hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ecec3f6592560ab1a3289897b1ade095ef8e8e3e | 108,916 | ipynb | Jupyter Notebook | notebooks/Ambient Seismic Noise/NoiseCorrelation.ipynb | krischer/seismo_live_build | e4e8e59d9bf1b020e13ac91c0707eb907b05b34f | [
"CC-BY-3.0"
] | 3 | 2020-07-11T10:01:39.000Z | 2020-12-16T14:26:03.000Z | notebooks/Ambient Seismic Noise/NoiseCorrelation.ipynb | krischer/seismo_live_build | e4e8e59d9bf1b020e13ac91c0707eb907b05b34f | [
"CC-BY-3.0"
] | null | null | null | notebooks/Ambient Seismic Noise/NoiseCorrelation.ipynb | krischer/seismo_live_build | e4e8e59d9bf1b020e13ac91c0707eb907b05b34f | [
"CC-BY-3.0"
] | 3 | 2020-11-11T05:05:41.000Z | 2022-03-12T09:36:24.000Z | 147.983696 | 53,536 | 0.854429 | [
[
[
"<div style='background-image: url(\"../share/images/header.svg\") ; padding: 0px ; background-size: cover ; border-radius: 5px ; height: 250px'>\n <div style=\"float: right ; margin: 50px ; padding: 20px ; background: rgba(255 , 255 , 255 , 0.7) ; width: 50% ; height: 150px\">\n <div style=\"position: relative ; top: 50% ; transform: translatey(-50%)\">\n <div style=\"font-size: xx-large ; font-weight: 900 ; color: rgba(0 , 0 , 0 , 0.8) ; line-height: 100%\">Ambient Seismic Noise Analysis</div>\n <div style=\"font-size: large ; padding-top: 20px ; color: rgba(0 , 0 , 0 , 0.5)\">Cross Correlation </div>\n </div>\n </div>\n</div>",
"_____no_output_____"
],
[
"In this tutorial you will try to reproduce one of the figures in Shapiro _et al._. To see which one, execute the second code block below. \n\nReference: *High-Resolution Surface-Wave Tomography from Ambient Seismic Noise*, Nikolai M. Shapiro, et al. **Science** 307, 1615 (2005);\nDOI: 10.1126/science.1108339\n\n##### Authors:\n* Celine Hadziioannou\n* Ashim Rijal\n---\n",
"_____no_output_____"
]
],
[
[
"# Configuration step (Please run it before the code!)\n\nimport numpy as np\nimport sys, obspy, os\nimport matplotlib.pyplot as plt\n\nfrom obspy.core import UTCDateTime\nfrom obspy.clients.fdsn import Client\nfrom obspy.geodetics import gps2dist_azimuth as gps2DistAzimuth # depends on obspy version; this is for v1.1.0\n#from obspy.core.util import gps2DistAzimuth\n\n#from PIL import Image\nimport requests\nfrom io import BytesIO\n\n%matplotlib inline\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"### In this notebook\nWe will reproduce figure B below. This figure compares: \n1) the seismogram from an event near station MLAC, recorded at station PHL (top)\n2) the \"Greens function\" obtained by correlating noise recorded at stations MLAC and PHL (center and bottom)\n\nAll bandpassed for periods between 5 - 10 seconds. \n\n<img src=\"https://raw.github.com/ashimrijal/NoiseCorrelation/master/data/shapiro_figure.png\">\n\n---",
"_____no_output_____"
],
[
"### 1. Read in noise data \nRead the noise data for station MLAC into a stream. \n\nThen, **read in** noise data for station PHL.\nAdd this to the stream created above.\n\nThese two data files contain 90 days of vertical component noise for each station.\n#### If you need data for more than 90 days, it can be downloaded form IRIS database. ###",
"_____no_output_____"
]
],
[
[
"# Shapiro et al. use noise data from MLAC and PHL stations\n\nnum_of_days = 90 # no of days of data: change if more than 90days of data is required\nif num_of_days <= 90:\n # get noise data for station MLAC\n stn = obspy.read('https://raw.github.com/ashimrijal/NoiseCorrelation/master/data/noise.CI.MLAC.LHZ.2004.294.2005.017.mseed')\n # get noise data for the station PHL and add it to the previous stream\n stn += obspy.read('https://raw.github.com/ashimrijal/NoiseCorrelation/master/data/noise.CI.PHL.LHZ.2004.294.2005.017.mseed')\n # if you have data stored locally, comment the stn = and stn += lines above\n # then uncomment the following 3 lines and adapt the path: \n # stn = obspy.read('./noise.CI.MLAC.LHZ.2004.294.2005.017.mseed')\n # stn += obspy.read('noise.CI.PHL.LHZ.2004.294.2005.017.mseed')\n # ste = obspy.read('event.CI.PHL.LHZ.1998.196.1998.196.mseed')\nelse:\n # download data from IRIS database\n client = Client(\"IRIS\") # client specification\n t1 = UTCDateTime(\"2004-10-20T00:00:00.230799Z\") # start UTC date/time\n t2 = t1+(num_of_days*86400) # end UTC date/time\n stn = client.get_waveforms(network=\"CI\", station=\"MLAC\",location=\"*\", channel=\"*\",\n starttime=t1, endtime=t2) # get data for MLAC\n stn += client.get_waveforms(network=\"CI\", station=\"PHL\", location=\"*\", channel=\"*\",\n starttime=t1, endtime=t2) # get data for PHL and add it to the previous stream",
"_____no_output_____"
]
],
[
[
"### 2. Preprocess noise ###\n***Preprocessing 1***\n* Just to be sure to keep a 'clean' original stream, first **copy** the noise stream with [st.copy()](https://docs.obspy.org/packages/autogen/obspy.core.stream.Stream.copy.html)\nThe copied stream is the stream you will use from now on. \n\n* In order to test the preprocessing without taking too long, it's also useful to first **trim** this copied noise data stream to just one or a few days. This can be done with [st.trim()](https://docs.obspy.org/packages/autogen/obspy.core.stream.Stream.trim.html), after defining your start- and endtime. \n\n\nMany processing functionalities are included in Obspy. For example, you can remove any (linear) trends with [st.detrend()](https://docs.obspy.org/packages/autogen/obspy.core.stream.Stream.detrend.html), and taper the edges with [st.taper()](https://docs.obspy.org/packages/autogen/obspy.core.stream.Stream.taper.html). \nDifferent types of filter are also available in [st.filter()](https://docs.obspy.org/packages/autogen/obspy.core.stream.Stream.filter.html). \n\n* first **detrend** the data. \n* next, apply a **bandpass filter** to select the frequencies with most noise energy. The secondary microseismic peak is roughly between 0.1 and 0.2 Hz. The primary microseismic peak between 0.05 and 0.1 Hz. Make sure to use a zero phase filter! *(specify argument zerophase=True)*",
"_____no_output_____"
]
],
[
[
"# Preprocessing 1\n\nstp = stn.copy() # copy stream\nt = stp[0].stats.starttime\nstp.trim(t, t + 4 * 86400) # shorten stream for quicker processing\n\nstp.detrend('linear') # remove trends using detrend\nstp.taper(max_percentage=0.05, type='cosine') # taper the edges\nstp.filter('bandpass', freqmin=0.1, freqmax=0.2, zerophase=True) # filter data of all traces in the streams",
"_____no_output_____"
]
],
[
[
"***Preprocessing 2***\n\nSome additional useful processing functions are provided in the following cell named **Functions**\n\n* For each trace in the stream, apply **spectral whitening** on the frequency range you chose before (either [0.1 0.2]Hz or [0.05 0.1]Hz), using function **``whiten``**. \n\n\n* For the **time normalization**, the simplest option is to use the one-bit normalization option provided in function **``normalize``**. \n\n* *Optional: play around with different normalization options, such as clipping to a certain number of standard deviations, or using the running absolute mean normalization.*\n\nA brief *desription of individual* **functions** (see the next cell) are as follows:\n\n1) **whiten**:\n\n spectral whitening of trace `tr` using a cosine tapered boxcar between `freqmin` and `freqmax`\n (courtesy Gaia Soldati & Licia Faenza, INGV)\n \n2) **correlateNoise**:\n \n correlate two stations, using slices of 'corrwin' seconds at a time correlations are also stacked. \n NB hardcoded: correlates 1st with 2nd station in the stream only signals are merged - any data gaps are\n filled with zeros.\n st : stream containing data from the two stations to correlate\n stations : list of stations\n corrwin : correlation window length\n returns 'corr' (all correlations) and 'stack' (averaged correlations)\n \n3) **plotStack**:\n\n plots stack of correlations with correct time axis\n st: stream containing noise (and station information)\n stack: array containing stack \n maxlag: maximum length of correlation to plot (in seconds)\n \n4) **plotXcorrEvent**:\n\n plot the noise correlation (MLAC, PHL) alongside the 1998 event signal \n st : event stream\n stn : noise stream\n stack : noise correlation array\n maxlag : maximum length of correlation, in seconds\n acausal : set to True to use acausal part (=negative times) of the correlation\n figurename : if a filename is specified, figure is saved in pdf format\n \n5) **Normalize**:\n\n Temporal normalization of the traces, most after Bensen 2007. NB. before this treatment, traces must be\n demeaned, detrended and filtered. Description of argument:\n\n norm_method=\"clipping\"\n signal is clipped to 'clip_factor' times the std\n clip_factor recommended: 1 (times std)\n \n norm_method=\"clipping_iter\"\n the signal is clipped iteratively: values above 'clip_factor * std' \n are divided by 'clip_weight'. until the whole signal is below \n 'clip_factor * std'\n clip_factor recommended: 6 (times std)\n \n \n norm_method=\"ramn\"\n running absolute mean normalization: a sliding window runs along the \n signal. The values within the window are used to calculate a \n weighting factor, and the center of the window is scaled by this \n factor. \n weight factor: w = np.mean(np.abs(tr.data[win]))/(2. * norm_win + 1) \n finally, the signal is tapered with a tukey window (alpha = 0.2).\n\n norm_win: running window length, in seconds.\n recommended: half the longest period\n\n norm_method=\"1bit\"\n only the sign of the signal is conserved\n \n6) **get_window**:\n\n Return tukey window of length N\n N: length of window\n alpha: alpha parameter in case of tukey window.\n 0 -> rectangular window\n 1 -> cosine taper\n returns: window (np.array)\n \nDoc of [scipy.signal.get_window](https://docs.scipy.org/doc/scipy-0.14.0/reference/generated/scipy.signal.get_window.html)",
"_____no_output_____"
]
],
[
[
"# Functions\n# collection of functions used in noise correlation processing\n\ndef normalize(tr, clip_factor=6, clip_weight=10, norm_win=None, norm_method=\"1bit\"): \n \n if norm_method == 'clipping':\n lim = clip_factor * np.std(tr.data)\n tr.data[tr.data > lim] = lim\n tr.data[tr.data < -lim] = -lim\n\n elif norm_method == \"clipping_iter\":\n lim = clip_factor * np.std(np.abs(tr.data))\n \n # as long as still values left above the waterlevel, clip_weight\n while tr.data[np.abs(tr.data) > lim] != []:\n tr.data[tr.data > lim] /= clip_weight\n tr.data[tr.data < -lim] /= clip_weight\n\n elif norm_method == 'ramn':\n lwin = tr.stats.sampling_rate * norm_win\n st = 0 # starting point\n N = lwin # ending point\n\n while N < tr.stats.npts:\n win = tr.data[st:N]\n\n w = np.mean(np.abs(win)) / (2. * lwin + 1)\n \n # weight center of window\n tr.data[st + lwin / 2] /= w\n\n # shift window\n st += 1\n N += 1\n\n # taper edges\n taper = get_window(tr.stats.npts)\n tr.data *= taper\n\n elif norm_method == \"1bit\":\n tr.data = np.sign(tr.data)\n tr.data = np.float32(tr.data)\n\n return tr\n\n\ndef get_window(N, alpha=0.2):\n\n window = np.ones(N)\n x = np.linspace(-1., 1., N)\n ind1 = (abs(x) > 1 - alpha) * (x < 0)\n ind2 = (abs(x) > 1 - alpha) * (x > 0)\n window[ind1] = 0.5 * (1 - np.cos(np.pi * (x[ind1] + 1) / alpha))\n window[ind2] = 0.5 * (1 - np.cos(np.pi * (x[ind2] - 1) / alpha))\n return window\n\n\ndef whiten(tr, freqmin, freqmax):\n \n nsamp = tr.stats.sampling_rate\n \n n = len(tr.data)\n if n == 1:\n return tr\n else: \n frange = float(freqmax) - float(freqmin)\n nsmo = int(np.fix(min(0.01, 0.5 * (frange)) * float(n) / nsamp))\n f = np.arange(n) * nsamp / (n - 1.)\n JJ = ((f > float(freqmin)) & (f<float(freqmax))).nonzero()[0]\n \n # signal FFT\n FFTs = np.fft.fft(tr.data)\n FFTsW = np.zeros(n) + 1j * np.zeros(n)\n\n # Apodization to the left with cos^2 (to smooth the discontinuities)\n smo1 = (np.cos(np.linspace(np.pi / 2, np.pi, nsmo+1))**2)\n FFTsW[JJ[0]:JJ[0]+nsmo+1] = smo1 * np.exp(1j * np.angle(FFTs[JJ[0]:JJ[0]+nsmo+1]))\n\n # boxcar\n FFTsW[JJ[0]+nsmo+1:JJ[-1]-nsmo] = np.ones(len(JJ) - 2 * (nsmo+1))\\\n * np.exp(1j * np.angle(FFTs[JJ[0]+nsmo+1:JJ[-1]-nsmo]))\n\n # Apodization to the right with cos^2 (to smooth the discontinuities)\n smo2 = (np.cos(np.linspace(0., np.pi/2., nsmo+1.))**2.)\n espo = np.exp(1j * np.angle(FFTs[JJ[-1]-nsmo:JJ[-1]+1]))\n FFTsW[JJ[-1]-nsmo:JJ[-1]+1] = smo2 * espo\n\n whitedata = 2. * np.fft.ifft(FFTsW).real\n \n tr.data = np.require(whitedata, dtype=\"float32\")\n\n return tr\n\n\ndef correlateNoise(st, stations, corrwin):\n\n print ('correlating stations', (stations[0], stations[1]))\n\n # initialize sliding timewindow (length = corrwin) for correlation\n # start 1 corrwin after the start to account for different stream lengths\n timewin = st.select(station=stations[1])[0].stats.starttime + corrwin\n\n # loop over timewindows \n # stop 1 corrwin before the end to account for different stream lengths\n while timewin < st.select(station=stations[0])[-1].stats.endtime - 2*corrwin:\n sig1 = st.select(station=stations[0]).slice(timewin, timewin+corrwin)\n sig1.merge(method=0, fill_value=0)\n sig2 = st.select(station=stations[1]).slice(timewin, timewin+corrwin)\n sig2.merge(method=0, fill_value=0)\n xcorr = np.correlate(sig1[0].data, sig2[0].data, 'same')\n\n try: \n # build array with all correlations\n corr = np.vstack((corr, xcorr))\n except: \n # if corr doesn't exist yet\n corr = xcorr\n \n # shift timewindow by one correlation window length\n timewin += corrwin\n\n # stack the correlations; normalize\n stack = np.sum(corr, 0)\n stack = stack / float((np.abs(stack).max())) \n print (\"...done\")\n\n return corr, stack\n\n\ndef plotStack(st, stack, maxlag, figurename=None):\n\n # define the time vector for the correlation (length of corr = corrwin + 1)\n limit = (len(stack) / 2.) * st[0].stats.delta\n timevec = np.arange(-limit, limit, st[0].stats.delta)\n\n plt.plot(timevec, stack, 'k')\n stations = list(set([_i.stats.station for _i in st]))\n plt.title(\"Stacked correlation between %s and %s\" % (stations[0], stations[1]))\n plt.xlim(-maxlag, maxlag)\n plt.xlabel('time [s]')\n\n if figurename is not None:\n fig.savefig(figurename, format=\"pdf\")\n else:\n plt.show()\n \n \ndef plotXcorrEvent(st, stn, stack, maxlag, acausal=False, figurename=None):\n\n eventtime = UTCDateTime(1998,7,15,4,53,21,0) # event near MLAC\n\n # station locations\n latP, lonP = 35.41, -120.55 # station PHL\n latM, lonM = 37.63, -118.84 # station MLAC\n latE, lonE = 37.55, -118.809 # event 1998\n \n # calculate distance between stations\n dist = gps2DistAzimuth(latP, lonP, latM, lonM)[0] # between PHL and MLAC\n distE = gps2DistAzimuth(latP, lonP, latE, lonE)[0] # between event and PHL\n #\n # CROSSCORRELATION\n # reverse stack to plot acausal part (= negative times of correlation)\n if acausal:\n stack = stack[::-1]\n \n # find center of stack\n c = int(np.ceil(len(stack)/2.) + 1)\n \n #cut stack to maxlag\n stack = stack[c - maxlag * int(np.ceil(stn[0].stats.sampling_rate)) : c + maxlag * int(np.ceil(stn[0].stats.sampling_rate))]\n \n # find new center of stack\n c2 = int(np.ceil(len(stack)/2.) + 1)\n\n # define time vector for cross correlation\n limit = (len(stack) / 2.) * stn[0].stats.delta\n timevec = np.arange(-limit, limit, stn[0].stats.delta)\n # define timevector: dist / t\n timevecDist = dist / timevec\n \n # EVENT\n ste = st.copy()\n st_PHL_e = ste.select(station='PHL')\n \n # cut down event trace to 'maxlag' seconds\n dt = len(stack[c2:])/stn[0].stats.sampling_rate #xcorrlength\n st_PHL_e[0].trim(eventtime, eventtime + dt)\n \n # create time vector for event signal\n # extreme values:\n limit = st_PHL_e[0].stats.npts * st_PHL_e[0].stats.delta\n timevecSig = np.arange(0, limit, st_PHL_e[0].stats.delta)\n\n # PLOTTING\n fig = plt.figure(figsize=(12.0, 6.0))\n ax1 = fig.add_subplot(2,1,1)\n ax2 = fig.add_subplot(2,1,2)\n\n # plot noise correlation\n ax1.plot(timevecDist[c2:], stack[c2:], 'k')\n ax1.set_title('Noise correlation between MLAC and PHL')\n\n # plot event near MLAC measured at PHL\n ax2.plot(distE/timevecSig, st_PHL_e[0].data / np.max(np.abs(st_PHL_e[0].data)), 'r')\n ax2.set_title('Event near MLAC observed at PHL')\n\n ax2.set_xlim((0, 8000))\n ax1.set_xlim((0, 8000))\n\n ax2.set_xlabel(\"group velocity [m/s]\")\n \n if figurename is not None:\n fig.savefig(figurename, format=\"pdf\")\n else:\n plt.show()",
"_____no_output_____"
]
],
[
[
"### Actual preprocessing happens here -- this can take a while!",
"_____no_output_____"
]
],
[
[
"# Preprocessing 2\nst = stp.copy() # copy stream\n\nfor tr in st:\n tr = normalize(tr, norm_method=\"1bit\")\n tr = whiten(tr, 0.1, 0.2)\nprint ('done!')",
"done!\n"
]
],
[
[
"#### Cross-correlation ####\nOnce you're happy with the preprocessing, you can calculate the **cross-correlation** using **``correlateNoise``** function. The cross-correlation are computed by slices of a few hours each (specified in *corrwin*). \n\n**For correlateNoise function**\n* input: stream, list of stations (here: ['MLAC', 'PHL']), slice length in seconds\n* output: all individual correlations, stack\n",
"_____no_output_____"
]
],
[
[
"# Cross-correlate\nxcorr, stack = correlateNoise(st, ['MLAC','PHL'], 7200)",
"correlating stations ('MLAC', 'PHL')\n...done\n"
]
],
[
[
"The resulting stack can be **plotted** with **``plotStack``** function. Since it doesn't make much sense to look at a 2 hour long correlation signal, you can decide to plot only the central part by specifying a ``maxlag`` (in seconds). ",
"_____no_output_____"
]
],
[
[
"# Plotting\n\nplotStack(st,stack,400)",
"_____no_output_____"
]
],
[
[
"If you're only working with a few days of noise (after trimming), this plot probably doesn't look very nice. You could go back to the code block named 'preprocessing 1', and keep a longer noise record (10 days works quite well already). ",
"_____no_output_____"
],
[
"#### Compare to event trace ####\nIn 1998, a M = 5.1 event occurred next to station MLAC. This event was recorded at PHL and we read this data.\n\n* **read** the event data to a separate stream",
"_____no_output_____"
]
],
[
[
"ste = obspy.read('https://raw.github.com/ashimrijal/NoiseCorrelation/master/data/event.CI.PHL.LHZ.1998.196.1998.196.mseed')\n# if data is stored locally, uncomment the following line and comment the line above:\n#ste = obspy.read('./event.CI.PHL.LHZ.1998.196.1998.196.mseed')",
"_____no_output_____"
]
],
[
[
"#### Preprocess event ####\n\nThe event signal should be processed in a similar way to the noise. \n\n* **detrend** in the same way as before\n* **bandpass filter** for the same frequencies as chosen above\n* apply **spectral whitening** to each trace in the event stream to ensure the spectrum is comparable to the noise. ",
"_____no_output_____"
]
],
[
[
"# Preprocessing\n\nste.detrend('linear')\nste.filter('bandpass', freqmin=0.1, freqmax=0.2, zerophase=True)\nfor tr in ste:\n tr = whiten(tr, 0.1, 0.2)",
"_____no_output_____"
]
],
[
[
"#### Plot! ####\n\nA plotting function is provided to plot both signals alongside: **``plotXcorrEvent``**. \n\n* input: event stream, noise stream, stack, maxlag",
"_____no_output_____"
]
],
[
[
"# Plotting\n\nplotXcorrEvent(ste, stn, stack, 400)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecec49c791b2eefd91f0be953fec86fb736f7cdd | 4,562 | ipynb | Jupyter Notebook | Notebooks/CSV_Creator.ipynb | CosmiQ/CometTS | a558df76d305712f7c46e88d98cd2159cdf8146e | [
"Apache-2.0"
] | 65 | 2018-01-12T17:18:44.000Z | 2022-03-24T20:58:40.000Z | Notebooks/CSV_Creator.ipynb | CosmiQ/ComeTS | a558df76d305712f7c46e88d98cd2159cdf8146e | [
"Apache-2.0"
] | 13 | 2019-02-25T22:40:51.000Z | 2019-11-23T17:41:18.000Z | Notebooks/CSV_Creator.ipynb | CosmiQ/ComeTS | a558df76d305712f7c46e88d98cd2159cdf8146e | [
"Apache-2.0"
] | 18 | 2018-03-09T04:18:00.000Z | 2021-07-21T09:53:57.000Z | 37.702479 | 525 | 0.65388 | [
[
[
"## CSV_Creator - Create a CSV that documents your raster data structure\nThe process works by first iterating through the folder structure and creating a .csv file that indicates where each raster is stored, the date of each image, optionally the number of observations (if using monthly or annual composite data), and optionally a mask band for each of your time series raster images. The workflow below walks through the usage of NPP VIIRS nighttime lights monthly composite data. These data are available for download here: https://ngdc.noaa.gov/eog/viirs/download_dnb_composites.html\n\nOnce downloaded, data should be stored in a hierarchical folder pattern that is common for remote sensing applications.\n\n VIIRSData/\n └── Lat075N_Long060W\n ├── SVDNB_npp_20120401-20120430_75N060W_vcmcfg_v10_c201605121456\n │ ├── SVDNB_npp_20120401-20120430_75N060W_vcmcfg_v10_c201605121456.avg_rade9\n │ ├── SVDNB_npp_20120401-20120430_75N060W_vcmcfg_v10_c201605121456.cf_cvg\n ├── SVDNB_npp_20120501-20120531_75N060W_vcmcfg_v10_c201605121458\n │ ├── SVDNB_npp_20120501-20120531_75N060W_vcmcfg_v10_c201605121458.avg_rade9\n │ ├── SVDNB_npp_20120501-20120531_75N060W_vcmcfg_v10_c201605121458.cf_cvg.tif\n ├── SVDNB_npp_20120601-20120630_75N060W_vcmcfg_v10_c201605121459\n\n ...\n \nIf you are using non-VIIRS data, this pattern will remain consistent regardless. Data should be organized into exclusive areas with identical extents. The most important element of this structure is that images on different dates are stored in their own separate subdirectory. Mask bands should be stored in the same subdirectory as your time series data.",
"_____no_output_____"
]
],
[
[
"## Import your packages, assumes you have already installed CometTS\nimport os\nfrom CometTS.CSV_It import csv_it",
"_____no_output_____"
]
],
[
[
"Optionally get the relative path to sample data below. This is NPP VIIRS nighttime monthly composite imagery.",
"_____no_output_____"
]
],
[
[
"input_path=os.path.abspath('')\ninput_path=os.path.join(input_path.split(\"Notebooks\")[0],\"CometTS/VIIRS_Sample\")",
"_____no_output_____"
]
],
[
[
"Specify paths and naming convetions below. Note the use of wildcards in TSdata, Observations, and Mask which will be used as a common search pattern to query your folders above and identify the appropriate files of interest.",
"_____no_output_____"
]
],
[
[
"#The input directory for the data. Specify directory directly for another directory.\ninput_dir = input_path\n# The naming pattern for your time-series data, if not necessary, just set at \"*\"\nTSdata = \"S*rade9*.tif\"\n# The number of observations that were aggregated per date-somewhat useful is working with monthly composite data\nObservations = \"S*cvg*.tif\"\n# The naming pattern for your masked data\nMask = \"S*cvg*.tif\"\n# The location of the date within your filename. Note python indexing of string here\nDateLoc = \"10:18\"",
"_____no_output_____"
]
],
[
[
"Create the final csv. This can then be fed to CometTS for visualization and analysis.",
"_____no_output_____"
]
],
[
[
"gdf_out = csv_it(input_dir, TSdata, Observations, Mask, DateLoc)\noutput = os.path.join(input_dir, 'Raster_List.csv')\ngdf_out.to_csv(output)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecec549a58660044b7a2b29ab56ab1f698f11163 | 594,980 | ipynb | Jupyter Notebook | 2.1 -- Demographics EDA.ipynb | np1919/DTCJ | 766d67dc73a0bc5ea59972c52cb9c7db81c43daf | [
"MIT"
] | null | null | null | 2.1 -- Demographics EDA.ipynb | np1919/DTCJ | 766d67dc73a0bc5ea59972c52cb9c7db81c43daf | [
"MIT"
] | null | null | null | 2.1 -- Demographics EDA.ipynb | np1919/DTCJ | 766d67dc73a0bc5ea59972c52cb9c7db81c43daf | [
"MIT"
] | null | null | null | 435.882784 | 409,400 | 0.932487 | [
[
[
"# Demographics\n\nDunnhumby: The Complete Journey\n\nNathaniel Poland\n\n\n*In this notebook we'll take a look at the `hh_demographic.csv` table; demographic information for 801 of the 2500 household answers collected by user survey*\n\n\n\n## Customer Focus\n\nThe project as a whole is based around understanding the purchase behaviour of customers of the grocery chain. We would be happy to have some sort of **customer profile**, which offers context to any findings regarding the purchase behaviour of any given `household_key`.\n\n\n### Bias of our sample data; is it representative of the underlying population?\n\nWe recognize the dangers of an **inherent sampling bias** in our data; the 2500 households for which we have transactions are already a subsection of our total population. We have to assume that we only have demographic information from those who opted to take the survey; but we still can't be sure what the selection process was.\n\nMoreover, this demographic **data was generated and entered by human input**.\n\n\n### Demographics or Transactions?\n\nI'd like to segment customers (households) into groups; this allows for targeted advertising through the recommender system we will bring online later.\n\nTo this end, does it make sense to have a customer profile based on how households **interacted with our stores through transactions**? Or, through **demographic labels**?\n\nAs far as supervised and unsupervised learning goes, we are searching for **viable target features** which clearly distinguish our customer segments from one another will be useful. ",
"_____no_output_____"
],
[
"## Business Summary/tl;dr",
"_____no_output_____"
],
[
"INPUTS/OBSERVATIONS:\n- 32% of 2500 total households are represented by the 801 `household_key`s in this data.\n - **These households represent 56% of total revenue in the `transaction_data.csv` file, a significant percentage.** \n\n- Sample Bias & Confusing Questions\n - Unknown values in marital status, homeowner status, kid description\n \n- **Households vary in size (number of members), potentially distorting our analysis**\n - Despite potentially representing distinct members of a household, we have a singular value for both the age and income columns\n \n- Many columns have poorly distributed classes; we can use pd.Categorical columns, or clean up the distributions by forcing binary labels (which is arbitrary, and might lead to biased, misleading, or uninformative results).",
"_____no_output_____"
],
[
"# EDA for `hh_demographic.csv`",
"_____no_output_____"
],
[
"## Importing Modules",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nplt.rcParams['figure.figsize']==(16,6)\nimport dtcj\n\nplt.rcParams['figure.figsize']=(16,6)\nplt.style.use('seaborn')\n\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"## Importing Data",
"_____no_output_____"
]
],
[
[
"demo = pd.read_csv('data/hh_demographic.csv')",
"_____no_output_____"
],
[
"print(demo.info())\ndemo.head(3)",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 801 entries, 0 to 800\nData columns (total 8 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 AGE_DESC 801 non-null object\n 1 MARITAL_STATUS_CODE 801 non-null object\n 2 INCOME_DESC 801 non-null object\n 3 HOMEOWNER_DESC 801 non-null object\n 4 HH_COMP_DESC 801 non-null object\n 5 HOUSEHOLD_SIZE_DESC 801 non-null object\n 6 KID_CATEGORY_DESC 801 non-null object\n 7 household_key 801 non-null int64 \ndtypes: int64(1), object(7)\nmemory usage: 50.2+ KB\nNone\n"
],
[
"demo.household_key.duplicated().sum()",
"_____no_output_____"
]
],
[
[
"- The table consists of 801 rows of demographic information, each for a unique `household_key`. \n\n- There are no null values\n\n- The data is largely in 'object' format; strings; the exception is the unique identifier, `household_key`.",
"_____no_output_____"
]
],
[
[
"### There are 8 columns in the table. Below are the skeleton value counts:\n\n# for col in demo.drop('household_key', axis=1):\n# print(demo[col].value_counts())",
"_____no_output_____"
]
],
[
[
"Further below we will plot this information in a more readable way.",
"_____no_output_____"
],
[
"# Data Dictionary for `hh_demographic.csv`\n`AGE_DESC`\nGood descriptive information, but poorly balanced classes.\n\n`MARITAL_STATUS_CODE` per the data dictionary available online:\n\n A = married\n B = single\n U = unknown\n\n`INCOME_DESC` Many distinct values, including with income ranges 100K+; perhaps due to double income.\n\n`HOMEOWNER_DESC`\nAround 1/4 unknown values; homeowner or renter.\n\n`HH_COMP_DESC`\nHousehold composition description; string labels for household type including gender for individuals. This column allows us to distinguish single parents with kids from couples.\n\n`HOUSEHOLD_SIZE_DESC`\nRanging from 1-5+.\n\n`KID_CATEGORY_DESC`\nAlmost three quarters None/unknown values.\n\n`household_key`\nUnique Identifier for each household",
"_____no_output_____"
]
],
[
[
"demo.household_key.duplicated().sum()\n# no duplicate household_keys",
"_____no_output_____"
]
],
[
[
"## Applying Categorical Ranks",
"_____no_output_____"
],
[
"Without losing any information at all, we can change the columns to pd.Categorical data type. This constructor allows us to 'rank' the category labels; this makes visualization more simple. By passing the `ordered=False` argument, we can prevent misinterpretations of our ranks as being sequentially ordered; for income, age, or household size we keep the order.",
"_____no_output_____"
]
],
[
[
"\ndef demo_map_categorical(demo):\n demo['AGE_DESC'] = pd.Categorical(demo['AGE_DESC'], ['19-24', '25-34','35-44', '45-54', '55-64', '65+',])\n\n demo['MARITAL_STATUS_CODE'] = pd.Categorical(demo['MARITAL_STATUS_CODE'].map({'A':'Married', 'B':'Single', 'U': 'Unknown'}), ['Married', 'Single', 'Unknown'], ordered=False)\n\n demo['INCOME_DESC']= pd.Categorical(demo['INCOME_DESC'], ['Under 15K','15-24K','25-34K', '35-49K', '50-74K','75-99K', \n '100-124K', '125-149K', '150-174K', '175-199K', '200-249K','250K+', ])\n\n demo['HOMEOWNER_DESC']= pd.Categorical(demo['HOMEOWNER_DESC'], ['Homeowner', 'Unknown', 'Renter', 'Probable Renter', 'Probable Owner'], ordered=False)\n\n demo['HH_COMP_DESC']=pd.Categorical(demo['HH_COMP_DESC'], ['Single Female', 'Single Male', '1 Adult Kids','2 Adults No Kids', '2 Adults Kids', 'Unknown',], ordered=False)\n demo['HOUSEHOLD_SIZE_DESC'] = pd.Categorical(demo['HOUSEHOLD_SIZE_DESC'], ['1', '2','3','4','5+',])\n demo['KID_CATEGORY_DESC']=pd.Categorical(demo['KID_CATEGORY_DESC'], ['1','2','3+','None/Unknown', ])\n \n return demo\n\ndemo_map_categorical(demo)",
"_____no_output_____"
]
],
[
[
"## Plotting Class Distributions for Demographics",
"_____no_output_____"
]
],
[
[
"def plot_demo_distributions():\n columns = 2\n rows = (len(demo.columns)-1)//columns + 1\n plt.subplots(rows, columns, figsize=(36, 24))\n plt.suptitle('Distribution of Columns', size=40)\n for idx, col in enumerate(demo.drop('household_key', axis=1).columns):\n data = dict(demo[col].value_counts())\n order = list(demo[col].cat.categories)\n plt.subplot(rows, columns, idx+1)\n plt.title(f'{col}', size=40)\n plt.xticks(fontsize=35, rotation=45)\n for x in order:\n plt.bar(x, data[x], color='blue')\n\n plt.tight_layout()\n plt.show()\nplot_demo_distributions()",
"_____no_output_____"
],
[
"dtcj.plot_pies(demo.drop('household_key', axis=1))",
"_____no_output_____"
]
],
[
[
"### Observations of Distributions",
"_____no_output_____"
],
[
"**`AGE_DESC`**\n\n- The households in our survey are leaning significantly towards the elderly age groups. **More than half the households for which we have demographic info report an age of 45+** \n\n**`HOMEOWNER_DESC`** \n\n- According to this data, **more than 63% of the households for which we have demographic info likely own homes**.\n\n**`HOUSEHOLD_SIZE_DESC`** \n- Seems to be the most complete and informative column about our household sizes here -- what I might guess is the most relevant factor with respect to comparisons with an RFM score. \n- **No NaN or Unknown values**\n\n**`HH_COMP_DESC`** \n\n- Creates some definition and variance in the data -- guided by someone with more experience, I might consider delving in here. \n - **'Unknown' values makes up 9% of the column**.\n - We could drop the 9% of rows, or impute their values somehow, based on a swag-y guess using the household size desc. \n - Taking it one step further, we could examine sales patterns for the households, to add a second decision boundary.\n- It seemingly separates Single male and female individuals -- as well as single-parent households with kids, which might be a point of confusion. \n\n\n**`KID_CATEGORY_DESC`** \n\n- Combined, **None and Unknown labels make up more than 2/3 of the column**\n- 31.3% of the households 'have kids' confirmed; compared with 28.4% of households in the 'HH_COMP_DESC' of size 3 or greater. \n\nUsing the recommender system class shown later in the project, we could examine households in these various categories and promote advertising based on their most recent purchases.",
"_____no_output_____"
],
[
"# `hh_demographic` Sales Totals",
"_____no_output_____"
]
],
[
[
"# import the full transactions table for a basic reference \ntransactions = pd.read_csv('data/transaction_data.csv')",
"_____no_output_____"
],
[
"# transactions = dtcj.load_merged()",
"_____no_output_____"
],
[
"# list of household_keys in hh_demographic\ndemo_hh = demo['household_key'].values\n\ndemo_sales = transactions[transactions['household_key'].isin(demo_hh)]['SALES_VALUE'].sum()\n\nnon_demo_sales = transactions[~transactions['household_key'].isin(demo_hh)]['SALES_VALUE'].sum()\n\ntotal_sales = demo_sales + non_demo_sales\n\nprint(f'{round(total_sales, 2)} is the total sales value across all households.')\nprint()\n\nprint(f'{round(demo_sales, 2)} is the total sales value of households with demographic information listed.')\nprint()\nprint(f'{round(non_demo_sales, 2)} is the total sales value of households NOT listed in hh_demographic.')\nprint()\n\nprint(f'{round((demo_sales/total_sales) * 100,2)}% is the percentage of sales accounted for\\nby households for which we have demographic information.')\nprint()\nprint(f'{round((non_demo_sales/total_sales) * 100,2)}% is the percentage of sales accounted for\\nby households for which we do not have demographic information.')",
"8057463.08 is the total sales value across all households.\n\n4497716.26 is the total sales value of households with demographic information listed.\n\n3559746.82 is the total sales value of households NOT listed in hh_demographic.\n\n55.82% is the percentage of sales accounted for\nby households for which we have demographic information.\n\n44.18% is the percentage of sales accounted for\nby households for which we do not have demographic information.\n"
]
],
[
[
"**Despite making up only 32% of our households, the customers for which we have demographic information account for ~56% of the total sales in the data** (This is consistent even after the data cleaning which happens in the following notebooks).\n\nWe knew that the customers in our data were all purportedly **frequent shoppers** at our grocery chain, making market segmentation more difficult in some senses. However, this subset of customers is valuable; within the context of our data, and likely outside it. \n\n**We should be cautious about extrapolating any analysis or interpretations onto customers of the grocery chain as a whole**.",
"_____no_output_____"
],
[
"### Notes about Household Size",
"_____no_output_____"
],
[
"Households have a varying number of members. This will almost certainly impact the sales total for each household, as well as the products they might want to receive advertisements for.\n\nWe might imagine that **households with more members will require more food and other goods**, and therefore that families (or otherwise larger households) have the potential to be our highest-grossing \"customers\" as is defined in the data. \n\n**We should be careful about assuming this equates to higher overall revenue** as there might be less families around than individuals. Still, since the adults are the ones making purchases for a family, **advertising dollars spent on the parent of a family is likely to have a more significant sales impact** than on an individual; that one parent would be purchasing goods for several other people. Individuals or couples could be at an inherent 'disadvantage' in terms of total monetary value or frequency of purchases, but we'll have to take a closer look. We look at sales analysis by demographic category in following notebooks.\n\nIn terms of total sales by all individuals versus all families, we should again take note of our sampling bias. **We have a select few households in our demographic information** (and a select 2500 in our transactions info!).\n\n**These class distributions and spending totals are not necessarily representative of the underlying population's demographic proportions or purchase behaviour.**",
"_____no_output_____"
],
[
"# Column Mappings",
"_____no_output_____"
],
[
"None of the transformations below are executed by the cells; they print the 'value counts'.",
"_____no_output_____"
]
],
[
[
"# AGE_DESC\ndemo['AGE_DESC'].map(\n{\n'19-24':\"0-34\",\n'25-34':\"0-34\",\n\"35-44\":\"35-44\",\n'45-54':\"45+\",\n'55-64':\"45+\",\n'65+':\"45+\",\n}).value_counts()\n# Splitting 45+ results in a much smaller fourth class, adding another class imbalance.\n# At least the other two categories are similar in size? \n# Are the two smaller classes the most 'variant'? in terms of their within-label spending, for our purposes??",
"_____no_output_____"
],
[
"# MARITAL_STATUS_CODE\ndemo['MARITAL_STATUS_CODE'].value_counts()\n\n# .map({'Married':1,\n# 'Single':2,\n# 'Unknown':0}).value_counts()",
"_____no_output_____"
],
[
"# INCOME_DESC\ndemo['INCOME_DESC'].map({'Under 15K': '0-49K',\n '175-199K': '100K+',\n '35-49K': '0-49K',\n '125-149K': '100K+',\n '150-174K': '100K+',\n '15-24K': '0-49K',\n '50-74K': '50-99K',\n '250K+': '100K+',\n '200-249K': '100K+',\n '100-124K': '100K+',\n '75-99K': '50-99K',\n '25-34K': '0-49K'}).value_counts()\n\n# demo['INCOME_DESC'].map({\n# 'Under 15K': 0,\n# '15-24K': 0,\n# '25-34K': 0,\n# '35-49K': 0,\n# '50-74K': 1,\n# '75-99K': 1,\n# '100-124K': 1,\n# '125-149K': 1,\n# '150-174K': 1, \n# '175-199K': 1, \n# '200-249K': 1,\n# '250K+': 1,\n# }).sum()",
"_____no_output_____"
],
[
"# HOMEOWNER_DESC\ndemo['HOMEOWNER_DESC'].map({\n\"Homeowner\":1, \n\"Unknown\":0, \n\"Renter\":0, \n\"Probable Renter\":0, \n\"Probable Owner\":1, \n}).value_counts()\n\n# Homeowner vs. Unknown/Renter",
"_____no_output_____"
],
[
"# HH_COMP_DESC -- consider dropping Unknown values?\ndemo['HH_COMP_DESC'].value_counts()",
"_____no_output_____"
],
[
"# HOUSEHOLD_SIZE_DESC series to type int; note we are changing 5+ to 5... (losing information here)\ndemo['HOUSEHOLD_SIZE_DESC'].map({'1':1, '2':2, '3':3, '4':4, '5+':5}).astype(int).value_counts()",
"_____no_output_____"
],
[
"# KID_CATEGORY_DESC\n# demo.drop('KID_CATEGORY_DESC', axis=1)",
"_____no_output_____"
]
],
[
[
"# Binary Mappings",
"_____no_output_____"
],
[
"## `single_couple_family`",
"_____no_output_____"
]
],
[
[
"demo['single_couple_family'] = demo['HOUSEHOLD_SIZE_DESC'].map({'1':1, '2':2, '3':3,'4':3,'5+':3})\ndemo['single_couple_family'] = pd.Categorical(demo['single_couple_family'], \n [1,2,3,])",
"_____no_output_____"
]
],
[
[
"## `single`",
"_____no_output_____"
]
],
[
[
"demo['single'] = np.where((demo['HH_COMP_DESC'] == 'Single Female') |\n (demo['HH_COMP_DESC'] == 'Single Male') |\n (demo['HOUSEHOLD_SIZE_DESC'] == '1'),\n 1, 0)",
"_____no_output_____"
]
],
[
[
"## `couple`",
"_____no_output_____"
]
],
[
[
"demo['couple'] = np.where((demo['HH_COMP_DESC'] == '2 Adults no Kids'),\n 1, 0)",
"_____no_output_____"
]
],
[
[
"## `has_kids`\n\n*Engineering a map for households with confirmed children*",
"_____no_output_____"
]
],
[
[
"# has_kids\ndemo['has_kids'] = np.where((demo['HH_COMP_DESC'] == '1 Adult Kids') |\n (demo['HH_COMP_DESC'] == '2 Adults Kids') |\n (demo['HOUSEHOLD_SIZE_DESC'].isin(['3', '4', '5+'])),\n 1, 0)\n\ndemo['has_kids'].value_counts()",
"_____no_output_____"
]
],
[
[
"Using the queries below we can confirm that:\n\nall entries labeled '1 Adult Kids' are now tagged appropriately as 'has_kids'",
"_____no_output_____"
]
],
[
[
"# demo[demo['HH_COMP_DESC']=='1 Adult Kids']",
"_____no_output_____"
]
],
[
[
"all entries labeled as '2 Adults No Kids' are appropriately NOT tagged as 'has_kids;'",
"_____no_output_____"
]
],
[
[
"# demo[demo['HH_COMP_DESC'] == '2 Adults No Kids']['has_kids'].sum()",
"_____no_output_____"
]
],
[
[
"## `age_45+`",
"_____no_output_____"
],
[
"I might be setting this boundary arbitrarily based on the class distributions of my test set.",
"_____no_output_____"
]
],
[
[
"# \ndemo['AGE_DESC'].map(\n{\n'19-24':0,\n'25-34':0,\n\"35-44\":0,\n'45-54':1,\n'55-64':1,\n'65+':1,\n}).value_counts()\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
ecec77d01146a00f81797d9b0e3701174a1789cb | 17,158 | ipynb | Jupyter Notebook | Intro-to-Python/07.ipynb | avicodingdojo/PyTutorial | d4b5ac4e2a0e7ddaf3cc52efcb5d63aefbd491ef | [
"CC-BY-3.0"
] | null | null | null | Intro-to-Python/07.ipynb | avicodingdojo/PyTutorial | d4b5ac4e2a0e7ddaf3cc52efcb5d63aefbd491ef | [
"CC-BY-3.0"
] | null | null | null | Intro-to-Python/07.ipynb | avicodingdojo/PyTutorial | d4b5ac4e2a0e7ddaf3cc52efcb5d63aefbd491ef | [
"CC-BY-3.0"
] | 1 | 2021-01-11T11:23:26.000Z | 2021-01-11T11:23:26.000Z | 23.731674 | 396 | 0.534211 | [
[
[
"<small><font style=\"font-size:6pt\"> <i>\nAll of these python notebooks are available at https://gitlab.erc.monash.edu.au/andrease/Python4Maths.git </i>\n</font></small>",
"_____no_output_____"
],
[
"# Classes",
"_____no_output_____"
],
[
"Variables, Lists, Dictionaries etc in python are objects. Without getting into the theory part of Object Oriented Programming, explanation of the concepts will be done along this tutorial.",
"_____no_output_____"
],
[
"A class is declared as follows",
"_____no_output_____"
],
[
"```python\nclass class_name:\n methods (functions)```\n",
"_____no_output_____"
]
],
[
[
"class FirstClass:\n \"This is an empty class\"\n pass",
"_____no_output_____"
]
],
[
[
"**pass** in python means do nothing. The string defines the documentation of the class, accessible via `help(FirstClass)`",
"_____no_output_____"
],
[
"Above, a class object named \"FirstClass\" is declared now consider a \"egclass\" which has all the characteristics of \"FirstClass\". So all you have to do is, equate the \"egclass\" to \"FirstClass\". In python jargon this is called as creating an instance. \"egclass\" is the instance of \"FirstClass\"",
"_____no_output_____"
]
],
[
[
"egclass = FirstClass()",
"_____no_output_____"
],
[
"type(egclass)",
"_____no_output_____"
],
[
"type(FirstClass)",
"_____no_output_____"
]
],
[
[
"Objects (instances of a class) can hold data. A variable in an object is also called a field or an attribute. To access a field use the notation `object.field`. For example:x",
"_____no_output_____"
]
],
[
[
"obj1 = FirstClass()\nobj2 = FirstClass()\nobj1.x = 5\nobj2.x = 6\nx = 7\nprint(\"x in object 1 =\",obj1.x,\"x in object 2=\",obj2.x,\"global x =\",x)",
"x in object 1 = 5 x in object 2= 6 global x = 7\n"
]
],
[
[
"Now let us add some \"functionality\" to the class. A function inside a class is called as a \"Method\" of that class",
"_____no_output_____"
]
],
[
[
"class Counter:\n def reset(self,init=0):\n self.count = init\n def getCount(self):\n self.count += 1\n return self.count\ncounter = Counter()\ncounter.reset(0)\nprint(\"one =\",counter.getCount(),\"two =\",counter.getCount(),\"three =\",counter.getCount())",
"one = 1 two = 2 three = 3\n"
]
],
[
[
"Note that the `reset()` and function and the `getCount()` method are callled with one less argument than they are declared with. The `self` argument is set by Python to the calling object. Here `counter.reset(0)` is equivalent to `Counter.reset(counter,0)`.\nUsing **self** as the name of the first argument of a method is simply a common convention. Python allows any name to be used.\n\nNote that here it would be better if we could initialise Counter objects immediately with a default value of `count` rather than having to call `reset()`. A constructor method is declared in Python with the special name `__init__`:",
"_____no_output_____"
]
],
[
[
"class FirstClass:\n def __init__(self,name,symbol):\n self.name = name\n self.symbol = symbol",
"_____no_output_____"
]
],
[
[
"Now that we have defined a function and added the \\_\\_init\\_\\_ method. We can create a instance of FirstClass which now accepts two arguments. ",
"_____no_output_____"
]
],
[
[
"eg1 = FirstClass('one',1)\neg2 = FirstClass('two',2)",
"_____no_output_____"
],
[
"print(eg1.name, eg1.symbol)\nprint(eg2.name, eg2.symbol)",
"one 1\ntwo 2\n"
]
],
[
[
"**dir( )** function comes very handy in looking into what the class contains and what all method it offers",
"_____no_output_____"
]
],
[
[
"print(\"Contents of Counter class:\",dir(Counter) )\nprint(\"Contents of counter object:\", dir(counter))",
"Contents of Counter class: ['__class__', '__delattr__', '__dict__', '__dir__', '__doc__', '__eq__', '__format__', '__ge__', '__getattribute__', '__gt__', '__hash__', '__init__', '__le__', '__lt__', '__module__', '__ne__', '__new__', '__reduce__', '__reduce_ex__', '__repr__', '__setattr__', '__sizeof__', '__str__', '__subclasshook__', '__weakref__', 'getCount', 'reset']\nContents of counter object: ['__class__', '__delattr__', '__dict__', '__dir__', '__doc__', '__eq__', '__format__', '__ge__', '__getattribute__', '__gt__', '__hash__', '__init__', '__le__', '__lt__', '__module__', '__ne__', '__new__', '__reduce__', '__reduce_ex__', '__repr__', '__setattr__', '__sizeof__', '__str__', '__subclasshook__', '__weakref__', 'count', 'getCount', 'reset']\n"
]
],
[
[
"**dir( )** of an instance also shows it's defined attributes so the object has the additional 'count' attribute. Note that Python defines several default methods for actions like comparison (`__le__` is $\\le$ operator). These and other special methods can be defined for classes to implement specific meanings for how object of that class should be compared, added, multiplied or the like.",
"_____no_output_____"
],
[
"Changing the FirstClass function a bit,",
"_____no_output_____"
],
[
"Just like global and local variables as we saw earlier, even classes have it's own types of variables.\n\nClass Attribute : attributes defined outside the method and is applicable to all the instances.\n\nInstance Attribute : attributes defined inside a method and is applicable to only that method and is unique to each instance.",
"_____no_output_____"
]
],
[
[
"class FirstClass:\n test = 'test'\n def __init__(self,n,s):\n self.name = n\n self.symbol = s",
"_____no_output_____"
]
],
[
[
"Here test is a class attribute and name is a instance attribute.",
"_____no_output_____"
]
],
[
[
"eg3 = FirstClass('Three',3)",
"_____no_output_____"
],
[
"print(eg3.test,eg3.name,eg3.symbol)",
"TEST Three 3\n"
]
],
[
[
"## Inheritance",
"_____no_output_____"
],
[
"There might be cases where a new class would have all the previous characteristics of an already defined class. So the new class can \"inherit\" the previous class and add it's own methods to it. This is called as inheritance.",
"_____no_output_____"
],
[
"Consider class SoftwareEngineer which has a method salary.",
"_____no_output_____"
]
],
[
[
"class SoftwareEngineer:\n def __init__(self,name,age):\n self.name = name\n self.age = age\n def salary(self, value):\n self.money = value\n print(self.name,\"earns\",self.money)",
"_____no_output_____"
],
[
"a = SoftwareEngineer('Kartik',26)",
"_____no_output_____"
],
[
"a.salary(40000)",
"Kartik earns 40000\n"
],
[
"[ name for name in dir(SoftwareEngineer) if not name.startswith(\"_\")]",
"_____no_output_____"
]
],
[
[
"Now consider another class Artist which tells us about the amount of money an artist earns and his artform.",
"_____no_output_____"
]
],
[
[
"class Artist:\n def __init__(self,name,age):\n self.name = name\n self.age = age\n def money(self,value):\n self.money = value\n print(self.name,\"earns\",self.money)\n def artform(self, job):\n self.job = job\n print(self.name,\"is a\", self.job)",
"_____no_output_____"
],
[
"b = Artist('Nitin',20)",
"_____no_output_____"
],
[
"b.money(50000)\nb.artform('Musician')",
"Nitin earns 50000\nNitin is a Musician\n"
],
[
"[ name for name in dir(b) if not name.startswith(\"_\")]",
"_____no_output_____"
]
],
[
[
"money method and salary method are the same. So we can generalize the method to salary and inherit the SoftwareEngineer class to Artist class. Now the artist class becomes,",
"_____no_output_____"
]
],
[
[
"class Artist(SoftwareEngineer):\n def artform(self, job):\n self.job = job\n print self.name,\"is a\", self.job",
"_____no_output_____"
],
[
"c = Artist('Nishanth',21)",
"_____no_output_____"
],
[
"dir(Artist)",
"_____no_output_____"
],
[
"c.salary(60000)\nc.artform('Dancer')",
"Nishanth earns 60000\nNishanth is a Dancer\n"
]
],
[
[
"Suppose say while inheriting a particular method is not suitable for the new class. One can override this method by defining again that method with the same name inside the new class.",
"_____no_output_____"
]
],
[
[
"class Artist(SoftwareEngineer):\n def artform(self, job):\n self.job = job\n print(self.name,\"is a\", self.job)\n def salary(self, value):\n self.money = value\n print(self.name,\"earns\",self.money)\n print(\"I am overriding the SoftwareEngineer class's salary method\")",
"_____no_output_____"
],
[
"c = Artist('Nishanth',21)",
"_____no_output_____"
],
[
"c.salary(60000)\nc.artform('Dancer')",
"Nishanth earns 60000\nI am overriding the SoftwareEngineer class's salary method\nNishanth is a Dancer\n"
]
],
[
[
"If the number of input arguments varies from instance to instance asterisk can be used as shown.",
"_____no_output_____"
]
],
[
[
"class NotSure:\n def __init__(self, *args):\n self.data = ' '.join(list(args)) ",
"_____no_output_____"
],
[
"yz = NotSure('I', 'Do' , 'Not', 'Know', 'What', 'To','Type')",
"_____no_output_____"
],
[
"yz.data",
"_____no_output_____"
]
],
[
[
"## Introspection\nWe have already seen the `dir()` function for working out what is in a class. Python has many facilities to make introspection easy (that is working out what is in a Python object or module). Some useful functions are **hasattr**, **getattr**, and **setattr**:",
"_____no_output_____"
]
],
[
[
"ns = NotSure('test')\nif hasattr(ns,'data'): # check if ns.data exists\n setattr(ns,'copy', # set ns.copy\n getattr(ns,'data')) # get ns.data\nprint('ns.copy =',ns.copy)",
"ns.copy = test\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecec7af9dd7d3b7f783dec07060a1b2c8569fa1d | 37,388 | ipynb | Jupyter Notebook | IntroToML-MissingValues.ipynb | aryanc55/MLIn10Minutes | ae22fc8e6fa99148604efdb359da75c354d7721b | [
"MIT"
] | 3 | 2020-05-17T18:49:40.000Z | 2020-05-19T16:45:15.000Z | IntroToML-MissingValues.ipynb | aryanc55/MLIn10Minutes | ae22fc8e6fa99148604efdb359da75c354d7721b | [
"MIT"
] | null | null | null | IntroToML-MissingValues.ipynb | aryanc55/MLIn10Minutes | ae22fc8e6fa99148604efdb359da75c354d7721b | [
"MIT"
] | null | null | null | 30.396748 | 387 | 0.445785 | [
[
[
"# Handling Missing Values",
"_____no_output_____"
],
[
"#### Let's get out dataset and important libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.experimental import enable_iterative_imputer\nfrom sklearn.impute import IterativeImputer\nfrom sklearn.ensemble import ExtraTreesRegressor\nfrom sklearn.linear_model import BayesianRidge\nfrom sklearn.impute import KNNImputer",
"_____no_output_____"
],
[
"# save filepath to variable for easier access\nmelbourne_file_path = '../hitchhikersGuideToMachineLearning/home-data-for-ml-course/train.csv'\n# read the data and store data in DataFrame titled melbourne_data\ntrain_data = pd.read_csv(melbourne_file_path) \n# print a summary of the data in Melbourne data\ntrain_data.head()\n",
"_____no_output_____"
]
],
[
[
"You can already see a few missing values in the first several rows. In the next step, you'll obtain a more comprehensive understanding of the missing values in the dataset.\n\n# Step 1: Preliminary investigation\n\nRun the code cell below without changes.",
"_____no_output_____"
]
],
[
[
"# Shape of training data (num_rows, num_columns)\nprint(train_data.shape)",
"(1460, 81)\n"
],
[
"# Number of missing values in each column of training data\nmissing_val_count_by_column = (train_data.isnull().sum())\nmissing_val_count_by_column[missing_val_count_by_column > 0]",
"_____no_output_____"
]
],
[
[
"Here is another piece of code used for same but with percentages too.",
"_____no_output_____"
]
],
[
[
"def missingcheck(data):\n total = data.isnull().sum().sort_values(ascending=False)\n percent_1 = data.isnull().sum()/data.isnull().count()*100\n percent_2 = (np.round(percent_1, 1)).sort_values(ascending=False)\n missing_data = pd.concat([total, percent_2], axis=1, keys=['Total', '%']) #ptr\n return missing_data",
"_____no_output_____"
],
[
"#just pass the data to the function\nmissingcheck(train_data)",
"_____no_output_____"
]
],
[
[
"### Also seggregate Numerical attributes and Categorical attributes\nBecuase it will affect the strategy we will follow for missing value treatment!\n#### Numerical data:\nThese data have meaning as a measurement, such as a person’s height, weight, IQ, or blood pressure; or they’re a count, such as the number of stock shares a person owns,how many teeth a dog has, or how many pages you can read of your favorite book before you fall asleep.\n\n#### Categorical data: \nCategorical data represent characteristics such as a person’s gender, marital status, hometown, or the types of movies they like. Categorical data can take on numerical values (such as “1” indicating male and “2” indicating female), but those numbers don’t have mathematical meaning",
"_____no_output_____"
]
],
[
[
"#Numerical data will generally have int64 or float64 representation while \n#Categorical may follow object datatype.\n\ntrain_data.dtypes",
"_____no_output_____"
]
],
[
[
"You can treat missing values seperately and later join them together! But to keep this notebook short and understandable I will continue only with numerical values.",
"_____no_output_____"
]
],
[
[
"train_data = train_data.select_dtypes(exclude=['object'])\n#train_data_Categorical = train_data.select_dtypes(include=['object'])\n",
"_____no_output_____"
]
],
[
[
"Select target variable and then Split the data in training and validation set",
"_____no_output_____"
]
],
[
[
"X = train_data.drop(['SalePrice'] , axis =1)\ny = train_data.SalePrice",
"_____no_output_____"
],
[
"#Splitting in training and Validation set\nX_train, X_valid, y_train, y_valid = train_test_split(X, y, train_size=0.8, test_size=0.2,random_state=0)",
"_____no_output_____"
]
],
[
[
"To compare different approaches to dealing with missing values, you'll use the same `score_dataset()` function from the tutorial. This function reports the [mean absolute error](https://en.wikipedia.org/wiki/Mean_absolute_error) (MAE) from a random forest model.",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestRegressor\nfrom sklearn.metrics import mean_absolute_error\n\n# Function for comparing different approaches\ndef score_dataset(X_train, X_valid, y_train, y_valid):\n model = RandomForestRegressor(n_estimators=100, random_state=0)\n model.fit(X_train, y_train)\n preds = model.predict(X_valid)\n return mean_absolute_error(y_valid, preds)",
"_____no_output_____"
]
],
[
[
"#### We are all set to LAUNCH!",
"_____no_output_____"
],
[
"# Step 2: Apply different Techniques\n\n",
"_____no_output_____"
],
[
"## 1. Drop Missing Values\nIn this step, you'll preprocess the data in `X_train` and `X_valid` to remove columns with missing values. Set the preprocessed DataFrames to `reduced_X_train` and `reduced_X_valid`, respectively. ",
"_____no_output_____"
]
],
[
[
"#get names of columns with missing values\ncols_with_missing = [col for col in X_train.columns\n if X_train[col].isnull().any()]\n# drop columns in training and validation data\nreduced_X_train = X_train.drop(cols_with_missing, axis=1)\nreduced_X_valid = X_valid.drop(cols_with_missing, axis=1)\n",
"_____no_output_____"
]
],
[
[
"If missing values are very less, you can also drop the whole row containing it.",
"_____no_output_____"
],
[
"Run the next code cell without changes to obtain the MAE for this approach.",
"_____no_output_____"
]
],
[
[
"print(\"MAE (Drop columns with missing values):\")\nprint(score_dataset(reduced_X_train, reduced_X_valid, y_train, y_valid))",
"MAE (Drop columns with missing values):\n17952.591404109586\n"
]
],
[
[
"## 2. Univariate Imputation\n###### Fancy words for replacinig missing values with mean, median, mode or some constant value!",
"_____no_output_____"
],
[
"One type of imputation algorithm is univariate, which imputes values in the i-th feature dimension using only non-missing values in that feature dimension (e.g. impute.SimpleImputer).\n",
"_____no_output_____"
],
[
"Use the next code cell to impute missing values with the mean value along each column. Set the preprocessed DataFrames to `imputed_X_train` and `imputed_X_valid`. Make sure that the column names match those in `X_train` and `X_valid`.",
"_____no_output_____"
]
],
[
[
"from sklearn.impute import SimpleImputer\n\n# imputation\nmy_imputer = SimpleImputer()\n\nimputed_X_train = pd.DataFrame(my_imputer.fit_transform(X_train))\n\nimputed_X_valid = pd.DataFrame(my_imputer.transform(X_valid))\n\n\n# imputation removed column names; put them back\nimputed_X_train.columns = X_train.columns\nimputed_X_valid.columns = X_valid.columns\n",
"_____no_output_____"
]
],
[
[
"One can use Mean, Median, Mode or Constant value to replace the missing data. Remeber I told you that our strategy for numerical and categorical attribute will be different. Observe yourself, if coloumn have categorical data like high, medium,low one can not use mean.",
"_____no_output_____"
],
[
"Run the next code cell without changes to obtain the MAE for this approach.",
"_____no_output_____"
]
],
[
[
"print(\"MAE (Imputation):\")\nprint(score_dataset(imputed_X_train, imputed_X_valid, y_train, y_valid))",
"MAE (Imputation):\n18250.608013698627\n"
]
],
[
[
"## 3. Multivariate Imputation\n###### Fancy words for predicting missing values with non-missing subset of data!",
"_____no_output_____"
],
[
"Multivariate imputation algorithms use the entire set of available feature dimensions to estimate the missing values (e.g. impute.IterativeImputer).",
"_____no_output_____"
]
],
[
[
"my_imputer = IterativeImputer(BayesianRidge())\n\n# \nimputed_X_train = pd.DataFrame(my_imputer.fit_transform(X_train))\n\nimputed_X_valid = pd.DataFrame(my_imputer.transform(X_valid))\n\n\n# imputation removed column names; put them back\nimputed_X_train.columns = X_train.columns\nimputed_X_valid.columns = X_valid.columns\n",
"_____no_output_____"
],
[
"print(\"MAE (Imputation):\")\nprint(score_dataset(imputed_X_train, imputed_X_valid, y_train, y_valid))",
"MAE (Imputation):\n17976.146438356165\n"
]
],
[
[
"##### Other estimators we can use \n#### BayesianRidge: regularized linear regression - default\n\n#### DecisionTreeRegressor: non-linear regression\n\n#### ExtraTreesRegressor: similar to missForest in R\n\n#### KNeighborsRegressor: comparable to other KNN imputation approaches",
"_____no_output_____"
],
[
"## 4. Nearest neighbors imputation\n###### Fancy word for KNN to select constant value for imputation\nNOTE: IterativeImputer KNeighbourNRegressor is different from this. ",
"_____no_output_____"
]
],
[
[
"my_imputer = KNNImputer(n_neighbors=2, weights=\"uniform\")\n\n\nimputed_X_train = pd.DataFrame(my_imputer.fit_transform(X_train))\n\nimputed_X_valid = pd.DataFrame(my_imputer.transform(X_valid))\n\n\n# imputation removed column names; put them back\nimputed_X_train.columns = X_train.columns\nimputed_X_valid.columns = X_valid.columns",
"_____no_output_____"
],
[
"print(\"MAE (Imputation):\")\nprint(score_dataset(imputed_X_train, imputed_X_valid, y_train, y_valid))",
"MAE (Imputation):\n18095.66794520548\n"
]
],
[
[
" ## 5. An Extension To Imputation\nImputation is the standard approach, and it usually works well. However, imputed values may by systematically above or below their actual values (which weren't collected in the dataset). Or rows with missing values may be unique in some other way. In that case, your model would make better predictions by considering which values were originally missing. Here's how it might look:",
"_____no_output_____"
]
],
[
[
"imputed_X_train_plus = X_train.copy()\nimputed_X_test_plus = X_valid.copy()\n\ncols_with_missing = (col for col in X_train.columns \n if X_train[col].isnull().any())\nfor col in cols_with_missing:\n imputed_X_train_plus[col + '_was_missing'] = imputed_X_train_plus[col].isnull()\n imputed_X_test_plus[col + '_was_missing'] = imputed_X_test_plus[col].isnull()\n#see what happend to the dataset\nimputed_X_train_plus.head()",
"_____no_output_____"
],
[
"# And now we Imputation\nmy_imputer = SimpleImputer()\nimputed_X_train_plus = my_imputer.fit_transform(imputed_X_train_plus)\nimputed_X_test_plus = my_imputer.transform(imputed_X_test_plus)\n\n",
"_____no_output_____"
]
],
[
[
"Run the next code cell without changes to obtain the MAE for this approach.",
"_____no_output_____"
]
],
[
[
"print(\"Mean Absolute Error from Imputation while Track What Was Imputed:\")\nprint(score_dataset(imputed_X_train_plus, imputed_X_test_plus, y_train, y_valid))",
"Mean Absolute Error from Imputation while Track What Was Imputed:\n18253.31479452055\n"
]
],
[
[
"### This whole can be also very easily done by Marking imputed values\nThe MissingIndicator transformer is useful to transform a dataset into corresponding binary matrix indicating the presence of missing values in the dataset. ",
"_____no_output_____"
]
],
[
[
"my_imputer = IterativeImputer(BayesianRidge())\n\n# \nimputed_X_train = pd.DataFrame(my_imputer.fit_transform(X_train))\n\nimputed_X_valid = pd.DataFrame(my_imputer.transform(X_valid))\n\n\n# imputation removed column names; put them back\nimputed_X_train.columns = X_train.columns\nimputed_X_valid.columns = X_valid.columns",
"_____no_output_____"
]
],
[
[
"## Step 3: Devising Ultimate Strategy ",
"_____no_output_____"
],
[
"### Comparing All Solutions\nIt is discussed in the [article](https://medium.com/@aryanc55/51b30be6eee8?source=friends_link&sk=493b75a2c315bcabd8a5a7f11f6ee68f)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecec856abb767cd1a48df4d50ca4e0ba5fcad832 | 88,654 | ipynb | Jupyter Notebook | model4/lgb_bystore_model4.ipynb | QingweiMeng1234/Kaggle_M5_Accuracy_Report | 2409b57271fcf6894852cdb388a892434bf32624 | [
"MIT"
] | null | null | null | model4/lgb_bystore_model4.ipynb | QingweiMeng1234/Kaggle_M5_Accuracy_Report | 2409b57271fcf6894852cdb388a892434bf32624 | [
"MIT"
] | null | null | null | model4/lgb_bystore_model4.ipynb | QingweiMeng1234/Kaggle_M5_Accuracy_Report | 2409b57271fcf6894852cdb388a892434bf32624 | [
"MIT"
] | null | null | null | 48.630828 | 1,865 | 0.510479 | [
[
[
"# General imports\nimport numpy as np\nimport pandas as pd\nimport os, sys, gc, time, warnings, pickle, psutil, random\n\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"# :seed to make all processes deterministic # type: int\ndef seed_everything(seed=0):\n random.seed(seed)\n np.random.seed(seed)",
"_____no_output_____"
],
[
"# Read data\ndef get_data_by_store(store):\n \n # Read and contact basic feature\n df = pd.concat([pd.read_pickle(BASE),\n pd.read_pickle(PRICE).iloc[:,2:],\n pd.read_pickle(CALENDAR).iloc[:,2:]],\n axis=1)\n \n # Leave only relevant store\n df = df[df['store_id']==store]\n\n # With memory limits we have to read \n # lags and mean encoding features\n # separately and drop items that we don't need.\n # As our Features Grids are aligned \n # we can use index to keep only necessary rows\n # Alignment is good for us as concat uses less memory than merge.\n df2 = pd.read_pickle(MEAN_ENC)[mean_features]\n df2 = df2[df2.index.isin(df.index)]\n \n df3 = pd.read_pickle(LAGS).iloc[:,3:]\n df3 = df3[df3.index.isin(df.index)]\n \n df = pd.concat([df, df2], axis=1)\n del df2 # to not reach memory limit \n \n df = pd.concat([df, df3], axis=1)\n del df3 # to not reach memory limit \n \n if store_id in ['CA_1', 'CA_2', 'CA_3','CA_4','TX_1','TX_2','TX_3']:\n remove_features = ['id','state_id','store_id','date','wm_yr_wk','d',TARGET,'cluster','snow_m']\n else:\n remove_features = ['id','state_id','store_id','date','wm_yr_wk','d',TARGET,'cluster']\n \n # Create features list\n features = [col for col in list(df) if col not in remove_features]\n df = df[['id','d',TARGET]+features]\n \n # Skipping first n rows\n df = df[df['d']>=START_TRAIN].reset_index(drop=True)\n \n return df, features\n\n# Recombine Test set after training\ndef get_base_test():\n base_test = pd.DataFrame()\n\n for store_id in STORES_IDS:\n temp_df = pd.read_pickle('test_'+store_id+str(VER)+'.pkl')\n temp_df['store_id'] = store_id\n base_test = pd.concat([base_test, temp_df]).reset_index(drop=True)\n \n return base_test\n\n\n########################### Helper to make dynamic rolling lags\n#################################################################################\ndef make_lag(LAG_DAY):\n lag_df = base_test[['id','d',TARGET]]\n col_name = 'sales_lag_'+str(LAG_DAY)\n lag_df[col_name] = lag_df.groupby(['id'])[TARGET].transform(lambda x: x.shift(LAG_DAY)).astype(np.float16)\n return lag_df[[col_name]]",
"_____no_output_____"
],
[
"def make_lag_roll(LAG_DAY,lag_df_new):\n \n lag_df = base_test[['id','d',TARGET]]\n \n lag_df=lag_df.sort_values(by=[\"d\"])\n \n for i in range(0,len(LAG_DAY)):\n\n shift_day = LAG_DAY[i][0]\n roll_wind = LAG_DAY[i][1]\n col_name = 'rolling_mean_tmp_'+str(shift_day)+'_'+str(roll_wind)\n lag_df[col_name] = (lag_df.groupby(['id'])[TARGET]).transform(lambda x: x.shift(shift_day).rolling(roll_wind).mean())\n lag_df_new=lag_df.drop(columns=[\"sales\"])\n return lag_df_new",
"_____no_output_____"
],
[
"import lightgbm as lgb\nlgb_params = {\n 'boosting_type': 'gbdt',\n 'objective': 'tweedie',\n 'tweedie_variance_power': 1.1,\n 'metric': 'rmse',\n 'subsample': 0.5,\n 'subsample_freq': 1,\n 'learning_rate': 0.03,\n \"lambda\":0.1,\n 'num_leaves': 2**11-1,\n 'min_data_in_leaf': 2**12-1,\n 'feature_fraction': 0.5,\n 'max_bin': 100,\n 'n_estimators': 1400,\n 'boost_from_average': False,\n 'verbose': -1,\n } \n\n\n\n# lgb_params ={\n# \"objective\" : \"tweedie\",\n# \"metric\" :\"rmse\",\n# \"force_row_wise\" : True,\n# \"learning_rate\" : 0.075,\n# \"sub_feature\" : 0.8,\n# \"sub_row\" : 0.75,\n# \"bagging_freq\" : 1,\n# \"lambda_l2\" : 0.1,\n# \"metric\": [\"rmse\"],\n# \"nthread\": -1,\n# \"tweedie_variance_power\":1.1,\n# 'verbosity': 1,\n# # 'num_iterations' : 1500,\n# 'num_leaves': 128,\n# \"min_data_in_leaf\": 104,\n# }\n\n\n\n\n# Let's look closer on params\n\n## 'boosting_type': 'gbdt'\n# we have 'goss' option for faster training\n# but it normally leads to underfit.\n# Also there is good 'dart' mode\n# but it takes forever to train\n# and model performance depends \n# a lot on random factor \n# https://www.kaggle.com/c/home-credit-default-risk/discussion/60921\n\n## 'objective': 'tweedie'\n# Tweedie Gradient Boosting for Extremely\n# Unbalanced Zero-inflated Data\n# https://arxiv.org/pdf/1811.10192.pdf\n# and many more articles about tweediie\n#\n# Strange (for me) but Tweedie is close in results\n# to my own ugly loss.\n# My advice here - make OWN LOSS function\n# https://www.kaggle.com/c/m5-forecasting-accuracy/discussion/140564\n# https://www.kaggle.com/c/m5-forecasting-accuracy/discussion/143070\n# I think many of you already using it (after poisson kernel appeared) \n# (kagglers are very good with \"params\" testing and tuning).\n# Try to figure out why Tweedie works.\n# probably it will show you new features options\n# or data transformation (Target transformation?).\n\n## 'tweedie_variance_power': 1.1\n# default = 1.5\n# set this closer to 2 to shift towards a Gamma distribution\n# set this closer to 1 to shift towards a Poisson distribution\n# my CV shows 1.1 is optimal \n# but you can make your own choice\n\n## 'metric': 'rmse'\n# Doesn't mean anything to us\n# as competition metric is different\n# and we don't use early stoppings here.\n# So rmse serves just for general \n# model performance overview.\n# Also we use \"fake\" validation set\n# (as it makes part of the training set)\n# so even general rmse score doesn't mean anything))\n# https://www.kaggle.com/c/m5-forecasting-accuracy/discussion/133834\n\n## 'subsample': 0.5\n# Serves to fight with overfit\n# this will randomly select part of data without resampling\n# Chosen by CV (my CV can be wrong!)\n# Next kernel will be about CV\n\n##'subsample_freq': 1\n# frequency for bagging\n# default value - seems ok\n\n## 'learning_rate': 0.03\n# Chosen by CV\n# Smaller - longer training\n# but there is an option to stop \n# in \"local minimum\"\n# Bigger - faster training\n# but there is a chance to\n# not find \"global minimum\" minimum\n\n## 'num_leaves': 2**11-1\n## 'min_data_in_leaf': 2**12-1\n# Force model to use more features\n# We need it to reduce \"recursive\"\n# error impact.\n# Also it leads to overfit\n# that's why we use small \n\n# 'max_bin': 100\n## l1, l2 regularizations\n# https://towardsdatascience.com/l1-and-l2-regularization-methods-ce25e7fc831c\n# Good tiny explanation\n# l2 can work with bigger num_leaves\n# but my CV doesn't show boost\n \n## 'n_estimators': 1400\n# CV shows that there should be\n# different values for each state/store.\n# Current value was chosen \n# for general purpose.\n# As we don't use any early stopings\n# careful to not overfit Public LB.\n\n##'feature_fraction': 0.5\n# LightGBM will randomly select \n# part of features on each iteration (tree).\n# We have maaaany features\n# and many of them are \"duplicates\"\n# and many just \"noise\"\n# good values here - 0.5-0.7 (by CV)\n\n## 'boost_from_average': False\n# There is some \"problem\"\n# to code boost_from_average for \n# custom loss\n# 'True' makes training faster\n# BUT carefull use it\n# https://github.com/microsoft/LightGBM/issues/1514",
"_____no_output_____"
],
[
"VER = 7 # Our model version\nSEED = 42 # We want all things\nseed_everything(SEED) # to be as deterministic \nlgb_params['seed'] = SEED # as possible\nN_CORES = psutil.cpu_count() # Available CPU cores\n\n\n#LIMITS and const\nTARGET = 'sales' # Our target\nSTART_TRAIN = 0 # We can skip some rows (Nans/faster training)\nEND_TRAIN = 1941 # End day of our train set, change this part for final\nP_HORIZON = 28 # Prediction horizon\n\n#FEATURES to remove\n## These features lead to overfit\n## or values not present in test set\nmean_features = ['enc_cat_id_mean','enc_cat_id_std',\n 'enc_dept_id_mean','enc_dept_id_std',\n 'enc_item_id_mean','enc_item_id_std'] \n\n#PATHS for Features\nBASE = 'grid_part_1.pkl'\nPRICE = 'grid_part_2.pkl'\nCALENDAR = 'grid_part_3.pkl'\nLAGS = 'lags_df_28_v3.pkl'\nMEAN_ENC = 'mean_encoding_df.pkl'\n\n\n# AUX(pretrained) Models paths\n\n#STORES ids\nSTORES_IDS = pd.read_csv('sales_train_evaluation.csv')['store_id']#change this part for final\nSTORES_IDS = list(STORES_IDS.unique())\n\n#SPLITS for lags creation\nSHIFT_DAY = 28\nN_LAGS = 15\nLAGS_SPLIT = [col for col in range(SHIFT_DAY,SHIFT_DAY+N_LAGS)]\nROLS_SPLIT = []\nfor i in [1,7,14]:\n for j in [7,14,28,56]:\n ROLS_SPLIT.append([i,j])",
"_____no_output_____"
],
[
"for store_id in STORES_IDS:\n print('Train', store_id)\n \n # Get grid for current store\n grid_df, features_columns = get_data_by_store(store_id)\n \n print(features_columns)\n # Masks for \n # Train (All data less than 1913)\n # \"Validation\" (Last 28 days - not real validation set)\n # Test (All data greater than 1913 day, \n # with some gap for recursive features)\n train_mask = grid_df['d']<=END_TRAIN\n valid_mask = train_mask&(grid_df['d']>(END_TRAIN-P_HORIZON))\n preds_mask = grid_df['d']>(END_TRAIN-100)\n \n # Apply masks and save lgb dataset as bin\n # to reduce memory spikes during dtype convertations\n # https://github.com/Microsoft/LightGBM/issues/1032\n # \"To avoid any conversions, you should always use np.float32\"\n # or save to bin before start training\n # https://www.kaggle.com/c/talkingdata-adtracking-fraud-detection/discussion/53773\n train_data = lgb.Dataset(grid_df[train_mask][features_columns], \n label=grid_df[train_mask][TARGET])\n \n \n valid_data = lgb.Dataset(grid_df[valid_mask][features_columns], \n label=grid_df[valid_mask][TARGET])\n \n # Saving part of the dataset for later predictions\n # Removing features that we need to calculate recursively \n grid_df = grid_df[preds_mask].reset_index(drop=True)\n keep_cols = [col for col in list(grid_df) if '_tmp_' not in col]\n grid_df = grid_df[keep_cols]\n grid_df.to_pickle('test_'+store_id+str(VER)+'.pkl')\n del grid_df\n gc.collect()\n \n # Launch seeder again to make lgb training 100% deterministic\n # with each \"code line\" np.random \"evolves\" \n # so we need (may want) to \"reset\" it\n seed_everything(SEED)\n estimator = lgb.train(lgb_params,\n train_data,\n valid_sets = [valid_data],\n verbose_eval = 100,\n )\n imp_type = \"gain\"\n features = estimator.feature_name()\n importances = estimator.feature_importance(imp_type)\n importance_df=pd.DataFrame(features,columns=['features'])\n importance_df['importances']=importances\n importance_df=importance_df.sort_values(by='importances', ascending=False)\n importance_df.to_csv(store_id+'_fe_imp_'+str(VER)+'.csv',index=False)\n del importance_df\n gc.collect()\n \n # Save model - it's not real '.bin' but a pickle file\n # estimator = lgb.Booster(model_file='model.txt')\n # can only predict with the best iteration (or the saving iteration)\n # pickle.dump gives us more flexibility\n # like estimator.predict(TEST, num_iteration=100)\n # num_iteration - number of iteration want to predict with, \n # NULL or <= 0 means use best iteration\n model_name = 'lgb_model_'+store_id+'_v'+str(VER)+'.bin'\n pickle.dump(estimator, open(model_name, 'wb'))\n\n # Remove temporary files and objects \n # to free some hdd space and ram memory\n # !rm train_data.bin\n del train_data, valid_data, estimator\n gc.collect()",
"Train CA_1\n['item_id', 'dept_id', 'cat_id', 'release', 'sell_price', 'price_max', 'price_min', 'price_std', 'price_mean', 'price_norm', 'price_rank_dept', 'price_nunique', 'item_nunique', 'price_momentum', 'price_momentum_m', 'price_momentum_y', 'temperature_high', 'temperature_con', 'rainfall_m', 'event_name_1', 'event_type_1', 'event_name_2', 'event_type_2', 'snap_CA', 'snap_TX', 'snap_WI', 'is_first_half_month', 'event_bef_weekend', 'event_after_weekend', 'NBA', 'event_attention_after', 'event_attention_bef', 'event_attention_sum', 'tm_d', 'tm_w', 'tm_m', 'tm_q', 'tm_y', 'tm_wm', 'tm_dw', 'tm_w_end', 'enc_cat_id_mean', 'enc_cat_id_std', 'enc_dept_id_mean', 'enc_dept_id_std', 'enc_item_id_mean', 'enc_item_id_std', 'sales_lag_28', 'sales_lag_29', 'sales_lag_30', 'sales_lag_31', 'sales_lag_32', 'sales_lag_33', 'sales_lag_34', 'sales_lag_35', 'sales_lag_36', 'sales_lag_37', 'sales_lag_38', 'sales_lag_39', 'sales_lag_40', 'sales_lag_41', 'sales_lag_42', 'rolling_mean_7', 'rolling_std_7', 'rolling_mean_14', 'rolling_std_14', 'rolling_mean_28', 'rolling_std_28', 'rolling_mean_56', 'rolling_std_56', 'rolling_mean_168', 'rolling_std_168', 'rolling_mean_tmp_1_7', 'rolling_mean_tmp_1_14', 'rolling_mean_tmp_1_28', 'rolling_mean_tmp_1_56', 'rolling_mean_tmp_7_7', 'rolling_mean_tmp_7_14', 'rolling_mean_tmp_7_28', 'rolling_mean_tmp_7_56', 'rolling_mean_tmp_14_7', 'rolling_mean_tmp_14_14', 'rolling_mean_tmp_14_28', 'rolling_mean_tmp_14_56', 'rolling_quantile_97_28', 'rolling_quantile_87.5_28', 'rolling_quantile_50_28', 'rolling_quantile_22.5_28', 'rolling_quantile_3_28', 'rolling_quantile_97_56', 'rolling_quantile_87.5_56', 'rolling_quantile_50_56', 'rolling_quantile_22.5_56', 'rolling_quantile_3_56', 'rolling_quantile_97_168', 'rolling_quantile_87.5_168', 'rolling_quantile_50_168', 'rolling_quantile_22.5_168', 'rolling_quantile_3_168']\n[100]\tvalid_0's rmse: 2.0187\n[200]\tvalid_0's rmse: 1.99223\n[300]\tvalid_0's rmse: 1.97922\n[400]\tvalid_0's rmse: 1.97017\n[500]\tvalid_0's rmse: 1.96347\n[600]\tvalid_0's rmse: 1.95705\n[700]\tvalid_0's rmse: 1.95223\n[800]\tvalid_0's rmse: 1.94645\n[900]\tvalid_0's rmse: 1.94099\n[1000]\tvalid_0's rmse: 1.93602\n[1100]\tvalid_0's rmse: 1.9307\n[1200]\tvalid_0's rmse: 1.92662\n[1300]\tvalid_0's rmse: 1.9223\n[1400]\tvalid_0's rmse: 1.91772\nTrain CA_2\n['item_id', 'dept_id', 'cat_id', 'release', 'sell_price', 'price_max', 'price_min', 'price_std', 'price_mean', 'price_norm', 'price_rank_dept', 'price_nunique', 'item_nunique', 'price_momentum', 'price_momentum_m', 'price_momentum_y', 'temperature_high', 'temperature_con', 'rainfall_m', 'event_name_1', 'event_type_1', 'event_name_2', 'event_type_2', 'snap_CA', 'snap_TX', 'snap_WI', 'is_first_half_month', 'event_bef_weekend', 'event_after_weekend', 'NBA', 'event_attention_after', 'event_attention_bef', 'event_attention_sum', 'tm_d', 'tm_w', 'tm_m', 'tm_q', 'tm_y', 'tm_wm', 'tm_dw', 'tm_w_end', 'enc_cat_id_mean', 'enc_cat_id_std', 'enc_dept_id_mean', 'enc_dept_id_std', 'enc_item_id_mean', 'enc_item_id_std', 'sales_lag_28', 'sales_lag_29', 'sales_lag_30', 'sales_lag_31', 'sales_lag_32', 'sales_lag_33', 'sales_lag_34', 'sales_lag_35', 'sales_lag_36', 'sales_lag_37', 'sales_lag_38', 'sales_lag_39', 'sales_lag_40', 'sales_lag_41', 'sales_lag_42', 'rolling_mean_7', 'rolling_std_7', 'rolling_mean_14', 'rolling_std_14', 'rolling_mean_28', 'rolling_std_28', 'rolling_mean_56', 'rolling_std_56', 'rolling_mean_168', 'rolling_std_168', 'rolling_mean_tmp_1_7', 'rolling_mean_tmp_1_14', 'rolling_mean_tmp_1_28', 'rolling_mean_tmp_1_56', 'rolling_mean_tmp_7_7', 'rolling_mean_tmp_7_14', 'rolling_mean_tmp_7_28', 'rolling_mean_tmp_7_56', 'rolling_mean_tmp_14_7', 'rolling_mean_tmp_14_14', 'rolling_mean_tmp_14_28', 'rolling_mean_tmp_14_56', 'rolling_quantile_97_28', 'rolling_quantile_87.5_28', 'rolling_quantile_50_28', 'rolling_quantile_22.5_28', 'rolling_quantile_3_28', 'rolling_quantile_97_56', 'rolling_quantile_87.5_56', 'rolling_quantile_50_56', 'rolling_quantile_22.5_56', 'rolling_quantile_3_56', 'rolling_quantile_97_168', 'rolling_quantile_87.5_168', 'rolling_quantile_50_168', 'rolling_quantile_22.5_168', 'rolling_quantile_3_168']\n[100]\tvalid_0's rmse: 1.95074\n[200]\tvalid_0's rmse: 1.89502\n[300]\tvalid_0's rmse: 1.87912\n[400]\tvalid_0's rmse: 1.87002\n[500]\tvalid_0's rmse: 1.8622\n[600]\tvalid_0's rmse: 1.85562\n[700]\tvalid_0's rmse: 1.84979\n[800]\tvalid_0's rmse: 1.84416\n[900]\tvalid_0's rmse: 1.83877\n[1000]\tvalid_0's rmse: 1.83345\n[1100]\tvalid_0's rmse: 1.82851\n[1200]\tvalid_0's rmse: 1.82346\n[1300]\tvalid_0's rmse: 1.81915\n[1400]\tvalid_0's rmse: 1.8149\nTrain CA_3\n['item_id', 'dept_id', 'cat_id', 'release', 'sell_price', 'price_max', 'price_min', 'price_std', 'price_mean', 'price_norm', 'price_rank_dept', 'price_nunique', 'item_nunique', 'price_momentum', 'price_momentum_m', 'price_momentum_y', 'temperature_high', 'temperature_con', 'rainfall_m', 'event_name_1', 'event_type_1', 'event_name_2', 'event_type_2', 'snap_CA', 'snap_TX', 'snap_WI', 'is_first_half_month', 'event_bef_weekend', 'event_after_weekend', 'NBA', 'event_attention_after', 'event_attention_bef', 'event_attention_sum', 'tm_d', 'tm_w', 'tm_m', 'tm_q', 'tm_y', 'tm_wm', 'tm_dw', 'tm_w_end', 'enc_cat_id_mean', 'enc_cat_id_std', 'enc_dept_id_mean', 'enc_dept_id_std', 'enc_item_id_mean', 'enc_item_id_std', 'sales_lag_28', 'sales_lag_29', 'sales_lag_30', 'sales_lag_31', 'sales_lag_32', 'sales_lag_33', 'sales_lag_34', 'sales_lag_35', 'sales_lag_36', 'sales_lag_37', 'sales_lag_38', 'sales_lag_39', 'sales_lag_40', 'sales_lag_41', 'sales_lag_42', 'rolling_mean_7', 'rolling_std_7', 'rolling_mean_14', 'rolling_std_14', 'rolling_mean_28', 'rolling_std_28', 'rolling_mean_56', 'rolling_std_56', 'rolling_mean_168', 'rolling_std_168', 'rolling_mean_tmp_1_7', 'rolling_mean_tmp_1_14', 'rolling_mean_tmp_1_28', 'rolling_mean_tmp_1_56', 'rolling_mean_tmp_7_7', 'rolling_mean_tmp_7_14', 'rolling_mean_tmp_7_28', 'rolling_mean_tmp_7_56', 'rolling_mean_tmp_14_7', 'rolling_mean_tmp_14_14', 'rolling_mean_tmp_14_28', 'rolling_mean_tmp_14_56', 'rolling_quantile_97_28', 'rolling_quantile_87.5_28', 'rolling_quantile_50_28', 'rolling_quantile_22.5_28', 'rolling_quantile_3_28', 'rolling_quantile_97_56', 'rolling_quantile_87.5_56', 'rolling_quantile_50_56', 'rolling_quantile_22.5_56', 'rolling_quantile_3_56', 'rolling_quantile_97_168', 'rolling_quantile_87.5_168', 'rolling_quantile_50_168', 'rolling_quantile_22.5_168', 'rolling_quantile_3_168']\n[100]\tvalid_0's rmse: 2.38494\n[200]\tvalid_0's rmse: 2.35115\n[300]\tvalid_0's rmse: 2.32814\n[400]\tvalid_0's rmse: 2.31258\n[500]\tvalid_0's rmse: 2.30208\n[600]\tvalid_0's rmse: 2.29496\n[700]\tvalid_0's rmse: 2.28906\n[800]\tvalid_0's rmse: 2.28341\n[900]\tvalid_0's rmse: 2.27768\n[1000]\tvalid_0's rmse: 2.27181\n[1100]\tvalid_0's rmse: 2.26639\n[1200]\tvalid_0's rmse: 2.26101\n[1300]\tvalid_0's rmse: 2.25602\n[1400]\tvalid_0's rmse: 2.25117\nTrain CA_4\n['item_id', 'dept_id', 'cat_id', 'release', 'sell_price', 'price_max', 'price_min', 'price_std', 'price_mean', 'price_norm', 'price_rank_dept', 'price_nunique', 'item_nunique', 'price_momentum', 'price_momentum_m', 'price_momentum_y', 'temperature_high', 'temperature_con', 'rainfall_m', 'event_name_1', 'event_type_1', 'event_name_2', 'event_type_2', 'snap_CA', 'snap_TX', 'snap_WI', 'is_first_half_month', 'event_bef_weekend', 'event_after_weekend', 'NBA', 'event_attention_after', 'event_attention_bef', 'event_attention_sum', 'tm_d', 'tm_w', 'tm_m', 'tm_q', 'tm_y', 'tm_wm', 'tm_dw', 'tm_w_end', 'enc_cat_id_mean', 'enc_cat_id_std', 'enc_dept_id_mean', 'enc_dept_id_std', 'enc_item_id_mean', 'enc_item_id_std', 'sales_lag_28', 'sales_lag_29', 'sales_lag_30', 'sales_lag_31', 'sales_lag_32', 'sales_lag_33', 'sales_lag_34', 'sales_lag_35', 'sales_lag_36', 'sales_lag_37', 'sales_lag_38', 'sales_lag_39', 'sales_lag_40', 'sales_lag_41', 'sales_lag_42', 'rolling_mean_7', 'rolling_std_7', 'rolling_mean_14', 'rolling_std_14', 'rolling_mean_28', 'rolling_std_28', 'rolling_mean_56', 'rolling_std_56', 'rolling_mean_168', 'rolling_std_168', 'rolling_mean_tmp_1_7', 'rolling_mean_tmp_1_14', 'rolling_mean_tmp_1_28', 'rolling_mean_tmp_1_56', 'rolling_mean_tmp_7_7', 'rolling_mean_tmp_7_14', 'rolling_mean_tmp_7_28', 'rolling_mean_tmp_7_56', 'rolling_mean_tmp_14_7', 'rolling_mean_tmp_14_14', 'rolling_mean_tmp_14_28', 'rolling_mean_tmp_14_56', 'rolling_quantile_97_28', 'rolling_quantile_87.5_28', 'rolling_quantile_50_28', 'rolling_quantile_22.5_28', 'rolling_quantile_3_28', 'rolling_quantile_97_56', 'rolling_quantile_87.5_56', 'rolling_quantile_50_56', 'rolling_quantile_22.5_56', 'rolling_quantile_3_56', 'rolling_quantile_97_168', 'rolling_quantile_87.5_168', 'rolling_quantile_50_168', 'rolling_quantile_22.5_168', 'rolling_quantile_3_168']\n"
],
[
"# Create Dummy DataFrame to store predictions\nall_preds = pd.DataFrame()\n\n# Join back the Test dataset with \n# a small part of the training data \n# to make recursive features\nbase_test = get_base_test()\n\n# Timer to measure predictions time \nmain_time = time.time()\n\n# Loop over each prediction day\n# As rolling lags are the most timeconsuming\n# we will calculate it for whole day\n\n\nfor PREDICT_DAY in range(1,29): \n print('Predict | Day:', PREDICT_DAY)\n start_time = time.time()\n\n # Make temporary grid to calculate rolling lags\n grid_df = base_test.copy()\n \n \n lag_df_new = pd.DataFrame()\n\n lag_df_new=make_lag_roll(ROLS_SPLIT,lag_df_new)\n\n\n grid_df = grid_df.merge(lag_df_new, on=['id','d'], how='left')\n\n\n for store_id in STORES_IDS:\n \n if store_id in ['CA_1', 'CA_2', 'CA_3','CA_4','TX_1','TX_2','TX_3']:\n MODEL_FEATURES = ['item_id', 'dept_id', 'cat_id', 'release', 'sell_price', 'price_max', 'price_min',\n 'price_std', 'price_mean', 'price_norm', 'price_rank_dept', 'price_nunique', \n 'item_nunique', 'price_momentum', 'price_momentum_m', 'price_momentum_y', \n 'temperature_high', 'temperature_con', 'rainfall_m', 'event_name_1', \n 'event_type_1', 'event_name_2', 'event_type_2', 'snap_CA', 'snap_TX', \n 'snap_WI', 'is_first_half_month', 'event_bef_weekend', 'event_after_weekend', \n 'NBA', 'event_attention_after', 'event_attention_bef', 'event_attention_sum', \n 'tm_d', 'tm_w', 'tm_m', 'tm_q', 'tm_y', 'tm_wm', 'tm_dw', 'tm_w_end', 'enc_cat_id_mean',\n 'enc_cat_id_std', 'enc_dept_id_mean', 'enc_dept_id_std', 'enc_item_id_mean', 'enc_item_id_std',\n 'sales_lag_28', 'sales_lag_29', 'sales_lag_30', 'sales_lag_31', 'sales_lag_32', 'sales_lag_33', \n 'sales_lag_34', 'sales_lag_35', 'sales_lag_36', 'sales_lag_37', 'sales_lag_38', 'sales_lag_39',\n 'sales_lag_40', 'sales_lag_41', 'sales_lag_42', 'rolling_mean_7', 'rolling_std_7', 'rolling_mean_14', \n 'rolling_std_14', 'rolling_mean_28', 'rolling_std_28', 'rolling_mean_56', 'rolling_std_56', 'rolling_mean_168',\n 'rolling_std_168', 'rolling_mean_tmp_1_7', 'rolling_mean_tmp_1_14', 'rolling_mean_tmp_1_28', 'rolling_mean_tmp_1_56', \n 'rolling_mean_tmp_7_7', 'rolling_mean_tmp_7_14', 'rolling_mean_tmp_7_28', 'rolling_mean_tmp_7_56', \n 'rolling_mean_tmp_14_7', 'rolling_mean_tmp_14_14', 'rolling_mean_tmp_14_28', 'rolling_mean_tmp_14_56',\n 'rolling_quantile_97_28', 'rolling_quantile_87.5_28', 'rolling_quantile_50_28', \n 'rolling_quantile_22.5_28', 'rolling_quantile_3_28', 'rolling_quantile_97_56', \n 'rolling_quantile_87.5_56', 'rolling_quantile_50_56', 'rolling_quantile_22.5_56',\n 'rolling_quantile_3_56', 'rolling_quantile_97_168', 'rolling_quantile_87.5_168', \n 'rolling_quantile_50_168', 'rolling_quantile_22.5_168', 'rolling_quantile_3_168']\n else:\n MODEL_FEATURES = ['item_id', 'dept_id', 'cat_id', 'release', 'sell_price', 'price_max', 'price_min',\n 'price_std', 'price_mean', 'price_norm', 'price_rank_dept', 'price_nunique', \n 'item_nunique', 'price_momentum', 'price_momentum_m', 'price_momentum_y', \n 'temperature_high', 'temperature_con', 'rainfall_m', 'snow_m','event_name_1', \n 'event_type_1', 'event_name_2', 'event_type_2', 'snap_CA', 'snap_TX', \n 'snap_WI', 'is_first_half_month', 'event_bef_weekend', 'event_after_weekend', \n 'NBA', 'event_attention_after', 'event_attention_bef', 'event_attention_sum', \n 'tm_d', 'tm_w', 'tm_m', 'tm_q', 'tm_y', 'tm_wm', 'tm_dw', 'tm_w_end', 'enc_cat_id_mean',\n 'enc_cat_id_std', 'enc_dept_id_mean', 'enc_dept_id_std', 'enc_item_id_mean', 'enc_item_id_std',\n 'sales_lag_28', 'sales_lag_29', 'sales_lag_30', 'sales_lag_31', 'sales_lag_32', 'sales_lag_33', \n 'sales_lag_34', 'sales_lag_35', 'sales_lag_36', 'sales_lag_37', 'sales_lag_38', 'sales_lag_39',\n 'sales_lag_40', 'sales_lag_41', 'sales_lag_42', 'rolling_mean_7', 'rolling_std_7', 'rolling_mean_14', \n 'rolling_std_14', 'rolling_mean_28', 'rolling_std_28', 'rolling_mean_56', 'rolling_std_56', 'rolling_mean_168',\n 'rolling_std_168', 'rolling_mean_tmp_1_7', 'rolling_mean_tmp_1_14', 'rolling_mean_tmp_1_28', 'rolling_mean_tmp_1_56', \n 'rolling_mean_tmp_7_7', 'rolling_mean_tmp_7_14', 'rolling_mean_tmp_7_28', 'rolling_mean_tmp_7_56', \n 'rolling_mean_tmp_14_7', 'rolling_mean_tmp_14_14', 'rolling_mean_tmp_14_28', 'rolling_mean_tmp_14_56',\n 'rolling_quantile_97_28', 'rolling_quantile_87.5_28', 'rolling_quantile_50_28', \n 'rolling_quantile_22.5_28', 'rolling_quantile_3_28', 'rolling_quantile_97_56', \n 'rolling_quantile_87.5_56', 'rolling_quantile_50_56', 'rolling_quantile_22.5_56',\n 'rolling_quantile_3_56', 'rolling_quantile_97_168', 'rolling_quantile_87.5_168', \n 'rolling_quantile_50_168', 'rolling_quantile_22.5_168', 'rolling_quantile_3_168']\n\n # Read all our models and make predictions\n # for each day/store pairs\n model_path = 'lgb_model_'+store_id+'_v'+str(VER)+'.bin' \n\n estimator = pickle.load(open(model_path, 'rb'))\n\n day_mask = base_test['d']==(END_TRAIN+PREDICT_DAY)\n store_mask = base_test['store_id']==store_id\n\n mask = (day_mask)&(store_mask)\n base_test[TARGET][mask] = estimator.predict(grid_df[mask][MODEL_FEATURES])\n\n # Make good column naming and add \n # to all_preds DataFrame\n temp_df = base_test[day_mask][['id',TARGET]]\n temp_df.columns = ['id','F'+str(PREDICT_DAY)]\n if 'id' in list(all_preds):\n all_preds = all_preds.merge(temp_df, on=['id'], how='left')\n else:\n all_preds = temp_df.copy()\n\n print('#'*10, ' %0.2f min round |' % ((time.time() - start_time) / 60),\n ' %0.2f min total |' % ((time.time() - main_time) / 60),\n ' %0.2f day sales |' % (temp_df['F'+str(PREDICT_DAY)].sum()))\n \n del temp_df, lag_df_new\n\nall_preds = all_preds.reset_index(drop=True)\nall_preds.head()",
"Predict | Day: 1\n########## 5.09 min round | 5.09 min total | 39994.80 day sales |\nPredict | Day: 2\n########## 5.27 min round | 10.35 min total | 37474.79 day sales |\nPredict | Day: 3\n########## 5.34 min round | 15.70 min total | 37025.19 day sales |\nPredict | Day: 4\n########## 5.06 min round | 20.75 min total | 37288.91 day sales |\nPredict | Day: 5\n########## 5.03 min round | 25.78 min total | 42811.64 day sales |\nPredict | Day: 6\n########## 5.02 min round | 30.80 min total | 50716.83 day sales |\nPredict | Day: 7\n########## 5.01 min round | 35.81 min total | 51343.54 day sales |\nPredict | Day: 8\n########## 5.01 min round | 40.82 min total | 45544.07 day sales |\nPredict | Day: 9\n########## 4.98 min round | 45.81 min total | 39022.93 day sales |\nPredict | Day: 10\n########## 5.25 min round | 51.06 min total | 44037.07 day sales |\nPredict | Day: 11\n########## 5.26 min round | 56.31 min total | 45627.60 day sales |\nPredict | Day: 12\n########## 5.29 min round | 61.61 min total | 51895.80 day sales |\nPredict | Day: 13\n########## 5.26 min round | 66.86 min total | 55282.09 day sales |\nPredict | Day: 14\n########## 5.00 min round | 71.86 min total | 56754.94 day sales |\nPredict | Day: 15\n########## 5.00 min round | 76.87 min total | 47900.21 day sales |\nPredict | Day: 16\n########## 5.08 min round | 81.95 min total | 43902.18 day sales |\nPredict | Day: 17\n########## 5.02 min round | 86.97 min total | 42706.16 day sales |\nPredict | Day: 18\n########## 5.06 min round | 92.03 min total | 45038.59 day sales |\nPredict | Day: 19\n########## 5.10 min round | 97.13 min total | 46792.58 day sales |\nPredict | Day: 20\n########## 5.04 min round | 102.17 min total | 58730.50 day sales |\nPredict | Day: 21\n########## 4.95 min round | 107.12 min total | 59805.52 day sales |\nPredict | Day: 22\n########## 4.95 min round | 112.07 min total | 46228.82 day sales |\nPredict | Day: 23\n########## 4.98 min round | 117.05 min total | 43256.29 day sales |\nPredict | Day: 24\n########## 5.01 min round | 122.05 min total | 45407.94 day sales |\nPredict | Day: 25\n########## 5.00 min round | 127.05 min total | 41940.34 day sales |\nPredict | Day: 26\n########## 5.00 min round | 132.05 min total | 47005.49 day sales |\nPredict | Day: 27\n########## 5.00 min round | 137.05 min total | 54524.10 day sales |\nPredict | Day: 28\n########## 4.98 min round | 142.03 min total | 50205.18 day sales |\n"
