________________________\nlambda_8 (Lambda) (None, 1, 1, 240) 0 swish_21[0][0] \n__________________________________________________________________________________________________\nconv2d_28 (Conv2D) (None, 1, 1, 10) 2410 lambda_8[0][0] \n__________________________________________________________________________________________________\nswish_22 (Swish) (None, 1, 1, 10) 0 conv2d_28[0][0] \n__________________________________________________________________________________________________\nconv2d_29 (Conv2D) (None, 1, 1, 240) 2640 swish_22[0][0] \n__________________________________________________________________________________________________\nactivation_8 (Activation) (None, 1, 1, 240) 0 conv2d_29[0][0] \n__________________________________________________________________________________________________\nmultiply_8 (Multiply) (None, 56, 56, 240) 0 activation_8[0][0] \n swish_21[0][0] \n__________________________________________________________________________________________________\nconv2d_30 (Conv2D) (None, 56, 56, 40) 9600 multiply_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_22 (BatchNo (None, 56, 56, 40) 160 conv2d_30[0][0] \n__________________________________________________________________________________________________\ndrop_connect_6 (DropConnect) (None, 56, 56, 40) 0 batch_normalization_22[0][0] \n__________________________________________________________________________________________________\nadd_6 (Add) (None, 56, 56, 40) 0 drop_connect_6[0][0] \n add_5[0][0] \n__________________________________________________________________________________________________\nconv2d_31 (Conv2D) (None, 56, 56, 240) 9600 add_6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_23 (BatchNo (None, 56, 56, 240) 960 conv2d_31[0][0] \n__________________________________________________________________________________________________\nswish_23 (Swish) (None, 56, 56, 240) 0 batch_normalization_23[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_9 (DepthwiseCo (None, 28, 28, 240) 6000 swish_23[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_24 (BatchNo (None, 28, 28, 240) 960 depthwise_conv2d_9[0][0] \n__________________________________________________________________________________________________\nswish_24 (Swish) (None, 28, 28, 240) 0 batch_normalization_24[0][0] \n__________________________________________________________________________________________________\nlambda_9 (Lambda) (None, 1, 1, 240) 0 swish_24[0][0] \n__________________________________________________________________________________________________\nconv2d_32 (Conv2D) (None, 1, 1, 10) 2410 lambda_9[0][0] \n__________________________________________________________________________________________________\nswish_25 (Swish) (None, 1, 1, 10) 0 conv2d_32[0][0] \n__________________________________________________________________________________________________\nconv2d_33 (Conv2D) (None, 1, 1, 240) 2640 swish_25[0][0] \n__________________________________________________________________________________________________\nactivation_9 (Activation) (None, 1, 1, 240) 0 conv2d_33[0][0] \n__________________________________________________________________________________________________\nmultiply_9 (Multiply) (None, 28, 28, 240) 0 activation_9[0][0] \n swish_24[0][0] \n__________________________________________________________________________________________________\nconv2d_34 (Conv2D) (None, 28, 28, 64) 15360 multiply_9[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_25 (BatchNo (None, 28, 28, 64) 256 conv2d_34[0][0] \n__________________________________________________________________________________________________\nconv2d_35 (Conv2D) (None, 28, 28, 384) 24576 batch_normalization_25[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_26 (BatchNo (None, 28, 28, 384) 1536 conv2d_35[0][0] \n__________________________________________________________________________________________________\nswish_26 (Swish) (None, 28, 28, 384) 0 batch_normalization_26[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_10 (DepthwiseC (None, 28, 28, 384) 9600 swish_26[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_27 (BatchNo (None, 28, 28, 384) 1536 depthwise_conv2d_10[0][0] \n__________________________________________________________________________________________________\nswish_27 (Swish) (None, 28, 28, 384) 0 batch_normalization_27[0][0] \n__________________________________________________________________________________________________\nlambda_10 (Lambda) (None, 1, 1, 384) 0 swish_27[0][0] \n__________________________________________________________________________________________________\nconv2d_36 (Conv2D) (None, 1, 1, 16) 6160 lambda_10[0][0] \n__________________________________________________________________________________________________\nswish_28 (Swish) (None, 1, 1, 16) 0 conv2d_36[0][0] \n__________________________________________________________________________________________________\nconv2d_37 (Conv2D) (None, 1, 1, 384) 6528 swish_28[0][0] \n__________________________________________________________________________________________________\nactivation_10 (Activation) (None, 1, 1, 384) 0 conv2d_37[0][0] \n__________________________________________________________________________________________________\nmultiply_10 (Multiply) (None, 28, 28, 384) 0 activation_10[0][0] \n swish_27[0][0] \n__________________________________________________________________________________________________\nconv2d_38 (Conv2D) (None, 28, 28, 64) 24576 multiply_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_28 (BatchNo (None, 28, 28, 64) 256 conv2d_38[0][0] \n__________________________________________________________________________________________________\ndrop_connect_7 (DropConnect) (None, 28, 28, 64) 0 batch_normalization_28[0][0] \n__________________________________________________________________________________________________\nadd_7 (Add) (None, 28, 28, 64) 0 drop_connect_7[0][0] \n batch_normalization_25[0][0] \n__________________________________________________________________________________________________\nconv2d_39 (Conv2D) (None, 28, 28, 384) 24576 add_7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_29 (BatchNo (None, 28, 28, 384) 1536 conv2d_39[0][0] \n__________________________________________________________________________________________________\nswish_29 (Swish) (None, 28, 28, 384) 0 batch_normalization_29[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_11 (DepthwiseC (None, 28, 28, 384) 9600 swish_29[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_30 (BatchNo (None, 28, 28, 384) 1536 depthwise_conv2d_11[0][0] \n__________________________________________________________________________________________________\nswish_30 (Swish) (None, 28, 28, 384) 0 batch_normalization_30[0][0] \n__________________________________________________________________________________________________\nlambda_11 (Lambda) (None, 1, 1, 384) 0 swish_30[0][0] \n__________________________________________________________________________________________________\nconv2d_40 (Conv2D) (None, 1, 1, 16) 6160 lambda_11[0][0] \n__________________________________________________________________________________________________\nswish_31 (Swish) (None, 1, 1, 16) 0 conv2d_40[0][0] \n__________________________________________________________________________________________________\nconv2d_41 (Conv2D) (None, 1, 1, 384) 6528 swish_31[0][0] \n__________________________________________________________________________________________________\nactivation_11 (Activation) (None, 1, 1, 384) 0 conv2d_41[0][0] \n__________________________________________________________________________________________________\nmultiply_11 (Multiply) (None, 28, 28, 384) 0 activation_11[0][0] \n swish_30[0][0] \n__________________________________________________________________________________________________\nconv2d_42 (Conv2D) (None, 28, 28, 64) 24576 multiply_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_31 (BatchNo (None, 28, 28, 64) 256 conv2d_42[0][0] \n__________________________________________________________________________________________________\ndrop_connect_8 (DropConnect) (None, 28, 28, 64) 0 batch_normalization_31[0][0] \n__________________________________________________________________________________________________\nadd_8 (Add) (None, 28, 28, 64) 0 drop_connect_8[0][0] \n add_7[0][0] \n__________________________________________________________________________________________________\nconv2d_43 (Conv2D) (None, 28, 28, 384) 24576 add_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_32 (BatchNo (None, 28, 28, 384) 1536 conv2d_43[0][0] \n__________________________________________________________________________________________________\nswish_32 (Swish) (None, 28, 28, 384) 0 batch_normalization_32[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_12 (DepthwiseC (None, 28, 28, 384) 9600 swish_32[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_33 (BatchNo (None, 28, 28, 384) 1536 depthwise_conv2d_12[0][0] \n__________________________________________________________________________________________________\nswish_33 (Swish) (None, 28, 28, 384) 0 batch_normalization_33[0][0] \n__________________________________________________________________________________________________\nlambda_12 (Lambda) (None, 1, 1, 384) 0 swish_33[0][0] \n__________________________________________________________________________________________________\nconv2d_44 (Conv2D) (None, 1, 1, 16) 6160 lambda_12[0][0] \n__________________________________________________________________________________________________\nswish_34 (Swish) (None, 1, 1, 16) 0 conv2d_44[0][0] \n__________________________________________________________________________________________________\nconv2d_45 (Conv2D) (None, 1, 1, 384) 6528 swish_34[0][0] \n__________________________________________________________________________________________________\nactivation_12 (Activation) (None, 1, 1, 384) 0 conv2d_45[0][0] \n__________________________________________________________________________________________________\nmultiply_12 (Multiply) (None, 28, 28, 384) 0 activation_12[0][0] \n swish_33[0][0] \n__________________________________________________________________________________________________\nconv2d_46 (Conv2D) (None, 28, 28, 64) 24576 multiply_12[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_34 (BatchNo (None, 28, 28, 64) 256 conv2d_46[0][0] \n__________________________________________________________________________________________________\ndrop_connect_9 (DropConnect) (None, 28, 28, 64) 0 batch_normalization_34[0][0] \n__________________________________________________________________________________________________\nadd_9 (Add) (None, 28, 28, 64) 0 drop_connect_9[0][0] \n add_8[0][0] \n__________________________________________________________________________________________________\nconv2d_47 (Conv2D) (None, 28, 28, 384) 24576 add_9[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_35 (BatchNo (None, 28, 28, 384) 1536 conv2d_47[0][0] \n__________________________________________________________________________________________________\nswish_35 (Swish) (None, 28, 28, 384) 0 batch_normalization_35[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_13 (DepthwiseC (None, 28, 28, 384) 9600 swish_35[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_36 (BatchNo (None, 28, 28, 384) 1536 depthwise_conv2d_13[0][0] \n__________________________________________________________________________________________________\nswish_36 (Swish) (None, 28, 28, 384) 0 batch_normalization_36[0][0] \n__________________________________________________________________________________________________\nlambda_13 (Lambda) (None, 1, 1, 384) 0 swish_36[0][0] \n__________________________________________________________________________________________________\nconv2d_48 (Conv2D) (None, 1, 1, 16) 6160 lambda_13[0][0] \n__________________________________________________________________________________________________\nswish_37 (Swish) (None, 1, 1, 16) 0 conv2d_48[0][0] \n__________________________________________________________________________________________________\nconv2d_49 (Conv2D) (None, 1, 1, 384) 6528 swish_37[0][0] \n__________________________________________________________________________________________________\nactivation_13 (Activation) (None, 1, 1, 384) 0 conv2d_49[0][0] \n__________________________________________________________________________________________________\nmultiply_13 (Multiply) (None, 28, 28, 384) 0 activation_13[0][0] \n swish_36[0][0] \n__________________________________________________________________________________________________\nconv2d_50 (Conv2D) (None, 28, 28, 64) 24576 multiply_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_37 (BatchNo (None, 28, 28, 64) 256 conv2d_50[0][0] \n__________________________________________________________________________________________________\ndrop_connect_10 (DropConnect) (None, 28, 28, 64) 0 batch_normalization_37[0][0] \n__________________________________________________________________________________________________\nadd_10 (Add) (None, 28, 28, 64) 0 drop_connect_10[0][0] \n add_9[0][0] \n__________________________________________________________________________________________________\nconv2d_51 (Conv2D) (None, 28, 28, 384) 24576 add_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_38 (BatchNo (None, 28, 28, 384) 1536 conv2d_51[0][0] \n__________________________________________________________________________________________________\nswish_38 (Swish) (None, 28, 28, 384) 0 batch_normalization_38[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_14 (DepthwiseC (None, 14, 14, 384) 3456 swish_38[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_39 (BatchNo (None, 14, 14, 384) 1536 depthwise_conv2d_14[0][0] \n__________________________________________________________________________________________________\nswish_39 (Swish) (None, 14, 14, 384) 0 batch_normalization_39[0][0] \n__________________________________________________________________________________________________\nlambda_14 (Lambda) (None, 1, 1, 384) 0 swish_39[0][0] \n__________________________________________________________________________________________________\nconv2d_52 (Conv2D) (None, 1, 1, 16) 6160 lambda_14[0][0] \n__________________________________________________________________________________________________\nswish_40 (Swish) (None, 1, 1, 16) 0 conv2d_52[0][0] \n__________________________________________________________________________________________________\nconv2d_53 (Conv2D) (None, 1, 1, 384) 6528 swish_40[0][0] \n__________________________________________________________________________________________________\nactivation_14 (Activation) (None, 1, 1, 384) 0 conv2d_53[0][0] \n__________________________________________________________________________________________________\nmultiply_14 (Multiply) (None, 14, 14, 384) 0 activation_14[0][0] \n swish_39[0][0] \n__________________________________________________________________________________________________\nconv2d_54 (Conv2D) (None, 14, 14, 128) 49152 multiply_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_40 (BatchNo (None, 14, 14, 128) 512 conv2d_54[0][0] \n__________________________________________________________________________________________________\nconv2d_55 (Conv2D) (None, 14, 14, 768) 98304 batch_normalization_40[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_41 (BatchNo (None, 14, 14, 768) 3072 conv2d_55[0][0] \n__________________________________________________________________________________________________\nswish_41 (Swish) (None, 14, 14, 768) 0 batch_normalization_41[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_15 (DepthwiseC (None, 14, 14, 768) 6912 swish_41[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_42 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_15[0][0] \n__________________________________________________________________________________________________\nswish_42 (Swish) (None, 14, 14, 768) 0 batch_normalization_42[0][0] \n__________________________________________________________________________________________________\nlambda_15 (Lambda) (None, 1, 1, 768) 0 swish_42[0][0] \n__________________________________________________________________________________________________\nconv2d_56 (Conv2D) (None, 1, 1, 32) 24608 lambda_15[0][0] \n__________________________________________________________________________________________________\nswish_43 (Swish) (None, 1, 1, 32) 0 conv2d_56[0][0] \n__________________________________________________________________________________________________\nconv2d_57 (Conv2D) (None, 1, 1, 768) 25344 swish_43[0][0] \n__________________________________________________________________________________________________\nactivation_15 (Activation) (None, 1, 1, 768) 0 conv2d_57[0][0] \n__________________________________________________________________________________________________\nmultiply_15 (Multiply) (None, 14, 14, 768) 0 activation_15[0][0] \n swish_42[0][0] \n__________________________________________________________________________________________________\nconv2d_58 (Conv2D) (None, 14, 14, 128) 98304 multiply_15[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_43 (BatchNo (None, 14, 14, 128) 512 conv2d_58[0][0] \n__________________________________________________________________________________________________\ndrop_connect_11 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_43[0][0] \n__________________________________________________________________________________________________\nadd_11 (Add) (None, 14, 14, 128) 0 drop_connect_11[0][0] \n batch_normalization_40[0][0] \n__________________________________________________________________________________________________\nconv2d_59 (Conv2D) (None, 14, 14, 768) 98304 add_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_44 (BatchNo (None, 14, 14, 768) 3072 conv2d_59[0][0] \n__________________________________________________________________________________________________\nswish_44 (Swish) (None, 14, 14, 768) 0 batch_normalization_44[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_16 (DepthwiseC (None, 14, 14, 768) 6912 swish_44[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_45 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_16[0][0] \n__________________________________________________________________________________________________\nswish_45 (Swish) (None, 14, 14, 768) 0 batch_normalization_45[0][0] \n__________________________________________________________________________________________________\nlambda_16 (Lambda) (None, 1, 1, 768) 0 swish_45[0][0] \n__________________________________________________________________________________________________\nconv2d_60 (Conv2D) (None, 1, 1, 32) 24608 lambda_16[0][0] \n__________________________________________________________________________________________________\nswish_46 (Swish) (None, 1, 1, 32) 0 conv2d_60[0][0] \n__________________________________________________________________________________________________\nconv2d_61 (Conv2D) (None, 1, 1, 768) 25344 swish_46[0][0] \n__________________________________________________________________________________________________\nactivation_16 (Activation) (None, 1, 1, 768) 0 conv2d_61[0][0] \n__________________________________________________________________________________________________\nmultiply_16 (Multiply) (None, 14, 14, 768) 0 activation_16[0][0] \n swish_45[0][0] \n__________________________________________________________________________________________________\nconv2d_62 (Conv2D) (None, 14, 14, 128) 98304 multiply_16[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_46 (BatchNo (None, 14, 14, 128) 512 conv2d_62[0][0] \n__________________________________________________________________________________________________\ndrop_connect_12 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_46[0][0] \n__________________________________________________________________________________________________\nadd_12 (Add) (None, 14, 14, 128) 0 drop_connect_12[0][0] \n add_11[0][0] \n__________________________________________________________________________________________________\nconv2d_63 (Conv2D) (None, 14, 14, 768) 98304 add_12[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_47 (BatchNo (None, 14, 14, 768) 3072 conv2d_63[0][0] \n__________________________________________________________________________________________________\nswish_47 (Swish) (None, 14, 14, 768) 0 batch_normalization_47[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_17 (DepthwiseC (None, 14, 14, 768) 6912 swish_47[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_48 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_17[0][0] \n__________________________________________________________________________________________________\nswish_48 (Swish) (None, 14, 14, 768) 0 batch_normalization_48[0][0] \n__________________________________________________________________________________________________\nlambda_17 (Lambda) (None, 1, 1, 768) 0 swish_48[0][0] \n__________________________________________________________________________________________________\nconv2d_64 (Conv2D) (None, 1, 1, 32) 24608 lambda_17[0][0] \n__________________________________________________________________________________________________\nswish_49 (Swish) (None, 1, 1, 32) 0 conv2d_64[0][0] \n__________________________________________________________________________________________________\nconv2d_65 (Conv2D) (None, 1, 1, 768) 25344 swish_49[0][0] \n__________________________________________________________________________________________________\nactivation_17 (Activation) (None, 1, 1, 768) 0 conv2d_65[0][0] \n__________________________________________________________________________________________________\nmultiply_17 (Multiply) (None, 14, 14, 768) 0 activation_17[0][0] \n swish_48[0][0] \n__________________________________________________________________________________________________\nconv2d_66 (Conv2D) (None, 14, 14, 128) 98304 multiply_17[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_49 (BatchNo (None, 14, 14, 128) 512 conv2d_66[0][0] \n__________________________________________________________________________________________________\ndrop_connect_13 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_49[0][0] \n__________________________________________________________________________________________________\nadd_13 (Add) (None, 14, 14, 128) 0 drop_connect_13[0][0] \n add_12[0][0] \n__________________________________________________________________________________________________\nconv2d_67 (Conv2D) (None, 14, 14, 768) 98304 add_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_50 (BatchNo (None, 14, 14, 768) 3072 conv2d_67[0][0] \n__________________________________________________________________________________________________\nswish_50 (Swish) (None, 14, 14, 768) 0 batch_normalization_50[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_18 (DepthwiseC (None, 14, 14, 768) 6912 swish_50[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_51 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_18[0][0] \n__________________________________________________________________________________________________\nswish_51 (Swish) (None, 14, 14, 768) 0 batch_normalization_51[0][0] \n__________________________________________________________________________________________________\nlambda_18 (Lambda) (None, 1, 1, 768) 0 swish_51[0][0] \n__________________________________________________________________________________________________\nconv2d_68 (Conv2D) (None, 1, 1, 32) 24608 lambda_18[0][0] \n__________________________________________________________________________________________________\nswish_52 (Swish) (None, 1, 1, 32) 0 conv2d_68[0][0] \n__________________________________________________________________________________________________\nconv2d_69 (Conv2D) (None, 1, 1, 768) 25344 swish_52[0][0] \n__________________________________________________________________________________________________\nactivation_18 (Activation) (None, 1, 1, 768) 0 conv2d_69[0][0] \n__________________________________________________________________________________________________\nmultiply_18 (Multiply) (None, 14, 14, 768) 0 activation_18[0][0] \n swish_51[0][0] \n__________________________________________________________________________________________________\nconv2d_70 (Conv2D) (None, 14, 14, 128) 98304 multiply_18[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_52 (BatchNo (None, 14, 14, 128) 512 conv2d_70[0][0] \n__________________________________________________________________________________________________\ndrop_connect_14 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_52[0][0] \n__________________________________________________________________________________________________\nadd_14 (Add) (None, 14, 14, 128) 0 drop_connect_14[0][0] \n add_13[0][0] \n__________________________________________________________________________________________________\nconv2d_71 (Conv2D) (None, 14, 14, 768) 98304 add_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_53 (BatchNo (None, 14, 14, 768) 3072 conv2d_71[0][0] \n__________________________________________________________________________________________________\nswish_53 (Swish) (None, 14, 14, 768) 0 batch_normalization_53[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_19 (DepthwiseC (None, 14, 14, 768) 6912 swish_53[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_54 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_19[0][0] \n__________________________________________________________________________________________________\nswish_54 (Swish) (None, 14, 14, 768) 0 batch_normalization_54[0][0] \n__________________________________________________________________________________________________\nlambda_19 (Lambda) (None, 1, 1, 768) 0 swish_54[0][0] \n__________________________________________________________________________________________________\nconv2d_72 (Conv2D) (None, 1, 1, 32) 24608 lambda_19[0][0] \n__________________________________________________________________________________________________\nswish_55 (Swish) (None, 1, 1, 32) 0 conv2d_72[0][0] \n__________________________________________________________________________________________________\nconv2d_73 (Conv2D) (None, 1, 1, 768) 25344 swish_55[0][0] \n__________________________________________________________________________________________________\nactivation_19 (Activation) (None, 1, 1, 768) 0 conv2d_73[0][0] \n__________________________________________________________________________________________________\nmultiply_19 (Multiply) (None, 14, 14, 768) 0 activation_19[0][0] \n swish_54[0][0] \n__________________________________________________________________________________________________\nconv2d_74 (Conv2D) (None, 14, 14, 128) 98304 multiply_19[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_55 (BatchNo (None, 14, 14, 128) 512 conv2d_74[0][0] \n__________________________________________________________________________________________________\ndrop_connect_15 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_55[0][0] \n__________________________________________________________________________________________________\nadd_15 (Add) (None, 14, 14, 128) 0 drop_connect_15[0][0] \n add_14[0][0] \n__________________________________________________________________________________________________\nconv2d_75 (Conv2D) (None, 14, 14, 768) 98304 add_15[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_56 (BatchNo (None, 14, 14, 768) 3072 conv2d_75[0][0] \n__________________________________________________________________________________________________\nswish_56 (Swish) (None, 14, 14, 768) 0 batch_normalization_56[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_20 (DepthwiseC (None, 14, 14, 768) 6912 swish_56[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_57 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_20[0][0] \n__________________________________________________________________________________________________\nswish_57 (Swish) (None, 14, 14, 768) 0 batch_normalization_57[0][0] \n__________________________________________________________________________________________________\nlambda_20 (Lambda) (None, 1, 1, 768) 0 swish_57[0][0] \n__________________________________________________________________________________________________\nconv2d_76 (Conv2D) (None, 1, 1, 32) 24608 lambda_20[0][0] \n__________________________________________________________________________________________________\nswish_58 (Swish) (None, 1, 1, 32) 0 conv2d_76[0][0] \n__________________________________________________________________________________________________\nconv2d_77 (Conv2D) (None, 1, 1, 768) 25344 swish_58[0][0] \n__________________________________________________________________________________________________\nactivation_20 (Activation) (None, 1, 1, 768) 0 conv2d_77[0][0] \n__________________________________________________________________________________________________\nmultiply_20 (Multiply) (None, 14, 14, 768) 0 activation_20[0][0] \n swish_57[0][0] \n__________________________________________________________________________________________________\nconv2d_78 (Conv2D) (None, 14, 14, 128) 98304 multiply_20[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_58 (BatchNo (None, 14, 14, 128) 512 conv2d_78[0][0] \n__________________________________________________________________________________________________\ndrop_connect_16 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_58[0][0] \n__________________________________________________________________________________________________\nadd_16 (Add) (None, 14, 14, 128) 0 drop_connect_16[0][0] \n add_15[0][0] \n__________________________________________________________________________________________________\nconv2d_79 (Conv2D) (None, 14, 14, 768) 98304 add_16[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_59 (BatchNo (None, 14, 14, 768) 3072 conv2d_79[0][0] \n__________________________________________________________________________________________________\nswish_59 (Swish) (None, 14, 14, 768) 0 batch_normalization_59[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_21 (DepthwiseC (None, 14, 14, 768) 19200 swish_59[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_60 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_21[0][0] \n__________________________________________________________________________________________________\nswish_60 (Swish) (None, 14, 14, 768) 0 batch_normalization_60[0][0] \n__________________________________________________________________________________________________\nlambda_21 (Lambda) (None, 1, 1, 768) 0 swish_60[0][0] \n__________________________________________________________________________________________________\nconv2d_80 (Conv2D) (None, 1, 1, 32) 24608 lambda_21[0][0] \n__________________________________________________________________________________________________\nswish_61 (Swish) (None, 1, 1, 32) 0 conv2d_80[0][0] \n__________________________________________________________________________________________________\nconv2d_81 (Conv2D) (None, 1, 1, 768) 25344 swish_61[0][0] \n__________________________________________________________________________________________________\nactivation_21 (Activation) (None, 1, 1, 768) 0 conv2d_81[0][0] \n__________________________________________________________________________________________________\nmultiply_21 (Multiply) (None, 14, 14, 768) 0 activation_21[0][0] \n swish_60[0][0] \n__________________________________________________________________________________________________\nconv2d_82 (Conv2D) (None, 14, 14, 176) 135168 multiply_21[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_61 (BatchNo (None, 14, 14, 176) 704 conv2d_82[0][0] \n__________________________________________________________________________________________________\nconv2d_83 (Conv2D) (None, 14, 14, 1056) 185856 batch_normalization_61[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_62 (BatchNo (None, 14, 14, 1056) 4224 conv2d_83[0][0] \n__________________________________________________________________________________________________\nswish_62 (Swish) (None, 14, 14, 1056) 0 batch_normalization_62[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_22 (DepthwiseC (None, 14, 14, 1056) 26400 swish_62[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_63 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_22[0][0] \n__________________________________________________________________________________________________\nswish_63 (Swish) (None, 14, 14, 1056) 0 batch_normalization_63[0][0] \n__________________________________________________________________________________________________\nlambda_22 (Lambda) (None, 1, 1, 1056) 0 swish_63[0][0] \n__________________________________________________________________________________________________\nconv2d_84 (Conv2D) (None, 1, 1, 44) 46508 lambda_22[0][0] \n__________________________________________________________________________________________________\nswish_64 (Swish) (None, 1, 1, 44) 0 conv2d_84[0][0] \n__________________________________________________________________________________________________\nconv2d_85 (Conv2D) (None, 1, 1, 1056) 47520 swish_64[0][0] \n__________________________________________________________________________________________________\nactivation_22 (Activation) (None, 1, 1, 1056) 0 conv2d_85[0][0] \n__________________________________________________________________________________________________\nmultiply_22 (Multiply) (None, 14, 14, 1056) 0 activation_22[0][0] \n swish_63[0][0] \n__________________________________________________________________________________________________\nconv2d_86 (Conv2D) (None, 14, 14, 176) 185856 multiply_22[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_64 (BatchNo (None, 14, 14, 176) 704 conv2d_86[0][0] \n__________________________________________________________________________________________________\ndrop_connect_17 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_64[0][0] \n__________________________________________________________________________________________________\nadd_17 (Add) (None, 14, 14, 176) 0 drop_connect_17[0][0] \n batch_normalization_61[0][0] \n__________________________________________________________________________________________________\nconv2d_87 (Conv2D) (None, 14, 14, 1056) 185856 add_17[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_65 (BatchNo (None, 14, 14, 1056) 4224 conv2d_87[0][0] \n__________________________________________________________________________________________________\nswish_65 (Swish) (None, 14, 14, 1056) 0 batch_normalization_65[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_23 (DepthwiseC (None, 14, 14, 1056) 26400 swish_65[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_66 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_23[0][0] \n__________________________________________________________________________________________________\nswish_66 (Swish) (None, 14, 14, 1056) 0 batch_normalization_66[0][0] \n__________________________________________________________________________________________________\nlambda_23 (Lambda) (None, 1, 1, 1056) 0 swish_66[0][0] \n__________________________________________________________________________________________________\nconv2d_88 (Conv2D) (None, 1, 1, 44) 46508 lambda_23[0][0] \n__________________________________________________________________________________________________\nswish_67 (Swish) (None, 1, 1, 44) 0 conv2d_88[0][0] \n__________________________________________________________________________________________________\nconv2d_89 (Conv2D) (None, 1, 1, 1056) 47520 swish_67[0][0] \n__________________________________________________________________________________________________\nactivation_23 (Activation) (None, 1, 1, 1056) 0 conv2d_89[0][0] \n__________________________________________________________________________________________________\nmultiply_23 (Multiply) (None, 14, 14, 1056) 0 activation_23[0][0] \n swish_66[0][0] \n__________________________________________________________________________________________________\nconv2d_90 (Conv2D) (None, 14, 14, 176) 185856 multiply_23[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_67 (BatchNo (None, 14, 14, 176) 704 conv2d_90[0][0] \n__________________________________________________________________________________________________\ndrop_connect_18 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_67[0][0] \n__________________________________________________________________________________________________\nadd_18 (Add) (None, 14, 14, 176) 0 drop_connect_18[0][0] \n add_17[0][0] \n__________________________________________________________________________________________________\nconv2d_91 (Conv2D) (None, 14, 14, 1056) 185856 add_18[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_68 (BatchNo (None, 14, 14, 1056) 4224 conv2d_91[0][0] \n__________________________________________________________________________________________________\nswish_68 (Swish) (None, 14, 14, 1056) 0 batch_normalization_68[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_24 (DepthwiseC (None, 14, 14, 1056) 26400 swish_68[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_69 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_24[0][0] \n__________________________________________________________________________________________________\nswish_69 (Swish) (None, 14, 14, 1056) 0 batch_normalization_69[0][0] \n__________________________________________________________________________________________________\nlambda_24 (Lambda) (None, 1, 1, 1056) 0 swish_69[0][0] \n__________________________________________________________________________________________________\nconv2d_92 (Conv2D) (None, 1, 1, 44) 46508 lambda_24[0][0] \n__________________________________________________________________________________________________\nswish_70 (Swish) (None, 1, 1, 44) 0 conv2d_92[0][0] \n__________________________________________________________________________________________________\nconv2d_93 (Conv2D) (None, 1, 1, 1056) 47520 swish_70[0][0] \n__________________________________________________________________________________________________\nactivation_24 (Activation) (None, 1, 1, 1056) 0 conv2d_93[0][0] \n__________________________________________________________________________________________________\nmultiply_24 (Multiply) (None, 14, 14, 1056) 0 activation_24[0][0] \n swish_69[0][0] \n__________________________________________________________________________________________________\nconv2d_94 (Conv2D) (None, 14, 14, 176) 185856 multiply_24[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_70 (BatchNo (None, 14, 14, 176) 704 conv2d_94[0][0] \n__________________________________________________________________________________________________\ndrop_connect_19 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_70[0][0] \n__________________________________________________________________________________________________\nadd_19 (Add) (None, 14, 14, 176) 0 drop_connect_19[0][0] \n add_18[0][0] \n__________________________________________________________________________________________________\nconv2d_95 (Conv2D) (None, 14, 14, 1056) 185856 add_19[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_71 (BatchNo (None, 14, 14, 1056) 4224 conv2d_95[0][0] \n__________________________________________________________________________________________________\nswish_71 (Swish) (None, 14, 14, 1056) 0 batch_normalization_71[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_25 (DepthwiseC (None, 14, 14, 1056) 26400 swish_71[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_72 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_25[0][0] \n__________________________________________________________________________________________________\nswish_72 (Swish) (None, 14, 14, 1056) 0 batch_normalization_72[0][0] \n__________________________________________________________________________________________________\nlambda_25 (Lambda) (None, 1, 1, 1056) 0 swish_72[0][0] \n__________________________________________________________________________________________________\nconv2d_96 (Conv2D) (None, 1, 1, 44) 46508 lambda_25[0][0] \n__________________________________________________________________________________________________\nswish_73 (Swish) (None, 1, 1, 44) 0 conv2d_96[0][0] \n__________________________________________________________________________________________________\nconv2d_97 (Conv2D) (None, 1, 1, 1056) 47520 swish_73[0][0] \n__________________________________________________________________________________________________\nactivation_25 (Activation) (None, 1, 1, 1056) 0 conv2d_97[0][0] \n__________________________________________________________________________________________________\nmultiply_25 (Multiply) (None, 14, 14, 1056) 0 activation_25[0][0] \n swish_72[0][0] \n__________________________________________________________________________________________________\nconv2d_98 (Conv2D) (None, 14, 14, 176) 185856 multiply_25[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_73 (BatchNo (None, 14, 14, 176) 704 conv2d_98[0][0] \n__________________________________________________________________________________________________\ndrop_connect_20 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_73[0][0] \n__________________________________________________________________________________________________\nadd_20 (Add) (None, 14, 14, 176) 0 drop_connect_20[0][0] \n add_19[0][0] \n__________________________________________________________________________________________________\nconv2d_99 (Conv2D) (None, 14, 14, 1056) 185856 add_20[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_74 (BatchNo (None, 14, 14, 1056) 4224 conv2d_99[0][0] \n__________________________________________________________________________________________________\nswish_74 (Swish) (None, 14, 14, 1056) 0 batch_normalization_74[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_26 (DepthwiseC (None, 14, 14, 1056) 26400 swish_74[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_75 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_26[0][0] \n__________________________________________________________________________________________________\nswish_75 (Swish) (None, 14, 14, 1056) 0 batch_normalization_75[0][0] \n__________________________________________________________________________________________________\nlambda_26 (Lambda) (None, 1, 1, 1056) 0 swish_75[0][0] \n__________________________________________________________________________________________________\nconv2d_100 (Conv2D) (None, 1, 1, 44) 46508 lambda_26[0][0] \n__________________________________________________________________________________________________\nswish_76 (Swish) (None, 1, 1, 44) 0 conv2d_100[0][0] \n__________________________________________________________________________________________________\nconv2d_101 (Conv2D) (None, 1, 1, 1056) 47520 swish_76[0][0] \n__________________________________________________________________________________________________\nactivation_26 (Activation) (None, 1, 1, 1056) 0 conv2d_101[0][0] \n__________________________________________________________________________________________________\nmultiply_26 (Multiply) (None, 14, 14, 1056) 0 activation_26[0][0] \n swish_75[0][0] \n__________________________________________________________________________________________________\nconv2d_102 (Conv2D) (None, 14, 14, 176) 185856 multiply_26[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_76 (BatchNo (None, 14, 14, 176) 704 conv2d_102[0][0] \n__________________________________________________________________________________________________\ndrop_connect_21 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_76[0][0] \n__________________________________________________________________________________________________\nadd_21 (Add) (None, 14, 14, 176) 0 drop_connect_21[0][0] \n add_20[0][0] \n__________________________________________________________________________________________________\nconv2d_103 (Conv2D) (None, 14, 14, 1056) 185856 add_21[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_77 (BatchNo (None, 14, 14, 1056) 4224 conv2d_103[0][0] \n__________________________________________________________________________________________________\nswish_77 (Swish) (None, 14, 14, 1056) 0 batch_normalization_77[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_27 (DepthwiseC (None, 14, 14, 1056) 26400 swish_77[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_78 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_27[0][0] \n__________________________________________________________________________________________________\nswish_78 (Swish) (None, 14, 14, 1056) 0 batch_normalization_78[0][0] \n__________________________________________________________________________________________________\nlambda_27 (Lambda) (None, 1, 1, 1056) 0 swish_78[0][0] \n__________________________________________________________________________________________________\nconv2d_104 (Conv2D) (None, 1, 1, 44) 46508 lambda_27[0][0] \n__________________________________________________________________________________________________\nswish_79 (Swish) (None, 1, 1, 44) 0 conv2d_104[0][0] \n__________________________________________________________________________________________________\nconv2d_105 (Conv2D) (None, 1, 1, 1056) 47520 swish_79[0][0] \n__________________________________________________________________________________________________\nactivation_27 (Activation) (None, 1, 1, 1056) 0 conv2d_105[0][0] \n__________________________________________________________________________________________________\nmultiply_27 (Multiply) (None, 14, 14, 1056) 0 activation_27[0][0] \n swish_78[0][0] \n__________________________________________________________________________________________________\nconv2d_106 (Conv2D) (None, 14, 14, 176) 185856 multiply_27[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_79 (BatchNo (None, 14, 14, 176) 704 conv2d_106[0][0] \n__________________________________________________________________________________________________\ndrop_connect_22 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_79[0][0] \n__________________________________________________________________________________________________\nadd_22 (Add) (None, 14, 14, 176) 0 drop_connect_22[0][0] \n add_21[0][0] \n__________________________________________________________________________________________________\nconv2d_107 (Conv2D) (None, 14, 14, 1056) 185856 add_22[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_80 (BatchNo (None, 14, 14, 1056) 4224 conv2d_107[0][0] \n__________________________________________________________________________________________________\nswish_80 (Swish) (None, 14, 14, 1056) 0 batch_normalization_80[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_28 (DepthwiseC (None, 7, 7, 1056) 26400 swish_80[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_81 (BatchNo (None, 7, 7, 1056) 4224 depthwise_conv2d_28[0][0] \n__________________________________________________________________________________________________\nswish_81 (Swish) (None, 7, 7, 1056) 0 batch_normalization_81[0][0] \n__________________________________________________________________________________________________\nlambda_28 (Lambda) (None, 1, 1, 1056) 0 swish_81[0][0] \n__________________________________________________________________________________________________\nconv2d_108 (Conv2D) (None, 1, 1, 44) 46508 lambda_28[0][0] \n__________________________________________________________________________________________________\nswish_82 (Swish) (None, 1, 1, 44) 0 conv2d_108[0][0] \n__________________________________________________________________________________________________\nconv2d_109 (Conv2D) (None, 1, 1, 1056) 47520 swish_82[0][0] \n__________________________________________________________________________________________________\nactivation_28 (Activation) (None, 1, 1, 1056) 0 conv2d_109[0][0] \n__________________________________________________________________________________________________\nmultiply_28 (Multiply) (None, 7, 7, 1056) 0 activation_28[0][0] \n swish_81[0][0] \n__________________________________________________________________________________________________\nconv2d_110 (Conv2D) (None, 7, 7, 304) 321024 multiply_28[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_82 (BatchNo (None, 7, 7, 304) 1216 conv2d_110[0][0] \n__________________________________________________________________________________________________\nconv2d_111 (Conv2D) (None, 7, 7, 1824) 554496 batch_normalization_82[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_83 (BatchNo (None, 7, 7, 1824) 7296 conv2d_111[0][0] \n__________________________________________________________________________________________________\nswish_83 (Swish) (None, 7, 7, 1824) 0 batch_normalization_83[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_29 (DepthwiseC (None, 7, 7, 1824) 45600 swish_83[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_84 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_29[0][0] \n__________________________________________________________________________________________________\nswish_84 (Swish) (None, 7, 7, 1824) 0 batch_normalization_84[0][0] \n__________________________________________________________________________________________________\nlambda_29 (Lambda) (None, 1, 1, 1824) 0 swish_84[0][0] \n__________________________________________________________________________________________________\nconv2d_112 (Conv2D) (None, 1, 1, 76) 138700 lambda_29[0][0] \n__________________________________________________________________________________________________\nswish_85 (Swish) (None, 1, 1, 76) 0 conv2d_112[0][0] \n__________________________________________________________________________________________________\nconv2d_113 (Conv2D) (None, 1, 1, 1824) 140448 swish_85[0][0] \n__________________________________________________________________________________________________\nactivation_29 (Activation) (None, 1, 1, 1824) 0 conv2d_113[0][0] \n__________________________________________________________________________________________________\nmultiply_29 (Multiply) (None, 7, 7, 1824) 0 activation_29[0][0] \n swish_84[0][0] \n__________________________________________________________________________________________________\nconv2d_114 (Conv2D) (None, 7, 7, 304) 554496 multiply_29[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_85 (BatchNo (None, 7, 7, 304) 1216 conv2d_114[0][0] \n__________________________________________________________________________________________________\ndrop_connect_23 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_85[0][0] \n__________________________________________________________________________________________________\nadd_23 (Add) (None, 7, 7, 304) 0 drop_connect_23[0][0] \n batch_normalization_82[0][0] \n__________________________________________________________________________________________________\nconv2d_115 (Conv2D) (None, 7, 7, 1824) 554496 add_23[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_86 (BatchNo (None, 7, 7, 1824) 7296 conv2d_115[0][0] \n__________________________________________________________________________________________________\nswish_86 (Swish) (None, 7, 7, 1824) 0 batch_normalization_86[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_30 (DepthwiseC (None, 7, 7, 1824) 45600 swish_86[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_87 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_30[0][0] \n__________________________________________________________________________________________________\nswish_87 (Swish) (None, 7, 7, 1824) 0 batch_normalization_87[0][0] \n__________________________________________________________________________________________________\nlambda_30 (Lambda) (None, 1, 1, 1824) 0 swish_87[0][0] \n__________________________________________________________________________________________________\nconv2d_116 (Conv2D) (None, 1, 1, 76) 138700 lambda_30[0][0] \n__________________________________________________________________________________________________\nswish_88 (Swish) (None, 1, 1, 76) 0 conv2d_116[0][0] \n__________________________________________________________________________________________________\nconv2d_117 (Conv2D) (None, 1, 1, 1824) 140448 swish_88[0][0] \n__________________________________________________________________________________________________\nactivation_30 (Activation) (None, 1, 1, 1824) 0 conv2d_117[0][0] \n__________________________________________________________________________________________________\nmultiply_30 (Multiply) (None, 7, 7, 1824) 0 activation_30[0][0] \n swish_87[0][0] \n__________________________________________________________________________________________________\nconv2d_118 (Conv2D) (None, 7, 7, 304) 554496 multiply_30[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_88 (BatchNo (None, 7, 7, 304) 1216 conv2d_118[0][0] \n__________________________________________________________________________________________________\ndrop_connect_24 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_88[0][0] \n__________________________________________________________________________________________________\nadd_24 (Add) (None, 7, 7, 304) 0 drop_connect_24[0][0] \n add_23[0][0] \n__________________________________________________________________________________________________\nconv2d_119 (Conv2D) (None, 7, 7, 1824) 554496 add_24[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_89 (BatchNo (None, 7, 7, 1824) 7296 conv2d_119[0][0] \n__________________________________________________________________________________________________\nswish_89 (Swish) (None, 7, 7, 1824) 0 batch_normalization_89[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_31 (DepthwiseC (None, 7, 7, 1824) 45600 swish_89[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_90 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_31[0][0] \n__________________________________________________________________________________________________\nswish_90 (Swish) (None, 7, 7, 1824) 0 batch_normalization_90[0][0] \n__________________________________________________________________________________________________\nlambda_31 (Lambda) (None, 1, 1, 1824) 0 swish_90[0][0] \n__________________________________________________________________________________________________\nconv2d_120 (Conv2D) (None, 1, 1, 76) 138700 lambda_31[0][0] \n__________________________________________________________________________________________________\nswish_91 (Swish) (None, 1, 1, 76) 0 conv2d_120[0][0] \n__________________________________________________________________________________________________\nconv2d_121 (Conv2D) (None, 1, 1, 1824) 140448 swish_91[0][0] \n__________________________________________________________________________________________________\nactivation_31 (Activation) (None, 1, 1, 1824) 0 conv2d_121[0][0] \n__________________________________________________________________________________________________\nmultiply_31 (Multiply) (None, 7, 7, 1824) 0 activation_31[0][0] \n swish_90[0][0] \n__________________________________________________________________________________________________\nconv2d_122 (Conv2D) (None, 7, 7, 304) 554496 multiply_31[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_91 (BatchNo (None, 7, 7, 304) 1216 conv2d_122[0][0] \n__________________________________________________________________________________________________\ndrop_connect_25 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_91[0][0] \n__________________________________________________________________________________________________\nadd_25 (Add) (None, 7, 7, 304) 0 drop_connect_25[0][0] \n add_24[0][0] \n__________________________________________________________________________________________________\nconv2d_123 (Conv2D) (None, 7, 7, 1824) 554496 add_25[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_92 (BatchNo (None, 7, 7, 1824) 7296 conv2d_123[0][0] \n__________________________________________________________________________________________________\nswish_92 (Swish) (None, 7, 7, 1824) 0 batch_normalization_92[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_32 (DepthwiseC (None, 7, 7, 1824) 45600 swish_92[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_93 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_32[0][0] \n__________________________________________________________________________________________________\nswish_93 (Swish) (None, 7, 7, 1824) 0 batch_normalization_93[0][0] \n__________________________________________________________________________________________________\nlambda_32 (Lambda) (None, 1, 1, 1824) 0 swish_93[0][0] \n__________________________________________________________________________________________________\nconv2d_124 (Conv2D) (None, 1, 1, 76) 138700 lambda_32[0][0] \n__________________________________________________________________________________________________\nswish_94 (Swish) (None, 1, 1, 76) 0 conv2d_124[0][0] \n__________________________________________________________________________________________________\nconv2d_125 (Conv2D) (None, 1, 1, 1824) 140448 swish_94[0][0] \n__________________________________________________________________________________________________\nactivation_32 (Activation) (None, 1, 1, 1824) 0 conv2d_125[0][0] \n__________________________________________________________________________________________________\nmultiply_32 (Multiply) (None, 7, 7, 1824) 0 activation_32[0][0] \n swish_93[0][0] \n__________________________________________________________________________________________________\nconv2d_126 (Conv2D) (None, 7, 7, 304) 554496 multiply_32[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_94 (BatchNo (None, 7, 7, 304) 1216 conv2d_126[0][0] \n__________________________________________________________________________________________________\ndrop_connect_26 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_94[0][0] \n__________________________________________________________________________________________________\nadd_26 (Add) (None, 7, 7, 304) 0 drop_connect_26[0][0] \n add_25[0][0] \n__________________________________________________________________________________________________\nconv2d_127 (Conv2D) (None, 7, 7, 1824) 554496 add_26[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_95 (BatchNo (None, 7, 7, 1824) 7296 conv2d_127[0][0] \n__________________________________________________________________________________________________\nswish_95 (Swish) (None, 7, 7, 1824) 0 batch_normalization_95[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_33 (DepthwiseC (None, 7, 7, 1824) 45600 swish_95[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_96 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_33[0][0] \n__________________________________________________________________________________________________\nswish_96 (Swish) (None, 7, 7, 1824) 0 batch_normalization_96[0][0] \n__________________________________________________________________________________________________\nlambda_33 (Lambda) (None, 1, 1, 1824) 0 swish_96[0][0] \n__________________________________________________________________________________________________\nconv2d_128 (Conv2D) (None, 1, 1, 76) 138700 lambda_33[0][0] \n__________________________________________________________________________________________________\nswish_97 (Swish) (None, 1, 1, 76) 0 conv2d_128[0][0] \n__________________________________________________________________________________________________\nconv2d_129 (Conv2D) (None, 1, 1, 1824) 140448 swish_97[0][0] \n__________________________________________________________________________________________________\nactivation_33 (Activation) (None, 1, 1, 1824) 0 conv2d_129[0][0] \n__________________________________________________________________________________________________\nmultiply_33 (Multiply) (None, 7, 7, 1824) 0 activation_33[0][0] \n swish_96[0][0] \n__________________________________________________________________________________________________\nconv2d_130 (Conv2D) (None, 7, 7, 304) 554496 multiply_33[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_97 (BatchNo (None, 7, 7, 304) 1216 conv2d_130[0][0] \n__________________________________________________________________________________________________\ndrop_connect_27 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_97[0][0] \n__________________________________________________________________________________________________\nadd_27 (Add) (None, 7, 7, 304) 0 drop_connect_27[0][0] \n add_26[0][0] \n__________________________________________________________________________________________________\nconv2d_131 (Conv2D) (None, 7, 7, 1824) 554496 add_27[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_98 (BatchNo (None, 7, 7, 1824) 7296 conv2d_131[0][0] \n__________________________________________________________________________________________________\nswish_98 (Swish) (None, 7, 7, 1824) 0 batch_normalization_98[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_34 (DepthwiseC (None, 7, 7, 1824) 45600 swish_98[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_99 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_34[0][0] \n__________________________________________________________________________________________________\nswish_99 (Swish) (None, 7, 7, 1824) 0 batch_normalization_99[0][0] \n__________________________________________________________________________________________________\nlambda_34 (Lambda) (None, 1, 1, 1824) 0 swish_99[0][0] \n__________________________________________________________________________________________________\nconv2d_132 (Conv2D) (None, 1, 1, 76) 138700 lambda_34[0][0] \n__________________________________________________________________________________________________\nswish_100 (Swish) (None, 1, 1, 76) 0 conv2d_132[0][0] \n__________________________________________________________________________________________________\nconv2d_133 (Conv2D) (None, 1, 1, 1824) 140448 swish_100[0][0] \n__________________________________________________________________________________________________\nactivation_34 (Activation) (None, 1, 1, 1824) 0 conv2d_133[0][0] \n__________________________________________________________________________________________________\nmultiply_34 (Multiply) (None, 7, 7, 1824) 0 activation_34[0][0] \n swish_99[0][0] \n__________________________________________________________________________________________________\nconv2d_134 (Conv2D) (None, 7, 7, 304) 554496 multiply_34[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_100 (BatchN (None, 7, 7, 304) 1216 conv2d_134[0][0] \n__________________________________________________________________________________________________\ndrop_connect_28 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_100[0][0] \n__________________________________________________________________________________________________\nadd_28 (Add) (None, 7, 7, 304) 0 drop_connect_28[0][0] \n add_27[0][0] \n__________________________________________________________________________________________________\nconv2d_135 (Conv2D) (None, 7, 7, 1824) 554496 add_28[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_101 (BatchN (None, 7, 7, 1824) 7296 conv2d_135[0][0] \n__________________________________________________________________________________________________\nswish_101 (Swish) (None, 7, 7, 1824) 0 batch_normalization_101[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_35 (DepthwiseC (None, 7, 7, 1824) 45600 swish_101[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_102 (BatchN (None, 7, 7, 1824) 7296 depthwise_conv2d_35[0][0] \n__________________________________________________________________________________________________\nswish_102 (Swish) (None, 7, 7, 1824) 0 batch_normalization_102[0][0] \n__________________________________________________________________________________________________\nlambda_35 (Lambda) (None, 1, 1, 1824) 0 swish_102[0][0] \n__________________________________________________________________________________________________\nconv2d_136 (Conv2D) (None, 1, 1, 76) 138700 lambda_35[0][0] \n__________________________________________________________________________________________________\nswish_103 (Swish) (None, 1, 1, 76) 0 conv2d_136[0][0] \n__________________________________________________________________________________________________\nconv2d_137 (Conv2D) (None, 1, 1, 1824) 140448 swish_103[0][0] \n__________________________________________________________________________________________________\nactivation_35 (Activation) (None, 1, 1, 1824) 0 conv2d_137[0][0] \n__________________________________________________________________________________________________\nmultiply_35 (Multiply) (None, 7, 7, 1824) 0 activation_35[0][0] \n swish_102[0][0] \n__________________________________________________________________________________________________\nconv2d_138 (Conv2D) (None, 7, 7, 304) 554496 multiply_35[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_103 (BatchN (None, 7, 7, 304) 1216 conv2d_138[0][0] \n__________________________________________________________________________________________________\ndrop_connect_29 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_103[0][0] \n__________________________________________________________________________________________________\nadd_29 (Add) (None, 7, 7, 304) 0 drop_connect_29[0][0] \n add_28[0][0] \n__________________________________________________________________________________________________\nconv2d_139 (Conv2D) (None, 7, 7, 1824) 554496 add_29[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_104 (BatchN (None, 7, 7, 1824) 7296 conv2d_139[0][0] \n__________________________________________________________________________________________________\nswish_104 (Swish) (None, 7, 7, 1824) 0 batch_normalization_104[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_36 (DepthwiseC (None, 7, 7, 1824) 45600 swish_104[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_105 (BatchN (None, 7, 7, 1824) 7296 depthwise_conv2d_36[0][0] \n__________________________________________________________________________________________________\nswish_105 (Swish) (None, 7, 7, 1824) 0 batch_normalization_105[0][0] \n__________________________________________________________________________________________________\nlambda_36 (Lambda) (None, 1, 1, 1824) 0 swish_105[0][0] \n__________________________________________________________________________________________________\nconv2d_140 (Conv2D) (None, 1, 1, 76) 138700 lambda_36[0][0] \n__________________________________________________________________________________________________\nswish_106 (Swish) (None, 1, 1, 76) 0 conv2d_140[0][0] \n__________________________________________________________________________________________________\nconv2d_141 (Conv2D) (None, 1, 1, 1824) 140448 swish_106[0][0] \n__________________________________________________________________________________________________\nactivation_36 (Activation) (None, 1, 1, 1824) 0 conv2d_141[0][0] \n__________________________________________________________________________________________________\nmultiply_36 (Multiply) (None, 7, 7, 1824) 0 activation_36[0][0] \n swish_105[0][0] \n__________________________________________________________________________________________________\nconv2d_142 (Conv2D) (None, 7, 7, 304) 554496 multiply_36[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_106 (BatchN (None, 7, 7, 304) 1216 conv2d_142[0][0] \n__________________________________________________________________________________________________\ndrop_connect_30 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_106[0][0] \n__________________________________________________________________________________________________\nadd_30 (Add) (None, 7, 7, 304) 0 drop_connect_30[0][0] \n add_29[0][0] \n__________________________________________________________________________________________________\nconv2d_143 (Conv2D) (None, 7, 7, 1824) 554496 add_30[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_107 (BatchN (None, 7, 7, 1824) 7296 conv2d_143[0][0] \n__________________________________________________________________________________________________\nswish_107 (Swish) (None, 7, 7, 1824) 0 batch_normalization_107[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_37 (DepthwiseC (None, 7, 7, 1824) 16416 swish_107[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_108 (BatchN (None, 7, 7, 1824) 7296 depthwise_conv2d_37[0][0] \n__________________________________________________________________________________________________\nswish_108 (Swish) (None, 7, 7, 1824) 0 batch_normalization_108[0][0] \n__________________________________________________________________________________________________\nlambda_37 (Lambda) (None, 1, 1, 1824) 0 swish_108[0][0] \n__________________________________________________________________________________________________\nconv2d_144 (Conv2D) (None, 1, 1, 76) 138700 lambda_37[0][0] \n__________________________________________________________________________________________________\nswish_109 (Swish) (None, 1, 1, 76) 0 conv2d_144[0][0] \n__________________________________________________________________________________________________\nconv2d_145 (Conv2D) (None, 1, 1, 1824) 140448 swish_109[0][0] \n__________________________________________________________________________________________________\nactivation_37 (Activation) (None, 1, 1, 1824) 0 conv2d_145[0][0] \n__________________________________________________________________________________________________\nmultiply_37 (Multiply) (None, 7, 7, 1824) 0 activation_37[0][0] \n swish_108[0][0] \n__________________________________________________________________________________________________\nconv2d_146 (Conv2D) (None, 7, 7, 512) 933888 multiply_37[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_109 (BatchN (None, 7, 7, 512) 2048 conv2d_146[0][0] \n__________________________________________________________________________________________________\nconv2d_147 (Conv2D) (None, 7, 7, 3072) 1572864 batch_normalization_109[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_110 (BatchN (None, 7, 7, 3072) 12288 conv2d_147[0][0] \n__________________________________________________________________________________________________\nswish_110 (Swish) (None, 7, 7, 3072) 0 batch_normalization_110[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_38 (DepthwiseC (None, 7, 7, 3072) 27648 swish_110[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_111 (BatchN (None, 7, 7, 3072) 12288 depthwise_conv2d_38[0][0] \n__________________________________________________________________________________________________\nswish_111 (Swish) (None, 7, 7, 3072) 0 batch_normalization_111[0][0] \n__________________________________________________________________________________________________\nlambda_38 (Lambda) (None, 1, 1, 3072) 0 swish_111[0][0] \n__________________________________________________________________________________________________\nconv2d_148 (Conv2D) (None, 1, 1, 128) 393344 lambda_38[0][0] \n__________________________________________________________________________________________________\nswish_112 (Swish) (None, 1, 1, 128) 0 conv2d_148[0][0] \n__________________________________________________________________________________________________\nconv2d_149 (Conv2D) (None, 1, 1, 3072) 396288 swish_112[0][0] \n__________________________________________________________________________________________________\nactivation_38 (Activation) (None, 1, 1, 3072) 0 conv2d_149[0][0] \n__________________________________________________________________________________________________\nmultiply_38 (Multiply) (None, 7, 7, 3072) 0 activation_38[0][0] \n swish_111[0][0] \n__________________________________________________________________________________________________\nconv2d_150 (Conv2D) (None, 7, 7, 512) 1572864 multiply_38[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_112 (BatchN (None, 7, 7, 512) 2048 conv2d_150[0][0] \n__________________________________________________________________________________________________\ndrop_connect_31 (DropConnect) (None, 7, 7, 512) 0 batch_normalization_112[0][0] \n__________________________________________________________________________________________________\nadd_31 (Add) (None, 7, 7, 512) 0 drop_connect_31[0][0] \n batch_normalization_109[0][0] \n__________________________________________________________________________________________________\nconv2d_151 (Conv2D) (None, 7, 7, 3072) 1572864 add_31[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_113 (BatchN (None, 7, 7, 3072) 12288 conv2d_151[0][0] \n__________________________________________________________________________________________________\nswish_113 (Swish) (None, 7, 7, 3072) 0 batch_normalization_113[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_39 (DepthwiseC (None, 7, 7, 3072) 27648 swish_113[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_114 (BatchN (None, 7, 7, 3072) 12288 depthwise_conv2d_39[0][0] \n__________________________________________________________________________________________________\nswish_114 (Swish) (None, 7, 7, 3072) 0 batch_normalization_114[0][0] \n__________________________________________________________________________________________________\nlambda_39 (Lambda) (None, 1, 1, 3072) 0 swish_114[0][0] \n__________________________________________________________________________________________________\nconv2d_152 (Conv2D) (None, 1, 1, 128) 393344 lambda_39[0][0] \n__________________________________________________________________________________________________\nswish_115 (Swish) (None, 1, 1, 128) 0 conv2d_152[0][0] \n__________________________________________________________________________________________________\nconv2d_153 (Conv2D) (None, 1, 1, 3072) 396288 swish_115[0][0] \n__________________________________________________________________________________________________\nactivation_39 (Activation) (None, 1, 1, 3072) 0 conv2d_153[0][0] \n__________________________________________________________________________________________________\nmultiply_39 (Multiply) (None, 7, 7, 3072) 0 activation_39[0][0] \n swish_114[0][0] \n__________________________________________________________________________________________________\nconv2d_154 (Conv2D) (None, 7, 7, 512) 1572864 multiply_39[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_115 (BatchN (None, 7, 7, 512) 2048 conv2d_154[0][0] \n__________________________________________________________________________________________________\ndrop_connect_32 (DropConnect) (None, 7, 7, 512) 0 batch_normalization_115[0][0] \n__________________________________________________________________________________________________\nadd_32 (Add) (None, 7, 7, 512) 0 drop_connect_32[0][0] \n add_31[0][0] \n__________________________________________________________________________________________________\nconv2d_155 (Conv2D) (None, 7, 7, 2048) 1048576 add_32[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_116 (BatchN (None, 7, 7, 2048) 8192 conv2d_155[0][0] \n__________________________________________________________________________________________________\nswish_116 (Swish) (None, 7, 7, 2048) 0 batch_normalization_116[0][0] \n__________________________________________________________________________________________________\nglobal_average_pooling2d_1 (Glo (None, 2048) 0 swish_116[0][0] \n__________________________________________________________________________________________________\nfinal_output (Dense) (None, 1) 2049 global_average_pooling2d_1[0][0] \n==================================================================================================\nTotal params: 28,515,569\nTrainable params: 2,049\nNon-trainable params: 28,513,520\n__________________________________________________________________________________________________\n"
],
[
"STEP_SIZE_TRAIN = train_generator.n//train_generator.batch_size\nSTEP_SIZE_VALID = valid_generator.n//valid_generator.batch_size\n\nhistory_warmup = model.fit_generator(generator=train_generator,\n steps_per_epoch=STEP_SIZE_TRAIN,\n validation_data=valid_generator,\n validation_steps=STEP_SIZE_VALID,\n epochs=WARMUP_EPOCHS,\n callbacks=callback_list,\n verbose=2).history",
"Epoch 1/5\n - 239s - loss: 1.7648 - acc: 0.3112 - val_loss: 1.6629 - val_acc: 0.2001\nEpoch 2/5\n - 228s - loss: 1.2572 - acc: 0.2916 - val_loss: 2.1284 - val_acc: 0.3302\nEpoch 3/5\n - 229s - loss: 1.2323 - acc: 0.3035 - val_loss: 3.2056 - val_acc: 0.3213\nEpoch 4/5\n - 229s - loss: 1.2271 - acc: 0.3029 - val_loss: 3.3689 - val_acc: 0.2768\nEpoch 5/5\n - 228s - loss: 1.1613 - acc: 0.3019 - val_loss: 1.8922 - val_acc: 0.3041\n"
]
],
[
[
"# Fine-tune the complete model",
"_____no_output_____"
]
],
[
[
"for layer in model.layers:\n layer.trainable = True\n\nes = EarlyStopping(monitor='val_loss', mode='min', patience=ES_PATIENCE, restore_best_weights=True, verbose=1)\ncosine_lr_2nd = WarmUpCosineDecayScheduler(learning_rate_base=LEARNING_RATE,\n total_steps=TOTAL_STEPS_2nd,\n warmup_learning_rate=0.0,\n warmup_steps=WARMUP_STEPS_2nd,\n hold_base_rate_steps=(3 * STEP_SIZE))\n\ncallback_list = [es, cosine_lr_2nd]\noptimizer = optimizers.Adam(lr=LEARNING_RATE)\nmodel.compile(optimizer=optimizer, loss='mean_squared_error', metrics=metric_list)\nmodel.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) (None, 224, 224, 3) 0 \n__________________________________________________________________________________________________\nconv2d_1 (Conv2D) (None, 112, 112, 48) 1296 input_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_1 (BatchNor (None, 112, 112, 48) 192 conv2d_1[0][0] \n__________________________________________________________________________________________________\nswish_1 (Swish) (None, 112, 112, 48) 0 batch_normalization_1[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_1 (DepthwiseCo (None, 112, 112, 48) 432 swish_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_2 (BatchNor (None, 112, 112, 48) 192 depthwise_conv2d_1[0][0] \n__________________________________________________________________________________________________\nswish_2 (Swish) (None, 112, 112, 48) 0 batch_normalization_2[0][0] \n__________________________________________________________________________________________________\nlambda_1 (Lambda) (None, 1, 1, 48) 0 swish_2[0][0] \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 1, 1, 12) 588 lambda_1[0][0] \n__________________________________________________________________________________________________\nswish_3 (Swish) (None, 1, 1, 12) 0 conv2d_2[0][0] \n__________________________________________________________________________________________________\nconv2d_3 (Conv2D) (None, 1, 1, 48) 624 swish_3[0][0] \n__________________________________________________________________________________________________\nactivation_1 (Activation) (None, 1, 1, 48) 0 conv2d_3[0][0] \n__________________________________________________________________________________________________\nmultiply_1 (Multiply) (None, 112, 112, 48) 0 activation_1[0][0] \n swish_2[0][0] \n__________________________________________________________________________________________________\nconv2d_4 (Conv2D) (None, 112, 112, 24) 1152 multiply_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_3 (BatchNor (None, 112, 112, 24) 96 conv2d_4[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_2 (DepthwiseCo (None, 112, 112, 24) 216 batch_normalization_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_4 (BatchNor (None, 112, 112, 24) 96 depthwise_conv2d_2[0][0] \n__________________________________________________________________________________________________\nswish_4 (Swish) (None, 112, 112, 24) 0 batch_normalization_4[0][0] \n__________________________________________________________________________________________________\nlambda_2 (Lambda) (None, 1, 1, 24) 0 swish_4[0][0] \n__________________________________________________________________________________________________\nconv2d_5 (Conv2D) (None, 1, 1, 6) 150 lambda_2[0][0] \n__________________________________________________________________________________________________\nswish_5 (Swish) (None, 1, 1, 6) 0 conv2d_5[0][0] \n__________________________________________________________________________________________________\nconv2d_6 (Conv2D) (None, 1, 1, 24) 168 swish_5[0][0] \n__________________________________________________________________________________________________\nactivation_2 (Activation) (None, 1, 1, 24) 0 conv2d_6[0][0] \n__________________________________________________________________________________________________\nmultiply_2 (Multiply) (None, 112, 112, 24) 0 activation_2[0][0] \n swish_4[0][0] \n__________________________________________________________________________________________________\nconv2d_7 (Conv2D) (None, 112, 112, 24) 576 multiply_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_5 (BatchNor (None, 112, 112, 24) 96 conv2d_7[0][0] \n__________________________________________________________________________________________________\ndrop_connect_1 (DropConnect) (None, 112, 112, 24) 0 batch_normalization_5[0][0] \n__________________________________________________________________________________________________\nadd_1 (Add) (None, 112, 112, 24) 0 drop_connect_1[0][0] \n batch_normalization_3[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_3 (DepthwiseCo (None, 112, 112, 24) 216 add_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_6 (BatchNor (None, 112, 112, 24) 96 depthwise_conv2d_3[0][0] \n__________________________________________________________________________________________________\nswish_6 (Swish) (None, 112, 112, 24) 0 batch_normalization_6[0][0] \n__________________________________________________________________________________________________\nlambda_3 (Lambda) (None, 1, 1, 24) 0 swish_6[0][0] \n__________________________________________________________________________________________________\nconv2d_8 (Conv2D) (None, 1, 1, 6) 150 lambda_3[0][0] \n__________________________________________________________________________________________________\nswish_7 (Swish) (None, 1, 1, 6) 0 conv2d_8[0][0] \n__________________________________________________________________________________________________\nconv2d_9 (Conv2D) (None, 1, 1, 24) 168 swish_7[0][0] \n__________________________________________________________________________________________________\nactivation_3 (Activation) (None, 1, 1, 24) 0 conv2d_9[0][0] \n__________________________________________________________________________________________________\nmultiply_3 (Multiply) (None, 112, 112, 24) 0 activation_3[0][0] \n swish_6[0][0] \n__________________________________________________________________________________________________\nconv2d_10 (Conv2D) (None, 112, 112, 24) 576 multiply_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_7 (BatchNor (None, 112, 112, 24) 96 conv2d_10[0][0] \n__________________________________________________________________________________________________\ndrop_connect_2 (DropConnect) (None, 112, 112, 24) 0 batch_normalization_7[0][0] \n__________________________________________________________________________________________________\nadd_2 (Add) (None, 112, 112, 24) 0 drop_connect_2[0][0] \n add_1[0][0] \n__________________________________________________________________________________________________\nconv2d_11 (Conv2D) (None, 112, 112, 144 3456 add_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_8 (BatchNor (None, 112, 112, 144 576 conv2d_11[0][0] \n__________________________________________________________________________________________________\nswish_8 (Swish) (None, 112, 112, 144 0 batch_normalization_8[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_4 (DepthwiseCo (None, 56, 56, 144) 1296 swish_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_9 (BatchNor (None, 56, 56, 144) 576 depthwise_conv2d_4[0][0] \n__________________________________________________________________________________________________\nswish_9 (Swish) (None, 56, 56, 144) 0 batch_normalization_9[0][0] \n__________________________________________________________________________________________________\nlambda_4 (Lambda) (None, 1, 1, 144) 0 swish_9[0][0] \n__________________________________________________________________________________________________\nconv2d_12 (Conv2D) (None, 1, 1, 6) 870 lambda_4[0][0] \n__________________________________________________________________________________________________\nswish_10 (Swish) (None, 1, 1, 6) 0 conv2d_12[0][0] \n__________________________________________________________________________________________________\nconv2d_13 (Conv2D) (None, 1, 1, 144) 1008 swish_10[0][0] \n__________________________________________________________________________________________________\nactivation_4 (Activation) (None, 1, 1, 144) 0 conv2d_13[0][0] \n__________________________________________________________________________________________________\nmultiply_4 (Multiply) (None, 56, 56, 144) 0 activation_4[0][0] \n swish_9[0][0] \n__________________________________________________________________________________________________\nconv2d_14 (Conv2D) (None, 56, 56, 40) 5760 multiply_4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_10 (BatchNo (None, 56, 56, 40) 160 conv2d_14[0][0] \n__________________________________________________________________________________________________\nconv2d_15 (Conv2D) (None, 56, 56, 240) 9600 batch_normalization_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_11 (BatchNo (None, 56, 56, 240) 960 conv2d_15[0][0] \n__________________________________________________________________________________________________\nswish_11 (Swish) (None, 56, 56, 240) 0 batch_normalization_11[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_5 (DepthwiseCo (None, 56, 56, 240) 2160 swish_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_12 (BatchNo (None, 56, 56, 240) 960 depthwise_conv2d_5[0][0] \n__________________________________________________________________________________________________\nswish_12 (Swish) (None, 56, 56, 240) 0 batch_normalization_12[0][0] \n__________________________________________________________________________________________________\nlambda_5 (Lambda) (None, 1, 1, 240) 0 swish_12[0][0] \n__________________________________________________________________________________________________\nconv2d_16 (Conv2D) (None, 1, 1, 10) 2410 lambda_5[0][0] \n__________________________________________________________________________________________________\nswish_13 (Swish) (None, 1, 1, 10) 0 conv2d_16[0][0] \n__________________________________________________________________________________________________\nconv2d_17 (Conv2D) (None, 1, 1, 240) 2640 swish_13[0][0] \n__________________________________________________________________________________________________\nactivation_5 (Activation) (None, 1, 1, 240) 0 conv2d_17[0][0] \n__________________________________________________________________________________________________\nmultiply_5 (Multiply) (None, 56, 56, 240) 0 activation_5[0][0] \n swish_12[0][0] \n__________________________________________________________________________________________________\nconv2d_18 (Conv2D) (None, 56, 56, 40) 9600 multiply_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_13 (BatchNo (None, 56, 56, 40) 160 conv2d_18[0][0] \n__________________________________________________________________________________________________\ndrop_connect_3 (DropConnect) (None, 56, 56, 40) 0 batch_normalization_13[0][0] \n__________________________________________________________________________________________________\nadd_3 (Add) (None, 56, 56, 40) 0 drop_connect_3[0][0] \n batch_normalization_10[0][0] \n__________________________________________________________________________________________________\nconv2d_19 (Conv2D) (None, 56, 56, 240) 9600 add_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_14 (BatchNo (None, 56, 56, 240) 960 conv2d_19[0][0] \n__________________________________________________________________________________________________\nswish_14 (Swish) (None, 56, 56, 240) 0 batch_normalization_14[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_6 (DepthwiseCo (None, 56, 56, 240) 2160 swish_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_15 (BatchNo (None, 56, 56, 240) 960 depthwise_conv2d_6[0][0] \n__________________________________________________________________________________________________\nswish_15 (Swish) (None, 56, 56, 240) 0 batch_normalization_15[0][0] \n__________________________________________________________________________________________________\nlambda_6 (Lambda) (None, 1, 1, 240) 0 swish_15[0][0] \n__________________________________________________________________________________________________\nconv2d_20 (Conv2D) (None, 1, 1, 10) 2410 lambda_6[0][0] \n__________________________________________________________________________________________________\nswish_16 (Swish) (None, 1, 1, 10) 0 conv2d_20[0][0] \n__________________________________________________________________________________________________\nconv2d_21 (Conv2D) (None, 1, 1, 240) 2640 swish_16[0][0] \n__________________________________________________________________________________________________\nactivation_6 (Activation) (None, 1, 1, 240) 0 conv2d_21[0][0] \n__________________________________________________________________________________________________\nmultiply_6 (Multiply) (None, 56, 56, 240) 0 activation_6[0][0] \n swish_15[0][0] \n__________________________________________________________________________________________________\nconv2d_22 (Conv2D) (None, 56, 56, 40) 9600 multiply_6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_16 (BatchNo (None, 56, 56, 40) 160 conv2d_22[0][0] \n__________________________________________________________________________________________________\ndrop_connect_4 (DropConnect) (None, 56, 56, 40) 0 batch_normalization_16[0][0] \n__________________________________________________________________________________________________\nadd_4 (Add) (None, 56, 56, 40) 0 drop_connect_4[0][0] \n add_3[0][0] \n__________________________________________________________________________________________________\nconv2d_23 (Conv2D) (None, 56, 56, 240) 9600 add_4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_17 (BatchNo (None, 56, 56, 240) 960 conv2d_23[0][0] \n__________________________________________________________________________________________________\nswish_17 (Swish) (None, 56, 56, 240) 0 batch_normalization_17[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_7 (DepthwiseCo (None, 56, 56, 240) 2160 swish_17[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_18 (BatchNo (None, 56, 56, 240) 960 depthwise_conv2d_7[0][0] \n__________________________________________________________________________________________________\nswish_18 (Swish) (None, 56, 56, 240) 0 batch_normalization_18[0][0] \n__________________________________________________________________________________________________\nlambda_7 (Lambda) (None, 1, 1, 240) 0 swish_18[0][0] \n__________________________________________________________________________________________________\nconv2d_24 (Conv2D) (None, 1, 1, 10) 2410 lambda_7[0][0] \n__________________________________________________________________________________________________\nswish_19 (Swish) (None, 1, 1, 10) 0 conv2d_24[0][0] \n__________________________________________________________________________________________________\nconv2d_25 (Conv2D) (None, 1, 1, 240) 2640 swish_19[0][0] \n__________________________________________________________________________________________________\nactivation_7 (Activation) (None, 1, 1, 240) 0 conv2d_25[0][0] \n__________________________________________________________________________________________________\nmultiply_7 (Multiply) (None, 56, 56, 240) 0 activation_7[0][0] \n swish_18[0][0] \n__________________________________________________________________________________________________\nconv2d_26 (Conv2D) (None, 56, 56, 40) 9600 multiply_7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_19 (BatchNo (None, 56, 56, 40) 160 conv2d_26[0][0] \n__________________________________________________________________________________________________\ndrop_connect_5 (DropConnect) (None, 56, 56, 40) 0 batch_normalization_19[0][0] \n__________________________________________________________________________________________________\nadd_5 (Add) (None, 56, 56, 40) 0 drop_connect_5[0][0] \n add_4[0][0] \n__________________________________________________________________________________________________\nconv2d_27 (Conv2D) (None, 56, 56, 240) 9600 add_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_20 (BatchNo (None, 56, 56, 240) 960 conv2d_27[0][0] \n__________________________________________________________________________________________________\nswish_20 (Swish) (None, 56, 56, 240) 0 batch_normalization_20[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_8 (DepthwiseCo (None, 56, 56, 240) 2160 swish_20[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_21 (BatchNo (None, 56, 56, 240) 960 depthwise_conv2d_8[0][0] \n__________________________________________________________________________________________________\nswish_21 (Swish) (None, 56, 56, 240) 0 batch_normalization_21[0][0] \n__________________________________________________________________________________________________\nlambda_8 (Lambda) (None, 1, 1, 240) 0 swish_21[0][0] \n__________________________________________________________________________________________________\nconv2d_28 (Conv2D) (None, 1, 1, 10) 2410 lambda_8[0][0] \n__________________________________________________________________________________________________\nswish_22 (Swish) (None, 1, 1, 10) 0 conv2d_28[0][0] \n__________________________________________________________________________________________________\nconv2d_29 (Conv2D) (None, 1, 1, 240) 2640 swish_22[0][0] \n__________________________________________________________________________________________________\nactivation_8 (Activation) (None, 1, 1, 240) 0 conv2d_29[0][0] \n__________________________________________________________________________________________________\nmultiply_8 (Multiply) (None, 56, 56, 240) 0 activation_8[0][0] \n swish_21[0][0] \n__________________________________________________________________________________________________\nconv2d_30 (Conv2D) (None, 56, 56, 40) 9600 multiply_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_22 (BatchNo (None, 56, 56, 40) 160 conv2d_30[0][0] \n__________________________________________________________________________________________________\ndrop_connect_6 (DropConnect) (None, 56, 56, 40) 0 batch_normalization_22[0][0] \n__________________________________________________________________________________________________\nadd_6 (Add) (None, 56, 56, 40) 0 drop_connect_6[0][0] \n add_5[0][0] \n__________________________________________________________________________________________________\nconv2d_31 (Conv2D) (None, 56, 56, 240) 9600 add_6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_23 (BatchNo (None, 56, 56, 240) 960 conv2d_31[0][0] \n__________________________________________________________________________________________________\nswish_23 (Swish) (None, 56, 56, 240) 0 batch_normalization_23[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_9 (DepthwiseCo (None, 28, 28, 240) 6000 swish_23[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_24 (BatchNo (None, 28, 28, 240) 960 depthwise_conv2d_9[0][0] \n__________________________________________________________________________________________________\nswish_24 (Swish) (None, 28, 28, 240) 0 batch_normalization_24[0][0] \n__________________________________________________________________________________________________\nlambda_9 (Lambda) (None, 1, 1, 240) 0 swish_24[0][0] \n__________________________________________________________________________________________________\nconv2d_32 (Conv2D) (None, 1, 1, 10) 2410 lambda_9[0][0] \n__________________________________________________________________________________________________\nswish_25 (Swish) (None, 1, 1, 10) 0 conv2d_32[0][0] \n__________________________________________________________________________________________________\nconv2d_33 (Conv2D) (None, 1, 1, 240) 2640 swish_25[0][0] \n__________________________________________________________________________________________________\nactivation_9 (Activation) (None, 1, 1, 240) 0 conv2d_33[0][0] \n__________________________________________________________________________________________________\nmultiply_9 (Multiply) (None, 28, 28, 240) 0 activation_9[0][0] \n swish_24[0][0] \n__________________________________________________________________________________________________\nconv2d_34 (Conv2D) (None, 28, 28, 64) 15360 multiply_9[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_25 (BatchNo (None, 28, 28, 64) 256 conv2d_34[0][0] \n__________________________________________________________________________________________________\nconv2d_35 (Conv2D) (None, 28, 28, 384) 24576 batch_normalization_25[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_26 (BatchNo (None, 28, 28, 384) 1536 conv2d_35[0][0] \n__________________________________________________________________________________________________\nswish_26 (Swish) (None, 28, 28, 384) 0 batch_normalization_26[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_10 (DepthwiseC (None, 28, 28, 384) 9600 swish_26[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_27 (BatchNo (None, 28, 28, 384) 1536 depthwise_conv2d_10[0][0] \n__________________________________________________________________________________________________\nswish_27 (Swish) (None, 28, 28, 384) 0 batch_normalization_27[0][0] \n__________________________________________________________________________________________________\nlambda_10 (Lambda) (None, 1, 1, 384) 0 swish_27[0][0] \n__________________________________________________________________________________________________\nconv2d_36 (Conv2D) (None, 1, 1, 16) 6160 lambda_10[0][0] \n__________________________________________________________________________________________________\nswish_28 (Swish) (None, 1, 1, 16) 0 conv2d_36[0][0] \n__________________________________________________________________________________________________\nconv2d_37 (Conv2D) (None, 1, 1, 384) 6528 swish_28[0][0] \n__________________________________________________________________________________________________\nactivation_10 (Activation) (None, 1, 1, 384) 0 conv2d_37[0][0] \n__________________________________________________________________________________________________\nmultiply_10 (Multiply) (None, 28, 28, 384) 0 activation_10[0][0] \n swish_27[0][0] \n__________________________________________________________________________________________________\nconv2d_38 (Conv2D) (None, 28, 28, 64) 24576 multiply_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_28 (BatchNo (None, 28, 28, 64) 256 conv2d_38[0][0] \n__________________________________________________________________________________________________\ndrop_connect_7 (DropConnect) (None, 28, 28, 64) 0 batch_normalization_28[0][0] \n__________________________________________________________________________________________________\nadd_7 (Add) (None, 28, 28, 64) 0 drop_connect_7[0][0] \n batch_normalization_25[0][0] \n__________________________________________________________________________________________________\nconv2d_39 (Conv2D) (None, 28, 28, 384) 24576 add_7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_29 (BatchNo (None, 28, 28, 384) 1536 conv2d_39[0][0] \n__________________________________________________________________________________________________\nswish_29 (Swish) (None, 28, 28, 384) 0 batch_normalization_29[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_11 (DepthwiseC (None, 28, 28, 384) 9600 swish_29[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_30 (BatchNo (None, 28, 28, 384) 1536 depthwise_conv2d_11[0][0] \n__________________________________________________________________________________________________\nswish_30 (Swish) (None, 28, 28, 384) 0 batch_normalization_30[0][0] \n__________________________________________________________________________________________________\nlambda_11 (Lambda) (None, 1, 1, 384) 0 swish_30[0][0] \n__________________________________________________________________________________________________\nconv2d_40 (Conv2D) (None, 1, 1, 16) 6160 lambda_11[0][0] \n__________________________________________________________________________________________________\nswish_31 (Swish) (None, 1, 1, 16) 0 conv2d_40[0][0] \n__________________________________________________________________________________________________\nconv2d_41 (Conv2D) (None, 1, 1, 384) 6528 swish_31[0][0] \n__________________________________________________________________________________________________\nactivation_11 (Activation) (None, 1, 1, 384) 0 conv2d_41[0][0] \n__________________________________________________________________________________________________\nmultiply_11 (Multiply) (None, 28, 28, 384) 0 activation_11[0][0] \n swish_30[0][0] \n__________________________________________________________________________________________________\nconv2d_42 (Conv2D) (None, 28, 28, 64) 24576 multiply_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_31 (BatchNo (None, 28, 28, 64) 256 conv2d_42[0][0] \n__________________________________________________________________________________________________\ndrop_connect_8 (DropConnect) (None, 28, 28, 64) 0 batch_normalization_31[0][0] \n__________________________________________________________________________________________________\nadd_8 (Add) (None, 28, 28, 64) 0 drop_connect_8[0][0] \n add_7[0][0] \n__________________________________________________________________________________________________\nconv2d_43 (Conv2D) (None, 28, 28, 384) 24576 add_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_32 (BatchNo (None, 28, 28, 384) 1536 conv2d_43[0][0] \n__________________________________________________________________________________________________\nswish_32 (Swish) (None, 28, 28, 384) 0 batch_normalization_32[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_12 (DepthwiseC (None, 28, 28, 384) 9600 swish_32[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_33 (BatchNo (None, 28, 28, 384) 1536 depthwise_conv2d_12[0][0] \n__________________________________________________________________________________________________\nswish_33 (Swish) (None, 28, 28, 384) 0 batch_normalization_33[0][0] \n__________________________________________________________________________________________________\nlambda_12 (Lambda) (None, 1, 1, 384) 0 swish_33[0][0] \n__________________________________________________________________________________________________\nconv2d_44 (Conv2D) (None, 1, 1, 16) 6160 lambda_12[0][0] \n__________________________________________________________________________________________________\nswish_34 (Swish) (None, 1, 1, 16) 0 conv2d_44[0][0] \n__________________________________________________________________________________________________\nconv2d_45 (Conv2D) (None, 1, 1, 384) 6528 swish_34[0][0] \n__________________________________________________________________________________________________\nactivation_12 (Activation) (None, 1, 1, 384) 0 conv2d_45[0][0] \n__________________________________________________________________________________________________\nmultiply_12 (Multiply) (None, 28, 28, 384) 0 activation_12[0][0] \n swish_33[0][0] \n__________________________________________________________________________________________________\nconv2d_46 (Conv2D) (None, 28, 28, 64) 24576 multiply_12[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_34 (BatchNo (None, 28, 28, 64) 256 conv2d_46[0][0] \n__________________________________________________________________________________________________\ndrop_connect_9 (DropConnect) (None, 28, 28, 64) 0 batch_normalization_34[0][0] \n__________________________________________________________________________________________________\nadd_9 (Add) (None, 28, 28, 64) 0 drop_connect_9[0][0] \n add_8[0][0] \n__________________________________________________________________________________________________\nconv2d_47 (Conv2D) (None, 28, 28, 384) 24576 add_9[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_35 (BatchNo (None, 28, 28, 384) 1536 conv2d_47[0][0] \n__________________________________________________________________________________________________\nswish_35 (Swish) (None, 28, 28, 384) 0 batch_normalization_35[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_13 (DepthwiseC (None, 28, 28, 384) 9600 swish_35[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_36 (BatchNo (None, 28, 28, 384) 1536 depthwise_conv2d_13[0][0] \n__________________________________________________________________________________________________\nswish_36 (Swish) (None, 28, 28, 384) 0 batch_normalization_36[0][0] \n__________________________________________________________________________________________________\nlambda_13 (Lambda) (None, 1, 1, 384) 0 swish_36[0][0] \n__________________________________________________________________________________________________\nconv2d_48 (Conv2D) (None, 1, 1, 16) 6160 lambda_13[0][0] \n__________________________________________________________________________________________________\nswish_37 (Swish) (None, 1, 1, 16) 0 conv2d_48[0][0] \n__________________________________________________________________________________________________\nconv2d_49 (Conv2D) (None, 1, 1, 384) 6528 swish_37[0][0] \n__________________________________________________________________________________________________\nactivation_13 (Activation) (None, 1, 1, 384) 0 conv2d_49[0][0] \n__________________________________________________________________________________________________\nmultiply_13 (Multiply) (None, 28, 28, 384) 0 activation_13[0][0] \n swish_36[0][0] \n__________________________________________________________________________________________________\nconv2d_50 (Conv2D) (None, 28, 28, 64) 24576 multiply_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_37 (BatchNo (None, 28, 28, 64) 256 conv2d_50[0][0] \n__________________________________________________________________________________________________\ndrop_connect_10 (DropConnect) (None, 28, 28, 64) 0 batch_normalization_37[0][0] \n__________________________________________________________________________________________________\nadd_10 (Add) (None, 28, 28, 64) 0 drop_connect_10[0][0] \n add_9[0][0] \n__________________________________________________________________________________________________\nconv2d_51 (Conv2D) (None, 28, 28, 384) 24576 add_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_38 (BatchNo (None, 28, 28, 384) 1536 conv2d_51[0][0] \n__________________________________________________________________________________________________\nswish_38 (Swish) (None, 28, 28, 384) 0 batch_normalization_38[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_14 (DepthwiseC (None, 14, 14, 384) 3456 swish_38[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_39 (BatchNo (None, 14, 14, 384) 1536 depthwise_conv2d_14[0][0] \n__________________________________________________________________________________________________\nswish_39 (Swish) (None, 14, 14, 384) 0 batch_normalization_39[0][0] \n__________________________________________________________________________________________________\nlambda_14 (Lambda) (None, 1, 1, 384) 0 swish_39[0][0] \n__________________________________________________________________________________________________\nconv2d_52 (Conv2D) (None, 1, 1, 16) 6160 lambda_14[0][0] \n__________________________________________________________________________________________________\nswish_40 (Swish) (None, 1, 1, 16) 0 conv2d_52[0][0] \n__________________________________________________________________________________________________\nconv2d_53 (Conv2D) (None, 1, 1, 384) 6528 swish_40[0][0] \n__________________________________________________________________________________________________\nactivation_14 (Activation) (None, 1, 1, 384) 0 conv2d_53[0][0] \n__________________________________________________________________________________________________\nmultiply_14 (Multiply) (None, 14, 14, 384) 0 activation_14[0][0] \n swish_39[0][0] \n__________________________________________________________________________________________________\nconv2d_54 (Conv2D) (None, 14, 14, 128) 49152 multiply_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_40 (BatchNo (None, 14, 14, 128) 512 conv2d_54[0][0] \n__________________________________________________________________________________________________\nconv2d_55 (Conv2D) (None, 14, 14, 768) 98304 batch_normalization_40[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_41 (BatchNo (None, 14, 14, 768) 3072 conv2d_55[0][0] \n__________________________________________________________________________________________________\nswish_41 (Swish) (None, 14, 14, 768) 0 batch_normalization_41[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_15 (DepthwiseC (None, 14, 14, 768) 6912 swish_41[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_42 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_15[0][0] \n__________________________________________________________________________________________________\nswish_42 (Swish) (None, 14, 14, 768) 0 batch_normalization_42[0][0] \n__________________________________________________________________________________________________\nlambda_15 (Lambda) (None, 1, 1, 768) 0 swish_42[0][0] \n__________________________________________________________________________________________________\nconv2d_56 (Conv2D) (None, 1, 1, 32) 24608 lambda_15[0][0] \n__________________________________________________________________________________________________\nswish_43 (Swish) (None, 1, 1, 32) 0 conv2d_56[0][0] \n__________________________________________________________________________________________________\nconv2d_57 (Conv2D) (None, 1, 1, 768) 25344 swish_43[0][0] \n__________________________________________________________________________________________________\nactivation_15 (Activation) (None, 1, 1, 768) 0 conv2d_57[0][0] \n__________________________________________________________________________________________________\nmultiply_15 (Multiply) (None, 14, 14, 768) 0 activation_15[0][0] \n swish_42[0][0] \n__________________________________________________________________________________________________\nconv2d_58 (Conv2D) (None, 14, 14, 128) 98304 multiply_15[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_43 (BatchNo (None, 14, 14, 128) 512 conv2d_58[0][0] \n__________________________________________________________________________________________________\ndrop_connect_11 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_43[0][0] \n__________________________________________________________________________________________________\nadd_11 (Add) (None, 14, 14, 128) 0 drop_connect_11[0][0] \n batch_normalization_40[0][0] \n__________________________________________________________________________________________________\nconv2d_59 (Conv2D) (None, 14, 14, 768) 98304 add_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_44 (BatchNo (None, 14, 14, 768) 3072 conv2d_59[0][0] \n__________________________________________________________________________________________________\nswish_44 (Swish) (None, 14, 14, 768) 0 batch_normalization_44[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_16 (DepthwiseC (None, 14, 14, 768) 6912 swish_44[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_45 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_16[0][0] \n__________________________________________________________________________________________________\nswish_45 (Swish) (None, 14, 14, 768) 0 batch_normalization_45[0][0] \n__________________________________________________________________________________________________\nlambda_16 (Lambda) (None, 1, 1, 768) 0 swish_45[0][0] \n__________________________________________________________________________________________________\nconv2d_60 (Conv2D) (None, 1, 1, 32) 24608 lambda_16[0][0] \n__________________________________________________________________________________________________\nswish_46 (Swish) (None, 1, 1, 32) 0 conv2d_60[0][0] \n__________________________________________________________________________________________________\nconv2d_61 (Conv2D) (None, 1, 1, 768) 25344 swish_46[0][0] \n__________________________________________________________________________________________________\nactivation_16 (Activation) (None, 1, 1, 768) 0 conv2d_61[0][0] \n__________________________________________________________________________________________________\nmultiply_16 (Multiply) (None, 14, 14, 768) 0 activation_16[0][0] \n swish_45[0][0] \n__________________________________________________________________________________________________\nconv2d_62 (Conv2D) (None, 14, 14, 128) 98304 multiply_16[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_46 (BatchNo (None, 14, 14, 128) 512 conv2d_62[0][0] \n__________________________________________________________________________________________________\ndrop_connect_12 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_46[0][0] \n__________________________________________________________________________________________________\nadd_12 (Add) (None, 14, 14, 128) 0 drop_connect_12[0][0] \n add_11[0][0] \n__________________________________________________________________________________________________\nconv2d_63 (Conv2D) (None, 14, 14, 768) 98304 add_12[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_47 (BatchNo (None, 14, 14, 768) 3072 conv2d_63[0][0] \n__________________________________________________________________________________________________\nswish_47 (Swish) (None, 14, 14, 768) 0 batch_normalization_47[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_17 (DepthwiseC (None, 14, 14, 768) 6912 swish_47[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_48 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_17[0][0] \n__________________________________________________________________________________________________\nswish_48 (Swish) (None, 14, 14, 768) 0 batch_normalization_48[0][0] \n__________________________________________________________________________________________________\nlambda_17 (Lambda) (None, 1, 1, 768) 0 swish_48[0][0] \n__________________________________________________________________________________________________\nconv2d_64 (Conv2D) (None, 1, 1, 32) 24608 lambda_17[0][0] \n__________________________________________________________________________________________________\nswish_49 (Swish) (None, 1, 1, 32) 0 conv2d_64[0][0] \n__________________________________________________________________________________________________\nconv2d_65 (Conv2D) (None, 1, 1, 768) 25344 swish_49[0][0] \n__________________________________________________________________________________________________\nactivation_17 (Activation) (None, 1, 1, 768) 0 conv2d_65[0][0] \n__________________________________________________________________________________________________\nmultiply_17 (Multiply) (None, 14, 14, 768) 0 activation_17[0][0] \n swish_48[0][0] \n__________________________________________________________________________________________________\nconv2d_66 (Conv2D) (None, 14, 14, 128) 98304 multiply_17[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_49 (BatchNo (None, 14, 14, 128) 512 conv2d_66[0][0] \n__________________________________________________________________________________________________\ndrop_connect_13 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_49[0][0] \n__________________________________________________________________________________________________\nadd_13 (Add) (None, 14, 14, 128) 0 drop_connect_13[0][0] \n add_12[0][0] \n__________________________________________________________________________________________________\nconv2d_67 (Conv2D) (None, 14, 14, 768) 98304 add_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_50 (BatchNo (None, 14, 14, 768) 3072 conv2d_67[0][0] \n__________________________________________________________________________________________________\nswish_50 (Swish) (None, 14, 14, 768) 0 batch_normalization_50[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_18 (DepthwiseC (None, 14, 14, 768) 6912 swish_50[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_51 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_18[0][0] \n__________________________________________________________________________________________________\nswish_51 (Swish) (None, 14, 14, 768) 0 batch_normalization_51[0][0] \n__________________________________________________________________________________________________\nlambda_18 (Lambda) (None, 1, 1, 768) 0 swish_51[0][0] \n__________________________________________________________________________________________________\nconv2d_68 (Conv2D) (None, 1, 1, 32) 24608 lambda_18[0][0] \n__________________________________________________________________________________________________\nswish_52 (Swish) (None, 1, 1, 32) 0 conv2d_68[0][0] \n__________________________________________________________________________________________________\nconv2d_69 (Conv2D) (None, 1, 1, 768) 25344 swish_52[0][0] \n__________________________________________________________________________________________________\nactivation_18 (Activation) (None, 1, 1, 768) 0 conv2d_69[0][0] \n__________________________________________________________________________________________________\nmultiply_18 (Multiply) (None, 14, 14, 768) 0 activation_18[0][0] \n swish_51[0][0] \n__________________________________________________________________________________________________\nconv2d_70 (Conv2D) (None, 14, 14, 128) 98304 multiply_18[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_52 (BatchNo (None, 14, 14, 128) 512 conv2d_70[0][0] \n__________________________________________________________________________________________________\ndrop_connect_14 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_52[0][0] \n__________________________________________________________________________________________________\nadd_14 (Add) (None, 14, 14, 128) 0 drop_connect_14[0][0] \n add_13[0][0] \n__________________________________________________________________________________________________\nconv2d_71 (Conv2D) (None, 14, 14, 768) 98304 add_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_53 (BatchNo (None, 14, 14, 768) 3072 conv2d_71[0][0] \n__________________________________________________________________________________________________\nswish_53 (Swish) (None, 14, 14, 768) 0 batch_normalization_53[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_19 (DepthwiseC (None, 14, 14, 768) 6912 swish_53[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_54 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_19[0][0] \n__________________________________________________________________________________________________\nswish_54 (Swish) (None, 14, 14, 768) 0 batch_normalization_54[0][0] \n__________________________________________________________________________________________________\nlambda_19 (Lambda) (None, 1, 1, 768) 0 swish_54[0][0] \n__________________________________________________________________________________________________\nconv2d_72 (Conv2D) (None, 1, 1, 32) 24608 lambda_19[0][0] \n__________________________________________________________________________________________________\nswish_55 (Swish) (None, 1, 1, 32) 0 conv2d_72[0][0] \n__________________________________________________________________________________________________\nconv2d_73 (Conv2D) (None, 1, 1, 768) 25344 swish_55[0][0] \n__________________________________________________________________________________________________\nactivation_19 (Activation) (None, 1, 1, 768) 0 conv2d_73[0][0] \n__________________________________________________________________________________________________\nmultiply_19 (Multiply) (None, 14, 14, 768) 0 activation_19[0][0] \n swish_54[0][0] \n__________________________________________________________________________________________________\nconv2d_74 (Conv2D) (None, 14, 14, 128) 98304 multiply_19[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_55 (BatchNo (None, 14, 14, 128) 512 conv2d_74[0][0] \n__________________________________________________________________________________________________\ndrop_connect_15 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_55[0][0] \n__________________________________________________________________________________________________\nadd_15 (Add) (None, 14, 14, 128) 0 drop_connect_15[0][0] \n add_14[0][0] \n__________________________________________________________________________________________________\nconv2d_75 (Conv2D) (None, 14, 14, 768) 98304 add_15[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_56 (BatchNo (None, 14, 14, 768) 3072 conv2d_75[0][0] \n__________________________________________________________________________________________________\nswish_56 (Swish) (None, 14, 14, 768) 0 batch_normalization_56[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_20 (DepthwiseC (None, 14, 14, 768) 6912 swish_56[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_57 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_20[0][0] \n__________________________________________________________________________________________________\nswish_57 (Swish) (None, 14, 14, 768) 0 batch_normalization_57[0][0] \n__________________________________________________________________________________________________\nlambda_20 (Lambda) (None, 1, 1, 768) 0 swish_57[0][0] \n__________________________________________________________________________________________________\nconv2d_76 (Conv2D) (None, 1, 1, 32) 24608 lambda_20[0][0] \n__________________________________________________________________________________________________\nswish_58 (Swish) (None, 1, 1, 32) 0 conv2d_76[0][0] \n__________________________________________________________________________________________________\nconv2d_77 (Conv2D) (None, 1, 1, 768) 25344 swish_58[0][0] \n__________________________________________________________________________________________________\nactivation_20 (Activation) (None, 1, 1, 768) 0 conv2d_77[0][0] \n__________________________________________________________________________________________________\nmultiply_20 (Multiply) (None, 14, 14, 768) 0 activation_20[0][0] \n swish_57[0][0] \n__________________________________________________________________________________________________\nconv2d_78 (Conv2D) (None, 14, 14, 128) 98304 multiply_20[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_58 (BatchNo (None, 14, 14, 128) 512 conv2d_78[0][0] \n__________________________________________________________________________________________________\ndrop_connect_16 (DropConnect) (None, 14, 14, 128) 0 batch_normalization_58[0][0] \n__________________________________________________________________________________________________\nadd_16 (Add) (None, 14, 14, 128) 0 drop_connect_16[0][0] \n add_15[0][0] \n__________________________________________________________________________________________________\nconv2d_79 (Conv2D) (None, 14, 14, 768) 98304 add_16[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_59 (BatchNo (None, 14, 14, 768) 3072 conv2d_79[0][0] \n__________________________________________________________________________________________________\nswish_59 (Swish) (None, 14, 14, 768) 0 batch_normalization_59[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_21 (DepthwiseC (None, 14, 14, 768) 19200 swish_59[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_60 (BatchNo (None, 14, 14, 768) 3072 depthwise_conv2d_21[0][0] \n__________________________________________________________________________________________________\nswish_60 (Swish) (None, 14, 14, 768) 0 batch_normalization_60[0][0] \n__________________________________________________________________________________________________\nlambda_21 (Lambda) (None, 1, 1, 768) 0 swish_60[0][0] \n__________________________________________________________________________________________________\nconv2d_80 (Conv2D) (None, 1, 1, 32) 24608 lambda_21[0][0] \n__________________________________________________________________________________________________\nswish_61 (Swish) (None, 1, 1, 32) 0 conv2d_80[0][0] \n__________________________________________________________________________________________________\nconv2d_81 (Conv2D) (None, 1, 1, 768) 25344 swish_61[0][0] \n__________________________________________________________________________________________________\nactivation_21 (Activation) (None, 1, 1, 768) 0 conv2d_81[0][0] \n__________________________________________________________________________________________________\nmultiply_21 (Multiply) (None, 14, 14, 768) 0 activation_21[0][0] \n swish_60[0][0] \n__________________________________________________________________________________________________\nconv2d_82 (Conv2D) (None, 14, 14, 176) 135168 multiply_21[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_61 (BatchNo (None, 14, 14, 176) 704 conv2d_82[0][0] \n__________________________________________________________________________________________________\nconv2d_83 (Conv2D) (None, 14, 14, 1056) 185856 batch_normalization_61[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_62 (BatchNo (None, 14, 14, 1056) 4224 conv2d_83[0][0] \n__________________________________________________________________________________________________\nswish_62 (Swish) (None, 14, 14, 1056) 0 batch_normalization_62[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_22 (DepthwiseC (None, 14, 14, 1056) 26400 swish_62[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_63 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_22[0][0] \n__________________________________________________________________________________________________\nswish_63 (Swish) (None, 14, 14, 1056) 0 batch_normalization_63[0][0] \n__________________________________________________________________________________________________\nlambda_22 (Lambda) (None, 1, 1, 1056) 0 swish_63[0][0] \n__________________________________________________________________________________________________\nconv2d_84 (Conv2D) (None, 1, 1, 44) 46508 lambda_22[0][0] \n__________________________________________________________________________________________________\nswish_64 (Swish) (None, 1, 1, 44) 0 conv2d_84[0][0] \n__________________________________________________________________________________________________\nconv2d_85 (Conv2D) (None, 1, 1, 1056) 47520 swish_64[0][0] \n__________________________________________________________________________________________________\nactivation_22 (Activation) (None, 1, 1, 1056) 0 conv2d_85[0][0] \n__________________________________________________________________________________________________\nmultiply_22 (Multiply) (None, 14, 14, 1056) 0 activation_22[0][0] \n swish_63[0][0] \n__________________________________________________________________________________________________\nconv2d_86 (Conv2D) (None, 14, 14, 176) 185856 multiply_22[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_64 (BatchNo (None, 14, 14, 176) 704 conv2d_86[0][0] \n__________________________________________________________________________________________________\ndrop_connect_17 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_64[0][0] \n__________________________________________________________________________________________________\nadd_17 (Add) (None, 14, 14, 176) 0 drop_connect_17[0][0] \n batch_normalization_61[0][0] \n__________________________________________________________________________________________________\nconv2d_87 (Conv2D) (None, 14, 14, 1056) 185856 add_17[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_65 (BatchNo (None, 14, 14, 1056) 4224 conv2d_87[0][0] \n__________________________________________________________________________________________________\nswish_65 (Swish) (None, 14, 14, 1056) 0 batch_normalization_65[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_23 (DepthwiseC (None, 14, 14, 1056) 26400 swish_65[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_66 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_23[0][0] \n__________________________________________________________________________________________________\nswish_66 (Swish) (None, 14, 14, 1056) 0 batch_normalization_66[0][0] \n__________________________________________________________________________________________________\nlambda_23 (Lambda) (None, 1, 1, 1056) 0 swish_66[0][0] \n__________________________________________________________________________________________________\nconv2d_88 (Conv2D) (None, 1, 1, 44) 46508 lambda_23[0][0] \n__________________________________________________________________________________________________\nswish_67 (Swish) (None, 1, 1, 44) 0 conv2d_88[0][0] \n__________________________________________________________________________________________________\nconv2d_89 (Conv2D) (None, 1, 1, 1056) 47520 swish_67[0][0] \n__________________________________________________________________________________________________\nactivation_23 (Activation) (None, 1, 1, 1056) 0 conv2d_89[0][0] \n__________________________________________________________________________________________________\nmultiply_23 (Multiply) (None, 14, 14, 1056) 0 activation_23[0][0] \n swish_66[0][0] \n__________________________________________________________________________________________________\nconv2d_90 (Conv2D) (None, 14, 14, 176) 185856 multiply_23[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_67 (BatchNo (None, 14, 14, 176) 704 conv2d_90[0][0] \n__________________________________________________________________________________________________\ndrop_connect_18 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_67[0][0] \n__________________________________________________________________________________________________\nadd_18 (Add) (None, 14, 14, 176) 0 drop_connect_18[0][0] \n add_17[0][0] \n__________________________________________________________________________________________________\nconv2d_91 (Conv2D) (None, 14, 14, 1056) 185856 add_18[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_68 (BatchNo (None, 14, 14, 1056) 4224 conv2d_91[0][0] \n__________________________________________________________________________________________________\nswish_68 (Swish) (None, 14, 14, 1056) 0 batch_normalization_68[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_24 (DepthwiseC (None, 14, 14, 1056) 26400 swish_68[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_69 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_24[0][0] \n__________________________________________________________________________________________________\nswish_69 (Swish) (None, 14, 14, 1056) 0 batch_normalization_69[0][0] \n__________________________________________________________________________________________________\nlambda_24 (Lambda) (None, 1, 1, 1056) 0 swish_69[0][0] \n__________________________________________________________________________________________________\nconv2d_92 (Conv2D) (None, 1, 1, 44) 46508 lambda_24[0][0] \n__________________________________________________________________________________________________\nswish_70 (Swish) (None, 1, 1, 44) 0 conv2d_92[0][0] \n__________________________________________________________________________________________________\nconv2d_93 (Conv2D) (None, 1, 1, 1056) 47520 swish_70[0][0] \n__________________________________________________________________________________________________\nactivation_24 (Activation) (None, 1, 1, 1056) 0 conv2d_93[0][0] \n__________________________________________________________________________________________________\nmultiply_24 (Multiply) (None, 14, 14, 1056) 0 activation_24[0][0] \n swish_69[0][0] \n__________________________________________________________________________________________________\nconv2d_94 (Conv2D) (None, 14, 14, 176) 185856 multiply_24[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_70 (BatchNo (None, 14, 14, 176) 704 conv2d_94[0][0] \n__________________________________________________________________________________________________\ndrop_connect_19 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_70[0][0] \n__________________________________________________________________________________________________\nadd_19 (Add) (None, 14, 14, 176) 0 drop_connect_19[0][0] \n add_18[0][0] \n__________________________________________________________________________________________________\nconv2d_95 (Conv2D) (None, 14, 14, 1056) 185856 add_19[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_71 (BatchNo (None, 14, 14, 1056) 4224 conv2d_95[0][0] \n__________________________________________________________________________________________________\nswish_71 (Swish) (None, 14, 14, 1056) 0 batch_normalization_71[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_25 (DepthwiseC (None, 14, 14, 1056) 26400 swish_71[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_72 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_25[0][0] \n__________________________________________________________________________________________________\nswish_72 (Swish) (None, 14, 14, 1056) 0 batch_normalization_72[0][0] \n__________________________________________________________________________________________________\nlambda_25 (Lambda) (None, 1, 1, 1056) 0 swish_72[0][0] \n__________________________________________________________________________________________________\nconv2d_96 (Conv2D) (None, 1, 1, 44) 46508 lambda_25[0][0] \n__________________________________________________________________________________________________\nswish_73 (Swish) (None, 1, 1, 44) 0 conv2d_96[0][0] \n__________________________________________________________________________________________________\nconv2d_97 (Conv2D) (None, 1, 1, 1056) 47520 swish_73[0][0] \n__________________________________________________________________________________________________\nactivation_25 (Activation) (None, 1, 1, 1056) 0 conv2d_97[0][0] \n__________________________________________________________________________________________________\nmultiply_25 (Multiply) (None, 14, 14, 1056) 0 activation_25[0][0] \n swish_72[0][0] \n__________________________________________________________________________________________________\nconv2d_98 (Conv2D) (None, 14, 14, 176) 185856 multiply_25[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_73 (BatchNo (None, 14, 14, 176) 704 conv2d_98[0][0] \n__________________________________________________________________________________________________\ndrop_connect_20 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_73[0][0] \n__________________________________________________________________________________________________\nadd_20 (Add) (None, 14, 14, 176) 0 drop_connect_20[0][0] \n add_19[0][0] \n__________________________________________________________________________________________________\nconv2d_99 (Conv2D) (None, 14, 14, 1056) 185856 add_20[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_74 (BatchNo (None, 14, 14, 1056) 4224 conv2d_99[0][0] \n__________________________________________________________________________________________________\nswish_74 (Swish) (None, 14, 14, 1056) 0 batch_normalization_74[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_26 (DepthwiseC (None, 14, 14, 1056) 26400 swish_74[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_75 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_26[0][0] \n__________________________________________________________________________________________________\nswish_75 (Swish) (None, 14, 14, 1056) 0 batch_normalization_75[0][0] \n__________________________________________________________________________________________________\nlambda_26 (Lambda) (None, 1, 1, 1056) 0 swish_75[0][0] \n__________________________________________________________________________________________________\nconv2d_100 (Conv2D) (None, 1, 1, 44) 46508 lambda_26[0][0] \n__________________________________________________________________________________________________\nswish_76 (Swish) (None, 1, 1, 44) 0 conv2d_100[0][0] \n__________________________________________________________________________________________________\nconv2d_101 (Conv2D) (None, 1, 1, 1056) 47520 swish_76[0][0] \n__________________________________________________________________________________________________\nactivation_26 (Activation) (None, 1, 1, 1056) 0 conv2d_101[0][0] \n__________________________________________________________________________________________________\nmultiply_26 (Multiply) (None, 14, 14, 1056) 0 activation_26[0][0] \n swish_75[0][0] \n__________________________________________________________________________________________________\nconv2d_102 (Conv2D) (None, 14, 14, 176) 185856 multiply_26[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_76 (BatchNo (None, 14, 14, 176) 704 conv2d_102[0][0] \n__________________________________________________________________________________________________\ndrop_connect_21 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_76[0][0] \n__________________________________________________________________________________________________\nadd_21 (Add) (None, 14, 14, 176) 0 drop_connect_21[0][0] \n add_20[0][0] \n__________________________________________________________________________________________________\nconv2d_103 (Conv2D) (None, 14, 14, 1056) 185856 add_21[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_77 (BatchNo (None, 14, 14, 1056) 4224 conv2d_103[0][0] \n__________________________________________________________________________________________________\nswish_77 (Swish) (None, 14, 14, 1056) 0 batch_normalization_77[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_27 (DepthwiseC (None, 14, 14, 1056) 26400 swish_77[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_78 (BatchNo (None, 14, 14, 1056) 4224 depthwise_conv2d_27[0][0] \n__________________________________________________________________________________________________\nswish_78 (Swish) (None, 14, 14, 1056) 0 batch_normalization_78[0][0] \n__________________________________________________________________________________________________\nlambda_27 (Lambda) (None, 1, 1, 1056) 0 swish_78[0][0] \n__________________________________________________________________________________________________\nconv2d_104 (Conv2D) (None, 1, 1, 44) 46508 lambda_27[0][0] \n__________________________________________________________________________________________________\nswish_79 (Swish) (None, 1, 1, 44) 0 conv2d_104[0][0] \n__________________________________________________________________________________________________\nconv2d_105 (Conv2D) (None, 1, 1, 1056) 47520 swish_79[0][0] \n__________________________________________________________________________________________________\nactivation_27 (Activation) (None, 1, 1, 1056) 0 conv2d_105[0][0] \n__________________________________________________________________________________________________\nmultiply_27 (Multiply) (None, 14, 14, 1056) 0 activation_27[0][0] \n swish_78[0][0] \n__________________________________________________________________________________________________\nconv2d_106 (Conv2D) (None, 14, 14, 176) 185856 multiply_27[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_79 (BatchNo (None, 14, 14, 176) 704 conv2d_106[0][0] \n__________________________________________________________________________________________________\ndrop_connect_22 (DropConnect) (None, 14, 14, 176) 0 batch_normalization_79[0][0] \n__________________________________________________________________________________________________\nadd_22 (Add) (None, 14, 14, 176) 0 drop_connect_22[0][0] \n add_21[0][0] \n__________________________________________________________________________________________________\nconv2d_107 (Conv2D) (None, 14, 14, 1056) 185856 add_22[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_80 (BatchNo (None, 14, 14, 1056) 4224 conv2d_107[0][0] \n__________________________________________________________________________________________________\nswish_80 (Swish) (None, 14, 14, 1056) 0 batch_normalization_80[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_28 (DepthwiseC (None, 7, 7, 1056) 26400 swish_80[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_81 (BatchNo (None, 7, 7, 1056) 4224 depthwise_conv2d_28[0][0] \n__________________________________________________________________________________________________\nswish_81 (Swish) (None, 7, 7, 1056) 0 batch_normalization_81[0][0] \n__________________________________________________________________________________________________\nlambda_28 (Lambda) (None, 1, 1, 1056) 0 swish_81[0][0] \n__________________________________________________________________________________________________\nconv2d_108 (Conv2D) (None, 1, 1, 44) 46508 lambda_28[0][0] \n__________________________________________________________________________________________________\nswish_82 (Swish) (None, 1, 1, 44) 0 conv2d_108[0][0] \n__________________________________________________________________________________________________\nconv2d_109 (Conv2D) (None, 1, 1, 1056) 47520 swish_82[0][0] \n__________________________________________________________________________________________________\nactivation_28 (Activation) (None, 1, 1, 1056) 0 conv2d_109[0][0] \n__________________________________________________________________________________________________\nmultiply_28 (Multiply) (None, 7, 7, 1056) 0 activation_28[0][0] \n swish_81[0][0] \n__________________________________________________________________________________________________\nconv2d_110 (Conv2D) (None, 7, 7, 304) 321024 multiply_28[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_82 (BatchNo (None, 7, 7, 304) 1216 conv2d_110[0][0] \n__________________________________________________________________________________________________\nconv2d_111 (Conv2D) (None, 7, 7, 1824) 554496 batch_normalization_82[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_83 (BatchNo (None, 7, 7, 1824) 7296 conv2d_111[0][0] \n__________________________________________________________________________________________________\nswish_83 (Swish) (None, 7, 7, 1824) 0 batch_normalization_83[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_29 (DepthwiseC (None, 7, 7, 1824) 45600 swish_83[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_84 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_29[0][0] \n__________________________________________________________________________________________________\nswish_84 (Swish) (None, 7, 7, 1824) 0 batch_normalization_84[0][0] \n__________________________________________________________________________________________________\nlambda_29 (Lambda) (None, 1, 1, 1824) 0 swish_84[0][0] \n__________________________________________________________________________________________________\nconv2d_112 (Conv2D) (None, 1, 1, 76) 138700 lambda_29[0][0] \n__________________________________________________________________________________________________\nswish_85 (Swish) (None, 1, 1, 76) 0 conv2d_112[0][0] \n__________________________________________________________________________________________________\nconv2d_113 (Conv2D) (None, 1, 1, 1824) 140448 swish_85[0][0] \n__________________________________________________________________________________________________\nactivation_29 (Activation) (None, 1, 1, 1824) 0 conv2d_113[0][0] \n__________________________________________________________________________________________________\nmultiply_29 (Multiply) (None, 7, 7, 1824) 0 activation_29[0][0] \n swish_84[0][0] \n__________________________________________________________________________________________________\nconv2d_114 (Conv2D) (None, 7, 7, 304) 554496 multiply_29[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_85 (BatchNo (None, 7, 7, 304) 1216 conv2d_114[0][0] \n__________________________________________________________________________________________________\ndrop_connect_23 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_85[0][0] \n__________________________________________________________________________________________________\nadd_23 (Add) (None, 7, 7, 304) 0 drop_connect_23[0][0] \n batch_normalization_82[0][0] \n__________________________________________________________________________________________________\nconv2d_115 (Conv2D) (None, 7, 7, 1824) 554496 add_23[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_86 (BatchNo (None, 7, 7, 1824) 7296 conv2d_115[0][0] \n__________________________________________________________________________________________________\nswish_86 (Swish) (None, 7, 7, 1824) 0 batch_normalization_86[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_30 (DepthwiseC (None, 7, 7, 1824) 45600 swish_86[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_87 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_30[0][0] \n__________________________________________________________________________________________________\nswish_87 (Swish) (None, 7, 7, 1824) 0 batch_normalization_87[0][0] \n__________________________________________________________________________________________________\nlambda_30 (Lambda) (None, 1, 1, 1824) 0 swish_87[0][0] \n__________________________________________________________________________________________________\nconv2d_116 (Conv2D) (None, 1, 1, 76) 138700 lambda_30[0][0] \n__________________________________________________________________________________________________\nswish_88 (Swish) (None, 1, 1, 76) 0 conv2d_116[0][0] \n__________________________________________________________________________________________________\nconv2d_117 (Conv2D) (None, 1, 1, 1824) 140448 swish_88[0][0] \n__________________________________________________________________________________________________\nactivation_30 (Activation) (None, 1, 1, 1824) 0 conv2d_117[0][0] \n__________________________________________________________________________________________________\nmultiply_30 (Multiply) (None, 7, 7, 1824) 0 activation_30[0][0] \n swish_87[0][0] \n__________________________________________________________________________________________________\nconv2d_118 (Conv2D) (None, 7, 7, 304) 554496 multiply_30[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_88 (BatchNo (None, 7, 7, 304) 1216 conv2d_118[0][0] \n__________________________________________________________________________________________________\ndrop_connect_24 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_88[0][0] \n__________________________________________________________________________________________________\nadd_24 (Add) (None, 7, 7, 304) 0 drop_connect_24[0][0] \n add_23[0][0] \n__________________________________________________________________________________________________\nconv2d_119 (Conv2D) (None, 7, 7, 1824) 554496 add_24[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_89 (BatchNo (None, 7, 7, 1824) 7296 conv2d_119[0][0] \n__________________________________________________________________________________________________\nswish_89 (Swish) (None, 7, 7, 1824) 0 batch_normalization_89[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_31 (DepthwiseC (None, 7, 7, 1824) 45600 swish_89[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_90 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_31[0][0] \n__________________________________________________________________________________________________\nswish_90 (Swish) (None, 7, 7, 1824) 0 batch_normalization_90[0][0] \n__________________________________________________________________________________________________\nlambda_31 (Lambda) (None, 1, 1, 1824) 0 swish_90[0][0] \n__________________________________________________________________________________________________\nconv2d_120 (Conv2D) (None, 1, 1, 76) 138700 lambda_31[0][0] \n__________________________________________________________________________________________________\nswish_91 (Swish) (None, 1, 1, 76) 0 conv2d_120[0][0] \n__________________________________________________________________________________________________\nconv2d_121 (Conv2D) (None, 1, 1, 1824) 140448 swish_91[0][0] \n__________________________________________________________________________________________________\nactivation_31 (Activation) (None, 1, 1, 1824) 0 conv2d_121[0][0] \n__________________________________________________________________________________________________\nmultiply_31 (Multiply) (None, 7, 7, 1824) 0 activation_31[0][0] \n swish_90[0][0] \n__________________________________________________________________________________________________\nconv2d_122 (Conv2D) (None, 7, 7, 304) 554496 multiply_31[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_91 (BatchNo (None, 7, 7, 304) 1216 conv2d_122[0][0] \n__________________________________________________________________________________________________\ndrop_connect_25 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_91[0][0] \n__________________________________________________________________________________________________\nadd_25 (Add) (None, 7, 7, 304) 0 drop_connect_25[0][0] \n add_24[0][0] \n__________________________________________________________________________________________________\nconv2d_123 (Conv2D) (None, 7, 7, 1824) 554496 add_25[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_92 (BatchNo (None, 7, 7, 1824) 7296 conv2d_123[0][0] \n__________________________________________________________________________________________________\nswish_92 (Swish) (None, 7, 7, 1824) 0 batch_normalization_92[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_32 (DepthwiseC (None, 7, 7, 1824) 45600 swish_92[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_93 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_32[0][0] \n__________________________________________________________________________________________________\nswish_93 (Swish) (None, 7, 7, 1824) 0 batch_normalization_93[0][0] \n__________________________________________________________________________________________________\nlambda_32 (Lambda) (None, 1, 1, 1824) 0 swish_93[0][0] \n__________________________________________________________________________________________________\nconv2d_124 (Conv2D) (None, 1, 1, 76) 138700 lambda_32[0][0] \n__________________________________________________________________________________________________\nswish_94 (Swish) (None, 1, 1, 76) 0 conv2d_124[0][0] \n__________________________________________________________________________________________________\nconv2d_125 (Conv2D) (None, 1, 1, 1824) 140448 swish_94[0][0] \n__________________________________________________________________________________________________\nactivation_32 (Activation) (None, 1, 1, 1824) 0 conv2d_125[0][0] \n__________________________________________________________________________________________________\nmultiply_32 (Multiply) (None, 7, 7, 1824) 0 activation_32[0][0] \n swish_93[0][0] \n__________________________________________________________________________________________________\nconv2d_126 (Conv2D) (None, 7, 7, 304) 554496 multiply_32[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_94 (BatchNo (None, 7, 7, 304) 1216 conv2d_126[0][0] \n__________________________________________________________________________________________________\ndrop_connect_26 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_94[0][0] \n__________________________________________________________________________________________________\nadd_26 (Add) (None, 7, 7, 304) 0 drop_connect_26[0][0] \n add_25[0][0] \n__________________________________________________________________________________________________\nconv2d_127 (Conv2D) (None, 7, 7, 1824) 554496 add_26[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_95 (BatchNo (None, 7, 7, 1824) 7296 conv2d_127[0][0] \n__________________________________________________________________________________________________\nswish_95 (Swish) (None, 7, 7, 1824) 0 batch_normalization_95[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_33 (DepthwiseC (None, 7, 7, 1824) 45600 swish_95[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_96 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_33[0][0] \n__________________________________________________________________________________________________\nswish_96 (Swish) (None, 7, 7, 1824) 0 batch_normalization_96[0][0] \n__________________________________________________________________________________________________\nlambda_33 (Lambda) (None, 1, 1, 1824) 0 swish_96[0][0] \n__________________________________________________________________________________________________\nconv2d_128 (Conv2D) (None, 1, 1, 76) 138700 lambda_33[0][0] \n__________________________________________________________________________________________________\nswish_97 (Swish) (None, 1, 1, 76) 0 conv2d_128[0][0] \n__________________________________________________________________________________________________\nconv2d_129 (Conv2D) (None, 1, 1, 1824) 140448 swish_97[0][0] \n__________________________________________________________________________________________________\nactivation_33 (Activation) (None, 1, 1, 1824) 0 conv2d_129[0][0] \n__________________________________________________________________________________________________\nmultiply_33 (Multiply) (None, 7, 7, 1824) 0 activation_33[0][0] \n swish_96[0][0] \n__________________________________________________________________________________________________\nconv2d_130 (Conv2D) (None, 7, 7, 304) 554496 multiply_33[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_97 (BatchNo (None, 7, 7, 304) 1216 conv2d_130[0][0] \n__________________________________________________________________________________________________\ndrop_connect_27 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_97[0][0] \n__________________________________________________________________________________________________\nadd_27 (Add) (None, 7, 7, 304) 0 drop_connect_27[0][0] \n add_26[0][0] \n__________________________________________________________________________________________________\nconv2d_131 (Conv2D) (None, 7, 7, 1824) 554496 add_27[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_98 (BatchNo (None, 7, 7, 1824) 7296 conv2d_131[0][0] \n__________________________________________________________________________________________________\nswish_98 (Swish) (None, 7, 7, 1824) 0 batch_normalization_98[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_34 (DepthwiseC (None, 7, 7, 1824) 45600 swish_98[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_99 (BatchNo (None, 7, 7, 1824) 7296 depthwise_conv2d_34[0][0] \n__________________________________________________________________________________________________\nswish_99 (Swish) (None, 7, 7, 1824) 0 batch_normalization_99[0][0] \n__________________________________________________________________________________________________\nlambda_34 (Lambda) (None, 1, 1, 1824) 0 swish_99[0][0] \n__________________________________________________________________________________________________\nconv2d_132 (Conv2D) (None, 1, 1, 76) 138700 lambda_34[0][0] \n__________________________________________________________________________________________________\nswish_100 (Swish) (None, 1, 1, 76) 0 conv2d_132[0][0] \n__________________________________________________________________________________________________\nconv2d_133 (Conv2D) (None, 1, 1, 1824) 140448 swish_100[0][0] \n__________________________________________________________________________________________________\nactivation_34 (Activation) (None, 1, 1, 1824) 0 conv2d_133[0][0] \n__________________________________________________________________________________________________\nmultiply_34 (Multiply) (None, 7, 7, 1824) 0 activation_34[0][0] \n swish_99[0][0] \n__________________________________________________________________________________________________\nconv2d_134 (Conv2D) (None, 7, 7, 304) 554496 multiply_34[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_100 (BatchN (None, 7, 7, 304) 1216 conv2d_134[0][0] \n__________________________________________________________________________________________________\ndrop_connect_28 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_100[0][0] \n__________________________________________________________________________________________________\nadd_28 (Add) (None, 7, 7, 304) 0 drop_connect_28[0][0] \n add_27[0][0] \n__________________________________________________________________________________________________\nconv2d_135 (Conv2D) (None, 7, 7, 1824) 554496 add_28[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_101 (BatchN (None, 7, 7, 1824) 7296 conv2d_135[0][0] \n__________________________________________________________________________________________________\nswish_101 (Swish) (None, 7, 7, 1824) 0 batch_normalization_101[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_35 (DepthwiseC (None, 7, 7, 1824) 45600 swish_101[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_102 (BatchN (None, 7, 7, 1824) 7296 depthwise_conv2d_35[0][0] \n__________________________________________________________________________________________________\nswish_102 (Swish) (None, 7, 7, 1824) 0 batch_normalization_102[0][0] \n__________________________________________________________________________________________________\nlambda_35 (Lambda) (None, 1, 1, 1824) 0 swish_102[0][0] \n__________________________________________________________________________________________________\nconv2d_136 (Conv2D) (None, 1, 1, 76) 138700 lambda_35[0][0] \n__________________________________________________________________________________________________\nswish_103 (Swish) (None, 1, 1, 76) 0 conv2d_136[0][0] \n__________________________________________________________________________________________________\nconv2d_137 (Conv2D) (None, 1, 1, 1824) 140448 swish_103[0][0] \n__________________________________________________________________________________________________\nactivation_35 (Activation) (None, 1, 1, 1824) 0 conv2d_137[0][0] \n__________________________________________________________________________________________________\nmultiply_35 (Multiply) (None, 7, 7, 1824) 0 activation_35[0][0] \n swish_102[0][0] \n__________________________________________________________________________________________________\nconv2d_138 (Conv2D) (None, 7, 7, 304) 554496 multiply_35[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_103 (BatchN (None, 7, 7, 304) 1216 conv2d_138[0][0] \n__________________________________________________________________________________________________\ndrop_connect_29 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_103[0][0] \n__________________________________________________________________________________________________\nadd_29 (Add) (None, 7, 7, 304) 0 drop_connect_29[0][0] \n add_28[0][0] \n__________________________________________________________________________________________________\nconv2d_139 (Conv2D) (None, 7, 7, 1824) 554496 add_29[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_104 (BatchN (None, 7, 7, 1824) 7296 conv2d_139[0][0] \n__________________________________________________________________________________________________\nswish_104 (Swish) (None, 7, 7, 1824) 0 batch_normalization_104[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_36 (DepthwiseC (None, 7, 7, 1824) 45600 swish_104[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_105 (BatchN (None, 7, 7, 1824) 7296 depthwise_conv2d_36[0][0] \n__________________________________________________________________________________________________\nswish_105 (Swish) (None, 7, 7, 1824) 0 batch_normalization_105[0][0] \n__________________________________________________________________________________________________\nlambda_36 (Lambda) (None, 1, 1, 1824) 0 swish_105[0][0] \n__________________________________________________________________________________________________\nconv2d_140 (Conv2D) (None, 1, 1, 76) 138700 lambda_36[0][0] \n__________________________________________________________________________________________________\nswish_106 (Swish) (None, 1, 1, 76) 0 conv2d_140[0][0] \n__________________________________________________________________________________________________\nconv2d_141 (Conv2D) (None, 1, 1, 1824) 140448 swish_106[0][0] \n__________________________________________________________________________________________________\nactivation_36 (Activation) (None, 1, 1, 1824) 0 conv2d_141[0][0] \n__________________________________________________________________________________________________\nmultiply_36 (Multiply) (None, 7, 7, 1824) 0 activation_36[0][0] \n swish_105[0][0] \n__________________________________________________________________________________________________\nconv2d_142 (Conv2D) (None, 7, 7, 304) 554496 multiply_36[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_106 (BatchN (None, 7, 7, 304) 1216 conv2d_142[0][0] \n__________________________________________________________________________________________________\ndrop_connect_30 (DropConnect) (None, 7, 7, 304) 0 batch_normalization_106[0][0] \n__________________________________________________________________________________________________\nadd_30 (Add) (None, 7, 7, 304) 0 drop_connect_30[0][0] \n add_29[0][0] \n__________________________________________________________________________________________________\nconv2d_143 (Conv2D) (None, 7, 7, 1824) 554496 add_30[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_107 (BatchN (None, 7, 7, 1824) 7296 conv2d_143[0][0] \n__________________________________________________________________________________________________\nswish_107 (Swish) (None, 7, 7, 1824) 0 batch_normalization_107[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_37 (DepthwiseC (None, 7, 7, 1824) 16416 swish_107[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_108 (BatchN (None, 7, 7, 1824) 7296 depthwise_conv2d_37[0][0] \n__________________________________________________________________________________________________\nswish_108 (Swish) (None, 7, 7, 1824) 0 batch_normalization_108[0][0] \n__________________________________________________________________________________________________\nlambda_37 (Lambda) (None, 1, 1, 1824) 0 swish_108[0][0] \n__________________________________________________________________________________________________\nconv2d_144 (Conv2D) (None, 1, 1, 76) 138700 lambda_37[0][0] \n__________________________________________________________________________________________________\nswish_109 (Swish) (None, 1, 1, 76) 0 conv2d_144[0][0] \n__________________________________________________________________________________________________\nconv2d_145 (Conv2D) (None, 1, 1, 1824) 140448 swish_109[0][0] \n__________________________________________________________________________________________________\nactivation_37 (Activation) (None, 1, 1, 1824) 0 conv2d_145[0][0] \n__________________________________________________________________________________________________\nmultiply_37 (Multiply) (None, 7, 7, 1824) 0 activation_37[0][0] \n swish_108[0][0] \n__________________________________________________________________________________________________\nconv2d_146 (Conv2D) (None, 7, 7, 512) 933888 multiply_37[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_109 (BatchN (None, 7, 7, 512) 2048 conv2d_146[0][0] \n__________________________________________________________________________________________________\nconv2d_147 (Conv2D) (None, 7, 7, 3072) 1572864 batch_normalization_109[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_110 (BatchN (None, 7, 7, 3072) 12288 conv2d_147[0][0] \n__________________________________________________________________________________________________\nswish_110 (Swish) (None, 7, 7, 3072) 0 batch_normalization_110[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_38 (DepthwiseC (None, 7, 7, 3072) 27648 swish_110[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_111 (BatchN (None, 7, 7, 3072) 12288 depthwise_conv2d_38[0][0] \n__________________________________________________________________________________________________\nswish_111 (Swish) (None, 7, 7, 3072) 0 batch_normalization_111[0][0] \n__________________________________________________________________________________________________\nlambda_38 (Lambda) (None, 1, 1, 3072) 0 swish_111[0][0] \n__________________________________________________________________________________________________\nconv2d_148 (Conv2D) (None, 1, 1, 128) 393344 lambda_38[0][0] \n__________________________________________________________________________________________________\nswish_112 (Swish) (None, 1, 1, 128) 0 conv2d_148[0][0] \n__________________________________________________________________________________________________\nconv2d_149 (Conv2D) (None, 1, 1, 3072) 396288 swish_112[0][0] \n__________________________________________________________________________________________________\nactivation_38 (Activation) (None, 1, 1, 3072) 0 conv2d_149[0][0] \n__________________________________________________________________________________________________\nmultiply_38 (Multiply) (None, 7, 7, 3072) 0 activation_38[0][0] \n swish_111[0][0] \n__________________________________________________________________________________________________\nconv2d_150 (Conv2D) (None, 7, 7, 512) 1572864 multiply_38[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_112 (BatchN (None, 7, 7, 512) 2048 conv2d_150[0][0] \n__________________________________________________________________________________________________\ndrop_connect_31 (DropConnect) (None, 7, 7, 512) 0 batch_normalization_112[0][0] \n__________________________________________________________________________________________________\nadd_31 (Add) (None, 7, 7, 512) 0 drop_connect_31[0][0] \n batch_normalization_109[0][0] \n__________________________________________________________________________________________________\nconv2d_151 (Conv2D) (None, 7, 7, 3072) 1572864 add_31[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_113 (BatchN (None, 7, 7, 3072) 12288 conv2d_151[0][0] \n__________________________________________________________________________________________________\nswish_113 (Swish) (None, 7, 7, 3072) 0 batch_normalization_113[0][0] \n__________________________________________________________________________________________________\ndepthwise_conv2d_39 (DepthwiseC (None, 7, 7, 3072) 27648 swish_113[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_114 (BatchN (None, 7, 7, 3072) 12288 depthwise_conv2d_39[0][0] \n__________________________________________________________________________________________________\nswish_114 (Swish) (None, 7, 7, 3072) 0 batch_normalization_114[0][0] \n__________________________________________________________________________________________________\nlambda_39 (Lambda) (None, 1, 1, 3072) 0 swish_114[0][0] \n__________________________________________________________________________________________________\nconv2d_152 (Conv2D) (None, 1, 1, 128) 393344 lambda_39[0][0] \n__________________________________________________________________________________________________\nswish_115 (Swish) (None, 1, 1, 128) 0 conv2d_152[0][0] \n__________________________________________________________________________________________________\nconv2d_153 (Conv2D) (None, 1, 1, 3072) 396288 swish_115[0][0] \n__________________________________________________________________________________________________\nactivation_39 (Activation) (None, 1, 1, 3072) 0 conv2d_153[0][0] \n__________________________________________________________________________________________________\nmultiply_39 (Multiply) (None, 7, 7, 3072) 0 activation_39[0][0] \n swish_114[0][0] \n__________________________________________________________________________________________________\nconv2d_154 (Conv2D) (None, 7, 7, 512) 1572864 multiply_39[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_115 (BatchN (None, 7, 7, 512) 2048 conv2d_154[0][0] \n__________________________________________________________________________________________________\ndrop_connect_32 (DropConnect) (None, 7, 7, 512) 0 batch_normalization_115[0][0] \n__________________________________________________________________________________________________\nadd_32 (Add) (None, 7, 7, 512) 0 drop_connect_32[0][0] \n add_31[0][0] \n__________________________________________________________________________________________________\nconv2d_155 (Conv2D) (None, 7, 7, 2048) 1048576 add_32[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_116 (BatchN (None, 7, 7, 2048) 8192 conv2d_155[0][0] \n__________________________________________________________________________________________________\nswish_116 (Swish) (None, 7, 7, 2048) 0 batch_normalization_116[0][0] \n__________________________________________________________________________________________________\nglobal_average_pooling2d_1 (Glo (None, 2048) 0 swish_116[0][0] \n__________________________________________________________________________________________________\nfinal_output (Dense) (None, 1) 2049 global_average_pooling2d_1[0][0] \n==================================================================================================\nTotal params: 28,515,569\nTrainable params: 28,342,833\nNon-trainable params: 172,736\n__________________________________________________________________________________________________\n"
],
[
"history = model.fit_generator(generator=train_generator,\n steps_per_epoch=STEP_SIZE_TRAIN,\n validation_data=valid_generator,\n validation_steps=STEP_SIZE_VALID,\n epochs=EPOCHS,\n callbacks=callback_list,\n verbose=2).history",
"Epoch 1/20\n - 523s - loss: 1.0489 - acc: 0.3302 - val_loss: 0.5248 - val_acc: 0.5987\nEpoch 2/20\n - 476s - loss: 0.8712 - acc: 0.3677 - val_loss: 0.4143 - val_acc: 0.7015\nEpoch 3/20\n - 477s - loss: 0.7568 - acc: 0.4041 - val_loss: 0.3843 - val_acc: 0.6943\nEpoch 4/20\n - 475s - loss: 0.6869 - acc: 0.4394 - val_loss: 0.3647 - val_acc: 0.6971\nEpoch 5/20\n - 475s - loss: 0.6534 - acc: 0.4575 - val_loss: 0.3472 - val_acc: 0.7615\nEpoch 6/20\n - 475s - loss: 0.6265 - acc: 0.4817 - val_loss: 0.3299 - val_acc: 0.7432\nEpoch 7/20\n - 475s - loss: 0.5936 - acc: 0.5018 - val_loss: 0.3711 - val_acc: 0.7232\nEpoch 8/20\n - 474s - loss: 0.5687 - acc: 0.5134 - val_loss: 0.3762 - val_acc: 0.7815\nEpoch 9/20\n - 475s - loss: 0.5618 - acc: 0.5245 - val_loss: 0.3382 - val_acc: 0.7727\nEpoch 10/20\n - 475s - loss: 0.5328 - acc: 0.5379 - val_loss: 0.3071 - val_acc: 0.7676\nEpoch 11/20\n - 474s - loss: 0.5065 - acc: 0.5565 - val_loss: 0.2722 - val_acc: 0.7860\nEpoch 12/20\n - 476s - loss: 0.4901 - acc: 0.5654 - val_loss: 0.2960 - val_acc: 0.7560\nEpoch 13/20\n - 475s - loss: 0.4508 - acc: 0.5967 - val_loss: 0.2791 - val_acc: 0.7832\nEpoch 14/20\n - 475s - loss: 0.4242 - acc: 0.6157 - val_loss: 0.2525 - val_acc: 0.7882\nEpoch 15/20\n - 474s - loss: 0.3841 - acc: 0.6432 - val_loss: 0.2581 - val_acc: 0.8021\nEpoch 16/20\n - 475s - loss: 0.3493 - acc: 0.6671 - val_loss: 0.2646 - val_acc: 0.7804\nEpoch 17/20\n - 474s - loss: 0.3251 - acc: 0.6839 - val_loss: 0.2561 - val_acc: 0.8105\nEpoch 18/20\n - 475s - loss: 0.2986 - acc: 0.7031 - val_loss: 0.2460 - val_acc: 0.8099\nEpoch 19/20\n - 474s - loss: 0.2807 - acc: 0.7182 - val_loss: 0.2567 - val_acc: 0.7982\nEpoch 20/20\n - 474s - loss: 0.2688 - acc: 0.7251 - val_loss: 0.2441 - val_acc: 0.8093\n"
],
[
"fig, (ax1, ax2) = plt.subplots(2, 1, sharex='col', figsize=(20, 6))\n\nax1.plot(cosine_lr_1st.learning_rates)\nax1.set_title('Warm up learning rates')\n\nax2.plot(cosine_lr_2nd.learning_rates)\nax2.set_title('Fine-tune learning rates')\n\nplt.xlabel('Steps')\nplt.ylabel('Learning rate')\nsns.despine()\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Model loss graph ",
"_____no_output_____"
]
],
[
[
"fig, (ax1, ax2) = plt.subplots(2, 1, sharex='col', figsize=(20, 14))\n\nax1.plot(history['loss'], label='Train loss')\nax1.plot(history['val_loss'], label='Validation loss')\nax1.legend(loc='best')\nax1.set_title('Loss')\n\nax2.plot(history['acc'], label='Train accuracy')\nax2.plot(history['val_acc'], label='Validation accuracy')\nax2.legend(loc='best')\nax2.set_title('Accuracy')\n\nplt.xlabel('Epochs')\nsns.despine()\nplt.show()",
"_____no_output_____"
],
[
"# Create empty arays to keep the predictions and labels\ndf_preds = pd.DataFrame(columns=['label', 'pred', 'set'])\ntrain_generator.reset()\nvalid_generator.reset()\n\n# Add train predictions and labels\nfor i in range(STEP_SIZE_TRAIN + 1):\n im, lbl = next(train_generator)\n preds = model.predict(im, batch_size=train_generator.batch_size)\n for index in range(len(preds)):\n df_preds.loc[len(df_preds)] = [lbl[index], preds[index][0], 'train']\n\n# Add validation predictions and labels\nfor i in range(STEP_SIZE_VALID + 1):\n im, lbl = next(valid_generator)\n preds = model.predict(im, batch_size=valid_generator.batch_size)\n for index in range(len(preds)):\n df_preds.loc[len(df_preds)] = [lbl[index], preds[index][0], 'validation']\n\ndf_preds['label'] = df_preds['label'].astype('int')",
"_____no_output_____"
],
[
"def classify(x):\n if x < 0.5:\n return 0\n elif x < 1.5:\n return 1\n elif x < 2.5:\n return 2\n elif x < 3.5:\n return 3\n return 4\n\n# Classify predictions\ndf_preds['predictions'] = df_preds['pred'].apply(lambda x: classify(x))\n\ntrain_preds = df_preds[df_preds['set'] == 'train']\nvalidation_preds = df_preds[df_preds['set'] == 'validation']",
"_____no_output_____"
]
],
[
[
"# Model Evaluation",
"_____no_output_____"
],
[
"## Confusion Matrix\n\n### Original thresholds",
"_____no_output_____"
]
],
[
[
"labels = ['0 - No DR', '1 - Mild', '2 - Moderate', '3 - Severe', '4 - Proliferative DR']\ndef plot_confusion_matrix(train, validation, labels=labels):\n train_labels, train_preds = train\n validation_labels, validation_preds = validation\n fig, (ax1, ax2) = plt.subplots(1, 2, sharex='col', figsize=(24, 7))\n train_cnf_matrix = confusion_matrix(train_labels, train_preds)\n validation_cnf_matrix = confusion_matrix(validation_labels, validation_preds)\n\n train_cnf_matrix_norm = train_cnf_matrix.astype('float') / train_cnf_matrix.sum(axis=1)[:, np.newaxis]\n validation_cnf_matrix_norm = validation_cnf_matrix.astype('float') / validation_cnf_matrix.sum(axis=1)[:, np.newaxis]\n\n train_df_cm = pd.DataFrame(train_cnf_matrix_norm, index=labels, columns=labels)\n validation_df_cm = pd.DataFrame(validation_cnf_matrix_norm, index=labels, columns=labels)\n\n sns.heatmap(train_df_cm, annot=True, fmt='.2f', cmap=\"Blues\",ax=ax1).set_title('Train')\n sns.heatmap(validation_df_cm, annot=True, fmt='.2f', cmap=sns.cubehelix_palette(8),ax=ax2).set_title('Validation')\n plt.show()\n\nplot_confusion_matrix((train_preds['label'], train_preds['predictions']), (validation_preds['label'], validation_preds['predictions']))",
"_____no_output_____"
]
],
[
[
"## Quadratic Weighted Kappa",
"_____no_output_____"
]
],
[
[
"def evaluate_model(train, validation):\n train_labels, train_preds = train\n validation_labels, validation_preds = validation\n print(\"Train Cohen Kappa score: %.3f\" % cohen_kappa_score(train_preds, train_labels, weights='quadratic'))\n print(\"Validation Cohen Kappa score: %.3f\" % cohen_kappa_score(validation_preds, validation_labels, weights='quadratic'))\n print(\"Complete set Cohen Kappa score: %.3f\" % cohen_kappa_score(np.append(train_preds, validation_preds), np.append(train_labels, validation_labels), weights='quadratic'))\n \nevaluate_model((train_preds['label'], train_preds['predictions']), (validation_preds['label'], validation_preds['predictions']))",
"Train Cohen Kappa score: 0.906\nValidation Cohen Kappa score: 0.912\nComplete set Cohen Kappa score: 0.906\n"
]
],
[
[
"## Apply model to test set and output predictions",
"_____no_output_____"
]
],
[
[
"def apply_tta(model, generator, steps=10):\n step_size = generator.n//generator.batch_size\n preds_tta = []\n for i in range(steps):\n generator.reset()\n preds = model.predict_generator(generator, steps=step_size)\n preds_tta.append(preds)\n\n return np.mean(preds_tta, axis=0)\n\npreds = apply_tta(model, test_generator, TTA_STEPS)\npredictions = [classify(x) for x in preds]\n\nresults = pd.DataFrame({'id_code':test['id_code'], 'diagnosis':predictions})\nresults['id_code'] = results['id_code'].map(lambda x: str(x)[:-4])",
"_____no_output_____"
],
[
"# Cleaning created directories\nif os.path.exists(train_dest_path):\n shutil.rmtree(train_dest_path)\nif os.path.exists(validation_dest_path):\n shutil.rmtree(validation_dest_path)\nif os.path.exists(test_dest_path):\n shutil.rmtree(test_dest_path)",
"_____no_output_____"
]
],
[
[
"# Predictions class distribution",
"_____no_output_____"
]
],
[
[
"fig = plt.subplots(sharex='col', figsize=(24, 8.7))\nsns.countplot(x=\"diagnosis\", data=results, palette=\"GnBu_d\").set_title('Test')\nsns.despine()\nplt.show()",
"_____no_output_____"
],
[
"results.to_csv('submission.csv', index=False)\ndisplay(results.head())",
"_____no_output_____"
]
],
[
[
"## Save model",
"_____no_output_____"
]
],
[
[
"model.save_weights('../working/effNetB5_img224.h5')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb5a89c945355b8f51be95c07769434d662fd11 | 8,608 | ipynb | Jupyter Notebook | notebooks/collision_avoidance/train_model.ipynb | vstoneofficial/jetbot-mecanum | cc161b888b3e6cccfde4ff9b653c97af66adb5c8 | [
"MIT"
] | null | null | null | notebooks/collision_avoidance/train_model.ipynb | vstoneofficial/jetbot-mecanum | cc161b888b3e6cccfde4ff9b653c97af66adb5c8 | [
"MIT"
] | null | null | null | notebooks/collision_avoidance/train_model.ipynb | vstoneofficial/jetbot-mecanum | cc161b888b3e6cccfde4ff9b653c97af66adb5c8 | [
"MIT"
] | null | null | null | 27.154574 | 208 | 0.556808 | [
[
[
"# Collision Avoidance - Train Model\n\nWelcome to this host side Jupyter Notebook! This should look familiar if you ran through the notebooks that run on the robot. In this notebook we'll train our image classifier to detect two classes\n``free`` and ``blocked``, which we'll use for avoiding collisions. For this, we'll use a popular deep learning library *PyTorch*",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.optim as optim\nimport torch.nn.functional as F\nimport torchvision\nimport torchvision.datasets as datasets\nimport torchvision.models as models\nimport torchvision.transforms as transforms",
"_____no_output_____"
]
],
[
[
"### Upload and extract dataset\n\nBefore you start, you should upload the ``dataset.zip`` file that you created in the ``data_collection.ipynb`` notebook on the robot.\n\nYou should then extract this dataset by calling the command below",
"_____no_output_____"
]
],
[
[
"!unzip -q dataset.zip",
"_____no_output_____"
]
],
[
[
"You should see a folder named ``dataset`` appear in the file browser.",
"_____no_output_____"
],
[
"### Create dataset instance",
"_____no_output_____"
],
[
"Now we use the ``ImageFolder`` dataset class available with the ``torchvision.datasets`` package. We attach transforms from the ``torchvision.transforms`` package to prepare the data for training. ",
"_____no_output_____"
]
],
[
[
"dataset = datasets.ImageFolder(\n 'dataset_20191212',\n transforms.Compose([\n transforms.ColorJitter(0.1, 0.1, 0.1, 0.1),\n transforms.Resize((224, 224)),\n transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])\n ])\n)",
"_____no_output_____"
]
],
[
[
"### Split dataset into train and test sets",
"_____no_output_____"
],
[
"Next, we split the dataset into *training* and *test* sets. The test set will be used to verify the accuracy of the model we train.",
"_____no_output_____"
]
],
[
[
"train_dataset, test_dataset = torch.utils.data.random_split(dataset, [len(dataset) - 50, 50])",
"_____no_output_____"
]
],
[
[
"### Create data loaders to load data in batches",
"_____no_output_____"
],
[
"We'll create two ``DataLoader`` instances, which provide utilities for shuffling data, producing *batches* of images, and loading the samples in parallel with multiple workers.",
"_____no_output_____"
]
],
[
[
"train_loader = torch.utils.data.DataLoader(\n train_dataset,\n batch_size=16,\n shuffle=True,\n num_workers=4\n)\n\ntest_loader = torch.utils.data.DataLoader(\n test_dataset,\n batch_size=16,\n shuffle=True,\n num_workers=4\n)",
"_____no_output_____"
]
],
[
[
"### Define the neural network\n\nNow, we define the neural network we'll be training. The *torchvision* package provides a collection of pre-trained models that we can use.\n\nIn a process called *transfer learning*, we can repurpose a pre-trained model (trained on millions of images) for a new task that has possibly much less data available.\n\nImportant features that were learned in the original training of the pre-trained model are re-usable for the new task. We'll use the ``alexnet`` model.",
"_____no_output_____"
]
],
[
[
"model = models.alexnet(pretrained=True)",
"_____no_output_____"
]
],
[
[
"The ``alexnet`` model was originally trained for a dataset that had 1000 class labels, but our dataset only has two class labels! We'll replace\nthe final layer with a new, untrained layer that has only two outputs. ",
"_____no_output_____"
]
],
[
[
"model.classifier[6] = torch.nn.Linear(model.classifier[6].in_features, 2)",
"_____no_output_____"
]
],
[
[
"Finally, we transfer our model for execution on the GPU",
"_____no_output_____"
]
],
[
[
"device = torch.device('cuda')\nmodel = model.to(device)",
"_____no_output_____"
]
],
[
[
"### Train the neural network\n\nUsing the code below we will train the neural network for 30 epochs, saving the best performing model after each epoch.\n\n> An epoch is a full run through our data.",
"_____no_output_____"
]
],
[
[
"NUM_EPOCHS = 30\nBEST_MODEL_PATH = 'best_model_191212.pth'\nbest_accuracy = 0.0\n\noptimizer = optim.SGD(model.parameters(), lr=0.001, momentum=0.9)\n\nfor epoch in range(NUM_EPOCHS):\n \n for images, labels in iter(train_loader):\n images = images.to(device)\n labels = labels.to(device)\n optimizer.zero_grad()\n outputs = model(images)\n loss = F.cross_entropy(outputs, labels)\n loss.backward()\n optimizer.step()\n \n test_error_count = 0.0\n for images, labels in iter(test_loader):\n images = images.to(device)\n labels = labels.to(device)\n outputs = model(images)\n test_error_count += float(torch.sum(torch.abs(labels - outputs.argmax(1))))\n \n test_accuracy = 1.0 - float(test_error_count) / float(len(test_dataset))\n print('%d: %f' % (epoch, test_accuracy))\n if test_accuracy > best_accuracy:\n torch.save(model.state_dict(), BEST_MODEL_PATH)\n best_accuracy = test_accuracy",
"0: 0.720000\n1: 0.620000\n2: 0.780000\n3: 0.720000\n4: 0.760000\n5: 0.700000\n6: 0.800000\n7: 0.760000\n8: 0.800000\n9: 0.700000\n10: 0.820000\n11: 0.800000\n12: 0.860000\n13: 0.820000\n14: 0.800000\n15: 0.740000\n16: 0.800000\n17: 0.780000\n18: 0.820000\n19: 0.760000\n20: 0.800000\n21: 0.800000\n22: 0.780000\n23: 0.840000\n24: 0.840000\n25: 0.780000\n26: 0.760000\n27: 0.780000\n28: 0.800000\n29: 0.760000\n"
]
],
[
[
"Once that is finished, you should see a file ``best_model.pth`` in the Jupyter Lab file browser. Select ``Right click`` -> ``Download`` to download the model to your workstation",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb5bc1a4f2a74fc9dd09a59aa9cd81950b78f11 | 17,870 | ipynb | Jupyter Notebook | Notebooks/Giuan/gradient_descent/2d.ipynb | alanoMartins/DIP_Nutes | e3580c5d0e269c6cd70f052bbd32d0e0ac19fe55 | [
"MIT"
] | null | null | null | Notebooks/Giuan/gradient_descent/2d.ipynb | alanoMartins/DIP_Nutes | e3580c5d0e269c6cd70f052bbd32d0e0ac19fe55 | [
"MIT"
] | null | null | null | Notebooks/Giuan/gradient_descent/2d.ipynb | alanoMartins/DIP_Nutes | e3580c5d0e269c6cd70f052bbd32d0e0ac19fe55 | [
"MIT"
] | 2 | 2018-08-13T18:47:55.000Z | 2018-08-13T18:49:15.000Z | 125.84507 | 15,144 | 0.886738 | [
[
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"def function(x):\n return x**2\ndef derivate_of_function(x):\n return 2*x\n\nlr = 0.3\n\naxes = list(range(-10, 11))\nfx = [function(x) for x in axes ]\n",
"_____no_output_____"
],
[
"x_history = []\nx = 9 #initial value (weight)\nn = 2",
"_____no_output_____"
],
[
"for i in range(n):\n gradient = (derivate_of_function(x) *-1) * lr\n loss = function(x)\n print(\"gradient: {}\".format(gradient))\n print(\"weight: {}\".format(x))\n print(\"loss: {}\".format(loss))\n x_history.append(x)\n #x = x - ( f'(x) * -1)\n x += gradient\n\n\nfx_history = [function(g) for g in x_history]\n\nplt.plot(axes, fx)\nplt.plot(x_history, fx_history , 'co')\nplt.show()",
"gradient: -5.3999999999999995\nweight: 9\nloss: 81\ngradient: -2.16\nweight: 3.6000000000000005\nloss: 12.960000000000004\n"
],
[
"# SGD\n# [1,1]\n# train\n# [1,0]\n# train\n# [0,0]\n# train\n# [0,1]\n# train",
"_____no_output_____"
],
[
"# mini-batch SGD\n# [1,1]\n# [1,0]\n# train\n# [0,0]\n# [0,1]\n# train",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb5c55fd40f337c07b4f77c79ebabf54496811e | 140,071 | ipynb | Jupyter Notebook | writeup.ipynb | samindaa/SFND_2D_Feature_Tracking | 75a34a7c63b39954cd5eb55fb747c9aa4d3c63be | [
"MIT"
] | null | null | null | writeup.ipynb | samindaa/SFND_2D_Feature_Tracking | 75a34a7c63b39954cd5eb55fb747c9aa4d3c63be | [
"MIT"
] | null | null | null | writeup.ipynb | samindaa/SFND_2D_Feature_Tracking | 75a34a7c63b39954cd5eb55fb747c9aa4d3c63be | [
"MIT"
] | null | null | null | 376.534946 | 51,784 | 0.941344 | [
[
[
"**Camera Based 2D Feature Tracking (Mid-Term)**\n\n*Saminda Abeyruwan*\n\nWe present herewith the analysis of the time it takes for keypoint detection and descriptor extraction.\n\nOur dataset is available in *stats_output.csv* file. We have used HARRIS, FAST, BRISK, ORB, AKAZE, and SIFT detectors\nand BRIEF, ORB, FREAK, AKAZE and SIFT descriptors. The matching configuration has been fixed. We have used the BF \napproach with the descriptor distance ratio set to 0.8.\n\nWe were not able to produce results for SIFT x BRIEF detector and descriptor combination. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"df = pd.read_csv(\"stats_output.csv\")",
"_____no_output_____"
]
],
[
[
"The following graphs shows the detector time in ms vs detectors aggregated across the dataset. The top three\ndetectors are (based on median):\n\n1. FAST\n2. ORB\n3. HARRIS",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(10, 10))\nsns.boxplot(x=\"det\", y=\"det_time_ms\", data=df)",
"_____no_output_____"
]
],
[
[
"The following graphs shows the descriptor time in ms vs descriptor aggregated across the dataset. The top three\ndescriptor are (based on median):\n\n1. FREAK\n2. ORB\n3. AKAZE",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(10, 10))\nsns.boxplot(x=\"des\", y=\"des_time_ms\", data=df)",
"_____no_output_____"
],
[
"df[\"det_des\"] = df[\"det\"] + \"_x_\" + df[\"des\"]\ndf[\"det_des_time_ms\"] = df.eval(\"det_time_ms+des_time_ms\")\n",
"_____no_output_____"
]
],
[
[
"Finally, we compare the total time in ms vs detectors cross descriptor combinations. As shown in the following figure,\nall FAST detectors cross descriptor combinations outperform all the other combinations. According to our results, the\nbest peforming detector and descriptor combinations are (based on median):\n\n1. FAST x FREAK\n2. FAST x ORB\n3. FAST x AKAZE\n\nIt is to be noted that we set *nfeatures=500* for ORB.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(16, 16))\nax = sns.boxplot(x=\"det_des\", y=\"det_des_time_ms\", data=df);\nax.set_xticklabels(ax.get_xticklabels(),rotation=90);\n",
"_____no_output_____"
]
],
[
[
"Following figure shows the number of keypoints detected per image.",
"_____no_output_____"
]
],
[
[
"sns.set_style(\"whitegrid\")\ng = sns.FacetGrid(df, col=\"det\", col_wrap=3, height=5)\ng.map(plt.scatter, \"run\", \"keypoints\", alpha=.7)\ng.add_legend();\n",
"_____no_output_____"
]
],
[
[
"Following figure shows the mean and standard deviation of the neighborhood sizes.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(10, 10))\nsns.catplot(x=\"det\", y=\"size_mu\", data=df);",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 10))\nsns.catplot(x=\"det\", y=\"size_std\", data=df);",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecb5d5b7ad058d9efa6fd29c4ee8f7157e4939ae | 2,900 | ipynb | Jupyter Notebook | docs/auto_examples/plot_coding_decoding_simulation.ipynb | xiaoxiaoyuwts/pyldpc | 55874f58c26f9dd1557f96ee8b701fb635ca534c | [
"BSD-3-Clause"
] | 1 | 2020-01-08T06:48:06.000Z | 2020-01-08T06:48:06.000Z | docs/auto_examples/plot_coding_decoding_simulation.ipynb | xiaoxiaoyuwts/pyldpc | 55874f58c26f9dd1557f96ee8b701fb635ca534c | [
"BSD-3-Clause"
] | null | null | null | docs/auto_examples/plot_coding_decoding_simulation.ipynb | xiaoxiaoyuwts/pyldpc | 55874f58c26f9dd1557f96ee8b701fb635ca534c | [
"BSD-3-Clause"
] | null | null | null | 32.222222 | 534 | 0.541724 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n# Coding - Decoding simulation of a random message\n\n\nThis example shows a simulation of the transmission of a binary message\nthrough a gaussian white noise channel with an LDPC coding and decoding system.\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom pyldpc import make_ldpc, decode, get_message, encode\nfrom matplotlib import pyplot as plt\n\nn = 30\nd_v = 2\nd_c = 3\nseed = np.random.RandomState(42)",
"_____no_output_____"
]
],
[
[
"First we create an LDPC code i.e a pair of decoding and coding matrices\nH and G. H is a regular parity-check matrix with d_v ones per row\nand d_c ones per column\n\n",
"_____no_output_____"
]
],
[
[
"H, G = make_ldpc(n, d_v, d_c, seed=seed, systematic=True, sparse=True)\n\nn, k = G.shape\nprint(\"Number of coded bits:\", k)",
"_____no_output_____"
]
],
[
[
"Now we simulate transmission for different levels of noise and\ncompute the percentage of errors using the bit-error-rate score\n\n",
"_____no_output_____"
]
],
[
[
"errors = []\nsnrs = np.linspace(-2, 10, 20)\nv = np.arange(k) % 2 # fixed k bits message\nn_trials = 50 # number of transmissions with different noise\nfor snr in snrs:\n error = 0.\n for ii in range(n_trials):\n y = encode(G, v, snr, seed=seed)\n d = decode(H, y, snr)\n x = get_message(G, d)\n error += abs(v - x).sum() / k\n errors.append(error / n_trials)\n\nplt.figure()\nplt.plot(snrs, errors, color=\"indianred\")\nplt.ylabel(\"Bit error rate\")\nplt.xlabel(\"SNR\")\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb5da0645c370e15629cbaa879675a6c789a5bb | 35,038 | ipynb | Jupyter Notebook | Untitled.ipynb | apaneser/World_Weather_Analysis | f586bb2c8bff871dd62f20271995fcddcc92fc2f | [
"MIT"
] | null | null | null | Untitled.ipynb | apaneser/World_Weather_Analysis | f586bb2c8bff871dd62f20271995fcddcc92fc2f | [
"MIT"
] | null | null | null | Untitled.ipynb | apaneser/World_Weather_Analysis | f586bb2c8bff871dd62f20271995fcddcc92fc2f | [
"MIT"
] | null | null | null | 60.51468 | 479 | 0.593013 | [
[
[
"# Dependencies and Setup\nimport requests\nimport gmaps\n\n# Import API key\nfrom config import g_key",
"_____no_output_____"
],
[
"# Set the parameters to search for a hotel in Paris.\nparams = {\n \"radius\": 5000,\n \"types\": \"lodging\",\n \"key\": g_key,\n \"location\": \"48.8566, 2.3522\"}\n# Use base URL to search for hotels in Paris.\nbase_url = \"https://maps.googleapis.com/maps/api/place/nearbysearch/json\"\n# Make request and get the JSON data from the search.\nhotels = requests.get(base_url, params=params).json()\n\nhotels",
"_____no_output_____"
],
[
"len(hotels['results'])",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecb5ee81b350ed55802c7e296cc2576bb2afd336 | 832,638 | ipynb | Jupyter Notebook | src/result/Calculate.ipynb | jbterrylin/GALGO-2.0 | 531e034b0d36a592e1e7cc75ed06c88a6d38c029 | [
"MIT"
] | null | null | null | src/result/Calculate.ipynb | jbterrylin/GALGO-2.0 | 531e034b0d36a592e1e7cc75ed06c88a6d38c029 | [
"MIT"
] | null | null | null | src/result/Calculate.ipynb | jbterrylin/GALGO-2.0 | 531e034b0d36a592e1e7cc75ed06c88a6d38c029 | [
"MIT"
] | null | null | null | 865.528067 | 63,340 | 0.936797 | [
[
[
"import os\nfrom os.path import exists\nimport pandas as pd\nimport numpy as np\nfrom statistics import mean\nimport matplotlib.pyplot as plt\nimport decimal\nplt.style.use('seaborn-whitegrid')\n\n# turn csv string to numbers\ndef fixFormat(df):\n df = df.drop( df[df['Repeat']=='Repeat'].index )\n \n if 'Repeat' in df.columns:\n df[\"Repeat\"] = df[\"Repeat\"].astype(int)\n if 'Generation' in df.columns:\n df[\"Generation\"] = df[\"Generation\"].astype(int)\n if 'FitnessEvaluateCountReachTarget' in df.columns:\n df[\"FitnessEvaluateCountReachTarget\"] = df[\"FitnessEvaluateCountReachTarget\"].astype(int)\n \n if 'F(x)' in df.columns:\n df[\"F(x)\"] = df[\"F(x)\"].astype(float)\n if wantFitness is False:\n df[\"F(x)\"] = -df[\"F(x)\"]\n \n if df.shape[1] > 3:\n for i in range(df.shape[1]-2):\n df.iloc[:,i+2] = df.iloc[:,i+2].astype(float)\n return df\n\n# get set of crossovers and benchmarks from folder\ndef getLists(files):\n crossovers = set()\n benchmarks = set()\n for file in files:\n crossovers.add(file.split(\"+\")[0])\n benchmarks.add(file.split(\"+\")[1].split(\".\")[0])\n return crossovers, benchmarks\n\n# showPlot(file, df, \"Generation\", \"F(x)\")\ndef showPlot(file, df, x, y):\n print(file)\n plt.plot(df[\"Generation\"], df[\"F(x)\"], 'o', color='black')\n plt.show()",
"_____no_output_____"
],
[
"# folders = [\"FrontRearCrossover\", \"HighDimensionalGeneticAlgorithmToolboxCrossover\",\"RingCrossover\", \"CollectiveCrossover\"]\n# stats = [\"mean\", \"max\", \"min\", \"var\", \"sum\", \"maxFitnessCount\"]\ntarget_id = 0\ntarget = [\n {\n \"folder\": \"RingCrossover\", \"stats\": [\"mean\", \"max\", \"min\"], \"wantFitness\": False, \n \"oriResult\": getRingCrossoverOriResult(\"RingCrossover\"), \"index\": {'max':'Worst', \"mean\": \"Average\", \"min\": \"Best\"}\n },\n {\n \"folder\": \"HighDimensionalGeneticAlgorithmToolboxCrossover\", \"stats\": [\"mean\", \"min\", \"var\"], \"wantFitness\": False, \n \"oriResult\": getHDGAOriResult(\"HighDimensionalGeneticAlgorithmToolboxCrossover\"), \n \"index\": {'mean':'Mean', \"min\": \"Min\", \"var\": \"Variance\"} },\n {\n \"folder\": \"CollectiveCrossover\", \"stats\": [\"mean\"], \"wantFitness\": False, \n \"oriResult\": getCollectiveCrossoverOriResult(\"CollectiveCrossover\"), \"index\": {'mean':'Mean'}\n },\n]\n\nfolder = target[target_id][\"folder\"]\nstats = target[target_id][\"stats\"]\nwantFitness = target[target_id][\"wantFitness\"]\noriResult = target[target_id][\"oriResult\"]\nindex = target[target_id][\"index\"]\n\n\ndef compareResultBetweenCrossover(df, crossover1, crossover2, stat):\n crossover1_win = 0\n crossover1_tie = 0\n for column in result.columns:\n if result.loc[crossover1, stat][column] < result.loc[crossover2, stat][column]:\n crossover1_win = crossover1_win + 1\n elif result.loc[crossover1, stat][column] == result.loc[crossover2, stat][column]:\n crossover1_tie = crossover1_tie + 1\n print(column)\n return crossover1_win, crossover1_tie, len(result.columns) - crossover1_win - crossover1_tie\n \n# process file not have FitnessEvaluateCountReachTarget in file name\nfor subdir, dirs, files in os.walk(folder):\n crossovers, benchmarks = getLists(files)\n iterables = [crossovers, stats]\n result = pd.DataFrame(columns = benchmarks, index = pd.MultiIndex.from_product(iterables, names=[\"crossover\", \"stat\"]))\n files = [file for file in files if 'FitnessEvaluateCountReachTarget' not in file]\n for file in files:\n crossover = file.split(\"+\")[0]\n benchmark = file.split(\"+\")[1].split(\".\")[0]\n df = pd.read_csv(folder+\"/\"+file, index_col=False)\n df = fixFormat(df)\n\n temp_result = {}\n temp_df = df.groupby(\"Generation\")[\"F(x)\"]\n temp_result[\"mean\"] = temp_df.mean()[df[\"Generation\"].max()]\n temp_result[\"max\"] = temp_df.max()[df[\"Generation\"].max()]\n temp_result[\"min\"] = temp_df.min()[df[\"Generation\"].max()]\n temp_result[\"var\"] = temp_df.var()[df[\"Generation\"].max()]\n temp_result[\"sum\"] = temp_df.sum()[df[\"Generation\"].max()]\n\n# if \"RingCrossover\" in file:\n# print(file)\n# for_graph = df.groupby(['Generation']).mean()[\"F(x)\"]\n# plt.plot(for_graph.index, for_graph)\n# plt.show()\n# plt.plot(for_graph.mean()[\"F(x)\"],index, for_graph.mean()[\"F(x)\"])\n# plt.plot(for_graph.mean()[\"F(x)\"],index, for_graph.max()[\"F(x)\"])\n# plt.plot(for_graph.mean()[\"F(x)\"],index, for_graph.min()[\"F(x)\"])\n# plt.scatter(df[\"Generation\"], df[\"F(x)\"])\n# plt.show()\n for stat in stats:\n if stat in temp_result.keys():\n result.loc[crossover, stat][benchmark] = temp_result[stat]\n\n for column in result.columns:\n fig, axes = plt.subplots(1, 2)\n # filter unrelated stat, rename to paper's stat (max -> Best...)\n a = oriResult[column][np.in1d(oriResult.index.get_level_values(1), stats)].rename(index=index, level=1)\n a = a.unstack().plot(kind='bar', title=column, ax=axes[0],figsize=(14,4.8))\n a.set_ylabel(\"Fitness value\" if wantFitness else \"f(x)\")\n b = result[column].rename(index=index, level=1).unstack().plot(kind='bar', title=column, ax=axes[1])\n b.set_ylabel(\"Fitness value\" if wantFitness else \"f(x)\")\n \n \n plt.savefig(column + \".jpg\", bbox_inches='tight')\n \n plt.show()\n \n# win, tie, lose = compareResultBetweenCrossover(df, \"CollectiveCrossover-30\", \"P1XO-30\", \"mean\")\n# print(str(win) + \" : \" + str(tie) + \" : \" + str(lose))\n# win, tie, lose = compareResultBetweenCrossover(df, \"CollectiveCrossover-30\", \"P2XO-30\", \"mean\")\n# print(str(win) + \" : \" + str(tie) + \" : \" + str(lose))",
