hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ecc87c6fbaced2cf02fb33ac7c6bc1813036022f | 286,600 | ipynb | Jupyter Notebook | doc/source/user_guide/dep_full_example.ipynb | sahahn/BPt | 1a2967f4ca3fa070b7417a4f59a218ae171daadd | [
"MIT"
] | 6 | 2020-11-06T15:45:28.000Z | 2022-03-08T19:15:35.000Z | doc/source/user_guide/dep_full_example.ipynb | sahahn/BPt | 1a2967f4ca3fa070b7417a4f59a218ae171daadd | [
"MIT"
] | 14 | 2020-10-20T13:55:23.000Z | 2022-01-25T17:36:07.000Z | docs/_sources/user_guide/dep_full_example.ipynb.txt | sahahn/BPt | 1a2967f4ca3fa070b7417a4f59a218ae171daadd | [
"MIT"
] | 2 | 2020-10-23T19:48:53.000Z | 2020-11-06T15:46:04.000Z | 128.866906 | 41,904 | 0.854648 | [
[
[
"# Predicting Substance Dependence from Multi-Site Data\n\nThis notebook explores an example using data from the ENIGMA Addiction Consortium. Within this notebook we will be trying to predict between participants with any drug dependence (alcohol, cocaine, etc...), vs. healthy controls. The data for this is sources from a number of individual studies from all around the world and with different scanners etc... making this a challenging problem with its own unique considerations. Structural FreeSurfer ROIs are used. The raw data cannot be made available due to data use agreements.\n\nThe key idea explored in this notebook is a particular tricky problem introduced by case-only sites, which are subject's data from site's with only case's. This introduces a confound where you cannot easily tell if the classifier is learning to predict site or the dependence status of interest.\n\nFeatured in this notebook as well are some helpful\ncode snippets for converting from BPt versions earlier than BPt 2.0 to valid BPt 2.0+ code.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport BPt as bp\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom warnings import simplefilter\nfrom sklearn.exceptions import ConvergenceWarning\nsimplefilter(\"ignore\", category=ConvergenceWarning)",
"/home/sage/anaconda3/envs/bpt/lib/python3.9/site-packages/nilearn/datasets/__init__.py:93: FutureWarning: Fetchers from the nilearn.datasets module will be updated in version 0.9 to return python strings instead of bytes and Pandas dataframes instead of Numpy arrays.\n warn(\"Fetchers from the nilearn.datasets module will be \"\n"
]
],
[
[
"### Loading / Preparing Data",
"_____no_output_____"
],
[
"As a general tip it can be useful to wrap something like a series of steps that load multiple DataFrames and merge them into a function. That said, it is often useful when first writing the function to try it taking advantage of the interactive-ness of the jupyter-notebook.",
"_____no_output_____"
]
],
[
[
"def load_base_df():\n '''Loads and merges a DataFrame from multiple raw files'''\n \n na_vals = [' ', ' ', 'nan', 'NaN']\n \n # Load first part of data\n d1 = pd.read_excel('/home/sage/Downloads/e1.xlsx', na_values=na_vals)\n d2 = pd.read_excel('/home/sage/Downloads/e2.xlsx', na_values=na_vals)\n df = pd.concat([d1, d2])\n\n df['Subject'] = df['Subject'].astype('str')\n df.rename({'Subject': 'subject'}, axis=1, inplace=True)\n df.set_index('subject', inplace=True)\n\n # Load second part\n df2 = pd.read_excel('/home/sage/Downloads/e3.xlsx', na_values=na_vals)\n df2['Subject ID'] = df2['Subject ID'].astype('str')\n df2.rename({'Subject ID': 'subject'}, axis=1, inplace=True)\n df2.set_index('subject', inplace=True)\n\n # Merge\n data = df2.merge(df, on='subject', how='outer')\n \n # Rename age and sex\n data = data.rename({'Sex_y': 'Sex', 'Age_y': 'Age'}, axis=1)\n \n # Remove subject name to obsficate\n data.index = list(range(len(data.index)))\n \n return data",
"_____no_output_____"
],
[
"df = load_base_df()\ndf.shape",
"_____no_output_____"
],
[
"# Cast to dataset\ndata = bp.Dataset(df)\ndata.verbose = 1\n\n# Drop non relevant columns\ndata.drop_cols_by_nan(threshold=.5, inplace=True)\ndata.drop_cols(scope='Dependent', inclusions='any drug', inplace=True)\ndata.drop_cols(exclusions=['Half', '30 days', 'Site ',\n 'Sex_', 'Age_', 'Primary Drug', 'ICV.'], inplace=True)\n\n# Set binary vars as categorical\ndata.auto_detect_categorical(inplace=True)\ndata.to_binary(scope='category', inplace=True)\nprint('to binary cols:', data.get_cols('category'))\n\n# Set target and drop any NaNs\ndata.set_role('Dependent any drug', 'target', inplace=True)\ndata.drop_nan_subjects('target', inplace=True)\n\n# Save this set of vars under scope covars\ndata = data.add_scope(['ICV', 'Sex', 'Age', 'Education', 'Handedness'], 'covars')\nprint('scope covars = ', data.get_cols('covars'))\n\n# Set site as non input\ndata = data.set_role('Site', 'non input')\ndata = data.ordinalize(scope='non input')\n\n# Drop subjects with too many NaN's and big outliers\ndata.drop_subjects_by_nan(threshold=.5, scope='all', inplace=True)\ndata.filter_outliers_by_std(n_std=10, scope='float', inplace=True)",
"Setting NaN threshold to: 1762.5\nDropped 20 Columns\nDropped 3 Columns\nDropped 36 Columns\nNum. categorical variables in dataset: 3\nto binary cols: ['Dependent any drug', 'Handedness', 'Sex']\nDropped 479 Rows\nscope covars = ['Age', 'Education', 'Handedness', 'ICV', 'Sex']\nSetting NaN threshold to: 82.5\nDropped 38 Rows\nDropped 5 Rows\n"
]
],
[
[
"**Legacy Equivilent pre BPt 2.0 loading code**\n\n```\nML = BPt_ML('Enigma_Alc',\n log_dr = None,\n n_jobs = 8)\n \nML.Set_Default_Load_Params(subject_id = 'subject',\n na_values = [' ', ' ', 'nan', 'NaN'],\n drop_na = .5)\n \nML.Load_Data(df=df,\n drop_keys = ['Unnamed:', 'Site', 'Half', 'PI', 'Dependent',\n 'Surface Area', 'Thickness', 'ICV', 'Subcortical',\n 'Sex', 'Age', 'Primary Drug', 'Education', 'Handedness'],\n inclusion_keys=None,\n unique_val_warn=None,\n clear_existing=True)\n\nML.Load_Targets(df=df,\n col_name = 'Dependent any drug',\n data_type = 'b')\n\nML.Load_Covars(df=df,\n col_name = ['ICV', 'Sex', 'Age'],\n drop_na = False,\n data_type = ['f', 'b', 'f'])\n\nML.Load_Covars(df = df,\n col_name = ['Education', 'Handedness'],\n data_type = ['f', 'b'],\n drop_na = False,\n filter_outlier_std = 10)\n\nML.Load_Strat(df=df,\n col_name=['Sex', 'Site'],\n binary_col=[True, False]\n )\n\nML.Prepare_All_Data()\n```",
"_____no_output_____"
],
[
"Let's take a look at what we prepared. We can see that visually the Dataset is grouped by role.",
"_____no_output_____"
],
[
"Let's plot some variables of interest",
"_____no_output_____"
]
],
[
[
"data.plot('target')",
"Dependent any drug: 3003 rows\n"
],
[
"data.plot('Site')",
"Site: 3003 rows\n"
],
[
"data.plot_bivar('Site', 'Dependent any drug')",
"Site: 3003 rows\nDependent any drug: 3003 rows\nPlotting 3003 overlap valid subjects.\n"
]
],
[
[
"That bi-variate plot is a little hard to read... we can make it bigger though easy enough. Let's say we also wanted to save it (we need to add show=False)",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(10, 10))\ndata.plot_bivar('Site', 'Dependent any drug', show=False)\nplt.savefig('site_by_drug.png', dpi=200)",
"Site: 3003 rows\nDependent any drug: 3003 rows\nPlotting 3003 overlap valid subjects.\n"
]
],
[
[
"Okay next, we are going to define a custom validation strategy to use, which is essentially going to preserve subjects within the same train and test fold based on site.\n\n\n**Legacy Code**\n```\nfrom BPt import CV\ngroup_site = CV(groups='Site')\n```",
"_____no_output_____"
]
],
[
[
"group_site = bp.CVStrategy(groups='Site')\ngroup_site",
"_____no_output_____"
]
],
[
[
"We then can use this CV strategy as an argument when defining the train test split.\n\n**Legacy Code**\n```\nML.Train_Test_Split(test_size =.2, cv=group_site)\n```",
"_____no_output_____"
]
],
[
[
"data = data.set_test_split(size=.2, cv_strategy=group_site, random_state=5)",
"Performing test split on: 3003 subjects.\nrandom_state: 5\nTest split size: 0.2\n\nPerformed train/test split\nTrain size: 2379\nTest size: 624\n"
],
[
"data.plot('target', subjects='train')\ndata.plot('target', subjects='test')",
"Dependent any drug: 2379 rows\n"
]
],
[
[
"### Running ML\n\nNext, we are going to evaluate some different machine learning models on the problem we have defined. Notably not much has changed here with respect to the old version, except some cosmetic changes like Problem_Spec to ProblemSpec. Also we have a new `evaluate` function instead of calling Evaluate via the ML object (`ML.Evaluate`).",
"_____no_output_____"
]
],
[
[
"# This just holds some commonly used values\nps = bp.ProblemSpec(subjects='train',\n scorer=['matthews', 'roc_auc', 'balanced_accuracy'],\n n_jobs=16)\nps",
"_____no_output_____"
],
[
"# Define a ModelPipeline to use with imputation, scaling and an elastic net\npipe = bp.ModelPipeline(imputers=[bp.Imputer(obj='mean', scope='float'),\n bp.Imputer(obj='median', scope='category')],\n scalers=bp.Scaler('standard'), \n model=bp.Model('elastic', params=1),\n param_search=bp.ParamSearch(\n search_type='DiscreteOnePlusOne', n_iter=64))\npipe.print_all()",
"ModelPipeline\n-------------\nimputers=\\\n[Imputer(obj='mean', scope='float'),\n Imputer(obj='median', scope='category')]\n\nscalers=\\\nScaler(obj='standard')\n\nmodel=\\\nModel(obj='elastic', params=1)\n\nparam_search=\\\nParamSearch(cv=CV(cv_strategy=CVStrategy()), n_iter=64,\n search_type='DiscreteOnePlusOne')\n\n"
]
],
[
[
"### 3-fold CV random splits\n\nLet's start by evaluating this model is a fairly naive way, just 3 folds of CV.",
"_____no_output_____"
]
],
[
[
"results = bp.evaluate(pipeline=pipe, dataset=data,\n problem_spec=ps, cv=bp.CV(splits=3))\nresults",
"Predicting target = Dependent any drug\nUsing problem_type = binary\nUsing scope = all (defining a total of 163 features).\nEvaluating 2379 total data points.\n"
]
],
[
[
"Notably this is likely going to give us overly optimistic results. Let's look at just the Site's drawn from the first evaluation fold to get a feel for why.",
"_____no_output_____"
]
],
[
[
"data.plot_bivar('Site', 'Dependent any drug', subjects=results.train_subjects[0])\ndata.plot_bivar('Site', 'Dependent any drug', subjects=results.val_subjects[0])",
"Site: 1586 rows\nDependent any drug: 1586 rows\nPlotting 1586 overlap valid subjects.\n"
]
],
[
[
"### Preserving Groups by Site\n\nWe can see that for example the 149 subjects from site 1 in the validation set had 269 subjects also from site 1 to potentially memorize site effects from! The confound is a site with only cases. One way we can account for this is through something we've already hinted at, using the group site cv from earlier. We can create a new CV object with this attribute.",
"_____no_output_____"
]
],
[
[
"site_cv = bp.CV(splits=3, cv_strategy=group_site)\nsite_cv",
"_____no_output_____"
],
[
"results = bp.evaluate(pipeline=pipe, dataset=data,\n problem_spec=ps, cv=site_cv)\nresults",
"_____no_output_____"
]
],
[
[
"We can now make the same plots as before, confirming now that the sites are different.",
"_____no_output_____"
]
],
[
[
"data.plot_bivar('Site', 'Dependent any drug', subjects=results.train_subjects[0])\ndata.plot_bivar('Site', 'Dependent any drug', subjects=results.val_subjects[0])",
"Site: 1471 rows\nDependent any drug: 1471 rows\nPlotting 1471 overlap valid subjects.\n"
]
],
[
[
"Another piece we might want to look at in this case is the weighted_mean_scores. That is, each of the splits has train and test sets of slightly different sizes, so how does the metric change if we weight it by the number of validation subjects in that fold",
"_____no_output_____"
]
],
[
[
"results.weighted_mean_scores",
"_____no_output_____"
],
[
"results = bp.evaluate(pipeline=pipe, dataset=data,\n problem_spec=ps, cv=3)\nresults",
"_____no_output_____"
]
],
[
[
"### Other options?\n\nWhat other options do we have to handle site with CV besides just the group splitting?\n\n1. Explicit leave-out-site CV: Where every train set is composed of all sites but one, and the validation set is the left out site.\n\n2. Use the train-only parameter in CV.\n\n3. Only evaluate subjects from sites with both cases and controls.\n\nLet's try option 1 first.",
"_____no_output_____"
],
[
"### Leave-out-site CV",
"_____no_output_____"
]
],
[
[
"# All we need to do for leave-out-site is specify that splits = Site\nleave_out_site_cv = bp.CV(splits='Site')\n\n# This will actually raise an error\ntry:\n results = bp.evaluate(pipeline=pipe, dataset=data,\n problem_spec=ps, cv=leave_out_site_cv)\nexcept Exception as exc:\n print(exc)",
"_____no_output_____"
]
],
[
[
"Uh oh looks like this option actually failed. What happened? Well basically since we have sites with only one class, we are trying to compute metrics that require two classes. Let's try again this time just using scorers that can work in the case of only one class.",
"_____no_output_____"
]
],
[
[
"results = bp.evaluate(pipeline=pipe, dataset=data, scorer='accuracy',\n problem_spec=ps, cv=leave_out_site_cv)\n\nresults.mean_scores, results.std_scores, results.weighted_mean_scores",
"_____no_output_____"
]
],
[
[
"The problem here is accuracy is likely a pretty bad metric, especially for the sites which have both cases and controls, but a skewed distribution. In general using the group preserving by Site is likely a better option.",
"_____no_output_____"
],
[
"### Train-only subjects\n\nOne optional way of handling these subjects from sites with only cases or only controls is to just never evaluate them. To instead always treat them as train subjects. We can notably then either use random splits or group preserving by CV.",
"_____no_output_____"
]
],
[
[
"def check_imbalanced(df):\n as_int = df['Dependent any drug'].astype('int')\n return np.sum(as_int) == len(as_int)\n\n# Apply function to check if each site has all the same target variable\nis_imbalanced = data.groupby('Site').apply(check_imbalanced)\n\n# Note these are internal values, not the original ones\nimbalanced_sites = np.flatnonzero(is_imbalanced)\n\n# We can specify just these subjects using the ValueSubset wrapper\nimbalanced_subjs = bp.ValueSubset('Site', values=imbalanced_sites, decode_values=False)",
"_____no_output_____"
],
[
"# Now define our new version of the group site cv\nsite_cv_tr_only = bp.CV(splits=3, cv_strategy=group_site,\n train_only_subjects=imbalanced_subjs)\n\n# And evaluate\nresults = bp.evaluate(pipeline=pipe, dataset=data,\n problem_spec=ps, cv=site_cv_tr_only)\nresults",
"_____no_output_____"
]
],
[
[
"The take-away here is maybe a little bleak. Essentially it is saying that some of the high scores we got earlier might have been in large part due to scores from the imbalanced sites.",
"_____no_output_____"
]
],
[
[
"# And we can also run a version with just random splits\ntr_only_cv = bp.CV(splits=3, train_only_subjects=imbalanced_subjs)\n\n# And evaluate\nresults = bp.evaluate(pipeline=pipe, dataset=data,\n problem_spec=ps, cv=tr_only_cv)\nresults",
"_____no_output_____"
]
],
[
[
"While this version with random splits does better, we are still notably ignoring the distribution of case to control in the sites with both cases and controls. Based on the group preserving results what it seems like is going on is that when subjects from the same site are split across validation folds, the classifier might be able to just memorize Site instead of the effect of interest. Lets use Site 29 as an example.\n\nLet's look at just fold 1 first.",
"_____no_output_____"
]
],
[
[
"# The train and val subjects from fold 0\ntr_subjs = results.train_subjects[0]\nval_subjs = results.val_subjects[0]\n\n# An object specifying just site 29's subjects\nsite29 = bp.ValueSubset('Site', 29, decode_values=True)\n\n# Get the intersection w/ intersection obj\nsite_29_tr = bp.Intersection([tr_subjs, site29])\nsite_29_val = bp.Intersection([val_subjs, site29])\n\n# Plot\ndata.plot_bivar('Site', 'Dependent any drug', subjects=bp.Intersection([tr_subjs, site29]))\ndata.plot_bivar('Site', 'Dependent any drug', subjects=bp.Intersection([val_subjs, site29]))\n\n# Grab just the subjects as actual index\nval_subjs = data.get_subjects(site_29_val)\n\n# Get a dataframe with the predictions made for just fold0\npreds_df_fold0 = results.get_preds_dfs()[0]\n\n# Let's see predictions for just these validation subjects\npreds_df_fold0.loc[val_subjs]",
"Site: 70 rows\nDependent any drug: 70 rows\nPlotting 70 overlap valid subjects.\n"
]
],
[
[
"We can see that it has learned to just predict 0 for every subject, which because of the imbalance is right most of the time, but really doesn't tell us if it has learned to predict site or substance dependence.",
"_____no_output_____"
],
[
"### Drop imbalanced subjects\nThe last option was just don't use the imbalanced subjects at all. We will do this by indexing only the balanced sites",
"_____no_output_____"
]
],
[
[
"# Note the ~ before the previously defined is_imbalanced\nbalanced_sites = np.flatnonzero(~is_imbalanced)\n\n# We can specify just these subjects using the ValueSubset wrapper\nbalanced_subjs = bp.ValueSubset('Site', values=balanced_sites, decode_values=False)",
"_____no_output_____"
],
[
"# Evaluate with site preserving cv from earlier but now using just the balanced train subjects\ntrain_just_balanced = bp.Intersection(['train', balanced_subjs])\n\nresults = bp.evaluate(pipeline=pipe, dataset=data,\n subjects=train_just_balanced,\n problem_spec=ps, cv=site_cv)\nresults",
"_____no_output_____"
]
],
[
[
"This actually seems to show that the imbalanced site confound with doesn't just corrupt cross validated results, but also actively impedes the model learning the real relationship. That we are better off just not using subjects with from sites with only cases. Yikes. \n\nLikewise, we can do the random split strategy again too.",
"_____no_output_____"
]
],
[
[
"results = bp.evaluate(pipeline=pipe, dataset=data,\n subjects=train_just_balanced,\n problem_spec=ps, cv=bp.CV(splits=3))\nresults",
"_____no_output_____"
]
],
[
[
"Also, we could run it with a leave-out-site CV. Now we can use the normal metrics.",
"_____no_output_____"
]
],
[
[
"results = bp.evaluate(pipeline=pipe, dataset=data,\n subjects=train_just_balanced,\n problem_spec=ps, cv=leave_out_site_cv)\nresults",
"_____no_output_____"
]
],
[
[
"### Results Summary\n\nTo summarize, of all of the above different iterations. Only the results from the versions where subjects from case-only sites are treated as train-only or dropped and evaluated group-preserving by site are likely un-biased! The leave-out-site CV results should be okay too, but let's skip those for now.\n\nTrain Only Scores:\nmean_scores = {'matthews': 0.024294039432900597, 'roc_auc': 0.5292721521624003, 'balanced_accuracy': 0.5116503104881627}\n\nDropped Scores:\nmean_scores = {'matthews': 0.23079311975258734, 'roc_auc': 0.6494071534373412, 'balanced_accuracy': 0.6141868842892168}\n\nBy these interm results we can with some confidence conclude dropping subjects is the way to go, unless we want to dig deeper...",
"_____no_output_____"
],
[
"### Disentangle Target and Site?\n\nCan information on dependance from case-only sites be some how disentangled from site?\n\nThis actually leads to a follow up a question. Namely is there any way to extract usable information from case-only site subjects? This question I actually investigated more deeply in https://onlinelibrary.wiley.com/doi/full/10.1002/hbm.25248 The idea of train-only subjects is actually from this paper, though it still has some important differences then the way we formulated it. \n\nThe key piece in the paper which we haven't tried to do yet is centered around the problem of the classifier learning site related information instead of dependence related information in the train-only setup. We essentially need a way of forcing the classifier to learn this. The way proposed in the paper proposes that maybe we can by restricting the features provided to the classifier, give it only features related to dependence and not those related to site. In order to do this, we need to add a feature selection step.\n\nThere are different ways to compose the problem, but we will specify the feature selection step as a seperate layer of nesting, importantly one that is evaluated according to the same site_cv_tr_only that we will be evaluating with. Remember the goal here is to select features which help us learn to predict useful information from case only sites, i.e., with feature only related to Dependence and not site!",
"_____no_output_____"
]
],
[
[
"# Same imputer and scaler\nimputers = [bp.Imputer(obj='mean', scope='float'),\n bp.Imputer(obj='median', scope='category')]\nscaler = bp.Scaler('standard')\n\n# This object will be used to set features as an array of binary hyper-parameters\nfeat_selector = bp.FeatSelector('selector', params=1)\n\n# This param search is responsible for optimizing the selected features from feat_selector\nparam_search = bp.ParamSearch('RandomSearch',\n n_iter=16,\n cv=site_cv_tr_only)\n\n# We create a nested elastic net model to optimize - no particular CV should be okay\nrandom_search = bp.ParamSearch('RandomSearch', n_iter=16)\nelastic_search = bp.Model('elastic', params=1,\n param_search=random_search)\n\n# Put it all together in a pipeline\nfs_pipe = bp.Pipeline(imputers + [scaler, feat_selector, elastic_search],\n param_search=param_search)",
"_____no_output_____"
]
],
[
[
"Note: This may take a while to evaluate as we now have two levels of parameters to optimize, e.g., for each choice\nof features, we need to train a nested elastic net.",
"_____no_output_____"
]
],
[
[
"results = bp.evaluate(pipeline=fs_pipe, dataset=data,\n problem_spec=ps, cv=site_cv_tr_only)\nresults",
"_____no_output_____"
]
],
[
[
"Unfortunetly this doesn't seem to work. Notably we still arn't quite following the method from the paper, which performs a meta-analysis on the results from the feature search to find overlapping subsets of features. As of the time of writing this example, a simmilar strategy has not been able to be easily integrated into BPt.\n\nThe above method still has a few problems, one being severe overfitting. Namely, because a number of subjects are reserved in train-only roles, the same small number of subjects are being used over and over in cross-validation. \n\nWe can confirm this intutiton by comparing the best score achieved internally for each set of features w/ how they score in the outer cross validation.",
"_____no_output_____"
]
],
[
[
"-results.estimators[0].best_score_, results.scores['matthews'][0]",
"_____no_output_____"
],
[
"-results.estimators[1].best_score_, results.scores['matthews'][1]",
"_____no_output_____"
],
[
"-results.estimators[2].best_score_, results.scores['matthews'][2]",
"_____no_output_____"
]
],
[
[
"These values are quite different to say the least. Part of the problem again is that such a small amount of data is being used to determine generalization. What if instead of a the 3-fold group preserving train only feature selection CV, we instead used a leave-out-site scheme on site with train only? I doubt it will help, but let's see...",
"_____no_output_____"
]
],
[
[
"leave_out_site_tr_only_cv = bp.CV(splits='Site',\n train_only_subjects=imbalanced_subjs)\n\nfs_pipe.param_search = bp.ParamSearch(\n 'RandomSearch', n_iter=16,\n cv=leave_out_site_tr_only_cv)\n\nresults = bp.evaluate(pipeline=fs_pipe, dataset=data,\n problem_spec=ps, cv=site_cv_tr_only)\nresults",
"_____no_output_____"
]
],
[
[
"This does a little better. But only slightly. What if we also used this leave-out site strategy for the outer loop? This would be to address the issue that the outer CV has the same problem as the inner, where the validation set is overly specific and small in some folds.",
"_____no_output_____"
]
],
[
[
"results = bp.evaluate(pipeline=fs_pipe,\n dataset=data,\n problem_spec=ps,\n cv=leave_out_site_tr_only_cv,\n eval_verbose=1)\nresults",
"Predicting target = Dependent any drug\nUsing problem_type = binary\nUsing scope = all defining an initial total of 163 features.\nEvaluating 2379 total data points.\n"
]
],
[
[
"Seeing the results for each site seperately with the verbose option really makes some of the problem clear, namely that a number of small sites don't seem to generalize well. This is simmilar to what we saw in the case of using leave-out-site with case-only subjects dropped, where we had a huge std in roc auc between folds.\n\nAnother potential problem with doing leave-out-site is that the way the underlying data was collected, different sites will have subjects with dependence to different substances. This can certainly cause issues, as say our classifier only learns to classify alcohol dependence (the strongest signal), then it will do poorly on sites for other substances. Which is good in one way, that it lets us know the classifier is not generalizing, but it doesn't solve the generalization problem.\n\nAnyways, that will end this example for now. Leaving the problem of how to best exploit information from case-only sites as an open question (though if interested, a more faithful reproduction of the method presented in https://onlinelibrary.wiley.com/doi/full/10.1002/hbm.25248 might prove to be fruitful. Specifically, in the end only using a few, maybe 3-4, features, which is notably quite different then what we are trying above, i.e., trying just 16 random sets of features).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecc87cc7341c10a97e32dfbd7d0d8fe4ec0c51e3 | 26,034 | ipynb | Jupyter Notebook | PY0101EN-2-2-Lists.ipynb | Aartithakur-Sudo/python-Cognitive | 2b096aebf7ae5a4eef5de9c55f93e261983f7da0 | [
"Apache-2.0"
] | null | null | null | PY0101EN-2-2-Lists.ipynb | Aartithakur-Sudo/python-Cognitive | 2b096aebf7ae5a4eef5de9c55f93e261983f7da0 | [
"Apache-2.0"
] | null | null | null | PY0101EN-2-2-Lists.ipynb | Aartithakur-Sudo/python-Cognitive | 2b096aebf7ae5a4eef5de9c55f93e261983f7da0 | [
"Apache-2.0"
] | null | null | null | 26.243952 | 921 | 0.507951 | [
[
[
"<center>\n <img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/IDSNlogo.png\" width=\"300\" alt=\"cognitiveclass.ai logo\" />\n</center>\n\n# Lists in Python\n\nEstimated time needed: **15** minutes\n\n## Objectives\n\nAfter completing this lab you will be able to:\n\n- Perform list operations in Python, including indexing, list manipulation and copy/clone list.\n",
"_____no_output_____"
],
[
"<h2>Table of Contents</h2>\n<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <ul>\n <li>\n <a href=\"#dataset\">About the Dataset</a>\n </li>\n <li>\n <a href=\"#list\">Lists</a>\n <ul>\n <li><a href=\"index\">Indexing</a></li>\n <li><a href=\"content\">List Content</a></li>\n <li><a href=\"op\">List Operations</a></li>\n <li><a href=\"co\">Copy and Clone List</a></li>\n </ul>\n </li>\n <li>\n <a href=\"#quiz\">Quiz on Lists</a>\n </li>\n </ul>\n \n</div>\n\n<hr>\n",
"_____no_output_____"
],
[
"<h2 id=\"#dataset\">About the Dataset</h2>\n",
"_____no_output_____"
],
[
"Imagine you received album recommendations from your friends and compiled all of the recommandations into a table, with specific information about each album.\n\nThe table has one row for each movie and several columns:\n\n- **artist** - Name of the artist\n- **album** - Name of the album\n- **released_year** - Year the album was released\n- **length_min_sec** - Length of the album (hours,minutes,seconds)\n- **genre** - Genre of the album\n- **music_recording_sales_millions** - Music recording sales (millions in USD) on [SONG://DATABASE](http://www.song-database.com/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01)\n- **claimed_sales_millions** - Album's claimed sales (millions in USD) on [SONG://DATABASE](http://www.song-database.com/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01)\n- **date_released** - Date on which the album was released\n- **soundtrack** - Indicates if the album is the movie soundtrack (Y) or (N)\n- **rating_of_friends** - Indicates the rating from your friends from 1 to 10\n <br>\n <br>\n\nThe dataset can be seen below:\n\n<font size=\"1\">\n<table font-size:xx-small>\n <tr>\n <th>Artist</th>\n <th>Album</th> \n <th>Released</th>\n <th>Length</th>\n <th>Genre</th> \n <th>Music recording sales (millions)</th>\n <th>Claimed sales (millions)</th>\n <th>Released</th>\n <th>Soundtrack</th>\n <th>Rating (friends)</th>\n </tr>\n <tr>\n <td>Michael Jackson</td>\n <td>Thriller</td> \n <td>1982</td>\n <td>00:42:19</td>\n <td>Pop, rock, R&B</td>\n <td>46</td>\n <td>65</td>\n <td>30-Nov-82</td>\n <td></td>\n <td>10.0</td>\n </tr>\n <tr>\n <td>AC/DC</td>\n <td>Back in Black</td> \n <td>1980</td>\n <td>00:42:11</td>\n <td>Hard rock</td>\n <td>26.1</td>\n <td>50</td>\n <td>25-Jul-80</td>\n <td></td>\n <td>8.5</td>\n </tr>\n <tr>\n <td>Pink Floyd</td>\n <td>The Dark Side of the Moon</td> \n <td>1973</td>\n <td>00:42:49</td>\n <td>Progressive rock</td>\n <td>24.2</td>\n <td>45</td>\n <td>01-Mar-73</td>\n <td></td>\n <td>9.5</td>\n </tr>\n <tr>\n <td>Whitney Houston</td>\n <td>The Bodyguard</td> \n <td>1992</td>\n <td>00:57:44</td>\n <td>Soundtrack/R&B, soul, pop</td>\n <td>26.1</td>\n <td>50</td>\n <td>25-Jul-80</td>\n <td>Y</td>\n <td>7.0</td>\n </tr>\n <tr>\n <td>Meat Loaf</td>\n <td>Bat Out of Hell</td> \n <td>1977</td>\n <td>00:46:33</td>\n <td>Hard rock, progressive rock</td>\n <td>20.6</td>\n <td>43</td>\n <td>21-Oct-77</td>\n <td></td>\n <td>7.0</td>\n </tr>\n <tr>\n <td>Eagles</td>\n <td>Their Greatest Hits (1971-1975)</td> \n <td>1976</td>\n <td>00:43:08</td>\n <td>Rock, soft rock, folk rock</td>\n <td>32.2</td>\n <td>42</td>\n <td>17-Feb-76</td>\n <td></td>\n <td>9.5</td>\n </tr>\n <tr>\n <td>Bee Gees</td>\n <td>Saturday Night Fever</td> \n <td>1977</td>\n <td>1:15:54</td>\n <td>Disco</td>\n <td>20.6</td>\n <td>40</td>\n <td>15-Nov-77</td>\n <td>Y</td>\n <td>9.0</td>\n </tr>\n <tr>\n <td>Fleetwood Mac</td>\n <td>Rumours</td> \n <td>1977</td>\n <td>00:40:01</td>\n <td>Soft rock</td>\n <td>27.9</td>\n <td>40</td>\n <td>04-Feb-77</td>\n <td></td>\n <td>9.5</td>\n </tr>\n</table></font>\n",
"_____no_output_____"
],
[
"<hr>\n",
"_____no_output_____"
],
[
"<h2 id=\"list\">Lists</h2>\n",
"_____no_output_____"
],
[
"<h3 id=\"index\">Indexing</h3>\n",
"_____no_output_____"
],
[
"We are going to take a look at lists in Python. A list is a sequenced collection of different objects such as integers, strings, and other lists as well. The address of each element within a list is called an <b>index</b>. An index is used to access and refer to items within a list.\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%202/images/ListsIndex.png\" width=\"1000\" />\n",
"_____no_output_____"
],
[
" To create a list, type the list within square brackets <b>[ ]</b>, with your content inside the parenthesis and separated by commas. Let’s try it!\n",
"_____no_output_____"
]
],
[
[
"# Create a list\n\nL = [\"Michael Jackson\", 10.1, 1982]\nL",
"_____no_output_____"
]
],
[
[
"We can use negative and regular indexing with a list :\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%202/images/ListsNeg.png\" width=\"1000\" />\n",
"_____no_output_____"
]
],
[
[
"# Print the elements on each index\n\nprint('the same element using negative and positive indexing:\\n Postive:',L[0],\n'\\n Negative:' , L[-3] )\nprint('the same element using negative and positive indexing:\\n Postive:',L[1],\n'\\n Negative:' , L[-2] )\nprint('the same element using negative and positive indexing:\\n Postive:',L[2],\n'\\n Negative:' , L[-1] )",
"the same element using negative and positive indexing:\n Postive: Michael Jackson \n Negative: Michael Jackson\nthe same element using negative and positive indexing:\n Postive: 10.1 \n Negative: 10.1\nthe same element using negative and positive indexing:\n Postive: 1982 \n Negative: 1982\n"
]
],
[
[
"<h3 id=\"content\">List Content</h3>\n",
"_____no_output_____"
],
[
"Lists can contain strings, floats, and integers. We can nest other lists, and we can also nest tuples and other data structures. The same indexing conventions apply for nesting: \n",
"_____no_output_____"
]
],
[
[
"# Sample List\n\n[\"Michael Jackson\", 10.1, 1982, [1, 2], (\"A\", 1)]",
"_____no_output_____"
]
],
[
[
"<h3 id=\"op\">List Operations</h3>\n",
"_____no_output_____"
],
[
" We can also perform slicing in lists. For example, if we want the last two elements, we use the following command:\n",
"_____no_output_____"
]
],
[
[
"# Sample List\n\nL = [\"Michael Jackson\", 10.1,1982,\"MJ\",1]\nL",
"_____no_output_____"
]
],
[
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%202/images/ListsSlice.png\" width=\"1000\">\n",
"_____no_output_____"
]
],
[
[
"# List slicing\n\nL[3:5]",
"_____no_output_____"
]
],
[
[
"We can use the method <code>extend</code> to add new elements to the list:\n",
"_____no_output_____"
]
],
[
[
"# Use extend to add elements to list\n\nL = [ \"Michael Jackson\", 10.2]\nL.extend(['pop', 10])\nL",
"_____no_output_____"
]
],
[
[
"Another similar method is <code>append</code>. If we apply <code>append</code> instead of <code>extend</code>, we add one element to the list:\n",
"_____no_output_____"
]
],
[
[
"# Use append to add elements to list\n\nL = [ \"Michael Jackson\", 10.2]\nL.append(['pop', 10])\nL",
"_____no_output_____"
]
],
[
[
" Each time we apply a method, the list changes. If we apply <code>extend</code> we add two new elements to the list. The list <code>L</code> is then modified by adding two new elements:\n",
"_____no_output_____"
]
],
[
[
"# Use extend to add elements to list\n\nL = [ \"Michael Jackson\", 10.2]\nL.extend(['pop', 10])\nL",
"_____no_output_____"
]
],
[
[
"If we append the list <code>['a','b']</code> we have one new element consisting of a nested list:\n",
"_____no_output_____"
]
],
[
[
"# Use append to add elements to list\n\nL.append(['a','b'])\nL",
"_____no_output_____"
]
],
[
[
"As lists are mutable, we can change them. For example, we can change the first element as follows:\n",
"_____no_output_____"
]
],
[
[
"# Change the element based on the index\n\nA = [\"disco\", 10, 1.2]\nprint('Before change:', A)\nA[0] = 'hard rock'\nprint('After change:', A)",
"Before change: ['disco', 10, 1.2]\nAfter change: ['hard rock', 10, 1.2]\n"
]
],
[
[
" We can also delete an element of a list using the <code>del</code> command:\n",
"_____no_output_____"
]
],
[
[
"# Delete the element based on the index\n\nprint('Before change:', A)\ndel(A[0])\nprint('After change:', A)",
"Before change: ['hard rock', 10, 1.2]\nAfter change: [10, 1.2]\n"
]
],
[
[
"We can convert a string to a list using <code>split</code>. For example, the method <code>split</code> translates every group of characters separated by a space into an element in a list:\n",
"_____no_output_____"
]
],
[
[
"# Split the string, default is by space\n\n'hard rock'.split()",
"_____no_output_____"
]
],
[
[
"We can use the split function to separate strings on a specific character. We pass the character we would like to split on into the argument, which in this case is a comma. The result is a list, and each element corresponds to a set of characters that have been separated by a comma: \n",
"_____no_output_____"
]
],
[
[
"# Split the string by comma\n\n'A,B,C,D'.split(',')",
"_____no_output_____"
]
],
[
[
"<h3 id=\"co\">Copy and Clone List</h3>\n",
"_____no_output_____"
],
[
"When we set one variable <b>B</b> equal to <b>A</b>; both <b>A</b> and <b>B</b> are referencing the same list in memory:\n",
"_____no_output_____"
]
],
[
[
"# Copy (copy by reference) the list A\n\nA = [\"hard rock\", 10, 1.2]\nB = A\nprint('A:', A)\nprint('B:', B)",
"A: ['hard rock', 10, 1.2]\nB: ['hard rock', 10, 1.2]\n"
]
],
[
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%202/images/ListsRef.png\" width=\"1000\" align=\"center\">\n",
"_____no_output_____"
],
[
"Initially, the value of the first element in <b>B</b> is set as hard rock. If we change the first element in <b>A</b> to <b>banana</b>, we get an unexpected side effect. As <b>A</b> and <b>B</b> are referencing the same list, if we change list <b>A</b>, then list <b>B</b> also changes. If we check the first element of <b>B</b> we get banana instead of hard rock:\n",
"_____no_output_____"
]
],
[
[
"# Examine the copy by reference\n\nprint('B[0]:', B[0])\nA[0] = \"banana\"\nprint('B[0]:', B[0])",
"_____no_output_____"
]
],
[
[
"This is demonstrated in the following figure: \n",
"_____no_output_____"
],
[
"<img src = \"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%202/images/ListsRefGif.gif\" width=\"1000\" />\n",
"_____no_output_____"
],
[
"You can clone list **A** by using the following syntax:\n",
"_____no_output_____"
]
],
[
[
"# Clone (clone by value) the list A\n\nB = A[:]\nB",
"_____no_output_____"
]
],
[
[
" Variable **B** references a new copy or clone of the original list; this is demonstrated in the following figure:\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%202/images/ListsVal.gif\" width=\"1000\" />\n",
"_____no_output_____"
],
[
"Now if you change <b>A</b>, <b>B</b> will not change: \n",
"_____no_output_____"
]
],
[
[
"print('B[0]:', B[0])\nA[0] = \"hard rock\"\nprint('B[0]:', B[0])",
"_____no_output_____"
]
],
[
[
"<h2 id=\"quiz\">Quiz on List</h2>\n",
"_____no_output_____"
],
[
"Create a list <code>a_list</code>, with the following elements <code>1</code>, <code>hello</code>, <code>[1,2,3]</code> and <code>True</code>. \n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nlst = [1,'hello',[1,2,3],True]\nprint(lst)",
"[1, 'hello', [1, 2, 3], True]\n"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\na_list = [1, 'hello', [1, 2, 3] , True]\na_list\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"Find the value stored at index 1 of <code>a_list</code>.\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nlst[1]",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\na_list[1]\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"Retrieve the elements stored at index 1, 2 and 3 of <code>a_list</code>.\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nlst[1:4]",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\na_list[1:4]\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"Concatenate the following lists <code>A = [1, 'a']</code> and <code>B = [2, 1, 'd']</code>:\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nA = [1, 'a']\nB = [2, 1, 'd']",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nA = [1, 'a'] \nB = [2, 1, 'd']\nA + B\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"<hr>\n<h2>The last exercise!</h2>\n<p>Congratulations, you have completed your first lesson and hands-on lab in Python. However, there is one more thing you need to do. The Data Science community encourages sharing work. The best way to share and showcase your work is to share it on GitHub. By sharing your notebook on GitHub you are not only building your reputation with fellow data scientists, but you can also show it off when applying for a job. Even though this was your first piece of work, it is never too early to start building good habits. So, please read and follow <a href=\"https://cognitiveclass.ai/blog/data-scientists-stand-out-by-sharing-your-notebooks/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\" target=\"_blank\">this article</a> to learn how to share your work.\n<hr>\n",
"_____no_output_____"
],
[
"## Author\n\n<a href=\"https://www.linkedin.com/in/joseph-s-50398b136/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\" target=\"_blank\">Joseph Santarcangelo</a>\n\n## Other contributors\n\n<a href=\"www.linkedin.com/in/jiahui-mavis-zhou-a4537814a\">Mavis Zhou</a>\n\n## Change Log\n\n| Date (YYYY-MM-DD) | Version | Changed By | Change Description |\n| ----------------- | ------- | ---------- | ---------------------------------- |\n| 2020-08-26 | 2.0 | Lavanya | Moved lab to course repo in GitLab |\n| | | | |\n| | | | |\n\n## <h3 align=\"center\"> © IBM Corporation 2020. All rights reserved. <h3/>\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ecc87fcfb454447a7593dbd8d4edab2244066197 | 4,455 | ipynb | Jupyter Notebook | notebooks/course/videos/mlm_processing.ipynb | yvr1037/transformers-of-Dian | 759a867e64c725e17a7aef37c78dd54b4c1fda81 | [
"Apache-2.0"
] | null | null | null | notebooks/course/videos/mlm_processing.ipynb | yvr1037/transformers-of-Dian | 759a867e64c725e17a7aef37c78dd54b4c1fda81 | [
"Apache-2.0"
] | null | null | null | notebooks/course/videos/mlm_processing.ipynb | yvr1037/transformers-of-Dian | 759a867e64c725e17a7aef37c78dd54b4c1fda81 | [
"Apache-2.0"
] | null | null | null | 28.375796 | 181 | 0.560494 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecc89beb30768f8a8a0c5207f1e98494545c9208 | 6,512 | ipynb | Jupyter Notebook | Code/para_dep_bru/Figure 2.ipynb | noninvertibility/noninvertibility | 6c7b1904b68e073edeb5ee02f4af9c8c71d1e437 | [
"Unlicense"
] | null | null | null | Code/para_dep_bru/Figure 2.ipynb | noninvertibility/noninvertibility | 6c7b1904b68e073edeb5ee02f4af9c8c71d1e437 | [
"Unlicense"
] | null | null | null | Code/para_dep_bru/Figure 2.ipynb | noninvertibility/noninvertibility | 6c7b1904b68e073edeb5ee02f4af9c8c71d1e437 | [
"Unlicense"
] | null | null | null | 32.56 | 160 | 0.498925 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\nfrom tqdm import tqdm_notebook\nfrom scipy.optimize import root\n\nimport warnings\n\nwarnings.filterwarnings('ignore')\n\n%matplotlib inline",
"_____no_output_____"
],
[
"default_tau = 0.15\ndefault_std = 2\ndefault_a = 1\ndefault_b = 2",
"_____no_output_____"
],
[
"def brusselator_euler(x, tau=default_tau, a=default_a, b=default_b):\n x_n = x[0] + tau * (x[0] ** 2 * x[1] - (b + 1) * x[0] + a)\n y_n = x[1] + tau * (-x[0] ** 2 * x[1] + b * x[0])\n return np.array([x_n, y_n])",
"_____no_output_____"
],
[
"def get_initial(a=default_a, b=default_b, std=default_std):\n mean = a, b/a\n return [np.random.normal(mean[0], std), np.random.normal(mean[1], std)]",
"_____no_output_____"
],
[
"def gen_states(tau=default_tau, iter_step=3000, a=default_a, b=default_b, std=default_std):\n initial = get_initial(a=a, b=b, std=std)\n states = [initial]\n for i in range(iter_step):\n state = brusselator_euler(x=states[-1], tau=tau, a=a, b=b)\n states.append(state)\n states = np.array(states)\n return states",
"_____no_output_____"
],
[
"def gen_states_true(tau=default_tau, iter_step=3000, a=default_a, b=default_b, std=default_std, max_allowed_step_size=1):\n states = gen_states(tau=tau, iter_step=iter_step, a=a, b=b, std=std)\n \n def check_bad(states):\n steps = np.diff(states, axis=0)\n return np.isnan(states).any()\n while check_bad(states):\n states = gen_states(tau=tau, iter_step=iter_step, a=a, b=b, std=std)\n return states",
"_____no_output_____"
],
[
"def J_curve0(x, tau=default_tau, a=default_a, b=default_b):\n y = (tau * (1 - tau) * x ** 2 + tau * (b + 1) - 1) / (2 * x * tau)\n return y",
"_____no_output_____"
],
[
"def analytical_preimages(states, tau=default_tau, a=default_a, b=default_b):\n x_n, y_n = states[:, 0], states[:, 1]\n A = tau - tau ** 2\n B = tau ** 2 - tau * x_n - tau * y_n\n C = tau * b + tau - 1\n D = x_n - tau * a\n preimages = []\n for i in range(states.shape[0]):\n x_pre = np.sort(np.roots([A, B[i], C, D[i]]))\n if np.abs(x_pre[1] - x_n[i]) > np.abs(x_pre[2] - x_n[i]):\n x_pre = x_pre[[0, 2, 1]]\n y_pre = (x_n[i] - x_pre + (b + 1) * tau * x_pre - tau * a) / (tau * x_pre ** 2)\n pre = np.vstack([x_pre, y_pre])\n preimages.append(pre)\n return np.array(preimages)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(12, 12), ncols=2, nrows=2)\nb = [2, 2.5, 3, 3.2]\nfor i in range(2):\n for j in range(2):\n indice_b = 2 * i + j\n states = gen_states_true(b=b[indice_b])\n states = states[-1000:, :]\n J0_x1 = np.linspace(-10, 0, 10000, endpoint=False)\n J0_x2 = np.linspace(10, 0, 10000, endpoint=False)[::-1]\n\n J0_y1 = J_curve0(J0_x1, b=b[indice_b])\n J0_y2 = J_curve0(J0_x2, b=b[indice_b])\n\n preimages = analytical_preimages(states, b=b[indice_b])\n\n ax[i, j].scatter(J0_x1, J0_y1, color='red', alpha=0.5, label=r'$J_0$', s=10, zorder=99)\n ax[i, j].scatter(J0_x2, J0_y2, color='red', alpha=0.5, s=10, zorder=99)\n\n ax[i, j].scatter(states[:-1, 0], states[:-1, 1], alpha=0.7, color='orange', label=r'$\\Gamma$, $F^{-1}(\\Gamma)$', s=10, zorder=99)\n pre1, pre2, pre3 = preimages[:, :, 0], preimages[:, :, 1], preimages[:, :, 2]\n real_1 = np.logical_and(*np.isreal(pre1).T)\n real_2 = np.logical_and(*np.isreal(pre2).T)\n real_3 = np.logical_and(*np.isreal(pre3).T)\n all_real = np.logical_and(np.logical_and(real_1, real_2), real_3)\n ax[i, j].scatter(pre3[all_real, 0], pre3[all_real, 1], alpha=0.7, color='green',label=r'$F^{-1}(\\Gamma)^{\\prime}$', s=10, zorder=50)\n ax[i, j].scatter(pre1[all_real, 0], pre1[all_real, 1], alpha=0.7, color='purple',label=r'$F^{-1}(\\Gamma)^{\\prime \\prime}$', s=10, zorder=50)\n\n if not i and not j:\n ax[i, j].legend(fontsize=16)\n\n ax[i, 0].set_ylabel('$y$')\n ax[0, j].set_xlabel('$x$')\n\n ax[i, j].set_xlim(-5, 10)\n ax[i, j].set_ylim(-5, 10)\n ax[i, j].set_title(r'$(a, b) = $ {}'.format((1, b[indice_b])), fontsize=20)\n\n ax[i, j].set_xticklabels(['' for x in ax[i, j].get_xticks()])\n ax[i, j].set_yticklabels(['' for y in ax[i, j].get_yticks()])\n\nplt.tight_layout()\n\n# plt.savefig('inv.pdf', bbox_inches='tight')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecc8d26d89b103e45b73601cd713f9cdb005b54f | 26,298 | ipynb | Jupyter Notebook | Regression/Assignmet_five/.ipynb_checkpoints/week-5-lasso-assignment-1-blank-checkpoint.ipynb | DibyashaPanda/Machine-Learning-Specialization | fcc59529d328e0fafa7ce49b336538b5ef96d81e | [
"MIT"
] | 25 | 2016-07-18T19:11:51.000Z | 2021-02-21T11:08:51.000Z | Regression/Assignmet_five/.ipynb_checkpoints/week-5-lasso-assignment-1-blank-checkpoint.ipynb | DibyashaPanda/Machine-Learning-Specialization | fcc59529d328e0fafa7ce49b336538b5ef96d81e | [
"MIT"
] | null | null | null | Regression/Assignmet_five/.ipynb_checkpoints/week-5-lasso-assignment-1-blank-checkpoint.ipynb | DibyashaPanda/Machine-Learning-Specialization | fcc59529d328e0fafa7ce49b336538b5ef96d81e | [
"MIT"
] | 30 | 2016-12-18T02:46:27.000Z | 2021-04-02T08:27:11.000Z | 32.8725 | 286 | 0.536923 | [
[
[
"# Regression Week 5: Feature Selection and LASSO (Interpretation)",
"_____no_output_____"
],
[
"In this notebook, you will use LASSO to select features, building on a pre-implemented solver for LASSO (using GraphLab Create, though you can use other solvers). You will:\n* Run LASSO with different L1 penalties.\n* Choose best L1 penalty using a validation set.\n* Choose best L1 penalty using a validation set, with additional constraint on the size of subset.\n\nIn the second notebook, you will implement your own LASSO solver, using coordinate descent. ",
"_____no_output_____"
],
[
"# Fire up graphlab create",
"_____no_output_____"
]
],
[
[
"import graphlab\ngraphlab.product_key.set_product_key(\"C0C2-04B4-D94B-70F6-8771-86F9-C6E1-E122\")",
"_____no_output_____"
]
],
[
[
"# Load in house sales data\n\nDataset is from house sales in King County, the region where the city of Seattle, WA is located.",
"_____no_output_____"
]
],
[
[
"sales = graphlab.SFrame('kc_house_data.gl/kc_house_data.gl')",
"[INFO] \u001b[1;32m1451721816 : INFO: (initialize_globals_from_environment:282): Setting configuration variable GRAPHLAB_FILEIO_ALTERNATIVE_SSL_CERT_FILE to /usr/local/lib/python2.7/dist-packages/certifi/cacert.pem\n\u001b[0m\u001b[1;32m1451721816 : INFO: (initialize_globals_from_environment:282): Setting configuration variable GRAPHLAB_FILEIO_ALTERNATIVE_SSL_CERT_DIR to \n\u001b[0mThis non-commercial license of GraphLab Create is assigned to [email protected] and will expire on December 06, 2016. For commercial licensing options, visit https://dato.com/buy/.\n\n[INFO] Start server at: ipc:///tmp/graphlab_server-29098 - Server binary: /usr/local/lib/python2.7/dist-packages/graphlab/unity_server - Server log: /tmp/graphlab_server_1451721816.log\n[INFO] GraphLab Server Version: 1.7.1\n"
]
],
[
[
"# Create new features",
"_____no_output_____"
],
[
"As in Week 2, we consider features that are some transformations of inputs.",
"_____no_output_____"
]
],
[
[
"from math import log, sqrt\nsales['sqft_living_sqrt'] = sales['sqft_living'].apply(sqrt)\nsales['sqft_lot_sqrt'] = sales['sqft_lot'].apply(sqrt)\nsales['bedrooms_square'] = sales['bedrooms']*sales['bedrooms']\n\n# In the dataset, 'floors' was defined with type string, \n# so we'll convert them to float, before creating a new feature.\nsales['floors'] = sales['floors'].astype(float) \nsales['floors_square'] = sales['floors']*sales['floors']",
"_____no_output_____"
]
],
[
[
"* Squaring bedrooms will increase the separation between not many bedrooms (e.g. 1) and lots of bedrooms (e.g. 4) since 1^2 = 1 but 4^2 = 16. Consequently this variable will mostly affect houses with many bedrooms.\n* On the other hand, taking square root of sqft_living will decrease the separation between big house and small house. The owner may not be exactly twice as happy for getting a house that is twice as big.",
"_____no_output_____"
],
[
"# Learn regression weights with L1 penalty",
"_____no_output_____"
],
[
"Let us fit a model with all the features available, plus the features we just created above.",
"_____no_output_____"
]
],
[
[
"all_features = ['bedrooms', 'bedrooms_square',\n 'bathrooms',\n 'sqft_living', 'sqft_living_sqrt',\n 'sqft_lot', 'sqft_lot_sqrt',\n 'floors', 'floors_square',\n 'waterfront', 'view', 'condition', 'grade',\n 'sqft_above',\n 'sqft_basement',\n 'yr_built', 'yr_renovated']",
"_____no_output_____"
]
],
[
[
"Applying L1 penalty requires adding an extra parameter (`l1_penalty`) to the linear regression call in GraphLab Create. (Other tools may have separate implementations of LASSO.) Note that it's important to set `l2_penalty=0` to ensure we don't introduce an additional L2 penalty.",
"_____no_output_____"
]
],
[
[
"model_all = graphlab.linear_regression.create(sales, target='price', features=all_features,\n validation_set=None, \n l2_penalty=0., l1_penalty=1e10)",
"PROGRESS: Linear regression:\nPROGRESS: --------------------------------------------------------\nPROGRESS: Number of examples : 21613\nPROGRESS: Number of features : 17\nPROGRESS: Number of unpacked features : 17\nPROGRESS: Number of coefficients : 18\nPROGRESS: Starting Accelerated Gradient (FISTA)\nPROGRESS: --------------------------------------------------------\nPROGRESS: +-----------+----------+-----------+--------------+--------------------+---------------+\nPROGRESS: | Iteration | Passes | Step size | Elapsed Time | Training-max_error | Training-rmse |\nPROGRESS: +-----------+----------+-----------+--------------+--------------------+---------------+\nPROGRESS: Tuning step size. First iteration could take longer than subsequent iterations.\nPROGRESS: | 1 | 2 | 0.000002 | 1.246046 | 6962915.603493 | 426631.749026 |\nPROGRESS: | 2 | 3 | 0.000002 | 1.273492 | 6843144.200219 | 392488.929838 |\nPROGRESS: | 3 | 4 | 0.000002 | 1.302511 | 6831900.032123 | 385340.166783 |\nPROGRESS: | 4 | 5 | 0.000002 | 1.328765 | 6847166.848958 | 384842.383767 |\nPROGRESS: | 5 | 6 | 0.000002 | 1.354595 | 6869667.895833 | 385998.458623 |\nPROGRESS: | 6 | 7 | 0.000002 | 1.381158 | 6847177.773672 | 380824.455891 |\nPROGRESS: +-----------+----------+-----------+--------------+--------------------+---------------+\nPROGRESS: TERMINATED: Iteration limit reached.\nPROGRESS: This model may not be optimal. To improve it, consider increasing `max_iterations`.\n"
]
],
[
[
"Find what features had non-zero weight.",
"_____no_output_____"
]
],
[
[
"model_all.get(\"coefficients\").print_rows(num_rows=18, num_columns=3)\nmodel_all['coefficients']['value'].nnz()\n",
"+------------------+-------+---------------+\n| name | index | value |\n+------------------+-------+---------------+\n| (intercept) | None | 196100.937806 |\n| bedrooms | None | 2181.57432107 |\n| bedrooms_square | None | 0.0 |\n| bathrooms | None | 17962.6966612 |\n| sqft_living | None | 34.1424656512 |\n| sqft_living_sqrt | None | 789.319789078 |\n| sqft_lot | None | 0.0 |\n| sqft_lot_sqrt | None | 0.0 |\n| floors | None | 3665.9308176 |\n| floors_square | None | 0.0 |\n| waterfront | None | 0.0 |\n| view | None | 11333.8410308 |\n| condition | None | 0.0 |\n| grade | None | 3578.90040044 |\n| sqft_above | None | 32.7432013718 |\n| sqft_basement | None | 12.7953811359 |\n| yr_built | None | 0.0 |\n| yr_renovated | None | 0.0 |\n+------------------+-------+---------------+\n[18 rows x 3 columns]\n\n"
]
],
[
[
"Note that a majority of the weights have been set to zero. So by setting an L1 penalty that's large enough, we are performing a subset selection. \n\n***QUIZ QUESTION***:\nAccording to this list of weights, which of the features have been chosen? ",
"_____no_output_____"
],
[
"# Selecting an L1 penalty",
"_____no_output_____"
],
[
"To find a good L1 penalty, we will explore multiple values using a validation set. Let us do three way split into train, validation, and test sets:\n* Split our sales data into 2 sets: training and test\n* Further split our training data into two sets: train, validation\n\nBe *very* careful that you use seed = 1 to ensure you get the same answer!",
"_____no_output_____"
]
],
[
[
"(training_and_validation, testing) = sales.random_split(.9,seed=1) # initial train/test split\n(training, validation) = training_and_validation.random_split(0.5, seed=1) # split training into train and validate",
"_____no_output_____"
]
],
[
[
"Next, we write a loop that does the following:\n* For `l1_penalty` in [10^1, 10^1.5, 10^2, 10^2.5, ..., 10^7] (to get this in Python, type `np.logspace(1, 7, num=13)`.)\n * Fit a regression model with a given `l1_penalty` on TRAIN data. Specify `l1_penalty=l1_penalty` and `l2_penalty=0.` in the parameter list.\n * Compute the RSS on VALIDATION data (here you will want to use `.predict()`) for that `l1_penalty`\n* Report which `l1_penalty` produced the lowest RSS on validation data.\n\nWhen you call `linear_regression.create()` make sure you set `validation_set = None`.\n\nNote: you can turn off the print out of `linear_regression.create()` with `verbose = False`",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndef get_RSS(prediction, output):\n residual = output - prediction\n # square the residuals and add them up\n RS = residual*residual\n RSS = RS.sum()\n return(RSS)\n\nfor l1_penalty in np.logspace(1, 7, num=13):\n A = 0 \n print l1_penalty\n model_all = graphlab.linear_regression.create(training, target='price', features=all_features,\n validation_set=None, verbose = False,\n l2_penalty=0, l1_penalty=l1_penalty)\n predictions=model_all.predict(validation)\n A = get_RSS(predictions,validation['price'])\n print A\n",
"10.0\n6.25766285142e+14\n31.6227766017\n6.25766285362e+14\n100.0\n6.25766286058e+14\n316.227766017\n6.25766288257e+14\n1000.0\n6.25766295212e+14\n3162.27766017\n6.25766317206e+14\n10000.0\n6.25766386761e+14\n31622.7766017\n6.25766606749e+14\n100000.0\n6.25767302792e+14\n316227.766017\n6.25769507644e+14\n1000000.0\n6.25776517727e+14\n3162277.66017\n6.25799062845e+14\n10000000.0\n6.25883719085e+14\n"
],
[
"model_all = graphlab.linear_regression.create(testing, target='price', features=all_features,\n validation_set=None, verbose = False,\n l2_penalty=0, l1_penalty=10)\nmodel_all['coefficients']['value'].nnz()",
"_____no_output_____"
]
],
[
[
"*** QUIZ QUESTIONS ***\n1. What was the best value for the `l1_penalty`?\n2. What is the RSS on TEST data of the model with the best `l1_penalty`?",
"_____no_output_____"
],
[
"***QUIZ QUESTION***\nAlso, using this value of L1 penalty, how many nonzero weights do you have?",
"_____no_output_____"
],
[
"# Limit the number of nonzero weights\n\nWhat if we absolutely wanted to limit ourselves to, say, 7 features? This may be important if we want to derive \"a rule of thumb\" --- an interpretable model that has only a few features in them.",
"_____no_output_____"
],
[
"In this section, you are going to implement a simple, two phase procedure to achive this goal:\n1. Explore a large range of `l1_penalty` values to find a narrow region of `l1_penalty` values where models are likely to have the desired number of non-zero weights.\n2. Further explore the narrow region you found to find a good value for `l1_penalty` that achieves the desired sparsity. Here, we will again use a validation set to choose the best value for `l1_penalty`.",
"_____no_output_____"
]
],
[
[
"max_nonzeros = 7",
"_____no_output_____"
]
],
[
[
"## Exploring the larger range of values to find a narrow range with the desired sparsity\n\nLet's define a wide range of possible `l1_penalty_values`:",
"_____no_output_____"
]
],
[
[
"l1_penalty_values = np.logspace(8, 10, num=20)",
"_____no_output_____"
]
],
[
[
"Now, implement a loop that search through this space of possible `l1_penalty` values:\n\n* For `l1_penalty` in `np.logspace(8, 10, num=20)`:\n * Fit a regression model with a given `l1_penalty` on TRAIN data. Specify `l1_penalty=l1_penalty` and `l2_penalty=0.` in the parameter list. When you call `linear_regression.create()` make sure you set `validation_set = None`\n * Extract the weights of the model and count the number of nonzeros. Save the number of nonzeros to a list.\n * *Hint: `model['coefficients']['value']` gives you an SArray with the parameters you learned. If you call the method `.nnz()` on it, you will find the number of non-zero parameters!* ",
"_____no_output_____"
]
],
[
[
"for l1_penalty in np.logspace(8, 10, num=20):\n A = 0\n predictions = 0\n print l1_penalty\n model_all = graphlab.linear_regression.create(training, target='price', features=all_features,\n validation_set=None, verbose = False,\n l2_penalty=0, l1_penalty=l1_penalty)\n predictions=model_all.predict(validation)\n A = get_RSS(predictions,validation['price'])\n print A\n print model_all['coefficients']['value'].nnz()",
"100000000.0\n6.27492659875e+14\n18\n127427498.57\n6.28210516771e+14\n18\n162377673.919\n6.29176689541e+14\n18\n206913808.111\n6.30650082719e+14\n18\n263665089.873\n6.32940229287e+14\n17\n335981828.628\n6.3626814023e+14\n17\n428133239.872\n6.41261198311e+14\n17\n545559478.117\n6.48983455376e+14\n17\n695192796.178\n6.60962217696e+14\n17\n885866790.41\n6.77261520728e+14\n16\n1128837891.68\n7.01046815867e+14\n15\n1438449888.29\n7.37850622829e+14\n15\n1832980710.83\n7.9616310964e+14\n13\n2335721469.09\n8.69018172894e+14\n12\n2976351441.63\n9.66925692362e+14\n10\n3792690190.73\n1.08186759232e+15\n6\n4832930238.57\n1.24492736032e+15\n5\n6158482110.66\n1.38416149024e+15\n3\n7847599703.51\n1.23079472046e+15\n1\n10000000000.0\n1.22915716064e+15\n1\n"
]
],
[
[
"Out of this large range, we want to find the two ends of our desired narrow range of `l1_penalty`. At one end, we will have `l1_penalty` values that have too few non-zeros, and at the other end, we will have an `l1_penalty` that has too many non-zeros. \n\nMore formally, find:\n* The largest `l1_penalty` that has more non-zeros than `max_nonzero` (if we pick a penalty smaller than this value, we will definitely have too many non-zero weights)\n * Store this value in the variable `l1_penalty_min` (we will use it later)\n* The smallest `l1_penalty` that has fewer non-zeros than `max_nonzero` (if we pick a penalty larger than this value, we will definitely have too few non-zero weights)\n * Store this value in the variable `l1_penalty_max` (we will use it later)\n\n\n*Hint: there are many ways to do this, e.g.:*\n* Programmatically within the loop above\n* Creating a list with the number of non-zeros for each value of `l1_penalty` and inspecting it to find the appropriate boundaries.",
"_____no_output_____"
]
],
[
[
"l1_penalty_min = 2976351441.63\nl1_penalty_max = 3792690190.73",
"_____no_output_____"
]
],
[
[
"***QUIZ QUESTIONS***\n\nWhat values did you find for `l1_penalty_min` and`l1_penalty_max`? ",
"_____no_output_____"
],
[
"## Exploring the narrow range of values to find the solution with the right number of non-zeros that has lowest RSS on the validation set \n\nWe will now explore the narrow region of `l1_penalty` values we found:",
"_____no_output_____"
]
],
[
[
"l1_penalty_values = np.linspace(l1_penalty_min,l1_penalty_max,20)",
"_____no_output_____"
]
],
[
[
"* For `l1_penalty` in `np.linspace(l1_penalty_min,l1_penalty_max,20)`:\n * Fit a regression model with a given `l1_penalty` on TRAIN data. Specify `l1_penalty=l1_penalty` and `l2_penalty=0.` in the parameter list. When you call `linear_regression.create()` make sure you set `validation_set = None`\n * Measure the RSS of the learned model on the VALIDATION set\n\nFind the model that the lowest RSS on the VALIDATION set and has sparsity *equal* to `max_nonzero`.",
"_____no_output_____"
]
],
[
[
"for l1_penalty in np.linspace(l1_penalty_min,l1_penalty_max,20):\n A = 0\n predictions = 0\n model_all = graphlab.linear_regression.create(training, target='price', features=all_features,\n validation_set=None, verbose = False,\n l2_penalty=0, l1_penalty=l1_penalty)\n predictions=model_all.predict(validation)\n A = get_RSS(predictions,validation['price'])\n if model_all['coefficients']['value'].nnz() <= 7:\n print l1_penalty\n print A\n print model_all['coefficients']['value'].nnz()",
"3448968612.16\n1.04693748875e+15\n7\n3491933809.48\n1.05114762561e+15\n7\n3534899006.8\n1.05599273534e+15\n7\n3577864204.12\n1.06079953176e+15\n7\n3620829401.45\n1.0657076895e+15\n6\n3663794598.77\n1.06946433543e+15\n6\n3706759796.09\n1.07350454959e+15\n6\n3749724993.41\n1.07763277558e+15\n6\n3792690190.73\n1.08186759232e+15\n6\n"
]
],
[
[
"***QUIZ QUESTIONS***\n1. What value of `l1_penalty` in our narrow range has the lowest RSS on the VALIDATION set and has sparsity *equal* to `max_nonzeros`?\n2. What features in this model have non-zero coefficients?",
"_____no_output_____"
]
],
[
[
"model_all = graphlab.linear_regression.create(training, target='price', features=all_features,\n validation_set=None, verbose = False,\n l2_penalty=0, l1_penalty=3448968612.16)\n# model_all['coefficients']['value'].nnz()\n# print np.linspace(l1_penalty_min,l1_penalty_max,20)[0]\nprint model_all['coefficients'].print_rows(num_rows=18, num_columns=3)\n ",
"2976351441.63\n+------------------+-------+---------------+\n| name | index | value |\n+------------------+-------+---------------+\n| (intercept) | None | 196100.937806 |\n| bedrooms | None | 2181.57432107 |\n| bedrooms_square | None | 0.0 |\n| bathrooms | None | 17962.6966612 |\n| sqft_living | None | 34.1424656512 |\n| sqft_living_sqrt | None | 789.319789078 |\n| sqft_lot | None | 0.0 |\n| sqft_lot_sqrt | None | 0.0 |\n| floors | None | 3665.9308176 |\n| floors_square | None | 0.0 |\n| waterfront | None | 0.0 |\n| view | None | 11333.8410308 |\n| condition | None | 0.0 |\n| grade | None | 3578.90040044 |\n| sqft_above | None | 32.7432013718 |\n| sqft_basement | None | 12.7953811359 |\n| yr_built | None | 0.0 |\n| yr_renovated | None | 0.0 |\n+------------------+-------+---------------+\n[18 rows x 3 columns]\n\nNone\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecc8dad428597d98651ba11a0aff22290e21e25d | 176,237 | ipynb | Jupyter Notebook | recommendation systems/.ipynb_checkpoints/Recommendations_with_IBM-checkpoint.ipynb | YueminLi/Recommendations-with-IBM | b0b085bbee53c8159553fa315423d1f1eac4ab63 | [
"MIT"
] | null | null | null | recommendation systems/.ipynb_checkpoints/Recommendations_with_IBM-checkpoint.ipynb | YueminLi/Recommendations-with-IBM | b0b085bbee53c8159553fa315423d1f1eac4ab63 | [
"MIT"
] | null | null | null | recommendation systems/.ipynb_checkpoints/Recommendations_with_IBM-checkpoint.ipynb | YueminLi/Recommendations-with-IBM | b0b085bbee53c8159553fa315423d1f1eac4ab63 | [
"MIT"
] | null | null | null | 58.12566 | 19,592 | 0.666137 | [
[
[
"# Recommendations with IBM\n\nIn this notebook, you will be putting your recommendation skills to use on real data from the IBM Watson Studio platform. \n\n\nYou may either submit your notebook through the workspace here, or you may work from your local machine and submit through the next page. Either way assure that your code passes the project [RUBRIC](https://review.udacity.com/#!/rubrics/2322/view). **Please save regularly.**\n\nBy following the table of contents, you will build out a number of different methods for making recommendations that can be used for different situations. \n\n\n## Table of Contents\n\nI. [Exploratory Data Analysis](#Exploratory-Data-Analysis)<br>\nII. [Rank Based Recommendations](#Rank)<br>\nIII. [User-User Based Collaborative Filtering](#User-User)<br>\nIV. [Content Based Recommendations (EXTRA - NOT REQUIRED)](#Content-Recs)<br>\nV. [Matrix Factorization](#Matrix-Fact)<br>\nVI. [Extras & Concluding](#conclusions)\n\nAt the end of the notebook, you will find directions for how to submit your work. Let's get started by importing the necessary libraries and reading in the data.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport sys\nsys.path.append('../test')\nimport project_tests as t\nimport pickle\n\n%matplotlib inline\n\ndf = pd.read_csv('../data/user-item-interactions.csv')\ndf_content = pd.read_csv('../data/articles_community.csv')\ndel df['Unnamed: 0']\ndel df_content['Unnamed: 0']\n\n# Show df to get an idea of the data\ndf.head()",
"_____no_output_____"
],
[
"# Show df_content to get an idea of the data\ndf_content.head()",
"_____no_output_____"
]
],
[
[
"### <a class=\"anchor\" id=\"Exploratory-Data-Analysis\">Part I : Exploratory Data Analysis</a>\n\nUse the dictionary and cells below to provide some insight into the descriptive statistics of the data.\n\n`1.` What is the distribution of how many articles a user interacts with in the dataset? Provide a visual and descriptive statistics to assist with giving a look at the number of times each user interacts with an article. ",
"_____no_output_____"
]
],
[
[
"df.groupby('email')['article_id'].nunique().median()",
"_____no_output_____"
],
[
"df[~df['email'].isnull()].groupby('email').size().max()",
"_____no_output_____"
],
[
"# Fill in the median and maximum number of user_article interactios below\n\nmedian_val = 3.0 # 50% of individuals interact with ____ number of articles or fewer.\nmax_views_by_user = 364.0 # The maximum number of user-article interactions by any 1 user is ______.",
"_____no_output_____"
],
[
"# boxplot of distribution\nplt.boxplot(df[~df['email'].isnull()].groupby('email').size())",
"_____no_output_____"
],
[
"# histogram of distribution\nplt.hist(df[~df['email'].isnull()].groupby('email').size())",
"_____no_output_____"
]
],
[
[
"`2.` Explore and remove duplicate articles from the **df_content** dataframe. ",
"_____no_output_____"
]
],
[
[
"# Find and explore duplicate articles\ndf_content['article_id'].nunique()",
"_____no_output_____"
],
[
"df_content.shape[0]",
"_____no_output_____"
],
[
"# Remove any rows that have the same article_id - only keep the first\ndf_content = df_content.drop_duplicates('article_id', keep='first')",
"_____no_output_____"
]
],
[
[
"`3.` Use the cells below to find:\n\n**a.** The number of unique articles that have an interaction with a user. \n**b.** The number of unique articles in the dataset (whether they have any interactions or not).<br>\n**c.** The number of unique users in the dataset. (excluding null values) <br>\n**d.** The number of user-article interactions in the dataset.",
"_____no_output_____"
]
],
[
[
"# The number of unique articles that have at least one interaction\n# get the array of article_id that is linked to null users\nnull_email = df[df['email'].isnull()]['article_id'].unique()",
"_____no_output_____"
],
[
"# get the number of unique articles that only have null values in users\nfor i in null_email:\n k = 0\n if df[df['article_id'] == i].shape[0] < 2:\n k += 1\n else:\n pass \nk ",
"_____no_output_____"
],
[
"unique_articles = df['article_id'].nunique() - k\nunique_articles",
"_____no_output_____"
],
[
"# The number of unique articles on the IBM platform\ndf_content['article_id'].nunique()",
"_____no_output_____"
],
[
"# The number of unique users\ndf['email'].nunique()",
"_____no_output_____"
],
[
"# The number of user-article interactions\ndf.shape",
"_____no_output_____"
],
[
"unique_articles = 714 # The number of unique articles that have at least one interaction\ntotal_articles = 1051 # The number of unique articles on the IBM platform\nunique_users = 5148 # The number of unique users\nuser_article_interactions = 45993 # The number of user-article interactions",
"_____no_output_____"
]
],
[
[
"`4.` Use the cells below to find the most viewed **article_id**, as well as how often it was viewed. After talking to the company leaders, the `email_mapper` function was deemed a reasonable way to map users to ids. There were a small number of null values, and it was found that all of these null values likely belonged to a single user (which is how they are stored using the function below).",
"_____no_output_____"
]
],
[
[
"max_views = df.groupby('article_id').size().max()\nmax_views",
"_____no_output_____"
],
[
"df.groupby(['article_id']).size().index[np.where(df.groupby('article_id').size() == 937)][0]",
"_____no_output_____"
],
[
"df[df['article_id'] == 1429.0]",
"_____no_output_____"
],
[
"most_viewed_article_id = str(1429.0) # The most viewed article in the dataset as a string with one value following the decimal \nmax_views = 937 # The most viewed article in the dataset was viewed how many times?",
"_____no_output_____"
],
[
"## No need to change the code here - this will be helpful for later parts of the notebook\n# Run this cell to map the user email to a user_id column and remove the email column\n\ndef email_mapper():\n coded_dict = dict()\n cter = 1\n email_encoded = []\n \n for val in df['email']:\n if val not in coded_dict:\n coded_dict[val] = cter\n cter+=1\n \n email_encoded.append(coded_dict[val])\n return email_encoded\n\nemail_encoded = email_mapper()\ndel df['email']\ndf['user_id'] = email_encoded\n\n# show header\ndf.head()",
"_____no_output_____"
],
[
"## If you stored all your results in the variable names above, \n## you shouldn't need to change anything in this cell\n\nsol_1_dict = {\n '`50% of individuals have _____ or fewer interactions.`': median_val,\n '`The total number of user-article interactions in the dataset is ______.`': user_article_interactions,\n '`The maximum number of user-article interactions by any 1 user is ______.`': max_views_by_user,\n '`The most viewed article in the dataset was viewed _____ times.`': max_views,\n '`The article_id of the most viewed article is ______.`': most_viewed_article_id,\n '`The number of unique articles that have at least 1 rating ______.`': unique_articles,\n '`The number of unique users in the dataset is ______`': unique_users,\n '`The number of unique articles on the IBM platform`': total_articles\n}\n\n# Test your dictionary against the solution\nt.sol_1_test(sol_1_dict)",
"It looks like you have everything right here! Nice job!\n"
]
],
[
[
"### <a class=\"anchor\" id=\"Rank\">Part II: Rank-Based Recommendations</a>\n\nUnlike in the earlier lessons, we don't actually have ratings for whether a user liked an article or not. We only know that a user has interacted with an article. In these cases, the popularity of an article can really only be based on how often an article was interacted with.\n\n`1.` Fill in the function below to return the **n** top articles ordered with most interactions as the top. Test your function using the tests below.",
"_____no_output_____"
]
],
[
[
"def get_top_articles(n, df=df):\n '''\n INPUT:\n n - (int) the number of top articles to return\n df - (pandas dataframe) df as defined at the top of the notebook \n \n OUTPUT:\n top_articles - (list) A list of the top 'n' article titles \n \n '''\n # get a list of top 'n' article_id\n top_list = df.groupby('article_id').size().sort_values(ascending=False).index[0:n]\n \n top_articles = []\n for i in top_list:\n top_articles.append(df[df['article_id'] == i]['title'].iloc[0])\n \n return top_articles # Return the top article titles from df (not df_content)\n\ndef get_top_article_ids(n, df=df):\n '''\n INPUT:\n n - (int) the number of top articles to return\n df - (pandas dataframe) df as defined at the top of the notebook \n \n OUTPUT:\n top_articles - (list) A list of the article ids associated with top 'n' article titles\n \n '''\n \n top_articles = df.groupby('article_id').size().sort_values(ascending=False).index[0:n]\n \n return top_articles # Return the top article ids",
"_____no_output_____"
],
[
"print(get_top_articles(10))\nprint(get_top_article_ids(10))",
"['use deep learning for image classification', 'insights from new york car accident reports', 'visualize car data with brunel', 'use xgboost, scikit-learn & ibm watson machine learning apis', 'predicting churn with the spss random tree algorithm', 'healthcare python streaming application demo', 'finding optimal locations of new store using decision optimization', 'apache spark lab, part 1: basic concepts', 'analyze energy consumption in buildings', 'gosales transactions for logistic regression model']\nFloat64Index([1429.0, 1330.0, 1431.0, 1427.0, 1364.0, 1314.0, 1293.0, 1170.0,\n 1162.0, 1304.0],\n dtype='float64', name='article_id')\n"
],
[
"# Test your function by returning the top 5, 10, and 20 articles\ntop_5 = get_top_articles(5)\ntop_10 = get_top_articles(10)\ntop_20 = get_top_articles(20)\n\n# Test each of your three lists from above\nt.sol_2_test(get_top_articles)",
"Your top_5 looks like the solution list! Nice job.\nYour top_10 looks like the solution list! Nice job.\nYour top_20 looks like the solution list! Nice job.\n"
]
],
[
[
"### <a class=\"anchor\" id=\"User-User\">Part III: User-User Based Collaborative Filtering</a>\n\n\n`1.` Use the function below to reformat the **df** dataframe to be shaped with users as the rows and articles as the columns. \n\n* Each **user** should only appear in each **row** once.\n\n\n* Each **article** should only show up in one **column**. \n\n\n* **If a user has interacted with an article, then place a 1 where the user-row meets for that article-column**. It does not matter how many times a user has interacted with the article, all entries where a user has interacted with an article should be a 1. \n\n\n* **If a user has not interacted with an item, then place a zero where the user-row meets for that article-column**. \n\nUse the tests to make sure the basic structure of your matrix matches what is expected by the solution.",
"_____no_output_____"
]
],
[
[
"# create the user-article matrix with 1's and 0's\n\ndef create_user_item_matrix(df):\n '''\n INPUT:\n df - pandas dataframe with article_id, title, user_id columns\n \n OUTPUT:\n user_item - user item matrix \n \n Description:\n Return a matrix with user ids as rows and article ids on the columns with 1 values where a user interacted with \n an article and a 0 otherwise\n '''\n # Get user_item matrix with size in intersectional cell\n user_item = df.groupby(['user_id', 'article_id']).size().unstack().fillna(0)\n user_item[user_item > 1] = 1\n \n return user_item # return the user_item matrix \n\nuser_item = create_user_item_matrix(df)",
"_____no_output_____"
],
[
"## Tests: You should just need to run this cell. Don't change the code.\nassert user_item.shape[0] == 5149, \"Oops! The number of users in the user-article matrix doesn't look right.\"\nassert user_item.shape[1] == 714, \"Oops! The number of articles in the user-article matrix doesn't look right.\"\nassert user_item.sum(axis=1)[1] == 36, \"Oops! The number of articles seen by user 1 doesn't look right.\"\nprint(\"You have passed our quick tests! Please proceed!\")",
"You have passed our quick tests! Please proceed!\n"
]
],
[
[
"`2.` Complete the function below which should take a user_id and provide an ordered list of the most similar users to that user (from most similar to least similar). The returned result should not contain the provided user_id, as we know that each user is similar to him/herself. Because the results for each user here are binary, it (perhaps) makes sense to compute similarity as the dot product of two users. \n\nUse the tests to test your function.",
"_____no_output_____"
]
],
[
[
"def find_similar_users(user_id, user_item=user_item):\n '''\n INPUT:\n user_id - (int) a user_id\n user_item - (pandas dataframe) matrix of users by articles: \n 1's when a user has interacted with an article, 0 otherwise\n \n OUTPUT:\n similar_users - (list) an ordered list where the closest users (largest dot product users)\n are listed first\n \n Description:\n Computes the similarity of every pair of users based on the dot product\n Returns an ordered\n \n '''\n # compute similarity of each user to the provided user\n dot_prod_user = user_item.dot(np.transpose(user_item))\n \n # sort by similarity and remove the own user's id\n similar_user = dot_prod_user.loc[user_id].sort_values(ascending=False).drop(user_id)\n \n # create list of just the ids\n most_similar_users = similar_user.index.values\n \n return most_similar_users # return a list of the users in order from most to least similar\n ",
"_____no_output_____"
],
[
"# Do a spot check of your function\nprint(\"The 10 most similar users to user 1 are: {}\".format(find_similar_users(1)[:10]))\nprint(\"The 5 most similar users to user 3933 are: {}\".format(find_similar_users(3933)[:5]))\nprint(\"The 3 most similar users to user 46 are: {}\".format(find_similar_users(46)[:3]))",
"The 10 most similar users to user 1 are: [3933 23 3782 203 4459 131 3870 46 4201 5041]\nThe 5 most similar users to user 3933 are: [ 1 23 3782 4459 203]\nThe 3 most similar users to user 46 are: [4201 23 3782]\n"
]
],
[
[
"`3.` Now that you have a function that provides the most similar users to each user, you will want to use these users to find articles you can recommend. Complete the functions below to return the articles you would recommend to each user. ",
"_____no_output_____"
]
],
[
[
"def get_article_names(article_ids, df=df):\n '''\n INPUT:\n article_ids - (list) a list of article ids\n df - (pandas dataframe) df as defined at the top of the notebook\n \n OUTPUT:\n article_names - (list) a list of article names associated with the list of article ids \n (this is identified by the title column)\n '''\n \n #article_names = df[df['article_id'].isin(article_ids)]['title'].unique()\n #article_names = df[df['article_id'].isin(article_ids)]['title'].drop_duplicates().values.tolist()\n article_names = []\n for article_id in article_ids:\n article_name = df[df['article_id']==float(article_id)]['title'].drop_duplicates().values\n article_names = np.concatenate([article_names, article_name])\n article_names = article_names.tolist()\n \n return article_names # Return the article names associated with list of article ids\n\n\ndef get_user_articles(user_id, user_item=user_item):\n '''\n INPUT:\n user_id - (int) a user id\n user_item - (pandas dataframe) matrix of users by articles: \n 1's when a user has interacted with an article, 0 otherwise\n \n OUTPUT:\n article_ids - (list) a list of the article ids seen by the user\n article_names - (list) a list of article names associated with the list of article ids \n \n Description:\n Provides a list of the article_ids and article titles that have been seen by a user\n '''\n # List of the article ids seen by the user\n #article_ids = np.where(user_item.loc[user_id] == 1)\n article_ids = user_item.loc[user_id][user_item.loc[user_id]==1].index.values\n #article_ids = user_item[user_item['user_id']==1]['articleid_column_name']\n \n # List of article names \n #article_names = df_content[df_content['article_id'].isin(article_ids)]['doc_full_name']\n #article_names = df[df['article_id'].isin(article_ids)]['title'].drop_duplicates().values.tolist()\n #article_names = get_article_names(article_ids, df)\n article_names = []\n for article_id in article_ids:\n article_name = df[df['article_id']==float(article_id)]['title'].drop_duplicates().values\n article_names = np.concatenate([article_names, article_name])\n article_names = article_names.tolist()\n \n return article_ids, article_names # return the ids and names\n\n\ndef user_user_recs(user_id, m=10):\n '''\n INPUT:\n user_id - (int) a user id\n m - (int) the number of recommendations you want for the user\n \n OUTPUT:\n recs - (list) a list of recommendations for the user\n \n Description:\n Loops through the users based on closeness to the input user_id\n For each user - finds articles the user hasn't seen before and provides them as recs\n Does this until m recommendations are found\n \n Notes:\n Users who are the same closeness are chosen arbitrarily as the 'next' user\n \n For the user where the number of recommended articles starts below m \n and ends exceeding m, the last items are chosen arbitrarily\n \n '''\n # Find similar users\n similar_neighbors = find_similar_users(user_id, user_item)\n \n # Get article_ids that have seen by the user\n user_articles = get_user_articles(user_id, user_item)[0]\n \n # Keep the recommended articles here\n recs = np.array([])\n \n # Go through the similar users and identify articles they have seen the user hasn't seen\n for neighbor in similar_neighbors:\n # Get article_ids that have seen by the neighbor\n neighbor_articles = get_user_articles(neighbor, user_item)[0]\n \n # Obtain recommendations for each neighbor\n new_recs = np.setdiff1d(neighbor_articles, user_articles, assume_unique=True)\n \n # Update recs with new recs\n recs = np.unique(np.concatenate([new_recs, recs], axis=0))\n \n # If we have enough recommendations exit the loop\n if len(recs) > m-1:\n break\n \n return recs[0:m] # return your recommendations for this user_id ",
"_____no_output_____"
],
[
"# Check Results\nget_article_names(user_user_recs(1, 10)) # Return 10 recommendations for user 1",
"_____no_output_____"
],
[
"# Test your functions here - No need to change this code - just run this cell\nassert set(get_article_names(['1024.0', '1176.0', '1305.0', '1314.0', '1422.0', '1427.0'])) == set(['using deep learning to reconstruct high-resolution audio', 'build a python app on the streaming analytics service', 'gosales transactions for naive bayes model', 'healthcare python streaming application demo', 'use r dataframes & ibm watson natural language understanding', 'use xgboost, scikit-learn & ibm watson machine learning apis']), \"Oops! Your the get_article_names function doesn't work quite how we expect.\"\nassert set(get_article_names(['1320.0', '232.0', '844.0'])) == set(['housing (2015): united states demographic measures', 'self-service data preparation with ibm data refinery', 'use the cloudant-spark connector in python notebook']), \"Oops! Your the get_article_names function doesn't work quite how we expect.\"\nassert set(get_user_articles(20)[0]) == set([1320.0, 232.0, 844.0])\nassert set(get_user_articles(20)[1]) == set(['housing (2015): united states demographic measures', 'self-service data preparation with ibm data refinery','use the cloudant-spark connector in python notebook'])\nassert set(get_user_articles(2)[0]) == set([1024.0, 1176.0, 1305.0, 1314.0, 1422.0, 1427.0])\nassert set(get_user_articles(2)[1]) == set(['using deep learning to reconstruct high-resolution audio', 'build a python app on the streaming analytics service', 'gosales transactions for naive bayes model', 'healthcare python streaming application demo', 'use r dataframes & ibm watson natural language understanding', 'use xgboost, scikit-learn & ibm watson machine learning apis'])\nprint(\"If this is all you see, you passed all of our tests! Nice job!\")",
"If this is all you see, you passed all of our tests! Nice job!\n"
]
],
[
[
"`4.` Now we are going to improve the consistency of the **user_user_recs** function from above. \n\n* Instead of arbitrarily choosing when we obtain users who are all the same closeness to a given user - choose the users that have the most total article interactions before choosing those with fewer article interactions.\n\n\n* Instead of arbitrarily choosing articles from the user where the number of recommended articles starts below m and ends exceeding m, choose articles with the articles with the most total interactions before choosing those with fewer total interactions. This ranking should be what would be obtained from the **top_articles** function you wrote earlier.",
"_____no_output_____"
]
],
[
[
"def get_top_sorted_users(user_id, df=df, user_item=user_item):\n '''\n INPUT:\n user_id - (int)\n df - (pandas dataframe) df as defined at the top of the notebook \n user_item - (pandas dataframe) matrix of users by articles: \n 1's when a user has interacted with an article, 0 otherwise\n \n \n OUTPUT:\n neighbors_df - (pandas dataframe) a dataframe with:\n neighbor_id - is a neighbor user_id\n similarity - measure of the similarity of each user to the provided user_id\n num_interactions - the number of articles viewed by the user - if a u\n \n Other Details - sort the neighbors_df by the similarity and then by number of interactions where \n highest of each is higher in the dataframe\n \n '''\n # Get similar neighbors to the provided user_id\n similar_neighbors = find_similar_users(user_id, user_item)\n \n # Get similarity of each neighbor to the provided user_id\n dot_prod_user = user_item.dot(np.transpose(user_item))\n neighbors_similarity = dot_prod_user.loc[user_id].sort_values(ascending=False).drop(user_id)\n \n # Get number of articles viewed by the neighbor\n num_interactions = df.groupby('user_id')['article_id'].size().sort_values(ascending=False).loc[similar_neighbors]\n \n # Get the dataframe\n neighbors_df = pd.DataFrame({'user_id': similar_neighbors,'similarity': neighbors_similarity, \n 'interactions': num_interactions}, columns=['user_id', 'similarity', 'interactions']).sort_values(by=['similarity', 'interactions'],ascending=False)\n \n return neighbors_df # Return the dataframe specified in the doc_string\n\n\ndef user_user_recs_part2(user_id, m=10):\n '''\n INPUT:\n user_id - (int) a user id\n m - (int) the number of recommendations you want for the user\n \n OUTPUT:\n recs - (list) a list of recommendations for the user by article id\n rec_names - (list) a list of recommendations for the user by article title\n \n Description:\n Loops through the users based on closeness to the input user_id\n For each user - finds articles the user hasn't seen before and provides them as recs\n Does this until m recommendations are found\n \n Notes:\n * Choose the users that have the most total article interactions \n before choosing those with fewer article interactions.\n\n * Choose articles with the articles with the most total interactions \n before choosing those with fewer total interactions. \n \n '''\n # Get similar neighbors to the provided user_id\n similar_neighbors = get_top_sorted_users(user_id, df, user_item).index.values\n \n # Get article_ids that have seen by the user\n user_articles = get_user_articles(user_id, user_item)[0]\n \n # Keep the recommended articles here\n recs = np.array([])\n \n # Go through the similar users and identify articles they have seen the user hasn't seen\n for neighbor in similar_neighbors:\n # Get article_ids that have seen by the neighbor\n neighbor_articles = get_user_articles(neighbor, user_item)[0]\n \n # Obtain recommendations for each neighbor\n new_recs = np.setdiff1d(neighbor_articles, user_articles, assume_unique=True)\n \n # Update recs with new recs\n recs = np.unique(np.concatenate([new_recs, recs], axis=0))\n \n # If we have enough recommendations exit the loop\n if len(recs) > m-1:\n break\n \n recs = recs[0:m]\n rec_names = get_article_names(recs)\n \n return recs, rec_names",
"_____no_output_____"
],
[
"# Quick spot check - don't change this code - just use it to test your functions\nrec_ids, rec_names = user_user_recs_part2(20, 10)\nprint(\"The top 10 recommendations for user 20 are the following article ids:\")\nprint(rec_ids)\nprint()\nprint(\"The top 10 recommendations for user 20 are the following article names:\")\nprint(rec_names)",
"The top 10 recommendations for user 20 are the following article ids:\n[ 12. 109. 125. 142. 164. 205. 302. 336. 362. 465.]\n\nThe top 10 recommendations for user 20 are the following article names:\n['timeseries data analysis of iot events by using jupyter notebook', 'dsx: hybrid mode', 'accelerate your workflow with dsx', 'learn tensorflow and deep learning together and now!', \"a beginner's guide to variational methods\", 'tensorflow quick tips', 'challenges in deep learning', 'neural networks for beginners: popular types and applications', 'statistics for hackers', 'introduction to neural networks, advantages and applications']\n"
]
],
[
[
"`5.` Use your functions from above to correctly fill in the solutions to the dictionary below. Then test your dictionary against the solution. Provide the code you need to answer each following the comments below.",
"_____no_output_____"
]
],
[
[
"### Tests with a dictionary of results\n\nuser1_most_sim = get_top_sorted_users(1, df, user_item).index.values[0] # Find the user that is most similar to user 1 \nuser131_10th_sim = get_top_sorted_users(131, df, user_item).index.values[9] # Find the 10th most similar user to user 131",
"_____no_output_____"
],
[
"## Dictionary Test Here\nsol_5_dict = {\n 'The user that is most similar to user 1.': user1_most_sim, \n 'The user that is the 10th most similar to user 131': user131_10th_sim,\n}\n\nt.sol_5_test(sol_5_dict)",
"This all looks good! Nice job!\n"
]
],
[
[
"`6.` If we were given a new user, which of the above functions would you be able to use to make recommendations? Explain. Can you think of a better way we might make recommendations? Use the cell below to explain a better method for new users.",
"_____no_output_____"
],
[
"If a totally new user was given, we can use functions from rank-based recommendations in Part II. We cannot use user-user collaborative filtering method from this part to make recommendations to this new user. Because when a new user is given, we have no records on this user and we will not be able to compare this user's records with any other existing users. Rank-based recommendations are recommendations based on the popularity of our items without examining collaborations among different users. Therefore, rank-based recommendation functions can be applied to make recommendations to the new user. ",
"_____no_output_____"
],
[
"`7.` Using your existing functions, provide the top 10 recommended articles you would provide for the a new user below. You can test your function against our thoughts to make sure we are all on the same page with how we might make a recommendation.",
"_____no_output_____"
]
],
[
[
"new_user = '0.0'\n\n# What would your recommendations be for this new user '0.0'? As a new user, they have no observed articles.\n# Provide a list of the top 10 article ids you would give to \nnew_user_recs = get_top_article_ids(10) # Your recommendations here ",
"_____no_output_____"
],
[
"new_user_recs",
"_____no_output_____"
],
[
"assert set(new_user_recs) == set([1314.0,1429.0,1293.0,1427.0,1162.0,1364.0,1304.0,1170.0,1431.0,1330.0]), \"Oops! It makes sense that in this case we would want to recommend the most popular articles, because we don't know anything about these users.\"\n\nprint(\"That's right! Nice job!\")",
"That's right! Nice job!\n"
]
],
[
[
"### <a class=\"anchor\" id=\"Content-Recs\">Part IV: Content Based Recommendations (EXTRA - NOT REQUIRED)</a>\n\nAnother method we might use to make recommendations is to perform a ranking of the highest ranked articles associated with some term. You might consider content to be the **doc_body**, **doc_description**, or **doc_full_name**. There isn't one way to create a content based recommendation, especially considering that each of these columns hold content related information. \n\n`1.` Use the function body below to create a content based recommender. Since there isn't one right answer for this recommendation tactic, no test functions are provided. Feel free to change the function inputs if you decide you want to try a method that requires more input values. The input values are currently set with one idea in mind that you may use to make content based recommendations. One additional idea is that you might want to choose the most popular recommendations that meet your 'content criteria', but again, there is a lot of flexibility in how you might make these recommendations.\n\n### This part is NOT REQUIRED to pass this project. However, you may choose to take this on as an extra way to show off your skills.",
"_____no_output_____"
]
],
[
[
"import re\nimport nltk\nfrom nltk.corpus import stopwords\nfrom nltk.stem.wordnet import WordNetLemmatizer\nfrom nltk.tokenize import word_tokenize\nfrom sklearn.feature_extraction.text import CountVectorizer\n\nnltk.download('punkt')\nnltk.download('stopwords')\nnltk.download('wordnet')",
"[nltk_data] Error loading punkt: <urlopen error [Errno 61] Connection\n[nltk_data] refused>\n[nltk_data] Error loading stopwords: <urlopen error [Errno 61]\n[nltk_data] Connection refused>\n[nltk_data] Error loading wordnet: <urlopen error [Errno 61]\n[nltk_data] Connection refused>\n"
],
[
"def tokenize(text):\n '''\n INPUT:\n text - text to be cleaned\n \n OUTPUT:\n tokens - cleaned tokens\n '''\n # Normalize case and remove punctuation\n text = re.sub(r\"[^a-zA-Z0-9]\", \" \", text.lower())\n \n # Tokenize text\n tokens = word_tokenize(text)\n \n # Lemmatize and remove stop words\n stop_words = stopwords.words(\"english\")\n lemmatizer = WordNetLemmatizer()\n tokens = [lemmatizer.lemmatize(word) for word in tokens if word not in stop_words]\n\n return tokens\n\ndef find_similar_docs(article_id):\n '''\n INPUT:\n article_id - an article id \n \n OUTPUT:\n similar_idx - an array of similar articles more than 75% percentile by index\n similar_docs - an array of similar articles more than 75% percentile by doc_full_name\n '''\n \n # Get article_ids in df that df_content does not include \n intsct = np.setdiff1d(df['article_id'].unique(), df_content['article_id'].unique())\n \n # Get the df subset that includes all articles existing in df but not in df_content\n df_intsct = df[df['article_id'].isin(intsct)][['article_id', 'title']].drop_duplicates()\n df_intsct.columns = ['article_id', 'doc_full_name']\n \n # Get the df_content subset of two columns: article_id and doc_full_name\n df_content_intsct = df_content[['article_id', 'doc_full_name']]\n \n # Concatenate df_intsct and df_content_intsct together\n df_ext = pd.concat([df_content_intsct, df_intsct])\n \n # Find the row of each article id\n doc_idx = np.where(df_ext['article_id'] == article_id)[0][0]\n \n # Initialize count vectorizer object\n vect = CountVectorizer(tokenizer=tokenize)\n # Transform doc_full_name tokens to a sparse matrix\n doc_full_name_temp = vect.fit_transform(df_ext['doc_full_name'].tolist())\n # Get a dot product of document_full_name tokens\n dot_prod_name = doc_full_name_temp.dot(np.transpose(doc_full_name_temp)).toarray()\n \n # Find similar article indices within the range of more than 75% percentile \n similar_idxs = np.where(dot_prod_name[doc_idx] > np.quantile(dot_prod_name[doc_idx], .75))[0]\n \n # Pull the article names based on the indices\n similar_docs = np.array(df_ext.iloc[similar_idxs, ]['doc_full_name'])\n \n return similar_idxs, similar_docs\n\n# create the user-article matrix with 1's and 0's\ndef create_user_item_matrix(df):\n '''\n INPUT:\n df - pandas dataframe with article_id, title, user_id columns\n \n OUTPUT:\n user_item - user item matrix \n \n Description:\n Return a matrix with user ids as rows and article ids on the columns with 1 values where a user interacted with \n an article and a 0 otherwise\n '''\n # Get user_item matrix with size in intersectional cell\n user_item = df.groupby(['user_id', 'article_id']).size().unstack().fillna(0)\n user_item[user_item > 1] = 1\n \n return user_item # return the user_item matrix \n\ndef make_content_recs(user_id, m, df):\n '''\n INPUT:\n user_id - (int) a user_id, the provided user we woule like to make recommendations for\n m - (int) the number of recommendations we would like to extract to the provided user\n \n OUTPUT:\n recs - (list) a list of recommendations for the user by article id\n rec_names - (list) a list of recommendations for the user by article title\n '''\n \n # Get user_item matrix\n user_item = create_user_item_matrix(df)\n \n # when we get an existing user_id\n if user_id in user_item.index.values.tolist():\n # Get article_ids that have seen by the user\n user_articles = get_user_articles(user_id, user_item)[0]\n\n recs = np.array([])\n for user_article in user_articles:\n rec_articles = find_similar_docs(user_article)[0]\n temp_recs = np.setdiff1d(user_articles, rec_articles)\n recs = np.unique(np.concatenate([temp_recs, recs], axis=0))\n\n if len(recs) > m-1:\n break\n\n recs = recs[0:m]\n rec_names = df_content[df_content['article_id'].isin(recs)]['doc_full_name'].drop_duplicates().values.tolist()\n \n # when we get a totally new user\n else:\n recs = get_top_article_ids(m)\n rec_names = get_top_articles(m)\n \n return recs, rec_names",
"_____no_output_____"
]
],
[
[
"`2.` Now that you have put together your content-based recommendation system, use the cell below to write a summary explaining how your content based recommender works. Do you see any possible improvements that could be made to your function? Is there anything novel about your content based recommender?\n### This part is NOT REQUIRED to pass this project. However, you may choose to take this on as an extra way to show off your skills.",
"_____no_output_____"
],
[
"My content based recommendation system is based on the similarities among document full name (the doc_full_name column from df_content data farme). I first extract this column and uses tokenize function to break the full title into tokens and complete a series of text cleaning steps including punctuation/stopwords removal, case normalization, and lemmatization. \n\nNext, in the find_similar_docs function, I find articles that rank more than the 75% percentile similarity with the given article. I convert the tokens obtained from the frist function to a sparse token-by-token matrix, with each article_id as the row and column indices and their similarities in the intersection cell between the row and column. A greater integer in the cell suggests a higher similarity between two articles. I extract articles that are beyond 75% percentile similarity to the given article. \n\nThe final function, make_content_recs, recommends similar articles to those articles seen by the provided user. I loop through every article seen by the provided user and recommend articles more than the 75% percentile similarity accordingly. \n\nMy content based recommender can recommend articles to users even though they have very few recordings or they have very few collaborations with other existing users. On the basis of content similarity, this system can provide recommendations to users who might not be sufficiently covered in user-user collaborative filtering method. One possible improvements to my content based recommender is to combine it with the user-user collaborative filtering method, i.e. choose articles with the most total interactions in addition to the 75% percentile similarity since those articles can be more popular among users. ",
"_____no_output_____"
],
[
"`3.` Use your content-recommendation system to make recommendations for the below scenarios based on the comments. Again no tests are provided here, because there isn't one right answer that could be used to find these content based recommendations.\n\n### This part is NOT REQUIRED to pass this project. However, you may choose to take this on as an extra way to show off your skills.",
"_____no_output_____"
]
],
[
[
"# make recommendations for a brand new user\nmake_content_recs(0, 10, df)\n\n# make a recommendations for a user who only has interacted with article id '1427.0'\nfind_similar_docs(1427.0)",
"_____no_output_____"
]
],
[
[
"### <a class=\"anchor\" id=\"Matrix-Fact\">Part V: Matrix Factorization</a>\n\nIn this part of the notebook, you will build use matrix factorization to make article recommendations to the users on the IBM Watson Studio platform.\n\n`1.` You should have already created a **user_item** matrix above in **question 1** of **Part III** above. This first question here will just require that you run the cells to get things set up for the rest of **Part V** of the notebook. ",
"_____no_output_____"
]
],
[
[
"# Load the matrix here\nuser_item_matrix = pd.read_pickle('../data/user_item_matrix.p')",
"_____no_output_____"
],
[
"# quick look at the matrix\nuser_item_matrix.head()",
"_____no_output_____"
]
],
[
[
"`2.` In this situation, you can use Singular Value Decomposition from [numpy](https://docs.scipy.org/doc/numpy-1.14.0/reference/generated/numpy.linalg.svd.html) on the user-item matrix. Use the cell to perform SVD, and explain why this is different than in the lesson.",
"_____no_output_____"
]
],
[
[
"# Perform SVD on the User-Item Matrix Here\nu, s, vt = np.linalg.svd(user_item_matrix) # use the built in to get the three matrices",
"_____no_output_____"
],
[
"u.shape, s.shape, vt.shape",
"_____no_output_____"
]
],
[
[
"The SVD output here is different because the current user_item_matrix is a sparse matrix with lots of 0s in its row and column intersection cells. All missing values in the original user_item_matrix have been filled by zeros. If we do not replace missing valaues with zeros, we will not be able to conduct the SVD function, or SVD will not converge. \nThe closed form solution for SVD consists of finding the eigenvalues and eigenvectors of AA' and A'A. With missing valuaes, we are not able to calculate either the AA' or the A'A, let alone their eigenvalues and eigenvectors. While filling missing values with zeros, we can proceed to use the built-in SVD function without encountering difficulties generating from missing values. ",
"_____no_output_____"
],
[
"`3.` Now for the tricky part, how do we choose the number of latent features to use? Running the below cell, you can see that as the number of latent features increases, we obtain a lower error rate on making predictions for the 1 and 0 values in the user-item matrix. Run the cell below to get an idea of how the accuracy improves as we increase the number of latent features.",
"_____no_output_____"
]
],
[
[
"num_latent_feats = np.arange(10,700+10,20)\nsum_errs = []\n\nfor k in num_latent_feats:\n # restructure with k latent features\n s_new, u_new, vt_new = np.diag(s[:k]), u[:, :k], vt[:k, :]\n \n # take dot product\n user_item_est = np.around(np.dot(np.dot(u_new, s_new), vt_new))\n \n # compute error for each prediction to actual value\n diffs = np.subtract(user_item_matrix, user_item_est)\n \n # total errors and keep track of them\n err = np.sum(np.sum(np.abs(diffs)))\n sum_errs.append(err)\n \n \nplt.plot(num_latent_feats, 1 - np.array(sum_errs)/df.shape[0]);\nplt.xlabel('Number of Latent Features');\nplt.ylabel('Accuracy');\nplt.title('Accuracy vs. Number of Latent Features');",
"_____no_output_____"
]
],
[
[
"`4.` From the above, we can't really be sure how many features to use, because simply having a better way to predict the 1's and 0's of the matrix doesn't exactly give us an indication of if we are able to make good recommendations. Instead, we might split our dataset into a training and test set of data, as shown in the cell below. \n\nUse the code from question 3 to understand the impact on accuracy of the training and test sets of data with different numbers of latent features. Using the split below: \n\n* How many users can we make predictions for in the test set? \n* How many users are we not able to make predictions for because of the cold start problem?\n* How many articles can we make predictions for in the test set? \n* How many articles are we not able to make predictions for because of the cold start problem?",
"_____no_output_____"
]
],
[
[
"df_train = df.head(40000)\ndf_test = df.tail(5993)\n\ndef create_user_item_matrix(df):\n '''\n INPUT:\n df - pandas dataframe with article_id, title, user_id columns\n \n OUTPUT:\n user_item - user item matrix \n \n Description:\n Return a matrix with user ids as rows and article ids on the columns with 1 values where a user interacted with \n an article and a 0 otherwise\n '''\n # Get user_item matrix with size in intersectional cell\n user_item = df.groupby(['user_id', 'article_id']).size().unstack().fillna(0)\n user_item[user_item > 1] = 1\n \n return user_item # return the user_item matrix \n\ndef create_test_and_train_user_item(df_train, df_test):\n '''\n INPUT:\n df_train - training dataframe\n df_test - test dataframe\n \n OUTPUT:\n user_item_train - a user-item matrix of the training dataframe \n (unique users for each row and unique articles for each column)\n user_item_test - a user-item matrix of the testing dataframe \n (unique users for each row and unique articles for each column)\n test_idx - all of the test user ids\n test_arts - all of the test article ids\n \n '''\n user_item_train = create_user_item_matrix(df_train)\n user_item_test = create_user_item_matrix(df_test)\n \n test_idx = user_item_test.index.values\n test_arts = user_item_test.columns.values\n \n return user_item_train, user_item_test, test_idx, test_arts\n\nuser_item_train, user_item_test, test_idx, test_arts = create_test_and_train_user_item(df_train, df_test)",
"_____no_output_____"
],
[
"user_item_test.shape[0]",
"_____no_output_____"
],
[
"user_item_test.shape[1]",
"_____no_output_____"
],
[
"user_item_test.index.values",
"_____no_output_____"
],
[
"user_item_train.index.values",
"_____no_output_____"
],
[
"# Get the same user_ids between train and test set\nintersection = np.intersect1d(user_item_test.index.values, user_item_train.index.values)\nintersection",
"_____no_output_____"
],
[
"# number of users who are both in train and test set, which is the number of users in the test set we can make predictions for\nintersection_num = np.where(user_item_test.index==intersection[-1])[0][0] - np.where(user_item_test.index==intersection[0])[0][0] + 1\nintersection_num",
"_____no_output_____"
],
[
"# number of users in the test set we are not able to make predictions for because of the cold start problem \nuser_item_test.shape[0] - intersection_num",
"_____no_output_____"
],
[
"# Get different article_ids between train and test set\nnp.setdiff1d(user_item_test.columns.values, user_item_train.columns.values)",
"_____no_output_____"
],
[
"# number of articles in the test set we are not able to make predictions for because of the cold start problem\nlen(np.setdiff1d(user_item_test.columns.values, user_item_train.columns.values))",
"_____no_output_____"
],
[
"# number of articles we can make predictions for\nuser_item_test.shape[1] - len(np.setdiff1d(user_item_test.columns.values, user_item_train.columns.values))",
"_____no_output_____"
],
[
"# Replace the values in the dictionary below\na = 662 \nb = 574 \nc = 20 \nd = 0 \n\n\nsol_4_dict = {\n 'How many users can we make predictions for in the test set?': c, \n 'How many users in the test set are we not able to make predictions for because of the cold start problem?': a, \n 'How many articles can we make predictions for in the test set?': b,\n 'How many articles in the test set are we not able to make predictions for because of the cold start problem?': d\n}\nt.sol_4_test(sol_4_dict)",
"Awesome job! That's right! All of the test movies are in the training data, but there are only 20 test users that were also in the training set. All of the other users that are in the test set we have no data on. Therefore, we cannot make predictions for these users using SVD.\n"
]
],
[
[
"`5.` Now use the **user_item_train** dataset from above to find U, S, and V transpose using SVD. Then find the subset of rows in the **user_item_test** dataset that you can predict using this matrix decomposition with different numbers of latent features to see how many features makes sense to keep based on the accuracy on the test data. This will require combining what was done in questions `2` - `4`.\n\nUse the cells below to explore how well SVD works towards making predictions for recommendations on the test data. ",
"_____no_output_____"
]
],
[
[
"# Fit SVD on the user_item_train_subset matrix\nu_train, s_train, vt_train = np.linalg.svd(user_item_train) # fit svd similar to above then use the cells below",
"_____no_output_____"
],
[
"u_train.shape, s_train.shape, vt_train.shape",
"_____no_output_____"
],
[
"num_latent_feats = np.arange(10,700+10,20)\nsum_errs = []\n\nfor k in num_latent_feats:\n # restructure with k latent features\n s_new, u_new, vt_new = np.diag(s_train[:k]), u_train[:, :k], vt_train[:k, :]\n \n # take dot product\n user_item_est = np.around(np.dot(np.dot(u_new, s_new), vt_new))\n \n # Get intersected user_ids that appear in both train and test set\n row_intsct = np.intersect1d(user_item_test.index.values, user_item_train.index.values)\n \n # Get intersected article_ids that appears in both train and test set\n column_intsct = np.intersect1d(user_item_test.columns.values, user_item_train.columns.values)\n \n # Get the subset of user_item_test that includes user_ids in row_intsct\n user_item_test_subset = user_item_test.loc[intersection[0]:intersection[-1]]\n \n # Get the indices of intersected user_ids\n row_idx = []\n for i in row_intsct:\n row_idx.append(user_item_train.index.get_loc(i))\n # Extract intersected rows from user_item_est\n user_item_est_subset = user_item_est[row_idx,:]\n \n # Get the indices of intersected article_ids\n col_idx = []\n for i in column_intsct:\n col_idx.append(user_item_train.columns.get_loc(i))\n # Extract intersected columns from user_item_est\n user_item_est_subset = user_item_est_subset[:,col_idx]\n \n # compute error for each prediction to actual value\n diffs = np.subtract(user_item_test_subset, user_item_est_subset)\n \n # total errors and keep track of them\n err = np.sum(np.sum(np.abs(diffs)))\n sum_errs.append(err)\n \n \nplt.plot(num_latent_feats, 1 - np.array(sum_errs)/df.shape[0]);\nplt.xlabel('Number of Latent Features');\nplt.ylabel('Accuracy');\nplt.title('Accuracy vs. Number of Latent Features');",
"_____no_output_____"
]
],
[
[
"`6.` Use the cell below to comment on the results you found in the previous question. Given the circumstances of your results, discuss what you might do to determine if the recommendations you make with any of the above recommendation systems are an improvement to how users currently find articles? ",
"_____no_output_____"
],
[
"The results I found from the previous question show that when we have a smaller number of latent features, the accuracy for our predictions is higher. This is counterintuitive since we assume that greater number of latent features can help us make better or more accurate predictions. Given the results, one possible solution is not to adopt SVD but instead to use a funkSVD method. FunkSVD method uses gradient descent to iterate through our sparse matrix and fill missing values with matrix factorization predictions. \n\nOne way to determine the accuracy of our recommendation system is to split our interaction matrix into training and testing set. We will first train our recommendation system using FunkSVD on training set. After some trail and error on the setting of the number of latent features and iterations, we will get a optimal recommender from training set and then we will apply this system to our testing set. One metric to measure the accuracy of our recommender is mean squared error (mse). A greater mse suggests a lower performance in model predictions. Another way to evaluate our recommender is to simply compare the actual value with the predicted value. \n\nAs for the cold start problem, we can combine rank based recommender (from Part II) or content based recommender (from Part IV) into our FunkSVD recommendation system. When we meet totally new users, we will make recommendations based on rank based or content based recommendation system. When we see existing users, we can make recommendations using the FunkSVD recommender. ",
"_____no_output_____"
],
[
"<a id='conclusions'></a>\n### Extras\nUsing your workbook, you could now save your recommendations for each user, develop a class to make new predictions and update your results, and make a flask app to deploy your results. These tasks are beyond what is required for this project. However, from what you learned in the lessons, you certainly capable of taking these tasks on to improve upon your work here!\n\n\n## Conclusion\n\n> Congratulations! You have reached the end of the Recommendations with IBM project! \n\n\n## Directions to Submit\n\n> Before you submit your project, you need to create a .html or .pdf version of this notebook in the workspace here. To do that, run the code cell below. If it worked correctly, you should get a return code of 0, and you should see the generated .html file in the workspace directory (click on the orange Jupyter icon in the upper left).\n\n> Alternatively, you can download this report as .html via the **File** > **Download as** submenu, and then manually upload it into the workspace directory by clicking on the orange Jupyter icon in the upper left, then using the Upload button.\n\n> Once you've done this, you can submit your project by clicking on the \"Submit Project\" button in the lower right here. This will create and submit a zip file with this .ipynb doc and the .html or .pdf version you created. Congratulations! ",
"_____no_output_____"
]
],
[
[
"from subprocess import call\ncall(['python', '-m', 'nbconvert', 'Recommendations_with_IBM.ipynb'])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
ecc8de9d19483d9e04e66b50af79b32b6542d0be | 6,822 | ipynb | Jupyter Notebook | Fake News Detection/Detecting Fake News.ipynb | rleary90/sturdy-octo-happiness | 0b11c8575fe984f632f52f4e326defab5a28fd2e | [
"MIT"
] | null | null | null | Fake News Detection/Detecting Fake News.ipynb | rleary90/sturdy-octo-happiness | 0b11c8575fe984f632f52f4e326defab5a28fd2e | [
"MIT"
] | null | null | null | Fake News Detection/Detecting Fake News.ipynb | rleary90/sturdy-octo-happiness | 0b11c8575fe984f632f52f4e326defab5a28fd2e | [
"MIT"
] | null | null | null | 27.959016 | 103 | 0.477866 | [
[
[
"import numpy as np\nimport pandas as pd\nimport itertools\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom sklearn.linear_model import PassiveAggressiveClassifier\nfrom sklearn.metrics import accuracy_score, confusion_matrix",
"_____no_output_____"
],
[
"#Read the data\ndf=pd.read_csv('news.csv')\n#Get shape and head\ndf.shape\ndf.head()",
"_____no_output_____"
],
[
"#DataFlair - Get the labels\nlabels=df.label\nlabels.head()",
"_____no_output_____"
],
[
"#DataFlair - Split the dataset\nx_train,x_test,y_train,y_test=train_test_split(df['text'], labels, test_size=0.2, random_state=7)",
"_____no_output_____"
],
[
"#DataFlair - Initialize a TfidfVectorizer\ntfidf_vectorizer=TfidfVectorizer(stop_words='english', max_df=0.7)\n#DataFlair - Fit and transform train set, transform test set\ntfidf_train=tfidf_vectorizer.fit_transform(x_train) \ntfidf_test=tfidf_vectorizer.transform(x_test)",
"_____no_output_____"
],
[
"#DataFlair - Initialize a PassiveAggressiveClassifier\npac=PassiveAggressiveClassifier(max_iter=50)\npac.fit(tfidf_train,y_train)\n#DataFlair - Predict on the test set and calculate accuracy\ny_pred=pac.predict(tfidf_test)\nscore=accuracy_score(y_test,y_pred)\nprint(f'Accuracy: {round(score*100,2)}%')",
"Accuracy: 92.58%\n"
],
[
"#DataFlair - Build confusion matrix\nconfusion_matrix(y_test,y_pred, labels=['FAKE','REAL'])",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecc8e239a2178c6f67f5f44c29375fb228155cf6 | 23,823 | ipynb | Jupyter Notebook | examples/optical_flow_sintel.ipynb | csbrown/pylomo | 377aa386427a32da8b42fe53aacbe3281fbf2bf6 | [
"MIT"
] | null | null | null | examples/optical_flow_sintel.ipynb | csbrown/pylomo | 377aa386427a32da8b42fe53aacbe3281fbf2bf6 | [
"MIT"
] | 1 | 2020-04-01T17:41:36.000Z | 2020-04-01T17:41:36.000Z | examples/optical_flow_sintel.ipynb | csbrown/pylomo | 377aa386427a32da8b42fe53aacbe3281fbf2bf6 | [
"MIT"
] | null | null | null | 40.309645 | 1,629 | 0.573186 | [
[
[
"import local_models.local_models\nimport local_models.algorithms\nimport local_models.utils\nimport local_models.linear_projections\nimport local_models.loggin\nimport local_models.TLS_models\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport sklearn.linear_model\nimport sklearn.cluster\nfrom importlib import reload\nfrom ml_battery.utils import cmap\nimport matplotlib as mpl\nimport sklearn.datasets\nimport sklearn.decomposition\nimport logging\nimport ml_battery.log\nimport time\nimport os\nimport mayavi\nimport mayavi.mlab\nimport matplotlib.image as img\n\n\nimport subprocess\n#on headless systems, tmux: \"Xvfb :1 -screen 0 1280x1024x24 -auth localhost\", then \"export DISPLAY=:1\" in the jupyter tmux\nmayavi.mlab.options.offscreen = True\n\n\n\nlogger = logging.getLogger(__name__)\n\n#reload(local_models.local_models)\n#reload(lm)\n#reload(local_models.loggin)\n#reload(local_models.TLS_models)\nnp.warnings.filterwarnings('ignore')\n",
"_____no_output_____"
],
[
"def import_shit():\n import local_models.local_models\n import local_models.algorithms\n import local_models.utils\n import local_models.linear_projections\n import local_models.loggin\n import local_models.TLS_models\n import numpy as np\n import logging\n import ml_battery.log\n\n logger = logging.getLogger(__name__)\n\n #reload(local_models.local_models)\n #reload(lm)\n #reload(local_models.loggin)\n #reload(local_models.TLS_models)\n np.warnings.filterwarnings('ignore')\n return logger",
"_____no_output_____"
],
[
"FRESH=True",
"_____no_output_____"
],
[
"kernel_names = {\n local_models.local_models.GaussianKernel: 'gaussian',\n local_models.local_models.TriCubeKernel: 'tricube'\n}",
"_____no_output_____"
],
[
"mpl.rcParams['figure.figsize'] = [8.0, 8.0]",
"_____no_output_____"
],
[
"def img2data(image):\n return np.concatenate((np.mgrid[0:1:image.shape[0]*1j, 0:1:image.shape[1]*1j], image.reshape(1,*image.shape)), axis=0)",
"_____no_output_____"
],
[
"import glob\nclass Everything(object):\n def __eq__(*args): return True\n\n# data is time, y, x, intensity\nimgs = []\nprevious_dims = Everything()\nfor i, im_path in enumerate(glob.glob(\"../data/sintel/training/clean/alley_1/*.png\")):\n image = np.average(img.imread(im_path), axis=2)\n dims = image.shape\n if not dims==previous_dims:\n raise BaseException(\"must all be same shape\")\n previous_dims = dims\n imgs.append(np.concatenate((np.ones([1] + list(image.shape[:2]))*i, img2data(image))))\ndata = np.stack(imgs, axis=3)\ndata[0,:,:,:] /= i\ngridder = local_models.utils.Grid2Vec()\ndata = gridder.fit_transform(data)",
"_____no_output_____"
],
[
"data.shape",
"_____no_output_____"
],
[
"KERNEL=local_models.local_models.GaussianKernel\nRUN = 1\n\nproject_dir = \"../data/sintel_alley1_{}_{:03d}\".format(kernel_names[KERNEL], RUN)\n#project_dir = \"../data/3dscene_{}\".format(kernel_names[KERNEL])\n\nos.makedirs(project_dir, exist_ok=1)\n\n",
"_____no_output_____"
],
[
"def read_flo(f_name):\n with open(f_name, 'rb') as f:\n magic = np.fromfile(f, np.float32, count=1)\n if 202021.25 != magic:\n raise BaseException(\"invalid flo file {}\".format(f_name))\n else:\n w = np.fromfile(f, np.int32, count=1)[0]\n h = np.fromfile(f, np.int32, count=1)[0]\n data = np.fromfile(f, np.float32, count=2*w*h)\n # Reshape data into 3D array (columns, rows, bands)\n data2D = np.resize(data, (h, w, 2))\n return data2D\n\ndef EPE(flo1, flo2):\n return np.average(np.linalg.norm(flo1-flo2, axis=2))\n ",
"_____no_output_____"
],
[
"linear_models = local_models.local_models.LocalModels(local_models.TLS_models.LinearODR_mD(3))\nlinear_models.fit(data)",
"_____no_output_____"
],
[
"BANDWIDTH = 0.03\npth = os.path.join(project_dir, \"bandwidth_{:08.03f}\".format(BANDWIDTH))\nos.makedirs(pth, exist_ok=1)",
"_____no_output_____"
],
[
"def converge_n_save(bandwidth, i, data):\n logger = import_shit()\n logger.info(\"stuff!\")\n for converged in local_models.algorithms.local_tls_shift_till_convergence(linear_models, data, tol=1e-8,\n kernel=KERNEL(bandwidth),\n report=False):\n pass\n logger.info(\"batch {:05d} complete\")\n np.savetxt(os.path.join(pth, \"converged{:08d}.dat\".format(i)), converged)\n return None\n\ndef batcher(data, size):\n for i in range(0, data.shape[0], size):\n batch = data[i:i+size]\n print(batch.shape)\n yield batch\n \nif FRESH:\n from joblib import Parallel, delayed\n convergededs = Parallel(n_jobs=32)(delayed(converge_n_save)(BANDWIDTH, i, batch) for i, batch in enumerate(batcher(data, 100)))\nelse:\n convergededs = []\n for i, bandwidth in enumerate(bandwidths):\n convergededs.append(np.loadtxt(os.path.join(pth, \"converged{:08.02f}.dat\".format(bandwidth))))\n",
"(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n(100, 4)\n"
],
[
"def mean_center(data, weights=None):\n return data - np.average(data, axis=0,weights=weights)",
"_____no_output_____"
],
[
"for i, bandwidth in enumerate(bandwidths):\n converged = convergededs[i]\n data_avg = np.average(converged, axis=0)\n data_std = np.std(converged, axis=0)\n figure = mayavi.mlab.figure(figure=None, bgcolor=(1,1,1), fgcolor=(0,0,0), engine=None, size=(1000, 500))\n #mayavi.mlab.surf(grid[0], grid[1], kde_wireframe/z_scale, colormap='Greys', opacity=1)\n nodes = mayavi.mlab.points3d(converged[:,0], converged[:,1], converged[:,2], \n scale_mode='scalar', colormap='gist_earth')\n nodes.glyph.scale_mode = 'scale_by_vector'\n nodes.mlab_source.dataset.point_data.vectors = np.ones((converged.shape[0],3))*(np.average(data_std)/400)\n nodes.mlab_source.dataset.point_data.scalars = (converged[:,1] - (data_avg[1]-3*data_std[1]))/(6*data_std[1])\n #bunodes = mayavi.mlab.points3d(data[:,0], data[:,1], data[:,2], scale_mode='scalar', color=(0,1,0))\n #bunodes.mlab_source.dataset.point_data.scalars = np.ones(data.shape[0])*0.1\n\n\n #mayavi.mlab.axes()\n\n #mayavi.mlab.view(views[0][1],views[0][0])\n mayavi.mlab.view(azimuth=180, elevation=80, distance=1*np.average(data_avg), focalpoint=(data_avg[0], data_avg[1], data_avg[2]))\n mayavi.mlab.move(forward=None, right=0.1*data_avg[1], up=-0.02*data_avg[0])\n title = \"converged_data_b{:08.02f}\".format(bandwidth)\n mayavi.mlab.savefig(os.path.join(pth, \"{}.png\".format(title)))\n mayavi.mlab.close(figure)",
"_____no_output_____"
],
[
"mayavi.mlab.close(all=True)",
"_____no_output_____"
],
[
"range_pct = 0.2\ngrid_steps = 20\ndata_mins, data_maxes, data_ranges = local_models.linear_projections.min_max_range(data)\ngraph_bounds = local_models.linear_projections.sane_graph_bounds(data_mins, data_maxes, data_ranges, range_pct)\n\ngrid_limits = tuple(map(lambda i: slice(graph_bounds[0,i], graph_bounds[1,i], grid_steps*1j), range(graph_bounds.shape[1])))\ngrid = np.mgrid[grid_limits]\ngridder = local_models.utils.Grid2Vec()\ngrid = gridder.fit_transform(grid)",
"_____no_output_____"
],
[
"for converged2 in local_models.algorithms.local_tls_shift_till_convergence(linear_models, grid, tol=1e-10,\n kernel=KERNEL(bandwidth),\n report=False):\n pass",
"_____no_output_____"
],
[
"converged2",
"_____no_output_____"
],
[
"cloud = pcl.PointCloud()\ncloud.from_array(converged2.astype('float32'))\n#visual = pcl.pcl_visualization.CloudViewing()\n#visual.ShowMonochromeCloud(cloud)",
"_____no_output_____"
],
[
"mayavi.mlab.figure(figure=None, bgcolor=(1,1,1), fgcolor=(0,0,0), engine=None, size=(800, 800))\n#mayavi.mlab.surf(grid[0], grid[1], kde_wireframe/z_scale, colormap='Greys', opacity=1)\nnodes = mayavi.mlab.points3d(converged2[:,0], converged2[:,1], converged2[:,2], scale_mode='scalar', color=(1,0,0))\nnodes.mlab_source.dataset.point_data.scalars = np.ones(converged2.shape[0])*0.1\nbunodes = mayavi.mlab.points3d(data[:,0], data[:,1], data[:,2], scale_mode='scalar', color=(0,1,0))\nbunodes.mlab_source.dataset.point_data.scalars = np.ones(data.shape[0])*0.1\n\n\n#mayavi.mlab.axes()\n\n#mayavi.mlab.view(views[0][1],views[0][0])\ndata_avg = np.average(converged2, axis=1)\nmayavi.mlab.view(azimuth=0, elevation=30, distance=1, focalpoint=(data_avg[0], data_avg[1], data_avg[2]))\ntitle = \"converged_g000{:05d}\".format(grid_steps)\nmayavi.mlab.savefig(os.path.join(project_dir, \"{}.png\".format(title)))\nmayavi.mlab.clf()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecc8f98a2928d869686894de463c3bcdf706e9eb | 16,218 | ipynb | Jupyter Notebook | src/homepage/workflow.ipynb | sosdocs/sos-docs | bb3caef5ef89518657bc74aabfc274902a101321 | [
"MIT"
] | null | null | null | src/homepage/workflow.ipynb | sosdocs/sos-docs | bb3caef5ef89518657bc74aabfc274902a101321 | [
"MIT"
] | null | null | null | src/homepage/workflow.ipynb | sosdocs/sos-docs | bb3caef5ef89518657bc74aabfc274902a101321 | [
"MIT"
] | null | null | null | 54.422819 | 326 | 0.648107 | [
[
[
"# <a id=\"notebook_intro_videos\"></a>Introductory Videos",
"_____no_output_____"
],
[
"<div class=\"container\">\n <div class=\"row\">\n <div class=\"col-sm-3\">\n <div class=\"embed-responsive embed-responsive-16by9\">\n <iframe width=\"560\" height=\"315\" src=\"https://www.youtube.com/embed/E5Eh7BVbbTM?wmode=opaque\" frameborder=\"0\" allowfullscreen></iframe>\n <div class=\"overlay-duration\">14:08</div>\n </div>\n <p>Basics of SoS workflow system</p>\n </div>\n <div class=\"col-sm-3\">\n <div class=\"embed-responsive embed-responsive-16by9\">\n <iframe width=\"560\" height=\"315\" src=\"https://www.youtube.com/embed/roTDvSXSPgU?wmode=opaque\" frameborder=\"0\" allowfullscreen></iframe>\n <div class=\"overlay-duration\">16:52</div>\n </div>\n <p>More on SoS workflow system</p>\n </div>\n <div class=\"col-sm-3\">\n <div class=\"embed-responsive embed-responsive-16by9\">\n <iframe width=\"560\" height=\"315\" src=\"https://www.youtube.com/embed/_LSQr3DHBpU?wmode=opaque\" frameborder=\"0\" allowfullscreen></iframe>\n <div class=\"overlay-duration\">16:16</div>\n </div>\n <p>Use SoS in SoS Notebook</p>\n </div>\n </div> \n</div>",
"_____no_output_____"
],
[
"# Documentation",
"_____no_output_____"
],
[
"Our documentation is hosted on our live server and can be edited directly. If you spot an error or feel like contributing to the document, please \n\n1. Click the <i class=\"fa fa-edit fa-2x\"></i> button to the top right corner of each tutorial\n2. Execute the notebook and edit it on our live server\n3. Enter `!create-pr <Shift-Enter>` in the console panel to send us a PR",
"_____no_output_____"
],
[
"# SoS Workflow",
"_____no_output_____"
],
[
"## The basics\n\n * [SoS workflow: a 20 minute overview](doc/user_guide/sos_overview.html)\n * [Variables and parameters](doc/user_guide/variable_and_parameter.html)\n * [Step input, output, and substeps](doc/user_guide/input_substeps.html)\n * [Basic SoS workflows](doc/user_guide/basic_workflow.html)\n * [Using SoS Workflow for daily data analysis](doc/user_guide/organize_scripts.html)",
"_____no_output_____"
],
[
"## Technical details\n\n### Syntax and data types\n * [Syntax and file formats](doc/user_guide/sos_syntax.html)\n * [Script format of function calls](doc/user_guide/script_format.html)\n * [The `parameter` statement](doc/user_guide/parameter_statement.html)\n * [Configuration files](doc/user_guide/config_files.html)\n * [SoS targets (`file_target`, `sos_variable`, `R_Library`, and more)](doc/user_guide/targets.html) \n * [The `sos_targets` data type](doc/user_guide/sos_targets.html)\n * [SoS functions (`get_output` and `expand_pattern`)](doc/user_guide/sos_functions.html)\n * [Extending SoS](doc/user_guide/extending_sos.html)\n \n\n### SoS Steps\n * [The `input` statement](doc/user_guide/input_statement.html)\n * [Named input](doc/user_guide/named_input.html)\n * [Input option `group_by`](doc/user_guide/group_by.html)\n * [Input option `for_each`](doc/user_guide/for_each.html)\n * [Input options `paired_with` and `group_with`](doc/user_guide/paired_group_with.html) \n * [Input option `pattern` and function `expand_pattern`](doc/user_guide/input_pattern.html) \n * [Input option `concurrent`](doc/user_guide/concurrent_substep.html) \n * [Dynamic input (`dynamic` target)](doc/user_guide/dynamic_input.html) \n * [The `output` statement](doc/user_guide/output_statement.html)\n * [Output option `group_by`](doc/user_guide/output_group_by.html)\n * [Output option `paired_with` and `group_with`](doc/user_guide/output_groups_vars.html)\n * [Dynamic output (`dynamic` target)](doc/user_guide/dynamic_output.html) \n * [The `depends` statement](doc/user_guide/depends_statement.html) \n * [SoS actions and common options](doc/user_guide/sos_actions.html)\n * [Command execution actions (actions `run`, `sh`, and `bash`)](doc/user_guide/shell_actions.html)\n * [Script execution actions (actions `Python`, `R`, `JavaScript` etc)](doc/user_guide/script_actions.html)\n * [Report generation actions (actions `report`, `pandoc`, and `RMarkdown`)](doc/user_guide/report_actions.html)\n * [Conditional actions (actions `warn_if`, `skip_if`, `done_if` and `fail_if`)](doc/user_guide/control_actions.html) \n * [Action `download`](doc/user_guide/download_actions.html)\n * [Running scripts in docker containers (Action option `container` and action `docker_build`)](doc/user_guide/docker.html)\n * [Running scripts in singularity (Action options `container` and `engine`, action `singularity_build`)](doc/user_guide/singularity.html)\n * [The `task` statement and option `-q`)](doc/user_guide/task_statement.html)\n * [Host configuration (`~/.sos/hosts.yml` and `sos remote` command)](doc/user_guide/host_setup.html) \n * [Task templates (options such as `mem`, `cores`, `walltime`, and `nodes`)](doc/user_guide/task_template.html) \n * [Task tags (option `tags`)](doc/user_guide/task_tags.html) \n * [Combining tasks (options `trunk_size` and `trunk_workers`)](doc/user_guide/trunk_size.html) \n * [Path translation and file synchronization (options `workdir`, `to_host` and `from_host`)](doc/user_guide/task_files.html)\n * [Working with remote files (`remote` target)](doc/user_guide/remote_target.html) \n \n### SoS Workflows\n * [Summary of step dependencies](doc/user_guide/step_dependencies.html)\n * [Named output (function `named_output`)](doc/user_guide/named_output.html) \n * [Output from another step (function `output_from`)](doc/user_guide/output_from.html) \n * [Explicit step and workflow dependency (target `sos_step`)](doc/user_guide/target_sos_step.html) \n * [Makefile-style pattern-matching rules (section option `provides`)](doc/user_guide/auxiliary_steps.html)\n * [Global and local variables](doc/user_guide/step_variables.html) \n * [Sharing variables across steps (step and task option `shared` and target `sos_variable`)](doc/user_guide/shared_variables.html) \n * [Introduction to SoS Workflows](doc/user_guide/sos_workflows.html) \n * [Process-oriented workflows](doc/user_guide/process_oriented.html) \n * [Outcome-oriented workflows](doc/user_guide/outcome_oriented.html)\n * [Mixed-style workflows](doc/user_guide/mixed_style.html) \n * [Data-flow style workflows](doc/user_guide/data_flow.html) \n * [Nested workflow (function `sos_run`)](doc/user_guide/nested_workflow.html)\n \n### Command line interface\n * [List of all sos commands and options](doc/user_guide/cli.html)\n * Execution of workflows \n * [Verbosity (option `-v`) and number of workers (option `-j`)](doc/user_guide/verbosity_and_jobs.html)\n * [Runtime signature (option `-s`)](doc/user_guide/signature.html)\n * [Error handling (option `-e`)](doc/user_guide/error_handling.html) \n * [Dependency tracing (option `-T`)](doc/user_guide/trace_dependency.html)\n * [Dryrun mode (option `-n`)](doc/user_guide/dryrun.html)\n * [Customized environments and remote execution of workflows (option `-r`)](doc/user_guide/remote_execution.html) \n * [Output DAG of workflows (option `-d`)](doc/user_guide/workflow_dag.html) \n * [Report generation (option `-p`)](doc/user_guide/workflow_summary.html) \n * Utilities \n * [Task management (commands `status`, `kill`, and `purge`)](doc/user_guide/task_management.html)\n * [Setup and test remote hosts (command `sos remote`)](doc/user_guide/sos_remote.html)\n * [Format conversion (command `sos convert` and `%convert`)](doc/user_guide/convert.html)",
"_____no_output_____"
],
[
"# Publication\n\nPlease cite the following publication if you use SoS workflow engine for your work:\n\n[**Script of Scripts: a pragmatic workflow system for daily computational research**](https://doi.org/10.1371/journal.pcbi.1006843)\n<small>Gao Wang and Bo Peng (2019) <em> PLoS Computational Biology</em>. doi: 10.1371/journal.pcbi.1006843</small>\n\nHere are a list of examples from the publication:\n\n* [Process_Oriented.sos](doc/examples/Process_Oriented.html) ([script](doc/examples/Process_Oriented.sos))<br>\n <small>Demonstration of a process_oriented forward-style workflow that executes steps in pre-sepecified order.</small>\n* [Process_Oriented_Modular.sos](doc/examples/Process_Oriented_Modular.html) ([script](doc/examples/Process_Oriented_Modular.sos))<br>\n <small>Same example as above with codes for core computations stored as separate scripts.</small>\n* [Outcome_Oriented.sos](doc/examples/Outcome_Oriented.html) ([script](doc/examples/Outcome_Oriented.sos))<br>\n <small>Demonstration of a outcome-oriented Makefile-style workflow that executes steps that are needed to generate specified outcome.</small>\n* [Outcome_Oriented_Step_Targets.sos](doc/examples/Outcome_Oriented_Step_Targets.html) ([script](doc/examples/Outcome_Oriented.sos))<br>\n <small>Demonstration of a outcome-oriented Makefile-style workflow that determines and executes dependencies by step targets instead of files.</small>\n* [Mixed_Style.sos](doc/examples/Mixed_Style.html) ([script](doc/examples/Mixed_Style.sos))<br>\n <small>Demonstration of a mixed-style workflow that executes steps of a forward-step workflow with dependencies produced by auxiliary steps.</small>\n* [Mixed_Style_Data_Flow.sos](doc/examples/Mixed_Style_Data_Flow.html) ([script](doc/examples/Mixed_Style_Data_Flow.sos))<br>\n <small>Demonstration of a mixed-style workflow that executes steps of a forward-step workflow with dependencies specified by named outputs.</small>\n* [Next-generation sequencing genotyping pipeline](doc/examples/WGS_Call.html) ([notebook](doc/examples/WGS_Call.ipynb))<br>\n <small>This is the SoS version of the <a href=\"http://www.htslib.org/workflow/#mapping_to_variant\">samtools workflow: WGS/WES Mapping to Variant Calls - Version 1.0</a></small>\n* [RNA Seq Differential Expression Analysis Workflow](doc/examples/RNASeqDE.html) ([notebook](doc/examples/RNASeqDE.ipynb) and [generated report](doc/examples/RNASeqDE_report.html))<br>\n <small>This is the SoS version of the <a href=\"https://www.bioconductor.org/help/workflows/rnaseqGene/\">RNA-seq workflow: gene-level exploratory analysis and differential expression</a> from bioconductor.</small>\n* [RNA Seq Normalization and Expression Residuals Analysis Workflow](doc/examples/RNASeqGTEx.html) ([notebook](doc/examples/RNASeqGTEx.ipynb) and [generated report](doc/examples/RNASeqGTEx_report.html))<br>\n <small>This is the SoS version of the RNA-seq preprocessing step of <a href=\"https://github.com/broadinstitute/gtex-pipeline/tree/63b13b8ced25cf8ab8e7a26f40a495e523630a9b/qtl\">eQTL discovery pipeline</a> from the <a href=\"https://www.gtexportal.org/home/\"> GTEx Consortium </a>, updated July 31, 2017.</small> \n\nPlease [follow instructions here](doc/examples/Workflow_Manuscript_Examples.html) to reproduce all but the last (due to data accessibility restrictions) examples.",
"_____no_output_____"
],
[
"# Example Scripts and workflow notebooks\n\nSoS scripts for the maintenance of SoS\n\n* [update_toc.sos](doc/examples/update_toc.html) ([script](doc/examples/update_toc.sos))<br>\n <small>A sos script that was used by SoS to update TOC of the wiki pages</small>\n* [release](doc/examples/release.html) ([script](doc/examples/release))<br>\n <small>Script to manage SoS website and release SoS to pip, docker hub, etc</small>\n\nPedagogical examples\n\n* [DEG Annotation with external task](doc/examples/DEG_Annotation_remote.html) ([notebook](doc/examples/DEG_Annotation_remote.ipynb))<br>\n <small>The same example but submit the R part of the workflow as an external task</small>\n* [Simple External Tasks](doc/examples/LineCount.html) ([notebook](doc/examples/LineCount.ipynb))<br>\n <small>An example to generate and execute a number of external tasks in Jupyter notebook. You can try to use option <code>trunk_size</code> to group the tasks. This example also demonstrates the use of option <code>shared</code> to collect results from tasks as variables</small>\n* [Remote Target](doc/examples/Remote_Target.html) ([notebook](doc/examples/Remote_Target.ipynb))<br>\n <small>An example on the analysis of targets that reside on remote servers using target <code>remote()</code> and step option <code>remote</code></small>\n \nReal-world applications\n\n* [Data preprocessing for TADA-A R analysis](https://github.com/TADA-A/TADA-A/blob/master/external_tools/mutation_annotation_pipeline.ipynb), for Liu et al 2018, American Journal of Human Genetics.\n* [Data preprocessing for MASH R analysis](https://github.com/stephenslab/gtexresults/blob/master/workflows/fastqtl_to_mash.ipynb) and [Multivariate eQTL analysis pipeline](https://gaow.github.io/mnm-gtex-v8/analysis/mashr_flashr_workflow.html), for Urbut et al 2018, Nature Genetics.",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecc928f7dbb124dade269f7bebdb555f74f25fc8 | 906,386 | ipynb | Jupyter Notebook | Twitter Sentiment Analysis.ipynb | nehanagle/twitter-sentiment | 275c2576ad474eb64a0182096f025c196f5d6f54 | [
"MIT"
] | 1 | 2021-05-08T07:47:32.000Z | 2021-05-08T07:47:32.000Z | Twitter Sentiment Analysis.ipynb | nehanagle/twitter-sentiment | 275c2576ad474eb64a0182096f025c196f5d6f54 | [
"MIT"
] | null | null | null | Twitter Sentiment Analysis.ipynb | nehanagle/twitter-sentiment | 275c2576ad474eb64a0182096f025c196f5d6f54 | [
"MIT"
] | 1 | 2020-10-04T10:06:05.000Z | 2020-10-04T10:06:05.000Z | 477.799684 | 275,168 | 0.934093 | [
[
[
"## Twitter Sentiment Analysis",
"_____no_output_____"
],
[
"The objective is to identify positive , hate and neutral speech in tweets. For the sake of simplicity, we say a tweet contains hate speech if it has a racist or sexiest sentiment associated with it.",
"_____no_output_____"
]
],
[
[
"## Importing required libraries \nimport re \nimport pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nfrom nltk.stem.porter import *\nfrom wordcloud import WordCloud\nimport nltk\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nimport gensim\n\nfrom tqdm import tqdm\ntqdm.pandas(desc=\"progress-bar\")\nfrom gensim.models.doc2vec import LabeledSentence\n\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import f1_score\n\nfrom sklearn import svm",
"D:\\Anaconda\\lib\\site-packages\\tqdm\\std.py:668: FutureWarning: The Panel class is removed from pandas. Accessing it from the top-level namespace will also be removed in the next version\n from pandas import Panel\n"
],
[
"#importing training and testing dataset from \ntrain_data= pd.read_csv(r'D:\\twiter_train.csv')\ntest_data= pd.read_csv(r'D:\\twiter_test.csv')",
"_____no_output_____"
],
[
"train_data.head(5)",
"_____no_output_____"
],
[
"test_data.head()",
"_____no_output_____"
]
],
[
[
"**Dataset consist of below field**\n\n* ID - Unique id of tweet\n* Label - sentiment of tweet\n* Tweet - text of the tweet",
"_____no_output_____"
]
],
[
[
"print(\"Total Number of tweet for training:\",train_data.shape[0])\nprint(\"Total Number of tweet for testing:\",test_data.shape[0])",
"Total Number of tweet for training: 31962\nTotal Number of tweet for testing: 17197\n"
]
],
[
[
"## Data Preprocessing",
"_____no_output_____"
]
],
[
[
"train_data[\"label\"].value_counts()",
"_____no_output_____"
],
[
"count_classes = pd.value_counts(train_data['label'], sort = True)\n\ncount_classes.plot(kind = 'bar', rot=0)\n\nplt.title(\"Label Distribution\")\n\n# plt.xticks(range(2), LABELS)\n\nplt.xlabel(\"Class\")\n\nplt.ylabel(\"Frequency\")",
"_____no_output_____"
]
],
[
[
"**Data set is imbalance as we have 7% of data with label 1 and 93% of data with label 0 **",
"_____no_output_____"
]
],
[
[
"#Combining training and testing data for data cleaning\nCombine= train_data.append(test_data, ignore_index=True)\n",
"_____no_output_____"
]
],
[
[
"* These Twitter handles are hardly giving any information about the nature of the tweet.\n* The punctuations, numbers and even special characters wouldn’t help in differentiating different kinds of tweets.\n* Most of the smaller words do not add much value. For example, ‘pdx’, ‘his’, ‘all’. So, we will try to remove them as well from our data.",
"_____no_output_____"
]
],
[
[
"Combine['tweet'] = Combine['tweet'].str.replace('@[\\w]*', '')\nCombine['tweet'] = Combine['tweet'].str.replace('[^a-zA-Z#\\s]', '')",
"_____no_output_____"
],
[
"Combine['tweet'] = Combine['tweet'].apply(lambda x:' '.join([w for w in x.split() if len(w)>3]))\nCombine.head()",
"_____no_output_____"
],
[
"#stemming\ntokenize_tweet= Combine['tweet'].apply(lambda x: x.split())\nstemmer=PorterStemmer()\ntokenize_tweet = tokenize_tweet.apply(lambda x:[stemmer.stem(i) for i in x])\n\n# Sentences\nfor i in range(len(tokenize_tweet)):\n tokenize_tweet[i]=' '.join(tokenize_tweet[i])\nCombine['tweet']=tokenize_tweet",
"_____no_output_____"
],
[
"Combine.head(10)",
"_____no_output_____"
]
],
[
[
"A wordcloud is a visualization wherein the most frequent words appear in large size and the less frequent words appear in smaller sizes.\n\nLet’s visualize all the words our data using the wordcloud plot.",
"_____no_output_____"
]
],
[
[
"All_Words= ' '.join([text for text in Combine['tweet']])\nwordcloud = WordCloud(width=800, height =500, random_state=21, max_font_size=110).generate(All_Words)\nplt.figure(figsize=(10,7))\nplt.imshow(wordcloud,interpolation=\"bilinear\")\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can see most of the words are positive or neutral. With happy and love being the most frequent ones. It doesn’t give us any idea about the words associated with the racist/sexist tweets. Hence, we will plot separate wordclouds for both the classes(racist/sexist or not) in our train data.",
"_____no_output_____"
],
[
"**Words in non racist/sexist tweets**",
"_____no_output_____"
]
],
[
[
"Z_Words= ' '.join([text for text in Combine['tweet'][Combine['label']==0]])\nwordcloud = WordCloud(width=800, height =500, random_state=21, max_font_size=110).generate(Z_Words)\nplt.figure(figsize=(10,7))\nplt.imshow(wordcloud,interpolation=\"bilinear\")\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can see most of the words are positive or neutral. With happy, smile, and love being the most frequent ones. Hence, most of the frequent words are compatible with the sentiment which is non racist/sexists tweets.",
"_____no_output_____"
],
[
"**Racist/Sexist Tweets**",
"_____no_output_____"
]
],
[
[
"ZZ_Words= ' '.join([text for text in Combine['tweet'][Combine['label']==1]])\nwordcloud = WordCloud(width=800, height =500, random_state=21, max_font_size=110).generate(ZZ_Words)\nplt.figure(figsize=(10,7))\nplt.imshow(wordcloud,interpolation=\"bilinear\")\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Most of the words have negative connotations. So, it seems we have a pretty good text data to work on. Next we will the hashtags/trends in our twitter data.",
"_____no_output_____"
],
[
"Hashtags in twitter are synonymous with the ongoing trends on twitter at any particular point in time. We should try to check whether these hashtags add any value to our sentiment analysis task, i.e., they help in distinguishing tweets into the different sentiments.",
"_____no_output_____"
]
],
[
[
"def extract_hashtag(x):\n hastag=[]\n for i in x:\n ht = re.findall(r\"#[\\w]*\",i)\n hastag.append(ht)\n return hastag\n\nhastag_pos = extract_hashtag(Combine['tweet'][Combine['label']==0])\nhastag_neg = extract_hashtag(Combine['tweet'][Combine['label']==1])\n\nHT_Pos = sum(hastag_pos,[])\nHT_Neg = sum(hastag_neg,[])",
"_____no_output_____"
]
],
[
[
"Now that we have prepared our lists of hashtags for both the sentiments, we can plot the top n hashtags. So, first let’s check the hashtags in the non-racist/sexist tweets.",
"_____no_output_____"
]
],
[
[
"a=nltk.FreqDist(HT_Pos)\nd = pd.DataFrame({'Hashtag': list(a.keys()),\n 'Count': list(a.values())})\n# selecting top 10 most frequent hashtags \nd = d.nlargest(columns=\"Count\", n = 11) \nplt.figure(figsize=(16,5))\nax= sns.barplot(data=d, x='Hashtag', y='Count')\nax.set(ylabel='Count')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"All these hashtags are positive and it makes sense. Let’s check the most frequent hashtags appearing in the racist/sexist tweets.",
"_____no_output_____"
]
],
[
[
"a=nltk.FreqDist(HT_Neg)\nd = pd.DataFrame({'Hashtag': list(a.keys()),\n 'Count': list(a.values())})\n# selecting top 10 most frequent hashtags \nd = d.nlargest(columns=\"Count\", n = 10) \nplt.figure(figsize=(16,5))\nax= sns.barplot(data=d, x='Hashtag', y='Count')\nax.set(ylabel='Count')\nplt.show()",
"_____no_output_____"
]
],
[
[
"As expected, most of the terms are negative with a few neutral terms as well. So, it’s not a bad idea to keep these hashtags in our data as they contain useful information. ",
"_____no_output_____"
],
[
"To analyze a preprocessed data, it needs to be converted into features.\n\n## Extracting Features",
"_____no_output_____"
],
[
"**Bag-of-Words Features**\n\nBag-of-Words is a method to represent text into numerical features. Consider a corpus (a collection of texts) called C of D documents {d1,d2…..dD} and N unique tokens extracted out of the corpus C. ",
"_____no_output_____"
]
],
[
[
"#Each tweet is converted into vector\nbow_vectorizer = CountVectorizer(max_df=0.90, min_df=2, max_features=2000, stop_words='english')\n# bag-of-words feature matrix\nbow = bow_vectorizer.fit_transform(Combine['tweet'])\nbow.shape",
"_____no_output_____"
]
],
[
[
"## TF-IDF Features\n\nThis is another method which is based on the frequency method but it is different to the bag-of-words approach in the sense that it takes into account, not just the occurrence of a word in a single document (or tweet) but in the entire corpus.\n\nTF-IDF works by penalizing the common words by assigning them lower weights while giving importance to words which are rare in the entire corpus but appear in good numbers in few documents.",
"_____no_output_____"
]
],
[
[
"tfidf_vectorizer = TfidfVectorizer(max_df=0.90, min_df=2, max_features=2000, stop_words='english')\n# TF-IDF feature matrix\ntfidf = tfidf_vectorizer.fit_transform(Combine['tweet'])\ntfidf.shape",
"_____no_output_____"
]
],
[
[
"## Word Vectorization (Word Embedding)\n\nWord vectorization is the process of mapping words to a set of real numbers or vectors. ",
"_____no_output_____"
]
],
[
[
"tokenized_tweet = Combine['tweet'].apply(lambda x: x.split())\nmodel_w2v = gensim.models.Word2Vec(tokenized_tweet,size =200, window=5, min_count=2 , sg= 1, hs =0, negative=10, workers=2, seed = 34)\nmodel_w2v.train(tokenized_tweet, total_examples= len(Combine['tweet']), epochs=20)",
"_____no_output_____"
],
[
"#words similar to dinner\nmodel_w2v.wv.most_similar(positive= \"dinner\")\n",
"_____no_output_____"
],
[
"#function to create a vector for each tweet by taking average of vectors of words present in the tweet\n\ndef word_vector(tokens, size):\n vec = np.zeros(size).reshape((1, size))\n count = 0.\n for word in tokens:\n try:\n vec += model_w2v[word].reshape((1, size))\n count += 1.\n except KeyError: # handling the case where the token is not in vocabulary\n \n continue\n if count != 0:\n vec /= count\n return vec\n\nwordvec_arrays = np.zeros((len(tokenized_tweet), 200))\n\nfor i in range(len(tokenized_tweet)):\n wordvec_arrays[i,:] = word_vector(tokenized_tweet[i], 200)\n \nwordvec_df = pd.DataFrame(wordvec_arrays)\nwordvec_df.shape",
"<ipython-input-23-6fe194e3ff16>:8: DeprecationWarning: Call to deprecated `__getitem__` (Method will be removed in 4.0.0, use self.wv.__getitem__() instead).\n vec += model_w2v[word].reshape((1, size))\n"
]
],
[
[
"## Doc2vec Embedding",
"_____no_output_____"
],
[
"Doc2Vec is unsupervised algorithm to generate vectors for sentences/paraghrap/documents. To implement Doc2Vec we have to labelize or tag each tokenize tweet with unique ID",
"_____no_output_____"
]
],
[
[
"def add_label(twt):\n output = []\n for i, s in zip(twt.index, twt):\n output.append(LabeledSentence(s, [\"tweet_\" + str(i)]))\n return output\nlabeled_tweets = add_label(tokenized_tweet)\nlabeled_tweets[:6]",
"<ipython-input-24-5203b6bc490a>:4: DeprecationWarning: Call to deprecated `LabeledSentence` (Class will be removed in 4.0.0, use TaggedDocument instead).\n output.append(LabeledSentence(s, [\"tweet_\" + str(i)]))\n"
],
[
"#Train Doc2Vec\nmodel_d2v = gensim.models.Doc2Vec(dm=1, # dm = 1 for ‘distributed memory’ model \n dm_mean=1, # dm = 1 for using mean of the context word vectors\n size=200, # no. of desired features\n window=5, # width of the context window\n negative=7, # if > 0 then negative sampling will be used\n min_count=5, # Ignores all words with total frequency lower than 2.\n workers=3, # no. of cores\n alpha=0.1, # learning rate\n seed = 23)\n\nmodel_d2v.build_vocab([i for i in tqdm(labeled_tweets)])",
"D:\\Anaconda\\lib\\site-packages\\gensim\\models\\doc2vec.py:319: UserWarning: The parameter `size` is deprecated, will be removed in 4.0.0, use `vector_size` instead.\n warnings.warn(\"The parameter `size` is deprecated, will be removed in 4.0.0, use `vector_size` instead.\")\n100%|████████████████████████████████████████████████████████████████████████| 49159/49159 [00:00<00:00, 353871.43it/s]\n"
],
[
"model_d2v.train(labeled_tweets, total_examples= len(Combine['tweet']), epochs=15)",
"_____no_output_____"
],
[
"#Preparing doc2vec feature set\ndocvec_arrays = np.zeros((len(tokenized_tweet), 200))\n\nfor i in range(len(Combine)):\n docvec_arrays[i,:] = model_d2v.docvecs[i].reshape((1,200))\n \ndocvec_df = pd.DataFrame(docvec_arrays)\ndocvec_df.head()",
"_____no_output_____"
]
],
[
[
"**Try to fit logistic Regression model on Bag of words Feature**",
"_____no_output_____"
]
],
[
[
"#BAG with Logistic Regression\n\ntrain_bow = bow[:31962,:]\ntest_bow = bow[31962:,:]\n\nxtrain_bow,xvalid_bow,ytrain,yvalid = train_test_split(train_bow, train_data['label'],random_state = 42,test_size = 0.3)\n",
"_____no_output_____"
],
[
"#Random Over Sampling\nfrom imblearn.over_sampling import BorderlineSMOTE, SMOTE, ADASYN, SMOTENC, RandomOverSampler\nros = RandomOverSampler(random_state=777)\nros_xtrain_bow, ros_ytrain_bow = ros.fit_sample(train_bow, train_data['label'])",
"_____no_output_____"
],
[
"ros_ytrain_bow.value_counts()",
"_____no_output_____"
],
[
"xtrain_bow,xvalid_bow,ytrain,yvalid = train_test_split(ros_xtrain_bow, ros_ytrain_bow,random_state = 42,test_size = 0.3)\nlreg = LogisticRegression()\nlreg.fit(xtrain_bow,ytrain)\nprediction =lreg.predict_proba(xvalid_bow)\nprediction_int = prediction[:,1]>=0.3\nprediction_int = prediction_int.astype(np.int)\nf1_score(yvalid, prediction_int)",
"D:\\Anaconda\\lib\\site-packages\\sklearn\\linear_model\\_logistic.py:762: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n n_iter_i = _check_optimize_result(\n"
],
[
"test_pred = lreg.predict_proba(test_bow)\ntest_pred_int = test_pred[:,1] >= 0.3\ntest_pred_int = test_pred_int.astype(np.int)\ntest_data['label']= test_pred_int\nsubmission = test_data[['id','tweet','label']]",
"_____no_output_____"
],
[
"submission.head()",
"_____no_output_____"
]
],
[
[
"**We will follow same step but with TFIDF feature**",
"_____no_output_____"
]
],
[
[
"\ntrain_tfidf = tfidf[:31962,:]\ntest_tfidf = tfidf[31962:,:]\n\nxtrain_tfidf,xvalid_tfidf,ytrain,yvalid = train_test_split(train_tfidf, train_data['label'],random_state = 42,test_size = 0.3)\nlreg.fit(xtrain_tfidf,ytrain)\nprediction = lreg.predict_proba(xvalid_tfidf)\nprediction_int = prediction[:,1] >= 0.3\nprediction_int = prediction_int.astype(np.int)\nf1_score(yvalid,prediction_int)",
"_____no_output_____"
],
[
"train_w2v = wordvec_df.iloc[:31962,:]\ntest_w2v = wordvec_df.iloc[31962:,:]\nxtrain_w2v = train_w2v.iloc[ytrain.index]\nxvalid_w2v = train_w2v.iloc[yvalid.index,:]\nlreg.fit(xtrain_w2v,ytrain)\nprediction = lreg.predict_proba(xvalid_w2v)\nprediction_int = prediction[:,1]>=0.3\nprediction_int = prediction_int.astype(np.int)\nf1_score(yvalid, prediction_int)",
"_____no_output_____"
],
[
"train_d2v = docvec_df.iloc[:31962,:]\ntest_d2v = docvec_df.iloc[31962:,:]\nxtrain_d2v = train_d2v.iloc[ytrain.index,:]\nxvalid_d2v = train_d2v.iloc[yvalid.index,:]\nlreg.fit(xtrain_d2v,ytrain)\nprediction = lreg.predict_proba(xvalid_d2v)\nprediction_int = prediction[:,1] >= 0.3\nprediction_int = prediction_int.astype(np.int)\nf1_score(yvalid,prediction_int)",
"_____no_output_____"
],
[
