hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ecb802d960633aa9034a6debcd53194cf18f1059 | 158,171 | ipynb | Jupyter Notebook | Project2.ipynb | ParasAlex/ETL_Project_2 | f93b88f326fdd6e460cdc63f89135bb97c2a9a82 | [
"MIT"
] | null | null | null | Project2.ipynb | ParasAlex/ETL_Project_2 | f93b88f326fdd6e460cdc63f89135bb97c2a9a82 | [
"MIT"
] | null | null | null | Project2.ipynb | ParasAlex/ETL_Project_2 | f93b88f326fdd6e460cdc63f89135bb97c2a9a82 | [
"MIT"
] | null | null | null | 49.754954 | 45,020 | 0.570964 | [
[
[
"#Import directories\nimport pandas as pd\nimport requests\n\n#get the data\nurl = \"https://fred.stlouisfed.org/graph/fredgraph.csv?bgcolor=%23e1e9f0&chart_type=line&drp=0&fo=open%20sans&graph_bgcolor=%23ffffff&height=450&mode=fred&recession_bars=on&txtcolor=%23444444&ts=12&tts=12&width=1168&nt=0&thu=0&trc=0&show_legend=yes&show_axis_titles=yes&show_tooltip=yes&id=UNRATE&scale=left&cosd=1948-01-01&coed=2021-08-01&line_color=%234572a7&link_values=false&line_style=solid&mark_type=none&mw=3&lw=2&ost=-99999&oet=99999&mma=0&fml=a&fq=Monthly&fam=avg&fgst=lin&fgsnd=2020-02-01&line_index=1&transformation=lin&vintage_date=2021-09-18&revision_date=2021-09-18&nd=1948-01-01\"\nc_df=pd.read_csv(url)\n\nurl2 = \"https://data.edd.ca.gov/api/views/e6gw-gvii/rows.csv?accessType=DOWNLOAD&bom=true&format=true\"\n\nd_df=pd.read_csv(url2)",
"_____no_output_____"
],
[
"#import addtional directories\nfrom sqlalchemy import create_engine\nfrom config import username, password\n#connect to database\nengine = create_engine(f'postgresql://{username}:{password}@localhost:5432/Project2')\nconnection = engine.connect()",
"_____no_output_____"
],
[
"#check table names\nengine.table_names()",
"<ipython-input-3-49982994423d>:2: SADeprecationWarning: The Engine.table_names() method is deprecated and will be removed in a future release. Please refer to Inspector.get_table_names(). (deprecated since: 1.4)\n engine.table_names()\n"
],
[
"#push US unenmployment Data csv to database\nc_df.to_sql(name='us', con=engine, if_exists='replace', index=False)",
"_____no_output_____"
],
[
"#push california unenmployment Data csv to database\nd_df.to_sql(name='ca', con=engine, if_exists='replace', index=False)",
"_____no_output_____"
],
[
"#confirm US unemployment Data\npd.read_sql_query('select * from us', con=engine)",
"_____no_output_____"
],
[
"#confirm CA unemployment Data\npd.read_sql_query('select * from CA', con=engine)",
"_____no_output_____"
],
[
"#get CA Data\nCA_data_df = pd.read_sql_query('select * from CA', con=engine)\n#get US data\nUS_data_df = pd.read_sql_query('select * from us', con=engine)",
"_____no_output_____"
],
[
"#Start filtering CA Data (cleaning data)\nCA_data_clean_df = CA_data_df.loc[CA_data_df[\"Area Type \"] == \"State\", :]\nCA_data_clean_df",
"_____no_output_____"
],
[
"#Addtional Cleaning\nCA_data_cleaner_df = CA_data_clean_df.loc[CA_data_df[\"Seasonally Adjusted (Y/N) \"] == \"N\", :]\nCA_data_cleaner_df",
"_____no_output_____"
],
[
"#filter out only Data and Unemployment Rate\nCA_Data_clean_short_df = CA_data_cleaner_df.loc[:, [\"Date\", \"Unemployment Rate \"]]\nCA_Data_clean_short_df",
"_____no_output_____"
],
[
"#check Data Types for CA Data\nprint(CA_Data_clean_short_df.dtypes)",
"Date object\nUnemployment Rate float64\ndtype: object\n"
],
[
"#Convert Data Types\nCA_Data_clean_short_df['Unemployment Rate '] = pd.to_numeric(CA_Data_clean_short_df['Unemployment Rate '])",
"_____no_output_____"
],
[
"#Make Conversion from Text to DateTime\nCA_Data_clean_short_df['Date'] = pd.to_datetime(CA_Data_clean_short_df['Date']).dt.date",
"_____no_output_____"
],
[
"#Multiply Unemployment Rate to unify with US Unemployment Data\nCA_Data_clean_short_df['Unemployment Rate '] = CA_Data_clean_short_df['Unemployment Rate '] * 100\nCA_Data_clean_short_df",
"_____no_output_____"
],
[
"#check US Unemployment Data Types\nUS_data_df['UNRATE'] = pd.to_numeric(US_data_df['UNRATE'])\nprint(US_data_df.dtypes)",
"DATE object\nUNRATE float64\ndtype: object\n"
],
[
"#Make Conversion from Text to DateTime\nUS_data_df['DATE'] = pd.to_datetime(US_data_df['DATE']).dt.date",
"_____no_output_____"
],
[
"#Rename Columns for US Unemployment Data (Match Headers with CA Unemployment Data)\nUS_data_df.rename(columns = {\"UNRATE\":\"Unemployment Rate\", \"DATE\":\"Date\"}, inplace=\"True\")\nUS_data_df.head(1)",
"_____no_output_____"
],
[
"#check Date Column\nlist(US_data_df[\"Date\"])",
"_____no_output_____"
],
[
"#Rename Headers\nCA_Data_clean_short_df.rename(columns = {\"Unemployment Rate \":\"Unemployment Rate\"}, inplace=\"True\")",
"_____no_output_____"
],
[
"#Final Check of CA Data\nCA_Data_clean_short_df",
"_____no_output_____"
],
[
"#Final Check of US Data\nUS_data_df",
"_____no_output_____"
],
[
"#check Date Column and Sort Values\nlist(CA_Data_clean_short_df.sort_values(by=\"Date\")[\"Date\"])",
"_____no_output_____"
],
[
"#Import Final Directories\nimport matplotlib.pyplot as plt\nimport pandas as pd",
"_____no_output_____"
],
[
"#Create the Chart plotting US and California Unemployment Data Side By Side *US in Orange, CA in Blue)\nfig, axis = plt.subplots(figsize=(8,5))\np1, = axis.plot(list(CA_Data_clean_short_df.sort_values(by=\"Date\")[\"Date\"]),list(CA_Data_clean_short_df.sort_values(by=\"Date\")[\"Unemployment Rate\"]),label=\"CA Unemployment\")\np2, = axis.plot(list(US_data_df[\"Date\"]),list(US_data_df[\"Unemployment Rate\"]),label=\"US Unemployment\")\nplt.xlabel('Unemployment Rate (%)')\nplt.ylabel('Year')\nplt.title('US versus California Unemployment Rates')\nplt.grid(True)\n\nplt.show",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb809e1f3c9a4699472c35454efcb0c3faf212a | 88,234 | ipynb | Jupyter Notebook | jupyter_notebooks/capturing/pico_final.ipynb | TomMasterThesis/sca_on_newhope | 3c72da72076119fb22a64e965c21d340f597cee1 | [
"Apache-2.0"
] | null | null | null | jupyter_notebooks/capturing/pico_final.ipynb | TomMasterThesis/sca_on_newhope | 3c72da72076119fb22a64e965c21d340f597cee1 | [
"Apache-2.0"
] | null | null | null | jupyter_notebooks/capturing/pico_final.ipynb | TomMasterThesis/sca_on_newhope | 3c72da72076119fb22a64e965c21d340f597cee1 | [
"Apache-2.0"
] | null | null | null | 143.469919 | 23,160 | 0.8856 | [
[
[
"## import libs",
"_____no_output_____"
]
],
[
[
"import ctypes\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom picosdk.ps2000 import ps2000 as ps\nfrom picosdk.functions import adc2mV, assert_pico2000_ok",
"_____no_output_____"
],
[
"from ctypes import byref, c_byte, c_int16, c_int32, sizeof\nfrom time import sleep\n\nfrom picosdk.ps2000 import ps2000\nfrom picosdk.functions import assert_pico2000_ok, adc2mV\nfrom picosdk.PicoDeviceEnums import picoEnum",
"_____no_output_____"
]
],
[
[
"## connect and setup",
"_____no_output_____"
]
],
[
[
"SAMPLES = 8_000\nOVERSAMPLING = 1\n\nwantedTimeInterval = 25_000",
"_____no_output_____"
],
[
"# Create status ready for use\nstatus = {}\n\n# Open 2000 series PicoScope\n# Returns handle to chandle for use in future API functions\nstatus[\"openUnit\"] = ps.ps2000_open_unit()\nassert_pico2000_ok(status[\"openUnit\"])\n\n# Create chandle for use\nchandle = ctypes.c_int16(status[\"openUnit\"])",
"_____no_output_____"
],
[
"# Set up channel A\n# handle\n# channel\n# enabled\n# coupling type\n# voltage range\n# analogue offset = 0 V\n\nvoltRangeA = ps.PS2000_VOLTAGE_RANGE['PS2000_100MV']\nstatus[\"setChA\"] = ps.ps2000_set_channel(\n chandle,\n picoEnum.PICO_CHANNEL['PICO_CHANNEL_A'],\n True,\n picoEnum.PICO_COUPLING['PICO_AC'],\n voltRangeA,\n )\n\nassert_pico2000_ok(status[\"setChA\"])",
"_____no_output_____"
],
[
"# Set up channel B\n# handle\n# channel\n# enabled\n# coupling type\n# voltage range\n# analogue offset = 0 V\n\nvoltRangeB = ps.PS2000_VOLTAGE_RANGE['PS2000_5V']\nstatus[\"setChB\"] = ps.ps2000_set_channel(\n chandle,\n picoEnum.PICO_CHANNEL['PICO_CHANNEL_B'],\n True,\n picoEnum.PICO_COUPLING['PICO_DC'],\n voltRangeB,\n )\n\nassert_pico2000_ok(status[\"setChB\"])",
"_____no_output_____"
],
[
"# Set up single trigger\n# handle\n# source\n# threshold in ADC counts (max adc count 32767) # evaluated values in bufferB\n# direction\n# delay = 0 s\n# auto Trigger = 1000 ms\nstatus[\"trigger\"] = ps.ps2000_set_trigger(chandle, \n picoEnum.PICO_CHANNEL['PICO_CHANNEL_B'], \n 10000, \n picoEnum.PICO_THRESHOLD_DIRECTION['PICO_RISING'], \n 0, \n 5000)\nassert_pico2000_ok(status[\"trigger\"])",
"_____no_output_____"
]
],
[
[
"## timebase information",
"_____no_output_____"
]
],
[
[
"def get_timebase(chandle, wantedTimeInterval):\n currentTimebase = 1\n\n oldTimeInterval = None\n timeInterval = c_int32(0)\n timeUnits = c_int16()\n maxSamples = c_int32()\n\n # handle\n # timebase\n # no_of_samples = maxSamples\n # pointer to time_interval = ctypes.byref(timeInterval)\n # pointer to time_units = ctypes.byref(timeUnits)\n # oversample = 1 = oversample\n # pointer to max_samples = ctypes.byref(maxSamplesReturn)\n while ps.ps2000_get_timebase(\n chandle,\n currentTimebase,\n 2000,\n byref(timeInterval),\n byref(timeUnits),\n 1,\n byref(maxSamples)) \\\n == 0 \\\n or timeInterval.value < wantedTimeInterval:\n\n currentTimebase += 1\n oldTimeInterval = timeInterval.value\n\n if currentTimebase.bit_length() > sizeof(c_int16) * 8:\n raise Exception('No appropriate timebase was identifiable')\n\n return currentTimebase - 1, oldTimeInterval, timeInterval\n\nget_timebase(chandle, wantedTimeInterval)",
"_____no_output_____"
]
],
[
[
"## capture",
"_____no_output_____"
]
],
[
[
"timebase_a, interval, timeInterval = get_timebase(chandle, wantedTimeInterval)\n\ncollection_time = c_int32()\n\n# Run block capture\n# handle\n# no_of_samples\n# timebase\n# oversample\n# pointer to time_indisposed_ms = ctypes.byref(timeIndisposedms)\nres = ps.ps2000_run_block(\n chandle,\n SAMPLES,\n timebase_a,\n OVERSAMPLING,\n byref(collection_time)\n)\nassert_pico2000_ok(res)\n\nwhile ps.ps2000_ready(chandle) == 0:\n sleep(0.1)\n\ntimes = (c_int32 * SAMPLES)()\n\nbufferA = (c_int16 * SAMPLES)()\nbufferB = (c_int16 * SAMPLES)()\n\noverflow = c_byte(0)\n\nstatus[\"timesValues\"] = ps.ps2000_get_times_and_values(\n chandle,\n byref(times),\n byref(bufferA),\n byref(bufferB),\n None,\n None,\n byref(overflow),\n 2,\n SAMPLES,\n)\nassert_pico2000_ok(status[\"timesValues\"])\n\nchannel_a_overflow = (overflow.value & 0b0000_0001) != 0",
"_____no_output_____"
]
],
[
[
"## visualize",
"_____no_output_____"
]
],
[
[
"# find maximum ADC count value\nmaxADC = ctypes.c_int16(32767)\n\n# convert ADC counts data to mV\nadc2mVChA = adc2mV(bufferA, voltRangeA, maxADC)\nadc2mVChB = adc2mV(bufferB, voltRangeB, maxADC)\n\n# Create time data\ntime = np.linspace(0, (SAMPLES) * timeInterval.value, SAMPLES)/1_000_000",
"_____no_output_____"
]
],
[
[
"#### channel A",
"_____no_output_____"
]
],
[
[
"# plot data from channel A and B\nplt.plot(time, adc2mVChA[:])\nplt.xlabel('Time (ms)')\nplt.ylabel('Voltage (mV)')\nif channel_a_overflow:\n plt.text(0.01, 0.01, 'Overflow present', color='red')\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### channel B",
"_____no_output_____"
]
],
[
[
"# plot data from channel A and B\nplt.plot(time, adc2mVChB[:], color='orange')\nplt.xlabel('Time (ms)')\nplt.ylabel('Voltage (mV)')\nif channel_a_overflow:\n plt.text(0.01, 0.01, 'Overflow present', color='red')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## capture multiple",
"_____no_output_____"
]
],
[
[
"# define function\ntimebase_a, interval, timeInterval = get_timebase(chandle, wantedTimeInterval)\n\ndef captureBlock(timebase_a, interval, timeInterval):\n\n collection_time = c_int32()\n\n # Run block capture\n # handle\n # no_of_samples\n # timebase\n # oversample\n # pointer to time_indisposed_ms = ctypes.byref(timeIndisposedms)\n res = ps.ps2000_run_block(\n chandle,\n SAMPLES,\n timebase_a,\n OVERSAMPLING,\n byref(collection_time)\n )\n assert_pico2000_ok(res)\n\n while ps.ps2000_ready(chandle) == 0:\n sleep(0.1)\n\n times = (c_int32 * SAMPLES)()\n\n bufferA = (c_int16 * SAMPLES)()\n bufferB = (c_int16 * SAMPLES)()\n\n overflow = c_byte(0)\n\n res = ps.ps2000_get_times_and_values(\n chandle,\n byref(times),\n byref(bufferA),\n byref(bufferB),\n None,\n None,\n byref(overflow),\n 2,\n SAMPLES,\n )\n assert_pico2000_ok(res)\n\n channel_a_overflow = (overflow.value & 0b0000_0001) != 0\n \n return bufferA, bufferB, channel_a_overflow\n\ncaptureBlock(timebase_a, interval, timeInterval)",
"_____no_output_____"
],
[
"# run multiple and average the trace to remove noise\n\navgTrace = None\n\nloops = 1000\nfor i in range(loops):\n bufferA, bufferB, channel_a_overflow = captureBlock(timebase_a, interval, timeInterval)\n \n trace = [bufferA[i] for i in range(len(bufferA)) if bufferB[i]>10_000] # 10,000 adc count as threshold\n \n if avgTrace == None:\n avgTrace = trace\n else:\n lastElt = min(len(avgTrace), len(trace))\n avgTrace = [avgTrace[i] + trace[i] for i in range(lastElt)]\n\navgTrace = [elt/loops for elt in avgTrace]\nprint(avgTrace[:10])",
"[30.166, 26.682, 12.207, 22.648, 33.072, 38.294, 47.277, 49.596, 58.295, 52.204]\n"
],
[
"# find maximum ADC count value\nmaxADC = ctypes.c_int16(32767)\n\n# convert ADC counts data to mV\nadc2mVtrace = adc2mV(avgTrace, voltRangeA, maxADC)\n\n# Create time data\ntime = np.linspace(0, (len(adc2mVtrace)) * timeInterval.value, len(adc2mVtrace))/1_000_000",
"_____no_output_____"
],
[
"# plot data\nplt.plot(time, adc2mVtrace[:])\nplt.xlabel('Time (ms)')\nplt.ylabel('Voltage (mV)')\n\nplt.show()",
"_____no_output_____"
],
[
"# plot data\nplt.plot(time, adc2mVtrace[:])\nplt.xlabel('Time (ms)')\nplt.ylabel('Voltage (mV)')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"## disconnect and print status",
"_____no_output_____"
]
],
[
[
"wait\n# Stop the scope\n# handle = chandle\nstatus[\"stop\"] = ps.ps2000_stop(chandle)\nassert_pico2000_ok(status[\"stop\"])\n\n# Close unitDisconnect the scope\n# handle = chandle\nstatus[\"close\"] = ps.ps2000_close_unit(chandle)\nassert_pico2000_ok(status[\"close\"])\n\n# display status returns\nprint(status)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb815e92af760c7aa08219f59ce0bdcfc6b0b1d | 5,276 | ipynb | Jupyter Notebook | 04_Python_Functions_examples/009_shuffle_a_deck_of_cards.ipynb | peterennis/90_Python_Examples | e2a5a4772ab47d8100b6f13713ea3bc9a25a1ee2 | [
"MIT"
] | 70 | 2021-07-02T07:56:45.000Z | 2022-03-19T04:13:31.000Z | 04_Python_Functions_examples/009_shuffle_a_deck_of_cards.ipynb | bbeella/90_Python_Examples | fbbb1f484b676648881f4287e8175ce9f6224a5a | [
"MIT"
] | null | null | null | 04_Python_Functions_examples/009_shuffle_a_deck_of_cards.ipynb | bbeella/90_Python_Examples | fbbb1f484b676648881f4287e8175ce9f6224a5a | [
"MIT"
] | 51 | 2021-10-30T10:16:28.000Z | 2022-03-19T04:11:05.000Z | 28.365591 | 208 | 0.571266 | [
[
[
"<small><small><i>\nAll the IPython Notebooks in this **Python Examples** series by Dr. Milaan Parmar are available @ **[GitHub](https://github.com/milaan9/90_Python_Examples)**\n</i></small></small>",
"_____no_output_____"
],
[
"# Python Program to Shuffle Deck of Cards\n\nIn this example, you'll learn to shuffle a deck of cards using random module.\n\nTo understand this example, you should have the knowledge of the following **[Python programming](https://github.com/milaan9/01_Python_Introduction/blob/main/000_Intro_to_Python.ipynb)** topics:\n\n* **[Python for Loop](https://github.com/milaan9/03_Python_Flow_Control/blob/main/005_Python_for_Loop.ipynb)**\n* **[Python Modules](https://github.com/milaan9/04_Python_Functions/blob/main/007_Python_Function_Module.ipynb)**\n* **[Python Random Module](https://github.com/milaan9/04_Python_Functions/blob/main/008_Python_Function_random_Module.ipynb)**\n* **[Python Programming Built-in Functions](https://github.com/milaan9/04_Python_Functions/tree/main/002_Python_Functions_Built_in)**",
"_____no_output_____"
]
],
[
[
"# Example 1: shuffle a deck of card\n\n# importing modules\nimport itertools, random\n\n# make a deck of cards\ndeck = list(itertools.product(range(1,14),['Spade','Heart','Diamond','Club']))\n\n# shuffle the cards\nrandom.shuffle(deck)\n\n# draw five cards\nprint(\"You've got:\")\nfor i in range(5):\n print(deck[i][0], \"of\", deck[i][1])\n \n'''\n>>Output/Runtime Test Cases:\n\nYou got:\n5 of Club\n12 of Club\n6 of Club\n9 of Diamond\n2 of Club\n'''",
"You got:\n5 of Club\n12 of Club\n6 of Club\n9 of Diamond\n2 of Club\n"
]
],
[
[
">**Note:** Run the program again to shuffle the cards.\n\n**Explanation:** \n\nIn the program, we used the **`product()`** function in **`itertools`** module to create a deck of cards. This function performs the Cartesian product of the two sequences.\n\nThe two sequences are numbers from 1 to 13 and the four suits. So, altogether we have **13 * 4 = 52** items in the deck with each card as a tuple. For example,\n\n```python\ndeck[0] = (1, 'Spade')\n```\n\nOur deck is ordered, so we shuffle it using the function **`shuffle()`** in **`random`** module.\n\nFinally, we draw the first five cards and display it to the user. We will get different output each time you run this program as shown in our two outputs.\n\nHere we have used the standard modules **`itertools`** and **`random`** that comes with Python.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb827f04110065e40c22636de93b8debb89a208 | 29,853 | ipynb | Jupyter Notebook | _build/jupyter_execute/ipynb/01a-introducao.ipynb | gcpeixoto/FMECD | 9bca72574c6630d1594396fffef31cfb8d58dec2 | [
"CC0-1.0"
] | null | null | null | _build/jupyter_execute/ipynb/01a-introducao.ipynb | gcpeixoto/FMECD | 9bca72574c6630d1594396fffef31cfb8d58dec2 | [
"CC0-1.0"
] | null | null | null | _build/jupyter_execute/ipynb/01a-introducao.ipynb | gcpeixoto/FMECD | 9bca72574c6630d1594396fffef31cfb8d58dec2 | [
"CC0-1.0"
] | null | null | null | 110.566667 | 1,111 | 0.743409 | [
[
[
"# Introdução à Ciência de Dados\n\n## Ciência de dados no século XXI\n\nA dinâmica do mundo globalizado elevou a importância dos dados e da informação a uma\nescala jamais vista na história humana em virtude da evolução exponencial dos recursos tecnológicos, dos meios de comunicação e, principalmente, da computação de alto desempenho. Está em vigor a *Era da Informação*, em que os dados são considerados a matéria-prima imprescindível.\n\nAssim como a terra era o recurso fundamental para a agricultura, e o ferro o era para a indústria, os dados tornaram-se um bem de valor inestimável para pessoas, empresas, governos e para a própria ciência. Com a expansão do fenômeno *Big Data*, diversos nichos do conhecimento começaram a eclodir trazendo consigo uma série de nomes elegantes, tais como *business intelligence*, *data analytics*, *data warehouse* e *data engineering*. \n\nApesar disso, *ciência de dados* desponta-se como o conceito mais razoável para denotar o aspecto científico dos dados. Em um contexto acadêmico, ela encontra-se na interseção de outras áreas do conhecimento e no cerne de uma cadeia maior envolvendo gestão de processos e o pensamento científico. \n\nÉ difícil estabelecer um modelo holístico unificado que traduza de maneira exata a capilaridade da ciência de dados nas atividades modernas. Diante disso, a Figura abaixo tenta ilustrar, para nossos propósitos, como a ciência de dados relaciona-se com outros domínios do conhecimento de maneira multidisciplinar. \n\n```{figure} ../figs/01/cycle-ds.png\n---\nwidth: 300px\nname: cycle-ds\n---\nModelo holístico da ciência de dados.\n```\n\n\nO diagrama mostrado na {numref}`cycle-ds` possui três camadas. A camada mais interna mostra como áreas do conhecimento tradicionais se intersectam para dar forma ao que chamamos de *ciência de dados*. Aqui, enfatizamos três grandes conjuntos: \n\n1. **Matemática/Estatística**, que fornece os modelos matemáticos e estatísticos fundamentais para estudo, análise e inferência de dados, aos quais se agregam as técnicas de aprendizagem de máquina;\n\n2. **Ciência da Computação/Engenharia de Software**, que fornece elementos básicos de hardware e software para projetar soluções de intercâmbio, armazenamento e segurança de dados, por exemplo. \n\n3. **Conhecimento do Domínio/Expertise**, que é o próprio ramo de aplicação do conhecimento que está sendo buscado através dos dados em questão, ao qual se aderem o *data reporting*, a inteligência de negócios, o *marketing* e a comunicação de dados em geral para suporte à tomada de decisões.\n\nA camada intermediária relaciona-se à gestão de processos da cadeia de dados, que envolvem governança, curadoria, armazenamento e reuso de dados, por exemplo, isto é, todos os aspectos relacionados à preservação, manutenção, destruição e compartilhamento de dados. \n\nNo invólucro mais externo, temos a camada relativa ao método científico de busca de soluções para um dado problema. Com alguma adaptação, os processos envolvidos nesta camada representam de maneira satisfatória tanto a ideia de *soluções dirigidas por dados* (*data-driven solutions*) amplamente utilizada em contextos empresariais e industriais, em que ferramentas inovadoras são construídas para entregar produtos e soluções especialmente voltadas a um segmento ou público particular com base em um cuidadoso mapeamento de clientes, quanto o compartilhamento e reprodutibilidade da pesquisa científica. Em linhas gerais, este ciclo contém os seguintes processos:\n\n1. **Definição do problema**, etapa em que uma \"grande pergunta\" é feita, a qual, a princípio, pode ser respondida ao se vasculhar um conjunto de dados específico.\n\n2. **Aquisição de dados**, etapa em que se coleta toda a informação relacionada ao problema lançado na etapa anterior.\n\n3. **Processamento de dados**, etapa em que os dados adquiridos são processados para análise. Nesta etapa realiza-se um verdadeiro tratamento dos dados (limpeza, formatação e organização).\n\n4. **Análise de dados**, etapa em que os dados são analisados e perscrutados por meio de técnicas de mineração, agrupamento e clusterização. Neste momento é que testes de hipótese e mecanismos de inferência são utilizados. \n\n5. **Descoberta de dados**, etapa em que descobertas são realizadas, tais como correlações entre variáveis, comportamentos distintivos e tendências claramente identificáveis, permitindo que conhecimento seja gerado a partir da informação. \n\n6. **Solução**, etapa final do ciclo na qual as descobertas podem ser convertidas em produtos e ativos de valor agregado para o domínio do problema proposto.",
"_____no_output_____"
],
[
"### O caso da COVID-19\n\nA pandemia causada pela COVID-19 que assolou o mundo recentemente pode ser tomada como um estudo de caso singular de aplicação do processo de análise de dados citado na seção anterior. Sob o ponto de vista científico, poderíamos levantar várias questões acerca do vírus no que diz respeito à velocidade de contágio, ao impacto em atividades econômicas, às alterações no comportamento social, entre outras. \n\nModelos epidemiológicos apontam que a interação entre pessoas é um dos principais mecanismos de transmissão viral. A partir dessa premissa e levando em consideração o nosso país, uma pergunta que poderíamos fazer a fim de nortear uma pesquisa em ciência de dados seria: _a taxa de contágio do vírus em pessoas vivendo próximas de um centro comercial localizado em uma zona rural é menor do do que em pessoas vivendo próximas de um centro comercial localizado em uma zona urbana?_. É evidente que, para responder uma pergunta como esta com precisão científica, necessitaríamos de definições e muitos dados. Como delimitaríamos a zona urbana? O centro comercial deveria ser caracterizado como um conjunto de lojas de pequeno porte? Feiras? Um local de comércio onde, diariamente, circulam 100 pessoas por hora? Além disso, neste caso, como faríamos para coletar as informações de que precisamos? No banco de dados do IBGE? No DATASUS?\n\nA aquisição de dados pode ser uma tarefa mais difícil do que se imagina. No caso em questão, certamente buscaríamos informações em bancos de dados do setor público, de secretarias municipais, de órgãos estaduais, até instituições especializadas em âmbito federal. Entretanto, no caso do Brasil, nem todas as regiões – quiçá o país inteiro – usufruem de bancos de dados amplos e precisos onde variáveis primárias necessárias para a análise de dados sejam facilmente obtidas. \n\nSupondo que tenhamos em mãos as informações de saúde acerca dos habitantes das zonas rural e urbana necessárias para nossa pesquisa sobre a COVID-19, o outro passo a tomar é o processamento dos dados. De que maneira o banco de dados se apresenta? Como uma infinidade de planilhas de Excel sem nenhuma formatação específica? Arquivos .csv estruturados e categorizados por faixa etária, município, densidade populacional? Toda a informação é hierárquica em arquivos HDF5? \n\nPara cada situação, devemos dispor de ferramentas específicas e adequadas para manipular, organizar, limpar e estruturar os dados. Todo este tratamento dos dados ocorre, em geral, por duas vias: soluções pré-existentes (programas, recursos, interfaces, frameworks, projetos _open source_ etc. já disponíveis no mercado ou na academia) ou soluções customizadas, criadas pelo cientista de dados para o atendimento de demandas específicas não cobertas pelas soluções pré-existentes. \n\nUma vez processados, os dados atingem uma condição minimamente razoável para serem escrutinados, isto é, analisados minuciosamente. Nesta fase, o intelecto de quem analisa os dados está a todo vapor, visto que um misto de conhecimento técnico, experiência, e criatividade são os ingredientes para realizar descobertas. Os dados são levados de um lado para outro, calculam-se expressões matemática aqui e acolá, testes estatísticos são feitos uma, duas, três, n vezes, até que conclusões surpreendentes podem aparecer. \n\nA propagação de um vírus é um fenômeno não linear suscetível a dinâmicas quase imprevisíveis. Portanto, ao procurarmos a resposta para uma pergunta difícil como a que pusemos acima, pode ser que descubramos padrões e tendências que sequer cogitávamos capazes de responder até mesmo perguntas para outros problemas. Poderíamos chegar à conclusão, por exemplo, que a taxa de contágio na zona urbana é afetada pelas características arquitetônicas do centro comercial: arejamento deficiente, corredores de movimentação estreitos, pontos de venda altamente concentrados, etc. \n\nAo final do ciclo, espera-se que respostas sejam obtidas para que soluções sejam propostas e decisões tomadas com responsabilidade. Quando o assunto é a saúde de pessoas, questões éticas e morais tornam-se extremamente sensíveis. O papel de cientistas e analistas de dados em situações particulares como a da COVID-19 é munir gestores e líderes com recomendações resultantes das evidências mostradas pelos dados. Todavia, é importante dizer que modelos matemáticos são estimativas da realidade e também possuem graus de falibilidade. Portanto, equilibrar as descobertas com o peso das decisões é essencial para o alcance de soluções adequadas. \n\nDiversos projetos focados em ciência e análise de dados focados no estudo da COVID-19 estão atualmente em curso no mundo. Um dos pioneiros foi o _Coronavirus Resource Center_ da _John Hopkins University_ [[CRC-JHU]](https://coronavirus.jhu.edu/map.html). Iniciativas no Brasil são as seguintes: _Observatório Covid-19 BR_ [[COVID19BR]](https://covid19br.github.io/index.html), _Observatório Covid-19 Fiocruz_ [[FIOCRUZ]](https://portal.fiocruz.br/observatorio-covid-19), CoronaVIS-UFRGS [[CoronaVIS-UFRGS]](https://covid19.ufrgs.dev/dashboard/#/dashboard), CovidBR-UFCG [[CovidBR-UFCG]](http://covid.lsi.ufcg.edu.br), entre outras. Na UFPB, destacamos a página do LEAPIG [[LEAPIG-UFPB]](http://www.de.ufpb.br/~leapig/projetos/covid_19.html#PB). Certamente, a COVID-19 deverá se consagrar como um dos maiores estudos de caso da história mundial para a ciência e análise de dados, haja vista o poder computacional de nossos dias.",
"_____no_output_____"
],
[
"### Cientista de dados x analista de dados x engenheiro de dados\n\nAs carreiras profissionais neste novo mundo dos dados converteram-se em muitas especialidades. Há três perfis, em particular, sobre os quais gostaríamos de comentar: _o cientista de dados_, o _analista de dados_ e o _engenheiro de dados_. Porém, antes de entrar nesta \"sopa de letrinhas\", vale a pena entender um pouco sobre como a ciência de dados, como um todo, é compreendida pelas pessoas mundo afora. \n\nNos Estados Unidos, um esforço conjunto entre representantes da universidade, do poder público, da indústria e de outros segmentos culminou na publicação especial No. 1500-1 (2015) do _National Institute of Standards and Technology_ (NIST), que definiu diversos conceitos relacionados à ciência de dados [[NIST 1500-1 (2015)]](https://bigdatawg.nist.gov/_uploadfiles/NIST.SP.1500-1.pdf). Segundo este documento, \n\n> _\"**Cientista de dados** é um profissional que tem conhecimentos suficientes sobre necessidades de negócio, domínio do conhecimento, além de possuir habilidades analíticas, de software e de engenharia de sistemas para gerir, de ponta a ponta, os processos envolvidos no ciclo de vida dos dados.\"_\n\nComo se vê, a identidade do cientista de dados é definida por uma interseção de competências. Todas essas competências estão distribuídas, de certa forma, nas três grandes áreas do conhecimento que citamos acima. Por outro lado, o que exatamente é a _ciência de dados_?\n\nDe acordo com o mesmo documento, \n\n> _\"**Ciência de dados** é a extração do conhecimento útil diretamente a partir de dados através de um processo de descoberta ou de formulação e teste de hipóteses.\"_ \n\nA partir disso, percebemos que _informação_ não é sinônimo de _conhecimento_. Para termos uma clareza melhor dessa distinção, basta refletirmos sobre nosso uso diário do celular. O número de mensagens de texto, de fotos, áudios e vídeos que trocamos com outras pessoas por meio de aplicativos de mensagem instantânea, redes sociais ou e-mail é gigantesco. Quantos de nós não passamos pela necessidade de apagar conteúdo salvo em nosso celular para liberar espaço! Às vezes, não temos ideia de quanta informação trocamos por minuto com três ou quatro colegas. A questão central é: de toda essa informação, que fração seria considerada útil? Isto é, o que poderíamos julgar como conhecimento aproveitável? A resposta talvez seja um incrível \"nada\"... \n\nPortanto, ter bastante informação à disposição não significa, necessariamente, possuir conhecimento. Da mesma forma que estudar para aprender é um exercício difícil para o cérebro, garimpar conhecimento em meio a um mar de informação é uma tarefa que exige paciência, análise, raciocínio dedutivo e criatividade. Por falar em análise de dados, vamos entender um pouco sobre o termo _analytics_, frequentemente utilizado no mercado de trabalho. \n\n_Analytics_ pode ser traduzido literalmente como \"análise\" e, segundo o documento NIST 1500-1, é definido como o \"processo de sintetizar conhecimento a partir da informação\". Diante disso, podemos dizer que \n\n> _\"**Analista de dados** é o profissional capaz de sintetizar conhecimento a partir da informação e convertê-lo em ativo exploráveis.\"_\n\nUma terceira vertente que surgiu com a evolução do _Big Data_ foi a _engenharia de dados_, que tem por objetivo projetar ferramentas, dispositivos e sistemas com robustez suficiente para lidar com a grande massa de dados em circulação. Podemos dizer que \n\n> _\"**Engenheiro(a) de dados** é o(a) profissional que explora recursos independentes para construir sistemas escaláveis capazes de armazenar, manipular e analisar dados com eficiência e e desenvolver novas arquiteturas sempre que a natureza do banco de dados exigi-las.\"_\n\nEmbora essas três especializações possuam características distintivas, elas são tratadas como partes de um corpo maior, que é a Ciência de Dados. O projeto [EDISON](https://edison-project.eu), coordenado pela Universidade de Amsterdã, Holanda, por exemplo, foi responsável por mapear definições e taxonomias para construir grupos profissionais em ciência de dados para ocuparem posições em centros de pesquisa e indústrias na Europa. De acordo com o _EDISON Data Science Framework_ [[EDSF]](https://edison-project.eu/sites/edison-project.eu/files/attached_files/node-5/edison2017poster02-dsp-profiles-v03.pdf), os grupos profissionais se dividem entre gerentes (CEOs, líderes de pesquisa), profissionais gerais (analista de negócios, engenheiros de dados etc.), profissionais de banco de dados (designer de computação em nuvem, designer de banco de dados etc.), profissionais de curadoria (bibliotecários, arquivistas etc.), profissionais técnicos (operadores de equipamentos, mantenedores de _warehouses_ etc.) e profissionais de apoio (suporte a usuários, alimentadores de sistemas, atendentes etc.).\n\n#### Quem faz o quê? \n\nResumimos a seguir as principais tarefas atribuídas a cientistas, analistas e engenheiros(as) de dados com base em artigos de canais especializados [[DataQuest]](https://www.dataquest.io/blog/data-analyst-data-scientist-data-engineer/), [[NCube]](https://ncube.com/blog/data-engineer-data-scientist-data-analyst-what-is-the-difference), [[Medium]](https://medium.com/@gdesantis7/decoding-the-data-scientist-51b353a01443), [[Data Science Academy]](http://datascienceacademy.com.br/blog/qual-a-diferenca-entre-cientista-de-dados-e-engenheiro-de-machine-learning/), [[Data Flair]](https://data-flair.training/blogs/data-scientist-vs-data-engineer-vs-data-analyst/). Uma característica importante entre os perfis diz respeito à organização dos dados. Enquanto cientistas e analistas de dados lidam com dados _estruturados_ – dados organizados e bem definidos que permitem fácil pesquisa –, engenheiros(as) de dados trabalham com dados _não estruturados_.\n\n##### Cientista de dados\n- Realiza o pré-processamento, a transformação e a limpeza dos dados;\n- Usa ferramentas de aprendizagem de máquina para descobrir padrões nos dados;\n- Aperfeiçoa e otimiza algoritmos de aprendizagem de máquina;\n- Formula questões de pesquisa com base em requisitos do domínio do conhecimento;\n \n##### Analista de dados\n- Analisa dados por meio de estatística descritiva;\n- Usa linguagens de consulta a banco de dados para recuperar e manipular a informação;\n- Confecciona relatórios usando visualização de dados; \n- Participa do processo de entendimento de negócios;\n\n##### Engenheiro(a) de dados\n- Desenvolve, constroi e mantém arquiteturas de dados;\n- Realiza testes de larga escala em plataformas de dados;\n- Manipula dados brutos e não estruturados;\n- Desenvolve _pipelines_ para modelagem, mineração e produção de dados\n- Cuida do suporte a cientistas e analistas de dados;\n\n\n#### Que ferramentas são usadas?\n\nAs ferramentas usadas por cada um desses profissionais são variadas e evoluem constantemente. Na lista a seguir, citamos algumas.\n\n##### Cientista de dados\n- R, Python, Hadoop, Ferramentas SQL (Oracle, PostgreSQL, MySQL etc.)\n- Álgebra, Estatística, Aprendizagem de Máquina\n- Ferramentas de visualização de dados\n \n##### Analista de dados\n- R, Python, \n- Excel, Pandas\n- Ferramentas de visualização de dados (Tableau, Infogram, PowerBi etc.)\n- Ferramentas para relatoria e comunicação \n\n##### Engenheiro(a) de dados\n- Ferramentas SQL e noSQL (Oracle NoSQL, MongoDB, Cassandra etc.)\n- Soluções ETL - Extract/Transform/Load (AWS Glue, xPlenty, Stitch etc.)\n- Python, Scala, Java etc.\n- Spark, Hadoop etc.\n\n",
"_____no_output_____"
],
[
"### Matemática por trás dos dados",
"_____no_output_____"
],
[
"No mundo real, lidamos com uma grande diversidade de dados, mas nem sempre percebemos como a Matemática atua por trás de cada pedacinho da informação. Ao longo da sua graduação em ciência de dados, você aprenderá conceitos abstratos novos e trabalhará com mais profundidade outros que já conhece, tais como vetor e matriz. \n\nVocê provavelmente já deve ter ouvido falar que o computador digital funciona com uma linguagem _binária_ cujas mensagens são todas codificadas como sequencias dos dígitos 0 e 1. Daí que vem o nome _bit_, um acrônimo para _binary digit_, ou dígito binário.\n\nEm termos de _bits_, a frase \"Ciência de dados é legal!\", por exemplo, é escrita como\n\n`1000011110100111101010110111011000111101001110000110000011001001100101\n1000001100100110000111001001101111111001110000011101001100000110110011001\n01110011111000011101100100001`. \n\nInteressante, não? Vejamos outros exemplos. \n\nNas aulas de Física, você aprendeu que um vetor possui uma origem e uma extremidade. Tanto a origem como a extremidade são \"pontos\" do espaço. Agora, imagine um plano cartesiano. Se a sua origem é o ponto $O = (0,0)$ e a sua extremidade é o ponto $B = (2,0)$, você pode traçar um vetor de $O$ a $B$ andando duas unidades para a direita. Claramente, este vetor estará sobre o eixo das abscissas. Imagine que você pudesse então usar cada unidade como se fosse uma \"caixa\" onde pudesse \"guardar\" uma informação sobre você. Ou seja, em $O = (0,0)$ você coloca seu nome, em $A = (0,1)$ a sua idade e em $B = (0,2)$ seu CPF. Você teria um \"vetor\" com 3 valores. Além disso, suponha que você pudesse fazer o mesmo para mais 9 pessoas de sua família repetindo este processo em outros 9 vetores paralelos ao primeiro. Você teria agora $3 + 9 \\times 3 = 3 + 27 = 30$ caixas para guardar informações. OK, e daí? \n\nO que acabamos de ilustrar é a ideia fundamental para estruturar tabelas, planilhas do Excel, ou _dataframes_ (que você aprenderá neste curso). Tudo isso são matrizes! Ou seja, informação organizada em linhas e colunas! Cada linha é como um vetor que contém 3 posições (são as colunas). Cada coluna são os registros que você coloca. Então, digamos que você tenha pensado na sua mãe como o próximo membro da família. O nome dela seria colocado na \"caixa\" que estaria no ponto $(1,0)$, a idade dela na \"caixa\" que estaria no ponto $(1,1)$ e o CPF dela na \"caixa\" que estaria no ponto $(1,2)$. Fazendo o com todos os demais membros, você vai concluiria que o CPF do 10o. membro da família deveria estar na \"caixa\" associada ao ponto $(9,9)$. \n\nPara um computador, vetores são chamados de _arrays_. Uma lista de coisas também pode ser comparada a um _array_. Note acima que a segunda coordenada do primeiro vetor (aquele que tem as caixas de informação a seu respeito) é sempre zero. Ela se mantém fixa. Isto significa que o dado assemelha-se a algo **unidimensional**. Isto é, basta que eu apenas faça uma contagem de elementos em uma direção. Isto é muito similar ao conjunto dos números inteiros positivos $\\mathbb{Z}_{+}$. \n\nQuando, porém, inserimos os vetores adicionais (as informações dos membros da sua família), a segunda coordenada também se altera. Isto significa que o dado assemelha-se a algo **bidimensional**. Ou seja, a contagem dos elementos ocorre em duas direções. Levando em conta um plano cartesiano com o eixo das ordenadas orientado para baixo e não para cima, como de costume, os números cresceriam não apenas para a direita, mas também para baixo. Logo, teríamos uma segunda contagem baseada em mais um conjunto de números inteiros positivos $\\mathbb{Z}_{+}$ independente do primeiro. De que estamos falando aqui? Estamos falando do conceito de _par ordenado_. Isto é, qualquer ponto $(x,y)$ com $x \\in \\mathbb{Z}_{+}$ e $y \\in \\mathbb{Z}_{+}$ é um \"local\" onde existiria uma caixinha onde podemos guardar informações de maneira independente. Uma matriz é exatamente isto.\n\nAs imagens vistas na televisão, as _selfies_ e fotografias que você faz com seu celular e as figuras neste livro podem todas ser descritas como matrizes. Cada elemento da matriz é identificado com uma posição $(x,y)$ ao qual damos o nome de _pixel_. Uma imagem é, por sua vez, uma pixelização. Porém, as imagens não são apenas \"endereços\" de pixels. Elas possuem cor, tons de cinza, ou são monocromáticas (preto e branco). As cores são representadas por \"canais\". E, acredite, cada canal é também uma matriz de dados! No final das contas, uma imagem colorida é uma \"matriz formada por outras matrizes\"! \n\nUma matriz formada a partir de outra matriz é um exemplo de dado **tridimensional**. Um exemplo disso são dados sequenciais, tais como um filme ou uma animação. O número de _frames per second_ (FPS), ou \"quadros por segundo\", é tão alta hoje em dia que nossa visão não é capaz de captar que, quando vamos ao cinema ou assistimos um filme pela TV ou no Youtube, o que vemos é exatamente a mudança rápida e sucessiva de vários \"quadros\" de imagens por segundo.\n\nComo você verá ao longo deste curso, muitos conceitos de Matemática que você aprendeu ao longo do Ensino Médio começarão a fazer mais sentido com as aplicações. ",
"_____no_output_____"
],
[
"## Ferramentas computacionais do curso\n\nNeste curso, usaremos Python 3.x (onde x é um número de versão) como linguagem de programação. Por se tratar de uma linguagem interpretada, interagir com ela é mais fácil do que uma linguagem compilada. Um conjunto mínimo de recursos para Python funcionar é composto do _core_ da linguagem, um terminal de comandos e um editor de texto. Enquanto programadores experientes usam menos recursos visuais, para efeito didático, usaremos interfaces mais amigáveis e interativas comprovadas como bons ambientes de aprendizagem. \n\n### _iPython_ e _Jupyter Notebook_ \n\nO [[iPython]](http://ipython.org) foi um projeto iniciado em 2001 para o desenvolvimento de um interpretador Python para melhorar a interatividade com a linguagem. Ele foi integrado como um _kernel_ (núcleo) no projeto [[Jupyter]](http://jupyter.org), desenvolvido em 2014, permitindo textos, códigos e elementos gráficos sejam integrados em cadernos interativos. _Jupyter notebooks_ são interfaces onde podemos executar códigos em diferentes linguagens desde que alteremos os _kernels_. A palavra _Jupyter_ é uma aglutinação das iniciais de _Julia_, _Python_ e _R_, que são as linguagens de programação mais usuais para ciência de dados.\n\n### *Anaconda* \n\nEm 2012, o projeto [[Anaconda]](https://www.anaconda.com) foi iniciado como objetivo de fornecer uma ferramenta completa para o trabalho com Python. Em 2020, já como uma empresa de ponta, ela tornou-se uma das pioneiras no fornecimento de plataformas individuais e empresariais para ciência de dados. Segundo a empresa, a [[Individual Edition]](https://www.anaconda.com/products/individual), que é a versão aberta para uso é a mais popular no mundo com mais de 20 milhões de usuários. Recomendamos que você siga as orientações de instalação desta versão. Uma vez instalada, basta lançar as ferramentas a partir do dashboard _Anaconda Navigator_.\n\n### *Jupyter Lab*\n\nUma ferramenta que melhorou a interatividade do Jupyter é o _Jupyter Lab_, que realiza um alto nível de integração. Este [[artigo]](https://blog.jupyter.org/jupyterlab-is-ready-for-users-5a6f039b8906) discute as características do Jupyter Lab, entre as quais vale citar o recurso de arrastar/soltar para reordenar células de cadernos e copiá-las entre cadernos.\n\n### *Binder* \n\nO projeto [[Binder]](https://mybinder.org) funciona como um servidor online baseada na tecnologia _Jupyter Hub_ para servir cadernos interativos online. Através do Binder, é possível executar códigos \"na nuvem\" sem a necessidade de instalações, porém as sessões são perdidas após o uso. \n\n### *Google Colab* \n\nO [[Google Colab]](http://colab.research.google.com), uma redução de _Colaboratory_, é uma ferramenta que possui características mistas entre o _Jupyter notebook_ e o _Binder_, porém permite que o usuário use a infra-estrutura de computação de alto desempenho (GPUs e TPUS) da Google. A vantagem é que usuários de contas Google podem sincronizar arquivos diretamente com o Google Drive. \n\n\n### Módulos principais\n\nNeste curso, o ecossistema de ferramentas torna-se pleno com a adição de alguns módulos que são considerados essenciais para a prática da ciência e análise de dados contemporânea: \n\n- *numpy* (*NUMeric PYthon*): o *numpy* serve para o trabalho de computação numérica, operando fundamentalmente com vetores, matrizes e ágebra linear.\n\n- *pandas* (*Python for Data Analysis*): é a biblioteca para análise de dados de Python, que opera *dataframes* com eficiência.\n\n- *sympy* (*SYMbolic PYthon*): é um módulo para trabalhar com matemática simbólica e cumpre o papel de um verdadeiro sistema algébrico computacional.\n\n- *matplotlib*: voltado para plotagem e visualização de dados, foi um dos primeiros módulos Python para este fim.\n\n- *scipy* (*SCIentific PYthon*): o *scipy* pode ser visto, na verdade, como um módulo mais amplo que integra os módulos anteriores. Em particular, ele é utilizado para cálculos de integração numérica, interpolação, otimização e estatística.\n\n- *seaborn*: é um módulo para visualização de dados baseado no *matplotlib*, porém com capacidades visuais melhores. \n\nA visualização de dados é um tema de suma importância para resultados da análise exploratória de dados em estatística. Um site recomendado para pesquisar as melhores ferramentas para análise de dados é o [[PyViz]](https://pyviz.org). ",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecb82b550822579640ec30d40dc5a7eaf3d2e22d | 6,985 | ipynb | Jupyter Notebook | proto/prototype_constrained_decoding_server.ipynb | chrishokamp/constrained_decoding | 187846cea4d2aeee6867781b8ceb04cd02d79a4e | [
"MIT"
] | 73 | 2017-04-25T16:38:23.000Z | 2022-02-21T21:39:50.000Z | proto/prototype_constrained_decoding_server.ipynb | Brucewuzhang/constrained_decoding | 187846cea4d2aeee6867781b8ceb04cd02d79a4e | [
"MIT"
] | 6 | 2017-04-24T13:07:38.000Z | 2020-03-12T08:58:01.000Z | proto/prototype_constrained_decoding_server.ipynb | Brucewuzhang/constrained_decoding | 187846cea4d2aeee6867781b8ceb04cd02d79a4e | [
"MIT"
] | 20 | 2017-06-16T08:11:50.000Z | 2021-12-06T01:36:41.000Z | 25.680147 | 127 | 0.536149 | [
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import json\nimport codecs\n\nfrom constrained_decoding import create_constrained_decoder\nfrom constrained_decoding.translation_model.nematus_tm import NematusTranslationModel",
"Using cuDNN version 5110 on context None\nMapped name None to device cuda: GeForce GTX TITAN X (0000:03:00.0)\n"
],
[
"def load_config(filename):\n # defaults -- params which are inputs to `nematus/translate.py`, but aren't part of the persisted *.json config\n translate_config = {\n \"return_alignment\": False\n }\n config = json.loads(codecs.open(filename, encoding='utf8').read())\n return dict(translate_config, **config)",
"_____no_output_____"
],
[
"# Working: test with hard-coded config and model paths\n\nconfigs = [\n '/media/1tb_drive/nematus_ape_experiments/amunmt_ape_pretrained/system/models/src-pe/model.npz.json'\n]\n\nmodels = [\n '/media/1tb_drive/nematus_ape_experiments/amunmt_ape_pretrained/system/models/src-pe/model.4-best.averaged.npz'\n]",
"_____no_output_____"
],
[
"# remember Nematus needs _encoded_ utf8\nconfigs = [load_config(f) for f in configs]\n\n# build ensembled TM\nnematus_tm = NematusTranslationModel(models, configs, model_weights=None)\n",
"Building f_init... Done\nBuilding f_next.. Done\n"
],
[
"model = nematus_tm",
"_____no_output_____"
],
[
"# from constrained_decoding.server import run_imt_server",
"_____no_output_____"
],
[
"decoder = create_constrained_decoder(nematus_tm)\n# app.decoders = {k: create_constrained_decoder(v) for k, v in models.items()}",
"_____no_output_____"
],
[
"# model_dict = {('en', 'de'): nematus_tm}\n\n# run_imt_server(models=model_dict)",
"_____no_output_____"
],
[
"# model = app.models[(source_lang, target_lang)]\n # TODO: create constrained decoders from all models\n# decoder = app.decoders[(source_lang, target_lang)]\n # WORKING: call the correct model\n # WORKING: remember we support multiple inputs for each model (i.e. each model may be an ensemble where sub-models\n # accept different inputs)\n\n # Note: for now we only support one source input\n\nlength_factor = 1.5\nbeam_size = 5\nn_best = 5\n\n \nsource_sentence = u'Help me .'\ninputs = [source_sentence]\n\nmapped_inputs = model.map_inputs(inputs)\nprint(mapped_inputs)\n\ninput_constraints = []\n# if constraints is not None:\n# input_constraints = model.map_constraints(constraints)\n\n# import ipdb; ipdb.set_trace()\nstart_hyp = model.start_hypothesis(mapped_inputs, input_constraints)\n\nsearch_grid = decoder.search(start_hyp=start_hyp, constraints=input_constraints,\n max_hyp_len=int(round(len(mapped_inputs[0][0]) * length_factor)),\n beam_size=beam_size)\n\nbest_output, best_alignments = decoder.best_n(search_grid, model.eos_token, n_best=n_best,\n return_model_scores=False, return_alignments=True,\n length_normalization=True)\n\nif n_best > 1:\n # start from idx 1 to cut off `None` at the beginning of the sequence\n # separate each n-best list with newline\n decoder_output = u'\\n'.join([u' '.join(s[0][1:]) for s in best_output]) + u'\\n\\n'\nelse:\n # start from idx 1 to cut off `None` at the beginning of the sequence\n decoder_output = u' '.join(best_output[0][1:])\n\n# Note alignments are always an n-best list (may be n=1)\n# if write_alignments is not None:\n# with codecs.open(write_alignments, 'a+', encoding='utf8') as align_out:\n# align_out.write(json.dumps([a.tolist() for a in best_alignments]) + u'\\n')\n",
"[array([[[1526],\n [ 732],\n [ 3],\n [ 0]]])]\n[[[1526]\n [ 732]\n [ 3]\n [ 0]]]\n"
],
[
"decoder_output",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb839e9066b96078b6280593e3e5e5a50a06b15 | 15,087 | ipynb | Jupyter Notebook | jupyter_russian/topic04_linear_models/topic4_linear_models_part3_regul_example.ipynb | salman394/AI-ml--course | 2ed3a1382614dd00184e5179026623714ccc9e8c | [
"Unlicense"
] | null | null | null | jupyter_russian/topic04_linear_models/topic4_linear_models_part3_regul_example.ipynb | salman394/AI-ml--course | 2ed3a1382614dd00184e5179026623714ccc9e8c | [
"Unlicense"
] | null | null | null | jupyter_russian/topic04_linear_models/topic4_linear_models_part3_regul_example.ipynb | salman394/AI-ml--course | 2ed3a1382614dd00184e5179026623714ccc9e8c | [
"Unlicense"
] | null | null | null | 34.603211 | 495 | 0.612912 | [
[
[
"<center>\n<img src=\"../../img/ods_stickers.jpg\">\n## Открытый курс по машинному обучению\n</center>\nАвтор материала: программист-исследователь Mail.ru Group, старший преподаватель Факультета Компьютерных Наук ВШЭ Юрий Кашницкий. Материал распространяется на условиях лицензии [Creative Commons CC BY-NC-SA 4.0](https://creativecommons.org/licenses/by-nc-sa/4.0/). Можно использовать в любых целях (редактировать, поправлять и брать за основу), кроме коммерческих, но с обязательным упоминанием автора материала.",
"_____no_output_____"
],
[
"# <center>Тема 4. Линейные модели классификации и регрессии\n## <center>Часть 3. Наглядный пример регуляризации логистической регрессии",
"_____no_output_____"
],
[
"\nВ 1 статье уже приводился пример того, как полиномиальные признаки позволяют линейным моделям строить нелинейные разделяющие поверхности. Покажем это в картинках.\n\nПосмотрим, как регуляризация влияет на качество классификации на наборе данных по тестированию микрочипов из курса Andrew Ng по машинному обучению. \nБудем использовать логистическую регрессию с полиномиальными признаками и варьировать параметр регуляризации C.\nСначала посмотрим, как регуляризация влияет на разделяющую границу классификатора, интуитивно распознаем переобучение и недообучение.\nПотом численно установим близкий к оптимальному параметр регуляризации с помощью кросс-валидации (`cross-validation`) и перебора по сетке (`GridSearch`). ",
"_____no_output_____"
]
],
[
[
"from __future__ import division, print_function\n\n# отключим всякие предупреждения Anaconda\nimport warnings\n\nwarnings.filterwarnings(\"ignore\")\n%matplotlib inline\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nfrom matplotlib import pyplot as plt\nfrom sklearn.linear_model import LogisticRegression, LogisticRegressionCV\nfrom sklearn.model_selection import GridSearchCV, StratifiedKFold, cross_val_score\nfrom sklearn.preprocessing import PolynomialFeatures",
"_____no_output_____"
]
],
[
[
"Загружаем данные с помощью метода `read_csv` библиотеки `pandas`. В этом наборе данных для 118 микрочипов (объекты) указаны результаты двух тестов по контролю качества (два числовых признака) и сказано, пустили ли микрочип в производство. Признаки уже центрированы, то есть из всех значений вычтены средние по столбцам. Таким образом, \"среднему\" микрочипу соответствуют нулевые значения результатов тестов. ",
"_____no_output_____"
]
],
[
[
"# загрузка данных\ndata = pd.read_csv(\n \"../../data/microchip_tests.txt\", header=None, names=(\"test1\", \"test2\", \"released\")\n)\n# информация о наборе данных\ndata.info()",
"_____no_output_____"
]
],
[
[
"Посмотрим на первые и последние 5 строк.",
"_____no_output_____"
]
],
[
[
"data.head(5)",
"_____no_output_____"
],
[
"data.tail(5)",
"_____no_output_____"
]
],
[
[
"Сохраним обучающую выборку и метки целевого класса в отдельных массивах NumPy.",
"_____no_output_____"
]
],
[
[
"X = data.iloc[:, :2].values\ny = data.iloc[:, 2].values",
"_____no_output_____"
]
],
[
[
"Отобразим данные. Красный цвет соответствует бракованным чипам, зеленый – нормальным. ",
"_____no_output_____"
]
],
[
[
"plt.scatter(X[y == 1, 0], X[y == 1, 1], c=\"green\", label=\"Выпущен\")\nplt.scatter(X[y == 0, 0], X[y == 0, 1], c=\"red\", label=\"Бракован\")\nplt.xlabel(\"Тест 1\")\nplt.ylabel(\"Тест 2\")\nplt.title(\"2 теста микрочипов\")\nplt.legend();",
"_____no_output_____"
]
],
[
[
"Определяем функцию для отображения разделяющей кривой классификатора",
"_____no_output_____"
]
],
[
[
"def plot_boundary(clf, X, y, grid_step=0.01, poly_featurizer=None):\n x_min, x_max = X[:, 0].min() - 0.1, X[:, 0].max() + 0.1\n y_min, y_max = X[:, 1].min() - 0.1, X[:, 1].max() + 0.1\n xx, yy = np.meshgrid(\n np.arange(x_min, x_max, grid_step), np.arange(y_min, y_max, grid_step)\n )\n\n # каждой точке в сетке [x_min, m_max]x[y_min, y_max]\n # ставим в соответствие свой цвет\n Z = clf.predict(poly_featurizer.transform(np.c_[xx.ravel(), yy.ravel()]))\n Z = Z.reshape(xx.shape)\n plt.contour(xx, yy, Z, cmap=plt.cm.Paired)",
"_____no_output_____"
]
],
[
[
"Полиномиальными признаками до степени $d$ для двух переменных $x_1$ и $x_2$ мы называем следующие:\n\n$$\\large \\{x_1^d, x_1^{d-1}x_2, \\ldots x_2^d\\} = \\{x_1^ix_2^j\\}_{i+j=d, i,j \\in \\mathbb{N}}$$\n\nНапример, для $d=3$ это будут следующие признаки:\n\n$$\\large 1, x_1, x_2, x_1^2, x_1x_2, x_2^2, x_1^3, x_1^2x_2, x_1x_2^2, x_2^3$$\n\nНарисовав треугольник Пифагора, Вы сообразите, сколько таких признаков будет для $d=4,5...$ и вообще для любого $d$.\nПопросту говоря, таких признаков экспоненциально много, и строить, скажем, для 100 признаков полиномиальные степени 10 может оказаться затратно (а более того, и не нужно). \n",
"_____no_output_____"
],
[
"Создадим объект `sklearn`, который добавит в матрицу $X$ полиномиальные признаки вплоть до степени 7.",
"_____no_output_____"
]
],
[
[
"poly = PolynomialFeatures(degree=7)\nX_poly = poly.fit_transform(X)",
"_____no_output_____"
],
[
"X_poly.shape",
"_____no_output_____"
]
],
[
[
"Обучим логистическую регрессию с параметром регуляризации $C = 10^{-2}$. Изобразим разделяющую границу.\nТакже проверим долю правильных ответов классификатора на обучающей выборке. Видим, что регуляризация оказалась \nслишком сильной, и модель \"недообучилась\".",
"_____no_output_____"
]
],
[
[
"C = 1e-2\nlogit = LogisticRegression(C=C, n_jobs=-1, random_state=17)\nlogit.fit(X_poly, y)\n\nplot_boundary(logit, X, y, grid_step=0.01, poly_featurizer=poly)\n\nplt.scatter(X[y == 1, 0], X[y == 1, 1], c=\"green\", label=\"Выпущен\")\nplt.scatter(X[y == 0, 0], X[y == 0, 1], c=\"red\", label=\"Бракован\")\nplt.xlabel(\"Тест 1\")\nplt.ylabel(\"Тест 2\")\nplt.title(\"2 теста микрочипов. Логит с C=0.01\")\nplt.legend()\n\nprint(\n \"Доля правильных ответов классификатора на обучающей выборке:\",\n round(logit.score(X_poly, y), 3),\n)",
"_____no_output_____"
]
],
[
[
"Увеличим $C$ до 1. Тем самым мы *ослабляем* регуляризацию, теперь в решении значния весов логистической регрессии могут оказаться больше (по модулю), чем в прошлом случае. ",
"_____no_output_____"
]
],
[
[
"C = 1\nlogit = LogisticRegression(C=C, n_jobs=-1, random_state=17)\nlogit.fit(X_poly, y)\n\nplot_boundary(logit, X, y, grid_step=0.005, poly_featurizer=poly)\n\nplt.scatter(X[y == 1, 0], X[y == 1, 1], c=\"green\", label=\"Выпущен\")\nplt.scatter(X[y == 0, 0], X[y == 0, 1], c=\"red\", label=\"Бракован\")\nplt.xlabel(\"Тест 1\")\nplt.ylabel(\"Тест 2\")\nplt.title(\"2 теста микрочипов. Логит с C=1\")\nplt.legend()\n\nprint(\n \"Доля правильных ответов классификатора на обучающей выборке:\",\n round(logit.score(X_poly, y), 3),\n)",
"_____no_output_____"
]
],
[
[
"Еще увеличим $C$ – до 10 тысяч. Теперь регуляризации явно недостаточно, и мы наблюдаем переобучение. Можно заметить, что в прошлом случае (при $C$=1 и \"гладкой\" границе) доля правильных ответов модели на обучающей выборке не намного ниже, чем в 3 случае, зато на новой выборке, можно себе представить, 2 модель сработает намного лучше. ",
"_____no_output_____"
]
],
[
[
"C = 1e4\nlogit = LogisticRegression(C=C, n_jobs=-1, random_state=17)\nlogit.fit(X_poly, y)\n\nplot_boundary(logit, X, y, grid_step=0.005, poly_featurizer=poly)\n\nplt.scatter(X[y == 1, 0], X[y == 1, 1], c=\"green\", label=\"Выпущен\")\nplt.scatter(X[y == 0, 0], X[y == 0, 1], c=\"red\", label=\"Бракован\")\nplt.xlabel(\"Тест 1\")\nplt.ylabel(\"Тест 2\")\nplt.title(\"2 теста микрочипов. Логит с C=10k\")\nplt.legend()\n\nprint(\n \"Доля правильных ответов классификатора на обучающей выборке:\",\n round(logit.score(X_poly, y), 3),\n)",
"_____no_output_____"
]
],
[
[
"Чтоб обсудить результаты, перепишем формулу для функционала, который оптимизируется в логистической регрессии, в таком виде:\n$$J(X,y,w) = \\mathcal{L} + \\frac{1}{C}||w||^2,$$\n\nгде\n - $\\mathcal{L}$ – логистическая функция потерь, просуммированная по всей выборке\n - $C$ – обратный коэффициент регуляризации (тот самый $C$ в `sklearn`-реализации `LogisticRegression`)",
"_____no_output_____"
],
[
"**Промежуточные выводы**:\n - чем больше параметр $C$, тем более сложные зависимости в данных может восстанавливать модель (интуитивно $C$ соответствует \"сложности\" модели (model capacity))\n - если регуляризация слишком сильная (малые значения $C$), то решением задачи минимизации логистической функции потерь может оказаться то, когда многие веса занулились или стали слишком малыми. Еще говорят, что модель недостаточно \"штрафуется\" за ошибки (то есть в функционале $J$ \"перевешивает\" сумма квадратов весов, а ошибка $\\mathcal{L}$ может быть относительно большой). В таком случае модель окажется *недообученной* (1 случай)\n - наоборот, если регуляризация слишком слабая (большие значения $C$), то решением задачи оптимизации может стать вектор $w$ с большими по модулю компонентами. В таком случае больший вклад в оптимизируемый функционал $J$ имеет $\\mathcal{L}$ и, вольно выражаясь, модель слишком \"боится\" ошибиться на объектах обучающей выборки, поэтому окажется *переобученной* (3 случай)\n - то, какое значение $C$ выбрать, сама логистическая регрессия \"не поймет\" (или еще говорят \"не выучит\"), то есть это не может быть определено решением оптимизационной задачи, которой является логистическая регрессия (в отличие от весов $w$). Так же точно, дерево решений не может \"само понять\", какое ограничение на глубину выбрать (за один процесс обучения). Поэтому $C$ – это *гиперпараметр* модели, который настраивается на кросс-валидации, как и *max_depth* для дерева.",
"_____no_output_____"
],
[
"**Настройка параметра регуляризации**",
"_____no_output_____"
],
[
"Теперь найдем оптимальное (в данном примере) значение параметра регуляризации $C$. Сделать это можно с помощью `LogisticRegressionCV` – перебора параметров по сетке с последующей кросс-валидацией. Этот класс создан специально для логистической регрессии (для нее известны эффективные алгоритмы перебора параметров), для произвольной модели мы бы использовали `GridSearchCV`, `RandomizedSearchCV` или, например, специальные алгоритмы оптимизации гиперпараметров, реализованные в `hyperopt`.",
"_____no_output_____"
]
],
[
[
"skf = StratifiedKFold(n_splits=5, shuffle=True, random_state=17)\n\nc_values = np.logspace(-2, 3, 500)\n\nlogit_searcher = LogisticRegressionCV(Cs=c_values, cv=skf, verbose=1, n_jobs=-1)\nlogit_searcher.fit(X_poly, y)",
"_____no_output_____"
],
[
"logit_searcher.C_",
"_____no_output_____"
]
],
[
[
"Посмотрим, как качество модели (доля правильных ответов на обучающей и валидационной выборках) меняется при изменении гиперпараметра $C$. ",
"_____no_output_____"
]
],
[
[
"plt.plot(c_values, np.mean(logit_searcher.scores_[1], axis=0))\nplt.xlabel(\"C\")\nplt.ylabel(\"Mean CV-accuracy\");",
"_____no_output_____"
]
],
[
[
"Выделим участок с \"лучшими\" значениями C.",
"_____no_output_____"
]
],
[
[
"plt.plot(c_values, np.mean(logit_searcher.scores_[1], axis=0))\nplt.xlabel(\"C\")\nplt.ylabel(\"Mean CV-accuracy\")\nplt.xlim((0, 10));",
"_____no_output_____"
]
],
[
[
"Такие кривые называются *валидационными*, и в `sklearn` для них их построения есть специальные методы.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb8409555e5f88d2d101e6c1331bedbb478671f | 77,271 | ipynb | Jupyter Notebook | experiments/speaker_verification/visualise_encoder_result.ipynb | onucharles/tensorized-rnn | 69fc031f1efe169ee88327d10bdf5e5bc24f03cf | [
"MIT"
] | 16 | 2020-11-19T16:10:17.000Z | 2021-12-02T13:31:30.000Z | experiments/speaker_verification/visualise_encoder_result.ipynb | kritostudent/tensorized-rnn | 69fc031f1efe169ee88327d10bdf5e5bc24f03cf | [
"MIT"
] | 2 | 2021-02-26T08:45:16.000Z | 2021-08-11T13:47:43.000Z | experiments/speaker_verification/visualise_encoder_result.ipynb | kritostudent/tensorized-rnn | 69fc031f1efe169ee88327d10bdf5e5bc24f03cf | [
"MIT"
] | 7 | 2021-01-21T11:24:31.000Z | 2022-03-21T08:59:08.000Z | 98.685824 | 31,068 | 0.805516 | [
[
[
"from comet_ml.api import API\nimport numpy as np\nimport matplotlib.pyplot as plt \nimport pandas as pd\nfrom scipy.stats import entropy\nfrom sympy.ntheory import factorint\nfrom sympy.utilities.iterables import multiset_partitions\nfrom pathlib import Path\n\ncomet_api = API(api_key=\"hRTMB66l59BMb9pPrRzLrbzwp\")",
"_____no_output_____"
],
[
"experiments = comet_api.get(workspace=\"tensorized-rnn\", project_name=\"speaker-verification\")\n\ndef fetch_val_eer(experiments, cores_result_dict):\n for exp in experiments:\n tags = exp.get_tags()\n tag = tags[-1]\n \n if tag in cores_result_dict.keys():\n # best validation eer is the last one recorded.\n best_val_eer_dict = exp.get_metrics(\"best_eer\")[-1]\n best_val_eer = float(best_val_eer_dict['metricValue'])\n # best_val_eer_step = int(best_val_eer_dict['step'])\n \n cores_result_dict[tag].append(best_val_eer)\n print(cores_result_dict)\n \n # average results of experiment repeats.\n cores_avg_result_dict = {}\n for tag, eer_list in cores_result_dict.items():\n print(\"Tag: {}. No of experiments: {}\".format(tag, len(eer_list)))\n cores_avg_result_dict[tag] = round(np.mean(eer_list) * 100, 2) \n \n return cores_avg_result_dict\n\n\ndef safely_get_parameter(api_exp, param_name):\n param_list = api_exp.get_parameters_summary(param_name)\n if len(param_list) == 0:\n raise ValueError(\"parameter '{}' was not found in experiment '{}'\"\n .format(param_name, api_exp.get_metadata()[\"experimentKey\"]))\n return param_list[\"valueCurrent\"]\n\n###----------FUNCTIONS from t3nsor library for getting auto shape of a tensor-train cores--------####\ndef _get_all_factors(n, d=3, mode='ascending'):\n p = _factorint2(n)\n if len(p) < d:\n p = p + [1, ] * (d - len(p))\n\n if mode == 'ascending':\n def prepr(x):\n return tuple(sorted([np.prod(_) for _ in x]))\n elif mode == 'descending':\n def prepr(x):\n return tuple(sorted([np.prod(_) for _ in x], reverse=True))\n\n elif mode == 'mixed':\n def prepr(x):\n x = sorted(np.prod(_) for _ in x)\n N = len(x)\n xf, xl = x[:N//2], x[N//2:]\n return tuple(_roundrobin(xf, xl))\n\n else:\n raise ValueError('Wrong mode specified, only {} are available'.format(MODES))\n\n raw_factors = multiset_partitions(p, d)\n clean_factors = [prepr(f) for f in raw_factors]\n clean_factors = list(set(clean_factors))\n return clean_factors\n\ndef _to_list(p):\n res = []\n for k, v in p.items():\n res += [k, ] * v\n return res\n\ndef _factorint2(p):\n return _to_list(factorint(p))\n\ndef auto_shape(n, d=3, criterion='entropy', mode='ascending'):\n factors = _get_all_factors(n, d=d, mode=mode)\n if criterion == 'entropy':\n weights = [entropy(f) for f in factors]\n elif criterion == 'var':\n weights = [-np.var(f) for f in factors]\n else:\n raise ValueError('Wrong criterion specified, only {} are available'.format(CRITERIONS))\n\n i = np.argmax(weights)\n return list(factors[i])\n###----------FUNCTIONS from t3nsor library for getting auto shape of a tensor-train cores--------####\n\ndef lstm_num_params(num_layers, hidden_size, input_size, bias=True, n_gates_and_cell=4):\n result = 0\n \n # layer 1\n result = n_gates_and_cell * ((input_size + hidden_size + int(bias)) * hidden_size)\n \n # for higher layers, input_size == hidden_size\n if num_layers > 1:\n num_layers_minus_1 = num_layers - 1\n result += num_layers_minus_1 * n_gates_and_cell * ((hidden_size + hidden_size + int(bias)) * hidden_size)\n \n return result\n\ndef tt_linear_num_params(in_size, out_size, n_cores, rank, bias=True):\n# print('in: {}, out: {}, rank: {}, cores: {}'.format(in_size, out_size, rank, n_cores))\n in_core_shape = auto_shape(in_size, n_cores)\n out_core_shape = auto_shape(out_size, n_cores)\n \n # number of parameters in a tt linear layer.\n assert len(in_core_shape) == len(out_core_shape)\n n_cores = len(in_core_shape)\n \n result = 0\n out_size = 1\n for i in range(n_cores):\n if i == 0 or i == n_cores - 1:\n result += rank * (in_core_shape[i]) * out_core_shape[i]\n else:\n result += rank**2 * (in_core_shape[i]) * out_core_shape[i]\n out_size *= out_core_shape[i]\n \n if bias:\n result += out_size\n \n# print('linear output ', result)\n return result\n\ndef tt_lstm_num_params(num_layers, in_size, hidden_size, n_cores, rank, bias=True, n_gates_and_cell = 4):\n result = 0\n \n # layer 1\n result = n_gates_and_cell * \\\n (tt_linear_num_params(in_size, hidden_size, n_cores, rank, False) \\\n + tt_linear_num_params(hidden_size, hidden_size, n_cores, rank, bias))\n \n # for higher layers, input_size == hidden_size\n if num_layers > 1:\n num_layers_minus_1 = num_layers - 1\n result += num_layers_minus_1 * n_gates_and_cell * \\\n (tt_linear_num_params(hidden_size, hidden_size, n_cores, rank, False) \\\n + tt_linear_num_params(hidden_size, hidden_size, n_cores, rank, bias))\n \n return result\n\ndef tt_lstm_num_params_with_concattrick(num_layers, in_size, hidden_size, n_cores, rank, bias=True, n_gates_and_cell = 4):\n # number of parameters in a tt lstm that uses concatenation trick.\n result = 0\n \n # layer 1\n result = (tt_linear_num_params(in_size, hidden_size * n_gates_and_cell, n_cores, rank, False) \n + tt_linear_num_params(hidden_size, hidden_size * n_gates_and_cell, n_cores, rank, bias))\n \n # for higher layers, input_size == hidden_size\n if num_layers > 1:\n num_layers_minus_1 = num_layers - 1\n result += num_layers_minus_1 * (tt_linear_num_params(hidden_size, hidden_size * n_gates_and_cell, n_cores, rank, False) \n + tt_linear_num_params(hidden_size, hidden_size * n_gates_and_cell, n_cores, rank, bias))\n \n return result",
"_____no_output_____"
]
],
[
[
"# Model size and parameters",
"_____no_output_____"
]
],
[
[
"hidden_sizes = [256, 512, 768]\nnum_layers = [1, 2, 3]\ninput_size = 4096\nemb_size = 256\nn_gates = 4\n\ndata = []\nfor hidden_size in hidden_sizes:\n row = []\n for layer_size in num_layers:\n num_linear_layer = ((hidden_size + 1) * emb_size)\n num_lstm_layers = lstm_num_params(layer_size, hidden_size, input_size, n_gates_and_cell=n_gates)\n row.append(num_lstm_layers + num_linear_layer)\n data.append(row)\nresult = pd.DataFrame(data, index=hidden_sizes, columns=num_layers)\nprint(\"No of parameters\")\nprint(result)",
"No of parameters\n 1 2 3\n256 4523264 5048576 5573888\n512 9570560 11669760 13768960\n768 15142144 19863808 24585472\n"
]
],
[
[
"## Evaluating no of parameters in TT-LSTM",
"_____no_output_____"
]
],
[
[
"input_size = 4096\nemb_size = 256\nn_cores_list = [2,3,4]\nranks_list = [1, 2, 3, 4, 5]\nn_gates = 4\n\ndef print_tt_lstm_options(num_layers, hidden_size, input_size, emb_size, n_cores_list, ranks_list):\n data = []\n for i in range(len(n_cores_list)):\n n_cores = n_cores_list[i]\n row = []\n for rank in ranks_list:\n num_lstm_layers = tt_lstm_num_params_with_concattrick(num_layers, input_size, hidden_size, \n n_cores, rank, bias=True, n_gates_and_cell=n_gates)\n num_linear_layer = tt_linear_num_params(hidden_size, emb_size, n_cores, rank, bias=True)\n row.append(num_lstm_layers + num_linear_layer)\n data.append(row)\n data = pd.DataFrame(data, index=n_cores_list, columns=ranks_list)\n return data\n\n#---------------\nhidden_size = 768\nnum_layers = 1\nprint(\"No of parameters ({}/{})\".format(num_layers, hidden_size))\nprint(print_tt_lstm_options(num_layers, hidden_size, input_size, emb_size, n_cores_list, ranks_list))\nprint(\"\")\n\n#---------------\nhidden_size = 256\nnum_layers = 3\nprint(\"No of parameters ({}/{})\".format(num_layers, hidden_size))\nprint(print_tt_lstm_options(num_layers, hidden_size, input_size, emb_size, n_cores_list, ranks_list))\nprint(\"\")\n\n#---------------\nhidden_size = 512\nnum_layers = 2\nprint(\"No of parameters ({}/{})\".format(num_layers, hidden_size))\nprint(print_tt_lstm_options(num_layers, hidden_size, input_size, emb_size, n_cores_list, ranks_list))\nprint(\"\")\n\nhidden_size = 512\nnum_layers = 1\nprint(\"No of parameters ({}/{})\".format(num_layers, hidden_size))\nprint(print_tt_lstm_options(num_layers, hidden_size, input_size, emb_size, n_cores_list, ranks_list))\nprint(\"\")",
"No of parameters (1/768)\n 1 2 3 4 5\n2 14592 25856 37120 48384 59648\n3 4640 6848 9952 13952 18848\n4 3824 4816 6304 8288 10768\n\nNo of parameters (3/256)\n 1 2 3 4 5\n2 13056 22784 32512 42240 51968\n3 5104 7904 11728 16576 22448\n4 4064 5536 7744 10688 14368\n\nNo of parameters (2/512)\n 1 2 3 4 5\n2 18944 33536 48128 62720 77312\n3 6112 9280 13856 19840 27232\n4 5088 6528 8672 11520 15072\n\nNo of parameters (1/512)\n 1 2 3 4 5\n2 11776 21248 30720 40192 49664\n3 3424 5440 8352 12160 16864\n4 2752 3648 4992 6784 9024\n\n"
]
],
[
[
"## Evaluating no of parameters in Naive TT-LSTM",
"_____no_output_____"
]
],
[
[
"input_size = 4096\nemb_size = 256\nn_cores_list = [2,3,4]\nranks_list = [1, 2, 3, 4, 5]\nn_gates = 4\n\ndef print_naive_tt_lstm_options(num_layers, hidden_size, input_size, emb_size, n_cores_list, ranks_list):\n data = []\n for i in range(len(n_cores_list)):\n n_cores = n_cores_list[i]\n row = []\n for rank in ranks_list:\n num_lstm_layers = tt_lstm_num_params(num_layers, input_size, hidden_size, \n n_cores, rank, bias=True, n_gates_and_cell=n_gates)\n num_linear_layer = tt_linear_num_params(hidden_size, emb_size, n_cores, rank, bias=True)\n row.append(num_lstm_layers + num_linear_layer)\n data.append(row)\n data = pd.DataFrame(data, index=n_cores_list, columns=ranks_list)\n return data\n\n#---------------\nhidden_size = 512\nnum_layers = 1\nprint(\"No of parameters ({}/{})\".format(num_layers, hidden_size))\nprint(print_naive_tt_lstm_options(num_layers, hidden_size, input_size, emb_size, n_cores_list, ranks_list))\nprint(\"\")",
"No of parameters (1/512)\n 1 2 3 4 5\n2 20480 38656 56832 75008 93184\n3 4768 8896 14688 22144 31264\n4 3472 5472 8304 11968 16464\n\n"
]
],
[
[
"# Evaluating no of parameters in linear layer using low-rank factorisation.",
"_____no_output_____"
]
],
[
[
"def low_rank_params(in_features, out_features, rank, bias):\n return ((in_features + int(bias)) * rank) + ((rank + int(bias)) * out_features)\n\nin_size = 256\nout_size = 256\nranks = 2 ** np.arange(1,7)\n\nprint(\"No of parameters (uncompressed):\", 256 * 256)\nprint(\"Rank\\tNum of Parms\")\nfor rank in ranks:\n print(\"{}\\t{}\".format(rank, low_rank_params(in_size, out_size, rank, True)))",
"No of parameters (uncompressed): 65536\nRank\tNum of Parms\n2\t1282\n4\t2308\n8\t4360\n16\t8464\n32\t16672\n64\t33088\n"
]
],
[
[
"# Plot experiment result",
"_____no_output_____"
]
],
[
[
"# dict with \"key: n-cores\", \"value: dict of (key: rank, value: test_eer)\"\n# experiments_result_dict = {2: {1: 0, 2: 0, 4: 0},\n# 3: {1: 0, 2: 0, 4: 0},\n# 4: {1: 0, 2: 0, 4: 0}}\nexperiments_result_dict = {2: {1: 0, 2: 0, 3:0, 4: 0},\n 3: {1: 0, 2: 0, 3:0, 4: 0},\n 4: {1: 0, 2: 0, 3:0, 4: 0}}\nexp_seed = 11\n\nfor exp in experiments: \n compression = exp.get_parameters_summary(\"compression\")[\"valueCurrent\"]\n if not compression == 'tt' : continue\n \n try:\n n_cores = int(safely_get_parameter(exp, \"n_cores\"))\n tt_rank = int(safely_get_parameter(exp, \"rank\"))\n seed = int(safely_get_parameter(exp, \"seed\"))\n best_val_eer = float(exp.get_metrics(\"test_EER\")[-1]['metricValue'])\n except Exception as e:\n print(e)\n continue\n\n if n_cores not in experiments_result_dict \\\n or tt_rank not in experiments_result_dict[n_cores] \\\n or seed != exp_seed:\n print(\"Skipping experiment: compression={}, n_cores={}, tt_rank={}, seed={}\".format(compression, n_cores, tt_rank, seed))\n continue\n experiments_result_dict[n_cores][tt_rank] = best_val_eer\nprint(experiments_result_dict)\n ",
"Skipping experiment: compression=tt, n_cores=2, tt_rank=5, seed=11\nSkipping experiment: compression=tt, n_cores=4, tt_rank=5, seed=11\nSkipping experiment: compression=tt, n_cores=3, tt_rank=5, seed=11\n{2: {1: 0.047115384616034435, 2: 0.04344512195077787, 3: 0.06874999999999927, 4: 0.06213942307541739}, 3: {1: 0.06093749999999991, 2: 0.053125000000000026, 3: 0.05468749999999987, 4: 0.053846153846863}, 4: {1: 0.05337284482744496, 2: 0.05576923076880846, 3: 0.05312499999999996, 4: 0.04278846153918008}}\n"
],
[
"%matplotlib qt\n\nplt.figure(dpi=100)\ninput_size = 40\nemb_size = 256\nhidden_size = 768\nnum_layers = 1\n\n# plot uncompressed experiment.\nplt.plot(2682112, 0.07333 * 100, \"D\", label=\"LSTM\", color='black', markersize=8)\n\n# # plot low-rank experiments\n# lr_param_counts = [low_rank_params(in_size, out_size, rank, True) for rank in [8, 16, 32, 64]]\n# print(\"LR params: \", lr_param_counts)\n# plt.plot(lr_param_counts, [.0647, 0.0587, 0.0433, 0.0511], \"-.D\", label=\"low-rank\")\n\n# plot tt experiments.\nfor k, v in experiments_result_dict.items():\n param_counts = []\n for rank in v.keys():\n num_lstm_layers = tt_lstm_num_params_with_concattrick(num_layers, input_size, hidden_size, k, rank, bias=True)\n num_linear_layer = tt_linear_num_params(hidden_size, emb_size, k, rank, bias=True)\n \n param_counts.append(num_lstm_layers + num_linear_layer)\n print(\"TT-{}cores params: {}\".format(k, param_counts))\n \n # plot all rank values for the current core, k.\n shape_str = \" $x$ \".join([str(s) for s in auto_shape(hidden_size, k)])\n values_in_perc = [val * 100 for val in v.values()]\n plt.plot(param_counts, values_in_perc, \"-o\", label=\"TT-LSTM \" + shape_str, linewidth=3, markersize=8)\nplt.xscale('log')\nplt.xlabel('# parameters', fontsize=14)\nplt.ylabel('EER (%)', fontsize=14)\nplt.ylim([0,20])\nplt.xticks(fontsize=13)\nplt.yticks(fontsize=13)\nplt.legend(fontsize=13, loc='upper center')\nplt.show()",
"TT-2cores params: [8176, 13024, 17872, 22720]\nTT-3cores params: [4104, 5392, 7192, 9504]\nTT-4cores params: [3668, 4312, 5260, 6512]\n"
],
[
"# plot the val EER for each rank.\nranks_result_dict = {'tt-cores4-rank2':[], 'tt-cores4-rank3':[], 'tt-cores4-rank4':[], 'tt-cores4-rank5':[], \n 'tt-cores4-rank6':[], 'tt-cores4-rank8':[]}\nranks_avg_result_dict = fetch_val_eer(experiments, ranks_result_dict)\nplt.figure()\nplt.plot(np.arange(2,8), ranks_avg_result_dict.values(), '-o')\nplt.xlabel('rank')\nplt.ylabel('val EER')\nplt.title('no of cores = 4')\nplt.show()",
"{'tt-cores4-rank2': [0.04134615384757199, 0.04238782051446542, 0.03906250000000022, 0.03906250000009872, 0.039583333334543266], 'tt-cores4-rank3': [0.03914262820514222, 0.04062500000000007], 'tt-cores4-rank4': [0.042187500000000024, 0.044991987178826826, 0.04658830275229364], 'tt-cores4-rank5': [0.04062500000000005, 0.04519230769182035, 0.039022943037974725], 'tt-cores4-rank6': [0.0452681107954545, 0.04311277932960928, 0.03906250000000891], 'tt-cores4-rank8': [0.04687500000000031, 0.0403125, 0.039062499999996725]}\nTag: tt-cores4-rank2. No of experiments: 5\nTag: tt-cores4-rank3. No of experiments: 2\nTag: tt-cores4-rank4. No of experiments: 3\nTag: tt-cores4-rank5. No of experiments: 3\nTag: tt-cores4-rank6. No of experiments: 3\nTag: tt-cores4-rank8. No of experiments: 3\n"
]
],
[
[
"# Visualising saved spectograms.",
"_____no_output_____"
]
],
[
[
"frame_fpath = r'D:\\Users\\Charley\\Documents\\Esperanza\\repos\\_experiments\\speech-model-compression\\speaker-verification\\_librispeech_train-clean-100_tisv\\LibriSpeech_train-clean-100_103\\1240_103-1240-0000.npy'\nframe_fpath = r'D:\\Users\\Charley\\Documents\\Esperanza\\repos\\_experiments\\speech-model-compression\\speaker-verification\\_librispeech_train-clean-100_tisv\\LibriSpeech_train-clean-100_1183\\128659_1183-128659-0000.npy'\nframes = np.load(frame_fpath).transpose()\nframes.shape\nplt.stem(y)\nplt.show()\n\n# import librosa\n# import IPython.display as ipd\n# wav_path = '777-126732-0006.wav'\n# y, sr = librosa.load(wav_path)\n# D = librosa.amplitude_to_db(np.abs(librosa.stft(y)), ref=np.max)\n# librosa.display.specshow(D, y_axis='linear')\n# plt.colorbar(format='%+2.0f dB')",
"_____no_output_____"
]
],
[
[
"# Learning curves and Data efficiency",
"_____no_output_____"
]
],
[
[
"from scipy.ndimage import median_filter\n\ndef get_learning_curve(exp_id, metric_name, step_limit=20000):\n myexp = comet_api.get_experiment_by_id(exp_id)\n \n train_eer = []\n train_step = []\n for dic in myexp.get_metrics(metric_name):\n cur_step = int(dic['step'])\n cur_train_eer = float(dic['metricValue'])\n if cur_step > step_limit:\n break\n train_eer.append(cur_train_eer)\n train_step.append(cur_step)\n\n return train_step, train_eer\n\ndef smooth_array(my_array, size=100):\n s_array = median_filter(my_array, size=size)\n return s_array\n \nlstm_train_steps, lstm_train_eer = get_learning_curve(\"445596754f414da8bef0377f7f0906d3\", \"train_EER\")\nlstm_val_steps, lstm_val_eer = get_learning_curve(\"445596754f414da8bef0377f7f0906d3\", \"val_EER\")\nttlstm_train_steps, ttlstm_train_eer = get_learning_curve(\"d6530fa27f11416298efa377b89b22ed\", \"train_EER\")\nttlstm_val_steps, ttlstm_val_eer = get_learning_curve(\"d6530fa27f11416298efa377b89b22ed\", \"val_EER\")\n\n# smooth curves\nlstm_train_eer, lstm_val_eer = smooth_array(lstm_train_eer, 150), smooth_array(lstm_val_eer, 10)\nttlstm_train_eer, ttlstm_val_eer = smooth_array(ttlstm_train_eer, 150), smooth_array(ttlstm_val_eer, 10)\n\n%matplotlib qt\nplt.figure(figsize=(20,6))\n\nplt.subplot(1,2,1)\nplt.plot(np.array(lstm_train_steps)/1000, np.array(lstm_train_eer) * 100, 'm-', label='LSTM (Train)' )\nplt.plot(np.array(lstm_val_steps)/1000, np.array(lstm_val_eer) * 100, 'm--', label='LSTM (Validation)' )\nplt.plot(np.array(ttlstm_train_steps)/1000, np.array(ttlstm_train_eer) * 100, 'b-', label='TT-LSTM (Train)' )\nplt.plot(np.array(ttlstm_val_steps)/1000, np.array(ttlstm_val_eer) * 100, 'b--', label='TT-LSTM (Validation)' )\nplt.ylabel('EER (%)', fontsize=14)\nplt.xlabel('Training steps (K)', fontsize=14)\nplt.xticks(fontsize=14)\nplt.yticks(fontsize=14)\nplt.yscale('log')\nplt.legend(fontsize=14)\n\n# Results of data efficency experiment.\nde_percs = np.array([0.2, 0.4, 0.6, 0.8, 1.0]) * 100\nde_lstm = np.array([0.1289, 0.1027, .0856, .0791, .0733]) * 100\nde_ttlstm = np.array([.1047, .0717, .0504, .0654, .0434]) * 100\n\nplt.subplot(1,2,2)\nplt.plot(de_percs, de_lstm, 'md-', label='LSTM')\nplt.plot(de_percs, de_ttlstm, 'bd-', label='TT-LSTM')\nplt.xlabel('% of training data used', fontsize=14)\nplt.ylabel('Test EER (%)', fontsize=14)\nplt.xticks(fontsize=14)\nplt.yticks(fontsize=14)\nplt.legend(fontsize=14)\nplt.tight_layout(pad=3.0, w_pad=20.0)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Gradients distribution",
"_____no_output_____"
]
],
[
[
"experiments = comet_api.get(workspace=\"tensorized-rnn\", project_name=\"speaker-verification-loggrad\")\nn_steps = 1000\n\ndef fetch_grads(exp_id, metric_name, step_limit=1000):\n myexp = comet_api.get_experiment_by_id(exp_id)\n \n gradients = []\n for dic in myexp.get_metrics(metric_name):\n if len(gradients) >= step_limit:\n break\n gradients.append(float(dic['metricValue']))\n\n return gradients\n\nnott_grads = np.array(fetch_grads(\"3545bf372e404dcf94fc57c9b9df3edf\", \"grad_norm\"))\ntt_grads = np.array(fetch_grads(\"3c9ada04c04c47fb91bd45c8fe7c7112\", \"grad_norm\"))\ntt_grads[tt_grads > 8] = 8\n\n# nottweights = np.ones_like(nott_grads) / len(nott_grads)\n# ttweights = np.ones_like(tt_grads) / len(tt_grads)\n\nfig, ax1 = plt.subplots()\n\ncolor = 'orchid'\nax1.set_xlabel('Gradient Value', fontsize=14)\nax1.set_ylabel('Counts - LSTM', fontsize=14)\nax1.tick_params(axis='y')\nax1.hist(nott_grads, bins=50, alpha=0.7, density=False, color=color, label='LSTM')\nax1.legend(loc='upper left', fontsize=14)\n\nax2 = ax1.twinx()\ncolor = 'tab:blue'\nax2.hist(tt_grads, bins=100, alpha=0.7, density=False, label='TT-LSTM')\nax2.set_ylabel('Counts - TT-LSTM', fontsize=14)\nax2.legend(fontsize=14)\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb84f3a062b2014e9f72ab9464b6b9a25d7a896 | 125,155 | ipynb | Jupyter Notebook | p2_continuous-control/old-attempts/Continuous_Control_workspace.ipynb | EternalSH/deep-reinforcement-learning | fd8cb04f37ae015135cea43aa7bfb17f61e25c35 | [
"MIT"
] | null | null | null | p2_continuous-control/old-attempts/Continuous_Control_workspace.ipynb | EternalSH/deep-reinforcement-learning | fd8cb04f37ae015135cea43aa7bfb17f61e25c35 | [
"MIT"
] | null | null | null | p2_continuous-control/old-attempts/Continuous_Control_workspace.ipynb | EternalSH/deep-reinforcement-learning | fd8cb04f37ae015135cea43aa7bfb17f61e25c35 | [
"MIT"
] | null | null | null | 136.931072 | 31,296 | 0.843346 | [
[
[
"# Continuous Control\n\n---\n\nYou are welcome to use this coding environment to train your agent for the project. Follow the instructions below to get started!\n\n### 1. Start the Environment\n\nRun the next code cell to install a few packages. This line will take a few minutes to run!",
"_____no_output_____"
]
],
[
[
"!pip -q install ./python",
"_____no_output_____"
]
],
[
[
"The environments corresponding to both versions of the environment are already saved in the Workspace and can be accessed at the file paths provided below. \n\nPlease select one of the two options below for loading the environment.",
"_____no_output_____"
]
],
[
[
"from unityagents import UnityEnvironment\nimport numpy as np\n\n# select this option to load version 1 (with a single agent) of the environment\n#env = UnityEnvironment(file_name='/data/Reacher_One_Linux_NoVis/Reacher_One_Linux_NoVis.x86_64')\n\n# select this option to load version 2 (with 20 agents) of the environment\nenv = UnityEnvironment(file_name='/data/Reacher_Linux_NoVis/Reacher.x86_64')",
"INFO:unityagents:\n'Academy' started successfully!\nUnity Academy name: Academy\n Number of Brains: 1\n Number of External Brains : 1\n Lesson number : 0\n Reset Parameters :\n\t\tgoal_speed -> 1.0\n\t\tgoal_size -> 5.0\nUnity brain name: ReacherBrain\n Number of Visual Observations (per agent): 0\n Vector Observation space type: continuous\n Vector Observation space size (per agent): 33\n Number of stacked Vector Observation: 1\n Vector Action space type: continuous\n Vector Action space size (per agent): 4\n Vector Action descriptions: , , , \n"
]
],
[
[
"Environments contain **_brains_** which are responsible for deciding the actions of their associated agents. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.",
"_____no_output_____"
]
],
[
[
"# get the default brain\nbrain_name = env.brain_names[0]\nbrain = env.brains[brain_name]\nprint(brain)",
"Unity brain name: ReacherBrain\n Number of Visual Observations (per agent): 0\n Vector Observation space type: continuous\n Vector Observation space size (per agent): 33\n Number of stacked Vector Observation: 1\n Vector Action space type: continuous\n Vector Action space size (per agent): 4\n Vector Action descriptions: , , , \n"
]
],
[
[
"### 2. Examine the State and Action Spaces\n\nRun the code cell below to print some information about the environment.",
"_____no_output_____"
]
],
[
[
"# reset the environment\nenv_info = env.reset(train_mode=True)[brain_name]\n\n# number of agents\nnum_agents = len(env_info.agents)\nprint('Number of agents:', num_agents)\n\n# size of each action\naction_size = brain.vector_action_space_size\nprint('Size of each action:', action_size)\n\n# examine the state space \nstates = env_info.vector_observations\nstate_size = states.shape[1]\nprint('There are {} agents. Each observes a state with length: {}'.format(states.shape[0], state_size))\nprint('The state for the first agent looks like:', states[0])",
"Number of agents: 20\nSize of each action: 4\nThere are 20 agents. Each observes a state with length: 33\nThe state for the first agent looks like: [ 0.00000000e+00 -4.00000000e+00 0.00000000e+00 1.00000000e+00\n -0.00000000e+00 -0.00000000e+00 -4.37113883e-08 0.00000000e+00\n 0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00\n 0.00000000e+00 0.00000000e+00 -1.00000000e+01 0.00000000e+00\n 1.00000000e+00 -0.00000000e+00 -0.00000000e+00 -4.37113883e-08\n 0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00\n 0.00000000e+00 0.00000000e+00 5.75471878e+00 -1.00000000e+00\n 5.55726624e+00 0.00000000e+00 1.00000000e+00 0.00000000e+00\n -1.68164849e-01]\n"
]
],
[
[
"### 3. Take Random Actions in the Environment\n\nIn the next code cell, you will learn how to use the Python API to control the agent and receive feedback from the environment.\n\nNote that **in this coding environment, you will not be able to watch the agents while they are training**, and you should set `train_mode=True` to restart the environment.",
"_____no_output_____"
]
],
[
[
"#env_info = env.reset(train_mode=True)[brain_name] # reset the environment \n#states = env_info.vector_observations # get the current state (for each agent)\n#scores = np.zeros(num_agents) # initialize the score (for each agent)\n#while True:\n# actions = np.random.randn(num_agents, action_size) # select an action (for each agent)\n# actions = np.clip(actions, -1, 1) # all actions between -1 and 1\n# env_info = env.step(actions)[brain_name] # send all actions to tne environment\n# next_states = env_info.vector_observations # get next state (for each agent)\n# rewards = env_info.rewards # get reward (for each agent)\n# dones = env_info.local_done # see if episode finished\n# scores += env_info.rewards # update the score (for each agent)\n# states = next_states # roll over states to next time step\n# if np.any(dones): # exit loop if episode finished\n# break\n#print('Total score (averaged over agents) this episode: {}'.format(np.mean(scores)))",
"_____no_output_____"
]
],
[
[
"When finished, you can close the environment.",
"_____no_output_____"
],
[
"### 4. It's Your Turn!\n\nNow it's your turn to train your own agent to solve the environment! A few **important notes**:\n- When training the environment, set `train_mode=True`, so that the line for resetting the environment looks like the following:\n```python\nenv_info = env.reset(train_mode=True)[brain_name]\n```\n- To structure your work, you're welcome to work directly in this Jupyter notebook, or you might like to start over with a new file! You can see the list of files in the workspace by clicking on **_Jupyter_** in the top left corner of the notebook.\n- In this coding environment, you will not be able to watch the agents while they are training. However, **_after training the agents_**, you can download the saved model weights to watch the agents on your own machine! ",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport random\nimport copy\nfrom collections import namedtuple, deque\n\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim",
"_____no_output_____"
],
[
"def hidden_init(layer):\n fan_in = layer.weight.data.size()[0]\n lim = 1. / np.sqrt(fan_in)\n return (-lim, lim)",
"_____no_output_____"
],
[
"class Actor(nn.Module):\n \"\"\"Actor (Policy) Model.\"\"\"\n\n def __init__(self, state_size, action_size, seed, fc1_units=400, fc2_units=300):\n \"\"\"Initialize parameters and build model.\n Params\n ======\n state_size (int): Dimension of each state\n action_size (int): Dimension of each action\n seed (int): Random seed\n fc1_units (int): Number of nodes in first hidden layer\n fc2_units (int): Number of nodes in second hidden layer\n \"\"\"\n super(Actor, self).__init__()\n \n #self.normalizer = nn.BatchNorm1d(state_size)\n self.seed = torch.manual_seed(seed)\n \n self.fc1 = nn.Linear(state_size, fc1_units)\n self.fc2 = nn.Linear(fc1_units, fc2_units)\n self.out = nn.Linear(fc2_units, action_size)\n \n self.reset_parameters()\n\n def reset_parameters(self):\n self.fc1.weight.data.uniform_(*hidden_init(self.fc1))\n self.fc2.weight.data.uniform_(*hidden_init(self.fc2))\n self.out.weight.data.uniform_(-3e-3, 3e-3)\n\n def forward(self, states):\n \"\"\"Build an actor (policy) network that maps states -> actions.\"\"\"\n x = states\n #x = self.normalizer(x)\n \n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n \n return torch.tanh(self.out(x))",
"_____no_output_____"
],
[
"class Critic(nn.Module):\n \"\"\"Critic (Value) Model.\"\"\"\n\n def __init__(self, state_size, action_size, seed, dropout=0.1, fcs1_units=400, fc2_units=300):\n \"\"\"Initialize parameters and build model.\n Params\n ======\n state_size (int): Dimension of each state\n action_size (int): Dimension of each action\n seed (int): Random seed\n fcs1_units (int): Number of nodes in the first hidden layer\n fc2_units (int): Number of nodes in the second hidden layer\n \"\"\"\n super(Critic, self).__init__()\n \n #self.normalizer = nn.BatchNorm1d(state_size)\n #self.dropout = nn.Dropout(p=dropout)\n self.seed = torch.manual_seed(seed)\n \n self.fcs1 = nn.Linear(state_size, fcs1_units)\n self.fc2 = nn.Linear(fcs1_units+action_size, fc2_units)\n self.fc3 = nn.Linear(fc2_units, 1)\n \n self.reset_parameters()\n\n def reset_parameters(self):\n self.fcs1.weight.data.uniform_(*hidden_init(self.fcs1))\n self.fc2.weight.data.uniform_(*hidden_init(self.fc2))\n self.fc3.weight.data.uniform_(-3e-3, 3e-3)\n\n def forward(self, states, actions):\n \"\"\"Build a critic (value) network that maps (state, action) pairs -> Q-values.\"\"\"\n xs = states\n #xs = self.normalizer(xs)\n xs = F.relu(self.fcs1(xs))\n \n x = torch.cat((xs, actions), dim=1)\n x = F.relu(self.fc2(x))\n #x = self.dropout(x)\n \n return self.fc3(x)",
"_____no_output_____"
],
[
"class OUNoise:\n \"\"\"Ornstein-Uhlenbeck process.\"\"\"\n\n def __init__(self, size, seed, mu=0., theta=0.15, sigma=0.2):\n \"\"\"Initialize parameters and noise process.\"\"\"\n self.mu = mu * np.ones(size)\n self.theta = theta\n self.sigma = sigma\n self.seed = random.seed(seed)\n self.reset()\n\n def reset(self):\n \"\"\"Reset the internal state (= noise) to mean (mu).\"\"\"\n self.state = copy.copy(self.mu)\n\n def sample(self):\n \"\"\"Update internal state and return it as a noise sample.\"\"\"\n x = self.state\n dx = self.theta * (self.mu - x) + self.sigma * np.array([random.random() for i in range(len(x))])\n self.state = x + dx\n return self.state",
"_____no_output_____"
],
[
"import torch\nimport numpy as np\nfrom collections import deque\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n\nprint(f\"Using device: {device}\")",
"Using device: cuda:0\n"
],
[
"class ReplayBuffer:\n \"\"\"Fixed-size buffer to store experience tuples.\"\"\"\n\n def __init__(self, action_size, buffer_size, batch_size, seed):\n \"\"\"Initialize a ReplayBuffer object.\n Params\n ======\n buffer_size (int): maximum size of buffer\n batch_size (int): size of each training batch\n \"\"\"\n self.action_size = action_size\n self.memory = deque(maxlen=buffer_size) # internal memory (deque)\n self.batch_size = batch_size\n self.experience = namedtuple(\"Experience\", field_names=[\"state\", \"action\", \"reward\", \"next_state\", \"done\"])\n self.seed = random.seed(seed)\n \n def add(self, state, action, reward, next_state, done):\n \"\"\"Add a new experience to memory.\"\"\"\n e = self.experience(state, action, reward, next_state, done)\n self.memory.append(e)\n \n def sample(self):\n \"\"\"Randomly sample a batch of experiences from memory.\"\"\"\n experiences = random.sample(self.memory, k=self.batch_size)\n\n states = torch.from_numpy(np.vstack([e.state for e in experiences if e is not None])).float().to(device)\n actions = torch.from_numpy(np.vstack([e.action for e in experiences if e is not None])).float().to(device)\n rewards = torch.from_numpy(np.vstack([e.reward for e in experiences if e is not None])).float().to(device)\n next_states = torch.from_numpy(np.vstack([e.next_state for e in experiences if e is not None])).float().to(device)\n dones = torch.from_numpy(np.vstack([e.done for e in experiences if e is not None]).astype(np.uint8)).float().to(device)\n\n return (states, actions, rewards, next_states, dones)\n\n def __len__(self):\n \"\"\"Return the current size of internal memory.\"\"\"\n return len(self.memory)\n",
"_____no_output_____"
],
[
"class Agent():\n \"\"\"Interacts with and learns from the environment.\"\"\"\n \n def __init__(self, device, state_size, action_size, random_seed, update_rate):\n \"\"\"Initialize an Agent object.\n \n Params\n ======\n state_size (int): dimension of each state\n action_size (int): dimension of each action\n random_seed (int): random seed\n \"\"\"\n self.state_size = state_size\n self.action_size = action_size\n self.agents_count = 20\n \n self.seed = random.seed(random_seed)\n self.update_rate = update_rate\n \n # Actor Network (w/ Target Network)\n self.actor_local = Actor(state_size, action_size, random_seed).to(device)\n self.actor_target = Actor(state_size, action_size, random_seed).to(device)\n self.actor_optimizer = optim.Adam(self.actor_local.parameters(), lr=LR_ACTOR)\n\n # Critic Network (w/ Target Network)\n self.critic_local = Critic(state_size, action_size, random_seed).to(device)\n self.critic_target = Critic(state_size, action_size, random_seed).to(device)\n self.critic_optimizer = optim.Adam(self.critic_local.parameters(), lr=LR_CRITIC, weight_decay=WEIGHT_DECAY)\n\n # Noise process\n self.noise = OUNoise(action_size, random_seed)\n\n # Replay memory\n self.memory = ReplayBuffer(action_size, BUFFER_SIZE, BATCH_SIZE, random_seed)\n \n def step(self, states, actions, rewards, next_states, dones, t):\n \"\"\"Save experience in replay memory, and use random sample from buffer to learn.\"\"\"\n # Save experience in replay memory\n for state, action, reward, next_state, done in zip(states, actions, rewards, next_states, dones):\n self.memory.add(state, action, reward, next_state, done)\n\n if t % self.update_rate == 0:\n # Learn, if enough samples are available in memory\n if len(self.memory) > BATCH_SIZE:\n experiences = self.memory.sample()\n self.learn(experiences, GAMMA) \n \n def act(self, states, add_noise=True):\n \"\"\"Returns actions for given state as per current policy.\"\"\"\n states = torch.from_numpy(states).float().to(device)\n self.actor_local.eval()\n \n with torch.no_grad():\n actions = self.actor_local(states).cpu().data.numpy()\n \n self.actor_local.train()\n \n if add_noise:\n actions += self.noise.sample()\n \n return np.clip(actions, -1, 1)\n\n def reset(self):\n self.noise.reset()\n\n def learn(self, experiences, gamma):\n \"\"\"Update policy and value parameters using given batch of experience tuples.\n Q_targets = r + γ * critic_target(next_state, actor_target(next_state))\n where:\n actor_target(state) -> action\n critic_target(state, action) -> Q-value\n\n Params\n ======\n experiences (Tuple[torch.Tensor]): tuple of (s, a, r, s', done) tuples \n gamma (float): discount factor\n \"\"\"\n states, actions, rewards, next_states, dones = experiences\n\n # ---------------------------- update critic ---------------------------- #\n # Get predicted next-state actions and Q values from target models\n actions_next = self.actor_target(next_states)\n Q_targets_next = self.critic_target(next_states, actions_next)\n # Compute Q targets for current states (y_i)\n Q_targets = rewards + (gamma * Q_targets_next * (1 - dones))\n # Compute critic loss\n Q_expected = self.critic_local(states, actions)\n critic_loss = F.mse_loss(Q_expected, Q_targets)\n # Minimize the loss\n self.critic_optimizer.zero_grad()\n critic_loss.backward()\n torch.nn.utils.clip_grad_norm_(self.critic_local.parameters(), 1)\n self.critic_optimizer.step()\n\n # ---------------------------- update actor ---------------------------- #\n # Compute actor loss\n actions_pred = self.actor_local(states)\n actor_loss = -self.critic_local(states, actions_pred).mean()\n # Minimize the loss\n self.actor_optimizer.zero_grad()\n actor_loss.backward()\n self.actor_optimizer.step()\n\n # ----------------------- update target networks ----------------------- #\n self.soft_update(self.critic_local, self.critic_target, TAU)\n self.soft_update(self.actor_local, self.actor_target, TAU) \n\n def soft_update(self, local_model, target_model, tau):\n \"\"\"Soft update model parameters.\n θ_target = τ*θ_local + (1 - τ)*θ_target\n\n Params\n ======\n local_model: PyTorch model (weights will be copied from)\n target_model: PyTorch model (weights will be copied to)\n tau (float): interpolation parameter \n \"\"\"\n for target_param, local_param in zip(target_model.parameters(), local_model.parameters()):\n target_param.data.copy_(tau*local_param.data + (1.0-tau)*target_param.data)",
"_____no_output_____"
],
[
"def ddpg(n_episodes, max_t_fn, window_size=100, print_every=10, early_break=False):\n scores_deque = deque(maxlen=window_size)\n scores_short = deque(maxlen=10)\n all_scores = []\n print('Starts training')\n for i_episode in range(1, n_episodes+1):\n agent.reset()\n \n env_info = env.reset(train_mode=True)[brain_name]\n states = env_info.vector_observations\n scores = np.zeros(20)\n print(i_episode)\n max_t = max_t_fn(i_episode)\n print(max_t)\n \n for t in range(max_t):\n actions = agent.act(states)\n env_info = env.step(actions)[brain_name]\n next_states = env_info.vector_observations\n rewards = env_info.rewards\n dones = env_info.local_done\n\n agent.step(states, actions, rewards, next_states, dones, t)\n states = next_states\n scores += rewards\n \n if early_break and np.any(dones):\n print(\"Early break at {}/{}\".format(t, max_t))\n break\n\n mean_score = np.mean(scores)\n scores_deque.append(mean_score)\n scores_short.append(mean_score)\n all_scores.append(mean_score) \n \n if i_episode % print_every == 0: \n print('Episode {}\\tScore: {:.2f}\\tAverage Score 10, 100: {:.2f} {:.2f}'.format(i_episode, mean_score, np.mean(scores_short), np.mean(scores_deque)))\n torch.save(agent.actor_local.state_dict(), 'checkpoint_actor.pth')\n torch.save(agent.critic_local.state_dict(), 'checkpoint_critic.pth')\n \n if np.mean(scores_deque)>=30.0:\n print('\\nEnvironment solved in {:d} episodes!\\tAverage Score: {:.3f}'.format(\n i_episode-100, np.mean(scores_window)))\n torch.save(agent.actor_local.state_dict(), 'checkpoint_actor.pth')\n torch.save(agent.critic_local.state_dict(), 'checkpoint_critic.pth')\n break\n \n return all_scores",
"_____no_output_____"
],
[
"BUFFER_SIZE = int(1e5) # replay buffer size\nBATCH_SIZE = 512 # minibatch size\nGAMMA = 0.999 # discount factor\nTAU = 1e-3 # for soft update of target parameters\nLR_ACTOR = 1e-4 # learning rate of the actor \nLR_CRITIC = 1e-4 # learning rate of the critic\nWEIGHT_DECAY = 0 # L2 weight decay\n\nagent = Agent(state_size=state_size, action_size=action_size, device=device, random_seed=2, update_rate=20)\nscores = ddpg(250, 2500, early_break = False)\n\n# plot the scores\nfig = plt.figure()\nax = fig.add_subplot(111)\nplt.plot(np.arange(len(scores)), scores)\nplt.ylabel('Score')\nplt.xlabel('Episode #')\nplt.show()",
"Starts training\nEpisode 10\tScore: 0.58\tAverage Score 10, 100: 0.42 0.42\nEpisode 20\tScore: 2.18\tAverage Score 10, 100: 1.69 1.05\nEpisode 30\tScore: 2.10\tAverage Score 10, 100: 1.97 1.36\nEpisode 40\tScore: 2.24\tAverage Score 10, 100: 2.06 1.53\nEpisode 50\tScore: 2.23\tAverage Score 10, 100: 2.26 1.68\nEpisode 60\tScore: 2.00\tAverage Score 10, 100: 2.27 1.78\nEpisode 70\tScore: 2.05\tAverage Score 10, 100: 2.29 1.85\nEpisode 80\tScore: 1.75\tAverage Score 10, 100: 2.29 1.91\nEpisode 90\tScore: 1.90\tAverage Score 10, 100: 2.48 1.97\nEpisode 100\tScore: 2.59\tAverage Score 10, 100: 2.85 2.06\nEpisode 110\tScore: 2.84\tAverage Score 10, 100: 2.96 2.31\nEpisode 120\tScore: 4.69\tAverage Score 10, 100: 3.93 2.54\nEpisode 130\tScore: 4.02\tAverage Score 10, 100: 4.18 2.76\nEpisode 140\tScore: 5.12\tAverage Score 10, 100: 4.47 3.00\nEpisode 150\tScore: 5.51\tAverage Score 10, 100: 4.95 3.27\nEpisode 160\tScore: 4.20\tAverage Score 10, 100: 4.68 3.51\nEpisode 170\tScore: 2.79\tAverage Score 10, 100: 3.98 3.68\nEpisode 180\tScore: 1.78\tAverage Score 10, 100: 2.39 3.69\nEpisode 190\tScore: 2.35\tAverage Score 10, 100: 2.25 3.66\nEpisode 200\tScore: 1.03\tAverage Score 10, 100: 1.85 3.56\nEpisode 210\tScore: 2.10\tAverage Score 10, 100: 1.79 3.45\nEpisode 220\tScore: 2.98\tAverage Score 10, 100: 2.98 3.35\nEpisode 230\tScore: 2.69\tAverage Score 10, 100: 2.73 3.21\nEpisode 240\tScore: 2.85\tAverage Score 10, 100: 2.48 3.01\nEpisode 250\tScore: 2.35\tAverage Score 10, 100: 2.29 2.74\n"
],
[
"BUFFER_SIZE = int(1e5) # replay buffer size\nBATCH_SIZE = 1024 # minibatch size\nGAMMA = 0.999 # discount factor\nTAU = 1e-3 # for soft update of target parameters\nLR_ACTOR = 1e-4 # learning rate of the actor \nLR_CRITIC = 1e-4 # learning rate of the critic\nWEIGHT_DECAY = 0 # L2 weight decay\n\nagent = Agent(state_size=state_size, action_size=action_size, device=device, random_seed=2, update_rate=20)\nscores = ddpg(250, 2500, early_break = False)\n\n# plot the scores\nfig = plt.figure()\nax = fig.add_subplot(111)\nplt.plot(np.arange(len(scores)), scores)\nplt.ylabel('Score')\nplt.xlabel('Episode #')\nplt.show()",
"Starts training\nEpisode 10\tScore: 0.69\tAverage Score 10, 100: 0.71 0.71\nEpisode 20\tScore: 1.75\tAverage Score 10, 100: 1.46 1.08\nEpisode 30\tScore: 1.25\tAverage Score 10, 100: 2.00 1.39\nEpisode 40\tScore: 1.48\tAverage Score 10, 100: 1.40 1.39\nEpisode 50\tScore: 2.77\tAverage Score 10, 100: 2.10 1.53\nEpisode 60\tScore: 1.98\tAverage Score 10, 100: 2.31 1.66\nEpisode 70\tScore: 3.05\tAverage Score 10, 100: 2.84 1.83\nEpisode 80\tScore: 3.68\tAverage Score 10, 100: 3.27 2.01\nEpisode 90\tScore: 2.78\tAverage Score 10, 100: 3.30 2.15\nEpisode 100\tScore: 5.93\tAverage Score 10, 100: 3.85 2.32\nEpisode 110\tScore: 3.36\tAverage Score 10, 100: 4.47 2.70\nEpisode 120\tScore: 1.82\tAverage Score 10, 100: 2.66 2.82\nEpisode 130\tScore: 1.69\tAverage Score 10, 100: 1.60 2.78\nEpisode 140\tScore: 1.03\tAverage Score 10, 100: 1.50 2.79\nEpisode 150\tScore: 1.89\tAverage Score 10, 100: 2.20 2.80\nEpisode 160\tScore: 1.95\tAverage Score 10, 100: 2.15 2.78\nEpisode 170\tScore: 3.83\tAverage Score 10, 100: 2.77 2.78\nEpisode 180\tScore: 2.79\tAverage Score 10, 100: 2.88 2.74\nEpisode 190\tScore: 2.54\tAverage Score 10, 100: 2.76 2.68\nEpisode 200\tScore: 2.12\tAverage Score 10, 100: 2.28 2.53\nEpisode 210\tScore: 1.17\tAverage Score 10, 100: 1.60 2.24\nEpisode 220\tScore: 0.62\tAverage Score 10, 100: 1.19 2.09\nEpisode 230\tScore: 0.40\tAverage Score 10, 100: 0.78 2.01\nEpisode 240\tScore: 0.29\tAverage Score 10, 100: 0.57 1.92\nEpisode 250\tScore: 0.84\tAverage Score 10, 100: 0.66 1.76\n"
],
[
"BUFFER_SIZE = int(1e5) # replay buffer size\nBATCH_SIZE = 1024 # minibatch size\nGAMMA = 0.999 # discount factor\nTAU = 1e-4 # for soft update of target parameters\nLR_ACTOR = 1e-4 # learning rate of the actor \nLR_CRITIC = 1e-4 # learning rate of the critic\nWEIGHT_DECAY = 0 # L2 weight decay\n\nagent = Agent(state_size=state_size, action_size=action_size, device=device, random_seed=2, update_rate=50)\nscores = ddpg(250, 2500, early_break = False)\n\n# plot the scores\nfig = plt.figure()\nax = fig.add_subplot(111)\nplt.plot(np.arange(len(scores)), scores)\nplt.ylabel('Score')\nplt.xlabel('Episode #')\nplt.show()",
"Starts training\nEpisode 10\tScore: 0.21\tAverage Score 10, 100: 0.20 0.20\nEpisode 20\tScore: 0.70\tAverage Score 10, 100: 0.99 0.59\nEpisode 30\tScore: 1.03\tAverage Score 10, 100: 1.00 0.73\nEpisode 40\tScore: 0.44\tAverage Score 10, 100: 0.54 0.68\nEpisode 50\tScore: 0.27\tAverage Score 10, 100: 0.47 0.64\nEpisode 60\tScore: 0.13\tAverage Score 10, 100: 0.16 0.56\nEpisode 70\tScore: 1.29\tAverage Score 10, 100: 0.52 0.56\nEpisode 80\tScore: 1.29\tAverage Score 10, 100: 1.03 0.61\nEpisode 90\tScore: 1.49\tAverage Score 10, 100: 1.38 0.70\nEpisode 100\tScore: 1.93\tAverage Score 10, 100: 1.49 0.78\nEpisode 110\tScore: 2.16\tAverage Score 10, 100: 1.86 0.94\nEpisode 120\tScore: 1.55\tAverage Score 10, 100: 1.49 0.99\nEpisode 130\tScore: 1.49\tAverage Score 10, 100: 1.55 1.05\nEpisode 140\tScore: 1.84\tAverage Score 10, 100: 1.70 1.17\nEpisode 150\tScore: 1.67\tAverage Score 10, 100: 1.62 1.28\nEpisode 160\tScore: 1.91\tAverage Score 10, 100: 1.75 1.44\nEpisode 170\tScore: 1.64\tAverage Score 10, 100: 1.80 1.57\nEpisode 180\tScore: 1.88\tAverage Score 10, 100: 1.79 1.64\nEpisode 190\tScore: 1.46\tAverage Score 10, 100: 1.69 1.67\nEpisode 200\tScore: 1.03\tAverage Score 10, 100: 1.31 1.66\nEpisode 210\tScore: 1.36\tAverage Score 10, 100: 1.59 1.63\nEpisode 220\tScore: 1.57\tAverage Score 10, 100: 1.78 1.66\nEpisode 230\tScore: 2.07\tAverage Score 10, 100: 1.71 1.67\nEpisode 240\tScore: 1.71\tAverage Score 10, 100: 1.92 1.70\nEpisode 250\tScore: 1.27\tAverage Score 10, 100: 1.84 1.72\n"
],
[
"BUFFER_SIZE = int(1e8) # replay buffer size\nBATCH_SIZE = 1024 # minibatch size\nGAMMA = 0.999 # discount factor\nTAU = 1e-4 # for soft update of target parameters\nLR_ACTOR = 1e-4 # learning rate of the actor \nLR_CRITIC = 1e-4 # learning rate of the critic\nWEIGHT_DECAY = 0 # L2 weight decay\n\nagent = Agent(state_size=state_size, action_size=action_size, device=device, random_seed=2, update_rate=1)\nscores = ddpg(250, lambda i: min(2500, 1000 + i * 100), early_break = False)\n\n# plot the scores\nfig = plt.figure()\nax = fig.add_subplot(111)\nplt.plot(np.arange(len(scores)), scores)\nplt.ylabel('Score')\nplt.xlabel('Episode #')\nplt.show()",
"Starts training\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb85750cb9c002f4843d4feccea57c7791d2e71 | 5,151 | ipynb | Jupyter Notebook | stimuli/.ipynb_checkpoints/generate_stimuli-checkpoint.ipynb | stefanuddenberg/Sound-Switch | 86296c8bb6e18c79a29f9db61f1b29a492c01216 | [
"MIT"
] | null | null | null | stimuli/.ipynb_checkpoints/generate_stimuli-checkpoint.ipynb | stefanuddenberg/Sound-Switch | 86296c8bb6e18c79a29f9db61f1b29a492c01216 | [
"MIT"
] | null | null | null | stimuli/.ipynb_checkpoints/generate_stimuli-checkpoint.ipynb | stefanuddenberg/Sound-Switch | 86296c8bb6e18c79a29f9db61f1b29a492c01216 | [
"MIT"
] | null | null | null | 29.267045 | 263 | 0.562415 | [
[
[
"# Generate stimuli for Sound-Switch Experiment 1 <a class=\"tocSkip\">\nC and G guitar chords generated [here](https://www.apronus.com/music/onlineguitar.htm) and subsequently recorded and amplified (25.98 db and 25.065 db respectively) using Audacity version 2.3.0 (using Effect --> Amplify) via Windows 10's Stereo Mix drivers.",
"_____no_output_____"
],
[
"# Imports",
"_____no_output_____"
]
],
[
[
"from pydub import AudioSegment\nfrom pydub.generators import Sine\nfrom pydub.playback import play\nimport random\nimport numpy as np\nimport os",
"_____no_output_____"
]
],
[
[
"# Functions",
"_____no_output_____"
]
],
[
[
"def generate_songs(path_prefix):\n for switch_probability in switch_probabilities:\n for exemplar in range(num_exemplars):\n # Begin with silence\n this_song = silence\n # Choose random tone to start with\n which_tone = round(random.random())\n for chunk in range(num_chunks):\n this_probability = random.random()\n\n # Change tones if necessary\n if this_probability < switch_probability:\n which_tone = 1 - which_tone\n\n this_segment = songs[which_tone][:chunk_size]\n\n # Add intervening silence\n this_song = this_song.append(silence, crossfade=crossfade_duration)\n # Add tone\n this_song = this_song.append(this_segment, crossfade=crossfade_duration)\n\n # Add final silence\n this_song.append(silence, crossfade=crossfade_duration)\n song_name = f\"{path_prefix}switch-{str(round(switch_probability,2))}_chunk-{str(chunk_size)}_C_G_alternating_{str(exemplar).zfill(2)}.mp3\"\n this_song.export(song_name, format=\"mp3\", bitrate=\"192k\")",
"_____no_output_____"
]
],
[
[
"# Stimulus Generation",
"_____no_output_____"
],
[
"## Guitar chords",
"_____no_output_____"
]
],
[
[
"songs = [\n AudioSegment.from_mp3(\"guitar_chords/guitar_C.mp3\"),\n AudioSegment.from_mp3(\"guitar_chords/guitar_G.mp3\"),\n]\n\nchunk_size = 500 # in ms\nnum_chunks = 20\ncrossfade_duration = 50 # in ms\nsilence_duration = 100 # in ms\nswitch_probabilities = np.linspace(0.1, 0.9, num=9)\nnum_exemplars = 10\nsilence = AudioSegment.silent(duration=silence_duration)\n# Generate the songs\ngenerate_songs(path_prefix=\"guitar_chords/\")",
"_____no_output_____"
]
],
[
[
"## Tones",
"_____no_output_____"
]
],
[
[
"# Create sine waves of given freqs\nfrequencies = [261.626, 391.995] # C4, G4\nsample_rate = 44100 # sample rate\nbit_depth = 16 # bit depth\n\n# Same params as above for guitar\nchunk_size = 500 # in ms\nnum_chunks = 20\ncrossfade_duration = 50 # in ms\nsilence_duration = 100 # in ms\nswitch_probabilities = np.linspace(0.1, 0.9, num=9)\nnum_exemplars = 10\nsilence = AudioSegment.silent(duration=silence_duration)\n\nsine_waves = []\nsongs = []\nfor i, frequency in enumerate(frequencies):\n sine_waves.append(Sine(frequency, sample_rate=sample_rate, bit_depth=bit_depth))\n #Convert waveform to audio_segment for playback and export\n songs.append(sine_waves[i].to_audio_segment(duration=chunk_size*2)) # just to make sure it's long enough\n\ngenerate_songs(path_prefix=\"pure_tones/\")",
"_____no_output_____"
]
],
[
[
"# Practice Stimulus\nJust choose one of the above stimuli to be a practice stimulus, and remake the stimuli so that it doesn't get repeated.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb86445eae08a97ab6117ae8405d292d95ff507 | 19,457 | ipynb | Jupyter Notebook | wb_hydra/Tables.ipynb | xuwangfmc/AkEL | cceae6c06b9d980aaadc9f387b82f6eb0e99ec7b | [
"BSD-3-Clause"
] | null | null | null | wb_hydra/Tables.ipynb | xuwangfmc/AkEL | cceae6c06b9d980aaadc9f387b82f6eb0e99ec7b | [
"BSD-3-Clause"
] | null | null | null | wb_hydra/Tables.ipynb | xuwangfmc/AkEL | cceae6c06b9d980aaadc9f387b82f6eb0e99ec7b | [
"BSD-3-Clause"
] | null | null | null | 34.869176 | 372 | 0.463895 | [
[
[
"<a href=\"https://colab.research.google.com/github/xuwangfmc/AkEL/blob/master/wb_hydra/Tables.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Tables\n\n该教程主要介绍如何利用Weights&Bias的Tables工具可视化IRIS数据集。\n",
"_____no_output_____"
],
[
"## 安装并导入wandb库",
"_____no_output_____"
]
],
[
[
"# Install Weights & Biases logging library\n!pip install wandb -qqq",
"\u001b[K |████████████████████████████████| 1.7 MB 7.6 MB/s \n\u001b[K |████████████████████████████████| 180 kB 67.6 MB/s \n\u001b[K |████████████████████████████████| 142 kB 75.3 MB/s \n\u001b[K |████████████████████████████████| 97 kB 6.4 MB/s \n\u001b[K |████████████████████████████████| 63 kB 1.5 MB/s \n\u001b[?25h Building wheel for subprocess32 (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for pathtools (setup.py) ... \u001b[?25l\u001b[?25hdone\n"
],
[
"# Import libraries\nimport wandb\nimport numpy as np\nimport pandas as pd\nfrom sklearn.datasets import load_iris",
"_____no_output_____"
]
],
[
[
"## 下载IRIS数据集",
"_____no_output_____"
]
],
[
[
"# Download a simple dataset\niris = load_iris()\n# Load it into a dataframe\niris_dataframe = pd.DataFrame(data=np.c_[iris['data'], iris['target']],\n columns=iris['feature_names'] + ['target'])",
"_____no_output_____"
]
],
[
[
"## 记录IRIS数据集并可视化",
"_____no_output_____"
]
],
[
[
"# Start a W&B run to log data\nwandb.init(project=\"Tables-Quickstart\")\n# Log the dataframe to visualize\nwandb.log({\"iris\": iris_dataframe})\n# Finish the run (useful in notebooks)\nwandb.finish()",
"_____no_output_____"
],
[
"%wandb charlesfrye/Tables-Quickstart -h 1024",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecb86bc0d7cc65fa6340a93f339615f72ff9f5a3 | 55,226 | ipynb | Jupyter Notebook | Chapter03/7 3D Plot.ipynb | anshulbhandari5/Jupyter-for-Data-Science | 7854e14c8b4a4c0e0da5f95a223807a938634fa7 | [
"MIT"
] | 19 | 2017-11-22T15:20:47.000Z | 2021-11-12T11:35:48.000Z | Chapter03/7 3D Plot.ipynb | anshulbhandari5/Jupyter-for-Data-Science | 7854e14c8b4a4c0e0da5f95a223807a938634fa7 | [
"MIT"
] | null | null | null | Chapter03/7 3D Plot.ipynb | anshulbhandari5/Jupyter-for-Data-Science | 7854e14c8b4a4c0e0da5f95a223807a938634fa7 | [
"MIT"
] | 16 | 2017-11-06T06:59:12.000Z | 2021-07-21T04:08:30.000Z | 531.019231 | 52,172 | 0.926792 | [
[
[
"%matplotlib inline\n\nimport pandas as pd\nimport numpy as np\nfrom mpl_toolkits.mplot3d import Axes3D\nimport matplotlib.pyplot as plt\n\ncolumn_names = ['mpg', 'cylinders', 'displacement', 'horsepower', 'weight', 'acceleration', 'year', 'origin', 'name']\ndf = pd.read_table('http://archive.ics.uci.edu/ml/machine-learning-databases/auto-mpg/auto-mpg.data', \\\n sep=r\"\\s+\", index_col=0, header=None, names = column_names)\nprint(df.head())\n\nfig = plt.figure()\nax = fig.add_subplot(111, projection='3d')\n\nxs = []\nys = []\nzs = []\nfor index, row in df.iterrows():\n xs.append(row['weight'])\n ys.append(index) #read_table uses first column as index\n zs.append(row['cylinders'])\n#print(xs)\n#print(ys)\n#print(zs)\nplt.xlim(min(xs), max(xs))\nplt.ylim(min(ys), max(ys))\nax.set_zlim(min(zs), max(zs))\n\nax.scatter(xs, ys, zs)\n\nax.set_xlabel('Weight')\nax.set_ylabel('MPG')\nax.set_zlabel('Cylinders')\n\nplt.show()",
" cylinders displacement horsepower weight acceleration year origin \\\nmpg \n18.0 8 307.0 130.0 3504.0 12.0 70 1 \n15.0 8 350.0 165.0 3693.0 11.5 70 1 \n18.0 8 318.0 150.0 3436.0 11.0 70 1 \n16.0 8 304.0 150.0 3433.0 12.0 70 1 \n17.0 8 302.0 140.0 3449.0 10.5 70 1 \n\n name \nmpg \n18.0 chevrolet chevelle malibu \n15.0 buick skylark 320 \n18.0 plymouth satellite \n16.0 amc rebel sst \n17.0 ford torino \n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
ecb86f521a94a2e0a6cb324d5daec279e2a56f4d | 1,417 | ipynb | Jupyter Notebook | release/songPathRnn/eval/config1/split_pos_neg.ipynb | plataKwon/KPRN | 248133e37b636ddd56e3c4c21a6a8510ab21e912 | [
"MIT"
] | 258 | 2019-01-22T02:58:17.000Z | 2022-03-31T06:41:15.000Z | release/songPathRnn/eval/config1/split_pos_neg.ipynb | jaemyoung/KPRN | 0a56255081ff01aed8de2f40ae6c949d43b702ee | [
"MIT"
] | 10 | 2019-02-28T15:18:52.000Z | 2020-12-17T09:20:44.000Z | release/songPathRnn/eval/config1/split_pos_neg.ipynb | jaemyoung/KPRN | 0a56255081ff01aed8de2f40ae6c949d43b702ee | [
"MIT"
] | 93 | 2019-02-13T01:56:38.000Z | 2022-03-24T03:43:44.000Z | 22.140625 | 75 | 0.52717 | [
[
[
"import codecs",
"_____no_output_____"
],
[
"pos_file_writer = codecs.open(\"pos_res.txt\", mode=\"w\")\nneg_file_writer = codecs.open(\"neg_res.txt\", mode=\"w\")\nfile_reader = codecs.open(\"test_combine_sorted.txt\", mode=\"r\")\nline = file_reader.readline()\nwhile line:\n line_list = line.strip().split(\"\\t\")\n if line_list[-2] == \"1\":\n pos_file_writer.write(line)\n else:\n neg_file_writer.write(line)\n line = file_reader.readline()\npos_file_writer.close()\nneg_file_writer.close()\nfile_reader.close()\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
ecb87256fb590b6e55737215a5c787a7c97465ca | 307,860 | ipynb | Jupyter Notebook | assignment1/svm.ipynb | e-soroush/cs231n | 0c6f18e6031f401e8a5c44f15f7fe2234d339efa | [
"MIT"
] | 2 | 2019-02-05T11:56:04.000Z | 2019-04-28T19:09:19.000Z | assignment1/svm.ipynb | e-soroush/cs231n | 0c6f18e6031f401e8a5c44f15f7fe2234d339efa | [
"MIT"
] | 1 | 2021-04-16T09:52:48.000Z | 2021-04-16T09:52:48.000Z | assignment1/svm.ipynb | e3oroush/cs231n | d3e7eb882f9cec70a6181c1b233d758800a01fa1 | [
"MIT"
] | null | null | null | 396.216216 | 181,132 | 0.930186 | [
[
[
"# Multiclass Support Vector Machine exercise\n\n*Complete and hand in this completed worksheet (including its outputs and any supporting code outside of the worksheet) with your assignment submission. For more details see the [assignments page](http://vision.stanford.edu/teaching/cs231n/assignments.html) on the course website.*\n\nIn this exercise you will:\n \n- implement a fully-vectorized **loss function** for the SVM\n- implement the fully-vectorized expression for its **analytic gradient**\n- **check your implementation** using numerical gradient\n- use a validation set to **tune the learning rate and regularization** strength\n- **optimize** the loss function with **SGD**\n- **visualize** the final learned weights\n",
"_____no_output_____"
]
],
[
[
"# Run some setup code for this notebook.\n\nimport random\nimport numpy as np\nfrom cs231n.data_utils import load_CIFAR10\nimport matplotlib.pyplot as plt\n\nfrom __future__ import print_function\n\n# This is a bit of magic to make matplotlib figures appear inline in the\n# notebook rather than in a new window.\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n# Some more magic so that the notebook will reload external python modules;\n# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"## CIFAR-10 Data Loading and Preprocessing",
"_____no_output_____"
]
],
[
[
"# Load the raw CIFAR-10 data.\ncifar10_dir = 'cs231n/datasets/cifar-10-batches-py'\n\n# Cleaning up variables to prevent loading data multiple times (which may cause memory issue)\ntry:\n del X_train, y_train\n del X_test, y_test\n print('Clear previously loaded data.')\nexcept:\n pass\n\nX_train, y_train, X_test, y_test = load_CIFAR10(cifar10_dir)\n\n# As a sanity check, we print out the size of the training and test data.\nprint('Training data shape: ', X_train.shape)\nprint('Training labels shape: ', y_train.shape)\nprint('Test data shape: ', X_test.shape)\nprint('Test labels shape: ', y_test.shape)",
"Training data shape: (50000, 32, 32, 3)\nTraining labels shape: (50000,)\nTest data shape: (10000, 32, 32, 3)\nTest labels shape: (10000,)\n"
],
[
"# Visualize some examples from the dataset.\n# We show a few examples of training images from each class.\nclasses = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']\nnum_classes = len(classes)\nsamples_per_class = 7\nfor y, cls in enumerate(classes):\n idxs = np.flatnonzero(y_train == y)\n idxs = np.random.choice(idxs, samples_per_class, replace=False)\n for i, idx in enumerate(idxs):\n plt_idx = i * num_classes + y + 1\n plt.subplot(samples_per_class, num_classes, plt_idx)\n plt.imshow(X_train[idx].astype('uint8'))\n plt.axis('off')\n if i == 0:\n plt.title(cls)\nplt.show()",
"_____no_output_____"
],
[
"# Split the data into train, val, and test sets. In addition we will\n# create a small development set as a subset of the training data;\n# we can use this for development so our code runs faster.\nnum_training = 49000\nnum_validation = 1000\nnum_test = 1000\nnum_dev = 500\n\n# Our validation set will be num_validation points from the original\n# training set.\nmask = range(num_training, num_training + num_validation)\nX_val = X_train[mask]\ny_val = y_train[mask]\n\n# Our training set will be the first num_train points from the original\n# training set.\nmask = range(num_training)\nX_train = X_train[mask]\ny_train = y_train[mask]\n\n# We will also make a development set, which is a small subset of\n# the training set.\nmask = np.random.choice(num_training, num_dev, replace=False)\nX_dev = X_train[mask]\ny_dev = y_train[mask]\n\n# We use the first num_test points of the original test set as our\n# test set.\nmask = range(num_test)\nX_test = X_test[mask]\ny_test = y_test[mask]\n\nprint('Train data shape: ', X_train.shape)\nprint('Train labels shape: ', y_train.shape)\nprint('Validation data shape: ', X_val.shape)\nprint('Validation labels shape: ', y_val.shape)\nprint('Test data shape: ', X_test.shape)\nprint('Test labels shape: ', y_test.shape)",
"Train data shape: (49000, 32, 32, 3)\nTrain labels shape: (49000,)\nValidation data shape: (1000, 32, 32, 3)\nValidation labels shape: (1000,)\nTest data shape: (1000, 32, 32, 3)\nTest labels shape: (1000,)\n"
],
[
"# Preprocessing: reshape the image data into rows\nX_train = np.reshape(X_train, (X_train.shape[0], -1))\nX_val = np.reshape(X_val, (X_val.shape[0], -1))\nX_test = np.reshape(X_test, (X_test.shape[0], -1))\nX_dev = np.reshape(X_dev, (X_dev.shape[0], -1))\n\n# As a sanity check, print out the shapes of the data\nprint('Training data shape: ', X_train.shape)\nprint('Validation data shape: ', X_val.shape)\nprint('Test data shape: ', X_test.shape)\nprint('dev data shape: ', X_dev.shape)",
"Training data shape: (49000, 3072)\nValidation data shape: (1000, 3072)\nTest data shape: (1000, 3072)\ndev data shape: (500, 3072)\n"
],
[
"# Preprocessing: subtract the mean image\n# first: compute the image mean based on the training data\nmean_image = np.mean(X_train, axis=0)\nprint(mean_image[:10]) # print a few of the elements\nplt.figure(figsize=(4,4))\nplt.imshow(mean_image.reshape((32,32,3)).astype('uint8')) # visualize the mean image\nplt.show()",
"[130.64189796 135.98173469 132.47391837 130.05569388 135.34804082\n 131.75402041 130.96055102 136.14328571 132.47636735 131.48467347]\n"
],
[
"# second: subtract the mean image from train and test data\nX_train -= mean_image\nX_val -= mean_image\nX_test -= mean_image\nX_dev -= mean_image",
"_____no_output_____"
],
[
"# third: append the bias dimension of ones (i.e. bias trick) so that our SVM\n# only has to worry about optimizing a single weight matrix W.\nX_train = np.hstack([X_train, np.ones((X_train.shape[0], 1))])\nX_val = np.hstack([X_val, np.ones((X_val.shape[0], 1))])\nX_test = np.hstack([X_test, np.ones((X_test.shape[0], 1))])\nX_dev = np.hstack([X_dev, np.ones((X_dev.shape[0], 1))])\n\nprint(X_train.shape, X_val.shape, X_test.shape, X_dev.shape)",
"(49000, 3073) (1000, 3073) (1000, 3073) (500, 3073)\n"
]
],
[
[
"## SVM Classifier\n\nYour code for this section will all be written inside **cs231n/classifiers/linear_svm.py**. \n\nAs you can see, we have prefilled the function `compute_loss_naive` which uses for loops to evaluate the multiclass SVM loss function. ",
"_____no_output_____"
]
],
[
[
"# Evaluate the naive implementation of the loss we provided for you:\nfrom cs231n.classifiers.linear_svm import svm_loss_naive\nimport time\n\n# generate a random SVM weight matrix of small numbers\nW = np.random.randn(3073, 10) * 0.0001 \n\nloss, grad = svm_loss_naive(W, X_dev, y_dev, 0.000005)\nprint('loss: %f' % (loss, ))",
"loss: 9.531909\n"
]
],
[
[
"The `grad` returned from the function above is right now all zero. Derive and implement the gradient for the SVM cost function and implement it inline inside the function `svm_loss_naive`. You will find it helpful to interleave your new code inside the existing function.\n\nTo check that you have correctly implemented the gradient correctly, you can numerically estimate the gradient of the loss function and compare the numeric estimate to the gradient that you computed. We have provided code that does this for you:",