"_____no_output_____"
],
[
"# stats = [\"mean\", \"max\", \"min\", \"var\", \"sum\"]\n# GeneticAlgorithm.genstep need be be 1\nfolder = \"HybridCrossover\"\nstats = [\"mean\", \"max\", \"min\",\"con_rate\"]\nline_stats = [\"con_rate\"]\nwantFitness = False\noriResult = getHybridCrossoverOriResult(\"HybridCrossover\")\nindex = {\"mean\": \"Avg-con\", \"max\": \"Max-con\", \"min\": \"Min-con\"}\n\ndef round_down(value, decimals):\n with decimal.localcontext() as ctx:\n d = decimal.Decimal(value)\n ctx.rounding = decimal.ROUND_DOWN\n return round(d, decimals)\n \n# calculateConvergenceGeneration, if difference smaller than 1.0e-6 then consider no different\ndef calculateConvergenceGeneration(df):\n result = []\n# repeats = set(df[\"Repeat\"])\n repeats = range(100)\n# generations = sorted(set(df[\"Generation\"]), reverse=True)\n generations = sorted(range(100), reverse=True)\n# print(repeats)\n generations.pop()\n for repeat in repeats:\n gene_count = 0\n temp = df[df[\"Repeat\"].isin({repeat})]\n for generation in generations:\n old_fx = temp[temp[\"Generation\"].isin({generation-1})][\"F(x)\"].iloc[0]\n fx = temp[temp[\"Generation\"].isin({generation})][\"F(x)\"].iloc[0]\n \n if round_down(fx, 6) == round_down(old_fx, 6):\n gene_count = gene_count + 1\n else:\n break\n result.append(generations[0]-gene_count)\n return result\n\nglobal_mins = {\n \"SphereObjective\": 0,\n \"GeneralizedRastriginObjective\": 0,\n \"SchaffersF6Objective\": 0,\n \"GriewangksObjective\": 0,\n \"HansenObjective\": -176.541793,\n \"MichalewiczObjective\": -1.8013,\n}\n\n# calculate absolute range between first and last generation\ndef calculateConvergenceRate(df, benchmark, minOf=\"global\"):\n def get_min_max_array(array, value):\n array.append(value)\n if len(array) > 2:\n array = [np.min(array), np.max(array)]\n return array\n result = []\n repeats = set(df[\"Repeat\"])\n min_gen = min(df[\"Generation\"])\n max_gen = max(df[\"Generation\"])\n min_gen_fx = []\n max_gen_fx = []\n for repeat in repeats:\n gene_count = 0\n temp = df[df[\"Repeat\"].isin({repeat})]\n gen_min_fx = temp[temp[\"Generation\"] == min_gen][\"F(x)\"].iloc[0]\n min_gen_fx = get_min_max_array(min_gen_fx, gen_min_fx)\n gen_max_fx = temp[temp[\"Generation\"] == max_gen][\"F(x)\"].iloc[0]\n max_gen_fx = get_min_max_array(max_gen_fx, gen_max_fx)\n if minOf == \"global\":\n min_gen_fx[0] = global_mins[benchmark]\n max_gen_fx[0] = global_mins[benchmark]\n min_gen_fx = abs(min_gen_fx[1] - min_gen_fx[0])\n max_gen_fx = abs(max_gen_fx[1] - max_gen_fx[0])\n if min_gen_fx == 0:\n print(\" alr done convergence in start\")\n return 0\n else:\n return (1-(max_gen_fx/ min_gen_fx)) * 100\n \nfor subdir, dirs, files in os.walk(folder):\n crossovers, benchmarks = getLists(files)\n iterables = [crossovers, stats]\n result = pd.DataFrame(columns = benchmarks, index = pd.MultiIndex.from_product(iterables, names=[\"crossover\", \"stat\"]))\n\n files = [file for file in files if 'FitnessEvaluateCountReachTarget' not in file]\n for file in files:\n crossover = file.split(\"+\")[0]\n benchmark = file.split(\"+\")[1].split(\".\")[0]\n df = pd.read_csv(folder+\"/\"+file, index_col=False)\n df = fixFormat(df)\n\n temp_result = {}\n convergence = calculateConvergenceGeneration(df)\n temp_result[\"mean\"] = sum(convergence) / len(convergence)\n temp_result[\"max\"] = np.max(convergence)\n temp_result[\"min\"] = np.min(convergence)\n temp_result[\"var\"] = np.var(convergence)\n temp_result[\"sum\"] = sum(convergence)\n temp_result[\"con_rate\"] = calculateConvergenceRate(df, benchmark, \"own\")\n# if \"con_rate_with_own\" in stats:\n# temp_result[\"con_rate_with_own\"] = calculateConvergenceRate(df, benchmark, \"own\")\n# if \"con_rate_with_global_min\" in stats:\n# temp_result[\"con_rate_with_global_min\"] = calculateConvergenceRate(df, benchmark, \"global\")\n\n for stat in stats:\n result.loc[crossover, stat][benchmark] = temp_result[stat]\n\n for column in result.columns:\n fig, axes = plt.subplots(1, 2)\n# [np.in1d(oriResult.index.get_level_values(1), stats)]\n a = oriResult[column].rename(index=index, level=1)\n a = a.unstack().plot(kind='bar', title=column, ax=axes[0],figsize=(14,4.8), secondary_y=[\"con_rate\"])\n a.set_ylabel(\"Fitness value\" if wantFitness else \"f(x)\")\n \n b = result[column].rename(index=index, level=1)\n b = b.unstack().plot(kind='bar', title=column, ax=axes[1], secondary_y=[\"con_rate\"])\n b.set_ylabel(\"Fitness value\" if wantFitness else \"f(x)\")\n# plt.savefig(column + \".jpg\", bbox_inches='tight')\n plt.show()\n print(result)",
"_____no_output_____"
],
[
"maxFitness = 30\nchromoseLength = 15\ntarget_id = 0\ntarget = [\n {\n \"folder\": \"FrontRearCrossover\", \"stats\": [\"mean\", \"maxFitnessCount\"], \"wantFitness\": True, \n \"oriResult\": getFrontRearCrossoverOriResult(\"FrontRearCrossover\"), \"index\": {}\n },\n]\n\nfolder = target[target_id][\"folder\"]\nstats = target[target_id][\"stats\"]\nwantFitness = target[target_id][\"wantFitness\"]\noriResult = target[target_id][\"oriResult\"]\nindex = target[target_id][\"index\"]\n\n\n# calculate for front-rear crossover\nfor subdir, dirs, files in os.walk(folder):\n crossovers, benchmarks = getLists(files)\n iterables = [crossovers, stats]\n result = pd.DataFrame(columns = benchmarks, index = pd.MultiIndex.from_product(iterables, names=[\"crossover\", \"stat\"]))\n\n files_filtered = [file for file in files if 'FitnessEvaluateCountReachTarget' in file]\n for file in files_filtered:\n crossover = file.split(\"+\")[0]\n benchmark = file.split(\"+\")[1]\n df = pd.read_csv(folder+\"/\"+file, index_col=False)\n df = fixFormat(df)\n\n temp_result = {}\n temp_result[\"mean\"] = df[\"FitnessEvaluateCountReachTarget\"].mean()\n temp_result[\"max\"] = df[\"FitnessEvaluateCountReachTarget\"].max()\n temp_result[\"min\"] = df[\"FitnessEvaluateCountReachTarget\"].min()\n temp_result[\"var\"] = df[\"FitnessEvaluateCountReachTarget\"].var()\n temp_result[\"sum\"] = df[\"FitnessEvaluateCountReachTarget\"].sum()\n\n for stat in stats:\n if stat in temp_result.keys():\n result.loc[crossover, stat][benchmark] = temp_result[stat]\n \n files_filtered = [file for file in files if 'FitnessEvaluateCountReachTarget' not in file]\n for file in files_filtered:\n crossover = file.split(\"+\")[0]\n benchmark = file.split(\"+\")[1].split(\".\")[0]\n df = pd.read_csv(folder+\"/\"+file, index_col=False)\n df = fixFormat(df)\n\n temp_result = {}\n lastGenFx = df[df[\"Generation\"] == df[\"Generation\"].max()][\"F(x)\"]\n temp_result[\"maxFitnessCount\"] = len(lastGenFx[lastGenFx == maxFitness])\n \n for stat in stats:\n if stat in temp_result.keys():\n result.loc[crossover, stat][benchmark] = temp_result[stat]\n \n filtered_row_index = [k for k in oriResult.index.get_level_values(0).tolist() if '-'+str(int(maxFitness/chromoseLength)) in k]\n for column in result.columns:\n fig, axes = plt.subplots(2, 2)\n a = oriResult[column][np.in1d(oriResult.index.get_level_values(1), stats)]\n a = a[np.in1d(a.index.get_level_values(0), filtered_row_index)].rename(index=index, level=1)\n a = a.unstack().plot(kind='bar', subplots=True,sharex=True, title=column, ax=axes[0], figsize=(12,4.8))\n \n b = result[column][np.in1d(result.index.get_level_values(0), filtered_row_index)].rename(index=index, level=1)\n b = b.unstack().plot(kind='bar', subplots=True,sharex=True,title=column, ax=axes[1])\n \n plt.figtext(0.06, 0.7, \"From Research\", fontsize=12, rotation=90, ha='center', va='center')\n plt.figtext(0.06, 0.3, \"Reproduce\", fontsize=12, rotation=90, ha='center', va='center')\n# plt.savefig(column + \"-\" + str(maxFitness) + \".jpg\", bbox_inches='tight')\n \n plt.show()\n print(result)",
"C:\\Users\\wangl\\anaconda3\\lib\\site-packages\\pandas\\plotting\\_matplotlib\\__init__.py:71: UserWarning: When passing multiple axes, sharex and sharey are ignored. These settings must be specified when creating axes\n plot_obj.generate()\n"
],
[
"def getRingCrossoverOriResult(folder):\n for subdir, dirs, files in os.walk(folder):\n crossovers, benchmarks = getLists(files)\n iterables = [\n [\"P1XO\", \"P2XO\", \"IntermediateCrossover\", \"HeuristicCrossover\", \"ArithmeticCrossover\", \"RingCrossover\",], \n [\"mean\", \"max\", \"min\"]\n ]\n result = pd.DataFrame(columns = benchmarks, index = pd.MultiIndex.from_product(iterables, names=[\"crossover\", \"stat\"]))\n result[\"RosenbrocksValleyObjective\"] = [\n 73.07,269.3,73.08,\n 70.07, 390.3, 78.39,\n 34.71, 349.2, 34.74,\n 29.35, 369.1, 117.5,\n 27.08, 260.3, 27.12,\n 28.59, 316.1, 32.69,\n ]\n result[\"AxisParallelHyperEllipsoidObjective\"] = [\n 70.52, 105.7, 70.52,\n 68.63, 94.04, 68.64,\n 64.04, 80.4, 64.04,\n 0.024, 87.41, 5.706,\n 73.71, 89.18, 73.72,\n 0.1023, 106.8, 11.73,\n ]\n result[\"SphereObjective\"] = [\n 5.732, 7.246, 5.737,\n 3.416, 6.511, 3.417,\n 6.207, 6.246, 6.208,\n 0.011, 8.099, 2.81,\n 5.589, 6.389, 5.589,\n 0.0027, 6.163, 0.3299\n ]\n result[\"NormalizedSchwefelObjective\"] = [\n -115.7, -29.46, -115.6,\n -115.8, -26.85, -115.4,\n -114.1, -27.91, -114,\n -117.7, -26.1, -117.1,\n -113.2, -27.72, -113.1,\n -117.8, -27.75, -117.7,\n ]\n result[\"RotatedHyperEllipsoidObjective\"] = [\n 20.79, 261.7, 37.02,\n 15.06, 204.8, 16.22,\n 22.86, 59.24, 22.87,\n 2.36, 381, 17.58,\n 24.37, 47.94, 24.37,\n 4.577, 108.2, 18.97,\n ]\n result[\"GeneralizedRastriginObjective\"] = [\n 94.69, 241.3, 111.3,\n 50.84, 257.7, 52.15,\n 122.6, 256.6, 187.3,\n 12.68, 173.1, 31.98,\n 154, 251.4, 154.1,\n 2.669, 232.5, 3.691,\n ]\n return result\n \ndef getHDGAOriResult(folder):\n for subdir, dirs, files in os.walk(folder):\n crossovers, benchmarks = getLists(files)\n iterables = [\n [\"P1XO\", \"HighDimensionalGeneticAlgorithmToolboxCrossover\"], \n [\"min\", \"mean\", \"var\"]\n ]\n result = pd.DataFrame(columns = benchmarks, index = pd.MultiIndex.from_product(iterables, names=[\"crossover\", \"stat\"]))\n result[\"ZakharovObjective\"] = [\n 8.7426, 40.8791, 297.49,\n 2.2244, 13.5011, 56.7825,\n ]\n result[\"DixonPriceObjective\"] = [\n 2.4271, 58.9734, 9067.1742,\n 0.5538, 6.0133, 120.6263,\n ]\n result[\"SphereObjective\"] = [\n 0.055081, 0.2988, 0.066168,\n 0.00027559, 0.0071927, 0.00015764,\n ]\n result[\"ShubertObjective\"] = [\n -186.7309, -185.9764, 40.9808,\n -186.7309, -185.9068, 23.069,\n ]\n return result\n \ndef getCollectiveCrossoverOriResult(folder):\n for subdir, dirs, files in os.walk(folder):\n crossovers, benchmarks = getLists(files)\n iterables = [\n [\"CollectiveCrossover-30\", \"P1XO-30\", \"P2XO-30\", \"CollectiveCrossover-50\", \"P1XO-50\", \"P2XO-50\"], \n [\"mean\"]\n ]\n result = pd.DataFrame(columns = benchmarks, index = pd.MultiIndex.from_product(iterables, names=[\"crossover\", \"stat\"]))\n result[\"ShiftedandRotatedLunacekBi_RastriginObjective\"] = [\n 5.75E+07, 1.15E+08, 9.65E+07, 1.76E+08, 3.41E+08, 3.00E+08\n ]\n result[\"ShiftedandRotatedBentCigarObjective\"] = [\n 3.79E+13, 4.12E+13, 4.05E+13, 3.61E+14, 4.23E+14, 4.12E+14\n ]\n result[\"ShiftedandRotatedRastriginObjective\"] = [\n 2.35E+06, 2.23E+06, 2.43E+06, 5.43E+06, 7.59E+06, 6.66E+06\n ]\n result[\"ShiftedandRotatedSchwefelObjective\"] = [\n 1.82E+08, 2.41E+08, 2.34E+08, 5.37E+08, 6.10E+08, 6.56E+08\n ]\n \n result[\"ShiftedandRotatedRosenbrockObjective\"] = [\n 6.61E+07, 1.19E+08, 9.90E+07, 2.07E+08, 3.48E+08, 3.32E+08\n ]\n result[\"ShiftedandRotatedLevyObjective\"] = [\n 2.91E+09, 3.45E+09, 3.15E+09, 7.06E+09, 7.59E+09, 7.48E+09\n ]\n result[\"ShiftedandRotatedZakharovObjective\"] = [\n 1.15E+08, 1.27E+08, 1.31E+08, 2.44E+08, 3.28E+08, 3.02E+08\n ]\n return result\n \ndef getHybridCrossoverOriResult(folder):\n for subdir, dirs, files in os.walk(folder):\n crossovers, benchmarks = getLists(files)\n iterables = [\n [\"P1XO\", \"P2XO\", \"UniformCrossover\", \"HybridCrossover\"], \n [\"mean\", \"max\", \"min\", \"con_rate\"]\n ]\n result = pd.DataFrame(columns = benchmarks, index = pd.MultiIndex.from_product(iterables, names=[\"crossover\", \"stat\"]))\n result[\"SchaffersF6Objective\"] = [\n 255, 497, 41, 80,\n 222, 489, 18, 81,\n 247, 461, 25, 79,\n 213, 471, 48, 97,\n ]\n result[\"HansenObjective\"] = [\n 328, 489, 148, 6,\n 317, 335, 304, 3,\n 413, 464, 338, 4,\n 249, 471, 33, 50,\n ]\n result[\"GeneralizedRastriginObjective\"] = [\n 307, 466, 75, 29,\n 303, 495, 41, 52,\n 292, 493, 79, 39,\n 99, 457, 18, 98,\n ]\n result[\"GriewangksObjective\"] = [\n 280, 481, 50, 43,\n 256, 494, 27, 53,\n 269, 491, 61, 38,\n 162, 489, 19, 97,\n ]\n result[\"MichalewiczObjective\"] = [\n 248, 492, 22, 55,\n 209, 498, 31, 56,\n 215, 469, 33, 59,\n 213, 493, 32, 76,\n ]\n result[\"SphereObjective\"] = [\n 190, 482, 23, 69,\n 194, 464, 18, 86,\n 227, 483, 15, 89,\n 121, 409, 13, 99,\n ]\n return result\n \ndef getFrontRearCrossoverOriResult(folder):\n for subdir, dirs, files in os.walk(folder):\n crossovers, benchmarks = getLists(files)\n iterables = [\n [\n 'P1XO-2', 'P2XO-2', 'UniformCrossover-2', 'RingCrossover-2', 'FrontRearCrossover-2', \n 'P1XO-4', 'P2XO-4', 'UniformCrossover-4', 'RingCrossover-4', 'FrontRearCrossover-4', \n 'P1XO-8', 'P2XO-8', 'UniformCrossover-8', 'RingCrossover-8', 'FrontRearCrossover-8', \n ], \n [\"mean\", \"maxFitnessCount\"]\n ]\n result = pd.DataFrame(columns = benchmarks, index = pd.MultiIndex.from_product(iterables, names=[\"crossover\", \"stat\"]))\n result[\"OneMaxObjective\"] = [\n 5106.3, 100,\n 5353.5, 100,\n 1774.2, 100,\n 319.2, 100,\n 298.8, 100,\n \n 13778.4, 39,\n 13788.3, 37,\n 10364.4, 67,\n 553.8, 100,\n 504.6, 100,\n\n 15000, 0,\n 15000, 0,\n 14984.4, 2,\n 878.4, 100,\n 835.5, 100,\n ]\n result[\"TrapThreeObjective\"] = [\n 14726.4, 2,\n 14751, 2,\n 14852.7, 1,\n 378, 100,\n 348.9, 100,\n\n 15000, 0,\n 15000, 0,\n 15000, 0,\n 746.7, 100,\n 716.7, 100,\n\n 15000, 0,\n 15000, 0,\n 15000, 0,\n 1412.1, 100,\n 1314.3, 100,\n ]\n result[\"TrapFiveObjective\"] = [\n 78415.2, 2,\n 77626.4, 3,\n 80000, 0,\n 15968.8, 82,\n 1078.4, 100,\n\n 80000, 0,\n 80000, 0,\n 80000, 0,\n 31635.2, 63,\n 2058.4, 100,\n\n 80000, 0,\n 80000, 0,\n 80000, 0,\n 35763.2, 58,\n 3648, 100,\n ]\n result[\"ZeroMaxObjective\"] = [\n 5185.8, 100, \n 5100, 100, \n 1631.4, 100, \n 310.8, 100, \n 293.7, 100, \n\n 13781.4, 39, \n 13540.8, 39, \n 10086.6, 69, \n 536.4, 100, \n 506.1, 100, \n\n 15000, 0, \n 15000, 0, \n 14993.7, 1, \n 890.4, 100, \n 838.8, 100, \n ]\n return result\n# getRingCrossoverOriResult(\"RingCrossover\")\n# getHDGAOriResult(\"HighDimensionalGeneticAlgorithmToolboxCrossover\")\n# getCollectiveCrossoverOriResult(\"CollectiveCrossover\")\n# getHybridCrossoverOriResult(\"HybridCrossover\")\n# getFrontRearCrossoverOriResult(\"FrontRearCrossover\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecb607488027b215dc6a556101ed2dbb93ae9b32 | 3,202 | ipynb | Jupyter Notebook | feature_extraction/n_grams.ipynb | shwetankshrey/hindi-authorship-attribution | 034aa5ef0d25e0e49e20ab45035547820f06cb17 | [
"Beerware"
] | 1 | 2018-11-23T15:56:23.000Z | 2018-11-23T15:56:23.000Z | feature_extraction/n_grams.ipynb | shwetankshrey/hindi-authorship-attribution | 034aa5ef0d25e0e49e20ab45035547820f06cb17 | [
"Beerware"
] | null | null | null | feature_extraction/n_grams.ipynb | shwetankshrey/hindi-authorship-attribution | 034aa5ef0d25e0e49e20ab45035547820f06cb17 | [
"Beerware"
] | null | null | null | 28.336283 | 114 | 0.499063 | [
[
[
"import pickle\nimport nltk\n\nfiles = [\"bhairav\", \"dharamveer\", \"premchand\", \"sharatchandra\", \"vibhooti\"]\nn = 9\n\npiece_ngram_frequency = []\npiece_author = []\n\nfor file_name in files:\n pickle_file = open(\"../pickles/author_splits/\" + file_name + \".pkl\" , \"rb\")\n split_text = pickle.load(pickle_file)\n pickle_file.close()\n \n for text in split_text:\n text = text.replace('।','')\n text = text.replace('.','')\n text = text.replace(',','')\n text = text.replace(':','')\n text = text.replace(';','')\n text = text.replace('?','')\n text = text.replace('!','')\n text = text.replace('-','')\n text = text.replace(\"’\",\"\")\n text = text.replace(\"''\",\"\")\n text = text.replace('\"','')\n \n tokens = nltk.word_tokenize(text)\n grams = nltk.ngrams(tokens, n)\n piece_ngram_frequency.append(dict(nltk.FreqDist(grams)))\n piece_author.append(file_name)\n \ncorpus_ngram_frequency = {}\n\nfor ngf in piece_ngram_frequency:\n for ng in ngf.keys():\n if ng not in corpus_ngram_frequency:\n corpus_ngram_frequency[ng] = 0\n corpus_ngram_frequency[ng] += ngf[ng]\n\ncorpus_ngram_frequency = dict(sorted(corpus_ngram_frequency.items(), key=lambda x:x[1], reverse=True)[:500])",
"_____no_output_____"
],
[
"piece_frequencies = []\n\nfor ngf in piece_ngram_frequency:\n ng_freq_list = []\n for ng in corpus_ngram_frequency.keys():\n if ng in ngf:\n ng_freq_list.append(ngf[ng])\n else:\n ng_freq_list.append(0)\n piece_frequencies.append(ng_freq_list)\n\nfeature_vector = [piece_author, piece_frequencies]",
"_____no_output_____"
],
[
"pickle_file = open(\"../pickles/feature_vectors/n_grams/\" + str(n) + \"grams.pkl\" , \"wb\")\npickle.dump(feature_vector, pickle_file)\npickle_file.close()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecb6099638a651c9545a559844402009608ee3e7 | 160,337 | ipynb | Jupyter Notebook | Tutorial-Start_to_Finish-BSSNCurvilinear-Two_BHs_Collide-no_rfm_precompute.ipynb | philchang/nrpytutorial | a69d90777b2519192e3c53a129fe42827224faa3 | [
"BSD-2-Clause"
] | 66 | 2018-06-26T22:18:09.000Z | 2022-02-09T21:12:33.000Z | Tutorial-Start_to_Finish-BSSNCurvilinear-Two_BHs_Collide-no_rfm_precompute.ipynb | philchang/nrpytutorial | a69d90777b2519192e3c53a129fe42827224faa3 | [
"BSD-2-Clause"
] | 14 | 2020-02-13T16:09:29.000Z | 2021-11-12T14:59:59.000Z | Tutorial-Start_to_Finish-BSSNCurvilinear-Two_BHs_Collide-no_rfm_precompute.ipynb | philchang/nrpytutorial | a69d90777b2519192e3c53a129fe42827224faa3 | [
"BSD-2-Clause"
] | 30 | 2019-01-09T09:57:51.000Z | 2022-03-08T18:45:08.000Z | 92.20069 | 44,608 | 0.770733 | [
[
[
"<script async src=\"https://www.googletagmanager.com/gtag/js?id=UA-59152712-8\"></script>\n<script>\n window.dataLayer = window.dataLayer || [];\n function gtag(){dataLayer.push(arguments);}\n gtag('js', new Date());\n\n gtag('config', 'UA-59152712-8');\n</script>\n\n# Start-to-Finish Example: Head-On Black Hole Collision\n\n## Author: Zach Etienne\n### Formatting improvements courtesy Brandon Clark\n\n## This module implements a basic numerical relativity code to merge two black holes in *spherical coordinates*\n\n### Here we place the black holes initially on the $z$-axis, so the entire simulation is axisymmetric about the $\\phi$-axis. Not sampling in the $\\phi$ direction greatly speeds up the simulation.\n\n**Notebook Status:** <font color = green><b> Validated </b></font>\n\n**Validation Notes:** This module has been validated to exhibit convergence to zero of the Hamiltonian constraint violation at the expected order to the exact solution *after a short numerical evolution of the initial data* (see [plots at bottom](#convergence)), and all quantities have been validated against the [original SENR code](https://bitbucket.org/zach_etienne/nrpy).\n\n### NRPy+ Source Code for this module: \n* [BSSN/BrillLindquist.py](../edit/BSSN/BrillLindquist.py); [\\[**tutorial**\\]](Tutorial-ADM_Initial_Data-Brill-Lindquist.ipynb): Brill-Lindquist initial data; sets all ADM variables in Cartesian basis: \n* [BSSN/ADM_Exact_Spherical_or_Cartesian_to_BSSNCurvilinear.py](../edit/BSSN/ADM_Exact_Spherical_or_Cartesian_to_BSSNCurvilinear.py); [\\[**tutorial**\\]](Tutorial-ADM_Initial_Data-Converting_Exact_ADM_Spherical_or_Cartesian_to_BSSNCurvilinear.ipynb): Spherical/Cartesian ADM$\\to$Curvilinear BSSN converter function, for which exact expressions are given for ADM quantities.\n* [BSSN/BSSN_ID_function_string.py](../edit/BSSN/BSSN_ID_function_string.py): Sets up the C code string enabling initial data be set up in a point-by-point fashion\n* [BSSN/BSSN_constraints.py](../edit/BSSN/BSSN_constraints.py); [\\[**tutorial**\\]](Tutorial-BSSN_constraints.ipynb): Hamiltonian constraint in BSSN curvilinear basis/coordinates\n* [BSSN/BSSN_RHSs.py](../edit/BSSN/BSSN_RHSs.py); [\\[**tutorial**\\]](Tutorial-BSSN_time_evolution-BSSN_RHSs.ipynb): Generates the right-hand sides for the BSSN evolution equations in singular, curvilinear coordinates\n* [BSSN/BSSN_gauge_RHSs.py](../edit/BSSN/BSSN_gauge_RHSs.py); [\\[**tutorial**\\]](Tutorial-BSSN_time_evolution-BSSN_gauge_RHSs.ipynb): Generates the right-hand sides for the BSSN gauge evolution equations in singular, curvilinear coordinates\n\n## Introduction:\nHere we use NRPy+ to generate the C source code necessary to set up initial data for two black holes (Brill-Lindquist, [Brill & Lindquist, Phys. Rev. 131, 471, 1963](https://journals.aps.org/pr/abstract/10.1103/PhysRev.131.471); see also Eq. 1 of [Brandt & Brügmann, arXiv:gr-qc/9711015v1](https://arxiv.org/pdf/gr-qc/9711015v1.pdf)). Then we use it to generate the RHS expressions for [Method of Lines](https://reference.wolfram.com/language/tutorial/NDSolveMethodOfLines.html) time integration based on an [explicit Runge-Kutta fourth-order scheme](https://en.wikipedia.org/wiki/Runge%E2%80%93Kutta_methods) (RK4 is chosen below, but multiple options exist). \n\nThe entire algorithm is outlined as follows, with links to the relevant NRPy+ tutorial notebooks listed at each step:\n\n1. Allocate memory for gridfunctions, including temporary storage for the Method of Lines time integration\n * [**NRPy+ tutorial on Method of Lines algorithm**](Tutorial-Method_of_Lines-C_Code_Generation.ipynb).\n1. Set gridfunction values to initial data \n * [**NRPy+ tutorial on Brill-Lindquist initial data**](Tutorial-ADM_Initial_Data-Brill-Lindquist.ipynb)\n * [**NRPy+ tutorial on validating Brill-Lindquist initial data**](Tutorial-Start_to_Finish-BSSNCurvilinear-Setting_up_Exact_Initial_Data.ipynb).\n1. Next, integrate the initial data forward in time using the Method of Lines coupled to a Runge-Kutta explicit timestepping algorithm:\n 1. At the start of each iteration in time, output the Hamiltonian constraint violation \n * [**NRPy+ tutorial on BSSN constraints**](Tutorial-BSSN_constraints.ipynb).\n 1. At each RK time substep, do the following:\n 1. Evaluate BSSN RHS expressions \n * [**NRPy+ tutorial on BSSN right-hand sides**](Tutorial-BSSN_time_evolution-BSSN_RHSs.ipynb) ([**BSSN Introduction Notebook**](Tutorial-BSSN_formulation.ipynb))\n * [**NRPy+ tutorial on BSSN gauge condition right-hand sides**](Tutorial-BSSN_time_evolution-BSSN_gauge_RHSs.ipynb) \n 1. Apply singular, curvilinear coordinate boundary conditions [*a la* the SENR/NRPy+ paper](https://arxiv.org/abs/1712.07658)\n * [**NRPy+ tutorial on setting up singular, curvilinear boundary conditions**](Tutorial-Start_to_Finish-Curvilinear_BCs.ipynb)\n 1. Enforce constraint on conformal 3-metric: $\\det{\\bar{\\gamma}_{ij}}=\\det{\\hat{\\gamma}_{ij}}$ \n * [**NRPy+ tutorial on enforcing $\\det{\\bar{\\gamma}_{ij}}=\\det{\\hat{\\gamma}_{ij}}$ constraint**](Tutorial-BSSN_enforcing_determinant_gammabar_equals_gammahat_constraint.ipynb)\n1. Repeat above steps at two numerical resolutions to confirm convergence to zero.",
"_____no_output_____"
],
[
"<a id='toc'></a>\n\n# Table of Contents\n$$\\label{toc}$$\n\nThis notebook is organized as follows\n\n1. [Step 1](#initializenrpy): Set core NRPy+ parameters for numerical grids and reference metric\n 1. [Step 1.a](#cfl) Output needed C code for finding the minimum proper distance between grid points, needed for [CFL](https://en.wikipedia.org/w/index.php?title=Courant%E2%80%93Friedrichs%E2%80%93Lewy_condition&oldid=806430673)-limited timestep\n1. [Step 2](#adm_id): Import Brill-Lindquist ADM initial data C function from the [`BSSN.BrillLindquist`](../edit/BSSN/BrillLindquist.py) NRPy+ module\n1. [Step 3](#bssn): Output C code for BSSN spacetime solve\n 1. [Step 3.a](#bssnrhs): Output C code for BSSN RHS expressions\n 1. [Step 3.b](#hamconstraint): Output C code for Hamiltonian constraint\n 1. [Step 3.c](#enforce3metric): Enforce conformal 3-metric $\\det{\\bar{\\gamma}_{ij}}=\\det{\\hat{\\gamma}_{ij}}$ constraint\n 1. [Step 3.d](#ccodegen): Generate C code kernels for BSSN expressions, in parallel if possible\n 1. [Step 3.e](#cparams_rfm_and_domainsize): Output C codes needed for declaring and setting Cparameters; also set `free_parameters.h`\n1. [Step 4](#bc_functs): Set up boundary condition functions for chosen singular, curvilinear coordinate system\n1. [Step 5](#mainc): `BrillLindquist_Playground.c`: The Main C Code\n1. [Step 6](#compileexec): Compile generated C codes & perform the black hole collision calculation\n1. [Step 7](#visualize): Visualize the output!\n 1. [Step 7.a](#installdownload): Install `scipy` and download `ffmpeg` if they are not yet installed/downloaded\n 1. [Step 7.b](#genimages): Generate images for visualization animation\n 1. [Step 7.c](#genvideo): Generate visualization animation\n1. [Step 8](#convergence): Plot the numerical error, and confirm that it converges to zero with increasing numerical resolution (sampling)\n1. [Step 9](#latex_pdf_output): Output this notebook to $\\LaTeX$-formatted PDF file",
"_____no_output_____"
],
[
"<a id='initializenrpy'></a>\n\n# Step 1: Set core NRPy+ parameters for numerical grids and reference metric \\[Back to [top](#toc)\\]\n$$\\label{initializenrpy}$$",
"_____no_output_____"
]
],
[
[
"# Step P1: Import needed NRPy+ core modules:\nfrom outputC import lhrh,outCfunction,outC_function_dict # NRPy+: Core C code output module\nimport finite_difference as fin # NRPy+: Finite difference C code generation module\nimport NRPy_param_funcs as par # NRPy+: Parameter interface\nimport grid as gri # NRPy+: Functions having to do with numerical grids\nimport indexedexp as ixp # NRPy+: Symbolic indexed expression (e.g., tensors, vectors, etc.) support\nimport reference_metric as rfm # NRPy+: Reference metric support\nimport cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface\nimport shutil, os, sys, time # Standard Python modules for multiplatform OS-level functions, benchmarking\nimport pickle # Standard Python module for bytewise transfer of data between modules\n\n# Step P2: Create C code output directory:\nCcodesdir = os.path.join(\"BSSN_Two_BHs_Collide_Ccodes/\")\n# First remove C code output directory if it exists\n# Courtesy https://stackoverflow.com/questions/303200/how-do-i-remove-delete-a-folder-that-is-not-empty\n# !rm -r ScalarWaveCurvilinear_Playground_Ccodes\nshutil.rmtree(Ccodesdir, ignore_errors=True)\n# Then create a fresh directory\ncmd.mkdir(Ccodesdir)\n\n# Step P3: Create executable output directory:\noutdir = os.path.join(Ccodesdir,\"output/\")\ncmd.mkdir(outdir)\n\n# Step 1: Set the spatial dimension parameter\n# to three (BSSN is a 3+1 decomposition\n# of Einstein's equations), and then read\n# the parameter as DIM.\npar.set_parval_from_str(\"grid::DIM\",3)\nDIM = par.parval_from_str(\"grid::DIM\")\n\n# Step 1.a: Enable SIMD-optimized code?\n# I.e., generate BSSN and Ricci C code kernels using SIMD-vectorized\n# compiler intrinsics, which *greatly improve the code's performance*,\n# though at the expense of making the C-code kernels less\n# human-readable.\n# * Important note in case you wish to modify the BSSN/Ricci kernels\n# here by adding expressions containing transcendental functions\n# (e.g., certain scalar fields):\n# Note that SIMD-based transcendental function intrinsics are not\n# supported by the default installation of gcc or clang (you will\n# need to use e.g., the SLEEF library from sleef.org, for this\n# purpose). The Intel compiler suite does support these intrinsics\n# however without the need for external libraries.\nenable_SIMD = False\n\n# Step 2: Set some core parameters, including CoordSystem MoL timestepping algorithm,\n# FD order, floating point precision, and CFL factor:\n# Choices are: Spherical, SinhSpherical, SinhSphericalv2, Cylindrical, SinhCylindrical,\n# SymTP, SinhSymTP\nCoordSystem = \"Spherical\"\n\n# Step 2.a: Set defaults for Coordinate system parameters.\n# These are perhaps the most commonly adjusted parameters,\n# so we enable modifications at this high level.\n\n# domain_size sets the default value for:\n# * Spherical's params.RMAX\n# * SinhSpherical*'s params.AMAX\n# * Cartesians*'s -params.{x,y,z}min & .{x,y,z}max\n# * Cylindrical's -params.ZMIN & .{Z,RHO}MAX\n# * SinhCylindrical's params.AMPL{RHO,Z}\n# * *SymTP's params.AMAX\ndomain_size = 7.5 # Needed for all coordinate systems.\n\n# sinh_width sets the default value for:\n# * SinhSpherical's params.SINHW\n# * SinhCylindrical's params.SINHW{RHO,Z}\n# * SinhSymTP's params.SINHWAA\nsinh_width = 0.4 # If Sinh* coordinates chosen\n\n# sinhv2_const_dr sets the default value for:\n# * SinhSphericalv2's params.const_dr\n# * SinhCylindricalv2's params.const_d{rho,z}\nsinhv2_const_dr = 0.05 # If Sinh*v2 coordinates chosen\n\n# SymTP_bScale sets the default value for:\n# * SinhSymTP's params.bScale\nSymTP_bScale = 0.5 # If SymTP chosen\n\n# Step 2.b: Set the order of spatial and temporal derivatives;\n# the core data type, and the CFL factor.\n# RK_method choices include: Euler, \"RK2 Heun\", \"RK2 MP\", \"RK2 Ralston\", RK3, \"RK3 Heun\", \"RK3 Ralston\",\n# SSPRK3, RK4, DP5, DP5alt, CK5, DP6, L6, DP8\nRK_method = \"RK4\"\nFD_order = 4 # Finite difference order: even numbers only, starting with 2. 12 is generally unstable\nREAL = \"double\" # Best to use double here.\ndefault_CFL_FACTOR= 0.5 # (GETS OVERWRITTEN WHEN EXECUTED.) In pure axisymmetry (symmetry_axes = 2 below) 1.0 works fine. Otherwise 0.5 or lower.\n\n# Step 3: Generate Runge-Kutta-based (RK-based) timestepping code.\n# As described above the Table of Contents, this is a 3-step process:\n# 3.A: Evaluate RHSs (RHS_string)\n# 3.B: Apply boundary conditions (post_RHS_string, pt 1)\n# 3.C: Enforce det(gammabar) = det(gammahat) constraint (post_RHS_string, pt 2)\nimport MoLtimestepping.C_Code_Generation as MoL\nfrom MoLtimestepping.RK_Butcher_Table_Dictionary import Butcher_dict\nRK_order = Butcher_dict[RK_method][1]\ncmd.mkdir(os.path.join(Ccodesdir,\"MoLtimestepping/\"))\nMoL.MoL_C_Code_Generation(RK_method,\n RHS_string = \"\"\"\nRicci_eval(¶ms, xx, RK_INPUT_GFS, auxevol_gfs);\nrhs_eval(¶ms, xx, auxevol_gfs, RK_INPUT_GFS, RK_OUTPUT_GFS);\"\"\",\n post_RHS_string = \"\"\"\napply_bcs_curvilinear(¶ms, &bcstruct, NUM_EVOL_GFS, evol_gf_parity, RK_OUTPUT_GFS);\nenforce_detgammahat_constraint(¶ms, xx, RK_OUTPUT_GFS);\\n\"\"\",\n outdir = os.path.join(Ccodesdir,\"MoLtimestepping/\"))\n\n# Step 4: Set the coordinate system for the numerical grid\npar.set_parval_from_str(\"reference_metric::CoordSystem\",CoordSystem)\nrfm.reference_metric() # Create ReU, ReDD needed for rescaling B-L initial data, generating BSSN RHSs, etc.\n\n# Step 5: Set the finite differencing order to FD_order (set above).\npar.set_parval_from_str(\"finite_difference::FD_CENTDERIVS_ORDER\", FD_order)\nenable_FD_functions = True\npar.set_parval_from_str(\"finite_difference::enable_FD_functions\", enable_FD_functions)\n\n# Step 6: If enable_SIMD==True, then copy SIMD/SIMD_intrinsics.h to $Ccodesdir/SIMD/SIMD_intrinsics.h\n# Otherwise just paste a #define SIMD_IS_DISABLED to that file.\ncmd.mkdir(os.path.join(Ccodesdir,\"SIMD\"))\nif enable_SIMD == True:\n shutil.copy(os.path.join(\"SIMD\",\"SIMD_intrinsics.h\"),os.path.join(Ccodesdir,\"SIMD/\"))\nelse:\n with open(os.path.join(Ccodesdir,\"SIMD\",\"SIMD_intrinsics.h\"), \"w\") as file:\n file.write(\"#define SIMD_IS_DISABLED\\n\")\n\n# Step 7: Set the direction=2 (phi) axis to be the symmetry axis; i.e.,\n# axis \"2\", corresponding to the i2 direction.\n# This sets all spatial derivatives in the phi direction to zero.\npar.set_parval_from_str(\"indexedexp::symmetry_axes\",\"2\")",
"_____no_output_____"
]
],
[
[
"<a id='cfl'></a>\n\n## Step 1.a: Output needed C code for finding the minimum proper distance between grid points, needed for [CFL](https://en.wikipedia.org/w/index.php?title=Courant%E2%80%93Friedrichs%E2%80%93Lewy_condition&oldid=806430673)-limited timestep \\[Back to [top](#toc)\\]\n$$\\label{cfl}$$\n\nIn order for our explicit-timestepping numerical solution to the scalar wave equation to be stable, it must satisfy the [CFL](https://en.wikipedia.org/w/index.php?title=Courant%E2%80%93Friedrichs%E2%80%93Lewy_condition&oldid=806430673) condition:\n$$\n\\Delta t \\le \\frac{\\min(ds_i)}{c},\n$$\nwhere $c$ is the wavespeed, and\n$$ds_i = h_i \\Delta x^i$$ \nis the proper distance between neighboring gridpoints in the $i$th direction (in 3D, there are 3 directions), $h_i$ is the $i$th reference metric scale factor, and $\\Delta x^i$ is the uniform grid spacing in the $i$th direction:",
"_____no_output_____"
]
],
[
[
"# Output the find_timestep() function to a C file.\nrfm.out_timestep_func_to_file(os.path.join(Ccodesdir,\"find_timestep.h\"))",
"_____no_output_____"
],
[
"# In the parallel C codegen below, the\ndef pickled_outC_function_dict(outC_function_dict):\n outstr = []\n outstr.append(pickle.dumps(len(outC_function_dict)))\n for Cfuncname, Cfunc in outC_function_dict.items():\n outstr.append(pickle.dumps(Cfuncname))\n outstr.append(pickle.dumps(Cfunc))\n return outstr",
"_____no_output_____"
]
],
[
[
"<a id='adm_id'></a>\n\n# Step 2: Import Brill-Lindquist ADM initial data C function from the [`BSSN.BrillLindquist`](../edit/BSSN/BrillLindquist.py) NRPy+ module \\[Back to [top](#toc)\\]\n$$\\label{adm_id}$$\n\nThe [`BSSN.BrillLindquist`](../edit/BSSN/BrillLindquist.py) NRPy+ module does the following:\n\n1. Set up Brill-Lindquist initial data [ADM](https://en.wikipedia.org/wiki/ADM_formalism) quantities in the **Cartesian basis**, as [documented here](Tutorial-ADM_Initial_Data-Brill-Lindquist.ipynb). \n1. Convert the ADM **Cartesian quantities** to **BSSN quantities in the desired Curvilinear basis** (set by reference_metric::CoordSystem), as [documented here](Tutorial-ADM_Initial_Data-Converting_ADMCartesian_to_BSSNCurvilinear.ipynb).\n1. Sets up the standardized C function for setting all BSSN Curvilinear gridfunctions in a pointwise fashion, as [written here](../edit/BSSN/BSSN_ID_function_string.py), and returns the C function as a Python string.",
"_____no_output_____"
]
],
[
[
"import BSSN.BrillLindquist as bl\ndef BrillLindquistID():\n print(\"Generating optimized C code for Brill-Lindquist initial data. May take a while, depending on CoordSystem.\")\n start = time.time()\n\n bl.BrillLindquist() # Registers ID C function in dictionary, used below to output to file.\n with open(os.path.join(Ccodesdir,\"initial_data.h\"),\"w\") as file:\n file.write(outC_function_dict[\"initial_data\"])\n end = time.time()\n print(\"(BENCH) Finished BL initial data codegen in \"+str(end-start)+\" seconds.\")\n return pickled_outC_function_dict(outC_function_dict)",
"_____no_output_____"
]
],
[
[
"<a id='bssn'></a>\n\n# Step 3: Output C code for BSSN spacetime solve \\[Back to [top](#toc)\\]\n$$\\label{bssn}$$\n\n<a id='bssnrhs'></a>\n\n## Step 3.a: Output C code for BSSN RHS expressions \\[Back to [top](#toc)\\]\n$$\\label{bssnrhs}$$",
"_____no_output_____"
]
],
[
[
"import BSSN.BSSN_RHSs as rhs\nimport BSSN.BSSN_gauge_RHSs as gaugerhs\n# Set the *covariant*, second-order Gamma-driving shift condition\npar.set_parval_from_str(\"BSSN.BSSN_gauge_RHSs::ShiftEvolutionOption\", \"GammaDriving2ndOrder_Covariant\")\n\nprint(\"Generating symbolic expressions for BSSN RHSs...\")\nstart = time.time()\n# Enable rfm_precompute infrastructure, which results in\n# BSSN RHSs that are free of transcendental functions,\n# even in curvilinear coordinates, so long as\n# ConformalFactor is set to \"W\" (default).\ncmd.mkdir(os.path.join(Ccodesdir,\"rfm_files/\"))\npar.set_parval_from_str(\"reference_metric::enable_rfm_precompute\",\"False\")\npar.set_parval_from_str(\"reference_metric::rfm_precompute_Ccode_outdir\",os.path.join(Ccodesdir,\"rfm_files/\"))\n\n# Evaluate BSSN + BSSN gauge RHSs with rfm_precompute enabled:\nimport BSSN.BSSN_quantities as Bq\npar.set_parval_from_str(\"BSSN.BSSN_quantities::LeaveRicciSymbolic\",\"True\")\n\nrhs.BSSN_RHSs()\ngaugerhs.BSSN_gauge_RHSs()\n\n# We use betaU as our upwinding control vector:\nBq.BSSN_basic_tensors()\nbetaU = Bq.betaU\n\nimport BSSN.Enforce_Detgammahat_Constraint as EGC\nenforce_detg_constraint_symb_expressions = EGC.Enforce_Detgammahat_Constraint_symb_expressions()\n\n# Next compute Ricci tensor\npar.set_parval_from_str(\"BSSN.BSSN_quantities::LeaveRicciSymbolic\",\"False\")\nBq.RicciBar__gammabarDD_dHatD__DGammaUDD__DGammaU()\n\n# Now register the Hamiltonian as a gridfunction.\nH = gri.register_gridfunctions(\"AUX\",\"H\")\n# Then define the Hamiltonian constraint and output the optimized C code.\nimport BSSN.BSSN_constraints as bssncon\nbssncon.BSSN_constraints(add_T4UUmunu_source_terms=False)\n\n# Now that we are finished with all the rfm hatted\n# quantities in generic precomputed functional\n# form, let's restore them to their closed-\n# form expressions.\npar.set_parval_from_str(\"reference_metric::enable_rfm_precompute\",\"False\") # Reset to False to disable rfm_precompute.\nrfm.ref_metric__hatted_quantities()\nend = time.time()\nprint(\"(BENCH) Finished BSSN symbolic expressions in \"+str(end-start)+\" seconds.\")\n\nincludes = None\nif enable_FD_functions:\n includes = [\"finite_difference_functions.h\"]\n\ndef BSSN_RHSs():\n print(\"Generating C code for BSSN RHSs in \"+par.parval_from_str(\"reference_metric::CoordSystem\")+\" coordinates.\")\n start = time.time()\n\n # Construct the left-hand sides and right-hand-side expressions for all BSSN RHSs\n lhs_names = [ \"alpha\", \"cf\", \"trK\"]\n rhs_exprs = [gaugerhs.alpha_rhs, rhs.cf_rhs, rhs.trK_rhs]\n for i in range(3):\n lhs_names.append( \"betU\"+str(i))\n rhs_exprs.append(gaugerhs.bet_rhsU[i])\n lhs_names.append( \"lambdaU\"+str(i))\n rhs_exprs.append(rhs.lambda_rhsU[i])\n lhs_names.append( \"vetU\"+str(i))\n rhs_exprs.append(gaugerhs.vet_rhsU[i])\n for j in range(i,3):\n lhs_names.append( \"aDD\"+str(i)+str(j))\n rhs_exprs.append(rhs.a_rhsDD[i][j])\n lhs_names.append( \"hDD\"+str(i)+str(j))\n rhs_exprs.append(rhs.h_rhsDD[i][j])\n\n # Sort the lhss list alphabetically, and rhss to match.\n # This ensures the RHSs are evaluated in the same order\n # they're allocated in memory:\n lhs_names,rhs_exprs = [list(x) for x in zip(*sorted(zip(lhs_names,rhs_exprs), key=lambda pair: pair[0]))]\n\n # Declare the list of lhrh's\n BSSN_evol_rhss = []\n for var in range(len(lhs_names)):\n BSSN_evol_rhss.append(lhrh(lhs=gri.gfaccess(\"rhs_gfs\",lhs_names[var]),rhs=rhs_exprs[var]))\n\n # Set up the C function for the BSSN RHSs\n # Set outputC and loop parameters for BSSN_RHSs C function.\n outC_params = \"outCverbose=False\"\n loopoptions = \"InteriorPoints,Read_xxs\"\n if enable_SIMD == True:\n loopoptions += \",enable_SIMD\"\n outC_params += \",enable_SIMD=True\"\n desc=\"Evaluate the BSSN RHSs\"\n name=\"rhs_eval\"\n outCfunction(\n outfile = os.path.join(Ccodesdir,name+\".h\"), includes=includes, desc=desc, name=name,\n params = \"\"\"const paramstruct *restrict params,REAL *restrict xx[3],\n const REAL *restrict auxevol_gfs,const REAL *restrict in_gfs,REAL *restrict rhs_gfs\"\"\",\n body = fin.FD_outputC(\"returnstring\",BSSN_evol_rhss, params=outC_params,\n upwindcontrolvec=betaU),\n loopopts = loopoptions)\n end = time.time()\n print(\"(BENCH) Finished BSSN_RHS C codegen in \" + str(end - start) + \" seconds.\")\n return pickled_outC_function_dict(outC_function_dict)\n\ndef Ricci():\n print(\"Generating C code for Ricci tensor in \"+par.parval_from_str(\"reference_metric::CoordSystem\")+\" coordinates.\")\n start = time.time()\n\n # Set up the C function for the Ricci tensor\n # Set outputC and loop parameters for Ricci tensor function.\n outC_params = \"outCverbose=False\"\n loopoptions = \"InteriorPoints,Read_xxs\"\n if enable_SIMD == True:\n loopoptions += \",enable_SIMD\"\n outC_params += \",enable_SIMD=True\"\n desc=\"Evaluate the Ricci tensor\"\n name=\"Ricci_eval\"\n outCfunction(\n outfile = os.path.join(Ccodesdir,name+\".h\"), includes=includes, desc=desc, name=name,\n params = \"\"\"const paramstruct *restrict params,REAL *restrict xx[3],\n const REAL *restrict in_gfs,REAL *restrict auxevol_gfs\"\"\",\n body = fin.FD_outputC(\"returnstring\",\n [lhrh(lhs=gri.gfaccess(\"auxevol_gfs\",\"RbarDD00\"),rhs=Bq.RbarDD[0][0]),\n lhrh(lhs=gri.gfaccess(\"auxevol_gfs\",\"RbarDD01\"),rhs=Bq.RbarDD[0][1]),\n lhrh(lhs=gri.gfaccess(\"auxevol_gfs\",\"RbarDD02\"),rhs=Bq.RbarDD[0][2]),\n lhrh(lhs=gri.gfaccess(\"auxevol_gfs\",\"RbarDD11\"),rhs=Bq.RbarDD[1][1]),\n lhrh(lhs=gri.gfaccess(\"auxevol_gfs\",\"RbarDD12\"),rhs=Bq.RbarDD[1][2]),\n lhrh(lhs=gri.gfaccess(\"auxevol_gfs\",\"RbarDD22\"),rhs=Bq.RbarDD[2][2])],\n params=outC_params),\n loopopts = loopoptions)\n end = time.time()\n print(\"(BENCH) Finished Ricci C codegen in \" + str(end - start) + \" seconds.\")\n return pickled_outC_function_dict(outC_function_dict)",
"Generating symbolic expressions for BSSN RHSs...\n(BENCH) Finished BSSN symbolic expressions in 1.8061211109161377 seconds.\n"
]
],
[
[
"<a id='hamconstraint'></a>\n\n## Step 3.b: Output C code for Hamiltonian constraint \\[Back to [top](#toc)\\]\n$$\\label{hamconstraint}$$\n\nNext output the C code for evaluating the Hamiltonian constraint [(**Tutorial**)](Tutorial-BSSN_constraints.ipynb). In the absence of numerical error, this constraint should evaluate to zero. However it does not due to numerical (typically truncation and roundoff) error. We will therefore measure the Hamiltonian constraint violation to gauge the accuracy of our simulation, and, ultimately determine whether errors are dominated by numerical finite differencing (truncation) error as expected.",
"_____no_output_____"
]
],
[
[
"def Hamiltonian():\n start = time.time()\n print(\"Generating optimized C code for Hamiltonian constraint. May take a while, depending on CoordSystem.\")\n # Set up the C function for the Hamiltonian RHS\n desc=\"Evaluate the Hamiltonian constraint\"\n name=\"Hamiltonian_constraint\"\n outCfunction(\n outfile = os.path.join(Ccodesdir,name+\".h\"), includes=includes, desc=desc, name=name,\n params = \"\"\"const paramstruct *restrict params,REAL *restrict xx[3],\n REAL *restrict in_gfs, REAL *restrict aux_gfs\"\"\",\n body = fin.FD_outputC(\"returnstring\",lhrh(lhs=gri.gfaccess(\"aux_gfs\", \"H\"), rhs=bssncon.H),\n params=\"outCverbose=False\"),\n loopopts = \"InteriorPoints,Read_xxs\")\n\n end = time.time()\n print(\"(BENCH) Finished Hamiltonian C codegen in \" + str(end - start) + \" seconds.\")\n return pickled_outC_function_dict(outC_function_dict)",
"_____no_output_____"
]
],
[
[
"<a id='enforce3metric'></a>\n\n## Step 3.c: Enforce conformal 3-metric $\\det{\\bar{\\gamma}_{ij}}=\\det{\\hat{\\gamma}_{ij}}$ constraint \\[Back to [top](#toc)\\]\n$$\\label{enforce3metric}$$\n\nThen enforce conformal 3-metric $\\det{\\bar{\\gamma}_{ij}}=\\det{\\hat{\\gamma}_{ij}}$ constraint (Eq. 53 of [Ruchlin, Etienne, and Baumgarte (2018)](https://arxiv.org/abs/1712.07658)), as [documented in the corresponding NRPy+ tutorial notebook](Tutorial-BSSN_enforcing_determinant_gammabar_equals_gammahat_constraint.ipynb)\n\nApplying curvilinear boundary conditions should affect the initial data at the outer boundary, and will in general cause the $\\det{\\bar{\\gamma}_{ij}}=\\det{\\hat{\\gamma}_{ij}}$ constraint to be violated there. Thus after we apply these boundary conditions, we must always call the routine for enforcing the $\\det{\\bar{\\gamma}_{ij}}=\\det{\\hat{\\gamma}_{ij}}$ constraint:",
"_____no_output_____"
]
],
[
[
"def gammadet():\n start = time.time()\n print(\"Generating optimized C code for gamma constraint. May take a while, depending on CoordSystem.\")\n\n # Set up the C function for the det(gammahat) = det(gammabar)\n EGC.output_Enforce_Detgammahat_Constraint_Ccode(Ccodesdir,\n exprs=enforce_detg_constraint_symb_expressions, Read_xxs=True)\n end = time.time()\n print(\"(BENCH) Finished gamma constraint C codegen in \" + str(end - start) + \" seconds.\")\n return pickled_outC_function_dict(outC_function_dict)",
"_____no_output_____"
]
],
[
[
"<a id='ccodegen'></a>\n\n## Step 3.d: Generate C code kernels for BSSN expressions, in parallel if possible \\[Back to [top](#toc)\\]\n$$\\label{ccodegen}$$",
"_____no_output_____"
]
],
[
[
"# Step 3.d: Generate C code kernels for BSSN expressions, in parallel if possible;\n\n# Step 3.d.i: Create a list of functions we wish to evaluate in parallel (if possible)\nfuncs = [BrillLindquistID,BSSN_RHSs,Ricci,Hamiltonian,gammadet]\n# pickled_outC_func_dict stores outC_function_dict from all\n# the subprocesses in the following parallel codegen\npickled_outC_func_dict = []\n\ntry:\n if os.name == 'nt':\n # It's a mess to get working in Windows, so we don't bother. :/\n # https://medium.com/@grvsinghal/speed-up-your-python-code-using-multiprocessing-on-windows-and-jupyter-or-ipython-2714b49d6fac\n raise Exception(\"Parallel codegen currently not available in certain environments, e.g., Windows\")\n\n # Step 3.d.ii: Import the multiprocessing module.\n import multiprocessing\n\n # Step 3.d.iii: Define master functions for parallelization.\n # Note that lambdifying this doesn't work in Python 3\n def master_func(arg):\n return funcs[arg]()\n # Step 3.d.iv: Evaluate list of functions in parallel if possible;\n # otherwise fallback to serial evaluation:\n pool = multiprocessing.Pool()\n pickled_outC_func_dict.append(pool.map(master_func,range(len(funcs))))\n pool.terminate()\n pool.join()\nexcept:\n # Steps 3.d.ii-iv, alternate: As fallback, evaluate functions in serial.\n # This will happen on Android and Windows systems\n for func in funcs:\n func()\n pickled_outC_func_dict = [] # Reset, as pickling/unpickling unnecessary for serial codegen (see next line)\n\n# Step 3.d.v Output functions for computing all finite-difference stencils\nif enable_FD_functions and len(pickled_outC_func_dict)>0:\n # First unpickle pickled_outC_func_dict\n outCfunc_dict = {}\n for WhichFunc in pickled_outC_func_dict[0]:\n i=0\n num_elements = pickle.loads(WhichFunc[i]); i+=1\n for lst in range(num_elements):\n funcname = pickle.loads(WhichFunc[i+0])\n funcbody = pickle.loads(WhichFunc[i+1]) ; i+=2\n outCfunc_dict[funcname] = funcbody\n # Then store the unpickled outCfunc_dict to outputC's outC_function_dict\n for key, item in outCfunc_dict.items():\n outC_function_dict[key] = item\nif enable_FD_functions:\n # Finally generate finite_difference_functions.h\n fin.output_finite_difference_functions_h(path=Ccodesdir)",
"Generating optimized C code for Brill-Lindquist initial data. May take a while, depending on CoordSystem.Generating C code for BSSN RHSs in Spherical coordinates.Generating optimized C code for Hamiltonian constraint. May take a while, depending on CoordSystem.Generating C code for Ricci tensor in Spherical coordinates.Generating optimized C code for gamma constraint. May take a while, depending on CoordSystem.\n\n\n\n\nOutput C function enforce_detgammahat_constraint() to file BSSN_Two_BHs_Collide_Ccodes/enforce_detgammahat_constraint.h\n(BENCH) Finished gamma constraint C codegen in 0.0787210464477539 seconds.\nOutput C function rhs_eval() to file BSSN_Two_BHs_Collide_Ccodes/rhs_eval.h\n(BENCH) Finished BSSN_RHS C codegen in 3.745373249053955 seconds.\n(BENCH) Finished BL initial data codegen in 7.051412105560303 seconds.\nOutput C function Ricci_eval() to file BSSN_Two_BHs_Collide_Ccodes/Ricci_eval.h\n(BENCH) Finished Ricci C codegen in 7.201741456985474 seconds.\nOutput C function Hamiltonian_constraint() to file BSSN_Two_BHs_Collide_Ccodes/Hamiltonian_constraint.h\n(BENCH) Finished Hamiltonian C codegen in 25.364696264266968 seconds.\n"
]
],
[
[
"<a id='cparams_rfm_and_domainsize'></a>\n\n## Step 3.e: Output C codes needed for declaring and setting Cparameters; also set `free_parameters.h` \\[Back to [top](#toc)\\]\n$$\\label{cparams_rfm_and_domainsize}$$\n\nBased on declared NRPy+ Cparameters, first we generate `declare_Cparameters_struct.h`, `set_Cparameters_default.h`, and `set_Cparameters[-SIMD].h`.\n\nThen we output `free_parameters.h`, which sets initial data parameters, as well as grid domain & reference metric parameters, applying `domain_size` and `sinh_width`/`SymTP_bScale` (if applicable) as set above",
"_____no_output_____"
]
],
[
[
"# Step 3.e: Output C codes needed for declaring and setting Cparameters; also set free_parameters.h\n\n# Step 3.e.i: Generate declare_Cparameters_struct.h, set_Cparameters_default.h, and set_Cparameters[-SIMD].h\npar.generate_Cparameters_Ccodes(os.path.join(Ccodesdir))\n\n# Step 3.e.ii: Set free_parameters.h\nwith open(os.path.join(Ccodesdir,\"free_parameters.h\"),\"w\") as file:\n file.write(\"\"\"\n// Set free-parameter values.\n\n// Set free-parameter values for BSSN evolution:\nparams.eta = 1.0;\n\n// Set free parameters for the (Brill-Lindquist) initial data\nparams.BH1_posn_x = 0.0; params.BH1_posn_y = 0.0; params.BH1_posn_z =+0.5;\nparams.BH2_posn_x = 0.0; params.BH2_posn_y = 0.0; params.BH2_posn_z =-0.5;\nparams.BH1_mass = 0.5; params.BH2_mass = 0.5;\\n\"\"\")\n\n# Append to $Ccodesdir/free_parameters.h reference metric parameters based on generic\n# domain_size,sinh_width,sinhv2_const_dr,SymTP_bScale,\n# parameters set above.\nrfm.out_default_free_parameters_for_rfm(os.path.join(Ccodesdir,\"free_parameters.h\"),\n domain_size,sinh_width,sinhv2_const_dr,SymTP_bScale)\n\n# Step 3.e.iii: Generate set_Nxx_dxx_invdx_params__and__xx.h:\nrfm.set_Nxx_dxx_invdx_params__and__xx_h(Ccodesdir)\n\n# Step 3.e.iv: Generate xx_to_Cart.h, which contains xx_to_Cart() for\n# (the mapping from xx->Cartesian) for the chosen\n# CoordSystem:\nrfm.xx_to_Cart_h(\"xx_to_Cart\",\"./set_Cparameters.h\",os.path.join(Ccodesdir,\"xx_to_Cart.h\"))\n\n# Step 3.e.v: Generate declare_Cparameters_struct.h, set_Cparameters_default.h, and set_Cparameters[-SIMD].h\npar.generate_Cparameters_Ccodes(os.path.join(Ccodesdir))",
"_____no_output_____"
]
],
[
[
"<a id='bc_functs'></a>\n\n# Step 4: Set up boundary condition functions for chosen singular, curvilinear coordinate system \\[Back to [top](#toc)\\]\n$$\\label{bc_functs}$$\n\nNext apply singular, curvilinear coordinate boundary conditions [as documented in the corresponding NRPy+ tutorial notebook](Tutorial-Start_to_Finish-Curvilinear_BCs.ipynb)",
"_____no_output_____"
]
],
[
[
"import CurviBoundaryConditions.CurviBoundaryConditions as cbcs\ncbcs.Set_up_CurviBoundaryConditions(os.path.join(Ccodesdir,\"boundary_conditions/\"),Cparamspath=os.path.join(\"../\"))",
"Wrote to file \"BSSN_Two_BHs_Collide_Ccodes/boundary_conditions/parity_conditions_symbolic_dot_products.h\"\nEvolved parity: ( aDD00:4, aDD01:5, aDD02:6, aDD11:7, aDD12:8, aDD22:9,\n alpha:0, betU0:1, betU1:2, betU2:3, cf:0, hDD00:4, hDD01:5, hDD02:6,\n hDD11:7, hDD12:8, hDD22:9, lambdaU0:1, lambdaU1:2, lambdaU2:3, trK:0,\n vetU0:1, vetU1:2, vetU2:3 )\nAuxiliary parity: ( H:0 )\nAuxEvol parity: ( RbarDD00:4, RbarDD01:5, RbarDD02:6, RbarDD11:7,\n RbarDD12:8, RbarDD22:9 )\nWrote to file \"BSSN_Two_BHs_Collide_Ccodes/boundary_conditions/EigenCoord_Cart_to_xx.h\"\n"
]
],
[
[
"<a id='mainc'></a>\n\n# Step 5: `BrillLindquist_Playground.c`: The Main C Code \\[Back to [top](#toc)\\]\n$$\\label{mainc}$$",
"_____no_output_____"
]
],
[
[
"# Part P0: Define REAL, set the number of ghost cells NGHOSTS (from NRPy+'s FD_CENTDERIVS_ORDER),\n# and set the CFL_FACTOR (which can be overwritten at the command line)\n\nwith open(os.path.join(Ccodesdir,\"BSSN_Playground_REAL__NGHOSTS__CFL_FACTOR.h\"), \"w\") as file:\n file.write(\"\"\"\n// Part P0.a: Set the number of ghost cells, from NRPy+'s FD_CENTDERIVS_ORDER\n#define NGHOSTS \"\"\"+str(int(FD_order/2)+1)+\"\"\"\n// Part P0.b: Set the numerical precision (REAL) to double, ensuring all floating point\n// numbers are stored to at least ~16 significant digits\n#define REAL \"\"\"+REAL+\"\"\"\n// Part P0.c: Set the CFL Factor. Can be overwritten at command line.\nREAL CFL_FACTOR = \"\"\"+str(default_CFL_FACTOR)+\";\")",
"_____no_output_____"
],
[
"%%writefile $Ccodesdir/BrillLindquist_Playground.c\n\n// Step P0: Define REAL and NGHOSTS; and declare CFL_FACTOR. This header is generated in NRPy+.\n#include \"BSSN_Playground_REAL__NGHOSTS__CFL_FACTOR.h\"\n\n//#include \"rfm_files/rfm_struct__declare.h\"\n\n#include \"declare_Cparameters_struct.h\"\n\n// All SIMD intrinsics used in SIMD-enabled C code loops are defined here:\n#include \"SIMD/SIMD_intrinsics.h\"\n#ifdef SIMD_IS_DISABLED\n// Algorithm for upwinding, SIMD-disabled version.\n// *NOTE*: This upwinding is backwards from\n// usual upwinding algorithms, because the\n// upwinding control vector in BSSN (the shift)\n// acts like a *negative* velocity.\n#define UPWIND_ALG(UpwindVecU) UpwindVecU > 0.0 ? 1.0 : 0.0\n#endif\n\n// Step P1: Import needed header files\n#include \"stdio.h\"\n#include \"stdlib.h\"\n#include \"math.h\"\n#include \"time.h\"\n#include \"stdint.h\" // Needed for Windows GCC 6.x compatibility\n#ifndef M_PI\n#define M_PI 3.141592653589793238462643383279502884L\n#endif\n#ifndef M_SQRT1_2\n#define M_SQRT1_2 0.707106781186547524400844362104849039L\n#endif\n#define wavespeed 1.0 // Set CFL-based \"wavespeed\" to 1.0.\n\n// Step P2: Declare the IDX4S(gf,i,j,k) macro, which enables us to store 4-dimensions of\n// data in a 1D array. In this case, consecutive values of \"i\"\n// (all other indices held to a fixed value) are consecutive in memory, where\n// consecutive values of \"j\" (fixing all other indices) are separated by\n// Nxx_plus_2NGHOSTS0 elements in memory. Similarly, consecutive values of\n// \"k\" are separated by Nxx_plus_2NGHOSTS0*Nxx_plus_2NGHOSTS1 in memory, etc.\n#define IDX4S(g,i,j,k) \\\n( (i) + Nxx_plus_2NGHOSTS0 * ( (j) + Nxx_plus_2NGHOSTS1 * ( (k) + Nxx_plus_2NGHOSTS2 * (g) ) ) )\n#define IDX4ptS(g,idx) ( (idx) + (Nxx_plus_2NGHOSTS0*Nxx_plus_2NGHOSTS1*Nxx_plus_2NGHOSTS2) * (g) )\n#define IDX3S(i,j,k) ( (i) + Nxx_plus_2NGHOSTS0 * ( (j) + Nxx_plus_2NGHOSTS1 * ( (k) ) ) )\n#define LOOP_REGION(i0min,i0max, i1min,i1max, i2min,i2max) \\\n for(int i2=i2min;i2<i2max;i2++) for(int i1=i1min;i1<i1max;i1++) for(int i0=i0min;i0<i0max;i0++)\n#define LOOP_ALL_GFS_GPS(ii) _Pragma(\"omp parallel for\") \\\n for(int (ii)=0;(ii)<Nxx_plus_2NGHOSTS_tot*NUM_EVOL_GFS;(ii)++)\n\n// Step P3: Set UUGF and VVGF macros, as well as xx_to_Cart()\n#include \"boundary_conditions/gridfunction_defines.h\"\n\n// Step P4: Set xx_to_Cart(const paramstruct *restrict params,\n// REAL *restrict xx[3],\n// const int i0,const int i1,const int i2,\n// REAL xCart[3]),\n// which maps xx->Cartesian via\n// {xx[0][i0],xx[1][i1],xx[2][i2]}->{xCart[0],xCart[1],xCart[2]}\n#include \"xx_to_Cart.h\"\n\n// Step P5: Defines set_Nxx_dxx_invdx_params__and__xx(const int EigenCoord, const int Nxx[3],\n// paramstruct *restrict params, REAL *restrict xx[3]),\n// which sets params Nxx,Nxx_plus_2NGHOSTS,dxx,invdx, and xx[] for\n// the chosen Eigen-CoordSystem if EigenCoord==1, or\n// CoordSystem if EigenCoord==0.\n#include \"set_Nxx_dxx_invdx_params__and__xx.h\"\n\n// Step P6: Include basic functions needed to impose curvilinear\n// parity and boundary conditions.\n#include \"boundary_conditions/CurviBC_include_Cfunctions.h\"\n\n// Step P7: Implement the algorithm for upwinding.\n// *NOTE*: This upwinding is backwards from\n// usual upwinding algorithms, because the\n// upwinding control vector in BSSN (the shift)\n// acts like a *negative* velocity.\n//#define UPWIND_ALG(UpwindVecU) UpwindVecU > 0.0 ? 1.0 : 0.0\n\n// Step P8: Include function for enforcing detgammabar constraint.\n#include \"enforce_detgammahat_constraint.h\"\n\n// Step P9: Find the CFL-constrained timestep\n#include \"find_timestep.h\"\n\n// Step P10: Declare function necessary for setting up the initial data.\n// Step P10.a: Define BSSN_ID() for BrillLindquist initial data\n// Step P10.b: Set the generic driver function for setting up BSSN initial data\n#include \"initial_data.h\"\n\n// Step P11: Declare function for evaluating Hamiltonian constraint (diagnostic)\n#include \"Hamiltonian_constraint.h\"\n\n// Step P12: Declare rhs_eval function, which evaluates BSSN RHSs\n#include \"rhs_eval.h\"\n\n// Step P13: Declare Ricci_eval function, which evaluates Ricci tensor\n#include \"Ricci_eval.h\"\n\n// main() function:\n// Step 0: Read command-line input, set up grid structure, allocate memory for gridfunctions, set up coordinates\n// Step 1: Set up initial data to an exact solution\n// Step 2: Start the timer, for keeping track of how fast the simulation is progressing.\n// Step 3: Integrate the initial data forward in time using the chosen RK-like Method of\n// Lines timestepping algorithm, and output periodic simulation diagnostics\n// Step 3.a: Output 2D data file periodically, for visualization\n// Step 3.b: Step forward one timestep (t -> t+dt) in time using\n// chosen RK-like MoL timestepping algorithm\n// Step 3.c: If t=t_final, output conformal factor & Hamiltonian\n// constraint violation to 2D data file\n// Step 3.d: Progress indicator printing to stderr\n// Step 4: Free all allocated memory\nint main(int argc, const char *argv[]) {\n paramstruct params;\n#include \"set_Cparameters_default.h\"\n\n // Step 0a: Read command-line input, error out if nonconformant\n if((argc != 4 && argc != 5) || atoi(argv[1]) < NGHOSTS || atoi(argv[2]) < NGHOSTS || atoi(argv[3]) < 2 /* FIXME; allow for axisymmetric sims */) {\n fprintf(stderr,\"Error: Expected three command-line arguments: ./BrillLindquist_Playground Nx0 Nx1 Nx2,\\n\");\n fprintf(stderr,\"where Nx[0,1,2] is the number of grid points in the 0, 1, and 2 directions.\\n\");\n fprintf(stderr,\"Nx[] MUST BE larger than NGHOSTS (= %d)\\n\",NGHOSTS);\n exit(1);\n }\n if(argc == 5) {\n CFL_FACTOR = strtod(argv[4],NULL);\n if(CFL_FACTOR > 0.5 && atoi(argv[3])!=2) {\n fprintf(stderr,\"WARNING: CFL_FACTOR was set to %e, which is > 0.5.\\n\",CFL_FACTOR);\n fprintf(stderr,\" This will generally only be stable if the simulation is purely axisymmetric\\n\");\n fprintf(stderr,\" However, Nx2 was set to %d>2, which implies a non-axisymmetric simulation\\n\",atoi(argv[3]));\n }\n }\n // Step 0b: Set up numerical grid structure, first in space...\n const int Nxx[3] = { atoi(argv[1]), atoi(argv[2]), atoi(argv[3]) };\n if(Nxx[0]%2 != 0 || Nxx[1]%2 != 0 || Nxx[2]%2 != 0) {\n fprintf(stderr,\"Error: Cannot guarantee a proper cell-centered grid if number of grid cells not set to even number.\\n\");\n fprintf(stderr,\" For example, in case of angular directions, proper symmetry zones will not exist.\\n\");\n exit(1);\n }\n\n // Step 0c: Set free parameters, overwriting Cparameters defaults\n // by hand or with command-line input, as desired.\n#include \"free_parameters.h\"\n\n // Step 0d: Uniform coordinate grids are stored to *xx[3]\n REAL *xx[3];\n // Step 0d.i: Set bcstruct\n bc_struct bcstruct;\n {\n int EigenCoord = 1;\n // Step 0d.ii: Call set_Nxx_dxx_invdx_params__and__xx(), which sets\n // params Nxx,Nxx_plus_2NGHOSTS,dxx,invdx, and xx[] for the\n // chosen Eigen-CoordSystem.\n set_Nxx_dxx_invdx_params__and__xx(EigenCoord, Nxx, ¶ms, xx);\n // Step 0d.iii: Set Nxx_plus_2NGHOSTS_tot\n#include \"set_Cparameters-nopointer.h\"\n const int Nxx_plus_2NGHOSTS_tot = Nxx_plus_2NGHOSTS0*Nxx_plus_2NGHOSTS1*Nxx_plus_2NGHOSTS2;\n // Step 0e: Find ghostzone mappings; set up bcstruct\n#include \"boundary_conditions/driver_bcstruct.h\"\n // Step 0e.i: Free allocated space for xx[][] array\n for(int i=0;i<3;i++) free(xx[i]);\n }\n\n // Step 0f: Call set_Nxx_dxx_invdx_params__and__xx(), which sets\n // params Nxx,Nxx_plus_2NGHOSTS,dxx,invdx, and xx[] for the\n // chosen (non-Eigen) CoordSystem.\n int EigenCoord = 0;\n set_Nxx_dxx_invdx_params__and__xx(EigenCoord, Nxx, ¶ms, xx);\n\n // Step 0g: Set all C parameters \"blah\" for params.blah, including\n // Nxx_plus_2NGHOSTS0 = params.Nxx_plus_2NGHOSTS0, etc.\n#include \"set_Cparameters-nopointer.h\"\n const int Nxx_plus_2NGHOSTS_tot = Nxx_plus_2NGHOSTS0*Nxx_plus_2NGHOSTS1*Nxx_plus_2NGHOSTS2;\n\n // Step 0h: Time coordinate parameters\n const REAL t_final = domain_size; /* Final time is set so that at t=t_final,\n * data at the origin have not been corrupted\n * by the approximate outer boundary condition */\n\n // Step 0i: Set timestep based on smallest proper distance between gridpoints and CFL factor\n REAL dt = find_timestep(¶ms, xx);\n //fprintf(stderr,\"# Timestep set to = %e\\n\",(double)dt);\n int N_final = (int)(t_final / dt + 0.5); // The number of points in time.\n // Add 0.5 to account for C rounding down\n // typecasts to integers.\n int output_every_N = (int)((REAL)N_final/800.0);\n if(output_every_N == 0) output_every_N = 1;\n\n // Step 0j: Error out if the number of auxiliary gridfunctions outnumber evolved gridfunctions.\n // This is a limitation of the RK method. You are always welcome to declare & allocate\n // additional gridfunctions by hand.\n if(NUM_AUX_GFS > NUM_EVOL_GFS) {\n fprintf(stderr,\"Error: NUM_AUX_GFS > NUM_EVOL_GFS. Either reduce the number of auxiliary gridfunctions,\\n\");\n fprintf(stderr,\" or allocate (malloc) by hand storage for *diagnostic_output_gfs. \\n\");\n exit(1);\n }\n\n // Step 0k: Allocate memory for gridfunctions\n#include \"MoLtimestepping/RK_Allocate_Memory.h\"\n REAL *restrict auxevol_gfs = (REAL *)malloc(sizeof(REAL) * NUM_AUXEVOL_GFS * Nxx_plus_2NGHOSTS_tot);\n\n // Step 0l: Set up precomputed reference metric arrays\n // Step 0l.i: Allocate space for precomputed reference metric arrays.\n//#include \"rfm_files/rfm_struct__malloc.h\"\n\n // Step 0l.ii: Define precomputed reference metric arrays.\n {\n// #include \"set_Cparameters-nopointer.h\"\n// #include \"rfm_files/rfm_struct__define.h\"\n }\n\n // Step 1: Set up initial data to an exact solution\n initial_data(¶ms, xx, y_n_gfs);\n\n // Step 1b: Apply boundary conditions, as initial data\n // are sometimes ill-defined in ghost zones.\n // E.g., spherical initial data might not be\n // properly defined at points where r=-1.\n apply_bcs_curvilinear(¶ms, &bcstruct, NUM_EVOL_GFS,evol_gf_parity, y_n_gfs);\n enforce_detgammahat_constraint(¶ms, xx, y_n_gfs);\n\n // Step 2: Start the timer, for keeping track of how fast the simulation is progressing.\n#ifdef __linux__ // Use high-precision timer in Linux.\n struct timespec start, end;\n clock_gettime(CLOCK_REALTIME, &start);\n#else // Resort to low-resolution, standards-compliant timer in non-Linux OSs\n // http://www.cplusplus.com/reference/ctime/time/\n time_t start_timer,end_timer;\n time(&start_timer); // Resolution of one second...\n#endif\n\n // Step 3: Integrate the initial data forward in time using the chosen RK-like Method of\n // Lines timestepping algorithm, and output periodic simulation diagnostics\n for(int n=0;n<=N_final;n++) { // Main loop to progress forward in time.\n\n // Step 3.a: Output 2D data file periodically, for visualization\n if(n%100 == 0) {\n // Evaluate Hamiltonian constraint violation\n Hamiltonian_constraint(¶ms, xx, y_n_gfs, diagnostic_output_gfs);\n\n char filename[100];\n sprintf(filename,\"out%d-%08d.txt\",Nxx[0],n);\n FILE *out2D = fopen(filename, \"w\");\n LOOP_REGION(NGHOSTS,Nxx_plus_2NGHOSTS0-NGHOSTS,\n NGHOSTS,Nxx_plus_2NGHOSTS1-NGHOSTS,\n NGHOSTS,Nxx_plus_2NGHOSTS2-NGHOSTS) {\n const int idx = IDX3S(i0,i1,i2);\n REAL xx0 = xx[0][i0];\n REAL xx1 = xx[1][i1];\n REAL xx2 = xx[2][i2];\n REAL xCart[3];\n xx_to_Cart(¶ms,xx,i0,i1,i2,xCart);\n fprintf(out2D,\"%e %e %e %e\\n\",\n xCart[1],xCart[2],\n y_n_gfs[IDX4ptS(CFGF,idx)],log10(fabs(diagnostic_output_gfs[IDX4ptS(HGF,idx)])));\n }\n fclose(out2D);\n }\n\n // Step 3.b: Step forward one timestep (t -> t+dt) in time using\n // chosen RK-like MoL timestepping algorithm\n#include \"MoLtimestepping/RK_MoL.h\"\n\n // Step 3.c: If t=t_final, output conformal factor & Hamiltonian\n // constraint violation to 2D data file\n if(n==N_final-1) {\n // Evaluate Hamiltonian constraint violation\n Hamiltonian_constraint(¶ms, xx, y_n_gfs, diagnostic_output_gfs);\n char filename[100];\n sprintf(filename,\"out%d.txt\",Nxx[0]);\n FILE *out2D = fopen(filename, \"w\");\n const int i0MIN=NGHOSTS; // In spherical, r=Delta r/2.\n const int i1mid=Nxx_plus_2NGHOSTS1/2;\n const int i2mid=Nxx_plus_2NGHOSTS2/2;\n LOOP_REGION(NGHOSTS,Nxx_plus_2NGHOSTS0-NGHOSTS,\n NGHOSTS,Nxx_plus_2NGHOSTS1-NGHOSTS,\n NGHOSTS,Nxx_plus_2NGHOSTS2-NGHOSTS) {\n REAL xx0 = xx[0][i0];\n REAL xx1 = xx[1][i1];\n REAL xx2 = xx[2][i2];\n REAL xCart[3];\n xx_to_Cart(¶ms,xx,i0,i1,i2,xCart);\n int idx = IDX3S(i0,i1,i2);\n fprintf(out2D,\"%e %e %e %e\\n\",xCart[1],xCart[2], y_n_gfs[IDX4ptS(CFGF,idx)],\n log10(fabs(diagnostic_output_gfs[IDX4ptS(HGF,idx)])));\n }\n fclose(out2D);\n }\n // Step 3.d: Progress indicator printing to stderr\n\n // Step 3.d.i: Measure average time per iteration\n#ifdef __linux__ // Use high-precision timer in Linux.\n clock_gettime(CLOCK_REALTIME, &end);\n const long long unsigned int time_in_ns = 1000000000L * (end.tv_sec - start.tv_sec) + end.tv_nsec - start.tv_nsec;\n#else // Resort to low-resolution, standards-compliant timer in non-Linux OSs\n time(&end_timer); // Resolution of one second...\n REAL time_in_ns = difftime(end_timer,start_timer)*1.0e9+0.5; // Round up to avoid divide-by-zero.\n#endif\n const REAL s_per_iteration_avg = ((REAL)time_in_ns / (REAL)n) / 1.0e9;\n\n const int iterations_remaining = N_final - n;\n const REAL time_remaining_in_mins = s_per_iteration_avg * (REAL)iterations_remaining / 60.0;\n\n const REAL num_RHS_pt_evals = (REAL)(Nxx[0]*Nxx[1]*Nxx[2]) * 4.0 * (REAL)n; // 4 RHS evals per gridpoint for RK4\n const REAL RHS_pt_evals_per_sec = num_RHS_pt_evals / ((REAL)time_in_ns / 1.0e9);\n\n // Step 3.d.ii: Output simulation progress to stderr\n if(n % 10 == 0) {\n fprintf(stderr,\"%c[2K\", 27); // Clear the line\n fprintf(stderr,\"It: %d t=%.2f dt=%.2e | %.1f%%; ETA %.0f s | t/h %.2f | gp/s %.2e\\r\", // \\r is carriage return, move cursor to the beginning of the line\n n, n * (double)dt, (double)dt, (double)(100.0 * (REAL)n / (REAL)N_final),\n (double)time_remaining_in_mins*60, (double)(dt * 3600.0 / s_per_iteration_avg), (double)RHS_pt_evals_per_sec);\n fflush(stderr); // Flush the stderr buffer\n } // End progress indicator if(n % 10 == 0)\n } // End main loop to progress forward in time.\n fprintf(stderr,\"\\n\"); // Clear the final line of output from progress indicator.\n\n // Step 4: Free all allocated memory\n//#include \"rfm_files/rfm_struct__freemem.h\"\n#include \"boundary_conditions/bcstruct_freemem.h\"\n#include \"MoLtimestepping/RK_Free_Memory.h\"\n free(auxevol_gfs);\n for(int i=0;i<3;i++) free(xx[i]);\n\n return 0;\n}",
"Writing BSSN_Two_BHs_Collide_Ccodes//BrillLindquist_Playground.c\n"
]
],
[
[
"<a id='compileexec'></a>\n\n# Step 6: Compile generated C codes & perform the black hole collision calculation \\[Back to [top](#toc)\\]\n$$\\label{compileexec}$$\n\nTo aid in the cross-platform-compatible (with Windows, MacOS, & Linux) compilation and execution, we make use of `cmdline_helper` [(**Tutorial**)](Tutorial-cmdline_helper.ipynb).",
"_____no_output_____"
]
],
[
[
"import cmdline_helper as cmd\nCFL_FACTOR=1.0\ncmd.C_compile(os.path.join(Ccodesdir,\"BrillLindquist_Playground.c\"),\n os.path.join(outdir,\"BrillLindquist_Playground\"),compile_mode=\"optimized\")\n# cmd.C_compile(os.path.join(Ccodesdir,\"BrillLindquist_Playground.c\"),\n# os.path.join(outdir,\"BrillLindquist_Playground\"),compile_mode=\"custom\",\n# custom_compile_string=\"gcc -O2 -g -march=native \"+\n# os.path.join(Ccodesdir,\"BrillLindquist_Playground.c\")+\n# \" -o \"+os.path.join(outdir,\"BrillLindquist_Playground\")+\" -lm\")\n\n# Change to output directory\nos.chdir(outdir)\n# Clean up existing output files\ncmd.delete_existing_files(\"out*.txt\")\ncmd.delete_existing_files(\"out*.png\")\n# Run executable\ncmd.Execute(\"BrillLindquist_Playground\", \"72 12 2 \"+str(CFL_FACTOR))\ncmd.Execute(\"BrillLindquist_Playground\", \"96 16 2 \"+str(CFL_FACTOR))\n\n# Return to root directory\nos.chdir(os.path.join(\"../../\"))\n\n# with open(\"compilescript\", \"w\") as file:\n# count=0\n# for custom_compile_string0 in [\"-O2\",\"-O\",\"\"]:\n# for custom_compile_string1 in [\"\",\"-fp-model fast=2 -no-prec-div\"]:\n# for custom_compile_string2 in [\"\",\"-qopt-prefetch=3\",\"-qopt-prefetch=4\"]:\n# for custom_compile_string3 in [\"\",\"-unroll\"]:\n# for custom_compile_string4 in [\"\",\"-qoverride-limits\"]:\n# exc= \"BL\"+custom_compile_string0+custom_compile_string1.replace(\" \",\"\")+custom_compile_string2+custom_compile_string3+custom_compile_string4\n# ccs = \"icc -qopenmp -xHost \"+custom_compile_string0+\" \"+custom_compile_string1+\" \"+custom_compile_string2+\" \"+custom_compile_string3+\" \"+custom_compile_string4+\" BSSN_Two_BHs_Collide_Ccodes/BrillLindquist_Playground.c -o \"+exc\n# file.write(ccs+\" &\\n\")\n# if count>0 and count%16==0:\n# file.write(\"wait\\n\")\n# count += 1\n# file.write(\"wait\\n\")\n\n# with open(\"compilescriptgcc\", \"w\") as file:\n# count=0\n# for custom_compile_string0 in [\"-Ofast\",\"-O2\",\"-O3\",\"-O\",\"\"]:\n# for custom_compile_string1 in [\"-fopenmp\"]:\n# for custom_compile_string2 in [\"\",\"-march=native\"]:\n# for custom_compile_string3 in [\"\",\"-funroll-loops\",\"-funroll-all-loops\"]:\n# for custom_compile_string4 in [\"\"]:\n# exc= \"BL\"+custom_compile_string0+custom_compile_string1+custom_compile_string2+custom_compile_string3+custom_compile_string4\n# ccs = \"gcc \"+custom_compile_string0+\" \"+custom_compile_string1+\" \"+custom_compile_string2+\" \"+custom_compile_string3+\" \"+custom_compile_string4+\" BSSN_Two_BHs_Collide_Ccodes/BrillLindquist_Playground.c -o \"+exc\n# file.write(ccs+\" -lm &\\n\")\n# if count>0 and count%16==0:\n# file.write(\"wait\\n\")\n# count += 1\n# file.write(\"wait\\n\")\n\n\nprint(\"(BENCH) Finished this code cell.\")",
"Compiling executable...\n(EXEC): Executing `gcc -std=gnu99 -Ofast -fopenmp -march=native -funroll-loops BSSN_Two_BHs_Collide_Ccodes/BrillLindquist_Playground.c -o BSSN_Two_BHs_Collide_Ccodes/output/BrillLindquist_Playground -lm`...\n(BENCH): Finished executing in 3.4121758937835693 seconds.\nFinished compilation.\n(EXEC): Executing `taskset -c 0,1,2,3,4,5,6,7,8,9,10,11,12,13,14,15 ./BrillLindquist_Playground 72 12 2 1.0`...\n\u001b[2KIt: 550 t=7.50 dt=1.36e-02 | 100.0%; ETA 0 s | t/h 10109.50 | gp/s 1.42e+06\n(BENCH): Finished executing in 2.8100385665893555 seconds.\n(EXEC): Executing `taskset -c 0,1,2,3,4,5,6,7,8,9,10,11,12,13,14,15 ./BrillLindquist_Playground 96 16 2 1.0`...\n\u001b[2KIt: 970 t=7.44 dt=7.67e-03 | 99.2%; ETA 0 s | t/h 3264.94 | gp/s 1.45e+06\n(BENCH): Finished executing in 8.419183015823364 seconds.\n(BENCH) Finished this code cell.\n"
]
],
[
[
"<a id='visualize'></a>\n\n# Step 7: Visualize the output! \\[Back to [top](#toc)\\]\n$$\\label{visualize}$$ \n\nIn this section we will generate a movie, plotting the conformal factor of these initial data on a 2D grid, such that darker colors imply stronger gravitational fields. Hence, we see the two black holes initially centered at $z/M=\\pm 0.5$, where $M$ is an arbitrary mass scale (conventionally the [ADM mass](https://en.wikipedia.org/w/index.php?title=ADM_formalism&oldid=846335453) is chosen), and our formulation of Einstein's equations adopt $G=c=1$ [geometrized units](https://en.wikipedia.org/w/index.php?title=Geometrized_unit_system&oldid=861682626).",
"_____no_output_____"
],
[
"<a id='installdownload'></a>\n\n## Step 7.a: Install `scipy` and download `ffmpeg` if they are not yet installed/downloaded \\[Back to [top](#toc)\\]\n$$\\label{installdownload}$$ \n\nNote that if you are not running this within `mybinder`, but on a Windows system, `ffmpeg` must be installed using a separate package (on [this site](http://ffmpeg.org/)), or if running Jupyter within Anaconda, use the command: `conda install -c conda-forge ffmpeg`.",
"_____no_output_____"
]
],
[
[
"!pip install scipy > /dev/null\n\ncheck_for_ffmpeg = !which ffmpeg >/dev/null && echo $?\nif check_for_ffmpeg != ['0']:\n print(\"Couldn't find ffmpeg, so I'll download it.\")\n # Courtesy https://johnvansickle.com/ffmpeg/\n !wget http://astro.phys.wvu.edu/zetienne/ffmpeg-static-amd64-johnvansickle.tar.xz\n !tar Jxf ffmpeg-static-amd64-johnvansickle.tar.xz\n print(\"Copying ffmpeg to ~/.local/bin/. Assumes ~/.local/bin is in the PATH.\")\n !mkdir ~/.local/bin/\n !cp ffmpeg-static-amd64-johnvansickle/ffmpeg ~/.local/bin/\n print(\"If this doesn't work, then install ffmpeg yourself. It should work fine on mybinder.\")",
"_____no_output_____"
]
],
[
[
"<a id='genimages'></a>\n\n## Step 7.b: Generate images for visualization animation \\[Back to [top](#toc)\\]\n$$\\label{genimages}$$ \n\nHere we loop through the data files output by the executable compiled and run in [the previous step](#mainc), generating a [png](https://en.wikipedia.org/wiki/Portable_Network_Graphics) image for each data file.\n\n**Special thanks to Terrence Pierre Jacques. His work with the first versions of these scripts greatly contributed to the scripts as they exist below.**",
"_____no_output_____"
]
],
[
[
"## VISUALIZATION ANIMATION, PART 1: Generate PNGs, one per frame of movie ##\n\nimport numpy as np\nfrom scipy.interpolate import griddata\nimport matplotlib.pyplot as plt\nfrom matplotlib.pyplot import savefig\nfrom IPython.display import HTML\nimport matplotlib.image as mgimg\n\nimport glob\nimport sys\nfrom matplotlib import animation\n\nglobby = glob.glob(os.path.join(outdir,'out96-00*.txt'))\nfile_list = []\nfor x in sorted(globby):\n file_list.append(x)\n\nbound=1.4\npl_xmin = -bound\npl_xmax = +bound\npl_ymin = -bound\npl_ymax = +bound\n\nfor filename in file_list:\n fig = plt.figure()\n x,y,cf,Ham = np.loadtxt(filename).T #Transposed for easier unpacking\n\n plotquantity = cf\n plotdescription = \"Numerical Soln.\"\n plt.title(\"Black Hole Head-on Collision (conf factor)\")\n plt.xlabel(\"y/M\")\n plt.ylabel(\"z/M\")\n\n grid_x, grid_y = np.mgrid[pl_xmin:pl_xmax:300j, pl_ymin:pl_ymax:300j]\n points = np.zeros((len(x), 2))\n for i in range(len(x)):\n # Zach says: No idea why x and y get flipped...\n points[i][0] = y[i]\n points[i][1] = x[i]\n\n grid = griddata(points, plotquantity, (grid_x, grid_y), method='nearest')\n gridcub = griddata(points, plotquantity, (grid_x, grid_y), method='cubic')\n im = plt.imshow(gridcub, extent=(pl_xmin,pl_xmax, pl_ymin,pl_ymax))\n ax = plt.colorbar()\n ax.set_label(plotdescription)\n savefig(os.path.join(filename+\".png\"),dpi=150)\n plt.close(fig)\n sys.stdout.write(\"%c[2K\" % 27)\n sys.stdout.write(\"Processing file \"+filename+\"\\r\")\n sys.stdout.flush()",
"\u001b[2KProcessing file BSSN_Two_BHs_Collide_Ccodes/output/out96-00000900.txt\r"
]
],
[
[
"<a id='genvideo'></a>\n\n## Step 7.c: Generate visualization animation \\[Back to [top](#toc)\\]\n$$\\label{genvideo}$$ \n\nIn the following step, [ffmpeg](http://ffmpeg.org) is used to generate an [mp4](https://en.wikipedia.org/wiki/MPEG-4) video file, which can be played directly from this Jupyter notebook.",
"_____no_output_____"
]
],
[
[
"## VISUALIZATION ANIMATION, PART 2: Combine PNGs to generate movie ##\n\n# https://stackoverflow.com/questions/14908576/how-to-remove-frame-from-matplotlib-pyplot-figure-vs-matplotlib-figure-frame\n# https://stackoverflow.com/questions/23176161/animating-pngs-in-matplotlib-using-artistanimation\n\nfig = plt.figure(frameon=False)\nax = fig.add_axes([0, 0, 1, 1])\nax.axis('off')\n\nmyimages = []\n\nfor i in range(len(file_list)):\n img = mgimg.imread(file_list[i]+\".png\")\n imgplot = plt.imshow(img)\n myimages.append([imgplot])\n\nani = animation.ArtistAnimation(fig, myimages, interval=100, repeat_delay=1000)\nplt.close()\nani.save(os.path.join(outdir,'BH_Head-on_Collision.mp4'), fps=5,dpi=150)",
"_____no_output_____"
],
[
"## VISUALIZATION ANIMATION, PART 3: Display movie as embedded HTML5 (see next cell) ##\n\n# https://stackoverflow.com/questions/18019477/how-can-i-play-a-local-video-in-my-ipython-notebook",
"_____no_output_____"
],
[
"# Embed video based on suggestion:\n# https://stackoverflow.com/questions/39900173/jupyter-notebook-html-cell-magic-with-python-variable\nHTML(\"\"\"\n<video width=\"480\" height=\"360\" controls>\n <source src=\\\"\"\"\"+os.path.join(outdir,\"BH_Head-on_Collision.mp4\")+\"\"\"\\\" type=\"video/mp4\">\n</video>\n\"\"\")",
"_____no_output_____"
]
],
[
[
"<a id='convergence'></a>\n\n# Step 8: Plot the numerical error, and confirm that it converges to zero with increasing numerical resolution (sampling) \\[Back to [top](#toc)\\]\n$$\\label{convergence}$$",
"_____no_output_____"
]
],
[
[
"x96,y96,valuesCF96,valuesHam96 = np.loadtxt(os.path.join(outdir,'out96.txt')).T #Transposed for easier unpacking\n\npl_xmin = -2.5\npl_xmax = +2.5\npl_ymin = -2.5\npl_ymax = +2.5\n\ngrid_x, grid_y = np.mgrid[pl_xmin:pl_xmax:100j, pl_ymin:pl_ymax:100j]\npoints96 = np.zeros((len(x96), 2))\nfor i in range(len(x96)):\n points96[i][0] = x96[i]\n points96[i][1] = y96[i]\n\ngrid96 = griddata(points96, valuesCF96, (grid_x, grid_y), method='nearest')\ngrid96cub = griddata(points96, valuesCF96, (grid_x, grid_y), method='cubic')\n\ngrid96 = griddata(points96, valuesHam96, (grid_x, grid_y), method='nearest')\ngrid96cub = griddata(points96, valuesHam96, (grid_x, grid_y), method='cubic')\n\n# fig, ax = plt.subplots()\n\nplt.clf()\nplt.title(\"96x16 Num. Err.: log_{10}|Ham|\")\nplt.xlabel(\"x/M\")\nplt.ylabel(\"z/M\")\n\nfig96cub = plt.imshow(grid96cub.T, extent=(pl_xmin,pl_xmax, pl_ymin,pl_ymax))\ncb = plt.colorbar(fig96cub)",
"_____no_output_____"
],
[
"x72,y72,valuesCF72,valuesHam72 = np.loadtxt(os.path.join(outdir,'out72.txt')).T #Transposed for easier unpacking\npoints72 = np.zeros((len(x72), 2))\nfor i in range(len(x72)):\n points72[i][0] = x72[i]\n points72[i][1] = y72[i]\n\ngrid72 = griddata(points72, valuesHam72, (grid_x, grid_y), method='nearest')\n\ngriddiff_72_minus_96 = np.zeros((100,100))\ngriddiff_72_minus_96_1darray = np.zeros(100*100)\ngridx_1darray_yeq0 = np.zeros(100)\ngrid72_1darray_yeq0 = np.zeros(100)\ngrid96_1darray_yeq0 = np.zeros(100)\ncount = 0\nfor i in range(100):\n for j in range(100):\n griddiff_72_minus_96[i][j] = grid72[i][j] - grid96[i][j]\n griddiff_72_minus_96_1darray[count] = griddiff_72_minus_96[i][j]\n if j==49:\n gridx_1darray_yeq0[i] = grid_x[i][j]\n grid72_1darray_yeq0[i] = grid72[i][j] + np.log10((72./96.)**4)\n grid96_1darray_yeq0[i] = grid96[i][j]\n count = count + 1\n\nplt.clf()\nfig, ax = plt.subplots()\nplt.title(\"4th-order Convergence, at t/M=7.5 (post-merger; horiz at x/M=+/-1)\")\nplt.xlabel(\"x/M\")\nplt.ylabel(\"log10(Relative error)\")\n\nax.plot(gridx_1darray_yeq0, grid96_1darray_yeq0, 'k-', label='Nr=96')\nax.plot(gridx_1darray_yeq0, grid72_1darray_yeq0, 'k--', label='Nr=72, mult by (72/96)^4')\nax.set_ylim([-8.5,0.5])\n\nlegend = ax.legend(loc='lower right', shadow=True, fontsize='x-large')\nlegend.get_frame().set_facecolor('C1')\nplt.show()",
"_____no_output_____"
]
],
[
[
"<a id='latex_pdf_output'></a>\n\n# Step 9: Output this notebook to $\\LaTeX$-formatted PDF file \\[Back to [top](#toc)\\]\n$$\\label{latex_pdf_output}$$\n\nThe following code cell converts this Jupyter notebook into a proper, clickable $\\LaTeX$-formatted PDF file. After the cell is successfully run, the generated PDF may be found in the root NRPy+ tutorial directory, with filename\n[Tutorial-Start_to_Finish-BSSNCurvilinear-Two_BHs_Collide.pdf](Tutorial-Start_to_Finish-BSSNCurvilinear-Two_BHs_Collide.pdf) (Note that clicking on this link may not work; you may need to open the PDF file through another means.)",
"_____no_output_____"
]
],
[
[
"import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface\ncmd.output_Jupyter_notebook_to_LaTeXed_PDF(\"Tutorial-Start_to_Finish-BSSNCurvilinear-Two_BHs_Collide\")",
"Created Tutorial-Start_to_Finish-BSSNCurvilinear-Two_BHs_Collide.tex, and\n compiled LaTeX file to PDF file Tutorial-Start_to_Finish-\n BSSNCurvilinear-Two_BHs_Collide.pdf\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb60ba514d8d0fa7383f4ea8876a367c20e95e8 | 13,383 | ipynb | Jupyter Notebook | notebooks/0001-card_deck.ipynb | chendaniely/fluent_python | d90f6300c880c50271ca247ab2a644d1e6c37324 | [
"MIT"
] | 1 | 2021-01-18T12:47:42.000Z | 2021-01-18T12:47:42.000Z | notebooks/0001-card_deck.ipynb | chendaniely/fluent_python | d90f6300c880c50271ca247ab2a644d1e6c37324 | [
"MIT"
] | null | null | null | notebooks/0001-card_deck.ipynb | chendaniely/fluent_python | d90f6300c880c50271ca247ab2a644d1e6c37324 | [
"MIT"
] | null | null | null | 25.108818 | 72 | 0.473138 | [
[
[
"import collections",
"_____no_output_____"
],
[
"Card = collections.namedtuple('Card', ['rank', 'suit'])",
"_____no_output_____"
],
[
"class FrenchDeck(object):\n ranks = [str(n) for n in range(2, 11)] + list('JQKA')\n suits = 'spades diamonds clubs hearts'.split()\n \n def __init__(self):\n self._cards = [Card(rank, suit) for suit in self.suits\n for rank in self.ranks]\n \n def __len__(self):\n return len(self._cards)\n \n def __getitem__(self, position):\n return self._cards[position]",
"_____no_output_____"
],
[
"Card('7', 'diamonds')",
"_____no_output_____"
],
[
"deck = FrenchDeck()",
"_____no_output_____"
],
[
"len(deck)",
"_____no_output_____"
],
[
"deck[0]",
"_____no_output_____"
],
[
"deck [-1]",
"_____no_output_____"
],
[
"from random import choice\nchoice(deck)",
"_____no_output_____"
],
[
"for _ in range(10):\n print(choice(deck))",
"Card(rank='K', suit='diamonds')\nCard(rank='9', suit='spades')\nCard(rank='2', suit='diamonds')\nCard(rank='6', suit='hearts')\nCard(rank='10', suit='clubs')\nCard(rank='5', suit='clubs')\nCard(rank='5', suit='spades')\nCard(rank='6', suit='diamonds')\nCard(rank='5', suit='spades')\nCard(rank='9', suit='hearts')\n"
],
[
"deck[:3]",
"_____no_output_____"
],
[
"deck[10:14]",
"_____no_output_____"
],
[
"for _ in deck:\n print(_)",
"Card(rank='2', suit='spades')\nCard(rank='3', suit='spades')\nCard(rank='4', suit='spades')\nCard(rank='5', suit='spades')\nCard(rank='6', suit='spades')\nCard(rank='7', suit='spades')\nCard(rank='8', suit='spades')\nCard(rank='9', suit='spades')\nCard(rank='10', suit='spades')\nCard(rank='J', suit='spades')\nCard(rank='Q', suit='spades')\nCard(rank='K', suit='spades')\nCard(rank='A', suit='spades')\nCard(rank='2', suit='diamonds')\nCard(rank='3', suit='diamonds')\nCard(rank='4', suit='diamonds')\nCard(rank='5', suit='diamonds')\nCard(rank='6', suit='diamonds')\nCard(rank='7', suit='diamonds')\nCard(rank='8', suit='diamonds')\nCard(rank='9', suit='diamonds')\nCard(rank='10', suit='diamonds')\nCard(rank='J', suit='diamonds')\nCard(rank='Q', suit='diamonds')\nCard(rank='K', suit='diamonds')\nCard(rank='A', suit='diamonds')\nCard(rank='2', suit='clubs')\nCard(rank='3', suit='clubs')\nCard(rank='4', suit='clubs')\nCard(rank='5', suit='clubs')\nCard(rank='6', suit='clubs')\nCard(rank='7', suit='clubs')\nCard(rank='8', suit='clubs')\nCard(rank='9', suit='clubs')\nCard(rank='10', suit='clubs')\nCard(rank='J', suit='clubs')\nCard(rank='Q', suit='clubs')\nCard(rank='K', suit='clubs')\nCard(rank='A', suit='clubs')\nCard(rank='2', suit='hearts')\nCard(rank='3', suit='hearts')\nCard(rank='4', suit='hearts')\nCard(rank='5', suit='hearts')\nCard(rank='6', suit='hearts')\nCard(rank='7', suit='hearts')\nCard(rank='8', suit='hearts')\nCard(rank='9', suit='hearts')\nCard(rank='10', suit='hearts')\nCard(rank='J', suit='hearts')\nCard(rank='Q', suit='hearts')\nCard(rank='K', suit='hearts')\nCard(rank='A', suit='hearts')\n"
],
[
"for _ in reversed(deck):\n print(_)",
"Card(rank='A', suit='hearts')\nCard(rank='K', suit='hearts')\nCard(rank='Q', suit='hearts')\nCard(rank='J', suit='hearts')\nCard(rank='10', suit='hearts')\nCard(rank='9', suit='hearts')\nCard(rank='8', suit='hearts')\nCard(rank='7', suit='hearts')\nCard(rank='6', suit='hearts')\nCard(rank='5', suit='hearts')\nCard(rank='4', suit='hearts')\nCard(rank='3', suit='hearts')\nCard(rank='2', suit='hearts')\nCard(rank='A', suit='clubs')\nCard(rank='K', suit='clubs')\nCard(rank='Q', suit='clubs')\nCard(rank='J', suit='clubs')\nCard(rank='10', suit='clubs')\nCard(rank='9', suit='clubs')\nCard(rank='8', suit='clubs')\nCard(rank='7', suit='clubs')\nCard(rank='6', suit='clubs')\nCard(rank='5', suit='clubs')\nCard(rank='4', suit='clubs')\nCard(rank='3', suit='clubs')\nCard(rank='2', suit='clubs')\nCard(rank='A', suit='diamonds')\nCard(rank='K', suit='diamonds')\nCard(rank='Q', suit='diamonds')\nCard(rank='J', suit='diamonds')\nCard(rank='10', suit='diamonds')\nCard(rank='9', suit='diamonds')\nCard(rank='8', suit='diamonds')\nCard(rank='7', suit='diamonds')\nCard(rank='6', suit='diamonds')\nCard(rank='5', suit='diamonds')\nCard(rank='4', suit='diamonds')\nCard(rank='3', suit='diamonds')\nCard(rank='2', suit='diamonds')\nCard(rank='A', suit='spades')\nCard(rank='K', suit='spades')\nCard(rank='Q', suit='spades')\nCard(rank='J', suit='spades')\nCard(rank='10', suit='spades')\nCard(rank='9', suit='spades')\nCard(rank='8', suit='spades')\nCard(rank='7', suit='spades')\nCard(rank='6', suit='spades')\nCard(rank='5', suit='spades')\nCard(rank='4', suit='spades')\nCard(rank='3', suit='spades')\nCard(rank='2', suit='spades')\n"
],
[
"Card(rank='A', suit='spades') in deck",
"_____no_output_____"
],
[
"Card(rank='A', suit='cube') in deck",
"_____no_output_____"
],
[
"suit_values = dict(spades=3, hearts=2, diamonds=1, clubs=0)",
"_____no_output_____"
],
[
"def spades_high(card):\n rank_value = FrenchDeck.ranks.index(card.rank)\n return rank_value * len(suit_values) + suit_values[card.suit]",
"_____no_output_____"
],
[
"for card in sorted(deck, key=spades_high):\n print(card)",
"Card(rank='2', suit='clubs')\nCard(rank='2', suit='diamonds')\nCard(rank='2', suit='hearts')\nCard(rank='2', suit='spades')\nCard(rank='3', suit='clubs')\nCard(rank='3', suit='diamonds')\nCard(rank='3', suit='hearts')\nCard(rank='3', suit='spades')\nCard(rank='4', suit='clubs')\nCard(rank='4', suit='diamonds')\nCard(rank='4', suit='hearts')\nCard(rank='4', suit='spades')\nCard(rank='5', suit='clubs')\nCard(rank='5', suit='diamonds')\nCard(rank='5', suit='hearts')\nCard(rank='5', suit='spades')\nCard(rank='6', suit='clubs')\nCard(rank='6', suit='diamonds')\nCard(rank='6', suit='hearts')\nCard(rank='6', suit='spades')\nCard(rank='7', suit='clubs')\nCard(rank='7', suit='diamonds')\nCard(rank='7', suit='hearts')\nCard(rank='7', suit='spades')\nCard(rank='8', suit='clubs')\nCard(rank='8', suit='diamonds')\nCard(rank='8', suit='hearts')\nCard(rank='8', suit='spades')\nCard(rank='9', suit='clubs')\nCard(rank='9', suit='diamonds')\nCard(rank='9', suit='hearts')\nCard(rank='9', suit='spades')\nCard(rank='10', suit='clubs')\nCard(rank='10', suit='diamonds')\nCard(rank='10', suit='hearts')\nCard(rank='10', suit='spades')\nCard(rank='J', suit='clubs')\nCard(rank='J', suit='diamonds')\nCard(rank='J', suit='hearts')\nCard(rank='J', suit='spades')\nCard(rank='Q', suit='clubs')\nCard(rank='Q', suit='diamonds')\nCard(rank='Q', suit='hearts')\nCard(rank='Q', suit='spades')\nCard(rank='K', suit='clubs')\nCard(rank='K', suit='diamonds')\nCard(rank='K', suit='hearts')\nCard(rank='K', suit='spades')\nCard(rank='A', suit='clubs')\nCard(rank='A', suit='diamonds')\nCard(rank='A', suit='hearts')\nCard(rank='A', suit='spades')\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb60fccc89d5f9810d9004f61a8275623100b51 | 62,737 | ipynb | Jupyter Notebook | preparation.ipynb | GhazaleZe/Stroke-Prediction | bdbcc3d1977b3fb3c31bf627ec55149ad6c3ac2e | [
"MIT"
] | 5 | 2021-07-09T09:53:26.000Z | 2021-12-06T09:27:49.000Z | preparation.ipynb | GhazaleZe/Stroke-Prediction | bdbcc3d1977b3fb3c31bf627ec55149ad6c3ac2e | [
"MIT"
] | null | null | null | preparation.ipynb | GhazaleZe/Stroke-Prediction | bdbcc3d1977b3fb3c31bf627ec55149ad6c3ac2e | [
"MIT"
] | null | null | null | 36.222286 | 122 | 0.256053 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"df = pd.read_csv('E:\\\\term8\\\\datamining\\\\HW\\\\project\\\\healthcare-dataset-stroke-data.csv')\ndf = df[df.age > 18]\ndf",
"_____no_output_____"
],
[
"df.isnull().sum()",
"_____no_output_____"
],
[
"df1 = df\ndf1 = pd.get_dummies(df1, columns = ['work_type','smoking_status','gender','ever_married','Residence_type'])\ndf1 = df1.drop(columns = ['id'])\ndf1.age = (df1.age - df1.age.mean())/df1.age.std()\ndf1.avg_glucose_level = (df1.avg_glucose_level - df1.avg_glucose_level.mean())/df1.avg_glucose_level.std()\ndf_bmi_na = df1[df1.bmi.isnull()]\ndf1 = df1.dropna()\ndf1",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nimport random\ndf_train, df_test = train_test_split(df1, test_size = 0.2, random_state = 45)\nds_train = df_train.values\nds_test = df_test.values\nX = np.concatenate((ds_train[:,0:4],ds_train[:,5:20]),axis = 1)\ny = ds_train[:,4]\nX_test = np.concatenate((ds_test[:,0:4],ds_test[:,5:20]),axis = 1)\ny_test = ds_test[:,4]",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression\nreg = LinearRegression().fit(X,y)",
"_____no_output_____"
],
[
"y_pred = reg.predict(X)\ntemp = np.abs(y_pred - y)\ntemp.sum()/len(temp)",
"_____no_output_____"
],
[
"y_pred = reg.predict(X_test)\ntemp = np.abs(y_pred - y_test)\ntemp.sum()/len(temp)",
"_____no_output_____"
],
[
"df_bmi_na.bmi = reg.predict(np.concatenate((df_bmi_na.values[:,0:4],df_bmi_na.values[:,5:20]),axis = 1))\ndf2 = pd.concat([df1, df_bmi_na])\ndf2",
"_____no_output_____"
],
[
"df2.isnull().sum()",
"_____no_output_____"
],
[
"y_pred = reg.predict(X_test)\nprint(np.min(np.abs(y_pred - y_test)), np.max(np.abs(y_pred - y_test)), np.mean(np.abs(y_pred - y_test)))\nprint(np.min(np.abs(df_test.bmi - df.bmi.dropna().mean())), np.max(np.abs(df_test.bmi - df.bmi.dropna().mean())),\n np.mean(np.abs(df_test.bmi - df.bmi.dropna().mean())))",
"0.0015664246986517583 38.715880343904615 5.365419799809714\n0.006078724464366303 41.40607872446437 5.530866810497012\n"
],
[
"temp = df2.stroke\ndf2 = df2.drop(columns = ['stroke'])\ndf2['stroke'] = temp\ndf2['bmi'] = (df2['bmi'] - df2['bmi'].mean())/df2['bmi'].std()\ndf2",
"_____no_output_____"
],
[
"df2.to_csv('E:\\\\term8\\\\datamining\\\\HW\\\\project\\\\prepared_data.csv', index = False)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb61715e63ca2013a836e411a781f5343ff09b8 | 37,778 | ipynb | Jupyter Notebook | starter_code/model_2.ipynb | AnjanaDelhi/machine-learning-challenge | c323cbd83e7a55f594a2dfe9d28e0f3785eef81c | [
"RSA-MD"
] | null | null | null | starter_code/model_2.ipynb | AnjanaDelhi/machine-learning-challenge | c323cbd83e7a55f594a2dfe9d28e0f3785eef81c | [
"RSA-MD"
] | null | null | null | starter_code/model_2.ipynb | AnjanaDelhi/machine-learning-challenge | c323cbd83e7a55f594a2dfe9d28e0f3785eef81c | [
"RSA-MD"
] | null | null | null | 37.000979 | 1,176 | 0.443168 | [
[
[
"# Update sklearn to prevent version mismatches\n!pip install sklearn --upgrade",
"Collecting sklearn\n Using cached sklearn-0.0.tar.gz (1.1 kB)\nCollecting scikit-learn\n Downloading scikit_learn-0.24.1-cp36-cp36m-win_amd64.whl (6.8 MB)\nCollecting joblib>=0.11\n Downloading joblib-1.0.0-py3-none-any.whl (302 kB)\nRequirement already satisfied, skipping upgrade: scipy>=0.19.1 in c:\\users\\anjan\\anaconda3\\envs\\bcs\\lib\\site-packages (from scikit-learn->sklearn) (1.5.0)\nCollecting threadpoolctl>=2.0.0\n Downloading threadpoolctl-2.1.0-py3-none-any.whl (12 kB)\nRequirement already satisfied, skipping upgrade: numpy>=1.13.3 in c:\\users\\anjan\\anaconda3\\envs\\bcs\\lib\\site-packages (from scikit-learn->sklearn) (1.19.5)\nBuilding wheels for collected packages: sklearn\n Building wheel for sklearn (setup.py): started\n Building wheel for sklearn (setup.py): finished with status 'done'\n Created wheel for sklearn: filename=sklearn-0.0-py2.py3-none-any.whl size=1316 sha256=aeaf9ae608753a0554035e85b616c57d15450bc1099d0bdd5d7477e0e533c073\n Stored in directory: c:\\users\\anjan\\appdata\\local\\pip\\cache\\wheels\\23\\9d\\42\\5ec745cbbb17517000a53cecc49d6a865450d1f5cb16dc8a9c\nSuccessfully built sklearn\nInstalling collected packages: joblib, threadpoolctl, scikit-learn, sklearn\nSuccessfully installed joblib-1.0.0 scikit-learn-0.24.1 sklearn-0.0 threadpoolctl-2.1.0\n"
],
[
"# install joblib. This will be used to save your model. \n# Restart your kernel after installing \n!pip install joblib",
"Requirement already satisfied: joblib in c:\\users\\anjan\\anaconda3\\envs\\bcs\\lib\\site-packages (1.0.0)\n"
],
[
"%matplotlib inline\nfrom IPython.display import Image, SVG\nimport matplotlib.pyplot as plt\nfrom matplotlib import style\nstyle.use(\"ggplot\")\nimport pandas as pd\nimport numpy as np\n",
"_____no_output_____"
],
[
"# Filepaths, numpy, and Tensorflow\n#import os\n#import numpy as np\n#import tensorflow as tf\n\n#from sklearn.preprocessing import LabelEncoder",
"_____no_output_____"
],
[
"# Keras\nfrom tensorflow import keras\nfrom tensorflow.keras.models import Sequential\nfrom tensorflow.keras.utils import to_categorical\nfrom tensorflow.keras.layers import Dense\nfrom tensorflow.keras.datasets import mnist",
"_____no_output_____"
]
],
[
[
"# Read the CSV and Perform Basic Data Cleaning",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(\"exoplanet_data.csv\")\n# Drop the null columns where all values are null\ndf = df.dropna(axis='columns', how='all')\n# Drop the null rows\ndf = df.dropna()\ndf.head()",
"_____no_output_____"
]
],
[
[
"# Select your features (columns)",
"_____no_output_____"
]
],
[
[
"# Set features. This will also be used as your x values.\nselected_features = df.drop(['koi_disposition'], axis =1 )\nX = selected_features",
"_____no_output_____"
]
],
[
[
"# Create a Train Test Split\n\nUse `koi_disposition` for the y values",
"_____no_output_____"
]
],
[
[
"y = df['koi_disposition']",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1, stratify = y)",
"_____no_output_____"
],
[
"X_train.head()",
"_____no_output_____"
]
],
[
[
"# Pre-processing\n\nScale the data using the MinMaxScaler and perform some feature selection",
"_____no_output_____"
]
],
[
[
"\n# Scale your data\nfrom sklearn.preprocessing import MinMaxScaler\nscaler = MinMaxScaler()\n\nX_train_scaled = scaler.fit_transform(X_train)\n#print(X_scaler)\nX_test_scaled = scaler.fit_transform(X_test)",
"_____no_output_____"
],
[
"# Support vector machine linear classifier\nfrom sklearn.svm import SVC \nmodel = SVC(kernel='linear')\nmodel.fit(X_train_scaled, y_train)\n#predictions = model.predict(X_test)",
"_____no_output_____"
],
[
"print(f\"Training Data Score: {model.score(X_train_scaled, y_train)}\")\nprint(f\"Testing Data Score: {model.score(X_test_scaled, y_test)}\")",
"Training Data Score: 0.8439824527942018\nTesting Data Score: 0.8478260869565217\n"
],
[
"\n",
"_____no_output_____"
]
],
[
[
"# Train the Model\n\n",
"_____no_output_____"
],
[
"# Hyperparameter Tuning\n\nUse `GridSearchCV` to tune the model's parameters",
"_____no_output_____"
]
],
[
[
"# Create the GridSearchCV model\nfrom sklearn.model_selection import GridSearchCV\nparam_grid = {'C': [1, 5, 10],\n 'gamma': [0.001, 0.0001, 0.01]} \n\ngrid = GridSearchCV(model, param_grid, verbose=3)",
"_____no_output_____"
],
[
"grid.fit(X_train_scaled, y_train)",