"train_d2v = docvec_df.iloc[:31962,:]\ntest_d2v = docvec_df.iloc[31962:,:]\nd2v = RandomOverSampler(random_state=777)\nd2v_xtrain, d2v_ytrain= d2v.fit_sample(train_d2v, train_data['label'])\nxtrain_d2v,xvalid_d2v,ytrain,yvalid = train_test_split(d2v_xtrain, d2v_ytrain,random_state = 42,test_size = 0.3)\nlreg.fit(xtrain_d2v,ytrain)\nprediction = lreg.predict_proba(xvalid_d2v)\nprediction_int = prediction[:,1] >= 0.3\nprediction_int = prediction_int.astype(np.int)\nf1_score(yvalid,prediction_int)",
"D:\\Anaconda\\lib\\site-packages\\sklearn\\linear_model\\_logistic.py:762: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n n_iter_i = _check_optimize_result(\n"
]
],
[
[
"**AS we can see f1 score is low for unbalance data and high for balance data, So it's good practice to build model on balance data**",
"_____no_output_____"
]
],
[
[
"#Random Forest\n\nfrom sklearn.ensemble import RandomForestClassifier\n\nrf = RandomForestClassifier(n_estimators = 600, random_state=11).fit(xtrain_bow, ytrain)\nprediction = rf.predict(xvalid_bow)\nf1_score(yvalid, prediction)",
"_____no_output_____"
],
[
"test_pred = rf.predict(test_bow)\ntest_data['label'] = test_pred\nsubmission = test_data[['id','tweet','label']]\nsubmission.head()",
"_____no_output_____"
],
[
"from xgboost import XGBClassifier\nxgb_model = XGBClassifier(max_depth=6, n_estimators= 1000).fit(xtrain_bow,ytrain)\nprediction = xgb_model.predict(xvalid_bow)\nf1_score(yvalid, prediction)\n\n ",
"_____no_output_____"
],
[
"test_pred = xgb_model.predict(test_bow)\ntest_data['label'] = test_pred\nsubmission = test_data[['id','tweet','label']]\nsubmission.head()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecc92cbc447bdd93ef675089e8f3eb21552bfa45 | 559,766 | ipynb | Jupyter Notebook | notebooks/Comp_Scatt_NeuralNetwork_Classification_All energies together.ipynb | chiarabadiali/comp_scatt_ML | 9a87dbcdff34e63e81439483e529b9404e5ff125 | [
"MIT"
] | null | null | null | notebooks/Comp_Scatt_NeuralNetwork_Classification_All energies together.ipynb | chiarabadiali/comp_scatt_ML | 9a87dbcdff34e63e81439483e529b9404e5ff125 | [
"MIT"
] | null | null | null | notebooks/Comp_Scatt_NeuralNetwork_Classification_All energies together.ipynb | chiarabadiali/comp_scatt_ML | 9a87dbcdff34e63e81439483e529b9404e5ff125 | [
"MIT"
] | null | null | null | 80.046618 | 24,836 | 0.509908 | [
[
[
"import pandas as pd \nimport numpy as np\nimport math\nimport keras\nimport tensorflow as tf\nimport progressbar\nimport os\nfrom os import listdir",
"_____no_output_____"
]
],
[
[
"## Print Dependencies\n\n\n\nDependences are fundamental to record the computational environment.",
"_____no_output_____"
]
],
[
[
"%load_ext watermark\n\n# python, ipython, packages, and machine characteristics\n%watermark -v -m -p pandas,keras,numpy,math,tensorflow,matplotlib,h5py\n\n# date\nprint (\" \")\n%watermark -u -n -t -z",
"Python implementation: CPython\nPython version : 3.8.8\nIPython version : 7.22.0\n\npandas : 1.2.3\nkeras : 2.4.3\nnumpy : 1.19.5\nmath : unknown\ntensorflow: 2.4.1\nmatplotlib: 3.4.0\nh5py : 2.10.0\n\nCompiler : Clang 12.0.0 (clang-1200.0.32.29)\nOS : Darwin\nRelease : 19.6.0\nMachine : x86_64\nProcessor : i386\nCPU cores : 8\nArchitecture: 64bit\n\n \nLast updated: Mon Apr 12 2021 13:15:06CEST\n\n"
]
],
[
[
"## Load of the data",
"_____no_output_____"
]
],
[
[
"from process import loaddata\n#low = low energy\n#high = high energy\nclass_data0_low = loaddata(\"../data_old/{}.csv\".format('probability'))\nclass_data0_high = loaddata(\"../data/{}.csv\".format('weight'))",
"_____no_output_____"
],
[
"class_data0_low = class_data0_low[class_data0_low[:,0] > 0.0001]\nclass_data0_high = class_data0_high[class_data0_high[:,0] > 0.0001]",
"_____no_output_____"
],
[
"data = []\n\nfor class_ in class_data0_low:\n if class_[0] > 0.3 and class_[0] < 0.45:\n for i in range(10):\n data.append(class_) \n if class_[0] > 0.45 and class_[0] < 0.6:\n for i in range(1000):\n data.append(class_) \n else: \n data.append(class_) ",
"_____no_output_____"
],
[
"len(data)\nclass_data_low = []\nclass_data_low = np.array(data)",
"_____no_output_____"
],
[
"data = []\ni = 0\nj = 0\nsaved = []\nfor class_ in class_data0_high:\n if j == 1000000:\n #print(class_[0])\n if class_[0] < 0.002: \n if i == 50:\n i = 0\n data.append(class_) \n else:\n i = i + 1\n if class_[0] > 0.003:\n for i in range(10):\n data.append(class_) \n if class_[0] > 0.005:\n for i in range(30):\n data.append(class_) \n if class_[0] > 0.007:\n for i in range(50):\n data.append(class_) \n else: \n data.append(class_) \n j = j + 1\n else: \n j = j + 1",
"_____no_output_____"
],
[
"len(data)\nclass_data_high = []\nclass_data_high = np.array(data)",
"_____no_output_____"
],
[
"np.random.shuffle(class_data_low)\ny_low = class_data_low[:,0]\nA = class_data_low\nprint(A[0])\nA[:,9] = A[:,13]\nprint(A[0])\nx_low = class_data_low[:,1:10]\nprint(x_low[0])\nprint(x_low.shape)",
"[ 3.17313364e-01 7.87977076e-01 -2.89899188e-01 6.99009964e-01\n -4.30383701e-01 1.40994990e-02 8.95661916e-02 2.46000000e+00\n 2.69999999e+09 1.55974671e-01 -9.33914234e-02 -3.34378385e-01\n -2.90375231e-01 3.81700462e-03]\n[ 3.17313364e-01 7.87977076e-01 -2.89899188e-01 6.99009964e-01\n -4.30383701e-01 1.40994990e-02 8.95661916e-02 2.46000000e+00\n 2.69999999e+09 3.81700462e-03 -9.33914234e-02 -3.34378385e-01\n -2.90375231e-01 3.81700462e-03]\n[ 7.87977076e-01 -2.89899188e-01 6.99009964e-01 -4.30383701e-01\n 1.40994990e-02 8.95661916e-02 2.46000000e+00 2.69999999e+09\n 3.81700462e-03]\n(535805, 9)\n"
],
[
"np.random.shuffle(class_data_high)\ny_high = class_data_high[:,0]\nA = class_data_high\nprint(A[0])\nA[:,9] = A[:,13]\nprint(A[0])\nx_high = class_data_high[:,1:10]\nprint(x_high[0])\nprint(x_high.shape)",
"[ 4.25059985e-04 -4.11999703e+01 4.22826150e+01 -9.96823156e+00\n 2.98617198e+01 -5.95334012e+01 -2.85761809e+01 3.87000000e+07\n 1.66200000e+07 -4.96133027e-01 -1.21340127e-01 4.35643161e-01\n 4.21174657e-01 2.82316972e-07]\n[ 4.25059985e-04 -4.11999703e+01 4.22826150e+01 -9.96823156e+00\n 2.98617198e+01 -5.95334012e+01 -2.85761809e+01 3.87000000e+07\n 1.66200000e+07 2.82316972e-07 -1.21340127e-01 4.35643161e-01\n 4.21174657e-01 2.82316972e-07]\n[-4.11999703e+01 4.22826150e+01 -9.96823156e+00 2.98617198e+01\n -5.95334012e+01 -2.85761809e+01 3.87000000e+07 1.66200000e+07\n 2.82316972e-07]\n(1, 9)\n"
],
[
"print(class_data_low.shape)\nprint(class_data0_low.shape)\nprint(class_data_high.shape)\nprint(class_data0_high.shape)",
"(535805, 14)\n(135911, 14)\n(1, 14)\n(1159974, 14)\n"
],
[
"x = np.vstack((x_low, x_high))\ny = np.hstack((y_low, y_high))",
"_____no_output_____"
],
[
"x.shape",
"_____no_output_____"
],
[
"y.shape",
"_____no_output_____"
],
[
"train_split = 0.75\ntrain_limit = int(len(y)*train_split)\nprint(\"Training sample: {0} \\nValuation sample: {1}\".format(train_limit, len(y)-train_limit))",
"Training sample: 401854 \nValuation sample: 133952\n"
],
[
"x_train = x[:train_limit]\nx_val = x[train_limit:]\n\ny_train = y[:train_limit]\ny_val = y[train_limit:]",
"_____no_output_____"
]
],
[
[
"## Model Build",
"_____no_output_____"
]
],
[
[
"from keras.models import Sequential\nfrom keras.layers.core import Dense\nimport keras.backend as K\nfrom keras import optimizers\nfrom keras import models\nfrom keras import layers\nfrom keras.layers.normalization import BatchNormalization",
"_____no_output_____"
],
[
"def build_model() :\n model = models.Sequential()\n model.add (BatchNormalization(input_dim = 9))\n model.add (layers.Dense (18 , activation = \"sigmoid\"))\n model.add (layers.Dense (18 , activation = \"relu\"))\n model.add (layers.Dense (1 , activation = \"sigmoid\"))\n model.compile(optimizer = \"adam\" , loss = 'mae' , metrics = [\"mape\"])\n return model",
"_____no_output_____"
],
[
"model = build_model ()\nhistory = model.fit ( x_train, y_train, epochs = 3000, batch_size = 10000000 , validation_data = (x_val, y_val) )\nmodel.save(\"../models/classifier/{}_noposition.h5\".format('probability'))",
"Epoch 1/3000\n1/1 [==============================] - 1s 1s/step - loss: 0.1273 - mape: 184.8909 - val_loss: 0.1384 - val_mape: 212.1995\nEpoch 2/3000\n1/1 [==============================] - 0s 135ms/step - loss: 0.1220 - mape: 180.8341 - val_loss: 0.1320 - val_mape: 205.7487\nEpoch 3/3000\n1/1 [==============================] - 0s 136ms/step - loss: 0.1174 - mape: 176.9396 - val_loss: 0.1263 - val_mape: 200.0144\nEpoch 4/3000\n1/1 [==============================] - 0s 137ms/step - loss: 0.1136 - mape: 173.2513 - val_loss: 0.1213 - val_mape: 194.8420\nEpoch 5/3000\n1/1 [==============================] - 0s 139ms/step - loss: 0.1107 - mape: 169.8372 - val_loss: 0.1175 - val_mape: 190.2351\nEpoch 6/3000\n1/1 [==============================] - 0s 141ms/step - loss: 0.1089 - mape: 166.7531 - val_loss: 0.1147 - val_mape: 186.1704\nEpoch 7/3000\n1/1 [==============================] - 0s 140ms/step - loss: 0.1080 - mape: 164.0465 - val_loss: 0.1131 - val_mape: 182.7531\nEpoch 8/3000\n1/1 [==============================] - 0s 137ms/step - loss: 0.1080 - mape: 161.7618 - val_loss: 0.1122 - val_mape: 179.9831\nEpoch 9/3000\n1/1 [==============================] - 0s 133ms/step - loss: 0.1083 - mape: 159.8563 - val_loss: 0.1117 - val_mape: 177.8474\nEpoch 10/3000\n1/1 [==============================] - 0s 140ms/step - loss: 0.1087 - mape: 158.3006 - val_loss: 0.1114 - val_mape: 176.2545\nEpoch 11/3000\n1/1 [==============================] - 0s 128ms/step - loss: 0.1090 - mape: 157.0620 - val_loss: 0.1112 - val_mape: 175.1471\nEpoch 12/3000\n1/1 [==============================] - 0s 137ms/step - loss: 0.1092 - mape: 156.1201 - val_loss: 0.1110 - val_mape: 174.4228\nEpoch 13/3000\n1/1 [==============================] - 0s 136ms/step - loss: 0.1092 - mape: 155.4290 - val_loss: 0.1109 - val_mape: 174.0296\nEpoch 14/3000\n1/1 [==============================] - 0s 134ms/step - loss: 0.1091 - mape: 154.9548 - val_loss: 0.1107 - val_mape: 173.9237\nEpoch 15/3000\n1/1 [==============================] - 0s 139ms/step - loss: 0.1088 - mape: 154.6687 - val_loss: 0.1106 - val_mape: 174.0729\nEpoch 16/3000\n1/1 [==============================] - 0s 138ms/step - loss: 0.1084 - mape: 154.5477 - val_loss: 0.1107 - val_mape: 174.5264\nEpoch 17/3000\n1/1 [==============================] - 0s 142ms/step - loss: 0.1079 - mape: 154.5682 - val_loss: 0.1108 - val_mape: 175.1052\nEpoch 18/3000\n1/1 [==============================] - 0s 138ms/step - loss: 0.1074 - mape: 154.7152 - val_loss: 0.1112 - val_mape: 175.8692\nEpoch 19/3000\n1/1 [==============================] - 0s 141ms/step - loss: 0.1068 - mape: 154.9713 - val_loss: 0.1117 - val_mape: 176.7566\nEpoch 20/3000\n1/1 [==============================] - 0s 135ms/step - loss: 0.1062 - mape: 155.3203 - val_loss: 0.1123 - val_mape: 177.7207\nEpoch 21/3000\n1/1 [==============================] - 0s 138ms/step - loss: 0.1057 - mape: 155.7266 - val_loss: 0.1131 - val_mape: 178.7229\nEpoch 22/3000\n1/1 [==============================] - 0s 138ms/step - loss: 0.1053 - mape: 156.1714 - val_loss: 0.1140 - val_mape: 179.6844\nEpoch 23/3000\n1/1 [==============================] - 0s 131ms/step - loss: 0.1050 - mape: 156.6086 - val_loss: 0.1149 - val_mape: 180.5573\nEpoch 24/3000\n1/1 [==============================] - 0s 131ms/step - loss: 0.1047 - mape: 157.0062 - val_loss: 0.1157 - val_mape: 181.2975\nEpoch 25/3000\n1/1 [==============================] - 0s 137ms/step - loss: 0.1046 - mape: 157.3360 - val_loss: 0.1164 - val_mape: 181.8515\nEpoch 26/3000\n1/1 [==============================] - 0s 142ms/step - loss: 0.1045 - mape: 157.5741 - val_loss: 0.1168 - val_mape: 182.1733\nEpoch 27/3000\n1/1 [==============================] - 0s 140ms/step - loss: 0.1044 - mape: 157.6782 - val_loss: 0.1170 - val_mape: 182.2490\nEpoch 28/3000\n1/1 [==============================] - 0s 137ms/step - loss: 0.1043 - mape: 157.6345 - val_loss: 0.1168 - val_mape: 182.0611\nEpoch 29/3000\n1/1 [==============================] - 0s 143ms/step - loss: 0.1042 - mape: 157.4242 - val_loss: 0.1164 - val_mape: 181.6362\nEpoch 30/3000\n1/1 [==============================] - 0s 134ms/step - loss: 0.1040 - mape: 157.0624 - val_loss: 0.1158 - val_mape: 181.0042\nEpoch 31/3000\n1/1 [==============================] - 0s 127ms/step - loss: 0.1037 - mape: 156.5644 - val_loss: 0.1150 - val_mape: 180.1900\nEpoch 32/3000\n1/1 [==============================] - 0s 135ms/step - loss: 0.1034 - mape: 155.9453 - val_loss: 0.1142 - val_mape: 179.2395\nEpoch 33/3000\n1/1 [==============================] - 0s 135ms/step - loss: 0.1031 - mape: 155.2275 - val_loss: 0.1133 - val_mape: 178.1955\nEpoch 34/3000\n1/1 [==============================] - 0s 132ms/step - loss: 0.1028 - mape: 154.4410 - val_loss: 0.1125 - val_mape: 177.1107\nEpoch 35/3000\n1/1 [==============================] - 0s 133ms/step - loss: 0.1025 - mape: 153.6228 - val_loss: 0.1117 - val_mape: 176.0527\nEpoch 36/3000\n1/1 [==============================] - 0s 128ms/step - loss: 0.1022 - mape: 152.8099 - val_loss: 0.1111 - val_mape: 175.0434\nEpoch 37/3000\n1/1 [==============================] - 0s 131ms/step - loss: 0.1020 - mape: 152.0253 - val_loss: 0.1106 - val_mape: 174.1335\nEpoch 38/3000\n1/1 [==============================] - 0s 131ms/step - loss: 0.1018 - mape: 151.3112 - val_loss: 0.1103 - val_mape: 173.3571\nEpoch 39/3000\n1/1 [==============================] - 0s 128ms/step - loss: 0.1016 - mape: 150.6893 - val_loss: 0.1100 - val_mape: 172.7265\nEpoch 40/3000\n1/1 [==============================] - 0s 134ms/step - loss: 0.1014 - mape: 150.1641 - val_loss: 0.1099 - val_mape: 172.4155\nEpoch 41/3000\n1/1 [==============================] - 0s 122ms/step - loss: 0.1012 - mape: 149.7415 - val_loss: 0.1098 - val_mape: 172.1102\nEpoch 42/3000\n1/1 [==============================] - 0s 133ms/step - loss: 0.1010 - mape: 149.4445 - val_loss: 0.1097 - val_mape: 171.9901\nEpoch 43/3000\n1/1 [==============================] - 0s 136ms/step - loss: 0.1008 - mape: 149.2704 - val_loss: 0.1097 - val_mape: 172.0403\nEpoch 44/3000\n1/1 [==============================] - 0s 142ms/step - loss: 0.1005 - mape: 149.2081 - val_loss: 0.1096 - val_mape: 172.2218\nEpoch 45/3000\n1/1 [==============================] - 0s 131ms/step - loss: 0.1003 - mape: 149.2369 - val_loss: 0.1097 - val_mape: 172.4887\nEpoch 46/3000\n1/1 [==============================] - 0s 127ms/step - loss: 0.1000 - mape: 149.3239 - val_loss: 0.1097 - val_mape: 172.7881\nEpoch 47/3000\n1/1 [==============================] - 0s 137ms/step - loss: 0.0997 - mape: 149.4369 - val_loss: 0.1098 - val_mape: 173.1031\nEpoch 48/3000\n1/1 [==============================] - 0s 127ms/step - loss: 0.0995 - mape: 149.5578 - val_loss: 0.1099 - val_mape: 173.3943\nEpoch 49/3000\n1/1 [==============================] - 0s 137ms/step - loss: 0.0993 - mape: 149.6599 - val_loss: 0.1100 - val_mape: 173.5821\nEpoch 50/3000\n1/1 [==============================] - 0s 131ms/step - loss: 0.0991 - mape: 149.6891 - val_loss: 0.1100 - val_mape: 173.6694\nEpoch 51/3000\n1/1 [==============================] - 0s 127ms/step - loss: 0.0989 - mape: 149.6448 - val_loss: 0.1100 - val_mape: 173.6328\nEpoch 52/3000\n1/1 [==============================] - 0s 131ms/step - loss: 0.0987 - mape: 149.5096 - val_loss: 0.1099 - val_mape: 173.4537\nEpoch 53/3000\n1/1 [==============================] - 0s 131ms/step - loss: 0.0985 - mape: 149.2693 - val_loss: 0.1098 - val_mape: 173.1484\nEpoch 54/3000\n1/1 [==============================] - 0s 139ms/step - loss: 0.0983 - mape: 148.9340 - val_loss: 0.1096 - val_mape: 172.7385\nEpoch 55/3000\n1/1 [==============================] - 0s 139ms/step - loss: 0.0981 - mape: 148.5188 - val_loss: 0.1093 - val_mape: 172.2540\nEpoch 56/3000\n1/1 [==============================] - 0s 136ms/step - loss: 0.0978 - mape: 148.0448 - val_loss: 0.1091 - val_mape: 171.7453\nEpoch 57/3000\n1/1 [==============================] - 0s 130ms/step - loss: 0.0976 - mape: 147.5498 - val_loss: 0.1089 - val_mape: 171.2401\nEpoch 58/3000\n1/1 [==============================] - 0s 133ms/step - loss: 0.0974 - mape: 147.0524 - val_loss: 0.1087 - val_mape: 170.7427\nEpoch 59/3000\n1/1 [==============================] - 0s 127ms/step - loss: 0.0972 - mape: 146.5561 - val_loss: 0.1086 - val_mape: 170.3112\nEpoch 60/3000\n"
],
[
"model.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nbatch_normalization (BatchNo (None, 9) 36 \n_________________________________________________________________\ndense (Dense) (None, 18) 180 \n_________________________________________________________________\ndense_1 (Dense) (None, 18) 342 \n_________________________________________________________________\ndense_2 (Dense) (None, 1) 19 \n=================================================================\nTotal params: 577\nTrainable params: 559\nNon-trainable params: 18\n_________________________________________________________________\n"
],
[
"import matplotlib.pyplot as plt\n\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\naccuracy = history.history['mape']\nval_accuracy = history.history['val_mape']\n\n\nepochs = range(1, len(loss) + 1)\nfig, ax1 = plt.subplots()\n\nl1 = ax1.plot(epochs, loss, 'bo', label='Training loss')\nvl1 = ax1.plot(epochs, val_loss, 'b', label='Validation loss')\nax1.set_title('Training and validation loss')\nax1.set_xlabel('Epochs')\nax1.set_ylabel('Loss (mae))')\n\nax2 = ax1.twinx()\nac2= ax2.plot(epochs, accuracy, 'o', c=\"red\", label='Training acc')\nvac2= ax2.plot(epochs, val_accuracy, 'r', label='Validation acc')\nax2.set_ylabel('mape')\n\nlns = l1 + vl1 + ac2 + vac2\nlabs = [l.get_label() for l in lns]\nax2.legend(lns, labs, loc=\"center right\")\nfig.tight_layout()\n#fig.savefig(\"acc+loss_drop.pdf\")\nfig.show()",
"<ipython-input-21-d79fc1320319>:29: UserWarning: Matplotlib is currently using module://ipykernel.pylab.backend_inline, which is a non-GUI backend, so cannot show the figure.\n fig.show()\n"
]
],
[
[
"## Probability density distribution",
"_____no_output_____"
]
],
[
[
"y0_low = class_data0_low[:,0]\nA0 = class_data0_low\nA0[:,9] = A0[:,13]\nx0_low = class_data0_low[:,1:10]",
"_____no_output_____"
],
[
"y_pred_low = model.predict(x0_low)",
"_____no_output_____"
],
[
"y0_high = class_data0_high[:,0]\nA0 = class_data0_high\nA0[:,9] = A0[:,13]\nx0_high = class_data0_high[:,1:10]",
"_____no_output_____"
],
[
"y_pred_high = model.predict(x0_high)",
"_____no_output_____"
],
[
"from matplotlib import pyplot\ny0_high = np.array(y0_high)\nbins = np.linspace(0, 0.6, 100)\npyplot.hist(y0_high, bins, color = 'indianred', alpha=0.5, label='Osiris')\npyplot.hist(y_pred_high, bins, color = 'mediumslateblue', alpha=0.5, label='NN')\npyplot.legend(loc='upper right')\npyplot.xlabel('Probability')\npyplot.yscale('log')\npyplot.title('Trained on ($p_e$, $p_{\\gamma}$, $\\omega_e$, $\\omega_{\\gamma}$, n)')\npyplot.show()",
"_____no_output_____"
],
[
"from matplotlib import pyplot\ny0_low = np.array(y0_low)\nbins = np.linspace(0, 0.1, 100)\npyplot.hist(y0_low, bins, color = 'indianred', alpha=0.5, label='Osiris')\npyplot.hist(y_pred_low, bins, color = 'mediumslateblue', alpha=0.5, label='NN')\npyplot.legend(loc='upper right')\npyplot.xlabel('Probability')\n#pyplot.yscale('log')\npyplot.title('Trained on ($p_e$, $p_{\\gamma}$, $\\omega_e$, $\\omega_{\\gamma}$, n)')\npyplot.show()",
"_____no_output_____"
],
[
"from matplotlib import pyplot\ny_low = np.array(y_low)\ny_high = np.array(y_high)\nbins = np.linspace(0, 0.6, 100)\npyplot.hist(y_low, bins, color = 'indianred', alpha=0.5, label='low ene')\npyplot.hist(y0_high, bins, color = 'mediumslateblue', alpha=0.5, label='high ene')\npyplot.legend(loc='upper right')\npyplot.xlabel('Probability')\n#pyplot.yscale('log')\npyplot.title('Data-sets comparison')\npyplot.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecc93bfbdd5e01b004bc0d1c94741b45147e1467 | 10,075 | ipynb | Jupyter Notebook | Secure 'Serverless' File Uploads with AWS Lambda, S3, and Zappa.ipynb | stratospark/zappa-s3-signature | f3a814263ac609aa291cd3a5bfe1327e08ff984d | [
"MIT"
] | 21 | 2017-02-17T04:31:18.000Z | 2021-02-16T20:05:11.000Z | Secure 'Serverless' File Uploads with AWS Lambda, S3, and Zappa.ipynb | stratospark/zappa-s3-signature | f3a814263ac609aa291cd3a5bfe1327e08ff984d | [
"MIT"
] | 3 | 2017-02-17T05:01:19.000Z | 2018-03-21T17:18:05.000Z | Secure 'Serverless' File Uploads with AWS Lambda, S3, and Zappa.ipynb | stratospark/zappa-s3-signature | f3a814263ac609aa291cd3a5bfe1327e08ff984d | [
"MIT"
] | 3 | 2017-08-10T08:04:09.000Z | 2018-09-29T05:06:09.000Z | 37.040441 | 340 | 0.588089 | [
[
[
"# Secure 'Serverless' File Uploads with AWS Lambda, S3, and Zappa",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"As I've been experimenting with [AWS Lambda](https://aws.amazon.com/lambda/), I've found the need to accept file uploads from the browser in order to kick off asynchronous Lambda functions. For example, allowing a user to directly upload in an S3 bucket from the browser, which would trigger a Lambda function for image processing.\n\nI decided to use the [Zappa](https://github.com/Miserlou/Zappa) framework, as it allows me to leverage my existing Python WSGI experience, while also providing a number of **awesome** features such as:\n\n* Access to powerful, prebuilt Python packages such as Numpy and scikit-learn\n* Automatic Let's Encrypt SSL registration and renewal\n* Automatic scheduled job to keep the Lambda function warm\n* Ability to invoke arbitrary Python functions within the Lambda execution environment (great for debugging)\n* Deploy bundles larger than 50 megs through a Slim Handler mechanism",
"_____no_output_____"
],
[
"This walkthrough will cover deploying an SSL-encrypted S3 signature microservice and integrating it with the browser-based [Fine Uploader](http://fineuploader.com/) component. In an upcoming post, I will show how to take the file uploads and process them with an additional Lambda function triggered by new files in an S3 bucket.",
"_____no_output_____"
],
[
"## Deploy Zappa Lambda Function for Signing S3 Requests",
"_____no_output_____"
],
[
"Here are the steps I took to create a secure file upload system in the cloud:\n\n* [Sign up for a domain using Namecheap](https://ap.www.namecheap.com/Profile/Tools/Affiliate)\n* Follow [these instructions](https://github.com/Miserlou/Zappa/blob/master/docs/domain_with_free_ssl_dns.md) to create a Route 53 Hosted Zone, update your domain DNS, and generate a Let's Encrypt account.key\n* Create S3 bucket to hold uploaded files, with the policy below. **Note: do not use periods in the bucket name if you want to be able to use SSL, as [explained here](http://stackoverflow.com/questions/39396634/fine-uploader-upload-to-s3-over-https-error)**",
"_____no_output_____"
],
[
"```\n{\n \"Version\": \"2008-10-17\",\n \"Id\": \"policy\",\n \"Statement\": [\n {\n \"Sid\": \"allow-public-put\",\n \"Effect\": \"Allow\",\n \"Principal\": {\n \"AWS\": \"*\"\n },\n \"Action\": \"s3:PutObject\",\n \"Resource\": \"arn:aws:s3:::BUCKET_NAME_HERE/*\"\n }\n ]\n}\n```",
"_____no_output_____"
],
[
"* Activate CORS for the S3 bucket. You may want to update the AllowedOrigin tag to limit the domains you are allowed to upload from.",
"_____no_output_____"
],
[
"```\n<?xml version=\"1.0\" encoding=\"UTF-8\"?>\n<CORSConfiguration xmlns=\"http://s3.amazonaws.com/doc/2006-03-01/\">\n <CORSRule>\n <AllowedOrigin>*</AllowedOrigin>\n <AllowedMethod>POST</AllowedMethod>\n <AllowedMethod>PUT</AllowedMethod>\n <AllowedMethod>DELETE</AllowedMethod>\n <MaxAgeSeconds>3000</MaxAgeSeconds>\n <ExposeHeader>ETag</ExposeHeader>\n <AllowedHeader>*</AllowedHeader>\n </CORSRule>\n</CORSConfiguration>\n```",
"_____no_output_____"
],
[
"* Optionally update the Lifecycle Rules for that bucket to automatically delete files after a certain period of time.\n* Create a new IAM user specifically to create a new set of keys with limited permissions for your Lambda function:",
"_____no_output_____"
],
[
"```\n{\n \"Version\": \"2012-10-17\",\n \"Statement\": [\n {\n \"Sid\": \"Stmt1486786154000\",\n \"Effect\": \"Allow\",\n \"Action\": [\n \"s3:PutObject\"\n ],\n \"Resource\": [\n \"arn:aws:s3:::BUCKET_NAME_HERE/*\"\n ]\n }\n ]\n}\n```",
"_____no_output_____"
],
[
"* Clone this Zappa project: `git clone https://github.com/stratospark/zappa-s3-signature`\n* Create a virtual environment for this project: `virtualenv myenv`. *Note, conda environments are currently unsupported, so I utilize a Docker container with a standard Python virtualenv*\n* Install packages: `pip install -r requirements.txt`. \n* Create an `s3-signature-config.json` file with the ACCESS_KEY and SECRET_KEY of the new User you created, for example:",
"_____no_output_____"
],
[
"```\n{\n \"ACCESS_KEY\": \"AKIAIHBBHGQSUN34COPA\",\n \"SECRET_KEY\": \"wJalrXUtnFEMI/K7MDENG+bPxRfiCYEXAMPLEKEY\"\n}\n```",
"_____no_output_____"
],
[
"* Upload `s3-signature-config.json` to an S3 bucket accessible by the Lambda function, used in **remote_env** config field\n* Update the *prod* section of `zappa_settings.json` with your **aws_region**, **s3_bucket**, **cors/allowed_origin**, **remote_env**, **domain**, and **lets_encrypt_key**\n* Deploy to AWS Lambda: `zappa deploy prod`\n* Enable SSL through Let's Encrypt: `zappa certify prod`",
"_____no_output_____"
],
[
"## Deploy HTML5/Javascript Fine Uploader Page",
"_____no_output_____"
],
[
"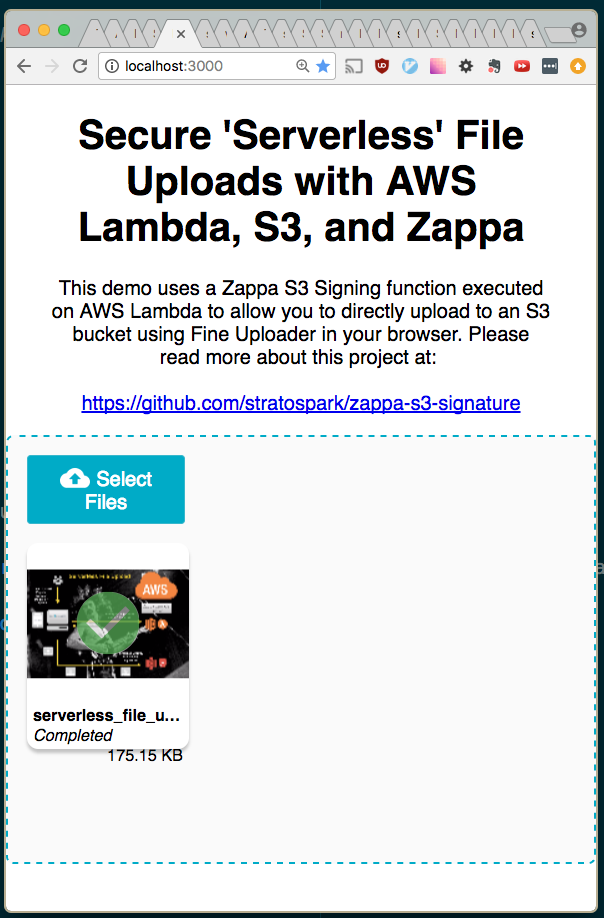",
"_____no_output_____"
],
[
"The following steps will allow you to host a static page that contains the Fine Uploader component. This is a very full-featured open-source component that has excellent S3 support. It also comes with pre-built UI components such as an Image Gallery, to help save time when developing prototypes.\n\nWe have deployed the AWS V4 Signature Lambda function in the previous section in order to take advantage of direct Browser -> S3 uploads.\n\nYou can deploy the HTML and Javascript files onto any server you have access to. However, as we have an opportunity to piggyback on existing AWS infrastructure, including SSL, we can just deploy a static site on S3.",
"_____no_output_____"
],
[
"* Clone the sample React Fine Uploader project: https://github.com/stratospark/react-fineuploader-s3-demo\n* Update the **request/endpoint**, **request/accessKey**, and **signature/endpoint** fields in the FineUploaderS3 constructor in App.js. Optionally update **objectProperties/region**. \n * For example, request/endpoint should be: `https://BUCKET_NAME.s3.amazonaws.com` ...\n **Note: the endpoints must not have trailing slashes or the signatures will not be valid!**\n* Run: ``npm build``. **Note: you need to add a `homepage` field to `package.json` if you will serve the pages at a location other than the root.**\n* Create S3 bucket and upload the contents of the build folder. **Note: once again, do not use periods in the name if you want to use HTTPS/SSL**\n* Make this S3 bucket a publically available static site. Also remember to set a policy like below:",
"_____no_output_____"
],
[
"```\n{\n \"Version\": \"2008-10-17\",\n \"Statement\": [\n {\n \"Sid\": \"PublicReadForGetBucketObjects\",\n \"Effect\": \"Allow\",\n \"Principal\": {\n \"AWS\": \"*\"\n },\n \"Action\": \"s3:GetObject\",\n \"Resource\": \"arn:aws:s3:::BUCKET_NAME/*\"\n }\n ]\n}\n```",
"_____no_output_____"
],
[
"* Access the Fine Uploader demo page in your browser, for example: https://stratospark-serverless-uploader.s3.amazonaws.com/index.html\n* Upload a file\n* Check your public uploads bucket",
"_____no_output_____"
],
[
"That's all!\n\n**Stay tuned for the next installment, where we take these uploaded files and run them through image processing, computer vision, and deep learning Lambda pipelines!**",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecc9452cd1de7170a6567f2046585e17c7a6d337 | 214,277 | ipynb | Jupyter Notebook | NoteBooks/.ipynb_checkpoints/Real Estate NN3-checkpoint.ipynb | kazimsanlav/RealEstate | 3abdb8a4a35b3975b8f6ad4b11b64cd73a97901a | [
"MIT"
] | null | null | null | NoteBooks/.ipynb_checkpoints/Real Estate NN3-checkpoint.ipynb | kazimsanlav/RealEstate | 3abdb8a4a35b3975b8f6ad4b11b64cd73a97901a | [
"MIT"
] | null | null | null | NoteBooks/.ipynb_checkpoints/Real Estate NN3-checkpoint.ipynb | kazimsanlav/RealEstate | 3abdb8a4a35b3975b8f6ad4b11b64cd73a97901a | [
"MIT"
] | 1 | 2020-07-08T18:41:21.000Z | 2020-07-08T18:41:21.000Z | 152.618946 | 20,948 | 0.88331 | [
[
[
"<center> <h1>Neural Network Model</h1> </center>",
"_____no_output_____"
],
[
"## 1. Reading data",
"_____no_output_____"
],
[
"We are reading data that we already prepared. We will look at the first few columns and we will choose 78. district to work on a small subset first. \nFinally we will use one hot encoding for categorical features then split the data for train and test.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport numpy as np\nimport warnings\nwarnings.filterwarnings('ignore')\n%matplotlib inline",
"_____no_output_____"
],
[
"fileName = 'C:/Users/kazIm/Desktop/projects/IE490/input/tubitak_data2_processesed2_outlier.csv'\ndf = pd.read_csv(fileName, sep = ',')",
"_____no_output_____"
],
[
"#preview data\ndf.head()",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 507 entries, 0 to 506\nData columns (total 9 columns):\nbagimsiz_bolum_kat 507 non-null float64\nyuzolcumu 507 non-null float64\nmevcut_alani 507 non-null float64\nadil_piyasa_degeri_yasal_durum 507 non-null float64\ntapunun_yasi 507 non-null float64\narea 507 non-null float64\nduration 507 non-null float64\nmahalle_kod_4673.0 507 non-null int64\nmahalle_kod_4676.0 507 non-null int64\ndtypes: float64(7), int64(2)\nmemory usage: 35.7 KB\n"
],
[
"mahalle = df[\"mahalle_kod\"]",
"_____no_output_____"
],
[
"df['mahalle_kod'].describe()",
"_____no_output_____"
],
[
"#we can drop yasal burut alani as it has almost 1 correlation with mevcut alan\ndf = df.drop('yasal_burut_alani',axis=1)",
"_____no_output_____"
],
[
"mahalle = df['mahalle_kod']",
"_____no_output_____"
]
],
[
[
"### One Hot Encoding for Categorical Variables",
"_____no_output_____"
]
],
[
[
"# df = pd.get_dummies(df, columns=[\"\"])\ndf = pd.get_dummies(df, columns=[\"mahalle_kod\"])",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"# split into input (X) and output (y) variables\nX = df.drop('adil_piyasa_degeri_yasal_durum',axis=1)\nY = df['adil_piyasa_degeri_yasal_durum']",
"_____no_output_____"
],
[
"print(X.shape)\nprint(Y.shape)",
"(507, 8)\n(507,)\n"
]
],
[
[
"## 2. Develop a Baseline Neural Network Model",
"_____no_output_____"
]
],
[
[
"from keras.models import Sequential\nfrom keras.layers import Dense\nfrom keras.callbacks import EarlyStopping\nfrom keras.wrappers.scikit_learn import KerasRegressor\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.model_selection import KFold, StratifiedKFold\nfrom sklearn.preprocessing import StandardScaler, MinMaxScaler\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.pipeline import Pipeline",
"Using TensorFlow backend.\n"
]
],
[
[
"The Keras wrappers require a function as an argument. This function that we must define is responsible for creating the neural network model to be evaluated.\n \nBelow we define the function to create the baseline model to be evaluated. It is a simple model that has a single fully connected hidden layer with the same number of neurons as input attributes (9). The network uses rectifier activation function (relu) for the hidden layer. No activation function is used for the output layer because it is a regression problem and we want to predict numerical values directly without transform.\n \nThe efficient ADAM optimization algorithm is used and a mean squared error loss function is optimized. ",
"_____no_output_____"
]
],
[
[
"seed = 42\nnp.random.seed(seed)\ndims = X.shape[1]\n# define base model\n \ndef baseline_model():\n\n # create model\n model = Sequential()\n model.add(Dense(dims, input_shape=(dims,), init = 'normal', activation='relu'))\n model.add(Dense(9, init = 'normal', activation='relu'))\n model.add(Dense(1, init = 'normal'))\n # compile model\n# sgd = SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)\n model.compile(loss='mse', optimizer = 'adam', metrics=['mape'])\n# model.summary()\n return model\n",
"_____no_output_____"
],
[
"# Function to plot history of model learning \ndef plot_history(network_history):\n \n plt.figure()\n plt.xlabel('Epochs')\n plt.ylabel('Loss')\n plt.plot(network_history.history['loss'])\n plt.plot(network_history.history['val_loss'])\n plt.legend(['Training', 'Validation'])\n\n plt.figure()\n plt.xlabel('Epochs')\n plt.ylabel('MAPE')\n plt.plot(network_history.history['mean_absolute_percentage_error'])\n plt.plot(network_history.history['val_mean_absolute_percentage_error'])\n plt.legend(['Training', 'Validation'])",
"_____no_output_____"
]
],
[
[
"We also initialize the random number generator with a constant random seed, a process we will repeat for each model evaluated in this tutorial. This is an attempt to ensure we compare models consistently.",
"_____no_output_____"
]
],
[
[
"# fix random seed for reproducibility\nnp.random.seed(seed)\n# evaluate model \nearly_stop = EarlyStopping(monitor='val_mean_absolute_percentage_error', patience=50, verbose=0)\nestimator = KerasRegressor(build_fn=baseline_model, epochs=1500, batch_size=128, verbose=0,\n callbacks=[early_stop])",
"_____no_output_____"
]
],
[
[
"**Train and Validation Split**",
"_____no_output_____"
]
],
[
[
"Xtrain, Xtest, Ytrain, Ytest = train_test_split( X, Y, test_size=0.20, random_state=42)",
"_____no_output_____"
],
[
"history = estimator.fit(Xtrain, Ytrain, validation_data=(Xtest, Ytest))",
"_____no_output_____"
],
[
"plot_history(history)",
"_____no_output_____"
],
[
"from sklearn.metrics import make_scorer\n\ndef mean_absolute_percentage_error(y_true, y_pred): \n y_true, y_pred = np.array(y_true), np.array(y_pred)\n try:\n return np.mean(np.abs((y_true - y_pred) / np.abs(y_true)) * 100)\n except:\n return np.mean((np.abs(y_true - y_pred)+0.001 ) / (np.abs(y_true)+0.001) * 100)\n\ndef absolute_twenty_percent_error_quartile(y_true, y_pred): \n \n pred = y_pred.reshape(-1,1) \n actual = y_true.reshape(-1,1)\n error = np.abs(pred-actual)/np.abs(actual)\n return (error[error<=0.2].shape[0]/actual.shape[0])*100\n \nmape = make_scorer(mean_absolute_percentage_error, greater_is_better=False)\ntwentyPercentErrorQuantile = make_scorer(absolute_twenty_percent_error_quartile, greater_is_better=True)",
"_____no_output_____"
]
],
[
[
"**All sample MAPE error and 20% error quartile score**",
"_____no_output_____"
]
],
[
[
"mean_absolute_percentage_error(estimator.predict(X),Y)",
"_____no_output_____"
],
[
"absolute_twenty_percent_error_quartile(estimator.predict(X),Y)",
"_____no_output_____"
]
],
[
[
"**Out of Sample sample MAPE error and 20% error quartile score**",
"_____no_output_____"
]
],
[
[
"mean_absolute_percentage_error(estimator.predict(Xtest),Ytest)",
"_____no_output_____"
],
[
"absolute_twenty_percent_error_quartile(estimator.predict(Xtest),Ytest)",
"_____no_output_____"
]
],
[
[
"** Cross Validation**",
"_____no_output_____"
],
[
"The final step is to evaluate this baseline model. We will use 3-fold cross validation to evaluate the model.",
"_____no_output_____"
],
[
"As it is not implemented before hand, we will write our Mean Absolute Percentage Error function and 20% error quartile function to use in `cross_val_score function` of scikit-learn library. ",
"_____no_output_____"
]
],
[
[
"kfold = StratifiedKFold(n_splits=3, random_state=seed)\nestimator2 = KerasRegressor(build_fn=baseline_model, epochs=1200, batch_size=128, verbose=0)\nresults = np.abs(cross_val_score(estimator2, X, Y, cv=kfold, scoring = mape))\nprint(results)\nprint()\nprint(\"Cross Validation Mean Absolute Percentage error: %.2f \" % (results.mean()))",
"[ 14.8476281 15.54637567 69.13683218]\n\nCross Validation Mean Absolute Percentage error: 33.18 \n"
],
[
"# kfold = StratifiedKFold(n_splits=3, random_state=seed)\n# results = np.abs(cross_val_score(estimator, X, Y, cv=kfold, scoring = twentyPercentErrorQuantile))\n# print(results)\n# print()\n# print(\"Cross Validation 20 percent error quartile: %.2f \" % (results.mean()))",
"_____no_output_____"
]
],
[
[
"## 3. Modeling The Standardized Dataset",
"_____no_output_____"
],
[
"An important concern with our house price dataset is that the input attributes all vary in their scales because they measure different quantities. \n\nIt is almost always good practice to prepare our data before modeling it using a neural network model for optimization algorithms' efficiency. \n\nContinuing on from the above baseline model, we can re-evaluate the same model using a scaled version (0-1) of the input dataset. \n\n** It is important that we should not use the test data for scaling process other wise it leads to information leakage from testset**\n",
"_____no_output_____"
]
],
[
[
"\n# evaluate model with standardized dataset\nfrom sklearn.preprocessing import StandardScaler\n\nscaler = StandardScaler().fit(X)\nX_ = scaler.transform(X)\nY_ = Y/1000\n",
"_____no_output_____"
],
[
"Xtrain, Xtest, Ytrain, Ytest = train_test_split( X_, Y_, test_size=0.20, random_state=42)",
"_____no_output_____"
],
[
"history = estimator.fit(Xtrain, Ytrain, validation_data=(Xtest, Ytest))",
"_____no_output_____"
],
[
"plot_history(history)",
"_____no_output_____"
]
],
[
[
"**All sample MAPE error and 20% error quartile score**",
"_____no_output_____"
]
],
[
[
"mean_absolute_percentage_error(estimator.predict(X_),Y_)",
"_____no_output_____"
],
[
"absolute_twenty_percent_error_quartile(estimator.predict(X_),Y_)",
"_____no_output_____"
]
],
[
[
"**Out of Sample sample MAPE error and 20% error quartile score**",
"_____no_output_____"
]
],
[
[
"mean_absolute_percentage_error(estimator.predict(Xtest),Ytest)",
"_____no_output_____"
],
[
"absolute_twenty_percent_error_quartile(estimator.predict(Xtest),Ytest)",
"_____no_output_____"
]
],
[
[
"A further extension would be to similarly apply a rescaling to the output variable such as normalizing it to the range of 0-1 and use a Sigmoid or similar activation function on the output layer to narrow output predictions to the same range.",
"_____no_output_____"
],
[
"## 4. Tune The Neural Network Topology",
"_____no_output_____"
],
[
"There are many concerns that can be optimized for a neural network model.\nPerhaps the most important one is the structure of the network itself, including the number of layers and the number of neurons in each layer. \n\nIn this section, we will evaluate two additional network topologies in an effort to further improve the performance of the model. We will look at both a deeper and a wider network topology.",
"_____no_output_____"
],
[
"### 4.1. Evaluate a Deeper Network Topology",
"_____no_output_____"
],
[
"One way to improve the performance a neural network is to add more layers. This might allow the model to extract and recombine higher order features embedded in the data. \n\nIn this section we will evaluate the effect of adding one more hidden layer to the model. This new layer will have about half the number of neurons. Because there are heuristics that suggest that number of neurons in the next layer should not exceed the half of the previuos layer's number of neurons.",
"_____no_output_____"
]
],
[
[
"# define the model\ndef larger_model():\n # create model\n model = Sequential()\n model.add(Dense(dims, input_shape=(dims,), init = 'normal', activation='relu'))\n model.add(Dense(9, init = 'normal', activation='relu'))\n model.add(Dense(4, kernel_initializer='normal', activation='relu'))\n model.add(Dense(1, init = 'normal'))\n # compile model\n# sgd = SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)\n model.compile(loss='mse', optimizer = 'adam', metrics=['mape'])\n# model.summary()\n return model",
"_____no_output_____"
]
],
[
[
"We will follow the same methodology as above;",
"_____no_output_____"
]
],
[
[
"# fix random seed for reproducibility\nnp.random.seed(seed)\n# evaluate model \nestimator = KerasRegressor(build_fn=larger_model, epochs=1500, batch_size=128, verbose=0,\n callbacks=[early_stop])",
"_____no_output_____"
],
[
"history = estimator.fit(Xtrain, Ytrain, validation_data=(Xtest, Ytest))",
"_____no_output_____"
],
[
"plot_history(history)",
"_____no_output_____"
]
],
[
[
"**Out of Sample sample MAPE error and 20% error quartile score**",
"_____no_output_____"
]
],
[
[
"mean_absolute_percentage_error(estimator.predict(Xtest),Ytest)",
"_____no_output_____"
],
[
"absolute_twenty_percent_error_quartile(estimator.predict(Xtest),Ytest)",
"_____no_output_____"
]
],
[
[
"### 4.2. Evaluate a Wider Network Topology",
"_____no_output_____"
],
[
"Another approach to increasing the representational capability of the model is to create a wider network. \nIn this section we evaluate the effect of keeping a shallow network architecture and nearly doubling the number of neurons in the one hidden layer. \nHere, we have increased the number of neurons in the hidden layer compared to the baseline model from 9 to 15.",
"_____no_output_____"
]
],
[
[
"# define wider model\ndef wider_model():\n # create model\n model = Sequential()\n model.add(Dense(dims, input_shape=(dims,), kernel_initializer='normal', activation='relu'))\n model.add(Dense(1, kernel_initializer='normal'))\n # compile model\n# sgd = SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)\n model.compile(loss='mse', optimizer = 'adam', metrics=['mape'])\n# model.summary()\n return model",
"_____no_output_____"
]
],
[
[
"We will follow the same methodology as above;",
"_____no_output_____"
]
],
[
[
"# fix random seed for reproducibility\nnp.random.seed(seed)\n# evaluate model \nestimator = KerasRegressor(build_fn=wider_model, epochs=1500, batch_size=128, verbose=0,\n callbacks=[early_stop])",
"_____no_output_____"
],
[
"history = estimator.fit(Xtrain, Ytrain, validation_data=(Xtest, Ytest))",
"_____no_output_____"
],
[
"plot_history(history)",
"_____no_output_____"
]
],
[
[
"**Out of Sample sample MAPE error and 20% error quartile score**",
"_____no_output_____"
]
],
[
[
"mean_absolute_percentage_error(estimator.predict(Xtest),Ytest)",
"_____no_output_____"
],
[
"absolute_twenty_percent_error_quartile(estimator.predict(Xtest),Ytest)",
"_____no_output_____"
]
],
[
[
"** We saw that deeper model is the best of the three.**",
"_____no_output_____"
],
[
"We may want to add another layer",
"_____no_output_____"
]
],
[
[
"# define the model\ndef larger_model2():\n # create model\n model = Sequential()\n model.add(Dense(dims, input_shape=(dims,), init = 'normal', activation='relu'))\n model.add(Dense(10, init = 'normal', activation='relu'))\n model.add(Dense(5, kernel_initializer='normal', activation='relu'))\n model.add(Dense(3, kernel_initializer='normal', activation='relu'))\n model.add(Dense(1, init = 'normal'))\n # compile model\n# sgd = SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)\n model.compile(loss='mse', optimizer = 'adam', metrics=['mape'])\n# model.summary()\n return model",
"_____no_output_____"
]
],
[
[
"We will follow the same methodology as above;",
"_____no_output_____"
]
],
[
[
"# fix random seed for reproducibility\nnp.random.seed(seed)\n# evaluate model \nearly_stop = EarlyStopping(monitor='val_mean_absolute_percentage_error', patience=100, verbose=0)\nestimator = KerasRegressor(build_fn=larger_model2, epochs=1500, batch_size=128, verbose=0,\n callbacks=[early_stop])",
"_____no_output_____"
],
[
"history = estimator.fit(Xtrain, Ytrain, validation_data=(Xtest, Ytest))",
"_____no_output_____"
],
[
"plot_history(history)",
"_____no_output_____"
]
],
[
[
"**Out of Sample sample MAPE error and 20% error quartile score**",
"_____no_output_____"
]
],
[
[
"mean_absolute_percentage_error(estimator.predict(Xtest),Ytest)",
"_____no_output_____"
],
[
"absolute_twenty_percent_error_quartile(estimator.predict(Xtest),Ytest)",
"_____no_output_____"
]
],
[
[
"** That did not improve our model ** ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
ecc94d14878dfb4421602fe1b80c1668ebdb5348 | 8,437 | ipynb | Jupyter Notebook | ballet/templates/project_template/{{cookiecutter.project_slug}}/notebooks/Analysis.ipynb | HDI-Project/fhub_core | 9667a47fbd8b4caf2e92118dc5357f34aae2098b | [
"MIT"
] | 19 | 2021-04-06T18:56:39.000Z | 2022-03-15T00:23:00.000Z | ballet/templates/project_template/{{cookiecutter.project_slug}}/notebooks/Analysis.ipynb | HDI-Project/ballet | 9667a47fbd8b4caf2e92118dc5357f34aae2098b | [
"MIT"
] | 52 | 2018-09-27T01:11:58.000Z | 2021-03-24T19:11:18.000Z | ballet/templates/project_template/{{cookiecutter.project_slug}}/notebooks/Analysis.ipynb | HDI-Project/ballet | 9667a47fbd8b4caf2e92118dc5357f34aae2098b | [
"MIT"
] | 3 | 2019-12-07T17:55:34.000Z | 2021-02-02T17:58:39.000Z | 34.157895 | 853 | 0.645372 | [
[
[
"# {{ cookiecutter.project_slug }} analysis",
"_____no_output_____"
],
[
"Welcome to the collaboration!\n\nSome of the steps you follow in this analysis are taken from the [Ballet Contributor Guide](https://ballet.github.io/ballet/contributor_guide.html), make sure to consult it for more information.",
"_____no_output_____"
]
],
[
[
"# some preliminaries...\nfrom ballet.util.log import enable as enable_logger\nenable_logger('ballet')\nenable_logger('{{ cookiecutter.package_slug }}')\nimport pandas as pd\npd.options.display.max_columns = None\npd.options.display.max_colwidth = None\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set()",
"_____no_output_____"
],
[
"# automagical client\nfrom ballet import b",
"_____no_output_____"
]
],
[
[
"## Explore the data",
"_____no_output_____"
]
],
[
[
"X_df, y_df = b.api.load_data()",
"_____no_output_____"
],
[
"X_df.head()",
"_____no_output_____"
],
[
"y_df.head()",
"_____no_output_____"
]
],
[
[
"## Explore existing features\n\nTo discover existing features, you can use the `discover` method of the `b` client, which returns information and summary statistics about each existing feature in a data frame. See its docstring for information on filtering features by different criteria.",
"_____no_output_____"
]
],
[
[
"discovery_df = b.discover()\ndiscovery_df",
"_____no_output_____"
]
],
[
[
"You can also use the `engineer_features` method of {{ cookiecutter.package_slug }}'s API, which is also exposed by `b`. The resulting object is a named tuple that allows you to access the transformed development dataset (feature matrix `X` and target `y`), the feature engineering pipeline (`pipeline`), the target encoder (`encoder`), and the set of existing features (`features`).",
"_____no_output_____"
]
],
[
[
"result = b.api.engineer_features(X_df, y_df)\nX_train, y_train = result.X, result.y\nprint('Number of existing features: ', len(result.features))\nprint('Number of columns in feature matrix: ', X_train.shape[1])",
"_____no_output_____"
]
],
[
[
"## Write a new feature",
"_____no_output_____"
],
[
"Now it's time to write your own feature!\n\n🚧 The content of the cell must be a standalone Python module, as it will be placed in an empty Python source file when it is submitted. This means that any imports or helper functions must be defined (or re-defined) within this cell, otherwise your submitted feature will fail to validate due to missing imports/helpers. 🚧 For example, if you use some functionality from numpy, then you must (re)import numpy within the code cell, even if you imported it elsewhere in the notebook.\n\nIf you have questions about feature engineering, see the [Feature Engineering Guide](https://ballet.github.io/ballet/feature_engineering_guide.html).",
"_____no_output_____"
]
],
[
[
"from ballet import Feature\n\ninput = None # TODO - str or list of str\ntransformer = None # TODO - function, transformer-like, or list thereof\nname = None # TODO - str\nfeature = Feature(input, transformer, name=name)",
"_____no_output_____"
]
],
[
[
"Let's play around with the feature we have created... what values does it extract?",
"_____no_output_____"
]
],
[
[
"feature_values = feature.as_feature_engineering_pipeline().fit_transform(X_df, y_df)\nfeature_values",
"_____no_output_____"
]
],
[
[
"## Test your feature\n\nYou probably want to make sure that your feature does not have bugs before you submit it to the upstream project.",
"_____no_output_____"
],
[
"This first command will check that your feature conforms to the feature API. (Assumes you have assigned your feature to a variable `feature`.) Even if your feature passes the tests here in the notebook, it may still fail to be validated, especially if you have not included all your imports/helpers in the code cell that you submit.",
"_____no_output_____"
]
],
[
[
"b.validate_feature_api(feature)",
"_____no_output_____"
]
],
[
[
"This second command will evaluate the ML performance of your feature. (Assumes you have assigned your feature to a variable feature.) Usually, this consists of computing the mutual information of your feature values with the target, conditional on the feature values extracted by the existing feature engineering pipeline. If this is above a certain threshold, your feature is determined to be contributing positively to the ML performance of the project. In this Demo, since we want to be nice to new collaborators, each and every feature will be accepted with respect to ML performance!",
"_____no_output_____"
]
],
[
[
"b.validate_feature_acceptance(feature)",
"_____no_output_____"
]
],
[
[
"## Submit your feature\n\nWhen you are ready to submit your feature, look for the \"Submit\" button in the right of your notebook toolbar. First, select the code cell that contains the feature you have written (you will have to scroll up). Then press the submit button, confirming that the feature code shown is what you want to submit. After submission, you will be shown a URL that takes you to the corresponding Pull Request that has been created.\n\nWhen you submit your feature, the source code you have written is extracted from the selected code cell. It is then sent to a app which does the job of structuring your feature as a pull request to the {{ cookiecutter.github_owner }}/{{ cookiecutter.project_slug }} repo, which it opens on your behalf. The pull request is validated by Travis CI, a continuous integration service, using the test suite defined by the ballet framework, testing the feature's API and running a streaming logical feature selection (SLFS) algorithm. If the feature is accepted, the GitHub app ballet-bot responds by automatically merging the pull request into the project. Otherwise, it automatically closes it. Similarly, after an accepted feature is merged, the CI service runs another ballet-defined script to prune redundant features using the same SLFS algorithm.",
"_____no_output_____"
],
[
"## Thanks for contributing\n\nIf you're reading this, you have already done some great data science work and contributed one (or more!) features to the upstream {{ cookiecutter.project_slug }} project.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecc953c3bce8f347ebb281820632a0d084d803e8 | 18,406 | ipynb | Jupyter Notebook | public/NLP/TextAnalysis.ipynb | sochan/CSIROAnnotatingText | 2c79445fa90297e1abf28581913d95a327ec611f | [
"Apache-2.0"
] | 1 | 2020-05-06T15:22:03.000Z | 2020-05-06T15:22:03.000Z | public/NLP/TextAnalysis.ipynb | sochan/CSIROAnnotatingText | 2c79445fa90297e1abf28581913d95a327ec611f | [
"Apache-2.0"
] | null | null | null | public/NLP/TextAnalysis.ipynb | sochan/CSIROAnnotatingText | 2c79445fa90297e1abf28581913d95a327ec611f | [
"Apache-2.0"
] | null | null | null | 45.112745 | 1,578 | 0.549332 | [
[
[
"import nltk\nnltk.download()",
"showing info https://raw.githubusercontent.com/nltk/nltk_data/gh-pages/index.xml\n"
],
[
"import nltk\nfrom nltk.corpus import stopwords\nset(stopwords.words('english'))",
"_____no_output_____"
],
[
"from nltk.corpus import stopwords \nfrom nltk.tokenize import word_tokenize \nfrom collections import Counter\n \nexample_sent = \"This is a sample sentence, showing off the stop words filtration.\"\nword_tokens = word_tokenize(example_sent)\n#s = open(\"C:\\sample.txt\").read()\n#word_tokens = word_tokenize(s) \n\n#stop_words = open(\"stopwords.txt\").read()\nstop_words = set(stopwords.words('english')) \n \nfiltered_sentence = [w for w in word_tokens if not w in stop_words] \n \nfiltered_sentence = [] \n\nfor w in word_tokens: \n if w not in stop_words and len(w)>2: \n filtered_sentence.append(w) \n\n\nprint(word_tokens) ",
"_____no_output_____"
],
[
"print(filtered_sentence)",
"_____no_output_____"
],
[
"counts = Counter(filtered_sentence)\nprint(counts)\n",
"Counter({'security': 3, 'risks': 2, 'assessment': 2, 'methods': 2, 'Security': 1, 'systems': 1, 'escalated': 1, 'past': 1, 'years': 1, 'due': 1, 'advancement': 1, 'information': 1, 'technology': 1, 'rapid': 1, 'increase': 1, 'number': 1, 'business': 1, 'reliance': 1, 'online': 1, 'platform': 1, 'outsourcing': 1, 'mitigate': 1, 'adequate': 1, 'planning': 1, 'address': 1, 'modern': 1, 'advanced': 1, 'threats': 1, 'replace': 1, 'efficient': 1, 'reliable': 1})\n"
],
[
"uni_words = set(filtered_sentence)\nfor word in uni_words:\n print(word)\n ",
"years\nadvanced\nincrease\noutsourcing\nreliable\npast\nsystems\nescalated\ninformation\ndue\nmitigate\nplatform\nmodern\naddress\nthreats\nassessment\nadvancement\nSecurity\nonline\nmethods\nadequate\ntechnology\nreliance\nreplace\nefficient\nplanning\nsecurity\nrisks\nrapid\nnumber\nbusiness\n"
],
[
"import json\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecc95aeb737d4dba0d2a71d465e46cde6278ad31 | 226,921 | ipynb | Jupyter Notebook | Code/8_1_Robust1_Remove_Deregulation.ipynb | zhoudanxie/sinclair-xie-sentiment | d53d7cea724b32ea69e9c47e8a3b7cec800c7d07 | [
"MIT"
] | null | null | null | Code/8_1_Robust1_Remove_Deregulation.ipynb | zhoudanxie/sinclair-xie-sentiment | d53d7cea724b32ea69e9c47e8a3b7cec800c7d07 | [
"MIT"
] | null | null | null | Code/8_1_Robust1_Remove_Deregulation.ipynb | zhoudanxie/sinclair-xie-sentiment | d53d7cea724b32ea69e9c47e8a3b7cec800c7d07 | [
"MIT"
] | null | null | null | 92.772281 | 149 | 0.29928 | [
[
[
"import numpy as np\nimport pandas as pd\nimport statsmodels.formula.api as sm\nimport re",
"_____no_output_____"
]
],
[
[
"## 1. Identify articles containing \"deregulat*\"",
"_____no_output_____"
]
],
[
[
"# Import sentiment scores for reg relevant articles\ndf=pd.read_csv('/home/ec2-user/SageMaker/New Uncertainty/Sentiment Analysis/RegRelevant_ArticleSentimentScores.csv')\nprint(df.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 493418 entries, 0 to 493417\nData columns (total 7 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ID 493418 non-null int64 \n 1 StartDate 493418 non-null object \n 2 Newspaper 493418 non-null object \n 3 UncertaintyScore 493418 non-null float64\n 4 GIscore 493418 non-null float64\n 5 LMscore 493418 non-null float64\n 6 LSDscore 493418 non-null float64\ndtypes: float64(4), int64(1), object(2)\nmemory usage: 26.4+ MB\nNone\n"
],
[
"df_regSentsExpand=pd.read_pickle('/home/ec2-user/SageMaker/New Uncertainty/Reg Relevance/RegSentsExpand_NounChunks3.pkl')\nprint(df_regSentsExpand.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 822737 entries, 0 to 822736\nData columns (total 6 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ID 822737 non-null object\n 1 RegSentsExpand 822737 non-null object\n 2 NounChunksMatch 822737 non-null int64 \n 3 NounChunkMatchWords 822737 non-null object\n 4 NounChunkMatchWordsFiltered 822737 non-null object\n 5 NounChunkMatchFiltered 822737 non-null int64 \ndtypes: int64(2), object(4)\nmemory usage: 37.7+ MB\nNone\n"
],
[
"df_regSentsExpand['ID']=df_regSentsExpand['ID'].astype('int64')",
"_____no_output_____"
],
[
"df=df.merge(df_regSentsExpand[['ID','RegSentsExpand']],on='ID',how='left')\nprint(df.info())",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 493418 entries, 0 to 493417\nData columns (total 8 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ID 493418 non-null int64 \n 1 StartDate 493418 non-null object \n 2 Newspaper 493418 non-null object \n 3 UncertaintyScore 493418 non-null float64\n 4 GIscore 493418 non-null float64\n 5 LMscore 493418 non-null float64\n 6 LSDscore 493418 non-null float64\n 7 RegSentsExpand 493418 non-null object \ndtypes: float64(4), int64(1), object(3)\nmemory usage: 33.9+ MB\nNone\n"
],
[
"# Identify reg sections with \"deregulat*\"\ndf['Dereg']=0\nfor i in range(0,len(df)):\n if len(re.findall(r'\\bderegulat[a-zA-Z]+\\b',df['RegSentsExpand'][i],flags=re.IGNORECASE))>0:\n df['Dereg'][i]=1\nprint(df['Dereg'].value_counts()) ",
"/home/ec2-user/SageMaker/.conda/envs/my_py/lib/python3.6/site-packages/ipykernel_launcher.py:5: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"\n"
],
[
"df_dereg=df[df['Dereg']==1].sort_values(['StartDate','Newspaper']).reset_index(drop=True)\nprint(df_dereg[['StartDate','Newspaper']])",
" StartDate Newspaper\n0 1985-01-01 Boston Globe\n1 1985-01-01 Boston Globe\n2 1985-01-01 Chicago Tribune\n3 1985-01-01 New York Times\n4 1985-01-02 New York Times\n... ... ...\n26062 2020-08-29 Wall Street Journal\n26063 2020-08-30 The Washington Post\n26064 2020-08-30 The Washington Post\n26065 2020-08-30 Wall Street Journal\n26066 2020-08-31 Wall Street Journal\n\n[26067 rows x 2 columns]\n"
]
],
[
[
"## 2. Clean data",
"_____no_output_____"
]
],
[
[
"df=df[df['Dereg']!=1].reset_index(drop=True)\nprint(df.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 467351 entries, 0 to 467350\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ID 467351 non-null int64 \n 1 StartDate 467351 non-null object \n 2 Newspaper 467351 non-null object \n 3 UncertaintyScore 467351 non-null float64\n 4 GIscore 467351 non-null float64\n 5 LMscore 467351 non-null float64\n 6 LSDscore 467351 non-null float64\n 7 RegSentsExpand 467351 non-null object \n 8 Dereg 467351 non-null int64 \ndtypes: float64(4), int64(2), object(3)\nmemory usage: 32.1+ MB\nNone\n"
],
[
"# Change data format\ndf['StartDate']=df['StartDate'].astype('datetime64[ns]')\ndf['Year']=df['StartDate'].dt.year\ndf['Month']=df['StartDate'].dt.month\ndf['Newspaper']=df['Newspaper'].astype('category')\nprint(df.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 467351 entries, 0 to 467350\nData columns (total 11 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ID 467351 non-null int64 \n 1 StartDate 467351 non-null datetime64[ns]\n 2 Newspaper 467351 non-null category \n 3 UncertaintyScore 467351 non-null float64 \n 4 GIscore 467351 non-null float64 \n 5 LMscore 467351 non-null float64 \n 6 LSDscore 467351 non-null float64 \n 7 RegSentsExpand 467351 non-null object \n 8 Dereg 467351 non-null int64 \n 9 Year 467351 non-null int64 \n 10 Month 467351 non-null int64 \ndtypes: category(1), datetime64[ns](1), float64(4), int64(4), object(1)\nmemory usage: 36.1+ MB\nNone\n"
],
[
"# Unique year-month dataframe\ndf_ym=df[['Year','Month']].drop_duplicates().sort_values(['Year','Month']).reset_index(drop=True).reset_index()\ndf_ym['YM']=df_ym['index']+1\ndf_ym=df_ym.drop('index',axis=1)\nprint(df_ym)",
" Year Month YM\n0 1985 1 1\n1 1985 2 2\n2 1985 3 3\n3 1985 4 4\n4 1985 5 5\n.. ... ... ...\n423 2020 4 424\n424 2020 5 425\n425 2020 6 426\n426 2020 7 427\n427 2020 8 428\n\n[428 rows x 3 columns]\n"
],
[
"df=df.merge(df_ym[['Year','Month','YM']],on=['Year','Month'],how='left').sort_values(['Year','Month']).reset_index(drop=True)\nprint(df.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 467351 entries, 0 to 467350\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ID 467351 non-null int64 \n 1 StartDate 467351 non-null datetime64[ns]\n 2 Newspaper 467351 non-null category \n 3 UncertaintyScore 467351 non-null float64 \n 4 GIscore 467351 non-null float64 \n 5 LMscore 467351 non-null float64 \n 6 LSDscore 467351 non-null float64 \n 7 RegSentsExpand 467351 non-null object \n 8 Dereg 467351 non-null int64 \n 9 Year 467351 non-null int64 \n 10 Month 467351 non-null int64 \n 11 YM 467351 non-null int64 \ndtypes: category(1), datetime64[ns](1), float64(4), int64(5), object(1)\nmemory usage: 39.7+ MB\nNone\n"
],
[
"print(df[['ID','UncertaintyScore','GIscore','LMscore','LSDscore']].head())",
" ID UncertaintyScore GIscore LMscore LSDscore\n0 292028440 0.000000 0.000000 -4.651163 -2.325581\n1 290815623 0.543478 4.347826 -1.630435 4.347826\n2 292029822 2.542373 1.694915 -0.847458 0.847458\n3 294225539 0.000000 6.140351 -0.877193 0.000000\n4 290808213 0.980392 -0.980392 -1.960784 -0.980392\n"
]
],
[
[
"## 3. Estimate sentiment and uncertainty indexes",
"_____no_output_____"
]
],
[
[
"# Function to estimate index (suppressing constant)\ndef estimate_index(var_name):\n FE_OLS=sm.ols(formula=var_name + ' ~ 0+C(YM)+C(Newspaper)',\n data=df).fit()\n print(FE_OLS.summary())\n\n FE_estimates=pd.DataFrame()\n FE_estimates[var_name+'Index']=FE_OLS.params[0:len(df_ym)]\n FE_estimates=FE_estimates.reset_index().rename(columns={'index':'FE'})\n FE_estimates['YM']=FE_estimates['FE'].str.split(\"[\",expand=True)[1].str.split(\"]\",expand=True)[0].astype('int64')\n \n return FE_estimates",
"_____no_output_____"
],
[
"# Uncertainty index\nUncertaintyIndex=estimate_index('UncertaintyScore')",
" OLS Regression Results \n==============================================================================\nDep. Variable: UncertaintyScore R-squared: 0.005\nModel: OLS Adj. R-squared: 0.004\nMethod: Least Squares F-statistic: 5.571\nDate: Tue, 18 May 2021 Prob (F-statistic): 2.38e-270\nTime: 19:39:27 Log-Likelihood: -6.3961e+05\nNo. Observations: 467351 AIC: 1.280e+06\nDf Residuals: 466917 BIC: 1.285e+06\nDf Model: 433 \nCovariance Type: nonrobust \n=======================================================================================================\n coef std err t P>|t| [0.025 0.975]\n-------------------------------------------------------------------------------------------------------\nC(YM)[1] 0.5934 0.035 16.772 0.000 0.524 0.663\nC(YM)[2] 0.5657 0.036 15.559 0.000 0.494 0.637\nC(YM)[3] 0.6157 0.032 19.048 0.000 0.552 0.679\nC(YM)[4] 0.6510 0.033 19.899 0.000 0.587 0.715\nC(YM)[5] 0.6524 0.032 20.222 0.000 0.589 0.716\nC(YM)[6] 0.6593 0.033 19.928 0.000 0.594 0.724\nC(YM)[7] 0.6407 0.034 18.714 0.000 0.574 0.708\nC(YM)[8] 0.6307 0.033 19.302 0.000 0.567 0.695\nC(YM)[9] 0.7112 0.034 20.718 0.000 0.644 0.779\nC(YM)[10] 0.6925 0.033 20.933 0.000 0.628 0.757\nC(YM)[11] 0.6679 0.035 18.917 0.000 0.599 0.737\nC(YM)[12] 0.6507 0.034 19.354 0.000 0.585 0.717\nC(YM)[13] 0.6745 0.032 20.976 0.000 0.611 0.738\nC(YM)[14] 0.6496 0.036 17.984 0.000 0.579 0.720\nC(YM)[15] 0.6592 0.034 19.178 0.000 0.592 0.727\nC(YM)[16] 0.5998 0.033 18.162 0.000 0.535 0.665\nC(YM)[17] 0.7091 0.033 21.652 0.000 0.645 0.773\nC(YM)[18] 0.7367 0.033 22.504 0.000 0.673 0.801\nC(YM)[19] 0.6340 0.034 18.794 0.000 0.568 0.700\nC(YM)[20] 0.6709 0.034 19.846 0.000 0.605 0.737\nC(YM)[21] 0.7596 0.034 22.124 0.000 0.692 0.827\nC(YM)[22] 0.6547 0.033 19.571 0.000 0.589 0.720\nC(YM)[23] 0.6584 0.033 19.836 0.000 0.593 0.723\nC(YM)[24] 0.6968 0.032 21.666 0.000 0.634 0.760\nC(YM)[25] 0.6868 0.032 21.515 0.000 0.624 0.749\nC(YM)[26] 0.7013 0.031 22.450 0.000 0.640 0.763\nC(YM)[27] 0.6773 0.029 23.324 0.000 0.620 0.734\nC(YM)[28] 0.6751 0.029 23.282 0.000 0.618 0.732\nC(YM)[29] 0.6790 0.031 22.219 0.000 0.619 0.739\nC(YM)[30] 0.6903 0.029 23.903 0.000 0.634 0.747\nC(YM)[31] 0.6758 0.029 23.589 0.000 0.620 0.732\nC(YM)[32] 0.6556 0.030 21.652 0.000 0.596 0.715\nC(YM)[33] 0.7240 0.031 23.715 0.000 0.664 0.784\nC(YM)[34] 0.7146 0.029 24.694 0.000 0.658 0.771\nC(YM)[35] 0.7271 0.030 24.099 0.000 0.668 0.786\nC(YM)[36] 0.6757 0.031 21.522 0.000 0.614 0.737\nC(YM)[37] 0.7415 0.029 25.600 0.000 0.685 0.798\nC(YM)[38] 0.7269 0.029 25.357 0.000 0.671 0.783\nC(YM)[39] 0.6522 0.029 22.597 0.000 0.596 0.709\nC(YM)[40] 0.6821 0.030 23.121 0.000 0.624 0.740\nC(YM)[41] 0.7564 0.029 26.432 0.000 0.700 0.812\nC(YM)[42] 0.6299 0.028 22.782 0.000 0.576 0.684\nC(YM)[43] 0.6566 0.030 21.895 0.000 0.598 0.715\nC(YM)[44] 0.6423 0.029 22.492 0.000 0.586 0.698\nC(YM)[45] 0.6332 0.029 22.119 0.000 0.577 0.689\nC(YM)[46] 0.6800 0.028 24.463 0.000 0.626 0.735\nC(YM)[47] 0.6977 0.029 24.373 0.000 0.642 0.754\nC(YM)[48] 0.6591 0.027 24.723 0.000 0.607 0.711\nC(YM)[49] 0.7426 0.028 26.836 0.000 0.688 0.797\nC(YM)[50] 0.7071 0.028 24.876 0.000 0.651 0.763\nC(YM)[51] 0.6533 0.026 25.228 0.000 0.603 0.704\nC(YM)[52] 0.7020 0.027 26.173 0.000 0.649 0.755\nC(YM)[53] 0.6421 0.027 23.820 0.000 0.589 0.695\nC(YM)[54] 0.6945 0.027 25.571 0.000 0.641 0.748\nC(YM)[55] 0.6480 0.028 23.509 0.000 0.594 0.702\nC(YM)[56] 0.6950 0.027 25.383 0.000 0.641 0.749\nC(YM)[57] 0.6711 0.028 23.896 0.000 0.616 0.726\nC(YM)[58] 0.6656 0.027 24.563 0.000 0.612 0.719\nC(YM)[59] 0.6143 0.027 23.008 0.000 0.562 0.667\nC(YM)[60] 0.6585 0.028 23.114 0.000 0.603 0.714\nC(YM)[61] 0.6749 0.028 24.273 0.000 0.620 0.729\nC(YM)[62] 0.6656 0.028 23.915 0.000 0.611 0.720\nC(YM)[63] 0.6498 0.026 25.168 0.000 0.599 0.700\nC(YM)[64] 0.6783 0.026 26.140 0.000 0.627 0.729\nC(YM)[65] 0.6691 0.027 24.787 0.000 0.616 0.722\nC(YM)[66] 0.6525 0.028 23.627 0.000 0.598 0.707\nC(YM)[67] 0.6806 0.026 25.794 0.000 0.629 0.732\nC(YM)[68] 0.6643 0.029 23.049 0.000 0.608 0.721\nC(YM)[69] 0.7006 0.028 25.248 0.000 0.646 0.755\nC(YM)[70] 0.6950 0.027 25.325 0.000 0.641 0.749\nC(YM)[71] 0.6650 0.028 23.772 0.000 0.610 0.720\nC(YM)[72] 0.6887 0.027 25.897 0.000 0.637 0.741\nC(YM)[73] 0.7181 0.028 25.826 0.000 0.664 0.773\nC(YM)[74] 0.6712 0.029 23.096 0.000 0.614 0.728\nC(YM)[75] 0.6157 0.029 21.435 0.000 0.559 0.672\nC(YM)[76] 0.6764 0.027 24.967 0.000 0.623 0.730\nC(YM)[77] 0.6877 0.025 27.285 0.000 0.638 0.737\nC(YM)[78] 0.6659 0.026 25.781 0.000 0.615 0.717\nC(YM)[79] 0.6758 0.026 25.679 0.000 0.624 0.727\nC(YM)[80] 0.6595 0.027 24.731 0.000 0.607 0.712\nC(YM)[81] 0.6850 0.027 25.192 0.000 0.632 0.738\nC(YM)[82] 0.6852 0.027 25.418 0.000 0.632 0.738\nC(YM)[83] 0.6340 0.027 23.526 0.000 0.581 0.687\nC(YM)[84] 0.6712 0.030 22.715 0.000 0.613 0.729\nC(YM)[85] 0.6515 0.028 23.254 0.000 0.597 0.706\nC(YM)[86] 0.6766 0.029 23.381 0.000 0.620 0.733\nC(YM)[87] 0.6730 0.027 24.743 0.000 0.620 0.726\nC(YM)[88] 0.6504 0.029 22.803 0.000 0.594 0.706\nC(YM)[89] 0.6841 0.030 23.181 0.000 0.626 0.742\nC(YM)[90] 0.6596 0.028 23.466 0.000 0.604 0.715\nC(YM)[91] 0.6953 0.029 24.125 0.000 0.639 0.752\nC(YM)[92] 0.6888 0.030 23.081 0.000 0.630 0.747\nC(YM)[93] 0.6103 0.030 20.593 0.000 0.552 0.668\nC(YM)[94] 0.5910 0.027 22.254 0.000 0.539 0.643\nC(YM)[95] 0.6539 0.029 22.322 0.000 0.596 0.711\nC(YM)[96] 0.6897 0.030 23.105 0.000 0.631 0.748\nC(YM)[97] 0.6531 0.030 21.851 0.000 0.594 0.712\nC(YM)[98] 0.6727 0.031 21.952 0.000 0.613 0.733\nC(YM)[99] 0.6432 0.029 22.104 0.000 0.586 0.700\nC(YM)[100] 0.6712 0.030 22.145 0.000 0.612 0.731\nC(YM)[101] 0.5915 0.029 20.273 0.000 0.534 0.649\nC(YM)[102] 0.6709 0.029 22.978 0.000 0.614 0.728\nC(YM)[103] 0.6527 0.031 21.204 0.000 0.592 0.713\nC(YM)[104] 0.6904 0.032 21.792 0.000 0.628 0.752\nC(YM)[105] 0.6577 0.029 22.711 0.000 0.601 0.714\nC(YM)[106] 0.6531 0.028 22.935 0.000 0.597 0.709\nC(YM)[107] 0.6261 0.029 21.758 0.000 0.570 0.683\nC(YM)[108] 0.5948 0.029 20.742 0.000 0.539 0.651\nC(YM)[109] 0.5868 0.029 20.340 0.000 0.530 0.643\nC(YM)[110] 0.6529 0.030 22.022 0.000 0.595 0.711\nC(YM)[111] 0.6980 0.028 24.834 0.000 0.643 0.753\nC(YM)[112] 0.6913 0.030 23.307 0.000 0.633 0.749\nC(YM)[113] 0.7511 0.029 26.185 0.000 0.695 0.807\nC(YM)[114] 0.6714 0.029 22.777 0.000 0.614 0.729\nC(YM)[115] 0.6759 0.030 22.760 0.000 0.618 0.734\nC(YM)[116] 0.6378 0.030 21.463 0.000 0.580 0.696\nC(YM)[117] 0.6438 0.029 21.940 0.000 0.586 0.701\nC(YM)[118] 0.6575 0.030 22.156 0.000 0.599 0.716\nC(YM)[119] 0.6758 0.029 23.054 0.000 0.618 0.733\nC(YM)[120] 0.6935 0.030 23.301 0.000 0.635 0.752\nC(YM)[121] 0.6643 0.035 19.172 0.000 0.596 0.732\nC(YM)[122] 0.6908 0.033 20.725 0.000 0.625 0.756\nC(YM)[123] 0.6942 0.033 21.350 0.000 0.630 0.758\nC(YM)[124] 0.6272 0.034 18.295 0.000 0.560 0.694\nC(YM)[125] 0.6210 0.034 18.064 0.000 0.554 0.688\nC(YM)[126] 0.5774 0.034 17.175 0.000 0.512 0.643\nC(YM)[127] 0.6532 0.033 19.557 0.000 0.588 0.719\nC(YM)[128] 0.6842 0.032 21.265 0.000 0.621 0.747\nC(YM)[129] 0.6304 0.035 18.061 0.000 0.562 0.699\nC(YM)[130] 0.5884 0.033 17.567 0.000 0.523 0.654\nC(YM)[131] 0.6638 0.034 19.719 0.000 0.598 0.730\nC(YM)[132] 0.6935 0.036 19.357 0.000 0.623 0.764\nC(YM)[133] 0.6029 0.036 16.821 0.000 0.533 0.673\nC(YM)[134] 0.6097 0.037 16.519 0.000 0.537 0.682\nC(YM)[135] 0.6382 0.034 18.568 0.000 0.571 0.706\nC(YM)[136] 0.5968 0.034 17.694 0.000 0.531 0.663\nC(YM)[137] 0.6006 0.034 17.654 0.000 0.534 0.667\nC(YM)[138] 0.6713 0.033 20.203 0.000 0.606 0.736\nC(YM)[139] 0.5950 0.035 17.233 0.000 0.527 0.663\nC(YM)[140] 0.6511 0.033 19.619 0.000 0.586 0.716\nC(YM)[141] 0.6585 0.036 18.297 0.000 0.588 0.729\nC(YM)[142] 0.6504 0.034 18.997 0.000 0.583 0.717\nC(YM)[143] 0.6931 0.035 19.905 0.000 0.625 0.761\nC(YM)[144] 0.7123 0.035 20.124 0.000 0.643 0.782\nC(YM)[145] 0.6688 0.036 18.685 0.000 0.599 0.739\nC(YM)[146] 0.6190 0.035 17.918 0.000 0.551 0.687\nC(YM)[147] 0.5781 0.034 17.022 0.000 0.512 0.645\nC(YM)[148] 0.6892 0.034 20.293 0.000 0.623 0.756\nC(YM)[149] 0.6578 0.034 19.560 0.000 0.592 0.724\nC(YM)[150] 0.6400 0.031 20.901 0.000 0.580 0.700\nC(YM)[151] 0.6565 0.034 19.426 0.000 0.590 0.723\nC(YM)[152] 0.6198 0.035 17.866 0.000 0.552 0.688\nC(YM)[153] 0.6536 0.034 19.072 0.000 0.586 0.721\nC(YM)[154] 0.5842 0.034 16.995 0.000 0.517 0.652\nC(YM)[155] 0.7048 0.035 20.273 0.000 0.637 0.773\nC(YM)[156] 0.6074 0.034 17.732 0.000 0.540 0.675\nC(YM)[157] 0.7108 0.034 20.756 0.000 0.644 0.778\nC(YM)[158] 0.6873 0.035 19.675 0.000 0.619 0.756\nC(YM)[159] 0.5680 0.032 17.848 0.000 0.506 0.630\nC(YM)[160] 0.7256 0.032 22.881 0.000 0.663 0.788\nC(YM)[161] 0.6886 0.032 21.684 0.000 0.626 0.751\nC(YM)[162] 0.6185 0.031 20.014 0.000 0.558 0.679\nC(YM)[163] 0.6562 0.032 20.754 0.000 0.594 0.718\nC(YM)[164] 0.5999 0.035 17.203 0.000 0.532 0.668\nC(YM)[165] 0.6641 0.033 20.285 0.000 0.600 0.728\nC(YM)[166] 0.6433 0.032 20.170 0.000 0.581 0.706\nC(YM)[167] 0.6468 0.033 19.551 0.000 0.582 0.712\nC(YM)[168] 0.6553 0.033 19.561 0.000 0.590 0.721\nC(YM)[169] 0.6767 0.035 19.545 0.000 0.609 0.745\nC(YM)[170] 0.7110 0.034 21.095 0.000 0.645 0.777\nC(YM)[171] 0.6943 0.031 22.185 0.000 0.633 0.756\nC(YM)[172] 0.6725 0.033 20.367 0.000 0.608 0.737\nC(YM)[173] 0.5888 0.032 18.591 0.000 0.527 0.651\nC(YM)[174] 0.5877 0.031 18.707 0.000 0.526 0.649\nC(YM)[175] 0.6041 0.030 19.842 0.000 0.544 0.664\nC(YM)[176] 0.6283 0.031 20.432 0.000 0.568 0.689\nC(YM)[177] 0.6468 0.031 20.867 0.000 0.586 0.708\nC(YM)[178] 0.6272 0.031 20.422 0.000 0.567 0.687\nC(YM)[179] 0.6870 0.033 20.987 0.000 0.623 0.751\nC(YM)[180] 0.6495 0.034 19.338 0.000 0.584 0.715\nC(YM)[181] 0.6730 0.033 20.238 0.000 0.608 0.738\nC(YM)[182] 0.6619 0.033 19.955 0.000 0.597 0.727\nC(YM)[183] 0.6316 0.030 21.028 0.000 0.573 0.690\nC(YM)[184] 0.6611 0.034 19.445 0.000 0.594 0.728\nC(YM)[185] 0.5777 0.031 18.529 0.000 0.517 0.639\nC(YM)[186] 0.6272 0.030 20.753 0.000 0.568 0.686\nC(YM)[187] 0.5622 0.033 16.885 0.000 0.497 0.627\nC(YM)[188] 0.6172 0.033 18.915 0.000 0.553 0.681\nC(YM)[189] 0.6499 0.030 21.407 0.000 0.590 0.709\nC(YM)[190] 0.5888 0.031 19.205 0.000 0.529 0.649\nC(YM)[191] 0.7025 0.032 22.188 0.000 0.640 0.765\nC(YM)[192] 0.6541 0.031 20.767 0.000 0.592 0.716\nC(YM)[193] 0.6624 0.031 21.375 0.000 0.602 0.723\nC(YM)[194] 0.5304 0.034 15.535 0.000 0.464 0.597\nC(YM)[195] 0.6743 0.030 22.618 0.000 0.616 0.733\nC(YM)[196] 0.6880 0.031 22.263 0.000 0.627 0.749\nC(YM)[197] 0.7033 0.029 23.907 0.000 0.646 0.761\nC(YM)[198] 0.6297 0.030 20.813 0.000 0.570 0.689\nC(YM)[199] 0.6637 0.030 21.807 0.000 0.604 0.723\nC(YM)[200] 0.6839 0.031 22.080 0.000 0.623 0.745\nC(YM)[201] 0.6714 0.034 19.634 0.000 0.604 0.738\nC(YM)[202] 0.6799 0.035 19.582 0.000 0.612 0.748\nC(YM)[203] 0.6990 0.035 20.242 0.000 0.631 0.767\nC(YM)[204] 0.7078 0.034 21.008 0.000 0.642 0.774\nC(YM)[205] 0.7034 0.032 22.209 0.000 0.641 0.765\nC(YM)[206] 0.6999 0.030 23.392 0.000 0.641 0.759\nC(YM)[207] 0.6758 0.030 22.470 0.000 0.617 0.735\nC(YM)[208] 0.6684 0.032 21.173 0.000 0.606 0.730\nC(YM)[209] 0.6319 0.029 21.733 0.000 0.575 0.689\nC(YM)[210] 0.6232 0.030 20.782 0.000 0.564 0.682\nC(YM)[211] 0.6425 0.028 22.965 0.000 0.588 0.697\nC(YM)[212] 0.7100 0.030 23.650 0.000 0.651 0.769\nC(YM)[213] 0.6298 0.031 20.392 0.000 0.569 0.690\nC(YM)[214] 0.6321 0.029 21.855 0.000 0.575 0.689\nC(YM)[215] 0.7100 0.030 23.744 0.000 0.651 0.769\nC(YM)[216] 0.6917 0.030 22.915 0.000 0.633 0.751\nC(YM)[217] 0.6685 0.029 22.696 0.000 0.611 0.726\nC(YM)[218] 0.7183 0.031 22.859 0.000 0.657 0.780\nC(YM)[219] 0.6829 0.031 22.037 0.000 0.622 0.744\nC(YM)[220] 0.6061 0.031 19.774 0.000 0.546 0.666\nC(YM)[221] 0.6265 0.030 20.552 0.000 0.567 0.686\nC(YM)[222] 0.6984 0.030 23.290 0.000 0.640 0.757\nC(YM)[223] 0.5675 0.030 18.781 0.000 0.508 0.627\nC(YM)[224] 0.5867 0.030 19.715 0.000 0.528 0.645\nC(YM)[225] 0.6236 0.028 22.014 0.000 0.568 0.679\nC(YM)[226] 0.6332 0.027 23.120 0.000 0.579 0.687\nC(YM)[227] 0.6537 0.028 23.596 0.000 0.599 0.708\nC(YM)[228] 0.6562 0.028 23.188 0.000 0.601 0.712\nC(YM)[229] 0.7093 0.030 23.405 0.000 0.650 0.769\nC(YM)[230] 0.6881 0.030 23.022 0.000 0.630 0.747\nC(YM)[231] 0.6340 0.029 21.568 0.000 0.576 0.692\nC(YM)[232] 0.6153 0.029 21.245 0.000 0.559 0.672\nC(YM)[233] 0.5959 0.030 19.882 0.000 0.537 0.655\nC(YM)[234] 0.6513 0.030 21.439 0.000 0.592 0.711\nC(YM)[235] 0.6072 0.031 19.514 0.000 0.546 0.668\nC(YM)[236] 0.6962 0.032 21.830 0.000 0.634 0.759\nC(YM)[237] 0.6533 0.030 21.714 0.000 0.594 0.712\nC(YM)[238] 0.6844 0.029 23.779 0.000 0.628 0.741\nC(YM)[239] 0.6778 0.031 21.542 0.000 0.616 0.739\nC(YM)[240] 0.6990 0.029 23.926 0.000 0.642 0.756\nC(YM)[241] 0.6276 0.032 19.890 0.000 0.566 0.689\nC(YM)[242] 0.6613 0.031 21.233 0.000 0.600 0.722\nC(YM)[243] 0.6724 0.029 23.337 0.000 0.616 0.729\nC(YM)[244] 0.7234 0.030 24.460 0.000 0.665 0.781\nC(YM)[245] 0.7337 0.031 23.372 0.000 0.672 0.795\nC(YM)[246] 0.7153 0.029 24.510 0.000 0.658 0.773\nC(YM)[247] 0.6488 0.032 20.451 0.000 0.587 0.711\nC(YM)[248] 0.7260 0.033 21.927 0.000 0.661 0.791\nC(YM)[249] 0.6884 0.033 20.984 0.000 0.624 0.753\nC(YM)[250] 0.6934 0.032 21.346 0.000 0.630 0.757\nC(YM)[251] 0.6526 0.033 19.717 0.000 0.588 0.717\nC(YM)[252] 0.5875 0.032 18.378 0.000 0.525 0.650\nC(YM)[253] 0.6774 0.033 20.672 0.000 0.613 0.742\nC(YM)[254] 0.6169 0.033 18.729 0.000 0.552 0.681\nC(YM)[255] 0.6613 0.032 20.691 0.000 0.599 0.724\nC(YM)[256] 0.6364 0.034 18.845 0.000 0.570 0.703\nC(YM)[257] 0.6054 0.033 18.130 0.000 0.540 0.671\nC(YM)[258] 0.6894 0.031 21.988 0.000 0.628 0.751\nC(YM)[259] 0.5777 0.034 17.031 0.000 0.511 0.644\nC(YM)[260] 0.6354 0.032 19.577 0.000 0.572 0.699\nC(YM)[261] 0.6947 0.033 21.090 0.000 0.630 0.759\nC(YM)[262] 0.6097 0.033 18.362 0.000 0.545 0.675\nC(YM)[263] 0.7032 0.033 21.444 0.000 0.639 0.767\nC(YM)[264] 0.6251 0.032 19.740 0.000 0.563 0.687\nC(YM)[265] 0.5908 0.033 18.028 0.000 0.527 0.655\nC(YM)[266] 0.7641 0.035 22.070 0.000 0.696 0.832\nC(YM)[267] 0.6541 0.030 21.510 0.000 0.594 0.714\nC(YM)[268] 0.6672 0.032 21.154 0.000 0.605 0.729\nC(YM)[269] 0.6621 0.032 20.561 0.000 0.599 0.725\nC(YM)[270] 0.6582 0.032 20.669 0.000 0.596 0.721\nC(YM)[271] 0.6727 0.033 20.532 0.000 0.609 0.737\nC(YM)[272] 0.6500 0.032 20.404 0.000 0.588 0.712\nC(YM)[273] 0.6750 0.032 21.275 0.000 0.613 0.737\nC(YM)[274] 0.7044 0.032 21.737 0.000 0.641 0.768\nC(YM)[275] 0.6597 0.033 20.247 0.000 0.596 0.724\nC(YM)[276] 0.6492 0.033 19.484 0.000 0.584 0.715\nC(YM)[277] 0.6991 0.034 20.675 0.000 0.633 0.765\nC(YM)[278] 0.6641 0.033 20.422 0.000 0.600 0.728\nC(YM)[279] 0.7514 0.030 24.809 0.000 0.692 0.811\nC(YM)[280] 0.7034 0.030 23.439 0.000 0.645 0.762\nC(YM)[281] 0.6862 0.033 20.765 0.000 0.621 0.751\nC(YM)[282] 0.6742 0.032 20.928 0.000 0.611 0.737\nC(YM)[283] 0.7253 0.031 23.482 0.000 0.665 0.786\nC(YM)[284] 0.7392 0.033 22.380 0.000 0.674 0.804\nC(YM)[285] 0.7990 0.029 27.882 0.000 0.743 0.855\nC(YM)[286] 0.7169 0.031 22.964 0.000 0.656 0.778\nC(YM)[287] 0.7096 0.035 20.553 0.000 0.642 0.777\nC(YM)[288] 0.6712 0.034 19.903 0.000 0.605 0.737\nC(YM)[289] 0.6714 0.035 19.449 0.000 0.604 0.739\nC(YM)[290] 0.6140 0.035 17.773 0.000 0.546 0.682\nC(YM)[291] 0.7346 0.032 22.786 0.000 0.671 0.798\nC(YM)[292] 0.6885 0.033 20.875 0.000 0.624 0.753\nC(YM)[293] 0.7310 0.035 21.138 0.000 0.663 0.799\nC(YM)[294] 0.7053 0.032 22.288 0.000 0.643 0.767\nC(YM)[295] 0.7583 0.033 23.077 0.000 0.694 0.823\nC(YM)[296] 0.7877 0.034 22.916 0.000 0.720 0.855\nC(YM)[297] 0.7603 0.033 23.202 0.000 0.696 0.824\nC(YM)[298] 0.7689 0.031 24.663 0.000 0.708 0.830\nC(YM)[299] 0.6926 0.035 19.599 0.000 0.623 0.762\nC(YM)[300] 0.7184 0.033 21.752 0.000 0.654 0.783\nC(YM)[301] 0.8282 0.027 30.514 0.000 0.775 0.881\nC(YM)[302] 0.8117 0.027 30.289 0.000 0.759 0.864\nC(YM)[303] 0.8131 0.024 34.595 0.000 0.767 0.859\nC(YM)[304] 0.8595 0.023 36.886 0.000 0.814 0.905\nC(YM)[305] 0.8080 0.022 36.761 0.000 0.765 0.851\nC(YM)[306] 0.7959 0.024 33.606 0.000 0.750 0.842\nC(YM)[307] 0.7800 0.024 32.078 0.000 0.732 0.828\nC(YM)[308] 0.7471 0.026 28.239 0.000 0.695 0.799\nC(YM)[309] 0.7466 0.025 30.393 0.000 0.698 0.795\nC(YM)[310] 0.7428 0.024 31.203 0.000 0.696 0.789\nC(YM)[311] 0.7068 0.026 27.377 0.000 0.656 0.757\nC(YM)[312] 0.6906 0.025 27.244 0.000 0.641 0.740\nC(YM)[313] 0.6934 0.025 27.287 0.000 0.644 0.743\nC(YM)[314] 0.6750 0.027 25.130 0.000 0.622 0.728\nC(YM)[315] 0.7377 0.023 31.608 0.000 0.692 0.783\nC(YM)[316] 0.6566 0.026 25.384 0.000 0.606 0.707\nC(YM)[317] 0.6350 0.026 24.547 0.000 0.584 0.686\nC(YM)[318] 0.6847 0.024 28.595 0.000 0.638 0.732\nC(YM)[319] 0.6766 0.026 25.537 0.000 0.625 0.729\nC(YM)[320] 0.7631 0.027 28.689 0.000 0.711 0.815\nC(YM)[321] 0.6891 0.025 27.511 0.000 0.640 0.738\nC(YM)[322] 0.7339 0.026 27.922 0.000 0.682 0.785\nC(YM)[323] 0.7195 0.026 27.192 0.000 0.668 0.771\nC(YM)[324] 0.7289 0.026 28.459 0.000 0.679 0.779\nC(YM)[325] 0.6989 0.027 26.256 0.000 0.647 0.751\nC(YM)[326] 0.7084 0.027 26.381 0.000 0.656 0.761\nC(YM)[327] 0.6869 0.026 26.030 0.000 0.635 0.739\nC(YM)[328] 0.7763 0.026 29.322 0.000 0.724 0.828\nC(YM)[329] 0.6935 0.026 26.957 0.000 0.643 0.744\nC(YM)[330] 0.7344 0.027 27.320 0.000 0.682 0.787\nC(YM)[331] 0.7250 0.026 27.814 0.000 0.674 0.776\nC(YM)[332] 0.7024 0.027 26.273 0.000 0.650 0.755\nC(YM)[333] 0.7013 0.029 24.199 0.000 0.644 0.758\nC(YM)[334] 0.6964 0.026 26.464 0.000 0.645 0.748\nC(YM)[335] 0.6903 0.026 26.628 0.000 0.640 0.741\nC(YM)[336] 0.6317 0.026 24.690 0.000 0.582 0.682\nC(YM)[337] 0.6950 0.024 28.398 0.000 0.647 0.743\nC(YM)[338] 0.6513 0.025 26.005 0.000 0.602 0.700\nC(YM)[339] 0.7060 0.025 28.532 0.000 0.658 0.755\nC(YM)[340] 0.7291 0.025 29.328 0.000 0.680 0.778\nC(YM)[341] 0.6803 0.024 28.060 0.000 0.633 0.728\nC(YM)[342] 0.6656 0.025 26.700 0.000 0.617 0.714\nC(YM)[343] 0.6529 0.024 26.886 0.000 0.605 0.701\nC(YM)[344] 0.6429 0.026 24.931 0.000 0.592 0.693\nC(YM)[345] 0.6913 0.026 26.245 0.000 0.640 0.743\nC(YM)[346] 0.7298 0.025 29.182 0.000 0.681 0.779\nC(YM)[347] 0.6730 0.025 26.782 0.000 0.624 0.722\nC(YM)[348] 0.6956 0.025 27.448 0.000 0.646 0.745\nC(YM)[349] 0.6181 0.025 24.636 0.000 0.569 0.667\nC(YM)[350] 0.6716 0.025 27.087 0.000 0.623 0.720\nC(YM)[351] 0.6677 0.024 28.218 0.000 0.621 0.714\nC(YM)[352] 0.6760 0.023 29.323 0.000 0.631 0.721\nC(YM)[353] 0.7251 0.023 31.744 0.000 0.680 0.770\nC(YM)[354] 0.6366 0.025 25.898 0.000 0.588 0.685\nC(YM)[355] 0.6979 0.025 28.438 0.000 0.650 0.746\nC(YM)[356] 0.6777 0.025 26.591 0.000 0.628 0.728\nC(YM)[357] 0.6872 0.026 26.945 0.000 0.637 0.737\nC(YM)[358] 0.6456 0.025 26.087 0.000 0.597 0.694\nC(YM)[359] 0.6188 0.025 25.174 0.000 0.571 0.667\nC(YM)[360] 0.6740 0.025 27.159 0.000 0.625 0.723\nC(YM)[361] 0.6482 0.025 25.440 0.000 0.598 0.698\nC(YM)[362] 0.6869 0.026 26.388 0.000 0.636 0.738\nC(YM)[363] 0.6535 0.025 26.407 0.000 0.605 0.702\nC(YM)[364] 0.6920 0.025 27.456 0.000 0.643 0.741\nC(YM)[365] 0.6517 0.025 25.934 0.000 0.602 0.701\nC(YM)[366] 0.6928 0.025 28.019 0.000 0.644 0.741\nC(YM)[367] 0.6313 0.025 25.399 0.000 0.583 0.680\nC(YM)[368] 0.6967 0.027 26.021 0.000 0.644 0.749\nC(YM)[369] 0.6750 0.026 25.716 0.000 0.624 0.726\nC(YM)[370] 0.6910 0.024 29.102 0.000 0.645 0.738\nC(YM)[371] 0.6366 0.025 25.358 0.000 0.587 0.686\nC(YM)[372] 0.6982 0.025 27.984 0.000 0.649 0.747\nC(YM)[373] 0.6826 0.025 27.154 0.000 0.633 0.732\nC(YM)[374] 0.7148 0.025 28.924 0.000 0.666 0.763\nC(YM)[375] 0.7344 0.023 32.637 0.000 0.690 0.779\nC(YM)[376] 0.7755 0.023 34.459 0.000 0.731 0.820\nC(YM)[377] 0.6930 0.023 29.949 0.000 0.648 0.738\nC(YM)[378] 0.7289 0.023 31.831 0.000 0.684 0.774\nC(YM)[379] 0.6479 0.024 26.852 0.000 0.601 0.695\nC(YM)[380] 0.6813 0.025 27.446 0.000 0.633 0.730\nC(YM)[381] 0.7232 0.024 30.018 0.000 0.676 0.770\nC(YM)[382] 0.6724 0.023 28.824 0.000 0.627 0.718\nC(YM)[383] 0.8169 0.024 34.243 0.000 0.770 0.864\nC(YM)[384] 0.7066 0.024 30.036 0.000 0.660 0.753\nC(YM)[385] 0.7537 0.023 32.347 0.000 0.708 0.799\nC(YM)[386] 0.7231 0.026 27.973 0.000 0.672 0.774\nC(YM)[387] 0.6770 0.024 28.252 0.000 0.630 0.724\nC(YM)[388] 0.6650 0.026 25.835 0.000 0.615 0.715\nC(YM)[389] 0.7032 0.027 25.744 0.000 0.650 0.757\nC(YM)[390] 0.7082 0.027 26.591 0.000 0.656 0.760\nC(YM)[391] 0.6599 0.027 24.487 0.000 0.607 0.713\nC(YM)[392] 0.6226 0.029 21.217 0.000 0.565 0.680\nC(YM)[393] 0.6867 0.029 23.522 0.000 0.629 0.744\nC(YM)[394] 0.7424 0.027 27.555 0.000 0.690 0.795\nC(YM)[395] 0.7401 0.029 25.234 0.000 0.683 0.798\nC(YM)[396] 0.6991 0.029 23.735 0.000 0.641 0.757\nC(YM)[397] 0.6469 0.029 22.696 0.000 0.591 0.703\nC(YM)[398] 0.6803 0.030 22.381 0.000 0.621 0.740\nC(YM)[399] 0.6731 0.027 25.156 0.000 0.621 0.726\nC(YM)[400] 0.6995 0.028 25.052 0.000 0.645 0.754\nC(YM)[401] 0.7450 0.029 25.735 0.000 0.688 0.802\nC(YM)[402] 0.7323 0.030 24.099 0.000 0.673 0.792\nC(YM)[403] 0.6472 0.031 20.927 0.000 0.587 0.708\nC(YM)[404] 0.6982 0.030 22.937 0.000 0.638 0.758\nC(YM)[405] 0.6617 0.032 20.923 0.000 0.600 0.724\nC(YM)[406] 0.7068 0.030 23.181 0.000 0.647 0.767\nC(YM)[407] 0.6841 0.031 22.132 0.000 0.624 0.745\nC(YM)[408] 0.7130 0.030 23.391 0.000 0.653 0.773\nC(YM)[409] 0.7451 0.031 23.934 0.000 0.684 0.806\nC(YM)[410] 0.6617 0.031 21.293 0.000 0.601 0.723\nC(YM)[411] 0.6670 0.029 23.285 0.000 0.611 0.723\nC(YM)[412] 0.5980 0.029 20.348 0.000 0.540 0.656\nC(YM)[413] 0.6477 0.030 21.952 0.000 0.590 0.706\nC(YM)[414] 0.5936 0.029 20.631 0.000 0.537 0.650\nC(YM)[415] 0.6602 0.030 21.925 0.000 0.601 0.719\nC(YM)[416] 0.6749 0.031 22.018 0.000 0.615 0.735\nC(YM)[417] 0.6427 0.028 23.046 0.000 0.588 0.697\nC(YM)[418] 0.6254 0.028 22.353 0.000 0.571 0.680\nC(YM)[419] 0.6554 0.030 22.080 0.000 0.597 0.714\nC(YM)[420] 0.7093 0.030 23.970 0.000 0.651 0.767\nC(YM)[421] 0.6842 0.030 23.063 0.000 0.626 0.742\nC(YM)[422] 0.6176 0.032 19.344 0.000 0.555 0.680\nC(YM)[423] 0.7250 0.031 23.380 0.000 0.664 0.786\nC(YM)[424] 0.7819 0.032 24.401 0.000 0.719 0.845\nC(YM)[425] 0.6841 0.030 22.685 0.000 0.625 0.743\nC(YM)[426] 0.6637 0.032 20.934 0.000 0.602 0.726\nC(YM)[427] 0.6601 0.031 21.081 0.000 0.599 0.721\nC(YM)[428] 0.6598 0.030 21.667 0.000 0.600 0.720\nC(Newspaper)[T.Chicago Tribune] 0.0438 0.006 6.854 0.000 0.031 0.056\nC(Newspaper)[T.Los Angeles Times] 0.0372 0.006 6.259 0.000 0.026 0.049\nC(Newspaper)[T.New York Times] 0.0807 0.006 13.598 0.000 0.069 0.092\nC(Newspaper)[T.The Washington Post] 0.0001 0.006 0.019 0.985 -0.012 0.012\nC(Newspaper)[T.USA Today] 0.0538 0.008 6.483 0.000 0.038 0.070\nC(Newspaper)[T.Wall Street Journal] 0.1159 0.005 21.732 0.000 0.105 0.126\n==============================================================================\nOmnibus: 180705.158 Durbin-Watson: 1.852\nProb(Omnibus): 0.000 Jarque-Bera (JB): 896272.078\nSkew: 1.830 Prob(JB): 0.00\nKurtosis: 8.713 Cond. No. 40.5\n==============================================================================\n\nNotes:\n[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.\n"
],
[
"# LM index\nLMindex=estimate_index('LMscore')",
" OLS Regression Results \n==============================================================================\nDep. Variable: LMscore R-squared: 0.006\nModel: OLS Adj. R-squared: 0.005\nMethod: Least Squares F-statistic: 6.471\nDate: Tue, 18 May 2021 Prob (F-statistic): 0.00\nTime: 19:40:10 Log-Likelihood: -1.1079e+06\nNo. Observations: 467351 AIC: 2.217e+06\nDf Residuals: 466917 BIC: 2.221e+06\nDf Model: 433 \nCovariance Type: nonrobust \n=======================================================================================================\n coef std err t P>|t| [0.025 0.975]\n-------------------------------------------------------------------------------------------------------\nC(YM)[1] -1.8737 0.096 -19.446 0.000 -2.063 -1.685\nC(YM)[2] -2.0267 0.099 -20.468 0.000 -2.221 -1.833\nC(YM)[3] -2.0357 0.088 -23.124 0.000 -2.208 -1.863\nC(YM)[4] -1.9367 0.089 -21.736 0.000 -2.111 -1.762\nC(YM)[5] -1.7965 0.088 -20.447 0.000 -1.969 -1.624\nC(YM)[6] -2.0423 0.090 -22.666 0.000 -2.219 -1.866\nC(YM)[7] -1.9601 0.093 -21.021 0.000 -2.143 -1.777\nC(YM)[8] -2.2932 0.089 -25.769 0.000 -2.468 -2.119\nC(YM)[9] -1.8816 0.093 -20.124 0.000 -2.065 -1.698\nC(YM)[10] -1.9579 0.090 -21.732 0.000 -2.134 -1.781\nC(YM)[11] -2.0943 0.096 -21.779 0.000 -2.283 -1.906\nC(YM)[12] -1.9389 0.092 -21.175 0.000 -2.118 -1.759\nC(YM)[13] -2.1495 0.088 -24.544 0.000 -2.321 -1.978\nC(YM)[14] -1.9483 0.098 -19.802 0.000 -2.141 -1.755\nC(YM)[15] -1.9604 0.094 -20.941 0.000 -2.144 -1.777\nC(YM)[16] -1.7708 0.090 -19.688 0.000 -1.947 -1.595\nC(YM)[17] -2.1952 0.089 -24.610 0.000 -2.370 -2.020\nC(YM)[18] -2.2028 0.089 -24.704 0.000 -2.378 -2.028\nC(YM)[19] -2.2797 0.092 -24.814 0.000 -2.460 -2.100\nC(YM)[20] -1.8695 0.092 -20.305 0.000 -2.050 -1.689\nC(YM)[21] -1.8816 0.094 -20.122 0.000 -2.065 -1.698\nC(YM)[22] -1.9940 0.091 -21.886 0.000 -2.173 -1.815\nC(YM)[23] -1.8523 0.090 -20.491 0.000 -2.030 -1.675\nC(YM)[24] -1.9952 0.088 -22.778 0.000 -2.167 -1.824\nC(YM)[25] -2.0649 0.087 -23.750 0.000 -2.235 -1.895\nC(YM)[26] -2.0417 0.085 -23.997 0.000 -2.209 -1.875\nC(YM)[27] -1.9889 0.079 -25.149 0.000 -2.144 -1.834\nC(YM)[28] -2.1958 0.079 -27.803 0.000 -2.351 -2.041\nC(YM)[29] -2.0616 0.083 -24.770 0.000 -2.225 -1.899\nC(YM)[30] -2.0727 0.079 -26.352 0.000 -2.227 -1.919\nC(YM)[31] -2.1512 0.078 -27.573 0.000 -2.304 -1.998\nC(YM)[32] -2.0959 0.082 -25.414 0.000 -2.258 -1.934\nC(YM)[33] -2.1890 0.083 -26.326 0.000 -2.352 -2.026\nC(YM)[34] -2.0520 0.079 -26.035 0.000 -2.206 -1.897\nC(YM)[35] -2.0571 0.082 -25.035 0.000 -2.218 -1.896\nC(YM)[36] -2.0069 0.086 -23.470 0.000 -2.174 -1.839\nC(YM)[37] -2.1650 0.079 -27.444 0.000 -2.320 -2.010\nC(YM)[38] -1.9122 0.078 -24.492 0.000 -2.065 -1.759\nC(YM)[39] -2.1842 0.079 -27.784 0.000 -2.338 -2.030\nC(YM)[40] -1.8830 0.080 -23.435 0.000 -2.040 -1.725\nC(YM)[41] -1.9706 0.078 -25.285 0.000 -2.123 -1.818\nC(YM)[42] -2.0703 0.075 -27.491 0.000 -2.218 -1.923\nC(YM)[43] -2.0320 0.082 -24.877 0.000 -2.192 -1.872\nC(YM)[44] -2.1677 0.078 -27.872 0.000 -2.320 -2.015\nC(YM)[45] -1.9841 0.078 -25.446 0.000 -2.137 -1.831\nC(YM)[46] -2.1783 0.076 -28.771 0.000 -2.327 -2.030\nC(YM)[47] -1.9427 0.078 -24.919 0.000 -2.096 -1.790\nC(YM)[48] -2.0917 0.073 -28.806 0.000 -2.234 -1.949\nC(YM)[49] -2.1289 0.075 -28.250 0.000 -2.277 -1.981\nC(YM)[50] -2.1911 0.077 -28.301 0.000 -2.343 -2.039\nC(YM)[51] -2.2421 0.071 -31.789 0.000 -2.380 -2.104\nC(YM)[52] -2.1584 0.073 -29.544 0.000 -2.302 -2.015\nC(YM)[53] -2.1972 0.073 -29.927 0.000 -2.341 -2.053\nC(YM)[54] -2.0056 0.074 -27.114 0.000 -2.151 -1.861\nC(YM)[55] -1.9414 0.075 -25.863 0.000 -2.089 -1.794\nC(YM)[56] -2.1267 0.075 -28.516 0.000 -2.273 -1.981\nC(YM)[57] -2.0182 0.076 -26.384 0.000 -2.168 -1.868\nC(YM)[58] -2.0549 0.074 -27.843 0.000 -2.200 -1.910\nC(YM)[59] -2.1218 0.073 -29.178 0.000 -2.264 -1.979\nC(YM)[60] -2.0387 0.078 -26.272 0.000 -2.191 -1.887\nC(YM)[61] -2.0562 0.076 -27.151 0.000 -2.205 -1.908\nC(YM)[62] -2.2175 0.076 -29.256 0.000 -2.366 -2.069\nC(YM)[63] -2.1295 0.070 -30.284 0.000 -2.267 -1.992\nC(YM)[64] -2.1250 0.071 -30.067 0.000 -2.263 -1.986\nC(YM)[65] -2.1388 0.074 -29.093 0.000 -2.283 -1.995\nC(YM)[66] -2.0367 0.075 -27.077 0.000 -2.184 -1.889\nC(YM)[67] -2.1334 0.072 -29.687 0.000 -2.274 -1.993\nC(YM)[68] -2.3319 0.078 -29.708 0.000 -2.486 -2.178\nC(YM)[69] -2.3483 0.076 -31.073 0.000 -2.496 -2.200\nC(YM)[70] -2.0801 0.075 -27.829 0.000 -2.227 -1.934\nC(YM)[71] -2.1796 0.076 -28.608 0.000 -2.329 -2.030\nC(YM)[72] -2.0382 0.072 -28.142 0.000 -2.180 -1.896\nC(YM)[73] -2.1407 0.076 -28.268 0.000 -2.289 -1.992\nC(YM)[74] -2.0961 0.079 -26.482 0.000 -2.251 -1.941\nC(YM)[75] -2.1717 0.078 -27.762 0.000 -2.325 -2.018\nC(YM)[76] -2.0572 0.074 -27.880 0.000 -2.202 -1.913\nC(YM)[77] -2.0175 0.069 -29.388 0.000 -2.152 -1.883\nC(YM)[78] -2.0430 0.070 -29.041 0.000 -2.181 -1.905\nC(YM)[79] -2.3308 0.072 -32.517 0.000 -2.471 -2.190\nC(YM)[80] -2.2698 0.073 -31.251 0.000 -2.412 -2.127\nC(YM)[81] -2.2184 0.074 -29.955 0.000 -2.363 -2.073\nC(YM)[82] -2.0356 0.073 -27.726 0.000 -2.180 -1.892\nC(YM)[83] -2.0775 0.073 -28.303 0.000 -2.221 -1.934\nC(YM)[84] -2.0216 0.080 -25.121 0.000 -2.179 -1.864\nC(YM)[85] -1.8925 0.076 -24.801 0.000 -2.042 -1.743\nC(YM)[86] -1.9672 0.079 -24.959 0.000 -2.122 -1.813\nC(YM)[87] -2.2058 0.074 -29.776 0.000 -2.351 -2.061\nC(YM)[88] -1.9226 0.078 -24.748 0.000 -2.075 -1.770\nC(YM)[89] -1.8476 0.080 -22.988 0.000 -2.005 -1.690\nC(YM)[90] -1.8600 0.077 -24.297 0.000 -2.010 -1.710\nC(YM)[91] -2.0337 0.078 -25.910 0.000 -2.188 -1.880\nC(YM)[92] -2.1113 0.081 -25.976 0.000 -2.271 -1.952\nC(YM)[93] -1.8525 0.081 -22.952 0.000 -2.011 -1.694\nC(YM)[94] -1.7583 0.072 -24.308 0.000 -1.900 -1.617\nC(YM)[95] -1.8324 0.080 -22.966 0.000 -1.989 -1.676\nC(YM)[96] -1.7268 0.081 -21.241 0.000 -1.886 -1.567\nC(YM)[97] -1.7300 0.081 -21.253 0.000 -1.890 -1.570\nC(YM)[98] -1.7759 0.083 -21.279 0.000 -1.939 -1.612\nC(YM)[99] -1.7718 0.079 -22.357 0.000 -1.927 -1.616\nC(YM)[100] -1.7154 0.083 -20.782 0.000 -1.877 -1.554\nC(YM)[101] -1.7795 0.079 -22.394 0.000 -1.935 -1.624\nC(YM)[102] -1.7853 0.080 -22.452 0.000 -1.941 -1.629\nC(YM)[103] -1.9264 0.084 -22.978 0.000 -2.091 -1.762\nC(YM)[104] -1.7949 0.086 -20.803 0.000 -1.964 -1.626\nC(YM)[105] -1.3658 0.079 -17.316 0.000 -1.520 -1.211\nC(YM)[106] -1.8142 0.078 -23.392 0.000 -1.966 -1.662\nC(YM)[107] -1.7663 0.078 -22.536 0.000 -1.920 -1.613\nC(YM)[108] -1.7120 0.078 -21.920 0.000 -1.865 -1.559\nC(YM)[109] -1.7153 0.079 -21.830 0.000 -1.869 -1.561\nC(YM)[110] -1.6463 0.081 -20.387 0.000 -1.805 -1.488\nC(YM)[111] -1.8806 0.077 -24.567 0.000 -2.031 -1.731\nC(YM)[112] -1.7968 0.081 -22.243 0.000 -1.955 -1.638\nC(YM)[113] -1.7630 0.078 -22.567 0.000 -1.916 -1.610\nC(YM)[114] -1.8803 0.080 -23.422 0.000 -2.038 -1.723\nC(YM)[115] -1.8718 0.081 -23.143 0.000 -2.030 -1.713\nC(YM)[116] -1.7710 0.081 -21.881 0.000 -1.930 -1.612\nC(YM)[117] -1.6945 0.080 -21.203 0.000 -1.851 -1.538\nC(YM)[118] -1.8542 0.081 -22.942 0.000 -2.013 -1.696\nC(YM)[119] -1.7422 0.080 -21.822 0.000 -1.899 -1.586\nC(YM)[120] -1.8481 0.081 -22.799 0.000 -2.007 -1.689\nC(YM)[121] -1.7890 0.094 -18.956 0.000 -1.974 -1.604\nC(YM)[122] -1.8664 0.091 -20.560 0.000 -2.044 -1.688\nC(YM)[123] -1.9411 0.089 -21.918 0.000 -2.115 -1.768\nC(YM)[124] -1.7154 0.093 -18.373 0.000 -1.898 -1.532\nC(YM)[125] -1.9061 0.094 -20.356 0.000 -2.090 -1.723\nC(YM)[126] -1.7851 0.092 -19.495 0.000 -1.965 -1.606\nC(YM)[127] -1.8818 0.091 -20.686 0.000 -2.060 -1.703\nC(YM)[128] -1.7957 0.088 -20.491 0.000 -1.967 -1.624\nC(YM)[129] -1.7755 0.095 -18.676 0.000 -1.962 -1.589\nC(YM)[130] -2.0349 0.091 -22.307 0.000 -2.214 -1.856\nC(YM)[131] -2.1023 0.092 -22.928 0.000 -2.282 -1.923\nC(YM)[132] -1.9794 0.098 -20.284 0.000 -2.171 -1.788\nC(YM)[133] -1.6961 0.098 -17.374 0.000 -1.887 -1.505\nC(YM)[134] -1.9784 0.101 -19.681 0.000 -2.175 -1.781\nC(YM)[135] -1.7550 0.094 -18.747 0.000 -1.938 -1.572\nC(YM)[136] -1.8395 0.092 -20.026 0.000 -2.020 -1.660\nC(YM)[137] -1.8824 0.093 -20.316 0.000 -2.064 -1.701\nC(YM)[138] -1.9562 0.091 -21.615 0.000 -2.134 -1.779\nC(YM)[139] -1.6863 0.094 -17.933 0.000 -1.871 -1.502\nC(YM)[140] -1.6786 0.090 -18.571 0.000 -1.856 -1.501\nC(YM)[141] -2.1501 0.098 -21.934 0.000 -2.342 -1.958\nC(YM)[142] -2.0447 0.093 -21.929 0.000 -2.227 -1.862\nC(YM)[143] -1.8724 0.095 -19.742 0.000 -2.058 -1.686\nC(YM)[144] -1.7537 0.096 -18.192 0.000 -1.943 -1.565\nC(YM)[145] -1.8297 0.097 -18.770 0.000 -2.021 -1.639\nC(YM)[146] -1.7048 0.094 -18.118 0.000 -1.889 -1.520\nC(YM)[147] -1.9697 0.092 -21.294 0.000 -2.151 -1.788\nC(YM)[148] -1.8008 0.092 -19.469 0.000 -1.982 -1.620\nC(YM)[149] -2.0389 0.092 -22.261 0.000 -2.218 -1.859\nC(YM)[150] -1.8819 0.083 -22.565 0.000 -2.045 -1.718\nC(YM)[151] -1.9229 0.092 -20.891 0.000 -2.103 -1.742\nC(YM)[152] -1.9220 0.094 -20.343 0.000 -2.107 -1.737\nC(YM)[153] -1.8217 0.093 -19.516 0.000 -2.005 -1.639\nC(YM)[154] -1.8232 0.094 -19.473 0.000 -2.007 -1.640\nC(YM)[155] -1.9650 0.095 -20.752 0.000 -2.151 -1.779\nC(YM)[156] -1.7647 0.093 -18.914 0.000 -1.948 -1.582\nC(YM)[157] -1.8935 0.093 -20.302 0.000 -2.076 -1.711\nC(YM)[158] -1.7076 0.095 -17.947 0.000 -1.894 -1.521\nC(YM)[159] -1.7349 0.087 -20.014 0.000 -1.905 -1.565\nC(YM)[160] -1.8101 0.086 -20.957 0.000 -1.979 -1.641\nC(YM)[161] -2.0496 0.086 -23.698 0.000 -2.219 -1.880\nC(YM)[162] -1.7641 0.084 -20.960 0.000 -1.929 -1.599\nC(YM)[163] -1.8776 0.086 -21.804 0.000 -2.046 -1.709\nC(YM)[164] -1.8462 0.095 -19.439 0.000 -2.032 -1.660\nC(YM)[165] -1.8210 0.089 -20.421 0.000 -1.996 -1.646\nC(YM)[166] -1.8716 0.087 -21.546 0.000 -2.042 -1.701\nC(YM)[167] -1.8451 0.090 -20.478 0.000 -2.022 -1.668\nC(YM)[168] -1.7887 0.091 -19.605 0.000 -1.968 -1.610\nC(YM)[169] -1.5683 0.094 -16.633 0.000 -1.753 -1.384\nC(YM)[170] -1.9220 0.092 -20.937 0.000 -2.102 -1.742\nC(YM)[171] -1.8070 0.085 -21.201 0.000 -1.974 -1.640\nC(YM)[172] -1.7217 0.090 -19.145 0.000 -1.898 -1.545\nC(YM)[173] -1.8876 0.086 -21.883 0.000 -2.057 -1.718\nC(YM)[174] -1.6333 0.086 -19.088 0.000 -1.801 -1.466\nC(YM)[175] -1.8661 0.083 -22.506 0.000 -2.029 -1.704\nC(YM)[176] -2.0198 0.084 -24.116 0.000 -2.184 -1.856\nC(YM)[177] -1.8659 0.084 -22.102 0.000 -2.031 -1.700\nC(YM)[178] -1.8076 0.084 -21.611 0.000 -1.972 -1.644\nC(YM)[179] -1.7418 0.089 -19.537 0.000 -1.917 -1.567\nC(YM)[180] -1.8407 0.091 -20.122 0.000 -2.020 -1.661\nC(YM)[181] -1.5275 0.091 -16.865 0.000 -1.705 -1.350\nC(YM)[182] -1.7872 0.090 -19.784 0.000 -1.964 -1.610\nC(YM)[183] -1.7599 0.082 -21.515 0.000 -1.920 -1.600\nC(YM)[184] -1.9053 0.093 -20.577 0.000 -2.087 -1.724\nC(YM)[185] -1.7494 0.085 -20.600 0.000 -1.916 -1.583\nC(YM)[186] -1.8578 0.082 -22.570 0.000 -2.019 -1.696\nC(YM)[187] -1.7942 0.091 -19.785 0.000 -1.972 -1.616\nC(YM)[188] -1.9489 0.089 -21.928 0.000 -2.123 -1.775\nC(YM)[189] -2.0839 0.083 -25.201 0.000 -2.246 -1.922\nC(YM)[190] -1.7775 0.084 -21.286 0.000 -1.941 -1.614\nC(YM)[191] -1.7255 0.086 -20.009 0.000 -1.895 -1.557\nC(YM)[192] -1.6854 0.086 -19.646 0.000 -1.854 -1.517\nC(YM)[193] -1.7002 0.084 -20.144 0.000 -1.866 -1.535\nC(YM)[194] -1.7346 0.093 -18.653 0.000 -1.917 -1.552\nC(YM)[195] -1.9969 0.081 -24.593 0.000 -2.156 -1.838\nC(YM)[196] -1.9305 0.084 -22.935 0.000 -2.095 -1.766\nC(YM)[197] -1.6686 0.080 -20.825 0.000 -1.826 -1.512\nC(YM)[198] -1.9129 0.082 -23.216 0.000 -2.074 -1.751\nC(YM)[199] -1.7924 0.083 -21.624 0.000 -1.955 -1.630\nC(YM)[200] -1.8882 0.084 -22.383 0.000 -2.054 -1.723\nC(YM)[201] -1.9288 0.093 -20.709 0.000 -2.111 -1.746\nC(YM)[202] -2.0187 0.095 -21.348 0.000 -2.204 -1.833\nC(YM)[203] -1.8747 0.094 -19.934 0.000 -2.059 -1.690\nC(YM)[204] -1.7936 0.092 -19.546 0.000 -1.973 -1.614\nC(YM)[205] -2.0486 0.086 -23.750 0.000 -2.218 -1.880\nC(YM)[206] -2.1085 0.081 -25.875 0.000 -2.268 -1.949\nC(YM)[207] -1.9792 0.082 -24.164 0.000 -2.140 -1.819\nC(YM)[208] -2.0134 0.086 -23.419 0.000 -2.182 -1.845\nC(YM)[209] -2.0999 0.079 -26.517 0.000 -2.255 -1.945\nC(YM)[210] -1.9263 0.082 -23.587 0.000 -2.086 -1.766\nC(YM)[211] -2.0208 0.076 -26.522 0.000 -2.170 -1.871\nC(YM)[212] -1.9557 0.082 -23.919 0.000 -2.116 -1.795\nC(YM)[213] -2.0188 0.084 -24.000 0.000 -2.184 -1.854\nC(YM)[214] -2.1052 0.079 -26.727 0.000 -2.260 -1.951\nC(YM)[215] -1.9016 0.081 -23.348 0.000 -2.061 -1.742\nC(YM)[216] -1.9253 0.082 -23.420 0.000 -2.086 -1.764\nC(YM)[217] -1.8741 0.080 -23.363 0.000 -2.031 -1.717\nC(YM)[218] -1.7705 0.086 -20.687 0.000 -1.938 -1.603\nC(YM)[219] -2.2690 0.084 -26.883 0.000 -2.434 -2.104\nC(YM)[220] -1.9136 0.083 -22.924 0.000 -2.077 -1.750\nC(YM)[221] -2.0132 0.083 -24.247 0.000 -2.176 -1.851\nC(YM)[222] -1.9436 0.082 -23.798 0.000 -2.104 -1.784\nC(YM)[223] -1.8693 0.082 -22.716 0.000 -2.031 -1.708\nC(YM)[224] -2.0295 0.081 -25.040 0.000 -2.188 -1.871\nC(YM)[225] -2.0073 0.077 -26.017 0.000 -2.158 -1.856\nC(YM)[226] -2.1146 0.075 -28.351 0.000 -2.261 -1.968\nC(YM)[227] -2.2514 0.075 -29.836 0.000 -2.399 -2.103\nC(YM)[228] -2.1784 0.077 -28.265 0.000 -2.330 -2.027\nC(YM)[229] -1.9788 0.083 -23.975 0.000 -2.141 -1.817\nC(YM)[230] -2.0641 0.081 -25.357 0.000 -2.224 -1.905\nC(YM)[231] -2.0364 0.080 -25.436 0.000 -2.193 -1.880\nC(YM)[232] -1.8943 0.079 -24.016 0.000 -2.049 -1.740\nC(YM)[233] -1.9470 0.082 -23.851 0.000 -2.107 -1.787\nC(YM)[234] -1.9922 0.083 -24.076 0.000 -2.154 -1.830\nC(YM)[235] -2.1355 0.085 -25.198 0.000 -2.302 -1.969\nC(YM)[236] -1.8850 0.087 -21.700 0.000 -2.055 -1.715\nC(YM)[237] -2.1122 0.082 -25.777 0.000 -2.273 -1.952\nC(YM)[238] -2.2773 0.078 -29.051 0.000 -2.431 -2.124\nC(YM)[239] -1.9251 0.086 -22.465 0.000 -2.093 -1.757\nC(YM)[240] -2.0308 0.080 -25.524 0.000 -2.187 -1.875\nC(YM)[241] -2.0927 0.086 -24.351 0.000 -2.261 -1.924\nC(YM)[242] -2.1504 0.085 -25.351 0.000 -2.317 -1.984\nC(YM)[243] -2.1621 0.078 -27.553 0.000 -2.316 -2.008\nC(YM)[244] -1.9183 0.081 -23.816 0.000 -2.076 -1.760\nC(YM)[245] -2.0624 0.085 -24.123 0.000 -2.230 -1.895\nC(YM)[246] -1.9763 0.079 -24.864 0.000 -2.132 -1.821\nC(YM)[247] -1.9144 0.086 -22.155 0.000 -2.084 -1.745\nC(YM)[248] -1.9507 0.090 -21.631 0.000 -2.127 -1.774\nC(YM)[249] -1.9103 0.089 -21.380 0.000 -2.085 -1.735\nC(YM)[250] -1.7971 0.088 -20.313 0.000 -1.970 -1.624\nC(YM)[251] -1.9777 0.090 -21.940 0.000 -2.154 -1.801\nC(YM)[252] -1.9140 0.087 -21.983 0.000 -2.085 -1.743\nC(YM)[253] -1.9430 0.089 -21.770 0.000 -2.118 -1.768\nC(YM)[254] -1.7946 0.090 -20.006 0.000 -1.970 -1.619\nC(YM)[255] -1.7753 0.087 -20.395 0.000 -1.946 -1.605\nC(YM)[256] -1.7497 0.092 -19.025 0.000 -1.930 -1.569\nC(YM)[257] -1.8879 0.091 -20.759 0.000 -2.066 -1.710\nC(YM)[258] -1.8480 0.085 -21.642 0.000 -2.015 -1.681\nC(YM)[259] -1.8888 0.092 -20.443 0.000 -2.070 -1.708\nC(YM)[260] -1.8567 0.088 -21.002 0.000 -2.030 -1.683\nC(YM)[261] -1.8646 0.090 -20.784 0.000 -2.040 -1.689\nC(YM)[262] -1.8995 0.090 -21.005 0.000 -2.077 -1.722\nC(YM)[263] -1.5880 0.089 -17.780 0.000 -1.763 -1.413\nC(YM)[264] -1.6874 0.086 -19.567 0.000 -1.856 -1.518\nC(YM)[265] -1.5295 0.089 -17.135 0.000 -1.704 -1.355\nC(YM)[266] -1.6974 0.094 -18.001 0.000 -1.882 -1.513\nC(YM)[267] -1.8168 0.083 -21.938 0.000 -1.979 -1.655\nC(YM)[268] -1.7821 0.086 -20.746 0.000 -1.950 -1.614\nC(YM)[269] -1.9495 0.088 -22.228 0.000 -2.121 -1.778\nC(YM)[270] -1.8973 0.087 -21.876 0.000 -2.067 -1.727\nC(YM)[271] -2.0819 0.089 -23.330 0.000 -2.257 -1.907\nC(YM)[272] -1.9819 0.087 -22.842 0.000 -2.152 -1.812\nC(YM)[273] -1.9589 0.086 -22.670 0.000 -2.128 -1.790\nC(YM)[274] -2.0130 0.088 -22.808 0.000 -2.186 -1.840\nC(YM)[275] -1.7815 0.089 -20.075 0.000 -1.955 -1.608\nC(YM)[276] -1.8135 0.091 -19.984 0.000 -1.991 -1.636\nC(YM)[277] -2.1563 0.092 -23.413 0.000 -2.337 -1.976\nC(YM)[278] -2.0426 0.089 -23.062 0.000 -2.216 -1.869\nC(YM)[279] -1.9757 0.082 -23.950 0.000 -2.137 -1.814\nC(YM)[280] -1.9005 0.082 -23.252 0.000 -2.061 -1.740\nC(YM)[281] -1.7905 0.090 -19.894 0.000 -1.967 -1.614\nC(YM)[282] -1.9295 0.088 -21.991 0.000 -2.102 -1.758\nC(YM)[283] -1.9472 0.084 -23.148 0.000 -2.112 -1.782\nC(YM)[284] -1.9576 0.090 -21.761 0.000 -2.134 -1.781\nC(YM)[285] -1.9783 0.078 -25.346 0.000 -2.131 -1.825\nC(YM)[286] -1.8762 0.085 -22.067 0.000 -2.043 -1.710\nC(YM)[287] -1.7287 0.094 -18.384 0.000 -1.913 -1.544\nC(YM)[288] -1.9207 0.092 -20.912 0.000 -2.101 -1.741\nC(YM)[289] -1.9296 0.094 -20.521 0.000 -2.114 -1.745\nC(YM)[290] -1.8651 0.094 -19.822 0.000 -2.050 -1.681\nC(YM)[291] -1.9246 0.088 -21.920 0.000 -2.097 -1.752\nC(YM)[292] -1.7695 0.090 -19.699 0.000 -1.946 -1.593\nC(YM)[293] -1.7685 0.094 -18.777 0.000 -1.953 -1.584\nC(YM)[294] -1.9202 0.086 -22.280 0.000 -2.089 -1.751\nC(YM)[295] -1.8090 0.089 -20.213 0.000 -1.984 -1.634\nC(YM)[296] -2.0105 0.094 -21.476 0.000 -2.194 -1.827\nC(YM)[297] -1.7422 0.089 -19.521 0.000 -1.917 -1.567\nC(YM)[298] -1.7981 0.085 -21.176 0.000 -1.965 -1.632\nC(YM)[299] -1.9271 0.096 -20.024 0.000 -2.116 -1.738\nC(YM)[300] -1.8536 0.090 -20.607 0.000 -2.030 -1.677\nC(YM)[301] -1.7864 0.074 -24.166 0.000 -1.931 -1.641\nC(YM)[302] -1.8937 0.073 -25.946 0.000 -2.037 -1.751\nC(YM)[303] -1.8800 0.064 -29.370 0.000 -2.005 -1.755\nC(YM)[304] -2.2933 0.063 -36.136 0.000 -2.418 -2.169\nC(YM)[305] -1.9876 0.060 -33.203 0.000 -2.105 -1.870\nC(YM)[306] -1.8809 0.065 -29.159 0.000 -2.007 -1.755\nC(YM)[307] -1.8054 0.066 -27.263 0.000 -1.935 -1.676\nC(YM)[308] -2.0580 0.072 -28.559 0.000 -2.199 -1.917\nC(YM)[309] -1.7813 0.067 -26.626 0.000 -1.912 -1.650\nC(YM)[310] -1.8490 0.065 -28.519 0.000 -1.976 -1.722\nC(YM)[311] -1.9180 0.070 -27.278 0.000 -2.056 -1.780\nC(YM)[312] -1.9136 0.069 -27.718 0.000 -2.049 -1.778\nC(YM)[313] -1.6541 0.069 -23.901 0.000 -1.790 -1.518\nC(YM)[314] -1.8091 0.073 -24.730 0.000 -1.952 -1.666\nC(YM)[315] -1.7985 0.064 -28.295 0.000 -1.923 -1.674\nC(YM)[316] -1.9449 0.070 -27.608 0.000 -2.083 -1.807\nC(YM)[317] -1.7694 0.070 -25.113 0.000 -1.908 -1.631\nC(YM)[318] -2.0613 0.065 -31.610 0.000 -2.189 -1.933\nC(YM)[319] -2.0421 0.072 -28.297 0.000 -2.184 -1.901\nC(YM)[320] -1.9509 0.072 -26.929 0.000 -2.093 -1.809\nC(YM)[321] -2.0506 0.068 -30.058 0.000 -2.184 -1.917\nC(YM)[322] -1.8758 0.072 -26.205 0.000 -2.016 -1.736\nC(YM)[323] -2.2570 0.072 -31.318 0.000 -2.398 -2.116\nC(YM)[324] -1.9214 0.070 -27.543 0.000 -2.058 -1.785\nC(YM)[325] -1.9562 0.072 -26.984 0.000 -2.098 -1.814\nC(YM)[326] -1.9804 0.073 -27.079 0.000 -2.124 -1.837\nC(YM)[327] -1.9493 0.072 -27.123 0.000 -2.090 -1.808\nC(YM)[328] -1.9804 0.072 -27.467 0.000 -2.122 -1.839\nC(YM)[329] -1.9715 0.070 -28.140 0.000 -2.109 -1.834\nC(YM)[330] -2.0136 0.073 -27.502 0.000 -2.157 -1.870\nC(YM)[331] -2.3773 0.071 -33.488 0.000 -2.516 -2.238\nC(YM)[332] -2.0372 0.073 -27.977 0.000 -2.180 -1.894\nC(YM)[333] -1.7852 0.079 -22.618 0.000 -1.940 -1.631\nC(YM)[334] -1.7359 0.072 -24.219 0.000 -1.876 -1.595\nC(YM)[335] -1.9715 0.071 -27.922 0.000 -2.110 -1.833\nC(YM)[336] -1.9948 0.070 -28.627 0.000 -2.131 -1.858\nC(YM)[337] -1.7629 0.067 -26.448 0.000 -1.894 -1.632\nC(YM)[338] -1.8341 0.068 -26.886 0.000 -1.968 -1.700\nC(YM)[339] -1.8726 0.067 -27.785 0.000 -2.005 -1.741\nC(YM)[340] -1.9528 0.068 -28.843 0.000 -2.086 -1.820\nC(YM)[341] -1.8248 0.066 -27.636 0.000 -1.954 -1.695\nC(YM)[342] -1.8050 0.068 -26.584 0.000 -1.938 -1.672\nC(YM)[343] -2.0428 0.066 -30.886 0.000 -2.172 -1.913\nC(YM)[344] -1.9894 0.070 -28.328 0.000 -2.127 -1.852\nC(YM)[345] -1.8562 0.072 -25.875 0.000 -1.997 -1.716\nC(YM)[346] -2.0564 0.068 -30.190 0.000 -2.190 -1.923\nC(YM)[347] -1.8036 0.068 -26.353 0.000 -1.938 -1.669\nC(YM)[348] -1.8986 0.069 -27.506 0.000 -2.034 -1.763\nC(YM)[349] -1.8341 0.068 -26.839 0.000 -1.968 -1.700\nC(YM)[350] -1.8621 0.068 -27.575 0.000 -1.994 -1.730\nC(YM)[351] -1.9359 0.064 -30.042 0.000 -2.062 -1.810\nC(YM)[352] -2.0273 0.063 -32.286 0.000 -2.150 -1.904\nC(YM)[353] -1.8408 0.062 -29.591 0.000 -1.963 -1.719\nC(YM)[354] -1.9379 0.067 -28.944 0.000 -2.069 -1.807\nC(YM)[355] -1.9155 0.067 -28.658 0.000 -2.046 -1.784\nC(YM)[356] -1.8879 0.069 -27.200 0.000 -2.024 -1.752\nC(YM)[357] -1.8781 0.069 -27.039 0.000 -2.014 -1.742\nC(YM)[358] -2.0309 0.067 -30.131 0.000 -2.163 -1.899\nC(YM)[359] -2.0292 0.067 -30.309 0.000 -2.160 -1.898\nC(YM)[360] -1.9418 0.068 -28.729 0.000 -2.074 -1.809\nC(YM)[361] -1.7911 0.069 -25.811 0.000 -1.927 -1.655\nC(YM)[362] -1.8508 0.071 -26.104 0.000 -1.990 -1.712\nC(YM)[363] -1.7016 0.067 -25.248 0.000 -1.834 -1.570\nC(YM)[364] -1.8260 0.069 -26.600 0.000 -1.961 -1.691\nC(YM)[365] -1.8625 0.068 -27.212 0.000 -1.997 -1.728\nC(YM)[366] -1.6624 0.067 -24.686 0.000 -1.794 -1.530\nC(YM)[367] -2.0238 0.068 -29.894 0.000 -2.156 -1.891\nC(YM)[368] -1.9516 0.073 -26.763 0.000 -2.095 -1.809\nC(YM)[369] -2.0307 0.071 -28.404 0.000 -2.171 -1.891\nC(YM)[370] -1.8090 0.065 -27.972 0.000 -1.936 -1.682\nC(YM)[371] -1.9391 0.068 -28.358 0.000 -2.073 -1.805\nC(YM)[372] -2.2172 0.068 -32.628 0.000 -2.350 -2.084\nC(YM)[373] -1.8348 0.068 -26.798 0.000 -1.969 -1.701\nC(YM)[374] -1.8923 0.067 -28.114 0.000 -2.024 -1.760\nC(YM)[375] -1.8787 0.061 -30.654 0.000 -1.999 -1.759\nC(YM)[376] -1.7605 0.061 -28.724 0.000 -1.881 -1.640\nC(YM)[377] -1.9509 0.063 -30.955 0.000 -2.074 -1.827\nC(YM)[378] -1.7924 0.062 -28.737 0.000 -1.915 -1.670\nC(YM)[379] -1.7958 0.066 -27.325 0.000 -1.925 -1.667\nC(YM)[380] -1.5882 0.068 -23.489 0.000 -1.721 -1.456\nC(YM)[381] -1.9362 0.066 -29.507 0.000 -2.065 -1.808\nC(YM)[382] -1.6597 0.064 -26.125 0.000 -1.784 -1.535\nC(YM)[383] -1.3446 0.065 -20.694 0.000 -1.472 -1.217\nC(YM)[384] -1.4899 0.064 -23.255 0.000 -1.615 -1.364\nC(YM)[385] -1.6680 0.063 -26.286 0.000 -1.792 -1.544\nC(YM)[386] -1.4565 0.070 -20.688 0.000 -1.594 -1.318\nC(YM)[387] -1.5257 0.065 -23.378 0.000 -1.654 -1.398\nC(YM)[388] -1.6804 0.070 -23.971 0.000 -1.818 -1.543\nC(YM)[389] -1.7000 0.074 -22.852 0.000 -1.846 -1.554\nC(YM)[390] -1.8428 0.073 -25.406 0.000 -1.985 -1.701\nC(YM)[391] -1.8248 0.073 -24.863 0.000 -1.969 -1.681\nC(YM)[392] -1.7486 0.080 -21.879 0.000 -1.905 -1.592\nC(YM)[393] -1.9082 0.080 -23.999 0.000 -2.064 -1.752\nC(YM)[394] -1.7249 0.073 -23.507 0.000 -1.869 -1.581\nC(YM)[395] -1.7055 0.080 -21.351 0.000 -1.862 -1.549\nC(YM)[396] -1.6461 0.080 -20.519 0.000 -1.803 -1.489\nC(YM)[397] -1.8311 0.078 -23.588 0.000 -1.983 -1.679\nC(YM)[398] -1.8560 0.083 -22.419 0.000 -2.018 -1.694\nC(YM)[399] -1.8947 0.073 -25.998 0.000 -2.037 -1.752\nC(YM)[400] -1.9127 0.076 -25.151 0.000 -2.062 -1.764\nC(YM)[401] -1.7648 0.079 -22.385 0.000 -1.919 -1.610\nC(YM)[402] -1.7651 0.083 -21.326 0.000 -1.927 -1.603\nC(YM)[403] -1.7826 0.084 -21.164 0.000 -1.948 -1.618\nC(YM)[404] -1.8995 0.083 -22.914 0.000 -2.062 -1.737\nC(YM)[405] -1.8720 0.086 -21.734 0.000 -2.041 -1.703\nC(YM)[406] -1.7620 0.083 -21.219 0.000 -1.925 -1.599\nC(YM)[407] -1.9639 0.084 -23.329 0.000 -2.129 -1.799\nC(YM)[408] -2.0853 0.083 -25.118 0.000 -2.248 -1.923\nC(YM)[409] -1.8343 0.085 -21.633 0.000 -2.000 -1.668\nC(YM)[410] -1.8871 0.085 -22.295 0.000 -2.053 -1.721\nC(YM)[411] -2.0358 0.078 -26.095 0.000 -2.189 -1.883\nC(YM)[412] -1.9309 0.080 -24.124 0.000 -2.088 -1.774\nC(YM)[413] -1.7436 0.080 -21.697 0.000 -1.901 -1.586\nC(YM)[414] -1.8198 0.078 -23.222 0.000 -1.973 -1.666\nC(YM)[415] -2.0775 0.082 -25.333 0.000 -2.238 -1.917\nC(YM)[416] -2.0817 0.083 -24.935 0.000 -2.245 -1.918\nC(YM)[417] -1.9689 0.076 -25.923 0.000 -2.118 -1.820\nC(YM)[418] -2.1178 0.076 -27.793 0.000 -2.267 -1.968\nC(YM)[419] -1.9295 0.081 -23.866 0.000 -2.088 -1.771\nC(YM)[420] -1.9922 0.081 -24.719 0.000 -2.150 -1.834\nC(YM)[421] -1.8362 0.081 -22.727 0.000 -1.995 -1.678\nC(YM)[422] -1.9797 0.087 -22.765 0.000 -2.150 -1.809\nC(YM)[423] -1.8316 0.084 -21.687 0.000 -1.997 -1.666\nC(YM)[424] -1.8132 0.087 -20.777 0.000 -1.984 -1.642\nC(YM)[425] -2.0082 0.082 -24.449 0.000 -2.169 -1.847\nC(YM)[426] -2.0498 0.086 -23.739 0.000 -2.219 -1.881\nC(YM)[427] -1.9540 0.085 -22.912 0.000 -2.121 -1.787\nC(YM)[428] -1.7740 0.083 -21.389 0.000 -1.937 -1.611\nC(Newspaper)[T.Chicago Tribune] -0.1285 0.017 -7.384 0.000 -0.163 -0.094\nC(Newspaper)[T.Los Angeles Times] -0.3958 0.016 -24.428 0.000 -0.428 -0.364\nC(Newspaper)[T.New York Times] -0.2543 0.016 -15.737 0.000 -0.286 -0.223\nC(Newspaper)[T.The Washington Post] -0.0949 0.016 -5.784 0.000 -0.127 -0.063\nC(Newspaper)[T.USA Today] -0.2710 0.023 -11.985 0.000 -0.315 -0.227\nC(Newspaper)[T.Wall Street Journal] -0.1796 0.015 -12.364 0.000 -0.208 -0.151\n==============================================================================\nOmnibus: 49915.355 Durbin-Watson: 1.808\nProb(Omnibus): 0.000 Jarque-Bera (JB): 100429.512\nSkew: -0.693 Prob(JB): 0.00\nKurtosis: 4.800 Cond. No. 40.5\n==============================================================================\n\nNotes:\n[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.\n"
],
[
"# GI index\nGIindex=estimate_index('GIscore')",
" OLS Regression Results \n==============================================================================\nDep. Variable: GIscore R-squared: 0.006\nModel: OLS Adj. R-squared: 0.005\nMethod: Least Squares F-statistic: 6.091\nDate: Tue, 18 May 2021 Prob (F-statistic): 1.21e-310\nTime: 19:40:53 Log-Likelihood: -1.3129e+06\nNo. Observations: 467351 AIC: 2.627e+06\nDf Residuals: 466917 BIC: 2.631e+06\nDf Model: 433 \nCovariance Type: nonrobust \n=======================================================================================================\n coef std err t P>|t| [0.025 0.975]\n-------------------------------------------------------------------------------------------------------\nC(YM)[1] 1.4162 0.149 9.479 0.000 1.123 1.709\nC(YM)[2] 1.3820 0.154 9.001 0.000 1.081 1.683\nC(YM)[3] 1.1571 0.137 8.476 0.000 0.890 1.425\nC(YM)[4] 1.4491 0.138 10.488 0.000 1.178 1.720\nC(YM)[5] 1.4950 0.136 10.974 0.000 1.228 1.762\nC(YM)[6] 1.1636 0.140 8.329 0.000 0.890 1.437\nC(YM)[7] 1.3957 0.145 9.653 0.000 1.112 1.679\nC(YM)[8] 0.9710 0.138 7.037 0.000 0.701 1.241\nC(YM)[9] 1.6370 0.145 11.291 0.000 1.353 1.921\nC(YM)[10] 1.3069 0.140 9.355 0.000 1.033 1.581\nC(YM)[11] 1.2440 0.149 8.343 0.000 0.952 1.536\nC(YM)[12] 1.5513 0.142 10.926 0.000 1.273 1.830\nC(YM)[13] 0.8631 0.136 6.356 0.000 0.597 1.129\nC(YM)[14] 1.1993 0.153 7.861 0.000 0.900 1.498\nC(YM)[15] 1.1742 0.145 8.089 0.000 0.890 1.459\nC(YM)[16] 1.5187 0.139 10.889 0.000 1.245 1.792\nC(YM)[17] 0.9380 0.138 6.782 0.000 0.667 1.209\nC(YM)[18] 0.9597 0.138 6.941 0.000 0.689 1.231\nC(YM)[19] 0.7161 0.142 5.027 0.000 0.437 0.995\nC(YM)[20] 1.2163 0.143 8.519 0.000 0.936 1.496\nC(YM)[21] 1.2272 0.145 8.464 0.000 0.943 1.511\nC(YM)[22] 1.0144 0.141 7.180 0.000 0.738 1.291\nC(YM)[23] 1.3915 0.140 9.927 0.000 1.117 1.666\nC(YM)[24] 1.0697 0.136 7.876 0.000 0.804 1.336\nC(YM)[25] 1.2176 0.135 9.032 0.000 0.953 1.482\nC(YM)[26] 1.2789 0.132 9.694 0.000 1.020 1.537\nC(YM)[27] 1.1734 0.123 9.569 0.000 0.933 1.414\nC(YM)[28] 1.0235 0.122 8.357 0.000 0.783 1.263\nC(YM)[29] 0.8678 0.129 6.724 0.000 0.615 1.121\nC(YM)[30] 0.9340 0.122 7.658 0.000 0.695 1.173\nC(YM)[31] 1.0785 0.121 8.915 0.000 0.841 1.316\nC(YM)[32] 1.2331 0.128 9.643 0.000 0.982 1.484\nC(YM)[33] 1.2041 0.129 9.339 0.000 0.951 1.457\nC(YM)[34] 0.9003 0.122 7.366 0.000 0.661 1.140\nC(YM)[35] 1.0330 0.127 8.108 0.000 0.783 1.283\nC(YM)[36] 1.1726 0.133 8.844 0.000 0.913 1.432\nC(YM)[37] 0.9192 0.122 7.515 0.000 0.679 1.159\nC(YM)[38] 1.2788 0.121 10.563 0.000 1.042 1.516\nC(YM)[39] 1.0126 0.122 8.307 0.000 0.774 1.252\nC(YM)[40] 1.4219 0.125 11.413 0.000 1.178 1.666\nC(YM)[41] 1.2173 0.121 10.073 0.000 0.980 1.454\nC(YM)[42] 1.2797 0.117 10.959 0.000 1.051 1.509\nC(YM)[43] 1.1131 0.127 8.789 0.000 0.865 1.361\nC(YM)[44] 1.0300 0.121 8.541 0.000 0.794 1.266\nC(YM)[45] 1.0381 0.121 8.586 0.000 0.801 1.275\nC(YM)[46] 1.0569 0.117 9.003 0.000 0.827 1.287\nC(YM)[47] 1.2647 0.121 10.462 0.000 1.028 1.502\nC(YM)[48] 1.0827 0.113 9.616 0.000 0.862 1.303\nC(YM)[49] 1.0889 0.117 9.318 0.000 0.860 1.318\nC(YM)[50] 0.8644 0.120 7.200 0.000 0.629 1.100\nC(YM)[51] 0.9400 0.109 8.595 0.000 0.726 1.154\nC(YM)[52] 1.0691 0.113 9.438 0.000 0.847 1.291\nC(YM)[53] 0.7392 0.114 6.493 0.000 0.516 0.962\nC(YM)[54] 0.9202 0.115 8.023 0.000 0.695 1.145\nC(YM)[55] 1.1639 0.116 9.999 0.000 0.936 1.392\nC(YM)[56] 0.7762 0.116 6.712 0.000 0.550 1.003\nC(YM)[57] 0.9037 0.119 7.619 0.000 0.671 1.136\nC(YM)[58] 1.3326 0.114 11.645 0.000 1.108 1.557\nC(YM)[59] 1.0724 0.113 9.510 0.000 0.851 1.293\nC(YM)[60] 1.1908 0.120 9.897 0.000 0.955 1.427\nC(YM)[61] 1.2271 0.117 10.450 0.000 0.997 1.457\nC(YM)[62] 0.7885 0.118 6.709 0.000 0.558 1.019\nC(YM)[63] 0.7245 0.109 6.644 0.000 0.511 0.938\nC(YM)[64] 1.0176 0.110 9.286 0.000 0.803 1.232\nC(YM)[65] 1.1091 0.114 9.730 0.000 0.886 1.333\nC(YM)[66] 1.3034 0.117 11.175 0.000 1.075 1.532\nC(YM)[67] 0.9107 0.111 8.173 0.000 0.692 1.129\nC(YM)[68] 0.8310 0.122 6.827 0.000 0.592 1.070\nC(YM)[69] 0.8432 0.117 7.196 0.000 0.614 1.073\nC(YM)[70] 0.9813 0.116 8.466 0.000 0.754 1.208\nC(YM)[71] 0.8759 0.118 7.414 0.000 0.644 1.107\nC(YM)[72] 1.1145 0.112 9.924 0.000 0.894 1.335\nC(YM)[73] 1.1550 0.117 9.836 0.000 0.925 1.385\nC(YM)[74] 0.9322 0.123 7.595 0.000 0.692 1.173\nC(YM)[75] 1.0248 0.121 8.449 0.000 0.787 1.263\nC(YM)[76] 1.1585 0.114 10.125 0.000 0.934 1.383\nC(YM)[77] 1.1266 0.106 10.584 0.000 0.918 1.335\nC(YM)[78] 1.3242 0.109 12.139 0.000 1.110 1.538\nC(YM)[79] 0.8513 0.111 7.659 0.000 0.633 1.069\nC(YM)[80] 0.7488 0.113 6.649 0.000 0.528 0.970\nC(YM)[81] 0.7214 0.115 6.283 0.000 0.496 0.947\nC(YM)[82] 0.9729 0.114 8.546 0.000 0.750 1.196\nC(YM)[83] 0.9244 0.114 8.122 0.000 0.701 1.147\nC(YM)[84] 1.2628 0.125 10.120 0.000 1.018 1.507\nC(YM)[85] 1.3090 0.118 11.063 0.000 1.077 1.541\nC(YM)[86] 1.1496 0.122 9.407 0.000 0.910 1.389\nC(YM)[87] 0.8954 0.115 7.795 0.000 0.670 1.120\nC(YM)[88] 1.1989 0.120 9.953 0.000 0.963 1.435\nC(YM)[89] 1.3088 0.125 10.502 0.000 1.065 1.553\nC(YM)[90] 1.1676 0.119 9.837 0.000 0.935 1.400\nC(YM)[91] 0.8357 0.122 6.866 0.000 0.597 1.074\nC(YM)[92] 0.8844 0.126 7.017 0.000 0.637 1.131\nC(YM)[93] 0.8437 0.125 6.741 0.000 0.598 1.089\nC(YM)[94] 1.7515 0.112 15.616 0.000 1.532 1.971\nC(YM)[95] 1.1155 0.124 9.017 0.000 0.873 1.358\nC(YM)[96] 1.4694 0.126 11.657 0.000 1.222 1.716\nC(YM)[97] 1.3034 0.126 10.326 0.000 1.056 1.551\nC(YM)[98] 1.0365 0.129 8.009 0.000 0.783 1.290\nC(YM)[99] 1.0957 0.123 8.917 0.000 0.855 1.337\nC(YM)[100] 1.3114 0.128 10.246 0.000 1.061 1.562\nC(YM)[101] 1.1873 0.123 9.636 0.000 0.946 1.429\nC(YM)[102] 1.2923 0.123 10.482 0.000 1.051 1.534\nC(YM)[103] 1.3041 0.130 10.032 0.000 1.049 1.559\nC(YM)[104] 1.3249 0.134 9.902 0.000 1.063 1.587\nC(YM)[105] 1.5924 0.122 13.021 0.000 1.353 1.832\nC(YM)[106] 1.3818 0.120 11.490 0.000 1.146 1.617\nC(YM)[107] 1.3717 0.122 11.287 0.000 1.134 1.610\nC(YM)[108] 1.6078 0.121 13.276 0.000 1.370 1.845\nC(YM)[109] 1.5598 0.122 12.802 0.000 1.321 1.799\nC(YM)[110] 1.3865 0.125 11.073 0.000 1.141 1.632\nC(YM)[111] 1.1955 0.119 10.072 0.000 0.963 1.428\nC(YM)[112] 1.5177 0.125 12.117 0.000 1.272 1.763\nC(YM)[113] 1.2356 0.121 10.200 0.000 0.998 1.473\nC(YM)[114] 1.3061 0.124 10.492 0.000 1.062 1.550\nC(YM)[115] 1.3454 0.125 10.727 0.000 1.100 1.591\nC(YM)[116] 1.5410 0.125 12.279 0.000 1.295 1.787\nC(YM)[117] 1.3713 0.124 11.066 0.000 1.128 1.614\nC(YM)[118] 1.3728 0.125 10.955 0.000 1.127 1.618\nC(YM)[119] 1.5051 0.124 12.158 0.000 1.262 1.748\nC(YM)[120] 1.3594 0.126 10.815 0.000 1.113 1.606\nC(YM)[121] 1.2430 0.146 8.494 0.000 0.956 1.530\nC(YM)[122] 1.3261 0.141 9.421 0.000 1.050 1.602\nC(YM)[123] 1.1875 0.137 8.648 0.000 0.918 1.457\nC(YM)[124] 1.3807 0.145 9.537 0.000 1.097 1.664\nC(YM)[125] 1.2938 0.145 8.911 0.000 1.009 1.578\nC(YM)[126] 1.5604 0.142 10.990 0.000 1.282 1.839\nC(YM)[127] 1.1601 0.141 8.225 0.000 0.884 1.437\nC(YM)[128] 1.2784 0.136 9.408 0.000 1.012 1.545\nC(YM)[129] 1.3157 0.147 8.926 0.000 1.027 1.605\nC(YM)[130] 1.2024 0.141 8.500 0.000 0.925 1.480\nC(YM)[131] 1.0171 0.142 7.154 0.000 0.738 1.296\nC(YM)[132] 1.2372 0.151 8.176 0.000 0.941 1.534\nC(YM)[133] 1.6262 0.151 10.742 0.000 1.329 1.923\nC(YM)[134] 0.9203 0.156 5.904 0.000 0.615 1.226\nC(YM)[135] 1.5126 0.145 10.420 0.000 1.228 1.797\nC(YM)[136] 1.3131 0.142 9.219 0.000 1.034 1.592\nC(YM)[137] 1.4114 0.144 9.824 0.000 1.130 1.693\nC(YM)[138] 1.4914 0.140 10.628 0.000 1.216 1.766\nC(YM)[139] 1.2658 0.146 8.682 0.000 0.980 1.552\nC(YM)[140] 1.3490 0.140 9.625 0.000 1.074 1.624\nC(YM)[141] 1.1705 0.152 7.701 0.000 0.873 1.468\nC(YM)[142] 1.1030 0.145 7.629 0.000 0.820 1.386\nC(YM)[143] 1.3249 0.147 9.009 0.000 1.037 1.613\nC(YM)[144] 1.3344 0.149 8.927 0.000 1.041 1.627\nC(YM)[145] 1.4851 0.151 9.825 0.000 1.189 1.781\nC(YM)[146] 1.5290 0.146 10.480 0.000 1.243 1.815\nC(YM)[147] 1.3816 0.143 9.633 0.000 1.100 1.663\nC(YM)[148] 1.4277 0.143 9.954 0.000 1.147 1.709\nC(YM)[149] 1.1666 0.142 8.214 0.000 0.888 1.445\nC(YM)[150] 1.6621 0.129 12.853 0.000 1.409 1.916\nC(YM)[151] 1.7044 0.143 11.942 0.000 1.425 1.984\nC(YM)[152] 1.4421 0.147 9.844 0.000 1.155 1.729\nC(YM)[153] 1.4636 0.145 10.112 0.000 1.180 1.747\nC(YM)[154] 1.5812 0.145 10.892 0.000 1.297 1.866\nC(YM)[155] 1.2867 0.147 8.764 0.000 0.999 1.574\nC(YM)[156] 1.5740 0.145 10.880 0.000 1.290 1.858\nC(YM)[157] 1.3884 0.145 9.600 0.000 1.105 1.672\nC(YM)[158] 1.7751 0.148 12.032 0.000 1.486 2.064\nC(YM)[159] 1.7892 0.134 13.311 0.000 1.526 2.053\nC(YM)[160] 1.4536 0.134 10.854 0.000 1.191 1.716\nC(YM)[161] 1.0289 0.134 7.672 0.000 0.766 1.292\nC(YM)[162] 1.6019 0.131 12.275 0.000 1.346 1.858\nC(YM)[163] 1.5584 0.134 11.671 0.000 1.297 1.820\nC(YM)[164] 1.6869 0.147 11.455 0.000 1.398 1.976\nC(YM)[165] 1.3108 0.138 9.480 0.000 1.040 1.582\nC(YM)[166] 1.3610 0.135 10.105 0.000 1.097 1.625\nC(YM)[167] 1.4147 0.140 10.126 0.000 1.141 1.689\nC(YM)[168] 1.4154 0.141 10.005 0.000 1.138 1.693\nC(YM)[169] 1.5373 0.146 10.514 0.000 1.251 1.824\nC(YM)[170] 0.9892 0.142 6.950 0.000 0.710 1.268\nC(YM)[171] 1.3180 0.132 9.973 0.000 1.059 1.577\nC(YM)[172] 1.4281 0.139 10.241 0.000 1.155 1.701\nC(YM)[173] 1.2475 0.134 9.327 0.000 0.985 1.510\nC(YM)[174] 1.4915 0.133 11.241 0.000 1.231 1.752\nC(YM)[175] 1.3016 0.129 10.124 0.000 1.050 1.554\nC(YM)[176] 1.0332 0.130 7.956 0.000 0.779 1.288\nC(YM)[177] 1.3996 0.131 10.692 0.000 1.143 1.656\nC(YM)[178] 1.4234 0.130 10.975 0.000 1.169 1.678\nC(YM)[179] 1.2497 0.138 9.040 0.000 0.979 1.521\nC(YM)[180] 1.5154 0.142 10.683 0.000 1.237 1.793\nC(YM)[181] 1.4438 0.140 10.281 0.000 1.169 1.719\nC(YM)[182] 1.5767 0.140 11.256 0.000 1.302 1.851\nC(YM)[183] 1.3567 0.127 10.697 0.000 1.108 1.605\nC(YM)[184] 1.2072 0.144 8.408 0.000 0.926 1.489\nC(YM)[185] 1.5218 0.132 11.557 0.000 1.264 1.780\nC(YM)[186] 1.6753 0.128 13.125 0.000 1.425 1.925\nC(YM)[187] 1.3797 0.141 9.812 0.000 1.104 1.655\nC(YM)[188] 1.1271 0.138 8.179 0.000 0.857 1.397\nC(YM)[189] 1.0600 0.128 8.267 0.000 0.809 1.311\nC(YM)[190] 1.3661 0.129 10.551 0.000 1.112 1.620\nC(YM)[191] 1.4088 0.134 10.536 0.000 1.147 1.671\nC(YM)[192] 1.6100 0.133 12.103 0.000 1.349 1.871\nC(YM)[193] 1.4214 0.131 10.860 0.000 1.165 1.678\nC(YM)[194] 1.4020 0.144 9.723 0.000 1.119 1.685\nC(YM)[195] 1.1924 0.126 9.471 0.000 0.946 1.439\nC(YM)[196] 1.2791 0.131 9.800 0.000 1.023 1.535\nC(YM)[197] 1.4309 0.124 11.518 0.000 1.187 1.674\nC(YM)[198] 1.2073 0.128 9.450 0.000 0.957 1.458\nC(YM)[199] 1.5666 0.129 12.189 0.000 1.315 1.818\nC(YM)[200] 1.1358 0.131 8.682 0.000 0.879 1.392\nC(YM)[201] 0.9274 0.144 6.421 0.000 0.644 1.210\nC(YM)[202] 0.8334 0.147 5.684 0.000 0.546 1.121\nC(YM)[203] 1.2634 0.146 8.663 0.000 0.978 1.549\nC(YM)[204] 1.2015 0.142 8.444 0.000 0.923 1.480\nC(YM)[205] 1.0196 0.134 7.623 0.000 0.757 1.282\nC(YM)[206] 1.1329 0.126 8.966 0.000 0.885 1.381\nC(YM)[207] 1.2099 0.127 9.526 0.000 0.961 1.459\nC(YM)[208] 0.9253 0.133 6.941 0.000 0.664 1.187\nC(YM)[209] 1.2754 0.123 10.387 0.000 1.035 1.516\nC(YM)[210] 1.2861 0.127 10.155 0.000 1.038 1.534\nC(YM)[211] 1.4592 0.118 12.351 0.000 1.228 1.691\nC(YM)[212] 1.5717 0.127 12.397 0.000 1.323 1.820\nC(YM)[213] 1.3180 0.130 10.105 0.000 1.062 1.574\nC(YM)[214] 1.2184 0.122 9.976 0.000 0.979 1.458\nC(YM)[215] 1.6545 0.126 13.100 0.000 1.407 1.902\nC(YM)[216] 1.3391 0.127 10.505 0.000 1.089 1.589\nC(YM)[217] 1.3860 0.124 11.142 0.000 1.142 1.630\nC(YM)[218] 1.4416 0.133 10.863 0.000 1.182 1.702\nC(YM)[219] 1.1313 0.131 8.645 0.000 0.875 1.388\nC(YM)[220] 1.5938 0.129 12.313 0.000 1.340 1.847\nC(YM)[221] 1.2366 0.129 9.605 0.000 0.984 1.489\nC(YM)[222] 1.3805 0.127 10.901 0.000 1.132 1.629\nC(YM)[223] 1.3321 0.128 10.440 0.000 1.082 1.582\nC(YM)[224] 1.4654 0.126 11.660 0.000 1.219 1.712\nC(YM)[225] 1.3020 0.120 10.883 0.000 1.067 1.536\nC(YM)[226] 1.2772 0.116 11.044 0.000 1.051 1.504\nC(YM)[227] 1.4934 0.117 12.764 0.000 1.264 1.723\nC(YM)[228] 1.4021 0.120 11.732 0.000 1.168 1.636\nC(YM)[229] 1.3870 0.128 10.838 0.000 1.136 1.638\nC(YM)[230] 1.3207 0.126 10.463 0.000 1.073 1.568\nC(YM)[231] 1.6168 0.124 13.024 0.000 1.373 1.860\nC(YM)[232] 1.3487 0.122 11.027 0.000 1.109 1.588\nC(YM)[233] 1.5427 0.127 12.188 0.000 1.295 1.791\nC(YM)[234] 1.2608 0.128 9.827 0.000 1.009 1.512\nC(YM)[235] 1.1976 0.131 9.114 0.000 0.940 1.455\nC(YM)[236] 1.4890 0.135 11.055 0.000 1.225 1.753\nC(YM)[237] 1.1908 0.127 9.372 0.000 0.942 1.440\nC(YM)[238] 0.9830 0.122 8.087 0.000 0.745 1.221\nC(YM)[239] 1.6371 0.133 12.321 0.000 1.377 1.898\nC(YM)[240] 1.6669 0.123 13.511 0.000 1.425 1.909\nC(YM)[241] 1.5204 0.133 11.409 0.000 1.259 1.782\nC(YM)[242] 1.2800 0.132 9.732 0.000 1.022 1.538\nC(YM)[243] 1.4801 0.122 12.164 0.000 1.242 1.719\nC(YM)[244] 1.5672 0.125 12.549 0.000 1.322 1.812\nC(YM)[245] 1.2993 0.133 9.801 0.000 1.039 1.559\nC(YM)[246] 1.6907 0.123 13.718 0.000 1.449 1.932\nC(YM)[247] 1.5805 0.134 11.796 0.000 1.318 1.843\nC(YM)[248] 1.3873 0.140 9.921 0.000 1.113 1.661\nC(YM)[249] 1.1475 0.139 8.282 0.000 0.876 1.419\nC(YM)[250] 1.6883 0.137 12.308 0.000 1.419 1.957\nC(YM)[251] 1.3468 0.140 9.636 0.000 1.073 1.621\nC(YM)[252] 1.6537 0.135 12.249 0.000 1.389 1.918\nC(YM)[253] 1.4962 0.138 10.811 0.000 1.225 1.767\nC(YM)[254] 1.4901 0.139 10.713 0.000 1.217 1.763\nC(YM)[255] 1.4937 0.135 11.066 0.000 1.229 1.758\nC(YM)[256] 1.4290 0.143 10.021 0.000 1.149 1.708\nC(YM)[257] 1.7062 0.141 12.099 0.000 1.430 1.983\nC(YM)[258] 1.4446 0.132 10.911 0.000 1.185 1.704\nC(YM)[259] 1.2952 0.143 9.041 0.000 1.014 1.576\nC(YM)[260] 1.5359 0.137 11.205 0.000 1.267 1.805\nC(YM)[261] 1.3374 0.139 9.614 0.000 1.065 1.610\nC(YM)[262] 1.3830 0.140 9.863 0.000 1.108 1.658\nC(YM)[263] 1.5498 0.138 11.190 0.000 1.278 1.821\nC(YM)[264] 1.7136 0.134 12.815 0.000 1.452 1.976\nC(YM)[265] 1.6392 0.138 11.844 0.000 1.368 1.910\nC(YM)[266] 1.4237 0.146 9.737 0.000 1.137 1.710\nC(YM)[267] 1.5254 0.128 11.878 0.000 1.274 1.777\nC(YM)[268] 1.5000 0.133 11.262 0.000 1.239 1.761\nC(YM)[269] 1.5237 0.136 11.204 0.000 1.257 1.790\nC(YM)[270] 1.3980 0.134 10.396 0.000 1.134 1.662\nC(YM)[271] 1.3314 0.138 9.622 0.000 1.060 1.603\nC(YM)[272] 1.3150 0.135 9.774 0.000 1.051 1.579\nC(YM)[273] 1.3291 0.134 9.919 0.000 1.066 1.592\nC(YM)[274] 1.2222 0.137 8.931 0.000 0.954 1.490\nC(YM)[275] 1.5741 0.138 11.439 0.000 1.304 1.844\nC(YM)[276] 1.4523 0.141 10.321 0.000 1.176 1.728\nC(YM)[277] 1.0009 0.143 7.009 0.000 0.721 1.281\nC(YM)[278] 1.2349 0.137 8.992 0.000 0.966 1.504\nC(YM)[279] 1.4947 0.128 11.686 0.000 1.244 1.745\nC(YM)[280] 1.0327 0.127 8.148 0.000 0.784 1.281\nC(YM)[281] 1.3680 0.140 9.803 0.000 1.095 1.642\nC(YM)[282] 1.1843 0.136 8.705 0.000 0.918 1.451\nC(YM)[283] 1.0692 0.130 8.197 0.000 0.814 1.325\nC(YM)[284] 1.4958 0.139 10.723 0.000 1.222 1.769\nC(YM)[285] 1.0937 0.121 9.037 0.000 0.856 1.331\nC(YM)[286] 1.2376 0.132 9.388 0.000 0.979 1.496\nC(YM)[287] 1.4283 0.146 9.796 0.000 1.143 1.714\nC(YM)[288] 1.0684 0.142 7.502 0.000 0.789 1.347\nC(YM)[289] 1.0078 0.146 6.913 0.000 0.722 1.294\nC(YM)[290] 1.3249 0.146 9.081 0.000 1.039 1.611\nC(YM)[291] 1.3996 0.136 10.281 0.000 1.133 1.666\nC(YM)[292] 1.5744 0.139 11.304 0.000 1.301 1.847\nC(YM)[293] 1.3684 0.146 9.369 0.000 1.082 1.655\nC(YM)[294] 1.3188 0.134 9.868 0.000 1.057 1.581\nC(YM)[295] 1.5010 0.139 10.816 0.000 1.229 1.773\nC(YM)[296] 1.2061 0.145 8.309 0.000 0.922 1.491\nC(YM)[297] 1.3159 0.138 9.509 0.000 1.045 1.587\nC(YM)[298] 1.3175 0.132 10.007 0.000 1.059 1.576\nC(YM)[299] 1.3024 0.149 8.728 0.000 1.010 1.595\nC(YM)[300] 1.1488 0.139 8.237 0.000 0.875 1.422\nC(YM)[301] 1.0157 0.115 8.861 0.000 0.791 1.240\nC(YM)[302] 1.1134 0.113 9.838 0.000 0.892 1.335\nC(YM)[303] 1.1652 0.099 11.740 0.000 0.971 1.360\nC(YM)[304] 0.5623 0.098 5.715 0.000 0.369 0.755\nC(YM)[305] 0.7057 0.093 7.603 0.000 0.524 0.888\nC(YM)[306] 0.6988 0.100 6.986 0.000 0.503 0.895\nC(YM)[307] 1.1423 0.103 11.125 0.000 0.941 1.344\nC(YM)[308] 1.0665 0.112 9.544 0.000 0.847 1.285\nC(YM)[309] 1.1928 0.104 11.498 0.000 0.989 1.396\nC(YM)[310] 1.0811 0.101 10.754 0.000 0.884 1.278\nC(YM)[311] 1.0497 0.109 9.627 0.000 0.836 1.263\nC(YM)[312] 1.3734 0.107 12.830 0.000 1.164 1.583\nC(YM)[313] 1.4062 0.107 13.104 0.000 1.196 1.616\nC(YM)[314] 1.0069 0.113 8.876 0.000 0.785 1.229\nC(YM)[315] 0.9631 0.099 9.772 0.000 0.770 1.156\nC(YM)[316] 0.7009 0.109 6.417 0.000 0.487 0.915\nC(YM)[317] 1.2962 0.109 11.864 0.000 1.082 1.510\nC(YM)[318] 1.0193 0.101 10.080 0.000 0.821 1.217\nC(YM)[319] 0.8560 0.112 7.650 0.000 0.637 1.075\nC(YM)[320] 0.9903 0.112 8.815 0.000 0.770 1.210\nC(YM)[321] 0.7541 0.106 7.129 0.000 0.547 0.961\nC(YM)[322] 0.8676 0.111 7.816 0.000 0.650 1.085\nC(YM)[323] 1.0383 0.112 9.291 0.000 0.819 1.257\nC(YM)[324] 0.9779 0.108 9.040 0.000 0.766 1.190\nC(YM)[325] 1.0746 0.112 9.560 0.000 0.854 1.295\nC(YM)[326] 1.3516 0.113 11.919 0.000 1.129 1.574\nC(YM)[327] 1.2659 0.111 11.360 0.000 1.047 1.484\nC(YM)[328] 1.3824 0.112 12.365 0.000 1.163 1.602\nC(YM)[329] 1.1230 0.109 10.337 0.000 0.910 1.336\nC(YM)[330] 0.9340 0.114 8.227 0.000 0.711 1.157\nC(YM)[331] 0.7766 0.110 7.055 0.000 0.561 0.992\nC(YM)[332] 1.1270 0.113 9.982 0.000 0.906 1.348\nC(YM)[333] 1.1662 0.122 9.529 0.000 0.926 1.406\nC(YM)[334] 1.1438 0.111 10.292 0.000 0.926 1.362\nC(YM)[335] 1.0399 0.109 9.499 0.000 0.825 1.255\nC(YM)[336] 1.2184 0.108 11.277 0.000 1.007 1.430\nC(YM)[337] 1.2278 0.103 11.879 0.000 1.025 1.430\nC(YM)[338] 1.2388 0.106 11.711 0.000 1.031 1.446\nC(YM)[339] 1.1959 0.105 11.443 0.000 0.991 1.401\nC(YM)[340] 1.0333 0.105 9.843 0.000 0.828 1.239\nC(YM)[341] 1.2327 0.102 12.040 0.000 1.032 1.433\nC(YM)[342] 1.3603 0.105 12.921 0.000 1.154 1.567\nC(YM)[343] 1.1169 0.103 10.890 0.000 0.916 1.318\nC(YM)[344] 1.1847 0.109 10.880 0.000 0.971 1.398\nC(YM)[345] 1.2059 0.111 10.841 0.000 0.988 1.424\nC(YM)[346] 1.0233 0.106 9.689 0.000 0.816 1.230\nC(YM)[347] 1.4031 0.106 13.221 0.000 1.195 1.611\nC(YM)[348] 1.3048 0.107 12.191 0.000 1.095 1.515\nC(YM)[349] 1.3584 0.106 12.820 0.000 1.151 1.566\nC(YM)[350] 1.0916 0.105 10.425 0.000 0.886 1.297\nC(YM)[351] 1.2308 0.100 12.317 0.000 1.035 1.427\nC(YM)[352] 1.3085 0.097 13.439 0.000 1.118 1.499\nC(YM)[353] 1.3013 0.096 13.490 0.000 1.112 1.490\nC(YM)[354] 1.2119 0.104 11.674 0.000 1.008 1.415\nC(YM)[355] 1.3165 0.104 12.703 0.000 1.113 1.520\nC(YM)[356] 1.4154 0.108 13.150 0.000 1.204 1.626\nC(YM)[357] 1.3398 0.108 12.440 0.000 1.129 1.551\nC(YM)[358] 1.1239 0.105 10.753 0.000 0.919 1.329\nC(YM)[359] 1.0080 0.104 9.710 0.000 0.805 1.212\nC(YM)[360] 1.1919 0.105 11.372 0.000 0.986 1.397\nC(YM)[361] 1.2597 0.108 11.707 0.000 1.049 1.471\nC(YM)[362] 1.1702 0.110 10.644 0.000 0.955 1.386\nC(YM)[363] 1.1661 0.105 11.158 0.000 0.961 1.371\nC(YM)[364] 1.2372 0.106 11.623 0.000 1.029 1.446\nC(YM)[365] 1.6512 0.106 15.558 0.000 1.443 1.859\nC(YM)[366] 1.4552 0.104 13.936 0.000 1.251 1.660\nC(YM)[367] 1.3726 0.105 13.076 0.000 1.167 1.578\nC(YM)[368] 1.0176 0.113 9.000 0.000 0.796 1.239\nC(YM)[369] 1.2752 0.111 11.503 0.000 1.058 1.492\nC(YM)[370] 1.3143 0.100 13.106 0.000 1.118 1.511\nC(YM)[371] 1.0557 0.106 9.957 0.000 0.848 1.263\nC(YM)[372] 1.0665 0.105 10.121 0.000 0.860 1.273\nC(YM)[373] 0.9439 0.106 8.891 0.000 0.736 1.152\nC(YM)[374] 1.1882 0.104 11.384 0.000 0.984 1.393\nC(YM)[375] 1.3401 0.095 14.101 0.000 1.154 1.526\nC(YM)[376] 1.4795 0.095 15.568 0.000 1.293 1.666\nC(YM)[377] 1.3273 0.098 13.582 0.000 1.136 1.519\nC(YM)[378] 1.2147 0.097 12.560 0.000 1.025 1.404\nC(YM)[379] 1.3869 0.102 13.610 0.000 1.187 1.587\nC(YM)[380] 1.4246 0.105 13.588 0.000 1.219 1.630\nC(YM)[381] 1.5826 0.102 15.554 0.000 1.383 1.782\nC(YM)[382] 1.5658 0.099 15.895 0.000 1.373 1.759\nC(YM)[383] 1.2025 0.101 11.935 0.000 1.005 1.400\nC(YM)[384] 1.4692 0.099 14.789 0.000 1.274 1.664\nC(YM)[385] 1.3235 0.098 13.451 0.000 1.131 1.516\nC(YM)[386] 1.5057 0.109 13.793 0.000 1.292 1.720\nC(YM)[387] 1.5823 0.101 15.636 0.000 1.384 1.781\nC(YM)[388] 1.2281 0.109 11.298 0.000 1.015 1.441\nC(YM)[389] 1.6354 0.115 14.177 0.000 1.409 1.862\nC(YM)[390] 1.4138 0.112 12.571 0.000 1.193 1.634\nC(YM)[391] 1.2747 0.114 11.201 0.000 1.052 1.498\nC(YM)[392] 1.3086 0.124 10.560 0.000 1.066 1.552\nC(YM)[393] 1.5679 0.123 12.718 0.000 1.326 1.810\nC(YM)[394] 1.3349 0.114 11.732 0.000 1.112 1.558\nC(YM)[395] 1.3964 0.124 11.274 0.000 1.154 1.639\nC(YM)[396] 1.5865 0.124 12.753 0.000 1.343 1.830\nC(YM)[397] 1.2857 0.120 10.681 0.000 1.050 1.522\nC(YM)[398] 1.0806 0.128 8.418 0.000 0.829 1.332\nC(YM)[399] 1.0575 0.113 9.358 0.000 0.836 1.279\nC(YM)[400] 1.4333 0.118 12.155 0.000 1.202 1.664\nC(YM)[401] 1.5247 0.122 12.472 0.000 1.285 1.764\nC(YM)[402] 1.3784 0.128 10.741 0.000 1.127 1.630\nC(YM)[403] 1.2031 0.131 9.212 0.000 0.947 1.459\nC(YM)[404] 1.2136 0.129 9.441 0.000 0.962 1.466\nC(YM)[405] 1.5807 0.134 11.835 0.000 1.319 1.842\nC(YM)[406] 1.4473 0.129 11.240 0.000 1.195 1.700\nC(YM)[407] 1.2348 0.131 9.460 0.000 0.979 1.491\nC(YM)[408] 1.0966 0.129 8.519 0.000 0.844 1.349\nC(YM)[409] 1.6643 0.131 12.658 0.000 1.407 1.922\nC(YM)[410] 1.7076 0.131 13.011 0.000 1.450 1.965\nC(YM)[411] 1.3941 0.121 11.524 0.000 1.157 1.631\nC(YM)[412] 1.6384 0.124 13.201 0.000 1.395 1.882\nC(YM)[413] 1.7651 0.125 14.165 0.000 1.521 2.009\nC(YM)[414] 1.4885 0.122 12.249 0.000 1.250 1.727\nC(YM)[415] 1.4438 0.127 11.354 0.000 1.195 1.693\nC(YM)[416] 1.2405 0.129 9.583 0.000 0.987 1.494\nC(YM)[417] 1.4442 0.118 12.262 0.000 1.213 1.675\nC(YM)[418] 1.4112 0.118 11.943 0.000 1.180 1.643\nC(YM)[419] 1.4153 0.125 11.290 0.000 1.170 1.661\nC(YM)[420] 1.3500 0.125 10.803 0.000 1.105 1.595\nC(YM)[421] 1.5670 0.125 12.508 0.000 1.321 1.813\nC(YM)[422] 1.2981 0.135 9.627 0.000 1.034 1.562\nC(YM)[423] 1.6299 0.131 12.446 0.000 1.373 1.887\nC(YM)[424] 1.8705 0.135 13.823 0.000 1.605 2.136\nC(YM)[425] 1.7561 0.127 13.788 0.000 1.506 2.006\nC(YM)[426] 1.4802 0.134 11.055 0.000 1.218 1.743\nC(YM)[427] 1.4868 0.132 11.244 0.000 1.228 1.746\nC(YM)[428] 1.5088 0.129 11.732 0.000 1.257 1.761\nC(Newspaper)[T.Chicago Tribune] -0.2815 0.027 -10.434 0.000 -0.334 -0.229\nC(Newspaper)[T.Los Angeles Times] -0.4685 0.025 -18.647 0.000 -0.518 -0.419\nC(Newspaper)[T.New York Times] -0.3261 0.025 -13.015 0.000 -0.375 -0.277\nC(Newspaper)[T.The Washington Post] -0.1893 0.025 -7.437 0.000 -0.239 -0.139\nC(Newspaper)[T.USA Today] -0.6491 0.035 -18.516 0.000 -0.718 -0.580\nC(Newspaper)[T.Wall Street Journal] -0.0204 0.023 -0.907 0.364 -0.065 0.024\n==============================================================================\nOmnibus: 10206.164 Durbin-Watson: 1.838\nProb(Omnibus): 0.000 Jarque-Bera (JB): 22009.468\nSkew: 0.085 Prob(JB): 0.00\nKurtosis: 4.050 Cond. No. 40.5\n==============================================================================\n\nNotes:\n[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.\n"
],
[
"# LSD index\nLSDindex=estimate_index('LSDscore')",
" OLS Regression Results \n==============================================================================\nDep. Variable: LSDscore R-squared: 0.006\nModel: OLS Adj. R-squared: 0.005\nMethod: Least Squares F-statistic: 6.773\nDate: Tue, 18 May 2021 Prob (F-statistic): 0.00\nTime: 19:41:38 Log-Likelihood: -1.2414e+06\nNo. Observations: 467351 AIC: 2.484e+06\nDf Residuals: 466917 BIC: 2.488e+06\nDf Model: 433 \nCovariance Type: nonrobust \n=======================================================================================================\n coef std err t P>|t| [0.025 0.975]\n-------------------------------------------------------------------------------------------------------\nC(YM)[1] 0.2298 0.128 1.792 0.073 -0.021 0.481\nC(YM)[2] 0.0147 0.132 0.112 0.911 -0.244 0.273\nC(YM)[3] -0.1378 0.117 -1.176 0.239 -0.367 0.092\nC(YM)[4] 0.0368 0.119 0.311 0.756 -0.196 0.269\nC(YM)[5] 0.2483 0.117 2.124 0.034 0.019 0.477\nC(YM)[6] -0.1340 0.120 -1.118 0.264 -0.369 0.101\nC(YM)[7] 0.0526 0.124 0.424 0.671 -0.191 0.296\nC(YM)[8] -0.2815 0.118 -2.377 0.017 -0.514 -0.049\nC(YM)[9] 0.2040 0.124 1.639 0.101 -0.040 0.448\nC(YM)[10] 0.1486 0.120 1.240 0.215 -0.086 0.384\nC(YM)[11] 0.0203 0.128 0.159 0.874 -0.230 0.271\nC(YM)[12] 0.2828 0.122 2.321 0.020 0.044 0.522\nC(YM)[13] -0.2938 0.117 -2.522 0.012 -0.522 -0.065\nC(YM)[14] -0.0759 0.131 -0.579 0.562 -0.332 0.181\nC(YM)[15] -0.0515 0.125 -0.414 0.679 -0.296 0.193\nC(YM)[16] 0.3578 0.120 2.989 0.003 0.123 0.592\nC(YM)[17] -0.2496 0.119 -2.103 0.035 -0.482 -0.017\nC(YM)[18] -0.1725 0.119 -1.454 0.146 -0.405 0.060\nC(YM)[19] -0.4938 0.122 -4.039 0.000 -0.733 -0.254\nC(YM)[20] 0.0581 0.123 0.474 0.636 -0.182 0.298\nC(YM)[21] 0.0805 0.124 0.647 0.518 -0.163 0.324\nC(YM)[22] -0.2170 0.121 -1.790 0.073 -0.455 0.021\nC(YM)[23] 0.2259 0.120 1.878 0.060 -0.010 0.462\nC(YM)[24] -0.1430 0.117 -1.227 0.220 -0.371 0.085\nC(YM)[25] -0.0727 0.116 -0.628 0.530 -0.299 0.154\nC(YM)[26] -0.2081 0.113 -1.838 0.066 -0.430 0.014\nC(YM)[27] 0.0472 0.105 0.448 0.654 -0.159 0.253\nC(YM)[28] -0.1838 0.105 -1.749 0.080 -0.390 0.022\nC(YM)[29] -0.2776 0.111 -2.507 0.012 -0.495 -0.061\nC(YM)[30] -0.2479 0.105 -2.369 0.018 -0.453 -0.043\nC(YM)[31] -0.0848 0.104 -0.817 0.414 -0.288 0.119\nC(YM)[32] -0.0167 0.110 -0.152 0.879 -0.232 0.198\nC(YM)[33] -0.2914 0.111 -2.634 0.008 -0.508 -0.075\nC(YM)[34] -0.2165 0.105 -2.064 0.039 -0.422 -0.011\nC(YM)[35] -0.0750 0.109 -0.686 0.493 -0.289 0.139\nC(YM)[36] -0.0754 0.114 -0.663 0.507 -0.298 0.148\nC(YM)[37] -0.1741 0.105 -1.658 0.097 -0.380 0.032\nC(YM)[38] 0.0361 0.104 0.347 0.728 -0.168 0.240\nC(YM)[39] -0.1203 0.105 -1.150 0.250 -0.325 0.085\nC(YM)[40] 0.0600 0.107 0.561 0.575 -0.150 0.270\nC(YM)[41] 0.1515 0.104 1.461 0.144 -0.052 0.355\nC(YM)[42] -0.1774 0.100 -1.770 0.077 -0.374 0.019\nC(YM)[43] 0.2266 0.109 2.085 0.037 0.014 0.440\nC(YM)[44] 0.0089 0.103 0.086 0.931 -0.194 0.212\nC(YM)[45] -0.0628 0.104 -0.605 0.545 -0.266 0.141\nC(YM)[46] -0.1399 0.101 -1.389 0.165 -0.337 0.057\nC(YM)[47] -0.1360 0.104 -1.311 0.190 -0.339 0.067\nC(YM)[48] -0.0414 0.097 -0.428 0.668 -0.231 0.148\nC(YM)[49] -0.1187 0.100 -1.184 0.236 -0.315 0.078\nC(YM)[50] -0.3386 0.103 -3.286 0.001 -0.540 -0.137\nC(YM)[51] -0.2962 0.094 -3.156 0.002 -0.480 -0.112\nC(YM)[52] -0.1340 0.097 -1.379 0.168 -0.325 0.057\nC(YM)[53] -0.2076 0.098 -2.125 0.034 -0.399 -0.016\nC(YM)[54] -0.1203 0.098 -1.223 0.221 -0.313 0.073\nC(YM)[55] 0.0292 0.100 0.292 0.770 -0.167 0.225\nC(YM)[56] -0.3079 0.099 -3.102 0.002 -0.502 -0.113\nC(YM)[57] -0.1245 0.102 -1.224 0.221 -0.324 0.075\nC(YM)[58] 0.1810 0.098 1.843 0.065 -0.011 0.373\nC(YM)[59] -0.0916 0.097 -0.947 0.344 -0.281 0.098\nC(YM)[60] 0.0960 0.103 0.929 0.353 -0.106 0.298\nC(YM)[61] -0.0130 0.101 -0.129 0.898 -0.210 0.185\nC(YM)[62] -0.3478 0.101 -3.448 0.001 -0.545 -0.150\nC(YM)[63] -0.2231 0.094 -2.384 0.017 -0.406 -0.040\nC(YM)[64] -0.2412 0.094 -2.565 0.010 -0.426 -0.057\nC(YM)[65] -0.1075 0.098 -1.098 0.272 -0.299 0.084\nC(YM)[66] 0.0717 0.100 0.717 0.474 -0.124 0.268\nC(YM)[67] -0.3080 0.096 -3.221 0.001 -0.495 -0.121\nC(YM)[68] -0.4124 0.104 -3.948 0.000 -0.617 -0.208\nC(YM)[69] -0.4309 0.101 -4.285 0.000 -0.628 -0.234\nC(YM)[70] -0.0266 0.099 -0.268 0.789 -0.222 0.168\nC(YM)[71] -0.1371 0.101 -1.352 0.176 -0.336 0.062\nC(YM)[72] -0.0010 0.096 -0.011 0.991 -0.190 0.188\nC(YM)[73] -0.0513 0.101 -0.509 0.611 -0.249 0.146\nC(YM)[74] 0.0477 0.105 0.453 0.650 -0.159 0.254\nC(YM)[75] 0.0183 0.104 0.175 0.861 -0.186 0.222\nC(YM)[76] 0.0304 0.098 0.310 0.757 -0.162 0.223\nC(YM)[77] -0.1784 0.091 -1.953 0.051 -0.357 0.001\nC(YM)[78] 0.0825 0.094 0.881 0.378 -0.101 0.266\nC(YM)[79] -0.2334 0.095 -2.447 0.014 -0.420 -0.046\nC(YM)[80] -0.2971 0.097 -3.074 0.002 -0.487 -0.108\nC(YM)[81] -0.2688 0.099 -2.728 0.006 -0.462 -0.076\nC(YM)[82] 0.0115 0.098 0.117 0.907 -0.180 0.203\nC(YM)[83] -0.1304 0.098 -1.335 0.182 -0.322 0.061\nC(YM)[84] -0.0267 0.107 -0.249 0.803 -0.237 0.183\nC(YM)[85] 0.1227 0.102 1.209 0.227 -0.076 0.322\nC(YM)[86] 0.0761 0.105 0.725 0.468 -0.129 0.282\nC(YM)[87] -0.2682 0.099 -2.721 0.007 -0.461 -0.075\nC(YM)[88] 0.1060 0.103 1.025 0.305 -0.097 0.309\nC(YM)[89] 0.1606 0.107 1.502 0.133 -0.049 0.370\nC(YM)[90] 0.0742 0.102 0.728 0.467 -0.125 0.274\nC(YM)[91] -0.1272 0.104 -1.218 0.223 -0.332 0.078\nC(YM)[92] -0.0997 0.108 -0.922 0.356 -0.312 0.112\nC(YM)[93] 0.2602 0.107 2.423 0.015 0.050 0.471\nC(YM)[94] 0.5569 0.096 5.786 0.000 0.368 0.746\nC(YM)[95] 0.1035 0.106 0.975 0.330 -0.105 0.312\nC(YM)[96] 0.3853 0.108 3.562 0.000 0.173 0.597\nC(YM)[97] 0.2716 0.108 2.508 0.012 0.059 0.484\nC(YM)[98] 0.1628 0.111 1.466 0.143 -0.055 0.380\nC(YM)[99] 0.1886 0.105 1.788 0.074 -0.018 0.395\nC(YM)[100] 0.4973 0.110 4.528 0.000 0.282 0.713\nC(YM)[101] 0.2737 0.106 2.588 0.010 0.066 0.481\nC(YM)[102] 0.1491 0.106 1.409 0.159 -0.058 0.356\nC(YM)[103] 0.1651 0.112 1.480 0.139 -0.054 0.384\nC(YM)[104] 0.2178 0.115 1.897 0.058 -0.007 0.443\nC(YM)[105] 0.5045 0.105 4.807 0.000 0.299 0.710\nC(YM)[106] 0.4480 0.103 4.341 0.000 0.246 0.650\nC(YM)[107] 0.4568 0.104 4.380 0.000 0.252 0.661\nC(YM)[108] 0.4817 0.104 4.635 0.000 0.278 0.685\nC(YM)[109] 0.4846 0.105 4.635 0.000 0.280 0.690\nC(YM)[110] 0.4282 0.107 3.985 0.000 0.218 0.639\nC(YM)[111] 0.0388 0.102 0.380 0.704 -0.161 0.238\nC(YM)[112] 0.2100 0.107 1.954 0.051 -0.001 0.421\nC(YM)[113] -0.0271 0.104 -0.261 0.794 -0.231 0.177\nC(YM)[114] 0.1127 0.107 1.055 0.292 -0.097 0.322\nC(YM)[115] 0.0203 0.108 0.189 0.850 -0.191 0.231\nC(YM)[116] 0.3823 0.108 3.549 0.000 0.171 0.593\nC(YM)[117] 0.3454 0.106 3.248 0.001 0.137 0.554\nC(YM)[118] 0.2477 0.108 2.303 0.021 0.037 0.458\nC(YM)[119] 0.3384 0.106 3.185 0.001 0.130 0.547\nC(YM)[120] 0.2693 0.108 2.497 0.013 0.058 0.481\nC(YM)[121] 0.3096 0.126 2.465 0.014 0.063 0.556\nC(YM)[122] 0.2948 0.121 2.441 0.015 0.058 0.532\nC(YM)[123] 0.0971 0.118 0.824 0.410 -0.134 0.328\nC(YM)[124] 0.3910 0.124 3.147 0.002 0.147 0.634\nC(YM)[125] 0.2366 0.125 1.899 0.058 -0.008 0.481\nC(YM)[126] 0.3440 0.122 2.824 0.005 0.105 0.583\nC(YM)[127] 0.0372 0.121 0.308 0.758 -0.200 0.274\nC(YM)[128] 0.1616 0.117 1.386 0.166 -0.067 0.390\nC(YM)[129] 0.0262 0.126 0.207 0.836 -0.222 0.274\nC(YM)[130] 0.1248 0.121 1.029 0.304 -0.113 0.363\nC(YM)[131] 0.0295 0.122 0.242 0.809 -0.210 0.269\nC(YM)[132] -0.0933 0.130 -0.718 0.473 -0.348 0.161\nC(YM)[133] 0.4424 0.130 3.406 0.001 0.188 0.697\nC(YM)[134] 0.0930 0.134 0.695 0.487 -0.169 0.355\nC(YM)[135] 0.3597 0.125 2.888 0.004 0.116 0.604\nC(YM)[136] 0.1892 0.122 1.548 0.122 -0.050 0.429\nC(YM)[137] 0.1199 0.123 0.972 0.331 -0.122 0.362\nC(YM)[138] 0.2202 0.120 1.828 0.068 -0.016 0.456\nC(YM)[139] 0.3593 0.125 2.872 0.004 0.114 0.605\nC(YM)[140] 0.2525 0.120 2.099 0.036 0.017 0.488\nC(YM)[141] -0.0461 0.130 -0.354 0.724 -0.302 0.210\nC(YM)[142] 0.0262 0.124 0.211 0.833 -0.217 0.269\nC(YM)[143] 0.2619 0.126 2.075 0.038 0.015 0.509\nC(YM)[144] 0.3783 0.128 2.949 0.003 0.127 0.630\nC(YM)[145] 0.2192 0.130 1.690 0.091 -0.035 0.473\nC(YM)[146] 0.4347 0.125 3.472 0.001 0.189 0.680\nC(YM)[147] 0.0400 0.123 0.325 0.745 -0.201 0.281\nC(YM)[148] 0.3030 0.123 2.462 0.014 0.062 0.544\nC(YM)[149] 0.0393 0.122 0.322 0.747 -0.200 0.278\nC(YM)[150] 0.2735 0.111 2.464 0.014 0.056 0.491\nC(YM)[151] 0.1122 0.122 0.916 0.360 -0.128 0.352\nC(YM)[152] 0.0492 0.126 0.391 0.696 -0.197 0.296\nC(YM)[153] 0.0485 0.124 0.390 0.696 -0.195 0.292\nC(YM)[154] 0.3500 0.125 2.809 0.005 0.106 0.594\nC(YM)[155] 0.1237 0.126 0.982 0.326 -0.123 0.371\nC(YM)[156] 0.4462 0.124 3.594 0.000 0.203 0.690\nC(YM)[157] 0.1437 0.124 1.158 0.247 -0.100 0.387\nC(YM)[158] 0.5110 0.127 4.036 0.000 0.263 0.759\nC(YM)[159] 0.4995 0.115 4.331 0.000 0.273 0.726\nC(YM)[160] 0.3021 0.115 2.628 0.009 0.077 0.527\nC(YM)[161] -0.0523 0.115 -0.455 0.649 -0.278 0.173\nC(YM)[162] 0.2639 0.112 2.356 0.018 0.044 0.483\nC(YM)[163] 0.2477 0.115 2.161 0.031 0.023 0.472\nC(YM)[164] 0.2052 0.126 1.624 0.104 -0.043 0.453\nC(YM)[165] 0.1866 0.119 1.572 0.116 -0.046 0.419\nC(YM)[166] 0.1996 0.116 1.727 0.084 -0.027 0.426\nC(YM)[167] 0.2556 0.120 2.132 0.033 0.021 0.491\nC(YM)[168] 0.4411 0.121 3.633 0.000 0.203 0.679\nC(YM)[169] 0.5246 0.125 4.181 0.000 0.279 0.771\nC(YM)[170] -0.1333 0.122 -1.092 0.275 -0.373 0.106\nC(YM)[171] 0.0616 0.113 0.543 0.587 -0.161 0.284\nC(YM)[172] 0.5292 0.120 4.422 0.000 0.295 0.764\nC(YM)[173] 0.4570 0.115 3.982 0.000 0.232 0.682\nC(YM)[174] 0.4089 0.114 3.592 0.000 0.186 0.632\nC(YM)[175] 0.1500 0.110 1.360 0.174 -0.066 0.366\nC(YM)[176] 0.0077 0.111 0.069 0.945 -0.211 0.226\nC(YM)[177] 0.3193 0.112 2.843 0.004 0.099 0.540\nC(YM)[178] 0.2593 0.111 2.330 0.020 0.041 0.477\nC(YM)[179] -0.0001 0.119 -0.001 0.999 -0.233 0.232\nC(YM)[180] 0.3961 0.122 3.254 0.001 0.157 0.635\nC(YM)[181] 0.4235 0.121 3.514 0.000 0.187 0.660\nC(YM)[182] 0.2797 0.120 2.327 0.020 0.044 0.515\nC(YM)[183] 0.2840 0.109 2.610 0.009 0.071 0.497\nC(YM)[184] 0.1429 0.123 1.160 0.246 -0.099 0.384\nC(YM)[185] 0.1784 0.113 1.579 0.114 -0.043 0.400\nC(YM)[186] 0.2856 0.110 2.607 0.009 0.071 0.500\nC(YM)[187] 0.1436 0.121 1.190 0.234 -0.093 0.380\nC(YM)[188] 0.1748 0.118 1.478 0.139 -0.057 0.407\nC(YM)[189] 0.0667 0.110 0.606 0.544 -0.149 0.282\nC(YM)[190] 0.2998 0.111 2.698 0.007 0.082 0.518\nC(YM)[191] 0.2345 0.115 2.044 0.041 0.010 0.459\nC(YM)[192] 0.5432 0.114 4.758 0.000 0.319 0.767\nC(YM)[193] 0.4258 0.112 3.792 0.000 0.206 0.646\nC(YM)[194] 0.4499 0.124 3.636 0.000 0.207 0.692\nC(YM)[195] 0.0385 0.108 0.356 0.721 -0.173 0.250\nC(YM)[196] 0.0923 0.112 0.824 0.410 -0.127 0.312\nC(YM)[197] 0.5224 0.107 4.900 0.000 0.313 0.731\nC(YM)[198] 0.1520 0.110 1.386 0.166 -0.063 0.367\nC(YM)[199] 0.4530 0.110 4.107 0.000 0.237 0.669\nC(YM)[200] 0.0686 0.112 0.611 0.541 -0.151 0.289\nC(YM)[201] -0.2906 0.124 -2.345 0.019 -0.534 -0.048\nC(YM)[202] -0.4870 0.126 -3.870 0.000 -0.734 -0.240\nC(YM)[203] -0.1588 0.125 -1.269 0.204 -0.404 0.086\nC(YM)[204] 0.1640 0.122 1.343 0.179 -0.075 0.403\nC(YM)[205] -0.1991 0.115 -1.735 0.083 -0.424 0.026\nC(YM)[206] -0.1228 0.108 -1.133 0.257 -0.335 0.090\nC(YM)[207] -0.1437 0.109 -1.319 0.187 -0.357 0.070\nC(YM)[208] -0.1493 0.114 -1.306 0.192 -0.374 0.075\nC(YM)[209] -0.0376 0.105 -0.356 0.722 -0.244 0.169\nC(YM)[210] -0.0390 0.109 -0.359 0.720 -0.252 0.174\nC(YM)[211] -0.0287 0.101 -0.283 0.777 -0.227 0.170\nC(YM)[212] 0.1977 0.109 1.817 0.069 -0.016 0.411\nC(YM)[213] 0.1318 0.112 1.178 0.239 -0.088 0.351\nC(YM)[214] -0.0700 0.105 -0.668 0.504 -0.275 0.135\nC(YM)[215] 0.2673 0.108 2.466 0.014 0.055 0.480\nC(YM)[216] 0.0351 0.109 0.321 0.748 -0.179 0.249\nC(YM)[217] 0.1261 0.107 1.182 0.237 -0.083 0.335\nC(YM)[218] 0.1948 0.114 1.710 0.087 -0.028 0.418\nC(YM)[219] -0.0729 0.112 -0.649 0.516 -0.293 0.147\nC(YM)[220] 0.2155 0.111 1.940 0.052 -0.002 0.433\nC(YM)[221] -0.0677 0.110 -0.612 0.540 -0.284 0.149\nC(YM)[222] 0.2182 0.109 2.008 0.045 0.005 0.431\nC(YM)[223] 0.1212 0.109 1.106 0.269 -0.093 0.336\nC(YM)[224] 0.1584 0.108 1.469 0.142 -0.053 0.370\nC(YM)[225] 0.0730 0.103 0.711 0.477 -0.128 0.274\nC(YM)[226] 0.0478 0.099 0.481 0.630 -0.147 0.242\nC(YM)[227] -0.1532 0.100 -1.526 0.127 -0.350 0.044\nC(YM)[228] -0.0478 0.103 -0.466 0.641 -0.249 0.153\nC(YM)[229] 0.0293 0.110 0.267 0.790 -0.186 0.245\nC(YM)[230] 0.0428 0.108 0.396 0.692 -0.169 0.255\nC(YM)[231] 0.1995 0.107 1.873 0.061 -0.009 0.408\nC(YM)[232] 0.1213 0.105 1.156 0.248 -0.084 0.327\nC(YM)[233] 0.2115 0.109 1.947 0.052 -0.001 0.424\nC(YM)[234] 0.1339 0.110 1.217 0.224 -0.082 0.350\nC(YM)[235] -0.1209 0.113 -1.072 0.284 -0.342 0.100\nC(YM)[236] 0.1504 0.116 1.301 0.193 -0.076 0.377\nC(YM)[237] -0.0405 0.109 -0.372 0.710 -0.254 0.173\nC(YM)[238] -0.2149 0.104 -2.060 0.039 -0.419 -0.010\nC(YM)[239] 0.1816 0.114 1.593 0.111 -0.042 0.405\nC(YM)[240] 0.2951 0.106 2.787 0.005 0.088 0.503\nC(YM)[241] -0.0120 0.114 -0.105 0.917 -0.236 0.212\nC(YM)[242] -0.1882 0.113 -1.667 0.096 -0.409 0.033\nC(YM)[243] -0.0557 0.104 -0.533 0.594 -0.260 0.149\nC(YM)[244] 0.1838 0.107 1.715 0.086 -0.026 0.394\nC(YM)[245] -0.0527 0.114 -0.463 0.643 -0.276 0.170\nC(YM)[246] 0.1302 0.106 1.231 0.218 -0.077 0.338\nC(YM)[247] 0.1222 0.115 1.063 0.288 -0.103 0.348\nC(YM)[248] 0.2685 0.120 2.238 0.025 0.033 0.504\nC(YM)[249] 0.0111 0.119 0.093 0.926 -0.222 0.244\nC(YM)[250] 0.1287 0.118 1.093 0.274 -0.102 0.359\nC(YM)[251] 0.1386 0.120 1.155 0.248 -0.097 0.374\nC(YM)[252] 0.2797 0.116 2.414 0.016 0.053 0.507\nC(YM)[253] 0.2395 0.119 2.017 0.044 0.007 0.472\nC(YM)[254] 0.2934 0.119 2.458 0.014 0.059 0.527\nC(YM)[255] 0.1022 0.116 0.882 0.378 -0.125 0.329\nC(YM)[256] 0.2511 0.122 2.052 0.040 0.011 0.491\nC(YM)[257] 0.0132 0.121 0.109 0.913 -0.224 0.250\nC(YM)[258] 0.3260 0.114 2.869 0.004 0.103 0.549\nC(YM)[259] 0.2548 0.123 2.073 0.038 0.014 0.496\nC(YM)[260] 0.3012 0.118 2.561 0.010 0.071 0.532\nC(YM)[261] 0.1716 0.119 1.438 0.150 -0.062 0.406\nC(YM)[262] 0.0244 0.120 0.203 0.839 -0.211 0.260\nC(YM)[263] 0.6247 0.119 5.257 0.000 0.392 0.858\nC(YM)[264] 0.4955 0.115 4.318 0.000 0.271 0.720\nC(YM)[265] 0.5432 0.119 4.574 0.000 0.310 0.776\nC(YM)[266] 0.3811 0.125 3.037 0.002 0.135 0.627\nC(YM)[267] 0.2925 0.110 2.655 0.008 0.077 0.509\nC(YM)[268] 0.2877 0.114 2.517 0.012 0.064 0.512\nC(YM)[269] 0.1061 0.117 0.909 0.363 -0.123 0.335\nC(YM)[270] 0.1258 0.115 1.090 0.276 -0.100 0.352\nC(YM)[271] -0.0175 0.119 -0.147 0.883 -0.250 0.215\nC(YM)[272] -0.1438 0.115 -1.245 0.213 -0.370 0.083\nC(YM)[273] 0.2090 0.115 1.818 0.069 -0.016 0.434\nC(YM)[274] 0.0058 0.117 0.049 0.961 -0.224 0.236\nC(YM)[275] 0.1525 0.118 1.291 0.197 -0.079 0.384\nC(YM)[276] 0.3833 0.121 3.174 0.002 0.147 0.620\nC(YM)[277] -0.0102 0.123 -0.083 0.934 -0.250 0.230\nC(YM)[278] 0.0719 0.118 0.610 0.542 -0.159 0.303\nC(YM)[279] 0.1700 0.110 1.548 0.122 -0.045 0.385\nC(YM)[280] 0.1899 0.109 1.746 0.081 -0.023 0.403\nC(YM)[281] 0.1140 0.120 0.952 0.341 -0.121 0.349\nC(YM)[282] 0.3038 0.117 2.602 0.009 0.075 0.533\nC(YM)[283] 0.0058 0.112 0.052 0.958 -0.214 0.225\nC(YM)[284] 0.0683 0.120 0.571 0.568 -0.166 0.303\nC(YM)[285] -0.1204 0.104 -1.160 0.246 -0.324 0.083\nC(YM)[286] 0.2911 0.113 2.573 0.010 0.069 0.513\nC(YM)[287] 0.4131 0.125 3.302 0.001 0.168 0.658\nC(YM)[288] 0.1448 0.122 1.185 0.236 -0.095 0.384\nC(YM)[289] 0.2271 0.125 1.815 0.070 -0.018 0.472\nC(YM)[290] 0.3809 0.125 3.042 0.002 0.136 0.626\nC(YM)[291] 0.3236 0.117 2.770 0.006 0.095 0.553\nC(YM)[292] 0.5310 0.120 4.443 0.000 0.297 0.765\nC(YM)[293] 0.4418 0.125 3.525 0.000 0.196 0.687\nC(YM)[294] 0.1094 0.115 0.954 0.340 -0.115 0.334\nC(YM)[295] 0.1567 0.119 1.316 0.188 -0.077 0.390\nC(YM)[296] 0.0276 0.125 0.222 0.824 -0.217 0.272\nC(YM)[297] 0.1584 0.119 1.334 0.182 -0.074 0.391\nC(YM)[298] 0.4240 0.113 3.753 0.000 0.203 0.645\nC(YM)[299] 0.2374 0.128 1.853 0.064 -0.014 0.488\nC(YM)[300] 0.2647 0.120 2.211 0.027 0.030 0.499\nC(YM)[301] -0.0033 0.098 -0.034 0.973 -0.196 0.189\nC(YM)[302] 0.2308 0.097 2.377 0.017 0.040 0.421\nC(YM)[303] 0.0975 0.085 1.144 0.253 -0.069 0.264\nC(YM)[304] -0.3091 0.084 -3.660 0.000 -0.475 -0.144\nC(YM)[305] -0.1051 0.080 -1.319 0.187 -0.261 0.051\nC(YM)[306] -0.0749 0.086 -0.872 0.383 -0.243 0.093\nC(YM)[307] 0.0820 0.088 0.930 0.352 -0.091 0.255\nC(YM)[308] 0.0715 0.096 0.746 0.456 -0.116 0.259\nC(YM)[309] 0.2550 0.089 2.865 0.004 0.081 0.430\nC(YM)[310] 0.1912 0.086 2.216 0.027 0.022 0.360\nC(YM)[311] 0.1690 0.094 1.807 0.071 -0.014 0.352\nC(YM)[312] 0.3270 0.092 3.559 0.000 0.147 0.507\nC(YM)[313] 0.2163 0.092 2.349 0.019 0.036 0.397\nC(YM)[314] 0.1962 0.097 2.016 0.044 0.005 0.387\nC(YM)[315] 0.0313 0.085 0.371 0.711 -0.134 0.197\nC(YM)[316] -0.0398 0.094 -0.424 0.671 -0.223 0.144\nC(YM)[317] 0.1847 0.094 1.970 0.049 0.001 0.368\nC(YM)[318] -0.1054 0.087 -1.214 0.225 -0.275 0.065\nC(YM)[319] -0.0433 0.096 -0.451 0.652 -0.231 0.145\nC(YM)[320] -0.0926 0.096 -0.960 0.337 -0.282 0.096\nC(YM)[321] -0.1107 0.091 -1.219 0.223 -0.289 0.067\nC(YM)[322] -0.0657 0.095 -0.690 0.490 -0.252 0.121\nC(YM)[323] -0.1139 0.096 -1.188 0.235 -0.302 0.074\nC(YM)[324] -0.0764 0.093 -0.823 0.411 -0.258 0.106\nC(YM)[325] 0.1377 0.096 1.427 0.153 -0.051 0.327\nC(YM)[326] 0.0878 0.097 0.902 0.367 -0.103 0.279\nC(YM)[327] 0.1541 0.096 1.612 0.107 -0.033 0.342\nC(YM)[328] 0.1935 0.096 2.017 0.044 0.005 0.382\nC(YM)[329] -0.0912 0.093 -0.978 0.328 -0.274 0.092\nC(YM)[330] -0.0491 0.097 -0.504 0.614 -0.240 0.142\nC(YM)[331] -0.2102 0.094 -2.225 0.026 -0.395 -0.025\nC(YM)[332] 0.0630 0.097 0.650 0.516 -0.127 0.253\nC(YM)[333] 0.2908 0.105 2.769 0.006 0.085 0.497\nC(YM)[334] 0.2980 0.095 3.125 0.002 0.111 0.485\nC(YM)[335] 0.1878 0.094 1.999 0.046 0.004 0.372\nC(YM)[336] 0.1633 0.093 1.761 0.078 -0.018 0.345\nC(YM)[337] 0.3118 0.089 3.516 0.000 0.138 0.486\nC(YM)[338] 0.4127 0.091 4.546 0.000 0.235 0.591\nC(YM)[339] 0.2144 0.090 2.391 0.017 0.039 0.390\nC(YM)[340] -0.0781 0.090 -0.867 0.386 -0.255 0.098\nC(YM)[341] 0.2846 0.088 3.239 0.001 0.112 0.457\nC(YM)[342] 0.2986 0.090 3.305 0.001 0.122 0.476\nC(YM)[343] 0.1181 0.088 1.341 0.180 -0.054 0.291\nC(YM)[344] 0.0651 0.093 0.697 0.486 -0.118 0.248\nC(YM)[345] 0.0142 0.095 0.149 0.881 -0.173 0.201\nC(YM)[346] -0.0439 0.091 -0.484 0.628 -0.222 0.134\nC(YM)[347] 0.2857 0.091 3.137 0.002 0.107 0.464\nC(YM)[348] 0.2269 0.092 2.470 0.013 0.047 0.407\nC(YM)[349] 0.2063 0.091 2.268 0.023 0.028 0.384\nC(YM)[350] 0.1106 0.090 1.231 0.218 -0.065 0.287\nC(YM)[351] 0.2164 0.086 2.523 0.012 0.048 0.384\nC(YM)[352] 0.1123 0.084 1.344 0.179 -0.051 0.276\nC(YM)[353] 0.1316 0.083 1.590 0.112 -0.031 0.294\nC(YM)[354] 0.2444 0.089 2.744 0.006 0.070 0.419\nC(YM)[355] 0.1079 0.089 1.213 0.225 -0.066 0.282\nC(YM)[356] 0.2071 0.092 2.242 0.025 0.026 0.388\nC(YM)[357] 0.2762 0.092 2.988 0.003 0.095 0.457\nC(YM)[358] 0.0766 0.090 0.854 0.393 -0.099 0.252\nC(YM)[359] -0.0124 0.089 -0.139 0.889 -0.187 0.162\nC(YM)[360] 0.1733 0.090 1.927 0.054 -0.003 0.350\nC(YM)[361] 0.1921 0.092 2.080 0.038 0.011 0.373\nC(YM)[362] 0.3109 0.094 3.295 0.001 0.126 0.496\nC(YM)[363] 0.2531 0.090 2.823 0.005 0.077 0.429\nC(YM)[364] 0.1730 0.091 1.894 0.058 -0.006 0.352\nC(YM)[365] 0.2883 0.091 3.165 0.002 0.110 0.467\nC(YM)[366] 0.4302 0.090 4.801 0.000 0.255 0.606\nC(YM)[367] 0.3268 0.090 3.628 0.000 0.150 0.503\nC(YM)[368] 0.2217 0.097 2.285 0.022 0.032 0.412\nC(YM)[369] 0.1601 0.095 1.683 0.092 -0.026 0.347\nC(YM)[370] 0.2621 0.086 3.046 0.002 0.093 0.431\nC(YM)[371] 0.1080 0.091 1.187 0.235 -0.070 0.286\nC(YM)[372] -0.0382 0.090 -0.422 0.673 -0.215 0.139\nC(YM)[373] -0.0281 0.091 -0.309 0.757 -0.207 0.150\nC(YM)[374] 0.0219 0.090 0.245 0.806 -0.154 0.197\nC(YM)[375] 0.0965 0.082 1.183 0.237 -0.063 0.256\nC(YM)[376] 0.3342 0.082 4.098 0.000 0.174 0.494\nC(YM)[377] 0.0686 0.084 0.819 0.413 -0.096 0.233\nC(YM)[378] 0.2510 0.083 3.024 0.002 0.088 0.414\nC(YM)[379] 0.3514 0.087 4.019 0.000 0.180 0.523\nC(YM)[380] 0.4673 0.090 5.194 0.000 0.291 0.644\nC(YM)[381] 0.2117 0.087 2.424 0.015 0.041 0.383\nC(YM)[382] 0.4736 0.085 5.602 0.000 0.308 0.639\nC(YM)[383] 0.3864 0.086 4.469 0.000 0.217 0.556\nC(YM)[384] 0.6950 0.085 8.152 0.000 0.528 0.862\nC(YM)[385] 0.4265 0.084 5.051 0.000 0.261 0.592\nC(YM)[386] 0.7551 0.094 8.060 0.000 0.571 0.939\nC(YM)[387] 0.6438 0.087 7.413 0.000 0.474 0.814\nC(YM)[388] 0.2833 0.093 3.037 0.002 0.100 0.466\nC(YM)[389] 0.4441 0.099 4.486 0.000 0.250 0.638\nC(YM)[390] 0.3400 0.097 3.523 0.000 0.151 0.529\nC(YM)[391] 0.2452 0.098 2.510 0.012 0.054 0.437\nC(YM)[392] 0.3533 0.106 3.322 0.001 0.145 0.562\nC(YM)[393] 0.2239 0.106 2.116 0.034 0.017 0.431\nC(YM)[394] 0.3087 0.098 3.162 0.002 0.117 0.500\nC(YM)[395] 0.6377 0.106 5.999 0.000 0.429 0.846\nC(YM)[396] 0.6177 0.107 5.786 0.000 0.408 0.827\nC(YM)[397] 0.4666 0.103 4.517 0.000 0.264 0.669\nC(YM)[398] 0.2624 0.110 2.382 0.017 0.046 0.478\nC(YM)[399] 0.1356 0.097 1.398 0.162 -0.055 0.326\nC(YM)[400] 0.3644 0.101 3.601 0.000 0.166 0.563\nC(YM)[401] 0.2561 0.105 2.441 0.015 0.051 0.462\nC(YM)[402] 0.2818 0.110 2.559 0.011 0.066 0.498\nC(YM)[403] 0.0378 0.112 0.338 0.736 -0.182 0.257\nC(YM)[404] 0.1539 0.110 1.395 0.163 -0.062 0.370\nC(YM)[405] 0.3811 0.115 3.325 0.001 0.156 0.606\nC(YM)[406] 0.3278 0.110 2.967 0.003 0.111 0.544\nC(YM)[407] 0.1968 0.112 1.757 0.079 -0.023 0.416\nC(YM)[408] 0.0651 0.110 0.590 0.555 -0.151 0.282\nC(YM)[409] 0.4022 0.113 3.565 0.000 0.181 0.623\nC(YM)[410] 0.3904 0.113 3.466 0.001 0.170 0.611\nC(YM)[411] 0.1244 0.104 1.198 0.231 -0.079 0.328\nC(YM)[412] 0.2425 0.107 2.277 0.023 0.034 0.451\nC(YM)[413] 0.3416 0.107 3.194 0.001 0.132 0.551\nC(YM)[414] 0.4918 0.104 4.716 0.000 0.287 0.696\nC(YM)[415] 0.0772 0.109 0.708 0.479 -0.137 0.291\nC(YM)[416] 0.0282 0.111 0.254 0.800 -0.190 0.246\nC(YM)[417] -0.0858 0.101 -0.849 0.396 -0.284 0.112\nC(YM)[418] 0.0787 0.101 0.776 0.438 -0.120 0.277\nC(YM)[419] 0.0879 0.108 0.817 0.414 -0.123 0.299\nC(YM)[420] 0.1836 0.107 1.712 0.087 -0.027 0.394\nC(YM)[421] 0.3648 0.108 3.393 0.001 0.154 0.575\nC(YM)[422] 0.2977 0.116 2.573 0.010 0.071 0.525\nC(YM)[423] 0.2757 0.112 2.454 0.014 0.055 0.496\nC(YM)[424] 0.4227 0.116 3.640 0.000 0.195 0.650\nC(YM)[425] 0.2727 0.109 2.495 0.013 0.058 0.487\nC(YM)[426] 0.4544 0.115 3.955 0.000 0.229 0.680\nC(YM)[427] 0.3168 0.113 2.792 0.005 0.094 0.539\nC(YM)[428] 0.2919 0.110 2.645 0.008 0.076 0.508\nC(Newspaper)[T.Chicago Tribune] -0.2979 0.023 -12.869 0.000 -0.343 -0.253\nC(Newspaper)[T.Los Angeles Times] -0.5335 0.022 -24.743 0.000 -0.576 -0.491\nC(Newspaper)[T.New York Times] -0.2227 0.022 -10.359 0.000 -0.265 -0.181\nC(Newspaper)[T.The Washington Post] -0.1513 0.022 -6.927 0.000 -0.194 -0.109\nC(Newspaper)[T.USA Today] -0.4924 0.030 -16.368 0.000 -0.551 -0.433\nC(Newspaper)[T.Wall Street Journal] -0.1609 0.019 -8.326 0.000 -0.199 -0.123\n==============================================================================\nOmnibus: 13646.158 Durbin-Watson: 1.834\nProb(Omnibus): 0.000 Jarque-Bera (JB): 34680.625\nSkew: -0.079 Prob(JB): 0.00\nKurtosis: 4.325 Cond. No. 40.5\n==============================================================================\n\nNotes:\n[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.\n"