"_____no_output_____"
]
],
[
[
"# Once you've implemented the gradient, recompute it with the code below\n# and gradient check it with the function we provided for you\n\n# Compute the loss and its gradient at W.\nloss, grad = svm_loss_naive(W, X_dev, y_dev, 0.0)\n\n# Numerically compute the gradient along several randomly chosen dimensions, and\n# compare them with your analytically computed gradient. The numbers should match\n# almost exactly along all dimensions.\nfrom cs231n.gradient_check import grad_check_sparse\nf = lambda w: svm_loss_naive(w, X_dev, y_dev, 0.0)[0]\ngrad_numerical = grad_check_sparse(f, W, grad)\n\n# do the gradient check once again with regularization turned on\n# you didn't forget the regularization gradient did you?\nloss, grad = svm_loss_naive(W, X_dev, y_dev, 5e1)\nf = lambda w: svm_loss_naive(w, X_dev, y_dev, 5e1)[0]\ngrad_numerical = grad_check_sparse(f, W, grad)",
"numerical: -15.559833 analytic: -15.559833, relative error: 2.008640e-12\nnumerical: 9.770812 analytic: 9.770812, relative error: 4.650541e-11\nnumerical: 22.314362 analytic: 22.314362, relative error: 6.529840e-12\nnumerical: 11.920512 analytic: 11.920512, relative error: 2.896529e-11\nnumerical: -12.288627 analytic: -12.288627, relative error: 1.774560e-11\nnumerical: 0.132092 analytic: 0.132092, relative error: 5.107410e-09\nnumerical: -4.391256 analytic: -4.391256, relative error: 1.180881e-10\nnumerical: -21.462669 analytic: -21.462669, relative error: 1.068976e-11\nnumerical: -35.229288 analytic: -35.229288, relative error: 4.005279e-12\nnumerical: -22.592814 analytic: -22.592814, relative error: 8.214018e-12\nnumerical: 3.238215 analytic: 3.238215, relative error: 5.796913e-12\nnumerical: 8.992705 analytic: 8.992705, relative error: 7.641765e-12\nnumerical: 4.396499 analytic: 4.396499, relative error: 3.526007e-11\nnumerical: -14.701621 analytic: -14.701621, relative error: 8.122674e-12\nnumerical: 8.288087 analytic: 8.288087, relative error: 4.498681e-11\nnumerical: 8.743911 analytic: 8.743911, relative error: 6.328061e-11\nnumerical: 27.497285 analytic: 27.497285, relative error: 1.841670e-11\nnumerical: 32.922066 analytic: 32.922066, relative error: 1.032834e-12\nnumerical: 14.873899 analytic: 14.873899, relative error: 6.536104e-12\nnumerical: 0.728542 analytic: 0.728542, relative error: 4.899022e-11\n"
]
],
[
[
"### Inline Question 1:\nIt is possible that once in a while a dimension in the gradcheck will not match exactly. What could such a discrepancy be caused by? Is it a reason for concern? What is a simple example in one dimension where a gradient check could fail? How would change the margin affect of the frequency of this happening? *Hint: the SVM loss function is not strictly speaking differentiable*\n\n**Your Answer:** *As in the hint has been said, hinge loss function has a `max` operator that in the vicinity of sink point, where x=0. The value of the analytic gradient could either be zero or one depending on which side you approximate; However, in the numerical gradient, if `h` wasn't selected small enough, the result might differ from analytical.*",
"_____no_output_____"
]
],
[
[
"# Next implement the function svm_loss_vectorized; for now only compute the loss;\n# we will implement the gradient in a moment.\ntic = time.time()\nloss_naive, grad_naive = svm_loss_naive(W, X_dev, y_dev, 0.000005)\ntoc = time.time()\nprint('Naive loss: %e computed in %fs' % (loss_naive, toc - tic))\n\nfrom cs231n.classifiers.linear_svm import svm_loss_vectorized\ntic = time.time()\nloss_vectorized, _ = svm_loss_vectorized(W, X_dev, y_dev, 0.000005)\ntoc = time.time()\nprint('Vectorized loss: %e computed in %fs' % (loss_vectorized, toc - tic))\n\n# The losses should match but your vectorized implementation should be much faster.\nprint('difference: %f' % (loss_naive - loss_vectorized))",
"Naive loss: 9.531909e+00 computed in 0.084203s\nVectorized loss: 9.531909e+00 computed in 0.002337s\ndifference: -0.000000\n"
],
[
"# Complete the implementation of svm_loss_vectorized, and compute the gradient\n# of the loss function in a vectorized way.\n\n# The naive implementation and the vectorized implementation should match, but\n# the vectorized version should still be much faster.\ntic = time.time()\n_, grad_naive = svm_loss_naive(W, X_dev, y_dev, 0.000005)\ntoc = time.time()\nprint('Naive loss and gradient: computed in %fs' % (toc - tic))\n\ntic = time.time()\n_, grad_vectorized = svm_loss_vectorized(W, X_dev, y_dev, 0.000005)\ntoc = time.time()\nprint('Vectorized loss and gradient: computed in %fs' % (toc - tic))\n\n# The loss is a single number, so it is easy to compare the values computed\n# by the two implementations. The gradient on the other hand is a matrix, so\n# we use the Frobenius norm to compare them.\ndifference = np.linalg.norm(grad_naive - grad_vectorized, ord='fro')\nprint('difference: %f' % difference)",
"Naive loss and gradient: computed in 0.091766s\nVectorized loss and gradient: computed in 0.002203s\ndifference: 0.000000\n"
]
],
[
[
"### Stochastic Gradient Descent\n\nWe now have vectorized and efficient expressions for the loss, the gradient and our gradient matches the numerical gradient. We are therefore ready to do SGD to minimize the loss.",
"_____no_output_____"
]
],
[
[
"# In the file linear_classifier.py, implement SGD in the function\n# LinearClassifier.train() and then run it with the code below.\nfrom cs231n.classifiers import LinearSVM\nsvm = LinearSVM()\ntic = time.time()\nloss_hist = svm.train(X_train, y_train, learning_rate=1e-7, reg=2.5e4,\n num_iters=1500, verbose=True)\ntoc = time.time()\nprint('That took %fs' % (toc - tic))",
"iteration 0 / 1500: loss 801.095113\niteration 100 / 1500: loss 290.180849\niteration 200 / 1500: loss 109.189572\niteration 300 / 1500: loss 42.841464\niteration 400 / 1500: loss 18.706714\niteration 500 / 1500: loss 10.540675\niteration 600 / 1500: loss 7.040347\niteration 700 / 1500: loss 5.610696\niteration 800 / 1500: loss 6.104535\niteration 900 / 1500: loss 5.497837\niteration 1000 / 1500: loss 5.449646\niteration 1100 / 1500: loss 5.566636\niteration 1200 / 1500: loss 5.454966\niteration 1300 / 1500: loss 4.900481\niteration 1400 / 1500: loss 5.134418\nThat took 2.497204s\n"
],
[
"# A useful debugging strategy is to plot the loss as a function of\n# iteration number:\nplt.plot(loss_hist)\nplt.xlabel('Iteration number')\nplt.ylabel('Loss value')\nplt.show()",
"_____no_output_____"
],
[
"# Write the LinearSVM.predict function and evaluate the performance on both the\n# training and validation set\ny_train_pred = svm.predict(X_train)\nprint('training accuracy: %f' % (np.mean(y_train == y_train_pred), ))\ny_val_pred = svm.predict(X_val)\nprint('validation accuracy: %f' % (np.mean(y_val == y_val_pred), ))",
"training accuracy: 0.367000\nvalidation accuracy: 0.373000\n"
],
[
"# Use the validation set to tune hyperparameters (regularization strength and\n# learning rate). You should experiment with different ranges for the learning\n# rates and regularization strengths; if you are careful you should be able to\n# get a classification accuracy of about 0.4 on the validation set.\nlearning_rates = [1e-7, 2e-7, 3e-7]\nregularization_strengths = [2.5e4, 1.5e4, 2e4, 1e4]\n\n# results is dictionary mapping tuples of the form\n# (learning_rate, regularization_strength) to tuples of the form\n# (training_accuracy, validation_accuracy). The accuracy is simply the fraction\n# of data points that are correctly classified.\nresults = {}\nbest_val = -1 # The highest validation accuracy that we have seen so far.\nbest_svm = None # The LinearSVM object that achieved the highest validation rate.\n\n################################################################################\n# TODO: #\n# Write code that chooses the best hyperparameters by tuning on the validation #\n# set. For each combination of hyperparameters, train a linear SVM on the #\n# training set, compute its accuracy on the training and validation sets, and #\n# store these numbers in the results dictionary. In addition, store the best #\n# validation accuracy in best_val and the LinearSVM object that achieves this #\n# accuracy in best_svm. #\n# #\n# Hint: You should use a small value for num_iters as you develop your #\n# validation code so that the SVMs don't take much time to train; once you are #\n# confident that your validation code works, you should rerun the validation #\n# code with a larger value for num_iters. #\n################################################################################\nnum_iter=1500\nfor lr in learning_rates:\n for reg in regularization_strengths:\n svm = LinearSVM()\n svm.train(X_train, y_train, learning_rate=lr, reg=reg,num_iters=num_iter, verbose=False)\n y_train_pred = svm.predict(X_train)\n train_acc=np.mean(y_train == y_train_pred)\n y_val_pred = svm.predict(X_val)\n val_acc=np.mean(y_val == y_val_pred)\n results[(lr,reg)]=(train_acc,val_acc)\n if val_acc > best_val:\n best_val=val_acc\n best_svm=svm\n \n \n################################################################################\n# END OF YOUR CODE #\n################################################################################\n \n# Print out results.\nfor lr, reg in sorted(results):\n train_accuracy, val_accuracy = results[(lr, reg)]\n print('lr %e reg %e train accuracy: %f val accuracy: %f' % (\n lr, reg, train_accuracy, val_accuracy))\n \nprint('best validation accuracy achieved during cross-validation: %f' % best_val)",
"lr 1.000000e-07 reg 1.000000e+04 train accuracy: 0.379796 val accuracy: 0.391000\nlr 1.000000e-07 reg 1.500000e+04 train accuracy: 0.381020 val accuracy: 0.387000\nlr 1.000000e-07 reg 2.000000e+04 train accuracy: 0.369694 val accuracy: 0.388000\nlr 1.000000e-07 reg 2.500000e+04 train accuracy: 0.371041 val accuracy: 0.384000\nlr 2.000000e-07 reg 1.000000e+04 train accuracy: 0.376653 val accuracy: 0.401000\nlr 2.000000e-07 reg 1.500000e+04 train accuracy: 0.367776 val accuracy: 0.370000\nlr 2.000000e-07 reg 2.000000e+04 train accuracy: 0.360653 val accuracy: 0.379000\nlr 2.000000e-07 reg 2.500000e+04 train accuracy: 0.359612 val accuracy: 0.371000\nlr 3.000000e-07 reg 1.000000e+04 train accuracy: 0.364306 val accuracy: 0.370000\nlr 3.000000e-07 reg 1.500000e+04 train accuracy: 0.349592 val accuracy: 0.376000\nlr 3.000000e-07 reg 2.000000e+04 train accuracy: 0.342776 val accuracy: 0.338000\nlr 3.000000e-07 reg 2.500000e+04 train accuracy: 0.357551 val accuracy: 0.369000\nbest validation accuracy achieved during cross-validation: 0.401000\n"
],
[
"# Visualize the cross-validation results\nimport math\nx_scatter = [math.log10(x[0]) for x in results]\ny_scatter = [math.log10(x[1]) for x in results]\n\n# plot training accuracy\nmarker_size = 100\ncolors = [results[x][0] for x in results]\nplt.subplot(2, 1, 1)\nplt.scatter(x_scatter, y_scatter, marker_size, c=colors)\nplt.colorbar()\nplt.xlabel('log learning rate')\nplt.ylabel('log regularization strength')\nplt.title('CIFAR-10 training accuracy')\n\n# plot validation accuracy\ncolors = [results[x][1] for x in results] # default size of markers is 20\nplt.subplot(2, 1, 2)\nplt.scatter(x_scatter, y_scatter, marker_size, c=colors)\nplt.colorbar()\nplt.xlabel('log learning rate')\nplt.ylabel('log regularization strength')\nplt.title('CIFAR-10 validation accuracy')\nplt.show()",
"_____no_output_____"
],
[
"# Evaluate the best svm on test set\ny_test_pred = best_svm.predict(X_test)\ntest_accuracy = np.mean(y_test == y_test_pred)\nprint('linear SVM on raw pixels final test set accuracy: %f' % test_accuracy)",
"linear SVM on raw pixels final test set accuracy: 0.372000\n"
],
[
"# Visualize the learned weights for each class.\n# Depending on your choice of learning rate and regularization strength, these may\n# or may not be nice to look at.\nw = best_svm.W[:-1,:] # strip out the bias\nw = w.reshape(32, 32, 3, 10)\nw_min, w_max = np.min(w), np.max(w)\nclasses = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']\nfor i in range(10):\n plt.subplot(2, 5, i + 1)\n \n # Rescale the weights to be between 0 and 255\n wimg = 255.0 * (w[:, :, :, i].squeeze() - w_min) / (w_max - w_min)\n plt.imshow(wimg.astype('uint8'))\n plt.axis('off')\n plt.title(classes[i])",
"_____no_output_____"
]
],
[
[
"### Inline question 2:\nDescribe what your visualized SVM weights look like, and offer a brief explanation for why they look they way that they do.\n\n**Your answer:** *These are learned template for each class by our linear SVM classifier. The car is a good representation of the red car with a blue window. Deer also looks like a brown deer located in the middle of a green woods. Horse, ship, and slightly ship also seem reasonable templates. However, other templates are not very good representation for their respective class*",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecb87389f4b8510019c2762b54e0f867a18ddd05 | 723 | ipynb | Jupyter Notebook | Pytorch/Pytorch_learn_by_dragen1860/lesson09-Broadcast广播机制.ipynb | tsuirak/skills | 22280be0870627c5dd84e069ec271aeeb6797831 | [
"MIT"
] | 362 | 2020-10-08T07:34:25.000Z | 2022-03-30T05:11:30.000Z | Pytorch/Pytorch_learn_by_dragen1860/lesson09-Broadcast广播机制.ipynb | tsuirak/skills | 22280be0870627c5dd84e069ec271aeeb6797831 | [
"MIT"
] | null | null | null | Pytorch/Pytorch_learn_by_dragen1860/lesson09-Broadcast广播机制.ipynb | tsuirak/skills | 22280be0870627c5dd84e069ec271aeeb6797831 | [
"MIT"
] | 238 | 2020-10-08T12:01:31.000Z | 2022-03-25T08:10:42.000Z | 16.431818 | 44 | 0.495159 | [
[
[
"# 参考PDF课件~",
"_____no_output_____"
],
[
"### $Broadcasting$\n\n- $Expand$\n\n- $without\\space copying\\space data$",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown"
]
] |
ecb8897cd799f0f34052b1e92562bb7429676845 | 5,625 | ipynb | Jupyter Notebook | examples/gallery/param/precedence.ipynb | andriyor/panel | e1d76f415df02ca90f54d42ebcaf42a1bcc65560 | [
"BSD-3-Clause"
] | 1,130 | 2019-11-23T09:53:37.000Z | 2022-03-31T11:30:07.000Z | examples/gallery/param/precedence.ipynb | andriyor/panel | e1d76f415df02ca90f54d42ebcaf42a1bcc65560 | [
"BSD-3-Clause"
] | 2,265 | 2019-11-20T17:09:09.000Z | 2022-03-31T22:09:38.000Z | examples/gallery/param/precedence.ipynb | datalayer-externals/holoviz-panel | 5e25cb09447d8edf0b316f130ee1318a2aeb880f | [
"BSD-3-Clause"
] | 215 | 2019-11-26T11:49:04.000Z | 2022-03-30T10:23:11.000Z | 40.178571 | 433 | 0.560711 | [
[
[
"import panel as pn\nimport param\n\npn.extension()",
"_____no_output_____"
]
],
[
[
"This example demonstrates how to order and hide widgets by means of the ``precedence`` and ``display_threshold`` attributes.\n\nEach ``Parameter`` object has a ``precedence`` attribute that is defined as follows in the documentation of ``param``:\n\n> ``precedence`` is a value, usually in the range 0.0 to 1.0, that allows the order of Parameters in a class to be defined (for e.g. in GUI menus). \n> A negative precedence indicates a parameter that should be hidden in e.g. GUI menus.\n\nA `Param` pane has a ``display_threshold`` attribute defaulting to 0 and defined as follows:\n\n> Parameters with precedence below this value are not displayed.\n\nThe interactive example below helps to understand how the interplay between these two attributes affects the display of widgets.\n\nThe ``PrecedenceTutorial`` class emulates a dummy app whose display we want to control and that consists of three input parameters, ``x``, ``y`` and ``z``. These parameters will be displayed by `panel` with their associated default widgets. Additionally, the class declares the four parameters that will control the dummy app display: ``x_precedence``, ``y_precedence`` and ``z_precedence`` and ``dummy_app_display_threshold``.",
"_____no_output_____"
]
],
[
[
"class Precedence(param.Parameterized):\n\n # Parameters of the dummy app.\n x = param.Number(precedence=-1)\n y = param.Boolean(precedence=3)\n z = param.String(precedence=2)\n\n # Parameters of the control app.\n x_precedence = param.Number(default=x.precedence, bounds=(-10, 10), step=1)\n y_precedence = param.Number(default=y.precedence, bounds=(-10, 10), step=1)\n z_precedence = param.Number(default=z.precedence, bounds=(-10, 10), step=1)\n dummy_app_display_threshold = param.Number(default=1, bounds=(-10, 10), step=1)\n \n def __init__(self):\n super().__init__()\n # Building the dummy app as a Param pane in here so that its ``display_threshold``\n # parameter can be accessed and linked via @param.depends(...).\n self.dummy_app = pn.Param(\n self.param,\n parameters=[\"x\", \"y\", \"z\"],\n widgets={\n \"x\": {\"background\": \"#fac400\"},\n \"y\": {\"background\": \"#07d900\"},\n \"z\": {\"background\": \"#00c0d9\"},\n },\n show_name=False\n )\n\n # Linking the two apps here.\n @param.depends(\"dummy_app_display_threshold\", \"x_precedence\", \"y_precedence\", \"z_precedence\", watch=True)\n def update_precedences_and_threshold(self):\n self.param.x.precedence = self.x_precedence\n self.param.y.precedence = self.y_precedence \n self.param.z.precedence = self.z_precedence \n self.dummy_app.display_threshold = self.dummy_app_display_threshold\n\nprecedence_model = Precedence()",
"_____no_output_____"
],
[
"# Bulding the control app as a Param pane too.\ncontrol_app = pn.Param(\n precedence_model.param,\n parameters=[\"x_precedence\", \"y_precedence\", \"z_precedence\", \"dummy_app_display_threshold\"],\n widgets={\n \"x_precedence\": {\"background\": \"#fac400\"},\n \"y_precedence\": {\"background\": \"#07d900\"},\n \"z_precedence\": {\"background\": \"#00c0d9\"},\n },\n show_name=False\n)",
"_____no_output_____"
],
[
"# Building the complete interactive example.\ninteractive_precedence_app = pn.Column(\n \"## Precedence Example\",\n \"Moving the sliders of the control app should update the display of the dummy app.\",\n pn.Row(\n pn.Column(\"**Control app**\", control_app),\n pn.Column(\"**Dummy app**\", precedence_model.dummy_app)\n )\n)\ninteractive_precedence_app",
"_____no_output_____"
],
[
"pn.template.FastListTemplate(site=\"Panel\", title=\"Parameter Precedence\", main_max_width=\"700px\",\n main=[\n pn.pane.Markdown(\"This example demonstrates how to order and hide widgets by means of the **``precedence`` and ``display_threshold``** parameter attributes.\", sizing_mode=\"stretch_width\"),\n interactive_precedence_app,\n ]).servable();",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecb890eac2820b3ce73dd8d14f060fd4a164c2ba | 6,617 | ipynb | Jupyter Notebook | aStar.ipynb | variousideas/tis | 9e44b58cb47ee0503caf1bedaee714062164abee | [
"MIT"
] | null | null | null | aStar.ipynb | variousideas/tis | 9e44b58cb47ee0503caf1bedaee714062164abee | [
"MIT"
] | null | null | null | aStar.ipynb | variousideas/tis | 9e44b58cb47ee0503caf1bedaee714062164abee | [
"MIT"
] | null | null | null | 25.45 | 112 | 0.445519 | [
[
[
"g는 first와 맨하튼\nh는 last와 맨하튼\n\n내 주변 4방향을 검사.\n- 벽, 밖 제외\n- close_list면 제외\n- open_list면 제외(?)\n- 남은 지점들을 열린 리스트에 추가\n\n열린 리스트에서\n- 가장 작은 F중 가장 앞에 있는 것은 닫힌 리스트로 넘기고 제거.\n\n닫힌 리스트에서\n- 가장 마지막 location을 지금 위치로.\n\n닫힌 리스트에서 도착점에 도달했다?\n종료.",
"_____no_output_____"
],
[
"maze = [[0, 0, 0, 0, -1, 0, 0, 0, 0, 0],\n [0, 0, 0, 0, -1, 0, 0, 0, 0, 0],\n [0, 0, 0, 0, -1, 0, 0, 0, 0, 0],\n [0, 0, 0, 0, -1, 0, 0, 0, 0, 0],\n [0, 0, 0, 0, -1, 0, 0, 0, 0, 0],\n [0, 0, 0, 0, 0, 0, 0, 0, 0, 0],\n [0, 0, 0, 0, -1, 0, 0, 0, 0, 0],\n [0, 0, 0, 0, -1, 0, 0, 0, 0, 0],\n [0, 0, 0, 0, -1, 0, 0, 0, 0, 0],\n [0, 0, 0, 0, 0, 0, 0, 0, 0, 0]]",
"_____no_output_____"
],
[
"class Path():\n def __init__(self):\n self.open_list = []\n self.open_list_info = []\n self.close_list = []\n self.close_list_info = []\n self.maze = []\n \n # 맨하튼 거리\n def distance(self, a, b):\n return (abs(a[0]-b[0])+abs(a[1]-b[1]))\n \n # 점의 유효성\n def is_valid(self, point):\n if (-1 < point[0] < len(self.maze[0])) and (-1 < point[1] < len(self.maze)):\n if self.maze[point[1]][point[0]] != -1:\n if point in self.open_list:\n return 0\n if point in self.close_list:\n return 0\n return 1\n return 0\n \n # 유효한 주변을 리스트로\n def list_around(self, point):\n tmp = [(point[0]-1,point[1]),(point[0]+1,point[1]),(point[0],point[1]-1),(point[0],point[1]+1)]\n result = []\n for i in tmp:\n if self.is_valid(i):\n result.append(i)\n return result",
"_____no_output_____"
],
[
"# 최초\nperfect = Path()\nperfect.maze = maze\nstart = (0,0)\nend = (9,0)\npoint = start",
"_____no_output_____"
],
[
"while point != end:\n # 경로\n list_valid = perfect.list_around(point)\n perfect.open_list += list_valid\n for i in list_valid:\n g = perfect.distance(start,i)\n h = perfect.distance(end,i)\n f = g + h\n perfect.open_list_info.append([i,g,h,f,point])\n\n # close_list에 삽입, open_list에 제거\n min_index = []\n for i in perfect.open_list_info:\n min_index.append(i[3])\n\n min_value = min(min_index)\n min_index = min_index.index(min_value)\n\n perfect.close_list.append(perfect.open_list[min_index])\n perfect.close_list_info.append(perfect.open_list_info[min_index])\n del perfect.open_list[min_index]\n del perfect.open_list_info[min_index]\n \n point = perfect.close_list[-1]",
"_____no_output_____"
],
[
"# 추출\nlast = len(perfect.close_list_info) - 1\nresult = []\nwhile perfect.close_list[last] != start:\n result.append(perfect.close_list[last])\n tmp = perfect.close_list_info[last][4]\n last = perfect.close_list.index(tmp)\nresult.append(perfect.close_list[last])\nresult.reverse()",
"_____no_output_____"
],
[
"copy = maze\nfor i in result:\n copy[i[1]][i[0]] = 2",
"_____no_output_____"
],
[
"maze",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb8a9141caa185423254c19206c3d2c5ce59e9d | 4,374 | ipynb | Jupyter Notebook | tutorials/W0D0_NeuroVideoSeries/W0D0_Tutorial10.ipynb | janeite/course-content | 2a3ba168c5bfb5fd5e8305fe3ae79465b0add52c | [
"CC-BY-4.0",
"BSD-3-Clause"
] | 2,294 | 2020-05-11T12:05:35.000Z | 2022-03-28T21:23:34.000Z | tutorials/W0D0_NeuroVideoSeries/W0D0_Tutorial10.ipynb | janeite/course-content | 2a3ba168c5bfb5fd5e8305fe3ae79465b0add52c | [
"CC-BY-4.0",
"BSD-3-Clause"
] | 629 | 2020-05-11T15:42:26.000Z | 2022-03-29T12:23:35.000Z | tutorials/W0D0_NeuroVideoSeries/W0D0_Tutorial10.ipynb | janeite/course-content | 2a3ba168c5bfb5fd5e8305fe3ae79465b0add52c | [
"CC-BY-4.0",
"BSD-3-Clause"
] | 917 | 2020-05-11T12:47:53.000Z | 2022-03-31T12:14:41.000Z | 26.834356 | 274 | 0.561728 | [
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W0D0_NeuroVideoSeries/W0D0_Tutorial10.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Stimulus Representation\n**Neuro Video Series**\n\n**By Neuromatch Academy**\n\n**Content creator**: Jens Kremkow\n\n**Content reviewers**: Jiaxin Tu, Tara van Viegen, Pooya Pakarian\n\n**Video editors, captioners, translators**: Maryam Ansari, Antony Puthussery, Tara van Viegen",
"_____no_output_____"
],
[
"**Our 2021 Sponsors, including Presenting Sponsor Facebook Reality Labs**\n\n<p align='center'><img src='https://github.com/NeuromatchAcademy/widgets/blob/master/sponsors.png?raw=True'/></p>",
"_____no_output_____"
],
[
"## Video",
"_____no_output_____"
]
],
[
[
"# @markdown\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=\"BV1d64y1t7mP\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=\"-BUM4JcT9WA\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"## Slides",
"_____no_output_____"
]
],
[
[
"# @markdown\nfrom IPython.display import IFrame\nurl = \"https://mfr.ca-1.osf.io/render?url=https://osf.io/45yju/?direct%26mode=render%26action=download%26mode=render\"\nIFrame(src=url, width=854, height=480)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb8b1f2d5aed62d0cc7e778fa3cc344b2903168 | 2,916 | ipynb | Jupyter Notebook | Untitled0.ipynb | ayberkkaraarslan/hu-bby162-2021 | a1cc8424dc487ceb27bb19fc45a0416df43c60d9 | [
"MIT"
] | null | null | null | Untitled0.ipynb | ayberkkaraarslan/hu-bby162-2021 | a1cc8424dc487ceb27bb19fc45a0416df43c60d9 | [
"MIT"
] | null | null | null | Untitled0.ipynb | ayberkkaraarslan/hu-bby162-2021 | a1cc8424dc487ceb27bb19fc45a0416df43c60d9 | [
"MIT"
] | null | null | null | 25.578947 | 237 | 0.441015 | [
[
[
"<a href=\"https://colab.research.google.com/github/ayberkkaraarslan/hu-bby162-2021/blob/main/Untitled0.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"#dosya okuma\n#Google Drive Bağlantısı\n'''\nfrom google.colab import drive\ndrive.mount('/gdrive')\n'''\n\ndosya =\"/content/drive/MyDrive/Adress.txt\"\n\nf = open (dosya, \"r\") \n\nfor line in f.readlines():\n print(line)\n\nf.close()\n",
"Ad, Soyad: Mustafa Ayberk Karaarslan\n\nE-mail: [email protected]\n"
],
[
"#dosya yazma\n\ndosya = \"/content/drive/MyDrive/adres.txt\"\n\nf = open(dosya, \"r\")\n\nf = open( dosya, 'w')\nf.write (\"Mustafa Ayberk\\n\")\nf.write (\"Karaarslan\\n\")\nf.write (\"[email protected]\")\nf.close()\n\nf = open(dosya, \"r\")\nfor line in f.readlines():\n print(line)\n\nf.close() ",
"Mustafa Ayberk\n\nKaraarslan\n\[email protected]\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
]
] |
ecb8ba8779be44b1371926062b178aed7542e046 | 51,331 | ipynb | Jupyter Notebook | docs/qcfractal/source/quickstart.ipynb | radical-cybertools/QCFractal | 676997ef4bc267ae9adfe0064843c42a5701dcf4 | [
"BSD-3-Clause"
] | null | null | null | docs/qcfractal/source/quickstart.ipynb | radical-cybertools/QCFractal | 676997ef4bc267ae9adfe0064843c42a5701dcf4 | [
"BSD-3-Clause"
] | null | null | null | docs/qcfractal/source/quickstart.ipynb | radical-cybertools/QCFractal | 676997ef4bc267ae9adfe0064843c42a5701dcf4 | [
"BSD-3-Clause"
] | null | null | null | 40.771247 | 2,027 | 0.550135 | [
[
[
"# Quickstart Tutorial\n\nThis tutorial will go over general QCFractal usage to give a feel for the ecosystem. \nIn this tutorial, we employ Snowflake, a simple QCFractal stack which runs on a local machine \nfor demonstration and exploration purposes.\n\n## Installation\n\nTo begin this quickstart tutorial, first install the QCArchive Snowflake environment from conda:\n\n```\nconda env create qcarchive/qcf-snowflake -n snowflake\nconda activate snowflake\n```\n\nIf you have a pre-existing environment with `qcfractal`, ensure that `rdkit` and `geometric` are installed from the `conda-forge` channel and `psi4` from the `psi4` channel.\n",
"_____no_output_____"
],
[
"## Importing QCFractal\n\nFirst let us import two items from the ecosystem:\n\n\n * [FractalSnowflakeHandler](http://docs.qcarchive.molssi.org/projects/qcfractal/en/latest/api/qcfractal.FractalSnowflakeHandler.html?highlight=FractalSnowflakeHandler) - This is a [FractalServer](https://qcarchivetutorials.readthedocs.io/projects/qcfractal/en/latest/api/qcfractal.FractalServer.html) that is temporary and is used for trying out new things.\n\n * `qcfractal.interface` is the [QCPortal](https://github.com/MolSSI/QCPortal) module, but if using QCFractal it is best to import it locally.\n \nTypically we alias `qcportal` as `ptl`. We will do the same for `qcfractal.interface` so that the code can be used anywhere.",
"_____no_output_____"
]
],
[
[
"from qcfractal import FractalSnowflakeHandler\nimport qcfractal.interface as ptl",
"_____no_output_____"
]
],
[
[
"We can now build a temporary server which acts just like a normal server, but we have a bit more direct control of it.\n\n**Warning!** All data is lost when this notebook shuts down! This is for demonstration purposes only!\nFor information about how to setup a permanent QCFractal server, see the [Setup Quickstart Guide](setup_quickstart.rst). ",
"_____no_output_____"
]
],
[
[
"server = FractalSnowflakeHandler()\nserver",
"_____no_output_____"
]
],
[
[
"We can then build a typical [FractalClient](http://docs.qcarchive.molssi.org/projects/qcportal/en/latest/client.html?highlight=fractalclient#portal-client) \nto automatically connect to this server using the [client()](http://docs.qcarchive.molssi.org/projects/qcfractal/en/latest/api/qcfractal.FractalSnowflakeHandler.html?highlight=FractalSnowflakeHandler#qcfractal.FractalSnowflakeHandler.client) helper command. \nNote that the server names and addresses are identical in both the server and client.",
"_____no_output_____"
]
],
[
[
"client = server.client()\nclient",
"_____no_output_____"
]
],
[
[
"## Adding and Querying data\n\nA server starts with no data, so let's add some! We can do this by adding a water molecule at a poor geometry from XYZ coordinates. \nNote that all internal QCFractal values are stored and used in atomic units; \nwhereas, the standard [Molecule.from_data()](http://docs.qcarchive.molssi.org/projects/qcelemental/en/latest/molecule.html?highlight=from_data#creation) assumes an input of Angstroms. \nWe can switch this back to Bohr by adding a `units` command in the text string. ",
"_____no_output_____"
]
],
[
[
"mol = ptl.Molecule.from_data(\"\"\"\nO 0 0 0\nH 0 0 2\nH 0 2 0\nunits bohr\n\"\"\")\nmol",
"_____no_output_____"
]
],
[
[
"We can then measure various aspects of this molecule to determine its shape. Note that the `measure` command will provide a distance, angle, or dihedral depending if 2, 3, or 4 indices are passed in.\n\nThis molecule is quite far from optimal, so let's run an geometry optimization!",
"_____no_output_____"
]
],
[
[
"print(mol.measure([0, 1]))\nprint(mol.measure([1, 0, 2]))",
"2.0\n90.0\n"
]
],
[
[
"## Evaluating a Geometry Optimization\n\nWe originally installed `psi4` and `geometric`, so we can use these programs to perform a geometry optimization. In QCFractal, we call a geometry optimization a `procedure`, where `procedure` is a generic term for a higher level operation that will run multiple individual quantum chemistry energy, gradient, or Hessian evaluations. Other `procedure` examples are finite-difference computations, n-body computations, and torsiondrives.\n\nWe provide a JSON-like input to the [client.add_procedure()](http://docs.qcarchive.molssi.org/projects/qcportal/en/latest/client-api.html?highlight=add_procedure#qcportal.FractalClient.add_procedure)\n command to specify the method, basis, and program to be used. \nThe `qc_spec` field is used in all procedures to determine the underlying quantum chemistry method behind the individual procedure.\nIn this way, we can use any program or method that returns a energy or gradient quantity to run our geometry optimization!\n(See also [add_compute()](http://docs.qcarchive.molssi.org/projects/qcportal/en/latest/client-api.html?highlight=add_procedure#qcportal.FractalClient.add_compute).)",
"_____no_output_____"
]
],
[
[
"spec = {\n \"keywords\": None,\n \"qc_spec\": {\n \"driver\": \"gradient\",\n \"method\": \"b3lyp-d3\",\n \"basis\": \"6-31g\",\n \"program\": \"psi4\"\n },\n}\n\n# Ask the server to compute a new computation\nr = client.add_procedure(\"optimization\", \"geometric\", spec, [mol])\nprint(r)\nprint(r.ids)",
"ComputeResponse(nsubmitted=1 nexisting=0)\n['5ce6f535bf1f5cb8ee9be73f']\n"
]
],
[
[
"We can see that we submitted a single task to be evaluated and the server has not seen this particular procedure before. \nThe `ids` field returns the unique `id` of the procedure. Different procedures will always have a unique `id`, while identical procedures will always return the same `id`. \nWe can submit the same procedure again to see this effect:",
"_____no_output_____"
]
],
[
[
"r2 = client.add_procedure(\"optimization\", \"geometric\", spec, [mol])\nprint(r)\nprint(r.ids)",
"ComputeResponse(nsubmitted=1 nexisting=0)\n['5ce6f535bf1f5cb8ee9be73f']\n"
]
],
[
[
"## Querying Procedures\n\nOnce a task is submitted, it will be placed in the compute queue and evaluated. In this particular case the [FractalSnowflakeHandler](http://docs.qcarchive.molssi.org/projects/qcfractal/en/latest/api/qcfractal.FractalSnowflakeHandler.html?highlight=FractalSnowflakeHandler) uses your local hardware to evaluate these jobs. We recommend avoiding large tasks!\n\nIn general, the server can handle anywhere between laptop-scale resources to many hundreds of thousands of concurrent cores at many physical locations. The amount of resources to connect is up to you and the amount of compute that you require.\n\nSince we did submit a very small job it is likely complete by now. Let us query this procedure from the server using its `id` like so:",
"_____no_output_____"
]
],
[
[
"proc = client.query_procedures(id=r.ids)[0]\nproc",
"_____no_output_____"
]
],
[
[
"This [OptimizationRecord](http://docs.qcarchive.molssi.org/projects/qcportal/en/latest/record-api.html?highlight=optimizationrecord#qcportal.models.OptimizationRecord) object has many different fields attached to it so that all quantities involved in the computation can be explored. For this example, let us pull the final molecule (optimized structure) and inspect the physical dimensions.\n\nNote: if the status does not say `COMPLETE`, these fields will not be available. Try querying the procedure again in a few seconds to see if the task completed in the background.",
"_____no_output_____"
]
],
[
[
"final_mol = proc.get_final_molecule()",
"_____no_output_____"
],
[
"print(final_mol.measure([0, 1]))\nprint(final_mol.measure([1, 0, 2]))\nfinal_mol",
"1.84413039713973\n108.31440065631584\n"
]
],
[
[
"This water molecule has bond length and angle dimensions much closer to expected values. We can also plot the optimization history to see how each step in the geometry optimization affected the results. Though the chart is not too impressive for this simple molecule, it is hopefully illuminating and is available for any geometry optimization ever completed.",
"_____no_output_____"
]
],
[
[
"proc.show_history()",
"_____no_output_____"
]
],
[
[
"## Collections\n\nSubmitting individual procedures or single quantum chemistry tasks is not typically done as it becomes hard to track individual tasks. To help resolve this, ``Collections`` are different ways of organizing standard computations so that many tasks can be referenced in a more human-friendly way. In this particular case, we will be exploring an intermolecular potential dataset.\n\nTo begin, we will create a new dataset and add a few intermolecular interactions to it.",
"_____no_output_____"
]
],
[
[
"ds = ptl.collections.ReactionDataset(\"My IE Dataset\", ds_type=\"ie\", client=client, default_program=\"psi4\")",
"_____no_output_____"
]
],
[
[
"We can construct a water dimer that has fragments used in the intermolecular computation with the `--` divider. A single water molecule with ghost atoms can be extracted like so:",
"_____no_output_____"
]
],
[
[
"water_dimer = ptl.Molecule.from_data(\"\"\"\nO 0.000000 0.000000 0.000000\nH 0.758602 0.000000 0.504284\nH 0.260455 0.000000 -0.872893\n--\nO 3.000000 0.500000 0.000000\nH 3.758602 0.500000 0.504284\nH 3.260455 0.500000 -0.872893\n\"\"\")\n\nwater_dimer.get_fragment(0, 1)",
"_____no_output_____"
]
],
[
[
"Many molecular entries can be added to this dataset where each is entry is a given intermolecular complex that is given a unique name. In addition, the `add_ie_rxn` method to can automatically fragment molecules. ",
"_____no_output_____"
]
],
[
[
"ds.add_ie_rxn(\"water dimer\", water_dimer)\nds.add_ie_rxn(\"helium dimer\", \"\"\"\nHe 0 0 -3\n--\nHe 0 0 3\n\"\"\")",
"_____no_output_____"
]
],
[
[
"Once the Collection is created it can be saved to the server so that it can always be retrived at a future date",
"_____no_output_____"
]
],
[
[
"ds.save()",
"_____no_output_____"
]
],
[
[
"The client can list all Collections currently on the server and retrive collections to be manipulated:",
"_____no_output_____"
]
],
[
[
"client.list_collections()",
"_____no_output_____"
],
[
"ds = client.get_collection(\"ReactionDataset\", \"My IE Dataset\")",
"_____no_output_____"
]
],
[
[
"## Computing with collections\n\nComputational methods can be applied to all of the reactions in the dataset with just a few simple lines:",
"_____no_output_____"
]
],
[
[
"ds.compute(\"B3LYP-D3\", \"def2-SVP\")",
"_____no_output_____"
]
],
[
[
"By default this collection evaluates the non-counterpoise corrected interaction energy which typically requires three computations per entry (the complex and each monomer). In this case we compute the B3LYP and -D3 additive correction separately, nominally 12 total computations. However the collection is smart enough to understand that each Helium monomer is identical and does not need to be computed twice, reducing the total number of computations to 10 as shown here. We can continue to compute additional methods. Again, this is being evaluated on your computer! Be careful of the compute requirements.",
"_____no_output_____"
]
],
[
[
"ds.compute(\"PBE-D3\", \"def2-SVP\")",
"_____no_output_____"
]
],
[
[
"A list of all methods that have been computed for this dataset can also be shown:",
"_____no_output_____"
]
],
[
[
"ds.list_history()",
"_____no_output_____"
]
],
[
[
"The above only shows what has been computed and does not pull this data from the server to your computer. To do so, the `get_history` command can be used:",
"_____no_output_____"
]
],
[
[
"ds.get_history()",
"_____no_output_____"
]
],
[
[
"Underlying the Collection is a pandas DataFrame which can show all results:",
"_____no_output_____"
]
],
[
[
"print(f\"DataFrame units: {ds.units}\")\nds.df",
"DataFrame units: kcal / mol\n"
]
],
[
[
"You can also visualize results and more!",
"_____no_output_____"
]
],
[
[
"ds.visualize([\"B3LYP-D3\", \"PBE-D3\"], \"def2-SVP\", bench=\"B3LYP/def2-svp\", kind=\"violin\")",
"_____no_output_____"
]
],
[
[
"This is only the very beginning of what you can do with QCFractal! Explore the documentation to learn more capabilities.\nIn particular, see the [next section](setup_quickstart.rst) for a quickstart guide on how to set up QCFractal in production.\n\nIf you like the project, consider starring us on [GitHub](https://github.com/MolSSI/QCFractal). If you have any questions, join our [Slack](https://join.slack.com/t/qcdb/shared_invite/enQtNDIzNTQ2OTExODk0LWM3OTgxN2ExYTlkMTlkZjA0OTExZDlmNGRlY2M4NWJlNDlkZGQyYWUxOTJmMzc3M2VlYzZjMjgxMDRkYzFmOTE) channel.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb8bbd9e259e0bfabd2409dc35bdb3c17330c17 | 1,843 | ipynb | Jupyter Notebook | Algorismica/4.2.ipynb | Javiooli/Uni | 2dd4d8e657222bf9723df6438303449182f5eb01 | [
"MIT"
] | null | null | null | Algorismica/4.2.ipynb | Javiooli/Uni | 2dd4d8e657222bf9723df6438303449182f5eb01 | [
"MIT"
] | null | null | null | Algorismica/4.2.ipynb | Javiooli/Uni | 2dd4d8e657222bf9723df6438303449182f5eb01 | [
"MIT"
] | null | null | null | 21.682353 | 137 | 0.488334 | [
[
[
"# Capítol 4 - Algorismes i Text",
"_____no_output_____"
],
[
"### 4.2 Alfabet aviació",
"_____no_output_____"
]
],
[
[
"def aviacio(cadena):\n \"\"\"\n Aquesta funció converteix una cadena d'entrada a l'alfabet fonètic.\n \n Parameters\n ----------\n cadena: string\n \n Returns\n -------\n traduccio: string\n \"\"\"\n alfabet = ['Alpha','Bravo','Charlie','Delta','Echo','Foxtrot','Golf','Hotel','India','Juliet','Kilo','Lima','Mike',\n 'November','Oscar','Papa','Quebec','Romeo','Sierra','Tango','Uniform','Victor','Whiskey','X-Ray','Yankee','Zulu']\n return [alfabet[ord(list(cadena.lower())[x])-97] for x in range(len(cadena))]",
"_____no_output_____"
],
[
"assert aviacio('YtH') == ['Yankee', 'Tango', 'Hotel']",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
ecb8e97466c1e8d6942815daadce50ca94da91df | 1,208 | ipynb | Jupyter Notebook | QiskitEducation/.ipynb_checkpoints/test-checkpoint.ipynb | veenaiyuri/qiskit-education | 0b58c8985b9377b4bc3404d205ea6235877a9778 | [
"Apache-2.0"
] | 6 | 2019-02-03T10:03:12.000Z | 2020-11-05T18:13:08.000Z | QiskitEducation/test.ipynb | quantumjim/qiskit-education | 285074042a5d595436e4c84395d8205ba668f43c | [
"Apache-2.0"
] | null | null | null | QiskitEducation/test.ipynb | quantumjim/qiskit-education | 285074042a5d595436e4c84395d8205ba668f43c | [
"Apache-2.0"
] | 2 | 2019-01-31T16:48:17.000Z | 2019-01-31T17:00:59.000Z | 19.803279 | 86 | 0.460265 | [
[
[
"from QiskitEducation import *",
"_____no_output_____"
],
[
"for alg in [QuantumAlgorithm(2,2),QuantumAlgorithm( [(2,'q')] , [(2,'c')] )]:\n \n alg.h(alg.q[0])\n alg.cx(alg.q[0],alg.q[1]) \n\n alg.measure(alg.q[0],alg.c[0])\n alg.measure(alg.q[1],alg.c[1]) \n\n print(alg.execute()['counts'])",
"{'11': 505, '00': 519}\n{'11': 506, '00': 518}\n"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
ecb8f9e8a94656490080606db842349b02b3dfe4 | 6,904 | ipynb | Jupyter Notebook | notebooks/Result-Analyse/Preprocessing-Differences/Semeval-preprocessing-differences.ipynb | geetickachauhan/relation-extraction | aa920449b20c7127954eaaaa05244e7fc379e018 | [
"MIT"
] | 19 | 2019-06-24T18:33:36.000Z | 2022-01-21T03:16:12.000Z | notebooks/Result-Analyse/Preprocessing-Differences/Semeval-preprocessing-differences.ipynb | geetickachauhan/relation-extraction | aa920449b20c7127954eaaaa05244e7fc379e018 | [
"MIT"
] | null | null | null | notebooks/Result-Analyse/Preprocessing-Differences/Semeval-preprocessing-differences.ipynb | geetickachauhan/relation-extraction | aa920449b20c7127954eaaaa05244e7fc379e018 | [
"MIT"
] | 11 | 2019-06-02T08:59:16.000Z | 2021-08-23T04:31:07.000Z | 26.452107 | 271 | 0.568511 | [
[
[
"# Differences that pre-processing cause to the baseline model in Semeval 2010 data",
"_____no_output_____"
],
[
"for reference, command that was run within scripts/ was ```CUDA_VISIBLE_DEVICES=<device_no> python main.py --<cross_validate/use_test> --dataset=semeval2010 --preprocessing_type=<entity_blinding/punct_digit/punct_stop_digit>```",
"_____no_output_____"
]
],
[
[
"from scipy.stats import ttest_rel",
"_____no_output_____"
]
],
[
[
"## First compare the cross validated score differences",
"_____no_output_____"
]
],
[
[
"baseline_test = 81.55 # Model ID 967c88e8-18c2-4a84-b73d-23c6bb33efd7 on harrison",
"_____no_output_____"
],
[
"# these are all results on the evaluation fold of the pickled files\nbaseline = [81.66, 82.26, 80.12, 78.82, 79.23, 80.74, 79.56, 81.45, 82.99, 81.63]\n# model ID ade5ce18-2eb0-4d2e-a04e-c727a09e5ef0\n# 80.85 +- 1.31\n\n# baseline = [80.54, 80.2, 79.25, 81.08, 80.69, 79.11, 80.75, 82.04, 80.78, 80.73]\n# # model ID 0b8525cc-7e0c-4afe-b952-c2e7636d61bd on harrison\n# # 80.52 +- 0.81",
"_____no_output_____"
],
[
"entity_blinding = [69.99, 71.33, 71.67, 69.07, 71.47, 70.6, 71.08, 72.54, 73.06, 72.25]\n# model ID 26667800-dc53-46e4-bbed-3a27cf89ef9e\n# 71.31 +- 1.14\n\n# entity_blinding = [71.52, 70.58, 70.42, 70.8, 70.22, 70.77, 71.46, 72.36, 70.06, 70.26]\n# # model ID a553fd1c-d872-456e-ab0c-145ab54cd3df on harrison",
"_____no_output_____"
],
[
"punct_digit = [81.12, 82.79, 80.26, 79.24, 80.05, 80.72, 80.19, 80.81, 83.48, 80.87]\n# model ID 52351631-bf32-4725-8fc0-6d2a4265ff7b\n# 80.95 +- 1.21\n\n# punct_digit = [81.21, 80.09, 79.43, 79.84, 79.34, 79.72, 78.68, 81.61, 80.3, 81.23]\n# # model ID 35dbfad3-3103-477e-8305-9e7bfd594822 on harrison",
"_____no_output_____"
],
[
"punct_stop_digit = [72.28, 72.65, 72.8, 70.4, 70.23, 72.0, 71.74, 69.19, 71.44, 73.38]\n# model ID 5b15d683-6355-4a8d-a303-0e35d29c2bca\n# 71.61 +- 1.25\n\n# punct_stop_digit = [69.14, 70.8, 69.82, 71.44, 70.69, 71.23, 70.08, 71.17, 72.43, 73.17]\n# # model ID 5100b4fa-4680-49be-9163-e32c42607e77 on harrison",
"_____no_output_____"
],
[
"ner_blinding = [82.58, 82.2, 79.69, 78.95, 80.61, 80.2, 80.53, 81.65, 81.46, 80.63]\n# model ID 90e10a46-3073-40d7-8319-1ecb88fa39f2\n# 80.85 +- 1.07\n\n# ner_blinding = [80.5, 79.71, 78.51, 80.34, 79.94, 78.97, 82.02, 80.43, 81.82, 81.37]\n# # model ID 2b1c26e4-c053-49c4-a2a7-0c9c61c527f2 on gray",
"_____no_output_____"
],
[
"ttest_rel(baseline, entity_blinding)",
"_____no_output_____"
],
[
"ttest_rel(baseline, punct_digit)",
"_____no_output_____"
],
[
"ttest_rel(baseline, punct_stop_digit)",
"_____no_output_____"
],
[
"ttest_rel(punct_digit, punct_stop_digit)",
"_____no_output_____"
],
[
"ttest_rel(baseline, ner_blinding)",
"_____no_output_____"
]
],
[
[
"In short, entity blinding is harmful for the semeval 2010 data, maybe because there is useful information within the entity names themselves. Removing punctuations and normalizing digits has no effect, but removing stop words is harmful. NER blinding has no effect.",
"_____no_output_____"
],
[
"Test score results for the above are (all model IDs on harrison):\n\n```81.55``` for baseline model with ID 967c88e8-18c2-4a84-b73d-23c6bb33efd7\n\n```72.73``` for entity blinding model ID 6217504a-4334-4e63-bcba-8e3646a963ea\n\n```81.23``` for punctuation removal and digit normalization with model ID d0305d80-51ee-4069-95d5-a86563df7fc6\n\n```72.92``` for punctuation and stop word removal and digit normalization with model ID 1331572b-b37a-4ad9-80fb-f49f9d5786d7\n\n```81.63``` for ner blinding with model ID 4d550ba5-08c0-4bc1-81f6-27fee75c61c8 on gray",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ecb9000adb129440aaeeb27b2580d9a22e69c307 | 6,306 | ipynb | Jupyter Notebook | nlp_topic_modelling.ipynb | Martins6/WebCrawler | 4a1cc86492d0266d6d317b356287aefb0a7d444a | [
"MIT"
] | null | null | null | nlp_topic_modelling.ipynb | Martins6/WebCrawler | 4a1cc86492d0266d6d317b356287aefb0a7d444a | [
"MIT"
] | null | null | null | nlp_topic_modelling.ipynb | Martins6/WebCrawler | 4a1cc86492d0266d6d317b356287aefb0a7d444a | [
"MIT"
] | null | null | null | 30.760976 | 670 | 0.552331 | [
[
[
"# Topic Modelling",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer\nfrom sklearn import decomposition\nimport nltk\nimport numpy as np\nimport json\nfrom pathlib import Path",
"_____no_output_____"
],
[
"corpus_list = []\nwith Path('crawled_data.jl').open() as f:\n for line in f:\n json_dict = json.loads(line)\n corpus_list.append(json_dict['corpus'])\ncorpus = np.array(corpus_list)\ncorpus[1]",
"_____no_output_____"
],
[
"nltk.download('stopwords')",
"[nltk_data] Downloading package stopwords to\n[nltk_data] /home/adriel_martins/nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n"
],
[
"stopwords = nltk.corpus.stopwords.words('portuguese')\nstopwords[1]\nvectorizer = CountVectorizer(stop_words=stopwords)\nvectors = vectorizer.fit_transform(corpus).todense() # (documents, vocab)\nvectors.shape #, vectors.nnz / vectors.shape[0], row_means.shape\nprint(len(corpus.data), vectors.shape)",
"76 (76, 4844)\n"
],
[
"vocab = np.array(vectorizer.get_feature_names())\nvocab[200:300]",
"_____no_output_____"
],
[
"num_top_words=8\n\ndef show_topics(a):\n top_words = lambda t: [vocab[i] for i in np.argsort(t)[:-num_top_words-1:-1]]\n topic_words = ([top_words(t) for t in a])\n return [' '.join(t) for t in topic_words]",
"_____no_output_____"
],
[
"m,n=vectors.shape\nd=5 # num topics\nclf = decomposition.NMF(n_components=d, random_state=1)\n\nW1 = clf.fit_transform(vectors)\nH1 = clf.components_\nshow_topics(H1)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb90205b0d4cbf68f6bf23e09b32aa9e14fc539 | 7,170 | ipynb | Jupyter Notebook | SQL/.ipynb_checkpoints/6. DQL - Data Query Language - parte 1-checkpoint.ipynb | alineat/SQL-e-Bancos-de-Dados | b6d017f8e0e656e04cf1068498dc59d62a243cbd | [
"Apache-2.0"
] | 1 | 2020-08-28T13:27:19.000Z | 2020-08-28T13:27:19.000Z | SQL/6. DQL - Data Query Language - parte 1.ipynb | alineat/SQL-e-Bancos-de-Dados | b6d017f8e0e656e04cf1068498dc59d62a243cbd | [
"Apache-2.0"
] | null | null | null | SQL/6. DQL - Data Query Language - parte 1.ipynb | alineat/SQL-e-Bancos-de-Dados | b6d017f8e0e656e04cf1068498dc59d62a243cbd | [
"Apache-2.0"
] | null | null | null | 40.508475 | 304 | 0.640725 | [
[
[
"# 1. Primeiras consultas no banco de dados\n",
"_____no_output_____"
],
[
"- como efetivamente consultar o banco de dados para extrair informações para os mais diversos propósitos?\n- uma das sub-linguagens mais importantes para o dia-a-dia do profissional de bancos de dados;\n- Uusado para obter listagens de registros simples, limitar as atualizações de dados e a remoção de dados, uso para consulta de registros relacionados;\n- em um banco de dados populado podemos extrair informações",
"_____no_output_____"
],
[
"- Categoria de subcomando da linguagem SQL que envolve a declaração de recuperação de dados (SELECT).\n- SELECT é uma declaração SQL que retorna um conjunto de resultados (já esperado) de registros de uma ou mais tabelas. Ela recupera zero ou mais linhas de uma ou mais tabelas-base, tabelas temporárias, funções ou visões em um banco de dados.\n",
"_____no_output_____"
],
[
"- Comandos básicos do SELECT: \n \n- SELECT: define quais colunas cou extrair\n- FROM: de onde estou extraindo, de quais tabelas estou extraindo os dados;\n- WHERE: cláusula onde as condições são atendidas;\n- GROUP BY: agrupa por nome, por estado, por situações específicas;\n- HAVING: condição de agrupamento. Ex: agrupar todas as pessoas que têm no mínimo 10 anos de idade; \n- ORDER BY: ordem em que o SELECT vai ser retornado. \n\n- WHERE, GROUP BY, HAVING e ORDER BY são opcionais. Tendo o SELECT * FROM tabela ele já executa.\n- No comando SELECT, quanto mais elementos, mais simplificado e detalhado será a sua busca.",
"_____no_output_____"
],
[
"- a ordem em que a consulta (query) é escrita não significa que será a mesma ordem que o banco de dados utilizará para executar o processamento. Ele pode executar primeiro o FROM, depois WHERE, GROUP BY...:\n \n- isso é só uma observação, não muda em nada no modo de executar as buscas\n",
"_____no_output_____"
],
[
" - A forma mais simples da declaração SELECT é a utilização junto ao elemento FROM (ex.: SELECT * FROM tabela - selecione todos os campos da tabela x, ai retorna todos os elementos ou SELECT nome_da_coluna, nome_da_coluna2 FROM tabela_X ), conforme mostrado abaixo;\n\n- Note que no \"select list\" há uma filtragem vertical, ou seja, retorna uma ou mais colunas de tabelas, mencionadas pela cláusula FROM.\n \n- Esse comando é simples, mas se eu tiver uma tabela com muitas linhas e eu só quiser saber quem tem o nome maria, por ex., eu não preciso receber de retorno todos os nomes da tabela. É por isso que o WHERE é importante, porque posso filtrar quais situações eu quero. \n- A ideia do SELECT é justamente simplificar o seu resultado. Uma busca bem feita é otimizada, o reusltado é melhor, mais prático, mais eficiente.\n",
"_____no_output_____"
],
[
"- Outros exemplos para SELECT simples:\n - ( * ) - Retorna todas as colunas da tabela exemploSQL: `SELECT * FROM exemploSQL` \n - ( coluna ) - Retorna a coluna específica, texto_curto_naonulo da tabela exemploSQL: `SELECT texto_curto_naonulo FROM exemploSQL`\n\n",
"_____no_output_____"
],
[
"- Podemos utilizar diversos operadores matemáticos para cálculo de valores. Veja os principais operadores:\n \n \n- quando retorno a coluna, posso usar funções matemáticas. Consigo otimizar a minha busca e economizar tempo.\n- P.S.: operadores possuem precedência entre si.\n",
"_____no_output_____"
],
[
"Ex. para SELECT simples e operadores:\n- Retorna o resultado das operações abaixo: \n\n`SELECT 20 + 20 / 5 FROM exemploSQL` \n`SELECT (20 + 20) / 5 FROM exemploSQL` \n`SELECT 20 + (20 / 5) FROM exemploSQL` \n\n`SELECT ( (10+2) / 2 ) * 0.3 ) % 2` \n\n`SELECT Nome, Salario * 1.07 FROM Funcionario` \n\n- Nota: O operador + se transforma em concatenador quando lidamos com string: SELECT ‘Hoje’ + ‘ ‘ + ‘é’ + ‘ terça-feira ’ + ‘ou’ + ‘ quinta-feira ’ \n- Dê uma olhada nas funções média, mínimo... Ao invés de fazer a função matemática na aplicação, você pode fazê-la diretamente no BD quando você estiver fazendo a consulta.\n",
"_____no_output_____"
],
[
"# 2. Como usar apelidos nas consultas de dados",
"_____no_output_____"
],
[
"Pode ser necessário darmos apelidos (Aliases) a colunas para facilitar o entendimento no retorno dos dados:\n- Apelidos na coluna utilizando a cláusula AS (não preciso imprimir no SELECT qtd, posso imprimir Quantidade ou qualquer outro nome que eu quiser. Vantagem: não preciso tratar na aplicação. Um campo SELECT bem feito, eu minimizo o trabalho na aplicação, ganho tempo e tenho um resultado melhor):\n \n\n- Também é possível realizar a mesma operação com =\n \n\n- Ou mesmo, sem a necessidade do AS (SQL entende que preco vai ser ValorProduto):\n \n\n",
"_____no_output_____"
],
[
"Também pode ser necessário darmos apelidos em tabelas, principalmente quando formos realizar joins: \n- Apelidos em tabelas com a cláusula AS:\n \n\n- Apelidos em tabelas sem AS:\n \n\n- Usando os apelidos no SELECT: \n \n- Vantagem: se eu tiver uma tabela com nome muito grande, na hora da consulta posso usar apelidos para simplificar esses nomes. Fica mais fácil de ler.",
"_____no_output_____"
],
[
"Exercício de consulta: http://sqlfiddle.com/#!9/caf268/6\nhttp://sqlfiddle.com/#!9/805bb7/1\nhttp://sqlfiddle.com/#!9/ded19e/2",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecb904989c6587c5e7f05a3e6cb440cf2de7e5b7 | 940,405 | ipynb | Jupyter Notebook | module4-classification-metrics/LS_DS_224_assignment.ipynb | austiezr/DS-Unit-2-Kaggle-Challenge | 82004fe1e2dbd857613f60dfe267abb6e947bf77 | [
"MIT"
] | null | null | null | module4-classification-metrics/LS_DS_224_assignment.ipynb | austiezr/DS-Unit-2-Kaggle-Challenge | 82004fe1e2dbd857613f60dfe267abb6e947bf77 | [
"MIT"
] | null | null | null | module4-classification-metrics/LS_DS_224_assignment.ipynb | austiezr/DS-Unit-2-Kaggle-Challenge | 82004fe1e2dbd857613f60dfe267abb6e947bf77 | [
"MIT"
] | null | null | null | 1,302.5 | 450,829 | 0.933087 | [
[
[
"<a href=\"https://colab.research.google.com/github/austiezr/DS-Unit-2-Kaggle-Challenge/blob/master/module4-classification-metrics/LS_DS_224_assignment.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"Lambda School Data Science\n\n*Unit 2, Sprint 2, Module 4*\n\n---",
"_____no_output_____"
],
[
"# Classification Metrics\n\n## Assignment\n- [x] If you haven't yet, [review requirements for your portfolio project](https://lambdaschool.github.io/ds/unit2), then submit your dataset.\n- [X] Plot a confusion matrix for your Tanzania Waterpumps model.\n- [x] Continue to participate in our Kaggle challenge. Every student should have made at least one submission that scores at least 70% accuracy (well above the majority class baseline).\n- [x] Submit your final predictions to our Kaggle competition. Optionally, go to **My Submissions**, and _\"you may select up to 1 submission to be used to count towards your final leaderboard score.\"_\n- [x] Commit your notebook to your fork of the GitHub repo.\n- [x] Read [Maximizing Scarce Maintenance Resources with Data: Applying predictive modeling, precision at k, and clustering to optimize impact](https://towardsdatascience.com/maximizing-scarce-maintenance-resources-with-data-8f3491133050), by Lambda DS3 student Michael Brady. His blog post extends the Tanzania Waterpumps scenario, far beyond what's in the lecture notebook.\n\n\n## Stretch Goals\n\n### Reading\n\n- [Attacking discrimination with smarter machine learning](https://research.google.com/bigpicture/attacking-discrimination-in-ml/), by Google Research, with interactive visualizations. _\"A threshold classifier essentially makes a yes/no decision, putting things in one category or another. We look at how these classifiers work, ways they can potentially be unfair, and how you might turn an unfair classifier into a fairer one. As an illustrative example, we focus on loan granting scenarios where a bank may grant or deny a loan based on a single, automatically computed number such as a credit score.\"_\n- [Notebook about how to calculate expected value from a confusion matrix by treating it as a cost-benefit matrix](https://github.com/podopie/DAT18NYC/blob/master/classes/13-expected_value_cost_benefit_analysis.ipynb)\n- [Visualizing Machine Learning Thresholds to Make Better Business Decisions](https://blog.insightdatascience.com/visualizing-machine-learning-thresholds-to-make-better-business-decisions-4ab07f823415)\n\n\n### Doing\n- [ ] Share visualizations in our Slack channel!\n- [x] RandomizedSearchCV / GridSearchCV, for model selection. (See module 3 assignment notebook)\n- [ ] Stacking Ensemble. (See module 3 assignment notebook)\n- [ ] More Categorical Encoding. (See module 2 assignment notebook)",
"_____no_output_____"
]
],
[
[
"!pip3 install category_encoders\n!pip3 install xgboost\nimport pandas as pd\nimport numpy as np\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.impute import SimpleImputer, KNNImputer\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.feature_selection import f_classif, mutual_info_classif\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.decomposition import PCA\nfrom sklearn.ensemble import RandomForestClassifier, BaggingClassifier, GradientBoostingClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.svm import SVC\nfrom sklearn.cluster import KMeans, MiniBatchKMeans, SpectralClustering\nfrom sklearn.mixture import GaussianMixture\nfrom category_encoders.target_encoder import TargetEncoder\nfrom category_encoders.woe import WOEEncoder\nfrom category_encoders import OneHotEncoder, OrdinalEncoder\nfrom sklearn.experimental import enable_iterative_imputer\nfrom sklearn.impute import IterativeImputer\nfrom scipy.stats import randint, uniform\nfrom sklearn.model_selection import RandomizedSearchCV\nfrom sklearn.linear_model import RidgeClassifierCV, RidgeClassifier\nfrom xgboost import XGBClassifier\nfrom sklearn.metrics import confusion_matrix",
"Collecting category_encoders\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/a0/52/c54191ad3782de633ea3d6ee3bb2837bda0cf3bc97644bb6375cf14150a0/category_encoders-2.1.0-py2.py3-none-any.whl (100kB)\n\r\u001b[K |███▎ | 10kB 19.2MB/s eta 0:00:01\r\u001b[K |██████▌ | 20kB 1.8MB/s eta 0:00:01\r\u001b[K |█████████▉ | 30kB 2.3MB/s eta 0:00:01\r\u001b[K |█████████████ | 40kB 1.7MB/s eta 0:00:01\r\u001b[K |████████████████▍ | 51kB 1.9MB/s eta 0:00:01\r\u001b[K |███████████████████▋ | 61kB 2.2MB/s eta 0:00:01\r\u001b[K |██████████████████████▉ | 71kB 2.4MB/s eta 0:00:01\r\u001b[K |██████████████████████████▏ | 81kB 2.6MB/s eta 0:00:01\r\u001b[K |█████████████████████████████▍ | 92kB 2.9MB/s eta 0:00:01\r\u001b[K |████████████████████████████████| 102kB 2.3MB/s \n\u001b[?25hRequirement already satisfied: pandas>=0.21.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.25.3)\nRequirement already satisfied: scipy>=0.19.0 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (1.4.1)\nRequirement already satisfied: statsmodels>=0.6.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.10.2)\nRequirement already satisfied: numpy>=1.11.3 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (1.17.5)\nRequirement already satisfied: patsy>=0.4.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.5.1)\nRequirement already satisfied: scikit-learn>=0.20.0 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.22.1)\nRequirement already satisfied: python-dateutil>=2.6.1 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.21.1->category_encoders) (2.6.1)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.21.1->category_encoders) (2018.9)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from patsy>=0.4.1->category_encoders) (1.12.0)\nRequirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.6/dist-packages (from scikit-learn>=0.20.0->category_encoders) (0.14.1)\nInstalling collected packages: category-encoders\nSuccessfully installed category-encoders-2.1.0\nRequirement already satisfied: xgboost in /usr/local/lib/python3.6/dist-packages (0.90)\nRequirement already satisfied: scipy in /usr/local/lib/python3.6/dist-packages (from xgboost) (1.4.1)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from xgboost) (1.17.5)\n"
],
[
"%%capture\nimport sys\n\n# If you're on Colab:\nif 'google.colab' in sys.modules:\n DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Kaggle-Challenge/master/data/'\n !pip install category_encoders==2.*\n\n# If you're working locally:\nelse:\n DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Kaggle-Challenge/master/data/'",
"_____no_output_____"
],
[
"train = pd.merge(pd.read_csv(DATA_PATH+'waterpumps/train_features.csv'), \n pd.read_csv(DATA_PATH+'waterpumps/train_labels.csv'))\ntest = pd.read_csv(DATA_PATH+'waterpumps/test_features.csv')\nsample_submission = pd.read_csv(DATA_PATH+'waterpumps/sample_submission.csv')\n\ntrain, val = train_test_split(train, test_size=len(test), \n stratify=train['status_group'], random_state=33)\n\ntrain.shape, val.shape, test.shape",
"_____no_output_____"
],
[
"def wrangle(X):\n X = X.copy()\n\n X['latitude'] = X['latitude'].replace(-2e-08, 0)\n \n cols_with_zeros = ['longitude', 'latitude', 'amount_tsh', 'construction_year', 'gps_height', 'population']\n # cols_with_zeros = ['longitude', 'latitude', 'amount_tsh', 'construction_year', 'gps_height']\n for col in cols_with_zeros:\n X[col] = X[col].replace(0, np.nan)\n \n cols_high_c = ['ward', 'subvillage', 'funder', 'installer', 'lga', 'wpt_name']\n\n for col in cols_high_c:\n top10 = X[col].value_counts()[:15].index\n X.loc[~X[col].isin(top10), col] = 'OTHER'\n\n X['date_recorded'] = pd.to_datetime(X['date_recorded'], infer_datetime_format=True)\n \n X['year_recorded'] = X['date_recorded'].dt.year\n X['month_recorded'] = X['date_recorded'].dt.month\n X['day_recorded'] = X['date_recorded'].dt.day\n X = X.drop(columns='date_recorded')\n \n X['years'] = X['year_recorded'] - X['construction_year']\n X['years_MISSING'] = X['years'].isnull()\n\n X = X.drop(columns=['quantity_group', 'payment', 'recorded_by', \n 'extraction_type_group', 'extraction_type_class'])\n \n return X\n\n\ntrain = wrangle(train)\nval = wrangle(val)\ntest = wrangle(test)",
"_____no_output_____"
],
[
"target = 'status_group'\n\nX_train = train.drop(columns=[target, 'id'])\ny_train = train[target]\nX_val = val.drop(columns=[target, 'id'])\ny_val = val[target]\nX_test = test.drop(columns=['id'])",
"_____no_output_____"
],
[
"pipe = make_pipeline(\n OrdinalEncoder(),\n IterativeImputer(random_state=33),\n XGBClassifier()\n )\n\n\npipe.fit(X_train, y_train)\naccuracy = pipe.score(X_val, y_val)\nprint(f'Train Accuracy: {pipe.score(X_train, y_train)}\\n')\nprint(f'Validation Accuracy: {accuracy}\\n')\n# output.eval_js('new Audio(\"http://noproblo.dayjo.org/ZeldaSounds/LOZ/LOZ_Secret.wav\").play()')",
"_____no_output_____"
],
[
"pipe = make_pipeline(\n OrdinalEncoder(),\n IterativeImputer(random_state=33),\n RandomForestClassifier(n_jobs=-1,\n max_features='auto',\n random_state=33)\n )\n\nparam_distributions = {\n 'iterativeimputer__max_iter': randint(10,500),\n 'iterativeimputer__initial_strategy': ['mean', 'median', 'most_frequent'],\n 'randomforestclassifier__n_estimators': randint(2, 600),\n 'randomforestclassifier__min_samples_leaf': randint(2, 100)\n}\n\nsearch = RandomizedSearchCV(\n pipe, \n param_distributions=param_distributions, \n n_iter=100, \n cv=5, \n scoring='accuracy', \n verbose=10, \n return_train_score=True, \n n_jobs=-1\n)\n\nsearch.fit(X_train, y_train);\nAudio(sound_file, autoplay=True)",
"Fitting 5 folds for each of 100 candidates, totalling 500 fits\n"
],
[
"pipe = make_pipeline(\n OrdinalEncoder(),\n IterativeImputer(random_state=33),\n RandomForestClassifier(n_jobs=-1,\n max_features='auto',\n random_state=33)\n )\n\nparam_distributions = {\n 'iterativeimputer__max_iter': randint(10,500),\n 'iterativeimputer__initial_strategy': ['mean', 'median', 'most_frequent'],\n 'randomforestclassifier__n_estimators': randint(2, 600),\n 'randomforestclassifier__min_samples_leaf': randint(2, 100)\n}\n\nsearch = RandomizedSearchCV(\n pipe, \n param_distributions=param_distributions, \n n_iter=300, \n cv=5, \n scoring='accuracy', \n verbose=10, \n return_train_score=True, \n n_jobs=-1\n)\n\nsearch.fit(X_train, y_train);\nAudio(sound_file, autoplay=True)",
"Fitting 5 folds for each of 100 candidates, totalling 500 fits\n"
],
[
"from IPython.display import Audio\nsound_file = './Desktop/Clarke.mp3'",
"_____no_output_____"
],
[
"search2.fit(X_train, y_train);",
"Fitting 5 folds for each of 300 candidates, totalling 1500 fits\n"
],
[
"pipe = search.best_estimator_",
"_____no_output_____"
],
[
"print('Best hyperparameters', search.best_params_)",
"Best hyperparameters {'iterativeimputer__initial_strategy': 'median', 'iterativeimputer__max_iter': 38, 'xgbclassifier__colsample_bytree': 0.35321164911458447, 'xgbclassifier__eval_metric': 'merror', 'xgbclassifier__gamma': 0, 'xgbclassifier__learning_rate': 0.05225778827907627, 'xgbclassifier__max_depth': 11, 'xgbclassifier__min_child_weight': 27, 'xgbclassifier__reg_alpha': 2, 'xgbclassifier__reg_lambda': 7, 'xgbclassifier__scale_pos_weight': 9, 'xgbclassifier__subsample': 0.8643134091331315}\n"
],
[
"accuracy = pipe.score(X_val, y_val)\nprint(f'Train Accuracy: {pipe.score(X_train, y_train)}\\n')\nprint(f'Validation Accuracy: {accuracy}\\n')",
"Train Accuracy: 0.922117135118334\n\nValidation Accuracy: 0.8074244323721966\n\n"
],
[
"pipe = make_pipeline(\n OrdinalEncoder(),\n IterativeImputer(random_state=33),\n RandomForestClassifier(n_jobs=-1,\n max_features='auto',\n random_state=33)\n )",
"_____no_output_____"
],
[
"submission = test.copy()\nsubmission['status_group'] = pipe.predict(X_test)\nsubmission = submission[['id', 'status_group']]\nsubmission.set_index('id', inplace=True)\nsubmission.head()\nsubmission.to_csv('submission.csv')",
"_____no_output_____"
],
[
"pipe.fit(X_train, y_train)\n\nconfusion_matrix(y_val, pipe.predict(X_val))",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb90e8fda4df0d491d41c5c38cabe767d94f7e6 | 4,120 | ipynb | Jupyter Notebook | ray-train/00-Ray-Train-Overview.ipynb | dmatrix/academy | dbcac28e08caf83a9d7937f3bcb2747d6f6e67ef | [
"Apache-2.0"
] | null | null | null | ray-train/00-Ray-Train-Overview.ipynb | dmatrix/academy | dbcac28e08caf83a9d7937f3bcb2747d6f6e67ef | [
"Apache-2.0"
] | null | null | null | ray-train/00-Ray-Train-Overview.ipynb | dmatrix/academy | dbcac28e08caf83a9d7937f3bcb2747d6f6e67ef | [
"Apache-2.0"
] | null | null | null | 40.392157 | 347 | 0.628398 | [
[
[
"# Ray Train - Overview\n\n© 2019-2022, Anyscale. All Rights Reserved\n\n\n\n## Watch Ray Summit 2021 on demand!\n\nRay Summit 2021 [virtual conference](https://www.anyscale.com/ray-summit-2021) was on June 22-24, 2021. We had an amazing lineup of luminar keynote speakers and breakout sessions on the Ray ecosystem, third-party Ray libraries, and applications of Ray in the real world.\n\nFor information about other online events, see [anyscale.com/events](https://anyscale.com/events).\n\n## About This Tutorial\n<img src=\"https://images.ctfassets.net/xjan103pcp94/7kS5Mc1LeZRPqVlItSGMNI/3b863c02e9d0569da038c69ec2537864/image_1.jpg\" width=\"800\" height=\"200\" alt=\"Ray Summit 2021\"/>\n</a>\n\n[Ray Train](https://docs.ray.io/en/latest/train/train.html), formerly known as **Ray SGD**, is a lightweight library for distributed deep learning, allowing you to scale up and speed up training for your deep learning models. Currently, Ray Train is available as beta (or experimental) in Ray 1.9 release, offering the following features:\n\n * Scale to multi-GPU and multi-node training with 0 code changes\n\n * Runs seamlessly on any cloud (AWS, GCP, Azure, Kubernetes, or on-prem)\n\n * Supports PyTorch, TensorFlow, and Horovod \n\n * Distributed data loading and hyperparameter tuning\n\n * Built-in loggers for TensorBoard and MLflow\n\nSee the instructions in the [README](../README.md) for setting up your environment to use this tutorial.\n\nGo [here](../Overview.ipynb) for an overview of all tutorials.",
"_____no_output_____"
],
[
"| | Lesson | Description |\n| :-- | :----- | :---------- |\n| 00 | [Ray Train Overview](00-Ray-Train-Overview.ipynb) | Overview of this tutorial. |\n| 01 | [Ray Train Quickstart ](01-Ray-Train-Quickstart.ipynb) | A quick start on PyTorch training on single worker. |\n| 02 | [Ray Train Quickstart Distributed](02-Ray-Train-Quickstart-Distributed.ipynb) |A quick start on PyTorch Distributed training on multiple workers . |\n| 03 | [Ray Train with PyTorch](03-Ray-Train-with-PyTorch.ipynb) | Use Ray Train Distributed API to train a linear model |\n| 04 | [Ray Train with PyTorch and FashionMNSIT](04-Ray-Train-with-PyTorch-FashionMNIST.ipynb) | Use Ray Train Distributed API to train a NN for FashionMNIST |\n| | [Ray Train Examples](https://docs.ray.io/en/latest/train/examples.html) | Explore examples for Ray Train with PyTorch, TensorFlow, and Horvod. |",
"_____no_output_____"
],
[
"## Getting Help\n\n* The [#tutorial channel](https://ray-distributed.slack.com/archives/C011ML23W5B) on the [Ray Slack](https://ray-distributed.slack.com). [Click here](https://forms.gle/9TSdDYUgxYs8SA9e8) to join.\n* [Email](mailto:[email protected])\n\nFind an issue? Please report it!\n\n* [GitHub issues](https://github.com/anyscale/academy/issues)",
"_____no_output_____"
],
[
"## Give Us Feedback!\n\nLet us know what you like and don't like about this HPO and Ray Tune tutorial.\n\n* [Survey](https://forms.gle/StzNufFyyDT3dapt8)",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecb91100090331d2a46910017095f120f2b52a54 | 1,738 | ipynb | Jupyter Notebook | Basic/handle_pandas_dataframe.ipynb | hayatochigi/AzureML | 6cd416ea29f0995ebfe6f34e7bae8830e5d8fb59 | [
"MIT"
] | null | null | null | Basic/handle_pandas_dataframe.ipynb | hayatochigi/AzureML | 6cd416ea29f0995ebfe6f34e7bae8830e5d8fb59 | [
"MIT"
] | null | null | null | Basic/handle_pandas_dataframe.ipynb | hayatochigi/AzureML | 6cd416ea29f0995ebfe6f34e7bae8830e5d8fb59 | [
"MIT"
] | null | null | null | 25.558824 | 97 | 0.552359 | [
[
[
"from azureml.core import Workspace, Datastore, Dataset\n\nws = Workspace.from_config()\naz_store = Datastore.get(ws, 'from_sdk')\naz_ds = Dataset.get_by_name(ws, 'Loan Applications Using SDK')\naz_default_store = ws.get_default_datastore()",
"_____no_output_____"
],
[
"# Load azureml dataste into the pandas dataframe\ndataset = az_ds.to_pandas_dataframe()\ndataset",
"_____no_output_____"
],
[
"# Upload datframe to azureml dataset\ndataset_df = dataset[['Married', 'Gender', 'Loan_Status']]\naz_ds_from_df = Dataset.Tabular.register_pandas_dataframe(dataframe = dataset_df,\n target=az_store,\n name='Loan Dataset from DataFrame')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecb914b1f973069a3bf0c04d3cfc7aac491b059e | 206,094 | ipynb | Jupyter Notebook | examples/notebooks/Pivoting Data from Wide to Long.ipynb | fireddd/pyjanitor | e76db2c619527e9f0b63d9aefcde11e3225937b8 | [
"MIT"
] | null | null | null | examples/notebooks/Pivoting Data from Wide to Long.ipynb | fireddd/pyjanitor | e76db2c619527e9f0b63d9aefcde11e3225937b8 | [
"MIT"
] | null | null | null | examples/notebooks/Pivoting Data from Wide to Long.ipynb | fireddd/pyjanitor | e76db2c619527e9f0b63d9aefcde11e3225937b8 | [
"MIT"
] | null | null | null | 69.368563 | 6,296 | 0.445602 | [
[
[
"# Pivot_Longer : One function to cover transformations from wide to long form.",
"_____no_output_____"
]
],
[
[
"import janitor\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"Unpivoting(reshaping data from wide to long form) in Pandas is executed either through [pd.melt](https://pandas.pydata.org/docs/reference/api/pandas.melt.html), [pd.wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html), or [pd.DataFrame.stack](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.stack.html). However, there are scenarios where a few more steps are required to massage the data into the long form that we desire. Take the dataframe below, copied from [Stack Overflow](https://stackoverflow.com/questions/64061588/pandas-melt-multiple-columns-to-tabulate-a-dataset#64062002): ",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(\n {\n \"id\": [1, 2, 3],\n \"M_start_date_1\": [201709, 201709, 201709],\n \"M_end_date_1\": [201905, 201905, 201905],\n \"M_start_date_2\": [202004, 202004, 202004],\n \"M_end_date_2\": [202005, 202005, 202005],\n \"F_start_date_1\": [201803, 201803, 201803],\n \"F_end_date_1\": [201904, 201904, 201904],\n \"F_start_date_2\": [201912, 201912, 201912],\n \"F_end_date_2\": [202007, 202007, 202007],\n }\n )\n\ndf",
"_____no_output_____"
]
],
[
[
"Below is a [beautiful solution](https://stackoverflow.com/a/64062027/7175713), from Stack Overflow : ",
"_____no_output_____"
]
],
[
[
"df1 = df.set_index('id')\ndf1.columns = df1.columns.str.split('_', expand=True)\ndf1 = (df1.stack(level=[0,2,3])\n .sort_index(level=[0,1], ascending=[True, False])\n .reset_index(level=[2,3], drop=True)\n .sort_index(axis=1, ascending=False)\n .rename_axis(['id','cod'])\n .reset_index())\n\ndf1",
"_____no_output_____"
]
],
[
[
"We propose an alternative, based on [pandas melt](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.melt.html) that abstracts the reshaping mechanism, allows the user to focus on the task, can be applied to other scenarios, and is chainable : ",
"_____no_output_____"
]
],
[
[
"result = df.pivot_longer(\n index=\"id\", \n names_to=(\"cod\", \".value\"), \n names_pattern=\"(M|F)_(start|end)_.+\", \n sort_by_appearance=True,\n )\n\nresult",
"_____no_output_____"
],
[
"df1.equals(result)",
"_____no_output_____"
]
],
[
[
"[pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) is not a new idea; it is a combination of ideas from R's [tidyr](https://tidyr.tidyverse.org/reference/pivot_longer.html) and [data.table](https://rdatatable.gitlab.io/data.table/) and is built on the powerful pandas' [melt](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.melt.html) function. \n\n ",
"_____no_output_____"
],
[
"[pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) can melt dataframes easily; It is just a wrapper around pandas' [melt](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.melt.html).\n\n[Source Data](https://pandas.pydata.org/pandas-docs/stable/user_guide/reshaping.html#reshaping-by-melt)",
"_____no_output_____"
]
],
[
[
"index = pd.MultiIndex.from_tuples([('person', 'A'), ('person', 'B')])\n\ndf = pd.DataFrame({'first': ['John', 'Mary'],\n 'last': ['Doe', 'Bo'],\n 'height': [5.5, 6.0],\n 'weight': [130, 150]},\n index=index)\n \ndf",
"_____no_output_____"
],
[
"df.pivot_longer(index=['first','last'])",
"_____no_output_____"
]
],
[
[
"If you want the data unpivoted in order of appearance, you can set `sort_by_appearance` to ``True``:",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index=['first','last'],\n sort_by_appearance = True\n )",
"_____no_output_____"
]
],
[
[
"If you wish to reuse the original index, you can set `ignore_index` to ``False``; note that the index labels will be repeated as necessary:",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index=['first','last'],\n ignore_index = False\n )",
"_____no_output_____"
]
],
[
[
"You can also unpivot MultiIndex columns, the same way you would with pandas' [melt](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.melt.html#pandas.melt):\n\n[Source Data](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.melt.html#pandas.melt)",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({'A': {0: 'a', 1: 'b', 2: 'c'},\n 'B': {0: 1, 1: 3, 2: 5},\n 'C': {0: 2, 1: 4, 2: 6}})\ndf.columns = [list('ABC'), list('DEF')]\n\ndf",
"_____no_output_____"
],
[
"df.pivot_longer(\n index = [(\"A\", \"D\")],\n values_to = \"num\"\n)",
"_____no_output_____"
],
[
"df.pivot_longer(\n index = [(\"A\", \"D\")],\n column_names = [(\"B\", \"E\")]\n)",
"_____no_output_____"
]
],
[
[
"And just like [melt](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.melt.html#pandas.melt), you can unpivot on a specific level, with `column_level`:",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index = \"A\",\n column_names = \"B\",\n column_level = 0\n)",
"_____no_output_____"
]
],
[
[
"Note that when unpivoting MultiIndex columns, you need to pass a list of tuples to the ``index`` or ``column_names`` parameters.\n\n\nAlso, if ``names_sep`` or ``names_pattern`` is not None, then unpivoting on MultiIndex columns is not supported.",
"_____no_output_____"
],
[
"You can dynamically select columns, using regular expressions with the `janitor.patterns` function (inspired by R's data.table's [patterns](https://rdatatable.gitlab.io/data.table/reference/patterns.html) function, and is really just a wrapper around `re.compile`), especially if it is a lot of column names, and you are *lazy* like me 😄",
"_____no_output_____"
]
],
[
[
"url = 'https://github.com/tidyverse/tidyr/raw/master/data-raw/billboard.csv'\ndf = pd.read_csv(url)\n\ndf",
"_____no_output_____"
],
[
"# unpivot all columns that start with 'wk'\ndf.pivot_longer(column_names = janitor.patterns(\"^(wk)\"), \n names_to='week')",
"_____no_output_____"
]
],
[
[
"You can also use [pyjanitor's](https://pyjanitor.readthedocs.io/) [select_columns](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.select_columns.html#janitor.select_columns) syntax:",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(column_names = \"wk*\", \n names_to = 'week')",
"_____no_output_____"
]
],
[
[
"[pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) can also unpivot paired columns. In this regard, it is like pandas' [wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html), but with more flexibility and power. Let's look at an example from pandas' [wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html) docs : ",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({\n 'famid': [1, 1, 1, 2, 2, 2, 3, 3, 3],\n 'birth': [1, 2, 3, 1, 2, 3, 1, 2, 3],\n 'ht1': [2.8, 2.9, 2.2, 2, 1.8, 1.9, 2.2, 2.3, 2.1],\n 'ht2': [3.4, 3.8, 2.9, 3.2, 2.8, 2.4, 3.3, 3.4, 2.9]\n})\n\ndf",
"_____no_output_____"
]
],
[
[
"In the data above, the `height`(ht) is paired with `age`(numbers). [pd.wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html) can handle this easily:",
"_____no_output_____"
]
],
[
[
"pd.wide_to_long(df, stubnames='ht', i=['famid', 'birth'], j='age')",
"_____no_output_____"
]
],
[
[
"Now let's see how [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) handles this:",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(index=['famid','birth'],\n names_to=('.value', 'age'),\n names_pattern=r\"(ht)(\\d)\")",
"_____no_output_____"
]
],
[
[
"The first observable difference is that [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) is method chainable, while [pd.wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html) is not. Now, let's learn more about the `.value` variable.\n\n\nWhen `.value` is used in `names_to`, a pairing is created between ``names_to`` and ``names_pattern``. For the example above, we get this pairing :\n\n {\".value\": (\"ht\"), \"age\": (\\d)} \n\nThis tells the [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) function to keep values associated with `.value`(`ht`) as the column name, while values not associated with `.value`, in this case, the numbers, will be collated under a new column ``age``. Internally, pandas `str.extract` is used to get the capturing groups before reshaping. This level of abstraction, we believe, allows the user to focus on the task, and get things done faster.\n\nNote that if you want the data returned in order of appearance you can set `sort_by_appearance` to `True`:\n",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index = ['famid','birth'],\n names_to = ('.value', 'age'),\n names_pattern = r\"(ht)(\\d)\", \n sort_by_appearance = True,\n )",
"_____no_output_____"
]
],
[
[
"Note that you are likely to get more speed when `sort_by_appearance` is ``False``.\n\nNote also that the values in the `age` column are of `object` dtype. You can change the dtype, using pandas' [astype](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.astype.html) method.",
"_____no_output_____"
],
[
"We've seen already that [pd.wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html) handles this already and very well, so why bother? Let's look at another scenario where [pd.wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html) would need a few more steps. [Source Data](https://community.rstudio.com/t/pivot-longer-on-multiple-column-sets-pairs/43958):",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(\n {\n \"off_loc\": [\"A\", \"B\", \"C\", \"D\", \"E\", \"F\"],\n \"pt_loc\": [\"G\", \"H\", \"I\", \"J\", \"K\", \"L\"],\n \"pt_lat\": [\n 100.07548220000001,\n 75.191326,\n 122.65134479999999,\n 124.13553329999999,\n 124.13553329999999,\n 124.01028909999998,\n ],\n \"off_lat\": [\n 121.271083,\n 75.93845266,\n 135.043791,\n 134.51128400000002,\n 134.484374,\n 137.962195,\n ],\n \"pt_long\": [\n 4.472089953,\n -144.387785,\n -40.45611048,\n -46.07156181,\n -46.07156181,\n -46.01594293,\n ],\n \"off_long\": [\n -7.188632000000001,\n -143.2288569,\n 21.242563,\n 40.937416999999996,\n 40.78472,\n 22.905889000000002,\n ],\n }\n)\n\ndf",
"_____no_output_____"
]
],
[
[
"We can unpivot with [pd.wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html) by first reorganising the columns : ",
"_____no_output_____"
]
],
[
[
"df1 = df.copy()\ndf1.columns = [\"_\".join(col.split(\"_\")[::-1])\n for col in df1.columns]\ndf1",
"_____no_output_____"
]
],
[
[
"Now, we can unpivot : ",
"_____no_output_____"
]
],
[
[
"pd.wide_to_long(\n df1.reset_index(),\n stubnames=[\"loc\", \"lat\", \"long\"],\n sep=\"_\",\n i=\"index\",\n j=\"set\",\n suffix=\".+\",\n)",
"_____no_output_____"
]
],
[
[
"We can get the same transformed dataframe, with less lines, using [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) :",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n names_to = [\"set\", \".value\"], \n names_pattern = \"(.+)_(.+)\"\n )",
"_____no_output_____"
],
[
"# Another way to see the pairings, \n# to see what is linked to `.value`, \n\n# names_to = [\"set\", \".value\"]\n# names_pattern = \"(.+)_(.+)\"\n# column _names = off_loc\n# off_lat\n# off_long",
"_____no_output_____"
]
],
[
[
"Again, the key here is the `.value` symbol. Pairing `names_to` with `names_pattern` and its results from [pd.str.extract](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.Series.str.extract.html), we get : \n\n set--> (.+) --> [off, pt] and \n .value--> (.+) --> [loc, lat, long] \n \nAll values associated with `.value`(loc, lat, long) remain as column names, while values not associated with `.value`(off, pt) are lumped into a new column ``set``. \n\nNotice that we did not have to reset the index - [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) takes care of that internally; [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) allows you to focus on what you want, so you can get it and move on.",
"_____no_output_____"
],
[
"Note that the unpivoting could also have been executed with `names_sep`:",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n names_to = [\"set\", \".value\"], \n names_sep = \"_\",\n ignore_index = False,\n sort_by_appearance = True\n )",
"_____no_output_____"
]
],
[
[
"Let's look at another example, from [Stack Overflow](https://stackoverflow.com/questions/45123924/convert-pandas-dataframe-from-wide-to-long/45124130) : ",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame([{'a_1': 2, 'ab_1': 3, \n 'ac_1': 4, 'a_2': 5, \n 'ab_2': 6, 'ac_2': 7}])\ndf",
"_____no_output_____"
]
],
[
[
"The data above requires extracting `a`, `ab` and `ac` from `1` and `2`. This is another example of a paired column. We could solve this using [pd.wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html); infact there is a very good solution from [Stack Overflow](https://stackoverflow.com/a/45124775/7175713)",
"_____no_output_____"
]
],
[
[
"df1 = df.copy()\ndf1['id'] = df1.index\npd.wide_to_long(df1, ['a','ab','ac'],i='id',j='num',sep='_')",
"_____no_output_____"
]
],
[
[
"Or you could simply pass the buck to [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) : ",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n names_to = ('.value', 'num'), \n names_sep = '_'\n )",
"_____no_output_____"
]
],
[
[
"In the solution above, we used the `names_sep` argument, as it is more convenient. A few more examples to get you familiar with the `.value` symbol.\n\n[Source Data](https://stackoverflow.com/questions/55403008/pandas-partial-melt-or-group-melt)",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame([[1,1,2,3,4,5,6],\n [2,7,8,9,10,11,12]], \n columns=['id', 'ax','ay','az','bx','by','bz'])\n\ndf",
"_____no_output_____"
],
[
"df.pivot_longer(\n index = 'id', \n names_to = ('name', '.value'), \n names_pattern = '(.)(.)'\n )",
"_____no_output_____"
]
],
[
[
"For the code above `.value` is paired with `x`, `y`, `z`(which become the new column names), while `a`, `b` are unpivoted into the `name` column. ",
"_____no_output_____"
],
[
"In the dataframe below, we need to unpivot the data, keeping only the suffix `hi`, and pulling out the number between `A` and `g`. [Source Data](https://stackoverflow.com/questions/35929985/melt-a-data-table-with-a-column-pattern)",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame([{'id': 1, 'A1g_hi': 2, \n 'A2g_hi': 3, 'A3g_hi': 4, \n 'A4g_hi': 5}])\ndf",
"_____no_output_____"
],
[
"df.pivot_longer(\n index = 'id', \n names_to = ['time','.value'], \n names_pattern = \"A(\\d)g_(hi)\")",
"_____no_output_____"
]
],
[
[
"Let's see an example where we have multiple values in a paired column, and we wish to split them into separate columns. [Source Data](https://stackoverflow.com/questions/64107566/how-to-pivot-longer-and-populate-with-fields-from-column-names-at-the-same-tim?noredirect=1#comment113369419_64107566) : ",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(\n {\n \"Sony | TV | Model | value\": {0: \"A222\", 1: \"A234\", 2: \"A4345\"},\n \"Sony | TV | Quantity | value\": {0: 5, 1: 5, 2: 4},\n \"Sony | TV | Max-quant | value\": {0: 10, 1: 9, 2: 9},\n \"Panasonic | TV | Model | value\": {0: \"T232\", 1: \"S3424\", 2: \"X3421\"},\n \"Panasonic | TV | Quantity | value\": {0: 1, 1: 5, 2: 1},\n \"Panasonic | TV | Max-quant | value\": {0: 10, 1: 12, 2: 11},\n \"Sanyo | Radio | Model | value\": {0: \"S111\", 1: \"S1s1\", 2: \"S1s2\"},\n \"Sanyo | Radio | Quantity | value\": {0: 4, 1: 2, 2: 4},\n \"Sanyo | Radio | Max-quant | value\": {0: 9, 1: 9, 2: 10},\n }\n)\n\ndf",
"_____no_output_____"
]
],
[
[
"The goal is to reshape the data into long format, with separate columns for `Manufacturer`(Sony,...), `Device`(TV, Radio), `Model`(S3424, ...), ``maximum quantity`` and ``quantity``. \n\nBelow is the [accepted solution](https://stackoverflow.com/a/64107688/7175713) on Stack Overflow :",
"_____no_output_____"
]
],
[
[
"df1 = df.copy()\n# Create a multiIndex column header\ndf1.columns = pd.MultiIndex.from_arrays(\n zip(*df1.columns.str.split(\"\\s?\\|\\s?\"))\n)\n\n# Reshape the dataframe using \n# `set_index`, `droplevel`, and `stack`\n(df1.stack([0, 1])\n .droplevel(1, axis=1)\n .set_index(\"Model\", append=True)\n .rename_axis([None, \"Manufacturer\", \"Device\", \"Model\"])\n .sort_index(level=[1, 2, 3])\n .reset_index()\n .drop(\"level_0\", axis=1)\n )\n",
"_____no_output_____"
]
],
[
[
"Or, we could use [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer), along with `.value` in `names_to` and a regular expression in `names_pattern` : ",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n names_to = (\"Manufacturer\", \"Device\", \".value\"),\n names_pattern = r\"(.+)\\|(.+)\\|(.+)\\|.*\",\n )",
"_____no_output_____"
]
],
[
[
"The cleanup (removal of whitespace in the column names) is left as an exercise for the reader.",
"_____no_output_____"
],
[
"What if we are interested in unpivoting only a part of the entire dataframe? [Source Data](https://stackoverflow.com/questions/63044119/converting-wide-format-data-into-long-format-with-multiple-indices-and-grouped-d)",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({'time': [1, 2, 3], \n 'factor': ['a','a','b'],\n 'variable1': [0,0,0],\n 'variable2': [0,0,1],\n 'variable3': [0,2,0],\n 'variable4': [2,0,1],\n 'variable5': [1,0,1],\n 'variable6': [0,1,1], \n 'O1V1': [0,0.2,-0.3],\n 'O1V2': [0,0.4,-0.9],\n 'O1V3': [0.5,0.2,-0.6],\n 'O1V4': [0.5,0.2,-0.6],\n 'O1V5': [0,0.2,-0.3],\n 'O1V6': [0,0.4,-0.9],\n 'O1V7': [0.5,0.2,-0.6],\n 'O1V8': [0.5,0.2,-0.6], \n 'O2V1': [0,0.5,0.3],\n 'O2V2': [0,0.2,0.9],\n 'O2V3': [0.6,0.1,-0.3],\n 'O2V4': [0.5,0.2,-0.6],\n 'O2V5': [0,0.5,0.3],\n 'O2V6': [0,0.2,0.9],\n 'O2V7': [0.6,0.1,-0.3],\n 'O2V8': [0.5,0.2,-0.6], \n 'O3V1': [0,0.7,0.4],\n 'O3V2': [0.9,0.2,-0.3],\n 'O3V3': [0.5,0.2,-0.7],\n 'O3V4': [0.5,0.2,-0.6],\n 'O3V5': [0,0.7,0.4],\n 'O3V6': [0.9,0.2,-0.3],\n 'O3V7': [0.5,0.2,-0.7],\n 'O3V8': [0.5,0.2,-0.6]})\ndf",
"_____no_output_____"
]
],
[
[
"What is the task? This is copied verbatim from the source:\n\n<blockquote>Each row of the data frame represents a time period. There are multiple 'subjects' being monitored, namely O1, O2, and O3. Each subject has 8 variables being measured. I need to convert this data into long format where each row contains the information for one subject at a given time period, but with only the first 4 subject variables, as well as the extra information about this time period in columns 2-4, but not columns 5-8.</blockquote>",
"_____no_output_____"
],
[
"Below is the accepted solution, using [wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html):",
"_____no_output_____"
]
],
[
[
"df1 = df.rename(columns={x: x[2:]+x[1:2] for x in df.columns[df.columns.str.startswith('O')]})\n\ndf1 = pd.wide_to_long(df1, i=['time', 'factor']+[f'variable{i}' for i in range(1,7)], \n j='id', stubnames=[f'V{i}' for i in range(1,9)], suffix='.*')\n\ndf1 = (df1.reset_index()\n .drop(columns=[f'V{i}' for i in range(5,9)]\n +[f'variable{i}' for i in range(3,7)]))\n\ndf1",
"_____no_output_____"
]
],
[
[
"We can abstract the details and focus on the task with [pivot_longer]([pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer)):",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index = slice(\"time\", \"variable2\"),\n column_names = janitor.patterns(\".+V[1-4]$\"),\n names_to = (\"id\", \".value\"),\n names_pattern = \".(.)(.+)$\",\n sort_by_appearance = True\n)",
"_____no_output_____"
]
],
[
[
"One more example on the `.value` symbol for paired columns [Source Data](https://stackoverflow.com/questions/59477686/python-pandas-melt-single-column-into-two-seperate) : ",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({'id': [1, 2], \n 'A_value': [50, 33], \n 'D_value': [60, 45]})\ndf",
"_____no_output_____"
],
[
"df.pivot_longer(\n index = 'id', \n names_to = ('value_type', '.value'), \n names_sep = '_'\n )",
"_____no_output_____"
]
],
[
[
"There are scenarios where we need to unpivot the data, and group values within the column names under new columns. The values in the columns will not become new column names, so we do not need the `.value` symbol. Let's see an example below: [Source Data](https://stackoverflow.com/questions/59550804/melt-column-by-substring-of-the-columns-name-in-pandas-python)",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({'subject': [1, 2],\n 'A_target_word_gd': [1, 11],\n 'A_target_word_fd': [2, 12],\n 'B_target_word_gd': [3, 13],\n 'B_target_word_fd': [4, 14],\n 'subject_type': ['mild', 'moderate']})\n\ndf",
"_____no_output_____"
]
],
[
[
"In the dataframe above, `A` and `B` represent conditions, while the suffixes `gd` and `fd` represent value types. We are not interested in the words in the middle (`_target_word`). We could solve it this way (this is the chosen solution, copied from [Stack Overflow](https://stackoverflow.com/a/59550967/7175713)) : ",
"_____no_output_____"
]
],
[
[
"new_df =(pd.melt(df,\n id_vars=['subject_type','subject'], \n var_name='abc')\n .sort_values(by=['subject', 'subject_type'])\n )\nnew_df['cond']=(new_df['abc']\n .apply(lambda x: (x.split('_'))[0])\n )\nnew_df['value_type']=(new_df\n .pop('abc')\n .apply(lambda x: (x.split('_'))[-1])\n )\nnew_df\n",
"_____no_output_____"
]
],
[
[
"Or, we could just pass the buck to [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) : ",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index = [\"subject\", \"subject_type\"],\n names_to = (\"cond\", \"value_type\"),\n names_pattern = \"([A-Z]).*(gd|fd)\",\n)\n",
"_____no_output_____"
]
],
[
[
"In the code above, we pass in the new names of the columns to `names_to`('cond', 'value_type'), and pass the groups to be extracted as a regular expression to `names_pattern`. ",
"_____no_output_____"
],
[
"Here's another example where [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) abstracts the process and makes reshaping easy.\n\n\nIn the dataframe below, we would like to unpivot the data and separate the column names into individual columns(`vault` should be in an `event` column, `2012` should be in a `year` column and `f` should be in a `gender` column). [Source Data](https://dcl-wrangle.stanford.edu/pivot-advanced.html)",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(\n {\n \"country\": [\"United States\", \"Russia\", \"China\"],\n \"vault_2012_f\": [\n 48.132,\n 46.366,\n 44.266,\n ],\n \"vault_2012_m\": [46.632, 46.866, 48.316],\n \"vault_2016_f\": [\n 46.866,\n 45.733,\n 44.332,\n ],\n \"vault_2016_m\": [45.865, 46.033, 45.0],\n \"floor_2012_f\": [45.366, 41.599, 40.833],\n \"floor_2012_m\": [45.266, 45.308, 45.133],\n \"floor_2016_f\": [45.999, 42.032, 42.066],\n \"floor_2016_m\": [43.757, 44.766, 43.799],\n }\n )\ndf\n",
"_____no_output_____"
]
],
[
[
"We could achieve this with a combination of [pd.melt](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.melt.html) and pandas string methods (or janitor's [deconcatenate_columns](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.deconcatenate_column.html#janitor.deconcatenate_column) method); or we could, again, pass the buck to [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) : ",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index = \"country\",\n names_to = [\"event\", \"year\", \"gender\"],\n names_sep = \"_\",\n values_to = \"score\",\n)",
"_____no_output_____"
]
],
[
[
"Again, if you want the data returned in order of appearance, you can turn on the `sort_by_appearance` parameter:",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index = \"country\",\n names_to = [\"event\", \"year\", \"gender\"],\n names_sep = \"_\",\n values_to = \"score\",\n sort_by_appearance = True\n)",
"_____no_output_____"
]
],
[
[
"One more feature that [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) offers is to pass a list of regular expressions to `names_pattern`. This comes in handy when one single regex cannot encapsulate similar columns for reshaping to long form. This idea is inspired by the [melt](https://rdatatable.gitlab.io/data.table/reference/melt.data.table.html) function in R's [data.table](https://rdatatable.gitlab.io/data.table/). A couple of examples should make this clear.\n\n[Source Data](https://stackoverflow.com/questions/61138600/tidy-dataset-with-pivot-longer-multiple-columns-into-two-columns)",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(\n [{'title': 'Avatar',\n 'actor_1': 'CCH_Pound…',\n 'actor_2': 'Joel_Davi…',\n 'actor_3': 'Wes_Studi',\n 'actor_1_FB_likes': 1000,\n 'actor_2_FB_likes': 936,\n 'actor_3_FB_likes': 855},\n {'title': 'Pirates_of_the_Car…',\n 'actor_1': 'Johnny_De…',\n 'actor_2': 'Orlando_B…',\n 'actor_3': 'Jack_Daven…',\n 'actor_1_FB_likes': 40000,\n 'actor_2_FB_likes': 5000,\n 'actor_3_FB_likes': 1000},\n {'title': 'The_Dark_Knight_Ri…',\n 'actor_1': 'Tom_Hardy',\n 'actor_2': 'Christian…',\n 'actor_3': 'Joseph_Gor…',\n 'actor_1_FB_likes': 27000,\n 'actor_2_FB_likes': 23000,\n 'actor_3_FB_likes': 23000},\n {'title': 'John_Carter',\n 'actor_1': 'Daryl_Sab…',\n 'actor_2': 'Samantha_…',\n 'actor_3': 'Polly_Walk…',\n 'actor_1_FB_likes': 640,\n 'actor_2_FB_likes': 632,\n 'actor_3_FB_likes': 530},\n {'title': 'Spider-Man_3',\n 'actor_1': 'J.K._Simm…',\n 'actor_2': 'James_Fra…',\n 'actor_3': 'Kirsten_Du…',\n 'actor_1_FB_likes': 24000,\n 'actor_2_FB_likes': 11000,\n 'actor_3_FB_likes': 4000},\n {'title': 'Tangled',\n 'actor_1': 'Brad_Garr…',\n 'actor_2': 'Donna_Mur…',\n 'actor_3': 'M.C._Gainey',\n 'actor_1_FB_likes': 799,\n 'actor_2_FB_likes': 553,\n 'actor_3_FB_likes': 284}]\n)\n\ndf",
"_____no_output_____"
]
],
[
[
"Above, we have a dataframe of movie titles, actors, and their facebook likes. It would be great if we could transform this into a long form, with just the title, the actor names, and the number of likes. Let's look at a possible solution : \n\nFirst, we reshape the columns, so that the numbers appear at the end.",
"_____no_output_____"
]
],
[
[
"df1 = df.copy()\npat = r\"(?P<actor>.+)_(?P<num>\\d)_(?P<likes>.+)\"\nrepl = lambda m: f\"\"\"{m.group('actor')}_{m.group('likes')}_{m.group('num')}\"\"\"\ndf1.columns = df1.columns.str.replace(pat, repl)\ndf1",
"_____no_output_____"
]
],
[
[
"Now, we can reshape, using [pd.wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html) :",
"_____no_output_____"
]
],
[
[
"pd.wide_to_long(df1, \n stubnames = ['actor', 'actor_FB_likes'], \n i = 'title', \n j = 'group', \n sep = '_')",
"_____no_output_____"
]
],
[
[
"We could attempt to solve it with [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer), using the `.value` symbol : ",
"_____no_output_____"
]
],
[
[
"df1.pivot_longer(\n index = 'title', \n names_to = (\".value\", \"group\"), \n names_pattern = \"(.+)_(\\d)$\"\n )",
"_____no_output_____"
]
],
[
[
"What if we could just get our data in long form without the massaging? We know our data has a pattern to it --> it either ends in a number or *likes*. Can't we take advantage of that? Yes, we can(I know, I know; it sounds like a campaign slogan 🤪)",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index = 'title',\n names_to = (\"actor\", \"num_likes\"),\n names_pattern = ('\\d$', 'likes$'),\n )",
"_____no_output_____"
]
],
[
[
"A pairing of `names_to` and `names_pattern` results in :\n\n {\"actor\": '\\d$', \"num_likes\": 'likes$'}\n \nThe first regex looks for columns that end with a number, while the other looks for columns that end with *likes*. [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) will then look for columns that end with a number and lump all the values in those columns under the `actor` column, and also look for columns that end with *like* and combine all the values in those columns into a new column -> `num_likes`. Underneath the hood, [numpy select](https://numpy.org/doc/stable/reference/generated/numpy.select.html) and [pd.Series.str.contains](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.Series.str.contains.html) are used to pull apart the columns into the new columns. \n\nAgain, it is about the goal; we are not interested in the numbers (1,2,3), we only need the names of the actors, and their facebook likes. [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) aims to give as much flexibility as possible, in addition to ease of use, to allow the end user focus on the task. \n\nLet's take a look at another example. [Source Data](https://stackoverflow.com/questions/60439749/pair-wise-melt-in-pandas-dataframe) :",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({'id': [0, 1],\n 'Name': ['ABC', 'XYZ'],\n 'code': [1, 2],\n 'code1': [4, np.nan],\n 'code2': ['8', 5],\n 'type': ['S', 'R'],\n 'type1': ['E', np.nan],\n 'type2': ['T', 'U']})\n\ndf",
"_____no_output_____"
]
],
[
[
"We cannot directly use [pd.wide_to_long](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.wide_to_long.html) here without some massaging, as there is no definite suffix(the first `code` does not have a suffix), neither can we use `.value` here, again because there is no suffix. However, we can see a pattern where some columns start with `code`, and others start with `type`. Let's see how [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) solves this, using a sequence of regular expressions in the ``names_pattern`` argument : ",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index = [\"id\", \"Name\"],\n names_to = (\"code_all\", \"type_all\"), \n names_pattern = (\"^code\", \"^type\")\n )",
"_____no_output_____"
]
],
[
[
"The key here is passing the right regular expression, and ensuring the names in `names_to` is paired with the right regex in `names_pattern`; as such, every column that starts with `code` will be included in the new `code_all` column; the same happens to the `type_all` column. Easy and flexible, right? \n\nLet's explore another example, from [Stack Overflow](https://stackoverflow.com/questions/12466493/reshaping-multiple-sets-of-measurement-columns-wide-format-into-single-columns) :",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(\n [\n {\n \"ID\": 1,\n \"DateRange1Start\": \"1/1/90\",\n \"DateRange1End\": \"3/1/90\",\n \"Value1\": 4.4,\n \"DateRange2Start\": \"4/5/91\",\n \"DateRange2End\": \"6/7/91\",\n \"Value2\": 6.2,\n \"DateRange3Start\": \"5/5/95\",\n \"DateRange3End\": \"6/6/96\",\n \"Value3\": 3.3,\n }\n ])\n\ndf",
"_____no_output_____"
]
],
[
[
"In the dataframe above, we need to reshape the data to have a start date, end date and value. For the `DateRange` columns, the numbers are embedded within the string, while for `value` it is appended at the end. One possible solution is to reshape the columns so that the numbers are at the end :",
"_____no_output_____"
]
],
[
[
"df1 = df.copy()\npat = r\"(?P<head>.+)(?P<num>\\d)(?P<tail>.+)\"\nrepl = lambda m: f\"\"\"{m.group('head')}{m.group('tail')}{m.group('num')}\"\"\"\ndf1.columns = df1.columns.str.replace(pat,repl)\ndf1",
"_____no_output_____"
]
],
[
[
"Now, we can unpivot:",
"_____no_output_____"
]
],
[
[
"pd.wide_to_long(df1, \n stubnames = ['DateRangeStart', \n 'DateRangeEnd', \n 'Value'],\n i = 'ID', \n j = 'num')",
"_____no_output_____"
]
],
[
[
"Using the `.value` symbol in pivot_longer:",
"_____no_output_____"
]
],
[
[
"df1.pivot_longer(\n index = 'ID', \n names_to = [\".value\",'num'], \n names_pattern = \"(.+)(\\d)$\"\n )",
"_____no_output_____"
]
],
[
[
"Or, we could allow pivot_longer worry about the massaging; simply pass to `names_pattern` a list of regular expressions that match what we are after : ",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index = 'ID', \n names_to = (\"DateRangeStart\", \"DateRangeEnd\", \"Value\"), \n names_pattern = (\"Start$\", \"End$\", \"^Value\")\n )",
"_____no_output_____"
]
],
[
[
"The code above looks for columns that end with *Start*(`Start$`), aggregates all the values in those columns into `DateRangeStart` column, looks for columns that end with *End*(`End$`), aggregates all the values within those columns into `DateRangeEnd` column, and finally looks for columns that start with *Value*(`^Value`), and aggregates the values in those columns into the `Value` column. Just know the patterns, and pair them accordingly. Again, the goal is a focus on the task, to make it simple for the end user.",
"_____no_output_____"
],
[
"Let's look at another example [Source Data](https://stackoverflow.com/questions/64316129/how-to-efficiently-melt-multiple-columns-using-the-module-melt-in-pandas/64316306#64316306) :",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({'Activity': ['P1', 'P2'],\n 'General': ['AA', 'BB'],\n 'm1': ['A1', 'B1'],\n 't1': ['TA1', 'TB1'],\n 'm2': ['A2', 'B2'],\n 't2': ['TA2', 'TB2'],\n 'm3': ['A3', 'B3'],\n 't3': ['TA3', 'TB3']})\n\ndf",
"_____no_output_____"
]
],
[
[
"This is a [solution](https://stackoverflow.com/a/64316306/7175713) provided by yours truly : ",
"_____no_output_____"
]
],
[
[
" (pd.wide_to_long(df, \n i = [\"Activity\", \"General\"], \n stubnames = [\"t\", \"m\"], \n j = \"number\")\n .set_axis([\"Task\", \"M\"], \n axis = \"columns\")\n .droplevel(-1)\n .reset_index()\n )",
"_____no_output_____"
]
],
[
[
"Or, we could use [pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer), abstract the details, and focus on the task : ",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index = ['Activity','General'], \n names_pattern = ['^m','^t'],\n names_to = ['M','Task']\n )",
"_____no_output_____"
]
],
[
[
"Alright, one last example : \n\n\n[Source Data](https://stackoverflow.com/questions/64159054/how-do-you-pivot-longer-columns-in-groups)",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({'Name': ['John', 'Chris', 'Alex'],\n 'activity1': ['Birthday', 'Sleep Over', 'Track Race'],\n 'number_activity_1': [1, 2, 4],\n 'attendees1': [14, 18, 100],\n 'activity2': ['Sleep Over', 'Painting', 'Birthday'],\n 'number_activity_2': [4, 5, 1],\n 'attendees2': [10, 8, 5]})\n\ndf",
"_____no_output_____"
]
],
[
[
"The task here is to unpivot the data, and group the data under three new columns (\"activity\", \"number_activity\", and \"attendees\"). \n\nWe can see that there is a pattern to the data; let's create a list of regular expressions that match the patterns and pass to ``names_pattern``:",
"_____no_output_____"
]
],
[
[
"df.pivot_longer(\n index = 'Name',\n names_to = ('activity','number_activity','attendees'), \n names_pattern = (\"^activity\",\"^number_activity\",\"^attendees\")\n )\n",
"_____no_output_____"
]
],
[
[
"Alright, let's look at one final example:\n\n\n[Source Data](https://stackoverflow.com/questions/60387077/reshaping-and-melting-dataframe-whilst-picking-up-certain-regex)",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({'Location': ['Madrid', 'Madrid', 'Rome', 'Rome'],\n 'Account': ['ABC', 'XYX', 'ABC', 'XYX'],\n 'Y2019:MTD:January:Expense': [4354, 769867, 434654, 632556456],\n 'Y2019:MTD:January:Income': [56456, 32556456, 5214, 46724423],\n 'Y2019:MTD:February:Expense': [235423, 6785423, 235423, 46588]})\n\ndf",
"_____no_output_____"
],
[
"df.pivot_longer(index = ['Location','Account'],\n names_to=(\"year\", \"month\", \".value\"),\n names_pattern=r\"Y(.+):MTD:(.{3}).+(Income|Expense)\",\n sort_by_appearance=True)\n\n",
"_____no_output_____"
]
],
[
[
"[pivot_longer](https://pyjanitor.readthedocs.io/reference/janitor.functions/janitor.pivot_longer.html#janitor.pivot_longer) does not solve all problems; no function does. Its aim is to make it easy to unpivot dataframes from wide to long form, while offering a lot of flexibility and power.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
ecb919b7b1cfeeecb03e54b56428e0ccad547eb2 | 51,316 | ipynb | Jupyter Notebook | Bash/bash-for-analytics-master/02-uso-interactivo.ipynb | wmsm/Notas-de-clase-Ciencia-de-los-datos | 77297343f035bde92728d611a160736a2762bd2d | [
"MIT"
] | null | null | null | Bash/bash-for-analytics-master/02-uso-interactivo.ipynb | wmsm/Notas-de-clase-Ciencia-de-los-datos | 77297343f035bde92728d611a160736a2762bd2d | [
"MIT"
] | null | null | null | Bash/bash-for-analytics-master/02-uso-interactivo.ipynb | wmsm/Notas-de-clase-Ciencia-de-los-datos | 77297343f035bde92728d611a160736a2762bd2d | [
"MIT"
] | 1 | 2020-03-02T02:27:52.000Z | 2020-03-02T02:27:52.000Z | 20.826299 | 515 | 0.497876 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecb91eb755b332758d8a8a16a0d8539023526d11 | 7,548 | ipynb | Jupyter Notebook | notebooks/evolving_results.ipynb | douglaswinstonr/GA-PDP | 152a756fa19a4276a89995a5f3ef697c7ec1d056 | [
"MIT"
] | 1 | 2020-05-21T23:16:14.000Z | 2020-05-21T23:16:14.000Z | notebooks/evolving_results.ipynb | douglaswinstonr/ga-pdp | 152a756fa19a4276a89995a5f3ef697c7ec1d056 | [
"MIT"
] | null | null | null | notebooks/evolving_results.ipynb | douglaswinstonr/ga-pdp | 152a756fa19a4276a89995a5f3ef697c7ec1d056 | [
"MIT"
] | null | null | null | 30.934426 | 168 | 0.570747 | [
[
[
"import auxiliary_tools\nfrom tqdm import tqdm \nfrom geopy.distance import geodesic",
"_____no_output_____"
],
[
"import array\nimport random\nimport numpy as np\nimport json\nimport pickle\nimport numpy\nfrom math import sqrt\nfrom deap import algorithms\nfrom deap import base\nfrom deap import benchmarks\nfrom deap.benchmarks.tools import diversity, convergence, hypervolume\nfrom deap import creator\nfrom deap import tools",
"_____no_output_____"
],
[
"def generate_individual(creator, route_requests, rand_dist_min, rand_dist_max):\n individual = []\n for request in route_requests:\n \n rand_distance = random.randint(rand_dist_min, rand_dist_max)/1000\n rand_angle = random.randint(1, 360)\n \n gene = geodesic(kilometers=rand_distance).destination(request, rand_angle)[:2]\n \n individual.append(gene)\n individual = np.array(individual)\n return creator.individual(individual)",
"_____no_output_____"
],
[
"def mutation(individual, mutation_probability, rand_dist_min, rand_dist_max):\n mutated_individual = []\n for gene in individual:\n if random.random() < mutation_probability:\n rand_distance = random.randint(rand_dist_min, rand_dist_max)/1000\n rand_angle = random.randint(1, 360)\n \n mutated_gene = geodesic(kilometers=rand_distance).destination(gene, rand_angle)[:2]\n mutated_individual.append( mutated_gene )\n else:\n mutated_individual.append( gene )\n return creator.individual(np.array(mutated_individual))",
"_____no_output_____"
],
[
"def crossover(individual_a, individual_b, crossover_probability):\n child_a = []\n child_b = []\n\n for i, (gene_a, gene_b) in enumerate(zip(individual_a, individual_b)):\n if random.random() < crossover_probability:\n child_a.append(gene_b)\n child_b.append(gene_a)\n else:\n child_a.append(gene_a)\n child_b.append(gene_b)\n\n return (creator.individual(np.array(child_a)), creator.individual(np.array(child_b)))",
"_____no_output_____"
],
[
"def client_fitness(route_requests, individual):\n c_fitness = []\n for i in range(len(route_requests)):\n request_r = route_requests[i]\n request_origin = [request_r[0], request_r[1]]\n vs_individual = individual[i]\n vs_destination = vs_individual\n c_fitness.append(auxiliary_tools.getGeoDistanceETA_OSRM(request_origin, vs_destination, 5005, 'walking'))\n fitness_value = np.sum([f[0] for f in c_fitness])\n return fitness_value\n\ndef operator_fitness(individual, penalty_const):\n ori_dest = [(first, second) for first, second in zip(individual, individual[1:])]\n penalty_sum = 0\n for pair in ori_dest:\n if max(pair[0] != pair[1]) == True:\n penalty_sum+=penalty_const\n o_fitness = []\n for od_r in ori_dest:\n o_fitness.append(auxiliary_tools.getGeoDistanceETA_OSRM(od_r[0], od_r[1], 5004, 'driving'))\n \n fitness_value = np.sum([f[0] for f in o_fitness]) + penalty_sum\n return fitness_value\n\ndef fitness(individual, route_requests, penalty_const):\n import time\n# start_time = time.time()\n from pexecute.thread import ThreadLoom\n loom = ThreadLoom(max_runner_cap=10)\n \n loom.add_function(client_fitness, [route_requests, individual], {})\n loom.add_function(operator_fitness, [individual, penalty_const], {})\n\n output = loom.execute()\n client_f = output[0]['output']\n operator_f = output[1]['output']\n# print(\"--- %s seconds ---\" % round(time.time() - start_time))\n return client_f, operator_f",
"_____no_output_____"
],
[
"penalty_const = auxiliary_tools.getPenaltyConst(2)\n\nroute_requests = auxiliary_tools.loadPrep(2, 1)\n\ncrossover_probability = 0.4\nmutation_probability = 0.5\n\nrand_dist_min = 0\nrand_dist_max = 500\n\npopulation_size = 25\nnumber_generations = 100\n\nidx_evol = 5",
"# proposals: 123402\n# requests 121427\n# rides 46616\n"
],
[
"import time\nstart_time = time.time()",
"_____no_output_____"
],
[
"creator.create(\"min_fitness\", base.Fitness, weights=(-1.0, -1.0))\ncreator.create(\"individual\", list, fitness=creator.min_fitness)\n\ntoolbox = base.Toolbox()\n\ntoolbox.register(\"create_individual\", generate_individual, creator, route_requests=route_requests, rand_dist_min=rand_dist_min, rand_dist_max=rand_dist_max)\ntoolbox.register(\"initialize_population\", tools.initRepeat, list, toolbox.create_individual)\n\ntoolbox.register(\"evaluate\", fitness, route_requests=route_requests, penalty_const=penalty_const)\ntoolbox.register(\"crossover\", crossover, crossover_probability=crossover_probability)\ntoolbox.register(\"mutate\", mutation, mutation_probability=mutation_probability, rand_dist_min=rand_dist_min, rand_dist_max=rand_dist_min)\n\ntoolbox.register(\"select\", tools.selNSGA2)",
"_____no_output_____"
],
[
"data_ga = pickle.load(open(\"../../data_ga_evol_1.pkl\", \"rb\"))",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb93bbe785e861168a15e6e33d692d75d080c93 | 218,798 | ipynb | Jupyter Notebook | notebooks/model_validation.ipynb | smoh/kinesis | 452940768f1cb7a93f97302867ae4f02d772229a | [
"MIT"
] | 6 | 2019-06-07T15:03:06.000Z | 2020-12-19T21:57:15.000Z | notebooks/model_validation.ipynb | smoh/kinesis | 452940768f1cb7a93f97302867ae4f02d772229a | [
"MIT"
] | 3 | 2019-08-08T21:19:58.000Z | 2019-09-02T16:28:57.000Z | notebooks/model_validation.ipynb | smoh/kinesis | 452940768f1cb7a93f97302867ae4f02d772229a | [
"MIT"
] | 2 | 2019-07-25T20:50:04.000Z | 2019-08-08T16:59:05.000Z | 172.28189 | 35,380 | 0.878989 | [
[
[
"# Code validation with mock clusters\n\nThis notebook demonstrates how to make mock data of a cluster with or without linear velocity gradient and how to do the fitting with the mock data.\n\nNotes on dependences\n- [gapipes](https://github.com/smoh/gapipes): for custom pandas accessor `g` to get covariance matrices, astropy coordinate objects..\n- [arviz](https://github.com/arviz-devs/arviz): for visualization",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nfrom matplotlib import colors\nimport pandas as pd\nimport numpy as np\nfrom astropy.table import Table\nimport astropy.units as u\nimport astropy.coordinates as coords\nimport arviz as az\n\n# project dependencies\nimport gapipes as gp\nimport kinesis as kn",
"_____no_output_____"
],
[
"np.random.seed(18324)",
"_____no_output_____"
]
],
[
[
"## basic cluster model",
"_____no_output_____"
],
[
"### Basic cluster model with no RVs - astrometric radial velocity",
"_____no_output_____"
]
],
[
[
"N = 150 # number of sources\nb0 = np.array([17.7, 41.2, 13.3]) # pc\nv0 = np.array([-6.32, 45.24, 5.30]) # [vx, vy, vz] in km/s\nsigv = 1. # dispersion, km/s\n\ncl = kn.Cluster(v0, sigv, b0=b0)\\\n .sample_sphere(N=N, Rmax=5)\\\n .observe(cov=np.eye(3)*0.01)",
"_____no_output_____"
],
[
"fitter = kn.Fitter(include_T=False)",
"INFO:kinesis.models:Reading model from disk\n"
],
[
"df = cl.members.observed.copy()\nfit = fitter.fit(df, sample=False)\nprint(f\"v0, sigv = {fit['v0']}, {fit['sigv']:.4f}\")\nprint(f\"diff from truth: {fit['v0']-v0}, {fit['sigv']-sigv:.4f}\")",
"v0, sigv = [-6.05218859 45.77023958 5.55129011], 0.9072\ndiff from truth: [0.26781141 0.53023958 0.25129011], -0.0928\n"
],
[
"stanfit = fitter.fit(df)",
"_____no_output_____"
],
[
"# workaround for a bug in arviz instantiating InferenceData\nd = stanfit.extract(permuted=False, pars=['v0', 'sigv'])\nd = {k:np.swapaxes(v, 0, 1) for k,v in d.items()}\nazfit = az.from_dict(d)",
"_____no_output_____"
],
[
"az.plot_forest(azfit, kind='ridgeplot', var_names=['sigv']);",
"_____no_output_____"
],
[
"stanfit.to_dataframe()['divergent__'].sum()",
"_____no_output_____"
],
[
"az.summary(azfit)",
"_____no_output_____"
],
[
"v0_diff = stanfit['v0'] - cl.v0[None,:]\n\nplt.figure(figsize=(4,3))\nplt.hist(v0_diff, bins=64, histtype='step', density=True, label=['x','y','z']);\nplt.xlabel(\"diff from truth [km/s]\");\nplt.legend(loc='upper left');\nplt.axvline(0, c='gray', lw=.5);",
"_____no_output_____"
],
[
"az.plot_posterior(azfit, var_names=['sigv'], ref_val=cl.sigmav, figsize=(4,3));",
"_____no_output_____"
],
[
"# plt.figure(figsize=(4,3))\n# g = np.einsum('ni,nij,nj->n', a-r['a_model'], np.linalg.inv(C), a-r['a_model'])\n\n# plt.hist(g, 32, density=True);\n# plt.hist(g, np.logspace(0, 4, 32), density=True);\n# plt.xscale('log');\n# plt.yscale('log')",
"_____no_output_____"
]
],
[
[
"### Basic cluster model with partial RVs",
"_____no_output_____"
]
],
[
[
"N = 150\nb0 = np.array([17.7, 41.2, 13.3]) # pc\nv0 = np.array([-6.32, 45.24, 5.30])\nsigv = 1.\n\ncl = kn.Cluster(v0, sigv, b0=b0)\\\n .sample_sphere(N=N, Rmax=5)\\\n .observe(cov=np.eye(3)*0.01)\n\n# Give random half of the stars RV with 0.5 km/s uncertainty\nNrv = int(N*0.5)\nrv_error = 0.5\nirand = np.random.choice(np.arange(N), size=Nrv, replace=False)\n\ndf = cl.members.observed.copy()\ndf['radial_velocity'] = np.random.normal(cl.members.truth['radial_velocity'].values, rv_error)\ndf['radial_velocity_error'] = rv_error",
"_____no_output_____"
],
[
"df = cl.members.observed.copy()\nfit = fitter.fit(df, sample=False)\nprint(f\"v0, sigv = {fit['v0']}, {fit['sigv']:.4f}\")\nprint(f\"diff from truth: {fit['v0']-v0}, {fit['sigv']-sigv:.4f}\")",
"v0, sigv = [-5.92897309 46.06913061 5.49316135], 0.9835\ndiff from truth: [0.39102691 0.82913061 0.19316135], -0.0165\n"
],
[
"stanfit_partialrv = fitter.fit(df)",
"_____no_output_____"
],
[
"# workaround for a bug in arviz instantiating InferenceData\nd = stanfit_partialrv.extract(permuted=False, pars=['v0', 'sigv'])\nd = {k:np.swapaxes(v, 0, 1) for k,v in d.items()}\nazfit_partialrv = az.from_dict(d)",
"_____no_output_____"
],
[
"v0_diff0 = stanfit['v0'] - cl.v0[None,:]\nv0_diff = stanfit_partialrv['v0'] - cl.v0[None,:]\n\nplt.figure(figsize=(4,3))\ncolor = ['tab:blue', 'tab:green', 'tab:orange']\nplt.hist(v0_diff0, bins=64, histtype='step', density=True, color=color, lw=.5);\nplt.hist(v0_diff, bins=64, histtype='step', density=True, color=color, label=['x','y','z'], lw=2);\nplt.xlabel(\"diff from truth [km/s]\");\nplt.legend(loc='upper left');\nplt.axvline(0, c='gray', lw=.5);",
"_____no_output_____"
],
[
"axs = az.plot_posterior(azfit, var_names=['sigv'], ref_val=cl.sigmav, figsize=(4,3))\naz.plot_posterior(azfit_partialrv, var_names=['sigv'], ax=axs);",
"_____no_output_____"
]
],
[
[
"## Cluster with rotation",
"_____no_output_____"
]
],
[
[
"N = 150\nb0 = np.array([17.7, 41.2, 13.3]) # pc\nv0 = np.array([-6.32, 45.24, 5.30])\nsigv = 1.\nomegas = [40., 20., 50.]\n\ncl = kn.Cluster(v0, sigv, b0=b0, omegas=omegas)\\\n .sample_sphere(N=N, Rmax=15)\\\n .observe(cov=np.eye(3)*0.001)\ncl0 = kn.Cluster(v0, sigv, b0=b0).sample_at(cl.members.truth.g.icrs)\n\n# Give random half of the stars RV with 0.5 km/s uncertainty\nNrv = int(N*0.5)\nrv_error = 0.5\nirand = np.random.choice(np.arange(N), size=Nrv, replace=False)\n\ndf = cl.members.observed.copy()\ndf['radial_velocity'] = np.random.normal(cl.members.truth['radial_velocity'].values, rv_error)\ndf['radial_velocity_error'] = rv_error",
"_____no_output_____"
],
[
"m = kn.Fitter(include_T=True)\nfit = m.fit(df, sample=False, b0=b0)",
"INFO:kinesis.models:Reading model from disk\n"
],
[
"print(f\"v0, sigv = {fit['v0']}, {fit['sigv']:.4f}\")\nprint(f\"diff from truth: {fit['v0']-v0}, {fit['sigv']-sigv:.4f}\")\nprint(f\"{fit['T_param']}\")\nprint(f\"{cl.T}\")",
"v0, sigv = [-6.382561 45.25883593 5.52632739], 0.9632\ndiff from truth: [-0.062561 0.01883593 0.22632739], -0.0368\n[[ -5.47942742 -48.81690422 14.15808424]\n [ 27.02856238 6.34378433 -29.12948134]\n [ -4.03730063 37.49138498 -3.81971577]]\n[[ 0. -50. 20.]\n [ 50. 0. -40.]\n [-20. 40. 0.]]\n"
],
[
"# omegax = 0.5*(r['T_param'][2, 1] - r['T_param'][1, 2])\n# omegay = 0.5*(r['T_param'][0, 2] - r['T_param'][2, 0])\n# omegaz = 0.5*(r['T_param'][1, 0] - r['T_param'][0, 1])\n\n# w1 = 0.5*(r['T_param'][2, 1] + r['T_param'][1, 2])\n# w2 = 0.5*(r['T_param'][0, 2] + r['T_param'][2, 0])\n# w3 = 0.5*(r['T_param'][1, 0] + r['T_param'][0, 1])\n# w4 = r['T_param'][0, 0]\n# w5 = r['T_param'][1, 1]\n# kappa = w4 + w5 + r['T_param'][2, 2]\n# print(omegax, omegay, omegaz)\n# print(w1, w2, w3)\n# print(w4, w5)\n# print(kappa)",
"_____no_output_____"
],
[
"stanfit = m.fit(df, b0=b0)",
"_____no_output_____"
],
[
"azfit = az.from_pystan(stanfit)",
"_____no_output_____"
],
[
"v0_diff = stanfit['v0'] - cl.v0[None,:]\n\nplt.figure(figsize=(4,3))\ncolor = ['tab:blue', 'tab:green', 'tab:orange']\nplt.hist(v0_diff, bins=64, histtype='step', density=True, color=color, label=['x','y','z'], lw=2);\nplt.xlabel(\"diff from truth [km/s]\");\nplt.legend(loc='upper left');\nplt.axvline(0, c='gray', lw=.5);",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(3, 3, figsize=(8, 8), sharex=True, sharey=True)\nfig.subplots_adjust(bottom=0.1, top=0.95, right=0.95, left=0.15)\nax = ax.ravel()\nfor cax, cT, truth in zip(ax, stanfit[\"T_param\"].reshape((-1, 9)).T, cl.T.ravel()):\n cax.hist(cT, bins=32, density=True, histtype=\"step\")\n cax.axvline(truth, c=\"k\")\n cax.axvline(0, c=\"gray\", lw=0.5)\nfig.text(0.55, 0.05, \"m/s/pc\", ha=\"center\", va=\"center\", size=20)\nfig.text(0.05, 0.55, \"Density\", ha=\"center\", va=\"center\", rotation=90, size=20)\nfig.savefig(\"mock_posterior_T.png\")",
"_____no_output_____"
]
],
[
[
"## Cluster with rotation - ideal case\n\nTest the most ideal case when the cluster has small dispersion and all velocities are measured to high precision.",
"_____no_output_____"
]
],
[
[
"N = 150\nb0 = np.array([17.7, 41.2, 13.3]) # pc\nv0 = np.array([-6.32, 45.24, 5.30])\nsigv = 0.1 # small dispersion\nomegas = [40., 20., 50.]\n\ncl = kn.Cluster(v0, sigv, b0=b0, omegas=omegas)\\\n .sample_sphere(N=N, Rmax=15)\\\n .observe(cov=np.eye(3)*0.001)\ncl0 = kn.Cluster(v0, sigv, b0=b0).sample_at(cl.members.truth.g.icrs)\n\n# Give all stars observed RV with small uncertainty\nNrv = int(N)\nrv_error = 0.1\nirand = np.random.choice(np.arange(N), size=Nrv, replace=False)\n\ndf = cl.members.observed.copy()\ndf['radial_velocity'] = np.random.normal(cl.members.truth['radial_velocity'].values, rv_error)\ndf['radial_velocity_error'] = rv_error",
"_____no_output_____"
],
[
"m = kn.Fitter(include_T=True)\nfit = m.fit(df, sample=False, b0=b0)",
"INFO:kinesis.models:Reading model from disk\n"
],
[
"print(f\"v0, sigv = {fit['v0']}, {fit['sigv']:.4f}\")\nprint(f\"diff from truth: {fit['v0']-v0}, {fit['sigv']-sigv:.4f}\")\nprint(f\"{fit['T_param']}\")\nprint(f\"{cl.T}\")",
"v0, sigv = [-6.31158325 45.23302118 5.30911615], 0.0974\ndiff from truth: [ 0.00841675 -0.00697882 0.00911615], -0.0026\n[[ -0.53514835 -49.6406874 18.48196561]\n [ 52.87740998 3.07364283 -38.98217046]\n [-19.87667794 42.07821215 2.22755235]]\n[[ 0. -50. 20.]\n [ 50. 0. -40.]\n [-20. 40. 0.]]\n"
],
[
"stanfit = m.fit(df, b0=b0)",
"WARNING:pystan:8 of 4000 iterations saturated the maximum tree depth of 10 (0.2%)\nWARNING:pystan:Run again with max_treedepth larger than 10 to avoid saturation\n"
],
[
"azfit = az.from_pystan(stanfit, coords={'x':['v1','v2','v3']}, dims={'v0':['x']})\nazfit.posterior",
"_____no_output_____"
],
[
"azfit.sample_stats",
"_____no_output_____"
],
[
"v0_diff = stanfit['v0'] - cl.v0[None,:]\n\nplt.figure(figsize=(4,3))\ncolor = ['tab:blue', 'tab:green', 'tab:orange']\nplt.hist(v0_diff, bins=64, histtype='step', density=True, color=color, label=['x','y','z'], lw=2);\nplt.xlabel(\"diff from truth [km/s]\");\nplt.legend(loc='upper left');\nplt.axvline(0, c='gray', lw=.5);",
"_____no_output_____"
],
[
"az.plot_posterior(azfit, var_names=['sigv']);",
"_____no_output_____"
],
[
"az.plot_posterior(azfit, var_names=['T_param'], coords={'T_param_dim_0':[0], 'T_param_dim_1':[0]});",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(3, 3, figsize=(8, 8), sharex=True, sharey=True)\nfig.subplots_adjust(bottom=0.1, top=0.95, right=0.95, left=0.15)\nax = ax.ravel()\nfor cax, cT, truth in zip(ax, stanfit[\"T_param\"].reshape((-1, 9)).T, cl.T.ravel()):\n cax.hist(cT, bins=32, density=True, histtype=\"step\")\n cax.axvline(truth, c=\"k\")\n cax.axvline(0, c=\"gray\", lw=0.5)\nfig.text(0.55, 0.05, \"m/s/pc\", ha=\"center\", va=\"center\", size=20)\nfig.text(0.05, 0.55, \"Density\", ha=\"center\", va=\"center\", rotation=90, size=20)\nfig.savefig(\"mock_posterior_T.png\")",
"_____no_output_____"
],
[
"az.summary(azfit, var_names=['v0', 'sigv', 'T_param'])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb940705e18b9d499a195a5e49f7bf1fff01cd0 | 511,294 | ipynb | Jupyter Notebook | Python for Data Science/Code with Matplotlib/Visualization/Exploring Indicator's Across Countries/Exploring Indicator's Across Countries.ipynb | innat-2k14/Data-Science-In-Python | 3d1731d2cd61c5dfd33776d830997c5c7d8841d1 | [
"Apache-2.0"
] | 11 | 2018-04-28T20:49:10.000Z | 2019-01-03T07:45:16.000Z | Python for Data Science/Code with Matplotlib/Visualization/Exploring Indicator's Across Countries/Exploring Indicator's Across Countries.ipynb | innat-2k14/Data-Science-In-Python | 3d1731d2cd61c5dfd33776d830997c5c7d8841d1 | [
"Apache-2.0"
] | 2 | 2021-03-19T08:14:38.000Z | 2021-06-08T20:34:36.000Z | Python for Data Science/Code with Matplotlib/Visualization/Exploring Indicator's Across Countries/Exploring Indicator's Across Countries.ipynb | innat-2k14/Data-Science-In-Python | 3d1731d2cd61c5dfd33776d830997c5c7d8841d1 | [