],
[
"# all the following is changed\n# replace validation part\ntrain_df = pd.read_csv('sales_train_evaluation.csv')\ntrain_df=train_df[['id','d_1914','d_1915','d_1916','d_1917','d_1918','d_1919','d_1920','d_1921','d_1922','d_1923',\n 'd_1924','d_1925','d_1926','d_1927','d_1928','d_1929','d_1930','d_1931','d_1932','d_1933',\n 'd_1934','d_1935','d_1936','d_1937','d_1938','d_1939','d_1940','d_1941']]",
"_____no_output_____"
],
[
"submission = pd.read_csv('sample_submission.csv')",
"_____no_output_____"
],
[
"train_df['id']=train_df['id'].str.replace('evaluation','validation')",
"_____no_output_____"
],
[
"train_df.columns=submission.columns",
"_____no_output_____"
],
[
"submission = submission[['id']]\nsub1 = submission.merge(train_df, on=['id'], how='left')",
"_____no_output_____"
],
[
"sub1=sub1[:30490]",
"_____no_output_____"
],
[
"sub2 = submission.merge(all_preds, on=['id'], how='left')",
"_____no_output_____"
],
[
"sub2=sub2[30490:]",
"_____no_output_____"
],
[
"final_sub=pd.concat([sub1,sub2],axis=0)",
"_____no_output_____"
],
[
"final_sub.head()",
"_____no_output_____"
],
[
"final_sub.tail()",
"_____no_output_____"
],
[
"final_sub.describe()",
"_____no_output_____"
],
[
"final_sub.to_csv('lgb_bystore_final4.csv',index=False)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecec8c335e1cc03f7df9f38d3ee040c34047b7c6 | 4,484 | ipynb | Jupyter Notebook | test/Models/energybalance_pkg/test/py/Soilevaporation.ipynb | cyrillemidingoyi/PyCropML | b866cc17374424379142d9162af985c1f87c74b6 | [
"MIT"
] | 5 | 2020-06-21T18:58:04.000Z | 2022-01-29T21:32:28.000Z | test/Models/energybalance_pkg/test/py/Soilevaporation.ipynb | cyrillemidingoyi/PyCropML | b866cc17374424379142d9162af985c1f87c74b6 | [
"MIT"
] | 27 | 2018-12-04T15:35:44.000Z | 2022-03-11T08:25:03.000Z | test/Models/energybalance_pkg/test/py/Soilevaporation.ipynb | cyrillemidingoyi/PyCropML | b866cc17374424379142d9162af985c1f87c74b6 | [
"MIT"
] | 7 | 2019-04-20T02:25:22.000Z | 2021-11-04T07:52:35.000Z | 40.035714 | 120 | 0.455397 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecec8d85f6097cc22ac58363897306d9dba1e593 | 63,939 | ipynb | Jupyter Notebook | code/analysis/Breast_Cancer/Supple_Fig12-Breast cancer.ipynb | WangPeng-Lab/scGCO_0910 | 0a2d89935b9067f5c31f43e1e2649e505800264b | [
"MIT"
] | 7 | 2018-12-19T15:18:07.000Z | 2021-06-22T00:52:22.000Z | code/analysis/Breast_Cancer/Supple_Fig12-Breast cancer.ipynb | WangPeng-Lab/scGCO_0910 | 0a2d89935b9067f5c31f43e1e2649e505800264b | [
"MIT"
] | 1 | 2021-06-28T13:04:58.000Z | 2021-06-29T07:32:22.000Z | code/analysis/Breast_Cancer/Supple_Fig12-Breast cancer.ipynb | WangPeng-Lab/scGCO_0910 | 0a2d89935b9067f5c31f43e1e2649e505800264b | [
"MIT"
] | 7 | 2018-12-06T15:45:47.000Z | 2020-09-14T02:06:38.000Z | 229.172043 | 43,800 | 0.919001 | [
[
[
"## Supplementary Figure 12 of Breast cancer",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\n# !pip install scikit-image\nfrom skimage import data\nfrom skimage.filters import threshold_multiotsu\nfrom skimage.filters import threshold_otsu\nimport sys\nsys.path.append('../../scGCO_code/')\n\n\nfrom scGCO_source import *\n\nimport matplotlib.pyplot as plt\nfrom matplotlib_venn import venn2,venn3",
"D:\\Annconda3\\envs\\spatial\\lib\\site-packages\\pysal\\lib\\weights\\util.py:19: UserWarning: geopandas not available. Some functionality will be disabled.\n warn('geopandas not available. Some functionality will be disabled.')\n"
]
],
[
[
"#### input data as cell x genes, (cell as xxy location)",
"_____no_output_____"
]
],
[
[
"from ast import literal_eval\nfrom matplotlib_venn import venn2,venn3\n\nfig,axis=plt.subplots(1,4,figsize=(15,3))\nfig.subplots_adjust(hspace=0.3, wspace=0.3) #,\n #top=0.925, right=0.925, bottom=0.075, left = 0.075)\ngco_genes=[]\ngco_array=[]\ngco_size=[]\n\nde_genes=[]\nde_array=[]\nde_size=[]\n\nspark_genes=[]\nspark_array=[]\nspark_size=[]\n\noverlap_gco_array=[]\noverlap_gco_size= []\n\noverlap_de_array = []\noverlap_de_size =[]\n\noverlap_spark_array=[]\noverlap_spark_size = []\nnn=0\n\nfor j in range(1,5):\n result_df=read_result_to_dataframe(\n '../../../results/BreastCancer/scGCO_results/Layers'+str(j)+'_result_df.csv')\n\n fdr_cut=0.01\n fdr_df_sct=result_df.loc[result_df.fdr< fdr_cut]\n count_gco=fdr_df_sct.index.values\n nn+= len(count_gco)\n gco_genes.extend(count_gco)\n gco_array.append(count_gco)\n gco_size.append(len(count_gco))\n\n spatialDE=pd.read_csv('../../../results/BreastCancer/SpatialDE_results/BC_{}_DE.csv'.format(j))\n count_de=spatialDE.g.values\n de_genes.extend(count_de)\n de_array.append(count_de)\n de_size.append(len(count_de))\n\n spark = pd.read_csv('../../../results/BreastCancer/SPARK_results/Layer{}_BC_spark.csv'.format(j))\n count_spark = spark.genes\n spark_genes.extend(count_spark)\n spark_array.append(count_spark)\n spark_size.append(len(count_spark))\n \n title='Layer'+str(j)\n y=(j-1)%4\n \n v=venn3(subsets=[set(count_gco),set(count_de),set(count_spark)],\n set_labels=['scGCO','spatialDE','SPARK'],\n ax=axis[y])\n axis[y].set_title(title)\n \n for text in v.subset_labels:\n text.set_fontsize(12)\n text.set_fontname('Arial')\n \n overlap_gco = (set(count_gco) & set(count_de)) |(set(count_gco) & set(count_spark))\n overlap_gco_array.append(overlap_gco)\n overlap_gco_size.append(len(overlap_gco))\n \n overlap_de = (set(count_de) & set(count_gco)) |(set(count_de) & set(count_spark))\n overlap_de_array.append(overlap_de)\n overlap_de_size.append(len(overlap_de))\n \n overlap_spark = (set(count_spark) & set(count_gco)) |(set(count_spark) & set(count_de))\n overlap_spark_array.append(overlap_spark)\n overlap_spark_size.append(len(overlap_spark))\n\nplt.show()",
"_____no_output_____"
],
[
"fig.savefig('../../../results/Figure/Supple_Fig12a-BC_venn.pdf')",
"_____no_output_____"
],
[
"from scipy import stats\nprint(stats.ttest_ind(gco_size, de_size))\n\nstats.ttest_ind(gco_size, spark_size)",
"Ttest_indResult(statistic=2.463998735260306, pvalue=0.04885371552165206)\n"
]
],
[
[
"## Supplementary Figure 12b - size ",
"_____no_output_____"
]
],
[
[
"y_mean= [np.mean(gco_size),np.mean(de_size),np.mean(spark_size)]\ny_error = [np.std(gco_size), np.std(de_size), np.std(spark_size)]\nprint('mean: ', y_mean)\nprint('sd: ', y_error)",
"mean: [167.75, 100.25, 271.25]\nsd: [31.877695964420013, 35.1452343853331, 45.42232380669223]\n"
],
[
"x=[1,2,3]\nxlabels= ['scGCO', 'spatialDE','SPARK']\ny_mean= [np.mean(gco_size),np.mean(de_size),np.mean(spark_size)]\ny_error = [np.std(gco_size), np.std(de_size), np.std(spark_size)]\ncolor = ['hotpink', 'limegreen','skyblue']\nplt.bar(x, y_mean, width=0.3, color=color, yerr=y_error, align='center',alpha=0.5, ecolor='gray',capsize=10, tick_label=xlabels)\n\nx_sca = np.array(x).repeat(4)\ny_sca = gco_size.copy()\ny_sca.extend(de_size)\ny_sca.extend(spark_size)\n\nplt.scatter(x_sca, y_sca, color='gray')\nplt.ylabel('Mean of identified SV genes')\nplt.title('Breast Cancer')\nplt.savefig('../../../results/Figure/Supple_Fig12b-BC-mean_SVGenes.pdf')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecec9594fa69c3398648ba3f1e28b8f809bab0a5 | 19,174 | ipynb | Jupyter Notebook | autopilot/model-explainability/explaining_customer_churn_model.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
] | 2 | 2021-03-31T21:10:44.000Z | 2021-04-03T04:27:26.000Z | autopilot/model-explainability/explaining_customer_churn_model.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
] | 1 | 2021-03-25T18:31:29.000Z | 2021-03-25T18:31:29.000Z | autopilot/model-explainability/explaining_customer_churn_model.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
] | 2 | 2021-09-25T08:40:23.000Z | 2021-11-08T03:01:52.000Z | 41.412527 | 855 | 0.657192 | [
[
[
"# Explaining Autopilot Models\n\nKernel `Python 3 (Data Science)` works well with this notebook.\n\n_This notebook was created and tested on an ml.m5.xlarge notebook instance._\n\n## Table of Contents\n\n1. [Introduction](#introduction)\n3. [Setup](#setup)\n4. [Local explanation with KernelExplainer](#Local)\n5. [KernelExplainer computation cost](#cost) \n6. [Global explanation with KernelExplainer](#global)\n7. [Conclusion](#conclusion)\n",
"_____no_output_____"
],
[
"---\n## Introduction<a name=\"introduction\"></a>\nMachine learning (ML) models have long been considered black boxes since predictions from these models are hard to interpret. While decision trees can be interpreted by observing the parameters learned by the models, it is generally difficult to get a clear picture.\n\nModel interpretation can be divided into local and global explanations. A local explanation considers a single sample and answers questions like: \"why the model predicts that customer A will stop using the product?\" or \"why the ML system refused John Doe a loan?\". Another interesting question is \"what should John Doe change in order to get the loan approved?\". On the contrary, global explanations aim at explaining the model itself and answer questions like \"which features are important for prediction?\". It is important to note that local explanations can be used to derive global explanations by averaging many samples. For further reading on interpretable ML, see the excellent book by [Christoph Molnar](https://christophm.github.io/interpretable-ml-book).\n\nIn this blog post, we will demonstrate the use of the popular model interpretation framework [SHAP](https://github.com/slundberg/shap) for both local and global interpretation.",
"_____no_output_____"
],
[
"### SHAP",
"_____no_output_____"
],
[
"[SHAP](https://github.com/slundberg/shap) is a game theoretic framework inspired by [Shapley Values](https://www.rand.org/pubs/papers/P0295.html) that provides local explanations for any model. SHAP has gained popularity in recent years, probably due to its strong theoretical basis. The SHAP package contains several algorithms that, given a sample and a model, derive the SHAP value for each of the model's input features. The SHAP value of a feature represents the feature's contribution to the model's prediction.\n\nTo explain models built by [Amazon SageMaker Autopilot](https://aws.amazon.com/sagemaker/autopilot/) we use SHAP's `KernelExplainer` which is a black box explainer. `KernelExplainer` is robust and can explain any model, thus can handle Autopilot's complex feature processing. `KernelExplainer` only requires that the model will support an inference functionality which, given a sample, will return the model's prediction for that sample. The prediction being the predicted value for regression and the class probability for classification.\n\nIt is worth noting that SHAP includes several other explainers such as `TreeExplainer` and `DeepExplainer` that are specific for decision forest and neural networks respectively. These are not black box explainers and require knowledge of the model structure and trained params. `TreeExplainer` and `DeepExplainer` are limited and currently can not support any feature processing.",
"_____no_output_____"
],
[
"---\n## Setup<a name=\"setup\"></a>\nIn this notebook we will start with a model built by SageMaker Autopilot which was already trained on a binary classification task. Please refer to this [notebook](https://github.com/awslabs/amazon-sagemaker-examples/blob/master/autopilot/autopilot_customer_churn.ipynb) to see how to create and train an Autopilot model. ",
"_____no_output_____"
]
],
[
[
"import boto3\nimport pandas as pd\nimport sagemaker\nfrom sagemaker import AutoML\nfrom datetime import datetime\nimport numpy as np\n\nregion = boto3.Session().region_name\nsession = sagemaker.Session()",
"_____no_output_____"
]
],
[
[
"Install SHAP",
"_____no_output_____"
]
],
[
[
"%conda install -c conda-forge shap",
"_____no_output_____"
],
[
"import shap\n\nfrom shap import KernelExplainer\nfrom shap import sample\nfrom scipy.special import expit\n\n# Initialize plugin to make plots interactive.\nshap.initjs()",
"_____no_output_____"
]
],
[
[
"### Create an inference endpoint<a name=\"Endpoint\"></a>\nCreating an inference endpoint for the trained Autopilot model. Skip this step if an endpoint with the argument `inference_response_keys` set as\n`['predicted_label', 'probability']` was already created.",
"_____no_output_____"
]
],
[
[
"automl_job_name = 'your-autopilot-job-that-exists'\nautoml_job = AutoML.attach(automl_job_name, sagemaker_session=session)\n\n# Endpoint name\nep_name = 'sagemaker-automl-' + datetime.now().strftime('%Y-%m-%d-%H-%M-%S')",
"_____no_output_____"
],
[
"# For classification response to work with SHAP we need the probability scores. This can be achieved by providing a list of keys for\n# response content. The order of the keys will dictate the content order in the response. This parameter is not needed for regression.\ninference_response_keys = ['predicted_label', 'probability']\n\n# Create the inference endpoint\nautoml_job.deploy(initial_instance_count=1, instance_type='ml.m5.2xlarge', inference_response_keys=inference_response_keys,\n endpoint_name=ep_name)",
"_____no_output_____"
]
],
[
[
"### Wrap Autopilot's endpoint with an estimator class.<a name=\"Endpoint\"></a>\nFor ease of use, we wrap the inference endpoint with a custom estimator class. Two inference functions are provided: `predict` which\nreturns the numeric prediction value to be used for regression and `predict_proba` which returns the class probability to be used for\nclassification.",
"_____no_output_____"
]
],
[
[
"from sagemaker.predictor import Predictor\n\n\nclass AutomlEstimator:\n def __init__(self, endpoint_name, sagemaker_session):\n self.predictor = Predictor(\n endpoint_name=endpoint_name,\n sagemaker_session=sagemaker_session,\n serializer=sagemaker.serializers.CSVSerializer(),\n content_type=\"text/csv\",\n accept=\"text/csv\"\n )\n \n def get_automl_response(self, x):\n if x.__class__.__name__ == 'ndarray':\n payload = \"\"\n for row in x:\n payload = payload + ','.join(map(str, row)) + '\\n'\n else:\n payload = x.to_csv(sep=',', header=False, index=False)\n return self.predictor.predict(payload).decode('utf-8')\n\n # Prediction function for regression\n def predict(self, x):\n response = self.get_automl_response(x)\n # we get the first column from the response array containing the numeric prediction value (or label in case of classification)\n response = np.array([x.split(',')[0] for x in response.split('\\n')[:-1]])\n return response\n\n # Prediction function for classification\n def predict_proba(self, x):\n \"\"\"Extract and return the probability score from the AutoPilot endpoint response.\"\"\"\n response = self.get_automl_response(x)\n # we get the second column from the response array containing the class probability\n response = np.array([x.split(',')[1] for x in response.split('\\n')[:-1]])\n return response.astype(float)",
"_____no_output_____"
]
],
[
[
"Create an instance of `AutomlEstimator`",
"_____no_output_____"
]
],
[
[
"automl_estimator = AutomlEstimator(endpoint_name=ep_name, sagemaker_session=session)",
"_____no_output_____"
]
],
[
[
"### Data\nIn this notebook we will use the same dataset as used in the [Customer Churn notebook.](https://github.com/awslabs/amazon-sagemaker-examples/blob/master/autopilot/autopilot_customer_churn.ipynb)\nPlease follow the \"Customer Churn\" notebook to download the dataset if it was not previously downloaded.\n\n### Background data\nKernelExplainer requires a sample of the data to be used as background data. KernelExplainer uses this data to simulate a feature being missing by replacing the feature value with a random value from the background. We use shap.sample to sample 50 rows from the dataset to be used as background data. Using more samples as background data will produce more accurate results but runtime will increase. Choosing background data is challenging, see the whitepapers: https://storage.googleapis.com/cloud-ai-whitepapers/AI%20Explainability%20Whitepaper.pdf and https://docs.seldon.io/projects/alibi/en/latest/methods/KernelSHAP.html#Runtime-considerations. Note that the clustering algorithms provided in shap only support numeric data. According to SHAP's documentation, a vector of zeros could be used as background data to produce reasonable results.",
"_____no_output_____"
]
],
[
[
"churn_data = pd.read_csv('../churn.txt')\ndata_without_target = churn_data.drop(columns=['Churn?'])\n\nbackground_data = sample(data_without_target, 50)",
"_____no_output_____"
]
],
[
[
"### Setup KernelExplainer",
"_____no_output_____"
],
[
"Next, we create the `KernelExplainer`. Note that since it's a black box explainer, `KernelExplainer` only requires a handle to the\npredict (or predict_proba) function and does not require any other information about the model. For classification it is recommended to\nderive feature importance scores in the log-odds space since additivity is a more natural assumption there thus we use `logit`. For\nregression `identity` should be used.",
"_____no_output_____"
]
],
[
[
"# Derive link function \nproblem_type = automl_job.describe_auto_ml_job(job_name=automl_job_name)['ResolvedAttributes']['ProblemType'] \nlink = \"identity\" if problem_type == 'Regression' else \"logit\"\n\n# the handle to predict_proba is passed to KernelExplainer since KernelSHAP requires the class probability\nexplainer = KernelExplainer(automl_estimator.predict_proba, background_data, link=link)",
"_____no_output_____"
]
],
[
[
"\nBy analyzing the background data `KernelExplainer` provides us with `explainer.expected_value` which is the model prediction with all features missing. Considering a customer for which we have no data at all (i.e. all features are missing) this should theoretically be the model prediction.",
"_____no_output_____"
]
],
[
[
"# Since expected_value is given in the log-odds space we convert it back to probability using expit which is the inverse function to logit\nprint('expected value =', expit(explainer.expected_value))",
"_____no_output_____"
]
],
[
[
"---\n## Local explanation with KernelExplainer<a name=\"local\"></a>\nWe will use `KernelExplainer` to explain the prediction of a single sample, the first sample in the dataset.",
"_____no_output_____"
]
],
[
[
"# Get the first sample\nx = data_without_target.iloc[0:1]\n\n# ManagedEndpoint can optionally auto delete the endpoint after calculating the SHAP values. To enable auto delete, use ManagedEndpoint(ep_name, auto_delete=True)\nfrom managed_endpoint import ManagedEndpoint\nwith ManagedEndpoint(ep_name) as mep:\n shap_values = explainer.shap_values(x, nsamples='auto', l1_reg='aic')",
"_____no_output_____"
]
],
[
[
"SHAP package includes many visualization tools. See below a `force_plot` which provides a good visualization for the SHAP values of a single sample",
"_____no_output_____"
]
],
[
[
"# Since shap_values are provided in the log-odds space, we convert them back to the probability space by using LogitLink\nshap.force_plot(explainer.expected_value, shap_values, x, link=link)",
"_____no_output_____"
]
],
[
[
"From the plot above we learn that the most influential feature is `VMail Message` which pushes the probability down by about 7%. It is\nimportant to note that `VMail Message = 25` makes the probability 7% lower in comparison to the notion of that feature being missing.\nSHAP values do not provide the information of how increasing or decreasing `VMail Message` will affect prediction.\n\nIn many cases we are interested only in the most influential features. By setting `l1_reg='num_features(5)'`, SHAP will provide non-zero scores for only the most influential 5 features.",
"_____no_output_____"
]
],
[
[
"with ManagedEndpoint(ep_name) as mep:\n shap_values = explainer.shap_values(x, nsamples='auto', l1_reg='num_features(5)')\nshap.force_plot(explainer.expected_value, shap_values, x, link=link)",
"_____no_output_____"
]
],
[
[
"---\n## KernelExplainer computation cost<a name=\"cost\"></a>\nKernelExplainer computation cost is dominated by the inference calls. In order to estimate SHAP values for a single sample, KernelExplainer calls the inference function twice: First, with the sample unaugmented. And second, with many randomly augmented instances of the sample. The number of augmented instances in our case is: 50 (#samples in the background data) * 2088 (nsamples = 'auto') = 104,400. So, in our case, the cost of running KernelExplainer for a single sample is roughly the cost of 104,400 inference calls.",
"_____no_output_____"
],
[
"---\n## Global explanation with KernelExplainer<a name=\"global\"></a>\nNext, we will use KernelExplainer to provide insight about the model as a whole. It is done by running KernelExplainer locally on 50 samples and aggregating the results",
"_____no_output_____"
]
],
[
[
"# Sample 50 random samples\nX = sample(data_without_target, 50)\n\n# Calculate SHAP values for these samples, and delete the endpoint\nwith ManagedEndpoint(ep_name, auto_delete=True) as mep:\n shap_values = explainer.shap_values(X, nsamples='auto', l1_reg='aic')",
"_____no_output_____"
]
],
[
[
"`force_plot` can be used to visualize SHAP values for many samples simultaneously by rotating the plot of each sample by 90 degrees and stacking the plots horizontally. The resulting plot is interactive and can be manually analyzed.",
"_____no_output_____"
]
],
[
[
"shap.force_plot(explainer.expected_value, shap_values, X, link=link)",
"_____no_output_____"
]
],
[
[
"`summary_plot` is another visualization tool displaying the mean absolute value of the SHAP values for each feature using a bar plot. Currently, `summary_plot` does not support link functions so the SHAP values are presented in the log-odds space (and not the probability space).",
"_____no_output_____"
]
],
[
[
"shap.summary_plot(shap_values, X, plot_type=\"bar\")",
"_____no_output_____"
]
],
[
[
"---\n## Conclusion<a name=\"conclusion\"></a>\n\nIn this post, we demonstrated how to use KernelSHAP to explain models created by Amazon SageMaker Autopilot both locally and globally. KernelExplainer is a robust black box explainer which requires only that the model will support an inference functionality which, given a sample, returns the model's prediction for that sample. This inference functionality was provided by wrapping Autopilot's inference endpoint with an estimator container.\n\nFor more about Amazon SageMaker Autopilot, please see [Amazon SageMaker Autopilot](https://aws.amazon.com/sagemaker/autopilot/).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecec98ddffb33f00f8b3e5bfe8615ea59c649526 | 8,602 | ipynb | Jupyter Notebook | nse day & time sorted.ipynb | PundirShivam/tradetest | 9f62740b685c017983e33e23eaa35141b13047f1 | [
"MIT"
] | null | null | null | nse day & time sorted.ipynb | PundirShivam/tradetest | 9f62740b685c017983e33e23eaa35141b13047f1 | [
"MIT"
] | null | null | null | nse day & time sorted.ipynb | PundirShivam/tradetest | 9f62740b685c017983e33e23eaa35141b13047f1 | [
"MIT"
] | 1 | 2021-12-15T13:06:37.000Z | 2021-12-15T13:06:37.000Z | 33.084615 | 337 | 0.532434 | [
[
[
"!pip install backtrader",
"Collecting backtrader\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/ab/09/472ad6e42d3a4c9fbd3e162d0a3f9d518ce9189ee8e964bd65ae5af67936/backtrader-1.9.70.122-py2.py3-none-any.whl (410kB)\n\u001b[K 100% |████████████████████████████████| 419kB 7.8MB/s \n\u001b[?25hInstalling collected packages: backtrader\nSuccessfully installed backtrader-1.9.70.122\n"
],
[
"import backtrader as bt\nimport matplotlib.pyplot as plt\nimport pandas as pd",
"_____no_output_____"
],
[
"from __future__ import (absolute_import, division, print_function,\n unicode_literals)\n\nimport datetime\nimport backtrader as bt\nimport backtrader.feeds as btfeeds\n\n# Import the backtrader platform\nimport backtrader as bt\n\n\n# Create a Stratey\nclass TestStrategy(bt.Strategy):\n\n def log(self, txt, dt=None):\n ''' Logging function for this strategy'''\n dt = dt or self.datas[0].datetime.date(0)\n t = self.datas[0].datetime.time(0)\n #print('%s, %s' % (dt.isoformat(), txt))\n print('%s, %s, %s' % (dt.isoformat(),t, txt))\n\n def __init__(self):\n # Keep a reference to the \"close\" line in the data[0] dataseries\n # Also, keeping a reference for volume \n self.dataclose = self.datas[0].close\n self.datavolume = self.datas[0].volume\n\n def next(self):\n # Simply log the closing price of the series from the reference\n #self.log('Close, %.2f at volume %d' % (self.dataclose[0],self.datavolume[0]))\n # modifying to log the close price of the day\n t = self.datas[0].datetime.time(0)\n if t == datetime.time(15, 30):\n self.log('Close @ eod , %.2f with volume @ eod %d ' % (self.dataclose[0],self.datavolume[0]))\n elif t== datetime.time(9,15):\n self.log('Close @ open , %.2f with volume @ open %d ' % (self.dataclose[0],self.datavolume[0]))\n\nif __name__ == '__main__':\n # Create a cerebro entity\n cerebro = bt.Cerebro()\n\n # Add a strategy\n cerebro.addstrategy(TestStrategy)\n\n # Datas are in a subfolder of the samples. Need to find where the script is\n # because it could have been called from anywhere\n #modpath = os.path.dirname(os.path.abspath(sys.argv[0]))\n #datapath = os.path.join(modpath, '../../datas/orcl-1995-2014.txt')\n #datapath='nse1.csv'\n # Create a Data Feed\n data = btfeeds.GenericCSVData(\n dataname='nse1.csv',\n timeframe=bt.TimeFrame.Minutes,\n fromdate=datetime.datetime(2018, 1, 1),\n todate=datetime.datetime(2018, 1, 4),\n nullvalue=0.0,\n\n dtformat=('%Y-%m-%d'),\n tmformat=('%H:%M:%S') , \n\n datetime=0,\n time=1,\n high=3,\n low=4,\n open=2,\n close=5,\n volume=6,\n openinterest=7\n)\n\n # Add the Data Feed to Cerebro\n cerebro.adddata(data)\n\n # Set our desired cash start\n cerebro.broker.setcash(100000.0)\n\n # Print out the starting conditions\n print('Starting Portfolio Value: %.2f' % cerebro.broker.getvalue())\n\n # Run over everything\n cerebro.run()\n\n # Print out the final result\n print('Final Portfolio Value: %.2f' % cerebro.broker.getvalue())",
"Starting Portfolio Value: 100000.00\n2018-01-01, 09:15:00, Close @ open , 10543.95 with volume @ open 93228 \n2018-01-01, 15:30:00, Close @ eod , 10490.00 with volume @ eod 300 \n2018-01-02, 09:15:00, Close @ open , 11080.00 with volume @ open 200478 \n2018-01-03, 09:15:00, Close @ open , 10509.80 with volume @ open 175202 \n2018-01-03, 15:30:00, Close @ eod , 10442.05 with volume @ eod 1500 \nFinal Portfolio Value: 100000.00\n"
],
[
"btfeeds.GenericCSVData?",
"_____no_output_____"
],
[
"import datetime\nt1=datetime.time(15,30,0)",
"_____no_output_____"
],
[
"t1",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ececb6a3ee7347c00d11df9da32b842f410ea9b4 | 3,600 | ipynb | Jupyter Notebook | pet_classifier.ipynb | Sturzgefahr/pet_breeds | a076b95c88387c53c868aa270c5f6b87f7b7e81f | [
"Apache-2.0"
] | null | null | null | pet_classifier.ipynb | Sturzgefahr/pet_breeds | a076b95c88387c53c868aa270c5f6b87f7b7e81f | [
"Apache-2.0"
] | null | null | null | pet_classifier.ipynb | Sturzgefahr/pet_breeds | a076b95c88387c53c868aa270c5f6b87f7b7e81f | [
"Apache-2.0"
] | null | null | null | 26.865672 | 680 | 0.563611 | [
[
[
"from fastai2.vision.all import *\nfrom fastai2.vision.widgets import *",
"_____no_output_____"
]
],
[
[
"# The Amazing Pet Classifier!",
"_____no_output_____"
],
[
"Upload your pet picture!\n\n----",
"_____no_output_____"
]
],
[
[
"path = Path()\nlearn_inf = load_learner(path/'export.pkl')\nbtn_upload = widgets.FileUpload()\nout_pl = widgets.Output()\nlbl_pred = widgets.Label()",
"_____no_output_____"
],
[
"def on_click(change):\n img = PILImage.create(btn_upload.data[-1])\n out_pl.clear_output()\n with out_pl: display(img.to_thumb(128,128))\n pred,pred_idx,probs = learn_inf.predict(img)\n lbl_pred.value = f'Prediction: {pred}; Probability: {probs[pred_idx]:.04f}'",
"_____no_output_____"
],
[
"btn_upload.observe(on_click, names=['data'])",
"_____no_output_____"
],
[
"display(VBox([widgets.Label('Select your pet!'), btn_upload, out_pl, lbl_pred]))",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ececd46572c1c94002530c9b180404c732d9a53a | 182,206 | ipynb | Jupyter Notebook | Week 3/1621583586_Quiz_Pekan_3.ipynb | jo-adithya/IM-Sanbercode-DataScience-Batch-24 | ade673f98377f85babb9e5e755c14558e745529b | [
"MIT"
] | null | null | null | Week 3/1621583586_Quiz_Pekan_3.ipynb | jo-adithya/IM-Sanbercode-DataScience-Batch-24 | ade673f98377f85babb9e5e755c14558e745529b | [
"MIT"
] | null | null | null | Week 3/1621583586_Quiz_Pekan_3.ipynb | jo-adithya/IM-Sanbercode-DataScience-Batch-24 | ade673f98377f85babb9e5e755c14558e745529b | [
"MIT"
] | null | null | null | 131.46176 | 101,192 | 0.809068 | [
[
[
"# **Quiz Pekan - 3**",
"_____no_output_____"
],
[
"# Soal 1 : Apakah harapan kalian saat mengikuti kelas Python Datascience dasar? ",
"_____no_output_____"
],
[
"Jawab:\n",
"_____no_output_____"
],
[
"\n\n---\n\nDapat menggunakan skill data analisis ini dalam kehidupan sehari-hari dan bisa mendapatkan pekerjaan yang berhubungan dengan data analisis. Saya juga ingin meneruskan lagi dari data science ini ke machine learning.\n\n---\n\n",
"_____no_output_____"
],
[
"\n## **Penjelasan Dataset**\nDataset-dataset berikut ini menunjukkan score happines dunia. Score happiness menggunakan data dari GallUp world poll. Data feature yang menjadi kata kunci di report ini adalah:\n\n* **Life Ladder** >> Ibarat sebuah tangga, pijakan 0 adalah dasar dan 10 adalah atas. Tangga atas menunjukkan kehidupan terbaik dan dasar tangga menunjukkan kehidupan terburuk \n* **Log GDP Per capita** >> nilai ekonomi yang dihasilkan setiap individu warga\n* **Social support** >> dukungan sosial seperti keluarga, teman, tetangga dll\n* **Healthy life expectancy at birth** >> rata-rata hidup dalam kesehatan yang baik\n* **Freedom to make life choices** >> Peluang tiap individu untuk melakukan aksi yang dipilih\n* **Generosity** >> Kemurahan hati\n* **Perceptions of corruption** >> Korupsi\n\n\nLink Download:\n\n[world-happiness-report.csv](https://drive.google.com/uc?export=download&id=14yujHboPMR5tc_n3btPcFW4fXgy4-SzL)\n\n[world-happiness-report-2021.csv](https://drive.google.com/uc?export=download&id=1gWCX58PyH0viABEMswzJ71sJ0v5KHco3)",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport seaborn as sns",
"_____no_output_____"
],
[
"#import semua data\n\ndf1 = pd.read_csv('world-happiness-report.csv')\ndf2 = pd.read_csv('world-happiness-report-2021.csv')\ndisplay(df1.head())\ndisplay(df2.head())",
"_____no_output_____"
]
],
[
[
"# Soal 2 : Tunjukkan summary statistik dari dataset world happiness 2021 seperti expected output berikut. \n\n\n\n* Tunjukkan summary statistik dari dataset df2\n\n\n\nHINT:\nTeman-teman perlu mendrop beberapa kolom yang tidak ditampilkan di tabel expected output dengan menggunakan atribut .drop() dari pandas DataFrame. Silahkan kunjungi referensi link berikut:\nhttps://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.drop.html",
"_____no_output_____"
]
],
[
[
"#code here\ndf2 = df2.drop(['Standard error of ladder score', 'upperwhisker', 'lowerwhisker', 'Ladder score in Dystopia', 'Explained by: Log GDP per capita', 'Explained by: Social support', 'Explained by: Healthy life expectancy', 'Explained by: Freedom to make life choices', 'Explained by: Generosity', 'Explained by: Perceptions of corruption'], axis=1)\ndisplay(df2.describe())",
"_____no_output_____"
]
],
[
[
"Expected output:\n\n",
"_____no_output_____"
],
[
"\n\n---\n\n\n\n---\n\n",
"_____no_output_____"
],
[
"# Soal 3: Happiness report negara Indonesia dari tahun 2006 - 2021\n\nOutput yang diharapkan adalah data negara Indonesia dari penggabungan dataset happiness report dan dataset happiness report 2021. Untuk sesuai dengan expected ouput, lakukan perintah-perintah berikut:\n\n\n1. Lakukan filtering dataset happiness report dari kolom ['Country name']== \n'Indonesia'\n - tambahkan atribut .reset_index(drop=True)\n - drop kolom 'Positive affect' dan 'Negative affect'\n\n2. Lakukan filtering dataset dari variabel no 2 hasil dropping beberapa kolom\n - tambahkan kolom ['year']=2021 pada dataset dari variabel no 2 hasil dropping beberapa kolom\n - filtering dataset tersebut menggunakan kolom ['Country name']== \n'Indonesia'\n - rename beberapa kolom dengan menggunakan atribut .rename() dari pandas, referensi dokumentasi : https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.rename.html\n { \n 'Ladder score':'Life Ladder',\n 'Logged GDP per capita':'Log GDP per capita',\n 'Healthy life expectancy':'Healthy life expectancy at birth'}\n\n - drop kolom 'Dystopia + residual' dan 'Regional indicator'\n\n3. Gabungkan data dari hasil poin 1 dan poin 2\n - gunakan pandas concat, .concat()\n - tambahkan atribut .reset_index(drop=True)\n - rename kolom {'year':'Year'}\n",
"_____no_output_____"
]
],
[
[
"# Code here\n\n# Poin 1\n# df = df1[df1['Country name'] == 'Indonesia'].reset_index(drop=True)\n# df = df.drop(['Positive affect', 'Negative affect'], axis=1)\n\n# Poin 2\n# df2['year'] = 2021\n# df2 = df2[df2['Country name'] == 'Indonesia']\n# df2.rename(columns={\n# 'Ladder score':'Life Ladder',\n# 'Logged GDP per capita':'Log GDP per capita',\n# 'Healthy life expectancy':'Healthy life expectancy at birth'\n# }, inplace=True)\n\n# Poin 3\n# df_total = pd.concat([df, df2], join='inner').reset_index(drop=True)\ndf_total.rename(columns={'year': 'Year'}, inplace=True)\ndisplay(df_total)",
"_____no_output_____"
]
],
[
[
"Expected output:\n\n",
"_____no_output_____"
],
[
"\n\n---\n\n\n\n---\n\n",
"_____no_output_____"
],
[
"# Soal 4: Visualisasi lineplot Life Ladder negara Indonesia\n\n* Gunakan library seaborn untuk plotting line plot kolom Life Ladder dari hasil penggabungan negara Indonesia tahun 2006-2021 (soal 3) seperti expected output, jika tidak bisa menyelesaikan soal 3 silahkan download data berikut \n[Download.csv](https://drive.google.com/uc?export=download&id=19g5oURnRNlbXPUEQQyvimoEx8DwqMPLW) \n* Berikan insight dari hasil visualisasi tersebut",
"_____no_output_____"
]
],
[
[
"#code here\nplt.figure(figsize=(12,4.5))\nwith sns.axes_style('whitegrid'):\n sns.lineplot(x='Year', y='Life Ladder', data=df_total, marker='o')\n plt.title(\"Indonesia's ladder score over the years\", fontweight=\"bold\")\n sns.despine(left=True)",
"_____no_output_____"
]
],
[
[
"Expected output:\n\n",
"_____no_output_____"
],
[
"#### insight here",
"_____no_output_____"
],
[
"\n\n---\n\n- Life ladder score di Indonesia naik drastis dari tahun 2008 ke 2009.\n- Life ladder score di Indonesia turun cukup drastis dari tahun 2014 ke 2015.\n- Setelah itu, dari tahun 2015 ke 2017 cukup stabil dan ada kenaikan sedikit dari tahun 2017 ke 2018.\n- Lalu dari tahun 2018 ke 2021 Life ladder score di Indonesia stabil di angka 5.3 - 5.4\n\n---\n\n",
"_____no_output_____"
],
[
"# Soal 5: Urutkan Ladder Score tertinggi dengan negara-negara Asia Tenggara seperti expected output\n\nUntuk mendapatkan expected output, lakukan perintah-perintah berikut:\n\n* Sorting data berdasarkan kolom 'Ladder Score' dari dataset world happiness 2021 dengan hanya menampilkan kolom ['Country name', 'Ladder score', 'Regional indicator']. Sorting data menggunakan sort_values() dari pandas, silahkan kunjungi dokumentasi berikut: https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.sort_values.html\n* Urutan pertama diperoleh dari hasil filtering dengan kondisi ['Ladder score']==['Ladder score'].max()\n\n* Urutan negara-negara Asia tenggara diperoleh dari hasil Filtering kolom ['Regional indicator'] == 'Southeast Asia'\n* Lakukan concatination antara data Urutan pertama dengan urutan negara Asia Tenggara menggunakan pd.concat()\n* Tambahkan kolom ['Rank'] yang berisi dari indeks hasil penggabungan\n* drop kolom 'Regional indicator'\n",
"_____no_output_____"
]
],
[
[
"#code here\ndf = df2.sort_values('Ladder score', ascending=False)[['Country name', 'Ladder score', 'Regional indicator']]\ndf['Rank'] = df.index + 1\ndf_asean = df[df['Regional indicator'] == 'Southeast Asia']\ndf_max = df[df['Ladder score'] == df['Ladder score'].max()]\ndf_result = pd.concat([df_max, df_asean]).reset_index(drop=True)\ndf_result.drop(['Regional indicator'], axis=1, inplace=True)\ndisplay(df_result)",
"_____no_output_____"
]
],
[
[
"Expected output:\n\n",
"_____no_output_____"
],
[
"\n\n---\n\n\n\n---\n\n",
"_____no_output_____"
],
[
"# Soal 6: Visualisasi Scatter plot\n\nBuatlah visualisasi scatter plot (perpaduan seaborn dan matplotlib) dari df2 antara GDP per capita ('Logged GDP per capita') dan skor korupsi ('Perceptions of corruptions') sesuai expected output dengan ketentuan berikut:\n\nGunakan kolom skor kebebasan beraksi ('Freedom to make life choices') sebagai ukuran pointnya kalikan suatu angka yang cukup besar agar terlihat.\n\nGunakan kolom skor kesehatan ('Healthy life expectancy') sebagai warna point, berikan cmap 'plasma'.\n\nGunakan regplot pada seaborn (sns.regplot) dengan argumen scatter=False dan color = 'black'",
"_____no_output_____"
]
],
[
[
"#code here\nsizes = df2['Freedom to make life choices'] * 500\nplt.figure(figsize=(12, 8))\nwith sns.axes_style('whitegrid'):\n sns.regplot(x='Logged GDP per capita', y='Perceptions of corruption', scatter=False, data=df2, color='black')\n plt.scatter(x=df2['Logged GDP per capita'], y=df2['Perceptions of corruption'], c=df2['Healthy life expectancy'], s=sizes, linewidth=0, alpha=0.6, cmap='plasma')\n plt.colorbar()\n plt.title('Corruption vs GDP percapita')",
"_____no_output_____"
]
],
[
[
"Expected output:\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecece74870e6ec9b04b0256f49df87314b7c7d3d | 52,716 | ipynb | Jupyter Notebook | docs/site/tutorials/a_swift_tour.ipynb | texasmichelle/swift | 37821ab7a57bc3013be20565a9a52f94145e2225 | [
"Apache-2.0"
] | null | null | null | docs/site/tutorials/a_swift_tour.ipynb | texasmichelle/swift | 37821ab7a57bc3013be20565a9a52f94145e2225 | [
"Apache-2.0"
] | null | null | null | docs/site/tutorials/a_swift_tour.ipynb | texasmichelle/swift | 37821ab7a57bc3013be20565a9a52f94145e2225 | [
"Apache-2.0"
] | null | null | null | 28.806557 | 501 | 0.571136 | [
[
[
"##### Adapted from the original [*A Swift Tour*](https://docs.swift.org/swift-book/GuidedTour/GuidedTour.html) on [Swift.org](https://swift.org) with modifications. The original content was authored by Apple Inc. Licensed under the [Creative Commons Attribution 4.0 International (CC BY 4.0) License](https://creativecommons.org/licenses/by/4.0/).",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/swift/tutorials/a_swift_tour\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/swift/blob/main/docs/site/tutorials/a_swift_tour.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/swift/blob/main/docs/site/tutorials/a_swift_tour.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"# A Swift Tour\n\nTradition suggests that the first program in a new language should print the words \"Hello, world!\" on the screen. In Swift, this can be done in a single line:",
"_____no_output_____"
]
],
[
[
"print(\"Hello, world!\")",
"_____no_output_____"
]
],
[
[
"If you have written code in C or Objective-C, this syntax looks familiar to you—in Swift, this line of code is a complete program. You don’t need to import a separate library for functionality like input/output or string handling. Code written at global scope is used as the entry point for the program, so you don’t need a `main()` function. You also don’t need to write semicolons at the end of every statement.\n\nThis tour gives you enough information to start writing code in Swift by showing you how to accomplish a variety of programming tasks. Don’t worry if you don’t understand something—everything introduced in this tour is explained in detail in the rest of this book.",
"_____no_output_____"
],
[
"## Simple Values\n\nUse `let` to make a constant and `var` to make a variable. The value of a constant doesn’t need to be known at compile time, but you must assign it a value exactly once. This means you can use constants to name a value that you determine once but use in many places.",
"_____no_output_____"
]
],
[
[
"var myVariable = 42\nmyVariable = 50\nlet myConstant = 42",
"_____no_output_____"
]
],
[
[
"A constant or variable must have the same type as the value you want to assign to it. However, you don’t always have to write the type explicitly. Providing a value when you create a constant or variable lets the compiler infer its type. In the example above, the compiler infers that `myVariable` is an integer because its initial value is an integer.\n\nIf the initial value doesn’t provide enough information (or if there is no initial value), specify the type by writing it after the variable, separated by a colon. Note: using `Double` instead of `Float` for floating point numbers gives you more precision, and is the default floating point type in Swift.",
"_____no_output_____"
]
],
[
[
"let implicitInteger = 70\nlet implicitDouble = 70.0\nlet explicitDouble: Double = 70",
"_____no_output_____"
],
[
"// Experiment:\n// Create a constant with an explicit type of `Float` and a value of 4.",
"_____no_output_____"
]
],
[
[
"Values are never implicitly converted to another type. If you need to convert a value to a different type, explicitly make an instance of the desired type.",
"_____no_output_____"
]
],
[
[
"let label = \"The width is \"\nlet width = 94\nprint(label + String(width))",
"_____no_output_____"
],
[
"// Experiment:\n// Try removing the conversion to `String` from the last line. What error do you get?",
"_____no_output_____"
]
],
[
[
"There’s an even simpler way to include values in strings: Write the value in parentheses, and write a backslash (`\\`) before the parentheses. For example:",
"_____no_output_____"
]
],
[
[
"let apples = 3\nprint(\"I have \\(apples) apples.\")",
"_____no_output_____"
],
[
"let oranges = 5\nprint(\"I have \\(apples + oranges) pieces of fruit.\")",
"_____no_output_____"
],
[
"// Experiment:\n// Use `\\()` to include a floating-point calculation in a string and to include someone's name in a\n// greeting.",
"_____no_output_____"
]
],
[
[
"Use three double quotation marks (`\"\"\"`) for strings that take up multiple lines. Indentation at the start of each quoted line is removed, as long as it matches the indentation of the closing quotation marks. For example:",
"_____no_output_____"
]
],
[
[
"let quotation = \"\"\"\n Even though there's whitespace to the left,\n the actual lines aren't indented.\n Except for this line.\n Double quotes (\") can appear without being escaped.\n\n I still have \\(apples + oranges) pieces of fruit.\n \"\"\"\nprint(quotation)",
"_____no_output_____"
]
],
[
[
"Create arrays and dictionaries using brackets (`[]`), and access their elements by writing the index or key in brackets. A comma is allowed after the last element.",
"_____no_output_____"
]
],
[
[
"var shoppingList = [\"catfish\", \"water\", \"tulips\", \"blue paint\"]\nshoppingList[1] = \"bottle of water\"\n \nvar occupations = [\n \"Malcolm\": \"Captain\",\n \"Kaylee\": \"Mechanic\",\n]\noccupations[\"Jayne\"] = \"Public Relations\"\nprint(occupations)",
"_____no_output_____"
]
],
[
[
"Arrays automatically grow as you add elements.",
"_____no_output_____"
]
],
[
[
"shoppingList.append(\"blue paint\")\nprint(shoppingList)",
"_____no_output_____"
]
],
[
[
"To create an empty array or dictionary, use the initializer syntax.",
"_____no_output_____"
]
],
[
[
"let emptyArray = [String]()\nlet emptyDictionary = [String: Float]()",
"_____no_output_____"
]
],
[
[
"If type information can be inferred, you can write an empty array as `[]` and an empty dictionary as `[:]` — for example, when you set a new value for a variable or pass an argument to a function.",
"_____no_output_____"
]
],
[
[
"shoppingList = []\noccupations = [:]",
"_____no_output_____"
]
],
[
[
"## Control Flow\n\nUse `if` and `switch` to make conditionals, and use `for`-`in`, `for`, `while`, and `repeat`-`while` to make loops. Parentheses around the condition or loop variable are optional. Braces around the body are required.",
"_____no_output_____"
]
],
[
[
"let individualScores = [75, 43, 103, 87, 12]\nvar teamScore = 0\nfor score in individualScores {\n if score > 50 {\n teamScore += 3\n } else {\n teamScore += 1\n }\n}\nprint(teamScore)",
"_____no_output_____"
]
],
[
[
"In an `if` statement, the conditional must be a Boolean expression—this means that code such as `if score { ... }` is an error, not an implicit comparison to zero.\n\nYou can use `if` and `let` together to work with values that might be missing. These values are represented as optionals. An optional value either contains a value or contains `nil` to indicate that a value is missing. Write a question mark (`?`) after the type of a value to mark the value as optional.",
"_____no_output_____"
]
],
[
[
"var optionalString: String? = \"Hello\"\nprint(optionalString == nil)",
"_____no_output_____"
],
[
"var optionalName: String? = \"John Appleseed\"\nvar greeting = \"Hello!\"\nif let name = optionalName {\n greeting = \"Hello, \\(name)\"\n}\nprint(greeting)",
"_____no_output_____"
],
[
"// Experiment:\n// Change `optionalName` to `nil`. What greeting do you get?\n// Add an `else` clause that sets a different greeting if `optionalName` is `nil`.",
"_____no_output_____"
]
],
[
[
"If the optional value is `nil`, the conditional is `false` and the code in braces is skipped. Otherwise, the optional value is unwrapped and assigned to the constant after `let`, which makes the unwrapped value available inside the block of code.\n\nAnother way to handle optional values is to provide a default value using the `??` operator. If the optional value is missing, the default value is used instead.",
"_____no_output_____"
]
],
[
[
"let nickName: String? = nil\nlet fullName: String = \"John Appleseed\"\nprint(\"Hi \\(nickName ?? fullName)\")",
"_____no_output_____"
]
],
[
[
"Switches support any kind of data and a wide variety of comparison operations—they aren’t limited to integers and tests for equality.",
"_____no_output_____"
]
],
[
[
"let vegetable = \"red pepper\"\nswitch vegetable {\ncase \"celery\":\n print(\"Add some raisins and make ants on a log.\")\ncase \"cucumber\", \"watercress\":\n print(\"That would make a good tea sandwich.\")\ncase let x where x.hasSuffix(\"pepper\"):\n print(\"Is it a spicy \\(x)?\")\ndefault:\n print(\"Everything tastes good in soup.\")\n}",
"_____no_output_____"
],
[
"// Experiment:\n// Try removing the default case. What error do you get?",
"_____no_output_____"
]
],
[
[
"Notice how `let` can be used in a pattern to assign the value that matched that part of a pattern to a constant.\n\nAfter executing the code inside the switch case that matched, the program exits from the switch statement. Execution doesn’t continue to the next case, so there is no need to explicitly break out of the switch at the end of each case’s code.\n\nYou use `for`-`in` to iterate over items in a dictionary by providing a pair of names to use for each key-value pair. Dictionaries are an unordered collection, so their keys and values are iterated over in an arbitrary order.",
"_____no_output_____"
]
],
[
[
"let interestingNumbers = [\n \"Prime\": [2, 3, 5, 7, 11, 13],\n \"Fibonacci\": [1, 1, 2, 3, 5, 8],\n \"Square\": [1, 4, 9, 16, 25],\n]\nvar largest = 0\nfor (kind, numbers) in interestingNumbers {\n for number in numbers {\n if number > largest {\n largest = number\n }\n }\n}\nprint(largest)",
"_____no_output_____"
],
[
"// Experiment:\n// Add another variable to keep track of which kind of number was the largest, as well as what that\n// largest number was.",
"_____no_output_____"
]
],
[
[
"Use `while` to repeat a block of code until a condition changes. The condition of a loop can be at the end instead, ensuring that the loop is run at least once.",
"_____no_output_____"
]
],
[
[
"var n = 2\nwhile n < 100 {\n n = n * 2\n}\n\nprint(n)",
"_____no_output_____"
],
[
"var m = 2\nrepeat {\n m = m * 2\n} while m < 100\n\nprint(m)",
"_____no_output_____"
]
],
[
[
"You can keep an index in a loop—either by using `..<` to make a range of indexes or by writing an explicit initialization, condition, and increment. These two loops do the same thing:",
"_____no_output_____"
]
],
[
[
"var total = 0\nfor i in 0..<4 {\n total += i\n}\n\nprint(total)",
"_____no_output_____"
]
],
[
[
"Use `..<` to make a range that omits its upper value, and use `...` to make a range that includes both values.",
"_____no_output_____"
],
[
"## Functions and Closures\n\nUse `func` to declare a function. Call a function by following its name with a list of arguments in parentheses. Use `->` to separate the parameter names and types from the function’s return type.",
"_____no_output_____"
]
],
[
[
"func greet(name: String, day: String) -> String {\n return \"Hello \\(name), today is \\(day).\"\n}\nprint(greet(name: \"Bob\", day: \"Tuesday\"))",
"_____no_output_____"
],
[
"// Experiment:\n// Remove the `day` parameter. Add a parameter to include today’s lunch special in the greeting.",
"_____no_output_____"
]
],
[
[
"By default, functions use their parameter names as labels for their arguments. Write a custom argument label before the parameter name, or write `_` to use no argument label.",
"_____no_output_____"
]
],
[
[
"func greet(_ person: String, on day: String) -> String {\n return \"Hello \\(person), today is \\(day).\"\n}\nprint(greet(\"John\", on: \"Wednesday\"))",
"_____no_output_____"
]
],
[
[
"Use a tuple to make a compound value—for example, to return multiple values from a function. The elements of a tuple can be referred to either by name or by number.",
"_____no_output_____"
]
],
[
[
"func calculateStatistics(scores: [Int]) -> (min: Int, max: Int, sum: Int) {\n var min = scores[0]\n var max = scores[0]\n var sum = 0\n \n for score in scores {\n if score > max {\n max = score\n } else if score < min {\n min = score\n }\n sum += score\n }\n \n return (min, max, sum)\n}\nlet statistics = calculateStatistics(scores: [5, 3, 100, 3, 9])\nprint(statistics.sum)\nprint(statistics.2)",
"_____no_output_____"
]
],
[
[
"Functions can be nested. Nested functions have access to variables that were declared in the outer function. You can use nested functions to organize the code in a function that is long or complex.",
"_____no_output_____"
]
],
[
[
"func returnFifteen() -> Int {\n var y = 10\n func add() {\n y += 5\n }\n add()\n return y\n}\nprint(returnFifteen())",
"_____no_output_____"
]
],
[
[
"Functions are a first-class type. This means that a function can return another function as its value.",
"_____no_output_____"
]
],
[
[
"func makeIncrementer() -> ((Int) -> Int) {\n func addOne(number: Int) -> Int {\n return 1 + number\n }\n return addOne\n}\nvar increment = makeIncrementer()\nprint(increment(7))",
"_____no_output_____"
]
],
[
[
"A function can take another function as one of its arguments.",
"_____no_output_____"
]
],
[
[
"func hasAnyMatches(list: [Int], condition: (Int) -> Bool) -> Bool {\n for item in list {\n if condition(item) {\n return true\n }\n }\n return false\n}\nfunc lessThanTen(number: Int) -> Bool {\n return number < 10\n}\nvar numbers = [20, 19, 7, 12]\nprint(hasAnyMatches(list: numbers, condition: lessThanTen))",
"_____no_output_____"
]
],
[
[
"Functions are actually a special case of closures: blocks of code that can be called later. The code in a closure has access to things like variables and functions that were available in the scope where the closure was created, even if the closure is in a different scope when it is executed—you saw an example of this already with nested functions. You can write a closure without a name by surrounding code with braces (`{}`). Use `in` to separate the arguments and return type from the body.",
"_____no_output_____"
]
],
[
[
"print(numbers.map({ (number: Int) -> Int in\n let result = 3 * number\n return result\n}))",
"_____no_output_____"
],
[
"// Experiment:\n// Rewrite the closure to return zero for all odd numbers.",
"_____no_output_____"
]
],
[
[
"You have several options for writing closures more concisely. When a closure’s type is already known, such as the callback for a delegate, you can omit the type of its parameters, its return type, or both. Single statement closures implicitly return the value of their only statement.",
"_____no_output_____"
]
],
[
[
"let mappedNumbers = numbers.map({ number in 3 * number })\nprint(mappedNumbers)",
"_____no_output_____"
]
],
[
[
"You can refer to parameters by number instead of by name—this approach is especially useful in very short closures. A closure passed as the last argument to a function can appear immediately after the parentheses. When a closure is the only argument to a function, you can omit the parentheses entirely.",
"_____no_output_____"
]
],
[
[
"let sortedNumbers = numbers.sorted { $0 > $1 }\nprint(sortedNumbers)",
"_____no_output_____"
]
],
[
[
"## Objects and Classes\n\nUse `class` followed by the class’s name to create a class. A property declaration in a class is written the same way as a constant or variable declaration, except that it is in the context of a class. Likewise, method and function declarations are written the same way.",
"_____no_output_____"
]
],
[
[
"class Shape {\n var numberOfSides = 0\n func simpleDescription() -> String {\n return \"A shape with \\(numberOfSides) sides.\"\n }\n}",
"_____no_output_____"
],
[
"// Experiment:\n// Add a constant property with `let`, and add another method that takes an argument.",
"_____no_output_____"
]
],
[
[
"Create an instance of a class by putting parentheses after the class name. Use dot syntax to access the properties and methods of the instance.",
"_____no_output_____"
]
],
[
[
"var shape = Shape()\nshape.numberOfSides = 7\nvar shapeDescription = shape.simpleDescription()",
"_____no_output_____"
]
],
[
[
"This version of the `Shape` class is missing something important: an initializer to set up the class when an instance is created. Use `init` to create one.",
"_____no_output_____"
]
],
[
[
"class NamedShape {\n var numberOfSides: Int = 0\n var name: String\n \n init(name: String) {\n self.name = name\n }\n \n func simpleDescription() -> String {\n return \"A shape with \\(numberOfSides) sides.\"\n }\n}",
"_____no_output_____"
]
],
[
[
"Notice how `self` is used to distinguish the `name` property from the `name` argument to the initializer. The arguments to the initializer are passed like a function call when you create an instance of the class. Every property needs a value assigned—either in its declaration (as with `numberOfSides`) or in the initializer (as with `name`).\n\nUse `deinit` to create a deinitializer if you need to perform some cleanup before the object is deallocated.\n\nSubclasses include their superclass name after their class name, separated by a colon. There is no requirement for classes to subclass any standard root class, so you can include or omit a superclass as needed.\n\nMethods on a subclass that override the superclass’s implementation are marked with `override`—overriding a method by accident, without `override`, is detected by the compiler as an error. The compiler also detects methods with `override` that don’t actually override any method in the superclass.",
"_____no_output_____"
]
],
[
[
"class Square: NamedShape {\n var sideLength: Double\n \n init(sideLength: Double, name: String) {\n self.sideLength = sideLength\n super.init(name: name)\n numberOfSides = 4\n }\n \n func area() -> Double {\n return sideLength * sideLength\n }\n \n override func simpleDescription() -> String {\n return \"A square with sides of length \\(sideLength).\"\n }\n}\nlet test = Square(sideLength: 5.2, name: \"my test square\")\nprint(test.area())\nprint(test.simpleDescription())",
"_____no_output_____"
],
[
"// Experiment:\n// - Make another subclass of `NamedShape` called `Circle` that takes a radius and a name as\n// arguments to its initializer.\n// - Implement an `area()` and a `simpleDescription()` method on the `Circle` class.",
"_____no_output_____"
]
],
[
[
"In addition to simple properties that are stored, properties can have a getter and a setter.",
"_____no_output_____"
]
],
[
[
"class EquilateralTriangle: NamedShape {\n var sideLength: Double = 0.0\n \n init(sideLength: Double, name: String) {\n self.sideLength = sideLength\n super.init(name: name)\n numberOfSides = 3\n }\n \n var perimeter: Double {\n get {\n return 3.0 * sideLength\n }\n set {\n sideLength = newValue / 3.0\n }\n }\n \n override func simpleDescription() -> String {\n return \"An equilateral triangle with sides of length \\(sideLength).\"\n }\n}\nvar triangle = EquilateralTriangle(sideLength: 3.1, name: \"a triangle\")\nprint(triangle.perimeter)\ntriangle.perimeter = 9.9\nprint(triangle.sideLength)",
"_____no_output_____"
]
],
[
[
"In the setter for `perimeter`, the new value has the implicit name `newValue`. You can provide an explicit name in parentheses after `set`.\n\nNotice that the initializer for the `EquilateralTriangle` class has three different steps:\n\n1. Setting the value of properties that the subclass declares.\n\n2. Calling the superclass’s initializer.\n\n3. Changing the value of properties defined by the superclass. Any additional setup work that uses methods, getters, or setters can also be done at this point.\n\nIf you don’t need to compute the property but still need to provide code that is run before and after setting a new value, use `willSet` and `didSet`. The code you provide is run any time the value changes outside of an initializer. For example, the class below ensures that the side length of its triangle is always the same as the side length of its square.",
"_____no_output_____"
]
],
[
[
"class TriangleAndSquare {\n var triangle: EquilateralTriangle {\n willSet {\n square.sideLength = newValue.sideLength\n }\n }\n var square: Square {\n willSet {\n triangle.sideLength = newValue.sideLength\n }\n }\n init(size: Double, name: String) {\n square = Square(sideLength: size, name: name)\n triangle = EquilateralTriangle(sideLength: size, name: name)\n }\n}\nvar triangleAndSquare = TriangleAndSquare(size: 10, name: \"another test shape\")\nprint(triangleAndSquare.square.sideLength)\nprint(triangleAndSquare.triangle.sideLength)\ntriangleAndSquare.square = Square(sideLength: 50, name: \"larger square\")\nprint(triangleAndSquare.triangle.sideLength)",
"_____no_output_____"
]
],
[
[
"When working with optional values, you can write `?` before operations like methods, properties, and subscripting. If the value before the `?` is `nil`, everything after the `?` is ignored and the value of the whole expression is `nil`. Otherwise, the optional value is unwrapped, and everything after the `?` acts on the unwrapped value. In both cases, the value of the whole expression is an optional value.",
"_____no_output_____"
]
],
[
[
"let optionalSquare: Square? = Square(sideLength: 2.5, name: \"optional square\")\nprint(optionalSquare?.sideLength)",
"_____no_output_____"
]
],
[
[
"## Enumerations and Structures\n\nUse `enum` to create an enumeration. Like classes and all other named types, enumerations can have methods associated with them.",
"_____no_output_____"
]
],
[
[
"enum Rank: Int {\n case ace = 1\n case two, three, four, five, six, seven, eight, nine, ten\n case jack, queen, king\n\n func simpleDescription() -> String {\n switch self {\n case .ace:\n return \"ace\"\n case .jack:\n return \"jack\"\n case .queen:\n return \"queen\"\n case .king:\n return \"king\"\n default:\n return String(self.rawValue)\n }\n }\n}\nlet ace = Rank.ace\nprint(ace)\nlet aceRawValue = ace.rawValue\nprint(aceRawValue)",
"_____no_output_____"
],
[
"// Experiment:\n// Write a function that compares two `Rank` values by comparing their raw values.",
"_____no_output_____"
]
],
[
[
"By default, Swift assigns the raw values starting at zero and incrementing by one each time, but you can change this behavior by explicitly specifying values. In the example above, Ace is explicitly given a raw value of `1`, and the rest of the raw values are assigned in order. You can also use strings or floating-point numbers as the raw type of an enumeration. Use the `rawValue` property to access the raw value of an enumeration case.",
"_____no_output_____"
],
[
"Use the `init?(rawValue:)` initializer to make an instance of an enumeration from a raw value. It returns either the enumeration case matching the raw value or `nil` if there is no matching `Rank`.",
"_____no_output_____"
]
],
[
[
"if let convertedRank = Rank(rawValue: 3) {\n let threeDescription = convertedRank.simpleDescription()\n}",
"_____no_output_____"
]
],
[
[
"The case values of an enumeration are actual values, not just another way of writing their raw values. In fact, in cases where there isn’t a meaningful raw value, you don’t have to provide one.",
"_____no_output_____"
]
],
[
[
"enum Suit {\n case spades, hearts, diamonds, clubs\n\n func simpleDescription() -> String {\n switch self {\n case .spades:\n return \"spades\"\n case .hearts:\n return \"hearts\"\n case .diamonds:\n return \"diamonds\"\n case .clubs:\n return \"clubs\"\n }\n }\n}\nlet hearts = Suit.hearts\nlet heartsDescription = hearts.simpleDescription()",
"_____no_output_____"
],
[
"// Experiment:\n// Add a `color()` method to `Suit` that returns \"black\" for spades and clubs, and returns \"red\" for\n// hearts and diamonds.",
"_____no_output_____"
]
],
[
[
"Notice the two ways that the `Hearts` case of the enumeration is referred to above: When assigning a value to the `hearts` constant, the enumeration case `Suit.Hearts` is referred to by its full name because the constant doesn’t have an explicit type specified. Inside the switch, the enumeration case is referred to by the abbreviated form `.Hearts` because the value of `self` is already known to be a suit. You can use the abbreviated form anytime the value’s type is already known.",
"_____no_output_____"
],
[
"If an enumeration has raw values, those values are determined as part of the declaration, which means every instance of a particular enumeration case always has the same raw value. Another choice for enumeration cases is to have values associated with the case—these values are determined when you make the instance, and they can be different for each instance of an enumeration case. You can think of the associated values as behaving like stored properties of the enumeration case instance.\n\nFor example, consider the case of requesting the sunrise and sunset times from a server. The server either responds with the requested information, or it responds with a description of what went wrong.",
"_____no_output_____"
]
],
[
[
"enum ServerResponse {\n case result(String, String)\n case failure(String)\n}\n\nlet success = ServerResponse.result(\"6:00 am\", \"8:09 pm\")\nlet failure = ServerResponse.failure(\"Out of cheese.\")\n\nswitch success {\ncase let .result(sunrise, sunset):\n print(\"Sunrise is at \\(sunrise) and sunset is at \\(sunset).\")\ncase let .failure(message):\n print(\"Failure... \\(message)\")\n}",
"_____no_output_____"
],
[
"// Experiment:\n// Add a third case to `ServerResponse` and to the switch.",
"_____no_output_____"
]
],
[
[
"Notice how the sunrise and sunset times are extracted from the `ServerResponse` value as part of matching the value against the switch cases.",
"_____no_output_____"
],
[
"Use `struct` to create a structure. Structures support many of the same behaviors as classes, including methods and initializers. One of the most important differences between structures and classes is that structures are always copied when they are passed around in your code, but classes are passed by reference.",
"_____no_output_____"
]
],
[
[
"struct Card {\n var rank: Rank\n var suit: Suit\n func simpleDescription() -> String {\n return \"The \\(rank.simpleDescription()) of \\(suit.simpleDescription())\"\n }\n}\nlet threeOfSpades = Card(rank: .three, suit: .spades)\nlet threeOfSpadesDescription = threeOfSpades.simpleDescription()",
"_____no_output_____"
],
[
"// Experiment:\n// Write a function that returns an array containing a full deck of cards, with one card of each\n// combination of rank and suit.",
"_____no_output_____"
]
],
[
[
"## Protocols and Extensions\n\nUse `protocol` to declare a protocol.",
"_____no_output_____"
]
],
[
[
"protocol ExampleProtocol {\n var simpleDescription: String { get }\n mutating func adjust()\n}",
"_____no_output_____"
]
],
[
[
"Classes, enumerations, and structs can all adopt protocols.",
"_____no_output_____"
]
],
[
[
"class SimpleClass: ExampleProtocol {\n var simpleDescription: String = \"A very simple class.\"\n var anotherProperty: Int = 69105\n func adjust() {\n simpleDescription += \" Now 100% adjusted.\"\n }\n}\nvar a = SimpleClass()\na.adjust()\nlet aDescription = a.simpleDescription\n \nstruct SimpleStructure: ExampleProtocol {\n var simpleDescription: String = \"A simple structure\"\n mutating func adjust() {\n simpleDescription += \" (adjusted)\"\n }\n}\nvar b = SimpleStructure()\nprint(b.adjust())\nprint(b.simpleDescription)",
"_____no_output_____"
],
[
"// Experiment:\n// Add another requirement to `ExampleProtocol`.\n// What changes do you need to make to `SimpleClass` and `SimpleStructure` so that they still\n// conform to the protocol?",
"_____no_output_____"
]
],
[
[
"Notice the use of the `mutating` keyword in the declaration of `SimpleStructure` to mark a method that modifies the structure. The declaration of `SimpleClass` doesn’t need any of its methods marked as mutating because methods on a class can always modify the class.\n\nUse `extension` to add functionality to an existing type, such as new methods and computed properties. You can use an extension to add protocol conformance to a type that is declared elsewhere, or even to a type that you imported from a library or framework.",
"_____no_output_____"
]
],
[
[
"extension Int: ExampleProtocol {\n public var simpleDescription: String {\n return \"The number \\(self)\"\n }\n public mutating func adjust() {\n self += 42\n }\n}\nprint(7.simpleDescription)",
"_____no_output_____"
],
[
"// Experiment:\n// Write an extension for the `Double` type that adds an `absoluteValue` property.",
"_____no_output_____"
]
],
[
[
"You can use a protocol name just like any other named type—for example, to create a collection of objects that have different types but that all conform to a single protocol. When you work with values whose type is a protocol type, methods outside the protocol definition are not available.",
"_____no_output_____"
]
],
[
[
"let protocolValue: ExampleProtocol = a\nprint(protocolValue.simpleDescription)",
"_____no_output_____"
],
[
"// Uncomment to see the error.\n// protocolValue.anotherProperty",
"_____no_output_____"
]
],
[
[
"Even though the variable `protocolValue` has a runtime type of `SimpleClass`, the compiler treats it as the given type of `ExampleProtocol`. This means that you can't accidentally access methods or properties that the class implements in addition to its protocol conformance.",
"_____no_output_____"
],
[
"## Error Handling\n\nYou represent errors using any type that adopts the `Error` protocol.",
"_____no_output_____"
]
],
[
[
"enum PrinterError: Error {\n case outOfPaper\n case noToner\n case onFire\n}",
"_____no_output_____"
]
],
[
[
"Use `throw` to throw an error and `throws` to mark a function that can throw an error. If you throw an error in a function, the function returns immediately and the code that called the function handles the error.",
"_____no_output_____"
]
],
[
[
"func send(job: Int, toPrinter printerName: String) throws -> String {\n if printerName == \"Never Has Toner\" {\n throw PrinterError.noToner\n }\n return \"Job sent\"\n}",
"_____no_output_____"
]
],
[
[
"There are several ways to handle errors. One way is to use `do-catch`. Inside the `do` block, you mark code that can throw an error by writing try in front of it. Inside the `catch` block, the error is automatically given the name `error` unless you give it a different name.",
"_____no_output_____"
]
],
[
[
"do {\n let printerResponse = try send(job: 1040, toPrinter: \"Bi Sheng\")\n print(printerResponse)\n} catch {\n print(error)\n}",
"_____no_output_____"
],
[
"// Experiment:\n// Change the printer name to `\"Never Has Toner\"`, so that the `send(job:toPrinter:)` function\n// throws an error.",
"_____no_output_____"
]
],
[
[
"You can provide multiple `catch` blocks that handle specific errors. You write a pattern after `catch` just as you do after `case` in a switch.",
"_____no_output_____"
]
],
[
[
"do {\n let printerResponse = try send(job: 1440, toPrinter: \"Gutenberg\")\n print(printerResponse)\n} catch PrinterError.onFire {\n print(\"I'll just put this over here, with the rest of the fire.\")\n} catch let printerError as PrinterError {\n print(\"Printer error: \\(printerError).\")\n} catch {\n print(error)\n}",
"_____no_output_____"
],
[
"// Experiment:\n// Add code to throw an error inside the `do` block.\n// What kind of error do you need to throw so that the error is handled by the first `catch` block?\n// What about the second and third blocks?",
"_____no_output_____"
]
],
[
[
"Another way to handle errors is to use `try?` to convert the result to an optional. If the function throws an error, the specific error is discarded and the result is `nil`. Otherwise, the result is an optional containing the value that the function returned.",
"_____no_output_____"
]
],
[
[
"let printerSuccess = try? send(job: 1884, toPrinter: \"Mergenthaler\")\nlet printerFailure = try? send(job: 1885, toPrinter: \"Never Has Toner\")",
"_____no_output_____"
]
],
[
[
"Use `defer` to write a block of code that is executed after all other code in the function, just before the function returns. The code is executed regardless of whether the function throws an error. You can use `defer` to write setup and cleanup code next to each other, even though they need to be executed at different times.",
"_____no_output_____"
]
],
[
[
"var fridgeIsOpen = false\nlet fridgeContent = [\"milk\", \"eggs\", \"leftovers\"]\n\nfunc fridgeContains(_ food: String) -> Bool {\n fridgeIsOpen = true\n defer {\n fridgeIsOpen = false\n }\n\n let result = fridgeContent.contains(food)\n return result\n}\nprint(fridgeContains(\"banana\"))\nprint(fridgeIsOpen)",
"_____no_output_____"
]
],
[
[
"## Generics\n\nWrite a name inside angle brackets to make a generic function or type.",
"_____no_output_____"
]
],
[
[
"func makeArray<Item>(repeating item: Item, numberOfTimes: Int) -> [Item] {\n var result = [Item]()\n for _ in 0..<numberOfTimes {\n result.append(item)\n }\n return result\n}\nprint(makeArray(repeating: \"knock\", numberOfTimes: 4))",
"_____no_output_____"
]
],
[
[
"You can make generic forms of functions and methods, as well as classes, enumerations, and structures.",
"_____no_output_____"
]
],
[
[
"// Reimplement the Swift standard library's optional type\nenum OptionalValue<Wrapped> {\n case none\n case some(Wrapped)\n}\nvar possibleInteger: OptionalValue<Int> = .none\npossibleInteger = .some(100)\nprint(possibleInteger)",
"_____no_output_____"
]
],
[
[
"Use `where` after the type name to specify a list of requirements — for example, to require the type to implement a protocol, to require two types to be the same, or to require a class to have a particular superclass.",
"_____no_output_____"
]
],
[
[
"func anyCommonElements<T: Sequence, U: Sequence>(_ lhs: T, _ rhs: U) -> Bool\n where T.Element: Equatable, T.Element == U.Element\n{\n for lhsItem in lhs {\n for rhsItem in rhs {\n if lhsItem == rhsItem {\n return true\n }\n }\n }\n return false\n}\nprint(anyCommonElements([1, 2, 3], [3]))",
"_____no_output_____"
]
],
[
[
"Writing `<T: Equatable>` is the same as writing `<T> ... where T: Equatable`.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ececece9dac051e77f71fedfb176258b8092690b | 22,239 | ipynb | Jupyter Notebook | notebooks/combine_data.ipynb | deepdialog/tfseg | f10e27e234b53fda52cd672489f7a3c0ce81a2f6 | [
"MIT"
] | 1 | 2021-11-09T08:22:19.000Z | 2021-11-09T08:22:19.000Z | notebooks/combine_data.ipynb | deepdialog/tfseg | f10e27e234b53fda52cd672489f7a3c0ce81a2f6 | [
"MIT"
] | null | null | null | notebooks/combine_data.ipynb | deepdialog/tfseg | f10e27e234b53fda52cd672489f7a3c0ce81a2f6 | [
"MIT"
] | 1 | 2022-01-24T10:05:15.000Z | 2022-01-24T10:05:15.000Z | 21.824338 | 92 | 0.420567 | [
[
[
"import os\nimport re\nfrom collections import Counter\nfrom tqdm import tqdm",
"_____no_output_____"
],
[
"def compose1(paths):\n if isinstance(paths, str):\n paths = [paths]\n def to_bi(line):\n segs = re.findall(r'([^/]+)/[a-zA-Z]+\\s+', line)\n x = []\n y = []\n for seg in segs:\n seg = list(seg)\n tag = ['I'] * len(seg)\n tag[0] = 'B'\n\n # if len(tag) == 1:\n # tag[0] = 'S'\n # elif len(tag) == 2:\n # tag[1] = 'E'\n # elif len(tag) >= 3:\n # tag[-1] = 'E'\n\n x += seg\n y += tag\n return x, y\n X, Y = [], []\n for path in paths:\n data = open(path).read()\n lines = data.split('\\n')\n for line in tqdm(lines):\n x, y = to_bi(line)\n X.append(x)\n Y.append(y)\n print(len(X), len(Y))\n return X, Y",
"_____no_output_____"
],
[
"def compose2(paths):\n if isinstance(paths, str):\n paths = [paths]\n def to_bi(line):\n segs = re.split(r'\\s+', line)\n x = []\n y = []\n for seg in segs:\n if seg:\n seg = list(seg)\n tag = ['I'] * len(seg)\n tag[0] = 'B'\n\n# if len(tag) == 1:\n# tag[0] = 'S'\n# elif len(tag) == 2:\n# tag[1] = 'E'\n# elif len(tag) >= 3:\n# tag[-1] = 'E'\n\n x += seg\n y += tag\n return x, y\n X, Y = [], []\n for path in paths:\n data = open(path).read()\n lines = data.split('\\n')\n for line in tqdm(lines):\n x, y = to_bi(line)\n X.append(x)\n Y.append(y)\n print(len(X), len(Y))\n return X, Y",
"_____no_output_____"
],
[
"def compose3(paths):\n if isinstance(paths, str):\n paths = [paths]\n def to_bi(line):\n segs = re.split(r'\\s+', line)\n x = []\n y = []\n for seg in segs:\n if seg:\n seg = list(seg)\n tag = ['I'] * len(seg)\n tag[0] = 'B'\n\n# if len(tag) == 1:\n# tag[0] = 'S'\n# elif len(tag) == 2:\n# tag[1] = 'E'\n# elif len(tag) >= 3:\n# tag[-1] = 'E'\n\n x += seg\n y += tag\n return x, y\n X, Y = [], []\n for path in paths:\n data = open(path).read()\n lines = data.split('\\n\\n')\n for line in tqdm(lines):\n line = [x.split('\\t')[1] for x in line.split('\\n') if '\\t' in x]\n line = ' '.join(line)\n if line:\n x, y = to_bi(line)\n X.append(x)\n Y.append(y)\n print(len(X), len(Y))\n return X, Y",
"_____no_output_____"
],
[
"def compose4(paths):\n if isinstance(paths, str):\n paths = [paths]\n def to_bi(line):\n segs = re.findall(r'([^/_]+)_[a-zA-Z]+\\s+', line)\n x = []\n y = []\n for seg in segs:\n seg = list(seg)\n tag = ['I'] * len(seg)\n tag[0] = 'B'\n\n# if len(tag) == 1:\n# tag[0] = 'S'\n# elif len(tag) == 2:\n# tag[1] = 'E'\n# elif len(tag) >= 3:\n# tag[-1] = 'E'\n\n x += seg\n y += tag\n return x, y\n X, Y = [], []\n for path in paths:\n data = open(path).read()\n lines = data.split('\\n')\n for line in tqdm(lines):\n x, y = to_bi(line)\n X.append(x)\n Y.append(y)\n print(len(X), len(Y))\n return X, Y",
"_____no_output_____"
],
[
"!ls '../multi-criteria-cws/data/other/cnc/'",
"dev.txt test.txt train.txt\r\n"
],
[
"cnc_data = compose1([\n '../multi-criteria-cws/data/other/cnc/train.txt',\n '../multi-criteria-cws/data/other/cnc/dev.txt',\n # '../multi-criteria-cws/data/other/cnc/test.txt'\n])",
"100%|██████████| 207002/207002 [00:04<00:00, 43899.93it/s]\n100%|██████████| 25876/25876 [00:00<00:00, 33378.53it/s]"
],
[
"!ls '../multi-criteria-cws/data/other/ctb/'",
"ctb6.dev.seg ctb6.test.seg ctb6.train.seg\r\n"
],
[
"ctb_data = compose2([\n '../multi-criteria-cws/data/other/ctb/ctb6.train.seg',\n '../multi-criteria-cws/data/other/ctb/ctb6.dev.seg',\n # '../multi-criteria-cws/data/other/ctb/ctb6.test.seg'\n])",
"100%|██████████| 24417/24417 [00:00<00:00, 46550.44it/s]\n100%|██████████| 1905/1905 [00:00<00:00, 54196.96it/s]"
],
[
"!ls '../multi-criteria-cws/data/other/sxu/'",
"test.txt train.txt\r\n"
],
[
"sxu_data = compose2([\n '../multi-criteria-cws/data/other/sxu/train.txt',\n # '../multi-criteria-cws/data/other/sxu/test.txt'\n])",
"100%|██████████| 17116/17116 [00:00<00:00, 42372.56it/s]"
],
[
"!ls '../multi-criteria-cws/data/other/udc/'",
"dev.conll test.conll train.conll\r\n"
],
[
"udc_data = compose3([\n '../multi-criteria-cws/data/other/udc/train.conll',\n '../multi-criteria-cws/data/other/udc/dev.conll',\n # '../multi-criteria-cws/data/other/udc/test.conll'\n])",
"100%|██████████| 3998/3998 [00:00<00:00, 32814.30it/s]\n100%|██████████| 501/501 [00:00<00:00, 34442.09it/s]"
],
[
"!ls '../multi-criteria-cws/data/other/wtb/'",
"dev.conll README.txt test.conll train.conll\r\n"
],
[
"wtb_data = compose3([\n '../multi-criteria-cws/data/other/wtb/train.conll',\n '../multi-criteria-cws/data/other/wtb/dev.conll',\n # '../multi-criteria-cws/data/other/wtb/test.conll'\n])",
"100%|██████████| 814/814 [00:00<00:00, 35901.53it/s]\n100%|██████████| 96/96 [00:00<00:00, 40974.17it/s]"
],
[
"!ls '../multi-criteria-cws/data/other/zx/'",
"dev.zhuxian.wordpos test.zhuxian.wordpos train.zhuxian.wordpos\r\n"
],
[
"zx_data = compose4([\n '../multi-criteria-cws/data/other/zx/train.zhuxian.wordpos',\n '../multi-criteria-cws/data/other/zx/dev.zhuxian.wordpos',\n # '../multi-criteria-cws/data/other/zx/test.zhuxian.wordpos'\n])",
"100%|██████████| 2374/2374 [00:00<00:00, 43368.31it/s]\n100%|██████████| 789/789 [00:00<00:00, 57174.30it/s]"
],
[
"!ls '../multi-criteria-cws/data/sighan2005/'",
"as_test_gold.utf8 cityu_test_gold.utf8 msr_test_gold.utf8 pku_test_gold.utf8\r\nas_training.utf8 cityu_training.utf8\t msr_training.utf8 pku_training.utf8\r\n"
],
[
"as_data = compose2([\n '../multi-criteria-cws/data/sighan2005/as_training.utf8',\n # '../multi-criteria-cws/data/sighan2005/as_test_gold.utf8'\n])",
"100%|██████████| 708954/708954 [00:08<00:00, 88074.38it/s] "
],
[
"!ls '../multi-criteria-cws/data/sighan2005/'",
"as_test_gold.utf8 cityu_test_gold.utf8 msr_test_gold.utf8 pku_test_gold.utf8\r\nas_training.utf8 cityu_training.utf8\t msr_training.utf8 pku_training.utf8\r\n"
],
[
"cityu_data = compose2([\n '../multi-criteria-cws/data/sighan2005/cityu_training.utf8',\n # '../multi-criteria-cws/data/sighan2005/cityu_test_gold.utf8'\n])",
"100%|██████████| 53020/53020 [00:01<00:00, 46528.82it/s]"
],
[
"!ls '../multi-criteria-cws/data/sighan2005/'",
"as_test_gold.utf8 cityu_test_gold.utf8 msr_test_gold.utf8 pku_test_gold.utf8\r\nas_training.utf8 cityu_training.utf8\t msr_training.utf8 pku_training.utf8\r\n"
],
[
"msr_data = compose2([\n '../multi-criteria-cws/data/sighan2005/msr_training.utf8',\n # '../multi-criteria-cws/data/sighan2005/msr_test_gold.utf8'\n])",
"100%|██████████| 86925/86925 [00:01<00:00, 46669.34it/s]"
],
[
"!ls '../multi-criteria-cws/data/sighan2005/'",
"as_test_gold.utf8 cityu_test_gold.utf8 msr_test_gold.utf8 pku_test_gold.utf8\r\nas_training.utf8 cityu_training.utf8\t msr_training.utf8 pku_training.utf8\r\n"
],
[
"pku_data = compose2([\n '../multi-criteria-cws/data/sighan2005/pku_training.utf8',\n # '../multi-criteria-cws/data/sighan2005/pku_test_gold.utf8'\n])",
"100%|██████████| 19057/19057 [00:00<00:00, 22718.62it/s]"
],
[
"zhyw = '../chn_segment_data/out.txt'",
"_____no_output_____"
],
[
"zhyw_data = compose1(zhyw)",
"100%|██████████| 616700/616700 [00:14<00:00, 42970.07it/s]"
],
[
"!ls '../OpenCorpus/ctb8.0/pos/'",
"dev.txt README.md test.txt train.txt\r\n"
],
[
"ctb8_data = compose1([\n '../OpenCorpus/ctb8.0/pos/train.txt',\n '../OpenCorpus/ctb8.0/pos/test.txt',\n # '../OpenCorpus/ctb8.0/pos/dev.txt'\n])",
"100%|██████████| 55680/55680 [00:00<00:00, 57327.48it/s]\n100%|██████████| 7775/7775 [00:00<00:00, 60790.41it/s]"
],
[
"data_sets = [\n cnc_data,\n ctb_data,\n sxu_data,\n udc_data,\n wtb_data,\n zx_data,\n as_data,\n cityu_data,\n msr_data,\n pku_data,\n zhyw_data,\n ctb8_data\n]",
"_____no_output_____"
],
[
"xy = {}\nfor d in data_sets:\n x, y = d\n for xx, yy in tqdm(zip(x, y)):\n if len(xx) <= 0:\n continue\n xx = tuple(xx)\n if xx not in xy:\n xy[xx] = yy\n else:\n b_old = Counter(xy[xx])['B']\n b_new = Counter(yy)['B']\n if b_old <= b_new:\n pass\n else:\n xy[xx] = yy",
"232878it [00:00, 268750.81it/s]\n26322it [00:00, 211621.42it/s]\n17116it [00:00, 203476.34it/s]\n4497it [00:00, 252146.05it/s]\n908it [00:00, 296768.33it/s]\n3163it [00:00, 249418.75it/s]\n708954it [00:01, 439042.57it/s]\n53020it [00:00, 241070.51it/s]\n86925it [00:00, 238438.19it/s]\n19057it [00:00, 123895.95it/s]\n616700it [00:04, 135197.69it/s]\n63455it [00:00, 162700.61it/s]\n"
],
[
"print(len(xy))",
"1513223\n"
],
[
"X, Y = [], []\nfor xx, yy in xy.items():\n X.append(xx)\n Y.append(yy)",
"_____no_output_____"
],
[
"print(\n sum([len(x) for x in X])\n)",
"42740494\n"
],
[
"print(\n sum([len(x) for x in Y])\n)",
"42740494\n"
],
[
"for x, y in tqdm(zip(X, Y)):\n assert len(x) == len(y)",
"1513223it [00:00, 2320490.44it/s]\n"
],
[
"import pickle",
"_____no_output_____"
],
[
"with open('x.pkl', 'wb') as fp:\n pickle.dump(X, fp)\nwith open('y.pkl', 'wb') as fp:\n pickle.dump(Y, fp)",
"_____no_output_____"
],
[
"!du -sh x.pkl\n!du -sh y.pkl",
"535M\tx.pkl\n94M\ty.pkl\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ececedcca32e3e06b3795796de97dbdb143f860c | 838 | ipynb | Jupyter Notebook | HelloGithub.ipynb | piotr14prywatny/dw_matrix | 8f655edb7e65b89de333864341d92a97666ecae7 | [
"MIT"
] | null | null | null | HelloGithub.ipynb | piotr14prywatny/dw_matrix | 8f655edb7e65b89de333864341d92a97666ecae7 | [
"MIT"
] | null | null | null | HelloGithub.ipynb | piotr14prywatny/dw_matrix | 8f655edb7e65b89de333864341d92a97666ecae7 | [
"MIT"
] | null | null | null | 838 | 838 | 0.688544 | [
[
[
"print(\"Hello Github\")",
"Hello Github\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
ececf953a8d0e3e3b5702495120e29ae76b769e4 | 54,729 | ipynb | Jupyter Notebook | notebooks/skipgram_torch.ipynb | ashishpapanai/pyprobml | 8eadb39af7ac4575f9d0b661168eb0e876c33dfc | [
"MIT"
] | null | null | null | notebooks/skipgram_torch.ipynb | ashishpapanai/pyprobml | 8eadb39af7ac4575f9d0b661168eb0e876c33dfc | [
"MIT"
] | 1 | 2021-04-19T12:25:26.000Z | 2021-04-19T12:25:26.000Z | notebooks/skipgram_torch.ipynb | ashishpapanai/pyprobml | 8eadb39af7ac4575f9d0b661168eb0e876c33dfc | [
"MIT"
] | null | null | null | 46.736977 | 18,379 | 0.523507 | [
[
[
"<a href=\"https://colab.research.google.com/github/probml/pyprobml/blob/master/notebooks/skipgram_torch.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"\n# Learning word emebddings using skipgram with negative sampling.\n\nBased on D2L 14.3 http://d2l.ai/chapter_natural-language-processing-pretraining/word-embedding-dataset.html and 14.4 of http://d2l.ai/chapter_natural-language-processing-pretraining/word2vec-pretraining.html.\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nnp.random.seed(seed=1)\nimport math\n\nimport os\nimport random\nimport torch\nfrom torch import nn\nfrom torch.nn import functional as F\n\n!mkdir figures # for saving plots\n\n!wget https://raw.githubusercontent.com/d2l-ai/d2l-en/master/d2l/torch.py -q -O d2l.py\nimport d2l",
"mkdir: cannot create directory ‘figures’: File exists\n"
]
],
[
[
"# Data\n\nWe use the Penn Tree Bank (PTB), which is a small but commonly-used corpus derived from the Wall Stree Journal.\n",
"_____no_output_____"
]
],
[
[
"d2l.DATA_HUB['ptb'] = (d2l.DATA_URL + 'ptb.zip',\n '319d85e578af0cdc590547f26231e4e31cdf1e42')\n\n#@save\ndef read_ptb():\n data_dir = d2l.download_extract('ptb')\n with open(os.path.join(data_dir, 'ptb.train.txt')) as f:\n raw_text = f.read()\n return [line.split() for line in raw_text.split('\\n')]\n\nsentences = read_ptb()\nf'# sentences: {len(sentences)}'",
"_____no_output_____"
]
],
[
[
"We make a vocabulary, replacing any word that occurs less than 10 times with unk.",
"_____no_output_____"
]
],
[
[
"vocab = d2l.Vocab(sentences, min_freq=10)\nf'vocab size: {len(vocab)}'",
"_____no_output_____"
]
],
[
[
"Mikolov suggested keeping word $w$ with probability\n$$\n\\sqrt{\\theta / f(w)}\n$$\nwhere $\\theta=10^{-4}$ is a threshold, and $f(w)=N(w)/N$ is the empirical frequency of word $w$.",
"_____no_output_____"
]
],
[
[
"def subsampling(sentences, vocab):\n # Map low frequency words into <unk>\n sentences = [[vocab.idx_to_token[vocab[tk]] for tk in line]\n for line in sentences]\n # Count the frequency for each word\n counter = d2l.count_corpus(sentences)\n num_tokens = sum(counter.values())\n\n # Return True if to keep this token during subsampling\n def keep(token):\n return (random.uniform(0, 1) < math.sqrt(\n 1e-4 / counter[token] * num_tokens))\n\n # Now do the subsampling\n return [[tk for tk in line if keep(tk)] for line in sentences]\n\nsubsampled = subsampling(sentences, vocab)",
"_____no_output_____"
]
],
[
[
"We compare the frequency of certain common and rare words in the original and subsampled data below.",
"_____no_output_____"
]
],
[
[
"def compare_counts(token):\n return (f'# of \"{token}\": '\n f'before={sum([line.count(token) for line in sentences])}, '\n f'after={sum([line.count(token) for line in subsampled])}')\n\nprint(compare_counts('the'))\nprint(compare_counts('join'))\n\n",
"# of \"the\": before=50770, after=2169\n# of \"join\": before=45, after=45\n"
]
],
[
[
"Let's tokenize the subsampled data.\n",
"_____no_output_____"
]
],
[
[
"corpus = [vocab[line] for line in subsampled]\n\nprint(corpus[0:3])\n",
"[[0], [392, 2115, 145, 18, 274], [140, 5277, 3054, 1580]]\n"
]
],
[
[
"## Extracting central target words and their contexts\n\nWe randomly sample a context length for each central word, up to some maximum length, and then extract all the context words as a list of lists.",
"_____no_output_____"
]
],
[
[
"def get_centers_and_contexts(corpus, max_window_size):\n centers, contexts = [], []\n for line in corpus:\n # Each sentence needs at least 2 words to form a \"central target word\n # - context word\" pair\n if len(line) < 2:\n continue\n centers += line\n for i in range(len(line)): # Context window centered at i\n window_size = random.randint(1, max_window_size)\n indices = list(\n range(max(0, i - window_size),\n min(len(line), i + 1 + window_size)))\n # Exclude the central target word from the context words\n indices.remove(i)\n contexts.append([line[idx] for idx in indices])\n return centers, contexts",
"_____no_output_____"
]
],
[
[
"Example. Suppose we have a corpus with 2 sentences of length 7 and 3, and we use a max context of size 2. Here are the centers and contexts.",
"_____no_output_____"
]
],
[
[
"tiny_dataset = [list(range(7)), list(range(7, 10))]\nprint('dataset', tiny_dataset)\nfor center, context in zip(*get_centers_and_contexts(tiny_dataset, 2)):\n print('center', center, 'has contexts', context)",
"dataset [[0, 1, 2, 3, 4, 5, 6], [7, 8, 9]]\ncenter 0 has contexts [1]\ncenter 1 has contexts [0, 2]\ncenter 2 has contexts [0, 1, 3, 4]\ncenter 3 has contexts [2, 4]\ncenter 4 has contexts [2, 3, 5, 6]\ncenter 5 has contexts [4, 6]\ncenter 6 has contexts [4, 5]\ncenter 7 has contexts [8, 9]\ncenter 8 has contexts [7, 9]\ncenter 9 has contexts [7, 8]\n"
]
],
[
[
"Extract context for the full dataset.",
"_____no_output_____"
]
],
[
[
"all_centers, all_contexts = get_centers_and_contexts(corpus, 5)\nf'# center-context pairs: {len(all_centers)}'",
"_____no_output_____"
]
],
[
[
"## Negative sampling\n\nFor speed, we define a sampling class that pre-computes 10,000 random indices from the weighted distribution, using a single call to `random.choices`, and then sequentially returns elements of this list. If we reach the end of the cache, we refill it.",
"_____no_output_____"
]
],
[
[
"class RandomGenerator:\n \"\"\"Draw a random int in [0, n] according to n sampling weights.\"\"\"\n def __init__(self, sampling_weights):\n self.population = list(range(len(sampling_weights)))\n self.sampling_weights = sampling_weights\n self.candidates = []\n self.i = 0\n\n def draw(self):\n if self.i == len(self.candidates):\n self.candidates = random.choices(self.population,\n self.sampling_weights, k=10000)\n self.i = 0\n self.i += 1\n return self.candidates[self.i - 1]",
"_____no_output_____"
]
],
[
[
"Example.",
"_____no_output_____"
]
],
[
[
"\ngenerator = RandomGenerator([2, 3, 4])\n[generator.draw() for _ in range(10)]",
"_____no_output_____"
]
],
[
[
"Now we generate $K$ negatives for each context. These are drawn from $p(w) \\propto \\text{freq}(w)^{0.75}$.",
"_____no_output_____"
]
],
[
[
"def get_negatives(all_contexts, corpus, K):\n counter = d2l.count_corpus(corpus)\n sampling_weights = [counter[i]**0.75 for i in range(len(counter))]\n all_negatives, generator = [], RandomGenerator(sampling_weights)\n for contexts in all_contexts:\n negatives = []\n while len(negatives) < len(contexts) * K:\n neg = generator.draw()\n # Noise words cannot be context words\n if neg not in contexts:\n negatives.append(neg)\n all_negatives.append(negatives)\n return all_negatives\n\nall_negatives = get_negatives(all_contexts, corpus, 5)",
"_____no_output_____"
]
],
[
[
"## Minibatching\n\nSuppose the $i$'th central word has $n_i$ contexts and $m_i$ noise words.\nSince $n_i+m_i$ might be different for each $i$ (due to edge effects), the minibatch will be ragged. To fix this, we pad to a maximum length $L$, and then create a validity mask of length $L$, where 0 means invalid location (to be ignored when computing the loss) and 1 means valid location. We assign the label vector to have $n_i$ 1's and $L-n_i$ 0's. (Some of these labels will be masked out.)\n",
"_____no_output_____"
]
],
[
[
"def batchify(data):\n max_len = max(len(c) + len(n) for _, c, n in data)\n centers, contexts_negatives, masks, labels = [], [], [], []\n for center, context, negative in data:\n cur_len = len(context) + len(negative)\n centers += [center]\n contexts_negatives += [context + negative + [0] * (max_len - cur_len)]\n masks += [[1] * cur_len + [0] * (max_len - cur_len)]\n labels += [[1] * len(context) + [0] * (max_len - len(context))]\n return (torch.tensor(centers).reshape(\n (-1, 1)), torch.tensor(contexts_negatives), torch.tensor(masks),\n torch.tensor(labels))",
"_____no_output_____"
]
],
[
[
"Example. We make a ragged minibatch with 2 examples, and then pad them to a standard size.",
"_____no_output_____"
]
],
[
[
"x_1 = (1, [2, 2], [3, 3, 3, 3])\nx_2 = (1, [2, 2, 2], [3, 3])\nbatch = batchify((x_1, x_2))\n\nnames = ['centers', 'contexts_negatives', 'masks', 'labels']\nfor name, data in zip(names, batch):\n print(name, '=', data)",
"centers = tensor([[1],\n [1]])\ncontexts_negatives = tensor([[2, 2, 3, 3, 3, 3],\n [2, 2, 2, 3, 3, 0]])\nmasks = tensor([[1, 1, 1, 1, 1, 1],\n [1, 1, 1, 1, 1, 0]])\nlabels = tensor([[1, 1, 0, 0, 0, 0],\n [1, 1, 1, 0, 0, 0]])\n"
]
],
[
[
"## Dataloader\n\nNow we put it altogether.",