],
[
"# Merge indexes\nsentimentIndex=df_ym.merge(UncertaintyIndex,on='YM',how='outer').\\\n merge(LMindex,on='YM',how='outer').\\\n merge(GIindex,on='YM',how='outer').\\\n merge(LSDindex,on='YM',how='outer').\\\n sort_values(['Year','Month'])\nprint(sentimentIndex.info())",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 428 entries, 0 to 427\nData columns (total 11 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Year 428 non-null int64 \n 1 Month 428 non-null int64 \n 2 YM 428 non-null int64 \n 3 FE_x 428 non-null object \n 4 UncertaintyScoreIndex 428 non-null float64\n 5 FE_y 428 non-null object \n 6 LMscoreIndex 428 non-null float64\n 7 FE_x 428 non-null object \n 8 GIscoreIndex 428 non-null float64\n 9 FE_y 428 non-null object \n 10 LSDscoreIndex 428 non-null float64\ndtypes: float64(4), int64(3), object(4)\nmemory usage: 40.1+ KB\nNone\n"
],
[
"sentimentIndex=sentimentIndex.drop(['FE_x','FE_y'],axis=1).\\\n rename(columns={'UncertaintyScoreIndex':'UncertaintyIndex','LMscoreIndex':'LMindex',\n 'GIscoreIndex':'GIindex','LSDscoreIndex':'LSDindex'})",
"_____no_output_____"
],
[
"print(sentimentIndex)",
" Year Month YM UncertaintyIndex LMindex GIindex LSDindex\n0 1985 1 1 0.593382 -1.873716 1.416218 0.229804\n1 1985 2 2 0.565654 -2.026707 1.382028 0.014728\n2 1985 3 3 0.615707 -2.035717 1.157062 -0.137810\n3 1985 4 4 0.651001 -1.936667 1.449065 0.036833\n4 1985 5 5 0.652354 -1.796473 1.494990 0.248346\n.. ... ... ... ... ... ... ...\n423 2020 4 424 0.781880 -1.813227 1.870549 0.422744\n424 2020 5 425 0.684124 -2.008187 1.756084 0.272658\n425 2020 6 426 0.663700 -2.049850 1.480211 0.454404\n426 2020 7 427 0.660098 -1.953976 1.486845 0.316831\n427 2020 8 428 0.659842 -1.774000 1.508842 0.291868\n\n[428 rows x 7 columns]\n"
],
[
"print(sentimentIndex.info())",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 428 entries, 0 to 427\nData columns (total 7 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Year 428 non-null int64 \n 1 Month 428 non-null int64 \n 2 YM 428 non-null int64 \n 3 UncertaintyIndex 428 non-null float64\n 4 LMindex 428 non-null float64\n 5 GIindex 428 non-null float64\n 6 LSDindex 428 non-null float64\ndtypes: float64(4), int64(3)\nmemory usage: 26.8 KB\nNone\n"
],
[
"# Export\nsentimentIndex.to_csv('/home/ec2-user/SageMaker/New Uncertainty/Sentiment Analysis/RegRelevant_MonthlySentimentIndex_Robust1.csv',index=False)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecc95ca02b7e34d072692536186c9492f5ca7336 | 131,165 | ipynb | Jupyter Notebook | scikit-learn-exercises.ipynb | bhrdwjvarun/SkLearn | c7c5046dbc621ce919b19fcdd1236a6c7fb7ba63 | [
"MIT"
] | null | null | null | scikit-learn-exercises.ipynb | bhrdwjvarun/SkLearn | c7c5046dbc621ce919b19fcdd1236a6c7fb7ba63 | [
"MIT"
] | null | null | null | scikit-learn-exercises.ipynb | bhrdwjvarun/SkLearn | c7c5046dbc621ce919b19fcdd1236a6c7fb7ba63 | [
"MIT"
] | null | null | null | 57.503288 | 26,120 | 0.70624 | [
[
[
"# Scikit-Learn Practice Exercises\n\nThis notebook offers a set of excercises for different tasks with Scikit-Learn.\n\nNotes:\n* There may be more than one different way to answer a question or complete an exercise. \n* Some skeleton code has been implemented for you.\n* Exercises are based off (and directly taken from) the quick [introduction to Scikit-Learn notebook](https://github.com/mrdbourke/zero-to-mastery-ml/blob/master/section-2-data-science-and-ml-tools/introduction-to-scikit-learn.ipynb).\n* Different tasks will be detailed by comments or text. Places to put your own code are defined by `###` (don't remove anything other than `###`).\n\nFor further reference and resources, it's advised to check out the [Scikit-Learn documnetation](https://scikit-learn.org/stable/user_guide.html).\n\nAnd if you get stuck, try searching for a question in the following format: \"how to do XYZ with Scikit-Learn\", where XYZ is the function you want to leverage from Scikit-Learn.\n\nSince we'll be working with data, we'll import Scikit-Learn's counterparts, Matplotlib, NumPy and pandas.\n\nLet's get started.",
"_____no_output_____"
]
],
[
[
"# Setup matplotlib to plot inline (within the notebook)\n%matplotlib inline\n\n# Import the pyplot module of Matplotlib as plt\nimport matplotlib.pyplot as plt\n\n# Import pandas under the abbreviation 'pd'\nimport pandas as pd\n\n# Import NumPy under the abbreviation 'np'\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"## End-to-end Scikit-Learn classification workflow\n\nLet's start with an end to end Scikit-Learn workflow.\n\nMore specifically, we'll:\n1. Get a dataset ready\n2. Prepare a machine learning model to make predictions\n3. Fit the model to the data and make a prediction\n4. Evaluate the model's predictions \n\nThe data we'll be using is [stored on GitHub](https://github.com/mrdbourke/zero-to-mastery-ml/tree/master/data). We'll start with [`heart-disease.csv`](https://raw.githubusercontent.com/mrdbourke/zero-to-mastery-ml/master/data/heart-disease.csv), a dataset which contains anonymous patient data and whether or not they have heart disease.\n\n**Note:** When viewing a `.csv` on GitHub, make sure it's in the raw format. For example, the URL should look like: https://raw.githubusercontent.com/mrdbourke/zero-to-mastery-ml/master/data/heart-disease.csv\n\n### 1. Getting a dataset ready",
"_____no_output_____"
]
],
[
[
"# Import the heart disease dataset and save it to a variable\n# using pandas and read_csv()\n# Hint: You can directly pass the URL of a csv to read_csv()\nheart_disease = pd.read_csv(\"data/heart-disease.csv\")\n\n# Check the first 5 rows of the data\nheart_disease.head()",
"_____no_output_____"
]
],
[
[
"Our goal here is to build a machine learning model on all of the columns except `target` to predict `target`.\n\nIn essence, the `target` column is our **target variable** (also called `y` or `labels`) and the rest of the other columns are our independent variables (also called `data` or `X`).\n\nAnd since our target variable is one thing or another (heart disease or not), we know our problem is a classification problem (classifying whether something is one thing or another).\n\nKnowing this, let's create `X` and `y` by splitting our dataframe up.",
"_____no_output_____"
]
],
[
[
"# Create X (all columns except target)\nX = heart_disease.drop(\"target\",axis=1)\n\n# Create y (only the target column)\ny = heart_disease[\"target\"]",
"_____no_output_____"
],
[
"y.head()",
"_____no_output_____"
]
],
[
[
"Now we've split our data into `X` and `y`, we'll use Scikit-Learn to split it into training and test sets.",
"_____no_output_____"
]
],
[
[
"# Import train_test_split from sklearn's model_selection module\nfrom sklearn.model_selection import train_test_split\n\n# Use train_test_split to split X & y into training and test sets\nX_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.2)",
"_____no_output_____"
],
[
"# View the different shapes of the training and test datasets\nX_train.shape, X_test.shape, y_train.shape, y_test.shape",
"_____no_output_____"
]
],
[
[
"What do you notice about the different shapes of the data?\n\nSince our data is now in training and test sets, we'll build a machine learning model to fit patterns in the training data and then make predictions on the test data.\n\nTo figure out which machine learning model we should use, you can refer to [Scikit-Learn's machine learning map](https://scikit-learn.org/stable/tutorial/machine_learning_map/index.html).\n\nAfter following the map, you decide to use the [`RandomForestClassifier`](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html).\n\n### 2. Preparing a machine learning model",
"_____no_output_____"
]
],
[
[
"# Import the RandomForestClassifier from sklearn's ensemble module\nfrom sklearn.ensemble import RandomForestClassifier\n\n# Instantiate an instance of RandomForestClassifier as clf\nclf = RandomForestClassifier()",
"_____no_output_____"
]
],
[
[
"Now you've got a `RandomForestClassifier` instance, let's fit it to the training data.\n\nOnce it's fit, we'll make predictions on the test data.\n\n### 3. Fitting a model and making predictions",
"_____no_output_____"
]
],
[
[
"# Fit the RandomForestClassifier to the training data\nclf.fit(X_train,y_train)",
"_____no_output_____"
],
[
"# Use the fitted model to make predictions on the test data and\n# save the predictions to a variable called y_preds\ny_preds = clf.predict(X_test)",
"_____no_output_____"
]
],
[
[
"### 4. Evaluating a model's predictions\n\nEvaluating predictions is as important making them. Let's check how our model did by calling the `score()` method on it and passing it the training (`X_train, y_train`) and testing data (`X_test, y_test`).",
"_____no_output_____"
]
],
[
[
"# Evaluate the fitted model on the training set using the score() function\nclf.score(X_train,y_train)",
"_____no_output_____"
],
[
"# Evaluate the fitted model on the test set using the score() function\nclf.score(X_test,y_test)",
"_____no_output_____"
]
],
[
[
"* How did you model go? \n* What metric does `score()` return for classifiers? \n* Did your model do better on the training dataset or test dataset?",
"_____no_output_____"
],
[
"## Experimenting with different classification models\n\nNow we've quickly covered an end-to-end Scikit-Learn workflow and since experimenting is a large part of machine learning, we'll now try a series of different machine learning models and see which gets the best results on our dataset.\n\nGoing through the [Scikit-Learn machine learning map](https://scikit-learn.org/stable/tutorial/machine_learning_map/index.html), we see there are a number of different classification models we can try (different models are in the green boxes).\n\nFor this exercise, the models we're going to try and compare are:\n* [LinearSVC](https://scikit-learn.org/stable/modules/svm.html#classification)\n* [KNeighborsClassifier](https://scikit-learn.org/stable/modules/neighbors.html) (also known as K-Nearest Neighbors or KNN)\n* [SVC](https://scikit-learn.org/stable/modules/svm.html#classification) (also known as support vector classifier, a form of [support vector machine](https://en.wikipedia.org/wiki/Support-vector_machine))\n* [LogisticRegression](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html) (despite the name, this is actually a classifier)\n* [RandomForestClassifier](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html) (an ensemble method and what we used above)\n\nWe'll follow the same workflow we used above (except this time for multiple models):\n1. Import a machine learning model\n2. Get it ready\n3. Fit it to the data and make predictions\n4. Evaluate the fitted model\n\n**Note:** Since we've already got the data ready, we can reuse it in this section.",
"_____no_output_____"
]
],
[
[
"# Import LinearSVC from sklearn's svm module\nfrom sklearn.svm import LinearSVC\n# Import KNeighborsClassifier from sklearn's neighbors module\nfrom sklearn.neighbors import NearestNeighbors\n\n# Import SVC from sklearn's svm module\nfrom sklearn.svm import SVC\n\n# Import LogisticRegression from sklearn's linear_model module\nfrom sklearn.linear_model import LogisticRegression\n\n# Note: we don't have to import RandomForestClassifier, since we already have",
"_____no_output_____"
]
],
[
[
"Thanks to the consistency of Scikit-Learn's API design, we can use virtually the same code to fit, score and make predictions with each of our models.\n\nTo see which model performs best, we'll do the following:\n1. Instantiate each model in a dictionary\n2. Create an empty results dictionary\n3. Fit each model on the training data\n4. Score each model on the test data\n5. Check the results\n\nIf you're wondering what it means to instantiate each model in a dictionary, see the example below.",
"_____no_output_____"
]
],
[
[
"# EXAMPLE: Instantiating a RandomForestClassifier() in a dictionary\nexample_dict = {\"RandomForestClassifier\": RandomForestClassifier()}\n\n# Create a dictionary called models which contains all of the classification models we've imported\n# Make sure the dictionary is in the same format as example_dict\n# The models dictionary should contain 5 models\nmodels = {\"LinearSVC\": LinearSVC(),\n \"SVC\": SVC(),\n \"LogisticRegression\": LogisticRegression(),\n \"RandomForestClassifier\": RandomForestClassifier()}\n\n\n# Create an empty dictionary called results \"KNN\": NearestNeighbors(),\nresults = {}",
"_____no_output_____"
]
],
[
[
"Since each model we're using has the same `fit()` and `score()` functions, we can loop through our models dictionary and, call `fit()` on the training data and then call `score()` with the test data.",
"_____no_output_____"
]
],
[
[
"# EXAMPLE: Looping through example_dict fitting and scoring the model\nexample_results = {}\nfor model_name, model in example_dict.items():\n model.fit(X_train, y_train)\n example_results[model_name] = model.score(X_test, y_test)\n\n# EXAMPLE: View the results\nexample_results ",
"_____no_output_____"
],
[
"# Loop through the models dictionary items, fitting the model on the training data\n# and appending the model name and model score on the test data to the results dictionary\nfor model_name, model in models.items():\n model.fit(X_train,y_train)\n results[model_name] = model.score(X_test,y_test)\n\n# View the results\nresults",
"C:\\Users\\bhaga\\anaconda3\\lib\\site-packages\\sklearn\\svm\\_base.py:976: ConvergenceWarning: Liblinear failed to converge, increase the number of iterations.\n warnings.warn(\"Liblinear failed to converge, increase \"\nC:\\Users\\bhaga\\anaconda3\\lib\\site-packages\\sklearn\\linear_model\\_logistic.py:762: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n n_iter_i = _check_optimize_result(\n"
]
],
[
[
"* Which model performed the best? \n* Do the results change each time you run the cell? \n* Why do you think this is?\n\nDue to the randomness of how each model finds patterns in the data, you might notice different results each time.\n\nWithout manually setting the random state using the `random_state` parameter of some models or using a NumPy random seed, every time you run the cell, you'll get slightly different results.\n\nLet's see this in effect by running the same code as the cell above, except this time setting a [NumPy random seed equal to 42](https://docs.scipy.org/doc/numpy-1.15.1/reference/generated/numpy.random.seed.html).",
"_____no_output_____"
]
],
[
[
"# Run the same code as the cell above, except this time set a NumPy random seed\n# equal to 42\nnp.random.seed(42)\n\nfor model_name, model in models.items():\n model.fit(X_train, y_train)\n results[model_name] = model.score(X_test, y_test)\n \nresults",
"C:\\Users\\bhaga\\anaconda3\\lib\\site-packages\\sklearn\\svm\\_base.py:976: ConvergenceWarning: Liblinear failed to converge, increase the number of iterations.\n warnings.warn(\"Liblinear failed to converge, increase \"\nC:\\Users\\bhaga\\anaconda3\\lib\\site-packages\\sklearn\\linear_model\\_logistic.py:762: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n n_iter_i = _check_optimize_result(\n"
]
],
[
[
"* Run the cell above a few times, what do you notice about the results? \n* Which model performs the best this time?\n* What happens if you add a NumPy random seed to the cell where you called `train_test_split()` (towards the top of the notebook) and then rerun the cell above?\n\nLet's make our results a little more visual.",
"_____no_output_____"
]
],
[
[
"# Create a pandas dataframe with the data as the values of the results dictionary,\n# the index as the keys of the results dictionary and a single column called accuracy.\n# Be sure to save the dataframe to a variable.\nresults_df = pd.DataFrame(results.values(), \n results.keys(), \n columns=[\"accuracy\"])\n\n# Create a bar plot of the results dataframe using plot.bar()\nresults_df.plot.bar();",
"_____no_output_____"
]
],
[
[
"Using `np.random.seed(42)` results in the `LogisticRegression` model perfoming the best (at least on my computer).\n\nLet's tune its hyperparameters and see if we can improve it.\n\n### Hyperparameter Tuning\n\nRemember, if you're ever trying to tune a machine learning models hyperparameters and you're not sure where to start, you can always search something like \"MODEL_NAME hyperparameter tuning\".\n\nIn the case of LogisticRegression, you might come across articles, such as [Hyperparameter Tuning Using Grid Search by Chris Albon](https://chrisalbon.com/machine_learning/model_selection/hyperparameter_tuning_using_grid_search/).\n\nThe article uses [`GridSearchCV`](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GridSearchCV.html) but we're going to be using [`RandomizedSearchCV`](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.RandomizedSearchCV.html).\n\nThe different hyperparameters to search over have been setup for you in `log_reg_grid` but feel free to change them.",
"_____no_output_____"
]
],
[
[
"# Different LogisticRegression hyperparameters\nlog_reg_grid = {\"C\": np.logspace(-4, 4, 20),\n \"solver\": [\"liblinear\"]}",
"_____no_output_____"
]
],
[
[
"Since we've got a set of hyperparameters we can import `RandomizedSearchCV`, pass it our dictionary of hyperparameters and let it search for the best combination.",
"_____no_output_____"
]
],
[
[
"# Setup np random seed of 42\nnp.random.seed(42)\n\n# Import RandomizedSearchCV from sklearn's model_selection module\nfrom sklearn.model_selection import RandomizedSearchCV\n\n# Setup an instance of RandomizedSearchCV with a LogisticRegression() estimator,\n# our log_reg_grid as the param_distributions, a cv of 5 and n_iter of 5.\nrs_log_reg = RandomizedSearchCV(estimator=models[\"LogisticRegression\"],\n param_distributions=log_reg_grid,\n cv=5,\n n_iter=5,\n verbose=2)\n\n# Fit the instance of RandomizedSearchCV\nrs_log_reg.fit(X_train,y_train)",
"Fitting 5 folds for each of 5 candidates, totalling 25 fits\n[CV] solver=liblinear, C=0.0001 ......................................\n"
]
],
[
[
"Once `RandomizedSearchCV` has finished, we can find the best hyperparmeters it found using the `best_params_` attributes.",
"_____no_output_____"
]
],
[
[
"# Find the best parameters of the RandomizedSearchCV instance using the best_params_ attribute\nrs_log_reg.best_params_",
"_____no_output_____"
],
[
"# Score the instance of RandomizedSearchCV using the test data\nrs_log_reg.score(X_test,y_test)",
"_____no_output_____"
]
],
[
[
"After hyperparameter tuning, did the models score improve? What else could you try to improve it? Are there any other methods of hyperparameter tuning you can find for `LogisticRegression`?\n\n### Classifier Model Evaluation\n\nWe've tried to find the best hyperparameters on our model using `RandomizedSearchCV` and so far we've only been evaluating our model using the `score()` function which returns accuracy. \n\nBut when it comes to classification, you'll likely want to use a few more evaluation metrics, including:\n* [**Confusion matrix**](https://www.dataschool.io/simple-guide-to-confusion-matrix-terminology/) - Compares the predicted values with the true values in a tabular way, if 100% correct, all values in the matrix will be top left to bottom right (diagnol line).\n* [**Cross-validation**](https://scikit-learn.org/stable/modules/cross_validation.html) - Splits your dataset into multiple parts and train and tests your model on each part and evaluates performance as an average. \n* [**Precision**](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.precision_score.html#sklearn.metrics.precision_score) - Proportion of true positives over total number of samples. Higher precision leads to less false positives.\n* [**Recall**](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.recall_score.html#sklearn.metrics.recall_score) - Proportion of true positives over total number of true positives and false positives. Higher recall leads to less false negatives.\n* [**F1 score**](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.f1_score.html#sklearn.metrics.f1_score) - Combines precision and recall into one metric. 1 is best, 0 is worst.\n* [**Classification report**](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.classification_report.html) - Sklearn has a built-in function called `classification_report()` which returns some of the main classification metrics such as precision, recall and f1-score.\n* [**ROC Curve**](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.roc_score.html) - [Receiver Operating Characterisitc](https://en.wikipedia.org/wiki/Receiver_operating_characteristic) is a plot of true positive rate versus false positive rate.\n* [**Area Under Curve (AUC)**](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.roc_auc_score.html) - The area underneath the ROC curve. A perfect model achieves a score of 1.0.\n\nBefore we get to these, we'll instantiate a new instance of our model using the best hyerparameters found by `RandomizedSearchCV`. ",
"_____no_output_____"
]
],
[
[
"# Instantiate a LogisticRegression classifier using the best hyperparameters from RandomizedSearchCV\nclf = LogisticRegression(solver = \"liblinear\", C = 0.23357214690901212)\n\n# Fit the new instance of LogisticRegression with the best hyperparameters on the training data \nrs_log_reg.fit(X_train,y_train)",
"Fitting 5 folds for each of 5 candidates, totalling 25 fits\n[CV] solver=liblinear, C=0.0001 ......................................\n[CV] ....................... solver=liblinear, C=0.0001, total= 0.0s\n[CV] solver=liblinear, C=0.0001 ......................................\n[CV] ....................... solver=liblinear, C=0.0001, total= 0.0s\n[CV] solver=liblinear, C=0.0001 ......................................\n[CV] ....................... solver=liblinear, C=0.0001, total= 0.0s\n[CV] solver=liblinear, C=0.0001 ......................................\n[CV] ....................... solver=liblinear, C=0.0001, total= 0.0s\n[CV] solver=liblinear, C=0.0001 ......................................\n[CV] ....................... solver=liblinear, C=0.0001, total= 0.0s\n[CV] solver=liblinear, C=0.012742749857031334 ........................\n[CV] ......... solver=liblinear, C=0.012742749857031334, total= 0.0s\n[CV] solver=liblinear, C=0.012742749857031334 ........................\n[CV] ......... solver=liblinear, C=0.012742749857031334, total= 0.0s\n[CV] solver=liblinear, C=0.012742749857031334 ........................\n[CV] ......... solver=liblinear, C=0.012742749857031334, total= 0.0s\n[CV] solver=liblinear, C=0.012742749857031334 ........................\n[CV] ......... solver=liblinear, C=0.012742749857031334, total= 0.0s\n[CV] solver=liblinear, C=0.012742749857031334 ........................\n[CV] ......... solver=liblinear, C=0.012742749857031334, total= 0.0s\n[CV] solver=liblinear, C=3792.690190732246 ...........................\n[CV] ............ solver=liblinear, C=3792.690190732246, total= 0.0s\n[CV] solver=liblinear, C=3792.690190732246 ...........................\n[CV] ............ solver=liblinear, C=3792.690190732246, total= 0.0s\n[CV] solver=liblinear, C=3792.690190732246 ...........................\n[CV] ............ solver=liblinear, C=3792.690190732246, total= 0.0s\n[CV] solver=liblinear, C=3792.690190732246 ...........................\n[CV] ............ solver=liblinear, C=3792.690190732246, total= 0.0s\n[CV] solver=liblinear, C=3792.690190732246 ...........................\n[CV] ............ solver=liblinear, C=3792.690190732246, total= 0.0s\n[CV] solver=liblinear, C=29.763514416313132 ..........................\n[CV] ........... solver=liblinear, C=29.763514416313132, total= 0.0s\n[CV] solver=liblinear, C=29.763514416313132 ..........................\n[CV] ........... solver=liblinear, C=29.763514416313132, total= 0.0s\n[CV] solver=liblinear, C=29.763514416313132 ..........................\n[CV] ........... solver=liblinear, C=29.763514416313132, total= 0.0s\n[CV] solver=liblinear, C=29.763514416313132 ..........................\n[CV] ........... solver=liblinear, C=29.763514416313132, total= 0.0s\n[CV] solver=liblinear, C=29.763514416313132 ..........................\n[CV] ........... solver=liblinear, C=29.763514416313132, total= 0.0s\n[CV] solver=liblinear, C=10000.0 .....................................\n[CV] ...................... solver=liblinear, C=10000.0, total= 0.0s\n[CV] solver=liblinear, C=10000.0 .....................................\n[CV] ...................... solver=liblinear, C=10000.0, total= 0.0s\n[CV] solver=liblinear, C=10000.0 .....................................\n"
]
],
[
[
"Now it's to import the relative Scikit-Learn methods for each of the classification evaluation metrics we're after.",
"_____no_output_____"
]
],
[
[
"# Import confusion_matrix and classification_report from sklearn's metrics module\nfrom sklearn.metrics import confusion_matrix,classification_report,precision_score,recall_score,f1_score,plot_roc_curve\n\n# Import precision_score, recall_score and f1_score from sklearn's metrics module\n\n\n# Import plot_roc_curve from sklearn's metrics module\n###",
"_____no_output_____"
]
],
[
[
"Evaluation metrics are very often comparing a model's predictions to some ground truth labels.\n\nLet's make some predictions on the test data using our latest model and save them to `y_preds`.",
"_____no_output_____"
]
],
[
[
"# Make predictions on test data and save them\ny_preds = rs_log_reg.predict(X_test)",
"_____no_output_____"
]
],
[
[
"Time to use the predictions our model has made to evaluate it beyond accuracy.",
"_____no_output_____"
]
],
[
[
"# Create a confusion matrix using the confusion_matrix function\nconfusion_matrix(y_test,y_preds)",
"_____no_output_____"
]
],
[
[
"**Challenge:** The in-built `confusion_matrix` function in Scikit-Learn produces something not too visual, how could you make your confusion matrix more visual?\n\nYou might want to search something like \"how to plot a confusion matrix\". Note: There may be more than one way to do this.",
"_____no_output_____"
]
],
[
[
"# Create a more visual confusion matrix\nimport seaborn as sns\nsns.set(font_scale=1.5)\nconf_mat = confusion_matrix(y_test,y_preds)\nsns.heatmap(conf_mat,annot=True);",
"_____no_output_____"
]
],
[
[
"How about a classification report?",
"_____no_output_____"
]
],
[
[
"# Create a classification report using the classification_report function\nprint(classification_report(y_test,y_preds))",
" precision recall f1-score support\n\n 0 0.88 0.84 0.86 25\n 1 0.89 0.92 0.90 36\n\n accuracy 0.89 61\n macro avg 0.88 0.88 0.88 61\nweighted avg 0.88 0.89 0.88 61\n\n"
]
],
[
[
"**Challenge:** Write down what each of the columns in this classification report are.\n\n* **Precision** - Indicates the proportion of positive identifications (model predicted class 1) which were actually correct. A model which produces no false positives has a precision of 1.0.\n* **Recall** - Indicates the proportion of actual positives which were correctly classified. A model which produces no false negatives has a recall of 1.0.\n* **F1 score** - A combination of precision and recall. A perfect model achieves an F1 score of 1.0.\n* **Support** - The number of samples each metric was calculated on.\n* **Accuracy** - The accuracy of the model in decimal form. Perfect accuracy is equal to 1.0.\n* **Macro avg** - Short for macro average, the average precision, recall and F1 score between classes. Macro avg doesn’t class imbalance into effort, so if you do have class imbalances, pay attention to this metric.\n* **Weighted avg** - Short for weighted average, the weighted average precision, recall and F1 score between classes. Weighted means each metric is calculated with respect to how many samples there are in each class. This metric will favour the majority class (e.g. will give a high value when one class out performs another due to having more samples).\n\nThe classification report gives us a range of values for precision, recall and F1 score, time to find these metrics using Scikit-Learn functions.",
"_____no_output_____"
]
],
[
[
"# Find the precision score of the model using precision_score()\nprecision_score(y_preds,y_test)",
"_____no_output_____"
],
[
"# Find the recall score\nrecall_score(y_preds,y_test)",
"_____no_output_____"
],
[
"# Find the F1 score\nf1_score(y_preds,y_test)",
"_____no_output_____"
]
],
[
[
"Confusion matrix: done.\nClassification report: done.\nROC (receiver operator characteristic) curve & AUC (area under curve) score: not done.\n\nLet's fix this.\n\nIf you're unfamiliar with what a ROC curve, that's your first challenge, to read up on what one is.\n\nIn a sentence, a [ROC curve](https://en.wikipedia.org/wiki/Receiver_operating_characteristic) is a plot of the true positive rate versus the false positive rate.\n\nAnd the AUC score is the area behind the ROC curve.\n\nScikit-Learn provides a handy function for creating both of these called [`plot_roc_curve()`](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.plot_roc_curve.html).",
"_____no_output_____"
]
],
[
[
"# Plot a ROC curve using our current machine learning model using plot_roc_curve\nclf.fit(X_train,y_train)\nplot_roc_curve(clf,X_test,y_test);",
"_____no_output_____"
]
],
[
[
"Beautiful! We've gone far beyond accuracy with a plethora extra classification evaluation metrics.\n\nIf you're not sure about any of these, don't worry, they can take a while to understand. That could be an optional extension, reading up on a classification metric you're not sure of.\n\nThe thing to note here is all of these metrics have been calculated using a single training set and a single test set. Whilst this is okay, a more robust way is to calculate them using [cross-validation](https://scikit-learn.org/stable/modules/cross_validation.html).\n\nWe can calculate various evaluation metrics using cross-validation using Scikit-Learn's [`cross_val_score()`](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.cross_val_score.html) function along with the `scoring` parameter.",
"_____no_output_____"
]
],
[
[
"# Import cross_val_score from sklearn's model_selection module\nfrom sklearn.model_selection import cross_val_score",
"_____no_output_____"
],
[
"# EXAMPLE: By default cross_val_score returns 5 values (cv=5).\ncross_val_score(clf, \n X, \n y, \n scoring=\"accuracy\",\n cv=5)",
"_____no_output_____"
],
[
"# EXAMPLE: Taking the mean of the returned values from cross_val_score \n# gives a cross-validated version of the scoring metric.\ncross_val_acc = np.mean(cross_val_score(clf,\n X,\n y,\n scoring=\"accuracy\",\n cv=5))\n\ncross_val_acc",
"_____no_output_____"
]
],
[
[
"In the examples, the cross-validated accuracy is found by taking the mean of the array returned by `cross_val_score()`.\n\nNow it's time to find the same for precision, recall and F1 score.",
"_____no_output_____"
]
],
[
[
"# Find the cross-validated precision\ncross_val_precision = np.mean(cross_val_score(clf,\n X,\n y,\n scoring=\"precision\",\n cv=5))\n\ncross_val_precision",
"_____no_output_____"
],
[
"# Find the cross-validated recall\ncross_val_recall = np.mean(cross_val_score(clf,\n X,\n y,\n scoring=\"recall\",\n cv=5))\n\ncross_val_recall",
"_____no_output_____"
],
[
"# Find the cross-validated F1 score\ncross_val_f1 = np.mean(cross_val_score(clf,\n X,\n y,\n scoring=\"f1\",\n cv=5))\n\ncross_val_f1",
"_____no_output_____"
]
],
[
[
"### Exporting and importing a trained model\n\nOnce you've trained a model, you may want to export it and save it to file so you can share it or use it elsewhere.\n\nOne method of exporting and importing models is using the joblib library.\n\nIn Scikit-Learn, exporting and importing a trained model is known as [model persistence](https://scikit-learn.org/stable/modules/model_persistence.html).",
"_____no_output_____"
]
],
[
[
"# Import the dump and load functions from the joblib library\n###",
"_____no_output_____"
],
[
"# Use the dump function to export the trained model to file\n###",
"_____no_output_____"
],
[
"# Use the load function to import the trained model you just exported\n# Save it to a different variable name to the origial trained model\n###\n\n# Evaluate the loaded trained model on the test data\n###",
"_____no_output_____"
]
],
[
[
"What do you notice about the loaded trained model results versus the original (pre-exported) model results?\n\n\n## Scikit-Learn Regression Practice\n\nFor the next few exercises, we're going to be working on a regression problem, in other words, using some data to predict a number.\n\nOur dataset is a [table of car sales](https://docs.google.com/spreadsheets/d/1LPEIWJdSSJYrfn-P3UQDIXbEn5gg-o6I7ExLrWTTBWs/edit?usp=sharing), containing different car characteristics as well as a sale price.\n\nWe'll use Scikit-Learn's built-in regression machine learning models to try and learn the patterns in the car characteristics and their prices on a certain group of the dataset before trying to predict the sale price of a group of cars the model has never seen before.\n\nTo begin, we'll [import the data from GitHub](https://raw.githubusercontent.com/mrdbourke/zero-to-mastery-ml/master/data/car-sales-extended-missing-data.csv) into a pandas DataFrame, check out some details about it and try to build a model as soon as possible.",
"_____no_output_____"
]
],
[
[
"# Read in the car sales data\ncar_sales = pd.read_csv(\"https://raw.githubusercontent.com/mrdbourke/zero-to-mastery-ml/master/data/car-sales-extended-missing-data.csv\")\n\n# View the first 5 rows of the car sales data\ncar_sales.head()",
"_____no_output_____"
],
[
"# Get information about the car sales DataFrame\ncar_sales.isna().sum()",
"_____no_output_____"
]
],
[
[
"Looking at the output of `info()`,\n* How many rows are there total?\n* What datatypes are in each column?\n* How many missing values are there in each column?",
"_____no_output_____"
]
],
[
[
"# Find number of missing values in each column\n###",
"_____no_output_____"
],
[
"# Find the datatypes of each column of car_sales\ncar_sales.dtypes",
"_____no_output_____"
]
],
[
[
"Knowing this information, what would happen if we tried to model our data as it is?\n\nLet's see.",
"_____no_output_____"
]
],
[
[
"# EXAMPLE: This doesn't work because our car_sales data isn't all numerical\nfrom sklearn.ensemble import RandomForestRegressor\ncar_sales_X, car_sales_y = car_sales.drop(\"Price\", axis=1), car_sales.Price\nrf_regressor = RandomForestRegressor().fit(car_sales_X, car_sales_y)",
"_____no_output_____"
]
],
[
[
"As we see, the cell above breaks because our data contains non-numerical values as well as missing data.\n\nTo take care of some of the missing data, we'll remove the rows which have no labels (all the rows with missing values in the `Price` column).",
"_____no_output_____"
]
],
[
[
"# Remove rows with no labels (NaN's in the Price column)\ncar_sales.dropna(subset=[\"Price\"],inplace=True)",
"_____no_output_____"
]
],
[
[
"### Building a pipeline\nSince our `car_sales` data has missing numerical values as well as the data isn't all numerical, we'll have to fix these things before we can fit a machine learning model on it.\n\nThere are ways we could do this with pandas but since we're practicing Scikit-Learn, we'll see how we might do it with the [`Pipeline`](https://scikit-learn.org/stable/modules/generated/sklearn.pipeline.Pipeline.html) class. \n\nBecause we're modifying columns in our dataframe (filling missing values, converting non-numerical data to numbers) we'll need the [`ColumnTransformer`](https://scikit-learn.org/stable/modules/generated/sklearn.compose.ColumnTransformer.html), [`SimpleImputer`](https://scikit-learn.org/stable/modules/generated/sklearn.impute.SimpleImputer.html) and [`OneHotEncoder`](https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.OneHotEncoder.html) classes as well.\n\nFinally, because we'll need to split our data into training and test sets, we'll import `train_test_split` as well.",
"_____no_output_____"
]
],
[
[
"# Import Pipeline from sklearn's pipeline module\nfrom sklearn.pipeline import Pipeline\n\n# Import ColumnTransformer from sklearn's compose module\nfrom sklearn.compose import ColumnTransformer\n \n# Import SimpleImputer from sklearn's impute module\nfrom sklearn.impute import SimpleImputer\n\n# Import OneHotEncoder from sklearn's preprocessing module\nfrom sklearn.preprocessing import OneHotEncoder\n\n# Import train_test_split from sklearn's model_selection module\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
]
],
[
[
"Now we've got the necessary tools we need to create our preprocessing `Pipeline` which fills missing values along with turning all non-numerical data into numbers.\n\nLet's start with the categorical features.",
"_____no_output_____"
]
],
[
[
"# Define different categorical features \ncategorical_features = [\"Make\", \"Colour\"]\n\n# Create categorical transformer Pipeline\ncategorical_transformer = Pipeline(steps=[\n # Set SimpleImputer strategy to \"constant\" and fill value to \"missing\"\n (\"imputer\", SimpleImputer(strategy=\"constant\", fill_value=\"missing\")),\n # Set OneHotEncoder to ignore the unknowns\n (\"onehot\", OneHotEncoder(handle_unknown=\"ignore\"))])",
"_____no_output_____"
]
],
[
[
"It would be safe to treat `Doors` as a categorical feature as well, however since we know the vast majority of cars have 4 doors, we'll impute the missing `Doors` values as 4.",
"_____no_output_____"
]
],
[
[
"# Define Doors features\ndoor_feature = [\"Doors\"]\n\n# Create Doors transformer Pipeline\ndoor_transformer = Pipeline(steps=[\n # Set SimpleImputer strategy to \"constant\" and fill value to 4\n (\"imputer\", SimpleImputer(strategy=\"constant\", fill_value=4))])",
"_____no_output_____"
]
],
[
[
"Now onto the numeric features. In this case, the only numeric feature is the `Odometer (KM)` column. Let's fill its missing values with the median.",
"_____no_output_____"
]
],
[
[
"# Define numeric features (only the Odometer (KM) column)\nnumeric_features = [\"Odometer (KM)\"]\n\n# Crearte numeric transformer Pipeline\nnumeric_transformer = Pipeline(steps=[\n # Set SimpleImputer strategy to fill missing values with the \"Median\"\n (\"imputer\", SimpleImputer(strategy=\"median\"))])",
"_____no_output_____"
]
],
[
[
"Time to put all of our individual transformer `Pipeline`'s into a single `ColumnTransformer` instance.",
"_____no_output_____"
]
],
[
[
"# Setup preprocessing steps (fill missing values, then convert to numbers)\npreprocessor = ColumnTransformer(\n transformers=[\n # Use the categorical_transformer to transform the categorical_features\n (\"cat\", categorical_transformer, categorical_features),\n # Use the door_transformer to transform the door_feature\n (\"door\", door_transformer, door_feature),\n # Use the numeric_transformer to transform the numeric_features\n (\"num\", numeric_transformer, numeric_features)])",
"_____no_output_____"
]
],
[
[
"Boom! Now our `preprocessor` is ready, time to import some regression models to try out.\n\nComparing our data to the [Scikit-Learn machine learning map](https://scikit-learn.org/stable/tutorial/machine_learning_map/index.html), we can see there's a handful of different regression models we can try.\n\n* [RidgeRegression](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Ridge.html)\n* [SVR(kernel=\"linear\")](https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVR.html) - short for Support Vector Regressor, a form form of support vector machine.\n* [SVR(kernel=\"rbf\")](https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVR.html) - short for Support Vector Regressor, a form of support vector machine.\n* [RandomForestRegressor](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestRegressor.html) - the regression version of RandomForestClassifier.",
"_____no_output_____"
]
],
[
[
"# Import Ridge from sklearn's linear_model module\nfrom sklearn.linear_model import Ridge\n\n# Import SVR from sklearn's svm module\nfrom sklearn.svm import SVR\n\n# Import RandomForestRegressor from sklearn's ensemble module\nfrom sklearn.ensemble import RandomForestRegressor",
"_____no_output_____"
]
],
[
[
"Again, thanks to the design of the Scikit-Learn library, we're able to use very similar code for each of these models.\n\nTo test them all, we'll create a dictionary of regression models and an empty dictionary for regression model results.",
"_____no_output_____"
]
],
[
[
"# Create dictionary of model instances, there should be 4 total key, value pairs\n# in the form {\"model_name\": model_instance}.\n# Don't forget there's two versions of SVR, one with a \"linear\" kernel and the\n# other with kernel set to \"rbf\".\nregression_models = {\"Ridge\": Ridge(),\n \"SVR_linear\":SVR(kernel=\"linear\"),\n \"SVR_rbf\": SVR(kernel=\"rbf\"),\n \"RandomForestRegressor\": RandomForestRegressor()}\n\n# Create an empty dictionary for the regression results\nregression_results = {}",
"_____no_output_____"
]
],
[
[
"Our regression model dictionary is prepared as well as an empty dictionary to append results to, time to get the data split into `X` (feature variables) and `y` (target variable) as well as training and test sets.\n\nIn our car sales problem, we're trying to use the different characteristics of a car (`X`) to predict its sale price (`y`).",
"_____no_output_____"
]
],
[
[
"# Create car sales X data (every column of car_sales except Price)\ncar_sales_X = car_sales.drop(\"Price\",axis=1)\n\n# Create car sales y data (the Price column of car_sales)\ncar_sales_y = car_sales[\"Price\"]",
"_____no_output_____"
],
[
"# Use train_test_split to split the car_sales_X and car_sales_y data into \n# training and test sets.\n# Give the test set 20% of the data using the test_size parameter.\n# For reproducibility set the random_state parameter to 42.\ncar_X_train, car_X_test, car_y_train, car_y_test = train_test_split(car_sales_X,\n car_sales_y,\n test_size=0.2,\n random_state=42)\n\n# Check the shapes of the training and test datasets\ncar_X_train.shape, car_X_test.shape, car_y_train.shape, car_y_test.shape",
"_____no_output_____"
]
],
[
[
"* How many rows are in each set?\n* How many columns are in each set?\n\nAlright, our data is split into training and test sets, time to build a small loop which is going to:\n1. Go through our `regression_models` dictionary\n2. Create a `Pipeline` which contains our `preprocessor` as well as one of the models in the dictionary\n3. Fits the `Pipeline` to the car sales training data\n4. Evaluates the target model on the car sales test data and appends the results to our `regression_results` dictionary",
"_____no_output_____"
]
],
[
[
"# Loop through the items in the regression_models dictionary\nfor model_name, model in regression_models.items():\n \n # Create a model Pipeline with a preprocessor step and model step\n model_pipeline = Pipeline(steps=[(\"preprocessor\", preprocessor),\n (\"model\", model)])\n \n # Fit the model Pipeline to the car sales training data\n print(f\"Fitting {model_name}...\")\n model_pipeline.fit(car_X_train, car_y_train)\n \n # Score the model Pipeline on the test data appending the model_name to the \n # results dictionary\n print(f\"Scoring {model_name}...\")\n regression_results[model_name] = model_pipeline.score(car_X_test,car_y_test)",
"Fitting Ridge...\nScoring Ridge...\nFitting SVR_linear...\nScoring SVR_linear...\nFitting SVR_rbf...\nScoring SVR_rbf...\nFitting RandomForestRegressor...\nScoring RandomForestRegressor...\n"
]
],
[
[
"Our regression models have been fit, let's see how they did!",
"_____no_output_____"
]
],
[
[
"# Check the results of each regression model by printing the regression_results\n# dictionary\nregression_results",
"_____no_output_____"
]
],
[
[
"* Which model did the best?\n* How could you improve its results?\n* What metric does the `score()` method of a regression model return by default?\n\nSince we've fitted some models but only compared them via the default metric contained in the `score()` method (R^2 score or coefficient of determination), let's take the `RidgeRegression` model and evaluate it with a few other [regression metrics](https://scikit-learn.org/stable/modules/model_evaluation.html#regression-metrics).\n\nSpecifically, let's find:\n1. **R^2 (pronounced r-squared) or coefficient of determination** - Compares your models predictions to the mean of the targets. Values can range from negative infinity (a very poor model) to 1. For example, if all your model does is predict the mean of the targets, its R^2 value would be 0. And if your model perfectly predicts a range of numbers it's R^2 value would be 1. \n2. **Mean absolute error (MAE)** - The average of the absolute differences between predictions and actual values. It gives you an idea of how wrong your predictions were.\n3. **Mean squared error (MSE)** - The average squared differences between predictions and actual values. Squaring the errors removes negative errors. It also amplifies outliers (samples which have larger errors).\n\nScikit-Learn has a few classes built-in which are going to help us with these, namely, [`mean_absolute_error`](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.mean_absolute_error.html), [`mean_squared_error`](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.mean_squared_error.html) and [`r2_score`](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.r2_score.html).",
"_____no_output_____"
]
],
[
[
"# Import mean_absolute_error from sklearn's metrics module\nfrom sklearn.metrics import mean_absolute_error,mean_squared_error,r2_score\n\n# Import mean_squared_error from sklearn's metrics module\n###\n\n# Import r2_score from sklearn's metrics module\n###",
"_____no_output_____"
]
],
[
[
"All the evaluation metrics we're concerned with compare a model's predictions with the ground truth labels. Knowing this, we'll have to make some predictions.\n\nLet's create a `Pipeline` with the `preprocessor` and a `Ridge()` model, fit it on the car sales training data and then make predictions on the car sales test data.",
"_____no_output_____"
]
],
[
[
"# Create RidgeRegression Pipeline with preprocessor as the \"preprocessor\" and\n# Ridge() as the \"model\".\nridge_pipeline = Pipeline(steps=[(\"preprocessor\", preprocessor),\n (\"model\", Ridge())])\n\n# Fit the RidgeRegression Pipeline to the car sales training data\nridge_pipeline.fit(car_X_train,car_y_train)\n\n# Make predictions on the car sales test data using the RidgeRegression Pipeline\ncar_y_preds = ridge_pipeline.predict(car_X_test)\n\n# View the first 50 predictions\ncar_y_preds[:50]",
"_____no_output_____"
]
],
[
[
"Nice! Now we've got some predictions, time to evaluate them. We'll find the mean squared error (MSE), mean absolute error (MAE) and R^2 score (coefficient of determination) of our model.",
"_____no_output_____"
]
],
[
[
"# EXAMPLE: Find the MSE by comparing the car sales test labels to the car sales predictions\nmse = mean_squared_error(car_y_test, car_y_preds)\n# Return the MSE\nmse",
"_____no_output_____"
],
[
"# Find the MAE by comparing the car sales test labels to the car sales predictions\nmae = mean_absolute_error(car_y_test, car_y_preds)\n# Return the MAE\nmae",
"_____no_output_____"
],
[
"# Find the R^2 score by comparing the car sales test labels to the car sales predictions\nr2 = r2_score(car_y_test, car_y_preds)\n# Return the R^2 score\nr2",
"_____no_output_____"
]
],
[
[
"Boom! Our model could potentially do with some hyperparameter tuning (this would be a great extension). And we could probably do with finding some more data on our problem, 1000 rows doesn't seem to be sufficient.\n\n* How would you export the trained regression model?",
"_____no_output_____"
],
[
"## Extensions\n\nYou should be proud. Getting this far means you've worked through a classification problem and regression problem using pure (mostly) Scikit-Learn (no easy feat!).\n\nFor more exercises, check out the [Scikit-Learn getting started documentation](https://scikit-learn.org/stable/getting_started.html). A good practice would be to read through it and for the parts you find interesting, add them into the end of this notebook.\n\nFinally, as always, remember, the best way to learn something new is to try it. And try it relentlessly. If you're unsure of how to do something, never be afraid to ask a question or search for something such as, \"how to tune the hyperparmaters of a scikit-learn ridge regression model\".",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ecc97238a20a1985dd36aa1304d21879ed154d92 | 768,425 | ipynb | Jupyter Notebook | jupyter/Experiment_unconstrained_problem_guru_pop01_rare01.ipynb | smnikolakaki/submodular-linear-cost-maximization | 98be3e79c11e4a36c253ed9a4800e6976b4aa3bf | [
"MIT"
] | null | null | null | jupyter/Experiment_unconstrained_problem_guru_pop01_rare01.ipynb | smnikolakaki/submodular-linear-cost-maximization | 98be3e79c11e4a36c253ed9a4800e6976b4aa3bf | [
"MIT"
] | null | null | null | jupyter/Experiment_unconstrained_problem_guru_pop01_rare01.ipynb | smnikolakaki/submodular-linear-cost-maximization | 98be3e79c11e4a36c253ed9a4800e6976b4aa3bf | [
"MIT"
] | null | null | null | 1,152.061469 | 382,212 | 0.951009 | [
[
[
"### Import packages",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport matplotlib\nimport matplotlib.pyplot as plt\nimport os\nimport sys\nimport dill\nimport yaml\nimport numpy as np\nimport pandas as pd\nimport ast\nimport collections\n\nimport seaborn as sns\nsns.set(style='ticks')",
"_____no_output_____"
]
],
[
[
"### Import submodular-optimization packages",
"_____no_output_____"
]
],
[
[
"sys.path.insert(0, \"/Users/smnikolakaki/GitHub/submodular-linear-cost-maximization/submodular_optimization/\")",
"_____no_output_____"
]
],
[
[
"### Visualizations directory",
"_____no_output_____"
]
],
[
[
"VIZ_DIR = os.path.abspath(\"/Users/smnikolakaki/GitHub/submodular-linear-cost-maximization/submodular_optimization/viz/\")",
"_____no_output_____"
]
],
[
[
"### Legends and style dictionary",
"_____no_output_____"
]
],
[
[
"legends = {\n \"distorted_greedy\":\"DistortedGreedy\",\n \"cost_scaled_lazy_exact_greedy\":\"CSLG\",\n \"unconstrained_linear\":\"OnlineCSG\",\n \"unconstrained_distorted_greedy\":\"UnconstrainedDistortedGreedy\",\n \"stochastic_distorted_greedy_0.01\":\"StochasticDistortedGreedy\",\n \"baseline_topk\": \"Top-k-Experts\",\n \"greedy\":\"Greedy\"\n}\n\nlegends = collections.OrderedDict(sorted(legends.items()))\n\nline_styles = {'distorted_greedy':':',\n 'cost_scaled_lazy_exact_greedy':'-',\n 'unconstrained_linear':'-',\n 'unconstrained_distorted_greedy':'-',\n 'stochastic_distorted_greedy_0.01':'-.',\n 'baseline_topk':'--',\n \"greedy\":\"--\"\n }\n\nline_styles = collections.OrderedDict(sorted(line_styles.items()))\n\nmarker_style = {'distorted_greedy':'s',\n 'cost_scaled_lazy_exact_greedy':'x',\n 'unconstrained_linear':'*',\n 'unconstrained_distorted_greedy':'+',\n 'stochastic_distorted_greedy_0.01':'o',\n 'baseline_topk':'d',\n \"greedy\":\"h\"\n }\n\nmarker_style = collections.OrderedDict(sorted(marker_style.items()))\n\nmarker_size = {'distorted_greedy':25,\n 'cost_scaled_lazy_exact_greedy':30,\n 'unconstrained_linear':25,\n 'unconstrained_distorted_greedy':25,\n 'stochastic_distorted_greedy_0.01':25,\n 'baseline_topk':22,\n \"greedy\":30\n }\n\nmarker_size = collections.OrderedDict(sorted(marker_size.items()))\n\nmarker_edge_width = {'distorted_greedy':6,\n 'cost_scaled_lazy_exact_greedy':10,\n 'unconstrained_linear':10,\n 'unconstrained_distorted_greedy':6,\n 'stochastic_distorted_greedy_0.01':6,\n 'baseline_topk':6,\n \"greedy\":6\n }\n\nmarker_edge_width = collections.OrderedDict(sorted(marker_edge_width.items()))\n\nline_width = {'distorted_greedy':5,\n 'cost_scaled_lazy_exact_greedy':5,\n 'unconstrained_linear':5,\n 'unconstrained_distorted_greedy':5,\n 'stochastic_distorted_greedy_0.01':5,\n 'baseline_topk':5,\n \"greedy\":5\n }\n\nline_width = collections.OrderedDict(sorted(line_width.items()))\n\nname_objective = \"Combined objective (g)\"\nfontsize = 53\nlegendsize = 42\nlabelsize = 53\nx_size = 20\ny_size = 16",
"_____no_output_____"
]
],
[
[
"### Plotting utilities",
"_____no_output_____"
]
],
[
[
"def set_style():\n # This sets reasonable defaults for font size for a paper\n sns.set_context(\"paper\") \n # Set the font to be serif\n sns.set(font='serif')#, rc={'text.usetex' : True}) \n # Make the background white, and specify the specific font family\n sns.set_style(\"white\", {\n \"font.family\": \"serif\",\n \"font.serif\": [\"Times\", \"Palatino\", \"serif\"]\n })\n # Set tick size for axes\n sns.set_style(\"ticks\", {\"xtick.major.size\": 6, \"ytick.major.size\": 6})",
"_____no_output_____"
],
[
"def set_size(fig, width=13, height=12):\n fig.set_size_inches(width, height)\n plt.tight_layout()",
"_____no_output_____"
],
[
"def save_fig(fig, filename):\n fig.savefig(os.path.join(VIZ_DIR, filename), dpi=600, format='pdf', bbox_inches='tight')",
"_____no_output_____"
]
],
[
[
"### Plots",
"_____no_output_____"
]
],
[
[
"df1 = pd.read_csv(\"/Users/smnikolakaki/GitHub/submodular-linear-cost-maximization/jupyter/experiment_00_guru_pop01_rare01_final.csv\",\n header=0,\n index_col=False)\n\ndf1.columns = ['Algorithm', 'sol', 'val', 'submodular_val', 'cost', 'runtime', 'lazy_epsilon',\n 'sample_epsilon','user_sample_ratio','scaling_factor','num_rare_skills','num_common_skills',\n 'num_popular_skills','num_sampled_skills','seed','k']\n\ndf2 = pd.read_csv(\"/Users/smnikolakaki/GitHub/submodular-linear-cost-maximization/jupyter/experiment_00_guru_pop01_rare01_greedy.csv\",\n header=0,\n index_col=False)\n\ndf2.columns = ['Algorithm', 'sol', 'val', 'submodular_val', 'cost', 'runtime', 'lazy_epsilon',\n 'sample_epsilon','user_sample_ratio','scaling_factor','num_rare_skills','num_common_skills',\n 'num_popular_skills','num_sampled_skills','seed','k']\n\nframes = []\nframes.append(df1)\nframes.append(df2)\n\ndf_final = pd.concat(frames)\ndf_final.to_csv(\"/Users/smnikolakaki/GitHub/submodular-linear-cost-maximization/jupyter/experiment_00_guru_pop01_rare01.csv\", index=False) ",
"_____no_output_____"
]
],
[
[
"#### Details\nOriginal marginal gain: $$g(e|S) = f(e|S) - w(e)$$\nScaled marginal gain: $$\\tilde{g}(e|S) = f(e|S) - 2w(e)$$\nDistorted marginal gain: $$\\tilde{g}(e|S) = (1-\\frac{\\gamma}{n})^{n-(i+1)}f(e|S) - w(e)$$ \n\n#### Algorithms:\n1. Cost Scaled Greedy: The algorithm performs iterations i = 0,...,n-1. In each iteration the algorithm selects the element that maximizes the scaled marginal gain. It adds the element to the solution if the original marginal gain of the element is >= 0. The algorithm returns a solution S: f(S) - w(S) >= (1/2)f(OPT) - w(OPT). The running time is O($n^2$).\n\n\n2. Cost Scaled Exact Lazy Greedy: The algorithm first initializes a max heap with all the elements. The key of each element is its scaled marginal gain and the value is the element id. If the scaled marginal gain of an element is < 0 the algorithm discards the element and never inserts it in the heap. Then, for 0,...,n-1 iterations the algorithm does the following: (i) pops the top element from the heap and computes its new scaled marginal gain, (ii) It checks the old scaled marginal gain of the next element in the heap, (iii) if the popped element's new scaled marginal gain is >= the next elements's old gain we return the popped element, otherwise if its new scaled marginal gain is >= 0 we reinsert the element to the heap and repeat step iii, otherwise we discard it and repeat step iii, (iv) if the returned element's original marginal gain is >= 0 we add it to the solution. The algorithm returns a solution S: f(S) - w(S) >= (1/2)f(OPT) - w(OPT). The running time is O($n^2$).\n\n\n3. Unconstrained Linear: The algorithm performs i = 0,...,n-1 iterations (one for each arriving element). For each element it adds it to the solution if its scaled marginal gain is > 0. The algorithm returns a solution S: f(S) - w(S) >= (1/2)f(OPT) - w(OPT). The running time is O($n$).\n\n\n4. Distorted Greedy: The algorithm performs i = 0,...,n-1 iterations. In each iteration the algorithm selects the element that maximizes the distorted marginal gain. It adds the element to the solution if the distorted marginal gain of the element is > 0. The algorithm returns a solution S: f(S) - w(S) >= (1-1/e)f(OPT) - w(OPT). The running time is O($n^2$). The algorithmic implementation is based on Algorithm 1 found [here](https://arxiv.org/pdf/1904.09354.pdf) for k=n.\n\n\n5. Stochastic Distorted Greedy: The algorithm performs i = 0,...,n-1 iterations. In each iteration the algorithm chooses a sample of s=log(1/ε) elements uniformly and independently and from this sample it selects the element that maximizes the distorted marginal gain. It adds the element to the solution if the distorted marginal gain of the element is > 0. We set $ε=0.01$. The algorithm returns a solution S: E[f(S) - w(S)] >= (1-1/e-ε)f(OPT) - w(OPT). The running time is O($n\\log{1/ε}$). The algorithmic implementation is based on Algorithm 2 found [here](https://arxiv.org/pdf/1904.09354.pdf) for k=n.\n\n\n6. Unconstrained Distorted Greedy: The algorithm performs i = 0,...,n-1 iterations. In each iteration the algorithm chooses a random single element uniformly. It adds the element to the solution if the distorted marginal gain of the element is > 0. The algorithm returns a solution S: E[f(S) - w(S)] >= (1-1/e)f(OPT) - w(OPT). The running time is O($n$).",
"_____no_output_____"
],
[
"#### Performance comparison",
"_____no_output_____"
]
],
[
[
"def plot_performance_comparison(df):\n \n palette = sns.color_palette(['#b30000','#dd8452', '#4c72b0','#ccb974',\n '#55a868', '#64b5cd', \n '#8172b3', '#937860', '#da8bc3', '#8c8c8c', \n '#ccb974', '#64b5cd'],7)\n \n ax = sns.lineplot(x='user_sample_ratio', y='val', data=df, \n style=\"Algorithm\",hue='Algorithm', ci='sd', \n mfc='none',palette=palette, dashes=False)\n i = 0\n for key, val in line_styles.items():\n ax.lines[i].set_linestyle(val)\n # ax.lines[i].set_color(colors[key])\n ax.lines[i].set_linewidth(line_width[key])\n ax.lines[i].set_marker(marker_style[key])\n ax.lines[i].set_markersize(marker_size[key])\n ax.lines[i].set_markeredgewidth(marker_edge_width[key])\n ax.lines[i].set_markeredgecolor(None)\n i += 1\n \n plt.yticks(np.arange(0, 45000, 5000))\n plt.xticks(np.arange(0, 1.1, 0.1))\n plt.xlabel('Expert sample fraction', fontsize=fontsize)\n plt.ylabel(name_objective, fontsize=fontsize)\n # plt.title('Performance comparison')\n fig = plt.gcf()\n figlegend = plt.legend([val for key,val in legends.items()],loc=3, bbox_to_anchor=(0., 1.02, 1., .102),\n ncol=2, mode=\"expand\", borderaxespad=0., frameon=False,prop={'size': legendsize})\n ax = plt.gca()\n plt.gca().tick_params(axis='y', labelsize=labelsize)\n plt.gca().tick_params(axis='x', labelsize=labelsize)\n return fig, ax",
"_____no_output_____"
],
[
"df = pd.read_csv(\"/Users/smnikolakaki/GitHub/submodular-linear-cost-maximization/jupyter/experiment_00_guru_pop01_rare01.csv\",\n header=0,\n index_col=False)\n\ndf.columns = ['Algorithm', 'sol', 'val', 'submodular_val', 'cost', 'runtime', 'lazy_epsilon',\n 'sample_epsilon','user_sample_ratio','scaling_factor','num_rare_skills','num_common_skills',\n 'num_popular_skills','num_sampled_skills','seed','k']\n\ndf = df[(df.Algorithm == 'distorted_greedy')\n # |(df.Algorithm == 'cost_scaled_greedy')\n |(df.Algorithm == 'cost_scaled_lazy_greedy')\n |(df.Algorithm == 'unconstrained_linear')\n |(df.Algorithm == 'unconstrained_distorted_greedy')\n |(df.Algorithm == 'stochastic_distorted_greedy_0.01')\n |(df.Algorithm == 'baseline_topk')\n |(df.Algorithm == 'greedy')\n ]\ndf0 = df[(df['sample_epsilon'].isnull()) | (df['sample_epsilon'] == 0.01)]\ndf0.sort_values(by ='Algorithm',inplace=True)\nset_style()\nfig, axes = plot_performance_comparison(df0)\nset_size(fig, x_size, y_size)\nsave_fig(fig,'score_unconstrained_guru_pop01_rare01.pdf')",
"_____no_output_____"
]
],
[
[
"#### Runtime comparison for different dataset sizes",
"_____no_output_____"
]
],
[
[
"legends = {\n \"distorted_greedy\":\"DistortedGreedy\",\n \"cost_scaled_greedy\":\"CSG\",\n \"cost_scaled_lazy_exact_greedy\":\"CSLG\",\n \"unconstrained_linear\":\"OnlineCSG\",\n \"unconstrained_distorted_greedy\":\"UnconstrainedDistortedGreedy\",\n \"stochastic_distorted_greedy_0.01\":\"StochasticDistortedGreedy\",\n \"baseline_topk\": \"Top-k-Experts\",\n \"greedy\":\"Greedy\"\n}\n\nlegends = collections.OrderedDict(sorted(legends.items()))\n\nline_styles = {'distorted_greedy':':',\n 'cost_scaled_greedy':'-',\n 'cost_scaled_lazy_exact_greedy':'-',\n 'unconstrained_linear':'-',\n 'unconstrained_distorted_greedy':'-',\n 'stochastic_distorted_greedy_0.01':'-.',\n 'baseline_topk':'--',\n \"greedy\":\"--\"\n }\n\nline_styles = collections.OrderedDict(sorted(line_styles.items()))\n\nmarker_style = {'distorted_greedy':'s',\n 'cost_scaled_greedy':'x',\n 'cost_scaled_lazy_exact_greedy':'x',\n 'unconstrained_linear':'*',\n 'unconstrained_distorted_greedy':'+',\n 'stochastic_distorted_greedy_0.01':'o',\n 'baseline_topk':'d',\n \"greedy\":\"h\"\n }\n\nmarker_style = collections.OrderedDict(sorted(marker_style.items()))\n\nmarker_size = {'distorted_greedy':25,\n 'cost_scaled_greedy':30,\n 'cost_scaled_lazy_exact_greedy':30,\n 'unconstrained_linear':25,\n 'unconstrained_distorted_greedy':25,\n 'stochastic_distorted_greedy_0.01':25,\n 'baseline_topk':22,\n \"greedy\":30\n }\n\nmarker_size = collections.OrderedDict(sorted(marker_size.items()))\n\nmarker_edge_width = {'distorted_greedy':6,\n 'cost_scaled_greedy':10,\n 'cost_scaled_lazy_exact_greedy':10,\n 'unconstrained_linear':6,\n 'unconstrained_distorted_greedy':6,\n 'stochastic_distorted_greedy_0.01':6,\n 'baseline_topk':6,\n \"greedy\":6\n }\n\nmarker_edge_width = collections.OrderedDict(sorted(marker_edge_width.items()))\n\nline_width = {'distorted_greedy':5,\n 'cost_scaled_greedy':5,\n 'cost_scaled_lazy_exact_greedy':5,\n 'unconstrained_linear':5,\n 'unconstrained_distorted_greedy':5,\n 'stochastic_distorted_greedy_0.01':5,\n 'baseline_topk':5,\n \"greedy\":5}\n\nline_width = collections.OrderedDict(sorted(line_width.items()))\n\n\nname_objective = \"Combined objective (g)\"\nfontsize = 53\nlegendsize = 42\nlabelsize = 53\nx_size = 20\ny_size = 16\n\ndef plot_performance_comparison(df):\n \n palette = sns.color_palette(['#b30000','#937860','#dd8452', '#4c72b0','#ccb974'\n ,'#55a868', '#64b5cd', \n '#8172b3', '#937860', '#da8bc3', '#8c8c8c', \n '#ccb974', '#64b5cd'],8)\n \n ax = sns.lineplot(x='user_sample_ratio', y='runtime', data=df, \n style=\"Algorithm\",hue='Algorithm', ci='sd', \n mfc='none',palette=palette, dashes=False)\n i = 0\n for key, val in line_styles.items():\n ax.lines[i].set_linestyle(val)\n # ax.lines[i].set_color(colors[key])\n ax.lines[i].set_linewidth(line_width[key])\n ax.lines[i].set_marker(marker_style[key])\n ax.lines[i].set_markersize(marker_size[key])\n ax.lines[i].set_markeredgewidth(marker_edge_width[key])\n ax.lines[i].set_markeredgecolor(None)\n i += 1\n \n# plt.yticks(np.arange(0, 45000, 5000))\n plt.xticks(np.arange(0, 1.1, 0.1))\n plt.xlabel('Expert sample fraction', fontsize=fontsize)\n plt.ylabel('Time (sec)', fontsize=fontsize)\n # plt.title('Performance comparison')\n fig = plt.gcf()\n figlegend = plt.legend([val for key,val in legends.items()],loc=3, bbox_to_anchor=(0., 1.02, 1., .102),\n ncol=2, mode=\"expand\", borderaxespad=0., frameon=False,prop={'size': legendsize})\n ax = plt.gca()\n plt.gca().tick_params(axis='y', labelsize=labelsize)\n plt.gca().tick_params(axis='x', labelsize=labelsize)\n \n a = plt.axes([.17, .43, .35, .3])\n ax2 = sns.lineplot(x='user_sample_ratio', y='runtime', data=df, \n hue='Algorithm', legend=False,\n mfc='none',palette=palette,label=False)\n \n i = 0\n for key, val in line_styles.items():\n ax2.lines[i].set_linestyle(val)\n # ax.lines[i].set_color(colors[key])\n ax2.lines[i].set_linewidth(2)\n ax2.lines[i].set_marker(marker_style[key])\n ax2.lines[i].set_markersize(12)\n ax2.lines[i].set_markeredgewidth(3)\n ax2.lines[i].set_markeredgecolor(None)\n i += 1\n \n ax2.set(ylim=(0, 2))\n ax2.set(xlim=(0, 1))\n ax2.set_ylabel('') \n ax2.set_xlabel('')\n \n # plt.gca().xaxis.set_major_formatter(mtick.FormatStrFormatter('%.1e'))\n # plt.gca().yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1e'))\n plt.gca().tick_params(axis='x', labelsize=22)\n plt.gca().tick_params(axis='y', labelsize=22)\n \n return fig, ax\n",
"_____no_output_____"
],
[
"df = pd.read_csv(\"/Users/smnikolakaki/GitHub/submodular-linear-cost-maximization/jupyter/experiment_00_guru_pop01_rare01.csv\",\n header=0,\n index_col=False)\n\ndf.columns = ['Algorithm', 'sol', 'val', 'submodular_val', 'cost', 'runtime', 'lazy_epsilon',\n 'sample_epsilon','user_sample_ratio','scaling_factor','num_rare_skills','num_common_skills',\n 'num_popular_skills','num_sampled_skills','seed','k']\n\ndf = df[(df.Algorithm == 'distorted_greedy')\n |(df.Algorithm == 'cost_scaled_greedy')\n |(df.Algorithm == 'cost_scaled_lazy_greedy')\n |(df.Algorithm == 'unconstrained_linear')\n |(df.Algorithm == 'unconstrained_distorted_greedy')\n |(df.Algorithm == 'stochastic_distorted_greedy_0.01')\n |(df.Algorithm == 'baseline_topk')\n |(df.Algorithm == 'greedy')\n ]\ndf0 = df[(df['sample_epsilon'].isnull()) | (df['sample_epsilon'] == 0.01)]\ndf0.sort_values(by ='Algorithm',inplace=True)\nset_style()\nfig, axes = plot_performance_comparison(df0)\nset_size(fig, x_size, y_size)\nsave_fig(fig,'time_unconstrained_guru_pop01_rare01.pdf')",
"/opt/anaconda3/envs/python3.6/lib/python3.6/site-packages/ipykernel_launcher.py:3: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.\n This is separate from the ipykernel package so we can avoid doing imports until\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecc977236ecf48f61e0816afefedeed5ec133c55 | 104,338 | ipynb | Jupyter Notebook | 04 - Classification/Batch1/Number_Classification/04- Classifying number.ipynb | madhu0309/Internship | 06a4cff28e0c3fc7b57e95c255e5ee770dff9716 | [
"MIT"
] | null | null | null | 04 - Classification/Batch1/Number_Classification/04- Classifying number.ipynb | madhu0309/Internship | 06a4cff28e0c3fc7b57e95c255e5ee770dff9716 | [
"MIT"
] | null | null | null | 04 - Classification/Batch1/Number_Classification/04- Classifying number.ipynb | madhu0309/Internship | 06a4cff28e0c3fc7b57e95c255e5ee770dff9716 | [
"MIT"
] | null | null | null | 119.65367 | 14,804 | 0.854272 | [
[
[
"# Classifier name ",
"_____no_output_____"
],
[
"Step -1 :import datascience libraries",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"**Step-2: Load Dataset**",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import load_digits",
"_____no_output_____"
],
[
"digits = load_digits()",
"_____no_output_____"
],
[
"digits.keys()",
"_____no_output_____"
],
[
"print(digits.DESCR)",
"Optical Recognition of Handwritten Digits Data Set\n===================================================\n\nNotes\n-----\nData Set Characteristics:\n :Number of Instances: 5620\n :Number of Attributes: 64\n :Attribute Information: 8x8 image of integer pixels in the range 0..16.\n :Missing Attribute Values: None\n :Creator: E. Alpaydin (alpaydin '@' boun.edu.tr)\n :Date: July; 1998\n\nThis is a copy of the test set of the UCI ML hand-written digits datasets\nhttp://archive.ics.uci.edu/ml/datasets/Optical+Recognition+of+Handwritten+Digits\n\nThe data set contains images of hand-written digits: 10 classes where\neach class refers to a digit.\n\nPreprocessing programs made available by NIST were used to extract\nnormalized bitmaps of handwritten digits from a preprinted form. From a\ntotal of 43 people, 30 contributed to the training set and different 13\nto the test set. 32x32 bitmaps are divided into nonoverlapping blocks of\n4x4 and the number of on pixels are counted in each block. This generates\nan input matrix of 8x8 where each element is an integer in the range\n0..16. This reduces dimensionality and gives invariance to small\ndistortions.\n\nFor info on NIST preprocessing routines, see M. D. Garris, J. L. Blue, G.\nT. Candela, D. L. Dimmick, J. Geist, P. J. Grother, S. A. Janet, and C.\nL. Wilson, NIST Form-Based Handprint Recognition System, NISTIR 5469,\n1994.\n\nReferences\n----------\n - C. Kaynak (1995) Methods of Combining Multiple Classifiers and Their\n Applications to Handwritten Digit Recognition, MSc Thesis, Institute of\n Graduate Studies in Science and Engineering, Bogazici University.\n - E. Alpaydin, C. Kaynak (1998) Cascading Classifiers, Kybernetika.\n - Ken Tang and Ponnuthurai N. Suganthan and Xi Yao and A. Kai Qin.\n Linear dimensionalityreduction using relevance weighted LDA. School of\n Electrical and Electronic Engineering Nanyang Technological University.\n 2005.\n - Claudio Gentile. A New Approximate Maximal Margin Classification\n Algorithm. NIPS. 2000.\n\n"
],
[
"X = digits.data # independent varible \ny = digits.target # dependent varible",
"_____no_output_____"
]
],
[
[
"each row is an image",
"_____no_output_____"
]
],
[
[
"print(X.shape)\nprint(X)",
"(1797, 64)\n[[ 0. 0. 5. ... 0. 0. 0.]\n [ 0. 0. 0. ... 10. 0. 0.]\n [ 0. 0. 0. ... 16. 9. 0.]\n ...\n [ 0. 0. 1. ... 6. 0. 0.]\n [ 0. 0. 2. ... 12. 0. 0.]\n [ 0. 0. 10. ... 12. 1. 0.]]\n"
],
[
"# Converting into binary image --> Thresholding\nX[X > 7] = X.max()\nX[X<= 7] = X.min()\n# Normalizing\nX = X / X.max()",
"_____no_output_____"
],
[
"X.shape, y.shape",
"_____no_output_____"
],
[
"img = X[0:1]",
"_____no_output_____"
],
[
"print(y[0:1])\nplt.imshow(img.reshape((8,8)),cmap = 'gray')\n",
"[0]\n"
]
],
[
[
"** Step 4 Splitting Data into traning and testing **",
"_____no_output_____"
]
],
[
[
"from sklearn.cross_validation import train_test_split",
"C:\\Users\\srikanth\\Anaconda\\lib\\site-packages\\sklearn\\cross_validation.py:41: DeprecationWarning: This module was deprecated in version 0.18 in favor of the model_selection module into which all the refactored classes and functions are moved. Also note that the interface of the new CV iterators are different from that of this module. This module will be removed in 0.20.\n \"This module will be removed in 0.20.\", DeprecationWarning)\n"
],
[
"x_train , x_test, y_train, y_test = train_test_split(X, y,\n test_size = 0.2,\n random_state = 0)",
"_____no_output_____"
],
[
"x_train.shape, x_test.shape, y_train.shape, y_test.shape",
"_____no_output_____"
]
],
[
[
"### Step 5: Building Machine Learning classifier or model",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.svm import SVC\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.ensemble import RandomForestClassifier",
"_____no_output_____"
],
[
"model_log = LogisticRegression(C = 10.0)\nmodel_knn = KNeighborsClassifier(n_neighbors=3)\nmodel_svm = SVC(C = 10.0,probability=True) \nmodel_dt = DecisionTreeClassifier()\nmodel_rf = RandomForestClassifier(n_estimators=100)",
"_____no_output_____"
]
],
[
[
"traning model",
"_____no_output_____"
]
],
[
[
"model_log.fit(x_train, y_train) # training model\nmodel_knn.fit(x_train, y_train) # training model\nmodel_svm.fit(x_train, y_train) # training model\nmodel_dt.fit(x_train, y_train) # training model\nmodel_rf.fit(x_train, y_train) # training model",
"_____no_output_____"
]
],
[
[
"**Step -6 :Evaluation model**",
"_____no_output_____"
]
],
[
[
"y_pred_log = model_log.predict(x_test) # we use this for evaluation\ny_pred_knn = model_knn.predict(x_test) # we use this for evaluation\ny_pred_svm = model_svm.predict(x_test) # we use this for evaluation\ny_pred_dt = model_dt.predict(x_test) # we use this for evaluation\ny_pred_rf = model_rf.predict(x_test) # we use this for evaluation\n",
"_____no_output_____"
]
],
[
[
"***classification metrics**",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import confusion_matrix, classification_report",
"_____no_output_____"
],
[
"cm_log = confusion_matrix(y_test, y_pred_log) # confusion matrix\ncm_knn = confusion_matrix(y_test, y_pred_knn) # confusion matrix\ncm_svm = confusion_matrix(y_test, y_pred_svm) # confusion matrix\ncm_dt = confusion_matrix(y_test, y_pred_dt) # confusion matrix\ncm_rf = confusion_matrix(y_test, y_pred_rf) # confusion matrix\n\ncr_log = classification_report(y_test, y_pred_log) # classification report\ncr_knn = classification_report(y_test, y_pred_knn) # classification report\ncr_svm = classification_report(y_test, y_pred_svm) # classification report\ncr_dt = classification_report(y_test, y_pred_dt) # classification report\ncr_rf = classification_report(y_test, y_pred_rf) # classification report",
"_____no_output_____"
],
[
"import seaborn as sns",
"_____no_output_____"
],
[
"sns.heatmap(cm_log,annot=True,cbar=None,cmap = 'summer')\nplt.title('Logistic Regression')\nplt.show()\nsns.heatmap(cm_knn,annot=True,cbar=None,cmap = 'spring')\nplt.title('K Nearest Neighbour')\nplt.show()\nsns.heatmap(cm_svm,annot=True,cbar=None,cmap = 'winter')\nplt.title('Support Vector Machine')\nplt.show()\nsns.heatmap(cm_dt,annot=True,cbar=None,cmap = 'cool')\nplt.title('Desicion Tree')\nplt.show()\nsns.heatmap(cm_rf,annot=True,cbar=None,cmap = 'autumn')\nplt.title('Random Forest')\nplt.show()",
"_____no_output_____"
],
[
"print('='*20+'Logistic Regression'+'='*20)\nprint(cr_log)\nprint('='*20+'KNearest Neighbour'+'='*20)\nprint(cr_knn)\nprint('='*20+'Support Vector Machine'+'='*20)\nprint(cr_svm)\nprint('='*20+'Descion Tree'+'='*20)\nprint(cr_dt)\nprint('='*20+'Random Forest'+'='*20)\nprint(cr_rf)",
"====================Logistic Regression====================\n precision recall f1-score support\n\n 0 1.00 1.00 1.00 27\n 1 0.86 0.86 0.86 35\n 2 0.94 0.94 0.94 36\n 3 0.93 0.90 0.91 29\n 4 0.82 0.93 0.87 30\n 5 0.92 0.90 0.91 40\n 6 0.96 0.98 0.97 44\n 7 0.97 0.95 0.96 39\n 8 0.91 0.77 0.83 39\n 9 0.84 0.93 0.88 41\n\navg / total 0.92 0.91 0.91 360\n\n====================KNearest Neighbour====================\n precision recall f1-score support\n\n 0 0.96 1.00 0.98 27\n 1 0.75 0.94 0.84 35\n 2 1.00 1.00 1.00 36\n 3 0.90 0.93 0.92 29\n 4 0.97 0.93 0.95 30\n 5 0.93 0.93 0.93 40\n 6 0.96 1.00 0.98 44\n 7 0.97 1.00 0.99 39\n 8 0.96 0.69 0.81 39\n 9 0.92 0.88 0.90 41\n\navg / total 0.93 0.93 0.93 360\n\n====================Support Vector Machine====================\n precision recall f1-score support\n\n 0 1.00 1.00 1.00 27\n 1 0.86 0.91 0.89 35\n 2 0.95 0.97 0.96 36\n 3 0.93 0.97 0.95 29\n 4 0.91 0.97 0.94 30\n 5 0.95 0.93 0.94 40\n 6 0.98 0.98 0.98 44\n 7 0.97 0.97 0.97 39\n 8 0.97 0.77 0.86 39\n 9 0.91 0.98 0.94 41\n\navg / total 0.94 0.94 0.94 360\n\n====================Descion Tree====================\n precision recall f1-score support\n\n 0 0.96 0.89 0.92 27\n 1 0.75 0.86 0.80 35\n 2 0.76 0.72 0.74 36\n 3 0.75 0.83 0.79 29\n 4 0.90 0.87 0.88 30\n 5 0.94 0.82 0.88 40\n 6 0.87 0.91 0.89 44\n 7 0.88 0.92 0.90 39\n 8 0.71 0.64 0.68 39\n 9 0.77 0.80 0.79 41\n\navg / total 0.83 0.82 0.82 360\n\n====================Random Forest====================\n precision recall f1-score support\n\n 0 1.00 0.96 0.98 27\n 1 0.89 0.97 0.93 35\n 2 1.00 0.97 0.99 36\n 3 1.00 0.93 0.96 29\n 4 0.97 0.97 0.97 30\n 5 0.95 0.93 0.94 40\n 6 0.98 1.00 0.99 44\n 7 0.95 1.00 0.97 39\n 8 1.00 0.87 0.93 39\n 9 0.89 0.98 0.93 41\n\navg / total 0.96 0.96 0.96 360\n\n"
]
],
[
[
"**Saving and loading model**",
"_____no_output_____"
]
],
[
[
"from sklearn.externals import joblib",
"_____no_output_____"
],
[
"joblib.dump(model_log,'number_rec_log.pkl')\njoblib.dump(model_knn,'number_rec_knn.pkl')\njoblib.dump(model_svm,'number_rec_svm.pkl')\njoblib.dump(model_dt,'number_rec_dt.pkl')\njoblib.dump(model_rf,'number_rec_rf.pkl')",
"_____no_output_____"
],
[
"classify = joblib.load('number_rec_rf.pkl')",
"_____no_output_____"
]
],
[
[
"**Tesing with new image**",
"_____no_output_____"
]
],
[
[
"import cv2",
"_____no_output_____"
],
[
"# Step 1 : Read image\nimg =cv2.imread('number2.jpg',0) # if you use zero it will convert into grayscale image\n# step 2: Thresholding\nret, thresh = cv2.threshold(img,127,255,cv2.THRESH_BINARY_INV)\n# step 3 : Resize image\nimg_re = cv2.resize(thresh,(8,8))\n# Step 4: reshape it to row matrix\ntest = img_re.reshape((1,64))\n# Step 5: Normalize\ntest = test/ test.max()\nplt.imshow(test,cmap ='gray')\nplt.show()",
"_____no_output_____"
],
[
"plt.imshow(img_re)",
"_____no_output_____"
],
[
"print('LogisticRegression',model_log.predict(test))\nprint('KNearest Neighbour', model_knn.predict(test))\nprint('Support Vector Machine', model_svm.predict(test))\nprint('Desicion Tree', model_dt.predict(test))\nprint('Random Forest',model_rf.predict(test))",
"LogisticRegression [2]\nKNearest Neighbour [2]\nSupport Vector Machine [2]\nDesicion Tree [2]\nRandom Forest [2]\n"
]
],
[
[
"# Real Time Number Detection",
"_____no_output_____"
]
],
[
[
"cap = cv2.VideoCapture(0)\n\nwhile True:\n _,img = cap.read()\n gray = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)\n blur = cv2.GaussianBlur(gray, (7,7),3)\n _,th3 = cv2.threshold(gray,100,255,cv2.THRESH_BINARY_INV)\n #th3 = cv2.adaptiveThreshold(blur,255,cv2.ADAPTIVE_THRESH_GAUSSIAN_C,cv2.THRESH_BINARY_INV,21,7)\n im2, contours, hierarchy = cv2.findContours(th3,cv2.RETR_TREE,cv2.CHAIN_APPROX_SIMPLE)\n areas = [cv2.contourArea(c) for c in contours]\n ix = np.where(np.array(areas) > 300)[0]\n result = np.array([1,0,0,0,0,0,0,0,0,0])\n for i in ix:\n cnt = contours[i]\n xr,yr,wr,hr = cv2.boundingRect(cnt)\n if xr< 20 :\n xr = 25\n\n\n if yr < 20:\n yr = 25\n\n\n cv2.rectangle(img,(xr-10,yr-10),(xr+wr+10,yr+hr+10), (0,255,0),2)\n roi = th3[yr-20:yr+hr+20, xr-20:xr+wr+20]\n \n roi_re=cv2.resize(roi,(8,8))\n g = roi_re.reshape(1,64).astype('float32')\n g = g/255.0\n \n \n result= model_rf.predict(g)\n #print(result)\n font = cv2.FONT_HERSHEY_SIMPLEX\n cv2.putText(img,'Number: '+str(result),(xr-10,yr-10), font, 0.4, (255,0,0), 1, cv2.LINE_AA)\n\n\n cv2.imshow('Threshold',th3)\n cv2.imshow('orginal',img)\n\n if cv2.waitKey(41) & 0xff == ord('q'):\n break\n \n \n \ncap.release() \ncv2.destroyAllWindows()",
"_____no_output_____"
],
[
"q",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecc97961ed46c7946f0a04fd149e6cf1265ac6f6 | 389 | ipynb | Jupyter Notebook | Hello World.ipynb | pid011/Hello-World | 0bbe19f30cab947907edb321f64db00ebf4fabb3 | [
"MIT"
] | 18 | 2019-03-24T02:54:43.000Z | 2020-08-16T12:09:04.000Z | Hello World.ipynb | pid011/Hello-World | 0bbe19f30cab947907edb321f64db00ebf4fabb3 | [
"MIT"
] | 32 | 2019-03-24T08:36:08.000Z | 2020-07-04T06:37:26.000Z | Hello World.ipynb | pid011/Hello-World | 0bbe19f30cab947907edb321f64db00ebf4fabb3 | [
"MIT"
] | 8 | 2020-10-07T08:03:47.000Z | 2021-10-10T12:13:10.000Z | 15.56 | 34 | 0.357326 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecc98507fe88453e864e2512b401e0fb6359430c | 96,858 | ipynb | Jupyter Notebook | week02_autodiff/seminar_pytorch.ipynb | kuynzereb/Practical_DL | c646a9a131941a3597a5ee7c8b1c756e676fe2ea | [
"MIT"
] | null | null | null | week02_autodiff/seminar_pytorch.ipynb | kuynzereb/Practical_DL | c646a9a131941a3597a5ee7c8b1c756e676fe2ea | [
"MIT"
] | null | null | null | week02_autodiff/seminar_pytorch.ipynb | kuynzereb/Practical_DL | c646a9a131941a3597a5ee7c8b1c756e676fe2ea | [
"MIT"
] | null | null | null | 76.993641 | 17,908 | 0.797064 | [
[
[
"# Pytorch basics\n\n\n\n__This notebook__ will teach you to use pytorch low-level core. You can install it [here](http://pytorch.org/). For high-level interface see the next notebook.\n\n__Pytorch feels__ differently than tensorflow/theano in almost every level. TensorFlow makes your code live in two \"worlds\" simultaneously: symbolic graphs and actual tensors. First you declare a symbolic \"recipe\" of how to get from inputs to outputs, then feed it with actual minibatches of data. In pytorch, __there's only one world__: all tensors have a numeric value.\n\nYou compute outputs on the fly without pre-declaring anything. The code looks exactly as in pure numpy with one exception: pytorch computes gradients for you. And can run stuff on GPU. And has a number of pre-implemented building blocks for your neural nets. [And a few more things.](https://medium.com/towards-data-science/pytorch-vs-tensorflow-spotting-the-difference-25c75777377b)\n\nAnd now we finally shut up and let pytorch do the talking.",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\nimport numpy as np\nimport torch\nprint(torch.__version__)",
"0.3.1.post2\n"
],
[
"# numpy world\n\nx = np.arange(16).reshape(4,4)\n\nprint(\"X :\\n%s\\n\" % x)\nprint(\"X.shape : %s\\n\" % (x.shape,))\nprint(\"add 5 :\\n%s\\n\" % (x + 5))\nprint(\"X*X^T :\\n%s\\n\" % np.dot(x,x.T))\nprint(\"mean over cols :\\n%s\\n\" % (x.mean(axis=-1)))\nprint(\"cumsum of cols :\\n%s\\n\" % (np.cumsum(x,axis=0)))",
"X :\n[[ 0 1 2 3]\n [ 4 5 6 7]\n [ 8 9 10 11]\n [12 13 14 15]]\n\nX.shape : (4, 4)\n\nadd 5 :\n[[ 5 6 7 8]\n [ 9 10 11 12]\n [13 14 15 16]\n [17 18 19 20]]\n\nX*X^T :\n[[ 14 38 62 86]\n [ 38 126 214 302]\n [ 62 214 366 518]\n [ 86 302 518 734]]\n\nmean over cols :\n[ 1.5 5.5 9.5 13.5]\n\ncumsum of cols :\n[[ 0 1 2 3]\n [ 4 6 8 10]\n [12 15 18 21]\n [24 28 32 36]]\n\n"
],
[
"# pytorch world\n\nx = np.arange(16).reshape(4,4)\n\nx = torch.from_numpy(x).type(torch.FloatTensor) #or torch.arange(0,16).view(4,4)\n\nprint (\"X :\\n%s\" % x)\nprint(\"X.shape : %s\\n\" % (x.shape,))\nprint (\"add 5 :\\n%s\" % (x+5))\nprint (\"X*X^T :\\n%s\" % torch.matmul(x,x.transpose(1,0)))\nprint (\"mean over cols :\\n%s\" % torch.mean(x,dim=-1))\nprint (\"cumsum of cols :\\n%s\" % torch.cumsum(x,dim=0))",
"X :\n\n 0 1 2 3\n 4 5 6 7\n 8 9 10 11\n 12 13 14 15\n[torch.FloatTensor of size 4x4]\n\nX.shape : torch.Size([4, 4])\n\nadd 5 :\n\n 5 6 7 8\n 9 10 11 12\n 13 14 15 16\n 17 18 19 20\n[torch.FloatTensor of size 4x4]\n\nX*X^T :\n\n 14 38 62 86\n 38 126 214 302\n 62 214 366 518\n 86 302 518 734\n[torch.FloatTensor of size 4x4]\n\nmean over cols :\n\n 1.5000\n 5.5000\n 9.5000\n 13.5000\n[torch.FloatTensor of size 4]\n\ncumsum of cols :\n\n 0 1 2 3\n 4 6 8 10\n 12 15 18 21\n 24 28 32 36\n[torch.FloatTensor of size 4x4]\n\n"
]
],
[
[
"## NumPy and Pytorch\n\nAs you can notice, pytorch allows you to hack stuff much the same way you did with numpy. No graph declaration, no placeholders, no sessions. This means that you can _see the numeric value of any tensor at any moment of time_. Debugging such code can be done with by printing tensors or using any debug tool you want (e.g. [gdb](https://wiki.python.org/moin/DebuggingWithGdb)).\n\nYou could also notice the a few new method names and a different API. So no, there's no compatibility with numpy [yet](https://github.com/pytorch/pytorch/issues/2228) and yes, you'll have to memorize all the names again. Get excited!\n\n\n\nFor example, \n* If something takes a list/tuple of axes in numpy, you can expect it to take *args in pytorch\n * `x.reshape([1,2,8]) -> x.view(1,2,8)`\n* You should swap _axis_ for _dim_ in operations like mean or cumsum\n * `x.sum(axis=-1) -> x.sum(dim=-1)`\n* most mathematical operations are the same, but types an shaping is different\n * `x.astype('int64') -> x.type(torch.LongTensor)`\n\nTo help you acclimatize, there's a [table](https://github.com/torch/torch7/wiki/Torch-for-Numpy-users) covering most new things. There's also a neat [documentation page](http://pytorch.org/docs/master/).\n\nFinally, if you're stuck with a technical problem, we recommend searching [pytorch forumns](https://discuss.pytorch.org/). Or just googling, which usually works just as efficiently. \n\nIf you feel like you almost give up, remember two things: __GPU__ an __free gradients__. Besides you can always jump back to numpy with x.numpy()",
"_____no_output_____"
],
[
"### Warmup: trigonometric knotwork\n_inspired by [this post](https://www.quora.com/What-are-the-most-interesting-equation-plots)_\n\nThere are some simple mathematical functions with cool plots. For one, consider this:\n\n$$ x(t) = t - 1.5 * cos( 15 t) $$\n$$ y(t) = t - 1.5 * sin( 16 t) $$\n",
"_____no_output_____"
]
],
[
[
"from ipywidgets import interact\nfrom ipywidgets import interactive\nimport ipywidgets as widgets",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n\nt = torch.linspace(-10, 10, steps = 10000)\n\n# compute x(t) and y(t) as defined above\nx = t - 1.5 * torch.cos(1 * t)\ny = t - 1.5 * torch.sin(2 * t)\n\nplt.plot(x.numpy(), y.numpy())\nplt.show()",
"_____no_output_____"
],
[
"def show_plot(a, b):\n t = torch.linspace(-10, 10, steps = 10000)\n\n # compute x(t) and y(t) as defined above\n x = t - 1.5 * torch.cos(a * t)\n y = t - 1.5 * torch.sin(b * t)\n\n plt.plot(x.numpy(), y.numpy())\n plt.show()",
"_____no_output_____"
],
[
"interact(show_plot, a=16, b=15);",
"_____no_output_____"
]
],
[
[
"if you're done early, try adjusting the formula and seing how it affects the function",
"_____no_output_____"
],
[
"```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n",
"_____no_output_____"
],
[
"## Automatic gradients\n\nAny self-respecting DL framework must do your backprop for you. Torch handles this with __`Variable`__s and the `autograd` module.\n\nThe general pipeline looks like this:\n* You create ```a = Variable(data, requires_grad=True)```\n* You define some differentiable `loss = whatever(a)`\n* Call `loss.backward()`\n* Gradients are now available as ```a.grads```\n\n__Here's an example:__ let's fit a linear regression on Boston house prices",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import load_boston\nboston = load_boston()\nplt.scatter(boston.data[:, -1], boston.target)\nplt.show()",
"_____no_output_____"
],
[
"from torch.autograd import Variable\nw = Variable(torch.zeros(2), requires_grad=True)\nb = Variable(torch.zeros(1), requires_grad=True)\n\n# cast data into torch variables\nx = Variable(torch.FloatTensor(boston.data[:,-1] / 10))\ny = Variable(torch.FloatTensor(boston.target))",
"_____no_output_____"
],
[
"y_pred = w[0] * x + w[1] * x**2 + b\nloss = torch.mean( (y_pred - y)**2 )\n\n# propagete gradients\nloss.backward()",
"_____no_output_____"
]
],
[
[
"The gradients are now stored in `.grad` of a variable.",
"_____no_output_____"
]
],
[
[
"print(\"dL/dw = \\n\", w.grad)\nprint(\"dL/db = \\n\", b.grad)",
"dL/dw = \n Variable containing:\n-47.3514\n-68.1231\n[torch.FloatTensor of size 2]\n\ndL/db = \n Variable containing:\n-45.0656\n[torch.FloatTensor of size 1]\n\n"
]
],
[
[
"If you compute gradient from multiple losses, the gradients will add up at variables, therefore it's useful to __zero the gradients__ between iteratons.",
"_____no_output_____"
]
],
[
[
"from IPython.display import clear_output\n\nfor i in range(300):\n\n y_pred = w[0] * x + w[1] * x**0.5 + b\n loss = torch.mean((y_pred - y)**2)\n loss.backward()\n\n w.data -= 0.05 * w.grad.data\n b.data -= 0.05 * b.grad.data\n \n #zero gradients\n w.grad.data.zero_()\n b.grad.data.zero_()\n \n # the rest of code is just bells and whistles\n if (i+1)%5==0:\n clear_output(True)\n plt.scatter(x.data.numpy(), y.data.numpy())\n plt.scatter(x.data.numpy(), y_pred.data.numpy(), color='orange', linewidth=5)\n plt.show()\n\n print(\"loss = \", loss.data.numpy()[0])\n if loss.data.numpy()[0] < 0.5:\n print(\"Done!\")\n break",
"_____no_output_____"
]
],
[
[
"__Bonus quest__: try implementing and writing some nonlinear regression. You can try quadratic features or some trigonometry, or a simple neural network. The only difference is that now you have more variables and a more complicated `y_pred`. ",
"_____no_output_____"
],
[
"```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n\n### Tensor vs Variable\n\n\n\n` `\n\nTensor and Variable Weasley were identical and mischievous twin abstractions living in pytorch. Brilliant pranksters, they make sure your code never runs successfully from the first attempt.\n\nSeriously though, Variable wraps around the torch tensor and allows you to compute gradients. Theoretically, Variable acts just like tensor for all intents and purposes. Practically, you will find yourself debugging tensor vs variable issues most of your first \n\nIdeally, you could wrap everything into Variable from the get-go an forget about Tensor. Hopefully you will once be able to do so __[upvote [this](https://github.com/pytorch/pytorch/issues/2228)]__. \n\nRight now you can't, but it's getting better with each update.\n\nThe good news is that you can always swap between the two seamlessly:\n* tensor to variable: `Variable(x)`\n* variable to tensor: `x.data`",
"_____no_output_____"
]
],
[
[
"from torch.autograd import Variable\n\nx = torch.arange(0,16).view(4,4).type(torch.IntTensor)\nx_var = Variable(x)\n\nprint (\"Result (tensors):\\n\", (x == 1) | (x % 3 == 0))\nprint (\"Result (variables):\\n\", (x_var == 1) | (x_var % 3==0))",
"Result (tensors):\n \n 1 1 0 1\n 0 0 1 0\n 0 1 0 0\n 1 0 0 1\n[torch.ByteTensor of size 4x4]\n\nResult (variables):\n Variable containing:\n 1 1 0 1\n 0 0 1 0\n 0 1 0 0\n 1 0 0 1\n[torch.ByteTensor of size 4x4]\n\n"
],
[
"sequence = torch.randn(1,8,10)\nfilters = torch.randn(2,8,3)\n\n#will work:\nprint(\"conv1d (variables):\\n\",torch.nn.functional.conv1d(Variable(sequence),Variable(filters)))\n\n#will not work:\ntry:\n print(\"conv1d (tensors):\")\n print(torch.nn.functional.conv1d(sequence,filters))\nexcept Exception as e:\n print (e)\n\n",
"conv1d (variables):\n Variable containing:\n(0 ,.,.) = \n -3.2865 -0.1808 -4.6515 -3.2173 -3.4818 4.4255 -5.0728 -3.2869\n -5.2591 4.2352 0.0535 3.6510 3.6506 -6.1675 -0.7387 0.0431\n[torch.FloatTensor of size 1x2x8]\n\nconv1d (tensors):\nargument 0 is not a Variable\n"
]
],
[
[
"# High-level pytorch\n\nSo far we've been dealing with low-level torch API. While it's absolutely vital for any custom losses or layers, building large neura nets in it is a bit clumsy.\n\nLuckily, there's also a high-level torch interface with a pre-defined layers, activations and training algorithms. \n\nWe'll cover them as we go through a simple image recognition problem: classifying letters into __\"A\"__ vs __\"B\"__.\n",
"_____no_output_____"
]
],
[
[
"from notmnist import load_notmnist\nX_train, y_train, X_test, y_test = load_notmnist(letters='AB')\nX_train, X_test = X_train.reshape([-1, 784]), X_test.reshape([-1, 784])\n\nprint(\"Train size = %i, test_size = %i\"%(len(X_train),len(X_test)))",
"Downloading data...\nExtracting ...\nParsing...\nfound broken img: ./notMNIST_small/A/RGVtb2NyYXRpY2FCb2xkT2xkc3R5bGUgQm9sZC50dGY=.png [it's ok if <10 images are broken]\nDone\nTrain size = 2808, test_size = 937\n"
],
[
"shift = 200\nfor i in [0,1]:\n plt.subplot(1,2,i+1)\n plt.imshow(X_train[shift+i].reshape([28,28]))\n plt.title(str(y_train[shift+i]))",
"_____no_output_____"
]
],
[
[
"Let's start with layers. The main abstraction here is __`torch.nn.Module`__",
"_____no_output_____"
]
],
[
[
"from torch import nn\nimport torch.nn.functional as F\n\nprint(nn.Module.__doc__)",
"Base class for all neural network modules.\n\n Your models should also subclass this class.\n\n Modules can also contain other Modules, allowing to nest them in\n a tree structure. You can assign the submodules as regular attributes::\n\n import torch.nn as nn\n import torch.nn.functional as F\n\n class Model(nn.Module):\n def __init__(self):\n super(Model, self).__init__()\n self.conv1 = nn.Conv2d(1, 20, 5)\n self.conv2 = nn.Conv2d(20, 20, 5)\n\n def forward(self, x):\n x = F.relu(self.conv1(x))\n return F.relu(self.conv2(x))\n\n Submodules assigned in this way will be registered, and will have their\n parameters converted too when you call .cuda(), etc.\n \n"
]
],
[
[
"There's a vast library of popular layers and architectures already built for ya'.\n\nThis is a binary classification problem, so we'll train a __Logistic Regression with sigmoid__.\n$$P(y_i | X_i) = \\sigma(W \\cdot X_i + b) ={ 1 \\over {1+e^{- [W \\cdot X_i + b]}} }$$\n",
"_____no_output_____"
]
],
[
[
"# create a network that stacks layers on top of each other\nmodel = nn.Sequential()\n\n# add first \"dense\" layer with 784 input units and 1 output unit. \nmodel.add_module('linear_1', nn.Linear(784, 512))\nmodel.add_module('batch_norm_1', nn.BatchNorm1d(512))\nmodel.add_module('relu_1', nn.ReLU())\n\nmodel.add_module('linear_2', nn.Linear(512, 512))\nmodel.add_module('batch_norm_2', nn.BatchNorm1d(512))\nmodel.add_module('relu_2', nn.ReLU())\n\nmodel.add_module('linear_3', nn.Linear(512, 1))\n\n# add softmax activation for probabilities. Normalize over axis 1\n# note: layer names must be unique\nmodel.add_module('sigmoid', nn.Sigmoid())",
"_____no_output_____"
],
[
"print(\"Weight shapes:\", [w.shape for w in model.parameters()])",
"Weight shapes: [torch.Size([512, 784]), torch.Size([512]), torch.Size([512]), torch.Size([512]), torch.Size([512, 512]), torch.Size([512]), torch.Size([512]), torch.Size([512]), torch.Size([1, 512]), torch.Size([1])]\n"
],
[
"# create dummy data with 3 samples and 784 features\nx = Variable(torch.FloatTensor(X_train[:3]))\ny = Variable(torch.FloatTensor(y_train[:3]))\n\n# compute outputs given inputs, both are variables\ny_predicted = model(x)[:, 0]\n\ny_predicted # display what we've got",
"_____no_output_____"
]
],
[
[
"Let's now define a loss function for our model.\n\nThe natural choice is to use binary crossentropy (aka logloss, negative llh):\n$$ L = {1 \\over N} \\underset{X_i,y_i} \\sum - [ y_i \\cdot log P(y_i | X_i) + (1-y_i) \\cdot log (1-P(y_i | X_i)) ]$$\n\n",
"_____no_output_____"
]
],
[
[
"EPS = 1e-3\ny_predicted = torch.clamp(y_predicted, EPS, 1. - EPS)\n\ncrossentropy = -(y * torch.log(y_predicted) + (1. - y) * torch.log(1. - y_predicted))\nloss = torch.mean(crossentropy)\n\nassert tuple(crossentropy.size()) == (3,), \"Crossentropy must be a vector with element per sample\"\nassert tuple(loss.size()) == (1,), \"Loss must be scalar. Did you forget the mean/sum?\"\nassert loss.data.numpy()[0] > 0, \"Crossentropy must non-negative, zero only for perfect prediction\"\nassert loss.data.numpy()[0] <= np.log(3), \"Loss is too large even for untrained model. Please double-check it.\"",
"_____no_output_____"
]
],
[
[
"When we trained Linear Regression above, we had to manually .zero_() gradients on both our variables. Imagine that code for a 50-layer network.\n\nAgain, to keep it from getting dirty, there's `torch.optim` module with pre-implemented algorithms:",
"_____no_output_____"
]
],
[
[
"opt = torch.optim.SGD(model.parameters(), lr=0.001)\n\n# here's how it's used:\nloss.backward() # add new gradients\nopt.step() # change weights\nopt.zero_grad() # clear gradients",
"_____no_output_____"
],
[
"# dispose of old variables to avoid bugs later\ndel x, y, y_predicted, loss",
"_____no_output_____"
]
],
[
[
"### Putting it all together",
"_____no_output_____"
]
],
[
[
"# create network again just in case\nmodel = nn.Sequential()\n\n# add first \"dense\" layer with 784 input units and 1 output unit. \nmodel.add_module('linear_1', nn.Linear(784, 512))\nmodel.add_module('batch_norm_1', nn.BatchNorm1d(512))\nmodel.add_module('relu_1', nn.ReLU())\n\nmodel.add_module('linear_2', nn.Linear(512, 512))\nmodel.add_module('batch_norm_2', nn.BatchNorm1d(512))\nmodel.add_module('relu_2', nn.ReLU())\n\nmodel.add_module('linear_3', nn.Linear(512, 1))\n\n# add softmax activation for probabilities. Normalize over axis 1\n# note: layer names must be unique\nmodel.add_module('sigmoid', nn.Sigmoid())",
"_____no_output_____"
],
[
"history = []\nlr = 0.001\nfor i in range(101):\n if i == 49:\n lr /= 3\n \n opt = torch.optim.Adam(model.parameters(), lr=lr)\n \n # sample 256 random images\n ix = np.random.randint(0, len(X_train), 256)\n x_batch = Variable(torch.FloatTensor(X_train[ix]))\n y_batch = Variable(torch.FloatTensor(y_train[ix]))\n \n # predict probabilities\n y_predicted = model(x_batch)[:, 0]\n \n # compute loss, just like before\n EPS = 1e-3\n y_predicted = torch.clamp(y_predicted, EPS, 1. - EPS)\n crossentropy = -(y_batch * torch.log(y_predicted) + (1. - y_batch) * torch.log(1. - y_predicted))\n loss = torch.mean(crossentropy)\n \n # compute gradients\n loss.backward()\n \n # optimizer step\n opt.step()\n \n # clear gradients\n opt.zero_grad()\n \n history.append(loss.data.numpy()[0])\n \n if i % 10 == 0:\n print(\"step #%i | mean loss = %.3f\" % (i, np.mean(history[-10:])))",
"step #0 | mean loss = 0.699\nstep #10 | mean loss = 0.180\nstep #20 | mean loss = 0.095\nstep #30 | mean loss = 0.066\nstep #40 | mean loss = 0.049\nstep #50 | mean loss = 0.047\nstep #60 | mean loss = 0.022\nstep #70 | mean loss = 0.020\nstep #80 | mean loss = 0.018\nstep #90 | mean loss = 0.013\nstep #100 | mean loss = 0.014\n"
],
[
"history = []\n\nfor i in range(101):\n \n # sample 256 random images\n ix = np.random.randint(0, len(X_train), 256)\n x_batch = Variable(torch.FloatTensor(X_train[ix]))\n y_batch = Variable(torch.FloatTensor(y_train[ix]))\n \n # predict probabilities\n y_predicted = model(x_batch)[:, 0]\n \n # compute loss, just like before\n EPS = 1e-3\n y_predicted = torch.clamp(y_predicted, EPS, 1. - EPS)\n crossentropy = -(y_batch * torch.log(y_predicted) + (1. - y_batch) * torch.log(1. - y_predicted))\n loss = torch.mean(crossentropy)\n \n # compute gradients\n loss.backward()\n \n # optimizer step\n opt.step()\n \n # clear gradients\n opt.zero_grad()\n \n history.append(loss.data.numpy()[0])\n \n if i % 10 == 0:\n print(\"step #%i | mean loss = %.3f\" % (i, np.mean(history[-10:])))",
"step #0 | mean loss = 0.670\nstep #10 | mean loss = 0.603\nstep #20 | mean loss = 0.398\nstep #30 | mean loss = 0.276\nstep #40 | mean loss = 0.188\nstep #50 | mean loss = 0.160\nstep #60 | mean loss = 0.117\nstep #70 | mean loss = 0.092\nstep #80 | mean loss = 0.089\nstep #90 | mean loss = 0.073\nstep #100 | mean loss = 0.062\n"
],
[
"history = []\n\nfor i in range(100):\n \n # sample 256 random images\n ix = np.random.randint(0, len(X_train), 256)\n x_batch = Variable(torch.FloatTensor(X_train[ix]))\n y_batch = Variable(torch.FloatTensor(y_train[ix]))\n \n # predict probabilities\n y_predicted = model(x_batch)[:, 0]\n \n # compute loss, just like before\n EPS = 1e-3\n y_predicted = torch.clamp(y_predicted, EPS, 1. - EPS)\n crossentropy = -(y_batch * torch.log(y_predicted) + (1. - y_batch) * torch.log(1. - y_predicted))\n loss = torch.mean(crossentropy)\n \n # compute gradients\n loss.backward()\n \n # optimizer step\n opt.step()\n \n # clear gradients\n opt.zero_grad()\n \n history.append(loss.data.numpy()[0])\n \n if i % 10 == 0:\n print(\"step #%i | mean loss = %.3f\" % (i, np.mean(history[-10:])))",
"step #0 | mean loss = 0.716\nstep #10 | mean loss = 0.689\nstep #20 | mean loss = 0.657\nstep #30 | mean loss = 0.628\nstep #40 | mean loss = 0.601\nstep #50 | mean loss = 0.574\nstep #60 | mean loss = 0.551\nstep #70 | mean loss = 0.531\nstep #80 | mean loss = 0.516\nstep #90 | mean loss = 0.493\n"
]
],
[
[
"__Debugging tips:__\n* make sure your model predicts probabilities correctly. Just print them and see what's inside.\n* don't forget _minus_ sign in the loss function! It's a mistake 99% ppl do at some point.\n* make sure you zero-out gradients after each step. Srsly:)\n* In general, pytorch's error messages are quite helpful, read 'em before you google 'em.\n* if you see nan/inf, print what happens at each iteration to find our where exactly it occurs.\n * If loss goes down and then turns nan midway through, try smaller learning rate. (Our current loss formula is unstable).\n",
"_____no_output_____"
],
[
"### Evaluation\n\nLet's see how our model performs on test data",
"_____no_output_____"
]
],
[
[
"# use your model to predict classes (0 or 1) for all test samples\npredicted_y_test = model(Variable(torch.FloatTensor(X_test)))[:, 0].data.numpy()\npredicted_y_test = predicted_y_test > 0.5\n \nassert isinstance(predicted_y_test, np.ndarray), \"please return np array, not %s\" % type(predicted_y_test)\nassert predicted_y_test.shape == y_test.shape, \"please predict one class for each test sample\"\nassert np.in1d(predicted_y_test, y_test).all(), \"please predict class indexes\"\n\naccuracy = np.mean(predicted_y_test == y_test)\n\nprint(\"Test accuracy: %.5f\" % accuracy)\nassert accuracy > 0.95, \"try training longer\"",
"Test accuracy: 0.95945\n"
]
],
[
[
"### Now what?\n\nMade it past this point? Seminar isn't over yet? ~~go to the nearest coffee~~ __We have a quest for you!__\n\nRemember adaptive optimizer ~~bulsh~~ guide we gave you during the lecture? Time to put that thing into action!\n\n#### 0. Stack more layers!\n\nYour model is linear. Which is... bad... because... deep learning. Try stacking several linear layers with nonlinearities inbetween. This should improve accuracy by another %%.\n\n#### 1. Momentum that matters\n\n* Find that `torch.optim.SGD(...)` thingy and make it into `torch.optim.SGD(..., momentum=0.5)` and see how it affects training\n* Try momentum={0.5, 0.9, 0.99} and see what's best\n* Try RMSprop and Adam for even greater performance.\n\n\n#### 2. Batch normalization\n\nIf you're done with that by the end of the seminar (in which case you are a robot), there's another thing from the lecture:\n * Add nn.BatchNorm1d(num_units) between intermediate linear layers and their nonlinearities.\n * This works better with deeper networks. \n * Consider setting `torch.nn.Linear(..., bias=False)` to avoid unnecessary parameters.",
"_____no_output_____"
],
[
"## More about pytorch:\n* Using torch on GPU and multi-GPU - [link](http://pytorch.org/docs/master/notes/cuda.html)\n* More tutorials on pytorch - [link](http://pytorch.org/tutorials/beginner/deep_learning_60min_blitz.html)\n* Pytorch examples - a repo that implements many cool DL models in pytorch - [link](https://github.com/pytorch/examples)\n* Practical pytorch - a repo that implements some... other cool DL models... yes, in pytorch - [link](https://github.com/spro/practical-pytorch)\n* And some more - [link](https://www.reddit.com/r/pytorch/comments/6z0yeo/pytorch_and_pytorch_tricks_for_kaggle/)\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecc98534b9420cc412f3b78df99e828f59eff3d8 | 20,871 | ipynb | Jupyter Notebook | 3d/pinn.ipynb | alxyok/burgers | 7080fe575d5cfc7914e3fcd650248974808897a7 | [
"MIT"
] | null | null | null | 3d/pinn.ipynb | alxyok/burgers | 7080fe575d5cfc7914e3fcd650248974808897a7 | [
"MIT"
] | null | null | null | 3d/pinn.ipynb | alxyok/burgers | 7080fe575d5cfc7914e3fcd650248974808897a7 | [
"MIT"
] | null | null | null | 35.018456 | 438 | 0.537923 | [
[
[
"MIT License\n\nCopyright (c) 2021 alxyok\n\nPermission is hereby granted, free of charge, to any person obtaining a copy\nof this software and associated documentation files (the \"Software\"), to deal\nin the Software without restriction, including without limitation the rights\nto use, copy, modify, merge, publish, distribute, sublicense, and/or sell\ncopies of the Software, and to permit persons to whom the Software is\nfurnished to do so, subject to the following conditions:\n\nThe above copyright notice and this permission notice shall be included in all\ncopies or substantial portions of the Software.\n\nTHE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\nIMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\nFITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\nAUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\nLIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\nOUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\nSOFTWARE.",
"_____no_output_____"
],
[
"# Solving the 3D Burger's equation",
"_____no_output_____"
],
[
"**FORGET ABOUT THIS CODE, IT'S UNTRACTABLE. FOR EDUCATIONAL PURPOSES ONLY.**\n\n**Let's have some fun! Untested fun even 😛 (It works, but what it really does no one knows for certain!)**",
"_____no_output_____"
],
[
"**Most of the content below is largely inspired from the work of [Raissi et al.](https://maziarraissi.github.io/PINNs/) as weel as [Liu et al.](https://www.sciencedirect.com/science/article/abs/pii/S0142727X21000527), please refer to those papers for a comprehensive theoretical understanding.**\n\nThe Burger's equation is one of the well-studied fundamental PDEs in that exhibit shocks, and for which an non-trivial analytical solution exists in the Physics litterature. A conjunction of factors (profusion of data, capable cheap hardware, and backprop) has lead to the resurection Deep Learning (DL) which has in turn paved the way for the development of scientific machine learning libraries such as TensorFlow and PyTorch. \n\nThose frameworks come with free auto-differentiation, a key tool for this lab which will enable the development of a self-supervised neural model based on residuals.\n\nWe'll use PyTorch, but TensorFlow + Keras could do just as fine. Be sure to check out [PyTorch Tutorials](https://pytorch.org/tutorials/) and [PyTorch API](https://pytorch.org/docs/1.9.0/), which are a great source of information. Also, [Stackoverflow](https://stackoverflow.com/questions/tagged/pytorch).",
"_____no_output_____"
]
],
[
[
"import this",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np\n\n# we'll use PyTorch, but TensorFlow + Keras could do just as fine\nimport torch\n\nfrom torch import nn ",
"_____no_output_____"
]
],
[
[
"## 1. Problem statement\n\nNote: we do not use the Hopf-Cole transformation that would allow for a simplified formula but instead use the raw explicit formulation of the problem. \n\nWe propose to solve the 3D nonlinear Burger's problem defined by the following set of equations:\n\n$u_t + u * u_x + v * u_y + w * u_z - \\frac{1}{R_e} (u_{xx} + u_{yy} + u_{zz}) = 0$\n\n$v_t + u * v_x + v * v_y + w * v_z - \\frac{1}{R_e} (v_{xx} + v_{yy} + v_{zz}) = 0$\n\n$w_t + u * w_x + v * w_y + w * w_z - \\frac{1}{R_e} (w_{xx} + w_{yy} + w_{zz}) = 0$\n\nin which $Re$ is the Reynolds number, which characterizes the fluid flow behavior in various situations, and under the initial condition and boundary conditions defined below. The space domain is $0 < x, y, z < 1$ and time domain is $t > 0$.\n\n$u(x, y, z, 0) = u_0(x, y, z) = sin(\\pi x) * cos(\\pi y) * cos(\\pi z)$\n\n$v(x, y, z, 0) = v_0(x, y, z) = sin(\\pi y) * cos(\\pi x) * cos(\\pi z)$\n\n$w(x, y, z, 0) = w_0(x, y, z) = sin(\\pi z) * cos(\\pi x) * cos(\\pi y)$\n\nas well as:\n\n$u(0, y, z, t) = u(1, y, z, t) = 0$\n\n$v(x, 0, z, t) = v(x, 1, z, t) = 0$\n\n$w(x, y, 0, t) = w(x, y, 1, t) = 0$",
"_____no_output_____"
],
[
"## 2. The resolution method\n\nWe will build an estimator and have it gradually converge to the 3-tuple solution $U = (u, v, w)$ thanks to a handcrafted loss function based on residuals, computed from original inputs $X = (x, y, z, t)$.\n\nWe define:\n\n* A neural model $pinn := U(x, y, z, t)$\n* An IC residual function $U0_{residual} := pinn(X, 0) - U0(X)$\n* A BC residuals function $Ulim_{residual} := U(0, t) = U(1, t) = 0$\n* A PDE residual function $f := U_t + U * U_{.} - \\frac{1}{R_e} * U_{..}$\n\nThe Physics constraint is a soft-constraint (based on the loss) built by summing the loss of all residuals $L = loss(U0) + loss(Ulim) + loss(f)$.",
"_____no_output_____"
],
[
"#### A few of the model's HParams",
"_____no_output_____"
]
],
[
[
"# number of samples in every dimension\nn = 4\ngrid_shape = (n, n, n, n)\n\ndtype = torch.float\n\n# reynolds number, try for a range of 10^p where p is an integer \nre: float = 100.\n\n# learning rate, classic\nlr = 1e-3",
"_____no_output_____"
]
],
[
[
"#### Helpers",
"_____no_output_____"
]
],
[
[
"def tuplify(X: torch.Tensor) -> tuple:\n \n x = X[:, 0:1]\n y = X[:, 1:2]\n z = X[:, 2:3]\n t = X[:, 3:4]\n \n return x, y, z, t\n\ndef meshify(X: torch.Tensor) -> torch.Tensor:\n x, y, z, t = tuplify(X)\n x, y, z, t = np.meshgrid(x, y, z, t)\n \n x = torch.tensor(x.reshape((-1, 1)))\n y = torch.tensor(y.reshape((-1, 1)))\n z = torch.tensor(z.reshape((-1, 1)))\n t = torch.tensor(t.reshape((-1, 1)))\n \n X = torch.squeeze(torch.stack((x, y, z, t), axis=1))\n \n return X",
"_____no_output_____"
]
],
[
[
"## 3. The actual implementation",
"_____no_output_____"
],
[
"#### a) IC residuals function\n\nFollowing the article specifications, we'll define the IC with a few cyclical functions.",
"_____no_output_____"
]
],
[
[
"def U0(X: torch.Tensor) -> torch.Tensor:\n \"\"\"Computes the IC as stated previously.\"\"\"\n \n # X = meshify(X)\n x, y, z, _ = tuplify(X)\n \n u_xyz0 = torch.squeeze(torch.sin(np.pi * x) * torch.cos(np.pi * y) * torch.cos(np.pi * z))\n v_xyz0 = torch.squeeze(torch.sin(np.pi * y) * torch.cos(np.pi * x) * torch.cos(np.pi * z))\n w_xyz0 = torch.squeeze(torch.sin(np.pi * z) * torch.cos(np.pi * x) * torch.cos(np.pi * y))\n \n U0_ = torch.stack((u_xyz0, v_xyz0, w_xyz0), axis=1)\n \n return U0_",
"_____no_output_____"
],
[
"def U0_residuals(X: torch.Tensor) -> torch.Tensor:\n \"\"\"Computes the residuals for the IC.\"\"\"\n \n return pinn(X) - U0(X)",
"_____no_output_____"
]
],
[
[
"#### b) BC residuals function\n\nResiduals on boundary is `0`.",
"_____no_output_____"
]
],
[
[
"def Ulim_residuals(X: torch.Tensor) -> torch.Tensor:\n \"\"\"Computes the residuals at the Boundary.\"\"\"\n \n return pinn(X) - 0.",
"_____no_output_____"
]
],
[
[
"#### c) PDE residuals function\n\nWe need to compute first-order and second-order derivatives of $U$ with respect to $X$. Currently, `torch.__version__ == 1.9.0`, it's a bit tricky, because we cannot filter out *a priori* part of terms that will end-up unused and thus computation is partly wasted. We can only filter *a posteriori*. There's probably some leverage at the DAG level *(Directed Acyclic Graph)*.\n\nPyTorch has a `torch.autograd.functional.hessian()` function but only of output scalars and not vectors so we can't use it.",
"_____no_output_____"
]
],
[
[
"def f(X: torch.Tensor) -> torch.Tensor:\n \"\"\"Computes the residuals from the PDE on the rest of the Domain.\"\"\"\n \n def first_order(X, second_order=False):\n \n U = pinn(X)\n u = torch.squeeze(U[:, 0:1])\n v = torch.squeeze(U[:, 1:2])\n w = torch.squeeze(U[:, 2:3])\n \n U_X = torch.autograd.functional.jacobian(pinn, X, create_graph=True)\n \n u_x = torch.diagonal(torch.squeeze(U_X[:, 0:1, :, 0:1]))\n u_y = torch.diagonal(torch.squeeze(U_X[:, 0:1, :, 1:2]))\n u_z = torch.diagonal(torch.squeeze(U_X[:, 0:1, :, 2:3]))\n u_t = torch.diagonal(torch.squeeze(U_X[:, 0:1, :, 3:4]))\n \n v_x = torch.diagonal(torch.squeeze(U_X[:, 1:2, :, 0:1]))\n v_y = torch.diagonal(torch.squeeze(U_X[:, 1:2, :, 1:2]))\n v_z = torch.diagonal(torch.squeeze(U_X[:, 1:2, :, 2:3]))\n v_t = torch.diagonal(torch.squeeze(U_X[:, 1:2, :, 3:4]))\n \n w_x = torch.diagonal(torch.squeeze(U_X[:, 2:3, :, 0:1]))\n w_y = torch.diagonal(torch.squeeze(U_X[:, 2:3, :, 1:2]))\n w_z = torch.diagonal(torch.squeeze(U_X[:, 2:3, :, 2:3]))\n w_t = torch.diagonal(torch.squeeze(U_X[:, 2:3, :, 3:4]))\n \n if second_order:\n return u, v, w, u_x, u_y, u_z, u_t, v_x, v_y, v_z, v_t, w_x, w_y, w_z, w_t\n \n return u_x, v_y, w_z\n \n # way sub-optimal, the first order jacobian should really be computed once\n # maybe pytorch is doing this lazy, but still, sub-optimal\n def second_order(X):\n U_XX = torch.autograd.functional.jacobian(first_order, X)\n \n u_xx = torch.diagonal(torch.squeeze(U_XX[0][:, :, 0:1]))\n v_xx = torch.diagonal(torch.squeeze(U_XX[1][:, :, 0:1]))\n w_xx = torch.diagonal(torch.squeeze(U_XX[2][:, :, 0:1]))\n \n u_yy = torch.diagonal(torch.squeeze(U_XX[0][:, :, 1:2]))\n v_yy = torch.diagonal(torch.squeeze(U_XX[1][:, :, 1:2]))\n w_yy = torch.diagonal(torch.squeeze(U_XX[2][:, :, 1:2]))\n \n u_zz = torch.diagonal(torch.squeeze(U_XX[0][:, :, 2:3]))\n v_zz = torch.diagonal(torch.squeeze(U_XX[1][:, :, 2:3]))\n w_zz = torch.diagonal(torch.squeeze(U_XX[2][:, :, 2:3]))\n \n return u_xx, u_yy, u_zz, v_xx, v_yy, v_zz, w_xx, w_yy, w_zz\n \n u, v, w, u_x, u_y, u_z, u_t, v_x, v_y, v_z, v_t, w_x, w_y, w_z, w_t = first_order(X, second_order=True)\n u_xx, u_yy, u_zz, v_xx, v_yy, v_zz, w_xx, w_yy, w_zz = second_order(X)\n \n u_ = u_t + u * u_x + v * u_y + w * u_z - re * (u_xx + u_yy + u_zz)\n v_ = v_t + u * v_x + v * v_y + w * v_z - re * (v_xx + v_yy + v_zz)\n w_ = w_t + u * w_x + v * w_y + w * w_z - re * (w_xx + w_yy + w_zz)\n \n U = torch.stack((u_, v_, w_), axis=1)\n \n return U",
"_____no_output_____"
]
],
[
[
"#### d) The total loss function\n\nSummed-up from all previously defined residuals. Given how input $X$ was produced, it contains both samples from main domain as well as singular values used to compute both IC and BC. We need to carefully route the computation to the right residual function.",
"_____no_output_____"
]
],
[
[
"def loss(X: torch.Tensor) -> torch.Tensor:\n \"\"\"Computes the loss based on all residual terms.\"\"\"\n \n x0 = X[:, 0:1] == 0.\n x1 = X[:, 0:1] == 1.\n xl_ = torch.logical_or(x0, x1)\n xl_ = torch.cat((xl_,) * 4, axis=1)\n xl = torch.masked_select(X, xl_).reshape(-1, 4)\n xl_residuals = torch.mean(torch.square(Ulim_residuals(xl)))\n \n y0 = X[:, 1:2] == 0.\n y1 = X[:, 1:2] == 1.\n yl_ = torch.logical_or(y0, y1)\n yl_ = torch.cat((yl_,) * 4, axis=1)\n yl = torch.masked_select(X, yl_).reshape(-1, 4)\n yl_residuals = torch.mean(torch.square(Ulim_residuals(yl)))\n \n z0 = X[:, 2:3] == 0.\n z1 = X[:, 2:3] == 1.\n zl_ = torch.logical_or(z0, z1)\n zl_ = torch.cat((zl_,) * 4, axis=1)\n zl = torch.masked_select(X, zl_).reshape(-1, 4)\n zl_residuals = torch.mean(torch.square(Ulim_residuals(zl)))\n \n t0_ = X[:, 3:4] == 0.\n t0_ = torch.cat((t0_,) * 4, axis=1)\n t0 = torch.masked_select(X, t0_).reshape(-1, 4)\n t0_residuals = torch.mean(torch.square(U0_residuals(t0)))\n \n or_ = torch.logical_or(t0_, torch.logical_or(zl_, torch.logical_or(xl_, yl_)))\n X_not = torch.logical_not(or_)\n X_ = torch.masked_select(X, X_not).reshape(-1, 4)\n f_residuals = torch.mean(torch.square(f(X_)))\n \n # final loss is simply the sum of residuals\n return torch.mean(torch.stack((\n xl_residuals,\n yl_residuals,\n zl_residuals,\n t0_residuals,\n f_residuals\n )))",
"_____no_output_____"
]
],
[
[
"#### e) Defining the model\n\n... as a simple straight-forward feed-forward MLP `depth=4` by `width=20` + `activation=nn.Tanh()` defined with PyTorch's sequential API.",
"_____no_output_____"
]
],
[
[
"# inputs: X = (x, y, z, t)\n# outputs: U = (u, v, w)\n\npinn = nn.Sequential(\n nn.Linear(4, 20, dtype=dtype),\n nn.Tanh(),\n nn.Linear(20, 20, dtype=dtype),\n nn.Tanh(),\n nn.Linear(20, 20, dtype=dtype),\n nn.Tanh(),\n nn.Linear(20, 20, dtype=dtype),\n nn.Tanh(),\n nn.Linear(20, 3, dtype=dtype),\n)",
"_____no_output_____"
]
],
[
[
"## 4. LET'S FIT",
"_____no_output_____"
],
[
"Let's start by sampling in both space and time, and create a 4D-meshgrid (main reason why all this is intractable).",
"_____no_output_____"
]
],
[
[
"x = torch.linspace(0.0, 1.0, steps=n, dtype=dtype).T\ny = torch.linspace(0.0, 1.0, steps=n, dtype=dtype).T\nz = torch.linspace(0.0, 1.0, steps=n, dtype=dtype).T\nt = torch.linspace(0.0, 1.0, steps=n, dtype=dtype).T\nX = torch.stack((x, y, z, t), axis=1)\n\nX = meshify(X)\n\nu0 = U0(X)[:, 0:1]\nv0 = U0(X)[:, 1:2]\nw0 = U0(X)[:, 2:3]",
"_____no_output_____"
]
],
[
[
"...and loop over epochs... And we're done!",
"_____no_output_____"
]
],
[
[
"def fit(X: torch.Tensor, \n epochs: int,\n lr: float = 1e-2):\n \"\"\"Implements the training loop.\"\"\"\n\n optimizer = torch.optim.SGD(pinn.parameters(), lr=lr)\n for epoch in range(epochs):\n \n optimizer.zero_grad()\n loss_ = loss(X)\n loss_.backward()\n optimizer.step()\n \n if epoch % 1000 == 0:\n print(f\"epoch: {epoch}, loss: {loss_}\")",
"_____no_output_____"
],
[
"fit(X, epochs=10000)",
"epoch: 0, loss: 0.11308014392852783\nepoch: 1000, loss: 0.029296875\nepoch: 2000, loss: 0.029296875\nepoch: 3000, loss: 0.029296875\n"
]
],
[
[
"**But let's forget about printing anything useful. Simply untractable.**\n\n**For a more realistic and less irrelevant example, checkout `../burger_1d`.**",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
ecc98553b0f95d94dafde4ecb373ce9e89c72ad9 | 23,144 | ipynb | Jupyter Notebook | data_analyses/code/Convert_Zooniverse.ipynb | LAAC-LSCP/zoo-babble-validation | efcc26cc994293b8fabf553a60833962b3cdc0f7 | [
"Apache-2.0"
] | null | null | null | data_analyses/code/Convert_Zooniverse.ipynb | LAAC-LSCP/zoo-babble-validation | efcc26cc994293b8fabf553a60833962b3cdc0f7 | [
"Apache-2.0"
] | 3 | 2021-01-28T17:39:20.000Z | 2021-02-09T10:43:51.000Z | data_analyses/code/Convert_Zooniverse.ipynb | LAAC-LSCP/zoo-babble-validation | efcc26cc994293b8fabf553a60833962b3cdc0f7 | [
"Apache-2.0"
] | null | null | null | 35.771252 | 257 | 0.458305 | [
[
[
"import sys, os\nimport numpy as np\nimport pandas as pd\nimport json",
"_____no_output_____"
],
[
"project_name = \"maturity-of-baby-sounds\"\nfolder=\"/Users/chiarasemenzin/Downloads/\"\nclassification_file = folder+\"maturity-of-baby-sounds-classifications_all.csv\"\n\nprint(classification_file)",
"/Users/chiarasemenzin/Downloads/maturity-of-baby-sounds-classifications_all.csv\n"
],
[
"classifications_all = pd.read_csv(classification_file,delimiter=\",\",encoding='utf-8')\nn_class = len(classifications_all)\n\nprint(\"File %s read with %d rows.\" % (classification_file, n_class))",
"File /Users/chiarasemenzin/Downloads/maturity-of-baby-sounds-classifications_all.csv read with 363443 rows.\n"
],
[
"classifications_all.head()",
"_____no_output_____"
],
[
"annot=classifications_all['annotations']\nsubj_data=classifications_all[\"subject_data\"]\nusr_id=classifications_all[\"user_id\"]\nids=classifications_all[\"subject_ids\"]\n\n\ni=0\ncount=0\ndata=[]\nfor v_,f_,id_ in zip(annot,subj_data,usr_id):\n #load string in json format\n jdata_v = json.loads(v_)\n jdata_f = json.loads(f_)\n # print(jdata_v,jdata_f)\n #read as dictionary\n values_dict=jdata_v[0]\n files_dict=jdata_f[str(ids[i])]\n #create dataset and convert to pandas df\n try:\n data.append([id_,files_dict[\"name\"],values_dict[\"value\"]])\n #Get around UTF-8-sig encoding issue\n except:\n try:\n data.append([id_,files_dict['\\ufeffname'],values_dict[\"value\"]])\n except:\n try:\n data.append([id_,files_dict['Name'],values_dict[\"value\"]])\n except:\n try:\n data.append([id_,files_dict['file'],values_dict[\"value\"]])\n except:\n #print(\"Fail\",id_,files_dict,values_dict[\"value\"])\n count+=1\n pass\n i+=1\n \nprint(\"There were {} files not parsed correctly\".format(count))\ndf = pd.DataFrame(data, columns = ['UserID', 'AudioData','Answer'])\ndf['Dataset'] = 'Maturity of Baby Sounds'\ndf['Question'] = 'Please Classify this sound'\ndf",
"There were 29 files not parsed correctly\n"
],
[
"def make_js(column):\n data_list=[]\n for s in column:\n js=json.loads(s)\n data_list.append(js)\n return data_list\n \njs=make_js(classifications_all['metadata'])\nfor i in js[0:10]:\n print(\"started at: {}\".format(i[\"started_at\"]))\n print(\"finished at: {} \\n\".format(i[\"finished_at\"]))",
"started at: 2019-10-03T14:00:16.137Z\nfinished at: 2019-10-03T14:00:24.916Z \n\nstarted at: 2019-10-03T14:24:59.219Z\nfinished at: 2019-10-03T14:25:03.498Z \n\nstarted at: 2019-10-03T14:25:05.405Z\nfinished at: 2019-10-03T14:25:07.388Z \n\nstarted at: 2019-10-03T14:25:08.767Z\nfinished at: 2019-10-03T14:25:11.787Z \n\nstarted at: 2019-10-03T14:25:15.511Z\nfinished at: 2019-10-03T14:25:30.330Z \n\nstarted at: 2019-10-03T14:25:15.511Z\nfinished at: 2019-10-03T14:25:48.345Z \n\nstarted at: 2019-10-03T14:25:52.382Z\nfinished at: 2019-10-03T14:25:55.811Z \n\nstarted at: 2019-10-03T14:27:15.047Z\nfinished at: 2019-10-03T14:27:17.642Z \n\nstarted at: 2019-10-03T14:27:19.160Z\nfinished at: 2019-10-03T14:27:21.048Z \n\nstarted at: 2019-10-04T09:46:10.546Z\nfinished at: 2019-10-04T09:46:20.098Z \n\n"
],
[
"json.loads(classifications_all['metadata'][0])",
"_____no_output_____"
],
[
"df.to_csv(\"zooniverse_data_all_final.csv\",na_rep='NULL')",
"_____no_output_____"
],
[
"users_all = classifications_all['user_name'].unique()\nn_users = len(users_all)\n\n# if the classification is from a classifier who isn't signed in, the user_name field has \"not-logged-in-[user_ip]\"\nis_unreg = np.array([q.startswith(\"not-logged-in\") for q in users_all])\nis_reg = np.invert(is_unreg)\n\nn_unreg = sum(is_unreg)\nn_reg = sum(is_reg)\n\nprint(\"%d classifications from %d classifiers, of which %d (%.0f percent) were signed-in and %d (%.0f percent) were not signed in.\\n\" % (n_class, n_users, n_reg, (float(n_reg)/float(n_users)*100.), n_unreg, (float(n_unreg)/float(n_users)*100.)))\n\nprint(\"Average classifications per user: %.1f\" % (float(n_class)/float(n_users)))",
"363443 classifications from 7864 classifiers, of which 4478 (57 percent) were signed-in and 3386 (43 percent) were not signed in.\n\nAverage classifications per user: 46.2\n"
],
[
"# use created_at to print date range for classifications\nprint(\"Classifications registered between %s and %s.\" % (classifications_all['created_at'][classifications_all.index[0]], classifications_all['created_at'][classifications_all.index[-1]]))",
"Classifications registered between 2019-10-03 14:00:23 UTC and 2020-09-14 04:47:37 UTC.\n"
],
[
"# print out the classification ID of the last classification (useful in some cases)\nprint(\"Latest classification ID in this file: %d\" % classifications_all['classification_id'][classifications_all.index[-1]])",
"Latest classification ID in this file: 273574526\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecc986aecd0bdb428ca963a14fceaab9b36528e3 | 60,701 | ipynb | Jupyter Notebook | example.ipynb | wiralyki/Bokeh_for_easy_map | 1b43365c3d6150e465be277a736d409394643f10 | [
"BSD-3-Clause"
] | 1 | 2020-12-10T10:14:16.000Z | 2020-12-10T10:14:16.000Z | example.ipynb | wiralyki/Bokeh_for_easy_map | 1b43365c3d6150e465be277a736d409394643f10 | [
"BSD-3-Clause"
] | 1 | 2020-07-19T12:05:57.000Z | 2020-07-19T12:05:57.000Z | example.ipynb | amauryval/gdf_2_bokeh | 1b43365c3d6150e465be277a736d409394643f10 | [
"BSD-3-Clause"
] | null | null | null | 89.794379 | 14,486 | 0.559431 | [
[
[
"# gdf 2 bokeh",
"_____no_output_____"
],
[
"## Import all required librairies",
"_____no_output_____"
]
],
[
[
"import geopandas as gpd\n\n\nfrom bokeh.plotting import output_notebook\nfrom bokeh.plotting import show\n\nfrom gdf2bokeh import Gdf2Bokeh\n\noutput_notebook()",
"_____no_output_____"
]
],
[
[
"## How to define style ?\n\nCheck bokeh documentation :\n \n* [bokeh marker style options](https://docs.bokeh.org/en/latest/docs/reference/models/markers.html) to style point features\n* [bokeh multi_line style options](https://docs.bokeh.org/en/latest/docs/reference/plotting.html?highlight=multi_polygons#bokeh.plotting.figure.Figure.multi_line) to style LineString and MultiLineString features\n* [bokeh multi_polygon style options](https://docs.bokeh.org/en/latest/docs/reference/plotting.html?highlight=multi_polygons#bokeh.plotting.figure.Figure.multi_polygons) to style polygon and multipolygons features",
"_____no_output_____"
],
[
"## first way",
"_____no_output_____"
],
[
"### Prepare input data from geojson and map them",
"_____no_output_____"
]
],
[
[
"\nlayers_to_add = [\n {\n # contains both Polygon and MultiPolygon features (Ugly but only for testing)\n \"input_gdf\": gpd.GeoDataFrame.from_file(\"tests/fixtures/multipolygons.geojson\"),\n \"legend\": \"MultiPolygons layer\", # required\n \"fill_color\": \"orange\", # bokeh multi_polygon style option\n },\n {\n \"input_gdf\": gpd.GeoDataFrame.from_file(\"tests/fixtures/polygons.geojson\"),\n \"legend\": \"Polygons layer\", # required\n \"fill_color\": \"red\", # bokeh multi_polygon style option\n \"line_color\": \"black\", # bokeh multi_polygon style option\n },\n {\n \"input_gdf\": gpd.GeoDataFrame.from_file(\"tests/fixtures/linestrings.geojson\"),\n \"legend\": \"name\", # we can use the attribute called 'name' containing name value (as usual on bokeh)\n \"color\": \"color\", # we can use the attribute called 'color' containing name color (as usual on bokeh)\n \"line_width\": 4 # bokeh multi_line style option\n },\n {\n # contains both LineString and MultiLineString features (Ugly but only for testing)\n \"input_gdf\": gpd.GeoDataFrame.from_file(\"tests/fixtures/multilinestrings.geojson\"),\n \"legend\": \"multilinestrings layer\", # required\n \"color\": \"blue\", # bokeh multi_line style option\n \"line_width\": 6 # bokeh multi_line style option\n\n },\n {\n \"input_gdf\": gpd.GeoDataFrame.from_file(\"tests/fixtures/points.geojson\"),\n \"legend\": \"points layer\", # required\n \"style\": \"square\", # required\n \"size\": 6, # bokeh marker style option\n \"fill_color\": \"red\", # bokeh marker style option\n \"line_color\": \"blue\", # bokeh marker style option\n },\n]",
"_____no_output_____"
]
],
[
[
"### Let's go to map our data",
"_____no_output_____"
]
],
[
[
"%%time\nmy_map = Gdf2Bokeh(\n \"My beautiful map\", # required: map title\n width=800, # optional: figure width, default 800\n height=600, # optional: figure width, default 600\n x_range=None, # optional: x_range, default None\n y_range=None, # optional: y_range, default None\n background_map_name=\"CARTODBPOSITRON\", # optional: background map name, default: CARTODBPOSITRON\n layers=layers_to_add # optional: bokeh layer to add from a list of dict contains geodataframe settings, see dict above\n)\nshow(my_map.figure)",
"_____no_output_____"
]
],
[
[
"## Second way",
"_____no_output_____"
]
],
[
[
"%%time\nmy_map = Gdf2Bokeh(\n \"My beautiful map v2\", # required: map title\n width=700, # optional: figure width, default 800\n height=800, # optional: figure width, default 600\n x_range=None, # optional: x_range, default None\n y_range=None, # optional: y_range, default None\n background_map_name=\"STAMEN_TERRAIN\", # optional: background map name, default: CARTODBPOSITRON\n)\n\nmy_map.add_points(\n gpd.GeoDataFrame.from_file(\"tests/fixtures/points.geojson\"),\n legend=\"points layer\", # required\n style=\"cross\", # optional, check list : https://docs.bokeh.org/en/latest/docs/reference/models/markers.html\n size=10, # bokeh marker style option\n fill_color=\"red\", # bokeh marker style option\n)\n\nmy_map.add_lines(\n gpd.GeoDataFrame.from_file(\"tests/fixtures/multilinestrings.geojson\"),\n legend=\"multilinestrings layer\", # required\n color=\"green\", # bokeh multi_line style option\n line_width=6 # bokeh multi_line style option\n)\n\nmy_map.add_lines(\n gpd.GeoDataFrame.from_file(\"tests/fixtures/linestrings.geojson\"),\n legend=\"linestrings layer\", # required\n color=\"orange\", # bokeh multi_line style option\n line_width=4 # bokeh multi_line style option\n)\n\nmy_map.add_polygons(\n gpd.GeoDataFrame.from_file(\"tests/fixtures/polygons.geojson\"),\n legend=\"Polygons layer\", # required\n fill_color=\"red\", # bokeh multi_polygon style option\n line_width=5, # bokeh multi_polygon style option\n line_color=\"yellow\" # bokeh multi_polygon style option\n)\n\nmy_map.add_polygons(\n gpd.GeoDataFrame.from_file(\"tests/fixtures/multipolygons.geojson\"),\n legend=\"MultiPolygons layer\", # required\n fill_color=\"blue\", # bokeh multi_polygon style option\n line_color=\"black\", # bokeh multi_polygon style option\n)\n\nshow(my_map.figure)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecc986f34792d073ead507350f3207690de9778c | 492,440 | ipynb | Jupyter Notebook | DCGAN_for_Image_Generation.ipynb | 3ZadeSSG/DCGAN-Face-Image-Generator | 9e66889de2792ae7d096d4141dbc904cd398db73 | [
"MIT"
] | 1 | 2021-01-19T14:53:23.000Z | 2021-01-19T14:53:23.000Z | DCGAN_for_Image_Generation.ipynb | 3ZadeSSG/DCGAN-Face-Image-Generator | 9e66889de2792ae7d096d4141dbc904cd398db73 | [
"MIT"
] | null | null | null | DCGAN_for_Image_Generation.ipynb | 3ZadeSSG/DCGAN-Face-Image-Generator | 9e66889de2792ae7d096d4141dbc904cd398db73 | [
"MIT"
] | null | null | null | 384.418423 | 335,164 | 0.908107 | [
[
[
"import pickle as pkl\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport time\nimport sys\n\nimport torch\nfrom torchvision import datasets\nfrom torchvision import transforms\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\n\n%matplotlib inline",
"_____no_output_____"
],
[
"DATA_DIR = \"processed_celeba_small/\"\nMODEL_CHECKPOINT_LOCATION = \"saved_models/\"\nTRAINING_LOGS_LOCATION = \"logs/\"\nIMAGE_OUTPUT_PATH = \"output/\"",
"_____no_output_____"
],
[
"device = \"cpu\"\nif torch.cuda.is_available():\n device = \"cuda\"\n \nprint(device)",
"cuda\n"
],
[
"def get_dataloader(batch_size, image_size, data_dir=DATA_DIR):\n \"\"\"\n Batch the neural network data using DataLoader\n :param batch_size: The size of each batch; the number of images in a batch\n :param img_size: The square size of the image data (x, y)\n :param data_dir: Directory where image data is located\n :return: DataLoader with batched data\n \"\"\"\n transformer = transforms.Compose([transforms.Resize(image_size),transforms.ToTensor()])\n dataset = datasets.ImageFolder(data_dir,transformer)\n data_loader = torch.utils.data.DataLoader(dataset,batch_size= batch_size, shuffle = True)\n \n return data_loader\n\n#=================================================\n# Convert image tensor to numpy and display it\n#=================================================\ndef imshow(img):\n npimg = img.numpy()\n plt.imshow(np.transpose(npimg, (1, 2, 0)))\n \n \n#=================================================\n# Put normalized image into given feature range\n#=================================================\ndef scale(x, feature_range=(-1,1)):\n min,max = feature_range\n x = (max-min)*x + min\n return x",
"_____no_output_____"
],
[
"#=================================================\n# Discriminator Module\n#=================================================\ndef conv(in_channels, out_channels, kernel_size, stride=2, padding=1, batch_norm=True):\n \"\"\"Creates a convolutional layer, with optional batch normalization.\n \"\"\"\n layers = []\n conv_layer = nn.Conv2d(in_channels=in_channels, out_channels= out_channels, \n kernel_size=kernel_size, stride=stride, padding=padding, bias=False)\n\n layers.append(conv_layer)\n\n if batch_norm:\n layers.append(nn.BatchNorm2d(out_channels))\n \n return nn.Sequential(*layers)\n\n\nclass Discriminator(nn.Module):\n\n def __init__(self, conv_dim):\n \"\"\"\n Initialize the Discriminator Module\n :param conv_dim: The depth of the first convolutional layer\n \"\"\"\n super(Discriminator, self).__init__()\n self.conv_dim = conv_dim\n self.conv1 = conv(3,conv_dim,4,batch_norm=False)\n self.conv2 = conv(conv_dim,conv_dim*2,4,batch_norm=True)\n self.conv3 = conv(conv_dim*2,conv_dim*4,4,batch_norm=True)\n self.conv4 = conv(conv_dim*4,conv_dim*8,4,batch_norm=True)\n self.fc1 = nn.Linear(conv_dim*8*2*2,1)\n self.dropout1 = nn.Dropout(0.2)\n \n def forward(self, x):\n \"\"\"\n Forward propagation of the neural network\n :param x: The input to the neural network \n :return: Discriminator logits; the output of the neural network\n \"\"\"\n # define feedforward behavior\n x = F.leaky_relu(self.conv1(x),0.2)\n x = F.leaky_relu(self.conv2(x),0.2)\n x = F.leaky_relu(self.conv3(x),0.2)\n x = F.leaky_relu(self.conv4(x),0.2)\n x = self.dropout1(x)\n x = x.view(-1,self.conv_dim*8*2*2)\n x = self.fc1(x)\n return x\n\n\n#=================================================\n# Generator Module\n#=================================================\n\ndef deconv(in_channels, out_channels, kernel_size, stride=2, padding=1, batch_norm=True):\n \"\"\"Creates a transposed-convolutional layer, with optional batch normalization.\n \"\"\"\n ## TODO: Complete this function\n ## create a sequence of transpose + optional batch norm layers\n layers = []\n trans_conv_layer = nn.ConvTranspose2d(in_channels=in_channels, out_channels=out_channels, \n kernel_size=kernel_size, stride=stride, padding=padding, bias=False)\n \n # append conv layer\n layers.append(trans_conv_layer)\n\n if batch_norm:\n # append batchnorm layer\n layers.append(nn.BatchNorm2d(out_channels))\n \n # using Sequential container\n return nn.Sequential(*layers)\n\n\n\nclass Generator(nn.Module):\n \n def __init__(self, z_size, conv_dim):\n \"\"\"\n Initialize the Generator Module\n :param z_size: The length of the input latent vector, z\n :param conv_dim: The depth of the inputs to the *last* transpose convolutional layer\n \"\"\"\n super(Generator, self).__init__()\n self.conv_dim = conv_dim\n self.fc1 = nn.Linear(z_size,conv_dim*8*2*2)\n self.conv1 = deconv(conv_dim*8,conv_dim*4,4,batch_norm=True)\n self.conv2 = deconv(conv_dim*4,conv_dim*2,4,batch_norm=True)\n self.conv3 = deconv(conv_dim*2,conv_dim,4,batch_norm=True)\n self.conv4 = deconv(conv_dim,3,4,batch_norm=False)\n self.dropout2 = nn.Dropout(0.2)\n \n def forward(self, x):\n \"\"\"\n Forward propagation of the neural network\n :param x: The input to the neural network \n :return: A 32x32x3 Tensor image as output\n \"\"\"\n x = self.fc1(x)\n x = x.view(-1,self.conv_dim*8,2,2)\n x = F.relu(self.conv1(x))\n x = self.dropout2(x)\n x = F.relu(self.conv2(x))\n x = self.dropout2(x)\n x = F.relu(self.conv3(x))\n x = self.dropout2(x)\n x = torch.tanh(self.conv4(x))\n \n return x",
"_____no_output_____"
],
[
"def weights_init_normal(m):\n \"\"\"\n Applies initial weights to certain layers in a model .\n The weights are taken from a normal distribution \n with mean = 0, std dev = 0.02.\n :param m: A module or layer in a network \n \"\"\"\n # classname will be something like:\n # `Conv`, `BatchNorm2d`, `Linear`, etc.\n classname = m.__class__.__name__\n \n #Apply initial weights to convolutional and linear layers\n if classname.find('Conv') != -1 or classname.find('Linear') != -1:\n nn.init.normal_(m.weight.data, mean=0, std=0.02)",
"_____no_output_____"
],
[
"def build_network(d_conv_dim, g_conv_dim, z_size):\n # define discriminator and generator\n D = Discriminator(d_conv_dim)\n G = Generator(z_size=z_size, conv_dim=g_conv_dim)\n\n # initialize model weights\n D.apply(weights_init_normal)\n G.apply(weights_init_normal)\n\n print(D)\n print()\n print(G)\n \n return D, G",
"_____no_output_____"
],
[
"#=====================================================\n# Calculating real and fake loss\n#=====================================================\ndef real_loss(D_out):\n '''Calculates how close discriminator outputs are to being real.\n param, D_out: discriminator logits\n return: real loss'''\n \n batch_size = D_out.size(0)\n \n labels = torch.ones(batch_size).to(device)\n \n criterion = nn.BCEWithLogitsLoss()\n\n \n loss = criterion(D_out.squeeze(), labels)\n \n return loss\n\ndef fake_loss(D_out):\n \n '''Calculates how close discriminator outputs are to being fake.\n param, D_out: discriminator logits\n return: fake loss'''\n \n batch_size = D_out.size(0)\n \n labels = torch.zeros(batch_size).to(device)\n \n criterion = nn.BCEWithLogitsLoss()\n\n \n loss = criterion(D_out.squeeze(), labels)\n \n return loss",
"_____no_output_____"
],
[
"def train(D, G, d_optimizer, g_optimizer, train_loader, start_epoch = 1, end_epoch = 1, print_every=100):\n '''Trains adversarial networks for some number of epochs\n param, D: the discriminator network\n param, G: the generator network\n param, n_epochs: number of epochs to train for\n param, print_every: when to print and record the models' losses\n return: D and G losses'''\n \n D.to(device)\n G.to(device)\n \n #log training time\n start_time = time.time()\n \n # keep track of loss and generated, \"fake\" samples\n samples = []\n losses = []\n \n len_train_loader = len(train_loader)\n\n # Get some fixed data for sampling. These are images that are held\n # constant throughout training, and allow us to inspect the model's performance\n sample_size=16\n fixed_z = np.random.uniform(-1, 1, size=(sample_size, z_size))\n fixed_z = torch.from_numpy(fixed_z).float().to(device)\n\n \n f = open(TRAINING_LOGS_LOCATION+\"training_log.txt\",'w')\n \n \n # epoch training loop\n for epoch in range(start_epoch, end_epoch+1):\n\n # batch training loop\n for batch_i, (real_images, _) in enumerate(train_loader):\n\n batch_size = real_images.size(0)\n real_images = scale(real_images).to(device)\n \n # ===============================================\n # YOUR CODE HERE: TRAIN THE NETWORKS\n # ===============================================\n # 1. Train the discriminator on real and fake images\n d_optimizer.zero_grad()\n \n d_real_out = D(real_images)\n d_real_loss = real_loss(d_real_out)\n \n fake_images = G(fixed_z)\n d_fake_out = D(fake_images)\n \n d_fake_loss = fake_loss(d_fake_out)\n d_loss = d_real_loss + d_fake_loss\n \n d_loss.backward()\n d_optimizer.step()\n \n # 2. Train the generator with an adversarial loss\n g_optimizer.zero_grad()\n \n fake_images = G(fixed_z)\n g_out = D(fake_images)\n \n g_loss = real_loss(g_out)\n g_loss.backward()\n g_optimizer.step()\n \n \n \n # Print some loss stats\n if batch_i % print_every == 0:\n # append discriminator loss and generator loss\n losses.append((d_loss.item(), g_loss.item()))\n \n stats = 'Epoch [%d/%d], Step: [%d/%d], \\t D_Loss: %.5f, G_Loss: %.5f, Time (mins): %.3f' % (epoch, end_epoch, batch_i, len_train_loader, d_loss.item(), g_loss.item(), (time.time()-start_time)/60)\n # print discriminator and generator loss\n #print('\\r' + stats, end=\"\")\n print(stats)\n\n sys.stdout.flush()\n f.write(stats + '\\n')\n f.flush()\n \n G.eval() # for generating samples\n samples_z = G(fixed_z)\n samples.append(samples_z)\n G.train() # back to training mode\n \n #print('\\r' + stats)\n save_checkpoint(G,MODEL_CHECKPOINT_LOCATION+'generator_'+str(epoch)+\".pth\")\n save_checkpoint(D,MODEL_CHECKPOINT_LOCATION+'discriminator_'+str(epoch)+\".pth\")\n\n # Save training generator samples\n with open('train_samples.pkl', 'wb') as f:\n pkl.dump(samples, f)\n \n # finally return losses\n f.close()\n print(\"============================================================================\")\n print(\"Total Training Time (hours): \"+str((time.time()-start_time)/3600))\n print(\"============================================================================\")\n \n return D, G, losses\n\ndef save_checkpoint(model,fileLocation):\n torch.save(model.state_dict(),fileLocation)\n\ndef view_samples(epoch, samples):\n fig, axes = plt.subplots(figsize=(16,4), nrows=2, ncols=8, sharey=True, sharex=True)\n for ax, img in zip(axes.flatten(), samples[epoch]):\n img = img.detach().cpu().numpy()\n img = np.transpose(img, (1, 2, 0))\n img = ((img + 1)*255 / (2)).astype(np.uint8)\n ax.xaxis.set_visible(False)\n ax.yaxis.set_visible(False)\n im = ax.imshow(img.reshape((32,32,3)))",
"_____no_output_____"
],
[
"batch_size = 64\nimg_size = 32\n\n\n# Create train loader\ntrain_loader = get_dataloader(batch_size,img_size)\n\n# Display some train samples\nimages, _ = iter(train_loader).next()\n\nfig = plt.figure(figsize=(20, 4))\nplot_size=20\nfor idx in np.arange(plot_size):\n ax = fig.add_subplot(2, plot_size/2, idx+1, xticks=[], yticks=[])\n imshow(images[idx])\n\n \n# Test scale\nimg = images[0]\nscaled_img = scale(img)\n\nprint('Min: ', scaled_img.min())\nprint('Max: ', scaled_img.max())",
"Min: tensor(-0.9059)\nMax: tensor(0.8118)\n"
],
[
"d_conv_dim = 32\ng_conv_dim = 32\nz_size = 100\n\nbeta1 = 0.2\nbeta2 = 0.09\n\nD, G = build_network(d_conv_dim, g_conv_dim, z_size)\n\nd_optimizer = optim.Adam(D.parameters(), 0.0005, [beta1, beta2])\ng_optimizer = optim.Adam(G.parameters(), 0.0005, [beta1, beta2])",
"Discriminator(\n (conv1): Sequential(\n (0): Conv2d(3, 32, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n )\n (conv2): Sequential(\n (0): Conv2d(32, 64, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv3): Sequential(\n (0): Conv2d(64, 128, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv4): Sequential(\n (0): Conv2d(128, 256, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (fc1): Linear(in_features=1024, out_features=1, bias=True)\n (dropout1): Dropout(p=0.2, inplace=False)\n)\n\nGenerator(\n (fc1): Linear(in_features=100, out_features=1024, bias=True)\n (conv1): Sequential(\n (0): ConvTranspose2d(256, 128, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): ConvTranspose2d(128, 64, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv3): Sequential(\n (0): ConvTranspose2d(64, 32, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(32, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv4): Sequential(\n (0): ConvTranspose2d(32, 3, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n )\n (dropout2): Dropout(p=0.2, inplace=False)\n)\n"
],
[
"start_epoch = 1\nend_epoch = 100\nprint_every = 400\n\nD, G, losses = train(D, G, d_optimizer, g_optimizer, \n train_loader, \n start_epoch = start_epoch, end_epoch = end_epoch, \n print_every = print_every)",