"Apache-2.0"
] | 7 | 2019-10-26T13:53:00.000Z | 2020-12-02T06:03:39.000Z | 521.196738 | 208,328 | 0.931016 | [
[
[
"# Data Source: https://www.kaggle.com/worldbank/world-development-indicators\n# Folder: 'world-development-indicators'",
"_____no_output_____"
]
],
[
[
"<br><p style=\"font-family: Arial; font-size:3.75em;color:purple; font-style:bold\">\nWorld Development Indicators</p><br><br>\n# Exploring Data Visualization Using Matplotlib",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport random\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"data = pd.read_csv('./world-development-indicators/Indicators.csv')\ndata.shape",
"_____no_output_____"
],
[
"countries = data['CountryName'].unique().tolist()\nindicators = data['IndicatorName'].unique().tolist()",
"_____no_output_____"
]
],
[
[
"This is a really large dataset, at least in terms of the number of rows. But with 6 columns, what does this hold?",
"_____no_output_____"
]
],
[
[
"data.head(2)",
"_____no_output_____"
]
],
[
[
"Looks like it has different indicators for different countries with the year and value of the indicator. ",
"_____no_output_____"
],
[
"We already saw how the USA's per-capita CO2 production related to other countries, let's see if we can find some more indicators in common between countries. \n\nTo have some fun, we've picked countries randomly but then stored our random results so you can rerun it with the same answers.",
"_____no_output_____"
]
],
[
[
"# Filter 1\n\n# Picks years of choice\nyearsFilter = [2010, 2011, 2012, 2013, 2014]",
"_____no_output_____"
],
[
"# Filter 2 \n\n# Pick 2 countries randomly\ncountryFilter = random.sample(countries, 2)",
"_____no_output_____"
],
[
"countryFilter",
"_____no_output_____"
],
[
"# Filter 3\n\n# Pick 1 Indicator randomly\nindicatorsFilter = random.sample(indicators, 1)\nindicatorsFilter",
"_____no_output_____"
]
],
[
[
"# Problem: We're missing data.\n# Not all countries have all indicators for all years\n\nTo solve this, we'll need to find two countries and two indicators for which we have data over this time range.",
"_____no_output_____"
]
],
[
[
"filterMesh = (data['CountryName'] == countryFilter[0]) & (data['IndicatorName'].isin(indicatorsFilter)) & (data['Year'].isin(yearsFilter))\ncountry1_data = data.loc[filterMesh]",
"_____no_output_____"
],
[
"len(country1_data)",
"_____no_output_____"
],
[
"filterMesh = (data['CountryName'] == countryFilter[1]) & (data['IndicatorName'].isin(indicatorsFilter)) & (data['Year'].isin(yearsFilter))\ncountry2_data = data.loc[filterMesh]",
"_____no_output_____"
],
[
"len(country2_data)",
"_____no_output_____"
]
],
[
[
"# So let's pick indicators and countries which have data over this time range\n\nThe code below will randomly pick countries and indicators until it finds two countries who have data for an indicator over this time frame. We used it to produce the fixed values you see later, feel free to play with this yourself!",
"_____no_output_____"
]
],
[
[
"filteredData1 = []\nfilteredData2 = []",
"_____no_output_____"
],
[
"'''\nPlot: \ncountryFilter: pick two countries, \nindicatorsFilter: pick an indicator, \nyearsFilter: plot for years in yearsFilter\n'''\n# problem - not all countries have all indicators so if you go to visualize, it'll have missing data.\n# randomly picking two indicators and countries, do these countries have valid data over those years.\n# brings up the discussion of missing data/ missing fields\n# until we find full data\n\nwhile(len(filteredData1) < len(yearsFilter)-1):\n # pick new indicator\n indicatorsFilter = random.sample(indicators, 1)\n countryFilter = random.sample(countries, 2)\n # how many rows are there that have this country name, this indicator, and this year. Mesh gives bool vector\n filterMesh = (data['CountryName'] == countryFilter[0]) & (data['IndicatorName'].isin(indicatorsFilter)) & (data['Year'].isin(yearsFilter))\n # which rows have this condition to be true?\n filteredData1 = data.loc[filterMesh]\n filteredData1 = filteredData1[['CountryName','IndicatorName','Year','Value']]\n\n # need to print this only when our while condition is true\n if(len(filteredData1) < len(yearsFilter)-1):\n print('Skipping ... %s since very few rows (%d) found' % (indicatorsFilter, len(filteredData1)))\n",
"Skipping ... ['PPG, other private creditors (NFL, current US$)'] since very few rows (0) found\n"
],
[
"# What did we pick eventually ?\nindicatorsFilter",
"_____no_output_____"
],
[
"len(filteredData1)",
"_____no_output_____"
],
[
"'''\nCountry 2\n'''\n\nwhile(len(filteredData2) < len(filteredData1)-1):\n filterMesh = (data['CountryName'] == countryFilter[1]) & (data['IndicatorName'].isin(indicatorsFilter)) & (data['Year'].isin(yearsFilter))\n filteredData2 = data.loc[filterMesh]\n filteredData2 = filteredData2[['CountryName','IndicatorName','Year','Value']]\n #pick new indicator\n old = countryFilter[1]\n countryFilter[1] = random.sample(countries, 1)[0]\n \n if(len(filteredData2) < len(filteredData1)-1):\n print('Skipping ... %s, since very few rows (%d) found' % (old, len(filteredData2)))",
"_____no_output_____"
],
[
"if len(filteredData1) < len(filteredData2):\n small = len(filteredData1)\nelse:\n small = len(filteredData2)",
"_____no_output_____"
],
[
"filteredData1=filteredData1[0:small]\nfilteredData2=filteredData2[0:small]",
"_____no_output_____"
],
[
"filteredData1",
"_____no_output_____"
],
[
"filteredData2",
"_____no_output_____"
]
],
[
[
"<br><p style=\"font-family: Arial; font-size:2.75em;color:blue; font-style:bold\">\nMatplotlib: Additional Examples</p><br>",
"_____no_output_____"
],
[
"## Example: Scatter Plot\n\nNow that we have the data for two countries for the same indicators, let's plot them using a scatterplot.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\n\nfig, axis = plt.subplots()\n# Grid lines, Xticks, Xlabel, Ylabel\n\naxis.yaxis.grid(True)\naxis.set_title(indicatorsFilter[0],fontsize=10)\naxis.set_xlabel(filteredData1['CountryName'].iloc[0],fontsize=10)\naxis.set_ylabel(filteredData2['CountryName'].iloc[0],fontsize=10)\n\nX = filteredData1['Value']\nY = filteredData2['Value']\n\naxis.scatter(X, Y)",
"_____no_output_____"
]
],
[
[
"## Example: Line Plot\n\nHere we'll plot the indicator over time for each country.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nfig, ax = plt.subplots(figsize=(20, 10))\n\nax.set_ylim(min(0,filteredData1['Value'].min()), 2*filteredData1['Value'].max())\nax.set_title('Indicator Name : ' + indicatorsFilter[0])\nax.plot(filteredData1['Year'], filteredData1['Value'] , 'r--', label=filteredData1['CountryName'].unique()) \n\n# Add the legend\nlegend = plt.legend(loc = 'upper center', \n shadow=True,\n prop={'weight':'roman','size':'xx-large'})\n\n# Rectangle around the legend\nframe = legend.get_frame()\nframe.set_facecolor('.95')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Let's plot country #2",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfig, ax = plt.subplots(figsize=(20, 10))\n\n# Adjust the lower and upper limit to bring the graph at center\nax.set_ylim(min(0,filteredData2['Value'].min()), 2*filteredData2['Value'].max())\n\nax.set_title('Indicator Name : ' + indicatorsFilter[0])\nax.plot(filteredData2['Year'], filteredData2['Value'] ,\n label=filteredData2['CountryName'].unique(),\n color=\"purple\", lw=1, ls='-', \n marker='s', markersize=20, \n markerfacecolor=\"yellow\", markeredgewidth=4, markeredgecolor=\"blue\") \n\n# Add the legend\nlegend = plt.legend(loc = 'upper left', \n shadow=True,\n prop={'weight':'roman','size':'xx-large'})\n\n# Rectangle around the legend\nframe = legend.get_frame()\nframe.set_facecolor('.95')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Example (random datasets)",
"_____no_output_____"
]
],
[
[
"from mpl_toolkits.mplot3d import Axes3D\nfrom matplotlib import cm\nimport matplotlib.pyplot as plt\nimport numpy as np\n\ncountof_angles = 36\ncountof_radii = 8\n\n# array - radii\narray_rad = np.linspace(0.125, 1.0, countof_radii)\n\n# array - angles\narray_ang = np.linspace(0, 2*np.pi, countof_angles, endpoint=False)\n\n# repeat all angles per radius\narray_ang = np.repeat(array_ang[...,np.newaxis], countof_radii, axis=1)\n\n# from polar (radii, angles) coords to cartesian (x, y) coords\nx = np.append(0, (array_rad*np.cos(array_ang)).flatten())\ny = np.append(0, (array_rad*np.sin(array_ang)).flatten())\n\n# saddle shaped surface\nz = np.sin(-x*y)\n\nfig = plt.figure(figsize=(20,10))\nax = fig.gca(projection='3d')\n\nax.plot_trisurf(x, y, z, cmap=cm.autumn, linewidth=0.2)\n\nplt.show()\nfig.savefig(\"vis_3d.png\")",
"_____no_output_____"
]
],
[
[
"# Example (random dataset)",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nn_points = 200\n\nradius = 2 * np.random.rand(n_points)\nangles = 2 * (np.pi) * np.random.rand(n_points)\narea = 400 * (radius**2) * np.random.rand(n_points)\n\ncolors = angles\n\nfig = plt.figure(figsize=(20,10))\nax = plt.subplot(111, polar=True)\n\nc = plt.scatter(angles, radius, c=colors, s=area, cmap=plt.cm.hsv)\nc.set_alpha(1.95)\n\nplt.show()\nfig.savefig(\"vis_bubbleplot.png\")",
"_____no_output_____"
]
],
[
[
"# Example 4: Box Plots (random datasets)",
"_____no_output_____"
]
],
[
[
"np.random.seed(452)\n\n# Three ararys of 100 points each\nA1 = np.random.normal(0, 1, 100)\nA2 = np.random.normal(0, 2, 100)\nA3 = np.random.normal(0, 1.5, 100)\n\n# Concatenate the three arrays\ndata = [ A1, A2, A3 ]\n\nfig, axes = plt.subplots(nrows=1, ncols=2, figsize=(20, 10))\n\n\n# Box plot: Notch Shape\nbplot1 = axes[1].boxplot(data,\n notch=True, \n vert=True, # vertical aligmnent\n patch_artist=True) # color\n\n# Box plot: Rectangular \nbplot2 = axes[0].boxplot(data,\n vert=True, # vertical aligmnent\n patch_artist=True) # color\n\n\n\n\ncolors = ['tomato', 'darkorchid', 'lime']\n# more colors here: http://matplotlib.org/examples/color/named_colors.html\n\nfor bplot in (bplot1, bplot2):\n for patch, color in zip(bplot['boxes'], colors):\n patch.set_facecolor(color)\n\n# Grid lines, Xticks, Xlabel, Ylabel\nfor axis in axes:\n axis.yaxis.grid(True)\n axis.set_xticks([y for y in range(len(data))], )\n axis.set_xlabel('Sample X-Label',fontsize=20)\n axis.set_ylabel('Sample Y-Label',fontsize=20)\n\n \n# Xtick labels\nplt.setp(axes, xticks=[y for y in range(len(data))],\n xticklabels=['X1', 'X2', 'X3'])\n\nplt.show()\nfig.savefig(\"vis_boxplot.png\")",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb9476ef1a8a92e72e7914cc0bb6dad4901575f | 7,059 | ipynb | Jupyter Notebook | Leituras de PDF.ipynb | cccadet/leiturapdf | 4a51af199f2ccdbb03e9b00ccd2e645dad4d90b4 | [
"MIT"
] | null | null | null | Leituras de PDF.ipynb | cccadet/leiturapdf | 4a51af199f2ccdbb03e9b00ccd2e645dad4d90b4 | [
"MIT"
] | null | null | null | Leituras de PDF.ipynb | cccadet/leiturapdf | 4a51af199f2ccdbb03e9b00ccd2e645dad4d90b4 | [
"MIT"
] | null | null | null | 29.911017 | 100 | 0.482788 | [
[
[
"#install ImageMagick pay attention for versions (x86_x64)\n# ftp://ftp.imagemagick.org/pub/ImageMagick/binaries/ \n# http://docs.wand-py.org/en/latest/guide/install.html#install-imagemagick-on-windows\n\n#install ghostscript\n\n# https://github.com/tesseract-ocr/tesseract/wiki/Downloads\n# https://github.com/tesseract-ocr/tesseract/wiki/Data-Files\n# https://www.quora.com/How-do-I-install-Tesseract-OCR-on-windows-10",
"_____no_output_____"
],
[
"#from __future__ import print_function\nfrom wand.image import Image\n\nimport pytesseract as ocr\nocr.pytesseract.tesseract_cmd = r'C:\\Program Files\\Tesseract4-OCR\\tesseract.exe'\nimport numpy as np\nimport cv2\n\nimport PIL\n\nimport os\n\nimport pandas as pd",
"_____no_output_____"
],
[
"def convert_image_to_text(temp_file): \n # tipando a leitura para os canais de ordem RGB\n #imagem = Image.open('png/teste-1.png').convert('RGB')\n imagem = PIL.Image.open(temp_file).convert('RGB')\n\n # convertendo em um array editável de numpy[x, y, CANALS]\n npimagem = np.asarray(imagem).astype(np.uint8) \n\n # diminuição dos ruidos antes da binarização\n npimagem[:, :, 0] = 0 # zerando o canal R (RED)\n npimagem[:, :, 2] = 0 # zerando o canal B (BLUE)\n\n # atribuição em escala de cinza\n im = cv2.cvtColor(npimagem, cv2.COLOR_RGB2GRAY) \n\n # aplicação da truncagem binária para a intensidade\n # pixels de intensidade de cor abaixo de 127 serão convertidos para 0 (PRETO)\n # pixels de intensidade de cor acima de 127 serão convertidos para 255 (BRANCO)\n # A atrubição do THRESH_OTSU incrementa uma análise inteligente dos nivels de truncagem\n ret, thresh = cv2.threshold(im, 127, 255, cv2.THRESH_BINARY | cv2.THRESH_OTSU) \n\n # reconvertendo o retorno do threshold em um objeto do tipo PIL.Image\n binimagem = PIL.Image.fromarray(thresh) \n\n # chamada ao tesseract OCR por meio de seu wrapper\n phrase = ocr.image_to_string(binimagem, lang='por')\n\n # impressão do resultado\n #print(phrase) \n return phrase",
"_____no_output_____"
],
[
"directory = 'pdf/'\n \nfor file in os.listdir(directory):\n filename = os.fsdecode(file)\n if filename.endswith(\".pdf\"): \n temp_file = os.path.join(directory, filename)\n with Image(filename=temp_file, resolution = 720) as img:\n pages = len(img.sequence)\n image = Image(\n width=img.width,\n height=img.height * pages\n )\n\n for i in range(pages):\n image.composite(\n img.sequence[i],\n top=img.height * i,\n left=0\n )\n filename += '.png'\n with image.convert('png') as converted:\n converted.save(filename='png/'+filename)\n #break",
"_____no_output_____"
],
[
"directory = 'png/'\n\ndf_conv = pd.DataFrame(columns=['file','text'])\n\nfor file in os.listdir(directory):\n \n filename = os.fsdecode(file)\n if filename.endswith(\".png\"): \n temp_file = os.path.join(directory, filename)\n text = convert_image_to_text(temp_file)\n d = {'file': [temp_file], 'text': [text]}\n df = pd.DataFrame(data=d)\n frames = [df_conv, df]\n df_conv = pd.concat(frames)",
"_____no_output_____"
],
[
"df_conv",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb9481160d6855df8e7c7e6141f1ab5eea46ed5 | 182,063 | ipynb | Jupyter Notebook | text+mining.ipynb | sayanb3/python_for_datascience | 47dab4e99d81307e6e5c9aa24beb6442b007e0f4 | [
"Apache-2.0"
] | 12 | 2017-05-21T06:39:57.000Z | 2018-09-16T18:18:40.000Z | text+mining.ipynb | sayanb3/python_for_datascience | 47dab4e99d81307e6e5c9aa24beb6442b007e0f4 | [
"Apache-2.0"
] | null | null | null | text+mining.ipynb | sayanb3/python_for_datascience | 47dab4e99d81307e6e5c9aa24beb6442b007e0f4 | [
"Apache-2.0"
] | 76 | 2018-10-06T13:05:58.000Z | 2021-03-31T14:46:15.000Z | 158.178106 | 109,074 | 0.533469 | [
[
[
"import nltk\n",
"_____no_output_____"
],
[
"#nltk.download()",
"_____no_output_____"
],
[
"from nltk.book import *\n",
"*** Introductory Examples for the NLTK Book ***\nLoading text1, ..., text9 and sent1, ..., sent9\nType the name of the text or sentence to view it.\nType: 'texts()' or 'sents()' to list the materials.\ntext1: Moby Dick by Herman Melville 1851\ntext2: Sense and Sensibility by Jane Austen 1811\ntext3: The Book of Genesis\ntext4: Inaugural Address Corpus\ntext5: Chat Corpus\ntext6: Monty Python and the Holy Grail\ntext7: Wall Street Journal\ntext8: Personals Corpus\ntext9: The Man Who Was Thursday by G . K . Chesterton 1908\n"
],
[
"texts()",
"text1: Moby Dick by Herman Melville 1851\ntext2: Sense and Sensibility by Jane Austen 1811\ntext3: The Book of Genesis\ntext4: Inaugural Address Corpus\ntext5: Chat Corpus\ntext6: Monty Python and the Holy Grail\ntext7: Wall Street Journal\ntext8: Personals Corpus\ntext9: The Man Who Was Thursday by G . K . Chesterton 1908\n"
],
[
"sents()",
"sent1: Call me Ishmael .\nsent2: The family of Dashwood had long been settled in Sussex .\nsent3: In the beginning God created the heaven and the earth .\nsent4: Fellow - Citizens of the Senate and of the House of Representatives :\nsent5: I have a problem with people PMing me to lol JOIN\nsent6: SCENE 1 : [ wind ] [ clop clop clop ] KING ARTHUR : Whoa there !\nsent7: Pierre Vinken , 61 years old , will join the board as a nonexecutive director Nov. 29 .\nsent8: 25 SEXY MALE , seeks attrac older single lady , for discreet encounters .\nsent9: THE suburb of Saffron Park lay on the sunset side of London , as red and ragged as a cloud of sunset .\n"
],
[
"!pip install textblob",
"Requirement already satisfied: textblob in c:\\users\\kogentix\\anaconda3\\lib\\site-packages\nRequirement already satisfied: nltk>=3.1 in c:\\users\\kogentix\\anaconda3\\lib\\site-packages (from textblob)\nRequirement already satisfied: six in c:\\users\\kogentix\\anaconda3\\lib\\site-packages (from nltk>=3.1->textblob)\n"
],
[
"!pip install tweepy",
"Requirement already satisfied: tweepy in c:\\users\\kogentix\\anaconda3\\lib\\site-packages\nRequirement already satisfied: requests-oauthlib>=0.4.1 in c:\\users\\kogentix\\anaconda3\\lib\\site-packages (from tweepy)\nRequirement already satisfied: six>=1.7.3 in c:\\users\\kogentix\\anaconda3\\lib\\site-packages (from tweepy)\nRequirement already satisfied: requests>=2.4.3 in c:\\users\\kogentix\\anaconda3\\lib\\site-packages (from tweepy)\nRequirement already satisfied: oauthlib>=0.6.2 in c:\\users\\kogentix\\anaconda3\\lib\\site-packages (from requests-oauthlib>=0.4.1->tweepy)\n"
],
[
"from urllib import request\nurl = \"http://www.gutenberg.org/files/2554/2554-0.txt\"\nresponse = request.urlopen(url)\nraw = response.read().decode('utf8')\ntype(raw)",
"_____no_output_____"
],
[
"len(raw)\n",
"_____no_output_____"
],
[
"raw[:75]\n",
"_____no_output_____"
],
[
"tokens = nltk.word_tokenize(raw)\ntype(tokens)",
"_____no_output_____"
],
[
"tokens[1:10]",
"_____no_output_____"
],
[
"!pip install textmining",
"Requirement already satisfied: textmining in c:\\users\\kogentix\\anaconda3\\lib\\site-packages\n"
]
],
[
[
"#!pip install stemmer\nFor Python 3 from https://stackoverflow.com/questions/15717752/python3-3-importerror-with-textmining-1-0\n\nConverting the textmining code to python3 solved the problem for me. To do so, I manually download the text mining package from here:\n\nhttps://pypi.python.org/pypi/textmining/1.0\n\nunzipped it:\n\nunzip textmining-1.0.zip\nconverted the folder to python 3:\n\n2to3 --output-dir=textmining-1.0_v3 -W -n textmining-1.0\nand installed it:\n\ncd textmining-1.0_v3\nsudo python3 setup.py install",
"_____no_output_____"
]
],
[
[
"import textmining\n",
"_____no_output_____"
],
[
"tdm = textmining.TermDocumentMatrix()\n",
"_____no_output_____"
],
[
"tdm.add_doc(raw)",
"_____no_output_____"
],
[
"for row in tdm.rows(cutoff=1):\n print(row)",
"['the', 'project', 'gutenberg', 'ebook', 'of', 'crime', 'and', 'punishment', 'by', 'fyodor', 'dostoevsky', 'this', 'is', 'for', 'use', 'anyone', 'anywhere', 'at', 'no', 'cost', 'with', 'almost', 'restrictions', 'whatsoever', 'you', 'may', 'copy', 'it', 'give', 'away', 'or', 're', 'under', 'terms', 'license', 'included', 'online', 'www', 'org', 'title', 'author', 'release', 'date', 'march', 'last', 'updated', 'october', 'language', 'english', 'character', 'set', 'encoding', 'utf', 'start', 'produced', 'john', 'bickers', 'dagny', 'translated', 'constance', 'garnett', 'translator', 's', 'preface', 'a', 'few', 'words', 'about', 'himself', 'help', 'reader', 'to', 'understand', 'his', 'work', 'was', 'son', 'doctor', 'parents', 'were', 'very', 'hard', 'working', 'deeply', 'religious', 'people', 'but', 'so', 'poor', 'that', 'they', 'lived', 'their', 'five', 'children', 'in', 'only', 'two', 'rooms', 'father', 'mother', 'spent', 'evenings', 'reading', 'aloud', 'generally', 'from', 'books', 'serious', 'though', 'always', 'sickly', 'delicate', 'came', 'out', 'third', 'final', 'examination', 'petersburg', 'school', 'engineering', 'there', 'he', 'had', 'already', 'begun', 'first', 'folk', 'story', 'published', 'poet', 'nekrassov', 'review', 'received', 'acclamations', 'shy', 'unknown', 'youth', 'found', 'instantly', 'something', 'celebrity', 'brilliant', 'successful', 'career', 'seemed', 'open', 'before', 'him', 'those', 'hopes', 'soon', 'dashed', 'arrested', 'neither', 'temperament', 'nor', 'conviction', 'revolutionist', 'one', 'little', 'group', 'young', 'men', 'who', 'met', 'together', 'read', 'fourier', 'proudhon', 'accused', 'taking', 'part', 'conversations', 'against', 'censorship', 'letter', 'byelinsky', 'gogol', 'knowing', 'intention', 'up', 'printing', 'press', 'nicholas', 'i', 'stern', 'just', 'man', 'as', 'maurice', 'baring', 'calls', 'enough', 'condemned', 'death', 'after', 'eight', 'months', 'imprisonment', 'twenty', 'others', 'taken', 'semyonovsky', 'square', 'be', 'shot', 'writing', 'brother', 'mihail', 'says', 'snapped', 'over', 'our', 'heads', 'made', 'us', 'put', 'on', 'white', 'shirts', 'worn', 'persons', 'thereupon', 'we', 'bound', 'threes', 'stakes', 'suffer', 'execution', 'being', 'row', 'concluded', 'minutes', 'life', 'me', 'thought', 'your', 'dear', 'ones', 'contrived', 'kiss', 'plestcheiev', 'dourov', 'next', 'bid', 'them', 'farewell', 'suddenly', 'troops', 'beat', 'tattoo', 'unbound', 'brought', 'back', 'upon', 'scaffold', 'informed', 'majesty', 'spared', 'lives', 'sentence', 'commuted', 'labour', 'prisoners', 'grigoryev', 'went', 'mad', 'untied', 'never', 'regained', 'sanity', 'intense', 'suffering', 'experience', 'left', 'lasting', 'stamp', 'mind', 'temper', 'led', 'end', 'accept', 'every', 'resignation', 'regard', 'blessing', 'own', 'case', 'constantly', 'recurs', 'subject', 'writings', 'describes', 'awful', 'agony', 'insists', 'cruelty', 'inflicting', 'such', 'torture', 'then', 'followed', 'four', 'years', 'penal', 'servitude', 'company', 'common', 'criminals', 'siberia', 'where', 'began', 'dead', 'house', 'some', 'service', 'disciplinary', 'battalion', 'shown', 'signs', 'obscure', 'nervous', 'disease', 'arrest', 'now', 'developed', 'into', 'violent', 'attacks', 'epilepsy', 'which', 'suffered', 'rest', 'fits', 'occurred', 'three', 'times', 'year', 'more', 'frequent', 'periods', 'great', 'strain', 'allowed', 'return', 'russia', 'started', 'journal', 'vremya', 'forbidden', 'through', 'misunderstanding', 'lost', 'wife', 'terrible', 'poverty', 'yet', 'took', 'payment', 'debts', 'another', 'epoch', 'within', 'also', 'prohibited', 'weighed', 'down', 'debt', 'family', 'dependent', 'forced', 'write', 'heart', 'breaking', 'speed', 'said', 'have', 'corrected', 'later', 'much', 'softened', 'tenderness', 'devotion', 'second', 'june', 'famous', 'speech', 'unveiling', 'monument', 'pushkin', 'moscow', 'extraordinary', 'demonstrations', 'love', 'honour', 'died', 'grave', 'vast', 'multitude', 'mourners', 'gave', 'hapless', 'funeral', 'king', 'still', 'probably', 'most', 'widely', 'writer', 'russian', 'critic', 'seeks', 'explain', 'feeling', 'inspired', 'ourselves', 'blood', 'bone', 'has', 'seen', 'than', 'insight', 'impresses', 'wisdom', 'seek', 'learn', 'how', 'live', 'all', 'other', 'gifts', 'nature', 'won', 'became', 'chapter', 'an', 'exceptionally', 'hot', 'evening', 'early', 'july', 'garret', 'lodged', 'place', 'walked', 'slowly', 'hesitation', 'towards', 'k', 'bridge', 'successfully', 'avoided', 'meeting', 'landlady', 'staircase', 'roof', 'high', 'storied', 'like', 'cupboard', 'room', 'provided', 'dinners', 'attendance', 'floor', 'below', 'time', 'obliged', 'pass', 'her', 'kitchen', 'door', 'invariably', 'stood', 'each', 'passed', 'sick', 'frightened', 'scowl', 'feel', 'ashamed', 'hopelessly', 'afraid', 'not', 'because', 'cowardly', 'abject', 'quite', 'contrary', 'past', 'been', 'overstrained', 'irritable', 'condition', 'verging', 'hypochondria', 'become', 'completely', 'absorbed', 'isolated', 'fellows', 'dreaded', 'crushed', 'anxieties', 'position', 'late', 'ceased', 'weigh', 'given', 'attending', 'matters', 'practical', 'importance', 'desire', 'do', 'nothing', 'any', 'could', 'real', 'terror', 'stopped', 'stairs', 'listen', 'trivial', 'irrelevant', 'gossip', 'pestering', 'demands', 'threats', 'complaints', 'rack', 'brains', 'excuses', 'prevaricate', 'lie', 'rather', 'would', 'creep', 'cat', 'slip', 'unseen', 'however', 'coming', 'street', 'acutely', 'aware', 'fears', 'want', 'attempt', 'thing', 'am', 'these', 'trifles', 'odd', 'smile', 'hm', 'yes', 'hands', 'lets', 'cowardice', 'axiom', 'interesting', 'know', 'what', 'are', 'new', 'step', 'uttering', 'word', 'fear', 'talking', 'too', 'chatter', 'perhaps', 've', 'learned', 'month', 'lying', 'days', 'my', 'den', 'thinking', 'jack', 'giant', 'killer', 'why', 'going', 'capable', 'simply', 'fantasy', 'amuse', 'myself', 'plaything', 'maybe', 'heat', 'airlessness', 'bustle', 'plaster', 'scaffolding', 'bricks', 'dust', 'special', 'stench', 'familiar', 'unable', 'get', 'town', 'summer', 'worked', 'painfully', 'overwrought', 'nerves', 'insufferable', 'pot', 'houses', 'particularly', 'numerous', 'drunken', 'whom', 'continually', 'although', 'day', 'completed', 'revolting', 'misery', 'picture', 'expression', 'profoundest', 'disgust', 'gleamed', 'moment', 'refined', 'face', 'way', 'handsome', 'above', 'average', 'height', 'slim', 'well', 'built', 'beautiful', 'dark', 'eyes', 'brown', 'hair', 'sank', 'deep', 'accurately', 'speaking', 'complete', 'blankness', 'along', 'observing', 'caring', 'observe', 'mutter', 'habit', 'confessed', 'moments', 'conscious', 'ideas', 'sometimes', 'tangle', 'weak', 'scarcely', 'tasted', 'food', 'badly', 'dressed', 'even', 'accustomed', 'shabbiness', 'rags', 'quarter', 'shortcoming', 'dress', 'created', 'surprise', 'owing', 'proximity', 'hay', 'market', 'number', 'establishments', 'bad', 'preponderance', 'trading', 'class', 'population', 'crowded', 'streets', 'alleys', 'types', 'various', 'figure', 'queer', 'caused', 'accumulated', 'bitterness', 'contempt', 'spite', 'fastidiousness', 'minded', 'least', 'different', 'matter', 'when', 'acquaintances', 'former', 'fellow', 'students', 'indeed', 'disliked', 'reason', 'somewhere', 'huge', 'waggon', 'dragged', 'heavy', 'dray', 'horse', 'shouted', 'drove', 'hey', 'german', 'hatter', 'bawling', 'top', 'voice', 'pointing', 'clutched', 'tremulously', 'hat', 'tall', 'round', 'zimmerman', 'rusty', 'age', 'torn', 'bespattered', 'brimless', 'bent', 'side', 'unseemly', 'fashion', 'shame', 'akin', 'overtaken', 'knew', 'muttered', 'confusion', 'worst', 'stupid', 'detail', 'might', 'spoil', 'whole', 'plan', 'noticeable', 'looks', 'absurd', 'makes', 'ought', 'wear', 'cap', 'sort', 'old', 'pancake', 'grotesque', 'nobody', 'wears', 'noticed', 'mile', 'off', 'remembered', 'remember', 'clue', 'business', 'should', 'conspicuous', 'possible', 'ruin', 'everything', 'far', 'go', 'many', 'steps', 'gate', 'lodging', 'exactly', 'seven', 'hundred', 'thirty', 'counted', 'once', 'dreams', 'faith', 'tantalising', 'hideous', 'daring', 'recklessness', 'look', 'differently', 'monologues', 'jeered', 'impotence', 'indecision', 'involuntarily', 'come', 'dream', 'exploit', 'attempted', 'did', 'realise', 'positively', 'rehearsal', 'excitement', 'grew', 'sinking', 'tremor', 'looked', 'canal', 'let', 'tiny', 'tenements', 'inhabited', 'kinds', 'tailors', 'locksmiths', 'cooks', 'germans', 'sorts', 'girls', 'picking', 'living', 'best', 'petty', 'clerks', 'etc', 'continual', 'gates', 'courtyards', 'keepers', 'employed', 'building', 'glad', 'meet', 'none', 'slipped', 'unnoticed', 'right', 'narrow', 'liked', 'surroundings', 'darkness', 'inquisitive', 'if', 'scared', 'somehow', 'really', 'asking', 'reached', 'fourth', 'storey', 'progress', 'barred', 'porters', 'engaged', 'moving', 'furniture', 'flat', 'occupied', 'clerk', 'civil', 'untenanted', 'except', 'woman', 'good', 'anyway', 'rang', 'bell', 'faint', 'tinkle', 'tin', 'copper', 'flats', 'bells', 'ring', 'forgotten', 'note', 'its', 'peculiar', 'remind', 'bring', 'clearly', 'terribly', 'while', 'opened', 'crack', 'eyed', 'visitor', 'evident', 'distrust', 'glittering', 'seeing', 'landing', 'she', 'bolder', 'wide', 'stepped', 'entry', 'partitioned', 'facing', 'silence', 'looking', 'inquiringly', 'diminutive', 'withered', 'sixty', 'sharp', 'malignant', 'nose', 'colourless', 'somewhat', 'grizzled', 'thickly', 'smeared', 'oil', 'wore', 'kerchief', 'thin', 'long', 'neck', 'hen', 'leg', 'knotted', 'flannel', 'rag', 'hung', 'flapping', 'shoulders', 'mangy', 'fur', 'cape', 'yellow', 'coughed', 'groaned', 'instant', 'must', 'gleam', 'mistrust', 'again', 'raskolnikov', 'student', 'here', 'ago', 'haste', 'half', 'bow', 'remembering', 'polite', 'sir', 'distinctly', 'keeping', 'inquiring', 'same', 'errand', 'continued', 'disconcerted', 'surprised', 'notice', 'uneasy', 'paused', 'hesitating', 'letting', 'front', 'paper', 'walls', 'geraniums', 'muslin', 'curtains', 'windows', 'brightly', 'lighted', 'setting', 'sun', 'will', 'shine', 'flashed', 'chance', 'rapid', 'glance', 'scanned', 'trying', 'arrangement', 'wood', 'consisted', 'sofa', 'wooden', 'oval', 'table', 'dressing', 'glass', 'fixed', 'between', 'chairs', 'penny', 'prints', 'frames', 'representing', 'damsels', 'birds', 'corner', 'light', 'burning', 'small', 'ikon', 'clean', 'polished', 'shone', 'lizaveta', 'speck', 'spiteful', 'widows', 'finds', 'cleanliness', 'stole', 'curious', 'cotton', 'curtain', 'leading', 'bed', 'chest', 'drawers', 'severely', 'standing', 'straight', 'pawn', 'drew', 'pocket', 'fashioned', 'silver', 'watch', 'engraved', 'globe', 'chain', 'steel', 'pledge', 'yesterday', 'interest', 'wait', 'please', 'sell', 'alyona', 'ivanovna', 'worth', 'anything', 'roubles', 'buy', 'jeweler', 'rouble', 'shall', 'redeem', 'getting', 'money', 'advance', 'cried', 'yourself', 'handed', 'angry', 'point', 'checked', 'nowhere', 'else', 'object', 'hand', 'roughly', 'fumbled', 'keys', 'disappeared', 'behind', 'alone', 'middle', 'listened', 'inquisitively', 'hear', 'unlocking', 'drawer', 'reflected', 'carries', 'bunch', 'key', 'big', 'notches', 'can', 't', 'strong', 'box', 'boxes', 'degrading', 'say', 'ten', 'copecks', 'take', 'fifteen', 'lent', 'owe', 'reckoning', 'altogether', 'dispute', 'hurry', 'wanted', 'bringing', 'valuable', 'cigarette', 'friend', 'broke', 'talk', 'bye', 'home', 'sister', 'asked', 'casually', 'passage', 'yours', 'oh', 'particular', 'quick', 'short', 'struck', 'god', 'loathsome', 'possibly', 'nonsense', 'rubbish', 'added', 'resolutely', 'atrocious', 'head', 'filthy', 'things', 'disgusting', 'exclamations', 'express', 'agitation', 'repulsion', 'oppress', 'pitch', 'definite', 'form', 'escape', 'wretchedness', 'pavement', 'regardless', 'passers', 'jostling', 'senses', 'close', 'tavern', 'entered', 'basement', 'abusing', 'supporting', 'mounted', 'without', 'stopping', 'think', 'till', 'felt', 'giddy', 'tormented', 'thirst', 'longed', 'drink', 'cold', 'beer', 'attributed', 'sudden', 'weakness', 'sat', 'sticky', 'dirty', 'ordered', 'eagerly', 'drank', 'glassful', 'easier', 'thoughts', 'clear', 'hopefully', 'worry', 'physical', 'derangement', 'piece', 'dry', 'bread', 'brain', 'stronger', 'clearer', 'firm', 'phew', 'utterly', 'scornful', 'reflection', 'cheerful', 'free', 'burden', 'gazed', 'friendly', 'dim', 'foreboding', 'happier', 'frame', 'normal', 'besides', 'consisting', 'girl', 'concertina', 'gone', 'departure', 'quiet', 'empty', 'appeared', 'artisan', 'drunk', 'extremely', 'sitting', 'companion', 'stout', 'grey', 'beard', 'full', 'skirted', 'coat', 'dropped', 'asleep', 'bench', 'sleep', 'cracking', 'fingers', 'arms', 'apart', 'upper', 'body', 'bounding', 'hummed', 'meaningless', 'refrain', 'recall', 'lines', 'fondly', 'loved', 'waking', 'walking', 'used', 'shared', 'enjoyment', 'silent', 'positive', 'hostility', 'manifestations', 'retired', 'government', 'sipping', 'ii', 'crowds', 'society', 'especially', 'weary', 'concentrated', 'gloomy', 'world', 'whatever', 'filthiness', 'stay', 'master', 'establishment', 'frequently', 'main', 'jaunty', 'tarred', 'boots', 'red', 'turn', 'tops', 'view', 'person', 'horribly', 'greasy', 'black', 'satin', 'waistcoat', 'cravat', 'iron', 'lock', 'counter', 'boy', 'fourteen', 'younger', 'lay', 'sliced', 'cucumber', 'pieces', 'dried', 'fish', 'chopped', 'smelling', 'insufferably', 'fumes', 'spirits', 'atmosphere', 'make', 'meetings', 'strangers', 'spoken', 'impression', 'distance', 'often', 'recalled', 'afterwards', 'ascribed', 'presentiment', 'repeatedly', 'partly', 'doubt', 'latter', 'staring', 'persistently', 'obviously', 'anxious', 'enter', 'conversation', 'including', 'keeper', 'showing', 'shade', 'condescending', 'station', 'culture', 'inferior', 'useless', 'converse', 'fifty', 'bald', 'medium', 'stoutly', 'bloated', 'drinking', 'greenish', 'tinge', 'swollen', 'eyelids', 'keen', 'reddish', 'chinks', 'strange', 'intelligence', 'madness', 'wearing', 'ragged', 'buttons', 'missing', 'buttoned', 'evidently', 'clinging', 'trace', 'respectability', 'crumpled', 'shirt', 'covered', 'spots', 'stains', 'protruded', 'canvas', 'moustache', 'unshaven', 'chin', 'stiff', 'greyish', 'brush', 'respectable', 'official', 'manner', 'restless', 'ruffled', 'drop', 'dejectedly', 'resting', 'elbows', 'stained', 'loudly', 'venture', 'honoured', 'engage', 'forasmuch', 'exterior', 'command', 'respect', 'admonishes', 'education', 'respected', 'conjunction', 'genuine', 'sentiments', 'titular', 'counsellor', 'rank', 'marmeladov', 'name', 'bold', 'inquire', 'studying', 'answered', 'grandiloquent', 'style', 'speaker', 'directly', 'addressed', 'momentary', 'actually', 'immediately', 'habitual', 'aversion', 'stranger', 'approached', 'approach', 'formerly', 'm', 'immense', 'tapped', 'forehead', 'self', 'approval', 'attended', 'institution', 'allow', 'got', 'staggered', 'jug', 'beside', 'sideways', 'spoke', 'fluently', 'boldly', 'occasionally', 'losing', 'thread', 'sentences', 'drawling', 'pounced', 'greedily', 'soul', 'solemnity', 'vice', 'true', 'saying', 'drunkenness', 'virtue', 'truer', 'beggary', 'retain', 'innate', 'nobility', 'chased', 'human', 'stick', 'swept', 'broom', 'humiliating', 'ready', 'humiliate', 'hence', 'mr', 'lebeziatnikov', 'beating', 'ask', 'question', 'simple', 'curiosity', 'ever', 'night', 'barge', 'neva', 'happened', 'mean', 'fifth', 'slept', 'filled', 'emptied', 'bits', 'fact', 'clothes', 'sticking', 'probable', 'undressed', 'washed', 'fat', 'nails', 'excite', 'general', 'languid', 'boys', 'fell', 'sniggering', 'innkeeper', 'apparently', 'purpose', 'funny', 'yawning', 'lazily', 'dignity', 'likely', 'acquired', 'flown', 'speeches', 'entering', 'develops', 'necessity', 'drunkards', 'sharply', 'kept', 'order', 'drinkers', 'try', 'justify', 'themselves', 'obtain', 'consideration', 'pronounced', 'don', 'aren', 'duty', 'addressing', 'exclusively', 'does', 'ache', 'worm', 'didn', 'excuse', 'petition', 'loan', 'fullest', 'sense', 'beforehand', 'instance', 'certainty', 'reputable', 'exemplary', 'citizen', 'knows', 'course', 'shan', 'pay', 'compassion', 'keeps', 'modern', 'explained', 'nowadays', 'science', 'itself', 'done', 'england', 'political', 'economy', 'since', 'absolutely', 'daughter', 'ticket', 'passport', 'parenthesis', 'certain', 'uneasiness', 'hurriedly', 'apparent', 'composure', 'both', 'guffawed', 'smiled', 'confounded', 'wagging', 'everyone', 'secret', 'humility', 'behold', 'strongly', 'dare', 'assert', 'pig', 'answer', 'orator', 'stolidly', 'increased', 'waiting', 'laughter', 'subside', 'lady', 'semblance', 'beast', 'katerina', 'spouse', 'officer', 'granted', 'scoundrel', 'noble', 'magnanimous', 'unjust', 'pulls', 'pity', 'repeat', 'declared', 'redoubled', 'hearing', 'vain', 'wish', 'fate', 'assented', 'fist', 'sold', 'stockings', 'shoes', 'less', 'mohair', 'shawl', 'present', 'property', 'mine', 'caught', 'winter', 'coughing', 'spitting', 'morning', 'scrubbing', 'cleaning', 'washing', 'child', 'tendency', 'consumption', 'suppose', 'find', 'sympathy', 'twice', 'despair', 'laid', 'raising', 'seem', 'trouble', 'unfolding', 'laughing', 'stock', 'idle', 'listeners', 'educated', 'daughters', 'noblemen', 'leaving', 'danced', 'dance', 'governor', 'personages', 'presented', 'gold', 'medal', 'certificate', 'merit', 'trunk', 'showed', 'tell', 'someone', 'honours', 'happy', 'condemn', 'blame', 'recollection', 'ashes', 'spirit', 'proud', 'determined', 'scrubs', 'floors', 'herself', 'eat', 'treated', 'disrespect', 'overlook', 'rudeness', 'hurt', 'feelings', 'blows', 'widow', 'married', 'smaller', 'husband', 'infantry', 'ran', 'exceedingly', 'fond', 'cards', 'paid', 'authentic', 'documentary', 'evidence', 'speaks', 'tears', 'throws', 'imagination', 'having', 'wild', 'remote', 'district', 'hopeless', 'ups', 'downs', 'equal', 'describing', 'relations', 'thrown', 'excessively', 'widower', 'offered', 'bear', 'sight', 'judge', 'extremity', 'calamities', 'distinguished', 'consented', 'weeping', 'sobbing', 'wringing', 'means', 'performed', 'duties', 'conscientiously', 'faithfully', 'touch', 'finger', 'fault', 'changes', 'office', 'wanderings', 'magnificent', 'capital', 'adorned', 'innumerable', 'monuments', 'obtained', 'situation', 'amalia', 'fyodorovna', 'lippevechsel', 'rent', 'lot', 'dirt', 'disorder', 'perfect', 'bedlam', 'meanwhile', 'grown', 'whilst', 'growing', 'speak', 'generous', 'spirited', 'tempered', 'sonia', 'fancy', 'effort', 'geography', 'universal', 'history', 'subjects', 'suitable', 'instruction', 'cyrus', 'persia', 'attained', 'maturity', 'romantic', 'book', 'lewes', 'physiology', 'recounted', 'extracts', 'address', 'account', 'private', 'earn', 'honest', 'farthings', 'talent', 'putting', 'ivan', 'ivanitch', 'klopstock', 'heard', 'dozen', 'linen', 'stamping', 'reviling', 'pretext', 'collars', 'pattern', 'askew', 'hungry', 'cheeks', 'flushed', 'warm', 'gets', 'crust', 'gentle', 'creature', 'soft', 'fair', 'pale', 'darya', 'frantsovna', 'evil', 'known', 'police', 'tried', 'jeer', 'mighty', 'precious', 'careful', 'driven', 'distraction', 'illness', 'crying', 'wound', 'cry', 'hunger', 'falls', 'six', 'o', 'clock', 'saw', 'nine', 'utter', 'picked', 'green', 'drap', 'de', 'dames', 'wall', 'shuddering', 'knees', 'kissing', 'feet', 'failed', 'cleared', 'throat', 'brief', 'pause', 'unfortunate', 'occurrence', 'information', 'intentioned', 'sofya', 'semyonovna', 'backed', 'making', 'highly', 'comes', 'mostly', 'comforts', 'gives', 'kapernaumovs', 'lodges', 'kapernaumov', 'lame', 'cleft', 'palate', 'palates', 'lifted', 'heaven', 'excellency', 'afanasyvitch', 'wax', 'lord', 'melteth', 'deceived', 'expectations', 'll', 'responsibility', 'kissed', 'reality', 'statesman', 'enlightened', 'returned', 'announced', 'd', 'receive', 'salary', 'heavens', 'party', 'revellers', 'sounds', 'hired', 'cracked', 'piping', 'singing', 'hamlet', 'noise', 'busy', 'comers', 'paying', 'attention', 'arrivals', 'talkative', 'recent', 'success', 'revive', 'radiance', 'attentively', 'weeks', 'mercy', 'kingdom', 'abuse', 'tiptoe', 'hushing', 'semyon', 'zaharovitch', 'tired', 'shh', 'coffee', 'boiled', 'cream', 'managed', 'decent', 'outfit', 'eleven', 'guess', 'fronts', 'uniform', 'splendid', 'cooked', 'courses', 'dinner', 'soup', 'salt', 'meat', 'radish', 'dreamed', 'dresses', 'visit', 'smartened', 'nicely', 'collar', 'cuffs', 'better', 'darling', 'helped', 'see', 'nap', 'quarrelled', 'degree', 'week', 'resist', 'hours', 'whispering', 'receiving', 'everybody', 'study', 'sure', 'services', 'propensity', 'foolish', 'promise', 'moreover', 'rely', 'gentleman', 'wantonness', 'sake', 'bragging', 'believes', 'amuses', 'fancies', 'earnings', 'forty', 'called', 'poppet', 'beauty', 'pinched', 'cheek', 'twitch', 'controlled', 'degraded', 'appearance', 'nights', 'poignant', 'bewildered', 'listener', 'intently', 'sensation', 'vexed', 'recovering', 'seems', 'worrying', 'stupidity', 'details', 'heavenly', 'fleeting', 'arrange', 'rescue', 'dishonour', 'restore', 'bosom', 'deal', 'excusable', 'raised', 'cunning', 'trick', 'thief', 'employment', 'egyptian', 'exchanged', 'garments', 'clenched', 'teeth', 'closed', 'leaned', 'heavily', 'elbow', 'minute', 'changed', 'assumed', 'slyness', 'affectation', 'bravado', 'glanced', 'laughed', 'pick', 'guffaw', 'quart', 'bought', 'earth', 'yonder', 'grieve', 'weep', 'hurts', 'needs', 'eh', 'keep', 'costs', 'smartness', 'pomatum', 'petticoats', 'starched', 'show', 'foot', 'puddle', 'sorry', 'pitied', 'near', 'shouts', 'oaths', 'listening', 'discharged', 'declaimed', 'arm', 'outstretched', 'crucified', 'cross', 'crucify', 'merry', 'tribulation', 'pint', 'sweet', 'sought', 'bottom', 'understood', 'consumptive', 'drunkard', 'earthly', 'undismayed', 'beastliness', 'forgiven', 'thee', 'thy', 'sins', 'thou', 'hast', 'forgive', 'wise', 'meek', 'summon', 'forth', 'ye', 'stand', 'unto', 'swine', 'image', 'mark', 'understanding', 'dost', 'believed', 'worthy', 'hold', 'fall', 'exhausted', 'helpless', 'oblivious', 'plunged', 'notion', 'talked', 'silly', 'fine', 'kozel', 'yard', 'wanting', 'meant', 'unsteadier', 'legs', 'paces', 'overcome', 'dismay', 'nearer', 'begin', 'pulling', 'bother', 'frightens', 'breathing', 'breathe', 'excited', 'pain', 'strike', 'relieves', 'cabinet', 'maker', 'lead', 'darker', 'nearly', 'grimy', 'ajar', 'candle', 'visible', 'entrance', 'littered', 'across', 'furthest', 'stretched', 'sheet', 'american', 'leather', 'holes', 'unpainted', 'uncovered', 'edge', 'smoldering', 'tallow', 'candlestick', 'practically', 'cupboards', 'divided', 'shouting', 'uproar', 'playing', 'tea', 'unceremonious', 'kind', 'flew', 'recognised', 'graceful', 'emaciated', 'hectic', 'flush', 'pacing', 'pressing', 'lips', 'parched', 'broken', 'gasps', 'glittered', 'fever', 'harsh', 'immovable', 'stare', 'flickering', 'sickening', 'certainly', 'window', 'rose', 'inner', 'clouds', 'tobacco', 'smoke', 'floated', 'youngest', 'curled', 'older', 'shaking', 'chemise', 'ancient', 'cashmere', 'pelisse', 'flung', 'bare', 'outgrown', 'barely', 'reaching', 'comfort', 'doing', 'whimpering', 'large', 'larger', 'thinness', 'watching', 'alarm', 'doorway', 'pushing', 'indifferently', 'wondering', 'decided', 'hers', 'further', 'outer', 'uttered', 'scream', 'ah', 'frenzy', 'criminal', 'monster', 'searching', 'submissively', 'obediently', 'held', 'facilitate', 'search', 'farthing', 'twelve', 'fury', 'seized', 'seconded', 'efforts', 'meekly', 'crawling', 'consolation', 'con', 'la', 'tion', 'ho', 'nou', 'shaken', 'fro', 'striking', 'ground', 'woke', 'control', 'trembling', 'screaming', 'rushed', 'fit', 'eldest', 'leaf', 'screamed', 'pointed', 'accursed', 'hastening', 'faces', 'peering', 'coarse', 'pipes', 'cigarettes', 'caps', 'thrust', 'figures', 'gowns', 'costumes', 'scantiness', 'diverted', 'sinister', 'shrill', 'outcry', 'amongst', 'hundredth', 'frighten', 'ordering', 'snatch', 'coppers', 'exchange', 'reflecting', 'impossible', 'dismissed', 'wave', 'wants', 'malignantly', 'bankrupt', 'risk', 'hunting', 'game', 'digging', 'morrow', 'hurrah', 'dug', 'wept', 'grows', 'wrong', 'race', 'mankind', 'prejudice', 'artificial', 'terrors', 'barriers', 'iii', 'waked', 'refreshed', 'bilious', 'ill', 'hatred', 'length', 'stricken', 'dusty', 'peeling', 'low', 'pitched', 'ease', 'knock', 'ceiling', 'rickety', 'painted', 'manuscripts', 'thick', 'untouched', 'clumsy', 'space', 'chintz', 'served', 'undressing', 'sheets', 'wrapped', 'overcoat', 'pillow', 'heaped', 'bolster', 'difficult', 'sink', 'lower', 'ebb', 'state', 'agreeable', 'tortoise', 'shell', 'servant', 'writhe', 'irritation', 'overtakes', 'monomaniacs', 'entirely', 'fortnight', 'sending', 'meals', 'expostulating', 'nastasya', 'cook', 'pleased', 'lodger', 'mood', 'sweeping', 'stray', 'cup', 'fairly', 'starving', 'teapot', 'stale', 'lumps', 'sugar', 'fumbling', 'handful', 'run', 'loaf', 'sausage', 'cheapest', 'pork', 'butcher', 'fetch', 'wouldn', 'cabbage', 'instead', 'saved', 'chatting', 'country', 'peasant', 'praskovya', 'pavlovna', 'complain', 'scowled', 'devil', 'straw', 'grinding', 'suit', 'fool', 'mistake', 'clever', 'sack', 'teach', 'sullenly', 'reluctantly', 'seriously', 'amused', 'inaudibly', 'quivering', 'articulate', 'lessons', 'quarrel', 'butter', 'replying', 'fortune', 'strangely', 'firmly', 'forgot', 'postman', 'greatly', 'province', 'r', 'turned', 'stabbed', 'leave', 'goodness', 'presence', 'quickly', 'sloping', 'handwriting', 'taught', 'delayed', 'weighing', 'ounces', 'rodya', 'wrote', 'distressed', 'awake', 'inevitable', 'dounia', 'hope', 'grief', 'university', 'pension', 'sent', 'borrowed', 'security', 'vassily', 'ivanovitch', 'vahrushin', 'merchant', 'hearted', 'send', 'thank', 'believe', 'able', 'congratulate', 'hasten', 'inform', 'guessed', 'separated', 'future', 'sufferings', 'hitherto', 'concealed', 'svidrigra', 'lovs', 'written', 'truth', 'walk', 'insulted', 'governess', 'deducted', 'throw', 'repaying', 'sum', 'chiefly', 'needed', 'savings', 'loves', 'svidriga', 'lov', 'rudely', 'disrespectful', 'jeering', 'remarks', 'painful', 'behaviour', 'marfa', 'petrovna', 'household', 'relapsing', 'regimental', 'habits', 'influence', 'bacchus', 'crazy', 'conceived', 'passion', 'beginning', 'horrified', 'flighty', 'considering', 'hoped', 'rude', 'sneering', 'hide', 'shameful', 'proposal', 'promising', 'inducements', 'offering', 'estate', 'abroad', 'imagine', 'spare', 'whose', 'suspicions', 'aroused', 'cause', 'rupture', 'scandal', 'reasons', 'endure', 'cases', 'fortitude', 'maintain', 'firmness', 'upsetting', 'communication', 'ended', 'unexpectedly', 'accidentally', 'overheard', 'imploring', 'garden', 'interpretation', 'threw', 'believing', 'scene', 'spot', 'refused', 'hour', 'orders', 'packed', 'plain', 'cart', 'pell', 'mell', 'folding', 'packing', 'shower', 'rain', 'drive', 'seventeen', 'versts', 'dared', 'unhappy', 'mortified', 'indignant', 'fill', 'sorrow', 'church', 'contemptuous', 'whispers', 'bowed', 'learnt', 'shopmen', 'intending', 'insult', 'smearing', 'landlord', 'slander', 'neighbourhood', 'gossiping', 'affairs', 'complaining', 'spread', 'surrounding', 'bore', 'endured', 'cheer', 'angel', 'cut', 'repented', 'unmistakable', 'proof', 'innocence', 'remained', 'refuse', 'personal', 'explanations', 'interviews', 'entreating', 'reproached', 'indignation', 'baseness', 'reminding', 'telling', 'infamous', 'torment', 'defenceless', 'nobly', 'touchingly', 'sobbed', 'cannot', 'servants', 'reputation', 'supposed', 'aback', 'convinced', 'sunday', 'cathedral', 'knelt', 'prayed', 'strength', 'trial', 'told', 'bitterly', 'fully', 'penitent', 'embraced', 'besought', 'delay', 'everywhere', 'shedding', 'asserted', 'flattering', 'behavior', 'copies', 'superfluous', 'several', 'driving', 'offence', 'precedence', 'therefore', 'turns', 'expected', 'arrived', 'assembled', 'opinion', 'unnecessary', 'succeeded', 'establishing', 'ignominy', 'affair', 'rested', 'indelible', 'disgrace', 'treating', 'harshly', 'families', 'treat', 'marked', 'event', 'fortunes', 'transformed', 'suitor', 'marry', 'arranged', 'consent', 'aggrieved', 'decision', 'judged', 'facts', 'pyotr', 'petrovitch', 'luzhin', 'distantly', 'related', 'active', 'match', 'expressing', 'acquaintance', 'properly', 'courteously', 'offer', 'begged', 'speedy', 'depended', 'posts', 'prepossessing', 'attractive', 'women', 'presentable', 'morose', 'conceited', 'beware', 'shortly', 'judging', 'hastily', 'warning', 'favourable', 'deliberate', 'avoid', 'forming', 'prejudices', 'mistaken', 'correct', 'indications', 'thoroughly', 'estimable', 'shares', 'expressed', 'convictions', 'rising', 'generation', 'opponent', 'likes', 'natured', 'resolute', 'sensible', 'patient', 'passionate', 'either', 'happiness', 'care', 'admitted', 'prudence', 'secure', 'defects', 'differences', 'happiest', 'marriages', 'regards', 'relies', 'relationship', 'honourable', 'straightforward', 'abrupt', 'outspoken', 'dowry', 'experienced', 'indebted', 'benefactor', 'add', 'politely', 'actual', 'phrases', 'meaning', 'design', 'smooth', 'deeds', 'perfectly', 'fervently', 'mentioned', 'legal', 'bureau', 'conducting', 'commercial', 'litigation', 'important', 'senate', 'greatest', 'agreed', 'definitely', 'consider', 'assured', 'benefit', 'providential', 'dreaming', 'ventured', 'cautious', 'secretary', 'relation', 'fitted', 'doubts', 'whether', 'studies', 'regular', 'becoming', 'associate', 'partner', 'law', 'agreement', 'share', 'plans', 'probability', 'realising', 'evasiveness', 'natural', 'persuaded', 'gain', 'coldly', 'breathed', 'helping', 'wasting', 'readily', 'assistance', 'charity', 'earned', 'agree', 'footing', 'enthusiasm', 'oneself', 'forward', 'womanish', 'wedding', 'invite', 'urge', 'remain', 'husbands', 'mothers', 'bit', 'independent', 'settle', 'joyful', 'news', 'embrace', 'separation', 'settled', 'depends', 'arrangements', 'ceremony', 'fast', 'joke', 'pen', 'bids', 'kisses', 'credit', 'improved', 'afanasy', 'trust', 'seventy', 'travelling', 'expenses', 'undertake', 'journey', 'conveyance', 'bags', 'conveyed', 'reckon', 'expense', 'arrival', 'halfpenny', 'calculated', 'ninety', 'railway', 'driver', 'readiness', 'travel', 'comfortably', 'events', 'beyond', 'prayers', 'creator', 'redeemer', 'visited', 'infidelity', 'pray', 'childhood', 'lisp', 'knee', 'warmly', 'pulcheria', 'wet', 'finished', 'distorted', 'bitter', 'wrathful', 'threadbare', 'pondered', 'violently', 'turmoil', 'cramped', 'stifled', 'craved', 'dread', 'direction', 'vassilyevsky', 'ostrov', 'prospect', 'noticing', 'muttering', 'astonishment', 'iv', 'chief', 'essential', 'irrevocably', 'marriage', 'alive', 'damned', 'anticipating', 'triumph', 'deceive', 'apologise', 'advice', 'post', 'holy', 'kazan', 'stands', 'bedroom', 'ascent', 'golgotha', 'finally', 'avdotya', 'romanovna', 'solid', 'impressive', 'holds', 'writes', 'observes', 'beats', 'marrying', 'descriptive', 'idea', 'favour', 'minds', 'need', 'simplicity', 'observations', 'angrily', 'angered', 'na', 'questions', 'discuss', 'conscience', 'prick', 'sacrificing', 'murdered', 'pursuing', 'whirling', 'wasn', 'bride', 'sacking', 'thousand', 'according', 'cloth', 'raise', 'partnership', 'mutual', 'luggage', 'fares', 'blossoming', 'fruits', 'stinginess', 'meanness', 'tone', 'foretaste', 'lavish', 'expect', 'guessing', 'deny', 'counting', 'knits', 'woollen', 'shawls', 'embroiders', 'ruining', 'generosity', 'schilleresque', 'hearts', 'goose', 'swan', 'inkling', 'shiver', 'until', 'deck', 'false', 'colours', 'puts', 'bet', 'anna', 'buttonhole', 'goes', 'dine', 'contractors', 'merchants', 'confound', 'wonder', 'bless', 'propounds', 'theory', 'superiority', 'wives', 'destitution', 'bounty', 'interview', 'understands', 'water', 'barter', 'moral', 'freedom', 'schleswig', 'holstein', 'denying', 'pill', 'spend', 'provinces', 'nigger', 'plantation', 'lett', 'degrade', 'binding', 'advantage', 'unalloyed', 'diamond', 'concubine', 'consenting', 'save', 'adores', 'amounts', 'necessary', 'peace', 'casuists', 'jesuitical', 'soothe', 'persuade', 'daylight', 'rodion', 'romanovitch', 'central', 'ensure', 'rich', 'prosperous', 'born', 'sacrifice', 'loving', 'partial', 'shrink', 'eternal', 'victim', 'lasts', 'measure', 'worse', 'viler', 'baser', 'bargain', 'luxuries', 'starvation', 'regret', 'curses', 'hidden', 'worried', 'sees', 'prevent', 'forbid', 'devote', 'borrow', 'millionaire', 'zeus', 'blind', 'knitting', 'shadow', 'fasting', 'happen', 'during', 'tortured', 'fretting', 'finding', 'confronting', 'aches', 'grip', 'rend', 'anguish', 'beginnings', 'waxed', 'gathered', 'matured', 'fearful', 'frenzied', 'fantastic', 'clamouring', 'insistently', 'burst', 'thunderclap', 'passively', 'unsolved', 'decide', 'humbly', 'stifle', 'giving', 'claim', 'activity', 'recurring', 'expecting', 'difference', 'mere', 'menacing', 'unfamiliar', 'shape', 'hammering', 'sit', 'seat', 'boulevard', 'adventure', 'objects', 'crossed', 'path', 'road', 'gradually', 'riveted', 'resentfully', 'bareheaded', 'parasol', 'gloves', 'waving', 'silky', 'material', 'awry', 'hooked', 'skirt', 'waist', 'hanging', 'loose', 'slanting', 'unsteadily', 'stumbling', 'staggering', 'overtook', 'extreme', 'exhaustion', 'closely', 'shocking', 'hardly', 'haired', 'sixteen', 'pretty', 'lifting', 'indecorously', 'sign', 'unconscious', 'unwilling', 'perplexity', 'frequented', 'stifling', 'deserted', 'impatiently', 'biding', 'unwelcome', 'moved', 'intentions', 'plump', 'fashionably', 'colour', 'moustaches', 'furious', 'longing', 'dandy', 'clenching', 'fists', 'spluttering', 'rage', 'sternly', 'scowling', 'haughty', 'cane', 'constable', 'gentlemen', 'fighting', 'public', 'soldierly', 'whiskers', 'catching', 'policeman', 'professional', 'somebody', 'unpractised', 'fight', 'eager', 'following', 'prevented', 'pretending', 'flash', 'easy', 'examine', 'sleepy', 'blankly', 'waved', 'call', 'cab', 'missy', 'ach', 'shook', 'shocked', 'sympathetic', 'job', 'handing', 'innocent', 'belongs', 'gentlefolk', 'ladies', 'pretensions', 'gentility', 'persisted', 'outrage', 'brute', 'fly', 'confined', 'halted', 'thoughtfully', 'wretches', 'avenue', 'eye', 'repeated', 'sighing', 'sting', 'revulsion', 'stared', 'ejaculated', 'gesture', 'madman', 'carried', 'murmured', 'interfere', 'devour', 'wretched', 'strayed', 'aimlessly', 'fix', 'forget', 'wake', 'anew', 'horrible', 'doors', 'frantsovnas', 'wind', 'slipping', 'sly', 'hospital', 'luck', 'taverns', 'wreck', 'eighteen', 'nineteen', 'ugh', 'percentage', 'chaste', 'interfered', 'scientific', 'consolatory', 'razumihin', 'wondered', 'comrades', 'remarkable', 'friends', 'aloof', 'welcome', 'gatherings', 'amusements', 'intensity', 'sparing', 'pride', 'reserve', 'superior', 'development', 'knowledge', 'beliefs', 'interests', 'beneath', 'unreserved', 'communicative', 'humoured', 'candid', 'depth', 'intelligent', 'simpleton', 'blackhaired', 'shaved', 'uproarious', 'reputed', 'festive', 'blow', 'gigantic', 'limit', 'powers', 'abstain', 'pranks', 'failure', 'unfavourable', 'circumstances', 'crush', 'lodge', 'extremes', 'resources', 'lighting', 'stove', 'declare', 'soundly', 'observed', 'annoy', 'v', 'lately', 'tidy', 'agitated', 'uneasily', 'seeking', 'significance', 'ordinary', 'action', 'rubbed', 'musing', 'spontaneously', 'calmly', 'determination', 'afresh', 'realised', 'jumping', 'homewards', 'loathing', 'hole', 'random', 'shudder', 'shivering', 'unconsciously', 'craving', 'distract', 'succeed', 'dropping', 'brooding', 'lesser', 'islands', 'greenness', 'freshness', 'restful', 'hemmed', 'closeness', 'pleasant', 'sensations', 'morbid', 'irritability', 'villa', 'among', 'foliage', 'fence', 'smartly', 'verandahs', 'balconies', 'running', 'gardens', 'flowers', 'longer', 'luxurious', 'carriages', 'horseback', 'watched', 'vanished', 'marmeladovs', 'passing', 'eating', 'vodka', 'ate', 'pie', 'effect', 'wineglassful', 'drowsiness', 'petrovsky', 'bushes', 'grass', 'singular', 'actuality', 'vividness', 'monstrous', 'images', 'artistically', 'consistent', 'dreamer', 'artist', 'turgenev', 'invented', 'memory', 'powerful', 'deranged', 'system', 'dreamt', 'birth', 'holiday', 'vividly', 'level', 'willow', 'copse', 'blur', 'horizon', 'crowd', 'hoarse', 'cling', 'track', 'winding', 'graveyard', 'stone', 'cupola', 'mass', 'grandmother', 'occasions', 'dish', 'tied', 'napkin', 'rice', 'pudding', 'raisins', 'stuck', 'unadorned', 'ikons', 'priest', 'whenever', 'religiously', 'reverently', 'holding', 'circumstance', 'attracted', 'festivity', 'gaily', 'townspeople', 'riff', 'raff', 'carts', 'usually', 'drawn', 'horses', 'laden', 'casks', 'wine', 'goods', 'manes', 'slow', 'pace', 'drawing', 'mountain', 'load', 'shafts', 'sorrel', 'peasants', 'nags', 'straining', 'utmost', 'wheels', 'mud', 'rut', 'cruelly', 'balala', 'ka', 'blue', 'coats', 'necked', 'fleshy', 'carrot', 'outbreak', 'mikolka', 'nag', 'mare', 'mates', 'leaping', 'seizing', 'reins', 'bay', 'matvey', 'kill', 'gallop', 'whip', 'preparing', 'relish', 'flog', 'jog', 'clambered', 'jokes', 'hauled', 'rosy', 'cheeked', 'beaded', 'headdress', 'nuts', 'drag', 'cartload', 'whips', 'tugged', 'galloping', 'move', 'struggled', 'gasping', 'shrinking', 'showered', 'hail', 'furiously', 'thrashed', 'appetite', 'draw', 'fun', 'tore', 'horror', 'tugging', 'falling', 'christian', 'meddle', 'choose', 'roar', 'roused', 'feebly', 'kicking', 'smiling', 'kick', 'lads', 'snatched', 'ribs', 'hit', 'song', 'joined', 'riotous', 'jingling', 'tambourine', 'whistling', 'whipped', 'choking', 'streaming', 'headed', 'disapproval', 'gasp', 'ferociously', 'shaft', 'brandished', 'swinging', 'sound', 'thud', 'thrash', 'voices', 'swung', 'spine', 'luckless', 'haunches', 'lurched', 'force', 'attacking', 'directions', 'measured', 'tough', 'admiring', 'spectator', 'axe', 'finish', 'frantically', 'stooped', 'crowbar', 'dealt', 'stunning', 'pull', 'bar', 'log', 'leapt', 'sticks', 'poles', 'dying', 'dealing', 'breath', 'butchered', 'bloodshot', 'brandishing', 'regretting', 'bleeding', 'jumped', 'shrieks', 'panting', 'brutal', 'choked', 'soaked', 'perspiration', 'tree', 'breaths', 'split', 'skull', 'tread', 'break', 'steal', 'tremble', 'spattered', 'profound', 'amazement', 'torturing', 'experiment', 'base', 'vile', 'couldn', 'flaw', 'reasoning', 'arithmetic', 'glowed', 'limb', 'easily', 'cast', 'relief', 'renounce', 'crossing', 'quietly', 'glowing', 'sky', 'fatigue', 'abscess', 'spell', 'sorcery', 'obsession', 'superstitiously', 'impressed', 'exceptional', 'predestined', 'turning', 'convenient', 'shortest', 'direct', 'unnecessarily', 'dozens', 'decisive', 'exert', 'gravest', 'destiny', 'tables', 'barrows', 'booths', 'shops', 'closing', 'clearing', 'wares', 'customers', 'pickers', 'costermongers', 'crowding', 'stinking', 'neighbouring', 'wandered', 'attract', 'attire', 'scandalising', 'alley', 'huckster', 'tapes', 'handkerchiefs', 'lingering', 'pawnbroker', 'previous', 'single', 'timid', 'submissive', 'idiotic', 'slave', 'bundle', 'earnestly', 'doubtfully', 'warmth', 'astonishing', 'fright', 'gabbled', 'lively', 'babe', 'interrupted', 'pondering', 'softly', 'miss', 'thrill', 'precisely', 'incapable', 'opportunity', 'greater', 'exactness', 'dangerous', 'inquiries', 'investigations', 'contemplated', 'vi', 'invited', 'reduced', 'selling', 'fetched', 'dealer', 'undertook', 'jobs', 'price', 'rule', 'superstitious', 'traces', 'superstition', 'ineradicable', 'disposed', 'mysterious', 'influences', 'coincidences', 'pokorev', 'harkov', 'chanced', 'articles', 'pawned', 'stones', 'parting', 'insurmountable', 'miserable', 'pecking', 'chicken', 'egg', 'played', 'billiards', 'mention', 'shake', 'expressly', 'rate', 'jew', 'lots', 'dealings', 'harpy', 'uncertain', 'value', 'article', 'percent', 'chattered', 'bondage', 'phenomenon', 'mending', 'cooking', 'sewing', 'charwoman', 'permission', 'movables', 'monastery', 'n', 'perpetuity', 'unmarried', 'awfully', 'uncouth', 'remarkably', 'outwards', 'battered', 'goatskin', 'amusement', 'skinned', 'soldier', 'strikingly', 'willing', 'queerness', 'assure', 'faintest', 'shuddered', 'hotly', 'joking', 'senseless', 'worthless', 'ailing', 'horrid', 'mischief', 'die', 'fresh', 'thousands', 'buried', 'hundreds', 'hospitals', 'humanity', 'wiped', 'corruption', 'decay', 'balance', 'existence', 'louse', 'beetle', 'harm', 'amputated', 'deserve', 'remarked', 'drown', 'ocean', 'speechifying', 'arguing', 'justice', 'youthful', 'forms', 'themes', 'discussion', 'conceiving', 'embryo', 'coincidence', 'preordained', 'guiding', 'hint', 'returning', 'stirring', 'occur', 'recollect', 'leaden', 'crushing', 'extraordinarily', 'difficulty', 'rousing', 'brew', 'sleeps', 'indignantly', 'ached', 'reply', 'offended', 'wrathfully', 'air', 'weakly', 'motioned', 'spoon', 'spoonfuls', 'mechanically', 'meal', 'haunted', 'fancied', 'africa', 'egypt', 'oasis', 'caravan', 'camels', 'peacefully', 'palms', 'around', 'circle', 'spring', 'flowed', 'gurgling', 'cool', 'wonderful', 'parti', 'coloured', 'sand', 'glistened', 'pulled', 'crept', 'stealthily', 'forgetfulness', 'prepared', 'stupefaction', 'feverish', 'distracted', 'preparations', 'energies', 'forgetting', 'thumping', 'noose', 'sew', 'rummaged', 'stuffed', 'unwashed', 'strip', 'couple', 'inches', 'folded', 'garment', 'ends', 'inside', 'armhole', 'sewed', 'outside', 'needle', 'ingenious', 'device', 'intended', 'carry', 'support', 'hang', 'handle', 'swing', 'designed', 'opening', 'smoothly', 'planed', 'size', 'thickness', 'courtyard', 'workshop', 'fastened', 'carefully', 'daintily', 'parcel', 'untie', 'divert', 'undo', 'knot', 'weight', 'stored', 'descend', 'thirteen', 'cautiously', 'noiselessly', 'deed', 'pruning', 'knife', 'resolved', 'peculiarity', 'resolutions', 'characteristic', 'agonising', 'inward', 'struggle', 'carrying', 'considered', 'uncertainty', 'renounced', 'unsettled', 'points', 'uncertainties', 'trifling', 'anxiety', 'neighbours', 'shop', 'scolding', 'doubtful', 'supposing', 'meantime', 'suspicion', 'grounds', 'unattainable', 'sometime', 'e', 'survey', 'cursing', 'analysis', 'casuistry', 'razor', 'rational', 'objections', 'resort', 'doggedly', 'slavishly', 'arguments', 'forcing', 'crimes', 'detected', 'obvious', 'conclusions', 'impossibility', 'concealing', 'power', 'childish', 'phenomenal', 'heedlessness', 'caution', 'eclipse', 'attacked', 'highest', 'perpetration', 'violence', 'shorter', 'individual', 'rise', 'accompanied', 'reaction', 'unimpaired', 'omit', 'process', 'conclusion', 'ahead', 'purely', 'difficulties', 'secondary', 'familiarised', 'minutest', 'preparation', 'decisions', 'upset', 'calculations', 'usual', 'absence', 'peep', 'basket', 'line', 'overwhelmed', 'gateway', 'assume', 'humiliated', 'anger', 'dull', 'animal', 'opposite', 'porter', 'shining', 'chunks', 'pockets', 'fails', 'helps', 'grin', 'sedately', 'awakening', 'curse', 'glancing', 'someway', 'yusupov', 'fountains', 'refreshing', 'squares', 'degrees', 'extended', 'field', 'mars', 'mihailovsky', 'palace', 'interested', 'towns', 'inclined', 'parts', 'smell', 'nastiness', 'walks', 'clutch', 'mentally', 'meets', 'lightning', 'dismiss', 'luckily', 'screening', 'quarrelling', 'quadrangular', 'throbbing', 'ascending', 'shut', 'painters', 'storeys', 'underneath', 'visiting', 'card', 'nailed', 'mistrustful', 'leaves', 'throbbed', 'ringing', 'suspicious', 'ear', 'peculiarly', 'distinct', 'rustle', 'secretly', 'hiding', 'soberly', 'impatience', 'recalling', 'clouded', 'latch', 'unfastened', 'vii', 'fearing', 'hoping', 'disarm', 'attempting', 'allowing', 'advanced', 'obey', 'uninvited', 'tongue', 'unloosed', 'promised', 'maliciously', 'mistrustfully', 'sneer', 'malice', 'elsewhere', 'recovered', 'restored', 'confidence', 'bathing', 'abruptly', 'articulating', 'failing', 'sounded', 'scanning', 'string', 'seconds', 'unbuttoned', 'freed', 'fearfully', 'numb', 'giddiness', 'vexation', 'lose', 'blunt', 'streaked', 'grease', 'plaited', 'rat', 'tail', 'horn', 'comb', 'nape', 'faintly', 'heap', 'gushed', 'overturned', 'starting', 'sockets', 'brow', 'contorted', 'convulsively', 'possession', 'faculties', 'collected', 'shrine', 'silk', 'patchwork', 'wadded', 'quilt', 'convulsive', 'tempted', 'terrifying', 'recover', 'bending', 'examining', 'pool', 'snap', 'hurried', 'touching', 'purse', 'crosses', 'cyprus', 'filigree', 'chamois', 'rim', 'unsuccessful', 'locks', 'mistakes', 'belong', 'bedstead', 'beds', 'sized', 'arched', 'lid', 'studded', 'notched', 'unlocked', 'brocade', 'lined', 'hareskin', 'wipe', 'sooner', 'pledges', 'unredeemed', 'redeemed', 'bracelets', 'chains', 'rings', 'pins', 'newspaper', 'tape', 'filling', 'trousers', 'undoing', 'parcels', 'moan', 'squatting', 'heels', 'waited', 'gazing', 'seeming', 'mouth', 'backing', 'twitched', 'piteously', 'babies', 'mouths', 'guard', 'motioning', 'snatching', 'gained', 'mastery', 'unexpected', 'murder', 'correctly', 'hopelessness', 'hideousness', 'absurdity', 'obstacles', 'commit', 'surged', 'dreaminess', 'bucket', 'bethought', 'blade', 'soap', 'saucer', 'rubbing', 'damp', 'wetted', 'overlooking', 'protecting', 'shock', 'awaited', 'rung', 'bolt', 'precaution', 'reflect', 'shrilly', 'patiently', 'noisily', 'downstairs', 'humming', 'tune', 'footsteps', 'suspect', 'significant', 'unhurried', 'mounting', 'higher', 'pursued', 'killed', 'rooted', 'neatly', 'hook', 'catch', 'instinct', 'crouched', 'panted', 'squeezing', 'tinkled', 'fastening', 'blank', 'damn', 'bawled', 'witch', 'enraged', 'authority', 'intimate', 'approaching', 'comer', 'koch', 'gambrinus', 'aie', 'sits', 'hag', 'hadn', 'shakes', 'shows', 'locked', 'clanks', 'proves', 'clanking', 'fasten', 'astonished', 'fainted', 'stop', 'dent', 'touched', 'gently', 'puffing', 'keyhole', 'tight', 'delirium', 'knocking', 'shout', 'swear', 'deserting', 'sentry', 'hurrying', 'flights', 'loud', 'mitka', 'blast', 'shriek', 'flight', 'deliverance', 'pail', 'paint', 'brushes', 'whisked', 'nick', 'bodies', 'murderer', 'escaping', 'upstairs', 'quicken', 'yards', 'fling', 'safety', 'risky', 'grain', 'weakened', 'drops', 'bank', 'dimly', 'farther', 'alarmed', 'recollected', 'problem', 'observation', 'fortunately', 'covering', 'chunk', 'scraps', 'shreds', 'swarming', 'dazed', 'oblivion', 'despairing', 'cries', 'leaped', 'dreadful', 'chill', 'limbs', 'fallen', 'threads', 'mistrusting', 'congealed', 'frayed', 'claspknife', 'tatters', 'stuffing', 'gleefully', 'bulged', 'whispered', 'reckoned', 'trinkets', 'unbearable', 'chair', 'consciousness', 'loop', 'rouse', 'concentration', 'simplest', 'surely', 'isn', 'distraught', 'perceptions', 'lining', 'triumphantly', 'sigh', 'sunlight', 'boot', 'sock', 'poked', 'tip', 'unwarily', 'ransack', 'burn', 'matches', 'icy', 'impulse', 'sleeping', 'banging', 'snoring', 'dog', 'ha', 'latched', 'retorted', 'bolting', 'stealing', 'discovered', 'unlatched', 'defiant', 'desperate', 'sealed', 'bottle', 'summons', 'downright', 'response', 'compassionately', 'grasped', 'tightly', 'treasure', 'hysterical', 'giggle', 'behave', 'discoloured', 'distinguish', 'seal', 'appear', 'superintendent', 'bewilderment', 'flinging', 'prayer', 'dustier', 'socks', 'conventional', 'relative', 'merely', 'surface', 'swam', 'decoy', 'mused', 'blurt', 'possessed', 'cynicism', 'mortar', 'finnish', 'pedlars', 'cabs', 'apt', 'bright', 'sunny', 'trepidation', 'averted', 'confess', 'steep', 'sloppy', 'kitchens', 'policemen', 'sexes', 'newly', 'decorated', 'slightest', 'unkempt', 'poorly', 'mourning', 'dictation', 'buxom', 'purplish', 'blotchy', 'brooch', 'freely', 'regain', 'urging', 'courage', 'calm', 'foolishness', 'carelessness', 'betray', 'dizzier', 'mobile', 'foppish', 'parted', 'combed', 'pomaded', 'scrubbed', 'french', 'foreigner', 'luise', 'purple', 'faced', 'venturing', 'ich', 'danke', 'trimmed', 'lace', 'balloon', 'smelt', 'scent', 'embarrassed', 'impudent', 'cringing', 'betrayed', 'jauntily', 'tossed', 'cockaded', 'skipped', 'curtsying', 'ecstasy', 'smallest', 'assistant', 'horizontally', 'features', 'expressive', 'insolence', 'askance', 'bearing', 'affronted', 'annihilated', 'summoned', 'faltered', 'recovery', 'due', 'tearing', 'papers', 'document', 'trembled', 'joy', 'indescribable', 'directed', 'shoulder', 'pleasure', 'kindly', 'splutter', 'inarticulately', 'smoking', 'satisfaction', 'unnatural', 'loudness', 'declaration', 'demanded', 'alexandr', 'grigorievitch', 'complaint', 'bird', 'explanation', 'u', 'writ', 'undertaking', 'conceal', 'creditor', 'liberty', 'proceed', 'legally', 'attested', 'assessor', 'zarnitsyn', 'tchebarov', 'hereupon', 'novice', 'fire', 'triumphant', 'overwhelming', 'danger', 'suppositions', 'surmises', 'questioning', 'instinctive', 'thunderstorm', 'fuming', 'wounded', 'smart', 'hussy', 'correction', 'warned', 'eleventh', 'wildly', 'unceremoniously', 'laugh', 'ilya', 'anxiously', 'storm', 'amiable', 'seductive', 'smiles', 'lavished', 'curtsied', 'incessantly', 'captain', 'pattered', 'peas', 'confidently', 'accent', 'dislike', 'tipsy', 'bottles', 'pianoforte', 'ganz', 'piano', 'manners', 'hitting', 'karl', 'henriette', 'slaps', 'ungentlemanly', 'squealing', 'fie', 'sein', 'rock', 'muss', 'damages', 'significantly', 'rapidly', 'slightly', 'literary', 'nice', 'authors', 'restaurant', 'eaten', 'satire', 'steamer', 'disgraceful', 'councillor', 'confectioner', 'criers', 'pfoo', 'deference', 'stumbled', 'backwards', 'nikodim', 'fomitch', 'curtsy', 'mincing', 'fluttered', 'thunder', 'hurricane', 'drawled', 'gentlemanly', 'nonchalance', 'protest', 'behaves', 'cad', 'powder', 'slight', 'daresay', 'affably', 'explosive', 'fires', 'boils', 'nickname', 'regiment', 'lieutenant', 'gratified', 'banter', 'sulky', 'pardon', 'mannered', 'shattered', 'x', 'exasperated', 'yourselves', 'rummaging', 'contemptuously', 'verbal', 'heedless', 'asks', 'waste', 'interposed', 'typhus', 'quarters', 'owed', 'takes', 'affecting', 'ours', 'tragic', 'gruffly', 'dictate', 'indifferent', 'cared', 'amazed', 'officers', 'nearest', 'dearest', 'everlasting', 'solitude', 'remoteness', 'sentimental', 'effusions', 'vanities', 'offices', 'sentenced', 'burnt', 'stirred', 'happening', 'appeal', 'outburst', 'brothers', 'sisters', 'conception', 'dictating', 'attend', 'pressed', 'nail', 'lodgings', 'released', 'contradicts', 'pestryakov', 'silversmith', 'contradiction', 'knocked', 'bolted', 'ass', 'interval', 'thanksgiving', 'noah', 'ark', 'maintained', 'reach', 'supported', 'yellowish', 'signing', 'settling', 'handkerchief', 'jerkily', 'upright', 'detain', 'faintness', 'brutes', 'mastered', 'peeped', 'decoration', 'remaining', 'pursuit', 'instructions', 'issued', 'rid', 'task', 'ekaterininsky', 'rafts', 'boats', 'moored', 'banks', 'sides', 'float', 'wandering', 'irrational', 'absent', 'forgetful', 'solitary', 'bush', 'judgment', 'destined', 'unwhitewashed', 'court', 'hoarding', 'parallel', 'fenced', 'smutty', 'shed', 'carriage', 'builder', 'carpenter', 'coal', 'workmen', 'drivers', 'scribbled', 'chalk', 'witticism', 'strictly', 'unhewn', 'pounds', 'unless', 'using', 'hollow', 'twist', 'scraped', 'edges', 'tracks', 'noiseless', 'hateful', 'whiskered', 'distractedly', 'circling', 'ungovernable', 'lies', 'despicably', 'fawned', 'folly', 'fawning', 'perplexed', 'deliberately', 'idiotically', 'undergone', 'agonies', 'undertaken', 'otherwise', 'jewel', 'grimly', 'gaining', 'immeasurable', 'obstinate', 'loathed', 'movements', 'gestures', 'spat', 'bitten', 'accord', 'busily', 'gown', 'slippers', 'comrade', 'whistled', 'sunk', 'pulse', 'delirious', 'spleen', 'threshold', 'insulting', 'kinder', 'cleverer', 'sweep', 'bookseller', 'heruvimov', 'lesson', 'publishing', 'issuing', 'manuals', 'circulation', 'titles', 'jove', 'fools', 'encourage', 'signatures', 'text', 'crudest', 'charlatanism', 'discusses', 'contribution', 'translating', 'expand', 'gorgeous', 'page', 'pays', 'signature', 'works', 'translation', 'whales', 'dullest', 'scandals', 'les', 'confessions', 'rousseau', 'radishchev', 'contradict', 'pens', 'spelling', 'secondly', 'adrift', 'change', 'laying', 'raving', 'farce', 'descending', 'stepping', 'nikolaevsky', 'unpleasant', 'incident', 'coachman', 'lash', 'hoofs', 'infuriated', 'railing', 'traffic', 'serves', 'pickpocket', 'profession', 'retreating', 'elderly', 'christ', 'beggar', 'alms', 'gift', 'doubtless', 'cloud', 'rare', 'chapel', 'pure', 'ornament', 'truly', 'spectacle', 'marvelled', 'vague', 'emotion', 'lifeless', 'sombre', 'enigmatic', 'perplexities', 'imagined', 'theories', 'pictures', 'amusing', 'wrung', 'problems', 'impressions', 'flying', 'upwards', 'vanishing', 'movement', 'coin', 'overdriven', 'greatcoat', 'dusk', 'howling', 'wailing', 'brutality', 'swooning', 'louder', 'shrieking', 'incoherently', 'beseeching', 'beaten', 'mercilessly', 'assailant', 'croak', 'indistinctly', 'thuds', 'topsy', 'turvy', 'staircases', 'lift', 'gripped', 'ice', 'numbed', 'continuing', 'moaning', 'groaning', 'slammed', 'exclaiming', 'disputing', 'calling', 'whisper', 'numbers', 'inmates', 'block', 'intolerable', 'infinite', 'plate', 'ascertaining', 'warrant', 'trudging', 'scrutinised', 'frowning', 'scrutiny', 'lasted', 'timidly', 'ears', 'outlet', 'clotted', 'fancying', 'earthenware', 'swallowing', 'sip', 'spilling', 'squabbling', 'discussing', 'threatened', 'plotted', 'mocked', 'bedside', 'fretted', 'moaned', 'throwing', 'streak', 'waisted', 'messenger', 'peeping', 'echoed', 'concluding', 'discussions', 'eyebrows', 'fatness', 'laziness', 'absurdly', 'bashful', 'stooping', 'cabin', 'pashenka', 'vrazumihin', 'shelopaev', 'seated', 'zossimov', 'examined', 'result', 'feeding', 'alexey', 'semyonovitch', 'mamma', 'request', 'presume', 'remittance', 'intelligible', 'remit', 'dreamily', 'scrawl', 'scribble', 'sweeter', 'treacle', 'witness', 'travels', 'earnest', 'bravo', 'potatoes', 'unreasoning', 'spoons', 'plates', 'pepper', 'mustard', 'beef', 'amiss', 'departed', 'strained', 'clumsily', 'spoonful', 'blowing', 'swallowed', 'faculty', 'mumbled', 'pour', 'poured', 'cups', 'steadily', 'principal', 'effective', 'resistance', 'repugnance', 'pushed', 'capriciously', 'pillows', 'raspberry', 'jam', 'raspberries', 'balancing', 'outspread', 'lump', 'decamped', 'rascally', 'punish', 'corners', 'harlamov', 'buch', 'muddles', 'kobelev', 'land', 'zametov', 'slyly', 'nikiforovna', 'mirth', 'explosion', 'uproot', 'locality', 'wished', 'shrieked', 'afforded', 'unspeakable', 'delight', 'unaccountable', 'natalya', 'yegorovna', 'delighted', 'essentially', 'loss', 'intellectually', 'metaphysical', 'symbolism', 'sprung', 'algebra', 'hid', 'planned', 'cherishing', 'fed', 'sensibly', 'retiring', 'starve', 'ins', 'outs', 'prospective', 'sensitive', 'listens', 'formal', 'demand', 'harmony', 'reigned', 'insisted', 'engaging', 'presenting', 'trusts', 'twinge', 'recognise', 'rave', 'countess', 'bulldog', 'krestovsky', 'island', 'whined', 'hunted', 'scented', 'bedecked', 'comforted', 'fringe', 'fascinated', 'bedclothes', 'twitching', 'eluded', 'mocking', 'grime', 'bah', 'mixing', 'helplessly', 'america', 'contained', 'gulped', 'quenching', 'flame', 'breast', 'incoherent', 'disconnected', 'nestled', 'replaced', 'sighed', 'tryst', 'uncle', 'untying', 'specially', 'cheap', 'presently', 'pettishly', 'oppose', 'fitting', 'proper', 'recommendation', 'tolstyakov', 'basin', 'hats', 'slavish', 'politeness', 'nest', 'boastful', 'specimens', 'headgear', 'palmerston', 'eighty', 'united', 'states', 'breeches', 'exhibited', 'pair', 'improvement', 'softer', 'smoother', 'seasons', 'insist', 'asparagus', 'january', 'purchase', 'buying', 'warmer', 'materials', 'autumn', 'lack', 'coherence', 'standard', 'luxury', 'fedyaev', 'satisfied', 'foreign', 'embassy', 'cash', 'stiffly', 'coated', 'hempen', 'fashionable', 'underclothes', 'rig', 'serve', 'sharmer', 'playful', 'purchases', 'sullen', 'puffy', 'shaven', 'flaxen', 'spectacles', 'spick', 'span', 'irreproachable', 'massive', 'nonchalant', 'studiously', 'tedious', 'depressed', 'aching', 'irritably', 'mushrooms', 'cucumbers', 'medicine', 'palais', 'cristal', 'disturb', 'nuisance', 'warming', 'herrings', 'stagnating', 'postmaster', 'porfiry', 'investigation', 'department', 'distant', 'teacher', 'musician', 'nodded', 'principles', 'springs', 'principle', 'delightful', 'bribes', 'praise', 'ways', 'shouldn', 'baked', 'onion', 'repel', 'improve', 'repelling', 'progressive', 'dullards', 'painter', 'mess', 'steam', 'haven', 'mixed', 'blurted', 'audibly', 'mended', 'flower', 'petals', 'scallops', 'obstinately', 'displeasure', 'prove', 'foo', 'stupidly', 'curiously', 'stir', 'busybody', 'offensive', 'leads', 'worship', 'murderers', 'logic', 'detained', 'swindler', 'buys', 'rotten', 'petrified', 'routine', 'introducing', 'method', 'psychological', 'data', 'interpret', 'tangible', 'dandling', 'accounted', 'pikestaff', 'dushkin', 'dram', 'jeweller', 'containing', 'rigamarole', 'journeyman', 'nikolay', 'quicker', 'rumours', 'taradiddle', 'receiver', 'stolen', 'cheat', 'trinket', 'dementyev', 'zara', 'sk', 'ryazan', 'drinks', 'painting', 'dmitri', 'village', 'glasses', 'spree', 'daybreak', 'stayed', 'finishing', 'tale', 'sober', 'peski', 'kolomensky', 'signed', 'darted', 'searched', 'cowshed', 'stable', 'adjoining', 'sash', 'beam', 'screeched', 'hardest', 'escort', 'folks', 'robbed', 'pavlovitch', 'guilt', 'literally', 'squeezed', 'swore', 'sport', 'escaped', 'hooks', 'undid', 'leaning', 'repeating', 'deduced', 'admit', 'fail', 'answers', 'kryukov', 'witnesses', 'blocking', 'thoroughfare', 'sworn', 'funniest', 'chasing', 'robbery', 'squeals', 'giggles', 'scuffling', 'axes', 'bloodshed', 'fiendish', 'booty', 'rolled', 'attracting', 'buts', 'constitutes', 'circumstantial', 'accounts', 'denied', 'irrefutable', 'conclusively', 'prosecution', 'guilty', 'excites', 'proved', 'reluctance', 'owner', 'defence', 'presumption', 'popped', 'conclusive', 'melodramatic', 'personage', 'portly', 'sour', 'countenance', 'undisguised', 'dishevelled', 'fixedly', 'deliberation', 'constrained', 'shifting', 'overawe', 'civilly', 'severity', 'emphasising', 'syllable', 'anticipated', 'pompous', 'nodding', 'prolonged', 'yawn', 'hunter', 'proceeded', 'operation', 'arouse', 'articulated', 'impressively', 'wholly', 'languidly', 'embarrassment', 'presumed', 'posted', 'chosen', 'suspiciously', 'nursing', 'invalid', 'yawned', 'familiarity', 'unaffected', 'shabby', 'introduced', 'shrugged', 'commenced', 'sojourning', 'purposely', 'elapse', 'tidings', 'impatient', 'fianc', 'applied', 'expectation', 'betrothed', 'permissible', 'proceeding', 'complacent', 'le', 'tailor', 'appropriate', 'stylish', 'respectfully', 'exquisite', 'lavender', 'louvain', 'predominated', 'charming', 'jacket', 'fawn', 'lightest', 'cambric', 'pink', 'stripes', 'suited', 'mutton', 'chop', 'hairdresser', 'inevitably', 'suggesting', 'unpleasing', 'repulsive', 'imposing', 'causes', 'hardened', 'determine', 'oddities', 'earlier', 'preoccupations', 'conjecture', 'bakaleyev', 'voskresensky', 'yushin', 'scandalous', 'replied', 'huffily', 'permanent', 'andrey', 'madame', 'ministry', 'inquiry', 'guardian', 'learns', 'novelties', 'reforms', 'views', 'criticism', 'practicality', 'divorced', 'fermenting', 'exists', 'honesty', 'brigands', 'shod', 'indulgence', 'abnormal', 'external', 'environment', 'accomplished', 'circulating', 'dreamy', 'literature', 'maturer', 'injurious', 'ridicule', 'hastened', 'interpose', 'superciliousness', 'economic', 'commonplace', 'neighbour', 'excessive', 'naked', 'proverb', 'hares', 'tells', 'rests', 'manage', 'remains', 'adds', 'organised', 'firmer', 'foundations', 'welfare', 'acquiring', 'wealth', 'solely', 'liberality', 'consequence', 'unhappily', 'hindered', 'idealism', 'sentimentality', 'wit', 'perceive', 'chattering', 'incessant', 'flow', 'commonplaces', 'blush', 'exhibit', 'acquirements', 'pardonable', 'unscrupulous', 'mire', 'suggest', 'continue', 'disavowal', 'closer', 'health', 'doesn', 'names', 'wrappers', 'practised', 'ruffian', 'boldness', 'coolness', 'supposition', 'inexperienced', 'foresee', 'jewels', 'ransacked', 'trunks', 'notes', 'rob', 'counsel', 'departing', 'intellectual', 'vanity', 'overcame', 'increase', 'classes', 'arson', 'strikes', 'strangest', 'increasing', 'proportionately', 'hears', 'robbing', 'mail', 'social', 'forge', 'banknotes', 'gang', 'captured', 'lottery', 'tickets', 'ringleaders', 'lecturer', 'motive', 'demoralisation', 'civilised', 'inveterate', 'impracticality', 'forging', 'exact', 'upshot', 'crutches', 'chewed', 'emancipation', 'serfs', 'morality', 'accordance', 'logically', 'advocating', 'follows', 'lip', 'superciliously', 'incitement', 'acceptance', 'reproach', 'crimson', 'distort', 'report', 'foundation', 'arrow', 'excellent', 'qualities', 'miles', 'misunderstand', 'misrepresent', 'fanciful', 'fixing', 'piercing', 'restrain', 'connection', 'hell', 'curve', 'tormenting', 'overtake', 'mustn', 'irritated', 'gather', 'respond', 'inflammation', 'thanks', 'lingered', 'panic', 'precise', 'spiritual', 'samovar', 'dizzy', 'savage', 'energy', 'wasted', 'barrel', 'organ', 'accompanying', 'crinoline', 'mantle', 'feather', 'coarsened', 'sang', 'copeck', 'grinder', 'music', 'aged', 'idly', 'startled', 'snow', 'lamps', 'recognising', 'gaping', 'corn', 'chandler', 'booth', 'christened', 'graciously', 'billiard', 'princesses', 'dense', 'thickest', 'inclination', 'groups', 'angle', 'sadovy', 'wander', 'buildings', 'indoor', 'entrances', 'din', 'tinkling', 'guitar', 'merriment', 'thronging', 'swearing', 'throng', 'husky', 'blackened', 'saloon', 'dancing', 'marking', 'falsetto', 'gloomily', 'trilled', 'singer', 'musical', 'compliment', 'bass', 'generals', 'snub', 'noses', 'jolly', 'sweetie', 'hesitated', 'duclida', 'pock', 'wench', 'bruises', 'thinks', 'ledge', 'tempest', 'eternity', 'newspapers', 'spacious', 'champagne', 'waiter', 'items', 'accident', 'spontaneous', 'combustion', 'shopkeeper', 'alcohol', 'additions', 'curly', 'humour', 'humouredly', 'aside', 'befriend', 'winked', 'places', 'pouring', 'fee', 'profit', 'slapping', 'workman', 'unwell', 'oughtn', 'mysteriously', 'twisted', 'winking', 'mayn', 'sixth', 'gymnasium', 'cock', 'sparrow', 'fibbing', 'deposition', 'depose', 'screwed', 'heeding', 'mock', 'stunned', 'thoughtful', 'melancholy', 'sipped', 'morsel', 'resumed', 'original', 'coiners', 'simpletons', 'blab', 'collapses', 'untrustworthy', 'casual', 'million', 'count', 'crash', 'shivers', 'forwards', 'seventh', 'stew', 'example', 'risked', 'miracle', 'gibing', 'spending', 'begins', 'mislead', 'frowned', 'enjoy', 'earnestness', 'fences', 'hundredweight', 'roll', 'tablecloth', 'frightening', 'assez', 'caus', 'element', 'rapture', 'irritating', 'stimulated', 'revived', 'stimulus', 'removed', 'unwittingly', 'revolution', 'blockhead', 'astounded', 'fiercely', 'idiot', 'tie', 'benevolence', 'benefits', 'feels', 'plainly', 'hindering', 'persecute', 'kindness', 'ungrateful', 'gloating', 'venomous', 'roared', 'babbling', 'posing', 'idiots', 'brood', 'plagiarists', 'spermaceti', 'ointment', 'lymph', 'veins', 'unlike', 'guests', 'weren', 'snug', 'patience', 'fought', 'tooth', 'potchinkov', 'anybody', 'sheer', 'babushkin', 'madmen', 'blunder', 'rail', 'weaker', 'sunset', 'gathering', 'twilight', 'attic', 'flashing', 'rays', 'darkening', 'circles', 'uncanny', 'sunken', 'parapet', 'drowning', 'current', 'inflated', 'thronged', 'spectators', 'afrosinya', 'tearfully', 'boat', 'pole', 'granite', 'embankment', 'sneezing', 'wiping', 'wailed', 'indifference', 'apathy', 'disgusted', 'depression', 'listlessly', 'prompting', 'framework', 'amaze', 'corpses', 'sill', 'papering', 'lilac', 'annoyed', 'rolling', 'elder', 'preening', 'prinking', 'tit', 'vassilitch', 'regarded', 'saturday', 'male', 'sex', 'female', 'fluffles', 'enthusiastically', 'sententiously', 'answering', 'agonisingly', 'alyoshka', 'shil', 'lazy', 'jerked', 'tones', 'funk', 'jeeringly', 'rogue', 'belt', 'roads', 'elegant', 'bridle', 'lantern', 'misfortune', 'commotion', 'flowing', 'mutilated', 'disfigured', 'injured', 'merciful', 'confirmed', 'belonged', 'awaiting', 'legitimate', 'volunteered', 'foremost', 'polenka', 'strove', 'motionless', 'toes', 'pouting', 'screen', 'relieve', 'thinner', 'brighter', 'papa', 'colonel', 'mihailovitch', 'cursed', 'ball', 'marshal', 'princess', 'bezzemelny', 'mend', 'tear', 'darn', 'cough', 'bigger', 'prince', 'schegolskoy', 'kammerjunker', 'mazurka', 'thanked', 'expressions', 'polya', 'lida', 'wash', 'vagabond', 'clout', 'terrified', 'despairingly', 'swoon', 'placed', 'biting', 'screams', 'induced', 'assuring', 'towel', 'oftener', 'uncleanliness', 'preferred', 'fastest', 'relapsed', 'dumb', 'rigidity', 'pin', 'lodgers', 'streamed', 'overflowed', 'gape', 'reproaches', 'awe', 'exempt', 'sincerest', 'disturbance', 'rushing', 'vent', 'wrath', 'quarrelsome', 'irresponsible', 'clasping', 'trampled', 'ludwigovna', 'beg', 'haughtily', 'despicable', 'flatterers', 'audible', 'warn', 'conduct', 'remembers', 'protectors', 'abandoned', 'connections', 'rapidity', 'eloquence', 'groan', 'recognition', 'oozed', 'sad', 'trickled', 'huskily', 'exclaimed', 'obeyed', 'favourite', 'barefoot', 'indicating', 'barefooted', 'relieved', 'bared', 'gashed', 'fractured', 'bruise', 'cruel', 'hoof', 'wheel', 'bleed', 'sacrament', 'exchanging', 'glances', 'confession', 'indistinct', 'kneel', 'kneeling', 'rhythmically', 'precision', 'afford', 'especial', 'cover', 'ceasing', 'denser', 'midst', 'decked', 'gutter', 'finery', 'unmistakably', 'betraying', 'gaudy', 'ridiculous', 'train', 'flaring', 'rakishly', 'tilted', 'admonition', 'succour', 'sin', 'madam', 'compensate', 'sweat', 'sousing', 'rinsing', 'drying', 'darning', 'forgiveness', 'peremptorily', 'propping', 'humiliation', 'downwards', 'bury', 'reverence', 'devoted', 'repay', 'jostled', 'lamplight', 'compared', 'pardoned', 'halfway', 'greeting', 'message', 'breathless', 'graver', 'vely', 'brushing', 'misfortunes', 'sedate', 'presents', 'grammar', 'scripture', 'kolya', 'ave', 'maria', 'hugged', 'enchanted', 'imaginary', 'phantoms', 'reign', 'defiantly', 'challenging', 'win', 'proudly', 'flagging', 'emergency', 'boyish', 'sally', 'animated', 'samovars', 'dishes', 'savouries', 'amount', 'liquor', 'perceptibly', 'affected', 'visitors', 'invaluable', 'introduce', 'stuff', 'brightened', 'considerable', 'afternoon', 'needn', 'thirdly', 'specialty', 'surgery', 'mental', 'diseases', 'hatched', 'bubble', 'thrashing', 'ticklish', 'fainting', 'testifies', 'convulsions', 'masterly', 'dumbfoundered', 'arrive', 'plying', 'greeted', 'thunderbolt', 'clasped', 'tottered', 'moans', 'trifle', 'dislocated', 'bend', 'gratitude', 'providence', 'competent', 'alexandrovna', 'consolations', 'revealed', 'insane', 'induce', 'content', 'presiding', 'distressing', 'interrupting', 'muddled', 'today', 'reporting', 'suspense', 'impetuously', 'hasty', 'tipsily', 'learning', 'crest', 'infamy', 'act', 'despot', 'irritate', 'staying', 'blackguard', 'beseech', 'quantities', 'imbibed', 'bordering', 'persuading', 'plainness', 'emphasise', 'vise', 'bony', 'paws', 'jump', 'eccentric', 'peculiarities', 'timorous', 'disposition', 'unbounded', 'considerably', 'reassured', 'twinkling', 'reports', 'jealous', 'strolled', 'unworthy', 'pailfuls', 'argument', 'argue', 'trash', 'preside', 'individualism', 'fuel', 'flames', 'privilege', 'creation', 'error', 'err', 'examples', 'invention', 'ideals', 'aims', 'liberalism', 'preparatory', 'prefer', 'transport', 'fount', 'purity', 'perfection', 'entreat', 'homage', 'sincerely', 'daren', 'barber', 'spy', 'speculator', 'skin', 'flint', 'buffoon', 'puppy', 'bullock', 'corridor', 'aid', 'depend', 'unhinged', 'consoled', 'probing', 'infatuation', 'justified', 'pensive', 'proportioned', 'reliant', 'quality', 'detract', 'grace', 'softness', 'resembled', 'described', 'lighter', 'healthy', 'pallor', 'radiant', 'vigour', 'projected', 'irregularity', 'gay', 'lighthearted', 'transfigured', 'quiver', 'insolent', 'retained', 'serenity', 'sensitiveness', 'sincere', 'preserve', 'retaining', 'grow', 'crow', 'wrinkles', 'projecting', 'underlip', 'emotional', 'yielding', 'barrier', 'deepest', 'subdued', 'knocks', 'grant', 'fetching', 'resuming', 'relying', 'desert', 'exhilarated', 'flattered', 'oracle', 'convincing', 'comforting', 'seriousness', 'consultation', 'display', 'remarking', 'dazzling', 'endeavoured', 'satisfactorily', 'origin', 'product', 'apprehensions', 'troubles', 'enlarge', 'theme', 'insanity', 'composed', 'exaggerated', 'monomania', 'branch', 'shocks', 'affable', 'blessings', 'entreaties', 'licking', 'struggling', 'failings', 'feeble', 'wretch', 'whims', 'slack', 'patients', 'modesty', 'bashfulness', 'melting', 'unholy', 'smitten', 'rot', 'curing', 'strum', 'performer', 'ma', 'tre', 'rubinstein', 'attraction', 'mathematics', 'teaching', 'integral', 'calculus', 'gaze', 'prussian', 'lords', 'perspired', 'hysterics', 'comfortable', 'reminded', 'anchorage', 'navel', 'fishes', 'essence', 'pancakes', 'savoury', 'pies', 'sighs', 'stoves', 'advantages', 'bedtime', 'troubled', 'confronted', 'unlooked', 'novel', 'befallen', 'fired', 'cares', 'bequeathed', 'thrice', 'jealousy', 'obligations', 'criticise', 'unguarded', 'thinkable', 'furnishing', 'justification', 'uncleanness', 'envious', 'noisy', 'braggart', 'cynical', 'juxtaposition', 'blushed', 'desperately', 'abasement', 'infamies', 'smoothed', 'cynic', 'sloven', 'offend', 'brushed', 'scrupulously', 'shave', 'stubbly', 'razors', 'negative', 'pothouse', 'admitting', 'essentials', 'dishonest', 'parlour', 'dormouse', 'cure', 'confessor', 'plenty', 'worries', 'monomaniac', 'mole', 'hill', 'realities', 'mystery', 'hypochondriac', 'frantic', 'chatterbox', 'annoyance', 'bonjour', 'vouchsafed', 'risen', 'awkwardly', 'host', 'friendliness', 'disguised', 'breakfast', 'invitation', 'disorderly', 'vigorously', 'stream', 'omitted', 'consequences', 'prokofitch', 'dislikes', 'naturally', 'inhumanly', 'callous', 'alternating', 'characters', 'reserved', 'hindrance', 'hasn', 'beneficial', 'compressed', 'transparent', 'scarf', 'belongings', 'queen', 'diffident', 'impartially', 'uncritically', 'decisively', 'crab', 'piqued', 'moody', 'capricious', 'disregarded', 'liking', 'ugly', 'inexplicable', 'briefly', 'rejoiced', 'tentatively', 'consternation', 'openly', 'blamed', 'intentionally', 'score', 'dejected', 'vulgar', 'deigned', 'disgustingly', 'crimsoned', 'faltering', 'emphatically', 'announcing', 'advise', 'dated', 'unforeseen', 'rendered', 'likewise', 'deprived', 'intrude', 'respects', 'herewith', 'imperative', 'gross', 'unprecedented', 'affront', 'occasion', 'personally', 'indispensable', 'anticipation', 'compelled', 'withdraw', 'assumption', 'belief', 'testimony', 'notorious', 'gravely', 'pains', 'respectful', 'humble', 'p', 'refusing', 'stratagem', 'enamelled', 'venetian', 'flutter', 'escorting', 'prison', 'inch', 'sumptuous', 'banquets', 'lev', 'es', 'dozed', 'blaming', 'omen', 'bruised', 'overjoyed', 'weaknesses', 'frown', 'distress', 'caressing', 'slam', 'cheerfully', 'follow', 'listless', 'brows', 'knitted', 'performing', 'restlessness', 'sling', 'bandage', 'dejection', 'zest', 'practise', 'sore', 'controlling', 'pressure', 'tentative', 'impress', 'elementary', 'fundamental', 'tending', 'produce', 'cured', 'stage', 'coincides', 'occupation', 'aim', 'sage', 'mystified', 'mockery', 'thanking', 'referring', 'weighs', 'candidly', 'penetration', 'unfeigned', 'thankful', 'ecstatic', 'unspoken', 'reconciliation', 'exaggerating', 'vigorous', 'impulses', 'delicately', 'vasya', 'rush', 'hug', 'potanchikov', 'plaintively', 'recollecting', 'annoying', 'preoccupied', 'inattentive', 'reconciled', 'rite', 'carefulness', 'actions', 'infrequently', 'madder', 'carelessly', 'meditating', 'unpardonable', 'crevez', 'chiens', 'si', 'vous', 'tes', 'pas', 'contents', 'sarcastically', 'praiseworthy', 'overstep', 'unhappier', 'twisting', 'constraint', 'h', 'encouraged', 'dreadfully', 'considerate', 'defending', 'harnessed', 'bath', 'undergoing', 'treatment', 'baths', 'bathe', 'regularly', 'stroke', 'convulsion', 'hush', 'deadly', 'perceptible', 'clutching', 'distrustfully', 'bows', 'surprising', 'liveliness', 'protested', 'blushing', 'expensive', 'unreasonably', 'nunnery', 'hunchback', 'intent', 'tomb', 'oppressive', 'companionship', 'endurance', 'urgent', 'drily', 'cease', 'mournfully', 'dryness', 'useful', 'vindictively', 'hate', 'evils', 'intend', 'honestly', 'expects', 'deceiving', 'limits', 'courtship', 'esteems', 'feminine', 'obstinacy', 'acting', 'basely', 'vileness', 'merciless', 'heroism', 'despotism', 'tyranny', 'committing', 'fuss', 'puzzled', 'surprises', 'lawyer', 'pretentious', 'uneducated', 'consulted', 'jargon', 'courts', 'documents', 'frivolous', 'apropos', 'threat', 'equivalent', 'abandon', 'disobedient', 'summoning', 'resent', 'animation', 'skill', 'coarser', 'disillusion', 'contemptible', 'dissension', 'eagerness', 'esteem', 'resolution', 'businesslike', 'inviting', 'concealment', 'deception', 'modestly', 'modest', 'shyness', 'retreat', 'confused', 'protesting', 'calumny', 'declaring', 'vaguely', 'fleetingly', 'pang', 'inconceivable', 'disturbing', 'falteringly', 'mitrofanievsky', 'stammered', 'cemetery', 'begs', 'lunch', 'irregular', 'angular', 'kindliness', 'childishness', 'coffin', 'fussily', 'dining', 'greet', 'await', 'attentive', 'courteous', 'discomfort', 'courtesy', 'brightness', 'jealously', 'portrait', 'egoist', 'store', 'sinks', 'breaks', 'incautiously', 'push', 'presentiments', 'introduces', 'slanderer', 'depart', 'secrets', 'managing', 'keepsake', 'quaking', 'splendidly', 'meditate', 'noted', 'broad', 'spotless', 'bones', 'abundant', 'remarkedly', 'preserved', 'preoccupation', 'inscribed', 'altered', 'resslich', 'pledged', 'solicitude', 'emphasis', 'mentioning', 'hullo', 'stake', 'range', 'incredulous', 'sceptical', 'impose', 'pooh', 'apologising', 'perceived', 'butterfly', 'flies', 'stung', 'writhing', 'sweetmeat', 'wriggling', 'schoolboy', 'shamefaced', 'romeo', 'fiend', 'suits', 'cleaned', 'unheard', 'guffawing', 'strode', 'gawky', 'awkward', 'peony', 'crestfallen', 'ferocious', 'amply', 'introduction', 'subdue', 'assuming', 'irresistibly', 'ferocity', 'naturalness', 'strengthened', 'crashing', 'crown', 'quoted', 'overdo', 'smashing', 'fragments', 'incredulity', 'unpleasantly', 'pleasantly', 'bursting', 'begging', 'sniffed', 'trodden', 'corpulence', 'prominent', 'nosed', 'ironical', 'watery', 'mawkish', 'blinking', 'eyelashes', 'embarrassing', 'coherent', 'charge', 'feign', 'funds', 'pecuniary', 'claiming', 'financial', 'irony', 'screwing', 'troubling', 'prize', 'vindictive', 'feigned', 'selfish', 'grasping', 'cent', 'inquired', 'ash', 'tray', 'ruthlessly', 'scattering', 'carpet', 'concerned', 'legibly', 'pencil', 'observant', 'changing', 'repress', 'backs', 'midnight', 'defiance', 'strangled', 'artfully', 'dryly', 'liberal', 'trivialities', 'boring', 'whirl', 'exasperation', 'disguise', 'tracking', 'pack', 'dogs', 'spit', 'play', 'mouse', 'despise', 'inexperience', 'nasty', 'unintentional', 'bluntly', 'wink', 'teasing', 'foresaw', 'thieves', 'cleverly', 'jovial', 'everyday', 'socialist', 'doctrine', 'abnormality', 'organisation', 'noticeably', 'pamphlets', 'phrase', 'normally', 'righteous', 'excluded', 'exist', 'developing', 'historical', 'mathematical', 'organise', 'sinless', 'instinctively', 'ugliness', 'rules', 'mechanics', 'retrograde', 'smells', 'india', 'rubber', 'servile', 'revolt', 'reducing', 'planning', 'passages', 'phalanstery', 'vital', 'skip', 'presupposes', 'possibilities', 'millions', 'reduce', 'easiest', 'solution', 'seductively', 'musn', 'pages', 'print', 'drum', 'punch', 'preliminary', 'violates', 'noteworthy', 'gravity', 'progressively', 'humbugging', 'gesticulating', 'dissembler', 'periodical', 'weekly', 'printed', 'amalgamated', 'concern', 'initial', 'editor', 'analysed', 'psychology', 'suggested', 'suggestion', 'breaches', 'intentional', 'distortion', 'submission', 'transgress', 'challenge', 'contention', 'stated', 'contend', 'morals', 'hinted', 'fulfilment', 'discoveries', 'kepler', 'newton', 'eliminate', 'legislators', 'leaders', 'lycurgus', 'solon', 'mahomet', 'napoleon', 'exception', 'transgressed', 'ancestors', 'sacred', 'bravely', 'majority', 'benefactors', 'carnage', 'submit', 'division', 'acknowledge', 'arbitrary', 'categories', 'reproduce', 'sub', 'divisions', 'distinguishing', 'category', 'conservative', 'abiding', 'vocation', 'destroyers', 'destruction', 'capacities', 'varied', 'corpse', 'wade', 'sanction', 'wading', 'dimensions', 'masses', 'fulfil', 'justly', 'pedestal', 'goal', 'rights', 'vive', 'guerre', 'ternelle', 'jerusalem', 'preceding', 'tirade', 'lazarus', 'executed', 'lifetime', 'attain', 'executing', 'remark', 'witty', 'exactitude', 'definition', 'adopt', 'branded', 'arises', 'member', 'imagines', 'happily', 'happens', 'wittier', 'arise', 'unfortunately', 'predisposition', 'obedience', 'playfulness', 'cow', 'unobserved', 'despised', 'reactionaries', 'grovelling', 'tendencies', 'castigate', 'conscientious', 'perform', 'chastise', 'acts', 'penitence', 'edifying', 'alarming', 'capacity', 'grades', 'unfailing', 'regularity', 'races', 'stocks', 'spark', 'independence', 'approximately', 'genius', 'geniuses', 'retort', 'mournful', 'unconcealed', 'persistent', 'discourteous', 'sarcasm', 'fanaticism', 'attitude', 'impertinence', 'remove', 'enterprise', 'tries', 'snare', 'protected', 'prisons', 'banishment', 'investigators', 'deserves', 'logical', 'prohibition', 'sadness', 'comparison', 'worldly', 'hardship', 'napoleons', 'intonation', 'amiably', 'officially', 'seemingly', 'ransacking', 'racking', 'nerve', 'trap', 'slapped', 'deuce', 'apologetically', 'refute', 'careless', 'brainless', 'mirage', 'ambiguous', 'floating', 'impudence', 'explaining', 'frankly', 'merest', 'insinuation', 'eve', 'severe', 'soles', 'reaumur', 'stomach', 'downhearted', 'condescended', 'squeeze', 'novices', 'flatly', 'examinations', 'suspects', 'simpler', 'knave', 'strangeness', 'frankness', 'aspects', 'striding', 'gritting', 'lemon', 'reassure', 'fold', 'stud', 'tangled', 'wrinkled', 'flabby', 'discontentedly', 'meditation', 'articulately', 'chilled', 'belfry', 'cigars', 'underground', 'strewn', 'shells', 'faded', 'oppression', 'pretended', 'withdrew', 'sprang', 'infinitesimal', 'build', 'pyramid', 'physically', 'standstill', 'permitted', 'storms', 'toulon', 'massacre', 'paris', 'forgets', 'army', 'wastes', 'expedition', 'jest', 'vilna', 'altars', 'flesh', 'bronze', 'pyramids', 'waterloo', 'skinny', 'hash', 'digest', 'inartistic', 'killing', 'socialists', 'industrious', 'brick', 'ech', 'sthetic', 'benevolent', 'fleshly', 'lusts', 'grand', 'aimed', 'measuring', 'calculating', 'lice', 'proposed', 'vulgarity', 'abjectness', 'prophet', 'sabre', 'steed', 'allah', 'commands', 'sets', 'battery', 'deigning', 'desires', 'moon', 'breathlessness', 'stagnant', 'beckoning', 'beckon', 'panes', 'stillness', 'flooded', 'moonlight', 'weaving', 'snapping', 'splinter', 'pane', 'plaintive', 'buzz', 'cloak', 'double', 'rows', 'huddled', 'persist', 'indefinitely', 'whitish', 'buzzed', 'oddly', 'arkady', 'exclamation', 'cherish', 'assist', 'concerning', 'prejudiced', 'wrongly', 'persecuted', 'proposals', 'et', 'nihil', 'humanum', 'proposing', 'elope', 'switzerland', 'cherished', 'promoting', 'frankest', 'bonne', 'unpleasantness', 'apprehension', 'medical', 'diagnosed', 'apoplexy', 'contribute', 'calamity', 'morally', 'switch', 'marks', 'bored', 'switches', 'instances', 'beings', 'linger', 'harmoniously', 'reactionary', 'beneficent', 'publicity', 'nobleman', 'golden', 'provoking', 'humane', 'adaptable', 'breeding', 'haughtiness', 'climate', 'lounging', 'consists', 'forests', 'meadows', 'revenue', 'officials', 'anatomy', 'clubs', 'dussauts', 'parades', 'sharper', 'poets', 'deteriorated', 'greek', 'nezhin', 'bargained', 'lawful', 'wedlock', 'iou', 'elect', 'restive', 'trapped', 'incompatible', 'restrained', 'sunrise', 'naples', 'sea', 'blames', 'north', 'j', 'ai', 'vin', 'mauvais', 'berg', 'passengers', 'meditatively', 'decently', 'approved', 'ghosts', 'plaire', 'malaya', 'vishera', 'ingenuously', 'talks', 'silliest', 'cigar', 'cookshop', 'aniska', 'dressmaker', 'serf', 'trained', 'gracious', 'tease', 'choice', 'rarely', 'y', 'filka', 'burial', 'pipe', 'revenge', 'scamp', 'sung', 'bowing', 'appears', 'unreal', 'worlds', 'completeness', 'organism', 'possibility', 'becomes', 'contact', 'dies', 'spiders', 'juster', 'enemies', 'abstract', 'formed', 'generously', 'imprudently', 'psychologically', 'idleness', 'depravity', 'depraved', 'arriving', 'determining', 'aunt', 'detest', 'dished', 'mediation', 'disinclined', 'ulterior', 'privileged', 'millionth', 'fraction', 'coolly', 'tiniest', 'formalities', 'intrusive', 'sharpers', 'svirbey', 'raphael', 'madonna', 'prilukov', 'album', 'viazemsky', 'landowner', 'persecuting', 'attentions', 'hallucination', 'phantom', 'cousin', 'mystify', 'punctually', 'welcoming', 'boiling', 'reeking', 'blew', 'slighted', 'emphatic', 'disobeyed', 'fatigued', 'unavoidable', 'national', 'railways', 'inconvenience', 'disheartening', 'hostile', 'sidelong', 'punctiliousness', 'sacks', 'flour', 'mute', 'recourse', 'item', 'acquainted', 'desirous', 'discovering', 'contributed', 'accelerate', 'characteristics', 'period', 'relapse', 'abjectly', 'vicious', 'specimen', 'exertions', 'sacrifices', 'involving', 'homicidal', 'hushed', 'sums', 'commissions', 'niece', 'deaf', 'hated', 'grudged', 'inquest', 'verdict', 'suicide', 'proceedings', 'outraged', 'established', 'trusted', 'statement', 'philip', 'abolition', 'serfdom', 'hanged', 'systematic', 'persecution', 'domestic', 'philosopher', 'behaved', 'astute', 'insinuating', 'renewed', 'debtor', 'substantial', 'sufficiency', 'insignificant', 'ephemeral', 'proposition', 'urged', 'propose', 'engagement', 'pique', 'desired', 'weighty', 'dignified', 'insistance', 'insults', 'goodwill', 'overstepped', 'esteeming', 'adoring', 'occupy', 'impertinent', 'thereby', 'existing', 'flushing', 'fidgeted', 'reproof', 'relished', 'outweigh', 'nevertheless', 'razsudkin', 'surname', 'misrepresenting', 'advantageous', 'conjugal', 'profitable', 'accusing', 'malicious', 'relied', 'correspondence', 'convince', 'considerately', 'instigation', 'approvingly', 'enlarged', 'alluded', 'falsehood', 'virtues', 'lofty', 'hinder', 'pleasures', 'intimacy', 'withdrawing', 'similar', 'compromises', 'delicacy', 'legacy', 'helplessness', 'entrusted', 'victims', 'quivered', 'dismissal', 'springing', 'intervened', 'stormed', 'restraining', 'conductor', 'implored', 'disregarding', 'reinstating', 'acted', 'recklessly', 'smashed', 'ending', 'overbearing', 'destitute', 'conceit', 'fatuity', 'insignificance', 'morbidly', 'admiration', 'gloated', 'valued', 'amassed', 'devices', 'superiors', 'sincerity', 'genuinely', 'ingratitude', 'groundlessness', 'contradicted', 'disbelieved', 'heroic', 'admired', 'admire', 'reap', 'flattery', 'undeservedly', 'unrecognised', 'unthinkable', 'voluptuous', 'amassing', 'brooded', 'virtuous', 'humbled', 'saviour', 'scenes', 'amorous', 'episodes', 'allurement', 'grateful', 'condescension', 'absolute', 'wider', 'fascination', 'wonders', 'aureole', 'ruins', 'clap', 'masterful', 'milksop', 'embracing', 'delivered', 'consciously', 'ton', 'insistent', 'omitting', 'ghostly', 'visitations', 'wishing', 'offers', 'designs', 'assumes', 'borrowing', 'disproportionate', 'soothing', 'ecstatically', 'accommodating', 'bothering', 'per', 'lend', 'unfold', 'publishers', 'booksellers', 'publications', 'publisher', 'european', 'languages', 'schwach', 'scuttling', 'saint', 'pots', 'hesitate', 'blockheads', 'untried', 'belonging', 'communicating', 'furnished', 'moderate', 'burying', 'dully', 'undertone', 'wicked', 'heartless', 'wits', 'lamp', 'penetrating', 'nervously', 'describe', 'soothed', 'whereabouts', 'gallery', 'inexpressibly', 'separate', 'barn', 'quadrangle', 'aslant', 'acute', 'disproportionately', 'obtuse', 'scratched', 'scrutinising', 'arbiter', 'destinies', 'theirs', 'stammer', 'stammers', 'hesitatingly', 'depths', 'champion', 'insatiable', 'feature', 'righteousness', 'canary', 'builds', 'native', 'boarding', 'superintend', 'hugs', 'tub', 'taste', 'pedlar', 'embroidered', 'grieved', 'pitilessly', 'insured', 'overburdened', 'imploringly', 'entreaty', 'rainy', 'ironically', 'paced', 'desperation', 'protect', 'malignance', 'unutterable', 'sobs', 'tearful', 'dishonourable', 'sinner', 'solemnly', 'destroyed', 'filth', 'loathe', 'saving', 'degradation', 'wiser', 'leap', 'monstrously', 'orphan', 'pitiful', 'unique', 'infrequent', 'exceptionalness', 'penetrated', 'madhouse', 'obscures', 'sceptic', 'iniquity', 'abyss', 'loathsomeness', 'forcibly', 'heaving', 'maniac', 'testament', 'unwillingly', 'gospel', 'sterner', 'requiem', 'maniacs', 'infectious', 'lunatic', 'breathlessly', 'named', 'bethany', 'unveil', 'stepmother', 'crazed', 'spasm', 'st', 'nineteenth', 'verse', 'jews', 'martha', 'mary', 'jesus', 'hadst', 'wilt', 'saith', 'resurrection', 'believeth', 'whosoever', 'liveth', 'believest', 'yea', 'art', 'passionately', 'reproduced', 'censure', 'disbelieving', 'blinded', 'unbelieving', 'cometh', 'cave', 'stinketh', 'hath', 'wouldest', 'shouldest', 'glory', 'hearest', 'thus', 'graveclothes', 'feverishly', 'harlot', 'analyse', 'infinitely', 'bade', 'hysterically', 'ant', 'chose', 'advertising', 'uninhabited', 'adjoined', 'enjoyed', 'pounce', 'elected', 'conflict', 'unmitigated', 'arrogant', 'vowed', 'upholstered', 'bookcase', 'genial', 'awkwardness', 'domain', 'tout', 'apologies', 'formally', 'proportions', 'dashes', 'avoiding', 'rebounding', 'repairs', 'repetition', 'incongruous', 'ineptitude', 'incautious', 'insolently', 'tradition', 'investigating', 'lawyers', 'attack', 'afar', 'fatal', 'crafty', 'contracted', 'broadened', 'disturbed', 'stress', 'addition', 'cackled', 'fidgeting', 'tickled', 'paralysis', 'bachelor', 'seed', 'c', 'est', 'rigueur', 'uncomfortable', 'babble', 'offending', 'exercise', 'sedentary', 'join', 'ranks', 'privy', 'councillors', 'skipping', 'interrogations', 'interrogator', 'interrogated', 'aptly', 'wittily', 'muddle', 'harping', 'reform', 'prisoner', 'rudest', 'disarming', 'felicitous', 'formality', 'chat', 'bounded', 'babbled', 'reverting', 'incoherence', 'gesticulations', 'elaborate', 'methods', 'adheres', 'precedent', 'instruct', 'publish', 'prematurely', 'mathematically', 'depriving', 'sevastopol', 'alma', 'enemy', 'siege', 'occurs', 'comic', 'cultivated', 'overlooked', 'measures', 'attractions', 'weave', 'provide', 'flop', 'swallow', 'scare', 'proofs', 'braced', 'ordeal', 'strangle', 'flecked', 'foam', 'policy', 'anyhow', 'harmless', 'chuckling', 'awaken', 'intellect', 'fascinate', 'austrian', 'hof', 'kriegsrath', 'military', 'cleverest', 'mack', 'surrendered', 'civilian', 'histories', 'missed', 'major', 'sharpest', 'calculation', 'reward', 'adornment', 'tricks', 'liable', 'incognito', 'flagrant', 'stuffy', 'lied', 'incomparably', 'betrays', 'paleness', 'questioner', 'brings', 'allegorical', 'allusions', 'psychologist', 'reflects', 'mirror', 'checking', 'murdering', 'prosecute', 'decanter', 'dined', 'wrongs', 'shivered', 'practice', 'imposed', 'unintentionally', 'imagining', 'acquitted', 'tut', 'studied', 'neglect', 'consult', 'lightheaded', 'rejected', 'penetrate', 'damnable', 'wily', 'tittered', 'genially', 'suspiciousness', 'disarmed', 'incensed', 'preserving', 'injury', 'reliable', 'delusions', 'imperiously', 'overhear', 'peremptory', 'mystification', 'paroxysm', 'obeying', 'friendship', 'chuckled', 'maddened', 'restraint', 'punchinello', 'retreated', 'rubbishly', 'priests', 'deputies', 'shove', 'deathly', 'crop', 'warder', 'instantaneously', 'birthday', 'comical', 'harassing', 'proving', 'vivisecting', 'witted', 'writers', 'collect', 'stupefied', 'amazing', 'imminent', 'sketchily', 'outlines', 'circumstantially', 'compromised', 'sleeve', 'connected', 'memorial', 'sinned', 'wronged', 'trade', 'prepare', 'hides', 'cuts', 'convict', 'detective', 'punching', 'scoundrels', 'scolded', 'confident', 'fateful', 'sobering', 'intensely', 'incredible', 'snake', 'gnawing', 'jaundice', 'fattish', 'sarcastic', 'impulsiveness', 'hitch', 'redecorated', 'tradesman', 'entertain', 'contract', 'forfeit', 'upholsterers', 'instalment', 'purchased', 'slay', 'promptly', 'trousseau', 'knick', 'knacks', 'jewellery', 'knopp', 'entertainment', 'nines', 'parsimony', 'ward', 'doings', 'legend', 'omniscient', 'approximate', 'progressives', 'nihilists', 'feared', 'transferring', 'patrons', 'anticipate', 'contingencies', 'allayed', 'doctrines', 'systems', 'pestered', 'expose', 'mic', 'scrofulous', 'attached', 'legion', 'animate', 'abortions', 'coxcombs', 'attach', 'vulgarise', 'caricature', 'duping', 'despising', 'expounding', 'darwinian', 'liar', 'propaganda', 'accepted', 'belauded', 'commune', 'christening', 'acquiesce', 'lover', 'praises', 'disdain', 'bonds', 'bundles', 'unmoved', 'entertaining', 'inferiority', 'incredibly', 'enlarging', 'clicking', 'beads', 'breach', 'discourse', 'console', 'promote', 'ceremonies', 'beggarly', 'feast', 'wines', 'obtaining', 'flustered', 'libel', 'permissable', 'defend', 'equality', 'convention', 'indirectly', 'enlightenment', 'terebyeva', 'community', 'conventionally', 'varents', 'communities', 'letters', 'fifteenth', 'regretted', 'distinguons', 'compulsory', 'voluntary', 'asset', 'dispose', 'assets', 'rejoice', 'yelled', 'disinterestedly', 'inappropriately', 'conditions', 'establish', 'broader', 'basis', 'reject', 'coarsely', 'crude', 'striving', 'chastity', 'lucky', 'mistakenly', 'deserving', 'reliance', 'inequality', 'debate', 'associations', 'france', 'inconvenient', 'tfoo', 'referred', 'privacy', 'cesspools', 'cesspool', 'nobler', 'snigger', 'heartily', 'protests', 'bantering', 'ignorance', 'rainbow', 'indecorous', 'sufficient', 'enable', 'ascertain', 'preternatural', 'comprehensible', 'comprehen', 'foreseeing', 'temporary', 'patronage', 'parent', 'term', 'credulous', 'subscription', 'outsiders', 'assisting', 'unsafe', 'jamaica', 'rum', 'madeira', 'celebrate', 'orphans', 'unfolded', 'ceremoniously', 'interrupt', 'verb', 'sympathise', 'eradicate', 'promotes', 'drawback', 'affection', 'legality', 'warhorse', 'trumpet', 'dictionary', 'corrective', 'opposing', 'avenging', 'sniggered', 'originated', 'disordered', 'deceased', 'suitably', 'compels', 'traditional', 'genteel', 'aristocratic', 'poorest', 'paroxysms', 'irresistible', 'intimidated', 'harassed', 'stages', 'doctors', 'affect', 'variety', 'lisbon', 'quantity', 'honey', 'purchasing', 'provisions', 'stranded', 'disposal', 'bazaar', 'pani', 'serviceable', 'invent', 'disillusioned', 'repulse', 'failures', 'keenly', 'jar', 'disaster', 'brightest', 'crockery', 'knives', 'forks', 'shapes', 'patterns', 'ribbons', 'justifiable', 'displeased', 'mistress', 'contented', 'inwardly', 'creatures', 'guest', 'exalted', 'adding', 'praised', 'cue', 'sharing', 'maidish', 'complained', 'threatening', 'harbour', 'governorship', 'spotty', 'abominably', 'immemorial', 'ages', 'commissariat', 'removing', 'feed', 'bred', 'loftily', 'seats', 'responsible', 'resented', 'professorship', 'apologised', 'suppressed', 'interspersing', 'uncontrollable', 'cuckoo', 'owl', 'patronising', 'sweeps', 'pan', 'starved', 'glee', 'nicety', 'baggage', 'provincial', 'nonentity', 'fray', 'skirts', 'paints', 'required', 'unpunctual', 'entr', 'jelly', 'fidget', 'choosing', 'flatter', 'gratify', 'striped', 'disdained', 'hospitality', 'poking', 'imbecile', 'draggletails', 'gulping', 'twelfth', 'disreputable', 'sole', 'shoe', 'gingerbread', 'vouchsafe', 'bounds', 'drubbing', 'heaved', 'alluding', 'tasting', 'hurting', 'peaceably', 'pierced', 'chemist', 'cabman', 'anecdotes', 'vater', 'aus', 'berlin', 'foreigners', 'stupider', 'punishing', 'addled', 'glaring', 'regaining', 'launched', 'alluring', 'armed', 'incontestably', 'adventuresses', 'fore', 'en', 'toutes', 'lettres', 'peaceful', 'teachers', 'frenchman', 'mangot', 'disdainfully', 'unaware', 'undoubted', 'ability', 'gentleness', 'tapping', 'misgivings', 'w', 'sche', 'dame', 'novels', 'laundry', 'maid', 'directress', 'appropriately', 'slut', 'poof', 'represent', 'resembling', 'amid', 'finn', 'lobster', 'squealed', 'milkman', 'lashed', 'johann', 'burgomeister', 'trample', 'vigilant', 'squabbles', 'stepdaughter', 'edging', 'thunderstruck', 'clamour', 'mademoiselle', 'admonishing', 'accuse', 'accusation', 'purposes', 'securities', 'confirm', 'advisability', 'primarily', 'allude', 'total', 'associated', 'donation', 'reproachfully', 'gott', 'der', 'barmherzige', 'pettifogging', 'eater', 'trashy', 'sovereign', 'tsar', 'meekness', 'parabola', 'arose', 'occasional', 'heartrending', 'wail', 'shelter', 'swaying', 'baby', 'audience', 'agonised', 'unrestrained', 'trustful', 'piteous', 'instigator', 'accomplice', 'lowered', 'compassionate', 'commiserating', 'hugging', 'madwoman', 'drowned', 'sighted', 'dumbfounded', 'stammering', 'nonsensical', 'riddles', 'oath', 'impudently', 'opposed', 'approve', 'effects', 'radical', 'decking', 'charitable', 'test', 'indelicate', 'kobilatnikov', 'treatise', 'recommend', 'piderit', 'wagner', 'reflections', 'winded', 'harangue', 'deduction', 'alas', 'vehemence', 'slandering', 'godless', 'propositions', 'murmurs', 'solved', 'suspected', 'scoundrelly', 'intrigue', 'recently', 'consequently', 'dividing', 'squandering', 'insinuations', 'unpardonably', 'estrange', 'revenging', 'interruptions', 'deliberating', 'implied', 'accusations', 'suggestions', 'tremendously', 'lajdak', 'polish', 'grasp', 'obstructing', 'unmasked', 'judges', 'infidels', 'agitators', 'atheists', 'motives', 'overbalancing', 'impunity', 'submissiveness', 'disappointment', 'murmur', 'throb', 'amidst', 'battle', 'unequal', 'raged', 'lamenting', 'commented', 'impelled', 'superficially', 'everlastingly', 'peevishly', 'roundabout', 'divine', 'grumbled', 'morosely', 'arrogance', 'incomplete', 'unendurable', 'paler', 'steeple', 'froze', 'immovably', 'infected', 'gripping', 'foreseen', 'miserably', 'recoiled', 'plunder', 'wearily', 'comprehend', 'enigmatically', 'coward', 'alike', 'mont', 'blanc', 'picturesque', 'monumental', 'sinful', 'sadly', 'drudge', 'centered', 'fared', 'decorously', 'inflicted', 'thorough', 'scale', 'harmful', 'brilliance', 'fees', 'sulkiness', 'spider', 'ceilings', 'cramp', 'candles', 'notebooks', 'waits', 'alter', 'whoever', 'despises', 'lawgiver', 'dares', 'creed', 'code', 'divined', 'stoop', 'needful', 'temptation', 'blasphemer', 'insistence', 'argued', 'headlong', 'web', 'sucking', 'committed', 'murders', 'hideously', 'defiled', 'expiate', 'destroy', 'knaves', 'supplication', 'unfair', 'shore', 'burdensome', 'immeasurably', 'cypress', 'perturbed', 'abused', 'sing', 'actors', 'tubercles', 'experiments', 'professor', 'incorrect', 'douches', 'tattered', 'continuous', 'sills', 'poison', 'surging', 'silently', 'vacantly', 'cutting', 'hardworking', 'fading', 'permanence', 'depending', 'rapping', 'frying', 'semyonova', 'principally', 'sunshine', 'flag', 'coaxed', 'appealed', 'scoffers', 'clapping', 'singers', 'turban', 'turk', 'costume', 'ostrich', 'fidelity', 'grinders', 'bereaved', 'drives', 'fatherless', 'tenez', 'droite', 'uninterrupted', 'castle', 'forsaken', 'inkpot', 'tongues', 'parlez', 'moi', 'fran', 'ais', 'judy', 'performance', 'impromptu', 'rehearse', 'nevsky', 'sings', 'hussar', 'cinq', 'sous', 'marlborough', 'va', 'lullaby', 'ne', 'sait', 'quand', 'reviendra', 'hips', 'monter', 'notre', 'menage', 'genteelly', 'bodice', 'widths', 'deformed', 'stupids', 'ceremonious', 'gentlewoman', 'grouse', 'stamped', 'basest', 'slandered', 'licence', 'sakes', 'troublesome', 'naughty', 'imps', 'flows', 'chokes', 'adopted', 'mouthed', 'sillies', 'och', 'rattle', 'glissez', 'basque', 'tap', 'du', 'diamanten', 'und', 'perlen', 'sch', 'nsten', 'augen', 'dchen', 'willst', 'mehr', 'invents', 'midday', 'vale', 'dagestan', 'rending', 'flood', 'caressingly', 'unconsciousness', 'asylum', 'enormously', 'foretold', 'fog', 'dreary', 'intervals', 'catastrophe', 'recollections', 'incidents', 'existed', 'prey', 'amounting', 'insensibility', 'immediate', 'irksome', 'permanently', 'city', 'reference', 'tacitly', 'satisfactory', 'institutions', 'server', 'incense', 'blessed', 'abnegation', 'interpreted', 'lonelier', 'mingle', 'restaurants', 'thoroughfares', 'songs', 'smote', 'requiring', 'greediness', 'fresher', 'calmer', 'reported', 'guzzling', 'bite', 'bout', 'wherever', 'conspirator', 'defended', 'companions', 'hypocrisy', 'resourcefulness', 'descended', 'hints', 'confessing', 'smallness', 'cramping', 'lethargy', 'suffocating', 'penned', 'feebler', 'fundamentally', 'riddle', 'te', 'drained', 'mortal', 'brigand', 'flinching', 'pernicious', 'tickling', 'dr', 'b', 'lungs', 'patting', 'careworn', 'unfairly', 'openness', 'disconcert', 'disdaining', 'wiles', 'proportion', 'extent', 'blurts', 'results', 'imperious', 'elements', 'magnanimity', 'efface', 'capitally', 'reproducing', 'rabbits', 'headstrong', 'sleepless', 'amateur', 'essays', 'mistiness', 'chord', 'vibrating', 'mist', 'incorruptible', 'prosecutor', 'notions', 'umsonst', 'blurting', 'reckless', 'formidable', 'bowled', 'bothered', 'semi', 'plausible', 'morgenfr', 'outsider', 'type', 'responsive', 'dances', 'stories', 'villages', 'attends', 'laughs', 'picks', 'believer', 'dissenter', 'wanderers', 'guidance', 'villagers', 'wilderness', 'fervour', 'sect', 'responds', 'juries', 'venerable', 'bible', 'authorities', 'mild', 'assaults', 'weapon', 'influencing', 'abjure', 'plausibly', 'renews', 'preached', 'bookish', 'precipice', 'tower', 'poses', 'recantation', 'sympathetically', 'scornfully', 'chase', 'capture', 'hare', 'confront', 'mug', 'rascal', 'notoriously', 'etiquette', 'surrender', 'shameless', 'lessen', 'aberration', 'abandoning', 'appearances', 'lessening', 'mitigation', 'expressively', 'shortened', 'bourgeois', 'grossly', 'torturer', 'entrails', 'safe', 'expiation', 'harden', 'majestic', 'proclaim', 'schiller', 'flunkey', 'freshen', 'manoeuvre', 'investigate', 'straws', 'irrevocable', 'owning', 'undoubtedly', 'deceitful', 'befriending', 'hovering', 'causing', 'dogging', 'transform', 'rash', 'root', 'intriguing', 'oppressed', 'overflowing', 'clarionet', 'violin', 'boom', 'turkish', 'mindedly', 'chorus', 'click', 'balls', 'tucked', 'tyrolese', 'hall', 'contralto', 'accompaniment', 'guttural', 'rhymes', 'katia', 'gulps', 'trailed', 'patriarchal', 'obsequious', 'cowards', 'philosophers', 'administrative', 'centre', 'declaim', 'mask', 'wonderfully', 'dainty', 'injure', 'derive', 'complex', 'glutton', 'club', 'gourmand', 'remnants', 'steak', 'cavalry', 'photographer', 'journalist', 'biography', 'gambler', 'challenged', 'philosophy', 'candour', 'founded', 'ember', 'extinguished', 'exceeds', 'moderation', 'exceed', 'prudent', 'shoot', 'parried', 'mystic', 'apparitions', 'pretend', 'preach', 'sthetics', 'idealist', 'brag', 'debtors', 'particulars', 'throughout', 'clove', 'swinishness', 'faithful', 'unwritten', 'fourthly', 'maidservants', 'fifthly', 'sixthly', 'reveal', 'dissolute', 'profligate', 'preconceived', 'opinions', 'woes', 'decorous', 'oraison', 'bre', 'tender', 'influenced', 'ardent', 'impressionable', 'reception', 'confide', 'betting', 'charged', 'refer', 'tales', 'idiocy', 'footman', 'satisfy', 'repellent', 'aspect', 'usefulness', 'cage', 'century', 'reigning', 'pro', 'consul', 'asia', 'minor', 'martyrdom', 'pincers', 'roots', 'ecstasies', 'visions', 'thirsting', 'suggests', 'divinity', 'phenomenally', 'parasha', 'wishes', 'exhortations', 'supplications', 'posed', 'hungering', 'resorted', 'subjection', 'resource', 'harder', 'discord', 'vestal', 'virgin', 'seduced', 'tactics', 'prostrate', 'shamelessly', 'resisted', 'unprincipled', 'treachery', 'yielded', 'unawares', 'triumphed', 'succumbed', 'spoiled', 'coarsest', 'convert', 'tremendous', 'epileptic', 'expresses', 'attorney', 'mockingly', 'epithet', 'bestowed', 'cher', 'ami', 'hi', 'wetting', 'annihilate', 'paralysed', 'serving', 'nephews', 'fascinating', 'curtseys', 'frock', 'unopened', 'bud', 'curls', 'lamb', 'charmer', 'flushes', 'delicious', 'rit', 'steals', 'scorches', 'sistine', 'diamonds', 'pearls', 'obedient', 'maiden', 'sensuality', 'haunts', 'renew', 'crippled', 'debauchery', 'reeked', 'odours', 'frightful', 'dens', 'cancan', 'specialist', 'vis', 'consoling', 'proffered', 'sensual', 'vertu', 'elle', 'se', 'nicher', 'outcries', 'bill', 'adventures', 'stimulating', 'ruder', 'adieu', 'mon', 'plaisir', 'actively', 'unearthed', 'mirthful', 'refrained', 'devoured', 'patroness', 'asylums', 'charmed', 'depositing', 'subscribing', 'suppressing', 'hotel', 'teaches', 'expound', 'latest', 'mischance', 'fare', 'coil', 'enrage', 'bond', 'hood', 'sensualist', 'lightly', 'tiresome', 'signalling', 'beloved', 'indiscretion', 'betrayal', 'meddling', 'apartments', 'conveniently', 'successive', 'glow', 'involuntary', 'secluded', 'brave', 'gasped', 'admission', 'combinations', 'misdeed', 'wrongdoing', 'galling', 'overweening', 'paltry', 'shaped', 'vivid', 'charm', 'apply', 'laws', 'une', 'th', 'orie', 'comme', 'autre', 'overstepping', 'remorse', 'russians', 'chaotic', 'terrace', 'supper', 'breadth', 'traditions', 'chronicle', 'fogeys', 'persevere', 'magazine', 'sprinkled', 'atone', 'passports', 'hem', 'exciting', 'uselessly', 'barricade', 'tormentor', 'assault', 'submitting', 'unbending', 'revolver', 'cocked', 'aha', 'alters', 'shooting', 'poisoned', 'oho', 'nightingale', 'kindle', 'bullet', 'grazed', 'wasp', 'temple', 'seize', 'pistol', 'loaded', 'stubborn', 'defined', 'suppliant', 'stubbornly', 'construction', 'charges', 'capsule', 'haunt', 'villain', 'tyrant', 'waiters', 'crooked', 'lanky', 'pine', 'vauxhall', 'clown', 'munich', 'entertained', 'teaspoon', 'waterfall', 'streams', 'flashes', 'drenched', 'locking', 'wonderingly', 'soaking', 'assigned', 'acknowledgments', 'receipts', 'alternatives', 'heedlessly', 'questioned', 'greetings', 'sends', 'perturbation', 'surmise', 'decrepit', 'wheeled', 'ejaculations', 'reinforced', 'patted', 'englishmen', 'fortunate', 'fedosya', 'sorrowful', 'mainland', 'roaring', 'waters', 'endless', 'adrianople', 'lights', 'veal', 'caf', 'chantant', 'planks', 'indistinguishable', 'upbraiding', 'shaded', 'occupants', 'inflamed', 'pose', 'smiting', 'sneeze', 'sheepish', 'befogged', 'dregs', 'attendant', 'wrapping', 'blanket', 'scratching', 'mice', 'reverie', 'trees', 'stormy', 'park', 'landscape', 'recommended', 'avenge', 'promises', 'damnation', 'dozing', 'zigzags', 'dart', 'draught', 'dampness', 'howled', 'dwelling', 'trinity', 'cottage', 'overgrown', 'fragrant', 'porch', 'wreathed', 'climbers', 'surrounded', 'roses', 'carpeted', 'rugs', 'plants', 'china', 'nosegays', 'narcissus', 'stalks', 'reluctant', 'balcony', 'freshly', 'chirruping', 'shroud', 'edged', 'frill', 'wreaths', 'carved', 'marble', 'wreath', 'rigid', 'profile', 'chiselled', 'unchildish', 'appalled', 'smirched', 'unmerited', 'unheeded', 'brutally', 'frost', 'daytime', 'cellar', 'blurs', 'cannon', 'resounded', 'signal', 'river', 'swirling', 'flooding', 'basements', 'cellars', 'rats', 'swim', 'ticking', 'touches', 'drip', 'mammy', 'bwoken', 'neglected', 'stockingless', 'lids', 'unchildlike', 'provocative', 'aglow', 'nightmare', 'overslept', 'notebook', 'milky', 'slippery', 'picturing', 'paths', 'passer', 'shutters', 'achilles', 'helmet', 'drowsy', 'perpetual', 'peevish', 'sourly', 'jewish', 'trigger', 'lagging', 'appallingly', 'exposure', 'speechless', 'foolishly', 'muddy', 'concerns', 'hatching', 'nudging', 'experiences', 'magazines', 'poems', 'manuscript', 'selfishness', 'fullness', 'dearly', 'vision', 'uncertainly', 'lustreless', 'doubted', 'expiating', 'noxious', 'insect', 'atonement', 'imbecility', 'spilt', 'crowned', 'capitol', 'clumsiness', 'sthetically', 'bombarding', 'symptom', 'manly', 'ivory', 'nun', 'confided', 'hardships', 'whimper', 'indiscriminately', 'viii', 'fellowship', 'fairest', 'angers', 'annoys', 'brutish', 'cooler', 'concentrate', 'symbol', 'grieving', 'nurse', 'resentful', 'procession', 'rebellious', 'retract', 'dawned', 'van', 'campany', 'incongruity', 'chanted', 'lachrymose', 'distasteful', 'jerky', 'unmixed', 'kindled', 'spreading', 'bliss', 'boozed', 'glimpse', 'shanties', 'mount', 'eggshells', 'spiral', 'privately', 'doorkeeper', 'seating', 'fairy', 'ro', 'rodionovitch', 'originality', 'talents', 'bun', 'covers', 'bigotry', 'youngster', 'boast', 'deterred', 'ascetic', 'monk', 'hermit', 'research', 'soars', 'livingstone', 'nihilist', 'almighty', 'ennobled', 'midwives', 'wenches', 'academy', 'immoderate', 'zeal', 'suicides', 'nil', 'pavlitch', 'listlessness', 'reeled', 'barking', 'grinned', 'hulloa', 'brokenly', 'epilogue', 'centres', 'fortress', 'adhered', 'confuse', 'soften', 'metal', 'minutely', 'voznesensky', 'uppermost', 'discover', 'truthful', 'versed', 'mania', 'hypochondriacal', 'robber', 'shallow', 'privation', 'heartfelt', 'repentance', 'exaggerate', 'incidentally', 'hypothesis', 'commits', 'thirteenth', 'rescued', 'investigated', 'extenuating', 'commission', 'thereafter', 'version', 'hinting', 'unsatisfactory', 'ardour', 'livelihood', 'emigrate', 'workers', 'convicts', 'despatched', 'fervent', 'anticipations', 'predicted', 'implicit', 'displayed', 'lectures', 'rescuing', 'conveyances', 'hangings', 'fatiguing', 'razumihins', 'clearest', 'description', 'minuteness', 'clearness', 'externally', 'shirking', 'sundays', 'holidays', 'barracks', 'unhealthy', 'plank', 'rug', 'inattention', 'visits', 'workshops', 'kilns', 'sheds', 'irtish', 'lightened', 'horrors', 'patched', 'trials', 'beetles', 'fetters', 'rough', 'borne', 'decree', 'objectless', 'strive', 'repent', 'raging', 'blunders', 'criticised', 'blundering', 'swarmed', 'clashed', 'independently', 'broadly', 'uninfluenced', 'sceptics', 'halt', 'inheriting', 'punished', 'criminality', 'falsity', 'crisis', 'attribute', 'prized', 'privations', 'tramps', 'ray', 'primeval', 'forest', 'tramp', 'sweetheart', 'downcast', 