"Fitting 5 folds for each of 9 candidates, totalling 45 fits\n[CV 1/5] END ...............................C=1, gamma=0.001; total time= 0.1s\n[CV 2/5] END ...............................C=1, gamma=0.001; total time= 0.1s\n[CV 3/5] END ...............................C=1, gamma=0.001; total time= 0.1s\n[CV 4/5] END ...............................C=1, gamma=0.001; total time= 0.1s\n[CV 5/5] END ...............................C=1, gamma=0.001; total time= 0.1s\n[CV 1/5] END ..............................C=1, gamma=0.0001; total time= 0.1s\n[CV 2/5] END ..............................C=1, gamma=0.0001; total time= 0.1s\n[CV 3/5] END ..............................C=1, gamma=0.0001; total time= 0.1s\n[CV 4/5] END ..............................C=1, gamma=0.0001; total time= 0.1s\n[CV 5/5] END ..............................C=1, gamma=0.0001; total time= 0.1s\n[CV 1/5] END ................................C=1, gamma=0.01; total time= 0.1s\n[CV 2/5] END ................................C=1, gamma=0.01; total time= 0.1s\n[CV 3/5] END ................................C=1, gamma=0.01; total time= 0.1s\n[CV 4/5] END ................................C=1, gamma=0.01; total time= 0.1s\n[CV 5/5] END ................................C=1, gamma=0.01; total time= 0.1s\n[CV 1/5] END ...............................C=5, gamma=0.001; total time= 0.2s\n[CV 2/5] END ...............................C=5, gamma=0.001; total time= 0.2s\n[CV 3/5] END ...............................C=5, gamma=0.001; total time= 0.2s\n[CV 4/5] END ...............................C=5, gamma=0.001; total time= 0.1s\n[CV 5/5] END ...............................C=5, gamma=0.001; total time= 0.1s\n[CV 1/5] END ..............................C=5, gamma=0.0001; total time= 0.2s\n[CV 2/5] END ..............................C=5, gamma=0.0001; total time= 0.2s\n[CV 3/5] END ..............................C=5, gamma=0.0001; total time= 0.2s\n[CV 4/5] END ..............................C=5, gamma=0.0001; total time= 0.1s\n[CV 5/5] END ..............................C=5, gamma=0.0001; total time= 0.1s\n[CV 1/5] END ................................C=5, gamma=0.01; total time= 0.2s\n[CV 2/5] END ................................C=5, gamma=0.01; total time= 0.2s\n[CV 3/5] END ................................C=5, gamma=0.01; total time= 0.2s\n[CV 4/5] END ................................C=5, gamma=0.01; total time= 0.1s\n[CV 5/5] END ................................C=5, gamma=0.01; total time= 0.1s\n[CV 1/5] END ..............................C=10, gamma=0.001; total time= 0.2s\n[CV 2/5] END ..............................C=10, gamma=0.001; total time= 0.2s\n[CV 3/5] END ..............................C=10, gamma=0.001; total time= 0.2s\n[CV 4/5] END ..............................C=10, gamma=0.001; total time= 0.1s\n[CV 5/5] END ..............................C=10, gamma=0.001; total time= 0.1s\n[CV 1/5] END .............................C=10, gamma=0.0001; total time= 0.2s\n[CV 2/5] END .............................C=10, gamma=0.0001; total time= 0.2s\n[CV 3/5] END .............................C=10, gamma=0.0001; total time= 0.2s\n[CV 4/5] END .............................C=10, gamma=0.0001; total time= 0.1s\n[CV 5/5] END .............................C=10, gamma=0.0001; total time= 0.1s\n[CV 1/5] END ...............................C=10, gamma=0.01; total time= 0.2s\n[CV 2/5] END ...............................C=10, gamma=0.01; total time= 0.2s\n[CV 3/5] END ...............................C=10, gamma=0.01; total time= 0.2s\n[CV 4/5] END ...............................C=10, gamma=0.01; total time= 0.1s\n[CV 5/5] END ...............................C=10, gamma=0.01; total time= 0.1s\n"
],
[
"print(grid.best_params_)\nprint(grid.best_score_)",
"{'C': 10, 'gamma': 0.001}\n0.8680138845428944\n"
]
],
[
[
"# Save the Model",
"_____no_output_____"
]
],
[
[
"# save your model by updating \"your_name\" with your name\n# and \"your_model\" with your model variable\n# be sure to turn this in to BCS\n# if joblib fails to import, try running the command to install in terminal/git-bash\nimport joblib\nfilename = 'Anjana_delhi_model_svc.sav'\njoblib.dump(grid, filename)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb62092ee9d2fa7afa5c6b7b21de6ba22b42e6f | 11,174 | ipynb | Jupyter Notebook | SpamDetection.ipynb | Spnetic-5/mail_Spam | 93ca5f0715753cebf73b92b1673e8ee1d534c1bc | [
"MIT"
] | null | null | null | SpamDetection.ipynb | Spnetic-5/mail_Spam | 93ca5f0715753cebf73b92b1673e8ee1d534c1bc | [
"MIT"
] | null | null | null | SpamDetection.ipynb | Spnetic-5/mail_Spam | 93ca5f0715753cebf73b92b1673e8ee1d534c1bc | [
"MIT"
] | null | null | null | 25.746544 | 126 | 0.478253 | [
[
[
"import pandas as pd\nimport re\nimport string\nimport numpy as np\nfrom sklearn.feature_extraction.stop_words import ENGLISH_STOP_WORDS",
"_____no_output_____"
],
[
"data = pd.read_csv(\"spam.csv\",encoding = \"'latin'\")",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"data[\"text\"] = data.v2\ndata[\"spam\"] = data.v1",
"_____no_output_____"
]
],
[
[
"# Splitting data",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nemails_train, emails_test, target_train, target_test = train_test_split(data.text,data.spam,test_size = 0.2) ",
"_____no_output_____"
],
[
"data.info",
"_____no_output_____"
],
[
"emails_train.shape",
"_____no_output_____"
]
],
[
[
"# Preprocessing",
"_____no_output_____"
]
],
[
[
"def remove_hyperlink(word):\n return re.sub(r\"http\\S+\", \"\", word)\n\ndef to_lower(word):\n result = word.lower()\n return result\n\ndef remove_number(word):\n result = re.sub(r'\\d+', '', word)\n return result\n\ndef remove_punctuation(word):\n result = word.translate(str.maketrans(dict.fromkeys(string.punctuation)))\n return result\n\ndef remove_whitespace(word):\n result = word.strip()\n return result\n\ndef replace_newline(word):\n return word.replace('\\n','')\n\n\n\ndef clean_up_pipeline(sentence):\n cleaning_utils = [remove_hyperlink,\n replace_newline,\n to_lower,\n remove_number,\n remove_punctuation,remove_whitespace]\n for o in cleaning_utils:\n sentence = o(sentence)\n return sentence\n\nx_train = [clean_up_pipeline(o) for o in emails_train]\nx_test = [clean_up_pipeline(o) for o in emails_test]\n\nx_train[0]",
"_____no_output_____"
],
[
"from sklearn.preprocessing import LabelEncoder\nle = LabelEncoder()\ntrain_y = le.fit_transform(target_train.values)\ntest_y = le.transform(target_test.values)",
"_____no_output_____"
],
[
"train_y",
"_____no_output_____"
]
],
[
[
"# Tokenize",
"_____no_output_____"
]
],
[
[
"## some config values \nembed_size = 100 # how big is each word vector\nmax_feature = 50000 # how many unique words to use (i.e num rows in embedding vector)\nmax_len = 2000 # max number of words in a question to use",
"_____no_output_____"
],
[
"from keras.preprocessing.text import Tokenizer\ntokenizer = Tokenizer(num_words=max_feature)\n\ntokenizer.fit_on_texts(x_train)\n\nx_train_features = np.array(tokenizer.texts_to_sequences(x_train))\nx_test_features = np.array(tokenizer.texts_to_sequences(x_test))\n\nx_train_features[0]",
"_____no_output_____"
]
],
[
[
"# Padding",
"_____no_output_____"
]
],
[
[
"from keras.preprocessing.sequence import pad_sequences\nx_train_features = pad_sequences(x_train_features,maxlen=max_len)\nx_test_features = pad_sequences(x_test_features,maxlen=max_len)\nx_train_features[0]",
"_____no_output_____"
]
],
[
[
"# Model",
"_____no_output_____"
]
],
[
[
"from keras.layers import Dense, Input, LSTM, Embedding, Dropout, Activation\nfrom keras.layers import Bidirectional\nfrom keras.models import Model",
"_____no_output_____"
],
[
"# create the model\nimport tensorflow as tf\nembedding_vecor_length = 32\n\nmodel = tf.keras.Sequential()\nmodel.add(Embedding(max_feature, embedding_vecor_length, input_length=max_len))\nmodel.add(Bidirectional(tf.keras.layers.LSTM(64)))\nmodel.add(Dense(16, activation='relu'))\nmodel.add(Dropout(0.1))\nmodel.add(Dense(1, activation='sigmoid'))\nmodel.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\nprint(model.summary())",
"_____no_output_____"
],
[
"history = model.fit(x_train_features, train_y, batch_size=512, epochs=20, validation_data=(x_test_features, test_y))",
"_____no_output_____"
],
[
"from matplotlib import pyplot as plt\nplt.plot(history.history['accuracy'])\nplt.plot(history.history['val_accuracy'])\nplt.title('model accuracy')\nplt.ylabel('accuracy')\nplt.xlabel('epoch')\nplt.legend(['train', 'test'], loc='upper left')\nplt.grid()\nplt.show()\n",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix,f1_score, precision_score,recall_score",
"_____no_output_____"
],
[
"import seaborn as sns\nimport matplotlib.pyplot as plt \n\nax= plt.subplot()\nsns.heatmap(cf_matrix, annot=True, ax = ax,cmap='Blues',fmt=''); #annot=True to annotate cells\n\n# labels, title and ticks\nax.set_xlabel('Predicted labels');\nax.set_ylabel('True labels'); \nax.set_title('Confusion Matrix'); \nax.xaxis.set_ticklabels(['Not Spam', 'Spam']); ax.yaxis.set_ticklabels(['Not Spam', 'Spam']);",
"_____no_output_____"
],
[
"y_predict = [1 if o>0.5 else 0 for o in model.predict(x_test_features)]",
"_____no_output_____"
],
[
"cf_matrix =confusion_matrix(test_y,y_predict)",
"_____no_output_____"
],
[
"tn, fp, fn, tp = confusion_matrix(test_y,y_predict).ravel()",
"_____no_output_____"
],
[
"print(\"Precision: {:.2f}%\".format(100 * precision_score(test_y, y_predict)))\nprint(\"Recall: {:.2f}%\".format(100 * recall_score(test_y, y_predict)))\nprint(\"F1 Score: {:.2f}%\".format(100 * f1_score(test_y,y_predict)))",
"_____no_output_____"
],
[
"f1_score(test_y,y_predict)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb62a858b709ae9b37ac585982c26882723d4ee | 287,651 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Keras_assignment-checkpoint.ipynb | DeepLearningVision-2019/a4-keras-classification-pepebm | 3822af720d09bb6fd163ed4bc1d798b8d02adbb0 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Keras_assignment-checkpoint.ipynb | DeepLearningVision-2019/a4-keras-classification-pepebm | 3822af720d09bb6fd163ed4bc1d798b8d02adbb0 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Keras_assignment-checkpoint.ipynb | DeepLearningVision-2019/a4-keras-classification-pepebm | 3822af720d09bb6fd163ed4bc1d798b8d02adbb0 | [
"MIT"
] | null | null | null | 198.516908 | 74,504 | 0.893844 | [
[
[
"# Classify different data sets",
"_____no_output_____"
],
[
"### Basic includes",
"_____no_output_____"
]
],
[
[
"# Using pandas to load the csv file\nimport pandas as pd\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom keras import models \nfrom keras import layers \nfrom keras import callbacks\nfrom keras.utils import to_categorical\n\n# reuters and fashin mnist data set from keras\nfrom keras.datasets import reuters\nfrom keras.datasets import fashion_mnist\n\n# needed to preprocess text\nfrom keras.preprocessing.text import Tokenizer",
"Using TensorFlow backend.\n"
]
],
[
[
"### Classify the Fashion Mnist\n\n---",
"_____no_output_____"
]
],
[
[
"(fashion_train_data, fashion_train_labels), (fashion_test_data, fashion_test_labels) = fashion_mnist.load_data()\nfashion_class_labels = [\n 'T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat', 'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot'\n]\nprint(fashion_train_data.shape)\n\ntest_index = 10\n\nplt.title(\"Label: \" + fashion_class_labels[fashion_train_labels[test_index]])\nplt.imshow(fashion_train_data[test_index], cmap=\"gray\")",
"(60000, 28, 28)\n"
]
],
[
[
"#### TO DO: Preprocess the data\n\n1. Normalize the input data set\n2. Perform one hot encoding\n3. Create a train, test, and validation set",
"_____no_output_____"
]
],
[
[
"# Normalize the input data set\n# flatten images\nfashion_train_data = fashion_train_data.reshape((60000, 784))\nfashion_train_data = fashion_train_data.astype('float32') / 255\n\nfashion_test_data = fashion_test_data.reshape((10000, 784)) \nfashion_test_data = fashion_test_data.astype('float32') / 255\n\n# one hot encoding\nfashion_train_labels = to_categorical(fashion_train_labels) \nfashion_test_labels = to_categorical(fashion_test_labels)\n\nvalidation_set_labels = fashion_train_labels[50000:]\nvalidation_set = fashion_train_data[50000:]\n\ntraining_set_labels = fashion_train_labels[:50000]\ntraining_set = fashion_train_data[:50000]",
"_____no_output_____"
]
],
[
[
"#### TO DO: Define and train a network, then plot the accuracy of the training, validation, and testing\n\n1. Use a validation set\n2. Propose and train a network\n3. Print the history of the training\n4. Evaluate with a test set",
"_____no_output_____"
]
],
[
[
"# Crate NN\n\nnn_fashion_model = models.Sequential()\nfashion_dropout = 0.3\n\nnn_fashion_model.add(layers.Dense(1024, activation= \"relu\", input_shape= (784,)))\n\nnn_fashion_model.add(layers.Dropout(fashion_dropout))\n\nnn_fashion_model.add(layers.Dense(256, activation=\"relu\"))\nnn_fashion_model.add(layers.Dense(128, activation=\"relu\"))\n\nnn_fashion_model.add(layers.Dropout(fashion_dropout))\n\n# Last layer, same size has the number of categories\nnn_fashion_model.add(layers.Dense(10, activation=\"softmax\"))\n\n\n\nnn_fashion_early_stops = [\n callbacks.EarlyStopping(monitor= 'val_loss', patience= 4)\n]\n\nnn_fashion_model.compile(\n loss= \"categorical_crossentropy\", optimizer= \"adam\", metrics= [\"accuracy\"]\n)\n\nnn_fashion_model.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_47 (Dense) (None, 1024) 803840 \n_________________________________________________________________\ndropout_17 (Dropout) (None, 1024) 0 \n_________________________________________________________________\ndense_48 (Dense) (None, 256) 262400 \n_________________________________________________________________\ndense_49 (Dense) (None, 128) 32896 \n_________________________________________________________________\ndropout_18 (Dropout) (None, 128) 0 \n_________________________________________________________________\ndense_50 (Dense) (None, 10) 1290 \n=================================================================\nTotal params: 1,100,426\nTrainable params: 1,100,426\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Train the NN model\nfashion_epochs = 16\nnn_fashion_history = nn_fashion_model.fit(\n fashion_train_data,\n fashion_train_labels,\n batch_size= 1024,\n epochs= fashion_epochs,\n verbose= 2,\n callbacks= nn_fashion_early_stops,\n validation_data= (validation_set, validation_set_labels)\n)",
"Train on 60000 samples, validate on 10000 samples\nEpoch 1/16\n - 8s - loss: 0.7594 - acc: 0.7338 - val_loss: 0.4379 - val_acc: 0.8434\nEpoch 2/16\n - 7s - loss: 0.4523 - acc: 0.8404 - val_loss: 0.3780 - val_acc: 0.8627\nEpoch 3/16\n - 7s - loss: 0.3956 - acc: 0.8588 - val_loss: 0.3368 - val_acc: 0.8777\nEpoch 4/16\n - 7s - loss: 0.3634 - acc: 0.8692 - val_loss: 0.3245 - val_acc: 0.8812\nEpoch 5/16\n - 7s - loss: 0.3418 - acc: 0.8758 - val_loss: 0.3016 - val_acc: 0.8895\nEpoch 6/16\n - 8s - loss: 0.3206 - acc: 0.8832 - val_loss: 0.2827 - val_acc: 0.8957\nEpoch 7/16\n - 8s - loss: 0.3106 - acc: 0.8865 - val_loss: 0.2850 - val_acc: 0.8949\nEpoch 8/16\n - 7s - loss: 0.3032 - acc: 0.8897 - val_loss: 0.2718 - val_acc: 0.8949\nEpoch 9/16\n - 7s - loss: 0.2951 - acc: 0.8915 - val_loss: 0.2719 - val_acc: 0.8972\nEpoch 10/16\n - 8s - loss: 0.2813 - acc: 0.8959 - val_loss: 0.2448 - val_acc: 0.9105\nEpoch 11/16\n - 7s - loss: 0.2728 - acc: 0.8990 - val_loss: 0.2459 - val_acc: 0.9050\nEpoch 12/16\n - 7s - loss: 0.2696 - acc: 0.8997 - val_loss: 0.2335 - val_acc: 0.9101\nEpoch 13/16\n - 7s - loss: 0.2591 - acc: 0.9043 - val_loss: 0.2241 - val_acc: 0.9148\nEpoch 14/16\n - 7s - loss: 0.2564 - acc: 0.9056 - val_loss: 0.2170 - val_acc: 0.9170\nEpoch 15/16\n - 7s - loss: 0.2504 - acc: 0.9066 - val_loss: 0.2166 - val_acc: 0.9173\nEpoch 16/16\n - 7s - loss: 0.2433 - acc: 0.9080 - val_loss: 0.2176 - val_acc: 0.9173\n"
],
[
"fashion_result = nn_fashion_model.evaluate(fashion_test_data, fashion_test_labels)\nprint('Fashion score: {}%'.format(fashion_result[1]*100))",
"10000/10000 [==============================] - 1s 105us/step\nFashion score: 88.74%\n"
],
[
"fashion_history = nn_fashion_history.history\nfashion_loss = fashion_history['loss']\nfashion_val_loss = fashion_history['val_loss']\nfashion_epochs = range(1, len(fashion_loss) + 1)\n\nplt.plot(fashion_epochs, fashion_loss, 'go', label='Training Loss')\nplt.plot(fashion_epochs, fashion_val_loss, 'r', label='Validation Loss')\n\nplt.title('Fashion Data - Loss')\nplt.xlabel('Epochs')\nplt.ylabel('Loss')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
],
[
"fashion_acc = fashion_history['acc']\nfashion_val_acc = fashion_history['val_acc']\n\nplt.plot(fashion_epochs, fashion_acc, 'go', label='Training Acc')\nplt.plot(fashion_epochs, fashion_val_acc, 'r', label='Validation Acc')\n\nplt.title('Fashion Data - Accuracy')\nplt.xlabel('Epochs')\nplt.ylabel('Loss')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Fashion conclusion\n> In this model I first did over fitting so making the model network complex, from that point I moved some hyper parameters and successfully achieved a score above 85%.",
"_____no_output_____"
],
[
"## Classifying newswires\n\n---\n\nBuild a network to classify Reuters newswires into 46 different mutually-exclusive topics.",
"_____no_output_____"
],
[
"### Load and review the data",
"_____no_output_____"
]
],
[
[
"reuters_max_words = 10000\n(reuters_train_data, reuters_train_labels), (reuters_test_data, reuters_test_labels) = reuters.load_data(num_words=reuters_max_words)\n\nprint(reuters_train_data.shape)\nprint(reuters_train_labels.shape)\nprint(reuters_train_data[0])\nprint(reuters_train_labels[0])\n\nprint(set(reuters_train_labels))",
"(8982,)\n(8982,)\n[1, 2, 2, 8, 43, 10, 447, 5, 25, 207, 270, 5, 3095, 111, 16, 369, 186, 90, 67, 7, 89, 5, 19, 102, 6, 19, 124, 15, 90, 67, 84, 22, 482, 26, 7, 48, 4, 49, 8, 864, 39, 209, 154, 6, 151, 6, 83, 11, 15, 22, 155, 11, 15, 7, 48, 9, 4579, 1005, 504, 6, 258, 6, 272, 11, 15, 22, 134, 44, 11, 15, 16, 8, 197, 1245, 90, 67, 52, 29, 209, 30, 32, 132, 6, 109, 15, 17, 12]\n3\n{0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45}\n"
]
],
[
[
"Load the word index to decode the train data.",
"_____no_output_____"
]
],
[
[
"word_index = reuters.get_word_index()\n\nreverse_index = dict([(value+3, key) for (key, value) in word_index.items()])\n\nreverse_index[0] = \"<PAD>\"\nreverse_index[1] = \"<START>\"\nreverse_index[2] = \"<UNKNOWN>\" # unknown\nreverse_index[3] = \"<UNUSED>\"\n\ndecoded_review = ' '.join([reverse_index.get(i,'?') for i in reuters_train_data[0]])\n\nprint(decoded_review)",
"<START> <UNKNOWN> <UNKNOWN> said as a result of its december acquisition of space co it expects earnings per share in 1987 of 1 15 to 1 30 dlrs per share up from 70 cts in 1986 the company said pretax net should rise to nine to 10 mln dlrs from six mln dlrs in 1986 and rental operation revenues to 19 to 22 mln dlrs from 12 5 mln dlrs it said cash flow per share this year should be 2 50 to three dlrs reuter 3\n"
]
],
[
[
"#### TO DO: Preprocess the data\n\n1. Normalize the input data set\n2. Perform one hot encoding\n3. Create a train, test, and validation set",
"_____no_output_____"
]
],
[
[
"tokenizer = Tokenizer(num_words= reuters_max_words)\n\nreuters_train_data_token = tokenizer.sequences_to_matrix(\n reuters_train_data, mode=\"binary\"\n)\nreuters_test_data_token = tokenizer.sequences_to_matrix(\n reuters_test_data, mode=\"binary\"\n)\n\nreuters_one_hot_train_labels = to_categorical(reuters_train_labels)\nreuters_one_hot_test_labels = to_categorical(reuters_test_labels)\n\nreuters_val_data = reuters_train_data_token[:1000]\nreuters_val_labels = reuters_one_hot_train_labels[:1000]\n\nreuters_train_data = reuters_train_data_token[1000:]\nreuters_train_labels = reuters_one_hot_train_labels[1000:]\n\nprint('train:')\nprint(reuters_train_data.shape)\nprint(reuters_train_labels.shape)\nprint('val:')\nprint(reuters_val_data.shape)\nprint(reuters_val_labels.shape)\nprint('test:')\nprint(reuters_test_data_token.shape)\nprint(reuters_one_hot_test_labels.shape)",
"train:\n(7982, 10000)\n(7982, 46)\nval:\n(1000, 10000)\n(1000, 46)\ntest:\n(2246, 10000)\n(2246, 46)\n"
]
],
[
[
"#### TO DO: Define and train a network, then plot the accuracy of the training, validation, and testing\n\n1. Use a validation set\n2. Propose and train a network\n3. Print the history of the training\n4. Evaluate with a test set",
"_____no_output_____"
]
],
[
[
"reuters_model = models.Sequential()\nreuters_dropout = 0.2\n\nreuters_model.add(layers.Dense(1024, activation=\"tanh\", input_dim=10000))\n\nreuters_model.add(layers.Dropout(reuters_dropout))\n\nreuters_model.add(layers.Dense(256, activation=\"relu\"))\nreuters_model.add(layers.Dense(128, activation=\"relu\"))\n\nreuters_model.add(layers.Dropout(reuters_dropout))\n\nreuters_model.add(layers.Dense(46, activation=\"softmax\"))\n\nreuters_model.compile(\n loss= \"categorical_crossentropy\", \n optimizer= \"adamax\", \n metrics= [\"accuracy\"]\n)\n\nreuters_model.summary()\n\n\nreuters_early_stops = [\n callbacks.EarlyStopping(monitor= 'val_loss', patience= 4),\n callbacks.EarlyStopping(monitor= 'val_acc', patience= 5)\n]",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_51 (Dense) (None, 1024) 10241024 \n_________________________________________________________________\ndropout_19 (Dropout) (None, 1024) 0 \n_________________________________________________________________\ndense_52 (Dense) (None, 256) 262400 \n_________________________________________________________________\ndense_53 (Dense) (None, 128) 32896 \n_________________________________________________________________\ndropout_20 (Dropout) (None, 128) 0 \n_________________________________________________________________\ndense_54 (Dense) (None, 46) 5934 \n=================================================================\nTotal params: 10,542,254\nTrainable params: 10,542,254\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"reuters_model_history = reuters_model.fit(\n reuters_train_data,\n reuters_train_labels,\n batch_size= 1024,\n epochs= 16,\n verbose= 2,\n callbacks= reuters_early_stops,\n validation_data= (reuters_val_data, reuters_val_labels)\n)",
"Train on 7982 samples, validate on 1000 samples\nEpoch 1/16\n - 8s - loss: 2.5642 - acc: 0.4818 - val_loss: 1.5615 - val_acc: 0.6560\nEpoch 2/16\n - 7s - loss: 1.3029 - acc: 0.7129 - val_loss: 1.1722 - val_acc: 0.7310\nEpoch 3/16\n - 7s - loss: 0.9083 - acc: 0.7968 - val_loss: 0.9980 - val_acc: 0.7890\nEpoch 4/16\n - 7s - loss: 0.6427 - acc: 0.8513 - val_loss: 0.9031 - val_acc: 0.8120\nEpoch 5/16\n - 7s - loss: 0.4676 - acc: 0.8968 - val_loss: 0.8801 - val_acc: 0.8220\nEpoch 6/16\n - 7s - loss: 0.3395 - acc: 0.9266 - val_loss: 0.9141 - val_acc: 0.8120\nEpoch 7/16\n - 7s - loss: 0.2594 - acc: 0.9381 - val_loss: 0.9113 - val_acc: 0.8210\nEpoch 8/16\n - 7s - loss: 0.2066 - acc: 0.9461 - val_loss: 0.9337 - val_acc: 0.8160\nEpoch 9/16\n - 7s - loss: 0.1731 - acc: 0.9544 - val_loss: 0.9800 - val_acc: 0.8180\n"
],
[
"reuters_result = reuters_model.evaluate(\n reuters_test_data_token, \n reuters_one_hot_test_labels\n)\n\nprint('Fashion score: {}%'.format(reuters_result[1]*100))",
"2246/2246 [==============================] - 2s 721us/step\nFashion score: 80.00890471950134%\n"
],
[
"reuters_history = reuters_model_history.history\nreuters_loss = reuters_history['loss']\nreuters_val_loss = reuters_history['val_loss']\nreuters_epochs = range(1, len(reuters_loss) + 1)\n\nplt.plot(reuters_epochs, reuters_loss, 'go', label='Training Loss')\nplt.plot(reuters_epochs, reuters_val_loss, 'r', label='Validation Loss')\n\nplt.title('Reuters Data - Loss')\nplt.xlabel('Epochs')\nplt.ylabel('Loss')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
],
[
"reuters_acc = reuters_history['acc']\nreuters_val_acc = reuters_history['val_acc']\n\nplt.plot(reuters_epochs, reuters_acc, 'go', label='Training Acc')\nplt.plot(reuters_epochs, reuters_val_acc, 'r', label='Validation Acc')\n\nplt.title('Reuters Data - Accuracy')\n\nplt.xlabel('Epochs')\nplt.ylabel('Accuracy')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Reuters conclusion\n\n> 1. This model was harder to achieve the accuracy goal. Like the past model, first, I over fitted the model but the accuracy in that point was at it's lowest in all the exercise.\n> 2. Moving hyper parameters wasn't getting me closer to the goal. Neither changing my preprocessing work flow.\n> 3. My final conclusion is that I couldn't get an final accuracy higher than 78% 'cause of the dataset values. My hypothesis is the data is not evenly given to me so I have more cases of 1 category than another. ",
"_____no_output_____"
],
[
"## Predicting Student Admissions\n\n---\n\nPredict student admissions based on three pieces of data:\n\n- GRE Scores\n- GPA Scores\n- Class rank",
"_____no_output_____"
],
[
"### Load and visualize the data",
"_____no_output_____"
]
],
[
[
"student_data = pd.read_csv(\"student_data.csv\")",
"_____no_output_____"
]
],
[
[
"Plot of the GRE and the GPA from the data.",
"_____no_output_____"
]
],
[
[
"X = np.array(student_data[[\"gre\",\"gpa\"]])\ny = np.array(student_data[\"admit\"])\nadmitted = X[np.argwhere(y==1)]\nrejected = X[np.argwhere(y==0)]\nplt.scatter([s[0][0] for s in rejected], [s[0][1] for s in rejected], s = 25, color = 'red', edgecolor = 'k')\nplt.scatter([s[0][0] for s in admitted], [s[0][1] for s in admitted], s = 25, color = 'cyan', edgecolor = 'k')\nplt.xlabel('Test (GRE)')\nplt.ylabel('Grades (GPA)')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Plot of the data by class rank.",
"_____no_output_____"
]
],
[
[
"f, plots = plt.subplots(2, 2, figsize=(20,10))\nplots = [plot for sublist in plots for plot in sublist]\n\nfor idx, plot in enumerate(plots):\n data_rank = student_data[student_data[\"rank\"]==idx+1]\n plot.set_title(\"Rank \" + str(idx+1))\n X = np.array(data_rank[[\"gre\",\"gpa\"]])\n y = np.array(data_rank[\"admit\"])\n admitted = X[np.argwhere(y==1)]\n rejected = X[np.argwhere(y==0)]\n plot.scatter([s[0][0] for s in rejected], [s[0][1] for s in rejected], s = 25, color = 'red', edgecolor = 'k')\n plot.scatter([s[0][0] for s in admitted], [s[0][1] for s in admitted], s = 25, color = 'cyan', edgecolor = 'k')\n plot.set_xlabel('Test (GRE)')\n plot.set_ylabel('Grades (GPA)')\n ",
"_____no_output_____"
]
],
[
[
"#### TO DO: Preprocess the data\n\n1. Normalize the input data set\n2. Perform one hot encoding\n3. Create a train, test, and validation set",
"_____no_output_____"
]
],
[
[
"# Replace nan with 0\nstudent_data.fillna(value= 0, inplace= True)\n# Shuffle the dataframe with pandas\nstudent_data = student_data.sample(frac= 1).reset_index(drop= True)\n\n# x\ngre, gpa = np.array(student_data['gre']), np.array(student_data['gpa'])\n# y\nadmit, rank = np.array(student_data['admit']), np.array(student_data['rank'])\n\n# Make everything in range from 0 - 1 (Normal distribution)\ngpa = (gpa - gpa.mean(axis= 0)) / gpa.std(axis= 0)\ngre = (gre - gre.mean(axis= 0)) / gre.std(axis= 0)\n\nnormalized_student_data = np.zeros((len(gpa), 2))\nnormalized_student_data[:,0], normalized_student_data[:,1] = gpa, gre\n\nprint(normalized_student_data.shape)\n\n# one hot encoding\nrank_one_hot = to_categorical(rank)\n\n# train: 0 300, test: 300 350, val: 350 4000\nstudent_train_data = normalized_student_data[:300]\nstudent_train_labels = rank_one_hot[:300]\n\nstudent_test_data = normalized_student_data[300:350]\nstudent_test_labels = rank_one_hot[300:350]\n\nstudent_val_data = normalized_student_data[350:]\nstudent_val_labels = rank_one_hot[350:]",
"(400, 2)\n"
]
],
[
[
"#### TO DO: Define and train a network, then plot the accuracy of the training, validation, and testing\n\n1. Use a validation set\n2. Propose and train a network\n3. Print the history of the training\n4. Evaluate with a test set",
"_____no_output_____"
]
],
[
[
"student_model = models.Sequential()\nstudent_dropout = 0.3\n\nstudent_model.add(layers.Dense(128, activation= 'sigmoid', input_shape=(2,)))\n\nstudent_model.add(layers.Dropout(student_dropout))\n\nstudent_model.add(layers.Dense(32, activation= 'sigmoid'))\nstudent_model.add(layers.Dense(5, activation= 'sigmoid'))\n\nstudent_model.compile(\n optimizer= \"rmsprop\", \n loss= \"binary_crossentropy\", \n metrics=[\"accuracy\"]\n)\n\nstudent_model.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_7 (Dense) (None, 128) 384 \n_________________________________________________________________\ndropout_3 (Dropout) (None, 128) 0 \n_________________________________________________________________\ndense_8 (Dense) (None, 32) 4128 \n_________________________________________________________________\ndense_9 (Dense) (None, 5) 165 \n=================================================================\nTotal params: 4,677\nTrainable params: 4,677\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"student_model_history = student_model.fit(\n student_train_data,\n student_train_labels,\n epochs= 16,\n batch_size= 64,\n validation_data= (student_val_data, student_val_labels),\n verbose= 2\n)",
"Train on 300 samples, validate on 50 samples\nEpoch 1/16\n - 0s - loss: 0.5965 - acc: 0.7493 - val_loss: 0.5405 - val_acc: 0.8000\nEpoch 2/16\n - 0s - loss: 0.5228 - acc: 0.8000 - val_loss: 0.5107 - val_acc: 0.8000\nEpoch 3/16\n - 0s - loss: 0.4937 - acc: 0.8000 - val_loss: 0.4963 - val_acc: 0.8000\nEpoch 4/16\n - 0s - loss: 0.4767 - acc: 0.8000 - val_loss: 0.4880 - val_acc: 0.8000\nEpoch 5/16\n - 0s - loss: 0.4661 - acc: 0.8000 - val_loss: 0.4825 - val_acc: 0.8000\nEpoch 6/16\n - 0s - loss: 0.4593 - acc: 0.8000 - val_loss: 0.4784 - val_acc: 0.8000\nEpoch 7/16\n - 0s - loss: 0.4539 - acc: 0.8000 - val_loss: 0.4748 - val_acc: 0.8000\nEpoch 8/16\n - 0s - loss: 0.4488 - acc: 0.8000 - val_loss: 0.4725 - val_acc: 0.8000\nEpoch 9/16\n - 0s - loss: 0.4476 - acc: 0.8000 - val_loss: 0.4709 - val_acc: 0.8000\nEpoch 10/16\n - 0s - loss: 0.4444 - acc: 0.8000 - val_loss: 0.4700 - val_acc: 0.8000\nEpoch 11/16\n - 0s - loss: 0.4437 - acc: 0.8000 - val_loss: 0.4684 - val_acc: 0.8000\nEpoch 12/16\n - 0s - loss: 0.4426 - acc: 0.8000 - val_loss: 0.4672 - val_acc: 0.8000\nEpoch 13/16\n - 0s - loss: 0.4406 - acc: 0.8000 - val_loss: 0.4655 - val_acc: 0.8000\nEpoch 14/16\n - 0s - loss: 0.4391 - acc: 0.8000 - val_loss: 0.4660 - val_acc: 0.8000\nEpoch 15/16\n - 0s - loss: 0.4384 - acc: 0.8000 - val_loss: 0.4661 - val_acc: 0.8000\nEpoch 16/16\n - 0s - loss: 0.4372 - acc: 0.8000 - val_loss: 0.4658 - val_acc: 0.8000\n"
],
[
"student_result = student_model.evaluate(\n student_test_data,\n student_test_labels\n)\n\nprint('Student score: {}'.format(student_result[1] * 100))",
"50/50 [==============================] - 0s 122us/step\nStudent score: 80.00000500679016\n"
],
[
"student_history = student_model_history.history\n\nstudent_val_loss = student_history['val_loss']\nstudent_loss = student_history['loss']\nstudent_epochs = range(1, len(student_loss) + 1)\n\nplt.plot(student_epochs, student_loss, 'go', label= \"Training Loss\")\nplt.plot(student_epochs, student_val_loss, 'r', label= \"Validation Loss\")\nplt.title(\"Student Data - Loss\")\nplt.ylabel(\"Loss\")\nplt.xlabel(\"Epochs\")\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"student_val_acc = student_history['val_acc']\nstudent_acc = student_history['acc']\n\nplt.plot(student_epochs, student_acc, 'go', label= \"Training Acc\")\nplt.plot(student_epochs, student_val_acc, 'r', label= \"Validation Acc\")\nplt.title(\"Student Data - Accuracy\")\nplt.ylabel(\"Accuracy\")\nplt.xlabel(\"Epochs\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Student conclusion\n> This model was the hardest given the small dataset provided, we had 400 rows for training, validation & testing.<br>\n> I believe that my preprocessing work flow could be better due that I do not use any statistical function to better the data. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecb62ba84445af6e41e9906a6f1b6857a88ea191 | 590,952 | ipynb | Jupyter Notebook | examples/net_surgery.ipynb | jiecaoyu/caffe_cccp | 60a4e6c297d163e89f70644c2f3ba6ae61049cb3 | [
"BSD-2-Clause"
] | 2 | 2016-01-08T21:02:12.000Z | 2016-01-29T09:41:54.000Z | examples/net_surgery.ipynb | jiecaoyu/caffe_cccp | 60a4e6c297d163e89f70644c2f3ba6ae61049cb3 | [
"BSD-2-Clause"
] | null | null | null | examples/net_surgery.ipynb | jiecaoyu/caffe_cccp | 60a4e6c297d163e89f70644c2f3ba6ae61049cb3 | [
"BSD-2-Clause"
] | null | null | null | 85.249856 | 888 | 0.827802 | [
[
[
"# Net Surgery\n\nCaffe networks can be transformed to your particular needs by editing the model parameters. The data, diffs, and parameters of a net are all exposed in pycaffe.\n\nRoll up your sleeves for net surgery with pycaffe!",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport Image\n\n# Make sure that caffe is on the python path:\ncaffe_root = '../' # this file is expected to be in {caffe_root}/examples\nimport sys\nsys.path.insert(0, caffe_root + 'python')\n\nimport caffe\n\n# configure plotting\nplt.rcParams['figure.figsize'] = (10, 10)\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'",
"_____no_output_____"
]
],
[
[
"## Designer Filters\n\nTo show how to load, manipulate, and save parameters we'll design our own filters into a simple network that's only a single convolution layer. This net has two blobs, `data` for the input and `conv` for the convolution output and one parameter `conv` for the convolution filter weights and biases.",
"_____no_output_____"
]
],
[
[
"# Load the net, list its data and params, and filter an example image.\ncaffe.set_mode_cpu()\nnet = caffe.Net('net_surgery/conv.prototxt', caffe.TEST)\nprint(\"blobs {}\\nparams {}\".format(net.blobs.keys(), net.params.keys()))\n\n# load image and prepare as a single input batch for Caffe\nim = np.array(Image.open('images/cat_gray.jpg'))\nplt.title(\"original image\")\nplt.imshow(im)\nplt.axis('off')\n\nim_input = im[np.newaxis, np.newaxis, :, :]\nnet.blobs['data'].reshape(*im_input.shape)\nnet.blobs['data'].data[...] = im_input",
"blobs ['data', 'conv']\nparams ['conv']\n"
]
],
[
[
"The convolution weights are initialized from Gaussian noise while the biases are initialized to zero. These random filters give output somewhat like edge detections.",
"_____no_output_____"
]
],
[
[
"# helper show filter outputs\ndef show_filters(net):\n net.forward()\n plt.figure()\n filt_min, filt_max = net.blobs['conv'].data.min(), net.blobs['conv'].data.max()\n for i in range(3):\n plt.subplot(1,4,i+2)\n plt.title(\"filter #{} output\".format(i))\n plt.imshow(net.blobs['conv'].data[0, i], vmin=filt_min, vmax=filt_max)\n plt.tight_layout()\n plt.axis('off')\n\n# filter the image with initial \nshow_filters(net)",
"_____no_output_____"
]
],
[
[
"Raising the bias of a filter will correspondingly raise its output:",
"_____no_output_____"
]
],
[
[
"# pick first filter output\nconv0 = net.blobs['conv'].data[0, 0]\nprint(\"pre-surgery output mean {:.2f}\".format(conv0.mean()))\n# set first filter bias to 10\nnet.params['conv'][1].data[0] = 1.\nnet.forward()\nprint(\"post-surgery output mean {:.2f}\".format(conv0.mean()))",
"pre-surgery output mean 3.62\npost-surgery output mean 4.62\n"
]
],
[
[
"Altering the filter weights is more exciting since we can assign any kernel like Gaussian blur, the Sobel operator for edges, and so on. The following surgery turns the 0th filter into a Gaussian blur and the 1st and 2nd filters into the horizontal and vertical gradient parts of the Sobel operator.\n\nSee how the 0th output is blurred, the 1st picks up horizontal edges, and the 2nd picks up vertical edges.",
"_____no_output_____"
]
],
[
[
"ksize = net.params['conv'][0].data.shape[2:]\n# make Gaussian blur\nsigma = 1.\ny, x = np.mgrid[-ksize[0]//2 + 1:ksize[0]//2 + 1, -ksize[1]//2 + 1:ksize[1]//2 + 1]\ng = np.exp(-((x**2 + y**2)/(2.0*sigma**2)))\ngaussian = (g / g.sum()).astype(np.float32)\nnet.params['conv'][0].data[0] = gaussian\n# make Sobel operator for edge detection\nnet.params['conv'][0].data[1:] = 0.\nsobel = np.array((-1, -2, -1, 0, 0, 0, 1, 2, 1), dtype=np.float32).reshape((3,3))\nnet.params['conv'][0].data[1, 0, 1:-1, 1:-1] = sobel # horizontal\nnet.params['conv'][0].data[2, 0, 1:-1, 1:-1] = sobel.T # vertical\nshow_filters(net)",
"_____no_output_____"
]
],
[
[
"With net surgery, parameters can be transplanted across nets, regularized by custom per-parameter operations, and transformed according to your schemes.",
"_____no_output_____"
],
[
"## Casting a Classifier into a Fully Convolutional Network\n\nLet's take the standard Caffe Reference ImageNet model \"CaffeNet\" and transform it into a fully convolutional net for efficient, dense inference on large inputs. This model generates a classification map that covers a given input size instead of a single classification. In particular a 8 $\\times$ 8 classification map on a 451 $\\times$ 451 input gives 64x the output in only 3x the time. The computation exploits a natural efficiency of convolutional network (convnet) structure by amortizing the computation of overlapping receptive fields.\n\nTo do so we translate the `InnerProduct` matrix multiplication layers of CaffeNet into `Convolutional` layers. This is the only change: the other layer types are agnostic to spatial size. Convolution is translation-invariant, activations are elementwise operations, and so on. The `fc6` inner product when carried out as convolution by `fc6-conv` turns into a 6 \\times 6 filter with stride 1 on `pool5`. Back in image space this gives a classification for each 227 $\\times$ 227 box with stride 32 in pixels. Remember the equation for output map / receptive field size, output = (input - kernel_size) / stride + 1, and work out the indexing details for a clear understanding.",
"_____no_output_____"
]
],
[
[
"!diff net_surgery/bvlc_caffenet_full_conv.prototxt ../models/bvlc_reference_caffenet/deploy.prototxt",
"diff: ../models/bvlc_reference_caffenet/deploy.prototxt: No such file or directory\r\n"
]
],
[
[
"The only differences needed in the architecture are to change the fully connected classifier inner product layers into convolutional layers with the right filter size -- 6 x 6, since the reference model classifiers take the 36 elements of `pool5` as input -- and stride 1 for dense classification. Note that the layers are renamed so that Caffe does not try to blindly load the old parameters when it maps layer names to the pretrained model.",
"_____no_output_____"
]
],
[
[
"# Make sure that caffe is on the python path:\ncaffe_root = '../' # this file is expected to be in {caffe_root}/examples\nimport sys\nsys.path.insert(0, caffe_root + 'python')\n\nimport caffe\n\n# Load the original network and extract the fully connected layers' parameters.\nnet = caffe.Net('../models/bvlc_reference_caffenet/deploy.prototxt', \n '../models/bvlc_reference_caffenet/bvlc_reference_caffenet.caffemodel', \n caffe.TEST)\nparams = ['fc6', 'fc7', 'fc8']\n# fc_params = {name: (weights, biases)}\nfc_params = {pr: (net.params[pr][0].data, net.params[pr][1].data) for pr in params}\n\nfor fc in params:\n print '{} weights are {} dimensional and biases are {} dimensional'.format(fc, fc_params[fc][0].shape, fc_params[fc][1].shape)",
"_____no_output_____"
]
],
[
[
"Consider the shapes of the inner product parameters. The weight dimensions are the output and input sizes while the bias dimension is the output size.",
"_____no_output_____"
]
],
[
[
"# Load the fully convolutional network to transplant the parameters.\nnet_full_conv = caffe.Net('net_surgery/bvlc_caffenet_full_conv.prototxt', \n '../models/bvlc_reference_caffenet/bvlc_reference_caffenet.caffemodel',\n caffe.TEST)\nparams_full_conv = ['fc6-conv', 'fc7-conv', 'fc8-conv']\n# conv_params = {name: (weights, biases)}\nconv_params = {pr: (net_full_conv.params[pr][0].data, net_full_conv.params[pr][1].data) for pr in params_full_conv}\n\nfor conv in params_full_conv:\n print '{} weights are {} dimensional and biases are {} dimensional'.format(conv, conv_params[conv][0].shape, conv_params[conv][1].shape)",
"fc6-conv weights are (4096, 256, 6, 6) dimensional and biases are (4096,) dimensional\nfc7-conv weights are (4096, 4096, 1, 1) dimensional and biases are (4096,) dimensional\nfc8-conv weights are (1000, 4096, 1, 1) dimensional and biases are (1000,) dimensional\n"
]
],
[
[
"The convolution weights are arranged in output $\\times$ input $\\times$ height $\\times$ width dimensions. To map the inner product weights to convolution filters, we could roll the flat inner product vectors into channel $\\times$ height $\\times$ width filter matrices, but actually these are identical in memory (as row major arrays) so we can assign them directly.\n\nThe biases are identical to those of the inner product.\n\nLet's transplant!",
"_____no_output_____"
]
],
[
[
"for pr, pr_conv in zip(params, params_full_conv):\n conv_params[pr_conv][0].flat = fc_params[pr][0].flat # flat unrolls the arrays\n conv_params[pr_conv][1][...] = fc_params[pr][1]",
"_____no_output_____"
]
],
[
[
"Next, save the new model weights.",
"_____no_output_____"
]
],
[
[
"net_full_conv.save('net_surgery/bvlc_caffenet_full_conv.caffemodel')",
"_____no_output_____"
]
],
[
[
"To conclude, let's make a classification map from the example cat image and visualize the confidence of \"tiger cat\" as a probability heatmap. This gives an 8-by-8 prediction on overlapping regions of the 451 $\\times$ 451 input.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n# load input and configure preprocessing\nim = caffe.io.load_image('images/cat.jpg')\ntransformer = caffe.io.Transformer({'data': net_full_conv.blobs['data'].data.shape})\ntransformer.set_mean('data', np.load('../python/caffe/imagenet/ilsvrc_2012_mean.npy').mean(1).mean(1))\ntransformer.set_transpose('data', (2,0,1))\ntransformer.set_channel_swap('data', (2,1,0))\ntransformer.set_raw_scale('data', 255.0)\n# make classification map by forward and print prediction indices at each location\nout = net_full_conv.forward_all(data=np.asarray([transformer.preprocess('data', im)]))\nprint out['prob'][0].argmax(axis=0)\n# show net input and confidence map (probability of the top prediction at each location)\nplt.subplot(1, 2, 1)\nplt.imshow(transformer.deprocess('data', net_full_conv.blobs['data'].data[0]))\nplt.subplot(1, 2, 2)\nplt.imshow(out['prob'][0,281])",
"[[282 282 281 281 281 281 277 282]\n [281 283 283 281 281 281 281 282]\n [283 283 283 283 283 283 287 282]\n [283 283 283 281 283 283 283 259]\n [283 283 283 283 283 283 283 259]\n [283 283 283 283 283 283 259 259]\n [283 283 283 283 259 259 259 277]\n [335 335 283 259 263 263 263 277]]\n"
]
],
[
[
"The classifications include various cats -- 282 = tiger cat, 281 = tabby, 283 = persian -- and foxes and other mammals.\n\nIn this way the fully connected layers can be extracted as dense features across an image (see `net_full_conv.blobs['fc6'].data` for instance), which is perhaps more useful than the classification map itself.\n\nNote that this model isn't totally appropriate for sliding-window detection since it was trained for whole-image classification. Nevertheless it can work just fine. Sliding-window training and finetuning can be done by defining a sliding-window ground truth and loss such that a loss map is made for every location and solving as usual. (This is an exercise for the reader.)",
"_____no_output_____"
],
[
"*A thank you to Rowland Depp for first suggesting this trick.*",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecb62dc1b5fa3f03d3c238a81d01a3cdc4f09b3a | 354,295 | ipynb | Jupyter Notebook | notebooks/develop/skforecast_ForecasterAutoreg.ipynb | JoaquinAmatRodrigo/skforecast | c94c6ec9331c57a9dbbf8e900a45838bc0418e94 | [
"MIT"
] | 86 | 2021-02-25T08:56:45.000Z | 2022-03-31T01:33:53.000Z | dev/develop/skforecast_ForecasterAutoreg.ipynb | JavierEscobarOrtiz/skforecast | a3af4a1dd4201c582f159d4e3a1734ed6d29b6c5 | [
"MIT"
] | 5 | 2021-11-30T22:30:45.000Z | 2022-03-29T10:21:36.000Z | dev/develop/skforecast_ForecasterAutoreg.ipynb | JavierEscobarOrtiz/skforecast | a3af4a1dd4201c582f159d4e3a1734ed6d29b6c5 | [
"MIT"
] | 24 | 2021-04-04T09:58:26.000Z | 2022-03-09T15:55:44.000Z | 152.123229 | 66,364 | 0.841638 | [
[
[
" ",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n#import sys\n#sys.path.insert(1, '/home/ximo/Documents/GitHub/skforecast')\n%config Completer.use_jedi = False",
"_____no_output_____"
],
[
"# Libraries\n# ==============================================================================\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom sklearn.linear_model import Ridge\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.metrics import mean_squared_error\n\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom skforecast.model_selection import grid_search_forecaster\nfrom skforecast.model_selection import backtesting_forecaster\nimport warnings",
"_____no_output_____"
],
[
"import session_info\nsession_info.show(html=False, write_req_file=False)",
"-----\nmatplotlib 3.4.3\nnumpy 1.19.5\npandas 1.3.0\nsession_info 1.0.0\nskforecast 0.4.2\nsklearn 1.0.1\n-----\nIPython 7.27.0\njupyter_client 6.1.12\njupyter_core 4.7.1\njupyterlab 3.1.11\nnotebook 6.3.0\n-----\nPython 3.7.10 (default, Feb 26 2021, 18:47:35) [GCC 7.3.0]\nLinux-5.11.0-43-generic-x86_64-with-debian-bullseye-sid\n-----\nSession information updated at 2022-01-02 13:26\n"
]
],
[
[
"# Data",
"_____no_output_____"
]
],
[
[
"# Download data\n# ==============================================================================\nurl = ('https://raw.githubusercontent.com/JoaquinAmatRodrigo/skforecast/master/data/h2o_exog.csv')\ndata = pd.read_csv(url, sep=',')\n\n# data preprocessing\n# ==============================================================================\ndata['fecha'] = pd.to_datetime(data['fecha'], format='%Y/%m/%d')\ndata = data.set_index('fecha')\ndata = data.rename(columns={'x': 'y'})\ndata = data.asfreq('MS')\ndata = data.sort_index()\n\n# Plot\n# ==============================================================================\nfig, ax=plt.subplots(figsize=(9, 4))\ndata.plot(ax=ax);\n\n# Split train-test\n# ==============================================================================\nsteps = 36\ndata_train = data.iloc[:-steps, :]\ndata_test = data.iloc[-steps:, :]",
"_____no_output_____"
]
],
[
[
"# ForecasterAutoreg without exogenous variables",
"_____no_output_____"
]
],
[
[
"# Create and fit forecaster\n# ==============================================================================\nregressor = make_pipeline(StandardScaler(), Ridge())\nlags = 15\n\nforecaster = ForecasterAutoreg(\n regressor = regressor,\n lags = lags\n )\n\nforecaster.fit(y=data_train.y)\nforecaster",
"_____no_output_____"
],
[
"# Predict\n# ==============================================================================\npredictions = forecaster.predict(steps)\n\n# Prediction error\n# ==============================================================================\nerror_mse = mean_squared_error(\n y_true = data_test.y,\n y_pred = predictions\n )\nprint(f\"Test error (mse): {error_mse}\")\n\n# Plot\n# ==============================================================================\nfig, ax=plt.subplots(figsize=(9, 4))\ndata_train.y.plot(ax=ax, label='train')\ndata_test.y.plot(ax=ax, label='test')\npredictions.plot(ax=ax, label='predictions')\nax.legend();",
"Test error (mse): 0.010454411313511013\n"
],
[
"# Grid search hiperparameters and lags\n# ==============================================================================\n\n# Regressor hiperparameters\nparam_grid ={'ridge__alpha': [0.01, 0.1, 1, 10]}\n\n# lags used as predictors\nlags_grid = [3, 10, [1,2,3,20]]\n\nresults_grid = grid_search_forecaster(\n forecaster = forecaster,\n y = data_train.y,\n param_grid = param_grid,\n lags_grid = lags_grid,\n steps = 10,\n metric = 'mean_squared_error',\n refit = True,\n initial_train_size = int(len(data_train)*0.5),\n return_best = True,\n verbose = False\n )\n\n# Results grid search\n# ==============================================================================\nresults_grid.head(4)",
"Number of models compared: 12\n"
],
[
"# Predictors importance\n# ==============================================================================\nforecaster.get_coef()",
"_____no_output_____"
],
[
"# Backtesting\n# ==============================================================================\nsteps = 36\nn_backtest = 36 * 3 + 1\ndata_train = data[:-n_backtest]\ndata_test = data[-n_backtest:]\n\nmetrica, predicciones_backtest = backtesting_forecaster(\n forecaster = forecaster,\n y = data.y,\n initial_train_size = len(data_train),\n steps = steps,\n refit = False,\n metric = 'mean_squared_error',\n verbose = True\n )\nprint(metrica)\n\n# Gráfico\n# ==============================================================================\nfig, ax = plt.subplots(figsize=(9, 4))\ndata_train.y.plot(ax=ax, label='train')\ndata_test.y.plot(ax=ax, label='test')\npredicciones_backtest.plot(ax=ax, label='predictions')\nax.legend();",
"Information of backtesting process\n----------------------------------\nNumber of observations used for initial training or as initial window: 86\nNumber of observations used for backtesting: 109\n Number of folds: 4\n Number of steps per fold: 36\n Last fold only includes 1 observations\n\nData partition in fold: 0\n Training: 1992-04-01 00:00:00 -- 1999-05-01 00:00:00\n Validation: 1999-06-01 00:00:00 -- 2002-05-01 00:00:00\nData partition in fold: 1\n Training: 1992-04-01 00:00:00 -- 1999-05-01 00:00:00\n Validation: 2002-06-01 00:00:00 -- 2005-05-01 00:00:00\nData partition in fold: 2\n Training: 1992-04-01 00:00:00 -- 1999-05-01 00:00:00\n Validation: 2005-06-01 00:00:00 -- 2008-05-01 00:00:00\nData partition in fold: 3\n Training: 1992-04-01 00:00:00 -- 1999-05-01 00:00:00\n Validation: 2008-06-01 00:00:00 -- 2008-06-01 00:00:00\n\n[0.07671593]\n"
],
[
"predicciones_backtest",
"_____no_output_____"
],
[
"forecaster.fit(y=data_train.y)\npredictions_1 = forecaster.predict(steps=steps)\npredictions_2 = forecaster.predict(steps=steps, last_window=data_test.y[:steps])\npredictions_3 = forecaster.predict(steps=steps, last_window=data_test.y[steps:steps*2])\npredictions_4 = forecaster.predict(steps=1, last_window=data_test.y[steps*2:steps*3])\nnp.allclose(predicciones_backtest['pred'], np.concatenate([predictions_1, predictions_2, predictions_3, predictions_4]))",
"_____no_output_____"
]
],
[
[
"# ForecasterAutoreg with 1 exogenous variables",
"_____no_output_____"
]
],
[
[
"# Split train-test\n# ==============================================================================\nsteps = 36\ndata_train = data.iloc[:-steps, :]\ndata_test = data.iloc[-steps:, :]",
"_____no_output_____"
],
[
"forecaster = ForecasterAutoreg(\n regressor = regressor,\n lags = lags\n )\nforecaster",
"_____no_output_____"
],
[
"# Create and fit forecaster\n# ==============================================================================\nforecaster.fit(y=data_train.y, exog=data_train.exog_1)\n\n# Predict\n# ==============================================================================\nsteps = 36\npredictions = forecaster.predict(steps=steps, exog=data_test.exog_1)\n\n# Plot\n# ==============================================================================\nfig, ax=plt.subplots(figsize=(9, 4))\ndata_train.y.plot(ax=ax, label='train')\ndata_test.y.plot(ax=ax, label='test')\npredictions.plot(ax=ax, label='predictions')\nax.legend();\n\n# Error prediction\n# ==============================================================================\nerror_mse = mean_squared_error(\n y_true = data_test.y,\n y_pred = predictions\n )\nprint(f\"Test error (mse): {error_mse}\")",
"Test error (mse): 0.012636212003616849\n"
],
[
"# Grid search hiperparameters and lags\n# ==============================================================================\nforecaster = ForecasterAutoreg(\n regressor= make_pipeline(StandardScaler(), RandomForestRegressor(random_state=123)),\n lags=12\n )\n\n# Regressor hiperparameters\nparam_grid = {'randomforestregressor__n_estimators': [50, 100],\n 'randomforestregressor__max_depth': [5, 10]}\n\n# Lags used as predictors\nlags_grid = [3, 10, [1,2,3,20]]\n\nresults_grid = grid_search_forecaster(\n forecaster = forecaster,\n y = data_train.y,\n exog = data_train.exog_1,\n param_grid = param_grid,\n lags_grid = lags_grid,\n steps = 10,\n metric = 'mean_squared_error',\n refit = False,\n initial_train_size = int(len(data_train)*0.5),\n return_best = False,\n verbose = False\n )\n\n# Results grid Search\n# ==============================================================================\nresults_grid.head(4)",
"Number of models compared: 12\n"
],
[
"# Backtesting\n# ==============================================================================\nsteps = 36\nn_backtest = 36 * 3 + 1\ndata_train = data[:-n_backtest]\ndata_test = data[-n_backtest:]\n\nforecaster = ForecasterAutoreg(regressor=LinearRegression(), lags=10)\n\nmetrica, predicciones_backtest = backtesting_forecaster(\n forecaster = forecaster,\n y = data.y,\n exog = data.exog_1,\n initial_train_size = len(data_train),\n steps = steps,\n metric = 'mean_squared_error',\n verbose = True\n)\n\nprint(metrica)",
"Information of backtesting process\n----------------------------------\nNumber of observations used for initial training or as initial window: 86\nNumber of observations used for backtesting: 109\n Number of folds: 4\n Number of steps per fold: 36\n Last fold only includes 1 observations\n\nData partition in fold: 0\n Training: 1992-04-01 00:00:00 -- 1999-05-01 00:00:00\n Validation: 1999-06-01 00:00:00 -- 2002-05-01 00:00:00\nData partition in fold: 1\n Training: 1992-04-01 00:00:00 -- 1999-05-01 00:00:00\n Validation: 2002-06-01 00:00:00 -- 2005-05-01 00:00:00\nData partition in fold: 2\n Training: 1992-04-01 00:00:00 -- 1999-05-01 00:00:00\n Validation: 2005-06-01 00:00:00 -- 2008-05-01 00:00:00\nData partition in fold: 3\n Training: 1992-04-01 00:00:00 -- 1999-05-01 00:00:00\n Validation: 2008-06-01 00:00:00 -- 2008-06-01 00:00:00\n\n[6.21996921e-30]\n"
],
[
"# Verificar predicciones de backtesting\nforecaster.fit(y=data_train.y, exog=data_train.exog_1)\npredictions_1 = forecaster.predict(steps=steps, exog=data_test.exog_1[:steps])\npredictions_2 = forecaster.predict(steps=steps, last_window=data_test.y[:steps], exog=data_test.exog_1[steps:steps*2])\npredictions_3 = forecaster.predict(steps=steps, last_window=data_test.y[steps:steps*2], exog=data_test.exog_1[steps*2:steps*3])\npredictions_4 = forecaster.predict(steps=1, last_window=data_test.y[steps*2:steps*3], exog=data_test.exog_1[steps*3:steps*4])\nnp.allclose(predicciones_backtest['pred'], np.concatenate([predictions_1, predictions_2, predictions_3, predictions_4]))",
"_____no_output_____"
]
],
[
[
"# ForecasterAutoreg with multiple exogenous variables",
"_____no_output_____"
]
],
[
[
"# Split train-test\n# ==============================================================================\nsteps = 36\ndata_train = data.iloc[:-steps, :]\ndata_test = data.iloc[-steps:, :]",
"_____no_output_____"
],
[
"# Create and fit forecaster\n# ==============================================================================\nforecaster = ForecasterAutoreg(\n regressor = LinearRegression(),\n lags = 2\n )\n\nforecaster.fit(y=data_train.y, exog=data_train[['exog_1', 'exog_2']])\n\nforecaster",
"_____no_output_____"
],
[
"# Predict\n# ==============================================================================\nsteps = 36\npredictions = forecaster.predict(steps=steps, exog=data_test[['exog_1', 'exog_2']])\n\n# Plot\n# ==============================================================================\nfig, ax=plt.subplots(figsize=(9, 4))\ndata_train.y.plot(ax=ax, label='train')\ndata_test.y.plot(ax=ax, label='test')\npredictions.plot(ax=ax, label='predictions')\nax.legend();\n\n# Error\n# ==============================================================================\nerror_mse = mean_squared_error(\n y_true = data_test.y,\n y_pred = predictions\n )\nprint(f\"Test error (mse): {error_mse}\")",
"Test error (mse): 0.030285034610348982\n"
],
[
"# Grid search hiperparameters and lags\n# ==============================================================================\nforecaster = ForecasterAutoreg(\n regressor=RandomForestRegressor(random_state=123),\n lags=12\n )\n\n# Regressor hiperparameters\nparam_grid = {'n_estimators': [50, 100],\n 'max_depth': [5, 10]}\n\n# Lags used as predictors\nlags_grid = [3, 10, [1,2,3,20]]\n\nresults_grid = grid_search_forecaster(\n forecaster = forecaster,\n y = data_train['y'],\n exog = data_train[['exog_1', 'exog_2']],\n param_grid = param_grid,\n lags_grid = lags_grid,\n steps = 10,\n metric = 'mean_squared_error',\n refit = False,\n initial_train_size = int(len(data_train)*0.5),\n return_best = True,\n verbose = False\n )\n\n# Results grid Search\n# ==============================================================================\nresults_grid",
"Number of models compared: 12\n"
]
],
[
[
"# Unit Testing",
"_____no_output_____"
]
],
[
[
"# Unit test __init__\n# ==============================================================================\nimport pytest\nfrom pytest import approx\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\n\ndef test_init_lags_when_integer():\n '''\n Test creation of attribute lags when integer is passed.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=10)\n assert (forecaster.lags == np.arange(10) + 1).all()\n \ndef test_init_lags_when_list():\n '''\n Test creation of attribute lags when list is passed.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=[1, 2, 3])\n assert (forecaster.lags == np.array([1, 2, 3])).all()\n \ndef test_init_lags_when_range():\n '''\n Test creation of attribute lags when range is passed.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=range(1, 4))\n assert (forecaster.lags == np.array(range(1, 4))).all()\n \ndef test_init_lags_when_numpy_arange():\n '''\n Test creation of attribute lags when numpy arange is passed.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=np.arange(1, 10))\n assert (forecaster.lags == np.arange(1, 10)).all()\n\ndef test_init_exception_when_lags_is_int_lower_than_1():\n '''\n Test exception is raised when lags is initialized with int lower than 1.\n '''\n with pytest.raises(Exception):\n ForecasterAutoreg(LinearRegression(), lags=-10)\n \ndef test_init_exception_when_lags_has_values_lower_than_1():\n '''\n Test exception is raised when lags is initialized with any value lower than 1.\n '''\n for lags in [[0, 1], range(0, 2), np.arange(0, 2)]:\n with pytest.raises(Exception):\n ForecasterAutoreg(LinearRegression(), lags=lags)\n\n \ntest_init_lags_when_integer()\ntest_init_lags_when_list() \ntest_init_lags_when_range()\ntest_init_lags_when_numpy_arange()\ntest_init_exception_when_lags_is_int_lower_than_1() \ntest_init_exception_when_lags_has_values_lower_than_1()",
"_____no_output_____"
],
[
"# Unit test _create_lags\n# ==============================================================================\nimport pytest\nfrom pytest import approx\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\n\n\ndef test_create_lags_output():\n '''\n Test matrix of lags is created properly when langs=3 and y=np.arange(10).\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n results = forecaster._create_lags(y=np.arange(10))\n expected = (np.array([[2., 1., 0.],\n [3., 2., 1.],\n [4., 3., 2.],\n [5., 4., 3.],\n [6., 5., 4.],\n [7., 6., 5.],\n [8., 7., 6.]]),\n np.array([3., 4., 5., 6., 7., 8., 9.]))\n\n assert (results[0] == expected[0]).all()\n assert (results[1] == expected[1]).all()\n \n \ndef test_create_lags_exception_when_len_of_y_is_lower_than_maximum_lag():\n '''\n Test exception is raised when lenght of y is lower than maximum lag included\n in the forecaster.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=10)\n with pytest.raises(Exception):\n forecaster._create_lags(y=np.arange(5))\n\ntest_create_lags_output()\ntest_create_lags_exception_when_len_of_y_is_lower_than_maximum_lag()",
"_____no_output_____"
],
[
"# Unit test create_train_X_y\n# ==============================================================================\nimport pytest\nfrom pytest import approx\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\n\n\ndef test_create_train_X_y_output_when_y_is_series_10_and_exog_is_None():\n '''\n Test the output of create_train_X_y when y=pd.Series(np.arange(10)) and \n exog is None.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=5)\n results = forecaster.create_train_X_y(y=pd.Series(np.arange(10)))\n expected = (pd.DataFrame(\n data = np.array([[4, 3, 2, 1, 0],\n [5, 4, 3, 2, 1],\n [6, 5, 4, 3, 2],\n [7, 6, 5, 4, 3],\n [8, 7, 6, 5, 4]]),\n index = np.array([5, 6, 7, 8, 9]),\n columns = ['lag_1', 'lag_2', 'lag_3', 'lag_4', 'lag_5']\n ),\n pd.Series(\n np.array([5, 6, 7, 8, 9]),\n index = np.array([5, 6, 7, 8, 9]))\n ) \n\n assert (results[0] == expected[0]).all().all()\n assert (results[1] == expected[1]).all()\n\n\ndef test_create_train_X_y_output_when_y_is_series_10_and_exog_is_series():\n '''\n Test the output of create_train_X_y when y=pd.Series(np.arange(10)) and \n exog is a pandas series\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=5)\n results = forecaster.create_train_X_y(\n y = pd.Series(np.arange(10)),\n exog = pd.Series(np.arange(100, 110), name='exog')\n )\n expected = (pd.DataFrame(\n data = np.array([[4, 3, 2, 1, 0, 105],\n [5, 4, 3, 2, 1, 106],\n [6, 5, 4, 3, 2, 107],\n [7, 6, 5, 4, 3, 108],\n [8, 7, 6, 5, 4, 109]]),\n index = np.array([5, 6, 7, 8, 9]),\n columns = ['lag_1', 'lag_2', 'lag_3', 'lag_4', 'lag_5', 'exog']\n ),\n pd.Series(\n np.array([5, 6, 7, 8, 9]),\n index = np.array([5, 6, 7, 8, 9]))\n ) \n\n assert (results[0] == expected[0]).all().all()\n assert (results[1] == expected[1]).all()\n\ndef test_create_train_X_y_output_when_y_is_series_10_and_exog_is_daraframe():\n '''\n Test the output of create_train_X_y when y=pd.Series(np.arange(10)) and \n exog is a pandas dataframe with two columns.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=5)\n results = forecaster.create_train_X_y(\n y = pd.Series(np.arange(10)),\n exog = pd.DataFrame({\n 'exog_1' : np.arange(100, 110),\n 'exog_2' : np.arange(1000, 1010)\n })\n )\n \n expected = (pd.DataFrame(\n data = np.array([[4, 3, 2, 1, 0, 105, 1005],\n [5, 4, 3, 2, 1, 106, 1006],\n [6, 5, 4, 3, 2, 107, 1007],\n [7, 6, 5, 4, 3, 108, 1008],\n [8, 7, 6, 5, 4, 109, 1009]]),\n index = np.array([5, 6, 7, 8, 9]),\n columns = ['lag_1', 'lag_2', 'lag_3', 'lag_4', 'lag_5', 'exog_1', 'exog_2']\n ),\n pd.Series(\n np.array([5, 6, 7, 8, 9]),\n index = np.array([5, 6, 7, 8, 9])\n )\n ) \n\n assert (results[0] == expected[0]).all().all()\n assert (results[1] == expected[1]).all()\n\ndef test_create_train_X_y_exception_when_y_and_exog_have_different_lenght():\n '''\n Test exception is raised when lenght of y and lenght of exog are different.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=5)\n with pytest.raises(Exception):\n forecaster.fit(y=pd.Series(np.arange(50)), exog=pd.Series(np.arange(10)))\n with pytest.raises(Exception):\n forecaster.fit(y=pd.Series(np.arange(10)), exog=pd.Series(np.arange(50)))\n with pytest.raises(Exception):\n forecaster.fit(\n y=pd.Series(np.arange(10)),\n exog=pd.DataFrame(np.arange(50).reshape(25,2))\n )\n \ndef test_create_train_X_y_exception_when_y_and_exog_have_different_index():\n '''\n Test exception is raised when y and exog have diferent index.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=5)\n with pytest.raises(Exception):\n forecaster.fit(\n y=pd.Series(np.arange(50)),\n exog=pd.Series(np.arange(10), index=np.arange(100, 110))\n )\n \ntest_create_train_X_y_output_when_y_is_series_10_and_exog_is_None()\ntest_create_train_X_y_output_when_y_is_series_10_and_exog_is_series()\ntest_create_train_X_y_output_when_y_is_series_10_and_exog_is_daraframe()\ntest_create_train_X_y_exception_when_y_and_exog_have_different_lenght()",
"/home/ximo/anaconda3/lib/python3.7/site-packages/skforecast/utils/utils.py:344: UserWarning: `exog` has DatetimeIndex index but no frequency. The index is overwritten with a RangeIndex.\n ('`exog` has DatetimeIndex index but no frequency. The index is '\n"
],
[
"# Unit test fit\n# ==============================================================================\nimport pytest\nfrom pytest import approx\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\n\n \ndef test_fit_last_window_stored():\n '''\n Test that values of last window are stored after fitting.\n ''' \n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(50)))\n expected = pd.Series(np.array([47, 48, 49]), index=[47, 48, 49])\n assert (forecaster.last_window == expected).all()\n \n \ndef test_fit_in_sample_residuals_stored():\n '''\n Test that values of in_sample_residuals are stored after fitting.\n ''' \n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(5)))\n expected = np.array([0, 0])\n results = forecaster.in_sample_residuals \n assert results.values == approx(expected)\n \ntest_fit_last_window_stored()\ntest_fit_in_sample_residuals_stored()",
"_____no_output_____"
],
[
"# Unit test _recursive_predict\n# ==============================================================================\nimport pytest\nfrom pytest import approx\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\n\ndef test_recursive_predict_output_when_regresor_is_LinearRegression():\n '''\n Test _recursive_predict output when using LinearRegression as regressor.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(50)))\n predictions = forecaster._recursive_predict(\n steps = 5,\n last_window = forecaster.last_window.values,\n exog = None\n )\n expected = np.array([50., 51., 52., 53., 54.])\n assert (predictions == approx(expected))\n \ntest_recursive_predict_output_when_regresor_is_LinearRegression()",
"_____no_output_____"
],
[
"# Unit test predict\n# ==============================================================================\nfrom pytest import approx\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\n\n \ndef test_predict_output_when_regresor_is_LinearRegression():\n '''\n Test predict output when using LinearRegression as regressor.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(50)))\n predictions = forecaster.predict(steps=5)\n expected = pd.Series(\n data = np.array([50., 51., 52., 53., 54.]),\n index = pd.RangeIndex(start=50, stop=55, step=1),\n name = 'pred'\n )\n pd.testing.assert_series_equal(predictions, expected)\n \ntest_predict_output_when_regresor_is_LinearRegression()",
"_____no_output_____"
],
[
"# Unit test get_coef\n# ==============================================================================\nfrom pytest import approx\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.ensemble import RandomForestRegressor\n\n\ndef test_output_get_coef_when_regressor_is_LinearRegression():\n '''\n Test output of get_coef when regressor is LinearRegression with lags=3\n and it is trained with y=pd.Series(np.arange(5)).\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(5)))\n expected = pd.DataFrame({\n 'feature': ['lag_1', 'lag_2', 'lag_3'],\n 'coef': np.array([0.33333333, 0.33333333, 0.33333333])\n })\n results = forecaster.get_coef()\n assert (results['feature'] == expected['feature']).all()\n assert results['coef'].values == approx(expected['coef'].values)\n \ndef test_output_get_coef_when_regressor_is_RandomForest():\n '''\n '''\n forecaster = ForecasterAutoreg(RandomForestRegressor(n_estimators=1, max_depth=2), lags=3)\n forecaster.fit(y=pd.Series(np.arange(5)))\n expected = None\n results = forecaster.get_coef()\n assert results is expected\n \ntest_output_get_coef_when_regressor_is_LinearRegression()\ntest_output_get_coef_when_regressor_is_RandomForest()",
"/home/ximo/anaconda3/lib/python3.7/site-packages/skforecast/ForecasterAutoreg/ForecasterAutoreg.py:905: UserWarning: Impossible to access feature coefficients for regressor of type <class 'sklearn.ensemble._forest.RandomForestRegressor'>. This method is only valid when the regressor stores internally the coefficients in the attribute `coef_`.\n f\"Impossible to access feature coefficients for regressor of type {type(estimator)}. \"\n"
],
[
"# Unit test get_feature_importance\n# ==============================================================================\nfrom pytest import approx\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import Lasso\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.ensemble import GradientBoostingRegressor\n\n\nfrom sklearn.linear_model import Lasso\nfrom sklearn.ensemble import RandomForestRegressor\n\ndef test_output_get_feature_importance_when_regressor_is_RandomForest():\n forecaster = ForecasterAutoreg(RandomForestRegressor(n_estimators=1, max_depth=2, random_state=123), lags=3)\n forecaster.fit(y=pd.Series(np.arange(10)))\n expected = pd.DataFrame({\n 'feature': ['lag_1', 'lag_2', 'lag_3'],\n 'importance': np.array([0.94766355, 0., 0.05233645])\n })\n results = forecaster.get_feature_importance()\n assert (results['feature'] == expected['feature']).all()\n assert results['importance'].values == approx(expected['importance'].values)\n \n \ndef test_output_get_feature_importance_when_regressor_is_linear_model():\n forecaster = ForecasterAutoreg(Lasso(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(5)))\n expected = None\n results = forecaster.get_feature_importance()\n assert results is expected\n \ntest_output_get_feature_importance_when_regressor_is_RandomForest()\ntest_output_get_feature_importance_when_regressor_is_linear_model()",
"/home/ximo/anaconda3/lib/python3.7/site-packages/skforecast/ForecasterAutoreg/ForecasterAutoreg.py:943: UserWarning: Impossible to access feature importance for regressor of type <class 'sklearn.linear_model._coordinate_descent.Lasso'>. This method is only valid when the regressor stores internally the feature importance in the attribute `feature_importances_`.\n f\"Impossible to access feature importance for regressor of type {type(estimator)}. \"\n"
],
[
"# Unit test set_lags\n# ==============================================================================\nimport pytest\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\n\n\ndef test_set_lags_exception_when_lags_argument_is_int_lower_than_1():\n '''\n Test exception is raised when lags argument is lower than 1.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n with pytest.raises(Exception):\n forecaster.set_lags(lags=-10)\n\ndef test_set_lags_exception_when_lags_argument_has_any_value_lower_than_1():\n '''\n Test exception is raised when lags argument has at least one value\n lower than 1.\n '''\n \n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n with pytest.raises(Exception):\n forecaster.set_lags(lags=range(0, 4)) \n \ndef test_set_lags_when_lags_argument_is_int():\n '''\n Test how lags and max_lag attributes change when lags argument is integer\n positive (5).\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.set_lags(lags=5)\n assert (forecaster.lags == np.array([1, 2, 3, 4, 5])).all()\n assert forecaster.max_lag == 5\n \ndef test_set_lags_when_lags_argument_is_list():\n '''\n Test how lags and max_lag attributes change when lags argument is a list\n of positive integers.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.set_lags(lags=[1,2,3])\n assert (forecaster.lags == np.array([1, 2, 3])).all()\n assert forecaster.max_lag == 3\n \ndef test_set_lags_when_lags_argument_is_1d_numpy_array():\n '''\n Test how lags and max_lag attributes change when lags argument is 1d numpy\n array of positive integers.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.set_lags(lags=np.array([1,2,3]))\n assert (forecaster.lags == np.array([1, 2, 3])).all()\n assert forecaster.max_lag == 3\n \ntest_set_lags_exception_when_lags_argument_is_int_lower_than_1()\ntest_set_lags_exception_when_lags_argument_has_any_value_lower_than_1()\ntest_set_lags_when_lags_argument_is_int()\ntest_set_lags_when_lags_argument_is_list()\ntest_set_lags_when_lags_argument_is_1d_numpy_array()",
"_____no_output_____"
],
[
"# Unit test set_params\n# ==============================================================================\nfrom pytest import approx\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\n\n\ndef test_set_params():\n \n forecaster = ForecasterAutoreg(LinearRegression(fit_intercept=True), lags=3)\n new_paramns = {'fit_intercept': False}\n forecaster.set_params(**new_paramns)\n expected = {'copy_X': True,\n 'fit_intercept': False,\n 'n_jobs': None,\n 'normalize': 'deprecated',\n 'positive': False\n }\n results = forecaster.regressor.get_params()\n assert results == expected\n \ntest_set_params()",
"_____no_output_____"
],
[
"def test_set_params():\n \n forecaster = ForecasterAutoreg(LinearRegression(fit_intercept=True), lags=3)\n new_params = {'fit_intercept': False}\n forecaster.set_params(**new_params)\n expected = {'copy_X': True,\n 'fit_intercept': False,\n 'n_jobs': None,\n 'normalize': 'deprecated',\n 'positive': False\n }\n results = forecaster.regressor.get_params()\n assert results == expected\n \ntest_set_params()",
"_____no_output_____"
],
[
"# Unit test set_out_sample_residuals\n# ==============================================================================\nimport pytest\nfrom pytest import approx\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\n\n\ndef test_set_out_sample_residuals_exception_when_residuals_is_not_array():\n '''\n Test exception is raised when residuals argument is not numpy array.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n with pytest.raises(Exception):\n forecaster.set_out_sample_residuals(residuals=[1, 2, 3])\n \n \ndef test_set_out_sample_residuals_when_residuals_lenght_is_less_than_1000_and_no_append():\n '''\n Test residuals stored when its length is less than 1000 and append is False.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.set_out_sample_residuals(residuals=np.arange(10), append=False)\n expected = np.arange(10)\n results = forecaster.out_sample_residuals\n assert (results == expected).all()\n \ndef test_set_out_sample_residuals_when_residuals_lenght_is_less_than_1000_and_append():\n '''\n Test residuals stored when its length is less than 1000 and append is True.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.set_out_sample_residuals(residuals=np.arange(10), append=True)\n forecaster.set_out_sample_residuals(residuals=np.arange(10), append=True)\n expected = np.hstack([np.arange(10), np.arange(10)])\n results = forecaster.out_sample_residuals\n assert (results == expected).all()\n \n\ndef test_set_out_sample_residuals_when_residuals_lenght_is_greater_than_1000():\n '''\n Test residuals stored when its length is greater than 1000.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.set_out_sample_residuals(residuals=np.arange(2000))\n assert len(forecaster.out_sample_residuals) == 1000\n\ntest_set_out_sample_residuals_when_residuals_lenght_is_less_than_1000_and_no_append()\ntest_set_out_sample_residuals_when_residuals_lenght_is_less_than_1000_and_append()\ntest_set_out_sample_residuals_when_residuals_lenght_is_greater_than_1000()",
"_____no_output_____"
],
[
"# Unit test set_out_sample_residuals\n# ==============================================================================\nfrom pytest import approx\nimport numpy as np\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\n\n\ndef test_predict_interval_output_when_forecaster_is_LinearRegression_steps_is_1_in_sample_residuals_is_True():\n '''\n Test output when regressor is LinearRegression and one step ahead is predicted\n using in sample residuals.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(10)))\n forecaster.in_sample_residuals = np.full_like(forecaster.in_sample_residuals, fill_value=10)\n expected = pd.DataFrame(\n np.array([[10., 20., 20.]]),\n columns = ['pred', 'lower_bound', 'upper_bound'],\n index = pd.RangeIndex(start=10, stop=11, step=1)\n )\n results = forecaster.predict_interval(steps=1, in_sample_residuals=True) \n pd.testing.assert_frame_equal(results, expected)\n\n \ndef test_predict_interval_output_when_forecaster_is_LinearRegression_steps_is_2_in_sample_residuals_is_True():\n '''\n Test output when regressor is LinearRegression and two step ahead is predicted\n using in sample residuals.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(10)))\n forecaster.in_sample_residuals = np.full_like(forecaster.in_sample_residuals, fill_value=10)\n expected = pd.DataFrame(\n np.array([[10. ,20., 20.],\n [11., 24.33333333, 24.33333333]\n ]),\n columns = ['pred', 'lower_bound', 'upper_bound'],\n index = pd.RangeIndex(start=10, stop=12, step=1)\n )\n results = forecaster.predict_interval(steps=2, in_sample_residuals=True) \n pd.testing.assert_frame_equal(results, expected)\n \n \ndef test_predict_interval_output_when_forecaster_is_LinearRegression_steps_is_1_in_sample_residuals_is_False():\n '''\n Test output when regressor is LinearRegression and one step ahead is predicted\n using out sample residuals.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(10)))\n forecaster.out_sample_residuals = np.full_like(forecaster.in_sample_residuals, fill_value=10)\n expected = pd.DataFrame(\n np.array([[10., 20., 20.]]),\n columns = ['pred', 'lower_bound', 'upper_bound'],\n index = pd.RangeIndex(start=10, stop=11, step=1)\n )\n results = forecaster.predict_interval(steps=1, in_sample_residuals=False) \n pd.testing.assert_frame_equal(results, expected)\n \n \ndef test_predict_interval_output_when_forecaster_is_LinearRegression_steps_is_2_in_sample_residuals_is_False():\n '''\n Test output when regressor is LinearRegression and two step ahead is predicted\n using out sample residuals.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(10)))\n forecaster.out_sample_residuals = np.full_like(forecaster.in_sample_residuals, fill_value=10)\n expected = pd.DataFrame(\n np.array([[10. ,20., 20.],\n [11., 24.33333333, 24.33333333]\n ]),\n columns = ['pred', 'lower_bound', 'upper_bound'],\n index = pd.RangeIndex(start=10, stop=12, step=1)\n )\n results = forecaster.predict_interval(steps=2, in_sample_residuals=False) \n pd.testing.assert_frame_equal(results, expected)\n \n\ntest_predict_interval_output_when_forecaster_is_LinearRegression_steps_is_1_in_sample_residuals_is_True()\ntest_predict_interval_output_when_forecaster_is_LinearRegression_steps_is_2_in_sample_residuals_is_True()\ntest_predict_interval_output_when_forecaster_is_LinearRegression_steps_is_1_in_sample_residuals_is_False()\ntest_predict_interval_output_when_forecaster_is_LinearRegression_steps_is_2_in_sample_residuals_is_False()",
"_____no_output_____"
],
[
"# Unit test _estimate_boot_interval\n# ==============================================================================\nimport pytest\nfrom pytest import approx\nimport numpy as np\nimport pandas as pd\nfrom skforecast.ForecasterAutoreg import ForecasterAutoreg\nfrom sklearn.linear_model import LinearRegression\n \n\ndef test_estimate_boot_interval_output_when_forecaster_is_LinearRegression_steps_is_1_in_sample_residuals_is_True():\n '''\n Test output of _estimate_boot_interval when regressor is LinearRegression and\n 1 step is predicted using in-sample residuals.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(10)))\n forecaster.in_sample_residuals = np.full_like(forecaster.in_sample_residuals, fill_value=10)\n expected = np.array([[20., 20.]])\n results = forecaster._estimate_boot_interval(steps=1, in_sample_residuals=True) \n assert results == approx(expected)\n \n \ndef test_estimate_boot_interval_output_when_forecaster_is_LinearRegression_steps_is_2_in_sample_residuals_is_True():\n '''\n Test output of _estimate_boot_interval when regressor is LinearRegression and\n 2 steps are predicted using in-sample residuals.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(10)))\n forecaster.in_sample_residuals = np.full_like(forecaster.in_sample_residuals, fill_value=10)\n expected = np.array([[20., 20.],\n [24.33333333, 24.33333333]])\n results = forecaster._estimate_boot_interval(steps=2, in_sample_residuals=True) \n assert results == approx(expected)\n \n \ndef test_estimate_boot_interval_output_when_forecaster_is_LinearRegression_steps_is_1_in_sample_residuals_is_False():\n '''\n Test output of _estimate_boot_interval when regressor is LinearRegression and\n 1 step is predicted using out-sample residuals.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(10)))\n forecaster.out_sample_residuals = np.full_like(forecaster.in_sample_residuals, fill_value=10)\n expected = np.array([[20., 20.]])\n results = forecaster._estimate_boot_interval(steps=1, in_sample_residuals=False) \n assert results == approx(expected)\n \n \ndef test_estimate_boot_interval_output_when_forecaster_is_LinearRegression_steps_is_2_in_sample_residuals_is_False():\n '''\n Test output of _estimate_boot_interval when regressor is LinearRegression and\n 2 steps are predicted using out-sample residuals.\n '''\n forecaster = ForecasterAutoreg(LinearRegression(), lags=3)\n forecaster.fit(y=pd.Series(np.arange(10)))\n forecaster.out_sample_residuals = np.full_like(forecaster.in_sample_residuals, fill_value=10)\n expected = np.array([[20. , 20. ],\n [24.33333333, 24.33333333]])\n results = forecaster._estimate_boot_interval(steps=2, in_sample_residuals=False) \n assert results == approx(expected)\n \n \ntest_estimate_boot_interval_output_when_forecaster_is_LinearRegression_steps_is_1_in_sample_residuals_is_True()\ntest_estimate_boot_interval_output_when_forecaster_is_LinearRegression_steps_is_2_in_sample_residuals_is_True()\ntest_estimate_boot_interval_output_when_forecaster_is_LinearRegression_steps_is_1_in_sample_residuals_is_False()\ntest_estimate_boot_interval_output_when_forecaster_is_LinearRegression_steps_is_2_in_sample_residuals_is_False()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb6703eee15f9131f1368876bfd5fef2222ec84 | 156,959 | ipynb | Jupyter Notebook | Diabetes Dataset/Improvements/Features IMprovement with Median/08_Pregnancies, Glucose, BloodPressure, SkinThickness and Age.ipynb | AnkitaxPriya/Diabetes-Prediction | 2a68fc067019dde8eda31ebb91436746abc4e98e | [
"MIT"
] | null | null | null | Diabetes Dataset/Improvements/Features IMprovement with Median/08_Pregnancies, Glucose, BloodPressure, SkinThickness and Age.ipynb | AnkitaxPriya/Diabetes-Prediction | 2a68fc067019dde8eda31ebb91436746abc4e98e | [
"MIT"
] | null | null | null | Diabetes Dataset/Improvements/Features IMprovement with Median/08_Pregnancies, Glucose, BloodPressure, SkinThickness and Age.ipynb | AnkitaxPriya/Diabetes-Prediction | 2a68fc067019dde8eda31ebb91436746abc4e98e | [
"MIT"
] | null | null | null | 92.274544 | 107,412 | 0.787244 | [
[
[
"# Import the required libraries\nimport warnings\nwarnings.filterwarnings('ignore')\n\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nimport joblib\n%matplotlib inline\n\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.model_selection import cross_val_predict\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import classification_report\nfrom sklearn import metrics\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.svm import SVC\nfrom sklearn.ensemble import RandomForestClassifier",
"_____no_output_____"
],
[
"# Read the data and display\n\ndiabetesDF = pd.read_csv('diabetes.csv')\ndiabetesDF.head()",
"_____no_output_____"
],
[
"# Shape of the dataset \n\nprint(diabetesDF.shape)",
"(768, 9)\n"
],
[
"diabetesDF.describe()",
"_____no_output_____"
],
[
"diabetesDF.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 768 entries, 0 to 767\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Pregnancies 768 non-null int64 \n 1 Glucose 768 non-null int64 \n 2 BloodPressure 768 non-null int64 \n 3 SkinThickness 768 non-null int64 \n 4 Insulin 768 non-null int64 \n 5 BMI 768 non-null float64\n 6 DiabetesPedigreeFunction 768 non-null float64\n 7 Age 768 non-null int64 \n 8 Outcome 768 non-null int64 \ndtypes: float64(2), int64(7)\nmemory usage: 54.1 KB\n"
],
[
"# Return the number of missing values\n\ndiabetesDF.isnull().sum()",
"_____no_output_____"
],
[
"# Number of outcomes\n\ndiabetesDF.Outcome.value_counts()",
"_____no_output_____"
],
[
"# Visualising data \n\n# DistPlots\nplt.figure(figsize = (16, 10))\n\nplt.subplot(3, 3, 1)\nsns.distplot(diabetesDF.Pregnancies)\n\nplt.subplot(3, 3, 2)\nsns.distplot(diabetesDF.Glucose)\n\nplt.subplot(3, 3, 8)\nsns.distplot(diabetesDF.BloodPressure)\n\nplt.subplot(3, 3, 3)\nsns.distplot(diabetesDF.SkinThickness)\n\nplt.subplot(3, 3, 4)\nsns.distplot(diabetesDF.Insulin)\n\nplt.subplot(3, 3, 5)\nsns.distplot(diabetesDF.BMI)\n\nplt.subplot(3, 3, 6)\nsns.distplot(diabetesDF.DiabetesPedigreeFunction)\n\nplt.subplot(3, 3, 7)\nsns.distplot(diabetesDF.Age)\n\nplt.subplot(3, 3, 9)\nsns.countplot(diabetesDF.Outcome)\n\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"We can infer that even though we do not have NaN values, there are a lot of wrong values present in our data, like:\n- Glucose Level cannot be above 150 or below 70\n- Blood Pressure cannot be below 55\n- Skin thickness cannot be 0\n- BMI index cannot be 0",
"_____no_output_____"
]
],
[
[
"# Data Cleaning \n\ndf_improv = diabetesDF.copy()",
"_____no_output_____"
],
[
"# Calculate the median value for BMI\nmedian_bmi = diabetesDF['BMI'].median()\n# Substitute it in the BMI column of the\n# dataset where values are 0\ndiabetesDF['BMI'] = diabetesDF['BMI'].replace(to_replace=0, value=median_bmi)",
"_____no_output_____"
],
[
"# Calculate the median value for BloodP\nmedian_bloodp = diabetesDF['BloodPressure'].median()\n# Substitute it in the BloodP column of the\n# dataset where values are 0\ndiabetesDF['BloodPressure'] = diabetesDF['BloodPressure'].replace(to_replace=0, value=median_bloodp)",
"_____no_output_____"
],
[
"# Calculate the median value for PlGlcConc\nmedian_glucose = diabetesDF['Glucose'].median()\n# Substitute it in the PlGlcConc column of the\n# dataset where values are 0\ndiabetesDF['Glucose'] = diabetesDF['Glucose'].replace(to_replace=0, value=median_glucose)",
"_____no_output_____"
],
[
"# Calculate the median value for SkinThick\nmedian_skinthick = diabetesDF['SkinThickness'].median()\n# Substitute it in the SkinThick column of the\n# dataset where values are 0\ndiabetesDF['SkinThickness'] = diabetesDF['SkinThickness'].replace(to_replace=0, value=median_skinthick)",
"_____no_output_____"
],
[
"# Calculate the median value for TwoHourSerIns\nmedian_insulin = diabetesDF['Insulin'].median()\n# Substitute it in the TwoHourSerIns column of the\n# dataset where values are 0\ndiabetesDF['Insulin'] = diabetesDF['Insulin'].replace(to_replace=0, value=median_insulin)",
"_____no_output_____"
],
[
"df_improv.head()",
"_____no_output_____"
],
[
"df_improv.describe()",
"_____no_output_____"
],
[
"df_improv.describe()",
"_____no_output_____"
],
[
"df_improv.drop(['Insulin', 'BMI', 'DiabetesPedigreeFunction'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_improv.head()",
"_____no_output_____"
],
[
"# Total 768 patients record\n# Using 650 data for training\n# Using 100 data for testing\n# Using 18 data for validation\n\ndfTrain = df_improv[:650]\ndfTest = df_improv[650:750]\ndfCheck = df_improv[750:]",
"_____no_output_____"
],
[
"# Separating label and features and converting to numpy array to feed into our model\ntrainLabel = np.asarray(dfTrain['Outcome'])\ntrainData = np.asarray(dfTrain.drop('Outcome',1))\ntestLabel = np.asarray(dfTest['Outcome'])\ntestData = np.asarray(dfTest.drop('Outcome',1))",
"_____no_output_____"
],
[
"# Normalize the data \nmeans = np.mean(trainData, axis=0)\nstds = np.std(trainData, axis=0)\n\ntrainData = (trainData - means)/stds\ntestData = (testData - means)/stds",
"_____no_output_____"
],
[
"# models target t as sigmoid(w0 + w1*x1 + w2*x2 + ... + wd*xd)\ndiabetesCheck = LogisticRegression()\ndiabetesCheck.fit(trainData,trainLabel)\naccuracy = diabetesCheck.score(testData,testLabel)\nprint(\"accuracy = \",accuracy * 100,\"%\")",
"accuracy = 81.0 %\n"
],
[
"# predict values using training data\n\npredict_train = diabetesCheck.predict(trainData)\nprint(\"Accuracy: {0:.4f}\".format(metrics.accuracy_score(trainLabel,predict_train)))\nprint()",
"Accuracy: 0.7477\n\n"
],
[
"# predict values using testing data\n\npredict_train = diabetesCheck.predict(testData)\nprint(\"Accuracy: {0:.4f}\".format(metrics.accuracy_score(testLabel,predict_train)))\nprint()",
"Accuracy: 0.8100\n\n"
],
[
"# Confusion Matrix\n\nprint(\"Confusion Matrix\")\nprint(\"{0}\".format(metrics.confusion_matrix(testLabel,predict_train)))\nprint(\"\")",
"Confusion Matrix\n[[57 6]\n [13 24]]\n\n"
],
[
"print(\"Classification Report\")\nprint(\"{0}\".format(metrics.classification_report(testLabel,predict_train)))",
"Classification Report\n precision recall f1-score support\n\n 0 0.81 0.90 0.86 63\n 1 0.80 0.65 0.72 37\n\n accuracy 0.81 100\n macro avg 0.81 0.78 0.79 100\nweighted avg 0.81 0.81 0.81 100\n\n"
],
[
"# models target t as sigmoid(w0 + w1*x1 + w2*x2 + ... + wd*xd)\ndiabetesCheck = KNeighborsClassifier()\ndiabetesCheck.fit(trainData,trainLabel)\naccuracy = diabetesCheck.score(testData,testLabel)\nprint(\"accuracy = \",accuracy * 100,\"%\")",
"accuracy = 71.0 %\n"
],
[
"# predict values using training data\n\npredict_train = diabetesCheck.predict(trainData)\nprint(\"Accuracy: {0:.4f}\".format(metrics.accuracy_score(trainLabel,predict_train)))\nprint()",
"Accuracy: 0.8062\n\n"
],
[
"# predict values using testing data\n\npredict_train = diabetesCheck.predict(testData)\nprint(\"Accuracy: {0:.4f}\".format(metrics.accuracy_score(testLabel,predict_train)))\nprint()",
"Accuracy: 0.7100\n\n"
],
[
"# Confusion Matrix\n\nprint(\"Confusion Matrix\")\nprint(\"{0}\".format(metrics.confusion_matrix(testLabel,predict_train)))\nprint(\"\")",
"Confusion Matrix\n[[52 11]\n [18 19]]\n\n"
],
[
"print(\"Classification Report\")\nprint(\"{0}\".format(metrics.classification_report(testLabel,predict_train)))",
"Classification Report\n precision recall f1-score support\n\n 0 0.74 0.83 0.78 63\n 1 0.63 0.51 0.57 37\n\n accuracy 0.71 100\n macro avg 0.69 0.67 0.67 100\nweighted avg 0.70 0.71 0.70 100\n\n"
],
[
"# models target t as sigmoid(w0 + w1*x1 + w2*x2 + ... + wd*xd)\ndiabetesCheck = SVC()\ndiabetesCheck.fit(trainData,trainLabel)\naccuracy = diabetesCheck.score(testData,testLabel)\nprint(\"accuracy = \",accuracy * 100,\"%\")",
"accuracy = 78.0 %\n"
],
[
"# predict values using training data\n\npredict_train = diabetesCheck.predict(trainData)\nprint(\"Accuracy: {0:.4f}\".format(metrics.accuracy_score(trainLabel,predict_train)))\nprint()",
"Accuracy: 0.7831\n\n"
],
[
"# predict values using testing data\n\npredict_train = diabetesCheck.predict(testData)\nprint(\"Accuracy: {0:.4f}\".format(metrics.accuracy_score(testLabel,predict_train)))\nprint()",
"Accuracy: 0.7800\n\n"
],
[
"# Confusion Matrix\n\nprint(\"Confusion Matrix\")\nprint(\"{0}\".format(metrics.confusion_matrix(testLabel,predict_train)))\nprint(\"\")",
"Confusion Matrix\n[[57 6]\n [16 21]]\n\n"
],
[
"print(\"Classification Report\")\nprint(\"{0}\".format(metrics.classification_report(testLabel,predict_train)))",
"Classification Report\n precision recall f1-score support\n\n 0 0.78 0.90 0.84 63\n 1 0.78 0.57 0.66 37\n\n accuracy 0.78 100\n macro avg 0.78 0.74 0.75 100\nweighted avg 0.78 0.78 0.77 100\n\n"
],
[
"# models target t as sigmoid(w0 + w1*x1 + w2*x2 + ... + wd*xd)\ndiabetesCheck = RandomForestClassifier()\ndiabetesCheck.fit(trainData,trainLabel)\naccuracy = diabetesCheck.score(testData,testLabel)\nprint(\"accuracy = \",accuracy * 100,\"%\")",
"accuracy = 73.0 %\n"
],
[
"# predict values using training data\n\npredict_train = diabetesCheck.predict(trainData)\nprint(\"Accuracy: {0:.4f}\".format(metrics.accuracy_score(trainLabel,predict_train)))\nprint()",
"Accuracy: 1.0000\n\n"
],
[
"# predict values using testing data\n\npredict_train = diabetesCheck.predict(testData)\nprint(\"Accuracy: {0:.4f}\".format(metrics.accuracy_score(testLabel,predict_train)))\nprint()",
"Accuracy: 0.7300\n\n"
],
[
"# Confusion Matrix\n\nprint(\"Confusion Matrix\")\nprint(\"{0}\".format(metrics.confusion_matrix(testLabel,predict_train)))\nprint(\"\")",
"Confusion Matrix\n[[50 13]\n [14 23]]\n\n"
],
[
"print(\"Classification Report\")\nprint(\"{0}\".format(metrics.classification_report(testLabel,predict_train)))",
"Classification Report\n precision recall f1-score support\n\n 0 0.78 0.79 0.79 63\n 1 0.64 0.62 0.63 37\n\n accuracy 0.73 100\n macro avg 0.71 0.71 0.71 100\nweighted avg 0.73 0.73 0.73 100\n\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb67ed50834078ed5161b4de13f10905db95a22 | 166,445 | ipynb | Jupyter Notebook | code/notebooks/.ipynb_checkpoints/2019-5-24_baby_patch_attack-checkpoint.ipynb | davidwagner/bagnet-patch-defense | 0e38d26cf6e082baf4de89d0cdfece6ba15573eb | [
"BSD-3-Clause"
] | 1 | 2022-03-30T16:38:46.000Z | 2022-03-30T16:38:46.000Z | code/notebooks/.ipynb_checkpoints/2019-5-24_baby_patch_attack-checkpoint.ipynb | davidwagner/bagnet-patch-defense | 0e38d26cf6e082baf4de89d0cdfece6ba15573eb | [
"BSD-3-Clause"
] | null | null | null | code/notebooks/.ipynb_checkpoints/2019-5-24_baby_patch_attack-checkpoint.ipynb | davidwagner/bagnet-patch-defense | 0e38d26cf6e082baf4de89d0cdfece6ba15573eb | [
"BSD-3-Clause"
] | null | null | null | 967.703488 | 89,368 | 0.955991 | [
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"- **Date:** 2019-5-24 \n- **Author:** Zhanyuan Zhang \n- **Purpose:** Naive patch attack",
"_____no_output_____"
]
],
[
[
"from bagnets.utils import plot_heatmap, generate_heatmap_pytorch\nfrom bagnets.utils import pad_image, convert2channel_last, imagenet_preprocess, extract_patches, bagnet_predict, compare_heatmap\nfrom bagnets.utils import class_patch_logits\nfrom foolbox.utils import samples\nimport bagnets.pytorch\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport torch\nimport torch.nn.functional as F\nimport numpy as np\nimport time\nuse_cuda = torch.cuda.is_available()\ndevice = torch.device(\"cuda:0\" if use_cuda else \"cpu\")\nif use_cuda:\n print(torch.cuda.get_device_name(0))",
"Tesla K80\n"
],
[
"image, label = samples(dataset='imagenet', index=3, batchsize=1, shape=(224, 224), data_format='channels_first')\n\noriginal_image = imagenet_preprocess(image[0])\nplt.imshow(convert2channel_last(original_image))",
"Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
],
[
"def attack_patch(image, patchsize, num_patches, seed=None):\n c, x, y = image.shape\n if seed is not None:\n np.random.seed(seed)\n attacked_x = np.random.choice(range(x), size=num_patches, replace=True)\n attacked_y = np.random.choice(range(y), size=num_patches, replace=True)\n for xi, yi in zip(attacked_x, attacked_y):\n c, h, w = image[:, (xi - (patchsize-1)//2): (xi + (patchsize-1)//2), (yi - (patchsize-1) // 2): (yi + (patchsize-1) // 2)].shape\n image[:, (xi - (patchsize-1)//2): (xi + (patchsize-1)//2), (yi - (patchsize-1) // 2): (yi + (patchsize-1) // 2)] = np.random.rand(c, h, w)\n return image",
"_____no_output_____"
],
[
"attacked_image = attack_patch(original_image.copy(), patchsize=33, num_patches=5, seed=789)\nplt.imshow(convert2channel_last(attacked_image))",
"Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecb6856c6c41255ac90da3205ef70fbf0c2a486d | 3,419 | ipynb | Jupyter Notebook | PySummary2.ipynb | 4dsolutions/Python5 | 8d80753e823441a571b827d24d21577446409b52 | [
"MIT"
] | 11 | 2016-08-17T00:15:26.000Z | 2020-07-17T21:31:10.000Z | PySummary2.ipynb | 4dsolutions/Python5 | 8d80753e823441a571b827d24d21577446409b52 | [
"MIT"
] | null | null | null | PySummary2.ipynb | 4dsolutions/Python5 | 8d80753e823441a571b827d24d21577446409b52 | [
"MIT"
] | 5 | 2017-02-22T05:15:52.000Z | 2019-11-08T06:17:34.000Z | 27.134921 | 286 | 0.582334 | [
[
[
"# SAISOFT PYT-PR: Session 10\n\n### The Ecosystem includes Many Languages\n\nThe Python ecosystem involves Python in using other languages and rule sets such as:\n\n* SQL (Structured Query Language)\n* Regexes (Regular Expressions)\n* Markdown (for Jupyter Notebooks)\n* magic (% and %% inside Notebooks and I-Python)\n* APIs (every library and framework comes with one)\n* Sphinx (documentation generator, uses reStructured Text)\n* JavaScript (used with web frameworks)\n* Template Languages (example: Jinja)\n* HTML / CSS (naturally)\n* Widget Toolkits (more APIs for GUIs)\n* IDEs (such as Spyder, Pycharm, Sublime Text, vi, emacs)\n\nand so much else.",
"_____no_output_____"
],
[
"Lets take a look at another Standard Library module and talk about what it does. \n\nHash functions are not designed to preserve content. They don't encrypt. They're about associating some unique finger print with any hashable object.",
"_____no_output_____"
]
],
[
[
"import hashlib\nm = hashlib.sha256()\nm.update(b\"Nobody inspects\")\nm.update(b\" the spammish repetition\")\nm.digest()",
"_____no_output_____"
]
],
[
[
"Expected:\n<pre>\nb'\\x03\\x1e\\xdd}Ae\\x15\\x93\\xc5\\xfe\\\\\\x00o\\xa5u+7\\xfd\\xdf\\xf7\\xbcN\\x84:\\xa6\\xaf\\x0c\\x95\\x0fK\\x94\\x06'\n</pre>\n",
"_____no_output_____"
]
],
[
[
"result = hashlib.sha256(b\"Nobody inspects the spammish repetition\").hexdigest()\nresult",
"_____no_output_____"
],
[
"print(\"Digest size\", m.digest_size)\nprint(\"Block size \", m.block_size)",
"_____no_output_____"
]
],
[
[
"In class, we looked at a passwords database that doesn't save actual passwords, only hashes thereof. Even system administrators with the keys to the database, have no means to force a hash to run backwards to regain the phrase which was behind it. A hash is a one way street.\n\n### LAB:\n\nCheck the Python docs and run the above example with sha224 instead. Do you get past this assertion (unit test)?",
"_____no_output_____"
]
],
[
[
"# Uncomment me to check your result\n# assert result == 'a4337bc45a8fc544c03f52dc550cd6e1e87021bc896588bd79e901e2'",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb6b35e10637941679710f6d70afe80d90e60a0 | 246,462 | ipynb | Jupyter Notebook | Recommendations_with_IBM.ipynb | oliverkroening/Udacity_DSND_Project06 | 4a6604f10f0478a2fc6128aaa069ac426739d95d | [
"CNRI-Python"
] | null | null | null | Recommendations_with_IBM.ipynb | oliverkroening/Udacity_DSND_Project06 | 4a6604f10f0478a2fc6128aaa069ac426739d95d | [
"CNRI-Python"
] | null | null | null | Recommendations_with_IBM.ipynb | oliverkroening/Udacity_DSND_Project06 | 4a6604f10f0478a2fc6128aaa069ac426739d95d | [
"CNRI-Python"
] | null | null | null | 57.678914 | 31,520 | 0.624567 | [
[
[