"Epoch [1/100], Step: [0/1406], \t D_Loss: 1.62923, G_Loss: 1.48748, Time (mins): 0.003\nEpoch [1/100], Step: [400/1406], \t D_Loss: 0.82422, G_Loss: 1.34349, Time (mins): 0.286\nEpoch [1/100], Step: [800/1406], \t D_Loss: 0.48212, G_Loss: 2.92239, Time (mins): 0.570\nEpoch [1/100], Step: [1200/1406], \t D_Loss: 0.48418, G_Loss: 2.57673, Time (mins): 0.851\nEpoch [2/100], Step: [0/1406], \t D_Loss: 0.65324, G_Loss: 1.03611, Time (mins): 0.996\nEpoch [2/100], Step: [400/1406], \t D_Loss: 1.05166, G_Loss: 4.03367, Time (mins): 1.273\nEpoch [2/100], Step: [800/1406], \t D_Loss: 0.19524, G_Loss: 5.81593, Time (mins): 1.551\nEpoch [2/100], Step: [1200/1406], \t D_Loss: 0.34252, G_Loss: 2.19171, Time (mins): 1.835\nEpoch [3/100], Step: [0/1406], \t D_Loss: 0.60448, G_Loss: 3.11102, Time (mins): 1.982\nEpoch [3/100], Step: [400/1406], \t D_Loss: 0.42410, G_Loss: 8.68229, Time (mins): 2.259\nEpoch [3/100], Step: [800/1406], \t D_Loss: 0.74379, G_Loss: 4.23086, Time (mins): 2.536\nEpoch [3/100], Step: [1200/1406], \t D_Loss: 0.16894, G_Loss: 4.81245, Time (mins): 2.817\nEpoch [4/100], Step: [0/1406], \t D_Loss: 0.75080, G_Loss: 4.49446, Time (mins): 2.959\nEpoch [4/100], Step: [400/1406], \t D_Loss: 0.18089, G_Loss: 8.99608, Time (mins): 3.238\nEpoch [4/100], Step: [800/1406], \t D_Loss: 0.20203, G_Loss: 3.88591, Time (mins): 3.518\nEpoch [4/100], Step: [1200/1406], \t D_Loss: 0.80955, G_Loss: 1.54910, Time (mins): 3.798\nEpoch [5/100], Step: [0/1406], \t D_Loss: 0.46898, G_Loss: 7.61716, Time (mins): 3.938\nEpoch [5/100], Step: [400/1406], \t D_Loss: 0.28722, G_Loss: 8.31571, Time (mins): 4.215\nEpoch [5/100], Step: [800/1406], \t D_Loss: 1.12729, G_Loss: 6.07726, Time (mins): 4.492\nEpoch [5/100], Step: [1200/1406], \t D_Loss: 0.49538, G_Loss: 4.81656, Time (mins): 4.767\nEpoch [6/100], Step: [0/1406], \t D_Loss: 0.07599, G_Loss: 2.51267, Time (mins): 4.909\nEpoch [6/100], Step: [400/1406], \t D_Loss: 0.18352, G_Loss: 6.20498, Time (mins): 5.185\nEpoch [6/100], Step: [800/1406], \t D_Loss: 0.78076, G_Loss: 3.98681, Time (mins): 5.468\nEpoch [6/100], Step: [1200/1406], \t D_Loss: 0.13739, G_Loss: 6.71653, Time (mins): 5.745\nEpoch [7/100], Step: [0/1406], \t D_Loss: 0.50472, G_Loss: 4.33761, Time (mins): 5.888\nEpoch [7/100], Step: [400/1406], \t D_Loss: 0.93689, G_Loss: 3.06699, Time (mins): 6.166\nEpoch [7/100], Step: [800/1406], \t D_Loss: 0.62873, G_Loss: 4.93864, Time (mins): 6.442\nEpoch [7/100], Step: [1200/1406], \t D_Loss: 0.08844, G_Loss: 5.91180, Time (mins): 6.721\nEpoch [8/100], Step: [0/1406], \t D_Loss: 0.10643, G_Loss: 3.46181, Time (mins): 6.866\nEpoch [8/100], Step: [400/1406], \t D_Loss: 2.54583, G_Loss: 5.30006, Time (mins): 7.142\nEpoch [8/100], Step: [800/1406], \t D_Loss: 2.65908, G_Loss: 5.11588, Time (mins): 7.422\nEpoch [8/100], Step: [1200/1406], \t D_Loss: 0.25555, G_Loss: 5.29794, Time (mins): 7.703\nEpoch [9/100], Step: [0/1406], \t D_Loss: 0.70065, G_Loss: 7.29642, Time (mins): 7.844\nEpoch [9/100], Step: [400/1406], \t D_Loss: 0.54909, G_Loss: 9.59770, Time (mins): 8.116\nEpoch [9/100], Step: [800/1406], \t D_Loss: 0.05401, G_Loss: 8.87507, Time (mins): 8.394\nEpoch [9/100], Step: [1200/1406], \t D_Loss: 0.02154, G_Loss: 2.90125, Time (mins): 8.674\nEpoch [10/100], Step: [0/1406], \t D_Loss: 0.28687, G_Loss: 5.99367, Time (mins): 8.814\nEpoch [10/100], Step: [400/1406], \t D_Loss: 0.06396, G_Loss: 4.22836, Time (mins): 9.088\nEpoch [10/100], Step: [800/1406], \t D_Loss: 0.86758, G_Loss: 7.20345, Time (mins): 9.364\nEpoch [10/100], Step: [1200/1406], \t D_Loss: 0.43713, G_Loss: 4.96830, Time (mins): 9.639\nEpoch [11/100], Step: [0/1406], \t D_Loss: 0.08118, G_Loss: 3.76172, Time (mins): 9.784\nEpoch [11/100], Step: [400/1406], \t D_Loss: 0.47412, G_Loss: 7.15460, Time (mins): 10.065\nEpoch [11/100], Step: [800/1406], \t D_Loss: 0.02389, G_Loss: 8.46076, Time (mins): 10.345\nEpoch [11/100], Step: [1200/1406], \t D_Loss: 0.71819, G_Loss: 0.60703, Time (mins): 10.620\nEpoch [12/100], Step: [0/1406], \t D_Loss: 0.03107, G_Loss: 10.10655, Time (mins): 10.761\nEpoch [12/100], Step: [400/1406], \t D_Loss: 1.65415, G_Loss: 6.84024, Time (mins): 11.042\nEpoch [12/100], Step: [800/1406], \t D_Loss: 1.01145, G_Loss: 9.71559, Time (mins): 11.324\nEpoch [12/100], Step: [1200/1406], \t D_Loss: 0.10922, G_Loss: 4.12881, Time (mins): 11.599\nEpoch [13/100], Step: [0/1406], \t D_Loss: 0.53332, G_Loss: 0.92668, Time (mins): 11.742\nEpoch [13/100], Step: [400/1406], \t D_Loss: 0.48411, G_Loss: 9.02356, Time (mins): 12.022\nEpoch [13/100], Step: [800/1406], \t D_Loss: 0.27534, G_Loss: 7.62958, Time (mins): 12.304\nEpoch [13/100], Step: [1200/1406], \t D_Loss: 0.56490, G_Loss: 6.88198, Time (mins): 12.583\nEpoch [14/100], Step: [0/1406], \t D_Loss: 0.22863, G_Loss: 5.32085, Time (mins): 12.728\nEpoch [14/100], Step: [400/1406], \t D_Loss: 0.24817, G_Loss: 8.53283, Time (mins): 13.006\nEpoch [14/100], Step: [800/1406], \t D_Loss: 0.02160, G_Loss: 9.70182, Time (mins): 13.283\nEpoch [14/100], Step: [1200/1406], \t D_Loss: 0.37994, G_Loss: 3.20336, Time (mins): 13.561\nEpoch [15/100], Step: [0/1406], \t D_Loss: 0.49415, G_Loss: 5.31060, Time (mins): 13.701\nEpoch [15/100], Step: [400/1406], \t D_Loss: 1.10601, G_Loss: 9.43853, Time (mins): 13.976\nEpoch [15/100], Step: [800/1406], \t D_Loss: 0.04235, G_Loss: 10.23702, Time (mins): 14.250\nEpoch [15/100], Step: [1200/1406], \t D_Loss: 0.03328, G_Loss: 5.87094, Time (mins): 14.525\nEpoch [16/100], Step: [0/1406], \t D_Loss: 1.44917, G_Loss: 3.93104, Time (mins): 14.668\nEpoch [16/100], Step: [400/1406], \t D_Loss: 0.04443, G_Loss: 9.11757, Time (mins): 14.945\nEpoch [16/100], Step: [800/1406], \t D_Loss: 0.06172, G_Loss: 7.59460, Time (mins): 15.222\nEpoch [16/100], Step: [1200/1406], \t D_Loss: 0.05135, G_Loss: 8.16017, Time (mins): 15.501\nEpoch [17/100], Step: [0/1406], \t D_Loss: 6.37874, G_Loss: 2.39830, Time (mins): 15.645\nEpoch [17/100], Step: [400/1406], \t D_Loss: 0.01966, G_Loss: 7.82355, Time (mins): 15.922\nEpoch [17/100], Step: [800/1406], \t D_Loss: 0.02082, G_Loss: 13.73861, Time (mins): 16.198\nEpoch [17/100], Step: [1200/1406], \t D_Loss: 0.04803, G_Loss: 9.15363, Time (mins): 16.474\nEpoch [18/100], Step: [0/1406], \t D_Loss: 0.33835, G_Loss: 4.83757, Time (mins): 16.615\nEpoch [18/100], Step: [400/1406], \t D_Loss: 0.08221, G_Loss: 8.72072, Time (mins): 16.895\nEpoch [18/100], Step: [800/1406], \t D_Loss: 0.80927, G_Loss: 4.99594, Time (mins): 17.169\nEpoch [18/100], Step: [1200/1406], \t D_Loss: 0.99249, G_Loss: 10.62824, Time (mins): 17.452\nEpoch [19/100], Step: [0/1406], \t D_Loss: 0.91890, G_Loss: 5.42726, Time (mins): 17.595\nEpoch [19/100], Step: [400/1406], \t D_Loss: 0.18544, G_Loss: 13.72453, Time (mins): 17.866\nEpoch [19/100], Step: [800/1406], \t D_Loss: 0.07727, G_Loss: 14.12419, Time (mins): 18.145\nEpoch [19/100], Step: [1200/1406], \t D_Loss: 0.25564, G_Loss: 8.22672, Time (mins): 18.421\nEpoch [20/100], Step: [0/1406], \t D_Loss: 0.00102, G_Loss: 8.67511, Time (mins): 18.563\nEpoch [20/100], Step: [400/1406], \t D_Loss: 1.28750, G_Loss: 1.62245, Time (mins): 18.840\nEpoch [20/100], Step: [800/1406], \t D_Loss: 0.56347, G_Loss: 7.08979, Time (mins): 19.114\nEpoch [20/100], Step: [1200/1406], \t D_Loss: 0.07799, G_Loss: 3.98289, Time (mins): 19.388\nEpoch [21/100], Step: [0/1406], \t D_Loss: 0.34973, G_Loss: 10.34963, Time (mins): 19.527\nEpoch [21/100], Step: [400/1406], \t D_Loss: 0.23270, G_Loss: 10.71995, Time (mins): 19.799\nEpoch [21/100], Step: [800/1406], \t D_Loss: 0.34064, G_Loss: 11.31845, Time (mins): 20.077\nEpoch [21/100], Step: [1200/1406], \t D_Loss: 0.28283, G_Loss: 7.95665, Time (mins): 20.352\nEpoch [22/100], Step: [0/1406], \t D_Loss: 3.25381, G_Loss: 3.39694, Time (mins): 20.492\nEpoch [22/100], Step: [400/1406], \t D_Loss: 0.01875, G_Loss: 6.30338, Time (mins): 20.768\nEpoch [22/100], Step: [800/1406], \t D_Loss: 0.01454, G_Loss: 6.96566, Time (mins): 21.043\nEpoch [22/100], Step: [1200/1406], \t D_Loss: 0.01241, G_Loss: 9.08419, Time (mins): 21.318\nEpoch [23/100], Step: [0/1406], \t D_Loss: 0.15016, G_Loss: 5.22657, Time (mins): 21.458\nEpoch [23/100], Step: [400/1406], \t D_Loss: 0.00977, G_Loss: 13.46030, Time (mins): 21.736\nEpoch [23/100], Step: [800/1406], \t D_Loss: 1.43188, G_Loss: 8.12532, Time (mins): 22.017\nEpoch [23/100], Step: [1200/1406], \t D_Loss: 0.06570, G_Loss: 11.13596, Time (mins): 22.292\nEpoch [24/100], Step: [0/1406], \t D_Loss: 0.19732, G_Loss: 6.95198, Time (mins): 22.441\nEpoch [24/100], Step: [400/1406], \t D_Loss: 3.72653, G_Loss: 1.27496, Time (mins): 22.721\nEpoch [24/100], Step: [800/1406], \t D_Loss: 0.07258, G_Loss: 7.09689, Time (mins): 22.995\nEpoch [24/100], Step: [1200/1406], \t D_Loss: 0.13008, G_Loss: 8.47567, Time (mins): 23.269\nEpoch [25/100], Step: [0/1406], \t D_Loss: 2.72627, G_Loss: 9.32195, Time (mins): 23.415\nEpoch [25/100], Step: [400/1406], \t D_Loss: 2.09079, G_Loss: 3.61501, Time (mins): 23.694\nEpoch [25/100], Step: [800/1406], \t D_Loss: 0.00639, G_Loss: 5.08444, Time (mins): 23.972\nEpoch [25/100], Step: [1200/1406], \t D_Loss: 0.17674, G_Loss: 10.34451, Time (mins): 24.252\nEpoch [26/100], Step: [0/1406], \t D_Loss: 0.01097, G_Loss: 13.99170, Time (mins): 24.394\nEpoch [26/100], Step: [400/1406], \t D_Loss: 0.32555, G_Loss: 14.83764, Time (mins): 24.672\nEpoch [26/100], Step: [800/1406], \t D_Loss: 0.02892, G_Loss: 12.04262, Time (mins): 24.949\nEpoch [26/100], Step: [1200/1406], \t D_Loss: 0.05855, G_Loss: 8.88636, Time (mins): 25.224\nEpoch [27/100], Step: [0/1406], \t D_Loss: 0.00038, G_Loss: 11.36292, Time (mins): 25.368\nEpoch [27/100], Step: [400/1406], \t D_Loss: 0.00438, G_Loss: 1.08798, Time (mins): 25.645\nEpoch [27/100], Step: [800/1406], \t D_Loss: 0.00083, G_Loss: 13.03282, Time (mins): 25.922\nEpoch [27/100], Step: [1200/1406], \t D_Loss: 0.01989, G_Loss: 13.06523, Time (mins): 26.196\nEpoch [28/100], Step: [0/1406], \t D_Loss: 0.19664, G_Loss: 5.53013, Time (mins): 26.338\nEpoch [28/100], Step: [400/1406], \t D_Loss: 0.21183, G_Loss: 5.81369, Time (mins): 26.612\nEpoch [28/100], Step: [800/1406], \t D_Loss: 0.20347, G_Loss: 8.03045, Time (mins): 26.886\nEpoch [28/100], Step: [1200/1406], \t D_Loss: 0.00152, G_Loss: 11.88147, Time (mins): 27.162\nEpoch [29/100], Step: [0/1406], \t D_Loss: 0.81517, G_Loss: 12.57764, Time (mins): 27.304\nEpoch [29/100], Step: [400/1406], \t D_Loss: 0.00055, G_Loss: 15.22765, Time (mins): 27.586\nEpoch [29/100], Step: [800/1406], \t D_Loss: 0.24010, G_Loss: 10.36880, Time (mins): 27.863\nEpoch [29/100], Step: [1200/1406], \t D_Loss: 0.01290, G_Loss: 11.39147, Time (mins): 28.138\nEpoch [30/100], Step: [0/1406], \t D_Loss: 1.30111, G_Loss: 6.12412, Time (mins): 28.281\nEpoch [30/100], Step: [400/1406], \t D_Loss: 0.07370, G_Loss: 12.12341, Time (mins): 28.557\nEpoch [30/100], Step: [800/1406], \t D_Loss: 0.05147, G_Loss: 7.21130, Time (mins): 28.841\nEpoch [30/100], Step: [1200/1406], \t D_Loss: 1.58338, G_Loss: 8.12551, Time (mins): 29.116\nEpoch [31/100], Step: [0/1406], \t D_Loss: 0.03262, G_Loss: 5.04587, Time (mins): 29.259\nEpoch [31/100], Step: [400/1406], \t D_Loss: 0.00791, G_Loss: 15.13551, Time (mins): 29.541\nEpoch [31/100], Step: [800/1406], \t D_Loss: 0.03908, G_Loss: 7.64325, Time (mins): 29.814\nEpoch [31/100], Step: [1200/1406], \t D_Loss: 0.19611, G_Loss: 7.83445, Time (mins): 30.094\nEpoch [32/100], Step: [0/1406], \t D_Loss: 0.00385, G_Loss: 4.60310, Time (mins): 30.238\nEpoch [32/100], Step: [400/1406], \t D_Loss: 0.15096, G_Loss: 4.26157, Time (mins): 30.512\nEpoch [32/100], Step: [800/1406], \t D_Loss: 0.00082, G_Loss: 10.31130, Time (mins): 30.792\nEpoch [32/100], Step: [1200/1406], \t D_Loss: 0.00082, G_Loss: 15.86946, Time (mins): 31.075\nEpoch [33/100], Step: [0/1406], \t D_Loss: 1.58002, G_Loss: 9.91054, Time (mins): 31.219\nEpoch [33/100], Step: [400/1406], \t D_Loss: 0.01624, G_Loss: 10.61663, Time (mins): 31.499\nEpoch [33/100], Step: [800/1406], \t D_Loss: 1.00883, G_Loss: 5.29529, Time (mins): 31.779\nEpoch [33/100], Step: [1200/1406], \t D_Loss: 0.61341, G_Loss: 10.83899, Time (mins): 32.057\nEpoch [34/100], Step: [0/1406], \t D_Loss: 0.00151, G_Loss: 12.20918, Time (mins): 32.201\nEpoch [34/100], Step: [400/1406], \t D_Loss: 0.16748, G_Loss: 13.06252, Time (mins): 32.478\nEpoch [34/100], Step: [800/1406], \t D_Loss: 0.00217, G_Loss: 10.20074, Time (mins): 32.763\nEpoch [34/100], Step: [1200/1406], \t D_Loss: 0.00116, G_Loss: 12.74970, Time (mins): 33.041\nEpoch [35/100], Step: [0/1406], \t D_Loss: 0.33473, G_Loss: 4.92111, Time (mins): 33.182\nEpoch [35/100], Step: [400/1406], \t D_Loss: 0.00026, G_Loss: 11.26232, Time (mins): 33.460\nEpoch [35/100], Step: [800/1406], \t D_Loss: 0.00616, G_Loss: 12.08147, Time (mins): 33.732\nEpoch [35/100], Step: [1200/1406], \t D_Loss: 0.00065, G_Loss: 12.97071, Time (mins): 34.011\nEpoch [36/100], Step: [0/1406], \t D_Loss: 0.03476, G_Loss: 11.52221, Time (mins): 34.155\nEpoch [36/100], Step: [400/1406], \t D_Loss: 0.22752, G_Loss: 7.80273, Time (mins): 34.429\nEpoch [36/100], Step: [800/1406], \t D_Loss: 0.68086, G_Loss: 10.05916, Time (mins): 34.705\nEpoch [36/100], Step: [1200/1406], \t D_Loss: 0.07435, G_Loss: 10.56988, Time (mins): 34.982\nEpoch [37/100], Step: [0/1406], \t D_Loss: 0.00844, G_Loss: 5.96709, Time (mins): 35.126\nEpoch [37/100], Step: [400/1406], \t D_Loss: 0.00617, G_Loss: 11.01734, Time (mins): 35.401\nEpoch [37/100], Step: [800/1406], \t D_Loss: 1.61468, G_Loss: 5.51086, Time (mins): 35.680\nEpoch [37/100], Step: [1200/1406], \t D_Loss: 0.03721, G_Loss: 13.62807, Time (mins): 35.956\nEpoch [38/100], Step: [0/1406], \t D_Loss: 0.11098, G_Loss: 4.15361, Time (mins): 36.099\nEpoch [38/100], Step: [400/1406], \t D_Loss: 0.38056, G_Loss: 4.99034, Time (mins): 36.377\nEpoch [38/100], Step: [800/1406], \t D_Loss: 0.00015, G_Loss: 13.92421, Time (mins): 36.653\nEpoch [38/100], Step: [1200/1406], \t D_Loss: 0.04218, G_Loss: 14.66880, Time (mins): 36.930\nEpoch [39/100], Step: [0/1406], \t D_Loss: 0.29051, G_Loss: 4.70696, Time (mins): 37.073\nEpoch [39/100], Step: [400/1406], \t D_Loss: 1.29758, G_Loss: 0.44890, Time (mins): 37.350\nEpoch [39/100], Step: [800/1406], \t D_Loss: 0.15416, G_Loss: 6.68668, Time (mins): 37.628\nEpoch [39/100], Step: [1200/1406], \t D_Loss: 0.00059, G_Loss: 13.08591, Time (mins): 37.909\nEpoch [40/100], Step: [0/1406], \t D_Loss: 0.00017, G_Loss: 14.23036, Time (mins): 38.056\nEpoch [40/100], Step: [400/1406], \t D_Loss: 0.04293, G_Loss: 5.57691, Time (mins): 38.330\nEpoch [40/100], Step: [800/1406], \t D_Loss: 0.00020, G_Loss: 12.78221, Time (mins): 38.608\nEpoch [40/100], Step: [1200/1406], \t D_Loss: 0.00093, G_Loss: 11.25627, Time (mins): 38.887\nEpoch [41/100], Step: [0/1406], \t D_Loss: 0.00235, G_Loss: 14.79993, Time (mins): 39.029\nEpoch [41/100], Step: [400/1406], \t D_Loss: 0.00017, G_Loss: 13.56823, Time (mins): 39.305\nEpoch [41/100], Step: [800/1406], \t D_Loss: 1.45324, G_Loss: 2.90974, Time (mins): 39.582\nEpoch [41/100], Step: [1200/1406], \t D_Loss: 0.28912, G_Loss: 3.54824, Time (mins): 39.863\nEpoch [42/100], Step: [0/1406], \t D_Loss: 0.27290, G_Loss: 8.02168, Time (mins): 40.007\nEpoch [42/100], Step: [400/1406], \t D_Loss: 0.00004, G_Loss: 16.11527, Time (mins): 40.284\nEpoch [42/100], Step: [800/1406], \t D_Loss: 0.00189, G_Loss: 8.32779, Time (mins): 40.559\nEpoch [42/100], Step: [1200/1406], \t D_Loss: 0.09282, G_Loss: 8.41294, Time (mins): 40.835\nEpoch [43/100], Step: [0/1406], \t D_Loss: 1.27821, G_Loss: 3.77602, Time (mins): 40.979\nEpoch [43/100], Step: [400/1406], \t D_Loss: 0.00168, G_Loss: 12.40047, Time (mins): 41.260\nEpoch [43/100], Step: [800/1406], \t D_Loss: 0.77468, G_Loss: 7.13143, Time (mins): 41.536\nEpoch [43/100], Step: [1200/1406], \t D_Loss: 0.32308, G_Loss: 15.69377, Time (mins): 41.812\nEpoch [44/100], Step: [0/1406], \t D_Loss: 0.02384, G_Loss: 6.01760, Time (mins): 41.954\nEpoch [44/100], Step: [400/1406], \t D_Loss: 0.00045, G_Loss: 14.03120, Time (mins): 42.232\nEpoch [44/100], Step: [800/1406], \t D_Loss: 0.02871, G_Loss: 9.73254, Time (mins): 42.508\nEpoch [44/100], Step: [1200/1406], \t D_Loss: 0.01433, G_Loss: 6.82285, Time (mins): 42.787\nEpoch [45/100], Step: [0/1406], \t D_Loss: 0.84353, G_Loss: 3.46978, Time (mins): 42.934\nEpoch [45/100], Step: [400/1406], \t D_Loss: 0.12687, G_Loss: 11.09457, Time (mins): 43.215\nEpoch [45/100], Step: [800/1406], \t D_Loss: 0.00001, G_Loss: 18.75487, Time (mins): 43.497\nEpoch [45/100], Step: [1200/1406], \t D_Loss: 0.04270, G_Loss: 4.60295, Time (mins): 43.778\nEpoch [46/100], Step: [0/1406], \t D_Loss: 0.00005, G_Loss: 18.15641, Time (mins): 43.921\nEpoch [46/100], Step: [400/1406], \t D_Loss: 2.18735, G_Loss: 7.56470, Time (mins): 44.193\nEpoch [46/100], Step: [800/1406], \t D_Loss: 0.98805, G_Loss: 8.68413, Time (mins): 44.469\nEpoch [46/100], Step: [1200/1406], \t D_Loss: 0.69373, G_Loss: 10.58309, Time (mins): 44.750\nEpoch [47/100], Step: [0/1406], \t D_Loss: 0.14883, G_Loss: 14.23434, Time (mins): 44.894\nEpoch [47/100], Step: [400/1406], \t D_Loss: 0.69993, G_Loss: 8.95093, Time (mins): 45.173\nEpoch [47/100], Step: [800/1406], \t D_Loss: 0.00003, G_Loss: 8.94532, Time (mins): 45.456\nEpoch [47/100], Step: [1200/1406], \t D_Loss: 6.84001, G_Loss: 1.03483, Time (mins): 45.733\nEpoch [48/100], Step: [0/1406], \t D_Loss: 0.21914, G_Loss: 5.99590, Time (mins): 45.875\nEpoch [48/100], Step: [400/1406], \t D_Loss: 0.15866, G_Loss: 14.54547, Time (mins): 46.153\nEpoch [48/100], Step: [800/1406], \t D_Loss: 0.00016, G_Loss: 10.15412, Time (mins): 46.435\nEpoch [48/100], Step: [1200/1406], \t D_Loss: 0.16788, G_Loss: 11.05639, Time (mins): 46.713\nEpoch [49/100], Step: [0/1406], \t D_Loss: 0.01852, G_Loss: 4.68856, Time (mins): 46.856\nEpoch [49/100], Step: [400/1406], \t D_Loss: 0.00835, G_Loss: 14.02534, Time (mins): 47.133\nEpoch [49/100], Step: [800/1406], \t D_Loss: 0.00008, G_Loss: 6.98730, Time (mins): 47.409\nEpoch [49/100], Step: [1200/1406], \t D_Loss: 0.01854, G_Loss: 13.49094, Time (mins): 47.684\nEpoch [50/100], Step: [0/1406], \t D_Loss: 8.50251, G_Loss: 3.90988, Time (mins): 47.825\nEpoch [50/100], Step: [400/1406], \t D_Loss: 0.34211, G_Loss: 8.57673, Time (mins): 48.101\nEpoch [50/100], Step: [800/1406], \t D_Loss: 0.00476, G_Loss: 13.43514, Time (mins): 48.381\nEpoch [50/100], Step: [1200/1406], \t D_Loss: 0.00699, G_Loss: 12.92156, Time (mins): 48.659\nEpoch [51/100], Step: [0/1406], \t D_Loss: 0.00000, G_Loss: 16.94420, Time (mins): 48.801\nEpoch [51/100], Step: [400/1406], \t D_Loss: 0.03187, G_Loss: 18.00760, Time (mins): 49.078\nEpoch [51/100], Step: [800/1406], \t D_Loss: 0.00682, G_Loss: 7.24392, Time (mins): 49.354\nEpoch [51/100], Step: [1200/1406], \t D_Loss: 0.00265, G_Loss: 10.26751, Time (mins): 49.629\nEpoch [52/100], Step: [0/1406], \t D_Loss: 0.02903, G_Loss: 13.34714, Time (mins): 49.774\nEpoch [52/100], Step: [400/1406], \t D_Loss: 0.00726, G_Loss: 16.10783, Time (mins): 50.049\nEpoch [52/100], Step: [800/1406], \t D_Loss: 0.00001, G_Loss: 8.84554, Time (mins): 50.329\nEpoch [52/100], Step: [1200/1406], \t D_Loss: 0.37717, G_Loss: 8.64758, Time (mins): 50.607\nEpoch [53/100], Step: [0/1406], \t D_Loss: 1.31888, G_Loss: 3.15899, Time (mins): 50.750\nEpoch [53/100], Step: [400/1406], \t D_Loss: 0.00001, G_Loss: 19.55876, Time (mins): 51.025\nEpoch [53/100], Step: [800/1406], \t D_Loss: 0.29935, G_Loss: 8.00058, Time (mins): 51.300\nEpoch [53/100], Step: [1200/1406], \t D_Loss: 0.00138, G_Loss: 18.76564, Time (mins): 51.577\nEpoch [54/100], Step: [0/1406], \t D_Loss: 0.00481, G_Loss: 8.03798, Time (mins): 51.719\nEpoch [54/100], Step: [400/1406], \t D_Loss: 0.10539, G_Loss: 7.10949, Time (mins): 51.996\nEpoch [54/100], Step: [800/1406], \t D_Loss: 0.00231, G_Loss: 12.54457, Time (mins): 52.273\nEpoch [54/100], Step: [1200/1406], \t D_Loss: 0.04789, G_Loss: 15.39933, Time (mins): 52.550\nEpoch [55/100], Step: [0/1406], \t D_Loss: 0.00133, G_Loss: 7.13262, Time (mins): 52.696\nEpoch [55/100], Step: [400/1406], \t D_Loss: 0.00005, G_Loss: 18.75164, Time (mins): 52.975\nEpoch [55/100], Step: [800/1406], \t D_Loss: 0.00126, G_Loss: 18.78413, Time (mins): 53.253\nEpoch [55/100], Step: [1200/1406], \t D_Loss: 0.00001, G_Loss: 18.67539, Time (mins): 53.531\nEpoch [56/100], Step: [0/1406], \t D_Loss: 0.39294, G_Loss: 4.68284, Time (mins): 53.673\nEpoch [56/100], Step: [400/1406], \t D_Loss: 0.01695, G_Loss: 10.60683, Time (mins): 53.948\nEpoch [56/100], Step: [800/1406], \t D_Loss: 0.00597, G_Loss: 8.28136, Time (mins): 54.232\nEpoch [56/100], Step: [1200/1406], \t D_Loss: 0.00263, G_Loss: 15.44516, Time (mins): 54.507\nEpoch [57/100], Step: [0/1406], \t D_Loss: 0.00022, G_Loss: 4.27467, Time (mins): 54.650\nEpoch [57/100], Step: [400/1406], \t D_Loss: 0.00499, G_Loss: 17.76367, Time (mins): 54.927\nEpoch [57/100], Step: [800/1406], \t D_Loss: 0.00912, G_Loss: 11.89055, Time (mins): 55.201\nEpoch [57/100], Step: [1200/1406], \t D_Loss: 0.46095, G_Loss: 11.11982, Time (mins): 55.478\nEpoch [58/100], Step: [0/1406], \t D_Loss: 0.46039, G_Loss: 9.26759, Time (mins): 55.622\nEpoch [58/100], Step: [400/1406], \t D_Loss: 0.07147, G_Loss: 16.20431, Time (mins): 55.899\nEpoch [58/100], Step: [800/1406], \t D_Loss: 0.00284, G_Loss: 16.98432, Time (mins): 56.177\nEpoch [58/100], Step: [1200/1406], \t D_Loss: 0.39063, G_Loss: 9.42444, Time (mins): 56.455\nEpoch [59/100], Step: [0/1406], \t D_Loss: 0.21898, G_Loss: 1.99430, Time (mins): 56.598\nEpoch [59/100], Step: [400/1406], \t D_Loss: 0.00011, G_Loss: 16.85331, Time (mins): 56.877\nEpoch [59/100], Step: [800/1406], \t D_Loss: 0.03677, G_Loss: 11.39615, Time (mins): 57.151\nEpoch [59/100], Step: [1200/1406], \t D_Loss: 0.06324, G_Loss: 12.87358, Time (mins): 57.433\nEpoch [60/100], Step: [0/1406], \t D_Loss: 0.00437, G_Loss: 9.21621, Time (mins): 57.575\nEpoch [60/100], Step: [400/1406], \t D_Loss: 0.02450, G_Loss: 9.91347, Time (mins): 57.851\nEpoch [60/100], Step: [800/1406], \t D_Loss: 0.34372, G_Loss: 8.01124, Time (mins): 58.126\nEpoch [60/100], Step: [1200/1406], \t D_Loss: 0.06054, G_Loss: 17.43484, Time (mins): 58.405\nEpoch [61/100], Step: [0/1406], \t D_Loss: 0.09720, G_Loss: 8.44616, Time (mins): 58.547\nEpoch [61/100], Step: [400/1406], \t D_Loss: 0.24036, G_Loss: 25.97115, Time (mins): 58.825\nEpoch [61/100], Step: [800/1406], \t D_Loss: 1.21867, G_Loss: 12.96243, Time (mins): 59.104\nEpoch [61/100], Step: [1200/1406], \t D_Loss: 0.02801, G_Loss: 19.30791, Time (mins): 59.380\nEpoch [62/100], Step: [0/1406], \t D_Loss: 0.07474, G_Loss: 6.59011, Time (mins): 59.523\nEpoch [62/100], Step: [400/1406], \t D_Loss: 1.00343, G_Loss: 6.67463, Time (mins): 59.800\nEpoch [62/100], Step: [800/1406], \t D_Loss: 0.00018, G_Loss: 12.43554, Time (mins): 60.079\nEpoch [62/100], Step: [1200/1406], \t D_Loss: 0.00561, G_Loss: 8.92353, Time (mins): 60.355\nEpoch [63/100], Step: [0/1406], \t D_Loss: 0.07968, G_Loss: 5.01382, Time (mins): 60.496\nEpoch [63/100], Step: [400/1406], \t D_Loss: 0.98821, G_Loss: 10.02040, Time (mins): 60.774\nEpoch [63/100], Step: [800/1406], \t D_Loss: 0.23045, G_Loss: 19.04301, Time (mins): 61.050\nEpoch [63/100], Step: [1200/1406], \t D_Loss: 0.00024, G_Loss: 16.65590, Time (mins): 61.331\nEpoch [64/100], Step: [0/1406], \t D_Loss: 2.75483, G_Loss: 3.96210, Time (mins): 61.473\nEpoch [64/100], Step: [400/1406], \t D_Loss: 0.02926, G_Loss: 7.97561, Time (mins): 61.748\nEpoch [64/100], Step: [800/1406], \t D_Loss: 0.00130, G_Loss: 20.99907, Time (mins): 62.031\nEpoch [64/100], Step: [1200/1406], \t D_Loss: 3.10942, G_Loss: 9.12002, Time (mins): 62.308\nEpoch [65/100], Step: [0/1406], \t D_Loss: 0.00357, G_Loss: 10.22406, Time (mins): 62.454\nEpoch [65/100], Step: [400/1406], \t D_Loss: 0.18300, G_Loss: 9.85074, Time (mins): 62.729\nEpoch [65/100], Step: [800/1406], \t D_Loss: 0.00000, G_Loss: 15.58737, Time (mins): 63.006\nEpoch [65/100], Step: [1200/1406], \t D_Loss: 0.00986, G_Loss: 6.63953, Time (mins): 63.285\nEpoch [66/100], Step: [0/1406], \t D_Loss: 1.82126, G_Loss: 5.06695, Time (mins): 63.426\nEpoch [66/100], Step: [400/1406], \t D_Loss: 0.02372, G_Loss: 12.98156, Time (mins): 63.703\nEpoch [66/100], Step: [800/1406], \t D_Loss: 14.58644, G_Loss: 9.68461, Time (mins): 63.988\nEpoch [66/100], Step: [1200/1406], \t D_Loss: 3.01473, G_Loss: 6.34464, Time (mins): 64.263\nEpoch [67/100], Step: [0/1406], \t D_Loss: 0.12075, G_Loss: 6.10638, Time (mins): 64.408\nEpoch [67/100], Step: [400/1406], \t D_Loss: 0.18791, G_Loss: 8.50696, Time (mins): 64.684\nEpoch [67/100], Step: [800/1406], \t D_Loss: 0.30296, G_Loss: 15.31750, Time (mins): 64.962\nEpoch [67/100], Step: [1200/1406], \t D_Loss: 0.00020, G_Loss: 10.33346, Time (mins): 65.239\nEpoch [68/100], Step: [0/1406], \t D_Loss: 0.00276, G_Loss: 10.13878, Time (mins): 65.381\nEpoch [68/100], Step: [400/1406], \t D_Loss: 0.04674, G_Loss: 14.08162, Time (mins): 65.661\nEpoch [68/100], Step: [800/1406], \t D_Loss: 0.07559, G_Loss: 9.97152, Time (mins): 65.940\nEpoch [68/100], Step: [1200/1406], \t D_Loss: 0.09686, G_Loss: 5.71794, Time (mins): 66.221\nEpoch [69/100], Step: [0/1406], \t D_Loss: 0.01119, G_Loss: 2.52922, Time (mins): 66.362\nEpoch [69/100], Step: [400/1406], \t D_Loss: 0.01386, G_Loss: 13.39199, Time (mins): 66.637\nEpoch [69/100], Step: [800/1406], \t D_Loss: 0.08788, G_Loss: 3.52077, Time (mins): 66.912\nEpoch [69/100], Step: [1200/1406], \t D_Loss: 0.13647, G_Loss: 14.44710, Time (mins): 67.185\nEpoch [70/100], Step: [0/1406], \t D_Loss: 0.10653, G_Loss: 7.56960, Time (mins): 67.328\nEpoch [70/100], Step: [400/1406], \t D_Loss: 0.00505, G_Loss: 13.55420, Time (mins): 67.609\nEpoch [70/100], Step: [800/1406], \t D_Loss: 0.01160, G_Loss: 12.51612, Time (mins): 67.882\nEpoch [70/100], Step: [1200/1406], \t D_Loss: 0.00886, G_Loss: 19.78920, Time (mins): 68.160\nEpoch [71/100], Step: [0/1406], \t D_Loss: 0.05317, G_Loss: 5.08094, Time (mins): 68.302\nEpoch [71/100], Step: [400/1406], \t D_Loss: 1.58699, G_Loss: 1.39797, Time (mins): 68.578\nEpoch [71/100], Step: [800/1406], \t D_Loss: 0.04144, G_Loss: 9.80083, Time (mins): 68.852\nEpoch [71/100], Step: [1200/1406], \t D_Loss: 0.00023, G_Loss: 15.44506, Time (mins): 69.132\nEpoch [72/100], Step: [0/1406], \t D_Loss: 1.03127, G_Loss: 11.96558, Time (mins): 69.276\nEpoch [72/100], Step: [400/1406], \t D_Loss: 0.04429, G_Loss: 9.76972, Time (mins): 69.551\nEpoch [72/100], Step: [800/1406], \t D_Loss: 0.03793, G_Loss: 7.44052, Time (mins): 69.829\nEpoch [72/100], Step: [1200/1406], \t D_Loss: 0.03382, G_Loss: 9.92828, Time (mins): 70.111\nEpoch [73/100], Step: [0/1406], \t D_Loss: 1.07288, G_Loss: 5.49714, Time (mins): 70.255\nEpoch [73/100], Step: [400/1406], \t D_Loss: 0.17048, G_Loss: 8.50917, Time (mins): 70.532\nEpoch [73/100], Step: [800/1406], \t D_Loss: 0.00002, G_Loss: 12.63051, Time (mins): 70.809\nEpoch [73/100], Step: [1200/1406], \t D_Loss: 0.00005, G_Loss: 21.18742, Time (mins): 71.086\nEpoch [74/100], Step: [0/1406], \t D_Loss: 0.20263, G_Loss: 7.50441, Time (mins): 71.229\nEpoch [74/100], Step: [400/1406], \t D_Loss: 0.00165, G_Loss: 13.32431, Time (mins): 71.505\nEpoch [74/100], Step: [800/1406], \t D_Loss: 0.05426, G_Loss: 3.12269, Time (mins): 71.781\nEpoch [74/100], Step: [1200/1406], \t D_Loss: 0.00403, G_Loss: 13.10350, Time (mins): 72.062\nEpoch [75/100], Step: [0/1406], \t D_Loss: 0.75536, G_Loss: 6.22852, Time (mins): 72.204\nEpoch [75/100], Step: [400/1406], \t D_Loss: 0.00608, G_Loss: 12.94872, Time (mins): 72.484\nEpoch [75/100], Step: [800/1406], \t D_Loss: 0.00089, G_Loss: 13.51840, Time (mins): 72.760\nEpoch [75/100], Step: [1200/1406], \t D_Loss: 0.00872, G_Loss: 12.24631, Time (mins): 73.039\nEpoch [76/100], Step: [0/1406], \t D_Loss: 0.01756, G_Loss: 12.86769, Time (mins): 73.182\nEpoch [76/100], Step: [400/1406], \t D_Loss: 0.37251, G_Loss: 13.42708, Time (mins): 73.457\nEpoch [76/100], Step: [800/1406], \t D_Loss: 0.04826, G_Loss: 6.72685, Time (mins): 73.731\nEpoch [76/100], Step: [1200/1406], \t D_Loss: 0.04493, G_Loss: 14.39716, Time (mins): 74.005\nEpoch [77/100], Step: [0/1406], \t D_Loss: 0.01316, G_Loss: 8.10184, Time (mins): 74.148\nEpoch [77/100], Step: [400/1406], \t D_Loss: 0.00001, G_Loss: 14.78357, Time (mins): 74.426\nEpoch [77/100], Step: [800/1406], \t D_Loss: 0.00000, G_Loss: 20.04281, Time (mins): 74.704\nEpoch [77/100], Step: [1200/1406], \t D_Loss: 0.00015, G_Loss: 6.99766, Time (mins): 74.980\nEpoch [78/100], Step: [0/1406], \t D_Loss: 0.00469, G_Loss: 8.87766, Time (mins): 75.121\nEpoch [78/100], Step: [400/1406], \t D_Loss: 0.35674, G_Loss: 8.19545, Time (mins): 75.396\nEpoch [78/100], Step: [800/1406], \t D_Loss: 0.00000, G_Loss: 23.92159, Time (mins): 75.677\nEpoch [78/100], Step: [1200/1406], \t D_Loss: 0.00006, G_Loss: 10.57607, Time (mins): 75.957\nEpoch [79/100], Step: [0/1406], \t D_Loss: 0.00022, G_Loss: 10.08823, Time (mins): 76.101\nEpoch [79/100], Step: [400/1406], \t D_Loss: 0.00157, G_Loss: 11.50863, Time (mins): 76.373\nEpoch [79/100], Step: [800/1406], \t D_Loss: 0.00000, G_Loss: 18.90618, Time (mins): 76.652\nEpoch [79/100], Step: [1200/1406], \t D_Loss: 0.00026, G_Loss: 4.51064, Time (mins): 76.927\nEpoch [80/100], Step: [0/1406], \t D_Loss: 0.00487, G_Loss: 10.00088, Time (mins): 77.070\nEpoch [80/100], Step: [400/1406], \t D_Loss: 0.00000, G_Loss: 18.85481, Time (mins): 77.345\nEpoch [80/100], Step: [800/1406], \t D_Loss: 0.00086, G_Loss: 2.85072, Time (mins): 77.620\nEpoch [80/100], Step: [1200/1406], \t D_Loss: 0.00008, G_Loss: 20.25375, Time (mins): 77.898\nEpoch [81/100], Step: [0/1406], \t D_Loss: 0.01581, G_Loss: 1.79758, Time (mins): 78.042\nEpoch [81/100], Step: [400/1406], \t D_Loss: 1.06213, G_Loss: 7.40349, Time (mins): 78.326\nEpoch [81/100], Step: [800/1406], \t D_Loss: 0.00001, G_Loss: 22.89689, Time (mins): 78.606\nEpoch [81/100], Step: [1200/1406], \t D_Loss: 0.00035, G_Loss: 20.36069, Time (mins): 78.879\nEpoch [82/100], Step: [0/1406], \t D_Loss: 0.40673, G_Loss: 5.81140, Time (mins): 79.021\nEpoch [82/100], Step: [400/1406], \t D_Loss: 0.07706, G_Loss: 5.79947, Time (mins): 79.300\nEpoch [82/100], Step: [800/1406], \t D_Loss: 0.25123, G_Loss: 25.24323, Time (mins): 79.585\nEpoch [82/100], Step: [1200/1406], \t D_Loss: 0.09584, G_Loss: 8.68277, Time (mins): 79.863\nEpoch [83/100], Step: [0/1406], \t D_Loss: 0.27774, G_Loss: 16.74648, Time (mins): 80.006\nEpoch [83/100], Step: [400/1406], \t D_Loss: 0.00720, G_Loss: 28.23718, Time (mins): 80.282\nEpoch [83/100], Step: [800/1406], \t D_Loss: 0.32978, G_Loss: 10.33164, Time (mins): 80.559\nEpoch [83/100], Step: [1200/1406], \t D_Loss: 10.04081, G_Loss: 10.62914, Time (mins): 80.835\nEpoch [84/100], Step: [0/1406], \t D_Loss: 0.00400, G_Loss: 5.95385, Time (mins): 80.978\nEpoch [84/100], Step: [400/1406], \t D_Loss: 0.67261, G_Loss: 11.95525, Time (mins): 81.257\nEpoch [84/100], Step: [800/1406], \t D_Loss: 0.00002, G_Loss: 19.04036, Time (mins): 81.534\nEpoch [84/100], Step: [1200/1406], \t D_Loss: 0.00685, G_Loss: 13.77893, Time (mins): 81.815\nEpoch [85/100], Step: [0/1406], \t D_Loss: 0.98654, G_Loss: 8.65013, Time (mins): 81.957\nEpoch [85/100], Step: [400/1406], \t D_Loss: 0.02424, G_Loss: 9.30856, Time (mins): 82.233\nEpoch [85/100], Step: [800/1406], \t D_Loss: 0.00004, G_Loss: 13.76115, Time (mins): 82.509\nEpoch [85/100], Step: [1200/1406], \t D_Loss: 0.32017, G_Loss: 14.01194, Time (mins): 82.786\nEpoch [86/100], Step: [0/1406], \t D_Loss: 0.00113, G_Loss: 8.28613, Time (mins): 82.929\nEpoch [86/100], Step: [400/1406], \t D_Loss: 0.02911, G_Loss: 19.34781, Time (mins): 83.207\nEpoch [86/100], Step: [800/1406], \t D_Loss: 0.00026, G_Loss: 18.79912, Time (mins): 83.484\nEpoch [86/100], Step: [1200/1406], \t D_Loss: 0.03142, G_Loss: 6.74821, Time (mins): 83.762\nEpoch [87/100], Step: [0/1406], \t D_Loss: 0.05239, G_Loss: 7.02435, Time (mins): 83.904\nEpoch [87/100], Step: [400/1406], \t D_Loss: 0.00720, G_Loss: 22.97920, Time (mins): 84.177\nEpoch [87/100], Step: [800/1406], \t D_Loss: 0.02646, G_Loss: 17.56147, Time (mins): 84.456\nEpoch [87/100], Step: [1200/1406], \t D_Loss: 0.51096, G_Loss: 10.65666, Time (mins): 84.735\nEpoch [88/100], Step: [0/1406], \t D_Loss: 0.16004, G_Loss: 8.92070, Time (mins): 84.876\nEpoch [88/100], Step: [400/1406], \t D_Loss: 3.72730, G_Loss: 2.85657, Time (mins): 85.158\nEpoch [88/100], Step: [800/1406], \t D_Loss: 0.34190, G_Loss: 7.00206, Time (mins): 85.430\nEpoch [88/100], Step: [1200/1406], \t D_Loss: 0.10335, G_Loss: 9.68768, Time (mins): 85.711\nEpoch [89/100], Step: [0/1406], \t D_Loss: 0.02333, G_Loss: 3.96704, Time (mins): 85.852\nEpoch [89/100], Step: [400/1406], \t D_Loss: 0.03063, G_Loss: 16.60397, Time (mins): 86.133\nEpoch [89/100], Step: [800/1406], \t D_Loss: 0.00001, G_Loss: 16.81374, Time (mins): 86.413\nEpoch [89/100], Step: [1200/1406], \t D_Loss: 3.68180, G_Loss: 3.01547, Time (mins): 86.691\nEpoch [90/100], Step: [0/1406], \t D_Loss: 0.97836, G_Loss: 6.46855, Time (mins): 86.834\nEpoch [90/100], Step: [400/1406], \t D_Loss: 0.00058, G_Loss: 9.47134, Time (mins): 87.108\nEpoch [90/100], Step: [800/1406], \t D_Loss: 0.00707, G_Loss: 16.09852, Time (mins): 87.387\nEpoch [90/100], Step: [1200/1406], \t D_Loss: 0.00000, G_Loss: 14.19717, Time (mins): 87.668\nEpoch [91/100], Step: [0/1406], \t D_Loss: 1.54525, G_Loss: 8.32228, Time (mins): 87.809\nEpoch [91/100], Step: [400/1406], \t D_Loss: 0.00033, G_Loss: 10.78172, Time (mins): 88.090\nEpoch [91/100], Step: [800/1406], \t D_Loss: 0.00101, G_Loss: 5.55548, Time (mins): 88.363\nEpoch [91/100], Step: [1200/1406], \t D_Loss: 0.00068, G_Loss: 18.89645, Time (mins): 88.640\nEpoch [92/100], Step: [0/1406], \t D_Loss: 0.00001, G_Loss: 14.95881, Time (mins): 88.782\nEpoch [92/100], Step: [400/1406], \t D_Loss: 0.07790, G_Loss: 18.96507, Time (mins): 89.058\nEpoch [92/100], Step: [800/1406], \t D_Loss: 0.00034, G_Loss: 23.63446, Time (mins): 89.335\nEpoch [92/100], Step: [1200/1406], \t D_Loss: 0.01090, G_Loss: 17.46540, Time (mins): 89.610\nEpoch [93/100], Step: [0/1406], \t D_Loss: 0.00230, G_Loss: 10.30249, Time (mins): 89.757\nEpoch [93/100], Step: [400/1406], \t D_Loss: 0.30676, G_Loss: 12.06446, Time (mins): 90.033\nEpoch [93/100], Step: [800/1406], \t D_Loss: 0.00154, G_Loss: 22.11076, Time (mins): 90.311\nEpoch [93/100], Step: [1200/1406], \t D_Loss: 0.38561, G_Loss: 7.50102, Time (mins): 90.587\nEpoch [94/100], Step: [0/1406], \t D_Loss: 1.52526, G_Loss: 9.03584, Time (mins): 90.730\nEpoch [94/100], Step: [400/1406], \t D_Loss: 0.00003, G_Loss: 18.66155, Time (mins): 91.008\nEpoch [94/100], Step: [800/1406], \t D_Loss: 4.03252, G_Loss: 11.56358, Time (mins): 91.284\nEpoch [94/100], Step: [1200/1406], \t D_Loss: 0.12738, G_Loss: 11.23191, Time (mins): 91.566\nEpoch [95/100], Step: [0/1406], \t D_Loss: 0.00216, G_Loss: 14.88111, Time (mins): 91.712\nEpoch [95/100], Step: [400/1406], \t D_Loss: 0.00001, G_Loss: 25.04624, Time (mins): 91.988\nEpoch [95/100], Step: [800/1406], \t D_Loss: 0.00002, G_Loss: 17.69450, Time (mins): 92.266\nEpoch [95/100], Step: [1200/1406], \t D_Loss: 0.00002, G_Loss: 20.65144, Time (mins): 92.544\nEpoch [96/100], Step: [0/1406], \t D_Loss: 0.11287, G_Loss: 7.17831, Time (mins): 92.688\nEpoch [96/100], Step: [400/1406], \t D_Loss: 1.87467, G_Loss: 6.55405, Time (mins): 92.964\nEpoch [96/100], Step: [800/1406], \t D_Loss: 0.00459, G_Loss: 14.68987, Time (mins): 93.247\nEpoch [96/100], Step: [1200/1406], \t D_Loss: 2.79662, G_Loss: 18.65228, Time (mins): 93.522\nEpoch [97/100], Step: [0/1406], \t D_Loss: 3.95189, G_Loss: 3.82688, Time (mins): 93.663\nEpoch [97/100], Step: [400/1406], \t D_Loss: 0.03929, G_Loss: 15.00077, Time (mins): 93.943\nEpoch [97/100], Step: [800/1406], \t D_Loss: 0.33522, G_Loss: 12.61992, Time (mins): 94.215\nEpoch [97/100], Step: [1200/1406], \t D_Loss: 0.00001, G_Loss: 17.03027, Time (mins): 94.494\nEpoch [98/100], Step: [0/1406], \t D_Loss: 0.00000, G_Loss: 17.04319, Time (mins): 94.637\nEpoch [98/100], Step: [400/1406], \t D_Loss: 0.00446, G_Loss: 10.39120, Time (mins): 94.913\nEpoch [98/100], Step: [800/1406], \t D_Loss: 0.00035, G_Loss: 26.22524, Time (mins): 95.188\nEpoch [98/100], Step: [1200/1406], \t D_Loss: 0.00081, G_Loss: 14.49038, Time (mins): 95.466\nEpoch [99/100], Step: [0/1406], \t D_Loss: 0.49141, G_Loss: 7.55526, Time (mins): 95.610\nEpoch [99/100], Step: [400/1406], \t D_Loss: 0.05990, G_Loss: 14.50757, Time (mins): 95.886\nEpoch [99/100], Step: [800/1406], \t D_Loss: 0.00401, G_Loss: 21.95535, Time (mins): 96.164\nEpoch [99/100], Step: [1200/1406], \t D_Loss: 0.00795, G_Loss: 14.51889, Time (mins): 96.441\nEpoch [100/100], Step: [0/1406], \t D_Loss: 0.35954, G_Loss: 8.25762, Time (mins): 96.583\nEpoch [100/100], Step: [400/1406], \t D_Loss: 0.00047, G_Loss: 15.00173, Time (mins): 96.861\nEpoch [100/100], Step: [800/1406], \t D_Loss: 0.00002, G_Loss: 8.68514, Time (mins): 97.143\nEpoch [100/100], Step: [1200/1406], \t D_Loss: 0.05498, G_Loss: 30.41126, Time (mins): 97.422\n============================================================================\nTotal Training Time (hours): 1.6261220439275106\n============================================================================\n"
],
[
"def plot_logs(fileLocation):\n \"\"\"\n Takes file location for log txt file and plots loss curve per 500 intervals\n \"\"\"\n d_loss = []\n g_loss = []\n step = []\n \n with open(fileLocation, \"r\") as f:\n myLogs = f.readlines()\n interval = 4\n count = 1\n for line in myLogs:\n if(count%interval == 0):\n d_loss.append(float(line.split(\",\")[2].strip().split(\" \")[1]))\n g_loss.append(float(line.split(\",\")[3].strip().split(\" \")[1]))\n step.append(count)\n \n count += 1\n \n \n fig, axs = plt.subplots(2, figsize=(15,10))\n fig.suptitle('Training Logs')\n axs[0].set_title(\"Discriminator Loss\")\n axs[1].set_title(\"Generator \")\n \n axs[0].plot(step, d_loss, color=\"g\",marker=\".\")\n axs[1].plot(step, g_loss, color=\"m\",marker=\".\")\n plt.show()\n \n print(d_loss[:10])\n print(g_loss[:10])\n ",
"_____no_output_____"
],
[
"plot_logs(TRAINING_LOGS_LOCATION+\"training_log.txt\")",
"_____no_output_____"
],
[
"# Load state dict from any of the checkpoints to generate new images\n\nD.load_state_dict(torch.load(MODEL_CHECKPOINT_LOCATION+'discriminator_2.pth'))\nG.load_state_dict(torch.load(MODEL_CHECKPOINT_LOCATION+'generator_2.pth'))",
"_____no_output_____"
],
[
"def generate_DCGAN_Images(D,G, num_images = 16):\n D.eval()\n G.eval()\n \n sample_size = num_images\n fixed_z = np.random.uniform(-1, 1, size=(sample_size, z_size))\n fixed_z = torch.from_numpy(fixed_z).float().to(device)\n \n result = G(fixed_z)\n print(result.size())\n\n #==========================================================\n # Save the generated image files and also return the list \n #==========================================================\n fileList = []\n imageList = []\n for i in range(result.size(0)):\n filename_start = str(time.time())\n\n image = result[i].detach().cpu().numpy()\n image = np.transpose(image, (1, 2, 0))\n image = ((image + 1)*255 / (2)).astype(np.uint8)\n image = image.reshape((32,32,3))\n\n dpi = 80\n figsize = (image.shape[1]/dpi, image.shape[0]/dpi)\n fig = plt.figure(figsize = figsize)\n ax = fig.add_axes([0,0,1,1])\n ax.axis('off')\n ax.imshow(image)\n\n output_file_name = IMAGE_OUTPUT_PATH+filename_start+\"_\"+str(i)+'.jpg'\n fig.savefig(output_file_name,dpi=dpi,transparent=True)\n fileList.append(output_file_name)\n \n plt.close(fig)\n \n imageList.append(image)\n\n return fileList, imageList\n ",
"_____no_output_____"
],
[
"fileList, result = generate_DCGAN_Images(D,G, num_images = 10)",
"torch.Size([10, 3, 32, 32])\n"
],
[
"plt.imshow(result[0])",
"_____no_output_____"
],
[
"print(fileList)",
"['output/1609689617.8505044_0.jpg', 'output/1609689617.8710802_1.jpg', 'output/1609689617.8828926_2.jpg', 'output/1609689617.8960388_3.jpg', 'output/1609689617.969398_4.jpg', 'output/1609689617.9820404_5.jpg', 'output/1609689617.9940927_6.jpg', 'output/1609689618.0063372_7.jpg', 'output/1609689618.0182002_8.jpg', 'output/1609689618.0301368_9.jpg']\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecc98e67d4bb3089397933a73ef285c246cc3a2c | 14,350 | ipynb | Jupyter Notebook | jupiter/AudioTestSampleGeneration.ipynb | hzuppur/Cotatron | bbf907d16de355adbb5f95f4306d98616a783215 | [
"BSD-3-Clause"
] | 1 | 2021-03-19T19:51:33.000Z | 2021-03-19T19:51:33.000Z | jupiter/AudioTestSampleGeneration.ipynb | hzuppur/Cotatron | bbf907d16de355adbb5f95f4306d98616a783215 | [
"BSD-3-Clause"
] | null | null | null | jupiter/AudioTestSampleGeneration.ipynb | hzuppur/Cotatron | bbf907d16de355adbb5f95f4306d98616a783215 | [
"BSD-3-Clause"
] | null | null | null | 65.825688 | 831 | 0.750105 | [
[
[
"# Generating audio sampels from model",
"_____no_output_____"
]
],
[
[
"import os\nimport gdown\nimport librosa\nimport argparse\nimport numpy as np\nimport IPython.display as ipd\nimport matplotlib.pyplot as plt\nfrom omegaconf import OmegaConf\nfrom matplotlib.colors import Normalize\nfrom collections import defaultdict \n\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport random\nfrom scipy.io.wavfile import write\n\nos.sys.path.append(\"../\")\nfrom synthesizer import Synthesizer\nfrom datasets.text import Language\nfrom melgan.generator import Generator\n\n\nhp_path = ['../config/global/default.yaml', '../config/vc/default.yaml']\n\nhp_global = OmegaConf.load(hp_path[0])\nhp_vc = OmegaConf.load(hp_path[1])\nhp = OmegaConf.merge(hp_global, hp_vc)\n\nparser = argparse.ArgumentParser()\nparser.add_argument('--config')\nhparams = parser.parse_args(['--config', hp_path])\n\ncheckpoint = torch.load('../chkpt/vc/cotatron_trained_vc/ff79670epoch=70.ckpt', map_location='cpu')\n\nmodel = Synthesizer(hparams)#.cuda()\nmodel.load_state_dict(checkpoint['state_dict'])\nmodel.eval()\nmodel.freeze()\n\nlang = Language(hp.data.lang, hp.data.text_cleaners)\n\nwith open('../datasets/metadata/estonian_metadata.txt', 'r') as f:\n metadata = f.readlines()",
"_____no_output_____"
],
[
"speaker_pairs_over_5000 = [('UT-uudised-Mari', 'EKI-ilukirjandus-Kylli'), ('UT-uudised-Albert', 'EKI-ilukirjandus-Meelis'), ('UT-uudised-Kalev', 'UT-uudised-Albert'), ('EKI-ilukirjandus-Meelis', 'UT-uudised-Mari')]\nspeaker_pairs_over_1000 = [('UT-uudised-Vesta', 'EKI-yksiklaused-Liivika'), ('EKI-yksiklaused-Kersti', 'ERR-uudised-Birgit_Itse'), ('ERR-uudised-Meelis_Kompus', 'ERR-uudised-Tarmo_Maiberg'), ('EKI-yksiklaused-Kylli', 'ERR-uudised-Vallo_Kelmsaar'), ('EKI-yksiklaused-Meelis', 'UT-uudised-Vesta')]\nspeaker_pairs_over_100 = [('44c80063991b3d8465541af15c082b89b8e1a2e3b37f0d296e25e790c49b30c768708bb1f58f810652a3d4d9cf3c04edb4b44971324a20c18c150aa1535a6032', '2546b52782d46a01500bff6bebb2527ce2a1ef0026bcfa399548cfac326f665f463d4ed6f3f10bef7131567212b9df4f41f24fc86c725b9b1425dd16ea7b6532'),('29a3279b66344d333c6ce542c44280d36128d716416c9396a0ed7cb24bcbdc7a6c23ea731443eb7d43cf2e969e162bf3db39abd6ee46851be5f923a5fc685a09', '21ef89b80bcdfd83dabc0c508af6c100e584112f2ba6fec9d4df12e9d6a793d7d11c63d936b74debb4a18206d5bb4dbf0f54a0edb696ebf3e69c7040ff24e406'), ('417b61ca0a5145ee60db908bd1d75a8499ed06d33e292e2aff6f380496ff7377e2a4630794a18c6de620aa65ddedf3ca2af40545176d831f89affae0bfca890b', 'fa7f67d93b2f3a6e685275897b5b67653df98a2880d1a8e4550274bb2420a4965e5561a92daf604000bedf67bd9958be1c8c8c1bd54322c265024890d56e51da')]\nspeaker_pairs_over_50 = [('a7c34b9164e1d6f5635846fd9e6fb261e978376e3494148c1ef8e1975a91dfbe58de1f2fecc96537475d1ded29574a21880628a15283d2a4283c3e9c15dd82d7','2e85b3674c07ef9a9d31e675f0f67b8e039a075fc6e424d8698716f14e16e649fdc5490e368729717440e649b7f74f860c82a6c3fb260fee4a89c803ff214f93'),('679437bfb82910388c0a490f96ee50d037744be0cc923793533ad62e25bb939a3230e5a889e607eea54cad0833efd2a1c9b075b7e247ed0074eb62d2dd6ea84f','2d9756ce06cc5f71e1e00febfb368d395e360f0fe2b4ae8196db8865bb2708b15e7a1d13d6636d911a22700be3a5861c9807ca5b1bd3dab74bab3f8a7c98b35f'),('682bb93d9bdb03118bfd77c33e34f320038dd83f1dad720287418320aa7c0255bf6f2781d9de539b15cf2cbe027ac3d01aea1791e62375880df8f9d34dc00524','6f8d70af77ed76d1c32cad4e55a58ae518bc04ae4c067023d2ef9a2a8ddaedc3f427ef8fb1d91a48bf068d4d9a9defe15157e8fe90c65189929d69e33e639a96')]\nspeaker_pairs_over_10 = [('aaf6d3b7f66fc4bdae0b79d5d085300049ff9daf8fa28ead6a94aad690f501b0fb685adfd1cb7b83b5c6f4c20d6b279101d274aee5e3697214e1f3b2acf832e4','f206871b24414909bbf9d48a5f8f05464ec2e253efe62fcff368fa7c9b4df02bf75453d2e1f4ca76a8894cd154d919b1757745b1ea611d773be27f4105cd7304'),('6e9719926a63b9ab51cbccdd82d4aef48c414b99d869a2260e7364bf1c037394d9e8ea836093de24b76687dc29ec77d3427907859c0065357b43430639651f47','429b98f8a1b7664c811f2b6a0c9a1892aee9e0e75b162822c4c91c733783704d3f4ea4d6a737d529fcc7cbd5d8e550fb50803e7bcd93502a4110cc56ddb65211'),('a8c589373b556b84b051459250704cd6a9f944ec056e40d7c0c3bdff9c037db7051e34f595127fa463a51e187bb31dfe1d3c70d87092dad68dee4ba17c63569f','d3ccdf976a828573eb84fb243a3b0d530c9032f980ee8a7c3c02cf8ca54d50894d573e04bc4523dcf3db0ee57a0fea315aa55562c04578207d01f8c6a33c96b3')]\nspeaker_pairs_under_10 = [('15092f32524ee8ee9ca3e356ebbfa2c6715b1ba76694e0a956d01d4633a8071715e91ac5441873409f3fd7230d91db6970eb835c3499afa2a04a6db88334e1b3','1a4845efd89d2badd7be6588c01c5d3a3b4a7011b112ea7fa39d6aca30450507585c9b77e478bf8d734ec147b2732333c4d4794e30d0b8bd2a4fd614de6c1cf0'),('3bb05133e24b5327b56e1c3e5abfa244270acce3d55246c182c3400976478a0e27b53573905efa2991a43e635254cf898c98fec140a256df0122d6ebd97573ae','420fc9fb82176ceaccb8ca54dd584195d16ef040758e8764092ca4c7e967a8b49271f307533fe9d1764dee341763bc0355678972a6555bc1280195d909978f5d'),('b51b1f340446794e314ccf7c8d7c02f42b8c302e891a49d598aa7c3b50c28a042643e77c35d6ceb2e7b1f2bcf873605bd9a2b3d4d93f14333a03beb200e4b288','c4f49a0dbe6ce71a85a94c1c1564e365cd0ba93420259b897064b4a5905815060db3b596b9c0916f598d098035d0880b682eb7fffb7c1254f7016f7e1eec62f1')]\n\ndef convert(source_input, target_audio_sample, target_id):\n text = source_input.split('|')[1]\n source_wavpath = '../data/' + source_input.split('|')[0]\n\n text_norm = torch.LongTensor(lang.text_to_sequence(text, hp.data.text_cleaners))\n text_norm = text_norm.unsqueeze(0)#.cuda()\n\n wav_source_original, sr = librosa.load(source_wavpath, sr=None, mono=True)\n wav_source_original *= (0.99 / np.max(np.abs(wav_source_original)))\n\n wav_target_sample, sr_sample = librosa.load(target_audio_sample, sr=None, mono=True)\n wav_target_sample *= (0.99 / np.max(np.abs(wav_target_sample)))\n\n assert sr == hp.audio.sampling_rate\n wav_source = torch.from_numpy(wav_source_original).view(1, 1, -1)#.cuda()\n mel_source = model.cotatron.audio2mel(wav_source)\n\n target_speaker = torch.LongTensor([hp.data.speakers.index(target_id)])#.cuda()\n\n with torch.no_grad():\n mel_s_t, alignment, residual = model.inference(text_norm, mel_source, target_speaker)\n\n melgan = Generator(80)#.cuda()\n melgan_ckpt = torch.load('melgan_libritts_g_only.ckpt', map_location='cpu')\n melgan.load_state_dict(melgan_ckpt['model_g'])\n melgan.eval()\n\n with torch.no_grad():\n audio_s_t = melgan(mel_s_t).squeeze().cpu().detach().numpy()\n \n return audio_s_t, wav_source_original, wav_target_sample\n\naudio_samples_path = \"../audioSampels/\"\n\ndef generate_audio_sampels(sampels_ids, name):\n print(f\"name: {name}\")\n print(f\"sampels_ids: {sampels_ids}\")\n \n # Create directory for the audio sampels\n current_dir = audio_samples_path + name\n os.mkdir(current_dir)\n \n sample = 1\n # Iterate over each pair\n for pair in sampels_ids:\n source_id = pair[0]\n target_id = pair[1]\n source_input = [i for i in metadata if i.split('|')[2].strip() == target_id][0]\n target_audio_sample ='../data/' + [i.split(\"|\")[0] for i in metadata if i.split('|')[2].strip() == source_id][0]\n # Convert source speaker to target speaker\n audio_s_t, wav_source_original, wav_target_sample = convert(source_input, target_audio_sample, target_id)\n \n write(f\"{current_dir}/Pair_{sample}_Converted.wav\", 22050, audio_s_t)\n write(f\"{current_dir}/Pair_{sample}_Source.wav\", 22050, wav_source_original)\n write(f\"{current_dir}/Pair_{sample}_Target.wav\", 22050, wav_target_sample)\n \n print(f\"Sample {sample} converted and writen to target dir\")\n sample += 1",
"_____no_output_____"
],
[
"generate_audio_sampels(speaker_pairs_over_5000, \"speaker_pairs_over_5000\")\ngenerate_audio_sampels(speaker_pairs_over_1000, \"speaker_pairs_over_1000\")\ngenerate_audio_sampels(speaker_pairs_over_100, \"speaker_pairs_over_100\")\ngenerate_audio_sampels(speaker_pairs_over_50, \"speaker_pairs_over_50\")\ngenerate_audio_sampels(speaker_pairs_over_10, \"speaker_pairs_over_10\")\ngenerate_audio_sampels(speaker_pairs_under_10, \"speaker_pairs_under_10\")",
"name: speaker_pairs_over_5000\nsampels_ids: [('UT-uudised-Mari', 'EKI-ilukirjandus-Kylli'), ('UT-uudised-Albert', 'EKI-ilukirjandus-Meelis'), ('UT-uudised-Kalev', 'UT-uudised-Albert'), ('EKI-ilukirjandus-Meelis', 'UT-uudised-Mari')]\nSample 1 converted and writen to target dir\nSample 2 converted and writen to target dir\nSample 3 converted and writen to target dir\nSample 4 converted and writen to target dir\nname: speaker_pairs_over_1000\nsampels_ids: [('UT-uudised-Vesta', 'EKI-yksiklaused-Liivika'), ('EKI-yksiklaused-Kersti', 'ERR-uudised-Birgit_Itse'), ('ERR-uudised-Meelis_Kompus', 'ERR-uudised-Tarmo_Maiberg'), ('EKI-yksiklaused-Kylli', 'ERR-uudised-Vallo_Kelmsaar'), ('EKI-yksiklaused-Meelis', 'UT-uudised-Vesta')]\nSample 1 converted and writen to target dir\nSample 2 converted and writen to target dir\nSample 3 converted and writen to target dir\nSample 4 converted and writen to target dir\nSample 5 converted and writen to target dir\nname: speaker_pairs_over_100\nsampels_ids: [('44c80063991b3d8465541af15c082b89b8e1a2e3b37f0d296e25e790c49b30c768708bb1f58f810652a3d4d9cf3c04edb4b44971324a20c18c150aa1535a6032', '2546b52782d46a01500bff6bebb2527ce2a1ef0026bcfa399548cfac326f665f463d4ed6f3f10bef7131567212b9df4f41f24fc86c725b9b1425dd16ea7b6532'), ('29a3279b66344d333c6ce542c44280d36128d716416c9396a0ed7cb24bcbdc7a6c23ea731443eb7d43cf2e969e162bf3db39abd6ee46851be5f923a5fc685a09', '21ef89b80bcdfd83dabc0c508af6c100e584112f2ba6fec9d4df12e9d6a793d7d11c63d936b74debb4a18206d5bb4dbf0f54a0edb696ebf3e69c7040ff24e406'), ('417b61ca0a5145ee60db908bd1d75a8499ed06d33e292e2aff6f380496ff7377e2a4630794a18c6de620aa65ddedf3ca2af40545176d831f89affae0bfca890b', 'fa7f67d93b2f3a6e685275897b5b67653df98a2880d1a8e4550274bb2420a4965e5561a92daf604000bedf67bd9958be1c8c8c1bd54322c265024890d56e51da')]\nSample 1 converted and writen to target dir\nSample 2 converted and writen to target dir\nSample 3 converted and writen to target dir\nname: speaker_pairs_over_50\nsampels_ids: [('a7c34b9164e1d6f5635846fd9e6fb261e978376e3494148c1ef8e1975a91dfbe58de1f2fecc96537475d1ded29574a21880628a15283d2a4283c3e9c15dd82d7', '2e85b3674c07ef9a9d31e675f0f67b8e039a075fc6e424d8698716f14e16e649fdc5490e368729717440e649b7f74f860c82a6c3fb260fee4a89c803ff214f93'), ('679437bfb82910388c0a490f96ee50d037744be0cc923793533ad62e25bb939a3230e5a889e607eea54cad0833efd2a1c9b075b7e247ed0074eb62d2dd6ea84f', '2d9756ce06cc5f71e1e00febfb368d395e360f0fe2b4ae8196db8865bb2708b15e7a1d13d6636d911a22700be3a5861c9807ca5b1bd3dab74bab3f8a7c98b35f'), ('682bb93d9bdb03118bfd77c33e34f320038dd83f1dad720287418320aa7c0255bf6f2781d9de539b15cf2cbe027ac3d01aea1791e62375880df8f9d34dc00524', '6f8d70af77ed76d1c32cad4e55a58ae518bc04ae4c067023d2ef9a2a8ddaedc3f427ef8fb1d91a48bf068d4d9a9defe15157e8fe90c65189929d69e33e639a96')]\nSample 1 converted and writen to target dir\nSample 2 converted and writen to target dir\nSample 3 converted and writen to target dir\nname: speaker_pairs_over_10\nsampels_ids: [('aaf6d3b7f66fc4bdae0b79d5d085300049ff9daf8fa28ead6a94aad690f501b0fb685adfd1cb7b83b5c6f4c20d6b279101d274aee5e3697214e1f3b2acf832e4', 'f206871b24414909bbf9d48a5f8f05464ec2e253efe62fcff368fa7c9b4df02bf75453d2e1f4ca76a8894cd154d919b1757745b1ea611d773be27f4105cd7304'), ('6e9719926a63b9ab51cbccdd82d4aef48c414b99d869a2260e7364bf1c037394d9e8ea836093de24b76687dc29ec77d3427907859c0065357b43430639651f47', '429b98f8a1b7664c811f2b6a0c9a1892aee9e0e75b162822c4c91c733783704d3f4ea4d6a737d529fcc7cbd5d8e550fb50803e7bcd93502a4110cc56ddb65211'), ('a8c589373b556b84b051459250704cd6a9f944ec056e40d7c0c3bdff9c037db7051e34f595127fa463a51e187bb31dfe1d3c70d87092dad68dee4ba17c63569f', 'd3ccdf976a828573eb84fb243a3b0d530c9032f980ee8a7c3c02cf8ca54d50894d573e04bc4523dcf3db0ee57a0fea315aa55562c04578207d01f8c6a33c96b3')]\nSample 1 converted and writen to target dir\n"
],
[
"speaker_counts = defaultdict(lambda: 0)\nfor i in metadata:\n speaker_counts[i.strip().split(\"|\")[2]] += 1\n \nspeaker_counts = dict(sorted(speaker_counts.items(), key=lambda item: item[1], reverse=True))",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecc9ab5e38b782755e52a3976b7a1a016b4bb657 | 19,449 | ipynb | Jupyter Notebook | tensorflow/chapter_deep-learning-computation/parameters.ipynb | RabbitBoiling/Zh-d2l-ai-LiMu-studying | f8926820d5a9b18f7c1f05a6b5f8310435431bdd | [
"MIT"
] | null | null | null | tensorflow/chapter_deep-learning-computation/parameters.ipynb | RabbitBoiling/Zh-d2l-ai-LiMu-studying | f8926820d5a9b18f7c1f05a6b5f8310435431bdd | [
"MIT"
] | null | null | null | tensorflow/chapter_deep-learning-computation/parameters.ipynb | RabbitBoiling/Zh-d2l-ai-LiMu-studying | f8926820d5a9b18f7c1f05a6b5f8310435431bdd | [
"MIT"
] | null | null | null | 23.208831 | 140 | 0.467582 | [
[
[
"# 参数管理\n\n在选择了架构并设置了超参数后,我们就进入了训练阶段。\n此时,我们的目标是找到使损失函数最小化的模型参数值。\n经过训练后,我们将需要使用这些参数来做出未来的预测。\n此外,有时我们希望提取参数,以便在其他环境中复用它们,\n将模型保存下来,以便它可以在其他软件中执行,\n或者为了获得科学的理解而进行检查。\n\n之前的介绍中,我们只依靠深度学习框架来完成训练的工作,\n而忽略了操作参数的具体细节。\n本节,我们将介绍以下内容:\n\n* 访问参数,用于调试、诊断和可视化。\n* 参数初始化。\n* 在不同模型组件间共享参数。\n\n(**我们首先看一下具有单隐藏层的多层感知机。**)\n",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\n\nnet = tf.keras.models.Sequential([\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(4, activation=tf.nn.relu),\n tf.keras.layers.Dense(1),\n])\n\nX = tf.random.uniform((2, 4))\nnet(X)",
"_____no_output_____"
]
],
[
[
"## [**参数访问**]\n\n我们从已有模型中访问参数。\n当通过`Sequential`类定义模型时,\n我们可以通过索引来访问模型的任意层。\n这就像模型是一个列表一样,每层的参数都在其属性中。\n如下所示,我们可以检查第二个全连接层的参数。\n",
"_____no_output_____"
]
],
[
[
"print(net.layers[2].weights)",
"[<tf.Variable 'dense_1/kernel:0' shape=(4, 1) dtype=float32, numpy=\narray([[ 0.89914334],\n [ 0.59173334],\n [-0.7819078 ],\n [-0.71155965]], dtype=float32)>, <tf.Variable 'dense_1/bias:0' shape=(1,) dtype=float32, numpy=array([0.], dtype=float32)>]\n"
]
],
[
[
"输出的结果告诉我们一些重要的事情:\n首先,这个全连接层包含两个参数,分别是该层的权重和偏置。\n两者都存储为单精度浮点数(float32)。\n注意,参数名称允许唯一标识每个参数,即使在包含数百个层的网络中也是如此。\n\n### [**目标参数**]\n\n注意,每个参数都表示为参数类的一个实例。\n要对参数执行任何操作,首先我们需要访问底层的数值。\n有几种方法可以做到这一点。有些比较简单,而另一些则比较通用。\n下面的代码从第二个全连接层(即第三个神经网络层)提取偏置,\n提取后返回的是一个参数类实例,并进一步访问该参数的值。\n",
"_____no_output_____"
]
],
[
[
"print(type(net.layers[2].weights[1]))\nprint(net.layers[2].weights[1])\nprint(tf.convert_to_tensor(net.layers[2].weights[1]))",
"<class 'tensorflow.python.ops.resource_variable_ops.ResourceVariable'>\n<tf.Variable 'dense_1/bias:0' shape=(1,) dtype=float32, numpy=array([0.], dtype=float32)>\ntf.Tensor([0.], shape=(1,), dtype=float32)\n"
]
],
[
[
"### [**一次性访问所有参数**]\n\n当我们需要对所有参数执行操作时,逐个访问它们可能会很麻烦。\n当我们处理更复杂的块(例如,嵌套块)时,情况可能会变得特别复杂,\n因为我们需要递归整个树来提取每个子块的参数。\n下面,我们将通过演示来比较访问第一个全连接层的参数和访问所有层。\n",
"_____no_output_____"
]
],
[
[
"print(net.layers[1].weights)\nprint(net.get_weights())",
"[<tf.Variable 'dense/kernel:0' shape=(4, 4) dtype=float32, numpy=\narray([[ 0.14004463, -0.26651722, 0.47146338, -0.4279218 ],\n [ 0.5441615 , 0.46101612, -0.0691905 , 0.21423548],\n [-0.16048867, 0.4943959 , 0.42357963, -0.19573313],\n [-0.70757675, 0.4728045 , 0.82884806, 0.857788 ]],\n dtype=float32)>, <tf.Variable 'dense/bias:0' shape=(4,) dtype=float32, numpy=array([0., 0., 0., 0.], dtype=float32)>]\n[array([[ 0.14004463, -0.26651722, 0.47146338, -0.4279218 ],\n [ 0.5441615 , 0.46101612, -0.0691905 , 0.21423548],\n [-0.16048867, 0.4943959 , 0.42357963, -0.19573313],\n [-0.70757675, 0.4728045 , 0.82884806, 0.857788 ]],\n dtype=float32), array([0., 0., 0., 0.], dtype=float32), array([[ 0.89914334],\n [ 0.59173334],\n [-0.7819078 ],\n [-0.71155965]], dtype=float32), array([0.], dtype=float32)]\n"
]
],
[
[
"这为我们提供了另一种访问网络参数的方式,如下所示。\n",
"_____no_output_____"
]
],
[
[
"net.get_weights()[1]",
"_____no_output_____"
]
],
[
[
"### [**从嵌套块收集参数**]\n\n让我们看看,如果我们将多个块相互嵌套,参数命名约定是如何工作的。\n我们首先定义一个生成块的函数(可以说是“块工厂”),然后将这些块组合到更大的块中。\n",
"_____no_output_____"
]
],
[
[
"def block1(name):\n return tf.keras.Sequential([\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(4, activation=tf.nn.relu)],\n name=name)\n\ndef block2():\n net = tf.keras.Sequential()\n for i in range(4):\n # 在这里嵌套\n net.add(block1(name=f'block-{i}'))\n return net\n\nrgnet = tf.keras.Sequential()\nrgnet.add(block2())\nrgnet.add(tf.keras.layers.Dense(1))\nrgnet(X)",
"_____no_output_____"
]
],
[
[
"[**设计了网络后,我们看看它是如何工作的。**]\n",
"_____no_output_____"
]
],
[
[
"print(rgnet.summary())",
"Model: \"sequential_1\"\n"
]
],
[
[
"因为层是分层嵌套的,所以我们也可以像通过嵌套列表索引一样访问它们。\n下面,我们访问第一个主要的块中、第二个子块的第一层的偏置项。\n",
"_____no_output_____"
]
],
[
[
"rgnet.layers[0].layers[1].layers[1].weights[1]",
"_____no_output_____"
]
],
[
[
"## 参数初始化\n\n知道了如何访问参数后,现在我们看看如何正确地初始化参数。\n我们在 :numref:`sec_numerical_stability`中讨论了良好初始化的必要性。\n深度学习框架提供默认随机初始化,\n也允许我们创建自定义初始化方法,\n满足我们通过其他规则实现初始化权重。\n",
"_____no_output_____"
],
[
"默认情况下,Keras会根据一个范围均匀地初始化权重矩阵,\n这个范围是根据输入和输出维度计算出的。\n偏置参数设置为0。\nTensorFlow在根模块和`keras.initializers`模块中提供了各种初始化方法。\n",
"_____no_output_____"
],
[
"### [**内置初始化**]\n\n让我们首先调用内置的初始化器。\n下面的代码将所有权重参数初始化为标准差为0.01的高斯随机变量,\n且将偏置参数设置为0。\n",
"_____no_output_____"
]
],
[
[
"net = tf.keras.models.Sequential([\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(\n 4, activation=tf.nn.relu,\n kernel_initializer=tf.random_normal_initializer(mean=0, stddev=0.01),\n bias_initializer=tf.zeros_initializer()),\n tf.keras.layers.Dense(1)])\n\nnet(X)\nnet.weights[0], net.weights[1]",
"_____no_output_____"
]
],
[
[
"我们还可以将所有参数初始化为给定的常数,比如初始化为1。\n",
"_____no_output_____"
]
],
[
[
"net = tf.keras.models.Sequential([\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(\n 4, activation=tf.nn.relu,\n kernel_initializer=tf.keras.initializers.Constant(1),\n bias_initializer=tf.zeros_initializer()),\n tf.keras.layers.Dense(1),\n])\n\nnet(X)\nnet.weights[0], net.weights[1]",
"_____no_output_____"
]
],
[
[
"我们还可以[**对某些块应用不同的初始化方法**]。\n例如,下面我们使用Xavier初始化方法初始化第一个神经网络层,\n然后将第三个神经网络层初始化为常量值42。\n",
"_____no_output_____"
]
],
[
[
"net = tf.keras.models.Sequential([\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(\n 4,\n activation=tf.nn.relu,\n kernel_initializer=tf.keras.initializers.GlorotUniform()),\n tf.keras.layers.Dense(\n 1, kernel_initializer=tf.keras.initializers.Constant(1)),\n])\n\nnet(X)\nprint(net.layers[1].weights[0])\nprint(net.layers[2].weights[0])",
"<tf.Variable 'dense_11/kernel:0' shape=(4, 4) dtype=float32, numpy=\narray([[ 0.07325727, -0.04950851, 0.11116493, -0.0877195 ],\n [ 0.38882798, 0.8527898 , 0.56546885, -0.53123504],\n [-0.4271114 , -0.00229436, -0.8124555 , -0.7082046 ],\n [ 0.7157926 , -0.49555257, -0.3923587 , 0.55925137]],\n dtype=float32)>\n<tf.Variable 'dense_12/kernel:0' shape=(4, 1) dtype=float32, numpy=\narray([[1.],\n [1.],\n [1.],\n [1.]], dtype=float32)>\n"
]
],
[
[
"### [**自定义初始化**]\n\n有时,深度学习框架没有提供我们需要的初始化方法。\n在下面的例子中,我们使用以下的分布为任意权重参数$w$定义初始化方法:\n\n$$\n\\begin{aligned}\n w \\sim \\begin{cases}\n U(5, 10) & \\text{ 可能性 } \\frac{1}{4} \\\\\n 0 & \\text{ 可能性 } \\frac{1}{2} \\\\\n U(-10, -5) & \\text{ 可能性 } \\frac{1}{4}\n \\end{cases}\n\\end{aligned}\n$$\n",
"_____no_output_____"
],
[
"在这里,我们定义了一个`Initializer`的子类,\n并实现了`__call__`函数。\n该函数返回给定形状和数据类型的所需张量。\n",
"_____no_output_____"
]
],
[
[
"class MyInit(tf.keras.initializers.Initializer):\n def __call__(self, shape, dtype=None):\n data=tf.random.uniform(shape, -10, 10, dtype=dtype)\n factor=(tf.abs(data) >= 5)\n factor=tf.cast(factor, tf.float32)\n return data * factor\n\nnet = tf.keras.models.Sequential([\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(\n 4,\n activation=tf.nn.relu,\n kernel_initializer=MyInit()),\n tf.keras.layers.Dense(1),\n])\n\nnet(X)\nprint(net.layers[1].weights[0])",
"<tf.Variable 'dense_13/kernel:0' shape=(4, 4) dtype=float32, numpy=\narray([[ 0. , 0. , 0. , 7.686863 ],\n [ 0. , -0. , 0. , 0. ],\n [ 0. , -8.358488 , -0. , 5.5009365],\n [-0. , -0. , -0. , -5.2141643]], dtype=float32)>\n"
]
],
[
[
"注意,我们始终可以直接设置参数。\n",
"_____no_output_____"
]
],
[
[
"net.layers[1].weights[0][:].assign(net.layers[1].weights[0] + 1)\nnet.layers[1].weights[0][0, 0].assign(42)\nnet.layers[1].weights[0]",
"_____no_output_____"
]
],
[
[
"## [**参数绑定**]\n\n有时我们希望在多个层间共享参数:\n我们可以定义一个稠密层,然后使用它的参数来设置另一个层的参数。\n",
"_____no_output_____"
]
],
[
[
"# tf.keras的表现有点不同。它会自动删除重复层\nshared = tf.keras.layers.Dense(4, activation=tf.nn.relu)\nnet = tf.keras.models.Sequential([\n tf.keras.layers.Flatten(),\n shared,\n shared,\n tf.keras.layers.Dense(1),\n])\n\nnet(X)\n# 检查参数是否不同\nprint(len(net.layers) == 3)",
"True\n"
]
],
[
[
"## 小结\n\n* 我们有几种方法可以访问、初始化和绑定模型参数。\n* 我们可以使用自定义初始化方法。\n\n## 练习\n\n1. 使用 :numref:`sec_model_construction` 中定义的`FancyMLP`模型,访问各个层的参数。\n1. 查看初始化模块文档以了解不同的初始化方法。\n1. 构建包含共享参数层的多层感知机并对其进行训练。在训练过程中,观察模型各层的参数和梯度。\n1. 为什么共享参数是个好主意?\n",
"_____no_output_____"
],
[
"[Discussions](https://discuss.d2l.ai/t/1830)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecc9b6227776010644a14ec0f5fa19ac104d0d96 | 434,841 | ipynb | Jupyter Notebook | homeproject.ipynb | mvcaro/ev_mode_classifier | 5488cc19354af27c4df35dd393e3c67c80a2cdf7 | [
"MIT"
] | null | null | null | homeproject.ipynb | mvcaro/ev_mode_classifier | 5488cc19354af27c4df35dd393e3c67c80a2cdf7 | [
"MIT"
] | null | null | null | homeproject.ipynb | mvcaro/ev_mode_classifier | 5488cc19354af27c4df35dd393e3c67c80a2cdf7 | [
"MIT"
] | null | null | null | 132.735348 | 82,008 | 0.816087 | [
[
[
"## Project planning\n\nGlancing at the problem description, this excercise is a binary classification problem - as what is needed is to predict if the vehicle driving mode is either electric or internal combustion powered\n\n- Exploratory analysis:\n - null data present \n - .describe(): max, mean, median, etc to have an overview of the data\n - Barplots, see distribution\n - Boxplots to identify magnitude differences and outlier\n\n- Create train and test set in 80:20 split. Using train set:\n - Variable correlation\n\n- Try different models and metrics\n\n\nsee (*) for \"if I had more time, I would...\" comment through the notebook\n\n",
"_____no_output_____"
]
],
[
[
"import os \nimport glob\nimport random\nimport seaborn as sns\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom datetime import datetime\nfrom pandas.plotting import scatter_matrix\nfrom zlib import crc32",
"_____no_output_____"
],
[
"# start random seed for results reproducibility\nrandom.seed(42)",
"_____no_output_____"
],
[
"# combine all csv files\n# Understand variables i.e. upper and lower bounds\nos.chdir('ds_recruitment_data')\nextension = 'csv'\nall_files = [i for i in glob.glob('*.{}'.format(extension))]\n\nall_vins = pd.concat([pd.read_csv(i) for i in all_files])\nall_vins.to_csv('all_vins.csv', index=False, encoding = 'utf-8-sig')\n#encoding = ‘utf-8-sig’ is added to overcome the issue when exporting ‘Non-English’ languages.",
"_____no_output_____"
],
[
"#all_vins\nall_vins_path = 'all_vins.csv'\ndf_vins = pd.read_csv(all_vins_path)\ndf_vins",
"_____no_output_____"
]
],
[
[
"#### translate driving mode from categorical to continuous vars ",
"_____no_output_____"
]
],
[
[
"df_vins['ev'] = df_vins['mode'].apply(lambda x: 1 if x == 'ev' else 0) # only for ev as this is our variable of interest\ndf_vins",
"_____no_output_____"
]
],
[
[
"## Exploratory analysis",
"_____no_output_____"
],
[
"#### view null data ",
"_____no_output_____"
]
],
[
[
"# understand null vals present\ndf_vins.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 242814 entries, 0 to 242813\nData columns (total 26 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 device_id 242814 non-null int64 \n 1 event_time 242814 non-null float64\n 2 event_received_time 242814 non-null int64 \n 3 ignition_status 242814 non-null bool \n 4 latitude 242814 non-null float64\n 5 longitude 242814 non-null float64\n 6 vin 242814 non-null object \n 7 deep_sleep 242814 non-null int64 \n 8 movement_sensor 242814 non-null int64 \n 9 gsm_signal_level 242814 non-null int64 \n 10 hybrid_batt_soc 242660 non-null float64\n 11 vehicle_speed 242660 non-null float64\n 12 fuel_Air_ratio 240834 non-null float64\n 13 hybrid_batt_current 240834 non-null float64\n 14 gps_speed 242814 non-null int64 \n 15 manifold_abs_pressure 240834 non-null float64\n 16 accel_pedal_position 240834 non-null float64\n 17 manifold_air_flow_rate 242660 non-null float64\n 18 gps_altitude 242814 non-null int64 \n 19 hybrid_batt_voltage 240834 non-null float64\n 20 gps_satellites 242814 non-null int64 \n 21 engine_speed 242660 non-null float64\n 22 adc_external_power_voltage 242814 non-null int64 \n 23 odometer 240834 non-null float64\n 24 mode 242814 non-null object \n 25 ev 242814 non-null int64 \ndtypes: bool(1), float64(13), int64(10), object(2)\nmemory usage: 46.5+ MB\n"
],
[
"# see how many nulls per column with nulls \nabove_0_missing = all_vins.isnull().sum() > 0\n\nall_vins.isnull().sum()[above_0_missing]",
"_____no_output_____"
]
],
[
[
"#### Understanding continuous variables:",
"_____no_output_____"
]
],
[
[
"df_desc = df_vins.describe()\ndf_desc",
"_____no_output_____"
]
],
[
[
"From df_dec (above) we can infer:\n\n##### questions: (*)\n- What is the difference between vehicle speed and gps speed?\n- why is ignition status = True when driving mode = ev?\n\n\n##### Dividing the variables into 4 categories:\n- continuous variables or vars_cont\n- categorical varibles or vars_cat\n- Variables that can be ignored as they are not representative, or vars_ignore\n- Variables that I am unsure that would have any impact, would need questioning for those who collected the dataset or PM; labelled as vars_unsure\n\n",
"_____no_output_____"
]
],
[
[
"#org\nvars_cont = ['hybrid_batt_soc', 'vehicle_speed', 'fuel_Air_ratio', 'hybrid_batt_current', 'accel_pedal_position', \n 'hybrid_batt_voltage', 'engine_speed', 'adc_external_power_voltage', 'manifold_air_flow_rate' ]\n\n# vars_cont = ['hybrid_batt_soc', 'hybrid_batt_current', 'hybrid_batt_voltage', 'engine_speed' ]\n\nvars_cat = ['ev']\nvars_ignore = ['device_id', 'event_time', 'event_received_time', 'ignition_status','latitude','longitude', 'vin','deep_sleep', \n 'movement_sensor', 'manifold_abs_pressure', 'odometer', 'gps_speed']\nvars_unsure = ['gsm_signal_level', 'gps_altitude', 'gps_satellites']\n\nvars_training = vars_cont + vars_cat\n\n# len(vars_cont) + len(vars_cat) + len(vars_ignore) + len(vars_unsure) # to ensure all cols accounted for ",
"_____no_output_____"
]
],
[
[
"#### Understanding data distribution ",
"_____no_output_____"
]
],
[
[
"df_vins[vars_cont].hist(figsize=(16,16))\n#plt.savefig('histograms.png')",
"_____no_output_____"
]
],
[
[
"From the histograms we can see that \nVariables that follow a fairly normal distribution:\n- hybrid_batt_soc\n- hybrid_batt_current\n- hybrid_batt_voltage\n\nVariables that need normalisation:\n\nskewed data:\n- fuel_Air_ratio \n- manifold_air_flow_rate\n- vehicle_speed\n- adc_external_power_voltage \n\nand\n- accel_pedal_position\n- engine_speed\n- odometer\n- gps_speed",
"_____no_output_____"
]
],
[
[
"# categorical data, only ev\n#sns.barplot(df_vins[vars_cat].value_counts().index)\n\nfor i in df_vins[vars_cat]:\n sns.barplot(df_vins[i].value_counts().index,df_vins[i].value_counts()).set_title(i)\n plt.show()\n",
"C:\\Users\\mvasque1\\Anaconda3\\envs\\dsenv\\lib\\site-packages\\seaborn\\_decorators.py:36: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n warnings.warn(\n"
],
[
"#df_vins.columns\n\n# scatter_matrix(df_vins[vars_cont],figsize=(12,8))",
"_____no_output_____"
]
],
[
[
"#### understanding value ranges & outliers\noverview of all ranges, show the different scales",
"_____no_output_____"
]
],
[
[
"df_vins[vars_cont].boxplot()\nplt.xticks(rotation=90)",
"_____no_output_____"
]
],
[
[
"and to investigate into more detail:",
"_____no_output_____"
]
],
[
[
"# add boxplots - as not normalised yet, plot in groups of similar units \n# from prev plot, show bar plots \n# for vals between 0 - 100\n\ndf_vins.boxplot(column = ['manifold_air_flow_rate', 'hybrid_batt_soc', \n 'vehicle_speed', 'accel_pedal_position'])\nplt.xticks(rotation=45)\n#plt.savefig('boxplot_g1'+ '.png', bbox_inches='tight')\nplt.show()\n\ndf_vins.boxplot(column = ['odometer', 'engine_speed'])\n#plt.savefig('boxplot_g2'+ '.png')\nplt.show() \n\ndf_vins.boxplot(column = ['adc_external_power_voltage'])\n#plt.savefig('boxplot_g3'+ '.png')\nplt.show() \n\ndf_vins.boxplot(column = ['hybrid_batt_current'])\n#plt.savefig('boxplot_g4'+ '.png')\nplt.show() \n\ndf_vins.boxplot(column = ['hybrid_batt_voltage'])\n#plt.savefig('boxplot_g5'+ '.png')\nplt.show() \n\ndf_vins.boxplot(column = ['fuel_Air_ratio'])\n#plt.savefig('boxplot_g6'+ '.png')\nplt.show() ",
"_____no_output_____"
]
],
[
[
"#### see geographical data",
"_____no_output_____"
]
],
[
[
"df_vins.plot(kind='scatter', x='longitude', y ='latitude', alpha=0.1)",
"_____no_output_____"
],
[
"import plotly.express as px\n\n#uncomment to see-\n# fig = px.scatter_geo(df_vins,lat='latitude',lon='longitude')\n# fig.update_layout(title = 'World map', title_x=0.5)\n# fig.show()",
"_____no_output_____"
]
],
[
[
"(*) If I had more time:\n- as part of data exploration, could see more accurately where the lats and lons are in the map\n- can this help us understand any seasonal behaviour? (i.e. electric vehicle mode not turning on properly during winter)\n- Coult data help us see behavioural use of ev vs ic per country? that can help inform business decisions for specific market areas",
"_____no_output_____"
],
[
"### To avoid \"data snooping bias\", create train and test sets\nNo further data exploration below this point with all the dataset, so only use train_set.\n\nTo ensure a stable train/test split even when the dataset is refreshed, we need to ensure the train and test data is not mixed, which can be achieved by computing the hash of each instance",
"_____no_output_____"
]
],
[
[
"def test_set_check(identifier, test_ratio):\n return crc32(np.int64(identifier)) & 0xffffffff < test_ratio * 2**32\n\ndef split_train_test_by_id(data, test_ratio, id_column):\n ids = data[id_column]\n in_test_set = ids.apply(lambda id_: test_set_check(id_, test_ratio))\n return data.loc[~in_test_set], data.loc[in_test_set]\n\ntrain_set, test_set = split_train_test_by_id(df_vins, 0.2, 'device_id')",
"_____no_output_____"
]
],
[
[
"#### Understand variable correlation",
"_____no_output_____"
]
],
[
[
"corr = train_set[vars_cont].corr()\ncorr",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(10,10)) \ndataplot = sns.heatmap(corr, cmap=\"YlGnBu\", annot=True)",
"_____no_output_____"
]
],
[
[
"#### Compare to dependant variable: if driving in ev mode ",
"_____no_output_____"
]
],
[
[
"# compare driving mode to other variables\n#temp_train_wout_ev = train_set.drop(['ev'],axis=1)\npd.pivot_table(train_set, index='ev', values = vars_cont)",
"_____no_output_____"
]
],
[
[
"#### data engineering results",
"_____no_output_____"
]
],
[
[
"#only use the features chosen\ntrain_set = train_set[vars_training]\ntrain_set",
"_____no_output_____"
]
],
[
[
"#### Identifying with missing data",
"_____no_output_____"
]
],
[
[
"above_0_missing = train_set.isnull().sum() > 0\n\ntrain_set.isnull().sum()[above_0_missing]",
"_____no_output_____"
]
],
[
[
"use SimpleImputer which uses the mean, and MinMaxScaler for feature scaling?\nMissing values will be filled with the median. The mean is not chosen as scatter graphs showed a lot of outliers ",
"_____no_output_____"
]
],
[
[
"from sklearn.impute import SimpleImputer\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import StandardScaler\n",
"_____no_output_____"
]
],
[
[
"below works",
"_____no_output_____"
]
],
[
[
"\n# vars_with_nans = ['hybrid_batt_soc', 'vehicle_speed','fuel_Air_ratio', 'hybrid_batt_current', \n# 'accel_pedal_position', 'manifold_air_flow_rate', 'hybrid_batt_voltage', 'engine_speed', 'odometer']\n\n# var_cont_pipeline = Pipeline(steps=[\n# ('impute', SimpleImputer(strategy='median')),\n# ('scale', MinMaxScaler())\n# ])\n\n\n# train_set_scaled = train_set.copy()\n# train_set_scaled[vars_cont] = var_cont_pipeline.fit_transform(train_set_scaled[vars_cont])\n# train_set_scaled",
"_____no_output_____"
]
],
[
[
"trying other stuff\n",
"_____no_output_____"
],
[
"#### Fill NaNs with median\n",
"_____no_output_____"
]
],
[
[
"imputer = SimpleImputer(missing_values=np.nan, strategy='median')\nimputer.fit_transform(train_set)\ntrain_set_filled = train_set.copy()\ntrain_set_filled[vars_cont] = imputer.fit_transform(train_set_filled[vars_cont])",
"_____no_output_____"
],
[
"train_set_filled",
"_____no_output_____"
]
],
[
[
"#### scale inputs",
"_____no_output_____"
]
],
[
[
"from sklearn.pipeline import Pipeline, make_pipeline\nfrom sklearn.preprocessing import PowerTransformer\nfrom sklearn.compose import ColumnTransformer\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.metrics import roc_auc_score\n\n\n# input arrays\nX_train, y_train = train_set_filled.drop('ev', axis=1), train_set_filled['ev']\n\n\ntest_set = test_set.dropna()\nX_test = test_set[vars_cont]\ny_test = test_set['ev']\n\nsc = StandardScaler()\n\nX_train = sc.fit_transform(X_train)\nX_test = sc.transform(X_test)\n",
"_____no_output_____"
],
[
"X_train_df = pd.DataFrame(X_train,columns=vars_cont)\nX_train_df.hist(figsize=(16,16))",
"_____no_output_____"
]
],
[
[
"(*) look more into data distributions\n\n\nskewed data:\n- fuel_Air_ratio \n- hybrid_batt_voltage\n- adc_external_power_voltage \n- manifold_air_flow_rate\n\n\nand\n- vehicle_speed\n- accel_pedal_position\n- engine_speed\n- odometer\n- gps_speed",
"_____no_output_____"
],
[
"## Model creation",
"_____no_output_____"
]
],
[
[
"from sklearn.naive_bayes import GaussianNB\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn import tree\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.svm import SVC\nfrom sklearn import svm\nfrom sklearn.metrics import confusion_matrix, classification_report\nfrom xgboost import XGBClassifier\n",
"_____no_output_____"
]
],
[
[
"###### LogisticRegression",
"_____no_output_____"
]
],
[
[
"lr = LogisticRegression(max_iter = 2000)\ncross_val = cross_val_score(lr, X_train,y_train,cv=5)\nprint(f'mean: {cross_val.mean()} | cross val: {cross_val} ')\n",
"mean: 0.9998944702636026 | cross val: [0.99988008 0.99990407 0.99988008 0.99990406 0.99990406] \n"
]
],
[
[
"##### Naive Bayes",
"_____no_output_____"
]
],
[
[
"nb = GaussianNB()\ncross_val = cross_val_score(nb, X_train, y_train, cv=5)\nprint(f'mean: {cross_val.mean()} | cross val: {cross_val} ')\n",
"mean: 0.9916631495590913 | cross val: [0.99155774 0.99165368 0.99196527 0.99138965 0.99174941] \n"
]
],
[
[
"##### Randomforrest classifier\n",
"_____no_output_____"
]
],
[
[
"rf = RandomForestClassifier(random_state = 1)\ncross_val = cross_val_score(rf, X_train, y_train,cv=5)\nprint(f'mean: {cross_val.mean()} | cross val: {cross_val} ')\n# rf.fit(X_train, y_train)\n\n# predict_rfc = rf.predict(X_test)",
"mean: 1.0 | cross val: [1. 1. 1. 1. 1.] \n"
]
],
[
[
"##### Decision tree",
"_____no_output_____"
]
],
[
[
"dtc = tree.DecisionTreeClassifier(random_state = 1)\ncross_val = cross_val_score(dtc, X_train,y_train,cv=5)\nprint(f'mean: {cross_val.mean()} | cross val: {cross_val} ')",
"mean: 1.0 | cross val: [1. 1. 1. 1. 1.] \n"
]
],
[
[
"###### XGBoost",
"_____no_output_____"
]
],
[
[
"xgb = XGBClassifier(random_state =1)\ncross_val = cross_val_score(xgb, X_train,y_train,cv=5)\nprint(f'mean: {cross_val.mean()} | cross val: {cross_val} ')",
"[10:24:26] WARNING: D:\\bld\\xgboost-split_1637426510059\\work\\src\\learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.\n"
]
],
[
[
"##### fit",
"_____no_output_____"
]
],
[
[
"lr.fit(X_train, y_train)\nnb.fit(X_train, y_train)\nrf.fit(X_train, y_train)\ndtc.fit(X_train, y_train)\nxgb.fit(X_train, y_train)",
"C:\\Users\\mvasque1\\Anaconda3\\envs\\dsenv\\lib\\site-packages\\xgboost\\sklearn.py:1224: UserWarning: The use of label encoder in XGBClassifier is deprecated and will be removed in a future release. To remove this warning, do the following: 1) Pass option use_label_encoder=False when constructing XGBClassifier object; and 2) Encode your labels (y) as integers starting with 0, i.e. 0, 1, 2, ..., [num_class - 1].\n warnings.warn(label_encoder_deprecation_msg, UserWarning)\n"
]
],
[
[
"##### predict",
"_____no_output_____"
]
],
[
[
"pred_lr = lr.predict(X_test)\npred_nb = nb.predict(X_test)\npred_rf = rf.predict(X_test)\npred_dtc = dtc.predict(X_test)\npred_xgb = xgb.predict(X_test)",
"_____no_output_____"
]
],
[
[
"#### perf",
"_____no_output_____"
]
],
[
[
"print('LogisticRegression\\n', confusion_matrix(y_test, pred_lr))\nprint('\\nBayes\\n', confusion_matrix(y_test, pred_nb))\nprint('\\nRandom Forrest\\n', confusion_matrix(y_test, pred_rf))\nprint('\\nDec trees\\n', confusion_matrix(y_test, pred_dtc))\nprint('\\nxgboost\\n', confusion_matrix(y_test, pred_xgb))\n\n",
"LogisticRegression\n [[21274 0]\n [ 0 13046]]\n\nBayes\n [[21274 0]\n [ 286 12760]]\n\nRandom Forrest\n [[21274 0]\n [ 0 13046]]\n\nDec trees\n [[21274 0]\n [ 0 13046]]\n\nxgboost\n [[21274 0]\n [ 0 13046]]\n"
],
[
"# roc ",
"_____no_output_____"
],
[
"from sklearn.model_selection import cross_val_predict\nfrom sklearn.metrics import roc_curve\n\n\ny_scores = cross_val_predict(lr, X_train, y_train, cv=3,\n method=\"decision_function\")\n\nfpr, tpr, thresholds = roc_curve(y_train, y_scores)\n\n\n\ndef plot_roc_curve(fpr, tpr, label=None):\n plt.plot(fpr, tpr, linewidth=2, label=label)\n plt.plot([0, 1], [0, 1], 'k--') # dashed diagonal \n #plt.axis([0, 1, 0, 1]) # Not shown in the book\n plt.xlabel('False Positive Rate (Fall-Out)', fontsize=16) # Not shown\n plt.ylabel('True Positive Rate (Recall)', fontsize=16) # Not shown\n plt.grid(True) # Not shown\n\nplt.figure(figsize=(8, 6)) # Not shown\nplot_roc_curve(fpr, tpr)\nplt.plot([4.837e-3, 4.837e-3], [0., 0.4368], \"r:\") # Not shown\nplt.plot([0.0, 4.837e-3], [0.4368, 0.4368], \"r:\") # Not shown\nplt.plot([4.837e-3], [0.4368], \"ro\") # Not shown\n#save_fig(\"roc_curve_plot\") # Not shown\nplt.show()",
"_____no_output_____"
]
],
[
[
"(*)\nAll models show no false negatives or false positives\nTrue negatives and true positives are correctly classified\n\nThe results, although promising, I am skepticall of the great performance. If I had more time, I would triple check I am not mixing training and test data and would add more performance metrics to ensure good model performance.\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecc9d2faec3f23c1a0a8fb1f86bfea4cca1cf01f | 1,338 | ipynb | Jupyter Notebook | test.ipynb | katierickelton/genomics_course | 21046ac7e9982c35dde8b684f295a678f058594d | [
"Apache-2.0"
] | null | null | null | test.ipynb | katierickelton/genomics_course | 21046ac7e9982c35dde8b684f295a678f058594d | [
"Apache-2.0"
] | null | null | null | test.ipynb | katierickelton/genomics_course | 21046ac7e9982c35dde8b684f295a678f058594d | [
"Apache-2.0"
] | null | null | null | 18.328767 | 94 | 0.518685 | [
[
[
"This is a test file for my GitHub repository.",
"_____no_output_____"
]
],
[
[
"2+2",
"_____no_output_____"
],
[
"print(\"My GitHub repository is called genomics_course on my account, katierickelton.\")",
"My GitHub repository is called genomics_course on my account, katierickelton.\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
]
] |
ecc9dd462d421a2b776f6b24b8e454f0c6960408 | 138,712 | ipynb | Jupyter Notebook | ML_Models/Regression/Simple_Regression/simple_regression_2.ipynb | lakshit2808/Machine-Learning-Notes | 1b7760c2626c36a7f62c5a474e9fdadb76cb023b | [
"MIT"
] | 2 | 2021-09-04T17:13:48.000Z | 2021-09-04T17:13:50.000Z | ML_Models/Regression/Simple_Regression/simple_regression_2.ipynb | lakshit2808/Machine-Learning-Notes | 1b7760c2626c36a7f62c5a474e9fdadb76cb023b | [
"MIT"
] | null | null | null | ML_Models/Regression/Simple_Regression/simple_regression_2.ipynb | lakshit2808/Machine-Learning-Notes | 1b7760c2626c36a7f62c5a474e9fdadb76cb023b | [
"MIT"
] | 1 | 2021-11-23T19:45:02.000Z | 2021-11-23T19:45:02.000Z | 166.521008 | 24,778 | 0.846185 | [
[
[
"<a href=\"https://colab.research.google.com/github/lakshit2808/Machine-Learning-Notes/blob/master/ML_Models/Regression/Simple_Regression/simple_regression_2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Simple Regression\n\n\n<img src='https://github.com/lakshit2808/Machine-Learning-Notes/blob/master/Resources/Images/LinearReg.jpg?raw=true' height = '800' width='400'>\n\n## 1. Importing Packages\n\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## 2. Reading Data",
"_____no_output_____"
]
],
[
[
"\ndf = pd.read_csv('/FuelConsumptionCo2.csv')\ndf.head()",
"_____no_output_____"
]
],
[
[
"## 3. Data Explortion",
"_____no_output_____"
]
],
[
[
"df.describe()",
"_____no_output_____"
],
[
"mdf = df[['ENGINESIZE' ,'CYLINDERS' , 'FUELCONSUMPTION_COMB', 'CO2EMISSIONS']]\nmdf.head(6)",
"_____no_output_____"
]
],
[
[
"## 4. Data Visualizatoin",
"_____no_output_____"
]
],
[
[
"mdf.hist()\nplt.show()",
"_____no_output_____"
],
[
"# ENGINESIZE vs CO2EMISSIONS\n\nplt.scatter(mdf.ENGINESIZE , mdf.CO2EMISSIONS , color='green')\nplt.title('ENGINESIZE vs CO2EMISSIONS')\nplt.xlabel('ENGINESIZE')\nplt.ylabel('CO2EMISSIONS ')\nplt.show()",
"_____no_output_____"
],
[
"# FUELCONSUMPTION_COMB vs CO2EMISSIONS\nplt.scatter(mdf.FUELCONSUMPTION_COMB , mdf.CO2EMISSIONS , color='red')\nplt.title('FUELCONSUMPTION_COMB vs CO2EMISSIONS')\nplt.xlabel('FUELCONSUMPTION_COMB')\nplt.ylabel('CO2EMISSIONS ')\nplt.show()",
"_____no_output_____"
],
[
"# CYLINDERS vs CO2EMISSIONS\nplt.scatter(mdf.CYLINDERS , mdf.CO2EMISSIONS , color='blue')\nplt.title('CYLINDERS vs CO2EMISSIONS')\nplt.xlabel('CYLINDERS')\nplt.ylabel('CO2EMISSIONS ')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 5. Train/Test Split",
"_____no_output_____"
]
],
[
[
"msk = np.random.rand(len(df)) < 0.8\ntrain = mdf[msk]\ntest = mdf[msk]\n",
"_____no_output_____"
]
],
[
[
"## 6. Simple Regression Model",
"_____no_output_____"
]
],
[
[
"plt.scatter(train.ENGINESIZE , train.CO2EMISSIONS , color='green')\nplt.title('ENGINESIZE vs CO2EMISSIONS')\nplt.xlabel('ENGINESIZE')\nplt.ylabel('CO2EMISSIONS ')\nplt.show()",
"_____no_output_____"
],
[
"from sklearn import linear_model\nregr = linear_model.LinearRegression()\ntrain_x = np.asanyarray(train[['ENGINESIZE']])\ntrain_y = np.asanyarray(train[['CO2EMISSIONS']])\n\nregr.fit(train_x , train_y)\nprint('Slope: {}'.format(regr.coef_[0][0]))\nprint('Intercept {}'.format(regr.intercept_[0]))",
"Slope: 39.325368270342835\nIntercept 124.58155385351395\n"
],
[
"plt.scatter(train.ENGINESIZE , train.CO2EMISSIONS , color='green')\nplt.plot(train_x , regr.coef_[0][0]*train_x + regr.intercept_[0] , '-r')\nplt.title('ENGINESIZE vs CO2EMISSIONS')\nplt.xlabel('ENGINESIZE')\nplt.ylabel('CO2EMISSIONS ')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 7. Evaluation\nWe compare the actual values and predicted values to calculate the accuracy of a regression model. Evaluation metrics provide a key role in the development of a model, as it provides insight to areas that require improvement.\n\nThere are different model evaluation metrics, lets use MSE here to calculate the accuracy of our model based on the test set:\n\n- Mean absolute error: It is the mean of the absolute value of the errors. This is the easiest of the metrics to understand since it’s just average error.\n- Mean Squared Error (MSE): Mean Squared Error (MSE) is the mean of the squared error. It’s more popular than Mean absolute error because the focus is geared more towards large errors. This is due to the squared term exponentially increasing larger errors in comparison to smaller ones.\n- Root Mean Squared Error (RMSE).\n- R-squared is not error, but is a popular metric for accuracy of your model. It represents how close the data are to the fitted regression line. The higher the R-squared, the better the model fits your data. Best possible score is 1.0 and it can be negative (because the model can be arbitrarily worse).",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import r2_score\n\ntest_x = np.asanyarray(test[['ENGINESIZE']])\ntest_y = np.asanyarray(test[['CO2EMISSIONS']])\ntest_y_ = regr.predict(test_x)\n\n# ERRORS\n\nprint('Mean Absolute Error: {}'.format(np.mean(test_y_ - test_y)))\nprint('Mean Square Error: {}'.format(np.mean((test_y_ - test_y)**2)))\nprint('R2 Score: {}'.format(r2_score(test_y , test_y_)))",
"Mean Absolute Error: -1.0496266792043686e-14\nMean Square Error: 973.5096581801105\nR2 Score: 0.7588594852300422\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecc9df289cc339afd5a2925f9842c48102dae1f8 | 20,996 | ipynb | Jupyter Notebook | hourly_model/.ipynb_checkpoints/XGB-checkpoint.ipynb | NREL/TrafficVolumeEstimation | 419b008b0da4809fbb13d65b4b1afa5572bda24e | [
"BSD-3-Clause"
] | null | null | null | hourly_model/.ipynb_checkpoints/XGB-checkpoint.ipynb | NREL/TrafficVolumeEstimation | 419b008b0da4809fbb13d65b4b1afa5572bda24e | [
"BSD-3-Clause"
] | null | null | null | hourly_model/.ipynb_checkpoints/XGB-checkpoint.ipynb | NREL/TrafficVolumeEstimation | 419b008b0da4809fbb13d65b4b1afa5572bda24e | [
"BSD-3-Clause"
] | 1 | 2021-08-04T19:13:30.000Z | 2021-08-04T19:13:30.000Z | 35.346801 | 139 | 0.536054 | [
[
[
"%matplotlib inline\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport math\nimport os\nimport random\nimport pickle\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.ensemble import GradientBoostingRegressor\nfrom sklearn.ensemble import GradientBoostingClassifier\nfrom sklearn.ensemble import AdaBoostRegressor\nfrom sklearn.ensemble import AdaBoostClassifier\nfrom sklearn.tree import DecisionTreeRegressor\nfrom sklearn.metrics import mean_squared_error, r2_score\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import roc_curve, auc\nfrom sklearn import preprocessing \nfrom xgboost import XGBRegressor\nfrom datetime import datetime\nfrom bayes_opt import BayesianOptimization\nrandom.seed(1234)",
"_____no_output_____"
]
],
[
[
"# Define Functions",
"_____no_output_____"
]
],
[
[
"# Performce cross validation using xgboost\ndef xgboostcv(X, y, fold, n_estimators, lr, depth, n_jobs, gamma, min_cw, subsample, colsample):\n uid = np.unique(fold)\n model_pred = np.zeros(X.shape[0])\n model_valid_loss = np.zeros(len(uid))\n model_train_loss = np.zeros(len(uid))\n for i in uid:\n x_valid = X[fold==i]\n x_train = X[fold!=i]\n y_valid = y[fold==i]\n y_train = y[fold!=i]\n model = XGBRegressor(n_estimators=n_estimators, learning_rate=lr, \n max_depth = depth, n_jobs = n_jobs, \n gamma = gamma, min_child_weight = min_cw,\n subsample = subsample, colsample_bytree = colsample, random_state=1234)\n model.fit(x_train, y_train)\n\n pred = model.predict(x_valid)\n model_pred[fold==i] = pred\n model_valid_loss[uid==i] = mean_squared_error(y_valid, pred)\n model_train_loss[uid==i] = mean_squared_error(y_train, model.predict(x_train))\n return {'pred':model_pred, 'valid_loss':model_valid_loss, 'train_loss':model_train_loss}\n\n# Compute MSE for xgboost cross validation\ndef xgboostcv_mse(n, p, depth, g, min_cw, subsample, colsample):\n model_cv = xgboostcv(X_train, y_train, fold_train, \n int(n)*100, 10**p, int(depth), n_nodes, \n 10**g, min_cw, subsample, colsample)\n MSE = mean_squared_error(y_train, model_cv['pred'])\n return -MSE\n\n# Display model performance metrics for each cv iteration\ndef cv_performance(model, y, fold):\n uid = np.unique(fold)\n pred = np.round(model['pred'])\n y = y.reshape(-1)\n model_valid_mse = np.zeros(len(uid))\n model_valid_mae = np.zeros(len(uid))\n model_valid_r2 = np.zeros(len(uid))\n for i in uid:\n pred_i = pred[fold==i]\n y_i = y[fold==i]\n model_valid_mse[uid==i] = mean_squared_error(y_i, pred_i)\n model_valid_mae[uid==i] = np.abs(pred_i-y_i).mean()\n model_valid_r2[uid==i] = r2_score(y_i, pred_i)\n \n results = pd.DataFrame(0, index=uid, \n columns=['valid_mse', 'valid_mae', 'valid_r2', \n 'valid_loss', 'train_loss'])\n results['valid_mse'] = model_valid_mse\n results['valid_mae'] = model_valid_mae\n results['valid_r2'] = model_valid_r2\n results['valid_loss'] = model['valid_loss']\n results['train_loss'] = model['train_loss']\n print(results)\n\n# Display overall model performance metrics\ndef cv_overall_performance(y, y_pred):\n overall_MSE = mean_squared_error(y, y_pred)\n overall_MAE = (np.abs(y_pred-y)).mean()\n overall_RMSE = np.sqrt(np.square(y_pred-y).mean())\n overall_R2 = r2_score(y, y_pred)\n print(\"XGB overall MSE: %0.4f\" %overall_MSE)\n print(\"XGB overall MAE: %0.4f\" %overall_MAE)\n print(\"XGB overall RMSE: %0.4f\" %overall_RMSE)\n print(\"XGB overall R^2: %0.4f\" %overall_R2) \n\n# Plot variable importance\ndef plot_importance(model, columns):\n importances = pd.Series(model.feature_importances_, index = columns).sort_values(ascending=False)\n n = len(columns)\n plt.figure(figsize=(10,15))\n plt.barh(np.arange(n)+0.5, importances)\n plt.yticks(np.arange(0.5,n+0.5), importances.index)\n plt.tick_params(axis='both', which='major', labelsize=22)\n plt.ylim([0,n])\n plt.gca().invert_yaxis()\n plt.savefig('variable_importance.png', dpi = 150)\n\n# Save xgboost model\ndef save(obj, path):\n pkl_fl = open(path, 'wb')\n pickle.dump(obj, pkl_fl)\n pkl_fl.close()\n\n# Load xgboost model\ndef load(path):\n f = open(path, 'rb')\n obj = pickle.load(f)\n f.close()\n return(obj)",
"_____no_output_____"
]
],
[
[
"# Parameter Values",
"_____no_output_____"
]
],
[
[
"# Set a few values\nvalidation_only = False # Whether test model with test data\nn_nodes = 96 # Number of computing nodes used for hyperparamter tunning\ntrained = False # If a trained model exits\ncols_drop = ['StationId', 'Date', 'PenRate', 'NumberOfLanes', 'Dir', 'FC', 'Month'] # Columns to be dropped\nif trained:\n params = load('params.dat')\n xgb_cv = load('xgb_cv.dat')\n xgb = load('xgb.dat')",
"_____no_output_____"
]
],
[
[
"# Read Data",
"_____no_output_____"
]
],
[
[
"if validation_only:\n raw_data_train = pd.read_csv(\"final_train_data.csv\")\n data = raw_data_train.drop(cols_drop, axis=1)\n if 'Dir' in data.columns:\n data[['Dir']] = data[['Dir']].astype('category')\n one_hot = pd.get_dummies(data[['Dir']])\n data = data.drop(['Dir'], axis = 1)\n data = data.join(one_hot)\n if 'FC' in data.columns:\n data[['FC']] = data[['FC']].astype('category')\n one_hot = pd.get_dummies(data[['FC']])\n data = data.drop(['FC'], axis = 1)\n data = data.join(one_hot)\n week_dict = {\"DayOfWeek\": {'Monday': 1, 'Tuesday': 2, 'Wednesday': 3, 'Thursday': 4, \n 'Friday': 5, 'Saturday': 6, 'Sunday': 7}}\n data = data.replace(week_dict)\n\n X = data.drop(['Volume', 'fold'], axis=1)\n X_col = X.columns\n y = data[['Volume']]\n\n fold_train = data[['fold']].values.reshape(-1)\n X_train = X.values\n y_train = y.values\n \nelse:\n raw_data_train = pd.read_csv(\"final_train_data.csv\")\n raw_data_test = pd.read_csv(\"final_test_data.csv\")\n raw_data_test1 = pd.DataFrame(np.concatenate((raw_data_test.values, np.zeros(raw_data_test.shape[0]).reshape(-1, 1)), axis=1),\n columns = raw_data_test.columns.append(pd.Index(['fold'])))\n raw_data = pd.DataFrame(np.concatenate((raw_data_train.values, raw_data_test1.values), axis=0), \n columns = raw_data_train.columns)\n\n data = raw_data.drop(cols_drop, axis=1)\n if 'Dir' in data.columns:\n data[['Dir']] = data[['Dir']].astype('category')\n one_hot = pd.get_dummies(data[['Dir']])\n data = data.drop(['Dir'], axis = 1)\n data = data.join(one_hot)\n if 'FC' in data.columns:\n data[['FC']] = data[['FC']].astype('category')\n one_hot = pd.get_dummies(data[['FC']])\n data = data.drop(['FC'], axis = 1)\n data = data.join(one_hot)\n week_dict = {\"DayOfWeek\": {'Monday': 1, 'Tuesday': 2, 'Wednesday': 3, 'Thursday': 4, \n 'Friday': 5, 'Saturday': 6, 'Sunday': 7}}\n data = data.replace(week_dict)\n\n X = data.drop(['Volume'], axis=1)\n y = data[['Volume']]\n\n X_train = X.loc[X.fold!=0, :]\n fold_train = X_train[['fold']].values.reshape(-1)\n X_col = X_train.drop(['fold'], axis = 1).columns\n X_train = X_train.drop(['fold'], axis = 1).values\n y_train = y.loc[X.fold!=0, :].values\n\n X_test = X.loc[X.fold==0, :]\n X_test = X_test.drop(['fold'], axis = 1).values\n y_test = y.loc[X.fold==0, :].values",
"_____no_output_____"
],
[
"X_col",
"_____no_output_____"
],
[
"# Explain variable names\nX_name_dict = {'Temp': 'Temperature', 'WindSp': 'Wind Speed', 'Precip': 'Precipitation', 'Snow': 'Snow', \n 'Long': 'Longitude', 'Lat': 'Latitude', 'NumberOfLanes': 'Number of Lanes', 'SpeedLimit': 'Speed Limit', \n 'FRC': 'TomTom FRC', 'DayOfWeek': 'Day of Week', 'Month': 'Month', 'Hour': 'Hour', \n 'AvgSp': 'Average Speed', 'ProbeCount': 'Probe Count', 'Dir_E': 'Direction(East)', \n 'Dir_N': 'Direction(North)', 'Dir_S': 'Direction(South)', 'Dir_W': 'Direction(West)', \n 'FC_3R': 'FHWA FC(3R)', 'FC_3U': 'FHWA FC(3U)', 'FC_4R': 'FHWA FC(4R)', 'FC_4U': 'FHWA FC(4U)', \n 'FC_5R': 'FHWA FC(5R)', 'FC_5U': 'FHWA FC(5U)', 'FC_7R': 'FHWA FC(7R)', 'FC_7U': 'FHWA FC(7U)'}",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"if validation_only == False:\n print(X_test.shape)",
"_____no_output_____"
]
],
[
[
"# Cross Validation & Hyperparameter Optimization",
"_____no_output_____"
]
],
[
[
"# Set hyperparameter ranges for Bayesian optimization \nxgboostBO = BayesianOptimization(xgboostcv_mse,\n {'n': (1, 10),\n 'p': (-4, 0),\n 'depth': (2, 10),\n 'g': (-3, 0),\n 'min_cw': (1, 10), \n 'subsample': (0.5, 1), \n 'colsample': (0.5, 1)\n })",
"_____no_output_____"
],
[
"# Use Bayesian optimization to tune hyperparameters\nimport time\nstart_time = time.time()\nxgboostBO.maximize(init_points=10, n_iter = 50)\nprint('-'*53)\nprint('Final Results')\nprint('XGBOOST: %f' % xgboostBO.max['target'])\nprint(\"--- %s seconds ---\" % (time.time() - start_time))",
"_____no_output_____"
],
[
"# Save the hyperparameters the yield the highest model performance\nparams = xgboostBO.max['params']\nsave(params, 'params.dat')\nparams",
"_____no_output_____"
],
[
"# Perform cross validation using the optimal hyperparameters\nxgb_cv = xgboostcv(X_train, y_train, fold_train, int(params['n'])*100, \n 10**params['p'], int(params['depth']), n_nodes, \n 10**params['g'], params['min_cw'], params['subsample'], params['colsample'])",
"_____no_output_____"
],
[
"# Display cv results for each iteration\ncv_performance(xgb_cv, y_train, fold_train)",
"_____no_output_____"
],
[
"# Display overall cv results\ncv_pred = xgb_cv['pred']\ncv_pred[cv_pred<0] = 0\ncv_overall_performance(y_train.reshape(-1), cv_pred)",
"_____no_output_____"
],
[
"# Save the cv results\nsave(xgb_cv, 'xgb_cv.dat')",
"_____no_output_____"
]
],
[
[
"# Model Test",
"_____no_output_____"
]
],
[
[
"# Build a xgboost using all the training data with the optimal hyperparameter\nxgb = XGBRegressor(n_estimators=int(params['n'])*100, learning_rate=10**params['p'], max_depth = int(params['depth']), \n n_jobs = n_nodes, gamma = 10**params['g'], min_child_weight = params['min_cw'], \n subsample = params['subsample'], colsample_bytree = params['colsample'], random_state=1234)\nxgb.fit(X_train, y_train)",
"_____no_output_____"
],
[
"# Test the trained model with test data\nif validation_only == False:\n y_pred = xgb.predict(X_test)\n y_pred[y_pred<0] = 0\n cv_overall_performance(y_test.reshape(-1), y_pred)",
"_____no_output_____"
],
[
"# Plot variable importance\ncol_names = [X_name_dict[i] for i in X_col]\nplot_importance(xgb, col_names)",
"_____no_output_____"
],
[
"# Save the trained xgboost model\nsave(xgb, 'xgb.dat')",
"_____no_output_____"
],
[
"# Produce cross validation estimates or estimates for test data\ntrain_data_pred = pd.DataFrame(np.concatenate((raw_data_train.values, cv_pred.reshape(-1, 1)), axis=1),\n columns = raw_data_train.columns.append(pd.Index(['PredVolume'])))\ntrain_data_pred.to_csv('train_data_pred.csv', index = False)\n\nif validation_only == False:\n test_data_pred = pd.DataFrame(np.concatenate((raw_data_test.values, y_pred.reshape(-1, 1)), axis=1),\n columns = raw_data_test.columns.append(pd.Index(['PredVolume'])))\n test_data_pred.to_csv('test_data_pred.csv', index = False)\n",
"_____no_output_____"
]
],
[
[
"# Plot Estimations vs. Observations",
"_____no_output_____"
]
],
[
[
"# Prepare data to plot estimated and observed values\nif validation_only:\n if trained:\n plot_df = pd.read_csv(\"train_data_pred.csv\")\n else:\n plot_df = train_data_pred\nelse:\n if trained:\n plot_df = pd.read_csv(\"test_data_pred.csv\")\n else:\n plot_df = test_data_pred \nplot_df = plot_df.sort_values(by=['StationId', 'Date', 'Dir', 'Hour'])\nplot_df = plot_df.set_index(pd.Index(range(plot_df.shape[0])))",
"_____no_output_____"
],
[
"# Define a function to plot estimated and observed values for a day\ndef plot_daily_estimate(frc):\n indices = plot_df.index[(plot_df.FRC == frc) & (plot_df.Hour == 0)].tolist()\n from_index = np.random.choice(indices, 1)[0]\n to_index = from_index + 23\n plot_df_sub = plot_df.loc[from_index:to_index, :]\n time = pd.date_range(plot_df_sub.Date.iloc[0] + ' 00:00:00', periods=24, freq='H')\n plt.figure(figsize=(20,10))\n plt.plot(time, plot_df_sub.PredVolume, 'b-', label='XGBoost', lw=2)\n plt.plot(time, plot_df_sub.Volume, 'r--', label='Observed', lw=3)\n plt.tick_params(axis='both', which='major', labelsize=24)\n plt.ylabel('Volume (vehs/hr)', fontsize=24)\n plt.xlabel(\"Time\", fontsize=24)\n plt.legend(loc='upper left', shadow=True, fontsize=24)\n plt.title('Station ID: {0}, MAE={1}, FRC = {2}'.format(\n plot_df_sub.StationId.iloc[0],\n round(np.abs(plot_df_sub.PredVolume-plot_df_sub.Volume).mean()),\n plot_df_sub.FRC.iloc[0]), fontsize=40)\n plt.savefig('frc_{0}.png'.format(frc), dpi = 150)\n return(plot_df_sub)",
"_____no_output_____"
],
[
"# Define a function to plot estimated and observed values for a week\ndef plot_weekly_estimate(frc):\n indices = plot_df.index[(plot_df.FRC == frc) & (plot_df.Hour == 0) & (plot_df.DayOfWeek == 'Monday')].tolist()\n from_index = np.random.choice(indices, 1)[0]\n to_index = from_index + 24*7-1\n plot_df_sub = plot_df.loc[from_index:to_index, :]\n time = pd.date_range(plot_df_sub.Date.iloc[0] + ' 00:00:00', periods=24*7, freq='H')\n plt.figure(figsize=(20,10))\n plt.plot(time, plot_df_sub.PredVolume, 'b-', label='XGBoost', lw=2)\n plt.plot(time, plot_df_sub.Volume, 'r--', label='Observed', lw=3)\n plt.tick_params(axis='both', which='major', labelsize=24)\n plt.ylabel('Volume (vehs/hr)', fontsize=24)\n plt.xlabel(\"Time\", fontsize=24)\n plt.legend(loc='upper left', shadow=True, fontsize=24)\n plt.title('Station ID: {0}, MAE={1}, FRC = {2}'.format(\n plot_df_sub.StationId.iloc[0],\n round(np.abs(plot_df_sub.PredVolume-plot_df_sub.Volume).mean()),\n plot_df_sub.FRC.iloc[0]), fontsize=40)\n plt.savefig('frc_{0}.png'.format(frc), dpi = 150)\n return(plot_df_sub)",
"_____no_output_____"
],
[
"# Plot estimated and observed values for a day\nfrc2_daily_plot = plot_daily_estimate(2)\nsave(frc2_daily_plot, 'frc2_daily_plot.dat')",
"_____no_output_____"
],
[
"# Plot estimated and observed values for a week\nfrc3_weekly_plot = plot_weekly_estimate(3)\nsave(frc3_daily_plot, 'frc3_daily_plot.dat')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecc9e11f9332ca0c2f7473213fb517d3ffd3e604 | 3,323 | ipynb | Jupyter Notebook | yolo_related/yolo_to_rcnn.ipynb | choucl/PCB-solder-joint-detection | 858760f249de828cd022f6a39fdf797c4bd5c693 | [
"MIT"
] | 5 | 2021-07-23T15:54:29.000Z | 2022-01-06T10:30:44.000Z | yolo_related/yolo_to_rcnn.ipynb | choucl/solder-joint-classfication | 858760f249de828cd022f6a39fdf797c4bd5c693 | [
"MIT"
] | null | null | null | yolo_related/yolo_to_rcnn.ipynb | choucl/solder-joint-classfication | 858760f249de828cd022f6a39fdf797c4bd5c693 | [
"MIT"
] | null | null | null | 28.646552 | 99 | 0.531748 | [
[
[
"from os import listdir\nfrom PIL import Image\nfrom os.path import isfile, join\n\n# origin_ann_url = '/home/user/PCB/slice_annotation/'\n# rcnn_ann_url = '/home/user/PCB/rcnn_ann.csv'\n# file_path = '/home/user/PCB/slice_img/'\n\norigin_ann_url = './slice_annotation/'\nrcnn_ann_url = './rcnn_ann.csv'\nfile_path = './slice_img/'",
"_____no_output_____"
],
[
"def yolo_rcnn_conversion(img_name, ann_strs):\n img_path = ''\n new_anns = ''\n if len(img_name) < 3:\n img_path = file_path + img_name + '.jpg'\n else:\n img_path = file_path + img_name + '.png'\n \n img = Image.open(img_path)\n w, h = img.size\n for ann_str in ann_strs:\n (class_name, center_x, center_y, width, height) = ann_str.split(' ')\n\n top_left_x = (float(center_x) * w) - (float(width) * w / 2)\n top_left_y = (float(center_y) * h) - (float(height) * h / 2)\n bottom_right_x = (float(center_x) * w) + (float(width) * w / 2)\n bottom_right_y = (float(center_y) * h) + (float(height) * h / 2)\n\n rcnn_ann = []\n rcnn_ann.append(img_path)\n rcnn_ann.append(str(top_left_x))\n rcnn_ann.append(str(top_left_y))\n rcnn_ann.append(str(bottom_right_x))\n rcnn_ann.append(str(bottom_right_y))\n rcnn_ann.append('normal' if class_name == '1' else 'defect')\n\n new_ann = ','.join(rcnn_ann)\n new_ann += '\\n'\n\n new_anns += new_ann\n return new_anns\n\n ",
"_____no_output_____"
],
[
"\nonlyfiles = [f for f in listdir(origin_ann_url) if isfile(join(origin_ann_url, f))]\nf = open(rcnn_ann_url, 'a+')\nfor filename in onlyfiles:\n ann_file = open(origin_ann_url+filename, 'r')\n anns = ann_file.readlines()\n file_num = filename[:-4]\n new_anns = yolo_rcnn_conversion(file_num, anns)\n f.write(new_anns)\nf.close()\n ",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecc9e2e25253ba890923c58cce61ee7948e2d580 | 38,539 | ipynb | Jupyter Notebook | 2nd-ML100Days/homework/D-030/Day_030_Leaf_Encoding.ipynb | qwerzxcv98/100Day-ML-Marathon | 3c86cd083b086a1e4b7a55a41b127909c7860f79 | [
"MIT"
] | 3 | 2019-08-22T15:19:11.000Z | 2019-08-24T00:54:54.000Z | 2nd-ML100Days/homework/D-030/Day_030_Leaf_Encoding.ipynb | magikerwin1993/100Day-ML-Marathon | 3c86cd083b086a1e4b7a55a41b127909c7860f79 | [
"MIT"
] | null | null | null | 2nd-ML100Days/homework/D-030/Day_030_Leaf_Encoding.ipynb | magikerwin1993/100Day-ML-Marathon | 3c86cd083b086a1e4b7a55a41b127909c7860f79 | [
"MIT"
] | 1 | 2019-07-18T01:52:04.000Z | 2019-07-18T01:52:04.000Z | 78.490835 | 23,380 | 0.754015 | [
[
[
"# 作業 : (Kaggle)鐵達尼生存預測",
"_____no_output_____"
],
[
"# [教學目標]\n- 以下用鐵達尼預測資料, 展示如何使用葉編碼, 並觀察預測效果\n- 因為只有分類問題比較適合葉編碼, 因此範例與作業都使用鐵達尼的資料(二元分類問題)",
"_____no_output_____"
],
[
"# [教學目標]\n- 了解葉編碼的寫作方式 : 使用梯度提升樹 (In[3]~In[5], Out[3]~Out[5])\n- 觀察葉編碼搭配邏輯斯迴歸後的效果 (In[6], Out[6], In[7], Out[7])",
"_____no_output_____"
]
],
[
[
"# 做完特徵工程前的所有準備\nimport pandas as pd\nimport numpy as np\nimport copy\nfrom sklearn.preprocessing import LabelEncoder, MinMaxScaler\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import GradientBoostingClassifier\n# 因為擬合(fit)與編碼(transform)需要分開, 因此不使用.get_dummy, 而採用 sklearn 的 OneHotEncoder\nfrom sklearn.preprocessing import OneHotEncoder\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import roc_curve\n\ndata_path = '../../data/'\ndf = pd.read_csv(data_path + 'titanic_train.csv')\n\ntrain_Y = df['Survived']\ndf = df.drop(['PassengerId', 'Survived'] , axis=1)\ndf.head()",
"_____no_output_____"
],
[
"# 因為需要把類別型與數值型特徵都加入, 故使用最簡版的特徵工程\nLEncoder = LabelEncoder()\nMMEncoder = MinMaxScaler()\nfor c in df.columns:\n df[c] = df[c].fillna(-1)\n if df[c].dtype == 'object':\n df[c] = LEncoder.fit_transform(list(df[c].values))\n df[c] = MMEncoder.fit_transform(df[c].astype(np.float64).values.reshape(-1, 1))\ndf.head()",
"_____no_output_____"
],
[
"train_X = df.values\n# 因為訓練邏輯斯迴歸時也要資料, 因此將訓練及切成三部分 train / val / test, 採用 test 驗證而非 k-fold 交叉驗證\n# train 用來訓練梯度提升樹, val 用來訓練邏輯斯迴歸, test 驗證效果\ntrain_X, test_X, train_Y, test_Y = train_test_split(train_X, train_Y, test_size=0.5)\ntrain_X, val_X, train_Y, val_Y = train_test_split(train_X, train_Y, test_size=0.5)",
"_____no_output_____"
],
[
"# 梯度提升樹調整參數並擬合後, 再將葉編碼 (*.apply) 結果做獨熱 / 邏輯斯迴歸\n# 調整參數的方式採用 RandomSearchCV 或 GridSearchCV, 以後的進度會再教給大家, 本次先直接使用調參結果\ngdbt = GradientBoostingClassifier(subsample=0.93, n_estimators=320, min_samples_split=0.1, min_samples_leaf=0.3, \n max_features=4, max_depth=4, learning_rate=0.16)\nonehot = OneHotEncoder()\nlr = LogisticRegression(solver='lbfgs', max_iter=1000)\n\ngdbt.fit(train_X, train_Y)\nonehot.fit(gdbt.apply(train_X)[:, :, 0])\nlr.fit(onehot.transform(gdbt.apply(val_X)[:, :, 0]), val_Y)",
"c:\\users\\qwerz\\miniconda3\\envs\\ml100\\lib\\site-packages\\sklearn\\preprocessing\\_encoders.py:371: FutureWarning: The handling of integer data will change in version 0.22. Currently, the categories are determined based on the range [0, max(values)], while in the future they will be determined based on the unique values.\nIf you want the future behaviour and silence this warning, you can specify \"categories='auto'\".\nIn case you used a LabelEncoder before this OneHotEncoder to convert the categories to integers, then you can now use the OneHotEncoder directly.\n warnings.warn(msg, FutureWarning)\n"
],
[
"gdbt.apply(train_X)[:, :, 0]",
"_____no_output_____"
],
[
"# 將梯度提升樹+葉編碼+邏輯斯迴歸結果輸出\npred_gdbt_lr = lr.predict_proba(onehot.transform(gdbt.apply(test_X)[:, :, 0]))[:, 1]\nfpr_gdbt_lr, tpr_gdbt_lr, _ = roc_curve(test_Y, pred_gdbt_lr)\n# 將梯度提升樹結果輸出\npred_gdbt = gdbt.predict_proba(test_X)[:, 1]\nfpr_gdbt, tpr_gdbt, _ = roc_curve(test_Y, pred_gdbt)",
"_____no_output_____"
],
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\n# 將結果繪圖\nplt.plot([0, 1], [0, 1], 'k--')\nplt.plot(fpr_gdbt, tpr_gdbt, label='GDBT')\nplt.plot(fpr_gdbt_lr, tpr_gdbt_lr, label='GDBT + LR')\nplt.xlabel('False positive rate')\nplt.ylabel('True positive rate')\nplt.title('ROC curve')\nplt.legend(loc='best')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# 作業1\n* 請對照範例,完成隨機森林的鐵達尼生存率預測,以及對應的葉編碼+邏輯斯迴歸\n\n# 作業2\n* 上述的結果,葉編碼是否有提高預測的正確性呢?",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecc9efc6e29bd4c86809a20bad8df0ed37d7d115 | 144,553 | ipynb | Jupyter Notebook | Home Assignments/IHA1/IHA1.ipynb | dddraco/DML | 12e7f62fb14fd0868c8d67c5b024a2ec7b63dc7a | [
"MIT"
] | null | null | null | Home Assignments/IHA1/IHA1.ipynb | dddraco/DML | 12e7f62fb14fd0868c8d67c5b024a2ec7b63dc7a | [
"MIT"
] | null | null | null | Home Assignments/IHA1/IHA1.ipynb | dddraco/DML | 12e7f62fb14fd0868c8d67c5b024a2ec7b63dc7a | [
"MIT"
] | null | null | null | 112.056589 | 40,756 | 0.84789 | [
[
[
"# IHA1 - Assignment",
"_____no_output_____"
],
[
"Welcome to the first individual home assignment! \n\nThis assignment consists of two parts:\n * Python and NumPy exercises\n * Build a deep neural network for forward propagation\n \nThe focus of this assignment is for you to gain practical knowledge with implementing forward propagation of deep neural networks without using any deep learning framework. You will also gain practical knowledge in two of Python's scientific libraries [NumPy](https://docs.scipy.org/doc/numpy-1.13.0/index.html) and [Matplotlib](https://matplotlib.org/devdocs/index.html). \n\nSkeleton code is provided for most tasks and every part you are expected to implement is marked with **TODO**\n\nWe expect you to search and learn by yourself any commands you think are useful for these tasks. Don't limit yourself to only what was taught in CL1. Use the help function, [stackoverflow](https://stackoverflow.com/), google, the [python documentation](https://docs.python.org/3.5/library/index.html) and the [NumPy](https://docs.scipy.org/doc/numpy-1.13.0/index.html) documentation to your advantage. \n\n**IMPORTANT NOTE**: The tests available are not exhaustive, meaning that if you pass a test you have avoided the most common mistakes, but it is still not guaranteed that you solution is 100% correct. \n\nLets start by importing the necessary libraries below",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom utils.tests.iha1Tests import *",
"_____no_output_____"
]
],
[
[
"## 1. Lists and arrays introduction\nFirst, we will warm up with a Python exercise and few NumPy exercises",
"_____no_output_____"
],
[
"### 1.1 List comprehensions\nExamine the code snippet provided below",
"_____no_output_____"
]
],
[
[
"myList = []\nfor i in range(25):\n if i % 2 == 0:\n myList.append(i**2)\n \nprint(myList)",
"[0, 4, 16, 36, 64, 100, 144, 196, 256, 324, 400, 484, 576]\n"
]
],
[
[
"This is not a very \"[pythonic](http://docs.python-guide.org/en/latest/writing/style/)\" way of writing. Lets re-write the code above using a [list comprehension](https://docs.python.org/2/tutorial/datastructures.html#list-comprehensions). The result will be less code, more readable and elegant. Your solution should be able to fit into one line of code.",
"_____no_output_____"
]
],
[
[
"myList = None # TODO\nprint(myList)",
"None\n"
],
[
"# sample output from cell above for reference",
"[0, 4, 16, 36, 64, 100, 144, 196, 256, 324, 400, 484, 576]\n"
]
],
[
[
"### 1.2 Numpy array vs numpy vectors\nRun the cell below to create a numpy array. ",
"_____no_output_____"
]
],
[
[
"myArr = np.array([1, 9, 25, 49, 81, 121, 169, 225, 289, 361, 441, 529])\nprint(myArr)\nprint(myArr.shape)",
"_____no_output_____"
]
],
[
[
"One of the core features of numpy is to efficiently perform linear algebra operations.\nThere are two types of one-dimensional representations in numpy: arrays of shape (x,) and vectors of shape (x,1)\n\nThe the above result indicates that **myArr** is an array of 12 elements with shape (12,). \n\nNumpy's arrays and vectors both have the type of `numpy.ndarray` but have in some cases different characteristics and it is important to separate the two types because it will save a lot of debugging time later on. Read more about numpy shapes [here](https://stackoverflow.com/a/22074424) \n\nRun the code below to see how the transpose operation behaves differently between an array and vector",
"_____no_output_____"
]
],
[
[
"# print the shape of an array and the shape of a transposed array\nprint('myArr is an array of shape:')\nprint(myArr.shape)\nprint('The transpose of myArr has the shape:')\nprint(myArr.T.shape)\n\n# print the shape of a vector and the transpose of a vector\nmyVec = myArr.reshape(12,1)\nprint('myVec is a vector of shape:')\nprint(myVec.shape)\nprint('The transpose of myVec has the shape:')\nprint(myVec.T.shape)",
"_____no_output_____"
]
],
[
[
"### 1.3 Numpy exercises\nNow run the cell below to create the numpy array `numbers` and then complete the exercises sequentially",
"_____no_output_____"
]
],
[
[
"numbers = np.arange(24)\nprint(numbers)",
"_____no_output_____"
],
[
"# TODO: reshape numbers into a 6x4 matrix\nprint(numbers)",
"_____no_output_____"
],
[
"# sample output from cell above for reference",
"[[ 0 1 2 3]\n [ 4 5 6 7]\n [ 8 9 10 11]\n [12 13 14 15]\n [16 17 18 19]\n [20 21 22 23]]\n"
],
[
"# test case\ntest_numpy_reshape(numbers)",
"_____no_output_____"
],
[
"# TODO: set the element of the last row of the last column to zero\n# Hint: Try what happends when indices are negative\nprint(numbers)",
"_____no_output_____"
],
[
"# sample output from cell above for reference",
"[[ 0 1 2 3]\n [ 4 5 6 7]\n [ 8 9 10 11]\n [12 13 14 15]\n [16 17 18 19]\n [20 21 22 0]]\n"
],
[
"# test case\ntest_numpy_neg_ix(numbers)",
"_____no_output_____"
],
[
"# TODO: set every element of the 0th row to 0\nprint(numbers)",
"_____no_output_____"
],
[
"# sample output from cell above for reference",
"[[ 0 0 0 0]\n [ 4 5 6 7]\n [ 8 9 10 11]\n [12 13 14 15]\n [16 17 18 19]\n [20 21 22 0]]\n"
],
[
"# test case\ntest_numpy_row_ix(numbers)",
"_____no_output_____"
],
[
"# TODO: append a 1x4 row vector of zeros to `numbers`, \n# resulting in a 7x4 matrix where the new row of zeros is the last row\n# Hint: A new matrix must be created in the procedure. Numpy arrays are not dynamic.\nprint(numbers)\nprint(numbers.shape)",
"_____no_output_____"
],
[
"# sample output from cell above for reference",
"[[ 0 0 0 0]\n [ 4 5 6 7]\n [ 8 9 10 11]\n [12 13 14 15]\n [16 17 18 19]\n [20 21 22 0]\n [ 0 0 0 0]]\n(7, 4)\n"
],
[
"# test case\ntest_numpy_append_row(numbers)",
"_____no_output_____"
],
[
"# TODO: set all elements above 10 to the value 1\nprint(numbers)",
"_____no_output_____"
],
[
"# sample output from cell above for reference",
"[[ 0 0 0 0]\n [ 4 5 6 7]\n [ 8 9 10 1]\n [ 1 1 1 1]\n [ 1 1 1 1]\n [ 1 1 1 0]\n [ 0 0 0 0]]\n"
],
[
"# test case\ntest_numpy_bool_matrix(numbers)",
"_____no_output_____"
],
[
"# TODO: compute the sum of every row and replace `numbers` with the answer\n# `numbers` will be a (7,) array as a result\nprint(numbers.shape)\nprint(numbers)",
"_____no_output_____"
],
[
"# sample output from cell above for reference",
"(7,)\n[ 0 22 28 4 4 3 0]\n"
],
[
"# test case\ntest_numpy_sum(numbers)",
"_____no_output_____"
]
],
[
[
"## 2 Building your deep neural network\nIt is time to start implementing your first feed-forward neural network. In this lab you will only focus on implementing the forward propagation procedure. \n\nWhen using a neural network, you can not forward propagate the entire dataset at once. Therefore, you divide the dataset into a number of sets/parts called batches. A batch will make up for the first dimension of every input to a layer and the notation `(BATCH_SIZE, NUM_FEATURES)` simply means the dimension of a batch of samples where every sample has `NUM_FEATURES` features",
"_____no_output_____"
],
[
"### 2.1 activation functions\nYou will start by defining a few activation functions that are later needed by the neural network.",
"_____no_output_____"
],
[
"#### 2.1.1 ReLU\nThe neural network will use the ReLU activation function in every layer except for the last. ReLU does element-wise comparison of the input matrix. For example, if the input is `X`, and `X[i,j] == 2` and `X[k,l] == -1`, then after applying ReLU, `X[i,j] == 2` and `X[k,l] == 0` should be true. \n\nThe formula for implementing ReLU for a single neuron $i$ is:\n\\begin{equation}\nrelu(z_i) = \n \\begin{cases}\n 0, & \\text{if}\\ z_i \\leq 0 \\\\\n z_i, & \\text{otherwise}\n \\end{cases}\n\\end{equation}\n\nNow implement `relu` in vectorized form",
"_____no_output_____"
]
],
[
[
"def relu(z):\n \"\"\" Implement the ReLU activation function\n \n Arguments:\n z - the input of the activation function. Has a type of `numpy.ndarray`\n \n Returns:\n a - the output of the activation function. Has a type of numpy.ndarray and the same shape as `z`\n \"\"\"\n \n a = None # TODO\n \n return a",
"_____no_output_____"
],
[
"# test case\ntest_relu(relu)",
"_____no_output_____"
]
],
[
[
"#### 2.1.2 Sigmoid\nThe sigmoid activation function is common for binary classification. This is because it squashes its input to the range [0,1]. \nImplement the activation function `sigmoid` using the formula: \n\\begin{equation}\n \\sigma(z) = \\frac{1}{1 + e^{-z}}\n\\end{equation}",
"_____no_output_____"
]
],
[
[
"def sigmoid(z):\n \"\"\" Implement the sigmoid activation function\n \n Arguments:\n z - the input of the activation function. Has a type of `numpy.ndarray`\n \n Returns:\n a - the output of the activation function. Has a type of `numpy.ndarray` and the same shape as `z`\n \"\"\"\n \n a = None # TODO\n \n return a",
"_____no_output_____"
],
[
"# test case\ntest_sigmoid(sigmoid)",
"_____no_output_____"
]
],
[
[
"#### 2.1.3 Visualization\nMake a plot using matplotlib to visualize the activation functions between the input interval [-3,3]. The plot should have the following properties\n * one plot should contain a visualization of both `ReLU` and `sigmoid`\n * x-axis: range of values between [-3,3], **hint**: np.linspace\n * y-axis: the value of the activation functions at a given input `x`\n * a legend explaining which line represents which activation function",
"_____no_output_____"
]
],
[
[
"# TODO: make a plot of ReLU and sigmoid values in the interval [-3,3]",
"_____no_output_____"
],
[
"# sample output from cell above for reference",
"_____no_output_____"
]
],
[
[
"#### 2.1.4 Softmax\nYou will use the softmax activation function / classifier as the final layer of your neural network later in the assignment. Implement `softmax` according the the formula below. The subtraction of the maximum value is there solely to avoid overflows in a practical implementation.\n\\begin{equation}\nsoftmax(z_i) = \\frac{e^{z_i - max(\\mathbf{z})}}{ \\sum^j e^{z_j - max(\\mathbf{z})}}\n\\end{equation}\n",
"_____no_output_____"
]
],
[
[
"def softmax(z):\n \"\"\" Implement the softmax activation function\n \n Arguments:\n z - the input of the activation function, shape (BATCH_SIZE, FEATURES) and type `numpy.ndarray`\n \n Returns:\n a - the output of the activation function, shape (BATCH_SIZE, FEATURES) and type umpy.ndarray\n \"\"\"\n \n a = None # TODO\n \n return a",
"_____no_output_____"
],
[
"# test case\ntest_softmax(softmax)",
"_____no_output_____"
]
],
[
[
"### 2.2 Initialize weights\nYou will implement a helper function that takes the shape of a layer as input, and returns an initialized weight matrix $\\mathbf{W}$ and bias vector $\\mathbf{b}$ as output. $\\mathbf{W}$ should be sampled from a normal distribution of mean 0 and standard deviation 2, and $\\mathbf{b}$ should be initialized to all zeros.",
"_____no_output_____"
]
],
[
[
"def initialize_weights(layer_shape):\n \"\"\" Implement initialization of the weight matrix and biases\n \n Arguments:\n layer_shape - a tuple of length 2, type (int, int), that determines the dimensions of the weight matrix: (input_dim, output_dim)\n \n Returns:\n w - a weight matrix with dimensions of `layer_shape`, (input_dim, output_dim), that is normally distributed with\n properties mu = 0, stddev = 2. Has a type of `numpy.ndarray`\n b - a vector of initialized biases with shape (1,output_dim), all of value zero. Has a type of `numpy.ndarray`\n \"\"\"\n w = None # TODO\n b = None # TODO\n \n return w, b",
"_____no_output_____"
],
[
"# test case\ntest_initialize_weights(initialize_weights)",
"_____no_output_____"
]
],
[
[
"### 2.3 Feed-forward neural network layer module\nTo build a feed-forward neural network of arbitrary depth you are going to define a neural network layer as a module that can be used to stack upon eachother. \n\nYour task is to complete the `Layer` class by following the descriptions in the comments. \n\nRecall the formula for forward propagation of an arbitrary layer $l$:\n\n\\begin{equation}\n\\mathbf{a}^{[l]} = g(\\mathbf{z}^{[l]}) = g(\\mathbf{a}^{[l-1]}\\mathbf{w}^{[l]} +\\mathbf{b}^{[l]})\n\\end{equation}\n\n$g$ is the activation function given by `activation_fn`, which can be relu, sigmoid or softmax. ",
"_____no_output_____"
]
],
[
[
"class Layer:\n \"\"\" \n TODO: Build a class called Layer that satisfies the descriptions of the methods\n Make sure to utilize the helper functions you implemented before\n \"\"\"\n \n def __init__(self, input_dim, output_dim, activation_fn=relu):\n \"\"\"\n Arguments:\n input_dim - the number of inputs of the layer. type int\n output_dim - the number of outputs of the layer. type int\n activation_fn - a reference to the activation function to use. Should be `relu` as a default\n possible values are the `relu`, `sigmoid` and `softmax` functions you implemented earlier.\n Has the type `function`\n \n Attributes:\n w - the weight matrix of the layer, should be initialized with `initialize_weights`\n and has the shape (INPUT_FEATURES, OUTPUT_FEATURES) and type `numpy.ndarray`\n b - the bias vector of the layer, should be initialized with `initialize_weights`\n and has the shape (1, OUTPUT_FEATURES) and type `numpy.ndarray`\n activation_fn - a reference to the activation function to use.\n Has the type `function`\n \"\"\"\n self.w, self.b = None, None # TODO\n \n self.activation_fn = None # TODO\n \n def forward_prop(self, a_prev):\n \"\"\" Implement the forward propagation module of the neural network layer\n Should use whatever activation function that `activation_fn` references to\n \n Arguments:\n a_prev - the input to the layer, which may be the data `X`, or the output from the previous layer.\n a_prev has the shape of (BATCH_SIZE, INPUT_FEATURES) and the type `numpy.ndarray`\n \n Returns:\n a - the output of the layer when performing forward propagation. Has the type `numpy.ndarray`\n \"\"\"\n \n a = None # TODO\n \n return a",
"_____no_output_____"
],
[
"# test case, be sure that you pass the previous activation function tests before running this test\ntest_layer(Layer, relu, sigmoid, softmax)",
"_____no_output_____"
]
],
[
[
"### 2.4 Logistic regression \nBinary logistic regression is a classifier where classification is performed by applying the sigmoid activation function to a linear combination of input values. You will now try out your neural network layer by utilizing it as a linear combination of input values and apply the sigmoid activation function to classify a simple problem. \n\nThe cell below defines a dataset of 5 points of either class `0` or class `1`. Your assignment is to: \n1. Create an instance of a `Layer` with sigmoid activation function \n2. Manually tune the weights `w` and `b` of your layer\n\nYou can use `test_logistic` to visually inspect how your classifier is performing.",
"_____no_output_____"
]
],
[
[
"# Run this cell to create the dataset\nX_s = np.array([[1, 2],\n [5, 3],\n [8, 8],\n [7, 5],\n [3, 6]])\nY_s = np.array([0,0,1,0,1])\n\ntest_logistic(X_s, Y_s)",
"_____no_output_____"
],
[
"# create an instance of layer\nl = Layer(2,1,sigmoid)\n\n# TODO: manually tune weights\nl.w = None\nl.b = None\n\n# testing your choice of weights with this function\ntest_logistic(X_s,Y_s,l,sigmoid)",
"_____no_output_____"
],
[
"# sample output from cell above for reference",
"_____no_output_____"
]
],
[
[
"### 2.5 Feed-forward neural network\nNow define the actual neural network class. It is an L-layer neural network, meaning that the number of layers and neurons in each layer is specified as input by the user. Once again, you will only focus on implementing the forward propagation part.\n\nRead the descriptions in the comments and complete the todos ",
"_____no_output_____"
]
],
[
[
"class NeuralNetwork:\n \"\"\" \n TODO: Implement an L-layer neural network class by utilizing the Layer module defined above \n Each layer should use `relu` activation function, except for the output layer, which should use `softmax`\n \"\"\"\n \n def __init__(self, input_n, layer_dims):\n \"\"\"\n Arguments:\n input_n - the number of inputs to the network. Should be the same as the length of a data sample\n Has type int\n layer_dims - a python list or tuple of the number of neurons in each layer. Layer `l` should have a weight matrix \n with the shape (`layer_dims[l-1]`, `layer_dims[l]`). \n `layer_dims[-1]` is the dimension of the output layer.\n Layer 1 should have the dimensions (`input_n`, `layer_dims[0]`).\n len(layer_dims) is the depth of the neural network\n Attributes:\n input_n - the number of inputs to the network. Has type int\n layers - a python list of each layer in the network. Each layer should use the `relu` activation function,\n except for the last layer, which should use `softmax`. \n Has type `list` containing layers of type `Layer`\n \"\"\"\n \n self.input_n = None # TODO\n self.layers = None # TODO\n \n def forward_prop(self, x):\n \"\"\" \n Implement the forward propagation procedure through the entire network, from input to output.\n You will now connect each layer's forward propagation function into a chain of layer-wise forward propagations.\n \n Arguments:\n x - the input data, which has the shape (BATCH_SIZE, NUM_FEATURES) and type `numpy.ndarray`\n \n Returns:\n a - the output of the last layer after forward propagating through the every layer in `layers`.\n Should have the dimension (BATCH_SIZE, layers[-1].w.shape[1]) and type `numpy.ndarray`\n \"\"\"\n a = None # TODO\n \n return a",
"_____no_output_____"
],
[
"# test case\ntest_neuralnetwork(NeuralNetwork)",
"_____no_output_____"
]
],
[
[
"## 3 Making predictions with a neural network\nIn practice, its common to load weights to your neural network that has already been trained. \nIn this section, you will create an instance of your neural network, load trained weights from disk, and perform predictions.\n\n### 3.1 Load weights from disk\nCreate an instance of `NeuralNetwork` with input size $28 \\times 28 = 784$, two hidden layers of size 100 and an output layer of size 10. Thereafter, load the weights contained in `./utils/ann_weights.npz` to your network.",
"_____no_output_____"
]
],
[
[
"ann = None # TODO: create instance of ann\n\n# load weights\nweights = np.load('./utils/ann_weights.npz')\nfor l in range(len(ann.layers)):\n ann.layers[l].w = weights['w' + str(l)]\n ann.layers[l].b = weights['b' + str(l)]",
"_____no_output_____"
]
],
[
[
"### 3.2 Prediction\nNow, implement the function `predict_and_correct` which does the following:\n1. Load `./utils/test_data.npz` from disk\n2. Extract test data `X` and `Y` from file\n2. Perform for every pair of data: \n a. plot the image `x` \n b. make a prediction using your neural network by forward propagating and picking the most probable class \n c. check wether the prediction is correct (compare with the ground truth number `y`) \n d. print the predicted label and wether it was correct or not ",
"_____no_output_____"
]
],
[
[
"def predict_and_correct(ann):\n \"\"\" Load test data from file and predict using your neural network. \n Make a prediction for ever data sample and print it along with wether it was a correct prediction or not\n \n Arguments:\n ann - the neural network to use for prediction. Has type `NeuralNetwork`\n \n Returns: # for test case purposes\n A `numpy.ndarray` of predicted classes (integers [0-9]) with shape (11,)\n \"\"\"\n data = np.load('./utils/test_data.npz_old')\n X, cls = data['X'], data['Y']\n \n cls_preds = None # TODO: make a predicted number for every image in X\n \n for i in range(len(X)):\n plt.imshow(X[i].reshape(28,28), cmap='gray')\n plt.show()\n correct = cls_preds[i] == cls[i]\n print('The prediction was {0}, it was {1}!'.format(cls_preds[i], 'correct' if correct else 'incorrect'))\n \n return cls_preds\n \ncls_pred = predict_and_correct(ann)",
"_____no_output_____"
],
[
"# final test case\ntest_predict_and_correct_answer(cls_pred)",
"_____no_output_____"
],
[
"# sample output from cell above for reference",
"_____no_output_____"
]
],
[
[
"## Congratulations!\nYou have successfully implemented a neural network from scratch using only NumPy! ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecc9f5a316a65c5a8f13845f4e91a39ae8339535 | 13,440 | ipynb | Jupyter Notebook | notebooks/PredictiveModelling2.ipynb | adarsh9780/titanic | a5f2953354b990fe4f41f9fa70b36cdfd745ec30 | [
"MIT"
] | null | null | null | notebooks/PredictiveModelling2.ipynb | adarsh9780/titanic | a5f2953354b990fe4f41f9fa70b36cdfd745ec30 | [
"MIT"
] | null | null | null | notebooks/PredictiveModelling2.ipynb | adarsh9780/titanic | a5f2953354b990fe4f41f9fa70b36cdfd745ec30 | [
"MIT"
] | null | null | null | 23.537653 | 105 | 0.529241 | [
[
[
"Author : Adarsh Maurya\nEmail : [email protected]\nproject: Titanic",
"_____no_output_____"
]
],
[
[
"# Hyper Parameter Optimaztion",
"_____no_output_____"
]
],
[
[
"import os\nimport numpy as np\nimport pandas as pd\n%matplotlib inline",
"_____no_output_____"
],
[
"data_path = os.path.join(os.pardir, 'data', 'processed')\n\ntest_data_path = os.path.join(data_path, 'test.csv')\ntrain_data_path = os.path.join(data_path, 'train.csv')",
"_____no_output_____"
],
[
"test_df = pd.read_csv(test_data_path, index_col=\"PassengerId\")\ntrain_df = pd.read_csv(train_data_path, index_col=\"PassengerId\")",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X = train_df.loc[:, :\"AgeState_Child\"].as_matrix().astype('float')\nY = train_df.Survived.ravel()",
"_____no_output_____"
],
[
"train_X, test_X, train_Y, test_Y = train_test_split(X, Y, test_size=0.2, random_state=0)",
"_____no_output_____"
],
[
"print(\"Shape of Input(training) : {}\".format(train_X.shape))\nprint(\"Shape of Output(training) : {}\".format(train_Y.shape))\nprint(\"Shape of Input(testing) : {}\".format(test_X.shape))\nprint(\"Shape of Output(testing) : {}\".format(test_Y.shape))",
"Shape of Input(training) : (712, 23)\nShape of Output(training) : (712,)\nShape of Input(testing) : (179, 23)\nShape of Output(testing) : (179,)\n"
],
[
"from sklearn.linear_model import LogisticRegression\n\nmodel_LR = LogisticRegression(random_state=0)",
"_____no_output_____"
],
[
"from sklearn.model_selection import GridSearchCV",
"_____no_output_____"
],
[
"parameters = {'C':[1.0, 10.0, 50.0, 100.0, 1000.0], 'penalty':['l1', 'l2']}\n\n# cv= K-fold, in this case k = 3\n# C = lambda in regularization\ngrid_search = GridSearchCV(model_LR, param_grid=parameters, cv=3)",
"_____no_output_____"
],
[
"grid_search.fit(train_X, train_Y)",
"_____no_output_____"
],
[
"grid_search.best_params_",
"_____no_output_____"
],
[
"print(\"Best score is : {}\".format(grid_search.best_score_))",
"Best score is : 0.824438202247191\n"
],
[
"print(\"Logistic Regression Testing Score : {}\".format(grid_search.score(test_X, test_Y)))",
"Logistic Regression Testing Score : 0.8268156424581006\n"
],
[
"def get_submission_file(model, filename):\n # create a test matrix with float values\n test_X = test_df.as_matrix().astype(\"float\")\n print(test_X.shape)\n # make prediction on the model\n predictions = model.predict(test_X)\n # create a dataframe to submit\n df_submit = pd.DataFrame({'PassengerId':test_df.index, 'Survived': predictions})\n # define submit path and save the data frame in csv format\n submit_path = os.path.join(os.pardir, \"data\", \"external\")\n submit_file_path = os.path.join(submit_path, filename)\n # index=False --> so that no extra column is added\n df_submit.to_csv(submit_file_path, index=False)",
"_____no_output_____"
],
[
"get_submission_file(grid_search, '03_LR_GridSearch.csv')",
"(418, 23)\n"
]
],
[
[
"## Feature Normalization & Feature Standardization",
"_____no_output_____"
]
],
[
[
"# Feature Standardization is similar to feature normalization.\n# In this case, instead of scaling our feature, e scale our distiribution of each feature\n# such that each one has a mean = 0, var = 1.0\n\nfrom sklearn.preprocessing import MinMaxScaler, StandardScaler",
"_____no_output_____"
]
],
[
[
"### Feature Normalization",
"_____no_output_____"
]
],
[
[
"scaler = MinMaxScaler()\nX_train_scaled = scaler.fit_transform(train_X)",
"_____no_output_____"
],
[
"# Check min and max value for column 1\nX_train_scaled[:, 0:].min(), X_train_scaled[:, 0:].max()",
"_____no_output_____"
],
[
"# Scale the test data\nX_test_scaled = scaler.fit_transform(test_X)",
"_____no_output_____"
]
],
[
[
"### Feature Standardization",
"_____no_output_____"
]
],
[
[
"distro_scaler = StandardScaler()\n\nX_train_scaled = distro_scaler.fit_transform(train_X)\nX_test_scaled = distro_scaler.fit_transform(test_X)",
"_____no_output_____"
]
],
[
[
"#### Create Model after standardization",
"_____no_output_____"
]
],
[
[
"model_LR = LogisticRegression(random_state=0)\n\nparameters = {'C':[1.0, 10.0, 50.0, 100.0, 1000.0], 'penalty':['l1', 'l2']}\n\n# cv= K-fold, in this case k = 3\n# C = lambda in regularization\ngrid_search = GridSearchCV(model_LR, param_grid=parameters, cv=3)\n\ngrid_search.fit(train_X, train_Y)",
"_____no_output_____"
],
[
"grid_search.best_score_",
"_____no_output_____"
],
[
"print(\"Logistic Regression Testing Score : {}\".format(grid_search.score(test_X, test_Y)))",
"Logistic Regression Testing Score : 0.8268156424581006\n"
]
],
[
[
"# Model Persistence",
"_____no_output_____"
]
],
[
[
"import pickle",
"_____no_output_____"
],
[
"model_file_path = os.path.join(os.pardir, 'models', 'LR_MODEL.pkl')",
"_____no_output_____"
],
[
"model_file_pickel = open(model_file_path, 'wb')",
"_____no_output_____"
],
[
"pickle.dump(grid_search, model_file_pickel)",
"_____no_output_____"
],
[
"model_file_pickel.close()",
"_____no_output_____"
]
],
[
[
"## Check Persisted file",
"_____no_output_____"
]
],
[
[
"model_file_pickel = open(model_file_path, 'rb')\n\ngrid_search_loaded = pickle.load(model_file_pickel)\n\nmodel_file_pickel.close()",
"_____no_output_____"
],
[
"grid_search_loaded",
"_____no_output_____"
],
[
"print(\"Logistic Score : {}\".format(grid_search_loaded.score(test_X, test_Y)))",
"Logistic Score : 0.8268156424581006\n"
]
],
[
[
"# -------> The End <--------",
"_____no_output_____"
]
]
] | [
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"raw"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecca1080c0809a07f18998757328f3469f0b7562 | 9,745 | ipynb | Jupyter Notebook | model-inference/decisionTree/experiments/model_transfer/year.ipynb | asu-cactus/netsdb | fe6ae0e1dcd07ea0b9a47ff4a657d5ed029c7af5 | [
"Apache-2.0"
] | 13 | 2022-01-17T16:14:26.000Z | 2022-03-30T02:06:04.000Z | model-inference/decisionTree/experiments/model_transfer/year.ipynb | asu-cactus/netsdb | fe6ae0e1dcd07ea0b9a47ff4a657d5ed029c7af5 | [
"Apache-2.0"
] | 1 | 2022-01-28T23:17:14.000Z | 2022-01-28T23:17:14.000Z | model-inference/decisionTree/experiments/model_transfer/year.ipynb | asu-cactus/netsdb | fe6ae0e1dcd07ea0b9a47ff4a657d5ed029c7af5 | [
"Apache-2.0"
] | 3 | 2022-01-18T02:13:53.000Z | 2022-03-06T19:28:19.000Z | 32.055921 | 88 | 0.39959 | [
[
[
"import pandas\nimport pickle\nimport joblib\nfrom sklearn.ensemble import RandomForestClassifier\n\nfilename = 'models/year/rf-10-8-6.pkl'\nloaded_model = joblib.load(filename)\n\nfrom sklearn.tree import export_graphviz\nfrom IPython.display import Image\nimport pydotplus\nimport graphviz\nimport os\n\nEstimators = loaded_model.estimators_\nfor index, model in enumerate(Estimators):\n filename = 'graphs/year/year_' + str(index) + '.pdf'\n outfilename = 'graphs/year/year_' + str(index) + '.txt'\n dot_data = export_graphviz(model)\n f = open(outfilename, 'w') \n f.write(dot_data) \n f.close()\n graph = pydotplus.graph_from_dot_data(dot_data)\n #Image(graph.create_png())\n #graph.write_pdf(filename)\n os.system('dot -Tpdf ' + outfilename + ' -o ' + filename)",
"_____no_output_____"
],
[
"import pandas as pd\nimport pickle\nimport joblib\n\ndf = pd.read_csv('datasets/year/YearPredictionMSD.txt')\ndf.head()",
"_____no_output_____"
],
[
"print (\"row,column\")\nprint (df.shape[0],df.shape[1])",
"row,column\n515344 91\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecca1d1fd09e0ef9ae801da5e00b588f973a17d9 | 8,425 | ipynb | Jupyter Notebook | 07 records.ipynb | AryaDevops/Python-for-DataScience | ab446f6b23c1b2fd6b57c94e5a2673234d7c29c5 | [
"MIT"
] | null | null | null | 07 records.ipynb | AryaDevops/Python-for-DataScience | ab446f6b23c1b2fd6b57c94e5a2673234d7c29c5 | [
"MIT"
] | null | null | null | 07 records.ipynb | AryaDevops/Python-for-DataScience | ab446f6b23c1b2fd6b57c94e5a2673234d7c29c5 | [
"MIT"
] | null | null | null | 20.107399 | 173 | 0.479169 | [
[
[
"## Records and Dates in NumPy\n\n** Creating and manipulating NumPy record arrays, where every row over 2D array can have mixed types.**\n\n** Working with NumPy datetime64 objects, which can encode a date and time.**",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"# Record array - is a hybrid of a list and a dictionary\n# A list because we can access the rows using a numerical index. And a dictionary since we access the columns by their names by full strings.\n\nreca = np.array([(1,(2.0,3.0),'hey'),(2,(3.5,4.0),'no')], dtype = [('x',np.int32),('y',np.float64,2),('z',np.str,4)])\n\n# Specify rows using numpy tuples.. datatypes given in a list of pairs\n",
"_____no_output_____"
],
[
"reca",
"_____no_output_____"
],
[
"reca[0] #first row",
"_____no_output_____"
],
[
"reca['x'] #first column",
"_____no_output_____"
],
[
"reca['x'][0] #to get single element",
"_____no_output_____"
],
[
"reca[0]['x'] #can also use opposite order",
"_____no_output_____"
]
],
[
[
" Record Arrays are often useful to represent non-homogeneous tabular data. And they can be read from this very conveniently using the Loadtxt and Genfromtxt functions.",
"_____no_output_____"
]
],
[
[
"#DATETIME64\n\n#64 = number of bits each such object takes in memory",
"_____no_output_____"
],
[
"np.datetime64('2015') #this object refers to year 2015",
"_____no_output_____"
],
[
"np.datetime64('2015-01')",
"_____no_output_____"
],
[
"np.datetime64('2015-02-03 12:00:00')",
"_____no_output_____"
],
[
"np.datetime64('2015-01-01') < np.datetime64('2015-04-03')",
"_____no_output_____"
],
[
"np.datetime64('2015-04-03') - np.datetime64('2015-01-01')\n\n#results in timedelta object",
"_____no_output_____"
],
[
"# timedelta objects can be added to datetimes\n\nnp.datetime64('2015-01-01') + np.timedelta64(92,'D')",
"_____no_output_____"
],
[
"np.datetime64('2015-01-01') + np.timedelta64(5,'h')",
"_____no_output_____"
],
[
"# Datetime64 objects can also be converted to numbers.\n\nnp.datetime64('2015-01-01').astype(float)",
"_____no_output_____"
],
[
"np.datetime64('1970-01-01').astype(float) #1970 is beginning standaard so returns 0",
"_____no_output_____"
],
[
"#To build a range of dates\n\nr = np.arange(np.datetime64('2016-02-01'),np.datetime64('2016-03-01'))",
"_____no_output_____"
],
[
"r",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecca3612a9aec51fd208683ff7ca7f11ec1ad4e4 | 30,572 | ipynb | Jupyter Notebook | Images Basics with OpenCV/Drawing On Images - Text & Polygons.ipynb | ayushjindal23/Python-For-Computer-Vision-With-OpenCv-And-Deep-Learning | 096dbba74f7fa3bd81bdd9a357c482f10b70e125 | [
"MIT"
] | null | null | null | Images Basics with OpenCV/Drawing On Images - Text & Polygons.ipynb | ayushjindal23/Python-For-Computer-Vision-With-OpenCv-And-Deep-Learning | 096dbba74f7fa3bd81bdd9a357c482f10b70e125 | [
"MIT"
] | null | null | null | Images Basics with OpenCV/Drawing On Images - Text & Polygons.ipynb | ayushjindal23/Python-For-Computer-Vision-With-OpenCv-And-Deep-Learning | 096dbba74f7fa3bd81bdd9a357c482f10b70e125 | [
"MIT"
] | null | null | null | 100.89769 | 8,080 | 0.874199 | [
[
[
"import cv2 as cv\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"blank_img = np.zeros(shape=(512,512,3),dtype=np.int16)",
"_____no_output_____"
],
[
"plt.imshow(blank_img)",
"_____no_output_____"
],
[
"font = cv.FONT_HERSHEY_SIMPLEX\ncv.putText(blank_img,text='Hello',org=(10,500),fontFace=font,fontScale=4,color=(255,255,255),thickness=3,lineType=cv.LINE_AA)\nplt.imshow(blank_img)",
"_____no_output_____"
],
[
"blank_img = np.zeros(shape=(512,512,3),dtype=np.int32)",
"_____no_output_____"
],
[
"plt.imshow(blank_img)",
"_____no_output_____"
],
[
"vertices = np.array([[100,300],[200,200],[400,300],[200,400]],dtype=np.int32)",
"_____no_output_____"
],
[
"vertices",
"_____no_output_____"
],
[
"vertices.shape #WE NEED IT 3-D",
"_____no_output_____"
],
[
"pts = vertices.reshape(-1,1,2) #TO ADD ANOTHER SET OF BRACES",
"_____no_output_____"
],
[
"pts",
"_____no_output_____"
],
[
"pts.shape #3-D",
"_____no_output_____"
],
[
"cv.polylines(blank_img,[pts],isClosed=True,color=(255,0,0),thickness=5)\nplt.imshow(blank_img)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecca43dfc0c2bc9cc5b6343420e5378f73edf6b4 | 691,901 | ipynb | Jupyter Notebook | identify_customer_segments_git.ipynb | avelicha/Unsupervised_Learning_DSND_Identify_Customer_Segments | 0b90712493044b6a2add61e98d8f9a4e9de609e8 | [
"MIT"
] | null | null | null | identify_customer_segments_git.ipynb | avelicha/Unsupervised_Learning_DSND_Identify_Customer_Segments | 0b90712493044b6a2add61e98d8f9a4e9de609e8 | [
"MIT"
] | null | null | null | identify_customer_segments_git.ipynb | avelicha/Unsupervised_Learning_DSND_Identify_Customer_Segments | 0b90712493044b6a2add61e98d8f9a4e9de609e8 | [
"MIT"
] | null | null | null | 183.576811 | 129,544 | 0.849581 | [
[
[
"# Unsupervised Learning: Identifying Customer Segments\n\nIn this project, we will apply unsupervised learning techniques to identify segments of the population that form the core customer base for a mail-order sales company in Germany. These segments can then be used to direct marketing campaigns towards audiences that will have the highest expected rate of returns. The data that we will use has been provided by Udacity's partners at Bertelsmann Arvato Analytics, and represents a real-life data science task.",
"_____no_output_____"
]
],
[
[
"# import libraries here; add more as necessary\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom sklearn.preprocessing import OneHotEncoder\nfrom sklearn.cluster import KMeans\nimport pprint\n\n# magic word for producing visualizations in notebook\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Step 0: Load the Data\n\nThere are four files associated with this project (not including this one):\n\n- `Udacity_AZDIAS_Subset.csv`: Demographics data for the general population of Germany; 891211 persons (rows) x 85 features (columns).\n- `Udacity_CUSTOMERS_Subset.csv`: Demographics data for customers of a mail-order company; 191652 persons (rows) x 85 features (columns).\n- `Data_Dictionary.md`: Detailed information file about the features in the provided datasets.\n- `AZDIAS_Feature_Summary.csv`: Summary of feature attributes for demographics data; 85 features (rows) x 4 columns\n\nEach row of the demographics files represents a single person, but also includes information outside of individuals, including information about their household, building, and neighborhood. We will use this information to cluster the general population into groups with similar demographic properties. Then, we will see how the people in the customers dataset fit into those created clusters. The hope here is that certain clusters are over-represented in the customers data, as compared to the general population; those over-represented clusters will be assumed to be part of the core userbase. This information can then be used for further applications, such as targeting for a marketing campaign.\n\nTo start off with, let's load in the demographics data for the general population into a pandas DataFrame, and do the same for the feature attributes summary. Note for all of the `.csv` data files in this project: they're semicolon (`;`) delimited, so we'll need an additional argument in our [`read_csv()`](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_csv.html) call to read in the data properly. Also, considering the size of the main dataset, it may take some time for it to load completely.\n\nOnce the dataset is loaded, we'll take a little bit of time just browsing the general structure of the dataset and feature summary file. We'll be getting deep into the innards of the cleaning in the first major step of the project, so gaining some general familiarity can help us get our bearings.",
"_____no_output_____"
]
],
[
[
"# Load in the general demographics data.\nazdias = pd.read_csv(\"Udacity_AZDIAS_Subset.csv\", delimiter=\";\")\n\n# Load in the feature summary file.\nfeat_info = pd.read_csv(\"AZDIAS_Feature_Summary.csv\", delimiter=\";\")",
"_____no_output_____"
],
[
"# Check the structure of the data after it's loaded (e.g. print the number of\n# rows and columns, print the first few rows).\nprint(\"AZDIAS General Population Dataset rows: \", azdias.shape[0], \"columns: \", azdias.shape[1])\nprint(\"AZDIAS Feature Summary Dataset rows: \", feat_info.shape[0], \"columns: \", feat_info.shape[1])",
"AZDIAS General Population Dataset rows: 891221 columns: 85\nAZDIAS Feature Summary Dataset rows: 85 columns: 4\n"
],
[
"print(\"*******First 20 rows of AZDIAS General Population Dataset*********\")\n# truth = azdias[\"CJT_GESAMTTYP\"] == 0\n# print(azdias[\"CJT_GESAMTTYP\"].dtype)\n# truth_data = azdias[truth]\n# print(truth.sum())\nazdias.head(20)",
"*******First 20 rows of AZDIAS General Population Dataset*********\n"
],
[
"print(\"*******First 20 rows of AZDIAS Feature Summary Dataset*********\")\nprint(feat_info.head(20))",
"*******First 20 rows of AZDIAS Feature Summary Dataset*********\n attribute information_level type missing_or_unknown\n0 AGER_TYP person categorical [-1,0]\n1 ALTERSKATEGORIE_GROB person ordinal [-1,0,9]\n2 ANREDE_KZ person categorical [-1,0]\n3 CJT_GESAMTTYP person categorical [0]\n4 FINANZ_MINIMALIST person ordinal [-1]\n5 FINANZ_SPARER person ordinal [-1]\n6 FINANZ_VORSORGER person ordinal [-1]\n7 FINANZ_ANLEGER person ordinal [-1]\n8 FINANZ_UNAUFFAELLIGER person ordinal [-1]\n9 FINANZ_HAUSBAUER person ordinal [-1]\n10 FINANZTYP person categorical [-1]\n11 GEBURTSJAHR person numeric [0]\n12 GFK_URLAUBERTYP person categorical []\n13 GREEN_AVANTGARDE person categorical []\n14 HEALTH_TYP person ordinal [-1,0]\n15 LP_LEBENSPHASE_FEIN person mixed [0]\n16 LP_LEBENSPHASE_GROB person mixed [0]\n17 LP_FAMILIE_FEIN person categorical [0]\n18 LP_FAMILIE_GROB person categorical [0]\n19 LP_STATUS_FEIN person categorical [0]\n"
]
],
[
[
"## Step 1: Preprocessing\n\n### Step 1.1: Assess Missing Data\n\nThe feature summary file contains a summary of properties for each demographics data column. We will use this file to make cleaning decisions during this stage of the project. First of all, we should assess the demographics data in terms of missing data.\n\n#### Step 1.1.1: Convert Missing Value Codes to NaNs\nThe fourth column of the feature attributes summary (loaded in above as `feat_info`) documents the codes from the data dictionary that indicate missing or unknown data. While the file encodes this as a list (e.g. `[-1,0]`), this will get read in as a string object. We'll need to do a little bit of parsing to make use of it to identify and clean the data. We'll onvert data that matches a 'missing' or 'unknown' value code into a numpy NaN value. We might want to see how much data takes on a 'missing' or 'unknown' code, and how much data is naturally missing, as a point of interest.",
"_____no_output_____"
]
],
[
[
"#Output data types of azdias features\nazdias_dtypes = azdias.dtypes\n\n#Check int64 azdias dataset columns\ncheck_int_dtypes_azdias = (azdias_dtypes == \"int64\")\nint_columns_azdias = azdias_dtypes[check_int_dtypes_azdias].index.values\n\n\n#Check str azdias dataset columns\ncheck_str_dtypes_azdias = (azdias_dtypes == \"object\")\nstr_columns_azdias = azdias_dtypes[check_str_dtypes_azdias].index.values\n\n#Check float64 azdias dataset columns\ncheck_float_dtypes_azdias = (azdias_dtypes == \"float64\")\nfloat_columns_azdias = azdias_dtypes[check_float_dtypes_azdias].index.values\n\n#Print number of columns of different data types\nprint(\"Number of int64 columns in azdias dataset: \", len(int_columns_azdias))\nprint(\"Number of float64 columns in azdias dataset: \", len(float_columns_azdias))\nprint(\"Number of str columns in azdias dataset: \", len(str_columns_azdias))",
"Number of int64 columns in azdias dataset: 32\nNumber of float64 columns in azdias dataset: 49\nNumber of str columns in azdias dataset: 4\n"
],
[
"print(\"*****What are the missing, unknown codes for string based columns in azdias dataset?******\")\nprint(\"\\n\")\nfor each_item in str_columns_azdias:\n print(feat_info[feat_info[\"attribute\"] == each_item])",
"*****What are the missing, unknown codes for string based columns in azdias dataset?******\n\n\n attribute information_level type missing_or_unknown\n55 OST_WEST_KZ building categorical [-1]\n attribute information_level type missing_or_unknown\n57 CAMEO_DEUG_2015 microcell_rr4 categorical [-1,X]\n attribute information_level type missing_or_unknown\n58 CAMEO_DEU_2015 microcell_rr4 categorical [XX]\n attribute information_level type missing_or_unknown\n59 CAMEO_INTL_2015 microcell_rr4 mixed [-1,XX]\n"
],
[
"#Create a dictionary whose keys are features and values are it's missing/unknown items\ndict_attr_to_missing_map = {}\n\nfor row_num in range(feat_info.shape[0]): # for every row in feat_info dataset\n if feat_info[\"missing_or_unknown\"][row_num] != str([]): # ignore no missing item features\n to_list = feat_info[\"missing_or_unknown\"][row_num].split(\",\") # split string to list\n to_list[0] = to_list[0][1:]\n to_list[-1] = to_list[-1][0:-1]\n if (feat_info[\"attribute\"][row_num] not in str_columns_azdias): #check if column data type is int or float\n dict_attr_to_missing_map[feat_info[\"attribute\"][row_num]] = list(map(int, to_list))\n else: \n dict_attr_to_missing_map[feat_info[\"attribute\"][row_num]] = to_list\n \nprint(len(dict_attr_to_missing_map))",
"75\n"
],
[
"# Identify missing or unknown data values in azdias and convert them to NaNs.\nazdias_copy = azdias.copy()\n\nfor each_key in dict_attr_to_missing_map.keys():\n azdias_copy[each_key].replace(to_replace = dict_attr_to_missing_map[each_key], value = np.nan, inplace=True)\n \nazdias_copy.head(20)",
"_____no_output_____"
]
],
[
[
"#### Step 1.1.2: Assess Missing Data in Each Column\n\nHow much missing data is present in each column? There are a few columns that are outliers in terms of the proportion of values that are missing. We'll use matplotlib's [`hist()`](https://matplotlib.org/api/_as_gen/matplotlib.pyplot.hist.html) function to visualize the distribution of missing value counts to find these columns. While some of these columns might have justifications for keeping or re-encoding the data, for this project let's just remove them from the dataframe. Check out the discussion below on how columns were dropped from the dataframe.",
"_____no_output_____"
]
],
[
[
"# Perform an assessment of how much missing data there is in each column of the\n# dataset.\nnull_perc_col_stats = azdias_copy.isnull().sum()\nnull_perc_col_stats = null_perc_col_stats*100/azdias.shape[0] #convert to percentage to avoid huge value differences\nnull_perc_col_stats.head()",
"_____no_output_____"
],
[
"# Investigate patterns in the amount of missing data in each column.\nplt.figure(figsize=(14, 6))\nplt.hist(null_perc_col_stats.values, 30, facecolor='g', alpha=0.75)\nplt.xlabel('Percentage of missing/unknown records', fontsize=14)\nplt.ylabel('Percentage of features', fontsize=14)\nplt.show()",
"_____no_output_____"
],
[
"# Remove the outlier columns from the dataset. (You'll perform other data\n# engineering tasks such as re-encoding and imputation later.)\ncolumns_being_dropped = null_perc_col_stats[null_perc_col_stats > 20].index.values\nprint(\"***Columns being dropped\")\nprint(columns_being_dropped)\nprint(\"***Remaining dataset shape\")\nazdias_no_outlier = azdias_copy.drop(columns_being_dropped, axis = 1)\nprint(azdias_no_outlier.shape)",
"***Columns being dropped\n['AGER_TYP' 'GEBURTSJAHR' 'TITEL_KZ' 'ALTER_HH' 'KK_KUNDENTYP'\n 'KBA05_BAUMAX']\n***Remaining dataset shape\n(891221, 79)\n"
]
],
[
[
"#### Discussion 1.1.2: Assess Missing Data in Each Column\n\nFrom the histogram, it is evident that except for a few columns, rest all have missing records < 20%.\nDigging deeper, we find that there are six outlier columns with high missing records which are printed above. Briefly going through their desrciption, it seemed like other columns could in some way capture the idea behind these columns (eg: GEBURTSJAHR which is the year of birth can be roughly captured using PRAEGENDE_JUGENDJAHRE column).\nConsidering we're removing < 10% of the columns, it should be fine. Having columns with few valid data points could potentially lead to skewed results. So they've been removed from further analysis.",
"_____no_output_____"
],
[
"#### Step 1.1.3: Assess Missing Data in Each Row\n\nNow, we'll perform a similar assessment for the rows of the dataset. How much data is missing in each row? As with the columns, we should see some groups of points that have a very different numbers of missing values. Let's divide the data into two subsets: one for data points that are above some threshold for missing values, and a second subset for points below that threshold.\n\nIn order to know what to do with the outlier rows, we should see if the distribution of data values on columns that are not missing data (or are missing very little data) are similar or different between the two groups. We'll select at least five of these columns and compare the distribution of values.\n\nDepending on what we observe in our comparison, this will have implications on how we approach our conclusions later in the analysis. If the distributions of non-missing features look similar between the data with many missing values and the data with few or no missing values, then we could argue that simply dropping those points from the analysis won't present a major issue. On the other hand, if the data with many missing values looks very different from the data with few or no missing values, then we should make a note on those data as special. We'll revisit these data later on. **Either way, let's continue our analysis for now using just the subset of the data with few or no missing values.**",
"_____no_output_____"
]
],
[
[
"# How much data is missing in each row of the dataset?\nnull_perc_row_stats = azdias_no_outlier.isnull().sum(axis=1)\nnull_perc_row_stats = null_perc_row_stats*100/azdias_no_outlier.shape[1] #convert to percentage to avoid huge value differences\nnull_perc_row_stats.head()",
"_____no_output_____"
],
[
"# Investigate patterns in the amount of missing data in each row.\nplt.figure(figsize=(14, 6))\nplt.hist(null_perc_row_stats.values, 30, facecolor='g', alpha=0.75)\nplt.xlabel('Percentage of missing/unknown features', fontsize=14)\nplt.ylabel('Percentage of rows', fontsize=14)\nplt.show()",
"_____no_output_____"
]
],
[
[
"From the above histogram, we see a clear distinction between low missing and high missing rows at 30%. Case could be made there's another demarkation at 10%. The decision comes down to how aggressive does one want to be about dropping data. We'll take our chances, go aggressive and split data using 10% demarkation.",
"_____no_output_____"
]
],
[
[
"# Write code to divide the data into two subsets based on the number of missing\n# values in each row.\nrows_high_missing = null_perc_row_stats[null_perc_row_stats > 10].index.values\nrows_low_missing = null_perc_row_stats[null_perc_row_stats < 10].index.values\nprint(rows_high_missing.shape, rows_low_missing.shape)\n#print(rows_high_missing[:2], rows_low_missing[:2])",
"(144112,) (747109,)\n"
],
[
"#Previous cell continued\nazdias_no_outlier_low_miss = azdias_no_outlier.drop(rows_high_missing)\nazdias_no_outlier_high_miss = azdias_no_outlier.drop(rows_low_missing)\nprint(azdias_no_outlier_low_miss.shape, azdias_no_outlier_high_miss.shape)",
"(747109, 79) (144112, 79)\n"
],
[
"# Compare the distribution of values for at least five columns where there are\n# no or few missing values, between the two subsets.\n\n#plot function\ndef barplot_by_side(data1, data2, feature): \n f, axes = plt.subplots(1, 2, figsize=(14, 2))\n sns.countplot(data1[feature], ax=axes[0])\n axes[0].set_title(\"high missing\")\n sns.countplot(data2[feature], ax=axes[1])\n axes[1].set_title(\"low missing\")\n f.show()\n\n# randomly select features\nnp.random.seed(38)\nfeatures_array = feat_info[\"attribute\"].values\nrandom_choice_features = np.random.choice(features_array, size=6)\n\n#get bar chart comparison for selected features\nfor each_feat in random_choice_features:\n barplot_by_side(azdias_no_outlier_high_miss, azdias_no_outlier_low_miss, each_feat)",
"/opt/conda/lib/python3.6/site-packages/matplotlib/figure.py:418: UserWarning: matplotlib is currently using a non-GUI backend, so cannot show the figure\n \"matplotlib is currently using a non-GUI backend, \"\n"
]
],
[
[
"#### Discussion 1.1.3: Assess Missing Data in Each Row\n\nBased on random selection & plotting of six features side-by-side for high missing rows & low missing rows data, we see that the distributions match reasonably well for 4 out of 6 features. So the idea of going aggressive with dropping \"missing row data above 10%\" wasn't a bad idea.",
"_____no_output_____"
],
[
"### Step 1.2: Select and Re-Encode Features\n\nChecking for missing data isn't the only way in which we can prepare a dataset for analysis. Since the unsupervised learning techniques to be used will only work on data that is encoded numerically, we need to make a few encoding changes or additional assumptions to be able to make progress. In addition, while almost all of the values in the dataset are encoded using numbers, not all of them represent numeric values. The third column of the feature summary (`feat_info`) gives a summary of types of measurement.\n- For numeric and interval data, these features can be kept without changes.\n- Most of the variables in the dataset are ordinal in nature. While ordinal values may technically be non-linear in spacing, let's make the simplifying assumption that the ordinal variables can be treated as being interval in nature (that is, kept without any changes).\n- Special handling may be necessary for the remaining two variable types: 'categorical', and 'mixed'.\n\nIn the first two parts of this sub-step, we will perform an investigation of the categorical and mixed-type features and make a decision on each of them, whether we will keep, drop, or re-encode each. Then, in the last part, we will create a new data frame with only the selected and engineered columns.",
"_____no_output_____"
]
],
[
[
"# How many features are there of each data type?\nfeat_info_mod = feat_info.set_index('attribute')\nfeat_info_mod.drop(labels = columns_being_dropped, inplace=True) #drop the removed features and run sum on that\ndata_types = feat_info_mod[\"type\"].unique()\n\nnum_data_types = {}\nfor each_type in data_types:\n count = (feat_info_mod[\"type\"] == each_type).sum()\n num_data_types[each_type] = count\n \nnum_data_types",
"_____no_output_____"
]
],
[
[
"#### Step 1.2.1: Re-Encode Categorical Features\n\nFor categorical data, we would ordinarily need to encode the levels as dummy variables. Depending on the number of categories, let's perform one of the following:\n- For binary (two-level) categoricals that take numeric values, let's keep them without needing to do anything.\n- For binary variables that takes on non-numeric values, let's either re-encode the values as numbers or create a dummy variable.\n- For multi-level categoricals (three or more values), we can either encode the values using multiple dummy variables or just drop them from the analysis.",
"_____no_output_____"
]
],
[
[
"# Assess categorical variables: which are binary, which are multi-level, and\n# which one needs to be re-encoded?\ncategorical_features = feat_info_mod[feat_info_mod[\"type\"] == \"categorical\"].index.values\n#azdias_subset_categorical = azdias_no_outlier_low_miss.loc[:,categorical_features]\n#azdias_subset_categorical.head()",
"_____no_output_____"
],
[
"for each_cat in categorical_features:\n num_of_uniq = azdias_no_outlier_low_miss.loc[:,each_cat].nunique()\n data_type = azdias_no_outlier_low_miss.loc[:,each_cat].dtype\n if num_of_uniq == 2 and data_type == \"object\":\n print(each_cat, \": binary non-numeric, Number of categories: \", num_of_uniq)\n elif num_of_uniq == 2:\n print(each_cat, \": binary numeric, Number of categories: \", num_of_uniq)\n else:\n print(each_cat, \": multi level, Number of categories: \", num_of_uniq)\n ",
"ANREDE_KZ : binary numeric, Number of categories: 2\nCJT_GESAMTTYP : multi level, Number of categories: 6\nFINANZTYP : multi level, Number of categories: 6\nGFK_URLAUBERTYP : multi level, Number of categories: 12\nGREEN_AVANTGARDE : binary numeric, Number of categories: 2\nLP_FAMILIE_FEIN : multi level, Number of categories: 11\nLP_FAMILIE_GROB : multi level, Number of categories: 5\nLP_STATUS_FEIN : multi level, Number of categories: 10\nLP_STATUS_GROB : multi level, Number of categories: 5\nNATIONALITAET_KZ : multi level, Number of categories: 3\nSHOPPER_TYP : multi level, Number of categories: 4\nSOHO_KZ : binary numeric, Number of categories: 2\nVERS_TYP : binary numeric, Number of categories: 2\nZABEOTYP : multi level, Number of categories: 6\nGEBAEUDETYP : multi level, Number of categories: 7\nOST_WEST_KZ : binary non-numeric, Number of categories: 2\nCAMEO_DEUG_2015 : multi level, Number of categories: 9\nCAMEO_DEU_2015 : multi level, Number of categories: 44\n"
],
[
"#Replacing binary non_numeric values in OST_WEST_KZ columns with numbers\nost_west_kz_elements = azdias_no_outlier_low_miss[\"OST_WEST_KZ\"].unique()\nazdias_no_outlier_low_miss_encoded = azdias_no_outlier_low_miss.copy()\nazdias_no_outlier_low_miss_encoded[\"OST_WEST_KZ\"].replace({ost_west_kz_elements[0]:0, ost_west_kz_elements[1]:1}, value = None, inplace=True)\n\n#Dropping columns LP_FAMILIE_FEIN, LP_STATUS_FEIN, CAMEO_DEU_2015 as they are redundant\nazdias_no_outlier_low_miss_encoded.drop([\"LP_FAMILIE_FEIN\", \"LP_STATUS_FEIN\", \"CAMEO_DEU_2015\"], axis=1, inplace=True)\n\n#Re-encoding multi level categorical features\ndrop_cat_columns = [0, 4, 5, 7, 11, 12, 15, 17] #column numbers generated using categorical_features array\nazdias_no_outlier_low_miss_encoded = pd.get_dummies(azdias_no_outlier_low_miss_encoded, columns = np.delete(categorical_features, drop_cat_columns))",
"_____no_output_____"
],
[
"#Printing some sample encoded features\nazdias_no_outlier_low_miss_encoded.iloc[:5,100:130]",
"_____no_output_____"
]
],
[
[
"#### Discussion 1.2.1: Re-Encode Categorical Features\n\nOut of the 18 categorical features, 4 are binary numeric, 1 is binary non-numeric and the rest 13 are multi level features.\n 1. As mentioned previously, binary numeric columns were left untouched. \n \n \n 2. The one binary non-numeric column was converted into numbers by replacing it's current values.\n \n \n 3. For multi-level features, three columns **LP_FAMILIE_FEIN, LP_STATUS_FEIN, CAMEO_DEU_2015** were dropped from further analysis as they are long, detailed forms of these columns **LP_FAMILIE_GROB, LP_STATUS_GROB, CAMEO_DEUG_2015**. The rest were re-encoded and the resultant dataset will be used further.",
"_____no_output_____"
],
[
"#### Step 1.2.2: Engineer Mixed-Type Features\n\nThere are a handful of features that are marked as \"mixed\" in the feature summary that require special treatment in order to be included in the analysis. We'll refer `Data_Dictionary.md` to get details on these features. Check out discussion section below for more info on how this task was handled.",
"_____no_output_____"
]
],
[
[
"#Investigating mixed features\nmixed_features = feat_info_mod[feat_info_mod[\"type\"] == \"mixed\"].index.values\nmixed_features",
"_____no_output_____"
],
[
"azdias_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE'].unique(), azdias_no_outlier_low_miss_encoded['CAMEO_INTL_2015'].unique()",
"_____no_output_____"
],
[
"# Investigate \"PRAEGENDE_JUGENDJAHRE\" and engineer two new variables.\n\n#Dictionaries to map current variable to two new variables\nmovement_dict = {(1, 3, 5, 8, 10, 12, 14): 0, (2, 4, 6, 7, 9, 11, 13, 15): 1}\ndecade_dict = {(1, 2): 0, (3, 4): 1, (5, 6, 7): 2, (8, 9): 3, (10, 11, 12, 13): 4, (14, 15): 5}\n\n#Create the two new columns\nazdias_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE_DECADE'] = azdias_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE']\nazdias_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE_MOVEMENT'] = azdias_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE']\n\n#modify the new columns\nfor keys, values in decade_dict.items():\n azdias_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE_DECADE'].replace(to_replace = keys, value = values, inplace=True)\n\nfor keys, values in movement_dict.items():\n azdias_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE_MOVEMENT'].replace(to_replace = keys, value = values, inplace=True)\n\n#printing some columns to check the mapping\nazdias_no_outlier_low_miss_encoded.loc[:,['PRAEGENDE_JUGENDJAHRE','PRAEGENDE_JUGENDJAHRE_DECADE','PRAEGENDE_JUGENDJAHRE_MOVEMENT']].tail(10)",
"_____no_output_____"
],
[
"# Investigate \"CAMEO_INTL_2015\" and engineer two new variables.\n\n# Convert strings to numeric values in \"CAMEO_INTL_2015\" column\nnum_converted = pd.to_numeric(azdias_no_outlier_low_miss_encoded[\"CAMEO_INTL_2015\"], downcast='integer', errors=\"coerce\")\nnum_converted.isnull().sum()\n\n#Create the two new columns\nazdias_no_outlier_low_miss_encoded[\"CAMEO_INTL_2015_WEALTH\"] = num_converted // 10\nazdias_no_outlier_low_miss_encoded[\"CAMEO_INTL_2015_LIFE_STAGE\"] = num_converted % 10\n\n#Printing some columns to check mapping\nazdias_no_outlier_low_miss_encoded.loc[:, [\"CAMEO_INTL_2015\",\"CAMEO_INTL_2015_WEALTH\", \"CAMEO_INTL_2015_LIFE_STAGE\"]].tail(10)",
"_____no_output_____"
],
[
"#Remove \"PRAEGENDE_JUGENDJAHRE\", \"CAMEO_INTL_2015\", \"WOHNLAGE\", \"PLZ8_BAUMAX\", \"LP_LEBENSPHASE_FEIN\", \"LP_LEBENSPHASE_GROB\" columns\nazdias_no_outlier_low_miss_encoded.drop(mixed_features, axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"#### Discussion 1.2.2: Engineer Mixed-Type Features\n1. Digging into the mixed features, we see that **WOHNLAGE & PLZ8_BAUMAX** almost feel like ordinal features but we cannot say that with 100% certainty, so they were dropped from further analysis.\n\n\n2. **LP_LEBENSPHASE_FEIN, LP_LEBENSPHASE_GROB** are detailed, concise forms respectively of ***Life Stage*** which are covered again in **PRAEGENDE_JUGENDJAHRE**. So they were dropped from further analysis.\n\n\n3. **PRAEGENDE_JUGENDJAHRE** combines information on three dimensions: generation by decade, movement (mainstream vs. avantgarde), and nation (east vs. west). As there aren't enough levels to disentangle east from west, two new variables were created to capture the other two dimensions: an interval-type variable for decade, and a binary variable for movement.\n\n\n4. **CAMEO_INTL_2015** combines information on two axes: wealth and life stage. As the data type is \"object\", first the data was converted to \"integer\" type and then split into two new columns using quotient and remainder operations.\n\n\n5. Finally the two original columns were removed from the dataset.\n\n\n",