'gulf', 'species', 'isolation', 'exiles', 'ignorant', 'churls', 'seminarists', 'hack', 'infidel', 'flinch', 'intervening', 'christmas', 'rolls', 'sweethearts', 'frail', 'gait', 'illnesses', 'easter', 'plague', 'europe', 'microbes', 'endowed', 'sufferers', 'infallible', 'peoples', 'infection', 'armies', 'soldiers', 'stabbing', 'devouring', 'trades', 'improvements', 'conflagrations', 'famine', 'involved', 'purify', 'grating', 'sentinel', 'pencilled', 'pound', 'alabaster', 'kiln', 'baking', 'tool', 'logs', 'steppe', 'bathed', 'specks', 'nomads', 'tents', 'abraham', 'flocks', 'contemplation', 'burnous', 'timidity', 'dawn', 'sources', 'religion', 'pester', 'aspirations', 'gradual', 'renewal', 'regeneration', 'initiation', 'file', 'txt', 'zip', 'files', 'formats', 'http', 'david', 'widger', 'editions', 'replace', 'renamed', 'creating', 'owns', 'copyright', 'distribute', 'royalties', 'copying', 'distributing', 'tm', 'electronic', 'concept', 'trademark', 'registered', 'ebooks', 'specific', 'complying', 'derivative', 'performances', 'modified', 'redistribution', 'mission', 'distribution', 'comply', 'available', 'section', 'redistributing', 'indicate', 'abide', 'access', 'refund', 'entity', 'paragraph', 'archive', 'pglaf', 'compilation', 'collection', 'located', 'displaying', 'based', 'references', 'compliance', 'format', 'govern', 'countries', 'constant', 'check', 'downloading', 'representations', 'status', 'links', 'prominently', 'accessed', 'viewed', 'copied', 'distributed', 'derived', 'contain', 'holder', 'providing', 'appearing', 'requirements', 'paragraphs', 'additional', 'linked', 'unlink', 'detach', 'redistribute', 'binary', 'nonproprietary', 'proprietary', 'processing', 'hypertext', 'vanilla', 'ascii', 'site', 'user', 'exporting', 'alternate', 'include', 'specified', 'viewing', 'reasonable', 'royalty', 'profits', 'calculate', 'applicable', 'taxes', 'donate', 'payments', 'periodic', 'tax', 'returns', 'donations', 'notifies', 'receipt', 'require', 'discontinue', 'f', 'replacement', 'defect', 'michael', 'hart', 'volunteers', 'employees', 'expend', 'identify', 'transcribe', 'proofread', 'despite', 'limited', 'inaccurate', 'corrupt', 'transcription', 'errors', 'infringement', 'defective', 'damaged', 'disk', 'computer', 'virus', 'codes', 'damage', 'equipment', 'warranty', 'disclaimer', 'disclaim', 'liability', 'remedies', 'negligence', 'strict', 'distributor', 'indirect', 'consequential', 'punitive', 'incidental', 'lieu', 'electronically', 'opportunities', 'warranties', 'merchantibility', 'fitness', 'disclaimers', 'exclusion', 'limitation', 'maximum', 'invalidity', 'unenforceability', 'provision', 'void', 'indemnity', 'indemnify', 'agent', 'employee', 'production', 'promotion', 'alteration', 'modification', 'deletions', 'synonymous', 'readable', 'widest', 'computers', 'obsolete', 'critical', 'goals', 'ensuring', 'generations', 'sections', 'non', 'educational', 'corporation', 'organized', 'mississippi', 'internal', 'ein', 'federal', 'identification', 'fundraising', 'contributions', 'deductible', 'melan', 'fairbanks', 'ak', 'scattered', 'locations', 'west', 'lake', 'ut', 'email', 'gregory', 'newby', 'executive', 'director', 'gbnewby', 'survive', 'licensed', 'machine', 'accessible', 'array', 'outdated', 'maintaining', 'irs', 'regulating', 'charities', 'paperwork', 'solicit', 'confirmation', 'solicitation', 'accepting', 'unsolicited', 'donors', 'international', 'gratefully', 'statements', 'swamp', 'staff', 'addresses', 'checks', 'originator', 'library', 'network', 'volunteer', 'necessarily', 'edition', 'pg', 'facility', 'includes', 'subscribe', 'newsletter']\n[8003, 93, 93, 10, 3928, 56, 7042, 9, 585, 4, 13, 733, 1461, 1686, 55, 77, 8, 2082, 703, 10, 1757, 217, 2, 3, 4118, 209, 14, 3463, 135, 266, 507, 68, 101, 30, 18, 3, 4, 6, 13, 5, 6, 1, 8, 3, 281, 2, 1, 6, 6, 36, 75, 1, 1, 16, 6, 3, 2, 2, 1, 1, 1, 4, 1765, 1, 4667, 56, 97, 538, 342, 85, 1, 5497, 179, 2116, 117, 2827, 20, 40, 4, 713, 466, 24, 18, 9, 7, 172, 1801, 908, 62, 3308, 803, 21, 226, 85, 117, 3254, 459, 244, 39, 88, 219, 23, 6, 33, 30, 8, 754, 17, 27, 445, 136, 8, 8, 242, 685, 37, 9, 4, 53, 17, 1, 814, 4900, 1582, 96, 30, 232, 2, 46, 2, 1, 1, 6, 34, 1, 4, 20, 8, 80, 17, 293, 1, 5, 4, 12, 185, 95, 319, 1592, 109, 17, 105, 5, 6, 7, 10, 29, 23, 1, 646, 290, 4, 113, 95, 408, 48, 66, 76, 2, 1, 6, 52, 65, 4, 78, 2, 65, 1, 4, 25, 4, 660, 2, 4, 1, 4405, 11, 283, 481, 1216, 1, 1, 3, 80, 9, 52, 228, 25, 33, 2, 55, 43, 80, 1, 12, 1141, 6, 14, 127, 2, 32, 5, 249, 147, 7, 232, 195, 188, 1489, 37, 4, 17, 19, 1, 461, 26, 1, 1, 19, 2, 102, 4, 10, 55, 139, 1071, 306, 677, 77, 25, 1, 16, 1, 1, 65, 1, 595, 3, 293, 1, 30, 1, 1, 79, 191, 149, 2, 10, 2, 4, 15, 14, 1, 3, 7, 1, 356, 47, 2, 167, 4, 1, 20, 38, 11, 138, 1, 2, 152, 10, 15, 84, 23, 121, 1, 18, 5, 178, 92, 6, 1, 34, 1, 1, 32, 10, 1, 2, 1, 293, 15, 472, 39, 30, 95, 4, 4, 17, 18, 9, 16, 209, 266, 47, 122, 405, 37, 1, 1, 20, 13, 2, 23, 9, 13, 566, 3, 404, 13, 4, 1, 343, 10, 45, 3, 12, 135, 65, 37, 416, 1, 1, 164, 3, 8, 25, 7, 29, 3, 1, 4, 148, 4, 66, 51, 48, 21, 112, 167, 8, 6, 199, 1, 29, 39, 1, 4, 302, 13, 48, 4, 18, 29, 134, 12, 1, 519, 1154, 2, 71, 183, 7, 1, 4, 58, 1, 2, 10, 1, 1, 4, 8, 27, 1, 75, 34, 31, 9, 9, 1, 1, 112, 2, 33, 1, 270, 33, 134, 1, 1, 12, 1, 1, 36, 78, 3, 14, 66, 1, 387, 106, 181, 1, 2, 4, 8, 8, 536, 63, 1323, 205, 2, 31, 133, 32, 40, 551, 4, 14, 56, 9, 1, 10, 5, 118, 102, 33, 10, 40, 3, 25, 2, 8, 29, 70, 32, 1, 30, 2, 455, 9, 285, 10, 2, 1, 36, 15, 385, 8, 42, 1829, 23, 227, 2, 175, 62, 76, 45, 57, 2, 67, 38, 10, 90, 1775, 154, 3, 1, 186, 25, 37, 576, 6, 15, 21, 1, 3, 41, 40, 8, 1, 7, 6, 19, 3, 57, 60, 16, 2, 56, 4, 15, 9, 10, 30, 601, 285, 179, 497, 32, 43, 50, 78, 58, 9, 8, 9, 2, 3, 5, 2, 2, 6, 3, 1, 34, 84, 575, 2, 6, 21, 3, 46, 81, 105, 1, 24, 1, 185, 11, 138, 546, 117, 13, 4, 85, 31, 283, 150, 2, 4, 1, 25, 530, 1231, 867, 111, 40, 12, 135, 28, 80, 505, 8, 209, 291, 12, 33, 68, 78, 791, 4, 81, 1, 2, 1, 445, 185, 20, 137, 4, 5, 160, 2, 32, 18, 1, 1, 1, 1, 3, 14, 43, 6, 18, 24, 207, 49, 12, 16, 9, 2, 13, 5, 9, 16, 39, 8, 32, 75, 26, 27, 286, 2, 8, 16, 8, 31, 1, 14, 5, 203, 5, 250, 259, 9, 29, 2, 5, 3, 335, 5, 7, 50, 246, 5, 46, 46, 24, 1, 62, 30, 2, 53, 3, 1, 9, 2, 11, 11, 13, 18, 37, 76, 4, 26, 42, 3, 14, 13, 42, 340, 5, 1, 31, 17, 1, 32, 5, 39, 11, 1, 26, 27, 15, 3, 34, 1, 1, 20, 1, 10, 14, 2, 3, 13, 16, 16, 12, 1, 4, 12, 49, 1, 2, 85, 47, 100, 439, 5, 12, 63, 6, 136, 5, 52, 44, 14, 4, 7, 19, 1, 9, 89, 10, 15, 27, 1, 3, 19, 84, 9, 7, 1, 37, 12, 117, 1, 1, 11, 15, 1, 1, 20, 78, 10, 12, 12, 1, 3, 130, 68, 20, 9, 44, 16, 164, 4, 121, 14, 8, 14, 17, 25, 58, 10, 33, 105, 210, 1, 5, 8, 2, 68, 2, 207, 46, 103, 5, 89, 219, 5, 56, 10, 168, 77, 368, 77, 48, 15, 39, 31, 38, 52, 33, 5, 348, 22, 19, 1, 15, 10, 1, 191, 7, 2, 4, 5, 1, 5, 480, 28, 2, 4, 497, 11, 35, 1, 19, 18, 4, 7, 293, 22, 205, 15, 1, 1, 2, 3, 1, 2, 3, 24, 7, 3, 45, 26, 4, 12, 5, 14, 6, 2, 1, 2, 7, 53, 31, 6, 18, 3, 222, 7, 22, 12, 13, 2, 604, 5, 29, 131, 32, 37, 19, 17, 6, 1, 15, 7, 27, 13, 77, 11, 56, 7, 1, 32, 203, 279, 35, 14, 25, 11, 4, 4, 10, 6, 3, 16, 34, 53, 57, 23, 3, 51, 42, 14, 104, 70, 12, 4, 32, 24, 2, 4, 48, 6, 1695, 1, 29, 18, 9, 2, 23, 65, 215, 10, 1, 1, 5, 9, 7, 9, 2, 25, 2, 5, 5, 3, 20, 7, 30, 182, 17, 3, 8, 1, 2, 6, 10, 1, 16, 1, 3, 4, 27, 2, 2, 46, 303, 6, 3, 410, 785, 44, 366, 72, 52, 100, 13, 13, 6, 58, 13, 30, 6, 179, 1, 23, 11, 32, 49, 39, 13, 9, 5, 22, 45, 9, 2, 3, 1, 20, 6, 15, 17, 12, 554, 3, 21, 26, 11, 25, 4, 53, 3, 22, 4, 77, 12, 1, 122, 9, 37, 35, 48, 10, 7, 1, 2, 1, 1, 3, 70, 60, 11, 25, 4, 22, 3, 7, 60, 1, 6, 1, 3, 2, 9, 11, 7, 3, 18, 59, 31, 9, 5, 90, 71, 7, 38, 58, 5, 19, 34, 1, 1, 8, 5, 15, 138, 39, 47, 59, 11, 16, 304, 33, 147, 95, 7, 1, 34, 196, 3, 65, 169, 9, 241, 128, 7, 49, 81, 6, 8, 52, 47, 167, 6, 3, 10, 3, 45, 96, 33, 34, 8, 84, 2, 3, 13, 1, 2, 17, 31, 2, 416, 1145, 25, 22, 5, 3, 297, 70, 36, 194, 23, 6, 3, 16, 22, 8, 32, 94, 14, 6, 13, 44, 44, 78, 55, 106, 132, 187, 5, 25, 21, 158, 11, 8, 51, 62, 114, 13, 18, 72, 6, 86, 12, 2, 191, 8, 111, 4, 9, 11, 7, 9, 2, 4, 11, 24, 34, 2, 23, 1, 7, 1, 14, 39, 38, 10, 1, 2, 4, 6, 228, 12, 207, 73, 172, 3, 5, 2, 12, 56, 45, 14, 3, 54, 13, 106, 4, 24, 8, 13, 10, 2, 13, 20, 61, 2, 25, 11, 4, 23, 8, 15, 19, 10, 3, 10, 1, 23, 1, 8, 5, 32, 10, 43, 20, 4, 2, 3, 3, 8, 60, 3, 106, 2, 81, 8, 14, 30, 24, 2, 76, 17, 57, 6, 8, 17, 7, 67, 1, 42, 26, 34, 10, 33, 3, 14, 36, 10, 6, 17, 1, 1, 2, 3, 7, 9, 2, 22, 5, 31, 68, 4, 7, 46, 15, 2, 1, 6, 15, 3, 8, 5, 28, 59, 5, 4, 25, 39, 17, 2, 43, 3, 2, 3, 3, 4, 1, 18, 35, 49, 1, 21, 53, 4, 5, 32, 2, 12, 2, 6, 18, 4, 36, 6, 16, 66, 1, 2, 7, 1, 4, 1, 2, 1, 3, 4, 5, 203, 3, 4, 21, 21, 15, 42, 15, 58, 4, 5, 1, 17, 51, 35, 18, 6, 27, 30, 14, 55, 11, 3, 10, 6, 3, 28, 3, 3, 14, 1, 17, 1, 7, 1, 1, 30, 1, 2, 5, 2, 6, 2, 1, 127, 8, 3, 19, 14, 1, 4, 1, 20, 3, 12, 2, 2, 15, 26, 3, 7, 1, 1, 2, 3, 10, 2, 1, 3, 12, 21, 13, 2, 1, 16, 3, 5, 10, 8, 13, 15, 21, 2, 2, 1, 4, 31, 1, 22, 9, 1, 11, 2, 2, 4, 7, 41, 50, 5, 4, 8, 112, 1, 6, 6, 30, 15, 3, 18, 30, 3, 6, 17, 8, 4, 5, 66, 7, 3, 11, 17, 1, 1, 3, 42, 151, 2, 3, 49, 2, 33, 1, 4, 2, 9, 8, 1, 1, 6, 4, 32, 2, 16, 52, 50, 1, 5, 1, 6, 3, 1, 2, 2, 15, 2, 1, 2, 6, 53, 1, 2, 86, 76, 27, 98, 124, 17, 19, 66, 98, 2, 15, 66, 90, 6, 10, 18, 2, 2, 99, 46, 2, 3, 4, 10, 15, 6, 4, 33, 1, 6, 51, 3, 4, 28, 35, 8, 2, 4, 21, 27, 2, 2, 2, 6, 1, 5, 2, 14, 67, 29, 1, 28, 5, 27, 7, 13, 10, 465, 12, 22, 34, 4, 104, 2, 1, 94, 45, 1, 1, 1, 26, 24, 40, 4, 1, 1, 7, 50, 169, 11, 39, 10, 10, 6, 27, 10, 7, 15, 88, 1, 5, 2, 72, 17, 50, 4, 4, 2, 76, 17, 31, 9, 3, 98, 1, 35, 3, 1, 73, 27, 1, 3, 3, 43, 1, 9, 56, 2, 1, 5, 54, 37, 2, 39, 2, 9, 216, 1, 20, 8, 16, 10, 2, 2, 2, 19, 11, 23, 6, 15, 4, 13, 15, 5, 13, 9, 6, 10, 25, 1, 15, 59, 15, 24, 34, 6, 8, 2, 61, 1, 2, 12, 55, 4, 11, 38, 98, 11, 27, 39, 31, 15, 39, 35, 1, 63, 2, 2, 3, 10, 5, 1, 24, 7, 12, 9, 4, 6, 18, 3, 7, 4, 6, 33, 239, 64, 1, 31, 2, 27, 6, 3, 9, 22, 8, 1, 4, 124, 15, 11, 3, 3, 5, 5, 25, 12, 15, 23, 2, 42, 1, 85, 18, 22, 6, 24, 1, 1, 40, 3, 51, 2, 8, 60, 10, 3, 11, 8, 1, 1, 10, 5, 8, 15, 2, 2, 12, 30, 31, 21, 1, 2, 4, 4, 24, 4, 8, 53, 3, 8, 1, 1, 12, 17, 11, 2, 52, 2, 5, 24, 1, 4, 1, 6, 6, 68, 3, 8, 4, 20, 6, 3, 17, 1, 23, 13, 2, 15, 118, 14, 5, 4, 402, 50, 24, 1, 3, 6, 4, 5, 1, 1, 1, 1, 1, 4, 23, 1, 1, 1, 1, 23, 40, 6, 8, 15, 3, 4, 28, 5, 2, 1, 158, 7, 24, 1, 1, 4, 4, 2, 1, 9, 13, 18, 21, 13, 6, 8, 23, 10, 8, 71, 4, 3, 10, 34, 79, 57, 5, 1, 11, 14, 8, 3, 42, 31, 4, 27, 3, 1, 39, 60, 49, 154, 12, 13, 19, 16, 3, 12, 3, 46, 4, 22, 14, 27, 13, 9, 12, 13, 17, 16, 1, 16, 1, 76, 71, 1, 54, 7, 19, 1, 2, 3, 5, 2, 10, 4, 4, 2, 2, 17, 9, 9, 2, 5, 16, 1, 8, 1, 274, 1, 19, 7, 2, 1, 21, 4, 107, 14, 7, 32, 14, 1, 14, 1, 4, 1, 20, 1, 11, 16, 2, 9, 35, 1, 4, 12, 6, 1, 1, 20, 8, 16, 4, 10, 7, 1, 12, 10, 17, 1, 8, 3, 2, 12, 5, 1, 10, 13, 1, 6, 12, 1, 1, 37, 19, 5, 4, 2, 4, 3, 34, 1, 4, 2, 3, 150, 9, 8, 386, 1, 9, 5, 20, 8, 19, 8, 6, 7, 7, 64, 4, 2, 13, 13, 21, 2, 43, 1, 36, 2, 2, 1, 9, 4, 15, 42, 4, 7, 2, 6, 1, 6, 1, 28, 6, 6, 11, 3, 39, 28, 13, 4, 36, 11, 8, 14, 1, 1, 6, 1, 2, 6, 8, 57, 1, 33, 15, 2, 11, 1, 3, 4, 2, 2, 10, 20, 11, 10, 8, 116, 25, 5, 3, 2, 1, 14, 48, 7, 5, 1, 10, 23, 2, 1, 7, 3, 5, 34, 79, 6, 8, 3, 1, 1, 55, 16, 2, 27, 5, 35, 5, 3, 34, 3, 1, 22, 1, 3, 29, 2, 1, 3, 2, 3, 4, 11, 30, 8, 8, 2, 1, 1, 10, 6, 6, 3, 12, 5, 32, 5, 3, 2, 16, 10, 34, 12, 1, 11, 4, 14, 1, 14, 6, 39, 19, 13, 7, 2, 4, 10, 39, 12, 15, 5, 27, 5, 37, 1, 20, 29, 14, 14, 6, 49, 9, 6, 4, 9, 7, 25, 6, 10, 1, 1, 1, 10, 3, 29, 2, 2, 26, 1, 24, 1, 18, 7, 6, 6, 2, 10, 3, 1, 1, 10, 1, 1, 4, 5, 1, 5, 22, 6, 10, 59, 1, 41, 22, 18, 2, 2, 3, 9, 2, 12, 56, 5, 39, 1, 4, 29, 4, 5, 7, 2, 3, 73, 72, 22, 10, 4, 6, 5, 11, 3, 4, 6, 35, 2, 3, 1, 1, 31, 11, 1, 1, 13, 22, 78, 3, 19, 2, 1, 21, 23, 30, 7, 3, 12, 36, 6, 31, 5, 17, 10, 96, 25, 24, 8, 6, 1, 1, 44, 1, 17, 7, 5, 22, 18, 1, 14, 2, 1, 5, 1, 7, 1, 1, 1, 5, 10, 3, 26, 13, 15, 38, 6, 49, 21, 3, 4, 10, 17, 6, 3, 18, 1, 10, 2, 3, 3, 11, 3, 1, 1, 1, 2, 6, 2, 3, 4, 3, 8, 1, 3, 5, 4, 10, 20, 2, 5, 22, 3, 1, 8, 2, 8, 1, 50, 2, 1, 13, 1, 42, 3, 8, 3, 2, 2, 2, 7, 13, 1, 1, 84, 15, 4, 9, 5, 1, 16, 5, 6, 7, 5, 1, 6, 1, 20, 3, 6, 9, 1, 8, 6, 5, 16, 11, 23, 1, 1, 23, 10, 10, 1, 27, 9, 1, 2, 11, 1, 8, 1, 2, 16, 7, 6, 1, 1, 89, 5, 17, 2, 9, 2, 1, 9, 16, 5, 1, 3, 1, 3, 2, 1, 65, 3, 3, 2, 1, 1, 7, 42, 2, 20, 13, 1, 26, 21, 9, 9, 3, 3, 18, 7, 6, 8, 42, 24, 18, 2, 5, 2, 7, 19, 3, 1, 12, 2, 18, 6, 1, 2, 12, 22, 20, 16, 1, 18, 7, 6, 175, 5, 70, 20, 24, 47, 2, 4, 6, 2, 12, 1, 127, 20, 12, 8, 10, 325, 34, 9, 18, 15, 52, 1, 4, 1, 18, 7, 3, 12, 29, 26, 165, 45, 2, 2, 8, 11, 2, 46, 4, 7, 6, 1, 4, 19, 45, 28, 11, 6, 1, 23, 1, 11, 5, 12, 2, 12, 210, 207, 6, 1, 5, 6, 14, 11, 78, 79, 2, 1, 1, 7, 14, 1, 12, 3, 9, 38, 2, 1, 6, 6, 9, 3, 13, 11, 1, 1, 1, 2, 3, 9, 31, 8, 20, 13, 5, 28, 7, 14, 10, 9, 20, 1, 6, 1, 4, 2, 13, 9, 5, 2, 3, 21, 2, 20, 7, 21, 19, 6, 70, 6, 2, 15, 20, 1, 1, 1, 2, 3, 16, 19, 3, 4, 14, 17, 1, 4, 3, 3, 9, 8, 2, 18, 20, 2, 5, 13, 2, 2, 7, 4, 1, 11, 3, 6, 2, 4, 3, 3, 7, 36, 1, 8, 14, 4, 31, 21, 14, 4, 4, 2, 3, 13, 2, 2, 33, 3, 2, 3, 1, 1, 3, 37, 6, 7, 7, 4, 29, 3, 3, 6, 8, 30, 8, 129, 9, 17, 1, 6, 2, 5, 10, 2, 1, 4, 2, 8, 2, 36, 6, 4, 1, 15, 8, 25, 14, 1, 28, 3, 28, 2, 1, 12, 9, 1, 12, 4, 3, 2, 8, 16, 1, 2, 3, 1, 15, 9, 5, 5, 14, 2, 31, 173, 287, 113, 1, 1, 5, 8, 3, 22, 6, 2, 14, 16, 1, 4, 2, 4, 4, 40, 1, 4, 6, 2, 3, 7, 8, 1, 5, 2, 17, 3, 7, 19, 4, 1, 12, 2, 4, 13, 12, 8, 7, 2, 5, 10, 4, 19, 16, 4, 42, 19, 39, 9, 4, 6, 2, 1, 1, 1, 3, 2, 2, 23, 3, 1, 1, 1, 5, 2, 6, 12, 3, 8, 10, 15, 10, 2, 8, 29, 1, 9, 28, 11, 2, 3, 1, 26, 4, 8, 13, 1, 33, 7, 14, 2, 10, 5, 3, 3, 10, 3, 5, 75, 5, 20, 6, 2, 5, 31, 21, 7, 12, 1, 7, 1, 23, 4, 12, 6, 4, 6, 2, 1, 9, 3, 3, 32, 4, 7, 9, 28, 5, 8, 6, 2, 18, 2, 6, 27, 3, 6, 4, 13, 6, 4, 17, 8, 8, 7, 11, 5, 8, 1, 3, 7, 1, 8, 17, 2, 1, 9, 7, 15, 1, 2, 2, 13, 4, 9, 3, 6, 2, 4, 2, 4, 2, 3, 5, 19, 8, 1, 1, 9, 1, 17, 6, 1, 4, 14, 124, 14, 16, 6, 15, 1, 1, 7, 15, 2, 4, 3, 1, 6, 14, 6, 5, 9, 20, 4, 9, 7, 19, 10, 4, 31, 12, 5, 2, 13, 9, 4, 7, 9, 6, 1, 6, 9, 1, 1, 14, 115, 115, 3, 3, 1, 8, 1, 2, 7, 1, 100, 9, 1, 66, 7, 2, 14, 2, 7, 30, 9, 20, 6, 4, 24, 3, 3, 35, 5, 1, 50, 9, 4, 10, 3, 3, 1, 1, 1, 3, 1, 5, 27, 3, 1, 18, 2, 10, 5, 1, 2, 4, 1, 1, 2, 1, 4, 1, 1, 2, 8, 7, 1, 12, 4, 4, 7, 1, 1, 9, 2, 1, 2, 13, 23, 5, 2, 21, 2, 2, 2, 1, 21, 4, 55, 2, 14, 11, 1, 1, 3, 1, 17, 4, 1, 1, 1, 1, 1, 12, 1, 1, 1, 2, 13, 1, 1, 19, 15, 1, 1, 2, 7, 7, 97, 86, 1, 1, 13, 1, 6, 10, 2, 3, 2, 3, 8, 1, 3, 19, 2, 1, 2, 1, 1, 9, 4, 21, 17, 10, 8, 3, 4, 5, 1, 1, 9, 2, 3, 1, 28, 23, 8, 3, 15, 1, 3, 1, 1, 20, 1, 1, 6, 1, 12, 2, 15, 1, 4, 9, 1, 1, 1, 16, 3, 1, 19, 4, 3, 2, 16, 7, 9, 4, 2, 3, 3, 73, 22, 6, 2, 4, 18, 8, 18, 9, 2, 1, 2, 4, 5, 5, 1, 12, 1, 1, 4, 2, 15, 9, 1, 1, 4, 4, 5, 9, 5, 6, 5, 48, 2, 6, 20, 4, 1, 21, 12, 10, 10, 1, 11, 9, 6, 1, 1, 37, 6, 1, 2, 10, 2, 8, 4, 5, 2, 3, 2, 12, 10, 4, 11, 3, 2, 14, 14, 20, 1, 6, 6, 28, 2, 7, 1, 6, 8, 13, 5, 11, 17, 9, 6, 2, 4, 7, 29, 9, 5, 30, 19, 1, 1, 10, 2, 13, 3, 1, 32, 1, 1, 8, 3, 4, 9, 2, 1, 10, 4, 2, 12, 52, 4, 1, 3, 29, 4, 9, 14, 21, 3, 1, 1, 20, 4, 5, 6, 15, 10, 1, 7, 12, 1, 11, 5, 13, 17, 6, 5, 1, 4, 1, 3, 4, 5, 2, 5, 1, 347, 15, 3, 3, 25, 2, 6, 1, 1, 3, 1, 20, 2, 5, 6, 4, 1, 8, 1, 1, 1, 7, 6, 1, 11, 3, 1, 2, 1, 1, 2, 16, 1, 3, 1, 2, 5, 6, 1, 21, 6, 5, 1, 2, 3, 8, 10, 3, 55, 3, 13, 9, 1, 10, 18, 9, 8, 19, 15, 4, 4, 3, 14, 9, 1, 17, 8, 2, 6, 15, 2, 8, 12, 9, 3, 3, 6, 11, 3, 1, 4, 1, 2, 2, 1, 1, 10, 5, 12, 6, 1, 33, 1, 1, 3, 1, 1, 32, 2, 7, 13, 2, 2, 1, 12, 4, 1, 15, 11, 15, 6, 3, 19, 1, 5, 5, 6, 3, 1, 1, 3, 6, 5, 1, 1, 1, 3, 1, 5, 14, 6, 3, 7, 5, 3, 2, 6, 10, 1, 1, 1, 1, 56, 5, 3, 5, 1, 5, 32, 2, 5, 3, 3, 3, 6, 3, 3, 2, 1, 8, 1, 2, 19, 2, 1, 2, 22, 9, 9, 2, 8, 2, 1, 1, 1, 6, 7, 10, 1, 1, 20, 3, 1, 6, 2, 10, 2, 4, 1, 2, 13, 1, 3, 11, 3, 3, 3, 3, 1, 1, 11, 3, 1, 1, 1, 1, 17, 4, 15, 6, 1, 8, 1, 2, 1, 27, 10, 6, 4, 6, 1, 1, 1, 4, 1, 3, 2, 1, 1, 2, 4, 2, 5, 7, 1, 12, 3, 11, 2, 3, 1, 5, 9, 4, 6, 7, 7, 26, 1, 8, 2, 1, 6, 2, 13, 1, 4, 18, 3, 1, 13, 2, 6, 11, 5, 1, 2, 1, 1, 2, 1, 3, 8, 2, 2, 1, 5, 2, 2, 23, 1, 3, 11, 2, 6, 3, 1, 2, 9, 6, 6, 2, 2, 1, 1, 58, 7, 3, 7, 2, 3, 1, 11, 5, 3, 4, 2, 5, 13, 2, 33, 1, 1, 1, 3, 4, 22, 1, 7, 3, 4, 6, 5, 2, 2, 2, 6, 1, 14, 5, 3, 3, 3, 11, 5, 4, 13, 14, 46, 1, 5, 2, 4, 1, 16, 5, 6, 2, 13, 22, 10, 4, 6, 2, 1, 1, 1, 1, 6, 6, 1, 55, 3, 1, 13, 1, 4, 5, 1, 1, 4, 5, 1, 2, 6, 5, 2, 1, 2, 1, 1, 2, 3, 1, 3, 3, 5, 1, 1, 6, 1, 1, 2, 14, 22, 10, 11, 4, 3, 4, 6, 11, 1, 7, 3, 8, 1, 7, 1, 33, 3, 25, 4, 1, 11, 9, 9, 8, 1, 10, 10, 2, 1, 7, 23, 4, 5, 3, 1, 2, 1, 4, 4, 1, 10, 1, 1, 9, 12, 8, 1, 1, 1, 5, 4, 3, 5, 9, 1, 13, 1, 1, 4, 3, 3, 8, 4, 2, 10, 3, 5, 1, 1, 5, 5, 35, 1, 4, 4, 2, 2, 1, 3, 1, 12, 1, 4, 9, 1, 1, 16, 2, 3, 1, 6, 3, 3, 2, 4, 3, 4, 1, 30, 3, 12, 9, 4, 8, 4, 1, 3, 3, 24, 23, 8, 10, 7, 1, 15, 4, 1, 1, 3, 5, 14, 1, 10, 1, 6, 3, 2, 2, 1, 1, 10, 6, 5, 1, 6, 1, 1, 2, 1, 1, 9, 4, 4, 6, 6, 2, 2, 6, 10, 4, 1, 3, 3, 6, 18, 17, 4, 47, 5, 3, 5, 5, 14, 1, 7, 33, 1, 3, 1, 1, 1, 1, 1, 6, 4, 6, 4, 1, 5, 5, 2, 8, 2, 1, 36, 3, 4, 4, 14, 4, 20, 3, 7, 2, 9, 3, 13, 1, 1, 3, 3, 4, 11, 3, 10, 1, 5, 12, 2, 1, 14, 2, 1, 1, 9, 16, 11, 24, 5, 3, 1, 8, 2, 1, 3, 1, 4, 2, 8, 35, 1, 4, 2, 2, 2, 1, 7, 2, 2, 2, 8, 7, 2, 1, 3, 13, 1, 1, 6, 11, 4, 8, 13, 9, 3, 1, 2, 9, 1, 7, 28, 4, 15, 4, 3, 5, 2, 30, 6, 3, 1, 28, 1, 4, 2, 2, 1, 5, 1, 1, 1, 2, 4, 3, 5, 2, 8, 5, 3, 3, 25, 11, 1, 1, 2, 1, 5, 3, 2, 4, 2, 6, 4, 8, 2, 2, 2, 9, 14, 2, 3, 5, 1, 1, 1, 1, 3, 13, 2, 12, 5, 3, 1, 13, 11, 18, 4, 5, 20, 3, 3, 19, 53, 3, 1, 14, 4, 1, 4, 1, 1, 4, 14, 1, 3, 2, 2, 1, 7, 1, 1, 1, 1, 2, 20, 3, 2, 4, 6, 2, 2, 3, 1, 1, 6, 3, 1, 1, 5, 1, 8, 2, 14, 7, 6, 1, 5, 9, 1, 1, 3, 5, 12, 15, 20, 4, 4, 5, 4, 15, 2, 16, 6, 3, 9, 5, 3, 1, 7, 3, 3, 1, 4, 6, 3, 8, 1, 12, 4, 6, 1, 3, 1, 9, 3, 8, 2, 15, 2, 3, 5, 4, 7, 10, 2, 1, 11, 4, 5, 11, 22, 2, 1, 1, 1, 1, 4, 1, 1, 1, 12, 5, 1, 1, 5, 1, 2, 3, 3, 13, 8, 3, 1, 8, 1, 2, 5, 1, 6, 1, 3, 9, 16, 3, 1, 15, 9, 20, 5, 1, 1, 2, 1, 2, 2, 5, 2, 4, 3, 1, 1, 1, 1, 1, 4, 2, 2, 1, 2, 10, 12, 2, 1, 1, 4, 20, 1, 3, 1, 2, 12, 1, 1, 3, 1, 7, 23, 19, 12, 20, 1, 2, 2, 1, 2, 13, 1, 5, 5, 2, 15, 50, 5, 1, 2, 4, 9, 6, 4, 2, 2, 1, 2, 3, 3, 4, 9, 3, 1, 2, 6, 3, 2, 1, 3, 6, 2, 1, 2, 20, 1, 1, 7, 14, 3, 1, 7, 7, 1, 42, 5, 2, 9, 25, 3, 2, 1, 7, 1, 4, 8, 21, 1, 2, 3, 5, 3, 10, 2, 21, 1, 2, 5, 4, 17, 1, 3, 25, 1, 4, 1, 2, 8, 7, 19, 1, 8, 6, 2, 1, 4, 31, 9, 9, 14, 2, 1, 7, 2, 17, 8, 1, 2, 3, 3, 1, 6, 1, 1, 2, 4, 33, 2, 12, 1, 8, 6, 4, 1, 2, 1, 8, 20, 6, 1, 12, 8, 4, 8, 4, 2, 1, 2, 3, 2, 1, 2, 3, 4, 2, 2, 7, 1, 15, 2, 3, 1, 4, 1, 1, 2, 3, 1, 41, 7, 6, 8, 54, 13, 1, 4, 2, 2, 13, 44, 1, 1, 2, 4, 3, 3, 3, 11, 2, 1, 2, 1, 3, 2, 1, 5, 3, 3, 1, 7, 67, 2, 2, 1, 3, 7, 2, 5, 9, 2, 8, 2, 1, 2, 3, 3, 5, 5, 7, 2, 6, 1, 5, 1, 14, 10, 6, 4, 5, 1, 3, 2, 8, 11, 4, 1, 1, 1, 1, 3, 1, 2, 2, 1, 1, 4, 14, 1, 1, 1, 29, 2, 2, 2, 4, 1, 2, 3, 15, 3, 4, 7, 1, 2, 1, 1, 1, 15, 1, 1, 4, 14, 4, 1, 12, 1, 1, 1, 11, 3, 2, 1, 10, 2, 9, 1, 1, 1, 1, 1, 1, 1, 4, 3, 6, 13, 5, 1, 6, 1, 2, 1, 2, 2, 10, 3, 13, 1, 4, 4, 5, 3, 3, 3, 3, 4, 5, 9, 11, 3, 16, 5, 16, 14, 6, 2, 15, 21, 18, 1, 1, 4, 11, 6, 1, 3, 1, 2, 2, 2, 6, 29, 20, 2, 2, 8, 1, 2, 2, 5, 1, 1, 2, 6, 1, 2, 20, 3, 7, 11, 1, 1, 5, 1, 1, 2, 6, 5, 2, 1, 2, 2, 10, 6, 37, 33, 5, 4, 1, 2, 2, 2, 2, 2, 9, 2, 1, 3, 1, 11, 4, 4, 1, 1, 3, 9, 3, 6, 1, 1, 4, 3, 1, 2, 4, 1, 5, 3, 7, 7, 21, 12, 3, 9, 4, 1, 1, 4, 1, 1, 1, 4, 1, 4, 2, 24, 24, 2, 1, 2, 5, 2, 1, 3, 2, 14, 1, 1, 4, 7, 3, 4, 10, 4, 1, 1, 2, 9, 2, 1, 2, 3, 1, 5, 7, 3, 5, 3, 9, 1, 2, 5, 10, 8, 1, 7, 4, 6, 5, 1, 2, 1, 1, 4, 4, 8, 6, 3, 2, 6, 5, 1, 6, 2, 1, 3, 3, 5, 5, 8, 7, 1, 2, 2, 2, 1, 4, 8, 3, 28, 2, 1, 10, 1, 1, 10, 1, 6, 2, 1, 1, 1, 11, 7, 1, 3, 3, 4, 9, 1, 1, 1, 1, 1, 3, 8, 2, 4, 4, 6, 1, 13, 3, 1, 1, 1, 1, 5, 15, 1, 6, 1, 7, 2, 7, 4, 7, 2, 2, 11, 3, 1, 1, 1, 13, 10, 1, 1, 1, 15, 1, 1, 3, 1, 5, 1, 1, 7, 5, 6, 2, 1, 2, 1, 2, 6, 1, 2, 4, 1, 11, 1, 1, 4, 1, 5, 7, 1, 3, 5, 3, 5, 7, 4, 2, 2, 1, 1, 5, 3, 2, 1, 5, 1, 3, 2, 3, 1, 3, 3, 29, 3, 3, 6, 1, 2, 3, 1, 3, 11, 4, 1, 2, 1, 1, 6, 11, 2, 2, 1, 1, 1, 1, 1, 4, 1, 2, 4, 1, 3, 35, 6, 1, 1, 1, 2, 1, 1, 1, 3, 1, 1, 13, 1, 23, 9, 5, 1, 2, 1, 1, 9, 8, 5, 3, 1, 1, 5, 1, 4, 1, 2, 1, 1, 3, 7, 2, 2, 2, 2, 4, 2, 5, 1, 4, 4, 3, 12, 9, 2, 4, 4, 2, 7, 5, 4, 2, 7, 1, 2, 6, 1, 1, 12, 1, 1, 3, 4, 3, 5, 2, 3, 5, 4, 4, 2, 5, 2, 2, 1, 2, 1, 1, 1, 1, 5, 3, 1, 1, 1, 2, 3, 1, 3, 1, 4, 17, 4, 1, 4, 4, 7, 4, 1, 1, 2, 5, 16, 1, 15, 23, 9, 1, 1, 6, 2, 2, 1, 1, 2, 3, 2, 1, 1, 1, 1, 1, 6, 1, 1, 5, 2, 2, 3, 1, 4, 1, 1, 2, 3, 3, 3, 12, 2, 1, 5, 87, 5, 3, 1, 1, 24, 20, 16, 2, 2, 2, 1, 7, 1, 1, 1, 1, 10, 4, 10, 2, 2, 1, 4, 2, 1, 1, 5, 3, 1, 6, 1, 3, 3, 2, 2, 4, 6, 2, 2, 3, 4, 1, 3, 4, 9, 1, 4, 2, 1, 1, 1, 1, 1, 1, 1, 4, 6, 3, 1, 1, 1, 3, 90, 1, 2, 5, 1, 1, 1, 3, 3, 1, 1, 2, 5, 1, 1, 22, 3, 4, 1, 1, 1, 4, 1, 11, 2, 1, 1, 5, 2, 1, 2, 2, 1, 1, 2, 1, 3, 2, 2, 13, 2, 1, 1, 1, 9, 5, 1, 1, 2, 4, 1, 2, 1, 1, 6, 2, 4, 2, 7, 1, 7, 1, 12, 1, 4, 9, 2, 1, 1, 4, 8, 4, 1, 1, 1, 5, 1, 11, 1, 2, 4, 1, 1, 2, 2, 5, 1, 1, 4, 3, 1, 5, 1, 1, 1, 1, 2, 2, 12, 15, 1, 1, 3, 2, 1, 1, 1, 8, 1, 1, 1, 1, 1, 2, 2, 4, 2, 1, 2, 1, 5, 4, 1, 2, 1, 1, 1, 6, 1, 2, 3, 1, 7, 1, 6, 1, 6, 6, 1, 1, 1, 1, 2, 1, 1, 1, 4, 2, 15, 1, 1, 4, 6, 6, 6, 1, 3, 1, 1, 1, 206, 3, 4, 4, 2, 1, 7, 6, 1, 8, 4, 3, 2, 16, 13, 1, 1, 2, 1, 1, 5, 2, 10, 2, 1, 39, 5, 11, 4, 2, 5, 2, 1, 5, 3, 13, 12, 5, 6, 4, 1, 5, 6, 3, 2, 3, 3, 1, 1, 1, 1, 1, 3, 5, 7, 2, 3, 2, 1, 1, 1, 8, 3, 1, 2, 1, 1, 65, 6, 6, 1, 1, 6, 1, 1, 1, 2, 1, 1, 1, 5, 49, 6, 4, 2, 6, 10, 3, 6, 3, 2, 2, 6, 5, 3, 1, 1, 2, 1, 1, 1, 2, 4, 4, 5, 1, 3, 9, 14, 10, 2, 2, 1, 2, 3, 4, 1, 30, 11, 4, 1, 12, 1, 2, 3, 2, 1, 4, 1, 1, 2, 1, 8, 1, 1, 2, 2, 1, 2, 5, 3, 2, 2, 3, 1, 15, 2, 9, 2, 9, 6, 1, 1, 4, 1, 4, 1, 1, 5, 1, 1, 2, 2, 3, 1, 1, 2, 4, 1, 3, 2, 1, 4, 3, 1, 1, 3, 1, 1, 10, 4, 1, 1, 9, 1, 6, 5, 5, 1, 7, 1, 4, 1, 7, 3, 2, 1, 1, 4, 1, 1, 9, 8, 2, 4, 17, 4, 1, 1, 4, 3, 2, 1, 4, 1, 1, 1, 1, 5, 2, 1, 1, 2, 2, 1, 4, 2, 2, 1, 1, 1, 1, 3, 1, 9, 1, 2, 1, 6, 1, 4, 8, 1, 1, 1, 13, 2, 5, 21, 15, 1, 7, 1, 1, 1, 1, 1, 4, 5, 1, 1, 5, 3, 1, 1, 1, 4, 4, 10, 1, 1, 5, 3, 1, 1, 2, 17, 1, 1, 4, 5, 7, 5, 1, 2, 1, 3, 2, 7, 3, 1, 3, 1, 2, 2, 2, 4, 9, 1, 13, 2, 2, 1, 2, 11, 4, 2, 1, 3, 1, 2, 1, 1, 1, 1, 1, 3, 2, 1, 6, 7, 25, 3, 1, 3, 1, 1, 2, 3, 2, 2, 3, 1, 1, 17, 7, 1, 1, 5, 10, 2, 1, 3, 1, 1, 3, 4, 1, 1, 2, 3, 11, 3, 1, 3, 1, 3, 2, 1, 2, 12, 1, 1, 2, 1, 1, 3, 1, 1, 1, 2, 2, 2, 4, 2, 1, 4, 8, 2, 1, 1, 6, 9, 1, 5, 30, 2, 6, 7, 2, 3, 2, 2, 1, 3, 14, 4, 5, 1, 3, 3, 13, 15, 1, 1, 1, 5, 3, 5, 4, 2, 5, 2, 3, 4, 11, 3, 13, 1, 3, 2, 10, 1, 2, 3, 3, 2, 2, 1, 4, 1, 1, 4, 2, 1, 1, 1, 1, 1, 3, 1, 1, 1, 2, 3, 3, 2, 1, 1, 2, 1, 1, 1, 2, 2, 1, 1, 1, 2, 2, 5, 5, 1, 1, 11, 1, 1, 1, 1, 1, 1, 2, 3, 2, 1, 4, 2, 1, 3, 1, 7, 1, 2, 5, 2, 1, 10, 4, 1, 10, 2, 1, 1, 1, 2, 3, 7, 2, 13, 1, 4, 5, 3, 10, 7, 1, 14, 2, 2, 2, 3, 3, 2, 2, 3, 7, 3, 1, 1, 1, 6, 1, 1, 1, 5, 8, 2, 1, 2, 7, 1, 3, 1, 1, 1, 2, 3, 7, 1, 1, 1, 1, 1, 3, 3, 7, 1, 4, 4, 2, 5, 2, 2, 1, 1, 1, 1, 1, 2, 2, 6, 4, 5, 1, 1, 1, 7, 1, 2, 3, 2, 2, 6, 1, 4, 2, 4, 6, 1, 1, 6, 4, 3, 1, 7, 1, 1, 1, 2, 1, 1, 1, 1, 1, 7, 5, 6, 2, 7, 3, 2, 5, 3, 1, 2, 3, 3, 1, 1, 6, 3, 2, 2, 6, 1, 2, 4, 1, 2, 1, 6, 4, 1, 1, 4, 1, 2, 3, 9, 1, 1, 1, 3, 6, 3, 3, 2, 1, 2, 3, 1, 1, 1, 2, 1, 1, 6, 3, 11, 1, 1, 1, 1, 5, 1, 1, 2, 3, 1, 1, 2, 4, 2, 1, 1, 3, 1, 1, 2, 1, 5, 1, 6, 2, 1, 2, 6, 1, 2, 1, 1, 11, 8, 4, 7, 6, 1, 1, 2, 45, 1, 4, 2, 1, 3, 2, 2, 2, 6, 7, 1, 4, 4, 1, 1, 1, 1, 4, 1, 39, 3, 4, 1, 1, 1, 2, 3, 1, 22, 4, 1, 1, 3, 2, 1, 2, 4, 1, 3, 1, 4, 1, 1, 5, 1, 2, 8, 1, 2, 19, 2, 1, 1, 5, 1, 3, 1, 3, 7, 1, 1, 1, 2, 4, 1, 10, 29, 3, 6, 1, 3, 9, 7, 2, 1, 10, 9, 4, 1, 1, 1, 1, 10, 1, 1, 5, 2, 4, 3, 3, 1, 4, 1, 1, 1, 1, 9, 1, 1, 2, 2, 2, 4, 15, 1, 3, 2, 1, 2, 1, 1, 2, 1, 2, 4, 1, 5, 2, 1, 1, 2, 12, 11, 1, 1, 1, 1, 1, 10, 12, 3, 4, 1, 1, 1, 1, 12, 1, 1, 1, 3, 1, 2, 7, 6, 1, 1, 4, 1, 1, 3, 6, 7, 1, 3, 1, 2, 1, 8, 1, 1, 16, 1, 1, 2, 1, 1, 1, 1, 1, 2, 3, 1, 1, 1, 1, 2, 3, 2, 4, 2, 5, 1, 2, 8, 9, 1, 6, 5, 1, 10, 4, 3, 4, 2, 1, 4, 2, 1, 1, 3, 1, 8, 1, 1, 2, 1, 2, 1, 1, 4, 8, 1, 1, 3, 1, 2, 6, 9, 5, 123, 1, 1, 7, 3, 1, 1, 1, 4, 8, 1, 1, 3, 1, 3, 1, 3, 1, 2, 10, 1, 5, 9, 2, 2, 1, 1, 1, 2, 1, 1, 3, 1, 1, 3, 6, 1, 1, 2, 2, 2, 5, 1, 2, 6, 1, 4, 1, 4, 1, 5, 1, 1, 2, 1, 1, 5, 3, 1, 4, 2, 2, 4, 1, 1, 3, 1, 1, 5, 1, 2, 3, 12, 2, 1, 1, 1, 4, 1, 2, 1, 1, 10, 1, 4, 9, 1, 2, 2, 2, 1, 1, 1, 2, 1, 1, 2, 1, 8, 2, 2, 2, 3, 1, 1, 1, 3, 1, 1, 3, 5, 2, 1, 1, 4, 4, 1, 4, 1, 2, 1, 1, 1, 1, 1, 3, 1, 1, 5, 4, 1, 2, 3, 3, 2, 1, 3, 2, 1, 2, 4, 1, 1, 1, 1, 1, 1, 1, 1, 2, 3, 5, 4, 11, 2, 1, 1, 1, 1, 3, 1, 3, 1, 3, 13, 1, 1, 5, 3, 2, 2, 1, 2, 1, 2, 1, 1, 1, 1, 1, 2, 2, 2, 1, 1, 3, 2, 1, 1, 2, 1, 6, 1, 1, 1, 1, 4, 1, 2, 1, 1, 2, 1, 8, 2, 2, 3, 1, 3, 3, 1, 1, 1, 4, 1, 2, 1, 1, 2, 1, 2, 1, 3, 2, 1, 5, 1, 1, 1, 4, 3, 1, 1, 2, 1, 1, 1, 1, 2, 2, 3, 1, 2, 2, 1, 4, 1, 7, 4, 2, 1, 1, 1, 6, 2, 9, 1, 10, 1, 3, 3, 3, 1, 1, 1, 1, 6, 3, 3, 3, 2, 2, 24, 1, 3, 1, 1, 1, 2, 2, 1, 10, 2, 4, 3, 1, 1, 4, 1, 3, 1, 1, 1, 1, 1, 1, 5, 3, 7, 4, 3, 1, 1, 1, 16, 6, 4, 5, 4, 3, 1, 2, 2, 3, 6, 1, 3, 1, 1, 1, 1, 1, 1, 5, 2, 1, 2, 1, 2, 9, 7, 3, 3, 4, 4, 1, 2, 2, 5, 3, 1, 4, 4, 1, 3, 1, 1, 1, 2, 1, 34, 2, 2, 1, 1, 1, 1, 3, 2, 1, 2, 1, 1, 11, 1, 1, 1, 15, 2, 4, 3, 6, 3, 1, 1, 6, 2, 2, 1, 2, 2, 1, 1, 1, 3, 1, 8, 1, 3, 1, 3, 5, 1, 1, 3, 3, 2, 1, 2, 1, 1, 1, 2, 1, 2, 2, 5, 1, 2, 1, 10, 3, 1, 1, 2, 3, 5, 2, 5, 1, 1, 6, 1, 1, 3, 3, 3, 1, 1, 1, 3, 1, 2, 2, 5, 1, 8, 2, 5, 1, 8, 2, 5, 1, 3, 1, 2, 1, 7, 1, 1, 2, 1, 1, 12, 4, 2, 2, 3, 2, 6, 1, 5, 4, 1, 1, 1, 1, 4, 2, 5, 1, 1, 3, 2, 4, 3, 1, 4, 1, 12, 1, 7, 1, 2, 1, 1, 1, 3, 2, 1, 2, 1, 2, 1, 1, 2, 1, 2, 2, 3, 1, 10, 1, 3, 1, 2, 1, 1, 1, 6, 5, 1, 3, 1, 2, 1, 3, 1, 2, 1, 16, 2, 3, 3, 5, 3, 2, 1, 4, 2, 1, 4, 3, 5, 2, 2, 2, 2, 1, 7, 1, 5, 1, 1, 1, 6, 1, 5, 5, 2, 1, 1, 7, 2, 4, 2, 2, 3, 1, 1, 1, 1, 1, 4, 2, 3, 1, 1, 1, 2, 1, 1, 2, 2, 8, 1, 1, 1, 2, 1, 5, 10, 1, 2, 1, 1, 3, 1, 1, 1, 11, 1, 2, 3, 5, 1, 1, 2, 2, 2, 2, 1, 1, 1, 2, 2, 4, 1, 1, 1, 2, 3, 3, 2, 1, 1, 1, 1, 3, 1, 5, 1, 1, 1, 1, 1, 1, 3, 2, 1, 1, 2, 1, 2, 2, 2, 1, 3, 1, 1, 1, 1, 8, 1, 1, 1, 1, 1, 8, 1, 1, 1, 4, 3, 1, 10, 1, 1, 2, 1, 2, 3, 1, 4, 2, 5, 1, 1, 1, 6, 5, 2, 1, 1, 1, 2, 8, 1, 2, 1, 3, 1, 1, 3, 2, 2, 2, 5, 2, 1, 2, 1, 2, 2, 1, 3, 1, 4, 4, 6, 2, 1, 4, 1, 1, 3, 2, 2, 1, 3, 1, 2, 1, 2, 1, 3, 1, 1, 9, 2, 3, 1, 10, 4, 1, 2, 1, 1, 2, 1, 4, 1, 1, 1, 2, 2, 1, 2, 1, 2, 2, 3, 3, 1, 1, 5, 6, 2, 1, 4, 1, 1, 4, 4, 3, 3, 3, 2, 2, 3, 1, 1, 1, 1, 4, 3, 5, 1, 4, 1, 1, 2, 12, 4, 3, 2, 1, 1, 1, 4, 3, 1, 2, 2, 1, 5, 1, 2, 1, 3, 2, 1, 2, 2, 1, 3, 11, 2, 2, 1, 3, 1, 1, 3, 1, 3, 1, 2, 1, 2, 1, 2, 2, 1, 5, 2, 2, 2, 2, 4, 2, 3, 1, 1, 5, 1, 1, 2, 3, 2, 1, 4, 2, 1, 4, 1, 3, 5, 1, 7, 1, 2, 3, 1, 4, 2, 1, 1, 1, 3, 1, 4, 2, 2, 5, 1, 4, 1, 1, 1, 1, 1, 2, 4, 1, 1, 1, 1, 1, 3, 1, 1, 1, 1, 3, 1, 2, 2, 2, 1, 1, 2, 6, 2, 8, 2, 3, 7, 1, 6, 2, 2, 3, 5, 2, 2, 3, 1, 1, 1, 1, 1, 1, 1, 8, 2, 2, 3, 2, 1, 2, 2, 2, 3, 1, 1, 1, 2, 7, 1, 1, 1, 1, 3, 1, 1, 4, 3, 1, 4, 6, 2, 2, 2, 4, 1, 1, 1, 1, 2, 3, 1, 1, 1, 3, 3, 1, 1, 4, 1, 1, 1, 1, 3, 2, 1, 1, 1, 1, 1, 2, 1, 2, 2, 1, 2, 2, 1, 1, 1, 4, 4, 2, 1, 2, 1, 2, 1, 5, 1, 2, 1, 3, 3, 3, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 5, 1, 2, 4, 1, 1, 3, 1, 1, 1, 5, 3, 1, 1, 5, 2, 1, 4, 1, 1, 1, 2, 2, 1, 1, 3, 4, 1, 1, 1, 2, 1, 2, 2, 3, 1, 1, 1, 2, 2, 2, 5, 1, 1, 1, 1, 1, 1, 1, 3, 11, 1, 1, 7, 4, 1, 1, 1, 3, 2, 2, 1, 2, 1, 1, 3, 3, 1, 1, 7, 2, 1, 1, 1, 2, 1, 1, 1, 1, 7, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 4, 2, 3, 3, 2, 1, 1, 1, 1, 2, 2, 3, 1, 1, 1, 5, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 2, 2, 3, 1, 1, 2, 1, 2, 1, 1, 1, 1, 2, 1, 1, 2, 1, 7, 1, 1, 1, 1, 2, 1, 1, 1, 2, 2, 3, 1, 3, 2, 2, 4, 2, 2, 1, 4, 2, 1, 4, 3, 3, 1, 1, 1, 2, 8, 3, 6, 2, 2, 2, 3, 3, 3, 1, 1, 5, 1, 5, 5, 2, 1, 2, 1, 1, 1, 2, 2, 1, 1, 1, 1, 1, 2, 1, 1, 1, 3, 4, 1, 1, 1, 3, 2, 1, 2, 5, 1, 1, 3, 2, 2, 1, 2, 1, 2, 1, 1, 1, 1, 1, 2, 1, 3, 1, 1, 1, 1, 1, 3, 1, 2, 2, 1, 1, 1, 2, 5, 1, 1, 2, 1, 2, 1, 1, 2, 1, 1, 1, 1, 4, 5, 1, 1, 2, 3, 2, 1, 2, 1, 6, 8, 1, 1, 3, 1, 1, 2, 1, 4, 1, 2, 4, 1, 2, 1, 2, 2, 4, 2, 1, 2, 2, 2, 3, 3, 1, 2, 1, 2, 1, 3, 1, 1, 2, 1, 2, 1, 1, 1, 1, 2, 1, 2, 1, 1, 1, 1, 1, 5, 3, 2, 3, 1, 2, 1, 1, 1, 2, 3, 2, 3, 1, 1, 1, 2, 1, 4, 1, 5, 2, 1, 2, 1, 1, 1, 1, 1, 1, 3, 2, 1, 6, 1, 1, 3, 1, 5, 2, 1, 1, 2, 1, 1, 2, 1, 1, 1, 2, 4, 2, 1, 1, 1, 3, 4, 4, 1, 8, 1, 1, 4, 2, 2, 1, 2, 3, 1, 3, 1, 1, 1, 4, 1, 2, 1, 1, 1, 1, 1, 3, 1, 2, 4, 2, 3, 1, 1, 1, 1, 1, 2, 2, 6, 1, 2, 1, 1, 1, 2, 1, 5, 2, 1, 4, 1, 1, 2, 1, 1, 1, 1, 1, 3, 2, 1, 1, 2, 1, 3, 3, 1, 1, 1, 6, 3, 2, 3, 2, 4, 6, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 1, 4, 1, 1, 3, 2, 3, 2, 3, 2, 1, 1, 2, 1, 1, 2, 7, 1, 1, 1, 4, 3, 1, 5, 1, 1, 1, 1, 1, 1, 2, 4, 2, 12, 1, 1, 2, 2, 1, 1, 2, 4, 2, 1, 3, 1, 4, 2, 1, 4, 3, 1, 1, 1, 2, 2, 1, 1, 1, 1, 1, 2, 2, 2, 3, 1, 3, 2, 3, 1, 2, 1, 1, 3, 1, 1, 3, 1, 2, 2, 1, 3, 6, 5, 4, 13, 2, 1, 5, 4, 2, 1, 1, 1, 1, 4, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 2, 1, 3, 3, 1, 3, 2, 1, 1, 2, 1, 2, 1, 4, 1, 1, 3, 1, 1, 3, 1, 1, 1, 1, 10, 2, 3, 1, 1, 1, 3, 1, 1, 1, 5, 1, 1, 1, 3, 1, 5, 5, 1, 6, 2, 1, 1, 1, 4, 2, 2, 1, 1, 1, 1, 3, 6, 2, 1, 1, 1, 1, 1, 1, 2, 2, 1, 1, 2, 1, 1, 1, 1, 4, 4, 1, 1, 6, 2, 2, 1, 4, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 2, 1, 1, 2, 1, 3, 2, 1, 3, 4, 1, 3, 2, 1, 12, 1, 1, 1, 4, 1, 1, 1, 1, 1, 2, 2, 2, 1, 2, 4, 1, 1, 1, 1, 5, 3, 1, 1, 2, 1, 5, 4, 1, 1, 2, 1, 3, 3, 1, 1, 3, 1, 2, 1, 1, 1, 3, 1, 3, 1, 1, 1, 1, 2, 4, 2, 2, 1, 1, 2, 2, 1, 2, 1, 3, 2, 2, 2, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 4, 1, 1, 1, 1, 2, 1, 2, 3, 2, 1, 2, 2, 1, 1, 3, 1, 3, 1, 1, 1, 5, 1, 1, 3, 1, 1, 1, 1, 1, 1, 4, 2, 4, 1, 4, 1, 4, 5, 1, 1, 5, 1, 2, 2, 1, 2, 2, 1, 1, 1, 1, 1, 1, 1, 2, 1, 3, 4, 1, 1, 2, 1, 1, 4, 1, 1, 1, 1, 1, 1, 1, 1, 4, 1, 1, 1, 1, 2, 2, 3, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 6, 4, 1, 2, 1, 1, 3, 3, 2, 2, 1, 1, 1, 1, 3, 1, 2, 2, 3, 2, 1, 1, 1, 2, 1, 2, 4, 1, 2, 1, 7, 2, 1, 2, 2, 2, 10, 1, 1, 1, 13, 1, 2, 1, 1, 1, 1, 1, 1, 2, 3, 2, 1, 1, 1, 3, 1, 2, 2, 2, 2, 2, 2, 1, 2, 1, 1, 2, 1, 1, 1, 3, 3, 1, 1, 2, 2, 1, 4, 1, 1, 1, 1, 2, 2, 1, 3, 2, 2, 1, 2, 3, 1, 1, 2, 1, 3, 1, 1, 1, 1, 2, 2, 1, 10, 1, 1, 2, 1, 2, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 2, 6, 6, 3, 1, 2, 2, 1, 2, 2, 3, 2, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 4, 2, 2, 6, 1, 4, 2, 3, 1, 1, 1, 1, 2, 1, 1, 2, 1, 1, 2, 1, 1, 1, 8, 1, 1, 3, 2, 1, 4, 1, 1, 2, 3, 1, 1, 1, 5, 1, 1, 5, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 1, 1, 1, 2, 1, 2, 1, 3, 2, 1, 3, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 3, 2, 3, 2, 5, 3, 3, 2, 2, 1, 1, 1, 3, 1, 1, 1, 1, 1, 1, 5, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 1, 1, 2, 2, 1, 1, 1, 1, 1, 1, 1, 7, 1, 1, 1, 1, 1, 2, 1, 2, 1, 2, 1, 1, 1, 1, 2, 1, 1, 3, 1, 4, 3, 1, 1, 2, 1, 1, 1, 1, 10, 2, 1, 1, 1, 1, 1, 1, 1, 2, 2, 1, 1, 3, 1, 1, 4, 1, 1, 4, 2, 1, 1, 1, 2, 1, 1, 3, 1, 1, 1, 2, 3, 2, 2, 1, 1, 1, 4, 1, 2, 1, 1, 1, 1, 3, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 3, 1, 1, 1, 5, 2, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 2, 3, 2, 1, 4, 1, 2, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 2, 1, 2, 1, 1, 2, 1, 2, 1, 1, 3, 1, 1, 1, 2, 1, 3, 1, 1, 1, 1, 1, 3, 2, 1, 1, 1, 1, 1, 4, 1, 1, 1, 2, 3, 2, 2, 1, 1, 1, 1, 1, 2, 1, 1, 1, 5, 2, 2, 1, 1, 1, 1, 1, 1, 1, 2, 1, 2, 1, 1, 2, 2, 1, 1, 1, 2, 1, 1, 7, 2, 5, 2, 2, 1, 1, 3, 1, 1, 1, 1, 1, 1, 2, 1, 1, 16, 1, 3, 1, 2, 1, 1, 1, 1, 1, 3, 1, 3, 1, 2, 1, 1, 1, 1, 2, 2, 1, 1, 3, 1, 1, 1, 1, 1, 4, 2, 1, 1, 2, 1, 2, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 1, 6, 6, 2, 3, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 2, 1, 1, 2, 1, 1, 1, 1, 1, 1, 2, 1, 1, 4, 1, 1, 1, 1, 1, 1, 1, 2, 2, 1, 2, 2, 2, 1, 2, 1, 1, 1, 1, 1, 1, 2, 1, 2, 2, 1, 1, 1, 1, 1, 6, 1, 1, 3, 1, 1, 3, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 2, 4, 2, 1, 1, 1, 1, 1, 1, 3, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 2, 1, 4, 4, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 3, 2, 1, 1, 2, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 1, 1, 4, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 2, 1, 4, 1, 2, 1, 3, 1, 2, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 6, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 1, 1, 2, 1, 1, 3, 1, 1, 1, 1, 1, 1, 1, 1, 1, 6, 1, 1, 1, 1, 1, 2, 2, 1, 1, 2, 2, 1, 1, 2, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 3, 1, 1, 1, 2, 1, 1, 1, 2, 1, 1, 7, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 3, 1, 1, 1, 1, 1, 1, 1, 2, 1, 3, 1, 1, 1, 1, 5, 1, 1, 1, 1, 1, 2, 1, 1, 1, 4, 2, 1, 1, 2, 1, 1, 2, 1, 1, 1, 2, 1, 1, 1, 1, 1, 4, 2, 1, 1, 1, 3, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 5, 1, 2, 1, 1, 2, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 3, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 5, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 2, 9, 2, 1, 1, 1, 1, 1, 3, 2, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 17, 3, 2, 1, 2, 2, 1, 1, 1, 3, 2, 1, 3, 2, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 2, 1, 3, 1, 1, 2, 1, 1, 1, 2, 1, 1, 3, 2, 1, 1, 3, 1, 1, 1, 1, 1, 1, 1, 3, 1, 2, 2, 1, 1, 1, 4, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 10, 1, 1, 1, 5, 1, 3, 2, 1, 1, 2, 2, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 3, 1, 1, 1, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 7, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 1, 1, 1, 1, 5, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 1, 1, 2, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 4, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 2, 1, 2, 1, 1, 1, 1, 4, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 2, 2, 1, 2, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 1, 2, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 2, 1, 1, 2, 2, 8, 1, 1, 4, 1, 1, 4, 2, 14, 6, 2, 4, 7, 57, 27, 2, 11, 2, 7, 1, 3, 3, 1, 1, 2, 4, 6, 6, 2, 7, 2, 1, 1, 10, 10, 3, 11, 13, 8, 1, 4, 4, 4, 2, 2, 5, 4, 1, 1, 1, 2, 1, 1, 4, 3, 2, 1, 1, 2, 4, 1, 2, 4, 4, 1, 4, 3, 4, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 4, 3, 1, 1, 1, 2, 1, 1, 3, 1, 1, 3, 1, 4, 3, 1, 6, 1, 15, 1, 2, 1, 1, 11, 5, 3, 2, 2, 6, 2, 1, 1, 1, 1, 1, 5, 1, 1, 1, 1, 1, 3, 1, 1, 2, 1, 1, 2, 3, 2, 3, 1, 3, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 1, 3, 1, 1, 1, 1, 3, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 2, 1, 1, 1, 1, 1, 2, 1, 1, 1, 3, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1]\n"
]
],
[
[
"also see https://stackoverflow.com/questions/15899861/efficient-term-document-matrix-with-nltk",
"_____no_output_____"
]
],
[
[
"type(row)",
"_____no_output_____"
],
[
"row[1:10]",
"_____no_output_____"
],
[
"import pandas as pd\nfrom sklearn.feature_extraction.text import CountVectorizer\n\n \nvec = CountVectorizer()\nX = vec.fit_transform(tokens)\ndf = pd.DataFrame(X.toarray(), columns=vec.get_feature_names())\nprint(df)",
"_____no_output_____"
],
[
"print(df)",
" 000 14 1500 1849 1859 1861 1864 1880 1887 20 ... zeus \\\n0 0 0 0 0 0 0 0 0 0 0 ... 0 \n1 0 0 0 0 0 0 0 0 0 0 ... 0 \n2 0 0 0 0 0 0 0 0 0 0 ... 0 \n3 0 0 0 0 0 0 0 0 0 0 ... 0 \n4 0 0 0 0 0 0 0 0 0 0 ... 0 \n5 0 0 0 0 0 0 0 0 0 0 ... 0 \n6 0 0 0 0 0 0 0 0 0 0 ... 0 \n7 0 0 0 0 0 0 0 0 0 0 ... 0 \n8 0 0 0 0 0 0 0 0 0 0 ... 0 \n9 0 0 0 0 0 0 0 0 0 0 ... 0 \n10 0 0 0 0 0 0 0 0 0 0 ... 0 \n11 0 0 0 0 0 0 0 0 0 0 ... 0 \n12 0 0 0 0 0 0 0 0 0 0 ... 0 \n13 0 0 0 0 0 0 0 0 0 0 ... 0 \n14 0 0 0 0 0 0 0 0 0 0 ... 0 \n15 0 0 0 0 0 0 0 0 0 0 ... 0 \n16 0 0 0 0 0 0 0 0 0 0 ... 0 \n17 0 0 0 0 0 0 0 0 0 0 ... 0 \n18 0 0 0 0 0 0 0 0 0 0 ... 0 \n19 0 0 0 0 0 0 0 0 0 0 ... 0 \n20 0 0 0 0 0 0 0 0 0 0 ... 0 \n21 0 0 0 0 0 0 0 0 0 0 ... 0 \n22 0 0 0 0 0 0 0 0 0 0 ... 0 \n23 0 0 0 0 0 0 0 0 0 0 ... 0 \n24 0 0 0 0 0 0 0 0 0 0 ... 0 \n25 0 0 0 0 0 0 0 0 0 0 ... 0 \n26 0 0 0 0 0 0 0 0 0 0 ... 0 \n27 0 0 0 0 0 0 0 0 0 0 ... 0 \n28 0 0 0 0 0 0 0 0 0 0 ... 0 \n29 0 0 0 0 0 0 0 0 0 0 ... 0 \n... ... .. ... ... ... ... ... ... ... .. ... ... \n257696 0 0 0 0 0 0 0 0 0 0 ... 0 \n257697 0 0 0 0 0 0 0 0 0 0 ... 0 \n257698 0 0 0 0 0 0 0 0 0 0 ... 0 \n257699 0 0 0 0 0 0 0 0 0 0 ... 0 \n257700 0 0 0 0 0 0 0 0 0 0 ... 0 \n257701 0 0 0 0 0 0 0 0 0 0 ... 0 \n257702 0 0 0 0 0 0 0 0 0 0 ... 0 \n257703 0 0 0 0 0 0 0 0 0 0 ... 0 \n257704 0 0 0 0 0 0 0 0 0 0 ... 0 \n257705 0 0 0 0 0 0 0 0 0 0 ... 0 \n257706 0 0 0 0 0 0 0 0 0 0 ... 0 \n257707 0 0 0 0 0 0 0 0 0 0 ... 0 \n257708 0 0 0 0 0 0 0 0 0 0 ... 0 \n257709 0 0 0 0 0 0 0 0 0 0 ... 0 \n257710 0 0 0 0 0 0 0 0 0 0 ... 0 \n257711 0 0 0 0 0 0 0 0 0 0 ... 0 \n257712 0 0 0 0 0 0 0 0 0 0 ... 0 \n257713 0 0 0 0 0 0 0 0 0 0 ... 0 \n257714 0 0 0 0 0 0 0 0 0 0 ... 0 \n257715 0 0 0 0 0 0 0 0 0 0 ... 0 \n257716 0 0 0 0 0 0 0 0 0 0 ... 0 \n257717 0 0 0 0 0 0 0 0 0 0 ... 0 \n257718 0 0 0 0 0 0 0 0 0 0 ... 0 \n257719 0 0 0 0 0 0 0 0 0 0 ... 0 \n257720 0 0 0 0 0 0 0 0 0 0 ... 0 \n257721 0 0 0 0 0 0 0 0 0 0 ... 0 \n257722 0 0 0 0 0 0 0 0 0 0 ... 0 \n257723 0 0 0 0 0 0 0 0 0 0 ... 0 \n257724 0 0 0 0 0 0 0 0 0 0 ... 0 \n257725 0 0 0 0 0 0 0 0 0 0 ... 0 \n\n zigzags zimmerman zip zossimov æsthetic æsthetically æsthetics \\\n0 0 0 0 0 0 0 0 \n1 0 0 0 0 0 0 0 \n2 0 0 0 0 0 0 0 \n3 0 0 0 0 0 0 0 \n4 0 0 0 0 0 0 0 \n5 0 0 0 0 0 0 0 \n6 0 0 0 0 0 0 0 \n7 0 0 0 0 0 0 0 \n8 0 0 0 0 0 0 0 \n9 0 0 0 0 0 0 0 \n10 0 0 0 0 0 0 0 \n11 0 0 0 0 0 0 0 \n12 0 0 0 0 0 0 0 \n13 0 0 0 0 0 0 0 \n14 0 0 0 0 0 0 0 \n15 0 0 0 0 0 0 0 \n16 0 0 0 0 0 0 0 \n17 0 0 0 0 0 0 0 \n18 0 0 0 0 0 0 0 \n19 0 0 0 0 0 0 0 \n20 0 0 0 0 0 0 0 \n21 0 0 0 0 0 0 0 \n22 0 0 0 0 0 0 0 \n23 0 0 0 0 0 0 0 \n24 0 0 0 0 0 0 0 \n25 0 0 0 0 0 0 0 \n26 0 0 0 0 0 0 0 \n27 0 0 0 0 0 0 0 \n28 0 0 0 0 0 0 0 \n29 0 0 0 0 0 0 0 \n... ... ... ... ... ... ... ... \n257696 0 0 0 0 0 0 0 \n257697 0 0 0 0 0 0 0 \n257698 0 0 0 0 0 0 0 \n257699 0 0 0 0 0 0 0 \n257700 0 0 0 0 0 0 0 \n257701 0 0 0 0 0 0 0 \n257702 0 0 0 0 0 0 0 \n257703 0 0 0 0 0 0 0 \n257704 0 0 0 0 0 0 0 \n257705 0 0 0 0 0 0 0 \n257706 0 0 0 0 0 0 0 \n257707 0 0 0 0 0 0 0 \n257708 0 0 0 0 0 0 0 \n257709 0 0 0 0 0 0 0 \n257710 0 0 0 0 0 0 0 \n257711 0 0 0 0 0 0 0 \n257712 0 0 0 0 0 0 0 \n257713 0 0 0 0 0 0 0 \n257714 0 0 0 0 0 0 0 \n257715 0 0 0 0 0 0 0 \n257716 0 0 0 0 0 0 0 \n257717 0 0 0 0 0 0 0 \n257718 0 0 0 0 0 0 0 \n257719 0 0 0 0 0 0 0 \n257720 0 0 0 0 0 0 0 \n257721 0 0 0 0 0 0 0 \n257722 0 0 0 0 0 0 0 \n257723 0 0 0 0 0 0 0 \n257724 0 0 0 0 0 0 0 \n257725 0 0 0 0 0 0 0 \n\n éternelle_ êtes \n0 0 0 \n1 0 0 \n2 0 0 \n3 0 0 \n4 0 0 \n5 0 0 \n6 0 0 \n7 0 0 \n8 0 0 \n9 0 0 \n10 0 0 \n11 0 0 \n12 0 0 \n13 0 0 \n14 0 0 \n15 0 0 \n16 0 0 \n17 0 0 \n18 0 0 \n19 0 0 \n20 0 0 \n21 0 0 \n22 0 0 \n23 0 0 \n24 0 0 \n25 0 0 \n26 0 0 \n27 0 0 \n28 0 0 \n29 0 0 \n... ... ... \n257696 0 0 \n257697 0 0 \n257698 0 0 \n257699 0 0 \n257700 0 0 \n257701 0 0 \n257702 0 0 \n257703 0 0 \n257704 0 0 \n257705 0 0 \n257706 0 0 \n257707 0 0 \n257708 0 0 \n257709 0 0 \n257710 0 0 \n257711 0 0 \n257712 0 0 \n257713 0 0 \n257714 0 0 \n257715 0 0 \n257716 0 0 \n257717 0 0 \n257718 0 0 \n257719 0 0 \n257720 0 0 \n257721 0 0 \n257722 0 0 \n257723 0 0 \n257724 0 0 \n257725 0 0 \n\n[257726 rows x 9860 columns]\n"
],
[
"df[['zossimov']]",
"_____no_output_____"
],
[
"type(tokens)",
"_____no_output_____"
],
[
"tokens2=pd.DataFrame(tokens)",
"_____no_output_____"
],
[
"tokens2.columns=['Words']",
"_____no_output_____"
],
[
"tokens2.head()",
"_____no_output_____"
],
[
"tokens2.Words.value_counts().head()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb94bade90585c0f7ec72640090203a5f103801 | 410,322 | ipynb | Jupyter Notebook | 3-Basic-visualization-of-2d-data.ipynb | adcroft/Analyzing-ESM-data-with-python | fc94d8f880d1f389c8f2d7a62b29cb687f3d772d | [
"MIT"
] | null | null | null | 3-Basic-visualization-of-2d-data.ipynb | adcroft/Analyzing-ESM-data-with-python | fc94d8f880d1f389c8f2d7a62b29cb687f3d772d | [
"MIT"
] | null | null | null | 3-Basic-visualization-of-2d-data.ipynb | adcroft/Analyzing-ESM-data-with-python | fc94d8f880d1f389c8f2d7a62b29cb687f3d772d | [
"MIT"
] | null | null | null | 1,189.33913 | 262,232 | 0.957075 | [
[
[
"# Basic visualization of 2d data\n\n## Synopsis\n\n- Import matplotlib.pyplot\n- Contour and shade 2d data",
"_____no_output_____"
]
],
[
[
"import netCDF4\nimport numpy",
"_____no_output_____"
]
],
[
[
"## pyplot from matplotlib\n\nNow let's visualize the data we now know how to read. We've already imported the netCDF4 package above. Now we will import pyplot which is a sub-package of matplotlib.\n\nWe could just import the package like this",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot",
"_____no_output_____"
]
],
[
[
"but to use it is rather cumbersome",
"_____no_output_____"
]
],
[
[
"matplotlib.pyplot.plot([1,3,2]);",
"_____no_output_____"
]
],
[
[
"So we will import pyplot with an alias \"plt\" as follows:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"Here, plt is a shorthand for pyplot that _is widely used_ but you can use whatever alias you like.\n\nNormally, we would put this import statement at the top of the notebook with the other imports.\n\nLet's look at the 2005 World Ocean Atlas temperature data on a 5$^\\circ$x5$^\\circ$ grid, served from IRIDL:",
"_____no_output_____"
]
],
[
[
"nc = netCDF4.Dataset('http://iridl.ldeo.columbia.edu/SOURCES/.NOAA/.NODC/.WOA05/.Grid-5x5/.Annual/.mn/.temperature/dods')",
"_____no_output_____"
]
],
[
[
"And let's load the coordinates and sea-surface temperature into variables:",
"_____no_output_____"
]
],
[
[
"lon = nc.variables['X'][:]\nlat = nc.variables['Y'][:]\nsst = nc.variables['temperature'][0,:,:]",
"_____no_output_____"
]
],
[
[
"Note that the `[:]` forced the reading of the data. Leaving the `[:]` out would have returned an object and deferred reading of the data which is generally considered better form. Here, I chose to force read so that the data is not repeatedly fetch across the Internet connection.\n\nThere are two main ways of looking at 2D data.\n\n1. Contours\n - `plt.contour()` Draw contours of constant values. Contours are colored by default.\n - `plt.contourf()` Draws bands of constant color between contours of constant value.\n \n Use `plt.clabel()` to label contours.\n2. Psuedo-color shading of value at data locations.\n - `plt.pcolor()` Colored pixels for each data value.\n - `plt.pcolormesh()` is optimized for quadrilateral data and thus faster. It also understands curvilinear grids better than pcolor().\n \n Use `plt.colorbar()` to add a color bar.\n\nHere are four variants using `plt.contour()` with different arguments (given in the title of each pane):",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(12,7)); # Makes figure large enough to see four panels (optional)\n\n# 1. Contour without coordinate value\nplt.subplot(221);\nplt.contour(sst);\nplt.xlabel('i index');\nplt.ylabel('j index');\nplt.title('Panel 1: plt.contour(sst)');\n\n# 2. As above but with contour labels\nplt.subplot(222);\nc = plt.contour(sst);\nplt.clabel(c);\nplt.xlabel('i index');\nplt.ylabel('j index');\nplt.title('Panel 2: plt.contour(sst) with contour labels');\n\n# 3. Contour with coordinates\nplt.subplot(223);\nc = plt.contour(lon, lat, sst);\nplt.clabel(c);\nplt.xlabel('Longitude ($^\\circ$E)');\nplt.ylabel('Latitude ($^\\circ$N)');\nplt.title('Panel 3: plt.contour(lon, lat, sst)');\n\n# 4. Contour with coordinates and specific contour levels\nplt.subplot(224);\nc = plt.contour(lon, lat, sst, levels=[0,5,10,15,20,25,30]);\nplt.clabel(c, fmt='%.1f');\nplt.xlabel('Longitude ($^\\circ$E)');\nplt.ylabel('Latitude ($^\\circ$N)');\nplt.title('Panel 4: Specifying levels=[0,5,10,15,20,25,30]) and label format');\n\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"And here are variants of `plt.pcolor()` and `plt.pcolormesh()`.\n\nIn the following examples you will not notice any difference between pcolor and pcolormesh but pcolormesh is the preferred method for i) efficiency and ii) flexibility with curvilinear grids.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(12,7)); # Makes figure large enough to see four panels (optional)\n\n# 1. Simple pcolor\nplt.subplot(221);\nplt.pcolor(sst);\nplt.colorbar();\nplt.xlabel('i index');\nplt.ylabel('j index');\nplt.title('Panel 1: plt.pcolor(sst)');\n\n# 2. Simple pcolormesh\nplt.subplot(222);\nplt.pcolormesh(sst);\nplt.colorbar();\nplt.xlabel('i index');\nplt.ylabel('j index');\nplt.title('Panel 2: plt.pcolormesh(sst)');\n\n# 3. pcolormesh with cell-centered coordinate value\nplt.subplot(223);\nplt.pcolormesh(lon, lat, sst);\nplt.colorbar();\nplt.xlabel('Longitude ($^\\circ$E)');\nplt.ylabel('Latitude ($^\\circ$N)');\nplt.title('Panel 3: plt.pcolormesh(lon, lat, sst) CAUTION!!!');\n\n# 4. pcolormesh with mesh coordinates value\nplt.subplot(224);\nmesh_lon = numpy.linspace(0,360,lon.shape[0]+1)\nmesh_lat = numpy.linspace(-90,90,lat.shape[0]+1)\nplt.pcolormesh(mesh_lon, mesh_lat, sst, vmin=0, vmax=30, cmap=plt.cm.cividis);\nplt.colorbar(extend='both');\nplt.xlabel('Longitude ($^\\circ$E)');\nplt.ylabel('Latitude ($^\\circ$N)');\nplt.title('Panel 4: plt.pcolormesh(mesh_lon, mesh_lat, sst, vmin=0, vmax=30)');\n\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"__VERY IMPORTANT NOTE__: In panel 3 above, we pass the coordinates of the data locations to pcolormesh and you should __notice that the top row and right column of values are not plotted!!!__ This is because pcolormesh expects the coordinates of the mesh, or cell boundaries, which should have one extra column and row of values than does the data. In panel 4, I make up appropriate mesh coordinates using `numpy.linspace()`.\n\nTo finish up, we can also combine pcolormesh and contour plots...",
"_____no_output_____"
]
],
[
[
"mesh_lon = numpy.linspace(0,360,lon.shape[0]+1)\nmesh_lat = numpy.linspace(-90,90,lat.shape[0]+1)\nplt.pcolormesh(mesh_lon, mesh_lat, sst, vmin=0, vmax=30, cmap=plt.cm.cividis);\nplt.colorbar(extend='both');\nc = plt.contour(lon, lat, sst, levels=[5,10,15,20,25], colors='w')\nplt.clabel(c, fmt='%.0f')\nplt.xlabel('Longitude ($^\\circ$E)');\nplt.ylabel('Latitude ($^\\circ$N)');",
"_____no_output_____"
]
],
[
[
"## Summary\n\n- Import matplotlib.pyplot with `import matplotlib.pyplot as plt`\n- Contour with `plt.contour()` and `plt.contourf()`\n- Shade with `plt.pcolormesh()`\n- Also used `plt.colorbar()`, `plt.xlabel()`, `plt.ylabel()`, `plt.clabel()`",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb9528aa3e5be8e939f97986a924b58094ae86d | 7,828 | ipynb | Jupyter Notebook | ComputeCCfromData.ipynb | swarnavoNIST/MQuIT | 24a5a7d1b191501592cbbbe1c2ccc9004a78320c | [
"MIT"
] | null | null | null | ComputeCCfromData.ipynb | swarnavoNIST/MQuIT | 24a5a7d1b191501592cbbbe1c2ccc9004a78320c | [
"MIT"
] | null | null | null | ComputeCCfromData.ipynb | swarnavoNIST/MQuIT | 24a5a7d1b191501592cbbbe1c2ccc9004a78320c | [
"MIT"
] | null | null | null | 27.758865 | 104 | 0.488631 | [
[
[
"import glob #filenames and pathnames utility\nimport os #operating sytem utility\nimport numpy as np\nimport pandas as pd\nimport pickle\nimport random as rand\nimport sys\nimport math\nfrom BA_C import BA\nfrom scipy.optimize import curve_fit",
"_____no_output_____"
],
[
"def linear(df,a,b):\n return (a*df + b)",
"_____no_output_____"
],
[
"\"\"\"data_directory: folder that contains the csv files for a method and replicate.\nfileset: Name of the csv files.\nreplicate_name: Part of the csv filename that identifies the replicate number.\n\"\"\"\n\ndata_directory = 'Examples/Flow_Chl_Rep1'\nfileset = 'Flow_Chl_1818.'\nreplicate_name = '_Rep1'\ncurrent_dir = os.getcwd()\nprint(current_dir)\nos.chdir(data_directory)",
"/Users/sns9/CodeX/MQuIT\n"
],
[
"def extract_data(data_set,index_set):\n extracted_data = []\n\n for i in index_set:\n try:\n extracted_data.append(data_set.values[i])\n except IndexError:\n print(i)\n sys.exit()\n\n return np.array(extracted_data)",
"_____no_output_____"
],
[
"response_files = glob.glob('*.csv')\n\nglob_max = -1\nglob_min = 1e12\n\ndatas = {}\nconclist = []\n\nsamples = 1e12\n\nfor f in response_files:\n if fileset in f:\n concname = f\n conc = concname.replace(fileset,'')\n conc = int(conc.replace(replicate_name+'.csv',''))\n \n if conc not in conclist:\n temp_d = pd.read_csv(f).to_numpy()[:,0]\n \n datas[int(conc)] = temp_d\n \n glob_max = max(glob_max,max(datas[conc]))\n glob_min = min(glob_min,min(datas[conc]))\n \n conclist.append(conc)\n \nconclist.sort()\nprint(glob_max,glob_min)\nconclist\n\n# Offset to shift all the expression values >= 1\noffset = -glob_min + 1\n\nbins = []\n\nfor conc in conclist:\n datas[conc] += offset\n \n \"\"\"Get the number of bins for each concentration determined by Freedman-Diaconis rule.\n Choice of linear scale (typically for RNAs) or log scale (typically for Proteins). \n \"\"\"\n \n hist, bin_edges = np.histogram(datas[conc],bins='fd')\n \n bins.append(len(hist))\n \nprint('bins: ',bins)",
"35875.059132225026 -284.7438512530847\nbins: [2947, 638, 1358, 1477, 1387, 505, 430, 392]\n"
],
[
"\"\"\"Freedman-Diaconis ('fd') is only a rule of thumb to estimate the number of bins. \nIt does not provide the exact number of bins required for accurately calculating \nchannel capacity. Start with a value for n_bins roughly 1/10th of the 'fd' estimates, \ncompute the channel capacity, then increase the n_bins and repeat the calculation.\n\"\"\"\nbin_set = [10,20,40,80,160]",
"_____no_output_____"
],
[
"of = open('C_summary.csv','w')\nprint('Bins,C',file=of)\nof.close()\n\ntry:\n os.chdir('response')\nexcept OSError:\n os.mkdir('response')\n os.chdir('response')",
"_____no_output_____"
],
[
"bao = BA()\n\n# Number of bins\n\nfor n_bins in bin_set:\n bin_edges = np.linspace(1.0,glob_max + offset,n_bins+1)\n \n # Compute and write probability transition matrix\n pdfs = np.zeros(shape=(len(conclist),n_bins))\n for j in range(0,len(conclist)):\n hist, bin_edges = np.histogram(datas[conc],bins=bin_edges)\n pdfs[j,:] = hist/np.sum(hist)\n \n np.savetxt('expressions.csv',pdfs,delimiter=',')\n \n # Compute probability transition matrix for subsampled data with replicates\n data_fractions = [1,2,5,10]\n n_reps = 5\n\n c_subsample = np.zeros(shape=(n_reps*len(data_fractions),2))\n idx = 0\n\n for df in data_fractions:\n for k in range(1,6):\n pdfs = np.zeros(shape=(len(conclist),n_bins))\n\n for i in range(0,len(conclist)):\n cs = conclist[i]\n darray = datas[cs]\n darray_list = list(darray)\n sample_size = int(len(darray_list)/df)\n\n d_sampled = rand.sample(darray_list,sample_size)\n\n hist, b_edges = np.histogram(np.array(d_sampled),bin_edges)\n\n pdfs[i,:] = hist/np.sum(hist)\n\n response_file = 'expressions'+str(int(df))+'_'+str(k)+'.csv'\n \n if n_bins!=bin_set[-1]:\n np.savetxt(response_file,pdfs,delimiter=',')\n \n \n # Compute channel capacity\n bao.set_response(pdfs)\n c, e, p = bao.get_CC()\n\n c_subsample[idx,0] = float(df)\n c_subsample[idx,1] = c\n \n idx += 1\n\n popt, pcov = curve_fit(linear, c_subsample[:,0], c_subsample[:,1])\n\n os.chdir('..')\n print(str(int(n_bins))+','+str(\"{:0.2f}\".format(popt[1])),file=open('C_summary.csv','a'))\n os.chdir('response')\n \n print(n_bins,popt[1])",
"10 0.8766649847714352\n20 1.1648080864496986\n40 1.389659695014482\n80 1.5800660239201518\n160 1.6482559272679644\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb9581c87f7edecde26ef199f3d136cc65fade5 | 3,969 | ipynb | Jupyter Notebook | kafka.ipynb | columbia-dsi/people_detection | 12854556b828dedd9be0c76a5b1f09706ebdddb4 | [
"MIT"
] | null | null | null | kafka.ipynb | columbia-dsi/people_detection | 12854556b828dedd9be0c76a5b1f09706ebdddb4 | [
"MIT"
] | 5 | 2019-04-27T04:08:34.000Z | 2019-10-28T11:41:04.000Z | kafka.ipynb | columbia-dsi/people_detection | 12854556b828dedd9be0c76a5b1f09706ebdddb4 | [
"MIT"
] | null | null | null | 25.941176 | 108 | 0.467372 | [
[
[
"import json\nfrom PIL import Image\nimport numpy as np\nfrom pykafka import KafkaClient\nfrom pykafka.common import OffsetType\n\ndef gen_client(hosts = \"127.0.0.1:9092\", topic_name = 'test'):\n client = KafkaClient(hosts=hosts)\n topic = client.topics[topic_name]\n return client, topic\n\ndef load_image_as_bytes(infilename) :\n img = Image.open( infilename )\n img.load()\n data = np.asarray( img, dtype=\"int32\" )\n np_array_to_list = data.tolist()\n b = bytes(json.dumps(np_array_to_list), 'utf-8')\n return b\n\ndef produce(topic, image_as_jsonified_bytes):\n with topic.get_sync_producer() as producer:\n producer.produce(image_as_jsonified_bytes)\n \ndef consume_latest(topic, LAST_N_MESSAGES = 1):\n consumer = topic.get_simple_consumer(\n auto_offset_reset=OffsetType.LATEST,\n reset_offset_on_start=True\n )\n offsets = [(p, op.next_offset - LAST_N_MESSAGES - 1) for p, op in consumer._partitions.items()]\n offsets = [(p, o) if o != -1 else (p, -2) for p, o in offsets]\n consumer.reset_offsets(offsets)\n return consumer.consume().value\n\ndef decode(incoming):\n as_json = json.loads(incoming.decode('utf-8'))\n arr = np.array(as_json)\n return arr",
"_____no_output_____"
],
[
"b = load_image_as_bytes('download.jpeg')\nclient, topic = gen_client()\nproduce(topic, b)",
"_____no_output_____"
],
[
"incoming = consume_latest(topic)\nif incoming is not None:\n received_image = decode(incoming)\n print(received_image)\nelse:\n print('No message received!')",
"[[[230 134 118]\n [230 134 118]\n [229 133 117]\n ...\n [220 133 116]\n [232 145 128]\n [209 121 107]]\n\n [[229 133 117]\n [229 133 117]\n [229 133 117]\n ...\n [227 140 130]\n [234 147 138]\n [216 129 120]]\n\n [[229 133 117]\n [229 133 117]\n [228 132 116]\n ...\n [194 108 109]\n [177 92 95]\n [154 69 74]]\n\n ...\n\n [[ 84 22 59]\n [ 92 28 65]\n [ 98 30 67]\n ...\n [127 43 67]\n [141 53 75]\n [149 60 78]]\n\n [[ 81 21 59]\n [ 87 25 62]\n [ 95 27 64]\n ...\n [140 55 78]\n [154 65 85]\n [163 71 86]]\n\n [[ 77 19 57]\n [ 86 24 63]\n [ 92 26 64]\n ...\n [150 62 84]\n [163 72 89]\n [170 76 90]]]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecb961fbe6ca8775385d51f9a14c9714b0b0e575 | 24,338 | ipynb | Jupyter Notebook | 09-Errors-and-Exceptions.ipynb | jkelley7/WhirlwindTourOfPythonJVP | f40b435dea823ad5f094d48d158cc8b8f282e9d5 | [
"CC0-1.0"
] | 1 | 2019-01-24T21:33:07.000Z | 2019-01-24T21:33:07.000Z | 09-Errors-and-Exceptions.ipynb | jkelley7/WhirlwindTourOfPythonJVP | f40b435dea823ad5f094d48d158cc8b8f282e9d5 | [
"CC0-1.0"
] | null | null | null | 09-Errors-and-Exceptions.ipynb | jkelley7/WhirlwindTourOfPythonJVP | f40b435dea823ad5f094d48d158cc8b8f282e9d5 | [
"CC0-1.0"
] | 1 | 2018-02-26T18:43:36.000Z | 2018-02-26T18:43:36.000Z | 31.363402 | 1,082 | 0.561098 | [
[
[
"*This notebook comes from [A Whirlwind Tour of Python](http://www.oreilly.com/programming/free/a-whirlwind-tour-of-python.csp) by Jake VanderPlas (OReilly Media, 2016). This content is licensed [CC0](https://github.com/jakevdp/WhirlwindTourOfPython/blob/master/LICENSE). The full notebook listing is available at https://github.com/jakevdp/WhirlwindTourOfPython.*",
"_____no_output_____"
],
[
"<!--NAVIGATION-->\n< [Defining and Using Functions](08-Defining-Functions.ipynb) | [Contents](Index.ipynb) | [Iterators](10-Iterators.ipynb) >",
"_____no_output_____"
],
[
"# Errors and Exceptions",
"_____no_output_____"
],
[
"No matter your skill as a programmer, you will eventually make a coding mistake.\nSuch mistakes come in three basic flavors:\n\n- *Syntax errors:* Errors where the code is not valid Python (generally easy to fix)\n- *Runtime errors:* Errors where syntactically valid code fails to execute, perhaps due to invalid user input (sometimes easy to fix)\n- *Semantic errors:* Errors in logic: code executes without a problem, but the result is not what you expect (often very difficult to track-down and fix)\n\nHere we're going to focus on how to deal cleanly with *runtime errors*.\nAs we'll see, Python handles runtime errors via its *exception handling* framework.",
"_____no_output_____"
],
[
"## Runtime Errors\n\nIf you've done any coding in Python, you've likely come across runtime errors.\nThey can happen in a lot of ways.\n\nFor example, if you try to reference an undefined variable:",
"_____no_output_____"
]
],
[
[
"print(Q)",
"_____no_output_____"
]
],
[
[
"Or if you try an operation that's not defined:",
"_____no_output_____"
]
],
[
[
"1 + 'abc'",
"_____no_output_____"
]
],
[
[
"Or you might be trying to compute a mathematically ill-defined result:",
"_____no_output_____"
]
],
[
[
"2 / 0",
"_____no_output_____"
]
],
[
[
"Or maybe you're trying to access a sequence element that doesn't exist:",
"_____no_output_____"
]
],
[
[
"L = [1, 2, 3]\nL[1000]",
"_____no_output_____"
]
],
[
[
"Note that in each case, Python is kind enough to not simply indicate that an error happened, but to spit out a *meaningful* exception that includes information about what exactly went wrong, along with the exact line of code where the error happened.\nHaving access to meanngful errors like this is immensely useful when trying to trace the root of problems in your code.",
"_____no_output_____"
],
[
"## Catching Exceptions: ``try`` and ``except``\nThe main tool Python gives you for handling runtime exceptions is the ``try``...``except`` clause.\nIts basic structure is this:",
"_____no_output_____"
]
],
[
[
"try:\n print(\"this gets executed first\")\nexcept:\n print(\"this gets executed only if there is an error\")",
"this gets executed first\n"
]
],
[
[
"Note that the second block here did not get executed: this is because the first block did not return an error.\nLet's put a problematic statement in the ``try`` block and see what happens:",
"_____no_output_____"
]
],
[
[
"try:\n print(\"let's try something:\")\n x = 1 / 0 # ZeroDivisionError\nexcept:\n print(\"something bad happened!\")",
"let's try something:\nsomething bad happened!\n"
]
],
[
[
"Here we see that when the error was raised in the ``try`` statement (in this case, a ``ZeroDivisionError``), the error was caught, and the ``except`` statement was executed.\n\nOne way this is often used is to check user input within a function or another piece of code.\nFor example, we might wish to have a function that catches zero-division and returns some other value, perhaps a suitably large number like $10^{100}$:",
"_____no_output_____"
]
],
[
[
"def safe_divide(a, b):\n try:\n return a / b\n except:\n return 1E100",
"_____no_output_____"
],
[
"safe_divide(1, 2)",
"_____no_output_____"
],
[
"safe_divide(2, 0)",
"_____no_output_____"
]
],
[
[
"There is a subtle problem with this code, though: what happens when another type of exception comes up? For example, this is probably not what we intended:",
"_____no_output_____"
]
],
[
[
"safe_divide (1, '2')",
"_____no_output_____"
]
],
[
[
"Dividing an integer and a string raises a ``TypeError``, which our over-zealous code caught and assumed was a ``ZeroDivisionError``!\nFor this reason, it's nearly always a better idea to catch exceptions *explicitly*:",
"_____no_output_____"
]
],
[
[
"def safe_divide(a, b):\n try:\n return a / b\n except ZeroDivisionError:\n return 1E100",
"_____no_output_____"
],
[
"safe_divide(1, 0)",
"_____no_output_____"
],
[
"safe_divide(1, '2')",
"_____no_output_____"
]
],
[
[
"We're now catching zero-division errors only, and letting all other errors pass through un-modified.",
"_____no_output_____"
],
[
"## Raising Exceptions: ``raise``\nWe've seen how valuable it is to have informative exceptions when using parts of the Python language.\nIt's equally valuable to make use of informative exceptions within the code you write, so that users of your code (foremost yourself!) can figure out what caused their errors.\n\nThe way you raise your own exceptions is with the ``raise`` statement. For example:",
"_____no_output_____"
]
],
[
[
"raise RuntimeError(\"my error message\")",
"_____no_output_____"
]
],
[
[
"As an example of where this might be useful, let's return to our ``fibonacci`` function that we defined previously:",
"_____no_output_____"
]
],
[
[
"def fibonacci(N):\n L = []\n a, b = 0, 1\n while len(L) < N:\n a, b = b, a + b\n L.append(a)\n return L",
"_____no_output_____"
]
],
[
[
"One potential problem here is that the input value could be negative.\nThis will not currently cause any error in our function, but we might want to let the user know that a negative ``N`` is not supported.\nErrors stemming from invalid parameter values, by convention, lead to a ``ValueError`` being raised:",
"_____no_output_____"
]
],
[
[
"def fibonacci(N):\n if N < 0:\n raise ValueError(\"N must be non-negative\")\n L = []\n a, b = 0, 1\n while len(L) < N:\n a, b = b, a + b\n L.append(a)\n return L",
"_____no_output_____"
],
[
"fibonacci(10)",
"_____no_output_____"
],
[
"fibonacci(-10)",
"_____no_output_____"
]
],
[
[
"Now the user knows exactly why the input is invalid, and could even use a ``try``...``except`` block to handle it!",
"_____no_output_____"
]
],
[
[
"N = -10\ntry:\n print(\"trying this...\")\n print(fibonacci(N))\nexcept ValueError:\n print(\"Bad value: need to do something else\")",
"trying this...\nBad value: need to do something else\n"
]
],
[
[
"## Diving Deeper into Exceptions\n\nBriefly, I want to mention here some other concepts you might run into.\nI'll not go into detail on these concepts and how and why to use them, but instead simply show you the syntax so you can explore more on your own.",
"_____no_output_____"
],
[
"### Accessing the error message\n\nSometimes in a ``try``...``except`` statement, you would like to be able to work with the error message itself.\nThis can be done with the ``as`` keyword:",
"_____no_output_____"
]
],
[
[
"try:\n x = 1 / 0\nexcept ZeroDivisionError as err:\n print(\"Error class is: \", type(err))\n print(\"Error message is:\", err)",
"Error class is: <class 'ZeroDivisionError'>\nError message is: division by zero\n"
]
],
[
[
"With this pattern, you can further customize the exception handling of your function.",
"_____no_output_____"
],
[
"### Defining custom exceptions\nIn addition to built-in exceptions, it is possible to define custom exceptions through *class inheritance*.\nFor instance, if you want a special kind of ``ValueError``, you can do this:",
"_____no_output_____"
]
],
[
[
"class MySpecialError(ValueError):\n pass\n\nraise MySpecialError(\"here's the message\")",
"_____no_output_____"
]
],
[
[
"This would allow you to use a ``try``...``except`` block that only catches this type of error:",
"_____no_output_____"
]
],
[
[
"try:\n print(\"do something\")\n raise MySpecialError(\"[informative error message here]\")\nexcept MySpecialError:\n print(\"do something else\")",
"do something\ndo something else\n"
]
],
[
[
"You might find this useful as you develop more customized code.",
"_____no_output_____"
],
[
"## ``try``...``except``...``else``...``finally``\nIn addition to ``try`` and ``except``, you can use the ``else`` and ``finally`` keywords to further tune your code's handling of exceptions.\nThe basic structure is this:",
"_____no_output_____"
]
],
[
[
"try:\n print(\"try something here\")\nexcept:\n print(\"this happens only if it fails\")\nelse:\n print(\"this happens only if it succeeds\")\nfinally:\n print(\"this happens no matter what\")",
"try something here\nthis happens only if it succeeds\nthis happens no matter what\n"
]
],
[
[
"The utility of ``else`` here is clear, but what's the point of ``finally``?\nWell, the ``finally`` clause really is executed *no matter what*: I usually see it used to do some sort of cleanup after an operation completes.",
"_____no_output_____"
],
[
"<!--NAVIGATION-->\n< [Defining and Using Functions](08-Defining-Functions.ipynb) | [Contents](Index.ipynb) | [Iterators](10-Iterators.ipynb) >",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecb967dfc90f8788459bc4da1dae75fed5215cff | 1,353 | ipynb | Jupyter Notebook | examples/linkcheck.ipynb | jtpio/pytest-check-links | 57aa513ad883c52537f27f6b233f9e77991e85e5 | [
"BSD-3-Clause"
] | null | null | null | examples/linkcheck.ipynb | jtpio/pytest-check-links | 57aa513ad883c52537f27f6b233f9e77991e85e5 | [
"BSD-3-Clause"
] | 1 | 2021-04-02T09:52:18.000Z | 2021-04-02T09:52:18.000Z | examples/linkcheck.ipynb | jtpio/pytest-check-links | 57aa513ad883c52537f27f6b233f9e77991e85e5 | [
"BSD-3-Clause"
] | null | null | null | 18.04 | 85 | 0.490022 | [
[
[
"a = 5",
"_____no_output_____"
],
[
"print(a)",
"5\n"
]
],
[
[
"\n<a href=\"https://jupyter.org\">real link</a>\n\n<a href=\"https://jupyter.org/404\">real link bad</a>\n\n\n\n\n[markdown internal](#hash)\n\n[markdown local good](./linkcheck.ipynb)\n\n[markdown local bad](doesntexist.html)\n\n[markdown absolute](/tmp)",
"_____no_output_____"
]
]
] | [
"code",
"markdown"
] | [
[
"code",
"code"
],
[
"markdown"
]
] |
ecb967e48eaf12feaa627ce320a9f320150d3d34 | 32,600 | ipynb | Jupyter Notebook | kipet_examples/Ex1_ode_sim_wk_stoi.ipynb | notesofdabbler/learn_kipet | abf0ed865b9f1dc4c82774b04da9154439292f15 | [
"MIT"
] | null | null | null | kipet_examples/Ex1_ode_sim_wk_stoi.ipynb | notesofdabbler/learn_kipet | abf0ed865b9f1dc4c82774b04da9154439292f15 | [
"MIT"
] | null | null | null | kipet_examples/Ex1_ode_sim_wk_stoi.ipynb | notesofdabbler/learn_kipet | abf0ed865b9f1dc4c82774b04da9154439292f15 | [
"MIT"
] | null | null | null | 73.589165 | 20,676 | 0.776043 | [
[
[
"#\n# Working through the example at \n# https://github.com/kwmcbride/kipet_examples/blob/master/examples/example_1/Ex_1_ode_sim.py\n#",
"_____no_output_____"
],
[
"from kipet import KipetModel\n\nimport matplotlib.pyplot as plt\nimport sys\nimport re\nimport numpy as np\nimport pandas as pd\nimport pyomo.environ as pyo\nfrom collections import defaultdict\n\n\nimport gen_rxnrates as rxn\n\nimport importlib\nimportlib.reload(rxn)",
"_____no_output_____"
],
[
"rxnlist = [\n {'rxnid':'r1', 'rxn':'A -> B', 'kf': 2.0, 'Keq': np.nan, 'Eaf': np.nan, 'Ear': np.nan},\n {'rxnid':'r2', 'rxn':'B -> C', 'kf': 0.2, 'Keq': np.nan, 'Eaf': np.nan, 'Ear': np.nan}\n]",
"_____no_output_____"
],
[
"rxndf = pd.DataFrame(rxnlist).set_index('rxnid')\nrxndf",
"_____no_output_____"
],
[
"rxndict = rxndf.to_dict()\nrxndict",
"_____no_output_____"
],
[
"kipet_model = KipetModel()\n\nr1 = kipet_model.new_reaction('reaction-1')",
"_____no_output_____"
],
[
"# only r1 object is needed. Others are for checking if rate expressions are correct\nr1, cmp, r_dict, rates = rxn.get_rxnrates(r1, rxndict, T = 325)",
"_____no_output_____"
],
[
"for r in rates.keys():\n print(rates[r])",
"k_r1[0]*A[0,0]\nk_r2[0]*B[0,0]\n"
],
[
"for c in r_dict.keys():\n print(c, \":\", r_dict[c])",
"A : - (k_r1[0]*A[0,0])\nB : k_r1[0]*A[0,0] - (k_r2[0]*B[0,0])\nC : k_r2[0]*B[0,0]\n"
],
[
"r1.parameters",
"_____no_output_____"
],
[
"r1.components",
"_____no_output_____"
],
[
"r1.components['A'].value = 1.0\nr1.components",
"_____no_output_____"
],
[
"# Create the model - simulations require times\nr1.set_times(0, 10)\n\nr1.settings.simulator.tee = True",
"_____no_output_____"
],
[
"r1.simulate()",
"Warning: No variance provided for model component A, it is being set to one\nWarning: No variance provided for model component B, it is being set to one\nWarning: No variance provided for model component C, it is being set to one\nFinished creating simulator\nIpopt 3.13.4: \n\n******************************************************************************\nThis program contains Ipopt, a library for large-scale nonlinear optimization.\n Ipopt is released as open source code under the Eclipse Public License (EPL).\n For more information visit https://github.com/coin-or/Ipopt\n******************************************************************************\n\nThis is Ipopt version 3.13.4, running with linear solver mumps.\nNOTE: Other linear solvers might be more efficient (see Ipopt documentation).\n\nNumber of nonzeros in equality constraint Jacobian...: 3303\nNumber of nonzeros in inequality constraint Jacobian.: 0\nNumber of nonzeros in Lagrangian Hessian.............: 0\n\nTotal number of variables............................: 903\n variables with only lower bounds: 0\n variables with lower and upper bounds: 0\n variables with only upper bounds: 0\nTotal number of equality constraints.................: 903\nTotal number of inequality constraints...............: 0\n inequality constraints with only lower bounds: 0\n inequality constraints with lower and upper bounds: 0\n inequality constraints with only upper bounds: 0\n\niter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls\n 0 0.0000000e+00 2.00e+00 0.00e+00 -1.0 0.00e+00 - 0.00e+00 0.00e+00 0\n 1 0.0000000e+00 7.77e-15 0.00e+00 -1.0 1.88e+00 - 1.00e+00 1.00e+00h 1\n\nNumber of Iterations....: 1\n\n (scaled) (unscaled)\nObjective...............: 0.0000000000000000e+00 0.0000000000000000e+00\nDual infeasibility......: 0.0000000000000000e+00 0.0000000000000000e+00\nConstraint violation....: 7.7715611723760958e-15 7.7715611723760958e-15\nComplementarity.........: 0.0000000000000000e+00 0.0000000000000000e+00\nOverall NLP error.......: 7.7715611723760958e-15 7.7715611723760958e-15\n\n\nNumber of objective function evaluations = 2\nNumber of objective gradient evaluations = 2\nNumber of equality constraint evaluations = 2\nNumber of inequality constraint evaluations = 0\nNumber of equality constraint Jacobian evaluations = 2\nNumber of inequality constraint Jacobian evaluations = 0\nNumber of Lagrangian Hessian evaluations = 1\nTotal CPU secs in IPOPT (w/o function evaluations) = 0.184\nTotal CPU secs in NLP function evaluations = 0.004\n\nEXIT: Optimal Solution Found.\n"
],
[
"plt.plot(r1.results.Z)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecb96ccb393622686369597dff6ec325459d6608 | 1,712 | ipynb | Jupyter Notebook | 11 - Introduction to Python/6_Functions/5_Combining Conditional Statements and Functions (3:06)/Combining Conditional Statements and Functions - Solution_Py2.ipynb | olayinka04/365-data-science-courses | 7d71215432f0ef07fd3def559d793a6f1938d108 | [
"Apache-2.0"
] | 3 | 2020-03-24T12:58:37.000Z | 2020-08-03T17:22:35.000Z | 11 - Introduction to Python/6_Functions/5_Combining Conditional Statements and Functions (3:06)/Combining Conditional Statements and Functions - Solution_Py2.ipynb | olayinka04/365-data-science-courses | 7d71215432f0ef07fd3def559d793a6f1938d108 | [
"Apache-2.0"
] | null | null | null | 11 - Introduction to Python/6_Functions/5_Combining Conditional Statements and Functions (3:06)/Combining Conditional Statements and Functions - Solution_Py2.ipynb | olayinka04/365-data-science-courses | 7d71215432f0ef07fd3def559d793a6f1938d108 | [
"Apache-2.0"
] | 1 | 2021-10-19T23:59:37.000Z | 2021-10-19T23:59:37.000Z | 20.878049 | 267 | 0.524533 | [
[
[
"## Combining Conditional Statements and Functions",
"_____no_output_____"
],
[
"*Suggested Answers follow (usually there are multiple ways to solve a problem in Python).*",
"_____no_output_____"
],
[
"Define a function, called **compare_the_two()**, with two arguments. If the first one is greater than the second one, let it print \"Greater\". If the second one is greater, it should print \"Less\". Let it print \"Equal\" if the two values are the same number.",
"_____no_output_____"
]
],
[
[
"def compare_the_two(x,y):\n if x > y:\n print \"Greater\"\n elif x < y:\n print \"Less\"\n else:\n print \"Equal\"",
"_____no_output_____"
],
[
"compare_the_two(10,10)",
"Equal\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
ecb96d26f3b0178d6a73df4fa04610f917de67fb | 2,872 | ipynb | Jupyter Notebook | examples/99.To be released, but working/4. SearchNet/1. milvus_test.ipynb | arita37/RecNN | a9ef42e2f9059dfcad0885cb1f5be034b3e5bba1 | [
"Apache-2.0"
] | 1 | 2020-10-08T13:49:54.000Z | 2020-10-08T13:49:54.000Z | examples/99.To be released, but working/4. SearchNet/1. milvus_test.ipynb | arita37/RecNN | a9ef42e2f9059dfcad0885cb1f5be034b3e5bba1 | [
"Apache-2.0"
] | null | null | null | examples/99.To be released, but working/4. SearchNet/1. milvus_test.ipynb | arita37/RecNN | a9ef42e2f9059dfcad0885cb1f5be034b3e5bba1 | [
"Apache-2.0"
] | null | null | null | 23.735537 | 264 | 0.529596 | [
[
[
"from milvus import Milvus, IndexType, MetricType, Status\n\n# == recnn ==\nimport sys\nsys.path.append(\"../../../\")\nimport recnn",
"/ssd/anaconda/envs/recnn/lib/python3.8/site-packages/tqdm/std.py:670: FutureWarning: The Panel class is removed from pandas. Accessing it from the top-level namespace will also be removed in the next version\n from pandas import Panel\n"
],
[
"\nframe_size = 10\nbatch_size = 1\n# embeddgings: https://drive.google.com/open?id=1EQ_zXBR3DKpmJR3jBgLvt-xoOvArGMsL\ndirs = recnn.data.env.DataPath(\n base=\"../../../data/\",\n embeddings=\"embeddings/ml20_pca128.pkl\",\n ratings=\"ml-20m/ratings.csv\",\n cache=\"cache/frame_env.pkl\", # cache will generate after you run\n use_cache=True\n)\nenv = recnn.data.env.FrameEnv(dirs, frame_size, batch_size)\n\nfrom recnn.data.db_con import MilvusConnection\nmilvus = MilvusConnection(env)\n\nimport numpy as np\nq_records = np.random.rand(3, 128)\n\nresults = milvus.search(q_records)",
"_____no_output_____"
],
[
"results",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecb9744a025b1fc3857c557d3f5a67b7d6816a0f | 5,015 | ipynb | Jupyter Notebook | Untitled.ipynb | MuhammadIbrahimUW/CharityVizApp | 2db35b00d5c1fc51f7cc024a63517a035605cefa | [
"MIT"
] | null | null | null | Untitled.ipynb | MuhammadIbrahimUW/CharityVizApp | 2db35b00d5c1fc51f7cc024a63517a035605cefa | [
"MIT"
] | null | null | null | Untitled.ipynb | MuhammadIbrahimUW/CharityVizApp | 2db35b00d5c1fc51f7cc024a63517a035605cefa | [
"MIT"
] | null | null | null | 42.142857 | 198 | 0.364506 | [
[
[
"Scoring Algorithm (based on most recent reporting year)",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n\n# import the data from the dataframe\ndf = pd.read_csv(\"expense data calcs.csv\").drop(['Unnamed: 0'],axis=1)\n\n# create",
" URL \\\n0 https://www.charitydata.ca/charity/full-name/0... \n1 https://www.charitydata.ca/charity/bethesda-ro... \n2 https://www.charitydata.ca/charity/cafe-commun... \n3 https://www.charitydata.ca/charity/camp-courag... \n4 https://www.charitydata.ca/charity/chants-libr... \n\n Name BRN \\\n0 full name 1 \n1 bethesda romanian church of god 875769515RR0001 \n2 cafe communautaire lentre gens de sainte adele... 892458076RR0001 \n3 camp courage the first responders society 858394364RR0001 \n4 chants libres compagnie lyrique de creation 140382375RR0001 \n\n Expense Table Reporting Year \\\n0 NaN NaN \n1 {'Expenditures by 6 Groups': [2020, 2019, 2018... 2020.0 \n2 {'Expenditures by 6 Groups': [2020, 2019, 2018... 2020.0 \n3 {'Expenditures by 6 Groups': [2019, 2018, 2017... 2019.0 \n4 {'Expenditures by 6 Groups': [2020, 2019, 2018... 2020.0 \n\n Total Expenditure Total Expenditure on Charitable Activities \\\n0 NaN NaN \n1 77309.0 75367.0 \n2 384979.0 341365.0 \n3 25277.0 23154.0 \n4 235640.0 149745.0 \n\n Total Expenditure on Management and Administration \\\n0 NaN \n1 1895.0 \n2 43614.0 \n3 2123.0 \n4 80493.0 \n\n Total Expenditures on Fundraising \\\n0 NaN \n1 0.0 \n2 0.0 \n3 0.0 \n4 5402.0 \n\n Total Expenditures on Political Activities Total Other Expenditures \\\n0 NaN NaN \n1 0.0 0.0 \n2 0.0 0.0 \n3 0.0 0.0 \n4 0.0 0.0 \n\n Total Amount of Gifts Made to All Qualified Donees \n0 NaN \n1 0.0 \n2 0.0 \n3 0.0 \n4 0.0 \n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
ecb9867cea1f95f8dde6781b2a0e29805e5796e5 | 9,784 | ipynb | Jupyter Notebook | reinforcement_learning/rl_hvac_ray_energyplus/train-hvac.ipynb | eugeneteoh/amazon-sagemaker-examples | 15c006d367d27371a407706953e2e962fe3bbe48 | [
"Apache-2.0"
] | 3 | 2020-04-07T00:58:53.000Z | 2020-08-24T04:28:13.000Z | reinforcement_learning/rl_hvac_ray_energyplus/train-hvac.ipynb | eugeneteoh/amazon-sagemaker-examples | 15c006d367d27371a407706953e2e962fe3bbe48 | [
"Apache-2.0"
] | 2 | 2020-09-26T01:31:38.000Z | 2020-10-07T22:23:56.000Z | reinforcement_learning/rl_hvac_ray_energyplus/train-hvac.ipynb | eugeneteoh/amazon-sagemaker-examples | 15c006d367d27371a407706953e2e962fe3bbe48 | [
"Apache-2.0"
] | 1 | 2022-03-28T09:18:00.000Z | 2022-03-28T09:18:00.000Z | 32.078689 | 455 | 0.551615 | [
[
[
"# Optimizing building HVAC with Amazon SageMaker RL",
"_____no_output_____"
]
],
[
[
"import sagemaker\nimport boto3\n\nfrom sagemaker.rl import RLEstimator\n\nfrom source.common.docker_utils import build_and_push_docker_image",
"_____no_output_____"
]
],
[
[
"## Initialize Amazon SageMaker",
"_____no_output_____"
]
],
[
[
"role = sagemaker.get_execution_role()\nsm_session = sagemaker.session.Session()\n\n# SageMaker SDK creates a default bucket. Change this bucket to your own bucket, if needed.\ns3_bucket = sm_session.default_bucket()\n\ns3_output_path = f\"s3://{s3_bucket}\"\nprint(f\"S3 bucket path: {s3_output_path}\")\nprint(f\"Role: {role}\")",
"_____no_output_____"
]
],
[
[
"### Configure the framework you want to use\n\nSet `framework` to `'tf'` or `'torch'` for TensorFlow or PyTorch, respectively.",
"_____no_output_____"
]
],
[
[
"job_name_prefix = \"energyplus-hvac-ray\"\n\nframework = \"tf\"",
"_____no_output_____"
]
],
[
[
"### Configure where training happens\n\nYou can train your RL training jobs using the SageMaker notebook instance or local notebook instance. In both of these scenarios, you can run the following in either local or SageMaker modes. The local mode uses the SageMaker Python SDK to run your code in a local container before deploying to SageMaker. This can speed up iterative testing and debugging while using the same familiar Python SDK interface. You just need to set `local_mode = True`.",
"_____no_output_____"
]
],
[
[
"# run in local_mode on this machine, or as a SageMaker TrainingJob?\nlocal_mode = False\n\nif local_mode:\n instance_type = \"local\"\nelse:\n # If on SageMaker, pick the instance type.\n instance_type = \"ml.g4dn.16xlarge\" # g4dn.16x large has 1 GPU and 64 cores\n\nif \"ml.p\" in instance_type or \"ml.g\" in instance_type:\n cpu_or_gpu = \"gpu\"\nelse:\n cpu_or_gpu = \"cpu\"",
"_____no_output_____"
]
],
[
[
"# Train your homogeneous scaling job here",
"_____no_output_____"
],
[
"### Edit the training code\n\nThe training code is written in the file `train-sagemaker-hvac.py` which is uploaded in the /source directory.\n\n*Note that ray will automatically set `\"ray_num_cpus\"` and `\"ray_num_gpus\"` in `_get_ray_config`*",
"_____no_output_____"
]
],
[
[
"!pygmentize source/train-sagemaker-hvac.py",
"_____no_output_____"
]
],
[
[
"### Train the RL model using the Python SDK Script mode\n\nWhen using SageMaker for distributed training, you can select a GPU or CPU instance. The RLEstimator is used for training RL jobs.\n\n1. Specify the source directory where the environment, presets and training code is uploaded.\n2. Specify the entry point as the training code\n3. Specify the image (CPU or GPU) to be used for the training environment.\n4. Define the training parameters such as the instance count, job name, S3 path for output and job name.\n5. Define the metrics definitions that you are interested in capturing in your logs. These can also be visualized in CloudWatch and SageMaker Notebooks.",
"_____no_output_____"
],
[
"#### Build custom docker image",
"_____no_output_____"
]
],
[
[
"# Build a custom docker image depending on if cpu or gpu is used\n\nsuffix = \"py37\" if framework == \"tf\" else \"py36\"\n\nrepository_short_name = f\"sagemaker-hvac-ray-{cpu_or_gpu}\"\ndocker_build_args = {\n \"CPU_OR_GPU\": cpu_or_gpu,\n \"AWS_REGION\": boto3.Session().region_name,\n \"FRAMEWORK\": framework,\n \"SUFFIX\": suffix,\n}\n\nimage_name = build_and_push_docker_image(repository_short_name, build_args=docker_build_args)\nprint(\"Using ECR image %s\" % image_name)",
"_____no_output_____"
],
[
"metric_definitions = [\n {\n \"Name\": \"training_iteration\",\n \"Regex\": \"training_iteration: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)\",\n },\n {\n \"Name\": \"episodes_total\",\n \"Regex\": \"episodes_total: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)\",\n },\n {\n \"Name\": \"num_steps_trained\",\n \"Regex\": \"num_steps_trained: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)\",\n },\n {\n \"Name\": \"timesteps_total\",\n \"Regex\": \"timesteps_total: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)\",\n },\n {\n \"Name\": \"training_iteration\",\n \"Regex\": \"training_iteration: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)\",\n },\n {\n \"Name\": \"episode_reward_max\",\n \"Regex\": \"episode_reward_max: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)\",\n },\n {\n \"Name\": \"episode_reward_mean\",\n \"Regex\": \"episode_reward_mean: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)\",\n },\n {\n \"Name\": \"episode_reward_min\",\n \"Regex\": \"episode_reward_min: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)\",\n },\n]",
"_____no_output_____"
]
],
[
[
"### Ray homogeneous scaling - Specify `train_instance_count` > 1\n\nHomogeneous scaling allows us to use multiple instances of the same type.\n\nSpot instances are unused EC2 instances that could be used at 90% discount compared to On-Demand prices (more information about spot instances can be found [here](https://aws.amazon.com/ec2/spot/?cards.sort-by=item.additionalFields.startDateTime&cards.sort-order=asc) and [here](https://docs.aws.amazon.com/sagemaker/latest/dg/model-managed-spot-training.html))\n\nTo use spot instances, set `train_use_spot_instances = True`. To use On-Demand instances, `train_use_spot_instances = False`.",
"_____no_output_____"
]
],
[
[
"hyperparameters = {\n # no. of days to simulate. Remember to adjust the dates in RunPeriod of\n # 'source/eplus/envs/buildings/MediumOffice/RefBldgMediumOfficeNew2004_Chicago.idf' to match simulation days.\n \"n_days\": 365,\n \"n_iter\": 50, # no. of training iterations\n \"algorithm\": \"PPO\", # only APEX_DDPG and PPO are tested\n \"multi_zone_control\": True, # if each zone temperature set point has to be independently controlled\n \"energy_temp_penalty_ratio\": 10,\n \"framework\": framework,\n}\n\n# Set additional training parameters\ntraining_params = {\n \"base_job_name\": job_name_prefix,\n \"train_instance_count\": 1,\n \"tags\": [{\"Key\": k, \"Value\": str(v)} for k, v in hyperparameters.items()],\n}\n\n# Defining the RLEstimator\nestimator = RLEstimator(\n entry_point=f\"train-sagemaker-hvac.py\",\n source_dir=\"source\",\n dependencies=[\"source/common/\"],\n image_uri=image_name,\n role=role,\n train_instance_type=instance_type,\n # train_instance_type='local',\n output_path=s3_output_path,\n metric_definitions=metric_definitions,\n hyperparameters=hyperparameters,\n **training_params,\n)\n\nestimator.fit(wait=True)\n\nprint(\" \")\nprint(estimator.latest_training_job.job_name)\nprint(\"type=\", instance_type, \"count=\", training_params[\"train_instance_count\"])\nprint(\" \")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb98e5e687f8bbc32e43e028ea3d8b97b6700cf | 382,987 | ipynb | Jupyter Notebook | notebooks/GBIF API documentation.ipynb | TheoLvs/batch8_ceebios_app | 4339809259ff362b1186d048e2a1182f2eb8e89f | [
"MIT"
] | null | null | null | notebooks/GBIF API documentation.ipynb | TheoLvs/batch8_ceebios_app | 4339809259ff362b1186d048e2a1182f2eb8e89f | [
"MIT"
] | null | null | null | notebooks/GBIF API documentation.ipynb | TheoLvs/batch8_ceebios_app | 4339809259ff362b1186d048e2a1182f2eb8e89f | [
"MIT"
] | null | null | null | 91.60177 | 94,574 | 0.571043 | [
[
[
"# Base Data Science snippet\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport os\nimport time\nfrom tqdm import tqdm_notebook\n\n%matplotlib inline\n%load_ext autoreload\n%autoreload 2",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"pd.set_option('display.max_rows', 500)\npd.set_option('display.max_columns', 500)",
"_____no_output_____"
]
],
[
[
"https://www.gbif.org/fr/developer/summary",
"_____no_output_____"
]
],
[
[
"import requests\n\ndef gbif_species(endpoint):\n url = f\"https://api.gbif.org/v1/species/{endpoint}\"\n print(url)\n r = requests.get(url)\n results = r.json()\n return results\n\ndef gbif_occurences(endpoint):\n url = f\"https://api.gbif.org/v1/occurrence/{endpoint}\"\n print(url)\n r = requests.get(url)\n results = r.json()\n return results",
"_____no_output_____"
]
],
[
[
"# Search",
"_____no_output_____"
]
],
[
[
"pd.DataFrame([x for x in gbif_species(\"search?q=vespa+ducalis\")[\"results\"] if (not \"taxonID\" in x) and (not \"nubKey\" in x)])",
"https://api.gbif.org/v1/species/search?q=vespa+ducalis\n"
],
[
"pd.DataFrame([x for x in gbif_species(\"search?q=blue+whale\")[\"results\"] if (not \"taxonID\" in x) and (not \"nubKey\" in x) and \"rank\" in x and x[\"rank\"] not in [\"VARIETY\",\"SUBSPECIES\"]])",
"https://api.gbif.org/v1/species/search?q=blue+whale\n"
]
],
[
[
"# Species data",
"_____no_output_____"
],
[
"## Images & media",
"_____no_output_____"
]
],
[
[
"vespa = \"119412046\"\nvespanub = \"1311418\"",
"_____no_output_____"
],
[
"gbif_species(f\"match?name=vespa&rank=GENUS&strict=true\")",
"https://api.gbif.org/v1/species/match?name=vespa&rank=GENUS&strict=true\n"
],
[
"gbif_species(f\"{1311418}\")",
"https://api.gbif.org/v1/species/1311418\n"
],
[
"gbif_species(f\"{1311418}/children\")[\"results\"]",
"https://api.gbif.org/v1/species/1311418/children\n"
],
[
"test = gbif_occurences(f\"search?limit=1&media_type=stillImage&taxon_key={vespanub}\")",
"https://api.gbif.org/v1/occurrence/search?limit=1&media_type=stillImage&taxon_key=1311418\n"
],
[
"test",
"_____no_output_____"
],
[
"test[\"results\"][0][\"media\"][][\"identifier\"]",
"_____no_output_____"
]
],
[
[
"## Parents",
"_____no_output_____"
]
],
[
[
"gbif(f\"{vespanub}/parents\")",
"https://api.gbif.org/v1/species/1311418/parents\n"
]
],
[
[
"## Occurence count",
"_____no_output_____"
]
],
[
[
"l = []\nfor i in range(40):\n r = requests.get(f\"https://api.gbif.org/v1/occurrence/count?taxonKey=1457&year={1980+i}\")\n results = r.json()\n l.append(results)\n",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"plt.plot(l)",
"_____no_output_____"
],
[
"import requests\n\nr = requests.get(\"https://api.gbif.org/v1/occurrence/count?taxonKey=4490&year=2010,2020\")\nresults = r.json()\nresults",
"_____no_output_____"
],
[
"import requests\n\nr = requests.get(\"https://api.gbif.org/v1/occurrence/count?taxonKey=4490&year=2000,2010\")\nresults = r.json()\nresults",
"_____no_output_____"
],
[
"decades = list(range(1900,2021,10))\ndecades",
"_____no_output_____"
],
[
"data = []\nindex = []\nfor i in tqdm_notebook(range(len(decades)-1)):\n t1,t2 = decades[i],decades[i+1]\n r = requests.get(f\"https://api.gbif.org/v1/occurrence/count?taxonKey=1&year={t1},{t2}\").json()\n index.append(f\"{t1}-{t2}\")\n data.append(r)",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:3: TqdmDeprecationWarning: This function will be removed in tqdm==5.0.0\nPlease use `tqdm.notebook.tqdm` instead of `tqdm.tqdm_notebook`\n This is separate from the ipykernel package so we can avoid doing imports until\n"
],
[
"data2 = []\nindex = []\nfor i in tqdm_notebook(range(len(decades)-1)):\n t1,t2 = decades[i],decades[i+1]\n r = requests.get(f\"https://api.gbif.org/v1/occurrence/count?taxonKey=2433451&year={t1},{t2}\").json()\n index.append(f\"{t1}-{t2}\")\n data2.append(r)",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:3: TqdmDeprecationWarning: This function will be removed in tqdm==5.0.0\nPlease use `tqdm.notebook.tqdm` instead of `tqdm.tqdm_notebook`\n This is separate from the ipykernel package so we can avoid doing imports until\n"
],
[
"df = pd.DataFrame({\"index\":index,\"all\":data,\"species\":data2})\ndf",
"_____no_output_____"
],
[
"df[\"species2\"] = df[\"species\"] * 851913774/df[\"all\"]",
"_____no_output_____"
],
[
"df.set_index(\"index\")[\"species2\"].plot(kind = \"bar\")",
"_____no_output_____"
],
[
"import requests\n\nr = requests.get(\"https://api.gbif.org/v1/occurrence/count?taxonKey=4490&year=2010,2020\")\nresults = r.json()\nresults",
"_____no_output_____"
],
[
"import requests\n\nr = requests.get(\"https://api.gbif.org/v1/occurrence/count?taxonKey=4490&year=2017,2020\")\nresults = r.json()\nresults",
"_____no_output_____"
]
],
[
[
"# Autosuggest",
"_____no_output_____"
]
],
[
[
"import requests\n\nr = requests.get(\"https://api.gbif.org/v1/species/suggest?q=dog\")\nresults = r.json()\nresults",
"_____no_output_____"
]
],
[
[
"# Core AC API",
"_____no_output_____"
],
[
"https://core.ac.uk/documentation/api/",
"_____no_output_____"
]
],
[
[
"import urllib.parse\n\nCORE_API = 'https://core.ac.uk:443/api-v2'\nAPI_KEY=\"cJmoVEila3gB0zCIM2q1vpZnsKjr9XdG\"\n\nparams = {\n 'page':1,\n 'pageSize':10,\n 'apiKey':API_KEY,\n 'citations':'true'\n}\n\nquery = \"title:vespa ducalis\"\nquery = urllib.parse.quote(query)\nquery",
"_____no_output_____"
],
[
"import requests\nfinal_url = f\"{CORE_API}/articles/search/{query}\"\nprint(final_url)\nr = requests.get(final_url)",
"https://core.ac.uk:443/api-v2/articles/search/title%3Avespa%20ducalis\n"
],
[
"r.json()",
"_____no_output_____"
],
[
"import requests\nquery=urllib.parse.quote('title: \"vespa\"')\nr = requests.get(f\"https://core.ac.uk:443/api-v2/articles/search/{query}?page=1&pageSize=10&metadata=true&fulltext=true&citations=true&similar=true&duplicate=false&urls=false&faithfulMetadata=false&apiKey=cJmoVEila3gB0zCIM2q1vpZnsKjr9XdG\").json()",
"_____no_output_____"
],
[
"pd.DataFrame(r[\"data\"])",
"_____no_output_____"
],
[
"for i in pd.DataFrame(r[\"data\"])[\"title\"]:\n print(i)\n print(\"\")",
"Asian hornet Vespa velutina nigrithorax venom: Evaluation and identification of the bioactive compound responsible for human keratinocyte protection against oxidative stress\n\nVirtual European Solar & Planetary Access (VESPA): a Planetary Science\n Virtual Observatory cornerstone\n\nFrom the reproduction biology to the foraging behaviour, towards the biological control of Vespa velutina in France\n\nVirtual European Solar & Planetary Access (VESPA): A Planetary Science Virtual Observatory Cornerstone\n\nOcular Lesions Other Than Stings Following Yellow-Legged Hornet ( Vespa velutina nigrithorax ) Projections, as Reported to French Poison Control Centers\n\nCharacterizing thermal tolerance in the invasive yellow-legged hornet (Vespa velutina nigrithorax): The first step toward a green control method\n\nDevelopment of an LC-MS multivariate nontargeted methodology for differential analysis of the peptide profile of Asian hornet venom (Vespa velutina nigrithorax): application to the investigation of the impact of collection period variation\n\nAsian hornet Vespa velutina nigrithorax venom: Evaluation and identification of the bioactive compound responsible for human keratinocyte protection against oxidative stress\n\nPopulation Genetic Structure of the Introduced North American Population of the Hornet, Vespa Crabro Germana Christ\n\nPopulation Genetic Structure of the Introduced North American Population of the Hornet, Vespa Crabro Germana Christ\n\n"
]
],
[
[
"## Maps",
"_____no_output_____"
],
[
"- https://api.gbif.org/v2/map/demo.html\n- http://api.gbif.org/v2/map/debug/ol/#",
"_____no_output_____"
]
],
[
[
"url = \"https://api.gbif.org/v2/map/occurrence/density/{z}/{x}/{y}@2x.png?srs=EPSG:4326&bin=hex&hexPerTile=105&year=1961,2013&taxonKey=2435098&style=classic.poly\"",
"_____no_output_____"
],
[
"r = requests.get(url)",
"_____no_output_____"
],
[
"r.content",
"_____no_output_____"
]
],
[
[
"# Publications",
"_____no_output_____"
]
],
[
[
"from pymongo import MongoClient",
"_____no_output_____"
],
[
"CONN_STR = f\"mongodb://{MONGO_USERNAME}:{MONGO_PASSWORD}@165.22.121.95:27017/\"\nprint(CONN_STR)\nclient = MongoClient(CONN_STR)",
"_____no_output_____"
],
[
"pubs = client[\"documents\"][\"posts\"]",
"_____no_output_____"
],
[
"pubs.count()",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:1: DeprecationWarning: count is deprecated. Use estimated_document_count or count_documents instead. Please note that $where must be replaced by $expr, $near must be replaced by $geoWithin with $center, and $nearSphere must be replaced by $geoWithin with $centerSphere\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"pubs.find({\"dict_species.gbif_id\":2440708})",
"_____no_output_____"
],
[
"cursor = pubs.find({}).limit(10)",
"_____no_output_____"
],
[
"articles = list(cursor)",
"_____no_output_____"
],
[
"articles",
"_____no_output_____"
],
[
"articles = []\nfor i in cursor:\n articles.append(i)",
"_____no_output_____"
],
[
"pubs.find_one({\"dict_species.rank\":\"species\"})",
"_____no_output_____"
]
],
[
[
"# Scrapping publications",
"_____no_output_____"
]
],
[
[
"r = requests.get(\"https://www.semanticscholar.org/search?q=vespa%20ducalis&sort=relevance\")",
"_____no_output_____"
],
[
"r.content",
"_____no_output_____"
]
],
[
[
"# Publications Database",
"_____no_output_____"
]
],
[
[
"import requests\n\nurl = \"http://165.22.121.95/documents/search/\"\n\ndef make_url(species):\n return f\"{url}{species}\"",
"_____no_output_____"
],
[
"r = requests.get(make_url(\"vespa ducalis\"))",
"_____no_output_____"
],
[
"r.json()[0]",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecb996b2d63eea76a217f743cfc3c46a4fdee9ca | 64,172 | ipynb | Jupyter Notebook | topic_10/lab12.ipynb | compstat-lmu/DeepLearningLMU | 4d571aea397568eaa9552b63a635df34e2aebb71 | [
"MIT"
] | 1 | 2021-03-22T19:20:43.000Z | 2021-03-22T19:20:43.000Z | topic_10/lab12.ipynb | jaeeun-n/DeepLearningLMU | 4d571aea397568eaa9552b63a635df34e2aebb71 | [
"MIT"
] | null | null | null | topic_10/lab12.ipynb | jaeeun-n/DeepLearningLMU | 4d571aea397568eaa9552b63a635df34e2aebb71 | [
"MIT"
] | 2 | 2020-11-26T13:08:07.000Z | 2021-02-15T16:33:37.000Z | 198.061728 | 32,436 | 0.909665 | [
[
[
"from random import sample\n\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.keras import layers\nfrom tensorflow.keras.datasets import mnist\nfrom tensorflow.python.keras.utils.np_utils import to_categorical",
"_____no_output_____"
]
],
[
[
"## Exercise 1",
"_____no_output_____"
],
[
"#### Load data",
"_____no_output_____"
]
],
[
[
"(x_train, y_train), (_, _) = mnist.load_data()\nx_train = (x_train - 127.5) / 127.5\ny_train = to_categorical(y_train)\ninput_size = 28 * 28\nx_train = x_train.reshape((x_train.shape[0], input_size))",
"_____no_output_____"
]
],
[
[
"#### Generator",
"_____no_output_____"
]
],
[
[
"optim = tf.optimizers.Adam(\n learning_rate=0.0002,\n beta_1=0.5,\n)\nnoise_size = 32\ngenerator = keras.Sequential(\n [\n layers.Dense(256, input_shape=(noise_size,)),\n layers.LeakyReLU(alpha=0.2),\n layers.Dense(512),\n layers.LeakyReLU(alpha=0.2),\n layers.Dense(1024),\n layers.LeakyReLU(alpha=0.2),\n layers.Dense(784, activation=\"tanh\"),\n ]\n)\n\ngenerator.compile(loss=\"mse\", optimizer=optim)",
"_____no_output_____"
]
],
[
[
"#### Discriminator",
"_____no_output_____"
]
],
[
[
"discriminator = keras.Sequential(\n [\n layers.Dense(1024, input_shape=(784,)),\n layers.LeakyReLU(alpha=0.2),\n layers.Dropout(0.3),\n layers.Dense(512),\n layers.LeakyReLU(alpha=0.2),\n layers.Dropout(0.3),\n layers.Dense(256),\n layers.LeakyReLU(alpha=0.2),\n layers.Dropout(0.3),\n layers.Dense(1, activation=\"sigmoid\"),\n ]\n)\n\ndiscriminator.compile(loss=\"binary_crossentropy\", optimizer=optim)",
"_____no_output_____"
]
],
[
[
"#### GAN",
"_____no_output_____"
]
],
[
[
"gan_input = layers.Input(shape=(noise_size,))\ngan_output = discriminator(generator(gan_input))\ngan = keras.Model(gan_input, gan_output)\ngan.compile(loss=\"binary_crossentropy\", optimizer=optim)",
"_____no_output_____"
]
],
[
[
"#### Training",
"_____no_output_____"
]
],
[
[
"batch_size = 128\n\ngenerator_losses = []\ndiscriminator_losses = []\ndloss = 0\ngloss = 0\n\nfor i in range(2500):\n\n for _ in range(5):\n batch_idx = sample(range(x_train.shape[0]), batch_size)\n batch_images = x_train[batch_idx, :]\n noise = np.random.normal(size=(batch_size, noise_size))\n generated = generator.predict(noise)\n dloss = discriminator.train_on_batch(\n np.vstack([batch_images, generated]),\n np.expand_dims(np.array([0.9] * batch_size + [0.1] * batch_size), 1),\n )\n\n noise = np.random.normal(size=(batch_size, noise_size))\n gloss = gan.train_on_batch(noise, np.expand_dims(np.array([1] * batch_size), 1))\n discriminator_losses.append(dloss)\n generator_losses.append(gloss)\n\n if i % 250 == 0:\n print(\n f\"Step {i}, generator losses: {np.mean(generator_losses[-100:])}, discriminator losses: {np.mean(discriminator_losses[-100:])},\"\n )",
"Step 0, generator losses: 0.9721341133117676, discriminator losses: 0.4704849123954773,\nStep 250, generator losses: 0.8033317816257477, discriminator losses: 0.5725460106134415,\nStep 500, generator losses: 0.8221080130338669, discriminator losses: 0.56315598487854,\nStep 750, generator losses: 0.8151345330476761, discriminator losses: 0.5601272004842758,\nStep 1000, generator losses: 0.8159313309192657, discriminator losses: 0.5595499545335769,\nStep 1250, generator losses: 0.807854653596878, discriminator losses: 0.5644290697574615,\nStep 1500, generator losses: 0.7996916097402572, discriminator losses: 0.5687394618988038,\nStep 1750, generator losses: 0.7872013276815415, discriminator losses: 0.5771817183494568,\nStep 2000, generator losses: 0.7831154292821885, discriminator losses: 0.578257549405098,\nStep 2250, generator losses: 0.7789176762104034, discriminator losses: 0.581608214378357,\n"
],
[
"assert 0.5 < dloss < 0.7 < gloss < 1.0",
"_____no_output_____"
],
[
"plt.figure()\nplt.plot(discriminator_losses, label=\"Discriminator\")\nplt.plot(generator_losses, label=\"Generator\")",
"_____no_output_____"
],
[
"n_images = 25\nnoise = np.random.normal(size=(n_images, noise_size))\ngenerated = generator.predict(noise)\ngenerated = np.clip(generated, 0, 1)\nfor i in range(n_images):\n # define subplot\n plt.subplot(5, 5, 1 + i)\n # turn off axis\n plt.axis(\"off\")\n # plot raw pixel data\n plt.imshow(generated[i, :].reshape(28, -1), cmap=\"gray\")\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecb9a473dcad86cf70c60cab25e8ba6c71dad2ad | 98,357 | ipynb | Jupyter Notebook | Mini_Tutorial_Python.ipynb | thalesvalente/machinelearning | 1a516121d2578614355f9231585e95e5afee7a05 | [
"MIT"
] | null | null | null | Mini_Tutorial_Python.ipynb | thalesvalente/machinelearning | 1a516121d2578614355f9231585e95e5afee7a05 | [
"MIT"
] | null | null | null | Mini_Tutorial_Python.ipynb | thalesvalente/machinelearning | 1a516121d2578614355f9231585e95e5afee7a05 | [
"MIT"
] | null | null | null | 58.650566 | 40,454 | 0.683276 | [
[
[
"<a href=\"https://colab.research.google.com/github/thalesvalente/machinelearning/blob/master/Mini_Tutorial_Python.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"#1 Mini tutorial python para iniciantes\n\nOlá! Hoje vamos iniciar os primeiros passos para criar códigos em python. A intenção deste material é fornecer direções e dicas básicas. Não ensinar de fato. Pode ser meio corrido, mas dúvidas podem ser tiradas com www.google.com. E claro, [email protected] ;). \nVamos ao que interessa. Aqui você terá a direção para iniciar seus estudos relacionados aos seguinte ítens.\n- Colab\n- Python (básico e as bibliotecas pandas, matplotlib e numpy)\n- Vetorização em python (não é nada disso que você está pensando :) Vamos com calma!)\n\nEntão, vamos ser rápidos! Em 6 horas totalmente dedicadas você poderá aprender a se localizar em tudo (não decorar) se focar! \nOriento vocês a não tentarem ler tudo pois não terão tempo! Como programadores, temos olhos de águia e temos a capacidade de identificar rápido os códigos (nossas presas) que precisamos. Então, utilize e devenvolva essa habilidade para capturar as informações de acordo com a necessidade. Let´s go!!!",
"_____no_output_____"
],
[
"## 1.1 Colab\nColab é uma ferramenta do google que já fornece um \"ambiente pronto\" e gratuito para você começar a desenvolver seus códigos python. Ou seja, você tem disponível:\n- Hadware (GPU, CPU, RAM e disco)\n- Bibliotecas pré-instaladas (matplot, numpy,, tensorflow, keras, opencv....lib $n$)\n- Um ambiente notebook jupyter\n- Integração com o google drive\n\n\n",
"_____no_output_____"
],
[
"###1.1.1 Hardware\n\nOs recursos de hardware disponíveis são limitados. No entanto, são bem satisfatórios para iniciar inclusive projetos com redes neurais.\nAbra o link abaixo e execute a página em modo playground. Executar em modo playground é uma forma de você executar códigos sem salvar dados ou realizar modificações permanentes no código. Para executar em modo playground faça:\n- clique no link > procure o botão \"Open in playground\" > clique > execute os códigos que desejar\n\nlink: https://colab.research.google.com/drive/151805XTDg--dgHb3-AXJCpnWaqRhop_2",
"_____no_output_____"
],
[
"###1.1.2 Bibliotecas pré-instaladas\n\nIsso mesmo. O Colab nos fornece diversas bibliotecas python. Isso significa que você pode pular os passos de instalação de um ambiente de programação e configuração. Para verificar as bibliotecas préintaladas, execute a célula de abaixo. \nVale ressaltar que você também pode instalar bibliotecas de sua preferência :), como ilustrado em https://colab.research.google.com/notebooks/snippets/importing_libraries.ipynb\n",
"_____no_output_____"
]
],
[
[
"!pip freeze",
"_____no_output_____"
]
],
[
[
"###1.1.3 Ambiente notebook jupyter\nVamos falar o básico que você precisa saber e o resto é ler documentação. Basicamente existem dois tipos de célula (markdowncells - bloco onde você escreve o que deseja)\n- Text: como esta célula, você pode inserir texto, fórmulas usando [mathjax](https://www.mathjax.org/#samples) (fórmulas matemáticas like a latex) e inserir figuras, links e etc usando html e outras ferramentas. Recomendo [este link](https://jupyter-notebook.readthedocs.io/en/latest/examples/Notebook/examples_index.html).\n- Code: onde você digita seu código e executa\n\nAs células podem ser executadas separadamente. No entanto, devem ser executadas em ordem caso o código contida em uma seja ´rérequisito para outra (exemplo: importar bibliotecas, métodos definidos em células anteriores, etc)",
"_____no_output_____"
],
[
"###1.1.4 Integração com o google drive\n\nIsso mesmo! Você pode usar seu drive pessoal para armazenar seus códigos.\nSegue o link que ensina como carregar imagens contidas no google drive. A mesma idéia serve para outros arquivos. Abra a [Seção 1.2](1cP5PfC83AQCThrBMrCZUh4T-BaA6zcH7#scrollTo=GFcO1obYVKwi) deste outro material :)",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\nimport io\n\ndrive.mount('/content/gdrive')",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3Aietf%3Awg%3Aoauth%3A2.0%3Aoob&scope=email%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdocs.test%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive.photos.readonly%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fpeopleapi.readonly&response_type=code\n\nEnter your authorization code:\n··········\nMounted at /content/gdrive\n"
]
],
[
[
"##1.2 Python (básico e as bibliotecas pandas, matplotlib e numpy)\n\nNesta seção serão apresentadas 2 tipos de direções principais:\n- Links úteis\n- Exemplos básicos\n\nVamos ser rápidos e práticos. Mãos a obra :)",
"_____no_output_____"
],
[
"###1.2.1 Links úteis\n\n- Exemplos básicos de Python\n - exemplos rápidos: [1](https://www.makeuseof.com/tag/basic-python-examples-learn-fast/)\n - exemplos mais completos: [2](https://www.geeksforgeeks.org/python-programming-examples/#simple) e [3](https://www.tutorialspoint.com/python_programming_examples/2)\n- Pandas\n - exemplos rápidos: [1- com exercícios e soluções](https://www.datacamp.com/community/tutorials/pandas-tutorial-dataframe-python) e [2](https://www.learndatasci.com/tutorials/python-pandas-tutorial-complete-introduction-for-beginners/) \n - [ler arquivos do excel](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_excel.html)\n - [documentação](https://pandas.pydata.org/pandas-docs/stable/getting_started/10min.html)\n- Matplotlib \n - [2D](https://matplotlib.org/tutorials/introductory/sample_plots.html)\n - [3D](https://matplotlib.org/mpl_toolkits/mplot3d/tutorial.html)\n- Numpy\n - exemplos rápidos: [1](https://docs.scipy.org/doc/numpy-1.13.0/user/quickstart.html) \n - [documentação](https://matplotlib.org/contents.html)\n- [Numpy + pandas](https://www.hackerearth.com/pt-br/practice/machine-learning/data-manipulation-visualisation-r-python/tutorial-data-manipulation-numpy-pandas-python/tutorial/)\n\nImportante: é imprescindível a utilização do numpy para fazer o trabalho. O numpy possui rotinas matemáticas otimizadas e também facilita muito trabalhar com vetores e matrizes. **CONSELHO: Não tente fazer as operações na \"mão\"!** \nVeremos mais detalhes na Seção sobre XXX",
"_____no_output_____"
],
[
"###1.2.2 Exemplos básicos\n\nVeremos alguns exemplos rapidamente. Lembre-se de verificar os links úteis caso tenha dúvidas!",
"_____no_output_____"
],
[
"#### 1.2.2.1 Importando os pacotes necessários...",
"_____no_output_____"
]
],
[
[
"# Importar e manipular dados\nimport pandas as pd \n\n# Manipular dados\nimport numpy as np\n\n# Ferramentas de visualizacao de dados\nfrom mpl_toolkits.mplot3d import Axes3D\nfrom mpl_toolkits.mplot3d.art3d import Poly3DCollection\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"#### 1.2.2.2 Abrindo arquivo .xls e importando os dados (Pandas)\nSão executados os seguintes passos:\n- ler arquivo .csv do google drive\n- exibir os dados lidos\n- exibir o tipo da variável que guarda os dados\n- exibir as colunas com seus respectivos tipos",
"_____no_output_____"
]
],
[
[
"#data = pd.read_csv(io.BytesIO('/content/gdrive/My Drive/Colab Notebooks/CG/all_1nm_data.xls'))\ndata = pd.read_excel('/content/gdrive/My Drive/Colab Notebooks/CG/all_1nm_data.xls', header=0, skiprows=3) ",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"type(data)",
"_____no_output_____"
],
[
"data.dtypes",
"_____no_output_____"
],
[
"data.columns",
"_____no_output_____"
]
],
[
[
"#### 1.2.2.3 Manipulando os dados (Pandas + Numpy)\nSão executados os seguintes passos:\n- acessar uma determinada coluna\n- acessar um determinado elemento de uma coluna\n- converter uma coluna para lista\n- acessar um elemento da lista",
"_____no_output_____"
]
],
[
[
"data['nm']",
"_____no_output_____"
],
[
"data['nm'][0]",
"_____no_output_____"
],
[
"col_1 = data['CIE A'].tolist()\ncol_1",
"_____no_output_____"
],
[
"col_1[0]",
"_____no_output_____"
]
],
[
[
"#### 1.2.2.4 Numpy - operações básicas ",
"_____no_output_____"
]
],
[
[
"a = np.array([[1, 1, 1], [2, 2, 2]])\nb = np.array([[3, 3, 3], [3, 3, 3]])",
"_____no_output_____"
],
[
"a[0]",
"_____no_output_____"
],
[
"a.shape",
"_____no_output_____"
],
[
"a.size",
"_____no_output_____"
],
[
"# Adição\n\na + b",
"_____no_output_____"
],
[
"# Multiplicação elemento a elemento\n\na*b",
"_____no_output_____"
],
[
"## Atenção: o operador * não serve para multiplicação matricial!\n\na*np.transpose(b)",
"_____no_output_____"
],
[
"# Multiplicação matricial\n\nnp.dot(a, np.transpose(b))",
"_____no_output_____"
],
[
"# Exponenciação\n\nprint(a)\nnp.power(a, 3)",
"[[1 1 1]\n [2 2 2]]\n"
],
[
"# Somar todos os elementos do array\n\nnp.sum(a)",
"_____no_output_____"
],
[
"# Fazer a média dos elementos do array\n\nnp.mean(a)",
"_____no_output_____"
]
],
[
[
"#### 1.2.2.5 Transformando o Dataframe do Pandas em um Numpy Array",
"_____no_output_____"
]
],
[
[
"np.array(data['nm'])",
"_____no_output_____"
],
[
"x_bar = np.array(data['x bar'])\nx_bar",
"_____no_output_____"
],
[
"nan_locs = np.isnan(x_bar)\nx_bar[nan_locs] = 0\nx_bar",
"_____no_output_____"
],
[
"np.where(x_bar == 0.0)",
"_____no_output_____"
]
],
[
[
"#### 1.2.2.6 Visualizando os dados (Matplotlib)",
"_____no_output_____"
],
[
"##### 1.2.2.6.1 2D",
"_____no_output_____"
]
],
[
[
"index = range(len(x_bar))\nindex",
"_____no_output_____"
],
[
"# Scatter plot\n\nplt.scatter(index, x_bar, c = 'b')\nplt.show()",
"_____no_output_____"
]
],
[
[
"##### 1.2.2.6.2 3D",
"_____no_output_____"
]
],
[
[
"fig = plt.figure()\nax = fig.add_subplot(111, projection='3d')\n\nax.set_xlabel('X Label')\nax.set_ylabel('Y Label')\nax.set_zlabel('Z Label')\n\nax.set_xlim3d((0, 1))\nax.set_ylim3d((0, 1))\nax.set_zlim3d((0, 1))\n\npoints = np.random.random((50, 3))\n\nfor i in points:\n ax.scatter(i[0], i[1], i[2], c = 'r', marker = ',')\n \nplt.show()",
"_____no_output_____"
]
],
[
[
"##1.3 Vetorização em python\n\nA vetorização é basicamente a arte de se livrar de laços (tipo for) em algoritmos. Utilizar vetorização não apenas \"enxuga\" o código como também nos permite aproveitar a paralelização da execução de códigos de forma transparente. Não precisamos nos preocupar com os detalhes da implementação, apenas usar. Nesta seção, iremos observar experimentalmente o porque a vetorização é imprescindível. Além disso, iremos também testar alguns exemplos para fixar a sua utilização.\n\nObserve o seguinte código abaixo. Este código aplica o produto escalar entre 2 vetores com 1 milhão de elementos. Observe a diferença de velocidade de execução utilizando um loop e a versão vetorizada.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport time\n\na = np.random.rand(1000000)\nb = np.random.rand(1000000)\n\ntic = time.time()\nc = np.dot(a,b)\ntac = time.time()\ndt1 = 1000*(tac-tic)\nprint(\"Versão vetorizada: \" + str((dt1)) + \" ms\")\n\ntic = time.time()\nfor i in range(1000000):\n c += a[i] * b[i]\ntac = time.time()\n\ndt2 = 1000*(tac-tic)\nprint(\"Versão loop for: \" + str((dt2)) + \" ms\")\nprint(\"A versão vetorizada é \" + str((dt2/dt1)) + \"x mais rápida!\")",
"[1 2 3 4]\nVersão vetorizada: 1.85608863831 ms\nVersão loop for: 630.362033844 ms\nA versão vetorizada é 339.61849711x mais rápida!\n"
]
],
[
[
"Agora vamos ao segundo exemplo. Dado um vetor de 100000 elementos, vamos executar os seguintes passos\n- aplicar as funções exponencial e logaritmo de cada elemento do vetor em um loop for\n- aplicar as funções exponencial, logaritmo e módulo de cada elemento do vetor utilizando vetorização",
"_____no_output_____"
]
],
[
[
"import math\nn = 100000\n\nv = np.random.rand(n)\nu = np.zeros(n)\n\ntic = time.time()\nfor i in range(n):\n math.exp(v[i])\n math.log(v[i])\n \ntac = time.time()\ndt1 = 1000*(tac-tic)\n\nprint(\"Versão loop for: \" + str((dt1)) + \" ms\")\n\n\ntic = time.time()\nu = np.exp(v)\nu = np.log(v)\nu = np.abs(v)\ntac = time.time()\ndt2 = 1000*(tac-tic)\nprint(\"Versão vetorizada: \" + str((dt2)) + \" ms\")\nprint(\"A versão vetorizada é \" + str((dt1/dt2)) + \"x mais rápida\")",
"Versão loop for: 72.8318691254 ms\nVersão vetorizada: 6.02197647095 ms\nA versão vetorizada é 12.0943463457x mais rápida\n"
]
],
[
[
"Espero que tenha ficado claro a importancia de utilizar a vetorização em python. Agora, você tem todas as ferramentas básicas para iniciar o seu trabalho. Boa sorte :)",
"_____no_output_____"
],
[
"# Créditos\n-----------------------------------------------------------------------------------------------------------------------------\n* @author Thales Valente\n* @contact: [email protected]\n* @date last version: 08/2019.\n* @version 1.0\n* @github link: https://github.com/thalesvalente/machinelearning/\n*\n* @Copyright/License\n* Educational Material\n* All the products under this license are free software: they can be used for both academic and commercial purposes at absolutely no cost.\n* There are no paperwork, no royalties, no GNU-like \"copyleft\" restrictions, either. Just download and use it.\n* They are licensed under the terms of the MIT license reproduced below, and so are compatible with GPL and also qualifies as Open Source software.\n* They are not in the public domain. The legal details are below.\n*\n* The spirit of this license is that you are free to use this material for any purpose at no cost without having to ask us.\n* The only requirement is that if you do use them, then you should give us credit by including the copyright notice below somewhere in your material.\n*\n* Copyright © 2019 Educational Material\n*\n* Permission is hereby granted, free of charge, to any person obtaining a copy of this material without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sub license, and/or sell copies of this Material, and to permit persons to whom the Material is furnished to do so, subject to the following conditions:\n*\n* The above copyright notice and this permission notice shall be included in all copies or suavlantial portions of the material.\n*\n* THE MATERIAL IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE MATERIAL OR THE USE OR OTHER DEALINGS IN THE MATERIAL.\n-----------------------------------------------------------------------------------------------------------------------------\n-----------------------------------------------------------------------------------------------------------------------------",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecb9c4224a422632b40de74bcf52deecffb409ad | 13,701 | ipynb | Jupyter Notebook | examples/bindingdb_deepdta/tutorial.ipynb | SheffieldAI/pykale | be7670941fb06835883c80477b26702d407017db | [
"MIT"
] | null | null | null | examples/bindingdb_deepdta/tutorial.ipynb | SheffieldAI/pykale | be7670941fb06835883c80477b26702d407017db | [
"MIT"
] | null | null | null | examples/bindingdb_deepdta/tutorial.ipynb | SheffieldAI/pykale | be7670941fb06835883c80477b26702d407017db | [
"MIT"
] | null | null | null | 36.342175 | 351 | 0.559302 | [
[
[
"# PyKale Tutorial: Drug-Target Interaction Prediction using DeepDTA\n\n| [Open In Colab](https://colab.research.google.com/github/pykale/pykale/blob/main/examples/bindingdb_deepdta/tutorial.ipynb) (click `Runtime` → `Run all (Ctrl+F9)` |\n\nIf using [Google Colab](https://colab.research.google.com), a free GPU can be enabled to save time via setting `Runtime` → `Change runtime type` → `Hardware accelerator: GPU`",
"_____no_output_____"
],
[
"## Introduction\nDrug-target interaction prediction is an important research area in the field of drug discovery. It refers to predicting the binding affinity between the given chemical compounds and protein targets. In this example we train a standard DeepDTA model as a baseline in BindingDB, a public, web-accessible dataset of measured binding affinities.\n\n### DeepDTA\n[DeepDTA](https://academic.oup.com/bioinformatics/article/34/17/i821/5093245) is the modeling of protein sequences and compound 1D representations with convolutional neural networks (CNNs). The whole architecture of DeepDTA is shown below.\n\n\n\n### Datasets\nWe construct **three datasets** from BindingDB distinguished by different affinity measurement metrics\n(**Kd, IC50 and Ki**). They are acquired from [Therapeutics Data Commons](https://tdcommons.ai/) (TDC), which is a collection of machine learning tasks spreading across different domains of therapeutics. The data statistics is shown below:\n\n| Metrics | Drugs | Targets | Pairs |\n| :----: | :----: | :----: | :----: |\n| Kd | 10,655 | 1,413 | 52,284 |\n| IC50 | 549,205 | 5,078 | 991,486 |\n| Ki | 174,662 | 3,070 | 375,032 |\n\nThis figure is the binding affinity distribution for the three datasets respectively, where the metric values (x-axis) have been transformed into log space.\n\nThis tutorial uses the (smallest) **Kd** dataset.\n\n## Setup\n\nThe first few blocks of code are necessary to set up the notebook execution environment and import the required modules, including PyKale.\n\nThis checks if the notebook is running on Google Colab and installs required packages.",
"_____no_output_____"
]
],
[
[
"if 'google.colab' in str(get_ipython()):\n print('Running on CoLab')\n !pip uninstall --yes imgaug && pip uninstall --yes albumentations && pip install git+https://github.com/aleju/imgaug.git\n !pip install rdkit-pypi torchaudio torch-scatter torch-sparse torch-cluster torch-spline-conv torch-geometric -f https://data.pyg.org/whl/torch-1.10.0+cu113.html \n !pip install git+https://github.com/pykale/pykale.git \n\n !git clone https://github.com/pykale/pykale.git\n %cd pykale/examples/bindingdb_deepdta\nelse:\n print('Not running on CoLab')",
"_____no_output_____"
]
],
[
[
"This imports required modules.",
"_____no_output_____"
]
],
[
[
"import pytorch_lightning as pl\nimport torch\nfrom config import get_cfg_defaults\nfrom model import get_model\nfrom pytorch_lightning.callbacks import ModelCheckpoint\nfrom pytorch_lightning.loggers import TensorBoardLogger\nfrom torch.utils.data import DataLoader, Subset\n\nfrom kale.loaddata.tdc_datasets import BindingDBDataset\nfrom kale.utils.seed import set_seed",
"_____no_output_____"
]
],
[
[
"## Configuration\n\nThe customized configuration used in this tutorial is stored in `./configs/tutorial.yaml`, this file overwrites defaults in `config.py` where a value is specified.\n\nFor saving time to run a whole pipeline in this tutorial, we sample small train/valid/test (8,000/1,000/1,000) subsets from the original BindingDB dataset.",
"_____no_output_____"
]
],
[
[
"cfg_path = \"./configs/tutorial.yaml\"\ntrain_subset_size, valid_subset_size, test_subset_size = 8000, 1000, 1000\n\ncfg = get_cfg_defaults()\ncfg.merge_from_file(cfg_path)\ncfg.freeze()\nprint(cfg)\n\nset_seed(cfg.SOLVER.SEED)",
"_____no_output_____"
]
],
[
[
"## Check if a GPU is available\n\nIf a CUDA GPU is available, this should be used to accelerate the training process. The code below checks and reports on this.\n",
"_____no_output_____"
]
],
[
[
"device = \"cuda\" if torch.cuda.is_available() else \"cpu\"\nprint(\"Using: \" + device)\ngpus = 1 if device == \"cuda\" else 0",
"_____no_output_____"
]
],
[
[
"## Select Datasets\n\nSource and target datasets are specified using the `BindingDBDataset()` function and loaded using the `DataLoader()` function.",
"_____no_output_____"
]
],
[
[
"train_dataset = BindingDBDataset(name=cfg.DATASET.NAME, split=\"train\", path=cfg.DATASET.PATH)\nvalid_dataset = BindingDBDataset(name=cfg.DATASET.NAME, split=\"valid\", path=cfg.DATASET.PATH)\ntest_dataset = BindingDBDataset(name=cfg.DATASET.NAME, split=\"test\", path=cfg.DATASET.PATH)\ntrain_size, valid_size, test_size = len(train_dataset), len(valid_dataset), len(test_dataset)\ntrain_sample_indices, valid_sample_indices, test_sample_indices = torch.randperm(train_size)[:train_subset_size].tolist(), torch.randperm(valid_size)[:valid_subset_size].tolist(), torch.randperm(test_size)[:test_subset_size].tolist()\ntrain_dataset, valid_dataset, test_dataset = Subset(train_dataset, train_sample_indices), Subset(valid_dataset, valid_sample_indices), Subset(test_dataset, test_sample_indices)",
"_____no_output_____"
],
[
"cfg.DATASET.PATH",
"_____no_output_____"
],
[
"train_loader = DataLoader(dataset=train_dataset, shuffle=True, batch_size=cfg.SOLVER.TRAIN_BATCH_SIZE)\nvalid_loader = DataLoader(dataset=valid_dataset, shuffle=True, batch_size=cfg.SOLVER.TEST_BATCH_SIZE)\ntest_loader = DataLoader(dataset=test_dataset, shuffle=True, batch_size=cfg.SOLVER.TEST_BATCH_SIZE)",
"_____no_output_____"
]
],
[
[
"## Setup model\n\nHere, we use the previously defined configuration and dataset to set up the model we will subsequently train.",
"_____no_output_____"
]
],
[
[
"model = get_model(cfg)",
"_____no_output_____"
]
],
[
[
"## Setup Logger\n\nA logger is used to store output generated during and after model training. This information can be used to assess the effectiveness of the training and to identify problems.",
"_____no_output_____"
]
],
[
[
"tb_logger = TensorBoardLogger(\"tb_logs\", name=cfg.DATASET.NAME)",
"_____no_output_____"
]
],
[
[
"## Setup Trainer\n\nA trainer object is used to determine and store model parameters. Here, one is configured with information on how a model should be trained, and what hardware will be used.",
"_____no_output_____"
]
],
[
[
"checkpoint_callback = ModelCheckpoint(monitor=\"valid_loss\", mode=\"min\")\ntrainer = pl.Trainer(min_epochs=cfg.SOLVER.MIN_EPOCHS, \n max_epochs=cfg.SOLVER.MAX_EPOCHS, \n gpus=gpus, logger=tb_logger, \n callbacks=[checkpoint_callback])",
"_____no_output_____"
]
],
[
[
"## Train Model\n\nOptimize model parameters using the trainer.",
"_____no_output_____"
]
],
[
[
"%time trainer.fit(model, train_dataloader=train_loader, val_dataloaders=valid_loader)",
"_____no_output_____"
]
],
[
[
"## Test Optimized Model\n\nCheck performance of model optimized with training data against test data which was not used in training.",
"_____no_output_____"
]
],
[
[
"trainer.test(test_dataloaders=test_loader)",
"_____no_output_____"
]
],
[
[
"You should get a test loss of $7.3\\cdots$ in root mean square error (RMSE). The target value ($y$) has a range of [-13, 20] (in log space). Thus, with only three epochs, we have learned to predict the target value with an RMSE of 7.3 over a range of [-13, 20].\n\nWe set the maximum epochs to 3 and extract a subset (8000/1000/1000) to save time in running this tutorial. You may change these settings. Setting the max epochs to 100 and using the full dataset will get a much better result (<1).",
"_____no_output_____"
],
[
"## Architecture\nBelow is the architecture of DeepDTA with default hyperparameters settings.\n\n<pre>\n==========================================================================================\nLayer (type:depth-idx) Output Shape Param #\n==========================================================================================\n├─CNNEncoder: 1-1 [256, 96] --\n| └─Embedding: 2-1 [256, 85, 128] 8,320\n| └─Conv1d: 2-2 [256, 32, 121] 21,792\n| └─Conv1d: 2-3 [256, 64, 114] 16,448\n| └─Conv1d: 2-4 [256, 96, 107] 49,248\n| └─AdaptiveMaxPool1d: 2-5 [256, 96, 1] --\n├─CNNEncoder: 1-2 [256, 96] --\n| └─Embedding: 2-6 [256, 1200, 128] 3,328\n| └─Conv1d: 2-7 [256, 32, 121] 307,232\n| └─Conv1d: 2-8 [256, 64, 114] 16,448\n| └─Conv1d: 2-9 [256, 96, 107] 49,248\n| └─AdaptiveMaxPool1d: 2-10 [256, 96, 1] --\n├─MLPDecoder: 1-3 [256, 1] --\n| └─Linear: 2-11 [256, 1024] 197,632\n| └─Dropout: 2-12 [256, 1024] --\n| └─Linear: 2-13 [256, 1024] 1,049,600\n| └─Dropout: 2-14 [256, 1024] --\n| └─Linear: 2-15 [256, 512] 524,800\n| └─Linear: 2-16 [256, 1] 513\n==========================================================================================\nTotal params: 2,244,609\nTrainable params: 2,244,609\nNon-trainable params: 0\nTotal mult-adds (M): 58.08\n==========================================================================================\nInput size (MB): 1.32\nForward/backward pass size (MB): 429.92\nParams size (MB): 8.98\nEstimated Total Size (MB): 440.21",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecb9d409706b4af3a05de65d68387ffe354a7000 | 2,850 | ipynb | Jupyter Notebook | CV/OpenCV/Learn_OpenCV_in_3_hours/tutorial/CHAPTER3.Resizing And Cropping.ipynb | tsuirak/skills | 22280be0870627c5dd84e069ec271aeeb6797831 | [
"MIT"
] | 362 | 2020-10-08T07:34:25.000Z | 2022-03-30T05:11:30.000Z | OpenCV/Learn_OpenCV_in_3_hours/tutorial/CHAPTER3.Resizing And Cropping.ipynb | fengxingying/skills | 38c437b0040a5353dc30d2ae418d6ca77a2a85cb | [
"MIT"
] | null | null | null | OpenCV/Learn_OpenCV_in_3_hours/tutorial/CHAPTER3.Resizing And Cropping.ipynb | fengxingying/skills | 38c437b0040a5353dc30d2ae418d6ca77a2a85cb | [
"MIT"
] | 238 | 2020-10-08T12:01:31.000Z | 2022-03-25T08:10:42.000Z | 16.964286 | 49 | 0.453684 | [
[
[
"# 笔记\n\n\n**OPENCV图像的规则**\n",
"_____no_output_____"
]
],
[
[
"import cv2",
"_____no_output_____"
]
],
[
[
"### Resizing",
"_____no_output_____"
]
],
[
[
"img=cv2.imread('./images/lambo.png')\nprint(img.shape)\n\ncv2.imshow(\"Image\",img)\ncv2.waitKey(0)\n\n# (高,宽,通道bgr)",
"(462, 623, 3)\n"
],
[
"# 调整大小\n\n# (宽,高)\nimgResize=cv2.resize(img,(300,200))\ncv2.imshow(\"Image Resize\",imgResize)\ncv2.waitKey(0)\nprint(imgResize.shape)",
"(200, 300, 3)\n"
],
[
"imgResize=cv2.resize(img,(1000,500))\ncv2.imshow(\"Image Resize\",imgResize)\ncv2.waitKey(0)\nprint(imgResize.shape)",
"(500, 1000, 3)\n"
]
],
[
[
"### Cropping ",
"_____no_output_____"
]
],
[
[
"# 裁剪\n\n# (高,宽)\nimgCropped=img[0:200,200:500]\ncv2.imshow(\"Image Cropped\",imgCropped)\ncv2.waitKey(0)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb9d7a4674c031fab37e4c38138f4af5763e8e7 | 30,840 | ipynb | Jupyter Notebook | _posts/python-v3/fundamentals/subplots/subplots.ipynb | bmb804/documentation | 57826d25e0afea7fff6a8da9abab8be2f7a4b48c | [
"CC-BY-3.0"
] | null | null | null | _posts/python-v3/fundamentals/subplots/subplots.ipynb | bmb804/documentation | 57826d25e0afea7fff6a8da9abab8be2f7a4b48c | [
"CC-BY-3.0"
] | null | null | null | _posts/python-v3/fundamentals/subplots/subplots.ipynb | bmb804/documentation | 57826d25e0afea7fff6a8da9abab8be2f7a4b48c | [
"CC-BY-3.0"