"_____no_output_____"
]
],
[
[
"def load_data_ptb(batch_size, max_window_size, num_noise_words):\n num_workers = d2l.get_dataloader_workers()\n sentences = read_ptb()\n vocab = d2l.Vocab(sentences, min_freq=10)\n subsampled = subsampling(sentences, vocab)\n corpus = [vocab[line] for line in subsampled]\n all_centers, all_contexts = get_centers_and_contexts(\n corpus, max_window_size)\n all_negatives = get_negatives(all_contexts, corpus, num_noise_words)\n\n class PTBDataset(torch.utils.data.Dataset):\n def __init__(self, centers, contexts, negatives):\n assert len(centers) == len(contexts) == len(negatives)\n self.centers = centers\n self.contexts = contexts\n self.negatives = negatives\n\n def __getitem__(self, index):\n return (self.centers[index], self.contexts[index],\n self.negatives[index])\n\n def __len__(self):\n return len(self.centers)\n\n dataset = PTBDataset(all_centers, all_contexts, all_negatives)\n\n data_iter = torch.utils.data.DataLoader(dataset, batch_size, shuffle=True,\n collate_fn=batchify,\n num_workers=num_workers)\n return data_iter, vocab",
"_____no_output_____"
]
],
[
[
"Let's print the first minibatch.",
"_____no_output_____"
]
],
[
[
"data_iter, vocab = load_data_ptb(512, 5, 5)\nfor batch in data_iter:\n for name, data in zip(names, batch):\n print(name, 'shape:', data.shape)\n break",
"/usr/local/lib/python3.7/dist-packages/torch/utils/data/dataloader.py:477: UserWarning: This DataLoader will create 4 worker processes in total. Our suggested max number of worker in current system is 2, which is smaller than what this DataLoader is going to create. Please be aware that excessive worker creation might get DataLoader running slow or even freeze, lower the worker number to avoid potential slowness/freeze if necessary.\n cpuset_checked))\n"
],
[
"batch_size, max_window_size, num_noise_words = 512, 5, 5\ndata_iter, vocab = d2l.load_data_ptb(batch_size, max_window_size,\n num_noise_words)",
"/usr/local/lib/python3.7/dist-packages/torch/utils/data/dataloader.py:477: UserWarning: This DataLoader will create 4 worker processes in total. Our suggested max number of worker in current system is 2, which is smaller than what this DataLoader is going to create. Please be aware that excessive worker creation might get DataLoader running slow or even freeze, lower the worker number to avoid potential slowness/freeze if necessary.\n cpuset_checked))\n"
]
],
[
[
"# Model\n\nThe model just has 2 embedding matrices, $U$ and $V$. The core computation is computing the logits, as shown below.\nThe center variable has the shape (batch size, 1), while the contexts_and_negatives variable has the shape (batch size, max_len). These get embedded into size $(B,1,E)$ and $(B,L,E)$. We permute the latter to $(B,E,L)$ and use [batch matrix multiplication](https://pytorch.org/docs/stable/generated/torch.bmm.html) to get $(B,1,L)$ matrix of inner products between each center's embedding and each context's embedding.\n",
"_____no_output_____"
]
],
[
[
"def skip_gram(center, contexts_and_negatives, embed_v, embed_u):\n v = embed_v(center)\n u = embed_u(contexts_and_negatives)\n pred = torch.bmm(v, u.permute(0, 2, 1))\n return pred",
"_____no_output_____"
]
],
[
[
"Example. Assume the vocab size is 20 and we use $E=4$ embedding dimensions.\nWe compute the logits for a minibatch of $B=2$ sequences, with max length $L=4$.",
"_____no_output_____"
]
],
[
[
"embed = nn.Embedding(num_embeddings=20, embedding_dim=4)\nprint(f'Parameter embedding_weight ({embed.weight.shape}, '\n 'dtype={embed.weight.dtype})')\n\nskip_gram(torch.ones((2, 1), dtype=torch.long),\n torch.ones((2, 4), dtype=torch.long), embed, embed).shape",
"Parameter embedding_weight (torch.Size([20, 4]), dtype={embed.weight.dtype})\n[torch.Size([2, 1, 4]), torch.Size([2, 4, 4]), torch.Size([2, 1, 4])]\n"
]
],
[
[
"# Loss\n\nWe use masked binary cross entropy loss.\n",
"_____no_output_____"
]
],
[
[
"class SigmoidBCELoss(nn.Module):\n \"BCEWithLogitLoss with masking on call.\"\n\n def __init__(self):\n super().__init__()\n\n def forward(self, inputs, target, mask=None):\n out = nn.functional.binary_cross_entropy_with_logits(\n inputs, target, weight=mask, reduction=\"none\")\n return out.mean(dim=1)\n\nloss = SigmoidBCELoss()",
"_____no_output_____"
]
],
[
[
"Different masks can lead to different results.",
"_____no_output_____"
]
],
[
[
"pred = torch.tensor([[.5] * 4] * 2)\nlabel = torch.tensor([[1., 0., 1., 0.]] * 2)\nmask = torch.tensor([[1, 1, 1, 1], [1, 1, 0, 0]])\nloss(pred, label, mask)",
"_____no_output_____"
]
],
[
[
"If we normalize by the number of valid masked entries, then predictions with the same per-token accuracy will score the same.",
"_____no_output_____"
]
],
[
[
"loss(pred, label, mask) / mask.sum(axis=1) * mask.shape[1]",
"_____no_output_____"
]
],
[
[
"# Training",
"_____no_output_____"
]
],
[
[
"embed_size = 100\nnet = nn.Sequential(\n nn.Embedding(num_embeddings=len(vocab), embedding_dim=embed_size),\n nn.Embedding(num_embeddings=len(vocab), embedding_dim=embed_size))\n",
"_____no_output_____"
],
[
"def train(net, data_iter, lr, num_epochs, device=d2l.try_gpu()):\n def init_weights(m):\n if type(m) == nn.Embedding:\n nn.init.xavier_uniform_(m.weight)\n\n net.apply(init_weights)\n net = net.to(device)\n optimizer = torch.optim.Adam(net.parameters(), lr=lr)\n animator = d2l.Animator(xlabel='epoch', ylabel='loss',\n xlim=[1, num_epochs])\n metric = d2l.Accumulator(2) # Sum of losses, no. of tokens\n for epoch in range(num_epochs):\n timer, num_batches = d2l.Timer(), len(data_iter)\n for i, batch in enumerate(data_iter):\n optimizer.zero_grad()\n center, context_negative, mask, label = [\n data.to(device) for data in batch]\n\n pred = skip_gram(center, context_negative, net[0], net[1])\n l = (\n loss(pred.reshape(label.shape).float(), label.float(), mask) /\n mask.sum(axis=1) * mask.shape[1])\n l.sum().backward()\n optimizer.step()\n metric.add(l.sum(), l.numel())\n if (i + 1) % (num_batches // 5) == 0 or i == num_batches - 1:\n animator.add(epoch + (i + 1) / num_batches,\n (metric[0] / metric[1],))\n print(f'loss {metric[0] / metric[1]:.3f}, '\n f'{metric[1] / timer.stop():.1f} tokens/sec on {str(device)}')",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"lr, num_epochs = 0.01, 5\ntrain(net, data_iter, lr, num_epochs)",
"loss 0.373, 243196.7 tokens/sec on cuda:0\n"
],
[
"",
"_____no_output_____"
]
],
[
[
"# Test\n\nWe find the $k$ nearest words to the query, where we measure similarity using cosine similarity\n$$\\text{sim} = \\frac{x^T y}{||x|| \\; ||y||}$$\n",
"_____no_output_____"
]
],
[
[
"def get_similar_tokens(query_token, k, embed):\n W = embed.weight.data\n x = W[vocab[query_token]]\n # Compute the cosine similarity. Add 1e-9 for numerical stability\n cos = torch.mv(\n W, x) / torch.sqrt(torch.sum(W * W, dim=1) * torch.sum(x * x) + 1e-9)\n topk = torch.topk(cos, k=k + 1)[1].cpu().numpy().astype('int32')\n for i in topk[1:]: # Remove the input words\n print(f'cosine sim={float(cos[i]):.3f}: {vocab.idx_to_token[i]}')\n",
"_____no_output_____"
],
[
"\nget_similar_tokens('chip', 3, net[0])",
"cosine sim=0.505: intel\ncosine sim=0.491: hewlett-packard\ncosine sim=0.485: chips\n"
],
[
"\nget_similar_tokens('president', 3, net[0])",
"cosine sim=0.606: chief\ncosine sim=0.581: vice\ncosine sim=0.528: e.\n"
],
[
"\nget_similar_tokens('dog', 3, net[0])",
"cosine sim=0.453: republican\ncosine sim=0.449: republicans\ncosine sim=0.445: athletics\n"
]
],
[
[
"# Pre-trained models\n\nFor better results, you should use a larger model that is trained on more data,\nsuch as those provided by the [Spacy library](https://spacy.io/usage/linguistic-features#vectors-similarity). For a demo, see [this script](https://github.com/probml/pyprobml/blob/master/scripts/word_embedding_spacy.py).",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
eced12ff97e0e3f373fc5d1a92bb76d84c1acc51 | 75,761 | ipynb | Jupyter Notebook | Note/RNN/sintest.ipynb | kotaronov27/KaggleSB | ff5d23a40e1898f29fe154d954515f3cd2a1daee | [
"MIT"
] | null | null | null | Note/RNN/sintest.ipynb | kotaronov27/KaggleSB | ff5d23a40e1898f29fe154d954515f3cd2a1daee | [
"MIT"
] | 1 | 2021-02-04T01:11:59.000Z | 2021-02-04T01:11:59.000Z | Note/RNN/sintest.ipynb | kotaronov27/KaggleSB | ff5d23a40e1898f29fe154d954515f3cd2a1daee | [
"MIT"
] | null | null | null | 301.836653 | 26,118 | 0.745898 | [
[
[
"# RNN デモ",
"_____no_output_____"
]
],
[
[
"# -*- coding: utf-8 -*-\n# Tensorflow 2.x\n\nimport pandas as pd\nimport numpy as np\nimport math\nimport random\n",
"_____no_output_____"
],
[
"%matplotlib inline\nrandom.seed(0)\n# 乱数の係数\nrandom_factor = 0.05\n# サイクルあたりのステップ数\nsteps_per_cycle = 80\n# 生成するサイクル数\nnumber_of_cycles = 50\n\ndf = pd.DataFrame(np.arange(steps_per_cycle * number_of_cycles + 1), columns=[\"t\"])\ndf[\"sin_t\"] = df.t.apply(lambda x: math.sin(x * (2 * math.pi / steps_per_cycle)+ random.uniform(-1.0, +1.0) * random_factor))\ndf[[\"sin_t\"]].head(steps_per_cycle * 2).plot()",
"_____no_output_____"
]
],
[
[
"## モデル作成",
"_____no_output_____"
]
],
[
[
"def _load_data(data, n_prev = 100): \n \"\"\"\n data should be pd.DataFrame()\n \"\"\"\n docX, docY = [], []\n for i in range(len(data)-n_prev):\n docX.append(data.iloc[i:i+n_prev])\n docY.append(data.iloc[i+n_prev])\n alsX = np.array(docX)\n alsY = np.array(docY)\n\n return alsX, alsY\n\ndef train_test_split(df, test_size=0.1, n_prev = 100): \n \"\"\"\n df should be np.array()\n This just splits data to training and testing parts\n \"\"\"\n ntrn = round(len(df) * (1 - test_size))\n ntrn = int(ntrn)\n x_train, y_train = _load_data(df.iloc[0:ntrn], n_prev)\n x_test, y_test = _load_data(df.iloc[ntrn:], n_prev)\n\n return (x_train, y_train), (x_test, y_test)",
"_____no_output_____"
],
[
"length_of_sequences = 100\ntmpdf = pd.DataFrame(df)\n(x_train, y_train), (x_test, y_test) = train_test_split(tmpdf, n_prev =length_of_sequences)\n\nprint(\"train=\")\nprint(x_train)\nprint(\"test=\")\nprint(x_test)",
"train=\n[[[ 0.00000000e+00 3.44353760e-02]\n [ 1.00000000e+00 1.04146063e-01]\n [ 2.00000000e+00 1.48584561e-01]\n ...\n [ 4.70000000e+01 -5.28242851e-01]\n [ 4.80000000e+01 -5.66440464e-01]\n [ 4.90000000e+01 -6.84277439e-01]]\n\n [[ 1.00000000e+00 1.04146063e-01]\n [ 2.00000000e+00 1.48584561e-01]\n [ 3.00000000e+00 2.09937587e-01]\n ...\n [ 4.80000000e+01 -5.66440464e-01]\n [ 4.90000000e+01 -6.84277439e-01]\n [ 5.00000000e+01 -7.28216569e-01]]\n\n [[ 2.00000000e+00 1.48584561e-01]\n [ 3.00000000e+00 2.09937587e-01]\n [ 4.00000000e+00 3.10089087e-01]\n ...\n [ 4.90000000e+01 -6.84277439e-01]\n [ 5.00000000e+01 -7.28216569e-01]\n [ 5.10000000e+01 -7.57016582e-01]]\n\n ...\n\n [[ 3.54800000e+03 8.20962668e-01]\n [ 3.54900000e+03 7.27295360e-01]\n [ 3.55000000e+03 6.93032043e-01]\n ...\n [ 3.59500000e+03 -3.37301107e-01]\n [ 3.59600000e+03 -3.12478690e-01]\n [ 3.59700000e+03 -2.21039817e-01]]\n\n [[ 3.54900000e+03 7.27295360e-01]\n [ 3.55000000e+03 6.93032043e-01]\n [ 3.55100000e+03 6.60091920e-01]\n ...\n [ 3.59600000e+03 -3.12478690e-01]\n [ 3.59700000e+03 -2.21039817e-01]\n [ 3.59800000e+03 -1.55778141e-01]]\n\n [[ 3.55000000e+03 6.93032043e-01]\n [ 3.55100000e+03 6.60091920e-01]\n [ 3.55200000e+03 6.12202015e-01]\n ...\n [ 3.59700000e+03 -2.21039817e-01]\n [ 3.59800000e+03 -1.55778141e-01]\n [ 3.59900000e+03 -1.21965561e-01]]]\ntest=\n[[[ 3.60100000e+03 6.85908116e-02]\n [ 3.60200000e+03 1.54667967e-01]\n [ 3.60300000e+03 2.64583589e-01]\n ...\n [ 3.64800000e+03 -5.55289655e-01]\n [ 3.64900000e+03 -6.63279314e-01]\n [ 3.65000000e+03 -7.08730567e-01]]\n\n [[ 3.60200000e+03 1.54667967e-01]\n [ 3.60300000e+03 2.64583589e-01]\n [ 3.60400000e+03 2.91042778e-01]\n ...\n [ 3.64900000e+03 -6.63279314e-01]\n [ 3.65000000e+03 -7.08730567e-01]\n [ 3.65100000e+03 -7.73120199e-01]]\n\n [[ 3.60300000e+03 2.64583589e-01]\n [ 3.60400000e+03 2.91042778e-01]\n [ 3.60500000e+03 3.49272947e-01]\n ...\n [ 3.65000000e+03 -7.08730567e-01]\n [ 3.65100000e+03 -7.73120199e-01]\n [ 3.65200000e+03 -8.33724652e-01]]\n\n ...\n\n [[ 3.94800000e+03 8.13775178e-01]\n [ 3.94900000e+03 7.85929341e-01]\n [ 3.95000000e+03 7.31436864e-01]\n ...\n [ 3.99500000e+03 -4.16984342e-01]\n [ 3.99600000e+03 -3.54502022e-01]\n [ 3.99700000e+03 -1.93910111e-01]]\n\n [[ 3.94900000e+03 7.85929341e-01]\n [ 3.95000000e+03 7.31436864e-01]\n [ 3.95100000e+03 6.21999588e-01]\n ...\n [ 3.99600000e+03 -3.54502022e-01]\n [ 3.99700000e+03 -1.93910111e-01]\n [ 3.99800000e+03 -1.27861754e-01]]\n\n [[ 3.95000000e+03 7.31436864e-01]\n [ 3.95100000e+03 6.21999588e-01]\n [ 3.95200000e+03 6.13421650e-01]\n ...\n [ 3.99700000e+03 -1.93910111e-01]\n [ 3.99800000e+03 -1.27861754e-01]\n [ 3.99900000e+03 -9.70259716e-02]]]\n"
]
],
[
[
"## 学習",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom keras.models import Sequential \nfrom keras.layers.core import Dense, Activation \nfrom keras.layers.recurrent import LSTM\nfrom keras.callbacks import EarlyStopping\n",
"_____no_output_____"
],
[
"in_out_neurons = 1\nhidden_neurons = 300\n\nmodel = Sequential() \nmodel.add(LSTM(hidden_neurons, batch_input_shape=(None, length_of_sequences, in_out_neurons), return_sequences=False)) \nmodel.add(Dense(in_out_neurons)) \nmodel.add(Activation(\"linear\")) \nmodel.compile(loss=\"mean_squared_error\", optimizer=\"rmsprop\")\n",
"_____no_output_____"
],
[
"\n#model.fit(x_train, y_train, batch_size=600, epochs=15, validation_split=0.05) \n# early stopping\nearly_stopping = EarlyStopping(monitor='val_loss', patience=2)\n\nmodel.fit(x_train, y_train, batch_size=600, epochs=15, validation_split=0.05, callbacks=[early_stopping]) ",
"Epoch 1/15\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
eced1997099abbba0501a5fb52e8e7e19ecc0b58 | 101,692 | ipynb | Jupyter Notebook | readme_generator.ipynb | haesleinhuepf/awesome-napari-plugins-that-are-not-on-the-napari-hub-yet | 6dfa5b8076949c2bb960e7881a0fdab5f4a33f0e | [
"CC0-1.0"
] | 6 | 2022-02-13T18:30:58.000Z | 2022-03-26T02:35:26.000Z | readme_generator.ipynb | haesleinhuepf/awesome-napari-plugins-that-are-not-on-the-napari-hub-yet | 6dfa5b8076949c2bb960e7881a0fdab5f4a33f0e | [
"CC0-1.0"
] | 1 | 2022-03-03T17:14:56.000Z | 2022-03-31T06:36:51.000Z | readme_generator.ipynb | haesleinhuepf/awesome-napari-plugins-that-are-not-on-the-napari-hub-yet | 6dfa5b8076949c2bb960e7881a0fdab5f4a33f0e | [
"CC0-1.0"
] | null | null | null | 61.706311 | 224 | 0.589525 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"template = \"\"\"\n### [{reponame}](https://github.com/{orgname}/{reponame})\n\n[](https://napari-hub.org/plugins/napari-mm)\n[](https://pypi.org/project/napari-brightness-contrast)\n\n[](https://codecov.io/gh/{orgname}/{reponame})\n[](https://github.com/{orgname}/{reponame}/actions)\n[](https://en.wikipedia.org/wiki/Software_release_life_cycle#Alpha)\n[](https://github.com/{orgname}/{reponame}/releases)\n\n{description}\n\n---\n\n\"\"\"",
"_____no_output_____"
],
[
"table = pd.read_csv(\"list.csv\")\ntable[\"order\"] = [t.lower().replace(\"napari-\",\"\") for t in table[\"github repository\"]]\ntable = table.sort_values(by=\"order\",ascending = True) \ntable",
"_____no_output_____"
],
[
"table.keys()",
"_____no_output_____"
],
[
"from IPython.display import display, Markdown, Latex\nimport numpy as np\n \noutput = \"\"\nfor index, r in table.iterrows():\n orgname = r[\"github organization\"]\n reponame = r[\"github repository\"]\n description = r[\"short description\"]\n \n text = template.format(\n orgname=orgname, \n reponame=reponame,\n description=description,\n )\n \n \n output = output + text\ndisplay(Markdown(output))\n\n\n\nwith open('readme.md', 'w') as f:\n \n with open('readme_header.md', 'r') as header:\n f.write(header.read())\n \n f.write(output)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
eced25ab9a50364cd1d6b4142a2b1e55cf574ebb | 56,257 | ipynb | Jupyter Notebook | project1/Not used/Energy OIX.ipynb | thomasnilsson/02805-social-graphs-2018 | 45199300e3b8ab85f5c051f3f9f473a229eb5253 | [
"MIT"
] | 1 | 2019-02-22T18:55:16.000Z | 2019-02-22T18:55:16.000Z | project1/Not used/Energy OIX.ipynb | thomasnilsson/02805-social-graphs-2018 | 45199300e3b8ab85f5c051f3f9f473a229eb5253 | [
"MIT"
] | null | null | null | project1/Not used/Energy OIX.ipynb | thomasnilsson/02805-social-graphs-2018 | 45199300e3b8ab85f5c051f3f9f473a229eb5253 | [
"MIT"
] | 1 | 2020-11-07T10:00:05.000Z | 2020-11-07T10:00:05.000Z | 282.698492 | 51,494 | 0.915779 | [
[
[
"import requests \nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"since2017 = requests.get(\"https://api.energidataservice.dk/datastore_search?resource_id=06380963-b7c6-46b7-aec5-173d15e4648b&limit=421564\").json()",
"_____no_output_____"
],
[
"d = since2017[\"result\"]['records']",
"_____no_output_____"
],
[
"df = pd.DataFrame(d)\ndf = df.fillna(0)",
"_____no_output_____"
],
[
"p17 = df[df.Minutes5DK < \"2017-12-31T23:59:59\"]\np17 = p17.drop([\"Minutes5UTC\", \"BornholmSE4\", \"_id\"], axis=1)",
"_____no_output_____"
],
[
"p17_dk1 = p17[p17.PriceArea == \"DK1\"]\np17_dk2 = p17[p17.PriceArea == \"DK1\"]",
"_____no_output_____"
],
[
"only2017 = df[df.Minutes5DK < \"2017-12-31T23:59:59\"]\nonly2017 = only2017.drop([\"Minutes5UTC\", \"BornholmSE4\", \"PriceArea\", \"_id\"], axis=1)",
"_____no_output_____"
],
[
"# Aggregate on timestamp on timestamp\naggregation_functions = {\n 'ExchangeGermany': 'sum', \n 'ExchangeGreatBelt': 'sum', \n 'ExchangeNetherlands' : 'sum',\n 'ExchangeNorway' : 'sum',\n 'ExchangeSweden' : 'sum',\n 'Minutes5DK' : 'sum',\n 'OffshoreWindPower' : 'sum',\n 'OnshoreWindPower' : 'sum',\n 'ProductionGe100MW' : 'sum',\n 'ProductionLt100MW' : 'sum',\n 'SolarPower' : 'sum',\n}\ng = only2017.groupby(\"Minutes5DK\").aggregate(aggregation_functions)",
"_____no_output_____"
],
[
"def export_data(dataframe, prefix):\n months = [\"jan\", \"feb\", \"mar\", \"apr\", \"may\", \"jun\", \"jul\", \"aug\", \"sep\", \"oct\", \"nov\", \"dec\"]\n for i in range(1, 13):\n start = \"2017-%s-01T00:00:00\" % str(i).zfill(2)\n end = \"2017-%s-31T23:59:59\" % str(i).zfill(2)\n month_df = dataframe[(start <= dataframe.Minutes5DK) & (dataframe.Minutes5DK <= end)]\n\n out = pd.DataFrame()\n out[\"solar\"] = month_df.SolarPower\n out[\"wind\"] = month_df.OffshoreWindPower + month_df.OnshoreWindPower\n out[\"energy\"] = month_df.loc[:, month_df.columns != 'Minutes5DK'].sum(axis=1)\n out.index.names = [\"time\"]\n\n out.to_csv(\"data/%s_%s.csv\" % (prefix, months[i-1]))",
"_____no_output_____"
],
[
"export_data(g, \"agg_2017\")",
"_____no_output_____"
],
[
"export_data(p17_dk1, \"dk1_2017\")",
"_____no_output_____"
],
[
"export_data(p17_dk1, \"dk2_2017\")",
"_____no_output_____"
],
[
"plt.figure(figsize=(20,7))\nplt.plot(time, solar)\nplt.plot(time, wind)\nplt.plot(time, energy)\nplt.legend([\"Solar\", \"Wind\", \"Energy\"])\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eced275d052e1129b7f6d2eaed86eac035f33570 | 17,930 | ipynb | Jupyter Notebook | backend/doc2vec-baselines/Generate-Doc2Vec-Model-From-Json/Doc2Vec_Generate_Doc2Vec_ModelTokenize.ipynb | WatVis/EDAssistant | a4be2849a65abcf2f81f9c01a2172ec67aa38853 | [
"BSD-3-Clause"
] | null | null | null | backend/doc2vec-baselines/Generate-Doc2Vec-Model-From-Json/Doc2Vec_Generate_Doc2Vec_ModelTokenize.ipynb | WatVis/EDAssistant | a4be2849a65abcf2f81f9c01a2172ec67aa38853 | [
"BSD-3-Clause"
] | null | null | null | backend/doc2vec-baselines/Generate-Doc2Vec-Model-From-Json/Doc2Vec_Generate_Doc2Vec_ModelTokenize.ipynb | WatVis/EDAssistant | a4be2849a65abcf2f81f9c01a2172ec67aa38853 | [
"BSD-3-Clause"
] | null | null | null | 31.511424 | 364 | 0.481651 | [
[
[
"# Perform Doc2Vec",
"_____no_output_____"
],
[
"# Read data",
"_____no_output_____"
],
[
"This notebook trains a doc2vec model on the entire notebook dataset. ",
"_____no_output_____"
],
[
"# Import modules",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport os\nimport gensim\nfrom gensim.models.doc2vec import TaggedDocument",
"_____no_output_____"
]
],
[
[
"# Read data",
"_____no_output_____"
]
],
[
[
"df = pd.read_json(\"../data/all-notebooks-tokenized.json\", orient=\"index\")\ndf['filename'] = df['filename'].astype(str)",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"### Ignore markdown, only consider code for now",
"_____no_output_____"
]
],
[
[
"subdf = df\nsubdf = df[df.cell_type == \"code\"] ",
"_____no_output_____"
]
],
[
[
"### Calculate the line number for each notebook",
"_____no_output_____"
]
],
[
[
"subdf['cell_num']=subdf.groupby(['filename']).cumcount()+1\nsubdf['filename_with_cellnum'] = subdf['filename'] + \"_\" + subdf['cell_num'].astype(str)\n\n# Convert list of strings to string\nsubdf['tokenized_source_str'] = subdf['tokenized_source'].astype(str)",
"<ipython-input-18-8a4e01e869df>:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n subdf['cell_num']=subdf.groupby(['filename']).cumcount()+1\n<ipython-input-18-8a4e01e869df>:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n subdf['filename_with_cellnum'] = subdf['filename'] + \"_\" + subdf['cell_num'].astype(str)\n<ipython-input-18-8a4e01e869df>:5: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n subdf['tokenized_source_str'] = subdf['tokenized_source'].astype(str)\n"
],
[
"subdf.head()",
"_____no_output_____"
]
],
[
[
"# Preprocessing\n## Generate input data from raw source",
"_____no_output_____"
]
],
[
[
"from gensim.utils import simple_preprocess\n\nclass MyDataframeCorpus(object):\n def __init__(self, source_df, text_col, tag_col):\n self.source_df = source_df\n self.text_col = text_col\n self.tag_col = tag_col\n\n def __iter__(self):\n for i, row in self.source_df.iterrows():\n yield TaggedDocument(words=simple_preprocess(row[self.text_col]), \n tags=[row[self.tag_col]])\n",
"_____no_output_____"
]
],
[
[
"## Train the model",
"_____no_output_____"
]
],
[
[
"train_corpus = MyDataframeCorpus(subdf, 'tokenized_source_str', 'filename_with_cellnum')",
"_____no_output_____"
],
[
"model = gensim.models.doc2vec.Doc2Vec(vector_size=768, epochs=40)",
"_____no_output_____"
],
[
"model.build_vocab(train_corpus)",
"_____no_output_____"
],
[
"model.train(train_corpus, total_examples=model.corpus_count, epochs=model.epochs)",
"_____no_output_____"
]
],
[
[
"## Test inference",
"_____no_output_____"
]
],
[
[
"testinput = \"import numpy as np\"\nvector = model.infer_vector(testinput.split(\" \"))\n# print(vector)",
"_____no_output_____"
],
[
"sims = model.docvecs.most_similar([vector])\nprint(sims)\n\nfor result in sims[0:3]:\n display(df[df.filename_with_cellnum == result[0]]['source'])",
"[('10506138_2', 0.8787171840667725), ('31005248_62', 0.876400887966156), ('11065888_1', 0.8746524453163147), ('4384070_7', 0.8730512857437134), ('32082014_34', 0.872079610824585), ('2604429_7', 0.8716046214103699), ('236052_1', 0.8713623285293579), ('38355971_1', 0.8710976839065552), ('22398214_10', 0.870916485786438), ('36992845_6', 0.87025386095047)]\n"
]
],
[
[
"# Save the model",
"_____no_output_____"
]
],
[
[
"model.save(\"../model/notebook-doc2vec-model-apr24.model\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
eced2ab51cb2a88298f33cb926e1d0f13404a5b3 | 21,332 | ipynb | Jupyter Notebook | notebooks/chap01.ipynb | wmmurrah/ModSimPy | eda2e79268de5f03eab63a00ea50b87329604d22 | [
"MIT"
] | null | null | null | notebooks/chap01.ipynb | wmmurrah/ModSimPy | eda2e79268de5f03eab63a00ea50b87329604d22 | [
"MIT"
] | null | null | null | notebooks/chap01.ipynb | wmmurrah/ModSimPy | eda2e79268de5f03eab63a00ea50b87329604d22 | [
"MIT"
] | null | null | null | 25.670277 | 357 | 0.546737 | [
[
[
"# Modeling and Simulation in Python\n\nChapter 1\n\nCopyright 2017 Allen Downey\n\nLicense: [Creative Commons Attribution 4.0 International](https://creativecommons.org/licenses/by/4.0)",
"_____no_output_____"
],
[
"## Jupyter\n\nWelcome to Modeling and Simulation, welcome to Python, and welcome to Jupyter.\n\nThis is a Jupyter notebook, which is a development environment where you can write and run Python code. Each notebook is divided into cells. Each cell contains either text (like this cell) or Python code.\n\n### Selecting and running cells\n\nTo select a cell, click in the left margin next to the cell. You should see a blue frame surrounding the selected cell.\n\nTo edit a code cell, click inside the cell. You should see a green frame around the selected cell, and you should see a cursor inside the cell.\n\nTo edit a text cell, double-click inside the cell. Again, you should see a green frame around the selected cell, and you should see a cursor inside the cell.\n\nTo run a cell, hold down SHIFT and press ENTER. \n\n* If you run a text cell, Jupyter typesets the text and displays the result.\n\n* If you run a code cell, it runs the Python code in the cell and displays the result, if any.\n\nTo try it out, edit this cell, change some of the text, and then press SHIFT-ENTER to run it.",
"_____no_output_____"
],
[
"### Adding and removing cells\n\nYou can add and remove cells from a notebook using the buttons in the toolbar and the items in the menu, both of which you should see at the top of this notebook.\n\nTry the following exercises:\n\n1. From the Insert menu select \"Insert cell below\" to add a cell below this one. By default, you get a code cell, as you can see in the pulldown menu that says \"Code\".\n\n2. In the new cell, add a print statement like `print('Hello')`, and run it.\n\n3. Add another cell, select the new cell, and then click on the pulldown menu that says \"Code\" and select \"Markdown\". This makes the new cell a text cell.\n\n4. In the new cell, type some text, and then run it.\n\n5. Use the arrow buttons in the toolbar to move cells up and down.\n\n6. Use the cut, copy, and paste buttons to delete, add, and move cells.\n\n7. As you make changes, Jupyter saves your notebook automatically, but if you want to make sure, you can press the save button, which looks like a floppy disk from the 1990s.\n\n8. Finally, when you are done with a notebook, select \"Close and Halt\" from the File menu.",
"_____no_output_____"
],
[
"### Using the notebooks\n\nThe notebooks for each chapter contain the code from the chapter along with addition examples, explanatory text, and exercises. I recommend you \n\n1. Read the chapter first to understand the concepts and vocabulary, \n2. Run the notebook to review what you learned and see it in action, and then\n3. Attempt the exercises.\n\nIf you try to work through the notebooks without reading the book, you're gonna have a bad time. The notebooks contain some explanatory text, but it is probably not enough to make sense if you have not read the book. If you are working through a notebook and you get stuck, you might want to re-read (or read!) the corresponding section of the book.",
"_____no_output_____"
],
[
"### Importing modsim\n\nThe following cell imports `modsim`, which is a collection of functions we will use throughout the book. Whenever you start the notebook, you will have to run the following cell. It does three things:\n\n1. It uses a Jupyter \"magic command\" to specify whether figures should appear in the notebook, or pop up in a new window.\n\n2. It configures Jupyter to display some values that would otherwise be invisible. \n\n3. It imports everything defined in `modsim`.\n\nSelect the following cell and press SHIFT-ENTER to run it.",
"_____no_output_____"
]
],
[
[
"# Configure Jupyter so figures appear in the notebook\n%matplotlib inline\n\n# Configure Jupyter to display the assigned value after an assignment\n%config InteractiveShell.ast_node_interactivity='last_expr_or_assign'\n\n# import functions from the modsim library\nfrom modsim import *\n\nprint('If this cell runs successfully, it produces no output other than this message.')",
"If this cell runs successfully, it produces no output other than this message.\n"
]
],
[
[
"The first time you run this on a new installation of Python, it might produce a warning message in pink. That's probably ok, but if you get a message that says `modsim.py depends on Python 3.7 features`, that means you have an older version of Python, and some features in `modsim.py` won't work correctly.\n\nIf you need a newer version of Python, I recommend installing Anaconda. You'll find more information in the preface of the book.",
"_____no_output_____"
],
[
"## The penny myth\n\nThe following cells contain code from the beginning of Chapter 1.\n\n`modsim` defines `UNITS`, which contains variables representing pretty much every unit you've ever heard of. It uses [Pint](https://pint.readthedocs.io/en/latest/), which is a Python library that provides tools for computing with units.\n\nThe following lines create new variables named `meter` and `second`.",
"_____no_output_____"
]
],
[
[
"meter = UNITS.meter",
"_____no_output_____"
],
[
"second = UNITS.second",
"_____no_output_____"
]
],
[
[
"To find out what other units are defined, type `UNITS.` (including the period) in the next cell and then press TAB. You should see a pop-up menu with a list of units.",
"_____no_output_____"
],
[
"Create a variable named `a` and give it the value of acceleration due to gravity.",
"_____no_output_____"
]
],
[
[
"a = 9.8 * meter / second**2",
"_____no_output_____"
]
],
[
[
"Create `t` and give it the value 4 seconds.",
"_____no_output_____"
]
],
[
[
"t = 4 * second",
"_____no_output_____"
]
],
[
[
"Compute the distance a penny would fall after `t` seconds with constant acceleration `a`. Notice that the units of the result are correct.",
"_____no_output_____"
]
],
[
[
"a * t**2 / 2",
"_____no_output_____"
]
],
[
[
"**Exercise**: Compute the velocity of the penny after `t` seconds. Check that the units of the result are correct.",
"_____no_output_____"
]
],
[
[
"78.4",
"_____no_output_____"
]
],
[
[
"**Exercise**: Why would it be nonsensical to add `a` and `t`? What happens if you try?",
"_____no_output_____"
],
[
"The error messages you get from Python are big and scary, but if you read them carefully, they contain a lot of useful information.\n\n1. Start from the bottom and read up.\n2. The last line usually tells you what type of error happened, and sometimes additional information.\n3. The previous lines are a \"traceback\" of what was happening when the error occurred. The first section of the traceback shows the code you wrote. The following sections are often from Python libraries.\n\nIn this example, you should get a `DimensionalityError`, which is defined by Pint to indicate that you have violated a rules of dimensional analysis: you cannot add quantities with different dimensions.\n\nBefore you go on, you might want to delete the erroneous code so the notebook can run without errors.",
"_____no_output_____"
],
[
"## Falling pennies\n\nNow let's solve the falling penny problem.\n\nSet `h` to the height of the Empire State Building:",
"_____no_output_____"
]
],
[
[
"h = 381 * meter",
"_____no_output_____"
]
],
[
[
"Compute the time it would take a penny to fall, assuming constant acceleration.\n\n$ a t^2 / 2 = h $\n\n$ t = \\sqrt{2 h / a}$",
"_____no_output_____"
]
],
[
[
"t = sqrt(2 * h / a)",
"_____no_output_____"
]
],
[
[
"Given `t`, we can compute the velocity of the penny when it lands.\n\n$v = a t$",
"_____no_output_____"
]
],
[
[
"v = a * t",
"_____no_output_____"
]
],
[
[
"We can convert from one set of units to another like this:",
"_____no_output_____"
]
],
[
[
"mile = UNITS.mile\nhour= UNITS.hour",
"_____no_output_____"
],
[
"v.to(mile/hour)",
"_____no_output_____"
]
],
[
[
"**Exercise:** Suppose you bring a 10 foot pole to the top of the Empire State Building and use it to drop the penny from `h` plus 10 feet.\n\nDefine a variable named `foot` that contains the unit `foot` provided by `UNITS`. Define a variable named `pole_height` and give it the value 10 feet.\n\nWhat happens if you add `h`, which is in units of meters, to `pole_height`, which is in units of feet? What happens if you write the addition the other way around?",
"_____no_output_____"
]
],
[
[
"# Solution goes here\nfoot = UNITS.feet\npole_height = 10 * foot",
"_____no_output_____"
],
[
"# Solution goes here\npole_ht_m = pole_height.to(meter)\nh + pole_ht_m",
"_____no_output_____"
],
[
"h + pole_height",
"_____no_output_____"
]
],
[
[
"**Exercise:** In reality, air resistance limits the velocity of the penny. At about 18 m/s, the force of air resistance equals the force of gravity and the penny stops accelerating.\n\nAs a simplification, let's assume that the acceleration of the penny is `a` until the penny reaches 18 m/s, and then 0 afterwards. What is the total time for the penny to fall 381 m?\n\nYou can break this question into three parts:\n\n1. How long until the penny reaches 18 m/s with constant acceleration `a`.\n2. How far would the penny fall during that time?\n3. How long to fall the remaining distance with constant velocity 18 m/s?\n\nSuggestion: Assign each intermediate result to a variable with a meaningful name. And assign units to all quantities!",
"_____no_output_____"
]
],
[
[
"# Solution goes here\nvmax = 18 *meter/second\n\nt_to_vmax = vmax/a",
"_____no_output_____"
],
[
"# Solution goes here\nd2max= a*t_to_vmax**2/2",
"_____no_output_____"
],
[
"# Solution goes here\nd_remain = h - d2max",
"_____no_output_____"
],
[
"# Solution goes here\n# v = d/t\nt_remain = d_remain/vmax",
"_____no_output_____"
],
[
"# Solution goes here\nt_total = t_to_vmax + t_remain",
"_____no_output_____"
]
],
[
[
"### Restart and run all\n\nWhen you change the contents of a cell, you have to run it again for those changes to have an effect. If you forget to do that, the results can be confusing, because the code you are looking at is not the code you ran.\n\nIf you ever lose track of which cells have run, and in what order, you should go to the Kernel menu and select \"Restart & Run All\". Restarting the kernel means that all of your variables get deleted, and running all the cells means all of your code will run again, in the right order.\n\n**Exercise:** Select \"Restart & Run All\" now and confirm that it does what you want.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
eced2afabcc1d41201377b294b357dd5e3d41535 | 7,712 | ipynb | Jupyter Notebook | spark/dbc_to_ipynb.ipynb | BenjaminLiuPenrose/Berkeley-CS-100 | 84fe1633ddaa808ccbf0b5c9f55762e6766c1d3e | [
"BSD-2-Clause"
] | null | null | null | spark/dbc_to_ipynb.ipynb | BenjaminLiuPenrose/Berkeley-CS-100 | 84fe1633ddaa808ccbf0b5c9f55762e6766c1d3e | [
"BSD-2-Clause"
] | null | null | null | spark/dbc_to_ipynb.ipynb | BenjaminLiuPenrose/Berkeley-CS-100 | 84fe1633ddaa808ccbf0b5c9f55762e6766c1d3e | [
"BSD-2-Clause"
] | null | null | null | 37.256039 | 595 | 0.571577 | [
[
[
"# ** Convert Databricks Exports to IPython Notebooks **\n#### This notebook should be uploaded to the VM just like other notebooks. In addition to uploading this notebook, you should upload your export from Databricks. The Databricks export should have the .dbc extension. To download your .dbc file from Databricks, click on the down arrow next to your root folder then hover the mouse over \"Export\" and click on \"DBC Archive\". You can also export a single file by clicking on the down arrow next to the file, hovering the mouse over \"Export\", and clicking on \"DBC Archive\".\n#### To perform the conversion, you should just run the cells below. If necessary, you can change the `fileLocation` in the first code cell below. The second code cell will list the contents of the .dbc file, so you can check that you're working with the correct file. The third code cell will list the .ipynb files that will be generated. Note that if the files already exist they will be overwritten. The fourth code cell will perform the conversion and overwrite any files, if necessary. The fifth code cell just cleans up the directory that was used for extracting the .dbc file.",
"_____no_output_____"
],
[
"#### ** Additional Details (Optional) **\n#### Note that this code does some processing tailored to the courses, like special handling of the \"baseDir\" directory which points to the course datasets, so it may do unexpected things to dbc files in rare cases. The notebook handles MathJax conversion, creates a do-nothing `display` function and a working `displayHTML` function. It also replaces `baseDir = <some path>` with `baseDir = 'data'`.\n#### The only requirement is IPython version 3, so this conversion can also be run outside of the Virtual Machine (VM) environment.",
"_____no_output_____"
]
],
[
[
"# Change this fileLocation if necessary\n# Subsequent downloads might be called 'Databricks (1).dbc', etc.\nfileLocation = 'Databricks.dbc'",
"_____no_output_____"
],
[
"# Extract dbc file\n\n# Cleanup from prior run\nimport shutil\ntry: shutil.rmtree('tmp_dbc')\nexcept OSError: pass\n\nimport zipfile\nimport os\ntry: os.mkdir('tmp_dbc')\nexcept OSError: pass\nwith zipfile.ZipFile(fileLocation, 'r') as z:\n z.extractall('tmp_dbc')\n\nprint '*** Contents from the .dbc file (usually one file or a directory) ***\\n'\nprint os.listdir('tmp_dbc')",
"_____no_output_____"
],
[
"# Find files to parse\nimport fnmatch\n\nfilesToParse = []\nfor root, dirNames, fileNames in os.walk('tmp_dbc'):\n for fileName in fnmatch.filter(fileNames, '*.python'):\n filesToParse.append((root, fileName))\n\ndef getIpynbName((path, fileName)):\n path = os.path.normpath(path)\n pathSplit = path.split(os.sep)[2:]\n baseDir = os.path.join(*pathSplit) if len(pathSplit) > 0 else '.'\n newFileName = os.path.splitext(fileName)[0] + '_export.ipynb'\n return os.path.join(baseDir, newFileName)\n\nprint \"*** Files to be created (relative to your current working directory) ***\"\nprint \"(Warning: files will be overwritten!)\\n\"\nfor path, fileName in filesToParse:\n print getIpynbName((path, fileName))",
"_____no_output_____"
],
[
"# Create the IPython Notebooks\n# Convert .python files to .ipynb files\nimport codecs\nfrom IPython import nbformat\nfrom IPython.nbformat.v3.nbpy import PyReader\nimport json\nimport re\n\n_header = u'# -*- coding: utf-8 -*-\\n# <nbformat>3.0</nbformat>\\n'\n_markdownCell = u'\\n\\n# <markdowncell>\\n\\n'\n_codeCell = u'\\n\\n# <codecell>\\n\\n'\n_firstCell = u\"\"\"# Increase compatibility with Databricks\nfrom IPython.display import display as idisplay, HTML\ndisplayHTML = lambda x: idisplay(HTML(x))\ndef display(*args, **kargs): pass\"\"\"\n\ndef convertToIpynb(fileToParse):\n \n with codecs.open(os.path.join(*fileToParse), encoding=\"utf-8\") as fp:\n jsonData = json.load(fp)\n commands = jsonData['commands']\n commandInfo = [(x['position'], x['command']) for x in commands]\n commandList = sorted(commandInfo)\n\n with codecs.open('tmp_ipynb.py', 'w', encoding=\"utf-8\") as fp:\n fp.write(_header)\n fp.write(_codeCell)\n fp.write(_firstCell)\n\n for position, command in commandList:\n if re.match(r'\\s*%md', command):\n command = re.sub(r'^\\s*%md', '', command, flags=re.MULTILINE)\n command = re.sub(r'(%\\(|\\)%)', '$', command)\n command = re.sub(r'(%\\[|\\]%)', '$$', command)\n\n fp.write(_markdownCell)\n asLines = command.split('\\n')\n command = '# ' + '\\n# '.join(asLines)\n else:\n command = re.sub(r'^\\s*baseDir\\s*=.*$', 'baseDir = \\'data\\'', \n command, flags=re.MULTILINE)\n fp.write(_codeCell)\n\n fp.write(command)\n\n outputName = getIpynbName(fileToParse)\n\n with codecs.open('tmp_ipynb.py', 'r', encoding=\"utf-8\") as intermediate:\n nb = PyReader().read(intermediate)\n\n os.remove('tmp_ipynb.py')\n baseDirectory = os.path.split(outputName)[0]\n\n if not os.path.isdir(baseDirectory):\n os.makedirs(baseDirectory)\n\n with codecs.open(outputName, 'w', encoding=\"utf-8\") as output:\n nbformat.write(nbformat.convert(nb, 4.0), output) \n print 'Created: {0}'.format(outputName)\n\nfor fileToParse in filesToParse:\n convertToIpynb(fileToParse)",
"_____no_output_____"
],
[
"# Cleanup\nimport shutil\ntry: shutil.rmtree('tmp_dbc')\nexcept OSError: pass",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
eced4906bcd6668b92653d87cf73ddb91eef6a25 | 621,668 | ipynb | Jupyter Notebook | utilities/snippets/generate-skewed-data.ipynb | jarokaz/drift-monitor | 43dce1bbc6eeb6a01e5a5d38aa60dc8f84e370e4 | [
"Apache-2.0"
] | 1 | 2021-02-25T03:58:56.000Z | 2021-02-25T03:58:56.000Z | utilities/snippets/generate-skewed-data.ipynb | jarokaz/drift-monitor | 43dce1bbc6eeb6a01e5a5d38aa60dc8f84e370e4 | [
"Apache-2.0"
] | null | null | null | utilities/snippets/generate-skewed-data.ipynb | jarokaz/drift-monitor | 43dce1bbc6eeb6a01e5a5d38aa60dc8f84e370e4 | [
"Apache-2.0"
] | null | null | null | 611.276303 | 96,729 | 0.672447 | [
[
[
"# Simulating Requests to an AI Prediction Deployed Model\n\n",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"try:\n from google.colab import auth\n auth.authenticate_user()\n print(\"Colab user is authenticated.\")\nexcept: pass",
"_____no_output_____"
],
[
"!pip install -U -q google-api-python-client\n!pip install -U -q pandas",
"_____no_output_____"
],
[
"import os\nfrom tensorflow import io as tf_io\nimport matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"PROJECT = 'sa-data-validation'\nBUCKET = 'sa-data-validation'\nGCS_DATA_LOCATION = 'gs://workshop-datasets/covertype/data_validation'\nREGION = 'us-central1'\nBUCKET = 'sa-data-validation'\nLOCAL_WORKSPACE = './workspace'\nLOCAL_DATA_DIR = os.path.join(LOCAL_WORKSPACE, 'data')\nLOCAL_DATA_FILE = os.path.join(LOCAL_DATA_DIR, 'train.csv')\nBQ_DATASET_NAME = 'data_validation'\nBQ_TABLE_NAME = 'covertype_classifier_logs'\nMODEL_NAME = 'covertype_classifier'\nVERSION_NAME = 'v2'\nMODEL_OUTPUT_KEY = 'probabilities'\nSIGNATURE_NAME = 'serving_default'",
"_____no_output_____"
],
[
"!gcloud config set project {PROJECT}",
"\u001b[1;31mERROR:\u001b[0m (gcloud.config.set) The project property must be set to a valid project ID, [{PROJECT}] is not a valid project ID.\nTo set your project, run:\n\n $ gcloud config set project PROJECT_ID\n\nor to unset it, run:\n\n $ gcloud config unset project\n"
]
],
[
[
"## 1. Download Data\n\nWe use the [covertype](https://archive.ics.uci.edu/ml/datasets/covertype) from UCI Machine Learning Repository. The task is to Predict forest cover type from cartographic variables only. \n\nThe dataset is preprocessed, split, and uploaded to uploaded to the `gs://workshop-datasets/covertype` public GCS location. \n\nWe use this version of the preprocessed dataset in this notebook. For more information, see [Cover Type Dataset](https://github.com/GoogleCloudPlatform/mlops-on-gcp/tree/master/datasets/covertype)",
"_____no_output_____"
]
],
[
[
"if tf_io.gfile.exists(LOCAL_WORKSPACE):\n print(\"Removing previous workspace artifacts...\")\n tf_io.gfile.rmtree(LOCAL_WORKSPACE)\n\nprint(\"Creating a new workspace...\")\ntf_io.gfile.makedirs(LOCAL_WORKSPACE)\ntf_io.gfile.makedirs(LOCAL_DATA_DIR)",
"_____no_output_____"
],
[
"!gsutil cp gs://workshop-datasets/covertype/data_validation/training/dataset.csv {LOCAL_DATA_FILE}\n!wc -l {LOCAL_DATA_FILE}",
"_____no_output_____"
],
[
"import pandas as pd\ndata = pd.read_csv(LOCAL_DATA_FILE)\nprint(\"Total number of records: {}\".format(len(data.index)))\ndata.sample(10)",
"Total number of records: 431009\n"
]
],
[
[
"## 2. Define Metadata",
"_____no_output_____"
]
],
[
[
"HEADER = ['Elevation', 'Aspect', 'Slope','Horizontal_Distance_To_Hydrology',\n 'Vertical_Distance_To_Hydrology', 'Horizontal_Distance_To_Roadways',\n 'Hillshade_9am', 'Hillshade_Noon', 'Hillshade_3pm',\n 'Horizontal_Distance_To_Fire_Points', 'Wilderness_Area', 'Soil_Type',\n 'Cover_Type']\n\nTARGET_FEATURE_NAME = 'Cover_Type'\n\nFEATURE_LABELS = ['0', '1', '2', '3', '4', '5', '6']\n\nNUMERIC_FEATURE_NAMES = ['Aspect', 'Elevation', 'Hillshade_3pm', \n 'Hillshade_9am', 'Hillshade_Noon', \n 'Horizontal_Distance_To_Fire_Points',\n 'Horizontal_Distance_To_Hydrology',\n 'Horizontal_Distance_To_Roadways','Slope',\n 'Vertical_Distance_To_Hydrology']\n\nCATEGORICAL_FEATURE_NAMES = ['Soil_Type', 'Wilderness_Area']\n\nFEATURE_NAMES = CATEGORICAL_FEATURE_NAMES + NUMERIC_FEATURE_NAMES\n\nHEADER_DEFAULTS = [[0] if feature_name in NUMERIC_FEATURE_NAMES + [TARGET_FEATURE_NAME] else ['NA'] \n for feature_name in HEADER]\n\nNUM_CLASSES = len(FEATURE_LABELS)",
"_____no_output_____"
]
],
[
[
"#### create a container for the serving data",
"_____no_output_____"
]
],
[
[
"serving_data = {\n '2020-04-01': None,\n '2020-04-02': None,\n '2020-04-03': None,\n '2020-04-04': None,\n '2020-04-05': None,\n '2020-04-06': None,\n}",
"_____no_output_____"
]
],
[
[
"## 3. Sampling Unskewed Data\n\n* Sample data for *three* consecutive dates: **01-04-20202**, **02-04-2020**, and **03-04-2020**\n* Each day has 1000 examples\n* No altering is applied\n\n",
"_____no_output_____"
]
],
[
[
"data_normal = data.sample(3000)",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(nrows=1, ncols=3, figsize=(20, 5))\ndata_normal['Aspect'].plot.hist(bins=15, ax=axes[0], title='Aspect')\ndata_normal['Wilderness_Area'].value_counts(normalize=True).plot.bar(ax=axes[1], title='Wilderness Area')\ndata_normal[TARGET_FEATURE_NAME].value_counts(normalize=True).plot.bar(ax=axes[2], title=TARGET_FEATURE_NAME)",
"_____no_output_____"
],
[
"normal_days_data_list =[\n data_normal[:1000],\n data_normal[1000:2000],\n data_normal[2000:]\n]\n",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(nrows=1, ncols=3, figsize=(20, 5))\nfor i, day_data in enumerate(normal_days_data_list):\n data_normal['Aspect'].plot.hist(bins=15, ax=axes[i])",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(nrows=1, ncols=3, figsize=(20, 5))\nfor i, day_data in enumerate(normal_days_data_list):\n data_normal['Wilderness_Area'].value_counts(normalize=True).plot.bar(ax=axes[i])",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(nrows=1, ncols=3, figsize=(20, 5))\nfor i, day_data in enumerate(normal_days_data_list):\n data_normal[TARGET_FEATURE_NAME].value_counts(normalize=True).plot.bar(ax=axes[i])",
"_____no_output_____"
],
[
"serving_data['2020-04-01'] = normal_days_data_list[0]\nserving_data['2020-04-02'] = normal_days_data_list[1]\nserving_data['2020-04-03'] = normal_days_data_list[2]",
"_____no_output_____"
]
],
[
[
"## 4. Preparing Skewed Data\nWe are going to introduce the following skews to the data:\n1. **Numerical Features**\n 1. *Elevation - Feature Skew*: Convert the unit of measure from meters to kilometers in 2020-04-05\n 2. *Aspect - Distribution Skew*: gradual decrease of the value\n\n2. **Categorical Features**\n 1. *Wilderness_Area - Feature Skew*: Adding a new category \"Others\" in 2020-04-05\n 2. *Wilderness_Area - Distribution Skew*: Gradual increase of of the frequency of \"Cache\" and \"Neota\" values\n3. **Target Features**: check the change of the distribution of predictied class labels",
"_____no_output_____"
]
],
[
[
"data_to_skew = data.sample(3000)\nserving_data['2020-04-04'] = data_to_skew[:1000]\nserving_data['2020-04-05'] = data_to_skew[1000:2000]\nserving_data['2020-04-06'] = data_to_skew[2000:]",
"_____no_output_____"
]
],
[
[
"### 4.1 Skew numerical features",
"_____no_output_____"
],
[
"#### 4.1.1 Elevation Feature Skew",
"_____no_output_____"
]
],
[
[
"serving_data['2020-04-05']['Elevation'] = serving_data['2020-04-05']['Elevation'] / 1000",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(20, 5))\nserving_data['2020-04-01']['Elevation'].plot.hist(bins=15, ax=axes[0], title='Elevation Meters')\nserving_data['2020-04-05']['Elevation'].plot.hist(bins=15, ax=axes[1], title='Elevation Kilometers')",
"_____no_output_____"
]
],
[
[
"#### 4.1.2 Aspect Distribution Skew",
"_____no_output_____"
]
],
[
[
"serving_data['2020-04-04']['Aspect'] = serving_data['2020-04-04']['Aspect'].apply(\n lambda value: value * np.random.uniform(0.90, 1) if value > 250 else value \n)\n\nserving_data['2020-04-05']['Aspect'] = serving_data['2020-04-05']['Aspect'].apply(\n lambda value: value * np.random.uniform(0.85, 1) if value > 250 else value \n)\n\nserving_data['2020-04-06']['Aspect'] = serving_data['2020-04-06']['Aspect'].apply(\n lambda value: value * np.random.uniform(0.80, 1) if value > 250 else value \n)",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(nrows=2, ncols=3, figsize=(20, 10))\nserving_data['2020-04-01']['Aspect'].plot.hist(bins=15, ax=axes[0, 0], title='Aspect - day 1')\nserving_data['2020-04-02']['Aspect'].plot.hist(bins=15, ax=axes[0, 1], title='Aspect - day 2')\nserving_data['2020-04-03']['Aspect'].plot.hist(bins=15, ax=axes[0, 2], title='Aspect - day 3')\nserving_data['2020-04-04']['Aspect'].plot.hist(bins=15, ax=axes[1, 0], title='Aspect - day 4')\nserving_data['2020-04-05']['Aspect'].plot.hist(bins=15, ax=axes[1, 1], title='Aspect - day 5')\nserving_data['2020-04-06']['Aspect'].plot.hist(bins=15, ax=axes[1, 2], title='Aspect - day 6')",
"_____no_output_____"
]
],
[
[
"### 4.2 Skew categorical features",
"_____no_output_____"
],
[
"#### 4.2.1 Wilderness Area Feature Skew\nAdding a new category \"Others\"\n",
"_____no_output_____"
]
],
[
[
"serving_data['2020-04-05']['Wilderness_Area'] = serving_data['2020-04-05']['Wilderness_Area'].apply(\n lambda value: 'Others' if np.random.uniform() <= 0.1 else value\n)",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(nrows=1, ncols=3, figsize=(20, 5))\nserving_data['2020-04-04']['Wilderness_Area'].value_counts(normalize=True).plot.bar(ax=axes[0], title='Wilderness Area - Normal')\nserving_data['2020-04-05']['Wilderness_Area'].value_counts(normalize=True).plot.bar(ax=axes[1], title='Wilderness Area - New Category')\nserving_data['2020-04-06']['Wilderness_Area'].value_counts(normalize=True).plot.bar(ax=axes[2], title='Wilderness Area - Normal')",
"_____no_output_____"
]
],
[
[
"#### 4.2.2 Wilderness Area Distribution Skew",
"_____no_output_____"
]
],
[
[
"serving_data['2020-04-04']['Wilderness_Area'] = serving_data['2020-04-04']['Wilderness_Area'].apply(\n lambda value: 'Neota' if value in ['Rawah', 'Commanche'] and np.random.uniform() <= 0.1 else value\n)\n\nserving_data['2020-04-05']['Wilderness_Area'] = serving_data['2020-04-05']['Wilderness_Area'].apply(\n lambda value: 'Neota' if value in ['Rawah', 'Commanche'] and np.random.uniform() <= 0.15 else value\n)\n\nserving_data['2020-04-06']['Wilderness_Area'] = serving_data['2020-04-06']['Wilderness_Area'].apply(\n lambda value: 'Neota' if value in ['Rawah', 'Commanche'] and np.random.uniform() <= 0.2 else value\n)",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(nrows=2, ncols=3, figsize=(20, 10))\nserving_data['2020-04-01']['Wilderness_Area'].value_counts(normalize=True).plot.bar(ax=axes[0, 0], title='Wilderness Area - day 1')\nserving_data['2020-04-02']['Wilderness_Area'].value_counts(normalize=True).plot.bar(ax=axes[0, 1], title='Wilderness Area - day 2')\nserving_data['2020-04-03']['Wilderness_Area'].value_counts(normalize=True).plot.bar(ax=axes[0, 2], title='Wilderness Area - day 3')\nserving_data['2020-04-04']['Wilderness_Area'].value_counts(normalize=True).plot.bar(ax=axes[1, 0], title='Wilderness Area - day 4')\nserving_data['2020-04-05']['Wilderness_Area'].value_counts(normalize=True).plot.bar(ax=axes[1, 1], title='Wilderness Area - day 5')\nserving_data['2020-04-06']['Wilderness_Area'].value_counts(normalize=True).plot.bar(ax=axes[1, 2], title='Wilderness Area - day 6')\n",
"_____no_output_____"
]
],
[
[
"## 5. Simulating serving workload",
"_____no_output_____"
],
[
"### 5.1 Implement the model API client",
"_____no_output_____"
]
],
[
[
"partitions = {}\nfor time_stamp, data in serving_data.items():\n instances = []\n for _, row in data.iterrows():\n instance = {}\n for column_name in data.columns:\n if column_name == TARGET_FEATURE_NAME: continue\n instance[column_name] = [str(row[column_name])] if column_name in CATEGORICAL_FEATURE_NAMES else [row[column_name]]\n instances.append(instance)\n partitions[time_stamp] = instances\n\n\n",
"_____no_output_____"
],
[
"path_root = '/home/jarekk/repos/drift-monitor/online_prediction_simulater/skewed_data'",
"_____no_output_____"
],
[
"import json\n\nfor partition in partitions.keys():\n print(partition)\n tf_io.gfile.makedirs(path_root + '/' +partition)",
"2020-04-01\n2020-04-02\n2020-04-03\n2020-04-04\n2020-04-05\n2020-04-06\n"
],
[
"for partition in partitions.keys():\n file_path = '{}/json-{}.txt'.format(path_root, partition)\n with open(file_path, 'w') as f:\n for instance in partitions[partition]:\n instance_json = json.dumps(instance)\n f.write(instance_json)\n f.write('\\n')\n\n ",
"_____no_output_____"
],
[
"import googleapiclient.discovery\nimport numpy as np\n\nservice = googleapiclient.discovery.build('ml', 'v1')\nname = 'projects/{}/models/{}/versions/{}'.format(PROJECT, MODEL_NAME, VERSION_NAME)\nprint(\"Service name: {}\".format(name))\n\ndef caip_predict(instances):\n \n serving_instances = []\n for instance in instances:\n serving_instances.append(\n {key: [value] for key, value in instance.items()})\n \n request_body={\n 'signature_name': SIGNATURE_NAME,\n 'instances': serving_instances}\n\n response = service.projects().predict(\n name=name,\n body=request_body\n\n ).execute()\n\n if 'error' in response:\n raise RuntimeError(response['error'])\n\n probability_list = [output[MODEL_OUTPUT_KEY] for output in response['predictions']]\n classes = [FEATURE_LABELS[int(np.argmax(probabilities))] for probabilities in probability_list]\n return classes",
"_____no_output_____"
]
],
[
[
"### 5.2 Prepare the request instances",
"_____no_output_____"
]
],
[
[
"def prepare_instances(serving_data):\n instances = []\n\n for request_timestamp, data in serving_data.items():\n for _, row in data.iterrows():\n instance = dict()\n for column_name in data.columns:\n if column_name == TARGET_FEATURE_NAME: continue\n instance[column_name] = str(row[column_name]) if column_name in CATEGORICAL_FEATURE_NAMES else row[column_name]\n instance['request_timestamp'] = request_timestamp\n instances.append(instance)\n\n return instances\n",
"_____no_output_____"
],
[
"import time\n\ndef simulate_requests():\n instances = prepare_instances(serving_data)\n\n print(\"Simulation started...\")\n print(\"--------------------\")\n print(\"Number of instances: {}\".format(len(instances)))\n\n for i, instance in enumerate(instances):\n predicted_class = caip_predict([instance])\n print(\".\", end='')\n \n if (i + 1) % 100 == 0:\n print()\n print(\"Sent {} requests.\".format(i + 1))\n\n #time.sleep(0.1)\n print(\"\")\n print(\"--------------------\")\n print(\"Simulation finised.\")\n ",
"_____no_output_____"
],
[
"simulate_requests()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
eced497aa43820e1d47be036725892c600691c42 | 8,930 | ipynb | Jupyter Notebook | Chapman/Ch6-Problem_6-31.ipynb | dietmarw/EK5312 | 33b31d953f782043db0264116881060c9f059731 | [
"Unlicense"
] | 12 | 2017-07-16T22:28:25.000Z | 2021-11-08T05:45:58.000Z | Chapman/Ch6-Problem_6-31.ipynb | dietmarw/EK5312 | 33b31d953f782043db0264116881060c9f059731 | [
"Unlicense"
] | null | null | null | Chapman/Ch6-Problem_6-31.ipynb | dietmarw/EK5312 | 33b31d953f782043db0264116881060c9f059731 | [
"Unlicense"
] | 7 | 2018-01-17T15:01:33.000Z | 2021-07-02T19:57:22.000Z | 22.325 | 731 | 0.469989 | [
[
[
"# Excercises Electric Machinery Fundamentals\n## Chapter 6",
"_____no_output_____"
],
[
"## Problem 6-31",
"_____no_output_____"
]
],
[
[
"%pylab notebook",
"Populating the interactive namespace from numpy and matplotlib\n"
]
],
[
[
"### Description",
"_____no_output_____"
],
[
"When it is necessary to stop an induction motor very rapidly, many induction motor controllers reverse the direction of rotation of the magnetic fields by switching any two stator leads. When the direction of rotation of the magnetic fields is reversed, the motor develops an induced torque opposite to the current direction of rotation, so it quickly stops and tries to start turning in the opposite direction. If power is removed from the stator circuit at the moment when the rotor speed goes through zero, then the motor has been stopped very rapidly. This technique for rapidly stopping an induction motor is called plugging . The motor of Problem 6-21 is running at rated conditions and is to be stopped by plugging.\n\n#### (a)\n * What is the slip s before plugging?\n \n#### (b)\n * What is the frequency of the rotor before plugging?\n \n#### (c)\n * What is the induced torque $\\tau_\\text{ind}$ before plugging?\n \n#### (d)\n * What is the slip s immediately after switching the stator leads?\n \n#### (e)\n * What is the frequency of the rotor immediately after switching the stator leads?\n \n#### (f)\n * What is the induced torque $\\tau_\\text{ind}$ immediately after switching the stator leads?",
"_____no_output_____"
]
],
[
[
"R1 = 0.54 # [Ohm]\nR2 = 0.488 # [Ohm]\nXm = 51.12 # [Ohm]\nX1 = 2.093 # [Ohm]\nX2 = 3.209 # [Ohm]\nPcore = 150 # [W]\nPf_w = 150 # [W]\nPmisc = 50 # [W]\nV = 460 # [V]\np = 4\nfse = 60 # [Hz]",
"_____no_output_____"
]
],
[
[
"### SOLUTION",
"_____no_output_____"
],
[
"#### (a)\nThe slip before plugging is (see [Problem 6-21](Problem_6-21.ipynb)).",
"_____no_output_____"
]
],
[
[
"s = 0.02\nprint('''\ns = {:.2f} \n========'''.format(s))",
"\ns = 0.02 \n========\n"
]
],
[
[
"#### (b)\nThe frequency of the rotor before plugging is:\n\n$$f_r = sf_s$$",
"_____no_output_____"
]
],
[
[
"fr = s * fse\nprint('''\nfr = {:.1f} Hz\n==========='''.format(fr))",
"\nfr = 1.2 Hz\n===========\n"
]
],
[
[
"#### (c)\nThe induced torque before plugging in the direction of motion is (see [Ch6-Problem 6-21](Ch6-Problem_6-21.ipynb)).",
"_____no_output_____"
]
],
[
[
"tau_ind = 39.2 # [Nm]\nprint('''\ntau_ind = {:.1f} Nm\n================='''.format(tau_ind))",
"\ntau_ind = 39.2 Nm\n=================\n"
]
],
[
[
"#### (d)\nAfter switching stator leads, the synchronous speed becomes –1800 r/min, while the mechanical speed initially remains 1764 r/min. \nTherefore, the slip becomes:\n\n$$s = \\frac{n_\\text{sync} - n_{m}}{n_\\text{sync}}$$",
"_____no_output_____"
]
],
[
[
"n_sync = -1800.0 # [r/min]\nn_m = 1764.0 # [r/min]\ns_after = (n_sync - n_m) / n_sync\nprint('''\ns_after = {:.2f}\n=============='''.format(s_after))",
"\ns_after = 1.98\n==============\n"
]
],
[
[
"#### (e)\nThe frequency of the rotor after plugging is\n\n$$f_r = sf_e$$",
"_____no_output_____"
]
],
[
[
"fr_after = s_after * fse\nprint('''\nfr_after = {:.1f} Hz\n==================='''.format(fr_after))",
"\nfr_after = 118.8 Hz\n===================\n"
]
],
[
[
"#### (f)\nThe equivalent circuit for this motor is:\n\n<img src=\"figs/Problem_6-31.jpg\" width=\"70%\">",
"_____no_output_____"
],
[
"The Thevenin equivalent of the input circuit is:\n\n$$Z_{TH} = \\frac{jX_M(R_1 + jX_1)}{R_1 + j(X_1 + X_M)}$$",
"_____no_output_____"
]
],
[
[
"Zth = (Xm*1j * (R1 + X1*1j)) / (R1 + (X1 + Xm)*1j)\nZth_angle = arctan(Zth.imag / Zth.real)\nprint('Zth = ({:.4f})Ω = {:.3f} Ω ∠{:.1f}°'\n .format(Zth, abs(Zth), Zth_angle/pi *180))",
"Zth = (0.4983+2.0157j)Ω = 2.076 Ω ∠76.1°\n"
]
],
[
[
"$$\\vec{V}_{TH} = \\frac{jX_M}{R_1 + j(X_1 +X_M)}\\vec{V}_\\phi$$",
"_____no_output_____"
]
],
[
[
"V_PHASE = V / sqrt(3)\nVTH = (Xm*1j) / (R1 + (X1 + Xm)*1j) * V_PHASE\nVTH_angle = arctan(VTH.imag / VTH.real)\nprint('VTH = {:.0f} V ∠{:.2f}°'\n .format(abs(VTH), VTH_angle/pi *180))",
"VTH = 255 V ∠0.58°\n"
]
],
[
[
"The induced torque immediately after switching the stator leads is:\n\n$$\\tau_\\text{ind} = \\frac{3V^2_{TH}R_2/s}{\\omega_\\text{sync}[(R_{TH} + R_2/s)^2 + (X_{TH} + X_2)^2]}$$",
"_____no_output_____"
]
],
[
[
"Rth = Zth.real\nXth = Zth.imag\nw_sync = abs(n_sync * (2*pi/60.0))\ntau_ind = ((3 * abs(VTH)**2 * R2/s_after) /\n (w_sync * ((Rth + R2/s_after)**2 + (Xth + X2)**2)))\nprint('''\ntau_ind = {:.1f} Nm, against the direction of motion\n================================================='''.format(tau_ind))",
"\ntau_ind = 9.2 Nm, against the direction of motion\n=================================================\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
eced562e2203829b7fb3473a1468d98d8456635e | 686,171 | ipynb | Jupyter Notebook | examples/Notebooks/flopy3_modpath7_create_simulation.ipynb | tomvansteijn/flopy | 06b0a71b9b69600d61c5233fd068946627e6cdad | [
"CC0-1.0",
"BSD-3-Clause"
] | null | null | null | examples/Notebooks/flopy3_modpath7_create_simulation.ipynb | tomvansteijn/flopy | 06b0a71b9b69600d61c5233fd068946627e6cdad | [
"CC0-1.0",
"BSD-3-Clause"
] | null | null | null | examples/Notebooks/flopy3_modpath7_create_simulation.ipynb | tomvansteijn/flopy | 06b0a71b9b69600d61c5233fd068946627e6cdad | [
"CC0-1.0",
"BSD-3-Clause"
] | null | null | null | 951.693481 | 283,280 | 0.951712 | [
[
[
"# FloPy\n\n## MODPATH 7 create simulation example\n\nThis notebook demonstrates how to create a simple forward and backward MODPATH 7 simulation using the `.create_mp7()` method. The notebooks also shows how to create subsets of endpoint output and plot MODPATH results on ModelMap objects.",
"_____no_output_____"
]
],
[
[
"import sys\nimport os\nimport numpy as np\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport platform\n\n# run installed version of flopy or add local path\ntry:\n import flopy\nexcept:\n fpth = os.path.abspath(os.path.join('..', '..'))\n sys.path.append(fpth)\n import flopy\n\nprint(sys.version)\nprint('numpy version: {}'.format(np.__version__))\nprint('matplotlib version: {}'.format(mpl.__version__))\nprint('flopy version: {}'.format(flopy.__version__))\n\nif not os.path.exists(\"data\"):\n os.mkdir(\"data\") ",
"3.7.7 (default, Mar 26 2020, 10:32:53) \n[Clang 4.0.1 (tags/RELEASE_401/final)]\nnumpy version: 1.19.2\nmatplotlib version: 3.3.0\nflopy version: 3.3.2\n"
],
[
"# define executable names\nmpexe = \"mp7\"\nmfexe = \"mf6\"\nif platform.system() == \"Windows\":\n mpexe += \".exe\"\n mfexe += \".exe\"",
"_____no_output_____"
]
],
[
[
"### Flow model data",
"_____no_output_____"
]
],
[
[
"nper, nstp, perlen, tsmult = 1, 1, 1., 1.\nnlay, nrow, ncol = 3, 21, 20\ndelr = delc = 500.\ntop = 400.\nbotm = [220., 200., 0.]\nlaytyp = [1, 0, 0]\nkh = [50., 0.01, 200.]\nkv = [10., 0.01, 20.]\nwel_loc = (2, 10, 9)\nwel_q = -150000.\nrch = 0.005\nriv_h = 320.\nriv_z = 317.\nriv_c = 1.e5",
"_____no_output_____"
],
[
"def get_nodes(locs):\n nodes = []\n for k, i, j in locs:\n nodes.append(k * nrow * ncol + i * ncol + j)\n return nodes",
"_____no_output_____"
]
],
[
[
"### MODPATH 7 using MODFLOW 6\n\n#### Create and run MODFLOW 6",
"_____no_output_____"
]
],
[
[
"ws = os.path.join('data', 'mp7_ex1_cs')\nnm = 'ex01_mf6'\n\n# Create the Flopy simulation object\nsim = flopy.mf6.MFSimulation(sim_name=nm, exe_name=mfexe,\n version='mf6', sim_ws=ws)\n\n# Create the Flopy temporal discretization object\npd = (perlen, nstp, tsmult)\ntdis = flopy.mf6.modflow.mftdis.ModflowTdis(sim, pname='tdis',\n time_units='DAYS', nper=nper,\n perioddata=[pd])\n\n# Create the Flopy groundwater flow (gwf) model object\nmodel_nam_file = '{}.nam'.format(nm)\ngwf = flopy.mf6.ModflowGwf(sim, modelname=nm,\n model_nam_file=model_nam_file, save_flows=True)\n\n# Create the Flopy iterative model solver (ims) Package object\nims = flopy.mf6.modflow.mfims.ModflowIms(sim, pname='ims', \n complexity='SIMPLE',\n outer_hclose=1e-6,\n inner_hclose=1e-6,\n rcloserecord=1e-6)\n\n# create gwf file\ndis = flopy.mf6.modflow.mfgwfdis.ModflowGwfdis(gwf, pname='dis', nlay=nlay,\n nrow=nrow, ncol=ncol,\n length_units='FEET',\n delr=delr, delc=delc,\n top=top,\n botm=botm)\n# Create the initial conditions package\nic = flopy.mf6.modflow.mfgwfic.ModflowGwfic(gwf, pname='ic', strt=top)\n\n# Create the node property flow package\nnpf = flopy.mf6.modflow.mfgwfnpf.ModflowGwfnpf(gwf, pname='npf',\n icelltype=laytyp, k=kh,\n k33=kv)\n\n\n# recharge\nflopy.mf6.modflow.mfgwfrcha.ModflowGwfrcha(gwf, recharge=rch)\n# wel\nwd = [(wel_loc, wel_q)]\nflopy.mf6.modflow.mfgwfwel.ModflowGwfwel(gwf, maxbound=1,\n stress_period_data={0: wd})\n# river\nrd = []\nfor i in range(nrow):\n rd.append([(0, i, ncol - 1), riv_h, riv_c, riv_z])\nflopy.mf6.modflow.mfgwfriv.ModflowGwfriv(gwf, stress_period_data={0: rd})\n# Create the output control package\nheadfile = '{}.hds'.format(nm)\nhead_record = [headfile]\nbudgetfile = '{}.cbb'.format(nm)\nbudget_record = [budgetfile]\nsaverecord = [('HEAD', 'ALL'),\n ('BUDGET', 'ALL')]\noc = flopy.mf6.modflow.mfgwfoc.ModflowGwfoc(gwf, pname='oc',\n saverecord=saverecord,\n head_filerecord=head_record,\n budget_filerecord=budget_record)\n\n# Write the datasets\nsim.write_simulation()\n# Run the simulation\nsuccess, buff = sim.run_simulation()\nassert success, 'mf6 model did not run'",
"writing simulation...\n writing simulation name file...\n writing simulation tdis package...\n writing ims package ims...\n writing model ex01_mf6...\n writing model name file...\n writing package dis...\n writing package ic...\n writing package npf...\n writing package rcha...\n writing package wel_0...\n writing package riv_0...\nINFORMATION: maxbound in ('gwf6', 'riv', 'dimensions') changed to 21 based on size of stress_period_data\n writing package oc...\nFloPy is using the following executable to run the model: /Users/jdhughes/.local/bin/mf6\n MODFLOW 6\n U.S. GEOLOGICAL SURVEY MODULAR HYDROLOGIC MODEL\n VERSION 6.2.0 10/22/2020\n\n MODFLOW 6 compiled Oct 25 2020 16:15:39 with IFORT compiler (ver. 19.0.5)\n\nThis software has been approved for release by the U.S. Geological \nSurvey (USGS). Although the software has been subjected to rigorous \nreview, the USGS reserves the right to update the software as needed \npursuant to further analysis and review. No warranty, expressed or \nimplied, is made by the USGS or the U.S. Government as to the \nfunctionality of the software and related material nor shall the \nfact of release constitute any such warranty. Furthermore, the \nsoftware is released on condition that neither the USGS nor the U.S. \nGovernment shall be held liable for any damages resulting from its \nauthorized or unauthorized use. Also refer to the USGS Water \nResources Software User Rights Notice for complete use, copyright, \nand distribution information.\n\n \n Run start date and time (yyyy/mm/dd hh:mm:ss): 2020/10/26 15:56:47\n \n Writing simulation list file: mfsim.lst\n Using Simulation name file: mfsim.nam\n \n Solving: Stress period: 1 Time step: 1\n \n Run end date and time (yyyy/mm/dd hh:mm:ss): 2020/10/26 15:56:47\n Elapsed run time: 0.029 Seconds\n \n\nWARNING REPORT:\n\n 1. NONLINEAR BLOCK VARIABLE 'OUTER_HCLOSE' IN FILE 'ex01_mf6.ims' WAS\n DEPRECATED IN VERSION 6.1.1. SETTING OUTER_DVCLOSE TO OUTER_HCLOSE VALUE.\n 2. LINEAR BLOCK VARIABLE 'INNER_HCLOSE' IN FILE 'ex01_mf6.ims' WAS\n DEPRECATED IN VERSION 6.1.1. SETTING INNER_DVCLOSE TO INNER_HCLOSE VALUE.\n Normal termination of simulation.\n"
]
],
[
[
"Get locations to extract data",
"_____no_output_____"
]
],
[
[
"nodew = get_nodes([wel_loc])\ncellids = gwf.riv.stress_period_data.get_data()[0]['cellid']\nnodesr = get_nodes(cellids)",
"_____no_output_____"
]
],
[
[
"#### Create and run MODPATH 7\n\nForward tracking",
"_____no_output_____"
]
],
[
[
"# create modpath files\nmpnamf = nm + '_mp_forward'\n\n# create basic forward tracking modpath simulation\nmp = flopy.modpath.Modpath7.create_mp7(modelname=mpnamf, trackdir='forward', flowmodel=gwf, model_ws=ws, \n rowcelldivisions=1, columncelldivisions=1, layercelldivisions=1,\n exe_name=mpexe)\n\n# write modpath datasets\nmp.write_input()\n\n# run modpath\nmp.run_model()",
"FloPy is using the following executable to run the model: /Users/jdhughes/.local/bin/mp7\n\nMODPATH Version 7.2.001 \nProgram compiled Oct 25 2020 16:19:48 with IFORT compiler (ver. 19.0.5) \n \n \nRun particle tracking simulation ...\nProcessing Time Step 1 Period 1. Time = 1.00000E+00 Steady-state flow \n"
]
],
[
[
"Backward tracking from well and river locations",
"_____no_output_____"
]
],
[
[
"# create modpath files\nmpnamb = nm + '_mp_backward'\n\n# create basic forward tracking modpath simulation\nmp = flopy.modpath.Modpath7.create_mp7(modelname=mpnamb, trackdir='backward', flowmodel=gwf, model_ws=ws, \n rowcelldivisions=5, columncelldivisions=5, layercelldivisions=5,\n nodes=nodew+nodesr, exe_name=mpexe)\n\n# write modpath datasets\nmp.write_input()\n\n# run modpath\nmp.run_model()",
"FloPy is using the following executable to run the model: /Users/jdhughes/.local/bin/mp7\n\nMODPATH Version 7.2.001 \nProgram compiled Oct 25 2020 16:19:48 with IFORT compiler (ver. 19.0.5) \n \n \nRun particle tracking simulation ...\nProcessing Time Step 1 Period 1. Time = 1.00000E+00 Steady-state flow \n"
]
],
[
[
"#### Load and Plot MODPATH 7 output\n\n##### Forward Tracking",
"_____no_output_____"
],
[
"Load forward tracking pathline data",
"_____no_output_____"
]
],
[
[
"fpth = os.path.join(ws, mpnamf + '.mppth')\np = flopy.utils.PathlineFile(fpth)\npw = p.get_destination_pathline_data(dest_cells=nodew)\npr = p.get_destination_pathline_data(dest_cells=nodesr)",
"_____no_output_____"
]
],
[
[
"Load forward tracking endpoint data",
"_____no_output_____"
]
],
[
[
"fpth = os.path.join(ws, mpnamf + '.mpend')\ne = flopy.utils.EndpointFile(fpth)",
"_____no_output_____"
]
],
[
[
"Get forward particles that terminate in the well",
"_____no_output_____"
]
],
[
[
"well_epd = e.get_destination_endpoint_data(dest_cells=nodew) ",
"_____no_output_____"
]
],
[
[
"Get particles that terminate in the river boundaries",
"_____no_output_____"
]
],
[
[
"riv_epd = e.get_destination_endpoint_data(dest_cells=nodesr)",
"_____no_output_____"
]
],
[
[
"Well and river forward tracking pathlines",
"_____no_output_____"
]
],
[
[
"colors = ['green', 'orange', 'red']",
"_____no_output_____"
],
[
"f, axes = plt.subplots(ncols=3, nrows=2, sharey=True, sharex=True, figsize=(15, 10))\naxes = axes.flatten()\n\nidax = 0\nfor k in range(nlay):\n ax = axes[idax]\n ax.set_aspect('equal')\n ax.set_title('Well pathlines - Layer {}'.format(k+1))\n mm = flopy.plot.PlotMapView(model=gwf, ax=ax)\n mm.plot_grid(lw=0.5)\n mm.plot_pathline(pw, layer=k, color=colors[k], lw=0.75)\n idax += 1\n\nfor k in range(nlay):\n ax = axes[idax]\n ax.set_aspect('equal')\n ax.set_title('River pathlines - Layer {}'.format(k+1))\n mm = flopy.plot.PlotMapView(model=gwf, ax=ax)\n mm.plot_grid(lw=0.5)\n mm.plot_pathline(pr, layer=k, color=colors[k], lw=0.75)\n idax += 1\n \nplt.tight_layout();",
"_____no_output_____"
]
],
[
[
"Forward tracking endpoints captured by the well and river",
"_____no_output_____"
]
],
[
[
"f, axes = plt.subplots(ncols=2, nrows=1, sharey=True, figsize=(10, 5))\naxes = axes.flatten()\n\nax = axes[0]\nax.set_aspect('equal')\nax.set_title('Well recharge area')\nmm = flopy.plot.PlotMapView(model=gwf, ax=ax)\nmm.plot_grid(lw=0.5)\nmm.plot_endpoint(well_epd, direction='starting', colorbar=True, shrink=0.5);\n\nax = axes[1]\nax.set_aspect('equal')\nax.set_title('River recharge area')\nmm = flopy.plot.PlotMapView(model=gwf, ax=ax)\nmm.plot_grid(lw=0.5)\nmm.plot_endpoint(riv_epd, direction='starting', colorbar=True, shrink=0.5);",
"_____no_output_____"
]
],
[
[
"##### Backward tracking\n\nLoad backward tracking pathlines",
"_____no_output_____"
]
],
[
[
"fpth = os.path.join(ws, mpnamb + '.mppth')\np = flopy.utils.PathlineFile(fpth)\npwb = p.get_destination_pathline_data(dest_cells=nodew)\nprb = p.get_destination_pathline_data(dest_cells=nodesr)",
"_____no_output_____"
]
],
[
[
"Load backward tracking endpoints",
"_____no_output_____"
]
],
[
[
"fpth = os.path.join(ws, mpnamb + '.mpend')\ne = flopy.utils.EndpointFile(fpth)\newb = e.get_destination_endpoint_data(dest_cells=nodew, source=True)\nerb = e.get_destination_endpoint_data(dest_cells=nodesr, source=True)",
"_____no_output_____"
]
],
[
[
"Well backward tracking pathlines",
"_____no_output_____"
]
],
[
[
"f, axes = plt.subplots(ncols=2, nrows=1, figsize=(10, 5))\n\nax = axes[0]\nax.set_aspect('equal')\nax.set_title('Well recharge area')\nmm = flopy.plot.PlotMapView(model=gwf, ax=ax)\nmm.plot_grid(lw=0.5)\nmm.plot_pathline(pwb, layer='all', color='blue', lw=0.5, linestyle=':', label='captured by wells')\nmm.plot_endpoint(ewb, direction='ending') #, colorbar=True, shrink=0.5);\n\nax = axes[1]\nax.set_aspect('equal')\nax.set_title('River recharge area')\nmm = flopy.plot.PlotMapView(model=gwf, ax=ax)\nmm.plot_grid(lw=0.5)\nmm.plot_pathline(prb, layer='all', color='green', lw=0.5, linestyle=':', label='captured by rivers')\n\nplt.tight_layout();",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
eced57acba0846477e8d8e88bea4c7b2c9c53172 | 22,080 | ipynb | Jupyter Notebook | Ch6_DealingWithOptionalData.ipynb | johnklee/fpu | eabc022e21a237b9bdf07628d1ccb4cedbd89347 | [
"MIT"
] | null | null | null | Ch6_DealingWithOptionalData.ipynb | johnklee/fpu | eabc022e21a237b9bdf07628d1ccb4cedbd89347 | [
"MIT"
] | 4 | 2020-03-24T17:00:40.000Z | 2021-06-01T23:41:07.000Z | Ch6_DealingWithOptionalData.ipynb | johnklee/fpu | eabc022e21a237b9bdf07628d1ccb4cedbd89347 | [
"MIT"
] | null | null | null | 48.849558 | 749 | 0.627989 | [
[
[
"# <font color='blue'>Preface</font>\nThis <a href='http://localhost/jforum/posts/list/4104.page'>Chapter</a> covers \n* The null reference, or “the billion-dollar mistake” \n* Alternatives to null references \n* Developing an Option data type for optional data \n* Applying functions to optional values \n* Composing optional values \n* Option use cases\n\n<b>Representing optional data in computer programs has always been a problem</b>. The concept of optional data is very simple in everyday life. Representing the absence of something when this something is contained in a container is easy—whatever it is, it can be represented by an empty container. An absence of apples can be represented by an empty apple basket. The absence of gasoline in a car can be visualized as an empty gas tank. Representing the absence of data in computer programs is more difficult. Most data is represented as a reference pointing to it, so the most obvious way to represent the absence of data is to use a pointer to nothing. This is what a null pointer is. \n\nIn Python, a variable is a pointer to a value. Variables may be created <b><a href='https://docs.python.org/2/library/constants.html#None'>None</a></b> , and they may then be changed to point to values. They can even be changed again to point to null if data is removed. To handle optional data, in this chapter, you’ll develop the class called <b><font color=\"blue\">Option</font></b>. The goal of this chapter is to learn how this kind of structure works in handling optional data.\n\n# <font color='blue'>Problems with the null pointer</font>\nOne of the most frequent bugs in imperative programs is the <font color='blue'><b>AttributeError</b></font>. This error is raised when an identifier is dereferenced and found to be pointing to nothing. In other words, some data is expected but is found missing. Such an identifier is said to be pointing to null. The null reference was invented in 1965 by Tony Hoare while he was designing the ALGOL object-oriented language. Here’s what he said 44 years later:\n",
"_____no_output_____"
],
[
"# <font color='blue'>The Option data type </font>\nThe <b><font color='blue'>Option</font></b> data type you’ll create in this chapter will be very similar to the List data type. Using an <b><font color='blue'>Option</font></b> type for optional data allows you to compose functions even when the data is absent (<font color='brown'>see figure 6.1</font>). It will be implemented as an abstract class, <b><font color='blue'>Option</font></b>, containing two private subclasses representing the presence and the absence of data. The subclass representing the absence of data will be called <b><font color='blue'>N</font></b> (<font color='brown'>Stands for None</font>), and the subclass representing the presence of data will be called Some. A Some will contain the corresponding data value.\n<img src='https://3.bp.blogspot.com/-55VlPQ6TLEk/WkuDRgPMWII/AAAAAAAAXII/Yj_WnnSeS4UvBMzDflzhwJoz4E8ecuZoQCLcBGAs/s1600/4104_f6-1.PNG'/>\n<b>Figure 6.1. Without the Option type, composing functions wouldn’t produce a function because the resulting program would potentially throw an Exception.</b>\n\n\n## <font color='green'>Getting a value from an Option</font>\nThe first thing you’ll need is a way to retrieve the value in an <b><font color='blue'>Option</font></b>. One frequent use case when data is missing is to use a default value. The <font color='blue'>getOrElse</font> method that will return either the contained value if it exists:",
"_____no_output_____"
]
],
[
[
"from fp import *\n\nprint(\"{}\".format(sys.version_info))\nprint(\"Retrieve default: {}\".format(Option.none().getOr('default value')))\nprint(\"Retrieve value: {}\".format(Option.some('some value').getOr('default value')))\n\ndef getDefault():\n print('api:getDefault is called')\n return 0\n\n# What if we only want api:getDefault to be called only when needed?\nprint(\"Retrieve: {}\".format(Option.some(123).getOr(getDefault())))\nprint(\"Retrieve: {}\".format(Option.none().getOr(getDefault())))",
"sys.version_info(major=2, minor=7, micro=12, releaselevel='final', serial=0)\nRetrieve default: default value\nRetrieve value: some value\napi:getDefault is called\nRetrieve: 123\napi:getDefault is called\nRetrieve: 0\n"
]
],
[
[
"Or you can use lazy evaluation for the <font color='blue'>getOrElse</font> method parameter:",
"_____no_output_____"
]
],
[
[
"print(\"Retrieve: {}\".format(Option.some(123).getOrElse(getDefault)))\nprint(\"Retrieve: {}\".format(Option.none().getOrElse(getDefault)))",
"Retrieve: 123\napi:getDefault is called\nRetrieve: 0\n"
]
],
[
[
"## <font color='green'>Applying functions to optional values</font>\nOne very important method in <b><font color='blue'>List</font></b> is the <font color='blue'>map</font> method, which allows you to apply a function from A to B to each element of a list of A, producing a list of B. Considering that an <b><font color='blue'>Option</font></b> is like a list containing at most one element, you can apply the same principle. ",
"_____no_output_____"
]
],
[
[
"some = Option.some(1)\nnone = Option.none()\ndouble = lambda e: e * 2\nprint(\"Double {}={}\".format(some, some.map(double)))\nprint(\"Double {}={}\".format(none, none.map(double)))",
"Double Some(1)=Some(2)\nDouble None()=None()\n"
]
],
[
[
"## <font color='green'>Dealing with Option composition</font>\nAs you’ll soon realize, functions from A to B aren’t the most common ones in functional programming. \nAt first you may have trouble getting acquainted with functions returning optional values. \nAfter all, it seems to involve extra work to wrap values in <b><font color='blue'>Some</font></b> instances and later retrieve these values. \nBut with further practice, you’ll see that these operations occur only rarely. \n<b>When chaining functions to build a complex computation, you’ll often start with a value that’s returned by some previous computation and pass the result to a new function without seeing the intermediate result. In other words, you’ll more often use functions from A to <font color='blue'>Option<B></font> than functions from A to B.</b> \n\n<font color='blue'>flatMap</font> instance method that takes as an argument a function from A to <b><font color='blue'>Option<B></font></b> and returns an <b><font color='blue'>Option<B></font></b>:",
"_____no_output_____"
]
],
[
[
"some = Option.some(1)\nnone = Option.none()\naddOne = lambda e: Option.some(e + 1)\ndouble = lambda e: Option.some(e * 2)\n\nprint(\"some.flatMap(addOne).flatMap(double)={}\".format(some.flatMap(addOne).flatMap(double)))\nprint(\"none.flatMap(addOne).flatMap(double)={}\".format(none.flatMap(addOne).flatMap(double)))",
"some.flatMap(addOne).flatMap(double)=Some(4)\nnone.flatMap(addOne).flatMap(double)=None()\n"
]
],
[
[
"Most of the case, there’s no need to “get” the value. Instead, you need to get the default value wrapped in <b><font color='blue'>Option</font></b> for chaining functions. Then <font color='blue'>orElse</font> will meet your need. For example:",
"_____no_output_____"
]
],
[
[
"some = Option.some(1)\nnone = Option.none()\ndef df():\n print('api:df is called')\n return Option.some(-1)\n\nprint(\"some.OrElse(df)={}\".format(some.orElse(df)))\nprint(\"none.OrElse(df)={}\".format(none.orElse(df)))",
"some.OrElse(df)=Some(1)\napi:df is called\nnone.OrElse(df)=Some(-1)\n"
]
],
[
[
"More useful, you can use <font color='blue'>filter</font> to pass in the filter function which will return the original <b><font color='blue'>Some</font></b> object if True; otherwise, <b><font color='blue'>N</font></b> is returned:",
"_____no_output_____"
]
],
[
[
"some1 = Option.some(1)\nsome2 = Option.some(2)\nnone = Option.none()\nbt1 = lambda e: e > 1 # Bigger than one\n\nprint(\"some1.filter(bt1)={}\".format(some1.filter(bt1)))\nprint(\"some2.filter(bt1)={}\".format(some2.filter(bt1)))\nprint(\"none.filter(bt1)={}\".format(none.filter(bt1)))",
"some1.filter(bt1)=None()\nsome2.filter(bt1)=Some(2)\nnone.filter(bt1)=None()\n"
]
],
[
[
"## <font color='green'>Option use cases</font>\nThe best way to use <b><font color='blue'>Option</font></b> is through composition. To do this, you must create all the necessary methods for all use cases. These use cases correspond to what you’d do with the value after testing that it’s not null. You could do one of the following: \n* Use the value as the input to another function \n* Apply an effect to the value \n* Use the value if it’s not null, or use a default value to apply a function or an effect\n\nBelow is a classis example in using <b><font color='blue'>Option</font></b>:",
"_____no_output_____"
]
],
[
[
"class TMap:\n def __init__(self):\n self.map = {}\n \n def get(self, k):\n if k in self.map:\n return Option.some(self.map[k])\n else:\n return Option.none()\n \n def put(self, k, v):\n self.map[k] = v\n return self\n \n def removeKey(self, k):\n if k in self.map:\n ov = self.map[k]\n del self.map[k]\n return Option.some(ov)\n else:\n return Option.none()\n \nclass Toon:\n def __init__(self, fn, sn, email=Option.none()):\n self.fn = fn\n self.sn = sn\n if isinstance(email, Option):\n self.email = email\n else:\n self.email = Option.some(email)\n \n def __str__(self):\n return \"{}/{} ({})\".format(self.fn, self.sn, email.getOrEalse(lambda: \"No data\"))\n\ntoons = TMap()\nfor k, t in [(\"Mickey\", Toon(\"Mickey\", \"Mouse\", \"[email protected]\")),\n (\"Minnie\", Toon(\"Minnie\", \"Mouse\")),\n (\"Donald\", Toon(\"Donald\", \"Duck\", \"[email protected]\"))]: \n toons.put(k, t)\n \ndef getEmail(e):\n return e.email\n\ndef dft():\n return \"No data\"\n\nmickey = toons.get(\"Mickey\").flatMap(getEmail)\nminnie = toons.get(\"Minnie\").flatMap(getEmail)\ngoofy = toons.get(\"Goofy\").flatMap(getEmail)\n\nprint(\"Mickey: {}\".format(mickey.getOrElse(dft)))\nprint(\"Minnie: {}\".format(minnie.getOrElse(dft)))\nprint(\"Goofy: {}\".format(goofy.getOrElse(dft)))",
"Mickey: [email protected]\nMinnie: No data\nGoofy: No data\n"
]
],
[
[
"# <font color='blue'>Miscellaneous utilities for Option</font>\nIn order to make <font color='blue'><b>Option</b></font> as useful as possible, you need to add some utility methods. Some of these methods are a must, and others are questionable because their use is not in the spirit of functional programming. You nevertheless must consider adding them. You may need a method to test whether an Option is a None or a Some. You may also need an equals method for comparing options, in which case you mustn’t forget to define a compatible hashCode method.\n\n## <font color='green'>Testing for Some or None</font>\nUntil now, you haven’t needed to test an option to know whether it was a <font color=blue><b>Some</b></font> or a <font color=blue><b>None</b></font>. Ideally, you should never have to do this. In practice, though, there are times when it’s simpler to use this trick than to resort to real functional techniques. For example, you defined the <i>map2</i> method as:\n",
"_____no_output_____"
]
],
[
[
"def map2(opa, opb, f):\n return opa.flatMap(lambda ax: opb.map(lambda bx: f(ax, bx)))\n\nopa = Option.some(1)\nopb = Option.some(2)\nmap2(opa, opb, lambda a, b: a + b).getOrElse(lambda e: 0)",
"_____no_output_____"
]
],
[
[
"This is very smart, and because you want to look smart, you might prefer this solution. But some may find the following version simpler to understand: ",
"_____no_output_____"
]
],
[
[
"def map2_v2(opa, opb, f):\n return Option.some(f(opa.getOrThrow(), opb.getOrThrow())) if opa.isSome and opb.isSome else opOption.none()\n\nmap2_v2(Option.some(1), Option.some(2), lambda a, b: a + b).getOrThrow()",
"_____no_output_____"
]
],
[
[
"If you want to test this code, you’ll have to define the <font color='blue'>isSome</font> method first, <b>but this is not to encourage you to use this nonfunctional technique</b>. You should always prefer the first form, but you should also understand fully the relation between the two forms. Besides, you’ll probably find yourself needing the <font color='blue'>isSome</font> method someday. \n\n## <font color='green'>equals and hashcode </font>\nMuch more important are the definitions of the <a href='https://docs.python.org/2/reference/datamodel.html#object.__eq__'>\\_\\_eq\\_\\_</a> and <a href='https://docs.python.org/2/reference/datamodel.html#object.__hash__'>\\_\\_hash\\_\\_</a> methods. As you know, these methods are strongly related and must be consistently defined. If equals is true for two instances of <font color='blue'><b>Option</b></font>, their hashcode methods should return the same value. (The inverse is not true. Objects having the same hashcode may not always be equal.) \n\n# <font color='blue'>How and when to use Option</font> \nHere’s how Brian Goetz, Java language architect at Oracle, answered a question about this subject on Stack Overflow. The question was “Should Java 8 getters return optional types?” Here is Brian Goetz’s answer: \nThe full discussion may be read at http://mng.bz/Rkk1. \n```python\nOf course, people will do what they want. But we did have a clear intention when adding this feature, and it was not to be a general purpose Maybe or Some type, as much as many people would have liked us to do so. Our intention was to provide a limited mechanism for library method return types where there needed to be a clear way to represent “no result” and using null for such was overwhelmingly likely to cause errors. \n\nFor example, you probably should never use it for something that returns an array of results, or a list of results; instead return an empty array or list. You should almost never use it as a field of something or a method parameter. I think routinely using it as a return value for getters would definitely be over-use. There’s nothing wrong with Optional that it should be avoided, it’s just not what many people wish it were, and accordingly we were fairly concerned about the risk of zealous over-use. \n````\nThis is a very important answer that deserves some reflection. First of all, and this might be the most important part, “people will do what they want.” Nothing to add here. Just do what you want. This doesn’t mean you should do whatever you want without thinking. But feel free to try every solution that comes to mind. You shouldn’t refrain from using Optional in a particular way just because it wasn’t intended to be used that way. Imagine the first man who ever thought about grabbing a stone to hit something with more strength. He had two options (pun intended!): refraining from doing it because stones had obviously not been intended to be used as hammers, or just trying it. \n\n## <font color='green'>When to use getOrThrow </font>\n<b>The correct advice is to avoid getOrThrow as much as possible. As a rule of thumb, each time you find yourself using this method outside of the <font color='blue'>Option</font> class, you should consider whether there’s another way to go</b>. Using <font color='blue'>getOrThrow</font> is exiting the functional safety of the <font color='blue'><b>Option</b></font> class. \n\nThe most important point is the original question: should getters return <font color='blue'><b>Option</b></font>? Generally, they shouldn’t, because properties should be final and initialized at declaration or in constructors, so there’s absolutely no need for getters to return <font color='blue'><b>Option</b></font>. (<font color='brown'>I must admit, however, that initializing fields in constructors doesn’t guarantee that access to properties is impossible before they’re initialized. This is a marginal problem that’s easily solved by making classes final, if possible.</font>) \n\n<b>But some properties might be optional.</b> For example, a person will always have a first name and a last name, but they might have no email. How can you represent this? By storing the property as an <font color='blue'><b>Option</b></font>. In such cases, the getter will have to return an <font color='blue'><b>Option</b></font>. Saying that “routinely using it as a return value for getters would definitely be over-use” is like saying that a property without a value should be set to null, and the corresponding getter should return null. This completely destroys the benefit of having <font color='blue'><b>Option</b></font>. \n\nWhat about methods that take <font color='blue'><b>Option</b></font> as their argument? In general, this should not occur. To compose methods returning <font color='blue'><b>Option</b></font>, you shouldn’t use methods that take <font color='blue'><b>Option</b></font> as their argument. For example, to compose the two following methods, you don’t need to change the methods to make them accept <font color='blue'><b>Option</b></font> as their argument: \n```python\ndef validate(String name):\n if name.startswith('J'):\n return Option.some(name)\n else:\n return Option.none()\n \ndef toUpper(name):\n return Option.some(name.upper())\n```\n\nThe functional way to compose these methods is as follows:\n```python\nOption.some('Ken').flatMap(validate).flatMap(toUpper).getOrElse(lambda: 'No name') # print 'No name'\nOption.some('John').flatMap(validate).flatMap(toUpper).getOrElse(lambda: 'No name') # print 'John'\n```",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eced5946a8769fa42852b89bfe44b00578268c89 | 1,391 | ipynb | Jupyter Notebook | Cap02/P-2_39.ipynb | IsraelBOrrico/montgomery_resolucao_python | eef4f7fb427c2dfd7ba0eb1fc3f9bf3c39a2de9f | [
"MIT"
] | null | null | null | Cap02/P-2_39.ipynb | IsraelBOrrico/montgomery_resolucao_python | eef4f7fb427c2dfd7ba0eb1fc3f9bf3c39a2de9f | [
"MIT"
] | null | null | null | Cap02/P-2_39.ipynb | IsraelBOrrico/montgomery_resolucao_python | eef4f7fb427c2dfd7ba0eb1fc3f9bf3c39a2de9f | [
"MIT"
] | null | null | null | 20.455882 | 166 | 0.548526 | [
[
[
"# Problem 39 from chapter 02\n***",
"_____no_output_____"
],
[
"### There are 10 operations to be done. But, five must be done first and other five must be done later. Between each set of five, they can be done in any order.",
"_____no_output_____"