"_____no_output_____"
],
[
"#### Step 1.2.3: Complete Feature Selection\n\nIn order to finish this step up, we need to make sure that our data frame now only has the columns that we want to keep. To summarize, the dataframe should consist of the following:\n- All numeric, interval, and ordinal type columns from the original dataset.\n- Binary categorical features (all numerically-encoded).\n- Engineered features from other multi-level categorical features and mixed features.",
"_____no_output_____"
],
[
"### Step 1.3: Create a Cleaning Function\n\nEven though we've finished cleaning up the general population demographics data, it's important to look ahead to the future and realize that we'll need to perform the same cleaning steps on the customer demographics data. In this substep, let's complete the function below to execute the main feature selection, encoding, and re-engineering steps we performed above. Then, when it comes to looking at the customer data in Step 3, we can just run this function on that DataFrame to get the trimmed dataset in a single step.",
"_____no_output_____"
]
],
[
[
"def clean_data(df):\n \"\"\"\n Perform feature trimming, re-encoding, and engineering for demographics\n data\n \n INPUT: Demographics DataFrame\n OUTPUT: Trimmed and cleaned demographics DataFrame\n \"\"\"\n \n df_copy = df.copy()\n #####print(df_copy.shape)\n\n # convert missing value codes into NaNs, ...\n for each_key in dict_attr_to_missing_map.keys():\n df_copy[each_key].replace(to_replace = dict_attr_to_missing_map[each_key], value = np.nan, inplace=True)\n #####print(df_copy.shape)\n \n # remove selected columns and rows, ...\n df_no_outlier = df_copy.drop(columns_being_dropped, axis = 1)\n #####print(df_no_outlier.shape)\n\n # How much data is missing in each row of the dataset?\n null_perc_row_stats_df = df_no_outlier.isnull().sum(axis=1)\n null_perc_row_stats_df = null_perc_row_stats_df*100/df_no_outlier.shape[1] #convert to percentage to avoid huge value differences\n rows_high_missing_df = null_perc_row_stats_df[null_perc_row_stats_df > 10].index.values\n df_no_outlier_low_miss = df_no_outlier.drop(rows_high_missing_df)\n #####print(df_no_outlier_low_miss.shape)\n\n # select, re-encode, and engineer column values.\n #Replacing binary non_numeric values in OST_WEST_KZ columns with numbers\n df_no_outlier_low_miss_encoded = df_no_outlier_low_miss.copy()\n df_no_outlier_low_miss_encoded[\"OST_WEST_KZ\"].replace({ost_west_kz_elements[0]:0, ost_west_kz_elements[1]:1}, value = None, inplace=True)\n\n #Dropping columns LP_FAMILIE_FEIN, LP_STATUS_FEIN, CAMEO_DEU_2015 as they are redundant\n df_no_outlier_low_miss_encoded.drop([\"LP_FAMILIE_FEIN\", \"LP_STATUS_FEIN\", \"CAMEO_DEU_2015\"], axis=1, inplace=True)\n #####print(df_no_outlier_low_miss_encoded.shape)\n #Re-encoding multi level categorical features\n df_no_outlier_low_miss_encoded = pd.get_dummies(df_no_outlier_low_miss_encoded, columns = np.delete(categorical_features, drop_cat_columns))\n #####print(df_no_outlier_low_miss_encoded.shape)\n\n # Investigate \"PRAEGENDE_JUGENDJAHRE\" and engineer two new variables.\n #Create the two new columns\n df_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE_DECADE'] = df_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE']\n df_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE_MOVEMENT'] = df_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE']\n\n #modify the new columns\n for keys, values in decade_dict.items():\n df_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE_DECADE'].replace(to_replace = keys, value = values, inplace=True)\n\n for keys, values in movement_dict.items():\n df_no_outlier_low_miss_encoded['PRAEGENDE_JUGENDJAHRE_MOVEMENT'].replace(to_replace = keys, value = values, inplace=True)\n \n #####print(df_no_outlier_low_miss_encoded.shape)\n \n # Investigate \"CAMEO_INTL_2015\" and engineer two new variables.\n # Convert strings to numeric values in \"CAMEO_INTL_2015\" column\n num_converted_df = pd.to_numeric(df_no_outlier_low_miss_encoded[\"CAMEO_INTL_2015\"], downcast='integer', errors=\"coerce\")\n\n #Create the two new columns\n df_no_outlier_low_miss_encoded[\"CAMEO_INTL_2015_WEALTH\"] = num_converted_df // 10\n df_no_outlier_low_miss_encoded[\"CAMEO_INTL_2015_LIFE_STAGE\"] = num_converted_df % 10\n #####print(df_no_outlier_low_miss_encoded.shape)\n\n #Printing some columns to check mapping\n #####print(df_no_outlier_low_miss_encoded.loc[:, [\"CAMEO_INTL_2015\",\"CAMEO_INTL_2015_WEALTH\", \"CAMEO_INTL_2015_LIFE_STAGE\"]].tail()) # Return the cleaned dataframe.\n \n #Remove \"PRAEGENDE_JUGENDJAHRE\", \"CAMEO_INTL_2015\", \"WOHNLAGE\", \"PLZ8_BAUMAX\", \"LP_LEBENSPHASE_FEIN\", \"LP_LEBENSPHASE_GROB\" columns\n df_no_outlier_low_miss_encoded.drop(mixed_features, axis=1, inplace=True)\n #####print(df_no_outlier_low_miss_encoded.shape)\n return df_no_outlier_low_miss_encoded",
"_____no_output_____"
]
],
[
[
"## Step 2: Feature Transformation\n\n### Step 2.1: Apply Feature Scaling\n\nBefore we apply dimensionality reduction techniques to the data, we need to perform feature scaling so that the principal component vectors are not influenced by the natural differences in scale for features. Starting from this part of the project, we'll want to keep an eye on the [API reference page for sklearn](http://scikit-learn.org/stable/modules/classes.html) to help navigate to all of the classes and functions that we'll need. In this substep, we'll need to check the following:\n\n- sklearn requires that data not have missing values in order for its estimators to work properly. So, before applying the scaler to our data, we need to make sure that we've cleaned the DataFrame of the remaining missing values. This can be as simple as just removing all data points with missing data, or applying an [Imputer](http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.Imputer.html) to replace all missing values.\n- For the actual scaling function, a [StandardScaler](http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.StandardScaler.html) instance is suggested, scaling each feature to mean 0 and standard deviation 1.\n- For these classes, we can make use of the `.fit_transform()` method to both fit a procedure to the data as well as apply the transformation to the data at the same time. We need to keep the fit sklearn objects handy, since we'll be applying them to the customer demographics data towards the end of the project.",
"_____no_output_____"
]
],
[
[
"no_null_rows = null_perc_row_stats[null_perc_row_stats == 0].index.values\nprint(\"% of number of rows with no null values: \", (no_null_rows.shape[0]*100/azdias_no_outlier_low_miss_encoded.shape[0]))",
"% of number of rows with no null values: 83.41607449515399\n"
],
[
"# If you've not yet cleaned the dataset of all NaN values, then investigate and\n# do that now.\nfrom sklearn.preprocessing import Imputer\nimp_median = Imputer(missing_values=np.nan, strategy='mean')\nazdias_clean_no_nan = imp_median.fit_transform(azdias_no_outlier_low_miss_encoded)",
"_____no_output_____"
],
[
"# Apply feature scaling to the general population demographics data.\nfrom sklearn.preprocessing import StandardScaler\nscaler_obj = StandardScaler()\nazdias_clean_no_nan_scaled = scaler_obj.fit_transform(azdias_clean_no_nan)\nazdias_clean_no_nan_scaled[:5, :5]",
"_____no_output_____"
]
],
[
[
"### Discussion 2.1: Apply Feature Scaling\n\nQuick analysis shows there are 83% of the remaining rows with no NaN values. That's pretty high but we need to keep in mind that the rest of the rows have < 10% of features missing which is quite low. So throwing them (~17%) away could prove too costly during training. So NaN values were replaced with mean of that column. And then StandardScaler was applied to scale the data and avoid dominance of a few features during training.",
"_____no_output_____"
],
[
"### Step 2.2: Perform Dimensionality Reduction\n\nOn our scaled data, we are now ready to apply dimensionality reduction techniques.\n\n- Let's use sklearn's [PCA](http://scikit-learn.org/stable/modules/generated/sklearn.decomposition.PCA.html) class to apply principal component analysis on the data, thus finding the vectors of maximal variance in the data. To start, let's not set any parameters (so all components are computed).\n- Check out the ratio of variance explained by each principal component as well as the cumulative variance explained. Based on what we find, let's select a value for the number of transformed features we'll retain for the clustering part of the project.\n- Once we've made a choice for the number of components to keep, we'll re-fit a PCA instance to perform the decided-on transformation.",
"_____no_output_____"
]
],
[
[
"# Apply PCA to the data.\nfrom sklearn.decomposition import PCA\npca_obj = PCA()\nazdias_clean_no_nan_scaled_pca = pca_obj.fit_transform(azdias_clean_no_nan_scaled)\nazdias_clean_no_nan_scaled_pca.shape",
"_____no_output_____"
],
[
"def scree_plot(pca):\n '''\n Creates a scree plot associated with the principal components \n \n INPUT: pca - the result of instantian of PCA in scikit learn\n \n OUTPUT:\n None\n '''\n num_components=len(pca.explained_variance_ratio_)\n ind = np.arange(num_components)\n vals = pca.explained_variance_ratio_\n \n plt.figure(figsize=(14, 8))\n plt.grid()\n ax = plt.subplot(111)\n cumvals = np.cumsum(vals)\n ax.bar(ind, vals)\n ax.plot(ind, cumvals)\n #for i in range(num_components):\n # ax.annotate(r\"%s%%\" % ((str(vals[i]*100)[:4])), (ind[i]+0.2, vals[i]), va=\"bottom\", ha=\"center\", fontsize=6)\n \n ax.xaxis.set_tick_params(width=0)\n ax.yaxis.set_tick_params(width=2, length=12)\n \n ax.set_xlabel(\"Principal Component\")\n ax.set_ylabel(\"Variance Explained (%)\")\n plt.title('Explained Variance Per Principal Component')",
"_____no_output_____"
],
[
"# Investigate the variance accounted for by each principal component.\nscree_plot(pca_obj)",
"/opt/conda/lib/python3.6/site-packages/matplotlib/cbook/deprecation.py:106: MatplotlibDeprecationWarning: Adding an axes using the same arguments as a previous axes currently reuses the earlier instance. In a future version, a new instance will always be created and returned. Meanwhile, this warning can be suppressed, and the future behavior ensured, by passing a unique label to each axes instance.\n warnings.warn(message, mplDeprecation, stacklevel=1)\n"
],
[
"# Re-apply PCA to the data while selecting for number of components to retain.\npca_obj = PCA(40)\nazdias_clean_no_nan_scaled_pca = pca_obj.fit_transform(azdias_clean_no_nan_scaled)\nazdias_clean_no_nan_scaled_pca.shape",
"_____no_output_____"
]
],
[
[
"### Discussion 2.2: Perform Dimensionality Reduction\nFrom the scree plot, we see that 70% of the variance is explained using 40 features which is reasonable to move ahead. We could go higher but the worry is it could lead to overfitting on training data.",
"_____no_output_____"
],
[
"### Step 2.3: Interpret Principal Components\n\nNow that we have our transformed principal components, it's a nice idea to check out the weight of each variable on the first few components to see if they can be interpreted in some fashion.\n\nAs a reminder, each principal component is a unit vector that points in the direction of highest variance (after accounting for the variance captured by earlier principal components). The further a weight is from zero, the more the principal component is in the direction of the corresponding feature. If two features have large weights of the same sign (both positive or both negative), then increases in one tend expect to be associated with increases in the other. To contrast, features with different signs can be expected to show a negative correlation: increases in one variable should result in a decrease in the other.\n\n- To investigate the features, we should map each weight to their corresponding feature name, then sort the features according to weight. The most interesting features for each principal component, then, will be those at the beginning and end of the sorted list. Using the data dictionary document can help us understand these most prominent features, their relationships, and what a positive or negative value on the principal component might indicate.\n- Let's investigate and interpret feature associations from the first three principal components in this substep. To help facilitate this, let's write a function that we can call at any time to print the sorted list of feature weights, for the *i*-th principal component. This might come in handy in the next step of the project, when we interpret the tendencies of the discovered clusters.",
"_____no_output_____"
]
],
[
[
"def weight_plot_map(inp_pca, n_comp):\n hahaha=pd.DataFrame(data = inp_pca.components_, columns = azdias_no_outlier_low_miss_encoded.columns.values).iloc[n_comp-1].sort_values(ascending=False)\n hahaha.plot(kind='bar', figsize=(30,4), fontsize=12)\n print(hahaha)",
"_____no_output_____"
],
[
"# Map weights for the first principal component to corresponding feature names\n# and then print the linked values, sorted by weight.\n# HINT: Try defining a function here or in a new cell that you can reuse in the\n# other cells.\nweight_plot_map(pca_obj, 1)\n",
"LP_STATUS_GROB_1.0 0.198658\nHH_EINKOMMEN_SCORE 0.187102\nPLZ8_ANTG3 0.186971\nCAMEO_INTL_2015_WEALTH 0.184444\nPLZ8_ANTG4 0.181426\nORTSGR_KLS9 0.158900\nEWDICHTE 0.157273\nFINANZ_SPARER 0.147076\nFINANZ_HAUSBAUER 0.141685\nFINANZTYP_1 0.131579\nKBA05_ANTG4 0.130184\nPLZ8_ANTG2 0.127267\nKBA05_ANTG3 0.118237\nANZ_HAUSHALTE_AKTIV 0.116600\nARBEIT 0.116061\nCAMEO_DEUG_2015_9 0.114685\nSEMIO_PFLICHT 0.112669\nRELAT_AB 0.109172\nPRAEGENDE_JUGENDJAHRE_DECADE 0.108794\nSEMIO_REL 0.107840\nSEMIO_RAT 0.095493\nSEMIO_TRADV 0.091896\nZABEOTYP_5 0.091880\nCAMEO_DEUG_2015_8 0.090571\nFINANZ_UNAUFFAELLIGER 0.081974\nFINANZ_ANLEGER 0.079209\nSEMIO_MAT 0.077217\nLP_FAMILIE_GROB_1.0 0.074712\nSEMIO_FAM 0.071860\nGEBAEUDETYP_3.0 0.068958\n ... \nCJT_GESAMTTYP_2.0 -0.061127\nWOHNDAUER_2008 -0.062903\nKBA13_ANZAHL_PKW -0.062991\nCAMEO_DEUG_2015_3 -0.064511\nCAMEO_DEUG_2015_4 -0.071454\nANZ_PERSONEN -0.073300\nSEMIO_ERL -0.074959\nNATIONALITAET_KZ_1.0 -0.075077\nSEMIO_LUST -0.080402\nCAMEO_DEUG_2015_2 -0.088937\nGEBAEUDETYP_1.0 -0.091098\nFINANZTYP_2 -0.094185\nZABEOTYP_1 -0.094351\nGEBAEUDETYP_RASTER -0.099653\nBALLRAUM -0.100080\nPRAEGENDE_JUGENDJAHRE_MOVEMENT -0.106425\nGREEN_AVANTGARDE -0.107465\nLP_STATUS_GROB_4.0 -0.113172\nCAMEO_INTL_2015_LIFE_STAGE -0.115165\nFINANZ_VORSORGER -0.115960\nLP_STATUS_GROB_5.0 -0.119608\nALTERSKATEGORIE_GROB -0.119835\nINNENSTADT -0.131654\nPLZ8_GBZ -0.137680\nKONSUMNAEHE -0.138924\nKBA05_GBZ -0.186279\nPLZ8_ANTG1 -0.187877\nKBA05_ANTG1 -0.196420\nFINANZ_MINIMALIST -0.208781\nMOBI_REGIO -0.209786\nName: 0, Length: 127, dtype: float64\n"
],
[
"# Map weights for the second principal component to corresponding feature names\n# and then print the linked values, sorted by weight.\nweight_plot_map(pca_obj, 2)\n",
"ALTERSKATEGORIE_GROB 0.223224\nFINANZ_VORSORGER 0.207461\nZABEOTYP_3 0.203349\nSEMIO_ERL 0.185919\nSEMIO_LUST 0.157013\nRETOURTYP_BK_S 0.155371\nW_KEIT_KIND_HH 0.125182\nFINANZ_HAUSBAUER 0.116510\nCJT_GESAMTTYP_2.0 0.102830\nPLZ8_ANTG3 0.098593\nPLZ8_ANTG4 0.094376\nFINANZTYP_5 0.094104\nEWDICHTE 0.090488\nORTSGR_KLS9 0.089819\nSEMIO_KRIT 0.089229\nCAMEO_INTL_2015_WEALTH 0.087250\nFINANZTYP_2 0.079125\nSEMIO_KAEM 0.078558\nSHOPPER_TYP_3.0 0.077621\nKBA05_ANTG4 0.073991\nARBEIT 0.069146\nCJT_GESAMTTYP_1.0 0.068130\nPLZ8_ANTG2 0.067319\nHH_EINKOMMEN_SCORE 0.067085\nANZ_HAUSHALTE_AKTIV 0.066377\nLP_FAMILIE_GROB_1.0 0.065106\nRELAT_AB 0.064881\nFINANZTYP_6 0.061593\nGFK_URLAUBERTYP_4.0 0.060890\nANREDE_KZ 0.057578\n ... \nSHOPPER_TYP_0.0 -0.061331\nLP_FAMILIE_GROB_4.0 -0.066404\nGFK_URLAUBERTYP_9.0 -0.070969\nSEMIO_SOZ -0.071906\nKONSUMNAEHE -0.072768\nINNENSTADT -0.073268\nANZ_PERSONEN -0.073810\nPLZ8_GBZ -0.075641\nZABEOTYP_1 -0.080149\nZABEOTYP_5 -0.081509\nFINANZTYP_3 -0.083527\nKBA05_ANTG1 -0.090971\nFINANZTYP_4 -0.093456\nKBA05_GBZ -0.096746\nPLZ8_ANTG1 -0.097832\nZABEOTYP_4 -0.099552\nMOBI_REGIO -0.100547\nFINANZTYP_1 -0.111288\nSEMIO_MAT -0.126731\nSEMIO_FAM -0.138299\nSEMIO_RAT -0.153837\nONLINE_AFFINITAET -0.168647\nSEMIO_KULT -0.173676\nFINANZ_ANLEGER -0.189897\nSEMIO_PFLICHT -0.197465\nSEMIO_TRADV -0.202611\nFINANZ_UNAUFFAELLIGER -0.207458\nFINANZ_SPARER -0.209150\nSEMIO_REL -0.213163\nPRAEGENDE_JUGENDJAHRE_DECADE -0.226454\nName: 1, Length: 127, dtype: float64\n"
],
[
"# Map weights for the third principal component to corresponding feature names\n# and then print the linked values, sorted by weight.\nweight_plot_map(pca_obj, 3)\n",
"SEMIO_VERT 0.327483\nSEMIO_SOZ 0.258983\nSEMIO_FAM 0.258336\nSEMIO_KULT 0.243967\nFINANZTYP_5 0.140133\nFINANZ_MINIMALIST 0.132838\nSHOPPER_TYP_0.0 0.127000\nZABEOTYP_1 0.108309\nSEMIO_REL 0.104724\nRETOURTYP_BK_S 0.090257\nW_KEIT_KIND_HH 0.080821\nSEMIO_MAT 0.076921\nFINANZ_VORSORGER 0.065030\nGREEN_AVANTGARDE 0.055584\nEWDICHTE 0.054876\nPRAEGENDE_JUGENDJAHRE_MOVEMENT 0.054823\nORTSGR_KLS9 0.054603\nZABEOTYP_6 0.051469\nPLZ8_ANTG4 0.049219\nPLZ8_ANTG3 0.048843\nSHOPPER_TYP_1.0 0.045319\nSEMIO_LUST 0.037813\nLP_STATUS_GROB_5.0 0.036983\nALTERSKATEGORIE_GROB 0.036663\nPLZ8_ANTG2 0.034389\nGFK_URLAUBERTYP_4.0 0.033248\nARBEIT 0.033187\nRELAT_AB 0.032846\nGEBAEUDETYP_3.0 0.029140\nKBA05_ANTG4 0.027850\n ... \nMOBI_REGIO -0.031327\nGFK_URLAUBERTYP_9.0 -0.031737\nZABEOTYP_5 -0.032170\nCJT_GESAMTTYP_6.0 -0.032363\nNATIONALITAET_KZ_3.0 -0.034528\nPLZ8_GBZ -0.037237\nONLINE_AFFINITAET -0.038365\nSEMIO_TRADV -0.038586\nGEBAEUDETYP_RASTER -0.038617\nSEMIO_PFLICHT -0.039893\nBALLRAUM -0.042497\nSHOPPER_TYP_3.0 -0.044751\nKONSUMNAEHE -0.045498\nFINANZ_HAUSBAUER -0.048565\nPLZ8_ANTG1 -0.048701\nINNENSTADT -0.049807\nLP_FAMILIE_GROB_3.0 -0.051640\nFINANZ_UNAUFFAELLIGER -0.062906\nZABEOTYP_4 -0.069280\nFINANZ_SPARER -0.069330\nPRAEGENDE_JUGENDJAHRE_DECADE -0.070919\nSHOPPER_TYP_2.0 -0.101846\nFINANZTYP_1 -0.106967\nFINANZ_ANLEGER -0.160311\nSEMIO_RAT -0.173746\nSEMIO_ERL -0.196491\nSEMIO_KRIT -0.266131\nSEMIO_DOM -0.294695\nSEMIO_KAEM -0.324172\nANREDE_KZ -0.352261\nName: 2, Length: 127, dtype: float64\n"
]
],
[
[
"### Discussion 2.3: Interpret Principal Components\n\n1. Component1 (**Financial status**): Positive heavy weights LP_STATUS_GROB_1.0, HH_EINKOMMEN_SCORE, CAMEO_INTL_2015_WEALTH are all related to financial status with higher number indicating poorer and lower number indicating richer which is why they are positively correlated. It's also proven by the fact that PLZ8_ANTG3, PLZ8_ANTG4 positively correlate with the previous features but negatively correlate with negative heavy weight PLZ8_ANTG1(1-2 person family homes indicating richer neighborhood).\n\n\n2. Component2 (**Age**): Looking at the heavy positive & negative weights, we can see that (PRAEGENDE_JUGENDJAHRE_DECADE), (SEMIO_ERL), -ve (RETOURTYP_BK_S), -ve (FINANZ_SPARER) all behave the same way, in other words **youth is more associated with being event oriented, crazy shoppers, financially not savvy** and the opposite for older population.\n\n\n3. Component3 (**Personality**): Most of the heavy weights in this component deal with personality type. Positive heavy weights (SEMIO_SOZ, SEMIO_FAM, SEMIO_VERT, SEMIO_KULT) show that people who are more socially minded also are more family minded, dreamful and culturally minded. On the other hand, people who have less of above mentioned personalities tend to be more SEMIO_RAT (rational), SEMIO_KAEM (combative), SEMIO_ERL(event oriented, SEMIO_KRIT (critical), SEMIO_DOM (dominant) as indicated by heavy negative weights.\n ",
"_____no_output_____"
],
[
"## Step 3: Clustering\n\n### Step 3.1: Apply Clustering to General Population\n\nWe've assessed and cleaned the demographics data, then scaled and transformed them. Now, it's time to see how the data clusters in the principal components space. In this substep, we will apply k-means clustering to the dataset and use the average within-cluster distances from each point to their assigned cluster's centroid to decide on a number of clusters to keep.\n\n- We'll use sklearn's [KMeans](http://scikit-learn.org/stable/modules/generated/sklearn.cluster.KMeans.html#sklearn.cluster.KMeans) class to perform k-means clustering on the PCA-transformed data.\n- Then, compute the average difference from each point to its assigned cluster's center. The KMeans object's `.score()` method can be useful here.\n- We'll perform the above two steps for a number of different cluster counts. That'll show us how the average distance decreases with an increasing number of clusters. However, each additional cluster provides a smaller net benefit. We'll use this fact to select a final number of clusters in which to group the data.\n- Once we've selected a final number of clusters to use, we'll re-fit a KMeans instance to perform the clustering operation. The cluster assignments for the general demographics data should also be obtained, since we'll be using them in the final Step 3.3.",
"_____no_output_____"
]
],
[
[
"# function that generates internal score given input data and number of clusters\ndef get_kmeans_score(data, center):\n '''\n returns the kmeans score regarding SSE for points to centers\n INPUT:\n data - the dataset you want to fit kmeans to\n center - the number of centers you want (the k value)\n OUTPUT:\n score - the SSE score for the kmeans model fit to the data\n '''\n #instantiate kmeans\n kmeans = KMeans(n_clusters=center)\n\n # Then fit the model to your data using the fit method\n model = kmeans.fit(data)\n \n # Obtain a score related to the model fit\n score = np.abs(model.score(data))\n \n return score",
"_____no_output_____"
],
[
"#generate scores for a bunch of clusters and plot them to identify ideal clsuters using elbow method\nscores = []\ncenters = list(range(1,11,2)) + [15, 20, 25]\n\nfor center in centers:\n scores.append(get_kmeans_score(azdias_clean_no_nan_scaled_pca, center))\n \nplt.plot(centers, scores, linestyle='--', marker='o', color='b');\nplt.xlabel('K');\nplt.ylabel('SSE');\nplt.title('SSE vs. K');",
"_____no_output_____"
],
[
"# Re-fit the k-means model with the selected number of clusters and obtain\n# cluster predictions for the general population demographics data.\nkmeans_gen_pop = KMeans(n_clusters=15)\nmodel_gen_pop = kmeans_gen_pop.fit(azdias_clean_no_nan_scaled_pca)\npredict_gen_pop = model_gen_pop.predict(azdias_clean_no_nan_scaled_pca)",
"_____no_output_____"
]
],
[
[
"### Discussion 3.1: Apply Clustering to General Population\n\nLooking at the score vs cluster number plot, we can see that the internal distance dropoff rate reduces significantly from 9 clusters which means that we will not gain too much benefit by adding more clusters. But to be conservative, let's go with the next data point, 15 clusters.",
"_____no_output_____"
],
[
"### Step 3.2: Apply All Steps to the Customer Data\n\nNow that we have clusters and cluster centers for the general population, it's time to see how the customer data maps on to those clusters. We're going to use the fits from the general population to clean, transform, and cluster the customer data. In the last step of the project, we will interpret how the general population fits apply to the customer data.\n\n- We'll apply the same feature wrangling, selection, and engineering steps to the customer demographics using the `clean_data()` function we created earlier. (we can assume that the customer demographics data has similar meaning behind missing data patterns as the general demographics data.)\n- We'll use the sklearn objects from the general demographics data, and apply their transformations to the customers data. That is, we should not be using a `.fit()` or `.fit_transform()` method to re-fit the old objects, nor should we be creating new sklearn objects! We'll carry the data through the feature scaling, PCA, and clustering steps, obtaining cluster assignments for all of the data in the customer demographics data.",
"_____no_output_____"
]
],
[
[
"# Load in the customer demographics data.\ncustomers = pd.read_csv(\"Udacity_CUSTOMERS_Subset.csv\", delimiter=\";\")",
"_____no_output_____"
],
[
"# Apply preprocessing, feature transformation, and clustering from the general\n# demographics onto the customer data, obtaining cluster predictions for the\n# customer demographics data.\ncustomers_clean = clean_data(customers)\n\n# Replace any missed columns in customer data with mean values from general data to get same shape\nmissing_columns_customer_data = azdias_no_outlier_low_miss_encoded.columns.difference(customers_clean.columns)\nfor each_missed_column in missing_columns_customer_data:\n customers_clean[each_missed_column] = azdias_no_outlier_low_miss_encoded[each_missed_column].mean()\n\n#remove nan\ncustomers_clean_no_nan = imp_median.transform(customers_clean)\n#apply feature scaling\ncustomers_clean_no_nan_scaled = scaler_obj.transform(customers_clean_no_nan)\n#apply pca feature transformation\ncustomers_clean_no_nan_scaled_pca = pca_obj.transform(customers_clean_no_nan_scaled)\n#predict using kmeans model for general population\npredict_customers = model_gen_pop.predict(customers_clean_no_nan_scaled_pca)",
"_____no_output_____"
]
],
[
[
"### Step 3.3: Compare Customer Data to Demographics Data\n\nAt this point, we have clustered data based on demographics of the general population of Germany, and seen how the customer data for a mail-order sales company maps onto those demographic clusters. In this final substep, we will compare the two cluster distributions to see where the strongest customer base for the company is.\n\nConsider the proportion of persons in each cluster for the general population, and the proportions for the customers. If we think the company's customer base to be universal, then the cluster assignment proportions should be fairly similar between the two. If there are only particular segments of the population that are interested in the company's products, then we should see a mismatch from one to the other. If there is a higher proportion of persons in a cluster for the customer data compared to the general population (e.g. 5% of persons are assigned to a cluster for the general population, but 15% of the customer data is closest to that cluster's centroid) then that suggests the people in that cluster to be a target audience for the company. On the other hand, the proportion of the data in a cluster being larger in the general population than the customer data (e.g. only 2% of customers closest to a population centroid that captures 6% of the data) suggests that group of persons to be outside of the target demographics.\n\nWe'll investigate the following points in this step:\n\n- Compute the proportion of data points in each cluster for the general population and the customer data. Visualizations will be useful here.\n- Which cluster or clusters are overrepresented in the customer dataset compared to the general population? Select at least one such cluster and infer what kind of people might be represented by that cluster.\n- Which cluster or clusters are underrepresented in the customer dataset compared to the general population, and what kinds of people are typified by these clusters?",
"_____no_output_____"
]
],
[
[
"uniq_gen, cnts_gen = np.unique(predict_gen_pop, return_counts = True)\nprint(\"Cluster number and corresponding % of general population\")\npprint.pprint(dict(zip(uniq_gen+1, cnts_gen*100/predict_gen_pop.shape[0])))",
"Cluster number and corresponding % of general population\n{1: 5.7177734440356094,\n 2: 6.6105481261770374,\n 3: 7.5169754346420667,\n 4: 7.3127214368987659,\n 5: 7.6559109848763702,\n 6: 4.9998059185473602,\n 7: 7.8293796487527256,\n 8: 6.4178051663144196,\n 9: 4.5041620432895337,\n 10: 7.1634794922829199,\n 11: 5.6555335299133054,\n 12: 8.0771346617427984,\n 13: 7.1921232377069479,\n 14: 8.9650907698876612,\n 15: 4.3815561049324794}\n"
],
[
"uniq_cust, cnts_cust = np.unique(predict_customers, return_counts = True)\nprint(\"Cluster number and corresponding % of customer population\")\npprint.pprint(dict(zip(uniq_cust+1, cnts_cust*100/predict_customers.shape[0])))",
"Cluster number and corresponding % of customer population\n{1: 1.5259280355550227,\n 2: 0.80268611300561354,\n 3: 2.1937089194840627,\n 4: 3.4895485921140397,\n 5: 0.58458932599848601,\n 6: 16.669789472895292,\n 7: 0.30353676542229086,\n 8: 43.477706910895094,\n 9: 3.4633170197935952,\n 10: 5.4381796787756604,\n 11: 6.4117457486116001,\n 12: 8.9157366949717822,\n 13: 1.9486310866616201,\n 14: 0.6497935200521634,\n 15: 4.125102115763676}\n"
],
[
"print(\"Cluster number and corresponding % (rounded to nearest integer) of general and customer population\")\nprint(\" No. Gen Cust\")\npprint.pprint(list(zip(uniq_gen+1, np.round_(cnts_gen*100/predict_gen_pop.shape[0]), np.round_(cnts_cust*100/predict_customers.shape[0]))))",
"Cluster number and corresponding % (rounded to nearest integer) of general and customer population\n No. Gen Cust\n[(1, 6.0, 2.0),\n (2, 7.0, 1.0),\n (3, 8.0, 2.0),\n (4, 7.0, 3.0),\n (5, 8.0, 1.0),\n (6, 5.0, 17.0),\n (7, 8.0, 0.0),\n (8, 6.0, 43.0),\n (9, 5.0, 3.0),\n (10, 7.0, 5.0),\n (11, 6.0, 6.0),\n (12, 8.0, 9.0),\n (13, 7.0, 2.0),\n (14, 9.0, 1.0),\n (15, 4.0, 4.0)]\n"
],
[
"#Vizualization of population distribution among clusters\nclust_pop_df = pd.DataFrame(data=cnts_gen*100/predict_gen_pop.shape[0], index = uniq_gen+1, columns = ['General Population'])\nclust_pop_df['Customer Population'] = (cnts_cust*100/predict_customers.shape[0])\nclust_pop_df.plot(kind='bar', figsize=(18,8), fontsize=18)",
"_____no_output_____"
],
[
"# Customer data (cluster = 8) first three component weights\ncustomers_clean_no_nan_scaled_pca[predict_customers == 7][:10,:3]",
"_____no_output_____"
],
[
"# Customer data (cluster = 6) first three component weights\ncustomers_clean_no_nan_scaled_pca[predict_customers == 5][:10,:3]\n",
"_____no_output_____"
],
[
"# Customer data (cluster = 14) first three component weights\ncustomers_clean_no_nan_scaled_pca[predict_customers == 13][:10,:3]",
"_____no_output_____"
],
[
"# Customer data (cluster = 5) first three component weights\ncustomers_clean_no_nan_scaled_pca[predict_customers == 4][:10,:3]",
"_____no_output_____"
]
],
[
[
"### Discussion 3.3: Compare Customer Data to Demographics Data\n\nThe above bar chart distribution of general & customer population shows that clusters 8 & 6 are over-represented in Customer data and clusters 14 & 5 are over-represented in General population data. \n 1. If we look at the first 3 principal component weights for clusters 8 & 6, we can see that they have heavy negative correlation with first component (aka FINANCIAL STATUS). The next two components don't seem to follow a pattern though cluster 8 has strong positive correlation with third component (aka PERSONALITY), and cluster 6 has positive correlation with second component (aka AGE).\n\n\n 2. Similarly, if we look at the first 3 principal component weights for clusters 14 & 5, we can see that the opposite is true. The first component (aka FINANCIAL STATUS) has a positive correlation with those clusters. The other two components do not seem to follow a pattern.\n \nConclusion: Based on the interpretation of principal components and what we see from the cluster data, FINANCIAL STATUS has a positive correlation with potential customer base. In other words, richer population has higher chance of signing up for mail order compared to poorer population. To further study the effects of AGE and PERSONALITY, these over-represented and under-represented clusters need to be separated out from main dataset and further clustering needs to be done on them.\n ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecca45c7f841586b71c591c1056df01f8b3488f0 | 6,512 | ipynb | Jupyter Notebook | notebooks/NER_conll_example.ipynb | JoaoLages/relational-transformers | 7bf401afe8e2b12531dc75f5402c2f9ffd99aaf7 | [
"MIT"
] | 30 | 2022-02-15T16:56:27.000Z | 2022-03-31T17:46:10.000Z | notebooks/NER_conll_example.ipynb | JoaoLages/relational-transformers | 7bf401afe8e2b12531dc75f5402c2f9ffd99aaf7 | [
"MIT"
] | null | null | null | notebooks/NER_conll_example.ipynb | JoaoLages/relational-transformers | 7bf401afe8e2b12531dc75f5402c2f9ffd99aaf7 | [
"MIT"
] | 2 | 2022-03-03T07:11:16.000Z | 2022-03-10T17:00:22.000Z | 28.561404 | 167 | 0.464834 | [
[
[
"!pip install datasets",
"_____no_output_____"
],
[
"from datasets import load_dataset\nimport ratransformers\n\n# Load dataset\ndataset = load_dataset('conll2003')\n\n# copied from https://huggingface.co/datasets/conll2003\npos_tag_to_id = {'\"': 0, \"''\": 1, '#': 2, '$': 3, '(': 4, ')': 5, ',': 6, '.': 7, ':': 8, '``': 9, 'CC': 10, 'CD': 11, 'DT': 12,\n 'EX': 13, 'FW': 14, 'IN': 15, 'JJ': 16, 'JJR': 17, 'JJS': 18, 'LS': 19, 'MD': 20, 'NN': 21, 'NNP': 22, 'NNPS': 23,\n 'NNS': 24, 'NN|SYM': 25, 'PDT': 26, 'POS': 27, 'PRP': 28, 'PRP$': 29, 'RB': 30, 'RBR': 31, 'RBS': 32, 'RP': 33,\n 'SYM': 34, 'TO': 35, 'UH': 36, 'VB': 37, 'VBD': 38, 'VBG': 39, 'VBN': 40, 'VBP': 41, 'VBZ': 42, 'WDT': 43,\n 'WP': 44, 'WP$': 45, 'WRB': 46}\n\nid_to_pos_tag = {v: k for k, v in pos_tag_to_id.items()}",
"Reusing dataset conll2003 (/home/ola/.cache/huggingface/datasets/conll2003/conll2003/1.0.0/63f4ebd1bcb7148b1644497336fd74643d4ce70123334431a3c053b7ee4e96ee)\n"
],
[
"from transformers import AutoModelForTokenClassification\n\n# Load ratransformer model and tokenizer\nratransformer = ratransformers.RATransformer(\n \"dslim/bert-base-NER\", \n relation_kinds=list(pos_tag_to_id),\n model_cls=AutoModelForTokenClassification\n)\nmodel = ratransformer.model\ntokenizer = ratransformer.tokenizer",
"_____no_output_____"
],
[
"dataset['train'][0]",
"_____no_output_____"
],
[
"from collections import defaultdict\n\n# Construct a map from span in text to POS_TAG\nword_relations = defaultdict(dict)\nspan_init = 0\nfor tok, pos_tag_id in zip(dataset['train'][0]['tokens'], dataset['train'][0]['pos_tags']):\n span = (span_init, span_init + len(tok))\n word_relations[span][span] = id_to_pos_tag[pos_tag_id]\n span_init = span_init + len(tok + ' ')\nword_relations",
"_____no_output_____"
],
[
"# encode \ntext = \" \".join(dataset['train'][0]['tokens'])\nencoding = tokenizer(\n text, \n return_tensors=\"pt\", \n input_relations=word_relations\n)\n\n# forward pass\noutputs = model(**encoding)\n\n# get labels ids and convert to label tags\nlabels = outputs.logits.argmax(-1)\ntokens_to_labels = [model.config.id2label[label_id.item()] for label_id in labels[0]]\n\n# print tokens with their predicted NER tags\nfor i, token_i_map in enumerate(encoding['offset_mapping'][0]):\n span = token_i_map.tolist()\n token = text[span[0]:span[1]]\n if token: # skip special tokens\n print(token, tokens_to_labels[i])",
"EU B-ORG\nrejects O\nGerman B-MISC\ncall O\nto O\nboycott O\nBritish B-MISC\nla O\nmb O\n. O\n"
]
],
[
[
"**Your model is now ready to be trained with relational information in the input!**\n\nCheck the standard procedure to train HuggingFace 🤗 models in [here](https://huggingface.co/docs/transformers/training).",
"_____no_output_____"
]
]
] | [
"code",
"markdown"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecca46af455dab06dbdd7d8c723877afa8210184 | 12,107 | ipynb | Jupyter Notebook | superviselySDK/help/jupyterlab_scripts/src/tutorials/06_inference_modes/inference_modes.ipynb | nicehuster/mmdetection-supervisely-person-datasets | ff1b57e16a71378510571dbb9cebfdb712656927 | [
"Apache-2.0"
] | 40 | 2019-05-05T08:08:18.000Z | 2021-10-17T00:07:58.000Z | superviselySDK/help/jupyterlab_scripts/src/tutorials/06_inference_modes/inference_modes.ipynb | nicehuster/mmdetection-supervisely-person-datasets | ff1b57e16a71378510571dbb9cebfdb712656927 | [
"Apache-2.0"
] | 8 | 2019-06-13T06:00:08.000Z | 2021-07-24T05:25:33.000Z | superviselySDK/help/jupyterlab_scripts/src/tutorials/06_inference_modes/inference_modes.ipynb | nicehuster/mmdetection-supervisely-person-datasets | ff1b57e16a71378510571dbb9cebfdb712656927 | [
"Apache-2.0"
] | 6 | 2019-07-30T06:36:27.000Z | 2021-06-03T11:57:36.000Z | 40.76431 | 740 | 0.573305 | [
[
[
"## Inference modes\n\nThe most usual approach of using neural networks for computer vision is to capture an image and feed the whole image to the neural network for inference. However, sometimes more complex logic is necessary, depending on the application. Some examples from our experience:\n* Crop off the boundaries of the image and only run inference on the inner part.\n* Run over the input image with a sliding window, run inference on each fragment and then aggregate to get the overall result.\n* In a pipeline setting, run inference only on specially selected regions. A typical example of this is first running a detection model on the whole image to get rough locations of the objects, and then run a segmentation model to find fine-grained contours on the objects *only within the locations found by the detector*.\n\nThe neural networks available out of the box in Supervisely all support the above *inferfence modes* with no code modifications. Only inference config for a given inference task need to be changed. Moreover, since this advanced logic is independent of the specific neural network being used, we have factored it out as a part of `supervisely_lib` SDK. It is trivial to integrate the available inference modes with your custom models also, as long as you rely on our `SingleImageInferenceBase` base class for inference. See [our guide on how to integrate your custom neural network with the Supervisely platform](https://github.com/supervisely/supervisely/blob/master/help/tutorials/02_custom_neural_net_plugin/custom_nn_plugin.md).\n\nIn this tutorial we will go over the examples of available inference modes for neural networks in Supervisely.\n\nOne important thing to keep in mind is that all the inference modes produce labels in the context of the *full original image*. So, for example, if the original image is cropped before passing it to the neural network, the inference results from",
"_____no_output_____"
],
[
"### Inference config structure\n\nWith all the Supervisely built-in neural networks, the inference mode settings and the settings of the specific network itself are decomposed into different sections of the overall inference task config as follows:\n```python\n{\n \"model\": {\n # Config of the specific neural network.\n # Detection thresholds, other runtime options.\n },\n \"mode\": {\n # Inference mode config. Options used here are general\n # and do not depend on the specific neural network being used.\n }\n}\n```",
"_____no_output_____"
],
[
"### Full image inference\n\nThe most basic inference mode is to pass the whole incoming image to the neural network for inference. Example config:\n\n```python\n{\n \"model\": {\n \"gpu_device\": 0 # Use GPU #0 for this model.\n },\n \"mode\": {\n\n # Inference mode name, mandatory field.\n # \"full_image\" is the most basic mode, where all the image is\n # passed on to the neural net.\n \"name\": \"full_image\",\n \n # Mode-specific settings. For example, here we describe how to\n # post-process the labels returned by the model depending on\n # the object class.\n\n \"model_classes\": {\n # This section is actually common to all of the inference modes.\n \n # Add a suffix to all of the class names returned by the model.\n # This helps distinguish model labels from original labels when\n # using a labeled dataset as input.\n \"add_suffix\": \"_unet\",\n \n # Save all the labels produced by the models. Alternatively,\n # here one can specify a whitelist of class names to be saved,\n # e.g. [\"car\", \"person\"].\n \"save_classes\": \"__all__\"\n }\n }\n}\n```",
"_____no_output_____"
],
[
"### ROI (fixed crop) inference\n\nSometimes we know that only a certain part of the input images is relevant for inference. For example, if a camera is fixed on the windshield of a car, we know that the top part of the image is the sky, where we are not going to have pedestrians, so it makes sense to only run the pedestrian detector on the bottom part of the image. In such cases we want to crop off a *fixed boundary* of the input image before passing it on to the neural network. This logic is handled by the `roi` inference mode.\n\n```python\n{\n \"model\": {},\n \"mode\": {\n\n # Crop the image boundaries before inference.\n \"name\": \"roi\",\n \n # Class renaming and filtering settings.\n # See \"Full image inference\" example for details.\n \"model_classes\": {\n }\n \n # Cropping settings.\n # How much to crop from every side of the image.\n \"bounds\": {\n # The amount can be specified in pixels or\n # as a percentage of the corresponding dimension.\n \"left\": \"100px\",\n \"right\": \"50px\",\n \"top\": \"20%\",\n \"bottom\": \"20%\"\n },\n\n # Whether to add a bounding box of the cropped region\n # of interest as a separate label in the results.\n \"save\": false,\n\n # If saving the cropped region bounding box, which\n # class name to use.\n \"class_name\": 'inference_roi'\n }\n}\n```",
"_____no_output_____"
],
[
"### Sliding window inference\n\n#### Segmentation\n\nIn segmentation sliding window mode, the per-pixels class scores are summed up over all the sliding windows affecting the given pixel, and then the class with the maximum total score is taken as a label. Example config:\n\n```python\n{\n \"model\": {},\n \"mode\": {\n\n # Go over the original image with a sliding window,\n # sum up per class segmentation scores and find the\n # class with the highest score for every pixel.\n \"name\": \"sliding_window\",\n \n # Class renaming and filtering settings.\n # See \"Full image inference\" example for details.\n \"model_classes\": {\n }\n \n # Sliding window parameters.\n \n # Width and height in pixels.\n # Cannot be larger than the original image.\n \"window\": {\n \"width\": 128,\n \"height\": 128,\n },\n\n # Minimum overlap for each dimension. The last\n # window in every dimension may have higher overlap\n # with the previous one if necessary to fit the whole\n # window within the original image.\n \"min_overlap\": {\n \"x\": 0,\n \"y\": 0,\n },\n \n # Whether to save each sliding window instance as a\n # bounding box rectangle.\n \"save\": false,\n\n # If saving the sliding window bounding boxes, which\n # class name to use.\n \"class_name\": 'sliding_window_bbox',\n }\n}\n```\n\n#### Detection\nWith sliding windows for detection, instead of summing up per-pixel class scores, we accumulate the detection results (as rectangular bounding boxes). Optionally, non-maximum suppression is done for the final results. All of the options of the segmentation sliding window config apply, and an extra section with non-maximum suppression config is added:\n\n```python\n{\n \"model\": {},\n \"mode\": {\n\n \"name\": \"sliding_window_det\",\n \n # All the sliding window options from the segmentation\n # sliding window config also apply here.\n \n \"nms_after\": {\n \n # Whether to run non-maximum suppression after accumulating\n # all the detection results from the sliding windows.\n \"enable\": true,\n \n # Intersection over union threshold above which the same-class\n # detection labels are considered to be significantly inersected\n # for non-maximum suppression.\n \"iou_threshold\": 0.2,\n \n # Tag name from which to read detection confidence by which we\n # rank the detections. This tag must be added by the model to\n # every detection label.\n \"confidence_tag_name\": \"confidence\"\n }\n }\n}\n\n```",
"_____no_output_____"
],
[
"### Pipelining - bounding boxes ROI inference\n\nIn this mode, the neural network inference is invoked on subimages, defined by bounding boxes of existing labels. Those existing labels may come both from a previous run of another neural network (which allows us to pipeline the models), or from manual annotations. Example config:\n\n```python\n{\n \"model\": {},\n \"mode\": {\n\n # Run inference within bounding boxes of existing labels.\n \"name\": \"bboxes\",\n \n # Class renaming and filtering settings.\n # See \"Full image inference\" example for details.\n \"model_classes\": {\n }\n \n # Filter the source bounding boxes by their object class name.\n # Use \"__all__\" to pass through all the classes or a whitelist\n # of classes like [\"car\", \"person\"].\n \"from_classes\": \"__all__\",\n \n # Padding settings for the source bounding boxes.\n \"padding\": {\n # Padding can be set in pixels or percents of the respective side\n \"left\": \"20px\",\n # Negative numbers mean crop instead of pad.\n \"right\": \"-30px\",\n \"top\": \"20%\",\n \"bottom\": \"10%\"\n },\n \n # Whether to save the bounding boxes that were used for selecting subimages\n # for inference.\n \"save\": true,\n\n # Because the input bounding boxes may have originated from multiple\n # object classes, we do not want to assign the same class name to them all.\n # Instead, append a given suffix to the original class name to get the\n # saved bounding box class name.\n \"add_suffix\": \"_input_bbox\"\n }\n}\n```",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecca48374788c640c2fd82d8f41576c985cc65bd | 9,920 | ipynb | Jupyter Notebook | software/preprocessing/old_notebooks/circle_detect.ipynb | luzgool/ReachMaster | e7ef5087fb4e5c55e0b680fc00391f2f8cdc5367 | [
"BSD-3-Clause-LBNL"
] | 2 | 2020-09-19T21:04:59.000Z | 2021-07-27T15:57:49.000Z | software/preprocessing/old_notebooks/circle_detect.ipynb | luzgool/ReachMaster | e7ef5087fb4e5c55e0b680fc00391f2f8cdc5367 | [
"BSD-3-Clause-LBNL"
] | 1 | 2021-02-16T21:53:00.000Z | 2021-02-16T21:53:00.000Z | software/preprocessing/old_notebooks/circle_detect.ipynb | luzgool/ReachMaster | e7ef5087fb4e5c55e0b680fc00391f2f8cdc5367 | [
"BSD-3-Clause-LBNL"
] | 4 | 2020-02-08T22:41:35.000Z | 2021-09-10T20:33:00.000Z | 28.181818 | 183 | 0.518448 | [
[
[
"import os \nimport sys\nimport numpy as np\nfrom skimage.viewer import ImageViewer\nimport re\nimport glob\nimport collections \nfrom sklearn.cluster import KMeans\n\nros_path = '/opt/ros/kinetic/lib/python2.7/dist-packages'\nif ros_path in sys.path:\n sys.path.remove('/opt/ros/kinetic/lib/python2.7/dist-packages')\nimport cv2\n",
"/home/pns/anaconda3/lib/python3.6/site-packages/skimage/viewer/utils/core.py:10: UserWarning: Recommended matplotlib backend is `Agg` for full skimage.viewer functionality.\n warn(\"Recommended matplotlib backend is `Agg` for full \"\n"
]
],
[
[
"# Make Video Footage with circles\n## Manually draw circles\n",
"_____no_output_____"
]
],
[
[
"path = '.'\nfiles= [file for file in os.listdir(path) if file.endswith('.png')]\ndef byFrame(img):\n return int(re.search(r'\\d+', img[18:22]).group())\nfiles.sort(key = byFrame)\n",
"_____no_output_____"
],
[
"#All circle Locations\ncirc_locs = [(672,358),(383,390),(412,555),(460,718),(660,696),(865,710),(890,545),(935,375)]\n#(675,532) for frame 192\n#(673,528) for frame 328\n#(658,520) for frame 444\n#(652,523) for frame 555\n#(650,525) for frame 753\n#(656,528) for frame 866\n#(678, 539) for frame 985\navg_center = (663,528)",
"_____no_output_____"
],
[
"def label_circles(locs, img_name):\n img = cv2.imread(img_name,1)\n #same color circles\n for loc in locs:\n cv2.circle(img, loc, 63,(0,0,255),5)\n #center circle different color \n cv2.circle(img, avg_center, 63, (255,0,0),5)\n# #Outer Circle \n# cv2.circle(img, avg_center, 106, (255,0,0),2)\n return img \ndef label_images(locs, img_files):\n labeled_imgs = collections.deque()\n for img_name in img_files:\n new_img = label_circles(locs, img_name)\n labeled_imgs.append(new_img)\n return labeled_imgs ",
"_____no_output_____"
],
[
"# labeled = label_images(circ_locs, files)",
"_____no_output_____"
],
[
"# height, width, layers = labeled[0].shape\n\n# fps = 60\n# video = cv2.VideoWriter('labeled_movement.avi', cv2.VideoWriter_fourcc(*'XVID'), fps, (width,height))\n# for i in range(len(labeled)): \n# video.write(labeled[i])\n# cv2.destroyAllWindows()\n# video.release()",
"_____no_output_____"
]
],
[
[
"# Hough Transform ",
"_____no_output_____"
]
],
[
[
"def find_circles(img_name):\n start_param = 30\n max_param = 50\n img = cv2.imread(img_name, 1)\n cimg = cv2.medianBlur(img,5)\n gray = cv2.cvtColor(cimg, cv2.COLOR_BGR2GRAY)\n circles = cv2.HoughCircles(gray,cv2.HOUGH_GRADIENT,1,10,param1=50,param2=start_param,minRadius=65,maxRadius=80)\n while ((circles is not None) and (circles.shape[1] > 1)):\n start_param = (start_param +max_param)/2\n circles = cv2.HoughCircles(gray,cv2.HOUGH_GRADIENT,1,10,param1=50,param2=start_param,minRadius=65,maxRadius=80)\n circ_tup = (-100, -100, -10)\n if circles is not None:\n circles = np.uint16(np.around(circles))\n for i in circles[0,:]:\n # draw the outer circle\n cv2.circle(img,(i[0],i[1]),i[2],(0,255,0),2)\n # draw the center of the circle\n cv2.circle(img,(i[0],i[1]),2,(0,0,255),3)\n circ_tup = (i[0],i[1],i[2])\n return img, circ_tup\ndef circles_all(img_array):\n labeled = collections.deque()\n for img in img_array:\n new_img,circ_tup = find_circles(img)\n labeled.append(new_img)\n return labeled",
"_____no_output_____"
],
[
"def find_center_radius(img_array,num_clusters):\n circ_list = []\n for img in img_array:\n new_img,circ_tup = find_circles(img)\n# cv2.imwrite(\"circle\" + str(byFrame(img))+\".png\", new_img)\n circ_list.append(circ_tup)\n circ_mat = np.array(circ_list)\n kmeans = KMeans(n_clusters=num_clusters, random_state=0).fit(circ_mat[:,:2])\n return circ_mat, kmeans\ndef calc_center_radius(circ_mat, kmeans, num_clusters):\n avg_radius = []\n for j in range(10):\n indices = np.where(kmeans.labels_ == j)\n radius = circ_m[indices,2]\n avg_radius.append(np.average(radius))\n return np.hstack((np.rint(kmeans.cluster_centers_),np.rint(avg_radius)[:,np.newaxis]))\ndef label_hough_circles(circles, img_name):\n img = cv2.imread(img_name,1)\n for idx in range(len(circles)):\n c = circles[idx]\n if (c > 0).all():\n if idx ==0:\n cv2.circle(img, (int(c[0]),int(c[1])), int(c[2]),(255,0,0),5)\n else:\n cv2.circle(img, (int(c[0]),int(c[1])), int(c[2]),(0,0,255),5)\n return img \ndef label_hough_images(circles, img_files):\n labeled_imgs = collections.deque()\n for img_name in img_files:\n new_img = label_hough_circles(circles, img_name)\n labeled_imgs.append(new_img)\n return labeled_imgs \n",
"_____no_output_____"
],
[
"#10 clusters because we move to 9 positions and 1 extra cluster to account for frames where no circles found.\ncirc_m, kmeans = find_center_radius(files,10)",
"_____no_output_____"
],
[
"circ_ = calc_center_radius(circ_m, kmeans, 10)\ncirc_",
"_____no_output_____"
]
],
[
[
"# Auto-Draw Circles",
"_____no_output_____"
]
],
[
[
"#first image to half way point of images\nlabeled_auto = label_hough_images(circ_, files[0:1186])",
"_____no_output_____"
],
[
"# labeled_auto[0]\n# viewer = ImageViewer(labeled_auto[0])\n# viewer.show()",
"_____no_output_____"
],
[
"height, width, layers = labeled_auto[0].shape\n\nfps = 60\nvideo = cv2.VideoWriter('auto_hough_movement.mov', cv2.VideoWriter_fourcc('m', 'p', '4', 'v'), fps, (width,height))\n# video = cv2.VideoWriter('auto_hough_movement.mov', cv2.VideoWriter_fourcc(*'XVID'), fps, (width,height))\nfor i in range(len(labeled_auto)): \n video.write(labeled_auto[i])\ncv2.destroyAllWindows()\nvideo.release()",
"_____no_output_____"
],
[
"for filename in glob.glob(\"circle*\"):\n os.remove(filename) ",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecca4e38c93f4c74e2ea7ac5a28e079e0f6e5be1 | 244,866 | ipynb | Jupyter Notebook | Multindex.ipynb | MKamyab1991/Pandas_class | 8e3d1db17943ca91cd559a3dfa18961cadacf65b | [
"MIT"
] | 2 | 2022-02-25T22:02:53.000Z | 2022-02-26T02:20:35.000Z | Multindex.ipynb | MKamyab1991/Pandas_class | 8e3d1db17943ca91cd559a3dfa18961cadacf65b | [
"MIT"
] | null | null | null | Multindex.ipynb | MKamyab1991/Pandas_class | 8e3d1db17943ca91cd559a3dfa18961cadacf65b | [
"MIT"
] | null | null | null | 39.744522 | 1,475 | 0.360242 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"bigmac = pd.read_csv(\"bigmac.csv\", parse_dates=[\"Date\"])\nbigmac.head()",
"_____no_output_____"
],
[
"bigmac.dtypes",
"_____no_output_____"
],
[
"bigmac.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 652 entries, 0 to 651\nData columns (total 3 columns):\nDate 652 non-null datetime64[ns]\nCountry 652 non-null category\nPrice in US Dollars 652 non-null float64\ndtypes: category(1), datetime64[ns](1), float64(1)\nmemory usage: 13.9 KB\n"
],
[
"bigmac[\"Country\"].nunique()",
"_____no_output_____"
],
[
"bigmac[\"Country\"] = bigmac[\"Country\"].astype(\"category\")",
"_____no_output_____"
],
[
"bigmac.set_index(keys = [\"Date\",\"Country\"], inplace=True)",
"_____no_output_____"
],
[
"bigmac.sort_index(inplace=True)",
"_____no_output_____"
],
[
"bigmac.index.names",
"_____no_output_____"
],
[
"type(bigmac.index)",
"_____no_output_____"
],
[
"bigmac = pd.read_csv(\"bigmac.csv\", parse_dates=[\"Date\"], index_col=[\"Date\",\"Country\"])\nbigmac.sort_index(inplace=True)\nbigmac.head()",
"_____no_output_____"
],
[
"bigmac.index.get_level_values(0)",
"_____no_output_____"
],
[
"bigmac.index.set_names([\"Date\",\"Country\"], inplace=True)",
"_____no_output_____"
],
[
"bigmac.head()",
"_____no_output_____"
],
[
"bigmac.sort_index(ascending=[True,False])",
"_____no_output_____"
],
[
"bigmac.loc[(\"2010-01-01\",\"Norway\"),\"Price in US Dollars\"]",
"_____no_output_____"
],
[
"bigmac.transpose()\n\nbigmac.head()",
"_____no_output_____"
],
[
"bigmac.ix[\"Price in US Dollars\"]",
"_____no_output_____"
],
[
"bigmac = pd.read_csv(\"bigmac.csv\", parse_dates=[\"Date\"], index_col=[\"Date\",\"Country\"])\nbigmac.sort_index(inplace=True)\nbigmac.head()",
"_____no_output_____"
],
[
"bigmac.swaplevel()",
"_____no_output_____"
],
[
"world = pd.read_csv(\"worldstats.csv\", index_col=[\"country\",\"year\"])\nworld.head()",
"_____no_output_____"
],
[
"world.stack()",
"_____no_output_____"
],
[
"world.info()",
"<class 'pandas.core.frame.DataFrame'>\nMultiIndex: 11211 entries, (Arab World, 2015) to (Zimbabwe, 1960)\nData columns (total 2 columns):\nPopulation 11211 non-null float64\nGDP 11211 non-null float64\ndtypes: float64(2)\nmemory usage: 210.5+ KB\n"
],
[
"world.head()",
"_____no_output_____"
],
[
"type(world.stack().to_frame())",
"_____no_output_____"
],
[
"s = world.stack()",
"_____no_output_____"
],
[
"s.unstack()",
"_____no_output_____"
],
[
"s.unstack().unstack().unstack()",
"_____no_output_____"
],
[
"s.unstack(2)\ns.unstack(\"year\")",
"_____no_output_____"
],
[
"s.unstack(level = [\"country\",\"year\"])",
"_____no_output_____"
],
[
"t = s.unstack(level = [\"year\"], fill_value = 0)",
"_____no_output_____"
],
[
"t.head()\n",
"_____no_output_____"
],
[
"sales = pd.read_csv(\"salesmen.csv\", parse_dates = [\"Date\"])\nsales[\"Salesman\"] = sales[\"Salesman\"].astype(\"category\")",
"_____no_output_____"
],
[
"sales.head()",
"_____no_output_____"
],
[
"sales.pivot(index=\"Date\", columns=\"Salesman\", values=\"Revenue\")",
"_____no_output_____"
],
[
"foods = pd.read_csv(\"foods.csv\")\nfoods.head()",
"_____no_output_____"
],
[
"foods.pivot_table(values=\"Spend\", index=[\"Gender\",\"Item\"], columns=\"City\", aggfunc=\"sum\")",
"_____no_output_____"
],
[
"sales = pd.read_csv(\"quarters.csv\")\nsales.head()",
"_____no_output_____"
],
[
"sales.melt(id_vars=\"Salesman\",var_name=\"Quater\", value_name=\"Revenue\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecca54f1afc2a93297a17de7281536cfe9765874 | 5,897 | ipynb | Jupyter Notebook | Pytorch_demos/basic_tutorail/2.4 Pretrained_CNN_Nets.ipynb | guofei1989/DL_Wiglet | c64832a4c6262bbf837cefe8326c2fd5443c0a8f | [
"MIT"
] | 1 | 2020-07-24T02:17:55.000Z | 2020-07-24T02:17:55.000Z | Pytorch_demos/basic_tutorail/2.4 Pretrained_CNN_Nets.ipynb | guofei1989/DL_Wiglet | c64832a4c6262bbf837cefe8326c2fd5443c0a8f | [
"MIT"
] | null | null | null | Pytorch_demos/basic_tutorail/2.4 Pretrained_CNN_Nets.ipynb | guofei1989/DL_Wiglet | c64832a4c6262bbf837cefe8326c2fd5443c0a8f | [
"MIT"
] | 1 | 2020-07-25T07:29:17.000Z | 2020-07-25T07:29:17.000Z | 32.401099 | 95 | 0.502459 | [
[
[
"介绍了pytorch中官方版的AlexNet、VGG、Inception和ResNet实现",
"_____no_output_____"
],
[
"### 1. AlexNet",
"_____no_output_____"
]
],
[
[
"import torchvision\nmodel = torchvision.models.alexnet(pretrained=False) #不下载预训练权重\nprint(model)",
"AlexNet(\n (features): Sequential(\n (0): Conv2d(3, 64, kernel_size=(11, 11), stride=(4, 4), padding=(2, 2))\n (1): ReLU(inplace)\n (2): MaxPool2d(kernel_size=3, stride=2, padding=0, dilation=1, ceil_mode=False)\n (3): Conv2d(64, 192, kernel_size=(5, 5), stride=(1, 1), padding=(2, 2))\n (4): ReLU(inplace)\n (5): MaxPool2d(kernel_size=3, stride=2, padding=0, dilation=1, ceil_mode=False)\n (6): Conv2d(192, 384, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (7): ReLU(inplace)\n (8): Conv2d(384, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (9): ReLU(inplace)\n (10): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (11): ReLU(inplace)\n (12): MaxPool2d(kernel_size=3, stride=2, padding=0, dilation=1, ceil_mode=False)\n )\n (avgpool): AdaptiveAvgPool2d(output_size=(6, 6))\n (classifier): Sequential(\n (0): Dropout(p=0.5)\n (1): Linear(in_features=9216, out_features=4096, bias=True)\n (2): ReLU(inplace)\n (3): Dropout(p=0.5)\n (4): Linear(in_features=4096, out_features=4096, bias=True)\n (5): ReLU(inplace)\n (6): Linear(in_features=4096, out_features=1000, bias=True)\n )\n)\n"
]
],
[
[
"### 2. VGG",
"_____no_output_____"
]
],
[
[
"model = torchvision.models.vgg16(pretrained=False) #不下载预训练权重\nprint(model)",
"VGG(\n (features): Sequential(\n (0): Conv2d(3, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (1): ReLU(inplace)\n (2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (3): ReLU(inplace)\n (4): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n (5): Conv2d(64, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (6): ReLU(inplace)\n (7): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (8): ReLU(inplace)\n (9): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n (10): Conv2d(128, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (11): ReLU(inplace)\n (12): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (13): ReLU(inplace)\n (14): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (15): ReLU(inplace)\n (16): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n (17): Conv2d(256, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (18): ReLU(inplace)\n (19): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (20): ReLU(inplace)\n (21): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (22): ReLU(inplace)\n (23): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n (24): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (25): ReLU(inplace)\n (26): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (27): ReLU(inplace)\n (28): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (29): ReLU(inplace)\n (30): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n )\n (avgpool): AdaptiveAvgPool2d(output_size=(7, 7))\n (classifier): Sequential(\n (0): Linear(in_features=25088, out_features=4096, bias=True)\n (1): ReLU(inplace)\n (2): Dropout(p=0.5)\n (3): Linear(in_features=4096, out_features=4096, bias=True)\n (4): ReLU(inplace)\n (5): Dropout(p=0.5)\n (6): Linear(in_features=4096, out_features=1000, bias=True)\n )\n)\n"
]
],
[
[
"### 3. Inception",
"_____no_output_____"
],
[
"### 4. ResNet",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecca6c94a6e2da5b1f270c8a29872997b5d0b05c | 173,350 | ipynb | Jupyter Notebook | Test Train Module.ipynb | crdietrich/sidewalkscanner | 62d167da930ea219f2cc638d7de29fa0a1bed65c | [
"MIT"
] | 1 | 2019-07-17T10:11:54.000Z | 2019-07-17T10:11:54.000Z | Test Train Module.ipynb | crdietrich/sidewalkscanner | 62d167da930ea219f2cc638d7de29fa0a1bed65c | [
"MIT"
] | null | null | null | Test Train Module.ipynb | crdietrich/sidewalkscanner | 62d167da930ea219f2cc638d7de29fa0a1bed65c | [
"MIT"
] | 1 | 2020-04-09T11:11:38.000Z | 2020-04-09T11:11:38.000Z | 245.886525 | 73,652 | 0.887182 | [
[
[
"import os",
"_____no_output_____"
],
[
"from tensorflow.python.client import device_lib\nprint(device_lib.list_local_devices())",
"[name: \"/device:CPU:0\"\ndevice_type: \"CPU\"\nmemory_limit: 268435456\nlocality {\n}\nincarnation: 13780878210071979643\n, name: \"/device:GPU:0\"\ndevice_type: \"GPU\"\nmemory_limit: 4951913267\nlocality {\n bus_id: 1\n links {\n }\n}\nincarnation: 10281240396222099732\nphysical_device_desc: \"device: 0, name: GeForce GTX 1060 6GB, pci bus id: 0000:02:00.0, compute capability: 6.1\"\n]\n"
],
[
"from keras import backend as K\nK.tensorflow_backend._get_available_gpus()",
"Using TensorFlow backend.\n"
],
[
"import train, plotting, config, models",
"_____no_output_____"
]
],
[
[
"#### 0. Description for Data Output",
"_____no_output_____"
]
],
[
[
"model_description = 'Model_A1_SDNET2018_20190321A'",
"_____no_output_____"
]
],
[
[
"#### 1. Build Model",
"_____no_output_____"
]
],
[
[
"reload(train)\nmodel = train.build_model_inceptionV3(frozen_layers=23, verbose=True)",
"_____no_output_____"
],
[
"# VGG16\nmodel = train.build_model_VGG16(verbose=True)",
"_____no_output_____"
],
[
"# Inception V3\nmodel = train.build_model_inceptionV3(frozen_layers=11, verbose=True)",
"_____no_output_____"
]
],
[
[
"# Custom Model vA1\nmodel = models.model_A1(verbose=True)",
"_____no_output_____"
]
],
[
[
"#### 2. Create Image Data Generators",
"_____no_output_____"
]
],
[
[
"train_gen, validation_gen, test_gen = train.build_generators()",
"Found 3400 images belonging to 2 classes.\nFound 1200 images belonging to 2 classes.\nFound 400 images belonging to 2 classes.\n"
]
],
[
[
"#### 3. Fit Model",
"_____no_output_____"
]
],
[
[
"history = train.train_model(model=model,\n train_generator=train_gen,\n validation_generator=validation_gen,\n model_description=model_description)",
"Epoch 1/100\n106/106 [==============================] - 21s 197ms/step - loss: 0.7296 - acc: 0.5395 - val_loss: 0.8071 - val_acc: 0.5574\n\nEpoch 00001: acc improved from -inf to 0.53950, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_01-0.56.hdf5\nEpoch 2/100\n106/106 [==============================] - 15s 142ms/step - loss: 0.6064 - acc: 0.6860 - val_loss: 0.6459 - val_acc: 0.6301\n\nEpoch 00002: acc improved from 0.53950 to 0.68735, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_02-0.63.hdf5\nEpoch 3/100\n106/106 [==============================] - 16s 150ms/step - loss: 0.5588 - acc: 0.7099 - val_loss: 0.9065 - val_acc: 0.6498\n\nEpoch 00003: acc improved from 0.68735 to 0.70873, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_03-0.65.hdf5\nEpoch 4/100\n106/106 [==============================] - 17s 156ms/step - loss: 0.5318 - acc: 0.7494 - val_loss: 0.7907 - val_acc: 0.5668\n\nEpoch 00004: acc improved from 0.70873 to 0.75030, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_04-0.57.hdf5\nEpoch 5/100\n106/106 [==============================] - 16s 154ms/step - loss: 0.5168 - acc: 0.7706 - val_loss: 0.7349 - val_acc: 0.5634\n\nEpoch 00005: acc improved from 0.75030 to 0.77078, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_05-0.56.hdf5\nEpoch 6/100\n106/106 [==============================] - 15s 146ms/step - loss: 0.4550 - acc: 0.8101 - val_loss: 1.1037 - val_acc: 0.6104\n\nEpoch 00006: acc improved from 0.77078 to 0.80968, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_06-0.61.hdf5\nEpoch 7/100\n106/106 [==============================] - 15s 146ms/step - loss: 0.4835 - acc: 0.7907 - val_loss: 0.9122 - val_acc: 0.5505\n\nEpoch 00007: acc did not improve from 0.80968\nEpoch 8/100\n106/106 [==============================] - 16s 148ms/step - loss: 0.4327 - acc: 0.8266 - val_loss: 0.9022 - val_acc: 0.5488\n\nEpoch 00008: acc improved from 0.80968 to 0.82542, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_08-0.55.hdf5\nEpoch 9/100\n106/106 [==============================] - 16s 153ms/step - loss: 0.4646 - acc: 0.8243 - val_loss: 1.3544 - val_acc: 0.5993\n\nEpoch 00009: acc did not improve from 0.82542\nEpoch 10/100\n106/106 [==============================] - 17s 159ms/step - loss: 0.4383 - acc: 0.8278 - val_loss: 1.0462 - val_acc: 0.5813\n\nEpoch 00010: acc improved from 0.82542 to 0.82749, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_10-0.58.hdf5\nEpoch 11/100\n106/106 [==============================] - 16s 149ms/step - loss: 0.4444 - acc: 0.8420 - val_loss: 1.0426 - val_acc: 0.6592\n\nEpoch 00011: acc improved from 0.82749 to 0.84175, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_11-0.66.hdf5\nEpoch 12/100\n106/106 [==============================] - 16s 146ms/step - loss: 0.4144 - acc: 0.8426 - val_loss: 1.2589 - val_acc: 0.5813\n\nEpoch 00012: acc improved from 0.84175 to 0.84323, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_12-0.58.hdf5\nEpoch 13/100\n106/106 [==============================] - 16s 148ms/step - loss: 0.4193 - acc: 0.8508 - val_loss: 1.6582 - val_acc: 0.5514\n\nEpoch 00013: acc improved from 0.84323 to 0.85065, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_13-0.55.hdf5\nEpoch 14/100\n106/106 [==============================] - 15s 143ms/step - loss: 0.4040 - acc: 0.8526 - val_loss: 1.0093 - val_acc: 0.5522\n\nEpoch 00014: acc improved from 0.85065 to 0.85333, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_14-0.55.hdf5\nEpoch 15/100\n106/106 [==============================] - 15s 144ms/step - loss: 0.4163 - acc: 0.8511 - val_loss: 0.8357 - val_acc: 0.6721\n\nEpoch 00015: acc did not improve from 0.85333\nEpoch 16/100\n106/106 [==============================] - 15s 145ms/step - loss: 0.4079 - acc: 0.8494 - val_loss: 0.9489 - val_acc: 0.5890\n\nEpoch 00016: acc did not improve from 0.85333\nEpoch 17/100\n106/106 [==============================] - 15s 146ms/step - loss: 0.3874 - acc: 0.8668 - val_loss: 1.3619 - val_acc: 0.5916\n\nEpoch 00017: acc improved from 0.85333 to 0.86758, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_17-0.59.hdf5\nEpoch 18/100\n106/106 [==============================] - 15s 144ms/step - loss: 0.4280 - acc: 0.8491 - val_loss: 1.1400 - val_acc: 0.5805\n\nEpoch 00018: acc did not improve from 0.86758\nEpoch 19/100\n106/106 [==============================] - 15s 145ms/step - loss: 0.4002 - acc: 0.8508 - val_loss: 1.2584 - val_acc: 0.5848\n\nEpoch 00019: acc did not improve from 0.86758\nEpoch 20/100\n106/106 [==============================] - 16s 147ms/step - loss: 0.4035 - acc: 0.8552 - val_loss: 0.7559 - val_acc: 0.6901\n\nEpoch 00020: acc did not improve from 0.86758\nEpoch 21/100\n106/106 [==============================] - 16s 150ms/step - loss: 0.3923 - acc: 0.8576 - val_loss: 1.1710 - val_acc: 0.5916\n\nEpoch 00021: acc did not improve from 0.86758\nEpoch 22/100\n106/106 [==============================] - 16s 151ms/step - loss: 0.3939 - acc: 0.8532 - val_loss: 1.7778 - val_acc: 0.6019\n\nEpoch 00022: acc did not improve from 0.86758\nEpoch 23/100\n106/106 [==============================] - 16s 147ms/step - loss: 0.4160 - acc: 0.8600 - val_loss: 0.8662 - val_acc: 0.6250\n\nEpoch 00023: acc did not improve from 0.86758\nEpoch 24/100\n106/106 [==============================] - 15s 145ms/step - loss: 0.4055 - acc: 0.8514 - val_loss: 1.8533 - val_acc: 0.4854\n\nEpoch 00024: acc did not improve from 0.86758\nEpoch 25/100\n106/106 [==============================] - 15s 142ms/step - loss: 0.3808 - acc: 0.8623 - val_loss: 1.1852 - val_acc: 0.5942\n\nEpoch 00025: acc did not improve from 0.86758\nEpoch 26/100\n106/106 [==============================] - 15s 145ms/step - loss: 0.3662 - acc: 0.8676 - val_loss: 0.9228 - val_acc: 0.6173\n\nEpoch 00026: acc did not improve from 0.86758\nEpoch 27/100\n106/106 [==============================] - 15s 141ms/step - loss: 0.4180 - acc: 0.8605 - val_loss: 0.7626 - val_acc: 0.7003\n\nEpoch 00027: acc did not improve from 0.86758\nEpoch 28/100\n106/106 [==============================] - 15s 141ms/step - loss: 0.3702 - acc: 0.8738 - val_loss: 1.3604 - val_acc: 0.5445\n\nEpoch 00028: acc improved from 0.86758 to 0.87292, saving model to C:\\Users\\colin\\Google_Drive\\ilocal\\data\\model_outputs\\Model_A1_SDNET2018_20190321A\\Model_A1_SDNET2018_20190321A_weights_improvement_28-0.54.hdf5\nEpoch 29/100\n106/106 [==============================] - 15s 143ms/step - loss: 0.4921 - acc: 0.8408 - val_loss: 0.9418 - val_acc: 0.6010\n\nEpoch 00029: acc did not improve from 0.87292\nEpoch 30/100\n106/106 [==============================] - 16s 154ms/step - loss: 0.3940 - acc: 0.8650 - val_loss: 1.0186 - val_acc: 0.6019\n\nEpoch 00030: acc did not improve from 0.87292\nEpoch 31/100\n106/106 [==============================] - 15s 141ms/step - loss: 0.3923 - acc: 0.8608 - val_loss: 1.4703 - val_acc: 0.5334\n\nEpoch 00031: acc did not improve from 0.87292\nEpoch 32/100\n106/106 [==============================] - 15s 142ms/step - loss: 0.3846 - acc: 0.8720 - val_loss: 1.1914 - val_acc: 0.5557\n\nEpoch 00032: acc did not improve from 0.87292\nEpoch 33/100\n"
],
[
"training_output_dir = (config.model_dir + os.path.sep + \n model_description + os.path.sep)\ntraining_output_dir",
"_____no_output_____"
]
],
[
[
"if isinstance(history, list):\n for k, k_history in enumerate(history):\n plotting.plot_history(k_history, k=k, folder_path=training_output_dir)\nelse:\n plotting.plot_history(history, folder_path=training_output_dir)",
"_____no_output_____"
]
],
[
[
"plotting.plot_history(history, description=model_description, folder_path=training_output_dir)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"raw",
"code",
"markdown",
"code",
"markdown",
"code",
"raw",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw",
"raw",
"raw"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"raw"
],
[
"code"
]
] |
ecca77510a66f60066337851f799382b848dfd36 | 102,755 | ipynb | Jupyter Notebook | analysis/cospar/working_019_venusbars.ipynb | rsbyrne/thesis | 44689b1d06c5b08fd44054313d7a6148b0e44a24 | [
"MIT"
] | null | null | null | analysis/cospar/working_019_venusbars.ipynb | rsbyrne/thesis | 44689b1d06c5b08fd44054313d7a6148b0e44a24 | [
"MIT"
] | null | null | null | analysis/cospar/working_019_venusbars.ipynb | rsbyrne/thesis | 44689b1d06c5b08fd44054313d7a6148b0e44a24 | [
"MIT"
] | 1 | 2021-06-10T14:35:54.000Z | 2021-06-10T14:35:54.000Z | 772.593985 | 99,332 | 0.943 | [
[
[
"from functools import reduce\nimport operator\n\nimport numpy as np\n\nfrom cospar import reader, F\nfrom everest.window import Canvas, DataChannel\n%matplotlib inline",
"_____no_output_____"
],
[
"conds = (\n F('inputs/aspect') == 1,\n F('inputs/f') == 1,\n F('inputs/tauRef') <= 1e6,\n F('inputs/tauRef') >= 1e5,\n F('inputs/temperatureField') == '_built_peaskauslu-thoesfthuec',\n )\ncut = reader[reduce(operator.__and__, conds)]\ndatas = reader[cut : ('inputs/tauRef', 'outputs/t', 'outputs/Nu')]",
"_____no_output_____"
],
[
"alltau, allt, allNu = [], [], []\nalls = []\ntauVals = np.log10(np.array(sorted(set(tau for tau, *_ in datas.values()))))\ntauDiff = np.diff(tauVals)\nsDict = dict(zip(tauVals, tauDiff))\nsDict[tauVals[-1]] = tauDiff[-1]\nssum = sum(sDict.values())\nfor tauRef, t, Nu in datas.values():\n tauRef = np.log10(tauRef)\n alltau.extend(np.full(t.shape, tauRef))\n allt.extend(t)\n allNu.extend(Nu)\n alls.extend(np.full(t.shape, sDict[tauRef] / ssum))\nalltau, allt, allNu = np.array(alltau), np.array(allt), np.array(allNu)\nalls = np.array(alls)",
"_____no_output_____"
],
[
"canvas = Canvas(size = (12, 6), colour = 'white', fill = 'black')\nax = canvas.make_ax(title = 'Nusselt number over time as a function of yield strength')\n\nax.scatter(\n DataChannel(allt, lims = (0, 0.45), label = 'Dimensionless time'),\n DataChannel(alltau, lims = (5, 6), label = 'Yield strength (log10)'),\n c = np.log10(allNu),\n cmap = 'magma',\n s = 1.1 * (alls * ax.inches[1] * 72) ** 2,\n alpha = 1.,\n marker = 2,\n )\n\nax.props.grid.colour = 'white'\n\ncanvas.show()",
"_____no_output_____"
],
[
"# conds = (\n# F('aspect') == 1,\n# F('f') == 1,\n# F('temperatureField') == '_built_oiskeaosle-woatihoo',\n# )\n# cut = reader[reduce(operator.__and__, conds)]\n# datas = reader[cut : ('tauRef', 't', 'Nu')]",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecca7affb96a9b3fd202ee724d57c2b3cc15f518 | 678,716 | ipynb | Jupyter Notebook | examples/Example01.ipynb | charlie9578/openstreetmap-mapping | d5c9cc5f702c683b191ee28cca054e786e90ea99 | [
"BSD-3-Clause"
] | null | null | null | examples/Example01.ipynb | charlie9578/openstreetmap-mapping | d5c9cc5f702c683b191ee28cca054e786e90ea99 | [
"BSD-3-Clause"
] | 2 | 2021-08-16T08:44:05.000Z | 2021-08-16T08:45:54.000Z | examples/Example01.ipynb | charlie9578/openstreetmap-mapping | d5c9cc5f702c683b191ee28cca054e786e90ea99 | [
"BSD-3-Clause"
] | null | null | null | 684.189516 | 354,465 | 0.572839 | [
[
[
"import requests\nimport json\nfrom types import SimpleNamespace\n\n\nimport openstreetmap_mapping as osm\n\nimport pandas as pd\n\nfrom bokeh.plotting import show, output_file\nfrom bokeh.io import output_notebook\noutput_notebook()",
"_____no_output_____"
],
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"class Tags(object):\n \n def __init__(self, *initial_data, **kwargs):\n \n for dictionary in initial_data:\n \n for key in dictionary:\n\n if isinstance(dictionary[key], dict):\n if len(dictionary[key])>1:\n setattr(self, 'value', key)\n setattr(self, key, Tags(dictionary[key]))\n else:\n setattr(self, key, Tags(dictionary[key]))\n else:\n setattr(self, key, dictionary[key])\n \n def __repr__(self):\n return self.value.replace(\"__\",\":\")\n \n def __str__(self):\n return self.value.replace(\"__\",\":\")",
"_____no_output_____"
],
[
"kvs = osm.toolkit.get_osm_kvs()",
"_____no_output_____"
],
[
"tags = Tags(kvs)",
"_____no_output_____"
],
[
"area_Guernsey = \"(49.4096, -2.6875,49.5194, -2.4901)\"\narea_Ealing = \"(51.487, -0.351, 51.5335, -0.2655)\"\narea_britishisles = \"(48.327, -12.063, 61.428, 1.78)\"\narea_Glasgow = \"(55.5683, -4.9507, 56.1318, -3.6639)\"\narea_Whitelee = \"(55.5, -4.4364, 55.7591, -4.1)\"\narea_Wales = \"(51.221, -5.559, 53.619, -2.845)\"\narea_Shetlands = \"(59.8034, -2.4527, 60.9331, -0.4257)\"\narea_London = \"(51.448, -0.2554, 51.6061, 0.0714)\"",
"_____no_output_____"
],
[
"df_windturbines = osm.toolkit.get_osm_data(key=tags.generator__method,tag=tags.generator__method.wind_turbine,area=area_Whitelee)",
"_____no_output_____"
],
[
"df_windturbines[\"generator:output:electricity\"] = df_windturbines[\"generator:output:electricity\"].fillna(\"Unknown\")\ndf_windturbines[\"generator:output:electricity\"]\ndf_windturbines.loc[df_windturbines[\"generator:output:electricity\"]==\"yes\",\"generator:output:electricity\"]=\"Unknown\"",
"_____no_output_____"
],
[
"output_file(\"WhiteleeWindFarm.html\")\n\ntooltips = {\"tooltips\":[(\"id\", \"@id\"),(\"key\", \"@key\"),(\"tag\", \"@tag\"),(\"name\", \"@name\"),(\"(lat,lon)\", \"@coordinates\")]}\n\nshow(osm.toolkit.plot_latlon(df_windturbines,kwargs_for_marker={\"legend_group\":\"generator:output:electricity\"}))",
"_____no_output_____"
],
[
"df_beach = osm.toolkit.get_osm_data(key=tags.natural,tag=tags.natural.beach,area=area_Guernsey)\n",
"_____no_output_____"
],
[
"df_beach['named'] = df_beach['name'].isna()\ndf_beach['named'] = df_beach['named'].map({False:\"Named beach\",True:\"Un-named beach\"})\ndf_beach['color'] = df_beach['named'].map({\"Named beach\":\"yellow\",\"Un-named beach\":\"red\"})",
"_____no_output_____"
],
[
"output_file(\"GuernseyBeaches.html\")\n\ntooltips = {\"tooltips\":[(\"id\", \"@id\"),(\"key\", \"@key\"),(\"tag\", \"@tag\"),(\"name\", \"@name\"),(\"(lat,lon)\", \"@coordinates\")]}\n\nshow(osm.toolkit.plot_latlon(df_beach,kwargs_for_figure=tooltips,kwargs_for_marker={\"legend_group\":\"named\",\"marker\":\"square_pin\",\"fill_color\":\"color\"}))",
"_____no_output_____"
],
[
"df_pubs = osm.toolkit.get_osm_data(key=tags.amenity,tag=tags.amenity.pub,area=area_Ealing)\ndf_bar = osm.toolkit.get_osm_data(key=tags.amenity,tag=tags.amenity.bar,area=area_Ealing)\n\ndf_drinks = pd.concat([df_pubs,df_bar])",
"_____no_output_____"
],
[
"output_file(\"EalingPubs.html\")\n\ntooltips = {\"tooltips\":[(\"id\", \"@id\"),(\"key\", \"@key\"),(\"tag\", \"@tag\"),(\"name\", \"@name\"),(\"(lat,lon)\", \"@coordinates\")]}\n\nshow(osm.toolkit.plot_latlon(df_drinks,kwargs_for_figure=tooltips,kwargs_for_marker={\"marker\":\"square_dot\"}))",
"_____no_output_____"
],
[
"df_castles = osm.toolkit.get_osm_data(key=tags.historic,tag=tags.historic.castle,area=area_Shetlands)\ndf_archaelogy = osm.toolkit.get_osm_data(key=tags.historic,tag=tags.historic.archaeological_site,area=area_Shetlands)\ndf_fort = osm.toolkit.get_osm_data(key=tags.historic,tag=tags.historic.fort,area=area_Shetlands)\ndf_ruins = osm.toolkit.get_osm_data(key=tags.historic,tag=tags.historic.ruins,area=area_Shetlands)\ndf_bunker = osm.toolkit.get_osm_data(key=tags.military,tag=tags.military.bunker,area=area_Shetlands)\n\ndf_history = pd.concat([df_castles,df_archaelogy,df_fort,df_ruins,df_bunker])\n\n",
"_____no_output_____"
],
[
"output_file(\"ShetlandsHistoric.html\")\n\ntooltips = {\"tooltips\":[(\"id\", \"@id\"),(\"key\", \"@key\"),(\"tag\", \"@tag\"),(\"name\", \"@name\"),(\"(lat,lon)\", \"@coordinates\")]}\n\np = osm.toolkit.plot_latlon(df_history,kwargs_for_figure=tooltips)\np.legend.location = \"top_left\"\n\nshow(p)",
"_____no_output_____"
],
[
"df_WelshCastles = osm.toolkit.get_osm_data(key=tags.historic,tag=tags.historic.castle,area=area_Wales)",
"_____no_output_____"
],
[
"df_WelshCastles['named'] = df_WelshCastles['name'].isna()\ndf_WelshCastles['named'] = df_WelshCastles['named'].map({False:\"Named castle\",True:\"Un-named castle\"})\ndf_WelshCastles['color'] = df_WelshCastles['named'].map({\"Named castle\":\"Red\",\"Un-named castle\":\"Green\"})",
"_____no_output_____"
],
[
"output_file(\"WelshCastles.html\")\n\ntooltips = {\"tooltips\":[(\"id\", \"@id\"),(\"key\", \"@key\"),(\"tag\", \"@tag\"),(\"name\", \"@name\"),(\"(lat,lon)\", \"@coordinates\")]}\n\nshow(osm.toolkit.plot_latlon(df_WelshCastles,kwargs_for_figure=tooltips,kwargs_for_marker={\"legend_group\":\"named\",\"fill_color\":\"color\",\"marker\":\"square_x\"}))",
"_____no_output_____"
],
[
"df_lighthouse = osm.toolkit.get_osm_data(key=tags.man_made,tag=tags.man_made.lighthouse,area=area_britishisles)\n",
"_____no_output_____"
],
[
"df_lighthouse[\"color\"] = df_lighthouse[\"seamark:light:1:colour\"]\ndf_lighthouse[\"color\"] = df_lighthouse[\"color\"].fillna(\"black\")\ndf_lighthouse.loc[df_lighthouse[\"color\"]==\"white;red\",\"color\"]=\"red\"\n\ndf_lighthouse[\"size\"] = df_lighthouse[\"seamark:light:1:range\"]\ndf_lighthouse[\"size\"] = pd.to_numeric(df_lighthouse[\"size\"])\ndf_lighthouse[\"size\"] = df_lighthouse[\"size\"].fillna(3)",
"_____no_output_____"
],
[
"output_file(\"BritIsles_LightHouses.html\")\n\ntooltips = {\"tooltips\":[(\"id\", \"@id\"),(\"key\", \"@key\"),(\"tag\", \"@tag\"),(\"name\", \"@name\"),(\"(lat,lon)\", \"@coordinates\")]}\n\nshow(osm.toolkit.plot_latlon(df_lighthouse,marker_size=\"size\",kwargs_for_figure=tooltips,kwargs_for_marker={\"marker\":\"circle\",\"fill_color\":\"color\"}))",
"_____no_output_____"
],
[
"df_mountains = osm.toolkit.get_osm_data(key=tags.natural,tag=tags.natural.peak,area=area_britishisles)\n",
"_____no_output_____"
],
[
"df_munros = df_mountains.copy()\ndf_munros['ele'] = pd.to_numeric(df_munros['ele'],errors='coerce')\ndf_munros = df_munros.dropna(subset = ['ele'])\n\ndf_munros['ele_ft'] = (df_munros['ele']*3.282).astype(int)\n\ndf_munros = df_munros[df_munros['ele_ft']>1000]\n\ndf_munros = df_munros.sort_values(by=['ele_ft'])\n\ndf_munros[\"size\"] = df_munros['ele_ft']/1000",
"_____no_output_____"
],
[
"output_file(\"BritishIsles_Munros.html\")\n\nkwargs_for_figure = {\"tooltips\":[(\"name\", \"@name\"),(\"height [ft]\", \"@ele_ft\"),(\"(lat,lon)\", \"@coordinates\")],\"sizing_mode\":\"scale_width\",}\n\nshow(osm.toolkit.plot_latlon(df_munros,marker_size=3,kwargs_for_figure=kwargs_for_figure,kwargs_for_marker={\"marker\":\"triangle\",\"fill_color\":\"white\",\"line_color\":\"white\"}))",
"_____no_output_____"
],
[
"df_stations = osm.toolkit.get_osm_data(key=tags.railway,tag=tags.railway.station,area=area_London)\n",
"_____no_output_____"
],
[
"df_stations[\"grouping\"] = df_stations[\"line\"].fillna(df_stations['network'])",
"_____no_output_____"
],
[
"df_stations['grouping'] = df_stations['grouping'].str.split(\";\",expand=True)[0]",
"_____no_output_____"
],
[
"output_file(\"London_Stations.html\")\n\ntooltips = {\"tooltips\":[(\"id\", \"@id\"),(\"key\", \"@key\"),(\"tag\", \"@tag\"),(\"name\", \"@name\"),(\"network\", \"@network\"),(\"line\", \"@line\"),(\"(lat,lon)\", \"@coordinates\")]}\n\np = osm.toolkit.plot_latlon(df_stations,kwargs_for_figure=tooltips,kwargs_for_marker={\"legend_group\":\"grouping\"})\n\np.legend.visible = False\n\nshow(p)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecca8c640a09e6fc656e7e5bc8360402e281b511 | 6,460 | ipynb | Jupyter Notebook | ipynb/Linear Regression.ipynb | YuanL12/DL-Pytorch | 5c0a9c700f3903c1abb6edc58efd868a1cbbe5f5 | [
"MIT"
] | null | null | null | ipynb/Linear Regression.ipynb | YuanL12/DL-Pytorch | 5c0a9c700f3903c1abb6edc58efd868a1cbbe5f5 | [
"MIT"
] | null | null | null | ipynb/Linear Regression.ipynb | YuanL12/DL-Pytorch | 5c0a9c700f3903c1abb6edc58efd868a1cbbe5f5 | [
"MIT"
] | null | null | null | 22.430556 | 109 | 0.484365 | [
[
[
"# Linear Regression",
"_____no_output_____"
],
[
"## Using torch.utils.data.DataLoader as an iterator to only obtain a minibatch-sized data every time\nsyntax:\n\n```\nfor X, y in data_iter:\n l = loss(net(X), y)\n```",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport torch\nfrom torch.utils import data\nfrom d2l import torch as d2l\n\ntrue_w = torch.tensor([2, -3.4])\ntrue_b = 4.2\nfeatures, labels = d2l.synthetic_data(true_w, true_b, 1000)",
"_____no_output_____"
],
[
"def load_array(data_arrays, batch_size, is_train=True): #@save\n \"\"\"Construct a PyTorch data iterator.\"\"\"\n dataset = data.TensorDataset(*data_arrays)\n return data.DataLoader(dataset, batch_size, shuffle=is_train)\n\nbatch_size = 10\ndata_iter = load_array((features, labels), batch_size)",
"_____no_output_____"
],
[
"next(iter(data_iter))",
"_____no_output_____"
]
],
[
[
"Create Layer",
"_____no_output_____"
]
],
[
[
"from torch import nn\nnet = nn.Sequential(nn.Linear(2, 1)) \n# 2 is the size of features\n# 1 is the size of output",
"_____no_output_____"
]
],
[
[
"Initialize weight and bias",
"_____no_output_____"
]
],
[
[
"net[0].weight.data.normal_(0, 0.01)\nnet[0].bias.data.fill_(0)",
"_____no_output_____"
]
],
[
[
"Define Loss function",
"_____no_output_____"
]
],
[
[
"loss = nn.MSELoss()",
"_____no_output_____"
]
],
[
[
"Optimizer (optimization method in math)",
"_____no_output_____"
]
],
[
[
"trainer = torch.optim.SGD(net.parameters(), lr=0.03) # learning rate is 0.03",
"_____no_output_____"
]
],
[
[
"Now, we are able to train:\n\nGenerate predictions by calling net(X) and calculate the loss l (the forward propagation).\n\nCalculate gradients by running the backpropagation.\n\nUpdate the model parameters by invoking our optimizer.",
"_____no_output_____"
]
],
[
[
"num_epochs = 3 # a epoch is a iteration over the whole data set\nfor epoch in range(num_epochs):\n for X, y in data_iter:\n l = loss(net(X), y) # calculate loss on a minibatch \n trainer.zero_grad() # zero out gradient because Pytorch will accumulate gradients by default\n l.backward() # backpropagation\n trainer.step() # update parameters\n l = loss(net(features), labels) # calculate current total loss\n print(f'epoch {epoch + 1}, loss {l:f}')",
"epoch 1, loss 0.000275\nepoch 2, loss 0.000105\nepoch 3, loss 0.000105\n"
]
],
[
[
"Print out error",
"_____no_output_____"
]
],
[
[
"w = net[0].weight.data\nprint('error in estimating w:', true_w - w.reshape(true_w.shape))\nb = net[0].bias.data\nprint('error in estimating b:', true_b - b)",
"error in estimating w: tensor([-0.0004, 0.0005])\nerror in estimating b: tensor([-0.0007])\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecca936094a4d1259ff760ae564672dc991520ce | 12,178 | ipynb | Jupyter Notebook | great_expectations/init_notebooks/pandas/validation_playground.ipynb | ag1011/great_expectations | 9e57a55d49a98442e59e5251b2ef87c5e3c90838 | [
"Apache-2.0"
] | 2 | 2020-03-04T19:35:57.000Z | 2020-04-13T21:06:02.000Z | great_expectations/init_notebooks/pandas/validation_playground.ipynb | ag1011/great_expectations | 9e57a55d49a98442e59e5251b2ef87c5e3c90838 | [
"Apache-2.0"
] | null | null | null | great_expectations/init_notebooks/pandas/validation_playground.ipynb | ag1011/great_expectations | 9e57a55d49a98442e59e5251b2ef87c5e3c90838 | [
"Apache-2.0"
] | null | null | null | 37.355828 | 446 | 0.663902 | [
[
[
"# Validation Playground\n\n**Watch** a [short tutorial video](https://greatexpectations.io/videos/getting_started/integrate_expectations) or **read** [the written tutorial](https://docs.greatexpectations.io/en/latest/tutorials/validate_data.html?utm_source=notebook&utm_medium=validate_data)\n\nWe'd love it if you **reach out for help on** the [**Great Expectations Slack Channel**](https://greatexpectations.io/slack)",
"_____no_output_____"
]
],
[
[
"import json\nimport great_expectations as ge\nfrom great_expectations.profile import ColumnsExistProfiler\nimport great_expectations.jupyter_ux\nfrom great_expectations.datasource.types import BatchKwargs\nfrom datetime import datetime",
"_____no_output_____"
]
],
[
[
"## 1. Get a DataContext\nThis represents your **project** that you just created using `great_expectations init`. [Read more in the tutorial](https://docs.greatexpectations.io/en/latest/tutorials/validate_data.html?utm_source=notebook&utm_medium=validate_data#get-a-datacontext-object)",
"_____no_output_____"
]
],
[
[
"context = ge.data_context.DataContext()",
"_____no_output_____"
]
],
[
[
"## 2. List the CSVs in your folder\n\nThe `DataContext` will now introspect your pandas `Datasource` and list the CSVs it finds. [Read more in the tutorial](https://docs.greatexpectations.io/en/latest/tutorials/validate_data.html?utm_source=notebook&utm_medium=validate_data#list-data-assets)",
"_____no_output_____"
]
],
[
[
"ge.jupyter_ux.list_available_data_asset_names(context)",
"_____no_output_____"
]
],
[
[
"## 3. Pick a csv and the expectation suite\n\nInternally, Great Expectations represents csvs and dataframes as `DataAsset`s and uses this notion to link them to `Expectation Suites`. [Read more in the tutorial](https://docs.greatexpectations.io/en/latest/tutorials/validate_data.html?utm_source=notebook&utm_medium=validate_data#pick-a-data-asset-and-expectation-suite)\n",
"_____no_output_____"
]
],
[
[
"data_asset_name = \"ONE_OF_THE_CSV_DATA_ASSET_NAMES_FROM_ABOVE\" # TODO: replace with your value!\nnormalized_data_asset_name = context.normalize_data_asset_name(data_asset_name)\nnormalized_data_asset_name",
"_____no_output_____"
]
],
[
[
"We recommend naming your first expectation suite for a table `warning`. Later, as you identify some of the expectations that you add to this suite as critical, you can move these expectations into another suite and call it `failure`. [Read more in the tutorial](https://docs.greatexpectations.io/en/latest/getting_started/pipeline_integration.html?utm_source=notebook&utm_medium=integrate_validation#choose-data-asset-and-expectation-suite)",
"_____no_output_____"
]
],
[
[
"expectation_suite_name = \"warning\" # TODO: replace with your value!",
"_____no_output_____"
]
],
[
[
"#### 3.a. If you don't have an expectation suite, let's create a simple one\n\nYou need expectations to validate your data. Expectations are grouped into Expectation Suites. \n\nIf you don't have an expectation suite for this data asset, the notebook's next cell will create a suite of very basic expectations, so that you have some expectations to play with. The expectation suite will have `expect_column_to_exist` expectations for each column.\n\nIf you created an expectation suite for this data asset, you can skip executing the next cell (if you execute it, it will do nothing).\n\nTo create a more interesting suite, open the [create_expectations.ipynb](create_expectations.ipynb) notebook.\n\n",
"_____no_output_____"
]
],
[
[
"try:\n context.get_expectation_suite(normalized_data_asset_name, expectation_suite_name)\nexcept great_expectations.exceptions.DataContextError:\n context.create_expectation_suite(data_asset_name=normalized_data_asset_name, expectation_suite_name=expectation_suite_name, overwrite_existing=True);\n batch_kwargs = context.yield_batch_kwargs(data_asset_name)\n batch = context.get_batch(normalized_data_asset_name, expectation_suite_name, batch_kwargs)\n ColumnsExistProfiler().profile(batch)\n batch.save_expectation_suite()\n expectation_suite = context.get_expectation_suite(normalized_data_asset_name, expectation_suite_name)\n context.build_data_docs()\n",
"_____no_output_____"
]
],
[
[
"## 4. Load a batch of data you want to validate\n\nTo learn more about `get_batch` with other data types (such as existing pandas dataframes, SQL tables or Spark), see [this tutorial](https://docs.greatexpectations.io/en/latest/tutorials/validate_data.html?utm_source=notebook&utm_medium=validate_data#load-a-batch-of-data-to-validate)\n",
"_____no_output_____"
]
],
[
[
"batch_kwargs = context.yield_batch_kwargs(data_asset_name)\nbatch = context.get_batch(normalized_data_asset_name, expectation_suite_name, batch_kwargs)\nbatch.head()",
"_____no_output_____"
]
],
[
[
"## 5. Get a pipeline run id\n\nGenerate a run id, a timestamp, or a meaningful string that will help you refer to validation results. We recommend they be chronologically sortable.\n[Read more in the tutorial](https://docs.greatexpectations.io/en/latest/getting_started/pipeline_integration.html?utm_source=notebook&utm_medium=validate_data#set-a-run-id)",
"_____no_output_____"
]
],
[
[
"# Let's make a simple sortable timestamp. Note this could come from your pipeline runner.\nrun_id = datetime.utcnow().isoformat().replace(\":\", \"\") + \"Z\"\nrun_id",
"_____no_output_____"
]
],
[
[
"## 6. Validate the batch\n\nThis is the \"workhorse\" of Great Expectations. Call it in your pipeline code after loading data and just before passing it to your computation.\n\n[Read more about the validate method in the tutorial](https://docs.greatexpectations.io/en/latest/tutorials/validate_data.html?utm_source=notebook&utm_medium=validate_data#validate-the-batch)\n",
"_____no_output_____"
]
],
[
[
"validation_result = batch.validate(run_id=run_id)\n\nif validation_result[\"success\"]:\n print(\"This data meets all expectations for {}\".format(str(data_asset_name)))\nelse:\n print(\"This data is not a valid batch of {}\".format(str(data_asset_name)))",
"_____no_output_____"
]
],
[
[
"## 6.a. OPTIONAL: Review the JSON validation results\n\nDon't worry - this blob of JSON is meant for machines. Continue on or skip this to see this in Data Docs!",
"_____no_output_____"
]
],
[
[
"# print(json.dumps(validation_result, indent=4))",
"_____no_output_____"
]
],
[
[
"## 7. Validation Operators\n\nThe `validate` method evaluates one batch of data against one expectation suite and returns a dictionary of validation results. This is sufficient when you explore your data and get to know Great Expectations.\nWhen deploying Great Expectations in a **real data pipeline, you will typically discover additional needs**:\n\n* validating a group of batches that are logically related\n* validating a batch against several expectation suites such as using a tiered pattern like `warning` and `failure`\n* doing something with the validation results (e.g., saving them for a later review, sending notifications in case of failures, etc.).\n\n`Validation Operators` provide a convenient abstraction for both bundling the validation of multiple expectation suites and the actions that should be taken after the validation.\n\n[Read more about Validation Operators in the tutorial](https://docs.greatexpectations.io/en/latest/tutorials/validate_data.html?utm_source=notebook&utm_medium=validate_data#save-validation-results)",
"_____no_output_____"
]
],
[
[
"# This is an example of invoking a validation operator that is configured by default in the great_expectations.yml file\n\nresults = context.run_validation_operator(\n assets_to_validate=[batch],\n run_id=run_id,\n validation_operator_name=\"action_list_operator\",\n)",
"_____no_output_____"
]
],
[
[
"## 8. View the Validation Results in Data Docs\n\nLet's now build and look at your Data Docs. These will now include an **data quality report** built from the `ValidationResults` you just created that helps you communicate about your data with both machines and humans.\n\n[Read more about Data Docs in the tutorial](https://docs.greatexpectations.io/en/latest/tutorials/validate_data.html?utm_source=notebook&utm_medium=validate_data#view-the-validation-results-in-data-docs)",
"_____no_output_____"
]
],
[
[
"context.open_data_docs()",
"_____no_output_____"
]
],
[
[
"## Congratulations! You ran Validations!\n\n## Next steps:\n\n### 1. Author more interesting Expectations\n\nHere we used some **extremely basic** `Expectations`. To really harness the power of Great Expectations you can author much more interesting and specific `Expectations` to protect your data pipelines and defeat pipeline debt. Go to [create_expectations.ipynb](create_expectations.ipynb) to see how!\n\n### 2. Explore the documentation & community\n\nYou are now among the elite data professionals who know how to build robust descriptions of your data and protections for pipelines and machine learning models. Join the [**Great Expectations Slack Channel**](https://greatexpectations.io/slack) to see how others are wielding these superpowers.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecca95728b7a0765e1c1aa7585a1a14b20ac9052 | 39,775 | ipynb | Jupyter Notebook | docs/examples/jupyter-notebooks/formatting_axes_etc.ipynb | ASmirnov-HORIS/lets-plot-kotlin | 9d0a73399684fa17f3b0d9fb4cf9c2dc8372cdce | [
"MIT"
] | null | null | null | docs/examples/jupyter-notebooks/formatting_axes_etc.ipynb | ASmirnov-HORIS/lets-plot-kotlin | 9d0a73399684fa17f3b0d9fb4cf9c2dc8372cdce | [
"MIT"
] | null | null | null | docs/examples/jupyter-notebooks/formatting_axes_etc.ipynb | ASmirnov-HORIS/lets-plot-kotlin | 9d0a73399684fa17f3b0d9fb4cf9c2dc8372cdce | [
"MIT"
] | null | null | null | 59.013353 | 2,345 | 0.591779 | [
[
[
"### Formatting labels on plots.\n\nIn Lets-Plot you can apply a formatting to:\n\n- axis break values.\n- labels displayed by `geomText()`.\n- tooltip text.\n- facet labels.\n\nUsing format string you can format values of numeric and date-time types.\n\nIn addition, you can use a *string template*.\n\nIn *string template* the value's format string is surrounded by curly braces: `\"... {.2f} ...\"`.\n\nAn empty placeholder `{}` is also allowed. In this case a default string representation will be shown. This is also applicable to categorical values.\n\nTo learn more about formatting templates see: [Formatting](https://github.com/JetBrains/lets-plot-kotlin/blob/master/docs/formats.md).",
"_____no_output_____"
]
],
[
[
"%useLatestDescriptors\n%use lets-plot\n%use krangl",
"_____no_output_____"
]
],
[
[
"### The US Unemployment Rates 2000-2016",
"_____no_output_____"
]
],
[
[
"var economics = DataFrame.readCSV(\"https://vincentarelbundock.github.io/Rdatasets/csv/ggplot2/economics.csv\")",
"_____no_output_____"
],
[
"import java.text.SimpleDateFormat\nval yyyyMmDd = SimpleDateFormat(\"yyyy-MM-dd\")\neconomics = economics.addColumn(\"date\") { it[\"date\"].map<String> {yyyyMmDd.parse(it)}}\neconomics.head(3)",
"_____no_output_____"
],
[
"import java.util.Date\nval startDate = yyyyMmDd.parse(\"2000-01-01\")\neconomics = economics.filterByRow { (it[\"date\"] as Date).compareTo(startDate) >= 0 }",
"_____no_output_____"
],
[
"val p = letsPlot(economics.toMap()){x=\"date\"; y=\"uempmed\"} + \n geomLine() +\n ylab(\"unemployment rate\") +\n ggsize(900, 400)",
"_____no_output_____"
]
],
[
[
"#### Default plot (no formatting)",
"_____no_output_____"
]
],
[
[
"p + scaleXDateTime()",
"_____no_output_____"
]
],
[
[
"#### Apply formatting to X and Y axis labels.\n\nUse the `format` parameter in `scaleXxx()`.\n\nNote that the text in tooltips is now also formatted.",
"_____no_output_____"
]
],
[
[
"p + scaleXDateTime(format=\"%b %Y\") + scaleYContinuous(format=\"{} %\")",
"_____no_output_____"
]
],
[
[
"#### Format axis labels for breaks specified manually\n",
"_____no_output_____"
]
],
[
[
"val breaks = listOf(\n yyyyMmDd.parse(\"2001-01-01\"), \n yyyyMmDd.parse(\"2016-01-01\")\n)\n\np + scaleXDateTime(format=\"%b %Y\", breaks=breaks) + scaleYContinuous(format=\"{} %\")",
"_____no_output_____"
]
],
[
[
"#### Configure tooltip's text and location.",
"_____no_output_____"
]
],
[
[
"letsPlot(economics.toMap()){x=\"date\"; y=\"uempmed\"} + \n geomLine(tooltips=layerTooltips()\n .line(\"Unemployment rate:|^y\")\n .color(\"black\")\n .anchor(\"top_center\")\n .minWidth(170)) + \n scaleXDateTime(format=\"%b %Y\") +\n scaleYContinuous(format=\"{} %\") +\n ylab(\"unemployment rate\") +\n ggsize(900, 400)",
"_____no_output_____"
]
],
[
[
"#### Format value shown in tooltip.",
"_____no_output_____"
]
],
[
[
"letsPlot(economics.toMap()){x=\"date\"; y=\"uempmed\"} +\n geomLine(tooltips=layerTooltips()\n .line(\"@uempmed % in @date\")\n .format(\"date\", \"%B %Y\")\n .color(\"black\")\n .anchor(\"top_left\")\n .minWidth(170)) +\n scaleXDateTime() +\n scaleYContinuous() +\n ylab(\"unemployment rate\") +\n ggsize(900, 400)",
"_____no_output_____"
]
],
[
[
"#### Add the unemployment rate mean.\n\nThe `geomText` label is formatted using the `labelFormat` parameter.",
"_____no_output_____"
]
],
[
[
"val unemploymentMean = economics[\"uempmed\"].mean()\n\n\nletsPlot(economics.toMap()){x=\"date\"; y=\"uempmed\"} + \n geomLine(tooltips=layerTooltips()\n .line(\"Unemployment rate:|^y\")\n .color(\"black\")\n .anchor(\"top_center\")\n .minWidth(170)) + \n geomHLine(yintercept=unemploymentMean, color=\"red\", linetype=\"dashed\", tooltips=tooltipsNone) +\n geomText(label=unemploymentMean.toString(), \n labelFormat=\"{.2f} %\",\n x=startDate.toInstant().toEpochMilli(), y=unemploymentMean!! + 0.5, \n color=\"red\") +\n scaleXDateTime(format=\"%b %Y\") + \n scaleYContinuous(format=\"{} %\") + \n ylab(\"unemployment rate\") +\n ggtitle(\"The US Unemployment Rates 2000-2016.\") +\n ggsize(900, 400)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecca984f5f66bed5c674e0de6b8b497854c81085 | 71,491 | ipynb | Jupyter Notebook | usage.ipynb | fkalaganis/graph_emd | 5c6715f9e85855b69d050dffb72c82010be84d0f | [
"CC-BY-4.0"
] | 2 | 2021-08-23T18:27:20.000Z | 2022-03-18T14:03:11.000Z | usage.ipynb | fkalaganis/graph_emd | 5c6715f9e85855b69d050dffb72c82010be84d0f | [
"CC-BY-4.0"
] | null | null | null | usage.ipynb | fkalaganis/graph_emd | 5c6715f9e85855b69d050dffb72c82010be84d0f | [
"CC-BY-4.0"
] | null | null | null | 114.3856 | 42,324 | 0.838119 | [
[
[
"# Introduction\n\n$\\newcommand{\\G}{\\mathcal{G}}$\n$\\newcommand{\\V}{\\mathcal{V}}$\n$\\newcommand{\\E}{\\mathcal{E}}$\n$\\newcommand{\\R}{\\mathbb{R}}$\n\nThis notebook shows how to apply the graph ConvNet ([paper] & [code]), for classifying graph signals (EEG). For this example we have a set of EEG signals (epochs) belonging to two different classes (passive vs attentive task). The SG.npy file correndonds to the $W$ matrix of our paper and is used as the basis to construct the spatiotemporal graph.\n\n[paper]: https://arxiv.org/abs/1606.09375\n[code]: https://github.com/mdeff/cnn_graph\n\nWe note that this just an exemplar usage scenario. The full code for the GCNN is provided at (and can be cloned) at [code].\n",
"_____no_output_____"
]
],
[
[
"from lib import models, graph, coarsening, utils\nimport tensorflow as tf\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom scipy.linalg import block_diag\nfrom sklearn import preprocessing\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# 1 Data\n\nFor the purpose of the demo, let's create a random data matrix $X \\in \\R^{n \\times d_x}$ and somehow infer a label $y_i = f(x_i)$.",
"_____no_output_____"
]
],
[
[
"X=np.load('total_graph_train_trials.npy')\nd = X.shape[1] # Dimensionality.\nn = X.shape[0] # Number of samples.\n#c = 5 # Number of feature communities.\ny=np.load('total_graph_train_labels.npy')\ny=y[0,:]\nprint('Graph Signal Dimensionality: '+str(d))\nprint('Number of Graph Signals: '+str(n))\nprint('Class imbalance: ', np.unique(y, return_counts=True)[1])\nprint(y.shape)\nprint(len(X))",
"Graph Signal Dimensionality: 24256\nNumber of Graph Signals: 474\nClass imbalance: [234 240]\n(474,)\n474\n"
]
],
[
[
"Then load training, validation and testing sets.",
"_____no_output_____"
]
],
[
[
"#min_max_scaler = preprocessing.MinMaxScaler()\n#X_train = min_max_scaler.fit_transform(X)\nX_train=X\nX_val = np.load('graph_val_trials.npy')\n#X_val = min_max_scaler.fit_transform(X_val)\nX_test = np.load('graph_test_trials.npy')\n#X_test = min_max_scaler.fit_transform(X_test)\n\ny_train = y\ny_val = np.load('val_labels.npy')\ny_val = y_val[0,:]\ny_test = np.load('test_labels.npy')\ny_test = y_test[0,:]\nprint('Class imbalance in Train Set: ', np.unique(y_train, return_counts=True)[1])\nprint('Class imbalance in Val Set: ', np.unique(y_val, return_counts=True)[1])\nprint('Class imbalance in Test Set: ', np.unique(y_test, return_counts=True)[1])\nn = X_train.shape[0]",
"Class imbalance in Train Set: [234 240]\nClass imbalance in Val Set: [5 5]\nClass imbalance in Test Set: [5 5]\n"
]
],
[
[
"If you choose to run the following cell then the GCNN will operate on the initial train set without the augmentation data (set the condition to \"True\"). The first 5/6 of the X_train contain the artificially generated epochs using the GEMD code (provided as Matlab scripts) with the last 1/6 of X_train contains the original training set.",
"_____no_output_____"
]
],
[
[
"if False:\n N=round(n/6)\n X_train=X[-N:]\n X_val = np.load('graph_val_trials.npy')\n X_test = np.load('graph_test_trials.npy')\n y_train = y[-N:]\n y_val = np.load('val_labels.npy')\n y_val = y_val[0,:]\n y_test = np.load('test_labels.npy')\n y_test = y_test[0,:]\n print('Class imbalance in Train Set: ', np.unique(y_train, return_counts=True)[1])\n print('Class imbalance in Val Set: ', np.unique(y_val, return_counts=True)[1])\n print('Class imbalance in Test Set: ', np.unique(y_test, return_counts=True)[1])\n",
"_____no_output_____"
]
],
[
[
"# 2 Spatiotemporal Graph Construction\n\nLet $\\mathcal{G}=(\\mathcal{V}, \\mathcal{E}, \\textbf{W})$ be a graph that expresses the topology of the EEG recording sensor array with $W \\in \\{0,1\\}^{E\\times E}$ and $E$ being the number of sensors. The spatiotemporal graph that expresses jointly the relationship of an EEG signal with $T$ time samples is defined through its adjacency matrix as \n\\begin{equation}\n\\label{eq:multiplex_graph}\n \\textbf{W}_{ST}=\\textbf{W}\\otimes \\textbf{I}_E + \\textbf{S}+\\textbf{S}^\\top\n\\end{equation}\nwhere ($\\otimes$) denotes the Kronecker product operator, $\\textbf{I}_E$ the $E \\times E$ identity matrix and $\\textbf{S}$ an $ET \\times ET$ matrix whose elements are calculated as $S_{i,j}=\\delta_{i,i+E}$ with $\\delta_{i,j}$ being the Kronecker delta.",
"_____no_output_____"
]
],
[
[
"SG=np.load('SG.npy')\ntemporal_dimension=int(d/SG.shape[0])\nspatial_dimension=int(SG.shape[0])\nSG=SG-np.eye(spatial_dimension,dtype=int)\nA=np.kron(np.eye(temporal_dimension,dtype=int),SG).astype(np.float32)\n\n#assert A.shape == (d, d)\n#print('d = |V| = {}, k|V| < |E| = {}'.format(d, A.nnz))\n#plt.spy(A, markersize=2, color='black');\nprint(temporal_dimension)",
"379\n"
],
[
"for i in range(spatial_dimension,d):\n j=i-spatial_dimension;\n A[i,j]=1\n A[j,i]=1",
"_____no_output_____"
],
[
"from scipy import sparse\nA=sparse.coo_matrix(A)\nassert A.shape == (d, d)\nprint('d = |V| = {}, k|V| < |E| = {}'.format(d, A.nnz))\nplt.spy(A, markersize=2, color='black');",
"d = |V| = 24256, k|V| < |E| = 200742\n"
]
],
[
[
"To be able to pool graph signals, we need first to coarsen the graph, i.e. to find which vertices to group together. At the end we'll have multiple graphs, like a pyramid, each at one level of resolution. The finest graph is where the input data lies, the coarsest graph is where the data at the output of the graph convolutional layers lie. That data, of reduced spatial dimensionality, can then be fed to a fully connected layer.\n\nThe parameter here is the number of times to coarsen the graph. Each coarsening approximately reduces the size of the graph by a factor two. Thus if you want a pooling of size 4 in the first layer followed by a pooling of size 2 in the second, you'll need to coarsen $\\log_2(4+2) = 3$ times.\n\nAfter coarsening we rearrange the vertices (and add fake vertices) such that pooling a graph signal is analog to pooling a 1D signal. See the [paper] for details.\n\n[paper]: https://arxiv.org/abs/1606.09375",
"_____no_output_____"
]
],
[
[
"graphs, perm = coarsening.coarsen(A, levels=4, self_connections=False)\n\nX_train = coarsening.perm_data(X_train, perm)\nX_val = coarsening.perm_data(X_val, perm)\nX_test = coarsening.perm_data(X_test, perm)",
"Layer 0: M_0 = |V| = 28272 nodes (4016 added),|E| = 100371 edges\nLayer 1: M_1 = |V| = 14136 nodes (1472 added),|E| = 64474 edges\nLayer 2: M_2 = |V| = 7068 nodes (489 added),|E| = 35301 edges\nLayer 3: M_3 = |V| = 3534 nodes (103 added),|E| = 18608 edges\nLayer 4: M_4 = |V| = 1767 nodes (0 added),|E| = 9404 edges\n"
]
],
[
[
"We finally need to compute the graph Laplacian $L$ for each of our graphs (the original and the coarsened versions), defined by their adjacency matrices $A$. The sole parameter here is the type of Laplacian, e.g. the combinatorial Laplacian, the normalized Laplacian or the random walk Laplacian.",
"_____no_output_____"
]
],
[
[
"L = [graph.laplacian(A, normalized=True) for A in graphs]\n#graph.plot_spectrum(L)",
"_____no_output_____"
]
],
[
[
"# 3 Graph ConvNet\n\nHere we apply the graph convolutional neural network to signals lying on graphs. After designing the architecture and setting the hyper-parameters, the model takes as inputs the data matrix $X$, the target $y$ and a list of graph Laplacians $L$, one per coarsening level.\n\nThe data, architecture and hyper-parameters are absolutely *not engineered to showcase performance*. Its sole purpose is to illustrate usage and functionality.",
"_____no_output_____"
]
],
[
[
"params = dict()\nparams['dir_name'] = 'demo'\nparams['num_epochs'] = 20\nparams['batch_size'] = round(n/6)\nparams['eval_frequency'] = 6\n\n# Building blocks.\nparams['filter'] = 'chebyshev5'\nparams['brelu'] = 'b1relu'\nparams['pool'] = 'mpool1'\n\n# Number of classes.\nC = y.max() + 1\nassert C == np.unique(y).size\n\n# Architecture.\nparams['F'] = [20, 20] # Number of graph convolutional filters.\nparams['K'] = [10, 10] # Polynomial orders.\nparams['p'] = [1, 2] # Pooling sizes.\nparams['M'] = [128, 64, C] # Output dimensionality of fully connected layers.\n\n# Optimization.\nparams['regularization'] = 5e-4\nparams['dropout'] = 0.95\nparams['learning_rate'] = 1e-3\nparams['decay_rate'] = 0.95\nparams['momentum'] = 0.9\nparams['decay_steps'] = n / params['batch_size']",
"_____no_output_____"
],
[
"tf.reset_default_graph()\nmodel = models.cgcnn(L, **params)\nprint(X_train.shape)\naccuracy, loss, t_step = model.fit(X_train, y_train, X_val, y_val)",
"NN architecture\n input: M_0 = 28272\n layer 1: cgconv1\n representation: M_0 * F_1 / p_1 = 28272 * 20 / 1 = 565440\n weights: F_0 * F_1 * K_1 = 1 * 20 * 10 = 200\n biases: F_1 = 20\n layer 2: cgconv2\n representation: M_1 * F_2 / p_2 = 28272 * 20 / 2 = 282720\n weights: F_1 * F_2 * K_2 = 20 * 20 * 10 = 4000\n biases: F_2 = 20\n layer 3: fc1\n representation: M_3 = 128\n weights: M_2 * M_3 = 282720 * 128 = 36188160\n biases: M_3 = 128\n layer 4: fc2\n representation: M_4 = 64\n weights: M_3 * M_4 = 128 * 64 = 8192\n biases: M_4 = 64\n layer 5: logits (softmax)\n representation: M_5 = 2.0\n weights: M_4 * M_5 = 64 * 2.0 = 128.0\n biases: M_5 = 2.0\nWARNING:tensorflow:From C:\\Users\\fkalaganis\\Anaconda3\\lib\\site-packages\\tensorflow\\python\\framework\\op_def_library.py:263: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\nWARNING:tensorflow:From C:\\Users\\fkalaganis\\Desktop\\cnn_graph\\lib\\models.py:969: calling dropout (from tensorflow.python.ops.nn_ops) with keep_prob is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `rate` instead of `keep_prob`. Rate should be set to `rate = 1 - keep_prob`.\nWARNING:tensorflow:From C:\\Users\\fkalaganis\\Desktop\\cnn_graph\\lib\\models.py:209: to_int64 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.cast instead.\n(474, 28272)\nstep 6 / 120 (epoch 1.00 / 20):\n learning_rate = 1.00e-03, loss_average = 3.40e+01\n"
]
],
[
[
"# 4 Evaluation\n\nWe often want to monitor:\n1. The convergence, i.e. the training loss and the classification accuracy on the validation set.\n2. The performance, i.e. the classification accuracy on the testing set (to be compared with the training set accuracy to spot overfitting).\n\nThe `model_perf` class in [utils.py](utils.py) can be used to compactly evaluate multiple models.",
"_____no_output_____"
]
],
[
[
"fig, ax1 = plt.subplots(figsize=(15, 5))\nax1.plot(accuracy, 'b.-')\nax1.set_ylabel('validation accuracy', color='b')\nax2 = ax1.twinx()\nax2.plot(loss, 'g.-')\nax2.set_ylabel('training loss', color='g')\nplt.show()",
"_____no_output_____"
],
[
"print('Time per step: {:.2f} ms'.format(t_step*1000))",
"Time per step: 1626.03 ms\n"
]
],
[
[
"Please keep in mind that some incosistencies may occur due to weight initialization and small validation/testing sets",
"_____no_output_____"
]
],
[
[
"res = model.evaluate(X_test, y_test)\nprint(res[0])",
"WARNING:tensorflow:From C:\\Users\\fkalaganis\\Anaconda3\\lib\\site-packages\\tensorflow\\python\\training\\saver.py:1266: checkpoint_exists (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse standard file APIs to check for files with this prefix.\nINFO:tensorflow:Restoring parameters from C:\\Users\\fkalaganis\\Desktop\\cnn_graph\\lib\\..\\checkpoints\\demo\\model-120\naccuracy: 80.00 (8 / 10), f1 (weighted): 80.00, loss: 5.60e+02\ntime: 3s (wall 1s)\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
eccaa66e28023359760c328cc9a957e59dafbc37 | 530,652 | ipynb | Jupyter Notebook | intro-to-pytorch/Part 7 - Loading Image Data (Exercises).ipynb | zarrinan/deep-learning-v2-pytorch | 63a8b0c5bf2ce2ed3e7b577bc5e44796ecb08c9c | [
"MIT"
] | null | null | null | intro-to-pytorch/Part 7 - Loading Image Data (Exercises).ipynb | zarrinan/deep-learning-v2-pytorch | 63a8b0c5bf2ce2ed3e7b577bc5e44796ecb08c9c | [
"MIT"
] | null | null | null | intro-to-pytorch/Part 7 - Loading Image Data (Exercises).ipynb | zarrinan/deep-learning-v2-pytorch | 63a8b0c5bf2ce2ed3e7b577bc5e44796ecb08c9c | [
"MIT"
] | null | null | null | 1,503.263456 | 506,503 | 0.950868 | [
[
[
"# Loading Image Data\n\nSo far we've been working with fairly artificial datasets that you wouldn't typically be using in real projects. Instead, you'll likely be dealing with full-sized images like you'd get from smart phone cameras. In this notebook, we'll look at how to load images and use them to train neural networks.\n\nWe'll be using a [dataset of cat and dog photos](https://www.kaggle.com/c/dogs-vs-cats) available from Kaggle. Here are a couple example images:\n\n<img src='assets/dog_cat.png'>\n\nWe'll use this dataset to train a neural network that can differentiate between cats and dogs. These days it doesn't seem like a big accomplishment, but five years ago it was a serious challenge for computer vision systems.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\nimport os\nimport sys\nimport matplotlib.pyplot as plt\n\nimport torch\nfrom torchvision import datasets, transforms\n",
"_____no_output_____"
]
],
[
[
"The easiest way to load image data is with `datasets.ImageFolder` from `torchvision` ([documentation](http://pytorch.org/docs/master/torchvision/datasets.html#imagefolder)). In general you'll use `ImageFolder` like so:\n\n```python\ndataset = datasets.ImageFolder('path/to/data', transform=transform)\n```\n\nwhere `'path/to/data'` is the file path to the data directory and `transform` is a list of processing steps built with the [`transforms`](http://pytorch.org/docs/master/torchvision/transforms.html) module from `torchvision`. ImageFolder expects the files and directories to be constructed like so:\n```\nroot/dog/xxx.png\nroot/dog/xxy.png\nroot/dog/xxz.png\n\nroot/cat/123.png\nroot/cat/nsdf3.png\nroot/cat/asd932_.png\n```\n\nwhere each class has it's own directory (`cat` and `dog`) for the images. The images are then labeled with the class taken from the directory name. So here, the image `123.png` would be loaded with the class label `cat`. You can download the dataset already structured like this [from here](https://s3.amazonaws.com/content.udacity-data.com/nd089/Cat_Dog_data.zip). I've also split it into a training set and test set.\n\n### Transforms\n\nWhen you load in the data with `ImageFolder`, you'll need to define some transforms. For example, the images are different sizes but we'll need them to all be the same size for training. You can either resize them with `transforms.Resize()` or crop with `transforms.CenterCrop()`, `transforms.RandomResizedCrop()`, etc. We'll also need to convert the images to PyTorch tensors with `transforms.ToTensor()`. Typically you'll combine these transforms into a pipeline with `transforms.Compose()`, which accepts a list of transforms and runs them in sequence. It looks something like this to scale, then crop, then convert to a tensor:\n\n```python\ntransform = transforms.Compose([transforms.Resize(255),\n transforms.CenterCrop(224),\n transforms.ToTensor()])\n\n```\n\nThere are plenty of transforms available, I'll cover more in a bit and you can read through the [documentation](http://pytorch.org/docs/master/torchvision/transforms.html). \n\n### Data Loaders\n\nWith the `ImageFolder` loaded, you have to pass it to a [`DataLoader`](http://pytorch.org/docs/master/data.html#torch.utils.data.DataLoader). The `DataLoader` takes a dataset (such as you would get from `ImageFolder`) and returns batches of images and the corresponding labels. You can set various parameters like the batch size and if the data is shuffled after each epoch.\n\n```python\ndataloader = torch.utils.data.DataLoader(dataset, batch_size=32, shuffle=True)\n```\n\nHere `dataloader` is a [generator](https://jeffknupp.com/blog/2013/04/07/improve-your-python-yield-and-generators-explained/). To get data out of it, you need to loop through it or convert it to an iterator and call `next()`.\n\n```python\n# Looping through it, get a batch on each loop \nfor images, labels in dataloader:\n pass\n\n# Get one batch\nimages, labels = next(iter(dataloader))\n```\n \n>**Exercise:** Load images from the `Cat_Dog_data/train` folder, define a few transforms, then build the dataloader.",
"_____no_output_____"
]
],
[
[
"\ntransform = transform = transforms.Compose([transforms.Resize(255), \n transforms.CenterCrop(224), \n transforms.RandomRotation(45), \n transforms.RandomHorizontalFlip(),\n transforms.ColorJitter(), \n \n transforms.Normalize([0.5, 0.5, 0.5], \n [0.5 , 0.5, 0.5]) \n)\n transforms.ToTensor(),])\ntrainset = datasets.ImageFolder('~/Downloads/Cat_Dog_Data/train', train=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)\n\ntestset = datasets.ImageFolder('~/Downloads/Cat_Dog_Data/test', train=False, transform=transform)\ntestloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)\n\n",
"_____no_output_____"
],
[
"# Run this to test your data loader\nimages, labels = next(iter(dataloader))\nhelper.imshow(images[0], normalize=False)",
"_____no_output_____"
]
],
[
[
"If you loaded the data correctly, you should see something like this (your image will be different):\n\n<img src='assets/cat_cropped.png' width=244>",
"_____no_output_____"
],
[
"## Data Augmentation\n\nA common strategy for training neural networks is to introduce randomness in the input data itself. For example, you can randomly rotate, mirror, scale, and/or crop your images during training. This will help your network generalize as it's seeing the same images but in different locations, with different sizes, in different orientations, etc.\n\nTo randomly rotate, scale and crop, then flip your images you would define your transforms like this:\n\n```python\ntrain_transforms = transforms.Compose([transforms.RandomRotation(30),\n transforms.RandomResizedCrop(224),\n transforms.RandomHorizontalFlip(),\n transforms.ToTensor(),\n transforms.Normalize([0.5, 0.5, 0.5], \n [0.5, 0.5, 0.5])])\n```\n\nYou'll also typically want to normalize images with `transforms.Normalize`. You pass in a list of means and list of standard deviations, then the color channels are normalized like so\n\n```input[channel] = (input[channel] - mean[channel]) / std[channel]```\n\nSubtracting `mean` centers the data around zero and dividing by `std` squishes the values to be between -1 and 1. Normalizing helps keep the network work weights near zero which in turn makes backpropagation more stable. Without normalization, networks will tend to fail to learn.\n\nYou can find a list of all [the available transforms here](http://pytorch.org/docs/0.3.0/torchvision/transforms.html). When you're testing however, you'll want to use images that aren't altered (except you'll need to normalize the same way). So, for validation/test images, you'll typically just resize and crop.\n\n>**Exercise:** Define transforms for training data and testing data below. Leave off normalization for now.",
"_____no_output_____"
]
],
[
[
"data_dir = '~/Downloads/Cat_Dog_Data'\n\n# TODO: Define transforms for the training data and testing data\ntrain_transforms = transforms.Compose([transforms.Resize(255), \n transforms.CenterCrop(224), \n transforms.RandomRotation(45), \n transforms.RandomHorizontalFlip(),\n transforms.ColorJitter(),\n transforms.ToTensor()\n ])\n\ntest_transforms = train_transforms\n\n\n# Pass transforms in here, then run the next cell to see how the transforms look\ntrain_data = datasets.ImageFolder(data_dir + '/train', transform=train_transforms)\ntest_data = datasets.ImageFolder(data_dir + '/test', transform=test_transforms)\n\ntrainloader = torch.utils.data.DataLoader(train_data, batch_size=32)\ntestloader = torch.utils.data.DataLoader(test_data, batch_size=32)",
"_____no_output_____"
],
[
"# change this to the trainloader or testloader \ndata_iter = iter(trainloader)\n\nimages, labels = next(data_iter)\nfig, axes = plt.subplots(figsize=(10,4), ncols=4)\nfor ii in range(4):\n ax = axes[ii]\n helper.imshow(images[ii], ax=ax, normalize=False)",
"_____no_output_____"
]
],
[
[
"Your transformed images should look something like this.\n\n<center>Training examples:</center>\n<img src='assets/train_examples.png' width=500px>\n\n<center>Testing examples:</center>\n<img src='assets/test_examples.png' width=500px>",
"_____no_output_____"
],
[
"At this point you should be able to load data for training and testing. Now, you should try building a network that can classify cats vs dogs. This is quite a bit more complicated than before with the MNIST and Fashion-MNIST datasets. To be honest, you probably won't get it to work with a fully-connected network, no matter how deep. These images have three color channels and at a higher resolution (so far you've seen 28x28 images which are tiny).\n\nIn the next part, I'll show you how to use a pre-trained network to build a model that can actually solve this problem.",
"_____no_output_____"
]
],
[
[
"# Optional TODO: Attempt to build a network to classify cats vs dogs from this dataset\nmodel = nn.Sequential(nn.Linear(784, 256),\n nn.ReLU(),\n nn.Linear(256, 128),\n nn.ReLU(),\n nn.Linear(128, 64),\n nn.ReLU(),\n nn.Linear(64, 10),\n nn.LogSoftmax(dim=1))\n\ncriterion = nn.NLLLoss()\noptimizer = optim.Adam(model.parameters(), lr=0.005)\n\nepochs = 3\nsteps = 0\n\ntrain_losses, test_losses = [], []\nfor e in range(epochs):\n running_loss = 0\n for images, labels in trainloader:\n \n images = images.view(images.shape[0], -1)\n optimizer.zero_grad()\n \n log_ps = model(images)\n loss = criterion(log_ps, labels)\n loss.backward()\n optimizer.step()\n \n running_loss += loss.item()\n \n else:\n ## TODO: Implement the validation pass and print out the validation accuracy\n test_loss = 0\n accuracy = 0\n with torch.no_grad(): \n for images, labels in testloader:\n log_ps = model(images)\n test_loss += criterion(log_ps, labels)\n \n ps = torch.exp(log_ps)\n top_p, top_class = ps.topk(1, dim=1)\n equals = top_class == labels.view(*top_class.shape)\n accuracy += torch.mean(equals.type(torch.FloatTensor))\n \n train_losses.append(running_loss/len(trainloader))\n test_losses.append(test_loss/len(testloader))\n print(f'Train loss: {running_loss/len(trainloader)}')\n print(f'Test loss: {test_loss/len(testloader)}')\n print(f'Accuracy: {accuracy/len(testloader)}')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
eccabc0fd9fd3d24aea23363bb873b6fcdd4e241 | 1,742 | ipynb | Jupyter Notebook | nbs/00_tests/blocks/tst.blocks.ipynb | Jaume-JCI/block-types | 868eb97afa41cd4fee1ec2e0a2a97489e21783e8 | [
"MIT"
] | null | null | null | nbs/00_tests/blocks/tst.blocks.ipynb | Jaume-JCI/block-types | 868eb97afa41cd4fee1ec2e0a2a97489e21783e8 | [
"MIT"
] | 37 | 2021-11-17T22:38:36.000Z | 2022-03-05T15:21:54.000Z | nbs/00_tests/blocks/tst.blocks.ipynb | Jaume-JCI/block-types | 868eb97afa41cd4fee1ec2e0a2a97489e21783e8 | [
"MIT"
] | null | null | null | 20.987952 | 71 | 0.552813 | [
[
[
"# hide\n# default_exp tests.blocks.test_blocks\nfrom nbdev.showdoc import *\nfrom dsblocks.utils.nbdev_utils import nbdev_setup, TestRunner\n\nnbdev_setup ()\ntst = TestRunner (targets=['dummy'])",
"_____no_output_____"
]
],
[
[
"# Test blocks",
"_____no_output_____"
]
],
[
[
"# export\nimport pytest\nimport numpy as np\nimport pandas as pd\nimport os\nimport joblib\nfrom IPython.display import display\n\nfrom sklearn.model_selection import KFold\n\nfrom dsblocks.blocks.blocks import *",
"_____no_output_____"
],
[
"#export\n#@pytest.fixture (name='example_people_data')\n#def example_people_data_fixture():\n# return example_people_data()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
eccac528d5ae5a6caaa3cc93a19ed575da00bb90 | 19,735 | ipynb | Jupyter Notebook | nbs/02_data.load.ipynb | aquietlife/fastai2 | af18d16aa5f7b7d31388697ab451cbb6b4104e02 | [
"Apache-2.0"
] | 1 | 2020-07-09T22:01:29.000Z | 2020-07-09T22:01:29.000Z | nbs/02_data.load.ipynb | jovsa/fastai2 | 9e270519fdeff7772b7aecbe96290c0c19c0907f | [
"Apache-2.0"
] | 1 | 2020-02-22T05:15:19.000Z | 2020-02-22T05:15:19.000Z | nbs/02_data.load.ipynb | akashpalrecha/fastai2 | 096d8c1a53c4490e5ccf98076a60dc30e6d0da43 | [
"Apache-2.0"
] | null | null | null | 31.475279 | 211 | 0.556676 | [
[
[
"#default_exp data.load",
"_____no_output_____"
],
[
"#export\nfrom fastai2.torch_basics import *\n\nfrom torch.utils.data.dataloader import _MultiProcessingDataLoaderIter,_SingleProcessDataLoaderIter,_DatasetKind\n_loaders = (_MultiProcessingDataLoaderIter,_SingleProcessDataLoaderIter)",
"_____no_output_____"
],
[
"from nbdev.showdoc import *",
"_____no_output_____"
],
[
"bs = 4\nletters = list(string.ascii_lowercase)",
"_____no_output_____"
]
],
[
[
"## DataLoader",
"_____no_output_____"
]
],
[
[
"#export\ndef _wif(worker_id):\n set_num_threads(1)\n info = get_worker_info()\n ds = info.dataset.d\n ds.nw,ds.offs = info.num_workers,info.id\n set_seed(info.seed)\n ds.wif()\n\nclass _FakeLoader:\n _IterableDataset_len_called,_auto_collation,collate_fn,drop_last,dataset_kind,_dataset_kind,_index_sampler = (\n None,False,noops,False,_DatasetKind.Iterable,_DatasetKind.Iterable,Inf.count)\n def __init__(self, d, pin_memory, num_workers, timeout):\n self.dataset,self.default,self.worker_init_fn = self,d,_wif\n store_attr(self, 'd,pin_memory,num_workers,timeout')\n\n def __iter__(self): return iter(self.d.create_batches(self.d.sample()))\n\n @property\n def multiprocessing_context(self): return (None,multiprocessing)[self.num_workers>0]\n\n @contextmanager\n def no_multiproc(self):\n old_nw = self.num_workers\n try:\n self.num_workers = 0\n yield self.d\n finally: self.num_workers = old_nw\n\n_collate_types = (ndarray, Tensor, typing.Mapping, str)",
"_____no_output_____"
],
[
"#export\ndef fa_collate(t):\n b = t[0]\n return (default_collate(t) if isinstance(b, _collate_types)\n else type(t[0])([fa_collate(s) for s in zip(*t)]) if isinstance(b, Sequence)\n else default_collate(t))",
"_____no_output_____"
],
[
"#e.g. x is int, y is tuple\nt = [(1,(2,3)),(1,(2,3))]\ntest_eq(fa_collate(t), default_collate(t))\ntest_eq(L(fa_collate(t)).map(type), [Tensor,tuple])\n\nt = [(1,(2,(3,4))),(1,(2,(3,4)))]\ntest_eq(fa_collate(t), default_collate(t))\ntest_eq(L(fa_collate(t)).map(type), [Tensor,tuple])\ntest_eq(L(fa_collate(t)[1]).map(type), [Tensor,tuple])",
"_____no_output_____"
],
[
"#export\ndef fa_convert(t):\n return (default_convert(t) if isinstance(t, _collate_types)\n else type(t)([fa_convert(s) for s in t]) if isinstance(t, Sequence)\n else default_convert(t))",
"_____no_output_____"
],
[
"t0 = array([1,2])\nt = [t0,(t0,t0)]\n\ntest_eq(fa_convert(t), default_convert(t))\ntest_eq(L(fa_convert(t)).map(type), [Tensor,tuple])",
"_____no_output_____"
],
[
"#export\nclass SkipItemException(Exception): pass",
"_____no_output_____"
],
[
"#export\n@funcs_kwargs\nclass DataLoader(GetAttr):\n _noop_methods = 'wif before_iter after_item before_batch after_batch after_iter'.split()\n for o in _noop_methods:\n exec(f\"def {o}(self, x=None, *args, **kwargs): return x\")\n _methods = _noop_methods + 'create_batches create_item create_batch retain \\\n get_idxs sample shuffle_fn do_batch create_batch'.split()\n _default = 'dataset'\n def __init__(self, dataset=None, bs=None, num_workers=0, pin_memory=False, timeout=0, batch_size=None,\n shuffle=False, drop_last=False, indexed=None, n=None, device=None, **kwargs):\n if batch_size is not None: bs = batch_size # PyTorch compatibility\n assert not (bs is None and drop_last)\n if indexed is None: indexed = dataset is not None and hasattr(dataset,'__getitem__')\n if n is None:\n try: n = len(dataset)\n except TypeError: pass\n store_attr(self, 'dataset,bs,shuffle,drop_last,indexed,n,pin_memory,timeout,device')\n self.rng,self.nw,self.offs = random.Random(),1,0\n self.fake_l = _FakeLoader(self, pin_memory, num_workers, timeout)\n\n def __len__(self):\n if self.n is None: raise TypeError\n if self.bs is None: return self.n\n return self.n//self.bs + (0 if self.drop_last or self.n%self.bs==0 else 1)\n\n def get_idxs(self):\n idxs = Inf.count if self.indexed else Inf.nones\n if self.n is not None: idxs = list(itertools.islice(idxs, self.n))\n if self.shuffle: idxs = self.shuffle_fn(idxs)\n return idxs\n\n def sample(self):\n idxs = self.get_idxs()\n return (b for i,b in enumerate(idxs) if i//(self.bs or 1)%self.nw==self.offs)\n\n def __iter__(self):\n self.randomize()\n self.before_iter()\n for b in _loaders[self.fake_l.num_workers==0](self.fake_l):\n if self.device is not None: b = to_device(b, self.device)\n yield self.after_batch(b)\n self.after_iter()\n if hasattr(self, 'it'): delattr(self, 'it')\n\n def create_batches(self, samps):\n self.it = iter(self.dataset) if self.dataset is not None else None\n res = filter(lambda o:o is not None, map(self.do_item, samps))\n yield from map(self.do_batch, self.chunkify(res))\n\n def new(self, dataset=None, cls=None, **kwargs):\n if dataset is None: dataset = self.dataset\n if cls is None: cls = type(self)\n cur_kwargs = dict(dataset=dataset, num_workers=self.fake_l.num_workers, pin_memory=self.pin_memory, timeout=self.timeout,\n bs=self.bs, shuffle=self.shuffle, drop_last=self.drop_last, indexed=self.indexed, device=self.device)\n for n in self._methods: cur_kwargs[n] = getattr(self, n)\n return cls(**merge(cur_kwargs, kwargs))\n\n @property\n def prebatched(self): return self.bs is None\n def do_item(self, s):\n try: return self.after_item(self.create_item(s))\n except SkipItemException: return None\n def chunkify(self, b): return b if self.prebatched else chunked(b, self.bs, self.drop_last)\n def shuffle_fn(self, idxs): return self.rng.sample(idxs, len(idxs))\n def randomize(self): self.rng = random.Random(self.rng.randint(0,2**32-1))\n def retain(self, res, b): return retain_types(res, b[0] if is_listy(b) else b)\n def create_item(self, s): return next(self.it) if s is None else self.dataset[s]\n def create_batch(self, b): return (fa_collate,fa_convert)[self.prebatched](b)\n def do_batch(self, b): return self.retain(self.create_batch(self.before_batch(b)), b)\n def one_batch(self):\n if self.n is not None and len(self)==0: raise ValueError(f'This DataLoader does not contain any batches')\n with self.fake_l.no_multiproc(): res = first(self)\n if hasattr(self, 'it'): delattr(self, 'it')\n return res",
"_____no_output_____"
]
],
[
[
"Override `item` and use the default infinite sampler to get a stream of unknown length (`stop()` when you want to stop the stream).",
"_____no_output_____"
]
],
[
[
"class RandDL(DataLoader):\n def create_item(self, s):\n r = random.random()\n return r if r<0.95 else stop()\n\nL(RandDL())",
"_____no_output_____"
],
[
"L(RandDL(bs=4, drop_last=True)).map(len)",
"_____no_output_____"
],
[
"dl = RandDL(bs=4, num_workers=4, drop_last=True)\nL(dl).map(len)",
"_____no_output_____"
],
[
"test_eq(dl.fake_l.num_workers, 4)\nwith dl.fake_l.no_multiproc(): \n test_eq(dl.fake_l.num_workers, 0)\n L(dl).map(len)\ntest_eq(dl.fake_l.num_workers, 4)",
"_____no_output_____"
],
[
"def _rand_item(s):\n r = random.random()\n return r if r<0.95 else stop()\n\nL(DataLoader(create_item=_rand_item))",
"_____no_output_____"
]
],
[
[
"If you don't set `bs`, then `dataset` is assumed to provide an iterator or a `__getitem__` that returns a batch.",
"_____no_output_____"
]
],
[
[
"ds1 = DataLoader(letters)\ntest_eq(L(ds1), letters)\ntest_eq(len(ds1), 26)\n\ntest_shuffled(L(DataLoader(letters, shuffle=True)), letters)\n\nds1 = DataLoader(letters, indexed=False)\ntest_eq(L(ds1), letters)\ntest_eq(len(ds1), 26)\n\nt2 = L(tensor([0,1,2]),tensor([3,4,5]))\nds2 = DataLoader(t2)\ntest_eq_type(L(ds2), t2)\n\nt3 = L(array([0,1,2]),array([3,4,5]))\nds3 = DataLoader(t3)\ntest_eq_type(L(ds3), t3.map(tensor))\n\nds4 = DataLoader(t3, create_batch=noop, after_iter=lambda: setattr(t3, 'f', 1))\ntest_eq_type(L(ds4), t3)\ntest_eq(t3.f, 1)",
"_____no_output_____"
]
],
[
[
"If you do set `bs`, then `dataset` is assumed to provide an iterator or a `__getitem__` that returns a single item of a batch.",
"_____no_output_____"
]
],
[
[
"def twoepochs(d): return ' '.join(''.join(o) for _ in range(2) for o in d)",
"_____no_output_____"
],
[
"ds1 = DataLoader(letters, bs=4, drop_last=True, num_workers=0)\ntest_eq(twoepochs(ds1), 'abcd efgh ijkl mnop qrst uvwx abcd efgh ijkl mnop qrst uvwx')\n\nds1 = DataLoader(letters,4,num_workers=2)\ntest_eq(twoepochs(ds1), 'abcd efgh ijkl mnop qrst uvwx yz abcd efgh ijkl mnop qrst uvwx yz')\n\nds1 = DataLoader(range(12), bs=4, num_workers=3)\ntest_eq_type(L(ds1), L(tensor([0,1,2,3]),tensor([4,5,6,7]),tensor([8,9,10,11])))\n\nds1 = DataLoader([str(i) for i in range(11)], bs=4, after_iter=lambda: setattr(t3, 'f', 2))\ntest_eq_type(L(ds1), L(['0','1','2','3'],['4','5','6','7'],['8','9','10']))\ntest_eq(t3.f, 2)\n\nit = iter(DataLoader(map(noop,range(20)), bs=4, num_workers=1))\ntest_eq_type([next(it) for _ in range(3)], [tensor([0,1,2,3]),tensor([4,5,6,7]),tensor([8,9,10,11])])",
"_____no_output_____"
],
[
"class SleepyDL(list):\n def __getitem__(self,i):\n time.sleep(random.random()/50)\n return super().__getitem__(i)\n\nt = SleepyDL(letters)\n\n%time test_eq(DataLoader(t, num_workers=0), letters)\n%time test_eq(DataLoader(t, num_workers=2), letters)\n%time test_eq(DataLoader(t, num_workers=4), letters)\n\ndl = DataLoader(t, shuffle=True, num_workers=1)\ntest_shuffled(L(dl), letters)\ntest_shuffled(L(dl), L(dl))",
"CPU times: user 3.83 ms, sys: 0 ns, total: 3.83 ms\nWall time: 223 ms\nCPU times: user 6.58 ms, sys: 15.6 ms, total: 22.2 ms\nWall time: 183 ms\nCPU times: user 13.5 ms, sys: 21.5 ms, total: 35 ms\nWall time: 126 ms\n"
],
[
"class SleepyQueue():\n \"Simulate a queue with varying latency\"\n def __init__(self, q): self.q=q\n def __iter__(self):\n while True:\n time.sleep(random.random()/100)\n try: yield self.q.get_nowait()\n except queues.Empty: return\n\nq = Queue()\nfor o in range(30): q.put(o)\nit = SleepyQueue(q)\n\n%time test_shuffled(L(DataLoader(it, num_workers=4)), range(30))",
"CPU times: user 7.89 ms, sys: 26.9 ms, total: 34.8 ms\nWall time: 89.4 ms\n"
],
[
"class A(TensorBase): pass\n\nfor nw in (0,2):\n t = A(tensor([1,2]))\n dl = DataLoader([t,t,t,t,t,t,t,t], bs=4, num_workers=nw)\n b = first(dl)\n test_eq(type(b), A)\n\n t = (A(tensor([1,2])),)\n dl = DataLoader([t,t,t,t,t,t,t,t], bs=4, num_workers=nw)\n b = first(dl)\n test_eq(type(b[0]), A)",
"_____no_output_____"
],
[
"class A(TensorBase): pass\nt = A(tensor(1,2))\n\ntdl = DataLoader([t,t,t,t,t,t,t,t], bs=4, num_workers=2, after_batch=to_device)\nb = first(tdl)\ntest_eq(type(b), A)\n\n# Unknown attributes are delegated to `dataset`\ntest_eq(tdl.pop(), tensor(1,2))",
"_____no_output_____"
]
],
[
[
"## Export -",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.export import notebook2script\nnotebook2script()",
"Converted 00_torch_core.ipynb.\nConverted 01_layers.ipynb.\nConverted 02_data.load.ipynb.\nConverted 03_data.core.ipynb.\nConverted 04_data.external.ipynb.\nConverted 05_data.transforms.ipynb.\nConverted 06_data.block.ipynb.\nConverted 07_vision.core.ipynb.\nConverted 08_vision.data.ipynb.\nConverted 09_vision.augment.ipynb.\nConverted 09b_vision.utils.ipynb.\nConverted 09c_vision.widgets.ipynb.\nConverted 10_tutorial.pets.ipynb.\nConverted 11_vision.models.xresnet.ipynb.\nConverted 12_optimizer.ipynb.\nConverted 13_learner.ipynb.\nConverted 13a_metrics.ipynb.\nConverted 14_callback.schedule.ipynb.\nConverted 14a_callback.data.ipynb.\nConverted 15_callback.hook.ipynb.\nConverted 15a_vision.models.unet.ipynb.\nConverted 16_callback.progress.ipynb.\nConverted 17_callback.tracker.ipynb.\nConverted 18_callback.fp16.ipynb.\nConverted 19_callback.mixup.ipynb.\nConverted 20_interpret.ipynb.\nConverted 20a_distributed.ipynb.\nConverted 21_vision.learner.ipynb.\nConverted 22_tutorial.imagenette.ipynb.\nConverted 23_tutorial.transfer_learning.ipynb.\nConverted 24_vision.gan.ipynb.\nConverted 30_text.core.ipynb.\nConverted 31_text.data.ipynb.\nConverted 32_text.models.awdlstm.ipynb.\nConverted 33_text.models.core.ipynb.\nConverted 34_callback.rnn.ipynb.\nConverted 35_tutorial.wikitext.ipynb.\nConverted 36_text.models.qrnn.ipynb.\nConverted 37_text.learner.ipynb.\nConverted 38_tutorial.ulmfit.ipynb.\nConverted 40_tabular.core.ipynb.\nConverted 41_tabular.data.ipynb.\nConverted 42_tabular.model.ipynb.\nConverted 43_tabular.learner.ipynb.\nConverted 45_collab.ipynb.\nConverted 50_datablock_examples.ipynb.\nConverted 60_medical.imaging.ipynb.\nConverted 65_medical.text.ipynb.\nConverted 70_callback.wandb.ipynb.\nConverted 71_callback.tensorboard.ipynb.\nConverted 72_callback.neptune.ipynb.\nConverted 97_test_utils.ipynb.\nConverted 99_pytorch_doc.ipynb.\nConverted index.ipynb.\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
eccac6eb446c7932d4d0eec6ae4bb78bca235b24 | 26,122 | ipynb | Jupyter Notebook | notebooks/03-ensemble.ipynb | giwankim/emnist | cd443076cc46895e8b7818d2177d3bce8fcd955f | [
"MIT"
] | null | null | null | notebooks/03-ensemble.ipynb | giwankim/emnist | cd443076cc46895e8b7818d2177d3bce8fcd955f | [
"MIT"
] | null | null | null | notebooks/03-ensemble.ipynb | giwankim/emnist | cd443076cc46895e8b7818d2177d3bce8fcd955f | [
"MIT"
] | null | null | null | 40.562112 | 8,496 | 0.592604 | [
[
[
"# Ensemble",
"_____no_output_____"
]
],
[
[
"%reload_ext autoreload\n%autoreload 2\n%matplotlib inline",
"_____no_output_____"
],
[
"import sys\nsys.path.insert(0, \"../src\")",
"_____no_output_____"
],
[
"import gc\nimport pathlib\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set()\n\nfrom sklearn import metrics\nfrom sklearn import model_selection\nfrom scipy.special import softmax\n\nimport torch\nimport torchcontrib\nfrom torch.optim.swa_utils import AveragedModel, SWALR, update_bn\n\nimport callbacks\nimport config\nimport dataset\nimport engine\nimport models\nimport utils\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"# df = pd.read_csv(config.DATA_PATH / \"train.csv\")\ndf = pd.read_csv(config.DATA_PATH / \"pl-spinal-ensemble10.csv\")\ndf.shape",
"_____no_output_____"
],
[
"nets = 10\ndevice = torch.device(config.DEVICE)\nEPOCHS = 200\nSEED = 42\nutils.seed_everything(SEED)",
"_____no_output_____"
],
[
"cnns = [None] * nets\nvalid_scores = []\n\nfor i in range(nets):\n print(\"#\" * 30)\n # DATA\n train_indices, valid_indices = model_selection.train_test_split(np.arange(len(df)), test_size=0.1, shuffle=True, stratify=df.digit)\n train_dataset = dataset.EMNISTDataset(df, train_indices)\n valid_dataset = dataset.EMNISTDataset(df, valid_indices)\n train_loader = torch.utils.data.DataLoader(train_dataset, batch_size=config.TRAIN_BATCH_SIZE, shuffle=True)\n valid_loader = torch.utils.data.DataLoader(valid_dataset, batch_size=config.TEST_BATCH_SIZE)\n \n # MODEL\n model = models.SpinalVGG().to(device)\n \n # OPTIMIZER\n optimizer = torch.optim.Adam(model.parameters(), lr=1e-2)\n # SCHEDULER\n scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(\n optimizer, mode='max', verbose=True, patience=10, factor=0.5,\n )\n# scheduler = torch.optim.lr_scheduler.CosineAnnealingLR(optimizer, T_max=EPOCHS)\n\n # STOCHASTIC WEIGHT AVERAGING\n swa_start = int(EPOCHS * 0.75)\n swa_scheduler = SWALR(\n optimizer, anneal_strategy=\"cos\", anneal_epochs=swa_start, swa_lr=5e-4\n )\n swa_model = AveragedModel(model)\n swa_model.to(device)\n\n # AMP\n scaler = torch.cuda.amp.GradScaler()\n \n # Loop\n for epoch in range(EPOCHS):\n # TRAIN ONE EPOCH\n engine.train(train_loader, model, optimizer, device, scaler)\n\n # VALIDATION\n predictions, targets = engine.evaluate(valid_loader, model, device)\n predictions = np.argmax(predictions, axis=1)\n accuracy = metrics.accuracy_score(targets, predictions)\n if epoch % 10 == 0:\n print(f\"Epoch={epoch}, Accuracy={accuracy:.5f}\")\n\n if epoch > swa_start:\n swa_model.update_parameters(model)\n swa_scheduler.step()\n else:\n scheduler.step(accuracy)\n\n # Warmup BN-layers\n swa_model = swa_model.cpu()\n update_bn(train_loader, swa_model)\n swa_model.to(device)\n\n # CV Score for SWA model\n valid_preds, valid_targs = engine.evaluate(valid_loader, swa_model, device)\n valid_predsb = np.argmax(valid_preds, axis=1)\n \n valid_accuracy = metrics.accuracy_score(valid_targs, valid_predsb)\n valid_scores.append(valid_accuracy)\n print(f\"CNN {i}, Validation accuracy of SWA model={valid_accuracy}\")\n \n cnns[i] = swa_model\n\n # CLEAN-UP\n del model, swa_model\n torch.cuda.empty_cache()\n gc.collect()\n \n break\n\nprint(f\"Average CV score={np.mean(valid_scores)}\")",
"##############################\nEpoch=0, Accuracy=0.69143\nEpoch=10, Accuracy=0.93071\nEpoch=20, Accuracy=0.94417\nEpoch=30, Accuracy=0.93868\nEpoch 37: reducing learning rate of group 0 to 5.0000e-03.\nEpoch=40, Accuracy=0.95414\nEpoch=50, Accuracy=0.94467\nEpoch=60, Accuracy=0.94716\nEpoch 62: reducing learning rate of group 0 to 2.5000e-03.\nEpoch=70, Accuracy=0.94865\nEpoch 78: reducing learning rate of group 0 to 1.2500e-03.\nEpoch=80, Accuracy=0.95065\nEpoch=90, Accuracy=0.94965\nEpoch 94: reducing learning rate of group 0 to 6.2500e-04.\nEpoch=100, Accuracy=0.95065\nEpoch 110: reducing learning rate of group 0 to 3.1250e-04.\nEpoch=110, Accuracy=0.95065\nEpoch=120, Accuracy=0.94915\nEpoch 126: reducing learning rate of group 0 to 1.5625e-04.\nEpoch=130, Accuracy=0.95015\nEpoch=140, Accuracy=0.94865\nEpoch 142: reducing learning rate of group 0 to 7.8125e-05.\nEpoch=150, Accuracy=0.94965\nEpoch=160, Accuracy=0.95065\nEpoch=170, Accuracy=0.94915\nEpoch=180, Accuracy=0.95015\nEpoch=190, Accuracy=0.95115\nCNN 0, Validation accuracy of SWA model=0.9481555333998006\nAverage CV score=0.9481555333998006\n"
]
],
[
[
"## PL",
"_____no_output_____"
]
],
[
[
"df_test = pd.read_csv(config.TEST_CSV)\ntest_dataset = dataset.EMNISTDataset(df_test, np.arange(len(df_test)), label=False)\ntest_loader = torch.utils.data.DataLoader(test_dataset, batch_size=config.TEST_BATCH_SIZE)",
"_____no_output_____"
],
[
"# BLENDED SOFTMAX OF MODELS\npreds = np.zeros((len(df_test), 10))\nfor model in cnns:\n preds += engine.evaluate(test_loader, model, device, target=False)\nprobs = softmax(preds, axis=1)\nprobs = np.max(probs, axis=1)\ndigits = np.argmax(preds, axis=1)",
"_____no_output_____"
],
[
"pl = df_test.copy()\npl[\"digit\"] = digits\npl[\"prob\"] = probs\npl.head()",
"_____no_output_____"
],
[
"pl = pl[pl.prob > 0.995]\nprint(f\"{len(pl)}/{len(df_test)}\")",
"20058/20480\n"
],
[
"pl.to_csv(config.DATA_PATH / f\"pl-spinal-ensemble{nets}.csv\", index=False)",
"_____no_output_____"
]
],
[
[
"## Inference",
"_____no_output_____"
]
],
[
[
"submission = pd.DataFrame({\"id\": df_test.id, \"digit\": digits})\nsubmission.to_csv(f\"../output/spinal-ensemble{nets}.csv\", index=False)\nsubmission.head()",
"_____no_output_____"
],
[
"sns.countplot(submission.digit);",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
eccad5a83f368e22c9e9e619864cc3d63d02a308 | 11,257 | ipynb | Jupyter Notebook | notebooks/ptm_animator_sample.ipynb | CADWRDeltaModeling/pydsm | 14e0a634e693ec8c5bbc026d8cfcd5b6da2a89a4 | [
"MIT"
] | 3 | 2020-05-11T22:55:34.000Z | 2021-05-19T19:49:11.000Z | notebooks/ptm_animator_sample.ipynb | CADWRDeltaModeling/pydsm | 14e0a634e693ec8c5bbc026d8cfcd5b6da2a89a4 | [
"MIT"
] | 6 | 2020-02-12T23:18:38.000Z | 2022-02-18T20:31:53.000Z | notebooks/ptm_animator_sample.ipynb | CADWRDeltaModeling/pydsm | 14e0a634e693ec8c5bbc026d8cfcd5b6da2a89a4 | [
"MIT"
] | 1 | 2021-07-07T18:55:44.000Z | 2021-07-07T18:55:44.000Z | 30.09893 | 195 | 0.580972 | [
[
[
"# PTM Animator Sample\n\nThis notebook shows how a PTM animation can be done with numpy, pandas, geopandas, shapely and holoviews. This is shared to be a useful tool while a more generalized tool is being built.\n\n**This sample will note be developed further and many of the functions here will be moved to pydsm modules**",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\n#import datashader.geo\nimport hvplot.pandas\nimport holoviews as hv\nfrom holoviews import opts\nhv.extension('bokeh')\nimport geopandas\nimport shapely",
"_____no_output_____"
]
],
[
[
"# Reads ptm animation binary file. needs swap to small endians\ndef load_anim_data(ptm_file):\n ptm_meta=np.core.records.fromfile(ptm_file,dtype=[('x','>h'), ('model_date','a9'), ('model_time','>h'), ('nparticles','>h')], shape=1)\n nparticles=ptm_meta.nparticles[0]\n ptm_data=np.core.records.fromfile(ptm_file,dtype=[('x','>h'), ('model_date','a9'), ('model_time','>h'), ('nparticles','>h'),('positions','(%d,6)>h'%nparticles)])\n ptm_data=ptm_data.byteswap().newbyteorder() # Needed for converting big endianess to small endianess\n return ptm_data",
"_____no_output_____"
]
],
[
[
"import pydsm\nfrom pydsm import ptm",
"_____no_output_____"
],
[
"ptm_file='D:/delta/dsm2_v8.2.0b1/studies/historical/output/anim_db.bin'\nptm_data=ptm.load_anim_data(ptm_file)\nprint('PTM Animation file has %d records'%len(ptm_data))\nprint('Animation starts at %s %04d'%(ptm_data[0].model_date.decode(\"utf-8\"),ptm_data[0].model_time))\nprint('Animation ends at %s %04d'%(ptm_data[-1].model_date.decode(\"utf-8\"),ptm_data[-1].model_time))\nprint('There are %d particles in this animation'%ptm_data[0].nparticles)",
"_____no_output_____"
],
[
"dfall=ptm.create_frame_for_anim_data(ptm_data)",
"_____no_output_____"
],
[
"import pydsm.hydroh5\nhydrofile='D:/delta/dsm2_v8.2.0b1/studies/historical/output/historical_v82.h5'\nhydro=pydsm.hydroh5.HydroH5(hydrofile)",
"_____no_output_____"
],
[
"chan_int2ext=pd.DataFrame(hydro.channel_index2number.items(),columns=['internal','external'],dtype=np.int32)\nchan_int2ext=chan_int2ext.assign(internal=chan_int2ext.internal+1)\nchan_int2ext.index=chan_int2ext.internal",
"_____no_output_____"
],
[
"dfallj=dfall.join(chan_int2ext,on='cid')\ndfallj=dfallj.fillna({'internal':-1,'external':-1})\ndfallj=dfallj.astype({'internal':np.int32,'external':np.int32})",
"_____no_output_____"
],
[
"#dsm2_chans=geopandas.read_file('maps/v8.2/DSM2_Channels.shp').to_crs(epsg=3857)\ndsm2_chans=geopandas.read_file('D:/delta/maps/v8.2/DSM2_Flowline_Segments.shp').to_crs(epsg=3857)\ndsm2_chan_geom_map={}\n# map from index+1 (numbers 1--> higher to )\nfor r in dsm2_chans.iterrows():\n chan=r[1]\n chan_index = hydro.channel_number2index[str(chan.channel_nu)]\n dsm2_chan_geom_map[chan_index+1] = chan.geometry",
"_____no_output_____"
],
[
"#dsm2_chans.index=dsm2_chans.ChannelNum\n#dfallj=dfallj.join(dsm2_chans['geometry'],on='external')",
"_____no_output_____"
],
[
"#dfallj[dfallj.id==1].external.hvplot()*dfallj[dfallj.id==10].external.hvplot()",
"_____no_output_____"
],
[
"def multindex_iloc(df, index):\n label = df.index.levels[0][index]\n return label,df.iloc[df.index.get_loc(label)]\ndef get_chan_geometry(channel_id):\n return dsm2_chan_geom_map[channel_id]\ndef interpolate_positions(dfn):\n vals=np.empty(len(dfn),dtype='object')\n i=0\n for r in dfn.iterrows():\n try:\n if r[1].cid > 0:\n geom=get_chan_geometry(r[1].cid)\n val=geom.interpolate(1.0-r[1].x/100.0,normalized=True)\n vals[i]=val\n i=i+1\n except:\n print('Exception for row: ',i, r)\n raise\n return vals\ndef get_particle_geopositions(time_index, dfall):\n dt,dfn = multindex_iloc(dfall,time_index) #\n #dfn=dfn.droplevel(0) # drop level as time is the slice \n dfn=dfall.loc[dt]\n #dfn['geometry'] = interpolate_positions(dfn) # add new column to copy of full slice so warning is ok\n dfn.assign(geometry=interpolate_positions(dfn))\n dfp=dfn[dfn.cid>0]\n gdf_merc=geopandas.GeoDataFrame(dfp,geometry='geometry',crs='EPSG:3857')\n #gdf_merc=gdf.to_crs(epsg=3857)\n x= np.fromiter((p.x for p in gdf_merc.geometry.values), dtype=np.float32)\n y = np.fromiter((p.y for p in gdf_merc.geometry.values), dtype=np.float32)\n dfp_xy = dfp.join([pd.DataFrame({'easting':x}), pd.DataFrame({'northing':y})])\n return dfp_xy",
"_____no_output_____"
],
[
"def cache_calcs(df):\n apos=interpolate_positions(df)\n #df_xy=df.assign('geometry',apos) # do we need the geometry column ?\n x=np.fromiter( (p.x if p else None for p in apos), dtype=np.float32)\n y=np.fromiter( (p.y if p else None for p in apos), dtype=np.float32)\n df_xy=df.assign(easting=x, northing=y)\n #save dfall_xy to cached parquet format or feather format\n return df_xy",
"_____no_output_____"
],
[
"#uncomment below to recalculate cached\ntry:\n dfall_xy = pd.read_pickle(ptm_file+'.pickle') # executed in 195ms\n #pd.read_parquet(ptm_file+'.parquet') # executed in 870 ms\n #pd.read_csv(ptm_file+'.csv') # executed in 4.30s,\nexcept: \n print('Failed to load cached calculated particle positions. Recalculating ...')\n dfall_xy=cache_calcs(dfall)\n dfall_xy.to_pickle(ptm_file+'.pickle') # executed in 469ms\n#dfall_xy.to_feather(ptm_file+'.feather') # failed to serialize multiindex\n#dfall_xy.to_parquet(ptm_file+'.parquet') # executed in 2.01s, 30 MB\n#dfall_xy.to_csv(ptm_file+'.csv') # executed in 29.6s, 348 MB\n#dfall_xy.to_pickle(ptm_file+'.pickle') # executed in 469ms, 138 MB",
"_____no_output_____"
],
[
"#dfall_xy.loc['2016-07-05']",
"_____no_output_____"
],
[
"#tlabel=dfall_xy.index.levels[0][500]\n#print(tlabel)",
"_____no_output_____"
],
[
"#dfp_xy=dfall_xy.loc[tlabel]\n#dfp_xy",
"_____no_output_____"
],
[
"import panel as pn\n# Tiles always in pseudo mercator epsg=3857\nb=dsm2_chans.to_crs(epsg=3857).geometry.bounds\nextents=(b.minx.min(),b.miny.min(),b.maxx.max(),b.maxy.max())\nmap=hv.element.tiles.OSM().opts(width=800,height=600)\nmap.extents=extents\n#display(map)\ndef particle_map(ti):\n tlabel=dfall_xy.index.levels[0][ti]\n dfp_xy=dfall_xy.loc[tlabel] \n ov=map*dfp_xy.hvplot.points(x='easting',y='northing',hover_cols=['id','cid']).opts(title=\"Time: %s\"%tlabel,framewise=False)\n return ov\ndmap=hv.DynamicMap(particle_map,kdims=['ti'])\ntime_index=dfall_xy.index.levels[0]\nanim_pane=dmap.redim.range(ti=(0,len(ptm_data)-1)).opts(framewise=True)\np=pn.panel(anim_pane)\ntitlePane=pn.pane.Markdown(''' # Particle Animation''')\nanim_panel=pn.Column(pn.Row(titlePane,*p[1]),p[0])\nanim_panel",
"_____no_output_____"
],
[
"#dsm2_chans.hvplot.line()",
"_____no_output_____"
],
[
"#anim_pane.select(ti=1500)",
"_____no_output_____"
],
[
"#pl1=dfall_xy[dfall_xy.id==373].cid.hvplot()\n#pl2=dfall_xy[dfall_xy.id==151].cid.hvplot()\n#pl1*pl2",
"_____no_output_____"
],
[
"#anim_pane.select(ti=500)",
"_____no_output_____"
],
[
"#from holoviews.streams import Selection1D\n#sel = Selection1D(source=anim_pane[0].Points.I)\n\n\n#print(anim_pane)\n\n#hv.DynamicMap(lambda index: dfall_xy[dfall_xy.index==id].hvplot(), streams=[sel])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccade14f836b23c961c243a67135e0ddb4fbbfe | 31,120 | ipynb | Jupyter Notebook | how-to-use-azureml/automated-machine-learning/forecasting-energy-demand/auto-ml-forecasting-energy-demand.ipynb | SivaReddyP/MachineLearningNotebooks | 3c9516797e67d8164fb5437224c6bb3c5b996182 | [
"MIT"
] | null | null | null | how-to-use-azureml/automated-machine-learning/forecasting-energy-demand/auto-ml-forecasting-energy-demand.ipynb | SivaReddyP/MachineLearningNotebooks | 3c9516797e67d8164fb5437224c6bb3c5b996182 | [
"MIT"
] | null | null | null | how-to-use-azureml/automated-machine-learning/forecasting-energy-demand/auto-ml-forecasting-energy-demand.ipynb | SivaReddyP/MachineLearningNotebooks | 3c9516797e67d8164fb5437224c6bb3c5b996182 | [
"MIT"
] | null | null | null | 45.497076 | 972 | 0.59608 | [
[
[
"Copyright (c) Microsoft Corporation. All rights reserved.\n\nLicensed under the MIT License.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# Automated Machine Learning\n_**Energy Demand Forecasting**_\n\n## Contents\n1. [Introduction](#Introduction)\n1. [Setup](#Setup)\n1. [Data](#Data)\n1. [Train](#Train)",
"_____no_output_____"
],
[
"## Introduction\nIn this example, we show how AutoML can be used to forecast a single time-series in the energy demand application area. \n\nMake sure you have executed the [configuration](../../../configuration.ipynb) before running this notebook.\n\nNotebook synopsis:\n1. Creating an Experiment in an existing Workspace\n2. Configuration and local run of AutoML for a simple time-series model\n3. View engineered features and prediction results\n4. Configuration and local run of AutoML for a time-series model with lag and rolling window features\n5. Estimate feature importance",
"_____no_output_____"
],
[
"## Setup\n",
"_____no_output_____"
]
],
[
[
"import azureml.core\nimport pandas as pd\nimport numpy as np\nimport logging\nimport warnings\n\n# Squash warning messages for cleaner output in the notebook\nwarnings.showwarning = lambda *args, **kwargs: None\n\nfrom azureml.core.workspace import Workspace\nfrom azureml.core.experiment import Experiment\nfrom azureml.train.automl import AutoMLConfig\nfrom matplotlib import pyplot as plt\nfrom sklearn.metrics import mean_absolute_error, mean_squared_error, r2_score",
"_____no_output_____"
]
],
[
[
"As part of the setup you have already created a <b>Workspace</b>. To run AutoML, you also need to create an <b>Experiment</b>. An Experiment corresponds to a prediction problem you are trying to solve, while a Run corresponds to a specific approach to the problem.",
"_____no_output_____"
]
],
[
[
"ws = Workspace.from_config()\n\n# choose a name for the run history container in the workspace\nexperiment_name = 'automl-energydemandforecasting'\n\nexperiment = Experiment(ws, experiment_name)\n\noutput = {}\noutput['SDK version'] = azureml.core.VERSION\noutput['Subscription ID'] = ws.subscription_id\noutput['Workspace'] = ws.name\noutput['Resource Group'] = ws.resource_group\noutput['Location'] = ws.location\noutput['Run History Name'] = experiment_name\npd.set_option('display.max_colwidth', -1)\noutputDf = pd.DataFrame(data = output, index = [''])\noutputDf.T",
"_____no_output_____"
]
],
[
[
"## Data\nWe will use energy consumption data from New York City for model training. The data is stored in a tabular format and includes energy demand and basic weather data at an hourly frequency. Pandas CSV reader is used to read the file into memory. Special attention is given to the \"timeStamp\" column in the data since it contains text which should be parsed as datetime-type objects. ",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv(\"nyc_energy.csv\", parse_dates=['timeStamp'])\ndata.head()",
"_____no_output_____"
]
],
[
[