] | null | null | null | 28.930582 | 358 | 0.490435 | [
[
[
"#### New to Plotly?\nPlotly's Python library is free and open source! [Get started](https://plot.ly/python/getting-started/) by downloading the client and [reading the primer](https://plot.ly/python/getting-started/).\n<br>You can set up Plotly to work in [online](https://plot.ly/python/getting-started/#initialization-for-online-plotting) or [offline](https://plot.ly/python/getting-started/#initialization-for-offline-plotting) mode, or in [jupyter notebooks](https://plot.ly/python/getting-started/#start-plotting-online).\n<br>We also have a quick-reference [cheatsheet](https://images.plot.ly/plotly-documentation/images/python_cheat_sheet.pdf) (new!) to help you get started!",
"_____no_output_____"
],
[
"#### Simple Subplot",
"_____no_output_____"
]
],
[
[
"from plotly import tools\nimport plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(\n x=[1, 2, 3],\n y=[4, 5, 6],\n mode='markers+text',\n text=['Text A', 'Text B', 'Text C'],\n textposition='bottom center'\n)\ntrace2 = go.Scatter(\n x=[20, 30, 40],\n y=[50, 60, 70],\n mode='markers+text',\n text=['Text D', 'Text E', 'Text F'],\n textposition='bottom center'\n)\n\nfig = tools.make_subplots(rows=1, cols=2)\n\nfig.append_trace(trace1, 1, 1)\nfig.append_trace(trace2, 1, 2)\n\nfig['layout'].update(height=600, width=800, title='i <3 annotations and subplots')\npy.iplot(fig, filename='simple-subplot-with-annotations')",
"This is the format of your plot grid:\n[ (1,1) x1,y1 ] [ (1,2) x2,y2 ]\n\n"
]
],
[
[
"#### Multiple Subplots",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(\n x=[1, 2, 3],\n y=[4, 5, 6]\n)\ntrace2 = go.Scatter(\n x=[20, 30, 40],\n y=[50, 60, 70],\n xaxis='x2',\n yaxis='y2'\n)\ntrace3 = go.Scatter(\n x=[300, 400, 500],\n y=[600, 700, 800],\n xaxis='x3',\n yaxis='y3'\n)\ntrace4 = go.Scatter(\n x=[4000, 5000, 6000],\n y=[7000, 8000, 9000],\n xaxis='x4',\n yaxis='y4'\n)\ndata = [trace1, trace2, trace3, trace4]\nlayout = go.Layout(\n xaxis=dict(\n domain=[0, 0.45]\n ),\n yaxis=dict(\n domain=[0, 0.45]\n ),\n xaxis2=dict(\n domain=[0.55, 1]\n ),\n xaxis3=dict(\n domain=[0, 0.45],\n anchor='y3'\n ),\n xaxis4=dict(\n domain=[0.55, 1],\n anchor='y4'\n ),\n yaxis2=dict(\n domain=[0, 0.45],\n anchor='x2'\n ),\n yaxis3=dict(\n domain=[0.55, 1]\n ),\n yaxis4=dict(\n domain=[0.55, 1],\n anchor='x4'\n )\n)\nfig = go.Figure(data=data, layout=layout)\npy.iplot(fig, filename='multiple-subplots')",
"_____no_output_____"
]
],
[
[
"#### Multiple Subplots with Titles",
"_____no_output_____"
]
],
[
[
"from plotly import tools\nimport plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(x=[1, 2, 3], y=[4, 5, 6])\ntrace2 = go.Scatter(x=[20, 30, 40], y=[50, 60, 70])\ntrace3 = go.Scatter(x=[300, 400, 500], y=[600, 700, 800])\ntrace4 = go.Scatter(x=[4000, 5000, 6000], y=[7000, 8000, 9000])\n\nfig = tools.make_subplots(rows=2, cols=2, subplot_titles=('Plot 1', 'Plot 2',\n 'Plot 3', 'Plot 4'))\n\nfig.append_trace(trace1, 1, 1)\nfig.append_trace(trace2, 1, 2)\nfig.append_trace(trace3, 2, 1)\nfig.append_trace(trace4, 2, 2)\n\nfig['layout'].update(height=600, width=600, title='Multiple Subplots' +\n ' with Titles')\n\npy.iplot(fig, filename='make-subplots-multiple-with-titles')",
"This is the format of your plot grid:\n[ (1,1) x1,y1 ] [ (1,2) x2,y2 ]\n[ (2,1) x3,y3 ] [ (2,2) x4,y4 ]\n\n"
]
],
[
[
"#### Simple Subplot with Annotations",
"_____no_output_____"
]
],
[
[
"from plotly import tools\nimport plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(\n x=[1, 2, 3],\n y=[4, 5, 6],\n mode='markers+text',\n text=['Text A', 'Text B', 'Text C'],\n textposition='bottom center'\n)\ntrace2 = go.Scatter(\n x=[20, 30, 40],\n y=[50, 60, 70],\n mode='markers+text',\n text=['Text D', 'Text E', 'Text F'],\n textposition='bottom center'\n)\n\nfig = tools.make_subplots(rows=1, cols=2)\n\nfig.append_trace(trace1, 1, 1)\nfig.append_trace(trace2, 1, 2)\n\nfig['layout'].update(height=600, width=800, title='i <3 annotations and subplots')\npy.iplot(fig, filename='simple-subplot-with-annotations')",
"This is the format of your plot grid:\n[ (1,1) x1,y1 ] [ (1,2) x2,y2 ]\n\n"
]
],
[
[
"#### Side by Side Subplot",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(\n x=[1, 2, 3],\n y=[4, 5, 6]\n)\ntrace2 = go.Scatter(\n x=[20, 30, 40],\n y=[50, 60, 70],\n xaxis='x2',\n yaxis='y2'\n)\ndata = [trace1, trace2]\nlayout = go.Layout(\n xaxis=dict(\n domain=[0, 0.7]\n ),\n xaxis2=dict(\n domain=[0.8, 1]\n ),\n yaxis2=dict(\n anchor='x2'\n )\n)\nfig = go.Figure(data=data, layout=layout)\npy.iplot(fig, filename='side-by-side-subplot')",
"_____no_output_____"
]
],
[
[
"#### Customizing Subplot Axes",
"_____no_output_____"
]
],
[
[
"from plotly import tools\nimport plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(x=[1, 2, 3], y=[4, 5, 6])\ntrace2 = go.Scatter(x=[20, 30, 40], y=[50, 60, 70])\ntrace3 = go.Scatter(x=[300, 400, 500], y=[600, 700, 800])\ntrace4 = go.Scatter(x=[4000, 5000, 6000], y=[7000, 8000, 9000])\n\nfig = tools.make_subplots(rows=2, cols=2, subplot_titles=('Plot 1', 'Plot 2',\n 'Plot 3', 'Plot 4'))\nfig.append_trace(trace1, 1, 1)\nfig.append_trace(trace2, 1, 2)\nfig.append_trace(trace3, 2, 1)\nfig.append_trace(trace4, 2, 2)\n\nfig['layout']['xaxis1'].update(title='xaxis 1 title')\nfig['layout']['xaxis2'].update(title='xaxis 2 title', range=[10, 50])\nfig['layout']['xaxis3'].update(title='xaxis 3 title', showgrid=False)\nfig['layout']['xaxis4'].update(title='xaxis 4 title', type='log')\n\nfig['layout']['yaxis1'].update(title='yaxis 1 title')\nfig['layout']['yaxis2'].update(title='yaxis 2 title', range=[40, 80])\nfig['layout']['yaxis3'].update(title='yaxis 3 title', showgrid=False)\nfig['layout']['yaxis4'].update(title='yaxis 4 title')\n\nfig['layout'].update(title='Customizing Subplot Axes')\n\npy.iplot(fig, filename='customizing-subplot-axes')",
"This is the format of your plot grid:\n[ (1,1) x1,y1 ] [ (1,2) x2,y2 ]\n[ (2,1) x3,y3 ] [ (2,2) x4,y4 ]\n\n"
]
],
[
[
"#### Subplots with Shared X-Axes",
"_____no_output_____"
]
],
[
[
"from plotly import tools\nimport plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(\n x=[0, 1, 2],\n y=[10, 11, 12]\n)\ntrace2 = go.Scatter(\n x=[2, 3, 4],\n y=[100, 110, 120],\n)\ntrace3 = go.Scatter(\n x=[3, 4, 5],\n y=[1000, 1100, 1200],\n)\nfig = tools.make_subplots(rows=3, cols=1, specs=[[{}], [{}], [{}]],\n shared_xaxes=True, shared_yaxes=True,\n vertical_spacing=0.001)\nfig.append_trace(trace1, 3, 1)\nfig.append_trace(trace2, 2, 1)\nfig.append_trace(trace3, 1, 1)\n\nfig['layout'].update(height=600, width=600, title='Stacked Subplots with Shared X-Axes')\npy.iplot(fig, filename='stacked-subplots-shared-xaxes')",
"This is the format of your plot grid:\n[ (1,1) x1,y1 ]\n[ (2,1) x1,y2 ]\n[ (3,1) x1,y3 ]\n\n"
]
],
[
[
"#### Subplots with Shared Y-Axes",
"_____no_output_____"
]
],
[
[
"from plotly import tools\nimport plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace0 = go.Scatter(\n x=[1, 2, 3],\n y=[2, 3, 4]\n)\ntrace1 = go.Scatter(\n x=[20, 30, 40],\n y=[5, 5, 5],\n)\ntrace2 = go.Scatter(\n x=[2, 3, 4],\n y=[600, 700, 800],\n)\ntrace3 = go.Scatter(\n x=[4000, 5000, 6000],\n y=[7000, 8000, 9000],\n)\n\nfig = tools.make_subplots(rows=2, cols=2, shared_yaxes=True)\n\nfig.append_trace(trace0, 1, 1)\nfig.append_trace(trace1, 1, 2)\nfig.append_trace(trace2, 2, 1)\nfig.append_trace(trace3, 2, 2)\n\nfig['layout'].update(height=600, width=600,\n title='Multiple Subplots with Shared Y-Axes')\npy.iplot(fig, filename='multiple-subplots-shared-yaxes')",
"This is the format of your plot grid:\n[ (1,1) x1,y1 ] [ (1,2) x2,y1 ]\n[ (2,1) x3,y2 ] [ (2,2) x4,y2 ]\n\n"
]
],
[
[
"#### Subplots with Shared Axes",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(\n x=[1, 2, 3],\n y=[2, 3, 4]\n)\ntrace2 = go.Scatter(\n x=[20, 30, 40],\n y=[5, 5, 5],\n xaxis='x2',\n yaxis='y'\n)\ntrace3 = go.Scatter(\n x=[2, 3, 4],\n y=[600, 700, 800],\n xaxis='x',\n yaxis='y3'\n)\ntrace4 = go.Scatter(\n x=[4000, 5000, 6000],\n y=[7000, 8000, 9000],\n xaxis='x4',\n yaxis='y4'\n)\ndata = [trace1, trace2, trace3, trace4]\nlayout = go.Layout(\n xaxis=dict(\n domain=[0, 0.45]\n ),\n yaxis=dict(\n domain=[0, 0.45]\n ),\n xaxis2=dict(\n domain=[0.55, 1]\n ),\n xaxis4=dict(\n domain=[0.55, 1],\n anchor='y4'\n ),\n yaxis3=dict(\n domain=[0.55, 1]\n ),\n yaxis4=dict(\n domain=[0.55, 1],\n anchor='x4'\n )\n)\nfig = go.Figure(data=data, layout=layout)\npy.iplot(fig, filename='shared-axes-subplots')",
"_____no_output_____"
]
],
[
[
"#### Stacked Subplots",
"_____no_output_____"
]
],
[
[
"from plotly import tools\nimport plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(\n x=[0, 1, 2],\n y=[10, 11, 12]\n)\ntrace2 = go.Scatter(\n x=[2, 3, 4],\n y=[100, 110, 120],\n)\ntrace3 = go.Scatter(\n x=[3, 4, 5],\n y=[1000, 1100, 1200],\n)\n\nfig = tools.make_subplots(rows=3, cols=1)\n\nfig.append_trace(trace3, 1, 1)\nfig.append_trace(trace2, 2, 1)\nfig.append_trace(trace1, 3, 1)\n\n\nfig['layout'].update(height=600, width=600, title='Stacked subplots')\npy.iplot(fig, filename='stacked-subplots')",
"This is the format of your plot grid:\n[ (1,1) x1,y1 ]\n[ (2,1) x2,y2 ]\n[ (3,1) x3,y3 ]\n\n"
]
],
[
[
"#### Stacked Subplots with a Shared X-Axis",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(\n x=[0, 1, 2],\n y=[10, 11, 12]\n)\ntrace2 = go.Scatter(\n x=[2, 3, 4],\n y=[100, 110, 120],\n yaxis='y2'\n)\ntrace3 = go.Scatter(\n x=[3, 4, 5],\n y=[1000, 1100, 1200],\n yaxis='y3'\n)\ndata = [trace1, trace2, trace3]\nlayout = go.Layout(\n yaxis=dict(\n domain=[0, 0.33]\n ),\n legend=dict(\n traceorder='reversed'\n ),\n yaxis2=dict(\n domain=[0.33, 0.66]\n ),\n yaxis3=dict(\n domain=[0.66, 1]\n )\n)\nfig = go.Figure(data=data, layout=layout)\npy.iplot(fig, filename='stacked-subplots-shared-x-axis')",
"_____no_output_____"
]
],
[
[
"#### Custom Sized Subplot with Subplot Titles",
"_____no_output_____"
]
],
[
[
"from plotly import tools\nimport plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace0 = go.Scatter(\n x=[1, 2],\n y=[1, 2]\n)\ntrace1 = go.Scatter(\n x=[1, 2],\n y=[1, 2]\n)\ntrace2 = go.Scatter(\n x=[1, 2, 3],\n y=[2, 1, 2]\n)\nfig = tools.make_subplots(rows=2, cols=2, specs=[[{}, {}], [{'colspan': 2}, None]],\n subplot_titles=('First Subplot','Second Subplot', 'Third Subplot'))\n\nfig.append_trace(trace0, 1, 1)\nfig.append_trace(trace1, 1, 2)\nfig.append_trace(trace2, 2, 1)\n\nfig['layout'].update(showlegend=False, title='Specs with Subplot Title')\npy.iplot(fig, filename='custom-sized-subplot-with-subplot-titles')",
"This is the format of your plot grid:\n[ (1,1) x1,y1 ] [ (1,2) x2,y2 ]\n[ (2,1) x3,y3 - ]\n\n"
]
],
[
[
"#### Multiple Custom Sized Subplots",
"_____no_output_____"
]
],
[
[
"from plotly import tools\nimport plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(x=[1, 2], y=[1, 2], name='(1,1)')\ntrace2 = go.Scatter(x=[1, 2], y=[1, 2], name='(1,2)')\ntrace3 = go.Scatter(x=[1, 2], y=[1, 2], name='(2,1)')\ntrace4 = go.Scatter(x=[1, 2], y=[1, 2], name='(3,1)')\ntrace5 = go.Scatter(x=[1, 2], y=[1, 2], name='(5,1)')\ntrace6 = go.Scatter(x=[1, 2], y=[1, 2], name='(5,2)')\n\nfig = tools.make_subplots(rows=5, cols=2,\n specs=[[{}, {'rowspan': 2}],\n [{}, None],\n [{'rowspan': 2, 'colspan': 2}, None],\n [None, None],\n [{}, {}]],\n print_grid=True)\n\nfig.append_trace(trace1, 1, 1)\nfig.append_trace(trace2, 1, 2)\nfig.append_trace(trace3, 2, 1)\nfig.append_trace(trace4, 3, 1)\nfig.append_trace(trace5, 5, 1)\nfig.append_trace(trace6, 5, 2)\n\nfig['layout'].update(height=600, width=600, title='specs examples')\npy.iplot(fig, filename='multiple-custom-sized-subplots')",
"This is the format of your plot grid:\n[ (1,1) x1,y1 ] [ (1,2) x2,y2 ]\n[ (2,1) x3,y3 ] | \n[ (3,1) x4,y4 - ]\n | | \n[ (5,1) x5,y5 ] [ (5,2) x6,y6 ]\n\n"
]
],
[
[
"### Dash Example",
"_____no_output_____"
],
[
"[Dash](https://plot.ly/products/dash/) is an Open Source Python library which can help you convert plotly figures into a reactive, web-based application. Below is a simple example of a dashboard created using Dash. Its [source code](https://github.com/plotly/simple-example-chart-apps/tree/master/dash-multiplesubplot) can easily be deployed to a PaaS.",
"_____no_output_____"
]
],
[
[
"from IPython.display import IFrame\nIFrame(src= \"https://dash-simple-apps.plotly.host/dash-multiplesubplot/\", width=\"100%\", height=\"950px\", frameBorder=\"0\")",
"_____no_output_____"
],
[
"from IPython.display import IFrame\nIFrame(src= \"https://dash-simple-apps.plotly.host/dash-multiplesubplot/code\", width=\"100%\", height=500, frameBorder=\"0\")",
"_____no_output_____"
]
],
[
[
"#### Reference\nAll of the x-axis properties are found here: https://plot.ly/python/reference/#XAxis\nAll of the y-axis properties are found here: https://plot.ly/python/reference/#YAxis",
"_____no_output_____"
]
],
[
[
"from IPython.display import display, HTML\n\ndisplay(HTML('<link href=\"//fonts.googleapis.com/css?family=Open+Sans:600,400,300,200|Inconsolata|Ubuntu+Mono:400,700\" rel=\"stylesheet\" type=\"text/css\" />'))\ndisplay(HTML('<link rel=\"stylesheet\" type=\"text/css\" href=\"http://help.plot.ly/documentation/all_static/css/ipython-notebook-custom.css\">'))\n\n! pip install git+https://github.com/plotly/publisher.git --upgrade\nimport publisher\npublisher.publish(\n 'subplots.ipynb', 'python/subplots/', 'Subplots | plotly',\n 'How to make subplots in python. Examples of stacked, custom-sized, gridded, and annotated subplts.',\n title = 'Python Subplots | Examples | Plotly',\n name = 'Subplots', has_thumbnail='true', thumbnail='thumbnail/subplots.jpg', \n language='python', page_type='example_index', redirect_from='ipython-notebooks/subplots/', \n display_as='file_settings', order=15,\n ipynb='~notebook_demo/269')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecb9d81907b5f6e9e659f3066e1aca2f4d6dfe04 | 17,177 | ipynb | Jupyter Notebook | docs/notebooks/Writing_custom_interpreters_in_Jax.ipynb | cdfreeman-google/jax | ca6f8186a36a8962845289ffc6baed3e96390f68 | [
"ECL-2.0",
"Apache-2.0"
] | 14 | 2021-04-24T03:26:39.000Z | 2022-01-28T14:25:13.000Z | docs/notebooks/Writing_custom_interpreters_in_Jax.ipynb | cdfreeman-google/jax | ca6f8186a36a8962845289ffc6baed3e96390f68 | [
"ECL-2.0",
"Apache-2.0"
] | 2 | 2021-06-08T22:19:08.000Z | 2021-06-08T22:19:49.000Z | docs/notebooks/Writing_custom_interpreters_in_Jax.ipynb | cdfreeman-google/jax | ca6f8186a36a8962845289ffc6baed3e96390f68 | [
"ECL-2.0",
"Apache-2.0"
] | null | null | null | 30.94955 | 420 | 0.598824 | [
[
[
"# Writing custom Jaxpr interpreters in JAX\n\n[](https://colab.research.google.com/github/google/jax/blob/master/docs/notebooks/Writing_custom_interpreters_in_Jax.ipynb)",
"_____no_output_____"
],
[
"JAX offers several composable function transformations (`jit`, `grad`, `vmap`,\netc.) that enable writing concise, accelerated code. \n\nHere we show how to add your own function transformations to the system, by writing a custom Jaxpr interpreter. And we'll get composability with all the other transformations for free.\n\n**This example uses internal JAX APIs, which may break at any time. Anything not in [the API Documentation](https://jax.readthedocs.io/en/latest/jax.html) should be assumed internal.**",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport jax\nimport jax.numpy as jnp\nfrom jax import jit, grad, vmap\nfrom jax import random",
"_____no_output_____"
]
],
[
[
"## What is JAX doing?",
"_____no_output_____"
],
[
"JAX provides a NumPy-like API for numerical computing which can be used as is, but JAX's true power comes from composable function transformations. Take the `jit` function transformation, which takes in a function and returns a semantically identical function but is lazily compiled by XLA for accelerators.",
"_____no_output_____"
]
],
[
[
"x = random.normal(random.PRNGKey(0), (5000, 5000))\ndef f(w, b, x):\n return jnp.tanh(jnp.dot(x, w) + b)\nfast_f = jit(f)",
"_____no_output_____"
]
],
[
[
"When we call `fast_f`, what happens? JAX traces the function and constructs an XLA computation graph. The graph is then JIT-compiled and executed. Other transformations work similarly in that they first trace the function and handle the output trace in some way. To learn more about Jax's tracing machinery, you can refer to the [\"How it works\"](https://github.com/google/jax#how-it-works) section in the README.",
"_____no_output_____"
],
[
"## Jaxpr tracer\n\nA tracer of special importance in Jax is the Jaxpr tracer, which records ops into a Jaxpr (Jax expression). A Jaxpr is a data structure that can be evaluated like a mini functional programming language and \nthus Jaxprs are a useful intermediate representation\nfor function transformation.",
"_____no_output_____"
],
[
"To get a first look at Jaxprs, consider the `make_jaxpr` transformation. `make_jaxpr` is essentially a \"pretty-printing\" transformation:\nit transforms a function into one that, given example arguments, produces a Jaxpr representation of its computation.\nAlthough we can't generally use the Jaxprs that it returns, it is useful for debugging and introspection.\nLet's use it to look at how some example Jaxprs\nare structured.",
"_____no_output_____"
]
],
[
[
"def examine_jaxpr(closed_jaxpr):\n jaxpr = closed_jaxpr.jaxpr\n print(\"invars:\", jaxpr.invars)\n print(\"outvars:\", jaxpr.outvars)\n print(\"constvars:\", jaxpr.constvars)\n for eqn in jaxpr.eqns:\n print(\"equation:\", eqn.invars, eqn.primitive, eqn.outvars, eqn.params)\n print()\n print(\"jaxpr:\", jaxpr)\n\ndef foo(x):\n return x + 1\nprint(\"foo\")\nprint(\"=====\")\nexamine_jaxpr(jax.make_jaxpr(foo)(5))\n\nprint()\n\ndef bar(w, b, x):\n return jnp.dot(w, x) + b + jnp.ones(5), x\nprint(\"bar\")\nprint(\"=====\")\nexamine_jaxpr(jax.make_jaxpr(bar)(jnp.ones((5, 10)), jnp.ones(5), jnp.ones(10)))",
"_____no_output_____"
]
],
[
[
"* `jaxpr.invars` - the `invars` of a Jaxpr are a list of the input variables to Jaxpr, analogous to arguments in Python functions\n* `jaxpr.outvars` - the `outvars` of a Jaxpr are the variables that are returned by the Jaxpr. Every Jaxpr has multiple outputs.\n* `jaxpr.constvars` - the `constvars` are a list of variables that are also inputs to the Jaxpr, but correspond to constants from the trace (we'll go over these in more detail later)\n* `jaxpr.eqns` - a list of equations, which are essentially let-bindings. Each equation is list of input variables, a list of output variables, and a *primitive*, which is used to evaluate inputs to produce outputs. Each equation also has a `params`, a dictionary of parameters.\n\nAll together, a Jaxpr encapsulates a simple program that can be evaluated with inputs to produce an output. We'll go over how exactly to do this later. The important thing to note now is that a Jaxpr is a data structure that can be manipulated and evaluated in whatever way we want.",
"_____no_output_____"
],
[
"### Why are Jaxprs useful?",
"_____no_output_____"
],
[
"Jaxprs are simple program representations that are easy to transform. And because Jax lets us stage out Jaxprs from Python functions, it gives us a way to transform numerical programs written in Python.",
"_____no_output_____"
],
[
"## Your first interpreter: `invert`",
"_____no_output_____"
],
[
"Let's try to implement a simple function \"inverter\", which takes in the output of the original function and returns the inputs that produced those outputs. For now, let's focus on simple, unary functions which are composed of other invertible unary functions.\n\nGoal:\n```python\ndef f(x):\n return jnp.exp(jnp.tanh(x))\nf_inv = inverse(f)\nassert jnp.allclose(f_inv(f(1.0)), 1.0)\n```\n\nThe way we'll implement this is by (1) tracing `f` into a Jaxpr, then (2) interpreting the Jaxpr *backwards*. While interpreting the Jaxpr backwards, for each equation we'll look up the primitive's inverse in a table and apply it.\n\n### 1. Tracing a function\n\nWe can't use `make_jaxpr` for this, because we need to pull out constants created during the trace to pass into the Jaxpr. However, we can write a function that does something very similar to `make_jaxpr`.",
"_____no_output_____"
]
],
[
[
"# Importing Jax functions useful for tracing/interpreting.\nimport numpy as np\nfrom functools import wraps\n\nfrom jax import core\nfrom jax import lax\nfrom jax._src.util import safe_map",
"_____no_output_____"
]
],
[
[
"This function first flattens its arguments into a list, which are the abstracted and wrapped as partial values. The `jax.make_jaxpr` function is used to then trace a function into a Jaxpr\nfrom a list of partial value inputs.",
"_____no_output_____"
]
],
[
[
"def f(x):\n return jnp.exp(jnp.tanh(x))\n\nclosed_jaxpr = jax.make_jaxpr(f)(jnp.ones(5))\nprint(closed_jaxpr)\nprint(closed_jaxpr.literals)",
"_____no_output_____"
]
],
[
[
"### 2. Evaluating a Jaxpr\n\n\nBefore we write a custom Jaxpr interpreter, let's first implement the \"default\" interpreter, `eval_jaxpr`, which evaluates the Jaxpr as-is, computing the same values that the original, un-transformed Python function would. \n\nTo do this, we first create an environment to store the values for each of the variables, and update the environment with each equation we evaluate in the Jaxpr.",
"_____no_output_____"
]
],
[
[
"def eval_jaxpr(jaxpr, consts, *args):\n # Mapping from variable -> value\n env = {}\n \n def read(var):\n # Literals are values baked into the Jaxpr\n if type(var) is core.Literal:\n return var.val\n return env[var]\n\n def write(var, val):\n env[var] = val\n\n # Bind args and consts to environment\n write(core.unitvar, core.unit)\n safe_map(write, jaxpr.invars, args)\n safe_map(write, jaxpr.constvars, consts)\n\n # Loop through equations and evaluate primitives using `bind`\n for eqn in jaxpr.eqns:\n # Read inputs to equation from environment\n invals = safe_map(read, eqn.invars) \n # `bind` is how a primitive is called\n outvals = eqn.primitive.bind(*invals, **eqn.params)\n # Primitives may return multiple outputs or not\n if not eqn.primitive.multiple_results: \n outvals = [outvals]\n # Write the results of the primitive into the environment\n safe_map(write, eqn.outvars, outvals) \n # Read the final result of the Jaxpr from the environment\n return safe_map(read, jaxpr.outvars) ",
"_____no_output_____"
],
[
"closed_jaxpr = jax.make_jaxpr(f)(jnp.ones(5))\neval_jaxpr(closed_jaxpr.jaxpr, closed_jaxpr.literals, jnp.ones(5))",
"_____no_output_____"
]
],
[
[
"Notice that `eval_jaxpr` will always return a flat list even if the original function does not.\n\nFurthermore, this interpreter does not handle `subjaxprs`, which we will not cover in this guide. You can refer to `core.eval_jaxpr` ([link](https://github.com/google/jax/blob/master/jax/core.py)) to see the edge cases that this interpreter does not cover.",
"_____no_output_____"
],
[
"### Custom `inverse` Jaxpr interpreter\n\nAn `inverse` interpreter doesn't look too different from `eval_jaxpr`. We'll first set up the registry which will map primitives to their inverses. We'll then write a custom interpreter that looks up primitives in the registry.\n\nIt turns out that this interpreter will also look similar to the \"transpose\" interpreter used in reverse-mode autodifferentiation [found here](https://github.com/google/jax/blob/master/jax/interpreters/ad.py#L141-L187).",
"_____no_output_____"
]
],
[
[
"inverse_registry = {}",
"_____no_output_____"
]
],
[
[
"We'll now register inverses for some of the primitives. By convention, primitives in Jax end in `_p` and a lot of the popular ones live in `lax`.",
"_____no_output_____"
]
],
[
[
"inverse_registry[lax.exp_p] = jnp.log\ninverse_registry[lax.tanh_p] = jnp.arctanh",
"_____no_output_____"
]
],
[
[
"`inverse` will first trace the function, then custom-interpret the Jaxpr. Let's set up a simple skeleton.",
"_____no_output_____"
]
],
[
[
"def inverse(fun):\n @wraps(fun)\n def wrapped(*args, **kwargs):\n # Since we assume unary functions, we won't\n # worry about flattening and\n # unflattening arguments\n closed_jaxpr = jax.make_jaxpr(fun)(*args, **kwargs)\n out = inverse_jaxpr(closed_jaxpr.jaxpr, closed_jaxpr.literals, *args)\n return out[0]\n return wrapped",
"_____no_output_____"
]
],
[
[
"Now we just need to define `inverse_jaxpr`, which will walk through the Jaxpr backward and invert primitives when it can.",
"_____no_output_____"
]
],
[
[
"def inverse_jaxpr(jaxpr, consts, *args):\n env = {}\n \n def read(var):\n if type(var) is core.Literal:\n return var.val\n return env[var]\n\n def write(var, val):\n env[var] = val\n # Args now correspond to Jaxpr outvars\n write(core.unitvar, core.unit)\n safe_map(write, jaxpr.outvars, args)\n safe_map(write, jaxpr.constvars, consts)\n\n # Looping backward\n for eqn in jaxpr.eqns[::-1]:\n # outvars are now invars \n invals = safe_map(read, eqn.outvars)\n if eqn.primitive not in inverse_registry:\n raise NotImplementedError(\"{} does not have registered inverse.\".format(\n eqn.primitive\n ))\n # Assuming a unary function \n outval = inverse_registry[eqn.primitive](*invals)\n safe_map(write, eqn.invars, [outval])\n return safe_map(read, jaxpr.invars)",
"_____no_output_____"
]
],
[
[
"That's it!",
"_____no_output_____"
]
],
[
[
"def f(x):\n return jnp.exp(jnp.tanh(x))\n\nf_inv = inverse(f)\nassert jnp.allclose(f_inv(f(1.0)), 1.0)",
"_____no_output_____"
]
],
[
[
"Importantly, you can trace through a Jaxpr interpreter.",
"_____no_output_____"
]
],
[
[
"jax.make_jaxpr(inverse(f))(f(1.))",
"_____no_output_____"
]
],
[
[
"That's all it takes to add a new transformation to a system, and you get composition with all the others for free! For example, we can use `jit`, `vmap`, and `grad` with `inverse`!",
"_____no_output_____"
]
],
[
[
"jit(vmap(grad(inverse(f))))((jnp.arange(5) + 1.) / 5.)",
"_____no_output_____"
]
],
[
[
"## Exercises for the reader\n\n* Handle primitives with multiple arguments where inputs are partially known, for example `lax.add_p`, `lax.mul_p`.\n* Handle `xla_call` and `xla_pmap` primitives, which will not work with both `eval_jaxpr` and `inverse_jaxpr` as written.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecb9e1579a33deeaf1ff33709119d6efe7db46e9 | 760,295 | ipynb | Jupyter Notebook | .ipynb_checkpoints/mod05-checkpoint.ipynb | theabc50111/machine_learning_tibame | af4926537220e23d974e1299030dda573e7eb429 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/mod05-checkpoint.ipynb | theabc50111/machine_learning_tibame | af4926537220e23d974e1299030dda573e7eb429 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/mod05-checkpoint.ipynb | theabc50111/machine_learning_tibame | af4926537220e23d974e1299030dda573e7eb429 | [
"MIT"
] | null | null | null | 116.30641 | 71,380 | 0.770705 | [
[
[
"# 探索性資料分析(EDA)",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\ndata_path = './data/house_train.csv'\ndf_train = pd.read_csv(data_path)\nprint(df_train.info())\nprint('-'*100)\nprint(df_train.describe())\nprint('-'*100)\nprint(df_train['主要建材'].nunique())\nprint('-'*100)\nprint(df_train['主要建材'].unique())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 69170 entries, 0 to 69169\nData columns (total 38 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 index 69170 non-null int64 \n 1 主要建材 62111 non-null object \n 2 主要用途 60013 non-null object \n 3 交易年月日 69170 non-null object \n 4 交易標的 69170 non-null object \n 5 交易筆棟數 69170 non-null object \n 6 備註 31058 non-null object \n 7 土地區段位置/建物區段門牌 69170 non-null object \n 8 土地移轉總面積(平方公尺) 69170 non-null float64\n 9 建物型態 69170 non-null object \n 10 建物現況格局-廳 69170 non-null int64 \n 11 建物現況格局-房 69170 non-null int64 \n 12 建物現況格局-衛 69170 non-null int64 \n 13 建物現況格局-隔間 69170 non-null object \n 14 建物移轉總面積(平方公尺) 69170 non-null float64\n 15 建築完成年月 54798 non-null float64\n 16 有無管理組織 69170 non-null object \n 17 移轉層次 61912 non-null object \n 18 編號 69170 non-null object \n 19 總價(元) 69170 non-null int64 \n 20 總樓層數 61964 non-null object \n 21 車位移轉總面積(平方公尺) 69170 non-null float64\n 22 車位總價(元) 69170 non-null int64 \n 23 車位類別 21450 non-null object \n 24 都市土地使用分區 68551 non-null object \n 25 鄉鎮市區 69170 non-null object \n 26 非都市土地使用分區 10 non-null object \n 27 非都市土地使用編定 0 non-null float64\n 28 num_of_bus_stations_in_100m 69166 non-null float64\n 29 income_avg 68808 non-null float64\n 30 income_var 68808 non-null float64\n 31 location_type 69166 non-null object \n 32 low_use_electricity 68808 non-null object \n 33 nearest_tarin_station 69166 non-null object \n 34 nearest_tarin_station_distance 69166 non-null float64\n 35 lat 69166 non-null float64\n 36 lng 69166 non-null float64\n 37 單價(元/平方公尺) 69170 non-null float64\ndtypes: float64(12), int64(6), object(20)\nmemory usage: 20.1+ MB\nNone\n----------------------------------------------------------------------------------------------------\n index 土地移轉總面積(平方公尺) 建物現況格局-廳 建物現況格局-房 建物現況格局-衛 \\\ncount 69170.000000 69170.000000 69170.000000 69170.000000 69170.000000 \nmean 34584.500000 57.365397 1.324967 2.099104 1.331907 \nstd 19967.803397 567.006678 0.937209 1.844211 1.279941 \nmin 0.000000 0.000000 0.000000 0.000000 0.000000 \n25% 17292.250000 11.520000 1.000000 1.000000 1.000000 \n50% 34584.500000 23.850000 2.000000 2.000000 1.000000 \n75% 51876.750000 37.000000 2.000000 3.000000 2.000000 \nmax 69169.000000 102554.980000 80.000000 168.000000 174.000000 \n\n 建物移轉總面積(平方公尺) 建築完成年月 總價(元) 車位移轉總面積(平方公尺) 車位總價(元) \\\ncount 69170.000000 5.479800e+04 6.917000e+04 69170.000000 6.917000e+04 \nmean 134.382342 8.511429e+05 2.798178e+07 12.091376 5.063665e+05 \nstd 472.970544 1.557311e+05 1.462158e+08 71.567118 2.210177e+06 \nmin 0.000000 8.000000e+00 0.000000e+00 0.000000 0.000000e+00 \n25% 52.960000 7.111020e+05 8.960000e+06 0.000000 0.000000e+00 \n50% 98.095000 8.508240e+05 1.563000e+07 0.000000 0.000000e+00 \n75% 154.797500 1.010426e+06 2.800000e+07 0.000000 0.000000e+00 \nmax 69125.530000 1.070130e+06 2.703400e+10 12188.160000 2.400000e+08 \n\n 非都市土地使用編定 num_of_bus_stations_in_100m income_avg income_var \\\ncount 0.0 69166.000000 68808.000000 68808.000000 \nmean NaN 2.237487 1298.164661 230.149715 \nstd NaN 1.576150 498.857620 250.098417 \nmin NaN 0.000000 682.000000 81.630000 \n25% NaN 1.000000 1005.000000 114.610000 \n50% NaN 2.000000 1133.000000 150.760000 \n75% NaN 3.000000 1446.000000 245.290000 \nmax NaN 10.000000 6598.000000 2403.410000 \n\n nearest_tarin_station_distance lat lng \\\ncount 69166.000000 69166.000000 69166.000000 \nmean 623.246573 25.055852 121.543798 \nstd 511.895384 0.036838 0.031904 \nmin 0.000000 24.965286 121.464657 \n25% 297.997594 25.032969 121.518965 \n50% 494.796560 25.051739 121.541236 \n75% 751.628239 25.077493 121.565418 \nmax 6007.085560 25.172812 121.624224 \n\n 單價(元/平方公尺) \ncount 6.917000e+04 \nmean 1.887246e+05 \nstd 2.774005e+05 \nmin 0.000000e+00 \n25% 1.248660e+05 \n50% 1.697065e+05 \n75% 2.244038e+05 \nmax 6.268571e+07 \n----------------------------------------------------------------------------------------------------\n17\n----------------------------------------------------------------------------------------------------\n[nan '鋼筋混凝土造' '鋼骨鋼筋混凝土造' '加強磚造' '木造' '見其他登記事項' '見使用執照' '磚造' '預力混凝土造'\n '鋼骨混凝土造' '土造' '鋼筋混凝土加強磚造' '土木造' '土磚石混合造' '壁式預鑄鋼筋混凝土造' '石造' '竹造' '鐵造']\n"
],
[
"df_train.head()",
"_____no_output_____"
]
],
[
[
"# 繪製圖表",
"_____no_output_____"
],
[
"#### 散布圖",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nplt.scatter(np.arange(len(df_train['建物現況格局-廳'])),df_train['建物現況格局-廳'])\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### 折線圖",
"_____no_output_____"
]
],
[
[
"x = np.linspace(0, 10, 1000)\nplt.plot(x,np.sin(x))\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### 直方圖",
"_____no_output_____"
]
],
[
[
"plt.hist(df_train['建物現況格局-廳'][df_train['建物現況格局-廳']<20])\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### 箱型圖",
"_____no_output_____"
]
],
[
[
"plt.boxplot(df_train['建物現況格局-廳'][df_train['建物現況格局-廳']<20])\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Heatmap",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nfrom matplotlib.font_manager import FontProperties\nprop = FontProperties(fname='C:\\\\Windows\\\\Fonts\\\\msjh.ttc')\nsns.set(font=prop.get_family())\nsns.set_style(\"whitegrid\",{\"font.sans-serif\":['Microsoft JhengHei']})\n#-----------------------設定中文字型-------------------\n\ncorr = df_train.corr()\nsns.heatmap(corr)\nplt.show()",
"_____no_output_____"
],
[
"corr['單價(元/平方公尺)']",
"_____no_output_____"
]
],
[
[
"#### 繪圖設定",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(10,8)) # 設定圖片大小\nplt.title('建物現況格局-廳-統計',fontsize=36)\nplt.xlabel('建物現況格局-廳',fontsize=16)\nplt.ylabel('次數',fontsize=16)\nplt.axis([0,10,0,38000])\nplt.hist(df_train['建物現況格局-廳'][df_train['建物現況格局-廳']<20],label = \"label1\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"# EDA整體流程",
"_____no_output_____"
],
[
"#### 讀取檔案",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\ndata_path = ['./data/house_train.csv','./data/house_test.csv']\ndf_train = pd.read_csv(data_path[0])\ndf_test = pd.read_csv(data_path[1])\n\n# 將答案抽離\ndf_train_Y = df_train['單價(元/平方公尺)']\ndf_train = df_train.drop(['單價(元/平方公尺)'] , axis=1)\n\n# 記得刪index\ndf_concat = pd.concat([df_train,df_test],ignore_index = True)\ndf_concat = df_concat.drop(['index'] , axis=1)\ndisplay(df_concat.tail())",
"_____no_output_____"
]
],
[
[
"#### 格式調整",
"_____no_output_____"
]
],
[
[
"print(df_concat.info())\nprint('-'*100)\nprint(df_concat.describe())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 70670 entries, 0 to 70669\nData columns (total 36 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 主要建材 63445 non-null object \n 1 主要用途 61307 non-null object \n 2 交易年月日 70670 non-null object \n 3 交易標的 70670 non-null object \n 4 交易筆棟數 70670 non-null object \n 5 備註 31743 non-null object \n 6 土地區段位置/建物區段門牌 70670 non-null object \n 7 土地移轉總面積(平方公尺) 70670 non-null float64\n 8 建物型態 70670 non-null object \n 9 建物現況格局-廳 70670 non-null int64 \n 10 建物現況格局-房 70670 non-null int64 \n 11 建物現況格局-衛 70670 non-null int64 \n 12 建物現況格局-隔間 70670 non-null object \n 13 建物移轉總面積(平方公尺) 70670 non-null float64\n 14 建築完成年月 55978 non-null float64\n 15 有無管理組織 70670 non-null object \n 16 移轉層次 63240 non-null object \n 17 編號 70670 non-null object \n 18 總價(元) 70670 non-null int64 \n 19 總樓層數 63289 non-null object \n 20 車位移轉總面積(平方公尺) 70670 non-null float64\n 21 車位總價(元) 70670 non-null int64 \n 22 車位類別 21937 non-null object \n 23 都市土地使用分區 70039 non-null object \n 24 鄉鎮市區 70670 non-null object \n 25 非都市土地使用分區 10 non-null object \n 26 非都市土地使用編定 0 non-null float64\n 27 num_of_bus_stations_in_100m 70666 non-null float64\n 28 income_avg 70301 non-null float64\n 29 income_var 70301 non-null float64\n 30 location_type 70666 non-null object \n 31 low_use_electricity 70301 non-null object \n 32 nearest_tarin_station 70666 non-null object \n 33 nearest_tarin_station_distance 70666 non-null float64\n 34 lat 70666 non-null float64\n 35 lng 70666 non-null float64\ndtypes: float64(11), int64(5), object(20)\nmemory usage: 19.4+ MB\nNone\n----------------------------------------------------------------------------------------------------\n 土地移轉總面積(平方公尺) 建物現況格局-廳 建物現況格局-房 建物現況格局-衛 建物移轉總面積(平方公尺) \\\ncount 70670.000000 70670.000000 70670.000000 70670.000000 70670.000000 \nmean 57.089100 1.324636 2.098373 1.331541 134.169339 \nstd 562.181164 0.937883 1.839681 1.275769 468.297146 \nmin 0.000000 0.000000 0.000000 0.000000 0.000000 \n25% 11.510000 1.000000 1.000000 1.000000 52.970000 \n50% 23.850000 2.000000 2.000000 1.000000 98.090000 \n75% 37.010000 2.000000 3.000000 2.000000 154.810000 \nmax 102554.980000 80.000000 168.000000 174.000000 69125.530000 \n\n 建築完成年月 總價(元) 車位移轉總面積(平方公尺) 車位總價(元) 非都市土地使用編定 \\\ncount 5.597800e+04 7.067000e+04 70670.000000 7.067000e+04 0.0 \nmean 8.512305e+05 2.793040e+07 12.108471 5.063280e+05 NaN \nstd 1.557748e+05 1.447541e+08 70.942234 2.197704e+06 NaN \nmin 8.000000e+00 0.000000e+00 0.000000 0.000000e+00 NaN \n25% 7.110300e+05 8.960000e+06 0.000000 0.000000e+00 NaN \n50% 8.509040e+05 1.562000e+07 0.000000 0.000000e+00 NaN \n75% 1.010508e+06 2.800000e+07 0.000000 0.000000e+00 NaN \nmax 1.070130e+06 2.703400e+10 12188.160000 2.400000e+08 NaN \n\n num_of_bus_stations_in_100m income_avg income_var \\\ncount 70666.000000 70301.000000 70301.000000 \nmean 2.238870 1297.724214 229.946104 \nstd 1.575739 497.892520 249.671501 \nmin 0.000000 682.000000 81.630000 \n25% 1.000000 1005.000000 114.460000 \n50% 2.000000 1133.000000 150.760000 \n75% 3.000000 1446.000000 245.290000 \nmax 10.000000 6598.000000 2403.410000 \n\n nearest_tarin_station_distance lat lng \ncount 70666.000000 70666.000000 70666.000000 \nmean 623.399032 25.055868 121.543811 \nstd 511.877028 0.036848 0.031931 \nmin 0.000000 24.965286 121.362342 \n25% 297.997594 25.032969 121.518950 \n50% 495.132682 25.051742 121.541236 \n75% 751.777726 25.077551 121.565418 \nmax 6588.758171 25.172812 121.624224 \n"
],
[
"# 只取 int64, float64 兩種數值型欄位, 存於 num_features 中, 其他類別存於notnum_features\nnum_features = []\nnotnum_features = []\nfor dtype, feature in zip(df_concat.dtypes, df_concat.columns):\n if dtype == 'float64' or dtype == 'int64':\n num_features.append(feature)\n else:\n notnum_features.append(feature)\n\nprint(f'lehgth of all featrues : {len(df_concat.columns)}\\n')\nprint(f'length of Numeric Features : {len(num_features)}\\n Numeric Features : {num_features}\\n')\nprint(f'length of Not Numeric Features : {len(notnum_features)}\\n Not Numeric Features : {notnum_features}')",
"lehgth of all featrues : 36\n\nlength of Numeric Features : 16\n Numeric Features : ['土地移轉總面積(平方公尺)', '建物現況格局-廳', '建物現況格局-房', '建物現況格局-衛', '建物移轉總面積(平方公尺)', '建築完成年月', '總價(元)', '車位移轉總面積(平方公尺)', '車位總價(元)', '非都市土地使用編定', 'num_of_bus_stations_in_100m', 'income_avg', 'income_var', 'nearest_tarin_station_distance', 'lat', 'lng']\n\nlength of Not Numeric Features : 20\n Not Numeric Features : ['主要建材', '主要用途', '交易年月日', '交易標的', '交易筆棟數', '備註', '土地區段位置/建物區段門牌', '建物型態', '建物現況格局-隔間', '有無管理組織', '移轉層次', '編號', '總樓層數', '車位類別', '都市土地使用分區', '鄉鎮市區', '非都市土地使用分區', 'location_type', 'low_use_electricity', 'nearest_tarin_station']\n"
]
],
[
[
"#### 偵測與填補遺失值",
"_____no_output_____"
]
],
[
[
"df_concat_pre = pd.DataFrame()\nfor feature in num_features:\n if df_concat[feature].isnull().any():\n print(f'{feature} has missing value')\n df_concat_pre[feature] = df_concat[feature].fillna(df_concat[feature].mean())\n else:\n df_concat_pre[feature] = df_concat[feature]\ndisplay(df_concat_pre.head())",
"建築完成年月 has missing value\n非都市土地使用編定 has missing value\nnum_of_bus_stations_in_100m has missing value\nincome_avg has missing value\nincome_var has missing value\nnearest_tarin_station_distance has missing value\nlat has missing value\nlng has missing value\n"
]
],
[
[
"#### 特徵選擇",
"_____no_output_____"
]
],
[
[
"df_concat['非都市土地使用編定']",
"_____no_output_____"
],
[
"df_corr_check = pd.DataFrame()\ndf_corr_check['非都市土地使用編定'] = df_concat['非都市土地使用編定']\ndf_corr_check['單價(元/平方公尺)'] = df_train_Y\nprint(df_corr_check.corr())",
" 非都市土地使用編定 單價(元/平方公尺)\n非都市土地使用編定 NaN NaN\n單價(元/平方公尺) NaN 1.0\n"
],
[
"df_concat_pre = df_concat_pre.drop(['非都市土地使用編定'] , axis=1)\nnum_features.remove('非都市土地使用編定')\ndisplay(df_concat_pre.head())",
"_____no_output_____"
]
],
[
[
"## 非數值型特徵處理",
"_____no_output_____"
],
[
"#### 時間格式特徵",
"_____no_output_____"
]
],
[
[
"df_concat['建築完成年月'].head()",
"_____no_output_____"
],
[
"import datetime\ndf_concat_pre['建築完成年'] = df_concat['建築完成年月'].apply(str)\ndf_concat_pre['建築完成年'] = df_concat_pre['建築完成年'].apply(lambda x: x[:-6])\ndf_concat_pre['建築完成年'][df_concat_pre['建築完成年']==''] = '0'\ndf_concat_pre['建築完成年'] = df_concat_pre['建築完成年'].apply(int)\ndf_concat_pre['建築完成年'][df_concat_pre['建築完成年']==0] = np.nan\ndf_concat_pre['建築完成年'] = df_concat_pre['建築完成年'].fillna(df_concat_pre['建築完成年'].mean()).apply(int)\ndf_concat_pre = df_concat_pre.drop(['建築完成年月'],axis=1) \nnum_features.remove('建築完成年月') ; num_features.append('建築完成年')\nprint(num_features)\ndf_concat_pre.head()",
"['土地移轉總面積(平方公尺)', '建物現況格局-廳', '建物現況格局-房', '建物現況格局-衛', '建物移轉總面積(平方公尺)', '總價(元)', '車位移轉總面積(平方公尺)', '車位總價(元)', 'num_of_bus_stations_in_100m', 'income_avg', 'income_var', 'nearest_tarin_station_distance', 'lat', 'lng', '建築完成年']\n"
],
[
"import math\ndf_date_temp = pd.DataFrame()\ndf_date_temp['transaction_datetime'] = df_concat['交易年月日'].apply(lambda x: datetime.datetime.strptime(x, '%Y-%m-%d %H:%M:%S'))\ndf_concat_pre['交易年'] = df_date_temp['transaction_datetime'].apply(lambda x: datetime.datetime.strftime(x, '%Y')).astype('int64')\ndf_concat_pre['交易月'] = df_date_temp['transaction_datetime'].apply(lambda x: datetime.datetime.strftime(x, '%m')).astype('int64')\ndf_concat_pre['交易月'] = df_concat_pre['交易月'].apply(lambda x:math.sin(x/4)) #以一季為一個週期\nnotnum_features.remove('交易年月日') ; num_features.extend(['交易年', '交易月']) \n\nprint(num_features)\nprint(notnum_features)\ndf_concat_pre.loc[:,['交易年','交易月']]",
"['土地移轉總面積(平方公尺)', '建物現況格局-廳', '建物現況格局-房', '建物現況格局-衛', '建物移轉總面積(平方公尺)', '總價(元)', '車位移轉總面積(平方公尺)', '車位總價(元)', 'num_of_bus_stations_in_100m', 'income_avg', 'income_var', 'nearest_tarin_station_distance', 'lat', 'lng', '建築完成年', '交易年', '交易月']\n['主要建材', '主要用途', '交易標的', '交易筆棟數', '備註', '土地區段位置/建物區段門牌', '建物型態', '建物現況格局-隔間', '有無管理組織', '移轉層次', '編號', '總樓層數', '車位類別', '都市土地使用分區', '鄉鎮市區', '非都市土地使用分區', 'location_type', 'low_use_electricity', 'nearest_tarin_station']\n"
],
[
"for feature in notnum_features:\n print(f'{feature}:\\n nan: {df_concat[feature].isnull().any()}\\n{df_concat[feature].unique()}')\n print('-'*50)",
"主要建材:\n nan: True\n[nan '鋼筋混凝土造' '鋼骨鋼筋混凝土造' '加強磚造' '木造' '見其他登記事項' '見使用執照' '磚造' '預力混凝土造'\n '鋼骨混凝土造' '土造' '鋼筋混凝土加強磚造' '土木造' '土磚石混合造' '壁式預鑄鋼筋混凝土造' '石造' '竹造' '鐵造']\n--------------------------------------------------\n主要用途:\n nan: True\n[nan '住家用' '商業用' '住商用' '見其他登記事項' '見使用執照' '工業用' '國民住宅' '農舍' '停車空間' '工商用'\n '住工用']\n--------------------------------------------------\n交易標的:\n nan: False\n['土地' '房地(土地+建物)' '房地(土地+建物)+車位' '建物']\n--------------------------------------------------\n交易筆棟數:\n nan: False\n['土地7建物0車位0' '土地3建物1車位0' '土地1建物1車位4' '土地1建物1車位0' '土地2建物1車位0' '土地1建物0車位0'\n '土地2建物1車位2' '土地1建物1車位2' '土地3建物2車位0' '土地2建物1車位1' '土地1建物2車位2' '土地1建物2車位1'\n '土地2建物1車位7' '土地3建物0車位0' '土地1建物1車位1' '土地5建物1車位0' '土地2建物1車位4' '土地4建物0車位0'\n '土地1建物1車位3' '土地1建物2車位0' '土地1建物1車位6' '土地1建物2車位3' '土地4建物1車位1' '土地2建物0車位0'\n '土地3建物1車位1' '土地2建物2車位3' '土地6建物1車位0' '土地7建物1車位0' '土地4建物1車位0' '土地0建物1車位0'\n '土地1建物3車位0' '土地0建物2車位0' '土地8建物1車位0' '土地0建物3車位0' '土地10建物0車位0' '土地0建物1車位4'\n '土地9建物1車位0' '土地6建物0車位0' '土地3建物1車位2' '土地11建物1車位0' '土地2建物1車位3' '土地2建物2車位0'\n '土地6建物2車位0' '土地5建物0車位0' '土地0建物1車位1' '土地1建物3車位3' '土地1建物1車位5' '土地1建物4車位0'\n '土地5建物1車位1' '土地1建物3車位2' '土地3建物2車位3' '土地8建物1車位1' '土地8建物2車位0' '土地4建物1車位4'\n '土地1建物1車位33' '土地4建物3車位0' '土地2建物3車位1' '土地12建物1車位0' '土地2建物4車位1' '土地5建物1車位5'\n '土地10建物1車位0' '土地0建物0車位0' '土地2建物2車位1' '土地2建物4車位0' '土地3建物3車位0' '土地2建物1車位6'\n '土地1建物1車位12' '土地1建物1車位7' '土地1建物3車位4' '土地2建物2車位2' '土地4建物1車位2' '土地0建物1車位2'\n '土地3建物1車位3' '土地4建物2車位4' '土地14建物0車位0' '土地9建物2車位0' '土地2建物3車位2' '土地11建物0車位0'\n '土地1建物2車位4' '土地1建物4車位20' '土地1建物4車位1' '土地1建物5車位0' '土地1建物1車位11' '土地2建物3車位0'\n '土地8建物0車位0' '土地3建物2車位1' '土地6建物1車位1' '土地4建物2車位0' '土地1建物1車位8' '土地1建物4車位4'\n '土地8建物1車位5' '土地5建物2車位0' '土地0建物4車位0' '土地19建物1車位0' '土地25建物0車位0'\n '土地2建物6車位95' '土地9建物0車位0' '土地2建物1車位8' '土地15建物0車位0' '土地2建物7車位0' '土地6建物1車位2'\n '土地6建物9車位2' '土地2建物1車位5' '土地1建物4車位2' '土地4建物2車位2' '土地5建物2車位1' '土地4建物1車位3'\n '土地6建物1車位4' '土地1建物7車位36' '土地0建物2車位11' '土地1建物6車位0' '土地14建物1車位0'\n '土地30建物0車位0' '土地24建物0車位0' '土地3建物1車位6' '土地4建物2車位1' '土地0建物1車位3' '土地3建物4車位0'\n '土地19建物0車位0' '土地7建物1車位4' '土地1建物1車位10' '土地7建物2車位2' '土地2建物2車位5'\n '土地17建物1車位1' '土地5建物1車位2' '土地12建物0車位0' '土地4建物4車位7' '土地4建物3車位1' '土地1建物4車位9'\n '土地1建物2車位5' '土地3建物1車位30' '土地20建物1車位0' '土地1建物8車位0' '土地9建物1車位3'\n '土地2建物28車位0' '土地27建物0車位0' '土地1建物52車位128' '土地3建物3車位1' '土地1建物3車位1'\n '土地1建物1車位86' '土地21建物0車位0' '土地16建物1車位0' '土地28建物1車位0' '土地12建物11車位0'\n '土地8建物3車位0' '土地0建物0車位1' '土地3建物5車位0' '土地10建物2車位0' '土地0建物2車位1' '土地0建物2車位2'\n '土地6建物11車位0' '土地1建物3車位7' '土地2建物3車位3' '土地2建物2車位4' '土地2建物9車位0' '土地2建物5車位3'\n '土地2建物12車位0' '土地9建物1車位1' '土地4建物2車位3' '土地32建物0車位0' '土地5建物3車位0' '土地6建物3車位0'\n '土地7建物1車位1' '土地1建物3車位8' '土地3建物6車位1' '土地58建物0車位0' '土地0建物5車位0' '土地1建物1車位15'\n '土地2建物3車位4' '土地1建物25車位306' '土地1建物11車位70' '土地1建物2車位6' '土地1建物28車位235'\n '土地8建物4車位0' '土地4建物12車位0' '土地2建物2車位8' '土地2建物2車位6' '土地1建物7車位16'\n '土地3建物7車位105' '土地1建物34車位71' '土地7建物10車位17' '土地3建物1車位4' '土地4建物2車位5'\n '土地3建物4車位4' '土地0建物13車位45' '土地15建物1車位0' '土地18建物1車位0' '土地31建物0車位0'\n '土地0建物15車位135' '土地4建物1車位12' '土地1建物3車位20' '土地18建物1車位1' '土地13建物0車位0'\n '土地1建物1車位16' '土地1建物2車位10' '土地16建物0車位0' '土地2建物2車位7' '土地1建物14車位0'\n '土地2建物1車位10' '土地1建物1車位40' '土地11建物3車位0' '土地3建物26車位99' '土地7建物2車位1'\n '土地1建物1車位9' '土地16建物1車位1' '土地6建物2車位2' '土地1建物3車位9' '土地13建物1車位0'\n '土地1建物7車位18' '土地5建物3車位2' '土地26建物0車位0' '土地22建物0車位0' '土地1建物4車位11'\n '土地5建物1車位3' '土地2建物6車位0' '土地17建物0車位0' '土地2建物6車位14' '土地1建物5車位5' '土地1建物7車位6'\n '土地20建物0車位0' '土地1建物5車位2' '土地5建物4車位0' '土地2建物8車位10' '土地3建物2車位4'\n '土地1建物5車位21' '土地10建物1車位1' '土地1建物5車位17' '土地7建物11車位0' '土地2建物5車位2'\n '土地17建物1車位0' '土地1建物3車位6' '土地1建物14車位28' '土地1建物15車位79' '土地1建物3車位12'\n '土地18建物0車位0' '土地0建物13車位16' '土地13建物2車位0' '土地1建物8車位2' '土地25建物1車位0'\n '土地7建物2車位6' '土地2建物7車位8' '土地2建物3車位6' '土地47建物0車位0' '土地1建物3車位16'\n '土地1建物8車位20' '土地25建物7車位4' '土地2建物4車位8' '土地0建物11車位7' '土地2建物5車位0'\n '土地3建物2車位2' '土地1建物7車位21' '土地1建物6車位12' '土地7建物2車位4' '土地1建物3車位11'\n '土地1建物2車位13' '土地2建物1車位13' '土地10建物5車位3' '土地8建物2車位1' '土地6建物4車位0'\n '土地2建物3車位75' '土地11建物2車位2' '土地4建物4車位0' '土地2建物12車位5' '土地2建物8車位5'\n '土地1建物4車位7' '土地1建物5車位18' '土地1建物7車位1' '土地1建物1車位30' '土地1建物3車位5'\n '土地11建物1車位2' '土地1建物3車位13' '土地1建物1車位21' '土地1建物12車位0' '土地0建物1車位5'\n '土地1建物4車位14' '土地1建物9車位49' '土地23建物0車位0' '土地5建物8車位0' '土地1建物2車位8'\n '土地1建物9車位16' '土地7建物3車位6' '土地1建物15車位0' '土地2建物42車位0' '土地1建物2車位12'\n '土地10建物2車位1' '土地5建物1車位10' '土地2建物3車位15' '土地3建物3車位45' '土地1建物5車位38'\n '土地2建物4車位4' '土地1建物2車位9' '土地1建物5車位1' '土地1建物4車位12' '土地1建物7車位34' '土地3建物1車位7'\n '土地1建物10車位140' '土地1建物30車位0' '土地2建物14車位4' '土地1建物3車位17' '土地1建物3車位21'\n '土地1建物10車位10' '土地6建物5車位0' '土地0建物0車位3' '土地0建物118車位0' '土地1建物5車位6'\n '土地1建物2車位18' '土地2建物6車位69' '土地0建物2車位6' '土地1建物16車位0' '土地1建物2車位15'\n '土地1建物12車位12' '土地13建物1車位1' '土地4建物5車位0' '土地30建物1車位1' '土地6建物6車位14'\n '土地1建物1車位18' '土地1建物7車位145' '土地3建物7車位69' '土地1建物7車位2' '土地3建物2車位6'\n '土地3建物4車位3' '土地1建物11車位76' '土地2建物1車位9' '土地1建物6車位4' '土地1建物1車位17'\n '土地1建物4車位8' '土地1建物6車位1' '土地13建物22車位0' '土地2建物15車位0' '土地1建物8車位34'\n '土地1建物55車位122' '土地18建物2車位0' '土地1建物7車位0' '土地1建物6車位10' '土地16建物1車位2'\n '土地1建物11車位0' '土地3建物1車位5' '土地13建物11車位0' '土地1建物4車位18' '土地1建物2車位16'\n '土地1建物4車位5' '土地2建物14車位173' '土地2建物3車位20' '土地1建物4車位3' '土地2建物4車位2']\n--------------------------------------------------\n備註:\n nan: True\n[nan '親友、員工或其他特殊關係間之交易。' '含增建或未登記建物。' ... '含增建或未登記建物。;另有頂樓增建1房2廳1衛'\n '1.地政士僅受託買賣案件申請登記,並未代理撰擬不動產買賣契約書,本成交資訊係由不動產經紀業或權利人自行提供屬實2.車位總價無法拆分各別車位價格'\n '102.4.29預售屋買賣過戶']\n--------------------------------------------------\n土地區段位置/建物區段門牌:\n nan: False\n['溪洲段二小段211~240地號' '臺北市中正區三元街172巷1弄1~30號' '臺北市南港區經貿二路235巷1~30號' ...\n '臺北市大同區甘州街31~60號' '臺北市文山區興隆路一段229巷14弄1~30號' '臺北市士林區文林路587巷1~30號']\n--------------------------------------------------\n建物型態:\n nan: False\n['其他' '公寓(5樓含以下無電梯)' '住宅大樓(11層含以上有電梯)' '透天厝' '華廈(10層含以下有電梯)' '辦公商業大樓'\n '套房(1房1廳1衛)' '店面(店鋪)' '廠辦']\n--------------------------------------------------\n建物現況格局-隔間:\n nan: False\n['有' '無']\n--------------------------------------------------\n有無管理組織:\n nan: False\n['無' '有']\n--------------------------------------------------\n移轉層次:\n nan: True\n[nan '一層' '十八層' '二層' '八層' '三層' '全' '六層' '十四層' '一層,平台' '四層' '一層,騎樓' '七層'\n '九層' '十五層' '十層' '十一層' '十二層' '五層' '地下層' '十六層' '一層,二層,見其他登記事項' '三層,陽台'\n '十四層,見其他登記事項' '一層,騎樓,地下層' '二十五層' '五層,六層' '十一層,十二層' '二十四層' '十三層'\n '二層,電梯樓梯間' '一層,地下一層' '十七層' '一層,地下層' '陽台,三層' '四層,陽台' '一層,二層' '陽台,六層'\n '四層,五層' '五層,電梯樓梯間' '二層,陽台' '平台,一層' '二十一層' '五層,陽台' '一層,二層,騎樓' '一層,停車場'\n '三層,四層,見其他登記事項,屋頂突出物' '三十二層' '一層,見使用執照' '二十層' '三層,四層' '三十一層' '五層,見其他登記事項'\n '四層,露台' '二層,露台' '三十九層' '九層,十層' '地下二層' '四層,露台,走廊' '地下一層' '一層,平台,騎樓'\n '二層,走廊' '四層,電梯樓梯間' '二十二層' '一層,見其他登記事項' '二十一層,二十二層' '六層,陽台' '九層,電梯樓梯間'\n '十九層' '六層,見其他登記事項' '十四層,屋頂突出物' '一層,露台' '防空避難室,見使用執照' '地下層,一層' '三層,露台'\n '二層,三層,四層' '十層,電梯樓梯間' '一層,騎樓,停車場,地下層' '四層,五層,夾層' '一層,二層,三層,屋頂突出物,地下一層'\n '二十三層' '三十層' '二十六層' '六層,電梯樓梯間' '一層,夾層' '一層,騎樓,停車場' '五層,四層' '十層,十一層'\n '十三層,十四層' '一層,二層,三層,四層,騎樓' '十二層,十三層' '六層,夾層' '三層,四層,夾層' '十二層,十三層,電梯樓梯間'\n '十二層,電梯樓梯間' '六層,七層' '二層,見使用執照' '陽台,二層' '三層,電梯樓梯間' '二十九層' '七層,電梯樓梯間'\n '四層,五層,三層' '電梯樓梯間,地下一層' '五層,夾層' '陽台,四層' '一層,走廊' '六層,陽台,見其他登記事項' '一層,陽台'\n '三層,四層,陽台' '八層,陽台' '一層,二層,三層' '十二層,屋頂突出物' '八層,九層' '六層,七層,夾層'\n '見其他登記事項,十一層' '二十二層,二十三層' '七層,夾層' '十七層,十八層,電梯樓梯間' '七層,陽台' '地下三層'\n '十五層,十六層,十七層' '二層,一層' '六層,走廊' '一層,二層,三層,騎樓' '二層,三層' '七層,走廊' '十四層,夾層'\n '地下一層,地下二層' '二層,三層,見其他登記事項' '四層,見使用執照' '二層,夾層' '十四層,十五層' '三層,四層,五層'\n '三層,四層,走廊' '一層,騎樓,地下一層' '見使用執照,四層' '二層,一層,騎樓' '十二層,陽台' '走廊,二層' '陽台,一層'\n '八層,電梯樓梯間' '十七層,十八層' '九層,陽台' '電梯樓梯間,三層' '一層,平台,地下層' '七層,八層' '露台,四層'\n '十層,陽台' '二層,騎樓' '地下二層,地下一層' '六層,見使用執照' '十一層,見其他登記事項' '一層,騎樓,停車場,通道,門廳'\n '一層,騎樓,夾層' '騎樓,一層,二層,三層,四層,五層,陽台' '二層,陽台,電梯樓梯間' '八層,夾層' '十一層,夾層'\n '陽台,電梯樓梯間,六層' '一層,電梯樓梯間' '十四層,陽台' '十層,見其他登記事項' '一層,夾層,走廊' '十六層,十七層'\n '一層,二層,三層,屋頂突出物,見其他登記事項,地下二層,地下一層,四層' '四層,見其他登記事項' '一層,門廳' '十二層,見其他登記事項'\n '二十七層' '十五層,十六層' '電梯樓梯間,十一層' '二層,三層,四層,電梯樓梯間' '十一層,陽台' '二層,平台' '十三層,夾層'\n '九層,夾層' '十層,走廊' '地下層,見使用執照' '二十八層' '電梯樓梯間,二層' '十二層,夾層' '陽台,五層' '陽台,屋頂突出物'\n '二十三層,二十四層' '一層,騎樓,露台' '二十八層,二十九層' '屋頂突出物' '電梯樓梯間,八層,陽台' '二十一層,見其他登記事項'\n '屋頂突出物,地下一層,一層,二層,三層,四層,五層,六層,七層,八層,九層,十層' '五層,露台' '電梯樓梯間,十四層,十五層'\n '八層,九層,十層' '地下三層,地下一層,地下二層' '四層,陽臺' '二十四層,電梯樓梯間' '騎樓,一層' '地下二層,地下三層'\n '四層,通道' '一層,二層,三層,四層' '四層,騎樓' '一層,平台,騎樓,地下層' '十三層,電梯樓梯間' '五層,見使用執照'\n '三層,四層,騎樓,一層,二層' '四層,五層,見其他登記事項' '露台,三層' '十一層,陽台,電梯樓梯間' '一層,二層,陽台,平台'\n '七層,見其他登記事項' '四層,陽台,電梯樓梯間' '一層,二層,三層,屋頂突出物' '防空避難室' '十層,夾層'\n '一層,二層,三層,四層,地下一層' '一層,露台,地下層' '三層,陽台,電梯樓梯間' '屋頂突出物,一層' '一層,停車場,通道'\n '六層,五層' '一層,通道' '七層,見使用執照' '二十層,二十一層' '七層,通道' '一層,二層,夾層' '地下三層,地下四層,地下二層'\n '八層,陽台,見使用執照' '地下一層,地下二層,地下三層,地下四層' '一層,二層,地下層' '見其他登記事項' '八層,見其他登記事項'\n '一層,二層,三層,四層,地下三層,屋頂突出物' '四層,五層,屋頂突出物' '電梯樓梯間,夾層,四層'\n '一層,二層,三層,四層,五層,六層,七層,八層,騎樓' '十七層,見其他登記事項' '一層,平台,走廊' '十層,電梯樓梯間,夾層'\n '十八層,十九層,電梯樓梯間' '一層,二層,屋頂突出物,地下一層,地下二層' '十一層,電梯樓梯間' '一層,騎樓,通道,電梯樓梯間'\n '十八層,電梯樓梯間' '三層,四層,見其他登記事項' '六層,電梯樓梯間,七層' '地下四層' '夾層' '一層,騎樓,通道' '十三層,陽台'\n '四層,五層,電梯樓梯間' '地下一層,見其他登記事項' '走廊,一層' '二層,通道' '十九層,夾層' '二十一層,二十層' '十五層,陽台'\n '十二層,走廊' '三層,見使用執照' '二層,陽臺' '八層,七層' '十四層,屋頂突出物,夾層' '一層,露台,走廊' '十六層,電梯樓梯間'\n '九層,見使用執照' '九層,露台' '一層,二層,地下一層' '一層,二層,三層,四層,五層,停車場,地下層' '陽台,電梯樓梯間,三層'\n '露台,二層' '二十五層,見其他登記事項' '十五層,電梯樓梯間,十四層' '陽台,十層' '防空避難室,騎樓,電梯樓梯間' '停車場,地下層'\n '十九層,屋頂突出物' '四層,平台' '夾層,一層' '一層,騎樓,見其他登記事項' '電梯樓梯間,地下層' '三層,電梯樓梯間,陽台'\n '一層,騎樓,見使用執照' '防空避難室,地下層' '三十三層' '地下二層,地下三層,地下四層,地下五層' '十層,九層'\n '四層,五層,二層,三層' '五層,通道' '一層,騎樓,走廊' '陽台,屋頂突出物,四層' '停車場,一層' '五層,陽台,見其他登記事項'\n '一層,夾層,地下一層' '九層,見其他登記事項' '三層,平台' '一層,二層,陽台,平台,騎樓' '四層,夾層'\n '一層,二層,三層,四層,五層,騎樓' '陽台,七層' '一層,二層,停車場' '一層,三層,四層,五層,六層,陽台,屋頂突出物,地下層'\n '六層,機械房' '十八層,十九層' '六層,通道' '一層,二層,三層,四層,五層,六層,七層' '三十七層,三十八層' '地下一層,一層'\n '騎樓,露台,一層' '騎樓,一層,平台' '電梯樓梯間,四層' '五層,電梯樓梯間,通道' '七層,露台' '五層,六層,電梯樓梯間'\n '二層,騎樓,一層' '夾層,十三層' '十一層,露台' '六層,露台' '三層,騎樓' '四層,走廊' '十四層,電梯樓梯間'\n '騎樓,一層,二層,三層,四層,陽台,平台' '一層,二層,屋頂突出物' '一層,地下層,見使用執照' '停車場' '八層,陽台,電梯樓梯間'\n '三層,通道' '三十五層' '十層,陽台,電梯樓梯間' '一層,二層,平台,騎樓,陽台' '三層,陽臺' '十一層,門廳'\n '八層,九層,一層,二層,三層,四層,五層,六層,七層,十層,十一層,陽台,騎樓,地下層' '十層,露台'\n '三層,四層,屋頂突出物,見其他登記事項' '騎樓,一層,夾層' '五層,走廊' '地下一層,地下三層' '二十八層,電梯樓梯間'\n '騎樓,夾層,一層' '電梯樓梯間,五層' '十七層,電梯樓梯間' '二十六層,電梯樓梯間,二十五層' '三層,四層,屋頂突出物'\n '見其他登記事項,十三層' '九層,八層' '夾層,一層,騎樓' '十二層,門廳' '二十六層,二十七層' '四層,五層,屋頂突出物,夾層'\n '四層,陽台,屋頂突出物' '屋頂突出物,四層' '露台,一層,走廊' '十九層,電梯樓梯間' '十四層,見使用執照'\n '防空避難室,停車場,見使用執照' '電梯樓梯間,十層,十一層'\n '一層,二層,三層,四層,五層,六層,七層,八層,屋頂突出物,騎樓,地下一層,地下二層,地下三層,地下四層,地下五層' '電梯樓梯間,九層'\n '防空避難室,地下層,見使用執照' '一層,停車場,騎樓' '五層,六層,陽台' '一層,陽台,騎樓' '一層,平台,停車場'\n '五層,陽台,電梯樓梯間' '十九層,二十層,電梯樓梯間' '走廊,五層' '騎樓,見使用執照,一層' '一層,二層,三層,四層,屋頂突出物'\n '六層,七層,屋頂突出物' '三層,屋頂突出物,一層,二層' '地下四層,地下二層,地下三層,地下一層' '六層,騎樓' '十二層,露台'\n '地下一層,地下二層,地下四層,地下三層' '三十八層' '電梯樓梯間,七層' '夾層,十九層' '一層,露台,地下一層'\n '三層,騎樓,一層,二層' '電梯樓梯間,六層' '十五層,電梯樓梯間' '屋頂突出物,二層,一層,見其他登記事項' '一層,二層,電梯樓梯間'\n '二十層,電梯樓梯間' '走廊,地下層' '八層,露台' '地下一層,地下二層,地下三層' '三層,四層,電梯樓梯間'\n '一層,二層,三層,四層,五層,六層,七層,八層,九層,十層,騎樓,地下層' '四層,三層' '五層,六層,七層' '九層,陽台,電梯樓梯間'\n '八層,走廊' '四層,陽台,走廊' '一層,地下一層,地下二層' '見其他登記事項,二十五層' '二層,陽台,走廊' '平台,騎樓,一層'\n '六層,七層,陽台' '停車場,地下二層' '一層,二層,三層,四層,陽台,平台,騎樓'\n '一層,二層,三層,四層,五層,六層,七層,地下一層,地下二層' '四層,五層,一層,二層,三層,騎樓' '一層,露台,騎樓,二層'\n '騎樓,一層,二層' '十一層,屋頂突出物,見其他登記事項' '三層,二層' '一層,地下四層' '四層,機械房'\n '騎樓,一層,二層,三層,四層,五層,六層,七層,八層' '見使用執照' '十七層,夾層' '防空避難室,見其他登記事項']\n--------------------------------------------------\n編號:\n nan: False\n['RPSOMLLKJHMFFEA57DA' 'RPVNMLLLLHLFFAA57DA' 'RPPNMLLKJIKFFDA86CA' ...\n 'RPPNMLKKQHKFFDA67CD' 'RPUPMLSKKIJFFEA48DA' 'RPPQMLLLJIMFFDA47CA']\n--------------------------------------------------\n總樓層數:\n nan: True\n[nan '四層' '二十一層' '十二層' '五層' '三層' '十三層' '十四層' '六層' '十層' '九層' '八層' '七層'\n '十九層' '十五層' '十一層' '十七層' '二層' '二十層' '十六層' '二十七層' '二十三層' '十八層' '三十三層'\n '二十四層' '二十六層' '一層' '二十二層' '四十二層' '二十五層' '三十五層' '二十九層' '二十八層' '三十層' '三十八層'\n '三十一層']\n--------------------------------------------------\n車位類別:\n nan: True\n[nan '坡道平面' '升降平面' '升降機械' '坡道機械' '塔式車位' '一樓平面' '其他']\n--------------------------------------------------\n都市土地使用分區:\n nan: True\n['其他' '住' '商' '工' '農' nan]\n--------------------------------------------------\n鄉鎮市區:\n nan: False\n['士林區' '中正區' '南港區' '文山區' '中山區' '北投區' '松山區' '內湖區' '大安區' '大同區' '信義區' '萬華區']\n--------------------------------------------------\n非都市土地使用分區:\n nan: True\n[nan '住宅區' '特定專用區']\n--------------------------------------------------\nlocation_type:\n nan: True\n['GEOMETRIC_CENTER' 'ROOFTOP' 'RANGE_INTERPOLATED' 'APPROXIMATE' nan]\n--------------------------------------------------\nlow_use_electricity:\n nan: True\n[nan '8.57%' '16.59%' '4.48%' '10.96%' '5.66%' '36.87%' '24.65%' '3.44%'\n '5.10%' '4.35%' '5.91%' '4.91%' '5.02%' '9.17%' '4.07%' '6.59%' '4.99%'\n '6.70%' '4.08%' '4.30%' '5.24%' '3.06%' '6.05%' '10.87%' '15.14%' '8.26%'\n '4.56%' '4.32%' '8.18%' '8.73%' '8.02%' '10.97%' '7.09%' '10.13%' '8.50%'\n '3.34%' '3.20%' '3.70%' '5.09%' '3.27%' '6.78%' '7.47%' '17.19%' '4.03%'\n '4.24%' '8.17%' '5.00%' '9.56%' '5.13%' '15.99%' '6.66%' '4.58%' '6.99%'\n '4.70%' '4.02%' '11.18%' '10.53%' '8.31%' '7.51%' '3.84%' '7.98%' '6.65%'\n '3.89%' '6.15%' '4.18%' '4.17%' '3.52%' '3.53%' '4.15%' '12.90%' '10.82%'\n '5.12%' '6.54%' '12.88%' '4.44%' '10.20%' '5.89%' '3.73%' '4.37%'\n '11.76%' '9.18%' '7.49%' '4.64%' '3.09%' '8.06%' '10.68%' '6.09%' '4.52%'\n '4.09%' '7.53%' '7.46%' '6.82%' '5.81%' '4.86%' '3.82%' '3.56%' '3.49%'\n '4.51%' '17.76%' '9.13%' '4.73%' '10.30%' '10.93%' '3.28%' '6.64%'\n '4.54%' '7.41%' '2.78%' '29.20%' '11.07%' '9.88%' '4.81%' '3.90%' '8.53%'\n '7.71%' '3.42%' '5.34%' '4.98%' '4.47%' '4.87%' '4.71%' '5.53%' '9.66%'\n '17.72%' '6.23%' '4.28%' '12.34%' '11.65%' '8.93%' '8.52%' '5.03%'\n '4.01%' '4.89%' '10.83%' '6.42%' '12.39%' '5.84%' '8.22%' '7.27%'\n '12.41%' '5.96%' '11.50%' '10.26%' '4.67%' '3.14%' '25.46%' '6.36%'\n '8.66%' '13.60%' '5.26%' '7.95%' '5.62%' '4.25%' '4.16%' '8.94%' '9.51%'\n '9.89%' '11.87%' '2.53%' '3.59%' '18.76%' '8.12%' '4.72%' '6.56%' '3.36%'\n '6.07%' '4.41%' '7.60%' '5.07%' '4.96%' '13.58%' '6.88%' '12.10%' '8.79%'\n '6.93%' '8.21%' '7.30%' '2.90%' '9.32%' '8.82%' '6.98%' '9.08%' '4.69%'\n '5.33%' '7.34%' '8.27%' '5.04%' '4.27%' '10.54%' '4.75%' '7.80%' '7.45%'\n '5.75%' '15.01%' '8.00%' '7.79%' '6.27%' '5.44%' '10.98%' '6.79%' '4.23%'\n '3.66%' '10.33%' '9.74%' '5.15%' '8.70%' '7.00%' '7.10%' '5.20%' '1.64%'\n '9.78%' '7.40%' '5.61%' '5.90%' '9.49%' '5.27%' '14.76%' '3.67%' '6.92%'\n '8.47%' '3.81%' '4.68%' '6.89%' '6.85%' '10.18%' '5.79%' '3.57%' '7.70%'\n '4.94%' '4.46%' '10.40%' '6.97%' '6.43%' '9.79%' '7.03%' '4.88%' '5.43%'\n '11.49%' '3.62%' '5.76%' '3.29%' '6.16%' '6.58%' '7.28%' '6.50%' '3.21%'\n '7.07%' '5.69%' '4.50%' '6.32%' '5.25%' '9.28%' '7.69%' '5.36%' '5.67%'\n '8.84%' '3.10%' '4.85%' '4.22%' '6.17%' '7.75%' '10.19%' '6.35%' '4.76%'\n '5.92%' '18.10%' '4.10%' '4.11%' '6.30%' '8.86%' '5.88%' '6.24%' '6.39%'\n '5.65%' '7.06%' '9.31%' '4.63%' '11.52%' '3.99%' '6.86%' '8.48%' '14.98%'\n '6.62%' '6.19%' '28.71%' '5.58%' '6.76%' '8.83%' '6.47%' '10.85%' '6.60%'\n '5.11%' '7.65%' '2.92%' '5.05%' '14.02%' '3.58%' '3.37%' '8.30%' '4.90%'\n '13.52%' '10.09%' '6.10%' '3.76%' '4.38%' '9.02%' '9.35%' '6.69%' '5.51%'\n '3.17%' '6.83%' '6.52%' '9.53%' '2.33%' '6.71%' '8.81%' '4.06%' '4.97%'\n '7.55%' '4.40%' '5.54%' '8.28%' '13.18%' '6.11%' '8.89%' '5.63%' '4.05%'\n '4.95%' '9.57%' '4.43%' '7.97%' '11.11%' '10.55%' '3.72%' '3.96%' '4.80%'\n '10.22%' '8.32%' '17.56%' '4.36%' '3.69%' '4.20%' '9.63%' '16.67%'\n '28.44%' '8.85%' '13.92%' '8.19%']\n--------------------------------------------------\nnearest_tarin_station:\n nan: True\n['小碧潭站' '小南門站' '南港軟體園區站' '景美站' '雙連站' '萬芳醫院站' '西門站' '台北小巨蛋站' '文德站' '國父紀念館站'\n '大安森林公園站' '芝山站' '木柵站' '北門站' '唭哩岸站' '南港站' '明德站' '新北投站' '葫洲站' '忠孝敦化站'\n '後山埤站' '內湖站' '台北101/世貿中心站' '東湖站' '石牌站' '關渡站' '麟光站' '大橋頭站' '松山站' '港墘站'\n '永春站' '大湖公園站' '臺北車站' '圓山站' '昆陽站' '科技大樓站' '中山國小站' '劍南路站' '中山國中站' '信義安和站'\n '忠孝新生站' '南港展覽館站' '南京三民站' '劍潭站' '善導寺站' '行天宮站' '大安站' '動物園站' '市政府站' '公館站'\n '奇岩站' '臺電大樓站' '古亭站' '辛亥站' '民權西路站' '萬隆站' '西湖站' '松山機場站' '大直站' '象山站' '復興崗站'\n '松江南京站' '三重站' '龍山寺站' '六張犁站' '中山站' '忠孝復興站' '南京復興站' '大坪林站' '中正紀念堂站' '萬芳社區站'\n '頂溪站' '忠義站' '三重國小站' '士林站' '東門站' '七張站' '三和國中站' '北投站' '臺大醫院站' '永安市場站'\n '三民高中站' nan '蘆洲站' '先嗇宮' '迴龍站']\n--------------------------------------------------\n"
]
],
[
[
"#### 特徵選取(domain knowledge)",
"_____no_output_____"
]
],
[
[
"notnum_features.remove('備註')\nnotnum_features.remove('編號')\nnotnum_features.remove('移轉層次')",
"_____no_output_____"
]
],
[
[
"#### 類別型特徵缺失值處理",
"_____no_output_____"
]
],
[
[
"df_concat_pre['主要建材'] = df_concat['主要建材']\ndf_concat_pre['主要建材'][df_concat_pre['主要建材'].isnull()] = '其他'\ndf_concat_pre['主要建材'][df_concat_pre['主要建材']=='見其他登記事項'] = '其他'\ndf_concat_pre['主要建材'][df_concat_pre['主要建材']=='見使用執照'] = '其他'\ndf_concat_pre['主要建材'].unique()",
"_____no_output_____"
],
[
"df_concat_pre['主要用途'] = df_concat['主要用途']\ndf_concat_pre['主要用途'][df_concat_pre['主要用途'].isnull()] = '其他'\ndf_concat_pre['主要用途'][df_concat_pre['主要用途']=='見其他登記事項'] = '其他'\ndf_concat_pre['主要用途'][df_concat_pre['主要用途']=='見使用執照'] = '其他'\ndf_concat_pre['主要用途'].unique()",
"_____no_output_____"
],
[
"df_concat_pre['車位類別'] = df_concat['車位類別']\ndf_concat_pre['車位類別'][df_concat_pre['車位類別'].isnull()] = '其他'\ndf_concat_pre['車位類別'].unique()",
"_____no_output_____"
],
[
"df_concat_pre['都市土地使用分區'] = df_concat['都市土地使用分區']\ndf_concat_pre['都市土地使用分區'][df_concat_pre['都市土地使用分區'].isnull()] = '其他'\ndf_concat_pre['都市土地使用分區'].unique()",
"_____no_output_____"
],
[
"df_concat_pre['非都市土地使用分區'] = df_concat['非都市土地使用分區']\ndf_concat_pre['非都市土地使用分區'][df_concat_pre['非都市土地使用分區'].isnull()] = '其他'\ndf_concat_pre['非都市土地使用分區'].unique()",
"_____no_output_____"
],
[
"df_concat_pre['location_type'] = df_concat['location_type']\ndf_concat_pre['location_type'][df_concat_pre['location_type'].isnull()] = 'other'\ndf_concat_pre['location_type'].unique()",
"_____no_output_____"
],
[
"df_concat_pre['nearest_tarin_station'] = df_concat['nearest_tarin_station']\ndf_concat_pre['nearest_tarin_station'][df_concat_pre['nearest_tarin_station'].isnull()] = '無'\ndf_concat_pre['nearest_tarin_station'].unique()",
"_____no_output_____"
]
],
[
[
"#### 文字類別資料轉數字",
"_____no_output_____"
]
],
[
[
"import re \n\ndf_concat_pre['交易筆棟數_土地']= df_concat['交易筆棟數'].apply(lambda x:int(re.findall(r'\\d+',x)[0]))\ndf_concat_pre['交易筆棟數_建物']= df_concat['交易筆棟數'].apply(lambda x:int(re.findall(r'\\d+',x)[1]))\ndf_concat_pre['交易筆棟數_車位']= df_concat['交易筆棟數'].apply(lambda x:int(re.findall(r'\\d+',x)[2]))\nnum_features.extend(['交易筆棟數_土地', '交易筆棟數_建物', '交易筆棟數_車位']) ; notnum_features.remove('交易筆棟數')\n\ndisplay(df_concat_pre.loc[:,['交易筆棟數_土地','交易筆棟數_建物','交易筆棟數_車位']])\ndisplay(df_concat['交易筆棟數'])",
"_____no_output_____"
],
[
"df_concat_pre['總樓層數'] = df_concat['總樓層數']\ncn_in = {np.nan:np.nan, '四層':4, '二十一層':21, '十二層':12, '五層':5, '三層':3, '十三層':13, \n '十四層':14, '六層':6, '十層':10,'九層':9, '八層':8, '七層':7, '十九層':19, '十五層':15, \n '十一層':11, '十七層':17, '二層':2, '二十層':20, '十六層':16,'二十七層':27, '二十三層':23, \n '十八層':18, '三十三層':33, '二十四層':24, '二十六層':26, '一層':1, '二十二層':20, \n '四十二層':42, '二十五層':25, '三十五層':35, '二十九層':29, '二十八層':28, '三十層':30, \n '三十八層':38, '三十一層':31}\ndf_concat_pre['總樓層數'] = df_concat_pre['總樓層數'].apply(lambda x : cn_in[x])\ndf_concat_pre['總樓層數'] = df_concat_pre['總樓層數'].fillna(int(df_concat_pre['總樓層數'].mean()))\nnum_features.append('總樓層數') ; notnum_features.remove('總樓層數')\n\nprint(df_concat_pre['總樓層數'].unique())",
"[ 9. 4. 21. 12. 5. 3. 13. 14. 6. 10. 8. 7. 19. 15. 11. 17. 2. 20.\n 16. 27. 23. 18. 33. 24. 26. 1. 42. 25. 35. 29. 28. 30. 38. 31.]\n"
],
[
"df_concat_pre['low_use_electricity'] = df_concat['low_use_electricity'] \ndf_concat_pre['low_use_electricity'][df_concat_pre['low_use_electricity'].isnull()] = 'empty' \ndf_concat_pre['low_use_electricity'] = \\\ndf_concat_pre['low_use_electricity'].apply(lambda x:x.replace('%',''))\ndf_concat_pre['low_use_electricity'][df_concat_pre['low_use_electricity']=='empty'] = np.nan\ndf_concat_pre['low_use_electricity'] = \\\ndf_concat_pre['low_use_electricity'].apply(float)\ndf_concat_pre['low_use_electricity'] = \\\ndf_concat_pre['low_use_electricity'].fillna(df_concat_pre['low_use_electricity'].mean())\nnum_features.append('low_use_electricity') ; notnum_features.remove('low_use_electricity')\ndf_concat_pre['low_use_electricity']",
"_____no_output_____"
]
],
[
[
"#### 路段整理(文字類型)",
"_____no_output_____"
]
],
[
[
"pattern = r'(.+路)(.+段)|(.+路)|(.+街)|(.+道)|(.+段)'\ndf_concat_pre['土地區段位置/建物區段門牌'] = df_concat['土地區段位置/建物區段門牌']\ndf_concat_pre['土地區段位置/建物區段門牌'] = \\\ndf_concat_pre['土地區段位置/建物區段門牌'].apply(lambda x : re.sub(r'臺北市..區','',x))\ndf_concat_pre['土地區段位置/建物區段門牌'] = \\\ndf_concat_pre['土地區段位置/建物區段門牌'].apply(lambda x : re.sub(r'\\w+鄰','',x))\nfor i,location in enumerate(df_concat_pre['土地區段位置/建物區段門牌']):\n if re.search(pattern,location) == None:\n print(df_concat['土地區段位置/建物區段門牌'][i],i) \n df_concat_pre = df_concat_pre.drop(i)\n df_concat = df_concat.drop(i)\n df_train_Y = df_train_Y.drop(i)\n\ndf_concat = df_concat.reset_index(drop=True) # drop後記得要重製index\ndf_concat_pre = df_concat_pre.reset_index(drop=True)\ndf_train_Y = df_train_Y.reset_index(drop=True) ",
"臺北市北投區光明賂61~90號 5901\n臺北市中山區龍江陸415巷1~30號 8200\n臺北市南港區南港陸斷12巷1弄1~30號 44914\n臺北市中山區241~270號 49456\n臺北市內湖區151~180號 56322\n"
],
[
"df_concat_pre['土地區段位置/建物區段門牌'] = \\\ndf_concat_pre['土地區段位置/建物區段門牌'].apply(lambda x : re.search(pattern,x).group())\ndf_concat_pre['土地區段位置/建物區段門牌']",
"_____no_output_____"
]
],
[
[
"#### 將剩餘類別型態資料填入",
"_____no_output_____"
]
],
[
[
"for item in notnum_features: \n if item not in df_concat_pre.columns:\n print(item)\n df_concat_pre[item] = df_concat[item]",
"交易標的\n建物型態\n建物現況格局-隔間\n有無管理組織\n鄉鎮市區\n"
]
],
[
[
"# 處理離群值",
"_____no_output_____"
]
],
[
[
"for i,f in enumerate(num_features):\n ori_series = df_concat_pre[f].copy()\n qt1 = ori_series.quantile(q=0.25)\n qt3 = ori_series.quantile(q=0.75)\n iqr = qt3-qt1\n ori_series[ori_series>(qt3 + 1.5*iqr)] = qt3 + 1.5*iqr\n ori_series[ori_series<(qt1 - 1.5*iqr)] = qt1 - 1.5*iqr\n #--------------------------------------------------------#\n plt.title(f) \n plt.boxplot(ori_series)\n plt.show()\n ",
"_____no_output_____"
]
],
[
[
"#### 去除偏態",
"_____no_output_____"
]
],
[
[
"df_concat_pre['交易筆棟數_建物'].describe()",
"_____no_output_____"
],
[
"plt.hist(df_concat_pre['交易筆棟數_建物'])\nplt.show()",
"_____no_output_____"
],
[
"df_concat_pre['車位總價(元)'] = np.log1p(df_concat_pre['車位總價(元)'] )\ndf_concat_pre['車位移轉總面積(平方公尺)'] = np.log1p(df_concat_pre['車位移轉總面積(平方公尺)'] )\ndf_concat_pre['交易筆棟數_建物'] = np.log1p(df_concat_pre['交易筆棟數_建物'])\ntrans = ['車位總價(元)','車位移轉總面積(平方公尺)','交易筆棟數_建物']\noutliers = [f for f in num_features if f not in trans]\noutliers",
"_____no_output_____"
],
[
"for i,f in enumerate(outliers):\n ori_series = df_concat_pre[f]\n qt1 = ori_series.quantile(q=0.25)\n qt3 = ori_series.quantile(q=0.75)\n iqr = qt3-qt1\n ori_series[ori_series>(qt3 + 1.5*iqr)] = qt3 + 1.5*iqr\n ori_series[ori_series<(qt1 - 1.5*iqr)] = qt1 - 1.5*iqr\n df_concat_pre[f] = ori_series\n #--------------------------------------------------------#\n plt.title(f) \n plt.boxplot(df_concat_pre[f])\n plt.show()",
"_____no_output_____"
]
],
[
[
"# Normalization",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import StandardScaler\n\n# split data\nsplit_point = len(df_train_Y)\n# loc include last one\ndf_train_norm = df_concat_pre.loc[:(split_point-1),num_features] # only num_feature need to be normalized\ndf_test_norm = df_concat_pre.loc[split_point:,num_features] # only num_feature need to be normalizedd\ns = StandardScaler()\nscaler = StandardScaler().fit(df_train_norm)\narray_train_norm = scaler.transform(df_train_norm)\narray_test_norm = scaler.transform(df_test_norm)\narray_concat = np.concatenate([array_train_norm,array_test_norm])\ndf_concat_pre_num = pd.DataFrame(array_concat,columns=num_features)\ndf_concat_pre = pd.concat([df_concat_pre_num,df_concat_pre.loc[:,notnum_features]],axis=1)\ndf_concat_pre.head()",
"_____no_output_____"
]
],
[
[
"# 檢查表格",
"_____no_output_____"
]
],
[
[
"print('Is there any null in table:',df_concat_pre.isnull().any().any()) #檢查是否有空值\nprint(f'df_concat_pre.columns : {len(df_concat_pre.columns)}')\nprint(f'num_features: {len(num_features)}, notnum_features: {len(notnum_features)}')",
"Is there any null in table: False\ndf_concat_pre.columns : 35\nnum_features: 22, notnum_features: 13\n"
]
],
[
[
"# label encoding",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import LabelEncoder\n\ndf_concat_pre_le = df_concat_pre.copy()\nfor c in notnum_features:\n df_concat_pre_le[c] = LabelEncoder().fit_transform(df_concat_pre_le[c])\nprint('Is there any null in table:',df_concat_pre_le.isnull().any().any()) #檢查是否有空值\ndf_concat_pre_le",
"Is there any null in table: False\n"
]
],
[
[
"# 將整理完的表格存出去",
"_____no_output_____"
]
],
[
[
"df_train_Y = pd.DataFrame(df_train_Y,columns=['單價(元/平方公尺)'])\n\n# split data\nsplit_point = len(df_train_Y)\ndf_train_clean = df_concat_pre_le[:split_point]\ndf_test_clean = df_concat_pre_le[split_point:]\ndf_train_clean = pd.concat([df_train_clean,df_train_Y],axis = 1)\ndf_train_clean",
"_____no_output_____"
],
[
"df_train_clean.to_csv('./data/output/house_train_clean.csv', index=False)\ndf_test_clean.to_csv('./data/output/house_test_clean.csv', index=False)",
"_____no_output_____"
],
[
"read_train_df = pd.read_csv('./data/output/house_train_clean.csv')\nprint(len(read_train_df))\nprint(len(read_train_df.columns))\nprint(read_train_df.columns)\ndisplay(read_train_df.iloc[:20])",
"69165\n36\nIndex(['土地移轉總面積(平方公尺)', '建物現況格局-廳', '建物現況格局-房', '建物現況格局-衛', '建物移轉總面積(平方公尺)',\n '總價(元)', '車位移轉總面積(平方公尺)', '車位總價(元)', 'num_of_bus_stations_in_100m',\n 'income_avg', 'income_var', 'nearest_tarin_station_distance', 'lat',\n 'lng', '建築完成年', '交易年', '交易月', '交易筆棟數_土地', '交易筆棟數_建物', '交易筆棟數_車位',\n '總樓層數', 'low_use_electricity', '主要建材', '主要用途', '交易標的', '土地區段位置/建物區段門牌',\n '建物型態', '建物現況格局-隔間', '有無管理組織', '車位類別', '都市土地使用分區', '鄉鎮市區', '非都市土地使用分區',\n 'location_type', 'nearest_tarin_station', '單價(元/平方公尺)'],\n dtype='object')\n"
]
],
[
[
"# one-hot encoding",
"_____no_output_____"
]
],
[
[
"df_concat_pre_onehot= pd.get_dummies(df_concat_pre)\ndf_concat_pre_onehot",
"_____no_output_____"
],
[
"split_point = len(df_train_Y)\ndf_train_clean_onehot = df_concat_pre_onehot[:split_point]\ndf_test_clean_onehot = df_concat_pre_onehot[split_point:]\ndf_train_clean_onehot = pd.concat([df_train_clean_onehot,df_train_Y],axis = 1)\ndf_train_clean_onehot",
"_____no_output_____"
],
[
"df_train_clean_onehot.to_csv('./data/output/house_train_clean_onehot.csv', index=False)\ndf_test_clean_onehot.to_csv('./data/output/house_test_clean_onehot.csv', index=False)",
"_____no_output_____"
],
[
"read_train_onehot_df = pd.read_csv('./data/output/house_train_clean_onehot.csv')\nprint(len(read_train_onehot_df))\nprint(len(read_train_onehot_df.columns))\nprint(read_train_onehot_df.columns)\ndisplay(read_train_onehot_df.iloc[:20])",
"69165\n1659\nIndex(['土地移轉總面積(平方公尺)', '建物現況格局-廳', '建物現況格局-房', '建物現況格局-衛', '建物移轉總面積(平方公尺)',\n '總價(元)', '車位移轉總面積(平方公尺)', '車位總價(元)', 'num_of_bus_stations_in_100m',\n 'income_avg',\n ...\n 'nearest_tarin_station_西門站', 'nearest_tarin_station_象山站',\n 'nearest_tarin_station_辛亥站', 'nearest_tarin_station_迴龍站',\n 'nearest_tarin_station_關渡站', 'nearest_tarin_station_雙連站',\n 'nearest_tarin_station_頂溪站', 'nearest_tarin_station_麟光站',\n 'nearest_tarin_station_龍山寺站', '單價(元/平方公尺)'],\n dtype='object', length=1659)\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecb9e9fb96bce3700601db0d68dfe9f79c8d4424 | 29,396 | ipynb | Jupyter Notebook | _ipynb_checkpoints/Section2.2-checkpoint.ipynb | simoto4/discover-r | 6dbabc662b6cb0e6c284a909b219d09490f5c8a9 | [
"CC-BY-4.0"
] | null | null | null | _ipynb_checkpoints/Section2.2-checkpoint.ipynb | simoto4/discover-r | 6dbabc662b6cb0e6c284a909b219d09490f5c8a9 | [
"CC-BY-4.0"
] | null | null | null | _ipynb_checkpoints/Section2.2-checkpoint.ipynb | simoto4/discover-r | 6dbabc662b6cb0e6c284a909b219d09490f5c8a9 | [
"CC-BY-4.0"
] | 2 | 2021-07-06T18:31:40.000Z | 2022-02-08T15:16:10.000Z | 27.810785 | 1,010 | 0.502755 | [
[
[
"## Subsetting Vectors\n\nIf we want to subset (or extract) one or several values from a vector, we must provide one or several indices in square brackets. For this example, we will use the state data, which is built into R and includes data related to the 50 states of the U.S.A. Type ?state to see the included datasets. state.name is a built in vector in R of all U.S. states:",
"_____no_output_____"
]
],
[
[
"state.name",
"_____no_output_____"
],
[
"state.name[1] #first element\nstate.name[13] #13th element",
"_____no_output_____"
]
],
[
[
"What would happen if your index was out of bounds?",
"_____no_output_____"
]
],
[
[
"state.name[51]",
"_____no_output_____"
]
],
[
[
"You can use the : colon to create a vector of consecutive numbers.",
"_____no_output_____"
]
],
[
[
"state.name[1:5] #first 5 elements\nstate.name[6:20] #elements 6-20 ",
"_____no_output_____"
]
],
[
[
"If the numbers are not consecutive, you must use the c() function:",
"_____no_output_____"
]
],
[
[
"state.name[c(1, 10, 20)]",
"_____no_output_____"
]
],
[
[
"We can also repeat the indices to create an object with more elements than the original one:",
"_____no_output_____"
]
],
[
[
"state.name[c(1, 2, 3, 2, 1, 3)]",
"_____no_output_____"
]
],
[
[
"> NOTE : R indices start at 1. Programming languages like Fortran, MATLAB, Julia, and R start counting at 1, because that’s what human beings typically do. Languages in the C family (including C++, Java, Perl, and Python) count from 0 because that’s simpler for computers to do.",
"_____no_output_____"
],
[
"## Conditional subsetting\nAnother common way of subsetting is by using a logical vector. TRUE will select the element with the same index, while FALSE will not:\n\nThe example below is greating a vector that contains the first 5 states (\"Alabama\",\"Alaska\",\"Arizona\",\"Arkansas\",\"California\"). The line below has changed the logical",
"_____no_output_____"
]
],
[
[
"five_states <- state.name[1:5]\nfive_states[c(TRUE, FALSE, TRUE, FALSE, TRUE)]",
"_____no_output_____"
]
],
[
[
"Typically, these logical vectors are not typed by hand, but are the output of other functions or logical tests. state.area is a vector of state areas in square miles. We can use the < operator to return a logical vector with TRUE for the indices that meet the condition:",
"_____no_output_____"
]
],
[
[
"state.area < 10000",
"_____no_output_____"
],
[
"state.area[state.area < 10000]",
"_____no_output_____"
]
],
[
[
"The first expression gives us a logical vector of length 50, where TRUE represents those states with areas less than 10,000 square miles. The second expression subsets state.name to include only those names where the value is TRUE.\n\nYou can also specify character values. state.region gives the region that each state belongs to:",
"_____no_output_____"
]
],
[
[
"state.region == \"Northeast\"",
"_____no_output_____"
],
[
"state.name[state.region == \"Northeast\"]",
"_____no_output_____"
]
],
[
[
"Again, a TRUE/FALSE index of all 50 states where the region is the Northeast, followed by a subset of state.name to return only those TRUE values.\n\nSometimes you need to do multiple logical tests (think Boolean logic). You can combine multiple tests using | (at least one of the conditions is true, OR) or & (both conditions are true, AND). Use help(Logic) to read the help file.",
"_____no_output_____"
]
],
[
[
"state.name[state.area < 10000 | state.region == \"Northeast\"]",
"_____no_output_____"
],
[
"state.name[state.area < 10000 & state.region == \"Northeast\"]",
"_____no_output_____"
]
],
[
[
"The first result includes both states with fewer than 10,000 sq. mi. and all states in the Northeast. New York, Pennsylvania, Delaware and Maine have areas with greater than 10,000 square miles, but are in the Northeastern U.S. Hawaii is not in the Northeast, but it has fewer than 10,000 square miles. The second result includes only states that are in the Northeast and have fewer than 10,000 sq. mi.\n\nR contains a number of operators you can use to compare values. Use help(Comparison) to read the R help file. Note that two equal signs (==) are used for evaluating equality (because one equals sign (=) is used for assigning variables).\n\nA common task is to search for certain strings in a vector. One could use the “or” operator | to test for equality to multiple values, but this can quickly become tedious. The function %in% allows you to test if any of the elements of a search vector are found:",
"_____no_output_____"
]
],
[
[
"west_coast <- c(\"California\", \"Oregon\", \"Washington\")\nstate.name[state.name == \"California\" | state.name == \"Oregon\" | state.name == \"Washington\"]",
"_____no_output_____"
],
[
"state.name %in% west_coast",
"_____no_output_____"
],
[
"state.name[state.name %in% west_coast]",
"_____no_output_____"
]
],
[
[
"## Missing Data\n\nAs R was designed to analyze datasets, it includes the concept of missing data (which is uncommon in other programming languages). Missing data are represented in vectors as NA. R functions have special actions when they encounter NA.\n\nWhen doing operations on numbers, most functions will return NA if the data you are working with include missing values. This feature makes it harder to overlook the cases where you are dealing with missing data. As we saw above, you can add the argument na.rm=TRUE to calculate the result while ignoring the missing values.",
"_____no_output_____"
]
],
[
[
"rooms <- c(2, 1, 1, NA, 4)\nmean(rooms)",
"_____no_output_____"
],
[
"max(rooms)",
"_____no_output_____"
],
[
"mean(rooms, na.rm = TRUE)",
"_____no_output_____"
],
[
"max(rooms, na.rm = TRUE)",
"_____no_output_____"
]
],
[
[
"If your data include missing values, you may want to become familiar with the functions is.na(), na.omit(), and complete.cases(). See below for examples.",
"_____no_output_____"
]
],
[
[
"## Use any() to check if any values are missing\nany(is.na(rooms))",
"_____no_output_____"
],
[
"## Use table() to tell you how many are missing vs. not missing\ntable(is.na(rooms))",
"_____no_output_____"
],
[
"## Identify those elements that are not missing values.\ncomplete.cases(rooms)",
"_____no_output_____"
],
[
"## Identify those elements that are missing values.\nis.na(rooms)",
"_____no_output_____"
],
[
"## Extract those elements that are not missing values.\nrooms[complete.cases(rooms)]",
"_____no_output_____"
]
],
[
[
"You can also use !is.na(rooms), which is exactly the same as complete.cases(rooms). The exclamation mark indicates logical negation.",
"_____no_output_____"
]
],
[
[
"!c(TRUE, FALSE)",
"_____no_output_____"
]
],
[
[
"How you deal with missing data in your analysis is a decision you will have to make–do you remove it entirely? Do you replace it with zeros? That will depend on your own methodological questions.",
"_____no_output_____"
],
[
"## Key Points\n - Use the assignment operator <- to assign values to objects. You can now manipulate that object in R\n - R contains a number of functions you use to do something with your data. Functions automate more complicated sets of commands. Many functions are predefined, or can be made available by importing R packages\n - A vector is a sequence of elements of the same type. All data in a vector must be of the same type–character, numeric (or double), integer, and logical. Create vectors with c(). Use [ ] to subset values from vectors.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecb9efe2f9fbcf49a012311ae60ca37eda191a94 | 514,619 | ipynb | Jupyter Notebook | CS421_LendingClub+ RF.ipynb | kanewu-smu/CS421-assignment | d07042b07c3923b4caa7a25c9ac141e558138472 | [
"MIT"
] | null | null | null | CS421_LendingClub+ RF.ipynb | kanewu-smu/CS421-assignment | d07042b07c3923b4caa7a25c9ac141e558138472 | [
"MIT"
] | null | null | null | CS421_LendingClub+ RF.ipynb | kanewu-smu/CS421-assignment | d07042b07c3923b4caa7a25c9ac141e558138472 | [
"MIT"
] | null | null | null | 168.341184 | 181,519 | 0.787544 | [
[
[
"## Setup",
"_____no_output_____"
]
],
[
[
"!pip install statsmodels monthdelta",
"Requirement already satisfied: statsmodels in c:\\users\\vridheep.2018\\anaconda3\\lib\\site-packages (0.13.0)\nRequirement already satisfied: monthdelta in c:\\users\\vridheep.2018\\anaconda3\\lib\\site-packages (0.9.1)\nRequirement already satisfied: scipy>=1.3 in c:\\users\\vridheep.2018\\anaconda3\\lib\\site-packages (from statsmodels) (1.6.0)\nRequirement already satisfied: numpy>=1.17 in c:\\users\\vridheep.2018\\anaconda3\\lib\\site-packages (from statsmodels) (1.19.2)\nRequirement already satisfied: patsy>=0.5.2 in c:\\users\\vridheep.2018\\anaconda3\\lib\\site-packages (from statsmodels) (0.5.2)\nRequirement already satisfied: pandas>=0.25 in c:\\users\\vridheep.2018\\anaconda3\\lib\\site-packages (from statsmodels) (1.1.5)\nRequirement already satisfied: six in c:\\users\\vridheep.2018\\anaconda3\\lib\\site-packages (from patsy>=0.5.2->statsmodels) (1.15.0)\nRequirement already satisfied: pytz>=2017.2 in c:\\users\\vridheep.2018\\anaconda3\\lib\\site-packages (from pandas>=0.25->statsmodels) (2018.5)\nRequirement already satisfied: python-dateutil>=2.7.3 in c:\\users\\vridheep.2018\\anaconda3\\lib\\site-packages (from pandas>=0.25->statsmodels) (2.7.3)\n"
],
[
"import numpy as np\nimport pandas as pd\nimport math\nimport statistics\nfrom datetime import datetime as dt\nimport sklearn\nfrom matplotlib import pyplot as plt\nimport seaborn as sns\nimport statsmodels.api as sm\n\n%matplotlib inline\n\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"## Import raw data",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv(\"loans_2007.csv\")",
"_____no_output_____"
]
],
[
[
"## Data cleaning",
"_____no_output_____"
],
[
"### Explore raw data",
"_____no_output_____"
],
[
"#### Data quantity",
"_____no_output_____"
]
],
[
[
"print(\"There are {} rows and {} columns\".format(data.shape[0], data.shape[1]))",
"There are 42538 rows and 52 columns\n"
],
[
"print(\"Number of unique values by column:\")\ndisplay(data.nunique())",
"Number of unique values by column:\n"
]
],
[
[
"#### Data quality",
"_____no_output_____"
]
],
[
[
"print(\"Columns with nulls:\")\nnum_nulls = data.isnull().sum()\ndisplay(num_nulls[num_nulls > 0])",
"Columns with nulls:\n"
]
],
[
[
"#### Summary statistics",
"_____no_output_____"
]
],
[
[
"pd.set_option('display.float_format', lambda x: f\"{x:.2f}\" if x % 1 != 0 else f\"{int(x):,}\")\ndata.describe()",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
]
],
[
[
"#### Distribution of target variable: loan_status",
"_____no_output_____"
]
],
[
[
"# Count plot of loan status\nplt.figure(figsize = (20, 6))\nplt.title(\"Count plot of loan_status categories\")\nax = sns.countplot(y = 'loan_status', data = data, orient = \"v\")",
"_____no_output_____"
]
],
[
[
"### Data cleaning",
"_____no_output_____"
],
[
"#### Keep only rows with loan_status \"Fully Paid\" (0) or \"Charged Off (1)",
"_____no_output_____"
]
],
[
[
"# Drop rows where loan_status is not \"Fully Paid\" or \"Charged Off\"\nold_len = len(data)\ndata = data[data.loan_status.isin([\"Fully Paid\", \"Charged Off\"])]\nprint(\"Original: {} rows. Dropped: {} rows. Remaining: {} rows.\".format(\n old_len, old_len - len(data), len(data)))\n\n# Convert loan_status to binary variable: default = 1 if loan_status = \"Charged Off\", else default = 0\ndata[\"default\"] = [\n 0 if status == \"Fully Paid\" else 1 for status in data.loan_status\n]\ndata.drop(\"loan_status\", axis=1, inplace=True)\ndata.head()",
"Original: 42538 rows. Dropped: 3768 rows. Remaining: 38770 rows.\n"
]
],
[
[
"#### Drop duplicate rows",
"_____no_output_____"
]
],
[
[
"# Drop duplicates\nold_len = len(data)\ndata.drop_duplicates(data.columns[:-3], keep=\"last\", inplace=True)\nprint(\"Original: {} rows. Dropped: {} rows. Remaining: {} rows.\".format(\n old_len, old_len - len(data), len(data)))",
"Original: 38770 rows. Dropped: 0 rows. Remaining: 38770 rows.\n"
]
],
[
[
"#### Drop rows with NA values",
"_____no_output_____"
]
],
[
[
"data[data.isnull().any(axis=1)]",
"_____no_output_____"
],
[
"# Drop duplicates\nold_len = len(data)\ndata.dropna(how = \"any\", inplace = True)\nprint(\"Original: {} rows. Dropped: {} rows. Remaining: {} rows.\".format(\n old_len, old_len - len(data), len(data)))",
"Original: 38770 rows. Dropped: 3222 rows. Remaining: 35548 rows.\n"
],
[
"data.head()",
"_____no_output_____"
]
],
[
[
"#### Drop columns that contain only 1 unique value\n\nThese columns do not add any information to each observation",
"_____no_output_____"
]
],
[
[
"# Drop columns with only 1 unique value\nold_len = len(data.columns)\nnum_unique = data.nunique()\ndata.drop(num_unique[num_unique <= 1].index, axis=1, inplace=True)\n\nprint(\n \"Original: {} columns. Dropped: {} columns. Remaining: {} columns.\".format(\n old_len, old_len - len(data.columns), len(data.columns)))\n\nprint(\"Dropped columns:\")\nfor col in num_unique[num_unique <= 1].index:\n print(col)\n",
"Original: 52 columns. Dropped: 11 columns. Remaining: 41 columns.\nDropped columns:\npymnt_plan\ninitial_list_status\nout_prncp\nout_prncp_inv\ncollections_12_mths_ex_med\npolicy_code\napplication_type\nacc_now_delinq\nchargeoff_within_12_mths\ndelinq_amnt\ntax_liens\n"
]
],
[
[
"#### Drop redundant or non-useful columns\n\nCertain columns contain information that are not useful for prediction, or redundant information that has been fully captured by another column.\n\n* `id`: arbitrary number assigned by Lending Club\n* `member_id`: same as `id`\n* `emp_title`: highly unstructured text data, not useful unless significant cleaning is performed\n* `title`: same as `emp_title`\n* `zip_code`: redundant since the `addr_state` column already captures all geographical information revealed by the first 3 digits of `zip_code`",
"_____no_output_____"
]
],
[
[
"# Drop redundant or non-useful columns\ndrop_cols = [\"id\", \"member_id\", \"emp_title\", \"title\", \"zip_code\"]\nold_len = len(data.columns)\ndata.drop(drop_cols, axis = 1, inplace = True)\nprint(\n \"Original: {} columns. Dropped: {} columns. Remaining: {} columns.\".format(\n old_len, old_len - len(data.columns), len(data.columns)))",
"Original: 41 columns. Dropped: 5 columns. Remaining: 36 columns.\n"
]
],
[
[
"#### Drop columns which contain information not available at application\n\nThis model aims to predict, at the point of loan application, whether a borrower would eventually default. Certain information would not be available at the point of loan application and may introduce lookahead bias and/or cause overfitting. Columns with such information are listed below, and will be removed.\n\n* `funded_amnt` and `funded_amnt_inv`: only known after the loan has already been funded\n* `total_pymnt` and `total_pymnt_inv`: only known after the loan has started to be paid off\n* `total_rec_prncp`, `total_rec_int`, and `total_rec_late_fee`: only known after the loan has started to be paid off\n* `recoveries` and `collection_recovery_fee`: only known after the loan has defaulted\n* `last_pymnt_d` and `last_pymnt_amnt`: only known after the loan has started to be paid off\n* `last_credit_pull_d`: only known after the loan has already been funded\n* `grade` and `sub_grade`: assigned by Lending Club after credit scoring, but not available at the point of application\n* `int_rate`: depends on `sub_grade`\n* `installment`: depends on `int_rate`\n\nOne particular column, `issue_d`, also contains information not available at application time (issue date is only known after funding has completed). However, according to [Lending Club](https://help.lendingclub.com/hc/en-us/articles/215492738-How-long-does-it-take-to-get-approved-), an average application takes around 7 days to be approved and funded. Thus the deviation between issue date and application date is likely to be small. Instead of removing the column, we can thus use `issue_d` as an approximate for time of application, which might contain useful information.",
"_____no_output_____"
]
],
[
[
"data[\"issue_d\"].astype('str')",
"_____no_output_____"
],
[
"# Drop columns with information not available at origination\ndrop_cols = [\n \"funded_amnt\", \"funded_amnt_inv\", \"total_pymnt\",\n \"total_pymnt_inv\", \"total_rec_prncp\", \"total_rec_int\",\n \"total_rec_late_fee\", \"recoveries\", \"collection_recovery_fee\",\n \"last_pymnt_d\", \"last_pymnt_amnt\", \"last_credit_pull_d\", \n \"grade\", \"sub_grade\", \"int_rate\", \"installment\"\n]\n\nold_len = len(data.columns)\ndata.drop(drop_cols, axis=1, inplace=True)\nprint(\n \"Original: {} columns. Dropped: {} columns. Remaining: {} columns.\".format(\n old_len, old_len - len(data.columns), len(data.columns)))\n\n\n# Use issue date as proxy for application time\ndata.rename({\"issue_d\": \"app_time\"}, axis = 1, inplace = True)\ndata[\"app_time\"] = pd.to_datetime(data.app_time.astype(str), format = \"%b-%Y\")",
"Original: 36 columns. Dropped: 16 columns. Remaining: 20 columns.\n"
]
],
[
[
"#### Re-format numeric columns\n\nSome numeric columns, e.g `term`, `revol_util` are formatted as text, and need to be re-formatted to float or integer type. Column `empl_length` contains inherently numeric data but is treated as categorical, thus we re-convert it to numeric type. ",
"_____no_output_____"
]
],
[
[
"# Re-format numeric columns\ndata.term = [int(str(term).strip().split(\" \")[0]) for term in data.term]\ndata.revol_util = [float(str(util[:-1])) for util in data.revol_util]\n\n# Map employment length to integers: < 1 year is mapped to 0, >= 10 is mapped to 10\ndata[\"emp_length\"] = [\"0 year\" if length == \"< 1 year\" \n else \"10 years\" if length == \"10+ years\"\n else length for length in data.emp_length]\n\ndata[\"emp_length\"] = [int(str(length).split(\" \")[0]) for length in data.emp_length]",
"_____no_output_____"
]
],
[
[
"## Data visualization",
"_____no_output_____"
],
[
"#### Univariate distribution of numeric columns\n\nObservations:\n* Many numeric columns appear to be right-skewed or resemble a lognormal distribution, e.g `loan_amnt`, `emp_length`, `open_acc`, `total_acc`, `revol_bal`, etc.\n* Some columns may potentially be highly correlated, e.g: `open_acc` and `total_acc`. \n* `Annual_income` appears to contain some extreme right ouliers. ",
"_____no_output_____"
]
],
[
[
"features = data._get_numeric_data().columns\n\nfor i in range(len(features)):\n if i % 4 == 0:\n fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(15, 5))\n ax1.hist(data[features[i]], bins = 50, rwidth = 0.5, label=features[i])\n ax1.legend(loc = \"best\")\n elif i % 4 == 1:\n ax2.hist(data[features[i]], bins = 50, rwidth = 0.5, label=features[i])\n ax2.legend(loc = \"best\")\n elif i % 4 == 2:\n ax3.hist(data[features[i]], bins = 50, rwidth = 0.5, label=features[i])\n ax3.legend(loc = \"best\")\n elif i % 4 == 3:\n ax4.hist(data[features[i]], bins = 50, rwidth = 0.5, label=features[i])\n ax4.legend(loc = \"best\")\n \n \nplt.show()",
"_____no_output_____"
]
],
[
[
"Looking at annual income without the outliers (the top 1% and bottom 1% is winsorized), the data appears to resemble a lognormal distribution as well.",
"_____no_output_____"
]
],
[
[
"from scipy.stats.mstats import winsorize\nincome_winsorized = winsorize(data.annual_inc, limits = (0.01, 0.01), inplace = False)\nplt.figure(figsize = (10, 6))\nplt.title(\"Histogram of annual income (winsorized top/bottom 1%)\")\nplt.hist(income_winsorized, bins = 50, rwidth = 0.5, label = \"Annual Income\")",
"_____no_output_____"
]
],
[
[
"#### Target variable distribution\n\nThe target variable is heavily imbalanced towards the non-defaults. Model training needs to account for this distribution, otherwise it is likely to result in models with high accuracy but low recall (high accuracy can be achieved trivially by predicting all as non-defaults). ",
"_____no_output_____"
]
],
[
[
"# Count plot of default status\nplt.figure(figsize = (10, 6))\nplt.title(\"Count of defaulted vs. non-default observations\")\nax = sns.countplot(x = 'default', data = data)\n\n# Display the absolute count and percentage of total loans in each category\nfor p in ax.patches:\n height = p.get_height()\n ax.text(p.get_x() + p.get_width() / 2., height / 2,\n '{} ({}%)'.format(height, round(100 * height / len(data), 2)),\n ha = \"center\")",
"_____no_output_____"
]
],
[
[
"#### Monthly loan count over time\n\nIncreasing trend in number of loans applied for each month, showing the increasing popularity of Lending Club as an avenue to access personal credit. ",
"_____no_output_____"
]
],
[
[
"# Plot monthly loan count over time\nnum_loans_by_time = data.groupby(\"app_time\")[\"default\"].count()\nplt.figure(figsize = (10, 6))\nplt.title(\"Number of loans over time\")\nplt.plot(num_loans_by_time)",
"_____no_output_____"
]
],
[
[
"#### Monthly average default rate over time\n\nExcluding an abnormally high period at the start, average default rate seems to remain stable at around 20% between 2007 and 2011. Unsurprisingly, given the steady increase in monthly loan count and stable default rate, total defaults increase steadily over time as well.",
"_____no_output_____"
]
],
[
[
"# Default rate over time\ntotal_default = data.groupby(\"app_time\")[\"default\"].sum()\nmean_default_rate = data.groupby(\"app_time\")[\"default\"].mean()\n\nfig, ax1 = plt.subplots(figsize = (10, 6))\n\nplt.title(\"Total defaults and mean default rate over time\")\nax1.set_xlabel('time')\nax1.set_ylabel('Total defaults', color = 'tab:red')\nax1.plot(total_default, color = 'tab:red')\nax1.tick_params(axis = 'y', labelcolor = 'tab:red')\n\nax2 = ax1.twinx() \n\nax2.set_ylabel('Mean default rate', color = 'tab:blue') \nax2.plot(mean_default_rate, color = 'tab:blue')\nax2.tick_params(axis = 'y', labelcolor = 'tab:blue')\n\nfig.tight_layout() \nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Average loan amount by purpose\n\nLoans for business were on average the highest among all documented purposes, followed by loans for housing needs (house purchase or home improvement) and financial needs (debt consolidation and credit cards). On the other end of the spectrum, discretionary expenses like vacation tend to have the lowest loan amounts.",
"_____no_output_____"
]
],
[
[
"loan_by_purpose = data.groupby(\"purpose\")[\"loan_amnt\"].mean().sort_values(ascending=False)\nplt.figure(figsize = (15, 6))\nplt.title(\"Average loan amount by purpose\")\nplt.barh(width = loan_by_purpose, y = loan_by_purpose.index)",
"_____no_output_____"
]
],
[
[
"#### Visualising default rate by state\n\nStates with highest default rates appear to be concentrated in the West Coast (e.g California), as well as South East region. Central states appear less likely to default.",
"_____no_output_____"
]
],
[
[
"import folium\nfrom IPython.display import HTML\n\n\ndefault_by_state = pd.DataFrame(data.groupby(\"addr_state\")[\"default\"].mean()).reset_index()\nstate_geo = r'https://gist.githubusercontent.com/datadave/108b5f382c838c3963d7/raw/3036216d894d49205948dbbfd562754ef3814785/us-states.json'\n\nmap = folium.Map(location=[40, -100], zoom_start=4)\n\nmap.choropleth(geo_data=state_geo, data=default_by_state,\n columns=['addr_state', 'default'],\n key_on='feature.id',\n threshold_scale = [0, 0.03, 0.06, 0.09, 0.12,\n 0.15, 0.18, 0.21, 0.24], \n fill_color=\"YlOrRd\", fill_opacity=0.75, line_opacity=0.5, \n legend_name='default rate') \nmap",
"_____no_output_____"
]
],
[
[
"## Feature engineering",
"_____no_output_____"
],
[
"#### Re-format datetime columns as time distance relative to a reference time point\n\nDatetime columns cannot be passed directly as features into a machine learning model. We thus re-format each datetime column as the time distance to a reference time point, i.e number of days / months / years passed since the reference point. \n\nApplication time, `app_time`, is re-formatted as the number of months passed since January 2007, which is the start of this dataset.\n\nEarliest credit line, `earliest_cr_line`, is re-formatted as the time distance (in months) to the application time. This potentially extracts more useful information such as: \"How long has the earliest credit line been in place, at the point of application?\". ",
"_____no_output_____"
]
],
[
[
"import monthdelta as md\n# Re-format earliest_cr_line as time distance relative to application time\ndata[\"earliest_cr_line\"] = pd.to_datetime(data[\"earliest_cr_line\"],\n format = \"%b-%Y\")\n\ndata[\"earliest_cr_line\"] = [\n md.monthmod(dt.date(data.iloc[i][\"earliest_cr_line\"]),\n dt.date(data.iloc[i][\"app_time\"]))[0].months\n for i in range(len(data))\n]\n\n\n# Re-format app_time as time distance relative to January 2007 (start of dataset)\nref_date = dt.date(dt(2007, 1, 1))\n\ndata[\"app_time\"] = [\n md.monthmod(ref_date, dt.date(data.iloc[i][\"app_time\"]))[0].months\n for i in range(len(data))\n]",
"_____no_output_____"
]
],
[
[
"#### Convert categorical columns to dummy variables\n\nColumn `addr_state` may contain useful information, but there are too many discrete values and we'd need to add too many dummy variables columns to use it for classification. The column is thus dropped instead. ",
"_____no_output_____"
]
],
[
[
"# Drop add_state column:\ndata.drop(\"addr_state\", axis = 1, inplace = True)\n\n# Map verification status to 0 or 1\ndata[\"verification_status\"] = [0 if status == \"Not Verified\" else 1 for status in data.verification_status]\n\n# Convert \"home_ownership\" and \"purpose\" to dummy variables\ndummy_cols = [\"home_ownership\", \"purpose\"]\ndata = pd.concat([data, pd.get_dummies(data[dummy_cols])], axis = 1)\ndata.drop(dummy_cols, axis = 1, inplace = True)",
"_____no_output_____"
]
],
[
[
"For each categorical variable converted to dummies, i.e `home_ownership` and `purpose`, one of the original categories must be removed to avoid multicollinearity issues, which would distort coefficients of linear models. ",
"_____no_output_____"
]
],
[
[
"# Remove one category of dummy variables\ndata.drop([\"home_ownership_OTHER\", \"purpose_other\"], axis =1, inplace = True)",
"_____no_output_____"
]
],
[
[
"## Model training",
"_____no_output_____"
]
],
[
[
"# Reorganize target variable to the end of dataframe\ndata[\"default2\"] = data[\"default\"]\ndata.drop(\"default\", axis = 1, inplace = True)\ndata.rename({\"default2\": \"default\"}, axis = 1, inplace = True)",
"_____no_output_____"
]
],
[
[
"### Train/test split\n\nTrain-test split must be done before feature selection to avoid using information from the eventual test set during the feature selection process, which may introduce unfair bias. We will use a stratified train-test split with 80% of the data in the training set, and 20% in the test set. As the dataset is highly imbalanced, a stratified split ensures that the proportion of defaults in the train and test set are similar. ",
"_____no_output_____"
],
[
"# New section",
"_____no_output_____"
]
],
[
[
"# Extract X and y columns:\nX = data.iloc[:, :-1]\ny = data.iloc[:, -1:]\n\n# Train/test split\nfrom sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, stratify = y,\n test_size = 0.2,\n random_state = 0)",
"_____no_output_____"
]
],
[
[
"### K-fold cross-validation\n\nInstead of a single train-test split, model performance can be better estimated using a technique called cross validation. In one popular approach, K-fold cross validation, the training set is further split into K equal non-intersecting, complementary subsets. In each iteration of training, the model can be trained on K-1 of the subsets, and validated on the remaining one (not used in training). The validation results from these K iterations can be averaged out to give a more robust, less biased estimate of model performance than in a single train-test split. \n\n",
"_____no_output_____"
],
[
"#### Generate K folds from training set\n\nWe select K = 5 for cross-validation, and each CV fold is generated using a stratified split similar to the train-test split earlier. This is to ensure that the ratio of defaults to non-defaults remain the same in our train and validation sets.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import StratifiedKFold\n# set random state for reproducibility\nskf = StratifiedKFold(n_splits = 5, shuffle=True, random_state=100)\nskf.split(X_train, y_train)\n\nX_train_train, X_validate = [], []\ny_train_train, y_validate = [], []\n\nfor train_index, validate_index in skf.split(X_train, y_train):\n X_train_train.append(X_train.iloc[list(train_index)])\n X_validate.append(X_train.iloc[list(validate_index)])\n y_train_train.append(y_train.iloc[list(train_index)])\n y_validate.append(y_train.iloc[list(validate_index)])",
"_____no_output_____"
]
],
[
[
"# random forest",
"_____no_output_____"
]
],
[
[
"from sklearn import tree, ensemble, model_selection, metrics\n\nX_train, X_test, y_train, y_test = model_selection.train_test_split(X, y, test_size=0.2, random_state=2019)\n\ndef my_linspace (min_value, max_value, steps):\n diff = max_value - min_value\n return np.linspace (min_value - 0.1 * diff, max_value + 0.1 * diff, steps)\n# steps = 200\n# x0 = my_linspace(min(X[:,0]), max(X[:,0]), steps)\n# x1 = my_linspace(min(X[:,1]), max(X[:,1]), steps)\n# xx0, xx1 = np.meshgrid(x0, x1)\n# mesh_data = np.c_[xx0.ravel(), xx1.ravel()]",
"_____no_output_____"
],
[
"rforest = ensemble.RandomForestClassifier(n_estimators = 10, max_features = 1, oob_score = True, random_state = 2019)\nrforest.fit(X_train, y_train)\n\n# plt.figure(figsize = (25, 10))\n# for i in range(rforest.n_estimators):\n# plt.subplot(2, 5, i+1)\n# mesh_prob = rforest.estimators_[i].predict_proba(mesh_data).reshape(steps, steps, 2)\n# plt.contourf(xx0, xx1, mesh_prob[:,:,0], 10, cmap=plt.cm.RdBu, alpha=0.3)\n# plt.scatter(X_train[:,0], X_train[:,1], c=y_train_color)\n# plt.scatter(X_test[:,0], X_test[:,1], c=y_test_color, marker='+')\n# plt.text(0, 1, rforest.estimators_[i].score(X_test, y_test))\n\n# plt.show()",
"_____no_output_____"
],
[
"from sklearn import metrics\n\ny_pred = rforest.predict(X_test)\n\nprint('Accuracy =', sklearn.metrics.accuracy_score(y_test, y_pred))\nprint('Precision:', sklearn.metrics.precision_score(y_test, y_pred))\nprint('Recall:',sklearn.metrics.recall_score(y_test, y_pred))\nprint('f1 score =', sklearn.metrics.f1_score(y_test, y_pred))",
"Accuracy = 0.8569620253164557\nPrecision: 0.26153846153846155\nRecall: 0.017241379310344827\nf1 score = 0.03235014272121789\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecb9f95e18f1aeff7abd7d89c5214381768734ab | 88,432 | ipynb | Jupyter Notebook | machine_learning_approach.ipynb | SAZZZO99/Toxic-Comment-Classification | 3354576362f501821a132a554f9e3db4b8adfee6 | [
"Apache-2.0"
] | null | null | null | machine_learning_approach.ipynb | SAZZZO99/Toxic-Comment-Classification | 3354576362f501821a132a554f9e3db4b8adfee6 | [
"Apache-2.0"
] | null | null | null | machine_learning_approach.ipynb | SAZZZO99/Toxic-Comment-Classification | 3354576362f501821a132a554f9e3db4b8adfee6 | [
"Apache-2.0"
] | null | null | null | 65.846612 | 19,254 | 0.68707 | [
[
[
"\n%matplotlib inline\nimport re\nimport matplotlib\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom sklearn.naive_bayes import MultinomialNB\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.multiclass import OneVsRestClassifier\nfrom nltk.corpus import stopwords\nstop_words = set(stopwords.words('english'))\nfrom sklearn.svm import LinearSVC\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.pipeline import Pipeline\nimport seaborn as sns",
"_____no_output_____"
],
[
"df = pd.read_csv(\"../input/nlp-coursework/1613373921-5e748a2d5fc288e9f69c5f86.csv\")",
"_____no_output_____"
],
[
"label_cols = ['toxic', 'severe_toxic', 'obscene', 'threat', 'insult', 'identity_hate']",
"_____no_output_____"
],
[
"df['none'] = 1-df[label_cols].max(axis=1)\ndf.describe()",
"_____no_output_____"
],
[
"df_toxic = df.drop(['id', 'comment_text'], axis=1)\ncounts = []\ncategories = list(df_toxic.columns.values)\nfor i in categories:\n counts.append((i, df_toxic[i].sum()))\ndf_stats = pd.DataFrame(counts, columns=['category', 'number_of_comments'])\ndf_stats",
"_____no_output_____"
],
[
"df_stats.plot(x='category', y='number_of_comments', kind='bar', legend=False, grid=True, figsize=(8, 5))\nplt.title(\"Number of comments per category\")\nplt.ylabel('# of Occurrences', fontsize=12)\nplt.xlabel('category', fontsize=12)",
"_____no_output_____"
],
[
"rowsums = df.iloc[:,2:].sum(axis=1)\nx=rowsums.value_counts()\n\n#plot\nplt.figure(figsize=(8,5))\nax = sns.barplot(x.index, x.values)\nplt.title(\"Multiple categories per comment\")\nplt.ylabel('# of Occurrences', fontsize=12)\nplt.xlabel('# of categories', fontsize=12)",
"/opt/conda/lib/python3.7/site-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
],
[
"lens = df.comment_text.str.len()\nlens.hist(bins = np.arange(0,5000,50))",
"_____no_output_____"
],
[
"print('Number of missing comments in comment text:')\ndf['comment_text'].isnull().sum()",
"Number of missing comments in comment text:\n"
],
[
"df['comment_text'][0]",
"_____no_output_____"
],
[
"categories = ['toxic', 'severe_toxic', 'obscene', 'threat', 'insult', 'identity_hate','none']",
"_____no_output_____"
],
[
"\ndef clean_text(text):\n text = text.lower()\n text = re.sub(r\"what's\", \"what is \", text)\n text = re.sub(r\"\\'s\", \" \", text)\n text = re.sub(r\"\\'ve\", \" have \", text)\n text = re.sub(r\"can't\", \"can not \", text)\n text = re.sub(r\"n't\", \" not \", text)\n text = re.sub(r\"i'm\", \"i am \", text)\n text = re.sub(r\"\\'re\", \" are \", text)\n text = re.sub(r\"\\'d\", \" would \", text)\n text = re.sub(r\"\\'ll\", \" will \", text)\n text = re.sub(r\"\\'scuse\", \" excuse \", text)\n text = re.sub('\\W', ' ', text)\n text = re.sub('\\s+', ' ', text)\n text = text.strip(' ')\n return text",
"_____no_output_____"
],
[
"df['comment_text'] = df['comment_text'].map(lambda com : clean_text(com))",
"_____no_output_____"
],
[
"df['comment_text'][0]",
"_____no_output_____"
],
[
"train, test = train_test_split(df, random_state=42, test_size=0.20, shuffle=True)",
"_____no_output_____"
],
[
"X_train = train.comment_text\nX_test = test.comment_text\nprint(X_train.shape)\nprint(X_test.shape)",
"(127656,)\n(31915,)\n"
],
[
"print(X_train)",
"140030 grandma terri should burn in trash grandma ter...\n159124 9 may 2009 utc it would be easiest if you were...\n60006 the objectivity of this discussion is doubtful...\n65432 shelly shock shelly shock is\n154979 i do not care refer to ong teng cheong talk pa...\n ... \n119879 redirect talk john loveday experimental physicist\n103694 back it up post the line here with the reference\n131932 i wo not stop that sometimes germanic equals g...\n146867 british bands i think you have mistaken scotti...\n121958 you are wrong justin thompson is mentioned in ...\nName: comment_text, Length: 127656, dtype: object\n"
],
[
"y_train = train.drop(['id', 'comment_text'], axis=1)\n#y_train.columns = [''] * len(y_train.columns)\ny_test = test.drop(['id', 'comment_text'], axis=1)\nprint(y_train)",
" toxic severe_toxic obscene threat insult identity_hate none\n140030 1 0 0 0 0 0 0\n159124 0 0 0 0 0 0 1\n60006 0 0 0 0 0 0 1\n65432 0 0 0 0 0 0 1\n154979 0 0 0 0 0 0 1\n... ... ... ... ... ... ... ...\n119879 0 0 0 0 0 0 1\n103694 0 0 0 0 0 0 1\n131932 1 0 0 0 0 0 0\n146867 0 0 0 0 0 0 1\n121958 0 0 0 0 0 0 1\n\n[127656 rows x 7 columns]\n"
],
[
"NB_pipeline = Pipeline([\n ('tfidf', TfidfVectorizer(stop_words=stop_words)),\n ('clf', OneVsRestClassifier(MultinomialNB(\n fit_prior=True, class_prior=None, alpha=1e-3))),\n ])",
"_____no_output_____"
],
[
"NB_pipeline = Pipeline([\n ('tfidf', TfidfVectorizer(stop_words=stop_words)),\n ('clf', OneVsRestClassifier(MultinomialNB(\n fit_prior=True, class_prior=None, alpha=1e-3))),\n ])\nfor category in categories:\n print('... Processing {}'.format(category))\n # train the model using X_dtm & y\n NB_pipeline.fit(X_train, train[category])\n # compute the testing accuracy\n prediction = NB_pipeline.predict(X_test)\n \n \n print('Test accuracy is {}'.format(accuracy_score(test[category], prediction)))",
"... Processing toxic\nTest accuracy is 0.9434435218549272\n... Processing severe_toxic\nTest accuracy is 0.9893153689487701\n... Processing obscene\nTest accuracy is 0.9671001096663011\n... Processing threat\nTest accuracy is 0.9975246749177503\n... Processing insult\nTest accuracy is 0.9639041203195989\n... Processing identity_hate\nTest accuracy is 0.9898167006109979\n... Processing none\nTest accuracy is 0.9417515274949083\n"
],
[
"\nSVC_pipeline = Pipeline([\n ('tfidf', TfidfVectorizer(stop_words=stop_words)),\n ('clf', OneVsRestClassifier(LinearSVC(), n_jobs=1)),\n ])",
"_____no_output_____"
],
[
"SVC_pipeline = Pipeline([\n ('tfidf', TfidfVectorizer(stop_words=stop_words)),\n ('clf', OneVsRestClassifier(LinearSVC(), n_jobs=1)),\n ])\nfor category in categories:\n print('... Processing {}'.format(category))\n # train the model using X_dtm & y\n SVC_pipeline.fit(X_train, train[category])\n # compute the testing accuracy\n prediction = SVC_pipeline.predict(X_test)\n print('Test accuracy is {}'.format(accuracy_score(test[category], prediction)))",
"... Processing toxic\nTest accuracy is 0.9602694657684474\n... Processing severe_toxic\nTest accuracy is 0.9906626977910074\n... Processing obscene\nTest accuracy is 0.9794454018486605\n... Processing threat\nTest accuracy is 0.9976813410621964\n... Processing insult\nTest accuracy is 0.9711107629641235\n... Processing identity_hate\nTest accuracy is 0.9920726930910231\n... Processing none\nTest accuracy is 0.9606767977440075\n"
],
[
"LogReg_pipeline = Pipeline([\n ('tfidf', TfidfVectorizer(stop_words=stop_words)),\n ('clf', OneVsRestClassifier(LogisticRegression(solver='sag', C=7), n_jobs=1)),\n ])\nfor category in categories:\n print('... Processing {}'.format(category))\n # train the model using X_dtm & y\n LogReg_pipeline.fit(X_train, train[category])\n # compute the testing accuracy\n prediction = LogReg_pipeline.predict(X_test)\n print('Test accuracy is {}'.format(accuracy_score(test[category], prediction)))",
"... Processing toxic\nTest accuracy is 0.9601127996240012\n... Processing severe_toxic\nTest accuracy is 0.9907253642487859\n... Processing obscene\nTest accuracy is 0.9791947360175466\n... Processing threat\nTest accuracy is 0.9976500078333073\n... Processing insult\nTest accuracy is 0.9708600971330096\n... Processing identity_hate\nTest accuracy is 0.9918846937176876\n... Processing none\nTest accuracy is 0.9603007989973367\n"
],
[
"from sklearn.tree import DecisionTreeClassifier\nfrom sklearn.ensemble import RandomForestClassifier",
"_____no_output_____"
],
[
"RanFor_pipeline = Pipeline([\n ('tfidf', TfidfVectorizer(stop_words=stop_words)),\n ('clf', OneVsRestClassifier(RandomForestClassifier(max_depth= 15, n_estimators=40), n_jobs=1)),\n ])\nfor category in categories:\n print('... Processing {}'.format(category))\n # train the model using X_dtm & y\n RanFor_pipeline.fit(X_train, train[category])\n # compute the testing accuracy\n prediction = RanFor_pipeline.predict(X_test)\n print('Test accuracy is {}'.format(accuracy_score(test[category], prediction)))",
"... Processing toxic\nTest accuracy is 0.9042456525144916\n... Processing severe_toxic\nTest accuracy is 0.989942033526555\n... Processing obscene\nTest accuracy is 0.9462635124549584\n... Processing threat\nTest accuracy is 0.9976813410621964\n... Processing insult\nTest accuracy is 0.9494281685727715\n... Processing identity_hate\nTest accuracy is 0.9907880307065643\n... Processing none\nTest accuracy is 0.8983863387122043\n"
],
[
"DecTree_pipeline = Pipeline([\n ('tfidf', TfidfVectorizer(stop_words=stop_words)),\n ('clf', OneVsRestClassifier(DecisionTreeClassifier(max_depth = 25), n_jobs=1)),\n ])\nfor category in categories:\n print('... Processing {}'.format(category))\n # train the model using X_dtm & y\n DecTree_pipeline.fit(X_train, train[category])\n # compute the testing accuracy\n prediction = DecTree_pipeline.predict(X_test)\n print('Test accuracy is {}'.format(accuracy_score(test[category], prediction)))",
"... Processing toxic\nTest accuracy is 0.9460128466238445\n... Processing severe_toxic\nTest accuracy is 0.9884380385398716\n... Processing obscene\nTest accuracy is 0.9772834090553032\n... Processing threat\nTest accuracy is 0.9973053423155256\n... Processing insult\nTest accuracy is 0.9652827823907254\n... Processing identity_hate\nTest accuracy is 0.991665361115463\n... Processing none\nTest accuracy is 0.944477518408272\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecba0c39bb785413cbe0cb4ae0f09621d0504040 | 145,070 | ipynb | Jupyter Notebook | chapter_deep-learning-basics/linear-regression-scratch.ipynb | femj007/d2l-zh | faa3e6230fd30d56c0400fe610e7f8396fa25f8b | [
"Apache-2.0"
] | null | null | null | chapter_deep-learning-basics/linear-regression-scratch.ipynb | femj007/d2l-zh | faa3e6230fd30d56c0400fe610e7f8396fa25f8b | [
"Apache-2.0"
] | null | null | null | chapter_deep-learning-basics/linear-regression-scratch.ipynb | femj007/d2l-zh | faa3e6230fd30d56c0400fe610e7f8396fa25f8b | [
"Apache-2.0"
] | null | null | null | 82.897143 | 275 | 0.550017 | [
[
[
"# 线性回归的从零开始实现\n\n在了解了线性回归的背景知识之后,现在我们可以动手实现它了。尽管强大的深度学习框架可以减少大量重复性工作,但若过于依赖它提供的便利,会导致我们很难深入理解深度学习是如何工作的。因此,本节将介绍如何只利用NDArray和`autograd`来实现一个线性回归的训练。\n\n首先,导入本节中实验所需的包或模块,其中的matplotlib包可用于作图,且设置成嵌入显示。",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nfrom IPython import display\nfrom matplotlib import pyplot as plt\nfrom mxnet import autograd, nd\nimport random",
"_____no_output_____"
]
],
[
[
"## 生成数据集\n\n我们构造一个简单的人工训练数据集,它可以使我们能够直观比较学到的参数和真实的模型参数的区别。设训练数据集样本数为1000,输入个数(特征数)为2。给定随机生成的批量样本特征$\\boldsymbol{X} \\in \\mathbb{R}^{1000 \\times 2}$,我们使用线性回归模型真实权重$\\boldsymbol{w} = [2, -3.4]^\\top$和偏差$b = 4.2$,以及一个随机噪音项$\\epsilon$来生成标签\n\n$$\\boldsymbol{y} = \\boldsymbol{X}\\boldsymbol{w} + b + \\epsilon,$$\n\n其中噪音项$\\epsilon$服从均值为0和标准差为0.01的正态分布。噪音(noise)代表了数据集中无意义的干扰。下面,让我们生成数据集。",
"_____no_output_____"
]
],
[
[
"num_inputs = 2\nnum_examples = 1000\ntrue_w = [2, -3.4]\ntrue_b = 4.2\nfeatures = nd.random.normal(scale=1, shape=(num_examples, num_inputs))\nlabels = true_w[0] * features[:, 0] + true_w[1] * features[:, 1] + true_b\nlabels += nd.random.normal(scale=0.01, shape=labels.shape)",
"_____no_output_____"
]
],
[
[
"注意到`features`的每一行是一个长度为2的向量,而`labels`的每一行是一个长度为1的向量(标量)。",
"_____no_output_____"
]
],
[
[
"features[0], labels[0]",
"_____no_output_____"
]
],
[
[
"通过生成第二个特征`features[:, 1]`和标签 `labels` 的散点图,我们可以更直观地观察两者间的线性关系。",
"_____no_output_____"
]
],
[
[
"def use_svg_display():\n # 用矢量图显示\n display.set_matplotlib_formats('svg')\n\ndef set_figsize(figsize=(3.5, 2.5)):\n use_svg_display()\n # 设置图的尺寸\n plt.rcParams['figure.figsize'] = figsize\n\nset_figsize()\nplt.scatter(features[:, 1].asnumpy(), labels.asnumpy(), 1);",
"_____no_output_____"
]
],
[
[
"我们将上面的`plt`作图函数以及`use_svg_display`和`set_figsize`函数定义在`d2lzh`包里。以后在作图时,我们将直接调用`d2lzh.plt`。由于`plt`在`d2lzh`包中是一个全局变量,我们在作图前只需要调用`d2lzh.set_figsize()`即可打印矢量图并设置图的尺寸。\n\n\n## 读取数据\n\n在训练模型的时候,我们需要遍历数据集并不断读取小批量数据样本。这里我们定义一个函数:它每次返回`batch_size`(批量大小)个随机样本的特征和标签。",
"_____no_output_____"
]
],
[
[
"# 本函数已保存在d2lzh包中方便以后使用\ndef data_iter(batch_size, features, labels):\n num_examples = len(features)\n indices = list(range(num_examples))\n random.shuffle(indices) # 样本的读取顺序是随机的\n for i in range(0, num_examples, batch_size):\n j = nd.array(indices[i: min(i + batch_size, num_examples)])\n yield features.take(j), labels.take(j) # take函数根据索引返回对应元素",
"_____no_output_____"
]
],
[
[
"让我们读取第一个小批量数据样本并打印。每个批量的特征形状为(10, 2),分别对应批量大小和输入个数;标签形状为批量大小。",
"_____no_output_____"
]
],
[
[
"batch_size = 10\n\nfor X, y in data_iter(batch_size, features, labels):\n print(X, y)\n break",
"\n[[ 2.6056926 0.58565795]\n [ 0.39822668 1.015803 ]\n [-0.09322714 2.28601 ]\n [ 0.42282835 0.22813949]\n [-0.87284005 0.13884856]\n [ 1.7203581 -1.6773723 ]\n [ 1.4569732 2.115476 ]\n [ 0.51380765 1.4481224 ]\n [ 0.167845 1.0191584 ]\n [-0.8624608 -0.4348689 ]]\n<NDArray 10x2 @cpu(0)> \n[ 7.4225383 1.5623367 -3.7693658 4.2868342 1.9889501 13.343663\n -0.08125029 0.29919574 1.0813215 3.9542494 ]\n<NDArray 10 @cpu(0)>\n"
]
],
[
[
"## 初始化模型参数\n\n我们将权重初始化成均值为0标准差为0.01的正态随机数,偏差则初始化成0。",
"_____no_output_____"
]
],
[
[
"w = nd.random.normal(scale=0.01, shape=(num_inputs, 1))\nb = nd.zeros(shape=(1,))",
"_____no_output_____"
]
],
[
[
"之后的模型训练中,我们需要对这些参数求梯度来迭代参数的值,因此我们需要创建它们的梯度。",
"_____no_output_____"
]
],
[
[
"w.attach_grad()\nb.attach_grad()",
"_____no_output_____"
]
],
[
[
"## 定义模型\n\n下面是线性回归的矢量计算表达式的实现。我们使用`dot`函数做矩阵乘法。",
"_____no_output_____"
]
],
[
[
"def linreg(X, w, b): # 本函数已保存在d2lzh包中方便以后使用\n return nd.dot(X, w) + b",
"_____no_output_____"
]
],
[
[
"## 定义损失函数\n\n我们使用上一节描述的平方损失来定义线性回归的损失函数。在实现中,我们需要把真实值`y`变形成预测值`y_hat`的形状。以下函数返回的结果也将和`y_hat`的形状相同。",
"_____no_output_____"
]
],
[
[
"def squared_loss(y_hat, y): # 本函数已保存在d2lzh包中方便以后使用\n return (y_hat - y.reshape(y_hat.shape)) ** 2 / 2",
"_____no_output_____"
]
],
[
[
"## 定义优化算法\n\n以下的`sgd`函数实现了上一节中介绍的小批量随机梯度下降算法。它通过不断迭代模型参数来优化损失函数。这里自动求梯度模块计算得来的梯度是一个批量样本的梯度和。我们将它除以批量大小来得到平均值。",
"_____no_output_____"
]
],
[
[
"def sgd(params, lr, batch_size): # 本函数已保存在d2lzh包中方便以后使用\n for param in params:\n param[:] = param - lr * param.grad / batch_size",
"_____no_output_____"
]
],
[
[
"## 训练模型\n\n在训练中,我们将多次迭代模型参数。在每次迭代中,我们根据当前读取的小批量数据样本(特征`X`和标签`y`),通过调用反向函数`backward`计算小批量随机梯度,并调用优化算法`sgd`迭代模型参数。由于我们之前设批量大小`batch_size`为10,每个小批量的损失`l`的形状为(10,1)。回忆一下[“自动求梯度”](../chapter_prerequisite/autograd.md)一节。由于变量`l`并不是一个标量,运行`l.backward()`将对`l`中元素求和得到新的变量,再求该变量有关模型参数的梯度。\n\n在一个迭代周期(epoch)中,我们将完整遍历一遍`data_iter`函数,并对训练数据集中所有样本都使用一次(假设样本数能够被批量大小整除)。这里的迭代周期个数`num_epochs`和学习率`lr`都是超参数,分别设3和0.03。在实践中,大多超参数都需要通过反复试错来不断调节。当迭代周期数设的越大时,虽然模型可能更有效,但是训练时间可能过长。而有关学习率对模型的影响,我们会在后面“优化算法”一章中详细介绍。",
"_____no_output_____"
]
],
[
[
"lr = 0.03\nnum_epochs = 3\nnet = linreg\nloss = squared_loss\n\nfor epoch in range(num_epochs): # 训练模型一共需要num_epochs个迭代周期\n # 在每一个迭代周期中,会使用训练数据集中所有样本一次(假设样本数能够被批量大小整除)。X\n # 和y分别是小批量样本的特征和标签\n for X, y in data_iter(batch_size, features, labels):\n with autograd.record():\n l = loss(net(X, w, b), y) # l是有关小批量X和y的损失\n l.backward() # 小批量的损失对模型参数求梯度\n sgd([w, b], lr, batch_size) # 使用小批量随机梯度下降迭代模型参数\n train_l = loss(net(features, w, b), labels)\n print('epoch %d, loss %f' % (epoch + 1, train_l.mean().asnumpy()))",
"epoch 1, loss 0.040678\n"
]
],
[
[
"训练完成后,我们可以比较学到的参数和用来生成训练集的真实参数。它们应该很接近。",
"_____no_output_____"
]
],
[
[
"true_w, w",
"_____no_output_____"
],
[
"true_b, b",
"_____no_output_____"
]
],
[
[
"## 小结\n\n* 可以看出,仅使用NDArray和`autograd`就可以很容易地实现一个模型。在接下来的章节中,我们会在此基础上描述更多深度学习模型,并介绍怎样使用更简洁的代码(例如下一节)来实现它们。\n\n\n## 练习\n\n* 为什么`squared_loss`函数中需要使用`reshape`函数?\n* 尝试使用不同的学习率,观察损失函数值的下降快慢。\n* 如果样本个数不能被批量大小整除,`data_iter`函数的行为会有什么变化?\n\n\n## 扫码直达[讨论区](https://discuss.gluon.ai/t/topic/743)\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
ecba15768e3f08032c8ffa08e36d6c997df577ab | 74,401 | ipynb | Jupyter Notebook | framework0.3.ipynb | gutouyu/IntrusionDetection | b56f3fbc04024c990ee01ae74f87302469621cf2 | [
"MIT"
] | 1 | 2019-04-06T09:21:54.000Z | 2019-04-06T09:21:54.000Z | framework0.3.ipynb | gutouyu/IntrusionDetection | b56f3fbc04024c990ee01ae74f87302469621cf2 | [
"MIT"
] | null | null | null | framework0.3.ipynb | gutouyu/IntrusionDetection | b56f3fbc04024c990ee01ae74f87302469621cf2 | [
"MIT"
] | null | null | null | 105.683239 | 49,708 | 0.807879 | [
[
[
"# # Deep Learning\n# \n# ## preprocessing training dataset\n\nimport os\nfrom MLP import MLP\nimport numpy as np\nimport pandas as pd\nfrom collections import Counter\nimport matplotlib.pyplot as plt\nfrom imblearn.over_sampling import SMOTE\nfrom sklearn.model_selection import train_test_split\n\nu'matplotlib inline'\nplt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\nu'load_ext autoreload'\nu'autoreload 2'",
"_____no_output_____"
],
[
"def read_data():\n input_file_dir = \"../datasets\"\n train_file_name = \"kddcup.data_10_percent.txt\"\n test_file_name = \"corrected.txt\"\n header_file_name = \"header.txt\"\n train_files = os.path.join(input_file_dir, train_file_name)\n test_files = os.path.join(input_file_dir, test_file_name)\n header_files = os.path.join(input_file_dir, header_file_name)\n with open(header_files, 'r') as f:\n header = f.readline().strip().split(',')\n train_dataset = pd.read_csv(train_files)\n test_dataset = pd.read_csv(test_files)\n train_dataset.columns = header\n test_dataset.columns = header\n return train_dataset, test_dataset\n\n\ndef labels_map(label):\n label = str(label).split('.')[0]\n \n DOS = ['apache2', 'back', 'land', 'mailbomb', 'neptune', 'pod', \n 'processtable', 'smurf', 'teardrop', 'udpstorm'] #DOS 10个\n PROBE = ['ipsweep', 'mscan', 'nmap', 'portsweep', 'saint', 'satan'] #PROBE\n U2R = ['buffer_overflow', 'httptunnel', 'loadmodule', 'perl', 'ps', 'rootkit', 'sqlattack', 'xterm'] #U2R\n R2L = ['ftp_write', 'guess_passwd', 'imap', 'multihop', 'named', 'phf', \n 'sendmail', 'snmpgetattack', 'snmpguess', 'spy', 'warezclient', \n 'warezmaster', 'worm', 'xlock', 'xsnoop']#R2L\n \n if label == 'normal':\n return 0\n if label in PROBE:\n return -1 \n if label in DOS:\n return DOS.index(label) + 1\n if label in U2R:\n return -1\n if label in R2L:\n return -1\n\ndef filter_labels(dataset):\n dataset['labels'] = dataset['labels'].apply(labels_map)\n #看看要保留哪些数据\n dataset = dataset[(dataset['labels']>-1)]\n return dataset\n\ndef split_valid_from_train(train_dataset, valid_size):\n # Method 1\n train_dataset, valid_dataset, _, _ = train_test_split(train_dataset, train_dataset['labels'], test_size=valid_size, random_state=None)\n \n # pandas中先重置index再打乱train. 否则只会调整各个行的顺序,而不会改变pandas的index\n # 重置\n train_dataset = train_dataset.reset_index(drop=True)\n # 打乱\n indexMask = np.arange(len(train_dataset))\n for i in xrange(10):\n np.random.shuffle(indexMask)\n train_dataset = train_dataset.iloc[indexMask]\n\n # Method 2\n #获取验证集\n # val_frac=0.25\n # valid_dataset_neg = train_dataset[(train_dataset['labels']==0)].sample(frac=val_frac)\n # valid_dataset_pos = train_dataset[(train_dataset['labels']==2)].sample(frac=val_frac)\n # valid_dataset = pd.concat([valid_dataset_neg, valid_dataset_pos], axis=0)\n # #train_dataset中分离出valid_dataset\n # train_dataset = train_dataset.select(lambda x: x not in valid_dataset.index, axis=0)\n return train_dataset, valid_dataset\n\ndef combine_train_valid_test(trainDF, validDF, testDF):\n all = pd.concat([trainDF, validDF, testDF], axis=0)\n return all, (trainDF.shape[0], validDF.shape[0], testDF.shape[0])\n\ndef process_data_features(all, handle_labels=True):\n if handle_labels:\n # 独热编码 labels\n labels_dummies = pd.get_dummies(all['labels'], prefix='label')\n all = pd.concat([all,labels_dummies], axis=1)\n all = all.drop(['labels'], axis=1)\n\n # 独热编码 protocol_type\n protocal_type_dummies = pd.get_dummies(all.protocol_type, prefix='protocol_type')\n all = pd.concat([all, protocal_type_dummies], axis=1)\n all = all.drop(['protocol_type'], axis=1)\n\n # 独热编码 flag\n flag_dummies = pd.get_dummies(all.flag, prefix='flag')\n all = pd.concat([all, flag_dummies], axis=1)\n all = all.drop(['flag'], axis=1)\n\n # 独热编码 Service 共有66个 暂时先去掉\n # all.service.value_counts()\n # service_dummies = pd.get_dummies(all.service, prefix='service')\n # all = pd.concat([all, service_dummies], axis=1)\n all = all.drop(['service'], axis=1)\n\n # 去中心化 src_bytes, dst_bytes\n all['src_bytes_norm'] = all.src_bytes - all.src_bytes.mean()\n all['dst_bytes_norm'] = all.dst_bytes - all.dst_bytes.mean()\n all = all.drop(['src_bytes'], axis=1)\n all = all.drop(['dst_bytes'], axis=1)\n\n return all.astype('float')\n\ndef recover_data_after_process_features(comb, num_comb, labels_list=[0,1,2,3]):\n #分离出Train Valid Test\n train_dataset_size, valid_dataset_size, test_dataset_size = num_comb\n sub_train_dataset = comb.iloc[:train_dataset_size, :].sample(frac=1)\n sub_valid_dataset = comb.iloc[train_dataset_size: train_dataset_size+valid_dataset_size, :].sample(frac=1)\n sub_test_dataset = comb.iloc[train_dataset_size+valid_dataset_size:, :].sample(frac=1)\n # 分离出 label\n total_labels = ['label_%d' % i for i in labels_list]\n print total_labels\n \n sub_train_labels = sub_train_dataset[total_labels]\n sub_valid_labels = sub_valid_dataset[total_labels]\n sub_test_labels = sub_test_dataset[total_labels]\n sub_train_dataset.drop(total_labels, axis=1, inplace=True)\n sub_valid_dataset.drop(total_labels, axis=1, inplace=True)\n sub_test_dataset.drop(total_labels, axis=1, inplace=True)\n data = {\n 'X_train': sub_train_dataset.as_matrix(),\n 'y_train': sub_train_labels.as_matrix(),\n 'X_val': sub_valid_dataset.as_matrix(),\n 'y_val': sub_valid_labels.as_matrix(),\n 'X_test': sub_test_dataset.as_matrix(),\n 'y_test': sub_test_labels.as_matrix()\n }\n for k, v in data.iteritems():\n print k, v.shape\n return data\n\n\n\ndef analysis_plot_loss_and_accuracy(model):\n print('Mean Accuracy( -top5 ) of train, valid, test: %.2f%%, %.2f%%, %.2f%%') % (100*np.mean(model.train_acc_history[-5:]),100*np.mean(model.val_acc_history[-5:]), 100*np.mean(model.test_acc_history[-5]))\n fig = plt.figure()\n ax = fig.add_subplot(2,1,1)\n ax.plot(range(len(model.loss_history)), model.loss_history, '-o')\n plt.plot()\n\n plt_len = len(model.train_acc_history)\n plt.subplot(2,1,2)\n plt.title('Accuracy')\n plt.plot(model.train_acc_history, '-o', label='train')\n plt.plot(model.test_acc_history, '-o', label='test')\n plt.plot(model.val_acc_history, '-o', label='valid')\n # plt.plot([90] * plt_len, 'k--')\n plt.xlabel('Iteration')\n plt.legend(loc='lower right')\n plt.gcf().set_size_inches(15,9)\n plt.show()\n\ndef accuracy(predictions, labels):\n return (100.0 * np.sum(np.argmax(predictions, 1) == np.argmax(labels, 1))\n / predictions.shape[0])\n\ndef analysis_confusion_matrix(X,y, model, header, verbose=True):\n y_pred = model.predict(X)\n m = len(header)\n y_true = np.argmax(y, axis=1)\n y_predIdx = np.argmax(y_pred, axis=1)\n mat = np.mat(np.zeros((m,m)))\n for real, pred in zip(y_true,y_predIdx):\n mat[real,pred] = mat[real,pred] + 1\n confMatrix = pd.DataFrame(data=mat, index= header, columns=header, dtype=int)\n if verbose:\n print ('Accuracy: %.2f%%' % accuracy(y_pred, y))\n# print confMatrix\n return confMatrix\n\ndef analysis_precision_recall(confMat):\n \"\"\"\n recall: 是相对于原始样本的, 检测率\n precision: 是相对于预测结果的, 精确率\n \"\"\"\n m = len(confMat)\n recall = np.diag(confMat) / np.sum(confMat, axis=1) #检测率\n precission = np.diag(confMat) / np.sum(confMat, axis=0) #精确率\n ret = pd.concat([recall, precission], axis=1)\n ret.columns=['recall', 'precission']\n return ret",
"_____no_output_____"
],
[
"# Prepare Data\ntrainDF, testDF = read_data()",
"_____no_output_____"
],
[
"trainDF = filter_labels(trainDF)\ntestDF = filter_labels(testDF)\ntestDF = testDF[(testDF['labels'] != 1) & (testDF['labels'] != 4) & (testDF['labels']!=7 ) & (testDF['labels']<10)]\ntrainDF, validDF = split_valid_from_train(trainDF, 0.25)",
"_____no_output_____"
],
[
"print Counter(trainDF['flag'])\nprint Counter(testDF['flag'])\nprint Counter(trainDF['protocol_type'])\nprint Counter(testDF['protocol_type'])",
"Counter({'SF': 281802, 'S0': 65148, 'REJ': 19055, 'RSTO': 395, 'RSTR': 90, 'S1': 45, 'S2': 11, 'S3': 5})\nCounter({'SF': 225663, 'REJ': 39640, 'S0': 17444, 'RSTO': 976, 'RSTR': 129, 'S1': 18, 'S2': 15, 'S3': 4, 'RSTOS0': 1})\nCounter({'icmp': 211929, 'tcp': 139419, 'udp': 15203})\nCounter({'icmp': 164556, 'tcp': 103226, 'udp': 16108})\n"