]
],
[
[
"from math import factorial",
"_____no_output_____"
],
[
"operations = factorial(5) * factorial(5)\nprint(f'The number of different production sequences that are possible is {operations}.')",
"The number of different production sequences that are possible is 14400.\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
eced5c7732f02fa6d509174832277370ed28459a | 214,533 | ipynb | Jupyter Notebook | notebooks/maml.ipynb | cclauss/jax | f17f2b976f05f218776fdb881797f3012a85c539 | [
"ECL-2.0",
"Apache-2.0"
] | null | null | null | notebooks/maml.ipynb | cclauss/jax | f17f2b976f05f218776fdb881797f3012a85c539 | [
"ECL-2.0",
"Apache-2.0"
] | null | null | null | notebooks/maml.ipynb | cclauss/jax | f17f2b976f05f218776fdb881797f3012a85c539 | [
"ECL-2.0",
"Apache-2.0"
] | null | null | null | 234.462295 | 51,842 | 0.888525 | [
[
[
"# MAML Tutorial with JAX\n\nEric Jang\n\nBlog post: https://blog.evjang.com/2019/02/maml-jax.html\n\n\n21 Feb 2019\n\nPedagogical tutorial for implementing Model-Agnostic Meta-Learning with JAX's awesome `grad` and `vmap` and `jit` operators.\n\n### Overview\n\nIn this notebook we'll go through:\n\n- how to take gradients, gradients of gradients.\n- how to fit a sinusoid function with a neural network (and do auto-batching with vmap)\n- how to implement MAML and check its numerics\n- how to implement MAML for sinusoid task (single-task objective, batching task instances).\n- extending MAML to handle batching at the task-level\n",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade -q https://storage.googleapis.com/jax-wheels/cuda$(echo $CUDA_VERSION | sed -e 's/\\.//' -e 's/\\..*//')/jaxlib-0.1.21-cp36-none-linux_x86_64.whl\n!pip install --upgrade -q jax",
"_____no_output_____"
],
[
"# import jax.numpy (almost-drop-in for numpy) and gradient operators.\nimport jax.numpy as np\nfrom jax import grad",
"_____no_output_____"
]
],
[
[
"## Gradients of Gradients\n\nJAX makes it easy to compute gradients of python functions. Here, we thrice-differentiate $e^x$ and $x^2$",
"_____no_output_____"
]
],
[
[
"f = lambda x : np.exp(x)\ng = lambda x : np.square(x)\nprint(grad(f)(1.)) # = e^{1}\nprint(grad(grad(f))(1.))\nprint(grad(grad(grad(f)))(1.))\n\nprint(grad(g)(2.)) # 2x = 4\nprint(grad(grad(g))(2.)) # x = 2\nprint(grad(grad(grad(g)))(2.)) # x = 0",
"2.7182817\n2.7182817\n2.7182817\n4.0\n2.0\n0.0\n"
]
],
[
[
"## Sinusoid Regression and vmap\n\nTo get you familiar with JAX syntax first, we'll optimize neural network params with fixed inputs on a mean-squared error loss to $f_\\theta(x) = sin(x)$.",
"_____no_output_____"
]
],
[
[
"from jax import vmap # for auto-vectorizing functions\nfrom functools import partial # for use with vmap\nfrom jax import jit # for compiling functions for speedup\nfrom jax import random # stax initialization uses jax.random\nfrom jax.experimental import stax # neural network library\nfrom jax.experimental.stax import Conv, Dense, MaxPool, Relu, Flatten, LogSoftmax # neural network layers\nimport matplotlib.pyplot as plt # visualization",
"_____no_output_____"
],
[
"# Use stax to set up network initialization and evaluation functions\nnet_init, net_apply = stax.serial(\n Dense(40), Relu,\n Dense(40), Relu,\n Dense(1)\n)\n\nrng = random.PRNGKey(0)\nin_shape = (-1, 1,)\nout_shape, net_params = net_init(rng, in_shape)",
"_____no_output_____"
],
[
"def loss(params, inputs, targets):\n # Computes average loss for the batch\n predictions = net_apply(params, inputs)\n return np.mean((targets - predictions)**2)",
"_____no_output_____"
],
[
"# batch the inference across K=100\nxrange_inputs = np.linspace(-5,5,100).reshape((100, 1)) # (k, 1)\ntargets = np.sin(xrange_inputs)\npredictions = vmap(partial(net_apply, net_params))(xrange_inputs)\nlosses = vmap(partial(loss, net_params))(xrange_inputs, targets) # per-input loss\nplt.plot(xrange_inputs, predictions, label='prediction')\nplt.plot(xrange_inputs, losses, label='loss')\nplt.plot(xrange_inputs, targets, label='target')\nplt.legend()",
"_____no_output_____"
],
[
"import numpy as onp\nfrom jax.experimental import optimizers\nfrom jax.tree_util import tree_multimap # Element-wise manipulation of collections of numpy arrays ",
"_____no_output_____"
],
[
"opt_init, opt_update, get_params = optimizers.adam(step_size=1e-2)\nopt_state = opt_init(net_params)\n\n# Define a compiled update step\n@jit\ndef step(i, opt_state, x1, y1):\n p = get_params(opt_state)\n g = grad(loss)(p, x1, y1)\n return opt_update(i, g, opt_state)\n\nfor i in range(100):\n opt_state = step(i, opt_state, xrange_inputs, targets)\nnet_params = get_params(opt_state)",
"_____no_output_____"
],
[
"# batch the inference across K=100\ntargets = np.sin(xrange_inputs)\npredictions = vmap(partial(net_apply, net_params))(xrange_inputs)\nlosses = vmap(partial(loss, net_params))(xrange_inputs, targets) # per-input loss\nplt.plot(xrange_inputs, predictions, label='prediction')\nplt.plot(xrange_inputs, losses, label='loss')\nplt.plot(xrange_inputs, targets, label='target')\nplt.legend()",
"_____no_output_____"
]
],
[
[
"## MAML: Optimizing for Generalization\n\nSuppose task loss function $\\mathcal{L}$ is defined with respect to model parameters $\\theta$, input features $X$, input labels $Y$. MAML optimizes the following:\n\n$\\mathcal{L}(\\theta - \\nabla \\mathcal{L}(\\theta, x_1, y_1), x_2, y_2)$\n\n$x_1, y_2$ and $x_2, y_2$ are identically distributed from $X, Y$. Therefore, MAML objective can be thought of as a differentiable cross-validation error (w.r.t. $x_2, y_2$) for a model that learns (via a single gradient descent step) from $x_1, y_1$. Minimizing cross-validation error provides an inductive bias on generalization.\n\nThe following toy example checks MAML numerics via parameter $x$ and input $y$.",
"_____no_output_____"
]
],
[
[
"# gradients of gradients test for MAML\n# check numerics\ng = lambda x, y : np.square(x) + y\nx0 = 2.\ny0 = 1.\nprint('grad(g)(x0) = {}'.format(grad(g)(x0, y0))) # 2x = 4\nprint('x0 - grad(g)(x0) = {}'.format(x0 - grad(g)(x0, y0))) # x - 2x = -2\ndef maml_objective(x, y):\n return g(x - grad(g)(x, y), y)\nprint('maml_objective(x,y)={}'.format(maml_objective(x0, y0))) # x**2 + 1 = 5\nprint('x0 - maml_objective(x,y) = {}'.format(x0 - grad(maml_objective)(x0, y0))) # x - (2x)",
"grad(g)(x0) = 4.0\nx0 - grad(g)(x0) = -2.0\nmaml_objective(x,y)=5.0\nx0 - maml_objective(x,y) = -2.0\n"
]
],
[
[
"## Sinusoid Task + MAML\n\n\nNow let's re-implement the Sinusoidal regression task from Chelsea Finn's [MAML paper](https://arxiv.org/abs/1703.03400).\n",
"_____no_output_____"
]
],
[
[
"alpha = .1\ndef inner_update(p, x1, y1):\n grads = grad(loss)(p, x1, y1)\n inner_sgd_fn = lambda g, state: (state - alpha*g)\n return tree_multimap(inner_sgd_fn, grads, p)\n\ndef maml_loss(p, x1, y1, x2, y2):\n p2 = inner_update(p, x1, y1)\n return loss(p2, x2, y2)",
"_____no_output_____"
],
[
"x1 = xrange_inputs\ny1 = targets\nx2 = np.array([0.])\ny2 = np.array([0.])\nmaml_loss(net_params, x1, y1, x2, y2)",
"_____no_output_____"
]
],
[
[
"Let's try minimizing the MAML loss (without batching across multiple tasks, which we will do in the next section)\n",
"_____no_output_____"
]
],
[
[
"opt_init, opt_update, get_params = optimizers.adam(step_size=1e-3) # this LR seems to be better than 1e-2 and 1e-4\nout_shape, net_params = net_init(rng, in_shape)\nopt_state = opt_init(net_params)\n\n@jit\ndef step(i, opt_state, x1, y1, x2, y2):\n p = get_params(opt_state)\n g = grad(maml_loss)(p, x1, y1, x2, y2)\n l = maml_loss(p, x1, y1, x2, y2)\n return opt_update(i, g, opt_state), l\nK=20\n\nnp_maml_loss = []\n\n# Adam optimization\nfor i in range(20000):\n # define the task\n A = onp.random.uniform(low=0.1, high=.5)\n phase = onp.random.uniform(low=0., high=np.pi)\n # meta-training inner split (K examples)\n x1 = onp.random.uniform(low=-5., high=5., size=(K,1))\n y1 = A * onp.sin(x1 + phase)\n # meta-training outer split (1 example). Like cross-validating with respect to one example.\n x2 = onp.random.uniform(low=-5., high=5.)\n y2 = A * onp.sin(x2 + phase)\n opt_state, l = step(i, opt_state, x1, y1, x2, y2)\n np_maml_loss.append(l)\n if i % 1000 == 0:\n print(i)\nnet_params = get_params(opt_state)",
"0\n1000\n2000\n3000\n4000\n5000\n6000\n7000\n8000\n9000\n10000\n11000\n12000\n13000\n14000\n15000\n16000\n17000\n18000\n19000\n"
],
[
"# batch the inference across K=100\ntargets = np.sin(xrange_inputs)\npredictions = vmap(partial(net_apply, net_params))(xrange_inputs)\nplt.plot(xrange_inputs, predictions, label='pre-update predictions')\nplt.plot(xrange_inputs, targets, label='target')\n\nx1 = onp.random.uniform(low=-5., high=5., size=(K,1))\ny1 = 1. * onp.sin(x1 + 0.)\n\nfor i in range(1,5):\n net_params = inner_update(net_params, x1, y1)\n predictions = vmap(partial(net_apply, net_params))(xrange_inputs)\n plt.plot(xrange_inputs, predictions, label='{}-shot predictions'.format(i))\nplt.legend()",
"_____no_output_____"
]
],
[
[
"## Batching Meta-Gradient Across Tasks\n\nKind of does the job but not that great. Let's reduce the variance of gradients in outer loop by averaging across a batch of tasks (not just one task at a time). \n\nvmap is awesome it enables nice handling of batching at two levels: inner-level \"intra-task\" batching, and outer level batching across tasks.\n\nFrom a software engineering perspective, it is nice because the \"task-batched\" MAML implementation simply re-uses code from the non-task batched MAML algorithm, without losing any vectorization benefits.",
"_____no_output_____"
]
],
[
[
"def sample_tasks(outer_batch_size, inner_batch_size):\n # Select amplitude and phase for the task\n As = []\n phases = []\n for _ in range(outer_batch_size): \n As.append(onp.random.uniform(low=0.1, high=.5))\n phases.append(onp.random.uniform(low=0., high=np.pi))\n def get_batch():\n xs, ys = [], []\n for A, phase in zip(As, phases):\n x = onp.random.uniform(low=-5., high=5., size=(inner_batch_size, 1))\n y = A * onp.sin(x + phase)\n xs.append(x)\n ys.append(y)\n return np.stack(xs), np.stack(ys)\n x1, y1 = get_batch()\n x2, y2 = get_batch()\n return x1, y1, x2, y2",
"_____no_output_____"
],
[
"outer_batch_size = 2\nx1, y1, x2, y2 = sample_tasks(outer_batch_size, 50)\nfor i in range(outer_batch_size):\n plt.scatter(x1[i], y1[i], label='task{}-train'.format(i))\nfor i in range(outer_batch_size):\n plt.scatter(x2[i], y2[i], label='task{}-val'.format(i))\nplt.legend()",
"_____no_output_____"
],
[
"x2.shape",
"_____no_output_____"
],
[
"opt_init, opt_update, get_params = optimizers.adam(step_size=1e-3)\nout_shape, net_params = net_init(rng, in_shape)\nopt_state = opt_init(net_params)\n\n# vmapped version of maml loss.\n# returns scalar for all tasks.\ndef batch_maml_loss(p, x1_b, y1_b, x2_b, y2_b):\n task_losses = vmap(partial(maml_loss, p))(x1_b, y1_b, x2_b, y2_b)\n return np.mean(task_losses)\n\n@jit\ndef step(i, opt_state, x1, y1, x2, y2):\n p = get_params(opt_state)\n g = grad(batch_maml_loss)(p, x1, y1, x2, y2)\n l = batch_maml_loss(p, x1, y1, x2, y2)\n return opt_update(i, g, opt_state), l\n\nnp_batched_maml_loss = []\nK=20\nfor i in range(20000):\n x1_b, y1_b, x2_b, y2_b = sample_tasks(4, K)\n opt_state, l = step(i, opt_state, x1_b, y1_b, x2_b, y2_b)\n np_batched_maml_loss.append(l)\n if i % 1000 == 0:\n print(i)\nnet_params = get_params(opt_state)",
"0\n1000\n2000\n3000\n4000\n5000\n6000\n7000\n8000\n9000\n10000\n11000\n12000\n13000\n14000\n15000\n16000\n17000\n18000\n19000\n"
],
[
"# batch the inference across K=100\ntargets = np.sin(xrange_inputs)\npredictions = vmap(partial(net_apply, net_params))(xrange_inputs)\nplt.plot(xrange_inputs, predictions, label='pre-update predictions')\nplt.plot(xrange_inputs, targets, label='target')\n\nx1 = onp.random.uniform(low=-5., high=5., size=(10,1))\ny1 = 1. * onp.sin(x1 + 0.)\n\nfor i in range(1,3):\n net_params = inner_update(net_params, x1, y1)\n predictions = vmap(partial(net_apply, net_params))(xrange_inputs)\n plt.plot(xrange_inputs, predictions, label='{}-shot predictions'.format(i))\nplt.legend()",
"_____no_output_____"
],
[
"# Comparison of maml_loss for task batch size = 1 vs. task batch size = 8\nplt.plot(onp.convolve(np_maml_loss, [.05]*20), label='task_batch=1')\nplt.plot(onp.convolve(np_batched_maml_loss, [.05]*20), label='task_batch=4')\nplt.ylim(0., 1e-1)\nplt.legend()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eced606c3ebe8abd2a7970149a7d5e94dc9c4f1c | 609,861 | ipynb | Jupyter Notebook | Cycle_GAN_for_Style_Transfer_(Image_to_image_translation).ipynb | ahmedsalahacc/Image-Style-Transfer-with-Cycle-GAN | 0bbf23914e1744a4c8dae2f503e0813c8e7d5e92 | [
"MIT"
] | 1 | 2022-03-01T07:54:23.000Z | 2022-03-01T07:54:23.000Z | Cycle_GAN_for_Style_Transfer_(Image_to_image_translation).ipynb | ahmedsalahacc/Image-Style-Transfer-with-Cycle-GAN | 0bbf23914e1744a4c8dae2f503e0813c8e7d5e92 | [
"MIT"
] | null | null | null | Cycle_GAN_for_Style_Transfer_(Image_to_image_translation).ipynb | ahmedsalahacc/Image-Style-Transfer-with-Cycle-GAN | 0bbf23914e1744a4c8dae2f503e0813c8e7d5e92 | [
"MIT"
] | null | null | null | 207.083531 | 327,664 | 0.896258 | [
[
[
"# Cycle GAN model for Image Style Transfer (Image to Image Translations)\n\nUsing summer2winter_yosemite dataset (you can find it as summer2winter_yosemite.zip)",
"_____no_output_____"
]
],
[
[
"import os\nimport numpy as np\nimport torch\nfrom torch.utils.data import DataLoader\nfrom torchvision.utils import make_grid\nimport torchvision.datasets as datasets\nimport torchvision.transforms as transforms\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"class Preprocessor:\n def __init__(self, image_class, train_dir, test_dir, image_size, batch_size):\n '''\n image_class -- str, refers to the class name of the images in which the first order parent containing them is named after\n train_dir -- str, refers to the directory containing the image_class directories (2nd order parents) of the training image\n test_dir -- str, refers to the directory containing the image_class directories (2nd order parents) of the test image\n image_size -- tuple of the size of the image\n batch_size -- int, number of images in a single patch\n '''\n self.image_class = image_class\n self.train_dir = train_dir\n self.test_dir = test_dir\n self.image_size = image_size\n self.batch_size = batch_size\n self.init_bool = False\n\n def get_data_loaders(self):\n # return the loaders if the loaders are already intialized\n if self.init_bool:\n return self.train_loader, self.test_loader\n\n # get position of images\n train_path = './' + self.train_dir\n train_path = os.path.join(train_path, self.image_class)\n test_path = './' + self.test_dir\n #test_path = os.path.join(test_path, self.image_class)\n\n # defining transform\n transform = transforms.Compose([transforms.Resize(self.image_size),\n transforms.ToTensor()])\n\n # define datasets\n training_set = datasets.ImageFolder(train_path, transform = transform)\n test_set = datasets.ImageFolder(test_path, transform = transform)\n\n # defining data loaders\n train_loader = DataLoader(dataset = training_set, batch_size = self.batch_size, shuffle=True)\n test_loader = DataLoader(dataset = test_set, batch_size = self.batch_size, shuffle = True)\n\n # declare as initialized\n self.train_loader = train_loader\n self.test_loader = test_loader\n self.init_bool = True\n\n return train_loader, test_loader\n\n def imshow(self, image):\n img = image.numpy()\n plt.imshow(np.transpose(img, (1,2,0))) # reshaping axes to visualize with np\n plt.show();\n\n def show_many_images(self, group='training', fig_size=(12,8)):\n '''\n show many images of the class\n group -- 'training' or 'testing'\n fig_size -- figure size\n '''\n images, _ = next(iter(self.train_loader)) if group == 'training' else\\\n next(iter(self.test_loader))\n fig = plt.figure(figsize=fig_size)\n self.imshow(make_grid(images))\n\n def scale(self,x, feature_scale=(-1,1)):\n minn, maxx = feature_scale\n x = x*(maxx - minn) - maxx\n return x",
"_____no_output_____"
],
[
"# let X be summer style and Y be winter style\nX_type = Preprocessor(image_class='summer', \n train_dir='summer2winter_yosemite', \n test_dir='summer2winter_yosemite/test_summer', \n image_size=128, \n batch_size=16)\n\nY_type = Preprocessor(image_class='winter', \n train_dir='summer2winter_yosemite', \n test_dir='summer2winter_yosemite/test_winter', \n image_size=128, \n batch_size=16)\n\ntrain_loader_X, test_loader_X = X_type.get_data_loaders()\ntrain_loader_Y, test_loader_Y = Y_type.get_data_loaders()",
"_____no_output_____"
],
[
"# sanity check show many images\nX_type.show_many_images()",
"_____no_output_____"
],
[
"# sanity check scale and show image\nimg, _ = iter(X_type.train_loader).next()\nimg = img[0]\nprint(img.shape)\nX_type.imshow(img)\n\nscaled_image = X_type.scale(img)\nX_type.imshow(scaled_image)",
"torch.Size([3, 128, 128])\n"
]
],
[
[
"## Defining the models\n### 1. Discrimintator\n\nBoth Discriminators DX and DY have the same properties\n",
"_____no_output_____"
]
],
[
[
"from torch import nn\nimport torch.nn.functional as F\n\nclass ConvSuper(nn.Module):\n '''\n This class acts as a base class with a function that is used in all children classes that are convolutional\n '''\n def _conv(self, in_channels, out_channels, kernel_size, stride=2, padding=1, batch_norm=True):\n ''' Creates a convolutional layer with optional batch normalization set to True.\n Used to downsample the image.\n '''\n\n layers = [nn.Conv2d(in_channels=in_channels, out_channels=out_channels,\n kernel_size=kernel_size, stride=stride, padding=padding, bias=False)]\n \n if batch_norm:\n layers.append(nn.BatchNorm2d(out_channels))\n\n return nn.Sequential(*layers)\n\n def _deconv(self, in_channels, out_channels, kernel_size, stride=2, padding=1, batch_norm=True):\n '''Creates a transpose convolutional layer with optional batch normalization.\n Used to upsample the image.\n '''\n\n layers = [nn.ConvTranspose2d(in_channels, out_channels, kernel_size, stride, \n padding, bias=False)]\n\n # optional batchnorm\n if batch_norm:\n layers.append(nn.BatchNorm2d(out_channels))\n \n return nn.Sequential(*layers)\n\nclass Discriminator(ConvSuper):\n def __init__(self, in_channels=3, conv_dim=64):\n super(Discriminator, self).__init__()\n \n # class attributes\n self.conv_dim = conv_dim\n self.in_channels = in_channels\n\n # defining the conv layers\n ### inspection #1--> depth = 3, w=128, h=128\n self.conv1 = self._conv(in_channels, conv_dim, kernel_size=4, batch_norm=False)\n ### inspection #2--> depth = 64, w=64, h=64\n self.conv2 = self._conv(conv_dim, 2*conv_dim, kernel_size=4)\n ### inspection #3--> depth = 128, w=32, h=32\n self.conv3 = self._conv(2*conv_dim, 4*conv_dim, kernel_size=4)\n ### inspection #4--> depth = 256, w=16, h=16\n self.conv4 = self._conv(4*conv_dim, 8*conv_dim, kernel_size=4)\n ### inspection #5--> depth = 512, w=8, h=8\n\n # The following conv layer will act as a classification layer\n self.conv5 = self._conv(8*conv_dim, 1, kernel_size=4, stride=1, batch_norm=False) \n ### inspection #6 (Final) --> depth = 1, w=1, h=1 \n\n def forward(self, X):\n # encoders\n out = F.relu(self.conv1(X))\n out = F.relu(self.conv2(out))\n out = F.relu(self.conv3(out))\n out = F.relu(self.conv4(out))\n\n # classifier\n out = F.relu(self.conv5(out))\n return out\n\nclass ResidualBlock(ConvSuper):\n '''Defines a residual block, adds input X to a convolutional layer that is applied to X\n with the same size input and output. The resblocks allow the model to learn effective transformation\n from a domian to another\n '''\n def __init__(self, conv_dim):\n # initializing super class\n super(ResidualBlock,self).__init__()\n\n # class attributes\n self.conv_dim = conv_dim\n\n # layers -- No change in dimentions\n self.conv1 = self._conv(in_channels=conv_dim, out_channels=conv_dim,\n kernel_size=3, stride=1, padding=1, batch_norm=True)\n self.conv2 = self._conv(in_channels=conv_dim, out_channels=conv_dim,\n kernel_size=3, stride=1, padding=1, batch_norm=True)\n\n def forward(self, X):\n '''\n feed forward function\n X -- input data\n '''\n out = F.relu(self.conv1(X))\n out = X + self.conv2(X)\n return X\n\nclass Generator(ConvSuper):\n def __init__(self, conv_dim=64, n_res_blocks=6):\n super(Generator, self).__init__()\n\n # class attributes\n self.conv_dim = conv_dim\n self.n_res_blocks = n_res_blocks\n\n ######## encoders ########\n ### inspection #1--> depth = 3, w=128, h=128\n self.conv1 = self._conv(3,conv_dim,4)\n ### inspection #2--> depth = 64, w=64, h=64\n self.conv2 = self._conv(conv_dim, conv_dim*2, 4)\n ### inspection #3--> depth = 128, w=32, h=32\n self.conv3 = self._conv(conv_dim*2, conv_dim*4, 4)\n ### inspection #4--> depth = 256, w=16, h=16\n\n ######## residual blocks ########\n self.res_blocks = self.__intialize_res(conv_dim*4, n_res_blocks)\n\n ######## decoder ########\n ### inspection #5--> depth = 256, w=16, h=16\n self.deconv1 = self._deconv(conv_dim*4, conv_dim*2, 4)\n ### inspection #6--> depth = 128, w=32, h=32 \n self.deconv2 = self._deconv(conv_dim*2, conv_dim, 4)\n ### inspection #7--> depth = 64, w=64, h=64 \n self.deconv3 = self._deconv(conv_dim, 3, 4, batch_norm=False) # no batch_norm\n ### inspection #8--> depth = 3, w=128, h=128\n\n def forward(self, X):\n '''Takes an image as an input and returns the transformed image as an output'''\n # encoders\n out = F.relu(self.conv1(X))\n out = F.relu(self.conv2(out))\n out = F.relu(self.conv3(out))\n\n # resblocks\n out = self.res_blocks(out)\n\n # decoders\n out = F.relu(self.deconv1(out))\n out = F.relu(self.deconv2(out))\n\n # tanh applied at the last layer\n out = F.tanh(self.deconv3(out))\n\n return out\n\n def __intialize_res(self,conv_dim, n_blocks=1):\n layers = []\n for i in range(n_blocks):\n layers.append(ResidualBlock(conv_dim))\n\n return nn.Sequential(*layers)",
"_____no_output_____"
],
[
"# sanity check for the discriminator class\nDX = Discriminator()\nout = DX(img.unsqueeze(0)).squeeze().squeeze()\nprint(out.shape)\n# delete to free memory\ndel DX\ndel out",
"torch.Size([7, 7])\n"
],
[
"# Sanity check for the generator class \nGX = Generator()\nout = GX(img.unsqueeze(0)).squeeze().squeeze();\nprint(out.shape)\n# delete to free memory\ndel GX\ndel out",
"torch.Size([3, 128, 128])\n"
]
],
[
[
"## Creating Model\n",
"_____no_output_____"
]
],
[
[
"from helpers import save_samples, checkpoint\nfrom tqdm import tqdm\n\nclass CycleGan:\n def __init__(self,g_conv_dim=64, d_conv_dim=64, n_res_blocks=6):\n self.create_CGAN_models(g_conv_dim, d_conv_dim, n_res_blocks)\n self.__print_models()\n\n def create_CGAN_models(self,g_conv_dim, d_conv_dim, n_res_blocks):\n '''Builds cycle gan generators and descriminator'''\n # Instintiate generators\n self.G_X2Y = Generator(g_conv_dim, n_res_blocks)\n self.G_Y2X = Generator(g_conv_dim, n_res_blocks)\n\n # Instantiate discriminatros\n self.D_X = Discriminator(3,d_conv_dim)\n self.D_Y = Discriminator(3,d_conv_dim)\n\n # config on GPU if available\n train_on_GPU = torch.cuda.is_available()\n if train_on_GPU:\n print(\"Configuring on GPU\")\n else:\n print(\"Configuring on CPU\")\n\n device = 'cuda' if train_on_GPU else 'cpu'\n self.G_X2Y.to(device)\n self.G_Y2X.to(device)\n self.D_X.to(device)\n self.D_Y.to(device)\n\n #return self.G_X2Y, self.G_Y2X, self.D_X, self.D_Y\n\n def train(self, trainloader_X, trainloader_Y, testloader_X, testloader_Y,\n g_optimizer, d_x_optimizer, d_y_optimizer, lambda_w = 10, epochs=1200, print_every=10, sample_every=100):\n '''\n trainloader_X: first domain train loader \n trainloader_Y: second domain train loader \n testloader_X: first domain test loader\n testloader_Y: second domain test loader\n g_optimizer: generator optimizer\n d_x_optimize: first domain discrminator optimizer \n d_y_optimizer: second domain discriminator optimizer\n lambda_w = 10: lambda weight of the cycle consistency loss with default value 10\n epochs=1200: training epochs with default value 1200 \n print_every=10: print inspection every (default=10) epochs \n sample_every=100: save sample every (default=100) epochs\n '''\n \n # keep track of the training losses through out time for visualization purposes\n losses = {\n 'D_X':[],\n 'D_Y':[],\n 'G': []\n }\n\n # get some fixed data from both domains X and Y, for sampling purposes\n # These images are held constants throughout the training that allows the inspection of the \n # model performance\n # take 1 image from both batches\n fixed_X = X_type.scale(next(iter(testloader_X))[0])\n fixed_Y = Y_type.scale(next(iter(testloader_Y))[0])\n\n # batches per epoch\n iter_X = iter(trainloader_X)\n iter_Y = iter(testloader_Y)\n batches_per_epoch = min(len(iter_X), len(iter_Y))\n\n for e in tqdm(range(1, epochs+1)):\n # reset iterations for each epoch\n if e % batches_per_epoch ==0:\n iter_X = iter(trainloader_X)\n iter_Y = iter(trainloader_Y)\n\n # get images\n X, _ = next(iter_X)\n X = X_type.scale(X)\n\n Y, _ = next(iter_Y)\n Y = Y_type.scale(Y)\n\n # check if GPU is available and move images to GPU accordingly\n device = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\n X = X.to(device)\n Y = Y.to(device)\n\n ##################################\n #### Train the Discriminators ####\n ##################################\n\n ## First: D_X real and fake loss components\n d_x_optimizer.zero_grad()\n\n # 1. Compute the discriminator losses on real images\n real_X = self.D_X(X)\n D_X_real_loss = self.__real_mse_loss(real_X)\n\n # 2. Generate fake images that look like the domain X from domain Y\n fake_X = self.G_Y2X(Y)\n\n # 3. Compute the fake losses for D_X\n fake_X = self.D_X(fake_X)\n D_X_fake_loss = self.__fake_mse_loss(fake_X)\n\n # 4. Compute the total loss and perform backpropagation\n D_X_loss = D_X_real_loss + D_X_fake_loss\n D_X_loss.backward()\n d_x_optimizer.step()\n\n\n ## Second: D_Y, real and fake components\n\n d_y_optimizer.zero_grad()\n\n # 1. Compute the discriminator losses on real images\n real_Y = self.D_Y(Y)\n D_Y_real_loss = self.__real_mse_loss(real_Y)\n\n # 2. Generate fake images that look like domain Y based on images from domain X\n fake_Y = self.G_X2Y(X)\n\n # 3. Compute fake loss for D_Y\n fake_Y = self.D_Y(fake_Y)\n D_Y_fake_loss = self.__fake_mse_loss(fake_Y)\n\n # 4. Compute total losses and perform backpropagation\n D_Y_loss = D_Y_real_loss + D_Y_fake_loss\n D_Y_loss.backward()\n d_y_optimizer.step()\n\n ##############################\n #### Train the Generators ####\n ##############################\n\n g_optimizer.zero_grad()\n\n #### First generate fake X images and reconstructed Y ###\n\n # 1. generate fake images that look like domain X\n fake_X = self.G_Y2X(Y)\n\n # 2. Compute the loss based on domain X\n out_X = self.D_X(fake_X)\n g_Y2X_loss = self.__real_mse_loss(out_X)\n\n # 3. Compute the reconstructed Y\n recon_Y = self.G_X2Y(fake_X)\n\n # 4. Compute the cycle consistency loss\n recon_Y_loss = self.__cycle_consistency_loss(Y, recon_Y, lambda_w)\n\n #### Second generate fake Y images and reconstructed X ###\n\n # 1. generate fake images that look like domain X\n fake_Y = self.G_X2Y(X)\n\n # 2. Compute the loss based on domain X\n out_Y = self.D_Y(fake_Y)\n g_X2Y_loss = self.__real_mse_loss(out_Y)\n\n # 3. Compute the reconstructed Y\n recon_X = self.G_Y2X(fake_Y)\n\n # 4. Compute the cycle consistency loss\n recon_X_loss = self.__cycle_consistency_loss(X, recon_X, lambda_w = lambda_w)\n\n ## addup all generator losses and perform backpropagation\n g_total_loss = g_X2Y_loss + g_Y2X_loss + recon_X_loss + recon_Y_loss\n g_total_loss.backward()\n g_optimizer.step()\n\n # Print the log info\n if e % print_every == 0:\n # append real and fake discriminator losses and the generator loss\n losses['D_X'].append(D_X_loss.item())\n losses['D_Y'].append(D_Y_loss.item())\n losses['G'].append(g_total_loss.item())\n #losses.append((D_X_loss.item(), D_Y_loss.item(), g_total_loss.item()))\n print('Epoch [{:5d}/{:5d}] | d_X_loss: {:6.4f} | d_Y_loss: {:6.4f} | g_total_loss: {:6.4f}'.format(\n e, epochs, D_X_loss.item(), D_Y_loss.item(), g_total_loss.item()))\n\n # Save the generated samples\n if e % sample_every == 0:\n self.G_Y2X.eval() # set generators to eval mode for sample generation\n self.G_X2Y.eval()\n save_samples(e, fixed_Y, fixed_X, self.G_Y2X, self.G_X2Y, batch_size=16)\n\n # return to training mode\n self.G_Y2X.train()\n self.G_X2Y.train()\n\n return losses\n\n def __real_mse_loss(self, D_out):\n return torch.mean((D_out-1)**2)\n\n def __fake_mse_loss(self, D_out):\n return torch.mean(D_out**2)\n\n def __cycle_consistency_loss(self, real_im, reconstructed_im, lambda_w):\n # calculate reconstruction loss\n recon_loss = torch.mean(torch.abs(real_im - reconstructed_im))\n\n # return weighted loss\n return lambda_w*recon_loss \n\n def __print_models(self):\n \"\"\"Prints model information for the generators and discriminators.\n \"\"\"\n print(\" G_X2Y \")\n print(\"-----------------------------------------------\")\n print(self.G_X2Y)\n print()\n\n print(\" G_Y2X \")\n print(\"-----------------------------------------------\")\n print(self.G_Y2X)\n print()\n\n print(\" D_X \")\n print(\"-----------------------------------------------\")\n print(self.D_X)\n print()\n\n print(\" D_Y \")\n print(\"-----------------------------------------------\")\n print(self.D_Y)\n print()\n\n# Creating Models\ncgan_model = CycleGan()",
"Configuring on GPU\n G_X2Y \n-----------------------------------------------\nGenerator(\n (conv1): Sequential(\n (0): Conv2d(3, 64, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(64, 128, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv3): Sequential(\n (0): Conv2d(128, 256, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (res_blocks): Sequential(\n (0): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (1): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (2): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (3): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (4): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (5): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n )\n (deconv1): Sequential(\n (0): ConvTranspose2d(256, 128, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (deconv2): Sequential(\n (0): ConvTranspose2d(128, 64, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (deconv3): Sequential(\n (0): ConvTranspose2d(64, 3, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n )\n)\n\n G_Y2X \n-----------------------------------------------\nGenerator(\n (conv1): Sequential(\n (0): Conv2d(3, 64, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(64, 128, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv3): Sequential(\n (0): Conv2d(128, 256, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (res_blocks): Sequential(\n (0): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (1): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (2): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (3): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (4): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (5): ResidualBlock(\n (conv1): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv2): Sequential(\n (0): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n )\n (deconv1): Sequential(\n (0): ConvTranspose2d(256, 128, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (deconv2): Sequential(\n (0): ConvTranspose2d(128, 64, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (deconv3): Sequential(\n (0): ConvTranspose2d(64, 3, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n )\n)\n\n D_X \n-----------------------------------------------\nDiscriminator(\n (conv1): Sequential(\n (0): Conv2d(3, 64, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n )\n (conv2): Sequential(\n (0): Conv2d(64, 128, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv3): Sequential(\n (0): Conv2d(128, 256, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv4): Sequential(\n (0): Conv2d(256, 512, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv5): Sequential(\n (0): Conv2d(512, 1, kernel_size=(4, 4), stride=(1, 1), padding=(1, 1), bias=False)\n )\n)\n\n D_Y \n-----------------------------------------------\nDiscriminator(\n (conv1): Sequential(\n (0): Conv2d(3, 64, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n )\n (conv2): Sequential(\n (0): Conv2d(64, 128, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv3): Sequential(\n (0): Conv2d(128, 256, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv4): Sequential(\n (0): Conv2d(256, 512, kernel_size=(4, 4), stride=(2, 2), padding=(1, 1), bias=False)\n (1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (conv5): Sequential(\n (0): Conv2d(512, 1, kernel_size=(4, 4), stride=(1, 1), padding=(1, 1), bias=False)\n )\n)\n\n"
],
[
"import torch.optim as optim\n\n# hyperparams for Adam optimizer\nlr=0.0002\nbeta1=0.5\nbeta2=0.999 # default value\n\ng_params = list(cgan_model.G_X2Y.parameters()) + list(cgan_model.G_Y2X.parameters()) # Get generator parameters\n\n# Create optimizers for the generators and discriminators\ng_optimizer = optim.Adam(g_params, lr, [beta1, beta2])\nd_x_optimizer = optim.Adam(cgan_model.D_X.parameters(), lr, [beta1, beta2])\nd_y_optimizer = optim.Adam(cgan_model.D_Y.parameters(), lr, [beta1, beta2])",
"_____no_output_____"
],
[
"n_epochs = 4000 # keep this small when testing if a model first works\n\nlosses = cgan_model.train(train_loader_X, train_loader_Y, \n test_loader_X, test_loader_Y, g_optimizer=g_optimizer, d_x_optimizer=d_x_optimizer,\n d_y_optimizer=d_y_optimizer,lambda_w = 10, epochs=n_epochs, print_every=100, sample_every=100)",
" 2%|▏ | 99/4000 [05:32<1:51:04, 1.71s/it]"
],
[
"%matplotlib inline\n\ndef visualize_losses(losses):\n '''\n losses ob of all the losses in the system.\n losses keys[\n 'D_X'\n 'D_Y'\n 'G'\n ]\n '''\n plt.title(\"Losses of the Cycle Gan Model\")\n plt.plot(losses['D_X'],label='Discriminator Summer')\n plt.plot(losses['D_Y'], label='Discriminator Winter')\n plt.plot(losses['G'], label = 'Generator')\n plt.legend()\n plt.show();\n\nvisualize_losses(losses)",
"_____no_output_____"
],
[
" def test_model(model, batch_size=16):\n g_loss = 0\n dx_loss = 0\n dy_loss = 0\n train_on_GPU = torch.cuda.is_available()\n if train_on_GPU:\n print(\"Configuring on GPU\")\n else:\n print(\"Configuring on CPU\")\n\n device = 'cuda' if train_on_GPU else 'cpu'\n model.D_X.eval()\n model.D_Y.eval()\n model.G_X2Y.eval()\n model.G_Y2X.eval() \n itr = 0\n with torch.no_grad(): \n for x, y in tqdm(zip(test_loader_X, test_loader_Y)):\n \n x,_ = x\n y,_ = y\n x = x.to(device)\n y = y.to(device)\n x = X_type.scale(x)\n y = Y_type.scale(y)\n #1. calculate the real error\n real_ims_res_x = model.D_X(x)\n real_ims_err_x = torch.mean((real_ims_res_x-1)**2)\n real_ims_res_y = model.D_Y(y)\n real_ims_err_y = torch.mean((real_ims_res_y-1)**2)\n\n #2. generate fake images from the batch\n fake_ims_x = model.G_Y2X(y)\n fake_ims_y = model.G_X2Y(x)\n fake_ims_err_x = torch.mean(model.D_X(fake_ims_y**2))\n fake_ims_err_y = torch.mean(model.D_Y(fake_ims_x**2))\n \n #3. calculate the Discriminator error\n d_error_x = fake_ims_err_x + real_ims_err_x\n d_error_y = fake_ims_err_x + real_ims_err_y\n \n #4. calculate the real error with these fakeimages\n recon_im_y = model.G_X2Y(fake_ims_x)\n recon_im_x = model.G_Y2X(fake_ims_y)\n recon_im_y_logits = model.D_Y(recon_im_y)\n recon_im_x_logits = model.D_X(recon_im_x)\n \n #5. calculate real adverserial error on the generated image\n adv_real_error_y = torch.mean((model.D_Y(fake_ims_x)-1)**2)\n adv_real_error_x = torch.mean((model.D_Y(fake_ims_y)-1)**2)\n \n #6. calculate the error of the generated image in discriminator\n recon_im_x_err = torch.mean((recon_im_x_logits-1)**2)\n recon_im_y_err = torch.mean((recon_im_y_logits-1)**2)\n \n #7. calculate cycle loss and add losses to the current loss\n dx_loss+= d_error_x/batch_size\n dy_loss+= d_error_y/batch_size\n ccl_x = torch.mean(torch.abs(x - recon_im_x))\n ccl_y = torch.mean(torch.abs(y - recon_im_y))\n g_loss += (ccl_x + ccl_y + recon_im_x_err + recon_im_y_err)/batch_size\n\n # save test samples\n save_samples(itr, y, x, model.G_Y2X, model.G_X2Y, batch_size=16, sample_dir='test_samples')\n\n # increment iteration\n itr+=1\n\n return dx_loss.item(), dy_loss.item(), g_loss.item()\n\n\nloss_dx, loss_dy, loss_g = test_model(cgan_model);\n\nprint()\nprint()\nprint(\"\\t\\t\\t Testing Loss\")\nprint()\nprint(\"Testing loss Discriminator X\\t:\\t\", loss_dx)\nprint(\"Testing loss Discriminator Y\\t:\\t\", loss_dy)\nprint(\"Testing loss Generators\\t\\t:\\t\", loss_g)",
"\r0it [00:00, ?it/s]"
]
],
[
[
"\n\n## Drawbacks \n1. Up-sampling with low resolution reduces the the quality of the image. May consider some other techniques (mentioned in the the Consideratons section).\n\n2. There are some images that are wrongly generated, seems like the model treats any kind of wood as a plant and hence, generates the green leaves in winter to summer generation, and vice versa.\n\n## Considerations\n\n1. Re-implementing the model with conditional GAN may lead to higher resolution\n2. Better tuning may lead to better results",
"_____no_output_____"
],
[
"#### Note that: Training samples are found in 'samples_cyclegan' folder and the test results are found in the 'test_samples' directory.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
eced65ff3e6647776513e00963684fa5d0b03c6a | 22,249 | ipynb | Jupyter Notebook | python_02_condition & loop.ipynb | hyeshinoh/Python_Study | 9f2cca2c5dea461e8324f1f294e36e154568b4b3 | [
"MIT"
] | null | null | null | python_02_condition & loop.ipynb | hyeshinoh/Python_Study | 9f2cca2c5dea461e8324f1f294e36e154568b4b3 | [
"MIT"
] | null | null | null | python_02_condition & loop.ipynb | hyeshinoh/Python_Study | 9f2cca2c5dea461e8324f1f294e36e154568b4b3 | [
"MIT"
] | 2 | 2018-11-10T16:03:37.000Z | 2020-12-05T11:47:35.000Z | 17.885048 | 108 | 0.409457 | [
[
[
"# **Python 02: Condition & Loop**",
"_____no_output_____"
],
[
"## 1. 입력 (Input)",
"_____no_output_____"
],
[
"### 1.1 문자열 입력",
"_____no_output_____"
]
],
[
[
"value = input(\"Insert string:\")\nprint(value, type(value))",
"hello <class 'str'>\n"
]
],
[
[
"### 1.2 숫자 입력\nint() 등으로 숫자형으로 변환해주어야 함",
"_____no_output_____"
]
],
[
[
"n = int(input(\"insert integer: \"))",
"insert integer: 3\n"
]
],
[
[
"### 1.3 여러 개의 값 입력",
"_____no_output_____"
]
],
[
[
"ls = input(\"insert list (sep=' '): \")\nls = ls.split(\" \")\nprint(type(ls), ls)",
"insert list (sep=' '): hyeshin byungwoo\n<class 'list'> ['hyeshin', 'byungwoo']\n"
]
],
[
[
"### 1.4 입력 받은 숫자 활용 반복문",
"_____no_output_____"
]
],
[
[
"n = int(input(\"number : \"))\nfor number in range(n):\n print(number, end = \" \")",
"number : 5\n0 1 2 3 4 "
]
],
[
[
"# 2. 출력 (print)",
"_____no_output_____"
],
[
"### 2.1 print",
"_____no_output_____"
]
],
[
[
"print(\"test\", \"page\") #한 칸 띄워 출력",
"test page\n"
]
],
[
[
"### 2.2 pprint\n- 딕셔너리와 같은 데이터 복잡할 때 가독성을 위해 정렬, 줄바꿈하여 출력",
"_____no_output_____"
]
],
[
[
"from pprint import pprint\n\ndic = {\"A\":90, \"B\":70, \"C\":100, \"D\":90, \"E\":70, \"F\":100, \"G\":90, \"H\":70, \"I\":100}\npprint(dic)",
"{'A': 90,\n 'B': 70,\n 'C': 100,\n 'D': 90,\n 'E': 70,\n 'F': 100,\n 'G': 90,\n 'H': 70,\n 'I': 100}\n"
]
],
[
[
"## 3. 조건 : if, elif, else",
"_____no_output_____"
],
[
"### 3.1 if",
"_____no_output_____"
],
[
"- condition이 True이면 code_1이 실행\n```\nif <condition>:\n <code_1>\n```",
"_____no_output_____"
]
],
[
[
"flag = True\nif flag:\n print(\":)\")",
":)\n"
]
],
[
[
"### 3.2 else",
"_____no_output_____"
]
],
[
[
"flag = True\nif flag:\n print(\"dss\")\nelse:\n print(\"data\")\nprint(\"done\")",
"dss\ndone\n"
]
],
[
[
"### 3.3 elif\n- 조건을 여러 개 줄 때",
"_____no_output_____"
]
],
[
[
"# 10-8:A / 7-4:B / 3-0: C\npoint = 3\nresult = \"\"\nif point >= 8:\n result = \"A\"\nelif point >= 4:\n result = \"B\"\nelse: \n result = \"C\"\n \nprint(result)\nprint(\"done\")",
"C\ndone\n"
]
],
[
[
"### 3.4 Quiz",
"_____no_output_____"
]
],
[
[
"# 숫자를 하나 입력 받아서 홀수이면 \"odd\", 짝수이면 \"even\"을 출력하는 코드 만들기\n\nuser_number = int(input(\"숫자를 입력해주세요: \"))\nif user_number % 2 == 0:\n print(\"even\")\nelse:\n print(\"odd\")",
"숫자를 입력해주세요: 4\neven\n"
]
],
[
[
"### 3.5 삼항연산\n- 문법: `(True) if (condition) else (False)`",
"_____no_output_____"
]
],
[
[
"user_number = int(input(\"숫자를 입력해주세요: \"))\nresult = \"even\" if user_number % 2 == 0 else \"odd\"\nprint(result)",
"숫자를 입력해주세요: 3\nodd\n"
]
],
[
[
"## 4. 반복 (loop) \n- while, for, list comprehension 반복문\n- break, continue",
"_____no_output_____"
],
[
"### 4.1 while",
"_____no_output_____"
],
[
"#### 문법",
"_____no_output_____"
]
],
[
[
"a = 3\nwhile a > 0:\n print(a)\n a -= 1\na",
"3\n2\n1\n"
]
],
[
[
"#### break\n- break 코드를 만나면 강제로 반복문이 종료",
"_____no_output_____"
]
],
[
[
"a = 3\nwhile a > 0:\n print(a)\n if a == 2:\n break\n a -= 1\na",
"3\n2\n"
]
],
[
[
"#### continue\n- continue 코드를 만나면 바로 반복문 구문으로 이동\n- continue 아래 구문이 실행되지 않음",
"_____no_output_____"
]
],
[
[
"a = 3\nwhile a > 0:\n print(a)\n if a == 2:\n a = 0\n continue\n a -= 1\na",
"3\n2\n"
],
[
"a = [1,2,3,4,\"q\"]\nidx = 0\nwhile True:\n data = a[idx]\n idx += 1\n if data == 'q':\n break\n elif data % 2 == 0:\n continue\n print(data)\nidx",
"1\n3\n"
]
],
[
[
"### 4.2 for문\n- range, zip, enumerate ",
"_____no_output_____"
],
[
"#### 문법",
"_____no_output_____"
]
],
[
[
"'''\nfor <value> in <list>:\n <code>\n'''",
"_____no_output_____"
],
[
"ls = [0, 1, 2, 3, 4]\nfor i in ls:\n print(i)",
"0\n1\n2\n3\n4\n"
]
],
[
[
"#### range\nlist를 간편하게 생성하기 위한 함수",
"_____no_output_____"
]
],
[
[
"ls = [0, 1, 2, 3, 4, 5]\nls",
"_____no_output_____"
],
[
"# 0~5 리스트를 만들고 싶을 때, 범위를 리스트 형태로 만들어 줄 수 있음\nprint(range(6))\nprint(list(range(6)))\nprint(list(range(2, 6)))",
"range(0, 6)\n[0, 1, 2, 3, 4, 5]\n[2, 3, 4, 5]\n"
],
[
"list(range(0,6,2)), list(range(6, 0, -1))",
"_____no_output_____"
],
[
"for i in range(5): \n print(\"dss\")",
"dss\ndss\ndss\ndss\ndss\n"
],
[
"for num in range(5):\n print(\"dss\", num)",
"dss 0\ndss 1\ndss 2\ndss 3\ndss 4\n"
],
[
"ls = [\"data\", \"science\", \"school\"]\nfor idx in range(len(ls)):\n print(ls[idx])",
"data\nscience\nschool\n"
],
[
"ls = [\"data\", \"science\", \"school\"]\nfor i in ls:\n print(i)",
"data\nscience\nschool\n"
]
],
[
[
"#### zip\n2개의 리스트를 key, value (dictionary)형태로 묶어주는 함수",
"_____no_output_____"
]
],
[
[
"subs = [\"korean\", \"english\", \"math\", \"science\"]\npoints = [100, 80, 90, 60]\n\nresult = {}\nfor k, v in zip(subs, points):\n result[k] = v # dict type으로 저장될 수 있다\nresult",
"_____no_output_____"
],
[
"# zip을 쓰지 않으면 이렇게 해야 함\nresult = {}\nfor idx in range(len(subs)):\n result[subs[idx]] = points[idx]\nresult\n ",
"_____no_output_____"
],
[
"# 가장 좋은 방법\nsubs = [\"korean\", \"english\", \"math\", \"science\"]\npoints = [100, 80, 90, 60]\n\nprint(tuple(zip(subs, points)))\nprint(dict(zip(subs, points)))",
"(('korean', 100), ('english', 80), ('math', 90), ('science', 60))\n{'korean': 100, 'english': 80, 'math': 90, 'science': 60}\n"
]
],
[
[
"#### enumerate\n리스트 형태의 데이터를 index와 value 값을 동시에 사용할수 있는 함수",
"_____no_output_____"
]
],
[
[
"ls = [\"math\", \"science\", \"korean\"]\nfor value in ls:\n print(value)",
"math\nscience\nkorean\n"
],
[
"ls = [\"math\", \"science\", \"korean\"]\n\nnum = 0\nfor value in ls:\n print(num, value)\n num += 1",
"0 math\n1 science\n2 korean\n"
],
[
"ls = [\"math\", \"science\", \"korean\"]\nfor num, value in enumerate(ls):\n print(num, value)",
"0 math\n1 science\n2 korean\n"
],
[
"ls = [\"math\", \"science\", \"korean\"]\nfiltered_ls = []\nfor num, value in enumerate(ls):\n if value ==\"science\":\n filtered_ls.append(num)\n print(num, value)\nfiltered_ls",
"0 math\n1 science\n2 korean\n"
]
],
[
[
"#### 퀴즈: for문\n",
"_____no_output_____"
]
],
[
[
"# numbers 안에 있는 숫자를 모두 더해서 result로 반환\nnumbers = [1, 3, 5, 7, 9]\nresult = 0\nfor i in numbers:\n result += i\nresult",
"_____no_output_____"
]
],
[
[
"### 4.3 list comprehension\n- 반복되어 출력되는 리스트 형태의 데이터를 만들어주는 문법\n- for문보다 속도가 빠름\n- `ls = [<vlaue> for <value> in <list> (if <condition>)]`\n- 간단하긴 하지만 사용 가능하다면 많이 쓰는 것이 좋은 문법",
"_____no_output_____"
]
],
[
[
"ls = [i for i in range(1, 6)]\nls",
"_____no_output_____"
],
[
"# 다른방법 1\nls = [1, 2, 3, 4, 5]\n\nls = []\nls.append(1)\nls.append(2)\nls.append(3)\nls.append(4)\nls.append(5)\n\n# 다른방법 2\nls = []\nfor num in range(1,6):\n ls.append(num)",
"_____no_output_____"
]
],
[
[
"#### list comprehension과 for문의 속도 비교",
"_____no_output_____"
]
],
[
[
"%%timeit\nls = []\nfor num in range(1, 10001):\n ls.append(num)\nlen(ls)",
"1.12 ms ± 40.3 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)\n"
],
[
"%%timeit\nls = [num for num in range(1, 10001)]\nlen(ls)",
"503 µs ± 29.5 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)\n"
]
],
[
[
"#### 삼항연산과 함께 사용",
"_____no_output_____"
]
],
[
[
"# 0 - 10까지 숫자를 출력하고 해당 수가 짝수인지 홀수 인지를 판별해서 odd / even 문자열을 추가하는 코드\n\nls = [str(num)+\": even\" if num % 2 == 0 else str(num)+\": odd\" for num in range(4)]\nls",
"_____no_output_____"
],
[
"ls = [str(num) + \": even\" if num % 2 == 0 else str(num) + \": odd\" for num in range(1, 11)]\nls",
"_____no_output_____"
]
],
[
[
"#### 조건문을 함께 사용",
"_____no_output_____"
]
],
[
[
"# 홀수만 출력되게 함\nls = [num for num in range(11) if num % 2 == 1]\nls",
"_____no_output_____"
]
],
[
[
"#### quiz",
"_____no_output_____"
]
],
[
[
"# 성이 lee인 사람의 성을 삭제하는 코드를 작성하세요\nnames = [\"kim dss\", \"lee python\", \"park data\", \"lee macpro\"]\n\nresult = [name.replace(\"lee \", \"\") for name in names]\n# result = [name.split()[1] if name.split(\"\")[0] = \"lee\" else name for name in names]\nprint(result)\n\nresult = [name for name in result if \"park \" not in name] \n# result = [name for name in result if name.startswith(\"park \") != True] \n# if name.split()[0] != \"park\"\nprint(result)\n\n# result 1: [\"kim dss\", \"python\", \"park data\", \"macpro\"]\n# result 2: [\"kim dss\", \"python\", \"macpro\"] - park 씨의 데이터 제거",
"['kim dss', 'python', 'park data', 'macpro']\n['kim dss', 'python', 'macpro']\n"
]
],
[
[
"#### 참고자료\n- 패스트캠퍼스, ⟪데이터사이언스스쿨 8기⟫ 수업자료",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eced69592d4665bb9a865c26d64bf35abb387d83 | 227,413 | ipynb | Jupyter Notebook | 0-Checking_Veras/Veras-Replica-Ctrl.ipynb | mdodici/trojan-WD-pollution | ec79a96f0d9517a53df4c82ca1be0d5d38f3346b | [
"MIT"
] | null | null | null | 0-Checking_Veras/Veras-Replica-Ctrl.ipynb | mdodici/trojan-WD-pollution | ec79a96f0d9517a53df4c82ca1be0d5d38f3346b | [
"MIT"
] | null | null | null | 0-Checking_Veras/Veras-Replica-Ctrl.ipynb | mdodici/trojan-WD-pollution | ec79a96f0d9517a53df4c82ca1be0d5d38f3346b | [
"MIT"
] | null | null | null | 405.370766 | 67,120 | 0.930312 | [
[
[
"import rebound\nimport reboundx\nimport numpy as np\nimport astropy.units as u\n\nfrom IPython.display import display, clear_output\nimport matplotlib.pyplot as plt\nfrom matplotlib import colors\nfrom scipy.interpolate import InterpolatedUnivariateSpline\n\nradeg = np.pi/180",
"_____no_output_____"
],
[
"###############\n# DEFINITIONS\n###############\n\n\ndef add_int(sim, number):\n a_rand = np.random.uniform(10,25,size=number)\n e_rand = np.random.normal(1,.2,size=number)\n e_rand = e_rand/100\n w_rand = np.random.uniform(0,2*np.pi,size=number)\n i_rand = np.random.uniform(0,10,size=number)*radeg\n f_rand = np.random.uniform(0,2*np.pi,size=number)\n \n for i in range(number):\n sem = a_rand[i]\n ecc = e_rand[i]\n icl = i_rand[i]\n Ome = w_rand[i]\n fan = f_rand[i]\n has = 'interior_{0}'.format(i)\n sim.add(m=0, primary=sim.particles['Sun'], a=sem, e=ecc, inc=icl, Omega=Ome, f=fan, hash=has)\n return\n\ndef add_ext(sim, number):\n a_rand = np.random.uniform(35,50,size=number)\n e_rand = np.random.normal(1,.2,size=number)\n e_rand = e_rand/100\n w_rand = np.random.uniform(0,2*np.pi,size=number)\n i_rand = np.random.uniform(0,10,size=number)*radeg\n f_rand = np.random.uniform(0,2*np.pi,size=number)\n \n for i in range(number):\n sem = a_rand[i]\n ecc = e_rand[i]\n icl = i_rand[i]\n Ome = w_rand[i]\n fan = f_rand[i]\n has = 'exterior_{0}'.format(i)\n sim.add(m=0, primary=sim.particles['Sun'], a=sem, e=ecc, inc=icl, Omega=Ome, f=fan, hash=has)\n return\n\n###########################\n###########################\n###########################\n\n### SIMULATION\n\n###########################\n###########################\n###########################\n\nfile = np.loadtxt('2solmass_track.txt')\nsol_t = file[700:,0]\nsol_m = file[700:,1]\nsol_l = file[700:,6]\n\nlog_l = InterpolatedUnivariateSpline(sol_t, sol_l,k=1)\nm_sol = InterpolatedUnivariateSpline(sol_t, sol_m,k=1)\n\nnum_int = 4\nnum_ext = num_int\nT0 = sol_t[107]\nt_tot = 10000\n\nNout = 10000\ntimes = np.linspace(0, t_tot, Nout)\nmtimes = m_sol(times + T0)\n\nM0 = 2\nnum_ast = num_int + num_ext\n\n\n##############\n# Setting up sim\n##############\n\n\nsim = rebound.Simulation()\n\nsim.add(m=M0,x=0, y=0, z=0, vx=0, vy=0, vz=0, hash='Sun')\nadd_int(sim, num_int)\nadd_ext(sim, num_ext)\n\nsim.integrator = \"whfast\"\nsim.dt = .5\nsim.testparticle_type = 0\nsim.track_energy_offset = 1\n#sim.ri_mercurius.hillfac = 5\n\nE0 = sim.calculate_energy()\nsim.move_to_com()\n\nps = sim.particles\nfig = rebound.OrbitPlot(sim, slices=True)\n\n\nx_sol = np.zeros(Nout); y_sol = np.zeros(Nout); z_sol = np.zeros(Nout)\nx_sol[0] = ps['Sun'].x\ny_sol[0] = ps['Sun'].y\nz_sol[0] = ps['Sun'].z\n\n\na_vals = np.zeros((num_ast, Nout))\ne_vals = np.zeros((num_ast, Nout))\ni_vals = np.zeros((num_ast, Nout))\npmvals = np.zeros((num_ast, Nout))\nomvals = np.zeros((num_ast, Nout))\no_vals = np.zeros((num_ast, Nout))\nlmvals = np.zeros((num_ast, Nout))\n\nenergy = np.zeros(Nout)\nrhill = np.zeros(Nout)\nmass = np.zeros(Nout)\nlums = np.zeros(Nout)\n\nx_vals = np.zeros((num_ast, Nout))\ny_vals = np.zeros((num_ast, Nout))\nz_vals = np.zeros((num_ast, Nout))\n\nfor moon in range(num_int):\n a_vals[moon,0] = ps['interior_{0}'.format(moon)].a\n e_vals[moon,0] = ps['interior_{0}'.format(moon)].e\n i_vals[moon,0] = ps['interior_{0}'.format(moon)].inc\n lmvals[moon,0] = ps['interior_{0}'.format(moon)].l\n pmvals[moon,0] = ps['interior_{0}'.format(moon)].pomega\n omvals[moon,0] = ps['interior_{0}'.format(moon)].Omega\n o_vals[moon,0] = ps['interior_{0}'.format(moon)].omega\n x_vals[moon,0] = ps['interior_{0}'.format(moon)].x\n y_vals[moon,0] = ps['interior_{0}'.format(moon)].y\n z_vals[moon,0] = ps['interior_{0}'.format(moon)].z\n\nfor moon in range(num_ext):\n a_vals[moon + num_int,0] = ps['exterior_{0}'.format(moon)].a\n e_vals[moon + num_int,0] = ps['exterior_{0}'.format(moon)].e\n i_vals[moon + num_int,0] = ps['exterior_{0}'.format(moon)].inc\n lmvals[moon + num_int,0] = ps['exterior_{0}'.format(moon)].l\n pmvals[moon + num_int,0] = ps['exterior_{0}'.format(moon)].pomega\n omvals[moon + num_int,0] = ps['exterior_{0}'.format(moon)].Omega\n o_vals[moon + num_int,0] = ps['exterior_{0}'.format(moon)].omega\n x_vals[moon + num_int,0] = ps['exterior_{0}'.format(moon)].x\n y_vals[moon + num_int,0] = ps['exterior_{0}'.format(moon)].y\n z_vals[moon + num_int,0] = ps['exterior_{0}'.format(moon)].z",
"_____no_output_____"
],
[
"%%time\n\n###########################\n###########################\n###########################\n\n### RUNNING\n\n###########################\n###########################\n###########################\n\nfor i, time in enumerate(times):\n sim.integrate(time)\n\n #ps['Sun'].m = starmass.interpolate(rebx, t=sim.t)\n sim.move_to_com()\n \n ###################\n # Tracking elements\n ###################\n\n mass[i] = ps['Sun'].m\n lums[i] = log_l(sim.t + T0)\n x_sol[i] = ps['Sun'].x\n y_sol[i] = ps['Sun'].y\n z_sol[i] = ps['Sun'].z\n \n energy[i] = sim.calculate_energy()\n\n for moon in range(num_int):\n a_vals[moon,i] = ps['interior_{0}'.format(moon)].a\n e_vals[moon,i] = ps['interior_{0}'.format(moon)].e\n i_vals[moon,i] = ps['interior_{0}'.format(moon)].inc\n lmvals[moon,i] = ps['interior_{0}'.format(moon)].l\n pmvals[moon,i] = ps['interior_{0}'.format(moon)].pomega\n omvals[moon,i] = ps['interior_{0}'.format(moon)].Omega\n o_vals[moon,i] = ps['interior_{0}'.format(moon)].omega\n x_vals[moon,i] = ps['interior_{0}'.format(moon)].x\n y_vals[moon,i] = ps['interior_{0}'.format(moon)].y\n z_vals[moon,i] = ps['interior_{0}'.format(moon)].z\n\n for moon in range(num_ext):\n a_vals[moon + num_int,i] = ps['exterior_{0}'.format(moon)].a\n e_vals[moon + num_int,i] = ps['exterior_{0}'.format(moon)].e\n i_vals[moon + num_int,i] = ps['exterior_{0}'.format(moon)].inc\n lmvals[moon + num_int,i] = ps['exterior_{0}'.format(moon)].l\n pmvals[moon + num_int,i] = ps['exterior_{0}'.format(moon)].pomega\n omvals[moon + num_int,i] = ps['exterior_{0}'.format(moon)].Omega\n o_vals[moon + num_int,i] = ps['exterior_{0}'.format(moon)].omega\n x_vals[moon + num_int,i] = ps['exterior_{0}'.format(moon)].x\n y_vals[moon + num_int,i] = ps['exterior_{0}'.format(moon)].y\n z_vals[moon + num_int,i] = ps['exterior_{0}'.format(moon)].z\n\ni_vals/= radeg\n\ndE = abs((energy - E0)/E0)",
"CPU times: user 7.14 s, sys: 62.5 ms, total: 7.2 s\nWall time: 7.2 s\n"
],
[
"a_i = a_vals; e_i = e_vals; i_i = i_vals; pm_i = pmvals; lm_i = lmvals; om_i = omvals; o_i = o_vals\nx_i = x_vals; y_i = y_vals; z_i = z_vals\nmsi = mass; lsi = lums; xsi = x_sol ; ysi = y_sol; zsi = z_sol\ndEi = dE; t_i = times\n\ni_dif = np.zeros_like(i_i)\ni_int = i_i[:,0]\nfor i in range(5000):\n i_dif[:,i] = i_i[:,i] - i_int",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(2,2,figsize=(14*.75,13*.75))\n\nax[0,0].plot(t_i[1:]/1e6,a_i.T[1:])\nax[0,0].set_ylabel(\"Semimajor Axis\")\n\nax[0,1].scatter(t_i[:],(o_i.T[:,0]),s=0.2)\nax[0,1].scatter(t_i[:],(om_i.T[:,0]),s=0.2)\nax[0,1].set_ylabel(r'$\\omega$ (Blue) and $\\Omega$ (Orange)')\nax[0,1].set_title(r'no yark')\n\nax[1,0].plot(t_i[1:]/1e6,i_i.T[1:])\nax[1,0].set_ylabel('inclination')\nax[1,0].set_xlabel('Time / Myr')\n\nax[1,1].plot(t_i[1:]/1e6,e_i.T[1:])\nax[1,1].set_ylabel('change in inclination')\nax[1,1].set_xlabel('Time / Myr')\n\nfig.tight_layout()\nfig.show()",
"_____no_output_____"
],
[
"def dadt(a, L, M):\n return 14.4 * (0.1/0.1) * (1/M)**0.5 * (1/20) * (1/3) * 1e-4/1e6 * np.power(10,L) / np.sqrt(a)\n\ndef dver(a, L, M):\n n = np.sqrt(4*np.pi**2*M/a**3)\n m = 3*(4/3)*np.pi*(10*100000)**3/1.9891e33\n R = 10/1.496e8\n c = 63197.8\n return -R**2 * np.power(10,L)*0.000235 / (16*c*m*n*a**2) * np.cos(i_i[0,0])\n\ndef didt(i,a,L,M):\n n = np.sqrt(4*np.pi**2*M/a**3)\n m = 3*(4/3)*np.pi*(10*100000)**3/1.9891e33\n R = 10/1.496e8\n c = 63197.8\n ",
"_____no_output_____"
],
[
"t0 = sol_t[107]\ntl = 3000000\nN_ = 3000000\nts = np.linspace(0, tl, N_)\ndt = ts[1] - ts[0]\nOm = om_i[0,0]\nprint(dt)\n\na_ = np.zeros(N_)\nav = np.zeros(N_)\ni_ = np.zeros(N_)\nl_ = np.zeros(N_)\nm_ = np.zeros(N_)\nda = np.zeros(N_)\ndv = np.zeros(N_)\ndi = np.zeros(N_)\n\na_[0] = 10\nav[0] = 10\ni_[0] = 2.5*radeg",
"1.0000003333334444\n"
],
[
"%%time\n\ni = 0 \nwhile i < N_-1:\n l_[i] = log_l(ts[i] + t0)\n m_[i] = 2\n \n at = dadt(a_[i],l_[i],m_[i])\n atv= dver(av[i],l_[i],m_[i])\n\n da[i+1] = at\n dv[i+1] = atv\n a_[i+1] = at*dt + a_[i]\n av[i+1] = atv*dt+ av[i]\n\n end = i\n i += 1\n \nprint(end)",
"2999998\nCPU times: user 1min 16s, sys: 31.2 ms, total: 1min 16s\nWall time: 1min 17s\n"
],
[
"fig, ax = plt.subplots(figsize=(10,5))\nax.plot(ts[1:]/1e6, a_[1:] - a_[0],zorder=10)\nax.plot(ts[1:]/1e6, av[1:] - av[0],zorder=10)\nax.plot(t_i[1:]/1e6, a_i.T[1:,0] - a_i.T[0,0],'k',ls='--')\nax.legend(['Leapfrog (Greenberg 2020)','Leapfrog (Veras 2015)', 'Rebound w/ Yark force'], loc='upper left')\nax.set_xlim(0.0001,3)\n#ax.set_ylim(0,0.08)\nax.set_yscale('log')\nax.set_xscale('log')\nax.set_xlabel('Time / Myr')\nax.set_ylabel(\"Cumulative change in a / au\")\nfig.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(3,figsize=(10,7),sharex=True,gridspec_kw={'height_ratios': [3, 1,1]})\nplt.subplots_adjust(hspace=0)\nax[0].plot(ts[:-1]/1e6, da[:-1],zorder=10)\nax[0].plot(ts[:-1]/1e6, dv[:-1],zorder=10)\nax[1].plot(ts[:-1]/1e6, l_[:-1],zorder=10,c='k')\nax[2].plot(ts[:-1]/1e6, m_[:-1],zorder=10,c='k')\nax[0].legend(['Leapfrog (Greenberg 2020)','Leapfrog (Veras 2015)'], loc='upper left')\nax[0].set_xlim(0,3)\nax[0].set_yscale('log')\nax[1].set_xlabel('Time / Myr')\nax[0].set_ylabel(\"da/dt (au/yr)\")\nax[1].set_ylabel(r\"log (L / L$_\\odot$)\")\nax[1].set_ylabel(r\"M$_\\star$ / M$_\\odot$\")\nfig.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(10,5))\nax.plot(ts[1:]/1e6, a_[1:],zorder=10)\nax.plot(ts[1:]/1e6, av[1:],zorder=10)\nax.plot(t_i[1:]/1e6, a_i.T[1:,0],'k',ls='--')\nax.legend(['Leapfrog (Greenberg 2020)','Leapfrog (Veras 2015)', 'Rebound w/ Yark force'], loc='upper left')\nax.set_xlim(0,3)\n#ax.set_ylim(0,0.08)\n#ax.set_yscale('log')\n#ax.set_xscale('log')\nax.set_xlabel('Time / Myr')\nax.set_ylabel(\"SMA / au\")\nfig.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eced70bbb8501c730355862716a1b58fe0bc4ee2 | 57,705 | ipynb | Jupyter Notebook | student_classification/PreProcessing_2.ipynb | hsdaniel86/machine_learning-classification | 3ba424196fe70b32c8ac094d15e1da9a5c49ec8e | [
"MIT"
] | null | null | null | student_classification/PreProcessing_2.ipynb | hsdaniel86/machine_learning-classification | 3ba424196fe70b32c8ac094d15e1da9a5c49ec8e | [
"MIT"
] | null | null | null | student_classification/PreProcessing_2.ipynb | hsdaniel86/machine_learning-classification | 3ba424196fe70b32c8ac094d15e1da9a5c49ec8e | [
"MIT"
] | null | null | null | 32.694051 | 116 | 0.273477 | [
[
[
"import numpy as np\nimport pandas as pd\nfrom sklearn.feature_selection import SelectKBest, SelectPercentile, chi2\nfrom sklearn.preprocessing import StandardScaler",
"_____no_output_____"
],
[
"# Loading Data\ndf = pd.read_csv(\"Data/data2.csv\")",
"_____no_output_____"
],
[
"# Viewing dataframe\ndf",
"_____no_output_____"
]
],
[
[
"Generating new variables",
"_____no_output_____"
]
],
[
[
"# compares variables and generates a new one\ndef compare_human(x):\n if x[\"DISAPPROVALS_BL\"] == 0 and x[\"DISAPPROVALS_EN\"] == 0:\n return(0)\n else:\n return(1)\ndf[\"DISAPPROVALS_HUMAN\"] = df.apply(compare_human, axis= 1)",
"_____no_output_____"
],
[
"def compare_exacts(x):\n if x[\"DISAPPROVALS_FM\"] == 0 and x[\"DISAPPROVALS_OM\"] == 0:\n return(0)\n else:\n return(1)\ndf[\"DISAPPROVALS_EXACTS\"] = df.apply(compare_exacts, axis= 1)",
"_____no_output_____"
],
[
"# Viewing new data frame\ndf",
"_____no_output_____"
],
[
"# Converting categorical variable into numeric variable\nvar = ['NOTE_BL_CAT', 'NOTE_EN_CAT', 'NOTE_FM_CAT', 'NOTE_OM_CAT']\ncount = 0\nfor i in var:\n df[count]= df[i].map({\"(-1, 2]\": 2,\n \"(2, 4]\": 4, \n \"(4, 6]\": 6,\n \"(6, 8]\": 8,\n \"(8, 10]\": 10})\n count += 1\ndf.rename(columns={0:'NOTE_BL_CAT_N', 1:'NOTE_EN_CAT_N', 2:'NOTE_FM_CAT_N', 3:'NOTE_OM_CAT_N'}, inplace=True) ",
"_____no_output_____"
],
[
"# checking data type \ndf.dtypes",
"_____no_output_____"
],
[
"# Arranging dataframe columns\na = ['DISAPPROVALS_BL', 'DISAPPROVALS_EN', 'DISAPPROVALS_HUMAN', 'DISAPPROVALS_FM','DISAPPROVALS_OM', \n 'DISAPPROVALS_EXACTS', 'NOTE_BL', 'NOTE_EN', 'NOTE_FM', 'NOTE_OM', 'NOTE_BL_CAT', 'NOTE_EN_CAT',\n 'NOTE_FM_CAT', 'NOTE_OM_CAT', 'NOTE_BL_CAT_N', 'NOTE_EN_CAT_N', 'NOTE_FM_CAT_N', 'NOTE_OM_CAT_N', \n 'ENGLISH', 'H_CLASS_PRES', 'ONLINE_TASKS', 'ABSENCES', 'PROFILE_N']\ndf = df.reindex(columns= a)",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.isna().sum()",
"_____no_output_____"
],
[
"# Saving data\ndf.to_csv(\"Data/data3.csv\", index= False)",
"_____no_output_____"
],
[
"# creating new dataframe and removing from categorical variables \ndf1 = df.drop(['NOTE_BL_CAT', 'NOTE_EN_CAT', 'NOTE_FM_CAT', 'NOTE_OM_CAT'], axis= 1)\ndf1",
"_____no_output_____"
],
[
"df1.shape",
"_____no_output_____"
],
[
"# Checking data type\ndf1.dtypes",
"_____no_output_____"
],
[
"df1.isna().sum()",
"_____no_output_____"
]
],
[
[
"<font color=\"blue\">Applying feature selection</font>",
"_____no_output_____"
]
],
[
[
"x = df1.iloc[:, 0:18]\ny = df1.PROFILE_N",
"_____no_output_____"
],
[
"x.shape",
"_____no_output_____"
],
[
"y.shape",
"_____no_output_____"
],
[
"feature_selection = pd.DataFrame(SelectKBest(chi2, k= 10).fit_transform(x, y))\nfeature_selection[\"TARGET\"]= y\nfeature_selection.head(15)",
"_____no_output_____"
],
[
"# saving feature selection in csv format\nfeature_selection.to_csv(\"Data/selection_variables.csv\", index= False)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
eced8311f5f70939085de870ca6f70317d0f0db5 | 10,957 | ipynb | Jupyter Notebook | session/quantization/quantize-generator-model.ipynb | AetherPrior/malaya | 45d37b171dff9e92c5d30bd7260b282cd0912a7d | [
"MIT"
] | 88 | 2021-01-06T10:01:31.000Z | 2022-03-30T17:34:09.000Z | session/quantization/quantize-generator-model.ipynb | AetherPrior/malaya | 45d37b171dff9e92c5d30bd7260b282cd0912a7d | [
"MIT"
] | 43 | 2021-01-14T02:44:41.000Z | 2022-03-31T19:47:42.000Z | session/quantization/quantize-generator-model.ipynb | AetherPrior/malaya | 45d37b171dff9e92c5d30bd7260b282cd0912a7d | [
"MIT"
] | 38 | 2021-01-06T07:15:03.000Z | 2022-03-19T05:07:50.000Z | 30.520891 | 600 | 0.541389 | [
[
[
"import os\nos.environ['CUDA_VISIBLE_DEVICES'] = ''",
"_____no_output_____"
],
[
"!wget https://f000.backblazeb2.com/file/malaya-model/v38/generator/base.pb\n!wget https://f000.backblazeb2.com/file/malaya-model/v38/generator/small.pb",
"--2020-11-16 11:22:24-- https://f000.backblazeb2.com/file/malaya-model/v38/generator/base.pb\nResolving f000.backblazeb2.com (f000.backblazeb2.com)... 104.153.233.177\nConnecting to f000.backblazeb2.com (f000.backblazeb2.com)|104.153.233.177|:443... connected.\nHTTP request sent, awaiting response... 200 \nLength: 1252668691 (1.2G) [application/octet-stream]\nSaving to: ‘base.pb’\n\nbase.pb 100%[===================>] 1.17G 13.0MB/s in 1m 46s \n\n2020-11-16 11:24:12 (11.3 MB/s) - ‘base.pb’ saved [1252668691/1252668691]\n\n--2020-11-16 11:24:12-- https://f000.backblazeb2.com/file/malaya-model/v38/generator/small.pb\nResolving f000.backblazeb2.com (f000.backblazeb2.com)... 104.153.233.177\nConnecting to f000.backblazeb2.com (f000.backblazeb2.com)|104.153.233.177|:443... connected.\nHTTP request sent, awaiting response... 200 \nLength: 355570391 (339M) [application/octet-stream]\nSaving to: ‘small.pb’\n\nsmall.pb 100%[===================>] 339.10M 11.2MB/s in 31s \n\n2020-11-16 11:24:45 (11.1 MB/s) - ‘small.pb’ saved [355570391/355570391]\n\n"
],
[
"import tensorflow as tf\nfrom tensorflow.tools.graph_transforms import TransformGraph\nfrom glob import glob\ntf.set_random_seed(0)",
"_____no_output_____"
],
[
"pbs = glob('*.pb')\npbs",
"_____no_output_____"
],
[
"import tensorflow_text\nimport tf_sentencepiece",
"_____no_output_____"
],
[
"transforms = ['add_default_attributes',\n 'remove_nodes(op=Identity, op=CheckNumerics, op=Dropout)',\n 'fold_constants(ignore_errors=true)',\n 'fold_batch_norms',\n 'fold_old_batch_norms',\n# 'quantize_weights(fallback_min=-10, fallback_max=10)',\n 'strip_unused_nodes',\n 'sort_by_execution_order']\n\nfor pb in pbs:\n input_graph_def = tf.GraphDef()\n with tf.gfile.FastGFile(pb, 'rb') as f:\n input_graph_def.ParseFromString(f.read())\n \n print(pb)\n \n transformed_graph_def = TransformGraph(input_graph_def, \n ['inputs'],\n ['SentenceTokenizer_1/SentenceTokenizer/SentencepieceDetokenizeOp'], transforms)\n \n with tf.gfile.GFile(f'{pb}.quantized', 'wb') as f:\n f.write(transformed_graph_def.SerializeToString())",
"WARNING:tensorflow:From <ipython-input-6-ed0c3462ae0b>:12: FastGFile.__init__ (from tensorflow.python.platform.gfile) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.gfile.GFile.\nsmall.pb\nbase.pb\n"
],
[
"def load_graph(frozen_graph_filename, **kwargs):\n with tf.gfile.GFile(frozen_graph_filename, 'rb') as f:\n graph_def = tf.GraphDef()\n graph_def.ParseFromString(f.read())\n\n # https://github.com/onnx/tensorflow-onnx/issues/77#issuecomment-445066091\n # to fix import T5\n for node in graph_def.node:\n if node.op == 'RefSwitch':\n node.op = 'Switch'\n for index in xrange(len(node.input)):\n if 'moving_' in node.input[index]:\n node.input[index] = node.input[index] + '/read'\n elif node.op == 'AssignSub':\n node.op = 'Sub'\n if 'use_locking' in node.attr:\n del node.attr['use_locking']\n elif node.op == 'AssignAdd':\n node.op = 'Add'\n if 'use_locking' in node.attr:\n del node.attr['use_locking']\n elif node.op == 'Assign':\n node.op = 'Identity'\n if 'use_locking' in node.attr:\n del node.attr['use_locking']\n if 'validate_shape' in node.attr:\n del node.attr['validate_shape']\n if len(node.input) == 2:\n node.input[0] = node.input[1]\n del node.input[1]\n\n with tf.Graph().as_default() as graph:\n tf.import_graph_def(graph_def)\n return graph",
"_____no_output_____"
],
[
"# g = load_graph('base.pb.quantized')\n# x = g.get_tensor_by_name('import/inputs:0')\n# logits = g.get_tensor_by_name('import/SentenceTokenizer_1/SentenceTokenizer/SentencepieceDetokenizeOp:0')",
"_____no_output_____"
],
[
"# x, x_len, logits",
"_____no_output_____"
],
[
"# test_sess = tf.InteractiveSession(graph = g)",
"_____no_output_____"
],
[
"# x",
"_____no_output_____"
],
[
"# %%time\n# test_sess.run(logits, feed_dict = {x: ['ringkasan: KUALA LUMPUR: Presiden Perancis Emmanuel Macron tidak menampakkan beliau seorang sosok yang bertamadun, selar Tun Dr Mahathir Mohamad menerusi kemas kini terbaharu di blognya. Bekas Perdana Menteri itu mendakwa, pemerintah tertinggi Perancis itu bersikap primitif kerana menuduh orang Islam terlibat dalam pembunuhan guru yang menghina Islam, malah menegaskan tindakan membunuh bukan ajaran Islam. Jelas Dr Mahathir, sejarah membuktikan bahawa orang Perancis pernah membunuh jutaan manusia, yang ramai mangsanya terdiri dari orang Islam.']})",
"_____no_output_____"
],
[
"# %%time\n# test_sess.run(logits, feed_dict = {x: [[1,2,3,3,4]], x_len: [[1,1,1,1,1]]})",
"_____no_output_____"
],
[
"quantized = glob('*.pb.quantized')\nquantized",
"_____no_output_____"
],
[
"!rm *.pb*",
"_____no_output_____"
],
[
"# converter = tf.compat.v1.lite.TFLiteConverter.from_frozen_graph(\n# graph_def_file='test.pb',\n# input_arrays=['Placeholder', 'Placeholder_1'],\n# input_shapes={'Placeholder' : [None, 512], 'Placeholder_1': [None, 512]},\n# output_arrays=['logits'],\n# )\n# # converter.allow_custom_ops=True",
"_____no_output_____"
],
[
"# converter.target_spec.supported_ops = [tf.lite.OpsSet.TFLITE_BUILTINS, tf.lite.OpsSet.SELECT_TF_OPS]\n# converter.target_spec.supported_types = [tf.float16]\n# converter.optimizations = [tf.lite.Optimize.DEFAULT]\n# converter.experimental_new_converter = True\n# tflite_model = converter.convert()",
"_____no_output_____"
],
[
"# converter.target_spec.supported_ops = [tf.lite.OpsSet.TFLITE_BUILTINS, \n# tf.lite.OpsSet.SELECT_TF_OPS]\n# converter.target_spec.supported_types = [tf.float16]\n# converter.optimizations = [tf.lite.Optimize.DEFAULT]\n# tflite_model = converter.convert()\n\n# with open('tiny-bert-sentiment-float16.tflite', 'wb') as f:\n# f.write(tflite_model)",
"_____no_output_____"
],
[
"# converter.target_spec.supported_ops = [tf.lite.OpsSet.TFLITE_BUILTINS, \n# tf.lite.OpsSet.SELECT_TF_OPS]\n# converter.optimizations = [tf.lite.Optimize.OPTIMIZE_FOR_SIZE]\n# tflite_model = converter.convert()\n\n# with open('tiny-bert-sentiment-hybrid.tflite', 'wb') as f:\n# f.write(tflite_model)",
"_____no_output_____"
],
[
"# interpreter = tf.lite.Interpreter(model_path='tiny-bert-sentiment-hybrid.tflite')\n# interpreter.allocate_tensors()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eced88318f6931b4192fd818ee565ab6d79225a5 | 23,740 | ipynb | Jupyter Notebook | courses/machine_learning/deepdive/08_image/mnist_linear.ipynb | KayvanShah1/training-data-analyst | 3f778a57b8e6d2446af40ca6063b2fd9c1b4bc88 | [
"Apache-2.0"
] | 6,140 | 2016-05-23T16:09:35.000Z | 2022-03-30T19:00:46.000Z | courses/machine_learning/deepdive/08_image/mnist_linear.ipynb | KayvanShah1/training-data-analyst | 3f778a57b8e6d2446af40ca6063b2fd9c1b4bc88 | [
"Apache-2.0"
] | 1,384 | 2016-07-08T22:26:41.000Z | 2022-03-24T16:39:43.000Z | courses/machine_learning/deepdive/08_image/mnist_linear.ipynb | KayvanShah1/training-data-analyst | 3f778a57b8e6d2446af40ca6063b2fd9c1b4bc88 | [
"Apache-2.0"
] | 5,110 | 2016-05-27T13:45:18.000Z | 2022-03-31T18:40:42.000Z | 48.252033 | 5,040 | 0.698863 | [
[
[
"# MNIST Image Classification with TensorFlow\n\nThis notebook demonstrates how to implement a simple linear image models on MNIST using tf.keras.\n<hr/>\nThis <a href=\"mnist_models.ipynb\">companion notebook</a> extends the basic harness of this notebook to a variety of models including DNN, CNN, dropout, pooling etc.",
"_____no_output_____"
]
],
[
[
"!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst",
"_____no_output_____"
],
[
"import numpy as np\nimport shutil\nimport os\nimport tensorflow.compat.v1 as tf\ntf.disable_v2_behavior()\nprint(tf.__version__)",
"WARNING:tensorflow:From /home/jupyter/.local/lib/python3.5/site-packages/tensorflow_core/python/compat/v2_compat.py:88: disable_resource_variables (from tensorflow.python.ops.variable_scope) is deprecated and will be removed in a future version.\nInstructions for updating:\nnon-resource variables are not supported in the long term\n2.1.0\n"
]
],
[
[
"## Exploring the data\n\nLet's download MNIST data and examine the shape. We will need these numbers ...",
"_____no_output_____"
]
],
[
[
"HEIGHT = 28\nWIDTH = 28\nNCLASSES = 10",
"_____no_output_____"
],
[
"# Get mnist data\nmnist = tf.keras.datasets.mnist\n\n(x_train, y_train), (x_test, y_test) = mnist.load_data()\n\n# Scale our features between 0 and 1\nx_train, x_test = x_train / 255.0, x_test / 255.0 \n\n# Convert labels to categorical one-hot encoding\ny_train = tf.keras.utils.to_categorical(y = y_train, num_classes = NCLASSES)\ny_test = tf.keras.utils.to_categorical(y = y_test, num_classes = NCLASSES)\n\nprint(\"x_train.shape = {}\".format(x_train.shape))\nprint(\"y_train.shape = {}\".format(y_train.shape))\nprint(\"x_test.shape = {}\".format(x_test.shape))\nprint(\"y_test.shape = {}\".format(y_test.shape))",
"x_train.shape = (60000, 28, 28)\ny_train.shape = (60000, 10)\nx_test.shape = (10000, 28, 28)\ny_test.shape = (10000, 10)\n"
],
[
"import matplotlib.pyplot as plt\nIMGNO = 12\nplt.imshow(x_test[IMGNO].reshape(HEIGHT, WIDTH));",
"_____no_output_____"
]
],
[
[
"## Define the model.\nLet's start with a very simple linear classifier. All our models will have this basic interface -- they will take an image and return probabilities.",
"_____no_output_____"
]
],
[
[
"# Build Keras Model Using Keras Sequential API\ndef linear_model():\n model = tf.keras.models.Sequential()\n model.add(tf.keras.layers.InputLayer(input_shape = [HEIGHT, WIDTH], name = \"image\"))\n model.add(tf.keras.layers.Flatten())\n model.add(tf.keras.layers.Dense(units = NCLASSES, activation = tf.nn.softmax, name = \"probabilities\"))\n return model",
"_____no_output_____"
]
],
[
[
"## Write Input Functions\n\nAs usual, we need to specify input functions for training, evaluation, and predicition.",
"_____no_output_____"
]
],
[
[
"# Create training input function\ntrain_input_fn = tf.estimator.inputs.numpy_input_fn(\n x = {\"image\": x_train},\n y = y_train,\n batch_size = 100,\n num_epochs = None,\n shuffle = True,\n queue_capacity = 5000\n )\n\n# Create evaluation input function\neval_input_fn = tf.estimator.inputs.numpy_input_fn(\n x = {\"image\": x_test},\n y = y_test,\n batch_size = 100,\n num_epochs = 1,\n shuffle = False,\n queue_capacity = 5000\n )\n\n# Create serving input function for inference\ndef serving_input_fn():\n placeholders = {\"image\": tf.placeholder(dtype = tf.float32, shape = [None, HEIGHT, WIDTH])}\n features = placeholders # as-is\n return tf.estimator.export.ServingInputReceiver(features = features, receiver_tensors = placeholders)",
"_____no_output_____"
]
],
[
[
"## Create train_and_evaluate function",
"_____no_output_____"
],
[
" tf.estimator.train_and_evaluate does distributed training.",
"_____no_output_____"
]
],
[
[
"def train_and_evaluate(output_dir, hparams):\n # Build Keras model\n model = linear_model()\n \n # Compile Keras model with optimizer, loss function, and eval metrics\n model.compile(\n optimizer = \"adam\",\n loss = \"categorical_crossentropy\",\n metrics = [\"accuracy\"])\n \n # Convert Keras model to an Estimator\n estimator = tf.keras.estimator.model_to_estimator(\n keras_model = model, \n model_dir = output_dir)\n\n # Set estimator's train_spec to use train_input_fn and train for so many steps\n train_spec = tf.estimator.TrainSpec(\n input_fn = train_input_fn,\n max_steps = hparams[\"train_steps\"])\n\n # Create exporter that uses serving_input_fn to create saved_model for serving\n exporter = tf.estimator.LatestExporter(\n name = \"exporter\", \n serving_input_receiver_fn = serving_input_fn)\n\n # Set estimator's eval_spec to use eval_input_fn and export saved_model\n eval_spec = tf.estimator.EvalSpec(\n input_fn = eval_input_fn,\n steps = None,\n exporters = exporter)\n\n # Run train_and_evaluate loop\n tf.estimator.train_and_evaluate(\n estimator = estimator, \n train_spec = train_spec, \n eval_spec = eval_spec)",
"_____no_output_____"
]
],
[
[
"This is the main() function",
"_____no_output_____"
]
],
[
[
"OUTDIR = \"mnist/learned\"\nshutil.rmtree(OUTDIR, ignore_errors = True) # start fresh each time\n\nhparams = {\"train_steps\": 1000, \"learning_rate\": 0.01}\ntrain_and_evaluate(OUTDIR, hparams)",
"INFO:tensorflow:Using default config.\nINFO:tensorflow:Using the Keras model provided.\nWARNING:tensorflow:From /home/jupyter/.local/lib/python3.5/site-packages/tensorflow_core/python/ops/init_ops.py:97: calling GlorotUniform.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nCall initializer instance with the dtype argument instead of passing it to the constructor\nWARNING:tensorflow:From /home/jupyter/.local/lib/python3.5/site-packages/tensorflow_core/python/ops/init_ops.py:97: calling Zeros.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nCall initializer instance with the dtype argument instead of passing it to the constructor\nINFO:tensorflow:Using config: {'_eval_distribute': None, '_session_creation_timeout_secs': 7200, '_save_summary_steps': 100, '_global_id_in_cluster': 0, '_service': None, '_experimental_distribute': None, '_num_ps_replicas': 0, '_cluster_spec': ClusterSpec({}), '_train_distribute': None, '_tf_random_seed': None, '_experimental_max_worker_delay_secs': None, '_protocol': None, '_log_step_count_steps': 100, '_model_dir': 'mnist/learned', '_is_chief': True, '_evaluation_master': '', '_keep_checkpoint_max': 5, '_device_fn': None, '_keep_checkpoint_every_n_hours': 10000, '_session_config': allow_soft_placement: true\ngraph_options {\n rewrite_options {\n meta_optimizer_iterations: ONE\n }\n}\n, '_save_checkpoints_secs': 600, '_task_id': 0, '_save_checkpoints_steps': None, '_task_type': 'worker', '_master': '', '_num_worker_replicas': 1}\nINFO:tensorflow:Not using Distribute Coordinator.\nINFO:tensorflow:Running training and evaluation locally (non-distributed).\nINFO:tensorflow:Start train and evaluate loop. The evaluate will happen after every checkpoint. Checkpoint frequency is determined based on RunConfig arguments: save_checkpoints_steps None or save_checkpoints_secs 600.\nWARNING:tensorflow:From /home/jupyter/.local/lib/python3.5/site-packages/tensorflow_core/python/training/training_util.py:236: Variable.initialized_value (from tensorflow.python.ops.variables) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse Variable.read_value. Variables in 2.X are initialized automatically both in eager and graph (inside tf.defun) contexts.\nWARNING:tensorflow:From /home/jupyter/.local/lib/python3.5/site-packages/tensorflow_estimator/python/estimator/inputs/queues/feeding_queue_runner.py:62: QueueRunner.__init__ (from tensorflow.python.training.queue_runner_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nTo construct input pipelines, use the `tf.data` module.\nWARNING:tensorflow:From /home/jupyter/.local/lib/python3.5/site-packages/tensorflow_estimator/python/estimator/inputs/queues/feeding_functions.py:500: add_queue_runner (from tensorflow.python.training.queue_runner_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nTo construct input pipelines, use the `tf.data` module.\nINFO:tensorflow:Calling model_fn.\nINFO:tensorflow:Done calling model_fn.\nINFO:tensorflow:Warm-starting with WarmStartSettings: WarmStartSettings(ckpt_to_initialize_from='mnist/learned/keras/keras_model.ckpt', vars_to_warm_start='.*', var_name_to_vocab_info={}, var_name_to_prev_var_name={})\nINFO:tensorflow:Warm-starting from: mnist/learned/keras/keras_model.ckpt\nINFO:tensorflow:Warm-starting variables only in TRAINABLE_VARIABLES.\nINFO:tensorflow:Warm-started 2 variables.\nINFO:tensorflow:Create CheckpointSaverHook.\nINFO:tensorflow:Graph was finalized.\nINFO:tensorflow:Running local_init_op.\nINFO:tensorflow:Done running local_init_op.\nWARNING:tensorflow:From /home/jupyter/.local/lib/python3.5/site-packages/tensorflow_core/python/training/monitored_session.py:906: start_queue_runners (from tensorflow.python.training.queue_runner_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nTo construct input pipelines, use the `tf.data` module.\nINFO:tensorflow:Saving checkpoints for 0 into mnist/learned/model.ckpt.\nINFO:tensorflow:loss = 2.3702068, step = 1\nINFO:tensorflow:global_step/sec: 340.867\nINFO:tensorflow:loss = 0.7732622, step = 101 (0.295 sec)\nINFO:tensorflow:global_step/sec: 384.292\nINFO:tensorflow:loss = 0.6145685, step = 201 (0.260 sec)\nINFO:tensorflow:global_step/sec: 391.351\nINFO:tensorflow:loss = 0.50422275, step = 301 (0.256 sec)\nINFO:tensorflow:global_step/sec: 397.311\nINFO:tensorflow:loss = 0.4027025, step = 401 (0.252 sec)\nINFO:tensorflow:global_step/sec: 391.875\nINFO:tensorflow:loss = 0.44188076, step = 501 (0.258 sec)\nINFO:tensorflow:global_step/sec: 386.557\nINFO:tensorflow:loss = 0.37594798, step = 601 (0.256 sec)\nINFO:tensorflow:global_step/sec: 389.035\nINFO:tensorflow:loss = 0.34926274, step = 701 (0.257 sec)\nINFO:tensorflow:global_step/sec: 400.38\nINFO:tensorflow:loss = 0.52942824, step = 801 (0.250 sec)\nINFO:tensorflow:global_step/sec: 391.907\nINFO:tensorflow:loss = 0.40796912, step = 901 (0.255 sec)\nINFO:tensorflow:Saving checkpoints for 1000 into mnist/learned/model.ckpt.\nINFO:tensorflow:Calling model_fn.\nINFO:tensorflow:Done calling model_fn.\nINFO:tensorflow:Starting evaluation at 2020-06-02T17:25:49Z\nINFO:tensorflow:Graph was finalized.\nINFO:tensorflow:Restoring parameters from mnist/learned/model.ckpt-1000\nINFO:tensorflow:Running local_init_op.\nINFO:tensorflow:Done running local_init_op.\nINFO:tensorflow:Inference Time : 0.65976s\nINFO:tensorflow:Finished evaluation at 2020-06-02-17:25:50\nINFO:tensorflow:Saving dict for global step 1000: acc = 0.9136, global_step = 1000, loss = 0.3200383\nINFO:tensorflow:Saving 'checkpoint_path' summary for global step 1000: mnist/learned/model.ckpt-1000\nINFO:tensorflow:Calling model_fn.\nINFO:tensorflow:Done calling model_fn.\nWARNING:tensorflow:From /home/jupyter/.local/lib/python3.5/site-packages/tensorflow_core/python/saved_model/signature_def_utils_impl.py:201: build_tensor_info (from tensorflow.python.saved_model.utils_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis function will only be available through the v1 compatibility library as tf.compat.v1.saved_model.utils.build_tensor_info or tf.compat.v1.saved_model.build_tensor_info.\nINFO:tensorflow:Signatures INCLUDED in export for Classify: None\nINFO:tensorflow:Signatures INCLUDED in export for Train: None\nINFO:tensorflow:Signatures INCLUDED in export for Regress: None\nINFO:tensorflow:Signatures INCLUDED in export for Eval: None\nINFO:tensorflow:Signatures INCLUDED in export for Predict: ['serving_default']\nINFO:tensorflow:Restoring parameters from mnist/learned/model.ckpt-1000\nINFO:tensorflow:Assets added to graph.\nINFO:tensorflow:No assets to write.\nINFO:tensorflow:SavedModel written to: mnist/learned/export/exporter/temp-b'1591118750'/saved_model.pb\nINFO:tensorflow:Loss for final step: 0.45906192.\n"
]
],
[
[
"I got:\n\n`Saving dict for global step 1000: categorical_accuracy = 0.9112, global_step = 1000, loss = 0.32516304`\n\nIn other words, we achieved 91.12% accuracy with the simple linear model!",
"_____no_output_____"
],
[
"<pre>\n# Copyright 2020 Google Inc. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n</pre>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
eced917f6d329963b20c4e5f023798d09d89d829 | 43,118 | ipynb | Jupyter Notebook | Project/LSTM model-real CV.ipynb | carloswert/MHC-Class-I-prediction-mice | e72714483b0c70169da66161e4cd5e89aeedb83a | [
"Apache-2.0"
] | null | null | null | Project/LSTM model-real CV.ipynb | carloswert/MHC-Class-I-prediction-mice | e72714483b0c70169da66161e4cd5e89aeedb83a | [
"Apache-2.0"
] | null | null | null | Project/LSTM model-real CV.ipynb | carloswert/MHC-Class-I-prediction-mice | e72714483b0c70169da66161e4cd5e89aeedb83a | [
"Apache-2.0"
] | null | null | null | 91.545648 | 1,487 | 0.630224 | [
[
[
"import numpy as np\nimport scipy as sc\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport keras\nimport tensorflow as tf\nfrom keras.callbacks import Callback, ModelCheckpoint\nfrom keras import backend as K\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Activation, Embedding, LSTM, BatchNormalization\nfrom keras.optimizers import SGD, Adam\nfrom sklearn.model_selection import train_test_split,GridSearchCV,cross_val_score\nfrom sklearn.metrics import confusion_matrix\nfrom keras.preprocessing.text import Tokenizer\nfrom keras.utils import to_categorical, Sequence\nfrom sklearn.utils import resample,class_weight, shuffle\nfrom keras.layers import Bidirectional\nfrom IPython.display import clear_output\nimport matplotlib.patches as mpatches\nfrom sklearn.metrics import roc_curve, auc, precision_recall_curve\nfrom keras.wrappers.scikit_learn import KerasClassifier\nfrom keras.constraints import max_norm\nfrom scipy.sparse import issparse\nimport types\nimport copy",
"_____no_output_____"
],
[
"#Statistical analysis of the physical properties of the 20 naturally occurring amino acids\ndef AminoAcidsEmb(string):\n AA_as10Factor= {\n 'A' :[-1.56 ,-1.67 ,-0.97 ,-0.27 ,-0.93 ,-0.78 ,-0.20 ,-0.08 ,0.21 ,-0.48 ],\n 'R' :[0.22 ,1.27 ,1.37 ,1.87 ,-1.70 ,0.46 ,0.92 ,-0.39 ,0.23 ,0.93 ],\n 'N' :[1.14 ,-0.07 ,-0.12 ,0.81 ,0.18 ,0.37 ,-0.09 ,1.23 ,1.10 ,-1.73 ],\n 'D' :[0.58 ,-0.22 ,-1.58 ,0.81 ,-0.92 ,0.15 ,-1.52 ,0.47 ,0.76 ,0.70 ],\n 'C' :[0.12 ,-0.89 ,0.45 ,-1.05 ,-0.71 ,2.41 ,1.52 ,-0.69 ,1.13 ,1.10 ],\n 'Q' :[-0.47 ,0.24 ,0.07 ,1.10 ,1.10 ,0.59 ,0.84 ,-0.71 ,-0.03 ,-2.33 ],\n 'E' :[-1.45 ,0.19 ,-1.61 ,1.17 ,-1.31 ,0.40 ,0.04 ,0.38 ,-0.35 ,-0.12 ],\n 'G' :[1.46 ,-1.96 ,-0.23 ,-0.16 ,0.10 ,-0.11 ,1.32 ,2.36 ,-1.66 ,0.46 ],\n 'H' :[-0.41 ,0.52 ,-0.28 ,0.28 ,1.61 ,1.01 ,-1.85 ,0.47 ,1.13 ,1.63 ],\n 'I' :[-0.73 ,-0.16 ,1.79 ,-0.77 ,-0.54 ,0.03 ,-0.83 ,0.51 ,0.66 ,-1.78 ],\n 'L' :[-1.04 ,0.00 ,-0.24 ,-1.10 ,-0.55 ,-2.05 ,0.96 ,-0.76 ,0.45 ,0.93 ],\n 'K' :[-0.34 ,0.82 ,-0.23 ,1.70 ,1.54 ,-1.62 ,1.15 ,-0.08 ,-0.48 ,0.60 ],\n 'M' :[-1.40 ,0.18 ,-0.42 ,-0.73 ,2.00 ,1.52 ,0.26 ,0.11 ,-1.27 ,0.27 ],\n 'F' :[-0.21 ,0.98 ,-0.36 ,-1.43 ,0.22 ,-0.81 ,0.67 ,1.10 ,1.71 ,-0.44 ],\n 'P' :[2.06 ,-0.33 ,-1.15 ,-0.75 ,0.88 ,-0.45 ,0.30 ,-2.30 ,0.74 ,-0.28 ],\n 'S' :[0.81 ,-1.08 ,0.16 ,0.42 ,-0.21 ,-0.43 ,-1.89 ,-1.15 ,-0.97 ,-0.23 ],\n 'T' :[0.26 ,-0.70 ,1.21 ,0.63 ,-0.10 ,0.21 ,0.24 ,-1.15 ,-0.56 ,0.19 ],\n 'W' :[0.30 ,2.10 ,-0.72 ,-1.57 ,-1.16 ,0.57 ,-0.48 ,-0.40 ,-2.30 ,-0.60 ],\n 'Y' :[1.38 ,1.48 ,0.80 ,-0.56 ,-0.00 ,-0.68 ,-0.31 ,1.03 ,-0.05 ,0.53 ],\n 'V' :[-0.74 ,-0.71 ,2.04 ,-0.40 ,0.50 ,-0.81 ,-1.07 ,0.06 ,-0.46 ,0.65 ],\n 'X' :[0.0]*10,\n '_' :[0.0]*10}\n ls=[]*10\n for item in string:\n if item in AA_as10Factor.keys():\n ls = ls+AA_as10Factor.get(item)\n embedding=np.reshape(np.array(ls),(-1,10))\n return embedding\n#(size of protein, dimensions or 21)\n\ndef Strings2Embed(array):\n arr = []\n for n in range(array.shape[0]): \n arr.append(AminoAcidsEmb(array[n]).T)\n arr=np.dstack(arr).T\n return arr\n\ndef BLOSUMSIM(string):\n #Obtain the BLOSUM62 Matrix\n string = list(string)\n if len(string)>1:\n num=np.random.randint(0,len(string)-1)\n char=string[num]\n else:\n char = string[0]\n num = 0\n with open(\"/Volumes/Maxtor/References/blosum62.txt\") as matrix_file:\n matrix = matrix_file.read()\n lines = matrix.strip().split('\\n')\n header = lines.pop(0)\n columns = header.split()\n matrix = {}\n for row in lines:\n entries = row.split()\n row_name = entries.pop(0)\n matrix[row_name] = {}\n if len(entries) != len(columns):\n raise Exception('Improper entry number in row')\n for column_name in columns:\n matrix[row_name][column_name] = int(entries.pop(0))\n #Retrieve the aa with highest similarity\n listaas = dict(map(reversed, matrix.get(char).items()))\n listprob = np.array(list(listaas.keys()))\n listprob = listprob[np.where(listprob>0)]\n prob = 1\n if listprob[1:].size > 0:\n nn = int(np.random.choice(listprob[1:],1))\n chrf = listaas.get(nn)\n prob = (np.exp(nn))/(sum(np.exp(listprob)))\n else:\n chrf = char\n string[num] = chrf\n string = \"\".join(string)\n return string, prob",
"_____no_output_____"
],
[
"class BalancedSequence(Sequence):\n \"\"\"Balancing input classes with augmentation possibility and setting the balancing fraction\n \"\"\"\n def __init__(self, X, y, batch_size, fracPos=0.5,val_split=0.1,isaug=False,naug=3,p_aug=0.5):\n self.X = X[0:int((1-val_split)*X.shape[0])]\n self.y = y[0:int((1-val_split)*y.shape[0])]\n self.batch_size = batch_size\n self.isaug = isaug\n self.naug = naug\n self.p_aug = p_aug\n self.pos_indices = np.where(self.y == 1)[0]\n self.neg_indices = np.where(self.y == 0)[0]\n self.X_Pos = self.X[self.pos_indices]\n self.n = min(len(self.pos_indices), len(self.neg_indices))\n if fracPos>(len(self.pos_indices)/(len(self.pos_indices)+len(self.neg_indices))):\n self.fracPos = fracPos\n else:\n self.fracPos = len(self.pos_indices)/(len(self.pos_indices)+len(self.neg_indices))\n self._index_array = None\n\n def __len__(self):\n # Reset batch after we are done with minority class.\n return int((self.n * (1/self.fracPos)) // self.batch_size)\n\n def on_epoch_end(self):\n # Reset batch after all minority indices are covered.\n self._index_array = None\n\n def __getitem__(self, batch_idx):\n if self._index_array is None:\n pos_indices = self.pos_indices.copy()\n neg_indices = self.neg_indices.copy()\n np.random.shuffle(pos_indices)\n np.random.shuffle(neg_indices)\n n_neg = int(np.floor(self.n*((1/self.fracPos)-1)))\n self._index_array = np.concatenate((pos_indices[:self.n], neg_indices[:n_neg]))\n np.random.shuffle(self._index_array)\n indices = self._index_array[batch_idx * self.batch_size: (batch_idx + 1) * self.batch_size]\n Xf = self.X[indices]\n Yf = self.y[indices]\n if self.isaug:\n for n in range(0,self.naug):\n new_pep, pp = BLOSUMSIM(self.X_Pos[np.random.randint(0,self.X_Pos.shape[0])])\n Xf = np.append(Xf,new_pep)\n if pp>=self.p_aug:\n Yf = np.append(Yf,1)\n else:\n Yf = np.append(Yf,0)\n indexx = np.arange(Xf.shape[0])\n np.random.shuffle(indexx)\n Xf = Xf[indexx]\n Yf = Yf[indexx]\n else:\n pass\n return Strings2Embed(Xf), Yf\n\nclass ValidationSet(Sequence):\n def __init__(self,X):\n self.X = X\n self.valToTake = int(X.shape[0])\n def __len__(self):\n return self.valToTake\n\n def __getitem__(self,batch_idx):\n return Strings2Embed(self.X)\n \n def on_epoch_end(self):\n pass\n\nclass CollectOutputAndTarget(Callback):\n def __init__(self):\n super(CollectOutputAndTarget, self).__init__()\n self.targets = [] # collect y_true batches\n self.outputs = [] # collect y_pred batches\n self.inputs= []\n\n # the shape of these 2 variables will change according to batch shape\n # to handle the \"last batch\", specify `validate_shape=False`\n self.var_y_true = tf.Variable(0., validate_shape=False)\n self.var_y_pred = tf.Variable(0., validate_shape=False)\n self.var_x = tf.Variable(0., validate_shape=False)\n\n def on_batch_end(self, batch, logs=None):\n # evaluate the variables and save them into lists\n self.targets.append(K.eval(self.var_y_true))\n self.outputs.append(K.eval(self.var_y_pred))\n if len(self.inputs)>1:\n print(np.array_equal(self.inputs[-1],self.validation_data[0]))\n self.inputs.append(self.validation_data[0])\n \nclass AccLossPlotter(Callback):\n \"\"\"Plot training Accuracy and Loss values on a Matplotlib graph. \n The graph is updated by the 'on_epoch_end' event of the Keras Callback class\n # Arguments\n graphs: list with some or all of ('acc', 'loss')\n save_graph: Save graph as an image on Keras Callback 'on_train_end' event \n \"\"\"\n\n def __init__(self, graphs=['acc', 'loss'], save_graph=True):\n self.graphs = graphs\n self.num_subplots = len(graphs)\n self.save_graph = save_graph\n\n\n def on_train_begin(self, logs={}):\n self.acc = []\n self.val_acc = []\n self.loss = []\n self.val_loss = []\n self.epoch_count = 0\n plt.ion()\n plt.show()\n\n\n def on_epoch_end(self, epoch, logs={}):\n self.epoch_count += 1\n self.val_acc.append(logs.get('val_acc'))\n self.acc.append(logs.get('acc'))\n self.loss.append(logs.get('loss'))\n self.val_loss.append(logs.get('val_loss'))\n epochs = [x for x in range(self.epoch_count)]\n\n count_subplots = 0\n \n if 'acc' in self.graphs:\n count_subplots += 1\n plt.subplot(self.num_subplots, 1, count_subplots)\n plt.title('Accuracy')\n plt.plot(epochs, self.val_acc, color='r')\n plt.plot(epochs, self.acc, color='b')\n plt.ylabel('accuracy')\n\n red_patch = mpatches.Patch(color='red', label='Test')\n blue_patch = mpatches.Patch(color='blue', label='Train')\n\n plt.legend(handles=[red_patch, blue_patch], loc=4)\n\n if 'loss' in self.graphs:\n count_subplots += 1\n plt.subplot(self.num_subplots, 1, count_subplots)\n plt.title('Loss')\n #plt.axis([0,100,0,5])\n plt.plot(epochs, self.val_loss, color='r')\n plt.plot(epochs, self.loss, color='b')\n plt.ylabel('loss')\n\n red_patch = mpatches.Patch(color='red', label='Test')\n blue_patch = mpatches.Patch(color='blue', label='Train')\n\n plt.legend(handles=[red_patch, blue_patch], loc=4)\n \n plt.draw()\n plt.pause(0.001)\n\n def on_train_end(self, logs={}):\n if self.save_graph:\n plt.savefig('training_acc_loss.png')\nplot_losses = AccLossPlotter()\n\ndef create_network(neurons=10):\n model = Sequential()\n model.add(Bidirectional(LSTM(neurons),input_shape=(9,10)))\n model.add(Dense(30, activation='relu'))\n model.add(Dropout(0.5))\n model.add(Dense(64, activation='relu'))\n model.add(Dropout(0.5))\n model.add(Dense(1, activation='sigmoid'))\n adam = Adam(lr=0.01, epsilon=None, decay=0.0, amsgrad=False)\n model.compile(loss='binary_crossentropy',\n optimizer=adam,\n metrics=['accuracy'])\n return model",
"_____no_output_____"
],
[