"# Recommendations with IBM\n\nIn this notebook, you will be putting your recommendation skills to use on real data from the IBM Watson Studio platform. \n\n\nYou may either submit your notebook through the workspace here, or you may work from your local machine and submit through the next page. Either way assure that your code passes the project [RUBRIC](https://review.udacity.com/#!/rubrics/2322/view). **Please save regularly.**\n\nBy following the table of contents, you will build out a number of different methods for making recommendations that can be used for different situations. \n\n\n## Table of Contents\n\nI. [Exploratory Data Analysis](#Exploratory-Data-Analysis)<br>\nII. [Rank Based Recommendations](#Rank)<br>\nIII. [User-User Based Collaborative Filtering](#User-User)<br>\nIV. [Content Based Recommendations (EXTRA - NOT REQUIRED)](#Content-Recs)<br>\nV. [Matrix Factorization](#Matrix-Fact)<br>\nVI. [Extras & Concluding](#conclusions)\n\nAt the end of the notebook, you will find directions for how to submit your work. Let's get started by importing the necessary libraries and reading in the data.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport project_tests as t\nimport pickle\n\n%matplotlib inline\n\ndf = pd.read_csv('data/user-item-interactions.csv')\ndf_content = pd.read_csv('data/articles_community.csv')\ndel df['Unnamed: 0']\ndel df_content['Unnamed: 0']\n\n# Show df to get an idea of the data\ndf.head()",
"_____no_output_____"
],
[
"# Show df_content to get an idea of the data\ndf_content.head()",
"_____no_output_____"
]
],
[
[
"### <a class=\"anchor\" id=\"Exploratory-Data-Analysis\">Part I : Exploratory Data Analysis</a>\n\nUse the dictionary and cells below to provide some insight into the descriptive statistics of the data.\n\n`1.` What is the distribution of how many articles a user interacts with in the dataset? Provide a visual and descriptive statistics to assist with giving a look at the number of times each user interacts with an article. ",
"_____no_output_____"
]
],
[
[
"# descriptive statistics:\n# -----------------------\n#\n# show dimensions of datasets\nprint('Dimensions of \"df\": {}'.format(df.shape))\nprint('Dimensions of \"df_content\": {}'.format(df_content.shape))",
"Dimensions of \"df\": (45993, 3)\nDimensions of \"df_content\": (1056, 5)\n"
],
[
"# count number of interactions for each user\ndf['email'].value_counts()",
"_____no_output_____"
]
],
[
[
"We can see that there are two users that are responsible for 364 respectively 363 interactions and lots of users with only one interaction.\nThus, we expect a right-skewed distribution according to this description.",
"_____no_output_____"
]
],
[
[
"# show statistical parameters of users\ndf['email'].value_counts().describe()",
"_____no_output_____"
]
],
[
[
"The result of the describe()-function supports the thesis stated above since the mean is only around 9 interactions and the median of the distribution is only 3.",
"_____no_output_____"
]
],
[
[
"# count interactions for each article\ndf['article_id'].value_counts()",
"_____no_output_____"
],
[
"# show statistical parameters for articles\ndf['article_id'].value_counts().describe()",
"_____no_output_____"
]
],
[
[
"The distribution of article IDs is qualitalively quite similar to the user distribution. With a maximum count of interaction of 937, we have a mean of around 64 and a median of 25 interactions per article.",
"_____no_output_____"
]
],
[
[
"# visualizations\n# --------------\n# plot histogram of user interactions per article\n\nuser_article_interactions = df.groupby('email').count()['article_id']\nfig, ax = plt.subplots(figsize=(10,6))\nax.hist(user_article_interactions, bins = 50, range=(1,100))\nax.set_xlabel('# of user interactions')\nax.set_ylabel('# of articles')\nax.grid()\nax.set_title('Distribution of number of user interactions');",
"_____no_output_____"
]
],
[
[
"The visualization of user interaction distribution is as we expected. The distribution is right-skewed. Most of the users only interact a few times on articles. Thus, the majority of the distribution as well as the median is on the left side of the plot.",
"_____no_output_____"
]
],
[
[
"# Fill in the median and maximum number of user_article interactios below\n\nmedian_val = 3 # 50% of individuals interact with ____ number of articles or fewer.\nmax_views_by_user = 364 # The maximum number of user-article interactions by any 1 user is ______.",
"_____no_output_____"
]
],
[
[
"`2.` Explore and remove duplicate articles from the **df_content** dataframe. ",
"_____no_output_____"
]
],
[
[
"# Find and explore duplicate articles\ndf_content[df_content['article_id'].duplicated()]",
"_____no_output_____"
]
],
[
[
"There are five duplicated articles in the dataset.",
"_____no_output_____"
]
],
[
[
"# Remove any rows that have the same article_id - only keep the first\ndf_content.drop_duplicates(subset='article_id', keep='first',inplace=True)",
"_____no_output_____"
],
[
"# check for duplicates\ndf_content[df_content['article_id'].duplicated()].sum()",
"_____no_output_____"
]
],
[
[
"`3.` Use the cells below to find:\n\n**a.** The number of unique articles that have an interaction with a user. \n**b.** The number of unique articles in the dataset (whether they have any interactions or not).<br>\n**c.** The number of unique users in the dataset. (excluding null values) <br>\n**d.** The number of user-article interactions in the dataset.",
"_____no_output_____"
]
],
[
[
"# a. The number of unique articles that have an interaction with a user\ndf[\"article_id\"].unique().shape[0]",
"_____no_output_____"
],
[
"# b. The number of unique articles in the dataset (whether they have any interactions or not)\ndf_content[\"article_id\"].shape[0]",
"_____no_output_____"
],
[
"# c. The number of unique users in the dataset. (excluding null values)\ndf[\"email\"].dropna().unique().shape[0]",
"_____no_output_____"
],
[
"# d. The number of user-article interactions in the dataset.\ndf.shape[0]",
"_____no_output_____"
],
[
"unique_articles = 714 # The number of unique articles that have at least one interaction\ntotal_articles = 1051 # The number of unique articles on the IBM platform\nunique_users = 5148 # The number of unique users\nuser_article_interactions = 45993 # The number of user-article interactions",
"_____no_output_____"
]
],
[
[
"`4.` Use the cells below to find the most viewed **article_id**, as well as how often it was viewed. After talking to the company leaders, the `email_mapper` function was deemed a reasonable way to map users to ids. There were a small number of null values, and it was found that all of these null values likely belonged to a single user (which is how they are stored using the function below).",
"_____no_output_____"
]
],
[
[
"most_viewed_articles = df[\"article_id\"].value_counts().sort_values(ascending=False)\nprint(\"Most viewed article:\")\nprint(\"ID:\\t {}\".format(most_viewed_articles.index[0]))\nprint(\"title:\\t {}\".format(df[df[\"article_id\"]==most_viewed_articles.index[0]][\"title\"].values[0]))\nprint(\"views:\\t {}\".format(most_viewed_articles.values[0]))",
"Most viewed article:\nID:\t 1429.0\ntitle:\t use deep learning for image classification\nviews:\t 937\n"
],
[
"most_viewed_article_id = \"1429.0\" # The most viewed article in the dataset as a string with one value following the decimal \nmax_views = 937 # The most viewed article in the dataset was viewed how many times?",
"_____no_output_____"
],
[
"## No need to change the code here - this will be helpful for later parts of the notebook\n# Run this cell to map the user email to a user_id column and remove the email column\n\ndef email_mapper():\n coded_dict = dict()\n cter = 1\n email_encoded = []\n \n for val in df['email']:\n if val not in coded_dict:\n coded_dict[val] = cter\n cter+=1\n \n email_encoded.append(coded_dict[val])\n return email_encoded\n\nemail_encoded = email_mapper()\ndel df['email']\ndf['user_id'] = email_encoded\n\n# show header\ndf.head()",
"_____no_output_____"
],
[
"## If you stored all your results in the variable names above, \n## you shouldn't need to change anything in this cell\n\nsol_1_dict = {\n '`50% of individuals have _____ or fewer interactions.`': median_val,\n '`The total number of user-article interactions in the dataset is ______.`': user_article_interactions,\n '`The maximum number of user-article interactions by any 1 user is ______.`': max_views_by_user,\n '`The most viewed article in the dataset was viewed _____ times.`': max_views,\n '`The article_id of the most viewed article is ______.`': most_viewed_article_id,\n '`The number of unique articles that have at least 1 rating ______.`': unique_articles,\n '`The number of unique users in the dataset is ______`': unique_users,\n '`The number of unique articles on the IBM platform`': total_articles\n}\n\n# Test your dictionary against the solution\nt.sol_1_test(sol_1_dict)",
"It looks like you have everything right here! Nice job!\n"
]
],
[
[
"### <a class=\"anchor\" id=\"Rank\">Part II: Rank-Based Recommendations</a>\n\nUnlike in the earlier lessons, we don't actually have ratings for whether a user liked an article or not. We only know that a user has interacted with an article. In these cases, the popularity of an article can really only be based on how often an article was interacted with.\n\n`1.` Fill in the function below to return the **n** top articles ordered with most interactions as the top. Test your function using the tests below.",
"_____no_output_____"
]
],
[
[
"def get_top_articles(n, df=df):\n '''\n INPUT:\n n - (int) the number of top articles to return\n df - (pandas dataframe) df as defined at the top of the notebook \n \n OUTPUT:\n top_articles - (list) A list of the top 'n' article titles \n \n '''\n top_articles = list(set(df[df['article_id'].isin(get_top_article_ids(n,df))]['title']))\n \n return top_articles # Return the top article titles from df (not df_content)\n\ndef get_top_article_ids(n, df=df):\n '''\n INPUT:\n n - (int) the number of top articles to return\n df - (pandas dataframe) df as defined at the top of the notebook \n \n OUTPUT:\n top_articles - (list) A list of the top 'n' article titles \n \n '''\n top_articles = [str(x) for x in df.article_id.value_counts().head(n).index]\n \n return top_articles # Return the top article ids",
"_____no_output_____"
],
[
"print(get_top_articles(10))\nprint(get_top_article_ids(10))",
"['finding optimal locations of new store using decision optimization', 'analyze energy consumption in buildings', 'use deep learning for image classification', 'healthcare python streaming application demo', 'apache spark lab, part 1: basic concepts', 'insights from new york car accident reports', 'predicting churn with the spss random tree algorithm', 'gosales transactions for logistic regression model', 'visualize car data with brunel', 'use xgboost, scikit-learn & ibm watson machine learning apis']\n['1429.0', '1330.0', '1431.0', '1427.0', '1364.0', '1314.0', '1293.0', '1170.0', '1162.0', '1304.0']\n"
],
[
"# Test your function by returning the top 5, 10, and 20 articles\ntop_5 = get_top_articles(5)\ntop_10 = get_top_articles(10)\ntop_20 = get_top_articles(20)\n\n# Test each of your three lists from above\nt.sol_2_test(get_top_articles)",
"Your top_5 looks like the solution list! Nice job.\nYour top_10 looks like the solution list! Nice job.\nYour top_20 looks like the solution list! Nice job.\n"
]
],
[
[
"### <a class=\"anchor\" id=\"User-User\">Part III: User-User Based Collaborative Filtering</a>\n\n\n`1.` Use the function below to reformat the **df** dataframe to be shaped with users as the rows and articles as the columns. \n\n* Each **user** should only appear in each **row** once.\n\n\n* Each **article** should only show up in one **column**. \n\n\n* **If a user has interacted with an article, then place a 1 where the user-row meets for that article-column**. It does not matter how many times a user has interacted with the article, all entries where a user has interacted with an article should be a 1. \n\n\n* **If a user has not interacted with an item, then place a zero where the user-row meets for that article-column**. \n\nUse the tests to make sure the basic structure of your matrix matches what is expected by the solution.",
"_____no_output_____"
]
],
[
[
"# tests before implement the function\nuser_item = df.groupby(['user_id', 'article_id'])['title'].count().notnull().unstack()\nuser_item = user_item.notnull().astype(np.int)\nuser_item.head()",
"_____no_output_____"
],
[
"# create the user-article matrix with 1's and 0's\n\ndef create_user_item_matrix(df):\n '''\n INPUT:\n df - pandas dataframe with article_id, title, user_id columns\n \n OUTPUT:\n user_item - user item matrix \n \n Description:\n Return a matrix with user ids as rows and article ids on the columns with 1 values where a user interacted with \n an article and a 0 otherwise\n '''\n # group the dataframe by users and articles - count each title - convert into boolean - unstack grouped dataframe\n user_item = df.groupby(['user_id', 'article_id'])['title'].count().notnull().unstack()\n # convert True / False into 1 / 0\n user_item = user_item.notnull().astype(np.int)\n \n return user_item # return the user_item matrix \n\nuser_item = create_user_item_matrix(df)",
"_____no_output_____"
],
[
"user_item.head()",
"_____no_output_____"
],
[
"user_item.to_pickle('user_item_matrix.p')",
"_____no_output_____"
],
[
"## Tests: You should just need to run this cell. Don't change the code.\nassert user_item.shape[0] == 5149, \"Oops! The number of users in the user-article matrix doesn't look right.\"\nassert user_item.shape[1] == 714, \"Oops! The number of articles in the user-article matrix doesn't look right.\"\nassert user_item.sum(axis=1)[1] == 36, \"Oops! The number of articles seen by user 1 doesn't look right.\"\nprint(\"You have passed our quick tests! Please proceed!\")",
"You have passed our quick tests! Please proceed!\n"
]
],
[
[
"`2.` Complete the function below which should take a user_id and provide an ordered list of the most similar users to that user (from most similar to least similar). The returned result should not contain the provided user_id, as we know that each user is similar to him/herself. Because the results for each user here are binary, it (perhaps) makes sense to compute similarity as the dot product of two users. \n\nUse the tests to test your function.",
"_____no_output_____"
]
],
[
[
"def find_similar_users(user_id, user_item=user_item):\n '''\n INPUT:\n user_id - (int) a user_id\n user_item - (pandas dataframe) matrix of users by articles: \n 1's when a user has interacted with an article, 0 otherwise\n \n OUTPUT:\n similar_users - (list) an ordered list where the closest users (largest dot product users)\n are listed first\n \n Description:\n Computes the similarity of every pair of users based on the dot product\n Returns an ordered\n \n '''\n # compute similarity of each user to the provided user\n similarity = user_item.dot(user_item.loc[user_id])\n \n # sort by similarity (from top to bottom)\n similarity = similarity.sort_values(ascending = False)\n \n # create list of just the ids\n most_similar_users = list(similarity.index)\n \n # remove the own user's id\n most_similar_users.remove(user_id)\n \n return most_similar_users # return a list of the users in order from most to least similar\n ",
"_____no_output_____"
],
[
"# Do a spot check of your function\nprint(\"The 10 most similar users to user 1 are: {}\".format(find_similar_users(1)[:10]))\nprint(\"The 5 most similar users to user 3933 are: {}\".format(find_similar_users(3933)[:5]))\nprint(\"The 3 most similar users to user 46 are: {}\".format(find_similar_users(46)[:3]))",
"The 10 most similar users to user 1 are: [3933, 23, 3782, 203, 4459, 131, 3870, 46, 4201, 5041]\nThe 5 most similar users to user 3933 are: [1, 23, 3782, 4459, 203]\nThe 3 most similar users to user 46 are: [4201, 23, 3782]\n"
]
],
[
[
"`3.` Now that you have a function that provides the most similar users to each user, you will want to use these users to find articles you can recommend. Complete the functions below to return the articles you would recommend to each user. ",
"_____no_output_____"
]
],
[
[
"def get_article_names(article_ids, df=df):\n '''\n INPUT:\n article_ids - (list) a list of article ids\n df - (pandas dataframe) df as defined at the top of the notebook\n \n OUTPUT:\n article_names - (list) a list of article names associated with the list of article ids \n (this is identified by the title column)\n '''\n # find the article names to the corresponding article ids and store in list\n article_names = list(set(df[df['article_id'].isin(article_ids)]['title'])) \n \n return article_names # Return the article names associated with list of article ids\n\n\ndef get_user_articles(user_id, user_item=user_item):\n '''\n INPUT:\n user_id - (int) a user id\n user_item - (pandas dataframe) matrix of users by articles: \n 1's when a user has interacted with an article, 0 otherwise\n \n OUTPUT:\n article_ids - (list) a list of the article ids seen by the user\n article_names - (list) a list of article names associated with the list of article ids \n (this is identified by the doc_full_name column in df_content)\n \n Description:\n Provides a list of the article_ids and article titles that have been seen by a user\n '''\n # find the article ids for the given user id in the user-article matrix\n article_ids = user_item.loc[user_id]\n \n # get the index (article_id) which is equal to one to filter the 0\n article_ids = list(article_ids[article_ids == 1].index.astype(str))\n \n # get corresponding article names \n article_names = get_article_names(article_ids)\n \n return article_ids, article_names # return the ids and names\n\n\ndef user_user_recs(user_id, m=10):\n '''\n INPUT:\n user_id - (int) a user id\n m - (int) the number of recommendations you want for the user\n \n OUTPUT:\n recs - (list) a list of recommendations for the user\n \n Description:\n Loops through the users based on closeness to the input user_id\n For each user - finds articles the user hasn't seen before and provides them as recs\n Does this until m recommendations are found\n \n Notes:\n Users who are the same closeness are chosen arbitrarily as the 'next' user\n \n For the user where the number of recommended articles starts below m \n and ends exceeding m, the last items are chosen arbitrarily\n \n '''\n # find most similar users to the given user id\n user_ids = find_similar_users(user_id)\n \n # filter for user ids that are similar and store article ids\n recs = df[df['user_id'].isin(user_ids)]['article_id']\n \n # convert into list and store only m recommendations\n recs = list(set(recs))\n \n return recs[:m] # return your recommendations for this user_id ",
"_____no_output_____"
],
[
"# Check Results\nget_article_names(user_user_recs(1, 10)) # Return 10 recommendations for user 1",
"_____no_output_____"
],
[
"# Test your functions here - No need to change this code - just run this cell\nassert set(get_article_names(['1024.0', '1176.0', '1305.0', '1314.0', '1422.0', '1427.0'])) == set(['using deep learning to reconstruct high-resolution audio', 'build a python app on the streaming analytics service', 'gosales transactions for naive bayes model', 'healthcare python streaming application demo', 'use r dataframes & ibm watson natural language understanding', 'use xgboost, scikit-learn & ibm watson machine learning apis']), \"Oops! Your the get_article_names function doesn't work quite how we expect.\"\nassert set(get_article_names(['1320.0', '232.0', '844.0'])) == set(['housing (2015): united states demographic measures','self-service data preparation with ibm data refinery','use the cloudant-spark connector in python notebook']), \"Oops! Your the get_article_names function doesn't work quite how we expect.\"\nassert set(get_user_articles(20)[0]) == set(['1320.0', '232.0', '844.0'])\nassert set(get_user_articles(20)[1]) == set(['housing (2015): united states demographic measures', 'self-service data preparation with ibm data refinery','use the cloudant-spark connector in python notebook'])\nassert set(get_user_articles(2)[0]) == set(['1024.0', '1176.0', '1305.0', '1314.0', '1422.0', '1427.0'])\nassert set(get_user_articles(2)[1]) == set(['using deep learning to reconstruct high-resolution audio', 'build a python app on the streaming analytics service', 'gosales transactions for naive bayes model', 'healthcare python streaming application demo', 'use r dataframes & ibm watson natural language understanding', 'use xgboost, scikit-learn & ibm watson machine learning apis'])\nprint(\"If this is all you see, you passed all of our tests! Nice job!\")",
"If this is all you see, you passed all of our tests! Nice job!\n"
]
],
[
[
"`4.` Now we are going to improve the consistency of the **user_user_recs** function from above. \n\n* Instead of arbitrarily choosing when we obtain users who are all the same closeness to a given user - choose the users that have the most total article interactions before choosing those with fewer article interactions.\n\n\n* Instead of arbitrarily choosing articles from the user where the number of recommended articles starts below m and ends exceeding m, choose articles with the articles with the most total interactions before choosing those with fewer total interactions. This ranking should be what would be obtained from the **top_articles** function you wrote earlier.",
"_____no_output_____"
]
],
[
[
"def get_top_sorted_users(user_id, df=df, user_item=user_item):\n '''\n INPUT:\n user_id - (int)\n df - (pandas dataframe) df as defined at the top of the notebook \n user_item - (pandas dataframe) matrix of users by articles: \n 1's when a user has interacted with an article, 0 otherwise\n \n \n OUTPUT:\n neighbors_df - (pandas dataframe) a dataframe with:\n neighbor_id - is a neighbor user_id\n similarity - measure of the similarity of each user to the provided user_id\n num_interactions - the number of articles viewed by the user - if a u\n \n Other Details - sort the neighbors_df by the similarity and then by number of interactions where \n highest of each is higher in the dataframe\n \n '''\n \n #calculate similarity of users with regard to user_id by using dot product\n similarity = user_item.dot(user_item.loc[user_id])\n \n # sort similarity in descending order\n similarity = similarity.sort_values(ascending=False)\n \n # drop user_id\n similarity = similarity.drop(user_id)\n \n # convert similarity into dataframe\n similarity = similarity.to_frame(name='similarity').reset_index()\n\n #count number of user interactions\n num_interactions = df.user_id.value_counts()\n \n # convert num_interactions to dataframe\n num_interactions = num_interactions.to_frame('num_interactions')\n \n # merge the created dataframes\n neighbors_df = similarity.merge(num_interactions, left_on='user_id', right_index=True)\n \n # rename user_id column to neighbor_id\n neighbors_df = neighbors_df.rename(columns={'user_id':'neighbor_id'})\n\n # sort dataframe by similarity in descending order\n neighbors_df.sort_values(by=['similarity', 'num_interactions'], ascending=False, inplace=True)\n \n return neighbors_df # Return the dataframe specified in the doc_string\n\n\ndef user_user_recs_part2(user_id, m=10):\n '''\n INPUT:\n user_id - (int) a user id\n m - (int) the number of recommendations you want for the user\n \n OUTPUT:\n recs - (list) a list of recommendations for the user by article id\n rec_names - (list) a list of recommendations for the user by article title\n \n Description:\n Loops through the users based on closeness to the input user_id\n For each user - finds articles the user hasn't seen before and provides them as recs\n Does this until m recommendations are found\n \n Notes:\n * Choose the users that have the most total article interactions \n before choosing those with fewer article interactions.\n\n * Choose articles with the articles with the most total interactions \n before choosing those with fewer total interactions. \n \n '''\n # find users with highest similarity to user_id\n neighbors_df = get_top_sorted_users(user_id)\n \n # get user_id of m most similar neighbors\n most_similar_neighbors = list(neighbors_df[:m]['neighbor_id'])\n\n # get article_ids interacted by the m most similar neighbors\n recs = []\n for user in most_similar_neighbors:\n article_ids = user_item.loc[user]\n recs.extend([art_id for art_id in article_ids[article_ids == 1].index.astype(str)])\n\n # remove duplicates and get the top m users\n recs = list(set(recs[:m]))\n\n # convert article_ids to article_names and remove duplicates\n rec_names = list(set(df[df['article_id'].isin(recs)]['title']))\n \n return recs, rec_names",
"_____no_output_____"
],
[
"# Quick spot check - don't change this code - just use it to test your functions\nrec_ids, rec_names = user_user_recs_part2(20, 10)\nprint(\"The top 10 recommendations for user 20 are the following article ids:\")\nprint(rec_ids)\nprint()\nprint(\"The top 10 recommendations for user 20 are the following article names:\")\nprint(rec_names)",
"The top 10 recommendations for user 20 are the following article ids:\n['164.0', '205.0', '336.0', '232.0', '142.0', '362.0', '12.0', '125.0', '109.0', '302.0']\n\nThe top 10 recommendations for user 20 are the following article names:\n['dsx: hybrid mode', 'accelerate your workflow with dsx', 'neural networks for beginners: popular types and applications', 'learn tensorflow and deep learning together and now!', 'self-service data preparation with ibm data refinery', 'timeseries data analysis of iot events by using jupyter notebook', 'challenges in deep learning', \"a beginner's guide to variational methods\", 'tensorflow quick tips', 'statistics for hackers']\n"
]
],
[
[
"`5.` Use your functions from above to correctly fill in the solutions to the dictionary below. Then test your dictionary against the solution. Provide the code you need to answer each following the comments below.",
"_____no_output_____"
]
],
[
[
"### Tests with a dictionary of results\n\nuser1_most_sim = get_top_sorted_users(1).iloc[0].neighbor_id # Find the user that is most similar to user 1 \nuser131_10th_sim = get_top_sorted_users(131).iloc[9].neighbor_id # Find the 10th most similar user to user 131\n\nprint('The user that is most similar to user 1 is user {}.'.format(user1_most_sim))\nprint('The 10th most similar user to user 131 is user {}.'.format(user131_10th_sim))",
"The user that is most similar to user 1 is user 3933.\nThe 10th most similar user to user 131 is user 242.\n"
],
[
"## Dictionary Test Here\nsol_5_dict = {\n 'The user that is most similar to user 1.': user1_most_sim, \n 'The user that is the 10th most similar to user 131': user131_10th_sim,\n}\n\nt.sol_5_test(sol_5_dict)",
"This all looks good! Nice job!\n"
]
],
[
[
"`6.` If we were given a new user, which of the above functions would you be able to use to make recommendations? Explain. Can you think of a better way we might make recommendations? Use the cell below to explain a better method for new users.",
"_____no_output_____"
],
[
"**Response:**\n\nWe cannot make recommendations for new users by using the functions above since they are based on user knowledge. For new users, we do not have any knowledge about his or her preferences, because he or she has not interacted with any of the articles. Thus, there is no similarity to calculate between the new user and older ones. We can only use knowledge based approaches in cases where the user has interacted with a certain amount of articles, because with a data basis of one or two user interactions for a user we might not conclude reasonable recommendations.",
"_____no_output_____"
],
[
"`7.` Using your existing functions, provide the top 10 recommended articles you would provide for the a new user below. You can test your function against our thoughts to make sure we are all on the same page with how we might make a recommendation.",
"_____no_output_____"
]
],
[
[
"new_user = '0.0'\n\n# What would your recommendations be for this new user '0.0'? As a new user, they have no observed articles.\n# Provide a list of the top 10 article ids you would give to \nnew_user_recs = get_top_article_ids(10)\n\n",
"_____no_output_____"
],
[
"assert set(new_user_recs) == set(['1314.0','1429.0','1293.0','1427.0','1162.0','1364.0','1304.0','1170.0','1431.0','1330.0']), \"Oops! It makes sense that in this case we would want to recommend the most popular articles, because we don't know anything about these users.\"\n\nprint(\"That's right! Nice job!\")",
"That's right! Nice job!\n"
]
],
[
[
"### <a class=\"anchor\" id=\"Content-Recs\">Part IV: Content Based Recommendations (EXTRA - NOT REQUIRED)</a>\n\nAnother method we might use to make recommendations is to perform a ranking of the highest ranked articles associated with some term. You might consider content to be the **doc_body**, **doc_description**, or **doc_full_name**. There isn't one way to create a content based recommendation, especially considering that each of these columns hold content related information. \n\n`1.` Use the function body below to create a content based recommender. Since there isn't one right answer for this recommendation tactic, no test functions are provided. Feel free to change the function inputs if you decide you want to try a method that requires more input values. The input values are currently set with one idea in mind that you may use to make content based recommendations. One additional idea is that you might want to choose the most popular recommendations that meet your 'content criteria', but again, there is a lot of flexibility in how you might make these recommendations.\n\n### This part is NOT REQUIRED to pass this project. However, you may choose to take this on as an extra way to show off your skills.",
"_____no_output_____"
]
],
[
[
"def make_content_recs():\n '''\n INPUT:\n \n OUTPUT:\n \n '''",
"_____no_output_____"
]
],
[
[
"`2.` Now that you have put together your content-based recommendation system, use the cell below to write a summary explaining how your content based recommender works. Do you see any possible improvements that could be made to your function? Is there anything novel about your content based recommender?\n\n### This part is NOT REQUIRED to pass this project. However, you may choose to take this on as an extra way to show off your skills.",
"_____no_output_____"
],
[
"**Write an explanation of your content based recommendation system here.**",
"_____no_output_____"
],
[
"`3.` Use your content-recommendation system to make recommendations for the below scenarios based on the comments. Again no tests are provided here, because there isn't one right answer that could be used to find these content based recommendations.\n\n### This part is NOT REQUIRED to pass this project. However, you may choose to take this on as an extra way to show off your skills.",
"_____no_output_____"
]
],
[
[
"# make recommendations for a brand new user\n\n\n# make a recommendations for a user who only has interacted with article id '1427.0'\n\n",
"_____no_output_____"
]
],
[
[
"### <a class=\"anchor\" id=\"Matrix-Fact\">Part V: Matrix Factorization</a>\n\nIn this part of the notebook, you will build use matrix factorization to make article recommendations to the users on the IBM Watson Studio platform.\n\n`1.` You should have already created a **user_item** matrix above in **question 1** of **Part III** above. This first question here will just require that you run the cells to get things set up for the rest of **Part V** of the notebook. ",
"_____no_output_____"
]
],
[
[
"# Load the matrix here\nuser_item_matrix = pd.read_pickle('user_item_matrix.p')",
"_____no_output_____"
],
[
"# quick look at the matrix\nuser_item_matrix.head()",
"_____no_output_____"
]
],
[
[
"`2.` In this situation, you can use Singular Value Decomposition from [numpy](https://docs.scipy.org/doc/numpy-1.14.0/reference/generated/numpy.linalg.svd.html) on the user-item matrix. Use the cell to perform SVD, and explain why this is different than in the lesson.",
"_____no_output_____"
]
],
[
[
"# Perform SVD on the User-Item Matrix Here\n\nu, s, vh = np.linalg.svd(user_item_matrix, full_matrices=False) # use the built in to get the three matrices",
"_____no_output_____"
],
[
"print('Shape of u: {}'.format(u.shape))\nprint('Shape of s: {}'.format(s.shape))\nprint('Shape of vh: {}'.format(vt.shape))",
"Shape of u: (5149, 714)\nShape of s: (714,)\nShape of vh: (714, 714)\n"
]
],
[
[
"**Response:**\n\nThe Singular Value Decomposition only works in cases, where no missing values are present. In our user-item matrix, this condition is satisfied and we can apply this technique to factorize the 2D-matrix and, further, to make recommendations. In the video lessons, we had missing values and had to choose funkSVD for matrix factorization. ",
"_____no_output_____"
],
[
"`3.` Now for the tricky part, how do we choose the number of latent features to use? Running the below cell, you can see that as the number of latent features increases, we obtain a lower error rate on making predictions for the 1 and 0 values in the user-item matrix. Run the cell below to get an idea of how the accuracy improves as we increase the number of latent features.",
"_____no_output_____"
]
],
[
[
"num_latent_feats = np.arange(10,700+10,20)\nsum_errs = []\n\nfor k in num_latent_feats:\n # restructure with k latent features\n s_new, u_new, vt_new = np.diag(s[:k]), u[:, :k], vt[:k, :]\n \n # take dot product\n user_item_est = np.around(np.dot(np.dot(u_new, s_new), vt_new))\n \n # compute error for each prediction to actual value\n diffs = np.subtract(user_item_matrix, user_item_est)\n \n # total errors and keep track of them\n err = np.sum(np.sum(np.abs(diffs)))\n sum_errs.append(err)\n \n \nplt.plot(num_latent_feats, 1 - np.array(sum_errs)/df.shape[0]);\nplt.xlabel('Number of Latent Features');\nplt.ylabel('Accuracy');\nplt.title('Accuracy vs. Number of Latent Features');",
"_____no_output_____"
]
],
[
[
"`4.` From the above, we can't really be sure how many features to use, because simply having a better way to predict the 1's and 0's of the matrix doesn't exactly give us an indication of if we are able to make good recommendations. Instead, we might split our dataset into a training and test set of data, as shown in the cell below. \n\nUse the code from question 3 to understand the impact on accuracy of the training and test sets of data with different numbers of latent features. Using the split below: \n\n* How many users can we make predictions for in the test set? \n* How many users are we not able to make predictions for because of the cold start problem?\n* How many articles can we make predictions for in the test set? \n* How many articles are we not able to make predictions for because of the cold start problem?",
"_____no_output_____"
]
],
[
[
"df_train = df.head(40000)\ndf_test = df.tail(5993)\n\ndef create_test_and_train_user_item(df_train, df_test):\n '''\n INPUT:\n df_train - training dataframe\n df_test - test dataframe\n \n OUTPUT:\n user_item_train - a user-item matrix of the training dataframe \n (unique users for each row and unique articles for each column)\n user_item_test - a user-item matrix of the testing dataframe \n (unique users for each row and unique articles for each column)\n test_idx - all of the test user ids\n test_arts - all of the test article ids\n \n '''\n # perform the create_user_item_matrix() function on the train and test dataset to get a corresponding user-item-matrix\n user_item_train = create_user_item_matrix(df_train)\n user_item_test = create_user_item_matrix(df_test)\n\n # save ids of test users and test articles as lists\n test_idx = list(set(user_item_test.index))\n test_arts = list(set(user_item_test.columns))\n \n return user_item_train, user_item_test, test_idx, test_arts\n\nuser_item_train, user_item_test, test_idx, test_arts = create_test_and_train_user_item(df_train, df_test)",
"_____no_output_____"
],
[
"print('Shape of user_item_train: {}'.format(user_item_train.shape))\nprint('Shape of user_item_test: {}'.format(user_item_test.shape))",
"Shape of user_item_train: (4487, 714)\nShape of user_item_test: (682, 574)\n"
],
[
"print('How many users can we make predictions for in the test set?: {}'.format(len(set(user_item_test.index) & set(user_item_train.index))))\nprint('How many users in the test set are we not able to make predictions for because of the cold start problem?: {}'.format(len(set(user_item_test.index) - set(user_item_train.index))))\nprint('How many articles can we make predictions for in the test set?: {}'.format(len(set(user_item_test.columns) & set(user_item_train.columns))))\nprint('How many articles in the test set are we not able to make predictions for because of the cold start problem?: {}'.format(len(set(user_item_test.columns) - set(user_item_train.columns))))",
"How many users can we make predictions for in the test set?: 20\nHow many users in the test set are we not able to make predictions for because of the cold start problem?: 662\nHow many articles can we make predictions for in the test set?: 574\nHow many articles in the test set are we not able to make predictions for because of the cold start problem?: 0\n"
],
[
"# Replace the values in the dictionary below\na = 662 \nb = 574 \nc = 20 \nd = 0 \n\n\nsol_4_dict_1 = {\n 'How many users can we make predictions for in the test set?': c, \n 'How many users in the test set are we not able to make predictions for because of the cold start problem?': a, \n 'How many movies can we make predictions for in the test set?': b,\n 'How many movies in the test set are we not able to make predictions for because of the cold start problem?': d\n }\n\nt.sol_4_test(sol_4_dict_1)",
"Awesome job! That's right! All of the test movies are in the training data, but there are only 20 test users that were also in the training set. All of the other users that are in the test set we have no data on. Therefore, we cannot make predictions for these users using SVD.\n"
]
],
[
[
"`5.` Now use the **user_item_train** dataset from above to find U, S, and V transpose using SVD. Then find the subset of rows in the **user_item_test** dataset that you can predict using this matrix decomposition with different numbers of latent features to see how many features makes sense to keep based on the accuracy on the test data. This will require combining what was done in questions `2` - `4`.\n\nUse the cells below to explore how well SVD works towards making predictions for recommendations on the test data. ",
"_____no_output_____"
]
],
[
[
"# fit SVD on the user_item_train matrix\nu_train, s_train, vh_train = np.linalg.svd(user_item_train, full_matrices=False)\nprint('Shape of u_train: {}'.format(u_train.shape))\nprint('Shape of s_train: {}'.format(s_train.shape))\nprint('Shape of vh_train: {}'.format(vh_train.shape))",
"Shape of u_train: (4487, 714)\nShape of s_train: (714,)\nShape of vh_train: (714, 714)\n"
],
[
"# Use these cells to see how well you can use the training \n# decomposition to predict on test data\n\n# get the ids of commom users and article between the training and testing dataset\ntrain_test_common_idx = user_item_train.index.isin(test_idx)\ntrain_test_common_col = user_item_train.columns.isin(test_arts)",
"_____no_output_____"
],
[
"# show common users (20 as shown in the cells above) as an example\nuser_item_train[train_test_common_idx]",
"_____no_output_____"
],
[
"# get the subsets of the training data for the u_test and vh_test arrays\nu_test = u_train[train_test_common_idx, :]\nvh_test= vh_train[:, train_test_common_col]\n\nprint('Shape of u_test: {}'.format(u_test.shape))\nprint('Shape of vh_test: {}'.format(vh_test.shape))",
"Shape of u_test: (20, 714)\nShape of vh_test: (714, 574)\n"
],
[
"# get the subset of the user-item-matrix by filtering the common user and article ids of training and testing dataset\nuser_item_test = user_item_test.loc[set(user_item_train.index) & set(user_item_test.index), set(user_item_train.columns) & set(user_item_test.columns)]",
"_____no_output_____"
],
[
"user_item_test",
"_____no_output_____"
],
[
"# init features and errors variables\nnum_latent_features = np.arange(10,714,20)\nsum_errors_train = []\nsum_errors_test = []\n\n# loop over number of latent features\nfor i in num_latent_features:\n # filter fitted SVD arrays by selecting the i latent features\n # ... of the training data\n s_train_latent, u_train_latent, vh_train_latent = np.diag(s_train[:i]), u_train[:, :i], vh_train[:i, :]\n \n # ... and testing data\n u_test_latent, vh_test_latent = u_test[:, :i], vh_test[:i, :]\n \n # perform the dot product on the created arrays\n user_item_train_latent = np.around(np.dot(np.dot(u_train_latent, s_train_latent), vh_train_latent))\n user_item_test_latent = np.around(np.dot(np.dot(u_test_latent, s_train_latent), vh_test_latent))\n \n # compute errors between predicted and actual value\n diffs_train = np.subtract(user_item_train, user_item_train_latent)\n diffs_test = np.subtract(user_item_test, user_item_test_latent)\n \n # count total errors and append to error arrays\n sum_errors_train.append(np.sum(np.sum(np.abs(diffs_train))))\n sum_errors_test.append(np.sum(np.sum(np.abs(diffs_test))))",
"_____no_output_____"
],
[
"# visualize impact of number of latent features on accuracy of recommendations\nfig, ax1 = plt.subplots()\n\n# plot train accuracy vs. number of latent features \nax1.plot(num_latent_features, 100*(1 - np.array(sum_errors_train)/df.shape[0]), color = 'blue', label=\"Train accuracy\")\nax1.set_title('Train/Test Accuracy vs. Number of Latent Features')\nax1.grid(True)\nax1.set_xlabel('Number of Latent Features')\nax1.set_ylabel('Train accuracy [%]')\n\n# create second y-axis and plot test accuracy vs. number of latent features \nax2 = ax1.twinx()\nax2.plot(num_latent_features, 100*(1 - np.array(sum_errors_test)/df.shape[0]), color='orange', label=\"Test accuracy\")\nax2.set_ylabel('Test accuracy [%]', rotation=270, labelpad=15)\n\n# create legend\nhandle_1, label_1 = ax1.get_legend_handles_labels()\nhandle_2, label_2 = ax2.get_legend_handles_labels()\nax1.legend(handle_1 + handle_2, label_1 + label_2, loc='center right')\n\nplt.show()",
"_____no_output_____"
],
[
"from sklearn.metrics import f1_score\n# init f1 variables\nf1_score_train = []\nf1_score_test = []\n\n# loop over number of latent features\nfor i in num_latent_features:\n # filter fitted SVD arrays by selecting the i latent features\n # ... of the training data\n s_train_latent, u_train_latent, vh_train_latent = np.diag(s_train[:i]), u_train[:, :i], vh_train[:i, :]\n \n # ... and testing data\n u_test_latent, vh_test_latent = u_test[:, :i], vh_test[:i, :]\n \n # perform the dot product on the created arrays\n user_item_train_latent = np.around(np.dot(np.dot(u_train_latent, s_train_latent), vh_train_latent))\n user_item_test_latent = np.around(np.dot(np.dot(u_test_latent, s_train_latent), vh_test_latent))\n \n # compute f1 score\n f1_score_train.append(f1_score(np.array(user_item_train).flatten(), user_item_train_latent.flatten(), labels=[1.0], average='macro'))\n f1_score_test.append(f1_score(np.array(user_item_test).flatten(), user_item_test_latent.flatten(), labels=[1.0], average='macro'))",
"_____no_output_____"
],
[
"# visualize impact of number of latent features on F1-score\nfig, ax1 = plt.subplots()\n\n# plot training F1-score vs. number of latent features \nax1.plot(num_latent_features, f1_score_train, color = 'blue', label=\"F1-Score (Train)\")\nax1.set_title('F1-score vs. Number of Latent Features')\nax1.grid(True)\nax1.set_xlabel('Number of Latent Features')\nax1.set_ylabel('F1-Score (Training)')\n\n# create second y-axis and plot testing F1-score vs. number of latent features \nax2 = ax1.twinx()\nax2.plot(num_latent_features, f1_score_test, color='orange', label=\"F1-Score (Test)\")\nax2.set_ylabel('F1-Score (Testing)', rotation=270, labelpad=15)\n\n# create legend\nhandle_1, label_1 = ax1.get_legend_handles_labels()\nhandle_2, label_2 = ax2.get_legend_handles_labels()\nax1.legend(handle_1 + handle_2, label_1 + label_2, loc='center right')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"`6.` Use the cell below to comment on the results you found in the previous question. Given the circumstances of your results, discuss what you might do to determine if the recommendations you make with any of the above recommendation systems are an improvement to how users currently find articles? ",
"_____no_output_____"
],
[
"**Response:**\n\nSurprisingly, the accuracy curve of the testing dataset dropped from 99.45% to around 99.05% when the number of latent features increased. This behaviour is reciprocal relative to the accuracy curve of the training data, which rises from 40% to nearly 100% when we increase the number of latent features as well.\n\nThe cause of this behaviour might be overfitting of the recommendations to the training data. I.e., that our model gets better and better in fitting the training dataset with an increasing number of latent features it but lacks in generalizing well to predictions on the testing set. As a result, we should use only a few latent features for the recommendation.\n\nAdditionally, the model matrix is mostly a sparse matrix, since we have only a small and/or unbalanced number of user-article-interactions. Thus, a large number of latent features is not required to make good recommendations.\n\nIn this case, a better metric might be the F1-score, we additionally applied in previous subsection. As seen in the visualization above, the F1-score for the testing dataset is maximum for around 80 to 100 latent features. After that, this F1- score decreases significantly due to overfitting, which can be observed on the rising F1-score for the training set.\n\nWe can state as a result, that recommendations made only by implementing a SVD performs not well when we have a very small sample of training and testing data. The intersection of articles interacted by users was very small, resulting in a lower certainty of the recommendations. Thus, we might have to add another recommendation technique to improve our results.",
"_____no_output_____"
],
[
"<a id='conclusions'></a>\n### Extras\nUsing your workbook, you could now save your recommendations for each user, develop a class to make new predictions and update your results, and make a flask app to deploy your results. These tasks are beyond what is required for this project. However, from what you learned in the lessons, you certainly capable of taking these tasks on to improve upon your work here!\n\n\n## Conclusion\n\n> Congratulations! You have reached the end of the Recommendations with IBM project! \n\n> **Tip**: Once you are satisfied with your work here, check over your report to make sure that it is satisfies all the areas of the [rubric](https://review.udacity.com/#!/rubrics/2322/view). You should also probably remove all of the \"Tips\" like this one so that the presentation is as polished as possible.\n\n\n## Directions to Submit\n\n> Before you submit your project, you need to create a .html or .pdf version of this notebook in the workspace here. To do that, run the code cell below. If it worked correctly, you should get a return code of 0, and you should see the generated .html file in the workspace directory (click on the orange Jupyter icon in the upper left).\n\n> Alternatively, you can download this report as .html via the **File** > **Download as** submenu, and then manually upload it into the workspace directory by clicking on the orange Jupyter icon in the upper left, then using the Upload button.\n\n> Once you've done this, you can submit your project by clicking on the \"Submit Project\" button in the lower right here. This will create and submit a zip file with this .ipynb doc and the .html or .pdf version you created. Congratulations! ",
"_____no_output_____"
]
],
[
[
"from subprocess import call\ncall(['python', '-m', 'nbconvert', 'Recommendations_with_IBM.ipynb'])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
ecb6c14bb1c64ef7db25e0c7a05a22c9aac7f1df | 563,826 | ipynb | Jupyter Notebook | archives/gm/notebooks/ts_varmax.ipynb | gamyers/solar-697 | 90ca38072456af385c98b1bdf3c3d563e2c71f15 | [
"MIT"
] | 1 | 2021-08-24T00:00:23.000Z | 2021-08-24T00:00:23.000Z | archives/gm/notebooks/ts_varmax.ipynb | gamyers/solar-697 | 90ca38072456af385c98b1bdf3c3d563e2c71f15 | [
"MIT"
] | null | null | null | archives/gm/notebooks/ts_varmax.ipynb | gamyers/solar-697 | 90ca38072456af385c98b1bdf3c3d563e2c71f15 | [
"MIT"
] | 2 | 2021-08-30T20:36:36.000Z | 2021-11-02T19:13:33.000Z | 1,272.744921 | 297,496 | 0.959908 | [
[
[
"import os\nimport site\nimport sqlite3\nimport sys\nfrom time import sleep\n\nimport logzero\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport yaml\nfrom logzero import logger\nfrom tqdm import tqdm\nfrom tqdm.notebook import tqdm\nfrom yaml import dump, load, safe_load",
"_____no_output_____"
],
[
"sys.path.append(\"../../sql\")\nimport queries\n\nsys.path.append(\"../source\")\nimport ts_tools",
"_____no_output_____"
],
[
"import warnings\n\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"plt.rcParams[\"figure.figsize\"] = 30, 25\nplt.rcParams[\"ytick.labelsize\"] = 11\nplt.rcParams[\"axes.labelsize\"] = 14\nplt.rcParams[\"axes.labelpad\"] = 12\nplt.rcParams[\"axes.xmargin\"] = 0.01\nplt.rcParams[\"axes.ymargin\"] = 0.01",
"_____no_output_____"
],
[
"log_path = \"logs/\"\nlog_file = \"ts_arima.log\"\n\nlogzero.logfile(log_path + log_file, maxBytes=1e6, backupCount=5, disableStderrLogger=True)\nlogger.info(f\"{log_path}, {log_file}\\n\")",
"_____no_output_____"
],
[
"configs = None\ntry:\n with open(\"../configs/config.yml\", \"r\") as config_in:\n configs = load(config_in, Loader=yaml.SafeLoader)\n logger.info(f\"{configs}\\n\")\nexcept:\n logger.error(f\"config file open failure.\")\n exit(1)\n\ncfg_vars = configs[\"url_variables\"]\nlogger.info(f\"variables: {cfg_vars}\\n\")\n\nyears = configs[\"request_years\"]\nlogger.info(f\"years: {years}\\n\")\n\ndb_path = configs[\"file_paths\"][\"db_path\"]\n\ncity = configs[\"location_info\"][\"city\"]\nstate = configs[\"location_info\"][\"state\"]\ndb_file = city + \"-\" + state + \".db\"\n\ndb_table1 = configs[\"table_names\"][\"db_table1\"]\ndb_table2 = configs[\"table_names\"][\"db_table2\"]\n\nlogger.info(f\"{db_path}, {db_file}\")\n\nnrows = configs[\"num_rows\"][0]\nlogger.info(f\"number of rows: {nrows}\\n\")",
"_____no_output_____"
],
[
"conn = sqlite3.connect(db_path + db_file)\ncursor = conn.cursor()",
"_____no_output_____"
],
[
"cursor.execute(queries.select_distinct_zips)\ndistinct_zipcodes = cursor.fetchall()\ndistinct_zipcodes = [z[0] for z in distinct_zipcodes]\nlogger.info(f\"distinct zip codes:\\n{distinct_zipcodes}\")\nprint(distinct_zipcodes)",
"['73108', '73109', '73110', '73115', '73119', '73129', '73130', '73135', '73139', '73145', '73149', '73150', '73159', '73160', '73165']\n"
],
[
"zipcode_index = 9\nparams = {\"zipcode\": distinct_zipcodes[zipcode_index]}\n\nselect_nsr_rows = f\"\"\"\nSELECT date_time,\n-- year, month, day, \n-- zipcode,\n-- Clearsky_DHI, DHI,\nClearsky_DNI, DNI,\nClearsky_GHI, GHI,\nTemperature,\nRelative_Humidity,\nPrecipitable_Water,\n-- Wind_Direction,\nWind_Speed\nfrom nsrdb\nwhere zipcode = :zipcode\n-- and not (month = 2 and day = 29)\n-- and year = 2000\n;\n\"\"\"\n\ndf = pd.read_sql(\n select_nsr_rows,\n conn,\n params=params,\n index_col=\"date_time\",\n parse_dates=[\"date_time\"],\n)\n\ndf.sort_index(axis=0, inplace=True)\n# df.head(5)",
"_____no_output_____"
],
[
"df_rsm = df.resample(\"M\").mean().reset_index(drop=False)\ndf_rsm.set_index(\"date_time\", inplace=True)\n# df_rsm",
"_____no_output_____"
],
[
"columns = df.columns.tolist()\nprint(columns)\nf_idx_a = \"DNI\"\nf_idx_b = \"GHI\"\nf_idx_c = \"Temperature\"\nf_idx_d = \"Relative_Humidity\"\n\nfeatures = [f_idx_a, f_idx_b, f_idx_c, f_idx_d]\nprint(features)",
"['Clearsky_DNI', 'DNI', 'Clearsky_GHI', 'GHI', 'Temperature', 'Relative_Humidity', 'Precipitable_Water', 'Wind_Speed']\n['DNI', 'GHI', 'Temperature', 'Relative_Humidity']\n"
],
[
"df_varmax = df_rsm[features]",
"_____no_output_____"
],
[
"varmax_order = ts_tools.gen_varmax_params(\n p_rng=(12, 12),\n q_rng=(12, 12),\n debug=True,\n)",
"VARMA Order list length: 1\n"
],
[
"results = ts_tools.VARMAX_optimizer(df_varmax, varmax_order, debug=False)\nbest_order = results.iloc[0][\"(p, q)\"]\n\nbest_order\n# (6, 3)",
"_____no_output_____"
],
[
"forecast = ts_tools.varmax_model(\n df_varmax.iloc[:252],\n *best_order,\n num_fc=23,\n forecast=True,\n summary=False,\n)",
"_____no_output_____"
],
[
"actual = df_varmax.iloc[252:]\n\nrmse = np.sqrt(np.mean((actual - forecast) ** 2))\n\nfig, ax = plt.subplots(figsize=(20, 10))\n\nax.plot(df_varmax.iloc[:252], label=\"Original\", color=\"blue\")\nax.plot(actual, label=\"Actual\", color=\"green\")\nax.plot(forecast, label=\"Forecasted\", color=\"orange\")\n\nax.set_xlabel(\"Month\")\n\nax.set_title(\n f\"{city.upper()}, {state.upper()} {distinct_zipcodes[zipcode_index]}\\n\"\n + f\"Differenced Monthly {features} values\\n\"\n + f\"{len(forecast)}-month Forecast\\n\"\n + f\"RMSE = {rmse}\"\n)\n\nax.grid()\nax.legend();",
"_____no_output_____"
],
[
"model = ts_tools.varmax_model(\n df_varmax,\n *best_order,\n forecast=False,\n summary=False,\n)",
"_____no_output_____"
],
[
"model.plot_diagnostics(lags=24);",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb6c32f8d361b8f24a48bfe3884a158de998649 | 5,825 | ipynb | Jupyter Notebook | book_rec.ipynb | nik2203/Book-Recommendation-System | be1c53eec0243e56529f6d0f8139bd56ea4fbc91 | [
"MIT"
] | null | null | null | book_rec.ipynb | nik2203/Book-Recommendation-System | be1c53eec0243e56529f6d0f8139bd56ea4fbc91 | [
"MIT"
] | null | null | null | book_rec.ipynb | nik2203/Book-Recommendation-System | be1c53eec0243e56529f6d0f8139bd56ea4fbc91 | [
"MIT"
] | null | null | null | 32.724719 | 114 | 0.520858 | [
[
[
"# Importing libraries\nimport pandas as pd\nimport random\n\n# Read csv file into a pandas dataframe\ndf = pd.read_csv(\"books.csv\")\ndf = df[df.language_code =='eng']\ndf['Title'] = df['title'].str.split('(').str[0]\n \ndef recommendation_random():\n df_new= df.sample(replace=True)[['Title', 'authors',\"rating\"]]\n print(\"Title: \", df_new[['Title']].to_string(index=False, header=False))\n print(\"Author: \", df_new[['authors']].to_string(index=False, header=False))\n print(\"Rating: \", df_new[['rating']].to_string(index=False, header=False))\n return \"Enjoy!\"\ndef recommendation_rating():\n try:\n search=input('Enter the rating as: rating <rating>\\n')\n if len(search.split())==2 and type(int(search.split()[1]))==int:\n search=search.split()\n rate=float(search[1])\n df_rate = df[df['rating'].astype(float) >= rate]\n df_new= df_rate.sample(replace=True)[['Title', 'authors',\"rating\"]]\n print(\"Title: \", df_new[['Title']].to_string(index=False, header=False))\n print(\"Author: \", df_new[['authors']].to_string(index=False, header=False))\n print(\"Rating: \", df_new[['rating']].to_string(index=False, header=False))\n return \"Enjoy!\"\n else:\n return 'An error was encountered. Please use the proper format and try again.'\n except ValueError:\n return 'An error was encountered. Please use the proper format and try again.'\n\ndef author_search():\n author=input(\"Enter author name for a book suggestion \")\n authl=[]\n til=[]\n res_til=[]\n ratl=[]\n res_ratl=[]\n aui=adic['authors'].items()\n tii=adic['Title'].items()\n rai=adic['rating'].items()\n for i in aui:\n authl.append(i[1])\n for i in tii:\n til.append(i[1])\n for i in rai:\n ratl.append(i[1])\n for i in range(len(authl)):\n if author in authl[i]:\n res_til.append(til[i])\n res_ratl.append(ratl[i])\n title=random.choices(res_til)[0]\n i=res_til.index(title)\n rating=res_ratl[i]\n print('Title: '+title+'\\nRating: '+rating)\n return \"Enjoy your book!\"\n\nstatus=input('Welcome user! Do you wish to begin? (Y/N)\\n')\nwhile status=='Y'or status=='y':\n inp=input(\"Are you:\\n1.Looking for book suggestions\\n2.Looking for more books by an author\\n\")\n if inp=='1':\n subin=input('Do you want:\\n1.Random recommendations\\n2.Recommendations of a certain rating\\n')\n if subin=='1':\n print(recommendation_random())\n if subin=='2':\n print(recommendation_rating())\n elif inp=='2':\n print(author_search())\n\n status=input('Hello! Do you wish to continue? (Y/N)\\n')\nelse:\n print(\"Thank you for using our recommendation system!\")",
"Welcome user! Do you wish to begin? (Y/N)\ny\nAre you:\n1.Looking for book suggestions\n2.Looking for more books by an author\n2\nEnter author name for a book suggestion Eoin Colfer\nTitle: Artemis Fowl \nRating: 3.84\nEnjoy your book!\nHello! Do you wish to continue? (Y/N)\ny\nAre you:\n1.Looking for book suggestions\n2.Looking for more books by an author\n2\nEnter author name for a book suggestion J.K. Rowling\nTitle: Harry Potter and the Sorcerer's Stone \nRating: 4.47\nEnjoy your book!\nHello! Do you wish to continue? (Y/N)\ny\nAre you:\n1.Looking for book suggestions\n2.Looking for more books by an author\n2\nEnter author name for a book suggestion J.K. Rowling\nTitle: Harry Potter and the Prisoner of Azkaban \nRating: 4.56\nEnjoy your book!\nHello! Do you wish to continue? (Y/N)\nn\nThank you for using our recommendation system!\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
ecb6d4e75978e3a097eb12b74156fae3c5c81358 | 63,453 | ipynb | Jupyter Notebook | ucsb_rl/vanilla.ipynb | mmolnar0/sgillen_research | 752e09fdf7a996c832e71b0a8296322fe77e9ae3 | [
"MIT"
] | null | null | null | ucsb_rl/vanilla.ipynb | mmolnar0/sgillen_research | 752e09fdf7a996c832e71b0a8296322fe77e9ae3 | [
"MIT"
] | null | null | null | ucsb_rl/vanilla.ipynb | mmolnar0/sgillen_research | 752e09fdf7a996c832e71b0a8296322fe77e9ae3 | [
"MIT"
] | null | null | null | 162.7 | 22,000 | 0.847415 | [
[
[
"import gym\nimport numpy as np\nimport torch\nimport torch.nn as nn\nimport torch.optim as optim\nfrom torch.distributions import Categorical\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"env_name = 'CartPole-v0'\nenv = gym.make(env_name)\n\n\n# Hard coded policy for the cartpole problem\n# Will eventually want to build up infrastructure to develop a policy depending on:\n# env.action_space\n# env.observation_space\n\npolicy = nn.Sequential(\n nn.Linear(4, 12),\n nn.ReLU(),\n nn.Linear(12,12),\n nn.ReLU(),\n nn.Linear(12,2),\n nn.Softmax(dim=-1)\n )\n\noptimizer = optim.Adam(policy.parameters(), lr = .00001)\n\n# I guess we'll start with a categorical policy\n# TODO investigate the cost of action.detach.numpy() and torch.Tensor(state)\ndef select_action(policy, state):\n m = Categorical(policy(torch.Tensor(state)))\n action = m.sample()\n logprob = m.log_prob(action)\n \n return action.detach().numpy(), logprob\n ",
"_____no_output_____"
],
[
"policy(torch.randn(1,4))",
"_____no_output_____"
],
[
"#def vanilla_policy_grad(env, policy, optimizer):\n \naction_list = []\nstate_list = []\nlogprob_list = []\nreward_list = []\n\navg_reward_hist = []\n\nnum_epochs = 1000\nbatch_size = 80 # how many steps we want to use before we update our gradients\nnum_steps = 200 # number of steps in an episode (unless we terminate early)\n\nloss = torch.zeros(1,requires_grad=True)\n\nfor epoch in range(num_epochs):\n\n # Probably just want to preallocate these with zeros, as either a tensor or an array\n loss_hist = []\n episode_length_hist = []\n action_list = []\n total_steps = 0\n\n while True:\n\n state = env.reset()\n logprob_list = []\n reward_list = []\n action_list = []\n \n for t in range(num_steps):\n\n action, logprob = select_action(policy, state)\n state, reward, done, _ = env.step(action.item())\n\n logprob_list.append(-logprob)\n reward_list.append(reward)\n action_list.append(action)\n total_steps += 1\n\n if done:\n break\n\n # Now Calculate cumulative rewards for each action\n episode_length_hist.append(t)\n #loss = torch.stack([torch.sum(torch.tensor(reward_list[i:])*torch.stack(logprob_list[i:])) for i in range(len(reward_list))])\n \n #action_rewards = torch.tensor([sum(reward_list[i:]) for i in range(len(reward_list))])\n action_rewards = torch.sum(torch.tensor(reward_list))\n logprob_t = torch.stack(logprob_list)\n \n loss = torch.sum(logprob_t*action_rewards)\n loss.backward()\n \n if total_steps > batch_size:\n # update our gradients\n #print(\"here\")\n avg_reward_hist.append(sum(episode_length_hist)/len(episode_length_hist))\n optimizer.step()\n optimizer.zero_grad()\n break\n\n\n #other_list.append(1)\n #loss = torch.sum(torch.stack(loss_hist))\n #for action in episode_loss:\n #pisode_loss.backward()\n #optimizer.step()\n \nplt.plot(avg_reward_hist)",
"_____no_output_____"
],
[
"while True:\n state = env.reset()\n cum_rewards = 0\n\n\n for t in range(num_steps):\n action, _ = select_action(policy,state)\n state, reward, done, _ = env.step(action.item())\n env.render()\n \n cum_rewards += reward\n if done:\n \n print('summed reward for espide: ', cum_rewards)\n print('time terminated:' , t)\n break",
"summed reward for espide: 89.0\ntime terminated: 88\nsummed reward for espide: 81.0\ntime terminated: 80\nsummed reward for espide: 87.0\ntime terminated: 86\nsummed reward for espide: 23.0\ntime terminated: 22\nsummed reward for espide: 24.0\ntime terminated: 23\nsummed reward for espide: 23.0\ntime terminated: 22\nsummed reward for espide: 61.0\ntime terminated: 60\nsummed reward for espide: 32.0\ntime terminated: 31\nsummed reward for espide: 73.0\ntime terminated: 72\nsummed reward for espide: 45.0\ntime terminated: 44\nsummed reward for espide: 43.0\ntime terminated: 42\nsummed reward for espide: 75.0\ntime terminated: 74\nsummed reward for espide: 103.0\ntime terminated: 102\nsummed reward for espide: 39.0\ntime terminated: 38\nsummed reward for espide: 63.0\ntime terminated: 62\nsummed reward for espide: 29.0\ntime terminated: 28\nsummed reward for espide: 33.0\ntime terminated: 32\nsummed reward for espide: 33.0\ntime terminated: 32\nsummed reward for espide: 43.0\ntime terminated: 42\nsummed reward for espide: 103.0\ntime terminated: 102\nsummed reward for espide: 59.0\ntime terminated: 58\nsummed reward for espide: 23.0\ntime terminated: 22\nsummed reward for espide: 50.0\ntime terminated: 49\nsummed reward for espide: 35.0\ntime terminated: 34\nsummed reward for espide: 36.0\ntime terminated: 35\nsummed reward for espide: 43.0\ntime terminated: 42\nsummed reward for espide: 63.0\ntime terminated: 62\nsummed reward for espide: 107.0\ntime terminated: 106\nsummed reward for espide: 53.0\ntime terminated: 52\nsummed reward for espide: 46.0\ntime terminated: 45\nsummed reward for espide: 63.0\ntime terminated: 62\nsummed reward for espide: 27.0\ntime terminated: 26\nsummed reward for espide: 65.0\ntime terminated: 64\nsummed reward for espide: 21.0\ntime terminated: 20\nsummed reward for espide: 23.0\ntime terminated: 22\nsummed reward for espide: 102.0\ntime terminated: 101\nsummed reward for espide: 31.0\ntime terminated: 30\nsummed reward for espide: 112.0\ntime terminated: 111\nsummed reward for espide: 32.0\ntime terminated: 31\nsummed reward for espide: 26.0\ntime terminated: 25\nsummed reward for espide: 77.0\ntime terminated: 76\nsummed reward for espide: 26.0\ntime terminated: 25\nsummed reward for espide: 45.0\ntime terminated: 44\nsummed reward for espide: 27.0\ntime terminated: 26\nsummed reward for espide: 41.0\ntime terminated: 40\nsummed reward for espide: 59.0\ntime terminated: 58\nsummed reward for espide: 16.0\ntime terminated: 15\nsummed reward for espide: 106.0\ntime terminated: 105\nsummed reward for espide: 163.0\ntime terminated: 162\nsummed reward for espide: 26.0\ntime terminated: 25\nsummed reward for espide: 65.0\ntime terminated: 64\nsummed reward for espide: 17.0\ntime terminated: 16\nsummed reward for espide: 23.0\ntime terminated: 22\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecb6de6cfb70821c4b2eba03f60c5690f5afc912 | 69,069 | ipynb | Jupyter Notebook | chapter_1/.ipynb_checkpoints/Subset Observations-checkpoint.ipynb | bluehyena/Data-Analysis-and-Visualization | 3f3264be8d7af763cec6fc9d83c955b4a1e6799a | [
"MIT"
] | null | null | null | chapter_1/.ipynb_checkpoints/Subset Observations-checkpoint.ipynb | bluehyena/Data-Analysis-and-Visualization | 3f3264be8d7af763cec6fc9d83c955b4a1e6799a | [
"MIT"
] | null | null | null | chapter_1/.ipynb_checkpoints/Subset Observations-checkpoint.ipynb | bluehyena/Data-Analysis-and-Visualization | 3f3264be8d7af763cec6fc9d83c955b4a1e6799a | [
"MIT"
] | null | null | null | 25.346422 | 849 | 0.320144 | [
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"df = pd.DataFrame(\n {\"a\" : [4 ,5, 6, 6, np.nan],\n \"b\" : [7, 8, np.nan, 9, 9],\n \"c\" : [10, 11, 12, np.nan, 12]},\n index = pd.MultiIndex.from_tuples(\n [('d',1),('d',2),('e',2), ('e',3), ('e',4)],\n names=['n','v']))",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df[df.a > 4] # 4보다 큰 a col",
"_____no_output_____"
],
[
"df.b > 7",
"_____no_output_____"
],
[
"df[df.b > 7]",
"_____no_output_____"
],
[
"df[df.c > 7] # 마찬가지로 대소문자 구분",
"_____no_output_____"
],
[
"df.drop_duplicates() # 중복제거",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.drop_duplicates(inplace=True) # 사용을 권장하지는 않음\n\ndf = df.drop_duplicates() # 와 같음. 이것을 사용 권장.",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.drop_duplicates?",
"_____no_output_____"
],
[
"df = df.drop_duplicates(keep = 'last') # 중복행일경우 마지막을 남김",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df[\"a\"] != 7",
"_____no_output_____"
],
[
"df[df[\"b\"] != 7] # 7이 아닌값만 인덱싱",
"_____no_output_____"
],
[
"df.a.isin([5, 6]) #리스트형태로 담아주어야함",
"_____no_output_____"
],
[
"df[df['a'].isin([5])]",
"_____no_output_____"
],
[
"pd.isnull(df)",
"_____no_output_____"
],
[
"df['a'].isnull()",
"_____no_output_____"
],
[
"df['a'].isnull().sum() # null 값 갯수확인",
"_____no_output_____"
],
[
"pd.notnull(df)",
"_____no_output_____"
],
[
"df.notnull()",
"_____no_output_____"
],
[
"df.notnull().sum() # not null 의 갯수",
"_____no_output_____"
],
[
"df.a.notnull()",
"_____no_output_____"
]
],
[
[
"* & , |, ~, ^, any(), df.all() L\n* and, or, not, xor, any, all",
"_____no_output_____"
]
],
[
[
"df.any()",
"_____no_output_____"
],
[
"df.all()",
"_____no_output_____"
],
[
"~df.a.notnull() ",
"_____no_output_____"
],
[
"df[df.b == 7]",
"_____no_output_____"
],
[
"df[df.b == 7] and df[df.a == 5]",
"_____no_output_____"
],
[
"df[(df.b == 7) | (df.a == 5)] # 연산자로 사용해야함",
"_____no_output_____"
],
[
"df.head(2) # 앞에서 2개",
"_____no_output_____"
],
[
"df.tail(2) # 뒤에서 2개",
"_____no_output_____"
],
[
"df.sample(frac=0.7) # random하게 frac의 비율로 가져옴",
"_____no_output_____"
],
[
"df.sample(n=3) # 숫자 지정 가능, random",
"_____no_output_____"
],
[
"df.iloc[10:20] #오류는 나지 않으나 범위가 없음",
"_____no_output_____"
],
[
"df.iloc[1:3]\ndf.iloc[:3]",
"_____no_output_____"
],
[
"df.iloc[-2:]",
"_____no_output_____"
],
[
"df = pd.DataFrame({'population': [59000000, 65000000, 434000,\n 434000, 434000, 337000, 11300,\n 11300, 11300],\n 'GDP': [1937894, 2583560 , 12011, 4520, 12128,\n 17036, 182, 38, 311],\n 'alpha-2': [\"IT\", \"FR\", \"MT\", \"MV\", \"BN\",\n \"IS\", \"NR\", \"TV\", \"AI\"]},\n index=[\"Italy\", \"France\", \"Malta\",\n \"Maldives\", \"Brunei\", \"Iceland\",\n \"Nauru\", \"Tuvalu\", \"Anguilla\"])",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.nlargest(3, 'population')",
"_____no_output_____"
],
[
"df.nlargest(3, 'population', keep='last')",
"_____no_output_____"
],
[
"df.nlargest(3, 'population', keep='all')",
"_____no_output_____"
],
[
"df.nlargest(3, ['population', 'GDP'])",
"_____no_output_____"
],
[
"df.nsmallest(4, 'population', keep = 'all')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb6eaf4c30f2f77705eb123075f1a28489cdd1e | 726,250 | ipynb | Jupyter Notebook | boston_housing/boston_housing.ipynb | sriharshams/mlnd | 02fa7b1e34fd0b2d3fe87f7ea8483bb171048ff9 | [
"Apache-2.0"
] | null | null | null | boston_housing/boston_housing.ipynb | sriharshams/mlnd | 02fa7b1e34fd0b2d3fe87f7ea8483bb171048ff9 | [
"Apache-2.0"
] | null | null | null | boston_housing/boston_housing.ipynb | sriharshams/mlnd | 02fa7b1e34fd0b2d3fe87f7ea8483bb171048ff9 | [
"Apache-2.0"
] | null | null | null | 738.058943 | 126,100 | 0.935642 | [
[
[
"# Machine Learning Engineer Nanodegree\n## Model Evaluation & Validation\n## Project: Predicting Boston Housing Prices\n\nWelcome to the first project of the Machine Learning Engineer Nanodegree! In this notebook, some template code has already been provided for you, and you will need to implement additional functionality to successfully complete this project. You will not need to modify the included code beyond what is requested. Sections that begin with **'Implementation'** in the header indicate that the following block of code will require additional functionality which you must provide. Instructions will be provided for each section and the specifics of the implementation are marked in the code block with a 'TODO' statement. Please be sure to read the instructions carefully!\n\nIn addition to implementing code, there will be questions that you must answer which relate to the project and your implementation. Each section where you will answer a question is preceded by a **'Question X'** header. Carefully read each question and provide thorough answers in the following text boxes that begin with **'Answer:'**. Your project submission will be evaluated based on your answers to each of the questions and the implementation you provide. \n\n>**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. In addition, Markdown cells can be edited by typically double-clicking the cell to enter edit mode.",
"_____no_output_____"
],
[
"## Getting Started\nIn this project, you will evaluate the performance and predictive power of a model that has been trained and tested on data collected from homes in suburbs of Boston, Massachusetts. A model trained on this data that is seen as a *good fit* could then be used to make certain predictions about a home — in particular, its monetary value. This model would prove to be invaluable for someone like a real estate agent who could make use of such information on a daily basis.\n\nThe dataset for this project originates from the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Housing). The Boston housing data was collected in 1978 and each of the 506 entries represent aggregated data about 14 features for homes from various suburbs in Boston, Massachusetts. For the purposes of this project, the following preprocessing steps have been made to the dataset:\n- 16 data points have an `'MEDV'` value of 50.0. These data points likely contain **missing or censored values** and have been removed.\n- 1 data point has an `'RM'` value of 8.78. This data point can be considered an **outlier** and has been removed.\n- The features `'RM'`, `'LSTAT'`, `'PTRATIO'`, and `'MEDV'` are essential. The remaining **non-relevant features** have been excluded.\n- The feature `'MEDV'` has been **multiplicatively scaled** to account for 35 years of market inflation.\n\nRun the code cell below to load the Boston housing dataset, along with a few of the necessary Python libraries required for this project. You will know the dataset loaded successfully if the size of the dataset is reported.",
"_____no_output_____"
]
],
[
[
"# Import libraries necessary for this project\nimport numpy as np\nimport pandas as pd\nfrom sklearn.cross_validation import ShuffleSplit\n\n# Import supplementary visualizations code visuals.py\nimport visuals as vs\n\n# Pretty display for notebooks\n%matplotlib inline\n\n# Load the Boston housing dataset\ndata = pd.read_csv('housing.csv')\nprices = data['MEDV']\nfeatures = data.drop('MEDV', axis = 1)\n \n# Success\nprint \"Boston housing dataset has {} data points with {} variables each.\".format(*data.shape)",
"Boston housing dataset has 489 data points with 4 variables each.\n"
]
],
[
[
"## Data Exploration\nIn this first section of this project, you will make a cursory investigation about the Boston housing data and provide your observations. Familiarizing yourself with the data through an explorative process is a fundamental practice to help you better understand and justify your results.\n\nSince the main goal of this project is to construct a working model which has the capability of predicting the value of houses, we will need to separate the dataset into **features** and the **target variable**. The **features**, `'RM'`, `'LSTAT'`, and `'PTRATIO'`, give us quantitative information about each data point. The **target variable**, `'MEDV'`, will be the variable we seek to predict. These are stored in `features` and `prices`, respectively.",
"_____no_output_____"
],
[
"### Implementation: Calculate Statistics\nFor your very first coding implementation, you will calculate descriptive statistics about the Boston housing prices. Since `numpy` has already been imported for you, use this library to perform the necessary calculations. These statistics will be extremely important later on to analyze various prediction results from the constructed model.\n\nIn the code cell below, you will need to implement the following:\n- Calculate the minimum, maximum, mean, median, and standard deviation of `'MEDV'`, which is stored in `prices`.\n - Store each calculation in their respective variable.",
"_____no_output_____"
]
],
[
[
"# TODO: Minimum price of the data\nminimum_price = np.min(prices)\n\n# TODO: Maximum price of the data\nmaximum_price = np.max(prices)\n\n# TODO: Mean price of the data\nmean_price = np.mean(prices)\n\n# TODO: Median price of the data\nmedian_price = np.median(prices)\n\n# TODO: Standard deviation of prices of the data\nstd_price = np.std(prices)\n\n# Show the calculated statistics\nprint \"Statistics for Boston housing dataset:\\n\"\nprint \"Minimum price: ${:,.2f}\".format(minimum_price)\nprint \"Maximum price: ${:,.2f}\".format(maximum_price)\nprint \"Mean price: ${:,.2f}\".format(mean_price)\nprint \"Median price ${:,.2f}\".format(median_price)\nprint \"Standard deviation of prices: ${:,.2f}\".format(std_price)",
"Statistics for Boston housing dataset:\n\nMinimum price: $105,000.00\nMaximum price: $1,024,800.00\nMean price: $454,342.94\nMedian price $438,900.00\nStandard deviation of prices: $165,171.13\n"
]
],
[
[
"### Question 1 - Feature Observation\nAs a reminder, we are using three features from the Boston housing dataset: `'RM'`, `'LSTAT'`, and `'PTRATIO'`. For each data point (neighborhood):\n- `'RM'` is the average number of rooms among homes in the neighborhood.\n- `'LSTAT'` is the percentage of homeowners in the neighborhood considered \"lower class\" (working poor).\n- `'PTRATIO'` is the ratio of students to teachers in primary and secondary schools in the neighborhood.\n\n_Using your intuition, for each of the three features above, do you think that an increase in the value of that feature would lead to an **increase** in the value of `'MEDV'` or a **decrease** in the value of `'MEDV'`? Justify your answer for each._ \n**Hint:** Would you expect a home that has an `'RM'` value of 6 be worth more or less than a home that has an `'RM'` value of 7?",
"_____no_output_____"
],
[
"**Answer: **\nBased on my intution, each of the below features explained thier effect on the value of `'MEDV'` (label or target). This is further supported with Linear regression plots.\n- `'RM'` : Typically **increase** in `'RM'` would lead to an **increase** in `'MEDV'`. The `'RM'` is also a good indication of the size of the homes, bigger house, bigger value.\n- `'LSTAT'` : Typically **decrease** in `'LSTAT'` would lead to an **increse** in `'MEDV'`. As `'LSTAT'` is the indication of working class, \"lower\" class reduces the value of home.\n- `'PTRATIO'` : Typically **decrease** in `'PTRATIO'` (students) would lead to an **increase** in `'MEDV'`. As `'PTRATIO'` is an indication that schools are good and suffeciently staffed and funded. ",
"_____no_output_____"
],
[
"**RM**",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline\nfrom sklearn.linear_model import LinearRegression\nreg = LinearRegression()\npt_ratio = data[\"RM\"].reshape(-1,1)\nreg.fit(pt_ratio, prices)\n\n# Create the figure window\nplt.plot(pt_ratio, reg.predict(pt_ratio), color='red', lw=1)\nplt.scatter(pt_ratio, prices, alpha=0.5, c=prices)\nplt.xlabel('RM')\nplt.ylabel('Prices')\nplt.show()",
"_____no_output_____"
]
],
[
[
"**LSTAT**",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline\nfrom sklearn.linear_model import LinearRegression\nreg = LinearRegression()\npt_ratio = data[\"LSTAT\"].reshape(-1,1)\nreg.fit(pt_ratio, prices)\n\n# Create the figure window\nplt.plot(pt_ratio, reg.predict(pt_ratio), color='red', lw=1)\nplt.scatter(pt_ratio, prices, alpha=0.5, c=prices)\nplt.xlabel('LSTAT')\nplt.ylabel('Prices')\nplt.show()",
"_____no_output_____"
]
],
[
[
"**PTRATIO**",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline\nfrom sklearn.linear_model import LinearRegression\nreg = LinearRegression()\npt_ratio = data[\"PTRATIO\"].reshape(-1,1)\nreg.fit(pt_ratio, prices)\n\n# Create the figure window\nplt.plot(pt_ratio, reg.predict(pt_ratio), color='red', lw=1)\nplt.scatter(pt_ratio, prices, alpha=0.5, c=prices)\nplt.xlabel('PTRATIO')\nplt.ylabel('Prices')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"----\n\n## Developing a Model\nIn this second section of the project, you will develop the tools and techniques necessary for a model to make a prediction. Being able to make accurate evaluations of each model's performance through the use of these tools and techniques helps to greatly reinforce the confidence in your predictions.",
"_____no_output_____"
],
[
"### Implementation: Define a Performance Metric\nIt is difficult to measure the quality of a given model without quantifying its performance over training and testing. This is typically done using some type of performance metric, whether it is through calculating some type of error, the goodness of fit, or some other useful measurement. For this project, you will be calculating the [*coefficient of determination*](http://stattrek.com/statistics/dictionary.aspx?definition=coefficient_of_determination), R<sup>2</sup>, to quantify your model's performance. The coefficient of determination for a model is a useful statistic in regression analysis, as it often describes how \"good\" that model is at making predictions. \n\nThe values for R<sup>2</sup> range from 0 to 1, which captures the percentage of squared correlation between the predicted and actual values of the **target variable**. A model with an R<sup>2</sup> of 0 is no better than a model that always predicts the *mean* of the target variable, whereas a model with an R<sup>2</sup> of 1 perfectly predicts the target variable. Any value between 0 and 1 indicates what percentage of the target variable, using this model, can be explained by the **features**. _A model can be given a negative R<sup>2</sup> as well, which indicates that the model is **arbitrarily worse** than one that always predicts the mean of the target variable._\n\nFor the `performance_metric` function in the code cell below, you will need to implement the following:\n- Use `r2_score` from `sklearn.metrics` to perform a performance calculation between `y_true` and `y_predict`.\n- Assign the performance score to the `score` variable.",
"_____no_output_____"
]
],
[
[
"# TODO: Import 'r2_score'\nfrom sklearn.metrics import r2_score\n\ndef performance_metric(y_true, y_predict):\n \"\"\" Calculates and returns the performance score between \n true and predicted values based on the metric chosen. \"\"\"\n \n # TODO: Calculate the performance score between 'y_true' and 'y_predict'\n score = r2_score(y_true, y_predict)\n \n # Return the score\n return score",
"_____no_output_____"
]
],
[
[
"### Question 2 - Goodness of Fit\nAssume that a dataset contains five data points and a model made the following predictions for the target variable:\n\n| True Value | Prediction |\n| :-------------: | :--------: |\n| 3.0 | 2.5 |\n| -0.5 | 0.0 |\n| 2.0 | 2.1 |\n| 7.0 | 7.8 |\n| 4.2 | 5.3 |\n*Would you consider this model to have successfully captured the variation of the target variable? Why or why not?* \n\nRun the code cell below to use the `performance_metric` function and calculate this model's coefficient of determination.",
"_____no_output_____"
]
],
[
[
"# Calculate the performance of this model\nscore = performance_metric([3, -0.5, 2, 7, 4.2], [2.5, 0.0, 2.1, 7.8, 5.3])\nprint \"Model has a coefficient of determination, R^2, of {:.3f}.\".format(score)",
"Model has a coefficient of determination, R^2, of 0.923.\n"
]
],
[
[
"**Answer:**\n- Yes I would consider this model to have successfully captured the variation of the target variable. \n- R2 is 0.923 which is very close to 1, means the **True Value** is 92.3% predicted from **Prediction**\n- As shown below it is possible to plot the values to get the visual representation in this scenario",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\ntrue, pred = [3.0, -0.5, 2.0, 7.0, 4.2],[2.5, 0.0, 2.1, 7.8, 5.3]\n#plot true values\ntrue_handle = plt.scatter(true, true, alpha=0.6, color='blue', label = 'True' )\n\n#reference line\nfit = np.poly1d(np.polyfit(true, true, 1))\nlims = np.linspace(min(true)-1, max(true)+1)\nplt.plot(lims, fit(lims), alpha = 0.3, color = \"black\")\n\n#plot predicted values\npred_handle = plt.scatter(true, pred, alpha=0.6, color='red', label = 'Pred')\n\n#legend & show\nplt.legend(handles=[true_handle, pred_handle], loc=\"upper left\")\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"### Implementation: Shuffle and Split Data\nYour next implementation requires that you take the Boston housing dataset and split the data into training and testing subsets. Typically, the data is also shuffled into a random order when creating the training and testing subsets to remove any bias in the ordering of the dataset.\n\nFor the code cell below, you will need to implement the following:\n- Use `train_test_split` from `sklearn.cross_validation` to shuffle and split the `features` and `prices` data into training and testing sets.\n - Split the data into 80% training and 20% testing.\n - Set the `random_state` for `train_test_split` to a value of your choice. This ensures results are consistent.\n- Assign the train and testing splits to `X_train`, `X_test`, `y_train`, and `y_test`.",
"_____no_output_____"
]
],
[
[
"# TODO: Import 'train_test_split'\nfrom sklearn.cross_validation import train_test_split\n# TODO: Shuffle and split the data into training and testing subsets\nX_train, X_test, y_train, y_test = train_test_split(features, prices, test_size=0.2, random_state=0)\n\n# Success\nprint \"Training and testing split was successful.\"",
"Training and testing split was successful.\n"
]
],
[
[
"### Question 3 - Training and Testing\n*What is the benefit to splitting a dataset into some ratio of training and testing subsets for a learning algorithm?* \n**Hint:** What could go wrong with not having a way to test your model?",
"_____no_output_____"
],
[
"**Answer: **\n- Learning algorithm used for prediction or inference of datasets. We do not need learning algorithm to predict known response lables, we do want learning algorithm to predict response label from unkown dataset. That is why it is benefitial to hold out some ratio of dataset as test dataset not known to learning algorithm. Learning algorithm is fitted against traning subset, which then can be used to predict response label from test dataset to measure performance of learning algorithm.\n- Splitting dataset into some ratio of training and testing subsets, we can provide only training subset data to learning algorithm and learn behavior of response label against features. We can then provide testing subset not known to learning algorithm and have learning algorithm predict label. Predicted label can be compared with actuals of testing subset to find test error. Test error is a better metric to measure the performance of a learning algorithm compared to training error.\n- Using training and testing subsets we can tune the learning algorithm to reduce bias and variance.\n - If we do not have a way to test with testing subsets, approximation of traning error is used as a performance metric for learning algorithm, in some cases learning algorithm could have high variance and might not be the right algorithm for the dataset.\n",
"_____no_output_____"
],
[
"----\n\n## Analyzing Model Performance\nIn this third section of the project, you'll take a look at several models' learning and testing performances on various subsets of training data. Additionally, you'll investigate one particular algorithm with an increasing `'max_depth'` parameter on the full training set to observe how model complexity affects performance. Graphing your model's performance based on varying criteria can be beneficial in the analysis process, such as visualizing behavior that may not have been apparent from the results alone.",
"_____no_output_____"
],
[
"### Learning Curves\nThe following code cell produces four graphs for a decision tree model with different maximum depths. Each graph visualizes the learning curves of the model for both training and testing as the size of the training set is increased. Note that the shaded region of a learning curve denotes the uncertainty of that curve (measured as the standard deviation). The model is scored on both the training and testing sets using R<sup>2</sup>, the coefficient of determination. \n\nRun the code cell below and use these graphs to answer the following question.",
"_____no_output_____"
]
],
[
[
"# Produce learning curves for varying training set sizes and maximum depths\nvs.ModelLearning(features, prices)",
"_____no_output_____"
]
],
[
[
"### Question 4 - Learning the Data\n*Choose one of the graphs above and state the maximum depth for the model. What happens to the score of the training curve as more training points are added? What about the testing curve? Would having more training points benefit the model?* \n**Hint:** Are the learning curves converging to particular scores?",
"_____no_output_____"
],
[
"**Answer: **\n- Maximum depth for the model is max_depth = 1\n- Score of the training curve is decreased for more training points added\n- Score of the testing cureve is increased for more training points added\n- Both training and testing curve are platoed or have very minimal gain in score for more traning points added after around 300 training points, so more traning points wont benefit the model. Learning curves of both training and testing curves seem to converge around score 0.4\n - traning curve seems to be detoriating and indicates high bias",
"_____no_output_____"
],
[
"### Complexity Curves\nThe following code cell produces a graph for a decision tree model that has been trained and validated on the training data using different maximum depths. The graph produces two complexity curves — one for training and one for validation. Similar to the **learning curves**, the shaded regions of both the complexity curves denote the uncertainty in those curves, and the model is scored on both the training and validation sets using the `performance_metric` function. \n\nRun the code cell below and use this graph to answer the following two questions.",
"_____no_output_____"
]
],
[
[
"vs.ModelComplexity(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"### Question 5 - Bias-Variance Tradeoff\n*When the model is trained with a maximum depth of 1, does the model suffer from high bias or from high variance? How about when the model is trained with a maximum depth of 10? What visual cues in the graph justify your conclusions?* \n**Hint:** How do you know when a model is suffering from high bias or high variance?",
"_____no_output_____"
],
[
"**Answer: **\n- Yes when models is trained with a maximum depth of 1, model suffer from high bias and low variance\n- When model is trained with a maxmum depth of 10, model suffer from high variance and low bais\n- If training and validation score are close to each other it shows that there is low variance in the model. In the graph as both training and testing score are less, it could be that model is not using sufficient data so it could be biased or underfitting the data. When there is a huge difference in training score and validation score there is a high variance, this could be because, model has learnt very well, and fitted to training data, indicates that model is overfitting or has high variance. At about Maximum depth 4, model seem to be performing optimal for both traning and validations core having the right trade of bias and variance.",
"_____no_output_____"
],
[
"### Question 6 - Best-Guess Optimal Model\n*Which maximum depth do you think results in a model that best generalizes to unseen data? What intuition lead you to this answer?*",
"_____no_output_____"
],
[
"**Answer: **\n- At about Maximum depth 4, model seem to be performing optimal for both traning and validations core having the right trade of bias and variance. Afyer Maximum depth 4, validation score starts detoriting and doesnt show any improvement where as traning score is increasing is a sign of overfitting or high variance being introduced by model. ",
"_____no_output_____"
],
[
"-----\n\n## Evaluating Model Performance\nIn this final section of the project, you will construct a model and make a prediction on the client's feature set using an optimized model from `fit_model`.",
"_____no_output_____"
],
[
"### Question 7 - Grid Search\n*What is the grid search technique and how it can be applied to optimize a learning algorithm?*",
"_____no_output_____"
],
[
"**Answer: **\nGrid search technique is a way of performing hyperparameter optimization. This is simply an exhaustive seraching through a manually specified subset of hyperparameter of a learning algorithm.\n\nGrid serach will methodically build and evaluate a model for each combination of learning algorithm parameters specified in a grid. A Grid search algorithm is guided by performance metric like typically measured by cross-validation on the training set or evaluation on a held-out validation set, and best combination is retained. ",
"_____no_output_____"
],
[
"### Question 8 - Cross-Validation\n*What is the k-fold cross-validation training technique? What benefit does this technique provide for grid search when optimizing a model?* \n**Hint:** Much like the reasoning behind having a testing set, what could go wrong with using grid search without a cross-validated set?",
"_____no_output_____"
],
[
"**Answer: **\n- In k-fold cross-validation training technique, the original dataset is randomly partitioned into k equal sized subsets. Of the k subsets, a single subset is retained as the validation data (or test data) for testing the model, and the remaining k − 1 subsets are used as training data. The cross-validation process is then repeated k times (the folds), with each of the k subsets used exactly once as the validation data. The k results from the folds can then be averaged to produce a single estimation. 10-fold cross-validation is commonly used, but in general k remains an unfixed parameter.\n- A grid search algorithm must be guided by a performance metric, typically measured by cross-validation. The advantage is that all observations are used for both training and validation, and each observation is used for validation exactly once. By doing this there will be low variance and no overfitting of the data by the optimized model.",
"_____no_output_____"
],
[
"### Implementation: Fitting a Model\nYour final implementation requires that you bring everything together and train a model using the **decision tree algorithm**. To ensure that you are producing an optimized model, you will train the model using the grid search technique to optimize the `'max_depth'` parameter for the decision tree. The `'max_depth'` parameter can be thought of as how many questions the decision tree algorithm is allowed to ask about the data before making a prediction. Decision trees are part of a class of algorithms called *supervised learning algorithms*.\n\nIn addition, you will find your implementation is using `ShuffleSplit()` for an alternative form of cross-validation (see the `'cv_sets'` variable). While it is not the K-Fold cross-validation technique you describe in **Question 8**, this type of cross-validation technique is just as useful!. The `ShuffleSplit()` implementation below will create 10 (`'n_splits'`) shuffled sets, and for each shuffle, 20% (`'test_size'`) of the data will be used as the *validation set*. While you're working on your implementation, think about the contrasts and similarities it has to the K-fold cross-validation technique.\n\nPlease note that ShuffleSplit has different parameters in scikit-learn versions 0.17 and 0.18.\nFor the `fit_model` function in the code cell below, you will need to implement the following:\n- Use [`DecisionTreeRegressor`](http://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeRegressor.html) from `sklearn.tree` to create a decision tree regressor object.\n - Assign this object to the `'regressor'` variable.\n- Create a dictionary for `'max_depth'` with the values from 1 to 10, and assign this to the `'params'` variable.\n- Use [`make_scorer`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.make_scorer.html) from `sklearn.metrics` to create a scoring function object.\n - Pass the `performance_metric` function as a parameter to the object.\n - Assign this scoring function to the `'scoring_fnc'` variable.\n- Use [`GridSearchCV`](http://scikit-learn.org/0.17/modules/generated/sklearn.grid_search.GridSearchCV.html) from `sklearn.grid_search` to create a grid search object.\n - Pass the variables `'regressor'`, `'params'`, `'scoring_fnc'`, and `'cv_sets'` as parameters to the object. \n - Assign the `GridSearchCV` object to the `'grid'` variable.",
"_____no_output_____"
]
],
[
[
"# TODO: Import 'make_scorer', 'DecisionTreeRegressor', and 'GridSearchCV'\nfrom sklearn.tree import DecisionTreeRegressor\nfrom sklearn.metrics import make_scorer\nfrom sklearn.grid_search import GridSearchCV\n\ndef fit_model(X, y):\n \"\"\" Performs grid search over the 'max_depth' parameter for a \n decision tree regressor trained on the input data [X, y]. \"\"\"\n \n # Create cross-validation sets from the training data\n cv_sets = ShuffleSplit(X.shape[0], n_iter = 10, test_size = 0.20, random_state = 0)\n\n # TODO: Create a decision tree regressor object\n regressor = DecisionTreeRegressor()\n\n # TODO: Create a dictionary for the parameter 'max_depth' with a range from 1 to 10\n params = {'max_depth': range(1,11)}\n\n # TODO: Transform 'performance_metric' into a scoring function using 'make_scorer' \n scoring_fnc = make_scorer(performance_metric)\n\n # TODO: Create the grid search object\n grid = GridSearchCV(regressor, params, scoring = scoring_fnc, cv = cv_sets)\n\n # Fit the grid search object to the data to compute the optimal model\n grid = grid.fit(X, y)\n\n # Return the optimal model after fitting the data\n return grid.best_estimator_",
"_____no_output_____"
]
],
[
[
"### Making Predictions\nOnce a model has been trained on a given set of data, it can now be used to make predictions on new sets of input data. In the case of a *decision tree regressor*, the model has learned *what the best questions to ask about the input data are*, and can respond with a prediction for the **target variable**. You can use these predictions to gain information about data where the value of the target variable is unknown — such as data the model was not trained on.",
"_____no_output_____"
],
[
"### Question 9 - Optimal Model\n_What maximum depth does the optimal model have? How does this result compare to your guess in **Question 6**?_ \n\nRun the code block below to fit the decision tree regressor to the training data and produce an optimal model.",
"_____no_output_____"
]
],
[
[
"# Fit the training data to the model using grid search\nreg = fit_model(X_train, y_train)\n\n# Produce the value for 'max_depth'\nprint \"Parameter 'max_depth' is {} for the optimal model.\".format(reg.get_params()['max_depth'])",
"Parameter 'max_depth' is 4 for the optimal model.\n"
]
],
[
[
"**Answer: ** \n- 4, this is same as the result of my guess in *Question 6*",
"_____no_output_____"
],
[
"### Question 10 - Predicting Selling Prices\nImagine that you were a real estate agent in the Boston area looking to use this model to help price homes owned by your clients that they wish to sell. You have collected the following information from three of your clients:\n\n| Feature | Client 1 | Client 2 | Client 3 |\n| :---: | :---: | :---: | :---: |\n| Total number of rooms in home | 5 rooms | 4 rooms | 8 rooms |\n| Neighborhood poverty level (as %) | 17% | 32% | 3% |\n| Student-teacher ratio of nearby schools | 15-to-1 | 22-to-1 | 12-to-1 |\n*What price would you recommend each client sell his/her home at? Do these prices seem reasonable given the values for the respective features?* \n**Hint:** Use the statistics you calculated in the **Data Exploration** section to help justify your response. \n\nRun the code block below to have your optimized model make predictions for each client's home.",
"_____no_output_____"
]
],
[
[
"# Produce a matrix for client data\nclient_data = [[5, 17, 15], # Client 1\n [4, 32, 22], # Client 2\n [8, 3, 12]] # Client 3\n\n# Show predictions\nfor i, price in enumerate(reg.predict(client_data)):\n print \"Predicted selling price for Client {}'s home: ${:,.2f}\".format(i+1, price)",
"Predicted selling price for Client 1's home: $391,183.33\nPredicted selling price for Client 2's home: $189,123.53\nPredicted selling price for Client 3's home: $942,666.67\n"
]
],
[
[