"We must now define the schema of this dataset. Every time-series must have a time column and a target. The target quantity is what will be eventually forecasted by a trained model. In this case, the target is the \"demand\" column. The other columns, \"temp\" and \"precip,\" are implicitly designated as features.",
"_____no_output_____"
]
],
[
[
"# Dataset schema\ntime_column_name = 'timeStamp'\ntarget_column_name = 'demand'",
"_____no_output_____"
]
],
[
[
"### Forecast Horizon\n\nIn addition to the data schema, we must also specify the forecast horizon. A forecast horizon is a time span into the future (or just beyond the latest date in the training data) where forecasts of the target quantity are needed. Choosing a forecast horizon is application specific, but a rule-of-thumb is that **the horizon should be the time-frame where you need actionable decisions based on the forecast.** The horizon usually has a strong relationship with the frequency of the time-series data, that is, the sampling interval of the target quantity and the features. For instance, the NYC energy demand data has an hourly frequency. A decision that requires a demand forecast to the hour is unlikely to be made weeks or months in advance, particularly if we expect weather to be a strong determinant of demand. We may have fairly accurate meteorological forecasts of the hourly temperature and precipitation on a the time-scale of a day or two, however.\n\nGiven the above discussion, we generally recommend that users set forecast horizons to less than 100 time periods (i.e. less than 100 hours in the NYC energy example). Furthermore, **AutoML's memory use and computation time increase in proportion to the length of the horizon**, so the user should consider carefully how they set this value. If a long horizon forecast really is necessary, it may be good practice to aggregate the series to a coarser time scale. \n\n\nForecast horizons in AutoML are given as integer multiples of the time-series frequency. In this example, we set the horizon to 48 hours.",
"_____no_output_____"
]
],
[
[
"max_horizon = 48",
"_____no_output_____"
]
],
[
[
"### Split the data into train and test sets\nWe now split the data into a train and a test set so that we may evaluate model performance. We note that the tail of the dataset contains a large number of NA values in the target column, so we designate the test set as the 48 hour window ending on the latest date of known energy demand. ",
"_____no_output_____"
]
],
[
[
"# Find time point to split on\nlatest_known_time = data[~pd.isnull(data[target_column_name])][time_column_name].max()\nsplit_time = latest_known_time - pd.Timedelta(hours=max_horizon)\n\n# Split into train/test sets\nX_train = data[data[time_column_name] <= split_time]\nX_test = data[(data[time_column_name] > split_time) & (data[time_column_name] <= latest_known_time)]\n\n# Move the target values into their own arrays \ny_train = X_train.pop(target_column_name).values\ny_test = X_test.pop(target_column_name).values",
"_____no_output_____"
]
],
[
[
"## Train\n\nWe now instantiate an AutoMLConfig object. This config defines the settings and data used to run the experiment. For forecasting tasks, we must provide extra configuration related to the time-series data schema and forecasting context. Here, only the name of the time column and the maximum forecast horizon are needed. Other settings are described below:\n\n|Property|Description|\n|-|-|\n|**task**|forecasting|\n|**primary_metric**|This is the metric that you want to optimize.<br> Forecasting supports the following primary metrics <br><i>spearman_correlation</i><br><i>normalized_root_mean_squared_error</i><br><i>r2_score</i><br><i>normalized_mean_absolute_error</i>\n|**iterations**|Number of iterations. In each iteration, Auto ML trains a specific pipeline on the given data|\n|**iteration_timeout_minutes**|Time limit in minutes for each iteration.|\n|**X**|(sparse) array-like, shape = [n_samples, n_features]|\n|**y**|(sparse) array-like, shape = [n_samples, ], targets values.|\n|**n_cross_validations**|Number of cross validation splits. Rolling Origin Validation is used to split time-series in a temporally consistent way.|",
"_____no_output_____"
]
],
[
[
"time_series_settings = {\n 'time_column_name': time_column_name,\n 'max_horizon': max_horizon\n}\n\nautoml_config = AutoMLConfig(task='forecasting',\n debug_log='automl_nyc_energy_errors.log',\n primary_metric='normalized_root_mean_squared_error',\n blacklist_models = ['ExtremeRandomTrees', 'AutoArima'],\n iterations=10,\n iteration_timeout_minutes=5,\n X=X_train,\n y=y_train,\n n_cross_validations=3,\n verbosity = logging.INFO,\n **time_series_settings)",
"_____no_output_____"
]
],
[
[
"Submitting the configuration will start a new run in this experiment. For local runs, the execution is synchronous. Depending on the data and number of iterations, this can run for a while. Parameters controlling concurrency may speed up the process, depending on your hardware.\n\nYou will see the currently running iterations printing to the console.",
"_____no_output_____"
]
],
[
[
"local_run = experiment.submit(automl_config, show_output=True)",
"_____no_output_____"
],
[
"local_run",
"_____no_output_____"
]
],
[
[
"### Retrieve the Best Model\nBelow we select the best pipeline from our iterations. The get_output method on automl_classifier returns the best run and the fitted model for the last fit invocation. There are overloads on get_output that allow you to retrieve the best run and fitted model for any logged metric or a particular iteration.",
"_____no_output_____"
]
],
[
[
"best_run, fitted_model = local_run.get_output()\nfitted_model.steps",
"_____no_output_____"
]
],
[
[
"### View the engineered names for featurized data\nBelow we display the engineered feature names generated for the featurized data using the time-series featurization.",
"_____no_output_____"
]
],
[
[
"fitted_model.named_steps['timeseriestransformer'].get_engineered_feature_names()",
"_____no_output_____"
]
],
[
[
"### Test the Best Fitted Model\n\nFor forecasting, we will use the `forecast` function instead of the `predict` function. There are two reasons for this.\n\nWe need to pass the recent values of the target variable `y`, whereas the scikit-compatible `predict` function only takes the non-target variables `X`. In our case, the test data immediately follows the training data, and we fill the `y` variable with `NaN`. The `NaN` serves as a question mark for the forecaster to fill with the actuals. Using the forecast function will produce forecasts using the shortest possible forecast horizon. The last time at which a definite (non-NaN) value is seen is the _forecast origin_ - the last time when the value of the target is known. \n\nUsing the `predict` method would result in getting predictions for EVERY horizon the forecaster can predict at. This is useful when training and evaluating the performance of the forecaster at various horizons, but the level of detail is excessive for normal use.",
"_____no_output_____"
]
],
[
[
"# Replace ALL values in y_pred by NaN. \n# The forecast origin will be at the beginning of the first forecast period\n# (which is the same time as the end of the last training period).\ny_query = y_test.copy().astype(np.float)\ny_query.fill(np.nan)\n# The featurized data, aligned to y, will also be returned.\n# This contains the assumptions that were made in the forecast\n# and helps align the forecast to the original data\ny_fcst, X_trans = fitted_model.forecast(X_test, y_query)",
"_____no_output_____"
],
[
"# limit the evaluation to data where y_test has actuals\ndef align_outputs(y_predicted, X_trans, X_test, y_test, predicted_column_name = 'predicted'):\n \"\"\"\n Demonstrates how to get the output aligned to the inputs\n using pandas indexes. Helps understand what happened if\n the output's shape differs from the input shape, or if\n the data got re-sorted by time and grain during forecasting.\n \n Typical causes of misalignment are:\n * we predicted some periods that were missing in actuals -> drop from eval\n * model was asked to predict past max_horizon -> increase max horizon\n * data at start of X_test was needed for lags -> provide previous periods\n \"\"\"\n df_fcst = pd.DataFrame({predicted_column_name : y_predicted})\n # y and X outputs are aligned by forecast() function contract\n df_fcst.index = X_trans.index\n \n # align original X_test to y_test \n X_test_full = X_test.copy()\n X_test_full[target_column_name] = y_test\n\n # X_test_full's does not include origin, so reset for merge\n df_fcst.reset_index(inplace=True)\n X_test_full = X_test_full.reset_index().drop(columns='index')\n together = df_fcst.merge(X_test_full, how='right')\n \n # drop rows where prediction or actuals are nan \n # happens because of missing actuals \n # or at edges of time due to lags/rolling windows\n clean = together[together[[target_column_name, predicted_column_name]].notnull().all(axis=1)]\n return(clean)\n\ndf_all = align_outputs(y_fcst, X_trans, X_test, y_test)\ndf_all.head()",
"_____no_output_____"
]
],
[
[
"Looking at `X_trans` is also useful to see what featurization happened to the data.",
"_____no_output_____"
]
],
[
[
"X_trans",
"_____no_output_____"
]
],
[
[
"### Calculate accuracy metrics\nFinally, we calculate some accuracy metrics for the forecast and plot the predictions vs. the actuals over the time range in the test set.",
"_____no_output_____"
]
],
[
[
"def MAPE(actual, pred):\n \"\"\"\n Calculate mean absolute percentage error.\n Remove NA and values where actual is close to zero\n \"\"\"\n not_na = ~(np.isnan(actual) | np.isnan(pred))\n not_zero = ~np.isclose(actual, 0.0)\n actual_safe = actual[not_na & not_zero]\n pred_safe = pred[not_na & not_zero]\n APE = 100*np.abs((actual_safe - pred_safe)/actual_safe)\n return np.mean(APE)",
"_____no_output_____"
],
[
"print(\"Simple forecasting model\")\nrmse = np.sqrt(mean_squared_error(df_all[target_column_name], df_all['predicted']))\nprint(\"[Test Data] \\nRoot Mean squared error: %.2f\" % rmse)\nmae = mean_absolute_error(df_all[target_column_name], df_all['predicted'])\nprint('mean_absolute_error score: %.2f' % mae)\nprint('MAPE: %.2f' % MAPE(df_all[target_column_name], df_all['predicted']))\n\n# Plot outputs\n%matplotlib inline\npred, = plt.plot(df_all[time_column_name], df_all['predicted'], color='b')\nactual, = plt.plot(df_all[time_column_name], df_all[target_column_name], color='g')\nplt.xticks(fontsize=8)\nplt.legend((pred, actual), ('prediction', 'truth'), loc='upper left', fontsize=8)\nplt.title('Prediction vs. Actual Time-Series')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"The distribution looks a little heavy tailed: we underestimate the excursions of the extremes. A normal-quantile transform of the target might help, but let's first try using some past data with the lags and rolling window transforms.\n",
"_____no_output_____"
],
[
"### Using lags and rolling window features",
"_____no_output_____"
],
[
"We did not use lags in the previous model specification. In effect, the prediction was the result of a simple regression on date, grain and any additional features. This is often a very good prediction as common time series patterns like seasonality and trends can be captured in this manner. Such simple regression is horizon-less: it doesn't matter how far into the future we are predicting, because we are not using past data. In the previous example, the horizon was only used to split the data for cross-validation.\n\nNow that we configured target lags, that is the previous values of the target variables, and the prediction is no longer horizon-less. We therefore must still specify the `max_horizon` that the model will learn to forecast. The `target_lags` keyword specifies how far back we will construct the lags of the target variable, and the `target_rolling_window_size` specifies the size of the rolling window over which we will generate the `max`, `min` and `sum` features.\n\nThis notebook uses the blacklist_models parameter to exclude some models that take a longer time to train on this dataset. You can choose to remove models from the blacklist_models list but you may need to increase the iteration_timeout_minutes parameter value to get results.",
"_____no_output_____"
]
],
[
[
"time_series_settings_with_lags = {\n 'time_column_name': time_column_name,\n 'max_horizon': max_horizon,\n 'target_lags': 12,\n 'target_rolling_window_size': 4\n}\n\nautoml_config_lags = AutoMLConfig(task='forecasting',\n debug_log='automl_nyc_energy_errors.log',\n primary_metric='normalized_root_mean_squared_error',\n blacklist_models=['ElasticNet','ExtremeRandomTrees','GradientBoosting','XGBoostRegressor'],\n iterations=10,\n iteration_timeout_minutes=10,\n X=X_train,\n y=y_train,\n n_cross_validations=3,\n verbosity=logging.INFO,\n **time_series_settings_with_lags)",
"_____no_output_____"
]
],
[
[
"We now start a new local run, this time with lag and rolling window featurization. AutoML applies featurizations in the setup stage, prior to iterating over ML models. The full training set is featurized first, followed by featurization of each of the CV splits. Lag and rolling window features introduce additional complexity, so the run will take longer than in the previous example that lacked these featurizations.",
"_____no_output_____"
]
],
[
[
"local_run_lags = experiment.submit(automl_config_lags, show_output=True)",
"_____no_output_____"
],
[
"best_run_lags, fitted_model_lags = local_run_lags.get_output()\ny_fcst_lags, X_trans_lags = fitted_model_lags.forecast(X_test, y_query)\ndf_lags = align_outputs(y_fcst_lags, X_trans_lags, X_test, y_test)\ndf_lags.head()",
"_____no_output_____"
],
[
"X_trans_lags",
"_____no_output_____"
],
[
"print(\"Forecasting model with lags\")\nrmse = np.sqrt(mean_squared_error(df_lags[target_column_name], df_lags['predicted']))\nprint(\"[Test Data] \\nRoot Mean squared error: %.2f\" % rmse)\nmae = mean_absolute_error(df_lags[target_column_name], df_lags['predicted'])\nprint('mean_absolute_error score: %.2f' % mae)\nprint('MAPE: %.2f' % MAPE(df_lags[target_column_name], df_lags['predicted']))\n\n# Plot outputs\n%matplotlib inline\npred, = plt.plot(df_lags[time_column_name], df_lags['predicted'], color='b')\nactual, = plt.plot(df_lags[time_column_name], df_lags[target_column_name], color='g')\nplt.xticks(fontsize=8)\nplt.legend((pred, actual), ('prediction', 'truth'), loc='upper left', fontsize=8)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### What features matter for the forecast?\nThe following steps will allow you to compute and visualize engineered feature importance based on your test data for forecasting. ",
"_____no_output_____"
],
[
"#### Setup the model explanations for AutoML models\nThe *fitted_model* can generate the following which will be used for getting the engineered and raw feature explanations using *automl_setup_model_explanations*:-\n1. Featurized data from train samples/test samples \n2. Gather engineered and raw feature name lists\n3. Find the classes in your labeled column in classification scenarios\n\nThe *automl_explainer_setup_obj* contains all the structures from above list. ",
"_____no_output_____"
]
],
[
[
"from azureml.train.automl.automl_explain_utilities import AutoMLExplainerSetupClass, automl_setup_model_explanations\nautoml_explainer_setup_obj = automl_setup_model_explanations(fitted_model, X=X_train.copy(), \n X_test=X_test.copy(), y=y_train, \n task='forecasting')",
"_____no_output_____"
]
],
[
[
"#### Initialize the Mimic Explainer for feature importance\nFor explaining the AutoML models, use the *MimicWrapper* from *azureml.explain.model* package. The *MimicWrapper* can be initialized with fields in *automl_explainer_setup_obj*, your workspace and a LightGBM model which acts as a surrogate model to explain the AutoML model (*fitted_model* here). The *MimicWrapper* also takes the *best_run* object where the raw and engineered explanations will be uploaded.",
"_____no_output_____"
]
],
[
[
"from azureml.explain.model.mimic.models.lightgbm_model import LGBMExplainableModel\nfrom azureml.explain.model.mimic_wrapper import MimicWrapper\nexplainer = MimicWrapper(ws, automl_explainer_setup_obj.automl_estimator, LGBMExplainableModel, \n init_dataset=automl_explainer_setup_obj.X_transform, run=best_run,\n features=automl_explainer_setup_obj.engineered_feature_names, \n feature_maps=[automl_explainer_setup_obj.feature_map])",
"_____no_output_____"
]
],
[
[
"#### Use Mimic Explainer for computing and visualizing engineered feature importance\nThe *explain()* method in *MimicWrapper* can be called with the transformed test samples to get the feature importance for the generated engineered features. You can also use *ExplanationDashboard* to view the dash board visualization of the feature importance values of the generated engineered features by AutoML featurizers.",
"_____no_output_____"
]
],
[
[
"engineered_explanations = explainer.explain(['local', 'global'], eval_dataset=automl_explainer_setup_obj.X_test_transform)\nprint(engineered_explanations.get_feature_importance_dict())\nfrom azureml.contrib.interpret.visualize import ExplanationDashboard\nExplanationDashboard(engineered_explanations, automl_explainer_setup_obj.automl_estimator, automl_explainer_setup_obj.X_test_transform)",
"_____no_output_____"
]
],
[
[
"#### Use Mimic Explainer for computing and visualizing raw feature importance\nThe *explain()* method in *MimicWrapper* can be again called with the transformed test samples and setting *get_raw* to *True* to get the feature importance for the raw features. You can also use *ExplanationDashboard* to view the dash board visualization of the feature importance values of the raw features.",
"_____no_output_____"
]
],
[
[
"raw_explanations = explainer.explain(['local', 'global'], get_raw=True, \n raw_feature_names=automl_explainer_setup_obj.raw_feature_names,\n eval_dataset=automl_explainer_setup_obj.X_test_transform)\nprint(raw_explanations.get_feature_importance_dict())\nfrom azureml.contrib.interpret.visualize import ExplanationDashboard\nExplanationDashboard(raw_explanations, automl_explainer_setup_obj.automl_pipeline, automl_explainer_setup_obj.X_test_raw)",
"_____no_output_____"
]
],
[
[
"Please go to the Azure Portal's best run to see the top features chart.\n\nThe informative features make all sorts of intuitive sense. Temperature is a strong driver of heating and cooling demand in NYC. Apart from that, the daily life cycle, expressed by `hour`, and the weekly cycle, expressed by `wday` drives people's energy use habits.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eccae806da2e05b0ff9b2decd1a74553ecec56e9 | 7,266 | ipynb | Jupyter Notebook | Week1/Dense Net.ipynb | Armos05/Ai-for-Medical-Diagnosis-Specialization | fab2a6799335beccdeb1255fba08a779909c2d8a | [
"MIT"
] | 1 | 2021-11-21T13:35:58.000Z | 2021-11-21T13:35:58.000Z | Week1/Dense Net.ipynb | Armos05/Ai-for-Medical-Diagnosis-Specialization | fab2a6799335beccdeb1255fba08a779909c2d8a | [
"MIT"
] | null | null | null | Week1/Dense Net.ipynb | Armos05/Ai-for-Medical-Diagnosis-Specialization | fab2a6799335beccdeb1255fba08a779909c2d8a | [
"MIT"
] | null | null | null | 27.011152 | 291 | 0.583127 | [
[
[
"## AI for Medicine Course 1 Week 1 lecture exercises",
"_____no_output_____"
],
[
"<a name=\"densenet\"></a>\n# Densenet\n\nIn this week's assignment, you'll be using a pre-trained Densenet model for image classification. \n\nDensenet is a convolutional network where each layer is connected to all other layers that are deeper in the network\n- The first layer is connected to the 2nd, 3rd, 4th etc.\n- The second layer is connected to the 3rd, 4th, 5th etc.\n\nLike this:\n\n<img src=\"densenet.png\" alt=\"U-net Image\" width=\"400\" align=\"middle\"/>\n\nFor a detailed explanation of Densenet, check out the source of the image above, a paper by Gao Huang et al. 2018 called [Densely Connected Convolutional Networks](https://arxiv.org/pdf/1608.06993.pdf).\n\nThe cells below are set up to provide an exploration of the Keras densenet implementation that you'll be using in the assignment. Run these cells to gain some insight into the network architecture. ",
"_____no_output_____"
]
],
[
[
"# Import Densenet from Keras\nfrom keras.applications.densenet import DenseNet121\nfrom keras.layers import Dense, GlobalAveragePooling2D\nfrom keras.models import Model\nfrom keras import backend as K",
"Using TensorFlow backend.\n"
]
],
[
[
"For your work in the assignment, you'll be loading a set of pre-trained weights to reduce training time.",
"_____no_output_____"
]
],
[
[
"# Create the base pre-trained model\nbase_model = DenseNet121(weights='./nih/densenet.hdf5', include_top=False);",
"WARNING:tensorflow:From /opt/conda/lib/python3.6/site-packages/tensorflow_core/python/ops/resource_variable_ops.py:1630: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.\nInstructions for updating:\nIf using Keras pass *_constraint arguments to layers.\nWARNING:tensorflow:From /opt/conda/lib/python3.6/site-packages/keras/backend/tensorflow_backend.py:4070: The name tf.nn.max_pool is deprecated. Please use tf.nn.max_pool2d instead.\n\nWARNING:tensorflow:From /opt/conda/lib/python3.6/site-packages/keras/backend/tensorflow_backend.py:4074: The name tf.nn.avg_pool is deprecated. Please use tf.nn.avg_pool2d instead.\n\n"
]
],
[
[
"View a summary of the model",
"_____no_output_____"
]
],
[
[
"# Print the model summary\nbase_model.summary()",
"_____no_output_____"
],
[
"# Print out the first five layers\nlayers_l = base_model.layers\n\nprint(\"First 5 layers\")\nlayers_l[0:5]",
"_____no_output_____"
],
[
"# Print out the last five layers\nprint(\"Last 5 layers\")\nlayers_l[-6:-1]",
"_____no_output_____"
],
[
"# Get the convolutional layers and print the first 5\nconv2D_layers = [layer for layer in base_model.layers \n if str(type(layer)).find('Conv2D') > -1]\nprint(\"The first five conv2D layers\")\nconv2D_layers[0:5]",
"_____no_output_____"
],
[
"# Print out the total number of convolutional layers\nprint(f\"There are {len(conv2D_layers)} convolutional layers\")",
"_____no_output_____"
],
[
"# Print the number of channels in the input\nprint(\"The input has 3 channels\")\nbase_model.input",
"_____no_output_____"
],
[
"# Print the number of output channels\nprint(\"The output has 1024 channels\")\nx = base_model.output\nx",
"_____no_output_____"
],
[
"# Add a global spatial average pooling layer\nx_pool = GlobalAveragePooling2D()(x)\nx_pool",
"_____no_output_____"
],
[
"# Define a set of five class labels to use as an example\nlabels = ['Emphysema', \n 'Hernia', \n 'Mass', \n 'Pneumonia', \n 'Edema']\nn_classes = len(labels)\nprint(f\"In this example, you want your model to identify {n_classes} classes\")",
"_____no_output_____"
],
[
"# Add a logistic layer the same size as the number of classes you're trying to predict\npredictions = Dense(n_classes, activation=\"sigmoid\")(x_pool)\nprint(f\"Predictions have {n_classes} units, one for each class\")\npredictions",
"_____no_output_____"
],
[
"# Create an updated model\nmodel = Model(inputs=base_model.input, outputs=predictions)",
"_____no_output_____"
],
[
"# Compile the model\nmodel.compile(optimizer='adam',\n loss='categorical_crossentropy')\n# (You'll customize the loss function in the assignment!)",
"_____no_output_____"
]
],
[
[
"#### This has been a brief exploration of the Densenet architecture you'll use in this week's graded assignment!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
eccaebd5c9d8f10f46a7dc4867346e8e830dc03b | 11,398 | ipynb | Jupyter Notebook | docs/sphinx/quickstart.ipynb | nalzok/BATS.py | d2c6c0bbec547fe48f13a62e23d41ff040b48796 | [
"MIT"
] | 2 | 2020-10-29T19:40:54.000Z | 2021-04-13T22:00:55.000Z | docs/sphinx/quickstart.ipynb | nalzok/BATS.py | d2c6c0bbec547fe48f13a62e23d41ff040b48796 | [
"MIT"
] | 3 | 2021-08-24T17:57:46.000Z | 2022-02-22T15:37:24.000Z | docs/sphinx/quickstart.ipynb | nalzok/BATS.py | d2c6c0bbec547fe48f13a62e23d41ff040b48796 | [
"MIT"
] | 1 | 2021-06-28T16:23:37.000Z | 2021-06-28T16:23:37.000Z | 24.724512 | 212 | 0.5279 | [
[
[
"# Quickstart Guide\n\nOnce you have [successfully installed](installation.html) `bats`, you can follow this guide to help you get started. You can download this guide as a Jupyter notebook [here](_sources/quickstart.ipynb).\n\nYou can find additional information and examples in the [tutorials](tutorials/index.html) and [examples](examples/index.html), and ultimately the [API reference](api.html).\n\nFirst, import `bats`:",
"_____no_output_____"
]
],
[
[
"import bats",
"_____no_output_____"
]
],
[
[
"## Simplicial Complexes and Homology\n\nBATS offers two implementations of simplicial complexes: `SimplicialComplex` and `LightSimplicialComplex`. While the internal representations differ, they both have the same interface which can be used.\n\n<div class=\"alert alert-warning\">\n\nSimplices in bats should generally be assumed to be *ordered*, meaning that\n `[0,1,2]` is not the same as `[1,2,0]`. If you want to use\n *unordered* simplices, you can either add vertices in sorted order, or use\n a sorting algorithm before adding simplices to complexes.\n\n</div>\n\nThe `add` method will add simplices, assuming that all faces have previously been added. The `add_recursive` method will recursively add faces as needed.\n",
"_____no_output_____"
]
],
[
[
"X = bats.SimplicialComplex()\nX.add_recursive([0,1,2])\nX.add_recursive([2,3])\nX.add([1,3])\n\nX.get_simplices()",
"_____no_output_____"
]
],
[
[
"Now let's compute homology",
"_____no_output_____"
]
],
[
[
"R = bats.reduce(X, bats.F2()) # F2 is coefficient field\n\nfor k in range(R.maxdim()):\n print(\"dim H_{}: {}\".format(k, R.hdim(k)))",
"dim H_0: 1\ndim H_1: 1\n"
]
],
[
[
"The output of `bats.reduce` is a `ReducedChainComplex` which holds information used to compute homology.\n\nFor `LightSimplicialComplex`, you need to provide an upper bound on the number of vertices and maximum simplex dimension.",
"_____no_output_____"
]
],
[
[
"n = 4 # number of vertices\nd = 2 # max simplex dimension\nX = bats.LightSimplicialComplex(n, d)\nX.add_recursive([0,1,2])\nX.add_recursive([2,3])\nX.add([1,3])\n\nX.get_simplices()",
"_____no_output_____"
],
[
"R = bats.reduce(X, bats.F2())\n\nfor k in range(R.maxdim()):\n print(\"dim H_{}: {}\".format(k, R.hdim(k)))",
"dim H_0: 1\ndim H_1: 1\n"
]
],
[
[
"## Persistent Homology\n\nYou can add simplices to a filtration by providing a parameter at which they first appear.",
"_____no_output_____"
]
],
[
[
"F = bats.FilteredSimplicialComplex()\nF.add_recursive(0.0, [0,1,2])\nF.add_recursive(1.0, [2, 3])\nF.add(2.0, [1,3])\n\nF.complex().get_simplices()",
"_____no_output_____"
]
],
[
[
"again, we can use the `reduce` function, but now we get a `ReducedFilteredChainComplex`",
"_____no_output_____"
]
],
[
[
"R = bats.reduce(F, bats.F2())\n\nfor k in range(R.maxdim()):\n for p in R.persistence_pairs(k):\n print(p)",
"0 : (0,inf) <0,-1>\n0 : (0,0) <1,0>\n0 : (0,0) <2,1>\n0 : (1,1) <3,3>\n1 : (0,0) <2,0>\n1 : (2,inf) <4,-1>\n"
]
],
[
[
"The output of `R.persistence_pairs(k)` is a vector of `PersistencePairs` for k-dimensional persistent homology. \n\nA `PersistencePair` includes 5 pieces of information:\n* The dimension of the homology class.\n* The birth and death parameters of the homology class.\n* The simplex indices responsible for birth and death.",
"_____no_output_____"
]
],
[
[
"p = R.persistence_pairs(1)[-1]\nprint(p)\nprint(p.dim(), p.birth(), p.death(), p.birth_ind(), p.death_ind(), sep=', ')",
"1 : (2,inf) <4,-1>\n1, 2.0, inf, 4, 18446744073709551615\n"
]
],
[
[
"infinite bars have a death index set to `2**64 - 1`",
"_____no_output_____"
],
[
"## Maps\n\nBATS makes dealing with maps between topological spaces and associated chain maps\nand induced maps on homology easy. The relevant class is a `CellularMap` which\nkeeps track of what cells in one complex map to what cells in another.\n\nWe'll just look at a wrapper for `CellularMap`, called `SimplcialMap` which can be used\nto extend a map on the vertex set of a `SimplicialComplex` to a map of simplices.\n\nFirst, we'll build identical simplicial complexes `X` and `Y` which are both cycle graphs\non four vertices.",
"_____no_output_____"
]
],
[
[
"X = bats.SimplicialComplex()\nX.add_recursive([0,1])\nX.add_recursive([1,2])\nX.add_recursive([2,3])\nX.add_recursive([0,3])\n\nY = X",
"_____no_output_____"
]
],
[
[
"We then build a simplicial map from X to Y which is extended from a reflection of the vertices.",
"_____no_output_____"
]
],
[
[
"f0 = [2, 1, 0, 3]\nF = bats.SimplicialMap(X, Y, f0)",
"_____no_output_____"
]
],
[
[
"The map is extended by sending vertex `i` in `X` to vertex\n`f0[i]` in `Y`. Next, we can apply the chain functor. We'll use\nF3 coefficients.",
"_____no_output_____"
]
],
[
[
"CX = bats.Chain(X, bats.F3())\nCY = bats.Chain(Y, bats.F3())\nCF = bats.Chain(F, bats.F3())",
"_____no_output_____"
]
],
[
[
"Finally, we can compute homology of the chain complexes and the induced maps.",
"_____no_output_____"
]
],
[
[
"RX = bats.reduce(CX)\nRY = bats.reduce(CY)\n\nfor k in range(RX.maxdim()+1):\n HFk = bats.InducedMap(CF, RX, RY, k)\n print(\"induced map in dimension {}:\".format(k))\n print(HFk.tolist())",
"induced map in dimension 0:\n[[1]]\ninduced map in dimension 1:\n[[2]]\n"
]
],
[
[
"The induced map in dimension 0 is the identity. The induced map in dimension 1 is multiplication by `2 = -1 mod 3`",
"_____no_output_____"
],
[
"## Zigzag Homology\n\nWe'll now compute a simple zigzag barcode, using the above example. We'll consider\na diagram with two (identical) spaces, connected by a single edge which applies\nthe reflection map in the above example.",
"_____no_output_____"
]
],
[
[
"D = bats.SimplicialComplexDiagram(2,1) # diagram with 2 nodes, 1 edge\nD.set_node(0, X)\nD.set_node(1, Y)\nD.set_edge(0, 0, 1, F) # edge 0: F maps from node 0 to node 1",
"_____no_output_____"
]
],
[
[
"We can now apply the `Chain` and `Hom` functors to obtain a diagram of homology vector spaces and maps between them",
"_____no_output_____"
]
],
[
[
"CD = bats.ChainFunctor(D, bats.F3())\n",
"_____no_output_____"
],
[
"HD = bats.Hom(CD, 1) # computes homology in dimension 1\nps = bats.barcode(HD, 1) # extracts barcode\nfor p in ps:\n print(p)",
"1 : (0,1) <0,0>\n"
]
],
[
[
"This indicates there is a 1-dimensional homology bar, which is born in the space\nwith index 0 and survives until the space with index 1. The `<0,0>` indicates which\ngenerators are associated with the homology class in the diagram.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
eccaf5489a0166f9e165f68f17b7c4089cd56c26 | 313,387 | ipynb | Jupyter Notebook | opc_python/gerkin/collaborative.ipynb | SelectricSimian/olfaction-prediction | fb47cb343cdfd5cd8b06b33161dfa8b82c0319c6 | [
"MIT"
] | 20 | 2015-08-21T08:40:01.000Z | 2022-01-14T22:25:21.000Z | opc_python/gerkin/collaborative.ipynb | SelectricSimian/olfaction-prediction | fb47cb343cdfd5cd8b06b33161dfa8b82c0319c6 | [
"MIT"
] | 5 | 2015-08-18T21:03:35.000Z | 2017-06-27T21:19:10.000Z | opc_python/gerkin/collaborative.ipynb | SelectricSimian/olfaction-prediction | fb47cb343cdfd5cd8b06b33161dfa8b82c0319c6 | [
"MIT"
] | 16 | 2015-08-10T19:53:25.000Z | 2020-07-02T15:28:25.000Z | 129.125258 | 95,466 | 0.769221 | [
[
[
"### Preliminaries",
"_____no_output_____"
]
],
[
[
"# Show all figures inline. \n%matplotlib inline\n\n# Add olfaction-prediction to the Python path. \nimport os\nimport sys\ncurr_path = os.getcwd()\ngerkin_path = os.path.split(curr_path)[0]\nolfaction_prediction_path = os.path.split(gerkin_path)[0]\nsys.path.append(olfaction_prediction_path)\nimport opc_python\n\n# Import numerical libraries. \nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# Import generic utility modules I wrote to load the data from the tab-delimited text files and to score predictions. \nfrom opc_python.utils import loading, scoring\n\n# Import the modules I wrote for actually shaping and fitting the data to the model. \nfrom opc_python.gerkin import dream,fit1,fit2,params",
"_____no_output_____"
]
],
[
[
"###Load the data",
"_____no_output_____"
]
],
[
[
"# Load the perceptual descriptors data. \nperceptual_headers, perceptual_obs_data = loading.load_perceptual_data('training')\nloading.format_leaderboard_perceptual_data()\n# Show the perceptual metadata types and perceptual descriptor names.\nprint(perceptual_headers)",
"['Compound Identifier', 'Odor', 'Replicate', 'Intensity', 'Dilution', 'subject #', 'INTENSITY/STRENGTH', 'VALENCE/PLEASANTNESS', 'BAKERY', 'SWEET', 'FRUIT', 'FISH', 'GARLIC', 'SPICES', 'COLD', 'SOUR', 'BURNT', 'ACID', 'WARM', 'MUSKY', 'SWEATY', 'AMMONIA/URINOUS', 'DECAYED', 'WOOD', 'GRASS', 'FLOWER', 'CHEMICAL']\n"
],
[
"# Show the metadata and perceptual descriptor values for the first compound.\nprint(perceptual_obs_data[1])",
"['126', '4-Hydroxybenzaldehyde', False, 'high', '1/10', '1', 37, 60, 0, 72, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]\n"
],
[
"num_descriptors = len(perceptual_headers[6:])\nnum_subjects = 49\nprint('There are %d different perceptual descriptors and %d different subjects.' % (num_descriptors,num_subjects))",
"There are 21 different perceptual descriptors and 49 different subjects.\n"
],
[
"# Load the molecular descriptors data. \nmolecular_headers, molecular_data = loading.load_molecular_data()\nprint(\"First ten molecular descriptor types are %s\" % molecular_headers[:10])\nprint(\"First ten descriptor values for the first compound are %s\" % molecular_data[0][:10])\ntotal_size = len(set([int(row[0]) for row in molecular_data]))\nprint(\"We have %d molecular descriptors for %d unique molecules.\" % \\\n (len(molecular_data[0])-1,total_size))",
"First ten molecular descriptor types are ['CID', 'complexity from pubmed', 'MW', 'AMW', 'Sv', 'Se', 'Sp', 'Si', 'Mv', 'Me']\nFirst ten descriptor values for the first compound are ['126', 93.1, 122.13, 8.142, 10.01, 15.305, 10.193, 16.664, 0.667, 1.02]\nWe have 4869 molecular descriptors for 476 unique molecules.\n"
],
[
"# Determine the size of the training set. \ntraining_size = len(set([int(row[0]) for row in perceptual_obs_data]))\nprint(\"We have perceptual data for %d unique molecules.\" % training_size)\nremaining_size = total_size - training_size\nprint (\"%d are left out for testing in the competition; half of these (%d) are used for the leaderboard.\" \\\n % (remaining_size,remaining_size/2))",
"We have perceptual data for 338 unique molecules.\n138 are left out for testing in the competition; half of these (69) are used for the leaderboard.\n"
],
[
"# Determine how many data points there, and how many of these are replicates. \nprint(\"There are %d rows in the perceptual data set (at least one for each subject and molecule)\" % len(perceptual_obs_data))\nprint(\"%d of these are replicates (same subject and molecules).\" % sum([x[2] for x in perceptual_obs_data]))",
"There are 35084 rows in the perceptual data set (at least one for each subject and molecule)\n1960 of these are replicates (same subject and molecules).\n"
],
[
"# Get all Chemical IDs and located the data directory. \nall_CIDs = sorted(loading.get_CIDs('training')+loading.get_CIDs('leaderboard')+loading.get_CIDs('testset'))\nDATA = '/Users/rgerkin/Dropbox/science/olfaction-prediction/data/'\nimport pandas",
"_____no_output_____"
],
[
"# Load the Episuite features. \nepisuite = pandas.read_table('%s/DREAM_episuite_descriptors.txt' % DATA)\nepisuite.iloc[:,49] = 1*(episuite.iloc[:,49]=='YES ')\nepisuite.iloc[:,49]\nepisuite = episuite.iloc[:,2:].as_matrix()\nprint(\"Episuite has %d features for %d molecules.\" % (episuite.shape[1],episuite.shape[0]))",
"Episuite has 62 features for 476 molecules.\n"
],
[
"# Load the Verbal descriptors (from chemical names).\nverbal = pandas.read_table('%s/name_features.txt' % DATA, sep='\\t', header=None)\nverbal = verbal.as_matrix()[:,1:]\nverbal.shape\nprint(\"Verbal has %d features for %d molecules.\" % (verbal.shape[1],verbal.shape[0]))",
"Verbal has 11786 features for 476 molecules.\n"
],
[
"# Load the Morgan features. \nmorgan = pandas.read_csv('%s/morgan_sim.csv' % DATA)\nmorgan = morgan.as_matrix()[:,1:]\nprint(\"Morgan has %d features for %d molecules.\" % (morgan.shape[1],morgan.shape[0]))",
"Morgan has 2437 features for 476 molecules.\n"
],
[
"# Start to load the NSPDK features. \nwith open('%s/derived/nspdk_r3_d4_unaug.svm' % DATA) as f:\n nspdk_dict = {}\n i = 0\n while True:\n x = f.readline()\n if(len(x)):\n key_vals = x.split(' ')[1:]\n for key_val in key_vals:\n key,val = key_val.split(':')\n if key in nspdk_dict:\n nspdk_dict[key][all_CIDs[i]] = val\n else:\n nspdk_dict[key] = {all_CIDs[i]:val}\n i+=1\n if i == len(all_CIDs):\n break\n else:\n break\nnspdk_dict = {key:value for key,value in nspdk_dict.items() if len(value)>1}",
"_____no_output_____"
],
[
"# Get the NSPDK features into the right format. \nnspdk = np.zeros((len(all_CIDs),len(nspdk_dict)))\nfor j,(feature,facts) in enumerate(nspdk_dict.items()):\n for CID,value in facts.items():\n i = all_CIDs.index(CID)\n nspdk[i,j] = value\nprint(\"NSPDK has %d features for %d molecules.\" % (nspdk.shape[1],nspdk.shape[0]))",
"NSPDK has 6163 features for 476 molecules.\n"
],
[
"# Load the NSPDK Gramian features. \n# These require a large file that is not on GitHub, but can be obtained separately. \nnspdk_gramian = pandas.read_table('%s/derived/nspdk_r3_d4_unaug_gramian.mtx' % DATA, delimiter=' ', header=None)\nnspdk_gramian = nspdk_gramian.as_matrix()[:len(all_CIDs),:]\nprint(\"NSPDK Gramian has %d features for %d molecules.\" % \\\n (nspdk_gramian.shape[1],nspdk_gramian.shape[0]))",
"NSPDK Gramian has 2437 features for 476 molecules.\n"
],
[
"# Add all these new features to the molecular data dict. \nmolecular_data_extended = molecular_data.copy()\nmdx = molecular_data_extended\nfor i,line in enumerate(molecular_data):\n CID = int(line[0])\n index = all_CIDs.index(CID)\n mdx[i] = line + list(episuite[index]) + list(morgan[index]) + list(nspdk[index]) + list(nspdk_gramian[index])\nprint(\"There are now %d total features.\" % (len(mdx[0])-1))",
"There are now 15968 total features.\n"
],
[
"4869+62+2437+6163+2437",
"_____no_output_____"
]
],
[
[
"### Create matrices",
"_____no_output_____"
]
],
[
[
"# Create the feature matrices from the feature dicts. \nX_training,good1,good2,means,stds,imputer = dream.make_X(mdx,\"training\")\nX_leaderboard_other,good1,good2,means,stds,imputer = dream.make_X(mdx,\"leaderboard\",target_dilution='high',good1=good1,good2=good2,means=means,stds=stds)\nX_leaderboard_int,good1,good2,means,stds,imputer = dream.make_X(mdx,\"leaderboard\",target_dilution=-3,good1=good1,good2=good2,means=means,stds=stds)\nX_testset_other,good1,good2,means,stds,imputer = dream.make_X(mdx,\"testset\",target_dilution='high',good1=good1,good2=good2,means=means,stds=stds)\nX_testset_int,good1,good2,means,stds,imputer = dream.make_X(mdx,\"testset\",target_dilution=-3,good1=good1,good2=good2,means=means,stds=stds)\nX_all,good1,good2,means,stds,imputer = dream.make_X(mdx,['training','leaderboard'],good1=good1,good2=good2,means=means,stds=stds)",
"The X matrix now has shape (676x13914) molecules by non-NaN good molecular descriptors\nThe X matrix now has shape (69x13914) molecules by non-NaN good molecular descriptors\nThe X matrix now has shape (69x13914) molecules by non-NaN good molecular descriptors\nThe X matrix now has shape (69x13914) molecules by non-NaN good molecular descriptors\nThe X matrix now has shape (69x13914) molecules by non-NaN good molecular descriptors\nThe X matrix now has shape (814x13914) molecules by non-NaN good molecular descriptors\n"
],
[
"# Create descriptor matrices for the training set. \n# One is done with median imputation, and the other by masking missing values. \nY_training_imp,imputer = dream.make_Y_obs('training',target_dilution=None,imputer='median')\nY_training_mask,imputer = dream.make_Y_obs('training',target_dilution=None,imputer='mask')",
"The Y['mean_std'] matrix now has shape (676x42) molecules by 2 x perceptual descriptors\nThe Y['subject'] dict now has 49 matrices of shape (676x21) molecules by perceptual descriptors, one for each subject\nThe Y['mean_std'] matrix now has shape (676x42) molecules by 2 x perceptual descriptors\nThe Y['subject'] dict now has 49 matrices of shape (676x21) molecules by perceptual descriptors, one for each subject\n"
],
[
"# Create descriptor matrices for the leaderboard set. \n# One is done with median imputation, and the other with no imputation \nY_leaderboard,imputer = dream.make_Y_obs('leaderboard',target_dilution='gold',imputer='mask')\nY_leaderboard_noimpute,_ = dream.make_Y_obs('leaderboard',target_dilution='gold',imputer=None)",
"The Y['mean_std'] matrix now has shape (69x42) molecules by 2 x perceptual descriptors\nThe Y['subject'] dict now has 49 matrices of shape (69x21) molecules by perceptual descriptors, one for each subject\nThe Y['mean_std'] matrix now has shape (69x42) molecules by 2 x perceptual descriptors\nThe Y['subject'] dict now has 49 matrices of shape (69x21) molecules by perceptual descriptors, one for each subject\n"
],
[
"# Create descriptor matrices for the combined training and leaderboard sets. \n# One is done with median imputation, and the other by masking missing values. \nY_all_imp,imputer = dream.make_Y_obs(['training','leaderboard'],target_dilution=None,imputer='median')\nY_all_mask,imputer = dream.make_Y_obs(['training','leaderboard'],target_dilution=None,imputer='mask')",
"The Y['mean_std'] matrix now has shape (814x42) molecules by 2 x perceptual descriptors\nThe Y['subject'] dict now has 49 matrices of shape (814x21) molecules by perceptual descriptors, one for each subject\nThe Y['mean_std'] matrix now has shape (814x42) molecules by 2 x perceptual descriptors\nThe Y['subject'] dict now has 49 matrices of shape (814x21) molecules by perceptual descriptors, one for each subject\n"
]
],
[
[
"### Data visualization and obtaining fit parameters",
"_____no_output_____"
]
],
[
[
"# Show the range of values for the molecular and perceptual descriptors. \nfig,axes = plt.subplots(1,2,figsize=(10,4))\nax = axes.flat\nax[0].hist(X_training.ravel())\nax[0].set_xlabel('Cube root transformed, N(0,1) normalized molecular descriptor values')\nax[1].hist(Y_training_imp['mean_std'][:21].ravel())\nax[1].set_xlabel('Perceptual descriptor subject-averaged values')\nfor ax_ in ax:\n ax_.set_yscale('log')\n ax_.set_ylabel('Count')\nplt.tight_layout()",
"_____no_output_____"
],
[
"import matplotlib\nmatplotlib.rcParams['font.size'] = 18\nplt.figure(figsize=(8,6))\nintensity = Y_leaderboard['mean_std'][:,0]\nintensity2 = -np.log(100/intensity - 1)\nintensity2 += 0.9*np.random.randn(69)\nintensity2 = 100/(1+np.exp(-intensity2))\nplt.scatter(intensity,intensity2)\nplt.xlabel('Intensity (predicted)')\nplt.ylabel('Intensity (actual)')\nplt.xlim(0,100)\nplt.ylim(0,100)\nplt.plot([0,100],[0,100],label='r = 0.75')\nplt.legend(loc=2)\nnp.corrcoef(intensity,intensity2)[0,1]",
"_____no_output_____"
],
[
"plt.figure(figsize=(8,6))\nintensity = Y_leaderboard['mean_std'][:,1]\nintensity2 = -np.log(100/intensity - 1)\nintensity2 += 0.55*np.random.randn(69)\nintensity2 = 100/(1+np.exp(-intensity2))\nplt.scatter(intensity,intensity2)\nplt.xlabel('Pleasantness (predicted)')\nplt.ylabel('Pleasantness (actual)')\nplt.xlim(0,100)\nplt.ylim(0,100)\nplt.plot([0,100],[0,100],label='r = 0.70')\nplt.legend(loc=2)\nnp.corrcoef(intensity,intensity2)[0,1]",
"_____no_output_____"
],
[
"# Plot stdev vs mean for each descriptor, and fit to a theoretically-motivated function. \n# These fit parameters will be used in the final model fit. \ndef f_transformation(x, k0=1.0, k1=1.0):\n return 100*(k0*(x/100)**(k1*0.5) - k0*(x/100)**(k1*2))\n \ndef sse(x, mean, stdev):\n predicted_stdev = f_transformation(mean, k0=x[0], k1=x[1])\n sse = np.sum((predicted_stdev - stdev)**2)\n return sse\n\nmatplotlib.rcParams['font.size'] = 12\nfig,axes = plt.subplots(3,7,sharex=True,sharey=True,figsize=(12,6))\nax = axes.flat\ntrans_params = {col:None for col in range(21)}\nfrom scipy.optimize import minimize\nfor col in range(len(ax)): \n Y_mean = Y_all_mask['mean_std'][:,col]\n Y_stdev = Y_all_mask['mean_std'][:,col+21]\n x = [1.0,1.0]\n res = minimize(sse, x, args=(Y_mean,Y_stdev), method='L-BFGS-B')\n trans_params[col] = res.x # We will use these for our transformations. \n ax[col].scatter(Y_mean,Y_stdev,s=0.1)\n x_ = np.linspace(0,100,100)\n ax[col].plot(x_,f_transformation(x_, k0=res.x[0], k1=res.x[1]))\n ax[col].set_title(perceptual_headers[col+6].split('/')[1 if col==1 else 0])\n ax[col].set_xlim(0,100)\n ax[col].set_ylim(0,60)\n if col == 17:\n ax[col].set_xlabel('Mean')\n if col == 7:\n ax[col].set_ylabel('StDev')\nplt.tight_layout()\nplt.savefig('3c.eps',format='eps')",
"_____no_output_____"
],
[
"plt.figure(figsize=(6,6))\nY_mean = Y_all_mask['mean_std'][:,0]\nY_stdev = Y_all_mask['mean_std'][:,0+21]\nplt.scatter(Y_mean,Y_stdev,color='black')\nplt.xlabel('Mean Rating',size=18)\nplt.ylabel('StDev of Rating',size=18)\nplt.xticks(np.arange(0,101,20),size=15)\nplt.yticks(np.arange(0,51,10),size=15)\nplt.xlim(0,100)\nplt.ylim(0,50)\nres = minimize(sse, x, args=(Y_mean,Y_stdev), method='L-BFGS-B')\nplt.plot(x_,f_transformation(x_, k0=res.x[0], k1=res.x[1]),color='cyan',linewidth=5)\nplt.title('INTENSITY',size=18)",
"_____no_output_____"
],
[
"# Load optimal parameters (obtained from extensive cross-validation).\ncols = range(42)\ndef get_params(i):\n return {col:params.best[col][i] for col in cols}\n\nuse_et = get_params(0)\nmax_features = get_params(1)\nmax_depth = get_params(2)\nmin_samples_leaf = get_params(3)\ntrans_weight = get_params(4)\nregularize = get_params(4)\nuse_mask = get_params(5)\nfor col in range(21):\n trans_weight[col] = trans_weight[col+21]",
"_____no_output_____"
]
],
[
[
"### Fitting and Generating Submission Files for challenge 2",
"_____no_output_____"
]
],
[
[
"# Fit training data. \n# Ignoring warning that arises if too few trees are used. \n# Ignore intensity score which is based on within-sample validation, \n# due to use of ExtraTreesClassifier.\nn_estimators = 1000\nrfcs_leaderboard,score,rs = fit2.rfc_final(X_training,Y_training_imp['mean_std'],\n Y_training_mask['mean_std'],max_features,\n min_samples_leaf,max_depth,use_et,use_mask,\n trans_weight,trans_params,\n n_estimators=n_estimators)",
"97.62% [------------------------------------------------- ]For subchallenge 2:\n\tScore = 9.71\n\tint_mean = 1.000\n\tint_sigma = 0.936\n\tple_mean = 0.699\n\tple_sigma = 0.237\n\tdec_mean = 0.516\n\tdec_sigma = 0.421\n"
],
[
"# Make challenge 2 leaderboard prediction files from the models. \nloading.make_prediction_files(rfcs_leaderboard,X_leaderboard_int,X_leaderboard_other,\n 'leaderboard',2,Y_test=Y_leaderboard_noimpute,\n write=True,trans_weight=trans_weight,trans_params=trans_params)",
"Score: 8.961307; rs = 0.641,0.573,0.569,0.431,0.117,0.499\nWrote to file with suffix \"1446422354\"\n"
],
[
"# Fit all available data. \n# Ignoring warning that arises if too few trees are used. \n# Ignore intensity score which is based on within-sample validation, \n# due to use of ExtraTreesClassifier.\nn_estimators = 500\nrfcs,score,rs = fit2.rfc_final(X_all,Y_all_imp['mean_std'],Y_all_mask['mean_std'],\n max_features,min_samples_leaf,max_depth,use_et,use_mask,\n trans_weight,trans_params,n_estimators=n_estimators)",
"97.62% [------------------------------------------------- ]For subchallenge 2:\n\tScore = 14.62\n\tint_mean = 1.000\n\tint_sigma = 0.938\n\tple_mean = 0.970\n\tple_sigma = 0.784\n\tdec_mean = 0.818\n\tdec_sigma = 0.734\n"
],
[
"feature_importances_matrix = np.array([rfcs[i].feature_importances_ for i in range(21)])\nfeature_importances_matrix.shape",
"_____no_output_____"
],
[
"episuite = pandas.read_table('%s/DREAM_episuite_descriptors.txt' % DATA)\nmorgan = pandas.read_csv('%s/morgan_sim.csv' % DATA)\nfeature_list = []\nfeature_list += ['dragon_'+x for x in molecular_headers[1:]]\nfeature_list += ['episuite_'+x for x in list(episuite)[2:]]\nfeature_list += ['morgan_'+x for x in list(morgan)[1:]]\nfeature_list += ['nspdk_'+x for x in nspdk_dict.keys()]\nfeature_list += ['nspdkgramian_'+str(x) for x in range(2437)]\nfeature_list += ['conc_absolute','conc_relative']\nfeature_list = [x for i,x in enumerate(feature_list) if i in good1]\nfeature_list = [x for i,x in enumerate(feature_list) if i in good2]\nprint(len(feature_list))",
"13914\n"
],
[
"feature_importances = pandas.DataFrame(data=feature_importances_matrix.T,index=feature_list,columns=perceptual_headers[6:])\nfeature_importances.head()\nfeature_importances.to_csv('feature_importances.csv')",
"_____no_output_____"
],
[
"# Make challenge 2 testset prediction files from the models. \nloading.make_prediction_files(rfcs,X_testset_int,X_testset_other,'testset',2,write=True,\n trans_weight=trans_weight,trans_params=trans_params)",
"Wrote to file with suffix \"1446435104\"\n"
],
[
"# Fit training data for subchallenge 1. \n# Ignoring warning that arises if too few trees are used. \n# Ignore intensity score which is based on within-sample validation, \n# due to use of ExtraTreesClassifier.\nn_estimators = 50\nrfcs_leaderboard,score,rs = fit1.rfc_final(X_training,Y_training_imp['subject'],max_features,\n min_samples_leaf,max_depth,use_et,\n Y_test=Y_leaderboard_noimpute['subject'],\n regularize=regularize,\n n_estimators=n_estimators)",
"98.00% [------------------------------------------------- ]For subchallenge 1:\n\tScore = 37.12\n\tint = 0.843\n\tple = 0.402\n\tdec = 0.182\n"
],
[
"# Make challenge 1 leaderboard prediction files from the models. \nloading.make_prediction_files(rfcs_leaderboard,X_leaderboard_int,X_leaderboard_other,\n 'leaderboard',1,Y_test=Y_leaderboard_noimpute,\n write=True,regularize=regularize)",
"Score: 27.753160; rs = 0.429,0.317,0.178\nWrote to file with suffix \"1446474725\"\n"
],
[
"# Fit all available data for subchallenge 1. \n# Ignoring warning that arises if too few trees are used. \n# Ignore intensity score which is based on within-sample validation, \n# due to use of ExtraTreesClassifier.\nrfcs1,score1,rs1 = fit1.rfc_final(X_all,Y_all_imp['subject'],max_features,\n min_samples_leaf,max_depth,use_et,\n regularize=regularize,\n n_estimators=n_estimators)",
"98.00% [------------------------------------------------- ]For subchallenge 1:\n\tScore = 37.33\n\tint = 0.841\n\tple = 0.406\n\tdec = 0.185\n"
],
[
"# Make challenge 1 testset prediction files from the models. \nloading.make_prediction_files(rfcs1,X_testset_int,X_testset_other,\n 'testset',1,write=True,regularize=regularize)",
"Wrote to file with suffix \"1446510971\"\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccb051554d29b5e295cf872593a2a5f9b65fd4c | 13,145 | ipynb | Jupyter Notebook | content/lessons/07/Class-Coding-Lab/CCL-Strings.ipynb | auramnar/Summer2018Learn2Code | 068b89215ad2b90f57a73511b0b1b6b126589c5f | [
"MIT"
] | null | null | null | content/lessons/07/Class-Coding-Lab/CCL-Strings.ipynb | auramnar/Summer2018Learn2Code | 068b89215ad2b90f57a73511b0b1b6b126589c5f | [
"MIT"
] | null | null | null | content/lessons/07/Class-Coding-Lab/CCL-Strings.ipynb | auramnar/Summer2018Learn2Code | 068b89215ad2b90f57a73511b0b1b6b126589c5f | [
"MIT"
] | null | null | null | 27.442589 | 910 | 0.539673 | [
[
[
"# In-Class Coding Lab: Strings\n\nThe goals of this lab are to help you to understand:\n\n- String slicing for substrings\n- How to use Python's built-in String functions in the standard library.\n- Tokenizing and Parsing Data\n- How to create user-defined functions to parse and tokenize strings\n\n\n# Strings\n\n## Strings are immutable sequences\n\nPython strings are immutable sequences.This means we cannot change them \"in part\" and there is impicit ordering. \n\nThe characters in a string are zero-based. Meaning the index of the first character is 0.\n\nWe can leverage this in a variety of ways.\n\nFor example:",
"_____no_output_____"
]
],
[
[
"x = input(\"Enter something: \")\nprint (\"You typed:\", x)\nprint (\"number of characters:\", len(x) )\nprint (\"First character is:\", x[0])\nprint (\"Last character is:\", x[-1])\n\n## They're sequences, so you can loop definately:\nprint(\"Printing one character at a time: \")\nfor ch in x:\n print(ch) # print a character at a time!\n ",
"Enter something: tony\nYou typed: tony\nnumber of characters: 4\nFirst character is: t\nLast character is: y\nPrinting one character at a time: \nt\no\nn\ny\n"
]
],
[
[
"## Slices as substrings\n\nPython lists and sequences use **slice notation** which is a clever way to get substring from a given string.\n\nSlice notation requires two values: A start index and the end index. The substring returned starts at the start index, and *ends at the position before the end index*. It ends at the position *before* so that when you slice a string into parts you know where you've \"left off\". \n\nFor example:",
"_____no_output_____"
]
],
[
[
"state = \"Mississippi\"\nprint (state[0:4]) # Miss\nprint (state[4:len(state)]) # issippi",
"Miss\nissippi\n"
]
],
[
[
"In this next example, play around with the variable `split` adjusting it to how you want the string to be split up. Re run the cell several times with different values to get a feel for what happens.",
"_____no_output_____"
]
],
[
[
"state = \"Mississippi\"\nsplit = 5 # TODO: play around with this number\nleft = state[0:split]\nright = state[split:len(state)]\nprint(left, right)",
"Missi ssippi\n"
]
],
[
[
"### Slicing from the beginning or to the end\n\nIf you omit the begin or end slice, Python will slice from the beginnning of the string or all the way to the end. So if you say `x[:5]` its the same as `x[0:5]`\n\nFor example:",
"_____no_output_____"
]
],
[
[
"state = \"Ohio\"\nprint(state[0:2], state[:2]) # same!\nprint(state[2:len(state)], state[2:]) # same\n",
"Oh Oh\nio io\n"
]
],
[
[
"### Now Try It!\n\nSplit the string `\"New Hampshire\"` into two sub-strings one containing `\"New\"` the other containing `\"Hampshire\"` (without the space).",
"_____no_output_____"
]
],
[
[
"## TODO: Write code here\nstate = \"New Hampshire\"\nstate.split()",
"_____no_output_____"
]
],
[
[
"## Python's built in String Functions\n\nPython includes several handy built-in string functions (also known as *methods* in object-oriented parlance). To get a list of available functions, use the `dir()` function on any string variable, or on the type `str` itself.\n",
"_____no_output_____"
]
],
[
[
"print ( dir(str))",
"['__add__', '__class__', '__contains__', '__delattr__', '__dir__', '__doc__', '__eq__', '__format__', '__ge__', '__getattribute__', '__getitem__', '__getnewargs__', '__gt__', '__hash__', '__init__', '__iter__', '__le__', '__len__', '__lt__', '__mod__', '__mul__', '__ne__', '__new__', '__reduce__', '__reduce_ex__', '__repr__', '__rmod__', '__rmul__', '__setattr__', '__sizeof__', '__str__', '__subclasshook__', 'capitalize', 'casefold', 'center', 'count', 'encode', 'endswith', 'expandtabs', 'find', 'format', 'format_map', 'index', 'isalnum', 'isalpha', 'isdecimal', 'isdigit', 'isidentifier', 'islower', 'isnumeric', 'isprintable', 'isspace', 'istitle', 'isupper', 'join', 'ljust', 'lower', 'lstrip', 'maketrans', 'partition', 'replace', 'rfind', 'rindex', 'rjust', 'rpartition', 'rsplit', 'rstrip', 'split', 'splitlines', 'startswith', 'strip', 'swapcase', 'title', 'translate', 'upper', 'zfill']\n"
]
],
[
[
"Let's suppose you want to learn how to use the `count` function. There are 2 ways you can do this.\n\n1. search the web for `python 3 str count` or\n1. bring up internal help `help(str.count)` \n\nBoth have their advantages and disadvanges. I would start with the second one, and only fall back to a web search when you can't figure it out from the Python documenation. \n\nHere's the documentation for `count`",
"_____no_output_____"
]
],
[
[
"help(str.count)",
"Help on method_descriptor:\n\ncount(...)\n S.count(sub[, start[, end]]) -> int\n \n Return the number of non-overlapping occurrences of substring sub in\n string S[start:end]. Optional arguments start and end are\n interpreted as in slice notation.\n\n"
]
],
[
[
"You'll notice in the help output it says S.count() this indicates this function is a method function. this means you invoke it like this `variable.count()`.\n\n### Now Try It\n\nTry to use the count() function method to count the number of `'i'`'s in the string `'Mississippi`:",
"_____no_output_____"
]
],
[
[
"state = 'Mississippi'\n#TODO: use state.count\nstate.count('i')",
"_____no_output_____"
]
],
[
[
"### TANGENT: The Subtle difference between function and method.\n\nYou'll notice sometimes we call our function alone, other times it's attached to a variable, as was the case in the example above. when we say `state.count('i')` the period (`.`) between the variable and function indicates this function is a *method function*. The key difference between a the two is a method is attached to a variable. To call a method function you must say `variable.function()` whereas when you call a function its just `function()`. The variable associated with the method call is usually part of the function's context.\n\nHere's an example:",
"_____no_output_____"
]
],
[
[
"name = \"Larry\"\nprint( len(name) ) # a function call len(name) stands on its own. Gets length of 'Larry'\nprint( name.__len__() ) # a method call name.__len__() does the name thing for its variable 'Larry'",
"5\n2\n"
]
],
[
[
"### Now Try It\n\nTry to figure out which built in string function to use to accomplish this task.\n\nWrite some code to find the text `'is'` in some text. The program shoud output the first position of `'is'` in the text. \n\nExamples:\n\n```\nWhen: text = 'Mississippi' then position = 1\nWhen: text = \"This is great\" then position = 2\nWhen: text = \"Burger\" then position = -1\n```",
"_____no_output_____"
]
],
[
[
"# TODO: Write your code here\ntext = input(\"Enter some text: \")\n\nis_location = text.find('is')\n\nprint(\"When: text = '\" + text + \"' then position =\", is_location)",
"Enter some text: This is it\nWhen: text = 'This is it' then position = 2\n"
]
],
[
[
"### Now Try It\n\n**Is that a URL?**\n\nTry to write a rudimentary URL checker. The program should input a text string and then use the `startswith` function to check if the string begins with `\"http://\"` or `\"https://\"` If it does we can assume it is a URL. ",
"_____no_output_____"
]
],
[
[
"## TODO: write code here:\nurl = input(\"Enter a URL?: \")\n\nif url.startswith('http://') or url.startswith('https://'):\n print(\"Yes! You've entered a valid URL.\")\nelse:\n print(\"Sorry. That is not a valid URL.\")",
"Enter a URL?: http://hello.com\nYes! You've entered a valid URL.\n"
],
[
"url = input(\"Enter a URL?: \")\n\nif url.startswith('http://') or url.startswith('https://'):\n print(\"Yes! You've entered a valid URL.\")\nelse:\n print(\"Sorry. That is not a valid URL.\")",
"Enter a URL?: https://secure.com\nYes! You've entered a valid URL.\n"
],
[
"url = input(\"Enter a URL?: \")\n\nif url.startswith('http://') or url.startswith('https://'):\n print(\"Yes! You've entered a valid URL.\")\nelse:\n print(\"Sorry. That is not a valid URL.\")",
"Enter a URL?: http\nSorry. That is not a valid URL.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
eccb095b08b9f1b72dbad91dc2524b187c3566b8 | 37,992 | ipynb | Jupyter Notebook | Chapter02/04_storage_benchmark/storage_benchmark.ipynb | gabrielmahia/pesAI | cb22627b2987ff58ae5ee1c05e21c913214c16cd | [
"MIT"
] | 944 | 2019-01-06T22:44:31.000Z | 2022-03-30T14:47:33.000Z | Chapter02/04_storage_benchmark/storage_benchmark.ipynb | VestiDev/Hands-On-Machine-Learning-for-Algorithmic-Trading-bookgit | 336b3e622c8f161fc1844a5d06ae30b75809aba3 | [
"MIT"
] | 14 | 2019-01-05T08:34:25.000Z | 2021-11-15T01:11:54.000Z | Chapter02/04_storage_benchmark/storage_benchmark.ipynb | VestiDev/Hands-On-Machine-Learning-for-Algorithmic-Trading-bookgit | 336b3e622c8f161fc1844a5d06ae30b75809aba3 | [
"MIT"
] | 598 | 2018-12-09T00:09:50.000Z | 2022-03-18T10:21:13.000Z | 33.532215 | 7,516 | 0.685407 | [
[
[
"from pathlib import Path\nimport pandas as pd\nimport numpy as np\nimport random\nimport string",
"_____no_output_____"
],
[
"results = {}",
"_____no_output_____"
]
],
[
[
"## Generate Test Data",
"_____no_output_____"
],
[
"The test `DataFrame` that can be configured to contain numerical or text data, or both. For the HDF5 library, we test both the fixed and table format. ",
"_____no_output_____"
]
],
[
[
"def generate_test_data(nrows=100000, numerical_cols=2000, text_cols=0, text_length=10):\n ncols = numerical_cols + text_cols\n s = \"\".join([random.choice(string.ascii_letters)\n for _ in range(text_length)])\n data = pd.concat([pd.DataFrame(np.random.random(size=(nrows, numerical_cols))),\n pd.DataFrame(np.full(shape=(nrows, text_cols), fill_value=s))],\n axis=1, ignore_index=True)\n data.columns = [str(i) for i in data.columns]\n return data",
"_____no_output_____"
],
[
"df = generate_test_data()\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 100000 entries, 0 to 99999\nColumns: 2000 entries, 0 to 1999\ndtypes: float64(2000)\nmemory usage: 1.5 GB\n"
]
],
[
[
"## Parquet",
"_____no_output_____"
],
[
"### Size",
"_____no_output_____"
]
],
[
[
"parquet_file = Path('test.parquet')",
"_____no_output_____"
],
[
"df.to_parquet(parquet_file)\nsize = parquet_file.stat().st_size",
"_____no_output_____"
]
],
[
[
"### Read",
"_____no_output_____"
]
],
[
[
"%%timeit -o\ndf = pd.read_parquet(parquet_file)",
"809 ms ± 14.2 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"read = _",
"_____no_output_____"
],
[
"parquet_file.unlink()",
"_____no_output_____"
]
],
[
[
"### Write",
"_____no_output_____"
]
],
[
[
"%%timeit -o\ndf.to_parquet(parquet_file)\nparquet_file.unlink()",
"12.1 s ± 32 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"write = _",
"_____no_output_____"
]
],
[
[
"### Results",
"_____no_output_____"
]
],
[
[
"results['parquet'] = {'read': read.all_runs, 'write': write.all_runs, 'size': size}",
"_____no_output_____"
]
],
[
[
"## HDF5",
"_____no_output_____"
]
],
[
[
"test_store = Path('index.h5')",
"_____no_output_____"
]
],
[
[
"### Fixed Format",
"_____no_output_____"
],
[
"#### Size",
"_____no_output_____"
]
],
[
[
"with pd.HDFStore(test_store) as store:\n store.put('file', df)\nsize = test_store.stat().st_size",
"_____no_output_____"
]
],
[
[
"#### Read",
"_____no_output_____"
]
],
[
[
"%%timeit -o\nwith pd.HDFStore(test_store) as store:\n store.get('file')",
"449 ms ± 9.4 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"read = _",
"_____no_output_____"
],
[
"test_store.unlink()",
"_____no_output_____"
]
],
[
[
"#### Write",
"_____no_output_____"
]
],
[
[
"%%timeit -o\nwith pd.HDFStore(test_store) as store:\n store.put('file', df)\ntest_store.unlink()",
"4.2 s ± 28.8 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"write = _",
"_____no_output_____"
]
],
[
[
"#### Results",
"_____no_output_____"
]
],
[
[
"results['hdf_fixed'] = {'read': read.all_runs, 'write': write.all_runs, 'size': size}",
"_____no_output_____"
]
],
[
[
"### Table Format",
"_____no_output_____"
],
[
"#### Size",
"_____no_output_____"
]
],
[
[
"with pd.HDFStore(test_store) as store:\n store.append('file', df, format='t')\nsize = test_store.stat().st_size ",
"_____no_output_____"
]
],
[
[
"#### Read",
"_____no_output_____"
]
],
[
[
"%%timeit -o\nwith pd.HDFStore(test_store) as store:\n df = store.get('file')",
"508 ms ± 9.14 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"read = _",
"_____no_output_____"
],
[
"test_store.unlink()",
"_____no_output_____"
]
],
[
[
"#### Write",
"_____no_output_____"
],
[
"Note that `write` in table format does not work with text data.",
"_____no_output_____"
]
],
[
[
"%%timeit -o\nwith pd.HDFStore(test_store) as store:\n store.append('file', df, format='t')\ntest_store.unlink() ",
"2.92 s ± 11.6 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"write = _",
"_____no_output_____"
]
],
[
[
"#### Results",
"_____no_output_____"
]
],
[
[
"results['hdf_table'] = {'read': read.all_runs, 'write': write.all_runs, 'size': size}",
"_____no_output_____"
]
],
[
[
"### Table Select",
"_____no_output_____"
],
[
"#### Size",
"_____no_output_____"
]
],
[
[
"with pd.HDFStore(test_store) as store:\n store.append('file', df, format='t', data_columns=['company', 'form'])\nsize = test_store.stat().st_size ",
"_____no_output_____"
]
],
[
[
"#### Read",
"_____no_output_____"
]
],
[
[
"company = 'APPLE INC'",
"_____no_output_____"
],
[
"# %%timeit\n# with pd.HDFStore(test_store) as store:\n# s = store.select('file', 'company = company')",
"_____no_output_____"
],
[
"# read = _",
"_____no_output_____"
],
[
"# test_store.unlink()",
"_____no_output_____"
]
],
[
[
"#### Write",
"_____no_output_____"
]
],
[
[
"# %%timeit\n# with pd.HDFStore(test_store) as store:\n# store.append('file', df, format='t', data_columns=['company', 'form'])\n# test_store.unlink() ",
"_____no_output_____"
],
[
"# write = _",
"_____no_output_____"
]
],
[
[
"#### Results",
"_____no_output_____"
]
],
[
[
"# results['hdf_select'] = {'read': read.all_runs, 'write': write.all_runs, 'size': size}",
"_____no_output_____"
]
],
[
[
"## CSV",
"_____no_output_____"
]
],
[
[
"test_csv = Path('test.csv')",
"_____no_output_____"
]
],
[
[
"### Size",
"_____no_output_____"
]
],
[
[
"df.to_csv(test_csv)\ntest_csv.stat().st_size",
"_____no_output_____"
]
],
[
[
"### Read",
"_____no_output_____"
]
],
[
[
"%%timeit -o\ndf = pd.read_csv(test_csv)",
"31.5 s ± 163 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"read = _",
"_____no_output_____"
],
[
"test_csv.unlink() ",
"_____no_output_____"
]
],
[
[
"### Write",
"_____no_output_____"
]
],
[
[
"%%timeit -o\ndf.to_csv(test_csv)\ntest_csv.unlink()",
"3min 7s ± 1.95 s per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"write = _",
"_____no_output_____"
]
],
[
[
"### Results",
"_____no_output_____"
]
],
[
[
"results['csv'] = {'read': read.all_runs, 'write': write.all_runs, 'size': size}",
"_____no_output_____"
]
],
[
[
"## Store Results",
"_____no_output_____"
]
],
[
[
"text_num = pd.concat([pd.DataFrame(data).mean().to_frame(f) for f, data in results.items()], axis=1).T\ntext_num[['read', 'write']].plot.barh();",
"_____no_output_____"
],
[
"df = pd.concat([pd.DataFrame(data).mean().to_frame(f) for f, data in results.items()], axis=1).T\n# df.to_csv('num_only.csv')\ndf[['read', 'write']].plot.barh();",
"_____no_output_____"
],
[
"# for f, data in results.items():\n# pd.DataFrame(data).to_csv('{}.csv'.format(f))",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
eccb0d49c3e284601dd6b418a357fc62f9b2114a | 8,792 | ipynb | Jupyter Notebook | code/data_munging.ipynb | HALtheWise/bayesian-ranking | d8c6cd394d53e999f5eec9ecf1f407da412f4c18 | [
"MIT"
] | 1 | 2021-09-15T07:20:23.000Z | 2021-09-15T07:20:23.000Z | code/data_munging.ipynb | HALtheWise/bayesian-ranking | d8c6cd394d53e999f5eec9ecf1f407da412f4c18 | [
"MIT"
] | null | null | null | code/data_munging.ipynb | HALtheWise/bayesian-ranking | d8c6cd394d53e999f5eec9ecf1f407da412f4c18 | [
"MIT"
] | null | null | null | 25.484058 | 88 | 0.352821 | [
[
[
"import pandas as pd\nfrom pathlib import Path",
"_____no_output_____"
]
],
[
[
"# Convert Blueprint data to HackMIT format",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(Path('../data/blueprint-rookie-data-raw.txt'), sep=' ')\nprint(len(df))\ndf.head()",
"235\n"
],
[
"df2 = df.copy()\nr = next(df.itertuples())\ndf2['winner'] = [r.TEAM2 if r.TEAM2wins else r.TEAM1 for r in df2.itertuples()]\ndf2['loser'] = [r.TEAM1 if r.TEAM2wins else r.TEAM2 for r in df2.itertuples()]\ndf2['annotator'] = 'na'\n\ndf2.head()",
"_____no_output_____"
],
[
"df3 = df2.drop('TEAM1 TEAM2 TEAM2wins'.split(), axis=1)\ndf3.head()",
"_____no_output_____"
],
[
"df3.to_csv(Path('../data/blueprint-rookie-data.csv'), index=False)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
eccb18133fd200e9642f48436d71be4d3f05900a | 5,535 | ipynb | Jupyter Notebook | notebooks/processing/webmap_prep.ipynb | carbonplan/trace | 5cf113891bdefa29c2afd4478dff099e0458c82c | [
"MIT"
] | 14 | 2021-02-15T22:40:52.000Z | 2022-02-24T15:25:28.000Z | notebooks/processing/webmap_prep.ipynb | carbonplan/trace | 5cf113891bdefa29c2afd4478dff099e0458c82c | [
"MIT"
] | 75 | 2021-02-11T17:57:42.000Z | 2022-03-22T00:47:57.000Z | notebooks/processing/webmap_prep.ipynb | carbonplan/trace | 5cf113891bdefa29c2afd4478dff099e0458c82c | [
"MIT"
] | 2 | 2021-09-28T01:51:19.000Z | 2021-11-22T21:32:35.000Z | 27.81407 | 111 | 0.557724 | [
[
[
"import xarray as xr\nimport pandas as pd\nimport rioxarray\nfrom ndpyramid import pyramid_reproject\nfrom carbonplan_data.utils import set_zarr_encoding\nfrom carbonplan_data.metadata import get_cf_global_attrs\nimport numpy as np",
"_____no_output_____"
],
[
"save_path = \"s3://carbonplan-climatetrace/v1.2/map/\"",
"_____no_output_____"
],
[
"path = \"s3://carbonplan-climatetrace/v1.2/results/global/3000m/raster_split.zarr\"\nv1_emissions = xr.open_zarr(path)",
"_____no_output_____"
],
[
"biomass = xr.open_zarr(\"s3://carbonplan-climatetrace/v1.2/results/global/3000m/raster_biomass.zarr\")\nbiomass = biomass.rename({\"time\": \"year\"})\nbiomass = biomass.assign_coords(\n {\"year\": np.arange(2014, 2021), \"lat\": v1_emissions.lat, \"lon\": v1_emissions.lon}\n)",
"_____no_output_____"
],
[
"v0_emissions = xr.open_zarr(\"s3://carbonplan-climatetrace/v0.4/global/3000m/raster_tot.zarr\").sel(\n year=slice(2015, 2020)\n)",
"_____no_output_____"
],
[
"v0_emissions = v0_emissions.assign_coords(v1_emissions.coords)",
"_____no_output_____"
],
[
"# hack to get hansen tile area\nvalid_mask = v0_emissions.emissions.sel(year=2018) > -1",
"_____no_output_____"
],
[
"v1_emissions = v1_emissions.fillna(0).where(valid_mask)\nbiomass = biomass.fillna(0).where(valid_mask)",
"_____no_output_____"
],
[
"full_ds = xr.Dataset(\n {\n \"biomass\": biomass[\"AGB\"],\n \"biomass-na-filled\": biomass[\"AGB_na_filled\"],\n \"emissions-v1\": v1_emissions[\"emissions_from_clearing\"]\n + v1_emissions[\"emissions_from_fire\"],\n \"sinks-v1\": v1_emissions[\"sinks\"],\n \"net-v1\": v1_emissions[\"sinks\"]\n + v1_emissions[\"emissions_from_clearing\"]\n + v1_emissions[\"emissions_from_fire\"],\n \"emissions-v0\": v0_emissions.sel(year=slice(2015, 2020))[\"emissions\"],\n }\n)",
"_____no_output_____"
],
[
"full_ds = xr.Dataset({\"variable\": full_ds.to_array(\"band\")}).rio.write_crs(\"EPSG:4326\")\nfull_ds = full_ds.rename({\"lat\": \"y\", \"lon\": \"x\"})",
"_____no_output_____"
],
[
"for dim in full_ds.dims.keys():\n if dim != \"band\":\n full_ds[dim] = full_ds[dim].astype(\"float32\")\nfor var in full_ds.data_vars:\n full_ds[var] = full_ds[var].astype(\"float32\")",
"_____no_output_____"
],
[
"# create the pyramid\ndt = pyramid_reproject(full_ds, levels=6, extra_dim=\"band\")",
"_____no_output_____"
],
[
"for child in dt.children:\n child.ds = child.ds.where(child.ds < 2e38)\n child.ds = set_zarr_encoding(\n child.ds, codec_config={\"id\": \"zlib\", \"level\": 1}, float_dtype=\"float32\"\n )\n child.ds = child.ds.chunk({\"x\": 128, \"y\": 128, \"band\": -1, \"year\": -1})\n child.ds[\"variable\"].attrs.clear()\ndt.attrs = get_cf_global_attrs()\n\n# write the pyramid to zarr\ndt.to_zarr(save_path + \"forest_carbon_web_data_v3.pyr\", consolidated=True)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccb1e772b3c227dd636bfaa2b80848423e65893 | 46,517 | ipynb | Jupyter Notebook | folium_demo.ipynb | HKuz/NHPython_Folium_Demo | 2ebb94a99c66aa442b8a9d9319e36edb231c683a | [
"MIT"
] | 1 | 2021-04-02T12:05:25.000Z | 2021-04-02T12:05:25.000Z | folium_demo.ipynb | HKuz/NHPython_Folium_Demo | 2ebb94a99c66aa442b8a9d9319e36edb231c683a | [
"MIT"
] | null | null | null | folium_demo.ipynb | HKuz/NHPython_Folium_Demo | 2ebb94a99c66aa442b8a9d9319e36edb231c683a | [
"MIT"
] | 1 | 2021-04-02T12:05:27.000Z | 2021-04-02T12:05:27.000Z | 231.427861 | 41,815 | 0.944257 | [
[
[
"# Folium Demo\n\nFor full documentation on how to install and use Folium, see [this page](https://python-visualization.github.io/folium/).",
"_____no_output_____"
]
],
[
[
"import requests\nimport folium\n\n%matplotlib inline",
"_____no_output_____"
],
[
"# Retrieve data on NH divesites from the divesites.com API\nnh_lat = \"43.998463\"\nnh_lon = \"-71.553859\"\n\nds_url = f\"http://api.divesites.com/?mode=sites&lat={nh_lat}&lng={nh_lon}&dist=100\"\nr_ds = requests.get(ds_url)\nds_json = r_ds.json()",
"_____no_output_____"
],
[
"len(ds_json['sites'])",
"_____no_output_____"
],
[
"# ds_json",
"_____no_output_____"
]
],
[
[
"The JSON top-level structure is:\n\n```py\n{'request': {...}\n 'sites': [{}, {}, ...]\n 'version': 1,\n 'loc': {'lat': '43.998463', 'lng': '-71.553859'},\n 'result': True}\n```\n\nThe dive sites are stored under the `sites` key as a list of objects, each with the form:\n\n```py\n{'currents': None,\n 'distance': '30.07',\n 'hazards': None,\n 'lat': '43.5958',\n 'name': 'Lake Winnipesaukee',\n 'water': None,\n 'marinelife': None,\n 'description': None,\n 'maxdepth': None,\n 'mindepth': None,\n 'predive': None,\n 'id': '16498',\n 'equipment': None,\n 'lng': '-71.3253'}\n```",
"_____no_output_____"
]
],
[
[
"# Helper function to display map\ndef display_map(m, filename):\n # Ensures the folium map will display in different\n # browsers when viewing notebook\n from IPython.display import IFrame\n m.save(filename)\n return IFrame(filename,\n width=\"100%\",\n height=\"600px\")",
"_____no_output_____"
],
[
"# Latitude and longitude to center map\nmap_lat = \"43.7\"\nmap_lon = \"-71.0\"",
"_____no_output_____"
],
[
"# Create a basemap\nm = folium.Map(location=[map_lat, map_lon], # Coords for the center of the map\n tiles=\"cartodbpositron\", # Background style - other free options available, see documentation\n zoom_start=8) # Higher numbers mean the maps is more zoomed in\n\n# Create and plot circles for each site\nfor site in ds_json['sites']:\n lat = site[\"lat\"]\n lon = site[\"lng\"]\n tt = site[\"name\"]\n \n folium.Circle(location=[lat, lon], # Coords for center of the circle\n radius=20, # Size of circle\n tooltip=tt).add_to(m) # Tooltip with name of dive site when user hovers\n\n# display_map(m, \"index.html\")\nm",
"_____no_output_____"
],
[
"m.save(\"index.html\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
eccb1e96106ca06491c51cb35378bdf775220708 | 180,570 | ipynb | Jupyter Notebook | demos/Examples -- 25-D Correlated Normal.ipynb | vandalt/dynesty | 2c5f1bbe5a0745c6625876f23ec6aa710c845fd4 | [
"MIT"
] | null | null | null | demos/Examples -- 25-D Correlated Normal.ipynb | vandalt/dynesty | 2c5f1bbe5a0745c6625876f23ec6aa710c845fd4 | [
"MIT"
] | null | null | null | demos/Examples -- 25-D Correlated Normal.ipynb | vandalt/dynesty | 2c5f1bbe5a0745c6625876f23ec6aa710c845fd4 | [
"MIT"
] | null | null | null | 537.410714 | 169,112 | 0.939863 | [
[
[
"# 25-D Correlated Normal",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
],
[
"First, let's set up some environmental dependencies. These just make the numerics easier and adjust some of the plotting defaults to make things more legible.",
"_____no_output_____"
]
],
[
[
"# Python 3 compatability\nfrom __future__ import division, print_function\nfrom six.moves import range\n\n# system functions that are always useful to have\nimport time, sys, os\n\n# basic numeric setup\nimport numpy as np\nfrom numpy import linalg\n\n# inline plotting\n%matplotlib inline\n\n# plotting\nimport matplotlib\nfrom matplotlib import pyplot as plt\n\n# seed the random number generator\nrstate = np.random.default_rng(21)",
"_____no_output_____"
],
[
"# re-defining plotting defaults\nfrom matplotlib import rcParams\nrcParams.update({'xtick.major.pad': '7.0'})\nrcParams.update({'xtick.major.size': '7.5'})\nrcParams.update({'xtick.major.width': '1.5'})\nrcParams.update({'xtick.minor.pad': '7.0'})\nrcParams.update({'xtick.minor.size': '3.5'})\nrcParams.update({'xtick.minor.width': '1.0'})\nrcParams.update({'ytick.major.pad': '7.0'})\nrcParams.update({'ytick.major.size': '7.5'})\nrcParams.update({'ytick.major.width': '1.5'})\nrcParams.update({'ytick.minor.pad': '7.0'})\nrcParams.update({'ytick.minor.size': '3.5'})\nrcParams.update({'ytick.minor.width': '1.0'})\nrcParams.update({'font.size': 30})",
"_____no_output_____"
],
[
"import dynesty",
"_____no_output_____"
]
],
[
[
"`dynesty` supports three tiers of sampling techniques: uniform sampling for low dimensional problems, random walks for low-to-moderate dimensional problems, and slice sampling for high-dimensional problems (with or without the use of gradients). Here we will quickly demonstrate that slice sampling is able to cope with high-dimensional problems using a 25-D correlated multivariate normal distribution.",
"_____no_output_____"
]
],
[
[
"ndim = 25 # number of dimensions\nC = np.identity(ndim) # set covariance to identity matrix\nC[C==0] = 0.4 # set off-diagonal terms (strongly correlated)\nCinv = linalg.inv(C) # precision matrix\nlnorm = -0.5 * (np.log(2 * np.pi) * ndim + np.log(linalg.det(C))) # ln(normalization)\n\n# 3-D correlated multivariate normal log-likelihood\ndef loglikelihood(x):\n \"\"\"Multivariate normal log-likelihood.\"\"\"\n return -0.5 * np.dot(x, np.dot(Cinv, x)) + lnorm\n\n# prior transform\ndef prior_transform(u):\n \"\"\"Transforms our unit cube samples `u` to a flat prior between -5. and 5. in each variable.\"\"\"\n return 5. * (2. * u - 1.)\n\n# gradient of log-likelihood *with respect to u*\ndef gradient(x):\n \"\"\"Multivariate normal log-likelihood gradient.\"\"\"\n dlnl_dv = -np.dot(Cinv, x) # standard gradient\n jac = np.diag(np.full_like(x, 10.)) # Jacobian\n return np.dot(jac, dlnl_dv) # transformed gradient\n\n# ln(evidence)\nlnz_truth = -ndim * np.log(10. * 0.999999426697)\nprint(lnz_truth)",
"-57.56461299227203\n"
]
],
[
[
"Since we know this is a unimodal case, we'll initialize our sampler in the `'single'` bounding mode. ",
"_____no_output_____"
]
],
[
[
"# multivariate slice sampling ('slice')\nsampler = dynesty.NestedSampler(loglikelihood, prior_transform, ndim, \n nlive=500, bound='single', \n sample='slice', slices=5, rstate=rstate)\nsampler.run_nested(dlogz=0.01)\nres = sampler.results",
"20502it [36:22, 9.40it/s, +500 | bound: 216 | nc: 1 | ncall: 12240254 | eff(%): 0.172 | loglstar: -inf < -21.346 < inf | logz: -57.705 +/- 0.236 | dlogz: 0.000 > 0.010]\n"
],
[
"# random slice sampling ('rslice')\nsampler = dynesty.NestedSampler(loglikelihood, prior_transform, ndim,\n nlive=500, bound='single', \n sample='rslice', slices=25,\n rstate=rstate)\nsampler.run_nested(dlogz=0.01)\nres2 = sampler.results",
"20692it [06:16, 55.00it/s, +500 | bound: 99 | nc: 1 | ncall: 2503029 | eff(%): 0.847 | loglstar: -inf < -20.831 < inf | logz: -57.570 +/- 0.234 | dlogz: 0.000 > 0.010]\n"
],
[
"# hamiltonian slice sampling ('hslice')\nsampler = dynesty.NestedSampler(loglikelihood, prior_transform, ndim,\n nlive=500, bound='single',\n sample='hslice', slices=5,\n gradient=gradient, rstate=rstate)\nsampler.run_nested(dlogz=0.01)\nres3 = sampler.results",
"20588it [23:56, 14.33it/s, +500 | bound: 206 | nc: 1 | ncall: 13006565 | eff(%): 0.162 | loglstar: -inf < -21.182 < inf | logz: -57.714 +/- 0.235 | dlogz: 0.000 > 0.010]\n"
]
],
[
[
"Now let's see how we do.",
"_____no_output_____"
]
],
[
[
"from dynesty import plotting as dyplot\n\nfig, axes = dyplot.runplot(res, color='navy', logplot=True,\n lnz_truth=lnz_truth, truth_color='black')\nfig, axes = dyplot.runplot(res2, color='darkorange', logplot=True, fig=(fig, axes))\nfig, axes = dyplot.runplot(res3, color='limegreen', logplot=True, fig=(fig, axes))\nfig.tight_layout()",
"_____no_output_____"
],
[
"from dynesty import utils as dyfunc\n\nprint('Mean:')\nprint('slice:', dyfunc.mean_and_cov(res.samples, \n np.exp(res.logwt-res.logz[-1]))[0])\nprint('rslice:', dyfunc.mean_and_cov(res2.samples, \n np.exp(res2.logwt-res2.logz[-1]))[0])\nprint('hslice:', dyfunc.mean_and_cov(res3.samples, \n np.exp(res3.logwt-res3.logz[-1]))[0])\n\nprint('\\nVariance:')\nprint('slice:', np.diagonal(dyfunc.mean_and_cov(res.samples, \n np.exp(res.logwt-res.logz[-1]))[1]))\nprint('rslice:', np.diagonal(dyfunc.mean_and_cov(res2.samples, \n np.exp(res2.logwt-res2.logz[-1]))[1]))\nprint('hslice:', np.diagonal(dyfunc.mean_and_cov(res3.samples, \n np.exp(res3.logwt-res3.logz[-1]))[1]))",
"Mean:\nslice: [ 0.00080291 0.00406672 -0.00493006 0.00768341 0.01168745 0.00257284\n -0.00608764 -0.01331321 0.00302164 0.00739829 0.00267557 0.00882297\n 0.01372292 0.00118094 -0.01034449 -0.01189181 0.00058856 0.01181997\n -0.00825656 -0.00345821 -0.01273608 0.00372074 -0.00297336 0.01811703\n -0.00099364]\nrslice: [-0.03315893 -0.00432107 0.00998467 -0.01663759 -0.0046303 -0.0283778\n -0.01667174 0.00259521 -0.02423891 -0.04306577 -0.02425065 -0.01529905\n -0.00842877 -0.01335346 0.01843598 -0.01925403 -0.00792745 -0.01670167\n -0.01072673 0.01449257 -0.03352311 0.00877791 0.01324813 -0.00673466\n 0.00528856]\nhslice: [-0.00962847 -0.00294799 -0.01239019 -0.01733799 -0.00946174 -0.00521269\n 0.0112683 -0.01446488 -0.01761637 -0.03406204 -0.02913731 0.00387279\n -0.01487945 -0.00976863 -0.01588725 -0.01137356 0.00906974 -0.0130511\n -0.00240558 0.01483746 -0.00522736 -0.01581743 0.00887628 -0.0120862\n 0.01468354]\n\nVariance:\nslice: [0.98333744 0.97947349 0.98637122 0.95107507 0.98989678 0.96456344\n 0.9900618 0.98027878 1.00134412 0.96352766 0.97268149 0.96338969\n 0.99440611 0.98819147 0.96996871 0.96982796 0.96706257 0.956757\n 0.96920808 0.99541159 0.99208384 0.96895823 0.98936159 0.97086107\n 0.96203984]\nrslice: [1.00730979 0.98051623 1.02145398 0.97744007 0.98316128 0.97575885\n 0.96103664 1.02153617 0.97999399 0.97382665 0.99328286 1.01714364\n 0.99644802 0.96118561 0.98855272 0.98605147 0.98784969 0.94578554\n 0.98621289 0.99277124 1.00480866 1.00002545 0.97275777 1.01207083\n 1.00697836]\nhslice: [1.00012333 0.99813877 0.94211363 0.98340967 0.99609071 0.97676216\n 0.98676971 0.97578611 1.01064289 1.00988348 0.98981543 1.00865836\n 0.98956519 0.95683679 0.99465799 0.96075738 0.99553791 0.98660242\n 0.97602937 0.99345186 0.99067044 0.99633264 0.9830718 0.99860424\n 0.95189784]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
eccb20a42e653baca15ea4bf390a3c2d217c074e | 6,537 | ipynb | Jupyter Notebook | training_notebook.ipynb | mkisantal/wing-displacement | 33c99e5f6191e6e1e2a5f546ee07142edc1fd1de | [
"MIT"
] | null | null | null | training_notebook.ipynb | mkisantal/wing-displacement | 33c99e5f6191e6e1e2a5f546ee07142edc1fd1de | [
"MIT"
] | null | null | null | training_notebook.ipynb | mkisantal/wing-displacement | 33c99e5f6191e6e1e2a5f546ee07142edc1fd1de | [
"MIT"
] | null | null | null | 31.427885 | 114 | 0.483861 | [
[
[
"# selecting GPU\n%env CUDA_DEVICE_ORDER=PCI_BUS_ID\n%env CUDA_VISIBLE_DEVICES=0\n%matplotlib notebook",
"_____no_output_____"
],
[
"from utils import WingDataset\n\nimport os\nimport time\nimport json\n\nimport torch\nfrom torch import nn\nfrom torchvision import models, transforms",
"_____no_output_____"
],
[
"# basic setup stuff\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\nrun_name = 'test_runs01/001_first_training'\nif not os.path.exists(run_name):\n os.makedirs(run_name)\n\nmodel_name = 'resnet18'\nbatch_size = 64\nepochs = 1\n\n# training loss\ncriterion = torch.nn.MSELoss().to(device)\n\n# model, replacing last fully connected layer\nmodel = models.resnet18(pretrained=True)\nnum_ftrs = model.fc.in_features\nmodel.fc = nn.Linear(num_ftrs, 1)\nmodel.to(device)\n\n# optimizer\noptimizer = torch.optim.Adam(model.parameters(), lr=1e-4)\n\n",
"_____no_output_____"
],
[
"# DATASETS, DATALOADERS\n\n# transforms: converting to pytorch tensor and normalization\n# later dataset autgmentation transforms can be added here, but be careful to consider label preservation\nimg_tr = transforms.Compose([transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406],\n [0.229, 0.224, 0.225])])\n\n# training and validation sets, by default 3 runs are used for test (see WingDataset), rest is training\ntrain_ds = WingDataset(test=False, transforms=img_tr)\nval_ds = WingDataset(test=True, transforms=img_tr)\n\nsets = {'train', 'val'}\ndsets = {'train': train_ds, 'val': val_ds}\n\ndataloaders = {x: torch.utils.data.DataLoader(dsets[x], batch_size=batch_size, shuffle=True,\n num_workers=8, drop_last=True)\n for x in sets}\ndataset_sizes = {x: len(dsets[x]) for x in sets}",
"_____no_output_____"
],
[
"def train_model(model, criterion, num_epochs, dataloaders, device, optimizer, start_epoch=0):\n since = time.time()\n history = {'train_loss': [],\n 'val_loss': [],\n 'mean_err': []}\n if start_epoch != 0:\n with open(os.path.join(run_name, 'history.json'), 'r') as fin:\n history = json.load(fin)\n \n \n for eph in range(start_epoch, start_epoch+num_epochs):\n \n for phase in ['train', 'val']:\n if phase == 'train':\n model.train() # Set model to training mode\n else:\n model.eval() # Set model to evaluate mode\n \n running_loss = 0.0\n errors = []\n \n for inputs, labels in dataloaders[phase]:\n inputs = inputs.to(device)\n labels = labels.to(device).float()\n optimizer.zero_grad()\n with torch.set_grad_enabled(phase == 'train'):\n outputs = model(inputs)\n loss = criterion(outputs, labels)\n \n if phase == 'train':\n loss.backward()\n optimizer.step()\n else:\n pass\n running_loss += loss.item() * inputs.size(0)\n \n \n epoch_loss = float(running_loss / dataset_sizes[phase]) \n if phase == 'train':\n history['train_loss'].append(epoch_loss)\n print(epoch_loss)\n else:\n history['val_loss'].append(epoch_loss)\n print('\\t\\t{}'.format(epoch_loss))\n \n if eph % 1 == 0:\n save_path = os.path.join(run_name, 'model_latest.pth')\n torch.save(model, save_path)\n \n with open(os.path.join(run_name, 'history.json'), 'w') as fout:\n json.dump(history, fout)\n \n time_elapsed = time.time() - since\n print('Training complete in {:.0f}m {:.0f}s'.format(\n time_elapsed // 60, time_elapsed % 60))\n return history, model",
"_____no_output_____"
],
[
"h, model = train_model(model, criterion, epochs, dataloaders, device, optimizer)",
"_____no_output_____"
],
[
"from matplotlib import pyplot as plt\nplt.plot(h['train_loss'], 'k')\nplt.plot(h['val_loss'], 'r')",
"_____no_output_____"
],
[
"# for inputs, labels in dataloaders['train']:\n# break",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccb2fb6081febe1fd7304dfca3c5ec8066ff7c6 | 132,712 | ipynb | Jupyter Notebook | PythonAdvanced/Prodvin_tems_python.ipynb | LubovFedoseeva/Learning | 2d0e87c31298186b4e8f0b7301207aea574e0d03 | [
"MIT"
] | 1 | 2021-12-08T19:41:57.000Z | 2021-12-08T19:41:57.000Z | PythonAdvanced/Prodvin_tems_python.ipynb | LubovFedoseeva/Learning | 2d0e87c31298186b4e8f0b7301207aea574e0d03 | [
"MIT"
] | null | null | null | PythonAdvanced/Prodvin_tems_python.ipynb | LubovFedoseeva/Learning | 2d0e87c31298186b4e8f0b7301207aea574e0d03 | [
"MIT"
] | null | null | null | 46.811993 | 21,068 | 0.533139 | [
[
[
"import pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"path = '/home/jupyter-l.fedoseeva-12/prodvin_tems_python/nyc.csv.zip'",
"_____no_output_____"
],
[
"nyc = pd.read_csv(path, compression='zip', encoding='ISO-8859-1')",
"_____no_output_____"
],
[
"nyc.head()",
"_____no_output_____"
],
[
"nyc['pickup_longitude'] = np.radians(nyc['pickup_longitude'])\nnyc['pickup_latitude'] = np.radians(nyc['pickup_latitude'])\nnyc['dropoff_longitude'] = np.radians(nyc['dropoff_longitude'])\nnyc['dropoff_latitude'] = np.radians(nyc['dropoff_latitude'])",
"_____no_output_____"
],
[
"nyc.head()",
"_____no_output_____"
],
[
"def haversine(pickup_latitude, pickup_longitude, dropoff_latitude, dropoff_longitude, radius=6371):\n \n lat_sin = np.sin((dropoff_latitude - pickup_latitude) / 2) **2\n lat_cos = np.cos(pickup_latitude) * np.cos(dropoff_latitude)\n long_sin = np.sin((dropoff_longitude - pickup_longitude) / 2) **2\n \n return 2 * radius * np.arcsin(np.sqrt(lat_sin + lat_cos * long_sin))",
"_____no_output_____"
],
[
"#latitude1, latitude2, long1, long2 = [0.710721, 0.710563, -0.022494, -1.288779]",
"_____no_output_____"
],
[
"#lat_sin = np.sin((latitude2 - latitude1) / 2) **2\n#lat_cos = np.cos(latitude2) * np.cos(latitude1)\n#long_sin = np.sin((long2 - long1) / 2) **2",
"_____no_output_____"
],
[
"#2 * 6371 * np.arcsin(np.sqrt(lat_sin + lat_cos * long_sin))",
"_____no_output_____"
],
[
"%%timeit\n\ndists = [ ]\nfor i, row in nyc.iterrows():\n dists.append(haversine(row.pickup_longitude, row.pickup_latitude, row.dropoff_longitude, row.dropoff_latitude))\nnyc['distance'] = dists",
"6.24 s ± 317 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"nyc.head()",
"_____no_output_____"
],
[
"import timeit",
"_____no_output_____"
],
[
"nyc(%timeit)",
"_____no_output_____"
]
],
[
[
"Второй вариант через itertuples",
"_____no_output_____"
]
],
[
[
"def haversine(pickup_latitude, pickup_longitude, dropoff_latitude, dropoff_longitude, radius=6371):\n \n lat_sin = np.sin((dropoff_latitude - pickup_latitude) / 2) **2\n lat_cos = np.cos(pickup_latitude) * np.cos(dropoff_latitude)\n long_sin = np.sin((dropoff_longitude - pickup_longitude) / 2) **2\n \n return 2 * radius * np.arcsin(np.sqrt(lat_sin + lat_cos * long_sin))",
"_____no_output_____"
],
[
"%%timeit\n\ndists_int = [ ]\nfor row in nyc.itertuples():\n dists_int.append(haversine(row.pickup_longitude, row.pickup_latitude, row.dropoff_longitude, row.dropoff_latitude))\nnyc['distance_int'] = dists_int",
"541 ms ± 21.2 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"nyc.head()",
"_____no_output_____"
]
],
[
[
"Третий способ через apply",
"_____no_output_____"
]
],
[
[
"%%timeit\n\nnyc['distance_apply'] = nyc.apply(lambda x: haversine(x['pickup_longitude'], x['pickup_latitude'], x['dropoff_longitude'], x['dropoff_latitude']), axis=1)",
"1.41 s ± 84.5 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"nyc.head()",
"_____no_output_____"
],
[
"longest = nyc.distance_apply.max()",
"_____no_output_____"
],
[
"sns.barplot(x=['itertuples', 'interrows', 'apply', 'simply', 'array'], y=[0.549, 6.39, 1.38, 0.00599, 0.00385])",
"_____no_output_____"
]
],
[
[
"Изменения в коде весьма незначительны: передаем функции на вход необходимые колонки, и получается магия!\ndf['distance'] = haversine(df['col1'], df['col2'], df['col3'], df['col4'])\nУбедимся, что векторизованные операции работают намного быстрее! Создайте функцию для подсчета расстояния \nhaversine, описанную в первом степе, и примените её к датасету nyc. Результат сохраните в колонку distance. ",
"_____no_output_____"
]
],
[
[
"def haversine(pickup_latitude, pickup_longitude, dropoff_latitude, dropoff_longitude, radius=6371):\n \n lat_sin = np.sin((dropoff_latitude - pickup_latitude) / 2) **2\n lat_cos = np.cos(pickup_latitude) * np.cos(dropoff_latitude)\n long_sin = np.sin((dropoff_longitude - pickup_longitude) / 2) **2\n \n return 2 * radius * np.arcsin(np.sqrt(lat_sin + lat_cos * long_sin))",
"_____no_output_____"
],
[
"nyc['distance'] = haversine(nyc['pickup_longitude'], nyc['pickup_latitude'], \n nyc['dropoff_longitude'], nyc['dropoff_latitude'])",
"_____no_output_____"
],
[
"nyc.head(10)",
"_____no_output_____"
],
[
"nyc['disctance2'] = haversine(nyc['pickup_longitude'], nyc['pickup_latitude'], \n nyc['dropoff_longitude'], nyc['dropoff_latitude'])",
"_____no_output_____"
],
[
"nyc.head(10)",
"_____no_output_____"
],
[
"%%timeit\n\nnyc['distance2'] = haversine(nyc['pickup_longitude'], nyc['pickup_latitude'], \n nyc['dropoff_longitude'], nyc['dropoff_latitude'])",
"5.99 ms ± 149 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)\n"
]
],
[
[
"В предыдущем степе мы передавали функции колонки формата Series, с чем связаны дополнительные вычислительные затраты, например, индексирование и проверка типа данных. Чтобы этого избежать, в качестве аргументов функции можно передать всё те же необходимые колонки, дополнительно указав .values, чтобы из Series получить массив NumPy (numpy array). В таком случае действия будут выполняться непосредственно над ndarrays. Такой способ можно использовать, когда отсутствие индексов и пр. не приведет к нарушению работы применяемой функции. ",
"_____no_output_____"
]
],
[
[
"nyc['pickup_longitude'] = np.radians(nyc['pickup_longitude'])\nnyc['pickup_latitude'] = np.radians(nyc['pickup_latitude'])\nnyc['dropoff_longitude'] = np.radians(nyc['dropoff_longitude'])\nnyc['dropoff_latitude'] = np.radians(nyc['dropoff_latitude'])",
"_____no_output_____"
],
[
"def haversine(pickup_latitude, pickup_longitude, dropoff_latitude, dropoff_longitude, radius=6371):\n \n lat_sin = np.sin((dropoff_latitude - pickup_latitude) / 2) **2\n lat_cos = np.cos(pickup_latitude) * np.cos(dropoff_latitude)\n long_sin = np.sin((dropoff_longitude - pickup_longitude) / 2) **2\n \n return 2 * radius * np.arcsin(np.sqrt(lat_sin + lat_cos * long_sin))",
"_____no_output_____"
],
[
"nyc['distance3'] = haversine(nyc['pickup_longitude'].values, nyc['pickup_latitude'].values, \n nyc['dropoff_longitude'].values, nyc['dropoff_latitude'].values)",
"_____no_output_____"
],
[
"%%timeit\n\nnyc['distance3'] = haversine(nyc['pickup_longitude'].values, nyc['pickup_latitude'].values, \n nyc['dropoff_longitude'].values, nyc['dropoff_latitude'].values)",
"3.85 ms ± 75.9 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)\n"
]
],
[
[
"Как вы уже заметили, произошло что-то странное: в качестве широты и долготы стоят нулевые значения, а расстояние — целых 8667 км! Неужели наш пассажир действительно уехал так далеко?\nПосмотрите, есть ли еще кейсы, где в качестве хотя бы одной из координат пункта назначения стоят нули, и в качестве ответа укажите количество таких случаев.",
"_____no_output_____"
]
],
[
[
"nyc[['dropoff_longitude', 'dropoff_latitude']].eq(0) \\\n .sum(1) \\\n .gt(0) \\\n .sum()",
"_____no_output_____"
]
],
[
[
"Постройте графики распределения переменных pickup_latitude, pickup_longitude, dropoff_latitude, dropoff_longitude, и для каждой переменной выберите только один наиболее подходящий вариант для дальнейшей фильтрации данных (в итоге – 4 галочки).",
"_____no_output_____"
]
],
[
[
"nyc.head(10)",
"_____no_output_____"
],
[
"nyc[['pickup_longitude', 'pickup_latitude', 'dropoff_longitude', 'dropoff_latitude']].hist(figsize=(12, 8))",
"_____no_output_____"
],
[
"sns.boxplot(nyc.pickup_longitude)",
"/opt/tljh/user/lib/python3.7/site-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variable as a keyword arg: x. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
]
],
[
[
"Критерии для фильтрации выбраны, остался последний шаг: отфильтровать имеющиеся данные и наконец-то найти самую дальнюю поездку.\nДля фильтрации используйте условия из предыдущего степа, а также добавьте еще одно ограничение для переменной distance. Предположим, максимальная удаленность пунктов друг от друга не должна превышать 50 километров, иначе такой заказ не будет принят системой, а наличие подобных результатов в таблице будет считаться ошибкой.\nВ качестве ответа укажите сумму, которую заплатил пассажир, проехавший наибольшее расстояние.",
"_____no_output_____"
]
],
[
[
"nyc.head(10)",
"_____no_output_____"
],
[
"nyc['pickup_longitude_2'] = np.radians(nyc['pickup_longitude'])\nnyc['pickup_latitude_2'] = np.radians(nyc['pickup_latitude'])\nnyc['dropoff_longitude_2'] = np.radians(nyc['dropoff_longitude'])\nnyc['dropoff_latitude_2'] = np.radians(nyc['dropoff_latitude'])",
"_____no_output_____"
],
[
"def haversine(pickup_latitude_2, pickup_longitude_2, dropoff_latitude_2, dropoff_longitude_2, radius=6371):\n \n lat_sin = np.sin((dropoff_latitude_2 - pickup_latitude_2) / 2) **2\n lat_cos = np.cos(pickup_latitude_2) * np.cos(dropoff_latitude_2)\n long_sin = np.sin((dropoff_longitude_2 - pickup_longitude_2) / 2) **2\n \n return 2 * radius * np.arcsin(np.sqrt(lat_sin + lat_cos * long_sin))",
"_____no_output_____"
],
[
"nyc['distance'] = haversine(nyc['pickup_longitude_2'].values, nyc['pickup_latitude_2'].values, \n nyc['dropoff_longitude_2'].values, nyc['dropoff_latitude_2'].values)",
"_____no_output_____"
],
[
"nyc.head(10)",
"_____no_output_____"
],
[
"nyc_filtered = nyc.query('(pickup_longitude < -60) and (pickup_latitude < 120) and (pickup_latitude > 10) and (dropoff_latitude > 10) and (dropoff_longitude < -60)')",
"_____no_output_____"
],
[
"nyc_filtered",
"_____no_output_____"
],
[
"nn = nyc_filtered.query('distance <= 50')",
"_____no_output_____"
],
[
"nn.query('distance == distance.max()')",
"_____no_output_____"
],
[
"final_nyc.distance.idxmax()",
"_____no_output_____"
],
[
"'(column1 == condition1) & (column2 == condition2)')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccb421d71172d4c24e62cfe457d8d7e5361fbf8 | 48,740 | ipynb | Jupyter Notebook | examples.ipynb | brando90/anatome | d5ea2a3d4c80054074b0b146f226d4f542f513fe | [
"MIT"
] | 42 | 2020-08-01T16:15:47.000Z | 2022-02-22T10:22:10.000Z | examples.ipynb | brando90/anatome | d5ea2a3d4c80054074b0b146f226d4f542f513fe | [
"MIT"
] | 27 | 2020-11-26T20:35:05.000Z | 2022-02-09T20:21:06.000Z | examples.ipynb | brando90/anatome | d5ea2a3d4c80054074b0b146f226d4f542f513fe | [
"MIT"
] | 5 | 2021-02-03T16:46:43.000Z | 2021-08-15T12:58:16.000Z | 164.107744 | 16,387 | 0.696204 | [
[
[
"import torch\nfrom torch.nn import functional as F\nfrom torchvision.models import resnet18\nfrom torchvision import transforms\nfrom torchvision.datasets import ImageFolder\nfrom torch.utils.data import DataLoader\n\nimport matplotlib.pyplot as plt\n\nbatch_size = 128\n\nmodel = resnet18(pretrained=True)\nimagenet = ImageFolder('~/.torch/data/imagenet/val', \n transforms.Compose([transforms.CenterCrop(224),transforms.ToTensor(),\n transforms.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225))]))\ndata = next(iter(DataLoader(imagenet, batch_size=batch_size, num_workers=8)))",
"_____no_output_____"
],
[
"model.eval()\nmodel.cuda()\ndata = data[0].cuda(), data[1].cuda()",
"_____no_output_____"
]
],
[
[
"## CCA\n\nMeasure CCA distance between trained and not trained models.",
"_____no_output_____"
]
],
[
[
"from anatome import CCAHook\nrandom_model = resnet18().cuda()\n\nhook1 = CCAHook(model, \"layer1.0.conv1\")\nhook2 = CCAHook(random_model, \"layer1.0.conv1\")\n\nwith torch.no_grad():\n model(data[0])\n random_model(data[0])\nhook1.distance(hook2, size=8)",
"_____no_output_____"
],
[
"# clear hooked tensors and caches\nhook1.clear()\nhook2.clear()\ntorch.cuda.empty_cache()",
"_____no_output_____"
]
],
[
[
"## Loss Landscape\n\nShow loss landscape in 1D space.",
"_____no_output_____"
]
],
[
[
"from anatome import landscape1d\nx, y = landscape1d(model, data, F.cross_entropy, x_range=(-1, 1), step_size=0.1)",
"100%|███████████████████████████████████████████| 21/21 [00:01<00:00, 13.02it/s]\n"
],
[
"plt.plot(x, y)",
"_____no_output_____"
],
[
"torch.cuda.empty_cache()",
"_____no_output_____"
]
],
[
[
"## Fourier Analysis\n\nShow a model's robustness.",
"_____no_output_____"
]
],
[
[
"from anatome import fourier_map\nmap = fourier_map(model, data, F.cross_entropy, 20, (6, 6), mean=(0.485, 0.456, 0.406), std=(0.229, 0.224, 0.225))",
"_____no_output_____"
],
[
"plt.imshow(map, interpolation='nearest')",
"_____no_output_____"
],
[
"torch.cuda.empty_cache()",
"_____no_output_____"
],
[
"from anatome import fourier_map\nmap = fourier_map(model, data, F.cross_entropy, 20, (6, 6), mean=(0.485, 0.456, 0.406), std=(0.229, 0.224, 0.225))",
"100%|███████████████████████████████████████████| 21/21 [00:03<00:00, 6.08it/s]\n"
],
[
"plt.imshow(map, interpolation='nearest')",
"_____no_output_____"
],
[
"torch.cuda.empty_cache()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccb4eac4b3e58972e7100c6456296203f2df6b9 | 9,194 | ipynb | Jupyter Notebook | Missions_to_Mars/mission_to_mars.ipynb | Trevor-Velez/web-scraping-challenge | 8407f62085f75647ee8a621f8eb7023a7b7f7380 | [
"ADSL"
] | null | null | null | Missions_to_Mars/mission_to_mars.ipynb | Trevor-Velez/web-scraping-challenge | 8407f62085f75647ee8a621f8eb7023a7b7f7380 | [
"ADSL"
] | null | null | null | Missions_to_Mars/mission_to_mars.ipynb | Trevor-Velez/web-scraping-challenge | 8407f62085f75647ee8a621f8eb7023a7b7f7380 | [
"ADSL"
] | null | null | null | 34.051852 | 908 | 0.512399 | [
[
[
"from splinter import Browser\nfrom bs4 import BeautifulSoup as bs\nimport time\nfrom webdriver_manager.chrome import ChromeDriverManager\nimport pandas as pd",
"_____no_output_____"
],
[
"def scrape_info():\n\n executable_path = {'executable_path': ChromeDriverManager().install()}\n browser = Browser('chrome', **executable_path, headless=False)\n\n\n url = \"https://redplanetscience.com/\"\n browser.visit(url)\n\n time.sleep(1)\n \n # Scrape page into Soup\n html = browser.html\n soup = bs(html, \"html.parser\")\n \n # Get the latest News Title\n news_title = soup.find('div', class_='content_title').text\n \n # Get the latest news paragraph\n news_p = soup.find('div', class_='article_teaser_body').text\n \n data = {\n \"news_title\": news_title,\n \"news_p\": news_p\n }\n \n browser.quit()\n \n return data",
"_____no_output_____"
],
[
"def scrape_info_2():\n\n executable_path = {'executable_path': ChromeDriverManager().install()}\n browser = Browser('chrome', **executable_path, headless=False)\n\n\n url = \"https://spaceimages-mars.com/\"\n browser.visit(url)\n\n time.sleep(1)\n \n # Scrape page into Soup\n html = browser.html\n soup = bs(html, \"html.parser\")\n \n # Get the feature image source\n featured_image_url = soup.find('img', class_='headerimage fade-in')['src']\n complete_url = url + featured_image_url\n \n browser.quit()\n \n return complete_url",
"_____no_output_____"
],
[
"def scrape_table():\n url = 'https://galaxyfacts-mars.com/'\n tables = pd.read_html(url)\n new_table = tables[0]\n new_table.columns=['Mars - Earth Comparison','Mars', 'Earth']\n new_table.set_index('Mars - Earth Comparison', inplace=True)\n df = new_table.iloc[1: , :]\n html_table = df.to_html()\n \n html_table = html_table.replace('\\n', ' ')\n \n return html_table",
"_____no_output_____"
],
[
"def scrape_info_3():\n\n executable_path = {'executable_path': ChromeDriverManager().install()}\n browser = Browser('chrome', **executable_path, headless=False)\n\n\n url = \"https://marshemispheres.com/\"\n browser.visit(url)\n\n time.sleep(1)\n \n\n xpath = '//*[@id=\"product-section\"]/div[2]/div[1]/div/a/h3'\n xpath2 = '//*[@id=\"product-section\"]/div[2]/div[2]/div/a/h3'\n xpath3 = '//*[@id=\"product-section\"]/div[2]/div[3]/div/a/h3'\n xpath4 = '//*[@id=\"product-section\"]/div[2]/div[4]/div/a/h3'\n info_dict = []\n xpath_list = [xpath, xpath2, xpath3, xpath4]\n \n for x in xpath_list:\n results = browser.find_by_xpath(x)\n click = results[0]\n click.click()\n\n html = browser.html\n soup = bs(html, \"html.parser\")\n\n hemi_title = soup.find('h2', class_='title').text\n full_image = soup.find('img', class_='wide-image')['src']\n full_image_url = url + full_image\n \n data = {\n \"title\": hemi_title,\n \"img_url\": full_image_url\n }\n info_dict.append(data)\n browser.back()\n\n \n browser.quit()\n \n return info_dict",
"_____no_output_____"
],
[
"def complete_scrape():\n \n first_scrape = scrape_info()\n second_scrape = scrape_info_2()\n third_scrape = scrape_table()\n fourth_scrape = scrape_info_3()\n \n result = {\n 'latest_news': first_scrape,\n 'featured_img': second_scrape,\n 'table_html': third_scrape,\n 'hemi_data': fourth_scrape\n }\n \n return result\n ",
"_____no_output_____"
],
[
"complete_scrape()",
"\n\n====== WebDriver manager ======\nCurrent google-chrome version is 92.0.4515\nGet LATEST driver version for 92.0.4515\nDriver [C:\\Users\\Trevor\\.wdm\\drivers\\chromedriver\\win32\\92.0.4515.107\\chromedriver.exe] found in cache\n\n\n====== WebDriver manager ======\nCurrent google-chrome version is 92.0.4515\nGet LATEST driver version for 92.0.4515\nDriver [C:\\Users\\Trevor\\.wdm\\drivers\\chromedriver\\win32\\92.0.4515.107\\chromedriver.exe] found in cache\n\n\n====== WebDriver manager ======\nCurrent google-chrome version is 92.0.4515\nGet LATEST driver version for 92.0.4515\nDriver [C:\\Users\\Trevor\\.wdm\\drivers\\chromedriver\\win32\\92.0.4515.107\\chromedriver.exe] found in cache\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccb50411f6cf5481e249279f17d7a6e19664a25 | 82,946 | ipynb | Jupyter Notebook | notebooks/04_More_Estimation.ipynb | uhasan1/ThinkBayes2 | c88a9fd66bcfa9f65d7af82520b5b09cc0449d4d | [
"MIT"
] | null | null | null | notebooks/04_More_Estimation.ipynb | uhasan1/ThinkBayes2 | c88a9fd66bcfa9f65d7af82520b5b09cc0449d4d | [
"MIT"
] | null | null | null | notebooks/04_More_Estimation.ipynb | uhasan1/ThinkBayes2 | c88a9fd66bcfa9f65d7af82520b5b09cc0449d4d | [
"MIT"
] | null | null | null | 73.664298 | 1,570 | 0.673149 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
eccb5fe2c5c78ccb8d9bbf9283a4531f80adfef3 | 2,358 | ipynb | Jupyter Notebook | bits_wilp/Ex2_Numpy_Q1.ipynb | deepak5998/Py | 5ae3bd9e8dcf3104a8ca7512911a1607f6c9ae20 | [
"MIT"
] | 726 | 2019-06-04T04:46:06.000Z | 2022-03-31T17:54:00.000Z | bits_wilp/Ex2_Numpy_Q1.ipynb | deepak5998/Py | 5ae3bd9e8dcf3104a8ca7512911a1607f6c9ae20 | [
"MIT"
] | 12 | 2019-06-05T14:21:35.000Z | 2021-04-17T05:11:01.000Z | bits_wilp/Ex2_Numpy_Q1.ipynb | deepak5998/Py | 5ae3bd9e8dcf3104a8ca7512911a1607f6c9ae20 | [
"MIT"
] | 118 | 2019-06-04T10:25:12.000Z | 2022-02-04T22:31:12.000Z | 25.085106 | 111 | 0.544529 | [
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"### Size of Numpy array in bytes",
"_____no_output_____"
]
],
[
[
"try:\n # feed integer array from user\n arr = list(map(int, input(\"Enter integers seperated by space. Press ENTER to end...\").split()))\n print(\"Given sequence is\")\n print(arr)\n # convert Python array to Numpy array\n np_array = np.array(arr, dtype=int)\n print(f\"Number of elements in the numpy array: {np_array.size}\")\n print(f\"Total bytes consumed by the numpy array: {np_array.nbytes}\")\n print(f\"Size in bytes of each element in the numpy array: {(np_array.nbytes)//(np_array.size)}\")\nexcept ValueError as e:\n print(\"ERROR: Please enter only integers !!!\")\n print(e)",
"Enter integers seperated by space. Press ENTER to end... 34 23 67 89\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
eccb6dbf5615f04d938d47898c54fd34e0f7626b | 1,944 | ipynb | Jupyter Notebook | doc/never-execute.ipynb | lamby/nbsphinx | 14c19c1f6521a42dfe5a3839199a2b85be0bffdf | [
"MIT"
] | null | null | null | doc/never-execute.ipynb | lamby/nbsphinx | 14c19c1f6521a42dfe5a3839199a2b85be0bffdf | [
"MIT"
] | 1 | 2022-02-19T19:11:39.000Z | 2022-02-19T19:11:39.000Z | doc/never-execute.ipynb | lamby/nbsphinx | 14c19c1f6521a42dfe5a3839199a2b85be0bffdf | [
"MIT"
] | 1 | 2022-02-19T19:09:41.000Z | 2022-02-19T19:09:41.000Z | 24.923077 | 152 | 0.561214 | [
[
[
"This notebook is part of the `nbsphinx` documentation: http://nbsphinx.readthedocs.io/.",
"_____no_output_____"
],
[
"# Explicitly Dis-/Enabling Notebook Execution\n\nIf you want to include a notebook without outputs and yet don't want `nbsphinx` to execute it for you, you can explicitly disable this feature.\n\nYou can do this globally by setting the following option in [conf.py](conf.py):\n\n```python\nnbsphinx_execute = 'never'\n```\n\nOr on a per-notebook basis by adding this to the notebook's JSON metadata:\n\n```json\n\"nbsphinx\": {\n \"execute\": \"never\"\n},\n```\n\nThere are three possible settings, `\"always\"`, `\"auto\"` and `\"never\"`.\nBy default (= `\"auto\"`), notebooks with no outputs are executed and notebooks with at least one output are not.\nAs always, per-notebook settings take precedence over the settings in `conf.py`.\n\nThis very notebook has its metadata set to `\"never\"`, therefore the following cell is not executed:",
"_____no_output_____"
]
],
[
[
"6 * 7",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
]
] |
eccb7c80b5e642ed6a6a78b3aaba1e1ce0715f39 | 139,976 | ipynb | Jupyter Notebook | Tidy-Tuesday-Insights/fish 2.ipynb | shawnd29/Tidy-Tuesday | faa06f214251227ab5afb253564350fc9e0c4dfe | [
"MIT"
] | null | null | null | Tidy-Tuesday-Insights/fish 2.ipynb | shawnd29/Tidy-Tuesday | faa06f214251227ab5afb253564350fc9e0c4dfe | [
"MIT"
] | null | null | null | Tidy-Tuesday-Insights/fish 2.ipynb | shawnd29/Tidy-Tuesday | faa06f214251227ab5afb253564350fc9e0c4dfe | [
"MIT"
] | null | null | null | 256.836697 | 47,818 | 0.756787 | [
[
[
"2021-10-12\t<br/>\nGlobal Seafood <br/>\nhttps://github.com/rfordatascience/tidytuesday/blob/master/data/2021/2021-10-12/readme.md",
"_____no_output_____"
],
[
"Wide-format bar plot - Completed <br/>\nFaceted plots - Completed ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport plotly.graph_objects as go\nimport plotly.io as pio\nimport plotly.express as px\npio.renderers.default='notebook'\nimport os",
"_____no_output_____"
],
[
"df =pd.read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2021/2021-10-12/global-fishery-catch-by-sector.csv')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df_10=df.tail(10)",
"_____no_output_____"
],
[
"wide_df = px.data.medals_wide()\n\nfig = px.bar(wide_df, x=\"nation\", y=[\"gold\", \"silver\", \"bronze\"], title=\"Wide-Form Input\")\nfig.show()",
"_____no_output_____"
],
[
"# Display Output in production\nimg_bytes = fig.to_image(format=\"png\")\nfrom IPython.display import Image\nImage(img_bytes)",
"_____no_output_____"
],
[
"communities=df_10.columns[3:]",
"_____no_output_____"
],
[
"df_10",
"_____no_output_____"
],
[
"cols=['Artisanal (small-scale commercial)','Discards','Industrial (large-scale commercial)','Recreational','Subsistence']",
"_____no_output_____"
],
[
" # ['stack', 'group', 'overlay', 'relative']\n#communities = ['Artisanal (small-scale commercial)','Discards','Industrial (large-scale commercial)\tRecreationa','Subsistence'] # or df_users_community.columns[3:]\n\nfig = px.bar(df_10, x='Year', y= cols, facet_col='Year', facet_col_wrap=5, barmode='group', \n title='A decade of fishing')\nfig.show()",
"_____no_output_____"
],
[
"# Display Output in production\nimg_bytes = fig.to_image(format=\"png\")\nfrom IPython.display import Image\nImage(img_bytes)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccb847dd2b4cfa5a789ab434fde2cf2128064d9 | 101,640 | ipynb | Jupyter Notebook | COMP90051-SML/Tutorials/Week4/worksheet04.ipynb | siriusctrl/UniPublic | 9df7f8bb9d1209de2af8ac4b5f57ada38587ad50 | [
"Apache-2.0"
] | 8 | 2021-03-14T14:19:10.000Z | 2021-07-13T12:35:26.000Z | COMP90051-SML/Tutorials/Week4/worksheet04.ipynb | siriusctrl/UniPublic | 9df7f8bb9d1209de2af8ac4b5f57ada38587ad50 | [
"Apache-2.0"
] | 2 | 2018-05-29T04:28:20.000Z | 2018-06-09T04:55:19.000Z | COMP90051-SML/Tutorials/Week4/worksheet04.ipynb | siriusctrl/UniPublic | 9df7f8bb9d1209de2af8ac4b5f57ada38587ad50 | [
"Apache-2.0"
] | 4 | 2019-01-10T10:30:33.000Z | 2019-05-30T09:33:20.000Z | 142.552595 | 20,012 | 0.852017 | [
[
[
"# COMP90051 Workshop 4\n## Logistic regression\n\n***\n\nIn this worksheet, we'll implement logistic regression from scratch using the iteratively reweighted least-squares (IRLS) algorithm presented in lectures. \nIn doing so, we'll see how logistic regression can be solved by iteratively performing weighted linear regression.\nFinally, we'll compare our IRLS implementation with gradient descent.\n\nFirstly, we'll import the relevant libraries (`numpy`, `matplotlib`, etc.).",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom scipy.special import expit\nsns.set_style('darkgrid')\nplt.rcParams['figure.dpi'] = 108\nRND_SEED = 0",
"_____no_output_____"
]
],
[
[
"## 1. Binary classification data\n\nLet's begin by generating some binary classification data.\nTo make it easy for us to visualise the results, we'll stick to a two-dimensional feature space.",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import make_classification\n\nX, y = make_classification(n_samples = 20, n_features = 2, n_informative=2, n_redundant=0, random_state=RND_SEED)\nX_b = np.column_stack((np.ones_like(y), X))\n\nplt.scatter(X[y==0,0], X[y==0,1], color='b', label=\"$y = 0$\")\nplt.scatter(X[y==1,0], X[y==1,1], color='r', label=\"$y = 1$\")\nplt.xlabel('$x_1$')\nplt.ylabel('$x_2$')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"***\n**Question:** What do you notice about the data? It is possible for logistic regression to achieve perfect accuracy on this data?\n***",
"_____no_output_____"
],
[
"## 2. Weighted linear regression\n\nBefore we can implement the IRLS algorithm for logistic regression, we'll need a function that can solve _weighted_ linear regression problems. \nThis is a minor extension of the material we covered in last week's worksheet.\nWhereas last week, each sample contributed equally to the empirical risk, we now allow the samples to contribute with different weights.\nSpecifically, the empirical risk becomes:\n\n$$\n\\hat{R}(\\mathbf{w}) = \\frac{1}{n} \\sum_{i = 1}^{n} m_i (y_i - \\mathbf{x}_i^\\top \\mathbf{w})^2\n$$\n\nwhere $m_i > 0$ is the weight of sample $i$. We assume the feature vectors $\\mathbf{x}_i$ have been prepended with \"1\" entries to incorporate a bias term.\n\n_\\[Aside: weighted linear regression can be used in applications where each sample has a different variance $\\sigma_i^2$ (e.g. measurement error). The best linear unbiased estimator is obtained by setting $m_i = \\frac{1}{\\sigma_i^2}$ assuming the variances are known.\\]_\n\nWeighted linear regression can also be solved analytically, by generalising the normal equation as follows:\n\n$$\n\\mathbf{w}^\\star = (\\mathbf{X}^\\top \\mathbf{M} \\mathbf{X})^{-1} \\mathbf{X}^\\top \\mathbf{M} \\mathbf{y},\n$$\n\nwhere $\\mathbf{X} = \\begin{pmatrix} \\mathbf{x}_1^\\top \\\\ \\vdots \\\\ \\mathbf{x}_n^\\top \\end{pmatrix}$, $\\mathbf{y} = \\begin{pmatrix} y_1 \\\\ \\vdots \\\\ y_n \\end{pmatrix}$ and $\\mathbf{M} = \\operatorname{diag}(m_1, \\ldots, m_n)$.\n\nBelow we define a function `fit_linear` that fits a weighted linear model given training data $\\mathbf{X}$, $\\mathbf{y}$ and sample weights $\\mathbf{m} = [m_1, \\ldots, m_n]^\\top$.",
"_____no_output_____"
]
],
[
[
"def fit_linear(X, y, sample_weight = None):\n \"\"\"Fits a linear regression model according to the given training data\n \n Parameters\n ----------\n X : array of shape (n_samples, n_features)\n Feature matrix. The matrix must contain a constant column to \n incorporate a non-zero bias.\n \n y : array of shape (n_samples,)\n Response relative to X\n \n sample_weight : numpy.ndarray of shape (n_samples,) default=None\n Weights that are assigned to individual samples.\n If not provided, then each sample is given unit weight.\n \n Returns\n -------\n weights : array of shape (n_features,)\n \"\"\"\n if sample_weight is not None:\n sqrt_sample_weight = np.sqrt(sample_weight)\n X = X * sqrt_sample_weight[:, np.newaxis]\n y = y * sqrt_sample_weight\n weights, _, _, _ = np.linalg.lstsq(X.T @ X, X.T @ y, rcond=None)\n return weights",
"_____no_output_____"
]
],
[
[
"## 3. Logistic regression via IRLS\n\nWe're now ready to start implementing the IRLS algorithm for logistic regression step-by-step.\n\nRecall that logistic regression assumes a linear relationship between the features $\\mathbf{x}$ and the log-odds of the event $Y = 1$:\n\n$$\n\\log \\frac{p(y = 1|\\mathbf{x})}{1 - p(y = 1|\\mathbf{x})} = \\mathbf{x}^\\top \\mathbf{w}.\n$$\n\nFrom a decision-theoretic point of view, we choose the weights vector $\\mathbf{w}$ to minimise the empirical risk under the log-loss (a.k.a. cross-entropy loss and logistic loss):\n\n$$\n\\hat{R}(\\mathbf{w}) = - \\frac{1}{n} \\sum_{i = 1}^{n} \\ell_\\mathrm{log}(y_i, \\mu_i(\\mathbf{w})),\n$$\n\nwhere $\\ell_\\mathrm{log}(y, \\mu) = y \\log \\mu + (1 - y) \\log (1 - \\mu)$ and $\\mu_i(\\mathbf{w}) := \\frac{1}{1 + \\exp( - \\mathbf{x}_i^\\top \\mathbf{w})}$.\n\nWe'll need to evaluate $\\hat{R}(\\mathbf{w})$ later on to generate convergence plots, so we define a function for this below.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import log_loss\n\ndef risk(X, y, w):\n \"\"\"Evaluate the empirical risk under the cross-entropy (logistic) loss\n \n Parameters\n ----------\n X : array of grad_risk(X, y, w)shape (n_samples, n_features)\n Feature matrix. The matrix must contain a constant column to \n incorporate a non-zero bias.\n \n y : array of shape (n_samples,)\n Response relative to X. Binary classes must be encoded as 0 and 1.\n \n w : array of shape (n_samples,)\n Weight vector.\n \n Returns\n -------\n risk : float\n \"\"\"\n mu = expit(X @ w)\n return log_loss(y, mu)",
"_____no_output_____"
]
],
[
[
"We'll also need to be able to compute the _gradient_ of the empirical risk $\\nabla_{\\mathbf{w}} \\hat{R}(\\mathbf{w}')$ in order to:\n\n* decide when we can stop the IRLS algorithm (we're close to optimality when $\\|\\nabla_{\\mathbf{w}} \\hat{R}(\\mathbf{w}')\\|_\\infty \\approx 0$)\n* implement gradient descent (later) as an alternative to IRLS\n\nIt's straightforward to show (using vector calculus) that:\n\n$$\n\\nabla_{\\mathbf{w}} \\hat{R}(\\mathbf{w}) = \\frac{1}{n} \\sum_{i = 1}^{n} (\\mu_i(\\mathbf{w}) - y_i)\\mathbf{x}_i = \\frac{1}{n} \\mathbf{X}^\\top (\\boldsymbol{\\mu} - \\mathbf{y}),\n$$\n\nwhere $\\boldsymbol{\\mu} = \\begin{pmatrix} \\mu_1(\\mathbf{w}) \\\\ \\vdots \\\\ \\mu_n(\\mathbf{w}) \\end{pmatrix}$.\n\n***\n**Exercise:** Complete the `grad_risk` function below, which computes $\\nabla_{\\mathbf{w}} \\hat{R}(\\mathbf{w})$ for a given weight vector $\\mathbf{w}$ and training data $\\mathbf{X}$ and $\\mathbf{y}$.\n***",
"_____no_output_____"
]
],
[
[
"def grad_risk(X, y, w):\n \"\"\"Evaluate the gradient of the empirical risk\n \n Parameters\n ----------\n X : array of shape (n_samples, n_features)\n Feature matrix. The matrix must contain a constant column to \n incorporate a non-zero bias.\n \n y : array of shape (n_samples,)\n Response relative to X. Binary classes must be encoded as 0 and 1.\n \n w : array of shape (n_samples,)\n Weight vector.\n \n Returns\n -------\n grad_w : array of shape (n_features,)\n \"\"\"\n mu = expit(X @ w)\n grad_w = (1/y.size) * X.T @ (mu-y) # fill in\n return grad_w\n\n# Test case\nif RND_SEED == 0:\n test_grad_risk_actual = grad_risk(X_b, y, np.ones(3))\n test_grad_risk_desired = np.array([0.11641865, -0.25260051, 0.20606407])\n np.testing.assert_allclose(test_grad_risk_actual, test_grad_risk_desired)",
"_____no_output_____"
]
],
[
[
"In lectures, we saw that the IRLS algorithm solves logistic regression by iteratively solving a series of weighted linear regression problems.\n\nAt iteration $t$, the weights vector $\\mathbf{w}_t$ is updated by solving the normal equation:\n\n$$\n\\mathbf{w}_{t + 1} = (\\mathbf{X}^\\top \\mathbf{M}_t \\mathbf{X})^{-1} \\mathbf{X}^\\top \\mathbf{M}_t \\mathbf{b}_t,\n$$\n\nwhere \n* $\\mathbf{b}_t = \\mathbf{X} \\mathbf{w}_t + \\mathbf{M}_t^{-1}(\\mathbf{y} - \\boldsymbol{\\mu}_t)$ is the linearised response (analogue of $\\mathbf{y}$),\n* $\\mathbf{M}_t = \\operatorname{diag}(\\boldsymbol{\\mu}_t (1 - \\boldsymbol{\\mu}_t))$ are the sample weights, and\n* $\\boldsymbol{\\mu}_t = [\\mu_{1}(\\mathbf{w}_t), \\ldots, \\mu_{n}(\\mathbf{w}_t)]^\\top$.\n\n***\n**Exercise:** Complete the `update_weight_irls` function below, which performs a single IRLS weight update.\n\n_Hint: you should use the previously-defined `fit_linear` function to solve the normal equation._\n***",
"_____no_output_____"
]
],
[
[
"def update_weight_irls(X, y, w):\n \"\"\"Performs a weight update using the IRLS algorithm\n \n Parameters\n ----------\n X : array of shape (n_samples, n_features)\n Feature matrix. The matrix must contain a constant column to \n incorporate a non-zero bias.\n \n y : array of shape (n_samples,)\n Response relative to X. Binary classes must be encoded as 0 and 1.\n \n w : array of shape (n_samples,)\n Current estimate of the weight vector.\n \n Returns\n -------\n w : array of shape (n_features,)\n Updated estimate of the weights vector.\n \"\"\"\n # Compute the sample weights and linearised response\n # fill in\n \n mu = expit(X @ w)\n M = np.diag(mu*(1-mu))\n b = X @ w + np.linalg.inv(M) @ (y - mu)\n # Fit a weighted linear regression model\n w = np.linalg.inv(X.T @ M @ X) @ X.T @ M @ b # fill in\n \n return w\n\n# Test case\nif RND_SEED == 0:\n test_update_weight_irls_actual = update_weight_irls(X_b, y, np.ones(3))\n test_update_weight_irls_desired = np.array([0.88475769, 2.08819846, 0.04110822])\n np.testing.assert_allclose(test_update_weight_irls_actual, test_update_weight_irls_desired)",
"_____no_output_____"
]
],
[
[
"Now that we've implemented `grad_risk` and `update_weight_irls`, we're very close to being able to run IRLS.\nWe just need to write some code to:\n\n* initialise the weight vector, and\n* iterate until the stopping criterion $\\| \\nabla_\\mathbf{w} \\hat{R}(\\mathbf{w}_t) \\|_\\infty \\leq \\mathtt{tol}$ is satisfied (or a max number of iterations is completed).\n\nWe do this in the `fit_logistic` function below. \nNote that we treat the `update_weight` function as a parameter, which defaults to IRLS. \nThis will allow us to reuse `fit_logistic` for gradient descent later on.",
"_____no_output_____"
]
],
[
[
"def fit_logistic(X, y, w_init=None, max_iter = 100, tol = 1e-4, \n update_weight = update_weight_irls, **kwargs):\n \"\"\"Fits a binary logistic regression model according to the given training \n data\n \n Parameters\n ----------\n X : array of shape (n_samples, n_features)\n Feature matrix. The matrix must contain a constant column to \n incorporate a non-zero bias.\n \n y : array of shape (n_samples,)\n Response relative to `X`. Binary classes must be encoded as 0 and 1.\n \n w_init : array of shape (n_features,) default=None\n Initial guess for the weights vector. Defaults to a vector of\n zeroes.\n \n max_iter : int, default=100\n Maximum number of iterations\n \n tol : float, default=1e-4\n Stop when the inf-norm of the gradient falls below this value.\n \n update_weight : callable, default=update_weight_irls\n Callable that performs a weight update. Must have signature \n (X, y, w, **kwargs). Defaults to IRLS.\n \n **kwargs : \n Keyword arguments passed to `update_weight`.\n \n Returns\n -------\n w_history : list of arrays of shape (n_features)\n History of weight vectors\n \"\"\"\n if w_init is None:\n # Default weight initialisation\n w_init = np.zeros(X.shape[1], dtype=float)\n \n # Store history of weights\n w_history = [w_init]\n w = w_init\n \n for t in range(max_iter):\n w = update_weight(X, y, w, **kwargs)\n w_history.append(w)\n \n # Check stopping criterion\n grad_inf = np.linalg.norm(grad_risk(X, y, w), ord=np.inf)\n if grad_inf <= tol:\n break\n \n print(\"Stopping after {} iterations\".format(t))\n print(\"Inf-norm of grad is {:.4g}\".format(grad_inf))\n \n return w_history",
"_____no_output_____"
]
],
[
[
"Let's run the algorithm on the 2D classification data we generated in Section 1 and visualise the result. Does the result look reasonable?",
"_____no_output_____"
]
],
[
[
"w_history_irls = fit_logistic(X_b, y, tol=1e-9)\n\ndef plot_decision_boundary(X, y, w):\n \"\"\"Plots the decision boundary of a logistic regression classifier defined \n by weights `w`\n \"\"\"\n fig, ax = plt.subplots()\n ax.scatter(X_b[y==0,1], X_b[y==0,2], color='b', label=\"$y = 0$\")\n ax.scatter(X_b[y==1,1], X_b[y==1,2], color='r', label=\"$y = 1$\")\n xlim, ylim = ax.get_xlim(), ax.get_ylim()\n ax.plot(list(xlim), [-w[0]/w[2] - w[1]/w[2] * x for x in xlim], ls = \"-\", color=\"k\")\n ax.set_xlim(xlim)\n ax.set_ylim(ylim)\n ax.set_xlabel('$x_1$')\n ax.set_ylabel('$x_2$')\n ax.set_title(\"Decision boundary\")\n ax.legend()\n plt.show()\n\nplot_decision_boundary(X_b, y, w_history_irls[-1])",
"Stopping after 6 iterations\nInf-norm of grad is 3.507e-12\n"
]
],
[
[
"We can also check the validity of our implementation by comparing with scikit-learn's implementation. Note that the scikit-learn implementation incorporates $L_2$ regularisation by default, so we need to switch it off by setting `penalty = 'none'`.",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\n\nclf = LogisticRegression(penalty='none')\nclf.fit(X, y)\nw_sklearn = np.r_[clf.intercept_, clf.coef_.squeeze()]\nprint(\"Weights according to IRLS: {}\".format(w_history_irls[-1]))\nprint(\"Weights according to scikit-learn: {}\".format(w_sklearn))",
"/Users/xinyaoniu/anaconda3/lib/python3.7/site-packages/sklearn/linear_model/logistic.py:432: FutureWarning: Default solver will be changed to 'lbfgs' in 0.22. Specify a solver to silence this warning.\n FutureWarning)\n"
]
],
[
[
"Let's take a look at the path taken by the IRLS algorithm to reach the optimal solution.\nWe plot the weight vectors at each iteration $\\mathbf{w}_0, \\mathbf{w}_1, \\ldots$ on top of contours of the empirical risk $\\hat{R}(\\mathbf{w})$. \nThe darker the shade, the lower the empirical risk.",
"_____no_output_____"
]
],
[
[
"def plot_iterates(X, y, w_history):\n \"\"\"Plots the path of iterates in weight space (excluding the bias)\"\"\"\n w_history = np.array(w_history)\n \n # Compute axes limits\n w12_max = w_history[:,1:].max()\n w12_min = w_history[:,1:].min()\n w12_ran = w12_max - w12_min\n border = 0.1\n \n # Compute objective on grid\n w12 = np.linspace(w12_min - border * w12_ran, w12_max + border * w12_ran, num=100)\n w1v, w2v = np.meshgrid(w12, w12)\n w12v = np.c_[w1v.ravel(), w2v.ravel()]\n z = np.array([risk(X_b, y, np.r_[w_history[-1][0], w12]) for w12 in w12v])\n z = z.reshape(w1v.shape)\n\n plt.contourf(w1v, w2v, z, cmap='gist_gray')\n plt.colorbar(label='Empirical risk')\n plt.plot(w_history[:,1], w_history[:,2], c='c', ls='--')\n plt.scatter(w_history[:,1], w_history[:,2], c='r', marker='.', label='Iterate')\n plt.xlabel('$w_1$')\n plt.ylabel('$w_2$')\n plt.legend()\n plt.title('Contour plot of iterates in weight space')\n plt.show()\n\nplot_iterates(X_b, y, w_history_irls)",
"_____no_output_____"
]
],
[
[
"## 4. Logistic regression via gradient descent\n\nFinally, let's compare the IRLS algorithm with gradient descent. \nTo do this, we can reuse the `fit_logistic` function defined earlier. \nWe just need to replace `update_weight_irls` with an analagous function for gradient descent.\n\nRecall that the weight update for gradient descent at iteration $t$ is given by:\n\n$$\n\\mathbf{w}_t = \\mathbf{w}_{t - 1} - \\eta \\nabla_\\mathbf{w} \\hat{R}(\\mathbf{w}_{t - 1})\n$$\n\nwhere $\\eta > 0$ is a learning rate parameter.\n***\n**Exercise:** Complete the `update_weight_gd` function below which implements the weight update for gradient descent.\n***",
"_____no_output_____"
]
],
[
[
"def update_weight_gd(X, y, w, **kwargs):\n \"\"\"Performs a gradient descent weight update\n \n Parameters\n ----------\n X : array of shape (n_samples, n_features)\n Feature matrix. The matrix must contain a constant column to \n incorporate a non-zero bias.\n \n y : array of shape (n_samples,)\n Response relative to X. Binary classes must be encoded as 0 and 1.\n \n w : array of shape (n_samples,)\n Current estimate of the weight vector.\n \n **kwargs : \n Keyword arguments.\n \n Returns\n -------\n w : array of shape (n_features,)\n Updated estimate of the weights vector.\n \"\"\"\n # Get learning rate from kwargs, defaulting to 1.0 if None\n eta = kwargs.get(\"eta\", 1)\n return w - eta*grad_risk(X, y, w) # fill in",
"_____no_output_____"
]
],
[
[
"Let's run our gradient descent implementation on the same classification data from before. \nWhat do you notice about the path taken by the gradient descent algorithm? \nHow does it compare with IRLS?",
"_____no_output_____"
]
],
[
[
"w_history_gd = fit_logistic(X_b, y, update_weight=update_weight_gd, max_iter=1000, eta = 5)\nplot_decision_boundary(X_b, y, w_history_gd[-1])\nplot_iterates(X_b, y, w_history_gd)",
"Stopping after 63 iterations\nInf-norm of grad is 9.395e-05\n"
]
],
[
[
"***\n## Bonus: Linearly separable case (optional)\n\n\n**Exercise:** What happens when you re-run the notebook with `RND_SEED = 90051`?",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eccb86e0da3270c3abce9cc5fc03ea5ecf69a71b | 15,743 | ipynb | Jupyter Notebook | docs/numpy-fft.ipynb | RoboticExplorationLab/micropython-ulab | b0679e6d16d87f5acb09dee690a71a54d4c2892b | [
"MIT"
] | 232 | 2019-10-30T02:47:59.000Z | 2022-03-29T13:35:42.000Z | docs/numpy-fft.ipynb | RoboticExplorationLab/micropython-ulab | b0679e6d16d87f5acb09dee690a71a54d4c2892b | [
"MIT"
] | 325 | 2019-10-25T00:27:29.000Z | 2022-03-16T19:47:45.000Z | docs/numpy-fft.ipynb | RoboticExplorationLab/micropython-ulab | b0679e6d16d87f5acb09dee690a71a54d4c2892b | [
"MIT"
] | 73 | 2019-11-04T19:31:22.000Z | 2022-03-10T03:11:41.000Z | 30.688109 | 328 | 0.525503 | [
[
[
"%pylab inline",
"Populating the interactive namespace from numpy and matplotlib\n"
]
],
[
[
"## Notebook magic",
"_____no_output_____"
]
],
[
[
"from IPython.core.magic import Magics, magics_class, line_cell_magic\nfrom IPython.core.magic import cell_magic, register_cell_magic, register_line_magic\nfrom IPython.core.magic_arguments import argument, magic_arguments, parse_argstring\nimport subprocess\nimport os",
"_____no_output_____"
],
[
"@magics_class\nclass PyboardMagic(Magics):\n @cell_magic\n @magic_arguments()\n @argument('-skip')\n @argument('-unix')\n @argument('-pyboard')\n @argument('-file')\n @argument('-data')\n @argument('-time')\n @argument('-memory')\n def micropython(self, line='', cell=None):\n args = parse_argstring(self.micropython, line)\n if args.skip: # doesn't care about the cell's content\n print('skipped execution')\n return None # do not parse the rest\n if args.unix: # tests the code on the unix port. Note that this works on unix only\n with open('/dev/shm/micropython.py', 'w') as fout:\n fout.write(cell)\n proc = subprocess.Popen([\"../../micropython/ports/unix/micropython\", \"/dev/shm/micropython.py\"], \n stdout=subprocess.PIPE, stderr=subprocess.PIPE)\n print(proc.stdout.read().decode(\"utf-8\"))\n print(proc.stderr.read().decode(\"utf-8\"))\n return None\n if args.file: # can be used to copy the cell content onto the pyboard's flash\n spaces = \" \"\n try:\n with open(args.file, 'w') as fout:\n fout.write(cell.replace('\\t', spaces))\n printf('written cell to {}'.format(args.file))\n except:\n print('Failed to write to disc!')\n return None # do not parse the rest\n if args.data: # can be used to load data from the pyboard directly into kernel space\n message = pyb.exec(cell)\n if len(message) == 0:\n print('pyboard >>>')\n else:\n print(message.decode('utf-8'))\n # register new variable in user namespace\n self.shell.user_ns[args.data] = string_to_matrix(message.decode(\"utf-8\"))\n \n if args.time: # measures the time of executions\n pyb.exec('import utime')\n message = pyb.exec('t = utime.ticks_us()\\n' + cell + '\\ndelta = utime.ticks_diff(utime.ticks_us(), t)' + \n \"\\nprint('execution time: {:d} us'.format(delta))\")\n print(message.decode('utf-8'))\n \n if args.memory: # prints out memory information \n message = pyb.exec('from micropython import mem_info\\nprint(mem_info())\\n')\n print(\"memory before execution:\\n========================\\n\", message.decode('utf-8'))\n message = pyb.exec(cell)\n print(\">>> \", message.decode('utf-8'))\n message = pyb.exec('print(mem_info())')\n print(\"memory after execution:\\n========================\\n\", message.decode('utf-8'))\n\n if args.pyboard:\n message = pyb.exec(cell)\n print(message.decode('utf-8'))\n\nip = get_ipython()\nip.register_magics(PyboardMagic)",
"_____no_output_____"
]
],
[
[
"## pyboard",
"_____no_output_____"
]
],
[
[
"import pyboard\npyb = pyboard.Pyboard('/dev/ttyACM0')\npyb.enter_raw_repl()",
"_____no_output_____"
],
[
"pyb.exit_raw_repl()\npyb.close()",
"_____no_output_____"
],
[
"%%micropython -pyboard 1\n\nimport utime\nimport ulab as np\n\ndef timeit(n=1000):\n def wrapper(f, *args, **kwargs):\n func_name = str(f).split(' ')[1]\n def new_func(*args, **kwargs):\n run_times = np.zeros(n, dtype=np.uint16)\n for i in range(n):\n t = utime.ticks_us()\n result = f(*args, **kwargs)\n run_times[i] = utime.ticks_diff(utime.ticks_us(), t)\n print('{}() execution times based on {} cycles'.format(func_name, n, (delta2-delta1)/n))\n print('\\tbest: %d us'%np.min(run_times))\n print('\\tworst: %d us'%np.max(run_times))\n print('\\taverage: %d us'%np.mean(run_times))\n print('\\tdeviation: +/-%.3f us'%np.std(run_times)) \n return result\n return new_func\n return wrapper\n\ndef timeit(f, *args, **kwargs):\n func_name = str(f).split(' ')[1]\n def new_func(*args, **kwargs):\n t = utime.ticks_us()\n result = f(*args, **kwargs)\n print('execution time: ', utime.ticks_diff(utime.ticks_us(), t), ' us')\n return result\n return new_func",
"\n"
]
],
[
[
"__END_OF_DEFS__",
"_____no_output_____"
],
[
"# numpy.fft\n\nFunctions related to Fourier transforms can be called by prepending them with `numpy.fft.`. The module defines the following two functions:\n\n1. [numpy.fft.fft](#fft)\n1. [numpy.fft.ifft](#ifft)\n\n`numpy`: https://docs.scipy.org/doc/numpy/reference/generated/numpy.fft.ifft.html\n\n## fft\n\nSince `ulab`'s `ndarray` does not support complex numbers, the invocation of the Fourier transform differs from that in `numpy`. In `numpy`, you can simply pass an array or iterable to the function, and it will be treated as a complex array:",
"_____no_output_____"
]
],
[
[
"fft.fft([1, 2, 3, 4, 1, 2, 3, 4])",
"_____no_output_____"
]
],
[
[
"**WARNING:** The array returned is also complex, i.e., the real and imaginary components are cast together. In `ulab`, the real and imaginary parts are treated separately: you have to pass two `ndarray`s to the function, although, the second argument is optional, in which case the imaginary part is assumed to be zero.\n\n**WARNING:** The function, as opposed to `numpy`, returns a 2-tuple, whose elements are two `ndarray`s, holding the real and imaginary parts of the transform separately. ",
"_____no_output_____"
]
],
[
[
"%%micropython -pyboard 1\n\nfrom ulab import numpy as np\n\nx = np.linspace(0, 10, num=1024)\ny = np.sin(x)\nz = np.zeros(len(x))\n\na, b = np.fft.fft(x)\nprint('real part:\\t', a)\nprint('\\nimaginary part:\\t', b)\n\nc, d = np.fft.fft(x, z)\nprint('\\nreal part:\\t', c)\nprint('\\nimaginary part:\\t', d)",
"real part:\t array([5119.996, -5.004663, -5.004798, ..., -5.005482, -5.005643, -5.006577], dtype=float)\r\n\r\nimaginary part:\t array([0.0, 1631.333, 815.659, ..., -543.764, -815.6588, -1631.333], dtype=float)\r\n\r\nreal part:\t array([5119.996, -5.004663, -5.004798, ..., -5.005482, -5.005643, -5.006577], dtype=float)\r\n\r\nimaginary part:\t array([0.0, 1631.333, 815.659, ..., -543.764, -815.6588, -1631.333], dtype=float)\r\n\n"
]
],
[
[
"## ifft\n\nThe above-mentioned rules apply to the inverse Fourier transform. The inverse is also normalised by `N`, the number of elements, as is customary in `numpy`. With the normalisation, we can ascertain that the inverse of the transform is equal to the original array.",
"_____no_output_____"
]
],
[
[
"%%micropython -pyboard 1\n\nfrom ulab import numpy as np\n\nx = np.linspace(0, 10, num=1024)\ny = np.sin(x)\n\na, b = np.fft.fft(y)\n\nprint('original vector:\\t', y)\n\ny, z = np.fft.ifft(a, b)\n# the real part should be equal to y\nprint('\\nreal part of inverse:\\t', y)\n# the imaginary part should be equal to zero\nprint('\\nimaginary part of inverse:\\t', z)",
"original vector:\t array([0.0, 0.009775016, 0.0195491, ..., -0.5275068, -0.5357859, -0.5440139], dtype=float)\n\nreal part of inverse:\t array([-2.980232e-08, 0.0097754, 0.0195494, ..., -0.5275064, -0.5357857, -0.5440133], dtype=float)\n\nimaginary part of inverse:\t array([-2.980232e-08, -1.451171e-07, 3.693752e-08, ..., 6.44871e-08, 9.34986e-08, 2.18336e-07], dtype=float)\n\n"
]
],
[
[
"Note that unlike in `numpy`, the length of the array on which the Fourier transform is carried out must be a power of 2. If this is not the case, the function raises a `ValueError` exception.",
"_____no_output_____"
],
[
"## Computation and storage costs",
"_____no_output_____"
],
[
"### RAM\n\nThe FFT routine of `ulab` calculates the transform in place. This means that beyond reserving space for the two `ndarray`s that will be returned (the computation uses these two as intermediate storage space), only a handful of temporary variables, all floats or 32-bit integers, are required. ",
"_____no_output_____"
],
[
"### Speed of FFTs\n\nA comment on the speed: a 1024-point transform implemented in python would cost around 90 ms, and 13 ms in assembly, if the code runs on the pyboard, v.1.1. You can gain a factor of four by moving to the D series \nhttps://github.com/peterhinch/micropython-fourier/blob/master/README.md#8-performance. ",
"_____no_output_____"
]
],
[
[
"%%micropython -pyboard 1\n\nfrom ulab import numpy as np\n\nx = np.linspace(0, 10, num=1024)\ny = np.sin(x)\n\n@timeit\ndef np_fft(y):\n return np.fft.fft(y)\n\na, b = np_fft(y)",
"execution time: 1985 us\n\n"
]
],
[
[
"The C implementation runs in less than 2 ms on the pyboard (we have just measured that), and has been reported to run in under 0.8 ms on the D series board. That is an improvement of at least a factor of four. ",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eccb8a43b6ad1c439f7c912be0fd616eacd4046b | 710,346 | ipynb | Jupyter Notebook | TransferLearning.ipynb | KiweeEu/transfer-learning-demo | 574ec05f67403b7e99ab7d1cb27a0e2ae9e5d4ea | [
"MIT"
] | 3 | 2020-09-18T14:16:27.000Z | 2021-02-09T21:30:02.000Z | TransferLearning.ipynb | KiweeEu/transfer-learning-demo | 574ec05f67403b7e99ab7d1cb27a0e2ae9e5d4ea | [
"MIT"
] | null | null | null | TransferLearning.ipynb | KiweeEu/transfer-learning-demo | 574ec05f67403b7e99ab7d1cb27a0e2ae9e5d4ea | [
"MIT"
] | null | null | null | 1,303.387156 | 664,996 | 0.95582 | [
[
[
"# Transfer Learning using Keras and EfficientNet\n\n**Author:** Serge Korzh, a data scientist at [Kiwee](https://kiwee.eu/)\n\nIn this notebook, we will train a classifier on the [Flowers image dataset](https://www.kaggle.com/olgabelitskaya/flower-color-images), but rather than building and training a [Convolutional Neural Network](https://developers.google.com/machine-learning/glossary/#convolutional-neural-network) model from scratch, we'll use [Google's EfficientNet](https://ai.googleblog.com/2019/05/efficientnet-improving-accuracy-and.html) model pre-trained on the [ImageNet dataset](http://www.image-net.org/) as a base. Essentially, this will transfer the knowledge accumulated during the training on a large image dataset with 1000 object classes to a similar, more specific problem – classifying flower images by species.",
"_____no_output_____"
],
[
"## Getting the dependencies\n\nLet's first get the [efficientnet](https://github.com/qubvel/efficientnet) package that includes the model architecture.",
"_____no_output_____"
]
],
[
[
"!pip install efficientnet==1.1.0",
"Collecting efficientnet==1.1.0\n Downloading efficientnet-1.1.0-py3-none-any.whl (18 kB)\nRequirement already satisfied: scikit-image in /opt/conda/lib/python3.7/site-packages (from efficientnet==1.1.0) (0.16.2)\nCollecting keras-applications<=1.0.8,>=1.0.7\n Downloading Keras_Applications-1.0.8-py3-none-any.whl (50 kB)\n\u001b[K |████████████████████████████████| 50 kB 3.5 MB/s eta 0:00:011\n\u001b[?25hRequirement already satisfied: scipy>=0.19.0 in /opt/conda/lib/python3.7/site-packages (from scikit-image->efficientnet==1.1.0) (1.4.1)\nRequirement already satisfied: matplotlib!=3.0.0,>=2.0.0 in /opt/conda/lib/python3.7/site-packages (from scikit-image->efficientnet==1.1.0) (3.2.1)\nRequirement already satisfied: networkx>=2.0 in /opt/conda/lib/python3.7/site-packages (from scikit-image->efficientnet==1.1.0) (2.4)\nRequirement already satisfied: pillow>=4.3.0 in /opt/conda/lib/python3.7/site-packages (from scikit-image->efficientnet==1.1.0) (7.1.2)\nRequirement already satisfied: imageio>=2.3.0 in /opt/conda/lib/python3.7/site-packages (from scikit-image->efficientnet==1.1.0) (2.8.0)\nRequirement already satisfied: PyWavelets>=0.4.0 in /opt/conda/lib/python3.7/site-packages (from scikit-image->efficientnet==1.1.0) (1.1.1)\nRequirement already satisfied: numpy>=1.9.1 in /opt/conda/lib/python3.7/site-packages (from keras-applications<=1.0.8,>=1.0.7->efficientnet==1.1.0) (1.18.5)\nRequirement already satisfied: h5py in /opt/conda/lib/python3.7/site-packages (from keras-applications<=1.0.8,>=1.0.7->efficientnet==1.1.0) (2.10.0)\nRequirement already satisfied: python-dateutil>=2.1 in /opt/conda/lib/python3.7/site-packages (from matplotlib!=3.0.0,>=2.0.0->scikit-image->efficientnet==1.1.0) (2.8.1)\nRequirement already satisfied: kiwisolver>=1.0.1 in /opt/conda/lib/python3.7/site-packages (from matplotlib!=3.0.0,>=2.0.0->scikit-image->efficientnet==1.1.0) (1.2.0)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /opt/conda/lib/python3.7/site-packages (from matplotlib!=3.0.0,>=2.0.0->scikit-image->efficientnet==1.1.0) (2.4.7)\nRequirement already satisfied: cycler>=0.10 in /opt/conda/lib/python3.7/site-packages (from matplotlib!=3.0.0,>=2.0.0->scikit-image->efficientnet==1.1.0) (0.10.0)\nRequirement already satisfied: decorator>=4.3.0 in /opt/conda/lib/python3.7/site-packages (from networkx>=2.0->scikit-image->efficientnet==1.1.0) (4.4.2)\nRequirement already satisfied: six in /opt/conda/lib/python3.7/site-packages (from h5py->keras-applications<=1.0.8,>=1.0.7->efficientnet==1.1.0) (1.15.0)\nInstalling collected packages: keras-applications, efficientnet\nSuccessfully installed efficientnet-1.1.0 keras-applications-1.0.8\n"
]
],
[
[
"## Imports\n\nIn this notebook we'll need:\n- `h5py` and `numpy` for data loading and manipulation\n- `tensorflow`, `keras` and `efficientnet` for model training\n- `matplotlib` for data visualisation. ",
"_____no_output_____"
]
],
[
[
"import h5py\nimport numpy as np\nimport tensorflow as tf\nfrom tensorflow import keras\n# import the tensorflow.keras version of efficientnet\nfrom efficientnet import tfkeras as efficientnet\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Loading data\n\nWe'll use a very small dataset of flower photos [taken from Kaggle](https://www.kaggle.com/olgabelitskaya/flower-color-images). \nRun the following cell to load the data:",
"_____no_output_____"
]
],
[
[
"# when running on Colab, this will download the dataset and save into runtime first\nimport sys\nif 'google.colab' in sys.modules:\n !wget https://github.com/KiweeEu/transfer-learning-demo/raw/master/data/flowers.h5 -P ./data\n\nflowers_data = h5py.File('./data/flowers.h5', 'r')\nimages_data = flowers_data['images'][:]\nlabels_data = flowers_data['labels'][:]",
"_____no_output_____"
]
],
[
[
"It's a good habit to check how data looks like before actually starting to work with it.\n\nLet's display basic information about the data to get a sense of what we're dealing with:",
"_____no_output_____"
]
],
[
[
"print(f'Images data shape: {images_data.shape}')\nprint(f'Labels data shape: {labels_data.shape}')\nprint(f'Images data min: {np.min(images_data)}, max: {np.max(images_data)}')\nprint(f'Images data type: {images_data.dtype}')\nprint(f'Labels data type: {labels_data.dtype}')\nprint(f'Unique labels: {np.unique(labels_data)}')\nprint(f'Number of unique labels: {len(np.unique(labels_data))}')",
"Images data shape: (210, 128, 128, 3)\nLabels data shape: (210,)\nImages data min: 0, max: 255\nImages data type: uint8\nLabels data type: uint8\nUnique labels: [0 1 2 3 4 5 6 7 8 9]\nNumber of unique labels: 10\n"
]
],
[
[
"We have 210 images in total belonging to 10 classes, and each image is 128×128×3 (where 3 is the three primary color channels – Red, Green, Blue). Each pixel is an integer from 0 to 255. Also, notice that the class is stored as an integer from 0 to 9.\n\nAll of this is very important, especially when we use external frameworks and pre-trained models, we have to make sure our data is structured properly. We'll touch on that topic later in the notebook.\n\nLet's also visualize a couple of images and the distribution of classes:",
"_____no_output_____"
]
],
[
[
"# Names of flower species are just for visualisation\nflower_names = [\n 'phlox','rose','calendula','iris','leucanthemum maximum',\n 'bellflower','viola','rudbeckia laciniata','peony','aquilegia'\n]\n\nplt.figure(figsize=(10,10))\nfor i in range(25):\n plt.subplot(5,5,i+1)\n plt.xticks([])\n plt.yticks([])\n plt.grid(False)\n plt.imshow(images_data[i])\n plt.xlabel(flower_names[labels_data[i]])\nplt.show()",
"_____no_output_____"
],
[
"unique, counts = np.unique(labels_data, return_counts=True)\nticks = np.arange(len(counts))\nplt.figure(figsize=(14,6))\nplt.grid(color='w', axis='y')\nplt.bar(ticks, counts)\nplt.xticks(ticks, flower_names, rotation=15)\nplt.title('Number of images per class')\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can see that the dataset is more or less balanced – there aren't any classes that have substantially more or fewer examples than others.\n\nLet's now shuffle the data and split it into two halves – one for training, and one for validation. Since we don't have a lot of data, let's split it equaly so that both training and in validation sets will have 105 images with ~10 images per class.",
"_____no_output_____"
]
],
[
[
"# Set a seed for reproducibility\nseed = 1\ndef shuffle(a, b, seed=None):\n rand = np.random.RandomState(seed)\n p = rand.permutation(a.shape[0])\n return a[p], b[p]\n\nx_data, y_data = shuffle(images_data, labels_data, seed=seed)\n\nsplit_idx = int(0.5*x_data.shape[0])\nx_train, y_train = x_data[:split_idx], y_data[:split_idx]\nx_val, y_val = x_data[split_idx:], y_data[split_idx:]",
"_____no_output_____"
]
],
[
[
"Now, let's get to the nitty gritties of working with Keras. As we discussed, it's important to provide data to the model in the right format. We'll use the [ImageDataGenerator](https://www.tensorflow.org/api_docs/python/tf/keras/preprocessing/image/ImageDataGenerator) class to do so. Let's go over its arguments:\n\n1. `rotation_range`, `width_shift_range`, `height_shift_range`, `horizontal_flip` – those are augmentation parameters. Augmentation helps us introduce more variability from a small amount of data. Instead of feeding the original images, ImageDataGenerator will randomly apply transformations such as rotation (up to 20 degrees), shift along X and Y axes, and horizontal flipping (mirroring) of the image.\n2. `data_format` – as you remember, the images have a format of 128×128×3 which corresponds to width×height×channels. The color channels are the last dimension. Thus we set the argument to `channels_last`.\n3. `preprocessing_function` – since we use EfficientNet with pre-trained weights, it means the model has been already trained with images formatted in a certain way. Fortunately, efficientnet package provides `preprocess_input` function that will format the data in the same way it was formatted during training on the ImageNet data.\n\nDuring validation, we don't want to alter the images because we want our model to perform well on the original ones. Thus, we'll create another ImageDataGenerator called `val_datagen` that omits augmentation.",
"_____no_output_____"
]
],
[
[
"train_datagen = keras.preprocessing.image.ImageDataGenerator(\n rotation_range=20,\n width_shift_range=0.1,\n height_shift_range=0.1,\n horizontal_flip=True,\n data_format='channels_last',\n preprocessing_function=efficientnet.preprocess_input\n)\n\nval_datagen = keras.preprocessing.image.ImageDataGenerator(\n data_format='channels_last',\n preprocessing_function=efficientnet.preprocess_input\n)",
"_____no_output_____"
]
],
[
[
"After defining the generators, we need to specify from where to take the data. We do so using the `.flow(x, y)` method of the generators. It will return a [Python Iterator](https://docs.python.org/3/howto/functional.html#iterators) object that represents a stream of batches of the processed data.",
"_____no_output_____"
]
],
[
[
"train_iterator = train_datagen.flow(x_train, y_train, batch_size=35)\nval_iterator = val_datagen.flow(x_val, y_val, batch_size=105)",
"_____no_output_____"
]
],
[
[
"## Model training\n\nOkay, now to the exciting part – training our classifier! First, we have to define the model architecture, and for that, let's create the `get_model` function. It will receive two parameters: `input_shape` (i.e., dimensions of an image) and `num_classes` (i.e., the output shape). Inside, we'll create `base_model` using the `EfficientNetB4` function. This is one of eight different EfficientNet models ranging from `B0` to `B7` that differ by size. The bigger the model, the more capacity it has for learning, but it also requires more computing power and memory.\n\nLet's go through the important arguments:\n1. `weights='noisy-student'` – specifies that we want to use the weights trained on ImageNet data and a large amount of unlabelled data using the novel [Noisy Student training approach](https://arxiv.org/pdf/1911.04252v4.pdf) (see the paper for more info).\n2. `pooling='avg'` - after passing the images through EfficientNet, the resulting values will be averaged by channel to form a single vector for each image.\n3. `include_top=False` – the pre-trained model also includes the layer that gives predictions for ImageNet classes. We don't want that, as we'll train our own top layer to produce class probabilities for the ten flower species. Thus, we have to set `include_top` to `False`.",
"_____no_output_____"
]
],
[
[
"def get_model(input_shape, num_classes):\n base_model = efficientnet.EfficientNetB4(\n weights='noisy-student',\n pooling='avg',\n include_top=False,\n input_shape=input_shape\n )\n \n # \"Freeze\" the base_model layers so that we don't backpropagate through them.\n # This effectively makes base_model a feature extractor.\n for layer in base_model.layers:\n layer.trainable = False\n\n # Create our own fully-conected top layer with num_classes outputs that takes base_model.output as an input.\n # We use the 'softmax' activation for computing class probabilities.\n x = base_model.output\n predictions = keras.layers.Dense(num_classes, activation='softmax')(x)\n\n # the Model class packages our architecture into one object\n model = keras.models.Model(inputs=base_model.input, outputs=predictions)\n return model",
"_____no_output_____"
]
],
[
[
"Let's now create the model. Running it for the first time might take some time, as has to download the weights. After that, we have to call `.compile()` on our model, specifying the following:\n1. `optimizer='adam'` – specifies that we want to use Adam optimization during training. It is a popular variant of stochastic gradient descent that speeds up convergence.\n2. `loss=keras.losses.sparse_categorical_crossentropy` – the loss function we want to use in this case is the [cross entropy loss](https://www.tensorflow.org/api_docs/python/tf/keras/losses/SparseCategoricalCrossentropy) used in categorical classification. It's called \"sparse\" since the true classes are specified as integers instead of as one-hot vectors.\n3. `metrics=['accuracy']` – sets which metrics to track during training. ",
"_____no_output_____"
]
],
[
[
"model = get_model(input_shape=(128, 128, 3), num_classes=10)\nmodel.compile(\n optimizer='adam',\n loss=keras.losses.sparse_categorical_crossentropy,\n metrics=['accuracy']\n)",
"Downloading data from https://github.com/qubvel/efficientnet/releases/download/v0.0.1/efficientnet-b4_noisy-student_notop.h5\n71680000/71678424 [==============================] - 5s 0us/step\n"
]
],
[
[
"Now, let's train the model by calling `.fit()`. All we need to pass is our iterator and the number of iterations (epochs).",
"_____no_output_____"
]
],
[
[
"model.fit(train_iterator, epochs=30)",
"Epoch 1/30\n3/3 [==============================] - 2s 542ms/step - loss: 2.3903 - accuracy: 0.1238\nEpoch 2/30\n3/3 [==============================] - 2s 598ms/step - loss: 2.0979 - accuracy: 0.2667\nEpoch 3/30\n3/3 [==============================] - 2s 639ms/step - loss: 1.7897 - accuracy: 0.4571\nEpoch 4/30\n3/3 [==============================] - 2s 603ms/step - loss: 1.7047 - accuracy: 0.4190\nEpoch 5/30\n3/3 [==============================] - 2s 523ms/step - loss: 1.6263 - accuracy: 0.4381\nEpoch 6/30\n3/3 [==============================] - 2s 504ms/step - loss: 1.4564 - accuracy: 0.5714\nEpoch 7/30\n3/3 [==============================] - 2s 624ms/step - loss: 1.3617 - accuracy: 0.5619\nEpoch 8/30\n3/3 [==============================] - 2s 555ms/step - loss: 1.3232 - accuracy: 0.7048\nEpoch 9/30\n3/3 [==============================] - 2s 565ms/step - loss: 1.1561 - accuracy: 0.7810\nEpoch 10/30\n3/3 [==============================] - 2s 537ms/step - loss: 1.1830 - accuracy: 0.6857\nEpoch 11/30\n3/3 [==============================] - 3s 934ms/step - loss: 1.0520 - accuracy: 0.7810\nEpoch 12/30\n3/3 [==============================] - 2s 678ms/step - loss: 0.9889 - accuracy: 0.7619\nEpoch 13/30\n3/3 [==============================] - 3s 999ms/step - loss: 1.0311 - accuracy: 0.7810\nEpoch 14/30\n3/3 [==============================] - 3s 1s/step - loss: 0.8745 - accuracy: 0.8095\nEpoch 15/30\n3/3 [==============================] - 2s 540ms/step - loss: 0.9230 - accuracy: 0.7619\nEpoch 16/30\n3/3 [==============================] - 2s 524ms/step - loss: 0.8571 - accuracy: 0.8000\nEpoch 17/30\n3/3 [==============================] - 2s 508ms/step - loss: 0.8386 - accuracy: 0.8095\nEpoch 18/30\n3/3 [==============================] - 2s 525ms/step - loss: 0.8887 - accuracy: 0.8190\nEpoch 19/30\n3/3 [==============================] - 1s 498ms/step - loss: 0.8230 - accuracy: 0.8000\nEpoch 20/30\n3/3 [==============================] - 2s 544ms/step - loss: 0.8531 - accuracy: 0.8190\nEpoch 21/30\n3/3 [==============================] - 2s 541ms/step - loss: 0.7563 - accuracy: 0.8571\nEpoch 22/30\n3/3 [==============================] - 2s 558ms/step - loss: 0.7024 - accuracy: 0.8952\nEpoch 23/30\n3/3 [==============================] - 2s 508ms/step - loss: 0.6735 - accuracy: 0.8667\nEpoch 24/30\n3/3 [==============================] - 2s 511ms/step - loss: 0.6588 - accuracy: 0.8381\nEpoch 25/30\n3/3 [==============================] - 2s 531ms/step - loss: 0.6383 - accuracy: 0.8952\nEpoch 26/30\n3/3 [==============================] - 2s 524ms/step - loss: 0.6709 - accuracy: 0.8952\nEpoch 27/30\n3/3 [==============================] - 2s 510ms/step - loss: 0.6360 - accuracy: 0.9333\nEpoch 28/30\n3/3 [==============================] - 2s 534ms/step - loss: 0.6943 - accuracy: 0.8476\nEpoch 29/30\n3/3 [==============================] - 2s 512ms/step - loss: 0.6475 - accuracy: 0.8857\nEpoch 30/30\n3/3 [==============================] - 2s 518ms/step - loss: 0.5661 - accuracy: 0.9429\n"