"class KerasClassifier(KerasClassifier):\n def fit(self, x, y, **kwargs):\n if self.build_fn is None:\n self.model = self.__call__(**self.filter_sk_params(self.__call__))\n elif (not isinstance(self.build_fn, types.FunctionType) and\n not isinstance(self.build_fn, types.MethodType)):\n self.model = self.build_fn(\n **self.filter_sk_params(self.build_fn.__call__))\n else:\n self.model = self.build_fn(**self.filter_sk_params(self.build_fn))\n\n loss_name = self.model.loss\n if hasattr(loss_name, '__name__'):\n loss_name = loss_name.__name__\n if loss_name == 'categorical_crossentropy' and len(y.shape) != 2:\n y = to_categorical(y)\n fit_args = copy.deepcopy(self.filter_sk_params(Sequential.fit))\n fit_args.update(kwargs)\n del fit_args['batch_size']\n #########################################################\n self.__history=self.model.fit_generator(BalancedSequence(x,y,self.sk_params[\"batch_size\"],fracPos=0.4,val_split=0,isaug=False,naug=3,p_aug=0.5),\n samples_per_epoch=x.shape[0],**fit_args)\n return self.__history\n def predict_proba(self, x,**kwargs):\n x = Strings2Embed(x)\n kwargs = self.filter_sk_params(Sequential.predict_proba, kwargs)\n probs = self.model.predict(x, **kwargs)\n # check if binary classification\n if probs.shape[1] == 1:\n # first column is probability of class 0 and second is of class 1\n probs = np.hstack([1 - probs, probs])\n return probs",
"_____no_output_____"
],
[
"data=pd.read_csv(\"/Volumes/Maxtor/firstrain.csv\",delimiter=\",\")\ninit=data['peptides'].values\nc=0\nfor word in init:\n init[c]=word[0:9]\n c+=1\nx=np.asarray(init)\ny=np.asarray(data['NB'])\nx_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.2, random_state = 0)\nx_test=Strings2Embed(x_test)\nneural_network = KerasClassifier(build_fn=create_network, \n epochs=100, \n batch_size=10, \n verbose=0)\nneurons = [25, 28, 30, 64, 70]\nparam_grid = dict(neurons=neurons)\ngrid=GridSearchCV(estimator=neural_network, param_grid=param_grid, n_jobs=1,scoring='roc_auc')\ngrid_result = grid.fit(x_train, y_train)\nprint(\"Best: %f using %s\" % (grid_result.best_score_, grid_result.best_params_))",
"/Users/carloswertcarvajal/anaconda3/lib/python3.7/site-packages/sklearn/model_selection/_split.py:2053: FutureWarning: You should specify a value for 'cv' instead of relying on the default value. The default value will change from 3 to 5 in version 0.22.\n warnings.warn(CV_WARNING, FutureWarning)\n/Users/carloswertcarvajal/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:22: UserWarning: Update your `fit_generator` call to the Keras 2 API: `fit_generator(<__main__...., epochs=100, verbose=0, steps_per_epoch=59)`\n/Users/carloswertcarvajal/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:22: UserWarning: Update your `fit_generator` call to the Keras 2 API: `fit_generator(<__main__...., epochs=100, verbose=0, steps_per_epoch=59)`\n/Users/carloswertcarvajal/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:22: UserWarning: Update your `fit_generator` call to the Keras 2 API: `fit_generator(<__main__...., epochs=100, verbose=0, steps_per_epoch=60)`\n/Users/carloswertcarvajal/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:22: UserWarning: Update your `fit_generator` call to the Keras 2 API: `fit_generator(<__main__...., epochs=100, verbose=0, steps_per_epoch=59)`\n/Users/carloswertcarvajal/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:22: UserWarning: Update your `fit_generator` call to the Keras 2 API: `fit_generator(<__main__...., epochs=100, verbose=0, steps_per_epoch=59)`\n"
],
[
"x_train.shape",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eced975711e054492a6cd515759313d5b120a9ba | 35,314 | ipynb | Jupyter Notebook | notebooks/robin_ue3/sfcrimec.ipynb | hhain/sdap17 | 8bd0b4cb60d6140141c834ffcac8835a888a0949 | [
"MIT"
] | null | null | null | notebooks/robin_ue3/sfcrimec.ipynb | hhain/sdap17 | 8bd0b4cb60d6140141c834ffcac8835a888a0949 | [
"MIT"
] | 1 | 2017-06-08T22:32:48.000Z | 2017-06-08T22:32:48.000Z | notebooks/robin_ue3/sfcrimec.ipynb | hhain/sdap17 | 8bd0b4cb60d6140141c834ffcac8835a888a0949 | [
"MIT"
] | null | null | null | 32.045372 | 1,490 | 0.45591 | [
[
[
"# Kaggle: San Francisco Crime Classification\n## Improvement as part of sdap17 excercise 3",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport pprint\nimport requests",
"_____no_output_____"
]
],
[
[
"## Exploration of the training data set",
"_____no_output_____"
]
],
[
[
"train_data = pd.read_csv(\"../../data/raw/train.csv\")\ntrain_data['Dates'] = pd.to_datetime(train_data['Dates'])\n\ntest_data = pd.read_csv(\"../../data/raw/test.csv\")\ntest_data['Dates'] = pd.to_datetime(test_data['Dates'])",
"_____no_output_____"
],
[
"len(train_data)",
"_____no_output_____"
],
[
"train_data.head()",
"_____no_output_____"
],
[
"crimes = train_data['Category'].unique()\npprint.pprint(\"Crimes: {}, #{}\".format(crimes, len(crimes)), indent=2)",
"(\"Crimes: ['WARRANTS' 'OTHER OFFENSES' 'LARCENY/THEFT' 'VEHICLE THEFT' \"\n \"'VANDALISM'\\n\"\n \" 'NON-CRIMINAL' 'ROBBERY' 'ASSAULT' 'WEAPON LAWS' 'BURGLARY'\\n\"\n \" 'SUSPICIOUS OCC' 'DRUNKENNESS' 'FORGERY/COUNTERFEITING' 'DRUG/NARCOTIC'\\n\"\n \" 'STOLEN PROPERTY' 'SECONDARY CODES' 'TRESPASS' 'MISSING PERSON' 'FRAUD'\\n\"\n \" 'KIDNAPPING' 'RUNAWAY' 'DRIVING UNDER THE INFLUENCE'\\n\"\n \" 'SEX OFFENSES FORCIBLE' 'PROSTITUTION' 'DISORDERLY CONDUCT' 'ARSON'\\n\"\n \" 'FAMILY OFFENSES' 'LIQUOR LAWS' 'BRIBERY' 'EMBEZZLEMENT' 'SUICIDE'\\n\"\n \" 'LOITERING' 'SEX OFFENSES NON FORCIBLE' 'EXTORTION' 'GAMBLING'\\n\"\n \" 'BAD CHECKS' 'TREA' 'RECOVERED VEHICLE' 'PORNOGRAPHY/OBSCENE MAT'], #39\")\n"
],
[
"train_data['Category'].value_counts()",
"_____no_output_____"
]
],
[
[
"# Generate time based features",
"_____no_output_____"
]
],
[
[
"def get_halfhour(minute):\n if minute < 30:\n return 0\n else:\n return 1\n\ndef get_daynight(hour):\n if 5 < hour and hour < 23:\n return 0\n else:\n return 1\n \ndef generate_time_features(times):\n minute_series = pd.Series([x.minute for x in times], name='minute')\n halfhour_series = pd.Series([get_halfhour(x.minute) for x in times], name='halfhour')\n hour_series = pd.Series([x.hour for x in times], name='hour')\n daynight_series = pd.Series([get_daynight(x.hour) for x in times], name='day_night')\n day_series = pd.Series([x.day for x in times], name='day')\n month_series = pd.Series([x.month for x in times], name='month')\n year_series = pd.Series([x.year for x in times], name='year')\n \n time_features = pd.concat([minute_series, halfhour_series, hour_series, daynight_series, day_series, month_series, year_series], axis=1)\n return time_features",
"_____no_output_____"
],
[
"times = train_data[\"Dates\"]",
"_____no_output_____"
],
[
"time_features = generate_time_features(times)\nprint(\"success\")",
"success\n"
],
[
"print(time_features)",
" minute halfhour hour day_night day month year\n0 53 1 23 1 13 5 2015\n1 53 1 23 1 13 5 2015\n2 33 1 23 1 13 5 2015\n3 30 1 23 1 13 5 2015\n4 30 1 23 1 13 5 2015\n5 30 1 23 1 13 5 2015\n6 30 1 23 1 13 5 2015\n7 30 1 23 1 13 5 2015\n8 0 0 23 1 13 5 2015\n9 0 0 23 1 13 5 2015\n10 58 1 22 0 13 5 2015\n11 30 1 22 0 13 5 2015\n12 30 1 22 0 13 5 2015\n13 6 0 22 0 13 5 2015\n14 0 0 22 0 13 5 2015\n15 0 0 22 0 13 5 2015\n16 0 0 22 0 13 5 2015\n17 55 1 21 0 13 5 2015\n18 40 1 21 0 13 5 2015\n19 30 1 21 0 13 5 2015\n20 30 1 21 0 13 5 2015\n21 17 0 21 0 13 5 2015\n22 11 0 21 0 13 5 2015\n23 11 0 21 0 13 5 2015\n24 10 0 21 0 13 5 2015\n25 0 0 21 0 13 5 2015\n26 0 0 21 0 13 5 2015\n27 0 0 21 0 13 5 2015\n28 0 0 21 0 13 5 2015\n29 56 1 20 0 13 5 2015\n... ... ... ... ... ... ... ...\n878019 37 1 2 1 6 1 2003\n878020 32 1 2 1 6 1 2003\n878021 24 0 2 1 6 1 2003\n878022 16 0 2 1 6 1 2003\n878023 15 0 2 1 6 1 2003\n878024 9 0 2 1 6 1 2003\n878025 6 0 2 1 6 1 2003\n878026 6 0 2 1 6 1 2003\n878027 0 0 2 1 6 1 2003\n878028 0 0 2 1 6 1 2003\n878029 54 1 1 1 6 1 2003\n878030 54 1 1 1 6 1 2003\n878031 50 1 1 1 6 1 2003\n878032 36 1 1 1 6 1 2003\n878033 30 1 1 1 6 1 2003\n878034 30 1 1 1 6 1 2003\n878035 55 1 0 1 6 1 2003\n878036 55 1 0 1 6 1 2003\n878037 55 1 0 1 6 1 2003\n878038 42 1 0 1 6 1 2003\n878039 40 1 0 1 6 1 2003\n878040 33 1 0 1 6 1 2003\n878041 31 1 0 1 6 1 2003\n878042 20 0 0 1 6 1 2003\n878043 20 0 0 1 6 1 2003\n878044 15 0 0 1 6 1 2003\n878045 1 0 0 1 6 1 2003\n878046 1 0 0 1 6 1 2003\n878047 1 0 0 1 6 1 2003\n878048 1 0 0 1 6 1 2003\n\n[878049 rows x 7 columns]\n"
]
],
[
[
"## Create grid for sector analysis",
"_____no_output_____"
]
],
[
[
"# outliers are all at position X = -120.5, Y = 90\n\ndef filter_x(x):\n if (x > -122):\n return -122.4483364\n else: \n return x\n \ndef filter_y(y):\n if y > 37.9:\n return 37.7563690\n else:\n return y",
"_____no_output_____"
],
[
"# take a look at the positions of our train data.\nmin_x_train = min([filter_x(x) for x in train_data[\"X\"]]) \nmax_x_train = max([filter_x(x) for x in train_data[\"X\"]]) \nmin_y_train = min([filter_y(y) for y in train_data[\"Y\"]]) \nmax_y_train = max([filter_y(y) for y in train_data[\"Y\"]]) \nprint(\"Min_X_train: \", min_x_train)\nprint(\"Max_X_train: \", max_x_train)\nprint(\"Min_Y_train: \", min_y_train)\nprint(\"Max_Y_train: \", max_y_train)",
"Min_X_train: -122.513642064\nMax_X_train: -122.364937494\nMin_Y_train: 37.7078790224\nMax_Y_train: 37.8199754923\n"
],
[
"# take a look at the positions of our test data.\nmin_x_test = min([filter_x(x) for x in test_data[\"X\"]]) \nmax_x_test = max([filter_x(x) for x in test_data[\"X\"]]) \nmin_y_test = min([filter_y(y) for y in test_data[\"Y\"]]) \nmax_y_test = max([filter_y(y) for y in test_data[\"Y\"]]) \nprint(\"Min_X_test: \", min_x_test)\nprint(\"Max_X_test: \", max_x_test)\nprint(\"Min_Y_test: \", min_y_test)\nprint(\"Max_Y_test: \", max_y_test) ",
"Min_X_test: -122.513642064\nMax_X_test: -122.364750704\nMin_Y_test: 37.7078790224\nMax_Y_test: 37.8206208381\n"
],
[
"# Final coordinates for grid that covers San Francisco.\nmin_x = -122.53\nmax_x = -122.35\nmin_y = 37.65\nmax_y = 37.84\n\ndif_x = max_x - min_x\ndif_y = max_y - min_y",
"_____no_output_____"
],
[
"# grid functions\n\ndef get_subregion_pos(subregion_id, min_x, min_y, dif_x, dif_y, x_sections, y_sections):\n x = subregion_id % x_sections\n x_pos = ((x + 1/2) / x_sections) * dif_x + min_x\n y = subregion_id // x_sections\n y_pos = ((y + 1/2) / y_sections) * dif_y + min_y\n return (x_pos, y_pos)\n\ndef get_subregion(pos_x, pos_y, min_x, min_y, dif_x, dif_y, x_sections, y_sections):\n x = pos_x - min_x\n x_sec = int(x_sections * x / dif_x)\n y = pos_y - min_y\n y_sec = int(y_sections * y / dif_y)\n return x_sec + x_sections * y_sec\n \ndef get_subregion_series(data, min_x, min_y, dif_x, dif_y):\n X_SECTIONS = 20\n Y_SECTIONS = 20\n subregion_list = []\n for i in range(len(data)):\n pos_x = data[\"X\"][i]\n pos_y = data[\"Y\"][i]\n subregion = get_subregion(pos_x, pos_y, min_x, min_y, dif_x, dif_y, X_SECTIONS, Y_SECTIONS)\n subregion_list.append(subregion)\n return pd.Series(subregion_list, name='subregion')",
"_____no_output_____"
],
[
"subregion_series = get_subregion_series(train_data, min_x, min_y, dif_x, dif_y)",
"_____no_output_____"
],
[
"# look at the numer of crimes in each subregion\nsubregion_series.value_counts()",
"_____no_output_____"
],
[
"# highest crime rate around union square\nget_subregion_pos(293, min_x, min_y, dif_x, dif_y, 20, 20)",
"_____no_output_____"
]
],
[
[
"## police station one hot encoding",
"_____no_output_____"
]
],
[
[
"# generate one hot encoding of police destricts\none_hot_police_destricts = pd.get_dummies(train_data[\"PdDistrict\"])",
"_____no_output_____"
],
[
"one_hot_police_destricts[\"NORTHERN\"]",
"_____no_output_____"
]
],
[
[
"## crime distribution per subregion",
"_____no_output_____"
]
],
[
[
"regions = subregion_series.unique()\ncrimes = train_data['Category'].unique()",
"_____no_output_____"
],
[
"# count crimes in each region\ncriminal_activity_local = {}\ncriminal_activity_overall = train_data[\"Category\"].value_counts()\nfor r in regions:\n criminal_activity_local[r] = {}\n criminal_activity_local[r][\"N\"] = 0\n for c in crimes:\n criminal_activity_local[r][c] = 0\nfor i, r in enumerate(subregion_series):\n criminal_activity_local[r][train_data[\"Category\"][i]] += 1\n criminal_activity_local[r][\"N\"] += 1",
"_____no_output_____"
],
[
"# union square\ncriminal_activity_local[293]",
"_____no_output_____"
],
[
"# global crime distribution\ndistribution_global = {}\nfor c in crimes:\n distribution_global[c] = criminal_activity_overall[c] / len(train_data)\nfor c in distribution_global:\n print(c, distribution_global[c])",
"WARRANTS 0.0480770435363\nOTHER OFFENSES 0.143707241851\nLARCENY/THEFT 0.199191616869\nVEHICLE THEFT 0.0612505680207\nVANDALISM 0.0509367928214\nNON-CRIMINAL 0.105123973719\nROBBERY 0.0261944378958\nASSAULT 0.0875532003339\nWEAPON LAWS 0.00974319200865\nBURGLARY 0.0418598506461\nSUSPICIOUS OCC 0.0357770466113\nDRUNKENNESS 0.00487444322583\nFORGERY/COUNTERFEITING 0.0120824692016\nDRUG/NARCOTIC 0.0614669568555\nSTOLEN PROPERTY 0.00517055426292\nSECONDARY CODES 0.0113718027126\nTRESPASS 0.00834349791413\nMISSING PERSON 0.0295985759337\nFRAUD 0.0189955230289\nKIDNAPPING 0.00266613822235\nRUNAWAY 0.00221627722371\nDRIVING UNDER THE INFLUENCE 0.00258299935425\nSEX OFFENSES FORCIBLE 0.00499744319508\nPROSTITUTION 0.00852344231358\nDISORDERLY CONDUCT 0.00491999877\nARSON 0.0017231384581\nFAMILY OFFENSES 0.000559194304646\nLIQUOR LAWS 0.00216730501373\nBRIBERY 0.000329138806604\nEMBEZZLEMENT 0.00132794411246\nSUICIDE 0.000578555410917\nLOITERING 0.0013951385401\nSEX OFFENSES NON FORCIBLE 0.000168555513417\nEXTORTION 0.000291555482667\nGAMBLING 0.000166277736208\nBAD CHECKS 0.000462388773292\nTREA 6.833331625e-06\nRECOVERED VEHICLE 0.00357383243988\nPORNOGRAPHY/OBSCENE MAT 2.50555492917e-05\n"
],
[
"# local crime distribution\ndistribution_local = {}\nsufficient_n = 500\nfor r in regions:\n distribution_local[r] = {}\n for c in crimes: \n if criminal_activity_local[r][\"N\"] >= sufficient_n:\n distribution_local[r][c] = criminal_activity_local[r][c] / criminal_activity_local[r][\"N\"]\n else:\n distribution_local[r][c] = distribution_global[c]",
"_____no_output_____"
],
[
"# crime distribution at union square\nprint(distribution_local[293])",
"{'WARRANTS': 0.07139216054516674, 'OTHER OFFENSES': 0.1293697850991334, 'LARCENY/THEFT': 0.2553591071284844, 'VEHICLE THEFT': 0.018444986987900088, 'VANDALISM': 0.02308641644085531, 'NON-CRIMINAL': 0.11219917903039733, 'ROBBERY': 0.026305905078743325, 'ASSAULT': 0.08097013924288358, 'WEAPON LAWS': 0.007310922115204035, 'BURGLARY': 0.03536071687280337, 'SUSPICIOUS OCC': 0.028157111045528937, 'DRUNKENNESS': 0.00618410109194323, 'FORGERY/COUNTERFEITING': 0.011147479408687254, 'DRUG/NARCOTIC': 0.10692726638585572, 'STOLEN PROPERTY': 0.005432887076436026, 'SECONDARY CODES': 0.006868242427494433, 'TRESPASS': 0.015064523918117672, 'MISSING PERSON': 0.01036943632119765, 'FRAUD': 0.02499128055160572, 'KIDNAPPING': 0.0019316931827328093, 'RUNAWAY': 0.00012073082392080058, 'DRIVING UNDER THE INFLUENCE': 0.0011536500952432055, 'SEX OFFENSES FORCIBLE': 0.004118262549298419, 'PROSTITUTION': 0.0019987658626888097, 'DISORDERLY CONDUCT': 0.005942639444101628, 'ARSON': 0.0007914576234808038, 'FAMILY OFFENSES': 0.0003756070077536018, 'LIQUOR LAWS': 0.002267056582512811, 'BRIBERY': 0.00016097443189440078, 'EMBEZZLEMENT': 0.002293885654495211, 'SUICIDE': 0.0004426796877096021, 'LOITERING': 0.0013012099911464063, 'SEX OFFENSES NON FORCIBLE': 4.0243607973600196e-05, 'EXTORTION': 0.00020121803986800096, 'GAMBLING': 0.00013414535991200063, 'BAD CHECKS': 0.0003353633997800016, 'TREA': 0.0, 'RECOVERED VEHICLE': 0.0014219408150672069, 'PORNOGRAPHY/OBSCENE MAT': 2.682907198240013e-05}\n"
],
[
"sum(distribution_local[293]",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eced97fc904487f2c4bd96fb7bd4b428dde5f049 | 903,735 | ipynb | Jupyter Notebook | Machine_failure_prediction.ipynb | Lakshmipriya-S/Machine_Failure_Prediction | 372f6e9a0f5de9978549a840c2169f146d12e0f4 | [
"Apache-2.0"
] | null | null | null | Machine_failure_prediction.ipynb | Lakshmipriya-S/Machine_Failure_Prediction | 372f6e9a0f5de9978549a840c2169f146d12e0f4 | [
"Apache-2.0"
] | null | null | null | Machine_failure_prediction.ipynb | Lakshmipriya-S/Machine_Failure_Prediction | 372f6e9a0f5de9978549a840c2169f146d12e0f4 | [
"Apache-2.0"
] | null | null | null | 219.779912 | 567,296 | 0.892701 | [
[
[
"# <center> Machine Failure Prediction",
"_____no_output_____"
],
[
"### Importing necessary libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nfrom sklearn.preprocessing import LabelEncoder\nfrom imblearn.over_sampling import RandomOverSampler\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"### Reading the input data",
"_____no_output_____"
]
],
[
[
"data=pd.read_csv('Maintenance.csv')\ndata.head()",
"_____no_output_____"
],
[
"data['TWF'].nunique()",
"_____no_output_____"
],
[
"data.shape",
"_____no_output_____"
],
[
"data['Product ID'].nunique()",
"_____no_output_____"
],
[
"data['Machine failure'].sum()",
"_____no_output_____"
],
[
"data.isnull().sum()",
"_____no_output_____"
],
[
"label=LabelEncoder()\ndata['Type']=label.fit_transform(data['Type'])\ndata.head()",
"_____no_output_____"
],
[
"data[data['Machine failure']==1]",
"_____no_output_____"
],
[
"data[data['Machine failure'] ==1][['TWF','HDF','PWF','OSF','RNF']].apply(pd.value_counts)",
"_____no_output_____"
],
[
"data['Machine failure'].value_counts().plot(kind='pie', autopct='%1.1f%%')\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,9))\nplt.subplot(1,5,1)\ndata['TWF'].value_counts().plot(kind='pie', autopct='%1.1f%%')\nplt.subplot(1,5,2)\ndata['HDF'].value_counts().plot(kind='pie', autopct='%1.1f%%')\nplt.subplot(1,5,3)\ndata['PWF'].value_counts().plot(kind='pie', autopct='%1.1f%%')\nplt.subplot(1,5,4)\ndata['OSF'].value_counts().plot(kind='pie', autopct='%1.1f%%')\nplt.subplot(1,5,5)\ndata['RNF'].value_counts().plot(kind='pie', autopct='%1.1f%%')\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"data1=data.drop(['UDI','Product ID','TWF','HDF','PWF','OSF','RNF','Machine failure'],axis=1)\nplt.subplot(3,3,1)\nplt.hist(data1['Type'])\nplt.subplot(3,3,2)\nplt.hist(data1['Air temperature [K]'])\nplt.subplot(3,3,3)\nplt.hist(data1['Process temperature [K]'])\nplt.subplot(3,3,4)\nplt.hist(data1['Rotational speed [rpm]'])\nplt.subplot(3,3,5)\nplt.hist(data1['Torque [Nm]'])\nplt.subplot(3,3,6)\nplt.hist(data1['Tool wear [min]'])\nplt.show()",
"_____no_output_____"
],
[
"data.dtypes",
"_____no_output_____"
],
[
"data1.describe()",
"_____no_output_____"
],
[
"data.corr()",
"_____no_output_____"
],
[
"sns.heatmap(data1)\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,10))\nsns.heatmap(data.corr(), annot=True)",
"_____no_output_____"
],
[
"def plot_pair():\n sns.pairplot(data=data.drop(['UDI', 'TWF', 'HDF', 'PWF', 'OSF', 'RNF'], axis=1).select_dtypes(include='number'),\n hue='Machine failure',\n #plot_kws={'s':6},\n corner=True\n )\n plt.show()\n\nplot_pair()",
"_____no_output_____"
],
[
"plt.subplot(3,3,1)\nsns.boxplot(data1['Type'])\nplt.subplot(3,3,2)\nplt.boxplot(data1['Air temperature [K]'])\nplt.subplot(3,3,3)\nplt.boxplot(data1['Process temperature [K]'])\nplt.subplot(3,3,4)\nplt.boxplot(data1['Rotational speed [rpm]'])\nplt.subplot(3,3,5)\nplt.boxplot(data1['Torque [Nm]'])\nplt.subplot(3,3,6)\nplt.boxplot(data1['Tool wear [min]'])\nplt.show()",
"/Users/lakshmipriya/opt/anaconda3/lib/python3.8/site-packages/seaborn/_decorators.py:36: FutureWarning: Pass the following variable as a keyword arg: x. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n warnings.warn(\n"
],
[
"data.columns",
"_____no_output_____"
],
[
"data=data.rename({'Air temperature [K]':'AT','Process temperature [K]':'PT','Rotational speed [rpm]':'RS',\n 'Torque [Nm]':'T','Tool wear [min]':'TW','Machine failure':'Machine_failure'},axis=1)\ndata.head()",
"_____no_output_____"
],
[
"temp = data.groupby(['Type','Machine_failure']).Type.count().unstack()\nt1 = temp.plot(kind = 'bar', stacked = True, \n title = 'Type: Working Machine vs failed machine', \n color = ['green','red'], alpha = .70)\nplt.xlabel('Quality - good, low, medium')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# <center> Feature engineering",
"_____no_output_____"
],
[
"### Label encoding and Random upsampling of data and Feature importance",
"_____no_output_____"
]
],
[
[
"ros = RandomOverSampler(random_state=42)\nfrom imblearn.over_sampling import SMOTE",
"_____no_output_____"
],
[
"x=data.drop(['Machine_failure'],axis=1)\ny=data.Machine_failure",
"_____no_output_____"
],
[
"x1,y1= ros.fit_resample(x, y)",
"_____no_output_____"
],
[
"len(y1), len(x1)",
"_____no_output_____"
],
[
"len(y),len(x)",
"_____no_output_____"
]
],
[
[
"### Upsampling With SMOTE",
"_____no_output_____"
]
],
[
[
"data2=data.drop(['Product ID'],axis=1)",
"_____no_output_____"
],
[
"smote = SMOTE()",
"_____no_output_____"
],
[
"X=data2.drop(['Machine_failure'],axis=1)\nY=data2.Machine_failure",
"_____no_output_____"
],
[
"x_smote, y_smote = smote.fit_resample(X, Y)\nlen(y_smote), len(x_smote)",
"_____no_output_____"
],
[
"x_smote",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"x_train,x_test,y_train,y_test=train_test_split(x_smote,y_smote,test_size=0.4,random_state=20)",
"_____no_output_____"
],
[
"X_train=x_train.drop(['UDI','TWF','HDF','OSF','PWF','RNF'],axis=1)\nX_test=x_test.drop(['UDI','TWF','HDF','OSF','PWF','RNF'],axis=1)",
"_____no_output_____"
]
],
[
[
"# <center> Model building",
"_____no_output_____"
],
[
"### 1) Logistic regression",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import confusion_matrix,accuracy_score, classification_report",
"_____no_output_____"
],
[
"log_reg=LogisticRegression()\nlog_reg.fit(x_train,y_train)",
"/Users/lakshmipriya/opt/anaconda3/lib/python3.8/site-packages/sklearn/linear_model/_logistic.py:763: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n n_iter_i = _check_optimize_result(\n"
],
[
"pred_log=log_reg.predict(x_test)",
"_____no_output_____"
],
[
"cm=confusion_matrix(pred_log,y_test)\naccuracy1=accuracy_score(pred_log,y_test)\nprint(cm,accuracy1)",
"[[3408 429]\n [ 444 3448]] 0.8870487773321257\n"
],
[
"classification_report(pred_log,y_test)",
"_____no_output_____"
],
[
"pred_log_train=log_reg.predict(x_train)\n\nclassification_report(pred_log_train,y_train)\n",
"_____no_output_____"
]
],
[
[
"### 2) SVM",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import SVC ",
"_____no_output_____"
],
[
"# kernel is linear\nsvc=SVC(gamma=0.22)\nsvc.fit(x_train,y_train)\n",
"_____no_output_____"
],
[
"svc_score=svc.score(x_test,y_test)\nsvc_score",
"_____no_output_____"
],
[
"# kernel is radial\nsvc2=SVC(gamma=0.22,kernel='rbf',C=1)\nsvc2.fit(x_train,y_train)\n",
"_____no_output_____"
],
[
"svc_score2=svc2.score(x_test,y_test)\nsvc_score2",
"_____no_output_____"
],
[
"svc_pred2=svc2.predict(x_test)\nclassification_report(svc_pred2,y_test)",
"_____no_output_____"
],
[
"cm=confusion_matrix(svc_pred2,y_test)\ncm",
"_____no_output_____"
],
[
"pred_svm_train=svc2.predict(x_train)\n\nclassification_report(pred_svm_train,y_train)\n\n",
"_____no_output_____"
],
[
"# kernel is sigmoid\nsvc3=SVC(gamma=0.22,kernel='sigmoid',C=1)\nsvc3.fit(x_train,y_train)",
"_____no_output_____"
],
[
"svc_score3=svc3.score(x_test,y_test)\nsvc_score3",
"_____no_output_____"
],
[
"svc_pred3=svc3.predict(x_test)",
"_____no_output_____"
],
[
"classification_report(svc_pred3,y_test)",
"/Users/lakshmipriya/opt/anaconda3/lib/python3.8/site-packages/sklearn/metrics/_classification.py:1248: UndefinedMetricWarning: Recall and F-score are ill-defined and being set to 0.0 in labels with no true samples. Use `zero_division` parameter to control this behavior.\n _warn_prf(average, modifier, msg_start, len(result))\n/Users/lakshmipriya/opt/anaconda3/lib/python3.8/site-packages/sklearn/metrics/_classification.py:1248: UndefinedMetricWarning: Recall and F-score are ill-defined and being set to 0.0 in labels with no true samples. Use `zero_division` parameter to control this behavior.\n _warn_prf(average, modifier, msg_start, len(result))\n/Users/lakshmipriya/opt/anaconda3/lib/python3.8/site-packages/sklearn/metrics/_classification.py:1248: UndefinedMetricWarning: Recall and F-score are ill-defined and being set to 0.0 in labels with no true samples. Use `zero_division` parameter to control this behavior.\n _warn_prf(average, modifier, msg_start, len(result))\n"
],
[
"# kernel is polynomial\n#svc4=SVC(gamma=0.22,kernel='poly',C=1)\n#svc4.fit(x_train,y_train)",
"_____no_output_____"
],
[
"#svc_score4=svc4.score(x_test,y_test)\n#svc_score4",
"_____no_output_____"
]
],
[
[
"### 3) Naive bayes",
"_____no_output_____"
]
],
[
[
"from sklearn.naive_bayes import MultinomialNB as MB\nfrom sklearn.naive_bayes import GaussianNB as GB\n",
"_____no_output_____"
],
[
"mult_nb=MB()\nmult_nb.fit(x_train,y_train)\nmult_nb.score(x_test,y_test)\n\n\n",
"_____no_output_____"
],
[
"pred_mult_nb=mult_nb.predict(x_test)\nclassification_report(pred_mult_nb,y_test)",
"_____no_output_____"
],
[
"cm=confusion_matrix(pred_mult_nb,y_test)\ncm",
"_____no_output_____"
],
[
"pred_mulnb_train=mult_nb.predict(x_train)\n\nclassification_report(pred_mulnb_train,y_train)\n\n\n",
"_____no_output_____"
],
[
"gaus_nb=GB()\ngaus_nb.fit(x_train,y_train)\ngaus_score=gaus_nb.score(x_test,y_test)\n\ngaus_pred=gaus_nb.predict(x_test)\naccuracy_gaus=np.mean(gaus_pred==y_test)\n\ngaus_score,accuracy_gaus",
"_____no_output_____"
],
[
"\nclassification_report(gaus_pred,y_test)",
"_____no_output_____"
],
[
"cm=confusion_matrix(gaus_pred,y_test)\ncm",
"_____no_output_____"
],
[
"pred_gausnb_train=gaus_nb.predict(x_train)\n\nclassification_report(pred_gausnb_train,y_train)\n\n\n\n",
"_____no_output_____"
]
],
[
[
"### K nearest neighbour",
"_____no_output_____"
]
],
[
[
"from sklearn.neighbors import KNeighborsClassifier",
"_____no_output_____"
],
[
"knn=KNeighborsClassifier(n_neighbors=3)\nknn.fit(x_train,y_train)",
"_____no_output_____"
],
[
"pred_knn=knn.predict(x_test)\naccuracy_knn=accuracy_score(y_test,pred_knn)",
"_____no_output_____"
],
[
"accuracy_knn",
"_____no_output_____"
],
[
"classification_report(pred_knn,y_test)",
"_____no_output_____"
],
[
"cm=confusion_matrix(pred_knn,y_test)\ncm",
"_____no_output_____"
],
[
"pred_knn_train=knn.predict(x_train)\n\nclassification_report(pred_knn_train,y_train)\n\n\n\n",
"_____no_output_____"
]
],
[
[
"### 4) Decision Tree ",
"_____no_output_____"
]
],
[
[
"from sklearn.tree import DecisionTreeClassifier",
"_____no_output_____"
],
[
"dec_tree=DecisionTreeClassifier(criterion='gini',max_depth=6)\ndec_tree.fit(x_train,y_train)",
"_____no_output_____"
],
[
"dec_tree.score(x_test,y_test)\n",
"_____no_output_____"
],
[
"pred_dec_tree=dec_tree.predict(x_test)\n\naccuracy_dec=accuracy_score(y_test,pred_dec_tree)",
"_____no_output_____"
],
[
"accuracy_dec",
"_____no_output_____"
],
[
"classification_report(pred_dec_tree,y_test)",
"_____no_output_____"
],
[
"cm=confusion_matrix(pred_dec_tree,y_test)\ncm",
"_____no_output_____"
],
[
"pred_Dectree_train=dec_tree.predict(x_train)\n\nclassification_report(pred_Dectree_train,y_train)\n\n\n\n\n",
"_____no_output_____"
]
],
[
[
"### Random forest",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier",
"_____no_output_____"
],
[
"ran_for=RandomForestClassifier(n_estimators=100,max_features=3)\nran_for.fit(x_train,y_train)",
"_____no_output_____"
],
[
"rf_score=ran_for.score(x_test,y_test)\nrf_score",
"_____no_output_____"
],
[
"pred_rf=ran_for.predict(x_test)\n\nclassification_report(pred_rf,y_test)",
"_____no_output_____"
],
[
"cm=confusion_matrix(pred_rf,y_test)\ncm",
"_____no_output_____"
],
[
"pred_rf_train=ran_for.predict(x_train)\n\nclassification_report(pred_rf_train,y_train)\n",
"_____no_output_____"
]
],
[
[
"### XGB",
"_____no_output_____"
]
],
[
[
"#pip install XGBoost ",
"_____no_output_____"
],
[
"import xgboost as xgb\n",
"_____no_output_____"
],
[
"from xgboost import XGBClassifier",
"_____no_output_____"
],
[
"model_xgb=XGBClassifier()",
"_____no_output_____"
],
[
"model_xgb.fit(X_train,y_train)\npred_xgb=model_xgb.predict(X_test)\nacc_xgb=accuracy_score(pred_xgb,y_test)\nacc_xgb",
"/Users/lakshmipriya/opt/anaconda3/lib/python3.8/site-packages/xgboost/sklearn.py:888: UserWarning: The use of label encoder in XGBClassifier is deprecated and will be removed in a future release. To remove this warning, do the following: 1) Pass option use_label_encoder=False when constructing XGBClassifier object; and 2) Encode your labels (y) as integers starting with 0, i.e. 0, 1, 2, ..., [num_class - 1].\n warnings.warn(label_encoder_deprecation_msg, UserWarning)\n"
],
[
"\n\nclassification_report(pred_xgb,y_test)",
"_____no_output_____"
],
[
"cm=confusion_matrix(pred_xgb,y_test)\ncm",
"_____no_output_____"
],
[
"pred_xgb_train=model_xgb.predict(x_train)\n\nclassification_report(pred_xgb_train,y_train)\n\n",
"_____no_output_____"
],
[
"list1=[0.98,0.98,0.98]\nlist2=[1.00,1.00,1.00]",
"_____no_output_____"
],
[
"fig = plt.figure(figsize =(10, 7))\nplt.subplot(1,2,1)\nplt.pie(list1, labels = ['Precision','Recall','Accuracy'],explode=(0.1,0.1,0.1),\n autopct='0.98')\n\nplt.subplot(1,2,2)\nplt.pie(list2, labels = ['Precision','Recall','Accuracy'],explode=(0.1,0.1,0.1),autopct='1.00')",
"_____no_output_____"
]
],
[
[
"### catboost",
"_____no_output_____"
]
],
[
[
"import catboost",
"_____no_output_____"
],
[
"from catboost import CatBoostClassifier",
"_____no_output_____"
],
[
"model_cat=CatBoostClassifier(verbose=0)\nmodel_cat.fit(x_train,y_train)\npred_cat=model_cat.predict(x_test)\nacc_cat=accuracy_score(pred_cat,y_test)\nacc_cat",
"_____no_output_____"
],
[
"a=classification_report(pred_cat,y_test)",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"cm=confusion_matrix(pred_cat,y_test)\ncm",
"_____no_output_____"
],
[
"pred_catboost_train=model_cat.predict(x_train)\n\nclassification_report(pred_catboost_train,y_train)\n\n\n",
"_____no_output_____"
]
],
[
[
"### summary",
"_____no_output_____"
]
],
[
[
"t={\"Models\":pd.Series([\"Logistic regression\", \"SVM\", \"Gaussian Naive bayes\",\"KNeighborsClassifier\",\"Decision tree\",\"Random forest\",\"XGB\",\"Cat boost\"]),\n \"Accuracy\":[accuracy1,svc_score2,gaus_score,accuracy_knn,accuracy_dec,rf_score,acc_xgb,acc_cat],\n#\"Precision\":[]\n }\ntable=pd.DataFrame(t)\ntable",
"_____no_output_____"
]
],
[
[
"### For deployment",
"_____no_output_____"
]
],
[
[
"import pickle\n\n",
"_____no_output_____"
],
[
"X_smote=x_smote.drop(['UDI','TWF','HDF','OSF','PWF','RNF'],axis=1)\nX_smote",
"_____no_output_____"
],
[
"x_smote.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 19322 entries, 0 to 19321\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 UDI 19322 non-null int64 \n 1 Type 19322 non-null int64 \n 2 AT 19322 non-null float64\n 3 PT 19322 non-null float64\n 4 RS 19322 non-null int64 \n 5 T 19322 non-null float64\n 6 TW 19322 non-null int64 \n 7 TWF 19322 non-null int64 \n 8 HDF 19322 non-null int64 \n 9 PWF 19322 non-null int64 \n 10 OSF 19322 non-null int64 \n 11 RNF 19322 non-null int64 \ndtypes: float64(3), int64(9)\nmemory usage: 1.8 MB\n"
],
[
"model_xgb_final=XGBClassifier()\nmodel_xgb_final.fit(X_smote,y_smote)",
"/Users/lakshmipriya/opt/anaconda3/lib/python3.8/site-packages/xgboost/sklearn.py:888: UserWarning: The use of label encoder in XGBClassifier is deprecated and will be removed in a future release. To remove this warning, do the following: 1) Pass option use_label_encoder=False when constructing XGBClassifier object; and 2) Encode your labels (y) as integers starting with 0, i.e. 0, 1, 2, ..., [num_class - 1].\n warnings.warn(label_encoder_deprecation_msg, UserWarning)\n"
],
[
"pickle.dump(model_xgb_final,open('xgb1.pkl','wb'))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
eced9a5af2a4a34bfcbda270b80abe8cc8e242c3 | 840,923 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Visualizing_state_effects-v2-checkpoint.ipynb | ozaltun/PM_COVID | a952211def983ba59f688ab5af3e79decca3a6ff | [
"Apache-2.0"
] | null | null | null | .ipynb_checkpoints/Visualizing_state_effects-v2-checkpoint.ipynb | ozaltun/PM_COVID | a952211def983ba59f688ab5af3e79decca3a6ff | [
"Apache-2.0"
] | null | null | null | .ipynb_checkpoints/Visualizing_state_effects-v2-checkpoint.ipynb | ozaltun/PM_COVID | a952211def983ba59f688ab5af3e79decca3a6ff | [
"Apache-2.0"
] | null | null | null | 1,415.695286 | 452,720 | 0.955112 | [
[
[
"import seaborn as sns",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"date_of_study = \"05-19-2020\"\nsave_root = \"/Users/ozaltun/Dropbox (MIT)/Project_coal_ng_mort/Notes/Harvard paper/{}\"\ndf = pd.read_csv(\"/Users/ozaltun/Dropbox (MIT)/Data/Health/covid/\"+date_of_study+\\\n \"_estimates.csv\", index_col=0)",
"_____no_output_____"
],
[
"df.date = df.date.str.slice(start=1)\ndf['date'] = pd.to_datetime(df['date'])\n#df['date'] = df.date.dt.strftime('%Y-%m-%d')",
"_____no_output_____"
],
[
"df[\"ci_indicator\"] = np.sign(df.lb) + np.sign(df.ub)\ndf[\"ci_indicator\"] = df[\"ci_indicator\"]/2\ndf[\"ci_indicator\"] = df[\"ci_indicator\"].astype(int)\ndf.head()",
"_____no_output_____"
],
[
"df.cf_name.unique()",
"_____no_output_____"
],
[
"coefficients = df.cf_name.unique()\nstates = [i for i in coefficients if 'factor(state)' in i]\nstates_map = {i:i[13:]for i in states}\n\ncoef_subset = ['mean_pm25', 'share.drive','share.PublicTransit','share.Home','percent.smokers',\\\n 'percent.obese','percent.uninsured','percent.obese','older_pecent','pct_blk',\\\n 'log(medianhousevalue)']\ncoef_subset_map = {'mean_pm25':'PM2.5', 'share.drive':'Share - driving','share.PublicTransit':'Share - public transit',\\\n 'share.Home':'Share - home','percent.smokers':'Percent smokers','percent.obese':'Percent Obese',\\\n 'percent.uninsured':'Percent Uninsured','percent.obese':'Percent Obese',\\\n 'older_pecent':'Percent old','pct_blk':'Percent Black', 'log(medianhousevalue)':'log(medianhousevalue)'}\nother_coefs = [i for i in coefficients if (i not in states) & (i not in coef_subset)]\nother_coefs_map = {i:i for i in other_coefs}\ncoef_map = {**states_map, **coef_subset_map, **other_coefs_map}\ndf.cf_name = df.cf_name.map(coef_map)\ndf.head()",
"_____no_output_____"
],
[
"states = list(states_map.values())\ncoef_subset = list(coef_subset_map.values())\nprint(states)\nprint(coef_subset)",
"['AR', 'AZ', 'CA', 'CO', 'CT', 'DE', 'FL', 'GA', 'IA', 'ID', 'IL', 'IN', 'KS', 'KY', 'LA', 'MA', 'MD', 'ME', 'MI', 'MN', 'MO', 'MS', 'MT', 'NC', 'ND', 'NE', 'NH', 'NJ', 'NM', 'NV', 'NY', 'OH', 'OK', 'OR', 'PA', 'RI', 'SC', 'SD', 'TN', 'TX', 'UT', 'VA', 'VT', 'WA', 'WI', 'WV', 'WY']\n['PM2.5', 'Share - driving', 'Share - public transit', 'Share - home', 'Percent smokers', 'Percent Obese', 'Percent Uninsured', 'Percent old', 'Percent Black', 'log(medianhousevalue)']\n"
],
[
"dates = df.date.unique()\nselected_dates = {'Early':dates[0], 'Mid':dates[int(dates.shape[0]/2)],'Latest':dates[dates.shape[0]-1]}\nselected_dates",
"_____no_output_____"
],
[
"df_levels = df.set_index(['type','date','cf_name'])",
"_____no_output_____"
],
[
"# df_levels.loc['with_states',:,states][df_levels.ci_indicator !=0]",
"_____no_output_____"
],
[
"fig, (ax, ax1) = plt.subplots(1,2, figsize=(30,15))\n\n# All\ndf_temp = df_levels.loc['with_states',:,states].reset_index().drop(columns=['type'])\nordered_states = df_temp[df_temp.date == '2020-05-16'].sort_values(by=['est'], ascending=False).cf_name.values\nfor i, state in enumerate(ordered_states):\n df_temp_state = df_temp[df_temp.cf_name == state].sort_values(by=['date'])\n ax.plot(df_temp_state['date'], df_temp_state['est'], label=state)\nax.set_xticks(dates[::5])\nax.legend(loc='upper left')\nax.set_xlabel('Day')\nax.set_ylabel('Coefficient')\n\n# Only statistically significant\ndf_temp = df_levels.loc['with_states',:,states].reset_index().drop(columns=['type'])\ndf_temp = df_temp[df_temp.ci_indicator !=0]\nordered_states = df_temp[df_temp.date == '2020-05-16'].sort_values(by=['est'], ascending=False).cf_name.values\nfor i, state in enumerate(ordered_states):\n df_temp_state = df_temp[df_temp.cf_name == state].sort_values(by=['date'])\n ax1.plot(df_temp_state['date'], df_temp_state['est'], label=state)\nax1.set_xticks(dates[::5])\nax1.legend(loc='upper left')\nax1.set_xlabel('Day')\nax1.set_ylabel('Coefficient')\nfig.savefig(save_root.format('line_states.png'), dpi=fig.dpi)\nplt.show()",
"_____no_output_____"
],
[
"rows = 5\nfig, ax = plt.subplots(rows,2, figsize=(30,30))\n\n# All\ndf_temp = df_levels.loc['with_states',:,coef_subset].reset_index().drop(columns=['type'])\nordered_coefs = df_temp[df_temp.date == '2020-05-16'].sort_values(by=['est'], ascending=False).cf_name.values\nfor i, coef in enumerate(ordered_coefs):\n df_temp_coefs = df_temp[df_temp.cf_name == coef].sort_values(by=['date'])\n ax[i %rows, int(i/rows)].plot(df_temp_coefs['date'], df_temp_coefs['est'], label=coef)\n ax[i %rows, int(i/rows)].fill_between(df_temp_coefs['date'], df_temp_coefs['lb'], df_temp_coefs['ub'],alpha=0.2)\n ax[i %rows, int(i/rows)].axhline(y=0, color='r', linestyle='--')\n ax[i %rows, int(i/rows)].set_xticks(dates[::5])\n ax[i %rows, int(i/rows)].legend(loc='upper left')\n ax[i %rows, int(i/rows)].set_xlabel('Day')\n ax[i %rows, int(i/rows)].set_ylabel('Coefficient')\n \n \nfig.savefig(save_root.format('line_coefs.png'), dpi=fig.dpi)\nplt.show()",
"_____no_output_____"
],
[
"fig, ax1 = plt.subplots(1,1, figsize=(30,15))\n# Only statistically significant\ndf_temp = df_levels.loc['with_states',:,coef_subset].reset_index().drop(columns=['type'])\ndf_temp = df_temp[df_temp.ci_indicator !=0]\ndates_subset = df_temp.date.unique()\n# ordered_coefs = df_temp[df_temp.date == '2020-05-16'].sort_values(by=['est'], ascending=False).cf_name.values\nfor i, coef in enumerate(ordered_coefs):\n df_temp_coefs = df_temp[df_temp.cf_name == coef].sort_values(by=['date'])\n ax1.plot(df_temp_coefs['date'], df_temp_coefs['est'],'-*', label=coef)\nax1.legend(loc='upper left')\nax1.set_xticks(dates_subset[::5])\n#ax1.set_xlim([datetime.date(2020, 4, 4), datetime.date(2020, 5, 16)])\n#ax1.set_xlim([dates[0], dates[-1]])\nax1.set_xlabel('Day')\nax1.set_ylabel('Coefficient')\nfig.savefig(save_root.format('line_sig_coefs.png'), dpi=fig.dpi)\nplt.show()",
"_____no_output_____"
],
[
"date_for_bar_graph = dates[-1]\n#df_temp = df[(df.type == 'with_states') & (df.date ==date_for_bar_graph)].drop(columns=['type', 'date'])\ndf_temp = df_levels.loc['with_states',date_for_bar_graph,states].reset_index().drop(columns=['type']).sort_values(by=['est'], ascending=False)\n\nfig, ax = plt.subplots(1,1, figsize=(40,20))\nax.bar(df_temp['cf_name'], df_temp['est'], yerr=(df_temp['ub']-df_temp['lb'])/2, align='center', alpha=0.5, ecolor='black', capsize=10)\n# ax.set_ylabel('Coefficient of Thermal Expansion ($\\degree C^{-1}$)')\nax.set_xticklabels(df_temp['cf_name'], rotation=65)\n# ax.set_title('Coefficent of Thermal Expansion (CTE) of Three Metals')\n# ax.yaxis.grid(True)\nfig.savefig(save_root.format('bar_states.png'), dpi=fig.dpi)\n# Save the figure and show\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eced9dbae963e1e131e1acafa974e3452ff3d9e1 | 333,414 | ipynb | Jupyter Notebook | _notebooks/06-Scraping.ipynb | M-Sender/cmps3160 | 54546d307f913b35caa45efe6c5528dadb8055f2 | [
"MIT"
] | null | null | null | _notebooks/06-Scraping.ipynb | M-Sender/cmps3160 | 54546d307f913b35caa45efe6c5528dadb8055f2 | [
"MIT"
] | null | null | null | _notebooks/06-Scraping.ipynb | M-Sender/cmps3160 | 54546d307f913b35caa45efe6c5528dadb8055f2 | [
"MIT"
] | null | null | null | 127.842791 | 200,121 | 0.747473 | [
[
[
"# Note you may have to install requests! pip3 install requests\n\nimport requests\n# These two things are for Pandas, it widens the notebook and lets us display data easily.\nfrom IPython.core.display import display, HTML\ndisplay(HTML(\"<style>.container { width:95% !important; }</style>\"))",
"_____no_output_____"
]
],
[
[
"## Simple API Call with Requests Library\n\nIt may be good to look at the reference documentation for the [requests library](https://2.python-requests.org/en/master/user/quickstart/).\n\nFirst, let's have a look at the [GitHub API](https://developer.github.com/v3/).",
"_____no_output_____"
]
],
[
[
"r = requests.get('https://api.github.com/users/nmattei', timeout=10)\nr.status_code",
"_____no_output_____"
],
[
"r.headers['content-type']",
"_____no_output_____"
],
[
"r.url",
"_____no_output_____"
],
[
"r.content",
"_____no_output_____"
],
[
"r.json()",
"_____no_output_____"
]
],
[
[
"## Looking at HTTP Requests\n\nWe'll try to get some data from Google. Note that this is kind of against the TOS and we **should not do it this way in general -- Google has very [specific rules on their site](https://developers.google.com/custom-search/v1/).**",
"_____no_output_____"
]
],
[
[
"params = {'q':'Tulane University'}\nr = requests.get('http://www.google.com/search', params = params, timeout=10)\nr.status_code",
"_____no_output_____"
],
[
"r.url",
"_____no_output_____"
],
[
"r.headers['content-type']",
"_____no_output_____"
],
[
"r.text",
"_____no_output_____"
]
],
[
[
"## More Complicated with Parameters\n\nWe'll look for some information from the [Apple ITunes API](https://affiliate.itunes.apple.com/resources/documentation/itunes-store-web-service-search-api/).",
"_____no_output_____"
]
],
[
[
"params = {'term' : \"the+meters\"}\nr = requests.get('https://itunes.apple.com/search', params=params, timeout=10)\nr.status_code",
"_____no_output_____"
],
[
"r.url",
"_____no_output_____"
],
[
"r.json()",
"_____no_output_____"
],
[
"r.url",
"_____no_output_____"
]
],
[
[
"We can do lots of parameters in the payload like [this](https://2.python-requests.org/en/master/user/quickstart/).",
"_____no_output_____"
]
],
[
[
"params = {'term' : \"the+meters\", 'entity' : 'album'}\nr = requests.get('https://itunes.apple.com/search', params=params, timeout=10)\nr.status_code\n",
"_____no_output_____"
],
[
"r.url",
"_____no_output_____"
],
[
"x = r.json()",
"_____no_output_____"
],
[
"type(x['results'][0])",
"_____no_output_____"
]
],
[
[
"## Converting the returned JSON to an object!",
"_____no_output_____"
]
],
[
[
"import json",
"_____no_output_____"
],
[
"data = json.loads(r.content)",
"_____no_output_____"
],
[
"data.keys()",
"_____no_output_____"
],
[
"type(data['results'])",
"_____no_output_____"
],
[
"type(data['results'][1])",
"_____no_output_____"
],
[
"data['results'][1]",
"_____no_output_____"
],
[
"data['results'][1].keys()",
"_____no_output_____"
]
],
[
[
"## Using Beautiful Soup to Parse a Webpage.\n\nThe [beautifulsoup4 documentation](https://www.crummy.com/software/BeautifulSoup/).",
"_____no_output_____"
]
],
[
[
"# Grab the course webpage.\nimport requests\nfrom bs4 import BeautifulSoup\n\nr = requests.get('https://nmattei.github.io/cmps3160/schedule/')\n\nroot = BeautifulSoup( r.content )",
"_____no_output_____"
],
[
"r.content",
"_____no_output_____"
],
[
"root.find(\"table\")",
"_____no_output_____"
],
[
"root.find(\"table\").findAll(\"a\")",
"_____no_output_____"
]
],
[
[
"## Trying out some Regular Expressions.",
"_____no_output_____"
]
],
[
[
"import re\n# Find the index in the raw HTML where we first mention CMPS3160\n\n# Note we use the r to make sure special flags get used correctly.\n\nr = requests.get('https://nmattei.github.io/cmps3160/syllabus/')\nmatch = re.search(r'CMPS 3160', r.text)\nprint(match.start())",
"460\n"
],
[
"r.text[390:500]",
"_____no_output_____"
],
[
"# Does the start match?\nmatch = re.match(r'CMPS 3160', r.text)\nprint(match)",
"None\n"
],
[
"# Iterate over all occurances and print a few characters.\nfor m in re.finditer(r'CMPS 3160', r.text):\n print(r.text[m.start()-50:m.start()+50])\n",
"rel=\"alternate\" type=\"application/rss+xml\" title=\"CMPS 3160 Intro. to Data Science - Intro to Data S\n-brand\" href=\"https://nmattei.github.io/cmps3160\">CMPS 3160 Intro. to Data Science</a></div>\n\n <d\nthe case of Labs and examples, via the <a href=\"\">CMPS 3160 Github Page</a>.</p>\n </li>\n <li>\n \n"
],
[
"# Find them all.\nmatch = re.findall(r'CMPS 3160', r.text)\nprint(match)",
"['CMPS 3160', 'CMPS 3160', 'CMPS 3160']\n"
],
[
"# More complicated RegExes - Groups\nregex = r'\\s*([Uu]niversity)\\s([Oo]f)\\s(\\w{3,})'\n\ntext = ''' The university of kentucky is the best\n basketball team and an ok university. and University of North CC\n The University Of Kentucky can be put in \n some weird capitalization and University of Ken spelled wrong'''\nm = re.search( regex, text)\nprint(m.groups())",
"('university', 'of', 'kentucky')\n"
],
[
"# Find all\nprint(re.findall(regex, text))",
"[('university', 'of', 'kentucky'), ('University', 'of', 'North'), ('University', 'Of', 'Kentucky'), ('University', 'of', 'Ken')]\n"
],
[
"# Named Groups.\nregex = r'\\s*([Uu]niversity)\\s([Oo]f)\\s(?P<school>\\w{3,})'\ntext = ''' The university of kentucky is the best University of Lousiana\n basketball team and an ok university.\n The University Of Kentucky can be put in \n some weird capitalization'''\nm = re.search( regex, text)\nprint(m.groupdict())\n",
"_____no_output_____"
],
[
"# Find all named groups\n\n# Named Groups.\nregex = r'\\s*([Uu]niversity)\\s([Oo]f)\\s(?P<school>\\w{3,})'\ntext = ''' The university of kentucky is the best\n basketball team and an ok university.\n The University Of Kentucky can be put in \n some weird capitalization. And Kentucky is much better than\n the University of Mississippi.'''\nfor m in re.finditer(regex, text):\n print(m.groupdict())\n",
"_____no_output_____"
],
[
"'abcabcabc'.replace('a', 'X')",
"_____no_output_____"
],
[
"text = 'I love Introduction to Data Science'\nre.sub(r'Data Science', r'Schmada Schmience', text) ",
"_____no_output_____"
],
[
"re.sub(r'(\\w+)\\s([Ss]cience)', r'\\2 \\1hmience', text) \n",
"_____no_output_____"
]
],
[
[
"## Downloadning All the ... PDFs from the course website.\n\nUsing beautiful soup and some regular expressions.",
"_____no_output_____"
]
],
[
[
"import re\nimport requests\nfrom bs4 import BeautifulSoup\nfrom urllib.parse import urlparse\nfrom urllib.parse import urljoin\nimport os\nimport pathlib",
"_____no_output_____"
],
[
"# HTTP GET request sent to the URL url\n# We're going to use last year's website as it's got direct links...\nr = requests.get( \"https://nmattei.github.io/cmps3160/schedule/\" )\n\n# Use BeautifulSoup to parse the GET response\nroot = BeautifulSoup( r.content )\nlnks = root.find(\"table\").findAll(\"a\")\nlnks",
"_____no_output_____"
]
],
[
[
"Let's do the easier one first and download all the `.ipynb` from the webpage. We'll get into why this is easier in a second...",
"_____no_output_____"
]
],
[
[
"# Cycle through the href for each anchor, checking\n# to see if it's an ipynb link or not\nnotebooks = []\nfor lnk in lnks:\n href = lnk['href']\n # If it's a PDF/PPTX link, queue a download \n if href.lower().endswith(('ipynb')):\n notebooks.append(href)\n print(\"{} is a Link to {}\".format(lnk.contents,lnk['href']))\nprint(notebooks)",
"_____no_output_____"
],
[
"# Download all the files to whatever you're running notebook from.\n\n# Be careful for href!\n\nfor i, href in enumerate(notebooks):\n print(\"Downloading... {}\".format(href))\n rd = requests.get(href, stream=True)\n \n # Write the downloaded object to a file -- first we should make a directory for it..\n outputdir = os.path.join(os.getcwd(), \"downloaded\")\n os.makedirs(outputdir, exist_ok=True)\n \n # Note because the href is a path we have to just get the filename!\n outfile = os.path.join(outputdir, href.split(\"/\")[-1])\n print(\"Writing: \",outfile)\n with open(outfile, 'wb') as f:\n f.write(rd.content)\n",
"_____no_output_____"
]
],
[
[
"Let's do this more complicated and try to grab all the PDF's...\n\nFirst thing to note is that the PDFs have it in the name but not the target and they're hosted on GOOGLE! -- so this doesn't really work :-(\n",
"_____no_output_____"
]
],
[
[
"# We can go check, we get a google drive directory...\n\nr = requests.get( \"https://drive.google.com/drive/u/1/folders/1uGrhWzhXbiqoChTK0fQXg340X319REks\" )\n\n# Use BeautifulSoup to parse the GET response\nroot = BeautifulSoup( r.content )\n#lnks = root.find(\"table\").findAll(\"a\")\n#lnks\nroot.content\n\n",
"_____no_output_____"
],
[
"# We have all google links so we need to check the tags to see if they contain PDF!\npdfs = []\nfor lnk in lnks:\n if 'pdf' in lnk.contents[0].lower():\n print(\"{} is a PDF Link to {}\".format(lnk.contents,lnk['href']))\n pdfs.append(lnk['href'])\nprint(pdfs)",
"_____no_output_____"
],
[
"# Note that google doens't make this easy... sorry, you have to do a little kung fu...\n# Format is: https://drive.google.com/u/1/uc?id=ID&export=download\ndownload_links = []\nfor c in pdfs:\n fid = c.split(\"/\")[-2]\n download_links.append(\"https://drive.google.com/u/1/uc?id={}&export=download\".format(fid))\nprint(download_links)\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecedbffac0dfef5f4598a4ef312637e8314053b5 | 33,745 | ipynb | Jupyter Notebook | misc/SL_construction.ipynb | weian312/ssd_detectors | 0eb88f06eb94b6c4a5f0058e55116a331b128da8 | [
"MIT"
] | 316 | 2018-06-11T07:29:01.000Z | 2022-02-12T12:26:26.000Z | misc/SL_construction.ipynb | weian312/ssd_detectors | 0eb88f06eb94b6c4a5f0058e55116a331b128da8 | [
"MIT"
] | 60 | 2018-08-25T23:11:14.000Z | 2021-08-30T16:57:55.000Z | misc/SL_construction.ipynb | weian312/ssd_detectors | 0eb88f06eb94b6c4a5f0058e55116a331b128da8 | [
"MIT"
] | 97 | 2018-08-22T12:26:12.000Z | 2021-12-03T04:25:29.000Z | 129.291188 | 27,024 | 0.834375 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# inter layer neighbors\n\nmap_w = 4; map_h = 6\n\nxy_pos = np.asanyarray(np.meshgrid(np.arange(map_w), np.arange(map_h))).reshape(2,-1).T\nxy = np.tile(xy_pos, (1,8))\nxy += np.array([-1,-1, 0,-1, +1,-1, \n -1, 0, +1, 0, \n -1,+1, 0,+1, +1,+1])\nvalide = (xy[:,0::2] >= 0) & (xy[:,0::2] < map_w) & (xy[:,1::2] >= 0) & (xy[:,1::2] < map_h)\nidxs = xy[:,1::2] * map_w + xy[:,0::2]\n\nfor i in [0, 10, -1]:\n print(idxs[i][valide[i]])\n f = np.zeros(map_w*map_h)\n for j in idxs[i][valide[i]]:\n f[j] = 1\n print(f.reshape((map_h,map_w)))",
"[1 4 5]\n[[ 0. 1. 0. 0.]\n [ 1. 1. 0. 0.]\n [ 0. 0. 0. 0.]\n [ 0. 0. 0. 0.]\n [ 0. 0. 0. 0.]\n [ 0. 0. 0. 0.]]\n[ 5 6 7 9 11 13 14 15]\n[[ 0. 0. 0. 0.]\n [ 0. 1. 1. 1.]\n [ 0. 1. 0. 1.]\n [ 0. 1. 1. 1.]\n [ 0. 0. 0. 0.]\n [ 0. 0. 0. 0.]]\n[18 19 22]\n[[ 0. 0. 0. 0.]\n [ 0. 0. 0. 0.]\n [ 0. 0. 0. 0.]\n [ 0. 0. 0. 0.]\n [ 0. 0. 1. 1.]\n [ 0. 0. 1. 0.]]\n"
],
[
"# cross layer neighbors\n\nmap_w_p = map_w * 2\nmap_h_p = map_h * 2\nxy = np.tile(xy_pos, (1,4)) * 2\nxy += np.array([0,0, 1,0,\n 0,1, 1,1])\nvalide = (xy[:,0::2] >= 0) & (xy[:,0::2] < map_w_p) & (xy[:,1::2] >= 0) & (xy[:,1::2] < map_h_p)\nidxs = xy[:,1::2] * map_w_p + xy[:,0::2]\n\nfor i in [0, 10, -1]:\n print(idxs[i][valide[i]])\n f = np.zeros(map_w_p*map_h_p)\n for j in idxs[i][valide[i]]:\n f[j] = 1\n print(f.reshape((map_h_p,map_w_p)))",
"[0 1 8 9]\n[[ 1. 1. 0. 0. 0. 0. 0. 0.]\n [ 1. 1. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]]\n[36 37 44 45]\n[[ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 1. 1. 0. 0.]\n [ 0. 0. 0. 0. 1. 1. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]]\n[86 87 94 95]\n[[ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 1. 1.]\n [ 0. 0. 0. 0. 0. 0. 1. 1.]]\n"
],
[
"# project point on line\n\np = np.random.randn(2)\nb = np.random.random()*2\na = np.random.random()*2\n\nx_p, y_p = p\n\n#y_p = a_p * x_p + b_p\na_p = -1/a\nb_p = y_p - a_p * x_p\nx_s = (b_p - b) / (a - a_p)\ny_s = a * x_s + b\n\nx_s = (y_p + 1/a * x_p - b) / (a + 1/a)\ny_s = a * x_s + b\n\nprint('a = %4.2f b = %4.2f a_p = %4.2f b_p = %4.2f ' % (a, b, a_p, b_p))\n\nplt.figure(figsize=[8]*2)\n\nplt.plot(x_p, y_p, 'or')\nx = np.array([-2,2])\nplt.plot(x, a*x + b)\nplt.plot(x, a_p*x + b_p)\nplt.plot(x_s, y_s, 'ob')\n\nplt.grid()\nplt.ylim([-2,2])\nplt.xlim([-2,2])\nplt.axes().set_aspect('equal', 'datalim')\nplt.show()",
"a = 1.63 b = 1.68 a_p = -0.61 b_p = 0.53 \n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
ecedcad6cd16b3aaf947a38845aeabc184291392 | 575,870 | ipynb | Jupyter Notebook | docs/manual.ipynb | dcherian/tracpy | 222db3e69f792dbc2732350bc7d266441d7b7a73 | [
"MIT"
] | null | null | null | docs/manual.ipynb | dcherian/tracpy | 222db3e69f792dbc2732350bc7d266441d7b7a73 | [
"MIT"
] | null | null | null | docs/manual.ipynb | dcherian/tracpy | 222db3e69f792dbc2732350bc7d266441d7b7a73 | [
"MIT"
] | null | null | null | 992.87931 | 317,773 | 0.940398 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecedcb7c858b86936d7c5c21adf18c6e920abf82 | 2,835 | ipynb | Jupyter Notebook | Lesson01/Activity01/Activity01-HandlingList.ipynb | RileyAfencl/DSC-540 | dffdabf11116aef2c25ac0ae1570594833734eca | [
"MIT"
] | 77 | 2018-09-26T05:50:24.000Z | 2022-03-21T08:25:30.000Z | Lesson01/Activity01/Activity01-HandlingList.ipynb | RileyAfencl/DSC-540 | dffdabf11116aef2c25ac0ae1570594833734eca | [
"MIT"
] | 2 | 2020-03-22T21:15:43.000Z | 2021-05-07T06:52:36.000Z | Lesson01/Activity01/Activity01-HandlingList.ipynb | RileyAfencl/DSC-540 | dffdabf11116aef2c25ac0ae1570594833734eca | [
"MIT"
] | 182 | 2018-10-16T05:55:06.000Z | 2022-03-26T21:47:26.000Z | 25.540541 | 263 | 0.588007 | [
[
[
"# Create a list of random numbers and then create another list from this one whose elements are divisible by three. Also repeat the experiment few times (at least three times) and calculate the arithimetic mean of the differenence of length of the two lists",
"_____no_output_____"
],
[
"### Task-1\n\nCreate a list of random numbers (at least 100 in length but we encourage you to play with the length)\n\n__Pay attention so that this list has as less number of duplicates as possible__",
"_____no_output_____"
]
],
[
[
"### Write your code here below this comment",
"_____no_output_____"
]
],
[
[
"### Task-2\n\nWrite a list comprehension to generate a second list from the one you just created. The condition of membership in the second list is divisibility by 3.",
"_____no_output_____"
]
],
[
[
"### Write your code bellow this comment",
"_____no_output_____"
]
],
[
[
"### Task-3\n\n- Use the `len` function to measure the length of the first list and the second list\n- Store both in two different variables\n- Calculate the difference of length between them",
"_____no_output_____"
]
],
[
[
"### Write your code below this comment",
"_____no_output_____"
]
],
[
[
"### Task-4\n\n- Pack `Task-2` and `Task-3` in a single while loop and perform them few times in such a way that at the end you have a list with difference of length\n- End the while loop when desired number of experiments are finished (at least three, please feel free to do more)\n- Calculate the arithmetic mean (common average) on the difference of length that you have. (How to sum all values of a list?)",
"_____no_output_____"
]
],
[
[
"### Write your code below this comment.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecedcd10e118f464f7ff46a4962503ad6ecdbff8 | 36,547 | ipynb | Jupyter Notebook | notebooks/twitter-analysis-dev.ipynb | jacob-heglund/socialsensing-jh | fd6d2d749f40fee46bee749ff868212bf117a747 | [
"BSD-2-Clause",
"MIT"
] | null | null | null | notebooks/twitter-analysis-dev.ipynb | jacob-heglund/socialsensing-jh | fd6d2d749f40fee46bee749ff868212bf117a747 | [
"BSD-2-Clause",
"MIT"
] | null | null | null | notebooks/twitter-analysis-dev.ipynb | jacob-heglund/socialsensing-jh | fd6d2d749f40fee46bee749ff868212bf117a747 | [
"BSD-2-Clause",
"MIT"
] | null | null | null | 44.138889 | 3,220 | 0.557665 | [
[
[
"# Development Notebook for Analyzing Sandy Twitter Data (from mdredze).",
"_____no_output_____"
]
],
[
[
"import sys\nimport os\nsys.path.append(os.path.abspath('../'))\n\nimport datetime as dt\nimport json\nimport numpy as np\nimport pandas as pd\nimport pymongo\nfrom twitterinfrastructure.tools import dump, output\n\nimport twitterinfrastructure.twitter_sandy as ts\nimport importlib\nimportlib.reload(ts)\n\n#os.chdir('../')\nprint(os.getcwd())",
"/Users/httran/Documents/projects/twitterinfrastructure\n"
]
],
[
[
"## Test pre-processing tweets.",
"_____no_output_____"
]
],
[
[
"# test tweet field names\ncollection = 'tweets_analysis'\ntweet_collection = 'tweets'\nzones_collection = 'taxi_zones'\nfields = ['_id', 'coordinates', 'created_at', 'entities', 'full_text', \n 'id_str', 'place']\ndb_name = 'sandy'\ndb_instance = 'mongodb://localhost:27017/'\n\nclient = pymongo.MongoClient(db_instance)\ndb = client[db_name]\n\nzones_iter = db[zones_collection].find()\nfor zone in zones_iter[0:1]:\n # query tweets within current taxi zone\n query_dict = {\n \"coordinates\": {\n \"$geoWithin\": {\n \"$geometry\": zone['geometry']\n }\n }\n }\n full_tweets = db[tweet_collection].find(query_dict)\n \nprint(full_tweets.count())\nfull_tweets = list(full_tweets)",
"1146\n"
],
[
"for full_tweet in full_tweets[0:5]:\n print(full_tweet.keys())\n print(full_tweet['entities']['hashtags'])\n print(full_tweet['text'])\n print('')",
"dict_keys(['_id', 'contributors', 'coordinates', 'created_at', 'entities', 'favorite_count', 'favorited', 'geo', 'id', 'id_str', 'in_reply_to_screen_name', 'in_reply_to_status_id', 'in_reply_to_status_id_str', 'in_reply_to_user_id', 'in_reply_to_user_id_str', 'is_quote_status', 'lang', 'place', 'retweet_count', 'retweeted', 'source', 'text', 'truncated', 'user'])\n[{'indices': [93, 109], 'text': 'reyesenterprise'}, {'indices': [110, 127], 'text': 'workhardplayhard'}]\nWith my dad Going to delaware with 10 of my dads trucks and 1 crane for emergency generators #reyesenterprise #workhardplayhard\n\ndict_keys(['_id', 'contributors', 'coordinates', 'created_at', 'entities', 'extended_entities', 'favorite_count', 'favorited', 'geo', 'id', 'id_str', 'in_reply_to_screen_name', 'in_reply_to_status_id', 'in_reply_to_status_id_str', 'in_reply_to_user_id', 'in_reply_to_user_id_str', 'is_quote_status', 'lang', 'place', 'possibly_sensitive', 'retweet_count', 'retweeted', 'source', 'text', 'truncated', 'user'])\n[]\nJust landed in Newark, next stop Berlin then on to Romania. Getting excited but could do without the 10.5 hour flight http://t.co/NMwHniu4\n\ndict_keys(['_id', 'contributors', 'coordinates', 'created_at', 'entities', 'favorite_count', 'favorited', 'geo', 'id', 'id_str', 'in_reply_to_screen_name', 'in_reply_to_status_id', 'in_reply_to_status_id_str', 'in_reply_to_user_id', 'in_reply_to_user_id_str', 'is_quote_status', 'lang', 'place', 'retweet_count', 'retweeted', 'source', 'text', 'truncated', 'user'])\n[{'indices': [3, 22], 'text': 'atlanticexpressbus'}]\nIn #atlanticexpressbus 315 inbound to NYC via Lincoln Tunnel. The driver is really aggressive in driving today!\n\ndict_keys(['_id', 'contributors', 'coordinates', 'created_at', 'entities', 'favorite_count', 'favorited', 'geo', 'id', 'id_str', 'in_reply_to_screen_name', 'in_reply_to_status_id', 'in_reply_to_status_id_str', 'in_reply_to_user_id', 'in_reply_to_user_id_str', 'is_quote_status', 'lang', 'place', 'retweet_count', 'retweeted', 'source', 'text', 'truncated', 'user'])\n[]\nI took today off, I won't be cutting!\n\ndict_keys(['_id', 'contributors', 'coordinates', 'created_at', 'entities', 'favorite_count', 'favorited', 'geo', 'id', 'id_str', 'in_reply_to_screen_name', 'in_reply_to_status_id', 'in_reply_to_status_id_str', 'in_reply_to_user_id', 'in_reply_to_user_id_str', 'is_quote_status', 'lang', 'place', 'retweet_count', 'retweeted', 'source', 'text', 'truncated', 'user'])\n[{'indices': [61, 76], 'text': 'hungryandtired'}]\nLanded at newark! Time to breath in that jersey air....mmmmm #hungryandtired\n\n"
],
[
"# test duplicate key error\n# due to tweets found in multiple zones?\ncollection = 'tweets_analysis'\ntweet_collection = 'tweets'\nzones_collection = 'taxi_zones'\nfields = ['_id', 'coordinates', 'created_at', 'entities', 'full_text', \n 'id_str', 'place']\ndb_name = 'sandy'\ndb_instance = 'mongodb://localhost:27017/'\n\nclient = pymongo.MongoClient(db_instance)\ndb = client[db_name]\n\ntweet = db[tweet_collection].find_one({'id_str': '263867240783425536'})\nprint(tweet)",
"{'_id': ObjectId('5b10b7000156950952b399d9'), 'contributors': None, 'coordinates': {'coordinates': [-73.95361427, 40.68187311], 'type': 'Point'}, 'created_at': 'Thu Nov 01 04:57:07 +0000 2012', 'entities': {'hashtags': [], 'symbols': [], 'urls': [], 'user_mentions': [{'id': 383956775, 'id_str': '383956775', 'indices': [0, 11], 'name': 'kash', 'screen_name': 'iamkashema'}]}, 'favorite_count': 0, 'favorited': False, 'geo': {'coordinates': [40.68187311, -73.95361427], 'type': 'Point'}, 'id': 263867240783425540, 'id_str': '263867240783425536', 'in_reply_to_screen_name': 'iamkashema', 'in_reply_to_status_id': None, 'in_reply_to_status_id_str': None, 'in_reply_to_user_id': 383956775, 'in_reply_to_user_id_str': '383956775', 'is_quote_status': False, 'lang': 'en', 'place': {'attributes': {}, 'bounding_box': {'coordinates': [[[-74.255641, 40.495865], [-73.699793, 40.495865], [-73.699793, 40.91533], [-74.255641, 40.91533]]], 'type': 'Polygon'}, 'contained_within': [], 'country': 'United States', 'country_code': 'US', 'full_name': 'New York, NY', 'id': '27485069891a7938', 'name': 'New York', 'place_type': 'admin', 'url': 'https://api.twitter.com/1.1/geo/id/27485069891a7938.json'}, 'retweet_count': 0, 'retweeted': False, 'source': '<a href=\"http://twitter.com/download/iphone\" rel=\"nofollow\">Twitter for iPhone</a>', 'text': \"@iamkashema I'm mad u didn't ring down my phone to find out. People, including my drivers was callin me all day for that. I hope u got thru\", 'truncated': False, 'user': {'contributors_enabled': False, 'created_at': 'Mon Mar 07 18:50:21 +0000 2011', 'default_profile': False, 'default_profile_image': False, 'description': 'IG: instadread SC:instadread', 'entities': {'description': {'urls': []}, 'url': {'urls': [{'display_url': 'facebook.com/ClydeAutoDealer', 'expanded_url': 'http://www.facebook.com/ClydeAutoDealer', 'indices': [0, 23], 'url': 'https://t.co/q9VdOBRRDh'}]}}, 'favourites_count': 219, 'follow_request_sent': False, 'followers_count': 290, 'following': False, 'friends_count': 588, 'geo_enabled': True, 'has_extended_profile': True, 'id': 262276763, 'id_str': '262276763', 'is_translation_enabled': False, 'is_translator': False, 'lang': 'en', 'listed_count': 6, 'location': 'East Coast', 'name': 'L Express', 'notifications': False, 'profile_background_color': '1A1B1F', 'profile_background_image_url': 'http://abs.twimg.com/images/themes/theme9/bg.gif', 'profile_background_image_url_https': 'https://abs.twimg.com/images/themes/theme9/bg.gif', 'profile_background_tile': False, 'profile_banner_url': 'https://pbs.twimg.com/profile_banners/262276763/1442383011', 'profile_image_url': 'http://pbs.twimg.com/profile_images/824637001131556864/3pNbHCUv_normal.jpg', 'profile_image_url_https': 'https://pbs.twimg.com/profile_images/824637001131556864/3pNbHCUv_normal.jpg', 'profile_link_color': '2FC2EF', 'profile_sidebar_border_color': '181A1E', 'profile_sidebar_fill_color': '252429', 'profile_text_color': '666666', 'profile_use_background_image': True, 'protected': False, 'screen_name': 'RideMyTaxi', 'statuses_count': 8243, 'time_zone': 'Quito', 'translator_type': 'none', 'url': 'https://t.co/q9VdOBRRDh', 'utc_offset': -18000, 'verified': False}}\n"
],
[
"# test duplicate key error\n# query zones intersecting with current tweet (matches 2)\nquery_dict = {\n \"geometry\": {\n \"$geoIntersects\": {\n \"$geometry\": {\n \"type\": \"Point\",\n \"coordinates\": tweet['coordinates']['coordinates'] \n }\n }\n }\n}\nzones = db[zones_collection].find(query_dict)\nprint(zones.count())\nfor zone in zones:\n print(zone['properties']['zone'])",
"2\nClinton Hill\nBedford\n"
],
[
"# test mongodb and pymongo datetime conversions\n# mongodb stores as UTC, but will display in MongoDB Compass based on current\n# timezone\nanalysis_collection = 'tweets_sandy'\ndb_name = 'sandy'\ndb_instance = 'mongodb://localhost:27017/'\nclient = pymongo.MongoClient(db_instance)\ndb = client[db_name]\n\ntweet = db[analysis_collection].find_one()\nutc_time = dt.datetime.strptime(tweet['created_at'], '%a %b %d %H:%M:%S %z %Y')\nprint('tweet[created_at]: ' + tweet['created_at'])\nprint('converted to utc_time:')\nprint(utc_time)\n\ndoc = {}\ndoc['datetimeUTC'] = utc_time\ndoc['timestampUNIX'] = int(\n utc_time.replace(tzinfo=dt.timezone.utc).timestamp() * 1000)\n#doc['date'] = utc_time.date()\n#doc['hour'] = utc_time.hour\nprint('document to be inserted:')\nprint(doc)\nprint('')\n\ncollection = 'datetime_test'\ndb.drop_collection(collection)\ndb[collection].insert_one(doc)\nresult1 = db[collection].aggregate([\n {\n \"$group\": {\n \"_id\": {\n \"month\": {\"$month\": \"$datetimeUTC\"},\n \"day\": {\"$dayOfMonth\": \"$datetimeUTC\"},\n \"year\": {\"$year\": \"$datetimeUTC\"},\n \"hour\": {\"$hour\": \"$datetimeUTC\"}\n },\n \"count\": {\"$sum\": 1}\n }\n }\n])\nprint('aggregation retrieved from collection:')\nprint(list(result1))",
"tweet[created_at]: Sun Oct 28 22:42:35 +0000 2012\nconverted to utc_time:\n2012-10-28 22:42:35+00:00\ndocument to be inserted:\n{'datetimeUTC': datetime.datetime(2012, 10, 28, 22, 42, 35, tzinfo=datetime.timezone.utc), 'timestampUNIX': 1351464155000}\n\naggregation retrieved from collection:\n[{'_id': {'month': 10, 'day': 28, 'year': 2012, 'hour': 22}, 'count': 1}]\n"
],
[
"# test create_analysis\nimport twitterinfrastructure.twitter_sandy as ts\nimport importlib\nimportlib.reload(ts)\n\ncollection = 'tweets_analysis'\ntweet_collection = 'tweets'\nnyisozones_collection = 'nyiso_zones'\ntaxizones_collection = 'taxi_zones'\nfields = ['_id', 'coordinates', 'created_at', 'entities', 'text', \n 'id_str', 'place']\ndb_name = 'sandy'\ndb_instance = 'mongodb://localhost:27017/'\nprogressbar = False\noverwrite = True\nverbose = 1\n\n# import_num, tokens, full_tweets = ts.create_analysis(collection=collection, \n# tweet_collection=tweet_collection, \n# nyisozones_collection=nyisozones_collection,\n# taxizones_collection=taxizones_collection,\n# fields=fields, db_name=db_name, \n# db_instance=db_instance, \n# progressbar=False, \n# overwrite=overwrite, verbose=verbose)\n\n# connect to db (creates if not exists)\nclient = pymongo.MongoClient(db_instance)\ndb = client[db_name]\n\n# ensure that nyisozones_collection and taxizones_collection exist\ncollections = db.collection_names()\nif (nyisozones_collection not in collections) or \\\n (taxizones_collection not in collections):\n output('{nyiso} or {taxi} collection not in database. No action '\n 'taken.'.format(nyiso=nyisozones_collection,\n taxi=taxizones_collection))\n\n# process and insert tweets\nfull_tweets = db[tweet_collection].find()\nif progressbar:\n tweets_iter = tqdm(full_tweets, total=full_tweets.count(),\n desc='tweets', leave=False)\nelse:\n tweets_iter = full_tweets\nfull_tweet = tweets_iter[0]\n# for full_tweet in tweets_iter:\n# remove extra fields\nif fields:\n tweet = {field: full_tweet[field] for field in fields}\nelse:\n tweet = full_tweet\n\n# identify and add nyiso zone, taxi zone, and taxi borough\nif tweet['coordinates'] is not None:\n query_dict = {\n \"geometry\": {\n \"$geoIntersects\": {\n \"$geometry\": tweet['coordinates']\n }\n }\n }\n nyiso_zone = db[nyisozones_collection].find_one(query_dict)\n if nyiso_zone:\n tweet['nyiso_zone'] = nyiso_zone['properties']['Zone']\n else:\n tweet['nyiso_zone'] = np.nan\n\n query_dict = {\n \"geometry\": {\n \"$geoIntersects\": {\n \"$geometry\": tweet['coordinates']\n }\n }\n }\n taxi_zone = db[taxizones_collection].find_one(query_dict)\n if taxi_zone:\n tweet['location_id'] = taxi_zone['properties']['LocationID']\n tweet['borough'] = taxi_zone['properties']['borough']\n else:\n tweet['location_id'] = np.nan\n tweet['borough'] = np.nan\nelse:\n # fails.append(tweet['id_str'])\n if verbose >= 2:\n output('Tweet skipped due to missing coordinates',\n 'create_analysis')\n\n",
"_____no_output_____"
],
[
"# query analysis collection\nclient = pymongo.MongoClient(db_instance)\ndb = client[db_name]\ntweets_analysis = db[collection].find()\nfor tweet in tweets_analysis:\n print(tweet['full_text'])\n print(tweet['tokens'])\n print(tweet['timestamp'])\n print('')\n \nprint(tokens)",
"@NOT_savinHOES Not r yu upp\n['r', 'yu', 'upp']\n1350882000.0\n\nWho's up?\n[\"who'\"]\n1350882000.0\n\n@augustushazel idk I'm just ugly or annoying or something\n['idk', \"i'm\", 'ugli', 'annoy', 'someth']\n1350882000.0\n\n@InYurMomsCovers your cool thou. You seem like one of those girls that you can talk about anything to. Not too many are like that\n['cool', 'thou', 'seem', 'like', 'one', 'girl', 'talk', 'anyth', 'mani', 'like']\n1350882001.0\n\n\"I suppose she has an appropriate costume for every activity...\" #ilovemaggiesmith #downtonseasonthree\n['suppos', 'appropri', 'costum', 'everi', 'activ', '...', '#ilovemaggiesmith', '#downtonseasonthre']\n1350882000.0\n\nHit and Run is so sad..\n['hit', 'run', 'sad', '..']\n1350882000.0\n\n10/22 @ 01:00 - Temperature 46.6F. Wind 1.5mph SW. Gust 3.8mph. Barometer 30.06in, Steady. Rain today 0.00in. Humidity 92%.\n['10/22', '01:00', 'temperatur', '46.6', 'F', 'wind', '1.5', 'mph', 'SW', 'gust', '3.8', 'mph', 'baromet', '30.06', 'steadi', 'rain', 'today', '0.00', 'humid', '92']\n1350882001.0\n\nA bitch nigga, that's that shit I don't like, nah\nSneak dissers, that's that shit I don't like\n['bitch', 'nigga', \"that'\", 'shit', 'like', 'nah', 'sneak', 'disser', \"that'\", 'shit', 'like']\n1350882001.0\n\n{'steadi', '1.5', 'wind', 'one', 'thou', '..', 'SW', 'hit', '...', '01:00', 'temperatur', 'sad', '30.06', 'cool', 'nigga', 'upp', 'mani', 'girl', 'bitch', 'costum', '92', \"that'\", 'humid', \"i'm\", 'everi', 'talk', 'annoy', '#downtonseasonthre', 'today', 'seem', 'shit', 'like', '#ilovemaggiesmith', 'baromet', 'appropri', 'nah', 'sneak', \"who'\", 'someth', 'F', 'activ', 'ugli', 'anyth', '10/22', '0.00', 'r', 'rain', 'run', 'gust', 'suppos', 'disser', 'mph', 'idk', '3.8', 'yu', '46.6'}\n"
],
[
"# test dump\ndump(['123', '456'], func_name='test')",
"_____no_output_____"
]
],
[
[
"## Test tweet keyword filters.",
"_____no_output_____"
]
],
[
[
"# query tweets containing specified tokens\ndb_instance = 'mongodb://localhost:27017/'\ndb_name = 'sandy_test'\ncollection = 'tweets_analysis_test'\n\ntokens = ['gust', 'humid', 'idk']\ntweets = ts.query_keyword(tokens=tokens, \n collection=collection, db_name=db_name, \n db_instance=db_instance, verbose=1)\n\nfor tweet in tweets:\n print(tweet['id_str'])\n print(tweet['full_text'])\n print(tweet['coordinates'])\n print(tweet.keys())\n print('')",
"2018-06-01 09:06:44 : Started query.\n\n2018-06-01 09:06:44 : Finished query. Returned 2 tweets.\n\n260244089985957888\n@augustushazel idk I'm just ugly or annoying or something\n{'coordinates': [-80.08961896, 42.09464892], 'type': 'Point'}\ndict_keys(['_id', 'coordinates', 'created_at', 'entities', 'full_text', 'id_str', 'place', 'location_id', 'borough', 'tokens'])\n\n260244093119102977\n10/22 @ 01:00 - Temperature 46.6F. Wind 1.5mph SW. Gust 3.8mph. Barometer 30.06in, Steady. Rain today 0.00in. Humidity 92%.\n{'coordinates': [-75.68444444, 39.695], 'type': 'Point'}\ndict_keys(['_id', 'coordinates', 'created_at', 'entities', 'full_text', 'id_str', 'place', 'location_id', 'borough', 'tokens'])\n\n"
],
[
"# query tweets containing specified hashtags\ndb_instance = 'mongodb://localhost:27017/'\ndb_name = 'sandy_test'\ncollection = 'tweets_analysis_test'\n\nhashtags = ['downtonseasonthree', 'sandy']\ntweets = ts.query_keyword(hashtags=hashtags, \n collection=collection, db_name=db_name, \n db_instance=db_instance, verbose=1)\n\nfor tweet in tweets:\n print(tweet['full_text'])\n print(tweet['entities']['hashtags'])\n print('')",
"2018-05-30 16:27:12 : Started query.\n\n2018-05-30 16:27:12 : Finished query. Returned 1 tweets.\n\n\"I suppose she has an appropriate costume for every activity...\" #ilovemaggiesmith #downtonseasonthree\n[{'indices': [65, 82], 'text': 'ilovemaggiesmith'}, {'indices': [83, 102], 'text': 'downtonseasonthree'}]\n\n"
],
[
"# query tweets containing specified tokens or hashtags\ndb_instance = 'mongodb://localhost:27017/'\ndb_name = 'sandy_test'\ncollection = 'tweets_analysis_test'\n\ntokens = ['gust', 'humid', 'idk']\nhashtags = ['test', 'downtonseasonthree']\ntweets = ts.query_keyword(tokens=tokens, hashtags=hashtags, \n collection=collection, db_name=db_name, \n db_instance=db_instance, verbose=1)\n\nfor tweet in tweets:\n print(tweet['full_text'])\n print('')",
"2018-05-30 16:27:33 : Started query.\n\n2018-05-30 16:27:33 : Finished query. Returned 3 tweets.\n\n@augustushazel idk I'm just ugly or annoying or something\n\n\"I suppose she has an appropriate costume for every activity...\" #ilovemaggiesmith #downtonseasonthree\n\n10/22 @ 01:00 - Temperature 46.6F. Wind 1.5mph SW. Gust 3.8mph. Barometer 30.06in, Steady. Rain today 0.00in. Humidity 92%.\n\n"
]
],
[
[
"## Test group by summaries",
"_____no_output_____"
]
],
[
[
"# test borough_day summary\n# can check datetime tz by finding first tweet in tweets_sandy\ntitle = 'sandy_test'\nanalysis_collection = 'tweets_sandy'\ndb_name = 'sandy'\ndb_instance = 'mongodb://localhost:27017/'\n\nclient = pymongo.MongoClient(db_instance)\ndb = client[db_name]\n\npipeline = [\n {\n \"$group\": {\n \"_id\": {\n \"year\": {\"$year\": \"$datetimeUTC\"},\n \"month\": {\"$month\": \"$datetimeUTC\"},\n \"day\": {\"$dayOfMonth\": \"$datetimeUTC\"},\n \"hour\": {\"$hour\": \"$datetimeUTC\"},\n \"borough\": \"$borough\"\n },\n \"datetimeUTC\": {\"$min\": {\"$dateFromParts\": {\n \"year\": {\"$year\": \"$datetimeUTC\"},\n \"month\": {\"$month\": \"$datetimeUTC\"},\n \"day\": {\"$dayOfMonth\": \"$datetimeUTC\"},\n \"hour\": {\"$hour\": \"$datetimeUTC\"}\n }}},\n \"borough\": {\"$first\": \"$borough\"},\n \"count\": {\"$sum\": 1}\n }\n },\n {\"$sort\": {\"datetimeUTC\": 1, \"_id.borough\": 1}}\n]\ngroups = list(db[analysis_collection].aggregate(pipeline))\n\n# convert to dataframe\n# groups = json.loads(df.to_json(orient='records', date_format='iso'))\ndf = pd.DataFrame(groups)\ndf['datetimeUTC'] = [datetime.tz_localize(tz='UTC') for datetime \n in df['datetimeUTC']]\nprint(df.head())\n\n# groups = list(db[analysis_collection].aggregate(pipeline))",
" _id borough count \\\n0 {'year': 2012, 'month': 10, 'day': 22, 'hour':... Brooklyn 1 \n1 {'year': 2012, 'month': 10, 'day': 22, 'hour':... Brooklyn 1 \n2 {'year': 2012, 'month': 10, 'day': 22, 'hour':... Manhattan 1 \n3 {'year': 2012, 'month': 10, 'day': 22, 'hour':... Manhattan 1 \n4 {'year': 2012, 'month': 10, 'day': 22, 'hour':... Queens 1 \n\n datetimeUTC \n0 2012-10-22 13:00:00+00:00 \n1 2012-10-22 15:00:00+00:00 \n2 2012-10-22 15:00:00+00:00 \n3 2012-10-22 16:00:00+00:00 \n4 2012-10-22 23:00:00+00:00 \n"
],
[
"# test datetime group summaries with pandas and tzone conversion\n\ntweet_count_filter = 5 # by day\n# startdate = pd.Timestamp(2012, 10, 28, 0, 0, 0) # inclusive\n# enddate = pd.Timestamp(2012, 11, 3, 0, 0, 0) # exclusive\nstartdate = pd.Timestamp('2012-10-28 00:00:00', \n tz='America/New_York') # inclusive\nenddate = pd.Timestamp('2012-11-03 00:00:00', \n tz='America/New_York') # exclusive\n\n# load sandy-related tweets filtered by dates\ndf_sandy = ts.mongod_to_df({}, collection='tweets_sandy')\ndf_sandy['datetimeUTC'] = [datetime.tz_localize(tz='UTC') for datetime\n in df_sandy['datetimeUTC']]\ndf_sandy['datetime'] = [datetime.tz_convert('America/New_York') for datetime\n in df_sandy['datetimeUTC']]\ndf_sandy = df_sandy.rename(columns={'location_id': 'zone'})\ndf_sandy = df_sandy[['datetime', 'zone', 'tokens']]\ndf_sandy = df_sandy.set_index('datetime')\ndf_sandy = df_sandy.sort_index()\ndf_sandy = df_sandy.loc[startdate:enddate]\nprint('[min, max] sandy tweets datetime: [' + \n str(min(df_sandy.index.get_level_values('datetime'))) + ', ' + \n str(max(df_sandy.index.get_level_values('datetime'))) + '].')\n# group by datetime and zone\ndf_sandy = df_sandy.reset_index()\ndf_sandy['datetime'] = df_sandy['datetime'].dt.date\ndf_sandy_group = df_sandy.groupby(['zone', 'datetime']).count()\ndf_sandy_group = df_sandy_group.rename(columns={'tokens': 'sandy-tweets'})\ndf_sandy_group = df_sandy_group[df_sandy_group['sandy-tweets'] >= tweet_count_filter] \nprint('[min, max] sandy tweets: [' + \n str(np.nanmin(df_sandy_group['sandy-tweets'])) + ', ' +\n str(np.nanmax(df_sandy_group['sandy-tweets'])) + '].')\nprint('')\ndf_sandy_group.head()",
"_____no_output_____"
]
],
[
[
"## Test heatmap",
"_____no_output_____"
]
],
[
[
"tweet_count_filter = 5\n#startdate = dt.datetime(2012, 10, 21, 0, 0, 0).isoformat()\n#enddate = dt.datetime(2012, 10, 28, 0, 0, 0).isoformat()\nstartdate = dt.datetime(2012, 10, 28, 0, 0, 0).isoformat()\nenddate = dt.datetime(2012, 11, 4, 0, 0, 0).isoformat()\nquery = {\n \"count\": {\"$gte\": tweet_count_filter},\n \"datetimeUTC\": {\"$lte\": enddate, \"$gte\": startdate}\n}\nprint(query)\ndf = ts.mongod_to_df(query, collection='borough_day_sandy')\n\n#daterange = ['10/22/2012', '10/27/2012']\ndaterange = ['10/28/2012', '11/03/2012']\n#daterange = None\n\n# update dtypes and columns \ndf['date'] = pd.to_datetime(df['datetimeUTC']).dt.date\ndf = df[['date', 'borough', 'count']]\n\n# build full dataframe (all dates and all boroughs initialized with nans)\nif daterange:\n dates = pd.date_range(start=daterange[0], end=daterange[1]).tolist()\n dates = [pd.Timestamp.to_pydatetime(date).date() for date in dates]\nelse:\n dates = df['date'].unique()\n#print(dates)\nboroughs = sorted(df['borough'].unique())\ndf_proc = pd.DataFrame({'date': [], 'borough': [], 'count': []})\nfor date in dates:\n for borough in boroughs:\n df_temp = pd.DataFrame({'date': date,\n 'borough': borough,\n 'count': [np.nan]})\n df_proc = df_proc.append(df_temp, ignore_index=True)\n\n# get matching indexes in df_proc of available data in df\nproc_indexes = [df_proc.index[(df_proc['date'] == date) & (\n df_proc['borough'] == borough)].tolist()[0]\n for date, borough in zip(list(df['date']),\n list(df['borough']))]\n\n# update df_proc with available data in df\ndf.index = proc_indexes\ndf_proc.loc[proc_indexes, ['count']] = df['count']\n\n# reformat and rename columns\ndf_proc['date'] = df_proc['date'].apply(lambda x: x.strftime('%m-%d'))\n\n# pivot dataframe for heat map visualization\ndf_pivot = df_proc.pivot('borough', 'date', 'count')\n\nprint(df[0:5])\nprint(df_proc[0:5])\nprint('')\nprint(df[-10::])\nprint(df_proc[-10::])",
"{'count': {'$gte': 5}, 'datetimeUTC': {'$lte': '2012-11-04T00:00:00', '$gte': '2012-10-28T00:00:00'}}\n date borough count\n0 2012-10-28 Bronx 146\n1 2012-10-28 Brooklyn 656\n2 2012-10-28 EWR 28\n3 2012-10-28 Manhattan 1436\n4 2012-10-28 Queens 498\n date borough count\n0 10-28 Bronx 146.0\n1 10-28 Brooklyn 656.0\n2 10-28 EWR 28.0\n3 10-28 Manhattan 1436.0\n4 10-28 Queens 498.0\n\n date borough count\n25 2012-11-01 Brooklyn 226\n26 2012-11-01 EWR 6\n27 2012-11-01 Manhattan 438\n28 2012-11-01 Queens 136\n29 2012-11-01 Staten Island 52\n30 2012-11-02 Bronx 13\n31 2012-11-02 Brooklyn 53\n33 2012-11-02 Manhattan 105\n34 2012-11-02 Queens 30\n35 2012-11-02 Staten Island 41\n date borough count\n32 11-02 EWR NaN\n33 11-02 Manhattan 105.0\n34 11-02 Queens 30.0\n35 11-02 Staten Island 41.0\n36 11-03 Bronx NaN\n37 11-03 Brooklyn NaN\n38 11-03 EWR NaN\n39 11-03 Manhattan NaN\n40 11-03 Queens NaN\n41 11-03 Staten Island NaN\n"
]
],
[
[
"## Code for test scripts",
"_____no_output_____"
]
],
[
[
"# test create_tweet_analysis\nimport sys\nimport os\nsys.path.append(os.path.abspath('../'))\n\nimport twitterinfrastructure.twitter_sandy as ts\nimport importlib\nimportlib.reload(ts)\n\n",
"[260244087901413376, 260244088203403264, 260244088161439744, 260244088819945472, 260244089080004609, 260244089985957888, 260244092527706112, 260244093119102977, 260244093257515008, 260244094939439105]\n"
]
],
[
[
"# Extras.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecedde79eb6b4bbd3dbce0d01a39b09a48e42e7e | 38,958 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Tarea3_RSlay_RGutierrez-checkpoint.ipynb | RodoSlay/Tareas_RSlay_RGutierrez | 80c764bb5e9a9dcb917e976fb278f28797cb67f4 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Tarea3_RSlay_RGutierrez-checkpoint.ipynb | RodoSlay/Tareas_RSlay_RGutierrez | 80c764bb5e9a9dcb917e976fb278f28797cb67f4 | [
"MIT"
] | 1 | 2020-09-17T23:09:36.000Z | 2020-09-17T23:09:36.000Z | .ipynb_checkpoints/Tarea3_RSlay_RGutierrez-checkpoint.ipynb | RodoSlay/Tareas_RSlay_RGutierrez | 80c764bb5e9a9dcb917e976fb278f28797cb67f4 | [
"MIT"
] | 1 | 2020-09-09T22:06:11.000Z | 2020-09-09T22:06:11.000Z | 19.097059 | 221 | 0.281816 | [
[
[
"<img style=\"float: left; margin: 30px 15px 15px 15px;\" src=\"https://pngimage.net/wp-content/uploads/2018/06/logo-iteso-png-5.png\" width=\"300\" height=\"500\" /> \n \n \n### <font color='navy'> Simulación de procesos financieros. \n\n**Nombres:** Rodolfo Slay y Romina Cortés.\n\n**Fecha:** 14 de Septiembre del 2020.\n\n**Expediente** : 715214 y 713415.\n**Profesor:** Oscar David Jaramillo Zuluaga.\n\n# Tarea 3: Clase 4",
"_____no_output_____"
],
[
"**Github:** [Repositorio de Tareas](https://github.com/RodoSlay/Tareas_RSlay_RGutierrez)",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"> ## Tarea 3: (Usando notebook de jupyter)** \n\n> Usando compresión de listas o funciones map(sino recuerda como funciona observar el siguiente enlace https://www.pythonforbeginners.com/lists/list-comprehensions-in-python/), resolver los siguientes ejercicios:\n\n>1. Resolver la siguiente ecuación recursiva usando funciones como se vió en clase\n$$ D_{n}=(n-1) D_{n-1}+(n-1) D_{n-2} \\quad n\\ge 3$$\ncon $D_1=0$ y $D_2 = 1$\n",
"_____no_output_____"
]
],
[
[
"#E1\ndef recursiva(n:\"Número de elementos\"):\n '''\n Esta función contiene el proceso para sacar n valores de Dn \n '''\n x = np.zeros(n-1)\n d1 = 0\n d2 = 1\n x[0] = d1\n x[1] = d2\n def llenar_vector(i):\n nonlocal x\n x[i-1] = (i - 1) * x[i-2] + (i-1) * x[i-3]\n [llenar_vector(i) for i in range(3,n)]\n return x\nrecursiva(7)",
"_____no_output_____"
],
[
"#E2\ndef rec(n:\"Número de elementos\"):\n '''\n Esta función contiene el proceso para sacar n valores de Dn \n '''\n x = np.zeros(n-1)\n d1 = 0\n d2 = 1\n x[0] = d1\n x[1] = d2\n def y(i):\n nonlocal x\n x[i-1] = (i - 1) * x[i-2] + (i-1) * x[i-3]\n [y(i) for i in range(3,n)]\n return x\nrec(7)",
"_____no_output_____"
]
],
[
[
">2. Count the number of spaces in the following string `variable = relaciónn requiere, para obtener el valor de un cierto término, el conocimiento de los dos anteriores`.\n",
"_____no_output_____"
]
],
[
[
"#E1\nvariable = \"relaciónn requiere, para obtener el valor de un cierto término, el conocimiento de los dos anteriores\"\nans = [i for i in variable if i == \" \"]\nlen(ans)",
"_____no_output_____"
],
[
"#E2\nvariable = \" relaciónn requiere, para obtener el valor de un cierto término, el conocimiento de los dos anteriores`.\"\nf1 = variable.count(\" \")\nf1",
"_____no_output_____"
]
],
[
[
">3. Remove all of the vowels in a string [make a list of the non-vowels].\n",
"_____no_output_____"
]
],
[
[
"#E1\nvariable = \"relación requiere, para obtener el valor de un cierto término, el conocimiento de los dos anteriores\"\nvocales = ('a', 'e', 'i', 'o', 'u', 'á', 'é', 'í', 'ó', 'ú')\n\nans = [i for i in variable if i not in vocales]\nans",
"_____no_output_____"
],
[
"#E2\ndef no_vocal(frase): \n vocales = ('a', 'e', 'i', 'o', 'u') \n f =\"\".join([x for x in frase if x.lower() not in vocales ]) \n # Print string without vowels \n print(f) ",
"_____no_output_____"
],
[
"no_vocal(\" relación requiere, para obtener el valor de un cierto término, el conocimiento de los dos anteriores\")",
" rlcón rqr, pr btnr l vlr d n crt térmn, l cncmnt d ls ds ntrrs\n"
]
],
[
[
">4. Find all of the words in a string that are less than 4 letters.\n",
"_____no_output_____"
]
],
[
[
"#E1\nvariable = \"relación requiere, para obtener el valor de un cierto término, el conocimiento de los dos anteriores\"\nvariable = variable.split()\n\nans = [i for i in variable if len(i) < 4]\nans",
"_____no_output_____"
],
[
"#E2\ntexto='relación requiere, para obtener el valor de un cierto término, el conocimiento de los dos anteriores'\n\ntexto = texto.split()\n\neliminar =list(filter(lambda x: len(x)<4, texto))\neliminar",
"_____no_output_____"
]
],
[
[
">5. Use a dictionary comprehension to count the length of each word in a sentence.\n",
"_____no_output_____"
]
],
[
[
"variable = \"relación requiere, para obtener el valor de un cierto término, el conocimiento de los dos anteriores\"\nvariable = variable.split()\n\nans = {i: len(i) for i in variable}\nans",
"_____no_output_____"
],
[
"#E2\nwords = ''.join([i for i in variable if i != ',']).split()\nprint({ word: len(word) for word in words })",
"{'relaciónn': 9, 'requiere': 8, 'para': 4, 'obtener': 7, 'el': 2, 'valor': 5, 'de': 2, 'un': 2, 'cierto': 6, 'término': 7, 'conocimiento': 12, 'los': 3, 'dos': 3, 'anteriores`.': 12}\n"
]
],
[
[
">6. Use a nested list comprehension to find all of the numbers from 1-1000 that are divisible by any singleM digit besides 1 (2-9). ",
"_____no_output_____"
]
],
[
[
"ans = [n for n in range(1,1001) if [d for d in range(2,9) if n%d == 0]]\nans",
"_____no_output_____"
],
[
"#E2\nnumeros=[i for i in range(1,1001) if [j for j in range(2,10) if i%j == 0]]\nnumeros",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecede1457c8c606a0264a2fe3c10b50a0e52f95b | 4,596 | ipynb | Jupyter Notebook | src_py/proof_of_concept.ipynb | ninja-nanz/CDT_dataviz | 133fef7f1d50b2d2191721dcd458b29ddcf94180 | [
"MIT"
] | null | null | null | src_py/proof_of_concept.ipynb | ninja-nanz/CDT_dataviz | 133fef7f1d50b2d2191721dcd458b29ddcf94180 | [
"MIT"
] | null | null | null | src_py/proof_of_concept.ipynb | ninja-nanz/CDT_dataviz | 133fef7f1d50b2d2191721dcd458b29ddcf94180 | [
"MIT"
] | null | null | null | 22.31068 | 96 | 0.495648 | [
[
[
"import time\nimport pandas as pd\nfrom zomatopy import Zomato\n#get_categories\n#get_city_ID\n#get_city_name\n#get_collections\n#get_cuisines\n#get_establishment_types\n#get_nearby_restaurants\n#get_restaurant\n#restaurant_search\n\nzconfig = {\"user_key\":\"2dde89df168a9311ecd9f1459126144c\"}",
"_____no_output_____"
]
],
[
[
"## READING DATA",
"_____no_output_____"
]
],
[
[
"zcities_df = pd.read_pickle('../results/zcities_df.pkl')\ncscities_df = pd.read_pickle('../results/cscities_df.pkl')",
"_____no_output_____"
],
[
"zcities_df.head()",
"_____no_output_____"
],
[
"ver = zcities_df[zcities_df.zcity_id==-9999]",
"_____no_output_____"
],
[
"ver",
"_____no_output_____"
],
[
"ver['json'][0]['location_suggestions'][0]['state_code']",
"_____no_output_____"
],
[
"len(zcities_df)",
"_____no_output_____"
],
[
"def zomato_cities_query(cscities_df, zconfig):\n zomato = Zomato(zconfig)\n \n city_queries = []\n zcity_ids = []\n jsons = []\n for i, row in cscities_df.iterrows():\n print(f\"city query {row.city_query}\")\n \n # Security saves of Zomato calls\n if i>0 and (i % 10 == 0):\n time.sleep(5)\n zcities_df = pd.DataFrame.from_dict({'zcity_id': zcity_ids, 'json': jsons, \n 'city_query': city_queries})\n zcities_df.to_pickle('../results/zcities_df.pkl')\n \n try:\n zcity_id = zomato.get_city_ID(city_name=row.city_query)\n\n city_queries += [row.city_query]\n zcity_ids += [zcity_id]\n jsons += [zomato.json]\n except:\n print(f\"failed city query {row.city_query}\")\n pass\n \n # Final save\n zcities_df = pd.DataFrame.from_dict({'zcity_id': zcity_ids, 'json': jsons, \n 'city_query': city_queries})\n zcities_df.to_pickle('../results/zcities_df.pkl')\n return zcities_df\n\nzcities_df = zomato_cities_query(cscities_df, zconfig)",
"_____no_output_____"
],
[
"zcities_df = pd.read_pickle('../results/zcities_df.pkl')",
"_____no_output_____"
],
[
"zcities_df.head()",
"_____no_output_____"
],
[
"len(zcities_df)",
"_____no_output_____"
],
[
"zomato = Zomato(zconfig)\nzomato.get_city_ID('Indianapolis')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecede7c66f20d5183335e8cf52409d82ed92d03d | 3,987 | ipynb | Jupyter Notebook | mix_random_effect/summary.ipynb | liangyy/idea-playground | 3e8130f67f3810a73c84898c1d869fc44c74a915 | [
"MIT"
] | null | null | null | mix_random_effect/summary.ipynb | liangyy/idea-playground | 3e8130f67f3810a73c84898c1d869fc44c74a915 | [
"MIT"
] | null | null | null | mix_random_effect/summary.ipynb | liangyy/idea-playground | 3e8130f67f3810a73c84898c1d869fc44c74a915 | [
"MIT"
] | null | null | null | 55.375 | 856 | 0.623276 | [
[
[
"## Issue of EM algorithm\n\nThe lack of analytical maximizer in M-step makes the EM algorithm not totally satisfiable in terms of the computational burden. Anyway, regarding the convergence, a numerical solver for M-step is not a huge issue since the improvement of likelihood is bounded by the improvement of $Q$ which we can always get some improvement by numerically optimizing the objective. \n\nNonetheless, a better solution is to use existing solver for M-step but I failed to find a one. **I am also waiting for an analytical result if it really exists!**\n\n### Using `lme4::lmer`\n\nThe straightforward idea is to use an out-of-box random effect solver. Well, the M-step problem can be formalized as a weighted random effect model (for $K = 2$).\n\n$$\\begin{align*}\n \\hat\\beta_{gwas} &= \\beta_{gene, 1} \\hat\\beta_{eqtl, 1} + \\beta_{gene, 2} \\hat\\beta_{eqtl, 2} + e \\\\\n \\hat\\beta_{eqtl, 1}, \\hat\\beta_{eqtl, 2} &= \\begin{cases}\n \\hat\\beta_{eqtl}, 0 \\\\\n 0, \\hat\\beta_{eqtl}\n \\end{cases} \\\\\n \\beta_{gene, 1} &\\sim \\mathcal{N}(0, \\sigma_1^2) \\\\\n \\beta_{gene, 2} &\\sim \\mathcal{N}(0, \\sigma_2^2) \\\\\n e &\\sim \\mathcal{N}(0, \\sigma^2)\n\\end{align*}$$\n\nBy doing this construction, it is almost solving the M-step problem and the only missing part is weight. Let $y = [\\hat\\beta_{gwas}, \\hat\\beta_{gwas}]$, $x_1 = [\\hat\\beta_{eqtl}, \\vec{0}], x_2 = [\\vec{0}, \\hat\\beta_{eqtl}], w = [w_{11}, \\cdots, w_{n1}, w_{12}, \\cdots, w_{n2}]$. The equivalent random effect model is\n\n$$\\begin{align*}\n y &= \\beta_{gwas, 1} x_1 + \\beta_{gwas, 2} x_2 + e \n\\end{align*}$$\n, with weighted likelihood \n$$\\begin{align*}\n lld &= \\sum_i \\log w_i \\Pr(y_i | x_1, x_2)\n\\end{align*}$$\n\nUnfortunately, I find no way to use weighted likelihood in `lme4::lmer`. The `weights` option in it is for modeling heteroscedasticity. Nonetheless, `lme4::lmer` becomes a validation of my own M-step solver by applying to the case with equal weights. See [`compare_to_lmer.html`](http://htmlpreview.github.io/?https://raw.githubusercontent.com/liangyy/idea-playground/master/mix_random_effect/compare_to_lmer.html). \n\n**I am waiting for a better alternative RE solver!**\n\n\n### EM solver and model performance\n\nSee [`run.html`](http://htmlpreview.github.io/?https://raw.githubusercontent.com/liangyy/idea-playground/master/mix_random_effect/run.html). Note that `idx` determines the initial value of EM algorithm. I generated 3 data sets with sample sizes 1e3, 1e4, 1e5. EM was run 10 times with 10 initial points. With hard work on implementing EM, it's time to see the performance of the model. Many times, $\\sigma_1^2$ is under-estimated to be really close to zero. The issue is that posterior $\\Pr(Z_i|\\theta)$ is largely determined by $\\hat\\beta_{gwas}^2$ and $\\hat\\beta_{eqtl} ^ 2$ contributes almost nothing. My guess to the solution of this issue is that if we can obtain multiple eQTLs for a gene (sharing the same $\\beta_{gene}$ is can be resolved since the pattern across all variants of that gene should share the same gene-level effect. \n\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