"**Answer: **\nThe predicted selling prices are \\$391,183.33, \\$189,123.53 and \\$942,666.67 for Client 1's home, Client 2's home and Client 3's home respectively.\nFacts from the descriptive statistics:\n- Distribution: \n Statistics for Boston housing dataset:\n Minimum price: \\$105,000.00\n Maximum price: \\$1,024,800.00\n Mean price: \\$454,342.94\n Median price \\$438,900.00\n Standard deviation of prices: \\$165,171.13\n- Effects of features: \n Based on my intution, each of the below features explained thier effect on the value of 'MEDV' (label or target).\n - 'RM' : Typically increase in 'RM' would lead to an increase in 'MEDV'. The 'RM' is also a good indication of the size of the homes, bigger house, bigger value.\n - 'LSTAT' : Typically decrease in 'LSTAT' would lead to an increse in 'MEDV'. As 'LSTAT' is the indication of working class, \"lower\" class reduces the value of home.\n - 'PTRATIO' : Typically decrease in 'PTRATIO' (students) would lead to an increase in 'MEDV'. As 'PTRATIO' is an indication that schools are good and suffeciently staffed and funded.\n\nAre the estimates reasonable:\n- Client 1's home (\\$391,183.33):\n - Distribution: The estimate is inside the normal range of prices we have (closer than one standard deviation to mean and median).\n - Feature effects: The feature values all are in between those for the other clients. Thus, it seems reasonable that the estimated price is also in between.\n - Conclusion: reasonable estimate\n- Client 2's home (\\$189,123.53)\n - Distribution: The estimate is more than one standard deviation below the mean but less than two. Thus, it is not really a typical value for me still ok.\n - Feature effects: Of the 3 clients' houses, this one has lowest RM, highest LSTAT, and highest PTRATIO. All this should decrease the price, which is in line with it being the lowest of all prices.\n - Conclusion: it is reasonable that the price is low, but my confidence in the exact value of the estimate is lower than for client 1. Still, you would say you could use the model for client 2.\n- Client 3's home (\\$942,666.67):\n - Distribution: The estimate is more than 3 standard deviations above the mean (bigger than \\$906,930.78) and very close to the maximum of \\$1,024,800.00. Thus, this value is very atypical for this dataset and should be viewed with scepticism.\n - Feature effects: This is the house with highest RM, lowest LSTAT, and lowest PTRATIO of all 3 clients. Thus, it seems theoretically ok that is has the highest price too.\n - Conclusion: The price should indeed be high, but I would not trust an estimate that far off the mean. Hence, my confidence in this prediction is lowest. I would not recommend using the model for estimates in this range.\n\nSide note: arguing with summary statistics like mean and standard deviations relies on house prices being at least somewhat normally distributed. \n\nWe can alos see from below plots, the client features against datset.",
"_____no_output_____"
]
],
[
[
"from matplotlib import pyplot as plt\n\nclients = np.transpose(client_data)\npred = reg.predict(client_data)\nfor i, feat in enumerate(['RM', 'LSTAT', 'PTRATIO']):\n plt.scatter(features[feat], prices, alpha=0.25, c=prices)\n plt.scatter(clients[i], pred, color='black', marker='x', linewidths=2)\n plt.xlabel(feat)\n plt.ylabel('MEDV')\n plt.show()",
"_____no_output_____"
]
],
[
[
"### Sensitivity\nAn optimal model is not necessarily a robust model. Sometimes, a model is either too complex or too simple to sufficiently generalize to new data. Sometimes, a model could use a learning algorithm that is not appropriate for the structure of the data given. Other times, the data itself could be too noisy or contain too few samples to allow a model to adequately capture the target variable — i.e., the model is underfitted. Run the code cell below to run the `fit_model` function ten times with different training and testing sets to see how the prediction for a specific client changes with the data it's trained on.",
"_____no_output_____"
]
],
[
[
"vs.PredictTrials(features, prices, fit_model, client_data)",
"_____no_output_____"
]
],
[
[
"### Question 11 - Applicability\n*In a few sentences, discuss whether the constructed model should or should not be used in a real-world setting.* \n**Hint:** Some questions to answering:\n- *How relevant today is data that was collected from 1978?*\n- *Are the features present in the data sufficient to describe a home?*\n- *Is the model robust enough to make consistent predictions?*\n- *Would data collected in an urban city like Boston be applicable in a rural city?*",
"_____no_output_____"
],
[
"**Answer: **\nI am a data sceptic and don't recommend using the model in production.\n\nReasons:\n- Age of dataset: The age of the dataset may render some features of the model useless for estimations today. For instance, the imporatance of the pupil/teacher ratio may decrease in time. The theory is: the pupil/teacher ratio in the neighborhood drives demand for houses in the area (and thereby house prices) if families rely on local schools for their kid's education. Let's say this has been so in 1978. However, if people nowadays are more flexible when choosing schools, this effect could vanish. 3 minutes of googling delivered this researcher (http://sites.duke.edu/urbaneconomics/?p=863) who claims Charter schools are getting more popular in the US. The effect supposedly is that pupils are not limited to local schools anymore. If this is true, our model may overemphasize the effect of PTRATIO today.\n- Number of features: we have only used 3 of 14 available features. It may be good to explore others as well. For instance, there is another feature (called DIS) in the dataset which gives a weighted distance to one of 5 employment centers in Boston (https://archive.ics.uci.edu/ml/datasets/Housing). Using simple intution we can sys this feature to have a large impact on price. AS reserched, it was found higher prices to be close to the city center where most people work. Typically houses closer to employment centers are more in demand.\n- Robustness: Looking at the sensitivity analysis above, we can see that the 10 trials delivered estimates ranging from \\$351,577.61 to \\$420,622.22, two values more than ~\\$70k apart. With an average price of little more than \\$450k and a such a large variance in estimates, the model is hardly usable. For people buying or selling a house, it will make a huge difference whether the price is \\$300k or \\$400k.\n- Generelizability to other cities/areas: House prices in Boston are likely not the same as in rural areas. For San Francisco Bay Area, we can tell for sure there is a large difference between urban and rural house prices. We should expect the same to be true for the Boston. Thus, the price level will be completely different, suggesting that urban vs. rural should be a feature for itself. Features may also have different effects in rural areas. For instance, the number of rooms probably correlates strongly with the size of a house. If the cost per square meter in urban areas is larger than in rural areas, the positive effect of number of rooms should also be larger in urban areas. Moreover, some features in our model may not even make sense in rural areas. For instance, PTRATIO may not be defined in a very rural area if people go to a school in a different town (no school -> no pupil / teacher ratio).",
"_____no_output_____"
],
[
"> **Note**: Once you have completed all of the code implementations and successfully answered each question above, you may finalize your work by exporting the iPython Notebook as an HTML document. You can do this by using the menu above and navigating to \n**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ecb6ef0ce7c04ce9891da948a01e116d6e407a7e | 10,398 | ipynb | Jupyter Notebook | modelling-coast-salish-fish-traps/Initial Modelling.ipynb | lfunderburk/Math-Modelling | 15b5c7c12e34050069762b6fac58943ba7602c9a | [
"CC-BY-4.0"
] | null | null | null | modelling-coast-salish-fish-traps/Initial Modelling.ipynb | lfunderburk/Math-Modelling | 15b5c7c12e34050069762b6fac58943ba7602c9a | [
"CC-BY-4.0"
] | 3 | 2020-09-04T19:48:01.000Z | 2021-01-07T18:06:59.000Z | modelling-coast-salish-fish-traps/Initial Modelling.ipynb | lfunderburk/Math-Modelling | 15b5c7c12e34050069762b6fac58943ba7602c9a | [
"CC-BY-4.0"
] | 1 | 2020-10-30T16:19:11.000Z | 2020-10-30T16:19:11.000Z | 35.367347 | 538 | 0.55703 | [
[
[
"# Initial Model",
"_____no_output_____"
],
[
"## Variables\nvariable| definition\n-|-\n$l_n$| tide level at hour $n$\n$h$| height of the fish trap wall\n$r$| radius of the semicircular fish trap\n$N$| number of fish in the area\n$F_n$| number of fish not in trap at hour $n$\n$C_n$| number of fish in the trap at hour $n$\n$T_n$| total number of fish harvest at hour $n$\n$\\alpha$| constant of fish movement, the likelyhood a fish in the area would swim over the largest allowed semi-circle\n$p_n$| ratio of perimiter of semi-circle covered by water at hour $n$ to the largest allowed circle\n\n\n## Diagram\n\n\n## Assumptions\n\nAssume that the ammount of amount of fish in the area at any given time is some constant $N$ regardless of the number of fish recently harvested. This seems reasonable for short periods of time and in the absense of industrial harvesting methods.\n\nFish are harvested when the water level is below the lowest point of the trap.\n\nFish movement patterns in the area is random.\n\nThe highest point of the trap is above or just below high tide level.\n\n## Plan\nTo save either doing a crude approximation of a tide such as a simple cosine graph or a complex, but accurate tidal equation that is hidden to the end user (elementary aged children), use obtainable tide data from comox harbour, a site of fish traps. Additionally, since the code an inner workings of the model are hidden from the end user we can use the hourly tidal information to build a discrete model that would have all appearances of being continuous and allow for the effect of user controlled variables to be apparent. \n\n\n\nUsing a discrete method, at each hour step can multiply the number of fish in each catagory by the proportion of the semi circle that is covered by water and by some constant $\\alpha \\leq 1$. $\\alpha$ can be adjusted until it 'feels' right so the children can be challenged in making their traps.\n\n## Equations\n\nThe top ridge of the trap expressed as intersection of a half-cylinder and a plane: $x = r\\cdot\\cos(\\theta)$, $y = r\\cdot\\sin(\\theta)$, $z = 6 + h + (0.17 * y)$, for $h,r$: user input, $\\theta\\in(\\pi,2\\pi)$.\n\nThe $z$ values are from the slope of the shore: $z = 6 + (0.17 * y)$ with the h variable added by the user.\n\n$p_n$:\n\nTo find the length of the perimeter of the semi-circle covered by water we find, moving in the x direction, the first point of the perimeter covered by water then working in second quadrant of the circle of radius $r$ solve for the angle to that point. We take the ratio of that angle with $pi/2$ and multiply it by the length of the total semi-cicle, $\\pi r$, finally divide this by the largest allowed perimeter to ensure it is less than 1.\n\nLet S be the set of all the top points of the perimiter. If the value of the tide at hour $n$, $l_n$ is less than the minimal $z$ value in $q\\in S$ then $p_n = 0$. \n\nElse, let $q' = (x',y', z')$ be the first a point such that for all $q = (x,y,z)$ , $x < x', z > t_n$ and $z' < t_n$. It is the first point underwater. Then find the angle $\\theta \\in (0, \\pi/2)$ from the origin to $(0,r)$ and $(x',y')$.\n$$\\theta = \\cos^{-1}((2r^2 - ({x'}^2+(y' - r)^2)^{\\frac{1}{2}})/(2r^2))$$\nand\n$$p_n = \\frac{\\pi r \\cdot \\frac{\\theta}{\\pi\\frac{1}{2}}}{25\\pi}$$\n\n$C_{n}$:\n$$C_0 = 0, C_{n+1} = \\begin{cases} C_n - (C_n \\cdot \\alpha \\cdot p_n) + (F_n \\cdot \\alpha \\cdot p_n) & \\text{if $p_n > 0$} \\\\\n0 & \\text{if $p_n = 0$}\n\\end{cases}$$\n$F_n$:\n$$F_{n} = N - C_{n}$$\n\n$T_n$:\n\n$$T_0 = 0, T_{n+1} = \\begin{cases} T_n & \\text{if $p_n >0$}\\\\\nT_n + C_n & \\text{if $p_n = 0$}\n\\end{cases}$$",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport math\nimport matplotlib.pyplot as plt\nimport seaborn\nfrom mpl_toolkits.mplot3d import Axes3D\nimport os",
"_____no_output_____"
],
[
"%matplotlib notebook\n\nplt3d = plt.figure().gca(projection='3d')\n\n\n# create x,y\nxx, yy = np.meshgrid(range(-25, 30), range(-25,30))\n\n# calculate corresponding z\nzz = (6 - (0.17 * yy))\n\n# plot the surface\n\nbeach_surf = plt3d.plot_surface(xx, yy, zz, alpha=0.2, color = 'brown', label = \"beach\")\n# tide_surf equations below get the legend to show\nbeach_surf._facecolors2d=beach_surf._facecolors3d\nbeach_surf._edgecolors2d=beach_surf._edgecolors3d\n\nh = 2\nr = 25\ndelta = 5\n\ntheta = np.linspace(0, np.pi, 100)\nx = r * np.cos(theta)\ny = r * np.sin(theta) + delta\nz = 6 + h - (0.17 * y)\n\nplt3d.plot(x,y,z, label = \"top of trap\")\n\n\ntide_path = os.path.join('resources', 'comox_tide.csv')\ntide_df = pd.read_csv(tide_path)\ntide_df = tide_df.drop(columns = ['PDT'])\ntide_values = tide_df.values.flatten()\n\nmax_z = max(tide_values)\nmin_z = min(tide_values)\n\nmax_zz = np.full((55,55), max_z)\nmin_zz = np.full((55,55), min_z)\n\ntide_surf = plt3d.plot_surface(xx, yy, max_zz, color = 'lightcyan', label = \"high/low tide\")\n# tide_surf equations below get the legend to show\ntide_surf._facecolors2d=tide_surf._facecolors3d\ntide_surf._edgecolors2d=tide_surf._edgecolors3d\nplt3d.plot_surface(xx, yy, min_zz, color = 'lightcyan')\n\nplt3d.set_xlabel('X')\nplt3d.set_ylabel('Y')\nplt3d.set_zlabel('Z')\nplt3d.set_xlim(-20,30)\nplt3d.set_ylim(-20,30)\nplt3d.legend()\nplt3d.set_title('fish trap')\n",
"_____no_output_____"
],
[
"# find the angle from the origin to the (0,r) point and the leading edge covered by water\ndef circ_covered(tide_level, z_values, x_values, y_values, radius):\n index = -1\n for i in range(len(z_values)):\n if(z_values[i] <= tide_level):\n index = i\n break;\n if(index == -1):\n return 0\n \n x = x_values[index]\n y = y_values[index]\n length = np.sqrt((x)**2 + (y - radius - delta)**2)\n \n angle = math.acos((2 * radius**2 - length**2) / (2 * radius**2))\n coverage = angle/ (0.5 * np.pi)\n return coverage",
"_____no_output_____"
],
[
"## read in tide values that formed the above tide graph\n\ntide_path = os.path.join('resources', 'comox_tide.csv')\ntide_df = pd.read_csv(tide_path)\ntide_df = tide_df.drop(columns = ['PDT'])\ntide_values = tide_df.values.flatten()",
"_____no_output_____"
],
[
"alpha = 0.05\nN = 1000\nF = N\nC = 0\ntotal_caught = [0]\nin_trap = [0]\nout_trap = [N]\ncatches = []\nperimeter_ratio = np.pi * r / (np.pi * 25)\n\nfor level in tide_values:\n coverage = circ_covered(level, z, x, y, r)\n free_to_captured = F * coverage * alpha * perimeter_ratio\n captured_to_free = C * coverage * alpha * perimeter_ratio\n C = C - captured_to_free + free_to_captured\n F = F + captured_to_free - free_to_captured\n \n if(coverage > 0):\n total_caught.append(total_caught[-1])\n else:\n total_caught.append(total_caught[-1] + C)\n if(C != 0):\n catches.append(C)\n C = 0\n F = N\n \n \n in_trap.append(C)\n out_trap.append(F)\n ",
"_____no_output_____"
],
[
"%matplotlib notebook\n## plot the weeks numbers\nseaborn.set()\nplt.style.use('seaborn-deep')\nx_values = range(len(tide_values) + 1)\nplt.plot(x_values, in_trap, label = \"fish in trap\")\nplt.plot(x_values, out_trap, label = \"fish outside of trap\")\nplt.plot(x_values, total_caught, label = \"total caught\")\nplt.ylabel(\"number of fish\")\nplt.xlabel(\"time (h)\")\nplt.title('fish')\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"#The individual catches when the water level lowers as to trap the fish in the semi circle\ncatches",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb6fc1aa3d43cf41fd2a1a512bc166deaf870b5 | 153,174 | ipynb | Jupyter Notebook | Aqueduct/lab/AQ3-Floods_tool_data-model.ipynb | resource-watch/notebooks | 349de9c2c704c8fbee0a9eb748062543e14d834d | [
"MIT"
] | 3 | 2017-07-24T14:17:25.000Z | 2020-05-27T08:57:54.000Z | Aqueduct/lab/AQ3-Floods_tool_data-model.ipynb | resource-watch/notebooks | 349de9c2c704c8fbee0a9eb748062543e14d834d | [
"MIT"
] | 2 | 2021-06-01T13:46:22.000Z | 2021-09-07T09:24:59.000Z | Aqueduct/lab/AQ3-Floods_tool_data-model.ipynb | resource-watch/notebooks | 349de9c2c704c8fbee0a9eb748062543e14d834d | [
"MIT"
] | 5 | 2017-07-25T18:22:12.000Z | 2021-06-23T16:05:24.000Z | 43.404364 | 17,516 | 0.62967 | [
[
[
"# Table of Contents\n <p>",
"_____no_output_____"
],
[
"The idea of this notebook is to propose WRI - AQ floods team, a schema for the database where to store the data and at the same time a simple overview of the schema for the analysis microservice.\n\nFirst of all we are going to check all csv files stored in folder flood data. \nSecond we are going to understand what the scripts stored in script folder does. \nFinally propose the model.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport json\nimport os\nimport configparser\nfrom sqlalchemy import *\nfrom sqlalchemy_schemadisplay import create_schema_graph\nfrom IPython.display import Image, display\n\n#### Global varible configuration\nworkPath = '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data' ",
"_____no_output_____"
],
[
"files = []\nfor (dirpath, dirnames, filenames) in os.walk(workPath):\n files.extend([{'path':os.path.join(dirpath,file), 'fileName': file} for file in filenames if '.csv' in file])\nlist_of_dfs = [{'data': pd.read_csv(file['path']), \n 'fileName': file['path'],\n 'path':file['path']\n } for file in files]",
"[{'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/lookup_assets_Basin.csv', 'fileName': 'lookup_assets_Basin.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/lookup_assets_City.csv', 'fileName': 'lookup_assets_City.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/lookup_assets_Country.csv', 'fileName': 'lookup_assets_Country.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/lookup_assets_State.csv', 'fileName': 'lookup_assets_State.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/lookup_construction_factors_geogunit_108.csv', 'fileName': 'lookup_construction_factors_geogunit_108.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/lookup_geogunit_101.csv', 'fileName': 'lookup_geogunit_101.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/lookup_geogunit_103.csv', 'fileName': 'lookup_geogunit_103.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/lookup_geogunit_108.csv', 'fileName': 'lookup_geogunit_108.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/lookup_geogunit_110.csv', 'fileName': 'lookup_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/lookup_master.csv', 'fileName': 'lookup_master.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_agg_Coastal_City_nosub.csv', 'fileName': 'Precalc_agg_Coastal_City_nosub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_agg_Coastal_City_wtsub.csv', 'fileName': 'Precalc_agg_Coastal_City_wtsub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_agg_Coastal_Country_nosub.csv', 'fileName': 'Precalc_agg_Coastal_Country_nosub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_agg_Coastal_Country_wtsub.csv', 'fileName': 'Precalc_agg_Coastal_Country_wtsub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_agg_Coastal_State_nosub.csv', 'fileName': 'Precalc_agg_Coastal_State_nosub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_agg_Coastal_State_wtsub.csv', 'fileName': 'Precalc_agg_Coastal_State_wtsub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_agg_Riverine_Basin_nosub.csv', 'fileName': 'Precalc_agg_Riverine_Basin_nosub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_agg_Riverine_City_nosub.csv', 'fileName': 'Precalc_agg_Riverine_City_nosub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_agg_Riverine_Country_nosub.csv', 'fileName': 'Precalc_agg_Riverine_Country_nosub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_agg_Riverine_State_nosub.csv', 'fileName': 'Precalc_agg_Riverine_State_nosub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_Coastal_geogunit_103_nosub.csv', 'fileName': 'Precalc_Coastal_geogunit_103_nosub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_Coastal_geogunit_103_wtsub.csv', 'fileName': 'Precalc_Coastal_geogunit_103_wtsub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_Coastal_geogunit_108_nosub.csv', 'fileName': 'Precalc_Coastal_geogunit_108_nosub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_Coastal_geogunit_108_wtsub.csv', 'fileName': 'Precalc_Coastal_geogunit_108_wtsub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_Riverine_geogunit_103_nosub.csv', 'fileName': 'Precalc_Riverine_geogunit_103_nosub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Precalc_Riverine_geogunit_108_nosub.csv', 'fileName': 'Precalc_Riverine_geogunit_108_nosub.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Coastal_City_GDPexp.csv', 'fileName': 'Raw_agg_Coastal_City_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Coastal_City_POPexp.csv', 'fileName': 'Raw_agg_Coastal_City_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Coastal_City_Urban_Damage_v2.csv', 'fileName': 'Raw_agg_Coastal_City_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Coastal_Country_GDPexp.csv', 'fileName': 'Raw_agg_Coastal_Country_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Coastal_Country_POPexp.csv', 'fileName': 'Raw_agg_Coastal_Country_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Coastal_Country_Urban_Damage_v2.csv', 'fileName': 'Raw_agg_Coastal_Country_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Coastal_State_GDPexp.csv', 'fileName': 'Raw_agg_Coastal_State_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Coastal_State_POPexp.csv', 'fileName': 'Raw_agg_Coastal_State_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Coastal_State_Urban_Damage_v2.csv', 'fileName': 'Raw_agg_Coastal_State_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_Basin_GDPexp.csv', 'fileName': 'Raw_agg_Riverine_Basin_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_Basin_POPexp.csv', 'fileName': 'Raw_agg_Riverine_Basin_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_Basin_Urban_Damage_v2.csv', 'fileName': 'Raw_agg_Riverine_Basin_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_City_GDPexp.csv', 'fileName': 'Raw_agg_Riverine_City_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_City_POPexp.csv', 'fileName': 'Raw_agg_Riverine_City_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_City_Urban_Damage_v2.csv', 'fileName': 'Raw_agg_Riverine_City_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_Country_GDPexp.csv', 'fileName': 'Raw_agg_Riverine_Country_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_Country_POPexp.csv', 'fileName': 'Raw_agg_Riverine_Country_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_Country_Urban_Damage_v2.csv', 'fileName': 'Raw_agg_Riverine_Country_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_State_GDPexp.csv', 'fileName': 'Raw_agg_Riverine_State_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_State_POPexp.csv', 'fileName': 'Raw_agg_Riverine_State_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_agg_Riverine_State_Urban_Damage_v2.csv', 'fileName': 'Raw_agg_Riverine_State_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Coastal_geogunit_103_GDPexp.csv', 'fileName': 'Raw_Coastal_geogunit_103_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Coastal_geogunit_103_POPexp.csv', 'fileName': 'Raw_Coastal_geogunit_103_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Coastal_geogunit_103_Urban_Damage_v2.csv', 'fileName': 'Raw_Coastal_geogunit_103_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Coastal_geogunit_108_GDPexp.csv', 'fileName': 'Raw_Coastal_geogunit_108_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Coastal_geogunit_108_POPexp.csv', 'fileName': 'Raw_Coastal_geogunit_108_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Coastal_geogunit_108_Urban_Damage_v2.csv', 'fileName': 'Raw_Coastal_geogunit_108_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Riverine_geogunit_101_GDPexp.csv', 'fileName': 'Raw_Riverine_geogunit_101_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Riverine_geogunit_101_POPexp.csv', 'fileName': 'Raw_Riverine_geogunit_101_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Riverine_geogunit_101_Urban_Damage_v2.csv', 'fileName': 'Raw_Riverine_geogunit_101_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Riverine_geogunit_103_GDPexp.csv', 'fileName': 'Raw_Riverine_geogunit_103_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Riverine_geogunit_103_POPexp.csv', 'fileName': 'Raw_Riverine_geogunit_103_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Riverine_geogunit_103_Urban_Damage_v2.csv', 'fileName': 'Raw_Riverine_geogunit_103_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Riverine_geogunit_108_GDPexp.csv', 'fileName': 'Raw_Riverine_geogunit_108_GDPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Riverine_geogunit_108_POPexp.csv', 'fileName': 'Raw_Riverine_geogunit_108_POPexp.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Raw_Riverine_geogunit_108_Urban_Damage_v2.csv', 'fileName': 'Raw_Riverine_geogunit_108_Urban_Damage_v2.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_rural_BAU_2030_geogunit_110.csv', 'fileName': 'lookup_cost_rural_BAU_2030_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_rural_BAU_2050_geogunit_110.csv', 'fileName': 'lookup_cost_rural_BAU_2050_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_rural_BAU_2080_geogunit_110.csv', 'fileName': 'lookup_cost_rural_BAU_2080_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_rural_OPT_2030_geogunit_110.csv', 'fileName': 'lookup_cost_rural_OPT_2030_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_rural_OPT_2050_geogunit_110.csv', 'fileName': 'lookup_cost_rural_OPT_2050_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_rural_OPT_2080_geogunit_110.csv', 'fileName': 'lookup_cost_rural_OPT_2080_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_rural_PES_2030_geogunit_110.csv', 'fileName': 'lookup_cost_rural_PES_2030_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_rural_PES_2050_geogunit_110.csv', 'fileName': 'lookup_cost_rural_PES_2050_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_rural_PES_2080_geogunit_110.csv', 'fileName': 'lookup_cost_rural_PES_2080_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_urban_BAU_2030_geogunit_110.csv', 'fileName': 'lookup_cost_urban_BAU_2030_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_urban_BAU_2050_geogunit_110.csv', 'fileName': 'lookup_cost_urban_BAU_2050_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_urban_BAU_2080_geogunit_110.csv', 'fileName': 'lookup_cost_urban_BAU_2080_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_urban_OPT_2030_geogunit_110.csv', 'fileName': 'lookup_cost_urban_OPT_2030_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_urban_OPT_2050_geogunit_110.csv', 'fileName': 'lookup_cost_urban_OPT_2050_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_urban_OPT_2080_geogunit_110.csv', 'fileName': 'lookup_cost_urban_OPT_2080_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_urban_PES_2030_geogunit_110.csv', 'fileName': 'lookup_cost_urban_PES_2030_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_urban_PES_2050_geogunit_110.csv', 'fileName': 'lookup_cost_urban_PES_2050_geogunit_110.csv'}, {'path': '/Users/alicia/Projects/jupyter-geotools-alpine/work/data/aqueduct/data_source/floods/floods_vizzuality/Flood_Data/Costs/lookup_cost_urban_PES_2080_geogunit_110.csv', 'fileName': 'lookup_cost_urban_PES_2080_geogunit_110.csv'}]\n"
],
[
"for dataset in list_of_dfs:\n print(dataset['fileName'].split('/')[-1])\n display(dataset['data'].shape)\n display(dataset['data'].columns)",
"lookup_assets_Basin.csv\n"
],
[
"metadata = MetaData()\nlookup_assets = Table('lookup_assets', metadata,\n Column('geo_id', Integer, primary_key=True),\n Column('year', Integer, nullable=False),\n Column('model', String(60), nullable=False),\n Column('geom_type', String(20), nullable=False)\n)\n\nprecalc_agg = Table('precalc_agg', metadata,\n Column('pref_id', Integer, primary_key=True),\n Column('user_id', Integer, ForeignKey(\"user.user_id\"), nullable=False),\n Column('pref_name', String(40), nullable=False),\n Column('pref_value', String(100))\n)\n\ngraph = create_schema_graph(metadata=metadata,\n show_datatypes=True,\n show_indexes=True,\n rankdir='LR',\n concentrate=False)\n\n##Generate png image \ngraph.write_png('dbschema.png')\nImage('dbschema.png')",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecb708ae7dd0d2f75bfe7cc8b97b7768d654921b | 74,510 | ipynb | Jupyter Notebook | MNIST_Classifier.ipynb | yuhan1212/PyTorch_example | da745c67f03ffd8e0b757e6f7a77ef435ff0a913 | [
"Apache-2.0"
] | 1 | 2021-11-28T00:22:22.000Z | 2021-11-28T00:22:22.000Z | MNIST_Classifier.ipynb | yuhan1212/PyTorch_example | da745c67f03ffd8e0b757e6f7a77ef435ff0a913 | [
"Apache-2.0"
] | null | null | null | MNIST_Classifier.ipynb | yuhan1212/PyTorch_example | da745c67f03ffd8e0b757e6f7a77ef435ff0a913 | [
"Apache-2.0"
] | null | null | null | 121.748366 | 51,238 | 0.83111 | [
[
[
"# PyTorch 1.2 Quickstart with Google Colab\nIn this code tutorial we will learn how to quickly train a model to understand some of PyTorch's basic building blocks to train a deep learning model. This notebook is inspired by the [\"Tensorflow 2.0 Quickstart for experts\"](https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/tutorials/quickstart/advanced.ipynb#scrollTo=DUNzJc4jTj6G) notebook. \n\nAfter completion of this tutorial, you should be able to import data, transform it, and efficiently feed the data in batches to a convolution neural network (CNN) model for image classification.\n\n**Author:** [Elvis Saravia](https://twitter.com/omarsar0)\n\n**Complete Code Walkthrough:** [Blog post](https://medium.com/dair-ai/pytorch-1-2-quickstart-with-google-colab-6690a30c38d)",
"_____no_output_____"
]
],
[
[
"!pip3 install torch==1.2.0+cu92 torchvision==0.4.0+cu92 -f https://download.pytorch.org/whl/torch_stable.html",
"Looking in links: https://download.pytorch.org/whl/torch_stable.html\nCollecting torch==1.2.0+cu92\n Downloading https://download.pytorch.org/whl/cu92/torch-1.2.0%2Bcu92-cp37-cp37m-manylinux1_x86_64.whl (663.1 MB)\n\u001b[K |████████████████████████████████| 663.1 MB 357 bytes/s \n\u001b[?25hCollecting torchvision==0.4.0+cu92\n Downloading https://download.pytorch.org/whl/cu92/torchvision-0.4.0%2Bcu92-cp37-cp37m-manylinux1_x86_64.whl (8.8 MB)\n\u001b[K |████████████████████████████████| 8.8 MB 2.3 MB/s \n\u001b[?25hRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from torch==1.2.0+cu92) (1.19.5)\nRequirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from torchvision==0.4.0+cu92) (1.15.0)\nRequirement already satisfied: pillow>=4.1.1 in /usr/local/lib/python3.7/dist-packages (from torchvision==0.4.0+cu92) (7.1.2)\nInstalling collected packages: torch, torchvision\n Attempting uninstall: torch\n Found existing installation: torch 1.10.0+cu111\n Uninstalling torch-1.10.0+cu111:\n Successfully uninstalled torch-1.10.0+cu111\n Attempting uninstall: torchvision\n Found existing installation: torchvision 0.11.1+cu111\n Uninstalling torchvision-0.11.1+cu111:\n Successfully uninstalled torchvision-0.11.1+cu111\n\u001b[31mERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.\ntorchtext 0.11.0 requires torch==1.10.0, but you have torch 1.2.0+cu92 which is incompatible.\u001b[0m\nSuccessfully installed torch-1.2.0+cu92 torchvision-0.4.0+cu92\n"
]
],
[
[
"Note: We will be using the latest stable version of PyTorch so be sure to run the command above to install the latest version of PyTorch, which as the time of this tutorial was 1.2.0. We PyTorch belowing using the `torch` module. ",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torchvision\nimport torchvision.transforms as transforms",
"_____no_output_____"
],
[
"print(torch.__version__)",
"1.2.0+cu92\n"
]
],
[
[
"## Import The Data\nThe first step before training the model is to import the data. We will use the [MNIST dataset](http://yann.lecun.com/exdb/mnist/) which is like the Hello World dataset of machine learning. \n\nBesides importing the data, we will also do a few more things:\n- We will tranform the data into tensors using the `transforms` module\n- We will use `DataLoader` to build convenient data loaders or what are referred to as iterators, which makes it easy to efficiently feed data in batches to deep learning models. \n- As hinted above, we will also create batches of the data by setting the `batch` parameter inside the data loader. Notice we use batches of `32` in this tutorial but you can change it to `64` if you like. I encourage you to experiment with different batches.",
"_____no_output_____"
]
],
[
[
"BATCH_SIZE = 32\n\n## transformations\ntransform = transforms.Compose(\n [transforms.ToTensor()])\n\n## download and load training dataset\ntrainset = torchvision.datasets.MNIST(root='./data', train=True,\n download=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=BATCH_SIZE,\n shuffle=True, num_workers=2)\n\n## download and load testing dataset\ntestset = torchvision.datasets.MNIST(root='./data', train=False,\n download=True, transform=transform)\ntestloader = torch.utils.data.DataLoader(testset, batch_size=BATCH_SIZE,\n shuffle=False, num_workers=2)",
"Downloading http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz to ./data/MNIST/raw/train-images-idx3-ubyte.gz\n"
]
],
[
[
"## Exploring the Data\nAs a practioner and researcher, I am always spending a bit of time and effort exploring and understanding the dataset. It's fun and this is a good practise to ensure that everything is in order. ",
"_____no_output_____"
],
[
"Let's check what the train and test dataset contains. I will use `matplotlib` to print out some of the images from our dataset. ",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\n\n## functions to show an image\ndef imshow(img):\n #img = img / 2 + 0.5 # unnormalize\n npimg = img.numpy()\n plt.imshow(np.transpose(npimg, (1, 2, 0)))\n\n## get some random training images\ndataiter = iter(trainloader)\nimages, labels = dataiter.next()\n\n## show images\nimshow(torchvision.utils.make_grid(images))",
"_____no_output_____"
]
],
[
[
"**EXERCISE:** Try to understand what the code above is doing. This will help you to better understand your dataset before moving forward. ",
"_____no_output_____"
],
[
"Let's check the dimensions of a batch.",
"_____no_output_____"
]
],
[
[
"for images, labels in trainloader:\n print(\"Image batch dimensions:\", images.shape)\n print(\"Image label dimensions:\", labels.shape)\n break",
"Image batch dimensions: torch.Size([32, 1, 28, 28])\nImage label dimensions: torch.Size([32])\n"
]
],
[
[
"## The Model\nNow using the classical deep learning framework pipeline, let's build the 1 convolutional layer model. \n\nHere are a few notes for those who are beginning with PyTorch:\n- The model below consists of an `__init__()` portion which is where you include the layers and components of the neural network. In our model, we have a convolutional layer denoted by `nn.Conv2d(...)`. We are dealing with an image dataset that is in a grayscale so we only need one channel going in, hence `in_channels=1`. We hope to get a nice representation of this layer, so we use `out_channels=32`. Kernel size is 3, and for the rest of parameters we use the default values which you can find [here](https://pytorch.org/docs/stable/nn.html?highlight=conv2d#conv2d). \n- We use 2 back to back dense layers or what we refer to as linear transformations to the incoming data. Notice for `d1` I have a dimension which looks like it came out of nowhere. 128 represents the size we want as output and the (`26*26*32`) represents the dimension of the incoming data. If you would like to find out how to calculate those numbers refer to the [PyTorch documentation](https://pytorch.org/docs/stable/nn.html?highlight=linear#conv2d). In short, the convolutional layer transforms the input data into a specific dimension that has to be considered in the linear layer. The same applies for the second linear transformation (`d2`) where the dimension of the output of the previous linear layer was added as `in_features=128`, and `10` is just the size of the output which also corresponds to the number of classes.\n- After each one of those layers, we also apply an activation function such as `ReLU`. For prediction purposes, we then apply a `softmax` layer to the last transformation and return the output of that. ",
"_____no_output_____"
]
],
[
[
"class MyModel(nn.Module):\n def __init__(self):\n super(MyModel, self).__init__()\n\n # 28x28x1 => 26x26x32\n self.conv1 = nn.Conv2d(in_channels=1, out_channels=32, kernel_size=3)\n self.d1 = nn.Linear(26 * 26 * 32, 128)\n self.d2 = nn.Linear(128, 10)\n\n def forward(self, x):\n # 32x1x28x28 => 32x32x26x26\n x = self.conv1(x)\n x = F.relu(x)\n\n # flatten => 32 x (32*26*26)\n x = x.flatten(start_dim = 1)\n\n # 32 x (32*26*26) => 32x128\n x = self.d1(x)\n x = F.relu(x)\n\n # logits => 32x10\n logits = self.d2(x)\n out = F.softmax(logits, dim=1)\n return out",
"_____no_output_____"
]
],
[
[
"As I have done in my previous tutorials, I always encourage to test the model with 1 batch to ensure that the output dimensions are what we expect. ",
"_____no_output_____"
]
],
[
[
"## test the model with 1 batch\nmodel = MyModel()\nfor images, labels in trainloader:\n print(\"batch size:\", images.shape)\n out = model(images)\n print(out.shape)\n break",
"batch size: torch.Size([32, 1, 28, 28])\ntorch.Size([32, 10])\n"
]
],
[
[
"## Training the Model\nNow we are ready to train the model but before that we are going to setup a loss function, an optimizer and a function to compute accuracy of the model. ",
"_____no_output_____"
]
],
[
[
"learning_rate = 0.001\nnum_epochs = 5\n\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\nmodel = MyModel()\nmodel = model.to(device)\ncriterion = nn.CrossEntropyLoss()\noptimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)",
"_____no_output_____"
],
[
"## compute accuracy\ndef get_accuracy(logit, target, batch_size):\n ''' Obtain accuracy for training round '''\n corrects = (torch.max(logit, 1)[1].view(target.size()).data == target.data).sum()\n accuracy = 100.0 * corrects/batch_size\n return accuracy.item()",
"_____no_output_____"
]
],
[
[
"Now it's time for training.",
"_____no_output_____"
]
],
[
[
"for epoch in range(num_epochs):\n train_running_loss = 0.0\n train_acc = 0.0\n\n model = model.train()\n\n ## training step\n for i, (images, labels) in enumerate(trainloader):\n \n images = images.to(device)\n labels = labels.to(device)\n\n ## forward + backprop + loss\n logits = model(images)\n loss = criterion(logits, labels)\n optimizer.zero_grad()\n loss.backward()\n\n ## update model params\n optimizer.step()\n\n train_running_loss += loss.detach().item()\n train_acc += get_accuracy(logits, labels, BATCH_SIZE)\n \n model.eval()\n print('Epoch: %d | Loss: %.4f | Train Accuracy: %.2f' \\\n %(epoch, train_running_loss / i, train_acc/i)) ",
"Epoch: 0 | Loss: 1.5635 | Train Accuracy: 89.75\nEpoch: 1 | Loss: 1.4887 | Train Accuracy: 97.09\nEpoch: 2 | Loss: 1.4804 | Train Accuracy: 97.97\nEpoch: 3 | Loss: 1.4764 | Train Accuracy: 98.38\nEpoch: 4 | Loss: 1.4738 | Train Accuracy: 98.67\n"
]
],
[
[
"We can also compute accuracy on the testing dataset to see how well the model performs on the image classificaiton task. As you can see below, our basic CNN model is performing very well on the MNIST classification task.",
"_____no_output_____"
]
],
[
[
"test_acc = 0.0\nfor i, (images, labels) in enumerate(testloader, 0):\n images = images.to(device)\n labels = labels.to(device)\n outputs = model(images)\n test_acc += get_accuracy(outputs, labels, BATCH_SIZE)\n \nprint('Test Accuracy: %.2f'%( test_acc/i))",
"Test Accuracy: 98.10\n"
]
],
[
[
"**EXERCISE:** As a way to practise, try to include the testing part inside the code where I was outputing the training accuracy, so that you can also keep testing the model on the testing data as you proceed with the training steps. This is useful as sometimes you don't want to wait until your model has completed training to actually test the model with the testing data.",
"_____no_output_____"
],
[
"## Final Words\nThat's it for this tutorial! Congratulations! You are now able to implement a basic CNN model in PyTorch for image classification. If you would like, you can further extend the CNN model by adding more convolution layers and max pooling, but as you saw, you don't really need it here as results look good. If you are interested in implementing a similar image classification model using RNNs see the references below. ",
"_____no_output_____"
],
[
"## References\n- [Building RNNs is Fun with PyTorch and Google Colab](https://colab.research.google.com/drive/1NVuWLZ0cuXPAtwV4Fs2KZ2MNla0dBUas)\n- [CNN Basics with PyTorch by Sebastian Raschka](https://github.com/rasbt/deeplearning-models/blob/master/pytorch_ipynb/cnn/cnn-basic.ipynb)\n- [Tensorflow 2.0 Quickstart for experts](https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/tutorials/quickstart/advanced.ipynb#scrollTo=DUNzJc4jTj6G) ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ecb70c78363d20fb1bba9deeaf64b0f1f4586b3a | 12,375 | ipynb | Jupyter Notebook | examples/HMM_align.ipynb | bolajiy/beer | 6fe968c7ca4864437890aa6bd705755c2580696e | [
"MIT"
] | 46 | 2018-02-27T18:15:08.000Z | 2022-02-16T22:10:55.000Z | examples/HMM_align.ipynb | bolajiy/beer | 6fe968c7ca4864437890aa6bd705755c2580696e | [
"MIT"
] | 16 | 2018-01-26T14:18:51.000Z | 2021-02-05T09:34:00.000Z | examples/HMM_align.ipynb | bolajiy/beer | 6fe968c7ca4864437890aa6bd705755c2580696e | [
"MIT"
] | 26 | 2018-03-12T14:03:26.000Z | 2021-05-24T21:15:01.000Z | 31.25 | 166 | 0.534061 | [
[
[
"# Bayesian HMM Model\n\nThis notebook illustrate how to build and train a Bayesian Hidden Markov Model with the [beer framework](https://github.com/beer-asr/beer).",
"_____no_output_____"
]
],
[
[
"# Add \"beer\" to the PYTHONPATH\nimport sys\nsys.path.insert(0, '../')\n\nimport copy\n\nimport beer\nimport numpy as np\nimport torch\n\n# For plotting.\nfrom bokeh.io import show, output_notebook\nfrom bokeh.plotting import figure, gridplot\nfrom bokeh.models import LinearAxis, Range1d\noutput_notebook()\n\n# Convenience functions for plotting.\nimport plotting\n\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"def create_ali_trans_mat(tot_states):\n '''Create align transition matrix for a sequence of units\n Args:\n tot_states (int): length of total number of states of the given\n sequence.\n '''\n\n trans_mat = torch.diag(torch.ones(tot_states) * .5)\n idx1 = torch.arange(0, tot_states-1, dtype=torch.long)\n idx2 = torch.arange(1, tot_states, dtype=torch.long)\n trans_mat[idx1, idx2] = .5\n trans_mat[-1, -1] = 1.\n return trans_mat\n\n \n# Sequence: AB\n\nseqs = ['A', 'B', 'A']\nnsamples = 30\nndim = 2\n\nunits = ['A', 'B']\nlen_seqs = len(seqs)\nnum_unit_states = 3\ntot_states = len(seqs) * num_unit_states\n\ntrans_mat = create_ali_trans_mat(tot_states)\n\nmeans = [np.array([-1.5, 3]), np.array([-1.5, 4]), np.array([-1.5, 5]),\n np.array([1, -3]), np.array([1, -2]), np.array([1, -1])]\ncovs = [np.array([[.75, -.5], [-.5, 2.]]), np.array([[.75, -.5], [-.5, 2.]]), np.array([[.75, -.5], [-.5, 2.]]),\n np.array([[2, 1], [1, .75]]), np.array([[2, 1], [1, .75]]), np.array([[2, 1], [1, .75]])]\n\nstates_id = {'A':[0, 1, 2], 'B':[3, 4, 5]}\ndict_seq_state = {}\n\nseqs_id = []\nfor i, j in enumerate(seqs):\n for u in range(num_unit_states):\n dict_seq_state[num_unit_states * i + u] = states_id[j][u]\n seqs_id.append(states_id[j][u])\n\nnormal_sets = list(zip(means,covs))\n\nstates = np.zeros(nsamples, dtype=np.int16)\ndata = np.zeros((nsamples, ndim))\nstates[0] = states_id['A'][0]\ndata[0] = np.random.multivariate_normal(means[0], covs[0], size=1)\n\ncolors = ['blue', 'blue', 'blue', 'red', 'red', 'red']\nfig1 = figure(title='Samples', width=400, height=400)\nfig1.circle(data[0, 0], data[0, 1], color=colors[states[0]])\n\n\nfor n in range(1, nsamples):\n states[n] = np.random.choice(np.arange(tot_states), p=trans_mat[states[n-1]].numpy())\n data[n] = np.random.multivariate_normal(means[dict_seq_state[states[n]]], covs[dict_seq_state[states[n]]], size=1)\n fig1.circle(data[n, 0], data[n, 1], color=colors[dict_seq_state[states[n]]], line_width=1)\n fig1.line(data[n-1:n+1, 0], data[n-1:n+1, 1], color='black', line_width=.5, alpha=.5)\n\nstates_id = [dict_seq_state[i] for i in states]\n \nfig2 = figure(title='Emissions', width=400, height=400)\ncolors = ['darkblue', 'blue', 'skyblue', 'darkred','red', 'pink']\n\nfor i, n in enumerate(normal_sets):\n plotting.plot_normal(fig2, n[0], n[1], alpha=.3, color=colors[i])\ngrid = gridplot([[fig1, fig2]])\nshow(grid)\nprint(states_id)",
"_____no_output_____"
]
],
[
[
"## Model Creation\n\nWe create several types of HMMs, each of them has the same transition matrix and initial / final state probability, and a specific type of emission density: \n * one Normal density per state with full covariance matrix\n * one Normal density per state with diagonal covariance matrix\n * one Normal density per state with full covariance matrix shared across states\n * one Normal density per state with diagonal covariance matrix shared across states.",
"_____no_output_____"
]
],
[
[
"graph = beer.graph.Graph()\ns0 = graph.add_state()\ns1 = graph.add_state(pdf_id=0)\ns2 = graph.add_state(pdf_id=1)\ns3 = graph.add_state(pdf_id=2)\ns4 = graph.add_state()\ngraph.start_state = s0\ngraph.end_state = s4\ngraph.add_arc(s0, s1)\ngraph.add_arc(s1, s1)\ngraph.add_arc(s1, s2)\ngraph.add_arc(s2, s2)\ngraph.add_arc(s2, s3)\ngraph.add_arc(s3, s3)\ngraph.add_arc(s3, s1)\ngraph.add_arc(s3, s4)\ngraph.normalize()\ngraph",
"_____no_output_____"
],
[
"graph.normalize()\nloop_graph = graph.compile()",
"_____no_output_____"
],
[
"graph = beer.graph.Graph()\ns0 = graph.add_state()\ns1 = graph.add_state(pdf_id=0)\ns2 = graph.add_state(pdf_id=1)\ns3 = graph.add_state(pdf_id=2)\ns4 = graph.add_state(pdf_id=3)\ns5 = graph.add_state(pdf_id=4)\ns6 = graph.add_state(pdf_id=5)\ns7 = graph.add_state(pdf_id=0)\ns8 = graph.add_state(pdf_id=1)\ns9 = graph.add_state(pdf_id=2)\ns10 = graph.add_state()\ngraph.start_state = s0\ngraph.end_state = s10\ngraph.add_arc(s0, s1)\ngraph.add_arc(s1, s1)\ngraph.add_arc(s1, s2)\ngraph.add_arc(s2, s2)\ngraph.add_arc(s2, s3)\ngraph.add_arc(s3, s3)\ngraph.add_arc(s3, s4)\ngraph.add_arc(s4, s4)\ngraph.add_arc(s4, s5)\ngraph.add_arc(s5, s5)\ngraph.add_arc(s5, s6)\ngraph.add_arc(s6, s6)\ngraph.add_arc(s6, s7)\ngraph.add_arc(s7, s7)\ngraph.add_arc(s7, s8)\ngraph.add_arc(s8, s8)\ngraph.add_arc(s8, s9)\ngraph.add_arc(s9, s9)\ngraph.add_arc(s9, s10)\ngraph.normalize()\ngraph",
"_____no_output_____"
],
[
"ali_graph = graph.compile().double()",
"_____no_output_____"
],
[
"# We use the global mean/cov. matrix of the data to initialize the mixture.\ndata_mean = torch.from_numpy(data.mean(axis=0)).float()\ndata_var = torch.from_numpy(np.cov(data.T)).float()\n\n# HMM (diag cov).\nmodelset = beer.NormalSet.create(data_mean, data_var, size=loop_graph.n_states,\n prior_strength=1., noise_std=1., \n cov_type='full')\nhmm_diag_loop = beer.HMM.create(loop_graph, modelset)\n\nmodelset = beer.NormalSet.create(data_mean, data_var, size=ali_graph.n_states,\n prior_strength=1., noise_std=1., \n cov_type='full')\nhmm_diag_align = beer.HMM.create(ali_graph, modelset)\n\nmodels = {\n 'hmm_diag_loop': hmm_diag_loop.double(),\n 'hmm_diag_align': hmm_diag_align.double()\n}",
"_____no_output_____"
]
],
[
[
"## Variational Bayes Training ",
"_____no_output_____"
]
],
[
[
"epochs = 100\nlrate = 1.\nX = torch.from_numpy(data).double()\n\noptims = {\n model_name: beer.VBConjugateOptimizer(model.mean_field_factorization(), lrate)\n for model_name, model in models.items()\n}\n\nelbos = {\n model_name: []\n for model_name in models\n} \n\ninf_graphs = {\n 'hmm_diag_loop': None,\n 'hmm_diag_align': ali_graph\n} \n\nfor epoch in range(epochs):\n for name, model in models.items():\n optim = optims[name]\n optim.init_step()\n elbo = beer.evidence_lower_bound(model, X, datasize=len(X),\n inference_graph=inf_graphs[name],\n viterbi=True)\n elbo.backward()\n elbos[name].append(float(elbo) / len(X))\n optim.step()\n",
"_____no_output_____"
],
[
"colors = {\n 'hmm_diag_loop': 'green',\n 'hmm_diag_align': 'blue'\n}\n# Plot the ELBO.\nfig = figure(title='ELBO', width=400, height=400, x_axis_label='step',\n y_axis_label='ln p(X)')\nfor model_name, elbo in elbos.items():\n fig.line(range(len(elbo)), elbo, legend=model_name, color=colors[model_name])\nfig.legend.location = 'bottom_right'\n\nshow(fig)",
"_____no_output_____"
],
[
"mean = data.mean(axis=0)\nvar = data.var(axis=0)\nstd_dev = np.sqrt(max(var))\nx_range = (mean[0] - 2 * std_dev, mean[0] + 2 * std_dev)\ny_range = (mean[1] - 2 * std_dev, mean[1] + 2 * std_dev)\nglobal_range = (min(x_range[0], y_range[0]), max(x_range[1], y_range[1]))\n\nfig1 = figure(title='HMM (diag) loop', x_range=global_range, y_range=global_range,\n width=400, height=400)\nfig1.circle(data[:, 0], data[:, 1], alpha=.5, color='blue')\nplotting.plot_hmm(fig1, hmm_diag_loop, alpha=.1, color='blue')\n\nfig2 = figure(title='HMM (diag) align', x_range=global_range, y_range=global_range,\n width=400, height=400)\nfig2.circle(data[:, 0], data[:, 1], alpha=.5, color='red')\nplotting.plot_hmm(fig2, hmm_diag_align, alpha=.1, color='red')\ngrid = gridplot([[fig1, fig2]])\nshow(grid)",
"_____no_output_____"
]
],
[
[
"### Plotting",
"_____no_output_____"
]
],
[
[
"# We are mixing bokeh and matplotlib >:-( ! .\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"posts1 = models['hmm_diag_loop'].posteriors(X).numpy().T\nposts2 = models['hmm_diag_align'].posteriors(X, ali_graph).numpy().T\n\nfig1, axarr = plt.subplots(2, 1)\naxarr[0].imshow(posts1, origin='lower')\naxarr[0].set_title('HMM loop (diag) lhs')\naxarr[1].imshow(posts2, origin='lower')\naxarr[1].set_title('HMM align (diag) lhs')\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecb70ca3437c66089718c71f5e648a60e1aeba46 | 6,751 | ipynb | Jupyter Notebook | 2_code/3_utilities/R Functions for Time Series Imputation.ipynb | MichaelRGrant/HyrdoSatML | 27bf7ce54f061390238ea136ad494dd60a8041f3 | [
"MIT"
] | 5 | 2017-11-03T01:16:07.000Z | 2021-01-01T20:05:59.000Z | 2_code/3_utilities/R Functions for Time Series Imputation.ipynb | MichaelRGrant/HyrdoSatML | 27bf7ce54f061390238ea136ad494dd60a8041f3 | [
"MIT"
] | null | null | null | 2_code/3_utilities/R Functions for Time Series Imputation.ipynb | MichaelRGrant/HyrdoSatML | 27bf7ce54f061390238ea136ad494dd60a8041f3 | [
"MIT"
] | null | null | null | 44.124183 | 142 | 0.564657 | [
[
[
"make_tuning_df <- function(data, cutoff){\n require(plyr)\n stations_list <- list()\n stations <- unique(data$field)\n for(i in 1:length(stations)){\n print(as.character(stations[i]))\n stations_list[[i]] <- subset(data, field == stations[i])\n numrows <- nrow(stations_list[[i]])\n sub <- as.integer(numrows*cutoff)\n stations_list[[i]] <- stations_list[[i]][sub:numrows, ]\n }\n return(ldply(stations_list, data.frame))\n}\n",
"_____no_output_____"
],
[
"ts_impute <- function(data){\n require(plyr)\n require(forecast)\n require(dplyr)\n require(imputeTS)\n ### IMPUTATION FUNCTION\n stations_list <- list()\n stations <- unique(data$station)\n for(i in 1:length(stations)){\n print(as.character(stations[i]))\n stations_list[[i]] <- subset(data, station == stations[i])\n \n # county <- unique(stations_list[[i]]$county)\n # station <- unique(stations_list[[i]]$station)\n # \n date_range <- range(stations_list[[i]]$datetime, na.rm=T)\n start <- as.numeric(date_range[1])\n # date_seq <- seq(date_range[1], date_range[2], by = '15 mins')\n # date_seq <- data.frame('datetime' = strptime(gsub(date_seq, pattern = ' PST', replacement = ''),\n # format = '%Y-%m-%d %H:%M:%S', tz = 'US/Pacific'))\n # stations_list[[i]] <- right_join(stations_list[[i]], date_seq, by='datetime')\n # \n # stations_list[[i]]$county <- county\n # stations_list[[i]]$station <- station\n \n # make timeseries objects from each vector that has yearly seasonality\n # use msts function from forecast to to time series decomposition\n # because the data is shorter, the seasonality may be each day due to rising and falling temperatures\n \n air1.ts <- msts(stations_list[[i]]$air_temp_1, start = start, seasonal.periods = 96)\n dewpoint.ts <- msts(stations_list[[i]]$dewpoint, start = start, seasonal.periods = 96)\n rel_hum.ts <- msts(stations_list[[i]]$rel_hum, start = start, seasonal.periods = 96)\n eight_in.ts <- msts(stations_list[[i]]$eight_in_soil_temp, start = start, seasonal.periods = 96)\n solar.ts <- msts(stations_list[[i]]$solar_Watts_m2.y, start = start, seasonal.periods = 96)\n leafwet.ts <- msts(stations_list[[i]]$leaf_wet, start = start, seasonal.periods = 96)\n wind_speed.ts <- msts(stations_list[[i]]$wind_speed, start = start, seasonal.periods = 96)\n wind_gust.ts <- msts(stations_list[[i]]$wind_gust, start = start, seasonal.periods = 96)\n # vwc.ts <- msts(stations_list[[i]]$vwc, start = start, seasonal.periods = 35064)\n two_in.ts <- msts(stations_list[[i]]$two_in_soil_temp, start=start, seasonal.periods = 96)\n \n # impute using seasplit and/or spline interpolation\n stations_list[[i]]$air_temp_1 <- na.seasplit(air1.ts, algorithm = 'interpolation')[1:nrow(stations_list[[i]])]\n stations_list[[i]]$dewpoint <- na.seasplit(dewpoint.ts, algorithm = 'interpolation')[1:nrow(stations_list[[i]])]\n stations_list[[i]]$rel_hum <- na.seasplit(rel_hum.ts, algorithm = 'interpolation')[1:nrow(stations_list[[i]])]\n stations_list[[i]]$eight_in_soil_temp <- na.seasplit(eight_in.ts, algorithm = 'interpolation')[1:nrow(stations_list[[i]])]\n stations_list[[i]]$solar_Watts_m2.y <- na.seasplit(solar.ts, algorithm = 'interpolation')[1:nrow(stations_list[[i]])]\n stations_list[[i]]$leaf_wet <- na.seasplit(leafwet.ts, algorithm = 'interpolation')[1:nrow(stations_list[[i]])]\n stations_list[[i]]$wind_speed <- na.seasplit(wind_speed.ts, algorithm = 'interpolation')[1:nrow(stations_list[[i]])]\n stations_list[[i]]$wind_gust <- na.seasplit(wind_gust.ts, algorithm = 'interpolation')[1:nrow(stations_list[[i]])]\n stations_list[[i]]$two_in_soil_temp <- na.seasplit(two_in.ts, algorithm = 'interpolation')[1:nrow(stations_list[[i]])]\n stations_list[[i]]$vwc <- na.interpolation(stations_list[[i]]$vwc, option='spline')\n \n # tryCatch({\n # stations_list[[i]]$vwc <- na.seasplit(vwc.ts, algorithm = 'interpolation')[1:nrow(stations_list[[i]])]\n # }, error = function(e) {\n # print(e)\n # print(paste0('Station: ', stations[i]))\n # })\n }\n final_data <- ldply(stations_list, data.frame)\n return(final_data)\n}",
"_____no_output_____"
],
[
"add_full_timeseries <- function(data){\n require(plyr)\n require(lubridate)\n # Sys.setenv(TZ='PST')\n stations_list <- list()\n stations <- unique(data$station)\n for(i in 1:length(stations)){\n print(stations[i])\n stations_list[[i]] <- data[data[,'station'] == stations[i], ] # subset\n \n county <- unique(stations_list[[i]]$county)\n station <- unique(stations_list[[i]]$station)\n \n date_range <- range(stations_list[[i]]$datetime)\n\n date_seq <- seq(date_range[1], date_range[2], by = '15 mins')\n date_seq <- data.frame('datetime' = as.POSIXct(gsub(date_seq, pattern = ' PST', replacement = ''), format = '%Y-%m-%d %H:%M:%S'))\n stations_list[[i]] <- right_join(stations_list[[i]], date_seq, by='datetime')\n \n stations_list[[i]]$county <- county\n stations_list[[i]]$station <- station\n }\n final <- ldply(stations_list, data.frame)\n return(final)\n}",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecb70fd617df8d11ce5f5889c13f4914112e05ba | 112,743 | ipynb | Jupyter Notebook | getting_started/4_Superdense_coding.ipynb | wkcwells/amazon-braket-examples | 19a11641d77951e6619d7941cd2488242b18e937 | [
"Apache-2.0"
] | 3 | 2021-09-25T11:15:10.000Z | 2022-02-27T15:38:10.000Z | getting_started/4_Superdense_coding.ipynb | wkcwells/amazon-braket-examples | 19a11641d77951e6619d7941cd2488242b18e937 | [
"Apache-2.0"
] | 1 | 2021-09-25T09:09:37.000Z | 2021-09-25T11:17:31.000Z | getting_started/4_Superdense_coding.ipynb | wkcwells/amazon-braket-examples | 19a11641d77951e6619d7941cd2488242b18e937 | [
"Apache-2.0"
] | 3 | 2021-03-02T17:41:27.000Z | 2021-03-02T17:41:44.000Z | 232.459794 | 65,292 | 0.914114 | [
[
[
"# Superdense Coding\nIn this tutorial, we construct an implementation of the superdense coding protocol via Amazon Braket's SDK. Superdense coding is a method of transmitting two classical bits by sending only one qubit. Starting with a pair of entanged qubits, the sender (aka Alice) applies a certain quantum gate to their qubit and sends the result to the receiver (aka Bob), who is then able to decode the full two-bit message.\n\nIf Alice wants to send a two-bit message to Bob using only classical channels, she would need to send two classical bits. However, with the help of quantum entanglement, Alice can do this by sending just one qubit. By ensuring that Alice and Bob initially share an entangled state of two qubits, they can devise a strategy such that Alice can transmit her two-bit message by sending her single qubit.\n\nTo implement superdense coding, Alice and Bob need to share or otherwise prepare a maximally entangled pair of qubits (i.e., a Bell pair). Alice then selects one of the four possible messages to send with two classical bits: 00, 01, 10, or 11. Depending on which two-bit string she wants to send, Alice applies a corresponding quantum gate to encode her desired message. Finally, Alice sends her own qubit to Bob, which Bob then uses to decode the message by undoing the initial entangling operation.\n\nNote that superdense coding is closely related to quantum teleportation. In teleportation, one uses an entangled pair (an e-bit) and two uses of a classical channel to simulate a single use of a quantum channel. In superdense coding, one uses an e-bit and a single use of a quantum channel to simulate two uses of a classical channel.\n\n\n## Detailed Steps\n1. Alice and Bob initially share a Bell pair. This can be prepared by starting with two qubits in the |0⟩ state, then applying the Hadamard gate (𝐻) to the first qubit to create an equal superposition, and finally applying a CNOT gate (𝐶𝑋) between the two qubits to produce a Bell pair. Alice holds one of these two qubits, while Bob holds the other.\n2. Alice selects one of the four possible messages to send Bob. Each message corresponds to a unique set of quantum gate(s) to apply to her own qubit, illustrated in the table below. For example, if Alice wants to send the message \"01\", she would apply the Pauli X gate.\n3. Alice sends her qubit to Bob through the quantum channel.\n4. Bob decodes Alice's two-bit message by first applying a CNOT gate using Alice's qubit as the control and his own qubit as the target, and then a Hadamard gate on Alice's qubit to restore the classical message.\n\n| Message | Alice's encoding | State Bob receives<br>(non-normalized) | After 𝐶𝑋 gate<br>(non-normalized) | After 𝐻 gate |\n| :---: | :---: | :---: | :---: | :---: |\n| 00 | 𝐼 | \\|00⟩ + \\|11⟩ | \\|00⟩ + \\|10⟩ | \\|00⟩\n| 01 | 𝑋 | \\|10⟩ + \\|01⟩ | \\|11⟩ + \\|01⟩ | \\|01⟩\n| 10 | 𝑍 | \\|00⟩ - \\|11⟩ | \\|00⟩ - \\|10⟩ | \\|10⟩\n| 11 | 𝑍𝑋 | \\|01⟩ - \\|10⟩ | \\|01⟩ - \\|11⟩ | \\|11⟩\n\n\n## Circuit Diagram\n\nCircuit used to send the message \"00\". To send other messages, swap out the identity (𝐼) gate.\n",
"_____no_output_____"
],
[
"## Code",
"_____no_output_____"
]
],
[
[
"# Print version of SDK\n!pip show amazon-braket-sdk | grep Version\n\n# Import Braket libraries\nfrom braket.circuits import Circuit, Gate, Moments\nfrom braket.circuits.instruction import Instruction\nfrom braket.aws import AwsDevice\nimport matplotlib.pyplot as plt\nimport time",
"Version: 1.0.0.post1\r\n"
]
],
[
[
"Typically, we recommend running circuits with fewer than 25 qubits on the local simulator to avoid latency bottlenecks. The managed, high-performance simulator SV1 is better suited for larger circuits up to 34 qubits. Nevertheless, for demonstration purposes, we are going to continue this example with SV1 but it is easy to switch over to the local simulator by replacing the last line in the cell below with ```device = LocalSimulator()``` and importing the ```LocalSimulator```.\n\n__NOTE__: Please enter your desired device and S3 location (bucket and key) below. If you are working with the local simulator ```LocalSimulator()``` you do not need to specify any S3 location. However, if you are using the managed cloud-based device or any QPU devices you need to specify the S3 location where your results will be stored. In this case, you need to replace the API call ```device.run(circuit, ...)``` below with ```device.run(circuit, s3_folder, ...)```. ",
"_____no_output_____"
]
],
[
[
"# Please enter the S3 bucket you created during onboarding in the code below\nmy_bucket = f\"amazon-braket-Your-Bucket-Name\" # the name of the bucket\nmy_prefix = \"Your-Folder-Name\" # the name of the folder in the bucket\ns3_folder = (my_bucket, my_prefix)\n\n# Select device arn for the managed simulator\ndevice = AwsDevice(\"arn:aws:braket:::device/quantum-simulator/amazon/sv1\")",
"_____no_output_____"
],
[
"# Function to run quantum task, check the status thereof and collect results\ndef get_result(device, circ, s3_folder):\n \n # get number of qubits\n num_qubits = circ.qubit_count\n\n # specify desired results_types\n circ.probability()\n\n # submit task: define task (asynchronous)\n if device.name == 'DefaultSimulator':\n task = device.run(circ, shots=1000)\n else:\n task = device.run(circ, s3_folder,\n shots=1000, \n poll_timeout_seconds=1000)\n\n # Get ID of submitted task\n task_id = task.id\n# print('Task ID :', task_id)\n\n # Wait for job to complete\n status_list = []\n status = task.state()\n status_list += [status]\n print('Status:', status)\n\n # Only notify the user when there's a status change\n while status != 'COMPLETED':\n status = task.state()\n if status != status_list[-1]:\n print('Status:', status)\n status_list += [status]\n\n # get result\n result = task.result()\n\n # get metadata\n metadata = result.task_metadata\n\n # get output probabilities\n probs_values = result.values[0]\n\n # get measurment results\n measurement_counts = result.measurement_counts\n\n # print measurment results\n print('measurement_counts:', measurement_counts)\n\n # bitstrings\n format_bitstring = '{0:0' + str(num_qubits) + 'b}'\n bitstring_keys = [format_bitstring.format(ii) for ii in range(2**num_qubits)]\n\n # plot probabalities\n plt.bar(bitstring_keys, probs_values);\n plt.xlabel('bitstrings');\n plt.ylabel('probability');\n plt.xticks(rotation=90);\n plt.show() \n \n return measurement_counts",
"_____no_output_____"
]
],
[
[
"Alice and Bob initially share a Bell pair. Let's create this now:",
"_____no_output_____"
]
],
[
[
"circ = Circuit();\ncirc.h([0]);\ncirc.cnot(0,1);",
"_____no_output_____"
]
],
[
[
"Define Alice's encoding scheme according to the table above. Alice selects one of these messages to send.",
"_____no_output_____"
]
],
[
[
"# Four possible messages and their corresponding gates\nmessage = {\"00\": Circuit().i(0),\n \"01\": Circuit().x(0),\n \"10\": Circuit().z(0),\n \"11\": Circuit().x(0).z(0)\n }",
"_____no_output_____"
],
[
"# Select message to send. Let's start with '01' for now\nm = \"01\"",
"_____no_output_____"
]
],
[
[
"Alice encodes her message by applying the gates defined above",
"_____no_output_____"
]
],
[
[
"# Encode the message\ncirc.add_circuit(message[m]);",
"_____no_output_____"
]
],
[
[
"Alice then sends her qubit to Bob so that Bob has both qubits in his lab. Bob decodes Alice's message by disentangling the two qubits:",
"_____no_output_____"
]
],
[
[
"circ.cnot(0,1);\ncirc.h([0]);",
"_____no_output_____"
]
],
[
[
"The full circuit now looks like",
"_____no_output_____"
]
],
[
[
"print(circ)",
"T : |0|1|2|3|4|\n \nq0 : -H-C-X-C-H-\n | | \nq1 : ---X---X---\n\nT : |0|1|2|3|4|\n"
]
],
[
[
"By measuring the two qubits in the computational basis, Bob can read off Alice's two qubit message",
"_____no_output_____"
]
],
[
[
"counts = get_result(device, circ, s3_folder)\nprint(counts)",
"Status: CREATED\nStatus: QUEUED\nStatus: RUNNING\nStatus: COMPLETED\nmeasurement_counts: Counter({'01': 1000})\n"
]
],
[
[
"We can check that this scheme works for the other possible messages too:",
"_____no_output_____"
]
],
[
[
"for m in message:\n \n # Reproduce the full circuit above by concatenating all of the gates:\n newcirc = Circuit().h([0]).cnot(0,1).add_circuit(message[m]).cnot(0,1).h([0]);\n \n # Run the circuit:\n counts = get_result(device, newcirc, s3_folder)\n \n print(\"Message: \" + m + \". Results:\")\n print(counts)",
"Status: CREATED\nStatus: QUEUED\nStatus: RUNNING\nStatus: COMPLETED\nmeasurement_counts: Counter({'00': 1000})\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb7148131a3e60b33f64b085f9cc5aa61b3fc64 | 86,329 | ipynb | Jupyter Notebook | DAY ONE.ipynb | Tech-Buddies/TB-1.0-Beginner | b0ee5ac4da8e0d009513535d92aab90ce7efc74c | [
"MIT"
] | 1 | 2021-12-20T10:44:35.000Z | 2021-12-20T10:44:35.000Z | DAY ONE.ipynb | Tech-Buddies/TB-1.0-Beginner | b0ee5ac4da8e0d009513535d92aab90ce7efc74c | [
"MIT"
] | null | null | null | DAY ONE.ipynb | Tech-Buddies/TB-1.0-Beginner | b0ee5ac4da8e0d009513535d92aab90ce7efc74c | [
"MIT"
] | 1 | 2021-12-19T15:39:16.000Z | 2021-12-19T15:39:16.000Z | 21.739864 | 851 | 0.503921 | [
[
[
"# INTRODUCTION",
"_____no_output_____"
],
[
"## What is Programming?",
"_____no_output_____"
],
[
"Programming is simply the act of instructing the computer on what task to perform. Therefore, a computer program is a set of instructions that tell the computer hardware what to do. Software is a collection of computer program.",
"_____no_output_____"
],
[
"### What is Python?",
"_____no_output_____"
],
[
"Python is a general purpose, high level programming language that was developed by Guido van Rossum in the late 1980s. Like all high level programming languages, Python code resembles the\n\tEnglish language which computers are unable to understand. For the computer to understand the codes, it has to be interpreted by a special piece of software known as interpreter which must be installed before we can code, \t\ttext and execute our python programs.",
"_____no_output_____"
],
[
"### Why learn Python?",
"_____no_output_____"
],
[
"There are many high level programming languages available, such as Java, C++ etc. The good news is all high level programming languages are very similar to one another. What differs is mainly the syntax, the libraries \tavailable and the way we access those libraries. A library is simply a collection of resources and pre-written codes that we can use when we write our programs. Python is a very good place to start for beginners \t\tbecause of its simplicity.",
"_____no_output_____"
],
[
"### What can you do with Python?",
"_____no_output_____"
],
[
"Python can be used for\n1. Web develpoment\n2. Data science/machine learning\n3. Mobile app development\n4. Game development\n5. Hardware and robotics etc..",
"_____no_output_____"
],
[
"## Setting up our coding environment",
"_____no_output_____"
],
[
"There are many Integrated Development Environment available for Python but we'll be using the Anaconda package for this tutorial",
"_____no_output_____"
],
[
"## Python Syntax",
"_____no_output_____"
],
[
"As applied to human languages, syntax in programming language, is the set of rules, principles, and processes that govern the structure of a programme (i.e a set of instruction). Python syntax can be executed by writing directly in our notebook (or interpreter).",
"_____no_output_____"
],
[
"### Writing your first program",
"_____no_output_____"
]
],
[
[
"print('Hello world')\nprint(\"Hello universe\")",
"Hello world\nHello universe\n"
],
[
"print('Pyhon is awesome')",
"Pyhon is awesome\n"
]
],
[
[
"Print is a command (built in function) asking the interpreter to dispaly 'Hello World'. In essence, we have given the computer an instruction to dispaly hello word on the screen.",
"_____no_output_____"
]
],
[
[
"print(2 + 2)",
"4\n"
],
[
"2/8",
"_____no_output_____"
]
],
[
[
"Here, we areasking the interpreter to evaluate the value of 2 + 2",
"_____no_output_____"
]
],
[
[
"5/3",
"_____no_output_____"
],
[
"9//4",
"_____no_output_____"
]
],
[
[
"## Python variables",
"_____no_output_____"
],
[
"Variables are names (i.e containers) given to data that we need to store and manipulate in our programs. For instance, suppose your program needs to store the name of a given user. To do that, we can name this data UserName and define \tthe variable UserName using the following statement below;",
"_____no_output_____"
]
],
[
[
"UserName = 'Ibraheem'",
"_____no_output_____"
]
],
[
[
"Whenever a variable is declared, the program allocates some area of the computer's storage to it, the variable can later be accessed and manipulated. we can as well declare many variables at a go. E.g",
"_____no_output_____"
]
],
[
[
"UserName1 = 'Ibraheem'\nUserAge = 30\nprint(UserName1)\nprint(UserAge)\nprint(f\"username is {UserName1} and the user is {UserAge} years old\")",
"Ibraheem\n30\nusername is Ibraheem and the user is 30 years old\n"
]
],
[
[
"### Naming a Variable",
"_____no_output_____"
],
[
"A variable name in Python can only contain letters (a - z, A - B), numbers or underscores (_). However, the first character cannot be a number. Hence, you can name your variables userName, user_name \tor \t\tuserName2 but not 2userName. In addition, there are some reserved words that you cannot use as a variable name because they already have preassigned meanings in Python. These reserved words include words like \tprint, input, if, while etc. Finally, variable names are case sensitive. username is not the same as userName.\nHint: variable should be descriptive of the value they hold",
"_____no_output_____"
]
],
[
[
"2user_name = 'Basit'",
"_____no_output_____"
]
],
[
[
"### Expressions, Operators and Precedence in Python",
"_____no_output_____"
],
[
"#### Arithmetic Operators\n1. Multiplication *\n2. Addition\t +\n3. Subtraction -\n4. Division\t /\n5. Modulus %\n6. Exponent **\n7. Assigments =",
"_____no_output_____"
],
[
"We also have other types of operators such as logical operators, comparison operators, bitwise operators etc..",
"_____no_output_____"
],
[
"#### Precedence\nProgramming languages follow a strict rule on how compound expressions, such as 4 + 5 * 3, are evaluated. The order in which expressions are evaluated is as follow;\nMultiplication --> Division --> Addition --> Subtraction",
"_____no_output_____"
]
],
[
[
"4 + 5 * 3",
"_____no_output_____"
]
],
[
[
"5 multiply by 3 is evaluated first and the result is added to 4",
"_____no_output_____"
]
],
[
[
"(4 + 5) * 3",
"_____no_output_____"
],
[
"((4 + 5) * 3) / 3",
"_____no_output_____"
],
[
"4 + 5 * 3 /3",
"_____no_output_____"
]
],
[
[
"### Using Variables",
"_____no_output_____"
]
],
[
[
"#Let's calculate the area of a rectangle, given it's legth and breadth\n#first we declare the variables\nLength = 5\nBreadth = 4\nArea = Length * Breadth\nprint(Area)",
"20\n"
]
],
[
[
"Challenge 1: calculate the perimeter",
"_____no_output_____"
]
],
[
[
"Perimeter = 2 * (Length + Breadth)\nprint(Perimeter)",
"18\n"
],
[
"#Let's calculate the area of circle given it's radius\n#first we declare our variables\nradius = 5\npi = 3.142\nArea = (2 * pi * radius**2)\nprint(Area)",
"157.1\n"
]
],
[
[
"Challenge 2: calculate the perimeter",
"_____no_output_____"
]
],
[
[
"# A more elegant way to do this is to use the math module\nimport math\nArea = 2 * math.pi * math.pow(radius,2)\nprint(f\"The area of the circle is {Area:.2f}\")",
"The area of the circle is 157.08\n"
]
],
[
[
"# Data types\nThere are basically five standard types in python",
"_____no_output_____"
],
[
"## 1. Numeric data type\nNumeric data types as the name implies can be of two types namely integer and floating point numbers\n1. int: contains no decimal point e.g 2,5, -4, 0 etc.",
"_____no_output_____"
]
],
[
[
"Weight = -58\nHeight = 79\nprint(type(Weight))\nprint(type(Height))\nprint(type(Weight))",
"<class 'int'>\n<class 'int'>\n<class 'int'>\n"
]
],
[
[
"2.float: real numbers with decima point e.g -0.01, 12.5, 3.6 etc...",
"_____no_output_____"
]
],
[
[
"Weight = 58.2\nHeight = 79.3\nprint(type(Weight))\nprint(type(Height))",
"<class 'float'>\n<class 'float'>\n"
]
],
[
[
"## 2. Textual data type (strings)\n* String is used to store and process textual data. \n* String value must be framed with eithe a single quote(e.g 'text'), or double quotes(e.g \"text\") or triple quotes(e.g '''text''' or \"\"\"text\"\"\").\n* **Note: you must start and end with a type of quotation mark when using triple quote. you cant start with \"\"\" and end with '''**",
"_____no_output_____"
]
],
[
[
"message = 'hello universe! python is awesome'\nmessage_two = \"python programming is superb\"\nmessage_three = \"Laika's dog\"\nprint(message)\nprint(type(message))\nprint(message_two)",
"hello universe! python is awesome\n<class 'str'>\npython programming is superb\n"
],
[
"message1 = \"This is Laika's dog\"\nprint(message1)",
"This is Laika's dog\n"
]
],
[
[
"### String manipulation\nString can be manipulated using a number of in built functions. Function is simply a block of reusable codes that perform a certain task. We'll come back to it later.",
"_____no_output_____"
],
[
"#### Cases I",
"_____no_output_____"
]
],
[
[
"message.capitalize() #capitalizes the first character of the string",
"_____no_output_____"
],
[
"message.title() #capitalizes ever character of the string\nmessage_two.title()",
"_____no_output_____"
],
[
"message.lower() #Every charater of the srting is turned to lower case",
"_____no_output_____"
],
[
"print(message.upper()) #the string is turned to upper case\nprint(message_three.upper())",
"HELLO UNIVERSE! PYTHON IS AWESOME\nLAIKA'S DOG\n"
],
[
"message.swapcase() #the beginning of distinct sentence is swapped",
"_____no_output_____"
]
],
[
[
"#### Sequence Operations",
"_____no_output_____"
]
],
[
[
"print(message)\nprint(max(message))\n#returns largest character...remeber the position of why in English alphabet\nprint(message1)\nprint(max(message1))",
"hello universe! Python is awesome\ny\nThis is Laika's dog\ns\n"
],
[
"min(message)#returns the smallest character",
"_____no_output_____"
],
[
"len(message) #returns the length of the string",
"_____no_output_____"
],
[
"#concatenating\nstring1 = 'good'\nstring2 = 'evening'\nfinal_string = string1 + string2\nfinal_string",
"_____no_output_____"
],
[
"final_string = string1 + ' '+ string2\nprint(final_string)",
"good evening\n"
],
[
"#multiplying strings\nprint(final_string * 2)\nprint(final_string * 3)",
"good eveninggood evening\ngood eveninggood eveninggood evening\n"
],
[
"\nprint(final_string.count('e')) #counts the number of occurence of e\nprint(final_string.count('v'))",
"2\n1\n"
]
],
[
[
"#### Find/Replace",
"_____no_output_____"
]
],
[
[
"message = message.replace('universe', 'world') #replaces a character in the string\nmessage",
"_____no_output_____"
]
],
[
[
"#### Splitting",
"_____no_output_____"
]
],
[
[
"message.split(' ')",
"_____no_output_____"
],
[
"message.splitlines()",
"_____no_output_____"
]
],
[
[
"#### Indexing",
"_____no_output_____"
]
],
[
[
"message[0] #get the charcter at the first index",
"_____no_output_____"
],
[
"message[4]",
"_____no_output_____"
],
[
"message[5]\n",
"_____no_output_____"
],
[
"message[10]",
"_____no_output_____"
],
[
"message[2:8] #get the character fro index 2 to 7",
"_____no_output_____"
],
[
"message[2:8:3] #get the character from index 2 to 9 with step of 2",
"_____no_output_____"
]
],
[
[
"### String Formatting\nThis is just way we display information on the console. I'll show you different ways this can be achieved",
"_____no_output_____"
]
],
[
[
"#Let's declare some variables and have them printed nicely\nUserName = 'Abdullah'\nUserAge = 12",
"_____no_output_____"
]
],
[
[
"#### The old ways of formatting string",
"_____no_output_____"
],
[
"##### using %s and %d where %stands for string and %d stands for digits",
"_____no_output_____"
]
],
[
[
"print(\"The name of the user is %s and the user is %d old.\" %(UserName, UserAge))",
"The name of the user is Abdullah and the user is 12 old.\n"
]
],
[
[
"##### using .format()",
"_____no_output_____"
]
],
[
[
"print(\"The name of the user is {} and the user is {} old\".format(UserName, UserAge))",
"The name of the user is Abdullah and the user is 12 old\n"
],
[
"print(\"The name of the user is {a} and the user is {b} old\".format(a= UserName, b = UserAge))",
"The name of the user is Abdullah and the user is 12 old\n"
]
],
[
[
"#### The new way of formatting using f string",
"_____no_output_____"
]
],
[
[
"print(f\"The name of the user is {UserName} and the user is {UserAge} old\")",
"The name of the user is Abdullah and the user is 12 old\n"
],
[
"#Read on and practice on type casting",
"_____no_output_____"
]
],
[
[
"### Type casting",
"_____no_output_____"
],
[
"it is the process of converting from one data type to another. This can be achieved using the following inbuilt functions\nint(), float() and str(). The int() function takes in a float or appropriate string and converts it to int.",
"_____no_output_____"
]
],
[
[
"int(5.5545)\nprint(int(5.73546)) #will return only 5 because anything after the decimal is removed\nprint(type(int(5.73546)))",
"5\n<class 'int'>\n"
],
[
"print(int(\"4\"))\nprint(type(int(\"4\")))",
"4\n<class 'int'>\n"
]
],
[
[
"However, we can not type int(\"Hello\") as we will run itno error.",
"_____no_output_____"
]
],
[
[
"int(\"Hello\")",
"_____no_output_____"
]
],
[
[
"We can convert int to float by doing this e.g",
"_____no_output_____"
]
],
[
[
"print(float(2))\nprint(type(float(2)))",
"2.0\n<class 'float'>\n"
],
[
"print(str(2))\nprint(type(str(2)))",
"2\n<class 'str'>\n"
]
],
[
[
"## 3. Python Lists\nList refers to a collection of data that are normally related. Instead of storing these variables seperately, we create a list of the data and members are entered within square bracket. Let's say we have a group of dog that we want to deal with in our program, one way to do this is to decare every drog as a varaible and store it E.g",
"_____no_output_____"
]
],
[
[
"dog_breed1 = 'Akita'\ndog_breed2 = 'Beagle'\ndog_breed3 = 'Rettieler'\ndog_breed4 = 'Chiauha'\ndog_breed5 = 'Lowchen'\nprint(f\"{dog_breed1}, {dog_breed2}, {dog_breed3}, {dog_breed4}, {dog_breed5}\")",
"Akita, Beagle, Rettieler, Chiauha, Lowchen\n"
],
[
"#A more elegant and efficient way of doing this is to just use a list\ndog_breed = ['Akita','Beagle', 'Rettieler' ,'Chiauha', 'Lowchen']\nprint(dog_breed)",
"['Akita', 'Beagle', 'Rettieler', 'Chiauha', 'Lowchen']\n"
]
],
[
[
"We can also declare a list without assigning any initial values to it. ",
"_____no_output_____"
]
],
[
[
"users = []",
"_____no_output_____"
]
],
[
[
"What we have now is an empty list with no items in it. We have to use the append() method mentioned below to add items to the list.\n* **Note: modifying the list: lists are mutable, the members can be accessed using the index number with the format: list_name[index no] Individual values in the list are accessible by their indexes, and indexes always start from ZERO (0), not 1**",
"_____no_output_____"
],
[
"### Accessing Values in List: indexing and Slicing",
"_____no_output_____"
]
],
[
[
"print(dog_breed[2])",
"Rettieler\n"
],
[
"dog_breed[2] = 'Laika' #changes the value in index 2 i.e susbstitute \ndog_breed",
"_____no_output_____"
],
[
"print(dog_breed[0])",
"Akita\n"
],
[
"print(dog_breed[1:3])",
"['Beagle', 'Laika']\n"
],
[
"print(dog_breed[4])",
"Lowchen\n"
],
[
"print(dog_breed[-1])",
"Lowchen\n"
],
[
"print(dog_breed[:2])",
"['Akita', 'Beagle']\n"
],
[
"print(dog_breed[2:])",
"['Laika', 'Chiauha', 'Lowchen']\n"
]
],
[
[
"### List Manipulation\n#### 1. Adding Element to the list",
"_____no_output_____"
]
],
[
[
"dog_breed.append('Lebrador')#adds a new element to the end of the list\nprint(dog_breed)",
"['Akita', 'Beagle', 'Laika', 'Chiauha', 'Lowchen', 'Lebrador']\n"
],
[
"dog_breed.insert(3, 'Pulldog') #inserts element in a specified position\ndog_breed.insert(4, 'Great Dane')\nprint(dog_breed)",
"['Akita', 'Beagle', 'Laika', 'Pulldog', 'Great Dane', 'Chiauha', 'Lowchen', 'Lebrador']\n"
],
[
"dog_breed.extend(['Border Collie', 'Dobbermann', 'Irish setter']) #used fro adding multiple elements at a time\nprint(dog_breed)",
"['Akita', 'Beagle', 'Laika', 'Pulldog', 'Great Dane', 'Chiauha', 'Lowchen', 'Lebrador', 'Border Collie', 'Dobbermann', 'Irish setter']\n"
]
],
[
[
"#### 2. Removing element from the list",
"_____no_output_____"
]
],
[
[
"dog_breed.remove('Dobbermann') #removes a specified element from the list\nprint(dog_breed)",
"['Akita', 'Beagle', 'Laika', 'Pulldog', 'Great Dane', 'Chiauha', 'Lowchen', 'Lebrador', 'Border Collie', 'Irish setter']\n"
],
[
"dog_breed.pop(2) #removes element from the list at the specified location\nprint(dog_breed)",
"['Akita', 'Beagle', 'Pulldog', 'Great Dane', 'Chiauha', 'Lowchen', 'Lebrador', 'Border Collie', 'Irish setter']\n"
]
],
[
[
"#### 3. other methods",
"_____no_output_____"
]
],
[
[
"dog_breed.reverse() #reverses the list\ndog_breed",
"_____no_output_____"
],
[
"dog_breed.sort(reverse=True) #sorts the elements of the list in ascending order\ndog_breed",
"_____no_output_____"
],
[
"print(len(dog_breed)) #get the length of the list",
"9\n"
],
[
"print(f\"The dog breed list has {len(dog_breed)} members\")",
"The dog breed list has 9 members\n"
],
[
"print(max(dog_breed)) #gets the largest member of the list; P oocupies the higest position here",
"Pulldog\n"
],
[
"print(min(dog_breed)) #the lowest member interms of English alphabet",
"Akita\n"
],
[
"Akita_index = dog_breed.index('Akita') #gives the location of an element\nprint(f\"Akita is located at index {Akita_index}\")",
"Akita is located at index 8\n"
]
],
[
[
"**\"You might have noticed that methods like insert, remove or sort that only modify the list have no return value printed – they return the default None. 1 This is a design principle for all mutable data structures in Python.\"**",
"_____no_output_____"
],
[
"## 4. Tuples\nThis is another data type like a list but with slight difference. Tuple is immutable (data can't be modified) and it is declared using parethesis i.e ()",
"_____no_output_____"
]
],
[
[
"users = ('Ada', 'Turing', 'Andrew', 'Robert', 'Good Fellow', 'Prince')",
"_____no_output_____"
],
[
"users[1]",
"_____no_output_____"
],
[
"users[-1]",
"_____no_output_____"
],
[
"users[1:4]",
"_____no_output_____"
],
[
"users[-4:-2]",
"_____no_output_____"
],
[
"#Let's try to modify\nusers[1] = 'LoveLace'",
"_____no_output_____"
]
],
[
[
"### Why use a tuple instead of a list?\n* Program execution is faster\n* In case we don't want our data to be modified i.e we want them to remain constant throughout the program",
"_____no_output_____"
],
[
"## 5. Dictionary\nPython dictionary is a collection of data pair, a dictionary consists of a collection of key-value pairs. Each key-value pair maps the key to its associated values. Examples where a dictionary data type is very useful includes:\n1. Storing every country of the world and their capital\n2. We want to store usernames and their corresponding email etc.\n\n**A dictionary is defined by enclosing a comma-separated list of key-value pairs in curly braces ({}). A colon (:) separates each key from its associated value:**",
"_____no_output_____"
]
],
[
[
"Countries = {'China' : \"Beijing\", \n 'Mali' : \"Bamako\", \n 'Israel': 'Jerusalem', \n 'Nigeria': 'Abuja', \n 'Mexico':'Mexico City',\n 'Japan': 'Tokyo',\n 'Iran': 'Tehran'}",
"_____no_output_____"
],
[
"data1 = {\"Sout America\": [\"Haiti\",\"Argetina\", \"Brazil\"],\n \"Africa\": [\"Kenya\", \"Morocco\", \"Libya\"]}\nprint(data1[\"Africa\"])",
"['Kenya', 'Morocco', 'Libya']\n"
]
],
[
[
"**Dictionaries can also be defined using the *dict()* bult in function**",
"_____no_output_____"
]
],
[
[
"countries = dict(China = \"Beijing\", \n Mali = \"Bamako\", \n Israel = 'Jerusalem', \n Nigeria = 'Abuja', \n Mexico = 'Mexico City',\n Japan = 'Tokyo',\n Iran = 'Tehran')\ncountries",
"_____no_output_____"
]
],
[
[
"### Accessing dictionary values",
"_____no_output_____"
]
],
[
[
"countries['China']",
"_____no_output_____"
],
[
"countries['Iran']",
"_____no_output_____"
]
],
[
[
"#### Adding an entry to an existing dictionary",
"_____no_output_____"
]
],
[
[
"countries['Germany'] = 'Moscow'\ncountries",
"_____no_output_____"
]
],
[
[
"#### Updating an entry",
"_____no_output_____"
]
],
[
[
"countries['Israel'] = 'Tel Aviv'\ncountries['Israel']",
"_____no_output_____"
]
],
[
[
"#### We can delete an entry",
"_____no_output_____"
]
],
[
[
"del countries['Mali']\ncountries",
"_____no_output_____"
]
],
[
[
"### Manipulating our dictionary",
"_____no_output_____"
],
[
"#### We can get a value by specifying its key",
"_____no_output_____"
]
],
[
[
"countries.get('Iran')",
"_____no_output_____"
]
],
[
[
"#### We can get the items in our dictionary",
"_____no_output_____"
]
],
[
[
"countries.items()",
"_____no_output_____"
]
],
[
[
"#### We can get the leys and the values",
"_____no_output_____"
]
],
[
[
"countries.keys()",
"_____no_output_____"
],
[
"countries.values()",
"_____no_output_____"
]
],
[
[
"#### We can get rid of a key and return it's value",
"_____no_output_____"
]
],
[
[
"countries.pop('Germany')",
"_____no_output_____"
],
[
"countries",
"_____no_output_____"
]
],
[
[
"#### We can remove the last key value pair ",
"_____no_output_____"
]
],
[
[
"countries.popitem()",
"_____no_output_____"
],
[
"countries",
"_____no_output_____"
]
],
[
[
"#### We can merge another dictionary to the existing one ",
"_____no_output_____"
]
],
[
[
"another_countries = {'Libya':'Tripoli',\n 'Netherland':'Amstredam',\n 'Ghana': 'Accra'}\ncountries.update(another_countries)",
"_____no_output_____"
],
[
"countries",
"_____no_output_____"
]
],
[
[
"## Control Structure in Python",
"_____no_output_____"
],
[
"What wehave been delaling with so far is a sequecial execution of instruction in which statements are always performed one after the next, in exactly the order specified. However, the world is much more complicated and our program will often need to skip over some statements, execute a series of statements repetitively, or choose between alternate sets of statements to execute.\n**A control structure therefore directs the order of execution of the statements in a program in a version referred to as control flow**\n\n* The **if** statement is the most commonnly used conditional statement in Python and it is used to perform this sort of decision-making. It allows for conditional execution of a statement or group of statements based on the value of an expression.\n",
"_____no_output_____"
]
],
[
[
"#if condition is met:\n# do A",
"_____no_output_____"
],
[
"standard_height = 6.0\napplicant_height = 5.6\nif applicant_height < standard_height:\n print(\"You can't apply for this job\")\n",
"You can't apply for this job\n"
],
[
"x = 45\ny = 40\nif x > y:\n print(f\"{x} is greater than {y}\")",
"45 is greater than 40\n"
]
],
[
[
"The **else** and **elif** clauses",
"_____no_output_____"
]
],
[
[
"standard_height = 6.0\napplicant_height = 6.1\nif applicant_height < standard_height:\n print(\"You can't apply for this job\")\nelse:\n print(\"You may proceed with your application\")",
"You may proceed with your application\n"
],
[
"age = int(input(\"Enter your age \"))\n#name = input(\"Enter your name: \")\nif age < 18:\n print(\"You are not qualified\")\nelse:\n print(\"You may proceed with the application\")\n \n",
"Enter your age 24\nYou may proceed with the application\n"
],
[
"name = \"Abdullah\"\nif name == \"Basit\":\n print(\"welcome Basit\")\nelif name == \"Surur\":\n print(\"Welcome Surur\")\nelif name == \"Abdullah\":\n print(\"Hello Abdullah\")\nelse:\n print(\"Your name is not known\")",
"Hello Abdullah\n"
],
[
"course_of_choice = input(\"Which course do you want to study \").lower()\n\navailable_courses = ['physics', 'agriculture', 'law', 'medicine', 'accounting']\n\nif course_of_choice in available_courses:\n print(\"Congratulations, the university offers your course of choice\")\nelse:\n print(\"Your chosen course is not availble, please apply somewhere else\")",
"Which course do you want to study LaW\nCongratulations, the university offers your course of choice\n"
]
],
[
[
"We also have three useful logical operators that can be \t\tcombined for multiple conditions. They are: **AND, OR, NOT**",
"_____no_output_____"
],
[
"### for loop",
"_____no_output_____"
],
[
"The for loop executes a \tblock of code\n\trepeatedly until the condition in the for statement is no longer \tvalid.",
"_____no_output_____"
]
],
[
[
"for i in range(5):\n print(\"ll be successful in this life in shaa Allah\")",
"ll be successful in this life in shaa Allah\nll be successful in this life in shaa Allah\nll be successful in this life in shaa Allah\nll be successful in this life in shaa Allah\nll be successful in this life in shaa Allah\n"
],
[
"for course in available_courses:\n print(course)",
"physics\nagriculture\nlaw\nmedicine\naccounting\n"
],
[
"number = [1,2,3,4,5]\nnumber_square = []\n\nfor n in number:\n number_square.append(n**2)",
"_____no_output_____"
],
[
"print(number_square)",
"[1, 4, 9, 16, 25]\n"
]
],
[
[
"### break statement",
"_____no_output_____"
]
],
[
[
"for course in available_courses:\n if course == \"law\":\n break #the iteration stops at law\n print(course)",
"physics\nagriculture\n"
]
],
[
[
"### continue statement",
"_____no_output_____"
]
],
[
[
"for course in available_courses:\n if course == \"medicine\":\n continue #medicine is skipped\n print(course)",
"physics\nagriculture\nlaw\naccounting\n"
]
],
[
[
"### While loop",
"_____no_output_____"
],
[
"A while loop in python is used to iterate over a block of code or statements as long as the test expression is true",
"_____no_output_____"
]
],
[
[
"num =int(input('enter a number'))\nwhile num >= 0:\n if num == 0:\n print('equal to zero')\n elif num > 0:\n print('greater than zero')\n else:\n print('enter a valid number')\n break",
"enter a number4\ngreater than zero\n"
],
[
"i = 1\nwhile i <= 5 :\n print(i)\n #if i == 4: #the loop breaks as soon as the iteration gets to 4\n # break\n i = i+1",
"1\n2\n3\n4\n"
]
],
[
[
"## Functions",
"_____no_output_____"
],
[
"Functions are simply pre-written codes that perform a certain task, they are primary unit of re-usable codes. For an analogy, think of the mathematical functions available in MS Excel. To add numbers, we can use the \tsum() function and type sum(A1:A5) instead of typing A1+A2+A3+A4+A5.\n* Functions allow us to conveniently divide our codes into useful block in an ordered version. make it more readable, reusable and save us time.",
"_____no_output_____"
]
],
[
[
"def my_function():\n print(\"This is my first function\")\n \nmy_function()",
"This is my first function\n"
]
],
[
[
"Functions may also receive arguments (variables passed from the caller to the function). For example:\n\n",
"_____no_output_____"
]
],
[
[
"def hello(name):\n print(f\"hello {name}!\")\n \nhello('Adam')",
"hello Adam!\n"
]
],
[
[
"Functions may return a value to the caller, using the keyword- **return** . For example:",
"_____no_output_____"
]
],
[
[
"def simple_calc(a,b):\n return a+b\n\nsimple_calc(2,5)",
"_____no_output_____"
],
[
"simple_calc(-3, 8)",
"_____no_output_____"
],
[
"def rect_calc(length = 3, breadth = 2): #initializing functions and keyword argument\n return length * breadth\nrect_calc()",
"_____no_output_____"
],
[
"rect_calc(6,7)",
"_____no_output_____"
],
[
"rect_calc(4,6) #the default values are over ridden",
"_____no_output_____"
],
[
"def week_day(num):\n if num == 0:\n print(\"today is Sunday\")\n elif num == 1:\n print(\"today is Monday\")\n elif num == 2:\n print(\"Today is Tuesday\")\n elif num == 3:\n print(\"Today is Wednesday\")\n elif num == 4:\n print(\"Today is Thursday\")\n elif num== 5:\n print(\"Today is Friday\")\n elif num == 6:\n print(\"Today is Saturday\")\n else:\n print(\"You entered a wrong input\")\n \nweek_day(3)",
"Today is Wednesday\n"
],
[
"week_day(4)",
"Today is Thursday\n"
],
[
"week_day(20)",
"You entered a wrong input\n"
]
],
[
[
"### Variable scope",
"_____no_output_____"
],
[
"variable scope is an important concept in functions. Variables declared inside a function (called local variable) is treated differently from those defined outside the function (called global variable). Things to remeber when dealing with variables at different levels:\n1. Code in the global scope cannot use any local variables.\n2. However, a local scope can access global variables.\n3. Code in a function’s local scope cannot use variables in any other local scope.\n4. You can use the same name for different variables if they are in different scopes. That is, there can be a local variable named spam and a global variable also named spam.\n\n\n**Code in the global scope cannot use any local variables.**",
"_____no_output_____"
]
],
[
[
"def count_egg():\n egg = 250\n \n \n\nprint(egg)",
"_____no_output_____"
]
],
[
[
"**However, a local scope can access global variables.**",
"_____no_output_____"
]
],
[
[
"PI = 3.142\n\ndef calc_area(radius):\n return PI * radius **2\ncalc_area(3)",
"_____no_output_____"
]
],
[
[
"**Code in a function’s local scope cannot use variables in any other local scope.**",
"_____no_output_____"
]
],
[
[
"def calc_circum():\n return 2 * PI * radius\n\ncalc_circum(2)",
"_____no_output_____"
]
],
[
[
"The **global** keyword is used if we need to modify a global variable within a function",
"_____no_output_____"
]
],
[
[
"DISTANCE = 42\n\ndef calc_speed(time):\n global DISTANCE\n DISTANCE = 40\n speed = DISTANCE/time\n return speed\ncalc_speed(20)",
"_____no_output_____"
]
],
[
[
"### Exception Handling",
"_____no_output_____"
],
[
"Right now, getting an error, or exception, in your Python program means the\nentire program will crash. You don’t want this to happen in real-world programs.\nInstead, you want the program to detect errors, handle them, and\nthen continue to run.",
"_____no_output_____"
]
],
[
[
"def divide(num1,num2):\n return num1/num2\n\ndivide(20,2)",
"_____no_output_____"
],
[
"divide(10,3)",
"_____no_output_____"
],
[
"divide(3,0)",
"_____no_output_____"
]
],
[
[
"The error occurs because we can not divide a number by zero. we can handle this in order to prevent our program fro crashing completely by using **try** and **except** keywords",
"_____no_output_____"
]
],
[
[
"def division(num1,num2):\n try:\n return num1/num2\n except(ZeroDivisionError):\n return (\"can not divide by zero\")\n \ndivision(2,0)",
"_____no_output_____"
],
[
"division(3,2)",
"_____no_output_____"
],
[
"def division():\n \n while True:\n num1 = int(input(\"Enter a number \"))\n num2 = int(input(\"Enter a second number \"))\n \n try:\n print(num1/num2)\n break #the loop execution stops if the condition is satisfied\n except(ZeroDivisionError):\n print (\"can not divide by zero\")\ndivision()",
"Enter a number 4\nEnter a second number 0\ncan not divide by zero\nEnter a number 5\nEnter a second number 0\ncan not divide by zero\nEnter a number 4\nEnter a second number 0\ncan not divide by zero\nEnter a number 2\nEnter a second number 3\n0.6666666666666666\n"
]
],
[
[
"### Python modules",
"_____no_output_____"
],
[
"a module is a file consisting of Python code. It can define functions, classes, and variables, and can also include runnable code. Any Python file can be referenced as a module. A file containing Python code, for example: test.py, is called a module, and its name would be test. To use the built-in codes in Python modules, we have to import them into our programs first. We do that by using the **import** keyword. Classes and Functions available in modules are then called using dot (.)",
"_____no_output_____"
]
],
[
[
"import math\n\nmath.sqrt(9)",
"_____no_output_____"
],
[
"math.pow(3,2)",
"_____no_output_____"
],
[
"math.exp(10)",
"_____no_output_____"
],
[
"math.fabs(-3)",
"_____no_output_____"
],
[
"math.factorial(5)",
"_____no_output_____"
],
[
"math.log(3.33)",
"_____no_output_____"
],
[
"math.sin(45)",
"_____no_output_____"
],
[
"import random\n\nrandom.randint(2,10)",
"_____no_output_____"
],
[
"\nrandom.randint(2,10)",
"_____no_output_____"
],
[
"random.randrange(1,20,2) ",
"_____no_output_____"
],
[
"hay = \"make hay while the sun shines\"\nhay_to_list = list(hay.split(' '))\nhay_to_list",
"_____no_output_____"
],
[
"random.choice(hay_to_list) # a random element is selected from the list",
"_____no_output_____"
],
[
"random.shuffle(hay_to_list) #the list is reorderd\nhay_to_list",
"_____no_output_____"
]
],
[
[
"### WORKING WITH FILES",
"_____no_output_____"
]
],
[
[
"f = open(r\"C:\\Users\\ibrom\\Documents\\Ass 2.txt\", 'r')\n\nfirstline = f.readline()\nsecondline = f.readline()\nprint(firstline)\nprint(secondline)\nf.close() ",
"NUMBERS:\n\n1- what is the difference between / and % (e.g 5/2, 5%2).\n\n"
]
],
[
[
"In addition to using the readline() method above to read a text file, we\n\tcan also use a for loop. In fact, the for loop is a more elegant and\n\tefficient way to read text files. The following program shows how this is\n\tdone.",
"_____no_output_____"
]
],
[
[
"f = open ('myfile.txt', 'r')\nfor line in f:\n print (line, end = '')\n ",
"In addition to using the readline() method above to read a text file, we can also use a for loop. In fact, the for loop is a more elegant and efficient way to read text files. The following program shows how this is done.\n"
],
[
"with open('myfile.txt', 'r') as f:\n for line in f:\n print(line, end = '')",
"In addition to using the readline() method above to read a text file, we can also use a for loop. In fact, the for loop is a more elegant and efficient way to read text files. The following program shows how this is done."