]
],
[
[
"After training for 30 epochs, you should achieve a loss below 0.7 and accuracy above 80%. Now, let's test the model on our validation set by calling `.evaluate()`:",
"_____no_output_____"
]
],
[
[
"val_loss, val_acc = model.evaluate(val_iterator)\nprint(f'Validation loss: {round(val_loss, 4)}\\nValidation accuracy: {round(val_acc*100, 4)}%')",
"1/1 [==============================] - 0s 5ms/step - loss: 0.6201 - accuracy: 0.8762\nValidation loss: 0.6201\nValidation accuracy: 87.619%\n"
]
],
[
[
"On the validation set, you should get accuracy in the range of 75% to 95%. Such a big uncertainty comes from the fact that we have an extremely small dataset. In a real-world project, I highly recommend using [cross-validation](https://developers.google.com/machine-learning/glossary#cross-validation) to test your models in such cases to get an accurate perspective on the model's performance.\n\n## Conclusion\n\nCongrats! Having only 210 images from 10 classes, we achieved quite a high accuracy by leveraging the power of transfer learning. Hopefully, this notebook will serve you as a base for applying this technique in your future machine learning projects. Good Luck!\n\n**P.S.** If you want to see a real-life example of how transfer learning can be used, check out our dog breed recognizer at [breedread.kiwee.eu](https://breedread.kiwee.eu/)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eccb924112dfe4f2fdfaff7ac99daaa7e71e455a | 197,614 | ipynb | Jupyter Notebook | 07_algorithmique/3_tris_selection_et_insertion.ipynb | efloti/cours-nsi-premiere | 5b05bc81e5f8d7df47bf5785068b4bf4d1e357bb | [
"CC0-1.0"
] | null | null | null | 07_algorithmique/3_tris_selection_et_insertion.ipynb | efloti/cours-nsi-premiere | 5b05bc81e5f8d7df47bf5785068b4bf4d1e357bb | [
"CC0-1.0"
] | null | null | null | 07_algorithmique/3_tris_selection_et_insertion.ipynb | efloti/cours-nsi-premiere | 5b05bc81e5f8d7df47bf5785068b4bf4d1e357bb | [
"CC0-1.0"
] | null | null | null | 218.841639 | 55,876 | 0.898261 | [
[
[
"# Algorithmes de tri par sélection et par insertion",
"_____no_output_____"
],
[
"**Trier** une collection indexée de données $A$ signifie *permuter* ses éléments de façon qu'on ait, pour tous index $i$ et $j$ valides: \n\n$$i< j\\implies A[i]\\leqslant A[j]\\qquad\\text{ou}\\qquad i<j\\implies A[i]\\geqslant A[j]$$",
"_____no_output_____"
],
[
"selon que $A$ est trié dans l'ordre **croissant** (*ascendent*) ou dans l'ordre **décroissant** (*descendent*). \n\n*Exemple*: Si $A=14,23,5,11,22,26$ alors, après tri dans l'ordre croisant, $A=5,11,14,22,23,26$",
"_____no_output_____"
],
[
"Bien sûr, encore faut-il que les objets $A[i]$ et $A[j]$ soit *comparables* et cela ne va pas toujours de soi: souvent les objets sont comparés par l'intermédiaire d'une ou de plusieurs **clé(s)**.\n\nIci, par simplicité, nous supposerons simplement que nos objets sont des *entiers* et que «trier» signifie précisément *trier dans l'ordre croissant*. Si ces conditions ne sont pas réunies, il suffira alors d'adapter l'algorithme comme vu précédemment. ",
"_____no_output_____"
],
[
"Bien que trier une collection de données puisse paraître ennuyeux, le besoin impérieux de fouiller de grandes masses de données (plusieurs millions voir milliards) rend ce problème incontournable voir central dans bien des cas: nous avons déjà vu que la *recherche d'un élément dans une collection non ordonnée* **est beaucoup, beaucoup plus longue** qu'une *recherche dichotomique* qui s'appuie sur le fait que la collection est triée...",
"_____no_output_____"
],
[
"## Tri par sélection",
"_____no_output_____"
],
[
"<a href=\"https://vimeo.com/429772628\">Vidéo d'accompagnement</a>",
"_____no_output_____"
],
[
"On part d'un tableau $A[1..n]$. L'idée est de:\n1. **sélectionner** le *minimum* de $A[1..n]$ puis l'*échanger* avec $A[1]$,\n2. recommencer la même chose avec le sous-tableau $A[2..n]$: sélectionner son minimum et l'échanger avec $A[2]$,\n3. recommencer avec $A[3..n]$ ... \n\nVoici un exemple pour une itération:",
"_____no_output_____"
],
[
"<img src=\"attachment:exemple_tri_selection.png\" />",
"_____no_output_____"
],
[
"### «Pseudo-code» de `Tri-Selection`\n\n**Donnée** un tableau $A$ non vide d'entier.\n\n**Problème**: modifier l'ordre de ses éléments de façon que $A$ soit trié dans l'ordre croissant.\n\n<pre><strong>Tri-Selection</strong>(A):\n n ← longueur(A)\n <strong>Pour</strong> i de 1 à n-1:\n imin ← <strong>Minimum</strong>(A,i,n) ⊳ Renvoie l'index du minimum de A[i..n]\n échanger A[i] et A[imin]\n</pre>",
"_____no_output_____"
],
[
"#### Exercice 1\n\n1. Détailler les différentes étapes de l'algorithme sur le tableau $A = 3,4,1,7,2$\n\n2. L'algorithme est-il encore correct si on remplace `n-1` par `n` dans la boucle Pour?\n__________",
"_____no_output_____"
],
[
"**1.**\n \n itération 1: A = 1, 4, 3, 7, 2\n itération 2: A = 1, 2, 3, 7, 4\n itération 3: A = 1, 2, 3, 7, 4\n itération 4: A = 1, 2, 3, 4, 7\n\n**2.** **Oui**: dans ce cas, à la dernière itération, on cherche le minimum d'un sous-tableau de taille 1. Comme la réponse est clairement que son minimum est son seul élément et qu'ensuite on l'échange avec lui-même, on s'aperçoit que cette étape, bien que correcte, est inutile.",
"_____no_output_____"
],
[
"#### Exercice 2\n\nRéécrire cet algorithme en remplaçant `Minimum` par le code correspondant puis l'implémenter en Python. Vérifier alors son bon fonctionnement avec:\n\n```python\ntab = [5, 12, 20, 36, 25, 22, 23]\ntri_insertion(tab)\nassert tab == [5, 12, 20, 22, 23, 25, 36]\n```\n____________",
"_____no_output_____"
],
[
"Il suffit d'utiliser l'algorithme de recherche du minimum et de l'adapter au contexte:\n\n<pre><strong>Tri-Selection</strong>(A):\n n ← longueur(A)\n <strong>Pour</strong> i de 1 à n-1:\n ⊳ début recherche du min\n imin ← i\n <strong>Pour</strong> j de i+1 à n:\n <strong>Si</strong> A[j] < A[imin]:\n imin ← j\n ⊳ fin recherche min\n échanger A[i] et A[imin]\n</pre>\n\n",
"_____no_output_____"
]
],
[
[
"def tri_insertion(A):\n \"tri le tableau A en place (ne renvoie rien) dans l'ordre croissant\"\n n = len(A)\n for i in range(n-1):\n imin = i\n for j in range(i+1, n):\n if A[j] < A[imin]:\n imin = j\n # échange dans le style de python\n A[imin], A[i] = A[i], A[imin]\n # échange dans le cas général:\n # tmp = A[imin]; A[imin] = A[i]; A[i] = tmp \n \ntab = [5, 12, 20, 36, 25, 22, 23]\ntri_insertion(tab)\nassert tab == [5, 12, 20, 22, 23, 25, 36]\n# Pourquoi «assert tri_insertion(tab) == [5, 12, 20, 22, 23, 25, 36]» ne fonctionnerait pas?",
"_____no_output_____"
]
],
[
[
"### Efficacité\n\nÀ la première itération, la recherche du minimum à un coût proportionnel à la taille du tableau $A[1..n]$ soit $n$.\n\nÀ la seconde, la recherche du minimum a lieu dans $A[2..n]$ et aura un coût proportionnel à $n-1$, etc.\n\nAinsi, on peut estimer que le coût total en temps est proportionnel à:\n\n$$n+(n-1)+\\cdots+1=\\dfrac{(n+1)n}{2}=\\dfrac{n^2}{2}+\\dfrac{n}{2}$$\n\nMais, lorsque $n$ est «grand», $n^2$ sera beaucoup, beaucoup plus grand que $n$. Par exemple si $n$ vaut mille alors $n^2$ vaut 1 million. Pour cette raison, on néglige $n$ dans le coût qui devient:$$\\dfrac{n^2}{2}$$\n\nMais $\\frac{1}{2}$ est un facteur constant, on peut donc dire que:\n> le coût de cet algorithme (pour $n$ grand) est proportionnel à $n^2$ ce qu'on note $O(n^2)$.",
"_____no_output_____"
],
[
"<a href=\"https://www.toptal.com/developers/sorting-algorithms/selection-sort\">Visualiser le tri par **sélection** \\[*Selection sort*\\]</a>",
"_____no_output_____"
],
[
"#### Exercice 3\n\nEn supposant que le **tri par sélection** prenne 2 secondes sur un tableau de longueur 10000, estimer le temps qu'il lui faudrait pour trier un tableau de longueur 1 million.\n__________",
"_____no_output_____"
],
[
"$O(n^2)$ signifie que le temps $T$ est proportionnel au carré de la taille du tableau $n$ donc\n$$T=k\\cdot n^2 \\qquad \\text{où }k\\text{ est le coefficient de proportionnalité}$$\n\nCherchons le coefficient de proportionnalité $k$: $$k\\cdot 10000^2=2s\\implies k=\\dfrac{2}{(10^4)^2}=\\dfrac{2}{10^8}$$\n\nAppliquons $T=k\\cdot n^2$ pour $n=10^6$ (un million): $$T=\\dfrac{2}{10^8}\\times (10^{6})^2=2\\times \\dfrac{10^{12}}{10^{8}}=2\\times 10^4=20000$$\n\nDonc il faudra $20000$ secondes soit **5 heures 33 minutes et 20 secondes**.\n\n**Note**: Observer que le calcul est similaire à «une règle de trois» en utilisant la colonne des carrés de $n$:\n\n$$\\begin{array}{c|c|c}\n\\text{temps}&n&n^2\\cr\n\\hline\n\\color{red}{2}&10^4&\\color{red}{10^8}\\cr\n\\hline\n\\color{red}{T}&10^6&\\color{red}{10^{12}}\n\\end{array}\\implies T=\\dfrac{2\\times 10^{12}}{10^8}$$\n\nÀ titre indicatif, voici le calcul des heures, minutes et secondes en python:",
"_____no_output_____"
]
],
[
[
"# Calcule du temps avec Python\nn1, n2 = 10000, 1000000\nm = 60 # en sec.\nh = 60 * m # en sec.\n\nT = 2 * n2**2 / n1**2\nT = int(T) # car la division donne un flottant.\nheure = T // h\nminute = (T % h) // m\nseconde = T % m \nprint(f\"{heure}h{minute}m{seconde}s\")",
"_____no_output_____"
]
],
[
[
"### Invariant de boucle et correction de l'algorithme",
"_____no_output_____"
],
[
"Observer qu'avec cette stratégie, au début d'une itération d'indice $i$:\n> le sous-tableau $A[1..(i-1)]$ est trié et tous ses éléments sont inférieurs ou égaux à ceux de $A[i..n]$.\n\n<img src=\"attachment:tri_selection.png\" />",
"_____no_output_____"
],
[
"**Initialisation**: $i=1$ et donc $A[1..(i-1)]=A[1..0]=\\emptyset$ est un sous-tableau vide: il est donc trivialement trié et tous ses éléments - *vu qu'il n'en a pas* - sont sont plus petits ou égaux à ceux $A[1..n]$ (car la condition est vide!).",
"_____no_output_____"
],
[
"**D'une itération à la suivante**: Si $i$ est l'indice de l'itération, l'hypothèse est que $A[1..(i-1)]$ est trié et tous ses éléments sont inférieurs ou égaux à ceux de $A[i..n]$.\n\nAprès les deux instructions de la boucle, $A[i]$ est inférieur ou égal à tous les éléments de $A[(i+1)..n]$ et il est aussi supérieur à tous ceux de $A[1..(i-1)]$ d'après l'hypothèse puisqu'il fait partie de $A[i..n]$. Donc $A[1..i]$ est clairement trié et tous ses éléments sont inférieures à ceux de $A[(i+1)..n]$ ce qui prouve l'invariant de boucle.",
"_____no_output_____"
],
[
"**Correction**: après la dernière itération, on a $i=(n-1)+1=n$ donc $A[1..(i-1)]=A[1..(n-1)]$ est trié et tous ses élément sont inférieurs à *celui* de $A[n..n]$ ce qui signifie clairement que $A[1..n]$ est trié!",
"_____no_output_____"
],
[
"## Tri par insertion",
"_____no_output_____"
],
[
"<a href=\"https://vimeo.com/430326154\">Vidéo d'accompagnement</a>",
"_____no_output_____"
],
[
"Ce tri s'inspire de celui qu'utilise un joueur de cartes: Dans sa main gauche il tient les cartes qu'il a déjà triées. Il en prend une nouvelle et la compare de la droite vers la gauche avec celles qui sont déjà triées, puis il insère cette carte au moment où il tombe sur une carte plus petite: il la place juste après elle et cela a pour effet de décaler toutes les cartes supérieures d'une position vers la droite.\n\n<img src=\"https://external-content.duckduckgo.com/iu/?u=https%3A%2F%2Finterstices.info%2Fupload%2Fcomplexite-algo%2Ftri_par_insertion.jpg&f=1&nofb=1\" />",
"_____no_output_____"
],
[
"**Supposons** disposer d'un sous-algorithme <code>Inserer(A,i)</code> qui, étant donné un tableau $A$ dont on suppose que $A[1..(i-1)]$ est trié (carte de la main gauche 4-5-10), insère - comme le joueur ci-dessus - l'élément $A[i]$ (carte 7) dans le sous-tableau $A[1..i]$ de façon qu'il soit trié.",
"_____no_output_____"
],
[
"#### «Pseudo code» de `Tri-Insertion`\n\n**Donnée** un tableau $A$ non vide d'entier.\n\n**Problème**: modifier l'ordre de ses éléments de façon que $A$ soit trié dans l'ordre croissant.\n\n<pre><strong>Tri-Insertion</strong>(A):\n <strong>Pour</strong> i de 2 à n:\n <strong>Inserer</strong>(A,i)\n</pre>",
"_____no_output_____"
],
[
"Simple ... non? Bien sûr, il faut encore préciser l'algorithme de <code>Inserer</code>... Nous le ferons un peu après.",
"_____no_output_____"
],
[
"#### Exercice 4\n\nDétailler les différentes étapes de `Tri-Insertion` sur le tableau $A = 3, 4, 1, 7, 2$.\n_________",
"_____no_output_____"
],
[
" tri insertion tri sélection\n ----------------- -----------------\n itération 1: A = 3, 4, 1, 7, 2 A = 1, 4, 3, 7, 2\n itération 2: A = 1, 3, 4, 7, 2 A = 1, 2, 3, 7, 4\n itération 3: A = 1, 3, 4, 7, 2 A = 1, 2, 3, 7, 4\n itération 4: A = 1, 2, 3, 4, 7 A = 1, 2, 3, 4, 7 ",
"_____no_output_____"
],
[
"La boucle **Pour** a l'invariant:\n\n> au début de l'itération $i$, le tableau $A[1..(i-1)]$ est trié.",
"_____no_output_____"
],
[
"#### Exercice 5\n\n1. En admettant l'invariant de boucle, prouver que l'algorithme est correct.\n\n2. Prouver cet invariant en deux étapes: \n - Il est vrai au début de la première itération - **initialisation**\n - S'il est vrai au début d'une itération, il est vrai au début de la suivante - **D'une itération à la suivante**\n____",
"_____no_output_____"
],
[
"**1. Correction de l'algorithme**: à la fin de la dernière itération $i=n+1$ (raison pour laquelle la boucle s'arrête!) et, d'après l'invariant de boucle, $A[1..(i-1)]=A[1..n]$ est trié: c'est précisément ce qu'on voulait.",
"_____no_output_____"
],
[
"**Initialisation**: Au début de la première itération, $i=2$ et le sous-tableau $A[1..(i-1)]=A[1..1]$ ne contient qu'un élément: il est donc «bêtement» trié!\n\n**D'une itération à la suivante**: nous supposons que $A[1..(i-1)]$ est trié ce qui est précisément la condition de bon fonctionnement de `Inserer`. Une fois qu'`Inserer` a fait son travail - on est au début de l'itération suivante - $A[1..i]$ est trié.\n\nCela démontre l'invariant de boucle.",
"_____no_output_____"
],
[
"### Sous-algorithme `Inserer`",
"_____no_output_____"
],
[
"Penser aux cartes à jouer qui représentent les éléments du tableau $A$: \n1. vous prenez $A[i]$, la carte n° $i$; les cartes $A[1],A[2],..,A[i-1]$ sont déjà triées,\n2. vous la comparez avec celle située le plus à droite $A[i-1]$ des cartes déjà triées,\n3. **si** $A[i]$ est plus grande, vous la placer tout à droite: donc elle conserve le numéro $i$,\n4. **sinon**: elle doit s'insérer parmi les cartes $A[1], A[2], ...,A[i-2]$ et la carte $A[i-1]$ devient la carte n°$i$ (pour laisser de la place) ... vous **recommencer donc à l'étape 2**. avec $A[i-2]$ cette fois...",
"_____no_output_____"
],
[
"Voici un exemple concret pour comprendre la stratégie de cet algorithme.\n\n<p style=\"text-align:center\"><img src=\"attachment:exemple_insertion.png\"/></p>",
"_____no_output_____"
],
[
"#### Exercice 6\n\nIllustrer simplement les étapes de l'insertion «à gauche» du dernier élément de $A = 1, 4, 5, 9, 2$.\n_____",
"_____no_output_____"
],
[
"<pre>début: 2 est mémorisé \nitération 1: A = 1, 4, 5, <span style=\"color: red\"/>9</span>, 9\nitération 2: A = 1, 4, <span style=\"color: red\"/>5</span>, 5, 9\nitération 3: A = 1, <span style=\"color: red\"/>4</span>, 4, 5, 9\nfin -> 2 remplace le nombre en rouge.\n</pre>",
"_____no_output_____"
],
[
"### «Pseudo code» du sous-algorithme `Inserer`",
"_____no_output_____"
],
[
"*Précondition*: $A[1..(i-1)]$ est trié dans l'ordre croissant et $A[i]$ existe.\n\n*Résultat*: le sous-tableau $A[1..i]$ est trié.\n\n<pre><strong>Inserer</strong>(A, i):\n1 x ← A[i]\n2 j ← i - 1\n3 <strong>Tant que</strong> j > 0 <strong>et</strong> A[j] > x:\n4 A[j+1] ← A[j]\n5 j ← j - 1\n ⊳ Fin Tant Que\n6 A[j+1] ← x\n</pre>",
"_____no_output_____"
],
[
"Hum ... ça paraît moins simple que l'insertion d'une carte ... un ordinateur est comme un tout «petit enfant»: il faut tout lui expliquer. Voici une vue d'ensemble sur deux cas possibles:\n\n<img src=\"attachment:insertion_vue_globale.png\" />",
"_____no_output_____"
],
[
"**Note**: Observer que $j$ n'est pas la position d'insertion mais celle de l'élément qu'on compare à $x$. Lorsque cet élément est plus petit que $x$, la boucle s'arrête et on l'insère à sa droite c'est-à-dire à la position $j+1$.",
"_____no_output_____"
],
[
"#### Exercice 7\n\nImplémenter le **tri par insertion** en Python puis tester le sur `tab = [3, 4, 1, 7, 2]`.\n____",
"_____no_output_____"
]
],
[
[
"def tri_insertion(A):\n n = len(A)\n for i in range(1, n):\n x = A[i]\n j = i - 1\n while j >= 0 and A[j] > x: # ou j > -1 ...\n A[j+1] = A[j]\n j -= 1\n A[j+1] = x\n \ntab = [3, 4, 1, 7, 2]\ntri_insertion(tab)\nassert tab == [1, 2, 3, 4, 7]",
"_____no_output_____"
]
],
[
[
"### Efficacité\n\nLe **cas le plus défavorable** se produit lorsque $A$ est trié dans l'ordre *décroissant* (cas où la «carte» à insérer est toujours plus petite que toutes celles déjà triées). En effet, dans ce cas la boucle d'insertion s'arrête lorsque la condition de boucle - $j>0$ - devient fausse (revoir le deuxième cas de la vue d'ensemble de l'algorithme): le nombre d'itération de la boucle est alors $i$ (variable de la boucle externe **Pour**)\n\nAinsi, pour chaque itération de la boucle **Pour** de variable $i$ , `inserer` produit $i$ itérations. Cela donne un coût total: $$2+3+\\cdots + n\\approx \\dfrac{n(n+1)}{2}$$\n\nC'est la même formule que pour le tri par sélection et nous savons déjà que cela signifie que l'algorithme est $O(n^2)$:\n> Dans **le cas le plus défavorable**, le temps d'exécution du **tri par insertion** est proportionnel au carré de la taille $n$ du tableau d'entrée.",
"_____no_output_____"
],
[
"<a href=\"https://www.toptal.com/developers/sorting-algorithms/insertion-sort\">Visualiser le tri par **insertion** \\[*Insertion sort*\\]</a>",
"_____no_output_____"
],
[
"### Invariant de boucle et correction\n\nLa boucle de `Inserer` a l'invariant suivant:\n> $A_g=A[1..j]$ et $A_d=A[(j+2)..i]$ sont triés et $A_g\\leqslant A_d$ (tous les éléments de $A_g$ sont inférieurs ou égaux à tous ceux de $A_d$) et $x<A_d$ (...).",
"_____no_output_____"
],
[
"<img src=\"attachment:invariant_inserer.png\"/>",
"_____no_output_____"
],
[
"Cet invariant n'est pas très agréable du fait qu'il contient quatre conditions. Supposons pour l'instant qu'il est vrai et montrons comment il permet de prouver la correction de cet algorithme.",
"_____no_output_____"
],
[
"**Preuve de la correction**: Si la boucle se termine, c'est que soit $j\\leqslant 0$ soit $A[j]\\leqslant x$:\n- $\\underline{j\\leqslant 0}$: ce n'est possible que si $j=0$ car $j$ est décrémenté d'une unité à chaque itération. L'invariant de boucle donne ici $A[(j+2)..i]=A[2..i]$ est trié et $x<A[2..i]$. Alors l'affectation (ligne 6) $A[j+1]=A[1]\\leftarrow x$ nous assure que $A[1..i]$ est trié.\n\n- $\\underline{A[j]\\leqslant x}$: l'invariant de boucle donne $A[1..j]$ et $A[(j+2)..i]$ sont triés et:\n$$A[1..j]\\leqslant A[j] \\leqslant x<A[(j+2)..i]$$\n \n Clairement, l'affectation $A[j+1]\\leftarrow x$ entraîne encore que $A[1..i]$ est trié.\n___",
"_____no_output_____"
],
[
"Reste à prouver que l'invariant de boucle lui-même... Vous pouvez l'admettre ou travailler la démonstration (un peu délicate) donnée ci-après pour les plus courageux.",
"_____no_output_____"
],
[
"**Initialisation**: au démarrage $j=i-1$ et $A[1..j]=A[1..(i-1)]=A_g$ et $A[(j+2)..i]=A[(i+1)..i]=A_d=\\emptyset$ sont triés: le premier car c'est la «pré-condition» de fonctionnement de `Inserer` et le second parcequ'il est vide. Du fait que $A_d$ est vide, les autres conditions sont satisfaites car il n'y a rien à satisfaire...\n\n**D'une itération à la suivante**: Donc on suppose que l'invariant de boucle est vrai au début de l'itération courante. On a donc \n$$j>0\\qquad \\text{et}\\qquad A[j] > x\\qquad \\text{(condition de boucle)}$$. \nOn doit vérifier que, *après que les lignes 4 et 5 ont été exécutées* (donc $j$ a été remplacé par $j-1$): \n\n$$A'_g=A[1..(j-1)]\\text{ et }A'_d=A[(j+1)..i]\\text{ sont triés et}\\\\ A'_g\\leqslant A'_d\\text{ et } x<A'_d$$\n\nAvant de démontrer les quatres affirmations de l'invariant de boucle, il est bon de noter quelques points:\n- $A[j+1]=A[j]$ d'après la ligne 4.\n- $A'_d=A[j+1]+A_d$ (concaténation) et donc $A'_d=A[j]+A_d$ d'après le point précédent.\n\nDémonstration des quatres affirmations de l'invariant de boucle:\n\n1. $A'_g=A[1..(j-1)]$ **est trié** soit parce qu'il est vide ($j=1$) soit comme sous-tableau de $A_g=A'_g+A[j-1]$ qui est trié par hypothèse.\n\n2. $A'_d=A[j]+A_d$ **est trié**. En effet, par hypothèse, $A_d$ est trié et $A[j]\\in A_g\\leqslant A_d$ par hypothèse.\n\n3. $A'_g\\leqslant A'_d$: En effet, du fait que $A_g=A'_g+A[j]$:\n$$\\left\\{\\begin{array}{l}A_g \\text{ trié}\\implies A'_g\\leqslant A[j]\\cr\nA_g \\leqslant A_d \\implies A[j]\\leqslant A_d \n\\end{array}\\right.\\implies A'_g\\leqslant A'_d=A[j]+A_d$$\n\n4. Comme $x<A_d$ (hyp.) et que $x<A[j]$ (condition de boucle) alors $x<A'_d$ puisque $A'_d=A[j]+A_d$.\n\nL'invariant de boucle est donc prouvé (ouf!)",
"_____no_output_____"
],
[
"## Comparaison des tris par sélection et insertion",
"_____no_output_____"
],
[
"<a href=\"https://vimeo.com/430327363\">Vidéo d'accompagnement</a>",
"_____no_output_____"
],
[
"Même si les deux algorithmes sont $O(n^2)$, l'insertion est plus économe que la séléction. En effet:\n\n**Séléction**: Trouver le minimum nécessite de parcourir *systématiquement* tout le sous-tableau droit (même s'il est déjà trié!): dans tous les cas, la boucle de parcours va jusqu'au bout!\n\n**Insertion**: La boucle d'insertion s'arrête dès que la position d'insertion est trouvée. très souvent, cela se produit bien avant d'avoir parcouru tout le sous-tableau. À l'extrême, si le tableau est déjà trié dans l'ordre croissant, la boucle d'insertion ne produit aucune itération.\n\n> En pratique, **le tri par insertion est plus rapide que le tri par sélection**.",
"_____no_output_____"
],
[
"**Ce sont des tris «en place»**: Il faut observer que ces deux algorithmes ne **renvoient rien**: ils modifient directement le tableau fourni en argument - on dit qu'ils **trient «sur place»**. En python, la méthode de liste `list.sort()` trie sur place, tandis que la fonction prédéfinie `sorted(<liste>)` *renvoie une nouvelle liste triée* sans modifier celle qui est fournie en argument.",
"_____no_output_____"
],
[
"#### Peut-on faire mieux? - O(n log n)",
"_____no_output_____"
],
[
"Nous n'avons fait qu'effleurer le thème des algorithmes de tri; il en existe beaucoup d'autres: \n> **tri bulle** (voir exercice 8) \\[ *bubble sort* \\], **tri par tas** \\[ *heap sort* \\], **tri fusion** \\[ *merge sort* \\], **tri «rapide»** \\[ *quicksort* \\]... pour n'en citer que quelques uns.",
"_____no_output_____"
],
[
"<a href=\"https://www.toptal.com/developers/sorting-algorithms\">Visualiser le fonctionnement de ces algorithmes</a>",
"_____no_output_____"
],
[
"Certains d'entre eux ont **une efficacité en $O(n\\log n)$** - ceux utilisés par les fonctions «sort» de python par exemple. $O(n\\log n)$ signifie que:\n\n> leur temps d'exécution est proportionnel à $n\\log n$ où $n$ est la taille de l'entrée.\n\n**Rappel**: $\\log n$ est approximativement égal à l'exposant $m$ de la puissance de $2$ voisine de $n$: \n$$\\text{si }n=2^m\\text{ alors }\\log n=m \\qquad(\\text{ ou }\\log 2^m=m)$$",
"_____no_output_____"
],
[
"Pour parvenir à cela, ils utilisent d'autres stratégies et notamment une technique appelée **récursivité** que vous découvrirez en Terminale.",
"_____no_output_____"
],
[
"#### Comprendre la différence entre O(n log n) et O(n^2)\n\nReprenons l'exemple de l'*exercice 3*.\n\nSupposez qu'un algorithme en $O(n\\log n)$ mette 2s pour trier un tableau de taille $10000=10^4$, pour un tableau de taille 1 million $=10^6$, il mettra **moins de** (règle de trois):\n\n$$\\begin{eqnarray}2\\times \\dfrac{10^6\\log 10^6}{10^4\\log 10^4}&\\approx& 2\\times 10^2\\times \\dfrac{20}{14}\\cr &<& 200\\times 2=400~\\text{secondes}={\\bf 6\\text{m}40\\text{s}}\\end{eqnarray}$$",
"_____no_output_____"
],
[
"Pour mémoire, dans une situation similaire de l'exercice 3, avec un algorithme en $O(n^2)$, on obtenait un temps de **5 heures et demi** environ...\n\nOn peut faire le calcul précis avec Python qui montre qu'on a été «pessismiste» dans notre calcul:\n```python\nfrom math import log2\n2 * (10**6 * log2(10**6))/(10**4 * log2(10**4)) # --> 300s = 5min!\n```",
"_____no_output_____"
],
[
"Pour mémoire, **du plus rapide au moins rapide** (𝑛 la taille de l'entrée de l'algorithme): $$O(1) < O(\\log n) < O(n) < O(n\\log n) < O(n^2)$$\nDe plus, $O(\\log n)$ est plus proche de $O(1)$ que de $O(n)$. De même, $O(n\\log n)$ est plus proche de $O(n \\log n)$ que de $O(n^2)$.",
"_____no_output_____"
],
[
"#### Exercice 8 - tri «bulle»\n\nLe tri «bulle», à chaque itération $i$, parcours le sous-tableau $A[1..i]$ de gauche à droite et permute les éléments *consécutifs* s'ils ne sont pas dans l'ordre croissant. Il fait ce travail sur $A[1..n]$, puis sur $A[1..(n-1)]$, puis sur $A[1..(n-2)]$ etc. Voici un exemple de fonctionnement pour $A=1,4,3,2$:\n\n$$\\text{sur }A[1..4]:\\qquad\\underline{1,4},3,2~\\rightarrow~ 1,\\underline{\\color{red}{4},\\color{red}{3}},2\\rightarrow 1,3,\\underline{\\color{red}{4},\\color{red}{2}}~\\rightarrow~1,3,2,{\\bf 4}\\\\\n\\text{sur } A[1..3]:\\qquad\\underline{1,3},2,{\\bf 4}~\\rightarrow~1,\\underline{\\color{red}{3},\\color{red}{2}},{\\bf 4}~\\rightarrow~1,2,{\\bf 3}, {\\bf 4}\\\\\n\\text{sur } A[1..2]:\\qquad \\underline{1,2},{\\bf 3}, {\\bf 4}~\\rightarrow~1,{\\bf 2},{\\bf 3},{\\bf 4}$$\n\n1. Implémenter le tri bulle en Python.\n\n2. Évaluer son efficacité.\n________",
"_____no_output_____"
]
],
[
[
"def tri_bulle(A):\n n = len(A)\n while n > 0:\n for j in range(n-1):\n if A[j] > A[j+1]:\n A[j], A[j+1] = A[j+1], A[j]\n n -= 1\n\nfrom random import randint\ntest = [randint(1,100) for i in range(10)]\nprint(test)\ntri_bulle(test)\nprint(test)",
"_____no_output_____"
]
],
[
[
"Son efficacité est $O(n^2)$ car la boucle externe parcourt (presque) toutes les positions $i$ du tableau en entrée et la boucle interne toutes celles du sous-tableau $A[1..i]$ comme pour le tri par sélection.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eccba3f4b46cb7d7ea66721b5450ede65d5cc2f5 | 137,965 | ipynb | Jupyter Notebook | Pre-processing/GZ1/Galaxy_zoo_datas_pre-processing.ipynb | MattiasEyh/Galaxy-Classification-CNN | c4d7de2be679ebad8e3725b36a22bb580d748678 | [
"MIT"
] | 2 | 2021-12-07T15:58:27.000Z | 2022-03-03T19:57:53.000Z | Pre-processing/GZ1/Galaxy_zoo_datas_pre-processing.ipynb | MattiasEyh/Galaxy-Classification-CNN | c4d7de2be679ebad8e3725b36a22bb580d748678 | [
"MIT"
] | null | null | null | Pre-processing/GZ1/Galaxy_zoo_datas_pre-processing.ipynb | MattiasEyh/Galaxy-Classification-CNN | c4d7de2be679ebad8e3725b36a22bb580d748678 | [
"MIT"
] | null | null | null | 212.580894 | 25,939 | 0.857413 | [
[
[
"# Clean and explore galaxy zoo datasets\nThis file details the data cleansing of the Galaxy Zoo dataset and explains the correlations found between the datasets features.\nMattias EYHERABIDE, Théo MOREL",
"_____no_output_____"
],
[
" <img src=\"../Pics/AM0002.png\" width=\"900\"> </br>\nPicture of AM0002 captured by Hubble telescope",
"_____no_output_____"
],
[
"## Abstract\n\n### Galaxy zoo\n...\n\n### Galaxies classification\n...",
"_____no_output_____"
],
[
"## Pre-processing\n....\n### Library imports",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"### Basic analysis",
"_____no_output_____"
]
],
[
[
"from IPython.core.display import display\n\ndef checkIfHasRowIncompatible(dataset, rowName1, rowName2):\n return dataset[dataset[rowName1] == 1][dataset[rowName2] == 1].shape[0] > 0\n\ndef VisualiseDataset(dataset):\n\n print(\"En-tête du dataset :\\n----------\\n\")\n display(dataset.head())\n\n print(\"Informations des types du dataset :\\n----------\\n\")\n display(dataset.info())\n\n print(\"\\n----------\\nTaille du dataset :\")\n display(dataset.shape)\n\n print(\"Informations du dataset :\\n----------\\n\")\n display(dataset.describe())\n\n print(\"Pourcentage de valeurs manquantes :\\n----------\\n\")\n display((df.isna().sum()/df.shape[0]).sort_values())\n\n print(\"Vérification si valeurs multiple à 1 incompatible :\\n----------\\n\")\n print(\"A ligne Spirale == Elliptique : {}\\n\".format(checkIfHasRowIncompatible(df, \"SPIRAL\", \"ELLIPTICAL\")))\n print(\"A ligne Spirale == Incertaine : {}\\n\".format(checkIfHasRowIncompatible(df, \"SPIRAL\", \"UNCERTAIN\")))\n print(\"A ligne Elliptique == Incertaine : {}\\n\".format(checkIfHasRowIncompatible(df, \"ELLIPTICAL\", \"UNCERTAIN\")))\n\n print(\"Type des valeurs :\\n----------\\n\")\n display(df.dtypes.value_counts().plot.pie())",
"_____no_output_____"
],
[
"data = pd.read_csv(\"../../Datas/GalaxyZoo1_DR_table2.csv\")\ndf = data.copy()\nVisualiseDataset(df)",
"En-tête du dataset :\n----------\n\n"
]
],
[
[
"'### Basic checklist\n- **Targets variables** : Spiral, Elliptical and Uncertain\n- **lines and columns** : (667944, 16)\n- **Type de variables** : Majortiée de numérique, le reste sont objet (ici des dates)\n- **Analyse des valeurs manquantes** : Aucunes valeurs manquantes",
"_____no_output_____"
]
],
[
[
"dfbiased = df.copy()\ndfbiased.drop(['OBJID', 'P_CS', 'P_EL', 'P_CW', 'P_MG', 'P_ACW', 'P_EDGE', 'RA', 'DEC', 'ELLIPTICAL', 'SPIRAL', 'UNCERTAIN'], axis=1, inplace=True)\nsns.histplot(dfbiased['NVOTE'])\n\nfor col in dfbiased.select_dtypes('double'):\n plt.figure()\n sns.distplot(df[col])\n",
"_____no_output_____"
]
],
[
[
"Supression des valeurs ou le nombre de vote est inférieur à 50.",
"_____no_output_____"
]
],
[
[
"shapeBase = len(df.index)\ndf = df[df.NVOTE >= 15]\ndf = df[df.P_DK > 0.05]\nshapeAfterModif = len(df.index)\n\nprint(\"Nombre de ligne suprimées : \")\nprint(shapeBase - shapeAfterModif)\n\ndf.shape",
"Nombre de ligne suprimées : \n327417\n"
],
[
"correlationDf = df.copy()\ncorrelationDf.drop(['OBJID', 'NVOTE', 'DEC', 'RA', 'SPIRAL', 'UNCERTAIN', 'ELLIPTICAL'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"sns.clustermap(correlationDf.corr())",
"_____no_output_____"
]
],
[
[
"Création d'un dataset final",
"_____no_output_____"
]
],
[
[
"print(\"Nombre de galaxies ayant le statut MERGED :\", df[\"P_MG\"][df[\"P_MG\"] > 0.4].count())",
"Nombre de galaxies ayant le statut MERGED : 6609\n"
],
[
"dfWithPreProcess = df.copy()\ndfWithPreProcess.drop(['NVOTE', 'P_EL', 'P_CW', 'P_ACW', 'P_EDGE', 'P_DK', 'P_CS', 'P_EL_DEBIASED', 'P_CS_DEBIASED'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"dfWithPreProcess['MERGED'] = np.where(df['P_MG'] > 0.5, 1, 0)\ndfWithPreProcess.head()",
"_____no_output_____"
],
[
"#Merge the three columns by using their name as type in the column \"TYPE\".\ncolumns = [\"MERGED\", \"SPIRAL\", \"ELLIPTICAL\", \"UNCERTAIN\"]\nfor name in columns: dfWithPreProcess.loc[dfWithPreProcess[name]==1, 'TYPE'] = name\n#Drop all columns not needed and each rows with type \"UNCERTAIN\"\ndfWithPreProcess.drop(index=dfWithPreProcess[dfWithPreProcess['TYPE'] == \"UNCERTAIN\"].index,\n columns=['P_MG', 'UNCERTAIN', \"MERGED\", \"ELLIPTICAL\", \"SPIRAL\"], inplace=True)\ndfWithPreProcess.to_csv('../Datas/dataWithPreProcess.csv', index=False)\ndfWithPreProcess.head()",
"_____no_output_____"
],
[
"dfShow = dfWithPreProcess[dfWithPreProcess[\"TYPE\"] == \"MERGED\"]\ndfShow.head()",
"_____no_output_____"
],
[
"dfShow = dfWithPreProcess[dfWithPreProcess[\"TYPE\"] == \"ELLIPTICAL\"]\ndfShow.head()",
"_____no_output_____"
],
[
"dfShow = dfWithPreProcess[dfWithPreProcess[\"TYPE\"] == \"SPIRAL\"]\ndfShow.head()",
"_____no_output_____"
],
[
"dfShow = dfWithPreProcess[dfWithPreProcess[\"TYPE\"] == \"UNKNOWN\"]\ndfShow.head()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccbe9973e97d22055cef862a55babc7ee00ea15 | 13,015 | ipynb | Jupyter Notebook | examples/.ipynb_checkpoints/contributor-classification-checkpoint.ipynb | b-hodges/augur | 9f1fbee96afaf8f9e7fa3a7c830f969b955482ef | [
"MIT"
] | null | null | null | examples/.ipynb_checkpoints/contributor-classification-checkpoint.ipynb | b-hodges/augur | 9f1fbee96afaf8f9e7fa3a7c830f969b955482ef | [
"MIT"
] | null | null | null | examples/.ipynb_checkpoints/contributor-classification-checkpoint.ipynb | b-hodges/augur | 9f1fbee96afaf8f9e7fa3a7c830f969b955482ef | [
"MIT"
] | null | null | null | 114.166667 | 1,369 | 0.677295 | [
[
[
"import ..ghdata\nimport pandas as pd\nimport numpy as np\n\nghtorrent = ghdata.GHTorrent(\"mysql+pymysql://msr:[email protected]/msr\")",
"_____no_output_____"
],
[
"cakephpid = ghtorrent.repoid(owner=\"cakephp\", repo=\"cakephp\")\nclassified_contributors = ghtorrent.classify_contributors(cakephpid)\nprint(classified_contributors)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
eccbec9f41addf5ceb11087a7b8a735600520156 | 44,003 | ipynb | Jupyter Notebook | examples/discretisation/ArbitraryDiscretiser_plus_MeanEncoder.ipynb | pradumna123/feature_engine | ebaa2cefa0bfdcb8b0eeafb88dd5d7b432ba8495 | [
"BSD-3-Clause"
] | 1 | 2022-03-13T11:40:59.000Z | 2022-03-13T11:40:59.000Z | examples/discretisation/ArbitraryDiscretiser_plus_MeanEncoder.ipynb | pradumna123/feature_engine | ebaa2cefa0bfdcb8b0eeafb88dd5d7b432ba8495 | [
"BSD-3-Clause"
] | 4 | 2021-09-13T07:22:51.000Z | 2021-12-28T12:18:05.000Z | examples/discretisation/ArbitraryDiscretiser_plus_MeanEncoder.ipynb | TremaMiguel/feature_engine | 117ea3061ec9cf65f9d012aff4875d2b88e8cf71 | [
"BSD-3-Clause"
] | 5 | 2021-09-10T15:34:08.000Z | 2022-03-06T07:28:07.000Z | 82.712406 | 17,456 | 0.779197 | [
[
[
"# ArbitraryDiscretiser + MeanEncoder\n\nThis is very useful for linear models, because by using discretisation + a monotonic encoding, we create monotonic variables with the target, from those that before were not originally. And this tends to help improve the performance of the linear model. ",
"_____no_output_____"
],
[
"## ArbitraryDiscretiser\n\nThe ArbitraryDiscretiser() divides continuous numerical variables into contiguous intervals arbitrarily defined by the user.\n\nThe user needs to enter a dictionary with variable names as keys, and a list of the limits of the intervals as values. For example {'var1': [0, 10, 100, 1000],'var2': [5, 10, 15, 20]}.\n\n<b>Note:</b> Check out the ArbitraryDiscretiser notebook to learn more about this transformer.",
"_____no_output_____"
],
[
"## MeanEncoder\n\nThe MeanEncoder() replaces the labels of the variables by the mean value of the target for that label. <br>For example, in the variable colour, if the mean value of the binary target is 0.5 for the label blue, then blue is replaced by 0.5\n\n<b>Note:</b> Read MeanEncoder notebook to know more about this transformer",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.pipeline import Pipeline\n\nfrom feature_engine.discretisation import ArbitraryDiscretiser\nfrom feature_engine.encoding import MeanEncoder\n\nplt.rcParams[\"figure.figsize\"] = [15,5]",
"_____no_output_____"
],
[
"# Load titanic dataset from OpenML\n\ndef load_titanic():\n data = pd.read_csv('https://www.openml.org/data/get_csv/16826755/phpMYEkMl')\n data = data.replace('?', np.nan)\n data['cabin'] = data['cabin'].astype(str).str[0]\n data['pclass'] = data['pclass'].astype('O')\n data['age'] = data['age'].astype('float').fillna(data.age.median())\n data['fare'] = data['fare'].astype('float').fillna(data.fare.median())\n data['embarked'].fillna('C', inplace=True)\n data.drop(labels=['boat', 'body', 'home.dest', 'name', 'ticket'], axis=1, inplace=True)\n return data",
"_____no_output_____"
],
[
"data = load_titanic()\ndata.head()",
"_____no_output_____"
],
[
"# let's separate into training and testing set\nX = data.drop(['survived'], axis=1)\ny = data.survived\n\nX_train, X_test, y_train, y_test = train_test_split(\n X, y, test_size=0.3, random_state=0)\n\nprint(\"X_train :\", X_train.shape)\nprint(\"X_test :\", X_test.shape)",
"X_train : (916, 8)\nX_test : (393, 8)\n"
],
[
"# we will transform two continuous variables\nX_train[[\"age\", 'fare']].hist(bins=30)\nplt.show()",
"_____no_output_____"
],
[
"# set up the discretiser\narb_disc = ArbitraryDiscretiser(\n binning_dict={'age': [0, 18, 30, 50, 100],\n 'fare': [-1, 20, 40, 60, 80, 600]},\n # returns values as categorical\n return_object=True)\n\n# set up the mean encoder\nmean_enc = MeanEncoder(variables=['age', 'fare'])\n\n# set up the pipeline\ntransformer = Pipeline(steps=[('ArbitraryDiscretiser', arb_disc),\n ('MeanEncoder', mean_enc),\n ])\n# train the pipeline\ntransformer.fit(X_train, y_train)",
"_____no_output_____"
],
[
"transformer.named_steps['ArbitraryDiscretiser'].binner_dict_",
"_____no_output_____"
],
[
"transformer.named_steps['MeanEncoder'].encoder_dict_",
"_____no_output_____"
],
[
"train_t = transformer.transform(X_train)\ntest_t = transformer.transform(X_test)\n\ntest_t.head()",
"_____no_output_____"
],
[
"# let's explore the monotonic relationship\nplt.figure(figsize=(7, 5))\npd.concat([test_t, y_test], axis=1).groupby(\"fare\")[\"survived\"].mean().plot()\nplt.title(\"Relationship between fare and target\")\nplt.xlabel(\"fare\")\nplt.ylabel(\"Mean of target\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can observe an almost linear relationship between the variable \"fare\" after the transformation and the target.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
eccbeca814bc950a0514593333c06b6b40810b90 | 317,875 | ipynb | Jupyter Notebook | opencv_camshift.ipynb | jihoon3327/openCV | 968a9233de6272ec514c9c26d1816731a7e9f0f1 | [
"MIT"
] | null | null | null | opencv_camshift.ipynb | jihoon3327/openCV | 968a9233de6272ec514c9c26d1816731a7e9f0f1 | [
"MIT"
] | null | null | null | opencv_camshift.ipynb | jihoon3327/openCV | 968a9233de6272ec514c9c26d1816731a7e9f0f1 | [
"MIT"
] | null | null | null | 2,464.147287 | 314,780 | 0.959711 | [
[
[
"## camshift\n#### \n#### 캠시프트란 : 추적하는 객체 그기가 변하더라도 검색 윈도우의 크키가 고정되어는 평균이동 알고리즘의 단점을 보안\n#### \n#### 캠시프트 동작 방법\n#### 1. 우선 평균 이동 알고리즘으로 이동위치 계산\n#### 2. 윈도우 크기를 조정\n#### 3. 특징 공간을 가장 잘 표현하는 타원 검출\n#### 4. 새로운 크기의 윈도우를 이용하여 다시 평균 이동 수행",
"_____no_output_____"
]
],
[
[
"import sys\nimport numpy as np\nimport cv2",
"_____no_output_____"
],
[
"#비디오 열기\n\ncap = cv2.VideoCapture(\"/Users/jungjihoon/Library/Mobile Documents/com~apple~CloudDocs/00_DataScience/민형기/camshift.avi\")\n\nif not cap.isOpened():\n print(\"video open failed\")\n sys.exit()\n \n#초기 사각형 영역 : (w, y, w, h)\nx, y, w, h = 135, 220, 100, 100\nrc = (x, y, w, h)\n\nret, frame = cap.read()\n\nif not ret:\n print('frame read failed')\n sys.exit()\n\n#관심영역 표기\nroi = frame[y:y+h, x:x+w]\nroi_hsv = cv2.cvtColor(roi, cv2.COLOR_BGR2HSV)\n\n# HS 히스토그램 계산\nchannels = [0, 1]\nranges = [0, 180, 0, 256]\nhist = cv2.calcHist([roi_hsv], channels, None, [90, 128], ranges)\n\n# Mean Shift 알고리즘 종료 기준 10번 반복할 동안 평균 1픽셀의 움직임이 없으면 종료\nterm_crit = (cv2.TERM_CRITERIA_EPS | cv2.TERM_CRITERIA_COUNT, 10, 1)\n\nwhile True:\n ret, frame = cap.read()\n \n if not ret:\n break\n \n # HS 히스토그램에 대한 역투영\n frame_hsv = cv2.cvtColor(frame, cv2.COLOR_BGR2HSV)\n backproj = cv2.calcBackProject([frame_hsv], channels, hist, ranges, 1)\n \n # Camshift\n ret, rc = cv2.CamShift(backproj, rc, term_crit)\n \n cv2.rectangle(frame, rc, (0,0,255), 2)\n cv2.ellipse(frame, ret, (0,255,0), 2)\n \n cv2.imshow('frame', frame)\n \n if cv2.waitKey(60) == 27:\n break\n \ncap.release()\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
eccbfaa918745adb74a003f518052c90aaa1f983 | 30,957 | ipynb | Jupyter Notebook | Python/03_Image_Details.ipynb | JenifferWuUCLA/pulmonary-nodules-SimpleITK | 19ca35f37195461ea9e8ce4ab1ddd2a5aec359be | [
"Apache-2.0"
] | 1 | 2018-01-12T08:03:53.000Z | 2018-01-12T08:03:53.000Z | Python/03_Image_Details.ipynb | JenifferWuUCLA/pulmonary-nodules-SimpleITK | 19ca35f37195461ea9e8ce4ab1ddd2a5aec359be | [
"Apache-2.0"
] | null | null | null | Python/03_Image_Details.ipynb | JenifferWuUCLA/pulmonary-nodules-SimpleITK | 19ca35f37195461ea9e8ce4ab1ddd2a5aec359be | [
"Apache-2.0"
] | 2 | 2018-04-13T07:47:12.000Z | 2021-03-17T02:41:17.000Z | 33.612378 | 439 | 0.600801 | [
[
[
"<h1 align=\"center\">Images</h1>\n\n<table width=\"100%\">\n<tr style=\"background-color: red;\"><td><font color=\"white\">SimpleITK conventions:</font></td></tr>\n<tr><td>\n<ul>\n<li>Image access is in x,y,z order, image.GetPixel(x,y,z) or image[x,y,z], with zero based indexing.</li>\n<li>If the output of an ITK filter has non-zero starting index, then the index will be set to 0, and the origin adjusted accordingly.</li>\n</ul>\n</td></tr>\n</table>\n\nThe unique feature of SimpleITK (derived from ITK) as a toolkit for image manipulation and analysis is that it views <b>images as physical objects occupying a bounded region in physical space</b>. In addition images can have different spacing between pixels along each axis, and the axes are not necessarily orthogonal. The following figure illustrates these concepts. \n\n<img src=\"ImageOriginAndSpacing.png\" style=\"width:700px\"/><br><br>\n\n\n### Pixel Types\n\nThe pixel type is represented as an enumerated type. The following is a table of the enumerated list.\n\n<table>\n <tr><td>sitkUInt8</td><td>Unsigned 8 bit integer</td></tr>\n <tr><td>sitkInt8</td><td>Signed 8 bit integer</td></tr>\n <tr><td>sitkUInt16</td><td>Unsigned 16 bit integer</td></tr>\n <tr><td>sitkInt16</td><td>Signed 16 bit integer</td></tr>\n <tr><td>sitkUInt32</td><td>Unsigned 32 bit integer</td></tr>\n <tr><td>sitkInt32</td><td>Signed 32 bit integer</td></tr>\n <tr><td>sitkUInt64</td><td>Unsigned 64 bit integer</td></tr>\n <tr><td>sitkInt64</td><td>Signed 64 bit integer</td></tr>\n <tr><td>sitkFloat32</td><td>32 bit float</td></tr>\n <tr><td>sitkFloat64</td><td>64 bit float</td></tr>\n <tr><td>sitkComplexFloat32</td><td>complex number of 32 bit float</td></tr>\n <tr><td>sitkComplexFloat64</td><td>complex number of 64 bit float</td></tr>\n <tr><td>sitkVectorUInt8</td><td>Multi-component of unsigned 8 bit integer</td></tr>\n <tr><td>sitkVectorInt8</td><td>Multi-component of signed 8 bit integer</td></tr>\n <tr><td>sitkVectorUInt16</td><td>Multi-component of unsigned 16 bit integer</td></tr>\n <tr><td>sitkVectorInt16</td><td>Multi-component of signed 16 bit integer</td></tr>\n <tr><td>sitkVectorUInt32</td><td>Multi-component of unsigned 32 bit integer</td></tr>\n <tr><td>sitkVectorInt32</td><td>Multi-component of signed 32 bit integer</td></tr>\n <tr><td>sitkVectorUInt64</td><td>Multi-component of unsigned 64 bit integer</td></tr>\n <tr><td>sitkVectorInt64</td><td>Multi-component of signed 64 bit integer</td></tr>\n <tr><td>sitkVectorFloat32</td><td>Multi-component of 32 bit float</td></tr>\n <tr><td>sitkVectorFloat64</td><td>Multi-component of 64 bit float</td></tr>\n <tr><td>sitkLabelUInt8</td><td>RLE label of unsigned 8 bit integers</td></tr>\n <tr><td>sitkLabelUInt16</td><td>RLE label of unsigned 16 bit integers</td></tr>\n <tr><td>sitkLabelUInt32</td><td>RLE label of unsigned 32 bit integers</td></tr>\n <tr><td>sitkLabelUInt64</td><td>RLE label of unsigned 64 bit integers</td></tr>\n</table>\n\nThere is also `sitkUnknown`, which is used for undefined or erroneous pixel ID's. It has a value of -1.\n\nThe 64-bit integer types are not available on all distributions. When not available the value is `sitkUnknown`.",
"_____no_output_____"
]
],
[
[
"import SimpleITK as sitk\n\nfrom __future__ import print_function\n\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport numpy as np\n\nfrom ipywidgets import interact, fixed\nimport os\n\nOUTPUT_DIR = 'Output'\n\n# Utility method that either downloads data from the MIDAS repository or\n# if already downloaded returns the file name for reading from disk (cached data).\n%run update_path_to_download_script\nfrom downloaddata import fetch_data as fdata",
"_____no_output_____"
]
],
[
[
"## Load your first image and display it",
"_____no_output_____"
]
],
[
[
"logo = sitk.ReadImage(fdata('SimpleITK.jpg'))\n\nplt.imshow(sitk.GetArrayViewFromImage(logo))\nplt.axis('off');",
"_____no_output_____"
]
],
[
[
"## Image Construction\n\nThere are a variety of ways to create an image. \n\nThe following components are required for a complete definition of an image:\n<ol>\n<li>Pixel type [fixed on creation, no default]: unsigned 32 bit integer, sitkVectorUInt8, etc., see list above.</li>\n<li> Sizes [fixed on creation, no default]: number of pixels/voxels in each dimension. This quantity implicitly defines the image dimension.</li>\n<li> Origin [default is zero]: coordinates of the pixel/voxel with index (0,0,0) in physical units (i.e. mm).</li>\n<li> Spacing [default is one]: Distance between adjacent pixels/voxels in each dimension given in physical units.</li>\n<li> Direction matrix [default is identity]: mapping, rotation, between direction of the pixel/voxel axes and physical directions.</li>\n</ol>\n\nInitial pixel/voxel values are set to zero.",
"_____no_output_____"
]
],
[
[
"image_3D = sitk.Image(256, 128, 64, sitk.sitkInt16)\nimage_2D = sitk.Image(64, 64, sitk.sitkFloat32)\nimage_2D = sitk.Image([32,32], sitk.sitkUInt32)\nimage_RGB = sitk.Image([128,64], sitk.sitkVectorUInt8, 3)",
"_____no_output_____"
]
],
[
[
"## Basic Image Attributes\n\nYou can change the image origin, spacing and direction. Making such changes to an image already containing data should be done cautiously. ",
"_____no_output_____"
]
],
[
[
"image_3D.SetOrigin((78.0, 76.0, 77.0))\nimage_3D.SetSpacing([0.5,0.5,3.0])\n\nprint(image_3D.GetOrigin())\nprint(image_3D.GetSize())\nprint(image_3D.GetSpacing())\nprint(image_3D.GetDirection())",
"_____no_output_____"
]
],
[
[
"Image dimension queries:",
"_____no_output_____"
]
],
[
[
"print(image_3D.GetDimension())\nprint(image_3D.GetWidth())\nprint(image_3D.GetHeight())\nprint(image_3D.GetDepth())",
"_____no_output_____"
]
],
[
[
"What is the depth of a 2D image?",
"_____no_output_____"
]
],
[
[
"print(image_2D.GetSize())\nprint(image_2D.GetDepth())",
"_____no_output_____"
]
],
[
[
"Pixel/voxel type queries: ",
"_____no_output_____"
]
],
[
[
"print(image_3D.GetPixelIDValue())\nprint(image_3D.GetPixelIDTypeAsString())\nprint(image_3D.GetNumberOfComponentsPerPixel())",
"_____no_output_____"
]
],
[
[
"What is the dimension and size of a Vector image and its data?",
"_____no_output_____"
]
],
[
[
"print(image_RGB.GetDimension())\nprint(image_RGB.GetSize())\nprint(image_RGB.GetNumberOfComponentsPerPixel())",
"_____no_output_____"
]
],
[
[
"## Accessing Pixels and Slicing\n\nThe Image class's member functions ``GetPixel`` and ``SetPixel`` provide an ITK-like interface for pixel access.",
"_____no_output_____"
]
],
[
[
"help(image_3D.GetPixel)",
"_____no_output_____"
],
[
"print(image_3D.GetPixel(0, 0, 0))\nimage_3D.SetPixel(0, 0, 0, 1)\nprint(image_3D.GetPixel(0, 0, 0))\n\n# This can also be done using Pythonic notation.\nprint(image_3D[0,0,1])\nimage_3D[0,0,1] = 2\nprint(image_3D[0,0,1])",
"_____no_output_____"
]
],
[
[
"Slicing of SimpleITK images returns a copy of the image data. \n\nThis is similar to slicing Python lists and differs from the \"view\" returned by slicing numpy arrays. ",
"_____no_output_____"
]
],
[
[
"# Brute force sub-sampling \nlogo_subsampled = logo[::2,::2]\n\n# Get the sub-image containing the word Simple\nsimple = logo[0:115,:]\n\n# Get the sub-image containing the word Simple and flip it\nsimple_flipped = logo[115:0:-1,:]\n\nn = 4\n\nplt.subplot(n,1,1)\nplt.imshow(sitk.GetArrayViewFromImage(logo))\nplt.axis('off');\n\nplt.subplot(n,1,2)\nplt.imshow(sitk.GetArrayViewFromImage(logo_subsampled))\nplt.axis('off');\n\nplt.subplot(n,1,3)\nplt.imshow(sitk.GetArrayViewFromImage(simple))\nplt.axis('off')\n\nplt.subplot(n,1,4)\nplt.imshow(sitk.GetArrayViewFromImage(simple_flipped))\nplt.axis('off');",
"_____no_output_____"
]
],
[
[
"Draw a square on top of the logo image:\nAfter running this cell, uncomment \"Version 3\" and see its effect.",
"_____no_output_____"
]
],
[
[
"# Version 0: get the numpy array and assign the value via broadcast - later on you will need to construct \n# a new image from the array \nlogo_pixels = sitk.GetArrayFromImage(logo)\nlogo_pixels[0:10,0:10] = [0,255,0]\n\n# Version 1: generates an error, the image slicing returns a new image and you cannot assign a value to an image \n#logo[0:10,0:10] = [255,0,0]\n\n# Version 2: image slicing returns a new image, so all assignments here will not have any effect on the original\n# 'logo' image\nlogo_subimage = logo[0:10, 0:10]\nfor x in range(0,10):\n for y in range(0,10):\n logo_subimage[x,y] = [255,0,0]\n\n# Version 3: modify the original image, iterate and assign a value to each pixel\n#for x in range(0,10):\n# for y in range(0,10):\n# logo[x,y] = [255,0,0]\n\n \nplt.subplot(2,1,1)\nplt.imshow(sitk.GetArrayViewFromImage(logo))\nplt.axis('off')\n\nplt.subplot(2,1,2)\nplt.imshow(logo_pixels)\nplt.axis('off');",
"_____no_output_____"
]
],
[
[
"## Conversion between numpy and SimpleITK\n\nSimpleITK and numpy indexing access is in opposite order! \n\nSimpleITK: image[x,y,z]<br>\nnumpy: image_numpy_array[z,y,x]",
"_____no_output_____"
],
[
"### From SimpleITK to numpy\n\nWe have two options for converting from SimpleITK to numpy:\n* GetArrayFromImage(): returns a copy of the image data. You can then freely modify the data as it has no effect on the original SimpleITK image.\n* GetArrayViewFromImage(): returns a view on the image data which is useful for display in a memory efficient manner. You cannot modify the data and __the view will be invalid if the original SimpleITK image is deleted__.",
"_____no_output_____"
]
],
[
[
"nda = sitk.GetArrayFromImage(image_3D)\nprint(image_3D.GetSize())\nprint(nda.shape)\n\nnda = sitk.GetArrayFromImage(image_RGB)\nprint(image_RGB.GetSize())\nprint(nda.shape)",
"_____no_output_____"
],
[
"gabor_image = sitk.GaborSource(size=[64,64], frequency=.03)\n# Getting a numpy array view on the image data doesn't copy the data\nnda_view = sitk.GetArrayViewFromImage(gabor_image)\nplt.imshow(nda_view, cmap=plt.cm.Greys_r)\nplt.axis('off');\n\n# Trying to assign a value to the array view will throw an exception\nnda_view[0,0] = 255",
"_____no_output_____"
]
],
[
[
"### From numpy to SimpleITK\n\nRemember to to set the image's origin, spacing, and possibly direction cosine matrix. The default values may not match the physical dimensions of your image.",
"_____no_output_____"
]
],
[
[
"nda = np.zeros((10,20,3))\n\n #if this is supposed to be a 3D gray scale image [x=3, y=20, z=10]\nimg = sitk.GetImageFromArray(nda)\nprint(img.GetSize())\n\n #if this is supposed to be a 2D color image [x=20,y=10]\nimg = sitk.GetImageFromArray(nda, isVector=True)\nprint(img.GetSize())",
"_____no_output_____"
]
],
[
[
"### Image operations\n\nSimpleITK supports basic arithmetic operations between images, <b>taking into account their physical space</b>.\n\nRepeatedly run this cell. Fix the error (comment out the SetDirection, then SetSpacing). Why doesn't the SetOrigin line cause a problem? How close do two physical attributes need to be in order to be considered equivalent?",
"_____no_output_____"
]
],
[
[
"img1 = sitk.Image(24,24, sitk.sitkUInt8)\nimg1[0,0] = 0\n\nimg2 = sitk.Image(img1.GetSize(), sitk.sitkUInt8)\nimg2.SetDirection([0,1,0.5,0.5])\nimg2.SetSpacing([0.5,0.8])\nimg2.SetOrigin([0.000001,0.000001])\nimg2[0,0] = 255\n\nimg3 = img1 + img2\nprint(img3[0,0])",
"_____no_output_____"
]
],
[
[
"## Reading and Writing\n\nSimpleITK can read and write images stored in a single file, or a set of files (e.g. DICOM series).\n\nImages stored in the DICOM format have a meta-data dictionary associated with them, which is populated with the DICOM tags. When a DICOM series is read as a single image, the meta-data information is not available since DICOM tags are specific to a file. If you need the meta-data, access the dictionary for each file by reading them separately. ",
"_____no_output_____"
],
[
"In the following cell, we read an image in JPEG format, and write it as PNG and BMP. File formats are deduced from the file extension. Appropriate pixel type is also set - you can override this and force a pixel type of your choice.",
"_____no_output_____"
]
],
[
[
"img = sitk.ReadImage(fdata('SimpleITK.jpg'))\nprint(img.GetPixelIDTypeAsString())\n\n# write as PNG and BMP\nsitk.WriteImage(img, os.path.join(OUTPUT_DIR, 'SimpleITK.png'))\nsitk.WriteImage(img, os.path.join(OUTPUT_DIR, 'SimpleITK.bmp'))",
"_____no_output_____"
]
],
[
[
"Read an image in JPEG format and cast the pixel type according to user selection.",
"_____no_output_____"
]
],
[
[
"# Several pixel types, some make sense in this case (vector types) and some are just show\n# that the user's choice will force the pixel type even when it doesn't make sense.\npixel_types = { 'sitkUInt8': sitk.sitkUInt8,\n 'sitkUInt16' : sitk.sitkUInt16,\n 'sitkFloat64' : sitk.sitkFloat64,\n 'sitkVectorUInt8' : sitk.sitkVectorUInt8,\n 'sitkVectorUInt16' : sitk.sitkVectorUInt16,\n 'sitkVectorFloat64' : sitk.sitkVectorFloat64}\n\ndef pixel_type_dropdown_callback(pixel_type, pixel_types_dict):\n #specify the file location and the pixel type we want\n img = sitk.ReadImage(fdata('SimpleITK.jpg'), pixel_types_dict[pixel_type])\n \n print(img.GetPixelIDTypeAsString())\n print(img[0,0])\n plt.imshow(sitk.GetArrayViewFromImage(img))\n plt.axis('off')\n \ninteract(pixel_type_dropdown_callback, pixel_type=list(pixel_types.keys()), pixel_types_dict=fixed(pixel_types)); ",
"_____no_output_____"
]
],
[
[
"Read a DICOM series and write it as a single mha file",
"_____no_output_____"
]
],
[
[
"data_directory = os.path.dirname(fdata(\"CIRS057A_MR_CT_DICOM/readme.txt\"))\nseries_ID = '1.2.840.113619.2.290.3.3233817346.783.1399004564.515'\n\n# Get the list of files belonging to a specific series ID.\nreader = sitk.ImageSeriesReader()\n# Use the functional interface to read the image series.\noriginal_image = sitk.ReadImage(reader.GetGDCMSeriesFileNames(data_directory, series_ID))\n\n# Write the image.\noutput_file_name_3D = os.path.join(OUTPUT_DIR, '3DImage.mha')\nsitk.WriteImage(original_image, output_file_name_3D)\n\n# Read it back again.\nwritten_image = sitk.ReadImage(output_file_name_3D)\n\n# Check that the original and written image are the same.\nstatistics_image_filter = sitk.StatisticsImageFilter()\nstatistics_image_filter.Execute(original_image - written_image)\n\n# Check that the original and written files are the same\nprint('Max, Min differences are : {0}, {1}'.format(statistics_image_filter.GetMaximum(), statistics_image_filter.GetMinimum()))",
"_____no_output_____"
]
],
[
[
"Write an image series as JPEG. The WriteImage function receives a volume and a list of images names and writes the volume according to the z axis. For a displayable result we need to rescale the image intensities (default is [0,255]) since the JPEG format requires a cast to the UInt8 pixel type.",
"_____no_output_____"
]
],
[
[
"sitk.WriteImage(sitk.Cast(sitk.RescaleIntensity(written_image), sitk.sitkUInt8), \n [os.path.join(OUTPUT_DIR, 'slice{0:03d}.jpg'.format(i)) for i in range(written_image.GetSize()[2])]) ",
"_____no_output_____"
]
],
[
[
"Select a specific DICOM series from a directory and only then load user selection.",
"_____no_output_____"
]
],
[
[
"data_directory = os.path.dirname(fdata(\"CIRS057A_MR_CT_DICOM/readme.txt\"))\n# Global variable 'selected_series' is updated by the interact function\nselected_series = ''\ndef DICOM_series_dropdown_callback(series_to_load, series_dictionary):\n global selected_series\n # Print some information about the series from the meta-data dictionary\n # DICOM standard part 6, Data Dictionary: http://medical.nema.org/medical/dicom/current/output/pdf/part06.pdf\n img = sitk.ReadImage(series_dictionary[series_to_load][0])\n tags_to_print = {'0010|0010': 'Patient name: ', \n '0008|0060' : 'Modality: ',\n '0008|0021' : 'Series date: ',\n '0008|0080' : 'Institution name: ',\n '0008|1050' : 'Performing physician\\'s name: '}\n for tag in tags_to_print:\n try:\n print(tags_to_print[tag] + img.GetMetaData(tag))\n except: # Ignore if the tag isn't in the dictionary\n pass\n selected_series = series_to_load \n\n# Directory contains multiple DICOM studies/series, store\n # in dictionary with key being the series ID\nreader = sitk.ImageSeriesReader()\nseries_file_names = {}\nseries_IDs = reader.GetGDCMSeriesIDs(data_directory)\n # Check that we have at least one series\nif series_IDs:\n for series in series_IDs:\n series_file_names[series] = reader.GetGDCMSeriesFileNames(data_directory, series)\n \n interact(DICOM_series_dropdown_callback, series_to_load=list(series_IDs), series_dictionary=fixed(series_file_names)); \nelse:\n print('Data directory does not contain any DICOM series.')",
"_____no_output_____"
],
[
"reader.SetFileNames(series_file_names[selected_series])\nimg = reader.Execute()\n# Display the image slice from the middle of the stack, z axis\nz = int(img.GetDepth()/2)\nplt.imshow(sitk.GetArrayViewFromImage(img)[z,:,:], cmap=plt.cm.Greys_r)\nplt.axis('off');",
"_____no_output_____"
]
],
[
[
"\n## Image Display\n\nWhile SimpleITK does not do visualization, it does contain a built in ``Show`` method. This function writes the image out to disk and than launches a program for visualization. By default it is configured to use <a href=\"http://imagej.nih.gov/ij/\">ImageJ</a>, because it readily supports many medical image formats and loads quickly. However, the ``Show`` visualization program is easily customizable by an environment variable:\n\n<ul>\n<li>SITK_SHOW_COMMAND: Viewer to use (<a href=\"http://www.itksnap.org\">ITK-SNAP</a>, <a href=\"http://www.slicer.org\">3D Slicer</a>...) </li>\n<li>SITK_SHOW_COLOR_COMMAND: Viewer to use when displaying color images.</li>\n<li>SITK_SHOW_3D_COMMAND: Viewer to use for 3D images.</li>\n</ul>",
"_____no_output_____"
]
],
[
[
"sitk.Show?",
"_____no_output_____"
]
],
[
[
"By converting into a numpy array, matplotlib can be used for visualization for integration into the scientific python environment. This is good for illustrative purposes, but is problematic when working with images that have a high dynamic range or non-isotropic spacing - most 3D medical images. \n\nWhen working with medical images it is recommended to visualize them using dedicated software such as the freely available 3D Slicer or ITK-SNAP.",
"_____no_output_____"
]
],
[
[
"mr_image = sitk.ReadImage(fdata('training_001_mr_T1.mha'))\nnpa = sitk.GetArrayViewFromImage(mr_image)\n\n# Display the image slice from the middle of the stack, z axis\nz = int(mr_image.GetDepth()/2)\nnpa_zslice = sitk.GetArrayViewFromImage(mr_image)[z,:,:]\n\n# Three plots displaying the same data, how do we deal with the high dynamic range?\nfig = plt.figure()\nfig.set_size_inches(15,30)\n\nfig.add_subplot(1,3,1)\nplt.imshow(npa_zslice)\nplt.title('default colormap')\nplt.axis('off')\n\nfig.add_subplot(1,3,2)\nplt.imshow(npa_zslice,cmap=plt.cm.Greys_r);\nplt.title('grey colormap')\nplt.axis('off')\n\nfig.add_subplot(1,3,3)\nplt.title('grey colormap,\\n scaling based on volumetric min and max values')\nplt.imshow(npa_zslice,cmap=plt.cm.Greys_r, vmin=npa.min(), vmax=npa.max())\nplt.axis('off');",
"_____no_output_____"
],
[
"# Display the image slice in the middle of the stack, x axis\n \nx = int(mr_image.GetWidth()/2)\n\nnpa_xslice = npa[:,:,x]\nplt.imshow(npa_xslice, cmap=plt.cm.Greys_r)\nplt.axis('off')\n\nprint('Image spacing: {0}'.format(mr_image.GetSpacing()))",
"_____no_output_____"
],
[
"# Collapse along the x axis\nextractSliceFilter = sitk.ExtractImageFilter() \nsize = list(mr_image.GetSize())\nsize[0] = 0\nextractSliceFilter.SetSize( size )\n \nindex = (x, 0, 0)\nextractSliceFilter.SetIndex(index)\nsitk_xslice = extractSliceFilter.Execute(mr_image)\n\n# Resample slice to isotropic\noriginal_spacing = sitk_xslice.GetSpacing()\noriginal_size = sitk_xslice.GetSize()\n\nmin_spacing = min(sitk_xslice.GetSpacing())\nnew_spacing = [min_spacing, min_spacing]\nnew_size = [int(round(original_size[0]*(original_spacing[0]/min_spacing))), \n int(round(original_size[1]*(original_spacing[1]/min_spacing)))]\nresampleSliceFilter = sitk.ResampleImageFilter()\n\n# Why is the image pixelated?\nsitk_isotropic_xslice = resampleSliceFilter.Execute(sitk_xslice, new_size, sitk.Transform(), sitk.sitkNearestNeighbor, sitk_xslice.GetOrigin(),\n new_spacing, sitk_xslice.GetDirection(), 0, sitk_xslice.GetPixelID())\n\nplt.imshow(sitk.GetArrayViewFromImage(sitk_isotropic_xslice), cmap=plt.cm.Greys_r)\nplt.axis('off')\nprint('Image spacing: {0}'.format(sitk_isotropic_xslice.GetSpacing()))",
"_____no_output_____"
]
],
[
[
"So if you really want to look at your images, use the sitk.Show command:",
"_____no_output_____"
]
],
[
[
"try:\n sitk.Show(mr_image)\nexcept RuntimeError:\n print('SimpleITK Show method could not find the viewer (ImageJ not installed or ' +\n 'environment variable pointing to non existant viewer).')",
"_____no_output_____"
]
],
[
[
"Use a different viewer by setting environment variable(s). Do this from within your Jupyter notebook using 'magic' functions, or set in a more permanent manner using your OS specific convention. ",
"_____no_output_____"
]
],
[
[
"%env SITK_SHOW_COMMAND /Applications/ITK-SNAP.app/Contents/MacOS/ITK-SNAP \n\ntry:\n sitk.Show(mr_image)\nexcept RuntimeError:\n print('SimpleITK Show method could not find the viewer (ITK-SNAP not installed or ' +\n 'environment variable pointing to non existant viewer).')",
"_____no_output_____"
],
[
"%env SITK_SHOW_COMMAND '/Applications/ImageJ/ImageJ.app/Contents/MacOS/JavaApplicationStub'\ntry:\n sitk.Show(mr_image)\nexcept RuntimeError:\n print('SimpleITK Show method could not find the viewer (ImageJ not installed or ' +\n 'environment variable pointing to non existant viewer).')",
"_____no_output_____"
],
[
"%env SITK_SHOW_COMMAND '/Applications/Slicer.app/Contents/MacOS/Slicer'\ntry:\n sitk.Show(mr_image)\nexcept RuntimeError:\n print('SimpleITK Show method could not find the viewer (Slicer not installed or ' +\n 'environment variable pointing to non existant viewer).')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
eccc13324c1b0abb9d2ab90208e1c899fa225c11 | 35,855 | ipynb | Jupyter Notebook | Day_2/Simple Linear Regression.ipynb | KushwahaDK/100DaysOfMLCode | 1f9b2250d2f11fa9170ba19dcccdceedd7764aff | [
"MIT"
] | null | null | null | Day_2/Simple Linear Regression.ipynb | KushwahaDK/100DaysOfMLCode | 1f9b2250d2f11fa9170ba19dcccdceedd7764aff | [
"MIT"
] | null | null | null | Day_2/Simple Linear Regression.ipynb | KushwahaDK/100DaysOfMLCode | 1f9b2250d2f11fa9170ba19dcccdceedd7764aff | [
"MIT"
] | 1 | 2021-09-30T10:14:25.000Z | 2021-09-30T10:14:25.000Z | 95.613333 | 11,086 | 0.845823 | [
[
[
"# Step 1 : Preprocessing Data ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\n# Import pyplot for visualisation of data\nfrom matplotlib import pyplot as plt",
"_____no_output_____"
],
[
"# Read dataset from csv\ndataset = pd.read_csv('studentscores.csv')",
"_____no_output_____"
],
[
"# To see the frst few elements of dataset as well as number of columns\ndataset.head()",
"_____no_output_____"
],
[
"# Check for any missing values in the dataframe\ndataset.isnull().values.any()",
"_____no_output_____"
],
[
"# Check for total number of Missing values\ndataset.isnull().sum()",
"_____no_output_____"
],
[
"# Assign X and Y values from Dataset\nX = dataset.iloc[:,:1].values # Only one column so index upto 1, also it ives a 2D array\nY = dataset.iloc[:,-1].values # Only last column so index -1",
"_____no_output_____"
],
[
"# Importing train test split function\nfrom sklearn.model_selection import train_test_split\n\n# splitting the data into train set and test set\n# random state to fix a random seed for shuffling\n# test_size will determine the ratio of split\nX_train, X_test , Y_train, Y_test = train_test_split(X, Y, random_state=42, test_size=0.2)",
"_____no_output_____"
]
],
[
[
"# Step 2 : Fitting Simple Linear Regression Model to the training data",
"_____no_output_____"
]
],
[
[
"# Import built in function for LinearRegression from sklearn library's linear_model module\nfrom sklearn.linear_model import LinearRegression",
"_____no_output_____"
],
[
"# Create an object of class LinearRegression\nlr = LinearRegression()",
"_____no_output_____"
],
[
"# Fit the training data X_train\nlr.fit(X_train,Y_train)",
"_____no_output_____"
]
],
[
[
"# Step 3: Predecting the Result",
"_____no_output_____"
]
],
[
[
"# Predicting the result with test data\nY_pred = lr.predict(X_test)",
"_____no_output_____"
]
],
[
[
"# Step 4: Visualization",
"_____no_output_____"
],
[
"## Visualising the Training results",
"_____no_output_____"
]
],
[
[
"# Creating a Scatter plot\nplt.scatter(X_train , Y_train, color = 'red')\n\n# NOTE: very important to write plt.show() to print graph on screen\nplt.show()",
"_____no_output_____"
],
[
"# creating the above scatter plot again and adding the trained model line in it.\nplt.scatter(X_train, Y_train, color='red')\n# Creating a line plot between actual and predicted training data\nplt.plot(X_train , lr.predict(X_train), color ='blue')\n# show the graph\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Visualizing the test results",
"_____no_output_____"
]
],
[
[
"# Create a scatter plot with test datasets\nplt.scatter(X_test, Y_test, color='red')\n\n# create a prediction model line\nplt.plot(X_test,Y_pred, color='blue')\n\n# Show the graph\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
eccc19d8fef3d997dcdfbcab8b1297ca1c6e0020 | 592,077 | ipynb | Jupyter Notebook | AgilePresentation/datamodel/combined_analysis.ipynb | ninoxcello/mscs710-project | d7059c01e9f80f5d8581dd04db69091db35f3481 | [
"MIT"
] | null | null | null | AgilePresentation/datamodel/combined_analysis.ipynb | ninoxcello/mscs710-project | d7059c01e9f80f5d8581dd04db69091db35f3481 | [
"MIT"
] | 18 | 2017-11-15T22:22:32.000Z | 2017-12-14T23:01:58.000Z | AgilePresentation/datamodel/combined_analysis.ipynb | ninoxcello/mscs710-project | d7059c01e9f80f5d8581dd04db69091db35f3481 | [
"MIT"
] | null | null | null | 1,062.974865 | 58,600 | 0.942011 | [
[
[
"# Data Exploration\n\nThis notebook is used to explore & visualize our dataset.",
"_____no_output_____"
]
],
[
[
"import numpy as np # Linear Alg\nimport pandas as pd # CSV file I/O & data processing\n\n# Visualization\nimport matplotlib.pyplot as plt\nimport matplotlib.dates as mdates\nimport seaborn as sns\nimport warnings \n\nfrom sklearn import preprocessing\nfrom matplotlib import style\n\nwarnings.filterwarnings(\"ignore\")\nstyle.use('ggplot')\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (12.0, 8.0)\n\nfrom subprocess import check_output",
"_____no_output_____"
],
[
"input_dir = '../../input'",
"_____no_output_____"
],
[
"currencies = {}\n\ncurrencies['bitcoin'] = pd.read_csv('{}/bitcoin_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['bitconnect'] = pd.read_csv('{}/bitconnect_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['dash'] = pd.read_csv('{}/dash_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['ethereum_classic'] = pd.read_csv('{}/ethereum_classic_price.csv'.format(input_dir), parse_dates=['Date'])\n\ncurrencies['ethereum'] = pd.read_csv('{}/ethereum_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['iota'] = pd.read_csv('{}/iota_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['litecoin'] = pd.read_csv('{}/litecoin_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['monero'] = pd.read_csv('{}/monero_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['nem'] = pd.read_csv('{}/nem_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['neo'] = pd.read_csv('{}/neo_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['numeraire'] = pd.read_csv('{}/numeraire_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['omisego'] = pd.read_csv('{}/omisego_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['qtum'] = pd.read_csv('{}/qtum_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['ripple'] = pd.read_csv('{}/ripple_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['stratis'] = pd.read_csv('{}/stratis_price.csv'.format(input_dir), parse_dates=['Date'])\ncurrencies['waves'] = pd.read_csv('{}/waves_price.csv'.format(input_dir), parse_dates=['Date'])",
"_____no_output_____"
],
[
"for c in currencies:\n plt.plot(currencies[c].index, currencies[c]['Close'], label=c)\nplt.legend(bbox_to_anchor=(1.05, 1))\nplt.show()",
"_____no_output_____"
],
[
"for c in currencies:\n df = pd.DataFrame(currencies[c])\n df = df[['Date' , 'Close']]\n \n df['Date_mpl'] = df['Date'].apply(lambda x: mdates.date2num(x)) \n fig, ax = plt.subplots(figsize=(6,4))\n sns.tsplot(df.Close.values, time=df.Date_mpl.values, alpha=0.8, color='b', ax=ax)\n ax.xaxis.set_major_locator(mdates.AutoDateLocator())\n ax.xaxis.set_major_formatter(mdates.DateFormatter('%Y.%m.%d'))\n fig.autofmt_xdate()\n plt.xlabel('Date', fontsize=12)\n plt.ylabel('Price in USD', fontsize=12)\n title_str = \"Closing price distribution of \" + c\n plt.title(title_str, fontsize=15)\n plt.show()",
"_____no_output_____"
],
[
"# Un-normalized data\nfor c in currencies:\n plt.plot(currencies[c].index, currencies[c]['Close'], label=c)\nplt.legend(bbox_to_anchor=(1.05, 1))\nplt.show()",
"_____no_output_____"
],
[
"currencies = {}\n\ncurrencies['bitcoin'] = pd.read_csv('{}/bitcoin_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['bitconnect'] = pd.read_csv('{}/bitconnect_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['dash'] = pd.read_csv('{}/dash_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['ethereum_classic'] = pd.read_csv('{}/ethereum_classic_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\n\ncurrencies['ethereum'] = pd.read_csv('{}/ethereum_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['iota'] = pd.read_csv('{}/iota_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['litecoin'] = pd.read_csv('{}/litecoin_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['monero'] = pd.read_csv('{}/monero_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['nem'] = pd.read_csv('{}/nem_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['neo'] = pd.read_csv('{}/neo_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['numeraire'] = pd.read_csv('{}/numeraire_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['omisego'] = pd.read_csv('{}/omisego_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['qtum'] = pd.read_csv('{}/qtum_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['ripple'] = pd.read_csv('{}/ripple_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['stratis'] = pd.read_csv('{}/stratis_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\ncurrencies['waves'] = pd.read_csv('{}/waves_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\n\n# currencies['bitcoin_cash'] = pd.read_csv('{}/bitcoin_cash_price.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\n# currencies['ethereum_data'] = pd.read_csv('{}/ethereum_dataset.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\n# currencies['bitcoin_data'] = pd.read_csv('{}/bitcoin_dataset.csv'.format(input_dir), parse_dates=['Date'], index_col=0)\n\nprint(len(currencies))",
"16\n"
],
[
"ohlc = ['Open', 'High', 'Low', 'Close']\n \ncurrencies_ohlc = {}\nfor i in currencies:\n currencies_ohlc[i] = currencies[i].drop([\"Volume\", \"Market Cap\"], axis=1)\n\ncurrencies_ohlc['bitcoin'].head()",
"_____no_output_____"
],
[
"normalized_currencies = {}\nfor i in currencies_ohlc:\n normalized_currencies[i] = preprocessing.normalize(currencies_ohlc[i], norm='l2')\n\n# normalized_df = {}\n# for i in normalized_currencies:\n# normalized_currencies[]\n# df = pd.DataFrame(normalized_currencies['bitcoin'], index=currencies_ohlc['bitcoin'].index)\n# df.head()\n\n# plt.plot(df[3], label='bitcoin')\n# plt.legend(bbox_to_anchor=(1.05, 1))\n# plt.show()\n# bitcoin_close = normalized_currencies['bitcoin']['Close']\n# bitcoin_close.head()\n# bitcoin_normalized = preprocessing.normalize(bitcoin_close[1], norm='l2')\n# bitcoin_normalized\n# normalized_currencies['bitcoin']['Close'].head()\n# for i in normalized_currencies:\n# for feat in ohlc:\n# normalized_currencies[i][feat] = preprocessing.normalize(currencies_ohlc[i][feat], norm='l2')\n \n# normalized_currencies['bitcoin'][\"Close\"].head()",
"_____no_output_____"
],
[
"\nfor c in normalized_currencies:\n plt.plot(normalized_currencies[c].index, currencies[c]['Close'], label=c)\nplt.legend(bbox_to_anchor=(1.05, 1))\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccc27c7df853d6e2bc4eb5b9c7b221ac9e23d40 | 13,488 | ipynb | Jupyter Notebook | notebooks/1.1-hz-Label_using_ Textblob.ipynb | JohnnyWang1998/ukraineRussiaTweetsSentimentAnalysis | 80132e1b453b4439c1610fc6b073dcce76307fb5 | [
"MIT"
] | null | null | null | notebooks/1.1-hz-Label_using_ Textblob.ipynb | JohnnyWang1998/ukraineRussiaTweetsSentimentAnalysis | 80132e1b453b4439c1610fc6b073dcce76307fb5 | [
"MIT"
] | null | null | null | notebooks/1.1-hz-Label_using_ Textblob.ipynb | JohnnyWang1998/ukraineRussiaTweetsSentimentAnalysis | 80132e1b453b4439c1610fc6b073dcce76307fb5 | [
"MIT"
] | null | null | null | 13,488 | 13,488 | 0.517126 | [
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"## Reading the data and removing columns that are not important. \ndf = pd.read_csv('labeled_sample_600.csv', sep = ',', encoding = 'latin-1')\ndf.head(5)",
"_____no_output_____"
],
[
"import re\nfrom bs4 import BeautifulSoup\nfrom html import unescape\n#Remove URLS\ndef remove_urls(x):\n cleaned_string = re.sub(r'(https|http)?:\\/\\/(\\w|\\.|\\/|\\?|\\=|\\&|\\%)*\\b', '', str(x), flags=re.MULTILINE)\n return cleaned_string",
"_____no_output_____"
],
[
"#Unescape characters\ndef unescape_stuff(x):\n soup = BeautifulSoup(unescape(x), 'lxml')\n return soup.text",
"_____no_output_____"
],
[
"#Remove emojis\ndef deEmojify(x):\n regrex_pattern = re.compile(pattern = \"[\"\n u\"\\U0001F600-\\U0001F64F\" # emoticons\n u\"\\U0001F300-\\U0001F5FF\" # symbols & pictographs\n u\"\\U0001F680-\\U0001F6FF\" # transport & map symbols\n u\"\\U0001F1E0-\\U0001F1FF\" # flags (iOS)\n \"]+\", flags = re.UNICODE)\n return regrex_pattern.sub(r'', x)",
"_____no_output_____"
],
[
"#Replace conservative whitespaces\ndef unify_whitespaces(x):\n cleaned_string = re.sub(' +', ' ', x)\n return cleaned_string ",
"_____no_output_____"
],
[
"#Remove unwanted symbols\ndef remove_symbols(x):\n cleaned_string = re.sub(r\"[^a-zA-Z0-9?!.,]+\", ' ', x)\n return cleaned_string ",
"_____no_output_____"
],
[
"df['text'] = df['text'].str.lower()\ndf['text'] = df['text'].apply(remove_urls)\ndf['text'] = df['text'].apply(unescape_stuff)\ndf['text'] = df['text'].apply(deEmojify)\ndf['text'] = df['text'].apply(remove_symbols)\ndf['text'] = df['text'].apply(unify_whitespaces)",
"_____no_output_____"
],
[
"df['text'].head()",
"_____no_output_____"
],
[
"pip install textblob",
"Collecting textblob\n Downloading textblob-0.17.1-py2.py3-none-any.whl (636 kB)\n\u001b[K |████████████████████████████████| 636 kB 3.5 MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied: nltk>=3.1 in /Users/huazhouliu/opt/anaconda3/lib/python3.9/site-packages (from textblob) (3.6.5)\nRequirement already satisfied: click in /Users/huazhouliu/opt/anaconda3/lib/python3.9/site-packages (from nltk>=3.1->textblob) (8.0.3)\nRequirement already satisfied: joblib in /Users/huazhouliu/opt/anaconda3/lib/python3.9/site-packages (from nltk>=3.1->textblob) (1.1.0)\nRequirement already satisfied: regex>=2021.8.3 in /Users/huazhouliu/opt/anaconda3/lib/python3.9/site-packages (from nltk>=3.1->textblob) (2021.8.3)\nRequirement already satisfied: tqdm in /Users/huazhouliu/opt/anaconda3/lib/python3.9/site-packages (from nltk>=3.1->textblob) (4.62.3)\nInstalling collected packages: textblob\nSuccessfully installed textblob-0.17.1\nNote: you may need to restart the kernel to use updated packages.\n"
],
[
"from textblob import TextBlob",
"_____no_output_____"
],
[
"#Determine the sentiments using polarity of textblob \naccuracy = 0\ncorrect = 0\nfor i in df.index:\n score = 0\n data = TextBlob(df['text'][i])\n if data.sentiment[0]>0.05:\n score = 2\n elif data.sentiment[0]<-0.05:\n score = 0 \n else:\n score = 1\n print(score, df['value'][i])\n if score == df['value'][i]: \n correct += 1\n else:\n pass\n",
"1 0\n0 2\n1 0\n1 0\n0 2\n1 0\n1 0\n1 1\n1 0\n1 1\n1 0\n2 0\n0 0\n1 0\n2 0\n0 0\n1 1\n2 0\n2 0\n1 0\n1 0\n2 0\n1 0\n2 1\n1 2\n2 2\n2 0\n1 2\n1 2\n2 0\n2 0\n2 0\n2 2\n1 0\n1 2\n1 0\n0 0\n0 0\n1 0\n2 2\n2 2\n2 0\n1 0\n2 2\n0 0\n1 1\n2 0\n1 0\n2 0\n2 0\n2 2\n0 0\n2 0\n2 2\n1 0\n2 2\n1 0\n2 2\n2 2\n1 0\n0 0\n2 2\n0 0\n0 0\n2 2\n2 2\n2 0\n1 0\n2 0\n2 0\n0 0\n0 0\n2 0\n0 0\n1 0\n2 2\n1 1\n2 1\n2 0\n0 0\n0 0\n1 0\n0 0\n1 0\n2 2\n1 0\n2 0\n0 0\n1 0\n2 0\n1 0\n1 0\n2 2\n1 2\n0 0\n2 2\n1 0\n0 0\n2 2\n0 2\n0 0\n1 0\n1 0\n1 0\n0 0\n2 0\n0 0\n2 2\n1 2\n1 2\n1 0\n1 0\n1 0\n1 0\n2 0\n0 0\n2 0\n0 2\n2 0\n1 1\n2 2\n2 1\n2 0\n1 1\n2 0\n0 0\n0 0\n2 0\n2 2\n1 2\n1 2\n0 0\n2 0\n1 1\n1 2\n0 1\n2 2\n1 2\n0 0\n1 2\n2 0\n2 2\n1 2\n2 2\n2 0\n1 2\n0 0\n0 0\n0 0\n2 1\n0 0\n2 2\n2 2\n2 2\n2 2\n0 0\n2 0\n2 0\n1 2\n2 2\n2 0\n2 0\n2 0\n1 0\n2 0\n2 0\n1 0\n1 0\n2 2\n2 0\n2 2\n0 2\n1 2\n2 2\n0 1\n1 0\n2 0\n1 2\n1 0\n0 1\n1 0\n1 0\n2 2\n2 0\n2 0\n1 1\n1 1\n2 0\n1 0\n2 0\n2 0\n1 2\n1 2\n1 2\n2 0\n1 0\n0 0\n2 0\n1 0\n1 2\n0 1\n1 1\n1 0\n2 0\n2 0\n2 1\n2 0\n0 1\n0 0\n1 1\n1 1\n2 0\n2 1\n1 2\n2 2\n2 2\n2 1\n1 1\n1 1\n1 1\n1 0\n2 2\n2 0\n2 0\n2 1\n1 1\n2 1\n0 0\n2 1\n1 0\n2 0\n1 0\n0 0\n1 2\n2 0\n1 1\n2 0\n1 1\n0 1\n2 1\n1 2\n2 1\n1 0\n1 1\n1 2\n1 2\n1 1\n0 1\n0 2\n1 1\n0 1\n1 1\n2 2\n2 2\n2 1\n0 0\n1 1\n0 0\n1 1\n2 1\n1 0\n1 1\n0 0\n2 1\n1 1\n1 1\n0 1\n1 1\n0 1\n1 1\n1 0\n0 1\n1 1\n0 0\n1 1\n1 0\n1 1\n1 1\n1 1\n1 1\n2 2\n1 1\n1 1\n0 0\n1 1\n1 1\n1 2\n0 1\n1 2\n0 1\n1 1\n0 1\n1 1\n1 2\n1 1\n2 2\n1 0\n0 0\n0 1\n1 1\n1 0\n2 2\n1 1\n2 1\n2 2\n1 1\n0 1\n1 1\n2 1\n2 1\n0 1\n1 0\n1 1\n1 0\n1 1\n1 1\n1 1\n1 0\n0 0\n1 0\n2 1\n1 0\n1 0\n2 0\n2 1\n2 1\n1 1\n2 0\n1 1\n1 2\n2 1\n1 0\n1 0\n2 0\n1 1\n1 1\n2 1\n2 1\n1 1\n1 1\n2 0\n2 0\n1 1\n2 1\n0 2\n1 2\n2 1\n1 1\n1 0\n1 2\n2 0\n1 1\n1 1\n2 2\n0 1\n1 0\n0 0\n2 1\n1 0\n0 0\n2 1\n2 1\n0 1\n1 1\n1 1\n0 0\n2 1\n2 0\n0 0\n2 0\n1 1\n1 1\n1 1\n2 1\n2 2\n2 2\n1 1\n2 2\n1 1\n1 0\n1 1\n2 2\n0 0\n0 0\n2 1\n0 0\n2 1\n1 1\n1 2\n0 0\n2 1\n1 0\n2 0\n2 1\n1 1\n2 1\n2 1\n2 1\n2 0\n1 1\n1 1\n2 2\n1 2\n2 0\n0 0\n2 0\n2 2\n1 0\n1 0\n0 0\n2 2\n1 0\n2 2\n1 2\n1 0\n2 1\n1 1\n1 0\n1 1\n1 0\n0 0\n0 0\n1 0\n0 0\n2 0\n1 1\n1 1\n0 0\n1 0\n1 1\n2 1\n1 2\n1 2\n1 1\n1 0\n1 1\n1 0\n1 1\n1 0\n1 1\n1 1\n2 0\n1 0\n1 0\n2 0\n1 1\n1 0\n2 1\n2 2\n2 2\n0 2\n1 0\n0 0\n1 0\n0 0\n2 2\n2 2\n0 0\n1 2\n1 0\n2 1\n0 0\n2 0\n1 2\n0 0\n1 2\n1 2\n1 2\n0 0\n2 2\n1 0\n0 0\n0 2\n2 2\n0 0\n0 0\n1 0\n2 0\n0 0\n2 2\n2 1\n1 0\n1 0\n0 0\n1 0\n1 0\n1 2\n1 2\n2 2\n1 2\n2 0\n2 2\n1 0\n2 2\n2 2\n0 1\n1 1\n1 2\n2 2\n2 0\n2 1\n2 2\n0 0\n1 0\n2 2\n2 0\n2 2\n1 2\n2 2\n1 0\n1 1\n1 0\n2 1\n1 0\n1 0\n2 2\n0 0\n1 0\n0 0\n2 2\n1 2\n1 2\n1 2\n2 2\n0 0\n2 2\n1 0\n2 0\n2 2\n1 2\n2 2\n2 2\n1 0\n1 0\n1 0\n0 0\n1 0\n1 2\n2 2\n2 2\n2 2\n2 2\n2 2\n0 0\n0 2\n1 2\n1 2\n1 0\n2 2\n1 2\n1 2\n1 2\n0 1\n2 2\n1 2\n1 0\n1 1\n2 2\n2 2\n0 0\n1 0\n2 2\n0 0\n1 2\n1 0\n1 1\n2 0\n1 0\n0 0\n1 0\n2 2\n0 0\n1 2\n0 0\n1 0\n1 0\n1 2\n1 0\n2 0\n2 2\n1 0\n2 2\n1 0\n2 2\n0 0\n0 0\n2 0\n2 0\n1 0\n2 2\n1 2\n1 2\n1 1\n1 2\n2 2\n2 0\n1 0\n1 0\n1 2\n1 0\n"
],
[
"accuracy = correct / len(df)",
"_____no_output_____"
],
[
"print(accuracy)",
"0.43166666666666664\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccc342ebc79d6cfcbc19e4e9db87e3c2a5013dd | 21,154 | ipynb | Jupyter Notebook | etl_notebook.ipynb | dsmatthew/sparkify_spark_aws | aa3d51f1c1f8ae74e2f965c20320ddce998590b5 | [
"OLDAP-2.5",
"OLDAP-2.7",
"OLDAP-2.3",
"OLDAP-2.8"
] | null | null | null | etl_notebook.ipynb | dsmatthew/sparkify_spark_aws | aa3d51f1c1f8ae74e2f965c20320ddce998590b5 | [
"OLDAP-2.5",
"OLDAP-2.7",
"OLDAP-2.3",
"OLDAP-2.8"
] | null | null | null | etl_notebook.ipynb | dsmatthew/sparkify_spark_aws | aa3d51f1c1f8ae74e2f965c20320ddce998590b5 | [
"OLDAP-2.5",
"OLDAP-2.7",
"OLDAP-2.3",
"OLDAP-2.8"
] | null | null | null | 33.053125 | 555 | 0.531672 | [
[
[
"# [DEV ONLY] Clean up workspace",
"_____no_output_____"
]
],
[
[
"!rm -r data_out",
"_____no_output_____"
],
[
"!mkdir data_out",
"_____no_output_____"
],
[
"import configparser\nfrom datetime import datetime\nimport os\n\nfrom pyspark.sql import SparkSession\nfrom pyspark.sql.functions import *\nfrom pyspark.sql.types import *",
"_____no_output_____"
],
[
"config = configparser.ConfigParser()\nconfig.read('dl.cfg')\n\nos.environ['AWS_ACCESS_KEY_ID'] = config['AWS']['AWS_ACCESS_KEY_ID']\nos.environ['AWS_SECRET_ACCESS_KEY'] = config['AWS']['AWS_SECRET_ACCESS_KEY']",
"_____no_output_____"
],
[
"def create_spark_session():\n spark = SparkSession \\\n .builder \\\n .config(\"spark.jars.packages\", \"org.apache.hadoop:hadoop-aws:2.7.0\") \\\n .getOrCreate()\n return spark\n\nspark = create_spark_session()",
"_____no_output_____"
]
],
[
[
"## SONG_DATA",
"_____no_output_____"
]
],
[
[
"def process_song_data(spark, input_data, output_data):\n '''\n Load and process the raw song data. Load JSON files and extract 'songs' and 'artists' tables.\n Store both tables via spark as parquet files.\n\n Parameters\n ----------\n spark : spark instance\n \n input_data : str\n Path to input data (json files)\n output_data : str\n Path to output data (parquet files) - could be local or S3 bucket.\n\n\n Return\n ------\n songs_table : PySpark dataframe\n Content of the dimensions \"songs\"\n artists_table : PySpark dataframe\n Content of the dimensions \"artists\"\n '''\n # get filepath to song data file\n song_data = input_data + '/song_data/*/*/*/*.json'\n \n # Predefined schema\n songSchema = StructType([\n StructField('num_songs', IntegerType())\n , StructField('artist_id', StringType())\n , StructField('artist_latitude', DoubleType())\n , StructField('artist_longitude', DoubleType())\n , StructField('artist_location', StringType())\n , StructField('artist_name', StringType())\n , StructField('song_id', StringType())\n , StructField('title', StringType())\n , StructField('duration', DoubleType())\n , StructField('year', IntegerType())\n ])\n \n # read song data file\n df = spark.read.json(song_data, schema=songSchema)\n if 's3' in input_data.lower():\n print('[SUCCESS] reading song data from AWS S3 bucket.')\n else:\n print('[SUCCESS] reading song data from local file system.')\n\n \n # extract columns to create songs table\n # cols for table 'songs': song_id, title, artist_id, year, duration --> only distinct values\n songs_table = df.select('song_id', 'title', 'artist_id', 'year', 'duration').dropDuplicates()\n \n # write songs table to parquet files partitioned by year and artist\n #songs_table.write.format('parquet').mode('overwrite').partitionBy('year', 'artist_id').saveAsTable(output_data + '/songs') # storing on a DBMS (e.g. Hadoop/Hive)\n songs_table.write.parquet(output_data + '/songs', mode='overwrite', partitionBy=['year', 'artist_id'])\n #songs_table.write.format('parquet').mode('append').partitionBy('year', 'artist_id').saveAsTable('songs')\n\n # extract columns to create artists table\n artists_table = df.select('artist_id', col('artist_name').alias('name'), col('artist_location').alias('location')\n , col('artist_latitude').alias('latitude'), col('artist_longitude').alias('longitude')).dropDuplicates()\n \n # write artists table to parquet files\n #artists_table.write.format('parquet').mode('overwrite').saveAsTable('artists') # storing on a DBMS (e.g. Hadoop/Hive)\n artists_table.write.parquet(output_data + '/artists', mode='overwrite')\n \n return songs_table, artists_table",
"_____no_output_____"
],
[
"def process_log_data(spark, input_data, output_data):\n '''\n Load and process the raw log data. Load JSON files and extract 'users', 'time' and 'songplays' tables.\n Store both tables via spark as parquet files.\n\n Parameters\n ----------\n spark : spark instance\n \n input_data : str\n Path to input data (json files)\n output_data : str\n Path to output data (parquet files) - could be local or S3 bucket.\n\n\n Return\n ------\n users_table : PySpark dataframe\n Content of the dimensions \"users\"\n time_table : PySpark dataframe\n Content of the dimensions \"time\"\n songplays_table: PySpark dataframe\n Content of the fact table \"songplays\"\n '''\n # get filepath to song data file\n song_data = input_data + '/song_data/*/*/*/*.json'\n #log_data = input_data + '/log-data/*.json'\n log_data = input_data + '/log_data/*/*/*.json'\n \n # Predefined schema\n logSchema = StructType([\n StructField('artist', StringType())\n , StructField('auth', StringType())\n , StructField('firstName', StringType())\n , StructField('gender', StringType())\n , StructField('itemInSession', IntegerType())\n , StructField('lastName', StringType())\n , StructField('length', DoubleType())\n , StructField('level', StringType())\n , StructField('location', StringType())\n , StructField('method', StringType())\n , StructField('page', StringType())\n , StructField('registration', StringType())\n , StructField('sessionId', IntegerType())\n , StructField('song', StringType())\n , StructField('status', IntegerType())\n , StructField('ts', IntegerType())\n , StructField('userAgent', IntegerType())\n , StructField('userId', IntegerType())\n ])\n \n # read song data file\n df = spark.read.json(log_data)\n if 's3' in input_data.lower():\n print('[SUCCESS] reading song data from AWS S3 bucket.')\n else:\n print('[SUCCESS] reading song data from local file system.')\n \n # filter by actions for song plays\n print('Dataframe unprocessd: ', str((df.count(), len(df.columns))))\n df = df.filter(lower(col('page')) == 'nextsong')\n print('Dataframe processed: ', str((df.count(), len(df.columns))))\n \n \n # extract columns to create songs table\n # cols for table 'users': user_id, first_name, last_name, gender, level --> distinct by 'user_id'\n users_table = df.select(col('userId').alias('user_id'), col('firstName').alias('first_name'), col('lastName').alias('last_name')\n , 'gender', 'level').dropDuplicates()\n \n \n # write users table to parquet files\n #users_table.write.format('parquet').mode('overwrite').saveAsTable('users') # storing on a DBMS (e.g. Hadoop/Hive)\n users_table.write.parquet(output_data + '/users', mode='overwrite')\n\n # create timestamp column from original timestamp column\n # cols for table 'time': start_time, hour, day, week, month, year, weekday\n #get_timestamp = \n time_table = df.select('ts').dropDuplicates() # get distinct timestamp\n \n # create datetime column from original timestamp column\n time_table = time_table.withColumn('start_time', to_timestamp(col('ts') / 1000))\n #get_datetime = udf()\n #df = \n \n # extract columns to create time table\n time_table = time_table.select('start_time', hour(col('start_time')).alias('hour'), dayofweek(col('start_time')).alias('day')\n , weekofyear(col('start_time')).alias('week'), month(col('start_time')).alias('month')\n , year(col('start_time')).alias('year'), weekofyear(col('start_time')).alias('weekday'))\n \n # write time table to parquet files partitioned by year and month\n #time_table.write.format('parquet').mode('overwrite').saveAsTable('time') # storing on a DBMS (e.g. Hadoop/Hive)\n time_table.write.parquet(output_data + '/time', mode='overwrite', partitionBy=['year', 'month']) # partition to spread the amount of data\n\n # read in song data to use for songplays table\n song_df = spark.read.json(song_data)\n #song_df = spark.read.load(output_data + '/songs', format='parquet')\n\n # extract columns from joined song and log datasets to create songplays table\n songplays_table = df.join(song_df, [df.artist == song_df.artist_name\n , df.song == song_df.title], how='inner')\n songplays_table = songplays_table.withColumn('start_time', to_timestamp(col('ts') / 1000)) # prepare timestamp\n songplays_table = songplays_table.join(time_table.select('start_time', 'month')\n , on=['start_time'], how='inner')\n songplays_table = songplays_table.withColumn('songplay_id', monotonically_increasing_id()) # generate an artificial ID\n\n # merge table parts & select columns\n # cols for 'songplays': songplay_id, start_time, user_id, level, song_id, artist_id, session_id, location, user_agent\n songplays_table = songplays_table.select('songplay_id', 'start_time', col('userId').alias('user_id'), 'level', 'song_id', 'artist_id'\n , col('sessionId').alias('session_id'), 'location', col('userAgent').alias('user_agent'), 'year', 'month')\n\n # write songplays table to parquet files partitioned by year and month\n songplays_table.write.parquet(output_data + '/songplays', mode='overwrite', partitionBy=['year', 'month'])\n if 's3' in output_data.lower():\n print('[SUCCESS] writing songplays data to AWS S3 bucket.')\n else:\n print('[SUCCESS] writing songplays data to local file system.')\n \n \n return users_table, time_table, songplays_table",
"_____no_output_____"
]
],
[
[
"## Execute functions",
"_____no_output_____"
]
],
[
[
"# input & output paths\n#input_data = 'data_in'\n#output_data = 'data_out'\n\ninput_data = 's3://udacity-dend'\noutput_data = 's3://dend-ms-project-4/data_out'\n\nvar_return1, var_return2 = process_song_data(spark, input_data, output_data)\nprint('nrows of:')\nprint('\\tsongs: ', var_return1.count())\nprint('\\tartists: ', var_return2.count())",
"_____no_output_____"
],
[
"var_return3, var_return4, var_return5 = process_log_data(spark, input_data, output_data)\nprint('nrows of:')\nprint('\\tusers: ', var_return3.count())\nprint('\\ttime: ', var_return4.count())\nprint('\\tsongsplays: ', var_return5.count())",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
eccc35347c1a9be48632a9d8f88d47aacee97c7c | 36,758 | ipynb | Jupyter Notebook | site/zh-cn/tutorials/customization/custom_training_walkthrough.ipynb | ilyaspiridonov/docs-l10n | a061a44e40d25028d0a4458094e48ab717d3565c | [
"Apache-2.0"
] | 1 | 2021-09-23T09:56:29.000Z | 2021-09-23T09:56:29.000Z | site/zh-cn/tutorials/customization/custom_training_walkthrough.ipynb | ilyaspiridonov/docs-l10n | a061a44e40d25028d0a4458094e48ab717d3565c | [
"Apache-2.0"
] | null | null | null | site/zh-cn/tutorials/customization/custom_training_walkthrough.ipynb | ilyaspiridonov/docs-l10n | a061a44e40d25028d0a4458094e48ab717d3565c | [
"Apache-2.0"
] | 1 | 2020-06-23T07:43:49.000Z | 2020-06-23T07:43:49.000Z | 32.586879 | 559 | 0.516187 | [
[
[
"##### Copyright 2018 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# 自定义训练: 演示",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://tensorflow.google.cn/tutorials/customization/custom_training_walkthrough\"><img src=\"https://tensorflow.google.cn/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/zh-cn/tutorials/customization/custom_training_walkthrough.ipynb\"><img src=\"https://tensorflow.google.cn/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs-l10n/blob/master/site/zh-cn/tutorials/customization/custom_training_walkthrough.ipynb\"><img src=\"https://tensorflow.google.cn/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/zh-cn/tutorials/customization/custom_training_walkthrough.ipynb\"><img src=\"https://tensorflow.google.cn/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"这个教程将利用机器学习的手段来对鸢尾花按照物种进行分类。本教程将利用 TensorFlow 来进行以下操作:\n1. 构建一个模型,\n2. 用样例数据集对模型进行训练,以及\n3. 利用该模型对未知数据进行预测。\n\n## TensorFlow 编程\n\n本指南采用了以下高级 TensorFlow 概念:\n\n* 使用 TensorFlow 默认的 [eager execution](https://tensorflow.google.cn/guide/eager) 开发环境,\n* 使用 [Datasets API](https://tensorflow.google.cn/guide/datasets) 导入数据,\n* 使用 TensorFlow 的 [Keras API](https://keras.io/getting-started/sequential-model-guide/) 来构建各层以及整个模型。\n\n本教程的结构同很多 TensorFlow 程序相似:\n\n1. 数据集的导入与解析\n2. 选择模型类型\n3. 对模型进行训练\n4. 评估模型效果\n5. 使用训练过的模型进行预测",
"_____no_output_____"
],
[
"## 环境的搭建",
"_____no_output_____"
],
[
"### 配置导入\n\n导入 TensorFlow 以及其他需要的 Python 库。 默认情况下,TensorFlow 用 [eager execution](https://tensorflow.google.cn/guide/eager) 来实时评估操作, 返回具体值而不是建立一个稍后执行的[计算图](https://tensorflow.google.cn/guide/graphs)。 如果您习惯使用 REPL 或 python 交互控制台, 对此您会感觉得心应手。",
"_____no_output_____"
]
],
[
[
"import os\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"print(\"TensorFlow version: {}\".format(tf.__version__))\nprint(\"Eager execution: {}\".format(tf.executing_eagerly()))",
"_____no_output_____"
]
],
[
[
"## 鸢尾花分类问题\n\n 想象一下,您是一名植物学家,正在寻找一种能够对所发现的每株鸢尾花进行自动归类的方法。机器学习可提供多种从统计学上分类花卉的算法。例如,一个复杂的机器学习程序可以根据照片对花卉进行分类。我们的要求并不高 - 我们将根据鸢尾花花萼和花瓣的长度和宽度对其进行分类。\n\n鸢尾属约有 300 个品种,但我们的程序将仅对下列三个品种进行分类:\n\n* 山鸢尾\n* 维吉尼亚鸢尾\n* 变色鸢尾\n\n<table>\n <tr><td>\n <img src=\"https://tensorflow.google.cn/images/iris_three_species.jpg\"\n alt=\"Petal geometry compared for three iris species: Iris setosa, Iris virginica, and Iris versicolor\">\n </td></tr>\n <tr><td align=\"center\">\n <b>Figure 1.</b> <a href=\"https://commons.wikimedia.org/w/index.php?curid=170298\">山鸢尾</a> (by <a href=\"https://commons.wikimedia.org/wiki/User:Radomil\">Radomil</a>, CC BY-SA 3.0), <a href=\"https://commons.wikimedia.org/w/index.php?curid=248095\">变色鸢尾</a>, (by <a href=\"https://commons.wikimedia.org/wiki/User:Dlanglois\">Dlanglois</a>, CC BY-SA 3.0), and <a href=\"https://www.flickr.com/photos/33397993@N05/3352169862\">维吉尼亚鸢尾</a> (by <a href=\"https://www.flickr.com/photos/33397993@N05\">Frank Mayfield</a>, CC BY-SA 2.0).<br/> \n </td></tr>\n</table>\n\n幸运的是,有人已经创建了一个包含有花萼和花瓣的测量值的[120 株鸢尾花的数据集](https://en.wikipedia.org/wiki/Iris_flower_data_set)。这是一个在入门级机器学习分类问题中经常使用的经典数据集。",
"_____no_output_____"
],
[
"## 导入和解析训练数据集\n\n下载数据集文件并将其转换为可供此 Python 程序使用的结构。\n\n### 下载数据集\n\n使用 [tf.keras.utils.get_file](https://tensorflow.google.cn/api_docs/python/tf/keras/utils/get_file) 函数下载训练数据集文件。该函数会返回下载文件的文件路径:",
"_____no_output_____"
]
],
[
[
"train_dataset_url = \"https://storage.googleapis.com/download.tensorflow.org/data/iris_training.csv\"\n\ntrain_dataset_fp = tf.keras.utils.get_file(fname=os.path.basename(train_dataset_url),\n origin=train_dataset_url)\n\nprint(\"Local copy of the dataset file: {}\".format(train_dataset_fp))",
"_____no_output_____"
]
],
[
[
"### 检查数据\n\n数据集 `iris_training.csv` 是一个纯文本文件,其中存储了逗号分隔值 (CSV) 格式的表格式数据.请使用 `head -n5` 命令查看前 5 个条目:",
"_____no_output_____"
]
],
[
[
"!head -n5 {train_dataset_fp}",
"_____no_output_____"
]
],
[
[
"我们可以从该数据集视图中注意到以下信息:\n1. 第一行是表头,其中包含数据集信息:\n\n* 共有 120 个样本。每个样本都有四个特征和一个标签名称,标签名称有三种可能。\n2. 后面的行是数据记录,每个[样本](https://developers.google.com/machine-learning/glossary/#example)各占一行,其中:\n * 前四个字段是[特征](https://developers.google.com/machine-learning/glossary/#feature): 这四个字段代表的是样本的特点。在此数据集中,这些字段存储的是代表花卉测量值的浮点数。\n * 最后一列是[标签](https://developers.google.com/machine-learning/glossary/#label):即我们想要预测的值。对于此数据集,该值为 0、1 或 2 中的某个整数值(每个值分别对应一个花卉名称)。\n\n我们用代码表示出来:",
"_____no_output_____"
]
],
[
[
"# CSV文件中列的顺序\ncolumn_names = ['sepal_length', 'sepal_width', 'petal_length', 'petal_width', 'species']\n\nfeature_names = column_names[:-1]\nlabel_name = column_names[-1]\n\nprint(\"Features: {}\".format(feature_names))\nprint(\"Label: {}\".format(label_name))",
"_____no_output_____"
]
],
[
[
"每个标签都分别与一个字符串名称(例如 “setosa” )相关联,但机器学习通常依赖于数字值。标签编号会映射到一个指定的表示法,例如:\n\n* `0` : 山鸢尾\n* `1` : 变色鸢尾\n* `2` : 维吉尼亚鸢尾\n\n如需详细了解特征和标签,请参阅 [《机器学习速成课程》的“机器学习术语”部分](https://developers.google.com/machine-learning/crash-course/framing/ml-terminology).",
"_____no_output_____"
]
],
[
[
"class_names = ['Iris setosa', 'Iris versicolor', 'Iris virginica']",
"_____no_output_____"
]
],
[
[
"### 创建一个 `tf.data.Dataset`\n\nTensorFlow的 [Dataset API](https://tensorflow.google.cn/guide/datasets) 可处理在向模型加载数据时遇到的许多常见情况。这是一种高阶 API ,用于读取数据并将其转换为可供训练使用的格式。如需了解详情,请参阅[数据集快速入门指南](https://tensorflow.google.cn/get_started/datasets_quickstart) \n\n\n由于数据集是 CSV 格式的文本文件,请使用 [make_csv_dataset](https://tensorflow.google.cn/api_docs/python/tf/data/experimental/make_csv_dataset) 函数将数据解析为合适的格式。由于此函数为训练模型生成数据,默认行为是对数据进行随机处理 (`shuffle=True, shuffle_buffer_size=10000`),并且无限期重复数据集(`num_epochs=None`)。 我们还设置了 [batch_size](https://developers.google.com/machine-learning/glossary/#batch_size) 参数:",
"_____no_output_____"
]
],
[
[
"batch_size = 32\n\ntrain_dataset = tf.data.experimental.make_csv_dataset(\n train_dataset_fp,\n batch_size,\n column_names=column_names,\n label_name=label_name,\n num_epochs=1)",
"_____no_output_____"
]
],
[
[
" `make_csv_dataset` 返回一个`(features, label)` 对构建的 `tf.data.Dataset` ,其中 `features` 是一个字典: `{'feature_name': value}`\n\n这些 `Dataset` 对象是可迭代的。 我们来看看下面的一些特征:",
"_____no_output_____"
]
],
[
[
"features, labels = next(iter(train_dataset))\n\nprint(features)",
"_____no_output_____"
]
],
[
[
"注意到具有相似特征的样本会归为一组,即分为一批。更改 `batch_size` 可以设置存储在这些特征数组中的样本数。\n\n绘制该批次中的几个特征后,就会开始看到一些集群现象:",
"_____no_output_____"
]
],
[
[
"plt.scatter(features['petal_length'],\n features['sepal_length'],\n c=labels,\n cmap='viridis')\n\nplt.xlabel(\"Petal length\")\nplt.ylabel(\"Sepal length\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"要简化模型构建步骤,请创建一个函数以将特征字典重新打包为形状为 `(batch_size, num_features)` 的单个数组。\n\n此函数使用 [tf.stack](https://tensorflow.google.cn/api_docs/python/tf/stack) 方法,该方法从张量列表中获取值,并创建指定维度的组合张量:",
"_____no_output_____"
]
],
[
[
"def pack_features_vector(features, labels):\n \"\"\"将特征打包到一个数组中\"\"\"\n features = tf.stack(list(features.values()), axis=1)\n return features, labels",
"_____no_output_____"
]
],
[
[
"然后使用 [tf.data.Dataset.map](https://tensorflow.google.cn/api_docs/python/tf/data/dataset/map) 方法将每个 `(features,label)` 对中的 `features` 打包到训练数据集中:",
"_____no_output_____"
]
],
[
[
"train_dataset = train_dataset.map(pack_features_vector)",
"_____no_output_____"
]
],
[
[
" `Dataset` 的特征元素被构成了形如 `(batch_size, num_features)` 的数组。我们来看看前几个样本:",
"_____no_output_____"
]
],
[
[
"features, labels = next(iter(train_dataset))\n\nprint(features[:5])",
"_____no_output_____"
]
],
[
[
"## 选择模型类型\n\n### 为何要使用模型?\n\n[模型](https://developers.google.com/machine-learning/crash-course/glossary#model)是指特征与标签之间的关系。对于鸢尾花分类问题,模型定义了花萼和花瓣测量值与预测的鸢尾花品种之间的关系。一些简单的模型可以用几行代数进行描述,但复杂的机器学习模型拥有大量难以汇总的参数。\n\n\n您能否在不使用机器学习的情况下确定四个特征与鸢尾花品种之间的关系?也就是说,您能否使用传统编程技巧(例如大量条件语句)创建模型?也许能,前提是反复分析该数据集,并最终确定花瓣和花萼测量值与特定品种的关系。对于更复杂的数据集来说,这会变得非常困难,或许根本就做不到。一个好的机器学习方法可为您确定模型。如果您将足够多的代表性样本馈送到正确类型的机器学习模型中,该程序便会为您找出相应的关系。\n\n### 选择模型\n\n我们需要选择要进行训练的模型类型。模型具有许多类型,挑选合适的类型需要一定的经验。本教程使用神经网络来解决鸢尾花分类问题。[神经网络](https://developers.google.com/machine-learning/glossary/#neural_network)可以发现特征与标签之间的复杂关系。神经网络是一个高度结构化的图,其中包含一个或多个[隐含层](https://developers.google.com/machine-learning/glossary/#hidden_layer)。每个隐含层都包含一个或多个[神经元](https://developers.google.com/machine-learning/glossary/#neuron)。 神经网络有多种类别,该程序使用的是密集型神经网络,也称为[全连接神经网络](https://developers.google.com/machine-learning/glossary/#fully_connected_layer) : 一个层中的神经元将从上一层中的每个神经元获取输入连接。例如,图 2 显示了一个密集型神经网络,其中包含 1 个输入层、2 个隐藏层以及 1 个输出层:\n\n<table>\n <tr><td>\n <img src=\"https://tensorflow.google.cn/images/custom_estimators/full_network.png\"\n alt=\"网络结构示意图: 输入层, 2 隐含层, 输出层\">\n </td></tr>\n <tr><td align=\"center\">\n <b>图 2.</b> 包含特征、隐藏层和预测的神经网络<br/> \n </td></tr>\n</table>\n\n当图 2 中的模型经过训练并获得无标签样本后,它会产生 3 个预测结果:相应鸢尾花属于指定品种的可能性。这种预测称为[推理](https://developers.google.com/machine-learning/crash-course/glossary#inference)。对于该示例,输出预测结果的总和是 1.0。在图 2 中,该预测结果分解如下:山鸢尾为 0.02,变色鸢尾为 0.95,维吉尼亚鸢尾为 0.03。这意味着该模型预测某个无标签鸢尾花样本是变色鸢尾的概率为 95%。",
"_____no_output_____"
],
[
"### 使用 Keras 创建模型\n\nTensorFlow [tf.keras](https://tensorflow.google.cn/api_docs/python/tf/keras) API 是创建模型和层的首选方式。通过该 API,您可以轻松地构建模型并进行实验,而将所有部分连接在一起的复杂工作则由 Keras 处理。\n\n[tf.keras.Sequential](https://tensorflow.google.cn/api_docs/python/tf/keras/Sequential) 模型是层的线性堆叠。该模型的构造函数会采用一系列层实例;在本示例中,采用的是 2 个[密集层](https://tensorflow.google.cn/api_docs/python/tf/keras/layers/Dense)(各自包含10个节点),以及 1 个输出层(包含 3 个代表标签预测的节点。第一个层的 `input_shape` 参数对应该数据集中的特征数量,它是一项必需参数:",
"_____no_output_____"
]
],
[
[
"model = tf.keras.Sequential([\n tf.keras.layers.Dense(10, activation=tf.nn.relu, input_shape=(4,)), # 需要给出输入的形式\n tf.keras.layers.Dense(10, activation=tf.nn.relu),\n tf.keras.layers.Dense(3)\n])",
"_____no_output_____"
]
],
[
[
"[激活函数](https://developers.google.com/machine-learning/crash-course/glossary#activation_function)可决定层中每个节点的输出形式。 这些非线性关系很重要,如果没有它们,模型将等同于单个层。[激活函数](https://tensorflow.google.cn/api_docs/python/tf/keras/activations)有很多种,但隐藏层通常使用 [ReLU](https://developers.google.com/machine-learning/crash-course/glossary#ReLU)。\n\n隐藏层和神经元的理想数量取决于问题和数据集。与机器学习的多个方面一样,选择最佳的神经网络形状需要一定的知识水平和实验基础。一般来说,增加隐藏层和神经元的数量通常会产生更强大的模型,而这需要更多数据才能有效地进行训练。",
"_____no_output_____"
],
[
"### 使用模型\n\n我们快速了解一下此模型如何处理一批特征:\n",
"_____no_output_____"
]
],
[
[
"predictions = model(features)\npredictions[:5]",
"_____no_output_____"
]
],
[
[
"在此示例中,每个样本针对每个类别返回一个 [logit](https://developers.google.com/machine-learning/crash-course/glossary#logits)。\n\n要将这些对数转换为每个类别的概率,请使用 [softmax](https://developers.google.com/machine-learning/crash-course/glossary#softmax) 函数:",
"_____no_output_____"
]
],
[
[
"tf.nn.softmax(predictions[:5])",
"_____no_output_____"
]
],
[
[
"对每个类别执行 `tf.argmax` 运算可得出预测的类别索引。不过,该模型尚未接受训练,因此这些预测并不理想。",
"_____no_output_____"
]
],
[
[
"print(\"Prediction: {}\".format(tf.argmax(predictions, axis=1)))\nprint(\" Labels: {}\".format(labels))",
"_____no_output_____"
]
],
[
[
"## 训练模型\n\n[训练](https://developers.google.com/machine-learning/crash-course/glossary#training) 是一个机器学习阶段,在此阶段中,模型会逐渐得到优化,也就是说,模型会了解数据集。目标是充分了解训练数据集的结构,以便对未见过的数据进行预测。如果您从训练数据集中获得了过多的信息,预测便会仅适用于模型见过的数据,但是无法泛化。此问题被称之为[过拟合](https://developers.google.com/machine-learning/crash-course/glossary#overfitting)—就好比将答案死记硬背下来,而不去理解问题的解决方式。\n\n鸢尾花分类问题是[监督式机器学习](https://developers.google.com/machine-learning/glossary/#supervised_machine_learning)的一个示例: 模型通过包含标签的样本加以训练。 而在[非监督式机器学习](https://developers.google.com/machine-learning/glossary/#unsupervised_machine_learning)中,样本不包含标签。相反,模型通常会在特征中发现一些规律。",
"_____no_output_____"
],
[
"### 定义损失和梯度函数\n\n在训练和评估阶段,我们都需要计算模型的[损失](https://developers.google.com/machine-learning/crash-course/glossary#loss)。 这样可以衡量模型的预测结果与预期标签有多大偏差,也就是说,模型的效果有多差。我们希望尽可能减小或优化这个值。\n\n我们的模型会使用 `tf.keras.losses.SparseCategoricalCrossentropy` 函数计算其损失,此函数会接受模型的类别概率预测结果和预期标签,然后返回样本的平均损失。",
"_____no_output_____"
]
],
[
[
"loss_object = tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True)",
"_____no_output_____"
],
[
"def loss(model, x, y):\n y_ = model(x)\n\n return loss_object(y_true=y, y_pred=y_)\n\n\nl = loss(model, features, labels)\nprint(\"Loss test: {}\".format(l))",
"_____no_output_____"
]
],
[
[
"使用 [tf.GradientTape](https://tensorflow.google.cn/api_docs/python/tf/GradientTape) 的前后关系来计算[梯度](https://developers.google.com/machine-learning/crash-course/glossary#gradient)以优化你的模型:",
"_____no_output_____"
]
],
[
[
"def grad(model, inputs, targets):\n with tf.GradientTape() as tape:\n loss_value = loss(model, inputs, targets)\n return loss_value, tape.gradient(loss_value, model.trainable_variables)",
"_____no_output_____"
]
],
[
[
"### 创建优化器\n\n[优化器](https://developers.google.com/machine-learning/crash-course/glossary#optimizer) 会将计算出的梯度应用于模型的变量,以使 `loss` 函数最小化。您可以将损失函数想象为一个曲面(见图 3),我们希望通过到处走动找到该曲面的最低点。梯度指向最高速上升的方向,因此我们将沿相反的方向向下移动。我们以迭代方式计算每个批次的损失和梯度,以在训练过程中调整模型。模型会逐渐找到权重和偏差的最佳组合,从而将损失降至最低。损失越低,模型的预测效果就越好。\n\n<table>\n <tr><td>\n <img src=\"https://cs231n.github.io/assets/nn3/opt1.gif\" width=\"70%\"\n alt=\"Optimization algorithms visualized over time in 3D space.\">\n </td></tr>\n <tr><td align=\"center\">\n <b>图 3.</b> 优化算法在三维空间中随时间推移而变化的可视化效果。<br/>(来源: <a href=\"http://cs231n.github.io/neural-networks-3/\">斯坦福大学 CS231n 课程</a>,MIT 许可证,Image credit: <a href=\"https://twitter.com/alecrad\">Alec Radford</a>)\n </td></tr>\n</table>\n\nTensorFlow有许多可用于训练的[优化算法](https://tensorflow.google.cn/api_guides/python/train)。此模型使用的是 [tf.train.GradientDescentOptimizer](https://tensorflow.google.cn/api_docs/python/tf/train/GradientDescentOptimizer) , 它可以实现[随机梯度下降法](https://developers.google.com/machine-learning/crash-course/glossary#gradient_descent)(SGD)。`learning_rate` 被用于设置每次迭代(向下行走)的步长。 这是一个 *超参数* ,您通常需要调整此参数以获得更好的结果。",
"_____no_output_____"
],
[
"我们来设置优化器:",
"_____no_output_____"
]
],
[
[
"optimizer = tf.keras.optimizers.Adam(learning_rate=0.01)",
"_____no_output_____"
]
],
[
[
"我们将使用它来计算单个优化步骤:",
"_____no_output_____"
]
],
[
[
"loss_value, grads = grad(model, features, labels)\n\nprint(\"Step: {}, Initial Loss: {}\".format(optimizer.iterations.numpy(),\n loss_value.numpy()))\n\noptimizer.apply_gradients(zip(grads, model.trainable_variables))\n\nprint(\"Step: {}, Loss: {}\".format(optimizer.iterations.numpy(),\n loss(model, features, labels).numpy()))",
"_____no_output_____"
]
],
[
[
"### 训练循环\n\n\n一切准备就绪后,就可以开始训练模型了!训练循环会将数据集样本馈送到模型中,以帮助模型做出更好的预测。以下代码块可设置这些训练步骤:\n\n1. 迭代每个周期。通过一次数据集即为一个周期。\n2. 在一个周期中,遍历训练 `Dataset` 中的每个样本,并获取样本的*特征*(`x`)和*标签*(`y`)。\n3. 根据样本的特征进行预测,并比较预测结果和标签。衡量预测结果的不准确性,并使用所得的值计算模型的损失和梯度。\n4. 使用 `optimizer` 更新模型的变量。\n5. 跟踪一些统计信息以进行可视化。\n6. 对每个周期重复执行以上步骤。\n\n\n`num_epochs` 变量是遍历数据集集合的次数。与直觉恰恰相反的是,训练模型的时间越长,并不能保证模型就越好。`num_epochs` 是一个可以调整的[超参数](https://developers.google.com/machine-learning/glossary/#hyperparameter)。选择正确的次数通常需要一定的经验和实验基础。",
"_____no_output_____"
]
],
[
[
"## Note: 使用相同的模型变量重新运行此单元\n\n# 保留结果用于绘制\ntrain_loss_results = []\ntrain_accuracy_results = []\n\nnum_epochs = 201\n\nfor epoch in range(num_epochs):\n epoch_loss_avg = tf.keras.metrics.Mean()\n epoch_accuracy = tf.keras.metrics.SparseCategoricalAccuracy()\n\n # Training loop - using batches of 32\n for x, y in train_dataset:\n # 优化模型\n loss_value, grads = grad(model, x, y)\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n\n # 追踪进度\n epoch_loss_avg(loss_value) # 添加当前的 batch loss\n # 比较预测标签与真实标签\n epoch_accuracy(y, model(x))\n\n # 循环结束\n train_loss_results.append(epoch_loss_avg.result())\n train_accuracy_results.append(epoch_accuracy.result())\n\n if epoch % 50 == 0:\n print(\"Epoch {:03d}: Loss: {:.3f}, Accuracy: {:.3%}\".format(epoch,\n epoch_loss_avg.result(),\n epoch_accuracy.result()))",
"_____no_output_____"
]
],
[
[
"### 可视化损失函数随时间推移而变化的情况",
"_____no_output_____"
],
[
"虽然输出模型的训练过程有帮助,但查看这一过程往往*更有帮助*。 [TensorBoard](https://tensorflow.google.cn/guide/summaries_and_tensorboard) 是与 TensorFlow 封装在一起的出色可视化工具,不过我们可以使用 `matplotlib` 模块创建基本图表。\n\n解读这些图表需要一定的经验,不过您确实希望看到*损失*下降且*准确率*上升。",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(2, sharex=True, figsize=(12, 8))\nfig.suptitle('Training Metrics')\n\naxes[0].set_ylabel(\"Loss\", fontsize=14)\naxes[0].plot(train_loss_results)\n\naxes[1].set_ylabel(\"Accuracy\", fontsize=14)\naxes[1].set_xlabel(\"Epoch\", fontsize=14)\naxes[1].plot(train_accuracy_results)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 评估模型的效果\n\n模型已经过训练,现在我们可以获取一些关于其效果的统计信息了。\n\n*评估* 指的是确定模型做出预测的效果。要确定模型在鸢尾花分类方面的效果,请将一些花萼和花瓣测量值传递给模型,并要求模型预测它们所代表的鸢尾花品种。然后,将模型的预测结果与实际标签进行比较。例如,如果模型对一半输入样本的品种预测正确,则 [准确率](https://developers.google.com/machine-learning/glossary/#accuracy) 为 `0.5` 。 图 4 显示的是一个效果更好一些的模型,该模型做出 5 次预测,其中有 4 次正确,准确率为 80%:\n\n<table cellpadding=\"8\" border=\"0\">\n <colgroup>\n <col span=\"4\" >\n <col span=\"1\" bgcolor=\"lightblue\">\n <col span=\"1\" bgcolor=\"lightgreen\">\n </colgroup>\n <tr bgcolor=\"lightgray\">\n <th colspan=\"4\">样本特征</th>\n <th colspan=\"1\">标签</th>\n <th colspan=\"1\" >模型预测</th>\n </tr>\n <tr>\n <td>5.9</td><td>3.0</td><td>4.3</td><td>1.5</td><td align=\"center\">1</td><td align=\"center\">1</td>\n </tr>\n <tr>\n <td>6.9</td><td>3.1</td><td>5.4</td><td>2.1</td><td align=\"center\">2</td><td align=\"center\">2</td>\n </tr>\n <tr>\n <td>5.1</td><td>3.3</td><td>1.7</td><td>0.5</td><td align=\"center\">0</td><td align=\"center\">0</td>\n </tr>\n <tr>\n <td>6.0</td> <td>3.4</td> <td>4.5</td> <td>1.6</td> <td align=\"center\">1</td><td align=\"center\" bgcolor=\"red\">2</td>\n </tr>\n <tr>\n <td>5.5</td><td>2.5</td><td>4.0</td><td>1.3</td><td align=\"center\">1</td><td align=\"center\">1</td>\n </tr>\n <tr><td align=\"center\" colspan=\"6\">\n <b>图 4.</b> 准确率为 80% 的鸢尾花分类器<br/> \n </td></tr>\n</table>",
"_____no_output_____"
],
[
"### 建立测试数据集\n\n评估模型与训练模型相似。最大的区别在于,样本来自一个单独的[测试集](https://developers.google.com/machine-learning/crash-course/glossary#test_set),而不是训练集。为了公正地评估模型的效果,用于评估模型的样本务必与用于训练模型的样本不同。\n\n测试 `Dataset` 的建立与训练 `Dataset` 相似。下载 CSV 文本文件并解析相应的值,然后对数据稍加随机化处理:",
"_____no_output_____"
]
],
[
[
"test_url = \"https://storage.googleapis.com/download.tensorflow.org/data/iris_test.csv\"\n\ntest_fp = tf.keras.utils.get_file(fname=os.path.basename(test_url),\n origin=test_url)",
"_____no_output_____"
],
[
"test_dataset = tf.data.experimental.make_csv_dataset(\n test_fp,\n batch_size,\n column_names=column_names,\n label_name='species',\n num_epochs=1,\n shuffle=False)\n\ntest_dataset = test_dataset.map(pack_features_vector)",
"_____no_output_____"
]
],
[
[
"### 根据测试数据集评估模型\n\n与训练阶段不同,模型仅评估测试数据的一个[周期](https://developers.google.com/machine-learning/glossary/#epoch)。在以下代码单元格中,我们会遍历测试集中的每个样本,然后将模型的预测结果与实际标签进行比较。这是为了衡量模型在整个测试集中的准确率。",
"_____no_output_____"
]
],
[
[
"test_accuracy = tf.keras.metrics.Accuracy()\n\nfor (x, y) in test_dataset:\n logits = model(x)\n prediction = tf.argmax(logits, axis=1, output_type=tf.int32)\n test_accuracy(prediction, y)\n\nprint(\"Test set accuracy: {:.3%}\".format(test_accuracy.result()))",
"_____no_output_____"
]
],
[
[
"例如,我们可以看到对于最后一批数据,该模型通常预测正确:",
"_____no_output_____"
]
],
[
[
"tf.stack([y,prediction],axis=1)",
"_____no_output_____"
]
],
[
[
"## 使用经过训练的模型进行预测\n\n我们已经训练了一个模型并“证明”它是有效的,但在对鸢尾花品种进行分类方面,这还不够。现在,我们使用经过训练的模型对 [无标签样本](https://developers.google.com/machine-learning/glossary/#unlabeled_example)(即包含特征但不包含标签的样本)进行一些预测。\n\n在现实生活中,无标签样本可能来自很多不同的来源,包括应用、CSV 文件和数据 Feed。暂时我们将手动提供三个无标签样本以预测其标签。回想一下,标签编号会映射到一个指定的表示法:\n\n* `0`: 山鸢尾\n* `1`: 变色鸢尾\n* `2`: 维吉尼亚鸢尾",
"_____no_output_____"
]
],
[
[
"predict_dataset = tf.convert_to_tensor([\n [5.1, 3.3, 1.7, 0.5,],\n [5.9, 3.0, 4.2, 1.5,],\n [6.9, 3.1, 5.4, 2.1]\n])\n\npredictions = model(predict_dataset)\n\nfor i, logits in enumerate(predictions):\n class_idx = tf.argmax(logits).numpy()\n p = tf.nn.softmax(logits)[class_idx]\n name = class_names[class_idx]\n print(\"Example {} prediction: {} ({:4.1f}%)\".format(i, name, 100*p))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
eccc3739565f9f88222964f0cde2273e492095cc | 114,832 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Space Analysis -checkpoint.ipynb | unmrds/cc-python | 65a02aefc6428e5ed6e8ba8be1a6c179331a1e57 | [
"Apache-2.0"
] | 2 | 2020-03-27T15:08:03.000Z | 2020-06-03T20:17:51.000Z | .ipynb_checkpoints/Space Analysis -checkpoint.ipynb | unmrds/cc-python | 65a02aefc6428e5ed6e8ba8be1a6c179331a1e57 | [
"Apache-2.0"
] | null | null | null | .ipynb_checkpoints/Space Analysis -checkpoint.ipynb | unmrds/cc-python | 65a02aefc6428e5ed6e8ba8be1a6c179331a1e57 | [
"Apache-2.0"
] | 2 | 2017-09-13T02:23:19.000Z | 2021-11-12T17:31:16.000Z | 153.930295 | 45,414 | 0.849424 | [
[
[
"# Space Analysis Demonstration\n\nWe are currently in the process of testing a strategy for capturing shelf space data and would like to use the growing dataset to better understand the time required to collect the data and perform some preliminary analyses of the data. Some questions that we would like to ask of this dataset include: \n\n* How many shelves can be measured per hour\n* How much shelf space is taken up by each (LC subject level) range in the dataset\n* How much shelf space is available within each range in the dataset\n\nIn support of answering these questions we need to do the following:\n\n* Import the data into the script for analysis\n* Aggregate the data by hour and count the number of shelves that were measured in each hour\n* Aggregate the occupied space for all of the shelves for each range\n* Calculate the total shelf space for each range",
"_____no_output_____"
],
[
"## 1. Import the data for the analysis\n\nImporting the data for the analysis includes the following sub-tasks:\n\n* Point to the local data file that containes the values that we want to analyze\n* Create a file-like object from which we can read the data\n* Read the data into a variable that we can use in our analyses",
"_____no_output_____"
]
],
[
[
"# Import a very useful and powerful module for interacting with tabular data\nimport pandas as pd\n\n# Install and import tabulate for generating tables for hardcopy reports\n!TABULATE_INSTALL=lib-only; pip install tabulate\nfrom tabulate import tabulate\n\n# Set up the report generation variables\nreport_file_name = 'report.md'\nreport_content = []\nprint(\"The generated report will be: \" + file_name)\n\nprint(\"The version of pandas is: \" + pd.__version__)\n\n# Define the location of the file of interest\nfile_path = \"\" # include a trailing \"/\" if not empty\nfile_name = \"Space_Analysis_Pilot_(Responses).csv\"\nfile_location = file_path + file_name\nprint(\"The file that will be read is: \" + file_location)\n\n# explicitly define the column names that will be associated with our table - this helps mitigate any\n# strangeness in the column names in the source CSV file\ncolumn_names=[\n \"timestamp\", \n \"building\", \n \"floor\", \n \"range\", \n \"section\", \n \"shelf\",\n \"leading_alpha\", \n \"leading_numeric\",\n \"ending_alpha\",\n \"ending_numeric\",\n \"occupied_in\",\n \"stacked\"\n]\n\n# Load the referenced file into a pandas dataframe for use in our analysis\nshelf_data = pd.read_csv(\n file_location,\n names = column_names,\n header = 0,\n usecols = ['timestamp','leading_alpha','ending_alpha','occupied_in'],\n skipinitialspace = True\n)\n\n# create a series of datatime values from the timestamps in the dataframe\n# attempt to coerce error generating values, if can't set value to NaT (missing)\nshelf_data['datetime'] = pd.to_datetime(shelf_data.loc[:,\"timestamp\"], errors='coerce')\n\n# fill the NA values in the leading and ending alpha fields with a symbol\nshelf_data.leading_alpha = shelf_data.leading_alpha.fillna('*')\nshelf_data.ending_alpha = shelf_data.ending_alpha.fillna('*')\nshelf_data.occupied_in.loc[(shelf_data.leading_alpha == \"*\")] = 0\n\n\n# drop the rows that are missing leading_alpha values\nshelf_data = shelf_data.loc[shelf_data.leading_alpha.notnull()]\n\n# strip any leading or trailing white space from the alpha fields\nshelf_data.leading_alpha.astype(str)\nshelf_data['leading_alpha'] = shelf_data.leading_alpha.str.strip()\nshelf_data['ending_alpha'] = shelf_data.ending_alpha.str.strip()\n\n# generate columns with alpha prefixes for higher-level grouping\nshelf_data['leading_prefix'] = shelf_data.leading_alpha.str[0]\nshelf_data['ending_prefix'] = shelf_data.ending_alpha.str[0]\n\n# add shelf capacity to each row in the table\nshelf_data['capacity'] = 35.5\n\nprint(\"\\nThe data types in the table are:\\n\"+str(shelf_data.dtypes))\nprint()\n#print(shelf_data)",
"Requirement already satisfied (use --upgrade to upgrade): tabulate in /opt/conda/lib/python3.5/site-packages\n\u001b[33mYou are using pip version 8.1.2, however version 9.0.1 is available.\nYou should consider upgrading via the 'pip install --upgrade pip' command.\u001b[0m\nThe generated report will be: Space_Analysis_Pilot_(Responses).csv\nThe version of pandas is: 0.18.1\nThe file that will be read is: Space_Analysis_Pilot_(Responses).csv\n\nThe data types in the table are:\ntimestamp object\nleading_alpha object\nending_alpha object\noccupied_in float64\ndatetime datetime64[ns]\nleading_prefix object\nending_prefix object\ncapacity float64\ndtype: object\n\n"
]
],
[
[
"## 2. Aggregate the data by hour and count the shelves for each hour\n\nThe following sub-tasks are required for this part of our analysis\n\n* Create a new column in the dataframe that contains a distinct value for each hour in the database\n* Group the individual shelves by hour label\n* Count the shelves for each hour\n* Plot the number of shelves per hour for the analysis period\n ",
"_____no_output_____"
]
],
[
[
"# create a dataframe that has a subset of rows that only contain valid datetime values\nclean_dates = shelf_data[shelf_data.datetime.notnull()]\nclean_dates = clean_dates.set_index('datetime')\n#clean_dates['hour'] = clean_dates['datetime'].strftime('%Y-%M-%d %H')\n\nclean_dates\n\n# resample the timestamps by hour and return the frequency distribution\nr = clean_dates.resample(\"1H\")\n\nr_count = r.count() # .loc[(r_count.timestamp > 0)]\n\nr_count.timestamp",
"_____no_output_____"
],
[
"r_count['timestamp'].plot()",
"_____no_output_____"
]
],
[
[
"## 3. Aggregate the occupied space for all of the shelves for each range\n\nThe following sub-tasks are required for this part of our analysis:\n\n* Total the occupied space for all of the whole shelves - i.e. those where the start and end range alpha values are the same\n* Estimate the occupied space for all of the partial shelves\n* Add these numbers up for an estimate of the total occupied space for each range",
"_____no_output_____"
]
],
[
[
"full_shelves = shelf_data.loc[(shelf_data.leading_alpha == shelf_data.ending_alpha)]\npartial_shelves = shelf_data.loc[(shelf_data.leading_alpha != shelf_data.ending_alpha)].loc[(shelf_data.leading_alpha.notnull())]\n\n# convert our column to numeric values instead of the string type it defaulted to - adding strings is not \n# what we want to do. Partial shelves are arbitrarily assigned half of the occupied value and the capacity. \npd.to_numeric(full_shelves['occupied_in'], errors=\"coerce\")\npartial_shelves['partial_occupied_in'] = pd.to_numeric(partial_shelves['occupied_in'], errors=\"coerce\") / 2\npartial_shelves['partial_capacity'] = pd.to_numeric(partial_shelves['capacity'], errors=\"coerce\") / 2\n\n\n#print(full_shelves[['leading_alpha','ending_alpha','occupied_in','capacity']])\n#print(partial_shelves[['leading_alpha','ending_alpha','partial_occupied_in','partial_capacity']])",
"_____no_output_____"
]
],
[
[
"## 4. Calculate occupied and total shelf space for each LC subject\n\nFor this we need to:\n\n* Group the individual shelf data by LC subject categories\n* Calculate the occupied and total capacity for each group\n* Calculate some derived values for reporting",
"_____no_output_____"
]
],
[
[
"# import a useful library for doing calculations on arrays - including our data table and its columns\nimport numpy as np\n\n# group our full and partial shelves by the leading_alpha subject string\nfull_shelves_by_range = full_shelves.groupby('leading_alpha')\npartial_shelves_by_range = partial_shelves.groupby('leading_alpha')\n\n# calculate the sum for all of the numeric columns for our full and partial shelves\nfull_ranges = full_shelves_by_range.aggregate(np.sum)\npartial_ranges = partial_shelves_by_range.aggregate(np.sum)\n\n# calculate some new columns of data for each range\ncombined_by_lc = pd.concat([full_ranges,partial_ranges], axis=1)\ncombined_by_lc['total_occupied_in'] = combined_by_lc[['occupied_in','partial_occupied_in']].sum(axis=1)\ncombined_by_lc['total_capacity_in'] = combined_by_lc[['capacity','partial_capacity']].sum(axis=1)\ncombined_by_lc['total_empty_in'] = combined_by_lc['total_capacity_in'] - combined_by_lc['total_occupied_in']\ncombined_by_lc['total_empty_pct'] = 100 * (1-combined_by_lc['total_occupied_in'] / combined_by_lc['total_capacity_in'])\n\n# now let's generate some plots for fun\n\nlc_plot = combined_by_lc['total_empty_in'].plot(kind=\"barh\",stacked=False,figsize={8,10}, title=\"Empty space (in) for each LC subject\")\nlc_fig = lc_plot.get_figure()\nlc_fig.savefig(\"lc_fig.png\")\n\nreport_content.append(\"\"\"\n\n\n\"\"\")\n\n# cumulative = combined_by_lc[['total_occupied_in','total_capacity_in','total_empty_in']].cumsum()\n# print(cumulative)\n\n#cumulative.plot(kind=\"barh\",stacked=True,figsize={8,10})",
"_____no_output_____"
]
],
[
[
"## 5. Repeat the above range-level analyses for the full top-level LC subject category prefixes",
"_____no_output_____"
]
],
[
[
"prefix_full_shelves = shelf_data.loc[(shelf_data.leading_prefix == shelf_data.ending_prefix)]\nprefix_partial_shelves = shelf_data.loc[(shelf_data.leading_prefix != shelf_data.ending_prefix)].loc[(shelf_data.leading_prefix.notnull())]\n\n# convert our column to numeric values instead of the string type it defaulted to - adding strings is not \n# what we want to do. Partial shelves are arbitrarily assigned half of the occupied value and the capacity. \npd.to_numeric(prefix_full_shelves['occupied_in'], errors=\"coerce\")\nprefix_partial_shelves['partial_occupied_in'] = pd.to_numeric(prefix_partial_shelves['occupied_in'], errors=\"coerce\") / 2\nprefix_partial_shelves['partial_capacity'] = pd.to_numeric(prefix_partial_shelves['capacity'], errors=\"coerce\") / 2\n\n\n#print(full_shelves[['leading_prefix','ending_prefix','occupied_in','capacity']])\n#print(partial_shelves[['leading_prefix','ending_prefix','partial_occupied_in','partial_capacity']])\n\n# import a useful library for doing calculations on arrays - including our data table and its columns\nimport numpy as np\n\n# group our full and partial shelves by the leading_alpha subject string\nprefix_full_shelves_by_range = prefix_full_shelves.groupby('leading_prefix')\nprefix_partial_shelves_by_range = prefix_partial_shelves.groupby('leading_prefix')\n\n# calculate the sum for all of the numeric columns for our full and partial shelves\nprefix_full_ranges = prefix_full_shelves_by_range.aggregate(np.sum)\nprefix_partial_ranges = prefix_partial_shelves_by_range.aggregate(np.sum)\n\n# calculate some new columns of data for each range\nprefix_combined_by_lc = pd.concat([prefix_full_ranges,prefix_partial_ranges], axis=1)\nprefix_combined_by_lc['total_occupied_in'] = prefix_combined_by_lc[['occupied_in','partial_occupied_in']].sum(axis=1)\nprefix_combined_by_lc['total_capacity_in'] = prefix_combined_by_lc[['capacity','partial_capacity']].sum(axis=1)\nprefix_combined_by_lc['total_empty_in'] = prefix_combined_by_lc['total_capacity_in'] - prefix_combined_by_lc['total_occupied_in']\nprefix_combined_by_lc['total_empty_pct'] = 100 * (1-prefix_combined_by_lc['total_occupied_in'] / prefix_combined_by_lc['total_capacity_in'])\n\n# now let's generate some plots for fun\n\nprefix_plot = prefix_combined_by_lc['total_empty_in'].plot(kind=\"barh\",stacked=False,figsize={8,10}, title=\"Empty space (in) for each LC subject\")\nprefix_fig = prefix_plot.get_figure()\nprefix_fig.savefig(\"prefix_fig.png\")\n\nreport_content.append(\"\"\"\n\n\n\"\"\")\n\n\n\n# prefix_cumulative = prefix_combined_by_lc[['total_occupied_in','total_capacity_in','total_empty_in']].cumsum()\n# print(cumulative)\n\n#cumulative.plot(kind=\"barh\",stacked=True,figsize={8,10})",
"_____no_output_____"
]
],
[
[
"## Generate a report with the results",
"_____no_output_____"
]
],
[
[
"report_content.append(\"\"\"\n# UNM College of University Libraries and Learning Sciences Shelf Space Analysis\n\nThis is where some intro text will go ...\n\"\"\")\n\n# calculate some overall totals for all LC subjects\nprefix_grand_total_capacity_in = prefix_combined_by_lc['total_capacity_in'].sum()\nprefix_grand_total_occupied_in = prefix_combined_by_lc['total_occupied_in'].sum()\nprefix_grand_total_empty_in = prefix_combined_by_lc['total_empty_in'].sum()\nprefix_grand_total_empty_pct = 100 * (prefix_grand_total_empty_in / prefix_grand_total_capacity_in)\n\n# print out the generated information\nprint(\"For Library of Congress Subject Prefixes\")\nprint(\"\\tTotal Capacity (in): \" + str(prefix_grand_total_capacity_in))\nprint(\"\\tTotal Occupied (in): \" + str(prefix_grand_total_occupied_in))\nprint(\"\\tTotal Empty Space (in): \" + str(prefix_grand_total_empty_in))\nprint(\"\\tTotal Empty Space (pct): \" + str(prefix_grand_total_empty_pct))\nprint()\n\n# write the information into the report\nreport_content.append(\"\"\"\n## Summary information for LC subject prefixes\n\nTotal Capacity (in): %s\\\\\nTotal Occupied (in): %s\\\\\nTotal Empty Space (in): %s\\\\\nTotal Empty Space (pct): %s\\\\\n\n\"\"\"%(\n prefix_grand_total_capacity_in,\n prefix_grand_total_occupied_in,\n prefix_grand_total_empty_in,\n prefix_grand_total_empty_pct\n ) \n)\n\n# calculate some overall totals for all LC subjects\ngrand_total_capacity_in = combined_by_lc['total_capacity_in'].sum()\ngrand_total_occupied_in = combined_by_lc['total_occupied_in'].sum()\ngrand_total_empty_in = combined_by_lc['total_empty_in'].sum()\ngrand_total_empty_pct = 100 * (grand_total_empty_in / grand_total_capacity_in)\n\n# print out the generated information\nprint(\"For Library of Congress Subjects\")\nprint(\"\\tTotal Capacity (in): \" + str(grand_total_capacity_in))\nprint(\"\\tTotal Occupied (in): \" + str(grand_total_occupied_in))\nprint(\"\\tTotal Empty Space (in): \" + str(grand_total_empty_in))\nprint(\"\\tTotal Empty Space (pct): \" + str(grand_total_empty_pct))\nprint()\n\n# write the information into the report\nreport_content.append(\"\"\"\n## Summary information for LC subjects\n\nTotal Capacity (in): %s\\\\\nTotal Occupied (in): %s\\\\\nTotal Empty Space (in): %s\\\\\nTotal Empty Space (pct): %s\\\\\n\n\"\"\"%(\n grand_total_capacity_in,\n grand_total_occupied_in,\n grand_total_empty_in,\n grand_total_empty_pct\n ) \n)\n",
"For Library of Congress Subject Prefixes\n\tTotal Capacity (in): 144786.75\n\tTotal Occupied (in): 129149.0\n\tTotal Empty Space (in): 15637.75\n\tTotal Empty Space (pct): 10.800539414\n\nFor Library of Congress Subjects\n\tTotal Capacity (in): 145532.25\n\tTotal Occupied (in): 129815.5\n\tTotal Empty Space (in): 15716.75\n\tTotal Empty Space (pct): 10.7994963316\n\n"
],
[
"# generate some tables for the report\nprefix_table = tabulate(prefix_combined_by_lc[['total_occupied_in','total_capacity_in','total_empty_in', 'total_empty_pct']], headers=[\"Occupied (in)\",\"Capacity (in)\",\"Empty (in)\",\"Empty (pct)\"])\nsubject_table = tabulate(combined_by_lc[['total_occupied_in','total_capacity_in','total_empty_in', 'total_empty_pct']], headers=[\"Occupied (in)\",\"Capacity (in)\",\"Empty (in)\",\"Empty (pct)\"])\n\nreport_content.append(\"\"\"\n## Summary Tables\n\n%s\n\nTable: LC Prefix Space Summary Table\n\n%s\n\nTable: LC Subject Space Summary Table\n\n\n\"\"\"%(prefix_table, subject_table))\n\n\nprint(prefix_table)\nprint(subject_table)",
" Occupied (in) Capacity (in) Empty (in) Empty (pct)\n-- --------------- --------------- ------------ -------------\n* 0 2130 2130 100\nA 4220 4828 608 12.5932\nB 57100 63403 6303 9.94117\nC 8464 9744.75 1280.75 13.143\nD 59365 64681 5316 8.2188\n Occupied (in) Capacity (in) Empty (in) Empty (pct)\n--- --------------- --------------- ------------ -------------\n* 0 2130 2130 100\nA 45 53.25 8.25 15.493\nAC 1214.5 1402.25 187.75 13.3892\nAE 1231 1420 189 13.3099\nAG 331.5 372.75 41.25 11.0664\nAI 49.5 53.25 3.75 7.04225\nAM 366 390.5 24.5 6.27401\nAP 207.5 230.75 23.25 10.0758\nAS 621 727.75 106.75 14.6685\nAY 78.5 88.75 10.25 11.5493\nAZ 180 213 33 15.493\nB 13594.5 15637.8 2043.25 13.0661\nBC 776 940.75 164.75 17.5126\nBD 2322.5 2680.25 357.75 13.3476\nBH 750 869.75 119.75 13.7683\nBJ 2166 2502.75 336.75 13.4552\nBL 6730 7508.25 778.25 10.3653\nBM 2034.5 2183.25 148.75 6.81324\nBP 1585 1721.75 136.75 7.9425\nBQ 1073 1171.5 98.5 8.40802\nBR 5478 6106 628 10.285\nBS 4828 5023.25 195.25 3.88693\nBT 3012 3195 183 5.7277\nBV 2500 2751.25 251.25 9.13221\nBX 10405 11289 884 7.83063\nC 36 53.25 17.25 32.3944\nCB 2039 2378.5 339.5 14.2737\nCC 1207.5 1366.75 159.25 11.6517\nCD 1330 1544.25 214.25 13.874\nCE 51 53.25 2.25 4.22535\nCJ 175 195.25 20.25 10.3713\nCN 106.5 124.25 17.75 14.2857\nCR 217 230.75 13.75 5.95883\nCS 699.5 798.75 99.25 12.4257\nCT 2726 3141.75 415.75 13.2331\nD 13744 14892.2 1148.25 7.71039\nDA 10809 11626.2 817.25 7.02935\nDAW 51 53.25 2.25 4.22535\nDB 1027.5 1082.75 55.25 5.10275\nDC 4652.5 5094.25 441.75 8.67154\nDD 3400 3567.75 167.75 4.70184\nDE 352 372.75 20.75 5.56673\nDF 1531.5 1686.25 154.75 9.17717\nDG 2861.5 3106.25 244.75 7.87928\nDH 247.5 266.25 18.75 7.04225\nDJ 121.5 124.25 2.75 2.21328\nDJK 233 266.25 33.25 12.4883\nDK 4562.5 5058.75 496.25 9.80974\nDL 399 443.75 44.75 10.0845\nDP 7748 8715.25 967.25 11.0984\nDQ 144 159.75 15.75 9.85915\nDR 932.5 1011.75 79.25 7.83296\nDS 6832 7455 623 8.35681\n"
],
[
"# generate the report\n\nwith open(report_file_name,\"w\") as f:\n for block in report_content:\n f.write(block)\n\npandoc_options = \"-o %s.pdf %s\"%(report_file_name,report_file_name)\n \nimport subprocess\nsubprocess.run(['pandoc', \"-o %s.pdf\"%(report_file_name), report_file_name])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
eccc4c589d73ce4de40763c9d30bfe233d4156e8 | 12,562 | ipynb | Jupyter Notebook | Labs/vision/01b - Image Classification.ipynb | hakimzulkufli/azure-cognitive-labs | b9ab095729b50f7d4c28f849a528f53449898e2d | [
"MIT"
] | 2 | 2020-07-22T05:14:54.000Z | 2020-11-28T11:26:08.000Z | Labs/vision/01b - Image Classification.ipynb | hakimzulkufli/azure-cognitive-labs | b9ab095729b50f7d4c28f849a528f53449898e2d | [
"MIT"
] | null | null | null | Labs/vision/01b - Image Classification.ipynb | hakimzulkufli/azure-cognitive-labs | b9ab095729b50f7d4c28f849a528f53449898e2d | [
"MIT"
] | 6 | 2020-07-18T09:16:35.000Z | 2020-11-28T12:22:15.000Z | 70.971751 | 512 | 0.646314 | [
[
[
"# Image Classification\n\nThe *Computer Vision* cognitive service provides useful pre-built models for working with images, but you'll often need to train your own model for computer vision. For example, suppose the Northwind Traders retail company wants to create an automated checkout system that identifies the grocery items customers want to buy based on an image taken by a camera at the checkout. To do this, you'll need to train a classification model that can classify the images to identify the item being purchased.\n\n<p style='text-align:center'><img src='./images/image-classification.jpg' alt='A robot holding a clipboard, classifying pictures of an apple, a banana, and an orange'/></p>\n\nIn Azure, you can use the ***Custom Vision*** cognitive service to train an image classification model based on existing images. There are two elements to creating an image classification solution. First, you must train a model to recognize different classes using existing images. Then, when the model is trained you must publish it as a service that can be consumed by applications.\n\n## Create a Custom Vision resource\n\nTo use the Custom Vision service, you need an Azure resource that you can use to *train* a model, and a resource with which you can *publish* it for applications to use. The resource for either (or both) tasks can be a general **Cognitive Services** resource, or a specific **Custom Vision** resource. You can use the same Cognitive Services resource for each of these tasks, or you can use different resources (in the same region) for each task to manage costs separately.\n\nUse the following instructions to create a new **Custom Vision** resource.\n\n1. In a new browser tab, open the Azure portal at [https://portal.azure.com](https://portal.azure.com), and sign in using the Microsoft account associated with your Azure subscription.\n2. Select the **+Create a resource** button, search for *custom vision*, and create a **Custom Vision** resource with the following settings:\n - **Create options**: Both\n - **Subscription**: *Your Azure subscription*\n - **Resource group**: *Create a new resource group with a unique name*\n - **Name**: *Enter a unique name*\n - **Training location**: *Choose any available region*\n - **Training pricing tier**: F0\n - **Prediction location**: *The same region as the training resource*\n - **Prediction pricing tier**: F0\n\n > **Note**: If you already have an F0 custom vision service in your subscription, select **S0** for this one.\n\n3. Wait for the resources to be created, and note that two Custom Vision resources are provisioned; one for training, an another for prediction. You can view these by navigating to the resource group where you created them.\n\n## Create a Custom Vision project\n\nTo train an object detection model, you need to create a Custom Vision project based on your training resource. To do this, you'll use the Custom Vision portal.\n\n1. Download and extract the training images from the [onedrive link](https://1drv.ms/u/s!AmjwdE_MMESkgalJABa30-OObS92dg?e=c7D4Wy).\n2. In another browser tab, open the Custom Vision portal at [https://customvision.ai](https://customvision.ai). If prompted, sign in using the Microsoft account associated with your Azure subscription and agree to the terms of service.\n3. In the Custom Vision portal, create a new project with the following settings:\n - **Name**: Grocery Checkout\n - **Description**: Image classification for groceries\n - **Resource**: *The Custom Vision resource you created previously*\n - **Project Types**: Classification\n - **Classification Types**: Multiclass (single tag per image)\n - **Domains**: Food\n4. Click **\\[+\\] Add images**, and select all of the files in the **golden eagle** folder you extracted previously. Then upload the image files, specifying the tag *golden eagle*, like this:\n <p style='text-align:center'><img src='./images/upload_golden_eagle.jpg' alt='Upload golden eagle with golden eagle tag'/></p>\n5. Repeat the previous step to upload the images in the **bald eagle** folder with the tag *bald eagle*.\n6. Explore the images you have uploaded in the Custom Vision project - there should be 15 images of each class, like this:\n <p style='text-align:center'><img src='./images/eagles.jpg' alt='Tagged images of Birds - 15 Golden Eagle, 15 Bald Eagles'/></p>\n7. In the Custom Vision project, above the images, click **Train** to train a classification model using the tagged images. Select the **Quick Training** option, and then wait for the training iteration to complete (this may take a minute or so).\n8. When the model iteration has been trained, review the *Precision*, *Recall*, and *AP* performance metrics - these measure the prediction accuracy of the classification model, and should all be high.\n\n## Test the model\n\nBefore publishing this iteration of the model for applications to use, you should test it.\n\n1. Above the performance metrics, click **Quick Test**.\n2. In the **Image URL** box, type `https://1drv.ms/u/s!AmjwdE_MMESkgalSz7OrMVWoGUk5tQ?e=8daIb3` and click ➔\n3. View the predictions returned by your model - the probability score for *golden eagle* should be the highest, like this:\n <p style='text-align:center'><img src='./images/test-eagle.jpg' alt='An image with a class prediction of golden eagle'/></p>\n4. Close the **Quick Test** window.\n\n## Publish and consume the image classification model\n\nNow you're ready to publish your trained model and use it from a client application.\n\n9. Click **🗸 Publish** to publish the trained model with the following settings:\n - **Model name**: groceries\n - **Prediction Resource**: *The prediction resource you created previously*.\n10. After publishing, click the *settings* (⚙) icon at the top right of the **Performance** page to view the project settings. Then, under **General** (on the left), copy the **Project Id** and paste it into the code cell below (replacing **YOUR_PROJECT_ID**).\n <p style='text-align:center'><img src='./images/cv_project_settings.jpg' alt='Project ID in project settings'/></p>\n\n> _**Note**: If you used a **Cognitive Services** resource instead of creating a **Custom Vision** resource at the beginning of this exercise, you can copy its key and endpoint from the right side of the project settings, paste it into the code cell below, and run it to see the results. Otherwise, continue completing the steps below to get the key and endpoint for your Custom Vision prediction resource._\n\n11. At the top left of the **Project Settings** page, click the *Projects Gallery* (👁) icon to return to the Custom Vision portal home page, where your project is now listed.\n12. On the Custom Vision portal home page, at the top right, click the *settings* (⚙) icon to view the settings for your Custom Vision service. Then, under **Resources**, expand your *prediction* resource (<u>not</u> the training resource) and copy its **Key** and **Endpoint** values to the code cell below, replacing **YOUR_KEY** and **YOUR_ENDPOINT**.\n <p style='text-align:center'><img src='./images/cv_settings.jpg' alt='Prediction resource key and endpoint in custom vision settings'/></p>\n13. Run the code cell below to set the variables to your project ID, key, and endpoint values.",
"_____no_output_____"
]
],
[
[
"project_id = 'YOUR_PROJECT_ID'\ncv_key = 'YOUR_KEY'\ncv_endpoint = 'YOUR_ENDPOINT'\n\nmodel_name = 'groceries' # this must match the model name you set when publishing your model iteration (it's case-sensitive)!\nprint('Ready to predict using model {} in project {}'.format(model_name, project_id))",
"_____no_output_____"
]
],
[
[
"Client applications can use the details above to connect to and your custom vision classification model.\n\nRun the following code cell to classifiy a selection of test images using your published model.\n\n> **Note**: Don't worry too much about the details of the code. It uses the Computer Vision SDK for Python to get a class prediction for each image in the /data/image-classification/test-fruit folder",
"_____no_output_____"
]
],
[
[
"from azure.cognitiveservices.vision.customvision.prediction import CustomVisionPredictionClient\nfrom msrest.authentication import ApiKeyCredentials\nimport matplotlib.pyplot as plt\nfrom PIL import Image\nimport os\n%matplotlib inline\n\n# Get the test images from the data/vision/test folder\ntest_folder = os.path.join('data', 'image-classification', 'test-eagle')\ntest_images = os.listdir(test_folder)\n\n# Create an instance of the prediction service\ncredentials = ApiKeyCredentials(in_headers={\"Prediction-key\": cv_key})\ncustom_vision_client = CustomVisionPredictionClient(endpoint=cv_endpoint, credentials=credentials)\n\n# Create a figure to display the results\nfig = plt.figure(figsize=(16, 8))\n\n# Get the images and show the predicted classes for each one\nprint('Classifying images in {} ...'.format(test_folder))\nfor i in range(len(test_images)):\n # Open the image, and use the custom vision model to classify it\n image_contents = open(os.path.join(test_folder, test_images[i]), \"rb\")\n classification = custom_vision_client.classify_image(project_id, model_name, image_contents.read())\n # The results include a prediction for each tag, in descending order of probability - get the first one\n prediction = classification.predictions[0].tag_name\n # Display the image with its predicted class\n img = Image.open(os.path.join(test_folder, test_images[i]))\n a=fig.add_subplot(len(test_images)/3, 3,i+1)\n a.axis('off')\n imgplot = plt.imshow(img)\n a.set_title(prediction)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Hopefully, your image classification model has correctly identified the birds in the images.\n\n## Learn more\n\nThe Custom Vision service offers more capabilities than we've explored in this exercise. For example, you can also use the Custom Vision service to create *object detection* models; which not only classify objects in images, but also identify *bounding boxes* that show the location of the object in the image.\n\nTo learn more about the Custom Vision cognitive service, view the [Custom Vision documentation](https://docs.microsoft.com/azure/cognitive-services/custom-vision-service/home)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eccc51d675abc0cf2450ff768941ea70c0849d32 | 24,195 | ipynb | Jupyter Notebook | experiments/multitask/old/mtbo_reizman.ipynb | ddceruti/summit | e4e6bbfc0e655c402436bde9fd1da9fb3fa6a286 | [
"MIT"
] | 60 | 2020-09-10T00:00:03.000Z | 2022-03-08T10:45:02.000Z | experiments/multitask/old/mtbo_reizman.ipynb | ddceruti/summit | e4e6bbfc0e655c402436bde9fd1da9fb3fa6a286 | [
"MIT"
] | 57 | 2020-09-07T11:06:15.000Z | 2022-02-16T16:30:48.000Z | experiments/multitask/old/mtbo_reizman.ipynb | ddceruti/summit | e4e6bbfc0e655c402436bde9fd1da9fb3fa6a286 | [
"MIT"
] | 12 | 2020-09-07T12:43:19.000Z | 2022-02-26T09:58:01.000Z | 40.527638 | 1,005 | 0.475015 | [
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"# MTBO Reizman Study",
"_____no_output_____"
]
],
[
[
"from summit.strategies import MTBO, Transform, LHS, Chimera\nfrom summit.benchmarks import ReizmanSuzukiEmulator, BaumgartnerCrossCouplingEmulator\nfrom summit.utils.dataset import DataSet\nfrom summit.domain import *\nimport summit\nimport pathlib\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"## Create DataSet",
"_____no_output_____"
]
],
[
[
"# Path to data\nsummit_path = pathlib.Path(summit.__file__).parents[0]\ndata_path = summit_path / \"benchmarks\" / \"experiment_emulator\" / \"data\"",
"_____no_output_____"
],
[
"# Import Reizman-Suzuki Data\ndatasets = [DataSet.read_csv(data_path / f\"reizman_suzuki_case{i}_train_test.csv\") for i in range(1,5)]\nfor i, dataset in enumerate(datasets):\n dataset[('task', 'METADATA')] = i\ndatasets[0].tail(5)",
"_____no_output_____"
],
[
"exp = ReizmanSuzukiEmulator(case=1)\nexp.domain",
"_____no_output_____"
],
[
"lhs = LHS(exp.domain)\nconditions = lhs.suggest_experiments(5)\nexp.run_experiments(conditions)",
"_____no_output_____"
],
[
"exp.reset()\nhierarchy = {\"yield\": {\"hierarchy\": 0, \"tolerance\": 0.5}, \"ton\": {\"hierarchy\": 1, \"tolerance\": 1}}\ntransform = Chimera(exp.domain, hierarchy=hierarchy)\nstrategy = MTBO(exp.domain, \n pretraining_data=datasets[1],\n transform=transform,\n categorical_method=\"one-hot\", \n task=0)\nr = summit.Runner(strategy=strategy, experiment=exp, max_iterations=10)\nr.run()",
"_____no_output_____"
],
[
"exp.pareto_plot(colorbar=True)",
"_____no_output_____"
],
[
"exp.data",
"_____no_output_____"
],
[
"new_exp = ReizmanSuzukiEmulator(case=2)\n# new_exp.train(dataset=datasets[1], verbose=False)",
"_____no_output_____"
],
[
"lhs = LHS(new_exp.domain)\nconditions = lhs.suggest_experiments(10)\nnew_exp.run_experiments(conditions)",
"_____no_output_____"
],
[
"pd_cn = BaumgartnerCrossCouplingEmulator()\npd_cn.domain",
"_____no_output_____"
],
[
"lhs = LHS(pd_cn.domain, categorical_method=\"descriptors\")\nconditions = lhs.suggest_experiments(10)\npd_cn.run_experiments(conditions)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccc5dd787d84f2c762c04e355870324eaae2b32 | 152,509 | ipynb | Jupyter Notebook | nlp/bert/fastai_hugginface_transformers.ipynb | martin-fabbri/colab-notebooks | 03658a7772fbe71612e584bbc767009f78246b6b | [
"Apache-2.0"
] | 8 | 2020-01-18T18:39:49.000Z | 2022-02-17T19:32:26.000Z | nlp/bert/fastai_hugginface_transformers.ipynb | martin-fabbri/colab-notebooks | 03658a7772fbe71612e584bbc767009f78246b6b | [
"Apache-2.0"
] | null | null | null | nlp/bert/fastai_hugginface_transformers.ipynb | martin-fabbri/colab-notebooks | 03658a7772fbe71612e584bbc767009f78246b6b | [
"Apache-2.0"
] | 6 | 2020-01-18T18:40:02.000Z | 2020-09-27T09:26:38.000Z | 47.629294 | 12,346 | 0.569934 | [
[
[
"<a href=\"https://colab.research.google.com/github/martin-fabbri/colab-notebooks/blob/master/nlp/bert/fastai_hugginface_transformers.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Fastai 2 HuggingFace Transormers",
"_____no_output_____"
]
],
[
[
"! [ -e /content ] && pip install -Uqq fastai\n!pip install -Uqq transformers",
"\u001b[K |████████████████████████████████| 194kB 5.2MB/s \n\u001b[K |████████████████████████████████| 61kB 4.2MB/s \n\u001b[K |████████████████████████████████| 1.4MB 5.8MB/s \n\u001b[K |████████████████████████████████| 890kB 42.6MB/s \n\u001b[K |████████████████████████████████| 2.9MB 44.1MB/s \n\u001b[?25h Building wheel for sacremoses (setup.py) ... \u001b[?25l\u001b[?25hdone\n"
]
],
[
[
"GPT2LMHeadModel -> Language Model",
"_____no_output_____"
]
],
[
[
"!nvidia-smi -L",
"GPU 0: Tesla V100-SXM2-16GB (UUID: GPU-29388e88-61d3-8576-61fb-3d92da8cd3ea)\n"
],
[
"#!cat /proc/cpuinfo",
"_____no_output_____"
],
[
"import torch\nfrom transformers import GPT2LMHeadModel\nfrom transformers import GPT2TokenizerFast\nfrom fastai.text.all import *\nfrom psutil import virtual_memory",
"_____no_output_____"
],
[
"ram_gb = virtual_memory().total / 1e9\nprint(f'Runtime has {ram_gb:.1f} gigabytes of available RAM')",
"Runtime has 13.7 gigabytes of available RAM\n"
],
[
"pretrained_weights = 'gpt2'\ntokenizer = GPT2TokenizerFast.from_pretrained(pretrained_weights)\nmodel = GPT2LMHeadModel.from_pretrained(pretrained_weights)",
"_____no_output_____"
],
[
"ram_gb = virtual_memory().total / 1e9\nprint(f'Runtime has {ram_gb:.1f} gigabytes of available RAM')",
"Runtime has 13.7 gigabytes of available RAM\n"
],
[
"ids = tokenizer.encode('Just another test text testing huggingfaces')\nids",
"_____no_output_____"
],
[
"tokenizer.decode(ids)",
"_____no_output_____"
],
[
"t = torch.LongTensor(ids)[None]\npreds = model.generate(t)",
"Setting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\n"
],
[
"preds.shape, preds[0]",
"_____no_output_____"
],
[
"tokenizer.decode(preds[0].numpy())",
"_____no_output_____"
],
[
"ids2 = tokenizer.encode('Deep Learning rules!')\nt2 = torch.LongTensor(ids2)[None]\npreds2 = model.generate(t2)\ntokenizer.decode(preds2[0].numpy())",
"Setting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\n"
],
[
"ids3 = tokenizer.encode('Machine Learning is everywhere.')\nt3 = torch.LongTensor(ids3)[None]\npreds3 = model.generate(t3)\ntokenizer.decode(preds3[0].numpy())",
"Setting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\n"
],
[
"path = untar_data(URLs.WIKITEXT_TINY)\npath.ls()",
"_____no_output_____"
],
[
"df_train = pd.read_csv(path/'train.csv', header=None)\ndf_valid = pd.read_csv(path/'test.csv', header=None)\nprint('df_train.shape ->', df_train.shape)\ndf_train.head()",
"df_train.shape -> (615, 1)\n"
],
[
"all_texts = np.concatenate([df_train[0].values, df_valid[0].values])",
"_____no_output_____"
]
],
[
[
"In a fastai `Transform` you can define:\n- an <code>encodes</code> method that is applied when you call the transform (a bit like the `forward` method in a `nn.Module`)\n- a <code>decodes</code> method that is applied when you call the `decode` method of the transform, if you need to decode anything for showing purposes (like converting ids to a text here)\n- a <code>setups</code> method that sets some inner state of the `Transform` (not needed here so we skip it)",
"_____no_output_____"
]
],
[
[
"class TransformersTokenizer(Transform):\n def __init__(self, tokenizer): \n self.tokenizer = tokenizer\n \n def encodes(self, x):\n toks = self.tokenizer.tokenize(x)\n toks = self.tokenizer.tokenize(x)\n return tensor(self.tokenizer.convert_tokens_to_ids(toks))\n return tensor(self.tokenizer.convert_tokens_to_ids(toks))\n\n def decodes(self, x):\n return TitledStr(self.tokenizer.decode(x.cpu().numpy()))",
"_____no_output_____"
],
[
"tok = tokenizer.tokenize('Just some simple text.')\ntok",
"_____no_output_____"
],
[
"tokenizer.convert_tokens_to_ids(tok)",
"_____no_output_____"
],
[
"tokenizer.encode('Just some simple text.')",
"_____no_output_____"
]
],
[
[
"Two comments on the code above:\n- in <code>encodes</code> we don't use the `tokenizer.encode` method since it does some additional preprocessing for the model after tokenizing and numericalizing (the part throwing a warning before). Here we don't need any post-processing so it's fine to skip it.\n- in <code>decodes</code> we return a `TitledStr` object and not just a plain string. That's a fastai class that adds a `show` method to the string, which will allow us to use all the fastai show methods.",
"_____no_output_____"
],
[
"You can then group your data with this `Transform` using a `TfmdLists`. It has an s in its name because it contains the training and validation set. We indicate the indices of the training set and the validation set with `splits` (here all the first indices until `len(df_train)` and then all the remaining indices):",
"_____no_output_____"
]
],
[
[
"splits = [range_of(df_train), list(range(len(df_train), len(all_texts)))]\ntls = TfmdLists(\n all_texts, \n TransformersTokenizer(tokenizer), # endode/decode -> GPT2TokenizerFast \n splits=splits, \n dl_type=LMDataLoader # Language Model DataLoader???\n)",
"_____no_output_____"
],
[
"len(range_of(df_train)), range_of(df_train)[:5]",
"_____no_output_____"
],
[
"len(all_texts)",
"_____no_output_____"
]
],
[
[
"We specify `dl_type=LMDataLoader` for when we will convert this `TfmdLists` to `DataLoaders`: we will use an `LMDataLoader` since we have a language modeling problem, not the usual fastai `TfmdDL`.\n\nIn a `TfmdLists` you can access the elements of the training or validation set quite easily:",
"_____no_output_____"
]
],
[
[
"tls.train[0], tls.valid[0]",
"_____no_output_____"
],
[
"tls.tfms(tls.train.items[0]).shape, tls.tfms(tls.valid.items[0]).shape",
"_____no_output_____"
],
[
"show_at(tls.train, 0)",