ecede910eeb168b46cfe1081b643a3159f4fe905 | 100,923 | ipynb | Jupyter Notebook | classifiers_comparative_analysis.ipynb | BSski/classifiers-comparative-analysis | b9dff73115b977f1973d153f02a8da8443380aa4 | [
"CNRI-Python"
] | null | null | null | classifiers_comparative_analysis.ipynb | BSski/classifiers-comparative-analysis | b9dff73115b977f1973d153f02a8da8443380aa4 | [
"CNRI-Python"
] | null | null | null | classifiers_comparative_analysis.ipynb | BSski/classifiers-comparative-analysis | b9dff73115b977f1973d153f02a8da8443380aa4 | [
"CNRI-Python"
] | null | null | null | 104.475155 | 15,962 | 0.812045 | [
[
[
"!pip install scikit-learn\nimport json\nimport string\nimport pprint\nimport glob\nimport itertools\nimport random\nimport numpy as np\nimport pandas as pd\nfrom sklearn.utils import shuffle\nfrom google.colab import drive\ndrive.mount('/content/drive')",
"Mounted at /content/drive\n"
],
[
"maps = glob.glob(\"drive/MyDrive/IAC/*.json\")",
"_____no_output_____"
],
[
"with open(maps[0]) as f:\n data = json.loads(f.read())",
"_____no_output_____"
],
[
"'''\n conclusionPremiseDict Create dictionary of pairs with an identifier with the following form: \n {id: {\"conclusion\": <SINGLE_CONCLUSION>, \"premises\":[<LIST_OF_PREMISES>]}}\n'''\ndef conclusionPremiseDict(premises, conclusions):\n pairs = {}\n for i, x in enumerate(conclusions):\n pairs[i] = {'conclusion':x, 'premises':[]}\n id_to = x['fromID']\n for p in premises:\n if p['toID'] == id_to:\n pairs[i]['premises'].append(p)\n \n return pairs\n\n\n'''\n aduPairs create list of ADU pairs containing connected conclusion and premise [[conclusion, premise]] \n'''\ndef aduPairs(edgePairs, nodesById):\n aduPair = []\n for pair in edgePairs.values():\n for p in pair['premises']:\n aduPair.append([nodesById[pair['conclusion']['toID']]['text'], nodesById[p['fromID']]['text']])\n return(aduPair)\n\n\n'''\n pairs creates conclusion - premise pairs for one map\n'''\ndef pairs(map):\n with open(map) as f:\n data = json.loads(f.read())\n #Creating nodesById dictionary which has nodeID as key and whole node as value for more efficient data extraction.\n nodesById = {}\n for _, node in enumerate(data['nodes']):\n nodesById[node['nodeID']] = node\n #Premises are nodes that have ingoing edges that are type 'RA' and outgoing edges that are type 'I'.\n premises = [x for x in data['edges'] if nodesById[x['fromID']]['type'] == 'I' and nodesById[x['toID']]['type'] == 'RA' ]\n\n #Conclusions are nodes that have ingoing edges that are type 'I' and outgoing edges that are type 'RA'.\n conclusions = [x for x in data['edges'] if nodesById[x['toID']]['type'] == 'I' and nodesById[x['fromID']]['type'] == 'RA' ]\n edgePairs = conclusionPremiseDict(premises, conclusions)\n adus = aduPairs(edgePairs, nodesById)\n return adus, conclusions, premises, nodesById\n\n\n'''\n comb makes combination of conclusions and premises lists and returns list of pairs that are not conclusion-premise pairs \n'''\ndef comb(conclusions, premises, l, nodesById):\n combList = [(x,y) for x in conclusions for y in premises] \n smallCombList = []\n for _ in range(l):\n p = random.choice(combList)\n smallCombList.append([nodesById[p[0]['toID']]['text'], nodesById[p[1]['fromID']]['text']])\n return smallCombList",
"_____no_output_____"
],
[
"'''\n truePairs is list of all conclusion-premise pairs; falsePairs is list od conclusion-premise non pairs\n'''\ntruePairs = []\nconclusions = []\npremises = []\nnodesById = {}\n\nfor m in maps:\n adus, c, p, n = pairs(m)\n truePairs.extend(adus)\n conclusions.extend(c)\n premises.extend(p)\n nodesById = {**nodesById, **n}\n\nfalsePairs = comb(conclusions, premises, len(truePairs), nodesById)",
"_____no_output_____"
],
[
"!pip install tensorflow_text\n!pip install simpleneighbors",
"_____no_output_____"
],
[
"import json\nimport os\nimport bokeh\nimport bokeh.models\nimport bokeh.plotting\nimport numpy as np\nimport os\nimport pandas as pd\nimport tensorflow.compat.v2 as tf\nimport tensorflow_hub as hub\nfrom tensorflow_text import SentencepieceTokenizer\nimport sklearn.metrics.pairwise\n\nfrom simpleneighbors import SimpleNeighbors\nfrom tqdm import tqdm\nfrom tqdm import trange",
"_____no_output_____"
],
[
"def get_similarity(embeddings_1, embeddings_2):\n \"\"\"Calculate cosine simiarity between two vectors.\n *[check similarity measure based on arccos]\n \n Args:\n - embeddings1, embeddings2 - vectors with sentence|word embeddings\n \n Returns:\n - sim, cosine - similarity measures\n \"\"\"\n cosine = sklearn.metrics.pairwise.cosine_similarity(embeddings_1, embeddings_2)\n sim = 1 - np.arccos(cosine)/np.pi\n return sim, cosine",
"_____no_output_____"
],
[
"def embed_text(text, model):\n \"\"\"Embed text as an embedding via universal sentence encoder model.\n Args:\n - text - text to encode\n - model - universal sentence encoder model \n\n Returns:\n - vector embedding of text\n \"\"\"\n return model(text)",
"_____no_output_____"
],
[
"# The 16-language multilingual module\nmodule_url = 'https://tfhub.dev/google/universal-sentence-encoder-multilingual-large/3' #@param ['https://tfhub.dev/google/universal-sentence-encoder-multilingual/3', 'https://tfhub.dev/google/universal-sentence-encoder-multilingual-large/3']\nmodel = hub.load(module_url)",
"_____no_output_____"
],
[
"def embed_pair(pair, model):\n \"\"\"Embed conclusion and premises from one pair.\n \n Args:\n - pair - dictionary of conclusion and premises\n \n Returns:\n - embed_conclusion - embedding of conclusion\n - embed_premises - embedding of premises\n \"\"\"\n embed_conclusion = embed_text(pair[0], model)\n embed_premise = embed_text(pair[1],model)\n return embed_conclusion, embed_premise",
"_____no_output_____"
],
[
"embed_conclusion, embed_premises = embed_pair(truePairs[0], model)",
"_____no_output_____"
],
[
"def getsimforpair(prem,conc):\n \"\"\"Given premise and conclusion returns similarity measure\n\n Args:\n - prem - embeded representation of the premise\n - conc - embeded representation of the conclusion\n Returns: \n - sim_pair - similarity measure\n \"\"\"\n sim_pair = get_similarity(prem,conc)\n return sim_pair",
"_____no_output_____"
],
[
"getsimforpair(embed_conclusion, embed_premises)",
"_____no_output_____"
],
[
"def loopallpairs(pairlist):\n \"\"\"Given list of pairs of premises and conclusions returns list of similarity measures for each pair in the corpus\n\n Args:\n - pairlist - list of pairs of premises and conclusions (raw text)\n Returns: \n - simlist - list of similarity measure for each pair (raw text)\n \"\"\"\n for pair in pairlist:\n conc = embed_text(pair[0], model)\n prem = embed_text(pair[1], model)\n simforpair = getsimforpair(prem,conc)\n simlist.append(simforpair)\n pairssimlist = zip(pairlist,simlist)\n return pairssimlist",
"_____no_output_____"
],
[
"def makesimlist(simlist,label,measure):\n \"\"\"\n Given list of list of similarity measure for each pairs makes list of similarity measurements plus labels\n \n Args:\n - simlist - list of similarity measure for each pair\n - label (0 false or 1 for true)\n - measure: 0 or 1\n \"\"\"\n simtable = pd.DataFrame(simlist)\n simlistsim = simtable[measure] \n simlistsim_flattened = [val for sublist in simlistsim for val in sublist]\n simtable = pd.DataFrame(simlistsim_flattened)\n simtable[\"label\"] = label\n return simtable",
"_____no_output_____"
],
[
"def loopallpairs(pairlist):\n \"\"\"Given list of pairs of premises and conclusions returns list of similarity measures for each pair in the corpus\n\n Args:\n - pairlist - list of pairs of premises and conclusions (raw text)\n Returns: \n - simlist - list of similarity measure for each pair (raw text)\n \"\"\"\n simlist = []\n for pair in pairlist:\n conc = embed_text(pair[0], model)\n prem = embed_text(pair[1], model)\n simforpair = getsimforpair(prem,conc)\n simlist.append(simforpair)\n return simlist",
"_____no_output_____"
],
[
"all_true_pairssimlist = loopallpairs(truePairs)",
"_____no_output_____"
],
[
"all_false_pairssimlist = loopallpairs(falsePairs)",
"_____no_output_____"
],
[
"all_false_pairssimlist[0]",
"_____no_output_____"
],
[
"all_true_pairssimlistSIMtable = makesimlist(all_true_pairssimlist,\"1\", 0)\nall_true_pairssimlistCOStable = makesimlist(all_true_pairssimlist,\"1\", 1)\nall_false_pairssimlistSIMtable = makesimlist(all_false_pairssimlist,\"0\", 0)\nall_false_pairssimlistCOStable = makesimlist(all_false_pairssimlist,\"0\", 1)\n\ndatabase = []\ndatabase = pd.concat([all_false_pairssimlistSIMtable, all_true_pairssimlistSIMtable], axis=0)\nfrom sklearn.utils import shuffle\ndatabase = shuffle(database)\n\n\n'''\n write data to .csv file\n'''\nmyFile = open('BK PROJEKT 2 SWĘDROWSKI.csv', 'w')\nwith myFile: \n myFields = ['similarity', 'feature']\n writer = csv.DictWriter(myFile, fieldnames=myFields) \n writer.writeheader()\n for i,j in zip(database[0], database['label']):\n writer.writerow({'similarity' : i, 'feature': j})",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\n\ntrain, test = train_test_split(database, test_size=0.2)",
"_____no_output_____"
],
[
"import tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.keras.layers import Activation, Dense\nfrom tensorflow.keras.optimizers import Adam\nfrom tensorflow.keras.metrics import categorical_crossentropy\nimport csv\nimport numpy as np\nfrom sklearn.metrics import confusion_matrix\nimport itertools\nimport matplotlib.pyplot as plt\nimport matplotlib as mpl\n\ntrainSamples = np.asarray(list(train[0]))\ntrainLabels = list(train['label'])\nfor i in range(len(trainLabels)):\n trainLabels[i] = float(trainLabels[i])\n\ntrainLabels = np.asarray(trainLabels)\n\ntestSamples = np.asarray(list(test[0]))\ntestLabels = list(test['label'])\nfor i in range(len(testLabels)):\n testLabels[i] = float(testLabels[i])\ntestLabels = np.asarray(testLabels)\n\nbb = []\nfor i in testLabels:\n bb.append(type(i))\ndd = set(bb)\nprint(dd)",
"{<class 'numpy.float64'>}\n"
],
[
"'''\n make model\n'''\nmodel = tf.keras.Sequential([\n Dense(units=64, input_shape=(1,), activation='relu'),\n Dense(units=64, activation='relu'),\n Dense(units=64, activation='relu'),\n Dense(units=64, activation='relu'),\n Dense(units=32, activation='relu'),\n Dense(units=2, activation='sigmoid')\n])\nmodel.summary()\n\n\n'''\n train model\n'''\nmodel.compile(optimizer=Adam(learning_rate=0.0001), loss='sparse_categorical_crossentropy', metrics=['accuracy'])\nhist = model.fit(x=trainSamples, y=trainLabels, validation_split = 0.03, batch_size=24, epochs=75, shuffle=True, verbose=1)\n\n\n'''\n predictions\n'''\npredictions = model.predict(x=testSamples, batch_size=1, verbose=0)\nroundedPredictions = np.argmax(predictions, axis=-1)\n\ncm = confusion_matrix(y_true = testLabels, y_pred = roundedPredictions)\n\ndef plot_confusion_matrix(cm, classes, normalize=False, title='Confusion Matrix,', cmap=mpl.cm.Greens):\n plt.imshow(cm, interpolation='nearest', cmap=cmap)\n plt.title(title)\n plt.colorbar()\n tick_marks = np.arange(len(classes))\n plt.xticks(tick_marks, classes, rotation=45)\n plt.yticks(tick_marks, classes)\n\n if normalize:\n cm = cm.astype('float') / cm.sum(axis=1)[:np.newaxis]\n print('Normalized confusiom matrix')\n else:\n print('Confusion matrix without normalization')\n\n thresh = cm.max() / 2.\n for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):\n plt.text(j, i, cm[i, j], \n horizontalalignment=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n\n plt.tight_layout()\n plt.ylabel('Prawdziwe')\n plt.xlabel('Przewidywane')\n plt.show()\n\ncm_plot_labels = ['nie para', 'para']\nplot_confusion_matrix(cm=cm, classes=cm_plot_labels, title=\"Macierz pomyłek\")\n\nprint(len(trainSamples), len(trainLabels))\nmodel.evaluate(testSamples, testLabels, batch_size=2)",
"_____no_output_____"
],
[
"plt.plot(hist.history['accuracy'])\nplt.title('Dokładność modelu')\nplt.ylabel('Dokładność')\nplt.xlabel('Epoka')\nplt.show()",
"_____no_output_____"
],
[
"#all_true_pairssimlistSIMtable.plot.hist(by=all_true_pairssimlistSIMtable[0], bins=10)",
"_____no_output_____"
],
[
"#all_false_pairssimlistSIMtable.plot.hist(by=all_false_pairssimlistSIMtable[0], bins=10)",
"_____no_output_____"
],
[
"#all_true_pairssimlistCOStable.plot.hist(by=all_true_pairssimlistCOStable[0], bins=10)",
"_____no_output_____"
],
[
"#all_false_pairssimlistCOStable.plot.hist(by=all_false_pairssimlistCOStable[0], bins=10)",
"_____no_output_____"
],
[
"bins = np.linspace(0, 1, 100)\nplt.hist(all_true_pairssimlistSIMtable[0], bins, alpha=0.5, label='prawdziwe pary')\nplt.hist(all_false_pairssimlistSIMtable[0], bins, alpha=0.5, label='fałszywe pary')\nplt.legend(loc='upper right')\nplt.title(\"Podobieństwo SIM\") \nplt.show()",
"_____no_output_____"
],
[
"bins = np.linspace(0, 1, 100)\nplt.hist(all_true_pairssimlistCOStable[0], bins, alpha=0.5, label='prawdziwe pary')\nplt.hist(all_false_pairssimlistCOStable[0], bins, alpha=0.5, label='fałszywe pary')\nplt.legend(loc='upper right')\nplt.title(\"Podobieństwo COS\") \nplt.show()",
"_____no_output_____"
],
[
"from sklearn import metrics\n\ntrainSamples_np = trainSamples.reshape(-1, 1)\ntestSamples_np = testSamples.reshape(-1, 1)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\n\nlr_model = LogisticRegression(solver='lbfgs')\nlr_model.fit(trainSamples_np, trainLabels)\n\npredictions = lr_model.predict(testSamples_np)\nmetrics.confusion_matrix(testLabels, predictions)\nprint(metrics.classification_report(testLabels,predictions))\nprint(metrics.accuracy_score(testLabels,predictions))\n\ncm = confusion_matrix(y_true = testLabels, y_pred = predictions)\nplot_confusion_matrix(cm=cm, classes=cm_plot_labels, title=\"Macierz pomyłek\")",
" precision recall f1-score support\n\n 0.0 0.74 0.87 0.80 429\n 1.0 0.85 0.70 0.77 438\n\n accuracy 0.79 867\n macro avg 0.79 0.79 0.78 867\nweighted avg 0.79 0.79 0.78 867\n\n0.7854671280276817\nConfusion matrix without normalization\n[[374 55]\n [131 307]]\n"
],
[
"from sklearn.svm import SVC\n\nsvc_model = SVC()\nsvc_model.fit(trainSamples_np, trainLabels)\n\npredictions = svc_model.predict(testSamples_np)\nmetrics.confusion_matrix(testLabels, predictions)\nprint(metrics.classification_report(testLabels,predictions))\nprint(metrics.accuracy_score(testLabels,predictions))\n\ncm = confusion_matrix(y_true = testLabels, y_pred = predictions)\nplot_confusion_matrix(cm=cm, classes=cm_plot_labels, title=\"Macierz pomyłek\")",
" precision recall f1-score support\n\n 0.0 0.74 0.88 0.80 429\n 1.0 0.86 0.69 0.77 438\n\n accuracy 0.79 867\n macro avg 0.80 0.79 0.78 867\nweighted avg 0.80 0.79 0.78 867\n\n0.7854671280276817\nConfusion matrix without normalization\n[[378 51]\n [135 303]]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecedf55dc3e0c7ea3e57bf23a7eacd3c2744d425 | 193,188 | ipynb | Jupyter Notebook | finch/tensorflow1/free_chat/chinese_lccc/main/transformer_train.ipynb | akshaykambleofficial/tensorflow-nlp | 7ea72939f29a9355a3e2b1315310c5dfa7cfb063 | [
"MIT"
] | 248 | 2019-07-18T05:59:03.000Z | 2022-03-29T21:57:24.000Z | finch/tensorflow1/free_chat/chinese_lccc/main/transformer_train.ipynb | akshaykambleofficial/tensorflow-nlp | 7ea72939f29a9355a3e2b1315310c5dfa7cfb063 | [
"MIT"
] | 5 | 2019-07-26T09:29:20.000Z | 2020-11-01T08:40:13.000Z | finch/tensorflow1/free_chat/chinese_lccc/main/transformer_train.ipynb | akshaykambleofficial/tensorflow-nlp | 7ea72939f29a9355a3e2b1315310c5dfa7cfb063 | [
"MIT"
] | 68 | 2019-07-25T06:59:58.000Z | 2022-03-22T06:44:30.000Z | 193,188 | 193,188 | 0.711618 | [
[
[
"from google.colab import drive\ndrive.mount('/content/gdrive')\nimport os\nos.chdir('/content/gdrive/My Drive/finch/tensorflow1/free_chat/chinese_lccc/main')",
"_____no_output_____"
],
[
"%tensorflow_version 1.x\n!pip install texar",
"_____no_output_____"
],
[
"import tensorflow as tf\nimport texar.tf as tx\n\nimport numpy as np\nimport pprint\nimport logging\nimport copy\n\nfrom pathlib import Path\nfrom texar.tf.modules import TransformerEncoder\n\nprint(\"TensorFlow Version\", tf.__version__)\nprint('GPU Enabled:', tf.test.is_gpu_available())",
"TensorFlow Version 1.15.2\nGPU Enabled: True\n"
],
[
"# stream data from text files\ndef data_generator(f_path, params):\n char2idx = params['char2idx']\n with open(f_path) as f:\n print('Reading', f_path)\n for line in f:\n line = line.rstrip()\n source, target = line.split('<SEP>')\n source = [char2idx.get(c, len(char2idx)) for c in list(source)]\n target = [char2idx.get(c, len(char2idx)) for c in list(target)]\n if len(source) > params['max_len']:\n source = source[:params['max_len']]\n if len(target) > params['max_len']:\n target = target[:params['max_len']]\n target_in = [1] + target\n target_out = target + [2]\n yield (source, (target_in, target_out))",
"_____no_output_____"
],
[
"def dataset(is_training, params):\n _shapes = ([None], ([None], [None]))\n _types = (tf.int32, (tf.int32, tf.int32))\n _pads = (0, (0, 0))\n \n if is_training:\n ds = tf.data.Dataset.from_generator(\n lambda: data_generator(params['train_path'], params),\n output_shapes = _shapes,\n output_types = _types,)\n ds = ds.shuffle(params['buffer_size'])\n ds = ds.padded_batch(params['batch_size'], _shapes, _pads)\n ds = ds.prefetch(tf.data.experimental.AUTOTUNE)\n else:\n ds = tf.data.Dataset.from_generator(\n lambda: data_generator(params['test_path'], params),\n output_shapes = _shapes,\n output_types = _types,)\n ds = ds.padded_batch(params['batch_size'], _shapes, _pads)\n ds = ds.prefetch(tf.data.experimental.AUTOTUNE)\n \n return ds",
"_____no_output_____"
],
[
"def clip_grads(loss):\n variables = tf.trainable_variables()\n pprint.pprint(variables)\n grads = tf.gradients(loss, variables)\n clipped_grads, _ = tf.clip_by_global_norm(grads, params['clip_norm'])\n return zip(clipped_grads, variables)\n\n\ndef rnn_cell():\n def cell_fn():\n cell = tf.nn.rnn_cell.LSTMCell(params['rnn_units'],\n initializer=tf.orthogonal_initializer())\n return cell\n if params['dec_layers'] > 1:\n cells = []\n for i in range(params['dec_layers']):\n if i == params['dec_layers'] - 1:\n cells.append(cell_fn())\n else:\n cells.append(tf.nn.rnn_cell.ResidualWrapper(cell_fn(), residual_fn=lambda i,o: tf.concat((i,o), -1)))\n return tf.nn.rnn_cell.MultiRNNCell(cells)\n else:\n return cell_fn()\n\n \ndef dec_cell(enc_out, enc_seq_len):\n attn = tf.contrib.seq2seq.BahdanauAttention(\n num_units = params['rnn_units'],\n memory = enc_out,\n memory_sequence_length = enc_seq_len)\n \n return tf.contrib.seq2seq.AttentionWrapper(\n cell = rnn_cell(),\n attention_mechanism = attn,\n attention_layer_size = params['rnn_units'])",
"_____no_output_____"
],
[
"class TiedDense(tf.layers.Layer):\n def __init__(self, tied_embed, out_dim):\n super().__init__()\n self.tied_embed = tied_embed\n self.out_dim = out_dim\n \n def build(self, input_shape):\n self.bias = self.add_weight(name='bias',\n shape=[self.out_dim],\n trainable=True)\n if params['rnn_units'] != 300:\n self.proj_W = self.add_weight(name='proj_W',\n shape=[params['rnn_units'], params['embed_dim']],\n trainable=True)\n self.proj_b = self.add_weight(name='proj_b',\n shape=[params['embed_dim']],\n trainable=True)\n super().build(input_shape)\n \n def call(self, inputs):\n if params['rnn_units'] != 300:\n inputs = params['activation'](tf.nn.bias_add(tf.matmul(inputs, self.proj_W), self.proj_b))\n x = tf.matmul(inputs, self.tied_embed, transpose_b=True)\n x = tf.nn.bias_add(x, self.bias)\n return x\n \n def compute_output_shape(self, input_shape):\n return input_shape[:-1].concatenate(self.out_dim)",
"_____no_output_____"
],
[
"def forward(words, labels, mode):\n words_len = tf.count_nonzero(words, 1, dtype=tf.int32)\n \n is_training = (mode == tf.estimator.ModeKeys.TRAIN)\n batch_sz = tf.shape(words)[0]\n \n \n with tf.variable_scope('Embedding'):\n embedding = tf.Variable(np.load('../vocab/char.npy'),\n dtype=tf.float32,\n name='fasttext_vectors')\n embedding = tf.concat([tf.zeros(shape=[1, params['embed_dim']]), embedding[1:, :]], axis=0)\n x = tf.nn.embedding_lookup(embedding, words)\n pos_embedder = tx.modules.SinusoidsPositionEmbedder(\n position_size = params['max_len'] + 1,\n hparams = config_model.position_embedder_hparams)\n x = (x * config_model.hidden_dim ** 0.5) + pos_embedder(sequence_length=words_len)\n\n\n with tf.variable_scope('Encoder'):\n encoder = TransformerEncoder(hparams=config_model.encoder)\n enc_out = encoder(inputs=x,\n sequence_length=words_len,\n mode=mode,)\n enc_state = tf.reduce_max(enc_out, axis=1)\n enc_state = tf.nn.rnn_cell.LSTMStateTuple(c=enc_state, h=enc_state)\n \n \n with tf.variable_scope('Decoder'):\n output_proj = TiedDense(embedding, len(params['char2idx'])+1)\n \n if is_training or (mode == tf.estimator.ModeKeys.EVAL):\n dec_inputs, dec_outputs = labels\n dec_seq_len = tf.count_nonzero(dec_inputs, 1, dtype=tf.int32)\n dec_inputs = tf.nn.embedding_lookup(embedding, dec_inputs)\n dec_inputs = tf.layers.dropout(dec_inputs, params['dropout_rate'], training=is_training)\n cell = dec_cell(enc_out, words_len)\n \n init_state = cell.zero_state(batch_sz, tf.float32).clone(\n cell_state=enc_state)\n \n helper = tf.contrib.seq2seq.TrainingHelper(\n inputs = dec_inputs,\n sequence_length = dec_seq_len,)\n decoder = tf.contrib.seq2seq.BasicDecoder(\n cell = cell,\n helper = helper,\n initial_state = init_state,\n output_layer = output_proj)\n decoder_output, _, _ = tf.contrib.seq2seq.dynamic_decode(\n decoder = decoder,\n maximum_iterations = tf.reduce_max(dec_seq_len))\n \n return decoder_output.rnn_output\n else:\n enc_out_t = tf.contrib.seq2seq.tile_batch(enc_out, params['beam_width'])\n enc_state_t = tf.contrib.seq2seq.tile_batch(enc_state, params['beam_width'])\n enc_seq_len_t = tf.contrib.seq2seq.tile_batch(words_len, params['beam_width'])\n \n cell = dec_cell(enc_out_t, enc_seq_len_t)\n \n init_state = cell.zero_state(batch_sz*params['beam_width'], tf.float32).clone(\n cell_state=enc_state_t)\n \n decoder = tf.contrib.seq2seq.BeamSearchDecoder(\n cell = cell,\n embedding = embedding,\n start_tokens = tf.tile(tf.constant([1], tf.int32), [batch_sz]),\n end_token = 2,\n initial_state = init_state,\n beam_width = params['beam_width'],\n output_layer = output_proj,\n length_penalty_weight = params['length_penalty'],)\n decoder_output, _, _ = tf.contrib.seq2seq.dynamic_decode(\n decoder = decoder,\n maximum_iterations = params['max_len'],)\n \n return decoder_output.predicted_ids[:, :, :params['top_k']]",
"_____no_output_____"
],
[
"def clr(step,\n initial_learning_rate,\n maximal_learning_rate,\n step_size,\n scale_fn,\n scale_mode,):\n step = tf.cast(step, tf.float32)\n \n initial_learning_rate = tf.convert_to_tensor(\n initial_learning_rate, name='initial_learning_rate')\n dtype = initial_learning_rate.dtype\n maximal_learning_rate = tf.cast(maximal_learning_rate, dtype)\n step_size = tf.cast(step_size, dtype)\n cycle = tf.floor(1 + step / (2 * step_size))\n x = tf.abs(step / step_size - 2 * cycle + 1)\n\n mode_step = cycle if scale_mode == 'cycle' else step\n\n return initial_learning_rate + (\n maximal_learning_rate - initial_learning_rate) * tf.maximum(\n tf.cast(0, dtype), (1 - x)) * scale_fn(mode_step)",
"_____no_output_____"
],
[
"def model_fn(features, labels, mode, params):\n logits_or_ids = forward(features, labels, mode)\n \n if mode == tf.estimator.ModeKeys.PREDICT:\n return tf.estimator.EstimatorSpec(mode, predictions=logits_or_ids)\n \n dec_inputs, dec_outputs = labels\n if (params['label_smoothing'] <= .0) or (mode == tf.estimator.ModeKeys.EVAL):\n loss_op = tf.contrib.seq2seq.sequence_loss(logits = logits_or_ids,\n targets = dec_outputs,\n weights = tf.to_float(tf.sign(dec_outputs)))\n else:\n loss_op = tf.losses.softmax_cross_entropy(onehot_labels = tf.one_hot(dec_outputs, len(params['char2idx'])+1),\n logits = logits_or_ids,\n weights = tf.to_float(tf.sign(dec_outputs)),\n label_smoothing = params['label_smoothing'],)\n \n if mode == tf.estimator.ModeKeys.TRAIN:\n global_step=tf.train.get_or_create_global_step()\n \n decay_lr = clr(\n step = global_step,\n initial_learning_rate = 1e-4,\n maximal_learning_rate = 8e-4,\n step_size = params['num_samples'] / params['batch_size'] // 2,\n scale_fn=lambda x: 1 / (2.0 ** (x - 1)),\n scale_mode = 'cycle',)\n \n train_op = tf.train.AdamOptimizer(decay_lr).apply_gradients(\n clip_grads(loss_op), global_step=global_step)\n \n hook = tf.train.LoggingTensorHook({'lr': decay_lr}, every_n_iter=100)\n \n return tf.estimator.EstimatorSpec(\n mode=mode, loss=loss_op, train_op=train_op, training_hooks=[hook],)\n \n if mode == tf.estimator.ModeKeys.EVAL:\n return tf.estimator.EstimatorSpec(mode=mode, loss=loss_op)",
"_____no_output_____"
],
[
"def get_vocab(f_path):\n word2idx = {}\n with open(f_path) as f:\n for i, line in enumerate(f):\n line = line.rstrip('\\n')\n word2idx[line] = i\n return word2idx",
"_____no_output_____"
],
[
"def pad(test_strs):\n max_len = max([len(test_str) for test_str in test_strs])\n for test_str in test_strs:\n if len(test_str) < max_len:\n test_str += ['<pad>'] * (max_len - len(test_str))\n\n\ndef unit_test(estimator):\n test_strs = [\n '你好',\n '早上好',\n '晚上好',\n '再见',\n '好久不见',\n '想死你了',\n '谢谢你',\n '爱你',\n '你好厉害啊',\n '你叫什么',\n '你几岁了',\n '现在几点',\n '今天天气怎么样',\n '你今天心情好吗',\n '我们现在在哪里',\n '讲个笑话',\n '你会几种语言呀',\n '你觉得我帅吗',\n '讨厌的周一',\n '好烦啊',\n '天气真好',\n '今天好冷',\n '今天好热',\n '下雨了',\n '风好大',\n '终于周五了',\n '我想去唱歌',\n ]\n test_strs = [list(test_str) for test_str in test_strs]\n pad(test_strs)\n test_arrs = [[params['char2idx'].get(c, len(params['char2idx'])) for c in test_str] for test_str in test_strs]\n predicted = list(estimator.predict(tf.estimator.inputs.numpy_input_fn(\n x = np.asarray(test_arrs), shuffle = False)))\n predicted = np.asarray(predicted)\n print('-'*12)\n print('unit test')\n for i, test_str in enumerate(test_strs):\n print('Q:', ' '.join([c for c in test_str if c != '<pad>']))\n for j in range(params['top_k']):\n sent = ' '.join([params['idx2char'].get(idx, '<unk>') for idx in predicted[i, :, j] if (idx != 0 and idx != 2)])\n print('A{}:'.format(j+1), sent)\n print()\n print('-'*12)",
"_____no_output_____"
],
[
"class config_model:\n hidden_dim = 300\n num_heads = 8\n dropout_rate = .2\n num_blocks = 3\n\n position_embedder_hparams = {\n 'dim': hidden_dim\n }\n\n encoder = {\n 'dim': hidden_dim,\n 'embedding_dropout': dropout_rate,\n 'residual_dropout': dropout_rate,\n 'num_blocks': num_blocks,\n 'initializer': {\n 'type': 'variance_scaling_initializer',\n 'kwargs': {\n 'scale': 1.0,\n 'mode': 'fan_avg',\n 'distribution': 'uniform',\n },\n },\n 'multihead_attention': {\n 'dropout_rate': dropout_rate,\n 'num_heads': num_heads,\n 'output_dim': hidden_dim,\n 'use_bias': True,\n },\n 'poswise_feedforward': {\n 'name': 'fnn',\n 'layers': [\n {\n 'type': 'Dense',\n 'kwargs': {\n 'name': 'conv1',\n 'units': hidden_dim * 4,\n 'activation': 'gelu',\n 'use_bias': True,\n },\n },\n {\n 'type': 'Dropout',\n 'kwargs': {\n 'rate': dropout_rate,\n }\n },\n {\n 'type': 'Dense',\n 'kwargs': {\n 'name': 'conv2',\n 'units': hidden_dim,\n 'use_bias': True,\n }\n }\n ],\n },\n }\n\n\nparams = {\n 'model_dir': '../model/transformer_rnn',\n 'train_path': '../data/train.txt',\n 'test_path': '../data/test.txt',\n 'vocab_path': '../vocab/char.txt',\n 'max_len': 30,\n 'embed_dim': config_model.hidden_dim,\n 'rnn_units': 300,\n 'dec_layers': 1,\n 'dropout_rate': .2,\n 'beam_width': 10,\n 'top_k': 3,\n 'length_penalty': .6,\n 'label_smoothing': .2,\n 'clip_norm': .1,\n 'num_samples': 5000000,\n 'buffer_size': 500000,\n 'batch_size': 64,\n 'num_patience': 5,\n}",
"_____no_output_____"
],
[
"params['char2idx'] = get_vocab(params['vocab_path'])\nparams['idx2char'] = {idx: char for char, idx in params['char2idx'].items()}\nprint(len(params['char2idx']), 'Chars')\n\n# Create directory if not exist\nPath(params['model_dir']).mkdir(exist_ok=True, parents=True)\n\n# Logging\nlogger = logging.getLogger('tensorflow')\nlogger.setLevel(logging.INFO)\n\n# Create an estimator\nestimator = tf.estimator.Estimator(\n model_fn=model_fn,\n model_dir=params['model_dir'],\n config=tf.estimator.RunConfig(\n save_checkpoints_steps = params['num_samples'] // params['batch_size'] + 1,\n keep_checkpoint_max = 2),\n params=params)\n\nbest_ppl = 10000.\ncount = 0\ntf.enable_eager_execution()\n\nwhile True:\n estimator.train(input_fn=lambda: dataset(is_training=True, params=params))\n\n unit_test(estimator)\n \n loss = estimator.evaluate(input_fn=lambda: dataset(is_training=False, params=params))['loss']\n ppl = np.exp(loss)\n print(\"Perplexity: {:.3f}\".format(ppl))\n\n if ppl < best_ppl:\n best_ppl = ppl\n count = 0\n else:\n count += 1\n print(\"Best Perplexity: {:.3f}\".format(best_ppl))\n\n if count == params['num_patience']:\n print(params['num_patience'], \"times not improve the best result, therefore stop training\")\n break",
"3042 Chars\nINFO:tensorflow:Using config: {'_model_dir': '../model/transformer_rnn', '_tf_random_seed': None, '_save_summary_steps': 100, '_save_checkpoints_steps': 78126, '_save_checkpoints_secs': None, '_session_config': allow_soft_placement: true\ngraph_options {\n rewrite_options {\n meta_optimizer_iterations: ONE\n }\n}\n, '_keep_checkpoint_max': 2, '_keep_checkpoint_every_n_hours': 10000, '_log_step_count_steps': 100, '_train_distribute': None, '_device_fn': None, '_protocol': None, '_eval_distribute': None, '_experimental_distribute': None, '_experimental_max_worker_delay_secs': None, '_session_creation_timeout_secs': 7200, '_service': None, '_cluster_spec': <tensorflow.python.training.server_lib.ClusterSpec object at 0x7f6e9eacef60>, '_task_type': 'worker', '_task_id': 0, '_global_id_in_cluster': 0, '_master': '', '_evaluation_master': '', '_is_chief': True, '_num_ps_replicas': 0, '_num_worker_replicas': 1}\nWARNING:tensorflow:From /tensorflow-1.15.2/python3.6/tensorflow_core/python/training/training_util.py:236: Variable.initialized_value (from tensorflow.python.ops.variables) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse Variable.read_value. Variables in 2.X are initialized automatically both in eager and graph (inside tf.defun) contexts.\nINFO:tensorflow:Calling model_fn.\nWARNING:tensorflow:From /tensorflow-1.15.2/python3.6/tensorflow_core/python/util/deprecation.py:507: calling count_nonzero (from tensorflow.python.ops.math_ops) with axis is deprecated and will be removed in a future version.\nInstructions for updating:\nreduction_indices is deprecated, use axis instead\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/texar/tf/module_base.py:83: The name tf.make_template is deprecated. Please use tf.compat.v1.make_template instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/texar/tf/modules/embedders/position_embedders.py:338: The name tf.mod is deprecated. Please use tf.math.mod instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/texar/tf/modules/encoders/transformer_encoders.py:142: The name tf.get_variable_scope is deprecated. Please use tf.compat.v1.get_variable_scope instead.\n\nWARNING:tensorflow:From /usr/lib/python3.6/pydoc.py:1595: The name tf.layers.Dense is deprecated. Please use tf.compat.v1.layers.Dense instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/texar/tf/core/layers.py:608: The name tf.layers.Layer is deprecated. Please use tf.compat.v1.layers.Layer instead.\n\nWARNING:tensorflow:From /usr/lib/python3.6/pydoc.py:1595: The name tf.layers.Dropout is deprecated. Please use tf.compat.v1.layers.Dropout instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/texar/tf/modules/encoders/transformer_encoders.py:317: dropout (from tensorflow.python.layers.core) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse keras.layers.dropout instead.\nWARNING:tensorflow:From /tensorflow-1.15.2/python3.6/tensorflow_core/python/layers/core.py:271: Layer.apply (from tensorflow.python.keras.engine.base_layer) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `layer.__call__` method instead.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/texar/tf/utils/transformer_utils.py:73: where (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/texar/tf/module_base.py:140: The name tf.get_collection is deprecated. Please use tf.compat.v1.get_collection instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/texar/tf/module_base.py:141: The name tf.GraphKeys is deprecated. Please use tf.compat.v1.GraphKeys instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/texar/tf/modules/networks/network_base.py:125: The name tf.layers.BatchNormalization is deprecated. Please use tf.compat.v1.layers.BatchNormalization instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/texar/tf/core/layers.py:1270: The name tf.erf is deprecated. Please use tf.math.erf instead.\n\nWARNING:tensorflow:From <ipython-input-6-33c7bca471bf>:12: LSTMCell.__init__ (from tensorflow.python.ops.rnn_cell_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis class is equivalent as tf.keras.layers.LSTMCell, and will be replaced by that in Tensorflow 2.0.\nWARNING:tensorflow:From /tensorflow-1.15.2/python3.6/tensorflow_core/python/ops/rnn_cell_impl.py:958: Layer.add_variable (from tensorflow.python.keras.engine.base_layer) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `layer.add_weight` method instead.\nWARNING:tensorflow:From /tensorflow-1.15.2/python3.6/tensorflow_core/python/ops/rnn_cell_impl.py:962: calling Zeros.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nCall initializer instance with the dtype argument instead of passing it to the constructor\nWARNING:tensorflow:Entity <bound method TiedDense.call of <__main__.TiedDense object at 0x7f6e903914a8>> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nWARNING: Entity <bound method TiedDense.call of <__main__.TiedDense object at 0x7f6e903914a8>> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nWARNING:tensorflow:From <ipython-input-10-1d70a7bd47f9>:15: to_float (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.cast` instead.\n[<tf.Variable 'Embedding/fasttext_vectors:0' shape=(3043, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/query/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/query/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/key/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/key/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/value/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/value/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/output/kernel:0' shape=(512, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/output/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/output/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/output/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/fnn/conv1/kernel:0' shape=(300, 1200) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/fnn/conv1/bias:0' shape=(1200,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/fnn/conv2/kernel:0' shape=(1200, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/fnn/conv2/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/query/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/query/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/key/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/key/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/value/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/value/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/output/kernel:0' shape=(512, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/output/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/output/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/output/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/fnn/conv1/kernel:0' shape=(300, 1200) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/fnn/conv1/bias:0' shape=(1200,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/fnn/conv2/kernel:0' shape=(1200, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/fnn/conv2/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/query/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/query/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/key/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/key/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/value/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/value/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/output/kernel:0' shape=(512, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/output/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/output/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/output/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/fnn/conv1/kernel:0' shape=(300, 1200) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/fnn/conv1/bias:0' shape=(1200,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/fnn/conv2/kernel:0' shape=(1200, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/fnn/conv2/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Decoder/memory_layer/kernel:0' shape=(300, 300) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/attention_wrapper/lstm_cell/kernel:0' shape=(900, 1200) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/attention_wrapper/lstm_cell/bias:0' shape=(1200,) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/attention_wrapper/bahdanau_attention/query_layer/kernel:0' shape=(300, 300) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/attention_wrapper/bahdanau_attention/attention_v:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/attention_wrapper/attention_layer/kernel:0' shape=(600, 300) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/tied_dense/bias:0' shape=(3043,) dtype=float32_ref>]\nINFO:tensorflow:Done calling model_fn.\nINFO:tensorflow:Create CheckpointSaverHook.\nINFO:tensorflow:Graph was finalized.\nINFO:tensorflow:Running local_init_op.\nINFO:tensorflow:Done running local_init_op.\nINFO:tensorflow:Saving checkpoints for 0 into ../model/transformer_rnn/model.ckpt.\nReading ../data/train.txt\nINFO:tensorflow:loss = 8.937515, step = 0\nINFO:tensorflow:lr = 1e-04\nINFO:tensorflow:global_step/sec: 8.71145\nINFO:tensorflow:loss = 6.517472, step = 100 (11.480 sec)\nINFO:tensorflow:lr = 0.00010179201 (11.481 sec)\nINFO:tensorflow:global_step/sec: 9.61915\nINFO:tensorflow:loss = 6.4360666, step = 200 (10.397 sec)\nINFO:tensorflow:lr = 0.000103584025 (10.397 sec)\nINFO:tensorflow:global_step/sec: 9.47261\nINFO:tensorflow:loss = 6.410827, step = 300 (10.555 sec)\nINFO:tensorflow:lr = 0.00010537608 (10.556 sec)\nINFO:tensorflow:global_step/sec: 9.76318\nINFO:tensorflow:loss = 6.240726, step = 400 (10.243 sec)\nINFO:tensorflow:lr = 0.000107168096 (10.244 sec)\nINFO:tensorflow:global_step/sec: 9.38388\nINFO:tensorflow:loss = 6.329311, step = 500 (10.656 sec)\nINFO:tensorflow:lr = 0.00010896011 (10.656 sec)\nINFO:tensorflow:global_step/sec: 9.68245\nINFO:tensorflow:loss = 6.216666, step = 600 (10.330 sec)\nINFO:tensorflow:lr = 0.000110752124 (10.331 sec)\nINFO:tensorflow:global_step/sec: 9.41577\nINFO:tensorflow:loss = 6.192756, step = 700 (10.618 sec)\nINFO:tensorflow:lr = 0.00011254418 (10.616 sec)\nINFO:tensorflow:global_step/sec: 9.78002\nINFO:tensorflow:loss = 6.1685157, step = 800 (10.227 sec)\nINFO:tensorflow:lr = 0.00011433619 (10.226 sec)\nINFO:tensorflow:global_step/sec: 9.49643\nINFO:tensorflow:loss = 6.175221, step = 900 (10.531 sec)\nINFO:tensorflow:lr = 0.000116128205 (10.531 sec)\nINFO:tensorflow:global_step/sec: 9.46536\nINFO:tensorflow:loss = 6.014949, step = 1000 (10.564 sec)\nINFO:tensorflow:lr = 0.000117920215 (10.564 sec)\nINFO:tensorflow:global_step/sec: 9.70079\nINFO:tensorflow:loss = 6.086758, step = 1100 (10.310 sec)\nINFO:tensorflow:lr = 0.00011971223 (10.310 sec)\nINFO:tensorflow:global_step/sec: 9.64372\nINFO:tensorflow:loss = 5.9653287, step = 1200 (10.368 sec)\nINFO:tensorflow:lr = 0.00012150429 (10.368 sec)\nINFO:tensorflow:global_step/sec: 9.50996\nINFO:tensorflow:loss = 5.899817, step = 1300 (10.516 sec)\nINFO:tensorflow:lr = 0.0001232963 (10.517 sec)\nINFO:tensorflow:global_step/sec: 9.67609\nINFO:tensorflow:loss = 5.9957976, step = 1400 (10.334 sec)\nINFO:tensorflow:lr = 0.0001250883 (10.335 sec)\nINFO:tensorflow:global_step/sec: 9.51666\nINFO:tensorflow:loss = 5.886746, step = 1500 (10.509 sec)\nINFO:tensorflow:lr = 0.00012688033 (10.508 sec)\nINFO:tensorflow:global_step/sec: 9.42603\nINFO:tensorflow:loss = 5.846593, step = 1600 (10.610 sec)\nINFO:tensorflow:lr = 0.00012867239 (10.610 sec)\nINFO:tensorflow:global_step/sec: 9.63493\nINFO:tensorflow:loss = 5.91529, step = 1700 (10.375 sec)\nINFO:tensorflow:lr = 0.0001304644 (10.377 sec)\nINFO:tensorflow:global_step/sec: 9.46334\nINFO:tensorflow:loss = 5.8557997, step = 1800 (10.570 sec)\nINFO:tensorflow:lr = 0.0001322564 (10.569 sec)\nINFO:tensorflow:global_step/sec: 9.35606\nINFO:tensorflow:loss = 5.9050403, step = 1900 (10.687 sec)\nINFO:tensorflow:lr = 0.00013404843 (10.688 sec)\nINFO:tensorflow:global_step/sec: 9.54904\nINFO:tensorflow:loss = 5.8140674, step = 2000 (10.474 sec)\nINFO:tensorflow:lr = 0.00013584044 (10.473 sec)\nINFO:tensorflow:global_step/sec: 9.35126\nINFO:tensorflow:loss = 5.8583922, step = 2100 (10.692 sec)\nINFO:tensorflow:lr = 0.0001376325 (10.692 sec)\nINFO:tensorflow:global_step/sec: 9.63218\nINFO:tensorflow:loss = 5.758723, step = 2200 (10.383 sec)\nINFO:tensorflow:lr = 0.0001394245 (10.382 sec)\nINFO:tensorflow:global_step/sec: 9.42249\nINFO:tensorflow:loss = 5.5649285, step = 2300 (10.610 sec)\nINFO:tensorflow:lr = 0.00014121651 (10.611 sec)\nINFO:tensorflow:global_step/sec: 9.43176\nINFO:tensorflow:loss = 5.705367, step = 2400 (10.606 sec)\nINFO:tensorflow:lr = 0.00014300854 (10.605 sec)\nINFO:tensorflow:global_step/sec: 9.35512\nINFO:tensorflow:loss = 5.627061, step = 2500 (10.688 sec)\nINFO:tensorflow:lr = 0.00014480058 (10.688 sec)\nINFO:tensorflow:global_step/sec: 9.30433\nINFO:tensorflow:loss = 5.874296, step = 2600 (10.746 sec)\nINFO:tensorflow:lr = 0.0001465926 (10.746 sec)\nINFO:tensorflow:global_step/sec: 9.62686\nINFO:tensorflow:loss = 5.717611, step = 2700 (10.389 sec)\nINFO:tensorflow:lr = 0.00014838461 (10.388 sec)\nINFO:tensorflow:global_step/sec: 9.42155\nINFO:tensorflow:loss = 5.7843623, step = 2800 (10.615 sec)\nINFO:tensorflow:lr = 0.00015017662 (10.616 sec)\nINFO:tensorflow:global_step/sec: 9.37644\nINFO:tensorflow:loss = 5.6821647, step = 2900 (10.663 sec)\nINFO:tensorflow:lr = 0.00015196865 (10.662 sec)\nINFO:tensorflow:global_step/sec: 9.7579\nINFO:tensorflow:loss = 5.754989, step = 3000 (10.250 sec)\nINFO:tensorflow:lr = 0.0001537607 (10.251 sec)\nINFO:tensorflow:global_step/sec: 9.52185\nINFO:tensorflow:loss = 5.6745124, step = 3100 (10.502 sec)\nINFO:tensorflow:lr = 0.00015555271 (10.499 sec)\nINFO:tensorflow:global_step/sec: 9.37107\nINFO:tensorflow:loss = 5.59257, step = 3200 (10.672 sec)\nINFO:tensorflow:lr = 0.00015734472 (10.673 sec)\nINFO:tensorflow:global_step/sec: 9.61674\nINFO:tensorflow:loss = 5.575217, step = 3300 (10.399 sec)\nINFO:tensorflow:lr = 0.00015913673 (10.399 sec)\nINFO:tensorflow:global_step/sec: 9.17963\nINFO:tensorflow:loss = 5.610762, step = 3400 (10.894 sec)\nINFO:tensorflow:lr = 0.00016092879 (10.896 sec)\nINFO:tensorflow:global_step/sec: 9.4759\nINFO:tensorflow:loss = 5.8031707, step = 3500 (10.554 sec)\nINFO:tensorflow:lr = 0.0001627208 (10.555 sec)\nINFO:tensorflow:global_step/sec: 9.22768\nINFO:tensorflow:loss = 5.616017, step = 3600 (10.835 sec)\nINFO:tensorflow:lr = 0.00016451282 (10.833 sec)\nINFO:tensorflow:global_step/sec: 9.45921\nINFO:tensorflow:loss = 5.503318, step = 3700 (10.575 sec)\nINFO:tensorflow:lr = 0.00016630483 (10.574 sec)\nINFO:tensorflow:global_step/sec: 9.65632\nINFO:tensorflow:loss = 5.469181, step = 3800 (10.354 sec)\nINFO:tensorflow:lr = 0.00016809686 (10.353 sec)\nINFO:tensorflow:global_step/sec: 9.63209\nINFO:tensorflow:loss = 5.605735, step = 3900 (10.381 sec)\nINFO:tensorflow:lr = 0.0001698889 (10.380 sec)\nINFO:tensorflow:global_step/sec: 9.78939\nINFO:tensorflow:loss = 5.615002, step = 4000 (10.218 sec)\nINFO:tensorflow:lr = 0.0001716809 (10.218 sec)\nINFO:tensorflow:global_step/sec: 9.52705\nINFO:tensorflow:loss = 5.3957057, step = 4100 (10.494 sec)\nINFO:tensorflow:lr = 0.00017347293 (10.496 sec)\nINFO:tensorflow:global_step/sec: 9.82795\nINFO:tensorflow:loss = 5.737772, step = 4200 (10.173 sec)\nINFO:tensorflow:lr = 0.00017526494 (10.173 sec)\nINFO:tensorflow:global_step/sec: 9.77213\nINFO:tensorflow:loss = 5.41721, step = 4300 (10.233 sec)\nINFO:tensorflow:lr = 0.000177057 (10.232 sec)\nINFO:tensorflow:global_step/sec: 9.5106\nINFO:tensorflow:loss = 5.3590026, step = 4400 (10.517 sec)\nINFO:tensorflow:lr = 0.000178849 (10.518 sec)\nINFO:tensorflow:global_step/sec: 9.76941\nINFO:tensorflow:loss = 5.5810437, step = 4500 (10.236 sec)\nINFO:tensorflow:lr = 0.00018064103 (10.235 sec)\nINFO:tensorflow:global_step/sec: 9.41166\nINFO:tensorflow:loss = 5.559284, step = 4600 (10.627 sec)\nINFO:tensorflow:lr = 0.00018243304 (10.626 sec)\nINFO:tensorflow:global_step/sec: 9.56269\nINFO:tensorflow:loss = 5.490581, step = 4700 (10.455 sec)\nINFO:tensorflow:lr = 0.00018422505 (10.455 sec)\nINFO:tensorflow:global_step/sec: 9.64462\nINFO:tensorflow:loss = 5.5523834, step = 4800 (10.370 sec)\nINFO:tensorflow:lr = 0.0001860171 (10.370 sec)\nINFO:tensorflow:global_step/sec: 9.65217\nINFO:tensorflow:loss = 5.3931236, step = 4900 (10.359 sec)\nINFO:tensorflow:lr = 0.00018780911 (10.360 sec)\nINFO:tensorflow:global_step/sec: 9.29587\nINFO:tensorflow:loss = 5.4739566, step = 5000 (10.757 sec)\nINFO:tensorflow:lr = 0.00018960114 (10.759 sec)\nINFO:tensorflow:global_step/sec: 9.58949\nINFO:tensorflow:loss = 5.5099225, step = 5100 (10.428 sec)\nINFO:tensorflow:lr = 0.00019139315 (10.426 sec)\nINFO:tensorflow:global_step/sec: 9.34559\nINFO:tensorflow:loss = 5.263151, step = 5200 (10.700 sec)\nINFO:tensorflow:lr = 0.0001931852 (10.699 sec)\nINFO:tensorflow:global_step/sec: 9.36656\nINFO:tensorflow:loss = 5.316375, step = 5300 (10.677 sec)\nINFO:tensorflow:lr = 0.00019497721 (10.678 sec)\nINFO:tensorflow:global_step/sec: 9.36721\nINFO:tensorflow:loss = 5.5502787, step = 5400 (10.676 sec)\nINFO:tensorflow:lr = 0.00019676922 (10.675 sec)\nINFO:tensorflow:global_step/sec: 9.64089\nINFO:tensorflow:loss = 5.6246333, step = 5500 (10.372 sec)\nINFO:tensorflow:lr = 0.00019856125 (10.372 sec)\nINFO:tensorflow:global_step/sec: 9.69617\nINFO:tensorflow:loss = 5.522617, step = 5600 (10.314 sec)\nINFO:tensorflow:lr = 0.00020035326 (10.314 sec)\nINFO:tensorflow:global_step/sec: 9.22217\nINFO:tensorflow:loss = 5.4793525, step = 5700 (10.843 sec)\nINFO:tensorflow:lr = 0.00020214531 (10.843 sec)\nINFO:tensorflow:global_step/sec: 9.59383\nINFO:tensorflow:loss = 5.429895, step = 5800 (10.425 sec)\nINFO:tensorflow:lr = 0.00020393732 (10.424 sec)\nINFO:tensorflow:global_step/sec: 9.47609\nINFO:tensorflow:loss = 5.4219847, step = 5900 (10.552 sec)\nINFO:tensorflow:lr = 0.00020572933 (10.553 sec)\nINFO:tensorflow:global_step/sec: 9.64629\nINFO:tensorflow:loss = 5.4989614, step = 6000 (10.364 sec)\nINFO:tensorflow:lr = 0.00020752136 (10.366 sec)\nINFO:tensorflow:global_step/sec: 9.47022\nINFO:tensorflow:loss = 5.4147143, step = 6100 (10.561 sec)\nINFO:tensorflow:lr = 0.00020931341 (10.560 sec)\nINFO:tensorflow:global_step/sec: 9.55096\nINFO:tensorflow:loss = 5.580407, step = 6200 (10.472 sec)\nINFO:tensorflow:lr = 0.00021110542 (10.472 sec)\nINFO:tensorflow:global_step/sec: 9.10851\nINFO:tensorflow:loss = 5.465265, step = 6300 (10.976 sec)\nINFO:tensorflow:lr = 0.00021289743 (10.977 sec)\nINFO:tensorflow:global_step/sec: 9.62073\nINFO:tensorflow:loss = 5.5079503, step = 6400 (10.396 sec)\nINFO:tensorflow:lr = 0.00021468944 (10.395 sec)\nINFO:tensorflow:global_step/sec: 9.4835\nINFO:tensorflow:loss = 5.3989377, step = 6500 (10.543 sec)\nINFO:tensorflow:lr = 0.00021648147 (10.544 sec)\nINFO:tensorflow:global_step/sec: 9.23903\nINFO:tensorflow:loss = 5.455261, step = 6600 (10.824 sec)\nINFO:tensorflow:lr = 0.00021827352 (10.824 sec)\nINFO:tensorflow:global_step/sec: 9.52242\nINFO:tensorflow:loss = 5.354864, step = 6700 (10.499 sec)\nINFO:tensorflow:lr = 0.00022006553 (10.499 sec)\nINFO:tensorflow:global_step/sec: 9.50308\nINFO:tensorflow:loss = 5.3888826, step = 6800 (10.525 sec)\nINFO:tensorflow:lr = 0.00022185754 (10.525 sec)\nINFO:tensorflow:global_step/sec: 9.42782\nINFO:tensorflow:loss = 5.4703174, step = 6900 (10.612 sec)\nINFO:tensorflow:lr = 0.00022364955 (10.610 sec)\nINFO:tensorflow:global_step/sec: 9.46152\nINFO:tensorflow:loss = 5.257609, step = 7000 (10.565 sec)\nINFO:tensorflow:lr = 0.00022544162 (10.566 sec)\nINFO:tensorflow:global_step/sec: 9.71523\nINFO:tensorflow:loss = 5.425678, step = 7100 (10.293 sec)\nINFO:tensorflow:lr = 0.00022723363 (10.293 sec)\nINFO:tensorflow:global_step/sec: 9.41054\nINFO:tensorflow:loss = 5.468957, step = 7200 (10.625 sec)\nINFO:tensorflow:lr = 0.00022902564 (10.625 sec)\nINFO:tensorflow:global_step/sec: 9.5384\nINFO:tensorflow:loss = 5.490213, step = 7300 (10.484 sec)\nINFO:tensorflow:lr = 0.00023081765 (10.485 sec)\nINFO:tensorflow:global_step/sec: 9.38962\nINFO:tensorflow:loss = 5.4084835, step = 7400 (10.650 sec)\nINFO:tensorflow:lr = 0.00023260966 (10.650 sec)\nINFO:tensorflow:global_step/sec: 9.44578\nINFO:tensorflow:loss = 5.2608895, step = 7500 (10.586 sec)\nINFO:tensorflow:lr = 0.00023440173 (10.586 sec)\nINFO:tensorflow:global_step/sec: 9.36414\nINFO:tensorflow:loss = 5.4410133, step = 7600 (10.681 sec)\nINFO:tensorflow:lr = 0.00023619374 (10.680 sec)\nINFO:tensorflow:global_step/sec: 9.55588\nINFO:tensorflow:loss = 5.4226823, step = 7700 (10.461 sec)\nINFO:tensorflow:lr = 0.00023798575 (10.461 sec)\nINFO:tensorflow:global_step/sec: 9.60524\nINFO:tensorflow:loss = 5.445272, step = 7800 (10.415 sec)\nINFO:tensorflow:lr = 0.00023977782 (10.416 sec)\nINFO:tensorflow:global_step/sec: 9.69397\nINFO:tensorflow:loss = 5.3613033, step = 7900 (10.315 sec)\nINFO:tensorflow:lr = 0.00024156983 (10.314 sec)\nINFO:tensorflow:global_step/sec: 9.31659\nINFO:tensorflow:loss = 5.3422837, step = 8000 (10.732 sec)\nINFO:tensorflow:lr = 0.00024336184 (10.733 sec)\nINFO:tensorflow:global_step/sec: 9.05782\nINFO:tensorflow:loss = 5.2976117, step = 8100 (11.041 sec)\nINFO:tensorflow:lr = 0.00024515385 (11.041 sec)\nINFO:tensorflow:global_step/sec: 9.30279\nINFO:tensorflow:loss = 5.369702, step = 8200 (10.750 sec)\nINFO:tensorflow:lr = 0.00024694586 (10.750 sec)\nINFO:tensorflow:global_step/sec: 9.33253\nINFO:tensorflow:loss = 5.3575172, step = 8300 (10.719 sec)\nINFO:tensorflow:lr = 0.00024873787 (10.719 sec)\nINFO:tensorflow:global_step/sec: 9.33383\nINFO:tensorflow:loss = 5.2213154, step = 8400 (10.708 sec)\nINFO:tensorflow:lr = 0.00025052993 (10.708 sec)\nINFO:tensorflow:global_step/sec: 9.49957\nINFO:tensorflow:loss = 5.300407, step = 8500 (10.529 sec)\nINFO:tensorflow:lr = 0.00025232194 (10.529 sec)\nINFO:tensorflow:global_step/sec: 9.51828\nINFO:tensorflow:loss = 5.315953, step = 8600 (10.506 sec)\nINFO:tensorflow:lr = 0.00025411395 (10.506 sec)\nINFO:tensorflow:global_step/sec: 9.24184\nINFO:tensorflow:loss = 5.412649, step = 8700 (10.818 sec)\nINFO:tensorflow:lr = 0.00025590602 (10.818 sec)\nINFO:tensorflow:global_step/sec: 9.24331\nINFO:tensorflow:loss = 5.3078456, step = 8800 (10.819 sec)\nINFO:tensorflow:lr = 0.00025769803 (10.820 sec)\nINFO:tensorflow:global_step/sec: 9.1363\nINFO:tensorflow:loss = 5.4023676, step = 8900 (10.947 sec)\nINFO:tensorflow:lr = 0.00025949004 (10.946 sec)\nINFO:tensorflow:global_step/sec: 8.98517\nINFO:tensorflow:loss = 5.483456, step = 9000 (11.130 sec)\nINFO:tensorflow:lr = 0.00026128205 (11.130 sec)\nINFO:tensorflow:global_step/sec: 9.49301\nINFO:tensorflow:loss = 5.274673, step = 9100 (10.532 sec)\nINFO:tensorflow:lr = 0.00026307406 (10.534 sec)\nINFO:tensorflow:global_step/sec: 9.47168\nINFO:tensorflow:loss = 5.335271, step = 9200 (10.560 sec)\nINFO:tensorflow:lr = 0.00026486607 (10.558 sec)\nINFO:tensorflow:global_step/sec: 9.68788\nINFO:tensorflow:loss = 5.4493904, step = 9300 (10.323 sec)\nINFO:tensorflow:lr = 0.00026665814 (10.322 sec)\nINFO:tensorflow:global_step/sec: 9.57234\nINFO:tensorflow:loss = 5.276461, step = 9400 (10.445 sec)\nINFO:tensorflow:lr = 0.00026845015 (10.446 sec)\nINFO:tensorflow:global_step/sec: 9.63206\nINFO:tensorflow:loss = 5.2798014, step = 9500 (10.383 sec)\nINFO:tensorflow:lr = 0.00027024216 (10.383 sec)\nINFO:tensorflow:global_step/sec: 9.16588\nINFO:tensorflow:loss = 5.3169374, step = 9600 (10.911 sec)\nINFO:tensorflow:lr = 0.00027203423 (10.911 sec)\nINFO:tensorflow:global_step/sec: 9.43001\nINFO:tensorflow:loss = 5.4106855, step = 9700 (10.603 sec)\nINFO:tensorflow:lr = 0.00027382624 (10.603 sec)\nINFO:tensorflow:global_step/sec: 9.23734\nINFO:tensorflow:loss = 5.334615, step = 9800 (10.824 sec)\nINFO:tensorflow:lr = 0.00027561825 (10.825 sec)\nINFO:tensorflow:global_step/sec: 9.48916\nINFO:tensorflow:loss = 5.2714796, step = 9900 (10.538 sec)\nINFO:tensorflow:lr = 0.00027741026 (10.539 sec)\nINFO:tensorflow:global_step/sec: 9.45148\nINFO:tensorflow:loss = 5.395485, step = 10000 (10.583 sec)\nINFO:tensorflow:lr = 0.00027920227 (10.581 sec)\nINFO:tensorflow:global_step/sec: 9.40397\nINFO:tensorflow:loss = 5.2520475, step = 10100 (10.634 sec)\nINFO:tensorflow:lr = 0.00028099428 (10.635 sec)\nINFO:tensorflow:global_step/sec: 9.40283\nINFO:tensorflow:loss = 5.1954203, step = 10200 (10.634 sec)\nINFO:tensorflow:lr = 0.0002827863 (10.633 sec)\nINFO:tensorflow:global_step/sec: 9.40064\nINFO:tensorflow:loss = 5.2193794, step = 10300 (10.641 sec)\nINFO:tensorflow:lr = 0.00028457836 (10.641 sec)\nINFO:tensorflow:global_step/sec: 9.34103\nINFO:tensorflow:loss = 5.309771, step = 10400 (10.703 sec)\nINFO:tensorflow:lr = 0.00028637037 (10.702 sec)\nINFO:tensorflow:global_step/sec: 9.19379\nINFO:tensorflow:loss = 5.3481083, step = 10500 (10.878 sec)\nINFO:tensorflow:lr = 0.00028816244 (10.878 sec)\nINFO:tensorflow:global_step/sec: 9.43225\nINFO:tensorflow:loss = 5.2674665, step = 10600 (10.601 sec)\nINFO:tensorflow:lr = 0.00028995445 (10.602 sec)\nINFO:tensorflow:global_step/sec: 9.05061\nINFO:tensorflow:loss = 5.3742795, step = 10700 (11.048 sec)\nINFO:tensorflow:lr = 0.00029174646 (11.048 sec)\nINFO:tensorflow:global_step/sec: 9.13986\nINFO:tensorflow:loss = 5.3197327, step = 10800 (10.943 sec)\nINFO:tensorflow:lr = 0.00029353847 (10.943 sec)\nINFO:tensorflow:global_step/sec: 8.81399\nINFO:tensorflow:loss = 5.2734423, step = 10900 (11.343 sec)\nINFO:tensorflow:lr = 0.00029533048 (11.343 sec)\nINFO:tensorflow:global_step/sec: 9.6329\nINFO:tensorflow:loss = 5.4017906, step = 11000 (10.381 sec)\nINFO:tensorflow:lr = 0.0002971225 (10.381 sec)\nINFO:tensorflow:global_step/sec: 9.39276\nINFO:tensorflow:loss = 5.2871704, step = 11100 (10.646 sec)\nINFO:tensorflow:lr = 0.0002989145 (10.646 sec)\nINFO:tensorflow:global_step/sec: 8.89523\nINFO:tensorflow:loss = 5.2185893, step = 11200 (11.246 sec)\nINFO:tensorflow:lr = 0.00030070657 (11.244 sec)\nINFO:tensorflow:global_step/sec: 9.39774\nINFO:tensorflow:loss = 5.3076186, step = 11300 (10.639 sec)\nINFO:tensorflow:lr = 0.00030249858 (10.640 sec)\nINFO:tensorflow:global_step/sec: 9.85811\nINFO:tensorflow:loss = 5.3705683, step = 11400 (10.146 sec)\nINFO:tensorflow:lr = 0.00030429062 (10.144 sec)\nINFO:tensorflow:global_step/sec: 9.47035\nINFO:tensorflow:loss = 5.3904276, step = 11500 (10.557 sec)\nINFO:tensorflow:lr = 0.00030608266 (10.559 sec)\nINFO:tensorflow:global_step/sec: 9.28909\nINFO:tensorflow:loss = 5.2577066, step = 11600 (10.765 sec)\nINFO:tensorflow:lr = 0.00030787467 (10.764 sec)\nINFO:tensorflow:global_step/sec: 9.84435\nINFO:tensorflow:loss = 5.3427906, step = 11700 (10.157 sec)\nINFO:tensorflow:lr = 0.00030966668 (10.157 sec)\nINFO:tensorflow:global_step/sec: 9.8187\nINFO:tensorflow:loss = 5.196344, step = 11800 (10.184 sec)\nINFO:tensorflow:lr = 0.0003114587 (10.187 sec)\nINFO:tensorflow:global_step/sec: 9.58391\nINFO:tensorflow:loss = 5.1877666, step = 11900 (10.437 sec)\nINFO:tensorflow:lr = 0.0003132507 (10.435 sec)\nINFO:tensorflow:global_step/sec: 9.5932\nINFO:tensorflow:loss = 5.273709, step = 12000 (10.423 sec)\nINFO:tensorflow:lr = 0.0003150427 (10.423 sec)\nINFO:tensorflow:global_step/sec: 9.53313\nINFO:tensorflow:loss = 5.275315, step = 12100 (10.490 sec)\nINFO:tensorflow:lr = 0.00031683478 (10.490 sec)\nINFO:tensorflow:global_step/sec: 9.24848\nINFO:tensorflow:loss = 5.347632, step = 12200 (10.811 sec)\nINFO:tensorflow:lr = 0.0003186268 (10.810 sec)\nINFO:tensorflow:global_step/sec: 9.76986\nINFO:tensorflow:loss = 5.1148157, step = 12300 (10.239 sec)\nINFO:tensorflow:lr = 0.00032041883 (10.240 sec)\nINFO:tensorflow:global_step/sec: 9.41562\nINFO:tensorflow:loss = 5.227973, step = 12400 (10.619 sec)\nINFO:tensorflow:lr = 0.00032221084 (10.619 sec)\nINFO:tensorflow:global_step/sec: 9.49199\nINFO:tensorflow:loss = 5.2387366, step = 12500 (10.534 sec)\nINFO:tensorflow:lr = 0.00032400287 (10.533 sec)\nINFO:tensorflow:global_step/sec: 9.57954\nINFO:tensorflow:loss = 5.0977798, step = 12600 (10.439 sec)\nINFO:tensorflow:lr = 0.00032579488 (10.440 sec)\nINFO:tensorflow:global_step/sec: 9.33589\nINFO:tensorflow:loss = 5.2063737, step = 12700 (10.710 sec)\nINFO:tensorflow:lr = 0.0003275869 (10.710 sec)\nINFO:tensorflow:global_step/sec: 9.10658\nINFO:tensorflow:loss = 5.295964, step = 12800 (10.991 sec)\nINFO:tensorflow:lr = 0.0003293789 (10.992 sec)\nINFO:tensorflow:global_step/sec: 9.52902\nINFO:tensorflow:loss = 5.3541036, step = 12900 (10.486 sec)\nINFO:tensorflow:lr = 0.00033117092 (10.485 sec)\nINFO:tensorflow:global_step/sec: 9.67691\nINFO:tensorflow:loss = 5.1890435, step = 13000 (10.332 sec)\nINFO:tensorflow:lr = 0.00033296298 (10.333 sec)\nINFO:tensorflow:global_step/sec: 9.59669\nINFO:tensorflow:loss = 5.0991793, step = 13100 (10.423 sec)\nINFO:tensorflow:lr = 0.000334755 (10.421 sec)\nINFO:tensorflow:global_step/sec: 9.23686\nINFO:tensorflow:loss = 5.2923245, step = 13200 (10.825 sec)\nINFO:tensorflow:lr = 0.00033654703 (10.825 sec)\nINFO:tensorflow:global_step/sec: 9.38213\nINFO:tensorflow:loss = 5.431452, step = 13300 (10.660 sec)\nINFO:tensorflow:lr = 0.00033833904 (10.660 sec)\nINFO:tensorflow:global_step/sec: 9.31188\nINFO:tensorflow:loss = 5.1751623, step = 13400 (10.741 sec)\nINFO:tensorflow:lr = 0.00034013108 (10.742 sec)\nINFO:tensorflow:global_step/sec: 9.24699\nINFO:tensorflow:loss = 5.3359776, step = 13500 (10.809 sec)\nINFO:tensorflow:lr = 0.0003419231 (10.809 sec)\nINFO:tensorflow:global_step/sec: 9.41265\nINFO:tensorflow:loss = 5.3060904, step = 13600 (10.627 sec)\nINFO:tensorflow:lr = 0.0003437151 (10.627 sec)\nINFO:tensorflow:global_step/sec: 9.25623\nINFO:tensorflow:loss = 5.199159, step = 13700 (10.801 sec)\nINFO:tensorflow:lr = 0.0003455071 (10.802 sec)\nINFO:tensorflow:global_step/sec: 9.47971\nINFO:tensorflow:loss = 5.248309, step = 13800 (10.551 sec)\nINFO:tensorflow:lr = 0.00034729912 (10.550 sec)\nINFO:tensorflow:global_step/sec: 9.4577\nINFO:tensorflow:loss = 5.3227744, step = 13900 (10.575 sec)\nINFO:tensorflow:lr = 0.0003490912 (10.574 sec)\nINFO:tensorflow:global_step/sec: 9.80108\nINFO:tensorflow:loss = 5.2083745, step = 14000 (10.201 sec)\nINFO:tensorflow:lr = 0.0003508832 (10.202 sec)\nINFO:tensorflow:global_step/sec: 9.12215\nINFO:tensorflow:loss = 5.0114703, step = 14100 (10.964 sec)\nINFO:tensorflow:lr = 0.00035267524 (10.963 sec)\nINFO:tensorflow:global_step/sec: 9.48393\nINFO:tensorflow:loss = 5.240639, step = 14200 (10.543 sec)\nINFO:tensorflow:lr = 0.00035446728 (10.544 sec)\nINFO:tensorflow:global_step/sec: 8.99056\nINFO:tensorflow:loss = 5.213593, step = 14300 (11.122 sec)\nINFO:tensorflow:lr = 0.0003562593 (11.121 sec)\nINFO:tensorflow:global_step/sec: 9.37065\nINFO:tensorflow:loss = 5.1901445, step = 14400 (10.670 sec)\nINFO:tensorflow:lr = 0.0003580513 (10.674 sec)\nINFO:tensorflow:global_step/sec: 9.02198\nINFO:tensorflow:loss = 5.0916905, step = 14500 (11.086 sec)\nINFO:tensorflow:lr = 0.0003598433 (11.083 sec)\nINFO:tensorflow:global_step/sec: 9.33291\nINFO:tensorflow:loss = 5.1945896, step = 14600 (10.715 sec)\nINFO:tensorflow:lr = 0.00036163532 (10.715 sec)\nINFO:tensorflow:global_step/sec: 9.47573\nINFO:tensorflow:loss = 5.136004, step = 14700 (10.552 sec)\nINFO:tensorflow:lr = 0.00036342733 (10.553 sec)\nINFO:tensorflow:global_step/sec: 9.61313\nINFO:tensorflow:loss = 5.096282, step = 14800 (10.404 sec)\nINFO:tensorflow:lr = 0.0003652194 (10.403 sec)\nINFO:tensorflow:global_step/sec: 9.48212\nINFO:tensorflow:loss = 5.1238246, step = 14900 (10.546 sec)\nINFO:tensorflow:lr = 0.0003670114 (10.546 sec)\nINFO:tensorflow:global_step/sec: 9.17427\nINFO:tensorflow:loss = 5.2331834, step = 15000 (10.900 sec)\nINFO:tensorflow:lr = 0.00036880345 (10.900 sec)\nINFO:tensorflow:global_step/sec: 9.52248\nINFO:tensorflow:loss = 5.3915863, step = 15100 (10.501 sec)\nINFO:tensorflow:lr = 0.0003705955 (10.500 sec)\nINFO:tensorflow:global_step/sec: 9.50163\nINFO:tensorflow:loss = 5.1143703, step = 15200 (10.523 sec)\nINFO:tensorflow:lr = 0.0003723875 (10.523 sec)\nINFO:tensorflow:global_step/sec: 9.51777\nINFO:tensorflow:loss = 5.182392, step = 15300 (10.506 sec)\nINFO:tensorflow:lr = 0.0003741795 (10.507 sec)\nINFO:tensorflow:global_step/sec: 9.58017\nINFO:tensorflow:loss = 5.202959, step = 15400 (10.440 sec)\nINFO:tensorflow:lr = 0.00037597152 (10.439 sec)\nINFO:tensorflow:global_step/sec: 9.61115\nINFO:tensorflow:loss = 5.3520365, step = 15500 (10.404 sec)\nINFO:tensorflow:lr = 0.00037776353 (10.405 sec)\nINFO:tensorflow:global_step/sec: 9.4878\nINFO:tensorflow:loss = 5.1896334, step = 15600 (10.540 sec)\nINFO:tensorflow:lr = 0.0003795556 (10.540 sec)\nINFO:tensorflow:global_step/sec: 9.52809\nINFO:tensorflow:loss = 5.2342257, step = 15700 (10.496 sec)\nINFO:tensorflow:lr = 0.0003813476 (10.495 sec)\nINFO:tensorflow:global_step/sec: 9.4086\nINFO:tensorflow:loss = 5.2121305, step = 15800 (10.629 sec)\nINFO:tensorflow:lr = 0.00038313962 (10.629 sec)\nINFO:tensorflow:global_step/sec: 9.38529\nINFO:tensorflow:loss = 5.181575, step = 15900 (10.655 sec)\nINFO:tensorflow:lr = 0.00038493166 (10.654 sec)\nINFO:tensorflow:global_step/sec: 9.37624\nINFO:tensorflow:loss = 5.264327, step = 16000 (10.666 sec)\nINFO:tensorflow:lr = 0.00038672367 (10.666 sec)\nINFO:tensorflow:global_step/sec: 8.98048\nINFO:tensorflow:loss = 5.263517, step = 16100 (11.134 sec)\nINFO:tensorflow:lr = 0.0003885157 (11.136 sec)\nINFO:tensorflow:global_step/sec: 9.37316\nINFO:tensorflow:loss = 5.229684, step = 16200 (10.670 sec)\nINFO:tensorflow:lr = 0.00039030772 (10.669 sec)\nINFO:tensorflow:global_step/sec: 9.4933\nINFO:tensorflow:loss = 5.246074, step = 16300 (10.534 sec)\nINFO:tensorflow:lr = 0.00039209973 (10.534 sec)\nINFO:tensorflow:global_step/sec: 9.10833\nINFO:tensorflow:loss = 5.3365836, step = 16400 (10.976 sec)\nINFO:tensorflow:lr = 0.00039389174 (10.976 sec)\nINFO:tensorflow:global_step/sec: 9.6444\nINFO:tensorflow:loss = 5.253062, step = 16500 (10.370 sec)\nINFO:tensorflow:lr = 0.00039568378 (10.370 sec)\nINFO:tensorflow:global_step/sec: 9.30588\nINFO:tensorflow:loss = 5.277682, step = 16600 (10.746 sec)\nINFO:tensorflow:lr = 0.00039747581 (10.748 sec)\nINFO:tensorflow:global_step/sec: 9.51747\nINFO:tensorflow:loss = 5.296861, step = 16700 (10.506 sec)\nINFO:tensorflow:lr = 0.00039926782 (10.505 sec)\nINFO:tensorflow:global_step/sec: 9.57023\nINFO:tensorflow:loss = 5.0729628, step = 16800 (10.450 sec)\nINFO:tensorflow:lr = 0.00040105986 (10.451 sec)\nINFO:tensorflow:global_step/sec: 9.68155\nINFO:tensorflow:loss = 5.2019506, step = 16900 (10.331 sec)\nINFO:tensorflow:lr = 0.00040285187 (10.332 sec)\nINFO:tensorflow:global_step/sec: 9.52325\nINFO:tensorflow:loss = 5.2418485, step = 17000 (10.496 sec)\nINFO:tensorflow:lr = 0.0004046439 (10.497 sec)\nINFO:tensorflow:global_step/sec: 9.47466\nINFO:tensorflow:loss = 5.156396, step = 17100 (10.557 sec)\nINFO:tensorflow:lr = 0.00040643592 (10.556 sec)\nINFO:tensorflow:global_step/sec: 9.43814\nINFO:tensorflow:loss = 5.28926, step = 17200 (10.593 sec)\nINFO:tensorflow:lr = 0.00040822793 (10.595 sec)\nINFO:tensorflow:global_step/sec: 9.30126\nINFO:tensorflow:loss = 5.2130136, step = 17300 (10.751 sec)\nINFO:tensorflow:lr = 0.00041001994 (10.748 sec)\nINFO:tensorflow:global_step/sec: 9.67383\nINFO:tensorflow:loss = 5.0762405, step = 17400 (10.338 sec)\nINFO:tensorflow:lr = 0.00041181198 (10.338 sec)\nINFO:tensorflow:global_step/sec: 9.60888\nINFO:tensorflow:loss = 5.1943226, step = 17500 (10.407 sec)\nINFO:tensorflow:lr = 0.00041360402 (10.408 sec)\nINFO:tensorflow:global_step/sec: 9.50219\nINFO:tensorflow:loss = 5.294518, step = 17600 (10.525 sec)\nINFO:tensorflow:lr = 0.00041539603 (10.527 sec)\nINFO:tensorflow:global_step/sec: 9.43169\nINFO:tensorflow:loss = 5.0493245, step = 17700 (10.603 sec)\nINFO:tensorflow:lr = 0.00041718807 (10.600 sec)\nINFO:tensorflow:global_step/sec: 9.67039\nINFO:tensorflow:loss = 5.2163715, step = 17800 (10.342 sec)\nINFO:tensorflow:lr = 0.00041898008 (10.342 sec)\nINFO:tensorflow:global_step/sec: 9.2621\nINFO:tensorflow:loss = 4.999476, step = 17900 (10.794 sec)\nINFO:tensorflow:lr = 0.0004207721 (10.795 sec)\nINFO:tensorflow:global_step/sec: 9.72487\nINFO:tensorflow:loss = 5.1670976, step = 18000 (10.284 sec)\nINFO:tensorflow:lr = 0.00042256413 (10.285 sec)\nINFO:tensorflow:global_step/sec: 9.45329\nINFO:tensorflow:loss = 5.088095, step = 18100 (10.580 sec)\nINFO:tensorflow:lr = 0.00042435614 (10.580 sec)\nINFO:tensorflow:global_step/sec: 9.84978\nINFO:tensorflow:loss = 5.1727524, step = 18200 (10.151 sec)\nINFO:tensorflow:lr = 0.00042614815 (10.150 sec)\nINFO:tensorflow:global_step/sec: 9.39427\nINFO:tensorflow:loss = 4.9821234, step = 18300 (10.644 sec)\nINFO:tensorflow:lr = 0.0004279402 (10.644 sec)\nINFO:tensorflow:global_step/sec: 9.62825\nINFO:tensorflow:loss = 5.046456, step = 18400 (10.386 sec)\nINFO:tensorflow:lr = 0.0004297322 (10.387 sec)\nINFO:tensorflow:global_step/sec: 9.32231\nINFO:tensorflow:loss = 5.0560246, step = 18500 (10.729 sec)\nINFO:tensorflow:lr = 0.00043152424 (10.728 sec)\nINFO:tensorflow:global_step/sec: 9.62334\nINFO:tensorflow:loss = 5.2404575, step = 18600 (10.391 sec)\nINFO:tensorflow:lr = 0.00043331628 (10.391 sec)\nINFO:tensorflow:global_step/sec: 9.65077\nINFO:tensorflow:loss = 5.1200423, step = 18700 (10.362 sec)\nINFO:tensorflow:lr = 0.0004351083 (10.363 sec)\nINFO:tensorflow:global_step/sec: 9.44334\nINFO:tensorflow:loss = 4.978124, step = 18800 (10.588 sec)\nINFO:tensorflow:lr = 0.0004369003 (10.587 sec)\nINFO:tensorflow:global_step/sec: 9.5336\nINFO:tensorflow:loss = 5.1311655, step = 18900 (10.489 sec)\nINFO:tensorflow:lr = 0.0004386923 (10.491 sec)\nINFO:tensorflow:global_step/sec: 9.7246\nINFO:tensorflow:loss = 5.125824, step = 19000 (10.283 sec)\nINFO:tensorflow:lr = 0.00044048435 (10.283 sec)\nINFO:tensorflow:global_step/sec: 8.98551\nINFO:tensorflow:loss = 5.180024, step = 19100 (11.127 sec)\nINFO:tensorflow:lr = 0.00044227636 (11.127 sec)\nINFO:tensorflow:global_step/sec: 9.40787\nINFO:tensorflow:loss = 5.2193055, step = 19200 (10.632 sec)\nINFO:tensorflow:lr = 0.0004440684 (10.632 sec)\nINFO:tensorflow:global_step/sec: 9.52289\nINFO:tensorflow:loss = 5.1205177, step = 19300 (10.500 sec)\nINFO:tensorflow:lr = 0.0004458604 (10.501 sec)\nINFO:tensorflow:global_step/sec: 9.24599\nINFO:tensorflow:loss = 5.092633, step = 19400 (10.816 sec)\nINFO:tensorflow:lr = 0.00044765242 (10.815 sec)\nINFO:tensorflow:global_step/sec: 9.2783\nINFO:tensorflow:loss = 5.195169, step = 19500 (10.775 sec)\nINFO:tensorflow:lr = 0.0004494445 (10.776 sec)\nINFO:tensorflow:global_step/sec: 9.66929\nINFO:tensorflow:loss = 5.181858, step = 19600 (10.345 sec)\nINFO:tensorflow:lr = 0.0004512365 (10.345 sec)\nINFO:tensorflow:global_step/sec: 9.71316\nINFO:tensorflow:loss = 5.076673, step = 19700 (10.295 sec)\nINFO:tensorflow:lr = 0.0004530285 (10.296 sec)\nINFO:tensorflow:global_step/sec: 9.45051\nINFO:tensorflow:loss = 5.0781593, step = 19800 (10.579 sec)\nINFO:tensorflow:lr = 0.00045482052 (10.577 sec)\nINFO:tensorflow:global_step/sec: 9.55697\nINFO:tensorflow:loss = 4.937835, step = 19900 (10.466 sec)\nINFO:tensorflow:lr = 0.00045661256 (10.467 sec)\nINFO:tensorflow:global_step/sec: 9.1631\nINFO:tensorflow:loss = 5.1505866, step = 20000 (10.913 sec)\nINFO:tensorflow:lr = 0.0004584046 (10.912 sec)\nINFO:tensorflow:global_step/sec: 9.4915\nINFO:tensorflow:loss = 5.133694, step = 20100 (10.536 sec)\nINFO:tensorflow:lr = 0.0004601966 (10.536 sec)\nINFO:tensorflow:global_step/sec: 9.57019\nINFO:tensorflow:loss = 5.06313, step = 20200 (10.448 sec)\nINFO:tensorflow:lr = 0.00046198862 (10.449 sec)\nINFO:tensorflow:global_step/sec: 9.79037\nINFO:tensorflow:loss = 5.0452394, step = 20300 (10.212 sec)\nINFO:tensorflow:lr = 0.00046378063 (10.213 sec)\nINFO:tensorflow:global_step/sec: 9.27914\nINFO:tensorflow:loss = 5.2335463, step = 20400 (10.779 sec)\nINFO:tensorflow:lr = 0.00046557267 (10.779 sec)\nINFO:tensorflow:global_step/sec: 9.13557\nINFO:tensorflow:loss = 5.10505, step = 20500 (10.947 sec)\nINFO:tensorflow:lr = 0.0004673647 (10.946 sec)\nINFO:tensorflow:global_step/sec: 9.16514\nINFO:tensorflow:loss = 5.094977, step = 20600 (10.908 sec)\nINFO:tensorflow:lr = 0.00046915672 (10.909 sec)\nINFO:tensorflow:global_step/sec: 9.38076\nINFO:tensorflow:loss = 5.1793923, step = 20700 (10.662 sec)\nINFO:tensorflow:lr = 0.00047094873 (10.661 sec)\nINFO:tensorflow:global_step/sec: 9.5493\nINFO:tensorflow:loss = 5.0512815, step = 20800 (10.473 sec)\nINFO:tensorflow:lr = 0.00047274074 (10.473 sec)\nINFO:tensorflow:global_step/sec: 9.49999\nINFO:tensorflow:loss = 5.065953, step = 20900 (10.524 sec)\nINFO:tensorflow:lr = 0.0004745328 (10.525 sec)\nINFO:tensorflow:global_step/sec: 9.53287\nINFO:tensorflow:loss = 5.0509553, step = 21000 (10.492 sec)\nINFO:tensorflow:lr = 0.0004763248 (10.493 sec)\nINFO:tensorflow:global_step/sec: 9.482\nINFO:tensorflow:loss = 5.1141, step = 21100 (10.546 sec)\nINFO:tensorflow:lr = 0.00047811682 (10.544 sec)\nINFO:tensorflow:global_step/sec: 9.48085\nINFO:tensorflow:loss = 5.178147, step = 21200 (10.549 sec)\nINFO:tensorflow:lr = 0.00047990883 (10.549 sec)\nINFO:tensorflow:global_step/sec: 9.05672\nINFO:tensorflow:loss = 5.056297, step = 21300 (11.041 sec)\nINFO:tensorflow:lr = 0.00048170084 (11.041 sec)\nINFO:tensorflow:global_step/sec: 9.70846\nINFO:tensorflow:loss = 5.2183027, step = 21400 (10.299 sec)\nINFO:tensorflow:lr = 0.0004834929 (10.300 sec)\nINFO:tensorflow:global_step/sec: 9.50661\nINFO:tensorflow:loss = 5.1443934, step = 21500 (10.520 sec)\nINFO:tensorflow:lr = 0.00048528492 (10.519 sec)\nINFO:tensorflow:global_step/sec: 9.26474\nINFO:tensorflow:loss = 5.140825, step = 21600 (10.792 sec)\nINFO:tensorflow:lr = 0.00048707693 (10.792 sec)\nINFO:tensorflow:global_step/sec: 9.38964\nINFO:tensorflow:loss = 5.0313034, step = 21700 (10.651 sec)\nINFO:tensorflow:lr = 0.0004888689 (10.651 sec)\nINFO:tensorflow:global_step/sec: 9.62222\nINFO:tensorflow:loss = 5.055342, step = 21800 (10.391 sec)\nINFO:tensorflow:lr = 0.000490661 (10.397 sec)\nINFO:tensorflow:global_step/sec: 9.53237\nINFO:tensorflow:loss = 5.0808406, step = 21900 (10.492 sec)\nINFO:tensorflow:lr = 0.000492453 (10.487 sec)\nINFO:tensorflow:global_step/sec: 9.74152\nINFO:tensorflow:loss = 5.1622124, step = 22000 (10.266 sec)\nINFO:tensorflow:lr = 0.00049424503 (10.265 sec)\nINFO:tensorflow:global_step/sec: 9.516\nINFO:tensorflow:loss = 5.157396, step = 22100 (10.509 sec)\nINFO:tensorflow:lr = 0.000496037 (10.510 sec)\nINFO:tensorflow:global_step/sec: 9.07583\nINFO:tensorflow:loss = 5.137361, step = 22200 (11.017 sec)\nINFO:tensorflow:lr = 0.00049782905 (11.016 sec)\nINFO:tensorflow:global_step/sec: 9.45514\nINFO:tensorflow:loss = 5.0328975, step = 22300 (10.577 sec)\nINFO:tensorflow:lr = 0.0004996211 (10.578 sec)\nINFO:tensorflow:global_step/sec: 9.51083\nINFO:tensorflow:loss = 5.121226, step = 22400 (10.512 sec)\nINFO:tensorflow:lr = 0.00050141313 (10.512 sec)\nINFO:tensorflow:global_step/sec: 9.33951\nINFO:tensorflow:loss = 4.971635, step = 22500 (10.710 sec)\nINFO:tensorflow:lr = 0.0005032051 (10.711 sec)\nINFO:tensorflow:global_step/sec: 9.64867\nINFO:tensorflow:loss = 5.090142, step = 22600 (10.364 sec)\nINFO:tensorflow:lr = 0.00050499715 (10.363 sec)\nINFO:tensorflow:global_step/sec: 9.67859\nINFO:tensorflow:loss = 5.094214, step = 22700 (10.332 sec)\nINFO:tensorflow:lr = 0.0005067892 (10.334 sec)\nINFO:tensorflow:global_step/sec: 9.09592\nINFO:tensorflow:loss = 5.1820188, step = 22800 (10.993 sec)\nINFO:tensorflow:lr = 0.00050858123 (10.994 sec)\nINFO:tensorflow:global_step/sec: 9.12329\nINFO:tensorflow:loss = 5.186921, step = 22900 (10.964 sec)\nINFO:tensorflow:lr = 0.0005103732 (10.959 sec)\nINFO:tensorflow:global_step/sec: 9.28302\nINFO:tensorflow:loss = 5.2035203, step = 23000 (10.771 sec)\nINFO:tensorflow:lr = 0.00051216525 (10.772 sec)\nINFO:tensorflow:global_step/sec: 9.33365\nINFO:tensorflow:loss = 4.9803834, step = 23100 (10.715 sec)\nINFO:tensorflow:lr = 0.00051395723 (10.714 sec)\nINFO:tensorflow:global_step/sec: 9.53787\nINFO:tensorflow:loss = 5.1692257, step = 23200 (10.485 sec)\nINFO:tensorflow:lr = 0.0005157493 (10.485 sec)\nINFO:tensorflow:global_step/sec: 9.36692\nINFO:tensorflow:loss = 5.1639504, step = 23300 (10.675 sec)\nINFO:tensorflow:lr = 0.0005175413 (10.676 sec)\nINFO:tensorflow:global_step/sec: 9.60574\nINFO:tensorflow:loss = 5.258109, step = 23400 (10.411 sec)\nINFO:tensorflow:lr = 0.00051933335 (10.410 sec)\nINFO:tensorflow:global_step/sec: 9.51388\nINFO:tensorflow:loss = 5.2187853, step = 23500 (10.510 sec)\nINFO:tensorflow:lr = 0.00052112533 (10.510 sec)\nINFO:tensorflow:global_step/sec: 9.13428\nINFO:tensorflow:loss = 5.10759, step = 23600 (10.949 sec)\nINFO:tensorflow:lr = 0.0005229174 (10.949 sec)\nINFO:tensorflow:global_step/sec: 9.40798\nINFO:tensorflow:loss = 5.1671615, step = 23700 (10.628 sec)\nINFO:tensorflow:lr = 0.0005247094 (10.628 sec)\nINFO:tensorflow:global_step/sec: 9.46928\nINFO:tensorflow:loss = 5.1048703, step = 23800 (10.561 sec)\nINFO:tensorflow:lr = 0.00052650145 (10.561 sec)\nINFO:tensorflow:global_step/sec: 8.96179\nINFO:tensorflow:loss = 5.1232705, step = 23900 (11.159 sec)\nINFO:tensorflow:lr = 0.0005282934 (11.159 sec)\nINFO:tensorflow:global_step/sec: 8.94983\nINFO:tensorflow:loss = 5.196185, step = 24000 (11.172 sec)\nINFO:tensorflow:lr = 0.00053008547 (11.172 sec)\nINFO:tensorflow:global_step/sec: 8.9806\nINFO:tensorflow:loss = 5.142331, step = 24100 (11.133 sec)\nINFO:tensorflow:lr = 0.0005318775 (11.134 sec)\nINFO:tensorflow:global_step/sec: 9.2065\nINFO:tensorflow:loss = 5.077751, step = 24200 (10.862 sec)\nINFO:tensorflow:lr = 0.00053366955 (10.863 sec)\nINFO:tensorflow:global_step/sec: 9.15252\nINFO:tensorflow:loss = 5.1835365, step = 24300 (10.927 sec)\nINFO:tensorflow:lr = 0.0005354615 (10.928 sec)\nINFO:tensorflow:global_step/sec: 9.12864\nINFO:tensorflow:loss = 5.1684585, step = 24400 (10.955 sec)\nINFO:tensorflow:lr = 0.00053725357 (10.953 sec)\nINFO:tensorflow:global_step/sec: 9.39152\nINFO:tensorflow:loss = 5.2003446, step = 24500 (10.647 sec)\nINFO:tensorflow:lr = 0.0005390456 (10.647 sec)\nINFO:tensorflow:global_step/sec: 9.26444\nINFO:tensorflow:loss = 5.1787515, step = 24600 (10.795 sec)\nINFO:tensorflow:lr = 0.00054083765 (10.795 sec)\nINFO:tensorflow:global_step/sec: 9.06221\nINFO:tensorflow:loss = 5.062696, step = 24700 (11.034 sec)\nINFO:tensorflow:lr = 0.0005426296 (11.034 sec)\nINFO:tensorflow:global_step/sec: 9.33903\nINFO:tensorflow:loss = 5.015447, step = 24800 (10.710 sec)\nINFO:tensorflow:lr = 0.00054442167 (10.711 sec)\nINFO:tensorflow:global_step/sec: 8.95611\nINFO:tensorflow:loss = 5.108074, step = 24900 (11.162 sec)\nINFO:tensorflow:lr = 0.00054621365 (11.165 sec)\nINFO:tensorflow:global_step/sec: 8.97971\nINFO:tensorflow:loss = 5.1784277, step = 25000 (11.137 sec)\nINFO:tensorflow:lr = 0.00054800574 (11.134 sec)\nINFO:tensorflow:global_step/sec: 8.8737\nINFO:tensorflow:loss = 5.0889997, step = 25100 (11.272 sec)\nINFO:tensorflow:lr = 0.0005497977 (11.271 sec)\nINFO:tensorflow:global_step/sec: 8.99304\nINFO:tensorflow:loss = 5.1338367, step = 25200 (11.118 sec)\nINFO:tensorflow:lr = 0.00055158976 (11.118 sec)\nINFO:tensorflow:global_step/sec: 9.20564\nINFO:tensorflow:loss = 5.049507, step = 25300 (10.863 sec)\nINFO:tensorflow:lr = 0.00055338175 (10.865 sec)\nINFO:tensorflow:global_step/sec: 8.86175\nINFO:tensorflow:loss = 5.004919, step = 25400 (11.285 sec)\nINFO:tensorflow:lr = 0.00055517384 (11.284 sec)\nINFO:tensorflow:global_step/sec: 8.75508\nINFO:tensorflow:loss = 5.2403455, step = 25500 (11.422 sec)\nINFO:tensorflow:lr = 0.0005569658 (11.422 sec)\nINFO:tensorflow:global_step/sec: 9.18257\nINFO:tensorflow:loss = 5.125975, step = 25600 (10.890 sec)\nINFO:tensorflow:lr = 0.00055875786 (10.890 sec)\nINFO:tensorflow:global_step/sec: 8.96184\nINFO:tensorflow:loss = 5.0116687, step = 25700 (11.157 sec)\nINFO:tensorflow:lr = 0.00056054984 (11.158 sec)\nINFO:tensorflow:global_step/sec: 8.52834\nINFO:tensorflow:loss = 5.0436788, step = 25800 (11.728 sec)\nINFO:tensorflow:lr = 0.0005623419 (11.726 sec)\nINFO:tensorflow:global_step/sec: 9.19879\nINFO:tensorflow:loss = 5.07081, step = 25900 (10.872 sec)\nINFO:tensorflow:lr = 0.0005641339 (10.874 sec)\nINFO:tensorflow:global_step/sec: 9.07916\nINFO:tensorflow:loss = 5.0617657, step = 26000 (11.013 sec)\nINFO:tensorflow:lr = 0.00056592596 (11.011 sec)\nINFO:tensorflow:global_step/sec: 9.15357\nINFO:tensorflow:loss = 5.0214477, step = 26100 (10.926 sec)\nINFO:tensorflow:lr = 0.00056771794 (10.926 sec)\nINFO:tensorflow:global_step/sec: 8.74457\nINFO:tensorflow:loss = 5.129234, step = 26200 (11.431 sec)\nINFO:tensorflow:lr = 0.00056951 (11.432 sec)\nINFO:tensorflow:global_step/sec: 9.20484\nINFO:tensorflow:loss = 5.0483747, step = 26300 (10.866 sec)\nINFO:tensorflow:lr = 0.000571302 (10.865 sec)\nINFO:tensorflow:global_step/sec: 9.0836\nINFO:tensorflow:loss = 5.135254, step = 26400 (11.011 sec)\nINFO:tensorflow:lr = 0.00057309406 (11.011 sec)\nINFO:tensorflow:global_step/sec: 9.65397\nINFO:tensorflow:loss = 5.09909, step = 26500 (10.356 sec)\nINFO:tensorflow:lr = 0.00057488604 (10.357 sec)\nINFO:tensorflow:global_step/sec: 9.27336\nINFO:tensorflow:loss = 5.0942545, step = 26600 (10.782 sec)\nINFO:tensorflow:lr = 0.0005766781 (10.781 sec)\nINFO:tensorflow:global_step/sec: 9.31149\nINFO:tensorflow:loss = 5.074828, step = 26700 (10.741 sec)\nINFO:tensorflow:lr = 0.00057847006 (10.742 sec)\nINFO:tensorflow:global_step/sec: 9.25461\nINFO:tensorflow:loss = 5.084673, step = 26800 (10.805 sec)\nINFO:tensorflow:lr = 0.00058026216 (10.805 sec)\nINFO:tensorflow:global_step/sec: 8.69425\nINFO:tensorflow:loss = 4.935843, step = 26900 (11.502 sec)\nINFO:tensorflow:lr = 0.00058205414 (11.501 sec)\nINFO:tensorflow:global_step/sec: 9.09111\nINFO:tensorflow:loss = 5.1584783, step = 27000 (10.999 sec)\nINFO:tensorflow:lr = 0.0005838462 (11.000 sec)\nINFO:tensorflow:global_step/sec: 9.30537\nINFO:tensorflow:loss = 5.091224, step = 27100 (10.745 sec)\nINFO:tensorflow:lr = 0.00058563816 (10.744 sec)\nINFO:tensorflow:global_step/sec: 9.42432\nINFO:tensorflow:loss = 4.955161, step = 27200 (10.614 sec)\nINFO:tensorflow:lr = 0.0005874302 (10.614 sec)\nINFO:tensorflow:global_step/sec: 9.34376\nINFO:tensorflow:loss = 4.995959, step = 27300 (10.702 sec)\nINFO:tensorflow:lr = 0.00058922224 (10.702 sec)\nINFO:tensorflow:global_step/sec: 8.91428\nINFO:tensorflow:loss = 5.2494087, step = 27400 (11.218 sec)\nINFO:tensorflow:lr = 0.0005910143 (11.219 sec)\nINFO:tensorflow:global_step/sec: 8.99617\nINFO:tensorflow:loss = 5.1566463, step = 27500 (11.116 sec)\nINFO:tensorflow:lr = 0.00059280626 (11.115 sec)\nINFO:tensorflow:global_step/sec: 8.97073\nINFO:tensorflow:loss = 5.157406, step = 27600 (11.148 sec)\nINFO:tensorflow:lr = 0.0005945983 (11.148 sec)\nINFO:tensorflow:global_step/sec: 9.24686\nINFO:tensorflow:loss = 5.03269, step = 27700 (10.813 sec)\nINFO:tensorflow:lr = 0.00059639034 (10.813 sec)\nINFO:tensorflow:global_step/sec: 8.96445\nINFO:tensorflow:loss = 4.99574, step = 27800 (11.155 sec)\nINFO:tensorflow:lr = 0.0005981824 (11.157 sec)\nINFO:tensorflow:global_step/sec: 9.02618\nINFO:tensorflow:loss = 5.0618687, step = 27900 (11.080 sec)\nINFO:tensorflow:lr = 0.00059997436 (11.079 sec)\nINFO:tensorflow:global_step/sec: 9.36297\nINFO:tensorflow:loss = 4.967255, step = 28000 (10.680 sec)\nINFO:tensorflow:lr = 0.0006017664 (10.680 sec)\nINFO:tensorflow:global_step/sec: 9.27127\nINFO:tensorflow:loss = 4.918425, step = 28100 (10.786 sec)\nINFO:tensorflow:lr = 0.00060355844 (10.786 sec)\nINFO:tensorflow:global_step/sec: 9.49182\nINFO:tensorflow:loss = 5.1641984, step = 28200 (10.535 sec)\nINFO:tensorflow:lr = 0.0006053505 (10.535 sec)\nINFO:tensorflow:global_step/sec: 9.3774\nINFO:tensorflow:loss = 5.039311, step = 28300 (10.664 sec)\nINFO:tensorflow:lr = 0.00060714246 (10.664 sec)\nINFO:tensorflow:global_step/sec: 9.19179\nINFO:tensorflow:loss = 5.066581, step = 28400 (10.877 sec)\nINFO:tensorflow:lr = 0.0006089345 (10.878 sec)\nINFO:tensorflow:global_step/sec: 9.48473\nINFO:tensorflow:loss = 5.068813, step = 28500 (10.545 sec)\nINFO:tensorflow:lr = 0.0006107265 (10.544 sec)\nINFO:tensorflow:global_step/sec: 8.85006\nINFO:tensorflow:loss = 4.9568076, step = 28600 (11.299 sec)\nINFO:tensorflow:lr = 0.0006125186 (11.299 sec)\nINFO:tensorflow:global_step/sec: 9.14199\nINFO:tensorflow:loss = 5.0653033, step = 28700 (10.940 sec)\nINFO:tensorflow:lr = 0.00061431056 (10.939 sec)\nINFO:tensorflow:global_step/sec: 9.46158\nINFO:tensorflow:loss = 5.0284724, step = 28800 (10.570 sec)\nINFO:tensorflow:lr = 0.0006161026 (10.569 sec)\nINFO:tensorflow:global_step/sec: 9.54863\nINFO:tensorflow:loss = 5.1366715, step = 28900 (10.472 sec)\nINFO:tensorflow:lr = 0.0006178946 (10.473 sec)\nINFO:tensorflow:global_step/sec: 9.43582\nINFO:tensorflow:loss = 5.264217, step = 29000 (10.597 sec)\nINFO:tensorflow:lr = 0.0006196867 (10.597 sec)\nINFO:tensorflow:global_step/sec: 9.48442\nINFO:tensorflow:loss = 5.1028385, step = 29100 (10.544 sec)\nINFO:tensorflow:lr = 0.00062147866 (10.545 sec)\nINFO:tensorflow:global_step/sec: 9.38207\nINFO:tensorflow:loss = 5.248551, step = 29200 (10.658 sec)\nINFO:tensorflow:lr = 0.0006232707 (10.658 sec)\nINFO:tensorflow:global_step/sec: 9.62767\nINFO:tensorflow:loss = 5.110166, step = 29300 (10.387 sec)\nINFO:tensorflow:lr = 0.0006250627 (10.386 sec)\nINFO:tensorflow:global_step/sec: 9.6329\nINFO:tensorflow:loss = 5.151551, step = 29400 (10.382 sec)\nINFO:tensorflow:lr = 0.0006268547 (10.382 sec)\nINFO:tensorflow:global_step/sec: 9.51864\nINFO:tensorflow:loss = 4.9802246, step = 29500 (10.505 sec)\nINFO:tensorflow:lr = 0.00062864675 (10.505 sec)\nINFO:tensorflow:global_step/sec: 9.01068\nINFO:tensorflow:loss = 5.120793, step = 29600 (11.098 sec)\nINFO:tensorflow:lr = 0.0006304388 (11.098 sec)\nINFO:tensorflow:global_step/sec: 9.1528\nINFO:tensorflow:loss = 5.0916967, step = 29700 (10.925 sec)\nINFO:tensorflow:lr = 0.0006322308 (10.925 sec)\nINFO:tensorflow:global_step/sec: 9.1005\nINFO:tensorflow:loss = 4.9674788, step = 29800 (10.989 sec)\nINFO:tensorflow:lr = 0.0006340228 (10.989 sec)\nINFO:tensorflow:global_step/sec: 9.53958\nINFO:tensorflow:loss = 5.112809, step = 29900 (10.482 sec)\nINFO:tensorflow:lr = 0.00063581485 (10.483 sec)\nINFO:tensorflow:global_step/sec: 9.47696\nINFO:tensorflow:loss = 5.2655168, step = 30000 (10.552 sec)\nINFO:tensorflow:lr = 0.0006376069 (10.550 sec)\nINFO:tensorflow:global_step/sec: 9.58024\nINFO:tensorflow:loss = 5.2323723, step = 30100 (10.440 sec)\nINFO:tensorflow:lr = 0.0006393989 (10.441 sec)\nINFO:tensorflow:global_step/sec: 9.1579\nINFO:tensorflow:loss = 5.038887, step = 30200 (10.916 sec)\nINFO:tensorflow:lr = 0.0006411909 (10.919 sec)\nINFO:tensorflow:global_step/sec: 9.4468\nINFO:tensorflow:loss = 4.993733, step = 30300 (10.589 sec)\nINFO:tensorflow:lr = 0.00064298295 (10.586 sec)\nINFO:tensorflow:global_step/sec: 9.4222\nINFO:tensorflow:loss = 5.165243, step = 30400 (10.609 sec)\nINFO:tensorflow:lr = 0.000644775 (10.611 sec)\nINFO:tensorflow:global_step/sec: 9.10203\nINFO:tensorflow:loss = 5.080005, step = 30500 (10.987 sec)\nINFO:tensorflow:lr = 0.000646567 (10.987 sec)\nINFO:tensorflow:global_step/sec: 9.37739\nINFO:tensorflow:loss = 5.001014, step = 30600 (10.665 sec)\nINFO:tensorflow:lr = 0.000648359 (10.666 sec)\nINFO:tensorflow:global_step/sec: 9.53468\nINFO:tensorflow:loss = 5.0422983, step = 30700 (10.488 sec)\nINFO:tensorflow:lr = 0.000650151 (10.488 sec)\nINFO:tensorflow:global_step/sec: 9.26078\nINFO:tensorflow:loss = 5.10322, step = 30800 (10.796 sec)\nINFO:tensorflow:lr = 0.0006519431 (10.797 sec)\nINFO:tensorflow:global_step/sec: 9.43497\nINFO:tensorflow:loss = 5.1736207, step = 30900 (10.602 sec)\nINFO:tensorflow:lr = 0.0006537351 (10.601 sec)\nINFO:tensorflow:global_step/sec: 8.9818\nINFO:tensorflow:loss = 5.086377, step = 31000 (11.132 sec)\nINFO:tensorflow:lr = 0.0006555271 (11.133 sec)\nINFO:tensorflow:global_step/sec: 9.2658\nINFO:tensorflow:loss = 5.021378, step = 31100 (10.793 sec)\nINFO:tensorflow:lr = 0.0006573191 (10.792 sec)\nINFO:tensorflow:global_step/sec: 9.16881\nINFO:tensorflow:loss = 4.9313107, step = 31200 (10.907 sec)\nINFO:tensorflow:lr = 0.0006591112 (10.907 sec)\nINFO:tensorflow:global_step/sec: 9.09478\nINFO:tensorflow:loss = 4.967325, step = 31300 (10.996 sec)\nINFO:tensorflow:lr = 0.00066090317 (10.996 sec)\nINFO:tensorflow:global_step/sec: 9.47929\nINFO:tensorflow:loss = 5.143935, step = 31400 (10.549 sec)\nINFO:tensorflow:lr = 0.0006626952 (10.550 sec)\nINFO:tensorflow:global_step/sec: 9.00435\nINFO:tensorflow:loss = 5.146677, step = 31500 (11.104 sec)\nINFO:tensorflow:lr = 0.0006644872 (11.104 sec)\nINFO:tensorflow:global_step/sec: 9.15019\nINFO:tensorflow:loss = 5.2024193, step = 31600 (10.926 sec)\nINFO:tensorflow:lr = 0.00066627923 (10.927 sec)\nINFO:tensorflow:global_step/sec: 9.58678\nINFO:tensorflow:loss = 4.9923096, step = 31700 (10.431 sec)\nINFO:tensorflow:lr = 0.00066807127 (10.431 sec)\nINFO:tensorflow:global_step/sec: 9.40868\nINFO:tensorflow:loss = 5.0396233, step = 31800 (10.630 sec)\nINFO:tensorflow:lr = 0.0006698633 (10.631 sec)\nINFO:tensorflow:global_step/sec: 9.33834\nINFO:tensorflow:loss = 5.1495657, step = 31900 (10.710 sec)\nINFO:tensorflow:lr = 0.0006716553 (10.709 sec)\nINFO:tensorflow:global_step/sec: 9.27637\nINFO:tensorflow:loss = 5.214168, step = 32000 (10.779 sec)\nINFO:tensorflow:lr = 0.0006734473 (10.780 sec)\nINFO:tensorflow:global_step/sec: 9.10232\nINFO:tensorflow:loss = 5.0548754, step = 32100 (10.988 sec)\nINFO:tensorflow:lr = 0.00067523937 (10.987 sec)\nINFO:tensorflow:global_step/sec: 8.94189\nINFO:tensorflow:loss = 5.0729666, step = 32200 (11.183 sec)\nINFO:tensorflow:lr = 0.0006770314 (11.182 sec)\nINFO:tensorflow:global_step/sec: 9.31504\nINFO:tensorflow:loss = 5.0461087, step = 32300 (10.736 sec)\nINFO:tensorflow:lr = 0.0006788234 (10.735 sec)\nINFO:tensorflow:global_step/sec: 9.21138\nINFO:tensorflow:loss = 5.0706806, step = 32400 (10.855 sec)\nINFO:tensorflow:lr = 0.0006806154 (10.856 sec)\nINFO:tensorflow:global_step/sec: 9.39348\nINFO:tensorflow:loss = 5.036418, step = 32500 (10.647 sec)\nINFO:tensorflow:lr = 0.0006824074 (10.646 sec)\nINFO:tensorflow:global_step/sec: 9.36836\nINFO:tensorflow:loss = 5.0860696, step = 32600 (10.671 sec)\nINFO:tensorflow:lr = 0.0006841995 (10.671 sec)\nINFO:tensorflow:global_step/sec: 9.22372\nINFO:tensorflow:loss = 5.1222463, step = 32700 (10.841 sec)\nINFO:tensorflow:lr = 0.0006859915 (10.843 sec)\nINFO:tensorflow:global_step/sec: 9.54108\nINFO:tensorflow:loss = 5.114647, step = 32800 (10.481 sec)\nINFO:tensorflow:lr = 0.0006877835 (10.481 sec)\nINFO:tensorflow:global_step/sec: 9.67795\nINFO:tensorflow:loss = 5.099014, step = 32900 (10.333 sec)\nINFO:tensorflow:lr = 0.0006895755 (10.332 sec)\nINFO:tensorflow:global_step/sec: 9.31083\nINFO:tensorflow:loss = 5.1770754, step = 33000 (10.741 sec)\nINFO:tensorflow:lr = 0.00069136755 (10.742 sec)\nINFO:tensorflow:global_step/sec: 9.5378\nINFO:tensorflow:loss = 5.069852, step = 33100 (10.485 sec)\nINFO:tensorflow:lr = 0.0006931596 (10.484 sec)\nINFO:tensorflow:global_step/sec: 9.67884\nINFO:tensorflow:loss = 5.210137, step = 33200 (10.333 sec)\nINFO:tensorflow:lr = 0.0006949516 (10.332 sec)\nINFO:tensorflow:global_step/sec: 9.53279\nINFO:tensorflow:loss = 4.9941797, step = 33300 (10.489 sec)\nINFO:tensorflow:lr = 0.0006967436 (10.490 sec)\nINFO:tensorflow:global_step/sec: 9.60949\nINFO:tensorflow:loss = 5.0948496, step = 33400 (10.406 sec)\nINFO:tensorflow:lr = 0.00069853564 (10.406 sec)\nINFO:tensorflow:global_step/sec: 9.79415\nINFO:tensorflow:loss = 5.22286, step = 33500 (10.210 sec)\nINFO:tensorflow:lr = 0.0007003277 (10.210 sec)\nINFO:tensorflow:global_step/sec: 9.53751\nINFO:tensorflow:loss = 4.872001, step = 33600 (10.484 sec)\nINFO:tensorflow:lr = 0.0007021197 (10.484 sec)\nINFO:tensorflow:global_step/sec: 9.55892\nINFO:tensorflow:loss = 5.152949, step = 33700 (10.465 sec)\nINFO:tensorflow:lr = 0.0007039117 (10.464 sec)\nINFO:tensorflow:global_step/sec: 9.48322\nINFO:tensorflow:loss = 5.1330466, step = 33800 (10.543 sec)\nINFO:tensorflow:lr = 0.00070570374 (10.544 sec)\nINFO:tensorflow:global_step/sec: 9.46137\nINFO:tensorflow:loss = 5.1620946, step = 33900 (10.567 sec)\nINFO:tensorflow:lr = 0.0007074958 (10.568 sec)\nINFO:tensorflow:global_step/sec: 9.46544\nINFO:tensorflow:loss = 5.143143, step = 34000 (10.567 sec)\nINFO:tensorflow:lr = 0.0007092878 (10.567 sec)\nINFO:tensorflow:global_step/sec: 9.0955\nINFO:tensorflow:loss = 5.0488315, step = 34100 (10.994 sec)\nINFO:tensorflow:lr = 0.0007110798 (10.994 sec)\nINFO:tensorflow:global_step/sec: 9.53646\nINFO:tensorflow:loss = 4.9134398, step = 34200 (10.486 sec)\nINFO:tensorflow:lr = 0.00071287184 (10.486 sec)\nINFO:tensorflow:global_step/sec: 9.59935\nINFO:tensorflow:loss = 5.149152, step = 34300 (10.418 sec)\nINFO:tensorflow:lr = 0.0007146638 (10.418 sec)\nINFO:tensorflow:global_step/sec: 9.02973\nINFO:tensorflow:loss = 5.0462427, step = 34400 (11.072 sec)\nINFO:tensorflow:lr = 0.00071645586 (11.076 sec)\nINFO:tensorflow:global_step/sec: 9.32217\nINFO:tensorflow:loss = 5.03478, step = 34500 (10.730 sec)\nINFO:tensorflow:lr = 0.0007182479 (10.726 sec)\nINFO:tensorflow:global_step/sec: 9.35269\nINFO:tensorflow:loss = 4.9798565, step = 34600 (10.692 sec)\nINFO:tensorflow:lr = 0.00072003994 (10.691 sec)\nINFO:tensorflow:global_step/sec: 9.51789\nINFO:tensorflow:loss = 5.096134, step = 34700 (10.505 sec)\nINFO:tensorflow:lr = 0.0007218319 (10.506 sec)\nINFO:tensorflow:global_step/sec: 9.52418\nINFO:tensorflow:loss = 4.980329, step = 34800 (10.501 sec)\nINFO:tensorflow:lr = 0.00072362396 (10.500 sec)\nINFO:tensorflow:global_step/sec: 9.24779\nINFO:tensorflow:loss = 5.10829, step = 34900 (10.811 sec)\nINFO:tensorflow:lr = 0.000725416 (10.811 sec)\nINFO:tensorflow:global_step/sec: 9.66491\nINFO:tensorflow:loss = 5.2325635, step = 35000 (10.348 sec)\nINFO:tensorflow:lr = 0.00072720804 (10.348 sec)\nINFO:tensorflow:global_step/sec: 9.48265\nINFO:tensorflow:loss = 5.120744, step = 35100 (10.546 sec)\nINFO:tensorflow:lr = 0.000729 (10.546 sec)\nINFO:tensorflow:global_step/sec: 9.3831\nINFO:tensorflow:loss = 5.130047, step = 35200 (10.655 sec)\nINFO:tensorflow:lr = 0.00073079206 (10.655 sec)\nINFO:tensorflow:global_step/sec: 9.0511\nINFO:tensorflow:loss = 5.078965, step = 35300 (11.051 sec)\nINFO:tensorflow:lr = 0.0007325841 (11.051 sec)\nINFO:tensorflow:global_step/sec: 9.425\nINFO:tensorflow:loss = 5.008522, step = 35400 (10.608 sec)\nINFO:tensorflow:lr = 0.0007343761 (10.609 sec)\nINFO:tensorflow:global_step/sec: 9.24663\nINFO:tensorflow:loss = 5.1093845, step = 35500 (10.820 sec)\nINFO:tensorflow:lr = 0.0007361681 (10.820 sec)\nINFO:tensorflow:global_step/sec: 9.31256\nINFO:tensorflow:loss = 4.984288, step = 35600 (10.733 sec)\nINFO:tensorflow:lr = 0.00073796016 (10.732 sec)\nINFO:tensorflow:global_step/sec: 9.46158\nINFO:tensorflow:loss = 5.137832, step = 35700 (10.572 sec)\nINFO:tensorflow:lr = 0.0007397522 (10.572 sec)\nINFO:tensorflow:global_step/sec: 9.40468\nINFO:tensorflow:loss = 5.016031, step = 35800 (10.632 sec)\nINFO:tensorflow:lr = 0.0007415442 (10.633 sec)\nINFO:tensorflow:global_step/sec: 9.00724\nINFO:tensorflow:loss = 5.049596, step = 35900 (11.102 sec)\nINFO:tensorflow:lr = 0.0007433362 (11.102 sec)\nINFO:tensorflow:global_step/sec: 9.46032\nINFO:tensorflow:loss = 5.1828265, step = 36000 (10.570 sec)\nINFO:tensorflow:lr = 0.00074512826 (10.569 sec)\nINFO:tensorflow:global_step/sec: 9.3646\nINFO:tensorflow:loss = 5.058272, step = 36100 (10.681 sec)\nINFO:tensorflow:lr = 0.00074692024 (10.682 sec)\nINFO:tensorflow:global_step/sec: 9.49169\nINFO:tensorflow:loss = 5.245429, step = 36200 (10.533 sec)\nINFO:tensorflow:lr = 0.0007487123 (10.533 sec)\nINFO:tensorflow:global_step/sec: 9.51424\nINFO:tensorflow:loss = 5.0098643, step = 36300 (10.513 sec)\nINFO:tensorflow:lr = 0.0007505043 (10.511 sec)\nINFO:tensorflow:global_step/sec: 9.2906\nINFO:tensorflow:loss = 5.0751667, step = 36400 (10.763 sec)\nINFO:tensorflow:lr = 0.00075229636 (10.765 sec)\nINFO:tensorflow:global_step/sec: 9.38383\nINFO:tensorflow:loss = 5.029735, step = 36500 (10.657 sec)\nINFO:tensorflow:lr = 0.00075408834 (10.656 sec)\nINFO:tensorflow:global_step/sec: 9.37703\nINFO:tensorflow:loss = 4.932591, step = 36600 (10.663 sec)\nINFO:tensorflow:lr = 0.0007558804 (10.663 sec)\nINFO:tensorflow:global_step/sec: 9.4795\nINFO:tensorflow:loss = 5.153864, step = 36700 (10.550 sec)\nINFO:tensorflow:lr = 0.0007576724 (10.550 sec)\nINFO:tensorflow:global_step/sec: 9.38389\nINFO:tensorflow:loss = 5.1201468, step = 36800 (10.658 sec)\nINFO:tensorflow:lr = 0.0007594644 (10.658 sec)\nINFO:tensorflow:global_step/sec: 9.46879\nINFO:tensorflow:loss = 5.0017695, step = 36900 (10.559 sec)\nINFO:tensorflow:lr = 0.00076125644 (10.560 sec)\nINFO:tensorflow:global_step/sec: 9.3874\nINFO:tensorflow:loss = 4.984927, step = 37000 (10.653 sec)\nINFO:tensorflow:lr = 0.0007630485 (10.652 sec)\nINFO:tensorflow:global_step/sec: 9.26789\nINFO:tensorflow:loss = 4.8914003, step = 37100 (10.790 sec)\nINFO:tensorflow:lr = 0.0007648405 (10.790 sec)\nINFO:tensorflow:global_step/sec: 9.33466\nINFO:tensorflow:loss = 4.935514, step = 37200 (10.713 sec)\nINFO:tensorflow:lr = 0.0007666325 (10.713 sec)\nINFO:tensorflow:global_step/sec: 8.86929\nINFO:tensorflow:loss = 5.0787344, step = 37300 (11.275 sec)\nINFO:tensorflow:lr = 0.00076842454 (11.279 sec)\nINFO:tensorflow:global_step/sec: 9.17307\nINFO:tensorflow:loss = 4.892041, step = 37400 (10.900 sec)\nINFO:tensorflow:lr = 0.0007702166 (10.897 sec)\nINFO:tensorflow:global_step/sec: 9.57276\nINFO:tensorflow:loss = 5.1717734, step = 37500 (10.447 sec)\nINFO:tensorflow:lr = 0.0007720086 (10.448 sec)\nINFO:tensorflow:global_step/sec: 9.39006\nINFO:tensorflow:loss = 5.0274525, step = 37600 (10.648 sec)\nINFO:tensorflow:lr = 0.0007738006 (10.647 sec)\nINFO:tensorflow:global_step/sec: 9.01223\nINFO:tensorflow:loss = 5.040253, step = 37700 (11.099 sec)\nINFO:tensorflow:lr = 0.00077559263 (11.099 sec)\nINFO:tensorflow:global_step/sec: 9.32665\nINFO:tensorflow:loss = 5.2148137, step = 37800 (10.723 sec)\nINFO:tensorflow:lr = 0.0007773846 (10.722 sec)\nINFO:tensorflow:global_step/sec: 9.31095\nINFO:tensorflow:loss = 4.983381, step = 37900 (10.738 sec)\nINFO:tensorflow:lr = 0.00077917665 (10.740 sec)\nINFO:tensorflow:global_step/sec: 9.27172\nINFO:tensorflow:loss = 5.139436, step = 38000 (10.788 sec)\nINFO:tensorflow:lr = 0.0007809687 (10.787 sec)\nINFO:tensorflow:global_step/sec: 9.22479\nINFO:tensorflow:loss = 4.9605513, step = 38100 (10.838 sec)\nINFO:tensorflow:lr = 0.00078276073 (10.837 sec)\nINFO:tensorflow:global_step/sec: 9.55009\nINFO:tensorflow:loss = 5.0494156, step = 38200 (10.473 sec)\nINFO:tensorflow:lr = 0.0007845527 (10.472 sec)\nINFO:tensorflow:global_step/sec: 9.16568\nINFO:tensorflow:loss = 5.0706954, step = 38300 (10.909 sec)\nINFO:tensorflow:lr = 0.00078634475 (10.909 sec)\nINFO:tensorflow:global_step/sec: 9.26769\nINFO:tensorflow:loss = 4.9262986, step = 38400 (10.788 sec)\nINFO:tensorflow:lr = 0.0007881368 (10.790 sec)\nINFO:tensorflow:global_step/sec: 9.38006\nINFO:tensorflow:loss = 4.9493513, step = 38500 (10.665 sec)\nINFO:tensorflow:lr = 0.00078992883 (10.663 sec)\nINFO:tensorflow:global_step/sec: 9.63205\nINFO:tensorflow:loss = 4.980843, step = 38600 (10.381 sec)\nINFO:tensorflow:lr = 0.0007917208 (10.381 sec)\nINFO:tensorflow:global_step/sec: 9.39848\nINFO:tensorflow:loss = 4.944447, step = 38700 (10.641 sec)\nINFO:tensorflow:lr = 0.00079351285 (10.647 sec)\nINFO:tensorflow:global_step/sec: 9.16364\nINFO:tensorflow:loss = 5.029889, step = 38800 (10.913 sec)\nINFO:tensorflow:lr = 0.00079530483 (10.907 sec)\nINFO:tensorflow:global_step/sec: 9.26539\nINFO:tensorflow:loss = 4.991285, step = 38900 (10.790 sec)\nINFO:tensorflow:lr = 0.00079709693 (10.791 sec)\nINFO:tensorflow:global_step/sec: 9.25618\nINFO:tensorflow:loss = 4.978191, step = 39000 (10.806 sec)\nINFO:tensorflow:lr = 0.0007988889 (10.804 sec)\nINFO:tensorflow:global_step/sec: 9.49369\nINFO:tensorflow:loss = 4.971673, step = 39100 (10.533 sec)\nINFO:tensorflow:lr = 0.00079931895 (10.534 sec)\nINFO:tensorflow:global_step/sec: 9.44335\nINFO:tensorflow:loss = 4.9710107, step = 39200 (10.589 sec)\nINFO:tensorflow:lr = 0.00079752697 (10.590 sec)\nINFO:tensorflow:global_step/sec: 9.38353\nINFO:tensorflow:loss = 5.0888605, step = 39300 (10.655 sec)\nINFO:tensorflow:lr = 0.00079573493 (10.657 sec)\nINFO:tensorflow:global_step/sec: 9.34901\nINFO:tensorflow:loss = 5.1406965, step = 39400 (10.699 sec)\nINFO:tensorflow:lr = 0.00079394295 (10.697 sec)\nINFO:tensorflow:global_step/sec: 9.40024\nINFO:tensorflow:loss = 5.0318465, step = 39500 (10.637 sec)\nINFO:tensorflow:lr = 0.0007921509 (10.638 sec)\nINFO:tensorflow:global_step/sec: 9.57672\nINFO:tensorflow:loss = 4.9905806, step = 39600 (10.440 sec)\nINFO:tensorflow:lr = 0.0007903589 (10.442 sec)\nINFO:tensorflow:global_step/sec: 9.23393\nINFO:tensorflow:loss = 5.020684, step = 39700 (10.832 sec)\nINFO:tensorflow:lr = 0.0007885669 (10.829 sec)\nINFO:tensorflow:global_step/sec: 9.67045\nINFO:tensorflow:loss = 4.956047, step = 39800 (10.340 sec)\nINFO:tensorflow:lr = 0.00078677485 (10.341 sec)\nINFO:tensorflow:global_step/sec: 9.50074\nINFO:tensorflow:loss = 5.0920815, step = 39900 (10.526 sec)\nINFO:tensorflow:lr = 0.00078498287 (10.526 sec)\nINFO:tensorflow:global_step/sec: 9.41762\nINFO:tensorflow:loss = 4.9771967, step = 40000 (10.617 sec)\nINFO:tensorflow:lr = 0.0007831908 (10.617 sec)\nINFO:tensorflow:global_step/sec: 9.4584\nINFO:tensorflow:loss = 5.0041213, step = 40100 (10.572 sec)\nINFO:tensorflow:lr = 0.00078139873 (10.574 sec)\nINFO:tensorflow:global_step/sec: 9.30685\nINFO:tensorflow:loss = 5.129474, step = 40200 (10.747 sec)\nINFO:tensorflow:lr = 0.00077960675 (10.745 sec)\nINFO:tensorflow:global_step/sec: 9.10269\nINFO:tensorflow:loss = 5.1569552, step = 40300 (10.985 sec)\nINFO:tensorflow:lr = 0.0007778147 (10.986 sec)\nINFO:tensorflow:global_step/sec: 9.20877\nINFO:tensorflow:loss = 5.0918455, step = 40400 (10.861 sec)\nINFO:tensorflow:lr = 0.00077602273 (10.860 sec)\nINFO:tensorflow:global_step/sec: 9.00773\nINFO:tensorflow:loss = 4.9962397, step = 40500 (11.098 sec)\nINFO:tensorflow:lr = 0.0007742307 (11.100 sec)\nINFO:tensorflow:global_step/sec: 9.2361\nINFO:tensorflow:loss = 5.024959, step = 40600 (10.829 sec)\nINFO:tensorflow:lr = 0.0007724387 (10.828 sec)\nINFO:tensorflow:global_step/sec: 9.43495\nINFO:tensorflow:loss = 5.0192513, step = 40700 (10.599 sec)\nINFO:tensorflow:lr = 0.0007706467 (10.598 sec)\nINFO:tensorflow:global_step/sec: 9.45791\nINFO:tensorflow:loss = 5.0989456, step = 40800 (10.574 sec)\nINFO:tensorflow:lr = 0.00076885463 (10.574 sec)\nINFO:tensorflow:global_step/sec: 9.43583\nINFO:tensorflow:loss = 5.1665564, step = 40900 (10.598 sec)\nINFO:tensorflow:lr = 0.00076706253 (10.598 sec)\nINFO:tensorflow:global_step/sec: 9.29103\nINFO:tensorflow:loss = 5.0380497, step = 41000 (10.760 sec)\nINFO:tensorflow:lr = 0.00076527055 (10.761 sec)\nINFO:tensorflow:global_step/sec: 9.33781\nINFO:tensorflow:loss = 4.9364643, step = 41100 (10.711 sec)\nINFO:tensorflow:lr = 0.0007634785 (10.710 sec)\nINFO:tensorflow:global_step/sec: 9.44074\nINFO:tensorflow:loss = 5.0375924, step = 41200 (10.590 sec)\nINFO:tensorflow:lr = 0.00076168653 (10.591 sec)\nINFO:tensorflow:global_step/sec: 9.11934\nINFO:tensorflow:loss = 5.2311244, step = 41300 (10.966 sec)\nINFO:tensorflow:lr = 0.0007598945 (10.968 sec)\nINFO:tensorflow:global_step/sec: 9.49467\nINFO:tensorflow:loss = 5.061126, step = 41400 (10.534 sec)\nINFO:tensorflow:lr = 0.0007581025 (10.531 sec)\nINFO:tensorflow:global_step/sec: 9.18987\nINFO:tensorflow:loss = 5.0643272, step = 41500 (10.882 sec)\nINFO:tensorflow:lr = 0.0007563105 (10.882 sec)\nINFO:tensorflow:global_step/sec: 9.25841\nINFO:tensorflow:loss = 5.047558, step = 41600 (10.801 sec)\nINFO:tensorflow:lr = 0.0007545185 (10.802 sec)\nINFO:tensorflow:global_step/sec: 9.57013\nINFO:tensorflow:loss = 4.957797, step = 41700 (10.448 sec)\nINFO:tensorflow:lr = 0.00075272645 (10.447 sec)\nINFO:tensorflow:global_step/sec: 9.60469\nINFO:tensorflow:loss = 4.9590044, step = 41800 (10.411 sec)\nINFO:tensorflow:lr = 0.00075093436 (10.411 sec)\nINFO:tensorflow:global_step/sec: 9.32542\nINFO:tensorflow:loss = 5.100118, step = 41900 (10.727 sec)\nINFO:tensorflow:lr = 0.0007491423 (10.726 sec)\nINFO:tensorflow:global_step/sec: 9.33525\nINFO:tensorflow:loss = 5.0650043, step = 42000 (10.713 sec)\nINFO:tensorflow:lr = 0.00074735034 (10.713 sec)\nINFO:tensorflow:global_step/sec: 9.32104\nINFO:tensorflow:loss = 4.912504, step = 42100 (10.726 sec)\nINFO:tensorflow:lr = 0.0007455583 (10.726 sec)\nINFO:tensorflow:global_step/sec: 9.55293\nINFO:tensorflow:loss = 5.0278497, step = 42200 (10.470 sec)\nINFO:tensorflow:lr = 0.0007437663 (10.469 sec)\nINFO:tensorflow:global_step/sec: 9.28278\nINFO:tensorflow:loss = 5.1773267, step = 42300 (10.772 sec)\nINFO:tensorflow:lr = 0.0007419743 (10.773 sec)\nINFO:tensorflow:global_step/sec: 9.45226\nINFO:tensorflow:loss = 5.0096893, step = 42400 (10.579 sec)\nINFO:tensorflow:lr = 0.0007401823 (10.579 sec)\nINFO:tensorflow:global_step/sec: 9.50569\nINFO:tensorflow:loss = 4.9839783, step = 42500 (10.518 sec)\nINFO:tensorflow:lr = 0.00073839026 (10.517 sec)\nINFO:tensorflow:global_step/sec: 9.37434\nINFO:tensorflow:loss = 4.950326, step = 42600 (10.667 sec)\nINFO:tensorflow:lr = 0.0007365983 (10.669 sec)\nINFO:tensorflow:global_step/sec: 9.42829\nINFO:tensorflow:loss = 5.3135276, step = 42700 (10.609 sec)\nINFO:tensorflow:lr = 0.0007348062 (10.611 sec)\nINFO:tensorflow:global_step/sec: 9.51614\nINFO:tensorflow:loss = 5.0509195, step = 42800 (10.510 sec)\nINFO:tensorflow:lr = 0.00073301414 (10.506 sec)\nINFO:tensorflow:global_step/sec: 9.70035\nINFO:tensorflow:loss = 4.912774, step = 42900 (10.307 sec)\nINFO:tensorflow:lr = 0.0007312221 (10.308 sec)\nINFO:tensorflow:global_step/sec: 9.53879\nINFO:tensorflow:loss = 4.9984446, step = 43000 (10.485 sec)\nINFO:tensorflow:lr = 0.0007294301 (10.484 sec)\nINFO:tensorflow:global_step/sec: 9.41552\nINFO:tensorflow:loss = 4.9727006, step = 43100 (10.621 sec)\nINFO:tensorflow:lr = 0.0007276381 (10.620 sec)\nINFO:tensorflow:global_step/sec: 9.34481\nINFO:tensorflow:loss = 5.032983, step = 43200 (10.701 sec)\nINFO:tensorflow:lr = 0.0007258461 (10.701 sec)\nINFO:tensorflow:global_step/sec: 9.57305\nINFO:tensorflow:loss = 5.0043697, step = 43300 (10.446 sec)\nINFO:tensorflow:lr = 0.00072405406 (10.447 sec)\nINFO:tensorflow:global_step/sec: 9.43392\nINFO:tensorflow:loss = 4.9424596, step = 43400 (10.599 sec)\nINFO:tensorflow:lr = 0.0007222621 (10.599 sec)\nINFO:tensorflow:global_step/sec: 9.50868\nINFO:tensorflow:loss = 5.039576, step = 43500 (10.514 sec)\nINFO:tensorflow:lr = 0.00072047004 (10.517 sec)\nINFO:tensorflow:global_step/sec: 9.34554\nINFO:tensorflow:loss = 5.0512376, step = 43600 (10.702 sec)\nINFO:tensorflow:lr = 0.00071867794 (10.700 sec)\nINFO:tensorflow:global_step/sec: 9.3316\nINFO:tensorflow:loss = 4.8776455, step = 43700 (10.718 sec)\nINFO:tensorflow:lr = 0.00071688596 (10.718 sec)\nINFO:tensorflow:global_step/sec: 9.42098\nINFO:tensorflow:loss = 4.9753275, step = 43800 (10.613 sec)\nINFO:tensorflow:lr = 0.0007150939 (10.613 sec)\nINFO:tensorflow:global_step/sec: 9.34695\nINFO:tensorflow:loss = 5.1619225, step = 43900 (10.696 sec)\nINFO:tensorflow:lr = 0.0007133019 (10.696 sec)\nINFO:tensorflow:global_step/sec: 9.66996\nINFO:tensorflow:loss = 4.897856, step = 44000 (10.344 sec)\nINFO:tensorflow:lr = 0.0007115099 (10.344 sec)\nINFO:tensorflow:global_step/sec: 9.31438\nINFO:tensorflow:loss = 4.9865513, step = 44100 (10.735 sec)\nINFO:tensorflow:lr = 0.00070971786 (10.734 sec)\nINFO:tensorflow:global_step/sec: 9.32455\nINFO:tensorflow:loss = 5.0448728, step = 44200 (10.726 sec)\nINFO:tensorflow:lr = 0.0007079259 (10.729 sec)\nINFO:tensorflow:global_step/sec: 9.48733\nINFO:tensorflow:loss = 5.148017, step = 44300 (10.540 sec)\nINFO:tensorflow:lr = 0.00070613384 (10.538 sec)\nINFO:tensorflow:global_step/sec: 9.52093\nINFO:tensorflow:loss = 4.945143, step = 44400 (10.501 sec)\nINFO:tensorflow:lr = 0.00070434186 (10.501 sec)\nINFO:tensorflow:global_step/sec: 9.47543\nINFO:tensorflow:loss = 5.114677, step = 44500 (10.554 sec)\nINFO:tensorflow:lr = 0.00070254976 (10.555 sec)\nINFO:tensorflow:global_step/sec: 9.50398\nINFO:tensorflow:loss = 4.9485087, step = 44600 (10.523 sec)\nINFO:tensorflow:lr = 0.0007007577 (10.523 sec)\nINFO:tensorflow:global_step/sec: 9.55528\nINFO:tensorflow:loss = 5.1555524, step = 44700 (10.467 sec)\nINFO:tensorflow:lr = 0.0006989657 (10.466 sec)\nINFO:tensorflow:global_step/sec: 9.40312\nINFO:tensorflow:loss = 5.0596046, step = 44800 (10.633 sec)\nINFO:tensorflow:lr = 0.0006971737 (10.634 sec)\nINFO:tensorflow:global_step/sec: 9.53255\nINFO:tensorflow:loss = 4.982494, step = 44900 (10.491 sec)\nINFO:tensorflow:lr = 0.00069538166 (10.491 sec)\nINFO:tensorflow:global_step/sec: 9.27382\nINFO:tensorflow:loss = 5.0182176, step = 45000 (10.781 sec)\nINFO:tensorflow:lr = 0.0006935897 (10.781 sec)\nINFO:tensorflow:global_step/sec: 9.39855\nINFO:tensorflow:loss = 5.0612426, step = 45100 (10.644 sec)\nINFO:tensorflow:lr = 0.00069179764 (10.644 sec)\nINFO:tensorflow:global_step/sec: 9.62145\nINFO:tensorflow:loss = 4.989537, step = 45200 (10.391 sec)\nINFO:tensorflow:lr = 0.00069000566 (10.392 sec)\nINFO:tensorflow:global_step/sec: 9.51908\nINFO:tensorflow:loss = 5.0123234, step = 45300 (10.505 sec)\nINFO:tensorflow:lr = 0.0006882136 (10.504 sec)\nINFO:tensorflow:global_step/sec: 9.51554\nINFO:tensorflow:loss = 4.9916244, step = 45400 (10.513 sec)\nINFO:tensorflow:lr = 0.0006864215 (10.513 sec)\nINFO:tensorflow:global_step/sec: 9.50763\nINFO:tensorflow:loss = 5.095473, step = 45500 (10.514 sec)\nINFO:tensorflow:lr = 0.00068462954 (10.515 sec)\nINFO:tensorflow:global_step/sec: 9.37476\nINFO:tensorflow:loss = 5.039061, step = 45600 (10.665 sec)\nINFO:tensorflow:lr = 0.0006828375 (10.665 sec)\nINFO:tensorflow:global_step/sec: 9.37142\nINFO:tensorflow:loss = 4.960917, step = 45700 (10.671 sec)\nINFO:tensorflow:lr = 0.00068104547 (10.670 sec)\nINFO:tensorflow:global_step/sec: 9.5341\nINFO:tensorflow:loss = 5.003217, step = 45800 (10.490 sec)\nINFO:tensorflow:lr = 0.0006792535 (10.491 sec)\nINFO:tensorflow:global_step/sec: 9.6098\nINFO:tensorflow:loss = 5.049335, step = 45900 (10.406 sec)\nINFO:tensorflow:lr = 0.00067746144 (10.406 sec)\nINFO:tensorflow:global_step/sec: 9.08646\nINFO:tensorflow:loss = 5.0135064, step = 46000 (11.003 sec)\nINFO:tensorflow:lr = 0.00067566946 (11.005 sec)\nINFO:tensorflow:global_step/sec: 8.75664\nINFO:tensorflow:loss = 5.2109594, step = 46100 (11.422 sec)\nINFO:tensorflow:lr = 0.0006738774 (11.421 sec)\nINFO:tensorflow:global_step/sec: 9.44446\nINFO:tensorflow:loss = 4.8870687, step = 46200 (10.587 sec)\nINFO:tensorflow:lr = 0.00067208544 (10.588 sec)\nINFO:tensorflow:global_step/sec: 9.28042\nINFO:tensorflow:loss = 4.961076, step = 46300 (10.776 sec)\nINFO:tensorflow:lr = 0.00067029335 (10.775 sec)\nINFO:tensorflow:global_step/sec: 9.67404\nINFO:tensorflow:loss = 4.972317, step = 46400 (10.337 sec)\nINFO:tensorflow:lr = 0.0006685013 (10.336 sec)\nINFO:tensorflow:global_step/sec: 9.38469\nINFO:tensorflow:loss = 5.1011596, step = 46500 (10.656 sec)\nINFO:tensorflow:lr = 0.0006667093 (10.657 sec)\nINFO:tensorflow:global_step/sec: 9.30863\nINFO:tensorflow:loss = 5.058347, step = 46600 (10.742 sec)\nINFO:tensorflow:lr = 0.0006649173 (10.742 sec)\nINFO:tensorflow:global_step/sec: 9.1271\nINFO:tensorflow:loss = 4.9873576, step = 46700 (10.957 sec)\nINFO:tensorflow:lr = 0.00066312525 (10.957 sec)\nINFO:tensorflow:global_step/sec: 9.22479\nINFO:tensorflow:loss = 4.923091, step = 46800 (10.839 sec)\nINFO:tensorflow:lr = 0.00066133327 (10.840 sec)\nINFO:tensorflow:global_step/sec: 9.53795\nINFO:tensorflow:loss = 5.0580025, step = 46900 (10.486 sec)\nINFO:tensorflow:lr = 0.0006595412 (10.486 sec)\nINFO:tensorflow:global_step/sec: 9.2316\nINFO:tensorflow:loss = 5.000795, step = 47000 (10.833 sec)\nINFO:tensorflow:lr = 0.00065774925 (10.834 sec)\nINFO:tensorflow:global_step/sec: 9.18839\nINFO:tensorflow:loss = 4.871485, step = 47100 (10.882 sec)\nINFO:tensorflow:lr = 0.0006559572 (10.882 sec)\nINFO:tensorflow:global_step/sec: 9.37196\nINFO:tensorflow:loss = 4.8876815, step = 47200 (10.670 sec)\nINFO:tensorflow:lr = 0.0006541651 (10.669 sec)\nINFO:tensorflow:global_step/sec: 9.21784\nINFO:tensorflow:loss = 5.043597, step = 47300 (10.848 sec)\nINFO:tensorflow:lr = 0.0006523731 (10.849 sec)\nINFO:tensorflow:global_step/sec: 9.28215\nINFO:tensorflow:loss = 5.1398096, step = 47400 (10.771 sec)\nINFO:tensorflow:lr = 0.0006505811 (10.771 sec)\nINFO:tensorflow:global_step/sec: 9.21442\nINFO:tensorflow:loss = 4.897675, step = 47500 (10.855 sec)\nINFO:tensorflow:lr = 0.0006487891 (10.856 sec)\nINFO:tensorflow:global_step/sec: 9.35004\nINFO:tensorflow:loss = 5.0621614, step = 47600 (10.694 sec)\nINFO:tensorflow:lr = 0.00064699707 (10.694 sec)\nINFO:tensorflow:global_step/sec: 9.58694\nINFO:tensorflow:loss = 5.076569, step = 47700 (10.432 sec)\nINFO:tensorflow:lr = 0.00064520503 (10.432 sec)\nINFO:tensorflow:global_step/sec: 9.32931\nINFO:tensorflow:loss = 5.0489826, step = 47800 (10.718 sec)\nINFO:tensorflow:lr = 0.00064341305 (10.718 sec)\nINFO:tensorflow:global_step/sec: 9.38456\nINFO:tensorflow:loss = 4.988774, step = 47900 (10.654 sec)\nINFO:tensorflow:lr = 0.000641621 (10.654 sec)\nINFO:tensorflow:global_step/sec: 9.39305\nINFO:tensorflow:loss = 5.1435757, step = 48000 (10.646 sec)\nINFO:tensorflow:lr = 0.000639829 (10.646 sec)\nINFO:tensorflow:global_step/sec: 9.53771\nINFO:tensorflow:loss = 4.9170194, step = 48100 (10.486 sec)\nINFO:tensorflow:lr = 0.00063803693 (10.488 sec)\nINFO:tensorflow:global_step/sec: 9.35327\nINFO:tensorflow:loss = 4.9841676, step = 48200 (10.692 sec)\nINFO:tensorflow:lr = 0.0006362449 (10.690 sec)\nINFO:tensorflow:global_step/sec: 9.45542\nINFO:tensorflow:loss = 5.089015, step = 48300 (10.575 sec)\nINFO:tensorflow:lr = 0.0006344529 (10.575 sec)\nINFO:tensorflow:global_step/sec: 9.48675\nINFO:tensorflow:loss = 5.1919518, step = 48400 (10.542 sec)\nINFO:tensorflow:lr = 0.0006326609 (10.542 sec)\nINFO:tensorflow:global_step/sec: 9.16907\nINFO:tensorflow:loss = 4.9903545, step = 48500 (10.907 sec)\nINFO:tensorflow:lr = 0.0006308689 (10.906 sec)\nINFO:tensorflow:global_step/sec: 9.37422\nINFO:tensorflow:loss = 5.088899, step = 48600 (10.667 sec)\nINFO:tensorflow:lr = 0.00062907685 (10.667 sec)\nINFO:tensorflow:global_step/sec: 9.48194\nINFO:tensorflow:loss = 4.8907027, step = 48700 (10.547 sec)\nINFO:tensorflow:lr = 0.0006272848 (10.548 sec)\nINFO:tensorflow:global_step/sec: 9.2032\nINFO:tensorflow:loss = 4.991426, step = 48800 (10.866 sec)\nINFO:tensorflow:lr = 0.00062549283 (10.865 sec)\nINFO:tensorflow:global_step/sec: 9.37849\nINFO:tensorflow:loss = 4.985582, step = 48900 (10.660 sec)\nINFO:tensorflow:lr = 0.0006237008 (10.660 sec)\nINFO:tensorflow:global_step/sec: 8.8565\nINFO:tensorflow:loss = 5.1335573, step = 49000 (11.293 sec)\nINFO:tensorflow:lr = 0.0006219087 (11.293 sec)\nINFO:tensorflow:global_step/sec: 9.43977\nINFO:tensorflow:loss = 4.9458632, step = 49100 (10.592 sec)\nINFO:tensorflow:lr = 0.0006201167 (10.592 sec)\nINFO:tensorflow:global_step/sec: 9.51817\nINFO:tensorflow:loss = 4.918289, step = 49200 (10.508 sec)\nINFO:tensorflow:lr = 0.0006183247 (10.507 sec)\nINFO:tensorflow:global_step/sec: 9.52134\nINFO:tensorflow:loss = 5.1120663, step = 49300 (10.502 sec)\nINFO:tensorflow:lr = 0.0006165327 (10.502 sec)\nINFO:tensorflow:global_step/sec: 9.35327\nINFO:tensorflow:loss = 5.1775713, step = 49400 (10.692 sec)\nINFO:tensorflow:lr = 0.00061474065 (10.693 sec)\nINFO:tensorflow:global_step/sec: 9.17388\nINFO:tensorflow:loss = 4.9629602, step = 49500 (10.899 sec)\nINFO:tensorflow:lr = 0.0006129486 (10.899 sec)\nINFO:tensorflow:global_step/sec: 9.58739\nINFO:tensorflow:loss = 5.0018997, step = 49600 (10.432 sec)\nINFO:tensorflow:lr = 0.00061115663 (10.432 sec)\nINFO:tensorflow:global_step/sec: 9.43341\nINFO:tensorflow:loss = 5.023855, step = 49700 (10.602 sec)\nINFO:tensorflow:lr = 0.0006093646 (10.603 sec)\nINFO:tensorflow:global_step/sec: 9.51348\nINFO:tensorflow:loss = 5.024961, step = 49800 (10.511 sec)\nINFO:tensorflow:lr = 0.0006075726 (10.509 sec)\nINFO:tensorflow:global_step/sec: 9.368\nINFO:tensorflow:loss = 4.97074, step = 49900 (10.672 sec)\nINFO:tensorflow:lr = 0.0006057805 (10.673 sec)\nINFO:tensorflow:global_step/sec: 9.59222\nINFO:tensorflow:loss = 5.1788244, step = 50000 (10.428 sec)\nINFO:tensorflow:lr = 0.0006039885 (10.426 sec)\nINFO:tensorflow:global_step/sec: 9.53275\nINFO:tensorflow:loss = 4.973164, step = 50100 (10.496 sec)\nINFO:tensorflow:lr = 0.0006021965 (10.497 sec)\nINFO:tensorflow:global_step/sec: 9.53317\nINFO:tensorflow:loss = 4.9693937, step = 50200 (10.481 sec)\nINFO:tensorflow:lr = 0.00060040446 (10.480 sec)\nINFO:tensorflow:global_step/sec: 9.65217\nINFO:tensorflow:loss = 4.950563, step = 50300 (10.363 sec)\nINFO:tensorflow:lr = 0.0005986125 (10.362 sec)\nINFO:tensorflow:global_step/sec: 9.58435\nINFO:tensorflow:loss = 4.9971285, step = 50400 (10.433 sec)\nINFO:tensorflow:lr = 0.00059682044 (10.435 sec)\nINFO:tensorflow:global_step/sec: 9.66644\nINFO:tensorflow:loss = 4.944786, step = 50500 (10.344 sec)\nINFO:tensorflow:lr = 0.0005950284 (10.344 sec)\nINFO:tensorflow:global_step/sec: 9.55486\nINFO:tensorflow:loss = 5.0195484, step = 50600 (10.466 sec)\nINFO:tensorflow:lr = 0.0005932364 (10.467 sec)\nINFO:tensorflow:global_step/sec: 9.52583\nINFO:tensorflow:loss = 4.9206963, step = 50700 (10.497 sec)\nINFO:tensorflow:lr = 0.0005914444 (10.495 sec)\nINFO:tensorflow:global_step/sec: 9.60777\nINFO:tensorflow:loss = 4.938217, step = 50800 (10.409 sec)\nINFO:tensorflow:lr = 0.0005896523 (10.411 sec)\nINFO:tensorflow:global_step/sec: 9.36609\nINFO:tensorflow:loss = 4.8422065, step = 50900 (10.677 sec)\nINFO:tensorflow:lr = 0.0005878603 (10.675 sec)\nINFO:tensorflow:global_step/sec: 9.52744\nINFO:tensorflow:loss = 5.123394, step = 51000 (10.498 sec)\nINFO:tensorflow:lr = 0.00058606826 (10.497 sec)\nINFO:tensorflow:global_step/sec: 9.28914\nINFO:tensorflow:loss = 5.1042643, step = 51100 (10.766 sec)\nINFO:tensorflow:lr = 0.0005842762 (10.766 sec)\nINFO:tensorflow:global_step/sec: 9.37764\nINFO:tensorflow:loss = 5.131436, step = 51200 (10.664 sec)\nINFO:tensorflow:lr = 0.00058248424 (10.664 sec)\nINFO:tensorflow:global_step/sec: 9.7\nINFO:tensorflow:loss = 4.88273, step = 51300 (10.308 sec)\nINFO:tensorflow:lr = 0.0005806922 (10.309 sec)\nINFO:tensorflow:global_step/sec: 9.60252\nINFO:tensorflow:loss = 5.05976, step = 51400 (10.414 sec)\nINFO:tensorflow:lr = 0.0005789002 (10.414 sec)\nINFO:tensorflow:global_step/sec: 9.59476\nINFO:tensorflow:loss = 5.0464835, step = 51500 (10.423 sec)\nINFO:tensorflow:lr = 0.0005771082 (10.423 sec)\nINFO:tensorflow:global_step/sec: 9.50306\nINFO:tensorflow:loss = 4.9827104, step = 51600 (10.522 sec)\nINFO:tensorflow:lr = 0.0005753162 (10.522 sec)\nINFO:tensorflow:global_step/sec: 9.38927\nINFO:tensorflow:loss = 5.0948005, step = 51700 (10.652 sec)\nINFO:tensorflow:lr = 0.0005735241 (10.652 sec)\nINFO:tensorflow:global_step/sec: 9.51707\nINFO:tensorflow:loss = 5.045607, step = 51800 (10.506 sec)\nINFO:tensorflow:lr = 0.00057173206 (10.506 sec)\nINFO:tensorflow:global_step/sec: 9.3261\nINFO:tensorflow:loss = 4.8977003, step = 51900 (10.722 sec)\nINFO:tensorflow:lr = 0.0005699401 (10.723 sec)\nINFO:tensorflow:global_step/sec: 9.1791\nINFO:tensorflow:loss = 4.9912663, step = 52000 (10.894 sec)\nINFO:tensorflow:lr = 0.00056814804 (10.894 sec)\nINFO:tensorflow:global_step/sec: 9.39772\nINFO:tensorflow:loss = 5.0285425, step = 52100 (10.640 sec)\nINFO:tensorflow:lr = 0.000566356 (10.639 sec)\nINFO:tensorflow:global_step/sec: 9.62507\nINFO:tensorflow:loss = 4.894455, step = 52200 (10.392 sec)\nINFO:tensorflow:lr = 0.000564564 (10.391 sec)\nINFO:tensorflow:global_step/sec: 9.70896\nINFO:tensorflow:loss = 5.096932, step = 52300 (10.301 sec)\nINFO:tensorflow:lr = 0.000562772 (10.300 sec)\nINFO:tensorflow:global_step/sec: 9.66523\nINFO:tensorflow:loss = 4.941601, step = 52400 (10.345 sec)\nINFO:tensorflow:lr = 0.00056098 (10.347 sec)\nINFO:tensorflow:global_step/sec: 9.52534\nINFO:tensorflow:loss = 4.811992, step = 52500 (10.498 sec)\nINFO:tensorflow:lr = 0.00055918796 (10.498 sec)\nINFO:tensorflow:global_step/sec: 9.35068\nINFO:tensorflow:loss = 4.9959965, step = 52600 (10.695 sec)\nINFO:tensorflow:lr = 0.00055739586 (10.695 sec)\nINFO:tensorflow:global_step/sec: 9.41247\nINFO:tensorflow:loss = 4.9717216, step = 52700 (10.621 sec)\nINFO:tensorflow:lr = 0.0005556039 (10.622 sec)\nINFO:tensorflow:global_step/sec: 9.58399\nINFO:tensorflow:loss = 5.0410056, step = 52800 (10.436 sec)\nINFO:tensorflow:lr = 0.00055381184 (10.438 sec)\nINFO:tensorflow:global_step/sec: 9.53823\nINFO:tensorflow:loss = 5.0049415, step = 52900 (10.483 sec)\nINFO:tensorflow:lr = 0.00055201986 (10.479 sec)\nINFO:tensorflow:global_step/sec: 9.46178\nINFO:tensorflow:loss = 5.153189, step = 53000 (10.572 sec)\nINFO:tensorflow:lr = 0.0005502278 (10.572 sec)\nINFO:tensorflow:global_step/sec: 9.38044\nINFO:tensorflow:loss = 5.045697, step = 53100 (10.659 sec)\nINFO:tensorflow:lr = 0.0005484358 (10.659 sec)\nINFO:tensorflow:global_step/sec: 9.31813\nINFO:tensorflow:loss = 4.9369597, step = 53200 (10.734 sec)\nINFO:tensorflow:lr = 0.0005466438 (10.734 sec)\nINFO:tensorflow:global_step/sec: 9.6857\nINFO:tensorflow:loss = 5.1337905, step = 53300 (10.320 sec)\nINFO:tensorflow:lr = 0.00054485176 (10.320 sec)\nINFO:tensorflow:global_step/sec: 9.61851\nINFO:tensorflow:loss = 5.0458665, step = 53400 (10.399 sec)\nINFO:tensorflow:lr = 0.0005430598 (10.399 sec)\nINFO:tensorflow:global_step/sec: 9.5056\nINFO:tensorflow:loss = 5.0202885, step = 53500 (10.520 sec)\nINFO:tensorflow:lr = 0.0005412677 (10.520 sec)\nINFO:tensorflow:global_step/sec: 9.49133\nINFO:tensorflow:loss = 5.0235763, step = 53600 (10.534 sec)\nINFO:tensorflow:lr = 0.00053947564 (10.537 sec)\nINFO:tensorflow:global_step/sec: 9.3808\nINFO:tensorflow:loss = 4.963329, step = 53700 (10.662 sec)\nINFO:tensorflow:lr = 0.00053768366 (10.660 sec)\nINFO:tensorflow:global_step/sec: 9.46908\nINFO:tensorflow:loss = 4.926022, step = 53800 (10.559 sec)\nINFO:tensorflow:lr = 0.0005358916 (10.561 sec)\nINFO:tensorflow:global_step/sec: 9.32461\nINFO:tensorflow:loss = 5.055513, step = 53900 (10.726 sec)\nINFO:tensorflow:lr = 0.0005340996 (10.724 sec)\nINFO:tensorflow:global_step/sec: 9.46242\nINFO:tensorflow:loss = 4.9468417, step = 54000 (10.569 sec)\nINFO:tensorflow:lr = 0.0005323076 (10.568 sec)\nINFO:tensorflow:global_step/sec: 9.21342\nINFO:tensorflow:loss = 5.1168594, step = 54100 (10.854 sec)\nINFO:tensorflow:lr = 0.00053051556 (10.854 sec)\nINFO:tensorflow:global_step/sec: 9.40069\nINFO:tensorflow:loss = 4.906956, step = 54200 (10.637 sec)\nINFO:tensorflow:lr = 0.0005287236 (10.637 sec)\nINFO:tensorflow:global_step/sec: 9.24311\nINFO:tensorflow:loss = 5.017212, step = 54300 (10.820 sec)\nINFO:tensorflow:lr = 0.00052693154 (10.819 sec)\nINFO:tensorflow:global_step/sec: 9.28552\nINFO:tensorflow:loss = 5.069296, step = 54400 (10.770 sec)\nINFO:tensorflow:lr = 0.00052513945 (10.769 sec)\nINFO:tensorflow:global_step/sec: 9.49994\nINFO:tensorflow:loss = 4.862296, step = 54500 (10.527 sec)\nINFO:tensorflow:lr = 0.00052334747 (10.527 sec)\nINFO:tensorflow:global_step/sec: 9.47938\nINFO:tensorflow:loss = 5.018501, step = 54600 (10.548 sec)\nINFO:tensorflow:lr = 0.0005215554 (10.549 sec)\nINFO:tensorflow:global_step/sec: 9.65287\nINFO:tensorflow:loss = 4.8488297, step = 54700 (10.360 sec)\nINFO:tensorflow:lr = 0.00051976345 (10.359 sec)\nINFO:tensorflow:global_step/sec: 9.33386\nINFO:tensorflow:loss = 4.833839, step = 54800 (10.715 sec)\nINFO:tensorflow:lr = 0.0005179714 (10.715 sec)\nINFO:tensorflow:global_step/sec: 9.21715\nINFO:tensorflow:loss = 4.930398, step = 54900 (10.847 sec)\nINFO:tensorflow:lr = 0.00051617937 (10.847 sec)\nINFO:tensorflow:global_step/sec: 9.2234\nINFO:tensorflow:loss = 4.9686055, step = 55000 (10.843 sec)\nINFO:tensorflow:lr = 0.0005143874 (10.843 sec)\nINFO:tensorflow:global_step/sec: 9.50335\nINFO:tensorflow:loss = 5.014664, step = 55100 (10.520 sec)\nINFO:tensorflow:lr = 0.00051259535 (10.521 sec)\nINFO:tensorflow:global_step/sec: 9.50403\nINFO:tensorflow:loss = 4.879487, step = 55200 (10.524 sec)\nINFO:tensorflow:lr = 0.00051080337 (10.524 sec)\nINFO:tensorflow:global_step/sec: 9.44257\nINFO:tensorflow:loss = 5.077122, step = 55300 (10.592 sec)\nINFO:tensorflow:lr = 0.00050901127 (10.591 sec)\nINFO:tensorflow:global_step/sec: 9.43012\nINFO:tensorflow:loss = 4.927007, step = 55400 (10.604 sec)\nINFO:tensorflow:lr = 0.0005072192 (10.606 sec)\nINFO:tensorflow:global_step/sec: 9.391\nINFO:tensorflow:loss = 5.031347, step = 55500 (10.648 sec)\nINFO:tensorflow:lr = 0.00050542725 (10.647 sec)\nINFO:tensorflow:global_step/sec: 9.33362\nINFO:tensorflow:loss = 4.995307, step = 55600 (10.714 sec)\nINFO:tensorflow:lr = 0.0005036352 (10.714 sec)\nINFO:tensorflow:global_step/sec: 9.42561\nINFO:tensorflow:loss = 5.0791025, step = 55700 (10.606 sec)\nINFO:tensorflow:lr = 0.0005018432 (10.608 sec)\nINFO:tensorflow:global_step/sec: 9.58455\nINFO:tensorflow:loss = 4.891101, step = 55800 (10.437 sec)\nINFO:tensorflow:lr = 0.0005000512 (10.434 sec)\nINFO:tensorflow:global_step/sec: 9.49456\nINFO:tensorflow:loss = 4.8779163, step = 55900 (10.531 sec)\nINFO:tensorflow:lr = 0.00049825915 (10.531 sec)\nINFO:tensorflow:global_step/sec: 9.55119\nINFO:tensorflow:loss = 4.991086, step = 56000 (10.470 sec)\nINFO:tensorflow:lr = 0.00049646717 (10.471 sec)\nINFO:tensorflow:global_step/sec: 9.55963\nINFO:tensorflow:loss = 4.9344687, step = 56100 (10.461 sec)\nINFO:tensorflow:lr = 0.0004946751 (10.460 sec)\nINFO:tensorflow:global_step/sec: 9.46301\nINFO:tensorflow:loss = 4.91139, step = 56200 (10.567 sec)\nINFO:tensorflow:lr = 0.00049288303 (10.567 sec)\nINFO:tensorflow:global_step/sec: 9.54473\nINFO:tensorflow:loss = 4.9695115, step = 56300 (10.478 sec)\nINFO:tensorflow:lr = 0.00049109105 (10.478 sec)\nINFO:tensorflow:global_step/sec: 9.63134\nINFO:tensorflow:loss = 4.8727584, step = 56400 (10.382 sec)\nINFO:tensorflow:lr = 0.000489299 (10.382 sec)\nINFO:tensorflow:global_step/sec: 9.52309\nINFO:tensorflow:loss = 5.0401974, step = 56500 (10.502 sec)\nINFO:tensorflow:lr = 0.00048750703 (10.502 sec)\nINFO:tensorflow:global_step/sec: 9.65266\nINFO:tensorflow:loss = 4.8206286, step = 56600 (10.356 sec)\nINFO:tensorflow:lr = 0.00048571502 (10.357 sec)\nINFO:tensorflow:global_step/sec: 9.64404\nINFO:tensorflow:loss = 4.914998, step = 56700 (10.371 sec)\nINFO:tensorflow:lr = 0.000483923 (10.372 sec)\nINFO:tensorflow:global_step/sec: 9.50445\nINFO:tensorflow:loss = 4.927148, step = 56800 (10.523 sec)\nINFO:tensorflow:lr = 0.00048213097 (10.522 sec)\nINFO:tensorflow:global_step/sec: 9.68784\nINFO:tensorflow:loss = 4.9782243, step = 56900 (10.321 sec)\nINFO:tensorflow:lr = 0.00048033896 (10.321 sec)\nINFO:tensorflow:global_step/sec: 9.50577\nINFO:tensorflow:loss = 4.9523335, step = 57000 (10.518 sec)\nINFO:tensorflow:lr = 0.00047854695 (10.518 sec)\nINFO:tensorflow:global_step/sec: 9.50627\nINFO:tensorflow:loss = 5.079739, step = 57100 (10.522 sec)\nINFO:tensorflow:lr = 0.00047675485 (10.521 sec)\nINFO:tensorflow:global_step/sec: 9.33996\nINFO:tensorflow:loss = 5.0048776, step = 57200 (10.707 sec)\nINFO:tensorflow:lr = 0.00047496284 (10.707 sec)\nINFO:tensorflow:global_step/sec: 9.66024\nINFO:tensorflow:loss = 4.9426284, step = 57300 (10.348 sec)\nINFO:tensorflow:lr = 0.00047317083 (10.349 sec)\nINFO:tensorflow:global_step/sec: 9.76629\nINFO:tensorflow:loss = 5.041515, step = 57400 (10.241 sec)\nINFO:tensorflow:lr = 0.00047137882 (10.241 sec)\nINFO:tensorflow:global_step/sec: 9.49702\nINFO:tensorflow:loss = 4.868841, step = 57500 (10.531 sec)\nINFO:tensorflow:lr = 0.0004695868 (10.530 sec)\nINFO:tensorflow:global_step/sec: 9.54386\nINFO:tensorflow:loss = 5.01951, step = 57600 (10.477 sec)\nINFO:tensorflow:lr = 0.0004677948 (10.477 sec)\nINFO:tensorflow:global_step/sec: 9.60844\nINFO:tensorflow:loss = 4.9338384, step = 57700 (10.408 sec)\nINFO:tensorflow:lr = 0.0004660028 (10.409 sec)\nINFO:tensorflow:global_step/sec: 9.30532\nINFO:tensorflow:loss = 4.9551077, step = 57800 (10.748 sec)\nINFO:tensorflow:lr = 0.00046421075 (10.747 sec)\nINFO:tensorflow:global_step/sec: 9.30418\nINFO:tensorflow:loss = 5.0550914, step = 57900 (10.746 sec)\nINFO:tensorflow:lr = 0.00046241874 (10.747 sec)\nINFO:tensorflow:global_step/sec: 9.43973\nINFO:tensorflow:loss = 4.955047, step = 58000 (10.596 sec)\nINFO:tensorflow:lr = 0.00046062664 (10.594 sec)\nINFO:tensorflow:global_step/sec: 9.3222\nINFO:tensorflow:loss = 5.0358024, step = 58100 (10.725 sec)\nINFO:tensorflow:lr = 0.00045883463 (10.729 sec)\nINFO:tensorflow:global_step/sec: 9.26154\nINFO:tensorflow:loss = 4.8850555, step = 58200 (10.797 sec)\nINFO:tensorflow:lr = 0.00045704262 (10.794 sec)\nINFO:tensorflow:global_step/sec: 9.51078\nINFO:tensorflow:loss = 4.9150257, step = 58300 (10.513 sec)\nINFO:tensorflow:lr = 0.0004552506 (10.512 sec)\nINFO:tensorflow:global_step/sec: 9.46567\nINFO:tensorflow:loss = 4.9813275, step = 58400 (10.567 sec)\nINFO:tensorflow:lr = 0.0004534586 (10.567 sec)\nINFO:tensorflow:global_step/sec: 9.51677\nINFO:tensorflow:loss = 4.9747343, step = 58500 (10.509 sec)\nINFO:tensorflow:lr = 0.0004516666 (10.509 sec)\nINFO:tensorflow:global_step/sec: 9.37885\nINFO:tensorflow:loss = 5.0838323, step = 58600 (10.661 sec)\nINFO:tensorflow:lr = 0.00044987458 (10.661 sec)\nINFO:tensorflow:global_step/sec: 9.44478\nINFO:tensorflow:loss = 4.9560633, step = 58700 (10.586 sec)\nINFO:tensorflow:lr = 0.00044808254 (10.587 sec)\nINFO:tensorflow:global_step/sec: 9.36644\nINFO:tensorflow:loss = 5.092565, step = 58800 (10.676 sec)\nINFO:tensorflow:lr = 0.00044629053 (10.675 sec)\nINFO:tensorflow:global_step/sec: 9.57847\nINFO:tensorflow:loss = 5.1015515, step = 58900 (10.443 sec)\nINFO:tensorflow:lr = 0.00044449844 (10.443 sec)\nINFO:tensorflow:global_step/sec: 9.66363\nINFO:tensorflow:loss = 4.9495497, step = 59000 (10.348 sec)\nINFO:tensorflow:lr = 0.00044270643 (10.348 sec)\nINFO:tensorflow:global_step/sec: 9.25838\nINFO:tensorflow:loss = 4.9359717, step = 59100 (10.799 sec)\nINFO:tensorflow:lr = 0.00044091442 (10.800 sec)\nINFO:tensorflow:global_step/sec: 9.69576\nINFO:tensorflow:loss = 5.07738, step = 59200 (10.315 sec)\nINFO:tensorflow:lr = 0.0004391224 (10.314 sec)\nINFO:tensorflow:global_step/sec: 9.55058\nINFO:tensorflow:loss = 4.9461327, step = 59300 (10.470 sec)\nINFO:tensorflow:lr = 0.0004373304 (10.470 sec)\nINFO:tensorflow:global_step/sec: 9.63653\nINFO:tensorflow:loss = 4.9056435, step = 59400 (10.377 sec)\nINFO:tensorflow:lr = 0.0004355384 (10.376 sec)\nINFO:tensorflow:global_step/sec: 9.62116\nINFO:tensorflow:loss = 5.0284085, step = 59500 (10.396 sec)\nINFO:tensorflow:lr = 0.00043374638 (10.397 sec)\nINFO:tensorflow:global_step/sec: 9.38683\nINFO:tensorflow:loss = 5.1237197, step = 59600 (10.651 sec)\nINFO:tensorflow:lr = 0.00043195437 (10.651 sec)\nINFO:tensorflow:global_step/sec: 9.33088\nINFO:tensorflow:loss = 5.0115285, step = 59700 (10.717 sec)\nINFO:tensorflow:lr = 0.00043016233 (10.717 sec)\nINFO:tensorflow:global_step/sec: 9.35386\nINFO:tensorflow:loss = 4.90229, step = 59800 (10.691 sec)\nINFO:tensorflow:lr = 0.00042837023 (10.691 sec)\nINFO:tensorflow:global_step/sec: 9.55297\nINFO:tensorflow:loss = 4.9314675, step = 59900 (10.470 sec)\nINFO:tensorflow:lr = 0.00042657822 (10.469 sec)\nINFO:tensorflow:global_step/sec: 9.66096\nINFO:tensorflow:loss = 4.955702, step = 60000 (10.351 sec)\nINFO:tensorflow:lr = 0.0004247862 (10.351 sec)\nINFO:tensorflow:global_step/sec: 9.70059\nINFO:tensorflow:loss = 4.94967, step = 60100 (10.307 sec)\nINFO:tensorflow:lr = 0.0004229942 (10.308 sec)\nINFO:tensorflow:global_step/sec: 9.20308\nINFO:tensorflow:loss = 4.909738, step = 60200 (10.868 sec)\nINFO:tensorflow:lr = 0.0004212022 (10.866 sec)\nINFO:tensorflow:global_step/sec: 9.69645\nINFO:tensorflow:loss = 4.9466395, step = 60300 (10.312 sec)\nINFO:tensorflow:lr = 0.00041941018 (10.311 sec)\nINFO:tensorflow:global_step/sec: 9.47731\nINFO:tensorflow:loss = 4.934656, step = 60400 (10.552 sec)\nINFO:tensorflow:lr = 0.00041761817 (10.554 sec)\nINFO:tensorflow:global_step/sec: 9.50505\nINFO:tensorflow:loss = 4.9714603, step = 60500 (10.520 sec)\nINFO:tensorflow:lr = 0.00041582616 (10.519 sec)\nINFO:tensorflow:global_step/sec: 9.40126\nINFO:tensorflow:loss = 5.0126266, step = 60600 (10.637 sec)\nINFO:tensorflow:lr = 0.00041403406 (10.637 sec)\nINFO:tensorflow:global_step/sec: 9.37801\nINFO:tensorflow:loss = 4.950481, step = 60700 (10.661 sec)\nINFO:tensorflow:lr = 0.00041224205 (10.662 sec)\nINFO:tensorflow:global_step/sec: 9.29794\nINFO:tensorflow:loss = 4.879242, step = 60800 (10.756 sec)\nINFO:tensorflow:lr = 0.00041045 (10.755 sec)\nINFO:tensorflow:global_step/sec: 9.71345\nINFO:tensorflow:loss = 4.974057, step = 60900 (10.295 sec)\nINFO:tensorflow:lr = 0.000408658 (10.297 sec)\nINFO:tensorflow:global_step/sec: 9.37829\nINFO:tensorflow:loss = 5.0180244, step = 61000 (10.664 sec)\nINFO:tensorflow:lr = 0.000406866 (10.664 sec)\nINFO:tensorflow:global_step/sec: 9.43858\nINFO:tensorflow:loss = 4.973854, step = 61100 (10.592 sec)\nINFO:tensorflow:lr = 0.00040507398 (10.593 sec)\nINFO:tensorflow:global_step/sec: 9.48033\nINFO:tensorflow:loss = 4.9414907, step = 61200 (10.548 sec)\nINFO:tensorflow:lr = 0.00040328197 (10.548 sec)\nINFO:tensorflow:global_step/sec: 9.44247\nINFO:tensorflow:loss = 5.125211, step = 61300 (10.593 sec)\nINFO:tensorflow:lr = 0.00040148996 (10.592 sec)\nINFO:tensorflow:global_step/sec: 9.5666\nINFO:tensorflow:loss = 5.022872, step = 61400 (10.451 sec)\nINFO:tensorflow:lr = 0.00039969795 (10.451 sec)\nINFO:tensorflow:global_step/sec: 9.49959\nINFO:tensorflow:loss = 5.071593, step = 61500 (10.527 sec)\nINFO:tensorflow:lr = 0.00039790585 (10.527 sec)\nINFO:tensorflow:global_step/sec: 9.35442\nINFO:tensorflow:loss = 4.858541, step = 61600 (10.690 sec)\nINFO:tensorflow:lr = 0.00039611384 (10.690 sec)\nINFO:tensorflow:global_step/sec: 9.48098\nINFO:tensorflow:loss = 5.007348, step = 61700 (10.551 sec)\nINFO:tensorflow:lr = 0.0003943218 (10.549 sec)\nINFO:tensorflow:global_step/sec: 9.1506\nINFO:tensorflow:loss = 5.004369, step = 61800 (10.925 sec)\nINFO:tensorflow:lr = 0.0003925298 (10.926 sec)\nINFO:tensorflow:global_step/sec: 9.38337\nINFO:tensorflow:loss = 5.111853, step = 61900 (10.658 sec)\nINFO:tensorflow:lr = 0.00039073778 (10.659 sec)\nINFO:tensorflow:global_step/sec: 9.1282\nINFO:tensorflow:loss = 4.9753876, step = 62000 (10.955 sec)\nINFO:tensorflow:lr = 0.00038894577 (10.953 sec)\nINFO:tensorflow:global_step/sec: 9.50418\nINFO:tensorflow:loss = 5.13612, step = 62100 (10.523 sec)\nINFO:tensorflow:lr = 0.00038715376 (10.524 sec)\nINFO:tensorflow:global_step/sec: 9.69161\nINFO:tensorflow:loss = 4.8964615, step = 62200 (10.318 sec)\nINFO:tensorflow:lr = 0.00038536175 (10.318 sec)\nINFO:tensorflow:global_step/sec: 9.43908\nINFO:tensorflow:loss = 4.858383, step = 62300 (10.592 sec)\nINFO:tensorflow:lr = 0.00038356974 (10.592 sec)\nINFO:tensorflow:global_step/sec: 9.5179\nINFO:tensorflow:loss = 5.02234, step = 62400 (10.509 sec)\nINFO:tensorflow:lr = 0.00038177765 (10.509 sec)\nINFO:tensorflow:global_step/sec: 9.29735\nINFO:tensorflow:loss = 5.062104, step = 62500 (10.755 sec)\nINFO:tensorflow:lr = 0.00037998563 (10.756 sec)\nINFO:tensorflow:global_step/sec: 9.37313\nINFO:tensorflow:loss = 4.941778, step = 62600 (10.670 sec)\nINFO:tensorflow:lr = 0.00037819362 (10.669 sec)\nINFO:tensorflow:global_step/sec: 9.20457\nINFO:tensorflow:loss = 4.950422, step = 62700 (10.865 sec)\nINFO:tensorflow:lr = 0.0003764016 (10.865 sec)\nINFO:tensorflow:global_step/sec: 9.48811\nINFO:tensorflow:loss = 4.9543242, step = 62800 (10.538 sec)\nINFO:tensorflow:lr = 0.00037460958 (10.538 sec)\nINFO:tensorflow:global_step/sec: 9.45625\nINFO:tensorflow:loss = 4.930625, step = 62900 (10.575 sec)\nINFO:tensorflow:lr = 0.00037281757 (10.575 sec)\nINFO:tensorflow:global_step/sec: 9.50433\nINFO:tensorflow:loss = 4.8656454, step = 63000 (10.521 sec)\nINFO:tensorflow:lr = 0.00037102556 (10.523 sec)\nINFO:tensorflow:global_step/sec: 9.39489\nINFO:tensorflow:loss = 4.959864, step = 63100 (10.643 sec)\nINFO:tensorflow:lr = 0.00036923354 (10.642 sec)\nINFO:tensorflow:global_step/sec: 9.35715\nINFO:tensorflow:loss = 4.9245005, step = 63200 (10.687 sec)\nINFO:tensorflow:lr = 0.00036744153 (10.687 sec)\nINFO:tensorflow:global_step/sec: 9.25415\nINFO:tensorflow:loss = 4.8904448, step = 63300 (10.805 sec)\nINFO:tensorflow:lr = 0.00036564944 (10.805 sec)\nINFO:tensorflow:global_step/sec: 9.49882\nINFO:tensorflow:loss = 5.053169, step = 63400 (10.531 sec)\nINFO:tensorflow:lr = 0.00036385743 (10.533 sec)\nINFO:tensorflow:global_step/sec: 9.36792\nINFO:tensorflow:loss = 5.0318637, step = 63500 (10.673 sec)\nINFO:tensorflow:lr = 0.00036206542 (10.672 sec)\nINFO:tensorflow:global_step/sec: 9.46675\nINFO:tensorflow:loss = 5.0811977, step = 63600 (10.562 sec)\nINFO:tensorflow:lr = 0.0003602734 (10.561 sec)\nINFO:tensorflow:global_step/sec: 9.09736\nINFO:tensorflow:loss = 5.030285, step = 63700 (10.993 sec)\nINFO:tensorflow:lr = 0.00035848137 (10.994 sec)\nINFO:tensorflow:global_step/sec: 9.36226\nINFO:tensorflow:loss = 5.0793304, step = 63800 (10.680 sec)\nINFO:tensorflow:lr = 0.00035668936 (10.680 sec)\nINFO:tensorflow:global_step/sec: 9.31215\nINFO:tensorflow:loss = 4.9413056, step = 63900 (10.742 sec)\nINFO:tensorflow:lr = 0.00035489735 (10.743 sec)\nINFO:tensorflow:global_step/sec: 9.16499\nINFO:tensorflow:loss = 5.0602775, step = 64000 (10.909 sec)\nINFO:tensorflow:lr = 0.00035310534 (10.909 sec)\nINFO:tensorflow:global_step/sec: 9.24164\nINFO:tensorflow:loss = 5.0007434, step = 64100 (10.822 sec)\nINFO:tensorflow:lr = 0.00035131333 (10.821 sec)\nINFO:tensorflow:global_step/sec: 9.4054\nINFO:tensorflow:loss = 4.8613544, step = 64200 (10.631 sec)\nINFO:tensorflow:lr = 0.00034952123 (10.631 sec)\nINFO:tensorflow:global_step/sec: 9.44425\nINFO:tensorflow:loss = 5.021824, step = 64300 (10.589 sec)\nINFO:tensorflow:lr = 0.00034772922 (10.590 sec)\nINFO:tensorflow:global_step/sec: 9.54381\nINFO:tensorflow:loss = 4.8927984, step = 64400 (10.477 sec)\nINFO:tensorflow:lr = 0.0003459372 (10.478 sec)\nINFO:tensorflow:global_step/sec: 9.53895\nINFO:tensorflow:loss = 4.904232, step = 64500 (10.486 sec)\nINFO:tensorflow:lr = 0.0003441452 (10.484 sec)\nINFO:tensorflow:global_step/sec: 9.34092\nINFO:tensorflow:loss = 4.911748, step = 64600 (10.705 sec)\nINFO:tensorflow:lr = 0.00034235316 (10.705 sec)\nINFO:tensorflow:global_step/sec: 9.52615\nINFO:tensorflow:loss = 5.046228, step = 64700 (10.496 sec)\nINFO:tensorflow:lr = 0.00034056115 (10.497 sec)\nINFO:tensorflow:global_step/sec: 9.33571\nINFO:tensorflow:loss = 4.869957, step = 64800 (10.715 sec)\nINFO:tensorflow:lr = 0.00033876914 (10.715 sec)\nINFO:tensorflow:global_step/sec: 9.18679\nINFO:tensorflow:loss = 4.893039, step = 64900 (10.880 sec)\nINFO:tensorflow:lr = 0.00033697713 (10.879 sec)\nINFO:tensorflow:global_step/sec: 9.39862\nINFO:tensorflow:loss = 4.911767, step = 65000 (10.642 sec)\nINFO:tensorflow:lr = 0.00033518512 (10.642 sec)\nINFO:tensorflow:global_step/sec: 9.37078\nINFO:tensorflow:loss = 4.999353, step = 65100 (10.671 sec)\nINFO:tensorflow:lr = 0.00033339302 (10.670 sec)\nINFO:tensorflow:global_step/sec: 9.57748\nINFO:tensorflow:loss = 4.7816143, step = 65200 (10.443 sec)\nINFO:tensorflow:lr = 0.000331601 (10.443 sec)\nINFO:tensorflow:global_step/sec: 9.20538\nINFO:tensorflow:loss = 4.998312, step = 65300 (10.863 sec)\nINFO:tensorflow:lr = 0.000329809 (10.862 sec)\nINFO:tensorflow:global_step/sec: 9.2367\nINFO:tensorflow:loss = 4.85855, step = 65400 (10.826 sec)\nINFO:tensorflow:lr = 0.00032801696 (10.826 sec)\nINFO:tensorflow:global_step/sec: 9.14618\nINFO:tensorflow:loss = 4.9665437, step = 65500 (10.934 sec)\nINFO:tensorflow:lr = 0.00032622495 (10.934 sec)\nINFO:tensorflow:global_step/sec: 9.45766\nINFO:tensorflow:loss = 4.899067, step = 65600 (10.572 sec)\nINFO:tensorflow:lr = 0.00032443294 (10.574 sec)\nINFO:tensorflow:global_step/sec: 9.35764\nINFO:tensorflow:loss = 4.946297, step = 65700 (10.687 sec)\nINFO:tensorflow:lr = 0.00032264093 (10.686 sec)\nINFO:tensorflow:global_step/sec: 9.2286\nINFO:tensorflow:loss = 5.0569944, step = 65800 (10.833 sec)\nINFO:tensorflow:lr = 0.00032084892 (10.835 sec)\nINFO:tensorflow:global_step/sec: 9.36854\nINFO:tensorflow:loss = 4.9371705, step = 65900 (10.677 sec)\nINFO:tensorflow:lr = 0.0003190569 (10.675 sec)\nINFO:tensorflow:global_step/sec: 9.62625\nINFO:tensorflow:loss = 5.1222367, step = 66000 (10.386 sec)\nINFO:tensorflow:lr = 0.0003172648 (10.387 sec)\nINFO:tensorflow:global_step/sec: 9.64598\nINFO:tensorflow:loss = 4.981133, step = 66100 (10.370 sec)\nINFO:tensorflow:lr = 0.0003154728 (10.369 sec)\nINFO:tensorflow:global_step/sec: 9.52698\nINFO:tensorflow:loss = 4.897505, step = 66200 (10.494 sec)\nINFO:tensorflow:lr = 0.0003136808 (10.494 sec)\nINFO:tensorflow:global_step/sec: 9.47894\nINFO:tensorflow:loss = 4.934471, step = 66300 (10.550 sec)\nINFO:tensorflow:lr = 0.00031188875 (10.551 sec)\nINFO:tensorflow:global_step/sec: 9.51108\nINFO:tensorflow:loss = 4.993697, step = 66400 (10.515 sec)\nINFO:tensorflow:lr = 0.00031009674 (10.515 sec)\nINFO:tensorflow:global_step/sec: 9.49395\nINFO:tensorflow:loss = 4.906064, step = 66500 (10.534 sec)\nINFO:tensorflow:lr = 0.00030830473 (10.534 sec)\nINFO:tensorflow:global_step/sec: 9.35772\nINFO:tensorflow:loss = 5.0655017, step = 66600 (10.687 sec)\nINFO:tensorflow:lr = 0.00030651272 (10.685 sec)\nINFO:tensorflow:global_step/sec: 9.62836\nINFO:tensorflow:loss = 5.0876303, step = 66700 (10.386 sec)\nINFO:tensorflow:lr = 0.0003047207 (10.386 sec)\nINFO:tensorflow:global_step/sec: 9.60331\nINFO:tensorflow:loss = 5.1032166, step = 66800 (10.411 sec)\nINFO:tensorflow:lr = 0.0003029287 (10.413 sec)\nINFO:tensorflow:global_step/sec: 9.5167\nINFO:tensorflow:loss = 4.8347807, step = 66900 (10.507 sec)\nINFO:tensorflow:lr = 0.0003011366 (10.507 sec)\nINFO:tensorflow:global_step/sec: 9.6098\nINFO:tensorflow:loss = 4.870035, step = 67000 (10.407 sec)\nINFO:tensorflow:lr = 0.0002993446 (10.406 sec)\nINFO:tensorflow:global_step/sec: 9.5727\nINFO:tensorflow:loss = 5.024679, step = 67100 (10.447 sec)\nINFO:tensorflow:lr = 0.0002975526 (10.448 sec)\nINFO:tensorflow:global_step/sec: 9.423\nINFO:tensorflow:loss = 5.0179296, step = 67200 (10.610 sec)\nINFO:tensorflow:lr = 0.00029576058 (10.611 sec)\nINFO:tensorflow:global_step/sec: 9.37353\nINFO:tensorflow:loss = 4.920677, step = 67300 (10.670 sec)\nINFO:tensorflow:lr = 0.00029396854 (10.669 sec)\nINFO:tensorflow:global_step/sec: 9.55465\nINFO:tensorflow:loss = 5.009587, step = 67400 (10.466 sec)\nINFO:tensorflow:lr = 0.00029217653 (10.468 sec)\nINFO:tensorflow:global_step/sec: 9.47831\nINFO:tensorflow:loss = 4.9350553, step = 67500 (10.550 sec)\nINFO:tensorflow:lr = 0.00029038452 (10.548 sec)\nINFO:tensorflow:global_step/sec: 9.74646\nINFO:tensorflow:loss = 4.977632, step = 67600 (10.259 sec)\nINFO:tensorflow:lr = 0.0002885925 (10.259 sec)\nINFO:tensorflow:global_step/sec: 9.43465\nINFO:tensorflow:loss = 5.0136166, step = 67700 (10.600 sec)\nINFO:tensorflow:lr = 0.0002868005 (10.598 sec)\nINFO:tensorflow:global_step/sec: 9.5977\nINFO:tensorflow:loss = 5.0190306, step = 67800 (10.419 sec)\nINFO:tensorflow:lr = 0.0002850084 (10.420 sec)\nINFO:tensorflow:global_step/sec: 9.34549\nINFO:tensorflow:loss = 4.88896, step = 67900 (10.700 sec)\nINFO:tensorflow:lr = 0.0002832164 (10.701 sec)\nINFO:tensorflow:global_step/sec: 9.46322\nINFO:tensorflow:loss = 4.9357834, step = 68000 (10.565 sec)\nINFO:tensorflow:lr = 0.00028142438 (10.566 sec)\nINFO:tensorflow:global_step/sec: 9.31911\nINFO:tensorflow:loss = 4.977519, step = 68100 (10.731 sec)\nINFO:tensorflow:lr = 0.00027963237 (10.729 sec)\nINFO:tensorflow:global_step/sec: 9.41641\nINFO:tensorflow:loss = 5.1117687, step = 68200 (10.621 sec)\nINFO:tensorflow:lr = 0.00027784036 (10.622 sec)\nINFO:tensorflow:global_step/sec: 9.5828\nINFO:tensorflow:loss = 5.0275726, step = 68300 (10.436 sec)\nINFO:tensorflow:lr = 0.00027604832 (10.436 sec)\nINFO:tensorflow:global_step/sec: 9.64405\nINFO:tensorflow:loss = 4.9940405, step = 68400 (10.370 sec)\nINFO:tensorflow:lr = 0.0002742563 (10.369 sec)\nINFO:tensorflow:global_step/sec: 9.19497\nINFO:tensorflow:loss = 4.910246, step = 68500 (10.875 sec)\nINFO:tensorflow:lr = 0.0002724643 (10.875 sec)\nINFO:tensorflow:global_step/sec: 9.37872\nINFO:tensorflow:loss = 4.858515, step = 68600 (10.662 sec)\nINFO:tensorflow:lr = 0.0002706723 (10.662 sec)\nINFO:tensorflow:global_step/sec: 9.80252\nINFO:tensorflow:loss = 4.936463, step = 68700 (10.199 sec)\nINFO:tensorflow:lr = 0.0002688802 (10.200 sec)\nINFO:tensorflow:global_step/sec: 9.47177\nINFO:tensorflow:loss = 4.845891, step = 68800 (10.561 sec)\nINFO:tensorflow:lr = 0.00026708818 (10.560 sec)\nINFO:tensorflow:global_step/sec: 9.57415\nINFO:tensorflow:loss = 4.938409, step = 68900 (10.442 sec)\nINFO:tensorflow:lr = 0.00026529617 (10.441 sec)\nINFO:tensorflow:global_step/sec: 9.68101\nINFO:tensorflow:loss = 4.938816, step = 69000 (10.333 sec)\nINFO:tensorflow:lr = 0.00026350416 (10.333 sec)\nINFO:tensorflow:global_step/sec: 9.64886\nINFO:tensorflow:loss = 5.0645933, step = 69100 (10.362 sec)\nINFO:tensorflow:lr = 0.00026171215 (10.362 sec)\nINFO:tensorflow:global_step/sec: 9.42143\nINFO:tensorflow:loss = 5.025646, step = 69200 (10.615 sec)\nINFO:tensorflow:lr = 0.0002599201 (10.615 sec)\nINFO:tensorflow:global_step/sec: 9.37213\nINFO:tensorflow:loss = 4.964944, step = 69300 (10.672 sec)\nINFO:tensorflow:lr = 0.0002581281 (10.671 sec)\nINFO:tensorflow:global_step/sec: 9.43661\nINFO:tensorflow:loss = 5.027821, step = 69400 (10.593 sec)\nINFO:tensorflow:lr = 0.0002563361 (10.593 sec)\nINFO:tensorflow:global_step/sec: 9.58868\nINFO:tensorflow:loss = 4.927606, step = 69500 (10.431 sec)\nINFO:tensorflow:lr = 0.00025454408 (10.434 sec)\nINFO:tensorflow:global_step/sec: 9.45007\nINFO:tensorflow:loss = 5.012537, step = 69600 (10.582 sec)\nINFO:tensorflow:lr = 0.00025275198 (10.580 sec)\nINFO:tensorflow:global_step/sec: 9.74233\nINFO:tensorflow:loss = 5.031917, step = 69700 (10.264 sec)\nINFO:tensorflow:lr = 0.00025095997 (10.265 sec)\nINFO:tensorflow:global_step/sec: 9.67259\nINFO:tensorflow:loss = 5.0179787, step = 69800 (10.338 sec)\nINFO:tensorflow:lr = 0.00024916796 (10.338 sec)\nINFO:tensorflow:global_step/sec: 9.60014\nINFO:tensorflow:loss = 4.998183, step = 69900 (10.417 sec)\nINFO:tensorflow:lr = 0.00024737595 (10.416 sec)\nINFO:tensorflow:global_step/sec: 9.80401\nINFO:tensorflow:loss = 5.025863, step = 70000 (10.199 sec)\nINFO:tensorflow:lr = 0.00024558394 (10.201 sec)\nINFO:tensorflow:global_step/sec: 9.33348\nINFO:tensorflow:loss = 4.792766, step = 70100 (10.715 sec)\nINFO:tensorflow:lr = 0.00024379193 (10.713 sec)\nINFO:tensorflow:global_step/sec: 9.44661\nINFO:tensorflow:loss = 4.921283, step = 70200 (10.586 sec)\nINFO:tensorflow:lr = 0.0002419999 (10.586 sec)\nINFO:tensorflow:global_step/sec: 9.24838\nINFO:tensorflow:loss = 4.991687, step = 70300 (10.813 sec)\nINFO:tensorflow:lr = 0.00024020788 (10.813 sec)\nINFO:tensorflow:global_step/sec: 9.79256\nINFO:tensorflow:loss = 4.8852735, step = 70400 (10.212 sec)\nINFO:tensorflow:lr = 0.00023841587 (10.214 sec)\nINFO:tensorflow:global_step/sec: 10.103\nINFO:tensorflow:loss = 4.789762, step = 70500 (9.897 sec)\nINFO:tensorflow:lr = 0.00023662377 (9.896 sec)\nINFO:tensorflow:global_step/sec: 10.0772\nINFO:tensorflow:loss = 4.9286113, step = 70600 (9.921 sec)\nINFO:tensorflow:lr = 0.00023483176 (9.920 sec)\nINFO:tensorflow:global_step/sec: 10.1454\nINFO:tensorflow:loss = 5.0289354, step = 70700 (9.858 sec)\nINFO:tensorflow:lr = 0.00023303975 (9.859 sec)\nINFO:tensorflow:global_step/sec: 10.0297\nINFO:tensorflow:loss = 4.8507133, step = 70800 (9.969 sec)\nINFO:tensorflow:lr = 0.00023124774 (9.970 sec)\nINFO:tensorflow:global_step/sec: 9.95442\nINFO:tensorflow:loss = 4.893291, step = 70900 (10.047 sec)\nINFO:tensorflow:lr = 0.00022945573 (10.046 sec)\nINFO:tensorflow:global_step/sec: 10.0049\nINFO:tensorflow:loss = 4.885566, step = 71000 (9.994 sec)\nINFO:tensorflow:lr = 0.00022766372 (9.993 sec)\nINFO:tensorflow:global_step/sec: 9.98131\nINFO:tensorflow:loss = 4.929348, step = 71100 (10.022 sec)\nINFO:tensorflow:lr = 0.00022587168 (10.023 sec)\nINFO:tensorflow:global_step/sec: 9.6712\nINFO:tensorflow:loss = 4.8488727, step = 71200 (10.340 sec)\nINFO:tensorflow:lr = 0.00022407967 (10.339 sec)\nINFO:tensorflow:global_step/sec: 10.0553\nINFO:tensorflow:loss = 4.949923, step = 71300 (9.943 sec)\nINFO:tensorflow:lr = 0.00022228766 (9.943 sec)\nINFO:tensorflow:global_step/sec: 9.79252\nINFO:tensorflow:loss = 4.901216, step = 71400 (10.213 sec)\nINFO:tensorflow:lr = 0.00022049557 (10.213 sec)\nINFO:tensorflow:global_step/sec: 9.91906\nINFO:tensorflow:loss = 4.825674, step = 71500 (10.082 sec)\nINFO:tensorflow:lr = 0.00021870356 (10.082 sec)\nINFO:tensorflow:global_step/sec: 9.67885\nINFO:tensorflow:loss = 4.869548, step = 71600 (10.330 sec)\nINFO:tensorflow:lr = 0.00021691155 (10.329 sec)\nINFO:tensorflow:global_step/sec: 9.89107\nINFO:tensorflow:loss = 4.974258, step = 71700 (10.110 sec)\nINFO:tensorflow:lr = 0.00021511954 (10.111 sec)\nINFO:tensorflow:global_step/sec: 9.72268\nINFO:tensorflow:loss = 4.9382033, step = 71800 (10.287 sec)\nINFO:tensorflow:lr = 0.00021332753 (10.286 sec)\nINFO:tensorflow:global_step/sec: 9.70116\nINFO:tensorflow:loss = 4.985545, step = 71900 (10.306 sec)\nINFO:tensorflow:lr = 0.00021153552 (10.307 sec)\nINFO:tensorflow:global_step/sec: 9.96855\nINFO:tensorflow:loss = 4.9777007, step = 72000 (10.033 sec)\nINFO:tensorflow:lr = 0.00020974349 (10.034 sec)\nINFO:tensorflow:global_step/sec: 10.0754\nINFO:tensorflow:loss = 5.077654, step = 72100 (9.928 sec)\nINFO:tensorflow:lr = 0.00020795148 (9.927 sec)\nINFO:tensorflow:global_step/sec: 9.83021\nINFO:tensorflow:loss = 4.855007, step = 72200 (10.171 sec)\nINFO:tensorflow:lr = 0.00020615946 (10.172 sec)\nINFO:tensorflow:global_step/sec: 10.0145\nINFO:tensorflow:loss = 4.9308124, step = 72300 (9.985 sec)\nINFO:tensorflow:lr = 0.00020436736 (9.984 sec)\nINFO:tensorflow:global_step/sec: 9.84289\nINFO:tensorflow:loss = 5.0317636, step = 72400 (10.161 sec)\nINFO:tensorflow:lr = 0.00020257535 (10.162 sec)\nINFO:tensorflow:global_step/sec: 9.59846\nINFO:tensorflow:loss = 4.929287, step = 72500 (10.418 sec)\nINFO:tensorflow:lr = 0.00020078334 (10.418 sec)\nINFO:tensorflow:global_step/sec: 9.33104\nINFO:tensorflow:loss = 4.75825, step = 72600 (10.717 sec)\nINFO:tensorflow:lr = 0.00019899133 (10.717 sec)\nINFO:tensorflow:global_step/sec: 10.0884\nINFO:tensorflow:loss = 4.926864, step = 72700 (9.909 sec)\nINFO:tensorflow:lr = 0.00019719932 (9.909 sec)\nINFO:tensorflow:global_step/sec: 10.1122\nINFO:tensorflow:loss = 4.856933, step = 72800 (9.892 sec)\nINFO:tensorflow:lr = 0.00019540731 (9.892 sec)\nINFO:tensorflow:global_step/sec: 10.2341\nINFO:tensorflow:loss = 4.9881806, step = 72900 (9.771 sec)\nINFO:tensorflow:lr = 0.0001936153 (9.771 sec)\nINFO:tensorflow:global_step/sec: 10.0102\nINFO:tensorflow:loss = 4.9389176, step = 73000 (9.989 sec)\nINFO:tensorflow:lr = 0.00019182327 (9.990 sec)\nINFO:tensorflow:global_step/sec: 10.1647\nINFO:tensorflow:loss = 4.991569, step = 73100 (9.839 sec)\nINFO:tensorflow:lr = 0.00019003126 (9.838 sec)\nINFO:tensorflow:global_step/sec: 10.1237\nINFO:tensorflow:loss = 4.8909016, step = 73200 (9.877 sec)\nINFO:tensorflow:lr = 0.00018823917 (9.878 sec)\nINFO:tensorflow:global_step/sec: 9.84481\nINFO:tensorflow:loss = 5.0041265, step = 73300 (10.156 sec)\nINFO:tensorflow:lr = 0.00018644714 (10.156 sec)\nINFO:tensorflow:global_step/sec: 10.0639\nINFO:tensorflow:loss = 5.0792747, step = 73400 (9.938 sec)\nINFO:tensorflow:lr = 0.00018465513 (9.939 sec)\nINFO:tensorflow:global_step/sec: 10.2123\nINFO:tensorflow:loss = 4.9866824, step = 73500 (9.791 sec)\nINFO:tensorflow:lr = 0.00018286312 (9.790 sec)\nINFO:tensorflow:global_step/sec: 10.0307\nINFO:tensorflow:loss = 4.9499493, step = 73600 (9.970 sec)\nINFO:tensorflow:lr = 0.00018107111 (9.970 sec)\nINFO:tensorflow:global_step/sec: 10.2258\nINFO:tensorflow:loss = 4.94972, step = 73700 (9.780 sec)\nINFO:tensorflow:lr = 0.0001792791 (9.779 sec)\nINFO:tensorflow:global_step/sec: 10.1027\nINFO:tensorflow:loss = 4.98489, step = 73800 (9.896 sec)\nINFO:tensorflow:lr = 0.00017748709 (9.897 sec)\nINFO:tensorflow:global_step/sec: 10.1063\nINFO:tensorflow:loss = 4.840899, step = 73900 (9.895 sec)\nINFO:tensorflow:lr = 0.00017569507 (9.897 sec)\nINFO:tensorflow:global_step/sec: 9.72925\nINFO:tensorflow:loss = 4.93603, step = 74000 (10.281 sec)\nINFO:tensorflow:lr = 0.00017390306 (10.278 sec)\nINFO:tensorflow:global_step/sec: 9.97716\nINFO:tensorflow:loss = 4.947037, step = 74100 (10.021 sec)\nINFO:tensorflow:lr = 0.00017211096 (10.021 sec)\nINFO:tensorflow:global_step/sec: 10.0941\nINFO:tensorflow:loss = 4.9884596, step = 74200 (9.908 sec)\nINFO:tensorflow:lr = 0.00017031893 (9.907 sec)\nINFO:tensorflow:global_step/sec: 10.0898\nINFO:tensorflow:loss = 4.981375, step = 74300 (9.910 sec)\nINFO:tensorflow:lr = 0.00016852692 (9.910 sec)\nINFO:tensorflow:global_step/sec: 9.93729\nINFO:tensorflow:loss = 5.059608, step = 74400 (10.066 sec)\nINFO:tensorflow:lr = 0.00016673491 (10.066 sec)\nINFO:tensorflow:global_step/sec: 10.0718\nINFO:tensorflow:loss = 4.946367, step = 74500 (9.928 sec)\nINFO:tensorflow:lr = 0.0001649429 (9.928 sec)\nINFO:tensorflow:global_step/sec: 10.0538\nINFO:tensorflow:loss = 4.967024, step = 74600 (9.945 sec)\nINFO:tensorflow:lr = 0.0001631509 (9.946 sec)\nINFO:tensorflow:global_step/sec: 9.94507\nINFO:tensorflow:loss = 4.9448195, step = 74700 (10.056 sec)\nINFO:tensorflow:lr = 0.00016135888 (10.055 sec)\nINFO:tensorflow:global_step/sec: 10.0953\nINFO:tensorflow:loss = 5.036145, step = 74800 (9.907 sec)\nINFO:tensorflow:lr = 0.00015956686 (9.910 sec)\nINFO:tensorflow:global_step/sec: 10.1335\nINFO:tensorflow:loss = 5.024201, step = 74900 (9.865 sec)\nINFO:tensorflow:lr = 0.00015777485 (9.865 sec)\nINFO:tensorflow:global_step/sec: 10.1867\nINFO:tensorflow:loss = 4.910201, step = 75000 (9.819 sec)\nINFO:tensorflow:lr = 0.00015598275 (9.816 sec)\nINFO:tensorflow:global_step/sec: 10.2466\nINFO:tensorflow:loss = 4.8420153, step = 75100 (9.761 sec)\nINFO:tensorflow:lr = 0.00015419074 (9.761 sec)\nINFO:tensorflow:global_step/sec: 10.0962\nINFO:tensorflow:loss = 4.831217, step = 75200 (9.903 sec)\nINFO:tensorflow:lr = 0.00015239873 (9.904 sec)\nINFO:tensorflow:global_step/sec: 10.1727\nINFO:tensorflow:loss = 4.9388647, step = 75300 (9.831 sec)\nINFO:tensorflow:lr = 0.0001506067 (9.830 sec)\nINFO:tensorflow:global_step/sec: 10.0675\nINFO:tensorflow:loss = 4.9850793, step = 75400 (9.933 sec)\nINFO:tensorflow:lr = 0.0001488147 (9.933 sec)\nINFO:tensorflow:global_step/sec: 10.2753\nINFO:tensorflow:loss = 4.8257756, step = 75500 (9.729 sec)\nINFO:tensorflow:lr = 0.00014702269 (9.730 sec)\nINFO:tensorflow:global_step/sec: 10.0116\nINFO:tensorflow:loss = 4.9931474, step = 75600 (9.990 sec)\nINFO:tensorflow:lr = 0.00014523068 (9.997 sec)\nINFO:tensorflow:global_step/sec: 9.89481\nINFO:tensorflow:loss = 4.9086366, step = 75700 (10.107 sec)\nINFO:tensorflow:lr = 0.00014343866 (10.099 sec)\nINFO:tensorflow:global_step/sec: 9.89529\nINFO:tensorflow:loss = 5.043158, step = 75800 (10.111 sec)\nINFO:tensorflow:lr = 0.00014164664 (10.112 sec)\nINFO:tensorflow:global_step/sec: 10.0947\nINFO:tensorflow:loss = 5.024129, step = 75900 (9.901 sec)\nINFO:tensorflow:lr = 0.00013985454 (9.900 sec)\nINFO:tensorflow:global_step/sec: 10.2464\nINFO:tensorflow:loss = 4.9341116, step = 76000 (9.757 sec)\nINFO:tensorflow:lr = 0.00013806253 (9.759 sec)\nINFO:tensorflow:global_step/sec: 9.97606\nINFO:tensorflow:loss = 4.783729, step = 76100 (10.026 sec)\nINFO:tensorflow:lr = 0.00013627052 (10.025 sec)\nINFO:tensorflow:global_step/sec: 9.91226\nINFO:tensorflow:loss = 4.995345, step = 76200 (10.089 sec)\nINFO:tensorflow:lr = 0.0001344785 (10.088 sec)\nINFO:tensorflow:global_step/sec: 10.2615\nINFO:tensorflow:loss = 4.944727, step = 76300 (9.743 sec)\nINFO:tensorflow:lr = 0.00013268649 (9.743 sec)\nINFO:tensorflow:global_step/sec: 9.99959\nINFO:tensorflow:loss = 4.956012, step = 76400 (10.004 sec)\nINFO:tensorflow:lr = 0.00013089448 (10.003 sec)\nINFO:tensorflow:global_step/sec: 9.87114\nINFO:tensorflow:loss = 4.8436537, step = 76500 (10.130 sec)\nINFO:tensorflow:lr = 0.00012910247 (10.129 sec)\nINFO:tensorflow:global_step/sec: 9.78018\nINFO:tensorflow:loss = 5.0111704, step = 76600 (10.224 sec)\nINFO:tensorflow:lr = 0.00012731046 (10.225 sec)\nINFO:tensorflow:global_step/sec: 10.2486\nINFO:tensorflow:loss = 5.05041, step = 76700 (9.757 sec)\nINFO:tensorflow:lr = 0.00012551843 (9.757 sec)\nINFO:tensorflow:global_step/sec: 10.0346\nINFO:tensorflow:loss = 5.1064954, step = 76800 (9.966 sec)\nINFO:tensorflow:lr = 0.00012372634 (9.966 sec)\nINFO:tensorflow:global_step/sec: 10.0762\nINFO:tensorflow:loss = 5.0424175, step = 76900 (9.926 sec)\nINFO:tensorflow:lr = 0.000121934325 (9.926 sec)\nINFO:tensorflow:global_step/sec: 10.1965\nINFO:tensorflow:loss = 4.9432592, step = 77000 (9.806 sec)\nINFO:tensorflow:lr = 0.000120142315 (9.806 sec)\nINFO:tensorflow:global_step/sec: 10.2105\nINFO:tensorflow:loss = 4.9125285, step = 77100 (9.795 sec)\nINFO:tensorflow:lr = 0.0001183503 (9.794 sec)\nINFO:tensorflow:global_step/sec: 9.99767\nINFO:tensorflow:loss = 4.88361, step = 77200 (10.000 sec)\nINFO:tensorflow:lr = 0.00011655829 (10.004 sec)\nINFO:tensorflow:global_step/sec: 10.0162\nINFO:tensorflow:loss = 4.9388685, step = 77300 (9.987 sec)\nINFO:tensorflow:lr = 0.00011476627 (9.983 sec)\nINFO:tensorflow:global_step/sec: 9.99087\nINFO:tensorflow:loss = 4.856239, step = 77400 (10.007 sec)\nINFO:tensorflow:lr = 0.00011297426 (10.008 sec)\nINFO:tensorflow:global_step/sec: 10.0051\nINFO:tensorflow:loss = 4.9585834, step = 77500 (9.995 sec)\nINFO:tensorflow:lr = 0.00011118225 (9.995 sec)\nINFO:tensorflow:global_step/sec: 9.89371\nINFO:tensorflow:loss = 5.098746, step = 77600 (10.105 sec)\nINFO:tensorflow:lr = 0.00010939023 (10.108 sec)\nINFO:tensorflow:global_step/sec: 10.0591\nINFO:tensorflow:loss = 5.009875, step = 77700 (9.944 sec)\nINFO:tensorflow:lr = 0.000107598134 (9.942 sec)\nINFO:tensorflow:global_step/sec: 9.96559\nINFO:tensorflow:loss = 4.862642, step = 77800 (10.036 sec)\nINFO:tensorflow:lr = 0.000105806124 (10.036 sec)\nINFO:tensorflow:global_step/sec: 10.0664\nINFO:tensorflow:loss = 5.0611267, step = 77900 (9.929 sec)\nINFO:tensorflow:lr = 0.00010401411 (9.929 sec)\nINFO:tensorflow:global_step/sec: 9.93765\nINFO:tensorflow:loss = 5.0704823, step = 78000 (10.066 sec)\nINFO:tensorflow:lr = 0.0001022221 (10.065 sec)\nINFO:tensorflow:global_step/sec: 10.1168\nINFO:tensorflow:loss = 4.9549403, step = 78100 (9.884 sec)\nINFO:tensorflow:lr = 0.00010043008 (9.884 sec)\nINFO:tensorflow:Saving checkpoints for 78125 into ../model/transformer_rnn/model.ckpt.\nINFO:tensorflow:Loss for final step: 4.9947433.\nWARNING:tensorflow:From /tensorflow-1.15.2/python3.6/tensorflow_estimator/python/estimator/inputs/queues/feeding_queue_runner.py:62: QueueRunner.__init__ (from tensorflow.python.training.queue_runner_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nTo construct input pipelines, use the `tf.data` module.\nWARNING:tensorflow:From /tensorflow-1.15.2/python3.6/tensorflow_estimator/python/estimator/inputs/queues/feeding_functions.py:500: add_queue_runner (from tensorflow.python.training.queue_runner_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nTo construct input pipelines, use the `tf.data` module.\nINFO:tensorflow:Calling model_fn.\nWARNING:tensorflow:Entity <bound method TiedDense.call of <__main__.TiedDense object at 0x7f6e1a554ba8>> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nWARNING: Entity <bound method TiedDense.call of <__main__.TiedDense object at 0x7f6e1a554ba8>> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nWARNING:tensorflow:From /tensorflow-1.15.2/python3.6/tensorflow_core/contrib/seq2seq/python/ops/beam_search_decoder.py:971: to_int64 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.cast` instead.\nWARNING:tensorflow:From /tensorflow-1.15.2/python3.6/tensorflow_core/contrib/seq2seq/python/ops/beam_search_decoder.py:1252: div (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nDeprecated in favor of operator or tf.math.divide.\nINFO:tensorflow:Done calling model_fn.\nINFO:tensorflow:Graph was finalized.\nINFO:tensorflow:Restoring parameters from ../model/transformer_rnn/model.ckpt-78125\nINFO:tensorflow:Running local_init_op.\nINFO:tensorflow:Done running local_init_op.\nWARNING:tensorflow:From /tensorflow-1.15.2/python3.6/tensorflow_core/python/training/monitored_session.py:882: start_queue_runners (from tensorflow.python.training.queue_runner_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nTo construct input pipelines, use the `tf.data` module.\n------------\nunit test\nQ: 你 好\nA1: 你 好\nA2: 你 好 !\nA3: 谢 谢\n\nQ: 早 上 好\nA1: 早 上 好\nA2: 早 安\nA3: 早 上 好 !\n\nQ: 晚 上 好\nA1: 晚 上 好\nA2: 晚 安\nA3: 晚 上 好 !\n\nQ: 再 见\nA1: 再 见\nA2: 好 的\nA3: 哈 哈\n\nQ: 好 久 不 见\nA1: 好 久 不 见\nA2: 好 久 不 见 了\nA3: 好 久 没 见 你 了\n\nQ: 想 死 你 了\nA1: 想 死 你 了\nA2: 我 也 想 你\nA3: 我 也 想 死\n\nQ: 谢 谢 你\nA1: 不 客 气\nA2: 谢 谢\nA3: 不 用 谢\n\nQ: 爱 你\nA1: 爱 你\nA2: 我 也 爱 你\nA3: 我 爱 你\n\nQ: 你 好 厉 害 啊\nA1: 好 厉 害\nA2: 哈 哈\nA3: 我 也 是\n\nQ: 你 叫 什 么\nA1: 你 猜\nA2: 不 知 道\nA3: 我 不 知 道\n\nQ: 你 几 岁 了\nA1: 你 猜 我 猜 不 猜\nA2: 你 猜\nA3: 你 猜 我 猜 你 猜 不 猜\n\nQ: 现 在 几 点\nA1: 1 2 点\nA2: 十 点 半\nA3: 不 知 道\n\nQ: 今 天 天 气 怎 么 样\nA1: 我 也 是\nA2: 今 天 下 雨 了\nA3: 我 也 是 这 么 想 的\n\nQ: 你 今 天 心 情 好 吗\nA1: 心 情 不 好\nA2: 我 也 是\nA3: 心 情 好\n\nQ: 我 们 现 在 在 哪 里\nA1: 在 哪 里\nA2: 你 在 哪 里\nA3: 在 哪 里 ?\n\nQ: 讲 个 笑 话\nA1: 笑 死 我 了\nA2: 哈 哈\nA3: 笑 死 了\n\nQ: 你 会 几 种 语 言 呀\nA1: 不 会 吧\nA2: 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈\nA3: 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈 哈\n\nQ: 你 觉 得 我 帅 吗\nA1: 我 觉 得 你 帅\nA2: 我 也 觉 得\nA3: 不 帅\n\nQ: 讨 厌 的 周 一\nA1: 我 也 是\nA2: 我 也 讨 厌\nA3: 我 也 喜 欢\n\nQ: 好 烦 啊\nA1: 我 也 是\nA2: 好 烦 啊\nA3: 怎 么 了\n\nQ: 天 气 真 好\nA1: 是 的\nA2: 是 啊\nA3: 嗯 嗯\n\nQ: 今 天 好 冷\nA1: 我 也 是\nA2: 好 冷\nA3: 好 冷 啊\n\nQ: 今 天 好 热\nA1: 我 也 是\nA2: 我 也 是 !\nA3: 今 天 下 雨 了\n\nQ: 下 雨 了\nA1: 下 雨 了\nA2: 我 也 是\nA3: 是 的\n\nQ: 风 好 大\nA1: 是 的\nA2: 哈 哈\nA3: 我 也 是\n\nQ: 终 于 周 五 了\nA1: 我 也 是\nA2: 哈 哈\nA3: 我 也 是 !\n\nQ: 我 想 去 唱 歌\nA1: 去 吧\nA2: 我 也 想 去\nA3: 来 啊 来 啊\n\n------------\nINFO:tensorflow:Calling model_fn.\nWARNING:tensorflow:Entity <bound method TiedDense.call of <__main__.TiedDense object at 0x7f6e3a1e2978>> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nWARNING: Entity <bound method TiedDense.call of <__main__.TiedDense object at 0x7f6e3a1e2978>> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nINFO:tensorflow:Done calling model_fn.\nINFO:tensorflow:Starting evaluation at 2020-08-28T07:38:35Z\nINFO:tensorflow:Graph was finalized.\nINFO:tensorflow:Restoring parameters from ../model/transformer_rnn/model.ckpt-78125\nINFO:tensorflow:Running local_init_op.\nINFO:tensorflow:Done running local_init_op.\nReading ../data/test.txt\nINFO:tensorflow:Finished evaluation at 2020-08-28-07:38:52\nINFO:tensorflow:Saving dict for global step 78125: global_step = 78125, loss = 3.7486782\nINFO:tensorflow:Saving 'checkpoint_path' summary for global step 78125: ../model/transformer_rnn/model.ckpt-78125\nPerplexity: 42.465\nBest Perplexity: 42.465\nINFO:tensorflow:Calling model_fn.\nWARNING:tensorflow:Entity <bound method TiedDense.call of <__main__.TiedDense object at 0x7f6e394b3b70>> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nWARNING: Entity <bound method TiedDense.call of <__main__.TiedDense object at 0x7f6e394b3b70>> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\n[<tf.Variable 'Embedding/fasttext_vectors:0' shape=(3043, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/query/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/query/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/key/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/key/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/value/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/value/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/output/kernel:0' shape=(512, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/attention/multihead_attention/output/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/output/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/output/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/fnn/conv1/kernel:0' shape=(300, 1200) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/fnn/conv1/bias:0' shape=(1200,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/fnn/conv2/kernel:0' shape=(1200, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_0/fnn/conv2/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/query/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/query/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/key/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/key/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/value/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/value/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/output/kernel:0' shape=(512, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/attention/multihead_attention/output/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/output/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/output/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/fnn/conv1/kernel:0' shape=(300, 1200) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/fnn/conv1/bias:0' shape=(1200,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/fnn/conv2/kernel:0' shape=(1200, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_1/fnn/conv2/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/query/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/query/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/key/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/key/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/value/kernel:0' shape=(300, 512) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/value/bias:0' shape=(512,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/output/kernel:0' shape=(512, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/attention/multihead_attention/output/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/output/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/output/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/fnn/conv1/kernel:0' shape=(300, 1200) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/fnn/conv1/bias:0' shape=(1200,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/fnn/conv2/kernel:0' shape=(1200, 300) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/layer_2/fnn/conv2/bias:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/LayerNorm/beta:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Encoder/transformer_encoder/LayerNorm/gamma:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Decoder/memory_layer/kernel:0' shape=(300, 300) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/attention_wrapper/lstm_cell/kernel:0' shape=(900, 1200) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/attention_wrapper/lstm_cell/bias:0' shape=(1200,) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/attention_wrapper/bahdanau_attention/query_layer/kernel:0' shape=(300, 300) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/attention_wrapper/bahdanau_attention/attention_v:0' shape=(300,) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/attention_wrapper/attention_layer/kernel:0' shape=(600, 300) dtype=float32_ref>,\n <tf.Variable 'Decoder/decoder/tied_dense/bias:0' shape=(3043,) dtype=float32_ref>]\nINFO:tensorflow:Done calling model_fn.\nINFO:tensorflow:Create CheckpointSaverHook.\nINFO:tensorflow:Graph was finalized.\nINFO:tensorflow:Restoring parameters from ../model/transformer_rnn/model.ckpt-78125\nWARNING:tensorflow:From /tensorflow-1.15.2/python3.6/tensorflow_core/python/training/saver.py:1069: get_checkpoint_mtimes (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse standard file utilities to get mtimes.\nINFO:tensorflow:Running local_init_op.\nINFO:tensorflow:Done running local_init_op.\nINFO:tensorflow:Saving checkpoints for 78125 into ../model/transformer_rnn/model.ckpt.\nReading ../data/train.txt\nINFO:tensorflow:loss = 4.951897, step = 78125\nINFO:tensorflow:lr = 0.000100008925\nINFO:tensorflow:global_step/sec: 8.39226\nINFO:tensorflow:loss = 4.996364, step = 78225 (11.919 sec)\nINFO:tensorflow:lr = 0.000100904974 (11.919 sec)\nINFO:tensorflow:global_step/sec: 9.52345\nINFO:tensorflow:loss = 4.9936595, step = 78325 (10.501 sec)\nINFO:tensorflow:lr = 0.00010180094 (10.501 sec)\nINFO:tensorflow:global_step/sec: 9.43821\nINFO:tensorflow:loss = 4.963275, step = 78425 (10.596 sec)\nINFO:tensorflow:lr = 0.00010269699 (10.596 sec)\nINFO:tensorflow:global_step/sec: 9.66368\nINFO:tensorflow:loss = 5.0328145, step = 78525 (10.345 sec)\nINFO:tensorflow:lr = 0.00010359304 (10.345 sec)\nINFO:tensorflow:global_step/sec: 9.51804\nINFO:tensorflow:loss = 5.0326567, step = 78625 (10.509 sec)\nINFO:tensorflow:lr = 0.000104489 (10.509 sec)\nINFO:tensorflow:global_step/sec: 9.50996\nINFO:tensorflow:loss = 4.865527, step = 78725 (10.512 sec)\nINFO:tensorflow:lr = 0.00010538505 (10.516 sec)\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecee05316917d5c700c0606b04ee4f3886c3bffe | 81,692 | ipynb | Jupyter Notebook | lab1/Part2_Music_Generation.ipynb | zhenbangt/introtodeeplearning | 7de1e19c579385a6eb2e1e4bbcde8b28bac13c30 | [
"MIT"
] | null | null | null | lab1/Part2_Music_Generation.ipynb | zhenbangt/introtodeeplearning | 7de1e19c579385a6eb2e1e4bbcde8b28bac13c30 | [
"MIT"
] | null | null | null | lab1/Part2_Music_Generation.ipynb | zhenbangt/introtodeeplearning | 7de1e19c579385a6eb2e1e4bbcde8b28bac13c30 | [
"MIT"
] | null | null | null | 50.614622 | 11,140 | 0.68686 | [
[
[
"<table align=\"center\">\n <td align=\"center\"><a target=\"_blank\" href=\"http://introtodeeplearning.com\">\n <img src=\"http://introtodeeplearning.com/images/colab/mit.png\" style=\"padding-bottom:5px;\" />\n Visit MIT Deep Learning</a></td>\n <td align=\"center\"><a target=\"_blank\" href=\"https://colab.research.google.com/github/aamini/introtodeeplearning/blob/master/lab1/Part2_Music_Generation.ipynb\">\n <img src=\"http://introtodeeplearning.com/images/colab/colab.png?v2.0\" style=\"padding-bottom:5px;\" />Run in Google Colab</a></td>\n <td align=\"center\"><a target=\"_blank\" href=\"https://github.com/aamini/introtodeeplearning/blob/master/lab1/Part2_Music_Generation.ipynb\">\n <img src=\"http://introtodeeplearning.com/images/colab/github.png\" height=\"70px\" style=\"padding-bottom:5px;\" />View Source on GitHub</a></td>\n</table>\n\n# Copyright Information",
"_____no_output_____"
]
],
[
[
"# Copyright 2020 MIT 6.S191 Introduction to Deep Learning. All Rights Reserved.\n#\n# Licensed under the MIT License. You may not use this file except in compliance\n# with the License. Use and/or modification of this code outside of 6.S191 must\n# reference:\n#\n# © MIT 6.S191: Introduction to Deep Learning\n# http://introtodeeplearning.com\n#",
"_____no_output_____"
]
],
[
[
"# Lab 1: Intro to TensorFlow and Music Generation with RNNs\n\n# Part 2: Music Generation with RNNs\n\nIn this portion of the lab, we will explore building a Recurrent Neural Network (RNN) for music generation. We will train a model to learn the patterns in raw sheet music in [ABC notation](https://en.wikipedia.org/wiki/ABC_notation) and then use this model to generate new music. ",
"_____no_output_____"
],
[
"## 2.1 Dependencies \nFirst, let's download the course repository, install dependencies, and import the relevant packages we'll need for this lab.",
"_____no_output_____"
]
],
[
[
"# Import Tensorflow 2.0\n# %tensorflow_version 2.x\nimport tensorflow as tf\n\nprint(tf.__version__)\n# Download and import the MIT 6.S191 package\n# !pip install mitdeeplearning\nimport mitdeeplearning as mdl\n\n# Import all remaining packages\nimport numpy as np\nimport os\nimport time\nimport functools\nfrom IPython import display as ipythondisplay\nfrom tqdm import tqdm\n\n# !apt-get install abcmidi timidity > /dev/null 2>&1\n\n# Check that we are using a GPU, if not switch runtimes\n# using Runtime > Change Runtime Type > GPU\nassert len(tf.config.list_physical_devices(\"GPU\")) > 0\n\ngpus = tf.config.experimental.list_physical_devices(\"GPU\")\nif gpus:\n try:\n # Currently, memory growth needs to be the same across GPUs\n for gpu in gpus:\n tf.config.experimental.set_memory_growth(gpu, True)\n logical_gpus = tf.config.experimental.list_logical_devices(\"GPU\")\n print(len(gpus), \"Physical GPUs,\", len(logical_gpus), \"Logical GPUs\")\n except RuntimeError as e:\n # Memory growth must be set before GPUs have been initialized\n print(e)",
"2.3.0\n1 Physical GPUs, 1 Logical GPUs\n"
]
],
[
[
"## 2.2 Dataset\n\n\n\nWe've gathered a dataset of thousands of Irish folk songs, represented in the ABC notation. Let's download the dataset and inspect it: \n",
"_____no_output_____"
]
],
[
[
"# Download the dataset\nsongs = mdl.lab1.load_training_data()\n\n# Print one of the songs to inspect it in greater detail!\nexample_song = songs[0]\nprint(\"\\nExample song: \")\nprint(example_song)",
"Found 816 songs in text\n\nExample song: \nX:2\nT:An Buachaill Dreoite\nZ: id:dc-hornpipe-2\nM:C|\nL:1/8\nK:G Major\nGF|DGGB d2GB|d2GF Gc (3AGF|DGGB d2GB|dBcA F2GF|!\nDGGB d2GF|DGGF G2Ge|fgaf gbag|fdcA G2:|!\nGA|B2BG c2cA|d2GF G2GA|B2BG c2cA|d2DE F2GA|!\nB2BG c2cA|d^cde f2 (3def|g2gf gbag|fdcA G2:|!\n"
]
],
[
[
"We can easily convert a song in ABC notation to an audio waveform and play it back. Be patient for this conversion to run, it can take some time.",
"_____no_output_____"
]
],
[
[
"# Convert the ABC notation to audio file and listen to it\nmdl.lab1.play_song(example_song)",
"_____no_output_____"
]
],
[
[
"One important thing to think about is that this notation of music does not simply contain information on the notes being played, but additionally there is meta information such as the song title, key, and tempo. How does the number of different characters that are present in the text file impact the complexity of the learning problem? This will become important soon, when we generate a numerical representation for the text data.",
"_____no_output_____"
]
],
[
[
"# Join our list of song strings into a single string containing all songs\nsongs_joined = \"\\n\\n\".join(songs)\n\n# Find all unique characters in the joined string\nvocab = sorted(set(songs_joined))\nprint(\"There are\", len(vocab), \"unique characters in the dataset\")",
"There are 83 unique characters in the dataset\n"
]
],
[
[
"## 2.3 Process the dataset for the learning task\n\nLet's take a step back and consider our prediction task. We're trying to train a RNN model to learn patterns in ABC music, and then use this model to generate (i.e., predict) a new piece of music based on this learned information. \n\nBreaking this down, what we're really asking the model is: given a character, or a sequence of characters, what is the most probable next character? We'll train the model to perform this task. \n\nTo achieve this, we will input a sequence of characters to the model, and train the model to predict the output, that is, the following character at each time step. RNNs maintain an internal state that depends on previously seen elements, so information about all characters seen up until a given moment will be taken into account in generating the prediction.",
"_____no_output_____"
],
[
"### Vectorize the text\n\nBefore we begin training our RNN model, we'll need to create a numerical representation of our text-based dataset. To do this, we'll generate two lookup tables: one that maps characters to numbers, and a second that maps numbers back to characters. Recall that we just identified the unique characters present in the text.",
"_____no_output_____"
]
],
[
[
"### Define numerical representation of text ###\n\n# Create a mapping from character to unique index.\n# For example, to get the index of the character \"d\",\n# we can evaluate `char2idx[\"d\"]`.\nchar2idx = {u: i for i, u in enumerate(vocab)}\n\n# Create a mapping from indices to characters. This is\n# the inverse of char2idx and allows us to convert back\n# from unique index to the character in our vocabulary.\nidx2char = np.array(vocab)",
"_____no_output_____"
]
],
[
[
"This gives us an integer representation for each character. Observe that the unique characters (i.e., our vocabulary) in the text are mapped as indices from 0 to `len(unique)`. Let's take a peek at this numerical representation of our dataset:",
"_____no_output_____"
]
],
[
[
"print(\"{\")\nfor char, _ in zip(char2idx, range(20)):\n print(\" {:4s}: {:3d},\".format(repr(char), char2idx[char]))\nprint(\" ...\\n}\")",
"{\n '\\n': 0,\n ' ' : 1,\n '!' : 2,\n '\"' : 3,\n '#' : 4,\n \"'\" : 5,\n '(' : 6,\n ')' : 7,\n ',' : 8,\n '-' : 9,\n '.' : 10,\n '/' : 11,\n '0' : 12,\n '1' : 13,\n '2' : 14,\n '3' : 15,\n '4' : 16,\n '5' : 17,\n '6' : 18,\n '7' : 19,\n ...\n}\n"
],
[
"### Vectorize the songs string ###\n\n\"\"\"TODO: Write a function to convert the all songs string to a vectorized\n (i.e., numeric) representation. Use the appropriate mapping\n above to convert from vocab characters to the corresponding indices.\n\n NOTE: the output of the `vectorize_string` function \n should be a np.array with `N` elements, where `N` is\n the number of characters in the input string\n\"\"\"\n\n\ndef vectorize_string(string):\n # TODO\n return np.array([char2idx[char] for char in string])\n\n\nvectorized_songs = vectorize_string(songs_joined)",
"_____no_output_____"
]
],
[
[
"We can also look at how the first part of the text is mapped to an integer representation:",
"_____no_output_____"
]
],
[
[
"print(\n \"{} ---- characters mapped to int ----> {}\".format(\n repr(songs_joined[:10]), vectorized_songs[:10]\n )\n)\n# check that vectorized_songs is a numpy array\nassert isinstance(\n vectorized_songs, np.ndarray\n), \"returned result should be a numpy array\"",
"'X:2\\nT:An B' ---- characters mapped to int ----> [49 22 14 0 45 22 26 69 1 27]\n"
]
],
[
[
"### Create training examples and targets\n\nOur next step is to actually divide the text into example sequences that we'll use during training. Each input sequence that we feed into our RNN will contain `seq_length` characters from the text. We'll also need to define a target sequence for each input sequence, which will be used in training the RNN to predict the next character. For each input, the corresponding target will contain the same length of text, except shifted one character to the right.\n\nTo do this, we'll break the text into chunks of `seq_length+1`. Suppose `seq_length` is 4 and our text is \"Hello\". Then, our input sequence is \"Hell\" and the target sequence is \"ello\".\n\nThe batch method will then let us convert this stream of character indices to sequences of the desired size.",
"_____no_output_____"
]
],
[
[
"### Batch definition to create training examples ###\n\n\ndef get_batch(vectorized_songs, seq_length, batch_size):\n # the length of the vectorized songs string\n n = vectorized_songs.shape[0] - 1\n # randomly choose the starting indices for the examples in the training batch\n idx = np.random.choice(n - seq_length, batch_size)\n\n \"\"\"TODO: construct a list of input sequences for the training batch\"\"\"\n input_batch = [vectorized_songs[i : i + seq_length] for i in idx] # TODO\n \"\"\"TODO: construct a list of output sequences for the training batch\"\"\"\n output_batch = [vectorized_songs[i + 1 : i + seq_length + 1] for i in idx] # TODO\n\n # x_batch, y_batch provide the true inputs and targets for network training\n x_batch = np.reshape(input_batch, [batch_size, seq_length])\n y_batch = np.reshape(output_batch, [batch_size, seq_length])\n return x_batch, y_batch\n\n\n# Perform some simple tests to make sure your batch function is working properly!\ntest_args = (vectorized_songs, 10, 2)\nif (\n not mdl.lab1.test_batch_func_types(get_batch, test_args)\n or not mdl.lab1.test_batch_func_shapes(get_batch, test_args)\n or not mdl.lab1.test_batch_func_next_step(get_batch, test_args)\n):\n print(\"======\\n[FAIL] could not pass tests\")\nelse:\n print(\"======\\n[PASS] passed all tests!\")",
"[PASS] test_batch_func_types\n[PASS] test_batch_func_shapes\n[PASS] test_batch_func_next_step\n======\n[PASS] passed all tests!\n"
]
],
[
[
"For each of these vectors, each index is processed at a single time step. So, for the input at time step 0, the model receives the index for the first character in the sequence, and tries to predict the index of the next character. At the next timestep, it does the same thing, but the RNN considers the information from the previous step, i.e., its updated state, in addition to the current input.\n\nWe can make this concrete by taking a look at how this works over the first several characters in our text:",
"_____no_output_____"
]
],
[
[
"x_batch, y_batch = get_batch(vectorized_songs, seq_length=5, batch_size=1)\n\nfor i, (input_idx, target_idx) in enumerate(\n zip(np.squeeze(x_batch), np.squeeze(y_batch))\n):\n print(\"Step {:3d}\".format(i))\n print(\" input: {} ({:s})\".format(input_idx, repr(idx2char[input_idx])))\n print(\" expected output: {} ({:s})\".format(target_idx, repr(idx2char[target_idx])))",
"Step 0\n input: 0 ('\\n')\n expected output: 38 ('M')\nStep 1\n input: 38 ('M')\n expected output: 22 (':')\nStep 2\n input: 22 (':')\n expected output: 28 ('C')\nStep 3\n input: 28 ('C')\n expected output: 82 ('|')\nStep 4\n input: 82 ('|')\n expected output: 0 ('\\n')\n"
]
],
[
[
"## 2.4 The Recurrent Neural Network (RNN) model",
"_____no_output_____"
],
[
"Now we're ready to define and train a RNN model on our ABC music dataset, and then use that trained model to generate a new song. We'll train our RNN using batches of song snippets from our dataset, which we generated in the previous section.\n\nThe model is based off the LSTM architecture, where we use a state vector to maintain information about the temporal relationships between consecutive characters. The final output of the LSTM is then fed into a fully connected [`Dense`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Dense) layer where we'll output a softmax over each character in the vocabulary, and then sample from this distribution to predict the next character. \n\nAs we introduced in the first portion of this lab, we'll be using the Keras API, specifically, [`tf.keras.Sequential`](https://www.tensorflow.org/api_docs/python/tf/keras/models/Sequential), to define the model. Three layers are used to define the model:\n\n* [`tf.keras.layers.Embedding`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Embedding): This is the input layer, consisting of a trainable lookup table that maps the numbers of each character to a vector with `embedding_dim` dimensions.\n* [`tf.keras.layers.LSTM`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/LSTM): Our LSTM network, with size `units=rnn_units`. \n* [`tf.keras.layers.Dense`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Dense): The output layer, with `vocab_size` outputs.\n\n\n<img src=\"https://raw.githubusercontent.com/aamini/introtodeeplearning/2019/lab1/img/lstm_unrolled-01-01.png\" alt=\"Drawing\"/>",
"_____no_output_____"
],
[
"### Define the RNN model\n\nNow, we will define a function that we will use to actually build the model.",
"_____no_output_____"
]
],
[
[
"def LSTM(rnn_units):\n return tf.keras.layers.LSTM(\n rnn_units,\n return_sequences=True,\n recurrent_initializer=\"glorot_uniform\",\n recurrent_activation=\"sigmoid\",\n stateful=True,\n )",
"_____no_output_____"
]
],
[
[
"The time has come! Fill in the `TODOs` to define the RNN model within the `build_model` function, and then call the function you just defined to instantiate the model!",
"_____no_output_____"
]
],
[
[