],
[
"# from sklearn.preprocessing import LabelEncoder\n# protocal_labelencoder = LabelEncoder()\n# flag_labelencoder = LabelEncoder()\n\n# testDF['protocol_type'] = protocal_labelencoder.fit_transform(testDF['protocol_type'])\n# testDF['flag'] = flag_labelencoder.fit_transform(testDF['flag']).astype(int)\n# testDF.drop('service', axis=1, inplace=True)\n\n# trainDF['protocol_type'] = protocal_labelencoder.transform(testDF['protocol_type'])\n# trainDF['flag'] = flag_labelencoder.transform(trainDF['flag']).astype(int)\n# trainDF.drop('service',axis=1, inplace=True)",
"_____no_output_____"
],
[
"mask_8 = trainDF['labels'] == 8\nmask_5 = trainDF['labels'] == 5\nmask_0 = trainDF['labels'] == 0\ntrainDF = pd.concat([trainDF[mask_8].sample(n=10000), \n trainDF[mask_5].sample(n=10000),\n trainDF[mask_0].sample(n=10000),\n trainDF[trainDF['labels'].isin([2,9,6,3])]], axis=0)\n\nprint Counter(trainDF['labels'])",
"Counter({0: 10000, 5: 10000, 8: 10000, 2: 1629, 9: 762, 6: 196, 3: 14})\n"
],
[
"count = Counter(trainDF['labels'])\nexpect_sample_size = 5000\nfor label in [2,9,6,3]:\n copy_cnt = expect_sample_size // count[label]\n copy_part = trainDF[trainDF['labels'] == label]\n for i in xrange(copy_cnt):\n trainDF = pd.concat([trainDF, copy_part], axis=0)\nCounter(trainDF['labels'])",
"_____no_output_____"
],
[
"combine, num_combine = combine_train_valid_test(trainDF, validDF, testDF)",
"_____no_output_____"
],
[
"combine = process_data_features(combine, True)",
"_____no_output_____"
],
[
"data = recover_data_after_process_features(combine, num_combine, [0,2,3,5,6,8,9])",
"['label_0', 'label_2', 'label_3', 'label_5', 'label_6', 'label_8', 'label_9']\nX_val (122184, 51)\nX_train (51958, 51)\nX_test (283890, 51)\ny_val (122184, 7)\ny_train (51958, 7)\ny_test (283890, 7)\n"
],
[
"# \"\"\"过采样\"\"\"\n# normal, back, land, neptune, pod, smurf, teardrop\n# DOS = np.array(DOS)[[0,2,3,5,6,8,9]].tolist()\n# sm = SMOTE(random_state=12)\n# X_train, y_train = sm.fit_sample(trainDF.drop('labels',axis=1), trainDF['labels'])\n# trainDF = pd.DataFrame(data=X_train, columns=trainDF.drop('labels',axis=1).columns)\n# y_train = pd.DataFrame(data=y_train, columns=['lables'])\n# trainDF = pd.concat([trainDF, y_train], axis=1)",
"_____no_output_____"
],
[
"# Model\ninput_dim = data['X_train'].shape[1]\noutput_dim = data['y_train'].shape[1]\nmodel = MLP(data, input_dim, [512],output_dim,\n learning_rate=3e-7, #1e-6\n dropout_prob=0.0,\n l2_strength=0.0,\n batch_size=200,\n num_epochs=20,\n print_every=200,\n verbose=True)\n\nmodel.train()",
"(Iteration 0 / 5180) train acc: 12.00%; val_acc: 0.47%; test_acc: 0.39%\n(Iteration 200 / 5180) train acc: 15.50%; val_acc: 2.70%; test_acc: 2.90%\n(Iteration 400 / 5180) train acc: 47.50%; val_acc: 25.11%; test_acc: 26.29%\n(Iteration 600 / 5180) train acc: 50.00%; val_acc: 29.23%; test_acc: 33.80%\n(Iteration 800 / 5180) train acc: 52.00%; val_acc: 32.18%; test_acc: 36.71%\n(Iteration 1000 / 5180) train acc: 64.00%; val_acc: 51.42%; test_acc: 75.62%\n(Iteration 1200 / 5180) train acc: 69.50%; val_acc: 52.55%; test_acc: 75.90%\n(Iteration 1400 / 5180) train acc: 83.50%; val_acc: 92.95%; test_acc: 96.30%\n(Iteration 1600 / 5180) train acc: 89.00%; val_acc: 91.73%; test_acc: 94.90%\n(Iteration 1800 / 5180) train acc: 90.50%; val_acc: 93.45%; test_acc: 96.42%\n(Iteration 2000 / 5180) train acc: 89.50%; val_acc: 94.01%; test_acc: 96.85%\n(Iteration 2200 / 5180) train acc: 92.50%; val_acc: 94.05%; test_acc: 96.80%\n(Iteration 2400 / 5180) train acc: 89.00%; val_acc: 94.42%; test_acc: 97.25%\n(Iteration 2600 / 5180) train acc: 94.00%; val_acc: 94.41%; test_acc: 97.08%\n(Iteration 2800 / 5180) train acc: 88.00%; val_acc: 93.63%; test_acc: 96.02%\n(Iteration 3000 / 5180) train acc: 92.00%; val_acc: 94.64%; test_acc: 97.13%\n(Iteration 3200 / 5180) train acc: 90.00%; val_acc: 94.41%; test_acc: 96.87%\n(Iteration 3400 / 5180) train acc: 81.50%; val_acc: 94.86%; test_acc: 97.01%\n(Iteration 3600 / 5180) train acc: 91.00%; val_acc: 94.49%; test_acc: 96.42%\n(Iteration 3800 / 5180) train acc: 89.00%; val_acc: 94.86%; test_acc: 96.88%\n(Iteration 4000 / 5180) train acc: 93.00%; val_acc: 95.22%; test_acc: 97.18%\n(Iteration 4200 / 5180) train acc: 91.50%; val_acc: 95.58%; test_acc: 97.44%\n(Iteration 4400 / 5180) train acc: 90.50%; val_acc: 95.43%; test_acc: 97.28%\n(Iteration 4600 / 5180) train acc: 93.50%; val_acc: 95.23%; test_acc: 97.03%\n(Iteration 4800 / 5180) train acc: 94.00%; val_acc: 95.42%; test_acc: 97.20%\n(Iteration 5000 / 5180) train acc: 94.50%; val_acc: 96.07%; test_acc: 97.83%\n"
],
[
"from sklearn.metrics import classification_report\nDOS = ['normal', 'apache2', 'back', 'land', 'mailbomb', 'neptune', 'pod', \n 'processtable', 'smurf', 'teardrop', 'udpstorm'] #DOS 10个\nDOS = np.array(DOS)[[0,2,3,5,6,8,9]].tolist()\n\ny_pred = model.predict(data['X_test'])\nprint classification_report(np.argmax(data['y_test'],axis=1), np.argmax(y_pred, axis=1), target_names=DOS)\nprint 'Accuracy %.2f' % accuracy(y_pred, data['y_test'])",
" precision recall f1-score support\n\n normal 0.99 0.93 0.96 60592\n back 0.69 1.00 0.82 1098\n land 0.01 0.78 0.02 9\n neptune 0.99 0.97 0.98 58001\n pod 0.07 0.82 0.14 87\n smurf 1.00 1.00 1.00 164091\n teardrop 0.00 0.42 0.00 12\n\navg / total 0.99 0.98 0.99 283890\n\nAccuracy 97.82\n"
],
[
"# Analysis\nDOS = ['normal', 'apache2', 'back', 'land', 'mailbomb', 'neptune', 'pod', \n 'processtable', 'smurf', 'teardrop', 'udpstorm'] #DOS 10个\nDOS = np.array(DOS)[[0,2,3,5,6,8,9]].tolist()\nATTACK = ['normal', 'probe', 'dos', 'u2r', 'r2l']\n\nconfMat = analysis_confusion_matrix(data['X_test'],data['y_test'], model, DOS)\nprint confMat\n\nanalysis_precision_recall(confMat)",
"Accuracy: 97.82%\n normal back land neptune pod smurf teardrop\nnormal 56235 494 652 437 818 313 1643\nback 2 1096 0 0 0 0 0\nland 2 0 7 0 0 0 0\nneptune 220 0 3 56381 0 0 1397\npod 0 0 4 6 71 6 0\nsmurf 83 0 0 12 58 163919 19\nteardrop 0 0 3 4 0 0 5\n"
],
[
"analysis_plot_loss_and_accuracy(model)",
"Mean Accuracy( -top5 ) of train, valid, test: 92.80%, 95.54%, 97.44%\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecba1767b4ea136db3947d4ef95d4761a65aa21d | 3,076 | ipynb | Jupyter Notebook | Accuracy_MSE_Calculation.ipynb | amcodec1/My_Practice_Notebooks | 27c17ec0f660651664e473b30936f77dd09f4362 | [
"MIT"
] | null | null | null | Accuracy_MSE_Calculation.ipynb | amcodec1/My_Practice_Notebooks | 27c17ec0f660651664e473b30936f77dd09f4362 | [
"MIT"
] | null | null | null | Accuracy_MSE_Calculation.ipynb | amcodec1/My_Practice_Notebooks | 27c17ec0f660651664e473b30936f77dd09f4362 | [
"MIT"
] | null | null | null | 20.92517 | 80 | 0.439207 | [
[
[
"import numpy as np\nyp = [0,2,1,3]\nye = [0,1,2,3]\ndef score_accuracy(yp,ye,metric):\n try:\n if (len(yp) != len(ye)):\n raise ValueError('Length unequal')\n else:\n if metric not in [\"accuracy\",\"mean_square_error\"]:\n raise ValueError('Metric not included')\n else:\n yp1 = np.array(yp)\n yp2 = np.array(ye)\n if metric == 'accuracy': \n yp3 = np.sum(yp1 == yp2)\n yp4 = (yp3*100.0)/len(yp)\n elif metric == 'mean_square_error':\n yp4 = float(np.sum(np.square(yp1 - yp2)))/len(yp) \n return yp4\n except:\n print ValueError",
"_____no_output_____"
],
[
"score_accuracy(yp,ye,'mean_square_error')",
"_____no_output_____"
],
[
"score_accuracy(yp,ye,'accuracy')",
"_____no_output_____"
],
[
"score_accuracy(yp,ye,'elephant')",
"<type 'exceptions.ValueError'>\n"
],
[
"yp = [0,2,1,3]\nye = [0,1,2,3]\ncount = 0\nfor idx, val1 in enumerate(yp):\n for jdx,val2 in enumerate(ye):\n if (idx == jdx) and (val1 == val2):\n count += 1\nprint float(count)/len(yp)",
"0.5\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecba2025e5091fb4ed6980f39a3fa497ce631cea | 202,672 | ipynb | Jupyter Notebook | class/7-tables-styling-performance.ipynb | ubco-cmps/data551_course | d5f7290f6c3bf35cd00acab0960d469c8da4e1c7 | [
"MIT"
] | null | null | null | class/7-tables-styling-performance.ipynb | ubco-cmps/data551_course | d5f7290f6c3bf35cd00acab0960d469c8da4e1c7 | [
"MIT"
] | null | null | null | class/7-tables-styling-performance.ipynb | ubco-cmps/data551_course | d5f7290f6c3bf35cd00acab0960d469c8da4e1c7 | [
"MIT"
] | null | null | null | 135.114667 | 97,116 | 0.65272 | [
[
[
"# 551 Lec 7 - Tables, styling, performance\n\nYou need to download this notebook to view images.",
"_____no_output_____"
],
[
"## Lecture learning goals\n\nBy the end of the lecture you will be able to:\n\n1. Create dash tables\n2. Use interactive dash table components in callbacks\n3. More readily implement best practices discussed before for layout and titles\n4. Cache callback values of expensive computations\n5. Recognize performance tweaks for plots such as WebGL, and datashader\n\n-------",
"_____no_output_____"
],
[
"## Dash tables\n\n### Creating tables with pagination, filters and sorting\n\nDash has tables that can filter and sort data,\nas well as send and respond to signals.\nlet's see how to create them!\n\n```python\nimport dash\nimport dash_table\nimport dash_bootstrap_components as dbc\nimport dash_html_components as html\nfrom dash.dependencies import Output, Input\nimport pandas as pd\nimport altair as alt\nfrom vega_datasets import data\n\n\ncars = data.cars()\n\napp = dash.Dash(__name__, external_stylesheets=[dbc.themes.BOOTSTRAP])\n\napp.layout = dbc.Container(\n dbc.Row([\n dbc.Col([\n dash_table.DataTable(\n id='table',\n columns=[{\"name\": col, \"id\": col} for col in cars.columns[:5]], \n data=cars.to_dict('records'),\n ),\n ])\n ])\n)\n\nif __name__ == '__main__':\n app.run_server(debug=True)\n```\n\nWe only pass the five first columns of the data frame to make it a bit easier to look at for our purposes.\nIn general,\nwe probably don't want to display massive amounts of data in this manner anyways.\n\n\n",
"_____no_output_____"
],
[
"Instead of viewing all the data in one long table,\nwe can enable pagination by specifying e.g. `page_size=10`.\n\n\n\n\nWe can enable sorting by setting `sort_action='native'`,\nwhich gives us small arrows to the click on the left side of each column header.\n\n\n\nFiltering can be enabled in the same manner \nby setting `filter_action='native'`.\n[Dash has it own filter syntax](https://dash.plotly.com/datatable/filtering),\nwhich is similar to what we have used previously with dataframes.\n\n\n\nTo make columns selectable\nwith a radio button,\nwe need to add three lines\n(also showing how to avoid making all columns selectable,\nwe're skipping `'name'` here:\n\n```python\ncolumns=[{\"name\": col, \"id\": col, 'selectable': True if col != 'Name' else False} for col in cars.columns[:5]], \ncolumn_selectable=\"single\",\nselected_columns=['Miles_per_Gallon'], \n```\n\n\nFinally,\nwe can make our table a bit prettier\nby styling it with the following segment,\nwhich color the rows in alternating dark and light colors.\n\n```python\nstyle_cell={'padding': '5px'},\nstyle_data_conditional=[{\n 'if': {'row_index': 'odd'},\n 'backgroundColor': 'rgb(248, 248, 248)'}],\n style_header={\n 'backgroundColor': 'rgb(230, 230, 230)',\n 'fontWeight': 'bold'}\n```\n\n\n\n\nOur entire app now looks like this:\n\n```python\nimport dash\nimport dash_table\nimport dash_bootstrap_components as dbc\nimport dash_html_components as html\nfrom dash.dependencies import Output, Input\nimport pandas as pd\nimport altair as alt\nfrom vega_datasets import data\n\n\ncars = data.cars()\n\napp = dash.Dash(__name__, external_stylesheets=[dbc.themes.BOOTSTRAP])\n\napp.layout = dbc.Container(\n dbc.Row([\n dbc.Col([\n dash_table.DataTable(\n id='table',\n columns=[{\"name\": col, \"id\": col, 'selectable': True if col != 'Name' else False} for col in cars.columns[:5]], \n data=cars.to_dict('records'),\n sort_action=\"native\",\n column_selectable=\"single\",\n selected_columns=['Miles_per_Gallon'], \n page_size=10,\n filter_action='native',\n style_cell={'padding': '5px'},\n style_data_conditional=[{\n 'if': {'row_index': 'odd'},\n 'backgroundColor': 'rgb(248, 248, 248)'}],\n style_header={\n 'backgroundColor': 'rgb(230, 230, 230)',\n 'fontWeight': 'bold'}\n ),\n ])\n ])\n)\n\nif __name__ == '__main__':\n app.run_server(debug=True)\n```",
"_____no_output_____"
],
[
"### Table callbacks\n\nTables have several emitted signals/events that we can listen for,\nthe most relevant in our case are the rows which change when we filter,\nand the column that is selected.\nFirst we add an empty div as a placeholder for the output area,\nbut we will not use it in the beginning.\nInstead,\nlet's just print the value\nwhich will show you that a list of columns is always returned.\n\n```python\[email protected](\n Output('output-div', \"children\"),\n Input('table', \"selected_columns\"))\ndef update_graphs(selected_column):\n print(selected_column)\n```\n\nThe other intersting signal,\nthe rows of the table,\nare a bit more cryptically named (`'derived_virtual_data'`).\n\n```python\[email protected](\n Output('output-div', \"children\"),\n Input('table', \"derived_virtual_data\"),\n Input('table', \"selected_columns\"))\ndef update_graphs(rows, selected_column):\n print(rows)\n```\n\nWhen we print the `rows`, \nyou can see that the format is a list with one dictionary per row.\nTo get this back to a dataframe,\nwe will use `pd.DataFrame(rows)`.\nIf you try filtering,\nyou will see that fewer rows are returned here.",
"_____no_output_____"
],
[
"### Controlling plots from the table callback\n\nWe can now use these callbacks to drive plotting events,\njust as we would have if we had used a dash core components widget.\nThe following would plot a histogram of the selected column and the filtered data points:\n\n```python\n# An Iframe with the 'histogram' ID is also needed\n\[email protected](\n Output('histogram', \"srcDoc\"),\n Input('table', \"derived_virtual_data\"),\n Input('table', \"selected_columns\"))\ndef update_graphs(rows, selected_column):\n chart = alt.Chart(pd.DataFrame(rows)).mark_bar().encode(\n alt.X(selected_column[0], bin=True),\n alt.Y('count()'))\n return chart.to_html()\n```\n\nHere is the entire app when plotting both a histogram and scatter plot\nbased on the state of the table.\n\n```python\nimport dash\nimport dash_table\nimport dash_bootstrap_components as dbc\nimport dash_html_components as html\nfrom dash.dependencies import Output, Input\nimport pandas as pd\nimport altair as alt\nfrom vega_datasets import data\n\n\ncars = data.cars()\n\napp = dash.Dash(__name__, external_stylesheets=[dbc.themes.BOOTSTRAP])\n\napp.layout = dbc.Container(\n dbc.Row([\n dbc.Col([\n dash_table.DataTable(\n id='table',\n columns=[{\"name\": col, \"id\": col, 'selectable': True if col != 'Name' else False} for col in cars.columns[:5]], \n data=cars.to_dict('records'),\n style_cell={'padding': '5px'},\n sort_action=\"native\",\n page_action='native',\n column_selectable=\"single\",\n selected_columns=['Miles_per_Gallon'], \n page_size= 10,\n filter_action='native',\n style_data_conditional=[{\n 'if': {'row_index': 'odd'},\n 'backgroundColor': 'rgb(248, 248, 248)'}],\n style_header={\n 'backgroundColor': 'rgb(230, 230, 230)',\n 'fontWeight': 'bold'}),\n html.Div(id='output-div'),\n html.Iframe(\n id='histogram',\n style={'border-width': '0', 'width': '100%', 'height': '400px'}\n )\n ])\n ])\n)\n\[email protected](\n Output('histogram', \"srcDoc\"),\n Input('table', \"derived_virtual_data\"),\n Input('table', \"selected_columns\"))\ndef update_graphs(rows, selected_column):\n chart = alt.Chart(pd.DataFrame(rows)).mark_bar().encode(\n alt.X(selected_column[0], bin=True),\n alt.Y('count()'))\n chart2 = alt.Chart(pd.DataFrame(rows)).mark_point().encode(\n alt.X(selected_column[0]),\n alt.Y('Horsepower'),\n tooltip='Name')\n return (chart | chart2).to_html()\n\nif __name__ == '__main__':\n app.run_server(debug=True\n```\n\n",
"_____no_output_____"
],
[
"## Updating a table based on a callback\n\nFor callbacks from other widgets or from plotly plots,\nyou can treat tables as any other output element\nand target their `columns` and `data` attributes to update them\nvia a callback function.\n\n```python\nimport dash\nimport dash_table\nimport dash_bootstrap_components as dbc\nimport dash_core_components as dcc\nfrom dash.dependencies import Output, Input\nfrom vega_datasets import data\n\n\ncars = data.cars()\n\napp = dash.Dash(__name__, external_stylesheets=[dbc.themes.BOOTSTRAP])\n\napp.layout = dbc.Container(\n dbc.Row([\n dbc.Col([\n dash_table.DataTable(id='table', page_size=5),\n dcc.Dropdown(\n id='dropdown',\n options=[{\"label\": col, \"value\": col} for col in cars.columns],\n multi=True,\n value=['Horsepower']\n ),\n ])\n ])\n)\n\[email protected](\n Output('table', 'data'),\n Output('table', 'columns'),\n Input('dropdown', 'value'))\ndef update_table(cols):\n columns=[{\"name\": col, \"id\": col} for col in cols]\n data=cars[cols].to_dict('records')\n return data, columns\n\nif __name__ == '__main__':\n app.run_server(debug=True)\n```\n\n",
"_____no_output_____"
],
[
"### Updating a table from an Altair plot\n\nFrom altair plots,\nthere is no way to update a dash table,\nbut if you just need to show a few numbers based on a plot selection,\nyou can [use `mark_text()`\nto update a plot area that looks like a table](https://altair-viz.github.io/gallery/scatter_linked_table.html).",
"_____no_output_____"
]
],
[
[
"import altair as alt\nfrom vega_datasets import data\n\nsource = data.cars()\n\n# Brush for selection\nbrush = alt.selection(type='interval')\n\n# Scatter Plot\npoints = alt.Chart(source).mark_point().encode(\n x='Horsepower:Q',\n y='Miles_per_Gallon:Q',\n color=alt.condition(brush, alt.value('steelblue'), alt.value('grey'))\n).add_selection(brush)\n\n# Base chart for data tables\nranked_text = alt.Chart(source).mark_text(align='right').encode(\n y=alt.Y('row_number:O',axis=None)\n).transform_window(\n row_number='row_number()'\n).transform_filter(\n brush\n).transform_window(\n rank='rank(row_number)'\n).transform_filter(\n alt.datum.rank<16\n)\n\n# Data Tables\nhorsepower = ranked_text.encode(text='Horsepower:N').properties(title=alt.TitleParams(text='Horsepower', align='right'))\nmpg = ranked_text.encode(text='Miles_per_Gallon:N').properties(title=alt.TitleParams(text='MPG', align='right'))\norigin = ranked_text.encode(text='Origin:N').properties(title=alt.TitleParams(text='Origin', align='right'))\ntext = alt.hconcat(horsepower, mpg, origin) # Combine data tables\n\n# Build chart\nalt.hconcat(\n points,\n text\n).resolve_legend(\n color=\"independent\"\n).configure_view(strokeWidth=0)",
"_____no_output_____"
]
],
[
[
"## Dash data tables in R\n\nIn R, we use `purrr` similar to how we did in a dropdown for creating the columns\nand the function `df_to_list` from `dashTable` to convert the dataframe to a dash table friendly format.\n\n```R\nlibrary(dash)\nlibrary(dashTable)\nlibrary(dplyr)\n\nmsleep <- ggplot2::msleep\n\napp <- Dash$new()\n\napp$layout(\n dashDataTable(\n id = \"table\",\n columns = msleep %>%\n colnames %>%\n purrr::map(function(col) list(name = col, id = col)),\n data = df_to_list(msleep),\n page_size = 5\n )\n)\n```\n\n\n",
"_____no_output_____"
],
[
"### Data Table callbacks in R\n\nThe most unusual aspect of this callback is the use of `unlist()`\nto convert the list of column names into a vector of column names\nthat is valid for indexing the dataframe.\n\n```\nlibrary(dash)\nlibrary(dashTable)\nlibrary(dplyr)\nlibrary(dashCoreComponents)\nlibrary(dashBootstrapComponents)\n\nmsleep <- ggplot2::msleep\n\napp <- Dash$new(external_stylesheets = dbcThemes$BOOTSTRAP)\n\napp$layout(\n dbcContainer(\n dbcCol(list(\n dashDataTable(\n id = \"table\",\n page_size = 5),\n dccDropdown(\n id = 'dropdown',\n options = msleep %>% \n colnames %>% \n purrr::map(function(col) list(label = col, value = col)),\n multi = T,\n value = list('name'))\n )\n )\n )\n)\n\napp$callback(\n list(output('table', 'data'),\n output('table', 'columns')),\n list(input('dropdown', 'value')),\n function(cols) {\n columns <- cols %>%\n purrr::map(function(col) list(name = col, id = col))\n data <- df_to_list(msleep %>% select(unlist(cols)))\n print(columns)\n list(data, columns)\n }\n)\n\napp$run_server(debug = T)\n```\n\n",
"_____no_output_____"
],
[
"## Styling",
"_____no_output_____"
],
[
"### Setting plot titles via cards\n\nYou can also put local widgets inside these cards to clearly separate them from the global widgets\nand to indicate which plot they control.\n\n```python\nimport dash\nimport dash_table\nimport dash_bootstrap_components as dbc\nimport dash_html_components as html\nfrom dash.dependencies import Output, Input\nimport pandas as pd\nimport altair as alt\nfrom vega_datasets import data\n\n\ncars = data.cars()\n\napp = dash.Dash(__name__, external_stylesheets=[dbc.themes.BOOTSTRAP])\n\napp.layout = dbc.Container(\n dbc.Row([\n dbc.Col([\n dash_table.DataTable(\n id='table',\n columns=[{\"name\": col, \"id\": col, 'selectable': True if col != 'Name' else False} for col in cars.columns[:5]], \n data=cars.to_dict('records'),\n style_cell={'padding': '5px'},\n sort_action=\"native\",\n page_action='native',\n column_selectable=\"single\",\n selected_columns=['Miles_per_Gallon'], \n page_size= 10,\n filter_action='native',\n style_data_conditional=[{\n 'if': {'row_index': 'odd'},\n 'backgroundColor': 'rgb(248, 248, 248)'}],\n style_header={\n 'backgroundColor': 'rgb(230, 230, 230)',\n 'fontWeight': 'bold'}),\n html.Br(),\n html.Br(),\n html.Br(),\n dbc.Row([\n dbc.Col([\n dbc.Card([\n dbc.CardHeader('Variable distrbution'),\n dbc.CardBody(\n html.Iframe(\n id='histogram',\n style={'border-width': '0', 'width': '100%', 'height': '400px'}))])]),\n dbc.Col([\n dbc.Card([\n dbc.CardHeader('Relation to Horsepower'),\n dbc.CardBody(\n html.Iframe(\n id='scatter',\n style={'border-width': '0', 'width': '100%', 'height': '400px'}))])])\n ])\n ])\n ])\n)\n\[email protected](\n Output('histogram', \"srcDoc\"),\n Input('table', \"derived_virtual_data\"),\n Input('table', \"selected_columns\"))\ndef update_histogram(rows, selected_column):\n chart = alt.Chart(pd.DataFrame(rows)).mark_bar().encode(\n alt.X(selected_column[0], bin=True),\n alt.Y('count()'))\n return chart.properties(width=320, height=320).to_html()\n\[email protected](\n Output('scatter', \"srcDoc\"),\n Input('table', \"derived_virtual_data\"),\n Input('table', \"selected_columns\"))\ndef update_scatter(rows, selected_column):\n chart2 = alt.Chart(pd.DataFrame(rows)).mark_point().encode(\n alt.X(selected_column[0]),\n alt.Y('Horsepower'),\n tooltip='Name')\n return chart2.properties(width=320, height=320).to_html()\n\nif __name__ == '__main__':\n app.run_server(debug=True)\n```\n\n",
"_____no_output_____"
],
[
"## Creating an appealing dashboard title\n\n```python\nimport dash\nimport dash_table\nimport dash_bootstrap_components as dbc\nimport dash_html_components as html\nfrom dash.dependencies import Output, Input\nimport pandas as pd\nimport altair as alt\nfrom vega_datasets import data\n\n\ncars = data.cars()\n\napp = dash.Dash(__name__, external_stylesheets=[dbc.themes.BOOTSTRAP])\n\napp.layout = dbc.Container(\n dbc.Row([\n dbc.Col([\n # dbc.Jumbotron(dbc.Container(html.H1('Layout demo', className='display-3'), fluid=True), fluid=True),\n html.H1('My splashboard demo',\n style={\n 'backgroundColor': 'steelblue',\n 'padding': 20,\n 'color': 'white',\n 'margin-top': 20,\n 'margin-bottom': 20,\n 'text-align': 'center',\n 'font-size': '48px',\n 'border-radius': 3}), #, 'width': 300}),\n # jumbotron,\n dash_table.DataTable(\n id='table',\n columns=[{\"name\": col, \"id\": col, 'selectable': True if col != 'Name' else False} for col in cars.columns[:5]], \n data=cars.to_dict('records'),\n style_cell={'padding': '5px'},\n sort_action=\"native\",\n page_action='native',\n column_selectable=\"single\",\n selected_columns=['Miles_per_Gallon'], \n page_size= 10,\n filter_action='native',\n style_data_conditional=[{\n 'if': {'row_index': 'odd'},\n 'backgroundColor': 'rgb(248, 248, 248)'}],\n style_header={\n 'backgroundColor': 'rgb(230, 230, 230)',\n 'fontWeight': 'bold'}),\n html.Br(),\n html.Br(),\n html.Br(),\n dbc.Row([\n dbc.Col([\n dbc.Card([\n dbc.CardHeader('Variable distrbution', style={'fontWeight': 'bold'}),\n dbc.CardBody(\n html.Iframe(\n id='histogram',\n style={'border-width': '0', 'width': '100%', 'height': '400px'}))])]),\n dbc.Col([\n dbc.Card([\n dbc.CardHeader('Relation to Horsepower', style={'fontWeight': 'bold'}),\n dbc.CardBody(\n html.Iframe(\n id='scatter',\n style={'border-width': '0', 'width': '100%', 'height': '400px'}))])])\n ])\n ])\n ])\n)\n\[email protected](\n Output('histogram', \"srcDoc\"),\n Input('table', \"derived_virtual_data\"),\n Input('table', \"selected_columns\"))\ndef update_histogram(rows, selected_column):\n chart = alt.Chart(pd.DataFrame(rows)).mark_bar().encode(\n alt.X(selected_column[0], bin=True),\n alt.Y('count()'))\n return chart.properties(width=320, height=320).to_html()\n\[email protected](\n Output('scatter', \"srcDoc\"),\n Input('table', \"derived_virtual_data\"),\n Input('table', \"selected_columns\"))\ndef update_scatter(rows, selected_column):\n chart2 = alt.Chart(pd.DataFrame(rows)).mark_point().encode(\n alt.X(selected_column[0]),\n alt.Y('Horsepower'),\n tooltip='Name')\n return chart2.properties(width=320, height=320).to_html()\n\nif __name__ == '__main__':\n app.run_server(debug=True)\n```\n\n",
"_____no_output_____"
],
[
"### Adding a sidebar for global widgets\n\n```python\nimport dash\nimport dash_table\nimport dash_bootstrap_components as dbc\nimport dash_core_components as dcc\nimport dash_html_components as html\nfrom dash.dependencies import Output, Input\nimport pandas as pd\nimport altair as alt\nfrom vega_datasets import data\n\n\ncars = data.cars()\n\napp = dash.Dash(__name__, external_stylesheets=[dbc.themes.BOOTSTRAP])\n\napp.layout = dbc.Container([\n dbc.Row([\n dbc.Col([\n html.H1('My splashboard demo',\n style={\n 'color': 'white',\n 'text-align': 'left',\n 'font-size': '48px',\n }) #, 'width': 300}),\n\n ], style={'backgroundColor': 'steelblue',\n 'border-radius': 3,\n 'padding': 15,\n 'margin-top': 20,\n 'margin-bottom': 20,\n 'margin-right': 15\n })\n \n ]),\n dbc.Row([\n dbc.Col([\n html.H5('Global controls'),\n html.Br(),\n dcc.Dropdown(),\n html.Br(),\n dcc.Dropdown(),\n html.Br(),\n dcc.Dropdown(),\n html.Br(),\n dcc.Dropdown(),\n ],\n md=2,\n style={\n 'background-color': '#e6e6e6',\n 'padding': 15,\n 'border-radius': 3}), \n dbc.Col([\n dash_table.DataTable(\n id='table',\n columns=[{\"name\": col, \"id\": col, 'selectable': True if col != 'Name' else False} for col in cars.columns[:5]], \n data=cars.to_dict('records'),\n style_cell={'padding': '5px'},\n sort_action=\"native\",\n page_action='native',\n column_selectable=\"single\",\n selected_columns=['Miles_per_Gallon'], \n page_size= 10,\n filter_action='native',\n style_data_conditional=[{\n 'if': {'row_index': 'odd'},\n 'backgroundColor': 'rgb(248, 248, 248)'}],\n style_header={\n 'backgroundColor': 'rgb(230, 230, 230)',\n 'fontWeight': 'bold'}),\n html.Br(),\n html.Br(),\n html.Br(),\n dbc.Row([\n dbc.Col([\n dbc.Card([\n dbc.CardHeader('Variable distrbution', style={'fontWeight': 'bold'}),\n dbc.CardBody(\n html.Iframe(\n id='histogram',\n style={'border-width': '0', 'width': '100%', 'height': '400px'}))])]),\n dbc.Col([\n dbc.Card([\n dbc.CardHeader('Relation to Horsepower', style={'fontWeight': 'bold'}),\n dbc.CardBody(\n html.Iframe(\n id='scatter',\n style={'border-width': '0', 'width': '100%', 'height': '400px'}))])])\n ])\n ])\n ]),\n html.Hr(),\n html.P('This dashboard was made by Joel, link to GitHub source. The data is from here and there (include links if appropriate), some copyright/license info')\n])\n\[email protected](\n Output('histogram', \"srcDoc\"),\n Input('table', \"derived_virtual_data\"),\n Input('table', \"selected_columns\"))\ndef update_histogram(rows, selected_column):\n chart = alt.Chart(pd.DataFrame(rows)).mark_bar().encode(\n alt.X(selected_column[0], bin=True),\n alt.Y('count()'))\n return chart.properties(width=320, height=320).to_html()\n\[email protected](\n Output('scatter', \"srcDoc\"),\n Input('table', \"derived_virtual_data\"),\n Input('table', \"selected_columns\"))\ndef update_scatter(rows, selected_column):\n chart2 = alt.Chart(pd.DataFrame(rows)).mark_point().encode(\n alt.X(selected_column[0]),\n alt.Y('Horsepower'),\n tooltip='Name')\n return chart2.properties(width=320, height=320).to_html()\n\nif __name__ == '__main__':\n app.run_server(debug=True)\n```\n\n",
"_____no_output_____"
],
[
"## Adding introductory text and references to code and data\n\n```python\nimport dash\nimport dash_table\nimport dash_bootstrap_components as dbc\nimport dash_core_components as dcc\nimport dash_html_components as html\nfrom dash.dependencies import Output, Input, State\nfrom datetime import datetime\nimport pandas as pd\nimport altair as alt\nfrom vega_datasets import data\n\n\ncars = data.cars()\n\napp = dash.Dash(__name__, external_stylesheets=[dbc.themes.BOOTSTRAP])\n\ncollapse = html.Div(\n [\n dbc.Button(\n \"Learn more\",\n id=\"collapse-button\",\n className=\"mb-3\",\n outline=False,\n style={'margin-top': '10px',\n 'width': '150px',\n 'background-color': 'white',\n 'color': 'steelblue'}\n ),\n ]\n)\n\n\[email protected](\n Output(\"collapse\", \"is_open\"),\n [Input(\"collapse-button\", \"n_clicks\")],\n [State(\"collapse\", \"is_open\")],\n)\ndef toggle_collapse(n, is_open):\n if n:\n return not is_open\n return is_open\n\napp.layout = dbc.Container([\n dbc.Row([\n dbc.Col([\n dbc.Row([\n dbc.Col([\n html.H1('My splashboard demo',\n style={\n 'color': 'white',\n 'text-align': 'left',\n 'font-size': '48px', #, 'width': 300}),\n }),\n dbc.Collapse(\n html.P(\"\"\"\n This dashboard is helping you understand x, y, and z, \n which are really important because a, b, c.\n Start using the dashboard by clicking on 1, 2, 3\n and pulling i, ii, and iii.\"\"\",\n style={'color': 'white', 'width': '50%'}),\n id=\"collapse\",\n ),\n\n ],\n md=10),\n dbc.Col([collapse])\n\n \n ])\n ], style={'backgroundColor': 'steelblue',\n 'border-radius': 3,\n 'padding': 15,\n 'margin-top': 20,\n 'margin-bottom': 20,\n 'margin-right': 15\n })\n \n ]),\n dbc.Row([\n dbc.Col([\n html.H5('Global controls'),\n html.Br(),\n dcc.Dropdown(),\n html.Br(),\n dcc.Dropdown(),\n html.Br(),\n dcc.Dropdown(),\n html.Br(),\n dcc.Dropdown(),\n ],\n md=2,\n style={\n 'background-color': '#e6e6e6',\n 'padding': 15,\n 'border-radius': 3}), \n dbc.Col([\n dash_table.DataTable(\n id='table',\n columns=[{\"name\": col, \"id\": col, 'selectable': True if col != 'Name' else False} for col in cars.columns[:5]], \n data=cars.to_dict('records'),\n style_cell={'padding': '5px'},\n sort_action=\"native\",\n page_action='native',\n column_selectable=\"single\",\n selected_columns=['Miles_per_Gallon'], \n page_size= 10,\n filter_action='native',\n style_data_conditional=[{\n 'if': {'row_index': 'odd'},\n 'backgroundColor': 'rgb(248, 248, 248)'}],\n style_header={\n 'backgroundColor': 'rgb(230, 230, 230)',\n 'fontWeight': 'bold'}),\n html.Br(),\n html.Br(),\n html.Br(),\n dbc.Row([\n dbc.Col([\n dbc.Card([\n dbc.CardHeader('Variable distrbution', style={'fontWeight': 'bold'}),\n dbc.CardBody(\n html.Iframe(\n id='histogram',\n style={'border-width': '0', 'width': '100%', 'height': '400px'}))])]),\n dbc.Col([\n dbc.Card([\n dbc.CardHeader('Relation to Horsepower', style={'fontWeight': 'bold'}),\n dbc.CardBody(\n html.Iframe(\n id='scatter',\n style={'border-width': '0', 'width': '100%', 'height': '400px'}))])])\n ])\n ])\n ]),\n html.Hr(),\n html.P(f'''\n This dashboard was made by Joel, link to GitHub source.\n The data is from here and there (include links if appropriate), some copyright/license info\n Mention when the dashboard was latest updated (and data if appropriate).\n This will show the date when you last resarted the server: {datetime.now().date()}\n ''')\n])\[email protected](\n Output('histogram', \"srcDoc\"),\n Input('table', \"derived_virtual_data\"),\n Input('table', \"selected_columns\"))\ndef update_histogram(rows, selected_column):\n chart = alt.Chart(pd.DataFrame(rows)).mark_bar().encode(\n alt.X(selected_column[0], bin=True),\n alt.Y('count()'))\n return chart.properties(width=300, height=300).to_html()\n\[email protected](\n Output('scatter', \"srcDoc\"),\n Input('table', \"derived_virtual_data\"),\n Input('table', \"selected_columns\"))\ndef update_scatter(rows, selected_column):\n chart2 = alt.Chart(pd.DataFrame(rows)).mark_point().encode(\n alt.X(selected_column[0]),\n alt.Y('Horsepower'),\n tooltip='Name')\n return chart2.properties(width=300, height=300).to_html()\n\nif __name__ == '__main__':\n app.run_server(debug=True)\n```\n\n",
"_____no_output_____"
],
[
"## Performance",
"_____no_output_____"
],
[
"### Loading data from binary formats\n\nIf you initial load times are slow,\nconsider storing the processed data (still keep a copy of the raw)\nas a binary file format.\n[According to the pandas docs,\nfeather and hdf5 are the fastest](https://pandas.pydata.org/pandas-docs/stable/user_guide/io.html#performance-considerations).\nIf you have complex data structures like nested lists,\nthese might not work,\nbut saving to a `pickle` (Python's native binary format)\nstill should and is also very fast.\n\nWhen working with `altair` and large data sets,\nyou can enable the altair data server like we did in 531\nin order to avoid writing all the data to each plot's json \nwhich can take a long time.\n[Uploading it to your server as a compatible json format (`data.to_json(path, orient='records'`)\nand reading directly from that path/URL is also a good approach](https://altair-viz.github.io/user_guide/faq.html?highlight=url#why-does-altair-lead-to-such-extremely-large-notebooks).",
"_____no_output_____"
],
[
"### Caching/memoization\n\nIf we have a time consuming computation step that generates an output,\nwe can save that output to the file system (or a database)\nso that next time we don't have to compute it again but just read it in.\nTrading space for time like this\nis called chaching or memoization.\n\nFirst let's create an app that simulates a timeconsuming step,\nby sleeping the process for a few seconds between each call.\nEvery time we select from the dropdown menu,\nthis app will do the time consuming process of reading in the data again,\nso we have to wait a couple of seconds for the output to refresh.\nIf we look at the timestamp in the `time` column,\nwe can see that it is always the same\nas the current time\n(because it is rerun each time the dropdown changes).\n\n```python\nimport time\nfrom datetime import datetime\n\nimport dash\nimport dash_core_components as dcc\nimport dash_html_components as html\nimport numpy as np\nimport pandas as pd\nfrom dash.dependencies import Input, Output\n\nexternal_stylesheets = ['https://codepen.io/chriddyp/pen/bWLwgP.css']\n\napp = dash.Dash(__name__, external_stylesheets=external_stylesheets)\n\ndef query_data():\n # Simulate a time consuming step with sleep\n time.sleep(2)\n df = pd.DataFrame(\n np.random.randint(0, 100, size=(100, 4)),\n columns=list('ABCD'))\n df['time'] = datetime.now().strftime('%H:%M:%S')\n return df.to_json(date_format='iso', orient='split')\n\n\ndef dataframe():\n return pd.read_json(query_data(), orient='split')\n\napp.layout = html.Div([\n html.H6(id='current-time'),\n dcc.Dropdown(\n id='live-dropdown',\n value='A',\n options=[{'label': i, 'value': i} for i in dataframe().columns]),\n html.Iframe(\n id='output-area',\n style={'border-width': '0', 'width': '100%', 'height': '400px'})])\n\n\[email protected](Output('output-area', 'srcDoc'),\n Output('current-time', 'children'),\n Input('live-dropdown', 'value'))\ndef update_live_graph(col):\n print(col)\n df = dataframe()\n current_time = f\"The current time is {datetime.now().strftime('%H:%M:%S')}\"\n return df[[col]].to_html(index=False), current_time\n\nif __name__ == '__main__':\n app.run_server(debug=True)\n```\n\n\n\nIdeally,\nwe would do most of our timeconsuming wrangling in the beginning when the page load\nor even better,\nbeforehand and save a processed dataset,\nbut there might be some wrangling or other time consuming process that can't be avoided.\nOften we don't need these process to update each time a selection is made,\nand we would be fine reusing the data from the selection made a few seconds ago.\nWe can set this up using the [flask-caching](https://flask-caching.readthedocs.io/en/latest/) package\n(`conda install flask-caching`) like this:\n\n```python\nfrom flask_caching import Cache\n\n\ncache = Cache(app.server, config={\n 'CACHE_TYPE': 'filesystem',\n 'CACHE_DIR': 'cache-directory'})\n\[email protected]()\ndef query_date():\n ...\n```\n\nThe decorator indicates which function to be cached,\nand the return value from this function will be save on the local filesystem\nin a directory name `cache-directory`.\nOur entire app now looks like this and you can see when you select a new dropdown value\nthat the time column will remain the same because it is not being rerun each time,\ninstead it is reusing data from an earlier run.\n\n```python\nimport time\nfrom datetime import datetime\n\nimport dash\nimport dash_core_components as dcc\nimport dash_html_components as html\nimport numpy as np\nimport pandas as pd\nfrom dash.dependencies import Input, Output\nfrom flask_caching import Cache\n\n\nexternal_stylesheets = ['https://codepen.io/chriddyp/pen/bWLwgP.css']\n\napp = dash.Dash(__name__, external_stylesheets=external_stylesheets)\n\ncache = Cache(app.server, config={\n 'CACHE_TYPE': 'filesystem',\n 'CACHE_DIR': 'cache-directory'})\n\[email protected]()\ndef query_data():\n # Simulate a time consuming step with sleep\n time.sleep(2)\n df = pd.DataFrame(\n np.random.randint(0, 100, size=(100, 4)),\n columns=list('ABCD'))\n df['time'] = datetime.now().strftime('%H:%M:%S')\n return df.to_json(date_format='iso', orient='split')\n\n\ndef dataframe():\n return pd.read_json(query_data(), orient='split')\n\napp.layout = html.Div([\n html.H6(id='current-time'),\n dcc.Dropdown(\n id='live-dropdown',\n value='A',\n options=[{'label': i, 'value': i} for i in dataframe().columns]),\n html.Iframe(\n id='output-area',\n style={'border-width': '0', 'width': '100%', 'height': '400px'})])\n\[email protected](Output('output-area', 'srcDoc'),\n Output('current-time', 'children'),\n Input('live-dropdown', 'value'))\ndef update_live_graph(col):\n print(col)\n df = dataframe()\n current_time = f\"The current time is {datetime.now().strftime('%H:%M:%S')}\"\n return df[[col]].to_html(index=False), current_time\n\nif __name__ == '__main__':\n app.run_server(debug=True)\n```\n\n\n\n\nWe could also add a timeout (in seconds) for how long the cache should be saved.\nFor example, if the dropdown is used after 20 seconds, \nthe function will run again.\n\n```\[email protected](timeout=20)\n```\n\nThere are [more details on caching in the dash docs](https://dash.plotly.com/performance).",
"_____no_output_____"
],
[
"## Entertaining the user\n\nAnother approach to dealing with slow callbacks is to clearly show the user that the page is loading.\n[Dash has built in support for different loading animations](https://dash.plotly.com/dash-core-components/loading)\nthat can be triggered by wrapping an output component in `dcc.Loading`.\n\n```python\nimport time\nfrom datetime import datetime\n\nimport dash\nimport dash_core_components as dcc\nimport dash_html_components as html\nimport numpy as np\nimport pandas as pd\nfrom dash.dependencies import Input, Output\n\n\nexternal_stylesheets = ['https://codepen.io/chriddyp/pen/bWLwgP.css']\n\napp = dash.Dash(__name__, external_stylesheets=external_stylesheets)\n\ndef query_data():\n # Simulate a time consuming step with sleep\n time.sleep(2)\n df = pd.DataFrame(\n np.random.randint(0, 100, size=(100, 4)),\n columns=list('ABCD'))\n df['time'] = datetime.now().strftime('%H:%M:%S')\n return df.to_json(date_format='iso', orient='split')\n\n\ndef dataframe():\n return pd.read_json(query_data(), orient='split')\n\napp.layout = html.Div([\n html.H6(id='current-time'),\n dcc.Dropdown(\n id='live-dropdown',\n value='A',\n options=[{'label': i, 'value': i} for i in dataframe().columns]),\n dcc.Loading(\n children=html.Iframe(\n id='output-area',\n style={'border-width': '0', 'width': '100%', 'height': '400px'}))])\n\[email protected](Output('output-area', 'srcDoc'),\n Output('current-time', 'children'),\n Input('live-dropdown', 'value'))\ndef update_live_graph(col):\n print(col)\n df = dataframe()\n current_time = f\"The current time is {datetime.now().strftime('%H:%M:%S')}\"\n return df[[col]].to_html(index=False), current_time\n\nif __name__ == '__main__':\n app.run_server(debug=True)\n```\n\nYou can change the type to any of `'graph'`, `'cube'`, `'circle'`, or `'dot'`.\nBut don't overdo it!\nYou don't want to give an not so serious imporession and undermine your carefully crafted visualizations,\nby having too many toy-like animations in your dashboards.\n\n",
"_____no_output_____"
],
[
"### Running on multiple workers\n\nYou can run your server on multiple threads locally via setting `app.run_server(processes=4)` or similar.\nThis might not work,\nand for running it in a production environment\nyou would use this instead in your `Procfile`: `gunicorn --workers 6 --threads 2 src/app:server --pythonpath=src`.\nI don't think there is an equivalent for dash R.\nYou can see [an example on this page for more details](https://dash.plotly.com/sharing-data-between-callbacks).\n\nNote that this is one of the reasons it is a bad idea to modify global variables inside a functions.\nThe dash docs explain why this can lead to unintended consequences and miscomputations in your app:\n\n> When Dash apps run across multiple workers, their memory is not shared. This means that if you modify a global variable in one callback, that modification will not be applied to the rest of the workers.\n>\n> https://dash.plotly.com/sharing-data-between-callbacks\n",
"_____no_output_____"
],
[
"### Better plot performance for many data points\n\n\nThese options are only available via Plotly, not Altair.\nFor Altair, [work is under way](https://github.com/vega/scalable-vega) to make it handle larger datasets,\nbut still not ready to use easily.",
"_____no_output_____"
],
[
"### WebGL for millions of points\n\nWebGL is a Javascript library that allows for GPU rendered graphics inside browsers.\nLeveraging this, it is possible to render millions of points without stuttering.\nWebGL backends are available for Holoviews, Bokeh, and Plotly\n([WebGL for Python](https://plotly.com/r/webgl-vs-svg/) and [WebGL for R](https://plotly.com/r/webgl-vs-svg/)).",
"_____no_output_____"
],
[
"### Datashader for billion of points\n\nThe Python package [Datashader](https://datashader.org/) works by rendering plots to images of lower resolution\nwhen you are looking at them from far away\n(since you cannot distinguish billions of points anyways),\nand then increases the resolution of the image as you zoom in\non an area (but then only renders the data set within the zoomed in region).\n\nDatashader is developed by the Anaconda and Holoviews team,\nand is available to use directly with Holoviews, Bokeh and [Plotly](https://plotly.com/python/datashader/) (in Python only).\nSince it creates an image it can also be used with any other library\nthat can show images (such as Altair),\nbut it is much easier and feature filled when used with a library that directly integrates it,\nlike the three mentioned above.\nDatashader can create beautiful heatmap images,\nand you [view some datashader examples in this gallery](https://plotly.com/python/datashader/),\ne.g. [this one about ship traffic along the US coast](https://examples.pyviz.org/ship_traffic/ship_traffic.html#ship-traffic-gallery-ship-traffic).\n\n\n\n#### rasterly for R\n\nPlotly has developed the [rasterly library to bring some of the functionality of datashader to R](https://z267xu.github.io/rasterly/).",
"_____no_output_____"
],
[
"## Attribution\n\nThese lecture notes were prepared by Dr. Joel Ostblom, a post-doctoral teaching fellow in the UBC Vancouver MDS program.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecba2272fefce76f23b40f8bf40544ee5480f8ea | 20,028 | ipynb | Jupyter Notebook | Classroom-Codes/Grid World - Value Iteration Algorithm.ipynb | adityajn105/Move37 | ed94508cd76b01f5f0655d2670dd35d8fceeceeb | [
"MIT"
] | 1 | 2020-09-12T14:29:38.000Z | 2020-09-12T14:29:38.000Z | Classroom-Codes/Grid World - Value Iteration Algorithm.ipynb | adityajn105/Move37 | ed94508cd76b01f5f0655d2670dd35d8fceeceeb | [
"MIT"
] | null | null | null | Classroom-Codes/Grid World - Value Iteration Algorithm.ipynb | adityajn105/Move37 | ed94508cd76b01f5f0655d2670dd35d8fceeceeb | [
"MIT"
] | null | null | null | 31.39185 | 267 | 0.404733 | [
[
[
"### Problem\n1. It contains 4 columns and 3 rows\n2. Position 1,4 has Queen\n3. Position 2,4 has firepit\n4. Position 2,2 has wall",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"\"\"\"\nOur Grid Object\n\"\"\"\nclass Grid:\n def __init__(self,width,height,start):\n self.width = width\n self.height = height\n self.i = start[0] #vertical axis\n self.j = start[1] #horizontal axis\n \n def set(self,actions,rewards,obey_prob):\n self.actions = actions # dict: (row,col): list of actions\n self.rewards = rewards # dict: (row,col): reward\n self.obey_prob = obey_prob\n \n def non_terminal_states(self):\n return self.actions.keys()\n \n def set_state(self,s):\n #change current stae\n self.i = s[0]\n self.j = s[1]\n \n def current_state(self):\n return (self.i,self.j)\n \n def is_terminal(self,s):\n return s not in self.actions\n \n def check_move(self,action):\n i = self.i\n j = self.j\n #check move validity\n if action in self.actions[(i,j)]:\n if action == \"U\":\n i-=1\n elif action == \"D\":\n i+=1\n elif action == \"R\":\n j+=1\n elif action == \"L\":\n j-=1\n \n reward = self.rewards.get((i,j),0)\n return ((i,j),reward)\n \n def get_transition_probs(self,action):\n # returns a list of (probability, reward, s') transition tuples\n probs = []\n state, reward = self.check_move(action)\n probs.append((self.obey_prob,reward,state))\n \n \"\"\"\n if obey prob is 0.8\n and if action is 'up' then \n 'up' has probabiltiy of 0.8\n 'left' has probability of 0.1\n 'right' has probability of 0.1\n \"\"\"\n disobey_prob = 1-self.obey_prob\n if disobey_prob <= 0:\n return probs\n if action == \"U\" or action == \"D\":\n state, reward = self.check_move(\"L\")\n probs.append((disobey_prob/2,reward,state))\n state, reward = self.check_move(\"R\")\n probs.append((disobey_prob/2,reward,state))\n elif action == \"L\" or action == \"R\":\n state, reward = self.check_move(\"U\")\n probs.append((disobey_prob/2,reward,state))\n state,reward = self.check_move(\"D\")\n probs.append((disobey_prob/2,reward,state))\n return probs\n \n def game_over(self):\n #return True if game is over else False\n #true if we are in state where no action are possible\n return (self.i,self.j) not in self.actions.keys()\n \n def all_states(self):\n # get all states\n # either a position that has possible next actions\n # or a position that yields a reward\n return set(self.actions.keys()) | set(self.rewards.keys())",
"_____no_output_____"
],
[
"\"\"\"\nInitialization of Grid\n\"\"\"\ndef standard_grid(obey_prob=0.1, step_cost = None):\n # lets define a grid that describes the reward for getting into each state\n # and possible actions at each state\n # the grid looks like this\n # x means you can't go there\n # s means start position\n # number means reward at that state\n # . . . 1\n # . x . -1\n # s . . .\n # obey_brob (float): the probability of obeying the command\n # step_cost (float): a penalty applied each step to minimize the number of moves (-0.1)\n g = Grid(3, 4, (2, 0))\n \n rewards = {(0,3):+1, (1,3):-1}\n start = (2,0)\n actions = {\n (0,0) : [\"R\",\"D\"],\n (0,1) : [\"L\",\"R\"],\n (0,2) : [\"L\",\"D\",\"R\"],\n (1,0) : [\"U\",\"D\"],\n (1,2) : [\"U\",\"D\",\"R\"],\n (2,0) : [\"U\",\"R\"],\n (2,1) : [\"L\",\"R\"],\n (2,2) : [\"L\",\"U\",\"R\"],\n (2,3) : [\"L\",\"U\"]\n }\n \n g.set(actions,rewards,obey_prob)\n if step_cost is not None:\n g.rewards.update({\n (0, 0): step_cost,\n (0, 1): step_cost,\n (0, 2): step_cost,\n (1, 0): step_cost,\n (1, 2): step_cost,\n (2, 0): step_cost,\n (2, 1): step_cost,\n (2, 2): step_cost,\n (2, 3): step_cost,\n })\n return g",
"_____no_output_____"
],
[
"\"\"\"\nDisplaying Results\n\"\"\"\ndef print_values(V, g):\n for i in range(g.width):\n print(\"---------------------------\")\n for j in range(g.height):\n v = V.get((i,j), 0)\n if v >= 0:\n print(\" %.2f|\" % v, end=\"\")\n else:\n print(\"%.2f|\" % v, end=\"\") # -ve sign takes up an extra space\n print(\"\")\n\ndef print_policy(P, g):\n for i in range(g.width):\n print(\"---------------------------\")\n for j in range(g.height):\n a = P.get((i,j), ' ')\n print(\" %s |\" % a, end=\"\")\n print(\"\")",
"_____no_output_____"
],
[
"\"\"\"\nValue-Iteration\n\"\"\"\nSMALL_ENOUGH = 1e-3\nGAMMA = 0.9\nALL_POSSIBLE_ACTIONS = [\"U\",\"D\",\"R\",\"L\"]\n\ndef best_action_value(grid,V,s):\n # finds the highest value action (max_a) from state s, returns the action and value\n best_a = None\n best_value = float('-inf')\n grid.set_state(s)\n \n #loop through all possible action to find the best possible actions\n for action in ALL_POSSIBLE_ACTIONS:\n transitions = grid.get_transition_probs(action)\n expected_v = 0\n expected_r = 0\n for (prob,r,state_prime) in transitions:\n expected_v += prob*V[state_prime]\n expected_r += prob*r\n v = expected_r + GAMMA* expected_v\n if v > best_value:\n best_value = v\n best_a = action\n return (best_a,best_value)\n\n\ndef calculate_values(grid):\n V = {}\n states = grid.all_states()\n for s in states:\n V[s] = 0\n \n #repeat until convergence\n #V[s] = max[a]{ sum[s',r] { p(s',r|s,a)[r + gamma*V[s']] } }\n while True:\n biggest_change = 0\n for s in grid.non_terminal_states():\n old_v = V[s]\n _,new_v = best_action_value(grid,V,s)\n V[s] = new_v\n biggest_change = max(biggest_change,np.abs(old_v-new_v))\n if biggest_change <= SMALL_ENOUGH:\n break;\n return V",
"_____no_output_____"
],
[
"def initialize_random_policy():\n # policy is a lookup table for state -> action\n # we'll randomly choose an action and update as we learn\n policy = {}\n for s in grid.non_terminal_states():\n policy[s] = np.random.choice(ALL_POSSIBLE_ACTIONS)\n return policy\n\n\ndef calculate_greedy_policy(grid, V):\n policy = initialize_random_policy()\n # find a policy that leads to optimal value function\n for s in policy.keys():\n grid.set_state(s)\n # loop through all possible actions to find the best current action\n best_a, _ = best_action_value(grid, V, s)\n policy[s] = best_a\n return policy",
"_____no_output_____"
],
[
"def print_game(grid):\n # print rewards\n print(\"rewards:\")\n print_values(grid.rewards, grid)\n\n # calculate accurate values for each square\n V = calculate_values(grid)\n\n # calculate the optimum policy based on our values\n policy = calculate_greedy_policy(grid, V)\n\n # our goal here is to verify that we get the same answer as with policy iteration\n print(\"\\nvalues:\")\n print_values(V, grid)\n print(\"\\npolicy:\")\n print_policy(policy, grid)",
"_____no_output_____"
],
[
"grid = standard_grid(obey_prob=1.0, step_cost=None)\nprint_game(grid)",
"rewards:\n---------------------------\n 0.00| 0.00| 0.00| 1.00|\n---------------------------\n 0.00| 0.00| 0.00|-1.00|\n---------------------------\n 0.00| 0.00| 0.00| 0.00|\n\nvalues:\n---------------------------\n 0.81| 0.90| 1.00| 0.00|\n---------------------------\n 0.73| 0.00| 0.90| 0.00|\n---------------------------\n 0.66| 0.73| 0.81| 0.73|\n\npolicy:\n---------------------------\n R | R | R | |\n---------------------------\n U | | U | |\n---------------------------\n U | R | U | L |\n"
]
],
[
[
"#### obey_prob = 1.0 | step_down = None\nMario is not drunk it will follow what it said, it is most simple one",
"_____no_output_____"
]
],
[
[
"grid = standard_grid(obey_prob=0.8, step_cost=None)\nprint_game(grid)",
"rewards:\n---------------------------\n 0.00| 0.00| 0.00| 1.00|\n---------------------------\n 0.00| 0.00| 0.00|-1.00|\n---------------------------\n 0.00| 0.00| 0.00| 0.00|\n\nvalues:\n---------------------------\n 0.72| 0.83| 0.94| 0.00|\n---------------------------\n 0.63| 0.00| 0.64| 0.00|\n---------------------------\n 0.54| 0.48| 0.53| 0.31|\n\npolicy:\n---------------------------\n R | R | R | |\n---------------------------\n U | | U | |\n---------------------------\n U | L | U | L |\n"
]
],
[
[
"#### obey_prob = 0.8 | step_down = None\nMario is little drunk, so it will try to avoid right path, so that chances of falling in pit is less",
"_____no_output_____"
]
],
[
[
"grid = standard_grid(obey_prob=0.5, step_cost=None)\nprint_game(grid)",
"rewards:\n---------------------------\n 0.00| 0.00| 0.00| 1.00|\n---------------------------\n 0.00| 0.00| 0.00|-1.00|\n---------------------------\n 0.00| 0.00| 0.00| 0.00|\n\nvalues:\n---------------------------\n 0.48| 0.63| 0.77| 0.00|\n---------------------------\n 0.39| 0.00| 0.44| 0.00|\n---------------------------\n 0.30| 0.24| 0.30| 0.20|\n\npolicy:\n---------------------------\n R | R | R | |\n---------------------------\n U | | L | |\n---------------------------\n U | L | U | D |\n"
]
],
[
[
"#### obey_prob = 0.5 | step_down = None\nMario is lot drunk, it will try to follow left path as it is safe. and on (1,2) it will start bumping into wall just to avoid pit. We can try to predict best move there\n* up = (0.5x0 + 0.5x0.9x0.77) + (0.25x0+ 0.25x0.9x.44) + (0.25x-1 + 0.25x0.9x0 ) = 0.3465+0.099-1 = -0.5545\n* left = (0.5x0 + 0.5x0.9x0.44 )+ (0.25x0+ 0.25x0.9x0.77) + (0.25x0+ 0.25x0.9x0.30) = 0.198+0.17325+0.0675 = 0.43875\n* So, Left seem to be more rewarding",
"_____no_output_____"
]
],
[
[
"grid = standard_grid(obey_prob=0.5, step_cost=-0.1)\nprint_game(grid)",
"rewards:\n---------------------------\n-0.10|-0.10|-0.10| 1.00|\n---------------------------\n-0.10| 0.00|-0.10|-1.00|\n---------------------------\n-0.10|-0.10|-0.10|-0.10|\n\nvalues:\n---------------------------\n-0.03| 0.27| 0.55| 0.00|\n---------------------------\n-0.21| 0.00|-0.10| 0.00|\n---------------------------\n-0.39|-0.50|-0.38|-0.57|\n\npolicy:\n---------------------------\n R | R | R | |\n---------------------------\n U | | U | |\n---------------------------\n U | R | U | D |\n"
]
],
[
[
"#### obey_prob = 0.5 | step_down = -0.1\nMario is still lot drunk but it will have to reach goal as soon as possible, it will try to follow left path as it is safe. This time mario will take a risk on (1,2). We can try to predict best move there\n* up = (0.5x-0.1 + 0.5x0.9x0.55) + (0.25x-.10+ 0.25x0.9x-.10) + (0.25x-1 + 0.25x0.9x0 ) = 0.2965-0.0475-0.25 = -0.001\n* left = (0.5x-0.1 + 0.5x0.9x-0.1 )+ (0.25x-0.1+ 0.25x0.9x0.55) + (0.25x-0.1+ 0.25x0.9x-0.38) = -0.095+0.09875-0.1105 = -0.10675\n* So, here up seems to be more rewarding\n* weird thing happened at (2,3), mario seem to prefer in stay in same state rather than moving, as staying is more rewarding.",
"_____no_output_____"
]
],
[
[
"grid = standard_grid(obey_prob=0.5, step_cost=-0.5)\nprint_game(grid)",
"rewards:\n---------------------------\n-0.50|-0.50|-0.50| 1.00|\n---------------------------\n-0.50| 0.00|-0.50|-1.00|\n---------------------------\n-0.50|-0.50|-0.50|-0.50|\n\nvalues:\n---------------------------\n-1.82|-0.82| 0.11| 0.00|\n---------------------------\n-2.40| 0.00|-0.74| 0.00|\n---------------------------\n-2.66|-2.28|-1.68|-1.45|\n\npolicy:\n---------------------------\n R | R | R | |\n---------------------------\n U | | U | |\n---------------------------\n R | R | U | U |\n"
]
],
[
[
"#### obey_prob = 0.5 | step_down = -0.5\nMario is still lot drunk but it will have to reach goal as soon as possible such that again a weired thing happened at (2,3), mario decided to commit sucide. Lets see why\n* up = ( 0.5x-1 + 0.5x0.9x0 )+( 0.25x-0.5 + 0.25x0.9x-1.68 )+(0.25x-0.5+0.25x0.9x-1.45) = -0.5-0.503-0.45125 = -1.45425\n* down = (0.5x-0.5+0.5x0.9x-1.45)+(0.25x-0.5+0.25x0.9x-1.45)+(0.25x-0.5+0.25x0.9x-1.68) = -0.9025-0.45125-0.503 =-1.85675\n* As it is clear, sucide is best option for mario",
"_____no_output_____"
]
],
[
[
"grid = standard_grid(obey_prob=1, step_cost=-2)\nprint_game(grid)",
"rewards:\n---------------------------\n-2.00|-2.00|-2.00| 1.00|\n---------------------------\n-2.00| 0.00|-2.00|-1.00|\n---------------------------\n-2.00|-2.00|-2.00|-2.00|\n\nvalues:\n---------------------------\n-2.99|-1.10| 1.00| 0.00|\n---------------------------\n-4.69| 0.00|-1.00| 0.00|\n---------------------------\n-6.15|-4.61|-2.90|-1.00|\n\npolicy:\n---------------------------\n R | R | R | |\n---------------------------\n U | | R | |\n---------------------------\n R | R | U | U |\n"
]
],
[
[
"#### obey_prob = 0.5 | step_down = -2\nMario is still lot drunk but it will have to reach goal as soon as possible and such that moving one extra tile is having more penalty which cant be compensated by any future reward. So here also mario prefers to die in some of the situations. Poor Mario!!!....",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecba313b8a99a2bcc8bc94ce100dc41963d6d28a | 4,130 | ipynb | Jupyter Notebook | content/03/02b_pandasVocab.ipynb | schemesmith/ledatascifi-2021 | f3a081c794655a2f560f8b72edd827ed5c90ada3 | [
"MIT"
] | null | null | null | content/03/02b_pandasVocab.ipynb | schemesmith/ledatascifi-2021 | f3a081c794655a2f560f8b72edd827ed5c90ada3 | [
"MIT"
] | null | null | null | content/03/02b_pandasVocab.ipynb | schemesmith/ledatascifi-2021 | f3a081c794655a2f560f8b72edd827ed5c90ada3 | [
"MIT"
] | null | null | null | 36.875 | 214 | 0.525908 | [
[
[
"# Pandas Vocab and the Shape of Data\n\nPandas is a library that helps you work with data! \n\nAt the top of your python code, load Pandas like this:\n```py\nimport pandas as pd\n```\nNow you can use pandas throughout your file via the `pd` object. \n\n\n\n## Pandas Vocab\n\n- The key object in the pandas library is that you put data into **dataframes**, which are like Excel spreadsheets\n- Variables are in columns (which have a **name** that identifies the column)\n- Observations are in rows (which have an **index** that identifies the row)\n - _In our \"Golden Rules\" chapter we used the term \"key\" (which I prefer), but pandas uses Index._\n- If you create an object with a single variable, pandas might store it as a **series** object\n- **\"Wide data\"** vs. **\"Long data\"**: See the [next section](#The-shape-of-data)",
"_____no_output_____"
],
[
"## The shape of data\n\n> _**The fundamental principle of database design is that the physical structure of a database should communicate its logical structure**_\n\nData can be logically stored in many ways. Let's start by showing one dataset, and three ways it can be stored.\n\n### Wide vs. Long (or Tall) Data\n\nHere is a **long dataset** - the \"key\" or \"index\" is the _combination of year and firm_:\n\n| Year | Firm | Sales | Profits |\n| :--- | :--- | :--- | :--- |\n| 2000 | Ford | 10 | 1 |\n| 2001 | Ford | 12 | 1 | \n| 2002 | Ford | 14 | 1 |\n| 2003 | Ford | 16 | 1 | \n| 2000 | GM | 11 | 0 |\n| 2001 | GM | 13 | 2 | \n| 2002 | GM | 13 | 0 | \n| 2003 | GM | 15 | 2 | \n\nThe exact same data, stored as a **wide dataset** - - the \"key\" or \"index\" is the _year_, and each variable is duplicated for each firm:\n\n| <br> Year | Sales <br> GM| <br> Ford | Profits <br> GM| <br> Ford |\n| :--- | :--- | :--- | :--- | :--- |\n| 2000 | 11 | 10 | 0 | 1 |\n| 2001 | 13 | 12 | 2 | 1 |\n| 2002 | 13 | 14 | 0 | 1 |\n| 2003 | 15 | 16 | 2 | 1 |\n\n```{admonition} \"MultiIndex\" in pandas\nNotice here how the variables have multiple levels for the variable name: level 0 is \"Sales\" which applies to the level 1 \"GM\" and \"Ford\". Thus, column 2 is Sales of GM and column 3 is Sales of Ford.\n\nThis combination of the two levels of the variable name is called a \"MultiIndex\" in pandas.\n```\n\nThe exact same data, stored as a **wide dataset** - - the \"key\" or \"index\" is the _firm_, and each variable is duplicated for each year:\n\n| <br> Firm | Sales <br> 2000 | <br> 2001 | <br> 2002 | <br> 2003 | Profits <br> 2000 | <br> 2001 | <br> 2002 | <br> 2003 | \n| :--- | :--- | :--- | :--- | :--- | :--- | :--- | :--- | :--- |\n| Ford | 10 | 12 | 14 | 16 | 1 | 1 | 1 | 1 |\n| GM | 11 | 13 | 13 | 15 | 0 | 2 | 0 | 2 |\n\n### Which shape should I use?\n\n**A, nuanced:** It depends on what you're using the data for!\n- I try to make my data \"tidy\" at the start of analysis. Tidy data is quicker to analyze!\n- Seaborn likes plotting long data, while pandas likes plotting wide data\n\n\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown"
]
] |
ecba4b6db75bb70067e72cd73dc4ba7d2a96ded4 | 104,148 | ipynb | Jupyter Notebook | male_2d_model.ipynb | puneat/Audio_Sentiment | 7129ffc7e5b57643c8e8698ca5dc37186fe7956e | [
"MIT"
] | 1 | 2020-07-01T19:24:51.000Z | 2020-07-01T19:24:51.000Z | male_2d_model.ipynb | puneat/Audio_Sentiment | 7129ffc7e5b57643c8e8698ca5dc37186fe7956e | [
"MIT"
] | null | null | null | male_2d_model.ipynb | puneat/Audio_Sentiment | 7129ffc7e5b57643c8e8698ca5dc37186fe7956e | [
"MIT"
] | 1 | 2020-06-20T19:04:50.000Z | 2020-06-20T19:04:50.000Z | 137.398417 | 48,822 | 0.809531 | [
[
[
"<a href=\"https://colab.research.google.com/github/puneat/Audio_Sentiment/blob/puneet/male_2d_model.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/gdrive', force_remount=True)",
"_____no_output_____"
],
[
"# Keras\nimport keras\nfrom keras import regularizers\nfrom keras.preprocessing import sequence\nfrom keras.preprocessing.text import Tokenizer\nfrom keras.preprocessing.sequence import pad_sequences\nfrom keras.models import Sequential, Model, model_from_json\nfrom keras.layers import Dense, Embedding, LSTM\nfrom keras.layers import Input, Flatten, Dropout, Activation, BatchNormalization\nfrom keras.layers import Conv1D, MaxPooling1D, AveragePooling1D\nfrom keras.utils import np_utils, to_categorical\nfrom keras.callbacks import (EarlyStopping, LearningRateScheduler,\n ModelCheckpoint, TensorBoard, ReduceLROnPlateau)\nfrom keras import losses, models, optimizers\nfrom keras.activations import relu, softmax\nfrom keras.layers import (Convolution2D, GlobalAveragePooling2D, BatchNormalization, Flatten, Dropout,\n GlobalMaxPool2D, MaxPool2D, concatenate, Activation, Input, Dense)\n\n# sklearn\nfrom sklearn.metrics import confusion_matrix, accuracy_score\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import LabelEncoder\n\n# Other \nfrom tqdm import tqdm, tqdm_pandas\nimport scipy\nfrom scipy.stats import skew\nimport librosa\nimport librosa.display\nimport json\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport tensorflow as tf\nfrom matplotlib.pyplot import specgram\nimport pandas as pd\nimport seaborn as sns\nimport glob \nimport os\nimport sys\nimport IPython.display as ipd # To play sound in the notebook\nimport warnings\n# ignore warnings \nif not sys.warnoptions:\n warnings.simplefilter(\"ignore\")",
"Using TensorFlow backend.\n/usr/local/lib/python3.6/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
],
[
"ref = pd.read_csv(\"/gdrive/My Drive/Audio_files/Combined_Dataframes/male_df.csv\")\nref.head()",
"_____no_output_____"
],
[
"result=np.load('/gdrive/My Drive/Audio_files/Combined_Dataframes/male_2d_aug.npy')",
"_____no_output_____"
],
[
"result.shape",
"_____no_output_____"
],
[
"frames=[ref,ref,ref,ref,ref,ref];\ndf_label=pd.concat(frames);",
"_____no_output_____"
],
[
"nclass = 7\ninp = Input(shape=(30,216,1)) #2D matrix of 30 MFCC bands by 216 audio length.\nx = Convolution2D(32, (4,10), padding=\"same\")(inp)\nx = BatchNormalization()(x)\nx = Activation(\"relu\")(x)\nx = MaxPool2D()(x)\nx = Dropout(rate=0.2)(x)\n \nx = Convolution2D(32, (4,10), padding=\"same\")(x)\nx = BatchNormalization()(x)\nx = Activation(\"relu\")(x)\nx = MaxPool2D()(x)\nx = Dropout(rate=0.2)(x)\n \nx = Convolution2D(32, (4,10), padding=\"same\")(x)\nx = BatchNormalization()(x)\nx = Activation(\"relu\")(x)\nx = MaxPool2D()(x)\nx = Dropout(rate=0.2)(x)\n \nx = Convolution2D(32, (4,10), padding=\"same\")(x)\nx = BatchNormalization()(x)\nx = Activation(\"relu\")(x)\nx = MaxPool2D()(x)\nx = Dropout(rate=0.2)(x)\n \nx = Flatten()(x)\nx = Dense(64)(x)\nx = Dropout(rate=0.2)(x)\nx = BatchNormalization()(x)\nx = Activation(\"relu\")(x)\nx = Dropout(rate=0.2)(x)\n \nout = Dense(nclass, activation=softmax)(x)\nmodel = models.Model(inputs=inp, outputs=out)\n \nopt = optimizers.Adam(0.001)\nmodel.compile(optimizer=opt, loss=losses.categorical_crossentropy, metrics=['acc'])",
"_____no_output_____"
],
[
"# Split between train and test \nX_train, X_test, y_train, y_test = train_test_split(result\n , df_label.labels\n , test_size=0.20\n , shuffle=True\n , random_state=42\n )",
"_____no_output_____"
],
[
"# one hot encode the target \nlb = LabelEncoder()\ny_train = np_utils.to_categorical(lb.fit_transform(y_train))\ny_test = np_utils.to_categorical(lb.fit_transform(y_test))",
"_____no_output_____"
],
[
"# Normalization as per the standard NN process\nmean = np.mean(X_train, axis=0)\nstd = np.std(X_train, axis=0)\n\nX_train = (X_train - mean)/std\nX_test = (X_test - mean)/std",
"_____no_output_____"
],
[
"model_history = model.fit(X_train, y_train, validation_data=(X_test, y_test), batch_size=100, verbose = 1, epochs=50)",
"Train on 24624 samples, validate on 6156 samples\nEpoch 1/50\n24624/24624 [==============================] - 33s 1ms/step - loss: 1.0014 - accuracy: 0.6285 - val_loss: 0.9475 - val_accuracy: 0.6535\nEpoch 2/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.9637 - accuracy: 0.6372 - val_loss: 1.0404 - val_accuracy: 0.5960\nEpoch 3/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.9227 - accuracy: 0.6559 - val_loss: 1.0225 - val_accuracy: 0.6200\nEpoch 4/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.8839 - accuracy: 0.6702 - val_loss: 0.8617 - val_accuracy: 0.6702\nEpoch 5/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.8659 - accuracy: 0.6793 - val_loss: 0.8534 - val_accuracy: 0.6824\nEpoch 6/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.8389 - accuracy: 0.6907 - val_loss: 0.8799 - val_accuracy: 0.6839\nEpoch 7/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.8170 - accuracy: 0.7007 - val_loss: 0.8234 - val_accuracy: 0.6979\nEpoch 8/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.7900 - accuracy: 0.7094 - val_loss: 0.8419 - val_accuracy: 0.6871\nEpoch 9/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.7726 - accuracy: 0.7162 - val_loss: 0.7533 - val_accuracy: 0.7305\nEpoch 10/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.7541 - accuracy: 0.7243 - val_loss: 0.7909 - val_accuracy: 0.7053\nEpoch 11/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.7302 - accuracy: 0.7310 - val_loss: 0.7900 - val_accuracy: 0.7238\nEpoch 12/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.7153 - accuracy: 0.7416 - val_loss: 0.7315 - val_accuracy: 0.7299\nEpoch 13/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.6944 - accuracy: 0.7420 - val_loss: 0.7320 - val_accuracy: 0.7351\nEpoch 14/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.6822 - accuracy: 0.7506 - val_loss: 0.6577 - val_accuracy: 0.7635\nEpoch 15/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.6689 - accuracy: 0.7547 - val_loss: 0.7001 - val_accuracy: 0.7438\nEpoch 16/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.6527 - accuracy: 0.7615 - val_loss: 0.6874 - val_accuracy: 0.7583\nEpoch 17/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.6458 - accuracy: 0.7636 - val_loss: 0.6456 - val_accuracy: 0.7641\nEpoch 18/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.6296 - accuracy: 0.7726 - val_loss: 0.6640 - val_accuracy: 0.7623\nEpoch 19/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.6328 - accuracy: 0.7715 - val_loss: 0.6338 - val_accuracy: 0.7727\nEpoch 20/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.6100 - accuracy: 0.7776 - val_loss: 0.6684 - val_accuracy: 0.7610\nEpoch 21/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.6064 - accuracy: 0.7801 - val_loss: 0.6382 - val_accuracy: 0.7781\nEpoch 22/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5899 - accuracy: 0.7828 - val_loss: 0.5988 - val_accuracy: 0.7875\nEpoch 23/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5835 - accuracy: 0.7894 - val_loss: 0.6241 - val_accuracy: 0.7755\nEpoch 24/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5838 - accuracy: 0.7860 - val_loss: 0.5576 - val_accuracy: 0.8075\nEpoch 25/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5728 - accuracy: 0.7921 - val_loss: 0.6009 - val_accuracy: 0.7859\nEpoch 26/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5661 - accuracy: 0.7937 - val_loss: 0.6012 - val_accuracy: 0.7927\nEpoch 27/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5576 - accuracy: 0.7971 - val_loss: 0.5815 - val_accuracy: 0.7930\nEpoch 28/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5472 - accuracy: 0.8051 - val_loss: 0.5924 - val_accuracy: 0.7904\nEpoch 29/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5425 - accuracy: 0.8018 - val_loss: 0.5417 - val_accuracy: 0.8155\nEpoch 30/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5396 - accuracy: 0.8059 - val_loss: 0.5846 - val_accuracy: 0.7895\nEpoch 31/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5287 - accuracy: 0.8083 - val_loss: 0.6007 - val_accuracy: 0.7930\nEpoch 32/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5248 - accuracy: 0.8106 - val_loss: 0.6049 - val_accuracy: 0.7939\nEpoch 33/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5097 - accuracy: 0.8169 - val_loss: 0.5318 - val_accuracy: 0.8173\nEpoch 34/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5085 - accuracy: 0.8150 - val_loss: 0.5260 - val_accuracy: 0.8163\nEpoch 35/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5163 - accuracy: 0.8112 - val_loss: 0.5038 - val_accuracy: 0.8259\nEpoch 36/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5057 - accuracy: 0.8189 - val_loss: 0.5340 - val_accuracy: 0.8202\nEpoch 37/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.5023 - accuracy: 0.8168 - val_loss: 0.5216 - val_accuracy: 0.8190\nEpoch 38/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4980 - accuracy: 0.8214 - val_loss: 0.5669 - val_accuracy: 0.8012\nEpoch 39/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4825 - accuracy: 0.8273 - val_loss: 0.5189 - val_accuracy: 0.8234\nEpoch 40/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4827 - accuracy: 0.8248 - val_loss: 0.5409 - val_accuracy: 0.8150\nEpoch 41/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4807 - accuracy: 0.8292 - val_loss: 0.5081 - val_accuracy: 0.8304\nEpoch 42/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4690 - accuracy: 0.8330 - val_loss: 0.5620 - val_accuracy: 0.8036\nEpoch 43/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4723 - accuracy: 0.8285 - val_loss: 0.5308 - val_accuracy: 0.8233\nEpoch 44/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4762 - accuracy: 0.8299 - val_loss: 0.5817 - val_accuracy: 0.8075\nEpoch 45/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4642 - accuracy: 0.8332 - val_loss: 0.5173 - val_accuracy: 0.8228\nEpoch 46/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4725 - accuracy: 0.8293 - val_loss: 0.6019 - val_accuracy: 0.7976\nEpoch 47/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4585 - accuracy: 0.8357 - val_loss: 0.4979 - val_accuracy: 0.8298\nEpoch 48/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4640 - accuracy: 0.8346 - val_loss: 0.5165 - val_accuracy: 0.8259\nEpoch 49/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4404 - accuracy: 0.8430 - val_loss: 0.5051 - val_accuracy: 0.8312\nEpoch 50/50\n24624/24624 [==============================] - 31s 1ms/step - loss: 0.4519 - accuracy: 0.8396 - val_loss: 0.6161 - val_accuracy: 0.7870\n"
],
[
"cd /gdrive/My Drive/Audio_files/",
"/gdrive/My Drive/Audio_files\n"
],
[
"# Save model and weights\nmodel_name = 'cnn2d_male_7870.h5'\nsave_dir = os.path.join(os.getcwd(), 'saved_models')\n\nif not os.path.isdir(save_dir):\n os.makedirs(save_dir)\nmodel_path = os.path.join(save_dir, model_name)\nmodel.save(model_path)\nprint('Save model and weights at %s ' % model_path)\n\n# Save the model to disk\nmodel_json = model.to_json()\nwith open(\"cnn2d_male_7870.json\", \"w\") as json_file:\n json_file.write(model_json)",
"Save model and weights at /gdrive/My Drive/Audio_files/saved_models/cnn2d_male_7870.h5 \n"
],
[
"# loading json and model architecture \njson_file = open('cnn2d_male_7870.json', 'r')\nloaded_model_json = json_file.read()\njson_file.close()\nloaded_model = model_from_json(loaded_model_json)\n\n# load weights into new model\nloaded_model.load_weights(\"saved_models/cnn2d_male_7870.h5\")\nprint(\"Loaded model from disk\")",
"Loaded model from disk\n"
],
[
"class get_results:\n \n def __init__(self, model_history, model ,X_test, y_test, labels):\n self.model_history = model_history\n self.model = model\n self.X_test = X_test\n self.y_test = y_test \n self.labels = labels\n\n def create_plot(self, model_history):\n '''Check the logloss of both train and validation, make sure they are close and have plateau'''\n plt.plot(model_history.history['loss'])\n plt.plot(model_history.history['val_loss'])\n plt.title('model loss')\n plt.ylabel('loss')\n plt.xlabel('epoch')\n plt.legend(['train', 'test'], loc='upper left')\n plt.show()\n\n def create_results(self, model):\n '''predict on test set and get accuracy results'''\n opt = optimizers.Adam(0.001)\n model.compile(loss='categorical_crossentropy', optimizer=opt, metrics=['accuracy'])\n score = model.evaluate(X_test, y_test, verbose=0)\n print(\"%s: %.2f%%\" % (model.metrics_names[1], score[1]*100))\n\n def confusion_results(self, X_test, y_test, labels, model):\n '''plot confusion matrix results'''\n preds = model.predict(X_test, \n batch_size=16, \n verbose=2)\n preds=preds.argmax(axis=1)\n preds = preds.astype(int).flatten()\n preds = (lb.inverse_transform((preds)))\n\n actual = y_test.argmax(axis=1)\n actual = actual.astype(int).flatten()\n actual = (lb.inverse_transform((actual)))\n\n classes = labels\n classes.sort() \n\n c = confusion_matrix(actual, preds)\n print_confusion_matrix(c, class_names = classes)\n \n def accuracy_results_gender(self, X_test, y_test, labels, model):\n '''Print out the accuracy score and confusion matrix heat map of the Gender classification results'''\n \n preds = model.predict(X_test, \n batch_size=16, \n verbose=2)\n preds=preds.argmax(axis=1)\n preds = preds.astype(int).flatten()\n preds = (lb.inverse_transform((preds)))\n\n actual = y_test.argmax(axis=1)\n actual = actual.astype(int).flatten()\n actual = (lb.inverse_transform((actual)))\n \n # print(accuracy_score(actual, preds))\n \n actual = pd.DataFrame(actual).replace({\n , 'male_angry':'male'\n , 'male_fear':'male'\n , 'male_happy':'male'\n , 'male_sad':'male'\n , 'male_surprise':'male'\n , 'male_neutral':'male'\n , 'male_disgust':'male'\n })\n preds = pd.DataFrame(preds).replace({\n , 'male_fear':'male'\n , 'male_happy':'male'\n , 'male_sad':'male'\n , 'male_surprise':'male'\n , 'male_neutral':'male'\n , 'male_disgust':'male'\n })\n\n classes = actual.loc[:,0].unique() \n classes.sort() \n\n c = confusion_matrix(actual, preds)\n print(accuracy_score(actual, preds))\n print_confusion_matrix(c, class_names = classes)\n \ndef print_confusion_matrix(confusion_matrix, class_names, figsize = (10,7), fontsize=14):\n '''Prints a confusion matrix, as returned by sklearn.metrics.confusion_matrix, as a heatmap.'''\n df_cm = pd.DataFrame(\n confusion_matrix, index=class_names, columns=class_names, \n )\n fig = plt.figure(figsize=figsize)\n try:\n heatmap = sns.heatmap(df_cm, annot=True, fmt=\"d\")\n except ValueError:\n raise ValueError(\"Confusion matrix values must be integers.\")\n\n heatmap.yaxis.set_ticklabels(heatmap.yaxis.get_ticklabels(), rotation=0, ha='right', fontsize=fontsize)\n heatmap.xaxis.set_ticklabels(heatmap.xaxis.get_ticklabels(), rotation=45, ha='right', fontsize=fontsize)\n plt.ylabel('True label')\n plt.xlabel('Predicted label')",
"_____no_output_____"
],
[
"results = get_results(model_history,model,X_test,y_test, ref.labels.unique())\nresults.create_plot(model_history)\nresults.create_results(model)\nresults.confusion_results(X_test, y_test, ref.labels.unique(), model)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecba515d3e5f2212c1d13d8286e9e532a38f802f | 1,543 | ipynb | Jupyter Notebook | analysis/variogram/gfed4_variogram.ipynb | akuhnregnier/empirical-fire-modelling | 4187f5bfce0595d98361a9264793c25607043047 | [
"MIT"
] | null | null | null | analysis/variogram/gfed4_variogram.ipynb | akuhnregnier/empirical-fire-modelling | 4187f5bfce0595d98361a9264793c25607043047 | [
"MIT"
] | null | null | null | analysis/variogram/gfed4_variogram.ipynb | akuhnregnier/empirical-fire-modelling | 4187f5bfce0595d98361a9264793c25607043047 | [
"MIT"
] | null | null | null | 19.782051 | 74 | 0.530784 | [
[
[
"import iris\nimport numpy as np\nfrom plot_mean_gfed4_variogram import gfed4_variogram\nfrom wildfires.analysis import cube_plotting\nfrom wildfires.data import GFEDv4\n\nfrom empirical_fire_modelling.utils import tqdm",
"_____no_output_____"
],
[
"cube = GFEDv4().cube[0]\n_ = cube_plotting(\n np.sqrt(iris.analysis.cartography.area_weights(cube)) / 1000,\n colorbar_kwargs={\"label\": \"approx. cell size (km)\"},\n)",
"_____no_output_____"
]
],
[
[
"#### Mean GFED4",
"_____no_output_____"
]
],
[
[
"gfed4_variogram(-1)",
"_____no_output_____"
],
[
"for i in tqdm(range(2)):\n gfed4_variogram(i)",
"_____no_output_____"
],
[
"for i in tqdm(range(-1, 36)):\n gfed4_variogram(i)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecba531e99d444c4d28c9567f12de6abd76948e2 | 92,462 | ipynb | Jupyter Notebook | image-processing-activity/image-as-numbers.ipynb | tengjuilin/textile | a9f15342442f01ba399306cca7d9ac79f28aa3e8 | [
"MIT"
] | null | null | null | image-processing-activity/image-as-numbers.ipynb | tengjuilin/textile | a9f15342442f01ba399306cca7d9ac79f28aa3e8 | [
"MIT"
] | null | null | null | image-processing-activity/image-as-numbers.ipynb | tengjuilin/textile | a9f15342442f01ba399306cca7d9ac79f28aa3e8 | [
"MIT"
] | null | null | null | 460.00995 | 87,692 | 0.947276 | [
[
[
"# Image as Numbers",
"_____no_output_____"
],
[
"### **Purpose**: To view an image from a computers perspective (as an array of numbers)\n\n",
"_____no_output_____"
],
[
"Created by: Hawley Helmbrecht\n\nCreation Date: 07/21/2020\n\nLast Update:",
"_____no_output_____"
],
[
"*Step 1: Necessary Imports*",
"_____no_output_____"
],
[
"In this step, we import all the packages that we will need later on in the document. \n\nWe decide to do this all at once rather than throughout because it is standard coding practice that helps create clean code.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom skimage import io #importing a specific module from scikit-imgae\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"*Step 2: User Inputs*",
"_____no_output_____"
],
[
"When creating Jupyter Notebooks for use by collaborators and experamentalists, I find it best practice to have a step near the top specifically for the imuts they need for the notebook.\n\nIn this case, the only input needed is the location of location of the cell.tif file included in this repository.",
"_____no_output_____"
]
],
[
[
"#replace the example path from my computer with the path to the image on your computer\n\ncell_im_location = '/Users/hhelmbre/Desktop/packages/textile/example_data/cell.tif'",
"_____no_output_____"
]
],
[
[
"Note: This example image is from Nance Lab's Andrea Joseph and is a NueN stain in the contralateral hemisphere of a p10 rat at 240x zoom",
"_____no_output_____"
],
[
"*Step 3: Reading in the image*",
"_____no_output_____"
],
[
"The image doesn't exist within the Jupyter Notebook until you actually \"read\" it into the notebooks data storage. To do that we use the scikit-image command io.imread\n\nGo ahead and check the imports to see that from skimage (scikit-image) we imported the io module",
"_____no_output_____"
]
],
[
[
"cell_im = io.imread(cell_im_location)",
"_____no_output_____"
]
],
[
[
"*Step 4: Viewing the Image*",
"_____no_output_____"
],
[
"For now don't worry about the [0,:,:] next to the image. But focus on the fact that matplotlib is able to show the image using the function 'imshow'. Where cmap is the color map we choose to apply and it has been set to gray.\n\nMatplotlib automatically includes x and y axis which designate in this case a numerical location for each pixel (0,0 in the top left and 500,500 in the bottom right). ",
"_____no_output_____"
]
],
[
[
"plt.imshow(cell_im[0,:,:], cmap='gray')",
"_____no_output_____"
]
],
[
[
"That's the end of this part of the activity! I hope you all are looking forward to the complete module!! :) ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecba544864e61fd4f9584ca22d802c8daae53ff2 | 98,408 | ipynb | Jupyter Notebook | 2_Training.ipynb | sdonatti/nd891-project-image-captioning | 80d2e56f31441b6c4a16a8866efdfebf5e90ee10 | [
"MIT"
] | null | null | null | 2_Training.ipynb | sdonatti/nd891-project-image-captioning | 80d2e56f31441b6c4a16a8866efdfebf5e90ee10 | [
"MIT"
] | null | null | null | 2_Training.ipynb | sdonatti/nd891-project-image-captioning | 80d2e56f31441b6c4a16a8866efdfebf5e90ee10 | [
"MIT"
] | null | null | null | 129.825858 | 53,140 | 0.75881 | [
[
[
"# Computer Vision Nanodegree\n\n## Project: Image Captioning\n\n---\n\nIn this notebook, you will train your CNN-RNN model.\n\nYou are welcome and encouraged to try out many different architectures and hyperparameters when searching for a good model.\n\nThis does have the potential to make the project quite messy! Before submitting your project, make sure that you clean up:\n- the code you write in this notebook. The notebook should describe how to train a single CNN-RNN architecture, corresponding to your final choice of hyperparameters. You should structure the notebook so that the reviewer can replicate your results by running the code in this notebook.\n- the output of the code cell in **Step 2**. The output should show the output obtained when training the model from scratch.\n\nThis notebook **will be graded**.\n\nFeel free to use the links below to navigate the notebook:\n- [Step 1](#step1): Training Setup\n- [Step 2](#step2): Train your Model\n- [Step 3](#step3): (Optional) Validate your Model",
"_____no_output_____"
],
[
"<a id='step1'></a>\n## Step 1: Training Setup\n\nIn this step of the notebook, you will customize the training of your CNN-RNN model by specifying hyperparameters and setting other options that are important to the training procedure. The values you set now will be used when training your model in **Step 2** below.\n\nYou should only amend blocks of code that are preceded by a `TODO` statement. **Any code blocks that are not preceded by a `TODO` statement should not be modified**.\n\n### Task #1\n\nBegin by setting the following variables:\n- `batch_size` - the batch size of each training batch. It is the number of image-caption pairs used to amend the model weights in each training step.\n- `vocab_threshold` - the minimum word count threshold. Note that a larger threshold will result in a smaller vocabulary, whereas a smaller threshold will include rarer words and result in a larger vocabulary.\n- `vocab_from_file` - a Boolean that decides whether to load the vocabulary from file.\n- `embed_size` - the dimensionality of the image and word embeddings.\n- `hidden_size` - the number of features in the hidden state of the RNN decoder.\n- `num_epochs` - the number of epochs to train the model. We recommend that you set `num_epochs=3`, but feel free to increase or decrease this number as you wish. [This paper](https://arxiv.org/pdf/1502.03044.pdf) trained a captioning model on a single state-of-the-art GPU for 3 days, but you'll soon see that you can get reasonable results in a matter of a few hours! (_But of course, if you want your model to compete with current research, you will have to train for much longer._)\n- `save_every` - determines how often to save the model weights. We recommend that you set `save_every=1`, to save the model weights after each epoch. This way, after the `i`th epoch, the encoder and decoder weights will be saved in the `models/` folder as `encoder-i.pkl` and `decoder-i.pkl`, respectively.\n- `print_every` - determines how often to print the batch loss to the Jupyter notebook while training. Note that you **will not** observe a monotonic decrease in the loss function while training - this is perfectly fine and completely expected! You are encouraged to keep this at its default value of `100` to avoid clogging the notebook, but feel free to change it.\n- `log_file` - the name of the text file containing - for every step - how the loss and perplexity evolved during training.\n\nIf you're not sure where to begin to set some of the values above, you can peruse [this paper](https://arxiv.org/pdf/1502.03044.pdf) and [this paper](https://arxiv.org/pdf/1411.4555.pdf) for useful guidance! **To avoid spending too long on this notebook**, you are encouraged to consult these suggested research papers to obtain a strong initial guess for which hyperparameters are likely to work best. Then, train a single model, and proceed to the next notebook (**3_Inference.ipynb**). If you are unhappy with your performance, you can return to this notebook to tweak the hyperparameters (and/or the architecture in **model.py**) and re-train your model.\n\n### Question 1\n\n**Question:** Describe your CNN-RNN architecture in detail. With this architecture in mind, how did you select the values of the variables in Task 1? If you consulted a research paper detailing a successful implementation of an image captioning model, please provide the reference.\n\n**Answer:**\n\n```\n----------------------------------------------------------------\n Encoder\n----------------------------------------------------------------\n Layer (type) Output Shape Param #\n================================================================\n Conv2d-1 [-1, 64, 112, 112] 9,408\n BatchNorm2d-2 [-1, 64, 112, 112] 128\n ReLU-3 [-1, 64, 112, 112] 0\n MaxPool2d-4 [-1, 64, 56, 56] 0\n Conv2d-5 [-1, 128, 56, 56] 8,192\n BatchNorm2d-6 [-1, 128, 56, 56] 256\n ReLU-7 [-1, 128, 56, 56] 0\n Conv2d-8 [-1, 128, 56, 56] 4,608\n BatchNorm2d-9 [-1, 128, 56, 56] 256\n ReLU-10 [-1, 128, 56, 56] 0\n Conv2d-11 [-1, 256, 56, 56] 32,768\n BatchNorm2d-12 [-1, 256, 56, 56] 512\n Conv2d-13 [-1, 256, 56, 56] 16,384\n BatchNorm2d-14 [-1, 256, 56, 56] 512\n ReLU-15 [-1, 256, 56, 56] 0\n Bottleneck-16 [-1, 256, 56, 56] 0\n Conv2d-17 [-1, 128, 56, 56] 32,768\n BatchNorm2d-18 [-1, 128, 56, 56] 256\n ReLU-19 [-1, 128, 56, 56] 0\n Conv2d-20 [-1, 128, 56, 56] 4,608\n BatchNorm2d-21 [-1, 128, 56, 56] 256\n ReLU-22 [-1, 128, 56, 56] 0\n Conv2d-23 [-1, 256, 56, 56] 32,768\n BatchNorm2d-24 [-1, 256, 56, 56] 512\n ReLU-25 [-1, 256, 56, 56] 0\n Bottleneck-26 [-1, 256, 56, 56] 0\n Conv2d-27 [-1, 128, 56, 56] 32,768\n BatchNorm2d-28 [-1, 128, 56, 56] 256\n ReLU-29 [-1, 128, 56, 56] 0\n Conv2d-30 [-1, 128, 56, 56] 4,608\n BatchNorm2d-31 [-1, 128, 56, 56] 256\n ReLU-32 [-1, 128, 56, 56] 0\n Conv2d-33 [-1, 256, 56, 56] 32,768\n BatchNorm2d-34 [-1, 256, 56, 56] 512\n ReLU-35 [-1, 256, 56, 56] 0\n Bottleneck-36 [-1, 256, 56, 56] 0\n Conv2d-37 [-1, 256, 56, 56] 65,536\n BatchNorm2d-38 [-1, 256, 56, 56] 512\n ReLU-39 [-1, 256, 56, 56] 0\n Conv2d-40 [-1, 256, 28, 28] 18,432\n BatchNorm2d-41 [-1, 256, 28, 28] 512\n ReLU-42 [-1, 256, 28, 28] 0\n Conv2d-43 [-1, 512, 28, 28] 131,072\n BatchNorm2d-44 [-1, 512, 28, 28] 1,024\n Conv2d-45 [-1, 512, 28, 28] 131,072\n BatchNorm2d-46 [-1, 512, 28, 28] 1,024\n ReLU-47 [-1, 512, 28, 28] 0\n Bottleneck-48 [-1, 512, 28, 28] 0\n Conv2d-49 [-1, 256, 28, 28] 131,072\n BatchNorm2d-50 [-1, 256, 28, 28] 512\n ReLU-51 [-1, 256, 28, 28] 0\n Conv2d-52 [-1, 256, 28, 28] 18,432\n BatchNorm2d-53 [-1, 256, 28, 28] 512\n ReLU-54 [-1, 256, 28, 28] 0\n Conv2d-55 [-1, 512, 28, 28] 131,072\n BatchNorm2d-56 [-1, 512, 28, 28] 1,024\n ReLU-57 [-1, 512, 28, 28] 0\n Bottleneck-58 [-1, 512, 28, 28] 0\n Conv2d-59 [-1, 256, 28, 28] 131,072\n BatchNorm2d-60 [-1, 256, 28, 28] 512\n ReLU-61 [-1, 256, 28, 28] 0\n Conv2d-62 [-1, 256, 28, 28] 18,432\n BatchNorm2d-63 [-1, 256, 28, 28] 512\n ReLU-64 [-1, 256, 28, 28] 0\n Conv2d-65 [-1, 512, 28, 28] 131,072\n BatchNorm2d-66 [-1, 512, 28, 28] 1,024\n ReLU-67 [-1, 512, 28, 28] 0\n Bottleneck-68 [-1, 512, 28, 28] 0\n Conv2d-69 [-1, 256, 28, 28] 131,072\n BatchNorm2d-70 [-1, 256, 28, 28] 512\n ReLU-71 [-1, 256, 28, 28] 0\n Conv2d-72 [-1, 256, 28, 28] 18,432\n BatchNorm2d-73 [-1, 256, 28, 28] 512\n ReLU-74 [-1, 256, 28, 28] 0\n Conv2d-75 [-1, 512, 28, 28] 131,072\n BatchNorm2d-76 [-1, 512, 28, 28] 1,024\n ReLU-77 [-1, 512, 28, 28] 0\n Bottleneck-78 [-1, 512, 28, 28] 0\n Conv2d-79 [-1, 512, 28, 28] 262,144\n BatchNorm2d-80 [-1, 512, 28, 28] 1,024\n ReLU-81 [-1, 512, 28, 28] 0\n Conv2d-82 [-1, 512, 14, 14] 73,728\n BatchNorm2d-83 [-1, 512, 14, 14] 1,024\n ReLU-84 [-1, 512, 14, 14] 0\n Conv2d-85 [-1, 1024, 14, 14] 524,288\n BatchNorm2d-86 [-1, 1024, 14, 14] 2,048\n Conv2d-87 [-1, 1024, 14, 14] 524,288\n BatchNorm2d-88 [-1, 1024, 14, 14] 2,048\n ReLU-89 [-1, 1024, 14, 14] 0\n Bottleneck-90 [-1, 1024, 14, 14] 0\n Conv2d-91 [-1, 512, 14, 14] 524,288\n BatchNorm2d-92 [-1, 512, 14, 14] 1,024\n ReLU-93 [-1, 512, 14, 14] 0\n Conv2d-94 [-1, 512, 14, 14] 73,728\n BatchNorm2d-95 [-1, 512, 14, 14] 1,024\n ReLU-96 [-1, 512, 14, 14] 0\n Conv2d-97 [-1, 1024, 14, 14] 524,288\n BatchNorm2d-98 [-1, 1024, 14, 14] 2,048\n ReLU-99 [-1, 1024, 14, 14] 0\n Bottleneck-100 [-1, 1024, 14, 14] 0\n Conv2d-101 [-1, 512, 14, 14] 524,288\n BatchNorm2d-102 [-1, 512, 14, 14] 1,024\n ReLU-103 [-1, 512, 14, 14] 0\n Conv2d-104 [-1, 512, 14, 14] 73,728\n BatchNorm2d-105 [-1, 512, 14, 14] 1,024\n ReLU-106 [-1, 512, 14, 14] 0\n Conv2d-107 [-1, 1024, 14, 14] 524,288\n BatchNorm2d-108 [-1, 1024, 14, 14] 2,048\n ReLU-109 [-1, 1024, 14, 14] 0\n Bottleneck-110 [-1, 1024, 14, 14] 0\n Conv2d-111 [-1, 512, 14, 14] 524,288\n BatchNorm2d-112 [-1, 512, 14, 14] 1,024\n ReLU-113 [-1, 512, 14, 14] 0\n Conv2d-114 [-1, 512, 14, 14] 73,728\n BatchNorm2d-115 [-1, 512, 14, 14] 1,024\n ReLU-116 [-1, 512, 14, 14] 0\n Conv2d-117 [-1, 1024, 14, 14] 524,288\n BatchNorm2d-118 [-1, 1024, 14, 14] 2,048\n ReLU-119 [-1, 1024, 14, 14] 0\n Bottleneck-120 [-1, 1024, 14, 14] 0\n Conv2d-121 [-1, 512, 14, 14] 524,288\n BatchNorm2d-122 [-1, 512, 14, 14] 1,024\n ReLU-123 [-1, 512, 14, 14] 0\n Conv2d-124 [-1, 512, 14, 14] 73,728\n BatchNorm2d-125 [-1, 512, 14, 14] 1,024\n ReLU-126 [-1, 512, 14, 14] 0\n Conv2d-127 [-1, 1024, 14, 14] 524,288\n BatchNorm2d-128 [-1, 1024, 14, 14] 2,048\n ReLU-129 [-1, 1024, 14, 14] 0\n Bottleneck-130 [-1, 1024, 14, 14] 0\n Conv2d-131 [-1, 512, 14, 14] 524,288\n BatchNorm2d-132 [-1, 512, 14, 14] 1,024\n ReLU-133 [-1, 512, 14, 14] 0\n Conv2d-134 [-1, 512, 14, 14] 73,728\n BatchNorm2d-135 [-1, 512, 14, 14] 1,024\n ReLU-136 [-1, 512, 14, 14] 0\n Conv2d-137 [-1, 1024, 14, 14] 524,288\n BatchNorm2d-138 [-1, 1024, 14, 14] 2,048\n ReLU-139 [-1, 1024, 14, 14] 0\n Bottleneck-140 [-1, 1024, 14, 14] 0\n Conv2d-141 [-1, 1024, 14, 14] 1,048,576\n BatchNorm2d-142 [-1, 1024, 14, 14] 2,048\n ReLU-143 [-1, 1024, 14, 14] 0\n Conv2d-144 [-1, 1024, 7, 7] 294,912\n BatchNorm2d-145 [-1, 1024, 7, 7] 2,048\n ReLU-146 [-1, 1024, 7, 7] 0\n Conv2d-147 [-1, 2048, 7, 7] 2,097,152\n BatchNorm2d-148 [-1, 2048, 7, 7] 4,096\n Conv2d-149 [-1, 2048, 7, 7] 2,097,152\n BatchNorm2d-150 [-1, 2048, 7, 7] 4,096\n ReLU-151 [-1, 2048, 7, 7] 0\n Bottleneck-152 [-1, 2048, 7, 7] 0\n Conv2d-153 [-1, 1024, 7, 7] 2,097,152\n BatchNorm2d-154 [-1, 1024, 7, 7] 2,048\n ReLU-155 [-1, 1024, 7, 7] 0\n Conv2d-156 [-1, 1024, 7, 7] 294,912\n BatchNorm2d-157 [-1, 1024, 7, 7] 2,048\n ReLU-158 [-1, 1024, 7, 7] 0\n Conv2d-159 [-1, 2048, 7, 7] 2,097,152\n BatchNorm2d-160 [-1, 2048, 7, 7] 4,096\n ReLU-161 [-1, 2048, 7, 7] 0\n Bottleneck-162 [-1, 2048, 7, 7] 0\n Conv2d-163 [-1, 1024, 7, 7] 2,097,152\n BatchNorm2d-164 [-1, 1024, 7, 7] 2,048\n ReLU-165 [-1, 1024, 7, 7] 0\n Conv2d-166 [-1, 1024, 7, 7] 294,912\n BatchNorm2d-167 [-1, 1024, 7, 7] 2,048\n ReLU-168 [-1, 1024, 7, 7] 0\n Conv2d-169 [-1, 2048, 7, 7] 2,097,152\n BatchNorm2d-170 [-1, 2048, 7, 7] 4,096\n ReLU-171 [-1, 2048, 7, 7] 0\n Bottleneck-172 [-1, 2048, 7, 7] 0\nAdaptiveAvgPool2d-173 [-1, 2048, 1, 1] 0\n Dropout-174 [-1, 2048] 0\n Linear-175 [-1, 512] 1,048,576\n BatchNorm1d-176 [-1, 512] 1,024\n================================================================\nTotal params: 24,029,504\nTrainable params: 1,049,600\nNon-trainable params: 22,979,904\n----------------------------------------------------------------\nInput size (MB): 0.57\nForward/backward pass size (MB): 361.80\nParams size (MB): 91.67\nEstimated Total Size (MB): 454.04\n----------------------------------------------------------------\n```\nThe encoder is based on the [ResNeXt-50 32x4d](http://pytorch.org/docs/stable/torchvision/models.html) model introduced by [Xie et al. 2017](http://arxiv.org/abs/1611.05431). The choice of this architecture is motivated by the results of [Bianco et al. 2018](http://arxiv.org/abs/1810.00736) and [Reddi et al. 2020](http://arxiv.org/abs/1911.02549) that indicate it has a better overall performance in comparison with [ResNet50](http://pytorch.org/docs/stable/torchvision/models.html) introduced by [He et al. 2015](https://arxiv.org/abs/1512.03385).\n\n```\n----------------------------------------------------------------\n Decoder\n----------------------------------------------------------------\n Layer (type) Output Shape Param #\n================================================================\n Embedding-1 [-1, 10, 512] 4,534,272\n GRU-2 [[-1, 11, 512], [-1, 1, 512]] 0\n Dropout-3 [-1, 11, 512] 0\n Linear-4 [-1, 11, 8856] 4,543,128\n================================================================\nTotal params: 9,077,400\nTrainable params: 9,077,400\nNon-trainable params: 0\n----------------------------------------------------------------\nInput size (MB): 0.04\nForward/backward pass size (MB): 21.17\nParams size (MB): 34.63\nEstimated Total Size (MB): 55.84\n----------------------------------------------------------------\n```\nThe decoder uses a single recurrent layer inspired by the architecture from [Vinyals et al. 2015](http://arxiv.org/abs/1411.4555). However, it replaces the LSTM with a [GRU](http://arxiv.org/abs/1406.1078) based on the image captioning results published by [Gu et al. 2017](http://arxiv.org/abs/1612.07086) and includes a Dropout layer motivated by the observations of [Eckhardt 2018](http://towardsdatascience.com/choosing-the-right-hyperparameters-for-a-simple-lstm-using-keras-f8e9ed76f046).\n\nThe values of `vocab_threshold`, `embed_size`, and `hidden_size` hyperparameters are set according to [Vinyals et al. 2015](http://arxiv.org/abs/1411.4555), while `batch_size` uses the same as [Xu et al. 2015](http://arxiv.org/abs/1502.03044). The `num_epochs` is chosen arbitrarily.\n\n### (Optional) Task #2\n\nNote that we have provided a recommended image transform `transform_train` for pre-processing the training images, but you are welcome (and encouraged!) to modify it as you wish. When modifying this transform, keep in mind that:\n- the images in the dataset have varying heights and widths, and\n- if using a pre-trained model, you must perform the corresponding appropriate normalization.\n\n### Question 2\n\n**Question:** How did you select the transform in `transform_train`? If you left the transform at its provided value, why do you think that it is a good choice for your CNN architecture?\n\n**Answer:**\n\nThe provided `transform_train` pipeline is already a good choice. It implements augmentation through random cropping, which is paired with resizing to provide better results, and horizontal flipping of the original images, which tends to maintain the object context unaltered (e.g., land and water are below the sky). It also normalizes the image channels with the mean and variance values from PyTorch model zoo, which are the same ones used to pre-train the feature extraction layers of the encoder. In comparison, `transform_val` and `transform_test` follow the same pipeline without augmentation.\n\n### Task #3\n\nNext, you will specify a Python list containing the learnable parameters of the model. For instance, if you decide to make all weights in the decoder trainable, but only want to train the weights in the embedding layer of the encoder, then you should set `params` to something like:\n```\nparams = list(decoder.parameters()) + list(encoder.embed.parameters())\n```\n\n### Question 3\n\n**Question:** How did you select the trainable parameters of your architecture? Why do you think this is a good choice?\n\n**Answer:**\n\nThe parameters of layers 1-173 of the encoder are pre-trained on [ImageNet](http://arxiv.org/abs/1409.0575) and layers 174-176 are re-trained on [COCO](http://arxiv.org/abs/1405.0312) to generate embedded image features using [fixed transfer-learning](http://cs231n.github.io/transfer-learning). The layers 1-4 of the decoder are re-trained on [COCO](http://arxiv.org/abs/1405.0312). This is a good idea because both datasets comprise a large collection of RGB images of diverse objects in multiple contexts and cycle/effort time is very limited for this project.\n\n### Task #4\n\nFinally, you will select an [optimizer](http://pytorch.org/docs/master/optim.html#torch.optim.Optimizer).\n\n### Question 4\n\n**Question:** How did you select the optimizer used to train your model?\n\n**Answer:**\n\nThe trainable parameters are optimized using [Adam](http://arxiv.org/abs/1412.6980) with an adaptive learning rate of $0.001$ following [Xu et al. 2014](http://arxiv.org/abs/1502.03044) design choice for [COCO](http://arxiv.org/abs/1405.0312).",
"_____no_output_____"
]
],
[
[
"import math\nimport multiprocessing\nimport os\n\nimport torch\nimport torch.nn as nn\nfrom torchvision import transforms\n\nfrom data_loader import get_loader\nfrom model import EncoderCNN, DecoderRNN\n\n# TODO #1: Select appropriate values for the Python variables below.\nbatch_size = 64 # batch size\nvocab_threshold = 5 # minimum word count threshold\nvocab_from_file = False # if True, load existing vocab file\nembed_size = 512 # dimensionality of image and word embeddings\nhidden_size = 512 # number of features in hidden state of the RNN decoder\nnum_epochs = 5 # number of training epochs\nsave_every = 1 # determines frequency of saving model weights\nprint_every = 1000 # determines window for printing average loss\nlog_file = 'training_log.txt' # name of file with saved training loss and perplexity\nmodel_dir = os.path.join('.', 'models') # path to save the trained CNN encoder and RNN decoder\n\n# (Optional) TODO #2: Amend the image transform below.\ntransform_train = transforms.Compose([transforms.Resize(256),\n transforms.RandomCrop(224),\n transforms.RandomHorizontalFlip(),\n transforms.ToTensor(),\n transforms.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225))])\n\n# Build data loader.\ndata_loader = get_loader(transform=transform_train,\n mode='train',\n batch_size=batch_size,\n vocab_threshold=vocab_threshold,\n vocab_from_file=vocab_from_file,\n num_workers=multiprocessing.cpu_count(),\n cocoapi_loc=os.path.join('..', '..', 'Data'))\n\n# The size of the vocabulary.\nvocab_size = len(data_loader.dataset.vocab)\n\n# Initialize the encoder and decoder.\nencoder = EncoderCNN(embed_size)\ndecoder = DecoderRNN(embed_size, hidden_size, vocab_size)\n\n# Move models to GPU if CUDA is available.\ndevice = torch.device('cuda' if torch.cuda.is_available() else 'cpu')\nencoder, decoder = encoder.to(device), decoder.to(device)\n\n# Define the loss function.\ncriterion = nn.CrossEntropyLoss().cuda() if torch.cuda.is_available() else nn.CrossEntropyLoss()\n\n# TODO #3: Specify the learnable parameters of the model.\nparams = list(decoder.parameters()) + list(encoder.embedding.parameters())\n\n# TODO #4: Define the optimizer.\noptimizer = torch.optim.Adam(params, lr=0.001)\n\n# Set the total number of training steps per epoch.\ntotal_step = math.ceil(len(data_loader.dataset.caption_lengths) / data_loader.batch_sampler.batch_size)\n\n# Get device name with gpu option.\nif device.type == 'cuda':\n print('Using: {}'.format(torch.cuda.get_device_name(0)))\n\n# Get device name with cpu option.\nelse:\n print('Using: {} with {} threads'.format(device, torch.get_num_threads()))",
"loading annotations into memory...\nDone (t=1.05s)\ncreating index...\nindex created!\n[0/414113] Tokenizing captions...\n[100000/414113] Tokenizing captions...\n[200000/414113] Tokenizing captions...\n[300000/414113] Tokenizing captions...\n[400000/414113] Tokenizing captions...\nloading annotations into memory...\nDone (t=1.06s)\ncreating index...\nindex created!\nObtaining caption lengths...\n"
]
],
[
[
"<a id='step2'></a>\n## Step 2: Train your Model\n\nOnce you have executed the code cell in **Step 1**, the training procedure below should run without issue.\n\nIt is completely fine to leave the code cell below as-is without modifications to train your model. However, if you would like to modify the code used to train the model below, you must ensure that your changes are easily parsed by your reviewer. In other words, make sure to provide appropriate comments to describe how your code works!\n\nYou may find it useful to load saved weights to resume training. In that case, note the names of the files containing the encoder and decoder weights that you'd like to load (`encoder_file` and `decoder_file`). Then you can load the weights by using the lines below:\n\n```python\n# Load pre-trained weights before resuming training.\nencoder.load_state_dict(torch.load(os.path.join('./models', encoder_file)))\ndecoder.load_state_dict(torch.load(os.path.join('./models', decoder_file)))\n```\n\nWhile trying out parameters, make sure to take extensive notes and record the settings that you used in your various training runs. In particular, you don't want to encounter a situation where you've trained a model for several hours but can't remember what settings you used :).\n\n### A Note on Tuning Hyperparameters\n\nTo figure out how well your model is doing, you can look at how the training loss and perplexity evolve during training - and for the purposes of this project, you are encouraged to amend the hyperparameters based on this information.\n\nHowever, this will not tell you if your model is overfitting to the training data, and, unfortunately, overfitting is a problem that is commonly encountered when training image captioning models.\n\nFor this project, you need not worry about overfitting. **This project does not have strict requirements regarding the performance of your model**, and you just need to demonstrate that your model has learned **_something_** when you generate captions on the test data. For now, we strongly encourage you to train your model for the suggested 3 epochs without worrying about performance; then, you should immediately transition to the next notebook in the sequence (**3_Inference.ipynb**) to see how your model performs on the test data. If your model needs to be changed, you can come back to this notebook, amend hyperparameters (if necessary), and re-train the model.\n\nThat said, if you would like to go above and beyond in this project, you can read about some approaches to minimizing overfitting in section 4.3.1 of [this paper](http://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=7505636). In the next (optional) step of this notebook, we provide some guidance for assessing the performance on the validation dataset.",
"_____no_output_____"
]
],
[
[
"import sys\n\nimport numpy as np\nimport torch.utils.data as data\n\n# Open the training log file.\nf = open(log_file, 'w')\n\n# Iterate over the training dataset.\nfor epoch in range(1, num_epochs+1):\n\n for i_step in range(1, total_step+1):\n\n # Randomly sample a caption length, and sample indices with that length.\n indices = data_loader.dataset.get_train_indices()\n # Create and assign a batch sampler to retrieve a batch with the sampled indices.\n new_sampler = data.sampler.SubsetRandomSampler(indices=indices)\n data_loader.batch_sampler.sampler = new_sampler\n\n # Obtain the batch.\n images, captions = next(iter(data_loader))\n\n # Move batch of images and captions to GPU if CUDA is available.\n images = images.to(device)\n captions = captions.to(device)\n\n # Zero the gradients.\n decoder.zero_grad()\n encoder.zero_grad()\n\n # Pass the inputs through the CNN-RNN model.\n features = encoder(images)\n outputs = decoder(features, captions)\n\n # Calculate the batch loss.\n loss = criterion(outputs.view(-1, vocab_size), captions.view(-1))\n\n # Backward pass.\n loss.backward()\n\n # Update the parameters in the optimizer.\n optimizer.step()\n\n # Get training statistics.\n message = 'Epoch [%d/%d], Step [%d/%d], Loss: %.4f, Perplexity: %5.4f'\n stats = message % (epoch, num_epochs, i_step, total_step, loss.item(), np.exp(loss.item()))\n\n # Print training statistics (on same line).\n print('\\r'+stats, end=\"\")\n sys.stdout.flush()\n\n # Print training statistics to file.\n f.write(stats + '\\n')\n f.flush()\n\n # Print training statistics (on different line).\n if i_step % print_every == 0:\n print('\\r'+stats)\n\n # Save the weights.\n if epoch % save_every == 0:\n torch.save(decoder.state_dict(), os.path.join(model_dir, 'decoder-{}.pkl'.format(epoch)))\n torch.save(encoder.state_dict(), os.path.join(model_dir, 'encoder-{}.pkl'.format(epoch)))\n\n# Close the training log file.\nf.close()",
"Epoch [1/5], Step [1000/6471], Loss: 2.7756, Perplexity: 16.0484\nEpoch [1/5], Step [2000/6471], Loss: 2.3028, Perplexity: 10.00190\nEpoch [1/5], Step [3000/6471], Loss: 2.4603, Perplexity: 11.7084\nEpoch [1/5], Step [4000/6471], Loss: 3.1333, Perplexity: 22.94858\nEpoch [1/5], Step [5000/6471], Loss: 2.3320, Perplexity: 10.2986\nEpoch [1/5], Step [6000/6471], Loss: 2.4834, Perplexity: 11.9823\nEpoch [2/5], Step [1000/6471], Loss: 2.1592, Perplexity: 8.66421\nEpoch [2/5], Step [2000/6471], Loss: 2.2683, Perplexity: 9.66316\nEpoch [2/5], Step [3000/6471], Loss: 1.9266, Perplexity: 6.86614\nEpoch [2/5], Step [4000/6471], Loss: 2.1426, Perplexity: 8.521547\nEpoch [2/5], Step [5000/6471], Loss: 2.0458, Perplexity: 7.735020\nEpoch [2/5], Step [6000/6471], Loss: 2.0632, Perplexity: 7.87098\nEpoch [3/5], Step [1000/6471], Loss: 2.0422, Perplexity: 7.70749\nEpoch [3/5], Step [2000/6471], Loss: 2.0533, Perplexity: 7.79340\nEpoch [3/5], Step [3000/6471], Loss: 1.9583, Perplexity: 7.08762\nEpoch [3/5], Step [4000/6471], Loss: 2.0349, Perplexity: 7.65176\nEpoch [3/5], Step [5000/6471], Loss: 2.3705, Perplexity: 10.7030\nEpoch [3/5], Step [6000/6471], Loss: 2.3840, Perplexity: 10.84794\nEpoch [4/5], Step [1000/6471], Loss: 2.2403, Perplexity: 9.39607\nEpoch [4/5], Step [2000/6471], Loss: 2.0256, Perplexity: 7.580578\nEpoch [4/5], Step [3000/6471], Loss: 2.0029, Perplexity: 7.41093\nEpoch [4/5], Step [4000/6471], Loss: 1.9883, Perplexity: 7.30297\nEpoch [4/5], Step [5000/6471], Loss: 2.0020, Perplexity: 7.40368\nEpoch [4/5], Step [6000/6471], Loss: 2.1606, Perplexity: 8.67600\nEpoch [5/5], Step [1000/6471], Loss: 1.9458, Perplexity: 6.99927\nEpoch [5/5], Step [2000/6471], Loss: 2.0815, Perplexity: 8.01646\nEpoch [5/5], Step [3000/6471], Loss: 1.8928, Perplexity: 6.63775\nEpoch [5/5], Step [4000/6471], Loss: 2.0464, Perplexity: 7.74031\nEpoch [5/5], Step [5000/6471], Loss: 2.0407, Perplexity: 7.69637\nEpoch [5/5], Step [6000/6471], Loss: 1.9787, Perplexity: 7.23308\nEpoch [5/5], Step [6471/6471], Loss: 1.7609, Perplexity: 5.81753"
]
],
[
[
"<a id='step3'></a>\n## Step 3: (Optional) Validate your Model\n\nTo assess potential overfitting, one approach is to assess performance on a validation set. If you decide to do this **optional** task, you are required to first complete all of the steps in the next notebook in the sequence (**3_Inference.ipynb**); as part of that notebook, you will write and test code (specifically, the `sample` method in the `DecoderRNN` class) that uses your RNN decoder to generate captions. That code will prove incredibly useful here.\n\nIf you decide to validate your model, please do not edit the data loader in **data_loader.py**. Instead, create a new file named **data_loader_val.py** containing the code for obtaining the data loader for the validation data. You can access:\n- the validation images at filepath `'/opt/cocoapi/images/train2014/'`, and\n- the validation image caption annotation file at filepath `'/opt/cocoapi/annotations/captions_val2014.json'`.\n\nThe suggested approach to validating your model involves creating a json file such as [this one](https://github.com/cocodataset/cocoapi/blob/master/results/captions_val2014_fakecap_results.json) containing your model's predicted captions for the validation images. Then, you can write your own script or use one that you [find online](https://github.com/tylin/coco-caption) to calculate the BLEU score of your model. You can read more about the BLEU score, along with other evaluation metrics (such as TEOR and Cider) in section 4.1 of [this paper](https://arxiv.org/pdf/1411.4555.pdf). For more information about how to use the annotation file, check out the [website](http://cocodataset.org/#download) for the COCO dataset.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nimport nltk\nfrom nltk.translate.meteor_score import meteor_score\n\n# Download the body of knowledge to use.\nnltk.download('wordnet')\n\n# Seed random generators.\ntorch.manual_seed(42)\nnp.random.seed(42)\n\n# Define a transform to pre-process the validation images.\ntransform_val = transforms.Compose([transforms.Resize(256),\n transforms.CenterCrop(224),\n transforms.ToTensor(),\n transforms.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225))])\n\n# Create the data loader.\ndata_loader = get_loader(transform=transform_val,\n mode='val',\n batch_size=1,\n num_workers=multiprocessing.cpu_count(),\n cocoapi_loc=os.path.join('..', '..', 'Data'))\n\n# Specify the saved image captioning models to load.\nencoder_files = [f for f in os.listdir(model_dir) if f.startswith('encoder') and f.endswith('.pkl')]\ndecoder_files = [f for f in os.listdir(model_dir) if f.startswith('decoder') and f.endswith('.pkl')]\nassert len(encoder_files) == len(decoder_files)\n\n# Validate the image captioning models.\nwith torch.no_grad():\n max_score = 0.0\n scores = torch.zeros(len(encoder_files))\n for idx in torch.arange(len(encoder_files)):\n\n # Load the trained weights.\n encoder.load_state_dict(torch.load(os.path.join(model_dir, encoder_files[idx])))\n decoder.load_state_dict(torch.load(os.path.join(model_dir, decoder_files[idx])))\n\n # Set the evaluation mode.\n encoder.eval()\n decoder.eval()\n\n # Move neural networks to GPU if CUDA is available.\n encoder, decoder = encoder.to(device), decoder.to(device)\n\n # Calculate the meteor_score of the image captioning model.\n for (image, caption) in data_loader:\n caption = [''.join(c) for c in caption]\n image = image.to(device)\n features = encoder(image).unsqueeze(1)\n output = decoder.sample(features)\n prediction = data_loader.dataset.clean_sentence(output)\n scores[idx] += meteor_score(str(caption), prediction)\n\n # Save best performing neural networks.\n scores[idx] /= len(data_loader.dataset)\n if max_score < scores[idx]:\n min_score = scores[idx]\n torch.save(encoder.state_dict(), os.path.join(model_dir, 'encoder-deploy.pkl'))\n torch.save(decoder.state_dict(), os.path.join(model_dir, 'decoder-deploy.pkl'))\n\n # Display the meteor_score of the image captioning model.\n print('Meteor Score of checkpoint {}: {:0.4f}'.format(idx+1, scores[idx]))\n\n# Plot all results.\nplt.figure(figsize=(24, 16))\nplt.plot(scores)",
"[nltk_data] Downloading package wordnet to /home/gdonatti/nltk_data...\n[nltk_data] Package wordnet is already up-to-date!\n"
]
],
[
[
"The validation results favor the encoder and decoder from the first checkpoint with a [$\\text{METEOR}_{c5}$](http://dl.acm.org/doi/10.5555/1626355.1626389) value of $27.9$, which overcomes the $25.2$ from humans and the $27.7$ from the top of the COCO 2015 [Captioning Leaderboard](http://cocodataset.org/#captions-leaderboard). The current [state-of-the-art](http://competitions.codalab.org/competitions/3221#results) is $29.9$ attained by Tsinghua-Samsung.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecba5d6ac1fd76e1bb571e5063094d540ba855bc | 14,485 | ipynb | Jupyter Notebook | Project/MACD_bootstrap.ipynb | yangtong951019/INFO7390_Final_Project | 7f2acbdcbd0aed48e25f34d305610df927a9df21 | [
"MIT"
] | 1 | 2020-12-19T20:57:54.000Z | 2020-12-19T20:57:54.000Z | Project/MACD_bootstrap.ipynb | yangtong951019/INFO7390_Final_Project | 7f2acbdcbd0aed48e25f34d305610df927a9df21 | [
"MIT"
] | null | null | null | Project/MACD_bootstrap.ipynb | yangtong951019/INFO7390_Final_Project | 7f2acbdcbd0aed48e25f34d305610df927a9df21 | [
"MIT"
] | null | null | null | 32.845805 | 135 | 0.387573 | [
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nfrom scipy import stats\nimport seaborn as sns\nimport scipy.stats as ss\nfrom pandas_datareader import DataReader\nfrom datetime import datetime\n\n# Make plots larger\nplt.rcParams['figure.figsize'] = (15, 9)",
"_____no_output_____"
],
[
"facebook = DataReader('FB', 'yahoo', datetime(2016,4,1), datetime(2019,9,1));\nfacebook.reset_index(inplace=True,drop=False)\nfacebook.set_index('Date')\nfacebook['OpenTmr'] = facebook['Open'].shift(-1)\nfacebook['OpenClose']= (facebook['Open']+ facebook['Close'])/2\nfacebook['HighLow'] = (facebook['High']+ facebook['Low'])/2\nfacebook['OCHL'] = (facebook['Open']+ facebook['Close']+facebook['High']+ facebook['Low'])/4\nfacebook.head()",
"_____no_output_____"
],
[
"N = facebook.shape[0] # total num days\nnum_boot = 300# total num bootstrap\nT= 250 # start day\nwindow = 200 # training period window",
"_____no_output_____"
],
[
"df = facebook[['Close']]\ndf.reset_index(level=0, inplace=True)\ndf.columns=['ds','y']\n\nX1 = df.y.ewm(span=12, adjust=False).mean()\nX2 = df.y.ewm(span=26, adjust=False).mean()\nX = X1 - X2\nY =facebook['OpenTmr'][-(N-(T+1)):].values\n#X = np.column_stack([np.ones((T,1)),X])\n#print(X)\n#print(Y.shape)\n#movAverage1= (movAvg) ",
"_____no_output_____"
],
[
"def MACD(X, Y):\n \n T = X.shape[0]\n #print(T)\n #mu = (facebook['Open '].mean(),facebook['Open '].mean(),facebook['Open '].mean()) \n #cov = [[1,0.75,-0.35],[0.75,1,0.9],[-0.35,0.9,1]]\n #F = np.random.multivariate_normal(mu,cov,T)\n #Sample for Y,X\n X = np.column_stack([np.ones((T,1)),X])\n #T = X.shape[0]\n N = X.shape\n #beta = np.array([0.56,2.53,2.05,1.78])\n #beta.shape=(N[1],1)\n #Y =X@beta+np.random.normal(0,1,(T,1))\n #Y=facebook['Open '].values\n #print(T)\n invXX = np.linalg.inv(X.transpose()@X)\n beta_hat = [email protected]()@Y\n y_hat = X@beta_hat\n \n residuals = Y-y_hat\n sigma2 = (1/T)*residuals.transpose()@residuals\n\n sigma = np.sqrt(sigma2)\n\n #variance - covariance of beta_hat\n varcov_beta_hat = (sigma2)*invXX\n std_beta_hat = np.sqrt(T*np.diag(varcov_beta_hat))\n\n R_square = 1-(residuals.transpose()@residuals)/(T*np.var(Y))\n\n adj_R_square = 1-(1-R_square)*(T-1)/(T - N[1])\n\n #Testing Coefficents:beta_i\n #Null Hypotesis\n\n t_stat = (beta_hat.transpose()-0)/std_beta_hat\n p_val = 1-ss.norm.cdf(t_stat)\n #print(p_val)\n #Test of joint significance\n F_stat= (beta_hat.transpose()@np.linalg.inv(varcov_beta_hat)@beta_hat/N[1])/(residuals.transpose()@residuals/(T-N[1]))\n p_val_F= 1 - ss.f.cdf(F_stat,N[1]-1, T-N[1])\n \n return beta_hat,y_hat",
"_____no_output_____"
],
[
"def bootstrap():\n T = 250\n #print(T)\n N = X.shape[0]\n #print(N)\n \n yhat_macd = np.zeros(N-(T+1))\n window = 200\n num_boost = 300 # increase\n \n for t in range(T+1,N):\n X_train = df.y.ewm(span=5, adjust=False).mean()[t-window:t-1]\n #X_train = np.column_stack([np.ones((len(X_train),1)),X_train])\n Y_train = facebook[['OpenTmr']][t-window:t-1].values\n #print(X_train.shape)\n #print(Y_train.shape)\n X_pred = df.y.ewm(span=5, adjust=False).mean()[t-1:t]\n X_pred = np.column_stack([np.ones((len(X_pred),1)),X_pred])\n \n yhat_train = MACD(X_train , Y_train)[1]\n res_train = Y_train - yhat_train\n \n y_pred_all = np.zeros(num_boost)\n \n for i in range (0,num_boost):\n #err = np.random.choice(res_train,(window-1, ),replace = True)\n err = res_train\n y_bstr = yhat_train + err\n beta_bstr = MACD(X_train,y_bstr)[0]\n #print(X_pred.shape)\n #print(beta_bstr.shape)\n \n y_pred_bstr = X_pred@beta_bstr\n y_pred_all[i] = y_pred_bstr\n \n y_pred_macd = y_pred_all.mean()\n \n yhat_macd[t-(T+1)] = y_pred_macd\n \n \n rmse_macd = np.sqrt(np.mean((Y[27:-1] - yhat_macd[27:-1])**2))\n \n return yhat_macd,rmse_macd",
"_____no_output_____"
],
[
"yhat_macd,rmse_macd = bootstrap()",
"_____no_output_____"
],
[
"rmse_macd",
"_____no_output_____"
],
[
"yhat_macd.shape",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecba62d04311aa0ecd9ece5c740b2b0aa04076da | 7,702 | ipynb | Jupyter Notebook | assignment_3/Task_4_Bonus.ipynb | antranx/ee604_assignments | bdd4313722157aad9548200e8872b51759e0e020 | [
"MIT"
] | 1 | 2020-11-22T15:09:01.000Z | 2020-11-22T15:09:01.000Z | assignment_3/Task_4_Bonus.ipynb | antranx/ee604_assignments | bdd4313722157aad9548200e8872b51759e0e020 | [
"MIT"
] | null | null | null | assignment_3/Task_4_Bonus.ipynb | antranx/ee604_assignments | bdd4313722157aad9548200e8872b51759e0e020 | [
"MIT"
] | null | null | null | 35.657407 | 489 | 0.585173 | [
[
[
"# Copyright 2020 IITK EE604A Image Processing. All Rights Reserved.\n# \n# Licensed under the MIT License. Use and/or modification of this code outside of EE604 must reference:\n#\n# © IITK EE604A Image Processing \n# https://github.com/ee604/ee604_assignments\n#\n# Author: Shashi Kant Gupta, Chiranjeev Prachand and Prof K. S. Venkatesh, Department of Electrical Engineering, IIT Kanpur",
"_____no_output_____"
]
],
[
[
"# Task 4 (Bonus Question): Removing Motion Blur\n\nIn this task, you guys have to help me restore a blurred image of a Diya scene, which I captured while traveling via my car. See the image below:\n\n\n\n<br>\nSince I only came to know about the image's low-quality next day and it's not possible to go back in time to capture that image again, the only possible option is to restore that blurred image. I know that my mobile phone has an excellent camera sensor and the only possible reason for the blurred image is due to my car's motion. I tried to simulate a similar situation and captured a photo of a templated image to help you.\n\n\n\n### Your Task\nUse the template image to restore the image of Diya scene. Your not allowed to use any external library specifically meant for restoration purpose. Follow along the following hints to achieve your target:\n* Follow the following restoration equation: $G(x, y) = F(x, y)H(x, y) + N(x, y)$; where G represents the blurred image and F original image.\n* Consider there is negligible noise while capturing the template image\n* Refer to numpy.fft module for most of the fourier analysis\n* Refer to lecture slides on Weiner Filtering. For additional reference, you can refer to [this](https://www.ee.columbia.edu/~xlx/ee4830/notes/lec7.pdf)\n\nThere are two images on which your algorithm will be tested. Try to achieve as clear images as possible for both of the cases. Note that both of the images may need different values for the parameter. For example, if you use Weiner Filter for restoration, the value of parameter $K = \\frac{S_\\eta}{S_f}$ can be different for both images. You are supposed to tune that manually. In your observations, explain why you needed/ not needed different parameter values for the two images.",
"_____no_output_____"
]
],
[
[
"%%bash\npip install git+https://github.com/ee604/ee604_plugins",
"_____no_output_____"
],
[
"# Importing required libraries\n\nimport cv2\nimport numpy as np\nfrom IPython.display import display\nfrom PIL import Image\nimport matplotlib.pyplot as plt\n\nfrom ee604_plugins import download_dataset\n\ndownload_dataset(assignment_no=3, task_no=4)",
"_____no_output_____"
],
[
"template_input = np.load(\"data/template_input.npy\")\ntemplate_output = np.load(\"data/template_output.npy\")\n\ndiya_motion_blur = np.load(\"data/diya_motion_blur.npy\")\ndiya_motion_blur_nd_noise = np.load(\"data/diya_motion_blur_nd_noise.npy\")",
"_____no_output_____"
],
[
"def restoreImage(img, template_input_img, template_output_img, param=None):\n ''' \n Inputs: \n + img - image which need to be restored\n + template_input_img - original image of the template\n + template_output_img - captured image of the template\n + param - a python dictionary of different parameter value that you will use.\n - https://www.tutorialspoint.com/python/python_dictionary.htm\n \n Ouputs:\n + out_img - restored image\n \n Allowed external package:\n + You are free to use any OpenCV/numpy module except any direct implementation meant for this task.\n \n '''\n out_img = np.copy(img)\n \n #############################\n # Start your code from here #\n #############################\n \n # Replace with your code...\n \n #############################\n # End your code here ########\n #############################\n \n return out_img",
"_____no_output_____"
],
[
"def plot_frame(gridx, gridy, subplot_id, img, name):\n plt.subplot(gridx, gridy, int(subplot_id))\n plt.imshow(img)\n plt.axis(\"off\")\n plt.title(name)",
"_____no_output_____"
],
[
"# Define your parameters for different images in this cell\n# Use the empty dictionary declared below inside your code to store the param values the two images\n\nparam_diya_motion_blur = {}\nparam_diya_motion_blur_nd_noise = {}\n\n#############################\n# Start your code from here #\n#############################\n\n# Replace with your code...\n\n#############################\n# End your code here ########\n#############################",
"_____no_output_____"
],
[
"restored = restoreImage(diya_motion_blur, template_input, template_output, param=param_diya_motion_blur)\nfig = plt.figure(figsize=(13, 9))\nplt.gray()\nplot_frame(1, 2, 1, diya_motion_blur, \"Captured Image\")\nplot_frame(1, 2, 2, restored, \"Restored Image\")\nfig.suptitle('Motion Blur', fontsize=18, y=0.79)\nplt.tight_layout()\nplt.show()\n\nprint(\"\\n\\n\")\n\nrestored = restoreImage(diya_motion_blur_nd_noise, template_input, template_output, param=param_diya_motion_blur_nd_noise)\nfig = plt.figure(figsize=(13, 9))\nplt.gray()\nplot_frame(1, 2, 1, diya_motion_blur, \"Captured Image\")\nplot_frame(1, 2, 2, restored, \"Restored Image\")\nfig.suptitle('Motion Blur and Noise', fontsize=18, y=0.79)\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecba6e36dd90475de323a06937a0bdf6c7e856f5 | 2,160 | ipynb | Jupyter Notebook | Module 02: Introduction to Theoretical Analysis/Exercises/E2.ipynb | ammarSherif/Analysis-and-Design-of-Algorithms-Tutorials | c45fa87ea20bc35000efb2420e58ed486196df14 | [
"MIT"
] | null | null | null | Module 02: Introduction to Theoretical Analysis/Exercises/E2.ipynb | ammarSherif/Analysis-and-Design-of-Algorithms-Tutorials | c45fa87ea20bc35000efb2420e58ed486196df14 | [
"MIT"
] | null | null | null | Module 02: Introduction to Theoretical Analysis/Exercises/E2.ipynb | ammarSherif/Analysis-and-Design-of-Algorithms-Tutorials | c45fa87ea20bc35000efb2420e58ed486196df14 | [
"MIT"
] | null | null | null | 24 | 206 | 0.544444 | [