" \n = 2013 – 14 York City F.C. season = \n \n The 2013 – 14 season was the <unk> season of competitive association football and 77th season in the Football League played by York City Football Club, a professional football club based in York, North Yorkshire, England. Their 17th @-@ place finish in 2012 – 13 meant it was their second consecutive season in League Two. The season ran from 1 July 2013 to 30 June 2014. \n Nigel Worthington, starting his first full season as York manager, made eight permanent summer signings. By the turn of the year York were only above the relegation zone on goal difference, before a 17 @-@ match unbeaten run saw the team finish in seventh @-@ place in the 24 @-@ team 2013 – 14 Football League Two. This meant York qualified for the play @-@ offs, and they were eliminated in the semi @-@ final by Fleetwood Town. York were knocked out of the 2013 – 14 FA Cup, Football League Cup and Football League Trophy in their opening round matches. \n 35 players made at least one appearance in nationally organised first @-@ team competition, and there were 12 different <unk>. Defender Ben Davies missed only five of the fifty @-@ two competitive matches played over the season. Wes Fletcher finished as leading scorer with 13 goals, of which 10 came in league competition and three came in the FA Cup. The winner of the <unk> of the Year award, voted for by the club's supporters, was <unk> Oyebanjo. \n \n = = Background and pre @-@ season = = \n \n The 2012 – 13 season was York City's first season back in the Football League, having won the Conference Premier play @-@ offs in 2011 – 12 after <unk> years in the Football Conference. Manager Gary Mills was sacked in March 2013 following an 11 @-@ match run without a victory, and was replaced by former Northern Ireland manager Nigel Worthington. Despite being in the relegation zone with three matches remaining, Worthington led the team to safety from relegation after a 1 – 0 win away to Dagenham & Redbridge on the final day of the season. York finished the season in 17th @-@ place in the 2012 – 13 League Two table. \n Following the previous season's conclusion Lee <unk>, Jon <unk>, Chris <unk>, Ben Everson, Scott Kerr, David <unk>, Patrick <unk>, Michael Potts, Jamie Reed and Jason Walker were released by York, while <unk> Blair departed for Fleetwood Town. David McGurk, <unk> Oyebanjo, Danny Parslow, Tom Platt and Chris Smith signed new contracts with the club. New players signed ahead of the start of the season were goalkeeper Chris <unk> on a season @-@ long loan from Blackpool, defender Ben Davies on loan from Preston North End, midfielders Craig Clay from Chesterfield and Lewis Montrose from Gillingham, winger <unk> Puri from St <unk> and strikers Ryan Bowman from Hereford United, Richard Cresswell from Sheffield United, Wes Fletcher from Burnley and Ryan Jarvis from Torquay United. Defender Mike Atkinson and striker Chris Dickinson entered the first @-@ team squad from the youth team after agreeing professional contracts. \n York retained the previous season's home and away kits. The home kit comprised red shirts with white sleeves, light blue shorts and white socks. The away kit included light blue shirts with white sleeves, white shorts and light blue socks. <unk> Health continued as shirt sponsors for the second successive season. \n \n = = Review = = \n \n \n = = = August = = = \n \n York began the season with a 1 – 0 home win over the previous season's play @-@ off finalists, Northampton Town, with <unk> Jarvis scoring the winning goal in the 90th @-@ minute. However, defeat came in York's match against Championship side Burnley in the first round of the League Cup, going down 4 – 0 at home. The team endured their first league defeat of the season in the following game after being beaten 2 – 0 away by Dagenham & Redbridge, the home team scoring in each half. York then held Hartlepool United to a 0 – 0 home draw, before being beaten 3 – 2 away by Bristol Rovers, in which Jarvis scored twice before John @-@ Joe O 'Toole scored the winning goal for the home team in the 67th @-@ minute. Two signings were made shortly before the transfer deadline ; defender George Taft was signed on a one @-@ month loan from Leicester City, while Middlesbrough midfielder Ryan Brobbel joined on a one @-@ month loan. <unk> John <unk>, who had been told he had no future with the club, departed after signing for FC Halifax Town. Jarvis gave York the lead away at Exeter City before Alan <unk> scored in each half to see the home team win 2 – 1. \n \n = = = September = = = \n \n York suffered their first home league defeat of the season after AFC Wimbledon won 2 – 0, with Michael Smith scoring in each half. Former Ipswich Town midfielder Josh Carson, who had a spell on loan with York the previous season, signed a contract until the end of 2013 – 14 and Sheffield United midfielder Elliott <unk> signed on a one @-@ month loan. Brobbel opened the scoring in the second minute of his home debut against Mansfield Town, although the away team went on to score twice to win 2 – 1. York's run of four defeats ended following a 1 – 1 draw away to Wycombe Wanderers, in which McGurk gave York the lead before the home team levelled through Dean Morgan. Taft was sent back to Leicester after he fell behind McGurk, Parslow and Smith in the pecking order for a central defensive berth. York achieved their first win since the opening day of the season after beating Portsmouth 4 – 2 at home, with Fletcher ( 2 ), Montrose and Jarvis scoring. \n \n = = = October = = = \n \n Defender Luke O 'Neill was signed from Burnley on a 28 @-@ day emergency loan. He made his debut in York's 3 – 0 win away at Torquay, which was the team's first successive win of the season. York were knocked out of the Football League Trophy in the second round after being beaten 3 – 0 at home by League One team Rotherham United, before their winning streak in the league was ended with a 3 – 0 defeat away to Newport County. York drew 2 – 2 away to Chesterfield, having taken a two @-@ goal lead through O 'Neill and Jarvis, before the home team fought back through Armand <unk> and Jay O <unk>. The team then hosted Fleetwood Town, and the visitors won 2 – 0 with goals scored in each half by Gareth Evans and <unk> Matt. Scunthorpe United were beaten 4 – 1 at home to end York's three @-@ match run without a win, with all the team's goals coming in the first half from Carson, Fletcher and Brobbel ( 2 ). \n \n = = = November = = = \n \n Bowman scored his first goals for York away to Cheltenham Town, as York twice fought back from behind to draw 2 – 2. York drew 3 – 3 away to Bristol Rovers to earn a first round replay in the FA Cup, taking the lead through Jarvis before Eliot Richards equalised for the home team. Carson scored a 30 yard volley to put York back in the lead, and after Bristol Rovers goals from Matt <unk> and Chris <unk>, Fletcher scored an 86th @-@ minute equaliser for York. Bowman scored with a header from an O 'Neill cross to open the scoring at home to Plymouth Argyle, which was the first goal the visitors had conceded in 500 minutes of action. However, Plymouth equalised 11 minutes later through <unk> <unk> and the match finished a 1 – 1 draw. York were knocked out of the FA Cup after losing 3 – 2 at home to Bristol Rovers in a first round replay ; the visitors were 3 – 0 up by 50 @-@ minutes before Fletcher pulled two back for York with a penalty and a long @-@ range strike. \n Defender Keith Lowe, of Cheltenham, and goalkeeper Nick Pope, of Charlton Athletic, were signed on loan until January 2014. They both played in York's first league defeat in four weeks, 2 – 1 away, to Southend United. <unk> <unk> gave Southend the lead early into the match and Bowman equalised for York with a low strike during the second half, before Luke Prosser scored the winning goal for the home side in stoppage time. With Pope preferred in goal, <unk> returned to Blackpool on his own accord, although his loan agreement would stay in place until January 2014. York then drew 0 – 0 away to Morecambe. After Pope was recalled from his loan by Charlton, York signed Wolverhampton Wanderers goalkeeper Aaron McCarey on loan until January 2014. McCarey kept a clean sheet in York's 0 – 0 home draw with Rochdale. \n \n = = = December = = = \n \n Cresswell retired from playing as a result of an eye complaint and a knee injury. York drew 1 – 1 away to Burton Albion, with an own goal scored by Shane <unk> @-@ <unk> giving York the lead in the 64th @-@ minute before the home team equalised eight minutes later through Billy <unk>. Atkinson was released after failing to force himself into the first team and signed for Scarborough Athletic, with whom he had been on loan. York drew 0 – 0 at home with second @-@ placed Oxford United, in which Carson came closest to scoring with a volley that <unk> across the face of the goal. This was followed by another draw after the match away to Accrington Stanley finished 1 – 1, with the home team <unk> 10 minutes after a Fletcher penalty had given York the lead in the 35th @-@ minute. Striker <unk> McDonald, who had been released by Peterborough United, was signed on a contract until the end of the season. York's last match of 2013 was a 2 – 1 defeat away at Bury, a result that ended York's run of consecutive draws at five. The home team were 2 – 0 up by the 19th @-@ minute, before Michael Coulson scored York's goal in the 73rd @-@ minute. This result meant York would begin 2014 in 22nd @-@ position in the table, only out of the relegation zone on goal difference. \n \n = = = January = = = \n \n Jarvis scored the only goal in York's first win since October 2013, a 1 – 0 home victory over Morecambe on New Year's Day. McCarey was recalled by Wolverhampton Wanderers due to an injury to one of their <unk>, while O 'Neill was recalled by Burnley to take part in their FA Cup match. York achieved back @-@ to @-@ back wins for the first time since October 2013 after Dagenham & Redbridge were beaten 3 – 1 at home, with Bowman opening the scoring in the second half before Fletcher scored twice. Adam Reed, who had a spell on loan with York in the previous season, was signed on a contract until the end of the season after parting company with Burton. Davies'loan was extended, while Brobbel and <unk> returned to their parent clubs. Cheltenham club captain Russell Penn, a midfielder, was signed on a two @-@ and @-@ a @-@ half @-@ year contract for an undisclosed fee. Lowe was subsequently signed permanently from Cheltenham on a two @-@ and @-@ a @-@ half @-@ year contract for an undisclosed fee. Having been allowed to leave the club on a free transfer, Ashley Chambers signed for Conference Premier club Cambridge United. \n York achieved three successive wins for the first time in 2013 – 14 after beating Northampton 2 – 0 away, with Bowman and Fletcher scoring in three @-@ second half minutes. Defender John McCombe was signed on a two @-@ and @-@ a @-@ half @-@ year contract following his release from Mansfield, before Clay and Jamal <unk> left York by mutual consent. Pope returned to York on loan from Charlton for the remainder of the season. York's run of wins ended with a 0 – 0 draw at home to Bristol Rovers, before their first defeat of the year came after losing 2 – 0 away to Hartlepool. Preston winger Will Hayhurst, a Republic of Ireland under @-@ 21 international, was signed on a one @-@ month loan. York fell to a successive defeat for the first time since September 2013 after being beaten 2 – 0 at home by Chesterfield. Shortly after the match, Smith left the club by mutual consent to pursue first @-@ team football. \n \n = = = February = = = \n \n Fletcher scored a 90th @-@ minute winner for York away to Fleetwood in a 2 – 1 win, a result that ended Fleetwood's five @-@ match unbeaten run. York then drew 0 – 0 at home to fellow mid @-@ table team Cheltenham, before beating Plymouth 4 – 0 away with goals from Fletcher, McCombe ( 2 ) and Carson as the team achieved successive away wins for the first time in 2013 – 14. York went without scoring for a fourth consecutive home match after drawing 0 – 0 with Southend. Having worn the <unk> since an injury to McGurk, Penn was appointed captain for the rest of the season, a position that had earlier been held by Smith and Parslow. \n \n = = = March = = = \n \n York achieved their first home win in five matches after beating Exeter 2 – 1, with first half goals scored by McCombe and Coulson. Hayhurst's loan was extended to the end of the season, having impressed in his six appearances for the club. Coulson scored again with the only goal, a 41st @-@ minute header, in York's 1 – 0 away win over AFC Wimbledon. Bowman scored the only goal with a 32nd @-@ minute penalty as York won 1 – 0 away against Mansfield, in which Fletcher missed the opportunity to extend the lead when his stoppage time penalty was saved by Alan Marriott. York moved one place outside the play @-@ offs with a 2 – 0 home win over Wycombe, courtesy of a second Bowman penalty in as many matches and a Carson goal from the edge of the penalty area. Coulson scored York's only goal in a 1 – 0 away win over struggling Portsmouth with a low volley in the fifth @-@ minute ; this result meant York moved into the play @-@ offs in seventh @-@ place with eight fixtures remaining. \n Striker Calvin Andrew, who had been released by Mansfield in January 2014, was signed on a contract for the remainder of the season. He made his debut as a substitute in York's 1 – 0 home win over bottom of the table Torquay, in which Hayhurst scored the only goal in the 11th @-@ minute with an 18 yard shot that <unk> off Aaron <unk>. Middlesbrough winger Brobbel rejoined on loan until the end of the season, following an injury to Carson. York's run of successive wins ended on six matches after a 0 – 0 home draw with Burton, and this result saw York drop out of the play @-@ offs in eighth @-@ place. With the team recording six wins and one draw in March 2014, including six clean sheets, Worthington was named League Two Manager of the Month. \n \n = = = April = = = \n \n Pope made a number of saves as York held league leaders Rochdale to a 0 – 0 away draw, with a point being enough to lift the team back into seventh @-@ place. York were prevented from equalling a club record of eight consecutive clean sheets when Accrington scored a stoppage time equaliser in a 1 – 1 home draw, in which York had taken earlier taken the lead with a Coulson penalty. A 1 – 0 win away win over Oxford, which was decided by a second half Coulson penalty, resulted in York moving one place above their opponents and back into seventh @-@ place. York consolidated their place in a play @-@ off position after beating Bury 1 – 0 at home with a fifth @-@ minute goal scored by Lowe from a Hayhurst corner. The result meant York opened up a five @-@ point lead over eighth @-@ placed Oxford with two fixtures remaining. A place in the League Two play @-@ offs was secured following a 1 – 0 win over Newport at home, in which Coulson scored the only goal in the 77th @-@ minute with a 25 yard free kick. Pope earned a nomination for League Two Player of the Month for April 2014, having conceded only one goal in five matches in that period. \n \n = = = May = = = \n \n The league season concluded with an away match against divisional runners @-@ up Scunthorpe ; having gone two goals down York fought back to draw 2 – 2 with goals scored by Brobbel and Andrew. This result meant York finished the season in seventh @-@ place in League Two, and would thus play fourth @-@ placed Fleetwood in the play @-@ off semi @-@ final on the back of a 17 @-@ match unbeaten run. York lost 1 – 0 to Fleetwood in the first leg at <unk> Crescent ; the goal came from former York player <unk> Blair in the 50th @-@ minute, who scored from close range after Antoni <unk>'s shot was blocked on the line. A 0 – 0 draw away to Fleetwood in the second leg meant York were eliminated 1 – 0 on aggregate, ending the prospect of a second promotion in three seasons. At an awards night held at York Racecourse, Oyebanjo was voted <unk> of the Year for 2013 – 14. \n \n = = Summary and aftermath = = \n \n York mostly occupied the bottom half of the table before the turn of the year, and dropped as low as 23rd in September 2013. During February 2014 the team broke into the top half of the table and with one match left were in sixth @-@ place. York's defensive record was the third best in League Two with 41 goals conceded, bettered only by Southend ( 39 ) and Chesterfield ( 40 ). Davies made the highest number of appearances over the season, appearing in 47 of York's 52 matches. Fletcher was York's top scorer in the league and in all competitions, with 10 league goals and 13 in total. He was the only player to reach double figures, and was followed by Jarvis with nine goals. \n After the season ended York released Tom Allan, Andrew, Dickinson, McDonald, Puri and Reed, while McGurk retired from professional football. Bowman and Oyebanjo left to sign for Torquay and Crawley Town respectively while Coulson signed a new contract with the club. York's summer signings included goalkeeper Jason <unk> from Tranmere Rovers, defenders <unk> <unk> from Dagenham, Marvin McCoy from Wycombe and Dave Winfield from Shrewsbury Town, midfielders <unk> <unk> from Mansfield, Anthony <unk> from Southend and Luke <unk> from Shrewsbury and striker Jake Hyde from <unk>. \n \n = = Match details = = \n \n League positions are sourced by <unk>, while the remaining information is referenced individually. \n \n = = = Football League Two = = = \n \n \n = = = League table ( part ) = = = \n \n \n = = = FA Cup = = = \n \n \n = = = League Cup = = = \n \n \n = = = Football League Trophy = = = \n \n \n = = = Football League Two play @-@ offs = = = \n \n \n = = <unk> = = \n \n \n = = = In = = = \n \n <unk> around club names denote the player's contract with that club had expired before he joined York. \n \n = = = Out = = = \n \n <unk> around club names denote the player joined that club after his York contract expired. \n \n = = = Loan in = = = \n \n \n = = = Loan out = = = \n \n \n = = Appearances and goals = = \n \n Source : \n Numbers in parentheses denote appearances as substitute. \n Players with names struck through and marked left the club during the playing season. \n Players with names in italics and marked * were on loan from another club for the whole of their season with York. \n Players listed with no appearances have been in the <unk> squad but only as unused <unk>. \n Key to positions : <unk> – <unk> ; <unk> – Defender ; <unk> – <unk> ; <unk> – Forward \n \n\n"
],
[
"show_at(tls.valid, 0)",
" \n = Tropical Storm <unk> ( 2008 ) = \n \n Tropical Storm <unk> was the tenth tropical storm of the 2008 Atlantic hurricane season. <unk> developed out of a strong tropical wave which moved off the African coast on August 31. The wave quickly became organized and was declared Tropical Depression Ten while located 170 mi ( 270 km ) to the south @-@ southeast of the Cape Verde Islands on September 2. The depression was quickly upgraded to Tropical Storm <unk> around noon the same day. Over the next several days, <unk> moved in a general west @-@ northwest direction and reached its peak intensity early on September 3. Strong wind shear, some due to the outflow of Hurricane Ike, and dry air caused the storm to weaken. On September 6, the combination of wind shear, dry air, and cooling waters caused <unk> to weaken into a tropical depression. <unk> deteriorated into a remnant low shortly after as convection continued to dissipate around the storm. The low ultimately dissipated while located 520 mi ( 835 km ) east of <unk> on September 10. However, the remnant moisture led to minor flooding on the island of St. Croix. \n \n = = Meteorological history = = \n \n Tropical Storm <unk> formed as a tropical wave that emerged off the west coast of Africa near the end of August 2008. It tracked south of Cape Verde and slowly developed, and on September 2 the disturbance became Tropical Depression Ten while located south @-@ southeast of the Cape Verde islands. As the depression became more organized, an eye @-@ like feature developed in the upper levels of the system. The depression was upgraded to Tropical Storm <unk> six hours after forming. <unk> was located in an area which was supportive for rapid intensification but was not forecast to intensify quickly. \n <unk> continued to intensify throughout the afternoon as the storm became more symmetrical. However, due to the location of the storm, there was a lack of accurate wind speed readings, and the National Hurricane Center was uncertain of its actual intensity. Despite the lack of wind shear around the storm, the center became slightly exposed and ceased further intensification. The storm was also heading into an area where shear was <unk> to significantly increase due to an upper @-@ level trough diving southward. Despite convection being partially removed from the center of <unk>, the storm intensified slightly in the early morning hours on September 3 as thunderstorm activity to the south of the center became more organized. The intensification was forecast to be short in duration as the trough to the north was deepening, causing the wind shear to the west to become stronger. \n <unk> reached its peak intensity of 65 mph ( 100 km / h ) around 8 a.m. ( <unk> ) as it continued to become more organized. However, there were indications that it had already begun to weaken. <unk> towards the north was becoming restricted and arc clouds began emanating from the storm, a sign that dry air was entering the system. During the afternoon hours, the structure of <unk> began to rapidly deteriorate as strong wind shear and dry air took their toll. By the late night, the center was almost completely exposed and only a band of convection persisted near the center. \n Despite continuing effects from the strong wind shear, a large, deep burst of convection formed in the northern <unk> of <unk>. The center was found to have shifted towards the new convection leading to an increase in intensity. The forecast showed a slight decrease in wind shear as <unk> continued westward and no change in intensity over the 5 @-@ day forecast was predicted. However, the convection decreased once more and the low became completely exposed by the late morning hours and <unk> weakened again. By the afternoon, the center of <unk> was only a <unk> of clouds, devoid of convection. During the overnight hours on September 4 into the morning of September 5, convection associated with <unk> began to <unk> somewhat, mostly to the north of the circulation, due to the strong <unk> wind shear. By mid @-@ morning, <unk> re @-@ intensified slightly due to the redevelopment of some convection. However, the redevelopment was short lived and wind shear again took its toll on <unk> by late morning. The convection around the system became <unk> from the center and <unk> weakened slightly. \n The weakening trend continued through the afternoon as the storm was being affected by strong <unk> shear. <unk> became almost fully devoid of any convection by mid @-@ afternoon and the storm weakened to 40 mph ( 65 km / h ), barely holding on to tropical storm status. <unk> regained a small amount of convection in the late night hours, but not enough to still be classified a tropical storm. Due to the lack of convection, <unk> was downgraded to a Tropical Depression at <unk> ( <unk> ) with winds of 35 mph ( 55 km / h ). Since there was no convection around the system, it would have normally been classified a remnant low but, due to the possibility of the storm <unk> over the next several days, it was considered a tropical depression. The next morning, <unk> was downgraded to a remnant low as strong wind shear and dry air caused the demise of the storm. No redevelopment was expected with <unk> as it began to move over colder waters and remain under strong wind shear until it dissipated. \n However, the remnant low associated with <unk> began to show signs of redevelopment during the afternoon on September 7. <unk> around the system increased significantly and the low was no longer exposed. On September 8, wind shear took over the system again. <unk> around the remnant low was torn away and the low was exposed once more. The National Hurricane Center did not state the chance of regeneration once the low became exposed. Finally, on September 9, wind shear and dry air led to the remnants of <unk> deteriorating into an open wave. However, on September 10, the remnants of <unk> redeveloped and global models picked up on the reformed system. Once more, the chance of regeneration was possible as the remnants of <unk> headed towards the Bahamas. However, on September 14, dry air and wind shear caused the remnants to dissipate entirely. \n \n = = Impact = = \n \n As <unk> passed to the south of the Cape Verde islands on September 2, outer rain bands produced minor rainfall, totaling around 0 @.@ 55 inches ( 14 mm ). There were no reports of damage or flooding from the rain and overall effects were minor. \n Several days after the low dissipated, the remnant moisture from <unk> brought showers and thunderstorms to St. Croix where up to 1 in ( 25 @.@ 4 mm ) of rain fell. The heavy rains led to minor street flooding and some urban flooding. No known damage was caused by the flood. \n \n\n"
]
],
[
[
"The fastai library expects the data to be assembled in a `DataLoaders` object (something that has a training and validation dataloader). We can get one by using the `dataloaders` method. We just have to specify a batch size and a sequence length. Since the GPT2 model was trained with sequences of size 1024, we use this sequence length (it's a stateless model, so it will change the perplexity if we use less):",
"_____no_output_____"
]
],
[
[
"bs, sl = 4, 1024\ndls = tls.dataloaders(bs=bs, seq_len=sl)",
"_____no_output_____"
]
],
[
[
"In fastai, as soon as we have a `DataLoaders`, we can use `show_batch` to have a look at the data (here texts for inputs, and the same text shifted by one token to the right for validation):",
"_____no_output_____"
]
],
[
[
"dls.show_batch(max_n=2)",
"_____no_output_____"
],
[
"def tokenize(text):\n toks = tokenizer.tokenize(text)\n return tensor(tokenizer.convert_tokens_to_ids(toks))\n\ntokenized = [tokenize(t) for t in progress_bar(all_texts)]",
"_____no_output_____"
]
],
[
[
"Now we change the previous `Tokenizer` like this:",
"_____no_output_____"
]
],
[
[
"class TransformersTokenizer(Transform):\n def __init__(self, tokenizer):\n self.tokenizer = tokenizer\n\n def encodes(self, x):\n return x if isinstance(x, Tensor) else tokenize(x)\n\n def decodes(self, x): \n return TitledStr(self.tokenizer.decode(x.cpu().numpy())) ",
"_____no_output_____"
],
[
"tls = TfmdLists(\n tokenized, \n TransformersTokenizer(tokenizer), splits=splits, dl_type=LMDataLoader\n)\ndls = tls.dataloaders(bs=bs, seq_len=sl)",
"_____no_output_____"
],
[
"dls.show_batch(max_n=2)",
"_____no_output_____"
]
],
[
[
"### Fine-tuning the model",
"_____no_output_____"
],
[
"The HuggingFace model will return a tuple in outputs, with the actual predictions and some additional activations (should we want to use them in some regularization scheme). To work inside the fastai training loop, we will need to drop those using a `Callback`: we use those to alter the behavior of the training loop.\n\nHere we need to write the event `after_pred` and replace `self.learn.pred` (which contains the predictions that will be passed to the loss function) by just its first element. In callbacks, there is a shortcut that lets you access any of the underlying `Learner` attributes so we can write `self.pred[0]` instead of `self.learn.pred[0]`. That shortcut only works for read access, not write, so we have to write `self.learn.pred` on the right side (otherwise we would set a `pred` attribute in the `Callback`).",
"_____no_output_____"
]
],
[
[
"class DropOutput(Callback):\n def after_pred(self):\n self.learn.pred = self.pred[0]",
"_____no_output_____"
]
],
[
[
"Of course we could make this a bit more complex and add some penalty to the loss using the other part of the tuple of predictions, like the `RNNRegularizer`.\n\nNow, we are ready to create our `Learner`, which is a fastai object grouping data, model and loss function and handles model training or inference. Since we are in a language model setting, we pass perplexity as a metric, and we need to use the callback we just defined. Lastly, we use mixed precision to save every bit of memory we can (and if you have a modern GPU, it will also make training faster):",
"_____no_output_____"
]
],
[
[
"learn = Learner(\n dls,\n model,\n loss_func=CrossEntropyLossFlat(),\n cbs=[DropOutput],\n metrics=Perplexity()\n).to_fp16()",
"_____no_output_____"
]
],
[
[
"We can check how good the model is without any fine-tuning step (spoiler alert, it's pretty good!)",
"_____no_output_____"
]
],
[
[
"learn.validate()",
"_____no_output_____"
]
],
[
[
"This lists the validation loss and metrics (so 26.6 as perplexity is kind of amazing).\n\nNow that we have a `Learner` we can use all the fastai training loop capabilities: learning rate finder, training with 1cycle etc... ",
"_____no_output_____"
]
],
[
[
"learn.lr_find()",
"_____no_output_____"
]
],
[
[
"The learning rate finder curve suggests picking something between 1e-4 and 1e-2.",
"_____no_output_____"
]
],
[
[
"learn.fit_one_cycle(1, 1e-4)",
"_____no_output_____"
]
],
[
[
"Now with just one epoch of fine-tuning and not much regularization, our model did not really improve since it was already amazing. To have a look at some generated texts, let's take a prompt that looks like a wikipedia article:",
"_____no_output_____"
]
],
[
[
"df_valid.head(1)",
"_____no_output_____"
]
],
[
[
"Article seems to begin with new line and the title between = signs, so we will mimic that:",
"_____no_output_____"
]
],
[
[
"prompt = \"\\n = Unicorn = \\n \\n A unicorn is a magical creature with a rainbow tail and a horn\"",
"_____no_output_____"
]
],
[
[
"The prompt needs to be tokenized and numericalized, so we use the same function as before to do this, before we use the `generate` method of the model.",
"_____no_output_____"
]
],
[
[
"prompt_ids = tokenizer.encode(prompt)\ninp = tensor(prompt_ids)[None].cuda()\ninp.shape",
"_____no_output_____"
],
[
"preds = learn.model.generate(inp, max_length=40, num_beams=5, temperature=1.5)",
"Setting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\n"
],
[
"tokenizer.decode(preds[0].cpu().numpy())",
"_____no_output_____"
],
[
"prompt = \"\\n = Pokemon = \\n \\n Pokemon also know as Pocket Monsters in Japan, is a Japanse media franchise\"\nprompt_ids = tokenizer.encode(prompt)\ninp = tensor(prompt_ids)[None].cuda()\npreds = learn.model.generate(inp, max_length=40, num_beams=5, temperature=1.5)\ntokenizer.decode(preds[0].cpu().numpy())",
"Setting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\n"
],
[
"prompt = \"\\n = NFL = \\n \\n The National Football League (NFL) is a professional American football league consisting of 32 teams, \"\nprompt_ids = tokenizer.encode(prompt)\ninp = tensor(prompt_ids)[None].cuda()\npreds = learn.model.generate(inp, max_length=40, num_beams=5, temperature=1.5)\ntokenizer.decode(preds[0].cpu().numpy())",
"Setting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\n"
],
[
"prompt = \"\\n = Julien = \\n \\n My name is Julien and I like to\"\nprompt_ids = tokenizer.encode(prompt)\ninp = tensor(prompt_ids)[None].cuda()\npreds = learn.model.generate(inp, max_length=40, num_beams=5, temperature=1.5)\ntokenizer.decode(preds[0].cpu().numpy())",
"Setting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccc671abb754f601cae68235b8dc43d7299f176 | 139,339 | ipynb | Jupyter Notebook | PolitiFactClassification.ipynb | ourDirection/FakeNews | 7ecd6b30acba281e791d8cd474ebaf8c178bb4df | [
"Apache-2.0"
] | null | null | null | PolitiFactClassification.ipynb | ourDirection/FakeNews | 7ecd6b30acba281e791d8cd474ebaf8c178bb4df | [
"Apache-2.0"
] | null | null | null | PolitiFactClassification.ipynb | ourDirection/FakeNews | 7ecd6b30acba281e791d8cd474ebaf8c178bb4df | [
"Apache-2.0"
] | null | null | null | 150.636757 | 34,996 | 0.847329 | [
[
[
"# Classification and prediction of PolitiFact Statements",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom sklearn.metrics import precision_score, accuracy_score, roc_curve, confusion_matrix\nfrom sklearn import preprocessing\nfrom sklearn.utils import shuffle\n\nimport itertools\nfrom sklearn import metrics\nimport seaborn as sns\n\n%matplotlib inline\nsns.set(style='whitegrid', palette='muted')",
"_____no_output_____"
],
[
"from data_util import get_fake_or_real_news, get_politifact, get_fact_fake\n\n# x_train, x_test, y_train, y_test, word_index, labels, num_classes, X, y = get_fake_or_real_news()\n# x_train, x_test, y_train, y_test, word_index, labels, num_classes, X, y = get_politifact()\n# x_train, x_test, y_train, y_test, word_index, labels, num_classes, X, y = get_fact_fake()\n\n\ndef to_label(cat):\n return categories[cat]\n##### RUMER TWITTER\n# categories = {0:'true', 1: 'false'}\n# data_train = pd.read_csv('data/RumerTwitter.csv', sep='\\t')\n# data_train['label'] = data_train['label'].apply(to_label)\n# X = data_train['text'].values\n# y = data_train['label'].values\n# labels = data_train['label'].unique()\n# print(data_train['label'].value_counts())\n# print(len(data_train['event_id'].unique()))\n# print(data_train['event_id'].unique())\n# data_train.tail()\n#####\n\n##### FakeNewsNet #####################################\n# def add_text(row):\n# return row['title_tokens'] + ' ' + row['text_tokens'] \n \n \n# categories = {'FakeNewsContent':'False', 'RealNewsContent':'True'}\n# data_train = pd.read_csv('data/fakeNewsNet.csv', sep='\\t')\n# data_train.dropna(how='any', inplace=True) \n# print(data_train['collected_by'].unique())\n# print(data_train['label'].unique())\n# print(data_train.isnull().values.any())\n# data_train['label'] = data_train['label'].apply(to_label)\n# data_train['document'] = data_train.apply(add_text, axis=1)\n# X = data_train['document'].values\n# y = data_train['label'].values\n# labels = data_train['label'].unique()\n# print(data_train['label'].value_counts())\n#################################################\n\n##### Unreliable News Data ##################################\ndata_train = pd.read_csv('data/newsfiles/fulltrain.csv', names=['label', 'text'])\ncategories = {1:\"Satire\",2:\"Hoax\",\n 3:\"Propaganda\",4:\"Trusted\"}\n\ndata_train['label'] = data_train['label'].apply(to_label)\nX = data_train['text'].values\ny = data_train['label'].values\nlabels = data_train['label'].unique()\nprint(data_train['label'].value_counts())\n############################\n\n# print(X[1] ,y[1])\nprint()\ndata_train.tail()\n",
"Propaganda 17870\nSatire 14047\nTrusted 9995\nHoax 6942\nName: label, dtype: int64\n\n"
],
[
"##### SNOPES\n# def label(row):\n# # print(row)\n# if str(row).lower() in ['true', 'mixture', 'mixture', 'mtrue']:\n# return 'true'\n# else:\n# return 'false'\n \n# data_train = pd.read_csv('data/snopes.tsv', sep='\\t')\n\n# data_train['label'] = data_train['lable']\n\n# print(data_train['label'].unique())\n# X = data_train['claim'].values.astype('U')\n# y = data_train['label'].values\n# #########################################\n\n# data_train = pd.read_csv('data/snopes.csv')\n# data_train = pd.DataFrame({'claim' : np.append(X, data_train['claim'].values.astype('U')), \n# 'label' : np.append(y, data_train['claim_label'].values)})\n\n# # data_train.dropna(how='any', inplace=True) \n# print(data_train['claim'].isnull().values.any())\n# data_train = data_train.drop_duplicates(['claim'], keep='last')\n# data_train['label'] = data_train['label'].apply(label)\n# labels = data_train['label'].unique()\n# print(data_train['label'].value_counts())\n# data_train.reset_index(drop=True, inplace=True)\n# X = data_train['claim'].values.astype('U')\n# y = data_train['label'].values\n# data_train.to_csv('data/snopes_processed.tsv', sep='\\t')\n# data_train.tail(100)\n",
"_____no_output_____"
],
[
"%%time\n\n\nTF_IDF = True\nclasses = labels\n\nif TF_IDF:\n# vect = TfidfVectorizer(norm='l2', lowercase=True, stop_words='english')\n vect = TfidfVectorizer(norm='l2', ngram_range=(1, 2), lowercase=True, stop_words='english')\n# vect = CountVectorizer(min_df=2,ngram_range=(1, 2)) # Count Vectorizer\n X = vect.fit_transform(X)\n\n# svd = TruncatedSVD(n_components=120)\n# X = svd.fit_transform(X)\n print('X.shape', X.shape) \n print(X[1])\nelse:\n X = data_train['glove'].values\n X = np.array(list(X), dtype=np.float)\n# X = preprocessing.normalize(X)\n print('X.shape', X.shape)",
"X.shape (48854, 7388647)\n (0, 326641)\t0.036607688245\n (0, 2491292)\t0.104417533529\n (0, 4456311)\t0.0389569461117\n (0, 5734826)\t0.017972309003\n (0, 1344014)\t0.0313887641587\n (0, 4482268)\t0.0439290980124\n (0, 7318933)\t0.0633074256704\n (0, 3056407)\t0.0823068460705\n (0, 5916472)\t0.0907734382803\n (0, 6135065)\t0.104533608098\n (0, 6719636)\t0.0366530184964\n (0, 1730422)\t0.0777167337457\n (0, 6314678)\t0.0942126446516\n (0, 6280237)\t0.0431510598897\n (0, 3655542)\t0.0474461834664\n (0, 7973)\t0.0329028643557\n (0, 4217007)\t0.0340594726862\n (0, 5940379)\t0.0689935132812\n (0, 5849505)\t0.143359796331\n (0, 5054847)\t0.0810016486168\n (0, 2294420)\t0.126780336686\n (0, 6406403)\t0.0450451642357\n (0, 1818864)\t0.0507194180492\n (0, 855649)\t0.0658562889161\n (0, 1209392)\t0.0474304693448\n :\t:\n (0, 7057714)\t0.123924825403\n (0, 3085509)\t0.128621702768\n (0, 4483215)\t0.128621702768\n (0, 3130964)\t0.123924825403\n (0, 5850144)\t0.112562972988\n (0, 2294962)\t0.123924825403\n (0, 6719327)\t0.128621702768\n (0, 2090693)\t0.128621702768\n (0, 5627765)\t0.114109790613\n (0, 3184698)\t0.128621702768\n (0, 3200093)\t0.112562972988\n (0, 3947217)\t0.128621702768\n (0, 6341840)\t0.128621702768\n (0, 2673689)\t0.128621702768\n (0, 5850450)\t0.128621702768\n (0, 3366216)\t0.0915685135673\n (0, 3412076)\t0.123924825403\n (0, 5049515)\t0.128621702768\n (0, 5136750)\t0.128621702768\n (0, 6828941)\t0.100844663989\n (0, 922008)\t0.118007459736\n (0, 2492324)\t0.114109790613\n (0, 5917262)\t0.111198583147\n (0, 2579663)\t0.128621702768\n (0, 1345152)\t0.101948729955\nCPU times: user 1min 21s, sys: 1.76 s, total: 1min 23s\nWall time: 1min 23s\n"
],
[
"# from os.path import expanduser, exists\n\n# embeddings_index = {}\n# KERAS_DATASETS_DIR = expanduser('~/.keras/datasets/')\n# GLOVE_FILE = 'glove.840B.300d.txt'\n\n# print(\"Processing\", GLOVE_FILE)\n\n# embeddings_index = {}\n# with open(KERAS_DATASETS_DIR + GLOVE_FILE, encoding='utf-8') as f:\n# for line in f:\n# values = line.split(' ')\n# word = values[0]\n# embedding = np.asarray(values[1:], dtype='float32')\n# embeddings_index[word] = embedding\n\n# print('Word embeddings: %d' % len(embeddings_index))\n\n# EMBEDDING_DIM = 300\n# vect = TfidfVectorizer(norm='l2')\n# vect.fit_transform(X)\n# print(len(vect.vocabulary_))\n\n# def sent2vec(s):\n# words = str(s).lower().split(' ')\n# M = []\n# for w in words:\n# try:\n# M.append(embeddings_index[w])\n# except:\n# continue\n# M = np.array(M)\n# v = M.sum(axis=0)\n# if type(v) != np.ndarray:\n# return np.random.uniform(EMBEDDING_DIM)\n \n# return np.mean(M, axis=0)\n# # return v / np.sqrt((v ** 2).sum())\n\n# data_train['glove'] = data_train['TokenizedContent'].apply(sent2vec)\n# data_train.to_csv('data/hansard_processed_all.csv')\n# data_train.tail()",
"_____no_output_____"
],
[
"def plot_confusion_matrix(cm, classes,\n normalize=False,\n title='Confusion matrix',\n cmap=plt.cm.Blues):\n \"\"\"\n This function prints and plots the confusion matrix.\n Normalization can be applied by setting `normalize=True`.\n \"\"\"\n if normalize:\n cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]\n print(\"Normalized confusion matrix\")\n else:\n print('Confusion matrix, without normalization')\n\n print(cm)\n\n plt.imshow(cm, interpolation='nearest', cmap=cmap)\n plt.title(title)\n plt.colorbar()\n tick_marks = np.arange(len(classes))\n plt.xticks(tick_marks, classes, rotation=45)\n plt.yticks(tick_marks, classes)\n\n fmt = '.2f' if normalize else 'd'\n thresh = cm.max() / 2.\n for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):\n plt.text(j, i, format(cm[i, j], fmt),\n horizontalalignment=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n\n plt.tight_layout()\n plt.ylabel('True label')\n plt.xlabel('Predicted label')",
"_____no_output_____"
],
[
"%%time\n\nfrom sklearn.model_selection import StratifiedKFold\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.model_selection import cross_val_predict\n\n# classifiers\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.neighbors.nearest_centroid import NearestCentroid\nfrom sklearn.naive_bayes import MultinomialNB\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.linear_model import SGDClassifier\nfrom sklearn.neural_network import MLPClassifier, BernoulliRBM\nfrom sklearn.naive_bayes import GaussianNB, BernoulliNB\nfrom sklearn.ensemble import AdaBoostClassifier\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.svm import SVC\nfrom sklearn.decomposition import TruncatedSVD\nfrom xgboost import XGBClassifier as XGBoostClassifier\nfrom sklearn.linear_model import PassiveAggressiveClassifier\n\n\nskf = StratifiedKFold(n_splits=2, random_state=42, shuffle=True)\n# print(skf) \nscore = []\n\n# print(X[9])\n\nclassifiers = {\n# 'KNeighbors' : KNeighborsClassifier(n_neighbors=3), \n# 'Centroid': NearestCentroid(), \n# 'MultinomialNB': MultinomialNB(), \n# 'XGBoostClassifier' : XGBoostClassifier(seed=42),\n# 'BernoulliNB' : BernoulliNB(),\n 'LogisticRegression' : LogisticRegression(C=15, random_state=42), \n 'PassiveAggressiveClassifier' : PassiveAggressiveClassifier(max_iter=50),\n# 'DecisionTreeClassifier' : DecisionTreeClassifier(max_depth=10, random_state=42), \n# 'SGDClassifier' : SGDClassifier(loss='hinge', penalty='l2', alpha=1e-3, random_state=42, max_iter=1000 ), \n# 'MLPClassifier' : MLPClassifier(hidden_layer_sizes=(30,30,30)), \n# 'AdaBoostClassifier' : AdaBoostClassifier(n_estimators=100), \n# 'RandomForestClassifier' : RandomForestClassifier(max_depth=20, random_state=42), \n 'SVC' : SVC(kernel='linear', C= 1000)\n}\n\nfor name, classifier in classifiers.items():\n print(name, classifier)\n# scores = cross_val_score(classifier, X, y, cv=skf)\n predicted = cross_val_predict(classifier, X, y, cv=skf)\n \n print(metrics.classification_report(y, predicted, \n target_names=classes, digits=3))\n print('metric accuracy Score' , metrics.accuracy_score(y, predicted) )\n# print(\"%s Accuracy: %0.3f (+/- %0.3f)\" % (name, scores.mean(), scores.std() * 2))\n\n CM = confusion_matrix(y_pred=predicted, y_true=y) \n # Plot normalized confusion matrix\n plt.figure()\n plot_confusion_matrix(CM, classes=classes, title='Confusion matrix')\n plt.show()\n print('-' * 100)",
"LogisticRegression LogisticRegression(C=15, class_weight=None, dual=False, fit_intercept=True,\n intercept_scaling=1, max_iter=100, multi_class='ovr', n_jobs=1,\n penalty='l2', random_state=42, solver='liblinear', tol=0.0001,\n verbose=0, warm_start=False)\n precision recall f1-score support\n\n Satire 0.980 0.933 0.956 6942\n Hoax 0.963 0.975 0.969 17870\n Propaganda 0.919 0.979 0.948 14047\n Trusted 0.977 0.900 0.937 9995\n\navg / total 0.956 0.955 0.955 48854\n\nmetric accuracy Score 0.954681295288\nConfusion matrix, without normalization\n[[ 6476 213 217 36]\n [ 74 17424 322 50]\n [ 27 146 13749 125]\n [ 32 305 667 8991]]\n"
]
],
[
[
"# Voting classifiers",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import VotingClassifier\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.linear_model import PassiveAggressiveClassifier\nfrom sklearn.svm import SVC\nfrom itertools import product\nfrom sklearn.linear_model import SGDClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, stratify=y, random_state=42)\n\nprint('X_train.shape', X_train.shape, 'y_train.shape', y_train.shape)\nprint('X_test.shape', X_test.shape, 'y_test.shape', y_test.shape)\n\nLogisticRegression = LogisticRegression(C=15, random_state=42) \nKNeighborsClassifier = KNeighborsClassifier(n_neighbors=3)\n# PassiveAggressiveClassifier = SGDClassifier(loss='hinge', penalty='l2', alpha=1e-3, random_state=42, max_iter=1000 )\nSVC = SVC(kernel='linear', probability=True)\n\neclf = VotingClassifier(estimators=[('LR', LogisticRegression), \n# ('PAC', PassiveAggressiveClassifier),\n ('SVC', SVC)\n# ('KNN', KNeighborsClassifier)\n ],\n voting='soft', weights=[2, 2])\n\nprint('Start fitting')\n\nLogisticRegression.fit(X_train, y_train)\n# PassiveAggressiveClassifier.fit(X_train, y_train)\nSVC.fit(X_train, y_train)\nKNeighborsClassifier.fit(X_train, y_train)\neclf.fit(X_train, y_train)\n\npredicted = eclf.predict(X_test)\n\nprint('finished fitting')\n\nscore = metrics.accuracy_score(y_test, predicted)\nprint(\"accuracy: %0.3f\" % score)\nprint(metrics.classification_report(y_test, predicted, \n target_names=classes, digits=3))\n \nCM = confusion_matrix(y_pred=predicted, y_true=y_test) \n# Plot normalized confusion matrix\nplt.figure()\nplot_confusion_matrix(CM, classes=classes, title='Confusion matrix')\nplt.show()\nprint('-' * 100)",
"_____no_output_____"
]
],
[
[
"# get informative features for classification",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import PassiveAggressiveClassifier\n\n# X = data_train['TokenizedContent'].values\n# print(len(X))\n# y = data_train['target']\n# y = pd.get_dummies(y).values\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, stratify=y, random_state=42)\n\nprint('X_train.shape', X_train.shape, 'y_train.shape', y_train.shape)\nprint('X_test.shape', X_test.shape, 'y_test.shape', y_test.shape)\n# print(y)\n\n\n# classifier = PassiveAggressiveClassifier(max_iter=50)\nclassifier = LogisticRegression(C=15, random_state=42)\n# classifier = SGDClassifier(loss='hinge', penalty='l2', alpha=1e-3, random_state=42, max_iter=1000 )\nclassifier.fit(X_train, y_train)\npred = classifier.predict(X_test)\n\nscore = metrics.accuracy_score(y_test, pred)\nprint(\"accuracy: %0.3f\" % score)\nprint(metrics.classification_report(y_test, pred, \n target_names=classes, digits=3))\n \nCM = confusion_matrix(y_pred=pred, y_true=y_test) \n# Plot normalized confusion matrix\nplt.figure()\nplot_confusion_matrix(CM, classes=classes, title='Confusion matrix')\nplt.show()\nprint('-' * 100)\n \ndef most_informative_feature_for_binary_classification(vectorizer, classifier, n=100):\n \"\"\"\n See: https://stackoverflow.com/a/26980472\n \n Identify most important features if given a vectorizer and binary classifier. Set n to the number\n of weighted features you would like to show. (Note: current implementation merely prints and does not \n return top classes.)\n \"\"\"\n\n class_labels = classifier.classes_\n feature_names = vectorizer.get_feature_names()\n topn_class1 = sorted(zip(classifier.coef_[0], feature_names))[:n]\n topn_class2 = sorted(zip(classifier.coef_[0], feature_names))[-n:]\n\n for coef, feat in topn_class1:\n print(class_labels[0], coef, feat)\n\n print()\n\n for coef, feat in reversed(topn_class2):\n print(class_labels[1], coef, feat)\n\n\nmost_informative_feature_for_binary_classification(vect, classifier, n=30)",
"_____no_output_____"
],
[
"from sklearn.model_selection import GridSearchCV\nfrom sklearn.metrics import classification_report\n# Split the dataset in two equal parts\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.5, stratify=y,\n random_state=42)\n\n# Set the parameters by cross-validation\ntuned_parameters = [{'kernel': ['rbf'], 'gamma': [1e-3, 1e-4],\n 'C': [1, 10, 100, 1000]},\n {'kernel': ['linear'], 'C': [1, 10, 100, 1000]}]\n\nscores = ['precision', 'recall']\n\nfor score in scores:\n print(\"# Tuning hyper-parameters for %s\" % score)\n print()\n\n clf = GridSearchCV(SVC(), tuned_parameters, cv=5,\n scoring='%s_macro' % score)\n clf.fit(X_train, y_train)\n\n print(\"Best parameters set found on development set:\")\n print()\n print(clf.best_params_)\n print()\n print(\"Grid scores on development set:\")\n print()\n means = clf.cv_results_['mean_test_score']\n stds = clf.cv_results_['std_test_score']\n for mean, std, params in zip(means, stds, clf.cv_results_['params']):\n print(\"%0.3f (+/-%0.03f) for %r\"\n % (mean, std * 2, params))\n print()\n\n print(\"Detailed classification report:\")\n print()\n print(\"The model is trained on the full development set.\")\n print(\"The scores are computed on the full evaluation set.\")\n print()\n y_true, y_pred = y_test, clf.predict(X_test)\n print(classification_report(y_true, y_pred))\n print()",
"_____no_output_____"
]
],
[
[
"# Topic Analysis",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\nfrom time import time\n\nfrom sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer\nfrom sklearn.decomposition import NMF, LatentDirichletAllocation\nfrom sklearn.datasets import fetch_20newsgroups\n\nn_samples = 2000\nn_features = 1000\nn_components = 10\nn_top_words = 20\n\n\ndef print_top_words(model, feature_names, n_top_words):\n for topic_idx, topic in enumerate(model.components_):\n message = \"Topic #%d: \" % topic_idx\n message += \" \".join([feature_names[i]\n for i in topic.argsort()[:-n_top_words - 1:-1]])\n print(message)\n print()\n\n\n# Load the 20 newsgroups dataset and vectorize it. We use a few heuristics\n# to filter out useless terms early on: the posts are stripped of headers,\n# footers and quoted replies, and common English words, words occurring in\n# only one document or in at least 95% of the documents are removed.\n\nprint(\"Loading dataset...\")\nt0 = time()\n# dataset = fetch_20newsgroups(shuffle=True, random_state=1,\n# remove=('headers', 'footers', 'quotes'))\n# data_samples = dataset.data[:n_samples]\ndata_samples = X\nprint(\"done in %0.3fs.\" % (time() - t0))\n\n# Use tf-idf features for NMF.\nprint(\"Extracting tf-idf features for NMF...\")\ntfidf_vectorizer = TfidfVectorizer(max_df=0.95, min_df=2,\n max_features=n_features,\n stop_words='english')\nt0 = time()\ntfidf = tfidf_vectorizer.fit_transform(data_samples)\nprint(\"done in %0.3fs.\" % (time() - t0))\n\n# Use tf (raw term count) features for LDA.\nprint(\"Extracting tf features for LDA...\")\ntf_vectorizer = CountVectorizer(max_df=0.95, min_df=2,\n max_features=n_features,\n stop_words='english')\nt0 = time()\ntf = tf_vectorizer.fit_transform(data_samples)\nprint(\"done in %0.3fs.\" % (time() - t0))\nprint()\n\n# Fit the NMF model\nprint(\"Fitting the NMF model (Frobenius norm) with tf-idf features, \"\n \"n_samples=%d and n_features=%d...\"\n % (n_samples, n_features))\nt0 = time()\nnmf = NMF(n_components=n_components, random_state=1,\n alpha=.1, l1_ratio=.5).fit(tfidf)\nprint(\"done in %0.3fs.\" % (time() - t0))\n\nprint(\"\\nTopics in NMF model (Frobenius norm):\")\ntfidf_feature_names = tfidf_vectorizer.get_feature_names()\nprint_top_words(nmf, tfidf_feature_names, n_top_words)\n\n# Fit the NMF model\nprint(\"Fitting the NMF model (generalized Kullback-Leibler divergence) with \"\n \"tf-idf features, n_samples=%d and n_features=%d...\"\n % (n_samples, n_features))\nt0 = time()\nnmf = NMF(n_components=n_components, random_state=1,\n beta_loss='kullback-leibler', solver='mu', max_iter=1000, alpha=.1,\n l1_ratio=.5).fit(tfidf)\nprint(\"done in %0.3fs.\" % (time() - t0))\n\nprint(\"\\nTopics in NMF model (generalized Kullback-Leibler divergence):\")\ntfidf_feature_names = tfidf_vectorizer.get_feature_names()\nprint_top_words(nmf, tfidf_feature_names, n_top_words)\n\nprint(\"Fitting LDA models with tf features, \"\n \"n_samples=%d and n_features=%d...\"\n % (n_samples, n_features))\nlda = LatentDirichletAllocation(n_components=n_components, max_iter=5,\n learning_method='online',\n learning_offset=50.,\n random_state=0)\nt0 = time()\nlda.fit(tf)\nprint(\"done in %0.3fs.\" % (time() - t0))\n\nprint(\"\\nTopics in LDA model:\")\ntf_feature_names = tf_vectorizer.get_feature_names()\nprint_top_words(lda, tf_feature_names, n_top_words)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
eccc6f0725d26e2b8e0e990db4367a96181ecceb | 5,774 | ipynb | Jupyter Notebook | pymc_examples/_numba_01.ipynb | danhphan/bayesian_inferences | d5976f7fde560b0d0bd47c0144d87724d4ac96b3 | [
"Apache-2.0"
] | null | null | null | pymc_examples/_numba_01.ipynb | danhphan/bayesian_inferences | d5976f7fde560b0d0bd47c0144d87724d4ac96b3 | [
"Apache-2.0"
] | null | null | null | pymc_examples/_numba_01.ipynb | danhphan/bayesian_inferences | d5976f7fde560b0d0bd47c0144d87724d4ac96b3 | [
"Apache-2.0"
] | null | null | null | 19.979239 | 192 | 0.480083 | [
[
[
"# !pip install numba",
"_____no_output_____"
],
[
"from numba import jit\nimport random",
"_____no_output_____"
],
[
"def monte_carlo_pi(nsamples):\n acc = 0\n for i in range(nsamples):\n x = random.random()\n y = random.random()\n if (x ** 2 + y ** 2) < 1.0:\n acc += 1\n return 4.0 * acc / nsamples",
"_____no_output_____"
],
[
"%%time\nmonte_carlo_pi(1000000)",
"CPU times: user 254 ms, sys: 0 ns, total: 254 ms\nWall time: 253 ms\n"
],
[
"monte_carlo_pi_jit = jit()(monte_carlo_pi)",
"_____no_output_____"
],
[
"%time monte_carlo_pi_jit(10000)",
"CPU times: user 73 µs, sys: 0 ns, total: 73 µs\nWall time: 75.6 µs\n"
],
[
"%time monte_carlo_pi_jit(1000000)",
"CPU times: user 8.08 ms, sys: 0 ns, total: 8.08 ms\nWall time: 7.9 ms\n"
],
[
"%pylab inline",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"from numba import jit, njit, vectorize",
"_____no_output_____"
],
[
"def original_func(input_list):\n output_list = np.zeros_like(input_list)\n for ii, item in enumerate(input_list):\n if item % 2 == 0:\n output_list[ii] = (2)\n else:\n output_list[ii] = (1)\n return output_list\n\n# test_array = list(range(100000))\ntest_array = np.arange(100000)",
"_____no_output_____"
],
[
"%time _ = original_func(test_array)",
"CPU times: user 46.4 ms, sys: 0 ns, total: 46.4 ms\nWall time: 45 ms\n"
],
[
"jitted_func = njit()(original_func)",
"_____no_output_____"
],
[
"%time _ = jitted_func(test_array)",
"CPU times: user 161 µs, sys: 0 ns, total: 161 µs\nWall time: 163 µs\n"
],
[
"@vectorize\ndef scalar_compu(num):\n if num % 2 == 0:\n return 2\n else:\n return 1",
"_____no_output_____"
],
[
"%time _ = scalar_compu(test_array)",
"CPU times: user 282 µs, sys: 81 µs, total: 363 µs\nWall time: 170 µs\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eccc70d632cc6d97daf13ec6654f964a71db3aec | 150,554 | ipynb | Jupyter Notebook | tutorial/PsyNeuLink Tutorial.ipynb | AlirezaFarnia/PsyNeuLink | c66f8248d1391830e76c97df4b644e12a02c2b73 | [
"Apache-2.0"
] | null | null | null | tutorial/PsyNeuLink Tutorial.ipynb | AlirezaFarnia/PsyNeuLink | c66f8248d1391830e76c97df4b644e12a02c2b73 | [
"Apache-2.0"
] | null | null | null | tutorial/PsyNeuLink Tutorial.ipynb | AlirezaFarnia/PsyNeuLink | c66f8248d1391830e76c97df4b644e12a02c2b73 | [
"Apache-2.0"
] | null | null | null | 100.503338 | 15,772 | 0.751086 | [
[
[
"# Welcome to PsyNeuLink\n\nPsyNeuLink is an integrated language and toolkit for creating cognitive models. It decreases the overhead required for cognitive modeling by providing standard building blocks (DDMS, Neural Nets, etc.) and the means to connect them together in a single environment. PsyNeuLink is designed to make the user think about computation in a \"mind/brain-like\" way while imposing minimal constraint on the type of models that can be implemented.",
"_____no_output_____"
],
[
"## How to get PsyNeuLink\n\nPsyNeuLink is compatible with python versions >= 3.5, and is available through PyPI:\n\n```python\npip install psyneulink\n```\nOr you can clone the github repo [here](https://github.com/PrincetonUniversity/PsyNeuLink). Download the package with the green \"Clone or download\" button on the right side of the page and \"Download ZIP.\" Open the version of this Tutorial in the cloned folder before continuing on.\n\n## Installation\n\nTo install the package, navigate to the cloned directory in a terminal, switch to your preferred python3 environment, then run the command __\"pip install .\"__ (make sure to include the period and to use the appropriate pip/pip3 command for python 3.5). All prerequisite packages will be automatically added to your enviroment.\n\nFor the curious, these are:\n* numpy\n* matplotlib\n* toposort\n* mpi4py\n* typecheck-decorator",
"_____no_output_____"
],
[
"\n## Tutorial Overview\n\nThis tutorial is meant to get you accustomed to the structure of PsyNeuLink and be able to construct basic models. Starting with a simple 1-to-1 transformation, we will build up to making the Stroop model from Cohen et al. (1990). Let's get started!",
"_____no_output_____"
],
[
"### Imports and file structure\n\nThe following code block will import the necessary components for basic neural network models in PsyNeuLink. In particular, we need tools for handling *[Compositions](https://princetonuniversity.github.io/PsyNeuLink/Composition.html)*, the set of specific *[mechanisms](https://princetonuniversity.github.io/PsyNeuLink/Mechanism.html)* that will make up our networks, and the *[projections](https://princetonuniversity.github.io/PsyNeuLink/Projection.html)* to connect them. We also import basic prerequisites and set up the jupyter environment for visualization.",
"_____no_output_____"
]
],
[
[
"!pip install psyneulink #run this command if opening in Google CoLaboratory \nimport psyneulink as pnl",
"Requirement already satisfied: psyneulink in /usr/local/lib/python3.6/dist-packages (0.5.3.2)\nRequirement already satisfied: typecheck-decorator==1.2 in /usr/local/lib/python3.6/dist-packages (from psyneulink) (1.2)\nRequirement already satisfied: toposort==1.4 in /usr/local/lib/python3.6/dist-packages (from psyneulink) (1.4)\nRequirement already satisfied: llvmlite in /usr/local/lib/python3.6/dist-packages (from psyneulink) (0.28.0)\nRequirement already satisfied: pillow in /usr/local/lib/python3.6/dist-packages (from psyneulink) (4.3.0)\nRequirement already satisfied: numpy<1.16 in /usr/local/lib/python3.6/dist-packages (from psyneulink) (1.15.4)\nRequirement already satisfied: olefile in /usr/local/lib/python3.6/dist-packages (from pillow->psyneulink) (0.46)\n"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Creating a mechanism\n\n*[Mechanisms](https://princetonuniversity.github.io/PsyNeuLink/Mechanism.html)* are the basic units of computation in PsyNeuLink. At their core is a parameterized *function* but they also contain the machinery to interact with input, output, control, and learning signals. Our first mechanism will perform a linear transformation on a scalar input. For now, we will initialize it by just specifying the *function* of the mechanism.",
"_____no_output_____"
]
],
[
[
"linear_transfer_mechanism = pnl.TransferMechanism(\n function=pnl.Linear(slope=1, intercept=0))",
"_____no_output_____"
]
],
[
[
"In this case, we didn't actually need to specify the slope and intercept as the function will default to reasonable values (if we didn't specify it would have defaulted to a slope of 1 and intercept of 0). The function above has two parameters, slope and intercept. If we wrote the equation as y = ax + b, a is the slope, b is the input, x is the input and y is the output. As a function we write this f(x) = ax + b. Note that you can change these parameter values of a=1 and b=0 to other numbers -- because parameters are variables within a function. \n\nSome transfer functions other than Linear that you could use are: Exponential, Logistic, or SoftMax. An Exponential function raises some input number to an exponent (e.g. squaring a number is an exponent of 2, and the square root of a number is an exponent of 1/2). The output of a Logistic function is bounded between 0 and 1, and we'll learn a bit more about it later in this tutorial. SoftMax is a more complex function that is often used in neural networks, and you don't need to understand how it works yet. \n\nNext let's try inputing the number 2 to our linear transfer mechanism...",
"_____no_output_____"
]
],
[
[
"linear_transfer_mechanism.execute([2])",
"_____no_output_____"
]
],
[
[
"Try reparamaterizing the mechanism (change the slope and/or intercept) and executing again before moving on... \n\nIf you change slope to 3, run the code by pressing play, then change the input value to 4, what output do you get? Why? Can you predict what will happen if you change the slope to 4 and intercept to 5 and run both cells again with an input of 3? \n\nAnother way of expressing this function is (slope x input) + intercept. \n\n### Logistic Function\n\nThe following cell plots a logistic function with the default parameters; gain = 1, bias = 0, offset = 0. ",
"_____no_output_____"
]
],
[
[
"logistic_transfer_demo = pnl.TransferMechanism(function=pnl.Logistic(gain=1, bias=0, offset=0))\nlogistic_transfer_demo.plot()",
"_____no_output_____"
]
],
[
[
"In the cell below you can plug a single number into this function and get an output value. Your input corresponds to a point on the x axis, and the output is the corresponding y value (height of the point on the curve above the x you specified). ",
"_____no_output_____"
]
],
[
[
"logistic_transfer_demo.execute([-2])",
"_____no_output_____"
]
],
[
[
"The logistic function is useful because it is bounded between 0 and 1. Gain determines how steep the central portion of the S curve is, with higher values being steeper. Bias shifts the curve left or right. You can turn the logistic function effectively into a step function that works as a threshhold by increasing gain. The step in the step function (where it crosses through 0.5 on the Y axis) is located on the X axis at (offset/gain) + bias.",
"_____no_output_____"
]
],
[
[
"logistic_transfer_offgain = pnl.TransferMechanism(function=pnl.Logistic(gain=5,offset=10, bias=0))\nlogistic_transfer_offgain.plot()",
"_____no_output_____"
]
],
[
[
"Negative values of gain mirror reverse the S curve accross the vertical axis, centered at the x value of (offset/gain)+bias. Below notice that offset/gain is -2 (10/-5), and at an X value of -2 the Y value is 0.5. ",
"_____no_output_____"
]
],
[
[
"logistic_transfer_invert = pnl.TransferMechanism(function=pnl.Logistic(gain=-5, bias=0, offset=10))\nlogistic_transfer_invert.plot()",
"_____no_output_____"
]
],
[
[
"### From Nodes to Graphs: Compositions\n\nGenerally with PsyNeuLink, you won't be executing mechanisms as stand-alone entities. Rather, mechanisms will be nodes in a graph, with Projections as edges of the graph, connecting nodes. We call the graph a Composition. \n\nThe simplest kind of Composition graph is one-dimensional: a linear progression from one node to the next. Information flowing through this graph will enter as input, be processed in the first mechanism, transfered via projection to the next mechanism, and so on. You can think of this with an analogy to digestion. Chew the food first, then swallow and it is \"projected\" to the stomach, then the stomach soaks food in digestive acid to further break it down, then the output is projected to the small intestine where nutrients are absorbed. Note that the order matters -- the small intestine wouldn't be effective if food hadn't been chewed and then broken down in the stompach. \n\nA Mechanism takes some input, performs a function, and delivers an output. The same is typically true of Compositions -- they take some input, perform multiple functions using multiple Mechanisms, and deliver some output. A powerful feature of this input-output architecture is that an entire Composition today can become a Mechanism tomorrow in a more complex Composition, and that Composition can become a Mechanisms in yet a more complex Composition, all the way up. [Note: this is true in principle, but PsyNeuLink is actively under development and such scaled up functionality is not all implemented.] \n\nThe main parameter when initializing a Composition is its pathway, which is the order in which the Mechanisms will execute. Of course, with only one Mechanism in our Composition, the list has just one element.\n\nTo better see how the Composition runs, we can also turn on output reporting. Reporting can happen at every level in PsyNeuLink and here we set the preference for the Mechanism.",
"_____no_output_____"
]
],
[
[
"comp_simplest = pnl.Composition()\ncomp_simplest.add_linear_processing_pathway(pathway = [linear_transfer_mechanism])\n \n\nlinear_transfer_mechanism.reportOutputPref = True\ncomp_simplest.show_graph(output_fmt = 'jupyter')",
"_____no_output_____"
],
[
"comp_simplest.run([4])",
"\n'TransferMechanism-14' executed:\n- input: 4.0\n- params:\n\tconvergence_criterion: 0.01\n\tconvergence_function: Distance Function-1\n\t\tintercept: 0\n\t\tslope: 1\n\tfunction: Linear Function-852\n\t\tintercept: 0\n\t\tslope: 1\n\tinitial_value: None\n\tintegration_rate: 0.5\n\tintegrator_mode: False\n\tmax_passes: 1000\n\tnoise: 0.\n- output: 2.0\n"
]
],
[
[
"Let's turn off the reporting and look at our process' output over a wider range of values.",
"_____no_output_____"
]
],
[
[
"linear_transfer_mechanism.reportOutputPref = False\n\nxVals = np.linspace(-3, 3, num=51) # create 51 points between -3 and +3\nyVals = np.zeros((51,))\nfor i in range(xVals.shape[0]):\n yVals[i] = comp_simplest.run([xVals[i]])[0]\n # Progress bar\n print(\"-\", end=\"\")\nplt.plot(xVals, yVals)\nplt.show()",
"---------------------------------------------------"
]
],
[
[
"Now let's put it all together and make a new transfer process, this time with a logistic activation function. We will also extend our mechanism by giving it two units (operating on a 1x2 matrix) rather than the default one (operating on a scalar).",
"_____no_output_____"
]
],
[
[
"# Create composition\ncomp_1x2 = pnl.Composition()\n\n# Create the mechanism\nlogistic_transfer_mechanism = pnl.TransferMechanism(default_variable=[0, 0],\n function=pnl.Logistic(gain=1,\n bias=0))\n# Place mechanism in composition\ncomp_1x2.add_linear_processing_pathway(pathway = [logistic_transfer_mechanism])\n\n\n# Iterate and plot\nxVals = np.linspace(-3, 3, num=51)\ny1Vals = np.zeros((51,))\ny2Vals = np.zeros((51,))\nfor i in range(xVals.shape[0]):\n # clarify why multiplying times 2\n output = comp_1x2.run([xVals[i], xVals[i] * 3])\n y1Vals[i] = output[0][0]\n y2Vals[i] = output[0][1]\n # Progress bar\n print(\"-\", end=\"\")\nplt.plot(xVals, y1Vals)\nplt.plot(xVals, y2Vals)\nplt.show()",
"---------------------------------------------------"
]
],
[
[
"The `default_variable` parameter serves a dual function. It specifies the dimensionality of the mechanism as well as providing the inputs that will be given in the absence of explicit input at runtime. You can also specify the dimensionality using \"size\", e.g. size=2 will also create default_variable=[0,0].",
"_____no_output_____"
]
],
[
[
"logistic_transfer_step = pnl.TransferMechanism(default_variable=[0, 0],\n function=pnl.Logistic(gain=100,\n offset=100))",
"_____no_output_____"
],
[
"logistic_transfer_step.execute([.9,1.1])",
"_____no_output_____"
]
],
[
[
"### Adding Projections\n\nTo connect Mechanisms together in a Composition, we need a way to link mechanisms together. This is done through *[Projections](https://princetonuniversity.github.io/PsyNeuLink/Projection.html)*. A projection takes a mechanism output, multiplies it by the projection's mapping matrix, and delivers the transformed value to the next mechanism in the Composition. ",
"_____no_output_____"
]
],
[
[
"# Create composition\ncomp_linlog = pnl.Composition()\n\n# Create mechanisms\nlinear_input_unit = pnl.TransferMechanism(function=pnl.Linear(slope=2, intercept=2))\nlogistic_output_unit = pnl.TransferMechanism(function=pnl.Logistic())\n\n# Place mechanism in composition\ncomp_linlog.add_linear_processing_pathway(pathway = [linear_input_unit, pnl.IDENTITY_MATRIX, logistic_output_unit])\n\n\n# Iterate and plot\nxVals = np.linspace(-3, 3, num=51)\nyVals = np.zeros((51,))\nfor i in range(xVals.shape[0]):\n yVals[i] = comp_linlog.run([xVals[i]])[0]\n # Progress bar\n print(\"-\", end=\"\")\nplt.plot(xVals, yVals)\nplt.show()",
"---------------------------------------------------"
]
],
[
[
"`IDENTITY_MATRIX` is a keyword that provides a projection from the unit preceding it to the unit following that creates a one-to-one output to input projection between the two. \n\nNow let's make our projection definition a bit more explicit.",
"_____no_output_____"
]
],
[
[
"# Create composition\ncomp_explicit = pnl.Composition()\n\n# Create mechanisms\nlinear_input_unit = pnl.TransferMechanism(function=pnl.Linear(slope=2, intercept=2), name=\"linear\")\nlogistic_output_unit = pnl.TransferMechanism(function=pnl.Logistic(), name=\"logistic\")\n\n# Create projection\nmapping_matrix = np.asarray([[1]])\nunit_mapping_projection = pnl.MappingProjection(sender=linear_input_unit,\n receiver=logistic_output_unit,\n matrix=mapping_matrix)\n\n# Place mechanisms and projections in composition\ncomp_explicit.add_linear_processing_pathway(pathway = [linear_input_unit, unit_mapping_projection, logistic_output_unit])\n\n\n# Iterate and plot\nxVals = np.linspace(-3, 3, num=51)\nyVals = np.zeros((51,))\nfor i in range(xVals.shape[0]):\n yVals[i] = comp_explicit.run([xVals[i]])[0]\n # Progress bar\n print(\"-\", end=\"\")\nplt.plot(xVals, yVals)\nplt.show()",
"---------------------------------------------------"
],
[
"comp_explicit.show_graph(output_fmt = 'jupyter')",
"_____no_output_____"
]
],
[
[
"This time we specified our mapping matrix (which is a 2-D numpy array) then explicitly initialized a *[MappingProjection](https://princetonuniversity.github.io/PsyNeuLink/MappingProjection.html)* with that matrix as well as its input and output mechanisms. Note: because we specified the input and output mechanisms in the projection itself, we didn't need to include it in the composition pathway as the composition would have infered its position from those parameters. Ultimately, however, this does the exact same thing as our keyword method above which is far less verbose for this common use case.",
"_____no_output_____"
],
[
"### Compositions\n\nThe highest level at which models are considered in PsyNeuLink is that of the *[Composition](https://princetonuniversity.github.io/PsyNeuLink/Composition.html)*. A composition is built out of one or more mechanisms connected by projections. This allows Composition graphs to be more complex than the strictly linear ones covered so far. Our first composition will consist of two input nodes that converge on a single output mechanism. We will be modelling competition between color naming and word reading in the Stroop task. [Recall that the Stroop task involves naming the ink color of words: when the word itself mismatches with the ink color it is written in (e.g. the word RED written in blue ink), that produces conflict and slower response times to name the ink color.] \n\n### Mini-Stroop Model",
"_____no_output_____"
]
],
[
[
"mini_stroop = pnl.Composition()\n\ncolors = pnl.TransferMechanism(default_variable=[0, 0], function=pnl.Linear,\n name=\"Colors\")\nwords = pnl.TransferMechanism(default_variable=[0, 0],\n function=pnl.Linear(slope=1.5), name=\"Words\")\nresponse = pnl.TransferMechanism(default_variable=[0, 0], function=pnl.Logistic,\n name=\"Response\")\n\n\n# Place mechanisms and projections in composition\nmini_stroop.add_linear_processing_pathway(pathway = [colors, response])\nmini_stroop.add_linear_processing_pathway(pathway = [words, response])\n\n\nmini_stroop.show_graph(output_fmt = 'jupyter')",
"_____no_output_____"
]
],
[
[
"An input of [1, 0] to ink is red ink, and [0, 1] is blue ink. \nAn input of [1, 0] to word is \"red\" and [0, 1] is \"blue\". \nIf the input values to ink and word are the same, that is a congruent trial. If the input values to the ink and word are different that is an incongruent trial. \n\nIn the output of the Composition, the first output value is the strength to respond red, and the second value is the strength to respond blue. \n\nIn the following two cells we will first specify a congruent trial (blue ink, word \"blue\") to see the output, and then specify an incongruent trial (red ink, word \"blue\"). ",
"_____no_output_____"
]
],
[
[
"input_allblue = {colors: [0, 1],\n words: [0, 1]}\nmini_stroop.run(input_allblue)",
"_____no_output_____"
],
[
"input_redblue = {colors: [1, 0], \n words: [0, 1]}\nmini_stroop.run(input_redblue)",
"_____no_output_____"
]
],
[
[
"### Pre-trained Complete Stroop Model\n\nLet's practice using compositions by recreating the more complex stroop model from Cohen et al (1990). Later we will train the network ourselves, but for now we will explicitly model the learned weights.",
"_____no_output_____"
]
],
[
[
"stroop_model = pnl.Composition()\n\nink_color = pnl.TransferMechanism(default_variable=[0, 0], function=pnl.Linear(),name=\"Ink_Color\")\nword = pnl.TransferMechanism(default_variable=[0, 0], function=pnl.Linear(),name=\"Word\")\ntask_demand = pnl.TransferMechanism(default_variable=[0, 0], function=pnl.Linear(),name=\"Task\")\n\nhidden_layer = pnl.TransferMechanism(default_variable=[0, 0, 0, 0],\n function=pnl.Logistic(bias=-4), name=\"Hidden\")\n\noutput_layer = pnl.TransferMechanism(default_variable=[[0, 0]], function=pnl.Linear(), name=\"Output\")\n\n\ncolor_mapping_matrix = np.asarray([[2.2, -2.2, 0, 0], [-2.2, 2.2, 0, 0]])\ncolor_projection = pnl.MappingProjection(sender=ink_color, receiver=hidden_layer,\n matrix=color_mapping_matrix)\nword_mapping_matrix = np.asarray([[0, 0, 2.6, -2.6], [0, 0, -2.6, 2.6]])\nword_projection = pnl.MappingProjection(sender=word, receiver=hidden_layer,\n matrix=word_mapping_matrix)\ntask_mapping_matrix = np.asarray([[4, 4, 0, 0], [0, 0, 4, 4]])\ntask_projection = pnl.MappingProjection(sender=task_demand, receiver=hidden_layer,\n matrix=task_mapping_matrix)\noutput_mapping_matrix = np.asarray(\n [[1.3, -1.3], [-1.3, 1.3], [2.5, -2.5], [-2.5, 2.5]])\noutput_projection = pnl.MappingProjection(sender=hidden_layer, receiver=output_layer,\n matrix=output_mapping_matrix)\n\n\nstroop_model.add_linear_processing_pathway(pathway = [ink_color, color_projection, hidden_layer, output_projection, output_layer])\nstroop_model.add_linear_processing_pathway(pathway = [word, word_projection, hidden_layer, output_projection, output_layer])\nstroop_model.add_linear_processing_pathway(pathway = [task_demand, task_projection, hidden_layer, output_projection, output_layer])\n\n\nink_color.reportOutputPref = True\nword.reportOutputPref = True\ntask_demand.reportOutputPref = True\nhidden_layer.reportOutputPref = True\n\nstroop_model.show_graph(output_fmt = 'jupyter')",
"_____no_output_____"
]
],
[
[
"In the next cell we will run the model with inputs. The \"ink_color\" and \"word\" are the same as the previous model, and the addition of task demand allows us to specify whether the task is to name the color of ink [0, 1], or to read the word [1, 0]. The output can be thought of as activation strengths of two possible responses [red, blue]. ",
"_____no_output_____"
]
],
[
[
"input_dict = {ink_color: [1, 0],\n word: [0, 1],\n task_demand: [1, 0]}\nstroop_model.run(input_dict)",
"\n'Ink_Color' mechanism executed:\n- input: [array([1., 0.])]\n- params:\n\tconvergence_criterion: 0.01\n\tconvergence_function: Distance Function-1\n\t\tintercept: 0.0\n\t\tslope: 1.0\n\tfunction: Linear Function-1038\n\t\tintercept: 0.0\n\t\tslope: 1.0\n\tinitial_value: None\n\tintegration_rate: 0.5\n\tintegrator_mode: False\n\tmax_passes: 1000\n\tnoise: 0.\n- output: 0.0 0.0\n\n'Word-1' mechanism executed:\n- input: [array([0., 1.])]\n- params:\n\tconvergence_criterion: 0.01\n\tconvergence_function: Distance Function-1\n\t\tintercept: 0.0\n\t\tslope: 1.0\n\tfunction: Linear Function-1048\n\t\tintercept: 0.0\n\t\tslope: 1.0\n\tinitial_value: None\n\tintegration_rate: 0.5\n\tintegrator_mode: False\n\tmax_passes: 1000\n\tnoise: 0.\n- output: 0.0 0.0\n\n'Task-2' mechanism executed:\n- input: [array([1., 0.])]\n- params:\n\tconvergence_criterion: 0.01\n\tconvergence_function: Distance Function-1\n\t\tintercept: 0.0\n\t\tslope: 1.0\n\tfunction: Linear Function-1058\n\t\tintercept: 0.0\n\t\tslope: 1.0\n\tinitial_value: None\n\tintegration_rate: 0.5\n\tintegrator_mode: False\n\tmax_passes: 1000\n\tnoise: 0.\n- output: 0.0 0.0\n\n'Hidden-1' mechanism executed:\n- input: [array([ 6.2, 1.8, -2.6, 2.6])]\n- params:\n\tconvergence_criterion: 0.01\n\tconvergence_function: Distance Function-1\n\t\tbias: -4\n\t\tgain: 1.0\n\t\toffset: 0.0\n\t\tscale: 1.0\n\t\tx_0: 0.0\n\tfunction: Logistic Function-19\n\t\tbias: -4\n\t\tgain: 1.0\n\t\toffset: 0.0\n\t\tscale: 1.0\n\t\tx_0: 0.0\n\tinitial_value: None\n\tintegration_rate: 0.5\n\tintegrator_mode: False\n\tmax_passes: 1000\n\tnoise: 0.\n- output: 0.018 0.018 0.018 0.018\n"
]
],
[
[
"To get a better sense of how the model works, try reverse engineering by changing each of the inputs (remember the options are only [1,0] or [0,1]) one at a time and running the model. Given 3 input states (Task, Word, Ink) with 2 options each, there will be 8 possibilities (2^3). \n\n",
"_____no_output_____"
],
[
"# Constructing Compositions\n\nAs shown in the Stroop models above, mechanisms are the building blocks in PsyNeuLink, and projections are how these building blocks get connected. The configuration of mechanisms connected by projections determines how information will flow and be processed within each model. Next we'll explore how to build models with different configurations of mechanisms and projections, along with generating diagrams that visualize these configurations. ",
"_____no_output_____"
]
],
[
[
"# Create composition\ncomp_line2 = pnl.Composition()\n\ninput_layer_cL2 = pnl.TransferMechanism(\n name='input_layer_cL2',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\noutput_layer_cL2 = pnl.TransferMechanism(\n name='output_layer_cL2',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\n# Place mechanisms in composition\ncomp_line2.add_linear_processing_pathway(pathway = [input_layer_cL2, output_layer_cL2])\n\n\ninput_layer_cL2.reportOutputPref = True\noutput_layer_cL2.reportOutputPref = True\n\ncomp_line2.show_graph(output_fmt = 'jupyter')",
"_____no_output_____"
],
[
"# Create composition\ncomp_line3 = pnl.Composition()\n\n# Create mechanisms\ninput_layer_cL3 = pnl.TransferMechanism(\n name='input_layer_cL3',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\n\nhidden_layer_cL3 = pnl.TransferMechanism(\n name='hidden_layer_cL3',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\noutput_layer_cL3 = pnl.TransferMechanism(\n name='output_layer_cL3',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\n\n\n# Place mechanisms in composition\ncomp_line3.add_linear_processing_pathway(pathway = [input_layer_cL3, hidden_layer_cL3, output_layer_cL3])\n\ninput_layer_cL3.reportOutputPref = True\nhidden_layer_cL3.reportOutputPref = True\noutput_layer_cL3.reportOutputPref = True\n\ncomp_line3.show_graph(output_fmt = 'jupyter')",
"_____no_output_____"
],
[
"# Create composition\ncomp_recur1 = pnl.Composition()\n\ninput_r1 = pnl.TransferMechanism(\n name='input_r1',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\n\nhidden_r1a = pnl.TransferMechanism(\n name='hidden_r1a',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\nhidden_r1b = pnl.TransferMechanism(\n name='hidden_r1b',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\noutput_r1 = pnl.TransferMechanism(\n name='output_r1',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\n\n# Place mechanisms in composition\ncomp_recur1.add_linear_processing_pathway(pathway = [input_r1, hidden_r1a, hidden_r1b, output_r1])\ncomp_recur1.add_linear_processing_pathway(pathway = [hidden_r1b, hidden_r1a])\n\n\ninput_r1.reportOutputPref = True\nhidden_r1a.reportOutputPref = True\nhidden_r1b.reportOutputPref = True\noutput_r1.reportOutputPref = True\n\ncomp_recur1.show_graph(output_fmt = 'jupyter')",
"_____no_output_____"
],
[
"# Create composition\ncomp_multihidden = pnl.Composition()\n\ninput_mh = pnl.TransferMechanism(\n name='input_mh',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\nhidden_mh_a = pnl.TransferMechanism(\n name='hidden_mh_a',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\nhidden_mh_b = pnl.TransferMechanism(\n name='hidden_mh_b',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\nhidden_mh_c = pnl.TransferMechanism(\n name='hidden_mh_c',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\noutput_mh = pnl.TransferMechanism(\n name='output_mh',\n function=pnl.Linear,\n default_variable=np.ones((4,)),\n)\n\n\n# Place mechanisms in composition\ncomp_multihidden.add_linear_processing_pathway(pathway = [input_mh, hidden_mh_a, output_mh])\ncomp_multihidden.add_linear_processing_pathway(pathway = [input_mh, hidden_mh_b, output_mh])\ncomp_multihidden.add_linear_processing_pathway(pathway = [input_mh, hidden_mh_c, output_mh])\n\n\n\ninput_mh.reportOutputPref = True\nhidden_mh_a.reportOutputPref = True\nhidden_mh_b.reportOutputPref = True\nhidden_mh_c.reportOutputPref = True\noutput_mh.reportOutputPref = True\n\ncomp_multihidden.show_graph(output_fmt = 'jupyter')",
"_____no_output_____"
]
],
[
[
"This is currently the end of the tutorial, but more content is being added weekly. For further examples, look to the Scripts folder inside your PsyNeuLink directory for a variety of functioning models.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
eccc8bb880c934b3a1af7289362c79623f3b9b79 | 6,260 | ipynb | Jupyter Notebook | notebooks/rust-101-chp2.ipynb | rarebreed/cogito | 6976bb817076da7fedd7ac97c52e4ea047c082a6 | [
"Apache-2.0"
] | null | null | null | notebooks/rust-101-chp2.ipynb | rarebreed/cogito | 6976bb817076da7fedd7ac97c52e4ea047c082a6 | [
"Apache-2.0"
] | 5 | 2022-02-20T15:57:55.000Z | 2022-02-20T21:59:23.000Z | notebooks/rust-101-chp2.ipynb | rarebreed/cogito | 6976bb817076da7fedd7ac97c52e4ea047c082a6 | [
"Apache-2.0"
] | null | null | null | 31.77665 | 130 | 0.444888 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
eccc8df8369811478df7a77d84c82b8e08b03b4b | 974,476 | ipynb | Jupyter Notebook | Employee_Retention_Prediction.ipynb | ManishSpk/Employee-Retention-Prediction | fffb2311ecbf25d613818e58e905f75ec054ce31 | [
"MIT"
] | null | null | null | Employee_Retention_Prediction.ipynb | ManishSpk/Employee-Retention-Prediction | fffb2311ecbf25d613818e58e905f75ec054ce31 | [
"MIT"
] | null | null | null | Employee_Retention_Prediction.ipynb | ManishSpk/Employee-Retention-Prediction | fffb2311ecbf25d613818e58e905f75ec054ce31 | [
"MIT"
] | null | null | null | 191.863753 | 404,066 | 0.832621 | [
[
[
"<a href=\"https://colab.research.google.com/github/ManishSpk/Employee-Retention-Prediction/blob/main/Employee_Retention_Prediction.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Mounted at /content/drive\n"
],
[
"employee_df = pd.read_csv('/content/drive/My Drive/Data Science Projects/Human_Resources.csv')",
"_____no_output_____"
],
[
"employee_df",
"_____no_output_____"
],
[
"employee_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1470 entries, 0 to 1469\nData columns (total 35 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Age 1470 non-null int64 \n 1 Attrition 1470 non-null object\n 2 BusinessTravel 1470 non-null object\n 3 DailyRate 1470 non-null int64 \n 4 Department 1470 non-null object\n 5 DistanceFromHome 1470 non-null int64 \n 6 Education 1470 non-null int64 \n 7 EducationField 1470 non-null object\n 8 EmployeeCount 1470 non-null int64 \n 9 EmployeeNumber 1470 non-null int64 \n 10 EnvironmentSatisfaction 1470 non-null int64 \n 11 Gender 1470 non-null object\n 12 HourlyRate 1470 non-null int64 \n 13 JobInvolvement 1470 non-null int64 \n 14 JobLevel 1470 non-null int64 \n 15 JobRole 1470 non-null object\n 16 JobSatisfaction 1470 non-null int64 \n 17 MaritalStatus 1470 non-null object\n 18 MonthlyIncome 1470 non-null int64 \n 19 MonthlyRate 1470 non-null int64 \n 20 NumCompaniesWorked 1470 non-null int64 \n 21 Over18 1470 non-null object\n 22 OverTime 1470 non-null object\n 23 PercentSalaryHike 1470 non-null int64 \n 24 PerformanceRating 1470 non-null int64 \n 25 RelationshipSatisfaction 1470 non-null int64 \n 26 StandardHours 1470 non-null int64 \n 27 StockOptionLevel 1470 non-null int64 \n 28 TotalWorkingYears 1470 non-null int64 \n 29 TrainingTimesLastYear 1470 non-null int64 \n 30 WorkLifeBalance 1470 non-null int64 \n 31 YearsAtCompany 1470 non-null int64 \n 32 YearsInCurrentRole 1470 non-null int64 \n 33 YearsSinceLastPromotion 1470 non-null int64 \n 34 YearsWithCurrManager 1470 non-null int64 \ndtypes: int64(26), object(9)\nmemory usage: 402.1+ KB\n"
],
[
"employee_df.describe()",
"_____no_output_____"
]
],
[
[
"VISUALIZE DATASET",
"_____no_output_____"
]
],
[
[
"# Let's replace 'Attritition' , 'overtime' , 'Over18' column with integers before performing any visualizations \nemployee_df['Attrition'] = employee_df['Attrition'].apply(lambda x:1 if x == 'Yes' else 0)\nemployee_df['Over18'] = employee_df['Over18'].apply(lambda x:1 if x == 'Y' else 0)\nemployee_df['OverTime'] = employee_df['OverTime'].apply(lambda x:1 if x == 'Yes' else 0)\n",
"_____no_output_____"
],
[
"employee_df",
"_____no_output_____"
],
[
"# Let's see if we have any missing data\nsns.heatmap(employee_df.isnull(), yticklabels = False, cbar = False, cmap = 'Blues')",
"_____no_output_____"
],
[
"employee_df.hist(bins = 30, figsize = (20,20), color = 'r')\n# Several features such as 'MonthlyIncome' and 'TotalWorkingYears' are tail heavy\n# It makes sense to drop 'EmployeeCount' and 'Standardhours' since they do not change from one employee to the other",
"_____no_output_____"
],
[
"# It makes sense to drop 'EmployeeCount' , 'Standardhours' and 'Over18' since they do not change from one employee to the other\n# Let's drop 'EmployeeNumber' as well\nemployee_df.drop(['EmployeeCount', 'StandardHours', 'Over18', 'EmployeeNumber'], axis = 1, inplace = True)",
"_____no_output_____"
],
[
"# Let's see how many employees left the company! \nleft_df = employee_df[employee_df['Attrition'] == 1]\nstayed_df = employee_df[employee_df['Attrition'] == 0]",
"_____no_output_____"
],
[
"# Count the number of employees who stayed and left\n# It seems that we are dealing with an imbalanced dataset \nprint(\"Total =\", len(employee_df))\n\nprint(\"Number of employees who left the company =\", len(left_df))\nprint(\"Percentage of employees who left the company =\", 1.*len(left_df)/len(employee_df)*100.0, \"%\")\n \nprint(\"Number of employees who did not leave the company (stayed) =\", len(stayed_df))\nprint(\"Percentage of employees who did not leave the company (stayed) =\", 1.*len(stayed_df)/len(employee_df)*100.0, \"%\")",
"Total = 1470\nNumber of employees who left the company = 237\nPercentage of employees who left the company = 16.122448979591837 %\nNumber of employees who did not leave the company (stayed) = 1233\nPercentage of employees who did not leave the company (stayed) = 83.87755102040816 %\n"
],
[
"left_df.describe()",
"_____no_output_____"
],
[
"stayed_df.describe()",
"_____no_output_____"
],
[
"correlations = employee_df.corr()\nf, ax = plt.subplots(figsize = (20,20))\nsns.heatmap(correlations, annot = True)",
"_____no_output_____"
],
[
"plt.figure(figsize = [25,12])\nsns.countplot(x = 'Age', hue = 'Attrition', data = employee_df)\n# Job level is strongly correlated with total working hours\n# Monthly income is strongly correlated with Job level\n# Monthly income is strongly correlated with total working hours\n# Age is stongly correlated with monthly income\n",
"_____no_output_____"
],
[
"plt.figure(figsize = [20,20])\nplt.subplot(411)\nsns.countplot(x= 'JobRole', hue = 'Attrition', data = employee_df)\nplt.subplot(412)\nsns.countplot(x= 'MaritalStatus', hue = 'Attrition', data = employee_df)\nplt.subplot(413)\nsns.countplot(x= 'JobInvolvement', hue = 'Attrition', data = employee_df)\nplt.subplot(414)\nsns.countplot(x= 'JobLevel', hue = 'Attrition', data = employee_df)",
"_____no_output_____"
],
[
"# Single employees tend to leave compared to married and divorced\n# Sales Representitives tend to leave compared to any other job \n# Less involved employees tend to leave the company \n# Less experienced (low job level) tend to leave the company ",
"_____no_output_____"
],
[
"# KDE (Kernel Density Estimate) is used for visualizing the Probability Density of a continuous variable. \n# KDE describes the probability density at different values in a continuous variable. \nplt.figure(figsize=(12,7))\n\nsns.kdeplot(left_df['DistanceFromHome'], label = 'Employees who left', shade = True, color = 'r')\nsns.kdeplot(stayed_df['DistanceFromHome'], label = 'Employees who Stayed', shade = True, color = 'b')\n\nplt.xlabel('Distance From Home')\nplt.legend()\n",
"_____no_output_____"
],
[
"plt.figure(figsize=(12,7))\n\nsns.kdeplot(left_df['YearsWithCurrManager'], label = 'Employees who left', shade = True, color = 'r')\nsns.kdeplot(stayed_df['YearsWithCurrManager'], label = 'Employees who Stayed', shade = True, color = 'b')\n\nplt.xlabel('Years With Current Manager')\nplt.legend()",
"_____no_output_____"
],
[
"\nplt.figure(figsize=(12,7))\n\nsns.kdeplot(left_df['TotalWorkingYears'], label = 'Employees who left', shade = True, color = 'r')\nsns.kdeplot(stayed_df['TotalWorkingYears'], label = 'Employees who Stayed', shade = True, color = 'b')\n\nplt.xlabel('Total Working Years')\nplt.legend()",
"_____no_output_____"
],
[
"# Let's see the Gender vs. Monthly Income\nsns.boxplot(x = 'MonthlyIncome', y = 'Gender', data = employee_df)",
"_____no_output_____"
],
[
"# Let's see the Gender vs. Monthly Income\nplt.figure(figsize=(15, 10))\nsns.boxplot(x = 'MonthlyIncome', y = 'JobRole', data = employee_df)",
"_____no_output_____"
]
],
[
[
"CREATE TESTING AND TRAINING DATASET & PERFORM DATA CLEANING",
"_____no_output_____"
]
],
[
[
"employee_df.head(3)",
"_____no_output_____"
],
[
"X_cat = employee_df[['BusinessTravel', 'Department', 'EducationField', 'Gender', 'JobRole', 'MaritalStatus' ]]",
"_____no_output_____"
],
[
"X_cat",
"_____no_output_____"
],
[
"from sklearn.preprocessing import OneHotEncoder\nonehotencoder = OneHotEncoder()\nX_cat = onehotencoder.fit_transform(X_cat).toarray()",
"_____no_output_____"
],
[
"X_cat.shape",
"_____no_output_____"
],
[
"X_cat = pd.DataFrame(X_cat) \nX_cat",
"_____no_output_____"
],
[
"# note that we dropped the target 'Atrittion'\nX_numerical = employee_df[['Age', 'DailyRate', 'DistanceFromHome',\t'Education', 'EnvironmentSatisfaction', 'HourlyRate', 'JobInvolvement',\t'JobLevel',\t'JobSatisfaction',\t'MonthlyIncome',\t'MonthlyRate',\t'NumCompaniesWorked',\t'OverTime',\t'PercentSalaryHike', 'PerformanceRating',\t'RelationshipSatisfaction',\t'StockOptionLevel',\t'TotalWorkingYears'\t,'TrainingTimesLastYear'\t, 'WorkLifeBalance',\t'YearsAtCompany'\t,'YearsInCurrentRole', 'YearsSinceLastPromotion',\t'YearsWithCurrManager']]\nX_numerical",
"_____no_output_____"
],
[
"X_all = pd.concat([X_cat, X_numerical], axis = 1)\nX_all",
"_____no_output_____"
],
[
"from sklearn.preprocessing import MinMaxScaler\nscaler = MinMaxScaler()\nX = scaler.fit_transform(X_all)",
"_____no_output_____"
],
[
"y = employee_df['Attrition']\ny",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X,y, test_size =0.25)",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"X_test.shape",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import accuracy_score\n\nmodel = LogisticRegression()\nmodel.fit(X_train, y_train)\n\ny_pred = model.predict(X_test)",
"_____no_output_____"
],
[
"y_pred",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix, classification_report\n\nprint(\"Accuracy {} %\".format( 100 * accuracy_score(y_pred, y_test)))",
"Accuracy 85.86956521739131 %\n"
],
[
"# Testing Set Performance\ncm = confusion_matrix(y_pred, y_test)\nsns.heatmap(cm, annot=True)\n",
"_____no_output_____"
],
[
"print(classification_report(y_test, y_pred))",
" precision recall f1-score support\n\n 0 0.88 0.96 0.92 307\n 1 0.64 0.34 0.45 61\n\n accuracy 0.86 368\n macro avg 0.76 0.65 0.68 368\nweighted avg 0.84 0.86 0.84 368\n\n"
]
],
[
[
"TRAIN AND EVALUATE A RANDOM FOREST CLASSIFIER",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\n\nmodel = RandomForestClassifier()\nmodel.fit(X_train, y_train)\n",
"_____no_output_____"
],
[
"y_pred = model.predict(X_test)",
"_____no_output_____"
],
[
"# Testing Set Performance\ncm = confusion_matrix(y_pred, y_test)\nsns.heatmap(cm, annot=True)",
"_____no_output_____"
],
[
"print(classification_report(y_test, y_pred))",
" precision recall f1-score support\n\n 0 0.85 0.99 0.92 307\n 1 0.73 0.13 0.22 61\n\n accuracy 0.85 368\n macro avg 0.79 0.56 0.57 368\nweighted avg 0.83 0.85 0.80 368\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
eccc95536621445c903455b7b39c9c2e84747f75 | 373,128 | ipynb | Jupyter Notebook | src/Task1/Report_task1.ipynb | Team-Horus/project-athena | 0b59c302cc8fb414607e871a2ae0bc0758ca7400 | [
"MIT"
] | null | null | null | src/Task1/Report_task1.ipynb | Team-Horus/project-athena | 0b59c302cc8fb414607e871a2ae0bc0758ca7400 | [
"MIT"
] | null | null | null | src/Task1/Report_task1.ipynb | Team-Horus/project-athena | 0b59c302cc8fb414607e871a2ae0bc0758ca7400 | [
"MIT"
] | null | null | null | 283.96347 | 13,452 | 0.913585 | [
[
[
"#Please Run this file in '../src' directory\n\nimport os\nimport sys\nmodule_path = os.path.abspath(os.path.join('../src'))\nif module_path not in sys.path:\n sys.path.append(module_path)\n \nimport numpy as np\nimport os\nfrom matplotlib import pyplot as plt",
"_____no_output_____"
]
],
[
[
"## (Optional) Prepare a smaller dataset for your experiment\n\n* python example: `tutorials/subsamples.py`\n* api: `utils.data.subsampling`",
"_____no_output_____"
]
],
[
[
"from utils.data import subsampling\nfrom utils.file import load_from_json\n\n# load the configurations for the experiment\ndata_configs = load_from_json(\"../src/configs/demo/data-mnist.json\")\noutput_root = \"../results\"\n# print(data_configs.get('bs_file'))\n\n\n# load the full-sized benign samples\nfile = os.path.join(data_configs.get('dir'), data_configs.get('bs_file'))\n# print('getcwd: ', os.getcwd())\n\n# print(file)\n# print(dir(file))\n# print(data_configs.get('dir'))\n# print(data_configs.get('label_file'))\nX_bs = np.load(file)\n\n# load the corresponding true labels\nfile = os.path.join(data_configs.get('dir'), data_configs.get('label_file'))\nprint(file)\nlabels = np.load(file)\n\n# get random subsamples\n# for MNIST, num_classes is 10\n# files \"subsamples-mnist-ratio_0.1-xxxxxx.npy\" and \"sublabels-mnist-ratio_0.1-xxxxxx.npy\"\n# will be generated and saved at \"/results\" folder, where \"xxxxxx\" are timestamps.\nsubsamples, sublabels = subsampling(data=X_bs,\n labels=labels,\n num_classes=10,\n filepath=output_root,\n filename='mnist')",
"Using TensorFlow backend.\nC:\\Users\\hezek\\.conda\\envs\\athena\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:526: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\nC:\\Users\\hezek\\.conda\\envs\\athena\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:527: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\nC:\\Users\\hezek\\.conda\\envs\\athena\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:528: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\nC:\\Users\\hezek\\.conda\\envs\\athena\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:529: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\nC:\\Users\\hezek\\.conda\\envs\\athena\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:530: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\nC:\\Users\\hezek\\.conda\\envs\\athena\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:535: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\n"
]
],
[
[
"## Update the json file accordingly\n\n1. Copy and paste the generated subsamples to the right place (i.e., defined by `\"dir\"` in `data-mnist.json`).\n2. In the `data-mnist.json`, replace the value of `\"bs_file\"` with the `\"subsamples-mnist-ratio_0.1-xxxxxx.npy\"` and the value of `\"label_file\"` with the `\"sublabels-mnist-ratio_0.1-xxxxxx.npy\"`.\n\n# Generate adversarial examples\nWe use `FGSM` as the example.\n* python example: `tutorials/craft_adversarial_examples.py`\n* main api: `attacks.attack.generate`\n* check tunable parameters for each attack in file `attacks/attack.py`.",
"_____no_output_____"
]
],
[
[
"# copied from tutorials/craft_adversarial_examples.py\ndef generate_ae(model, data, labels, attack_configs, save=False, output_dir=None):\n \"\"\"\n Generate adversarial examples\n :param model: WeakDefense. The targeted model.\n :param data: array. The benign samples to generate adversarial for.\n :param labels: array or list. The true labels.\n :param attack_configs: dictionary. Attacks and corresponding settings.\n :param save: boolean. True, if save the adversarial examples.\n :param output_dir: str or path. Location to save the adversarial examples.\n It cannot be None when save is True.\n :return:\n \"\"\"\n img_rows, img_cols = data.shape[1], data.shape[2]\n num_attacks = attack_configs.get(\"num_attacks\")\n data_loader = (data, labels)\n\n if len(labels.shape) > 1:\n labels = np.asarray([np.argmax(p) for p in labels])\n\n # generate attacks one by one\n for id in range(num_attacks):\n key = \"configs{}\".format(id)\n data_adv = generate(model=model,\n data_loader=data_loader,\n attack_args=attack_configs.get(key)\n )\n # predict the adversarial examples\n predictions = model.predict(data_adv)\n predictions = np.asarray([np.argmax(p) for p in predictions])\n\n err = error_rate(y_pred=predictions, y_true=labels)\n print(\">>> error rate:\", err)\n\n # plotting some examples\n num_plotting = min(data.shape[0], 2)\n for i in range(num_plotting):\n img = data_adv[i].reshape((img_rows, img_cols))\n plt.imshow(img, cmap='gray')\n title = '{}: {}->{}'.format(attack_configs.get(key).get(\"description\"),\n labels[i],\n predictions[i]\n )\n plt.title(title)\n plt.show()\n plt.close()\n\n # save the adversarial example\n if save:\n if output_dir is None:\n raise ValueError(\"Cannot save images to a none path.\")\n # save with a random name\n file = os.path.join(output_dir, \"{}.npy\".format(attack_configs.get(key).get(\"description\")))\n# file = os.path.join(output_dir, \"{}.npy\".format(time.monotonic()))\n print(\"Save the adversarial examples to file [{}].\".format(file))\n np.save(file, data_adv)",
"_____no_output_____"
],
[
"import time\nfrom utils.model import load_lenet\nfrom utils.metrics import error_rate\nfrom attacks.attack import generate\n\n# loading experiment configurations\nmodel_configs = load_from_json(\"../src/configs/demo/model-mnist.json\")\ndata_configs = load_from_json(\"../src/configs/demo/data-mnist.json\")\nattack_configs = load_from_json(\"../src/configs/demo/attack-zk-mnist.json\")\n\n# load the targeted model\nmodel_file = os.path.join(model_configs.get(\"dir\"), model_configs.get(\"um_file\"))\ntarget = load_lenet(file=model_file, wrap=True)\n\n# load the benign samples\n\ndata_file = os.path.join(data_configs.get('dir'), data_configs.get('bs_file'))\ndata_bs = np.load(data_file)\n# load the corresponding true labels\nlabel_file = os.path.join(data_configs.get('dir'), data_configs.get('label_file'))\nlabels = np.load(label_file)\n\n# generate AEs\n# in this project, we generate AEs for all benign samples\ndata_bs = data_bs[:]\nlabels = labels[:]\n# let save=True and specify an output folder to save the generated AEs\ngenerate_ae(model=target, data=data_bs, labels=labels, attack_configs=attack_configs,save = True, output_dir = \"../results\")",
">>> Loading model [../models/cnn/model-mnist-cnn-clean.h5]...\nWARNING:tensorflow:From C:\\Users\\hezek\\.conda\\envs\\athena\\lib\\site-packages\\tensorflow\\python\\ops\\resource_variable_ops.py:435: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\nWARNING:tensorflow:From C:\\Users\\hezek\\.conda\\envs\\athena\\lib\\site-packages\\tensorflow\\python\\ops\\math_ops.py:3066: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.cast instead.\n>>> Generating FGSM examples.\n>>> error rate: 0.143\n"
]
],
[
[
"## Update the generated adversarial examples in json\n\n* Either add an item for the generated AEs. e.g., assume that we named the generated AE as `\"fgsm_eps0.3.npy\"`, then add the item as below example, then get your AE list by `data_configs.get(\"task1_aes\")`.\n\n```\n\"task1_aes\" : [\n \"fgsm_eps0.3.npy\"\n ]\n\n```\n\n\n2. Or, create a new json file similar to `\"data-mnist.json\"`, and replace the whole list for `\"ae_files\"` with your own list.\n\n# Evaluate the generated AEs\n* python example: `tutorials/eval_model.py`\n* api: `utils.metrics.error_rate`",
"_____no_output_____"
]
],
[
[
"from utils.model import load_pool\nfrom utils.metrics import error_rate, get_corrections\nfrom models.athena import Ensemble, ENSEMBLE_STRATEGY\n\n# copied from tutorials/eval_model.py\ndef evaluate(trans_configs, model_configs,\n data_configs, save=False, output_dir=None):\n \"\"\"\n Apply transformation(s) on images.\n :param trans_configs: dictionary. The collection of the parameterized transformations to test.\n in the form of\n { configsx: {\n param: value,\n }\n }\n The key of a configuration is 'configs'x, where 'x' is the id of corresponding weak defense.\n :param model_configs: dictionary. Defines model related information.\n Such as, location, the undefended model, the file format, etc.\n :param data_configs: dictionary. Defines data related information.\n Such as, location, the file for the true labels, the file for the benign samples,\n the files for the adversarial examples, etc.\n :param save: boolean. Save the transformed sample or not.\n :param output_dir: path or str. The location to store the transformed samples.\n It cannot be None when save is True.\n :return:\n \"\"\"\n # Load the baseline defense (PGD-ADT model)\n baseline = load_lenet(file=model_configs.get('pgd_trained'), trans_configs=None,\n use_logits=False, wrap=False)\n\n # get the undefended model (UM)\n file = os.path.join(model_configs.get('dir'), model_configs.get('um_file'))\n undefended = load_lenet(file=file,\n trans_configs=trans_configs.get('configs0'),\n wrap=True)\n print(\">>> um:\", type(undefended))\n\n # load weak defenses into a pool\n pool, _ = load_pool(trans_configs=trans_configs,\n model_configs=model_configs,\n active_list=True,\n wrap=True)\n # create an AVEP ensemble from the WD pool\n wds = list(pool.values())\n print(\">>> wds:\", type(wds), type(wds[0]))\n ensemble = Ensemble(classifiers=wds, strategy=ENSEMBLE_STRATEGY.AVEP.value)\n\n # load the benign samples\n bs_file = os.path.join(data_configs.get('dir'), data_configs.get('bs_file'))\n x_bs = np.load(bs_file)\n img_rows, img_cols = x_bs.shape[1], x_bs.shape[2]\n\n # load the corresponding true labels\n label_file = os.path.join(data_configs.get('dir'), data_configs.get('label_file'))\n labels = np.load(label_file)\n\n # get indices of benign samples that are correctly classified by the targeted model\n print(\">>> Evaluating UM on [{}], it may take a while...\".format(bs_file))\n pred_bs = undefended.predict(x_bs)\n corrections = get_corrections(y_pred=pred_bs, y_true=labels)\n\n # Evaluate AEs.\n results = {}\n eval_result = {}\n# ae_list = data_configs.get('ae_files')\n ae_list = data_configs.get('task1_aes')\n \n for i in ae_list:\n ae_file = os.path.join(data_configs.get('dir'), i)\n# print(type(ae_file))\n# print(ae_file)\n #print(ae_list[4])\n x_adv = np.load(ae_file)\n\n # evaluate the undefended model on the AE\n print(\">>> Evaluating UM on [{}], it may take a while...\".format(ae_file))\n pred_adv_um = undefended.predict(x_adv)\n err_um = error_rate(y_pred=pred_adv_um, y_true=labels, correct_on_bs=corrections)\n # track the result\n results['UM'] = err_um\n if 'UM' not in eval_result:\n eval_result['UM'] = [err_um]\n else:\n eval_result['UM'].append(err_um)\n\n # evaluate the ensemble on the AE\n print(\">>> Evaluating ensemble on [{}], it may take a while...\".format(ae_file))\n pred_adv_ens = ensemble.predict(x_adv)\n err_ens = error_rate(y_pred=pred_adv_ens, y_true=labels, correct_on_bs=corrections)\n # track the result\n results['Ensemble'] = err_ens\n if 'Ensemble' not in eval_result:\n eval_result['Ensemble'] = [err_ens]\n else:\n eval_result['Ensemble'].append(err_ens)\n\n # evaluate the baseline on the AE\n print(\">>> Evaluating baseline model on [{}], it may take a while...\".format(ae_file))\n pred_adv_bl = baseline.predict(x_adv)\n err_bl = error_rate(y_pred=pred_adv_bl, y_true=labels, correct_on_bs=corrections)\n # track the result\n results['PGD-ADT'] = err_bl\n if 'PGD-ADT' not in eval_result:\n eval_result['PGD-ADT'] = [err_bl]\n else:\n eval_result['PGD-ADT'].append(err_bl)\n\n # TODO: collect and dump the evaluation results to file(s) such that you can analyze them later.\n print(\">>> Evaluations on [{}]:\\n{}\".format(ae_file, results))\n print(eval_result)\n return eval_result\n",
"_____no_output_____"
],
[
"# load experiment configurations\ntrans_configs = load_from_json(\"../src/configs/demo/athena-mnist.json\")\nmodel_configs = load_from_json(\"../src/configs/demo/model-mnist.json\")\ndata_configs = load_from_json(\"../src/configs/demo/data-mnist.json\")\n\noutput_root = \"../results\"\n\n# evaluate\neval_result = evaluate(trans_configs=trans_configs,\n model_configs=model_configs,\n data_configs=data_configs,\n save=True,\n output_dir=output_root)",
">>> Loading model [../models/baseline/advTrained-mnist-adtC.h5]...\n>>> Loading model [../models/cnn/model-mnist-cnn-clean.h5]...\n>>> um: <class 'models.keras.WeakDefense'>\n>>> Loading model [../models/cnn/model-mnist-cnn-flip_horizontal.h5]...\n>>> Loading model [../models/cnn/model-mnist-cnn-affine_both_stretch.h5]...\n>>> Loading model [../models/cnn/model-mnist-cnn-morph_gradient.h5]...\n>>> Loaded 3 models.\n>>> wds: <class 'list'> <class 'models.keras.WeakDefense'>\n>>> Evaluating UM on [../data/subsamples-mnist-ratio_0.1-3357.156.npy], it may take a while...\n>>> Evaluating UM on [../data/FGSM_eps0.07.npy], it may take a while...\n>>> Evaluating ensemble on [../data/FGSM_eps0.07.npy], it may take a while...\n>>> Evaluating baseline model on [../data/FGSM_eps0.07.npy], it may take a while...\n>>> Evaluations on [../data/FGSM_eps0.07.npy]:\n{'UM': 0.13157894736842105, 'Ensemble': 0.00708502024291498, 'PGD-ADT': 0.015182186234817813}\n>>> Evaluating UM on [../data/FGSM_eps0.10.npy], it may take a while...\n>>> Evaluating ensemble on [../data/FGSM_eps0.10.npy], it may take a while...\n>>> Evaluating baseline model on [../data/FGSM_eps0.10.npy], it may take a while...\n>>> Evaluations on [../data/FGSM_eps0.10.npy]:\n{'UM': 0.24493927125506074, 'Ensemble': 0.012145748987854251, 'PGD-ADT': 0.022267206477732792}\n>>> Evaluating UM on [../data/FGSM_eps0.15.npy], it may take a while...\n>>> Evaluating ensemble on [../data/FGSM_eps0.15.npy], it may take a while...\n>>> Evaluating baseline model on [../data/FGSM_eps0.15.npy], it may take a while...\n>>> Evaluations on [../data/FGSM_eps0.15.npy]:\n{'UM': 0.521255060728745, 'Ensemble': 0.032388663967611336, 'PGD-ADT': 0.043522267206477734}\n>>> Evaluating UM on [../data/FGSM_eps0.20.npy], it may take a while...\n>>> Evaluating ensemble on [../data/FGSM_eps0.20.npy], it may take a while...\n>>> Evaluating baseline model on [../data/FGSM_eps0.20.npy], it may take a while...\n>>> Evaluations on [../data/FGSM_eps0.20.npy]:\n{'UM': 0.7267206477732794, 'Ensemble': 0.09109311740890688, 'PGD-ADT': 0.07692307692307693}\n>>> Evaluating UM on [../data/FGSM_eps0.25.npy], it may take a while...\n>>> Evaluating ensemble on [../data/FGSM_eps0.25.npy], it may take a while...\n>>> Evaluating baseline model on [../data/FGSM_eps0.25.npy], it may take a while...\n>>> Evaluations on [../data/FGSM_eps0.25.npy]:\n{'UM': 0.832995951417004, 'Ensemble': 0.3259109311740891, 'PGD-ADT': 0.1214574898785425}\n>>> Evaluating UM on [../data/FGSM_eps0.30.npy], it may take a while...\n>>> Evaluating ensemble on [../data/FGSM_eps0.30.npy], it may take a while...\n>>> Evaluating baseline model on [../data/FGSM_eps0.30.npy], it may take a while...\n>>> Evaluations on [../data/FGSM_eps0.30.npy]:\n{'UM': 0.8866396761133604, 'Ensemble': 0.6771255060728745, 'PGD-ADT': 0.21963562753036436}\n>>> Evaluating UM on [../data/PGD_eps0.07.npy], it may take a while...\n>>> Evaluating ensemble on [../data/PGD_eps0.07.npy], it may take a while...\n>>> Evaluating baseline model on [../data/PGD_eps0.07.npy], it may take a while...\n>>> Evaluations on [../data/PGD_eps0.07.npy]:\n{'UM': 0.2783400809716599, 'Ensemble': 0.009109311740890687, 'PGD-ADT': 0.016194331983805668}\n>>> Evaluating UM on [../data/PGD_eps0.10.npy], it may take a while...\n>>> Evaluating ensemble on [../data/PGD_eps0.10.npy], it may take a while...\n>>> Evaluating baseline model on [../data/PGD_eps0.10.npy], it may take a while...\n>>> Evaluations on [../data/PGD_eps0.10.npy]:\n{'UM': 0.667004048582996, 'Ensemble': 0.013157894736842105, 'PGD-ADT': 0.02327935222672065}\n>>> Evaluating UM on [../data/PGD_eps0.15.npy], it may take a while...\n>>> Evaluating ensemble on [../data/PGD_eps0.15.npy], it may take a while...\n>>> Evaluating baseline model on [../data/PGD_eps0.15.npy], it may take a while...\n>>> Evaluations on [../data/PGD_eps0.15.npy]:\n{'UM': 0.9463562753036437, 'Ensemble': 0.0354251012145749, 'PGD-ADT': 0.05060728744939271}\n>>> Evaluating UM on [../data/PGD_eps0.20.npy], it may take a while...\n>>> Evaluating ensemble on [../data/PGD_eps0.20.npy], it may take a while...\n>>> Evaluating baseline model on [../data/PGD_eps0.20.npy], it may take a while...\n>>> Evaluations on [../data/PGD_eps0.20.npy]:\n{'UM': 0.9868421052631579, 'Ensemble': 0.0708502024291498, 'PGD-ADT': 0.06983805668016195}\n>>> Evaluating UM on [../data/PGD_eps0.25.npy], it may take a while...\n>>> Evaluating ensemble on [../data/PGD_eps0.25.npy], it may take a while...\n>>> Evaluating baseline model on [../data/PGD_eps0.25.npy], it may take a while...\n>>> Evaluations on [../data/PGD_eps0.25.npy]:\n{'UM': 0.9878542510121457, 'Ensemble': 0.14979757085020243, 'PGD-ADT': 0.11639676113360324}\n>>> Evaluating UM on [../data/PGD_eps0.30.npy], it may take a while...\n>>> Evaluating ensemble on [../data/PGD_eps0.30.npy], it may take a while...\n>>> Evaluating baseline model on [../data/PGD_eps0.30.npy], it may take a while...\n>>> Evaluations on [../data/PGD_eps0.30.npy]:\n{'UM': 0.9878542510121457, 'Ensemble': 0.30465587044534415, 'PGD-ADT': 0.1811740890688259}\n>>> Evaluating UM on [../data/BIM_eps0.07.npy], it may take a while...\n>>> Evaluating ensemble on [../data/BIM_eps0.07.npy], it may take a while...\n>>> Evaluating baseline model on [../data/BIM_eps0.07.npy], it may take a while...\n>>> Evaluations on [../data/BIM_eps0.07.npy]:\n{'UM': 0.4797570850202429, 'Ensemble': 0.010121457489878543, 'PGD-ADT': 0.01720647773279352}\n>>> Evaluating UM on [../data/BIM_eps0.10.npy], it may take a while...\n>>> Evaluating ensemble on [../data/BIM_eps0.10.npy], it may take a while...\n>>> Evaluating baseline model on [../data/BIM_eps0.10.npy], it may take a while...\n>>> Evaluations on [../data/BIM_eps0.10.npy]:\n{'UM': 0.9200404858299596, 'Ensemble': 0.015182186234817813, 'PGD-ADT': 0.02834008097165992}\n>>> Evaluating UM on [../data/BIM_eps0.15.npy], it may take a while...\n>>> Evaluating ensemble on [../data/BIM_eps0.15.npy], it may take a while...\n>>> Evaluating baseline model on [../data/BIM_eps0.15.npy], it may take a while...\n>>> Evaluations on [../data/BIM_eps0.15.npy]:\n{'UM': 0.9878542510121457, 'Ensemble': 0.05060728744939271, 'PGD-ADT': 0.05668016194331984}\n>>> Evaluating UM on [../data/BIM_eps0.20.npy], it may take a while...\n>>> Evaluating ensemble on [../data/BIM_eps0.20.npy], it may take a while...\n>>> Evaluating baseline model on [../data/BIM_eps0.20.npy], it may take a while...\n>>> Evaluations on [../data/BIM_eps0.20.npy]:\n{'UM': 0.9878542510121457, 'Ensemble': 0.1508097165991903, 'PGD-ADT': 0.1062753036437247}\n>>> Evaluating UM on [../data/BIM_eps0.25.npy], it may take a while...\n>>> Evaluating ensemble on [../data/BIM_eps0.25.npy], it may take a while...\n>>> Evaluating baseline model on [../data/BIM_eps0.25.npy], it may take a while...\n>>> Evaluations on [../data/BIM_eps0.25.npy]:\n{'UM': 0.9878542510121457, 'Ensemble': 0.37955465587044535, 'PGD-ADT': 0.19939271255060728}\n>>> Evaluating UM on [../data/BIM_eps0.30.npy], it may take a while...\n>>> Evaluating ensemble on [../data/BIM_eps0.30.npy], it may take a while...\n>>> Evaluating baseline model on [../data/BIM_eps0.30.npy], it may take a while...\n>>> Evaluations on [../data/BIM_eps0.30.npy]:\n{'UM': 0.9878542510121457, 'Ensemble': 0.6477732793522267, 'PGD-ADT': 0.3694331983805668}\n{'UM': [0.13157894736842105, 0.24493927125506074, 0.521255060728745, 0.7267206477732794, 0.832995951417004, 0.8866396761133604, 0.2783400809716599, 0.667004048582996, 0.9463562753036437, 0.9868421052631579, 0.9878542510121457, 0.9878542510121457, 0.4797570850202429, 0.9200404858299596, 0.9878542510121457, 0.9878542510121457, 0.9878542510121457, 0.9878542510121457], 'Ensemble': [0.00708502024291498, 0.012145748987854251, 0.032388663967611336, 0.09109311740890688, 0.3259109311740891, 0.6771255060728745, 0.009109311740890687, 0.013157894736842105, 0.0354251012145749, 0.0708502024291498, 0.14979757085020243, 0.30465587044534415, 0.010121457489878543, 0.015182186234817813, 0.05060728744939271, 0.1508097165991903, 0.37955465587044535, 0.6477732793522267], 'PGD-ADT': [0.015182186234817813, 0.022267206477732792, 0.043522267206477734, 0.07692307692307693, 0.1214574898785425, 0.21963562753036436, 0.016194331983805668, 0.02327935222672065, 0.05060728744939271, 0.06983805668016195, 0.11639676113360324, 0.1811740890688259, 0.01720647773279352, 0.02834008097165992, 0.05668016194331984, 0.1062753036437247, 0.19939271255060728, 0.3694331983805668]}\n"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np\n\n\nFGSM_err = {}\nPGD_err = {}\nBIM_err = {}\n#for FGSM\nFGSM_err['UM'] = eval_result['UM'][:6]\nFGSM_err['Ensemble'] = eval_result['Ensemble'][:6]\nFGSM_err['PGD-ADT'] = eval_result['PGD-ADT'][:6]\nprint(FGSM_err['UM'])\nprint(FGSM_err['Ensemble'])\nprint(FGSM_err['PGD-ADT'])\n\n# for PGD\nPGD_err['UM'] = eval_result['UM'][6:12]\nPGD_err['Ensemble'] = eval_result['Ensemble'][6:12]\nPGD_err['PGD-ADT'] = eval_result['PGD-ADT'][6:12]\nprint(PGD_err['UM'])\nprint(PGD_err['Ensemble'])\nprint(PGD_err['PGD-ADT'])\n\n# for BIM\nBIM_err['UM'] = eval_result['UM'][12:]\nBIM_err['Ensemble'] = eval_result['Ensemble'][12:]\nBIM_err['PGD-ADT'] = eval_result['PGD-ADT'][12:]\nprint(BIM_err['UM'])\nprint(BIM_err['Ensemble'])\nprint(BIM_err['PGD-ADT'])\n\nw = 0.2\nx = [0.07, 0.10, 0.15, 0.20, 0.25, 0.30] #epsilon values for attack\n\n# FGSM Plot\nbar1 = np.arange(len(x))\nbar2 = [i+w for i in bar1]\nbar3 = [i+2*w for i in bar1]\nplt.bar(bar1, FGSM_err['UM'], w , label='Undefended Model')\nplt.bar(bar2, FGSM_err['Ensemble'], w , label='Ensemble')\nplt.bar(bar3, FGSM_err['PGD-ADT'], w , label='PGD-ADT')\n\nplt.xlabel('Epsilon')\nplt.ylabel('Error rate')\nplt.title('Error-rate vs Epsilon values for FGSM')\nplt.xticks(bar1+w, x)\nplt.legend()\nplt.show()\n\n#For PGD Plot of Error rates\nbar1 = np.arange(len(x))\nbar2 = [i+w for i in bar1]\nbar3 = [i+2*w for i in bar1]\nplt.bar(bar1, PGD_err['UM'], w , label='Undefended Model')\nplt.bar(bar2, PGD_err['Ensemble'], w , label='Ensemble')\nplt.bar(bar3, PGD_err['PGD-ADT'], w , label='PGD-ADT')\n\nplt.xlabel('Epsilon')\nplt.ylabel('Error rate')\nplt.title('Error-rate vs Epsilon values for PGD Attack')\nplt.xticks(bar1+w, x)\nplt.legend()\nplt.show()\n\n#For BIM Plot of Error rates\nbar1 = np.arange(len(x))\nbar2 = [i+w for i in bar1]\nbar3 = [i+2*w for i in bar1]\nplt.bar(bar1, BIM_err['UM'], w , label='Undefended Model')\nplt.bar(bar2, BIM_err['Ensemble'], w , label='Ensemble')\nplt.bar(bar3, BIM_err['PGD-ADT'], w , label='PGD-ADT')\n\nplt.xlabel('Epsilon')\nplt.ylabel('Error rate')\nplt.title('Error-rate vs Epsilon values for BIM Attack')\nplt.xticks(bar1+w, x)\nplt.legend()\nplt.show()",
"[0.13157894736842105, 0.24493927125506074, 0.521255060728745, 0.7267206477732794, 0.832995951417004, 0.8866396761133604]\n[0.00708502024291498, 0.012145748987854251, 0.032388663967611336, 0.09109311740890688, 0.3259109311740891, 0.6771255060728745]\n[0.015182186234817813, 0.022267206477732792, 0.043522267206477734, 0.07692307692307693, 0.1214574898785425, 0.21963562753036436]\n[0.2783400809716599, 0.667004048582996, 0.9463562753036437, 0.9868421052631579, 0.9878542510121457, 0.9878542510121457]\n[0.009109311740890687, 0.013157894736842105, 0.0354251012145749, 0.0708502024291498, 0.14979757085020243, 0.30465587044534415]\n[0.016194331983805668, 0.02327935222672065, 0.05060728744939271, 0.06983805668016195, 0.11639676113360324, 0.1811740890688259]\n[0.4797570850202429, 0.9200404858299596, 0.9878542510121457, 0.9878542510121457, 0.9878542510121457, 0.9878542510121457]\n[0.010121457489878543, 0.015182186234817813, 0.05060728744939271, 0.1508097165991903, 0.37955465587044535, 0.6477732793522267]\n[0.01720647773279352, 0.02834008097165992, 0.05668016194331984, 0.1062753036437247, 0.19939271255060728, 0.3694331983805668]\n"
]
],
[
[
"# Group Name: Team Horus\n\nGroup Members (Git-hub Usernames): \n 1. Withana (Rasika) Jayarathna (Rasika-prog)\n 2. Kaveh Shariati (kavehshariati)\n 3. Joshua Ojih (ojihjo)\n 4. Olajide Bamidele (42n8dzydoo)\n\n\n\n### Attacks Used and Settings: FGSM, PGD and BIM\nThe following instances of Epsilon settings were used to generate Adversarial Examples for the three types of attacks done (listed above): \n 1. Epsilon 0.07\n 2. Epsilon 0.10\n 3. Epsilon 0.15\n 4. Epsilon 0.20\n 5. Epsilon 0.25\n 6. Epsilon 0.30\n\nAdditionally, the following default settings were used to generate the PGD and BIM Adversarial Examples:\n 1. Epsilon step (eps_step) = (0.1 * Epsilon)\n 2. Maximum iteration = 100",
"_____no_output_____"
],
[
"# Members contribution:\nAll members contributed to the success of task1.\n\nRasika and Olajide contributed to tuning the codes and generating BIM Adversarial Examples. \nJoshua and Kaveh contributed to tuning the codes to generate the FGSM and PGD Adversarial Examples\n\nPlotting of Results was done by Olajide and all team members performed the analysis together.\n\n## Debugging the Code (the evaluate function in eval_model.py)\nInitially, the evaluate function only evaluates the error rate for just one instance of Adversarial Examples. \nThis was debugged by iterating over all the instances of the Adversarial Examples (AEs) using a \"for-loop\".\n\nAll team members were involved in debugging the codes to calculate the error rate for each Adversarial Examples (AEs) generated.",
"_____no_output_____"
],
[
"# Analysis of Results\n\nThe 18 Adversarial Examples (AEs) generated were tested on the Athena model using the AVEP Ensemble strategy and the results were plotted above.\n\n# Observations\n1. At low values of Epsilon (for values of epsilon < 0.20), the undefended model has a large Error-rate. The Athena model performed better than the state-of-the-art PGD-ADT. The model recorded lesser error rate compared to both the undefended model and the PGD-ADT\n\n\n2. At larger values of Epsilon (epsilon > 0.15), the Adversarial Examples were able to fool the Athena model and larger error rates were observed. However, the PGD-ADT performed better than the Athena model for these epsilon values. ",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ecccb700415f4b3bbc4e9eab699893e105fc2258 | 100,532 | ipynb | Jupyter Notebook | Docking_0608.ipynb | ChrizZhuang/marginalized_graph_kernel_protein | 5c0d0c36f66be174521b86fb11d53933dfa6bc1e | [
"MIT"
] | null | null | null | Docking_0608.ipynb | ChrizZhuang/marginalized_graph_kernel_protein | 5c0d0c36f66be174521b86fb11d53933dfa6bc1e | [
"MIT"
] | null | null | null | Docking_0608.ipynb | ChrizZhuang/marginalized_graph_kernel_protein | 5c0d0c36f66be174521b86fb11d53933dfa6bc1e | [
"MIT"
] | null | null | null | 105.60084 | 16,574 | 0.760126 | [
[
[
"<a href=\"https://colab.research.google.com/github/ChrizZhuang/marginalized_graph_kernel_protein/blob/main/Docking_0608.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!nvidia-smi",
"Tue Jun 8 01:01:30 2021 \n+-----------------------------------------------------------------------------+\n| NVIDIA-SMI 465.27 Driver Version: 460.32.03 CUDA Version: 11.2 |\n|-------------------------------+----------------------+----------------------+\n| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |\n| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |\n| | | MIG M. |\n|===============================+======================+======================|\n| 0 Tesla T4 Off | 00000000:00:04.0 Off | 0 |\n| N/A 42C P8 10W / 70W | 0MiB / 15109MiB | 0% Default |\n| | | N/A |\n+-------------------------------+----------------------+----------------------+\n \n+-----------------------------------------------------------------------------+\n| Processes: |\n| GPU GI CI PID Type Process name GPU Memory |\n| ID ID Usage |\n|=============================================================================|\n| No running processes found |\n+-----------------------------------------------------------------------------+\n"
],
[
"#!pip install pymatgen==2020.12.31\n!pip install pymatgen==2019.11.11\n!pip install --pre graphdot\n!pip install gdown",
"Requirement already satisfied: pymatgen==2019.11.11 in /usr/local/lib/python3.7/dist-packages (2019.11.11)\nRequirement already satisfied: palettable>=3.1.1 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (3.3.0)\nRequirement already satisfied: pydispatcher>=2.0.5 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (2.0.5)\nRequirement already satisfied: ruamel.yaml>=0.15.6 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (0.17.7)\nRequirement already satisfied: spglib>=1.9.9.44 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (1.16.1)\nRequirement already satisfied: networkx>=2.2 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (2.5.1)\nRequirement already satisfied: numpy>=1.14.3 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (1.19.5)\nRequirement already satisfied: tabulate in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (0.8.9)\nRequirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (2.23.0)\nRequirement already satisfied: pandas in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (1.1.5)\nRequirement already satisfied: scipy>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (1.4.1)\nRequirement already satisfied: sympy in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (1.7.1)\nRequirement already satisfied: matplotlib>=1.5 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (3.2.2)\nRequirement already satisfied: monty>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11) (2021.5.9)\nRequirement already satisfied: ruamel.yaml.clib>=0.1.2; platform_python_implementation == \"CPython\" and python_version < \"3.10\" in /usr/local/lib/python3.7/dist-packages (from ruamel.yaml>=0.15.6->pymatgen==2019.11.11) (0.2.2)\nRequirement already satisfied: decorator<5,>=4.3 in /usr/local/lib/python3.7/dist-packages (from networkx>=2.2->pymatgen==2019.11.11) (4.4.2)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->pymatgen==2019.11.11) (3.0.4)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->pymatgen==2019.11.11) (2.10)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->pymatgen==2019.11.11) (2020.12.5)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->pymatgen==2019.11.11) (1.24.3)\nRequirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.7/dist-packages (from pandas->pymatgen==2019.11.11) (2.8.1)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.7/dist-packages (from pandas->pymatgen==2019.11.11) (2018.9)\nRequirement already satisfied: mpmath>=0.19 in /usr/local/lib/python3.7/dist-packages (from sympy->pymatgen==2019.11.11) (1.2.1)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=1.5->pymatgen==2019.11.11) (2.4.7)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=1.5->pymatgen==2019.11.11) (0.10.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=1.5->pymatgen==2019.11.11) (1.3.1)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.7/dist-packages (from python-dateutil>=2.7.3->pandas->pymatgen==2019.11.11) (1.15.0)\nRequirement already satisfied: graphdot in /usr/local/lib/python3.7/dist-packages (0.8a17)\nRequirement already satisfied: numpy>=1.17 in /usr/local/lib/python3.7/dist-packages (from graphdot) (1.19.5)\nRequirement already satisfied: scipy>=1.3.0 in /usr/local/lib/python3.7/dist-packages (from graphdot) (1.4.1)\nRequirement already satisfied: sympy>=1.3 in /usr/local/lib/python3.7/dist-packages (from graphdot) (1.7.1)\nRequirement already satisfied: networkx>=2.4 in /usr/local/lib/python3.7/dist-packages (from graphdot) (2.5.1)\nRequirement already satisfied: pandas>=0.24 in /usr/local/lib/python3.7/dist-packages (from graphdot) (1.1.5)\nRequirement already satisfied: pymatgen==2019.11.11 in /usr/local/lib/python3.7/dist-packages (from graphdot) (2019.11.11)\nRequirement already satisfied: numba>=0.51.0 in /usr/local/lib/python3.7/dist-packages (from graphdot) (0.51.2)\nRequirement already satisfied: kahypar>=1.1.4 in /usr/local/lib/python3.7/dist-packages (from graphdot) (1.1.6)\nRequirement already satisfied: mendeleev>=0.7 in /usr/local/lib/python3.7/dist-packages (from graphdot) (0.7.0)\nRequirement already satisfied: treelib>=1.6.1 in /usr/local/lib/python3.7/dist-packages (from graphdot) (1.6.1)\nRequirement already satisfied: ase>=3.17 in /usr/local/lib/python3.7/dist-packages (from graphdot) (3.21.1)\nRequirement already satisfied: pycuda>=2019 in /usr/local/lib/python3.7/dist-packages (from graphdot) (2021.1)\nRequirement already satisfied: tqdm>=4.55 in /usr/local/lib/python3.7/dist-packages (from graphdot) (4.61.0)\nRequirement already satisfied: mpmath>=0.19 in /usr/local/lib/python3.7/dist-packages (from sympy>=1.3->graphdot) (1.2.1)\nRequirement already satisfied: decorator<5,>=4.3 in /usr/local/lib/python3.7/dist-packages (from networkx>=2.4->graphdot) (4.4.2)\nRequirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.7/dist-packages (from pandas>=0.24->graphdot) (2.8.1)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.7/dist-packages (from pandas>=0.24->graphdot) (2018.9)\nRequirement already satisfied: palettable>=3.1.1 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11->graphdot) (3.3.0)\nRequirement already satisfied: monty>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11->graphdot) (2021.5.9)\nRequirement already satisfied: ruamel.yaml>=0.15.6 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11->graphdot) (0.17.7)\nRequirement already satisfied: pydispatcher>=2.0.5 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11->graphdot) (2.0.5)\nRequirement already satisfied: spglib>=1.9.9.44 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11->graphdot) (1.16.1)\nRequirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11->graphdot) (2.23.0)\nRequirement already satisfied: matplotlib>=1.5 in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11->graphdot) (3.2.2)\nRequirement already satisfied: tabulate in /usr/local/lib/python3.7/dist-packages (from pymatgen==2019.11.11->graphdot) (0.8.9)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.7/dist-packages (from numba>=0.51.0->graphdot) (57.0.0)\nRequirement already satisfied: llvmlite<0.35,>=0.34.0.dev0 in /usr/local/lib/python3.7/dist-packages (from numba>=0.51.0->graphdot) (0.34.0)\nRequirement already satisfied: six<2.0.0,>=1.15.0 in /usr/local/lib/python3.7/dist-packages (from mendeleev>=0.7->graphdot) (1.15.0)\nRequirement already satisfied: colorama<0.5.0,>=0.4.4 in /usr/local/lib/python3.7/dist-packages (from mendeleev>=0.7->graphdot) (0.4.4)\nRequirement already satisfied: pyfiglet<0.9,>=0.8.post1 in /usr/local/lib/python3.7/dist-packages (from mendeleev>=0.7->graphdot) (0.8.post1)\nRequirement already satisfied: Pygments<3.0.0,>=2.8.0 in /usr/local/lib/python3.7/dist-packages (from mendeleev>=0.7->graphdot) (2.9.0)\nRequirement already satisfied: SQLAlchemy<2.0.0,>=1.3.23 in /usr/local/lib/python3.7/dist-packages (from mendeleev>=0.7->graphdot) (1.4.15)\nRequirement already satisfied: future in /usr/local/lib/python3.7/dist-packages (from treelib>=1.6.1->graphdot) (0.16.0)\nRequirement already satisfied: pytools>=2011.2 in /usr/local/lib/python3.7/dist-packages (from pycuda>=2019->graphdot) (2021.2.7)\nRequirement already satisfied: appdirs>=1.4.0 in /usr/local/lib/python3.7/dist-packages (from pycuda>=2019->graphdot) (1.4.4)\nRequirement already satisfied: mako in /usr/local/lib/python3.7/dist-packages (from pycuda>=2019->graphdot) (1.1.4)\nRequirement already satisfied: ruamel.yaml.clib>=0.1.2; platform_python_implementation == \"CPython\" and python_version < \"3.10\" in /usr/local/lib/python3.7/dist-packages (from ruamel.yaml>=0.15.6->pymatgen==2019.11.11->graphdot) (0.2.2)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->pymatgen==2019.11.11->graphdot) (2.10)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->pymatgen==2019.11.11->graphdot) (2020.12.5)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->pymatgen==2019.11.11->graphdot) (3.0.4)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->pymatgen==2019.11.11->graphdot) (1.24.3)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=1.5->pymatgen==2019.11.11->graphdot) (1.3.1)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=1.5->pymatgen==2019.11.11->graphdot) (0.10.0)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=1.5->pymatgen==2019.11.11->graphdot) (2.4.7)\nRequirement already satisfied: greenlet!=0.4.17; python_version >= \"3\" in /usr/local/lib/python3.7/dist-packages (from SQLAlchemy<2.0.0,>=1.3.23->mendeleev>=0.7->graphdot) (1.1.0)\nRequirement already satisfied: importlib-metadata; python_version < \"3.8\" in /usr/local/lib/python3.7/dist-packages (from SQLAlchemy<2.0.0,>=1.3.23->mendeleev>=0.7->graphdot) (4.0.1)\nRequirement already satisfied: MarkupSafe>=0.9.2 in /usr/local/lib/python3.7/dist-packages (from mako->pycuda>=2019->graphdot) (2.0.1)\nRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.7/dist-packages (from importlib-metadata; python_version < \"3.8\"->SQLAlchemy<2.0.0,>=1.3.23->mendeleev>=0.7->graphdot) (3.4.1)\nRequirement already satisfied: typing-extensions>=3.6.4; python_version < \"3.8\" in /usr/local/lib/python3.7/dist-packages (from importlib-metadata; python_version < \"3.8\"->SQLAlchemy<2.0.0,>=1.3.23->mendeleev>=0.7->graphdot) (3.7.4.3)\nRequirement already satisfied: gdown in /usr/local/lib/python3.7/dist-packages (3.6.4)\nRequirement already satisfied: tqdm in /usr/local/lib/python3.7/dist-packages (from gdown) (4.61.0)\nRequirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from gdown) (2.23.0)\nRequirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from gdown) (1.15.0)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->gdown) (3.0.4)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->gdown) (1.24.3)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->gdown) (2020.12.5)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->gdown) (2.10)\n"
],
[
"%matplotlib inline\nimport io\nimport sys\nsys.path.append('/usr/local/lib/python3.6/site-packages/')\nimport os\nimport urllib\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nimport graphdot\nfrom graphdot import Graph\nfrom graphdot.graph.adjacency import AtomicAdjacency\nfrom graphdot.graph.reorder import rcm\nfrom graphdot.kernel.marginalized import MarginalizedGraphKernel # https://graphdot.readthedocs.io/en/latest/apidoc/graphdot.kernel.marginalized.html\nfrom graphdot.kernel.marginalized.starting_probability import Uniform\nfrom graphdot.model.gaussian_process import (\n GaussianProcessRegressor,\n LowRankApproximateGPR\n)\nfrom graphdot.kernel.fix import Normalization\nimport graphdot.microkernel as uX\nimport ase.io\n\n# for getting all file names into a list under a directory \nfrom os import listdir\n# for getting file names that match certain pattern\nimport glob\nimport time",
"_____no_output_____"
],
[
"from google.colab import drive\ndrive.mount('/content/gdrive', force_remount=True)",
"Mounted at /content/gdrive\n"
],
[
"#cd gdrive/MyDrive/Google\\ Colab/Covid-Data\n%cd gdrive/MyDrive/Covid-Data/",
"/content/gdrive/.shortcut-targets-by-id/1wtzMcocuK8kPsz8K0ktjCZPkv567W6M2/Covid-Data\n"
],
[
"!pwd",
"/content/gdrive/.shortcut-targets-by-id/1wtzMcocuK8kPsz8K0ktjCZPkv567W6M2/Covid-Data\n"
],
[
"#!ls",
"_____no_output_____"
],
[
"#files = ['uncharged_NSP15_6W01_A_3_H.Orderable_zinc_db_enaHLL.2col.csv.1.xz']\n#files = [f for f in listdir('/content/gdrive/.shortcut-targets-by-id/1wtzMcocuK8kPsz8K0ktjCZPkv567W6M2/Covid-Data')]\nfiles = glob.glob(\"uncharged_NSP15_6W01_A_3_H.Orderable_zinc_db_enaHLL.2col.csv.*.xz\")\n#print(files[0:5])\n# concatenate all files into 1 dataset\ndataset = pd.DataFrame()\nfor i in range(len(files[0:5])):\n data = pd.read_pickle(files[i])\n dataset = pd.concat([dataset, data])\n#frames = [pd.read_pickle(f) for f in files]\n#dataset = pd.concat(frames)",
"_____no_output_____"
],
[
"len(dataset)",
"_____no_output_____"
],
[
"target = 'energy'\nN_train = len(dataset)//2\nN_test = len(dataset)//2",
"_____no_output_____"
],
[
"np.random.seed(0)\n# select train and test data\ntrain_sel = np.random.choice(len(dataset), N_train, replace=False)\ntest_sel = np.random.choice(np.setxor1d(np.arange(len(dataset)), train_sel), N_test, replace=False)\ntrain = dataset.iloc[train_sel]\ntest = dataset.iloc[test_sel]",
"_____no_output_____"
],
[
"#uX.SquareExponential?",
"_____no_output_____"
],
[
"gpr = GaussianProcessRegressor(\n # kernel is the covariance function of the gaussian process (GP)\n kernel=Normalization( # kernel equals to normalization -> normalizes a kernel using the cosine of angle formula, k_normalized(x,y) = k(x,y)/sqrt(k(x,x)*k(y,y))\n # graphdot.kernel.fix.Normalization(kernel), set kernel as marginalized graph kernel, which is used to calculate the similarity between 2 graphs\n # implement the random walk-based graph similarity kernel as Kashima, H., Tsuda, K., & Inokuchi, A. (2003). Marginalized kernels between labeled graphs. ICML\n MarginalizedGraphKernel( \n # node_kernel - A kernelet that computes the similarity between individual nodes\n # uX - graphdot.microkernel - microkernels are positive-semidefinite functions between individual nodes and edges of graphs\n node_kernel=uX.Additive( # addition of kernal matrices: sum of k_a(X_a, Y_a) cross for a in features\n # uX.Constant - a kernel that returns a constant value, always mutlipled with other microkernels as an adjustable weight\n # c, the first input arg. as 0.5, (0.01, 10) the lower and upper bounds of c that is allowed to vary during hyperpara. optimizartion\n # uX.KroneckerDelta - a kronecker delta returns 1 when two features are equal and return h (the first input arg here, which is 0.5 in this case) otherwise\n # (0.1, 0.9) the lower and upper bounds that h is allowed to vary during hyperpara. optimization\n aromatic=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.5,(0.1, 0.9)), # the 2nd element of graphdot.graph.Graph.nodes\n atomic_number=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.8,(0.1, 0.9)), # the 3rd element of graphdot.graph.Graph.nodes\n # uX.SquareExponential - Equ. 26 in the paper\n # input arg. length_sacle is a float32, set as 1 in this case, which correspond to approx. 1 of the kernal value. \n # This is used to determins how quicklys should the kernel decay to zero.\n charge=uX.Constant(0.5, (0.01, 10.0)) * uX.SquareExponential(1.0), # the 4th element of graphdot.graph.Graph.nodes\n chiral=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.5,(0.1, 0.9)), # the 5th element of graphdot.graph.Graph.nodes\n hcount=uX.Constant(0.5, (0.01, 10.0)) * uX.SquareExponential(1.0), # the 6th element of graphdot.graph.Graph.nodes\n hybridization=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.5,(0.1, 0.9)), # the 7th element of graphdot.graph.Graph.nodes\n # uX.Convolution - a convolutional microkernel which averages evaluations of a base microkernel between pairs pf elememts of two variable-length feature sequences\n # uX.KroneckerDelta as the base kernel\n ring_list=uX.Constant(0.5, (0.01, 100.0)) * uX.Convolution(uX.KroneckerDelta(0.5,(0.1, 0.9))) # the 8th element of graphdot.graph.Graph.nodes\n ).normalized,\n # edge_kernel - A kernelet that computes the similarity between individual edge\n edge_kernel=uX.Additive(\n aromatic=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.5,(0.1, 0.9)), # the 3rd element of graphdot.graph.Graph.nodes\n conjugated=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.5,(0.1, 0.9)), # the 4th element of graphdot.graph.Graph.nodes\n order=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.8,(0.1, 0.9)), # the 5th element of graphdot.graph.Graph.nodes\n ring_stereo=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.8,(0.1, 0.9)), # the 6th element of graphdot.graph.Graph.nodes\n stereo=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.8,(0.1, 0.9)) # the 7th element of graphdot.graph.Graph.nodes\n ).normalized,\n p=Uniform(1.0, p_bounds='fixed'), # the strating probability of the random walk on each node\n q=0.05 # the probability for the random walk to stop during each step\n )\n ),\n alpha=1e-4, # value added to the diagonal of the kernel matrix during fitting\n optimizer=True, # default optimizer of L-BFGS-B based on scipy.optimize.minimize\n normalize_y=True, # normalize the y values so taht the means and variance is 0 and 1, repsectively. Will be reversed when predicions are returned\n regularization='+', # alpha (1e-4 in this case) is added to the diagonals of the kernal matrix\n)",
"the total time consumption is 0.0034666061401367188.\n"
],
[
"#gpr.fit(train.graphs, train[target], repeat=3, verbose=True)\n# using the molecular graph to predict the energy\nstart_time = time.time()\ngpr.fit(train.graphs, train[target], repeat=5, verbose=True)\nend_time = time.time()\nprint(\"the total time consumption is \" + str(end_time - start_time) + \".\")",
"_____no_output_____"
],
[
"gpr.kernel.hyperparameters",
"_____no_output_____"
],
[
"mu = gpr.predict(train.graphs)",
"_____no_output_____"
],
[
"plt.scatter(train[target], mu)\nplt.show()",
"_____no_output_____"
],
[
"print('Training set')\nprint('MAE:', np.mean(np.abs(train[target] - mu)))\nprint('RMSE:', np.std(train[target] - mu))",
"Training set\nMAE: 0.2074369392067814\nRMSE: 0.3195501910711287\n"
],
[
"mu_test = gpr.predict(test.graphs)",
"_____no_output_____"
],
[
"plt.scatter(test[target], mu_test)\nplt.show()",
"_____no_output_____"
],
[
"print('Test set')\nprint('MAE:', np.mean(np.abs(test[target] - mu_test)))\nprint('RMSE:', np.std(test[target] - mu_test))",
"Test set\nMAE: 1.2461703788060545\nRMSE: 1.6040210737240486\n"
]
],
[
[
"Work\u0001on the kernel. Find a kernel that trains and predicts well.",
"_____no_output_____"
]
],
[
[
"gpr2 = GaussianProcessRegressor(\n kernel=Normalization(\n MarginalizedGraphKernel(\n node_kernel=uX.Additive(\n aromatic=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.5,(0.1, 0.9)),\n atomic_number=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.8,(0.1, 0.9)),\n charge=uX.Constant(0.5, (0.01, 10.0)) * uX.SquareExponential(1.0),\n chiral=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.5,(0.1, 0.9)),\n hcount=uX.Constant(0.5, (0.01, 10.0)) * uX.SquareExponential(1.0),\n hybridization=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.5,(0.1, 0.9)),\n ring_list=uX.Constant(0.5, (0.01, 100.0)) * uX.Convolution(uX.KroneckerDelta(0.5,(0.1, 0.9)))\n ).normalized,\n edge_kernel=uX.Additive(\n aromatic=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.5,(0.1, 0.9)),\n conjugated=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.5,(0.1, 0.9)),\n order=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.8,(0.1, 0.9)),\n ring_stereo=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.8,(0.1, 0.9)),\n stereo=uX.Constant(0.5, (0.01, 10.0)) * uX.KroneckerDelta(0.8,(0.1, 0.9))\n ).normalized,\n p=Uniform(1.0, p_bounds='fixed'),\n q=0.05\n )\n ),\n alpha=1e-2, #different from gpr in alpha where gpr's alpha is 1e-4\n optimizer=True,\n normalize_y=True,\n regularization='+',\n)",
"_____no_output_____"
],
[
"#gpr2.fit(train.graphs, train[target], repeat=3, verbose=True)\ngpr2.fit(train.graphs, train[target], repeat=1, verbose=True)",
"| logP| dlogP| y^T.K.y| log|K| | Cond(K)| GPU time| CPU time|\n|------------|------------|------------|------------|------------|----------|----------|\n| 1.667e+05| 1.928e+05| 1.7773e+05| -11034| 2.2742e+05| 1.3e+02| 1.7|\n| 1.5585e+05| 2.2171e+05| 1.6671e+05| -10857| 2.2218e+05| 1.3e+02| 1.7|\n| 1.3711e+05| 2.4873e+05| 1.4756e+05| -10447| 2.0803e+05| 1.2e+02| 1.8|\n| 1.3286e+05| 2.3627e+05| 1.432e+05| -10338| 2.0336e+05| 1.1e+02| 1.7|\n| 1.2977e+05| 2.2744e+05| 1.4002e+05| -10249| 1.9714e+05| 1.1e+02| 1.7|\n| 1.3006e+05| 2.1429e+05| 1.4032e+05| -10258| 2.0119e+05| 1e+02| 1.7|\n| 1.2923e+05| 2.2103e+05| 1.3946e+05| -10238| 1.9906e+05| 1.1e+02| 1.7|\n| 1.2888e+05| 2.2097e+05| 1.3911e+05| -10228| 1.9865e+05| 1.1e+02| 1.7|\n| 1.2843e+05| 2.2338e+05| 1.3864e+05| -10211| 1.9582e+05| 1.1e+02| 1.7|\n| 1.2837e+05| 2.2244e+05| 1.3858e+05| -10212| 1.969e+05| 1.1e+02| 1.7|\n| 1.2836e+05| 2.2225e+05| 1.3857e+05| -10212| 1.9696e+05| 1.1e+02| 1.7|\n| 1.2831e+05| 2.2155e+05| 1.3852e+05| -10211| 1.9715e+05| 1.1e+02| 1.7|\n| 1.2826e+05| 2.2184e+05| 1.3847e+05| -10209| 1.9704e+05| 1.1e+02| 1.7|\n| 1.2821e+05| 2.2181e+05| 1.3841e+05| -10207| 1.9671e+05| 1.1e+02| 1.7|\n| 1.2819e+05| 2.2165e+05| 1.384e+05| -10207| 1.9664e+05| 1.1e+02| 1.8|\n| 1.2818e+05| 2.2125e+05| 1.3838e+05| -10207| 1.9671e+05| 1.1e+02| 1.8|\n| 1.2817e+05| 2.2114e+05| 1.3838e+05| -10207| 1.9673e+05| 1.1e+02| 1.7|\n| 1.2815e+05| 2.2077e+05| 1.3836e+05| -10207| 1.9677e+05| 1.1e+02| 1.7|\n| 1.2812e+05| 2.185e+05| 1.3833e+05| -10209| 1.9695e+05| 1.1e+02| 1.7|\n| 1.2809e+05| 2.178e+05| 1.383e+05| -10209| 1.97e+05| 1.1e+02| 1.7|\n| 1.2807e+05| 2.1764e+05| 1.3828e+05| -10208| 1.9695e+05| 1.1e+02| 1.7|\n| 1.2805e+05| 2.1597e+05| 1.3826e+05| -10209| 1.9693e+05| 1.1e+02| 1.7|\n| 1.2805e+05| 2.172e+05| 1.3825e+05| -10208| 1.9694e+05| 1.1e+02| 1.7|\n| 1.2804e+05| 2.1693e+05| 1.3825e+05| -10208| 1.9694e+05| 1.1e+02| 1.8|\n| 1.2804e+05| 2.1674e+05| 1.3825e+05| -10208| 1.9694e+05| 1.1e+02| 1.7|\n| 1.2804e+05| 2.1691e+05| 1.3825e+05| -10208| 1.9694e+05| 1.1e+02| 1.7|\n| 1.2805e+05| 2.1694e+05| 1.3825e+05| -10208| 1.9694e+05| 1.1e+02| 1.7|\n| 1.2804e+05| 2.1694e+05| 1.3825e+05| -10208| 1.9694e+05| 1.1e+02| 1.7|\n| 1.2804e+05| 2.1694e+05| 1.3825e+05| -10208| 1.9694e+05| 1.1e+02| 1.7|\n| 1.2804e+05| 2.1694e+05| 1.3825e+05| -10208| 1.9694e+05| 1.1e+02| 1.8|\n| 1.2805e+05| 2.1694e+05| 1.3825e+05| -10208| 1.9694e+05| 1.1e+02| 1.7|\nOptimization result:\n fun: 128045.20193517115\n hess_inv: <25x25 LbfgsInvHessProduct with dtype=float64>\n jac: array([ 21.67035135, 5.42797631, 0.91584794, 6.62684845,\n 636.88297304, 10.22203408, 0. , 6.13155297,\n 0.80318432, -45.79780911, 0. , 9.32446636,\n 2.06137402, 7.94609186, 767.483899 , -8.19180354,\n 92.93654292, 7.92503084, 118.62629465, 3.36719643,\n 38.71995733, 7.96834618, 0. , -11.08475724,\n 7.2860016 ])\n message: b'CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH'\n nfev: 31\n nit: 21\n status: 0\n success: True\n x: array([-9.21034037, -4.51710969, -1.98267626, 1.72521606, -2.30258509,\n -4.60517019, 0. , -4.60517019, -1.8070945 , 2.30258509,\n -2.88031384, -3.6019383 , -2.30258509, 1.91177723, -2.30258509,\n 0.28746812, -2.30258509, 0.54884256, -2.30258509, -0.5686784 ,\n -2.30258509, -4.35637808, -0.22314355, -2.55717239, -2.21179129])\n"
],
[
"mu = gpr2.predict(train.graphs)\nplt.scatter(train[target], mu)\nplt.show()",
"_____no_output_____"
],
[
"print('Training set')\nprint('MAE:', np.mean(np.abs(train[target] - mu)))\nprint('RMSE:', np.std(train[target] - mu))",
"Training set\nMAE: 0.7238389142657052\nRMSE: 0.9634943206325994\n"
],
[
"mu_test = gpr2.predict(test.graphs)\nplt.scatter(test[target], mu_test)\nplt.show()",
"_____no_output_____"
],
[
"print('Test set')\nprint('MAE:', np.mean(np.abs(test[target] - mu_test)))\nprint('RMSE:', np.std(test[target] - mu_test))",
"Test set\nMAE: 0.9561539409612109\nRMSE: 1.2284268143181998\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecccb87d0a9fdd042c8cd3fec7f1f513cf805818 | 940,440 | ipynb | Jupyter Notebook | notebooks/preprocessing.ipynb | emolinaro/scRNAseq-AAE | 7c654514fa9061e8c2f0397f2a1d2ea9c4125977 | [
"MIT"
] | 1 | 2019-08-17T13:00:13.000Z | 2019-08-17T13:00:13.000Z | notebooks/preprocessing.ipynb | emolinaro/scRNAseq-AAE | 7c654514fa9061e8c2f0397f2a1d2ea9c4125977 | [
"MIT"
] | 5 | 2019-08-17T13:00:02.000Z | 2022-02-10T00:13:02.000Z | notebooks/preprocessing.ipynb | emolinaro/scRNAseq-AAE | 7c654514fa9061e8c2f0397f2a1d2ea9c4125977 | [
"MIT"
] | null | null | null | 643.255814 | 322,888 | 0.949988 | [
[
[
"import scanpy as sc\nimport numpy as np\nimport os, sys\n\nos.makedirs('../data', exist_ok=True)\nos.makedirs('../data/proc', exist_ok=True)",
"_____no_output_____"
],
[
"import warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
]
],
[
[
"# Download datasets",
"_____no_output_____"
],
[
"## PBMC 3k",
"_____no_output_____"
]
],
[
[
"os.makedirs('../data/pbmc3k/', exist_ok=True)\n!wget http://cf.10xgenomics.com/samples/cell-exp/1.1.0/pbmc3k/pbmc3k_filtered_gene_bc_matrices.tar.gz -O ../data/pbmc3k/pbmc3k_filtered_gene_bc_matrices.tar.gz\n!cd ../data/pbmc3k/; tar -xzf pbmc3k_filtered_gene_bc_matrices.tar.gz; rm pbmc3k_filtered_gene_bc_matrices.tar.gz\n",
"_____no_output_____"
],
[
"data_path = '../data/pbmc3k/filtered_gene_bc_matrices/hg19'\nresults_file = '../data/proc/pbmc3k.h5ad'\nresults_file2 = '../data/proc/pbmc3k.tfrecord'",
"_____no_output_____"
],
[
"gene_up = 2500\npercent_mito_up = 0.05\nn_pcs = 40\nresolution = 1.0",
"_____no_output_____"
],
[
"marker_genes = ['IL7R', 'CD79A', 'MS4A1', 'CD8A', 'CD8B', 'LYZ', 'CD14',\n 'LGALS3', 'S100A8', 'GNLY', 'NKG7', 'KLRB1',\n 'FCGR3A', 'MS4A7', 'FCER1A', 'CST3', 'PPBP']",
"_____no_output_____"
],
[
"#Louvain Markers Cell Type\n#0 IL7R, CD4 T cells\n#1 CD14, LYZ CD14+ Monocytes\n#2 MS4A1 B cells\n#3 CD8A, CD8 T cells\n#4 GNLY, NKG7 NK cells\n#5 FCGR3A, MS4A7 FCGR3A+ Monocytes\n#6 FCER1A, CST3 Dendritic Cells\n#7 PPBP Megakaryocytes",
"_____no_output_____"
],
[
"new_cluster_names = [\n 'CD4 T', 'CD14+ Monocytes',\n 'B', 'CD8 T',\n 'NK', 'FCGR3A+ Monocytes',\n 'Dendritic', 'Megakaryocytes']",
"_____no_output_____"
]
],
[
[
"## PBMC 8k",
"_____no_output_____"
]
],
[
[
"os.makedirs('../data/pbmc8k/', exist_ok=True)\n!wget http://cf.10xgenomics.com/samples/cell-exp/2.1.0/pbmc8k/pbmc8k_filtered_gene_bc_matrices.tar.gz -O ../data/pbmc8k/pbmc8k_filtered_gene_bc_matrices.tar.gz\n!cd ../data/pbmc8k/; tar -xzf pbmc8k_filtered_gene_bc_matrices.tar.gz; rm pbmc8k_filtered_gene_bc_matrices.tar.gz\n",
"_____no_output_____"
],
[
"data_path = '../data/pbmc8k/filtered_gene_bc_matrices/GRCh38/'\nresults_file = '../data/proc/pbmc8k.h5ad'\nresults_file2 = '../data/proc/pbmc8k.tfrecord'",
"_____no_output_____"
],
[
"gene_up = 2500\npercent_mito_up = 0.05\nn_pcs = 20\nresolution = 0.7",
"_____no_output_____"
],
[
"marker_genes = ['IL7R','CD4' ,'CD8A', 'CD8B', \n 'LYZ', 'CD14',\n 'MS4A1', \n 'CD79A',\n 'LGALS3', 'S100A8', \n 'GNLY', 'NKG7', 'KLRB1',\n 'FCGR3A', 'MS4A7', \n 'FCER1A', 'CST3', \n ]",
"_____no_output_____"
],
[
"new_cluster_names = [\n 'CD4+_T-Cells',\n 'CD14+_Monocytes',\n 'CD8+_T-Cells',\n 'CD4+_T-Cells ',\n 'B_cells',\n 'NK',\n 'B_cells ',\n 'CD4+_T-Cells ',\n 'NK ',\n 'NK ',\n 'FCGR3A+_Monocytes',\n 'Dendritic_cells',\n 'Megakaryocytes']",
"_____no_output_____"
]
],
[
[
"## PBMC 10k",
"_____no_output_____"
]
],
[
[
"os.makedirs('../data/pbmc10k/', exist_ok=True)\n!wget http://cf.10xgenomics.com/samples/cell-exp/3.0.0/pbmc_10k_v3/pbmc_10k_v3_filtered_feature_bc_matrix.tar.gz -O ../data/pbmc10k/pbmc_10k_v3_filtered_feature_bc_matrix.tar.gz\n!cd ../data/pbmc10k/; tar -xzf pbmc_10k_v3_filtered_feature_bc_matrix.tar.gz; rm pbmc_10k_v3_filtered_feature_bc_matrix.tar.gz\n",
"--2019-09-24 09:41:36-- http://cf.10xgenomics.com/samples/cell-exp/3.0.0/pbmc_10k_v3/pbmc_10k_v3_filtered_feature_bc_matrix.tar.gz\nResolving cf.10xgenomics.com (cf.10xgenomics.com)... 143.204.247.100, 143.204.247.31, 143.204.247.106, ...\nConnecting to cf.10xgenomics.com (cf.10xgenomics.com)|143.204.247.100|:80... "
],
[
"os.makedirs('../data/proc', exist_ok=True)\ndata_path = '../data/pbmc10k/filtered_feature_bc_matrix'\nresults_file = '../data/proc/pbmc10k.h5ad'\nresults_file2 = '../data/proc/pbmc10k.tfrecord'",
"_____no_output_____"
],
[
"gene_up = 4500\npercent_mito_up = 0.3\nn_pcs = 40\nresolution = 0.6",
"_____no_output_____"
],
[
"marker_genes = ['IL7R', 'CD79A', 'MS4A1', 'TRAC', 'CD8A', 'CD8B','CD4', 'LYZ', 'CD14',\n 'LGALS3', 'S100A8','CD14', 'GNLY', 'NKG7', 'KLRB1',\n 'FCGR3A', 'MS4A7','IL6R', 'FCER1A', 'CST3', 'PPBP', 'IL3RA', 'CD40']",
"_____no_output_____"
],
[
"new_cluster_names = [\n 'CD14+ Monocytes', \n 'Double negative T cells',\n 'CD14+ Monocytes__', \n 'Double negative T cells__',\n 'Mature B cell',\n 'CD8 Effector', \n 'NK cells','Plasma cell','CD8 Effector__','FCGR3A+ Monocytes','CD8 Naive','Megakaryocytes','Immature B cell','CD14+ Monocytes______','Dendritic cells',\n 'CD8 Effector____','pDC','Dendritic cells__']",
"_____no_output_____"
]
],
[
[
"## CLUSTER1-8k-SPLATTER",
"_____no_output_____"
]
],
[
[
"data_path = '/work/SPLATTER-DATA-8k/CLUSTER1-8k-SPLATTER'\nresults_file = '../data/proc/cluster1-8k-splatter.h5ad'\nresults_file2 = '../data/proc/cluster1-8k-splatter.tfrecord'",
"_____no_output_____"
],
[
"gene_up = 14000\nn_pcs = 50\nresolution = 0.6",
"_____no_output_____"
]
],
[
[
"## CLUSTER3-8k-SPLATTER",
"_____no_output_____"
]
],
[
[
"data_path = '/work/SPLATTER-DATA-8k/CLUSTER3-8k-SPLATTER'\nresults_file = '../data/proc/cluster3-8k-splatter.h5ad'\nresults_file2 = '../data/proc/cluster3-8k-splatter.tfrecord'",
"_____no_output_____"
],
[
"gene_up = 14000\nn_pcs = 50\nresolution = 0.6",
"_____no_output_____"
]
],
[
[
"## CLUSTER5-8k-SPLATTER",
"_____no_output_____"
]
],
[
[
"data_path = '/work/SPLATTER-DATA-8k/CLUSTER5-8k-SPLATTER'\nresults_file = '../data/proc/cluster5-8k-splatter.h5ad'\nresults_file2 = '../data/proc/cluste5-8k-splatter.tfrecord'",
"_____no_output_____"
],
[
"gene_up = 14000\nn_pcs = 50\nresolution = 0.6",
"_____no_output_____"
]
],
[
[
"## CLUSTER12-8k-SPLATTER",
"_____no_output_____"
]
],
[
[
"data_path = '/work/SPLATTER-DATA-8k/CLUSTER12-8k-SPLATTER'\nresults_file = '../data/proc/cluster12-8k-splatter.h5ad'\nresults_file2 = '../data/proc/cluste12-8k-splatter.tfrecord'",
"_____no_output_____"
],
[
"gene_up = 14000\nn_pcs = 50\nresolution = 0.6",
"_____no_output_____"
]
],
[
[
"# Data preprocessing",
"_____no_output_____"
]
],
[
[
"sc.settings.verbosity = 3 # verbosity: errors (0), warnings (1), info (2), hints (3)\nsc.logging.print_versions()",
"scanpy==1.4.4.post1 anndata==0.6.22.post1 umap==0.3.10 numpy==1.16.0 scipy==1.3.0 pandas==0.25.0 scikit-learn==0.20.3 statsmodels==0.10.2 python-igraph==0.7.1 louvain==0.6.1\n"
],
[
"adata = sc.read_10x_mtx(\n data_path, # the directory with the `.mtx` file\n var_names='gene_symbols', # use gene symbols for the variable names (variables-axis index)\n cache=True) # write a cache file for faster subsequent reading",
"... writing an h5ad cache file to speedup reading next time\n"
],
[
"sc.settings.set_figure_params(dpi=80)",
"_____no_output_____"
],
[
"adata.var_names_make_unique() # this is unnecessary if using 'gene_ids'",
"_____no_output_____"
],
[
"# Show those genes that yield the highest fraction of counts in each single cells, across all cells.\nsc.pl.highest_expr_genes(adata, n_top=20)\n\n\n# Basic filtering\nsc.pp.filter_cells(adata, min_genes=200)\nsc.pp.filter_genes(adata, min_cells=3)",
"normalizing by total count per cell\n finished (0:00:01): normalized adata.X and added 'n_counts', counts per cell before normalization (adata.obs)\n"
],
[
"mito_genes = adata.var_names.str.startswith('MT-')\n# for each cell compute fraction of counts in mito genes vs. all genes\n# the `.A1` is only necessary as X is sparse (to transform to a dense array after summing)\nadata.obs['percent_mito'] = np.sum(\n adata[:, mito_genes].X, axis=1).A1 / np.sum(adata.X, axis=1).A1\n# add the total counts per cell as observations-annotation to adata\nadata.obs['n_counts'] = adata.X.sum(axis=1).A1",
"_____no_output_____"
],
[
"sc.pl.violin(adata, ['n_genes', 'n_counts', 'percent_mito'],\n jitter=0.4, multi_panel=True)",
"_____no_output_____"
],
[
"sc.pl.scatter(adata, x='n_counts', y='percent_mito')\nsc.pl.scatter(adata, x='n_counts', y='n_genes')",
"_____no_output_____"
],
[
"adata = adata[adata.obs['n_genes'] < gene_up, :] \n# adata = adata[adata.obs['percent_mito'] < percent_mito_up, :]",
"_____no_output_____"
],
[
"# Data in log scale\nsc.pp.normalize_per_cell(adata, counts_per_cell_after=1e4)\nsc.pp.log1p(adata)\n\nadata.raw = adata",
"normalizing by total count per cell\n"
],
[
"sc.pp.highly_variable_genes(adata, min_mean=0.0125, max_mean=3, min_disp=0.5)\nsc.pl.highly_variable_genes(adata)",
"extracting highly variable genes\n finished (0:00:08)\n--> added\n 'highly_variable', boolean vector (adata.var)\n 'means', float vector (adata.var)\n 'dispersions', float vector (adata.var)\n 'dispersions_norm', float vector (adata.var)\n"
],
[
"adata = adata[:, adata.var['highly_variable']]",
"_____no_output_____"
],
[
"# sc.pp.regress_out(adata, ['n_counts', 'percent_mito'])\nsc.pp.regress_out(adata, ['n_counts'])",
"regressing out ['n_counts']\n sparse input is densified and may lead to high memory use\n finished (0:02:16)\n"
],
[
"sc.pp.scale(adata, max_value=10)",
"_____no_output_____"
],
[
"sc.tl.pca(adata, svd_solver='arpack')",
"computing PCA with n_comps = 50\ncomputing PCA on highly variable genes\n finished (0:00:11)\n"
],
[
"sc.pl.pca(adata, color='GENE6815')",
"_____no_output_____"
],
[
"sc.pl.pca_variance_ratio(adata, log=True)",
"_____no_output_____"
],
[
"sc.pp.neighbors(adata, n_neighbors=10, n_pcs=n_pcs)",
"computing neighbors\n using 'X_pca' with n_pcs = 50\n finished: added to `.uns['neighbors']`\n 'distances', distances for each pair of neighbors\n 'connectivities', weighted adjacency matrix (0:00:04)\n"
],
[
"sc.tl.umap(adata)",
"computing UMAP\n finished: added\n 'X_umap', UMAP coordinates (adata.obsm) (0:00:44)\n"
],
[
"sc.pl.umap(adata, color=['GENE6815'])",
"_____no_output_____"
],
[
"sc.tl.louvain(adata, resolution=resolution)",
"running Louvain clustering\n using the \"louvain\" package of Traag (2017)\n finished: found 12 clusters and added\n 'louvain', the cluster labels (adata.obs, categorical) (0:00:00)\n"
],
[
"sc.pl.umap(adata, color=['louvain'])",
"_____no_output_____"
],
[
"# sc.tl.rank_genes_groups(adata, 'louvain', method='t-test')\n# sc.pl.rank_genes_groups(adata, n_genes=25, sharey=False)",
"_____no_output_____"
],
[
"# adata.var_names[adata.var_names.str.startswith('IG')]",
"_____no_output_____"
],
[
"# sc.pl.stacked_violin(adata, groupby='louvain', var_names=marker_genes);",
"_____no_output_____"
],
[
"try:\n sc.pl.stacked_violin(adata, groupby='louvain', var_names=marker_genes)\nexcept:\n print(\"marker genes not defined for this dataset\")",
"marker genes not defined for this dataset\n"
],
[
"try:\n adata.rename_categories('louvain', new_cluster_names)\nexcept:\n print(\"new cluster names not defined for this dataset\")",
"new cluster names not defined for this dataset\n"
],
[
"adata.obs.louvain = [ i.split('_')[0] for i in adata.obs.louvain]\n\ndel adata.uns['louvain_colors']",
"_____no_output_____"
],
[
"sc.pl.umap(adata, color='louvain', legend_loc='on data', title='', frameon=False)",
"... storing 'louvain' as categorical\n"
],
[
"adata.write_h5ad(results_file, compression='gzip')",
"_____no_output_____"
],
[
"# export file in TFRecords format\n\nsys.path.append('../src')\nfrom utils import export_to_tfrecord\n\nexport_to_tfrecord(results_file2, adata)",
"WARNING: Logging before flag parsing goes to stderr.\nW1211 10:16:57.442955 140078085113664 __init__.py:690] \n\n TensorFlow's `tf-nightly` package will soon be updated to TensorFlow 2.0.\n\n Please upgrade your code to TensorFlow 2.0:\n * https://www.tensorflow.org/beta/guide/migration_guide\n\n Or install the latest stable TensorFlow 1.X release:\n * `pip install -U \"tensorflow==1.*\"`\n\n Otherwise your code may be broken by the change.\n\n \nW1211 10:16:57.451815 140078085113664 module_wrapper.py:137] From ../src/utils.py:134: The name tf.python_io.TFRecordWriter is deprecated. Please use tf.io.TFRecordWriter instead.\n\n"
],
[
"adata",
"_____no_output_____"
],
[
"adata.var['gene_ids']",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.