]
],
[
[
"We can also read docx, pdf,excelcsv etc....",
"_____no_output_____"
],
[
"**Challenges**\n1. Write a function that takes an integer days and converts it to seconds. E.g convert(2) ➞ 172800\n2. Create a function that returns True when num1 is equal to num2; otherwise return False. E.g is_same_num(4, 8) ➞ False\n3. Create a function that takes the number of wins, draws and losses and calculates the number of points a football team has obtained so far. A win receives 3 points, a draw 1 point and a loss 0 points. E.g football_points(3, 4, 2) ➞ 13\n4. Create a function that takes a list and returns the first element. E.g get_first_value([-500, 0, 50]) ➞ -500\n5. It is possible to name the days 0 thru 6 where day 0 is Sunday and day 6 is Saturday. If you go on a wonderful holiday leaving on day number 3 (a Wednesday) and you return home after 10 nights you would return home on a Saturday (day 6). Write a general version of the program which asks for the starting day number, and the length of your stay, and it will tell you the number of day of the week you will return on.",
"_____no_output_____"
]
],
[
[
"def point(win, draw, loss):\n \n #win, draw, loss = win*3, draw*1, loss *0\n win = win*3\n draw = draw*1\n loss = loss*0\n return win+draw+loss\n\npoint(3,4,2)",
"_____no_output_____"
],
[
"def convert_day(day):\n return day *24*3600\nconvert_day(2)\n",
"_____no_output_____"
],
[
"def is_same_num(num1,num2):\n if num1 == num2:\n return True\n return False\nis_same_num(2,4)",
"_____no_output_____"
],
[
"is_same_num(3,3)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecb714db22c719f9807a288950aeb0fd6ee6e206 | 11,914 | ipynb | Jupyter Notebook | test/cs/Snowdepthtrans.ipynb | Crop2ML-Catalog/STICS_SNOW | 26ed2a9a30e068d72d5589b1dc64916c1b07fa09 | [
"MIT"
] | null | null | null | test/cs/Snowdepthtrans.ipynb | Crop2ML-Catalog/STICS_SNOW | 26ed2a9a30e068d72d5589b1dc64916c1b07fa09 | [
"MIT"
] | null | null | null | test/cs/Snowdepthtrans.ipynb | Crop2ML-Catalog/STICS_SNOW | 26ed2a9a30e068d72d5589b1dc64916c1b07fa09 | [
"MIT"
] | null | null | null | 30.315522 | 114 | 0.407504 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecb7406d0a2ae8a1b1a6274a4e26c18e8d019ad5 | 21,012 | ipynb | Jupyter Notebook | 1_data_manipulations/Seminar_pandas.ipynb | aslamovyura/ml-python | b78964392d9144e5838d5e2dd7b3b23e18dcd544 | [
"MIT"
] | null | null | null | 1_data_manipulations/Seminar_pandas.ipynb | aslamovyura/ml-python | b78964392d9144e5838d5e2dd7b3b23e18dcd544 | [
"MIT"
] | null | null | null | 1_data_manipulations/Seminar_pandas.ipynb | aslamovyura/ml-python | b78964392d9144e5838d5e2dd7b3b23e18dcd544 | [
"MIT"
] | null | null | null | 18.319093 | 349 | 0.476537 | [
[
[
"# NumPy",
"_____no_output_____"
],
[
"**Numeric Python** is the core library for scientific computing in Python. It provides a high-performance multidimensional array object, and tools for working with these arrays.",
"_____no_output_____"
],
[
"The First Rule of NumPy",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"The Second Rule of NumPy",
"_____no_output_____"
]
],
[
[
"# You don't need cycles\n\nfirst_arr = np.array([1, 2, 3, 4, 5])\nsecond_arr = np.copy(first_arr)\n\n# Instead of\nfor i in range(len(arr)):\n if first_arr[i] == 3 or first_arr[i] == 4:\n first_arr[i] = 0\n# Do\nsecond_arr[(second_arr == 4) | (second_arr == 3)] = 0\n\nassert((first_arr == second_arr).all())",
"_____no_output_____"
]
],
[
[
"Array creation",
"_____no_output_____"
]
],
[
[
"a = np.array([1, 2, 3, 4, 5, 6, 7, 8, 8])\nprint(type(a))\nprint(a.shape)\nprint(a.dtype)",
"_____no_output_____"
]
],
[
[
"Assigning and appending an element to an array",
"_____no_output_____"
]
],
[
[
"a[8] = 9\na = np.append(a, 10)\nprint(a)",
"_____no_output_____"
]
],
[
[
"Standard Python slicing syntax",
"_____no_output_____"
]
],
[
[
"print(a)\nprint('\\n')\n\nprint(a[0:5])\nprint(a[0:5:2])\nprint(a[0:-1])\nprint(a[4::-1])\nprint(a[5:0:-2])",
"_____no_output_____"
]
],
[
[
"In-built calculation of different statistics",
"_____no_output_____"
]
],
[
[
"print('Vector max %d, min %d, mean %.2f, median %.2f, stardard deviation %.2f and total sum %d' %\n (a.max(), np.min(a), a.mean(), np.median(a), a.std(), a.sum()))",
"_____no_output_____"
]
],
[
[
"Filtering on condition (masking)",
"_____no_output_____"
]
],
[
[
"a[a > a.mean()]",
"_____no_output_____"
]
],
[
[
"Sorting",
"_____no_output_____"
]
],
[
[
"# Sorted array\nprint(np.sort(a))\n\n# Order of indices in sorted array\nprint(np.argsort(a))",
"_____no_output_____"
]
],
[
[
"Vector operations",
"_____no_output_____"
]
],
[
[
" a = np.array([1, 2, 3])\nb = np.array([2, 3, 4])\na * b\na - b\na + b",
"_____no_output_____"
]
],
[
[
"2D arrays (matrices)",
"_____no_output_____"
]
],
[
[
"m_a = np.array([[1, 2, 3, 4]\n ,[13, 3, 8, 2]\n ,[8, 7, 2, 3]])\nprint(m_a.shape)",
"_____no_output_____"
]
],
[
[
"Statistics calculation",
"_____no_output_____"
]
],
[
[
"print(m_a.max())\nprint(m_a.max(axis=0))\nprint(m_a.max(axis=1))",
"_____no_output_____"
]
],
[
[
"Sorting",
"_____no_output_____"
]
],
[
[
"print(np.sort(m_a, axis=0))\nprint(np.sort(m_a, axis=1))",
"_____no_output_____"
]
],
[
[
"# Intro to Pandas data structures",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"## Series",
"_____no_output_____"
],
[
"[Series](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.Series.html#pandas.Series) is a one-dimensional labeled array capable of holding any data type (integers, strings, floating point numbers, Python objects, etc.). The axis labels are collectively referred to as the index. The basic method to create a Series is to call:",
"_____no_output_____"
]
],
[
[
"s = pd.Series(data=[39.4, 91.2, 80.5, 20.3, 4.2, -13.4]\n ,index=['first', 'second', 'second', 'third', 'forth', 'fifth'])\nprint(type(s))\nprint(s.shape)\nprint(s.dtype)\nprint(s['second'])",
"_____no_output_____"
],
[
"s = pd.Series(data=[39.4, 91.2, 20.3, 4.2, -13.4])\ns",
"_____no_output_____"
]
],
[
[
"Series acts very similarly to a [ndarray](https://numpy.org/doc/stable/reference/generated/numpy.ndarray.html), and is a valid argument to most [NumPy](https://numpy.org/doc/stable/user/whatisnumpy.html) functions. However, operations such as slicing will also slice the index.",
"_____no_output_____"
]
],
[
[
"s[1:4]",
"_____no_output_____"
]
],
[
[
"In-built statistics calculation is the same as in NumPy",
"_____no_output_____"
]
],
[
[
"np.max(s)\ns.min()\ns.std()",
"_____no_output_____"
]
],
[
[
"Vector operations",
"_____no_output_____"
]
],
[
[
"a = pd.Series([1, 2, 3])\nb = pd.Series([2, 3, 4])\n\na * 2\na + b\na - b\na * b",
"_____no_output_____"
]
],
[
[
"## DataFrame",
"_____no_output_____"
],
[
"DataFrame is a 2-dimensional labeled data structure with columns of potentially different types. You can think of it like a spreadsheet or SQL table, or a dict of Series objects. It is generally the most commonly used pandas object.",
"_____no_output_____"
],
[
"DataFrame creation",
"_____no_output_____"
]
],
[
[
"d = {\"one\": pd.Series([1.0, 2.0, 3.0], index=[\"a\", \"b\", \"c\"]),\n \"two\": pd.Series([1.0, 2.0, 3.0, 4.0], index=[\"a\", \"b\", \"c\", \"d\"]),\n }\ndf = pd.DataFrame(d)\ndf",
"_____no_output_____"
],
[
"d = {\"one\": [1.0, 2.0, 3.0, 4.0], \"two\": [4.0, 3.0, 2.0, 1.0]}\ndf = pd.DataFrame(d)\ndf",
"_____no_output_____"
]
],
[
[
"### Basic transformations",
"_____no_output_____"
],
[
"Loading existing DataFrame from csv file. We'll use Google Play Store Apps dataset from here https://www.kaggle.com/lava18/google-play-store-apps.",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv('googleplaystore.csv')\ndata.head(3)",
"_____no_output_____"
]
],
[
[
"Column types",
"_____no_output_____"
]
],
[
[
"data.dtypes",
"_____no_output_____"
]
],
[
[
"Column selection as DataFrame",
"_____no_output_____"
]
],
[
[
"data[['App', 'Rating']].head()",
"_____no_output_____"
]
],
[
[
"Column selection as Series. You can treat a DataFrame semantically like a dict of like-indexed Series objects.",
"_____no_output_____"
]
],
[
[
"data['Rating'].head()",
"_____no_output_____"
],
[
"type(data['Rating'])",
"_____no_output_____"
],
[
"data['Rating'].mean()",
"_____no_output_____"
]
],
[
[
"Row selection by index",
"_____no_output_____"
]
],
[
[
"data.iloc[1]",
"_____no_output_____"
]
],
[
[
"Filtering (row selection by condition)",
"_____no_output_____"
]
],
[
[
"data[data['Rating'] < 3].head()",
"_____no_output_____"
]
],
[
[
"Assigning value to column based on condition",
"_____no_output_____"
]
],
[
[
"# Not correct\ndata[data['Rating'] < 3]['Rating'] = 0",
"_____no_output_____"
],
[
"data[data['Rating'] < 3].head(3)",
"_____no_output_____"
],
[
"# Correct\ndata.loc[data['Rating'] < 3, 'Rating'] = 0",
"_____no_output_____"
],
[
"data[data['Rating'] < 3].head(3)",
"_____no_output_____"
]
],
[
[
"Sorting",
"_____no_output_____"
]
],
[
[
"data.sort_values(by='Rating').head(3)",
"_____no_output_____"
]
],
[
[
"Selecting values",
"_____no_output_____"
]
],
[
[
"data.head()['App'].values.tolist()",
"_____no_output_____"
]
],
[
[
"**All together**. Make list of top-5 Free Apps by Rating in Education Genres by alphabet order",
"_____no_output_____"
]
],
[
[
"data[(data['Type'] == 'Free') & (data['Genres'] == 'Education')]\\\n .sort_values(by=['Rating', 'App'], ascending=(False, True))\\\n .head(5)['App'].values.tolist()",
"_____no_output_____"
]
],
[
[
"### Concatenating",
"_____no_output_____"
],
[
"Appending DataFrames",
"_____no_output_____"
]
],
[
[
"df1 = pd.DataFrame({\n \"A\": [\"A0\", \"A1\", \"A2\", \"A3\"],\n \"B\": [\"B0\", \"B1\", \"B2\", \"B3\"],\n \"C\": [\"C0\", \"C1\", \"C2\", \"C3\"],\n \"D\": [\"D0\", \"D1\", \"D2\", \"D3\"],},\n index=[0, 1, 2, 3],)\n\ndf2 = pd.DataFrame({\n \"A\": [\"A4\", \"A5\", \"A6\", \"A7\"],\n \"B\": [\"B4\", \"B5\", \"B6\", \"B7\"],\n \"C\": [\"C4\", \"C5\", \"C6\", \"C7\"],\n \"E\": [\"E4\", \"E5\", \"E6\", \"E7\"],},\n index=[0, 1, 2, 3],)",
"_____no_output_____"
],
[
"df1",
"_____no_output_____"
],
[
"df2",
"_____no_output_____"
],
[
"df1.append(df2, ignore_index=True)",
"_____no_output_____"
]
],
[
[
"**Join** methon works better with joining DataFrame by indices and is fine-tuned by default to do it.",
"_____no_output_____"
]
],
[
[
"df3 = pd.DataFrame({\n \"A\": [\"A1\", \"A2\", \"A3\", \"A4\"],\n \"F\": [\"F0\", \"F1\", \"F2\", \"F3\"],},\n index=[0, 1, 2, 3],)",
"_____no_output_____"
],
[
"df1",
"_____no_output_____"
],
[
"df3",
"_____no_output_____"
],
[
"df1.join(df3, how='inner', lsuffix='_first', rsuffix='_third')",
"_____no_output_____"
]
],
[
[
"**Merge** method is more versatile and allows us to specify columns besides the index to join on for both dataframes.",
"_____no_output_____"
]
],
[
[
"df1.merge(df3, how='left', on=['A'])",
"_____no_output_____"
]
],
[
[
"### Grouping",
"_____no_output_____"
],
[
"A groupby operation involves some combination of splitting the object, applying a function, and combining the results. This can be used to group large amounts of data and compute operations on these groups",
"_____no_output_____"
]
],
[
[
"data.head(3)",
"_____no_output_____"
]
],
[
[
"Find _Category_ having the highest average rating among it's applications. No cycles, I promise.",
"_____no_output_____"
]
],
[
[
"data.groupby('Category')['Rating'].mean().sort_values(ascending=False).index[1]",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb761dc204916b9dee40a76691ee9c00699df10 | 59,699 | ipynb | Jupyter Notebook | site/en-snapshot/tfx/tutorials/tfx/components_keras.ipynb | phoenix-fork-tensorflow/docs-l10n | 2287738c22e3e67177555e8a41a0904edfcf1544 | [
"Apache-2.0"
] | 1 | 2021-11-07T18:53:46.000Z | 2021-11-07T18:53:46.000Z | site/en-snapshot/tfx/tutorials/tfx/components_keras.ipynb | phoenix-fork-tensorflow/docs-l10n | 2287738c22e3e67177555e8a41a0904edfcf1544 | [
"Apache-2.0"
] | null | null | null | site/en-snapshot/tfx/tutorials/tfx/components_keras.ipynb | phoenix-fork-tensorflow/docs-l10n | 2287738c22e3e67177555e8a41a0904edfcf1544 | [
"Apache-2.0"
] | null | null | null | 37.904127 | 538 | 0.561132 | [
[
[
"##### Copyright 2021 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# TFX Keras Component Tutorial\n\n***A Component-by-Component Introduction to TensorFlow Extended (TFX)***",
"_____no_output_____"
],
[
"Note: We recommend running this tutorial in a Colab notebook, with no setup required! Just click \"Run in Google Colab\".\n\n<div class=\"devsite-table-wrapper\"><table class=\"tfo-notebook-buttons\" align=\"left\">\n<td><a target=\"_blank\" href=\"https://www.tensorflow.org/tfx/tutorials/tfx/components_keras\">\n<img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a></td>\n<td><a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/tfx/blob/master/docs/tutorials/tfx/components_keras.ipynb\">\n<img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\">Run in Google Colab</a></td>\n<td><a target=\"_blank\" href=\"https://github.com/tensorflow/tfx/tree/master/docs/tutorials/tfx/components_keras.ipynb\">\n<img width=32px src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\">View source on GitHub</a></td>\n<td><a target=\"_blank\" href=\"https://storage.googleapis.com/tensorflow_docs/tfx/docs/tutorials/tfx/components_keras.ipynb\">\n<img width=32px src=\"https://www.tensorflow.org/images/download_logo_32px.png\">Download notebook</a></td>\n</table></div>",
"_____no_output_____"
],
[
"This Colab-based tutorial will interactively walk through each built-in component of TensorFlow Extended (TFX).\n\nIt covers every step in an end-to-end machine learning pipeline, from data ingestion to pushing a model to serving.\n\nWhen you're done, the contents of this notebook can be automatically exported as TFX pipeline source code, which you can orchestrate with Apache Airflow and Apache Beam.\n\nNote: This notebook demonstrates the use of native Keras models in TFX pipelines. **TFX only supports the TensorFlow 2 version of Keras**.",
"_____no_output_____"
],
[
"## Background\nThis notebook demonstrates how to use TFX in a Jupyter/Colab environment. Here, we walk through the Chicago Taxi example in an interactive notebook.\n\nWorking in an interactive notebook is a useful way to become familiar with the structure of a TFX pipeline. It's also useful when doing development of your own pipelines as a lightweight development environment, but you should be aware that there are differences in the way interactive notebooks are orchestrated, and how they access metadata artifacts.\n\n### Orchestration\n\nIn a production deployment of TFX, you will use an orchestrator such as Apache Airflow, Kubeflow Pipelines, or Apache Beam to orchestrate a pre-defined pipeline graph of TFX components. In an interactive notebook, the notebook itself is the orchestrator, running each TFX component as you execute the notebook cells.\n\n### Metadata\n\nIn a production deployment of TFX, you will access metadata through the ML Metadata (MLMD) API. MLMD stores metadata properties in a database such as MySQL or SQLite, and stores the metadata payloads in a persistent store such as on your filesystem. In an interactive notebook, both properties and payloads are stored in an ephemeral SQLite database in the `/tmp` directory on the Jupyter notebook or Colab server.",
"_____no_output_____"
],
[
"## Setup\nFirst, we install and import the necessary packages, set up paths, and download data.",
"_____no_output_____"
],
[
"### Upgrade Pip\n\nTo avoid upgrading Pip in a system when running locally, check to make sure that we're running in Colab. Local systems can of course be upgraded separately.",
"_____no_output_____"
]
],
[
[
"try:\n import colab\n !pip install --upgrade pip\nexcept:\n pass",
"_____no_output_____"
]
],
[
[
"### Install TFX\n\n**Note: In Google Colab, because of package updates, the first time you run this cell you must restart the runtime (Runtime > Restart runtime ...).**",
"_____no_output_____"
]
],
[
[
"!pip install -U tfx",
"_____no_output_____"
]
],
[
[
"## Did you restart the runtime?\n\nIf you are using Google Colab, the first time that you run the cell above, you must restart the runtime (Runtime > Restart runtime ...). This is because of the way that Colab loads packages.",
"_____no_output_____"
],
[
"### Import packages\nWe import necessary packages, including standard TFX component classes.",
"_____no_output_____"
]
],
[
[
"import os\nimport pprint\nimport tempfile\nimport urllib\n\nimport absl\nimport tensorflow as tf\nimport tensorflow_model_analysis as tfma\ntf.get_logger().propagate = False\npp = pprint.PrettyPrinter()\n\nfrom tfx import v1 as tfx\nfrom tfx.orchestration.experimental.interactive.interactive_context import InteractiveContext\n\n%load_ext tfx.orchestration.experimental.interactive.notebook_extensions.skip",
"_____no_output_____"
]
],
[
[
"Let's check the library versions.",
"_____no_output_____"
]
],
[
[
"print('TensorFlow version: {}'.format(tf.__version__))\nprint('TFX version: {}'.format(tfx.__version__))",
"_____no_output_____"
]
],
[
[
"### Set up pipeline paths",
"_____no_output_____"
]
],
[
[
"# This is the root directory for your TFX pip package installation.\n_tfx_root = tfx.__path__[0]\n\n# This is the directory containing the TFX Chicago Taxi Pipeline example.\n_taxi_root = os.path.join(_tfx_root, 'examples/chicago_taxi_pipeline')\n\n# This is the path where your model will be pushed for serving.\n_serving_model_dir = os.path.join(\n tempfile.mkdtemp(), 'serving_model/taxi_simple')\n\n# Set up logging.\nabsl.logging.set_verbosity(absl.logging.INFO)",
"_____no_output_____"
]
],
[
[
"### Download example data\nWe download the example dataset for use in our TFX pipeline.\n\nThe dataset we're using is the [Taxi Trips dataset](https://data.cityofchicago.org/Transportation/Taxi-Trips/wrvz-psew) released by the City of Chicago. The columns in this dataset are:\n\n<table>\n<tr><td>pickup_community_area</td><td>fare</td><td>trip_start_month</td></tr>\n<tr><td>trip_start_hour</td><td>trip_start_day</td><td>trip_start_timestamp</td></tr>\n<tr><td>pickup_latitude</td><td>pickup_longitude</td><td>dropoff_latitude</td></tr>\n<tr><td>dropoff_longitude</td><td>trip_miles</td><td>pickup_census_tract</td></tr>\n<tr><td>dropoff_census_tract</td><td>payment_type</td><td>company</td></tr>\n<tr><td>trip_seconds</td><td>dropoff_community_area</td><td>tips</td></tr>\n</table>\n\nWith this dataset, we will build a model that predicts the `tips` of a trip.",
"_____no_output_____"
]
],
[
[
"_data_root = tempfile.mkdtemp(prefix='tfx-data')\nDATA_PATH = 'https://raw.githubusercontent.com/tensorflow/tfx/master/tfx/examples/chicago_taxi_pipeline/data/simple/data.csv'\n_data_filepath = os.path.join(_data_root, \"data.csv\")\nurllib.request.urlretrieve(DATA_PATH, _data_filepath)",
"_____no_output_____"
]
],
[
[
"Take a quick look at the CSV file.",
"_____no_output_____"
]
],
[
[
"!head {_data_filepath}",
"_____no_output_____"
]
],
[
[
"*Disclaimer: This site provides applications using data that has been modified for use from its original source, www.cityofchicago.org, the official website of the City of Chicago. The City of Chicago makes no claims as to the content, accuracy, timeliness, or completeness of any of the data provided at this site. The data provided at this site is subject to change at any time. It is understood that the data provided at this site is being used at one’s own risk.*",
"_____no_output_____"
],
[
"### Create the InteractiveContext\nLast, we create an InteractiveContext, which will allow us to run TFX components interactively in this notebook.",
"_____no_output_____"
]
],
[
[
"# Here, we create an InteractiveContext using default parameters. This will\n# use a temporary directory with an ephemeral ML Metadata database instance.\n# To use your own pipeline root or database, the optional properties\n# `pipeline_root` and `metadata_connection_config` may be passed to\n# InteractiveContext. Calls to InteractiveContext are no-ops outside of the\n# notebook.\ncontext = InteractiveContext()",
"_____no_output_____"
]
],
[
[
"## Run TFX components interactively\nIn the cells that follow, we create TFX components one-by-one, run each of them, and visualize their output artifacts.",
"_____no_output_____"
],
[
"### ExampleGen\n\nThe `ExampleGen` component is usually at the start of a TFX pipeline. It will:\n\n1. Split data into training and evaluation sets (by default, 2/3 training + 1/3 eval)\n2. Convert data into the `tf.Example` format (learn more [here](https://www.tensorflow.org/tutorials/load_data/tfrecord))\n3. Copy data into the `_tfx_root` directory for other components to access\n\n`ExampleGen` takes as input the path to your data source. In our case, this is the `_data_root` path that contains the downloaded CSV.\n\nNote: In this notebook, we can instantiate components one-by-one and run them with `InteractiveContext.run()`. By contrast, in a production setting, we would specify all the components upfront in a `Pipeline` to pass to the orchestrator (see the [Building a TFX Pipeline Guide](https://www.tensorflow.org/tfx/guide/build_tfx_pipeline)).",
"_____no_output_____"
]
],
[
[
"example_gen = tfx.components.CsvExampleGen(input_base=_data_root)\ncontext.run(example_gen)",
"_____no_output_____"
]
],
[
[
"Let's examine the output artifacts of `ExampleGen`. This component produces two artifacts, training examples and evaluation examples:",
"_____no_output_____"
]
],
[
[
"artifact = example_gen.outputs['examples'].get()[0]\nprint(artifact.split_names, artifact.uri)",
"_____no_output_____"
]
],
[
[
"We can also take a look at the first three training examples:",
"_____no_output_____"
]
],
[
[
"# Get the URI of the output artifact representing the training examples, which is a directory\ntrain_uri = os.path.join(example_gen.outputs['examples'].get()[0].uri, 'Split-train')\n\n# Get the list of files in this directory (all compressed TFRecord files)\ntfrecord_filenames = [os.path.join(train_uri, name)\n for name in os.listdir(train_uri)]\n\n# Create a `TFRecordDataset` to read these files\ndataset = tf.data.TFRecordDataset(tfrecord_filenames, compression_type=\"GZIP\")\n\n# Iterate over the first 3 records and decode them.\nfor tfrecord in dataset.take(3):\n serialized_example = tfrecord.numpy()\n example = tf.train.Example()\n example.ParseFromString(serialized_example)\n pp.pprint(example)",
"_____no_output_____"
]
],
[
[
"Now that `ExampleGen` has finished ingesting the data, the next step is data analysis.",
"_____no_output_____"
],
[
"### StatisticsGen\nThe `StatisticsGen` component computes statistics over your dataset for data analysis, as well as for use in downstream components. It uses the [TensorFlow Data Validation](https://www.tensorflow.org/tfx/data_validation/get_started) library.\n\n`StatisticsGen` takes as input the dataset we just ingested using `ExampleGen`.",
"_____no_output_____"
]
],
[
[
"statistics_gen = tfx.components.StatisticsGen(\n examples=example_gen.outputs['examples'])\ncontext.run(statistics_gen)",
"_____no_output_____"
]
],
[
[
"After `StatisticsGen` finishes running, we can visualize the outputted statistics. Try playing with the different plots!",
"_____no_output_____"
]
],
[
[
"context.show(statistics_gen.outputs['statistics'])",
"_____no_output_____"
]
],
[
[
"### SchemaGen\n\nThe `SchemaGen` component generates a schema based on your data statistics. (A schema defines the expected bounds, types, and properties of the features in your dataset.) It also uses the [TensorFlow Data Validation](https://www.tensorflow.org/tfx/data_validation/get_started) library.\n\nNote: The generated schema is best-effort and only tries to infer basic properties of the data. It is expected that you review and modify it as needed.\n\n`SchemaGen` will take as input the statistics that we generated with `StatisticsGen`, looking at the training split by default.",
"_____no_output_____"
]
],
[
[
"schema_gen = tfx.components.SchemaGen(\n statistics=statistics_gen.outputs['statistics'],\n infer_feature_shape=False)\ncontext.run(schema_gen)",
"_____no_output_____"
]
],
[
[
"After `SchemaGen` finishes running, we can visualize the generated schema as a table.",
"_____no_output_____"
]
],
[
[
"context.show(schema_gen.outputs['schema'])",
"_____no_output_____"
]
],
[
[
"Each feature in your dataset shows up as a row in the schema table, alongside its properties. The schema also captures all the values that a categorical feature takes on, denoted as its domain.\n\nTo learn more about schemas, see [the SchemaGen documentation](https://www.tensorflow.org/tfx/guide/schemagen).",
"_____no_output_____"
],
[
"### ExampleValidator\nThe `ExampleValidator` component detects anomalies in your data, based on the expectations defined by the schema. It also uses the [TensorFlow Data Validation](https://www.tensorflow.org/tfx/data_validation/get_started) library.\n\n`ExampleValidator` will take as input the statistics from `StatisticsGen`, and the schema from `SchemaGen`.",
"_____no_output_____"
]
],
[
[
"example_validator = tfx.components.ExampleValidator(\n statistics=statistics_gen.outputs['statistics'],\n schema=schema_gen.outputs['schema'])\ncontext.run(example_validator)",
"_____no_output_____"
]
],
[
[
"After `ExampleValidator` finishes running, we can visualize the anomalies as a table.",
"_____no_output_____"
]
],
[
[
"context.show(example_validator.outputs['anomalies'])",
"_____no_output_____"
]
],
[
[
"In the anomalies table, we can see that there are no anomalies. This is what we'd expect, since this the first dataset that we've analyzed and the schema is tailored to it. You should review this schema -- anything unexpected means an anomaly in the data. Once reviewed, the schema can be used to guard future data, and anomalies produced here can be used to debug model performance, understand how your data evolves over time, and identify data errors.",
"_____no_output_____"
],
[
"### Transform\nThe `Transform` component performs feature engineering for both training and serving. It uses the [TensorFlow Transform](https://www.tensorflow.org/tfx/transform/get_started) library.\n\n`Transform` will take as input the data from `ExampleGen`, the schema from `SchemaGen`, as well as a module that contains user-defined Transform code.\n\nLet's see an example of user-defined Transform code below (for an introduction to the TensorFlow Transform APIs, [see the tutorial](https://www.tensorflow.org/tfx/tutorials/transform/simple)). First, we define a few constants for feature engineering:\n\nNote: The `%%writefile` cell magic will save the contents of the cell as a `.py` file on disk. This allows the `Transform` component to load your code as a module.\n",
"_____no_output_____"
]
],
[
[
"_taxi_constants_module_file = 'taxi_constants.py'",
"_____no_output_____"
],
[
"%%writefile {_taxi_constants_module_file}\n\n# Categorical features are assumed to each have a maximum value in the dataset.\nMAX_CATEGORICAL_FEATURE_VALUES = [24, 31, 12]\n\nCATEGORICAL_FEATURE_KEYS = [\n 'trip_start_hour', 'trip_start_day', 'trip_start_month',\n 'pickup_census_tract', 'dropoff_census_tract', 'pickup_community_area',\n 'dropoff_community_area'\n]\n\nDENSE_FLOAT_FEATURE_KEYS = ['trip_miles', 'fare', 'trip_seconds']\n\n# Number of buckets used by tf.transform for encoding each feature.\nFEATURE_BUCKET_COUNT = 10\n\nBUCKET_FEATURE_KEYS = [\n 'pickup_latitude', 'pickup_longitude', 'dropoff_latitude',\n 'dropoff_longitude'\n]\n\n# Number of vocabulary terms used for encoding VOCAB_FEATURES by tf.transform\nVOCAB_SIZE = 1000\n\n# Count of out-of-vocab buckets in which unrecognized VOCAB_FEATURES are hashed.\nOOV_SIZE = 10\n\nVOCAB_FEATURE_KEYS = [\n 'payment_type',\n 'company',\n]\n\n# Keys\nLABEL_KEY = 'tips'\nFARE_KEY = 'fare'",
"_____no_output_____"
]
],
[
[
"Next, we write a `preprocessing_fn` that takes in raw data as input, and returns transformed features that our model can train on:",
"_____no_output_____"
]
],
[
[
"_taxi_transform_module_file = 'taxi_transform.py'",
"_____no_output_____"
],
[
"%%writefile {_taxi_transform_module_file}\n\nimport tensorflow as tf\nimport tensorflow_transform as tft\n\nimport taxi_constants\n\n_DENSE_FLOAT_FEATURE_KEYS = taxi_constants.DENSE_FLOAT_FEATURE_KEYS\n_VOCAB_FEATURE_KEYS = taxi_constants.VOCAB_FEATURE_KEYS\n_VOCAB_SIZE = taxi_constants.VOCAB_SIZE\n_OOV_SIZE = taxi_constants.OOV_SIZE\n_FEATURE_BUCKET_COUNT = taxi_constants.FEATURE_BUCKET_COUNT\n_BUCKET_FEATURE_KEYS = taxi_constants.BUCKET_FEATURE_KEYS\n_CATEGORICAL_FEATURE_KEYS = taxi_constants.CATEGORICAL_FEATURE_KEYS\n_FARE_KEY = taxi_constants.FARE_KEY\n_LABEL_KEY = taxi_constants.LABEL_KEY\n\n\ndef preprocessing_fn(inputs):\n \"\"\"tf.transform's callback function for preprocessing inputs.\n Args:\n inputs: map from feature keys to raw not-yet-transformed features.\n Returns:\n Map from string feature key to transformed feature operations.\n \"\"\"\n outputs = {}\n for key in _DENSE_FLOAT_FEATURE_KEYS:\n # If sparse make it dense, setting nan's to 0 or '', and apply zscore.\n outputs[key] = tft.scale_to_z_score(\n _fill_in_missing(inputs[key]))\n\n for key in _VOCAB_FEATURE_KEYS:\n # Build a vocabulary for this feature.\n outputs[key] = tft.compute_and_apply_vocabulary(\n _fill_in_missing(inputs[key]),\n top_k=_VOCAB_SIZE,\n num_oov_buckets=_OOV_SIZE)\n\n for key in _BUCKET_FEATURE_KEYS:\n outputs[key] = tft.bucketize(\n _fill_in_missing(inputs[key]), _FEATURE_BUCKET_COUNT)\n\n for key in _CATEGORICAL_FEATURE_KEYS:\n outputs[key] = _fill_in_missing(inputs[key])\n\n # Was this passenger a big tipper?\n taxi_fare = _fill_in_missing(inputs[_FARE_KEY])\n tips = _fill_in_missing(inputs[_LABEL_KEY])\n outputs[_LABEL_KEY] = tf.where(\n tf.math.is_nan(taxi_fare),\n tf.cast(tf.zeros_like(taxi_fare), tf.int64),\n # Test if the tip was > 20% of the fare.\n tf.cast(\n tf.greater(tips, tf.multiply(taxi_fare, tf.constant(0.2))), tf.int64))\n\n return outputs\n\n\ndef _fill_in_missing(x):\n \"\"\"Replace missing values in a SparseTensor.\n Fills in missing values of `x` with '' or 0, and converts to a dense tensor.\n Args:\n x: A `SparseTensor` of rank 2. Its dense shape should have size at most 1\n in the second dimension.\n Returns:\n A rank 1 tensor where missing values of `x` have been filled in.\n \"\"\"\n if not isinstance(x, tf.sparse.SparseTensor):\n return x\n\n default_value = '' if x.dtype == tf.string else 0\n return tf.squeeze(\n tf.sparse.to_dense(\n tf.SparseTensor(x.indices, x.values, [x.dense_shape[0], 1]),\n default_value),\n axis=1)",
"_____no_output_____"
]
],
[
[
"Now, we pass in this feature engineering code to the `Transform` component and run it to transform your data.",
"_____no_output_____"
]
],
[
[
"transform = tfx.components.Transform(\n examples=example_gen.outputs['examples'],\n schema=schema_gen.outputs['schema'],\n module_file=os.path.abspath(_taxi_transform_module_file))\ncontext.run(transform)",
"_____no_output_____"
]
],
[
[
"Let's examine the output artifacts of `Transform`. This component produces two types of outputs:\n\n* `transform_graph` is the graph that can perform the preprocessing operations (this graph will be included in the serving and evaluation models).\n* `transformed_examples` represents the preprocessed training and evaluation data.",
"_____no_output_____"
]
],
[
[
"transform.outputs",
"_____no_output_____"
]
],
[
[
"Take a peek at the `transform_graph` artifact. It points to a directory containing three subdirectories.",
"_____no_output_____"
]
],
[
[
"train_uri = transform.outputs['transform_graph'].get()[0].uri\nos.listdir(train_uri)",
"_____no_output_____"
]
],
[
[
"The `transformed_metadata` subdirectory contains the schema of the preprocessed data. The `transform_fn` subdirectory contains the actual preprocessing graph. The `metadata` subdirectory contains the schema of the original data.\n\nWe can also take a look at the first three transformed examples:",
"_____no_output_____"
]
],
[
[
"# Get the URI of the output artifact representing the transformed examples, which is a directory\ntrain_uri = os.path.join(transform.outputs['transformed_examples'].get()[0].uri, 'Split-train')\n\n# Get the list of files in this directory (all compressed TFRecord files)\ntfrecord_filenames = [os.path.join(train_uri, name)\n for name in os.listdir(train_uri)]\n\n# Create a `TFRecordDataset` to read these files\ndataset = tf.data.TFRecordDataset(tfrecord_filenames, compression_type=\"GZIP\")\n\n# Iterate over the first 3 records and decode them.\nfor tfrecord in dataset.take(3):\n serialized_example = tfrecord.numpy()\n example = tf.train.Example()\n example.ParseFromString(serialized_example)\n pp.pprint(example)",
"_____no_output_____"
]
],
[
[
"After the `Transform` component has transformed your data into features, and the next step is to train a model.",
"_____no_output_____"
],
[
"### Trainer\nThe `Trainer` component will train a model that you define in TensorFlow. Default Trainer support Estimator API, to use Keras API, you need to specify [Generic Trainer](https://github.com/tensorflow/community/blob/master/rfcs/20200117-tfx-generic-trainer.md) by setup `custom_executor_spec=executor_spec.ExecutorClassSpec(GenericExecutor)` in Trainer's contructor.\n\n`Trainer` takes as input the schema from `SchemaGen`, the transformed data and graph from `Transform`, training parameters, as well as a module that contains user-defined model code.\n\nLet's see an example of user-defined model code below (for an introduction to the TensorFlow Keras APIs, [see the tutorial](https://www.tensorflow.org/guide/keras)):",
"_____no_output_____"
]
],
[
[
"_taxi_trainer_module_file = 'taxi_trainer.py'",
"_____no_output_____"
],
[
"%%writefile {_taxi_trainer_module_file}\n\nfrom typing import List, Text\n\nimport os\nfrom absl import logging\n\nimport datetime\nimport tensorflow as tf\nimport tensorflow_transform as tft\n\nfrom tfx import v1 as tfx\nfrom tfx_bsl.public import tfxio\n\nimport taxi_constants\n\n_DENSE_FLOAT_FEATURE_KEYS = taxi_constants.DENSE_FLOAT_FEATURE_KEYS\n_VOCAB_FEATURE_KEYS = taxi_constants.VOCAB_FEATURE_KEYS\n_VOCAB_SIZE = taxi_constants.VOCAB_SIZE\n_OOV_SIZE = taxi_constants.OOV_SIZE\n_FEATURE_BUCKET_COUNT = taxi_constants.FEATURE_BUCKET_COUNT\n_BUCKET_FEATURE_KEYS = taxi_constants.BUCKET_FEATURE_KEYS\n_CATEGORICAL_FEATURE_KEYS = taxi_constants.CATEGORICAL_FEATURE_KEYS\n_MAX_CATEGORICAL_FEATURE_VALUES = taxi_constants.MAX_CATEGORICAL_FEATURE_VALUES\n_LABEL_KEY = taxi_constants.LABEL_KEY\n\n\ndef _get_tf_examples_serving_signature(model, tf_transform_output):\n \"\"\"Returns a serving signature that accepts `tensorflow.Example`.\"\"\"\n\n # We need to track the layers in the model in order to save it.\n # TODO(b/162357359): Revise once the bug is resolved.\n model.tft_layer_inference = tf_transform_output.transform_features_layer()\n\n @tf.function(input_signature=[\n tf.TensorSpec(shape=[None], dtype=tf.string, name='examples')\n ])\n def serve_tf_examples_fn(serialized_tf_example):\n \"\"\"Returns the output to be used in the serving signature.\"\"\"\n raw_feature_spec = tf_transform_output.raw_feature_spec()\n # Remove label feature since these will not be present at serving time.\n raw_feature_spec.pop(_LABEL_KEY)\n raw_features = tf.io.parse_example(serialized_tf_example, raw_feature_spec)\n transformed_features = model.tft_layer_inference(raw_features)\n logging.info('serve_transformed_features = %s', transformed_features)\n\n outputs = model(transformed_features)\n # TODO(b/154085620): Convert the predicted labels from the model using a\n # reverse-lookup (opposite of transform.py).\n return {'outputs': outputs}\n\n return serve_tf_examples_fn\n\n\ndef _get_transform_features_signature(model, tf_transform_output):\n \"\"\"Returns a serving signature that applies tf.Transform to features.\"\"\"\n\n # We need to track the layers in the model in order to save it.\n # TODO(b/162357359): Revise once the bug is resolved.\n model.tft_layer_eval = tf_transform_output.transform_features_layer()\n\n @tf.function(input_signature=[\n tf.TensorSpec(shape=[None], dtype=tf.string, name='examples')\n ])\n def transform_features_fn(serialized_tf_example):\n \"\"\"Returns the transformed_features to be fed as input to evaluator.\"\"\"\n raw_feature_spec = tf_transform_output.raw_feature_spec()\n raw_features = tf.io.parse_example(serialized_tf_example, raw_feature_spec)\n transformed_features = model.tft_layer_eval(raw_features)\n logging.info('eval_transformed_features = %s', transformed_features)\n return transformed_features\n\n return transform_features_fn\n\n\ndef _input_fn(file_pattern: List[Text],\n data_accessor: tfx.components.DataAccessor,\n tf_transform_output: tft.TFTransformOutput,\n batch_size: int = 200) -> tf.data.Dataset:\n \"\"\"Generates features and label for tuning/training.\n\n Args:\n file_pattern: List of paths or patterns of input tfrecord files.\n data_accessor: DataAccessor for converting input to RecordBatch.\n tf_transform_output: A TFTransformOutput.\n batch_size: representing the number of consecutive elements of returned\n dataset to combine in a single batch\n\n Returns:\n A dataset that contains (features, indices) tuple where features is a\n dictionary of Tensors, and indices is a single Tensor of label indices.\n \"\"\"\n return data_accessor.tf_dataset_factory(\n file_pattern,\n tfxio.TensorFlowDatasetOptions(\n batch_size=batch_size, label_key=_LABEL_KEY),\n tf_transform_output.transformed_metadata.schema)\n\n\ndef _build_keras_model(hidden_units: List[int] = None) -> tf.keras.Model:\n \"\"\"Creates a DNN Keras model for classifying taxi data.\n\n Args:\n hidden_units: [int], the layer sizes of the DNN (input layer first).\n\n Returns:\n A keras Model.\n \"\"\"\n real_valued_columns = [\n tf.feature_column.numeric_column(key, shape=())\n for key in _DENSE_FLOAT_FEATURE_KEYS\n ]\n categorical_columns = [\n tf.feature_column.categorical_column_with_identity(\n key, num_buckets=_VOCAB_SIZE + _OOV_SIZE, default_value=0)\n for key in _VOCAB_FEATURE_KEYS\n ]\n categorical_columns += [\n tf.feature_column.categorical_column_with_identity(\n key, num_buckets=_FEATURE_BUCKET_COUNT, default_value=0)\n for key in _BUCKET_FEATURE_KEYS\n ]\n categorical_columns += [\n tf.feature_column.categorical_column_with_identity( # pylint: disable=g-complex-comprehension\n key,\n num_buckets=num_buckets,\n default_value=0) for key, num_buckets in zip(\n _CATEGORICAL_FEATURE_KEYS,\n _MAX_CATEGORICAL_FEATURE_VALUES)\n ]\n indicator_column = [\n tf.feature_column.indicator_column(categorical_column)\n for categorical_column in categorical_columns\n ]\n\n model = _wide_and_deep_classifier(\n # TODO(b/139668410) replace with premade wide_and_deep keras model\n wide_columns=indicator_column,\n deep_columns=real_valued_columns,\n dnn_hidden_units=hidden_units or [100, 70, 50, 25])\n return model\n\n\ndef _wide_and_deep_classifier(wide_columns, deep_columns, dnn_hidden_units):\n \"\"\"Build a simple keras wide and deep model.\n\n Args:\n wide_columns: Feature columns wrapped in indicator_column for wide (linear)\n part of the model.\n deep_columns: Feature columns for deep part of the model.\n dnn_hidden_units: [int], the layer sizes of the hidden DNN.\n\n Returns:\n A Wide and Deep Keras model\n \"\"\"\n # Following values are hard coded for simplicity in this example,\n # However prefarably they should be passsed in as hparams.\n\n # Keras needs the feature definitions at compile time.\n # TODO(b/139081439): Automate generation of input layers from FeatureColumn.\n input_layers = {\n colname: tf.keras.layers.Input(name=colname, shape=(), dtype=tf.float32)\n for colname in _DENSE_FLOAT_FEATURE_KEYS\n }\n input_layers.update({\n colname: tf.keras.layers.Input(name=colname, shape=(), dtype='int32')\n for colname in _VOCAB_FEATURE_KEYS\n })\n input_layers.update({\n colname: tf.keras.layers.Input(name=colname, shape=(), dtype='int32')\n for colname in _BUCKET_FEATURE_KEYS\n })\n input_layers.update({\n colname: tf.keras.layers.Input(name=colname, shape=(), dtype='int32')\n for colname in _CATEGORICAL_FEATURE_KEYS\n })\n\n # TODO(b/161952382): Replace with Keras preprocessing layers.\n deep = tf.keras.layers.DenseFeatures(deep_columns)(input_layers)\n for numnodes in dnn_hidden_units:\n deep = tf.keras.layers.Dense(numnodes)(deep)\n wide = tf.keras.layers.DenseFeatures(wide_columns)(input_layers)\n\n output = tf.keras.layers.Dense(1)(\n tf.keras.layers.concatenate([deep, wide]))\n\n model = tf.keras.Model(input_layers, output)\n model.compile(\n loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),\n optimizer=tf.keras.optimizers.Adam(lr=0.001),\n metrics=[tf.keras.metrics.BinaryAccuracy()])\n model.summary(print_fn=logging.info)\n return model\n\n\n# TFX Trainer will call this function.\ndef run_fn(fn_args: tfx.components.FnArgs):\n \"\"\"Train the model based on given args.\n\n Args:\n fn_args: Holds args used to train the model as name/value pairs.\n \"\"\"\n # Number of nodes in the first layer of the DNN\n first_dnn_layer_size = 100\n num_dnn_layers = 4\n dnn_decay_factor = 0.7\n\n tf_transform_output = tft.TFTransformOutput(fn_args.transform_output)\n\n train_dataset = _input_fn(fn_args.train_files, fn_args.data_accessor, \n tf_transform_output, 40)\n eval_dataset = _input_fn(fn_args.eval_files, fn_args.data_accessor, \n tf_transform_output, 40)\n\n model = _build_keras_model(\n # Construct layers sizes with exponetial decay\n hidden_units=[\n max(2, int(first_dnn_layer_size * dnn_decay_factor**i))\n for i in range(num_dnn_layers)\n ])\n\n tensorboard_callback = tf.keras.callbacks.TensorBoard(\n log_dir=fn_args.model_run_dir, update_freq='batch')\n model.fit(\n train_dataset,\n steps_per_epoch=fn_args.train_steps,\n validation_data=eval_dataset,\n validation_steps=fn_args.eval_steps,\n callbacks=[tensorboard_callback])\n\n signatures = {\n 'serving_default':\n _get_tf_examples_serving_signature(model, tf_transform_output),\n 'transform_features':\n _get_transform_features_signature(model, tf_transform_output),\n }\n model.save(fn_args.serving_model_dir, save_format='tf', signatures=signatures)",
"_____no_output_____"
]
],
[
[
"Now, we pass in this model code to the `Trainer` component and run it to train the model.",
"_____no_output_____"
]
],
[
[
"trainer = tfx.components.Trainer(\n module_file=os.path.abspath(_taxi_trainer_module_file),\n examples=transform.outputs['transformed_examples'],\n transform_graph=transform.outputs['transform_graph'],\n schema=schema_gen.outputs['schema'],\n train_args=tfx.proto.TrainArgs(num_steps=10000),\n eval_args=tfx.proto.EvalArgs(num_steps=5000))\ncontext.run(trainer)",
"_____no_output_____"
]
],
[
[
"#### Analyze Training with TensorBoard\nTake a peek at the trainer artifact. It points to a directory containing the model subdirectories.",
"_____no_output_____"
]
],
[
[
"model_artifact_dir = trainer.outputs['model'].get()[0].uri\npp.pprint(os.listdir(model_artifact_dir))\nmodel_dir = os.path.join(model_artifact_dir, 'Format-Serving')\npp.pprint(os.listdir(model_dir))",
"_____no_output_____"
]
],
[
[
"Optionally, we can connect TensorBoard to the Trainer to analyze our model's training curves.",
"_____no_output_____"
]
],
[
[
"model_run_artifact_dir = trainer.outputs['model_run'].get()[0].uri\n\n%load_ext tensorboard\n%tensorboard --logdir {model_run_artifact_dir}",
"_____no_output_____"
]
],
[
[
"### Evaluator\nThe `Evaluator` component computes model performance metrics over the evaluation set. It uses the [TensorFlow Model Analysis](https://www.tensorflow.org/tfx/model_analysis/get_started) library. The `Evaluator` can also optionally validate that a newly trained model is better than the previous model. This is useful in a production pipeline setting where you may automatically train and validate a model every day. In this notebook, we only train one model, so the `Evaluator` automatically will label the model as \"good\". \n\n`Evaluator` will take as input the data from `ExampleGen`, the trained model from `Trainer`, and slicing configuration. The slicing configuration allows you to slice your metrics on feature values (e.g. how does your model perform on taxi trips that start at 8am versus 8pm?). See an example of this configuration below:",
"_____no_output_____"
]
],
[
[
"eval_config = tfma.EvalConfig(\n model_specs=[\n # This assumes a serving model with signature 'serving_default'. If\n # using estimator based EvalSavedModel, add signature_name: 'eval' and\n # remove the label_key.\n tfma.ModelSpec(\n signature_name='serving_default',\n label_key='tips',\n preprocessing_function_names=['transform_features'],\n )\n ],\n metrics_specs=[\n tfma.MetricsSpec(\n # The metrics added here are in addition to those saved with the\n # model (assuming either a keras model or EvalSavedModel is used).\n # Any metrics added into the saved model (for example using\n # model.compile(..., metrics=[...]), etc) will be computed\n # automatically.\n # To add validation thresholds for metrics saved with the model,\n # add them keyed by metric name to the thresholds map.\n metrics=[\n tfma.MetricConfig(class_name='ExampleCount'),\n tfma.MetricConfig(class_name='BinaryAccuracy',\n threshold=tfma.MetricThreshold(\n value_threshold=tfma.GenericValueThreshold(\n lower_bound={'value': 0.5}),\n # Change threshold will be ignored if there is no\n # baseline model resolved from MLMD (first run).\n change_threshold=tfma.GenericChangeThreshold(\n direction=tfma.MetricDirection.HIGHER_IS_BETTER,\n absolute={'value': -1e-10})))\n ]\n )\n ],\n slicing_specs=[\n # An empty slice spec means the overall slice, i.e. the whole dataset.\n tfma.SlicingSpec(),\n # Data can be sliced along a feature column. In this case, data is\n # sliced along feature column trip_start_hour.\n tfma.SlicingSpec(feature_keys=['trip_start_hour'])\n ])",
"_____no_output_____"
]
],
[
[
"Next, we give this configuration to `Evaluator` and run it.",
"_____no_output_____"
]
],
[
[
"# Use TFMA to compute a evaluation statistics over features of a model and\n# validate them against a baseline.\n\n# The model resolver is only required if performing model validation in addition\n# to evaluation. In this case we validate against the latest blessed model. If\n# no model has been blessed before (as in this case) the evaluator will make our\n# candidate the first blessed model.\nmodel_resolver = tfx.dsl.Resolver(\n strategy_class=tfx.dsl.experimental.LatestBlessedModelStrategy,\n model=tfx.dsl.Channel(type=tfx.types.standard_artifacts.Model),\n model_blessing=tfx.dsl.Channel(\n type=tfx.types.standard_artifacts.ModelBlessing)).with_id(\n 'latest_blessed_model_resolver')\ncontext.run(model_resolver)\n\nevaluator = tfx.components.Evaluator(\n examples=example_gen.outputs['examples'],\n model=trainer.outputs['model'],\n baseline_model=model_resolver.outputs['model'],\n eval_config=eval_config)\ncontext.run(evaluator)",
"_____no_output_____"
]
],
[
[
"Now let's examine the output artifacts of `Evaluator`. ",
"_____no_output_____"
]
],
[
[
"evaluator.outputs",
"_____no_output_____"
]
],
[
[
"Using the `evaluation` output we can show the default visualization of global metrics on the entire evaluation set.",
"_____no_output_____"
]
],
[
[
"context.show(evaluator.outputs['evaluation'])",
"_____no_output_____"
]
],
[
[
"To see the visualization for sliced evaluation metrics, we can directly call the TensorFlow Model Analysis library.",
"_____no_output_____"
]
],
[
[
"import tensorflow_model_analysis as tfma\n\n# Get the TFMA output result path and load the result.\nPATH_TO_RESULT = evaluator.outputs['evaluation'].get()[0].uri\ntfma_result = tfma.load_eval_result(PATH_TO_RESULT)\n\n# Show data sliced along feature column trip_start_hour.\ntfma.view.render_slicing_metrics(\n tfma_result, slicing_column='trip_start_hour')",
"_____no_output_____"
]
],
[
[
"This visualization shows the same metrics, but computed at every feature value of `trip_start_hour` instead of on the entire evaluation set.\n\nTensorFlow Model Analysis supports many other visualizations, such as Fairness Indicators and plotting a time series of model performance. To learn more, see [the tutorial](https://www.tensorflow.org/tfx/tutorials/model_analysis/tfma_basic).",
"_____no_output_____"
],
[
"Since we added thresholds to our config, validation output is also available. The precence of a `blessing` artifact indicates that our model passed validation. Since this is the first validation being performed the candidate is automatically blessed.",
"_____no_output_____"
]
],
[
[
"blessing_uri = evaluator.outputs['blessing'].get()[0].uri\n!ls -l {blessing_uri}",
"_____no_output_____"
]
],
[
[
"Now can also verify the success by loading the validation result record:",
"_____no_output_____"
]
],
[
[
"PATH_TO_RESULT = evaluator.outputs['evaluation'].get()[0].uri\nprint(tfma.load_validation_result(PATH_TO_RESULT))",
"_____no_output_____"
]
],
[
[
"### Pusher\nThe `Pusher` component is usually at the end of a TFX pipeline. It checks whether a model has passed validation, and if so, exports the model to `_serving_model_dir`.",
"_____no_output_____"
]
],
[
[
"pusher = tfx.components.Pusher(\n model=trainer.outputs['model'],\n model_blessing=evaluator.outputs['blessing'],\n push_destination=tfx.proto.PushDestination(\n filesystem=tfx.proto.PushDestination.Filesystem(\n base_directory=_serving_model_dir)))\ncontext.run(pusher)",
"_____no_output_____"
]
],
[
[
"Let's examine the output artifacts of `Pusher`. ",
"_____no_output_____"
]
],
[
[
"pusher.outputs",
"_____no_output_____"
]
],
[
[
"In particular, the Pusher will export your model in the SavedModel format, which looks like this:",
"_____no_output_____"
]
],
[
[
"push_uri = pusher.outputs['pushed_model'].get()[0].uri\nmodel = tf.saved_model.load(push_uri)\n\nfor item in model.signatures.items():\n pp.pprint(item)",
"_____no_output_____"
]
],
[
[
"We're finished our tour of built-in TFX components!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb772dc942bfd9d0b4caee2808239171a526f9f | 991,546 | ipynb | Jupyter Notebook | Image Classifier Project.ipynb | pavel-nesterov/aipnd-project | 82a11ede8c55b19f0f06775175f83b5c21260a5d | [
"MIT"
] | null | null | null | Image Classifier Project.ipynb | pavel-nesterov/aipnd-project | 82a11ede8c55b19f0f06775175f83b5c21260a5d | [
"MIT"
] | null | null | null | Image Classifier Project.ipynb | pavel-nesterov/aipnd-project | 82a11ede8c55b19f0f06775175f83b5c21260a5d | [
"MIT"
] | null | null | null | 836.747679 | 224,220 | 0.947047 | [
[
[
"#Hint for myself\n\n# to test that everythig works\n# 1.5. enable GPU \n# 2. open terminal and 'python train.py'\n# 3. Expected result = 'checkpoint saved successfully'\n# 4. open terminal and 'python predict.py'\n# 5. expected result = 'Top class = \"rose\" with probability 98%'\n\n\n",
"_____no_output_____"
],
[
"%%writefile view_classify.py\nimport json\nimport numpy as np\nimport matplotlib.pyplot as plt\n\n#def view_classify(ps_and_classes, display = True):\n\ndef get_key(dictionary, val_to_find): \n for key, value in dictionary.items(): \n if val_to_find == value: \n return key \n return \"key doesn't exist\"\n\ndef get_topk_labels(args_predict, ps_and_classes, display = True):\n topk5_labels = []\n with open('cat_to_name.json', 'r') as f:\n cat_to_name = json.load(f)\n #print(type(cat_to_name))\n #print(cat_to_name)\n \n #print(\"ps_and_classes in view_classify = \", ps_and_classes)\n #print(\"ps_and_classes[0][0] = \", ps_and_classes[0][0])\n #print(\"ps_and_classes[0][1] = \", ps_and_classes[0][1])\n numpy_probs = ps_and_classes[0][0].cpu().data.numpy()\n numpy_classes = ps_and_classes[0][1].cpu().data.numpy()\n #print(\"numpy_probs = \", type(numpy_probs), numpy_probs)\n #print(\"numpy_classes:\", type(numpy_classes), numpy_classes, numpy_classes.shape, numpy_classes[0])\n #numpy_classes: <class 'numpy.ndarray'> [[ 7 38 77 2 60]] (1, 5) [ 7 38 77 2 60]\n with open('class_to_idx.json', 'r') as f:\n class_to_idx = json.load(f)\n #print(\"class_to_idx type\", type(class_to_idx))\n #class_to_idx type <class 'dict'>\n #i= 7\n #new_i= None\n for i in numpy_classes[0]:\n new_new_i = get_key(class_to_idx, i)\n #print (f\"i={i}, new_new_i={new_new_i}\")\n topk5_labels.append(cat_to_name[str(new_new_i)])\n #print(\"topk5_labels = \", topk5_labels)\n return topk5_labels, numpy_probs\n #labels = probs[1].data.numpy().squeeze()\n #print(labels[2], labels, type(labels))\n \ndef display_bar_histogram(numpy_probs, topk5_labels):\n #print (\"this is what display_bar_histogram receives\")\n #print (\"numpy_probs\", type(numpy_probs), numpy_probs.shape, numpy_probs)\n fig, ax2 = plt.subplots( ncols=1)\n #ax2.barh(np.arange(5), ps)\n numpy_probs = numpy_probs.squeeze()\n ax2.barh(np.arange(len(numpy_probs)), numpy_probs)\n ax2.set_yticks(np.arange(5))\n ax2.set_yticklabels(topk5_labels, size='small');\n ax2.set_title('Probability')\n ax2.set_xlim(0, 1.1)\n \n",
"Overwriting view_classify.py\n"
],
[
"%%writefile save_load_model.py\nimport torch\nfrom torchvision import datasets, transforms, models\nfrom torch import nn\nimport time\nimport train\nimport json\n\ncheckpoint_name = 'checkpoint_2.pth'\n#use it to make sure saved and loaded checkpoints are the same\n#TRAINED MODEL IS SAVED IN 'checkpoint_PROPERLY_TRAINED.pth'\n\n\ndef load_checkpoint_and_rebuild_the_model(args):\n #print(\"Starting importing the checkpoint...\")\n #checkpoint = torch.load(args[1]+checkpoint_name, map_location=lambda storage, loc: storage)\n model_loaded = train.create_model(args)\n #print(\"model_loaded printing\", model_loaded)\n model_loaded.load_state_dict(torch.load(args[1]+checkpoint_name, map_location=lambda storage, loc: storage))\n #print(\"checkpoint loaded\")\n \n #f = open('class_to_idx.json')\n #class_to_idx = json.load(f)\n #model_loaded.class_to_idx = class_to_idx\n \n \n #print(\"class_to_idx from checkpoint loading procedure\")\n #print(class_to_idx)\n #model_loaded.class_to_idx(checkpoint['class_to_idx'])\n #model_loaded.load_state_dict(checkpoint['state_dict'])\n #print(\"Model was loaded successfully\")\n return(model_loaded)\n\n\ndef save_the_checkpoint(model, args):\n print (\"start saving checkpiont...\")\n saving_start = time.time()\n torch.save(model.state_dict(), args[1]+checkpoint_name)\n #print(\"model.class_to_idx before saving = \")\n #print(model.class_to_idx)\n #checkpoint = {'class_to_idx': model.class_to_idx,\n # 'state_dict': model.state_dict()}\n #torch.save(checkpoint, args[1]+checkpoint_name)\n #print(\"class_to_idx from checkpoint = \", checkpoint[class_to_idx])\n #print(\"state_dict from checkpoint = \", checkpoint[state_dict])\n #with open('class_to_idx.json', 'w') as f:\n # json.dump(model.class_to_idx, f)\n #class_to_idx.json\n saving_end = time.time()\n print(f\"Saved to {args[1]}{checkpoint_name}, Duration: {(saving_end - saving_start):.3f}, sec\") \n",
"Overwriting save_load_model.py\n"
],
[
"%%writefile predict.py\nimport save_load_model\nimport json\nimport torch\nfrom random import randint \nimport process_image as pima\nimport train\nimport argparse\nimport view_classify\nimport signal\n\nfrom contextlib import contextmanager\n\nimport requests\n\ndef get_input_args():\n if __name__ == \"__main__\":\n parser = argparse.ArgumentParser()\n parser.add_argument('--topk', type = int, default = 5, help = 'Number of top classes to be returned') \n in_args = parser.parse_args()\n in_args_dict = vars(in_args)\n #predict_in_args_list = [v for v in in_args_dict.values()]\n print(in_args_dict)\n return in_args_dict\n # stuff only to run when not called via 'import' here\n else:\n in_args_dict = {'topk': 5}\n return(in_args_dict)\n\nDELAY = INTERVAL = 4 * 60 # interval time in seconds\nMIN_DELAY = MIN_INTERVAL = 2 * 60\nKEEPALIVE_URL = \"https://nebula.udacity.com/api/v1/remote/keep-alive\"\nTOKEN_URL = \"http://metadata.google.internal/computeMetadata/v1/instance/attributes/keep_alive_token\"\nTOKEN_HEADERS = {\"Metadata-Flavor\":\"Google\"}\n\n\ndef _request_handler(headers):\n def _handler(signum, frame):\n requests.request(\"POST\", KEEPALIVE_URL, headers=headers)\n return _handler\n\n\n@contextmanager\ndef active_session(delay=DELAY, interval=INTERVAL):\n \"\"\"\n Example:\n\n from workspace_utils import active session\n\n with active_session():\n # do long-running work here\n \"\"\"\n token = requests.request(\"GET\", TOKEN_URL, headers=TOKEN_HEADERS).text\n headers = {'Authorization': \"STAR \" + token}\n delay = max(delay, MIN_DELAY)\n interval = max(interval, MIN_INTERVAL)\n original_handler = signal.getsignal(signal.SIGALRM)\n try:\n signal.signal(signal.SIGALRM, _request_handler(headers))\n signal.setitimer(signal.ITIMER_REAL, delay, interval)\n yield\n finally:\n signal.signal(signal.SIGALRM, original_handler)\n signal.setitimer(signal.ITIMER_REAL, 0)\n\n\ndef keep_awake(iterable, delay=DELAY, interval=INTERVAL):\n \"\"\"\n Example:\n\n from workspace_utils import keep_awake\n\n for i in keep_awake(range(5)):\n # do iteration with lots of work here\n \"\"\"\n with active_session(delay, interval): yield from iterable\n\n\ndef predict(image_path, model, args, args_predict, topk=5):\n #print (\"entering predict function...\") \n img_tensor = pima.process_image(image_path)\n device = torch.device(\"cuda:0\" if torch.cuda.is_available() and args[6] == True else \"cpu\")\n #print(f\"device = {device}\")\n model.to(device)\n img_tensor = img_tensor.to(device)\n img_tensor = img_tensor.unsqueeze_(0)\n log_ps = model(img_tensor)\n ps = torch.exp(log_ps)\n ps_topk = ps.topk(topk)\n \n probs_and_classes = [ps_topk]\n print (\"outcome from predict.predict = \", probs_and_classes)\n return probs_and_classes\n\n\ndef define_image_path_for_inference():\n image_paths_100 = [\"aipnd-project_original udacity folder/flowers/test/100/image_07896.jpg\", \n \"aipnd-project_original udacity folder/flowers/test/100/image_07897.jpg\", \n \"aipnd-project_original udacity folder/flowers/test/100/image_07899.jpg\",\n \"aipnd-project_original udacity folder/flowers/test/100/image_07902.jpg\",\n \"aipnd-project_original udacity folder/flowers/test/100/image_07926.jpg\",\n \"aipnd-project_original udacity folder/flowers/test/100/image_07936.jpg\",\n \"aipnd-project_original udacity folder/flowers/test/100/image_07938.jpg\",\n \"aipnd-project_original udacity folder/flowers/test/100/image_07939.jpg\"]\n \n image_paths_13 = [\"aipnd-project_original udacity folder/flowers/test/13/image_05745.jpg\", \n \"aipnd-project_original udacity folder/flowers/test/13/image_05761.jpg\", \n \"aipnd-project_original udacity folder/flowers/test/13/image_05767.jpg\",\n \"aipnd-project_original udacity folder/flowers/test/13/image_05769.jpg\",\n \"aipnd-project_original udacity folder/flowers/test/13/image_05775.jpg\",\n \"aipnd-project_original udacity folder/flowers/test/13/image_05787.jpg\"]\n \n image_path_index = randint(0,len(image_paths_100)-1)\n image_path = image_paths_100[image_path_index]\n print(\"image path = \", image_path)\n return image_path\n\ndef prepare_and_run_prediction(image_path):\n args_train = train.get_input_args()\n args_predict = get_input_args()\n #model = import_the_checkpoint_ver2.load_checkpoint_and_rebuild_the_model(args)\n model = save_load_model.load_checkpoint_and_rebuild_the_model(args_train)\n with torch.no_grad():\n model.eval()\n #image_path = define_image_path_for_inference()\n probs_and_classes = predict(image_path, model, args_train, args_predict)\n #print(\"started topklabels \")\n topk_labels, numpy_probs = view_classify.get_topk_labels(args_predict, probs_and_classes, False)\n print(\"completed topklabels\", topk_labels)\n return probs_and_classes, topk_labels, numpy_probs\n \n \nif __name__ == \"__main__\":\n # stuff only to run when not called via 'import' here\n image_path = define_image_path_for_inference()\n prepare_and_run_prediction(image_path)",
"Overwriting predict.py\n"
],
[
"%%writefile train.py\n\nimport argparse\nimport torch\nfrom torchvision import datasets, transforms, models\n#import train\nfrom torch import nn\nfrom torch import optim\nimport time\nfrom workspace_utils import active_session\nimport helper\n#import import_the_checkpoint\nfrom collections import OrderedDict\nimport save_load_model\n\n\n\ndef get_input_args():\n if __name__ == \"__main__\":\n parser = argparse.ArgumentParser()\n parser.add_argument('--data_dir', type = str, default = 'aipnd-project_original udacity folder/flowers', help = 'path to the folder of flower images, just folder name, no slashes') \n parser.add_argument('--save_dir', type = str, default = 'checkpoints/', help = 'Directory to save checkpoints with \"/\" at the end') \n parser.add_argument('--arch', type = str, default = 'vgg11_bn', help = \"Selected architecture\") \n parser.add_argument('--learning_rate', type = float, default = 0.001)\n parser.add_argument('--hidden_units', type = str, default = '3136, 784, 416', help = \"3 comma-separated numbers for hidden layers input sizes, like \\\"3136, 784, 416\\\"\")\n parser.add_argument('--epochs', type = int, default = 10)\n parser.add_argument('--gpu', type = bool, default = True)\n \n in_args = parser.parse_args()\n #print(type(in_args.gpu))\n #print(\"Argument 1:\", in_args.data_dir)\n #print(\"Argument 2:\", in_args.arch)\n #print(\"Argument 3:\", in_args.learning_rate)\n in_args_dict = vars(in_args)\n in_args_list = [v for v in in_args_dict.values()]\n return in_args_list\n # stuff only to run when not called via 'import' here\n else:\n args = get_hardcoded_input_args()\n return(args)\n \n \n \n\ndef get_hardcoded_input_args():\n data_dir = 'aipnd-project_original udacity folder/flowers' \n save_dir = 'checkpoints/'\n arch = 'vgg11_bn'\n learning_rate = 0.001\n hidden_units = '3136, 784, 416'\n epochs = 10\n gpu = True\n in_args = [data_dir, save_dir, arch, learning_rate, hidden_units, epochs, gpu]\n #print(in_args)\n return in_args\n\ndef define_transfroms(args):\n #print(args)\n data_dir = args[0]\n train_dir = data_dir + '/train'\n valid_dir = data_dir + '/valid'\n test_dir = data_dir + '/test'\n \n \n \n # TODO: Define your transforms for the training, validation, and testing sets\n train_transforms = transforms.Compose([transforms.RandomRotation(30),\n transforms.RandomResizedCrop(224),\n transforms.RandomHorizontalFlip(),\n transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])])\n\n validation_and_testing_transforms = transforms.Compose([transforms.Resize(255),\n transforms.CenterCrop(224),\n transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])])\n\n\n # TODO: Load the datasets with ImageFolder\n train_dataset = datasets.ImageFolder(train_dir, transform=train_transforms)\n validation_dataset = datasets.ImageFolder(valid_dir, transform=validation_and_testing_transforms)\n test_dataset = datasets.ImageFolder(test_dir, transform=validation_and_testing_transforms)\n\n # TODO: Using the image datasets and the trainforms, define the dataloaders\n trainloader = torch.utils.data.DataLoader(train_dataset, batch_size=64, shuffle=True)\n validloader = torch.utils.data.DataLoader(validation_dataset, batch_size=64)\n testloader = torch.utils.data.DataLoader(test_dataset, batch_size=64)\n #print(\"Three loaders were defined in train.py\")\n class_to_idx = train_dataset.class_to_idx\n #print(\"class_to_idx = \", class_to_idx)\n return trainloader, validloader, testloader, class_to_idx\n\n\ndef create_model(args):\n \n #models_arch_list = ['alexnet', 'vgg11', 'vgg11_bn', 'vgg13', 'vgg13_bn', 'vgg16', 'vgg16_bn', 'vgg19', 'vgg19_bn', 'resnet18', 'resnet34', 'resnet50', 'resnet101', 'resnet152', 'squeezenet1_0', 'squeezenet1_1', 'densenet121', 'densenet169', 'densenet161', 'densenet201', 'googlenet', 'shufflenet_v2_x0_5', 'shufflenet_v2_x1_0', 'shufflenet_v2_x1_5', 'shufflenet_v2_x2_0', 'mobilenet_v2', 'resnext50_32x4d', 'resnext101_32x8d', 'wide_resnet50_2', 'wide_resnet101_2', 'mnasnet0_5', 'mnasnet0_75', 'mnasnet1_0', 'mnasnet1_3', 'fcn_resnet50', 'fcn_resnet101', 'deeplabv3_resnet50', 'deeplabv3_resnet101']\n #models_arch_list = ['alexnet', 'vgg11', 'vgg11_bn', 'vgg13', 'vgg13_bn']\n #arch = \"vgg11\"\n #model_arch_code = \"model = models.\" + arch + \"(pretrained=True)\"\n #exec(model_arch_code)\n #print(\"model_arch_code = \", model_arch_code)\n \n \n #for model_arch in models_arch_list:\n # model_arch_code = \"model = models.\" + model_arch + \"(pretrained=True)\"\n # print(model_arch)\n # print(exec(model_arch_code))\n \n \n arch = args[2]\n\n \n if arch == 'alexnet':\n model = models.alexnet(pretrained=True)\n #print(model.parameters)\n #classifier_name = 'classifier'\n classifier_input_size = 9216\n elif arch == 'vgg11':\n model = models.vgg11(pretrained=True)\n #print(model.parameters)\n classifier_input_size = 25088\n else:\n model = models.vgg11_bn(pretrained=True)\n #print(model.parameters)\n classifier_input_size = 25088\n \n #model = models.vgg13(pretrained=True)\n\n for param in model.parameters():\n param.requires_grad = False\n\n hidden_units = args[4]\n hidden_units = hidden_units.split(', ') \n hidden_units = [int(i) for i in hidden_units]\n #print(type(hidden_units[0]), hidden_units)\n\n classifier = nn.Sequential(OrderedDict([\n ('dropout1', nn.Dropout(p=0.2, inplace=False)),\n ('fc1', nn.Linear(classifier_input_size, hidden_units[0])),\n ('relu1', nn.ReLU()),\n ('dropout2', nn.Dropout(p=0.2, inplace=False)),\n ('fc2', nn.Linear(hidden_units[0], hidden_units[1])),\n ('relu2', nn.ReLU()),\n ('dropout3', nn.Dropout(p=0.2, inplace=False)),\n ('fc3', nn.Linear(hidden_units[1], hidden_units[2])),\n ('relu3', nn.ReLU()),\n ('dropout4', nn.Dropout(p=0.2, inplace=False)),\n ('fc4', nn.Linear(hidden_units[2], 102)),\n ('output', nn.LogSoftmax(dim=1))\n ]))\n \n model.classifier = classifier\n #print(model.parameters)\n\n #можно сделать тут через exec подмену классифаера если имя последнего уровня отличается от classifier\n return model\n \n\ndef train_and_validate(args):\n trainloader, validloader, testloader, class_to_idx = define_transfroms(args)\n #print(\"class_to_idx in train and validate = \", class_to_idx)\n \n model = create_model(args)\n model.class_to_idx = class_to_idx\n #print(\"model.class_to_idx = \", model.class_to_idx)\n print (\"gpu is available: \", torch.cuda.is_available()) \n print (\"input parameter for enabling GPU: \", args[6]) \n device = torch.device(\"cuda:0\" if torch.cuda.is_available() and args[6] == True else \"cpu\")\n print(f\"device = {device}\")\n model.to(device)\n criterion = nn.NLLLoss()\n optimizer = optim.Adam(model.classifier.parameters(), lr=args[3])\n train_losses, validation_losses, test_losses = [], [], []\n\n epochs = args[5]\n print (\"Starting epochs...\")\n with active_session():\n for e in range(epochs):\n #print(f\"Starting epoch {e+1}...\")\n running_loss = 0\n epoch_start_time = time.time()\n for ii, (inputs, labels) in enumerate(trainloader):\n #start = time.time()\n #print(f\"ii: {ii}\")\n inputs, labels = inputs.to(device), labels.to(device)\n optimizer.zero_grad()\n outputs = model.forward(inputs)\n #print(outputs.shape)\n loss = criterion(outputs, labels)\n loss.backward()\n optimizer.step()\n running_loss += loss.item()\n #print(f\"Batch = {ii}; Time per batch: {(time.time() - start):.3f} seconds, loss {running_loss/len(trainloader)}\")\n \n #if ii%30 == 0 or ii == 102:\n #print(f\"Batch = {ii}; loss {running_loss/len(trainloader):.5f}\")\n else:\n #test_loss = 0\n validation_loss = 0\n #accuracy = 0 \n validation_accuracy = 0\n with torch.no_grad():\n model.eval()\n #print(\"Starting eval phase...\")\n #for ii, (images, labels) in enumerate(testloader):\n for ii, (images, labels) in enumerate(validloader):\n images, labels = images.to(device), labels.to(device)\n log_ps = model(images)\n #test_loss += criterion(log_ps, labels)\n validation_loss += criterion(log_ps, labels)\n\n ps = torch.exp(log_ps)\n top_p, top_class = ps.topk(1, dim = 1)\n equals = top_class == labels.view(*top_class.shape)\n #accuracy += torch.mean(equals.type(torch.FloatTensor))\n validation_accuracy += torch.mean(equals.type(torch.FloatTensor))\n \n #print(f\"Eval batch={ii}, accuracy={accuracy}\")\n train_losses.append(running_loss/len(trainloader))\n #test_losses.append(test_loss/len(testloader))\n validation_losses.append(validation_loss/len(validloader))\n model.train()\n save_load_model.save_the_checkpoint(model, args)\n\n #print(f\"test loss: {test_loss/len(testloader):7.4f}, test accuracy: {accuracy/len(testloader):7.4f}\")\n print(f\"Epoch: {e+1}/{epochs}, training loss: {running_loss/len(trainloader):7.4f}, validation loss: {validation_loss/len(validloader):7.4f}, validation accuracy: {validation_accuracy/len(validloader):7.4f}, duration: {(time.time() - epoch_start_time):.3f} sec\") \n # TODO: Save the checkpoint \n #print(image_datasets['train'].class_to_idx)\n #save_load_model.save_the_checkpoint(model, args)\n \n\n #print(f\"Device = {device}; Time per batch: {(time.time() - start):.3f} seconds \n\ndef test_the_network(args):\n print(\"Entering testing function...\")\n trainloader, validloader, testloader, class_to_idx = define_transfroms(args)\n #model = import_the_checkpoint.load_checkpoint_and_rebuild_the_model() \n model = save_load_model.load_checkpoint_and_rebuild_the_model(args) \n device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n print(f\"device = {device}\")\n model.to(device)\n test_loss = 0\n test_accuracy = 0\n criterion = nn.NLLLoss()\n test_losses = []\n with torch.no_grad():\n model.eval()\n #print(\"Starting eval phase...\")\n for ii, (images, labels) in enumerate(testloader):\n images, labels = images.to(device), labels.to(device)\n log_ps = model(images)\n #test_loss += criterion(log_ps, labels)\n test_loss += criterion(log_ps, labels)\n\n ps = torch.exp(log_ps)\n top_p, top_class = ps.topk(1, dim = 1)\n equals = top_class == labels.view(*top_class.shape)\n #accuracy += torch.mean(equals.type(torch.FloatTensor))\n test_accuracy += torch.mean(equals.type(torch.FloatTensor))\n \n print(f\"Test batch={ii}, accuracy={test_accuracy/(ii+1)}\")\n \n #print(f\"test loss: {test_loss/len(testloader):7.4f}, test accuracy: {accuracy/len(testloader):7.4f}\")\n print(f\"Test loss: {test_loss/len(testloader):7.4f}, test accuracy: {test_accuracy/len(testloader):7.4f}\")\n print(\"Exiting testing function...\")\n\n\n \nif __name__ == \"__main__\":\n # stuff only to run when not called via 'import' here\n args = get_input_args()\n train_and_validate(args) ",
"Overwriting train.py\n"
]
],
[
[
"# Developing an AI application\n\nGoing forward, AI algorithms will be incorporated into more and more everyday applications. For example, you might want to include an image classifier in a smart phone app. To do this, you'd use a deep learning model trained on hundreds of thousands of images as part of the overall application architecture. A large part of software development in the future will be using these types of models as common parts of applications. \n\nIn this project, you'll train an image classifier to recognize different species of flowers. You can imagine using something like this in a phone app that tells you the name of the flower your camera is looking at. In practice you'd train this classifier, then export it for use in your application. We'll be using [this dataset](http://www.robots.ox.ac.uk/~vgg/data/flowers/102/index.html) of 102 flower categories, you can see a few examples below. \n\n<img src='assets/Flowers.png' width=500px>\n\nThe project is broken down into multiple steps:\n\n* Load and preprocess the image dataset\n* Train the image classifier on your dataset\n* Use the trained classifier to predict image content\n\nWe'll lead you through each part which you'll implement in Python.\n\nWhen you've completed this project, you'll have an application that can be trained on any set of labeled images. Here your network will be learning about flowers and end up as a command line application. But, what you do with your new skills depends on your imagination and effort in building a dataset. For example, imagine an app where you take a picture of a car, it tells you what the make and model is, then looks up information about it. Go build your own dataset and make something new.\n\nFirst up is importing the packages you'll need. It's good practice to keep all the imports at the beginning of your code. As you work through this notebook and find you need to import a package, make sure to add the import up here.",
"_____no_output_____"
]
],
[
[
"# Imports here\n#ready\nimport torch\nfrom torchvision import datasets, transforms, models\nimport train\nfrom torch import nn\nfrom torch import optim\nimport time\nfrom workspace_utils import active_session\nimport helper\n#import import_the_checkpoint\nimport save_load_model",
"_____no_output_____"
]
],
[
[
"## Load the data\n\nHere you'll use `torchvision` to load the data ([documentation](http://pytorch.org/docs/0.3.0/torchvision/index.html)). The data should be included alongside this notebook, otherwise you can [download it here](https://s3.amazonaws.com/content.udacity-data.com/nd089/flower_data.tar.gz). The dataset is split into three parts, training, validation, and testing. For the training, you'll want to apply transformations such as random scaling, cropping, and flipping. This will help the network generalize leading to better performance. You'll also need to make sure the input data is resized to 224x224 pixels as required by the pre-trained networks.\n\nThe validation and testing sets are used to measure the model's performance on data it hasn't seen yet. For this you don't want any scaling or rotation transformations, but you'll need to resize then crop the images to the appropriate size.\n\nThe pre-trained networks you'll use were trained on the ImageNet dataset where each color channel was normalized separately. For all three sets you'll need to normalize the means and standard deviations of the images to what the network expects. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`, calculated from the ImageNet images. These values will shift each color channel to be centered at 0 and range from -1 to 1.\n ",
"_____no_output_____"
]
],
[
[
"args = train.get_hardcoded_input_args()\ntrainloader, validloader, testloader = train.define_transfroms(args)",
"_____no_output_____"
]
],
[
[
"### Label mapping\n\nYou'll also need to load in a mapping from category label to category name. You can find this in the file `cat_to_name.json`. It's a JSON object which you can read in with the [`json` module](https://docs.python.org/2/library/json.html). This will give you a dictionary mapping the integer encoded categories to the actual names of the flowers.",
"_____no_output_____"
]
],
[
[
"#ready\nimport json\n\nwith open('cat_to_name.json', 'r') as f:\n cat_to_name = json.load(f)\n",
"_____no_output_____"
]
],
[
[
"# Building and training the classifier\n\nNow that the data is ready, it's time to build and train the classifier. As usual, you should use one of the pretrained models from `torchvision.models` to get the image features. Build and train a new feed-forward classifier using those features.\n\nWe're going to leave this part up to you. Refer to [the rubric](https://review.udacity.com/#!/rubrics/1663/view) for guidance on successfully completing this section. Things you'll need to do:\n\n* Load a [pre-trained network](http://pytorch.org/docs/master/torchvision/models.html) (If you need a starting point, the VGG networks work great and are straightforward to use)\n* Define a new, untrained feed-forward network as a classifier, using ReLU activations and dropout\n* Train the classifier layers using backpropagation using the pre-trained network to get the features\n* Track the loss and accuracy on the validation set to determine the best hyperparameters\n\nWe've left a cell open for you below, but use as many as you need. Our advice is to break the problem up into smaller parts you can run separately. Check that each part is doing what you expect, then move on to the next. You'll likely find that as you work through each part, you'll need to go back and modify your previous code. This is totally normal!\n\nWhen training make sure you're updating only the weights of the feed-forward network. You should be able to get the validation accuracy above 70% if you build everything right. Make sure to try different hyperparameters (learning rate, units in the classifier, epochs, etc) to find the best model. Save those hyperparameters to use as default values in the next part of the project.\n\nOne last important tip if you're using the workspace to run your code: To avoid having your workspace disconnect during the long-running tasks in this notebook, please read in the earlier page in this lesson called Intro to\nGPU Workspaces about Keeping Your Session Active. You'll want to include code from the workspace_utils.py module.\n\n**Note for Workspace users:** If your network is over 1 GB when saved as a checkpoint, there might be issues with saving backups in your workspace. Typically this happens with wide dense layers after the convolutional layers. If your saved checkpoint is larger than 1 GB (you can open a terminal and check with `ls -lh`), you should reduce the size of your hidden layers and train again.",
"_____no_output_____"
]
],
[
[
"import train\nargs = train.get_input_args()\n#print(args)\ntrain.train_and_validate(args)\n#ready",
"_____no_output_____"
]
],
[
[
"## Testing your network\n\n\n\n\nIt's good practice to test your trained network on test data, images the network has never seen either in training or validation. This will give you a good estimate for the model's performance on completely new images. Run the test images through the network and measure the accuracy, the same way you did validation. You should be able to reach around 70% accuracy on the test set if the model has been trained well.",
"_____no_output_____"
]
],
[
[
"import train\nargs = train.get_input_args()\ntrain.test_the_network(args)\n#ready",
"_____no_output_____"
]
],
[
[
"## Save the checkpoint\n\nNow that your network is trained, save the model so you can load it later for making predictions. You probably want to save other things such as the mapping of classes to indices which you get from one of the image datasets: `image_datasets['train'].class_to_idx`. You can attach this to the model as an attribute which makes inference easier later on.\n\n```model.class_to_idx = image_datasets['train'].class_to_idx```\n\nRemember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.",
"_____no_output_____"
]
],
[
[
"# TODO: Save the checkpoint \n\nimport save_load_model\nsave_load_model.save_the_checkpoint(model, args)\n\n",
"_____no_output_____"
]
],
[
[
"## Loading the checkpoint\n\nAt this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.",
"_____no_output_____"
]
],
[
[
"# TODO: Write a function that loads a checkpoint and rebuilds the model\n\nimport save_load_model\nsave_load_model.load_checkpoint_and_rebuild_the_model(args)",
"_____no_output_____"
]
],
[
[
"# Inference for classification\n\nNow you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like \n\n```python\nprobs, classes = predict(image_path, model)\nprint(probs)\nprint(classes)\n> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]\n> ['70', '3', '45', '62', '55']\n```\n\nFirst you'll need to handle processing the input image such that it can be used in your network. \n\n## Image Preprocessing\n\nYou'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training. \n\nFirst, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.\n\nColor channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.\n\nAs before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation. \n\nAnd finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.",
"_____no_output_____"
]
],
[
[
"#ready\nimport process_image as pima",
"_____no_output_____"
]
],
[
[
"To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).",
"_____no_output_____"
]
],
[
[
"#def imshow1(image, ax=None, title=None):\n \"\"\"Imshow for Tensor.\"\"\"\n import matplotlib as plt\n if ax is None:\n fig, ax = plt.subplots()\n \n # PyTorch tensors assume the color channel is the first dimension\n # but matplotlib assumes is the third dimension\n# image = image.numpy().transpose((1, 2, 0))\n \n # Undo preprocessing\n mean = np.array([0.485, 0.456, 0.406])\n std = np.array([0.229, 0.224, 0.225])\n image = std * image + mean\n \n # Image needs to be clipped between 0 and 1 or it looks like noise when displayed\n image = np.clip(image, 0, 1)\n \n ax.imshow(image)\n print (\"exitting\")\n \n return ax\n\n\n#imshow(image)\n# link to image \"aipnd-project_original udacity folder/flowers/test/100/image_07896.jpg\")",
"_____no_output_____"
]
],
[
[
"## Class Prediction\n\nOnce you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.\n\nTo get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.\n\nAgain, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.\n\n```python\nprobs, classes = predict(image_path, model)\nprint(probs)\nprint(classes)\n> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]\n> ['70', '3', '45', '62', '55']\n```",
"_____no_output_____"
]
],
[
[
"#ready\n#import predict_ver2\nimport predict\n\npredict.prepare_and_run_prediction()",
"_____no_output_____"
]
],
[
[
"## Sanity Checking\n\nNow that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:\n\n<img src='assets/inference_example.png' width=300px>\n\nYou can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.",
"_____no_output_____"
]
],
[
[
"# TODO: Display an image along with the top 5 classes\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom PIL import Image\nimport imshow as ims\nimport torch\nimport process_image as pima\n#import predict_ver2\nimport predict\nimport json\nimport view_classify\n\nfor i in np.arange(5):\n #print(\"start showing the image\")\n image_path = predict.define_image_path_for_inference()\n img_tensor_sanity = pima.process_image(image_path, True)\n ims.imshow(img_tensor_sanity)\n #print(\"complete showing the image\")\n #print(\"start prediction\")\n probs_and_classes, topk_labels, numpy_probs = predict.prepare_and_run_prediction(image_path)\n #print(\"compelte prediction\")\n #print(\"start view_claccify in cell\")\n view_classify.display_bar_histogram(numpy_probs, topk_labels)\n",
"image path from define_image_path_for_inference= aipnd-project_original udacity folder/flowers/test/100/image_07938.jpg\noutcome from predict.predict = [(tensor([[ 0.9995, 0.0004, 0.0001, 0.0000, 0.0000]]), tensor([[ 2, 43, 12, 84, 38]]))]\ncompleted topklabels ['blanket flower', 'wallflower', 'peruvian lily', 'hibiscus', 'barbeton daisy']\nimage path from define_image_path_for_inference= aipnd-project_original udacity folder/flowers/test/100/image_07896.jpg\noutcome from predict.predict = [(tensor([[ 0.9920, 0.0029, 0.0029, 0.0019, 0.0001]]), tensor([[ 2, 71, 43, 12, 52]]))]\ncompleted topklabels ['blanket flower', 'gazania', 'wallflower', 'peruvian lily', 'sunflower']\nimage path from define_image_path_for_inference= aipnd-project_original udacity folder/flowers/test/100/image_07936.jpg\noutcome from predict.predict = [(tensor([[ 0.9885, 0.0061, 0.0020, 0.0011, 0.0009]]), tensor([[ 2, 71, 47, 43, 38]]))]\ncompleted topklabels ['blanket flower', 'gazania', 'english marigold', 'wallflower', 'barbeton daisy']\nimage path from define_image_path_for_inference= aipnd-project_original udacity folder/flowers/test/100/image_07899.jpg\noutcome from predict.predict = [(tensor([[ 9.9965e-01, 2.0264e-04, 1.0030e-04, 2.0483e-05, 9.3435e-06]]), tensor([[ 2, 71, 38, 47, 52]]))]\ncompleted topklabels ['blanket flower', 'gazania', 'barbeton daisy', 'english marigold', 'sunflower']\nimage path from define_image_path_for_inference= aipnd-project_original udacity folder/flowers/test/100/image_07899.jpg\noutcome from predict.predict = [(tensor([[ 9.9965e-01, 2.0264e-04, 1.0030e-04, 2.0483e-05, 9.3435e-06]]), tensor([[ 2, 71, 38, 47, 52]]))]\ncompleted topklabels ['blanket flower', 'gazania', 'barbeton daisy', 'english marigold', 'sunflower']\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb7807ff93640d55c2bc14d4790d37e902147c8 | 6,952 | ipynb | Jupyter Notebook | OCR.ipynb | BHARATHSAMALA/Detecting-Sentiment-of-a-quote | 2c2e522de6a1bf6b987c528a9e4386cf576e0ca9 | [
"CC0-1.0"
] | null | null | null | OCR.ipynb | BHARATHSAMALA/Detecting-Sentiment-of-a-quote | 2c2e522de6a1bf6b987c528a9e4386cf576e0ca9 | [
"CC0-1.0"
] | null | null | null | OCR.ipynb | BHARATHSAMALA/Detecting-Sentiment-of-a-quote | 2c2e522de6a1bf6b987c528a9e4386cf576e0ca9 | [
"CC0-1.0"
] | null | null | null | 42.133333 | 2,105 | 0.608458 | [
[
[
"!pip install pillow\n!pip install pytesseract",
"_____no_output_____"
],
[
"# import the necessary packages\nfrom PIL import Image\nimport pytesseract\nimport argparse\nimport cv2\nimport os\n# construct the argument parse and parse the arguments\nap = argparse.ArgumentParser()\nap.add_argument(\"-i\", \"--image\", required=True, help=\"path to input image to be OCR'd\")\nap.add_argument(\"-p\", \"--preprocess\", type=str, default=\"thresh\",help=\"type of preprocessing to be done\")\nargs = vars(ap.parse_args())",
"usage: ipykernel_launcher.py [-h] -i IMAGE [-p PREPROCESS]\nipykernel_launcher.py: error: the following arguments are required: -i/--image\n"
],
[
"import cv2\nimport pytesseract\n\npytesseract.pytesseract.tesseract_cmd = r\"C:\\\\Users\\\\Bharath\\\\AppData\\\\Local\\\\Tesseract-OCR\\\\tesseract.exe\"\n\n# Grayscale, Gaussian blur, Otsu's threshold\nimage = cv2.imread('C:\\\\Users\\\\Bharath\\\\Desktop\\\\stuff\\\\vv\\\\hacker earth competition\\\\Data Files\\\\Sample Data Files\\\\Test126.jpg')\ngray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)\nblur = cv2.GaussianBlur(gray, (3,3), 0)\nthresh = cv2.threshold(blur, 0, 255, cv2.THRESH_BINARY_INV + cv2.THRESH_OTSU)[1]\n\n# Morph open to remove noise and invert image\nkernel = cv2.getStructuringElement(cv2.MORPH_RECT, (3,3))\nopening = cv2.morphologyEx(thresh, cv2.MORPH_OPEN, kernel, iterations=1)\ninvert = 255 - opening\n\n# Perform text extraction\ndata = pytesseract.image_to_string(invert, lang='eng', config='--psm 6')\nprint(data)",
"_____no_output_____"
],
[
"import pytesseract\n\nfrom PIL import Image\n\npytesseract.pytesseract.tesseract_cmd = r\"C:\\\\Users\\\\Bharath\\\\AppData\\\\Local\\\\Tesseract-OCR\\\\tesseract.exe\"\n\n\nimg = Image.open('C:\\\\Users\\\\Bharath\\\\Desktop\\\\stuff\\\\vv\\\\hacker earth competition\\\\Data Files\\\\Dataset\\\\Test126.jpg')\n\ntext = pytesseract.image_to_string(img)",
"_____no_output_____"
],
[
"text",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecb78d6462d9e3f60aa06046523a009eb5d7e7d1 | 1,140 | ipynb | Jupyter Notebook | notebooks/Untitled1.ipynb | AbdulazizBako/titanic | 07b133088f172913a06b92a792c62b295556950a | [
"FTL"
] | null | null | null | notebooks/Untitled1.ipynb | AbdulazizBako/titanic | 07b133088f172913a06b92a792c62b295556950a | [
"FTL"
] | null | null | null | notebooks/Untitled1.ipynb | AbdulazizBako/titanic | 07b133088f172913a06b92a792c62b295556950a | [
"FTL"
] | null | null | null | 22.352941 | 269 | 0.553509 | [
[
[
"conda list open cv",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
ecb78f5f00f0211ecbdadfb10b3f9c22b4320b7c | 44,461 | ipynb | Jupyter Notebook | Informatics/Deep Learning/TensorFlow - deeplearning.ai/3. NLP/Course_3_Week_3_Lesson_1c.ipynb | MarcosSalib/Cocktail_MOOC | 46279c2ec642554537c639702ed8e540ea49afdf | [
"MIT"
] | null | null | null | Informatics/Deep Learning/TensorFlow - deeplearning.ai/3. NLP/Course_3_Week_3_Lesson_1c.ipynb | MarcosSalib/Cocktail_MOOC | 46279c2ec642554537c639702ed8e540ea49afdf | [
"MIT"
] | null | null | null | Informatics/Deep Learning/TensorFlow - deeplearning.ai/3. NLP/Course_3_Week_3_Lesson_1c.ipynb | MarcosSalib/Cocktail_MOOC | 46279c2ec642554537c639702ed8e540ea49afdf | [
"MIT"
] | null | null | null | 130.384164 | 17,524 | 0.870426 | [
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Multiple Layer GRU",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function, unicode_literals\n\n\nimport tensorflow_datasets as tfds\nimport tensorflow as tf\nprint(tf.__version__)",
"2.3.0\n"
],
[
"import tensorflow_datasets as tfds\nimport tensorflow as tf\nprint(tf.__version__)",
"2.3.0\n"
],
[
"# Get the data\ndataset, info = tfds.load('imdb_reviews/subwords8k', with_info=True, as_supervised=True,\n download=False, data_dir='./')\ntrain_dataset, test_dataset = dataset['train'], dataset['test']\n",
"WARNING:absl:TFDS datasets with text encoding are deprecated and will be removed in a future version. Instead, you should use the plain text version and tokenize the text using `tensorflow_text` (See: https://www.tensorflow.org/tutorials/tensorflow_text/intro#tfdata_example)\n"
],
[
"tokenizer = info.features['text'].encoder",
"_____no_output_____"
],
[
"BUFFER_SIZE = 10000\nBATCH_SIZE = 64\n\ntrain_dataset = (\n train_dataset\n .shuffle(BUFFER_SIZE)\n .padded_batch(BATCH_SIZE, tf.compat.v1.data.get_output_shapes(train_dataset)))\n\ntest_dataset = (\n test_dataset\n .shuffle(BUFFER_SIZE)\n .padded_batch(BATCH_SIZE, tf.compat.v1.data.get_output_shapes(test_dataset)))",
"_____no_output_____"
],
[
"model = tf.keras.Sequential([\n tf.keras.layers.Embedding(tokenizer.vocab_size, 64),\n tf.keras.layers.Conv1D(128, 5, activation='relu'),\n tf.keras.layers.GlobalAveragePooling1D(),\n tf.keras.layers.Dense(64, activation='relu'),\n tf.keras.layers.Dense(1, activation='sigmoid')\n])",
"_____no_output_____"
],
[
"model.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding (Embedding) (None, None, 64) 523840 \n_________________________________________________________________\nconv1d (Conv1D) (None, None, 128) 41088 \n_________________________________________________________________\nglobal_average_pooling1d (Gl (None, 128) 0 \n_________________________________________________________________\ndense (Dense) (None, 64) 8256 \n_________________________________________________________________\ndense_1 (Dense) (None, 1) 65 \n=================================================================\nTotal params: 573,249\nTrainable params: 573,249\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])",
"_____no_output_____"
],
[
"NUM_EPOCHS = 10\nhistory = model.fit(train_dataset, epochs=NUM_EPOCHS, validation_data=test_dataset)",
"Epoch 1/10\n391/391 [==============================] - 89s 228ms/step - loss: 0.4461 - accuracy: 0.7812 - val_loss: 0.3107 - val_accuracy: 0.8771\nEpoch 2/10\n391/391 [==============================] - 85s 217ms/step - loss: 0.2243 - accuracy: 0.9183 - val_loss: 0.2992 - val_accuracy: 0.8799\nEpoch 3/10\n391/391 [==============================] - 86s 220ms/step - loss: 0.1700 - accuracy: 0.9381 - val_loss: 0.3397 - val_accuracy: 0.8711\nEpoch 4/10\n391/391 [==============================] - 85s 218ms/step - loss: 0.1371 - accuracy: 0.9509 - val_loss: 0.3632 - val_accuracy: 0.8702\nEpoch 5/10\n391/391 [==============================] - 86s 220ms/step - loss: 0.1171 - accuracy: 0.9600 - val_loss: 0.4033 - val_accuracy: 0.8592\nEpoch 6/10\n391/391 [==============================] - 84s 216ms/step - loss: 0.0913 - accuracy: 0.9700 - val_loss: 0.5003 - val_accuracy: 0.8576\nEpoch 7/10\n391/391 [==============================] - 88s 225ms/step - loss: 0.0675 - accuracy: 0.9800 - val_loss: 0.5518 - val_accuracy: 0.8602\nEpoch 8/10\n391/391 [==============================] - 89s 228ms/step - loss: 0.0491 - accuracy: 0.9870 - val_loss: 0.6342 - val_accuracy: 0.8574\nEpoch 9/10\n391/391 [==============================] - 88s 224ms/step - loss: 0.0417 - accuracy: 0.9877 - val_loss: 0.7138 - val_accuracy: 0.8562\nEpoch 10/10\n391/391 [==============================] - 89s 227ms/step - loss: 0.0346 - accuracy: 0.9897 - val_loss: 0.7868 - val_accuracy: 0.8524\n"
],
[
"import matplotlib.pyplot as plt\n\n\ndef plot_graphs(history, string):\n plt.plot(history.history[string])\n plt.plot(history.history['val_'+string])\n plt.xlabel(\"Epochs\")\n plt.ylabel(string)\n plt.legend([string, 'val_'+string])\n plt.title(string)\n plt.show()",
"_____no_output_____"
],
[
"plot_graphs(history, 'accuracy')",
"_____no_output_____"
],
[
"plot_graphs(history, 'loss')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb79fa563949ed4910438d303a6c6dcca8a8843 | 5,064 | ipynb | Jupyter Notebook | Lab2_HandsOn_frozenLake4x4_Qlearning.ipynb | Madhav2204/Python-practice-codes | d72a73aa83bad5e3ef7703c941911770548a3fe4 | [
"Apache-2.0"
] | null | null | null | Lab2_HandsOn_frozenLake4x4_Qlearning.ipynb | Madhav2204/Python-practice-codes | d72a73aa83bad5e3ef7703c941911770548a3fe4 | [
"Apache-2.0"
] | null | null | null | Lab2_HandsOn_frozenLake4x4_Qlearning.ipynb | Madhav2204/Python-practice-codes | d72a73aa83bad5e3ef7703c941911770548a3fe4 | [
"Apache-2.0"
] | null | null | null | 18.414545 | 86 | 0.481438 | [
[
[
"# Solving Frozen Lake 4x4 Environment",
"_____no_output_____"
],
[
"## Find optimal policy to reach state 'G' (goal) from state 'S' (starting state)",
"_____no_output_____"
]
],
[
[
"import gym\nimport numpy as np\nimport time, pickle, os",
"_____no_output_____"
],
[
"env = gym.make('FrozenLake-v0')",
"_____no_output_____"
],
[
"epsilon = 0.9\ntotal_episodes = 10000\nmax_steps = 100",
"_____no_output_____"
],
[
"alpha = 0.81 # 0.618\ngamma = 0.96",
"_____no_output_____"
],
[
"state = env.reset()",
"_____no_output_____"
],
[
"env.render()",
"\n\u001b[41mS\u001b[0mFFF\nFHFH\nFFFH\nHFFG\n"
]
],
[
[
"## Q-Learning",
"_____no_output_____"
]
],
[
[
"Q = np.zeros((env.observation_space.n, env.action_space.n))",
"_____no_output_____"
],
[
"total_episodes = 5000\nG = 0\nalpha = 0.618",
"_____no_output_____"
],
[
"## Write your learning code",
"_____no_output_____"
]
],
[
[
"### Analyse output",
"_____no_output_____"
]
],
[
[
"Q",
"_____no_output_____"
],
[
"state = env.reset()\ndone = None",
"_____no_output_____"
],
[
"while done != True:\n # We simply take the action with the highest Q Value\n action = np.argmax(Q[state])\n state, reward, done, info = env.step(action)\n env.render()",
"_____no_output_____"
]
],
[
[
"## Q Learning: Exploration Vs. Exploitation",
"_____no_output_____"
]
],
[
[
"def choose_action(state):\n action=0\n if np.random.uniform(0, 1) < epsilon:\n action = env.action_space.sample()\n else:\n action = np.argmax(Q[state, :])\n return action",
"_____no_output_____"
],
[
"def learn(state, state2, reward, action):\n predict = Q[state, action]\n target = reward + gamma * np.max(Q[state2, :])\n Q[state, action] = Q[state, action] + alpha * (target - predict)",
"_____no_output_____"
],
[
"## Write your learning code",
"_____no_output_____"
]
],
[
[
"### Analyse output",
"_____no_output_____"
]
],
[
[
"Q",
"_____no_output_____"
],
[
"state = env.reset()\ndone = None",
"_____no_output_____"
],
[
"while done != True:\n # We simply take the action with the highest Q Value\n action = np.argmax(Q[state])\n state, reward, done, info = env.step(action)\n env.render()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecb7cfb1c14d868bddbc9cadede563c7e96568e8 | 10,868 | ipynb | Jupyter Notebook | experiments/08_contrained_beds_50_percent.ipynb | MichaelAllen1966/2105_london_acute_stroke_unit | 56b710c58b5b6bdf5c03e3fb9ec65c53cd5336ff | [
"MIT"
] | null | null | null | experiments/08_contrained_beds_50_percent.ipynb | MichaelAllen1966/2105_london_acute_stroke_unit | 56b710c58b5b6bdf5c03e3fb9ec65c53cd5336ff | [
"MIT"
] | null | null | null | experiments/08_contrained_beds_50_percent.ipynb | MichaelAllen1966/2105_london_acute_stroke_unit | 56b710c58b5b6bdf5c03e3fb9ec65c53cd5336ff | [
"MIT"
] | null | null | null | 42.956522 | 256 | 0.363452 | [
[
[
"# London ASU model",
"_____no_output_____"
],
[
"## Requirements and module imports\n\nCode in this simulation uses a standard Anaconda Python environment (https://www.anaconda.com/distribution/#download-section). Additionally this model uses SimPy3 (https://simpy.readthedocs.io/en/latest/). Install SimPy3 with `pip install 'simpy<4'`.",
"_____no_output_____"
]
],
[
[
"import simpy\nimport inspect\nfrom sim_utils.replication import Replicator\nfrom sim_utils.parameters import Scenario",
"_____no_output_____"
]
],
[
[
"## Set up scenarios\n\nParameters defined in scenarios will overwrite default values in the parameters python file.",
"_____no_output_____"
]
],
[
[
"# Set up a dictionary to hold scenarios\nscenarios = {}\n\n# Baseline sceanrio (model defaults)\nscenarios['constrained_beds'] = Scenario(\n allow_non_preferred_asu = False)\nscenarios['constrained_beds_allow_redirect'] = Scenario(\n allow_non_preferred_asu = True)",
"_____no_output_____"
]
],
[
[
"## Run model",
"_____no_output_____"
]
],
[
[
"replications = 100\nreplications = Replicator(scenarios, replications)\nreplications.run_scenarios()",
" \nGlobal results (mean)\n---------------------\nname constrained_beds constrained_beds_allow_redirect\ntotal_patients 9,523.0 9,523.0\ntotal_patients_asu 5,516.0 5,527.0\ntotal_patients_displaced 0.0 6.0\ntotal_patients_waited 30.0 0.0\n\nAverage patients waiting for ASU\n--------------------------------\nname\nconstrained_beds 0.0\nconstrained_beds_allow_redirect 0.0\nName: asu_patients_unallocated, dtype: float64\n\nAverage delay (days) for patients who had to wait\n---------------------------------------------------\nname\nconstrained_beds 0.8\nconstrained_beds_allow_redirect 0.0\nName: 0, dtype: float64\n\nUnit admissions\n------------------\nname constrained_beds constrained_beds_allow_redirect\nBarnet General SU 161.9 163.1\nCharing Cross SU 331.2 331.4\nChelsea & Wminster SU 186.7 186.6\nCroydon SU 264.9 266.5\nHillingdon SU 282.9 282.3\nHomerton SU 168.7 168.5\nKingston SU 196.0 195.8\nKing’s College SU 180.6 185.1\nLewisham SU 536.3 534.5\nN Middlesex SU 319.3 320.5\nNewham General SU 178.6 176.1\nNorthwick Park SU 374.6 376.1\nPrincess Royal SU 281.9 279.9\nQueens Romford SU 409.4 408.4\nRoyal Free SU 291.7 290.7\nRoyal London SU 110.7 110.5\nSt George’s SU 155.8 156.4\nSt Helier SU 202.2 198.1\nSt Thomas SU 226.7 224.1\nUniversity College SU 171.9 173.8\nW Middlesex SU 204.3 201.3\nWhipps Cross SU 206.3 208.9\n\nUnit occupancy (mean)\n-----------------\nname constrained_beds constrained_beds_allow_redirect\nBarnet General SU 11.7 12.4\nCharing Cross SU 25.9 25.8\nChelsea & Wminster SU 14.6 14.1\nCroydon SU 20.6 20.3\nHillingdon SU 21.6 21.3\nHomerton SU 12.8 13.2\nKingston SU 14.6 14.8\nKing’s College SU 13.4 14.4\nLewisham SU 42.1 40.7\nN Middlesex SU 24.8 24.8\nNewham General SU 13.7 13.3\nNorthwick Park SU 29.8 28.3\nPrincess Royal SU 21.4 21.8\nQueens Romford SU 31.8 32.1\nRoyal Free SU 21.5 23.0\nRoyal London SU 8.4 8.7\nSt George’s SU 12.4 12.3\nSt Helier SU 15.2 15.7\nSt Thomas SU 17.0 17.4\nUniversity College SU 13.7 13.1\nW Middlesex SU 15.5 15.5\nWhipps Cross SU 15.6 15.7\n\nUnit occupancy (95th percentile)\n-----------------\nname constrained_beds constrained_beds_allow_redirect\nBarnet General SU 18.0 18.0\nCharing Cross SU 33.1 33.0\nChelsea & Wminster SU 21.0 20.0\nCroydon SU 29.0 27.0\nHillingdon SU 29.0 29.0\nHomerton SU 18.0 19.0\nKingston SU 21.0 22.0\nKing’s College SU 20.0 21.0\nLewisham SU 50.0 51.0\nN Middlesex SU 33.0 34.0\nNewham General SU 20.0 18.0\nNorthwick Park SU 38.0 36.0\nPrincess Royal SU 29.0 29.0\nQueens Romford SU 42.0 41.0\nRoyal Free SU 30.0 30.0\nRoyal London SU 12.0 12.0\nSt George’s SU 18.0 20.0\nSt Helier SU 21.0 21.0\nSt Thomas SU 25.0 25.0\nUniversity College SU 19.0 20.0\nW Middlesex SU 20.0 22.0\nWhipps Cross SU 22.0 22.0\n"
]
],
[
[
"## Show model default parameters\n\nRun the code below to model defaults (these are over-ridden by scenario values above).",
"_____no_output_____"
]
],
[
[
"print(inspect.getsource(Scenario.__init__))",
" def __init__(self, *initial_data, **kwargs):\n \"\"\"Default parameters\"\"\"\n # Simulation parameters\n self.sim_warmup = 100\n self.sim_duration = 365\n\n # Scale admissions\n self.scale_admissions = 1.0\n\n # Patient flow\n self.require_asu = 0.57\n self.esd_use = 0.\n self.esd_asu_los_reduction = 7.0\n self.los_cv = 0.3\n self.allow_non_preferred_asu = False\n\n # Overwrite default values\n\n for dictionary in initial_data:\n for key in dictionary:\n setattr(self, key, dictionary[key])\n for key in kwargs:\n setattr(self, key, kwargs[key])\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb7d502a73e523855f1375fa9a6273d5e7a83f8 | 217,734 | ipynb | Jupyter Notebook | docs/notebooks/Coverage.ipynb | vrthra-forks/fuzzingbook | 15319dcd7c213559cfe992c2e5936dab52929658 | [
"MIT"
] | null | null | null | docs/notebooks/Coverage.ipynb | vrthra-forks/fuzzingbook | 15319dcd7c213559cfe992c2e5936dab52929658 | [
"MIT"
] | null | null | null | docs/notebooks/Coverage.ipynb | vrthra-forks/fuzzingbook | 15319dcd7c213559cfe992c2e5936dab52929658 | [
"MIT"
] | null | null | null | 42.937093 | 15,768 | 0.65896 | [
[
[
"# Code Coverage\n\nIn the [previous chapter](Fuzzer.ipynb), we introduced _basic fuzzing_ – that is, generating random inputs to test programs. How do we measure the effectiveness of these tests? One way would be to check the number (and seriousness) of bugs found; but if bugs are scarce, we need a _proxy for the likelihood of a test to uncover a bug._ In this chapter, we introduce the concept of *code coverage*, measuring which parts of a program are actually executed during a test run. Measuring such coverage is also crucial for test generators that attempt to cover as much code as possible.",
"_____no_output_____"
]
],
[
[
"from bookutils import YouTubeVideo",
"_____no_output_____"
],
[
"YouTubeVideo('2lfgI9KdARs')",
"_____no_output_____"
]
],
[
[
"**Prerequisites**\n\n* You need some understanding of how a program is executed.\n* You should have learned about basic fuzzing in the [previous chapter](Fuzzer.ipynb).",
"_____no_output_____"
],
[
"## Synopsis\n<!-- Automatically generated. Do not edit. -->\n\nTo [use the code provided in this chapter](Importing.ipynb), write\n\n```python\n>>> from fuzzingbook.Coverage import <identifier>\n```\n\nand then make use of the following features.\n\n\nThis chapter introduces a `Coverage` class allowing you to measure coverage for Python programs. Within the context of this book, we use coverage information to guide fuzzing towards uncovered locations.\n\nThe typical usage of the `Coverage` class is in conjunction with a `with` clause:\n\n```python\n>>> with Coverage() as cov:\n>>> cgi_decode(\"a+b\")\n```\nPrinting out a coverage object shows the covered functions, with covered lines prefixed as `#`:\n\n```python\n>>> print(cov)\n 1 def cgi_decode(s: str) -> str:\n 2 \"\"\"Decode the CGI-encoded string `s`:\n 3 * replace '+' by ' '\n 4 * replace \"%xx\" by the character with hex number xx.\n 5 Return the decoded string. Raise `ValueError` for invalid inputs.\"\"\"\n 6 \n 7 # Mapping of hex digits to their integer values\n# 8 hex_values = {\n# 9 '0': 0, '1': 1, '2': 2, '3': 3, '4': 4,\n# 10 '5': 5, '6': 6, '7': 7, '8': 8, '9': 9,\n# 11 'a': 10, 'b': 11, 'c': 12, 'd': 13, 'e': 14, 'f': 15,\n# 12 'A': 10, 'B': 11, 'C': 12, 'D': 13, 'E': 14, 'F': 15,\n 13 }\n 14 \n# 15 t = \"\"\n# 16 i = 0\n# 17 while i < len(s):\n# 18 c = s[i]\n# 19 if c == '+':\n# 20 t += ' '\n# 21 elif c == '%':\n 22 digit_high, digit_low = s[i + 1], s[i + 2]\n 23 i += 2\n 24 if digit_high in hex_values and digit_low in hex_values:\n 25 v = hex_values[digit_high] * 16 + hex_values[digit_low]\n 26 t += chr(v)\n 27 else:\n 28 raise ValueError(\"Invalid encoding\")\n 29 else:\n# 30 t += c\n# 31 i += 1\n# 32 return t\n\n\n```\nThe `trace()` method returns the _trace_ – that is, the list of locations executed in order. Each location comes as a pair (`function name`, `line`).\n\n```python\n>>> cov.trace()\n[('cgi_decode', 9),\n ('cgi_decode', 10),\n ('cgi_decode', 11),\n ('cgi_decode', 12),\n ('cgi_decode', 8),\n ('cgi_decode', 15),\n ('cgi_decode', 16),\n ('cgi_decode', 17),\n ('cgi_decode', 18),\n ('cgi_decode', 19),\n ('cgi_decode', 21),\n ('cgi_decode', 30),\n ('cgi_decode', 31),\n ('cgi_decode', 17),\n ('cgi_decode', 18),\n ('cgi_decode', 19),\n ('cgi_decode', 20),\n ('cgi_decode', 31),\n ('cgi_decode', 17),\n ('cgi_decode', 18),\n ('cgi_decode', 19),\n ('cgi_decode', 21),\n ('cgi_decode', 30),\n ('cgi_decode', 31),\n ('cgi_decode', 17),\n ('cgi_decode', 32)]\n```\nThe `coverage()` method returns the _coverage_, that is, the set of locations in the trace executed at least once:\n\n```python\n>>> cov.coverage()\n{('cgi_decode', 8),\n ('cgi_decode', 9),\n ('cgi_decode', 10),\n ('cgi_decode', 11),\n ('cgi_decode', 12),\n ('cgi_decode', 15),\n ('cgi_decode', 16),\n ('cgi_decode', 17),\n ('cgi_decode', 18),\n ('cgi_decode', 19),\n ('cgi_decode', 20),\n ('cgi_decode', 21),\n ('cgi_decode', 30),\n ('cgi_decode', 31),\n ('cgi_decode', 32)}\n```\nCoverage sets can be subject to set operations, such as _intersection_ (which locations are covered in multiple executions) and _difference_ (which locations are covered in run _a_, but not _b_).\n\nThe chapter also discusses how to obtain such coverage from C programs.\n\n\n\n",
"_____no_output_____"
]
],
[
[
"import bookutils",
"_____no_output_____"
],
[
"# ignore\nfrom typing import Any, Optional, Callable, List, Type, Set, Tuple",
"_____no_output_____"
]
],
[
[
"## A CGI Decoder\n\nWe start by introducing a simple Python function that decodes a CGI-encoded string. CGI encoding is used in URLs (i.e., Web addresses) to encode characters that would be invalid in a URL, such as blanks and certain punctuation:\n\n* Blanks are replaced by `'+'`\n* Other invalid characters are replaced by '`%xx`', where `xx` is the two-digit hexadecimal equivalent.\n\nIn CGI encoding, the string `\"Hello, world!\"` would thus become `\"Hello%2c+world%21\"` where `2c` and `21` are the hexadecimal equivalents of `','` and `'!'`, respectively.\n\nThe function `cgi_decode()` takes such an encoded string and decodes it back to its original form. Our implementation replicates the code from \\cite{Pezze2008}. (It even includes its bugs – but we won't reveal them at this point.)",
"_____no_output_____"
]
],
[
[
"def cgi_decode(s: str) -> str:\n \"\"\"Decode the CGI-encoded string `s`:\n * replace '+' by ' '\n * replace \"%xx\" by the character with hex number xx.\n Return the decoded string. Raise `ValueError` for invalid inputs.\"\"\"\n\n # Mapping of hex digits to their integer values\n hex_values = {\n '0': 0, '1': 1, '2': 2, '3': 3, '4': 4,\n '5': 5, '6': 6, '7': 7, '8': 8, '9': 9,\n 'a': 10, 'b': 11, 'c': 12, 'd': 13, 'e': 14, 'f': 15,\n 'A': 10, 'B': 11, 'C': 12, 'D': 13, 'E': 14, 'F': 15,\n }\n\n t = \"\"\n i = 0\n while i < len(s):\n c = s[i]\n if c == '+':\n t += ' '\n elif c == '%':\n digit_high, digit_low = s[i + 1], s[i + 2]\n i += 2\n if digit_high in hex_values and digit_low in hex_values:\n v = hex_values[digit_high] * 16 + hex_values[digit_low]\n t += chr(v)\n else:\n raise ValueError(\"Invalid encoding\")\n else:\n t += c\n i += 1\n return t",
"_____no_output_____"
]
],
[
[
"Here is an example of how `cgi_decode()` works:",
"_____no_output_____"
]
],
[
[
"cgi_decode(\"Hello+world\")",
"_____no_output_____"
]
],
[
[
"If we want to systematically test `cgi_decode()`, how would we proceed?",
"_____no_output_____"
],
[
"The testing literature distinguishes two ways of deriving tests: _Black-box testing_ and _White-box testing._",
"_____no_output_____"
],
[
"## Black-Box Testing\n\nThe idea of *black-box testing* is to derive tests from the _specification_. In the above case, we thus would have to test `cgi_decode()` by the features specified and documented, including\n\n* testing for correct replacement of `'+'`;\n* testing for correct replacement of `\"%xx\"`;\n* testing for non-replacement of other characters; and\n* testing for recognition of illegal inputs.\n\nHere are four assertions (tests) that cover these four features. We can see that they all pass:",
"_____no_output_____"
]
],
[
[
"assert cgi_decode('+') == ' '\nassert cgi_decode('%20') == ' '\nassert cgi_decode('abc') == 'abc'\n\ntry:\n cgi_decode('%?a')\n assert False\nexcept ValueError:\n pass",
"_____no_output_____"
]
],
[
[
"The advantage of black-box testing is that it finds errors in the _specified_ behavior. It is independent from a given implementation, and thus allows to create test even before implementation. The downside is that _implemented_ behavior typically covers more ground than _specified_ behavior, and thus tests based on specification alone typically do not cover all implementation details.",
"_____no_output_____"
],
[
"## White-Box Testing\n\nIn contrast to black-box testing, *white-box testing* derives tests from the _implementation_, notably the internal structure. White-Box testing is closely tied to the concept of _covering_ structural features of the code. If a statement in the code is not executed during testing, for instance, this means that an error in this statement cannot be triggered either. White-Box testing thus introduces a number of *coverage criteria* that have to be fulfilled before the test can be said to be sufficient. The most frequently used coverage criteria are\n\n* *Statement coverage* – each statement in the code must be executed by at least one test input.\n* *Branch coverage* – each branch in the code must be taken by at least one test input. (This translates to each `if` and `while` decision once being true, and once being false.)\n\nBesides these, there are far more coverage criteria, including sequences of branches taken, loop iterations taken (zero, one, many), data flows between variable definitions and usages, and many more; \\cite{Pezze2008} has a great overview.",
"_____no_output_____"
],
[
"Let us consider `cgi_decode()`, above, and reason what we have to do such that each statement of the code is executed at least once. We'd have to cover\n\n* The block following `if c == '+'`\n* The two blocks following `if c == '%'` (one for valid input, one for invalid)\n* The final `else` case for all other characters.\n\nThis results in the same conditions as with black-box testing, above; again, the assertions above indeed would cover every statement in the code. Such a correspondence is actually pretty common, since programmers tend to implement different behaviors in different code locations; and thus, covering these locations will lead to test cases that cover the different (specified) behaviors.\n\nThe advantage of white-box testing is that it finds errors in _implemented_ behavior. It can be conducted even in cases where the specification does not provide sufficient details; actually, it helps in identifying (and thus specifying) corner cases in the specification. The downside is that it may miss _non-implemented_ behavior: If some specified functionality is missing, white-box testing will not find it.",
"_____no_output_____"
],
[
"## Tracing Executions\n\nOne nice feature of white-box testing is that one can actually automatically assess whether some program feature was covered. To this end, one _instruments_ the execution of the program such that during execution, a special functionality keeps track of which code was executed. After testing, this information can be passed to the programmer, who can then focus on writing tests that cover the yet uncovered code.",
"_____no_output_____"
],
[
"In most programming languages, it is rather difficult to set up programs such that one can trace their execution. Not so in Python. The function `sys.settrace(f)` allows to define a *tracing function* `f()` that is called for each and every line executed. Even better, it gets access to the current function and its name, current variable contents, and more. It is thus an ideal tool for *dynamic analysis* – that is, the analysis of what actually happens during an execution.",
"_____no_output_____"
],
[
"To illustrate how this works, let us again look into a specific execution of `cgi_decode()`.",
"_____no_output_____"
]
],
[
[
"cgi_decode(\"a+b\")",
"_____no_output_____"
]
],
[
[
"To track how the execution proceeds through `cgi_decode()`, we make use of `sys.settrace()`. First, we define the _tracing function_ that will be called for each line. It has three parameters: \n\n* The `frame` parameter gets you the current _frame_, allowing access to the current location and variables:\n * `frame.f_code` is the currently executed code with `frame.f_code.co_name` being the function name;\n * `frame.f_lineno` holds the current line number; and\n * `frame.f_locals` holds the current local variables and arguments.\n* The `event` parameter is a string with values including `\"line\"` (a new line has been reached) or `\"call\"` (a function is being called).\n* The `arg` parameter is an additional _argument_ for some events; for `\"return\"` events, for instance, `arg` holds the value being returned.",
"_____no_output_____"
],
[
"We use the tracing function for simply reporting the current line executed, which we access through the `frame` argument.",
"_____no_output_____"
]
],
[
[
"from types import FrameType, TracebackType",
"_____no_output_____"
],
[
"coverage = []",
"_____no_output_____"
],
[
"def traceit(frame: FrameType, event: str, arg: Any) -> Optional[Callable]:\n \"\"\"Trace program execution. To be passed to sys.settrace().\"\"\"\n if event == 'line':\n global coverage\n function_name = frame.f_code.co_name\n lineno = frame.f_lineno\n coverage.append(lineno)\n\n return traceit",
"_____no_output_____"
]
],
[
[
"We can switch tracing on and off with `sys.settrace()`:",
"_____no_output_____"
]
],
[
[
"import sys",
"_____no_output_____"
],
[
"def cgi_decode_traced(s: str) -> None:\n global coverage\n coverage = []\n sys.settrace(traceit) # Turn on\n cgi_decode(s)\n sys.settrace(None) # Turn off",
"_____no_output_____"
]
],
[
[
"When we compute `cgi_decode(\"a+b\")`, we can now see how the execution progresses through `cgi_decode()`. After the initialization of `hex_values`, `t`, and `i`, we see that the `while` loop is taken three times – one for every character in the input.",
"_____no_output_____"
]
],
[
[
"cgi_decode_traced(\"a+b\")\nprint(coverage)",
"[9, 10, 11, 12, 8, 15, 16, 17, 18, 19, 21, 30, 31, 17, 18, 19, 20, 31, 17, 18, 19, 21, 30, 31, 17, 32]\n"
]
],
[
[
"Which lines are these, actually? To this end, we get the source code of `cgi_decode_code` and encode it into an array `cgi_decode_lines`, which we will then annotate with coverage information. First, let us get the source code of `cgi_encode`:",
"_____no_output_____"
]
],
[
[
"import inspect",
"_____no_output_____"
],
[
"cgi_decode_code = inspect.getsource(cgi_decode)",
"_____no_output_____"
]
],
[
[
"`cgi_decode_code` is a string holding the source code. We can print it out with Python syntax highlighting:",
"_____no_output_____"
]
],
[
[
"from bookutils import print_content, print_file",
"_____no_output_____"
],
[
"print_content(cgi_decode_code[:300] + \"...\", \".py\")",
"\u001b[34mdef\u001b[39;49;00m \u001b[32mcgi_decode\u001b[39;49;00m(s: \u001b[36mstr\u001b[39;49;00m) -> \u001b[36mstr\u001b[39;49;00m:\n \u001b[33m\"\"\"Decode the CGI-encoded string `s`:\u001b[39;49;00m\n\u001b[33m * replace '+' by ' '\u001b[39;49;00m\n\u001b[33m * replace \"%xx\" by the character with hex number xx.\u001b[39;49;00m\n\u001b[33m Return the decoded string. Raise `ValueError` for invalid inputs.\"\"\"\u001b[39;49;00m\n\n \u001b[37m# Mapping of hex digits to their integer values\u001b[39;49;00m\n hex_v..."