[
[
"# Exercise 2: Prove by Contradiction",
"_____no_output_____"
],
[
"CSCI304: Analysis and Design of Algorithms<br>\nNile University<br>\nAmmar Sherif<br>\nModule 02",
"_____no_output_____"
],
[
"## Problem Statement",
"_____no_output_____"
],
[
"$$\\boxed{ n \\text{ is not prime} \\implies \\exists_{k \\vert n} \\quad k \\leq \\sqrt{n} } $$",
"_____no_output_____"
],
[
"$$a \\vert b \\iff \\frac{b}{a} = k \\in \\mathbb{Z}$$\n$$3 \\vert 6 \\text{ is }\\textbf{True}\\text{ because } \\frac{6}{3} = 2 \\in \\mathbb{Z}$$\n$$3 \\nmid 4 \\text{ because } \\frac{4}{3} = \\boxed{1.33 \\not\\in \\mathbb{Z}}$$",
"_____no_output_____"
],
[
"## Proof",
"_____no_output_____"
],
[
"To submit your proof here, you have multiple options as follows:\n- Write your proof here in the notebook using LaTeX [*Preferred*]\n- Write your proof in any other application, any word processing application, and take a screenshot of your proof; then, put it here as an image\n- Write your proof by hand, and take a photo of your solution and put it here as an image [**least preferred**]<br>Make sure your write is clear enough; we would not return to ask for a clarification.",
"_____no_output_____"
],
[
"[write your proof here]",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecba70abd5ee107f782f519e19644f491a57e926 | 53,219 | ipynb | Jupyter Notebook | homeworks/hw06/hw6_MachineLearning.ipynb | cwakamiya/ieor135 | 084490380f265225927d11b43d948c1206b0aab8 | [
"Apache-2.0"
] | null | null | null | homeworks/hw06/hw6_MachineLearning.ipynb | cwakamiya/ieor135 | 084490380f265225927d11b43d948c1206b0aab8 | [
"Apache-2.0"
] | null | null | null | homeworks/hw06/hw6_MachineLearning.ipynb | cwakamiya/ieor135 | 084490380f265225927d11b43d948c1206b0aab8 | [
"Apache-2.0"
] | null | null | null | 28.277896 | 421 | 0.423552 | [
[
[
"# Data-X Fall 2018: Homework 06 \n\n### Machine Learning\n\n**Authors:** Sana Iqbal (Part 1, 2, 3)\n\n\nIn this homework, you will do some exercises with prediction. Please note that not all tests are visible. There will be further tests on gradescope. \n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport otter ",
"_____no_output_____"
],
[
" # machine learning libraries\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.svm import SVC, LinearSVC\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.ensemble import AdaBoostClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.linear_model import Perceptron\nfrom sklearn.linear_model import SGDClassifier\nfrom sklearn.tree import DecisionTreeClassifier\n#import xgboost as xgb",
"_____no_output_____"
],
[
"grader = otter.Notebook()",
"_____no_output_____"
]
],
[
[
"## Part 1",
"_____no_output_____"
],
[
"\n\n#### 1.a Read __`diabetesdata.csv`__ file into a pandas dataframe. \nAbout the data: __\n\n1. __TimesPregnant__: Number of times pregnant \n2. __glucoseLevel__: Plasma glucose concentration a 2 hours in an oral glucose tolerance test \n3. __BP__: Diastolic blood pressure (mm Hg) \n5. __insulin__: 2-Hour serum insulin (mu U/ml) \n6. __BMI__: Body mass index (weight in kg/(height in m)^2) \n7. __pedigree__: Diabetes pedigree function \n8. __Age__: Age (years) \n9. __IsDiabetic__: 0 if not diabetic or 1 if diabetic) \n\n\n\n\n\n\n",
"_____no_output_____"
]
],
[
[
"#Read data & print it\ndata = pd.read_csv(\"diabetesdata.csv\")\nprint(data)",
" TimesPregnant glucoseLevel BP insulin BMI Pedigree Age \\\n0 6 148.0 72 0 33.6 0.627 50.0 \n1 1 NaN 66 0 26.6 0.351 31.0 \n2 8 183.0 64 0 23.3 0.672 NaN \n3 1 NaN 66 94 28.1 0.167 21.0 \n4 0 137.0 40 168 43.1 2.288 33.0 \n.. ... ... .. ... ... ... ... \n763 10 101.0 76 180 32.9 0.171 63.0 \n764 2 122.0 70 0 36.8 0.340 27.0 \n765 5 121.0 72 112 26.2 0.245 30.0 \n766 1 126.0 60 0 30.1 0.349 47.0 \n767 1 93.0 70 0 30.4 0.315 23.0 \n\n IsDiabetic \n0 1 \n1 0 \n2 1 \n3 0 \n4 1 \n.. ... \n763 0 \n764 0 \n765 0 \n766 1 \n767 0 \n\n[768 rows x 8 columns]\n"
],
[
"grader.check('q1a')",
"_____no_output_____"
]
],
[
[
"#### 1.b Calculate the percentage of NaN values in each column.",
"_____no_output_____"
]
],
[
[
"# NullsPerColumn should be a dataframe that includes a column called 'Percentage Null'. The values are between 0 and 1.\nNullsPerColumn = data.isnull().sum() / len(data)\nNullsPerColumn = pd.DataFrame(NullsPerColumn).rename(columns= {0: \"Percentage Null\"})\nNullsPerColumn",
"_____no_output_____"
],
[
"grader.check('q1b')",
"_____no_output_____"
]
],
[
[
"**1.c Calculate the TOTAL percent of ROWS with NaN values in the dataframe (make sure values are floats).**",
"_____no_output_____"
]
],
[
[
"# Return values between 0 and 1. \nPercentNull = data.isnull().any(axis = 1).sum()/float(len(data))\nPercentNull",
"_____no_output_____"
]
],
[
[
"**1.d Split __`data`__ into __`train_df`__ and __`test_df`__ with 15% test split.**\n",
"_____no_output_____"
]
],
[
[
"#split values\nfrom sklearn.model_selection import train_test_split\n\nnumTest = round(0.15*len(data),0)\ntrain_df, test_df = data.loc[numTest+1:,:], data.loc[:numTest,:]",
"_____no_output_____"
]
],
[
[
"**1.e Replace the Nan values in __`train_df`__ and __`test_df`__ with the mean of EACH feature.**",
"_____no_output_____"
]
],
[
[
"train_df = train_df.apply(lambda x: x.fillna(x.mean()),axis=0)",
"_____no_output_____"
],
[
"train_df.isna().sum()",
"_____no_output_____"
],
[
"train_df = train_df.apply(lambda x: x.fillna(x.mean()),axis=0)\ntest_df = test_df.apply(lambda x: x.fillna(x.mean()),axis=0)\n\n#train_test_split(data.loc[:,\"TimesPregnant\":\"Age\"], data[\"IsDiabetic\"], test_size = 0.15)",
"_____no_output_____"
],
[
"grader.check('q1e')",
"_____no_output_____"
]
],
[
[
"**1.f Split __`train_df`__ & __`test_df`__ into __`X_train`__, __`Y_train`__ and __`X_test`__, __`Y_test`__. __`Y_train`__ and __`Y_test`__ should only have the column we are trying to predict, __`IsDiabetic`__.**",
"_____no_output_____"
]
],
[
[
"X_train = train_df.loc[:,'TimesPregnant':'Age']\nY_train = train_df.loc[:,'IsDiabetic']\nX_test = test_df.loc[:,'TimesPregnant':'Age']\nY_test = test_df.loc[:,'IsDiabetic']\n\n[X_train.shape, Y_train.shape, X_test.shape,Y_test.shape]",
"_____no_output_____"
],
[
"grader.check('q1f')",
"_____no_output_____"
]
],
[
[
"**1.g Use this dataset to train perceptron, logistic regression and random forest models using 15% test split. Report training and test accuracies.**",
"_____no_output_____"
]
],
[
[
"# Logistic Regression\n\nlogreg = LogisticRegression(solver = \"newton-cg\", multi_class = 'ovr') # use newton-cg solver and 'ovr' for multi_class\nlogreg.fit(X_train, Y_train)\nlogreg_train_acc = logreg.score(X_train, Y_train)\nlogreg_test_acc = logreg.score(X_test, Y_test)\nprint ('logreg training acuracy= ',logreg_train_acc)\nprint('logreg test accuracy= ',logreg_test_acc)",
"logreg training acuracy= 0.7806748466257669\nlogreg test accuracy= 0.7672413793103449\n"
],
[
"# Perceptron\n\nperceptron = Perceptron()\nperceptron.fit(X_train, Y_train)\nperceptron_train_acc = perceptron.score(X_train, Y_train)\nperceptron_test_acc = perceptron.score(X_test, Y_test)\nprint ('perceptron training acuracy= ',perceptron_train_acc)\nprint('perceptron test accuracy= ',perceptron_test_acc)",
"perceptron training acuracy= 0.6809815950920245\nperceptron test accuracy= 0.603448275862069\n"
],
[
"# Adaboost\nadaboost = AdaBoostClassifier()\nadaboost.fit(X_train, Y_train)\nadaboost_train_acc = adaboost.score(X_train, Y_train)\nadaboost_test_acc = adaboost.score(X_test, Y_test)\nprint ('adaboost training acuracy= ',adaboost_train_acc)\nprint('adaboost test accuracy= ',adaboost_test_acc)",
"adaboost training acuracy= 0.8174846625766872\nadaboost test accuracy= 0.75\n"
],
[
"# Random Forest\n\nrandom_forest = RandomForestClassifier()\nrandom_forest.fit(X_train, Y_train)\nrandom_forest_train_acc = random_forest.score(X_train, Y_train)\nrandom_forest_test_acc = random_forest.score(X_test, Y_test)\nprint('random_forest training acuracy= ',random_forest_train_acc)\nprint('random_forest test accuracy= ',random_forest_test_acc)",
"C:\\Users\\Chad\\.conda\\envs\\data-x\\lib\\site-packages\\sklearn\\ensemble\\forest.py:245: FutureWarning: The default value of n_estimators will change from 10 in version 0.20 to 100 in 0.22.\n \"10 in version 0.20 to 100 in 0.22.\", FutureWarning)\n"
]
],
[
[
"**1.h Is mean imputation is the best type of imputation to use? Why or why not? What are some other ways to impute the data?**",
"_____no_output_____"
]
],
[
[
"better = False # [boolean] change this to be True if you believe mean imputation is the best imputation to use, and False otherwise\nexplanation = \"The best type of imputation to use depends on the distribution of the feature. If the data appears normally distributed, then using the mean would be a good choice. However, sometimes it may be best to just remove rows with NaN or replace them with zeros. In other cases it may be better to use the median or some type of algorithm such as KNN.\" # [string] write why or why not here, and other ways",
"_____no_output_____"
]
],
[
[
"## Part 2",
"_____no_output_____"
],
[
"\n__2.a Add columns __`BMI_band`__ & __`Pedigree_band`__ to a new dataframe called __`data2`__ by cutting __`BMI`__ & __`Pedigree`__ into 3 intervals. PRINT the first 5 rows of __`data2`__.\n",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\n\ndata2 = data.copy()\ndata2['BMI_band'] = pd.cut(data2['BMI'], bins = 3)\ndata2['Pedigree_band'] = pd.cut(data2['Pedigree'], bins = 3)\ndata2.head(5)",
"_____no_output_____"
]
],
[
[
"__2.b Print the category intervals for __`BMI_band`__ & __`Pedigree_band`.",
"_____no_output_____"
]
],
[
[
"data2['BMI_band'].values.unique()",
"_____no_output_____"
],
[
"data2['Pedigree_band'].values.unique()",
"_____no_output_____"
],
[
"print('BMI_Band_Interval: ' + \"[(22.367, 44.733], (-0.001, 22.367], (44.733, 67.1]]\")\nprint('Pedigree_Band_Interval: ' + '[(0.077, 0.859], (1.639, 2.42], (0.859, 1.639]]')\n ",
"BMI_Band_Interval: [(22.367, 44.733], (-0.001, 22.367], (44.733, 67.1]]\nPedigree_Band_Interval: [(0.077, 0.859], (1.639, 2.42], (0.859, 1.639]]\n"
]
],
[
[
"__2.c Group __`data`__ by __`Pedigree_band`__ & determine ratio of diabetic in each band.__",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\n \ngroupedData2 = data2.groupby(\"Pedigree_band\").agg({'IsDiabetic':'sum'})\ngroupedData2['Count'] = data2.groupby(['Pedigree_band'])['IsDiabetic'].count()\n\npedigree_DiabeticRatio = pd.DataFrame(groupedData2['IsDiabetic']/ groupedData2['Count'], columns = ['IsDiabetic'])\npedigree_DiabeticRatio",
"_____no_output_____"
],
[
"\ngroupedData2 = data2[['IsDiabetic','Pedigree_band']].groupby(\"Pedigree_band\").mean()\ngroupedData2",
"_____no_output_____"
]
],
[
[
"__2.d Group __`data`__ by __`BMI_band`__ & determine ratio of diabetic in each band.__",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\n\ngroupedData2 = data2.groupby(\"BMI_band\").agg({'IsDiabetic':'sum'})\ngroupedData2['Count'] = data2.groupby(['BMI_band'])['IsDiabetic'].count()\n\nBMI_DiabeticRatio = pd.DataFrame(groupedData2['IsDiabetic']/ groupedData2['Count'], columns = ['IsDiabetic'])\nBMI_DiabeticRatio",
"_____no_output_____"
]
],
[
[
"__2.e Convert these features - 'BP','insulin','BMI' and 'Pedigree' into categorical values by mapping different bands of values of these features to integers 0,1,2 in a dataframe called `data3`.__ \n \nHINT: USE pd.cut with bin=3 to create 3 bins\n\n",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\n\ndata3 = data2.copy()\ndata3['BP'] = pd.cut(data3['BP'], bins = 3, labels = [0,1,2])\ndata3['insulin'] = pd.cut(data3['insulin'], bins = 3, labels = [0,1,2])\ndata3['BMI'] = pd.cut(data3['BMI'], bins = 3, labels = [0,1,2])\ndata3['Pedigree'] = pd.cut(data3['Pedigree'], bins = 3, labels = [0,1,2])\n#data3.drop(columns = ['BMI_band','Pedigree_band'], inplace = True)\ndata3.head()",
"_____no_output_____"
]
],
[
[
"\n__2.f Now consider the original dataset again, instead of generalizing the NAN values with the mean of the feature we will try assigning values to NANs based on some hypothesis. For example for age we assume that the relation between BMI and BP of people is a reflection of the age group. We can have 9 types of BMI and BP relations and our aim is to find the median age of each of that group:__\n\nYour Age guess matrix will look like this: \n\n| BMI | 0 | 1 | 2 |\n|-----|-------------|------------- |----- |\n| BP | | | |\n| 0 | a00 | a01 | a02 |\n| 1 | a10 | a11 | a12 |\n| 2 | a20 | a21 | a22 |\n\n\n__Create a guess_matrix for NaN values of *'Age'* ( using 'BMI' and 'BP') and *'glucoseLevel'* (using 'BP' and 'Pedigree') for the given dataset and assign values accordingly to the NaNs in 'Age' or *'glucoseLevel'* .__\n\n\nRefer to how we guessed age in the titanic notebook in the class.\n\n",
"_____no_output_____"
]
],
[
[
"guess_matrix_age= np.zeros((3,3),dtype=float) \nguess_matrix_age\nguess_matrix_glucoseLevel= np.zeros((3,3),dtype=float) \nguess_matrix_glucoseLevel",
"_____no_output_____"
],
[
"# YOUR CODE HERE\n\n# Fill the NA's for the Age columns\n# with \"qualified guesses\"\n\nfor i in range(0, 3):\n for j in range(0,3):\n guess_df = data3[(data3['BMI'] == i)&(data3['BP'] == j)]['Age'].dropna()\n \n # Extract the median age for this group\n # (less sensitive) to outliers\n age_guess = guess_df.median()\n \n guess_matrix_age[i,j] = age_guess \n\nfor i in range(0, 3):\n for j in range(0, 3):\n data3.loc[ (data3.Age.isnull()) & (data3.BMI == i) & (data3.BP == j),'Age'] = guess_matrix_age[i,j]\n\ndata3['Age'] = data3['Age']\n\nprint(guess_matrix_age)",
"[[24.5 25. 55.5]\n [29.5 29. 37. ]\n [33. 32. 31. ]]\n"
],
[
"for i in range(0, 3):\n for j in range(0,3):\n guess_df = data3[(data3['BP'] == i)&(data3['Pedigree'] == j)]['glucoseLevel'].dropna()\n \n # Extract the median age for this group\n # (less sensitive) to outliers\n glucoseLevel_guess = guess_df.median()\n \n # Convert random age float to int\n guess_matrix_glucoseLevel[i,j] = glucoseLevel_guess\n \nguess_matrix_glucoseLevel",
"_____no_output_____"
],
[
"#guess_matrix_glucoseLevel == np.array([[115. , 127.5, 137. ],[112. , 115.5, 149. ],[133. , 129.5, 159.5]])",
"_____no_output_____"
],
[
"grader.check('q2f')",
"_____no_output_____"
]
],
[
[
"\n\n__2.g Now, convert 'glucoseLevel' and 'Age' features also to categorical variables of 4 categories each in a dataframe called `data4`. PRINT the head of `data4`__\n\n\n\n\n\n\n",
"_____no_output_____"
]
],
[
[
"#Replace NaN's with estimated values\nfor i in range(0, 3):\n for j in range(0, 3):\n data3.loc[ (data3.Age.isnull()) & (data3.BMI == i) & (data3.BP == j),'Age'] = guess_matrix_age[i,j]\n\ndata3['Age'] = data3['Age']\n\nfor i in range(0, 3):\n for j in range(0, 3):\n data3.loc[ (data3.glucoseLevel.isnull()) & (data3.BP == i) & (data3.Pedigree == j),'glucoseLevel'] = guess_matrix_glucoseLevel[i,j]\n\ndata3['glucoseLevel'] = data3['glucoseLevel']",
"_____no_output_____"
],
[
"data3.isna().sum()",
"_____no_output_____"
],
[
"# YOUR CODE HERE\ndata4 = data3.copy()\ndata4['glucoseLevel'] = pd.cut(data4['glucoseLevel'], bins = 4, labels = [0,1,2,3])\ndata4['Age'] = pd.cut(data4['Age'], bins = 4, labels = [0,1,2,3])\ndata4.head()",
"_____no_output_____"
],
[
"data4.isna().sum()",
"_____no_output_____"
],
[
"grader.check('q2g')",
"_____no_output_____"
]
],
[
[
"__2.h Use this dataset (with all features in categorical form) to train perceptron, logistic regression and random forest models using 15% test split. Report training and test accuracies.__\n",
"_____no_output_____"
]
],
[
[
"numTest = round(0.15*len(data4),0)\n\ntrain_df, test_df = data4.loc[numTest+1:,:], data4.loc[:numTest,:]\nX_train = train_df.loc[:,'TimesPregnant':'Age']\nY_train = train_df.loc[:,'IsDiabetic']\nX_test = test_df.loc[:,'TimesPregnant':'Age']\nY_test= test_df.loc[:,'IsDiabetic']\nX_train.shape, Y_train.shape, X_test.shape",
"_____no_output_____"
],
[
"#Logistic Regression\nlogreg = LogisticRegression(solver = \"newton-cg\", multi_class = 'ovr') # use newton-cg solver and 'ovr' for multi_class\nlogreg.fit(X_train, Y_train)\nlogreg_train_acc = logreg.score(X_train, Y_train)\nlogreg_test_acc = logreg.score(X_test, Y_test)\nprint ('logreg training acuracy= ',logreg_train_acc)\nprint('logreg test accuracy= ',logreg_test_acc)",
"logreg training acuracy= 0.7515337423312883\nlogreg test accuracy= 0.6896551724137931\n"
],
[
"# Perceptron\nperceptron = Perceptron()\nperceptron.fit(X_train, Y_train)\nperceptron_train_acc = perceptron.score(X_train, Y_train)\nperceptron_test_acc = perceptron.score(X_test, Y_test)\nprint ('perceptron training acuracy= ',perceptron_train_acc)\nprint('perceptron test accuracy= ',perceptron_test_acc)",
"perceptron training acuracy= 0.5352760736196319\nperceptron test accuracy= 0.5172413793103449\n"
],
[
"# Random Forest\nrandom_forest = RandomForestClassifier()\nrandom_forest.fit(X_train, Y_train)\nrandom_forest_train_acc = random_forest.score(X_train, Y_train)\nrandom_forest_test_acc = random_forest.score(X_test, Y_test)\nprint('random_forest training acuracy= ',random_forest_train_acc)\nprint('random_forest test accuracy= ',random_forest_test_acc)",
"C:\\Users\\Chad\\.conda\\envs\\data-x\\lib\\site-packages\\sklearn\\ensemble\\forest.py:245: FutureWarning: The default value of n_estimators will change from 10 in version 0.20 to 100 in 0.22.\n \"10 in version 0.20 to 100 in 0.22.\", FutureWarning)\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecba80324621ee6b011fe025f6960082e1423b3a | 10,117 | ipynb | Jupyter Notebook | M3-competition. Arima model.ipynb | olchas/meum_project | 2dc94a85f0a942c669ff7f168e62326fd92829b1 | [
"MIT"
] | null | null | null | M3-competition. Arima model.ipynb | olchas/meum_project | 2dc94a85f0a942c669ff7f168e62326fd92829b1 | [
"MIT"
] | null | null | null | M3-competition. Arima model.ipynb | olchas/meum_project | 2dc94a85f0a942c669ff7f168e62326fd92829b1 | [
"MIT"
] | null | null | null | 42.330544 | 178 | 0.569339 | [
[
[
"import argparse\nimport os\nimport time\nfrom math import sqrt\n\nimport numpy as np\nimport pandas as pd\nfrom functools import reduce\nfrom sklearn import preprocessing\nfrom sklearn.metrics import mean_squared_error\n\nfrom ArimaModel import ArimaModel",
"_____no_output_____"
],
[
"input_file = 'data/M3C.xls'\ncategories = ['MICRO', 'INDUSTRY', 'MACRO', 'FINANCE', 'DEMOGRAPHIC', 'OTHER']\nfrequencies = ['Year', 'Quart', 'Month', 'Other']\nforecast_type = 'full' #or 'one_step'\nvalidation_criteria = 'cross' #or 'aic'\nseasonal_arima_model = False",
"_____no_output_____"
],
[
"def load_data(input_file, categories, frequency):\n\n m3c_file = pd.ExcelFile(input_file)\n\n sheet_name = 'M3' + frequency\n\n m3c_month_df = m3c_file.parse(sheet_name)\n\n # strip unnecessary spaces from 'Category' column\n m3c_month_df['Category'] = m3c_month_df['Category'].apply(lambda x: x.strip())\n\n return m3c_month_df[m3c_month_df['Category'].isin(categories)]\n\ndef next_series_generator(data_df, test_data_size):\n\n scaler = preprocessing.MinMaxScaler()\n\n for _, row in data_df.iterrows():\n # series_size incremented for the purpose of range\n series_size = row['N'] + 1\n # conversion to type float64 to silence the warning when fitting scaler\n train_data = row.loc[range(1, series_size - test_data_size)].values.astype(np.float64)\n test_data = row.loc[range(series_size - test_data_size, series_size)].values.astype(np.float64)\n\n # normalise data to range [0, 1]\n scaler.fit(train_data.reshape(-1, 1))\n train_data = scaler.transform(train_data.reshape(1, -1)).tolist()[0]\n test_data = scaler.transform(test_data.reshape(1, -1)).tolist()[0]\n\n yield train_data, test_data, scaler",
"_____no_output_____"
],
[
"for frequency in frequencies:\n for category in categories:\n data_df = load_data(input_file, [category], frequency)\n\n if frequency == 'Year':\n test_data_size = 6\n data_frequency = 1\n elif frequency == 'Quart':\n test_data_size = 8\n data_frequency = 4\n elif frequency == 'Month':\n test_data_size = 18\n data_frequency = 12\n else:\n test_data_size = 8\n data_frequency = 0\n\n if not seasonal_arima_model:\n data_frequency = 0\n\n predicted_data_matrix = []\n expected_data_matrix = []\n\n normalized_forecast_data_matrix = []\n normalized_test_data_matrix = []\n\n # list of dicts of parameters of each trained model\n parameters_dicts = []\n\n rmse_prediction = []\n rmse_forecast = []\n rmse_total = []\n\n training_time = 0\n testing_time = 0\n\n for train_data, test_data, scaler in next_series_generator(data_df, test_data_size):\n training_start = time.time()\n model = ArimaModel(train_data, validation_criteria, data_frequency)\n training_end = time.time()\n\n prediction, forecast = model.make_prediction(test_data, forecast_type)\n testing_end = time.time()\n\n training_time += training_end - training_start\n testing_time += testing_end - training_end\n parameters_dicts.append(model.get_parameters())\n\n # rmse per series\n rmse_prediction.append(sqrt(mean_squared_error(train_data, prediction)))\n rmse_forecast.append(sqrt(mean_squared_error(test_data, forecast)))\n rmse_total.append(sqrt(mean_squared_error(train_data + test_data, prediction + forecast)))\n\n normalized_forecast_data_matrix.append(forecast)\n normalized_test_data_matrix.append(test_data)\n\n # denormalization of data\n predicted_data = scaler.inverse_transform([prediction + forecast]).tolist()[0]\n expected_data = scaler.inverse_transform([train_data + test_data]).tolist()[0]\n\n predicted_data_matrix.append(predicted_data)\n expected_data_matrix.append(expected_data)\n\n # names of data columns in form of integers\n data_columns = list(range(1, max(len(x) for x in predicted_data_matrix) + 1))\n series_index = list(range(len(predicted_data_matrix)))\n series_length = [len(x) for x in predicted_data_matrix]\n\n predicted_data_df = pd.DataFrame(predicted_data_matrix, columns=data_columns)\n predicted_data_df['series'] = series_index\n predicted_data_df['N'] = series_length\n predicted_data_df['NF'] = test_data_size\n\n # take parameters names from first dict\n parameters_names = list(parameters_dicts[0].keys())\n for parameter in parameters_names:\n predicted_data_df[parameter] = [x[parameter] for x in parameters_dicts]\n\n predicted_data_df['rmse_prediction'] = rmse_prediction\n predicted_data_df['rmse_forecast'] = rmse_forecast\n predicted_data_df['rmse_total'] = rmse_total\n\n # change the order of columns\n output_columns = ['series', 'N', 'NF'] + parameters_names + ['rmse_prediction', 'rmse_forecast', 'rmse_total'] \\\n + data_columns\n predicted_data_df = predicted_data_df[output_columns]\n\n expected_data_df = pd.DataFrame(expected_data_matrix, columns=data_columns)\n expected_data_df['series'] = series_index\n expected_data_df['N'] = series_length\n expected_data_df['NF'] = test_data_size\n\n output_columns = ['series', 'N', 'NF'] + data_columns\n expected_data_df = expected_data_df[output_columns]\n\n # calculate error separately for each month of forecast\n rmse_per_month = []\n for i in range(test_data_size):\n month_forecast = [x[i] for x in normalized_forecast_data_matrix]\n month_expected = [x[i] for x in normalized_test_data_matrix]\n rmse_per_month.append(sqrt(mean_squared_error(month_expected, month_forecast)))\n\n # calculate total error of all forecasts\n all_forecast = reduce(lambda x, y: x + y, normalized_forecast_data_matrix)\n all_test = reduce(lambda x, y: x + y, normalized_test_data_matrix)\n\n total_rmse = sqrt(mean_squared_error(all_test, all_forecast))\n\n output_dir = '{frequency}_{category}_{forecast_type}_{validation_criteria}'.format(\n frequency=frequency, category=category,\n forecast_type=forecast_type, validation_criteria=validation_criteria)\n \n if not os.path.isdir(output_dir):\n os.makedirs(output_dir)\n\n normalized_forecast_data_df = pd.DataFrame(normalized_forecast_data_matrix)\n normalized_test_dat_df = pd.DataFrame(normalized_test_data_matrix)\n\n normalized_forecast_data_df.to_csv(os.path.join(output_dir, 'normalized_forecast.tsv'), sep='\\t', index=False)\n normalized_test_dat_df.to_csv(os.path.join(output_dir, 'normalized_test_data.tsv'), sep='\\t', index=False)\n\n predicted_data_df.to_csv(os.path.join(output_dir, 'predictions.tsv'), sep='\\t', index=False)\n expected_data_df.to_csv(os.path.join(output_dir, 'expected.tsv'), sep='\\t', index=False)\n\n with open(os.path.join(output_dir, 'rmse_file.tsv'), 'w') as f:\n f.write('\\t'.join([str(x) for x in rmse_per_month]))\n f.write('\\n' + str(total_rmse))\n\n mean_training_time = training_time / len(predicted_data_df)\n mean_testing_time = testing_time / len(predicted_data_df)\n\n with open(os.path.join(output_dir, 'time.tsv'), 'w') as f:\n f.write('mean_training_time\\tmean_testing_time\\n{training_time}\\t{testing_time}\\n'.format(training_time=mean_training_time, testing_time=mean_testing_time))",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
ecba81947dcd941837c624238203c4e529506b68 | 131,076 | ipynb | Jupyter Notebook | notebooks/regression1e.ipynb | eacunafer/Data-Mining-Machine-Learning-subgraduado- | 17aacb44c37ff7b6365ab3c8b0c0ab93ad685f92 | [
"MIT"
] | 15 | 2018-02-18T15:34:50.000Z | 2021-11-22T07:05:07.000Z | notebooks/regression1e.ipynb | eacunafer/Data-Mining-Machine-Learning-subgraduado- | 17aacb44c37ff7b6365ab3c8b0c0ab93ad685f92 | [
"MIT"
] | null | null | null | notebooks/regression1e.ipynb | eacunafer/Data-Mining-Machine-Learning-subgraduado- | 17aacb44c37ff7b6365ab3c8b0c0ab93ad685f92 | [
"MIT"
] | 8 | 2018-02-22T20:07:46.000Z | 2021-02-19T21:37:29.000Z | 127.879024 | 25,084 | 0.849522 | [
[
[
"## Data Mining and Machine Learning\n## Linear Regression and Correlation \n### Edgar Acuna\n#### March 2021",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport warnings\nwarnings.filterwarnings('ignore')\nimport matplotlib.pyplot as plt\nfrom scipy import stats \nimport seaborn as sns\nimport statsmodels.formula.api as sm\nimport plotnine\nfrom plotnine import *\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Example 1: Predicting number of cars sold according to the seller's year of experience ",
"_____no_output_____"
]
],
[
[
"years=[3,4,6,7,8,12,15,20,22,26]\nventas=[9,12,16,19,23,27,34,37,40,45]",
"_____no_output_____"
]
],
[
[
"Drawing the scatterplot",
"_____no_output_____"
]
],
[
[
"#Haciendo el scatterplot\nplt.scatter(years,ventas)",
"_____no_output_____"
]
],
[
[
"Finding the correlation using numpy",
"_____no_output_____"
]
],
[
[
"#Hallando la correlacion usando numpy\nnp.corrcoef(years,ventas)[0][1]",
"_____no_output_____"
]
],
[
[
"Comentario: Muy buena relacion lineal entre years y ventas. Cuando los anos de experiencia aumentan, las ventas tienden a aumentar",
"_____no_output_____"
],
[
"Finding the coefficients of the regression Line using numpy",
"_____no_output_____"
]
],
[
[
"slope, intercepto= np.poly1d(np.polyfit(years, ventas, 1))\nprint(intercepto, slope)",
"7.661413319776308 1.5072021691238777\n"
],
[
"#Preparando el dataframe para usar pandas\ndata=[years,ventas]\ndata=np.transpose(data)\ndf=pd.DataFrame(data,columns=['years','ventas'])\ndf",
"_____no_output_____"
],
[
"#Calculando la correlacion con pandas\ndf.corr()[\"years\"][\"ventas\"]",
"_____no_output_____"
]
],
[
[
"Nota: Pandas no hace regresion lineal\n\nFinding correlation and regression using scipy.stats",
"_____no_output_____"
]
],
[
[
"#usando stats de scipy muestra la correlacion y su p-value\nstats.pearsonr(years,ventas)",
"_____no_output_____"
],
[
"ec=stats.linregress(years,ventas)\nprint('slope=', ec[0], 'intercepto=', ec[1])",
"slope= 1.5072021691238775 intercepto= 7.661413319776305\n"
],
[
"# Hallando la regresion con statmodels\nresult = sm.ols(formula=\"ventas ~ years\", data=df).fit()\nprint(result.params)",
"Intercept 7.661413\nyears 1.507202\ndtype: float64\n"
],
[
"print(result.summary())",
" OLS Regression Results \n==============================================================================\nDep. Variable: ventas R-squared: 0.967\nModel: OLS Adj. R-squared: 0.963\nMethod: Least Squares F-statistic: 237.8\nDate: Wed, 10 Mar 2021 Prob (F-statistic): 3.11e-07\nTime: 14:47:15 Log-Likelihood: -21.720\nNo. Observations: 10 AIC: 47.44\nDf Residuals: 8 BIC: 48.05\nDf Model: 1 \nCovariance Type: nonrobust \n==============================================================================\n coef std err t P>|t| [0.025 0.975]\n------------------------------------------------------------------------------\nIntercept 7.6614 1.417 5.405 0.001 4.393 10.930\nyears 1.5072 0.098 15.421 0.000 1.282 1.733\n==============================================================================\nOmnibus: 0.770 Durbin-Watson: 0.965\nProb(Omnibus): 0.680 Jarque-Bera (JB): 0.671\nSkew: 0.469 Prob(JB): 0.715\nKurtosis: 2.144 Cond. No. 27.5\n==============================================================================\n\nWarnings:\n[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.\n"
],
[
"#Trazando la linea de regresion sobre el plot de puntos\nx=years\ny=ventas\nplt.scatter(x,y)\nplt.plot(x, np.poly1d(np.polyfit(x, y, 1))(x),color='red')\n\nplt.show()",
"_____no_output_____"
],
[
"sns.regplot(x='years',y='ventas',data=df,ci=False)",
"_____no_output_____"
],
[
"#usando ggplot con la libreria plotnine\n(ggplot(df, aes(x='years',y='ventas'))+geom_point()+geom_smooth(method=\"lm\",se=False))",
"_____no_output_____"
],
[
"#Hallando los resultados completos de la regresion de ventas versus years\nresult = sm.ols(formula=\"ventas ~ years\", data=df).fit()\nprint(result.params)",
"Intercept 7.661413\nyears 1.507202\ndtype: float64\n"
],
[
"print(result.summary())",
" OLS Regression Results \n==============================================================================\nDep. Variable: ventas R-squared: 0.967\nModel: OLS Adj. R-squared: 0.963\nMethod: Least Squares F-statistic: 237.8\nDate: Wed, 10 Mar 2021 Prob (F-statistic): 3.11e-07\nTime: 14:47:16 Log-Likelihood: -21.720\nNo. Observations: 10 AIC: 47.44\nDf Residuals: 8 BIC: 48.05\nDf Model: 1 \nCovariance Type: nonrobust \n==============================================================================\n coef std err t P>|t| [0.025 0.975]\n------------------------------------------------------------------------------\nIntercept 7.6614 1.417 5.405 0.001 4.393 10.930\nyears 1.5072 0.098 15.421 0.000 1.282 1.733\n==============================================================================\nOmnibus: 0.770 Durbin-Watson: 0.965\nProb(Omnibus): 0.680 Jarque-Bera (JB): 0.671\nSkew: 0.469 Prob(JB): 0.715\nKurtosis: 2.144 Cond. No. 27.5\n==============================================================================\n\nWarnings:\n[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.\n"
],
[
"#Finding the Mean Sqaure error\nresult.mse_resid",
"_____no_output_____"
]
],
[
[
"### Example 2: Predicting price of houses according to their area",
"_____no_output_____"
]
],
[
[
"#Ejemplo 2\n#df = pd.read_csv('http://academic.uprm.edu/eacuna/casas.txt',sep='\\s+')\ndf = pd.read_table('http://academic.uprm.edu/eacuna/casas.txt',sep=\",\",delim_whitespace=True)\ndf.head()",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 15 entries, 0 to 14\nData columns (total 2 columns):\n # Column Non-Null Count Dtype\n--- ------ -------------- -----\n 0 area 15 non-null int64\n 1 precio 15 non-null int64\ndtypes: int64(2)\nmemory usage: 368.0 bytes\n"
],
[
"# Haciendp el scatterplot\nplt.figure(figsize=(10,8))\n# Create a scatterplOT\nplt.scatter(df['area'], df['precio'])\n# Chart title\nplt.title('Precio versus areas de casas')\n# y label\nplt.ylabel('Precio')\n# x label\nplt.xlabel('Area')\nplt.show()",
"_____no_output_____"
],
[
"# Hallando la correlacion\ndf.corr()['area']['precio']",
"_____no_output_____"
],
[
"#Trazando la linea de regresion junto con los data points normalizados por escala\nx=df['area']/1000\ny=df['precio']/1000\nslope, intercepto= np.poly1d(np.polyfit(x,y, 1))\nprint(intercepto, slope)\nplt.scatter(x,y)\nplt.plot(x, np.poly1d(np.polyfit(x, y, 1))(x))",
"73.16774838085122 38.52307116035072\n"
],
[
"#usando ggplot con la libreria plotnine\n(ggplot(df/1000, aes(x='area',y='precio'))+geom_point()+geom_smooth(method=\"lm\",se=False))",
"_____no_output_____"
],
[
"df=df/1000\ndf.head()",
"_____no_output_____"
],
[
"#Hallando los resultados completos de la regresion de precio versus area\nresult = sm.ols(formula=\"precio ~ area\", data=df).fit()\nprint(result.params)\nprint(result.summary())",
"Intercept 73.167748\narea 38.523071\ndtype: float64\n OLS Regression Results \n==============================================================================\nDep. Variable: precio R-squared: 0.736\nModel: OLS Adj. R-squared: 0.716\nMethod: Least Squares F-statistic: 36.33\nDate: Wed, 10 Mar 2021 Prob (F-statistic): 4.25e-05\nTime: 15:26:39 Log-Likelihood: -59.922\nNo. Observations: 15 AIC: 123.8\nDf Residuals: 13 BIC: 125.3\nDf Model: 1 \nCovariance Type: nonrobust \n==============================================================================\n coef std err t P>|t| [0.025 0.975]\n------------------------------------------------------------------------------\nIntercept 73.1677 12.674 5.773 0.000 45.787 100.549\narea 38.5231 6.391 6.028 0.000 24.716 52.330\n==============================================================================\nOmnibus: 1.859 Durbin-Watson: 2.241\nProb(Omnibus): 0.395 Jarque-Bera (JB): 1.006\nSkew: 0.631 Prob(JB): 0.605\nKurtosis: 2.877 Cond. No. 8.53\n==============================================================================\n\nWarnings:\n[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.\n"
],
[
"r2 = sm.ols(formula=\"precio ~ area\", data=df/1000).fit().rsquared\nprint (r2)",
"0.736472782770762\n"
],
[
"mse = sm.ols(formula=\"precio ~ area\", data=df/1000).fit().mse_resid\nprint (mse)",
"0.00019931441863104698\n"
],
[
"result.predict(pd.DataFrame({'area': [2.6,3.8]}))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecba876bb5170d142ba6fb1ae324d76e2f3c2388 | 9,094 | ipynb | Jupyter Notebook | trainings/data science academy/python fundamentals for data analysis/2 - loops, conditionals, methods and functions/6 - exercises-loops-conditions.ipynb | Phelipe-Sempreboni/python | 8ad6d18df728d89e5b036759b3bdbec9c4d08f8a | [
"MIT"
] | null | null | null | trainings/data science academy/python fundamentals for data analysis/2 - loops, conditionals, methods and functions/6 - exercises-loops-conditions.ipynb | Phelipe-Sempreboni/python | 8ad6d18df728d89e5b036759b3bdbec9c4d08f8a | [
"MIT"
] | null | null | null | trainings/data science academy/python fundamentals for data analysis/2 - loops, conditionals, methods and functions/6 - exercises-loops-conditions.ipynb | Phelipe-Sempreboni/python | 8ad6d18df728d89e5b036759b3bdbec9c4d08f8a | [
"MIT"
] | null | null | null | 21.966184 | 156 | 0.46987 | [
[
[
"# <font color='blue'>Data Science Academy - Python Fundamentos - Capítulo 3</font>\n\n## Download: http://github.com/dsacademybr",
"_____no_output_____"
]
],
[
[
"# Versão da Linguagem Python\nfrom platform import python_version\nprint('Versão da Linguagem Python Usada Neste Jupyter Notebook:', python_version())",
"Versão da Linguagem Python Usada Neste Jupyter Notebook: 3.9.7\n"
]
],
[
[
"## Exercícios - Loops e Condiconais",
"_____no_output_____"
]
],
[
[
"# Exercício 1 - Crie uma estrutura que pergunte ao usuário qual o dia da semana. Se o dia for igual a Domingo ou \n# igual a sábado, imprima na tela \"Hoje é dia de descanso\", caso contrário imprima na tela \"Você precisa trabalhar!\"\n\npergunta1 = input('Que dia da semana é hoje ?')\n\nif pergunta1 == 'Sabádo' or pergunta1 == 'Domingo':\n \n print('Hoje é dia de descanso!')\nelse:\n \n print('Você precisa trabalhar!')",
"Que dia da semana é hoje ?Sabádo\nHoje é dia de descanso!\n"
],
[
"# Exercício 2 - Crie uma lista de 5 frutas e verifique se a fruta 'Morango' faz parte da lista\n\nlist_fruits = ['Abacaxi', 'Laranja', 'Melância', 'Morango', 'Ameixa']\n\nfor fruit in list_fruits:\n \n if fruit == 'Morango':\n \n print('A fruta morango existe na lista!')",
"A fruta morango existe na lista!\n"
],
[
"# Exercício 3 - Crie uma tupla de 4 elementos, multiplique cada elemento da tupla por 2 e guarde os resultados em uma \n# lista\n\ntuple_numbers = (2, 3, 4, 5)\n\nlist_numbers = []\n\nfor numbers in tuple_numbers:\n \n calcule = numbers * 2\n \n list_numbers.append(calcule)\n \nprint(list_numbers)",
"[4, 6, 8, 10]\n"
],
[
"# Exercício 4 - Crie uma sequência de números pares entre 100 e 150 e imprima na tela\n\nfor itens in range(100, 151, 2):\n \n print(itens)",
"100\n102\n104\n106\n108\n110\n112\n114\n116\n118\n120\n122\n124\n126\n128\n130\n132\n134\n136\n138\n140\n142\n144\n146\n148\n150\n"
],
[
"# Exercício 5 - Crie uma variável chamada temperatura e atribua o valor 40. Enquanto temperatura for maior que 35, \n# imprima as temperaturas na tela\n\ntemperatura = 40\n\nwhile temperatura >= 35:\n \n print(temperatura)\n \n temperatura -= 1",
"40\n39\n38\n37\n36\n35\n"
],
[
"# Exercício 6 - Crie uma variável chamada contador = 0. Enquanto counter for menor que 100, imprima os valores na tela,\n# mas quando for encontrado o valor 23, interrompa a execução do programa\n\ncontador = 0\n\nwhile contador <= 100:\n \n if contador == 23:\n \n break\n \n else:\n \n print(contador)\n \n contador += 1",
"0\n1\n2\n3\n4\n5\n6\n7\n8\n9\n10\n11\n12\n13\n14\n15\n16\n17\n18\n19\n20\n21\n22\n"
],
[
"# Exercício 7 - Crie uma lista vazia e uma variável com valor 4. Enquanto o valor da variável for menor ou igual a 20, \n# adicione à lista, apenas os valores pares e imprima a lista\n\nlist_values = []\n\nnumbers = 4\n\nwhile numbers <= 20:\n \n if numbers <= 20:\n \n list_values.append(numbers)\n \n else:\n \n break\n \n numbers += 1\n \nprint(list_values)",
"[4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20]\n"
],
[
"# Exercício 8 - Transforme o resultado desta função range em uma lista: range(5, 45, 2)\n\nnums = range(5, 45, 2)\n\nconvert = list(nums)\n\nprint(convert)\n\nprint(type(convert))",
"[5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43]\n<class 'list'>\n"
],
[
"# Exercício 9 - Faça a correção dos erros no código abaixo e execute o programa. Dica: são 3 erros.\n\ntemperatura = float(input('Qual a temperatura? '))\n\nif temperatura > 30:\n \n print('Vista roupas leves.')\n\nelse:\n \n print('Busque seus casacos.')",
"Qual a temperatura? 29.9\nBusque seus casacos.\n"
],
[
"# Exercício 10 - Faça um programa que conte quantas vezes a letra \"r\" aparece na frase abaixo. Use um placeholder na \n# sua instrução de impressão\n\n# “É melhor, muito melhor, contentar-se com a realidade; se ela não é tão brilhante como os sonhos, tem pelo menos a \n# vantagem de existir.” (Machado de Assis)\n\nfrase = \"É melhor, muito melhor, contentar-se com a realidade; se ela não é tão brilhante como os sonhos, tem pelo menos a vantagem de existir.\" \n\ncount = 0\n\nfor caracter in frase:\n \n if caracter == 'r':\n \n count += 1\n \nprint(\"O caracter r aparece %s vezes na frase.\" %(count))",
"6\n"
]
],
[
[
"# Fim",
"_____no_output_____"
],
[
"### Obrigado\n\n### Visite o Blog da Data Science Academy - <a href=\"http://blog.dsacademy.com.br\">Blog DSA</a>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ecba963cf2dd17acf42a806dd05eda80dbfff71d | 218,200 | ipynb | Jupyter Notebook | Visualizing_Function_Space_Similarity_MediumCNN.ipynb | ayulockin/LossLandscape | 36fb29c3f7a840ccad50c2a81786d232749946bc | [
"Apache-2.0"
] | 43 | 2020-07-25T15:24:43.000Z | 2022-02-23T04:34:55.000Z | Visualizing_Function_Space_Similarity_MediumCNN.ipynb | ayulockin/LossLandscape | 36fb29c3f7a840ccad50c2a81786d232749946bc | [
"Apache-2.0"
] | null | null | null | Visualizing_Function_Space_Similarity_MediumCNN.ipynb | ayulockin/LossLandscape | 36fb29c3f7a840ccad50c2a81786d232749946bc | [
"Apache-2.0"
] | 5 | 2020-10-22T23:49:21.000Z | 2022-01-31T09:39:15.000Z | 57.345598 | 31,446 | 0.678332 | [
[
[
"<a href=\"https://colab.research.google.com/github/ayulockin/LossLandscape/blob/master/Visualizing_Function_Space_Similarity_MediumCNN.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Setups, Imports and Installations",
"_____no_output_____"
]
],
[
[
"## This is so that I can save my models.\nfrom google.colab import drive\ndrive.mount('gdrive')",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&response_type=code&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly\n\nEnter your authorization code:\n··········\nMounted at gdrive\n"
],
[
"%%capture\n!pip install wandb",
"_____no_output_____"
],
[
"import tensorflow as tf\n\nfrom tensorflow import keras\nfrom tensorflow.keras.datasets import cifar10\nfrom tensorflow.keras.applications import resnet50",
"_____no_output_____"
],
[
"import os\nos.environ[\"TF_DETERMINISTIC_OPS\"] = \"1\"\n\nimport numpy as np\nfrom numpy.linalg import norm\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.mplot3d import Axes3D\n\n%matplotlib inline\n\nimport seaborn as sns\nsns.set()\n\nfrom tqdm.notebook import tqdm_notebook\nfrom sklearn.manifold import TSNE",
"/usr/local/lib/python3.6/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
],
[
"import wandb\nfrom wandb.keras import WandbCallback\n\nwandb.login()",
"_____no_output_____"
]
],
[
[
"# Get Trained Models",
"_____no_output_____"
],
[
"(For now I am using models trained from SmallCNN.)",
"_____no_output_____"
]
],
[
[
"ROOT_PATH = 'gdrive/My Drive/LossLandscape/'\n## MediumCNN model checkpoint for each epoch(total 40 epochs)\nMODEL_PATH = ROOT_PATH+'MediumCNN_CheckpointID_1/'\n## MediumCNN models with different initialization(total 10 models)\nINDEPENDENT_MODEL_PATH = ROOT_PATH+'MediumIndependentSolutions/'\n## MediumCNN models with different initialization(total 3 models) with all checkpoints saved\nINDEPENDENT_MODEL_PATH_tsne = ROOT_PATH+'MediumIndependentSolutions_tsne/'",
"_____no_output_____"
],
[
"same_model_ckpts = os.listdir(MODEL_PATH)\nindependent_models = os.listdir(INDEPENDENT_MODEL_PATH)\nindependent_models_tsne = os.listdir(INDEPENDENT_MODEL_PATH_tsne)\n\n\nprint(len(same_model_ckpts))\nprint(len(independent_models))\nprint(len(independent_models_tsne))",
"40\n10\n3\n"
],
[
"# https://stackoverflow.com/a/2669120/7636462\nimport re \n\ndef sorted_nicely(l): \n \"\"\" Sort the given iterable in the way that humans expect.\"\"\" \n convert = lambda text: int(text) if text.isdigit() else text \n alphanum_key = lambda key: [ convert(c) for c in re.split('([0-9]+)', key) ] \n return sorted(l, key = alphanum_key)",
"_____no_output_____"
],
[
"same_model_ckpts = sorted_nicely(same_model_ckpts)\nsame_model_ckpts[:5]",
"_____no_output_____"
],
[
"independent_models = sorted_nicely(independent_models)\nindependent_models[:5]",
"_____no_output_____"
]
],
[
[
"# Get Dataset and Prepare",
"_____no_output_____"
],
[
"#### CIFAR-10",
"_____no_output_____"
]
],
[
[
"(x_train, y_train), (x_test, y_test) = cifar10.load_data()\n\ny_train = y_train.flatten()\ny_test = y_test.flatten()\n\nCLASS_NAMES = (\"airplane\", \"automobile\", \"bird\", \"cat\", \"deer\", \"dog\", \"frog\", \"horse\", \"ship\", \"truck\")\nprint(x_train.shape, y_train.shape, x_test.shape, y_test.shape)",
"Downloading data from https://www.cs.toronto.edu/~kriz/cifar-10-python.tar.gz\n170500096/170498071 [==============================] - 2s 0us/step\n(50000, 32, 32, 3) (50000,) (10000, 32, 32, 3) (10000,)\n"
]
],
[
[
"#### Dataloader",
"_____no_output_____"
]
],
[
[
"AUTO = tf.data.experimental.AUTOTUNE\nBATCH_SIZE = 128\nIMG_SHAPE = 32\n\ntestloader = tf.data.Dataset.from_tensor_slices((x_test, y_test))\n\ndef preprocess_image(image, label):\n img = tf.cast(image, tf.float32)\n img = img/255.\n\n return img, label\n\ntestloader = (\n testloader\n .map(preprocess_image, num_parallel_calls=AUTO)\n .batch(BATCH_SIZE)\n .prefetch(AUTO)\n)",
"_____no_output_____"
]
],
[
[
"# Similarity of Functions Within Initialized Trajectories",
"_____no_output_____"
],
[
"## Disagreement of predictions \n\n(Refer figure 2(b) from paper)",
"_____no_output_____"
]
],
[
[
"# Given model and test data, return true_labels and predictions.\ndef evaluate(test_dataloader, model):\n true_labels = []\n pred_labels = []\n \n for imgs, labels in iter(test_dataloader):\n preds = model.predict(imgs)\n \n true_labels.extend(labels)\n pred_labels.extend(np.argmax(preds, axis=1))\n\n return np.array(true_labels), np.array(pred_labels)",
"_____no_output_____"
],
[
"predictions = []\nfor i in tqdm_notebook(range(40)):\n # load model\n model = tf.keras.models.load_model(MODEL_PATH+same_model_ckpts[i])\n # get predictions for model\n _, preds = evaluate(testloader, model)\n\n predictions.append(preds)",
"_____no_output_____"
],
[
"empty_arr = np.zeros(shape=(40,40))\n\nfor i in tqdm_notebook(range(40)):\n preds1 = predictions[i]\n for j in range(i, 40):\n preds2 = predictions[j]\n \n # compute dissimilarity\n dissimilarity_score = 1-np.sum(np.equal(preds1, preds2))/10000 \n \n empty_arr[i][j] = dissimilarity_score\n if i is not j:\n empty_arr[j][i] = dissimilarity_score\n\ndissimilarity_coeff = empty_arr[::-1]",
"_____no_output_____"
],
[
"plt.figure(figsize=(9,8))\nsns.heatmap(dissimilarity_coeff, cmap='RdBu_r');\nplt.xticks([5,10,15,20,25,30,35],[5,10,15,20,25,30,35]);\nplt.yticks([5,10,15,20,25,30,35],[35,30,25,20,15,10,5]);\n\nplt.savefig('prediction_disagreement.png')",
"_____no_output_____"
]
],
[
[
"## Cosine Similarity of weights\n\n(Refer figure 2(a) from paper)",
"_____no_output_____"
]
],
[
[
"# Get the weights of the input model. \ndef get_model_weights(model):\n model_weights = []\n # iterate through model layers.\n for layer in model.layers:\n # grab weights of that layer\n weights = layer.get_weights() # list\n # check if layer got triainable weights\n if len(weights)==0:\n continue\n\n # discard biases term, wrap with ndarray, flatten weights\n model_weights.extend(np.array(weights[0]).flatten())\n\n return np.array(model_weights)",
"_____no_output_____"
],
[
"weights_of_models = []\nfor i in tqdm_notebook(range(40)):\n # load model\n model = tf.keras.models.load_model(MODEL_PATH+same_model_ckpts[i])\n # get predictions for model\n weights = get_model_weights(model)\n\n weights_of_models.append(weights)",
"_____no_output_____"
],
[
"empty_arr = np.zeros(shape=(40,40))\n\nfor i in tqdm_notebook(range(40)):\n weights1 = weights_of_models[i]\n for j in range(i, 40):\n weights2 = weights_of_models[j]\n \n # compute cosine similarity of weights\n cos_sim = np.dot(weights1, weights2)/(norm(weights1)*norm(weights2))\n \n empty_arr[i][j] = cos_sim\n if i is not j:\n empty_arr[j][i] = cos_sim\n\ncos_sim_coeff = empty_arr[::-1]",
"_____no_output_____"
],
[
"plt.figure(figsize=(9,8))\nsns.heatmap(cos_sim_coeff, cmap='RdBu_r');\nplt.xticks([5,10,15,20,25,30,35],[5,10,15,20,25,30,35]);\nplt.yticks([5,10,15,20,25,30,35],[35,30,25,20,15,10,5]);\n\nplt.savefig('functional_similarity.png')",
"_____no_output_____"
],
[
"wandb.init(entity='authors', project='loss-landscape', id='medium_cnn_same_model_investigations')\nwandb.log({'prediction_disagreement': wandb.Image('prediction_disagreement.png')})\nwandb.log({'functional_similarity': wandb.Image('functional_similarity.png')})",
"_____no_output_____"
]
],
[
[
"# Similarity of Functions Across Randomly Initialized Trajectories",
"_____no_output_____"
],
[
"## Disagreement of predictions\n\n(Refer figure 3(a) from paper)",
"_____no_output_____"
]
],
[
[
"predictions = []\nfor i in tqdm_notebook(range(10)):\n # load model\n model = tf.keras.models.load_model(INDEPENDENT_MODEL_PATH+independent_models[i])\n # get predictions for model\n _, preds = evaluate(testloader, model)\n\n predictions.append(preds)",
"_____no_output_____"
],
[
"empty_arr = np.zeros(shape=(10,10))\n\nfor i in tqdm_notebook(range(10)):\n preds1 = predictions[i]\n for j in range(i, 10):\n preds2 = predictions[j]\n \n # compute dissimilarity\n dissimilarity_score = 1-np.sum(np.equal(preds1, preds2))/10000 \n \n empty_arr[i][j] = dissimilarity_score\n if i is not j:\n empty_arr[j][i] = dissimilarity_score\n\ndissimilarity_coeff = empty_arr[::-1]",
"_____no_output_____"
],
[
"plt.figure(figsize=(9,8))\nsns.heatmap(dissimilarity_coeff, cmap='RdBu_r');\nplt.xticks([0,2,4,6,8],[0,2,4,6,8]);\nplt.yticks([0,2,4,6,8],[8,6,4,2,1]);\n\nplt.savefig('independent_prediction_disagreement.png')",
"_____no_output_____"
]
],
[
[
"## Cosine Similarity of weights\n\n(Refer figure 3(a) from paper)",
"_____no_output_____"
]
],
[
[
"# Get the weights of the input model. \ndef get_model_weights(model):\n model_weights = []\n # iterate through model layers.\n for layer in model.layers:\n # grab weights of that layer\n weights = layer.get_weights() # list\n # check if layer got triainable weights\n if len(weights)==0:\n continue\n\n # discard biases term, wrap with ndarray, flatten weights\n model_weights.extend(np.array(weights[0]).flatten())\n\n return np.array(model_weights)",
"_____no_output_____"
],
[
"weights_of_models = []\nfor i in tqdm_notebook(range(10)):\n # load model\n model = tf.keras.models.load_model(INDEPENDENT_MODEL_PATH+independent_models[i])\n # get predictions for model\n weights = get_model_weights(model)\n\n weights_of_models.append(weights)",
"_____no_output_____"
],
[
"empty_arr = np.zeros(shape=(10,10))\n\nfor i in tqdm_notebook(range(10)):\n weights1 = weights_of_models[i]\n for j in range(i, 10):\n weights2 = weights_of_models[j]\n \n # compute cosine similarity of weights\n cos_sim = np.dot(weights1, weights2)/(norm(weights1)*norm(weights2))\n \n empty_arr[i][j] = cos_sim\n if i is not j:\n empty_arr[j][i] = cos_sim\n\ncos_sim_coeff = empty_arr[::-1]",
"_____no_output_____"
],
[
"plt.figure(figsize=(9,8))\nsns.heatmap(cos_sim_coeff, cmap='RdBu_r');\nplt.xticks([0,2,4,6,8],[0,2,4,6,8]);\nplt.yticks([0,2,4,6,8],[8,6,4,2,1]);\n\nplt.savefig('independent_functional_similarity.png')",
"_____no_output_____"
],
[
"wandb.init(entity='authors', project='loss-landscape', id='medium_cnn_independent_model_investigations')\nwandb.log({'independent_prediction_disagreement': wandb.Image('independent_prediction_disagreement.png')})\nwandb.log({'independent_functional_similarity': wandb.Image('independent_functional_similarity.png')})",
"_____no_output_____"
]
],
[
[
"# Investigating Load Landscape",
"_____no_output_____"
]
],
[
[
"NUM_EXAMPLES = 1024\nNUM_CLASSES = 10\ntest_tsne_ds = testloader.unbatch().take(NUM_EXAMPLES).batch(BATCH_SIZE)",
"_____no_output_____"
],
[
"# Given model and test data, return true_labels and predictions.\ndef evaluate_tsne(test_dataloader, model):\n true_labels = []\n pred_labels = []\n \n for imgs, labels in iter(test_dataloader):\n preds = model.predict(imgs)\n \n true_labels.extend(labels)\n pred_labels.extend(preds) ## change here \n\n return np.array(true_labels), np.array(pred_labels)",
"_____no_output_____"
],
[
"NUM_TRAJECTORIES = 3\n\npredictions_for_tsne = []\n\nfor i in tqdm_notebook(range(NUM_TRAJECTORIES)):\n subdir = independent_models_tsne[i]\n model_files = os.listdir(INDEPENDENT_MODEL_PATH_tsne+subdir)\n model_files = sorted_nicely(model_files)\n predictions = []\n for model_file in model_files:\n try:\n # if model_file=='small_cnn_checkpoint_5.h5': # this check is because this model checkpoint didn't serialize properly.\n # continue\n model = tf.keras.models.load_model(INDEPENDENT_MODEL_PATH_tsne+subdir+'/'+model_file)\n except:\n print(INDEPENDENT_MODEL_PATH_tsne+subdir+'/'+model_file)\n continue\n\n _, preds = evaluate_tsne(test_tsne_ds, model)\n\n predictions.append(preds)\n \n predictions = np.array(predictions)\n print(predictions.shape)\n predictions_for_tsne.append(predictions)\n\n# convert list to array\npredictions_for_tsne = np.array(predictions_for_tsne)\nprint('[INFO] shape of predictions tensor: ', predictions_for_tsne.shape)\n\n# reshape the tensor \nreshaped_predictions_for_tsne = predictions_for_tsne.reshape([-1, NUM_EXAMPLES*NUM_CLASSES])\nprint('[INFO] shape of reshaped tensor: ', reshaped_predictions_for_tsne.shape)",
"_____no_output_____"
],
[
"# initialize tsne object\ntsne = TSNE(n_components=2)\n# compute tsne\nprediction_embed = tsne.fit_transform(reshaped_predictions_for_tsne)\nprint('[INFO] Shape of embedded tensor: ', prediction_embed.shape)\n# reshape\ntrajectory_embed = prediction_embed.reshape([NUM_TRAJECTORIES, -1, 2])\nprint('[INFO] Shape of reshaped tensor: ', trajectory_embed.shape)",
"[INFO] Shape of embedded tensor: (120, 2)\n[INFO] Shape of reshaped tensor: (3, 40, 2)\n"
],
[
"# Plot\nplt.figure(constrained_layout=True, figsize=(6,6))\n\ncolors_list=['r', 'b', 'g']\nlabels_list = ['traj_{}'.format(i) for i in range(NUM_TRAJECTORIES)]\nfor i in range(NUM_TRAJECTORIES):\n plt.plot(trajectory_embed[i,:,0],trajectory_embed[i,:,1],color = colors_list[i], alpha = 0.8,linestyle = \"\", marker = \"o\")\n plt.plot(trajectory_embed[i,:,0],trajectory_embed[i,:,1],color = colors_list[i], alpha = 0.3,linestyle = \"-\", marker = \"\")\n plt.plot(trajectory_embed[i,0,0],trajectory_embed[i,0,1],color = colors_list[i], alpha = 1.0,linestyle = \"\", marker = \"*\")\n\nplt.savefig('loss_landscape_2d.png')",
"_____no_output_____"
],
[
"wandb.init(entity='authors', project='loss-landscape', id='medium_cnntrajectory_investigation')\nwandb.log({'loss_landscape_2d': wandb.Image('loss_landscape_2d.png')})",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecbabddc76ca27a795eb6dc2dd4217899f17af12 | 10,213 | ipynb | Jupyter Notebook | Data-Modeling-with-Postgres/test.ipynb | naderAsadi/Udacity-Data-Engineering-Projects | d12c42b3260379a470abd244f98a1fd5b32718f7 | [
"MIT"
] | 4 | 2020-10-03T18:14:20.000Z | 2021-11-01T08:15:32.000Z | Data-Modeling-with-Postgres/test.ipynb | naderAsadi/Udacity-Data-Engineering-Projects | d12c42b3260379a470abd244f98a1fd5b32718f7 | [
"MIT"
] | null | null | null | Data-Modeling-with-Postgres/test.ipynb | naderAsadi/Udacity-Data-Engineering-Projects | d12c42b3260379a470abd244f98a1fd5b32718f7 | [
"MIT"
] | null | null | null | 76.789474 | 1,531 | 0.673455 | [
[
[
"%load_ext sql",
"_____no_output_____"
],
[
"!sql postgresql://student:[email protected]/sparkifydb",
"zsh:1: command not found: sql\n"
],
[
"%sql SELECT * FROM songplays LIMIT 5;",
"_____no_output_____"
],
[
"%sql SELECT * FROM users LIMIT 5;",
"_____no_output_____"
],
[
"%sql SELECT * FROM songs LIMIT 5;",
"_____no_output_____"
],
[
"%sql SELECT * FROM artists LIMIT 5;",
"_____no_output_____"
],
[
"%sql SELECT * FROM time LIMIT 5;",
"_____no_output_____"
]
],
[
[
"## REMEMBER: Restart this notebook to close connection to `sparkifydb`\nEach time you run the cells above, remember to restart this notebook to close the connection to your database. Otherwise, you won't be able to run your code in `create_tables.py`, `etl.py`, or `etl.ipynb` files since you can't make multiple connections to the same database (in this case, sparkifydb).",
"_____no_output_____"
]
]
] | [
"code",
"markdown"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecbac750ac552b143bd28305c26bfac87f944f04 | 1,218 | ipynb | Jupyter Notebook | Notebooks/Ileum_Lamina_Propria_Immunocytes.ipynb | lshh125/geo-crawler | d1e180a16060c3dc297e2d142458ed47106e490e | [
"MIT"
] | null | null | null | Notebooks/Ileum_Lamina_Propria_Immunocytes.ipynb | lshh125/geo-crawler | d1e180a16060c3dc297e2d142458ed47106e490e | [
"MIT"
] | null | null | null | Notebooks/Ileum_Lamina_Propria_Immunocytes.ipynb | lshh125/geo-crawler | d1e180a16060c3dc297e2d142458ed47106e490e | [
"MIT"
] | null | null | null | 23.423077 | 260 | 0.497537 | [
[
[
"<a href=\"https://colab.research.google.com/github/lshh125/geo-crawler/blob/master/Notebooks/Ileum_Lamina_Propria_Immunocytes.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Test",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
]
] |
ecbaccb7439183f47fc4df9b68237603f5da073c | 697 | ipynb | Jupyter Notebook | Notebooks/Test.ipynb | oaramos/Data_Mural_Project | 90b63bcbb7b7e1c2ca5b588c5876cc882b6f6235 | [
"CC0-1.0"
] | null | null | null | Notebooks/Test.ipynb | oaramos/Data_Mural_Project | 90b63bcbb7b7e1c2ca5b588c5876cc882b6f6235 | [
"CC0-1.0"
] | null | null | null | Notebooks/Test.ipynb | oaramos/Data_Mural_Project | 90b63bcbb7b7e1c2ca5b588c5876cc882b6f6235 | [
"CC0-1.0"
] | null | null | null | 16.595238 | 42 | 0.512195 | [
[
[
"hello",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
ecbad1cbad1a2a81c3acf397a3f702df9fa7ecdc | 48,535 | ipynb | Jupyter Notebook | Repositorio/20191213 - Examen Ordinario/.ipynb_checkpoints/examen_practica_ESP_sol-checkpoint.ipynb | juanvecino/Python_LAB | e43a13d74ba688783fca8924e4eb5f52e84e7d49 | [
"MIT"
] | 3 | 2020-11-08T16:47:02.000Z | 2020-12-20T19:45:13.000Z | Repositorio/20191213 - Examen Ordinario/.ipynb_checkpoints/examen_practica_ESP_sol-checkpoint.ipynb | juanvecino/Python_LAB | e43a13d74ba688783fca8924e4eb5f52e84e7d49 | [
"MIT"
] | null | null | null | Repositorio/20191213 - Examen Ordinario/.ipynb_checkpoints/examen_practica_ESP_sol-checkpoint.ipynb | juanvecino/Python_LAB | e43a13d74ba688783fca8924e4eb5f52e84e7d49 | [
"MIT"
] | null | null | null | 105.054113 | 11,712 | 0.84403 | [
[
[
"# Examen Final\n# Fundamentos de la Informática\n# 13-Diciembre-2019\n\n# Tiempo: 2h15",
"_____no_output_____"
],
[
"## *Rellena la siguiente información*:\n\n### Nombre completo:\n### Grupo:",
"_____no_output_____"
],
[
"## Ej1. Lectura de ficheros (2 ptos.)\n\nJunto al enunciado se han distribuido dos ficheros \"Meses.txt\" y \"Descuentos.txt\". \"Meses.txt\" contiene los diferentes meses del año y un identificador de descuento para cada uno. \"Descuentos.txt\" contiene el importe exacto de cada descuento.\n\nSe pide:\n\n1. Leer el fichero \"Meses.txt\" y generar un diccionario llamado \"dic_month\" en el que las claves sean los nombres de los meses y los valores el identificador del descuento:\n\n {'Enero': 'd1',\n 'Febrero': 'd3',....}\n\n2. Leer el fichero \"Descuentos.txt\" y generar un diccionario llamado \"dic_discount\" en el que las claves sean los identificadores de descuento y el valor sea el valor numérico de dicho descuento:\n\n {'d0': 0.0, \n 'd1': 10.0,....}\n",
"_____no_output_____"
]
],
[
[
"dic_month ={}\n\nwith open(\"Meses.txt\", \"r\") as f:\n header= f.readline().split(\";\")\n for row in f:\n id_mes, mes, desc_id =row.split(\";\")\n dic_month[mes] = desc_id.replace('\\n','')\n\ndic_month",
"_____no_output_____"
],
[
"dic_discount ={}\n\nwith open(\"Descuentos.txt\", \"r\") as f:\n header= f.readline().split(\"^\")\n for row in f:\n desc_id, desc, value =row.split(\"^\")\n dic_discount[desc_id] = float(value)\n\ndic_discount",
"_____no_output_____"
],
[
"print(list(dic_month.items()))",
"[('Enero', 'd1'), ('Febrero', 'd3'), ('Marzo', 'd0'), ('Abril', 'd2'), ('Mayo', 'd1'), ('Junio', 'd3'), ('Julio', 'd0'), ('Agosto', 'd2'), ('Septiembre', 'd3'), ('Octubre', 'd4'), ('Noviembre', 'd5'), ('Diciembre', 'd2')]\n"
]
],
[
[
"## Ej2. Combinar diccionarios (2 ptos.)\n\nUtilizar los diccionarios generados en el apartado anterior (si no se ha conseguido, definirlos a mano) para construir un nuevo diccionario llamado \"dic_final\" en el que las claves sean el nombre de los meses del año y el valor sea una tupla en el que el primer elemento sea el identificador del descuento y el segundo elemento sea el importe exacto de cada descuento:\n\n {'Enero': (\"d1\", 10.0),\n 'Febrero': (\"d3\", 25.0) ,....}",
"_____no_output_____"
]
],
[
[
"dic_final={}\nfor mes,desc_id in dic_month.items():\n dic_final[mes]=(desc_id, dic_discount[desc_id])\ndic_final ",
"_____no_output_____"
]
],
[
[
"## Ej3. Salvar a fichero (1 pto.)\n\nUtilizar el diccionario generado en el apartado anterior (si no se ha conseguido, definirlo a mano), para salvar su información en un fichero de tipo CSV llamado \"Mes-Descuentos.csv\", cuya primera línea debe incluir una cabecera con los nombres \"mes\",\"id_descuento\",\"valor\". Y el separador debe ser el caracter \"|\":\n\n mes|id_descuento|valor\n Enero|d1|10.0\n Febrero|d3|25.0",
"_____no_output_____"
]
],
[
[
"with open(\"Mes-Descuentos.csv\", \"w\") as f:\n f.write(\"mes|id_descuento|valor\\n\")\n for mes,(id_d,valor) in dic_final.items():\n f.write(\"|\".join([mes,id_d,str(valor)+'\\n']))",
"_____no_output_____"
]
],
[
[
"## Ej4. Genera cualquier polinomio (2 ptos.)\n\nPreguntar al usuario por el grado del polinomio que quiere pintar. A continuacion, el código debe preguntar al usuario por tantos coeficientes como grado tenga el polinomio. El código debe funcionar para cualquier grado (no es necesario chequear si el usuario introduce grados negativos o letras o símbolos). Al final, el código debe imprimir la función solicitada por el usuario:\n\n\n Introduzca el grado del polinomio: 3\n - Introduzca el coeficiente grado 0:0\n - Introduzca el coeficiente grado 1:1\n - Introduzca el coeficiente grado 2:2\n - Introduzca el coeficiente grado 3:3\n El polinomio solicitado es: \n 3x^3+2x^2+1x^1+0",
"_____no_output_____"
]
],
[
[
"grado=int(input(\"Introduzca el grado del polinomio: \"))\ncoeficientes=[]\nfor i in range(grado+1):\n tmp=int(input(\" - Introduzca el coeficiente grado \"+str(i)+\":\"))\n coeficientes.append(tmp)\nprint(\"El polinomio solicitado es: \")\n\nprint(\"f(x)=\",end=\"\")\nfor i in range(grado+1):\n if i!=grado:\n print(str(coeficientes[grado-i])+\"x^\"+str(grado-i)+\"+\",end=\"\")\n else:\n print(coeficientes[grado-i])",
"Introduzca el grado del polinomio: 3\n - Introduzca el coeficiente grado 0:0\n - Introduzca el coeficiente grado 1:1\n - Introduzca el coeficiente grado 2:2\n - Introduzca el coeficiente grado 3:3\nEl polinomio solicitado es: \nf(x)=3x^3+2x^2+1x^1+0\n"
]
],
[
[
"## Ej5. Calcula el polinomio (2 ptos.)\n\nConstruye una función que responda a esta difinición:\n\n def apply_polynomio(ejex,coeficientes):\n ...\n ...\n return ejey\n \nDónde la variable \"ejex\" reciba una lista de valores y la variable \"coeficientes\" reciba la lista de coeficientes seleccionados por el usuario (no es necesario haber realizado el apartado anterior para hacer este ejercicio).\nLa variable devuelta \"ejey\" debe ser una lista de valores resultado de aplicar el polinomio definido por los coeficientes a cada uno de los valores de la lista \"ejex\".",
"_____no_output_____"
]
],
[
[
"import numpy as np\ndef apply_polynomio(ejex,coeficientes):\n ejey=[]\n for x in ejex:\n value=0\n for i in range(len(coeficientes)):\n if i==0:\n value=value+coeficientes[i]\n else:\n value=value+coeficientes[i]*np.power(x,i)\n ejey.append(value)\n return ejey",
"_____no_output_____"
],
[
"ejex=[0,1,2,3]\nejey=apply_polynomio([0,1,2,3],[1,2,3,4])\nejey",
"_____no_output_____"
]
],
[
[
"## Ej6. Visualiza el polinomio (1 pto.)\n\nConstruye una función con la siguiente definición:\n \n def plot_polynomio(ejex,ejey,kind):\n\nQue visualice la función descrita por las listas de valores \"ejex\" y \"ejey\". El tipo de gráfico utilizado dependerá del parámetro \"kind\" que puede tomar los siguientes valores:\n\n kind=\"line\"\n kind=\"scatter\"\n kind=\"bar\"\n \nCuando la varibale \"kind\" tome el valor \"line\" imprimirá el polinomio con una línea contínua negra utilizando cruces para las marcas. \nCuando la varible \"kind\" tome el valor \"scatter\" imprimirá el polinomio como una sucesión de puntos con forma circular de color rojo. \nCuando la variable \"kind\" tome el valor \"bar\" imprimirá el polinomio como un diagrama de barras verticales de color magenta.\nCuando la variable \"kind\" no reciba ningún valor por parte del usuario, por defecto, tomará el valor \"line\".\nCuando la variable \"kind\" reciba un valor no reconocible, por defecto, tomará el valor \"line\"",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\ndef plot_polynomio(ejex,ejey,kind=\"line\"):\n if kind==\"scatter\":\n plt.scatter(ejex,ejey,marker=\"o\",c=\"r\")\n elif kind==\"bar\":\n plt.bar(ejex,ejey,color=\"magenta\",alpha=0.5)\n else:\n plt.plot(ejex,ejey,\"k-+\")",
"_____no_output_____"
],
[
"plot_polynomio([0,1,2,3],[1, 10, 49, 142])",
"_____no_output_____"
],
[
"plot_polynomio([0,1,2,3],[1, 10, 49, 142],kind=\"cualquiercosa\")",
"_____no_output_____"
],
[
"plot_polynomio([0,1,2,3],[1, 10, 49, 142],\"scatter\")",
"_____no_output_____"
],
[
"plot_polynomio([0,1,2,3],[1, 10, 49, 142],\"bar\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecbad3a6cea06703dc4f5335559b7ce5361c0cf4 | 715,691 | ipynb | Jupyter Notebook | dmu1/dmu1_ml_Lockman-SWIRE/1.6_SWIRE.ipynb | djbsmith/dmu_products | 4a6e1496a759782057c87ab5a65763282f61c497 | [
"MIT"
] | null | null | null | dmu1/dmu1_ml_Lockman-SWIRE/1.6_SWIRE.ipynb | djbsmith/dmu_products | 4a6e1496a759782057c87ab5a65763282f61c497 | [
"MIT"
] | null | null | null | dmu1/dmu1_ml_Lockman-SWIRE/1.6_SWIRE.ipynb | djbsmith/dmu_products | 4a6e1496a759782057c87ab5a65763282f61c497 | [
"MIT"
] | null | null | null | 1,463.580777 | 215,546 | 0.945284 | [
[
[
"# Lockman SWIRE master catalogue\n## Preparation of Spitzer datafusion SWIRE data\n\nThe Spitzer catalogues were produced by the datafusion team are available in `dmu0_DataFusion-Spitzer`.\nLucia told that the magnitudes are aperture corrected.\n\nIn the catalouge, we keep:\n\nWe keep:\n- The internal identifier (this one is only in HeDaM data);\n- The position;\n- The fluxes in aperture 2 (1.9 arcsec) for IRAC bands.\n- The Kron flux;\n- The stellarity in each band\n\nA query of the position in the Spitzer heritage archive show that the ELAIS-N1 images were observed in 2004. Let's take this as epoch.\n\nWe do not use the MIPS fluxes as they will be extracted on MIPS maps using XID+.",
"_____no_output_____"
]
],
[
[
"from herschelhelp_internal import git_version\nprint(\"This notebook was run with herschelhelp_internal version: \\n{}\".format(git_version()))",
"This notebook was run with herschelhelp_internal version: \n44f1ae0 (Thu Nov 30 18:27:54 2017 +0000)\n"
],
[
"%matplotlib inline\n#%config InlineBackend.figure_format = 'svg'\n\nimport matplotlib.pyplot as plt\nplt.rc('figure', figsize=(10, 6))\n\nfrom collections import OrderedDict\nimport os\n\nfrom astropy import units as u\nfrom astropy.coordinates import SkyCoord\nfrom astropy.table import Column, Table\nimport numpy as np\n\nfrom herschelhelp_internal.flagging import gaia_flag_column\nfrom herschelhelp_internal.masterlist import nb_astcor_diag_plot, remove_duplicates\nfrom herschelhelp_internal.utils import astrometric_correction, flux_to_mag",
"_____no_output_____"
],
[
"OUT_DIR = os.environ.get('TMP_DIR', \"./data_tmp\")\ntry:\n os.makedirs(OUT_DIR)\nexcept FileExistsError:\n pass\n\nRA_COL = \"swire_ra\"\nDEC_COL = \"swire_dec\"",
"_____no_output_____"
]
],
[
[
"## I - Column selection",
"_____no_output_____"
]
],
[
[
"imported_columns = OrderedDict({\n 'internal_id': \"swire_intid\",\n 'ra_spitzer': \"swire_ra\",\n 'dec_spitzer': \"swire_dec\",\n 'flux_ap2_36': \"f_ap_swire_irac1\",\n 'uncf_ap2_36': \"ferr_ap_swire_irac1\",\n 'flux_kr_36': \"f_swire_irac1\",\n 'uncf_kr_36': \"ferr_swire_irac1\",\n 'stell_36': \"swire_stellarity_irac1\",\n 'flux_ap2_45': \"f_ap_swire_irac2\",\n 'uncf_ap2_45': \"ferr_ap_swire_irac2\",\n 'flux_kr_45': \"f_swire_irac2\",\n 'uncf_kr_45': \"ferr_swire_irac2\",\n 'stell_45': \"swire_stellarity_irac2\",\n 'flux_ap2_58': \"f_ap_irac_i3\",\n 'uncf_ap2_58': \"ferr_ap_irac_i3\",\n 'flux_kr_58': \"f_irac_i3\",\n 'uncf_kr_58': \"ferr_irac_i3\",\n 'stell_58': \"swire_stellarity_irac3\",\n 'flux_ap2_80': \"f_ap_irac_i4\",\n 'uncf_ap2_80': \"ferr_ap_irac_i4\",\n 'flux_kr_80': \"f_irac_i4\",\n 'uncf_kr_80': \"ferr_irac_i4\",\n 'stell_80': \"swire_stellarity_irac4\",\n })\n\n\ncatalogue = Table.read(\"../../dmu0/dmu0_DataFusion-Spitzer/data/DF-SWIRE_Lockman-SWIRE.fits\")[list(imported_columns)]\nfor column in imported_columns:\n catalogue[column].name = imported_columns[column]\n\nepoch = 2004\n\n# Clean table metadata\ncatalogue.meta = None",
"_____no_output_____"
],
[
"# Adding magnitude and band-flag columns\nfor col in catalogue.colnames:\n if col.startswith('f_'):\n errcol = \"ferr{}\".format(col[1:])\n \n magnitude, error = flux_to_mag(\n np.array(catalogue[col])/1.e6, np.array(catalogue[errcol])/1.e6)\n # Note that some fluxes are 0.\n \n catalogue.add_column(Column(magnitude, name=\"m{}\".format(col[1:])))\n catalogue.add_column(Column(error, name=\"m{}\".format(errcol[1:])))\n \n # Band-flag column\n if \"ap\" not in col:\n catalogue.add_column(Column(np.zeros(len(catalogue), dtype=bool), name=\"flag{}\".format(col[1:])))\n",
"/opt/herschelhelp_internal/herschelhelp_internal/utils.py:76: RuntimeWarning: invalid value encountered in log10\n magnitudes = 2.5 * (23 - np.log10(fluxes)) - 48.6\n"
],
[
"catalogue[:10].show_in_notebook()",
"_____no_output_____"
]
],
[
[
"## II - Removal of duplicated sources",
"_____no_output_____"
],
[
"We remove duplicated objects from the input catalogues.",
"_____no_output_____"
]
],
[
[
"SORT_COLS = ['ferr_ap_swire_irac1', 'ferr_ap_swire_irac2', 'ferr_ap_irac_i3', 'ferr_ap_irac_i4']\nFLAG_NAME = \"swire_flag_cleaned\"\n\nnb_orig_sources = len(catalogue)\n\ncatalogue = remove_duplicates(catalogue, RA_COL, DEC_COL, sort_col=SORT_COLS,flag_name=FLAG_NAME)\n\nnb_sources = len(catalogue)\n\nprint(\"The initial catalogue had {} sources.\".format(nb_orig_sources))\nprint(\"The cleaned catalogue has {} sources ({} removed).\".format(nb_sources, nb_orig_sources - nb_sources))\nprint(\"The cleaned catalogue has {} sources flagged as having been cleaned\".format(np.sum(catalogue[FLAG_NAME])))",
"/opt/anaconda3/envs/herschelhelp_internal/lib/python3.6/site-packages/astropy/table/column.py:1096: MaskedArrayFutureWarning: setting an item on a masked array which has a shared mask will not copy the mask and also change the original mask array in the future.\nCheck the NumPy 1.11 release notes for more information.\n ma.MaskedArray.__setitem__(self, index, value)\n"
]
],
[
[
"## III - Astrometry correction\n\nWe match the astrometry to the Gaia one. We limit the Gaia catalogue to sources with a g band flux between the 30th and the 70th percentile. Some quick tests show that this give the lower dispersion in the results.",
"_____no_output_____"
]
],
[
[
"gaia = Table.read(\"../../dmu0/dmu0_GAIA/data/GAIA_Lockman-SWIRE.fits\")\ngaia_coords = SkyCoord(gaia['ra'], gaia['dec'])",
"_____no_output_____"
],
[
"nb_astcor_diag_plot(catalogue[RA_COL], catalogue[DEC_COL], \n gaia_coords.ra, gaia_coords.dec)",
"_____no_output_____"
],
[
"delta_ra, delta_dec = astrometric_correction(\n SkyCoord(catalogue[RA_COL], catalogue[DEC_COL]),\n gaia_coords\n)\n\nprint(\"RA correction: {}\".format(delta_ra))\nprint(\"Dec correction: {}\".format(delta_dec))",
"RA correction: -0.04179821406751216 arcsec\nDec correction: -0.11961620856055788 arcsec\n"
],
[
"catalogue[RA_COL] += delta_ra.to(u.deg)\ncatalogue[DEC_COL] += delta_dec.to(u.deg)",
"_____no_output_____"
],
[
"nb_astcor_diag_plot(catalogue[RA_COL], catalogue[DEC_COL], \n gaia_coords.ra, gaia_coords.dec)",
"_____no_output_____"
]
],
[
[
"## IV - Flagging Gaia objects",
"_____no_output_____"
]
],
[
[
"catalogue.add_column(\n gaia_flag_column(SkyCoord(catalogue[RA_COL], catalogue[DEC_COL]), epoch, gaia)\n)",
"_____no_output_____"
],
[
"GAIA_FLAG_NAME = \"swire_flag_gaia\"\n\ncatalogue['flag_gaia'].name = GAIA_FLAG_NAME\nprint(\"{} sources flagged.\".format(np.sum(catalogue[GAIA_FLAG_NAME] > 0)))",
"29668 sources flagged.\n"
]
],
[
[
"## V - Flagging objects near bright stars",
"_____no_output_____"
],
[
"# VI - Saving to disk",
"_____no_output_____"
]
],
[
[
"catalogue.write(\"{}/SWIRE.fits\".format(OUT_DIR), overwrite=True)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
ecbad693f57c6db1e906864aa6ffe0388c11c894 | 34,237 | ipynb | Jupyter Notebook | dda_analysis/test.ipynb | KopelmanLab/au_nanosnake_dda | a40db4fca076df86f1862bc69e67c6241f90f743 | [
"MIT"
] | null | null | null | dda_analysis/test.ipynb | KopelmanLab/au_nanosnake_dda | a40db4fca076df86f1862bc69e67c6241f90f743 | [
"MIT"
] | null | null | null | dda_analysis/test.ipynb | KopelmanLab/au_nanosnake_dda | a40db4fca076df86f1862bc69e67c6241f90f743 | [
"MIT"
] | null | null | null | 70.157787 | 20,644 | 0.703391 | [
[
[
"import pandas as pd\nimport numpy as np\nimport re\n\nwith open('Au_evap') as fp:\n for blank in range(0,3):\n fp.readline()\n wave = []\n Re = []\n Im = []\n for line in fp:\n inter = line.rstrip().split('\\t')\n inter = list(chain(*[text.split(' ') for text in inter]))\n ary = [line for line in inter if line]\n \n ary = np.array(ary[0:3]).astype(np.float)\n wave.append(float(ary[0]))\n Re.append(float(ary[1]))\n Im.append(float(ary[2]))\n\n\n df = pd.DataFrame({'wave': wave, 'Re': Re, 'Im': Im})\n\ndf",
"_____no_output_____"
],
[
"from matplotlib import pyplot as plt\n%matplotlib inline\n\nplt.plot(df['wave'], df['Re'], label=\"Real\")\nplt.plot(df['wave'], df['Im'], label=\"Imaginary\")\nplt.plot(df['wave'], np.sqrt(df['Re']*df['Re'] + df['Im']*df['Im']), label=\"Magnitude\")\n\nplt.legend()\nplt.title('Polarizability vs. Wavelength')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
ecbadb6f2ef4122ccbca9e547fce8aa1bdfc77e9 | 1,920 | ipynb | Jupyter Notebook | gQuant/plugins/simple_example/notebooks/plugin_example.ipynb | t-triobox/gQuant | 6ee3ba104ce4c6f17a5755e7782298902d125563 | [
"Apache-2.0"
] | null | null | null | gQuant/plugins/simple_example/notebooks/plugin_example.ipynb | t-triobox/gQuant | 6ee3ba104ce4c6f17a5755e7782298902d125563 | [
"Apache-2.0"
] | null | null | null | gQuant/plugins/simple_example/notebooks/plugin_example.ipynb | t-triobox/gQuant | 6ee3ba104ce4c6f17a5755e7782298902d125563 | [
"Apache-2.0"
] | null | null | null | 21.098901 | 120 | 0.526042 | [
[
[
"import json\nfrom greenflow.dataframe_flow import TaskGraph\ntaskGraph=TaskGraph.load_taskgraph('./simple_plugin.gq.yaml')\ntaskGraph.draw()",
"_____no_output_____"
],
[
"taskGraph.run(formated=True)",
"\n"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
ecbb01d6b55570f37894d15df995fa7de849294b | 8,986 | ipynb | Jupyter Notebook | assignment2/.ipynb_checkpoints/Dropout-checkpoint.ipynb | zhongnanf/cs231n_2016_hws | d7943535e1d3c96ea6fd38badc28701852b9b056 | [
"MIT"
] | null | null | null | assignment2/.ipynb_checkpoints/Dropout-checkpoint.ipynb | zhongnanf/cs231n_2016_hws | d7943535e1d3c96ea6fd38badc28701852b9b056 | [
"MIT"
] | null | null | null | assignment2/.ipynb_checkpoints/Dropout-checkpoint.ipynb | zhongnanf/cs231n_2016_hws | d7943535e1d3c96ea6fd38badc28701852b9b056 | [
"MIT"
] | null | null | null | 31.41958 | 352 | 0.571222 | [
[
[
"# Dropout\nDropout [1] is a technique for regularizing neural networks by randomly setting some features to zero during the forward pass. In this exercise you will implement a dropout layer and modify your fully-connected network to optionally use dropout.\n\n[1] Geoffrey E. Hinton et al, \"Improving neural networks by preventing co-adaptation of feature detectors\", arXiv 2012",
"_____no_output_____"
]
],
[
[
"# As usual, a bit of setup\n\nimport time\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom cs231n.classifiers.fc_net import *\nfrom cs231n.data_utils import get_CIFAR10_data\nfrom cs231n.gradient_check import eval_numerical_gradient, eval_numerical_gradient_array\nfrom cs231n.solver import Solver\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n# for auto-reloading external modules\n# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython\n%load_ext autoreload\n%autoreload 2\n\ndef rel_error(x, y):\n \"\"\" returns relative error \"\"\"\n return np.max(np.abs(x - y) / (np.maximum(1e-8, np.abs(x) + np.abs(y))))",
"run the following from the cs231n directory and try again:\npython setup.py build_ext --inplace\nYou may also need to restart your iPython kernel\n"
],
[
"# Load the (preprocessed) CIFAR10 data.\n\ndata = get_CIFAR10_data()\nfor k, v in data.iteritems():\n print '%s: ' % k, v.shape",
"_____no_output_____"
]
],
[
[
"# Dropout forward pass\nIn the file `cs231n/layers.py`, implement the forward pass for dropout. Since dropout behaves differently during training and testing, make sure to implement the operation for both modes.\n\nOnce you have done so, run the cell below to test your implementation.",
"_____no_output_____"
]
],
[
[
"x = np.random.randn(500, 500) + 10\n\nfor p in [0.3, 0.6, 0.75]:\n out, _ = dropout_forward(x, {'mode': 'train', 'p': p})\n out_test, _ = dropout_forward(x, {'mode': 'test', 'p': p})\n\n print 'Running tests with p = ', p\n print 'Mean of input: ', x.mean()\n print 'Mean of train-time output: ', out.mean()\n print 'Mean of test-time output: ', out_test.mean()\n print 'Fraction of train-time output set to zero: ', (out == 0).mean()\n print 'Fraction of test-time output set to zero: ', (out_test == 0).mean()\n print",
"_____no_output_____"
]
],
[
[
"# Dropout backward pass\nIn the file `cs231n/layers.py`, implement the backward pass for dropout. After doing so, run the following cell to numerically gradient-check your implementation.",
"_____no_output_____"
]
],
[
[
"x = np.random.randn(10, 10) + 10\ndout = np.random.randn(*x.shape)\n\ndropout_param = {'mode': 'train', 'p': 0.8, 'seed': 123}\nout, cache = dropout_forward(x, dropout_param)\ndx = dropout_backward(dout, cache)\ndx_num = eval_numerical_gradient_array(lambda xx: dropout_forward(xx, dropout_param)[0], x, dout)\n\nprint 'dx relative error: ', rel_error(dx, dx_num)",
"_____no_output_____"
]
],
[
[
"# Fully-connected nets with Dropout\nIn the file `cs231n/classifiers/fc_net.py`, modify your implementation to use dropout. Specificially, if the constructor the the net receives a nonzero value for the `dropout` parameter, then the net should add dropout immediately after every ReLU nonlinearity. After doing so, run the following to numerically gradient-check your implementation.",
"_____no_output_____"
]
],
[
[
"N, D, H1, H2, C = 2, 15, 20, 30, 10\nX = np.random.randn(N, D)\ny = np.random.randint(C, size=(N,))\n\nfor dropout in [0, 0.25, 0.5]:\n print 'Running check with dropout = ', dropout\n model = FullyConnectedNet([H1, H2], input_dim=D, num_classes=C,\n weight_scale=5e-2, dtype=np.float64,\n dropout=dropout, seed=123)\n\n loss, grads = model.loss(X, y)\n print 'Initial loss: ', loss\n\n for name in sorted(grads):\n f = lambda _: model.loss(X, y)[0]\n grad_num = eval_numerical_gradient(f, model.params[name], verbose=False, h=1e-5)\n print '%s relative error: %.2e' % (name, rel_error(grad_num, grads[name]))\n print",
"_____no_output_____"
]
],
[
[
"# Regularization experiment\nAs an experiment, we will train a pair of two-layer networks on 500 training examples: one will use no dropout, and one will use a dropout probability of 0.75. We will then visualize the training and validation accuracies of the two networks over time.",
"_____no_output_____"
]
],
[
[
"# Train two identical nets, one with dropout and one without\n\nnum_train = 500\nsmall_data = {\n 'X_train': data['X_train'][:num_train],\n 'y_train': data['y_train'][:num_train],\n 'X_val': data['X_val'],\n 'y_val': data['y_val'],\n}\n\nsolvers = {}\ndropout_choices = [0, 0.75]\nfor dropout in dropout_choices:\n model = FullyConnectedNet([500], dropout=dropout)\n print dropout\n\n solver = Solver(model, small_data,\n num_epochs=25, batch_size=100,\n update_rule='adam',\n optim_config={\n 'learning_rate': 5e-4,\n },\n verbose=True, print_every=100)\n solver.train()\n solvers[dropout] = solver",
"_____no_output_____"
],
[
"# Plot train and validation accuracies of the two models\n\ntrain_accs = []\nval_accs = []\nfor dropout in dropout_choices:\n solver = solvers[dropout]\n train_accs.append(solver.train_acc_history[-1])\n val_accs.append(solver.val_acc_history[-1])\n\nplt.subplot(3, 1, 1)\nfor dropout in dropout_choices:\n plt.plot(solvers[dropout].train_acc_history, 'o', label='%.2f dropout' % dropout)\nplt.title('Train accuracy')\nplt.xlabel('Epoch')\nplt.ylabel('Accuracy')\nplt.legend(ncol=2, loc='lower right')\n \nplt.subplot(3, 1, 2)\nfor dropout in dropout_choices:\n plt.plot(solvers[dropout].val_acc_history, 'o', label='%.2f dropout' % dropout)\nplt.title('Val accuracy')\nplt.xlabel('Epoch')\nplt.ylabel('Accuracy')\nplt.legend(ncol=2, loc='lower right')\n\nplt.gcf().set_size_inches(15, 15)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Question\nExplain what you see in this experiment. What does it suggest about dropout?",
"_____no_output_____"
],
[
"# Answer\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ecbb056e7160fd2f5dce52806912ea0def337089 | 45,106 | ipynb | Jupyter Notebook | examples/tutorials/04_Introduction_to_Graph_Convolutions.ipynb | micimize/deepchem | 651df9f9d8e6f1b33d3af8f0be251e9e8095fe88 | [
"MIT"
] | null | null | null | examples/tutorials/04_Introduction_to_Graph_Convolutions.ipynb | micimize/deepchem | 651df9f9d8e6f1b33d3af8f0be251e9e8095fe88 | [
"MIT"
] | null | null | null | examples/tutorials/04_Introduction_to_Graph_Convolutions.ipynb | micimize/deepchem | 651df9f9d8e6f1b33d3af8f0be251e9e8095fe88 | [
"MIT"
] | 1 | 2022-03-11T00:10:23.000Z | 2022-03-11T00:10:23.000Z | 63.799151 | 8,942 | 0.71618 | [
[
[
"# Tutorial Part 4: Introduction to Graph Convolutions\n\nIn the previous sections of the tutorial, we learned about `Dataset` and `Model` objects. We learned how to load some data into DeepChem from files on disk and also learned some basic facts about molecular data handling. We then dove into some basic deep learning architectures. However, until now, we stuck with vanilla deep learning architectures and didn't really consider how to handle deep architectures specifically engineered to work with life science data.\n\nIn this tutorial, we'll change that by going a little deeper and learn about \"graph convolutions.\" These are one of the most powerful deep learning tools for working with molecular data. The reason for this is that molecules can be naturally viewed as graphs.\n\n\n\nNote how standard chemical diagrams of the sort we're used to from high school lend themselves naturally to visualizing molecules as graphs. In the remainder of this tutorial, we'll dig into this relationship in significantly more detail. This will let us get an in-the guts understanding of how these systems work.\n\n## Colab\n\nThis tutorial and the rest in this sequence are designed to be done in Google colab. If you'd like to open this notebook in colab, you can use the following link.\n\n[](https://colab.research.google.com/github/deepchem/deepchem/blob/master/examples/tutorials/04_Introduction_to_Graph_Convolutions.ipynb)\n\n## Setup\n\nTo run DeepChem within Colab, you'll need to run the following cell of installation commands. This will take about 5 minutes to run to completion and install your environment.",
"_____no_output_____"
]
],
[
[
"!curl -Lo conda_installer.py https://raw.githubusercontent.com/deepchem/deepchem/master/scripts/colab_install.py\nimport conda_installer\nconda_installer.install()\n!/root/miniconda/bin/conda info -e",
" % Total % Received % Xferd Average Speed Time Time Time Current\n Dload Upload Total Spent Left Speed\n100 3490 100 3490 0 0 18177 0 --:--:-- --:--:-- --:--:-- 18082\n"
],
[
"!pip install --pre deepchem\nimport deepchem\ndeepchem.__version__",
"Requirement already satisfied: deepchem in /usr/local/lib/python3.6/dist-packages (2.4.0rc1.dev20200908171924)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from deepchem) (1.18.5)\nRequirement already satisfied: pandas in /usr/local/lib/python3.6/dist-packages (from deepchem) (1.0.5)\nRequirement already satisfied: joblib in /usr/local/lib/python3.6/dist-packages (from deepchem) (0.16.0)\nRequirement already satisfied: scikit-learn in /usr/local/lib/python3.6/dist-packages (from deepchem) (0.22.2.post1)\nRequirement already satisfied: scipy in /usr/local/lib/python3.6/dist-packages (from deepchem) (1.4.1)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.6/dist-packages (from pandas->deepchem) (2018.9)\nRequirement already satisfied: python-dateutil>=2.6.1 in /usr/local/lib/python3.6/dist-packages (from pandas->deepchem) (2.8.1)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.6/dist-packages (from python-dateutil>=2.6.1->pandas->deepchem) (1.15.0)\n"
]
],
[
[
"Ok now that we have our environment installed, we can actually import the core `GraphConvModel` that we'll use through this tutorial.",
"_____no_output_____"
]
],
[
[
"import deepchem as dc\nfrom deepchem.models.graph_models import GraphConvModel",
"_____no_output_____"
]
],
[
[
"Now, let's use the MoleculeNet suite to load the Tox21 dataset. We need to make sure to process the data in a way that graph convolutional networks can use for that, we make sure to set the featurizer option to 'GraphConv'. The MoleculeNet call will return a training set, a validation set, and a test set for us to use. The call also returns `transformers`, a list of data transformations that were applied to preprocess the dataset. (Most deep networks are quite finicky and require a set of data transformations to ensure that training proceeds stably.)",
"_____no_output_____"
]
],
[
[
"# Load Tox21 dataset\ntox21_tasks, tox21_datasets, transformers = dc.molnet.load_tox21(featurizer='GraphConv', reload=False)\ntrain_dataset, valid_dataset, test_dataset = tox21_datasets",
"smiles_field is deprecated and will be removed in a future version of DeepChem.Use feature_field instead.\n/usr/local/lib/python3.6/dist-packages/deepchem/data/data_loader.py:162: FutureWarning: featurize() is deprecated and has been renamed to create_dataset().featurize() will be removed in DeepChem 3.0\n \"featurize() will be removed in DeepChem 3.0\", FutureWarning)\n"
]
],
[
[
"Let's now train a graph convolutional network on this dataset. DeepChem has the class `GraphConvModel` that wraps a standard graph convolutional architecture underneath the hood for user convenience. Let's instantiate an object of this class and train it on our dataset.",
"_____no_output_____"
]
],
[
[
"n_tasks = len(tox21_tasks)\nmodel = GraphConvModel(n_tasks, batch_size=50, mode='classification')\n\nnum_epochs = 10\nlosses = []\nfor i in range(num_epochs):\n loss = model.fit(train_dataset, nb_epoch=1)\n print(\"Epoch %d loss: %f\" % (i, loss))\n losses.append(loss)",
"/usr/local/lib/python3.6/dist-packages/tensorflow/python/framework/indexed_slices.py:432: UserWarning: Converting sparse IndexedSlices to a dense Tensor of unknown shape. This may consume a large amount of memory.\n \"Converting sparse IndexedSlices to a dense Tensor of unknown shape. \"\n"
]
],
[
[
"Let's plot these losses so we can take a look at how the loss changes over the process of training.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plot\n\nplot.ylabel(\"Loss\")\nplot.xlabel(\"Epoch\")\nx = range(num_epochs)\ny = losses\nplot.scatter(x, y)\nplot.show()",
"_____no_output_____"
]
],
[
[
"We see that the losses fall nicely and give us stable learning.\n\nLet's try to evaluate the performance of the model we've trained. For this, we need to define a metric, a measure of model performance. `dc.metrics` holds a collection of metrics already. For this dataset, it is standard to use the ROC-AUC score, the area under the receiver operating characteristic curve (which measures the tradeoff between precision and recall). Luckily, the ROC-AUC score is already available in DeepChem. \n\nTo measure the performance of the model under this metric, we can use the convenience function `model.evaluate()`.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nmetric = dc.metrics.Metric(dc.metrics.roc_auc_score, np.mean)\n\nprint(\"Evaluating model\")\ntrain_scores = model.evaluate(train_dataset, [metric], transformers)\nprint(\"Training ROC-AUC Score: %f\" % train_scores[\"mean-roc_auc_score\"])\nvalid_scores = model.evaluate(valid_dataset, [metric], transformers)\nprint(\"Validation ROC-AUC Score: %f\" % valid_scores[\"mean-roc_auc_score\"])",
"Evaluating model\nTraining ROC-AUC Score: 0.887089\nValidation ROC-AUC Score: 0.778292\n"
]
],
[
[
"What's going on under the hood? Could we build GraphConvModel ourselves? Of course! Let's first understand the inputs to the model and generate the relevant data.\n\nConceptually, graph convolutions just require the structure of the molecule in question and a vector of features for every atom that describes the local chemical environment.\n\n`atom_features` holds a feature vector of length 75 for each atom. The other inputs are required to support minibatching in TensorFlow. `degree_slice` is an indexing convenience that makes it easy to locate atoms from all molecules with a given degree. `membership` determines the membership of atoms in molecules (atom `i` belongs to molecule membership[i]). `deg_adjs` is a list that contains adjacency lists grouped by atom degree. For more details, check out the [code](https://github.com/deepchem/deepchem/blob/master/deepchem/feat/mol_graphs.py).\n\nFollowing code creates a Python generator that given a batch of data generates the lists of inputs, labels, and weights whose values are Numpy arrays. We will use for this step of training.",
"_____no_output_____"
]
],
[
[
"from deepchem.metrics import to_one_hot\nfrom deepchem.feat.mol_graphs import ConvMol\n\ndef data_generator(dataset, predict=False, pad_batches=True):\n for ind, (X_b, y_b, w_b, ids_b) in enumerate(\n dataset.iterbatches(\n batch_size, pad_batches=pad_batches, deterministic=True)):\n multiConvMol = ConvMol.agglomerate_mols(X_b)\n inputs = [multiConvMol.get_atom_features(), multiConvMol.deg_slice, np.array(multiConvMol.membership)]\n for i in range(1, len(multiConvMol.get_deg_adjacency_lists())):\n inputs.append(multiConvMol.get_deg_adjacency_lists()[i])\n labels = [to_one_hot(y_b.flatten(), 2).reshape(-1, n_tasks, 2)]\n weights = [w_b]\n yield (inputs, labels, weights)",
"_____no_output_____"
]
],
[
[
"Now let's create the `Keras model` and [keras layers](https://keras.io/api/layers/) of the model.\n\nDeepChem already provides wrapper around keras layers to build graph convolutional model. We are going to apply following layers from DeepChem.\n\n- `GraphConv` layer: This layer implements the graph convolution. The graph convolution combines per-node feature vectures in a nonlinear fashion with the feature vectors for neighboring nodes. This \"blends\" information in local neighborhoods of a graph.\n\n- `GraphPool` layer: This layer does a max-pooling over the feature vectors of atoms in a neighborhood. You can think of this layer as analogous to a max-pooling layer for 2D convolutions but which operates on graphs instead. \n\n- `GraphGather`: Many graph convolutional networks manipulate feature vectors per graph-node. For a molecule for example, each node might represent an atom, and the network would manipulate atomic feature vectors that summarize the local chemistry of the atom. However, at the end of the application, we will likely want to work with a molecule level feature representation. This layer creates a graph level feature vector by combining all the node-level feature vectors.\n\nApart from this we are going to apply standard neural network layers such as [Dense](https://keras.io/api/layers/core_layers/dense/), [BatchNormalization](https://keras.io/api/layers/normalization_layers/batch_normalization/) and [Softmax](https://keras.io/api/layers/activation_layers/softmax/) layer.",
"_____no_output_____"
]
],
[
[
"from deepchem.models.layers import GraphConv, GraphPool, GraphGather\nimport tensorflow as tf\nimport tensorflow.keras.layers as layers\n\nbatch_size = 50\n\nclass MyKerasModel(tf.keras.Model):\n\n def __init__(self):\n super(MyKerasModel, self).__init__()\n self.gc1 = GraphConv(128, activation_fn=tf.nn.tanh)\n self.batch_norm1 = layers.BatchNormalization()\n self.gp1 = GraphPool()\n\n self.gc2 = GraphConv(128, activation_fn=tf.nn.tanh)\n self.batch_norm2 = layers.BatchNormalization()\n self.gp2 = GraphPool()\n\n self.dense1 = layers.Dense(256, activation=tf.nn.tanh)\n self.batch_norm3 = layers.BatchNormalization()\n self.readout = GraphGather(batch_size=batch_size, activation_fn=tf.nn.tanh)\n\n self.dense2 = layers.Dense(n_tasks*2)\n self.logits = layers.Reshape((n_tasks, 2))\n self.softmax = layers.Softmax()\n\n def call(self, inputs):\n gc1_output = self.gc1(inputs)\n batch_norm1_output = self.batch_norm1(gc1_output)\n gp1_output = self.gp1([batch_norm1_output] + inputs[1:])\n\n gc2_output = self.gc2([gp1_output] + inputs[1:])\n batch_norm2_output = self.batch_norm1(gc2_output)\n gp2_output = self.gp2([batch_norm2_output] + inputs[1:])\n\n dense1_output = self.dense1(gp2_output)\n batch_norm3_output = self.batch_norm3(dense1_output)\n readout_output = self.readout([batch_norm3_output] + inputs[1:])\n\n logits_output = self.logits(self.dense2(readout_output))\n return self.softmax(logits_output)",
"_____no_output_____"
]
],
[
[
"Let's now create the DeepChem model which will be a wrapper around the keras model that we just created. \n\nDeepChem models provide useful utilities on top of the keras model. We will also specify the loss function so the model know the objective to minimize.",
"_____no_output_____"
]
],
[
[
"loss = dc.models.losses.CategoricalCrossEntropy()\nmodel = dc.models.KerasModel(MyKerasModel(), loss=loss)",
"_____no_output_____"
]
],
[
[
"Now, we can train the model using `fit_generator(generator)` which will use the generator we've defined to train the model.",
"_____no_output_____"
]
],
[
[
"num_epochs = 10\nlosses = []\nfor i in range(num_epochs):\n loss = model.fit_generator(data_generator(train_dataset))\n print(\"Epoch %d loss: %f\" % (i, loss))\n losses.append(loss)",
"/usr/local/lib/python3.6/dist-packages/tensorflow/python/framework/indexed_slices.py:432: UserWarning: Converting sparse IndexedSlices to a dense Tensor of unknown shape. This may consume a large amount of memory.\n \"Converting sparse IndexedSlices to a dense Tensor of unknown shape. \"\n"
]
],
[
[
"Let's now plot these losses and take a quick look.",
"_____no_output_____"
]
],
[
[
"plot.title(\"Keras Version\")\nplot.ylabel(\"Loss\")\nplot.xlabel(\"Epoch\")\nx = range(num_epochs)\ny = losses\nplot.scatter(x, y)\nplot.show()",
"_____no_output_____"
]
],
[
[
"Now that we have trained our graph convolutional method, let's evaluate its performance. We again have to use our defined generator to evaluate model performance.",
"_____no_output_____"
]
],
[
[
"metric = dc.metrics.Metric(dc.metrics.roc_auc_score, np.mean)\n\ndef reshape_y_pred(y_true, y_pred):\n \"\"\"\n GraphConv always pads batches, so we need to remove the predictions\n for the padding samples. Also, it outputs two values for each task\n (probabilities of positive and negative), but we only want the positive\n probability.\n \"\"\"\n n_samples = len(y_true)\n return y_pred[:n_samples, :, 1]\n \n\nprint(\"Evaluating model\")\ntrain_predictions = model.predict_on_generator(data_generator(train_dataset, predict=True))\ntrain_predictions = reshape_y_pred(train_dataset.y, train_predictions)\ntrain_scores = metric.compute_metric(train_dataset.y, train_predictions, train_dataset.w)\nprint(\"Training ROC-AUC Score: %f\" % train_scores)\n\nvalid_predictions = model.predict_on_generator(data_generator(valid_dataset, predict=True))\nvalid_predictions = reshape_y_pred(valid_dataset.y, valid_predictions)\nvalid_scores = metric.compute_metric(valid_dataset.y, valid_predictions, valid_dataset.w)\nprint(\"Valid ROC-AUC Score: %f\" % valid_scores)",
"Evaluating model\nTraining ROC-AUC Score: 0.776245\nValid ROC-AUC Score: 0.702370\n"
]
],
[
[
"Success! The model we've constructed behaves nearly identically to `GraphConvModel`. If you're looking to build your own custom models, you can follow the example we've provided here to do so. We hope to see exciting constructions from your end soon!",
"_____no_output_____"
],
[
"# Congratulations! Time to join the Community!\n\nCongratulations on completing this tutorial notebook! If you enjoyed working through the tutorial, and want to continue working with DeepChem, we encourage you to finish the rest of the tutorials in this series. You can also help the DeepChem community in the following ways:\n\n## Star DeepChem on [GitHub](https://github.com/deepchem/deepchem)\nThis helps build awareness of the DeepChem project and the tools for open source drug discovery that we're trying to build.\n\n## Join the DeepChem Gitter\nThe DeepChem [Gitter](https://gitter.im/deepchem/Lobby) hosts a number of scientists, developers, and enthusiasts interested in deep learning for the life sciences. Join the conversation!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecbb149bd6121cdf5c129d5da12591ff70231ef3 | 134,076 | ipynb | Jupyter Notebook | notebooks/2. Bayesian Networks.ipynb | sean578/pgmpy_notebook | 102549516322f47bd955fa165c8ae1d6786bfa77 | [
"MIT"
] | 326 | 2015-03-12T15:14:17.000Z | 2022-03-24T15:02:44.000Z | notebooks/2. Bayesian Networks.ipynb | sean578/pgmpy_notebook | 102549516322f47bd955fa165c8ae1d6786bfa77 | [
"MIT"
] | 48 | 2015-03-06T09:42:17.000Z | 2022-03-18T13:22:40.000Z | notebooks/2. Bayesian Networks.ipynb | sean578/pgmpy_notebook | 102549516322f47bd955fa165c8ae1d6786bfa77 | [
"MIT"
] | 215 | 2015-02-11T13:28:22.000Z | 2022-01-17T08:58:17.000Z | 169.501896 | 54,584 | 0.869731 | [
[
[
"# Bayesian Network",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image",
"_____no_output_____"
]
],
[
[
"## Bayesian Models\n1. What are Bayesian Models\n2. Independencies in Bayesian Networks\n3. How is Bayesian Model encoding the Joint Distribution\n4. How we do inference from Bayesian models\n5. Types of methods for inference",
"_____no_output_____"
],
[
"### 1. What are Bayesian Models\nA Bayesian network, Bayes network, belief network, Bayes(ian) model or probabilistic directed acyclic graphical model is a probabilistic graphical model (a type of statistical model) that represents a set of random variables and their conditional dependencies via a directed acyclic graph (DAG). Bayesian networks are mostly used when we want to represent causal relationship between the random variables. Bayesian Networks are parameterized using Conditional Probability Distributions (CPD). Each node in the network is parameterized using $ P(node | Pa(node)) $ where $ Pa(node) $ represents the parents of node in the network.\n\nWe can take the example of the student model:",
"_____no_output_____"
]
],
[
[
"Image('../images/2/student_full_param.png')",
"_____no_output_____"
]
],
[
[
"In pgmpy we define the network structure and the CPDs separately and then associate them with the structure. Here's an example for defining the above model:",
"_____no_output_____"
]
],
[
[
"from pgmpy.models import BayesianModel\nfrom pgmpy.factors.discrete import TabularCPD\n\n# Defining the model structure. We can define the network by just passing a list of edges.\nmodel = BayesianModel([('D', 'G'), ('I', 'G'), ('G', 'L'), ('I', 'S')])\n\n# Defining individual CPDs.\ncpd_d = TabularCPD(variable='D', variable_card=2, values=[[0.6], [0.4]])\ncpd_i = TabularCPD(variable='I', variable_card=2, values=[[0.7], [0.3]])\n\n# The representation of CPD in pgmpy is a bit different than the CPD shown in the above picture. In pgmpy the colums\n# are the evidences and rows are the states of the variable. So the grade CPD is represented like this:\n#\n# +---------+---------+---------+---------+---------+\n# | diff | intel_0 | intel_0 | intel_1 | intel_1 |\n# +---------+---------+---------+---------+---------+\n# | intel | diff_0 | diff_1 | diff_0 | diff_1 |\n# +---------+---------+---------+---------+---------+\n# | grade_0 | 0.3 | 0.05 | 0.9 | 0.5 |\n# +---------+---------+---------+---------+---------+\n# | grade_1 | 0.4 | 0.25 | 0.08 | 0.3 |\n# +---------+---------+---------+---------+---------+\n# | grade_2 | 0.3 | 0.7 | 0.02 | 0.2 |\n# +---------+---------+---------+---------+---------+\n\ncpd_g = TabularCPD(variable='G', variable_card=3, \n values=[[0.3, 0.05, 0.9, 0.5],\n [0.4, 0.25, 0.08, 0.3],\n [0.3, 0.7, 0.02, 0.2]],\n evidence=['I', 'D'],\n evidence_card=[2, 2])\n\ncpd_l = TabularCPD(variable='L', variable_card=2, \n values=[[0.1, 0.4, 0.99],\n [0.9, 0.6, 0.01]],\n evidence=['G'],\n evidence_card=[3])\n\ncpd_s = TabularCPD(variable='S', variable_card=2,\n values=[[0.95, 0.2],\n [0.05, 0.8]],\n evidence=['I'],\n evidence_card=[2])\n\n# Associating the CPDs with the network\nmodel.add_cpds(cpd_d, cpd_i, cpd_g, cpd_l, cpd_s)\n\n# check_model checks for the network structure and CPDs and verifies that the CPDs are correctly \n# defined and sum to 1.\nmodel.check_model()",
"_____no_output_____"
],
[
"# CPDs can also be defined using the state names of the variables. If the state names are not provided\n# like in the previous example, pgmpy will automatically assign names as: 0, 1, 2, ....\n\ncpd_d_sn = TabularCPD(variable='D', variable_card=2, values=[[0.6], [0.4]], state_names={'D': ['Easy', 'Hard']})\ncpd_i_sn = TabularCPD(variable='I', variable_card=2, values=[[0.7], [0.3]], state_names={'I': ['Dumb', 'Intelligent']})\ncpd_g_sn = TabularCPD(variable='G', variable_card=3, \n values=[[0.3, 0.05, 0.9, 0.5],\n [0.4, 0.25, 0.08, 0.3],\n [0.3, 0.7, 0.02, 0.2]],\n evidence=['I', 'D'],\n evidence_card=[2, 2],\n state_names={'G': ['A', 'B', 'C'],\n 'I': ['Dumb', 'Intelligent'],\n 'D': ['Easy', 'Hard']})\n\ncpd_l_sn = TabularCPD(variable='L', variable_card=2, \n values=[[0.1, 0.4, 0.99],\n [0.9, 0.6, 0.01]],\n evidence=['G'],\n evidence_card=[3],\n state_names={'L': ['Bad', 'Good'],\n 'G': ['A', 'B', 'C']})\n\ncpd_s_sn = TabularCPD(variable='S', variable_card=2,\n values=[[0.95, 0.2],\n [0.05, 0.8]],\n evidence=['I'],\n evidence_card=[2],\n state_names={'S': ['Bad', 'Good'],\n 'I': ['Dumb', 'Intelligent']})\n\n# These defined CPDs can be added to the model. Since, the model already has CPDs associated to variables, it will\n# show warning that pmgpy is now replacing those CPDs with the new ones.\nmodel.add_cpds(cpd_d_sn, cpd_i_sn, cpd_g_sn, cpd_l_sn, cpd_s_sn)\nmodel.check_model()",
"WARNING:root:Replacing existing CPD for D\nWARNING:root:Replacing existing CPD for I\nWARNING:root:Replacing existing CPD for G\nWARNING:root:Replacing existing CPD for L\nWARNING:root:Replacing existing CPD for S\n"
],
[
"# We can now call some methods on the BayesianModel object.\nmodel.get_cpds()",
"_____no_output_____"
],
[
"# Printing a CPD which doesn't have state names defined.\nprint(cpd_g)",
"+------+------+------+------+------+\n| I | I(0) | I(0) | I(1) | I(1) |\n+------+------+------+------+------+\n| D | D(0) | D(1) | D(0) | D(1) |\n+------+------+------+------+------+\n| G(0) | 0.3 | 0.05 | 0.9 | 0.5 |\n+------+------+------+------+------+\n| G(1) | 0.4 | 0.25 | 0.08 | 0.3 |\n+------+------+------+------+------+\n| G(2) | 0.3 | 0.7 | 0.02 | 0.2 |\n+------+------+------+------+------+\n"
],
[
"# Printing a CPD with it's state names defined.\nprint(model.get_cpds('G'))",
"+------+---------+---------+----------------+----------------+\n| I | I(Dumb) | I(Dumb) | I(Intelligent) | I(Intelligent) |\n+------+---------+---------+----------------+----------------+\n| D | D(Easy) | D(Hard) | D(Easy) | D(Hard) |\n+------+---------+---------+----------------+----------------+\n| G(A) | 0.3 | 0.05 | 0.9 | 0.5 |\n+------+---------+---------+----------------+----------------+\n| G(B) | 0.4 | 0.25 | 0.08 | 0.3 |\n+------+---------+---------+----------------+----------------+\n| G(C) | 0.3 | 0.7 | 0.02 | 0.2 |\n+------+---------+---------+----------------+----------------+\n"
],
[
"model.get_cardinality('G')",
"_____no_output_____"
]
],
[
[
"### 2. Independencies in Bayesian Networks\n\nIndependencies implied by the network structure of a Bayesian Network can be categorized in 2 types:\n1. __Local Independencies:__ Any variable in the network is independent of its non-descendents given its parents. Mathematically it can be written as: $$ (X \\perp NonDesc(X) | Pa(X) $$\nwhere $ NonDesc(X) $ is the set of variables which are not descendents of $ X $ and $ Pa(X) $ is the set of variables which are parents of $ X $.\n\n2. __Global Independencies:__ For discussing global independencies in Bayesian Networks we need to look at the various network structures possible. \nStarting with the case of 2 nodes, there are only 2 possible ways for it to be connected:",
"_____no_output_____"
]
],
[
[
"Image('../images/2/two_nodes.png')",
"_____no_output_____"
]
],
[
[
"In the above two cases it is fairly obvious that change in any of the node will affect the other. For the first case we can take the example of $ difficulty \\rightarrow grade $. If we increase the difficulty of the course the probability of getting a higher grade decreases. For the second case we can take the example of $ SAT \\leftarrow Intel $. Now if we increase the probability of getting a good score in SAT that would imply that the student is intelligent, hence increasing the probability of $ i_1 $. Therefore in both the cases shown above any change in the variables leads to change in the other variable.\n\nNow, there are four possible ways of connection between 3 nodes: ",
"_____no_output_____"
]
],
[
[
"Image('../images/2/three_nodes.png')",
"_____no_output_____"
]
],
[
[
"Now in the above cases we will see the flow of influence from $ A $ to $ C $ under various cases.\n\n1. __Causal:__ In the general case when we make any changes in the variable $ A $, it will have effect of variable $ B $ (as we discussed above) and this change in $ B $ will change the values in $ C $. One other possible case can be when $ B $ is observed i.e. we know the value of $ B $. So, in this case any change in $ A $ won't affect $ B $ since we already know the value. And hence there won't be any change in $ C $ as it depends only on $ B $. Mathematically we can say that: $ (A \\perp C | B) $.\n2. __Evidential:__ Similarly in this case also observing $ B $ renders $ C $ independent of $ A $. Otherwise when $ B $ is not observed the influence flows from $ A $ to $ C $. Hence $ (A \\perp C | B) $.\n3. __Common Evidence:__ This case is a bit different from the others. When $ B $ is not observed any change in $ A $ reflects some change in $ B $ but not in $ C $. Let's take the example of $ D \\rightarrow G \\leftarrow I $. In this case if we increase the difficulty of the course the probability of getting a higher grade reduces but this has no effect on the intelligence of the student. But when $ B $ is observed let's say that the student got a good grade. Now if we increase the difficulty of the course this will increase the probability of the student to be intelligent since we already know that he got a good grade. Hence in this case $ (A \\perp C) $ and $ ( A \\not\\perp C | B) $. This structure is also commonly known as V structure.\n4. __Common Cause:__ The influence flows from $ A $ to $ C $ when $ B $ is not observed. But when $ B $ is observed and change in $ A $ doesn't affect $ C $ since it's only dependent on $ B $. Hence here also $ ( A \\perp C | B) $. \n\nLet's not see a few examples for finding the independencies in a newtork using pgmpy:",
"_____no_output_____"
]
],
[
[
"# Getting the local independencies of a variable.\nmodel.local_independencies('G')",
"_____no_output_____"
],
[
"# Getting all the local independencies in the network.\nmodel.local_independencies(['D', 'I', 'S', 'G', 'L'])",
"_____no_output_____"
],
[
"# Active trail: For any two variables A and B in a network if any change in A influences the values of B then we say\n# that there is an active trail between A and B.\n# In pgmpy active_trail_nodes gives a set of nodes which are affected (i.e. correlated) by any \n# change in the node passed in the argument.\nmodel.active_trail_nodes('D')",
"_____no_output_____"
],
[
"model.active_trail_nodes('D', observed='G')",
"_____no_output_____"
]
],
[
[
"### 3. How is this Bayesian Network representing the Joint Distribution over the variables ?\nTill now we just have been considering that the Bayesian Network can represent the Joint Distribution without any proof. Now let's see how to compute the Joint Distribution from the Bayesian Network.\n\nFrom the chain rule of probabiliy we know that: \n$$ P(A, B) = P(A | B) * P(B) $$\n\nNow in this case: \n$$ P(D, I, G, L, S) = P(L| S, G, D, I) * P(S | G, D, I) * P(G | D, I) * P(D | I) * P(I) $$\n\nApplying the local independence conditions in the above equation we will get: \n$$ P(D, I, G, L, S) = P(L|G) * P(S|I) * P(G| D, I) * P(D) * P(I) $$\n\nFrom the above equation we can clearly see that the Joint Distribution over all the variables is just the product of all the CPDs in the network. Hence encoding the independencies in the Joint Distribution in a graph structure helped us in reducing the number of parameters that we need to store.",
"_____no_output_____"
],
[
"### 4. Inference in Bayesian Models\nTill now we discussed just about representing Bayesian Networks. Now let's see how we can do inference in a Bayesian Model and use it to predict values over new data points for machine learning tasks. In this section we will consider that we already have our model. We will talk about constructing the models from data in later parts of this tutorial.\n\nIn inference we try to answer probability queries over the network given some other variables. So, we might want to know the probable grade of an intelligent student in a difficult class given that he scored good in SAT. So for computing these values from a Joint Distribution we will have to reduce over the given variables that is $ I = 1 $, $ D = 1 $, $ S = 1 $ and then marginalize over the other variables that is $ L $ to get $ P(G | I=1, D=1, S=1) $.\nBut carrying on marginalize and reduce operation on the complete Joint Distribution is computationaly expensive since we need to iterate over the whole table for each operation and the table is exponential is size to the number of variables. But in Graphical Models we exploit the independencies to break these operations in smaller parts making it much faster.\n\nOne of the very basic methods of inference in Graphical Models is __Variable Elimination__.",
"_____no_output_____"
],
[
"#### Variable Elimination\nWe know that:\n\n$ P(D, I, G, L, S) = P(L|G) * P(S|I) * P(G|D, I) * P(D) * P(I) $\n\nNow let's say we just want to compute the probability of G. For that we will need to marginalize over all the other variables.\n\n$ P(G) = \\sum_{D, I, L, S} P(D, I, G, L, S) $\n$ P(G) = \\sum_{D, I, L, S} P(L|G) * P(S|I) * P(G|D, I) * P(D) * P(I) $\n$ P(G) = \\sum_D \\sum_I \\sum_L \\sum_S P(L|G) * P(S|I) * P(G|D, I) * P(D) * P(I) $\n\nNow since not all the conditional distributions depend on all the variables we can push the summations inside:\n\n$ P(G) = \\sum_D \\sum_I \\sum_L \\sum_S P(L|G) * P(S|I) * P(G|D, I) * P(D) * P(I) $ \n$ P(G) = \\sum_D P(D) \\sum_I P(G|D, I) * P(I) \\sum_S P(S|I) \\sum_L P(L|G) $\n\nSo, by pushing the summations inside we have saved a lot of computation because we have to now iterate over much smaller tables.\n\nLet's take an example for inference using Variable Elimination in pgmpy:",
"_____no_output_____"
]
],
[
[
"from pgmpy.inference import VariableElimination\ninfer = VariableElimination(model)\ng_dist = infer.query(['G'])\nprint(g_dist)",
"Finding Elimination Order: : 100%|██████████| 4/4 [00:00<00:00, 1210.13it/s]\nEliminating: I: 100%|██████████| 4/4 [00:00<00:00, 240.56it/s]"
]
],
[
[
"There can be cases in which we want to compute the conditional distribution let's say $ P(G | D=0, I=1) $. In such cases we need to modify our equations a bit:\n\n$ P(G | D=0, I=1) = \\sum_L \\sum_S P(L|G) * P(S| I=1) * P(G| D=0, I=1) * P(D=0) * P(I=1) $\n$ P(G | D=0, I=1) = P(D=0) * P(I=1) * P(G | D=0, I=1) * \\sum_L P(L | G) * \\sum_S P(S | I=1) $\n\nIn pgmpy we will just need to pass an extra argument in the case of conditional distributions:",
"_____no_output_____"
]
],
[
[
"print(infer.query(['G'], evidence={'D': 'Easy', 'I': 'Intelligent'}))",
"Finding Elimination Order: : 100%|██████████| 2/2 [00:00<00:00, 552.57it/s]\nEliminating: S: 100%|██████████| 2/2 [00:00<00:00, 326.68it/s]"
]
],
[
[
"#### Predicting values from new data points\nPredicting values from new data points is quite similar to computing the conditional probabilities. We need to query for the variable that we need to predict given all the other features. The only difference is that rather than getting the probabilitiy distribution we are interested in getting the most probable state of the variable.\n\nIn pgmpy this is known as MAP query. Here's an example:",
"_____no_output_____"
]
],
[
[
"infer.map_query(['G'])",
"Finding Elimination Order: : 100%|██████████| 4/4 [00:00<00:00, 1073.12it/s]\nEliminating: I: 100%|██████████| 4/4 [00:00<00:00, 273.20it/s]\n"
],
[
"infer.map_query(['G'], evidence={'D': 'Easy', 'I': 'Intelligent'})",
"Finding Elimination Order: : 100%|██████████| 2/2 [00:00<00:00, 417.30it/s]\nEliminating: S: 100%|██████████| 2/2 [00:00<00:00, 219.08it/s]\n"
],
[
"infer.map_query(['G'], evidence={'D': 'Easy', 'I': 'Intelligent', 'L': 'Good', 'S': 'Good'})",
"Finding Elimination Order: : : 0it [00:00, ?it/s]\n0it [00:00, ?it/s]\n"
]
],
[
[
"### 5. Other methods for Inference\nEven though exact inference algorithms like Variable Elimination optimize the inference task, it is still computationally quite expensive in the case of large models. For such cases we can use approximate algorithms like Message Passing Algorithms, Sampling Algorithms etc. We will talk about a few other exact and approximate algorithms in later parts of the tutorial.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecbb1ca71cdf93a8084977ddf1ef47e9056d7273 | 436,497 | ipynb | Jupyter Notebook | Neural_Networks/test 1 neuron logloss.ipynb | ValeriyaArt/MachineLearning_Pet_projects | c45cd8715059379994d73ec3c88ce7e4f9c565c8 | [
"Apache-2.0"
] | null | null | null | Neural_Networks/test 1 neuron logloss.ipynb | ValeriyaArt/MachineLearning_Pet_projects | c45cd8715059379994d73ec3c88ce7e4f9c565c8 | [
"Apache-2.0"
] | null | null | null | Neural_Networks/test 1 neuron logloss.ipynb | ValeriyaArt/MachineLearning_Pet_projects | c45cd8715059379994d73ec3c88ce7e4f9c565c8 | [
"Apache-2.0"
] | null | null | null | 436,497 | 436,497 | 0.942648 | [
[
[
"# Обучение нейрона с помощью функции потерь LogLoss",
"_____no_output_____"
],
[
"<h3 style=\"text-align: center;\"><b>Нейрон с сигмоидой</b></h3>",
"_____no_output_____"
],
[
"Снова рассмотрим нейрон с сигмоидой, то есть $$f(x) = \\sigma(x)=\\frac{1}{1+e^{-x}}$$ ",
"_____no_output_____"
],
[
"Ранее мы установили, что **обучение нейрона с сигмоидой с квадратичной функцией потерь**: \n\n$$MSE(w, x) = \\frac{1}{2n}\\sum_{i=1}^{n} (\\hat{y_i} - y_i)^2 = \\frac{1}{2n}\\sum_{i=1}^{n} (\\sigma(w \\cdot x_i) - y_i)^2$$ \n\nгде $w \\cdot x_i$ - скалярное произведение, а $\\sigma(w \\cdot x_i) =\\frac{1}{1+e^{-w \\cdot x_i}} $ - сигмоида -- **неэффективно**, то есть мы увидели, что даже за большое количество итераций нейрон предсказывает плохо.",
"_____no_output_____"
],
[
"Давайте ещё раз взглянем на формулу для градиентного спуска от функции потерь $MSE$ по весам нейрона:",
"_____no_output_____"
],
[
"$$ \\frac{\\partial MSE}{\\partial w} = \\frac{1}{n} X^T (\\sigma(w \\cdot X) - y)\\sigma(w \\cdot X)(1 - \\sigma(w \\cdot X))$$",
"_____no_output_____"
],
[
"А теперь смотрим на график сигмоиды:",
"_____no_output_____"
],
[
"<img src=\"https://cdn-images-1.medium.com/max/1200/1*IDAnCFoeXqWL7F4u9MJMtA.png\" width=500px height=350px>",
"_____no_output_____"
],
[
"**Её значения: числа от 0 до 1.**",
"_____no_output_____"
],
[
"Если получше проанализировать формулу, то теперь можно заметить, что, поскольку сигмоида принимает значения между 0 и 1 (а значит (1-$\\sigma$) тоже принимает значения от 0 до 1), то мы умножаем $X^T$ на столбец $(\\sigma(w \\cdot X) - y)$ из чисел от -1 до 1, а потом ещё на столбцы $\\sigma(w \\cdot X)$ и $(1 - \\sigma(w \\cdot X))$ из чисел от 0 до 1. Таким образом в лучшем случае $\\frac{\\partial{Loss}}{\\partial{w}}$ будет столбцом из чисел, порядок которых максимум 0.01 (в среднем, понятно, что если сигмоида выдаёт все 0, то будет 0, если все 1, то тоже 0). После этого мы умножаем на шаг градиентного спуска, который обычно порядка 0.001 или 0.1 максимум. То есть мы вычитаем из весов числа порядка ~0.0001. Медленновато спускаемся, не правда ли? Это называют **проблемой затухающих градиентов**.",
"_____no_output_____"
],
[
"Чтобы избежать эту проблему в задачах классификации, в которых моделью является нейрон с сигмоидной функцией активации, предсказывающий \"вероятности\" принадлженостей к классамиспользуют **LogLoss**: ",
"_____no_output_____"
],
[
"$$J(\\hat{y}, y) = -\\frac{1}{n} \\sum_{i=1}^n y_i \\log(\\hat{y_i}) + (1 - y_i) \\log(1 - \\hat{y_i}) = -\\frac{1}{n} \\sum_{i=1}^n y_i \\log(\\sigma(w \\cdot x_i)) + (1 - y_i) \\log(1 - \\sigma(w \\cdot x_i))$$",
"_____no_output_____"
],
[
"где, как и прежде, $y$ - столбец $(n, 1)$ из истинных значений классов, а $\\hat{y}$ - столбец $(n, 1)$ из предсказаний нейрона.",
"_____no_output_____"
]
],
[
[
"from matplotlib import pyplot as plt\nfrom matplotlib.colors import ListedColormap\nimport numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"def loss(y_pred, y):\n return -np.mean(y * np.log(y_pred) + (1 - y) * np.log(1 - y_pred))",
"_____no_output_____"
]
],
[
[
"Отметим, что сейчас речь идёт именно о **бинарной классификации (на два класса)**, в многоклассовой классификации используется функция потерь под названием *кросс-энтропия*, которая является обобщением LogLoss'а на случай нескольких классов.",
"_____no_output_____"
],
[
"Почему же теперь всё будет лучше? Раньше была проблема умножения маленьких чисел в градиенте. Давайте посмотрим, что теперь:",
"_____no_output_____"
],
[
"* Для веса $w_j$:",
"_____no_output_____"
],
[
"$$ \\frac{\\partial Loss}{\\partial w_j} = \n-\\frac{1}{n} \\sum_{i=1}^n \\left(\\frac{y_i}{\\sigma(w \\cdot x_i)} - \\frac{1 - y_i}{1 - \\sigma(w \\cdot x_i)}\\right)(\\sigma(w \\cdot x_i))_{w_j}' = -\\frac{1}{n} \\sum_{i=1}^n \\left(\\frac{y_i}{\\sigma(w \\cdot x_i)} - \\frac{1 - y_i}{1 - \\sigma(w \\cdot x_i)}\\right)\\sigma(w \\cdot x_i)(1 - \\sigma(w \\cdot x_i))x_{ij} = $$\n$$-\\frac{1}{n} \\sum_{i=1}^n \\left(y_i - \\sigma(w \\cdot x_i)\\right)x_{ij}$$",
"_____no_output_____"
],
[
"* Градиент $Loss$'а по вектору весов -- это вектор, $j$-ая компонента которого равна $\\frac{\\partial Loss}{\\partial w_j}$ (помним, что весов всего $m$):",
"_____no_output_____"
],
[
"$$\\begin{align}\n \\frac{\\partial Loss}{\\partial w} &= \\begin{bmatrix}\n -\\frac{1}{n} \\sum_{i=1}^n \\left(y_i - \\sigma(w \\cdot x_i)\\right)x_{i1} \\\\\n -\\frac{1}{n} \\sum_{i=1}^n \\left(y_i - \\sigma(w \\cdot x_i)\\right)x_{i2} \\\\\n \\vdots \\\\\n -\\frac{1}{n} \\sum_{i=1}^n \\left(y_i - \\sigma(w \\cdot x_i)\\right)x_{im}\n \\end{bmatrix}\n\\end{align}=\\frac{1}{n} X^T \\left(\\hat{y} - y\\right)$$",
"_____no_output_____"
],
[
"По аналогии с $w_j$ выведите формулу для свободного члена (bias'а) $b$ (*hint*: можно считать, что при нём есть признак $x_{i0}=1$ на всех $i$):",
"_____no_output_____"
],
[
"Получили новое правило для обновления $w$ и $b$. ",
"_____no_output_____"
]
],
[
[
"def sigmoid(x):\n \"\"\"Сигмоидальная функция\"\"\"\n return 1 / (1 + np.exp(-x))",
"_____no_output_____"
]
],
[
[
"Реализуйте нейрон с функцией потерь LogLoss:",
"_____no_output_____"
]
],
[
[
"class Neuron:\n \n def __init__(self, w=None, b=0):\n \"\"\"\n :param: w -- вектор весов\n :param: b -- смещение\n \"\"\"\n # пока что мы не знаем размер матрицы X, а значит не знаем, сколько будет весов\n self.w = w\n self.b = b\n \n \n def activate(self, x):\n return sigmoid(x)\n \n \n def forward_pass(self, X):\n \"\"\"\n Эта функция рассчитывает ответ нейрона при предъявлении набора объектов\n :param: X -- матрица объектов размера (n, m), каждая строка - отдельный объект\n :return: вектор размера (n, 1) из нулей и единиц с ответами перцептрона \n \"\"\"\n # реализуйте forward_pass\n n = X.shape[0]\n y_pred = np.zeros((n, 1))\n y_pred = self.activate(X @ self.w.reshape(X.shape[1], 1) + self.b)\n return y_pred.reshape(-1, 1)\n \n \n def backward_pass(self, X, y, y_pred, learning_rate=0.1):\n \"\"\"\n Обновляет значения весов нейрона в соответствие с этим объектом\n :param: X -- матрица объектов размера (n, m)\n y -- вектор правильных ответов размера (n, 1)\n learning_rate - \"скорость обучения\" (символ alpha в формулах выше)\n В этом методе ничего возвращать не нужно, только правильно поменять веса\n с помощью градиентного спуска.\n \"\"\"\n # тут нужно обновить веса по формулам, написанным выше\n n = len(y)\n y = np.array(y).reshape(-1, 1)\n sigma = self.activate(X @ self.w + self.b)\n self.w = self.w - learning_rate * (X.T @ (sigma - y)) / n\n self.b = self.b - learning_rate * np.mean(sigma - y)\n \n \n def fit(self, X, y, num_epochs=5000):\n \"\"\"\n Спускаемся в минимум\n :param: X -- матрица объектов размера (n, m)\n y -- вектор правильных ответов размера (n, 1)\n num_epochs -- количество итераций обучения\n :return: J_values -- вектор значений функции потерь\n \"\"\"\n self.w = np.zeros((X.shape[1], 1)) # столбец (m, 1)\n self.b = 0 # смещение\n loss_values = [] # значения функции потерь на различных итерациях обновления весов\n \n for i in range(num_epochs):\n # предсказания с текущими весами\n y_pred = self.forward_pass(X)\n # считаем функцию потерь с текущими весами\n loss_values.append(loss(y_pred, y))\n # обновляем веса по формуле градиентного спуска\n self.backward_pass(X, y, y_pred)\n\n return loss_values",
"_____no_output_____"
]
],
[
[
"<h3 style=\"text-align: center;\"><b>Тестирование</b></h3>",
"_____no_output_____"
],
[
"Протестируем нейрон, обученный с новой функцией потерь, на тех же данных, что и в предыдущем ноутбуке:",
"_____no_output_____"
],
[
"**Проверка forward_pass()**",
"_____no_output_____"
]
],
[
[
"w = np.array([1., 2.]).reshape(2, 1)\nb = 2.\nX = np.array([[1., 3.],\n [2., 4.],\n [-1., -3.2]])\n\nneuron = Neuron(w, b)\ny_pred = neuron.forward_pass(X)\nprint(\"y_pred = \" + str(y_pred))",
"y_pred = [[0.99987661]\n [0.99999386]\n [0.00449627]]\n"
]
],
[
[
"**Проверка backward_pass()**",
"_____no_output_____"
]
],
[
[
"y = np.array([1, 0, 1]).reshape(3, 1)",
"_____no_output_____"
],
[
"neuron.backward_pass(X, y, y_pred)\n\nprint(\"w = \" + str(neuron.w))\nprint(\"b = \" + str(neuron.b))",
"w = [[0.9001544 ]\n [1.76049276]]\nb = 1.9998544421863216\n"
]
],
[
[
"Проверьте на наборах данных \"яблоки и груши\" и \"голос\".",
"_____no_output_____"
]
],
[
[
"data_apples_pears = pd.read_csv('apples_pears.csv')",
"_____no_output_____"
],
[
"data_apples_pears.head()",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 8))\r\nplt.scatter(data_apples_pears.iloc[:, 0], data_apples_pears.iloc[:, 1], c=data_apples_pears['target'], cmap='rainbow')\r\nplt.title('Яблоки и груши', fontsize=15)\r\nplt.xlabel('симметричность', fontsize=14)\r\nplt.ylabel('желтизна', fontsize=14)\r\nplt.show();",
"_____no_output_____"
],
[
"X = data_apples_pears.iloc[:,:2].values # матрица объекты-признаки\r\ny = data_apples_pears['target'].values.reshape((-1, 1)) # классы (столбец из нулей и единиц)",
"_____no_output_____"
],
[
"%%time\r\nneuron = Neuron(w=np.random.rand(X.shape[1], 1), b=np.random.rand(1))\r\nlosses = neuron.fit(X, y, num_epochs=10000)\r\n\r\nplt.figure(figsize=(10, 8))\r\nplt.plot(losses)\r\nplt.title('Функция потерь', fontsize=15)\r\nplt.xlabel('номер итерации', fontsize=14)\r\nplt.ylabel('$LogLoss(\\hat{y}, y)$', fontsize=14)\r\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 8))\r\nplt.scatter(data_apples_pears.iloc[:, 0], data_apples_pears.iloc[:, 1], c=np.array(neuron.forward_pass(X) > 0.7).ravel(), cmap='spring')\r\nplt.title('Яблоки и груши', fontsize=15)\r\nplt.xlabel('симметричность', fontsize=14)\r\nplt.ylabel('желтизна', fontsize=14)\r\nplt.show();",
"_____no_output_____"
],
[
"y_pred = np.array(neuron.forward_pass(X) > 0.7).ravel()\r\nfrom sklearn.metrics import accuracy_score\r\nprint('Точность (доля правильных ответов, из 100%) нашего нейрона: {:.3f} %'.format(\r\n accuracy_score(y, y_pred) * 100))",
"Точность (доля правильных ответов, из 100%) нашего нейрона: 98.800 %\n"
],
[
"data_voice = pd.read_csv(\"voice.csv\")\r\ndata_voice['label'] = data_voice['label'].apply(lambda x: 1 if x == 'male' else 0)",
"_____no_output_____"
],
[
"data_voice.head()",
"_____no_output_____"
],
[
"# Чтобы перемешать данные. Изначально там сначала идут все мужчины, потом все женщины\r\ndata_voice = data_voice.sample(frac=1)",
"_____no_output_____"
],
[
"X_train = data_voice.iloc[:int(len(data_voice)*0.7), :-1] # матрица объекты-признаки\r\ny_train = data_voice.iloc[:int(len(data_voice)*0.7), -1] # истинные значения пола (мужчина/женщина)\r\n\r\nX_test = data_voice.iloc[int(len(data_voice)*0.7):, :-1] # матрица объекты-признаки\r\ny_test = data_voice.iloc[int(len(data_voice)*0.7):, -1] # истинные значения пола (мужчина/женщина)",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler",
"_____no_output_____"
],
[
"scaler = StandardScaler()\r\nX_train = scaler.fit_transform(X_train.values)\r\nX_test = scaler.transform(X_test.values)",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 8))\r\nplt.scatter(X_test[:, 0], X_test[:, 1], c=y_test, cmap='rainbow')\r\nplt.title('Мужские и женские голоса', fontsize=15)\r\nplt.show();",
"_____no_output_____"
],
[
"neuron = Neuron(w=np.random.rand(X.shape[1], 1), b=np.random.rand(1))\r\nlosses = neuron.fit(X_train, y_train.values);",
"_____no_output_____"
],
[
"y_pred = neuron.forward_pass(X_test)\r\ny_pred = (y_pred > 0.5).astype(int)",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\r\nprint('Точность (доля правильных ответов, из 100%) нашего нейрона: {:.3f} %'.format(\r\n accuracy_score(y_test, y_pred) * 100))",
"Точность (доля правильных ответов, из 100%) нашего нейрона: 97.476 %\n"
],
[
"plt.figure(figsize=(10, 8))\r\nplt.scatter(X_test[:, 0], X_test[:, 1], c=y_pred, cmap='spring')\r\nplt.title('Мужские и женские голоса', fontsize=15)\r\nplt.show();",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecbb2445ee09782069f0af8c31d9bde8bc4dedbd | 292,105 | ipynb | Jupyter Notebook | univariate,multivarite,bivariate analysis EDA.ipynb | ashwiniagandhi/IRIS_Decision-tree-_random_forest_Univariate_byvairate_multivariate | 74b4a679c0dbadaff2cd64c7b5bc73107dd8c4f4 | [
"BSD-3-Clause"
] | null | null | null | univariate,multivarite,bivariate analysis EDA.ipynb | ashwiniagandhi/IRIS_Decision-tree-_random_forest_Univariate_byvairate_multivariate | 74b4a679c0dbadaff2cd64c7b5bc73107dd8c4f4 | [
"BSD-3-Clause"
] | null | null | null | univariate,multivarite,bivariate analysis EDA.ipynb | ashwiniagandhi/IRIS_Decision-tree-_random_forest_Univariate_byvairate_multivariate | 74b4a679c0dbadaff2cd64c7b5bc73107dd8c4f4 | [
"BSD-3-Clause"
] | null | null | null | 122.578682 | 155,336 | 0.780877 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"df=pd.read_csv('https://raw.githubusercontent.com/uiuc-cse/data-fa14/gh-pages/data/iris.csv')",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
]
],
[
[
"# Univariate analysis",
"_____no_output_____"
]
],
[
[
"df_setosa=df.loc[df['species']=='setosa']#take only one feature is sepal_legth\ndf_setosa",
"_____no_output_____"
],
[
"df_virginica=df.loc[df['species']=='virginica']\ndf_versicolor=df.loc[df['species']=='versicolor']\ndf_versicolor",
"_____no_output_____"
],
[
"df_virginica",
"_____no_output_____"
],
[
"plt.plot(df_setosa['sepal_length'],np.zeros_like(df_setosa['sepal_length']),'o')\n\nplt.xlabel('Petal length')\nplt.show()",
"_____no_output_____"
],
[
"plt.plot(df_virginica['sepal_length'],np.zeros_like(df_virginica['sepal_length']),'o')\nplt.plot(df_versicolor['sepal_length'],np.zeros_like(df_versicolor['sepal_length']),'o')\nplt.plot(df_setosa['sepal_length'],np.zeros_like(df_setosa['sepal_length']))\nplt.xlabel('Petal length')\nplt.show()",
"_____no_output_____"
],
[
"plt.plot(df_virginica['sepal_length'],np.zeros_like(df_virginica['sepal_length']),'o')\nplt.plot(df_versicolor['sepal_length'],np.zeros_like(df_versicolor['sepal_length']),'o')\nplt.plot(df_setosa['sepal_length'],np.zeros_like(df_setosa['sepal_length']),'o')\nplt.xlabel('Petal length')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Bivariate analysis",
"_____no_output_____"
]
],
[
[
"sns.FacetGrid(df,hue=\"species\",size=5).map(plt.scatter,\"sepal_length\",\"sepal_width\").add_legend();\nplt.show()#its take 2 features petal length,sepal width#what feature to categories is species,",
"C:\\logs\\lib\\site-packages\\seaborn\\axisgrid.py:230: UserWarning: The `size` paramter has been renamed to `height`; please update your code.\n warnings.warn(msg, UserWarning)\n"
],
[
"sns.FacetGrid(df,hue=\"species\",size=5).map(plt.scatter,\"petal_length\",\"sepal_width\").add_legend();\nplt.show()#its take 2 features petal length,sepal width#what feature to categories is species,",
"C:\\logs\\lib\\site-packages\\seaborn\\axisgrid.py:230: UserWarning: The `size` paramter has been renamed to `height`; please update your code.\n warnings.warn(msg, UserWarning)\n"
]
],
[
[
"# multivariate analysis",
"_____no_output_____"
]
],
[
[
"sns.pairplot(df,hue=\"species\",size=3)#all fetaures are takesepal legth,width,petal legth,width",
"C:\\logs\\lib\\site-packages\\seaborn\\axisgrid.py:2065: UserWarning: The `size` parameter has been renamed to `height`; pleaes update your code.\n warnings.warn(msg, UserWarning)\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecbb28319e74830cf41bc832e0b3ec91485876aa | 62,838 | ipynb | Jupyter Notebook | 3.4-classifying-movie-reviews.ipynb | data-science-team/keras-books-notebooks | d5eaf7d93904c81dcbd3318340994ac75f5983bc | [
"MIT"
] | null | null | null | 3.4-classifying-movie-reviews.ipynb | data-science-team/keras-books-notebooks | d5eaf7d93904c81dcbd3318340994ac75f5983bc | [
"MIT"
] | null | null | null | 3.4-classifying-movie-reviews.ipynb | data-science-team/keras-books-notebooks | d5eaf7d93904c81dcbd3318340994ac75f5983bc | [
"MIT"
] | 1 | 2019-06-20T08:02:58.000Z | 2019-06-20T08:02:58.000Z | 62.775225 | 17,796 | 0.754305 | [
[
[
"import keras\nkeras.__version__",
"Using TensorFlow backend.\n"
]
],
[
[
"# 영화 리뷰 분류: 이진 분류 예제\n\n이 노트북은 [케라스 창시자에게 배우는 딥러닝](https://tensorflow.blog/케라스-창시자에게-배우는-딥러닝/) 책의 3장 4절의 코드 예제입니다. 책에는 더 많은 내용과 그림이 있습니다. 이 노트북에는 소스 코드에 관련된 설명만 포함합니다. 이 노트북의 설명은 케라스 버전 2.2.2에 맞추어져 있습니다. 케라스 최신 버전이 릴리스되면 노트북을 다시 테스트하기 때문에 설명과 코드의 결과가 조금 다를 수 있습니다.\n\n----\n\n2종 분류 또는 이진 분류는 아마도 가장 널리 적용된 머신 러닝 문제일 것입니다. 이 예제에서 리뷰 텍스트를 기반으로 영화 리뷰를 긍정과 부정로 분류하는 법을 배우겠습니다.",
"_____no_output_____"
],
[
"## IMDB 데이터셋\n\n인터넷 영화 데이터베이스로부터 가져온 양극단의 리뷰 50,000개로 이루어진 IMDB 데이터셋을 사용하겠습니다. 이 데이터셋은 훈련 데이터 25,000개와 테스트 데이터 25,000개로 나뉘어 있고 각각 50%는 부정, 50%는 긍정 리뷰로 구성되어 있습니다.\n\n왜 훈련 데이터와 테스트 데이터를 나눌까요? 같은 데이터에서 머신 러닝 모델을 훈련하고 테스트해서는 절대 안 되기 때문입니다! 모델이 훈련 데이터에서 잘 작동한다는 것이 처음 만난 데이터에서도 잘 동작한다는 것을 보장하지 않습니다. 중요한 것은 새로운 데이터에 대한 모델의 성능입니다(사실 훈련 데이터의 레이블은 이미 알고 있기 때문에 이를 예측하는 모델은 필요하지 않습니다). 예를 들어 모델이 훈련 샘플과 타깃 사이의 매핑을 모두 외워버릴 수 있습니다. 이런 모델은 처음 만나는 데이터에서 타깃을 예측하는 작업에는 쓸모가 없습니다. 다음 장에서 이에 대해 더 자세히 살펴보겠습니다.\n\nMNIST 데이터셋처럼 IMDB 데이터셋도 케라스에 포함되어 있습니다. 이 데이터는 전처리되어 있어 각 리뷰(단어 시퀀스)가 숫자 시퀀스로 변환되어 있습니다. 여기서 각 숫자는 사전에 있는 고유한 단어를 나타냅니다.\n\n다음 코드는 데이터셋을 로드합니다(처음 실행하면 17MB 정도의 데이터가 컴퓨터에 다운로드됩니다):",
"_____no_output_____"
]
],
[
[
"from keras.datasets import imdb\n\n(train_data, train_labels), (test_data, test_labels) = imdb.load_data(num_words=10000)",
"_____no_output_____"
]
],
[
[
"매개변수 `num_words=10000`은 훈련 데이터에서 가장 자주 나타나는 단어 10,000개만 사용하겠다는 의미입니다. 드물게 나타나는 단어는 무시하겠습니다. 이렇게 하면 적절한 크기의 벡터 데이터를 얻을 수 있습니다.\n\n변수 `train_data`와 `test_data`는 리뷰의 목록입니다. 각 리뷰는 단어 인덱스의 리스트입니다(단어 시퀀스가 인코딩된 것입니다). `train_labels`와 `test_labels`는 부정을 나타내는 0과 긍정을 나타내는 1의 리스트입니다:",
"_____no_output_____"
]
],
[
[
"train_data[0]",
"_____no_output_____"
],
[
"train_labels[0]",
"_____no_output_____"
]
],
[
[
"가장 자주 등장하는 단어 10,000개로 제한했기 때문에 단어 인덱스는 10,000을 넘지 않습니다:",
"_____no_output_____"
]
],
[
[
"max([max(sequence) for sequence in train_data])",
"_____no_output_____"
]
],
[
[
"재미 삼아 이 리뷰 데이터 하나를 원래 영어 단어로 어떻게 바꾸는지 보겠습니다:",
"_____no_output_____"
]
],
[
[
"# word_index는 단어와 정수 인덱스를 매핑한 딕셔너리입니다\nword_index = imdb.get_word_index()\n# 정수 인덱스와 단어를 매핑하도록 뒤집습니다\nreverse_word_index = dict([(value, key) for (key, value) in word_index.items()])\n# 리뷰를 디코딩합니다. \n# 0, 1, 2는 '패딩', '문서 시작', '사전에 없음'을 위한 인덱스이므로 3을 뺍니다\ndecoded_review = ' '.join([reverse_word_index.get(i - 3, '?') for i in train_data[0]])",
"_____no_output_____"
],
[
"decoded_review",
"_____no_output_____"
]
],
[
[
"## 데이터 준비\n\n신경망에 숫자 리스트를 주입할 수는 없습니다. 리스트를 텐서로 바꾸는 두 가지 방법이 있습니다:\n\n* 같은 길이가 되도록 리스트에 패딩을 추가하고 `(samples, sequence_length)` 크기의 정수 텐서로 변환합니다. 그다음 이 정수 텐서를 다룰 수 있는 층을 신경망의 첫 번째 층으로 사용합니다(`Embedding` 층을 말하며 나중에 자세히 다루겠습니다).\n* 리스트를 원-핫 인코딩하여 0과 1의 벡터로 변환합니다. 예를 들면 시퀀스 `[3, 5]`를 인덱스 3과 5의 위치는 1이고 그 외는 모두 0인 10,000차원의 벡터로 각각 변환합니다. 그다음 부동 소수 벡터 데이터를 다룰 수 있는 `Dense` 층을 신경망의 첫 번째 층으로 사용합니다.\n\n여기서는 두 번째 방식을 사용하고 이해를 돕기 위해 직접 데이터를 원-핫 벡터로 만들겠습니다:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndef vectorize_sequences(sequences, dimension=10000):\n # 크기가 (len(sequences), dimension))이고 모든 원소가 0인 행렬을 만듭니다\n results = np.zeros((len(sequences), dimension))\n for i, sequence in enumerate(sequences):\n results[i, sequence] = 1. # results[i]에서 특정 인덱스의 위치를 1로 만듭니다\n return results\n\n# 훈련 데이터를 벡터로 변환합니다\nx_train = vectorize_sequences(train_data)\n# 테스트 데이터를 벡터로 변환합니다\nx_test = vectorize_sequences(test_data)",
"_____no_output_____"
]
],
[
[
"이제 샘플은 다음과 같이 나타납니다:",
"_____no_output_____"
]
],
[
[
"x_train[0]",
"_____no_output_____"
]
],
[
[
"레이블은 쉽게 벡터로 바꿀 수 있습니다:",
"_____no_output_____"
]
],
[
[
"# 레이블을 벡터로 바꿉니다\ny_train = np.asarray(train_labels).astype('float32')\ny_test = np.asarray(test_labels).astype('float32')",
"_____no_output_____"
]
],
[
[
"이제 신경망에 주입할 데이터가 준비되었습니다.",
"_____no_output_____"
],
[
"## 신경망 모델 만들기\n\n입력 데이터가 벡터이고 레이블은 스칼라(1 또는 0)입니다. 아마 앞으로 볼 수 있는 문제 중에서 가장 간단할 것입니다. 이런 문제에 잘 작동하는 네트워크 종류는 `relu` 활성화 함수를 사용한 완전 연결 층(즉, `Dense(16, activation='relu')`)을 그냥 쌓은 것입니다.\n\n`Dense` 층에 전달한 매개변수(16)는 은닉 유닛의 개수입니다. 하나의 은닉 유닛은 층이 나타내는 표현 공간에서 하나의 차원이 됩니다. 2장에서 `relu` 활성화 함수를 사용한 `Dense` 층을 다음과 같은 텐서 연산을 연결하여 구현하였습니다:\n\n`output = relu(dot(W, input) + b)`\n\n16개의 은닉 유닛이 있다는 것은 가중치 행렬 `W`의 크기가 `(input_dimension, 16)`이라는 뜻입니다. 입력 데이터와 `W`를 점곱하면 입력 데이터가 16 차원으로 표현된 공간으로 투영됩니다(그리고 편향 벡터 `b`를 더하고 `relu` 연산을 적용합니다). 표현 공간의 차원을 '신경망이 내재된 표현을 학습할 때 가질 수 있는 자유도'로 이해할 수 있습니다. 은닉 유닛을 늘리면 (표현 공간을 더 고차원으로 만들면) 신경망이 더욱 복잡한 표현을 학습할 수 있지만 계산 비용이 커지고 원치 않은 패턴을 학습할 수도 있습니다(훈련 데이터에서는 성능이 향상되지만 테스트 데이터에서는 그렇지 않은 패턴입니다).\n\n`Dense` 층을 쌓을 때 두 가진 중요한 구조상의 결정이 필요합니다:\n\n* 얼마나 많은 층을 사용할 것인가\n* 각 층에 얼마나 많은 은닉 유닛을 둘 것인가\n\n4장에서 이런 결정을 하는 데 도움이 되는 일반적인 원리를 배우겠습니다. 당분간은 저를 믿고 선택한 다음 구조를 따라 주세요.\n\n* 16개의 은닉 유닛을 가진 두 개의 은닉층\n* 현재 리뷰의 감정을 스칼라 값의 예측으로 출력하는 세 번째 층\n\n중간에 있는 은닉층은 활성화 함수로 `relu`를 사용하고 마지막 층은 확률(0과 1 사이의 점수로, 어떤 샘플이 타깃 '1'일 가능성이 높다는 것은 그 리뷰가 긍정일 가능성이 높다는 것을 의미합니다)을 출력하기 위해 시그모이드 활성화 함수를 사용합니다. `relu`는 음수를 0으로 만드는 함수입니다. 시그모이드는 임의의 값을 [0, 1] 사이로 압축하므로 출력 값을 확률처럼 해석할 수 있습니다.",
"_____no_output_____"
],
[
"다음이 이 신경망의 모습입니다:\n\n",
"_____no_output_____"
],
[
"다음은 이 신경망의 케라스 구현입니다. 이전에 보았던 MNIST 예제와 비슷합니다:",
"_____no_output_____"
]
],
[
[
"from keras import models\nfrom keras import layers\n\nmodel = models.Sequential()\nmodel.add(layers.Dense(16, activation='relu', input_shape=(10000,)))\nmodel.add(layers.Dense(16, activation='relu'))\nmodel.add(layers.Dense(1, activation='sigmoid'))",
"_____no_output_____"
]
],
[
[
"마지막으로 손실 함수와 옵티마이저를 선택해야 합니다. 이진 분류 문제이고 신경망의 출력이 확률이기 때문에(네트워크의 끝에 시그모이드 활성화 함수를 사용한 하나의 유닛으로 된 층을 놓았습니다), `binary_crossentropy` 손실이 적합합니다. 이 함수가 유일한 선택은 아니고 예를 들어 `mean_squared_error`를 사용할 수도 있습니다. 확률을 출력하는 모델을 사용할 때는 크로스엔트로피가 최선의 선택입니다. 크로스엔트로피는 정보 이론 분야에서 온 개념으로 확률 분포 간의 차이를 측정합니다. 여기에서는 원본 분포와 예측 분포 사이를 측정합니다.\n\n다음은 `rmsprop` 옵티마이저와 `binary_crossentropy` 손실 함수로 모델을 설정하는 단계입니다. 훈련하는 동안 정확도를 사용해 모니터링하겠습니다.",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='rmsprop',\n loss='binary_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"케라스에 `rmsprop`, `binary_crossentropy`, `accuracy`가 포함되어 있기 때문에 옵티마이저, 손실 함수, 측정 지표를 문자열로 지정하는 것이 가능합니다. 이따금 옵티마이저의 매개변수를 바꾸거나 자신만의 손실 함수, 측정 함수를 전달해야 할 경우가 있습니다. 전자의 경우에는 옵티마이저 파이썬 클래스를 사용해 객체를 직접 만들어 `optimizer` 매개변수에 전달하면 됩니다:",
"_____no_output_____"
]
],
[
[
"from keras import optimizers\n\nmodel.compile(optimizer=optimizers.RMSprop(lr=0.001),\n loss='binary_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"후자의 경우는 `loss`와 `metrics` 매개변수에 함수 객체를 전달하면 됩니다:",
"_____no_output_____"
]
],
[
[
"from keras import losses\nfrom keras import metrics\n\nmodel.compile(optimizer=optimizers.RMSprop(lr=0.001),\n loss=losses.binary_crossentropy,\n metrics=[metrics.binary_accuracy])",
"_____no_output_____"
],
[
"model.compile(optimizer='rmsprop',\n loss='binary_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"## 훈련 검증\n\n훈련하는 동안 처음 본 데이터에 대한 모델의 정확도를 측정하기 위해서는 원본 훈련 데이터에서 10,000의 샘플을 떼어서 검증 세트를 만들어야 합니다:",
"_____no_output_____"
]
],
[
[
"x_val = x_train[:10000]\npartial_x_train = x_train[10000:]\n\ny_val = y_train[:10000]\npartial_y_train = y_train[10000:]",
"_____no_output_____"
]
],
[
[
"이제 모델을 512개 샘플씩 미니 배치를 만들어 20번의 에포크 동안 훈련시킵니다(`x_train`과 `y_train` 텐서에 있는 모든 샘플에 대해 20번 반복합니다). 동시에 따로 떼어 놓은 10,000개의 샘플에서 손실과 정확도를 측정할 것입니다. 이렇게 하려면 `validation_data` 매개변수에 검증 데이터를 전달해야 합니다:",
"_____no_output_____"
]
],
[
[
"history = model.fit(partial_x_train,\n partial_y_train,\n epochs=20,\n batch_size=512,\n validation_data=(x_val, y_val))",
"Train on 15000 samples, validate on 10000 samples\nEpoch 1/20\n15000/15000 [==============================] - 2s 116us/step - loss: 0.5084 - acc: 0.7813 - val_loss: 0.3797 - val_acc: 0.8684\nEpoch 2/20\n15000/15000 [==============================] - 1s 70us/step - loss: 0.3004 - acc: 0.9047 - val_loss: 0.3004 - val_acc: 0.8897\nEpoch 3/20\n15000/15000 [==============================] - 1s 65us/step - loss: 0.2179 - acc: 0.9285 - val_loss: 0.3085 - val_acc: 0.8711\nEpoch 4/20\n15000/15000 [==============================] - 1s 69us/step - loss: 0.1750 - acc: 0.9437 - val_loss: 0.2840 - val_acc: 0.8832\nEpoch 5/20\n15000/15000 [==============================] - 1s 67us/step - loss: 0.1427 - acc: 0.9543 - val_loss: 0.2841 - val_acc: 0.8872\nEpoch 6/20\n15000/15000 [==============================] - 1s 67us/step - loss: 0.1150 - acc: 0.9650 - val_loss: 0.3166 - val_acc: 0.8772\nEpoch 7/20\n15000/15000 [==============================] - 1s 67us/step - loss: 0.0980 - acc: 0.9705 - val_loss: 0.3127 - val_acc: 0.8846\nEpoch 8/20\n15000/15000 [==============================] - 1s 66us/step - loss: 0.0807 - acc: 0.9763 - val_loss: 0.3859 - val_acc: 0.8649\nEpoch 9/20\n15000/15000 [==============================] - 1s 67us/step - loss: 0.0661 - acc: 0.9821 - val_loss: 0.3635 - val_acc: 0.8782\nEpoch 10/20\n15000/15000 [==============================] - 1s 69us/step - loss: 0.0561 - acc: 0.9853 - val_loss: 0.3843 - val_acc: 0.8792\nEpoch 11/20\n15000/15000 [==============================] - 1s 68us/step - loss: 0.0439 - acc: 0.9893 - val_loss: 0.4153 - val_acc: 0.8779\nEpoch 12/20\n15000/15000 [==============================] - 1s 59us/step - loss: 0.0381 - acc: 0.9921 - val_loss: 0.4525 - val_acc: 0.8690\nEpoch 13/20\n15000/15000 [==============================] - 1s 59us/step - loss: 0.0300 - acc: 0.9928 - val_loss: 0.4698 - val_acc: 0.8729\nEpoch 14/20\n15000/15000 [==============================] - 1s 66us/step - loss: 0.0247 - acc: 0.9945 - val_loss: 0.5023 - val_acc: 0.8726\nEpoch 15/20\n15000/15000 [==============================] - 1s 68us/step - loss: 0.0175 - acc: 0.9979 - val_loss: 0.5342 - val_acc: 0.8693\nEpoch 16/20\n15000/15000 [==============================] - 1s 68us/step - loss: 0.0149 - acc: 0.9983 - val_loss: 0.5710 - val_acc: 0.8698\nEpoch 17/20\n15000/15000 [==============================] - 1s 69us/step - loss: 0.0151 - acc: 0.9971 - val_loss: 0.6025 - val_acc: 0.8697\nEpoch 18/20\n15000/15000 [==============================] - 1s 65us/step - loss: 0.0075 - acc: 0.9996 - val_loss: 0.6782 - val_acc: 0.8633\nEpoch 19/20\n15000/15000 [==============================] - 1s 62us/step - loss: 0.0117 - acc: 0.9975 - val_loss: 0.6693 - val_acc: 0.8673\nEpoch 20/20\n15000/15000 [==============================] - 1s 68us/step - loss: 0.0041 - acc: 0.9999 - val_loss: 0.6942 - val_acc: 0.8658\n"
]
],
[
[
"CPU를 사용해도 에포크마다 2초가 걸리지 않습니다. 전체 훈련은 20초 이상 걸립니다. 에포크가 끝날 때마다 10,000개의 검증 샘플 데이터에서 손실과 정확도를 계산하기 때문에 약간씩 지연됩니다.\n\n`model.fit()` 메서드는 `History` 객체를 반환합니다. 이 객체는 훈련하는 동안 발생한 모든 정보를 담고 있는 딕셔너리인 `history` 속성을 가지고 있습니다. 한 번 확인해 보죠:",
"_____no_output_____"
]
],
[
[
"history_dict = history.history\nhistory_dict.keys()",
"_____no_output_____"
]
],
[
[
"이 딕셔너리는 훈련과 검증하는 동안 모니터링할 측정 지표당 하나씩 모두 네 개의 항목을 담고 있습니다. 맷플롯립을 사용해 훈련과 검증 데이터에 대한 손실과 정확도를 그려 보겠습니다:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"acc = history.history['acc']\nval_acc = history.history['val_acc']\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs = range(1, len(acc) + 1)\n\n# ‘bo’는 파란색 점을 의미합니다\nplt.plot(epochs, loss, 'bo', label='Training loss')\n# ‘b’는 파란색 실선을 의미합니다\nplt.plot(epochs, val_loss, 'b', label='Validation loss')\nplt.title('Training and validation loss')\nplt.xlabel('Epochs')\nplt.ylabel('Loss')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
],
[
"plt.clf() # 그래프를 초기화합니다\nacc = history_dict['acc']\nval_acc = history_dict['val_acc']\n\nplt.plot(epochs, acc, 'bo', label='Training acc')\nplt.plot(epochs, val_acc, 'b', label='Validation acc')\nplt.title('Training and validation accuracy')\nplt.xlabel('Epochs')\nplt.ylabel('Accuracy')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"점선은 훈련 손실과 정확도이고 실선은 검증 손실과 정확도입니다. 신경망의 무작위한 초기화 때문에 사람마다 결과거 조금 다를 수 있습니다.\n\n여기에서 볼 수 있듯이 훈련 손실이 에포크마다 감소하고 훈련 정확도는 에포크마다 증가합니다. 경사 하강법 최적화를 사용했을 때 반복마다 최소화되는 것이 손실이므로 기대했던 대로입니다. 검증 손실과 정확도는 이와 같지 않습니다. 4번째 에포크에서 그래프가 역전되는 것 같습니다. 이것이 훈련 세트에서 잘 작동하는 모델이 처음 보는 데이터에 잘 작동하지 않을 수 있다고 앞서 언급한 경고의 한 사례입니다. 정확한 용어로 말하면 과대적합되었다고 합니다. 2번째 에포크 이후부터 훈련 데이터에 과도하게 최적화되어 훈련 데이터에 특화된 표현을 학습하므로 훈련 세트 이외의 데이터에는 일반화되지 못합니다.\n\n이런 경우에 과대적합을 방지하기 위해서 3번째 에포크 이후에 훈련을 중지할 수 있습니다. 일반적으로 4장에서 보게 될 과대적합을 완화하는 다양한 종류의 기술을 사용할 수 있습니다.\n\n처음부터 다시 새로운 신경망을 4번의 에포크 동안만 훈련하고 테스트 데이터에서 평가해 보겠습니다:",
"_____no_output_____"
]
],
[
[
"model = models.Sequential()\nmodel.add(layers.Dense(16, activation='relu', input_shape=(10000,)))\nmodel.add(layers.Dense(16, activation='relu'))\nmodel.add(layers.Dense(1, activation='sigmoid'))\n\nmodel.compile(optimizer='rmsprop',\n loss='binary_crossentropy',\n metrics=['accuracy'])\n\nmodel.fit(x_train, y_train, epochs=4, batch_size=512)\nresults = model.evaluate(x_test, y_test)",
"Epoch 1/4\n25000/25000 [==============================] - 1s 47us/step - loss: 0.4749 - acc: 0.8217\nEpoch 2/4\n25000/25000 [==============================] - 1s 42us/step - loss: 0.2658 - acc: 0.9097\nEpoch 3/4\n25000/25000 [==============================] - 1s 42us/step - loss: 0.1982 - acc: 0.9299\nEpoch 4/4\n25000/25000 [==============================] - 1s 42us/step - loss: 0.1679 - acc: 0.9404\n25000/25000 [==============================] - 1s 45us/step\n"
],
[
"results",
"_____no_output_____"
]
],
[
[
"아주 단순한 방식으로도 87%의 정확도를 달성했습니다. 최고 수준의 기법을 사용하면 95%에 가까운 성능을 얻을 수 있습니다.",
"_____no_output_____"
],
[
"## 훈련된 모델로 새로운 데이터에 대해 예측하기\n\n모델을 훈련시킨 후에 이를 실전 환경에서 사용하고 싶을 것입니다. `predict` 메서드를 사용해서 어떤 리뷰가 긍정일 확률을 예측할 수 있습니다:",
"_____no_output_____"
]
],
[
[
"model.predict(x_test)",
"_____no_output_____"
]
],
[
[
"여기에서처럼 이 모델은 어떤 샘플에 대해 확신을 가지고 있지만(0.99 또는 그 이상, 0.01 또는 그 이하) 어떤 샘플에 대해서는 확신이 부족합니다(0.6, 0.4). ",
"_____no_output_____"
],
[
"## 추가 실험\n\n* 여기에서는 두 개의 은닉층을 사용했습니다. 한 개 또는 세 개의 은닉층을 사용하고 검증과 테스트 정확도에 어떤 영향을 미치는지 확인해 보세요.\n* 층의 은닉 유닛을 추가하거나 줄여 보세요: 32개 유닛, 64개 유닛 등\n* `binary_crossentropy` 대신에 `mse` 손실 함수를 사용해 보세요.\n* `relu` 대신에 `tanh` 활성화 함수(초창기 신경망에서 인기 있었던 함수입니다)를 사용해 보세요.\n\n다음 실험을 진행하면 여기에서 선택한 구조가 향상의 여지는 있지만 어느 정도 납득할 만한 수준이라는 것을 알게 것입니다!",
"_____no_output_____"
],
[
"## 정리\n\n다음은 이 예제에서 배운 것들입니다:\n\n* 원본 데이터를 신경망에 텐서로 주입하기 위해서는 꽤 많은 전처리가 필요합니다. 단어 시퀀스는 이진 벡터로 인코딩될 수 있고 다른 인코딩 방식도 있습니다.\n* `relu` 활성화 함수와 함께 `Dense` 층을 쌓은 네트워크는 (감성 분류를 포함하여) 여러 종류의 문제에 적용할 수 있어서 앞으로 자주 사용하게 될 것입니다.\n* (출력 클래스가 두 개인) 이진 분류 문제에서 네트워크는 하나의 유닛과 `sigmoid` 활성화 함수를 가진 `Dense` 층으로 끝나야 합니다. 이 신경망의 출력은 확률을 나타내는 0과 1 사이의 스칼라 값입니다.\n* 이진 분류 문제에서 이런 스칼라 시그모이드 출력에 대해 사용할 손실 함수는 `binary_crossentropy`입니다.\n* `rmsprop` 옵티마이저는 문제에 상관없이 일반적으로 충분히 좋은 선택입니다. 걱정할 거리가 하나 줄은 셈입니다.\n* 훈련 데이터에 대해 성능이 향상됨에 따라 신경망은 과대적합되기 시작하고 이전에 본적 없는 데이터에서는 결과가 점점 나빠지게 됩니다. 항상 훈련 세트 이외의 데이터에서 성능을 모니터링해야 합니다.",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ecbb2b69900e61327793161751d569eb35a82da0 | 562,121 | ipynb | Jupyter Notebook | code/3D plots in matplotlib.ipynb | pawjast/medium | 574528488f5c1e02974dcdf71a070dd947e64a8f | [
"MIT"
] | null | null | null | code/3D plots in matplotlib.ipynb | pawjast/medium | 574528488f5c1e02974dcdf71a070dd947e64a8f | [
"MIT"
] | null | null | null | code/3D plots in matplotlib.ipynb | pawjast/medium | 574528488f5c1e02974dcdf71a070dd947e64a8f | [
"MIT"
] | null | null | null | 1,283.381279 | 112,196 | 0.960585 | [
[
[
"# Checking software version",
"_____no_output_____"
]
],
[
[
"import matplotlib\nprint(matplotlib.__version__)",
"3.2.2\n"
],
[
"!python -V",
"Python 3.8.3\n"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"From `matplotlib` version 3.2.0 onwards it's no longer necessary to explicitly impotrt the `mpl_toolkits.mplot3d` utility to access 3D plotting capability.\n\nhttps://matplotlib.org/stable/tutorials/toolkits/mplot3d.html",
"_____no_output_____"
],
[
"# 3D plots in Matplotlib\n\nContent:\n\nTypes of plot:\n\n* scatter plot \n* line plot\n* surface plot\n* wireframe plot\n* contour plot\n * filled contour plot",
"_____no_output_____"
],
[
"# Create empty 3D figure\n\nIn order to create any plot using `matplotlib` we always follow the same steps. First, we create a `figure` object and then we create `axes` object.",
"_____no_output_____"
]
],
[
[
"# 2D\nfig = plt.figure(facecolor='palegreen')\nax = fig.add_subplot()\nax.set_title(type(ax));",
"_____no_output_____"
]
],
[
[
"There are 3 ways to create an axes:\n\n* [fig.add_subplot](https://matplotlib.org/stable/api/figure_api.html?highlight=add_subplot#matplotlib.figure.Figure.add_subplot)\n* [fig.add_axes()](https://matplotlib.org/stable/api/figure_api.html?highlight=figure#matplotlib.figure.Figure.add_axes)\n* [fig.subplots()](https://matplotlib.org/stable/api/figure_api.html#matplotlib.figure.Figure.subplots)",
"_____no_output_____"
],
[
"### Create empty 3D `axis` using `projection` method\n\nWhen creating a 3D plot, we follow the same workflow of creating the `figure` and then `axis`. However, for the 3D plot, we need to specify the `projection` parameter when creating an `axis`.\n\n* [fig.add_subplot](https://matplotlib.org/stable/api/figure_api.html?highlight=add_subplot#matplotlib.figure.Figure.add_subplot)",
"_____no_output_____"
]
],
[
[
"# 3d\nfig = plt.figure(facecolor='palegreen')\nax = fig.add_subplot(projection='3d')\nax.set_title(type(ax));",
"_____no_output_____"
]
],
[
[
"**Note:** the value of `projection` must be lowercase '3d'. Upper case '3D' will produce an error.",
"_____no_output_____"
],
[
"* [fig.add_axes()](https://matplotlib.org/stable/api/figure_api.html?highlight=figure#matplotlib.figure.Figure.add_axes)",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(facecolor='palegreen')\nax = fig.add_axes([0, 0 , 1, 1],\n projection='3d'\n )\nax.set_title(type(ax));",
"_____no_output_____"
]
],
[
[
"* [plt.subplots()](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.subplots.html#matplotlib-pyplot-subplots)",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(subplot_kw={'projection': '3d'})\nax.set_title(type(ax));",
"_____no_output_____"
]
],
[
[
"# Types of plot\n\nAll of the examples below we'll be using the `fig.add_subplot()` method for creating an axis\n\n## Scatter\n### Scatter with 1 data point",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(16, 9),\n facecolor='palegreen'\n )\nax = fig.add_subplot(projection='3d')\nax.scatter(0.5, 0.5, 0.5)\n\n# Settings\nax.set_xlim(0, 1)\nax.set_ylim(0, 1)\nax.set_zlim(0, 1)\nax.set_title('Scatter with 1 data point')\nax.set_xlabel('x-axis')\nax.set_ylabel('y-axis')\nax.set_zlabel('z_axis');",
"_____no_output_____"
]
],
[
[
"### Scatter with multiple data points: example 1",
"_____no_output_____"
]
],
[
[
"# Data\nx = [0.25, 0.5, 0.75]\ny = [0.25, 0.5, 0.75]\nz = [0.25, 0.5, 0.75]\n\nfig = plt.figure(figsize=(16, 9),\n facecolor='palegreen'\n )\nax = fig.add_subplot(projection='3d')\nax.scatter(x, y, z)\n\n# Settings\nax.set_xlim(0, 1)\nax.set_ylim(0, 1)\nax.set_zlim(0, 1)\nax.set_title('Scatter with multiple data points')\nax.set_xlabel('x-axis')\nax.set_ylabel('y-axis')\nax.set_zlabel('z_axis');",
"_____no_output_____"
]
],
[
[
"It can be noticed that some points are faded away. To remove this effect, we need to set the `depth shade` paramter to `False`:",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(16, 9),\n facecolor='palegreen'\n )\nax = fig.add_subplot(projection='3d')\nax.scatter(x, y, z,\n depthshade=False)\n\n# Settings\nax.set_xlim(0, 1)\nax.set_ylim(0, 1)\nax.set_zlim(0, 1)\nax.set_title('Scatter with multiple data points')\nax.set_xlabel('x-axis')\nax.set_ylabel('y-axis')\nax.set_zlabel('z_axis');",
"_____no_output_____"
]
],
[
[
"### Scatter with multiple data points: example 1\n3D scatter plot will take any argument a regular 2D scatter plot would take. For more options, see:\n\n* [ax.scatter()](https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.scatter.html#matplotlib-axes-axes-scatter)\n\nLet's create a plot with more data points and customize them a bit:",
"_____no_output_____"
]
],
[
[
"# Data\nnp\nx = np.random.random(30)\ny = np.random.random(30)\nz = np.random.random(30)\n\nfig = plt.figure(figsize=(16, 9),\n facecolor='palegreen'\n )\nax = fig.add_subplot(projection='3d')\nax.scatter(x, y, z,\n c='g',\n edgecolor='r',\n linewidth=2,\n s=100,\n marker='D'\n )\n\n# Settings\nax.set_xlim(0, 1)\nax.set_ylim(0, 1)\nax.set_zlim(0, 1)\nax.set_title('Scatter with multiple data points')\nax.set_xlabel('x-axis')\nax.set_ylabel('y-axis')\nax.set_zlabel('z_axis');",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecbb47cc5df06749557e14d24681548bd476c80c | 7,872 | ipynb | Jupyter Notebook | notebooks/Canopy_netSWLW_APAR_fluxes.ipynb | bbuman/Land | b0f3a390eb17330abbfe1a6ddffefdad2c7353ff | [
"Apache-2.0"
] | 49 | 2020-05-06T19:15:17.000Z | 2022-03-05T23:17:26.000Z | notebooks/Canopy_netSWLW_APAR_fluxes.ipynb | bbuman/Land | b0f3a390eb17330abbfe1a6ddffefdad2c7353ff | [
"Apache-2.0"
] | 35 | 2020-05-07T16:15:18.000Z | 2021-11-23T04:04:16.000Z | notebooks/Canopy_netSWLW_APAR_fluxes.ipynb | bbuman/Land | b0f3a390eb17330abbfe1a6ddffefdad2c7353ff | [
"Apache-2.0"
] | 10 | 2020-09-28T17:50:14.000Z | 2022-03-27T21:24:34.000Z | 23.63964 | 191 | 0.54408 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecbb4b5536861c514b14c8f60798a7420d12865a | 73,072 | ipynb | Jupyter Notebook | BehavioralAnalyses/BehavPerfExtract_fromRawBehavSheet.ipynb | alexgonzl/TreeMazeAnalyses | a834dc6b59beffe6bce59cdd9749b761fab3fe08 | [
"MIT"
] | null | null | null | BehavioralAnalyses/BehavPerfExtract_fromRawBehavSheet.ipynb | alexgonzl/TreeMazeAnalyses | a834dc6b59beffe6bce59cdd9749b761fab3fe08 | [
"MIT"
] | null | null | null | BehavioralAnalyses/BehavPerfExtract_fromRawBehavSheet.ipynb | alexgonzl/TreeMazeAnalyses | a834dc6b59beffe6bce59cdd9749b761fab3fe08 | [
"MIT"
] | null | null | null | 419.954023 | 35,448 | 0.938855 | [
[
[
"%matplotlib notebook\n%matplotlib inline\n\nimport numpy as np\nimport pandas as pd\nimport matplotlib\nimport matplotlib.pyplot as plt\n\n\nfont = {'family' : 'sans-serif',\n 'size' : 20}\n\nplt.rc('font', **font)",
"_____no_output_____"
],
[
"dataDir = '/Users/alexgonzalez/Google Drive/PostDoc/Data/'\nsubjIDs = ['Li','Ne']\nstartIDs = [47,196]\nendIDs = [308,238]",
"_____no_output_____"
],
[
"cnt=0\n\nfor subj in subjIDs:\n\n df = pd.read_csv(dataDir+subj+'_Perf.csv')\n df=df[df.Experiment!='T3i']\n x=df.loc[startIDs[cnt]:endIDs[cnt],['trial number','correct trials','pct correct']]\n x=x.dropna()\n x = x.reset_index(drop=True)\n x=x.apply(pd.to_numeric, errors='ignore')\n\n nAvg =3\n y=x.rolling(nAvg,center=True).sum()/nAvg\n \n f, (ax1, ax2) = plt.subplots(2, 1, sharex=True,figsize=(8,8))\n \n ax1.plot(y['trial number'],'rd-',linewidth=3)\n ax1.grid(True)\n ax1.set_ylabel('Number of Trials',color='r')\n ax1.tick_params('y', colors='r')\n ax1.set(ylim=(0,250))\n #ax1.set(title='Behavioral Performance')\n\n ax1b = ax1.twinx()\n ax1b.plot(y['pct correct'],'ko-',linewidth=3)\n ax1b.set(ylim=(0,100))\n ax1b.set_ylabel('Percent Correct')\n plt.show()\n fig.savefig(dataDir+subj+'BehavPerf_1.pdf',bbox_inches='tight')\n cnt+=1\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecbb6d37a3cdba8068dce7764e83487a2a6296ef | 68,168 | ipynb | Jupyter Notebook | module03_research_data_in_python/03_02answer_earthquake_exercise.ipynb | mmlchang/rse-course | 86baedd4b949109efad12534ec15a9a4698e259e | [
"CC-BY-3.0"
] | null | null | null | module03_research_data_in_python/03_02answer_earthquake_exercise.ipynb | mmlchang/rse-course | 86baedd4b949109efad12534ec15a9a4698e259e | [
"CC-BY-3.0"
] | null | null | null | module03_research_data_in_python/03_02answer_earthquake_exercise.ipynb | mmlchang/rse-course | 86baedd4b949109efad12534ec15a9a4698e259e | [
"CC-BY-3.0"
] | null | null | null | 141.134576 | 57,588 | 0.892486 | [
[
[
"# Solution: the biggest Earthquake in the UK this Century",
"_____no_output_____"
],
[
"## Download the data",
"_____no_output_____"
]
],
[
[
"import requests\n\nquakes = requests.get(\n \"http://earthquake.usgs.gov/fdsnws/event/1/query.geojson\",\n params={\n \"starttime\": \"2000-01-01\",\n \"maxlatitude\": \"58.723\",\n \"minlatitude\": \"50.008\",\n \"maxlongitude\": \"1.67\",\n \"minlongitude\": \"-9.756\",\n \"minmagnitude\": \"1\",\n \"endtime\": \"2018-10-11\",\n \"orderby\": \"time-asc\",\n },\n)",
"_____no_output_____"
]
],
[
[
"## Parse the data as JSON",
"_____no_output_____"
]
],
[
[
"import json",
"_____no_output_____"
],
[
"quakes.text[0:200]",
"_____no_output_____"
],
[
"requests_json = json.loads(quakes.text)",
"_____no_output_____"
]
],
[
[
"Note that the `requests` library has native JSON support, so you could do this instead:\n`requests_json = quakes.json()`",
"_____no_output_____"
],
[
"## Investigate the data to discover how it is structured",
"_____no_output_____"
],
[
"There is no foolproof way of doing this. A good first step is to see the type of our data!",
"_____no_output_____"
]
],
[
[
"type(requests_json)",
"_____no_output_____"
]
],
[
[
"Now we can navigate through this dictionary to see how the information is stored in the nested dictionaries and lists. The `keys` method can indicate what kind of information each dictionary holds, and the `len` function tells us how many entries are contained in a list. How you explore is up to you!",
"_____no_output_____"
]
],
[
[
"requests_json.keys()",
"_____no_output_____"
],
[
"type(requests_json[\"features\"])",
"_____no_output_____"
],
[
"len(requests_json[\"features\"])",
"_____no_output_____"
],
[
"requests_json[\"features\"][0]",
"_____no_output_____"
],
[
"requests_json[\"features\"][0].keys()",
"_____no_output_____"
]
],
[
[
"It looks like the coordinates are in the `geometry` section and the magnitude is in the `properties` section.",
"_____no_output_____"
]
],
[
[
"requests_json[\"features\"][0][\"geometry\"]",
"_____no_output_____"
],
[
"requests_json[\"features\"][0][\"properties\"].keys()",
"_____no_output_____"
],
[
"requests_json[\"features\"][0][\"properties\"][\"mag\"]",
"_____no_output_____"
]
],
[
[
"## Find the largest quake",
"_____no_output_____"
]
],
[
[
"quakes = requests_json[\"features\"]",
"_____no_output_____"
],
[
"largest_so_far = quakes[0]\nfor quake in quakes:\n if quake[\"properties\"][\"mag\"] > largest_so_far[\"properties\"][\"mag\"]:\n largest_so_far = quake\nlargest_so_far[\"properties\"][\"mag\"]",
"_____no_output_____"
],
[
"lat = largest_so_far[\"geometry\"][\"coordinates\"][1]\nlong = largest_so_far[\"geometry\"][\"coordinates\"][0]\nprint(\"Latitude: {} Longitude: {}\".format(lat, long))",
"Latitude: 52.52 Longitude: -2.15\n"
]
],
[
[
"## Get a map at the point of the quake",
"_____no_output_____"
]
],
[
[
"import requests\n\n\ndef request_map_at(lat, long, satellite=True, zoom=10, size=(400, 400)):\n base = \"https://static-maps.yandex.ru/1.x/?\"\n\n params = dict(\n z=zoom,\n size=\"{},{}\".format(size[0], size[1]),\n ll=\"{},{}\".format(long, lat),\n l=\"sat\" if satellite else \"map\",\n lang=\"en_US\",\n )\n\n return requests.get(base, params=params)",
"_____no_output_____"
],
[
"map_png = request_map_at(lat, long, zoom=10, satellite=False)",
"_____no_output_____"
]
],
[
[
"## Display the map",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\n\nImage(map_png.content)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecbb74a8f33c0e6ea02bac19e9a120ae196dc364 | 20,135 | ipynb | Jupyter Notebook | site/el/guide/keras/masking_and_padding.ipynb | gmb-ftcont/docs-l10n | 8b24263ca37dbf5cb4b0c15070a3d32c7284729d | [
"Apache-2.0"
] | 1 | 2020-08-05T05:52:57.000Z | 2020-08-05T05:52:57.000Z | site/el/guide/keras/masking_and_padding.ipynb | gmb-ftcont/docs-l10n | 8b24263ca37dbf5cb4b0c15070a3d32c7284729d | [
"Apache-2.0"
] | null | null | null | site/el/guide/keras/masking_and_padding.ipynb | gmb-ftcont/docs-l10n | 8b24263ca37dbf5cb4b0c15070a3d32c7284729d | [
"Apache-2.0"
] | null | null | null | 39.403131 | 507 | 0.555798 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# \"Επικάλυψη\"(masking) και \"γέμισμα\"(padding) με την Keras\n\n<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/guide/keras/masking_and_padding\">\n <img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />\n Άνοιγμα στο TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/el/guide/keras/masking_and_padding.ipynb\">\n <img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />\n Εκτέλεση στο Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs-l10n/blob/master/site/el/guide/keras/masking_and_padding.ipynb\">\n <img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />\n Προβολή πηγαίου στο GitHub</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/el/guide/keras/masking_and_padding.ipynb\">\n <img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />\n Λήψη \"σημειωματάριου\"</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"Note: Η κοινότητα του TensorFlow έχει μεταφράσει αυτά τα έγγραφα. Καθότι οι μεταφράσεις αυτές αποτελούν την καλύτερη δυνατή προσπάθεια , δεν υπάρχει εγγύηση ότι θα παραμείνουν ενημερωμένες σε σχέση με τα [επίσημα Αγγλικά έγγραφα](https://www.tensorflow.org/?hl=en).\nΑν έχετε υποδείξεις για βελτίωση των αρχείων αυτών , δημιουργήστε ένα pull request στο [tensorflow/docs](https://github.com/tensorflow/docs) GitHub repository . Για να συμμετέχετε στη σύνταξη ή στην αναθεώρηση των μεταφράσεων της κοινότητας , επικοινωνήστε με το [[email protected] list](https://groups.google.com/a/tensorflow.org/forum/#!forum/docs).",
"_____no_output_____"
],
[
"# Διάταξη\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nimport tensorflow as tf\n\nfrom tensorflow.keras import layers",
"_____no_output_____"
]
],
[
[
"# \"Γέμισμα\" σειριακών δεδομένων\n\nΚατά την επεξεργασία ακολουθιών δεδομένων, είναι σύνηθες ορισμένα δείγματα να έχουν διαφορετικό μήκος. Για παράδειγμα (το κείμενο ομαδοποιείται κατά λέξεις) :\n\n```\n[\n [\"The\", \"weather\", \"will\", \"be\", \"nice\", \"tomorrow\"],\n [\"How\", \"are\", \"you\", \"doing\", \"today\"],\n [\"Hello\", \"world\", \"!\"]\n]\n```\nΜετά από λεξιλογική αναζήτηση , τα δεδομένα μπορούν να εμφανιστούν ως ακέραιοι :\n\n```\n[\n [83, 91, 1, 645, 1253, 927],\n [73, 8, 3215, 55, 927],\n [71, 1331, 4231]\n]\n```\n\nΤο παραπάνω πρόκειται για μία δισδιάστατη λίστα όπου ορισμένα δείγματα έχουν μήκος 6,5, και 3 αντίστοιχα. Επειδή τα δεδομένα εισόδου για ένα μοντέλο μάθησης εις βάθος πρέπει να είναι ένας τανυστής(π.χ `batch_size, 6 , vocab_size` στην παραπάνω περίπτωση), δείγματα τα οποία είναι μικρότερα από το μεγαλύτερο στοιχείο , πρέπει να \"γεμίσουν\" με κάποια συμβολική τιμή υποκατάστασης (εναλλακτικά, κάποιος μπορεί να \"κόψει\" τα μεγάλα δείγματα αντί να \"γεμίσει\" τα μικρότερα σε μήκος δείγματα).\n\nΗ Keras παρέχει ένα API μέσω του οποίου μπορείτε εύκολα να \"κόψετε\" και να \"γεμίσετε\" τις ακολουθίες δεδομένων : `tf.keras.preprocessing.sequence.pad_sequences`",
"_____no_output_____"
]
],
[
[
"raw_inputs = [\n [83, 91, 1, 645, 1253, 927],\n [73, 8, 3215, 55, 927],\n [711, 632, 71]\n]\n\n# By default, this will pad using 0s; it is configurable via the\n# \"value\" parameter.\n# Note that you could \"pre\" padding (at the beginning) or\n# \"post\" padding (at the end).\n# We recommend using \"post\" padding when working with RNN layers\n# (in order to be able to use the \n# CuDNN implementation of the layers).\npadded_inputs = tf.keras.preprocessing.sequence.pad_sequences(raw_inputs,\n padding='post')\n\nprint(padded_inputs)",
"_____no_output_____"
]
],
[
[
"# \"Επικάλυψη\"\n\nΤώρα που όλα τα δείγματα έχουν όμοιο μήκος , το μοντέλο πρέπει κάπως να ενημερωθεί ότι μέρος των δεδομένων είναι στην πραγματικότητα \"γέμισμα\" και πρέπει να αγνοηθεί. Ο μηχανισμός αυτός λέγεται \"επικάλυψη\".\n\nΥπάρχουν τρείς τρόποι εισαγωγής \"επικάλυψης\" στα μοντέλα Keras :\n\n* Προσθήκη ενός επιπέδου `keras.layers.Masking`.\n* Ρύθμιση ενός επιπέδου `keras.layers.Embedding` με `mask_zero=True.\n* Πέρασμα ενός ορίσματος `mask` χειροκίνητα , όταν καλούνται επίπεδα που το υποστηρίζουν (π.χ RNN επίπεδα).",
"_____no_output_____"
],
[
"# Επίπεδα \"επικάλυψης\" : ενσωμάτωση και \"επικάλυψη\"\n\nΣτο παρασκήνιο, αυτά τα επίπεδα θα δημιουργήσουν έναν τανιστή επικάλυψης(2D τανιστής με μορφή `(batch, sequence_length())`,και θα τον ενσωματώσουν στον τανιστή εξόδου,τον οποίο επιστρέφει το επίπεδο `Επικάλυψης` ή `Ενσωμάτωσης`.\n",
"_____no_output_____"
]
],
[
[
"embedding = layers.Embedding(input_dim=5000, output_dim=16, mask_zero=True)\nmasked_output = embedding(padded_inputs)\n\nprint(masked_output._keras_mask)",
"_____no_output_____"
],
[
"masking_layer = layers.Masking()\n# Simulate the embedding lookup by expanding the 2D input to 3D,\n# with embedding dimension of 10.\nunmasked_embedding = tf.cast(\n tf.tile(tf.expand_dims(padded_inputs, axis=-1), [1, 1, 10]),\n tf.float32)\n\nmasked_embedding = masking_layer(unmasked_embedding)\nprint(masked_embedding._keras_mask)",
"_____no_output_____"
]
],
[
[
"Η μάσκα είναι ένας δισδιάστατος boolean τανιστής με μορφή `(batch_size , sequence_length)`,όπου κάθε `Ψευδής` καταχώριση υποδεικνύει ότι το αντίστοιχο χρονικό σημείο θα πρέπει να αγνοηθεί κατά την επεξεργασία. ",
"_____no_output_____"
],
[
"# Αναπαραγωγή επικάλυψης στα Functional API και Sequential API\n\nΌταν χρησιμοποιείται το Functional ή το Sequential API , μία επικάλυψη που έχει δημιουργειθεί από ένα επίπεδο `Ενσωμάτωσης` ή `Επικάλυψης` θα αναπαραχθεί εντός του δικτύου για κάθε επίπεδο το οποίο είναι σε θέση να τη χρησιμοποιήσει(π.χ RNN επίπεδα). Η επικάλυψη που αντιστοιχεί σε μία είσοδο περνά σε κάθε επίπεδο που μπορεί να τη χρησιμοποιήσει - αυτό γίνεται αυτόματα από την Keras -.\n\nΣημειώνεται ότι στη μέθοδο `call` μίας υποκλάσσης μοντέλου ή επιπέδου , οι μάσκες δεν αναπαράγωνται αυτόματα , οπότε θα χρειαστεί να περάσει χειροκίνητα το όρισμα `mask` σε όποιο επίπεδο το χρειάζεται. Δείτε παρακάτω για λεπτομέρειες.\n\nΠ.χ , στο ακόλουθο Sequential μοντέλο , το επίπεδο `LSTM` θα δεχτεί αυτόματα τη μάσκα , οπότε θα αγνοήσει τις \"γεμισμένες\" τιμές :",
"_____no_output_____"
]
],
[
[
"model = tf.keras.Sequential([\n layers.Embedding(input_dim=5000, output_dim=16, mask_zero=True),\n layers.LSTM(32),\n])",
"_____no_output_____"
]
],
[
[
"Όμοια , για το Functional API μοντέλο : ",
"_____no_output_____"
]
],
[
[
"inputs = tf.keras.Input(shape=(None,), dtype='int32')\nx = layers.Embedding(input_dim=5000, output_dim=16, mask_zero=True)(inputs)\noutputs = layers.LSTM(32)(x)\n\nmodel = tf.keras.Model(inputs, outputs)",
"_____no_output_____"
]
],
[
[
"# Πέρασμα τανιστών \"επικάλυψης\" απευθείας σε επίπεδο\n\nΕπίπεδα τα οποία μπορούν να διαχειριστούν μάσκες(όπως το `LSTM`) έχουν όρισμα `mask` στην μέθοδο `__call__` τους.\n\nΣτο μεταξύ , επίπεδα τα οποία παράγουν μάσκες (π.χ `Ενσωμάτωσης`) εκθέτουν την μέθοδο `compute_mask(input, previous_mask)` την οποία μπορείτε να καλέσετε.\n\nΕπομένως, μπορείτε να κάνετε κάτι τέτοιο :\n",
"_____no_output_____"
]
],
[
[
"class MyLayer(layers.Layer):\n \n def __init__(self, **kwargs):\n super(MyLayer, self).__init__(**kwargs)\n self.embedding = layers.Embedding(input_dim=5000, output_dim=16, mask_zero=True)\n self.lstm = layers.LSTM(32)\n \n def call(self, inputs):\n x = self.embedding(inputs)\n # Note that you could also prepare a `mask` tensor manually.\n # It only needs to be a boolean tensor\n # with the right shape, i.e. (batch_size, timesteps).\n mask = self.embedding.compute_mask(inputs)\n output = self.lstm(x, mask=mask) # The layer will ignore the masked values\n return output\n\nlayer = MyLayer()\nx = np.random.random((32, 10)) * 100\nx = x.astype('int32')\nlayer(x)",
"_____no_output_____"
]
],
[
[
"# Υποστήριξη masking σε δικά σας layers \n\nΜερικές φορές , μπορεί να χρειαστεί να γράψετε επίπεδα τα οποία παράγουν μία μάσκα (π.χ `Ενσωμάτωσης`) ή επίπεδα τα οποία θα χρειάζεται να τροποποιούν την τρέχουσα μάσκα.\n\nΓια παράδειγμα, κάθε επίπεδο το οποίο παράγει έναν τανιστή με διαφορετική διάσταση χρόνου από ότι στην είσοδο του , όπως το επίπεδο `Συγχώνευση` (Concatenate) το οποίο συγχωνεύει στη διάσταση του χρόνου, θα χρειάζεται να τροποποιεί την τρέχουσα μάσκα έτσι ώστε τα χαμηλότερα επίπεδα να μπορούν να λάβουν υπ'όψιν τις κατάλληλες χρονοσημάνσεις.\n\nΓια να γίνει το παραπάνω,το επίπεδο σας πρέπει να υλοποιεί την μέθοδο `layer.compute_mask()`, η οποία παράγει μία νέα μάσκα , με βάση τη δοσμένη είσοδο και την τρέχουσα μάσκα\n\nΤα περισσοτερα επίπεδα δεν τροποποιούν τη διάσταση του χρόνου , επομένως δεν χρειάζεται να ανησυχείτε για την επικάλυψη. Η προεπιλεγμένη συμπεριφορά της `compute_mask()` είναι απλά να περνάει την τρέχουσα μάσκα στο δίκτυο , όταν αυτό χρειάζεται.\nΑκολουθεί παράδειγμα με το επίπεδο `TemporalSplit` το οποίο χρειάζεται να τροποποιήσει την τρέχουσα μάσκα: ",
"_____no_output_____"
]
],
[
[
"class TemporalSplit(tf.keras.layers.Layer):\n \"\"\"Split the input tensor into 2 tensors along the time dimension.\"\"\"\n\n def call(self, inputs):\n # Expect the input to be 3D and mask to be 2D, split the input tensor into 2\n # subtensors along the time axis (axis 1).\n return tf.split(inputs, 2, axis=1)\n \n def compute_mask(self, inputs, mask=None):\n # Also split the mask into 2 if it presents.\n if mask is None:\n return None\n return tf.split(mask, 2, axis=1)\n\nfirst_half, second_half = TemporalSplit()(masked_embedding)\nprint(first_half._keras_mask)\nprint(second_half._keras_mask)",
"_____no_output_____"
]
],
[
[
"Άλλο ένα παράδειγμα , ενός επιπέδου `CustomEmbedding` το οποίο είναι ικανό να παράγει μάσκα από τις δοσμένες τιμές εισόδου :",
"_____no_output_____"
]
],
[
[
"class CustomEmbedding(tf.keras.layers.Layer):\n \n def __init__(self, input_dim, output_dim, mask_zero=False, **kwargs):\n super(CustomEmbedding, self).__init__(**kwargs)\n self.input_dim = input_dim\n self.output_dim = output_dim\n self.mask_zero = mask_zero\n \n def build(self, input_shape):\n self.embeddings = self.add_weight(\n shape=(self.input_dim, self.output_dim),\n initializer='random_normal',\n dtype='float32')\n \n def call(self, inputs):\n return tf.nn.embedding_lookup(self.embeddings, inputs)\n \n def compute_mask(self, inputs, mask=None):\n if not self.mask_zero:\n return None\n return tf.not_equal(inputs, 0)\n \n \nlayer = CustomEmbedding(10, 32, mask_zero=True)\nx = np.random.random((3, 10)) * 9\nx = x.astype('int32')\n\ny = layer(x)\nmask = layer.compute_mask(x)\n\nprint(mask)",
"_____no_output_____"
]
],
[
[
"# Συγγραφή επιπέδων που χρειάζονται πληροφορίες της μάσκας\n\nΜερικά επίπεδα είναι *καταναλωτές* μασκών · δέχονται ένα όρισμα `mask` στην `call` και το χρησιμοποιούν ώστε να προσδιορίσουν αν θα αγνοήσουν συγκεκριμένες χρονοσημάνσεις.\nΓια να γράψετε είναι τέτοιο επίπεδο , μπορείτε απλά να προσθέσετε το όρισμα `mask=None` στην υπογραφή της μεθόδου `call`. Η μάσκα που συσχετίζεται με τις εισόδους θα περάσει στο επίπεδο σας όποτε είναι διαθέσιμη.\n\n```python\nclass MaskConsumer(tf.keras.layers.Layer):\n \n def call(self, inputs, mask=None):\n ...\n```\n",
"_____no_output_____"
],
[
"# Σύνοψη\n\nΑυτά αποτελούν όσα χρειάζεστε για το masking στην Keras . Για να συνοψίσουμε :\n\n* \"Masking\" είναι ο τρόπος ,με τον οποίο τα επίπεδα είναι ικανά να γνωρίζουν πότε να αγνοήσουν συγκεκριμένες χρονοσημάνσεις σε ακολουθίες δεδομένων.\n* Μερικά επίπεδα είναι γεννήτορες μασκών : το `Ενσωμάτωση` μπορεί να παράγει μάσκα από τιμές εισόδου (αν `mask_zero=True`), ομοιώς και το επίπεδο `Masking`.\n* Μερικά επίπεδα είναι καταναλωτές μασκών : \"εκθέτουν\" ένα όρισμα `mask` στην μέθοδο `__call__` τους . Αυτή είναι η περίπτωση των RNN δικτύων.\n* Στα Functional και Sequential API's , οι πληροφορίες επικάλυψης διαχέονται αυτόματα.\n* Κατά την σύνταξη υποκλάσσεων μοντέλων ή όταν χρησιμοποιούνται επίπεδα κατά αυτοτελή τρόπο , το όρισμα `mask` πρέπει να περνάει στα επίπεδα χειροκίνητα.\n* Μπορείτε εύκολα να συντάξετε επίπεδα τα οποία να τροποποιούν την τρέχουσα μάσκα, να παράγουν μία νέα μάσκα ,ή να καταναλώνουν την μάσκα που συσχετίζεται με την είσοδο. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecbb7de35d588428b3be7316b97612f1f8f62533 | 98,074 | ipynb | Jupyter Notebook | PS_Workspace/content_type_predictions.ipynb | wdolesin/Neuron_X | 4cc17582f20a972a96b2d34b8233b8474996e0e4 | [
"MIT"
] | null | null | null | PS_Workspace/content_type_predictions.ipynb | wdolesin/Neuron_X | 4cc17582f20a972a96b2d34b8233b8474996e0e4 | [
"MIT"
] | null | null | null | PS_Workspace/content_type_predictions.ipynb | wdolesin/Neuron_X | 4cc17582f20a972a96b2d34b8233b8474996e0e4 | [
"MIT"
] | null | null | null | 40.392916 | 7,053 | 0.346555 | [
[
[
"<a href=\"https://colab.research.google.com/github/puru444/Neuron_X/blob/main/content_type_predictions.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"# Imports\nimport pandas as pd\nimport numpy as np\nfrom pathlib import Path\nimport tensorflow as tf\nfrom tensorflow.keras.layers import Dense\nfrom tensorflow.keras.models import Sequential\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import StandardScaler,OneHotEncoder",
"_____no_output_____"
],
[
"# Upload the \"content_type.xlxs\" file into Colab, then store in a Pandas DataFrame\n\nfrom google.colab import files\nuploaded = files.upload()\n\ndf_content_type = pd.read_excel(\n (\"content_type.xlsx\"))\n\ndisplay(df_content_type.head(500))\ndisplay(df_content_type.tail(500))",
"_____no_output_____"
],
[
"df_content_type = df_content_type.dropna()",
"_____no_output_____"
],
[
"\n# Review the data types associated with the columns\ndf_content_type.dtypes",
"_____no_output_____"
],
[
"# Create a list of categorical variables \ncategorical_variables = list(df_content_type.dtypes[df_content_type.dtypes == \"object\"].index)\n\n# Display the categorical variables list\ncategorical_variables",
"_____no_output_____"
],
[
"#np.any(np.isnan(mat))",
"_____no_output_____"
],
[
"#np.all(np.isfinite(mat))",
"_____no_output_____"
],
[
"# Create a OneHotEncoder instance\nenc = OneHotEncoder(sparse=False)",
"_____no_output_____"
],
[
"# Encode the categorcal variables using OneHotEncoder\nencoded_data = enc.fit_transform(df_content_type[categorical_variables])",
"_____no_output_____"
],
[
"# Create a DataFrame with the encoded variables\nencoded_df = pd.DataFrame(\n encoded_data,\n columns = enc.get_feature_names(categorical_variables)\n)\n\n# Display the DataFrame\ndisplay(encoded_df.head())",
"_____no_output_____"
],
[
"# Add the numerical variables from the original DataFrame to the one-hot encoding DataFrame\nencoded_df = pd.concat([\n df_content_type.drop(columns=categorical_variables),\n encoded_df,\n], axis=1)\n\n# Review the Dataframe\nencoded_df.head()",
"_____no_output_____"
],
[
"# Define the target set y using the IS_SUCCESSFUL column\ny = encoded_df[\"Content_Type\"]\n\n# Display a sample of y\ny[:5]",
"_____no_output_____"
],
[
"# Define features set X by selecting all columns but IS_SUCCESSFUL\nX = encoded_df.drop(columns=[\"Content_Type\"])\n\n# Review the features DataFrame\ndisplay(X.head())\ndisplay(X.tail())",
"_____no_output_____"
],
[
"\n# Split the preprocessed data into a training and testing dataset\n# Assign the function a random_state equal to 1\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)",
"_____no_output_____"
],
[
"# Create a StandardScaler instance\nscaler = StandardScaler()\n\n# Fit the scaler to the features training dataset\nX_scaler = scaler.fit(X_train)\n\n# Fit the scaler to the features training dataset\nX_train_scaled = X_scaler.transform(X_train)\nX_test_scaled = X_scaler.transform(X_test)",
"_____no_output_____"
],
[
"# Define the the number of inputs (features) to the model\nnumber_input_features = X_train_scaled.shape[1]\n\n# Review the number of features\nnumber_input_features",
"_____no_output_____"
],
[
"# Define the number of neurons in the output layer\nnumber_output_neurons = 1\n\n# Review the number of neurons in the output layer\nnumber_output_neurons",
"_____no_output_____"
],
[
"# Define the number of hidden nodes for the first hidden layer\nhidden_nodes_layer1 = (number_input_features + number_output_neurons) // 2\n\n# Review the number hidden nodes in the first layer\nhidden_nodes_layer1",
"_____no_output_____"
],
[
"# Define the number of hidden nodes for the second hidden layer\nhidden_nodes_layer2 = (hidden_nodes_layer1 + number_output_neurons) // 2\n\n# Review the number hidden nodes in the second layer\nhidden_nodes_layer2",
"_____no_output_____"
],
[
"\n# Create the Sequential model instance\nnn = Sequential()\n\n\n# Add the first hidden layer\nnn.add(Dense(\n units=hidden_nodes_layer1, \n input_dim=number_input_features, \n activation=\"relu\")\n)\n\n\n# Add the second hidden layer\nnn.add(Dense(\n units=hidden_nodes_layer2, \n activation=\"relu\")\n)\n\n\n# Add the output layer to the model specifying the number of output neurons and activation function\nnn.add(Dense(\n 1, \n activation=\"sigmoid\")\n)",
"_____no_output_____"
],
[
"# Display the Sequential model summary\nnn.summary()",
"Model: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_3 (Dense) (None, 21) 903 \n_________________________________________________________________\ndense_4 (Dense) (None, 11) 242 \n_________________________________________________________________\ndense_5 (Dense) (None, 1) 12 \n=================================================================\nTotal params: 1,157\nTrainable params: 1,157\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile the Sequential model\nnn.compile(loss=\"binary_crossentropy\", optimizer=\"adam\", metrics=[\"accuracy\"])",
"_____no_output_____"
],
[
"# Fit the model using 50 epochs and the training data\nfit_model = nn.fit(X_train_scaled, y_train, epochs=50)",
"Epoch 1/50\n25/25 [==============================] - 1s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 2/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 3/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 4/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 5/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 6/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 7/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 8/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 9/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 10/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 11/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 12/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 13/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 14/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 15/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 16/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 17/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 18/50\n25/25 [==============================] - 0s 3ms/step - loss: nan - accuracy: 0.5386\nEpoch 19/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 20/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 21/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 22/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 23/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 24/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 25/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 26/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 27/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 28/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 29/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 30/50\n25/25 [==============================] - 0s 3ms/step - loss: nan - accuracy: 0.5386\nEpoch 31/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 32/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 33/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 34/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 35/50\n25/25 [==============================] - 0s 3ms/step - loss: nan - accuracy: 0.5386\nEpoch 36/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 37/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 38/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 39/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 40/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 41/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 42/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 43/50\n25/25 [==============================] - 0s 3ms/step - loss: nan - accuracy: 0.5386\nEpoch 44/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 45/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 46/50\n25/25 [==============================] - 0s 3ms/step - loss: nan - accuracy: 0.5386\nEpoch 47/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 48/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 49/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\nEpoch 50/50\n25/25 [==============================] - 0s 2ms/step - loss: nan - accuracy: 0.5386\n"
],
[
"# Evaluate the model loss and accuracy metrics using the evaluate method and the test data\nmodel_loss, model_accuracy = nn.evaluate(X_test_scaled, y_test, verbose=2)\n\n# Display the model loss and accuracy results\nprint(f\"Loss: {model_loss}, Accuracy: {model_accuracy}\")",
"9/9 - 0s - loss: nan - accuracy: 0.5115\nLoss: nan, Accuracy: 0.5115384459495544\n"
],
[
"# Define the the number of inputs (features) to the model\nnumber_input_features = len(X_train.iloc[0])\n\n# Review the number of features\nnumber_input_features",
"_____no_output_____"
],
[
"# Define the number of neurons in the output layer\nnumber_output_neurons_A1 = 1\n",
"_____no_output_____"
],
[
"# Define the number of hidden nodes for the first hidden layer\nhidden_nodes_layer1_A1 = (number_input_features + number_output_neurons_A1) // 2\n\n# Review the number of hidden nodes in the first layer\nhidden_nodes_layer1_A1",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecbb7ef0f4ab2d04e29ffab3f99b7ffe1a4a9134 | 194,175 | ipynb | Jupyter Notebook | analisis_univariado.ipynb | juan-pascual/data-science-coderhouse | bb13f592f7080ca62c754159de474fead1cf8575 | [
"MIT"
] | 1 | 2022-02-09T17:39:54.000Z | 2022-02-09T17:39:54.000Z | analisis_univariado.ipynb | juan-pascual/data-science-coderhouse | bb13f592f7080ca62c754159de474fead1cf8575 | [
"MIT"
] | 7 | 2022-02-09T17:39:06.000Z | 2022-02-24T17:48:45.000Z | analisis_univariado.ipynb | juan-pascual/data-science-coderhouse | bb13f592f7080ca62c754159de474fead1cf8575 | [
"MIT"
] | 1 | 2022-02-18T00:12:45.000Z | 2022-02-18T00:12:45.000Z | 113.685597 | 45,548 | 0.809929 | [
[
[
"# CoderHouse - Grupo 7 - Analisis Univariado\n\n* <b>Profesor:</b> David Romero Acosta\n\n* <b>Tutoria:</b> Hector Ponce Schwarz\n\n<b>Alumnos:</b>\n* Juan Pascual\n* Lucas Ariel Saavedra\n* Bartolome Oscar Meritello\n* Jose Mornaghi\n\nArrancamos el analisis importando todo lo que necesitamos y desactivamos los carteles de Warnings. Tambien, creamos las funciones que vamos a usar en todo el notebook para ahorrar codigo.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport warnings\nimport numpy as np\nfrom sklearn.impute import KNNImputer\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"data_directory = \"./data/\"\nstroke_dataset = pd.read_csv(data_directory + \"stroke.csv\")\nstroke_dataset = stroke_dataset.drop(\"id\", axis=1) #Sacamos este metadato\n\ndef plot_numerical_serie(dataset, column_name):\n fig,axes = plt.subplots(nrows=1,ncols=2,dpi=120,figsize = (8,4))\n\n plot0=sns.distplot(dataset[dataset[column_name]!=0][column_name],ax=axes[0],color='green')\n # axes[0].yaxis.set_major_formatter(FormatStrFormatter('%.3f'))\n axes[0].set_title(f'Distribution of {column_name}',fontdict={'fontsize':8})\n axes[0].set_xlabel(f'{column_name} Class',fontdict={'fontsize':7})\n axes[0].set_ylabel('Count/Dist.',fontdict={'fontsize':7})\n plt.tight_layout()\n\n plot1=sns.boxplot(dataset[dataset[column_name]!=0][column_name],ax=axes[1],orient='v')\n axes[1].set_title('Numerical Summary',fontdict={'fontsize':8})\n axes[1].set_xlabel(column_name,fontdict={'fontsize':7})\n axes[1].set_ylabel(column_name,fontdict={'fontsize':7})\n plt.tight_layout()\n\n plt.show()\n\ndef plot_pie(dataset, group_by_variable, output_variable):\n comparison = dataset.groupby(group_by_variable).count()[output_variable]\n plt.pie(x=comparison, labels=comparison.index, autopct='%1.2f%%')\n plt.title(f'{group_by_variable} comparison')\n plt.show()\n\ndef frequency_dataframe(dataset, variable):\n frec = dataset[variable].value_counts()\n frec_df = pd.DataFrame(frec)\n\n frec_df.rename(columns={variable:'Frec_abs'},inplace=True)\n\n Frec_abs_val = frec_df[\"Frec_abs\"].values\n acum = []\n valor_acum = 0\n for i in Frec_abs_val:\n valor_acum = valor_acum + i\n acum.append(valor_acum)\n\n frec_df[\"frec_abs_acum\"] = acum\n\n frec_df[\"frec_rel_%\"] = round(100 * frec_df[\"Frec_abs\"]/len(stroke_dataset.smoking_status),4)\n\n Frec_rel_val = frec_df[\"frec_rel_%\"].values\n\n acum = []\n\n valor_acum = 0\n\n for i in Frec_rel_val:\n valor_acum = valor_acum + i\n acum.append(valor_acum)\n\n frec_df[\"frec_rel_%_acum\"] = acum\n \n return frec_df\n \nstroke_dataset",
"_____no_output_____"
],
[
"stroke_dataset.shape",
"_____no_output_____"
]
],
[
[
"## Datos imputados",
"_____no_output_____"
]
],
[
[
"stroke_dataset.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 5110 entries, 0 to 5109\nData columns (total 11 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 gender 5110 non-null object \n 1 age 5110 non-null float64\n 2 hypertension 5110 non-null int64 \n 3 heart_disease 5110 non-null int64 \n 4 ever_married 5110 non-null object \n 5 work_type 5110 non-null object \n 6 Residence_type 5110 non-null object \n 7 avg_glucose_level 5110 non-null float64\n 8 bmi 4909 non-null float64\n 9 smoking_status 5110 non-null object \n 10 stroke 5110 non-null int64 \ndtypes: float64(3), int64(3), object(5)\nmemory usage: 439.3+ KB\n"
],
[
"stroke_dataset.isna().sum()",
"_____no_output_____"
]
],
[
[
"Observamos que hay valores de BMI (Indice de masa corporal) que no tenemos disponibles. Veamos que descripciones estadísticas obtenemos y luego imputamos los datos faltantes usando el metodo de KNN.",
"_____no_output_____"
]
],
[
[
"stroke_dataset.describe().T",
"_____no_output_____"
],
[
"imputador = KNNImputer(n_neighbors=3, weights=\"uniform\")\nstroke_dataset['imputed_bmi'] = imputador.fit_transform(stroke_dataset[['bmi']])\nstroke_dataset.isna().sum()",
"_____no_output_____"
],
[
"stroke_dataset.describe()[[\"bmi\", \"imputed_bmi\"]].T",
"_____no_output_____"
]
],
[
[
"Imputar no nos afecta el análisis y no perdemos filas. Guardamos el dataset para usarlo a futuro.",
"_____no_output_____"
]
],
[
[
"stroke_dataset = stroke_dataset.drop(\"bmi\", axis = 1)\nstroke_dataset = stroke_dataset.rename(columns={'imputed_bmi': 'bmi'})\nstroke_dataset = stroke_dataset.drop(3116)\nstroke_dataset.to_csv(data_directory + \"stroke_imputed.csv\", index=False)\nstroke_dataset",
"_____no_output_____"
]
],
[
[
"## Análisis Univariado\n\nHacemos una descripción de cada una de las variables cuantitativas haciendo un histograma y un boxplot a cada una. A las cualitativas hicimos un grafico Pie.",
"_____no_output_____"
]
],
[
[
"stroke_dataset.bmi.describe()",
"_____no_output_____"
],
[
"plot_numerical_serie(stroke_dataset, 'bmi')",
"_____no_output_____"
]
],
[
[
"Los analisis de bmi indican que los valores sobre 35 es obesidad extrema. Hay que analizar en el dataset si puede que algunos valores esten mal ingresados o no tengan sentido.",
"_____no_output_____"
]
],
[
[
"plot_pie(stroke_dataset, 'gender', 'stroke')",
"_____no_output_____"
],
[
"plot_pie(stroke_dataset, 'hypertension', 'stroke')",
"_____no_output_____"
],
[
"plot_numerical_serie(stroke_dataset, 'age')",
"_____no_output_____"
],
[
"plot_numerical_serie(stroke_dataset, 'avg_glucose_level')",
"_____no_output_____"
]
],
[
[
"# Distribución de Frecuencias",
"_____no_output_____"
]
],
[
[
"frequency_dataframe(stroke_dataset, 'smoking_status')",
"_____no_output_____"
],
[
"frequency_dataframe(stroke_dataset, 'gender')",
"_____no_output_____"
],
[
"frequency_dataframe(stroke_dataset, 'hypertension')",
"_____no_output_____"
],
[
"frequency_dataframe(stroke_dataset, 'heart_disease')",
"_____no_output_____"
],
[
"frequency_dataframe(stroke_dataset, 'ever_married')",
"_____no_output_____"
],
[
"frequency_dataframe(stroke_dataset, 'work_type')",
"_____no_output_____"
],
[
"frequency_dataframe(stroke_dataset, 'Residence_type')",
"_____no_output_____"
],
[
"frequency_dataframe(stroke_dataset, 'stroke')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecbb85836c804df80fb2892ac7dc8c627f152827 | 90,600 | ipynb | Jupyter Notebook | lectures/ml/overfitting/04_reducir_overfitting.ipynb | dannyowen1/mat281_portfolio_dannyowen1 | c7dbcfc5f7724d8bef1d48d2381f9c065a6d6f20 | [
"MIT"
] | null | null | null | lectures/ml/overfitting/04_reducir_overfitting.ipynb | dannyowen1/mat281_portfolio_dannyowen1 | c7dbcfc5f7724d8bef1d48d2381f9c065a6d6f20 | [
"MIT"
] | null | null | null | lectures/ml/overfitting/04_reducir_overfitting.ipynb | dannyowen1/mat281_portfolio_dannyowen1 | c7dbcfc5f7724d8bef1d48d2381f9c065a6d6f20 | [
"MIT"
] | null | null | null | 87.0317 | 52,268 | 0.788256 | [
[
[
"# Reducir el overfitting\n\n\nAlgunas de las técnicas que podemos utilizar para reducir el overfitting, son:\n\n* Recolectar más datos.\n* Introducir una penalización a la complejidad con alguna técnica de regularización.\n* Utilizar modelos ensamblados.\n* Utilizar validación cruzada.\n* Optimizar los parámetros del modelo con *grid search*.\n* Reducir la dimensión de los datos.\n* Aplicar técnicas de selección de atributos.\n\n\nVeremos ejemplos de algunos métodos para reducir el sobreajuste (overfitting).",
"_____no_output_____"
],
[
"## Validación cruzada\n\nLa **validación cruzada** se inicia mediante el fraccionamiento de un conjunto de datos en un número $k$ de particiones (generalmente entre 5 y 10) llamadas *pliegues*.\n\nLa validación cruzada luego itera entre los datos de *evaluación* y *entrenamiento* $k$ veces, de un modo particular. En cada iteración de la validación cruzada, un *pliegue* diferente se elige como los datos de *evaluación*. En esta iteración, los otros *pliegues* $k-1$ se combinan para formar los datos de *entrenamiento*. Por lo tanto, en cada iteración tenemos $(k-1) / k$ de los datos utilizados para el *entrenamiento* y $1 / k$ utilizado para la *evaluación*.\n\nCada iteración produce un modelo, y por lo tanto una estimación del rendimiento de la *generalización*, por ejemplo, una estimación de la precisión. Una vez finalizada la validación cruzada, todos los ejemplos se han utilizado sólo una vez para *evaluar* pero $k -1$ veces para *entrenar*. En este punto tenemos estimaciones de rendimiento de todos los *pliegues* y podemos calcular la media y la desviación estándar de la precisión del modelo. \n\n\n<img src=\"./images/validacion_cruzada.png\" width=\"550\" height=\"650\" align=\"center\"/>\n\n",
"_____no_output_____"
],
[
"Veamos un ejemplo en python, ocupando el conjunto de datos **make_classification**.",
"_____no_output_____"
]
],
[
[
"# librerias \n\nimport pandas as pd\nimport numpy as np \nimport matplotlib.pyplot as plt \nimport seaborn as sns \nfrom sklearn.model_selection import train_test_split\nfrom sklearn.datasets import make_classification\nfrom sklearn.svm import SVC\nfrom sklearn.tree import DecisionTreeClassifier,DecisionTreeRegressor\nimport random\n\nrandom.seed(1982) # semilla\n\n# graficos incrustados\n%matplotlib inline\n\n# parametros esteticos de seaborn\nsns.set_palette(\"deep\", desat=.6)\nsns.set_context(rc={\"figure.figsize\": (12, 4)})",
"_____no_output_____"
],
[
"# Ejemplo en python - árboles de decisión\n# dummy data con 100 atributos y 2 clases\nX, y = make_classification(10000, 100, n_informative=3, n_classes=2,\n random_state=1982)\n\n# separ los datos en train y eval\nx_train, x_eval, y_train, y_eval = train_test_split(X, y, test_size=0.35, \n train_size=0.65,\n random_state=1982)\n\n# Grafico de ajuste del árbol de decisión\ntrain_prec = []\neval_prec = []\nmax_deep_list = list(range(2, 20))",
"_____no_output_____"
],
[
"# Ejemplo cross-validation\nfrom sklearn.model_selection import cross_validate,StratifiedKFold\n\n# creando pliegues\n\nskf = StratifiedKFold(n_splits=20)\nprecision = []\nmodel = DecisionTreeClassifier(criterion='entropy', max_depth=5)\n\nskf.get_n_splits(x_train, y_train)\nfor k, (train_index, test_index) in enumerate(skf.split(X, y)):\n X_train, X_test = X[train_index], X[test_index]\n y_train, y_test = y[train_index], y[test_index]\n model.fit(X_train,y_train) \n score = model.score(X_test,y_test)\n precision.append(score)\n print('Pliegue: {0:}, Dist Clase: {1:}, Prec: {2:.3f}'.format(k+1,\n np.bincount(y_train), score))\n ",
"Pliegue: 1, Dist Clase: [4763 4737], Prec: 0.928\nPliegue: 2, Dist Clase: [4763 4737], Prec: 0.914\nPliegue: 3, Dist Clase: [4763 4737], Prec: 0.914\nPliegue: 4, Dist Clase: [4763 4737], Prec: 0.938\nPliegue: 5, Dist Clase: [4763 4737], Prec: 0.922\nPliegue: 6, Dist Clase: [4763 4737], Prec: 0.938\nPliegue: 7, Dist Clase: [4763 4737], Prec: 0.924\nPliegue: 8, Dist Clase: [4762 4738], Prec: 0.938\nPliegue: 9, Dist Clase: [4762 4738], Prec: 0.936\nPliegue: 10, Dist Clase: [4762 4738], Prec: 0.908\nPliegue: 11, Dist Clase: [4762 4738], Prec: 0.936\nPliegue: 12, Dist Clase: [4762 4738], Prec: 0.938\nPliegue: 13, Dist Clase: [4762 4738], Prec: 0.934\nPliegue: 14, Dist Clase: [4762 4738], Prec: 0.920\nPliegue: 15, Dist Clase: [4762 4738], Prec: 0.930\nPliegue: 16, Dist Clase: [4762 4738], Prec: 0.928\nPliegue: 17, Dist Clase: [4762 4738], Prec: 0.924\nPliegue: 18, Dist Clase: [4762 4738], Prec: 0.926\nPliegue: 19, Dist Clase: [4762 4738], Prec: 0.936\nPliegue: 20, Dist Clase: [4762 4738], Prec: 0.920\n"
]
],
[
[
"En este ejemplo, utilizamos el iterador `StratifiedKFold` que nos proporciona Scikit-learn. Este iterador es una versión mejorada de la validación cruzada, ya que cada *pliegue* va a estar estratificado para mantener las proporciones entre las *clases* del conjunto de datos original, lo que suele dar mejores estimaciones del sesgo y la varianza del modelo. \n\nTambién podríamos utilizar `cross_val_score` que ya nos proporciona los resultados de la precisión que tuvo el modelo en cada *pliegue*.",
"_____no_output_____"
]
],
[
[
"# Ejemplo con cross_val_score\nfrom sklearn.model_selection import cross_val_score\n\n# separ los datos en train y eval\nx_train, x_eval, y_train, y_eval = train_test_split(X, y, test_size=0.35, \n train_size=0.65,\n random_state=1982)\n\n\nmodel = DecisionTreeClassifier(criterion='entropy',\n max_depth=5)\n\n\nprecision = cross_val_score(estimator=model,\n X=x_train,\n y=y_train,\n cv=20)",
"_____no_output_____"
],
[
"precision = [round(x,2) for x in precision]\nprint('Precisiones: {} '.format(precision))\nprint('Precision promedio: {0: .3f} +/- {1: .3f}'.format(np.mean(precision),\n np.std(precision)))",
"Precisiones: [0.93, 0.94, 0.92, 0.94, 0.93, 0.9, 0.92, 0.94, 0.94, 0.93, 0.94, 0.92, 0.91, 0.9, 0.94, 0.94, 0.93, 0.93, 0.93, 0.94] \nPrecision promedio: 0.928 +/- 0.013\n"
]
],
[
[
"### Más datos y curvas de aprendizaje\n\n* Muchas veces, reducir el Sobreajuste es tan fácil como conseguir más datos, dame más datos y te predeciré el futuro!. \n* En la vida real nunca es una tarea tan sencilla conseguir más datos. \n* Una técnica para reducir el sobreajuste son las *curvas de aprendizaje*, las cuales grafican la precisión en función del tamaño de los datos de entrenamiento. \n\n<img alt=\"Curva de aprendizaje\" title=\"Curva de aprendizaje\" src=\"./images/curva_aprendizaje.png\" width=\"600\" height=\"600\" >",
"_____no_output_____"
],
[
"Para graficar las curvas de aprendizaje es necesario ocupar el comando de sklearn llamado `learning_curve`.",
"_____no_output_____"
]
],
[
[
"# Ejemplo Curvas de aprendizaje\nfrom sklearn.model_selection import learning_curve\n\ntrain_sizes, train_scores, test_scores = learning_curve(\n estimator=model,\n X=x_train,\n y=y_train, \n train_sizes=np.linspace(0.1, 1.0, 20),\n cv=10,\n n_jobs=-1\n )\n\n# calculo de metricas\ntrain_mean = np.mean(train_scores, axis=1)\ntrain_std = np.std(train_scores, axis=1)\ntest_mean = np.mean(test_scores, axis=1)\ntest_std = np.std(test_scores, axis=1)",
"_____no_output_____"
]
],
[
[
"Veamos que el comando `learning_curve` va creando conjunto de datos, pero de distintos tamaños.",
"_____no_output_____"
]
],
[
[
"# tamano conjunto de entrenamiento\nfor k in range(len(train_sizes)):\n print('Tamaño Conjunto {}: {}'.format(k+1,train_sizes[k]))",
"Tamaño Conjunto 1: 585\nTamaño Conjunto 2: 862\nTamaño Conjunto 3: 1139\nTamaño Conjunto 4: 1416\nTamaño Conjunto 5: 1693\nTamaño Conjunto 6: 1970\nTamaño Conjunto 7: 2247\nTamaño Conjunto 8: 2524\nTamaño Conjunto 9: 2801\nTamaño Conjunto 10: 3078\nTamaño Conjunto 11: 3356\nTamaño Conjunto 12: 3633\nTamaño Conjunto 13: 3910\nTamaño Conjunto 14: 4187\nTamaño Conjunto 15: 4464\nTamaño Conjunto 16: 4741\nTamaño Conjunto 17: 5018\nTamaño Conjunto 18: 5295\nTamaño Conjunto 19: 5572\nTamaño Conjunto 20: 5850\n"
]
],
[
[
"Finalmente, graficamos las precisiones tanto para el conjunto de entranamiento como de evaluación para los distintos conjuntos de datos generados. ",
"_____no_output_____"
]
],
[
[
"# graficando las curvas\nplt.figure(figsize=(12,8))\n\nplt.plot(train_sizes, train_mean, color='r', marker='o', markersize=5,\n label='entrenamiento')\nplt.fill_between(train_sizes, train_mean + train_std, \n train_mean - train_std, alpha=0.15, color='r')\nplt.plot(train_sizes, test_mean, color='b', linestyle='--', \n marker='s', markersize=5, label='evaluacion')\nplt.fill_between(train_sizes, test_mean + test_std, \n test_mean - test_std, alpha=0.15, color='b')\nplt.grid()\nplt.title('Curva de aprendizaje')\nplt.legend(loc='upper right')\nplt.xlabel('Cant de ejemplos de entrenamiento')\nplt.ylabel('Precision')\nplt.show()",
"_____no_output_____"
]
],
[
[
"En este gráfico podemos concluir que:\n\n* Con pocos datos la precisión entre los datos de entrenamiento y los de evaluación son muy distintas y luego a medida que la cantidad de datos va aumentando, el modelo puede generalizar mucho mejor y las precisiones se comienzan a emparejar. \n\n\n* Este gráfico también puede ser importante a la hora de decidir invertir en la obtención de más datos, ya que por ejemplo nos indica que a partir las 2500 muestras, el modelo ya no gana mucha más precisión a pesar de obtener más datos.",
"_____no_output_____"
],
[
"## Optimización de parámetros con Grid Search\n\nLa mayoría de los modelos de Machine Learning cuentan con varios parámetros para ajustar su comportamiento, por lo tanto, otra alternativa que tenemos para reducir el Sobreajuste es optimizar estos parámetros por medio de un proceso conocido como **grid search** e intentar encontrar la combinación ideal que nos proporcione mayor precisión.\n\nEl enfoque que utiliza *grid search* es bastante simple, se trata de una búsqueda exhaustiva por el paradigma de fuerza bruta en el que se especifica una lista de valores para diferentes parámetros, y la computadora evalúa el rendimiento del modelo para cada combinación de éstos parámetros para obtener el conjunto óptimo que nos brinda el mayor rendimiento. \n\n\n<img alt=\"Curva de aprendizaje\" title=\"Curva de aprendizaje\" src=\"images/hyper.png\" width=\"600\" height=\"500\" >",
"_____no_output_____"
]
],
[
[
"# Ejemplo de grid search con SVM.\nfrom sklearn.model_selection import GridSearchCV\n\n# creación del modelo\nmodel = DecisionTreeClassifier()\n\n# rango de parametros\nrango_criterion = ['gini','entropy']\nrango_max_depth =np.array( [4,5,6,7,8,9,10,11,12,15,20,30,40,50,70,90,120,150])\nparam_grid = dict(criterion=rango_criterion, max_depth=rango_max_depth)\nparam_grid",
"_____no_output_____"
],
[
"# aplicar greed search\n\ngs = GridSearchCV(estimator=model, \n param_grid=param_grid, \n scoring='accuracy',\n cv=5,\n n_jobs=-1)\n\ngs = gs.fit(x_train, y_train)",
"_____no_output_____"
],
[
"# imprimir resultados\nprint(gs.best_score_)\nprint(gs.best_params_)",
"0.9329230769230769\n{'criterion': 'entropy', 'max_depth': 6}\n"
],
[
"# utilizando el mejor modelo\nmejor_modelo = gs.best_estimator_\nmejor_modelo.fit(x_train, y_train)\nprint('Precisión: {0:.3f}'.format(mejor_modelo.score(x_eval, y_eval)))",
"Precisión: 0.938\n"
]
],
[
[
"En este ejemplo, primero utilizamos el objeto `GridSearchCV` que nos permite realizar *grid search* junto con validación cruzada, luego comenzamos a ajustar el modelo con las diferentes combinaciones de los valores de los parámetros `criterion` y `max_depth`. Finalmente imprimimos el mejor resultado de precisión y los valores de los parámetros que utilizamos para obtenerlos; por último utilizamos este mejor modelo para realizar las predicciones con los datos de *evaluación*. \n\nPodemos ver que la precisión que obtuvimos con los datos de evaluación es casi idéntica a la que nos indicó *grid search*, lo que indica que el modelo *generaliza* muy bien.\n",
"_____no_output_____"
],
[
"## Reducción de dimensionalidad\n\nLa **reducción de dimensiones** es frecuentemente usada como una etapa de preproceso en el entrenamiento de\nsistemas, y consiste en escoger un subconjunto de\nvariables, de tal manera, que el espacio de características\nquede óptimamente reducido de acuerdo a un criterio de\nevaluación, cuyo fin es distinguir el subconjunto que\nrepresenta mejor el espacio inicial de entrenamiento.\n\nComo cada característica que se incluye en el análisis,\npuede incrementar el costo y el tiempo de proceso de los\nsistemas, hay una fuerte motivación para diseñar e\nimplementar sistemas con pequeños conjuntos de\ncaracterísticas. Sin dejar de lado, que al mismo tiempo,\nhay una opuesta necesidad de incluir un conjunto\nsuficiente de características para lograr un alto\nrendimiento. \n\n\nLa reducción de dimensionalidad se puede separar en dos tipos: **Extracción de atributos** y **Selección de aributos**.\n\n### Extracción de atributos\n\nLa **extracción de atributos** comienza a partir de un conjunto inicial de datos medidos y crea valores derivados (características) destinados a ser informativos y no redundantes, lo que facilita los pasos de aprendizaje y generalización posteriores, y en algunos casos conduce a a mejores interpretaciones humanas. \n\nCuando los datos de entrada a un algoritmo son demasiado grandes para ser procesados y se sospecha que son redundantes (por ejemplo, la misma medición en pies y metros, o la repetitividad de las imágenes presentadas como píxeles), entonces se puede transformar en un conjunto reducido de características (también denominado un vector de características). \n\n\nEstos algoritmos fueron analizados con profundidad en la sección de **Análisis no supervisados - Reducción de la dimensionalidad**.",
"_____no_output_____"
],
[
"### Selección de atributos\n\nProceso por el cual seleccionamos un subconjunto de atributos (representados por cada una de las columnas en un datasetde forma tabular) que son más relevantes para la construcción del modelo predictivo sobre el que estamos trabajando. \n\n\nEl objetivo de la selección de atributos es : \n* mejorar la capacidad predictiva de nuestro modelo, \n* proporcionando modelos predictivos más rápidos y eficientes, \n* proporcionar una mejor comprensión del proceso subyacente que generó los datos. \n \n \nLos métodos de selección de atributos se pueden utilizar para identificar y eliminar los atributos innecesarios, irrelevantes y redundantes que no contribuyen a la exactitud del modelo predictivo o incluso puedan disminuir su precisión. \n\n\n",
"_____no_output_____"
],
[
"**Algoritmos para selección de atributos**\n\nPodemos encontrar dos clases generales de algoritmos de [selección de atributos](https://en.wikipedia.org/wiki/Feature_selection): los métodos de filtrado, y los métodos empaquetados.\n\n* **Métodos de filtrado**: Estos métodos aplican una medida estadística para asignar una puntuación a cada atributo. Los atributos luego son clasificados de acuerdo a su puntuación y son, o bien seleccionados para su conservación o eliminados del conjunto de datos. Los métodos de filtrado son a menudo [univariantes](https://en.wikipedia.org/wiki/Univariate_analysis) y consideran a cada atributo en forma independiente, o con respecto a la variable dependiente.\n * Ejemplos : [prueba de Chi cuadrado](https://es.wikipedia.org/wiki/Prueba_%CF%87%C2%B2), [prueba F de Fisher](https://es.wikipedia.org/wiki/Prueba_F_de_Fisher), [ratio de ganancia de información](https://en.wikipedia.org/wiki/Information_gain_ratio) y los [coeficientes de correlación](https://es.wikipedia.org/wiki/Correlaci%C3%B3n).\n \n\n* **Métodos empaquetados**: Estos métodos consideran la selección de un conjunto de atributos como un problema de búsqueda, en donde las diferentes combinaciones son evaluadas y comparadas. Para hacer estas evaluaciones se utiliza un modelo predictivo y luego se asigna una puntuación a cada combinación basada en la precisión del modelo.\n * Un ejemplo de este método es el algoritmo de eliminación recursiva de atributos.",
"_____no_output_____"
],
[
"Un método popular en sklearn es el método **SelectKBest**, el cual selecciona las características de acuerdo con las $k$ puntuaciones más altas (de acuerdo al criterio escogido). \n\nPara entender este conceptos, transformemos el conjunto de datos anterior a formato pandas DataFrame.",
"_____no_output_____"
]
],
[
[
"# Datos\nX, y = make_classification(10000, 100, n_informative=3, n_classes=2,\n random_state=1982)\n\ndf = pd.DataFrame(X)\ndf.columns = [f'V{k}' for k in range(1,X.shape[1]+1)]\ndf['y']=y\ndf.head()",
"_____no_output_____"
]
],
[
[
"Comencemos con un simple algoritmo [univariante](https://en.wikipedia.org/wiki/Univariate_analysis) que aplica el método de filtrado. Para esto vamos a utilizar los objetos `SelectKBest` y `f_classif` del paquete `sklearn.feature_selection`.\n\nEste algoritmo selecciona a los mejores atributos basándose en una prueba estadística [univariante](https://en.wikipedia.org/wiki/Univariate_analysis). Al objeto `SelectKBest` le pasamos la prueba estadística que vamos a a aplicar, en este caso una [prueba F](https://es.wikipedia.org/wiki/Prueba_F_de_Fisher) definida por el objeto `f_classif`, junto con el número de atributos a seleccionar. El algoritmo va a aplicar la prueba a todos los atributos y va a seleccionar los que mejor resultado obtuvieron.\n",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_selection import SelectKBest\nfrom sklearn.feature_selection import f_classif",
"_____no_output_____"
],
[
"# Separamos las columnas objetivo\nx_training = df.drop(['y',], axis=1)\ny_training = df['y']\n\n# Aplicando el algoritmo univariante de prueba F.\nk = 15 # número de atributos a seleccionar\ncolumnas = list(x_training.columns.values)\nseleccionadas = SelectKBest(f_classif, k=k).fit(x_training, y_training)",
"_____no_output_____"
],
[
"catrib = seleccionadas.get_support()\natributos = [columnas[i] for i in list(catrib.nonzero()[0])]\natributos",
"_____no_output_____"
]
],
[
[
"Como podemos ver, el algoritmo nos seleccionó la cantidad de atributos que le indicamos; en este ejemplo decidimos seleccionar solo 15; obviamente, cuando armemos nuestro modelo final vamos a tomar un número mayor de atributos. Ahora se procederá a comparar los resultados de entrenar un modelo en particular con todas las variables y el subconjunto de variables seleccionadas.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.linear_model import LogisticRegression\nfrom metrics_classification import summary_metrics",
"_____no_output_____"
],
[
"%%timeit\n# Entrenamiento con todas las variables \nX = df.drop('y',axis = 1)\nY = df['y']\n\n# split dataset\nX_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2, random_state = 2) \n\n# Creando el modelo\nrlog = LogisticRegression()\nrlog.fit(X_train, Y_train) # ajustando el modelo\n\npredicciones = rlog.predict(X_test)\n\ndf_pred = pd.DataFrame({\n 'y':Y_test,\n 'yhat':predicciones\n})\n\ndf_s1 = summary_metrics(df_pred).assign(name = 'Todas las variables')",
"82.4 ms ± 6.51 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)\n"
],
[
"%%timeit\n# Entrenamiento con las variables seleccionadas\nX = df[atributos]\nY = df['y']\n\n# split dataset\nX_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2, random_state = 2) \n\n# Creando el modelo\nrlog = LogisticRegression()\nrlog.fit(X_train, Y_train) # ajustando el modelo\n\npredicciones = rlog.predict(X_test)\n\ndf_pred = pd.DataFrame({\n 'y':Y_test,\n 'yhat':predicciones\n})\n\ndf_s2 = summary_metrics(df_pred).assign(name = 'Variables Seleccionadas')",
"60.4 ms ± 8.62 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)\n"
]
],
[
[
"Juntando ambos resultados:",
"_____no_output_____"
]
],
[
[
"# juntar resultados en formato dataframe\npd.concat([df_s1,df_s2])",
"_____no_output_____"
]
],
[
[
"Las métricas para ambos casos son parecidas y el tiempo de ejecución del modelo con menos variable resulta ser menor (lo cual era algo esperable). Lo cual nos muestra que trabajando con menos variables, se puede captar las características más relevante del problema, y en la medida que se trabaje con más datos, las mejoras a nivel de capacidad de cómputo tendrán un mejor desempeño.",
"_____no_output_____"
],
[
"## Referencia\n\n1. [K-Fold Cross Validation](https://medium.com/datadriveninvestor/k-fold-cross-validation-6b8518070833)\n2. [Cross Validation and Grid Search for Model Selection in Python](https://stackabuse.com/cross-validation-and-grid-search-for-model-selection-in-python/)\n3. [Feature selection for supervised models using SelectKBest](https://www.kaggle.com/jepsds/feature-selection-using-selectkbest?utm_campaign=News&utm_medium=Community&utm_source=DataCamp.com)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecbb892678ea2e300567b75cbb5798f9db0cfe2f | 17,503 | ipynb | Jupyter Notebook | notebooks/course/chapter7/section2_tf.ipynb | yvr1037/transformers-of-Dian | 759a867e64c725e17a7aef37c78dd54b4c1fda81 | [
"Apache-2.0"
] | null | null | null | notebooks/course/chapter7/section2_tf.ipynb | yvr1037/transformers-of-Dian | 759a867e64c725e17a7aef37c78dd54b4c1fda81 | [
"Apache-2.0"
] | null | null | null | notebooks/course/chapter7/section2_tf.ipynb | yvr1037/transformers-of-Dian | 759a867e64c725e17a7aef37c78dd54b4c1fda81 | [
"Apache-2.0"
] | null | null | null | 25.589181 | 186 | 0.483174 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecbbaa1f2bce13908070daa13fab17abb54f88a9 | 130,592 | ipynb | Jupyter Notebook | jupyter_test/.ipynb_checkpoints/Stock_Price_Sample-checkpoint.ipynb | seunghyun-lee/ML-02_stock_prediction | c5984118e63478861b501d93346fea951b8c1d1a | [
"MIT"
] | null | null | null | jupyter_test/.ipynb_checkpoints/Stock_Price_Sample-checkpoint.ipynb | seunghyun-lee/ML-02_stock_prediction | c5984118e63478861b501d93346fea951b8c1d1a | [
"MIT"
] | null | null | null | jupyter_test/.ipynb_checkpoints/Stock_Price_Sample-checkpoint.ipynb | seunghyun-lee/ML-02_stock_prediction | c5984118e63478861b501d93346fea951b8c1d1a | [
"MIT"
] | null | null | null | 267.606557 | 61,152 | 0.913027 | [
[
[
"# import packages\nImport all of necessary pakages we need, which include NunPy, Pandas, Matplotlib, Scikit-learn.",
"_____no_output_____"
]
],
[
[
"# import essencial packages\nimport numpy as np\nimport pandas as pd\n\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nimport seaborn as seabornInstance\nfrom datetime import datetime, timedelta\n\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.linear_model import LinearRegression, Ridge, Lasso\nfrom sklearn import metrics",
"_____no_output_____"
]
],
[
[
"# Loading Finance Dateset - Disney from YahooFinance\nUse Pandas to read in the data and set the independent variable to be the Date. and see the first 5 rows data.",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('DIS.csv')\n# converts string to datetime\ndf['Date'] = pd.to_datetime(df.Date, format='%Y-%m-%d')\n# set the index of dataset to date column\ndf.set_index('Date', inplace=True)\ndf.tail()",
"_____no_output_____"
]
],
[
[
"Let's take a look at how the prices looks like each day since the beginning up to this point with the date on the x-axis and the price on the y-axis.",
"_____no_output_____"
]
],
[
[
"df['Adj Close'].plot(label='DIS', figsize=(16,8), title='Adjusted Closing Price', grid=True)",
"_____no_output_____"
]
],
[
[
"# Preprocessing & Cross Validation",
"_____no_output_____"
]
],
[
[
"# number of how many days to forecast\nforecast_out = 30\n# Add new label here, we want to predict the 'Adj Close'\ndf['Prediction'] = df['Adj Close'].shift(-forecast_out)\nx = np.array(df.drop(['Prediction'], 1))\n# Scale the x so that eveyone can have the same distribution for linear regression\n# x = preprocessing.scale(x)\n# Finally wa want to find data se\n# x_lately = x[-forecast_out:]\nx = x[:-forecast_out]\n# separate label and identify it as y\ny = np.array(df['Prediction'])\ny = y[:-forecast_out]",
"_____no_output_____"
]
],
[
[
"# Training set & data set for test\nWe split 80% of the data to training set while 20% of the data to test set using below code.",
"_____no_output_____"
]
],
[
[
"x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.2)",
"_____no_output_____"
]
],
[
[
"# Model Generation\nNow lets train our model.",
"_____no_output_____"
]
],
[
[
"# train model\nlinear_model = LinearRegression()\nlinear_model.fit(x_train, y_train)\nprint(linear_model.coef_, linear_model.intercept_)\n\nridge_model = Ridge()\nridge_model.fit(x_train, y_train)\n\nlasso_model = Lasso()\nlasso_model.fit(x_train, y_train)",
"[-1.64471965e-01 -5.23999137e-02 1.57313798e-01 -5.49909950e-01\n 1.59157185e+00 5.91703323e-09] 5.5497244628740106\n"
]
],
[
[
"# Evaluation\nthe regression model has to find the most optimal coefficients for all the attributes. To see what coefficients our regression model has chosen, execute the following script:",
"_____no_output_____"
]
],
[
[
"# each models score\nlinear_model_score = linear_model.score(x_test, y_test)\nprint('LinearModel Score:', linear_model_score)\n\nridge_model_score = ridge_model.score(x_test, y_test)\nprint('RidgeModel Score:', ridge_model_score)\n\nlasso_model_score = lasso_model.score(x_test, y_test)\nprint('LassoModel Score:', lasso_model_score)",
"LinearModel Score: 0.9674065173546317\nRidgeModel Score: 0.967408030170158\nLassoModel Score: 0.9672436582484243\n"
]
],
[
[
"# Prediction\nlet's do prediction on test data.",
"_____no_output_____"
]
],
[
[
"x_forecast = np.array(df.drop(['Prediction'], 1))[-forecast_out:]\nlinear_model_forecast_prediction = linear_model.predict(x_forecast)\nridge_model_forecast_prediction = ridge_model.predict(x_forecast)\nlasso_model_forecast_prediction = lasso_model.predict(x_forecast)\nx_forecast",
"_____no_output_____"
]
],
[
[
"# Plotting the Prediction\nBased on the forecast, we will visualize the plot with our existing historical data. This will help us visualize how the model fares to predict future stocks pricing.",
"_____no_output_____"
]
],
[
[
"predicted_dates = []\nrecent_date = df.index.max()\nfor i in range(forecast_out):\n recent_date += timedelta(days=1)\n predicted_dates.append(recent_date)\n \ndf = df[-365:]\n# plt.title('Adjusted Closing Price')\n# plt.xlabel('Date')\n# plt.ylabel('Price')\nplt.figure(figsize=(16,8))\nplt.grid(True)\nplt.plot(df.index, df['Adj Close'], label='Disney')\nplt.plot(predicted_dates, linear_model_forecast_prediction, label='Linear Regression')\nplt.plot(predicted_dates, ridge_model_forecast_prediction, label='Ridge')\nplt.plot(predicted_dates, lasso_model_forecast_prediction, label='Lasso')\nplt.legend()",
"_____no_output_____"
]
],
[
[
"참고:\nhttps://tbacking.com/2017/08/18/%EC%88%9C%ED%99%98-%EC%8B%A0%EA%B2%BD%EB%A7%9D-lstm-%ED%99%9C%EC%9A%A9-%EC%A3%BC%EA%B0%80-%EC%98%88%EC%B8%A1/\nhttps://m.blog.naver.com/wideeyed/221160038616\nhttps://github.com/kairess/stock_crypto_price_prediction/blob/master/stock_samsung.ipynb\nhttps://github.com/kdsian/STOCK/blob/master/RNN_STOCK_seq_result_0717_rev4.ipynb\nhttps://nbviewer.jupyter.org/github/rayryeng/make-money-ml-course/blob/master/week2/Week_2_Make_Money_with_Machine_Learning_Homework.ipynb",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecbbaad7b5ddb517001a6c95de782b699811d054 | 25,819 | ipynb | Jupyter Notebook | Basic_python/Analisis_de_planetas.ipynb | levraines/Portfolio | b134a6334cae34796a8de7d44b667413272166b6 | [
"CC0-1.0"
] | null | null | null | Basic_python/Analisis_de_planetas.ipynb | levraines/Portfolio | b134a6334cae34796a8de7d44b667413272166b6 | [
"CC0-1.0"
] | null | null | null | Basic_python/Analisis_de_planetas.ipynb | levraines/Portfolio | b134a6334cae34796a8de7d44b667413272166b6 | [
"CC0-1.0"
] | 1 | 2020-05-28T00:32:06.000Z | 2020-05-28T00:32:06.000Z | 28.880313 | 114 | 0.374066 | [
[
[
"## Datos de planetas",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nimport pandas.util.testing as tm",
"_____no_output_____"
],
[
"planets = sns.load_dataset('planets')",
"_____no_output_____"
],
[
"planets.shape",
"_____no_output_____"
],
[
"planets.head()",
"_____no_output_____"
],
[
"planets.dropna().describe()",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"planets.distance.sum()",
"_____no_output_____"
]
],
[
[
"$$\\frac{\\sum_{i=1}^n | x_i -\\bar{x}|}{n}$$",
"_____no_output_____"
]
],
[
[
"planets.distance.mad() # Desviacion media absoluta",
"_____no_output_____"
],
[
"planets.distance.var() # varianza",
"_____no_output_____"
],
[
"planets.distance.std() # desviacion estandar",
"_____no_output_____"
],
[
"planets.groupby('method')",
"_____no_output_____"
],
[
"planets.groupby('method')['orbital_period'].median() # mediana de cada uno de los metodos",
"_____no_output_____"
],
[
"planets.groupby('method')['year'].count() # para saber cuantos se descubrieron segun el anno",
"_____no_output_____"
],
[
"for (method, group) in planets.groupby('method'):\n print(\"{0:30s} shape = {1}\".format(method, group.shape))",
"Astrometry shape = (2, 6)\nEclipse Timing Variations shape = (9, 6)\nImaging shape = (38, 6)\nMicrolensing shape = (23, 6)\nOrbital Brightness Modulation shape = (3, 6)\nPulsar Timing shape = (5, 6)\nPulsation Timing Variations shape = (1, 6)\nRadial Velocity shape = (553, 6)\nTransit shape = (397, 6)\nTransit Timing Variations shape = (4, 6)\n"
],
[
"planets.groupby('method')['year'].describe() # se puede saber desde cuando se han usado esas tecnicas con\n# los anos en que se descubrieron los planetas. ",
"_____no_output_____"
],
[
"decade = 10*(planets['year']//10)",
"_____no_output_____"
],
[
"decade = decade.astype(str) + 's'",
"_____no_output_____"
],
[
"decade",
"_____no_output_____"
],
[
"decade.name = \"decade\"",
"_____no_output_____"
],
[
"planets.groupby(['method', decade])['number'].sum().unstack().fillna(0) # para desapilarlos",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecbbab393627a4cd2c45a1859a4eab5611bc6155 | 7,163 | ipynb | Jupyter Notebook | solutions/solutions_05.ipynb | scriptotek/programming-for-librarians | ba9048d6a3e90e206092355509d969cf938e8aaa | [
"BSD-2-Clause"
] | 2 | 2020-02-10T09:52:15.000Z | 2021-09-18T21:41:26.000Z | solutions/solutions_05.ipynb | scriptotek/programming-for-librarians | ba9048d6a3e90e206092355509d969cf938e8aaa | [
"BSD-2-Clause"
] | 3 | 2020-06-12T08:04:32.000Z | 2020-06-15T08:43:14.000Z | solutions/solutions_05.ipynb | scriptotek/programming-for-librarians | ba9048d6a3e90e206092355509d969cf938e8aaa | [
"BSD-2-Clause"
] | null | null | null | 34.109524 | 303 | 0.586207 | [
[
[
"### <span style=\"color:green\"> Exercise: multiple hits </span>\n\nThe code below is supposed to show a warning for temperatures above 70, but there is a bug.\nFind two different ways to fix the code, so that the warning is displayed.\n\n#### <span style=\"color:blue\"> Solution 1: multiple hits </span>\n\nChange the last test to `if`.",
"_____no_output_____"
]
],
[
[
"temperature = 120\n\nif temperature > 0:\n print(\"it's warm\")\nelif temperature <= 0:\n print(\"it's freezing\")\nif temperature > 70:\n print(\"WARNING: dangerously hot\")",
"_____no_output_____"
]
],
[
[
"#### <span style=\"color:blue\"> Solution 2: multiple hits </span>\n\nChange the order of the tests.\nNote that this version produces different output, here only one line is printed.\n\n\nThe exercise doesn't specify whether only the warning is supposed to be displayed. Sometimes we need to clarify vague requirements before we can write the program. ",
"_____no_output_____"
]
],
[
[
"temperature = 120\n\nif temperature > 70:\n print(\"WARNING: dangerously hot\")\nelif temperature > 0:\n print(\"it's warm\")\nelif temperature <= 0:\n print(\"it's freezing\")",
"_____no_output_____"
]
],
[
[
"### <span style=\"color:green\"> Exercise: Boolean Operators </span>\n\nAgain we look at the temperature test.\nThis time, use a Boolean operator to fix this test so that the warning is displayed.\n\n#### <span style=\"color:blue\"> Solution : Boolean Operators </span>\n\nThe solution below uses two conditions for the message *it’s warm* to be printed out, by using the boolean operator `and`.",
"_____no_output_____"
]
],
[
[
"temperature = 120\n\nif temperature > 0 and temperature <= 70:\n print(\"it's warm\")\nelif temperature <= 0:\n print(\"it's freezing\")\nelif temperature > 70:\n print(\"WARNING: dangerously hot\")",
"_____no_output_____"
]
],
[
[
"### <span style=\"color:green\">Case Law Exercise: Count Dissenting Opinions</span>\n\nIn the code below, we loop through a list of cases from the Case Law Api, then\nloop through the opinions for each of those cases. Each `opinion` has a `\"type\"`\nfield which describes if it's a majority opinion, dissenting opinion or concurring opinion. \nFirst, try to run the code below to check if you can print out the value of this field for each opinion:\n```\n...\n```\nNow, try to modify the code below to count the number of dissenting opinions by using an `if` test with `opinion[\"type\"]`.\nIf you find a dissent, you will need to increase the variable `dissent_count`:\n\n#### <span style=\"color:blue\"> Solution : Count Dissenting Opinions </span>\n\nA common way of counting in programming is by using a *counter* variable. Simply set the counter variable to zero in the beginning of the program, and then increase the variable by adding one each time the criteria is met. In this case, an if-test is needed to check that the `type` is `dissent`.",
"_____no_output_____"
]
],
[
[
"import requests\nimport json\n\nURL = \"https://api.case.law/v1/cases/?jurisdiction=ill&full_case=true&decision_date_min=2011-01-01&page_size=20\"\ndata = requests.get(URL).json()\n\ndissent_count = 0\n\ncases = data[\"results\"]\nfor case in cases:\n opinions = case[\"casebody\"][\"data\"][\"opinions\"]\n for opinion in opinions:\n # Your code here:\n if opinion[\"type\"] == \"dissent\":\n dissent_count = dissent_count + 1\n\nprint(\"Number of dissents:\", dissent_count)",
"_____no_output_____"
]
],
[
[
"### <span style=\"color:green\"> Library Data Exercise: Count Fulltext Documents </span>\n\nIn the code below, we loop through a list of items from the National Library API.\nEach `item` has a dictionary `accessInfo`, containing a key `isDigital`.\nThe corresponding value is a Boolean which is `True` if the document is available digitally in fulltext.\nFirst, try to run the code below to check if you can print out the value of `isDigital` for each item:\n```\n...\n```\nNow, try to modify the code below to count the number of dissenting opinions by using an `if` test with `opinion[\"type\"]`.\nIf you find a dissent, you will need to increase the variable `dissent_count`.\n\n#### <span style=\"color:blue\"> Solution: Count Fulltext Documents </span>\n\nA common way of counting in programming is by using a *counter* variable. Simply set the counter variable to zero in the beginning of the program, and then increase the variable by adding one each time the criteria is met. In this case, an if-test is needed to check that `isDigital` is `True`.\nHere, we don't need to use a comparator, since the value of `isDigital` is already a Boolean.",
"_____no_output_____"
]
],
[
[
"import requests\nimport json\n\nURL = \"https://api.nb.no/catalog/v1/items?size=20&filter=mediatype:b%C3%B8ker&q=Bing,Jon\"\ndata = requests.get(URL).json()\nembedded = data['_embedded']\nitems = embedded['items']\n\nfulltext_count = 0\n\nfor item in items:\n accessInfo = item['accessInfo']\n isDigital = accessInfo['isDigital']\n if isDigital:\n fulltext_count = fulltext_count + 1\n\nprint('Number of fulltext documents found:', fulltext_count)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecbbac202cdc2449617b5ce0c514a86435e7d930 | 123,700 | ipynb | Jupyter Notebook | Explore Notebooks/Big_o_notation_and_sorting_algorithms-2072.ipynb | George-Kibe/Data-Science | 1e4e4c1fff3d4c3ccd2e2821e73c113ad83472d3 | [
"MIT"
] | null | null | null | Explore Notebooks/Big_o_notation_and_sorting_algorithms-2072.ipynb | George-Kibe/Data-Science | 1e4e4c1fff3d4c3ccd2e2821e73c113ad83472d3 | [
"MIT"
] | null | null | null | Explore Notebooks/Big_o_notation_and_sorting_algorithms-2072.ipynb | George-Kibe/Data-Science | 1e4e4c1fff3d4c3ccd2e2821e73c113ad83472d3 | [
"MIT"
] | null | null | null | 113.59045 | 46,664 | 0.849838 | [
[
[
"# Big-O Notation & Sorting Algorithms\n\n© Explore Data Science Academy",
"_____no_output_____"
],
[
"## Learning Objectives\nBy the end of this train, you should be able to:\n\n* Understand the concepts of big-O Notation,\n* Understand the concepts of Sorting Algorithms and their complexity, and\n* Write pseudocode for sorting algorithms.\n\n## Outline\nIn this train we will:\n\n* Explain computational complexity and big-O Notation, and\n* Work through several sorting algorithms to characterise their complexity.",
"_____no_output_____"
],
[
"## Complexity and Big-O Notation\n\nBig-O notation is a formal mathematical language which helps us define the performance or complexity of a given algorithm. It is defined as the asymptotic upper limit of a function. In plain English, this means it is a notation which helps us know what the maximum space (storage) or time (speed) requirements are when running a piece of code. This notation helps us predict worst-case performance, and allows for various algorithms to be compared. \n\nWhen looking at big-O notation, there are two aspects to its syntax.\n\nTo help us understand the syntax of big-O notation, let's say that we characterise an algorithm as being $O(x)$ in nature. Here the $O$ refers to the 'order' of the algorithm, and the quantity inside the brackets ($x$ in our case) is this associated growth rate or order. We often express this growth rate in terms of $n$, or the *number of elements* upon which the algorithm needs to act. \n\nWe can use the figure below to help us visually compare the growth rates for some of the complexity categories described by Big O notation. As seen, the order of an algorithm can lead to significantly different complexities being realised with only a small number of input elements. ",
"_____no_output_____"
],
[
"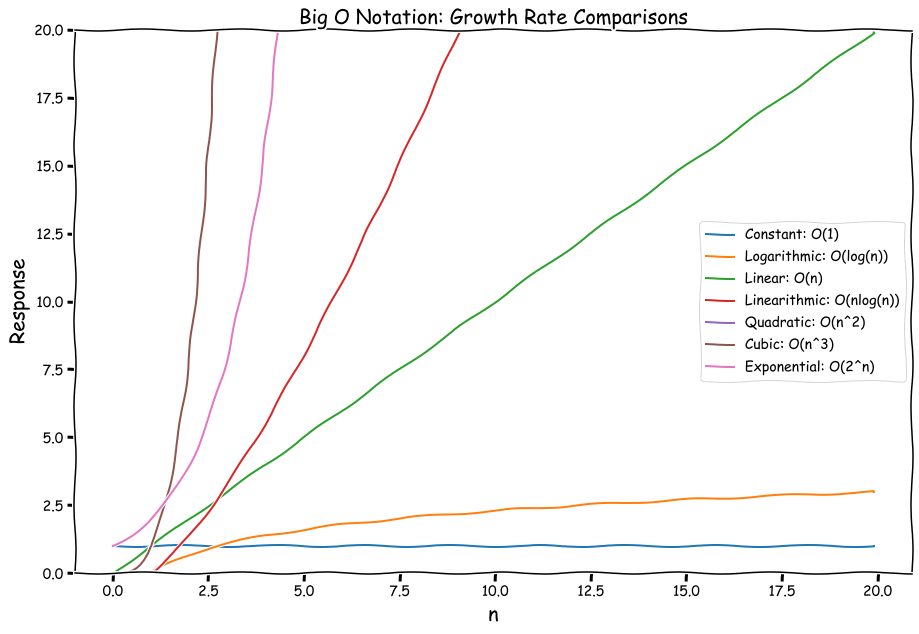",
"_____no_output_____"
],
[
"To see the relevance of big-O notation, consider how often there is more than one way to solve a problem. We need to learn how to compare the performance of different algorithms and choose the most efficient way to solve the problem. While analysing an algorithm, we mostly consider time complexity and space complexity. \n\n* The **Time complexity** of an algorithm represents the amount of time an algorithm takes to complete and is dependent on the size of the input.\n\n* The **Space complexity** of an algorithm represents the amount of space or memory an algorithm requires during operation and is dependent on the size of the input.\n\nTo gain an understanding of the basics of big-O notation, let's work through a few examples of the most common growth rates. \n\nWe first import the packages we'll need to compare these methods:",
"_____no_output_____"
]
],
[
[
"import random\nfrom time import time\nfrom timeit import timeit\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport warnings\nwarnings.filterwarnings('ignore')\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"Note: the following implementations were obtained from [A beginner guide of Big O notation.](https://sites.google.com/site/mingchangliu1980/algorithms/a-beginner-guide-of-big-o-notation)",
"_____no_output_____"
],
[
"### O(1)\n\nO(1), named Constant Time, represents an algorithm which will always execute in the same time, or space, independant of the size of the input.",
"_____no_output_____"
]
],
[
[
"def is_first_element_null(elements):\n return elements[0] == None",
"_____no_output_____"
]
],
[
[
"In this example above, regardless of the number of elements we pass to our function, we will always require a constant number of operations to index and return the element. ",
"_____no_output_____"
],
[
"### O(N)\n\nO(N), named Linear Time, represents an algorithm whose performance will grow linearly and in direct proportion to the size of the input. \n\nThe example below also demonstrates how big-O favours the worst-case performance scenario. A matching string could be found during any iteration of the `for loop` and the function would return early, but big-O will always assume the upper limit where the algorithm will perform the maximum number of iterations.",
"_____no_output_____"
]
],
[
[
"def contains_value(elements, string_value):\n \"\"\"Run through all elements in the list and compare to string_value.\"\"\"\n for e in elements:\n if e == string_value:\n return True\n return False",
"_____no_output_____"
]
],
[
[
"### O(N$^2$)\n\nO(N$^2$), known as Quadratic Time, represents an algorithm whose performance is directly proportional to the square of the size of the input. \n\nThis is common with algorithms that involve nested iterations over the data set. Deeper nested iterations will result in O(N$^3$), O(N$^4$), etc.",
"_____no_output_____"
]
],
[
[
"def contains_duplicates(elements):\n \"\"\"Check if any element in a list occurs more than once\"\"\"\n for i, e1 in enumerate(elements):\n for j, e2 in enumerate(elements):\n \"\"\"return a true if the elements indices are different and the elements are the same\"\"\"\n if ((i != j) & (e1 == e2)):\n return True\n return False",
"_____no_output_____"
]
],
[
[
"### O(2$^N$)\n\nO(2$^N$), named Exponential Time, denotes an algorithm whose growth doubles with each addition to the input data set. The growth curve of an O(2$^N$) function is exponential - starting off shallow, then rising meteorically. \n\nAn example of an O(2$^N$) function is the recursive calculation of Fibonacci numbers:",
"_____no_output_____"
]
],
[
[
"def fibonacci(number):\n \"\"\"\n The Fibonacci sequence is characterized by the fact that every number \n after the first two is the sum of the two preceding ones \n \"\"\"\n if number <= 1:\n return 1\n return fibonacci(number - 2) + fibonacci(number - 1)",
"_____no_output_____"
]
],
[
[
"### O(logN)\n\nLogarithms are slightly trickier to explain. O(logN) means that time increases linearly whilst _n_ increases exponentially. This complexity occurs with \"divide and conquer\" algorithms like binary search as seen in the figure below.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"The recursion continues until the array examined consists of only one element. Courtesy of Luke Francl.",
"_____no_output_____"
]
],
[
[
"def binary_search(elements, string_val):\n\n if len(elements) == 1:\n return 0 if elements[0] == string_val else None\n \n mid = len(elements) // 2\n if string_val == elements[mid]:\n return mid\n \n if string_val < elements[mid]:\n return binary_search(elements[:mid], string_val)\n else:\n return mid + binary_search(elements[mid:], string_val)",
"_____no_output_____"
],
[
"binary_search([1, 2, 3, 4, 5, 6, 7, 8, 9, 10], 5)",
"_____no_output_____"
]
],
[
[
"## Sorting Algorithms\n\nA sorting algorithm is an algorithm that puts elements of a list in a certain logical order. \n\nHere let's cover some of the more common sorting algorithms.\n\n### Bubble Sort\n\nBubble sort is a basic sorting algorithm that is the relatively simple to understand. The basic idea is to 'bubble' up the largest (or smallest) element within a list to its beginning, then the 2nd largest element and the 3rd and so on to the end of the list. Each bubble up takes a full sweep through the list. The following implementation of bubble sort was obtained from [Teach Yourself Python](www.teachyourselfpython.com/)",
"_____no_output_____"
],
[
"```python\n# Pseudo Code\nprocedure bubble_sort( input A --> which is a list of sortable items )\n n = length(A)\n repeat \n swapped = false\n for i = 1 to n-1 inclusive do\n # if this pair is out of order\n if A[i-1] > A[i] then\n # swap them and remember something changed\n swap( A[i-1], A[i] )\n swapped = true\n end if\n end for\n until not swapped\nend procedure\n```",
"_____no_output_____"
]
],
[
[
"def bubble_sort(items):\n \"\"\" Implementation of bubble sort \"\"\"\n out = items.copy() # in place protection on items\n for i in range(len(out)):\n for j in range(len(out)-1-i):\n if out[j] > out[j+1]:\n out[j], out[j+1] = out[j+1], out[j] # Swap!\n \n return out",
"_____no_output_____"
]
],
[
[
"### Insertion Sort\n\nThe Insertion Sort algorithm works by taking elements from an unsorted list and inserting them at the right place in a new sorted list. The sorted list is empty in the beginning. Since the total number of elements in the new and old list stays the same, we can use the same list to represent the sorted and the unsorted sections. \n\nImplementation for insertion sort can be found at [Geeks for geeks](https://www.geeksforgeeks.org/insertion-sort/)",
"_____no_output_____"
],
[
"```python\n# Pseudo Code\ni = 1\nwhile i < length(A)\n j = i\n while j > 0 and A[j-1] > A[j]\n swap A[j] and A[j-1]\n j = j - 1\n end while\n i = i + 1\nend while\n```",
"_____no_output_____"
]
],
[
[
"def insertion_sort(items):\n \"\"\" Implementation of insertion sort \"\"\"\n new_list = [items[0]] \n for x in items[1:]:\n i = 0\n while (i < len(new_list)) and (x > new_list[i]):\n i += 1\n new_list.insert(i, x) \n return new_list",
"_____no_output_____"
]
],
[
[
"### Merge Sort\n\nMerge Sort is a parallelizable algorithm which works by first repeatedly dividing an unsorted list into sub-lists; breaking-down its elements until each is placed within an individual sub-list. A recursive process is then followed to merge neighbouring sublists together in an ordered manner, ultimately yielding a fully sorted list. \n\nNOTE: Two functions, `merge` and `merge_sort` are often used to implement the merge sort algorithm. ",
"_____no_output_____"
],
[
"```python\n# Pseudo Code\nfunction merge_sort(list m)\n # Base case. A list of zero or one elements is sorted, by definition.\n if length of m <= 1 then\n return m\n\n # Recursive case. First, divide the list into equal-sized sublists\n # consisting of the first half and second half of the list.\n # This assumes lists start at index 0.\n left starts as an empty list\n right starts as an empty list\n for each x with index i in m do\n if i < (length of m)/2 then\n left = left + x\n else\n right = right + x\n\n # Recursively sort both sublists.\n left = merge_sort(left)\n right = merge_sort(right)\n\n # Then merge the now-sorted sublists.\n return merge(left, right)\n```",
"_____no_output_____"
],
[
"``` python\nfunction merge(left, right)\n result starts as an empty list\n\n while left is not empty and right is not empty do\n if left[0] <= right[0] then\n result = result + left[0] \n left = left[1:]\n else\n result = result + right[0]\n right = right[1:]\n\n # Either left or right may have elements left; consume them.\n # (Only one of the following loops will actually be entered.)\n while left is not empty do\n result = result + left[0]\n left = left[1:]\n \n while right is not empty do\n result = result + right[0]\n right = right[1:]\n return result\n```",
"_____no_output_____"
]
],
[
[
"def merge(A, B): \n \"\"\" The merge function used in merge sort \"\"\"\n new_list = []\n while len(A) > 0 and len(B) > 0:\n if A[0] < B[0]:\n new_list.append(A[0])\n A.pop(0)\n else:\n new_list.append(B[0])\n B.pop(0)\n \n if len(A) == 0:\n new_list = new_list + B \n if len(B) == 0:\n new_list = new_list + A\n \n return new_list \n \n\ndef merge_sort(items):\n \"\"\" Implementation of merge sort \"\"\"\n len_i = len(items)\n if len_i == 1:\n return items \n \n mid_point = int(len_i / 2)\n i1 = merge_sort(items[:mid_point])\n i2 = merge_sort(items[mid_point:]) \n \n return merge(i1, i2)",
"_____no_output_____"
]
],
[
[
"### Quick Sort\n\nThe Quick Sort algorithm works by first selecting a pivot element from an unsorted list. It then creates two lists, one containing elements less than the pivot and the other containing elements higher than the pivot. It then sorts the two lists and joins them with the pivot in between. \n\nImplementation for insertion sort can be found at [Geeks for geeks](https://www.geeksforgeeks.org/quick-sort/)",
"_____no_output_____"
],
[
"```Python\n# Pseudo Code\nfunction quick_sort(arr, low_index, high_index)\n\n # The pivot element is always to the right of a joined list\n pivot_element = arr[high_index]\n \n # Index of the smaller element\n i = low_index - 1 \n \n for j = low_index to high_index -1 inclusive do\n # If current element is smaller than the pivot \n if arr[j] < pivot_element \n i++ #Increase the index of the smaller element\n swap arr[i] and arr[j]\n \n swap arr[i + 1] and arr[high_index]\n return (i + 1)\n```",
"_____no_output_____"
]
],
[
[
"def quick_sort(items, index=-1):\n \"\"\" Implementation of quick sort \"\"\"\n len_i = len(items)\n\n if len_i <= 1:\n return items\n\n pivot = items[index]\n small = []\n large = []\n dup = []\n for i in items:\n if i < pivot:\n small.append(i)\n elif i > pivot:\n large.append(i)\n elif i == pivot:\n dup.append(i)\n\n small = quick_sort(small)\n large = quick_sort(large)\n\n return small + dup + large\n",
"_____no_output_____"
]
],
[
[
"### Exercise: Heap Sort\n\nThis implementation uses the built-in heap data structures in Python. To truly understand Heap sort, one must implement the heap_sort() function themselves.\n\nSee if you can code the heapsort algorithm yourself. This is likely to be a challenge.",
"_____no_output_____"
]
],
[
[
"def buildHeap(lista, n):\n for i in range(n//2 - 1, -1, -1):\n heapify(lista, n, i)\n\ndef heapify(lista, n, i):\n largest = i \n left = (2 * i) + 1 \n right = (2 * i) + 2 \n\n if left < n and lista[largest] < lista[left]:\n largest = left\n\n if right < n and lista[largest] < lista[right]:\n largest = right\n\n if largest != i:\n lista[i], lista[largest] = lista[largest], lista[i] \n heapify(lista, n, largest) \ndef heap_sort(items):\n \"\"\"Your implementation of heap sort\"\"\"\n n = len(items)\n buildHeap(items, n)\n \n for i in range(n-1, 0, -1):\n items[i], items[0] = items[0], items[i]\n heapify(items, i, 0)\n # your code here\n return",
"_____no_output_____"
],
[
"# unit test for your heap_sort function\n# this should return True if you're heap_sort was programmed correctly\nitems = [1, -3, 2, 0, 3, -2, -1]\nheap_sort(items) == [-3, -2, -1, 0, 1, 2, 3]",
"_____no_output_____"
]
],
[
[
"### Comparison of sorting algorithms\nLet's visualise the run-time results of these sorting algorithms. Before running the following code blocks, spend some time considering which one you think will run the fastest, and why.\n\nWe first define some helper functions to help us with our visualisation:",
"_____no_output_____"
]
],
[
[
"def get_time(fn, num_loops=10):\n \"\"\"Record the average execution of an input function\"\"\"\n output = []\n for n in range(num_loops):\n start = time()\n if type(fn) == str:\n eval(fn)\n else:\n fn\n end = time()\n output.append(end-start)\n output = np.array(output).mean()\n return output",
"_____no_output_____"
],
[
"def get_all_times(fn=[], items=[], num_loops=100):\n \"\"\"Record the individual average execution time for an arrray of input functions\"\"\"\n output = np.zeros((len(fn), len(items)))\n for x, f in enumerate(fn): \n for y, i in enumerate(items): \n output[x][y] = get_time('{:s}({:s})'.format(f, str(i)), num_loops=num_loops)\n return output",
"_____no_output_____"
]
],
[
[
"We now define our experimental parameters. We start out by sorting lists containing 1000 random elements: ",
"_____no_output_____"
]
],
[
[
"num_items = 1000\nrandom_items = [random.randint(-100, 100) for c in range(num_items)]\nordered = sorted(random_items)\nreverse_sort = ordered[::-1]",
"_____no_output_____"
]
],
[
[
"We will compare the following algorithms: \n * No sort required --> 'sorted'\n * Bubble sort --> 'bubble_sort'\n * Insertion sort --> 'insertion_sort'\n * Merge sort --> 'merge_sort'\n * Quick sort --> 'quick_sort'\n * Heap sort --> 'heap_sort'",
"_____no_output_____"
]
],
[
[
"fn = ['sorted', 'bubble_sort', 'insertion_sort', 'merge_sort', 'quick_sort', 'heap_sort'] ",
"_____no_output_____"
],
[
"items = ['random_items', 'ordered', 'reverse_sort']",
"_____no_output_____"
]
],
[
[
"With everything set up, let's run our experiment. Note that depending on the specifications of your machine, this may take a while to run. ",
"_____no_output_____"
]
],
[
[
"compare = get_all_times(fn=fn, items=items)",
"_____no_output_____"
]
],
[
[
"Let's plot out collected results:",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(3, 1, figsize=(10,10), gridspec_kw={'hspace': 0.3})\nmax_out = compare.max() * 1.2\nmin_out = 0\nfor i, a, f in zip(range(len(items)), ax, items): \n a.bar(x=range(len(fn)), height=compare[:,i])\n a.set_title(f)\n a.set_ylim(bottom=min_out, top=max_out)\n a.set_xticklabels([''] + fn, ha=\"center\")\n a.set_ylabel('time (s)')\n for j, h in zip(range(len(fn)), compare[:, i]):\n a.text(j, h + 0.01, '%.5f' %h, ha='center')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Even though we're speaking about millisecond run-times, we can still see how inefficient the bubble-sort algorithm is compared to its peers, having on average the worst performance. \n\nWhat results did you obtain from the heap sort algorithm, and were these in line with your initial intuition? ",
"_____no_output_____"
],
[
"### Performance characteristics as input size increases\nLet's now see how the performance changes as we vary the number of elements over multiple runs. We perform this for lists in the range of 1 to 5000 elements:",
"_____no_output_____"
]
],
[
[
"n_runs = [1, 5, 10, 25, 50, 100, 250, 500, 1000, 2000, 3000, 4000, 5000]\nn_random = [[random.randint(-100, 100) for c in range(n)] for n in n_runs]",
"_____no_output_____"
]
],
[
[
"Again, the following code may take some time to run. ",
"_____no_output_____"
]
],
[
[
"scale = get_all_times(fn=fn, items=n_random, num_loops=1)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(15,10))\nmax_out = compare.max() * 1.2\nmin_out = 0\nfor i, f in enumerate(fn): \n ax.plot(n_runs, scale[i], label=f)\n ax.legend()\n ax.set_xlabel('Number of elements')\n ax.set_ylabel('Execution time (s)')\nfig.show()",
"_____no_output_____"
]
],
[
[
"We once again get a similar intuition of the complexity of the various sorting algorithms, with bubble sort being significantly more expensive compared to the rest. This result aligns with theory, as the naive versions of the bubble sort algorithm typically have $O(n^2)$ time complexity. \n\nAlternatively, the quick sort algorithm is seen to have one of the lowest computational costs. Again the theory has our backs here, with the average time complexity of this algorithm being linearithmic with $O(n \\log n)$. ",
"_____no_output_____"
],
[
"## Conclusion\n\nUnderstanding the time and space complexity of your algorithm will ensure that you create solutions that are not only accurate but are efficient as well. This train only presents a simple introduction into these concepts and the reader is encouraged to read up further on these concepts.",
"_____no_output_____"
],
[
"## Appendix\n\n- [Basic sorting algorithms in python\n](http://danishmujeeb.com/blog/2014/01/basic-sorting-algorithms-implemented-in-python/?lipi=urn%3Ali%3Apage%3Ad_flagship3_feed%3BCu6nF%2FUCRO2Yz9VwNtpJOg%3D%3D) ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ecbbb0eff15f52ec4fa54d5726c972ea8efc3bf9 | 7,705 | ipynb | Jupyter Notebook | docs/source/CLP/solved/Fasting Diet (Solved).ipynb | ffraile/operations-research-notebooks | a65a8cccdc4496d4cda045dabd1366156b5948d0 | [
"MIT"
] | null | null | null | docs/source/CLP/solved/Fasting Diet (Solved).ipynb | ffraile/operations-research-notebooks | a65a8cccdc4496d4cda045dabd1366156b5948d0 | [
"MIT"
] | null | null | null | docs/source/CLP/solved/Fasting Diet (Solved).ipynb | ffraile/operations-research-notebooks | a65a8cccdc4496d4cda045dabd1366156b5948d0 | [
"MIT"
] | null | null | null | 38.143564 | 283 | 0.606489 | [
[
[
"# Fasting Diet\n## Problem I\nYou have read an article about the benefits of fasting. This article publishes a series of findings around the fasting diet method. Specifically, the article states that: \n\n- Decreasing the number of meals to three, two heavier meals and a light meal in between has significant benefits for the health \n\n- The study reveals that the optimal amount of daily calories for an adult is higher than 1800 \n\n- and also lower than 2200 calories\n\n- The study also reveals that the light meal should not represent more than 10% of the number of daily calories\n\n- And that the last meal should not provide more than 700 calories\n\n\n\nYou think you can design a Linear Programming Problem where the decision variables represent the calories of each meal to find and optimal diet that fulfils all these requirements, while at the same time provide the maximum number of calories. For such a model, define:\n\n**a.** The objective function (1 point)\n\nMaximise the calories intake in the three mails: \n$max z = x_1 + x_2+x_3$ \n\nwhere:\n - $x_1$: Calories of first meal\n - $x_2$: Calories of second meal (light meal)\n - $x_3$: Calories of third meal\n \n**b.** The constraints (2 points)\nNow, given the results of the study: \n\nOptimal daily amount\n\n$x_1+x_2+x_3 \\geq 1800$\n$x_1+x_2+x_3 \\leq 2200$\n\nCalories of light meal (less than 10% of total calories):\n$x_2 \\leq 0.1(x_1+x_2+x_3)$\n\nCalories of third meal (less than 700):\n$x_3 \\leq 700$\n\n**c.** The dual problem (2 points) \nWe can write the problem as: \n$max z = x_1 + x_2+x_3 \\\\\ns.t. \\\\\nx_1+x_2+x_3 \\geq 1800 \\\\\nx_1+x_2+x_3 \\leq 2200 \\\\\n-0.1·x_1 + 0.9·x_2 -0.1·x_3 \\leq 0 \\\\\n0·x_1 + 0·x_2 + 1·x_3 \\leq 700$\n \nHence, the dual becomes: \n\n$max_z = 1800·u_1 + 2200·u_2 + 0·u_3 + 700·u_4 \ns.t \\\\\nu_1+u_2-u_3+0·u_4 \\leq 1\nu_1 + u_2 + 0.9·u_3 + 0·u_4 \\leq 1 \nu_1 + u_2 -0.1·u_3 + u_3 \\leq 1\n$\n\n## Problem II\nYou are the production manager of a company. You have used the following programming model to define the optimal production plan. The problem is expressed in the standard form.\nObjective function:\n\nMax z = 3X1+4X2-MA1+0S2+0S3\nsubject to:\nX1+0X2+A1+0S2+0S3=18\n20X1+40X2+0A1+S2+0S3=1000\n26.67X1+20X2+0A1+0S2+S3=800\n*The slack variables are indexed in order of appearance.\n\nThe intern submitted the following data describing the solution obtained by a solver. This solver provides the solution of both the primal and the dual and a sensitivity analysis in a table, but your intern handled you the following handwritten report:\n\nX1 ->\nSolution value: 18\nReduced Cost: ?\nStatus: basic\nObjective coefficient: ?\nObjective Lower Bound: -Inf\nObjective Upper Bound: 5.334\n\nX2 ->\nSolution value: 15.997\nReduced Cost: ?\nStatus: ?\nObjective coefficient: 4\nObjective Lower Bound: 2.2497\nObjective Upper Bound: Inf\n\nA1->\nRHS= 18\nSlack = 0\nSense = ?\nShadow price= -2.334\nMin RHS= 17.9964\nMax RHS= 29.9963\n\nS2->\nRHS= 1000\nSlack = 0.12\nSense = less or equal\nShadow price= 0\nMin RHS= 999.\nMax RHS= Inf\n\nS3->\nRHS= ?\nSlack = ?\nSense = less or equal\nShadow price= 0\nMin RHS= 480.06\nMax RHS= 800.06\n\n\n**a.** You analise the solution and notice that there are some missing values noted as ? (You are going to fire this intern!). Write down the values missing in the solution. Motivate your response in each case (2 points)\n\nFor decision variable X1\nSince the value is non zero (it is a basic decision variable), by complementary slackness, the reduced cost is 0. \nThe objective function indicates that the objective coefficient is 3. \n\nFor decision variable X2\nAlso, by complementary slackness, the reduced cost is 0 and since the value is greater than zero, the decision variable is basic. \n\nFor slack, A1\nIf we note the coefficient in the objective function, (-M) we know that this is an artifical variable that must be always 0 and the sense is 'equal'.\n\nFor S3, \nThe Right Hand Side for the third constraint is 800 looking at the problem. \n\n\nWe can calculate the slack by substituting the values of X1 and X2 in the expression: \n\n26.67X1+20X2+0A1+0S2+S3=800\n26.67·18 + 20·15.997 + S3 = 800\nS3 = 0\n\nWe could have also argue that since the solution is a Feasible Basic Solution, only 3 decision variables (in this case X1, X2, and S2) can be basic (as many as constraints) and therefore S3 must be non-basic (0).\n\n**b.** Which of the two decision variables has a larger contribution to the optimal solution? Motivate your response (1 point). \n\nIf we multiply the value times the objective function: \n3·X1 = 3·18 = 54\n4·X2 = 4·15.997 = 63,998\nClearly X2 has a larger contribution to the objective function\n\n**c.** The first constraint determines the value of one of the decision variables (X1). What would happen to the objective function if the right hand side of this constraint changes to 19? and to 31? Motivate your response (1 point)\n\nThe maximum RHS for the first constraint is 29.993. If the RHS changes to 19, the basic decision variables would remain the same, although the values in the optimal solution might change. If the values changes to 31 then the basic decision variables would certainly change. \n\n\n**d.** What would happen to the solution if the objective function coefficient of X1 changes to 5? And what if the coefficient of X2 changes to 3? Motivate your response (1 point)\n\nThe Upper bound for the objective coefficient of X1 is 5.334 and therefore, the solution would not change if the objective 5 increased to 5. \n\nThe lower bound for the objective coefficient of X2 is 2.2497 and therefore, the soultion would change if the coefficient decreases down to 3.\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.