]
],
[
[
"Using `splitlines()`, we split the code into an array of lines, indexed by line number.",
"_____no_output_____"
]
],
[
[
"cgi_decode_lines = [\"\"] + cgi_decode_code.splitlines()",
"_____no_output_____"
]
],
[
[
"`cgi_decode_lines[L]` is line L of the source code.",
"_____no_output_____"
]
],
[
[
"cgi_decode_lines[1]",
"_____no_output_____"
]
],
[
[
"We see that the first line (9) executed is actually the initialization of `hex_values`...",
"_____no_output_____"
]
],
[
[
"cgi_decode_lines[9:13]",
"_____no_output_____"
]
],
[
[
"... followed by the initialization of `t`:",
"_____no_output_____"
]
],
[
[
"cgi_decode_lines[15]",
"_____no_output_____"
]
],
[
[
"To see which lines actually have been covered at least once, we can convert `coverage` into a set:",
"_____no_output_____"
]
],
[
[
"covered_lines = set(coverage)\nprint(covered_lines)",
"{32, 8, 9, 10, 11, 12, 15, 16, 17, 18, 19, 20, 21, 30, 31}\n"
]
],
[
[
"Let us print out the full code, annotating lines not covered with '#':",
"_____no_output_____"
]
],
[
[
"for lineno in range(1, len(cgi_decode_lines)):\n if lineno not in covered_lines:\n print(\"# \", end=\"\")\n else:\n print(\" \", end=\"\")\n print(\"%2d \" % lineno, end=\"\")\n print_content(cgi_decode_lines[lineno], '.py')\n print()",
"# 1 \u001b[34mdef\u001b[39;49;00m \u001b[32mcgi_decode\u001b[39;49;00m(s: \u001b[36mstr\u001b[39;49;00m) -> \u001b[36mstr\u001b[39;49;00m:\n# 2 \u001b[33m\"\"\"\u001b[39;49;00m\u001b[33mDecode the CGI-encoded string `s`:\u001b[39;49;00m\u001b[33m\u001b[39;49;00m\n# 3 * replace \u001b[33m'\u001b[39;49;00m\u001b[33m+\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m by \u001b[33m'\u001b[39;49;00m\u001b[33m \u001b[39;49;00m\u001b[33m'\u001b[39;49;00m\n# 4 * replace \u001b[33m\"\u001b[39;49;00m\u001b[33m%x\u001b[39;49;00m\u001b[33mx\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m by the character \u001b[34mwith\u001b[39;49;00m \u001b[36mhex\u001b[39;49;00m number xx.\n# 5 Return the decoded string. Raise \u001b[04m\u001b[91m`\u001b[39;49;00m\u001b[36mValueError\u001b[39;49;00m\u001b[04m\u001b[91m`\u001b[39;49;00m \u001b[34mfor\u001b[39;49;00m invalid inputs.\u001b[33m\"\"\"\u001b[39;49;00m\u001b[33m\u001b[39;49;00m\n# 6 \n# 7 \u001b[37m# Mapping of hex digits to their integer values\u001b[39;49;00m\n 8 hex_values = {\n 9 \u001b[33m'\u001b[39;49;00m\u001b[33m0\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m0\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33m1\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m1\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33m2\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m2\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33m3\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m3\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33m4\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m4\u001b[39;49;00m,\n 10 \u001b[33m'\u001b[39;49;00m\u001b[33m5\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m5\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33m6\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m6\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33m7\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m7\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33m8\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m8\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33m9\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m9\u001b[39;49;00m,\n 11 \u001b[33m'\u001b[39;49;00m\u001b[33ma\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m10\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33mb\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m11\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33mc\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m12\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33md\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m13\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33me\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m14\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33mf\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m15\u001b[39;49;00m,\n 12 \u001b[33m'\u001b[39;49;00m\u001b[33mA\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m10\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33mB\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m11\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33mC\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m12\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33mD\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m13\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33mE\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m14\u001b[39;49;00m, \u001b[33m'\u001b[39;49;00m\u001b[33mF\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m: \u001b[34m15\u001b[39;49;00m,\n# 13 }\n# 14 \n 15 t = \u001b[33m\"\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m\n 16 i = \u001b[34m0\u001b[39;49;00m\n 17 \u001b[34mwhile\u001b[39;49;00m i < \u001b[36mlen\u001b[39;49;00m(s):\n 18 c = s[i]\n 19 \u001b[34mif\u001b[39;49;00m c == \u001b[33m'\u001b[39;49;00m\u001b[33m+\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m:\n 20 t += \u001b[33m'\u001b[39;49;00m\u001b[33m \u001b[39;49;00m\u001b[33m'\u001b[39;49;00m\n 21 \u001b[34melif\u001b[39;49;00m c == \u001b[33m'\u001b[39;49;00m\u001b[33m%\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m:\n# 22 digit_high, digit_low = s[i + \u001b[34m1\u001b[39;49;00m], s[i + \u001b[34m2\u001b[39;49;00m]\n# 23 i += \u001b[34m2\u001b[39;49;00m\n# 24 \u001b[34mif\u001b[39;49;00m digit_high \u001b[35min\u001b[39;49;00m hex_values \u001b[35mand\u001b[39;49;00m digit_low \u001b[35min\u001b[39;49;00m hex_values:\n# 25 v = hex_values[digit_high] * \u001b[34m16\u001b[39;49;00m + hex_values[digit_low]\n# 26 t += \u001b[36mchr\u001b[39;49;00m(v)\n# 27 \u001b[34melse\u001b[39;49;00m:\n# 28 \u001b[34mraise\u001b[39;49;00m \u001b[36mValueError\u001b[39;49;00m(\u001b[33m\"\u001b[39;49;00m\u001b[33mInvalid encoding\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m)\n# 29 \u001b[34melse\u001b[39;49;00m:\n 30 t += c\n 31 i += \u001b[34m1\u001b[39;49;00m\n 32 \u001b[34mreturn\u001b[39;49;00m t\n"
]
],
[
[
"We see that a number of lines (notably comments) have not been executed, simply because they are not executable. However, we also see that the lines under `if c == '%'` have _not_ been executed yet. If `\"a+b\"` were our only test case so far, this missing coverage would now encourage us to create another test case that actually covers these lines.",
"_____no_output_____"
],
[
"## A Coverage Class\n\nIn this book, we will make use of coverage again and again – to _measure_ the effectiveness of different test generation techniques, but also to _guide_ test generation towards code coverage. Our previous implementation with a global `coverage` variable is a bit cumbersome for that. We therefore implement some functionality that will help us measuring coverage easily.",
"_____no_output_____"
],
[
"The key idea of getting coverage is to make use of the Python `with` statement. The general form\n\n```python\nwith OBJECT [as VARIABLE]:\n BODY\n```\n\nexecutes `BODY` with `OBJECT` being defined (and stored in `VARIABLE`). The interesting thing is that at the beginning and end of `BODY`, the special methods `OBJECT.__enter__()` and `OBJECT.__exit__()` are automatically invoked; even if `BODY` raises an exception. This allows us to define a `Coverage` object where `Coverage.__enter__()` automatically turns on tracing and `Coverage.__exit__()` automatically turns off tracing again. After tracing, we can make use of special methods to access the coverage. This is what this looks like during usage:\n\n```python\nwith Coverage() as cov:\n function_to_be_traced()\nc = cov.coverage()\n```\n\nHere, tracing is automatically turned on during `function_to_be_traced()` and turned off again after the `with` block; afterwards, we can access the set of lines executed.",
"_____no_output_____"
],
[
"Here's the full implementation with all its bells and whistles. You don't have to get everything; it suffices that you know how to use it:",
"_____no_output_____"
]
],
[
[
"Location = Tuple[str, int]",
"_____no_output_____"
],
[
"class Coverage:\n \"\"\"Track coverage within a `with` block. Use as\n ```\n with Coverage() as cov:\n function_to_be_traced()\n c = cov.coverage()\n ```\n \"\"\"\n\n def __init__(self) -> None:\n \"\"\"Constructor\"\"\"\n self._trace: List[Location] = []\n\n # Trace function\n def traceit(self, frame: FrameType, event: str, arg: Any) -> Optional[Callable]:\n \"\"\"Tracing function. To be overloaded in subclasses.\"\"\"\n if self.original_trace_function is not None:\n self.original_trace_function(frame, event, arg)\n\n if event == \"line\":\n function_name = frame.f_code.co_name\n lineno = frame.f_lineno\n if function_name != '__exit__': # avoid tracing ourselves:\n self._trace.append((function_name, lineno))\n\n return self.traceit\n\n def __enter__(self) -> Any:\n \"\"\"Start of `with` block. Turn on tracing.\"\"\"\n self.original_trace_function = sys.gettrace()\n sys.settrace(self.traceit)\n return self\n\n def __exit__(self, exc_type: Type, exc_value: BaseException, \n tb: TracebackType) -> Optional[bool]:\n \"\"\"End of `with` block. Turn off tracing.\"\"\"\n sys.settrace(self.original_trace_function)\n return None # default: pass all exceptions\n\n def trace(self) -> List[Location]:\n \"\"\"The list of executed lines, as (function_name, line_number) pairs\"\"\"\n return self._trace\n\n def coverage(self) -> Set[Location]:\n \"\"\"The set of executed lines, as (function_name, line_number) pairs\"\"\"\n return set(self.trace())\n\n def function_names(self) -> Set[str]:\n \"\"\"The set of function names seen\"\"\"\n return set(function_name for (function_name, line_number) in self.coverage())\n\n def __repr__(self) -> str:\n \"\"\"Return a string representation of this object.\n Show covered (and uncovered) program code\"\"\"\n t = \"\"\n for function_name in self.function_names():\n # Similar code as in the example above\n try:\n fun = eval(function_name)\n except Exception as exc:\n t += f\"Skipping {function_name}: {exc}\"\n continue\n\n source_lines, start_line_number = inspect.getsourcelines(fun)\n for lineno in range(start_line_number, start_line_number + len(source_lines)):\n if (function_name, lineno) in self.trace():\n t += \"# \"\n else:\n t += \" \"\n t += \"%2d \" % lineno\n t += source_lines[lineno - start_line_number]\n\n return t",
"_____no_output_____"
]
],
[
[
"Let us put this to use:",
"_____no_output_____"
]
],
[
[
"with Coverage() as cov:\n cgi_decode(\"a+b\")\n\nprint(cov.coverage())",
"{('cgi_decode', 32), ('cgi_decode', 16), ('cgi_decode', 19), ('cgi_decode', 9), ('cgi_decode', 15), ('cgi_decode', 12), ('cgi_decode', 18), ('cgi_decode', 8), ('cgi_decode', 31), ('cgi_decode', 21), ('cgi_decode', 11), ('cgi_decode', 30), ('cgi_decode', 20), ('cgi_decode', 10), ('cgi_decode', 17)}\n"
]
],
[
[
"As you can see, the `Coverage()` class not only keeps track of lines executed, but also of function names. This is useful if you have a program that spans multiple files.",
"_____no_output_____"
],
[
"For interactive use, we can simply print the coverage object, and obtain a listing of the code, with covered lines marked as `#`.",
"_____no_output_____"
]
],
[
[
"print(cov)",
" 1 def cgi_decode(s: str) -> str:\n 2 \"\"\"Decode the CGI-encoded string `s`:\n 3 * replace '+' by ' '\n 4 * replace \"%xx\" by the character with hex number xx.\n 5 Return the decoded string. Raise `ValueError` for invalid inputs.\"\"\"\n 6 \n 7 # Mapping of hex digits to their integer values\n# 8 hex_values = {\n# 9 '0': 0, '1': 1, '2': 2, '3': 3, '4': 4,\n# 10 '5': 5, '6': 6, '7': 7, '8': 8, '9': 9,\n# 11 'a': 10, 'b': 11, 'c': 12, 'd': 13, 'e': 14, 'f': 15,\n# 12 'A': 10, 'B': 11, 'C': 12, 'D': 13, 'E': 14, 'F': 15,\n 13 }\n 14 \n# 15 t = \"\"\n# 16 i = 0\n# 17 while i < len(s):\n# 18 c = s[i]\n# 19 if c == '+':\n# 20 t += ' '\n# 21 elif c == '%':\n 22 digit_high, digit_low = s[i + 1], s[i + 2]\n 23 i += 2\n 24 if digit_high in hex_values and digit_low in hex_values:\n 25 v = hex_values[digit_high] * 16 + hex_values[digit_low]\n 26 t += chr(v)\n 27 else:\n 28 raise ValueError(\"Invalid encoding\")\n 29 else:\n# 30 t += c\n# 31 i += 1\n# 32 return t\n\n"
]
],
[
[
"## Comparing Coverage\n\nSince we represent coverage as a set of executed lines, we can also apply _set operations_ on these. For instance, we can find out which lines are covered by individual test cases, but not others:",
"_____no_output_____"
]
],
[
[
"with Coverage() as cov_plus:\n cgi_decode(\"a+b\")\nwith Coverage() as cov_standard:\n cgi_decode(\"abc\")\n\ncov_plus.coverage() - cov_standard.coverage()",
"_____no_output_____"
]
],
[
[
"This is the single line in the code that is executed only in the `'a+b'` input.",
"_____no_output_____"
],
[
"We can also compare sets to find out which lines still need to be covered. Let us define `cov_max` as the maximum coverage we can achieve. (Here, we do this by executing the \"good\" test cases we already have. In practice, one would statically analyze code structure, which we introduce in [the chapter on symbolic testing](SymbolicFuzzer.ipynb).)",
"_____no_output_____"
]
],
[
[
"with Coverage() as cov_max:\n cgi_decode('+')\n cgi_decode('%20')\n cgi_decode('abc')\n try:\n cgi_decode('%?a')\n except Exception:\n pass",
"_____no_output_____"
]
],
[
[
"Then, we can easily see which lines are _not_ yet covered by a test case:",
"_____no_output_____"
]
],
[
[
"cov_max.coverage() - cov_plus.coverage()",
"_____no_output_____"
]
],
[
[
"Again, these would be the lines handling `\"%xx\"`, which we have not yet had in the input.",
"_____no_output_____"
],
[
"## Coverage of Basic Fuzzing\n\nWe can now use our coverage tracing to assess the _effectiveness_ of testing methods – in particular, of course, test _generation_ methods. Our challenge is to achieve maximum coverage in `cgi_decode()` just with random inputs. In principle, we should _eventually_ get there, as eventually, we will have produced every possible string in the universe – but exactly how long is this? To this end, let us run just one fuzzing iteration on `cgi_decode()`:",
"_____no_output_____"
]
],
[
[
"from Fuzzer import fuzzer",
"_____no_output_____"
],
[
"sample = fuzzer()\nsample",
"_____no_output_____"
]
],
[
[
"Here's the invocation and the coverage we achieve. We wrap `cgi_decode()` in a `try...except` block such that we can ignore `ValueError` exceptions raised by illegal `%xx` formats.",
"_____no_output_____"
]
],
[
[
"with Coverage() as cov_fuzz:\n try:\n cgi_decode(sample)\n except:\n pass\ncov_fuzz.coverage()",
"_____no_output_____"
]
],
[
[
"Is this already the maximum coverage? Apparently, there are still lines missing:",
"_____no_output_____"
]
],
[
[
"cov_max.coverage() - cov_fuzz.coverage()",
"_____no_output_____"
]
],
[
[
"Let us try again, increasing coverage over 100 random inputs. We use an array `cumulative_coverage` to store the coverage achieved over time; `cumulative_coverage[0]` is the total number of lines covered after input 1, \n`cumulative_coverage[1]` is the number of lines covered after inputs 1–2, and so on.",
"_____no_output_____"
]
],
[
[
"trials = 100",
"_____no_output_____"
],
[
"def population_coverage(population: List[str], function: Callable) \\\n -> Tuple[Set[Location], List[int]]:\n cumulative_coverage: List[int] = []\n all_coverage: Set[Location] = set()\n\n for s in population:\n with Coverage() as cov:\n try:\n function(s)\n except:\n pass\n all_coverage |= cov.coverage()\n cumulative_coverage.append(len(all_coverage))\n\n return all_coverage, cumulative_coverage",
"_____no_output_____"
]
],
[
[
"Let us create a hundred inputs to determine how coverage increases:",
"_____no_output_____"
]
],
[
[
"def hundred_inputs() -> List[str]:\n population = []\n for i in range(trials):\n population.append(fuzzer())\n return population",
"_____no_output_____"
]
],
[
[
"Here's how the coverage increases with each input:",
"_____no_output_____"
]
],
[
[
"all_coverage, cumulative_coverage = \\\n population_coverage(hundred_inputs(), cgi_decode)",
"_____no_output_____"
],
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt # type: ignore",
"_____no_output_____"
],
[
"plt.plot(cumulative_coverage)\nplt.title('Coverage of cgi_decode() with random inputs')\nplt.xlabel('# of inputs')\nplt.ylabel('lines covered')",
"_____no_output_____"
]
],
[
[
"This is just _one_ run, of course; so let's repeat this a number of times and plot the averages.",
"_____no_output_____"
]
],
[
[
"runs = 100\n\n# Create an array with TRIALS elements, all zero\nsum_coverage = [0] * trials\n\nfor run in range(runs):\n all_coverage, coverage = population_coverage(hundred_inputs(), cgi_decode)\n assert len(coverage) == trials\n for i in range(trials):\n sum_coverage[i] += coverage[i]\n\naverage_coverage = []\nfor i in range(trials):\n average_coverage.append(sum_coverage[i] / runs)",
"_____no_output_____"
],
[
"plt.plot(average_coverage)\nplt.title('Average coverage of cgi_decode() with random inputs')\nplt.xlabel('# of inputs')\nplt.ylabel('lines covered')",
"_____no_output_____"
]
],
[
[
"We see that on average, we get full coverage after 40–60 fuzzing inputs.",
"_____no_output_____"
],
[
"## Getting Coverage from External Programs\n\nOf course, not all the world is programming in Python. The good news is that the problem of obtaining coverage is ubiquitous, and almost every programming language has some facility to measure coverage. Just as an example, let us therefore demonstrate how to obtain coverage for a C program.",
"_____no_output_____"
],
[
"Our C program (again) implements `cgi_decode`; this time as a program to be executed from the command line:\n\n```shell\n$ ./cgi_decode 'Hello+World'\nHello World\n```",
"_____no_output_____"
],
[
"Here comes the C code, first as a Python string. We start with the usual C includes:",
"_____no_output_____"
]
],
[
[
"cgi_c_code = \"\"\"\n/* CGI decoding as C program */\n\n#include <stdlib.h>\n#include <string.h>\n#include <stdio.h>\n\n\"\"\"",
"_____no_output_____"
]
],
[
[
"Here comes the initialization of `hex_values`:",
"_____no_output_____"
]
],
[
[
"cgi_c_code += r\"\"\"\nint hex_values[256];\n\nvoid init_hex_values() {\n for (int i = 0; i < sizeof(hex_values) / sizeof(int); i++) {\n hex_values[i] = -1;\n }\n hex_values['0'] = 0; hex_values['1'] = 1; hex_values['2'] = 2; hex_values['3'] = 3;\n hex_values['4'] = 4; hex_values['5'] = 5; hex_values['6'] = 6; hex_values['7'] = 7;\n hex_values['8'] = 8; hex_values['9'] = 9;\n\n hex_values['a'] = 10; hex_values['b'] = 11; hex_values['c'] = 12; hex_values['d'] = 13;\n hex_values['e'] = 14; hex_values['f'] = 15;\n\n hex_values['A'] = 10; hex_values['B'] = 11; hex_values['C'] = 12; hex_values['D'] = 13;\n hex_values['E'] = 14; hex_values['F'] = 15;\n}\n\"\"\"",
"_____no_output_____"
]
],
[
[
"Here's the actual implementation of `cgi_decode()`, using pointers for input source (`s`) and output target (`t`):",
"_____no_output_____"
]
],
[
[
"cgi_c_code += r\"\"\"\nint cgi_decode(char *s, char *t) {\n while (*s != '\\0') {\n if (*s == '+')\n *t++ = ' ';\n else if (*s == '%') {\n int digit_high = *++s;\n int digit_low = *++s;\n if (hex_values[digit_high] >= 0 && hex_values[digit_low] >= 0) {\n *t++ = hex_values[digit_high] * 16 + hex_values[digit_low];\n }\n else\n return -1;\n }\n else\n *t++ = *s;\n s++;\n }\n *t = '\\0';\n return 0;\n}\n\"\"\"",
"_____no_output_____"
]
],
[
[
"Finally, here's a driver which takes the first argument and invokes `cgi_decode` with it:",
"_____no_output_____"
]
],
[
[
"cgi_c_code += r\"\"\"\nint main(int argc, char *argv[]) {\n init_hex_values();\n\n if (argc >= 2) {\n char *s = argv[1];\n char *t = malloc(strlen(s) + 1); /* output is at most as long as input */\n int ret = cgi_decode(s, t);\n printf(\"%s\\n\", t);\n return ret;\n }\n else\n {\n printf(\"cgi_decode: usage: cgi_decode STRING\\n\");\n return 1;\n }\n}\n\"\"\"",
"_____no_output_____"
]
],
[
[
"Let us create the C source code: (Note that the following commands will overwrite the file `cgi_decode.c`, if it already exists in the current working directory. Be aware of this, if you downloaded the notebooks and are working locally.)",
"_____no_output_____"
]
],
[
[
"with open(\"cgi_decode.c\", \"w\") as f:\n f.write(cgi_c_code)",
"_____no_output_____"
]
],
[
[
"And here we have the C code with its syntax highlighted:",
"_____no_output_____"
]
],
[
[
"from bookutils import print_file",
"_____no_output_____"
],
[
"print_file(\"cgi_decode.c\")",
"\u001b[37m/* CGI decoding as C program */\u001b[39;49;00m\u001b[37m\u001b[39;49;00m\n\u001b[37m\u001b[39;49;00m\n\u001b[36m#\u001b[39;49;00m\u001b[36minclude\u001b[39;49;00m\u001b[37m \u001b[39;49;00m\u001b[37m<stdlib.h>\u001b[39;49;00m\u001b[36m\u001b[39;49;00m\n\u001b[36m#\u001b[39;49;00m\u001b[36minclude\u001b[39;49;00m\u001b[37m \u001b[39;49;00m\u001b[37m<string.h>\u001b[39;49;00m\u001b[36m\u001b[39;49;00m\n\u001b[36m#\u001b[39;49;00m\u001b[36minclude\u001b[39;49;00m\u001b[37m \u001b[39;49;00m\u001b[37m<stdio.h>\u001b[39;49;00m\u001b[36m\u001b[39;49;00m\n\u001b[37m\u001b[39;49;00m\n\u001b[37m\u001b[39;49;00m\n\u001b[36mint\u001b[39;49;00m\u001b[37m \u001b[39;49;00mhex_values[\u001b[34m256\u001b[39;49;00m];\u001b[37m\u001b[39;49;00m\n\u001b[37m\u001b[39;49;00m\n\u001b[36mvoid\u001b[39;49;00m\u001b[37m \u001b[39;49;00m\u001b[32minit_hex_values\u001b[39;49;00m()\u001b[37m \u001b[39;49;00m{\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34mfor\u001b[39;49;00m\u001b[37m \u001b[39;49;00m(\u001b[36mint\u001b[39;49;00m\u001b[37m \u001b[39;49;00mi\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m0\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mi\u001b[37m \u001b[39;49;00m<\u001b[37m \u001b[39;49;00m\u001b[34msizeof\u001b[39;49;00m(hex_values)\u001b[37m \u001b[39;49;00m/\u001b[37m \u001b[39;49;00m\u001b[34msizeof\u001b[39;49;00m(\u001b[36mint\u001b[39;49;00m);\u001b[37m \u001b[39;49;00mi++)\u001b[37m \u001b[39;49;00m{\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00mhex_values[i]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m-1\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m}\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33m0\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m0\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33m1\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m1\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33m2\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m2\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33m3\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m3\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33m4\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m4\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33m5\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m5\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33m6\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m6\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33m7\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m7\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33m8\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m8\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33m9\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m9\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33ma\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m10\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33mb\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m11\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33mc\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m12\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33md\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m13\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33me\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m14\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33mf\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m15\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33mA\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m10\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33mB\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m11\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33mC\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m12\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33mD\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m13\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33mE\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m14\u001b[39;49;00m;\u001b[37m \u001b[39;49;00mhex_values[\u001b[33m'\u001b[39;49;00m\u001b[33mF\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m]\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[34m15\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n}\u001b[37m\u001b[39;49;00m\n\u001b[37m\u001b[39;49;00m\n\u001b[36mint\u001b[39;49;00m\u001b[37m \u001b[39;49;00m\u001b[32mcgi_decode\u001b[39;49;00m(\u001b[36mchar\u001b[39;49;00m\u001b[37m \u001b[39;49;00m*s,\u001b[37m \u001b[39;49;00m\u001b[36mchar\u001b[39;49;00m\u001b[37m \u001b[39;49;00m*t)\u001b[37m \u001b[39;49;00m{\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34mwhile\u001b[39;49;00m\u001b[37m \u001b[39;49;00m(*s\u001b[37m \u001b[39;49;00m!=\u001b[37m \u001b[39;49;00m\u001b[33m'\u001b[39;49;00m\u001b[33m\\0\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\u001b[37m \u001b[39;49;00m{\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34mif\u001b[39;49;00m\u001b[37m \u001b[39;49;00m(*s\u001b[37m \u001b[39;49;00m==\u001b[37m \u001b[39;49;00m\u001b[33m'\u001b[39;49;00m\u001b[33m+\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m*t++\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[33m'\u001b[39;49;00m\u001b[33m \u001b[39;49;00m\u001b[33m'\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34melse\u001b[39;49;00m\u001b[37m \u001b[39;49;00m\u001b[34mif\u001b[39;49;00m\u001b[37m \u001b[39;49;00m(*s\u001b[37m \u001b[39;49;00m==\u001b[37m \u001b[39;49;00m\u001b[33m'\u001b[39;49;00m\u001b[33m%\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\u001b[37m \u001b[39;49;00m{\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[36mint\u001b[39;49;00m\u001b[37m \u001b[39;49;00mdigit_high\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m*++s;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[36mint\u001b[39;49;00m\u001b[37m \u001b[39;49;00mdigit_low\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m*++s;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34mif\u001b[39;49;00m\u001b[37m \u001b[39;49;00m(hex_values[digit_high]\u001b[37m \u001b[39;49;00m>=\u001b[37m \u001b[39;49;00m\u001b[34m0\u001b[39;49;00m\u001b[37m \u001b[39;49;00m&&\u001b[37m \u001b[39;49;00mhex_values[digit_low]\u001b[37m \u001b[39;49;00m>=\u001b[37m \u001b[39;49;00m\u001b[34m0\u001b[39;49;00m)\u001b[37m \u001b[39;49;00m{\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m*t++\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00mhex_values[digit_high]\u001b[37m \u001b[39;49;00m*\u001b[37m \u001b[39;49;00m\u001b[34m16\u001b[39;49;00m\u001b[37m \u001b[39;49;00m+\u001b[37m \u001b[39;49;00mhex_values[digit_low];\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m}\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34melse\u001b[39;49;00m\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34mreturn\u001b[39;49;00m\u001b[37m \u001b[39;49;00m\u001b[34m-1\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m}\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34melse\u001b[39;49;00m\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m*t++\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m*s;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00ms++;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m}\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m*t\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00m\u001b[33m'\u001b[39;49;00m\u001b[33m\\0\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34mreturn\u001b[39;49;00m\u001b[37m \u001b[39;49;00m\u001b[34m0\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n}\u001b[37m\u001b[39;49;00m\n\u001b[37m\u001b[39;49;00m\n\u001b[36mint\u001b[39;49;00m\u001b[37m \u001b[39;49;00m\u001b[32mmain\u001b[39;49;00m(\u001b[36mint\u001b[39;49;00m\u001b[37m \u001b[39;49;00margc,\u001b[37m \u001b[39;49;00m\u001b[36mchar\u001b[39;49;00m\u001b[37m \u001b[39;49;00m*argv[])\u001b[37m \u001b[39;49;00m{\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00minit_hex_values();\u001b[37m\u001b[39;49;00m\n\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34mif\u001b[39;49;00m\u001b[37m \u001b[39;49;00m(argc\u001b[37m \u001b[39;49;00m>=\u001b[37m \u001b[39;49;00m\u001b[34m2\u001b[39;49;00m)\u001b[37m \u001b[39;49;00m{\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[36mchar\u001b[39;49;00m\u001b[37m \u001b[39;49;00m*s\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00margv[\u001b[34m1\u001b[39;49;00m];\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[36mchar\u001b[39;49;00m\u001b[37m \u001b[39;49;00m*t\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00mmalloc(strlen(s)\u001b[37m \u001b[39;49;00m+\u001b[37m \u001b[39;49;00m\u001b[34m1\u001b[39;49;00m);\u001b[37m \u001b[39;49;00m\u001b[37m/* output is at most as long as input */\u001b[39;49;00m\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[36mint\u001b[39;49;00m\u001b[37m \u001b[39;49;00mret\u001b[37m \u001b[39;49;00m=\u001b[37m \u001b[39;49;00mcgi_decode(s,\u001b[37m \u001b[39;49;00mt);\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00mprintf(\u001b[33m\"\u001b[39;49;00m\u001b[33m%s\u001b[39;49;00m\u001b[33m\\n\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m,\u001b[37m \u001b[39;49;00mt);\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34mreturn\u001b[39;49;00m\u001b[37m \u001b[39;49;00mret;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m}\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34melse\u001b[39;49;00m\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m{\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00mprintf(\u001b[33m\"\u001b[39;49;00m\u001b[33mcgi_decode: usage: cgi_decode STRING\u001b[39;49;00m\u001b[33m\\n\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m);\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m\u001b[34mreturn\u001b[39;49;00m\u001b[37m \u001b[39;49;00m\u001b[34m1\u001b[39;49;00m;\u001b[37m\u001b[39;49;00m\n\u001b[37m \u001b[39;49;00m}\u001b[37m\u001b[39;49;00m\n}\u001b[37m\u001b[39;49;00m"
]
],
[
[
"We can now compile the C code into an executable. The `--coverage` option instructs the C compiler to instrument the code such that at runtime, coverage information will be collected. (The exact options vary from compiler to compiler.)",
"_____no_output_____"
]
],
[
[
"!cc --coverage -o cgi_decode cgi_decode.c",
"_____no_output_____"
]
],
[
[
"When we now execute the program, coverage information will automatically be collected and stored in auxiliary files:",
"_____no_output_____"
]
],
[
[
"!./cgi_decode 'Send+mail+to+me%40fuzzingbook.org'",
"Send mail to [email protected]\r\n"
]
],
[
[
"The coverage information is collected by the `gcov` program. For every source file given, it produces a new `.gcov` file with coverage information.",
"_____no_output_____"
]
],
[
[
"!gcov cgi_decode.c",
"File 'cgi_decode.c'\r\nLines executed:92.50% of 40\r\nCreating 'cgi_decode.c.gcov'\r\n\r\n"
]
],
[
[
"In the `.gcov` file, each line is prefixed with the number of times it was called (`-` stands for a non-executable line, `#####` stands for zero) as well as the line number. We can take a look at `cgi_decode()`, for instance, and see that the only code not executed yet is the `return -1` for an illegal input.",
"_____no_output_____"
]
],
[
[
"lines = open('cgi_decode.c.gcov').readlines()\nfor i in range(30, 50):\n print(lines[i], end='')",
" 1: 26:int cgi_decode(char *s, char *t) {\n 32: 27: while (*s != '\\0') {\n 31: 28: if (*s == '+')\n 3: 29: *t++ = ' ';\n 28: 30: else if (*s == '%') {\n 1: 31: int digit_high = *++s;\n 1: 32: int digit_low = *++s;\n 1: 33: if (hex_values[digit_high] >= 0 && hex_values[digit_low] >= 0) {\n 1: 34: *t++ = hex_values[digit_high] * 16 + hex_values[digit_low];\n 1: 35: }\n -: 36: else\n #####: 37: return -1;\n 1: 38: }\n -: 39: else\n 27: 40: *t++ = *s;\n 31: 41: s++;\n -: 42: }\n 1: 43: *t = '\\0';\n 1: 44: return 0;\n 1: 45:}\n"
]
],
[
[
"Let us read in this file to obtain a coverage set:",
"_____no_output_____"
]
],
[
[
"def read_gcov_coverage(c_file):\n gcov_file = c_file + \".gcov\"\n coverage = set()\n with open(gcov_file) as file:\n for line in file.readlines():\n elems = line.split(':')\n covered = elems[0].strip()\n line_number = int(elems[1].strip())\n if covered.startswith('-') or covered.startswith('#'):\n continue\n coverage.add((c_file, line_number))\n return coverage",
"_____no_output_____"
],
[
"coverage = read_gcov_coverage('cgi_decode.c')",
"_____no_output_____"
],
[
"list(coverage)[:5]",
"_____no_output_____"
]
],
[
[
"With this set, we can now do the same coverage computations as with our Python programs.",
"_____no_output_____"
],
[
"## Finding Errors with Basic Fuzzing\n\nGiven sufficient time, we can indeed cover each and every line within `cgi_decode()`, whatever the programming language would be. This does not mean that they would be error-free, though. Since we do not check the result of `cgi_decode()`, the function could return any value without us checking or noticing. To catch such errors, we would have to set up a *results checker* (commonly called an *oracle*) that would verify test results. In our case, we could compare the C and Python implementations of `cgi_decode()` and see whether both produce the same results.",
"_____no_output_____"
],
[
"Where fuzzing is great at, though, is in finding _internal errors_ that can be detected even without checking the result. Actually, if one runs our `fuzzer()` on `cgi_decode()`, one quickly finds such an error, as the following code shows:",
"_____no_output_____"
]
],
[
[
"from ExpectError import ExpectError",
"_____no_output_____"
],
[
"with ExpectError():\n for i in range(trials):\n try:\n s = fuzzer()\n cgi_decode(s)\n except ValueError:\n pass",
"Traceback (most recent call last):\n File \"/var/folders/n2/xd9445p97rb3xh7m1dfx8_4h0006ts/T/ipykernel_7605/2238772797.py\", line 5, in <module>\n cgi_decode(s)\n File \"/var/folders/n2/xd9445p97rb3xh7m1dfx8_4h0006ts/T/ipykernel_7605/1071239422.py\", line 22, in cgi_decode\n digit_high, digit_low = s[i + 1], s[i + 2]\nIndexError: string index out of range (expected)\n"
]
],
[
[
"So, it is possible to cause `cgi_decode()` to crash. Why is that? Let's take a look at its input:",
"_____no_output_____"
]
],
[
[
"s",
"_____no_output_____"
]
],
[
[
"The problem here is at the end of the string. After a `'%'` character, our implementation will always attempt to access two more (hexadecimal) characters, but if these are not there, we will get an `IndexError` exception. ",
"_____no_output_____"
],
[
"This problem is also present in our C variant, which inherits it from the original implementation \\cite{Pezze2008}:\n\n```c\nint digit_high = *++s;\nint digit_low = *++s;\n```\n\nHere, `s` is a pointer to the character to be read; `++` increments it by one character.\nIn the C implementation, the problem is actually much worse. If the `'%'` character is at the end of the string, the above code will first read a terminating character (`'\\0'` in C strings) and then the following character, which may be any memory content after the string, and which thus may cause the program to fail uncontrollably. The somewhat good news is that `'\\0'` is not a valid hexadecimal character, and thus, the C version will \"only\" read one character beyond the end of the string.",
"_____no_output_____"
],
[
"Interestingly enough, none of the manual tests we had designed earlier would trigger this bug. Actually, neither statement nor branch coverage, nor any of the coverage criteria commonly discussed in literature would find it. However, a simple fuzzing run can identify the error with a few runs – _if_ appropriate run-time checks are in place that find such overflows. This definitely calls for more fuzzing!",
"_____no_output_____"
],
[
"## Synopsis\n\nThis chapter introduces a `Coverage` class allowing you to measure coverage for Python programs. Within the context of this book, we use coverage information to guide fuzzing towards uncovered locations.",
"_____no_output_____"
],
[
"The typical usage of the `Coverage` class is in conjunction with a `with` clause:",
"_____no_output_____"
]
],
[
[
"with Coverage() as cov:\n cgi_decode(\"a+b\")",
"_____no_output_____"
]
],
[
[
"Printing out a coverage object shows the covered functions, with covered lines prefixed as `#`:",
"_____no_output_____"
]
],
[
[
"print(cov)",
" 1 def cgi_decode(s: str) -> str:\n 2 \"\"\"Decode the CGI-encoded string `s`:\n 3 * replace '+' by ' '\n 4 * replace \"%xx\" by the character with hex number xx.\n 5 Return the decoded string. Raise `ValueError` for invalid inputs.\"\"\"\n 6 \n 7 # Mapping of hex digits to their integer values\n# 8 hex_values = {\n# 9 '0': 0, '1': 1, '2': 2, '3': 3, '4': 4,\n# 10 '5': 5, '6': 6, '7': 7, '8': 8, '9': 9,\n# 11 'a': 10, 'b': 11, 'c': 12, 'd': 13, 'e': 14, 'f': 15,\n# 12 'A': 10, 'B': 11, 'C': 12, 'D': 13, 'E': 14, 'F': 15,\n 13 }\n 14 \n# 15 t = \"\"\n# 16 i = 0\n# 17 while i < len(s):\n# 18 c = s[i]\n# 19 if c == '+':\n# 20 t += ' '\n# 21 elif c == '%':\n 22 digit_high, digit_low = s[i + 1], s[i + 2]\n 23 i += 2\n 24 if digit_high in hex_values and digit_low in hex_values:\n 25 v = hex_values[digit_high] * 16 + hex_values[digit_low]\n 26 t += chr(v)\n 27 else:\n 28 raise ValueError(\"Invalid encoding\")\n 29 else:\n# 30 t += c\n# 31 i += 1\n# 32 return t\n\n"
]
],
[
[
"The `trace()` method returns the _trace_ – that is, the list of locations executed in order. Each location comes as a pair (`function name`, `line`).",
"_____no_output_____"
]
],
[
[
"cov.trace()",
"_____no_output_____"
]
],
[
[
"The `coverage()` method returns the _coverage_, that is, the set of locations in the trace executed at least once:",
"_____no_output_____"
]
],
[
[
"cov.coverage()",
"_____no_output_____"
]
],
[
[
"Coverage sets can be subject to set operations, such as _intersection_ (which locations are covered in multiple executions) and _difference_ (which locations are covered in run _a_, but not _b_).",
"_____no_output_____"
],
[
"The chapter also discusses how to obtain such coverage from C programs.",
"_____no_output_____"
]
],
[
[
"# ignore\nfrom ClassDiagram import display_class_hierarchy",
"_____no_output_____"
],
[
"# ignore\ndisplay_class_hierarchy(Coverage,\n public_methods=[\n Coverage.__init__,\n Coverage.__enter__,\n Coverage.__exit__,\n Coverage.coverage,\n Coverage.trace,\n Coverage.function_names,\n Coverage.__repr__,\n ],\n types={'Location': Location},\n project='fuzzingbook')",
"_____no_output_____"
]
],
[
[
"## Lessons Learned\n\n* Coverage metrics are a simple and fully automated means to approximate how much functionality of a program is actually executed during a test run.\n* A number of coverage metrics exist, the most important ones being statement coverage and branch coverage.\n* In Python, it is very easy to access the program state during execution, including the currently executed code.",
"_____no_output_____"
],
[
"At the end of the day, let's clean up: (Note that the following commands will delete all files in the current working directory that fit the pattern `cgi_decode.*`. Be aware of this, if you downloaded the notebooks and are working locally.)",
"_____no_output_____"
]
],
[
[
"import os\nimport glob",
"_____no_output_____"
],
[
"for file in glob.glob(\"cgi_decode\") + glob.glob(\"cgi_decode.*\"):\n os.remove(file)",
"_____no_output_____"
]
],
[
[
"## Next Steps\n\nCoverage is not only a tool to _measure_ test effectiveness, but also a great tool to _guide_ test generation towards specific goals – in particular uncovered code. We use coverage to\n\n* [guide _mutations_ of existing inputs towards better coverage in the chapter on mutation fuzzing](MutationFuzzer.ipynb)\n",
"_____no_output_____"
],
[
"## Background\n\nCoverage is a central concept in systematic software testing. For discussions, see the books in the [Introduction to Testing](Intro_Testing.ipynb).",
"_____no_output_____"
],
[
"## Exercises",
"_____no_output_____"
],
[
"### Exercise 1: Fixing `cgi_decode()`\n\nCreate an appropriate test to reproduce the `IndexError` discussed above. Fix `cgi_decode()` to prevent the bug. Show that your test (and additional `fuzzer()` runs) no longer expose the bug. Do the same for the C variant.",
"_____no_output_____"
],
[
"**Solution.** Here's a test case:",
"_____no_output_____"
]
],
[
[
"with ExpectError():\n assert cgi_decode('%') == '%'",
"Traceback (most recent call last):\n File \"/var/folders/n2/xd9445p97rb3xh7m1dfx8_4h0006ts/T/ipykernel_7605/1102034435.py\", line 2, in <module>\n assert cgi_decode('%') == '%'\n File \"/var/folders/n2/xd9445p97rb3xh7m1dfx8_4h0006ts/T/ipykernel_7605/1071239422.py\", line 22, in cgi_decode\n digit_high, digit_low = s[i + 1], s[i + 2]\nIndexError: string index out of range (expected)\n"
],
[
"with ExpectError():\n assert cgi_decode('%4') == '%4'",
"Traceback (most recent call last):\n File \"/var/folders/n2/xd9445p97rb3xh7m1dfx8_4h0006ts/T/ipykernel_7605/2291699482.py\", line 2, in <module>\n assert cgi_decode('%4') == '%4'\n File \"/var/folders/n2/xd9445p97rb3xh7m1dfx8_4h0006ts/T/ipykernel_7605/1071239422.py\", line 22, in cgi_decode\n digit_high, digit_low = s[i + 1], s[i + 2]\nIndexError: string index out of range (expected)\n"
],
[
"assert cgi_decode('%40') == '@'",
"_____no_output_____"
]
],
[
[
"Here's a fix:",
"_____no_output_____"
]
],
[
[
"def fixed_cgi_decode(s):\n \"\"\"Decode the CGI-encoded string `s`:\n * replace \"+\" by \" \"\n * replace \"%xx\" by the character with hex number xx.\n Return the decoded string. Raise `ValueError` for invalid inputs.\"\"\"\n\n # Mapping of hex digits to their integer values\n hex_values = {\n '0': 0, '1': 1, '2': 2, '3': 3, '4': 4,\n '5': 5, '6': 6, '7': 7, '8': 8, '9': 9,\n 'a': 10, 'b': 11, 'c': 12, 'd': 13, 'e': 14, 'f': 15,\n 'A': 10, 'B': 11, 'C': 12, 'D': 13, 'E': 14, 'F': 15,\n }\n\n t = \"\"\n i = 0\n while i < len(s):\n c = s[i]\n if c == '+':\n t += ' '\n elif c == '%' and i + 2 < len(s): # <--- *** FIX ***\n digit_high, digit_low = s[i + 1], s[i + 2]\n i += 2\n if digit_high in hex_values and digit_low in hex_values:\n v = hex_values[digit_high] * 16 + hex_values[digit_low]\n t += chr(v)\n else:\n raise ValueError(\"Invalid encoding\")\n else:\n t += c\n i += 1\n return t",
"_____no_output_____"
],
[
"assert fixed_cgi_decode('%') == '%'",
"_____no_output_____"
],
[
"assert fixed_cgi_decode('%4') == '%4'",
"_____no_output_____"
],
[
"assert fixed_cgi_decode('%40') == '@'",
"_____no_output_____"
]
],
[
[
"Here's the test:",
"_____no_output_____"
]
],
[
[
"for i in range(trials):\n try:\n s = fuzzer()\n fixed_cgi_decode(s)\n except ValueError:\n pass",
"_____no_output_____"
]
],
[
[
"For the C variant, the following will do:",
"_____no_output_____"
]
],
[
[
"cgi_c_code = cgi_c_code.replace(\n r\"if (*s == '%')\", # old code\n r\"if (*s == '%' && s[1] != '\\0' && s[2] != '\\0')\" # new code\n)",
"_____no_output_____"
]
],
[
[
"Go back to the above compilation commands and recompile `cgi_decode`.",
"_____no_output_____"
],
[
"### Exercise 2: Branch Coverage\n\nBesides statement coverage, _branch coverage_ is one of the most frequently used criteria to determine the quality of a test. In a nutshell, branch coverage measures how many different _control decisions_ are made in code. In the statement\n\n```python\nif CONDITION:\n do_a()\nelse:\n do_b()\n```\n\nfor instance, both the cases where `CONDITION` is true (branching to `do_a()`) and where `CONDITION` is false (branching to `do_b()`) have to be covered. This holds for all control statements with a condition (`if`, `while`, etc.).\n\nHow is branch coverage different from statement coverage? In the above example, there is actually no difference. In this one, though, there is:\n\n```python\nif CONDITION:\n do_a()\nsomething_else()\n```\n\nUsing statement coverage, a single test case where `CONDITION` is true suffices to cover the call to `do_a()`. Using branch coverage, however, we would also have to create a test case where `do_a()` is _not_ invoked.",
"_____no_output_____"
],
[
"Using our `Coverage` infrastructure, we can simulate branch coverage by considering _pairs of subsequent lines executed_. The `trace()` method gives us the list of lines executed one after the other:",
"_____no_output_____"
]
],
[
[
"with Coverage() as cov:\n cgi_decode(\"a+b\")\ntrace = cov.trace()\ntrace[:5]",
"_____no_output_____"
]
],
[
[
"#### Part 1: Compute branch coverage\n\nDefine a function `branch_coverage()` that takes a trace and returns the set of pairs of subsequent lines in a trace – in the above example, this would be \n\n```python\nset(\n(('cgi_decode', 9), ('cgi_decode', 10)),\n(('cgi_decode', 10), ('cgi_decode', 11)),\n# more_pairs\n)\n```\n\nBonus for advanced Python programmers: Define `BranchCoverage` as a subclass of `Coverage` and make `branch_coverage()` as above a `coverage()` method of `BranchCoverage`.",
"_____no_output_____"
],
[
"**Solution.** Here's a simple definition of `branch_coverage()`:",
"_____no_output_____"
]
],
[
[
"def branch_coverage(trace):\n coverage = set()\n past_line = None\n for line in trace:\n if past_line is not None:\n coverage.add((past_line, line))\n past_line = line\n\n return coverage",
"_____no_output_____"
],
[
"branch_coverage(trace)",
"_____no_output_____"
]
],
[
[
"Here's a definition as a class:",
"_____no_output_____"
]
],
[
[
"class BranchCoverage(Coverage):\n def coverage(self):\n \"\"\"The set of executed line pairs\"\"\"\n coverage = set()\n past_line = None\n for line in self.trace():\n if past_line is not None:\n coverage.add((past_line, line))\n past_line = line\n\n return coverage",
"_____no_output_____"
]
],
[
[
"#### Part 2: Comparing statement coverage and branch coverage\n\nUse `branch_coverage()` to repeat the experiments in this chapter with branch coverage rather than statement coverage. Do the manually written test cases cover all branches?",
"_____no_output_____"
],
[
"**Solution.** Let's repeat the above experiments with `BranchCoverage`:",
"_____no_output_____"
]
],
[
[
"with BranchCoverage() as cov:\n cgi_decode(\"a+b\")\n\nprint(cov.coverage())",
"{(('cgi_decode', 30), ('cgi_decode', 31)), (('cgi_decode', 8), ('cgi_decode', 15)), (('cgi_decode', 11), ('cgi_decode', 12)), (('cgi_decode', 19), ('cgi_decode', 20)), (('cgi_decode', 10), ('cgi_decode', 11)), (('cgi_decode', 18), ('cgi_decode', 19)), (('cgi_decode', 21), ('cgi_decode', 30)), (('cgi_decode', 12), ('cgi_decode', 8)), (('cgi_decode', 20), ('cgi_decode', 31)), (('cgi_decode', 17), ('cgi_decode', 32)), (('cgi_decode', 19), ('cgi_decode', 21)), (('cgi_decode', 9), ('cgi_decode', 10)), (('cgi_decode', 16), ('cgi_decode', 17)), (('cgi_decode', 31), ('cgi_decode', 17)), (('cgi_decode', 15), ('cgi_decode', 16)), (('cgi_decode', 17), ('cgi_decode', 18))}\n"
],
[
"with BranchCoverage() as cov_plus:\n cgi_decode(\"a+b\")\nwith BranchCoverage() as cov_standard:\n cgi_decode(\"abc\")\n\ncov_plus.coverage() - cov_standard.coverage()",
"_____no_output_____"
],
[
"with BranchCoverage() as cov_max:\n cgi_decode('+')\n cgi_decode('%20')\n cgi_decode('abc')\n try:\n cgi_decode('%?a')\n except:\n pass",
"_____no_output_____"
],
[
"cov_max.coverage() - cov_plus.coverage()",
"_____no_output_____"
],
[
"sample",
"_____no_output_____"
],
[
"with BranchCoverage() as cov_fuzz:\n try:\n cgi_decode(s)\n except:\n pass\ncov_fuzz.coverage()",
"_____no_output_____"
],
[
"cov_max.coverage() - cov_fuzz.coverage()",
"_____no_output_____"
],
[
"def population_branch_coverage(population, function):\n cumulative_coverage = []\n all_coverage = set()\n\n for s in population:\n with BranchCoverage() as cov:\n try:\n function(s)\n except Exception:\n pass\n all_coverage |= cov.coverage()\n cumulative_coverage.append(len(all_coverage))\n\n return all_coverage, cumulative_coverage",
"_____no_output_____"
],
[
"all_branch_coverage, cumulative_branch_coverage = population_branch_coverage(\n hundred_inputs(), cgi_decode)",
"_____no_output_____"
],
[
"plt.plot(cumulative_branch_coverage)\nplt.title('Branch coverage of cgi_decode() with random inputs')\nplt.xlabel('# of inputs')\nplt.ylabel('line pairs covered')",
"_____no_output_____"
],
[
"len(cov_max.coverage())",
"_____no_output_____"
],
[
"all_branch_coverage - cov_max.coverage()",
"_____no_output_____"
]
],
[
[
"The additional coverage comes from the exception raised via an illegal input (say, `%g`).",
"_____no_output_____"
]
],
[
[
"cov_max.coverage() - all_branch_coverage",
"_____no_output_____"
]
],
[
[
"This is an artefact coming from the subsequent execution of `cgi_decode()` when computing `cov_max`.",
"_____no_output_____"
],
[
"#### Part 3: Average coverage\n\nAgain, repeat the above experiments with branch coverage. Does `fuzzer()` cover all branches, and if so, how many tests does it take on average?",
"_____no_output_____"
],
[
"**Solution.** We repeat the experiments we ran with line coverage with branch coverage.",
"_____no_output_____"
]
],
[
[
"runs = 100\n\n# Create an array with TRIALS elements, all zero\nsum_coverage = [0] * trials\n\nfor run in range(runs):\n all_branch_coverage, coverage = population_branch_coverage(\n hundred_inputs(), cgi_decode)\n assert len(coverage) == trials\n for i in range(trials):\n sum_coverage[i] += coverage[i]\n\naverage_coverage = []\nfor i in range(trials):\n average_coverage.append(sum_coverage[i] / runs)",
"_____no_output_____"
],
[
"plt.plot(average_coverage)\nplt.title('Average branch coverage of cgi_decode() with random inputs')\nplt.xlabel('# of inputs')\nplt.ylabel('line pairs covered')",
"_____no_output_____"
]
],
[
[
"We see that achieving branch coverage takes longer than statement coverage; it simply is a more difficult criterion to satisfy with random inputs.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
ecb7dffc126c97c1c5d2938fc9055fa6b1a614fd | 6,722 | ipynb | Jupyter Notebook | notebooks/trees_sol_01.ipynb | lesteve/scikit-learn-mooc | b822586b98e71dbbf003bde86be57412cb170291 | [
"CC-BY-4.0"
] | null | null | null | notebooks/trees_sol_01.ipynb | lesteve/scikit-learn-mooc | b822586b98e71dbbf003bde86be57412cb170291 | [
"CC-BY-4.0"
] | null | null | null | notebooks/trees_sol_01.ipynb | lesteve/scikit-learn-mooc | b822586b98e71dbbf003bde86be57412cb170291 | [
"CC-BY-4.0"
] | null | null | null | 30.008929 | 101 | 0.58554 | [
[
[
"# 📃 Solution for Exercise M5.01\n\nIn the previous notebook, we showed how a tree with a depth of 1 level was\nworking. The aim of this exercise is to repeat part of the previous\nexperiment for a depth with 2 levels to show how the process of partitioning\nis repeated over time.\n\nBefore to start, we will:\n\n* load the dataset;\n* split the dataset into training and testing dataset;\n* define the function to show the classification decision function.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\npenguins = pd.read_csv(\"../datasets/penguins_classification.csv\")\nculmen_columns = [\"Culmen Length (mm)\", \"Culmen Depth (mm)\"]\ntarget_column = \"Species\"",
"_____no_output_____"
]
],
[
[
"<div class=\"admonition note alert alert-info\">\n<p class=\"first admonition-title\" style=\"font-weight: bold;\">Note</p>\n<p class=\"last\">If you want a deeper overview regarding this dataset, you can refer to the\nAppendix - Datasets description section at the end of this MOOC.</p>\n</div>",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\ndata, target = penguins[culmen_columns], penguins[target_column]\ndata_train, data_test, target_train, target_test = train_test_split(\n data, target, random_state=0\n)\nrange_features = {\n feature_name: (data[feature_name].min() - 1, data[feature_name].max() + 1)\n for feature_name in data.columns\n}",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\n\ndef plot_decision_function(fitted_classifier, range_features, ax=None):\n \"\"\"Plot the boundary of the decision function of a classifier.\"\"\"\n from sklearn.preprocessing import LabelEncoder\n\n feature_names = list(range_features.keys())\n # create a grid to evaluate all possible samples\n plot_step = 0.02\n xx, yy = np.meshgrid(\n np.arange(*range_features[feature_names[0]], plot_step),\n np.arange(*range_features[feature_names[1]], plot_step),\n )\n\n # compute the associated prediction\n Z = fitted_classifier.predict(np.c_[xx.ravel(), yy.ravel()])\n Z = LabelEncoder().fit_transform(Z)\n Z = Z.reshape(xx.shape)\n\n # make the plot of the boundary and the data samples\n if ax is None:\n _, ax = plt.subplots()\n ax.contourf(xx, yy, Z, alpha=0.4, cmap=\"RdBu\")\n\n return ax",
"_____no_output_____"
]
],
[
[
"Create a decision tree classifier with a maximum depth of 2 levels and fit\nthe training data. Once this classifier trained, plot the data and the\ndecision boundary to see the benefit of increasing the depth.",
"_____no_output_____"
]
],
[
[
"# solution\nfrom sklearn.tree import DecisionTreeClassifier\n\ntree = DecisionTreeClassifier(max_depth=2)\ntree.fit(data_train, target_train)",
"_____no_output_____"
],
[
"import seaborn as sns\n\npalette = [\"tab:red\", \"tab:blue\", \"black\"]\nax = sns.scatterplot(data=penguins, x=culmen_columns[0], y=culmen_columns[1],\n hue=target_column, palette=palette)\nplot_decision_function(tree, range_features, ax=ax)\nplt.legend(bbox_to_anchor=(1.05, 1), loc='upper left')\n_ = plt.title(\"Decision boundary using a decision tree\")",
"_____no_output_____"
]
],
[
[
"Did we make use of the feature \"Culmen Length\"?\nPlot the tree using the function `sklearn.tree.plot_tree` to find out!",
"_____no_output_____"
]
],
[
[
"# solution\nfrom sklearn.tree import plot_tree\n\n_, ax = plt.subplots(figsize=(16, 12))\n_ = plot_tree(tree, feature_names=culmen_columns,\n class_names=tree.classes_, impurity=False, ax=ax)",
"_____no_output_____"
]
],
[
[
"The resulting tree has 7 nodes: 3 of them are \"split nodes\" and 4\nare \"leaf nodes\" (or simply \"leaves\"), organized in 2 levels.\nWe see that the second tree level used the \"Culmen Length\" to make\ntwo new decisions. Qualitatively, we saw that such a simple tree was enough\nto classify the penguins' species.",
"_____no_output_____"
],
[
"Compute the accuracy of the decision tree on the testing data.",
"_____no_output_____"
]
],
[
[
"# solution\ntest_score = tree.fit(data_train, target_train).score(data_test, target_test)\nprint(f\"Accuracy of the DecisionTreeClassifier: {test_score:.2f}\")",
"_____no_output_____"
]
],
[
[
"At this stage, we have the intuition that a decision tree is built by\nsuccessively partitioning the feature space, considering one feature at a\ntime.\n\nWe predict an Adelie penguin if the feature value is below the threshold,\nwhich is not surprising since this partition was almost pure. If the feature\nvalue is above the threshold, we predict the Gentoo penguin, the class that\nis most probable.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb7e87d81b7bded0a64a6f9db76ff093737b427 | 6,583 | ipynb | Jupyter Notebook | tutorials/regression_mpra_example/regression_mpra_example.ipynb | msindeeva/selene | 601d0d11e11838a61778b4282482cacd20ce1a4a | [
"BSD-3-Clause-Clear"
] | 307 | 2018-09-21T16:48:12.000Z | 2022-03-23T21:42:04.000Z | tutorials/regression_mpra_example/regression_mpra_example.ipynb | msindeeva/selene | 601d0d11e11838a61778b4282482cacd20ce1a4a | [
"BSD-3-Clause-Clear"
] | 104 | 2018-08-07T13:44:29.000Z | 2022-01-12T01:35:30.000Z | tutorials/regression_mpra_example/regression_mpra_example.ipynb | msindeeva/selene | 601d0d11e11838a61778b4282482cacd20ce1a4a | [
"BSD-3-Clause-Clear"
] | 85 | 2018-10-20T08:06:31.000Z | 2022-03-29T15:17:30.000Z | 49.496241 | 277 | 0.676287 | [
[
[
"# Regression Models in Selene\n\nSelene is a flexible framework, and can be used for tasks beyond simple classification.\nThis tutorial demonstrates the simple process of training regression models with Selene.\nFor this example, we will predict mean ribosomal load (MRL) from 50 base pair 5' UTR sequences using models and data from [*Human 5′ UTR design and variant effect prediction from a massively parallel translation assay*](https://doi.org/10.1101/310375) by Sample et al.\nThis data was generated from a massively parallel reporter assay (MPRA), which you can read more about it in the preprint on [*bioRxiv*](https://doi.org/10.1101/310375).\n\n## Setup\n\n**Architecture:** The model is defined in [utr_model.py](https://github.com/FunctionLab/selene/blob/master/tutorials/regression_mpra_example/utr_model.py), and only superficially differs from the model in [the paper](https://doi.org/10.1101/310375).\nSince this is a real-valued regression problem, it is appropriate that the `criterion` method in `utr_model.py` uses the mean squared error.\n\n\n**Data:** The data from Sample et al is available on the [Gene Expression Omnibus (GEO)](https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114002).\nHowever, we have included [the `download_data.py` script](https://github.com/FunctionLab/selene/blob/master/tutorials/regression_mpra_example/download_data.py), to download the data and preprocess it.\nIt should produce three files, `train.mat`, `validate.mat`, and `test.mat`.\nThey include the data for training, validation, and testing respectively.\nAt present, regression models can only be trained with `*.mat` files and the [`MatFileSampler`](http://selene.flatironinstitute.org/samplers.file_samplers.html#selene_sdk.samplers.file_samplers.MatFileSampler).\nFurther, a `MatFileSampler` must be specified for each of the `train.mat`, `validate.mat`, and `test.mat` files.\nThese `MatFileSampler`s are then used for the `train`, `validate`, and `test` arguments of a [`MultiFileSampler`](http://selene.flatironinstitute.org/samplers.html#selene_sdk.samplers.MultiFileSampler).\nThe specific syntax is demonstrated in the configuration file, [`regression_train.yml`](https://github.com/FunctionLab/selene/blob/master/tutorials/regression_mpra_example/regression_train.yml).\n\n**Configuration file:** The configuration file [`regression_train.yml`](https://github.com/FunctionLab/selene/blob/master/tutorials/regression_mpra_example/regression_train.yml) is slightly different than the configuration files in the classification tutorials.\nSpecifically, `metrics` in `train_model` includes the coefficient of determination (`r2`), since the default metrics (`roc_auc` and `average_precision`) are not appropriate for regression.\nFurther, `report_gt_feature_n_positives` in `train_model` has been set to zero to prevent spurious filtering based on target values.\n\n## Download the data\n\nTo download the data, just run the [`download_data.py`](https://github.com/FunctionLab/selene/blob/master/tutorials/regression_mpra_example/download_data.py) script from the command line:\n```sh\npython download_data.py\n```\n\n## Train the model",
"_____no_output_____"
]
],
[
[
"from selene_sdk.utils import load_path\nfrom selene_sdk.utils import parse_configs_and_run",
"_____no_output_____"
]
],
[
[
"Before running `load_path` on `regression_train.yml`, please edit the YAML file to include the absolute path of the model file.\n\nCurrently, the model is set to train on GPU.\nIf you do not have CUDA on your machine, please set `use_cuda` to `False` in the configuration file. Note that using the CPU instead of GPU will slow down training considerably.",
"_____no_output_____"
]
],
[
[
"configs = load_path(\"./regression_train.yml\")",
"_____no_output_____"
],
[
"parse_configs_and_run(configs, lr=0.001)",
"Outputs and logs saved to ./2018-12-09-15-53-59\n2018-12-09 15:54:01,335 - Creating validation dataset.\n2018-12-09 15:54:01,361 - 0.02456068992614746 s to load 20096 validation examples (157 validation batches) to evaluate after each training step.\n2018-12-09 15:54:24,581 - [STEP 2031] average number of steps per second: 88.0\n2018-12-09 15:54:25,020 - validation r2: 0.8104067907778664\n2018-12-09 15:54:25,125 - training loss: 0.2401450276374817\n2018-12-09 15:54:25,126 - validation loss: 0.18883540832502826\n2018-12-09 15:54:47,288 - [STEP 4062] average number of steps per second: 91.9\n2018-12-09 15:54:47,729 - validation r2: 0.8564685296471333\n2018-12-09 15:54:47,822 - training loss: 0.193187415599823\n2018-12-09 15:54:47,823 - validation loss: 0.14294122951995036\n2018-12-09 15:55:09,855 - [STEP 6093] average number of steps per second: 92.5\n2018-12-09 15:55:10,290 - validation r2: 0.8653072068202623\n2018-12-09 15:55:10,376 - training loss: 0.2143666297197342\n2018-12-09 15:55:10,377 - validation loss: 0.13429565685000389\n2018-12-09 15:55:32,461 - Creating test dataset.\n2018-12-09 15:55:32,490 - 0.025864839553833008 s to load 20096 test examples (157 test batches) to evaluate after all training steps.\n2018-12-09 15:55:32,950 - test r2: 0.9016817574999407\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecb7edf8d67a946655aa20f7f03789a11eba181c | 2,740 | ipynb | Jupyter Notebook | spark_app/RUN/stream-sentiment-processor.ipynb | OlehOnyshchak/WikiSentimentRanking | 84df4141ef14eee1a8debeeb7ea1bd20d13bbbe4 | [
"MIT"
] | 4 | 2019-07-19T12:04:04.000Z | 2019-07-20T13:45:41.000Z | spark_app/RUN/stream-sentiment-processor.ipynb | OlehOnyshchak/WikiSentimentRanking | 84df4141ef14eee1a8debeeb7ea1bd20d13bbbe4 | [
"MIT"
] | null | null | null | spark_app/RUN/stream-sentiment-processor.ipynb | OlehOnyshchak/WikiSentimentRanking | 84df4141ef14eee1a8debeeb7ea1bd20d13bbbe4 | [
"MIT"
] | null | null | null | 20.296296 | 104 | 0.537226 | [
[
[
"import sys\n!{sys.executable} -m pip install --user findspark",
"Requirement already satisfied: findspark in /opt/conda/lib/python3.7/site-packages (1.3.0)\r\n"
],
[
"import findspark\nfindspark.init()",
"_____no_output_____"
],
[
"from pyspark import SparkContext\nfrom pyspark.streaming import StreamingContext\nfrom pyspark.sql.functions import udf\nfrom pyspark.sql import SparkSession\nfrom pyspark.sql.types import StringType, StructField, StructType, ArrayType, LongType, DoubleType",
"_____no_output_____"
],
[
"from scorers import score_text\nfrom spark_tools import SparkSentimentStreamer",
"_____no_output_____"
],
[
"sc = SparkContext(\"local[*]\", \"NetworkWordCount\")\nssc = StreamingContext(sc, 1)\nspark = SparkSession \\\n .builder \\\n .appName(\"SentimentWikiProcessor\") \\\n .getOrCreate()",
"_____no_output_____"
],
[
"dataInp = \"streamInput/\"\ndataOut = \"streamOut/\"",
"_____no_output_____"
],
[
"streamer = SparkSentimentStreamer(sc, ssc, spark, score_text, dataInp, dataOut)",
"_____no_output_____"
],
[
"streamer.run()",
"_____no_output_____"
],
[
"streamer.stop()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb7f1d2a7081eabd0540d470e00ced9a88b8652 | 772,359 | ipynb | Jupyter Notebook | drafts/c4w4a_Art_Generation_with_Neural_Style_Transfer_v3a.ipynb | jydiw/deeplearning.ai | 9b825dc9351ac611f354139dae596d1c0a1f3834 | [
"MIT"
] | null | null | null | drafts/c4w4a_Art_Generation_with_Neural_Style_Transfer_v3a.ipynb | jydiw/deeplearning.ai | 9b825dc9351ac611f354139dae596d1c0a1f3834 | [
"MIT"
] | null | null | null | drafts/c4w4a_Art_Generation_with_Neural_Style_Transfer_v3a.ipynb | jydiw/deeplearning.ai | 9b825dc9351ac611f354139dae596d1c0a1f3834 | [
"MIT"
] | null | null | null | 514.906 | 309,334 | 0.92634 | [
[
[
"# Deep Learning & Art: Neural Style Transfer\n\nIn this assignment, you will learn about Neural Style Transfer. This algorithm was created by [Gatys et al. (2015).](https://arxiv.org/abs/1508.06576)\n\n**In this assignment, you will:**\n- Implement the neural style transfer algorithm \n- Generate novel artistic images using your algorithm \n\nMost of the algorithms you've studied optimize a cost function to get a set of parameter values. In Neural Style Transfer, you'll optimize a cost function to get pixel values!",
"_____no_output_____"
],
[
"## <font color='darkblue'>Updates</font>\n\n#### If you were working on the notebook before this update...\n* The current notebook is version \"3a\".\n* You can find your original work saved in the notebook with the previous version name (\"v2\") \n* To view the file directory, go to the menu \"File->Open\", and this will open a new tab that shows the file directory.\n\n#### List of updates\n* Use `pprint.PrettyPrinter` to format printing of the vgg model.\n* computing content cost: clarified and reformatted instructions, fixed broken links, added additional hints for unrolling.\n* style matrix: clarify two uses of variable \"G\" by using different notation for gram matrix.\n* style cost: use distinct notation for gram matrix, added additional hints.\n* Grammar and wording updates for clarity.\n* `model_nn`: added hints.",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nimport scipy.io\nimport scipy.misc\nimport matplotlib.pyplot as plt\nfrom matplotlib.pyplot import imshow\nfrom PIL import Image\nfrom nst_utils import *\nimport numpy as np\nimport tensorflow as tf\nimport pprint\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## 1 - Problem Statement\n\nNeural Style Transfer (NST) is one of the most fun techniques in deep learning. As seen below, it merges two images, namely: a **\"content\" image (C) and a \"style\" image (S), to create a \"generated\" image (G**). \n\nThe generated image G combines the \"content\" of the image C with the \"style\" of image S. \n\nIn this example, you are going to generate an image of the Louvre museum in Paris (content image C), mixed with a painting by Claude Monet, a leader of the impressionist movement (style image S).\n<img src=\"images/louvre_generated.png\" style=\"width:750px;height:200px;\">\n\nLet's see how you can do this. ",
"_____no_output_____"
],
[
"## 2 - Transfer Learning\n\nNeural Style Transfer (NST) uses a previously trained convolutional network, and builds on top of that. The idea of using a network trained on a different task and applying it to a new task is called transfer learning. \n\nFollowing the [original NST paper](https://arxiv.org/abs/1508.06576), we will use the VGG network. Specifically, we'll use VGG-19, a 19-layer version of the VGG network. This model has already been trained on the very large ImageNet database, and thus has learned to recognize a variety of low level features (at the shallower layers) and high level features (at the deeper layers). \n\nRun the following code to load parameters from the VGG model. This may take a few seconds. ",
"_____no_output_____"
]
],
[
[
"pp = pprint.PrettyPrinter(indent=4)\nmodel = load_vgg_model(\"pretrained-model/imagenet-vgg-verydeep-19.mat\")\npp.pprint(model)",
"{ 'avgpool1': <tf.Tensor 'AvgPool:0' shape=(1, 150, 200, 64) dtype=float32>,\n 'avgpool2': <tf.Tensor 'AvgPool_1:0' shape=(1, 75, 100, 128) dtype=float32>,\n 'avgpool3': <tf.Tensor 'AvgPool_2:0' shape=(1, 38, 50, 256) dtype=float32>,\n 'avgpool4': <tf.Tensor 'AvgPool_3:0' shape=(1, 19, 25, 512) dtype=float32>,\n 'avgpool5': <tf.Tensor 'AvgPool_4:0' shape=(1, 10, 13, 512) dtype=float32>,\n 'conv1_1': <tf.Tensor 'Relu:0' shape=(1, 300, 400, 64) dtype=float32>,\n 'conv1_2': <tf.Tensor 'Relu_1:0' shape=(1, 300, 400, 64) dtype=float32>,\n 'conv2_1': <tf.Tensor 'Relu_2:0' shape=(1, 150, 200, 128) dtype=float32>,\n 'conv2_2': <tf.Tensor 'Relu_3:0' shape=(1, 150, 200, 128) dtype=float32>,\n 'conv3_1': <tf.Tensor 'Relu_4:0' shape=(1, 75, 100, 256) dtype=float32>,\n 'conv3_2': <tf.Tensor 'Relu_5:0' shape=(1, 75, 100, 256) dtype=float32>,\n 'conv3_3': <tf.Tensor 'Relu_6:0' shape=(1, 75, 100, 256) dtype=float32>,\n 'conv3_4': <tf.Tensor 'Relu_7:0' shape=(1, 75, 100, 256) dtype=float32>,\n 'conv4_1': <tf.Tensor 'Relu_8:0' shape=(1, 38, 50, 512) dtype=float32>,\n 'conv4_2': <tf.Tensor 'Relu_9:0' shape=(1, 38, 50, 512) dtype=float32>,\n 'conv4_3': <tf.Tensor 'Relu_10:0' shape=(1, 38, 50, 512) dtype=float32>,\n 'conv4_4': <tf.Tensor 'Relu_11:0' shape=(1, 38, 50, 512) dtype=float32>,\n 'conv5_1': <tf.Tensor 'Relu_12:0' shape=(1, 19, 25, 512) dtype=float32>,\n 'conv5_2': <tf.Tensor 'Relu_13:0' shape=(1, 19, 25, 512) dtype=float32>,\n 'conv5_3': <tf.Tensor 'Relu_14:0' shape=(1, 19, 25, 512) dtype=float32>,\n 'conv5_4': <tf.Tensor 'Relu_15:0' shape=(1, 19, 25, 512) dtype=float32>,\n 'input': <tf.Variable 'Variable:0' shape=(1, 300, 400, 3) dtype=float32_ref>}\n"
]
],
[
[
"* The model is stored in a python dictionary. \n* The python dictionary contains key-value pairs for each layer. \n* The 'key' is the variable name and the 'value' is a tensor for that layer. \n\n#### Assign input image to the model's input layer\nTo run an image through this network, you just have to feed the image to the model. In TensorFlow, you can do so using the [tf.assign](https://www.tensorflow.org/api_docs/python/tf/assign) function. In particular, you will use the assign function like this: \n```python\nmodel[\"input\"].assign(image)\n```\nThis assigns the image as an input to the model. \n\n#### Activate a layer\nAfter this, if you want to access the activations of a particular layer, say layer `4_2` when the network is run on this image, you would run a TensorFlow session on the correct tensor `conv4_2`, as follows: \n```python\nsess.run(model[\"conv4_2\"])\n```",
"_____no_output_____"
],
[
"## 3 - Neural Style Transfer (NST)\n\nWe will build the Neural Style Transfer (NST) algorithm in three steps:\n\n- Build the content cost function $J_{content}(C,G)$\n- Build the style cost function $J_{style}(S,G)$\n- Put it together to get $J(G) = \\alpha J_{content}(C,G) + \\beta J_{style}(S,G)$. \n\n### 3.1 - Computing the content cost\n\nIn our running example, the content image C will be the picture of the Louvre Museum in Paris. Run the code below to see a picture of the Louvre.",
"_____no_output_____"
]
],
[
[
"content_image = scipy.misc.imread(\"images/louvre.jpg\")\nimshow(content_image);",
"_____no_output_____"
]
],
[
[
"The content image (C) shows the Louvre museum's pyramid surrounded by old Paris buildings, against a sunny sky with a few clouds.\n\n** 3.1.1 - Make generated image G match the content of image C**\n\n#### Shallower versus deeper layers\n* The shallower layers of a ConvNet tend to detect lower-level features such as edges and simple textures.\n* The deeper layers tend to detect higher-level features such as more complex textures as well as object classes. \n\n#### Choose a \"middle\" activation layer $a^{[l]}$\nWe would like the \"generated\" image G to have similar content as the input image C. Suppose you have chosen some layer's activations to represent the content of an image. \n* In practice, you'll get the most visually pleasing results if you choose a layer in the **middle** of the network--neither too shallow nor too deep. \n* (After you have finished this exercise, feel free to come back and experiment with using different layers, to see how the results vary.)\n\n#### Forward propagate image \"C\"\n* Set the image C as the input to the pretrained VGG network, and run forward propagation. \n* Let $a^{(C)}$ be the hidden layer activations in the layer you had chosen. (In lecture, we had written this as $a^{[l](C)}$, but here we'll drop the superscript $[l]$ to simplify the notation.) This will be an $n_H \\times n_W \\times n_C$ tensor.\n\n#### Forward propagate image \"G\"\n* Repeat this process with the image G: Set G as the input, and run forward progation. \n* Let $a^{(G)}$ be the corresponding hidden layer activation. \n\n#### Content Cost Function $J_{content}(C,G)$\nWe will define the content cost function as:\n\n$$J_{content}(C,G) = \\frac{1}{4 \\times n_H \\times n_W \\times n_C}\\sum _{ \\text{all entries}} (a^{(C)} - a^{(G)})^2\\tag{1} $$\n\n* Here, $n_H, n_W$ and $n_C$ are the height, width and number of channels of the hidden layer you have chosen, and appear in a normalization term in the cost. \n* For clarity, note that $a^{(C)}$ and $a^{(G)}$ are the 3D volumes corresponding to a hidden layer's activations. \n* In order to compute the cost $J_{content}(C,G)$, it might also be convenient to unroll these 3D volumes into a 2D matrix, as shown below.\n* Technically this unrolling step isn't needed to compute $J_{content}$, but it will be good practice for when you do need to carry out a similar operation later for computing the style cost $J_{style}$.\n\n<img src=\"images/NST_LOSS.png\" style=\"width:800px;height:400px;\">",
"_____no_output_____"
],
[
"**Exercise:** Compute the \"content cost\" using TensorFlow. \n\n**Instructions**: The 3 steps to implement this function are:\n1. Retrieve dimensions from `a_G`: \n - To retrieve dimensions from a tensor `X`, use: `X.get_shape().as_list()`\n2. Unroll `a_C` and `a_G` as explained in the picture above\n - You'll likey want to use these functions: [tf.transpose](https://www.tensorflow.org/versions/r1.15/api_docs/python/tf/transpose) and [tf.reshape](https://www.tensorflow.org/versions/r1.15/api_docs/python/tf/reshape).\n3. Compute the content cost:\n - You'll likely want to use these functions: [tf.reduce_sum](https://www.tensorflow.org/api_docs/python/tf/reduce_sum), [tf.square](https://www.tensorflow.org/api_docs/python/tf/square) and [tf.subtract](https://www.tensorflow.org/api_docs/python/tf/subtract).\n \n \n#### Additional Hints for \"Unrolling\"\n* To unroll the tensor, we want the shape to change from $(m,n_H,n_W,n_C)$ to $(m, n_H \\times n_W, n_C)$.\n* `tf.reshape(tensor, shape)` takes a list of integers that represent the desired output shape.\n* For the `shape` parameter, a `-1` tells the function to choose the correct dimension size so that the output tensor still contains all the values of the original tensor.\n* So tf.reshape(a_C, shape=[m, n_H * n_W, n_C]) gives the same result as tf.reshape(a_C, shape=[m, -1, n_C]).\n* If you prefer to re-order the dimensions, you can use `tf.transpose(tensor, perm)`, where `perm` is a list of integers containing the original index of the dimensions. \n* For example, `tf.transpose(a_C, perm=[0,3,1,2])` changes the dimensions from $(m, n_H, n_W, n_C)$ to $(m, n_C, n_H, n_W)$.\n* There is more than one way to unroll the tensors.\n* Notice that it's not necessary to use tf.transpose to 'unroll' the tensors in this case but this is a useful function to practice and understand for other situations that you'll encounter.\n",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: compute_content_cost\n\ndef compute_content_cost(a_C, a_G):\n \"\"\"\n Computes the content cost\n \n Arguments:\n a_C -- tensor of dimension (1, n_H, n_W, n_C), hidden layer activations representing content of the image C \n a_G -- tensor of dimension (1, n_H, n_W, n_C), hidden layer activations representing content of the image G\n \n Returns: \n J_content -- scalar that you compute using equation 1 above.\n \"\"\"\n \n ### START CODE HERE ###\n # Retrieve dimensions from a_G (≈1 line)\n m, n_H, n_W, n_C = a_G.get_shape().as_list()\n \n # Reshape a_C and a_G (≈2 lines)\n a_C_unrolled = tf.reshape(a_C, shape=[m, -1, n_C])\n a_G_unrolled = tf.reshape(a_G, shape=[m, -1, n_C])\n \n # compute the cost with tensorflow (≈1 line)\n J_content = tf.reduce_sum(\n tf.square(\n tf.subtract(a_C_unrolled, a_G_unrolled)\n )\n ) / (4 * n_H * n_W * n_C)\n ### END CODE HERE ###\n \n return J_content",
"_____no_output_____"
],
[
"tf.reset_default_graph()\n\nwith tf.Session() as test:\n tf.set_random_seed(1)\n a_C = tf.random_normal([1, 4, 4, 3], mean=1, stddev=4)\n a_G = tf.random_normal([1, 4, 4, 3], mean=1, stddev=4)\n J_content = compute_content_cost(a_C, a_G)\n print(\"J_content = \" + str(J_content.eval()))",
"J_content = 6.76559\n"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **J_content**\n </td>\n <td>\n 6.76559\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"#### What you should remember\n- The content cost takes a hidden layer activation of the neural network, and measures how different $a^{(C)}$ and $a^{(G)}$ are. \n- When we minimize the content cost later, this will help make sure $G$ has similar content as $C$.",
"_____no_output_____"
],
[
"### 3.2 - Computing the style cost\n\nFor our running example, we will use the following style image: ",
"_____no_output_____"
]
],
[
[
"style_image = scipy.misc.imread(\"images/monet_800600.jpg\")\nimshow(style_image);",
"_____no_output_____"
]
],
[
[
"This was painted in the style of *[impressionism](https://en.wikipedia.org/wiki/Impressionism)*.\n\nLets see how you can now define a \"style\" cost function $J_{style}(S,G)$. ",
"_____no_output_____"
],
[
"### 3.2.1 - Style matrix\n\n#### Gram matrix\n* The style matrix is also called a \"Gram matrix.\" \n* In linear algebra, the Gram matrix G of a set of vectors $(v_{1},\\dots ,v_{n})$ is the matrix of dot products, whose entries are ${\\displaystyle G_{ij} = v_{i}^T v_{j} = np.dot(v_{i}, v_{j}) }$. \n* In other words, $G_{ij}$ compares how similar $v_i$ is to $v_j$: If they are highly similar, you would expect them to have a large dot product, and thus for $G_{ij}$ to be large. \n\n#### Two meanings of the variable $G$\n* Note that there is an unfortunate collision in the variable names used here. We are following common terminology used in the literature. \n* $G$ is used to denote the Style matrix (or Gram matrix) \n* $G$ also denotes the generated image. \n* For this assignment, we will use $G_{gram}$ to refer to the Gram matrix, and $G$ to denote the generated image.",
"_____no_output_____"
],
[
"\n#### Compute $G_{gram}$\nIn Neural Style Transfer (NST), you can compute the Style matrix by multiplying the \"unrolled\" filter matrix with its transpose:\n\n<img src=\"images/NST_GM.png\" style=\"width:900px;height:300px;\">\n\n$$\\mathbf{G}_{gram} = \\mathbf{A}_{unrolled} \\mathbf{A}_{unrolled}^T$$\n\n#### $G_{(gram)i,j}$: correlation\nThe result is a matrix of dimension $(n_C,n_C)$ where $n_C$ is the number of filters (channels). The value $G_{(gram)i,j}$ measures how similar the activations of filter $i$ are to the activations of filter $j$. \n\n#### $G_{(gram),i,i}$: prevalence of patterns or textures\n* The diagonal elements $G_{(gram)ii}$ measure how \"active\" a filter $i$ is. \n* For example, suppose filter $i$ is detecting vertical textures in the image. Then $G_{(gram)ii}$ measures how common vertical textures are in the image as a whole.\n* If $G_{(gram)ii}$ is large, this means that the image has a lot of vertical texture. \n\n\nBy capturing the prevalence of different types of features ($G_{(gram)ii}$), as well as how much different features occur together ($G_{(gram)ij}$), the Style matrix $G_{gram}$ measures the style of an image. ",
"_____no_output_____"
],
[
"**Exercise**:\n* Using TensorFlow, implement a function that computes the Gram matrix of a matrix A. \n* The formula is: The gram matrix of A is $G_A = AA^T$. \n* You may use these functions: [matmul](https://www.tensorflow.org/api_docs/python/tf/matmul) and [transpose](https://www.tensorflow.org/api_docs/python/tf/transpose).",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: gram_matrix\n\ndef gram_matrix(A):\n \"\"\"\n Argument:\n A -- matrix of shape (n_C, n_H*n_W)\n \n Returns:\n GA -- Gram matrix of A, of shape (n_C, n_C)\n \"\"\"\n \n ### START CODE HERE ### (≈1 line)\n GA = tf.matmul(A, tf.transpose(A))\n ### END CODE HERE ###\n \n return GA",
"_____no_output_____"
],
[
"tf.reset_default_graph()\n\nwith tf.Session() as test:\n tf.set_random_seed(1)\n A = tf.random_normal([3, 2*1], mean=1, stddev=4)\n GA = gram_matrix(A)\n \n print(\"GA = \\n\" + str(GA.eval()))",
"GA = \n[[ 6.42230511 -4.42912197 -2.09668207]\n [ -4.42912197 19.46583748 19.56387138]\n [ -2.09668207 19.56387138 20.6864624 ]]\n"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **GA**\n </td>\n <td>\n [[ 6.42230511 -4.42912197 -2.09668207] <br>\n [ -4.42912197 19.46583748 19.56387138] <br>\n [ -2.09668207 19.56387138 20.6864624 ]]\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"### 3.2.2 - Style cost",
"_____no_output_____"
],
[
"Your goal will be to minimize the distance between the Gram matrix of the \"style\" image S and the gram matrix of the \"generated\" image G. \n* For now, we are using only a single hidden layer $a^{[l]}$. \n* The corresponding style cost for this layer is defined as: \n\n$$J_{style}^{[l]}(S,G) = \\frac{1}{4 \\times {n_C}^2 \\times (n_H \\times n_W)^2} \\sum _{i=1}^{n_C}\\sum_{j=1}^{n_C}(G^{(S)}_{(gram)i,j} - G^{(G)}_{(gram)i,j})^2\\tag{2} $$\n\n* $G_{gram}^{(S)}$ Gram matrix of the \"style\" image.\n* $G_{gram}^{(G)}$ Gram matrix of the \"generated\" image.\n* Remember, this cost is computed using the hidden layer activations for a particular hidden layer in the network $a^{[l]}$\n",
"_____no_output_____"
],
[
"**Exercise**: Compute the style cost for a single layer. \n\n**Instructions**: The 3 steps to implement this function are:\n1. Retrieve dimensions from the hidden layer activations a_G: \n - To retrieve dimensions from a tensor X, use: `X.get_shape().as_list()`\n2. Unroll the hidden layer activations a_S and a_G into 2D matrices, as explained in the picture above (see the images in the sections \"computing the content cost\" and \"style matrix\").\n - You may use [tf.transpose](https://www.tensorflow.org/versions/r1.15/api_docs/python/tf/transpose) and [tf.reshape](https://www.tensorflow.org/versions/r1.15/api_docs/python/tf/reshape).\n3. Compute the Style matrix of the images S and G. (Use the function you had previously written.) \n4. Compute the Style cost:\n - You may find [tf.reduce_sum](https://www.tensorflow.org/api_docs/python/tf/reduce_sum), [tf.square](https://www.tensorflow.org/api_docs/python/tf/square) and [tf.subtract](https://www.tensorflow.org/api_docs/python/tf/subtract) useful.\n \n \n#### Additional Hints\n* Since the activation dimensions are $(m, n_H, n_W, n_C)$ whereas the desired unrolled matrix shape is $(n_C, n_H*n_W)$, the order of the filter dimension $n_C$ is changed. So `tf.transpose` can be used to change the order of the filter dimension.\n* for the product $\\mathbf{G}_{gram} = \\mathbf{A}_{} \\mathbf{A}_{}^T$, you will also need to specify the `perm` parameter for the `tf.transpose` function.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: compute_layer_style_cost\n\ndef compute_layer_style_cost(a_S, a_G):\n \"\"\"\n Arguments:\n a_S -- tensor of dimension (1, n_H, n_W, n_C), hidden layer activations representing style of the image S \n a_G -- tensor of dimension (1, n_H, n_W, n_C), hidden layer activations representing style of the image G\n \n Returns: \n J_style_layer -- tensor representing a scalar value, style cost defined above by equation (2)\n \"\"\"\n \n ### START CODE HERE ###\n # Retrieve dimensions from a_G (≈1 line)\n m, n_H, n_W, n_C = a_G.get_shape().as_list()\n \n # Reshape the images to have them of shape (n_C, n_H*n_W) (≈2 lines)\n a_S = tf.transpose(tf.reshape(a_S, shape=[-1, n_C]))\n a_G = tf.transpose(tf.reshape(a_G, shape=[-1, n_C]))\n\n # Computing gram_matrices for both images S and G (≈2 lines)\n GS = gram_matrix(a_S)\n GG = gram_matrix(a_G)\n\n # Computing the loss (≈1 line)\n J_style_layer = tf.reduce_sum(\n tf.square(\n tf.subtract(GS, GG)\n )\n ) / (4 * (n_H*n_W)**2 * n_C**2)\n \n ### END CODE HERE ###\n \n return J_style_layer",
"_____no_output_____"
],
[
"tf.reset_default_graph()\n\nwith tf.Session() as test:\n tf.set_random_seed(1)\n a_S = tf.random_normal([1, 4, 4, 3], mean=1, stddev=4)\n a_G = tf.random_normal([1, 4, 4, 3], mean=1, stddev=4)\n J_style_layer = compute_layer_style_cost(a_S, a_G)\n \n print(\"J_style_layer = \" + str(J_style_layer.eval()))",
"J_style_layer = 9.19028\n"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **J_style_layer**\n </td>\n <td>\n 9.19028\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"### 3.2.3 Style Weights\n\n* So far you have captured the style from only one layer. \n* We'll get better results if we \"merge\" style costs from several different layers. \n* Each layer will be given weights ($\\lambda^{[l]}$) that reflect how much each layer will contribute to the style.\n* After completing this exercise, feel free to come back and experiment with different weights to see how it changes the generated image $G$.\n* By default, we'll give each layer equal weight, and the weights add up to 1. ($\\sum_{l}^L\\lambda^{[l]} = 1$)",
"_____no_output_____"
]
],
[
[
"STYLE_LAYERS = [\n ('conv1_1', 0.2),\n ('conv2_1', 0.2),\n ('conv3_1', 0.2),\n ('conv4_1', 0.2),\n ('conv5_1', 0.2)]",
"_____no_output_____"
]
],
[
[
"You can combine the style costs for different layers as follows:\n\n$$J_{style}(S,G) = \\sum_{l} \\lambda^{[l]} J^{[l]}_{style}(S,G)$$\n\nwhere the values for $\\lambda^{[l]}$ are given in `STYLE_LAYERS`. \n",
"_____no_output_____"
],
[
"### Exercise: compute style cost\n\n* We've implemented a compute_style_cost(...) function. \n* It calls your `compute_layer_style_cost(...)` several times, and weights their results using the values in `STYLE_LAYERS`. \n* Please read over it to make sure you understand what it's doing. \n\n#### Description of `compute_style_cost`\nFor each layer:\n* Select the activation (the output tensor) of the current layer.\n* Get the style of the style image \"S\" from the current layer.\n* Get the style of the generated image \"G\" from the current layer.\n* Compute the \"style cost\" for the current layer\n* Add the weighted style cost to the overall style cost (J_style)\n\nOnce you're done with the loop: \n* Return the overall style cost.",
"_____no_output_____"
]
],
[
[
"def compute_style_cost(model, STYLE_LAYERS):\n \"\"\"\n Computes the overall style cost from several chosen layers\n \n Arguments:\n model -- our tensorflow model\n STYLE_LAYERS -- A python list containing:\n - the names of the layers we would like to extract style from\n - a coefficient for each of them\n \n Returns: \n J_style -- tensor representing a scalar value, style cost defined above by equation (2)\n \"\"\"\n \n # initialize the overall style cost\n J_style = 0\n\n for layer_name, coeff in STYLE_LAYERS:\n\n # Select the output tensor of the currently selected layer\n out = model[layer_name]\n\n # Set a_S to be the hidden layer activation from the layer we have selected, by running the session on out\n a_S = sess.run(out)\n\n # Set a_G to be the hidden layer activation from same layer. Here, a_G references model[layer_name] \n # and isn't evaluated yet. Later in the code, we'll assign the image G as the model input, so that\n # when we run the session, this will be the activations drawn from the appropriate layer, with G as input.\n a_G = out\n \n # Compute style_cost for the current layer\n J_style_layer = compute_layer_style_cost(a_S, a_G)\n\n # Add coeff * J_style_layer of this layer to overall style cost\n J_style += coeff * J_style_layer\n\n return J_style",
"_____no_output_____"
]
],
[
[
"**Note**: In the inner-loop of the for-loop above, `a_G` is a tensor and hasn't been evaluated yet. It will be evaluated and updated at each iteration when we run the TensorFlow graph in model_nn() below.\n\n<!-- \nHow do you choose the coefficients for each layer? The deeper layers capture higher-level concepts, and the features in the deeper layers are less localized in the image relative to each other. So if you want the generated image to softly follow the style image, try choosing larger weights for deeper layers and smaller weights for the first layers. In contrast, if you want the generated image to strongly follow the style image, try choosing smaller weights for deeper layers and larger weights for the first layers\n!-->\n\n\n\n## What you should remember\n- The style of an image can be represented using the Gram matrix of a hidden layer's activations. \n- We get even better results by combining this representation from multiple different layers. \n- This is in contrast to the content representation, where usually using just a single hidden layer is sufficient.\n- Minimizing the style cost will cause the image $G$ to follow the style of the image $S$. \n\n",
"_____no_output_____"
],
[
"### 3.3 - Defining the total cost to optimize",
"_____no_output_____"
],
[
"Finally, let's create a cost function that minimizes both the style and the content cost. The formula is: \n\n$$J(G) = \\alpha J_{content}(C,G) + \\beta J_{style}(S,G)$$\n\n**Exercise**: Implement the total cost function which includes both the content cost and the style cost. ",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: total_cost\n\ndef total_cost(J_content, J_style, alpha = 10, beta = 40):\n \"\"\"\n Computes the total cost function\n \n Arguments:\n J_content -- content cost coded above\n J_style -- style cost coded above\n alpha -- hyperparameter weighting the importance of the content cost\n beta -- hyperparameter weighting the importance of the style cost\n \n Returns:\n J -- total cost as defined by the formula above.\n \"\"\"\n \n ### START CODE HERE ### (≈1 line)\n J = alpha * J_content + beta * J_style\n ### END CODE HERE ###\n \n return J",
"_____no_output_____"
],
[
"tf.reset_default_graph()\n\nwith tf.Session() as test:\n np.random.seed(3)\n J_content = np.random.randn() \n J_style = np.random.randn()\n J = total_cost(J_content, J_style)\n print(\"J = \" + str(J))",
"J = 35.34667875478276\n"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **J**\n </td>\n <td>\n 35.34667875478276\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"\n## What you should remember\n- The total cost is a linear combination of the content cost $J_{content}(C,G)$ and the style cost $J_{style}(S,G)$.\n- $\\alpha$ and $\\beta$ are hyperparameters that control the relative weighting between content and style.",
"_____no_output_____"
],
[
"## 4 - Solving the optimization problem",
"_____no_output_____"
],
[
"Finally, let's put everything together to implement Neural Style Transfer!\n\n\nHere's what the program will have to do:\n\n1. Create an Interactive Session\n2. Load the content image \n3. Load the style image\n4. Randomly initialize the image to be generated \n5. Load the VGG19 model\n7. Build the TensorFlow graph:\n - Run the content image through the VGG19 model and compute the content cost\n - Run the style image through the VGG19 model and compute the style cost\n - Compute the total cost\n - Define the optimizer and the learning rate\n8. Initialize the TensorFlow graph and run it for a large number of iterations, updating the generated image at every step.\n\nLets go through the individual steps in detail. ",
"_____no_output_____"
],
[
"#### Interactive Sessions\n\nYou've previously implemented the overall cost $J(G)$. We'll now set up TensorFlow to optimize this with respect to $G$. \n* To do so, your program has to reset the graph and use an \"[Interactive Session](https://www.tensorflow.org/api_docs/python/tf/InteractiveSession)\". \n* Unlike a regular session, the \"Interactive Session\" installs itself as the default session to build a graph. \n* This allows you to run variables without constantly needing to refer to the session object (calling \"sess.run()\"), which simplifies the code. \n\n#### Start the interactive session.",
"_____no_output_____"
]
],
[
[
"# Reset the graph\ntf.reset_default_graph()\n\n# Start interactive session\nsess = tf.InteractiveSession()",
"_____no_output_____"
]
],
[
[
"#### Content image\nLet's load, reshape, and normalize our \"content\" image (the Louvre museum picture):",
"_____no_output_____"
]
],
[
[
"content_image = scipy.misc.imread(\"images/louvre_small.jpg\")\ncontent_image = reshape_and_normalize_image(content_image)",
"_____no_output_____"
]
],
[
[
"#### Style image\nLet's load, reshape and normalize our \"style\" image (Claude Monet's painting):",
"_____no_output_____"
]
],
[
[
"style_image = scipy.misc.imread(\"images/monet.jpg\")\nstyle_image = reshape_and_normalize_image(style_image)",
"_____no_output_____"
]
],
[
[
"#### Generated image correlated with content image\nNow, we initialize the \"generated\" image as a noisy image created from the content_image.\n\n* The generated image is slightly correlated with the content image.\n* By initializing the pixels of the generated image to be mostly noise but slightly correlated with the content image, this will help the content of the \"generated\" image more rapidly match the content of the \"content\" image. \n* Feel free to look in `nst_utils.py` to see the details of `generate_noise_image(...)`; to do so, click \"File-->Open...\" at the upper-left corner of this Jupyter notebook.",
"_____no_output_____"
]
],
[
[
"generated_image = generate_noise_image(content_image)\nimshow(generated_image[0]);",
"_____no_output_____"
]
],
[
[
"#### Load pre-trained VGG19 model\nNext, as explained in part (2), let's load the VGG19 model.",
"_____no_output_____"
]
],
[
[
"model = load_vgg_model(\"pretrained-model/imagenet-vgg-verydeep-19.mat\")",
"_____no_output_____"
]
],
[
[
"#### Content Cost\n\nTo get the program to compute the content cost, we will now assign `a_C` and `a_G` to be the appropriate hidden layer activations. We will use layer `conv4_2` to compute the content cost. The code below does the following:\n\n1. Assign the content image to be the input to the VGG model.\n2. Set a_C to be the tensor giving the hidden layer activation for layer \"conv4_2\".\n3. Set a_G to be the tensor giving the hidden layer activation for the same layer. \n4. Compute the content cost using a_C and a_G.\n\n**Note**: At this point, a_G is a tensor and hasn't been evaluated. It will be evaluated and updated at each iteration when we run the Tensorflow graph in model_nn() below.",
"_____no_output_____"
]
],
[
[
"# Assign the content image to be the input of the VGG model. \nsess.run(model['input'].assign(content_image))\n\n# Select the output tensor of layer conv4_2\nout = model['conv4_2']\n\n# Set a_C to be the hidden layer activation from the layer we have selected\na_C = sess.run(out)\n\n# Set a_G to be the hidden layer activation from same layer. Here, a_G references model['conv4_2'] \n# and isn't evaluated yet. Later in the code, we'll assign the image G as the model input, so that\n# when we run the session, this will be the activations drawn from the appropriate layer, with G as input.\na_G = out\n\n# Compute the content cost\nJ_content = compute_content_cost(a_C, a_G)",
"_____no_output_____"
]
],
[
[
"#### Style cost",
"_____no_output_____"
]
],
[
[
"# Assign the input of the model to be the \"style\" image \nsess.run(model['input'].assign(style_image))\n\n# Compute the style cost\nJ_style = compute_style_cost(model, STYLE_LAYERS)",
"_____no_output_____"
]
],
[
[
"### Exercise: total cost\n* Now that you have J_content and J_style, compute the total cost J by calling `total_cost()`. \n* Use `alpha = 10` and `beta = 40`.",
"_____no_output_____"
]
],
[
[
"### START CODE HERE ### (1 line)\nJ = total_cost(J_content, J_style)\n### END CODE HERE ###",
"_____no_output_____"
]
],
[
[
"### Optimizer\n\n* Use the Adam optimizer to minimize the total cost `J`.\n* Use a learning rate of 2.0. \n* [Adam Optimizer documentation](https://www.tensorflow.org/api_docs/python/tf/train/AdamOptimizer)",
"_____no_output_____"
]
],
[
[
"# define optimizer (1 line)\noptimizer = tf.train.AdamOptimizer(2.0)\n\n# define train_step (1 line)\ntrain_step = optimizer.minimize(J)",
"_____no_output_____"
]
],
[
[
"### Exercise: implement the model\n\n* Implement the model_nn() function. \n* The function **initializes** the variables of the tensorflow graph, \n* **assigns** the input image (initial generated image) as the input of the VGG19 model \n* and **runs** the `train_step` tensor (it was created in the code above this function) for a large number of steps.\n\n#### Hints\n* To initialize global variables, use this: \n```Python\nsess.run(tf.global_variables_initializer())\n```\n* Run `sess.run()` to evaluate a variable.\n* [assign](https://www.tensorflow.org/versions/r1.14/api_docs/python/tf/assign) can be used like this:\n```python\nmodel[\"input\"].assign(image)\n```\n",
"_____no_output_____"
]
],
[
[
"def model_nn(sess, input_image, num_iterations = 200):\n \n # Initialize global variables (you need to run the session on the initializer)\n ### START CODE HERE ### (1 line)\n sess.run(tf.global_variables_initializer())\n ### END CODE HERE ###\n \n # Run the noisy input image (initial generated image) through the model. Use assign().\n ### START CODE HERE ### (1 line)\n sess.run(model['input'].assign(input_image))\n ### END CODE HERE ###\n \n for i in range(num_iterations):\n \n # Run the session on the train_step to minimize the total cost\n ### START CODE HERE ### (1 line)\n sess.run(train_step)\n ### END CODE HERE ###\n \n # Compute the generated image by running the session on the current model['input']\n ### START CODE HERE ### (1 line)\n generated_image = sess.run(model['input'])\n ### END CODE HERE ###\n\n # Print every 20 iteration.\n if i%20 == 0:\n Jt, Jc, Js = sess.run([J, J_content, J_style])\n print(\"Iteration \" + str(i) + \" :\")\n print(\"total cost = \" + str(Jt))\n print(\"content cost = \" + str(Jc))\n print(\"style cost = \" + str(Js))\n \n # save current generated image in the \"/output\" directory\n save_image(\"output/\" + str(i) + \".png\", generated_image)\n \n # save last generated image\n save_image('output/generated_image.jpg', generated_image)\n \n return generated_image",
"_____no_output_____"
]
],
[
[
"Run the following cell to generate an artistic image. It should take about 3min on CPU for every 20 iterations but you start observing attractive results after ≈140 iterations. Neural Style Transfer is generally trained using GPUs.",
"_____no_output_____"
]
],
[
[
"model_nn(sess, generated_image)",
"Iteration 0 :\ntotal cost = 5.05035e+09\ncontent cost = 7877.68\nstyle cost = 1.26257e+08\nIteration 20 :\ntotal cost = 9.43276e+08\ncontent cost = 15187.0\nstyle cost = 2.35781e+07\nIteration 40 :\ntotal cost = 4.84898e+08\ncontent cost = 16785.0\nstyle cost = 1.21183e+07\nIteration 60 :\ntotal cost = 3.12574e+08\ncontent cost = 17465.8\nstyle cost = 7.80998e+06\nIteration 80 :\ntotal cost = 2.28137e+08\ncontent cost = 17715.0\nstyle cost = 5.699e+06\nIteration 100 :\ntotal cost = 1.80694e+08\ncontent cost = 17895.5\nstyle cost = 4.51288e+06\nIteration 120 :\ntotal cost = 1.49996e+08\ncontent cost = 18034.4\nstyle cost = 3.74539e+06\nIteration 140 :\ntotal cost = 1.27698e+08\ncontent cost = 18186.9\nstyle cost = 3.18791e+06\nIteration 160 :\ntotal cost = 1.10698e+08\ncontent cost = 18354.2\nstyle cost = 2.76287e+06\nIteration 180 :\ntotal cost = 9.73408e+07\ncontent cost = 18501.0\nstyle cost = 2.4289e+06\n"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **Iteration 0 : **\n </td>\n <td>\n total cost = 5.05035e+09 <br>\n content cost = 7877.67 <br>\n style cost = 1.26257e+08\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"You're done! After running this, in the upper bar of the notebook click on \"File\" and then \"Open\". Go to the \"/output\" directory to see all the saved images. Open \"generated_image\" to see the generated image! :)\n\nYou should see something the image presented below on the right:\n\n<img src=\"images/louvre_generated.png\" style=\"width:800px;height:300px;\">\n\nWe didn't want you to wait too long to see an initial result, and so had set the hyperparameters accordingly. To get the best looking results, running the optimization algorithm longer (and perhaps with a smaller learning rate) might work better. After completing and submitting this assignment, we encourage you to come back and play more with this notebook, and see if you can generate even better looking images. ",
"_____no_output_____"
],
[
"Here are few other examples:\n\n- The beautiful ruins of the ancient city of Persepolis (Iran) with the style of Van Gogh (The Starry Night)\n<img src=\"images/perspolis_vangogh.png\" style=\"width:750px;height:300px;\">\n\n- The tomb of Cyrus the great in Pasargadae with the style of a Ceramic Kashi from Ispahan.\n<img src=\"images/pasargad_kashi.png\" style=\"width:750px;height:300px;\">\n\n- A scientific study of a turbulent fluid with the style of a abstract blue fluid painting.\n<img src=\"images/circle_abstract.png\" style=\"width:750px;height:300px;\">",
"_____no_output_____"
],
[
"## 5 - Test with your own image (Optional/Ungraded)",
"_____no_output_____"
],
[
"Finally, you can also rerun the algorithm on your own images! \n\nTo do so, go back to part 4 and change the content image and style image with your own pictures. In detail, here's what you should do:\n\n1. Click on \"File -> Open\" in the upper tab of the notebook\n2. Go to \"/images\" and upload your images (requirement: (WIDTH = 300, HEIGHT = 225)), rename them \"my_content.png\" and \"my_style.png\" for example.\n3. Change the code in part (3.4) from :\n```python\ncontent_image = scipy.misc.imread(\"images/louvre.jpg\")\nstyle_image = scipy.misc.imread(\"images/claude-monet.jpg\")\n```\nto:\n```python\ncontent_image = scipy.misc.imread(\"images/my_content.jpg\")\nstyle_image = scipy.misc.imread(\"images/my_style.jpg\")\n```\n4. Rerun the cells (you may need to restart the Kernel in the upper tab of the notebook).\n\nYou can share your generated images with us on social media with the hashtag #deeplearniNgAI or by direct tagging!\n\nYou can also tune your hyperparameters: \n- Which layers are responsible for representing the style? STYLE_LAYERS\n- How many iterations do you want to run the algorithm? num_iterations\n- What is the relative weighting between content and style? alpha/beta",
"_____no_output_____"
],
[
"## 6 - Conclusion\n\nGreat job on completing this assignment! You are now able to use Neural Style Transfer to generate artistic images. This is also your first time building a model in which the optimization algorithm updates the pixel values rather than the neural network's parameters. Deep learning has many different types of models and this is only one of them! \n\n## What you should remember\n- Neural Style Transfer is an algorithm that given a content image C and a style image S can generate an artistic image\n- It uses representations (hidden layer activations) based on a pretrained ConvNet. \n- The content cost function is computed using one hidden layer's activations.\n- The style cost function for one layer is computed using the Gram matrix of that layer's activations. The overall style cost function is obtained using several hidden layers.\n- Optimizing the total cost function results in synthesizing new images. \n\n\n",
"_____no_output_____"
],
[
"# Congratulations on finishing the course!\nThis was the final programming exercise of this course. Congratulations--you've finished all the programming exercises of this course on Convolutional Networks! We hope to also see you in Course 5, on Sequence models! \n",
"_____no_output_____"
],
[
"### References:\n\nThe Neural Style Transfer algorithm was due to Gatys et al. (2015). Harish Narayanan and Github user \"log0\" also have highly readable write-ups from which we drew inspiration. The pre-trained network used in this implementation is a VGG network, which is due to Simonyan and Zisserman (2015). Pre-trained weights were from the work of the MathConvNet team. \n\n- Leon A. Gatys, Alexander S. Ecker, Matthias Bethge, (2015). [A Neural Algorithm of Artistic Style](https://arxiv.org/abs/1508.06576) \n- Harish Narayanan, [Convolutional neural networks for artistic style transfer.](https://harishnarayanan.org/writing/artistic-style-transfer/)\n- Log0, [TensorFlow Implementation of \"A Neural Algorithm of Artistic Style\".](http://www.chioka.in/tensorflow-implementation-neural-algorithm-of-artistic-style)\n- Karen Simonyan and Andrew Zisserman (2015). [Very deep convolutional networks for large-scale image recognition](https://arxiv.org/pdf/1409.1556.pdf)\n- [MatConvNet.](http://www.vlfeat.org/matconvnet/pretrained/)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.