"### Defining the RNN Model ###\n\n\"\"\"TODO: Add LSTM and Dense layers to define the RNN model using the Sequential API.\"\"\"\n\n\ndef build_model(vocab_size, embedding_dim, rnn_units, batch_size):\n model = tf.keras.Sequential(\n [\n # Layer 1: Embedding layer to transform indices into dense vectors\n # of a fixed embedding size\n tf.keras.layers.Embedding(\n vocab_size, embedding_dim, batch_input_shape=[batch_size, None]\n ),\n # Layer 2: LSTM with `rnn_units` number of units.\n # TODO: Call the LSTM function defined above to add this layer.\n LSTM(rnn_units),\n # Layer 3: Dense (fully-connected) layer that transforms the LSTM output\n # into the vocabulary size.\n # TODO: Add the Dense layer.\n tf.keras.layers.Dense(vocab_size),\n ]\n )\n\n return model\n\n\n# Build a simple model with default hyperparameters. You will get the\n# chance to change these later.\nmodel = build_model(len(vocab), embedding_dim=256, rnn_units=1024, batch_size=32)",
"_____no_output_____"
]
],
[
[
"### Test out the RNN model\n\nIt's always a good idea to run a few simple checks on our model to see that it behaves as expected. \n\nFirst, we can use the `Model.summary` function to print out a summary of our model's internal workings. Here we can check the layers in the model, the shape of the output of each of the layers, the batch size, etc.",
"_____no_output_____"
]
],
[
[
"model.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding (Embedding) (32, None, 256) 21248 \n_________________________________________________________________\nlstm (LSTM) (32, None, 1024) 5246976 \n_________________________________________________________________\ndense (Dense) (32, None, 83) 85075 \n=================================================================\nTotal params: 5,353,299\nTrainable params: 5,353,299\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"We can also quickly check the dimensionality of our output, using a sequence length of 100. Note that the model can be run on inputs of any length.",
"_____no_output_____"
]
],
[
[
"x, y = get_batch(vectorized_songs, seq_length=100, batch_size=32)\npred = model(x)\nprint(\"Input shape: \", x.shape, \" # (batch_size, sequence_length)\")\nprint(\"Prediction shape: \", pred.shape, \"# (batch_size, sequence_length, vocab_size)\")",
"Input shape: (32, 100) # (batch_size, sequence_length)\nPrediction shape: (32, 100, 83) # (batch_size, sequence_length, vocab_size)\n"
]
],
[
[
"### Predictions from the untrained model\n\nLet's take a look at what our untrained model is predicting.\n\nTo get actual predictions from the model, we sample from the output distribution, which is defined by a `softmax` over our character vocabulary. This will give us actual character indices. This means we are using a [categorical distribution](https://en.wikipedia.org/wiki/Categorical_distribution) to sample over the example prediction. This gives a prediction of the next character (specifically its index) at each timestep.\n\nNote here that we sample from this probability distribution, as opposed to simply taking the `argmax`, which can cause the model to get stuck in a loop.\n\nLet's try this sampling out for the first example in the batch.",
"_____no_output_____"
]
],
[
[
"sampled_indices = tf.random.categorical(pred[0], num_samples=1)\nsampled_indices = tf.squeeze(sampled_indices, axis=-1).numpy()\nsampled_indices",
"_____no_output_____"
]
],
[
[
"We can now decode these to see the text predicted by the untrained model:",
"_____no_output_____"
]
],
[
[
"print(\"Input: \\n\", repr(\"\".join(idx2char[x[0]])))\nprint()\nprint(\"Next Char Predictions: \\n\", repr(\"\".join(idx2char[sampled_indices])))",
"Input: \n 'age dced|]!\\n\\nX:13\\nT:Blackberry Blossom\\nZ: id:dc-reel-14\\nM:C\\nL:1/8\\nK:G Major\\nge|dBAc BGG2|dBBA B2ge|d'\n\nNext Char Predictions: \n 'LIB<RkTu,V\\n58|V:r<jbWT[[OdZL0Cq5]YTr7DhSeZ4j\\'(1C93jra^IS\"V(:CO3I#aWXQym[8^\\'LwKN/SjW2K,9EFeRdQzQP#4m8'\n"
]
],
[
[
"As you can see, the text predicted by the untrained model is pretty nonsensical! How can we do better? We can train the network!",
"_____no_output_____"
],
[
"## 2.5 Training the model: loss and training operations\n\nNow it's time to train the model!\n\nAt this point, we can think of our next character prediction problem as a standard classification problem. Given the previous state of the RNN, as well as the input at a given time step, we want to predict the class of the next character -- that is, to actually predict the next character. \n\nTo train our model on this classification task, we can use a form of the `crossentropy` loss (negative log likelihood loss). Specifically, we will use the [`sparse_categorical_crossentropy`](https://www.tensorflow.org/api_docs/python/tf/keras/losses/sparse_categorical_crossentropy) loss, as it utilizes integer targets for categorical classification tasks. We will want to compute the loss using the true targets -- the `labels` -- and the predicted targets -- the `logits`.\n\nLet's first compute the loss using our example predictions from the untrained model: ",
"_____no_output_____"
]
],
[
[
"### Defining the loss function ###\n\n\"\"\"TODO: define the loss function to compute and return the loss between\n the true labels and predictions (logits). Set the argument from_logits=True.\"\"\"\n\n\ndef compute_loss(labels, logits):\n loss = tf.keras.losses.sparse_categorical_crossentropy(\n labels, logits, from_logits=True\n ) # TODO\n return loss\n\n\n\"\"\"TODO: compute the loss using the true next characters from the example batch \n and the predictions from the untrained model several cells above\"\"\"\nexample_batch_loss = compute_loss(y, pred) # TODO\n\nprint(\"Prediction shape: \", pred.shape, \" # (batch_size, sequence_length, vocab_size)\")\nprint(\"scalar_loss: \", example_batch_loss.numpy().mean())",
"Prediction shape: (32, 100, 83) # (batch_size, sequence_length, vocab_size)\nscalar_loss: 4.4182186\n"
]
],
[
[
"Now, we are ready to define our training operation -- the optimizer and duration of training -- and use this function to train the model. You will experiment with the choice of optimizer and the duration for which you train your models, and see how these changes affect the network's output. Some optimizers you may like to try are [`Adam`](https://www.tensorflow.org/api_docs/python/tf/keras/optimizers/Adam?version=stable) and [`Adagrad`](https://www.tensorflow.org/api_docs/python/tf/keras/optimizers/Adagrad?version=stable).\n\nFirst, we will instantiate a new model and an optimizer. Then, we will use the [`tf.GradientTape`](https://www.tensorflow.org/api_docs/python/tf/GradientTape) method to perform the backpropagation operations. \n\nWe will also generate a print-out of the model's progress through training, which will help us easily visualize whether or not we are minimizing the loss.",
"_____no_output_____"
],
[
"Let's start by defining some hyperparameters for training the model. To start, we have provided some reasonable values for some of the parameters. It is up to you to use what we've learned in class to help optimize the parameter selection here!",
"_____no_output_____"
]
],
[
[
"### Define optimizer and training operation ###\n\n### Hyperparameter setting and optimization ###\n\ndef build_model(vocab_size, embedding_dim, rnn_units, batch_size):\n model = tf.keras.Sequential(\n [\n # Layer 1: Embedding layer to transform indices into dense vectors\n # of a fixed embedding size\n tf.keras.layers.Embedding(\n vocab_size, embedding_dim, batch_input_shape=[batch_size, None]\n ),\n # Layer 2: LSTM with `rnn_units` number of units.\n # TODO: Call the LSTM function defined above to add this layer.\n LSTM(rnn_units),\n # Layer 3: Dense (fully-connected) layer that transforms the LSTM output\n # into the vocabulary size.\n # TODO: Add the Dense layer.\n LSTM(256),\n tf.keras.layers.Dense(vocab_size),\n ]\n )\n\n return model\n\n\n# Optimization parameters:\nnum_training_iterations = 20000 # Increase this to train longer\nbatch_size = 4 # Experiment between 1 and 64\nseq_length = 250 # Experiment between 50 and 500\nlearning_rate = 1e-3 # Experiment between 1e-5 and 1e-1\n\n# Model parameters:\nvocab_size = len(vocab)\nembedding_dim = 256\nrnn_units = 2048 # Experiment between 1 and 2048\n\n# Checkpoint location:\ncheckpoint_dir = \"./training_checkpoints\"\ncheckpoint_prefix = os.path.join(checkpoint_dir, \"my_ckpt\")\n\n\n\"\"\"TODO: instantiate a new model for training using the `build_model`\n function and the hyperparameters created above.\"\"\"\nmodel = build_model(vocab_size, embedding_dim, rnn_units, batch_size)\n\n\"\"\"TODO: instantiate an optimizer with its learning rate.\n Checkout the tensorflow website for a list of supported optimizers.\n https://www.tensorflow.org/api_docs/python/tf/keras/optimizers/\n Try using the Adam optimizer to start.\"\"\"\noptimizer = tf.keras.optimizers.Adam(learning_rate) # TODO\n\n\[email protected]\ndef train_step(x, y):\n # Use tf.GradientTape()\n with tf.GradientTape() as tape:\n\n \"\"\"TODO: feed the current input into the model and generate predictions\"\"\"\n y_hat = model(x)\n\n \"\"\"TODO: compute the loss!\"\"\"\n loss = compute_loss(y, y_hat)\n\n # Now, compute the gradients\n \"\"\"TODO: complete the function call for gradient computation. \n Remember that we want the gradient of the loss with respect all \n of the model parameters. \n HINT: use `model.trainable_variables` to get a list of all model\n parameters.\"\"\"\n grads = tape.gradient(loss, model.trainable_variables)\n\n # Apply the gradients to the optimizer so it can update the model accordingly\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n return loss\n\n\n##################\n# Begin training!#\n##################\n\nhistory = []\nplotter = mdl.util.PeriodicPlotter(sec=2, xlabel=\"Iterations\", ylabel=\"Loss\")\nif hasattr(tqdm, \"_instances\"):\n tqdm._instances.clear() # clear if it exists\n\nfor iter in tqdm(range(num_training_iterations)):\n\n # Grab a batch and propagate it through the network\n x_batch, y_batch = get_batch(vectorized_songs, seq_length, batch_size)\n loss = train_step(x_batch, y_batch)\n\n # Update the progress bar\n history.append(loss.numpy().mean())\n plotter.plot(history)\n\n # Update the model with the changed weights!\n if iter % 100 == 0:\n model.save_weights(checkpoint_prefix)\n\n# Save the trained model and the weights\nmodel.save_weights(checkpoint_prefix)",
"_____no_output_____"
]
],
[
[
"## 2.6 Generate music using the RNN model\n\nNow, we can use our trained RNN model to generate some music! When generating music, we'll have to feed the model some sort of seed to get it started (because it can't predict anything without something to start with!).\n\nOnce we have a generated seed, we can then iteratively predict each successive character (remember, we are using the ABC representation for our music) using our trained RNN. More specifically, recall that our RNN outputs a `softmax` over possible successive characters. For inference, we iteratively sample from these distributions, and then use our samples to encode a generated song in the ABC format.\n\nThen, all we have to do is write it to a file and listen!",
"_____no_output_____"
],
[
"### Restore the latest checkpoint\n\nTo keep this inference step simple, we will use a batch size of 1. Because of how the RNN state is passed from timestep to timestep, the model will only be able to accept a fixed batch size once it is built. \n\nTo run the model with a different `batch_size`, we'll need to rebuild the model and restore the weights from the latest checkpoint, i.e., the weights after the last checkpoint during training:",
"_____no_output_____"
]
],
[
[
"\"\"\"TODO: Rebuild the model using a batch_size=1\"\"\"\nmodel = build_model(vocab_size, embedding_dim, rnn_units, batch_size=1)\n\n# Restore the model weights for the last checkpoint after training\nmodel.load_weights(tf.train.latest_checkpoint(checkpoint_dir))\nmodel.build(tf.TensorShape([1, None]))\n\nmodel.summary()",
"Model: \"sequential_3\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding_3 (Embedding) (1, None, 256) 21248 \n_________________________________________________________________\nlstm_5 (LSTM) (1, None, 2048) 18882560 \n_________________________________________________________________\nlstm_6 (LSTM) (1, None, 256) 2360320 \n_________________________________________________________________\ndense_3 (Dense) (1, None, 83) 21331 \n=================================================================\nTotal params: 21,285,459\nTrainable params: 21,285,459\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"Notice that we have fed in a fixed `batch_size` of 1 for inference.",
"_____no_output_____"
],
[
"### The prediction procedure\n\nNow, we're ready to write the code to generate text in the ABC music format:\n\n* Initialize a \"seed\" start string and the RNN state, and set the number of characters we want to generate.\n\n* Use the start string and the RNN state to obtain the probability distribution over the next predicted character.\n\n* Sample from multinomial distribution to calculate the index of the predicted character. This predicted character is then used as the next input to the model.\n\n* At each time step, the updated RNN state is fed back into the model, so that it now has more context in making the next prediction. After predicting the next character, the updated RNN states are again fed back into the model, which is how it learns sequence dependencies in the data, as it gets more information from the previous predictions.\n\n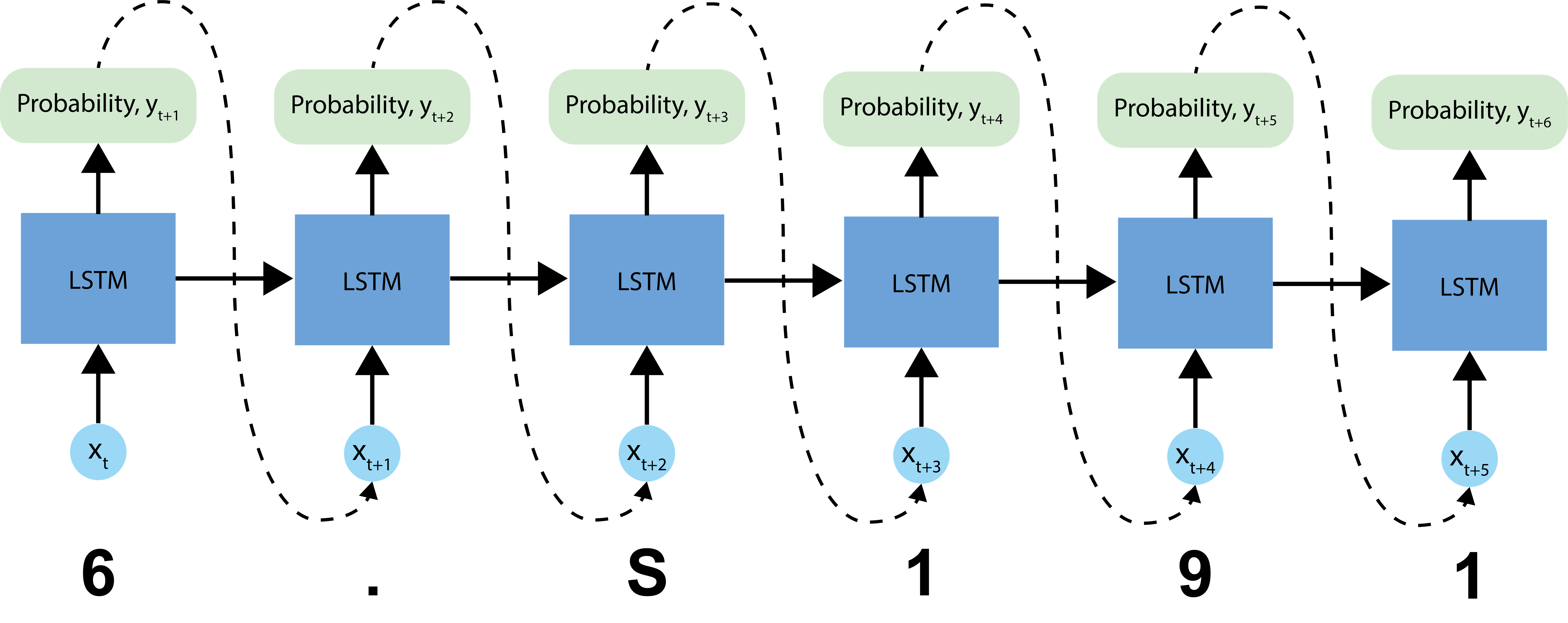\n\nComplete and experiment with this code block (as well as some of the aspects of network definition and training!), and see how the model performs. How do songs generated after training with a small number of epochs compare to those generated after a longer duration of training?",
"_____no_output_____"
]
],
[
[
"### Prediction of a generated song ###\n\n\ndef generate_text(model, start_string, generation_length=1000):\n # Evaluation step (generating ABC text using the learned RNN model)\n\n \"\"\"TODO: convert the start string to numbers (vectorize)\"\"\"\n input_eval = [char2idx[i] for i in start_string]\n input_eval = tf.expand_dims(input_eval, 0)\n\n # Empty string to store our results\n text_generated = []\n\n # Here batch size == 1\n model.reset_states()\n tqdm._instances.clear()\n\n for i in tqdm(range(generation_length)):\n \"\"\"TODO: evaluate the inputs and generate the next character predictions\"\"\"\n predictions = model(input_eval)\n # Remove the batch dimension (1,n into n)\n predictions = tf.squeeze(predictions, 0)\n\n \"\"\"TODO: use a multinomial distribution to sample\"\"\"\n predicted_id = tf.random.categorical(predictions, num_samples=1)[-1, 0].numpy()\n\n # Pass the prediction along with the previous hidden state\n # as the next inputs to the model\n input_eval = tf.expand_dims([predicted_id], 0)\n\n \"\"\"TODO: add the predicted character to the generated text!\"\"\"\n # Hint: consider what format the prediction is in vs. the output\n text_generated.append(idx2char[predicted_id])\n\n return start_string + \"\".join(text_generated)",
"_____no_output_____"
],
[
"\"\"\"TODO: Use the model and the function defined above to generate ABC format text of length 1000!\n As you may notice, ABC files start with \"X\" - this may be a good start string.\"\"\"\ngenerated_text = generate_text(model, start_string=\"X\", generation_length=2000) # TODO\n# generated_text = generate_text('''TODO''', start_string=\"X\", generation_length=1000)",
"100%|█████████████████████████████████████████████████████████████████████████████| 2000/2000 [00:08<00:00, 249.32it/s]\n"
],
[
"generated_text",
"_____no_output_____"
]
],
[
[
"### Play back the generated music!\n\nWe can now call a function to convert the ABC format text to an audio file, and then play that back to check out our generated music! Try training longer if the resulting song is not long enough, or re-generating the song!",
"_____no_output_____"
]
],
[
[
"### Play back generated songs ###\n\ngenerated_songs = mdl.lab1.extract_song_snippet(generated_text)\n\nfor i, song in enumerate(generated_songs):\n # Synthesize the waveform from a song\n waveform = mdl.lab1.play_song(song)\n\n # If its a valid song (correct syntax), lets play it!\n if waveform:\n print(\"Generated song\", i)\n ipythondisplay.display(waveform)",
"Found 7 songs in text\n"
],
[
"songs = extract_song_snippet(generated_text)",
"Found 8 songs in text\n"
],
[
"for i, song in enumerate(songs):\n play_song(song,str(i))",
"_____no_output_____"
],
[
"import os\nimport regex as re\nimport subprocess\nimport urllib\nimport numpy as np\nimport tensorflow as tf\nimport numpy as np\n\nfrom IPython.display import Audio\n\n\ncwd = os.path.dirname('./')\n\ndef load_training_data():\n with open(os.path.join(cwd, \"data\", \"irish.abc\"), \"r\") as f:\n text = f.read()\n songs = extract_song_snippet(text)\n return songs\n\ndef extract_song_snippet(text):\n pattern = '(^|\\n\\n)(.*?)\\n\\n'\n search_results = re.findall(pattern, text, overlapped=True, flags=re.DOTALL)\n songs = [song[1] for song in search_results]\n print(\"Found {} songs in text\".format(len(songs)))\n return songs\n\ndef save_song_to_abc(song, ext, filename=\"tmp\"):\n filename =filename+ext\n save_name = \"{}.abc\".format(filename)\n with open(save_name, \"w\") as f:\n f.write(song)\n return filename\n\ndef abc2wav(abc_file):\n path_to_tool = os.path.join(cwd, 'bin', 'abc2wav')\n cmd = \"{} {}\".format(path_to_tool, abc_file)\n return os.system(cmd)\n\ndef play_wav(wav_file):\n return Audio(wav_file)\n\ndef play_song(song,ext=\"\"):\n basename = save_song_to_abc(song, ext)\n ret = abc2wav(basename+'.abc')\n if ret == 0: #did not suceed\n return play_wav(basename+'.wav')\n return None\n\ndef play_generated_song(generated_text):\n songs = extract_song_snippet(generated_text)\n if len(songs) == 0:\n print(\"No valid songs found in generated text. Try training the \\\n model longer or increasing the amount of generated music to \\\n ensure complete songs are generated!\")\n\n for song in songs:\n play_song(song)\n print(\"None of the songs were valid, try training longer to improve \\\n syntax.\")\n\ndef test_batch_func_types(func, args):\n ret = func(*args)\n assert len(ret) == 2, \"[FAIL] get_batch must return two arguments (input and label)\"\n assert type(ret[0]) == np.ndarray, \"[FAIL] test_batch_func_types: x is not np.array\"\n assert type(ret[1]) == np.ndarray, \"[FAIL] test_batch_func_types: y is not np.array\"\n print(\"[PASS] test_batch_func_types\")\n return True\n\ndef test_batch_func_shapes(func, args):\n dataset, seq_length, batch_size = args\n x, y = func(*args)\n correct = (batch_size, seq_length)\n assert x.shape == correct, \"[FAIL] test_batch_func_shapes: x {} is not correct shape {}\".format(x.shape, correct)\n assert y.shape == correct, \"[FAIL] test_batch_func_shapes: y {} is not correct shape {}\".format(y.shape, correct)\n print(\"[PASS] test_batch_func_shapes\")\n return True\n\ndef test_batch_func_next_step(func, args):\n x, y = func(*args)\n assert (x[:,1:] == y[:,:-1]).all(), \"[FAIL] test_batch_func_next_step: x_{t} must equal y_{t-1} for all t\"\n print(\"[PASS] test_batch_func_next_step\")\n return True\n\ndef test_custom_dense_layer_output(y):\n true_y = np.array([[0.2697859, 0.45750418, 0.66536945]],dtype='float32')\n assert tf.shape(y).numpy().tolist() == list(true_y.shape), \"[FAIL] output is of incorrect shape. expected {} but got {}\".format(true_y.shape, y.numpy().shape)\n np.testing.assert_almost_equal(y.numpy(), true_y, decimal=7, err_msg=\"[FAIL] output is of incorrect value. expected {} but got {}\".format(y.numpy(), true_y), verbose=True)\n print(\"[PASS] test_custom_dense_layer_output\")\n return True\n",
"_____no_output_____"
]
],
[
[
"## 2.7 Experiment and **get awarded for the best songs**!!\n\nCongrats on making your first sequence model in TensorFlow! It's a pretty big accomplishment, and hopefully you have some sweet tunes to show for it.\n\nIf you want to go further, try to optimize your model and submit your best song! Tweet us at [@MITDeepLearning](https://twitter.com/MITDeepLearning) or [email us](mailto:[email protected]) a copy of the song (if you don't have Twitter), and we'll give out prizes to our favorites! \n\nConsider how you may improve your model and what seems to be most important in terms of performance. Here are some ideas to get you started:\n\n* How does the number of training epochs affect the performance?\n* What if you alter or augment the dataset? \n* Does the choice of start string significantly affect the result? \n\nHave fun and happy listening!\n\n\n\n\n\n",
"_____no_output_____"
]
],
[
[
"# Example submission by a previous 6.S191 student (credit: Christian Adib) \n\n%%html\n<blockquote class=\"twitter-tweet\"><a href=\"https://twitter.com/AdibChristian/status/1090030964770783238?ref_src=twsrc%5Etfw\">January 28, 2019</a></blockquote> \n<script async src=\"https://platform.twitter.com/widgets.js\" charset=\"utf-8\"></script>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecee1a92807242a85a17522c2fdd3c61756b3ab1 | 47,772 | ipynb | Jupyter Notebook | A01-Grid-Generation/grid-generation.ipynb | fuqianyin/numericalmethods | e8ac8e4c378fa1c6ed9cfd67a89bcc12567f658d | [
"MIT"
] | 1 | 2020-10-18T19:40:22.000Z | 2020-10-18T19:40:22.000Z | A01-Grid-Generation/grid-generation.ipynb | fuqianyin/numericalmethods | e8ac8e4c378fa1c6ed9cfd67a89bcc12567f658d | [
"MIT"
] | null | null | null | A01-Grid-Generation/grid-generation.ipynb | fuqianyin/numericalmethods | e8ac8e4c378fa1c6ed9cfd67a89bcc12567f658d | [
"MIT"
] | null | null | null | 129.113514 | 12,740 | 0.877397 | [
[
[
"# Non-uniform structured grid\n\nIn fluid mechanics and heat transfer, the solution often contains regions of large gradients and regions of small gradients. In cases where both regions are well localized, computational efficiency favors clustering computational nodes in regions of large gradients and mapping regions of small gradients with coarse meshes. This notebook described a common method to define a non-uniform grid for a symmetrical problem, with the finest resolution at the two ends of the domain, e.g. channel flow.",
"_____no_output_____"
],
[
"## Grid transformation\n\nConsider a domain $[-H/2,+H/2]$ discretized with $N$ points. The location of computational nodes on a uniform grid is defined as:\n\n$$\n\\tilde{y}_j = -h + j\\Delta \\text{ with }\\Delta = \\frac{2h}{N-1}\\text{ and }j\\in[0,N-1] \n$$\nwhere $h=H/2$.\n\nThe common approach to create a non-uniform grid is to operate a transform function over the uniform grid. For a channel flow, one such function is:\n\n$$\ny_j = h\\frac{\\tanh\\gamma \\tilde{y}_j}{\\tanh\\gamma h}\n$$\n\nThe coefficient $\\gamma$ controls the stretching of the grid, in this case the minimum mesh size, which is at the wall.",
"_____no_output_____"
],
[
"### Python set-up and useful functions",
"_____no_output_____"
]
],
[
[
"%matplotlib inline \n# plots graphs within the notebook\n\nfrom IPython.display import display,Image, Latex\nfrom sympy.interactive import printing\nprinting.init_printing(use_latex='mathjax')\nfrom IPython.display import clear_output\n\nimport time\n\nimport numericaltools as numtools\n\n\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport math\nimport scipy.constants as sc\nimport h5py\n\n\nimport sympy as sym\n\n\n\n\nclass PDF(object):\n def __init__(self, pdf, size=(200,200)):\n self.pdf = pdf\n self.size = size\n\n def _repr_html_(self):\n return '<iframe src={0} width={1[0]} height={1[1]}></iframe>'.format(self.pdf, self.size)\n\n def _repr_latex_(self):\n return r'\\includegraphics[width=1.0\\textwidth]{{{0}}}'.format(self.pdf)\n\nclass ListTable(list):\n \"\"\" Overridden list class which takes a 2-dimensional list of \n the form [[1,2,3],[4,5,6]], and renders an HTML Table in \n IPython Notebook. \"\"\"\n \n def _repr_html_(self):\n html = [\"<table>\"]\n for row in self:\n html.append(\"<tr>\")\n \n for col in row:\n html.append(\"<td>{0}</td>\".format(col))\n \n html.append(\"</tr>\")\n html.append(\"</table>\")\n return ''.join(html)\n \nfont = {'family' : 'serif',\n #'color' : 'black',\n 'weight' : 'normal',\n 'size' : 16,\n }\nfontlabel = {'family' : 'serif',\n #'color' : 'black',\n 'weight' : 'normal',\n 'size' : 16,\n }\n\n\nfrom matplotlib.ticker import FormatStrFormatter\nplt.rc('font', **font)\n",
"_____no_output_____"
]
],
[
[
"## Example\n\nCreate a grid for $H=2$, $N=33$, and $\\Delta_{min}=0.001$\n\n\n\n",
"_____no_output_____"
]
],
[
[
"H = 2\nN = 33\nDeltamin = 0.0001\ngamma_guess = 2\ny,gamma = numtools.stretched_mesh(H,N,Deltamin,gamma_guess)\nprint(\"Stretching coefficient gamma: %1.4e\" %gamma)\nplt.plot(y,'o')\nplt.show()\nplt.plot(0.5*(y[1:]+y[:-1]),y[1:]-y[:-1],'o')\nplt.show()\n\n",
"Stretching coefficient gamma: 4.8624e+00\n"
]
],
[
[
"## Grid generation at fixed $\\gamma$\n\n$H=2$, $N=257$, $\\gamma = 2.6$",
"_____no_output_____"
]
],
[
[
"H = 2.\nh = H/2\nN =257\ngamma = 2.6\ny_uni = np.linspace(-h,h,N)\ny = h*np.tanh(gamma*y_uni)/np.tanh(gamma*h)\nplt.plot(y,'o')\nplt.show()\nplt.plot(0.5*(y[1:]+y[:-1]),y[1:]-y[:-1],'o')\nplt.show()\nprint(\"Deltamin = %1.4e\" %(y[1]-y[0]))",
"_____no_output_____"
]
],
[
[
"## Generate a bunch of grids with different $N$ but constant $\\Delta_{min}$",
"_____no_output_____"
]
],
[
[
"H = 2\ndeltamin = 5e-4\ngamma_guess = 2.\nN_array = np.array([33,65,129,257,513,1025],dtype=int)\ngamma_array = np.zeros(len(N_array))\nj = 0\nfor N in N_array:\n y,gamma_array[j] = numtools.stretched_mesh(H,N,Deltamin,gamma_guess)\n gamma_guess = gamma_array[j]\n print(\"for N=%4i, gamma=%1.4f\" %(N,gamma_array[j]))\n \n j += 1\n \nprint(gamma_array)\n ",
"for N= 33, gamma=4.8624\nfor N= 65, gamma=4.3730\nfor N= 129, gamma=3.9348\nfor N= 257, gamma=3.5145\nfor N= 513, gamma=3.0970\nfor N=1025, gamma=2.6734\n[4.86238469 4.37303735 3.93484222 3.51452064 3.09698058 2.67343703]\n"
]
],
[
[
"## Generate a bunch of grids at constant $N$ and varying $\\Delta_{min}$",
"_____no_output_____"
]
],
[
[
"Deltamin_array = np.array([1e-2,5e-3,1e-3,5e-4,1e-4])\nH = 2 \nN = 129\ngamma_guess = 2\nfor Deltamin in Deltamin_array:\n y,gamma = numtools.stretched_mesh(H,N,Deltamin,gamma_guess)\n print(\"For Dmin=%1.1e, gamma=%1.4f\" %(Deltamin,gamma))",
"For Dmin=1.0e-02, gamma=0.8639\nFor Dmin=5.0e-03, gamma=1.4658\nFor Dmin=1.0e-03, gamma=2.5568\nFor Dmin=5.0e-04, gamma=2.9842\nFor Dmin=1.0e-04, gamma=3.9348\n"
],
[
"2/129",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecee3164833f47f6df7f263dbdd8f92aba753869 | 34,961 | ipynb | Jupyter Notebook | Functional Programming/First-Class Functions/05 - Function Introspection.ipynb | Rafiei-tayebe/AP4002 | 460422af73b1a14ddb7441331c59f0af66bcdeea | [
"MIT"
] | null | null | null | Functional Programming/First-Class Functions/05 - Function Introspection.ipynb | Rafiei-tayebe/AP4002 | 460422af73b1a14ddb7441331c59f0af66bcdeea | [
"MIT"
] | null | null | null | Functional Programming/First-Class Functions/05 - Function Introspection.ipynb | Rafiei-tayebe/AP4002 | 460422af73b1a14ddb7441331c59f0af66bcdeea | [
"MIT"
] | 2 | 2022-02-20T18:33:32.000Z | 2022-03-30T12:47:08.000Z | 22.410897 | 293 | 0.445296 | [
[
[
"### Function Introspection",
"_____no_output_____"
]
],
[
[
"def fact(n: \"some non-negative integer\") -> \"n! or 0 if n < 0\":\n \"\"\"Calculates the factorial of a non-negative integer n\n \n If n is negative, returns 0.\n \"\"\"\n if n < 0:\n return 0\n elif n <= 1:\n return 1\n else:\n return n * fact(n-1)",
"_____no_output_____"
]
],
[
[
"We can see all the attributes that belong to a function using the **dir** function:",
"_____no_output_____"
]
],
[
[
"dir(fact)",
"_____no_output_____"
],
[
"fact.__annotations__",
"_____no_output_____"
],
[
"class A:\n c = 2\n def __init__(self):\n self.a = 1\n self.b = 2",
"_____no_output_____"
],
[
"aa = A()\nprint(aa.a, aa.b, aa.c)",
"1 2 2\n"
],
[
"aa.d = 12\nprint(aa.d)",
"12\n"
]
],
[
[
"Since functions are objects, we can add attributes to a function:",
"_____no_output_____"
]
],
[
[
"fact.short_description = \"factorial function\"",
"_____no_output_____"
],
[
"print(fact.short_description)",
"factorial function\n"
]
],
[
[
"We can see our **short_description** attribute, as well as some attributes we have seen before: **__annotations__** and **__doc__**:",
"_____no_output_____"
]
],
[
[
"print(fact.__doc__)",
"Calculates the factorial of a non-negative integer n\n \n If n is negative, returns 0.\n \n"
],
[
"fact.__annotations__",
"_____no_output_____"
]
],
[
[
"We'll revisit some of these attributes later in this course, but let's take a look at a few here:",
"_____no_output_____"
]
],
[
[
"def my_func(a, b=2, c=3, *, kw1, kw2=2, **kwargs):\n pass",
"_____no_output_____"
]
],
[
[
"Let's assign my_func to another variable:",
"_____no_output_____"
]
],
[
[
"f = my_func",
"_____no_output_____"
],
[
"type(f)",
"_____no_output_____"
]
],
[
[
"The **__name__** attribute holds the function's name:",
"_____no_output_____"
]
],
[
[
"my_func.__name__",
"_____no_output_____"
],
[
"f.__name__",
"_____no_output_____"
]
],
[
[
"The **__defaults__** attribute is a tuple containing any positional parameter defaults:",
"_____no_output_____"
]
],
[
[
"my_func.__defaults__",
"_____no_output_____"
],
[
"my_func.__kwdefaults__",
"_____no_output_____"
]
],
[
[
"Let's create a function with some local variables:",
"_____no_output_____"
]
],
[
[
"def my_func(a, b=1, *args, **kwargs):\n i = 10\n b = min(i, b)\n return a * b",
"_____no_output_____"
],
[
"my_func('a', 100)",
"_____no_output_____"
]
],
[
[
"The **__code__** attribute contains a **code** object:",
"_____no_output_____"
]
],
[
[
"my_func.__code__ # dunder",
"_____no_output_____"
]
],
[
[
"This **code** object itself has various properties:",
"_____no_output_____"
]
],
[
[
"dir(my_func.__code__)",
"_____no_output_____"
]
],
[
[
"Attribute **__co_varnames__** is a tuple containing the parameter names and local variables:",
"_____no_output_____"
]
],
[
[
"my_func.__code__.co_varnames",
"_____no_output_____"
]
],
[
[
"Attribute **co_argcount** returns the number of arguments (minus any \\* and \\*\\* args)",
"_____no_output_____"
]
],
[
[
"my_func.__code__.co_argcount",
"_____no_output_____"
]
],
[
[
"#### The **inspect** module",
"_____no_output_____"
],
[
"It is much easier to use the **inspect** module!",
"_____no_output_____"
]
],
[
[
"import inspect",
"_____no_output_____"
],
[
"inspect.isfunction(my_func)",
"_____no_output_____"
]
],
[
[
"By the way, there is a difference between a function and a method! A method is a function that is bound to some object:",
"_____no_output_____"
]
],
[
[
"inspect.ismethod(my_func)",
"_____no_output_____"
]
],
[
[
"#### Introspecting Callable Code",
"_____no_output_____"
],
[
"We can get back the source code of our function using the **getsource()** method:",
"_____no_output_____"
]
],
[
[
"print(inspect.getsource(fact))",
"def fact(n: \"some non-negative integer\") -> \"n! or 0 if n < 0\":\n \"\"\"Calculates the factorial of a non-negative integer n\n \n If n is negative, returns 0.\n \"\"\"\n if n < 0:\n return 0\n elif n <= 1:\n return 1\n else:\n return n * fact(n-1)\n\n"
],
[
"import requests\nprint(inspect.getsource(requests.get))",
"def get(url, params=None, **kwargs):\n r\"\"\"Sends a GET request.\n\n :param url: URL for the new :class:`Request` object.\n :param params: (optional) Dictionary, list of tuples or bytes to send\n in the query string for the :class:`Request`.\n :param \\*\\*kwargs: Optional arguments that ``request`` takes.\n :return: :class:`Response <Response>` object\n :rtype: requests.Response\n \"\"\"\n\n kwargs.setdefault('allow_redirects', True)\n return request('get', url, params=params, **kwargs)\n\n"
]
],
[
[
"We can also find out where the function was defined:",
"_____no_output_____"
]
],
[
[
"inspect.getmodule(fact)",
"_____no_output_____"
],
[
"inspect.getmodule(print)",
"_____no_output_____"
],
[
"import math",
"_____no_output_____"
],
[
"inspect.getmodule(math.sin)",
"_____no_output_____"
],
[
"# setting up variable\ni = 10\n\n# comment line 1\n# comment line 2\ndef my_func(a, b=1):\n # comment inside my_func\n pass",
"_____no_output_____"
],
[
"inspect.getcomments(my_func)",
"_____no_output_____"
],
[
"print(inspect.getcomments(my_func))",
"# comment line 1\n# comment line 2\n\n"
]
],
[
[
"for more info: https://docs.python.org/3/library/inspect.html",
"_____no_output_____"
],
[
"#### Introspecting Callable Signatures",
"_____no_output_____"
]
],
[
[
"# TODO: Provide implementation\ndef my_func(a: 'a string', \n b: int = 1, \n *args: 'additional positional args', \n kw1: 'first keyword-only arg', \n kw2: 'second keyword-only arg' = 10,\n **kwargs: 'additional keyword-only args') -> str:\n \"\"\"does something\n or other\"\"\"\n pass",
"_____no_output_____"
],
[
"import inspect\ninspect.signature(my_func)",
"_____no_output_____"
],
[
"type(inspect.signature(my_func))",
"_____no_output_____"
],
[
"sig = inspect.signature(my_func)",
"_____no_output_____"
],
[
"dir(sig)",
"_____no_output_____"
],
[
"str(sig)",
"_____no_output_____"
],
[
"sig.parameters",
"_____no_output_____"
],
[
"sig.parameters['a']",
"_____no_output_____"
],
[
"sig.parameters['b'].annotation",
"_____no_output_____"
],
[
"sig.parameters['a']._name",
"_____no_output_____"
],
[
"sig.parameters.keys()",
"_____no_output_____"
],
[
"sig.parameters.values()",
"_____no_output_____"
],
[
"for param_name, param in sig.parameters.items():\n print(param_name, param)",
"a a: 'a string'\nb b: int = 1\nargs *args: 'additional positional args'\nkw1 kw1: 'first keyword-only arg'\nkw2 kw2: 'second keyword-only arg' = 10\nkwargs **kwargs: 'additional keyword-only args'\n"
],
[
"def print_info(f: \"callable\") -> None:\n print(f.__name__)\n print('=' * len(f.__name__), end='\\n\\n')\n \n print(f'{inspect.getcomments(f)}\\n{f.__doc__}\\n')\n \n print(f'Inputs\\n------')\n \n sig = inspect.signature(f)\n for param in sig.parameters.values():\n print('Name:', param.name)\n print('Default:', param.default)\n print('Annotation:', param.annotation)\n print('Kind:', param.kind)\n print('--------------------------\\n')\n \n print('\\n\\nOutput\\n------')\n print(sig.return_annotation)",
"_____no_output_____"
],
[
"import pandas\nprint_info(pandas.DataFrame.sort_values)",
"sort_values\n===========\n\n# ----------------------------------------------------------------------\n# Sorting\n# TODO: Just move the sort_values doc here.\n\n\nSort by the values along either axis.\n\nParameters\n----------\n by : str or list of str\n Name or list of names to sort by.\n\n - if `axis` is 0 or `'index'` then `by` may contain index\n levels and/or column labels.\n - if `axis` is 1 or `'columns'` then `by` may contain column\n levels and/or index labels.\naxis : {0 or 'index', 1 or 'columns'}, default 0\n Axis to be sorted.\nascending : bool or list of bool, default True\n Sort ascending vs. descending. Specify list for multiple sort\n orders. If this is a list of bools, must match the length of\n the by.\ninplace : bool, default False\n If True, perform operation in-place.\nkind : {'quicksort', 'mergesort', 'heapsort'}, default 'quicksort'\n Choice of sorting algorithm. See also ndarray.np.sort for more\n information. `mergesort` is the only stable algorithm. For\n DataFrames, this option is only applied when sorting on a single\n column or label.\nna_position : {'first', 'last'}, default 'last'\n Puts NaNs at the beginning if `first`; `last` puts NaNs at the\n end.\nignore_index : bool, default False\n If True, the resulting axis will be labeled 0, 1, …, n - 1.\n\n .. versionadded:: 1.0.0\n\nkey : callable, optional\n Apply the key function to the values\n before sorting. This is similar to the `key` argument in the\n builtin :meth:`sorted` function, with the notable difference that\n this `key` function should be *vectorized*. It should expect a\n ``Series`` and return a Series with the same shape as the input.\n It will be applied to each column in `by` independently.\n\n .. versionadded:: 1.1.0\n\nReturns\n-------\nDataFrame or None\n DataFrame with sorted values or None if ``inplace=True``.\n\nSee Also\n--------\nDataFrame.sort_index : Sort a DataFrame by the index.\nSeries.sort_values : Similar method for a Series.\n\nExamples\n--------\n>>> df = pd.DataFrame({\n... 'col1': ['A', 'A', 'B', np.nan, 'D', 'C'],\n... 'col2': [2, 1, 9, 8, 7, 4],\n... 'col3': [0, 1, 9, 4, 2, 3],\n... 'col4': ['a', 'B', 'c', 'D', 'e', 'F']\n... })\n>>> df\n col1 col2 col3 col4\n0 A 2 0 a\n1 A 1 1 B\n2 B 9 9 c\n3 NaN 8 4 D\n4 D 7 2 e\n5 C 4 3 F\n\nSort by col1\n\n>>> df.sort_values(by=['col1'])\n col1 col2 col3 col4\n0 A 2 0 a\n1 A 1 1 B\n2 B 9 9 c\n5 C 4 3 F\n4 D 7 2 e\n3 NaN 8 4 D\n\nSort by multiple columns\n\n>>> df.sort_values(by=['col1', 'col2'])\n col1 col2 col3 col4\n1 A 1 1 B\n0 A 2 0 a\n2 B 9 9 c\n5 C 4 3 F\n4 D 7 2 e\n3 NaN 8 4 D\n\nSort Descending\n\n>>> df.sort_values(by='col1', ascending=False)\n col1 col2 col3 col4\n4 D 7 2 e\n5 C 4 3 F\n2 B 9 9 c\n0 A 2 0 a\n1 A 1 1 B\n3 NaN 8 4 D\n\nPutting NAs first\n\n>>> df.sort_values(by='col1', ascending=False, na_position='first')\n col1 col2 col3 col4\n3 NaN 8 4 D\n4 D 7 2 e\n5 C 4 3 F\n2 B 9 9 c\n0 A 2 0 a\n1 A 1 1 B\n\nSorting with a key function\n\n>>> df.sort_values(by='col4', key=lambda col: col.str.lower())\n col1 col2 col3 col4\n0 A 2 0 a\n1 A 1 1 B\n2 B 9 9 c\n3 NaN 8 4 D\n4 D 7 2 e\n5 C 4 3 F\n\nNatural sort with the key argument,\nusing the `natsort <https://github.com/SethMMorton/natsort>` package.\n\n>>> df = pd.DataFrame({\n... \"time\": ['0hr', '128hr', '72hr', '48hr', '96hr'],\n... \"value\": [10, 20, 30, 40, 50]\n... })\n>>> df\n time value\n0 0hr 10\n1 128hr 20\n2 72hr 30\n3 48hr 40\n4 96hr 50\n>>> from natsort import index_natsorted\n>>> df.sort_values(\n... by=\"time\",\n... key=lambda x: np.argsort(index_natsorted(df[\"time\"]))\n... )\n time value\n0 0hr 10\n3 48hr 40\n2 72hr 30\n4 96hr 50\n1 128hr 20\n\n\nInputs\n------\nName: self\nDefault: <class 'inspect._empty'>\nAnnotation: <class 'inspect._empty'>\nKind: POSITIONAL_OR_KEYWORD\n--------------------------\n\nName: by\nDefault: <class 'inspect._empty'>\nAnnotation: <class 'inspect._empty'>\nKind: POSITIONAL_OR_KEYWORD\n--------------------------\n\nName: axis\nDefault: 0\nAnnotation: <class 'inspect._empty'>\nKind: POSITIONAL_OR_KEYWORD\n--------------------------\n\nName: ascending\nDefault: True\nAnnotation: <class 'inspect._empty'>\nKind: POSITIONAL_OR_KEYWORD\n--------------------------\n\nName: inplace\nDefault: False\nAnnotation: <class 'inspect._empty'>\nKind: POSITIONAL_OR_KEYWORD\n--------------------------\n\nName: kind\nDefault: quicksort\nAnnotation: <class 'inspect._empty'>\nKind: POSITIONAL_OR_KEYWORD\n--------------------------\n\nName: na_position\nDefault: last\nAnnotation: <class 'inspect._empty'>\nKind: POSITIONAL_OR_KEYWORD\n--------------------------\n\nName: ignore_index\nDefault: False\nAnnotation: <class 'inspect._empty'>\nKind: POSITIONAL_OR_KEYWORD\n--------------------------\n\nName: key\nDefault: None\nAnnotation: ValueKeyFunc\nKind: POSITIONAL_OR_KEYWORD\n--------------------------\n\n\n\nOutput\n------\n<class 'inspect._empty'>\n"
],
[
"print_info(my_func)",
"my_func\n=======\n\n# TODO: Provide implementation\n\ndoes something\n or other\n\nInputs\n------\nName: a\nDefault: <class 'inspect._empty'>\nAnnotation: a string\nKind: POSITIONAL_OR_KEYWORD\n--------------------------\n\nName: b\nDefault: 1\nAnnotation: <class 'int'>\nKind: POSITIONAL_OR_KEYWORD\n--------------------------\n\nName: args\nDefault: <class 'inspect._empty'>\nAnnotation: additional positional args\nKind: VAR_POSITIONAL\n--------------------------\n\nName: kw1\nDefault: <class 'inspect._empty'>\nAnnotation: first keyword-only arg\nKind: KEYWORD_ONLY\n--------------------------\n\nName: kw2\nDefault: 10\nAnnotation: second keyword-only arg\nKind: KEYWORD_ONLY\n--------------------------\n\nName: kwargs\nDefault: <class 'inspect._empty'>\nAnnotation: additional keyword-only args\nKind: VAR_KEYWORD\n--------------------------\n\n\n\nOutput\n------\n<class 'str'>\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecee33b50d40b98cdddcc9aa5a80ac137cff20b7 | 27,875 | ipynb | Jupyter Notebook | examples/structured_data/ipynb/tabtransformer.ipynb | bn1b/keras-io | 5517b37c2032ad8dc7821c1259fa7bc8545f822b | [
"Apache-2.0"
] | null | null | null | examples/structured_data/ipynb/tabtransformer.ipynb | bn1b/keras-io | 5517b37c2032ad8dc7821c1259fa7bc8545f822b | [
"Apache-2.0"
] | null | null | null | examples/structured_data/ipynb/tabtransformer.ipynb | bn1b/keras-io | 5517b37c2032ad8dc7821c1259fa7bc8545f822b | [
"Apache-2.0"
] | null | null | null | 33.665459 | 142 | 0.58583 | [
[
[
"# Structured data learning with TabTransformer\n\n**Author:** [Khalid Salama](https://www.linkedin.com/in/khalid-salama-24403144/)<br>\n**Date created:** 2022/01/18<br>\n**Last modified:** 2022/01/18<br>\n**Description:** Using contextual embeddings for structured data classification.",
"_____no_output_____"
],
[
"## Introduction\n\nThis example demonstrates how to do structured data classification using\n[TabTransformer](https://arxiv.org/abs/2012.06678), a deep tabular data modeling\narchitecture for supervised and semi-supervised learning.\nThe TabTransformer is built upon self-attention based Transformers.\nThe Transformer layers transform the embeddings of categorical features\ninto robust contextual embeddings to achieve higher predictive accuracy.\n\nThis example should be run with TensorFlow 2.7 or higher,\nas well as [TensorFlow Addons](https://www.tensorflow.org/addons/overview),\nwhich can be installed using the following command:\n\n```python\npip install -U tensorflow-addons\n```\n\n## Setup",
"_____no_output_____"
]
],
[
[
"import math\nimport numpy as np\nimport pandas as pd\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.keras import layers\nimport tensorflow_addons as tfa\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Prepare the data\n\nThis example uses the\n[United States Census Income Dataset](https://archive.ics.uci.edu/ml/datasets/census+income)\nprovided by the\n[UC Irvine Machine Learning Repository](https://archive.ics.uci.edu/ml/index.php).\nThe task is binary classification\nto predict whether a person is likely to be making over USD 50,000 a year.\n\nThe dataset includes 48,842 instances with 14 input features: 5 numerical features and 9 categorical features.\n\nFirst, let's load the dataset from the UCI Machine Learning Repository into a Pandas\nDataFrame:",
"_____no_output_____"
]
],
[
[
"CSV_HEADER = [\n \"age\",\n \"workclass\",\n \"fnlwgt\",\n \"education\",\n \"education_num\",\n \"marital_status\",\n \"occupation\",\n \"relationship\",\n \"race\",\n \"gender\",\n \"capital_gain\",\n \"capital_loss\",\n \"hours_per_week\",\n \"native_country\",\n \"income_bracket\",\n]\n\ntrain_data_url = (\n \"https://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.data\"\n)\ntrain_data = pd.read_csv(train_data_url, header=None, names=CSV_HEADER)\n\ntest_data_url = (\n \"https://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.test\"\n)\ntest_data = pd.read_csv(test_data_url, header=None, names=CSV_HEADER)\n\nprint(f\"Train dataset shape: {train_data.shape}\")\nprint(f\"Test dataset shape: {test_data.shape}\")",
"_____no_output_____"
]
],
[
[
"Remove the first record (because it is not a valid data example) and a trailing 'dot' in the class labels.",
"_____no_output_____"
]
],
[
[
"test_data = test_data[1:]\ntest_data.income_bracket = test_data.income_bracket.apply(\n lambda value: value.replace(\".\", \"\")\n)",
"_____no_output_____"
]
],
[
[
"Now we store the training and test data in separate CSV files.",
"_____no_output_____"
]
],
[
[
"train_data_file = \"train_data.csv\"\ntest_data_file = \"test_data.csv\"\n\ntrain_data.to_csv(train_data_file, index=False, header=False)\ntest_data.to_csv(test_data_file, index=False, header=False)",
"_____no_output_____"
]
],
[
[
"## Define dataset metadata\n\nHere, we define the metadata of the dataset that will be useful for reading and parsing\nthe data into input features, and encoding the input features with respect to their types.",
"_____no_output_____"
]
],
[
[
"# A list of the numerical feature names.\nNUMERIC_FEATURE_NAMES = [\n \"age\",\n \"education_num\",\n \"capital_gain\",\n \"capital_loss\",\n \"hours_per_week\",\n]\n# A dictionary of the categorical features and their vocabulary.\nCATEGORICAL_FEATURES_WITH_VOCABULARY = {\n \"workclass\": sorted(list(train_data[\"workclass\"].unique())),\n \"education\": sorted(list(train_data[\"education\"].unique())),\n \"marital_status\": sorted(list(train_data[\"marital_status\"].unique())),\n \"occupation\": sorted(list(train_data[\"occupation\"].unique())),\n \"relationship\": sorted(list(train_data[\"relationship\"].unique())),\n \"race\": sorted(list(train_data[\"race\"].unique())),\n \"gender\": sorted(list(train_data[\"gender\"].unique())),\n \"native_country\": sorted(list(train_data[\"native_country\"].unique())),\n}\n# Name of the column to be used as instances weight.\nWEIGHT_COLUMN_NAME = \"fnlwgt\"\n# A list of the categorical feature names.\nCATEGORICAL_FEATURE_NAMES = list(CATEGORICAL_FEATURES_WITH_VOCABULARY.keys())\n# A list of all the input features.\nFEATURE_NAMES = NUMERIC_FEATURE_NAMES + CATEGORICAL_FEATURE_NAMES\n# A list of column default values for each feature.\nCOLUMN_DEFAULTS = [\n [0.0] if feature_name in NUMERIC_FEATURE_NAMES + [WEIGHT_COLUMN_NAME] else [\"NA\"]\n for feature_name in CSV_HEADER\n]\n# The name of the target feature.\nTARGET_FEATURE_NAME = \"income_bracket\"\n# A list of the labels of the target features.\nTARGET_LABELS = [\" <=50K\", \" >50K\"]",
"_____no_output_____"
]
],
[
[
"## Configure the hyperparameters\n\nThe hyperparameters includes model architecture and training configurations.",
"_____no_output_____"
]
],
[
[
"LEARNING_RATE = 0.001\nWEIGHT_DECAY = 0.0001\nDROPOUT_RATE = 0.2\nBATCH_SIZE = 265\nNUM_EPOCHS = 15\n\nNUM_TRANSFORMER_BLOCKS = 3 # Number of transformer blocks.\nNUM_HEADS = 4 # Number of attention heads.\nEMBEDDING_DIMS = 16 # Embedding dimensions of the categorical features.\nMLP_HIDDEN_UNITS_FACTORS = [\n 2,\n 1,\n] # MLP hidden layer units, as factors of the number of inputs.\nNUM_MLP_BLOCKS = 2 # Number of MLP blocks in the baseline model.",
"_____no_output_____"
]
],
[
[
"## Implement data reading pipeline\n\nWe define an input function that reads and parses the file, then converts features\nand labels into a[`tf.data.Dataset`](https://www.tensorflow.org/guide/datasets)\nfor training or evaluation.",
"_____no_output_____"
]
],
[
[
"target_label_lookup = layers.StringLookup(\n vocabulary=TARGET_LABELS, mask_token=None, num_oov_indices=0\n)\n\n\ndef prepare_example(features, target):\n target_index = target_label_lookup(target)\n weights = features.pop(WEIGHT_COLUMN_NAME)\n return features, target_index, weights\n\n\ndef get_dataset_from_csv(csv_file_path, batch_size=128, shuffle=False):\n dataset = tf.data.experimental.make_csv_dataset(\n csv_file_path,\n batch_size=batch_size,\n column_names=CSV_HEADER,\n column_defaults=COLUMN_DEFAULTS,\n label_name=TARGET_FEATURE_NAME,\n num_epochs=1,\n header=False,\n na_value=\"?\",\n shuffle=shuffle,\n ).map(prepare_example, num_parallel_calls=tf.data.AUTOTUNE, deterministic=False)\n return dataset.cache()\n",
"_____no_output_____"
]
],
[
[
"## Implement a training and evaluation procedure",
"_____no_output_____"
]
],
[
[
"\ndef run_experiment(\n model,\n train_data_file,\n test_data_file,\n num_epochs,\n learning_rate,\n weight_decay,\n batch_size,\n):\n\n optimizer = tfa.optimizers.AdamW(\n learning_rate=learning_rate, weight_decay=weight_decay\n )\n\n model.compile(\n optimizer=optimizer,\n loss=keras.losses.BinaryCrossentropy(),\n metrics=[keras.metrics.BinaryAccuracy(name=\"accuracy\")],\n )\n\n train_dataset = get_dataset_from_csv(train_data_file, batch_size, shuffle=True)\n validation_dataset = get_dataset_from_csv(test_data_file, batch_size)\n\n print(\"Start training the model...\")\n history = model.fit(\n train_dataset, epochs=num_epochs, validation_data=validation_dataset\n )\n print(\"Model training finished\")\n\n _, accuracy = model.evaluate(validation_dataset, verbose=0)\n\n print(f\"Validation accuracy: {round(accuracy * 100, 2)}%\")\n\n return history\n",
"_____no_output_____"
]
],
[
[
"## Create model inputs\n\nNow, define the inputs for the models as a dictionary, where the key is the feature name,\nand the value is a `keras.layers.Input` tensor with the corresponding feature shape\nand data type.",
"_____no_output_____"
]
],
[
[
"\ndef create_model_inputs():\n inputs = {}\n for feature_name in FEATURE_NAMES:\n if feature_name in NUMERIC_FEATURE_NAMES:\n inputs[feature_name] = layers.Input(\n name=feature_name, shape=(), dtype=tf.float32\n )\n else:\n inputs[feature_name] = layers.Input(\n name=feature_name, shape=(), dtype=tf.string\n )\n return inputs\n",
"_____no_output_____"
]
],
[
[
"## Encode features\n\nThe `encode_inputs` method returns `encoded_categorical_feature_list` and `numerical_feature_list`.\nWe encode the categorical features as embeddings, using a fixed `embedding_dims` for all the features,\nregardless their vocabulary sizes. This is required for the Transformer model.",
"_____no_output_____"
]
],
[
[
"\ndef encode_inputs(inputs, embedding_dims):\n\n encoded_categorical_feature_list = []\n numerical_feature_list = []\n\n for feature_name in inputs:\n if feature_name in CATEGORICAL_FEATURE_NAMES:\n\n # Get the vocabulary of the categorical feature.\n vocabulary = CATEGORICAL_FEATURES_WITH_VOCABULARY[feature_name]\n\n # Create a lookup to convert string values to an integer indices.\n # Since we are not using a mask token nor expecting any out of vocabulary\n # (oov) token, we set mask_token to None and num_oov_indices to 0.\n lookup = layers.StringLookup(\n vocabulary=vocabulary,\n mask_token=None,\n num_oov_indices=0,\n output_mode=\"int\",\n )\n\n # Convert the string input values into integer indices.\n encoded_feature = lookup(inputs[feature_name])\n\n # Create an embedding layer with the specified dimensions.\n embedding = layers.Embedding(\n input_dim=len(vocabulary), output_dim=embedding_dims\n )\n\n # Convert the index values to embedding representations.\n encoded_categorical_feature = embedding(encoded_feature)\n encoded_categorical_feature_list.append(encoded_categorical_feature)\n\n else:\n\n # Use the numerical features as-is.\n numerical_feature = tf.expand_dims(inputs[feature_name], -1)\n numerical_feature_list.append(numerical_feature)\n\n return encoded_categorical_feature_list, numerical_feature_list\n",
"_____no_output_____"
]
],
[
[
"## Implement an MLP block",
"_____no_output_____"
]
],
[
[
"\ndef create_mlp(hidden_units, dropout_rate, activation, normalization_layer, name=None):\n\n mlp_layers = []\n for units in hidden_units:\n mlp_layers.append(normalization_layer),\n mlp_layers.append(layers.Dense(units, activation=activation))\n mlp_layers.append(layers.Dropout(dropout_rate))\n\n return keras.Sequential(mlp_layers, name=name)\n",
"_____no_output_____"
]
],
[
[
"## Experiment 1: a baseline model\n\nIn the first experiment, we create a simple multi-layer feed-forward network.",
"_____no_output_____"
]
],
[
[
"\ndef create_baseline_model(\n embedding_dims, num_mlp_blocks, mlp_hidden_units_factors, dropout_rate\n):\n\n # Create model inputs.\n inputs = create_model_inputs()\n # encode features.\n encoded_categorical_feature_list, numerical_feature_list = encode_inputs(\n inputs, embedding_dims\n )\n # Concatenate all features.\n features = layers.concatenate(\n encoded_categorical_feature_list + numerical_feature_list\n )\n # Compute Feedforward layer units.\n feedforward_units = [features.shape[-1]]\n\n # Create several feedforwad layers with skip connections.\n for layer_idx in range(num_mlp_blocks):\n features = create_mlp(\n hidden_units=feedforward_units,\n dropout_rate=dropout_rate,\n activation=keras.activations.gelu,\n normalization_layer=layers.LayerNormalization(epsilon=1e-6),\n name=f\"feedforward_{layer_idx}\",\n )(features)\n\n # Compute MLP hidden_units.\n mlp_hidden_units = [\n factor * features.shape[-1] for factor in mlp_hidden_units_factors\n ]\n # Create final MLP.\n features = create_mlp(\n hidden_units=mlp_hidden_units,\n dropout_rate=dropout_rate,\n activation=keras.activations.selu,\n normalization_layer=layers.BatchNormalization(),\n name=\"MLP\",\n )(features)\n\n # Add a sigmoid as a binary classifer.\n outputs = layers.Dense(units=1, activation=\"sigmoid\", name=\"sigmoid\")(features)\n model = keras.Model(inputs=inputs, outputs=outputs)\n return model\n\n\nbaseline_model = create_baseline_model(\n embedding_dims=EMBEDDING_DIMS,\n num_mlp_blocks=NUM_MLP_BLOCKS,\n mlp_hidden_units_factors=MLP_HIDDEN_UNITS_FACTORS,\n dropout_rate=DROPOUT_RATE,\n)\n\nprint(\"Total model weights:\", baseline_model.count_params())\nkeras.utils.plot_model(baseline_model, show_shapes=True, rankdir=\"LR\")",
"_____no_output_____"
]
],
[
[
"Let's train and evaluate the baseline model:",
"_____no_output_____"
]
],
[
[
"history = run_experiment(\n model=baseline_model,\n train_data_file=train_data_file,\n test_data_file=test_data_file,\n num_epochs=NUM_EPOCHS,\n learning_rate=LEARNING_RATE,\n weight_decay=WEIGHT_DECAY,\n batch_size=BATCH_SIZE,\n)",
"_____no_output_____"
]
],
[
[
"The baseline linear model achieves ~81% validation accuracy.",
"_____no_output_____"
],
[
"## Experiment 2: TabTransformer\n\nThe TabTransformer architecture works as follows:\n\n1. All the categorical features are encoded as embeddings, using the same `embedding_dims`.\nThis means that each value in each categorical feature will have its own embedding vector.\n2. A column embedding, one embedding vector for each categorical feature, is added (point-wise) to the categorical feature embedding.\n3. The embedded categorical features are fed into a stack of Transformer blocks.\nEach Transformer block consists of a multi-head self-attention layer followed by a feed-forward layer.\n3. The outputs of the final Transformer layer, which are the *contextual embeddings* of the categorical features,\nare concatenated with the input numerical features, and fed into a final MLP block.\n4. A `softmax` classifer is applied at the end of the model.\n\nThe [paper](https://arxiv.org/abs/2012.06678) discusses both addition and concatenation of the column embedding in the\n*Appendix: Experiment and Model Details* section.\nThe architecture of TabTransformer is shown below, as presented in the paper.\n\n",
"_____no_output_____"
]
],
[
[
"\ndef create_tabtransformer_classifier(\n num_transformer_blocks,\n num_heads,\n embedding_dims,\n mlp_hidden_units_factors,\n dropout_rate,\n use_column_embedding=False,\n):\n\n # Create model inputs.\n inputs = create_model_inputs()\n # encode features.\n encoded_categorical_feature_list, numerical_feature_list = encode_inputs(\n inputs, embedding_dims\n )\n # Stack categorical feature embeddings for the Tansformer.\n encoded_categorical_features = tf.stack(encoded_categorical_feature_list, axis=1)\n # Concatenate numerical features.\n numerical_features = layers.concatenate(numerical_feature_list)\n\n # Add column embedding to categorical feature embeddings.\n if use_column_embedding:\n num_columns = encoded_categorical_features.shape[1]\n column_embedding = layers.Embedding(\n input_dim=num_columns, output_dim=embedding_dims\n )\n column_indices = tf.range(start=0, limit=num_columns, delta=1)\n encoded_categorical_features = encoded_categorical_features + column_embedding(\n column_indices\n )\n\n # Create multiple layers of the Transformer block.\n for block_idx in range(num_transformer_blocks):\n # Create a multi-head attention layer.\n attention_output = layers.MultiHeadAttention(\n num_heads=num_heads,\n key_dim=embedding_dims,\n dropout=dropout_rate,\n name=f\"multihead_attention_{block_idx}\",\n )(encoded_categorical_features, encoded_categorical_features)\n # Skip connection 1.\n x = layers.Add(name=f\"skip_connection1_{block_idx}\")(\n [attention_output, encoded_categorical_features]\n )\n # Layer normalization 1.\n x = layers.LayerNormalization(name=f\"layer_norm1_{block_idx}\", epsilon=1e-6)(x)\n # Feedforward.\n feedforward_output = create_mlp(\n hidden_units=[embedding_dims],\n dropout_rate=dropout_rate,\n activation=keras.activations.gelu,\n normalization_layer=layers.LayerNormalization(epsilon=1e-6),\n name=f\"feedforward_{block_idx}\",\n )(x)\n # Skip connection 2.\n x = layers.Add(name=f\"skip_connection2_{block_idx}\")([feedforward_output, x])\n # Layer normalization 2.\n encoded_categorical_features = layers.LayerNormalization(\n name=f\"layer_norm2_{block_idx}\", epsilon=1e-6\n )(x)\n\n # Flatten the \"contextualized\" embeddings of the categorical features.\n categorical_features = layers.Flatten()(encoded_categorical_features)\n # Apply layer normalization to the numerical features.\n numerical_features = layers.LayerNormalization(epsilon=1e-6)(numerical_features)\n # Prepare the input for the final MLP block.\n features = layers.concatenate([categorical_features, numerical_features])\n\n # Compute MLP hidden_units.\n mlp_hidden_units = [\n factor * features.shape[-1] for factor in mlp_hidden_units_factors\n ]\n # Create final MLP.\n features = create_mlp(\n hidden_units=mlp_hidden_units,\n dropout_rate=dropout_rate,\n activation=keras.activations.selu,\n normalization_layer=layers.BatchNormalization(),\n name=\"MLP\",\n )(features)\n\n # Add a sigmoid as a binary classifer.\n outputs = layers.Dense(units=1, activation=\"sigmoid\", name=\"sigmoid\")(features)\n model = keras.Model(inputs=inputs, outputs=outputs)\n return model\n\n\ntabtransformer_model = create_tabtransformer_classifier(\n num_transformer_blocks=NUM_TRANSFORMER_BLOCKS,\n num_heads=NUM_HEADS,\n embedding_dims=EMBEDDING_DIMS,\n mlp_hidden_units_factors=MLP_HIDDEN_UNITS_FACTORS,\n dropout_rate=DROPOUT_RATE,\n)\n\nprint(\"Total model weights:\", tabtransformer_model.count_params())\nkeras.utils.plot_model(tabtransformer_model, show_shapes=True, rankdir=\"LR\")",
"_____no_output_____"
]
],
[
[
"Let's train and evaluate the TabTransformer model:",
"_____no_output_____"
]
],
[
[
"history = run_experiment(\n model=tabtransformer_model,\n train_data_file=train_data_file,\n test_data_file=test_data_file,\n num_epochs=NUM_EPOCHS,\n learning_rate=LEARNING_RATE,\n weight_decay=WEIGHT_DECAY,\n batch_size=BATCH_SIZE,\n)",
"_____no_output_____"
]
],
[
[
"The TabTransformer model achieves ~85% validation accuracy.\nNote that, with the default parameter configurations, both the baseline and the TabTransformer\nhave similar number of trainable weights: 109,629 and 92,151 respectively, and both use the same training hyperparameters.",
"_____no_output_____"
],
[
"## Conclusion\n\nTabTransformer significantly outperforms MLP and recent\ndeep networks for tabular data while matching the performance of tree-based ensemble models.\nTabTransformer can be learned in end-to-end supervised training using labeled examples.\nFor a scenario where there are a few labeled examples and a large number of unlabeled\nexamples, a pre-training procedure can be employed to train the Transformer layers using unlabeled data.\nThis is followed by fine-tuning of the pre-trained Transformer layers along with\nthe top MLP layer using the labeled data.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecee3d96b5dd89bf5cf39ece60467f33929bed58 | 709,228 | ipynb | Jupyter Notebook | NotebookExamples/csharp/Samples/HousingML.ipynb | 0xblack/try | 02e383d598f6a62b5000304e569cc896a79e813d | [
"MIT"
] | null | null | null | NotebookExamples/csharp/Samples/HousingML.ipynb | 0xblack/try | 02e383d598f6a62b5000304e569cc896a79e813d | [
"MIT"
] | null | null | null | NotebookExamples/csharp/Samples/HousingML.ipynb | 0xblack/try | 02e383d598f6a62b5000304e569cc896a79e813d | [
"MIT"
] | 1 | 2021-09-06T03:58:12.000Z | 2021-09-06T03:58:12.000Z | 1,353.48855 | 467,216 | 0.711912 | [
[
[
"#r \"nuget:Microsoft.ML,1.4.0-preview\"\n#r \"nuget:Microsoft.ML.AutoML\"\n#r \"nuget:Microsoft.Data.DataFrame,0.1.1-e190920-1\"",
"_____no_output_____"
],
[
"using Microsoft.Data;\nusing XPlot.Plotly;",
"_____no_output_____"
],
[
"using Microsoft.AspNetCore.Html;\nFormatter<DataFrame>.Register((df, writer) =>\n{\n var headers = new List<IHtmlContent>();\n headers.Add(th(i(\"index\")));\n headers.AddRange(df.Columns.Select(c => (IHtmlContent) th(c)));\n var rows = new List<List<IHtmlContent>>();\n var take = 20;\n for (var i = 0; i < Math.Min(take, df.RowCount); i++)\n {\n var cells = new List<IHtmlContent>();\n cells.Add(td(i));\n foreach (var obj in df[i])\n {\n cells.Add(td(obj));\n }\n rows.Add(cells);\n }\n \n var t = table(\n thead(\n headers),\n tbody(\n rows.Select(\n r => tr(r))));\n \n writer.Write(t);\n}, \"text/html\");",
"_____no_output_____"
],
[
"using System.IO;\nusing System.Net.Http;\nstring housingPath = \"housing.csv\";\nif (!File.Exists(housingPath))\n{\n var contents = new HttpClient()\n .GetStringAsync(\"https://raw.githubusercontent.com/ageron/handson-ml2/master/datasets/housing/housing.csv\").Result;\n \n File.WriteAllText(\"housing.csv\", contents);\n}",
"_____no_output_____"
],
[
"var housingData = DataFrame.ReadCsv(housingPath);\nhousingData",
"_____no_output_____"
],
[
"housingData.Description()",
"_____no_output_____"
],
[
"Chart.Plot(\n new Graph.Histogram()\n {\n x = housingData[\"median_house_value\"],\n nbinsx = 20\n }\n)\n",
"_____no_output_____"
],
[
"var chart = Chart.Plot(\n new Graph.Scatter()\n {\n x = housingData[\"longitude\"],\n y = housingData[\"latitude\"],\n mode = \"markers\",\n marker = new Graph.Marker()\n {\n color = housingData[\"median_house_value\"],\n colorscale = \"Jet\"\n }\n }\n);\n\nchart.Width = 600;\nchart.Height = 600;\ndisplay(chart);",
"_____no_output_____"
],
[
"static T[] Shuffle<T>(T[] array)\n{\n Random rand = new Random();\n for (int i = 0; i < array.Length; i++)\n {\n int r = i + rand.Next(array.Length - i);\n T temp = array[r];\n array[r] = array[i];\n array[i] = temp;\n }\n return array;\n}\n\nvar randomIndices = Shuffle(Enumerable.Range(0, (int)housingData.RowCount).ToArray());\nvar testSize = (int)(housingData.RowCount * .1);\nvar trainRows = randomIndices[testSize..];\nvar testRows = randomIndices[..testSize];\n\nvar housing_train = housingData[trainRows];\nvar housing_test = housingData[testRows];\n\ndisplay(housing_train.RowCount);\ndisplay(housing_test.RowCount);",
"_____no_output_____"
],
[
"using Microsoft.ML;\nusing Microsoft.ML.Data;\nusing Microsoft.ML.AutoML;",
"_____no_output_____"
],
[
"%%time\n\nvar mlContext = new MLContext();\n\nvar experiment = mlContext.Auto().CreateRegressionExperiment(maxExperimentTimeInSeconds: 15);\nvar result = experiment.Execute(housing_train, labelColumnName:\"median_house_value\");",
"_____no_output_____"
],
[
"var scatters = result.RunDetails.Where(d => d.ValidationMetrics != null).GroupBy( \n r => r.TrainerName,\n (name, details) => new Graph.Scatter()\n {\n name = name,\n x = details.Select(r => r.RuntimeInSeconds),\n y = details.Select(r => r.ValidationMetrics.MeanAbsoluteError),\n mode = \"markers\",\n marker = new Graph.Marker() { size = 12 }\n });\n\nvar chart = Chart.Plot(scatters);\nchart.WithXTitle(\"Training Time\");\nchart.WithYTitle(\"Error\");\ndisplay(chart);\n\nConsole.WriteLine($\"Best Trainer:{result.BestRun.TrainerName}\");",
"_____no_output_____"
],
[
"var testResults = result.BestRun.Model.Transform(housing_test);\n\nvar trueValues = testResults.GetColumn<float>(\"median_house_value\");\nvar predictedValues = testResults.GetColumn<float>(\"Score\");\n\nvar predictedVsTrue = new Graph.Scatter()\n{\n x = trueValues,\n y = predictedValues,\n mode = \"markers\",\n};\n\nvar maximumValue = Math.Max(trueValues.Max(), predictedValues.Max());\n\nvar perfectLine = new Graph.Scatter()\n{\n x = new[] {0, maximumValue},\n y = new[] {0, maximumValue},\n mode = \"lines\",\n};\n\nvar chart = Chart.Plot(new[] {predictedVsTrue, perfectLine });\nchart.WithXTitle(\"True Values\");\nchart.WithYTitle(\"Predicted Values\");\nchart.WithLegend(false);\nchart.Width = 600;\nchart.Height = 600;\ndisplay(chart);",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecee44ccfa72be0f80e7f9cbbbcd69eca83d5f67 | 2,878 | ipynb | Jupyter Notebook | notebooks/neural_ode.ipynb | ali-ramadhan/neural-differential-equation-climate-parameterizations | cc0a9a8e740a0d5654d1844a6d2ce4e096ae9412 | [
"MIT"
] | null | null | null | notebooks/neural_ode.ipynb | ali-ramadhan/neural-differential-equation-climate-parameterizations | cc0a9a8e740a0d5654d1844a6d2ce4e096ae9412 | [
"MIT"
] | 4 | 2019-12-17T19:37:03.000Z | 2020-02-08T15:57:23.000Z | notebooks/neural_ode.ipynb | ali-ramadhan/6S898-climate-parameterization | cc0a9a8e740a0d5654d1844a6d2ce4e096ae9412 | [
"MIT"
] | 1 | 2022-01-17T12:54:21.000Z | 2022-01-17T12:54:21.000Z | 25.469027 | 93 | 0.504517 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecee4788ff8f0c0f50548c8f3188ff5bb1c0368b | 954,156 | ipynb | Jupyter Notebook | notebooks/data-notebooks/earth-observation/sentinel/S5P_NO2_NRTI_plot.ipynb | Mahir-Sparkess/ceda-notebooks | 618f473eb36acf3e6d2154e25d582688f03425aa | [
"BSD-2-Clause"
] | null | null | null | notebooks/data-notebooks/earth-observation/sentinel/S5P_NO2_NRTI_plot.ipynb | Mahir-Sparkess/ceda-notebooks | 618f473eb36acf3e6d2154e25d582688f03425aa | [
"BSD-2-Clause"
] | null | null | null | notebooks/data-notebooks/earth-observation/sentinel/S5P_NO2_NRTI_plot.ipynb | Mahir-Sparkess/ceda-notebooks | 618f473eb36acf3e6d2154e25d582688f03425aa | [
"BSD-2-Clause"
] | null | null | null | 4,240.693333 | 946,172 | 0.964537 | [
[
[
"import netCDF4 as nc4\nfrom netCDF4 import Dataset\nimport numpy as np\nimport os\nimport matplotlib.pyplot as plt\nimport matplotlib.pyplot as plt\nfrom matplotlib.colors import LogNorm\nfrom mpl_toolkits.basemap import Basemap",
"Libaries properly loaded. Ready to start\n"
],
[
"# Sentinel 5P file I want to process\n#s5p_file = '/neodc/sentinel5p/data/L2_NO2/v1.3/2020/04/26/S5P_NRTI_L2__NO2____20200426T123448_20200426T123948_13139_01_010302_20200426T131747.nc'\ns5p_file = '/neodc/sentinel5p/data/L2_NO2/v1.3/2020/01/01/S5P_OFFL_L2__NO2____20200101T110146_20200101T124316_11493_01_010302_20200103T041218.nc'\n\nfile = Dataset(s5p_file, mode='r')",
"_____no_output_____"
],
[
"print(file)",
"<class 'netCDF4._netCDF4.Dataset'>\nroot group (NETCDF4 data model, file format HDF5):\n Conventions: CF-1.7\n institution: KNMI\n source: Sentinel 5 precursor, TROPOMI, space-borne remote sensing, L2\n summary: TROPOMI/S5P NO2 1-Orbit L2 Swath 7x3.5km\n tracking_id: 029dc346-4505-471e-a81a-1f1c7a2786b4\n id: S5P_OFFL_L2__NO2____20200101T110146_20200101T124316_11493_01_010302_20200103T041218\n time_reference: 2020-01-01T00:00:00Z\n time_reference_days_since_1950: 25567\n time_reference_julian_day: 2458849.5\n time_reference_seconds_since_1970: 1577836800\n time_coverage_start: 2020-01-01T11:23:20Z\n time_coverage_end: 2020-01-01T12:21:44Z\n time_coverage_duration: PT3504.402S\n time_coverage_resolution: PT0.840S\n orbit: 11493\n references: http://www.tropomi.eu/data-products/nitrogen-dioxide\n processor_version: 1.3.2\n keywords_vocabulary: AGU index terms, http://publications.agu.org/author-resource-center/index-terms/\n keywords: 0345 Pollution, Urban and Regional; 0365 Troposphere, Composition and Chemistry; 0368 Troposphere, Constituent Transport and Chemistry; 3360 Remote Sensing; 3363 Stratospheric Dynamics\n standard_name_vocabulary: NetCDF Climate and Forecast Metadata Conventions Standard Name Table (v29, 08 July 2015), http://cfconventions.org/standard-names.html\n naming_authority: nl.knmi\n cdm_data_type: Swath\n date_created: 2020-01-03T04:12:39Z\n creator_name: The Sentinel 5 Precursor TROPOMI Level 2 products are developed with funding from the European Space Agency (ESA), the Netherlands Space Office (NSO), the Belgian Science Policy Office, the German Aerospace Center (DLR) and the Bayerisches Staatsministerium für Wirtschaft und Medien, Energie und Technologie (StMWi).\n creator_url: http://www.tropomi.eu\n creator_email: [email protected]\n project: Sentinel 5 precursor/TROPOMI\n geospatial_lat_min: -89.95969\n geospatial_lat_max: 81.79367\n geospatial_lon_min: 179.99991\n geospatial_lon_max: -179.99933\n license: No conditions apply\n platform: S5P\n sensor: TROPOMI\n spatial_resolution: 7x3.5km2\n cpp_compiler_version: g++ (GCC) 4.8.5 20150623 (Red Hat 4.8.5-11)\n cpp_compiler_flags: -g -O2 -fPIC -std=c++11 -W -Wall -Wno-ignored-qualifiers -Wno-write-strings -Wno-unused-variable -DTROPNLL2DP\n f90_compiler_version: GNU Fortran (GCC) 4.8.5 20150623 (Red Hat 4.8.5-11)\n f90_compiler_flags: -gdwarf-3 -O2 -fPIC -cpp -ffpe-trap=invalid -fno-range-check -frecursive -fimplicit-none -ffree-line-length-none -DTROPNLL2DP -Wuninitialized -Wtabs\n build_date: 2019-04-05T12:04:00Z\n revision_control_identifier: \n geolocation_grid_from_band: 4\n identifier_product_doi: 10.5270/S5P-s4ljg54\n identifier_product_doi_authority: http://dx.doi.org/\n algorithm_version: 1.3.0\n title: TROPOMI/S5P NO2 1-Orbit L2 Swath 7x3.5km\n product_version: 1.1.0\n Status_MET_2D: Nominal\n Status_NISE__: Nominal\n Status_CTMFCT: Nominal\n history: 2020-01-03 04:24:53 f_s5pops tropnll2dp /mnt/data1/storage_offl_l2/cache_offl_l2/WORKING-608406617/JobOrder.608406574.xml; 2020-01-10 13:21:24 TM5-MP-DOMINO offline\n processing_status: OFFL-processing nominal product\n date_modified: 2020-01-10T13:21:24Z\n dimensions(sizes): \n variables(dimensions): \n groups: PRODUCT, METADATA\n\n"
],
[
"lons = file.groups['PRODUCT'].variables['longitude'][:][0,:,:]\nlats = file.groups['PRODUCT'].variables['latitude'][:][0,:,:]\nno2 = file.groups['PRODUCT'].variables['nitrogendioxide_tropospheric_column_precision'][0,:,:]\nprint(lons.shape)\nprint (lats.shape)\nprint (no2.shape)\n\n\nno2_units = file.groups['PRODUCT'].variables['nitrogendioxide_tropospheric_column_precision'].units",
"(4173, 450)\n(4173, 450)\n(4173, 450)\n"
],
[
"plt.figure(figsize=(48,24))\n\nlon_0 = lons.mean()\nlat_0 = lats.mean()\n\n#m = Basemap(width=5000000,height=3500000,\n# resolution='l',projection='stere',\\\n# lat_ts=40,lat_0=lat_0,lon_0=lon_0)\n\nm = Basemap(resolution='l',projection='mill',lon_0=0)\n\nxi, yi = m(lons, lats)\n\n# Plot Data\ncs = m.pcolor(xi,yi,np.squeeze(no2),norm=LogNorm(), cmap='jet')\n\n# Add Grid Lines\nm.drawparallels(np.arange(-80., 81., 10.), labels=[1,0,0,0], fontsize=10)\nm.drawmeridians(np.arange(-180., 181., 10.), labels=[0,0,0,1], fontsize=10)\n\n# Add Coastlines, States, and Country Boundaries\nm.drawcoastlines()\nm.drawstates()\nm.drawcountries()\n\n# Add Colorbar\ncbar = m.colorbar(cs, location='bottom', pad=\"10%\")\ncbar.set_label(no2_units)\n\n# Add Title\nplt.title('NO2 in atmosphere')\n\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecee5002826fa0598e847e4e7b31b8387aeb33e4 | 38,721 | ipynb | Jupyter Notebook | Competition-Solutions/Text/#ZindiWeekendz Learning- To Vaccinate or Not to Vaccinate/Solution 2/ToVaccineOrNotVaccine.ipynb | ZindiAfrica/Natural-Language-Processing-NLP- | 41763b83677f1a4853af397a34d8a82fa9ac45fc | [
"MIT"
] | null | null | null | Competition-Solutions/Text/#ZindiWeekendz Learning- To Vaccinate or Not to Vaccinate/Solution 2/ToVaccineOrNotVaccine.ipynb | ZindiAfrica/Natural-Language-Processing-NLP- | 41763b83677f1a4853af397a34d8a82fa9ac45fc | [
"MIT"
] | null | null | null | Competition-Solutions/Text/#ZindiWeekendz Learning- To Vaccinate or Not to Vaccinate/Solution 2/ToVaccineOrNotVaccine.ipynb | ZindiAfrica/Natural-Language-Processing-NLP- | 41763b83677f1a4853af397a34d8a82fa9ac45fc | [
"MIT"
] | null | null | null | 29.004494 | 171 | 0.408667 | [
[
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"_____no_output_____"
]
],
[
[
"#Requirements",
"_____no_output_____"
]
],
[
[
"%%writefile setup.sh\n\nexport CUD_HOME=/usr/local/cuda-10.1\ngit clone https://github.com/NVIDIA/apex\npip install -v --no-cache-dir --global-option=\"--cpp_ext\" --global-option=\"--cuda_ext\" ./apex",
"Overwriting setup.sh\n"
],
[
"#For apex\n# !sh setup.sh",
"_____no_output_____"
],
[
"!pip install transformers --quiet",
"\u001b[K |████████████████████████████████| 573kB 2.7MB/s \n\u001b[K |████████████████████████████████| 1.0MB 8.6MB/s \n\u001b[K |████████████████████████████████| 890kB 16.2MB/s \n\u001b[K |████████████████████████████████| 3.7MB 25.6MB/s \n\u001b[?25h Building wheel for sacremoses (setup.py) ... \u001b[?25l\u001b[?25hdone\n"
]
],
[
[
"#Imports",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.simplefilter('ignore')",
"_____no_output_____"
],
[
"import os\nimport re\nimport gc\nimport sys\nimport random\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom tqdm.notebook import tqdm",
"_____no_output_____"
],
[
"from sklearn.model_selection import KFold, StratifiedKFold\nfrom sklearn.metrics import mean_squared_error",
"_____no_output_____"
],
[
"from keras.utils import to_categorical",
"Using TensorFlow backend.\n"
],
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torch.utils.data import Dataset, DataLoader",
"_____no_output_____"
],
[
"import transformers\nfrom transformers import (AutoTokenizer, AutoModel, AdamW, AutoConfig, get_linear_schedule_with_warmup)",
"_____no_output_____"
]
],
[
[
"#Envs",
"_____no_output_____"
]
],
[
[
"seed = 2020 # for reproductibility\nrandom.seed(seed)\nnp.random.seed(seed)\ntorch.manual_seed(seed)\n\nif torch.cuda.is_available(): \n torch.cuda.manual_seed(seed)\n torch.cuda.manual_seed_all(seed)\n torch.backends.cudnn.deterministic = True\n torch.backends.cudnn.benchmark = False",
"_____no_output_____"
],
[
"os.makedirs('MODELS/', exist_ok=True)",
"_____no_output_____"
]
],
[
[
"#Utilities",
"_____no_output_____"
]
],
[
[
"def process_prediction(preds):\n r'''\n This function helps us go from a classifiaction\n problem to a regression one.\n The regression values range are in [-1, 1].\n '''\n\n final_preds = []\n for pred in preds:\n argmax = np.argmax(pred, axis=0)\n if argmax == 0: final_preds.append( -1*pred[0] )\n elif argmax == 1: final_preds.append( 0 )\n else: final_preds.append( pred[2] )\n \n return final_preds\n\n\ndef rmse(true, pred):\n return np.sqrt(mean_squared_error(true, pred))",
"_____no_output_____"
],
[
"def convert_examples(tweets, tokenizer, max_length): \n bep = tokenizer.batch_encode_plus(tweets, add_special_tokens=True, max_length=max_length, \n pad_to_max_length=True, return_token_type_ids=True,\n return_attention_masks=True)\n \n return bep['input_ids'], bep['token_type_ids'], bep['attention_mask']",
"_____no_output_____"
],
[
"class EarlyStopping:\n def __init__(self, patience=3, mode='min'):\n self.step = 0\n self.patience = patience\n self.mode = mode\n self.stop = False\n self.loss = np.inf\n\n def update(self, loss):\n if self.loss < loss:\n self.step += 1\n else: \n self.step = 0\n self.loss = loss\n \n if self.step == self.patience: self.stop = True ",
"_____no_output_____"
],
[
"class VaccineOrNotModel(nn.Module):\n def __init__(self, model_name):\n super(VaccineOrNotModel, self).__init__()\n\n config = AutoConfig.from_pretrained(model_name)\n self.model = AutoModel.from_pretrained(model_name, config=config)\n self.dropout = nn.Dropout(0.1)\n self.classifier = nn.Linear(config.hidden_size, 3)\n nn.init.xavier_normal_(self.classifier.weight)\n \n def forward(self, input_ids, attention_mask=None, token_type_ids=None):\n _, out = self.model(\n input_ids,\n attention_mask=attention_mask,\n token_type_ids=token_type_ids\n )\n\n out = self.dropout(out)\n logits = self.classifier(out)\n logits = F.sigmoid(logits)\n\n return logits",
"_____no_output_____"
],
[
"class VaccineOrNotDataset(Dataset):\n def __init__(self, input_ids, att_masks, token_ids, labels=None, task='train', classes=3):\n self.input_ids = input_ids\n self.att_masks = att_masks\n self.token_ids = token_ids\n self.labels = labels\n self.c = classes\n self.task = task\n\n def __len__(self):\n return len(self.input_ids)\n\n def __getitem__(self, id):\n out = {\n 'input_ids': torch.tensor(self.input_ids[id]),\n 'token_type_ids': torch.tensor(self.token_ids[id]),\n 'attention_mask': torch.tensor(self.att_masks[id]),\n }\n\n if self.task=='train': \n out.update(\n {\n 'label': torch.tensor(to_categorical(self.labels[id]+1, self.c), dtype=torch.float), #The former labels are in [-1, 1], so we add +1 to be in [0, 2]\n }\n )\n\n return out",
"_____no_output_____"
],
[
"def training_fn(model, opt, scheduler, criterion, train_ds, bs, device='cuda', n_labels=3):\n train_dl = DataLoader(train_ds, bs)\n\n avg_rmse = 0\n avg_loss = 0\n\n pbar = tqdm(train_dl, desc='Training ...')\n model.train()\n\n for i, data in enumerate(pbar):\n input_ids = data['input_ids'].to(device)\n attention_mask = data['attention_mask'].to(device)\n token_type_ids = data['token_type_ids'].to(device)\n labels = data['label'].to(device)\n\n opt.zero_grad()\n\n logits = model(input_ids=input_ids,\n attention_mask=attention_mask,\n token_type_ids=token_type_ids)\n \n loss = criterion(logits, labels)\n custom_loss = rmse( labels.cpu().detach().numpy().argmax(1)-1, process_prediction(logits.cpu().detach().numpy()) )\n\n avg_loss += loss.item()\n avg_rmse += custom_loss\n\n loss.backward()\n\n opt.step()\n scheduler.step()\n\n pbar.set_postfix(loss=avg_loss/(i+1), rmse=avg_rmse/(i+1))\n pbar.update()\n\n return avg_rmse/len(train_dl)",
"_____no_output_____"
],
[
"def eval_fn(model, criterion, valid_ds, eval_bs, device='cuda', n_labels=3):\n valid_dl = DataLoader(valid_ds, eval_bs)\n\n avg_rmse = 0\n avg_loss = 0\n\n pbar = tqdm(valid_dl, desc='Evaluation ...')\n model.eval()\n\n for i, data in enumerate(pbar):\n input_ids = data['input_ids'].to(device)\n attention_mask = data['attention_mask'].to(device)\n token_type_ids = data['token_type_ids'].to(device)\n labels = data['label'].to(device)\n\n logits = model(input_ids=input_ids,\n attention_mask=attention_mask,\n token_type_ids=token_type_ids\n )\n \n\n loss = criterion(logits, labels)\n \n custom_loss = rmse( labels.cpu().detach().numpy().argmax(1)-1, process_prediction(logits.cpu().detach().numpy()) )\n\n avg_loss += loss.item()\n avg_rmse += custom_loss\n\n pbar.set_postfix(loss=avg_loss/(i+1), rmse=avg_rmse/(i+1))\n pbar.update()\n\n return avg_rmse/len(valid_dl)",
"_____no_output_____"
],
[
"def predict(input_ids, mask, token_ids, MODELS, bs=8, n_labels=3):\n test_ds = VaccineOrNotDataset(input_ids, mask, token_ids, task='test')\n testloader = DataLoader(test_ds, bs, shuffle=False)\n\n predictions_proba = []\n\n out = None\n\n for data in tqdm(testloader):\n x = data['input_ids'].to(device)\n\n for i in range(n_folds):\n if i == 0: out = MODELS[i](input_ids=x)\n else: out += MODELS[i](input_ids=x)\n\n out /= n_folds\n out_probas = out.cpu().detach().numpy()\n\n predictions_proba += out_probas.tolist()\n\n return predictions_proba\n",
"_____no_output_____"
],
[
"def predict_by_fold(model, input_ids, mask, token_ids, bs=8, n_labels=3):\n test_ds = VaccineOrNotDataset(input_ids, mask, token_ids, task='test')\n testloader = DataLoader(test_ds, bs, shuffle=False)\n\n predictions_proba = []\n\n out = None\n\n for data in tqdm(testloader):\n x = data['input_ids'].to(device)\n \n out = model(input_ids=x)\n\n out /= n_folds\n out_probas = out.cpu().detach().numpy()\n\n predictions_proba += out_probas.tolist()\n\n return predictions_proba\n",
"_____no_output_____"
],
[
"def run_fold(fold, epochs, splits, bs, eval_bs, lr, model_name, path='MODELS/'):\n tr, vr = splits\n train_ds = VaccineOrNotDataset(np.take(tr_ids, tr, axis=0), np.take(tr_masks,tr, axis=0),\n np.take(tr_tokens,tr, axis=0), np.take(train_labels,tr, axis=0)) \n valid_ds = VaccineOrNotDataset(np.take(tr_ids, vr, axis=0), np.take(tr_masks,vr, axis=0),\n np.take(tr_tokens,vr, axis=0), np.take(train_labels,vr, axis=0)) \n\n train_dl = DataLoader(train_ds, bs)\n valid_dl = DataLoader(valid_ds, eval_bs)\n\n device='cuda'\n\n model = VaccineOrNotModel(model_name)\n model.to(device)\n\n num_train_steps = int(len(train_ds) / bs*epochs)\n\n param_optimizer = list(model.named_parameters())\n no_decay = [\"bias\", \"LayerNorm.bias\", \"LayerNorm.weight\"]\n optimizer_parameters = [\n {'params': [p for n, p in param_optimizer if not any(nd in n for nd in no_decay)], 'weight_decay': 0.001},\n {'params': [p for n, p in param_optimizer if any(nd in n for nd in no_decay)], 'weight_decay': 0.0},\n ]\n\n opt = AdamW(optimizer_parameters, lr=lr)\n scheduler = get_linear_schedule_with_warmup(\n opt, num_warmup_steps=0, num_training_steps=num_train_steps\n )\n\n criterion = nn.BCELoss()\n es = EarlyStopping(patience=3)\n\n best_rmse = np.inf\n\n for epoch in tqdm(range(epochs)):\n training_fn(model, opt, scheduler, criterion, train_ds, bs, device)\n score = eval_fn(model, criterion, valid_ds, eval_bs, device)\n es.update(score)\n\n if score < best_rmse:\n best_rmse = score\n torch.save(model.state_dict(), f'{path}model_state_dict_{fold}.bin')\n\n if es.stop:\n print(\"Early Stopping triggered\")\n return best_rmse\n \n return best_rmse",
"_____no_output_____"
]
],
[
[
"#Reading the Data",
"_____no_output_____"
]
],
[
[
"path = 'drive/My Drive/Zindi/#ZindiWeekendz/'",
"_____no_output_____"
],
[
"train = pd.read_csv(path+'Train.csv')\ntest = pd.read_csv(path+'Test.csv')\nsample = pd.read_csv(path+'SampleSubmission.csv')",
"_____no_output_____"
]
],
[
[
"#Cleaning and Tokenization",
"_____no_output_____"
]
],
[
[
"model_name = 'roberta-large'",
"_____no_output_____"
],
[
"tokenizer = AutoTokenizer.from_pretrained(model_name)",
"_____no_output_____"
],
[
"train['safe_text'] = train['safe_text'].apply(str)\ntest['safe_text'] = test['safe_text'].apply(str)",
"_____no_output_____"
],
[
"train['safe_text'] = train['safe_text'].apply(str.lower)\ntest['safe_text'] = test['safe_text'].apply(str.lower)",
"_____no_output_____"
],
[
"train['safe_text'] = train['safe_text'].apply(lambda x: x.replace('&', ' '))\ntest['safe_text'] = test['safe_text'].apply(lambda x: x.replace('&', ' '))",
"_____no_output_____"
],
[
"train['safe_text'] = train['safe_text'].apply(lambda x: x.replace('<user>', ' '))\ntest['safe_text'] = test['safe_text'].apply(lambda x: x.replace('<user>', ' '))\n\ntrain['safe_text'] = train['safe_text'].apply(lambda x: x.replace('<url>', ' '))\ntest['safe_text'] = test['safe_text'].apply(lambda x: x.replace('<url>', ' '))",
"_____no_output_____"
],
[
"train['safe_text'] = train['safe_text'].apply(lambda x: x.replace('#', ' '))\ntest['safe_text'] = test['safe_text'].apply(lambda x: x.replace('#', ' '))",
"_____no_output_____"
],
[
"train['safe_text'] = train['safe_text'].apply(lambda x: x.strip('.').strip())\ntest['safe_text'] = test['safe_text'].apply(lambda x: x.strip('.').strip())",
"_____no_output_____"
],
[
"train.drop(index=[4798, 4799], inplace=True)\ntrain.reset_index(drop=True, inplace=True)",
"_____no_output_____"
],
[
"train.head()",
"_____no_output_____"
],
[
"train_text = train['safe_text'].values\ntrain_labels = train['label'].values\n\ntest_text = test['safe_text'].values",
"_____no_output_____"
],
[
"tr_ids, tr_tokens, tr_masks = convert_examples(train_text, tokenizer, max_length=34)",
"_____no_output_____"
],
[
"te_ids, te_tokens, te_masks = convert_examples(test_text, tokenizer, max_length=34)",
"_____no_output_____"
]
],
[
[
"#Training",
"_____no_output_____"
]
],
[
[
"#These Hyperparameters could be tune\nepochs = 10\nn_folds = 10\nbs = 32\neval_bs = 4\nlr = 3e-5\ndevice = 'cuda'",
"_____no_output_____"
],
[
"skf = StratifiedKFold(n_folds, shuffle=True, random_state=seed)\nall_folds = list(skf.split(tr_ids, train_labels))",
"_____no_output_____"
],
[
"cv_score = 0\nfor fold in range(n_folds):\n splits = all_folds[fold]\n\n s = run_fold(fold, epochs, splits, bs, eval_bs, lr, model_name=model_name)\n cv_score += s/n_folds\nprint(\"Avg rmse : \", cv_score)",
"_____no_output_____"
]
],
[
[
"#Prediction",
"_____no_output_____"
]
],
[
[
"#Just to cool down the memory :)\ndel train,tokenizer,tr_ids,tr_masks,tr_tokens\ngc.collect()",
"_____no_output_____"
],
[
"preds = []\n\nfor fold in range(n_folds):\n model = VaccineOrNotModel(model_name)\n model.to(device)\n model.load_state_dict(torch.load(f'MODELS/model_state_dict_{fold}.bin'))\n model.eval()\n\n preds.append( predict_on_fold(model, te_ids, te_masks, te_tokens, bs=8) )\n\n del model\n gc.collect()",
"_____no_output_____"
],
[
"predictions_proba = np.sum(preds, axis=0)",
"_____no_output_____"
],
[
"predictions = process_prediction(predictions_proba) #Final step",
"_____no_output_____"
]
],
[
[
"#Submission",
"_____no_output_____"
]
],
[
[
"subs = test[['tweet_id']]\nsubs['target'] = predictions\nsubs.columns = ['ID', 'target']\nsubs.head()",
"_____no_output_____"
],
[
"subs.describe()",
"_____no_output_____"
],
[
"subs.to_csv(f'arch_{model_name}_folds_{n_folds}_epochs_{epochs}_bs_{bs}_lr_{lr}_cv_{cv_score}.csv', index=False)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecee624efe1de2f32afa195af0fb9d851bd4255e | 353,966 | ipynb | Jupyter Notebook | Credit_Risk_Prediction.ipynb | Jhou23/credit-risk-prediction | c01cdf64d177369bb4958ea42aba8c330394cc7d | [
"MIT"
] | null | null | null | Credit_Risk_Prediction.ipynb | Jhou23/credit-risk-prediction | c01cdf64d177369bb4958ea42aba8c330394cc7d | [
"MIT"
] | null | null | null | Credit_Risk_Prediction.ipynb | Jhou23/credit-risk-prediction | c01cdf64d177369bb4958ea42aba8c330394cc7d | [
"MIT"
] | null | null | null | 292.29232 | 66,886 | 0.897631 | [
[
[
"**Import Dataset & Packages**",
"_____no_output_____"
]
],
[
[
"import pandas as pd \nimport numpy as np \nfrom datetime import datetime\nimport datetime as parent_date\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n#!pip install sweetviz # install the package if you haven't \nimport sweetviz\nimport statsmodels.api as sm\nfrom scipy.stats import chi2_contingency\nfrom sklearn.ensemble import ExtraTreesClassifier\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.metrics import roc_auc_score, roc_curve\nfrom sklearn.metrics import confusion_matrix",
"/usr/local/lib/python3.6/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
],
[
"df = pd.read_csv('/content/drive/My Drive/Colab Notebooks/Data set/Propensity to Pay_Collections Data.csv')\ndf_org = pd.read_csv('/content/drive/My Drive/Colab Notebooks/Data set/cleaned_data.csv')\ndf_org['Amount'] = df_clean['Amount'].copy()\ndf_org = df_org.iloc[:,:18].copy() # for some reason, the org dataset I got has wireld column (ie: unnamed column 18)",
"/usr/local/lib/python3.6/dist-packages/IPython/core/interactiveshell.py:2718: DtypeWarning: Columns (8) have mixed types.Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
]
],
[
[
"# Initial Data Cleaning & Feature Engineering for EDA",
"_____no_output_____"
],
[
"**Data Cleaning & Feature Engineering**",
"_____no_output_____"
]
],
[
[
"### 1. Rearrange the order of origal dataset, so that the dataset has more consecutive logical meaning\ncolumn_order = ['Cust_Num','Customer Group','ZIPCODE','Region','City','Customer Name','Age Of Customer(Months)','Age Of Customer(Year)',\n 'DocumentNo','Clrng doc.','Payment Method description','Doc. Date','Net due dt','Pstng Date','Clearing Date',\n 'Amount','Payment Term','Days Overdue (Delay)']\ndf_org = df_org.reindex(columns=column_order)\n\n### 2. reform the datatime columns \n# Payment Term = Net Due Date - Psting Date \ndf_org['Doc. Date'] = pd.to_datetime(df_org['Doc. Date'])\ndf_org['Net due dt'] = pd.to_datetime(df_org['Net due dt'])\ndf_org['Pstng Date'] = pd.to_datetime(df_org['Pstng Date'])\n# 2-1: Clearing Date = Net Due Date - Days Overdue (Delay)\nholder = []\nfor date, delay in zip(df_org['Net due dt'],df_org['Days Overdue (Delay)']):\n result = date + parent_date.timedelta(days=delay)\n holder.append(result)\n\ndf_org['Clearing Date'] = holder\n\n### 3. Create a new column: Is_Pay_Delay (binary variables to indicate whether this payment is delayed or not)\nholder = []\nfor delay in df_org['Days Overdue (Delay)']:\n if delay <= 0: holder.append(0)\n else: holder.append(1)\n\ndf_org['Is_Pay_Delay'] = holder\n\n### 4: Handle Zipcode\n# The zipcode we have is psedo zipcode, so we cannot get the exact geo info (may be for privacy reason)\n# So, I picked the top 6 occurance zipcode and put the rest in 'Other' Category. The occurancy as follow: \n# AX0032 - 10219\n# AX0014 - 9712\n# AX0028 - 7838\n# AX0048 - 6367\n# AX0025 - 3935\n# AX0030 - 2637\n# Other - 5133\nfor index, zip in enumerate(df_org['ZIPCODE']):\n if zip not in ['AX0032','AX0014','AX0028','AX0048','AX0025','AX0030']: \n df_org.iloc[index,2] = 'Other'\n\n### 5: Handle Region Code\n# Same logic as zipcode\n# AA125 - 10219\n# AA113 - 9712\n# AA121 - 8054\n# AA127 - 6367\n# AA120 - 3983\n# AA123 - 2637\n# Other - 4869\nfor index, zip in enumerate(df_org['Region']):\n if zip not in ['AA125','AA113','AA121','AA127','AA120','AA123']: \n df_org.iloc[index,3] = 'Other'\n\n### 6: Handle City Code\n# Same logic as zipcode\n# AA42 - 10219\n# AA24 - 9712\n# AA38 - 7838\n# AA57 - 6367\n# AA35 - 3935\n# AA40 - 2637\n# Other - 5133\nfor index, zip in enumerate(df_org['City']):\n if zip not in ['AA42','AA24','AA38','AA57','AA35','AA40']: \n df_org.iloc[index,4] = 'Other'\n\n### 7. Drop the following columns. Customer Name is not useful since those name are psedo names. \n# Age of the Customer in Year is overlapping with Age of Customer in Month. (One thing to note: when transfer year to month, it is not accurate)\n# ie: first row: Month as 34 which is 2 Year and 10 Months but the same year reprentation is 2 year. \n# Clrng doc: not useful\nto_drop = ['Customer Name','Age Of Customer(Year)','Clrng doc.']\ndf_org.drop(columns=to_drop,inplace=True)\n\n### 8: Since Date column has no practical use case in prediciton and all info is in the df, let z drop them\nto_drop = ['Doc. Date','Net due dt','Pstng Date','Clearing Date']\ndf_org.drop(columns=to_drop,inplace=True)\n\n### 9: Create a new column indicate the total number of delay for a specific customer and aggregate into the documents level \nto_merge = df_org.groupby('Cust_Num')['Is_Pay_Delay'].agg('sum').reset_index()\nNum = df_org['Cust_Num'].copy().reset_index()\nHolder = Num.merge(to_merge).sort_values('index')\ntotal_delay = Holder['Is_Pay_Delay'].values\ndf_org['Customer_Total_Delay'] = total_delay\n\n### 10: Drop Customer Group Column since there are only 1 value in there as Wholeseller\ndf_org.drop(labels='Customer Group',axis=1,inplace=True)\n\n### 11: Categorize the Payment Method\n# ie: Put Regulatory, Regulatory 1, Regulatory 2 into Regulatory type\n# Since Direct Payment is mojority, so turn this column into 2 level binary value\ndf_org.loc[df_org['Payment Method description'].isin(['Direct Debits 2','Direct Debits 1']),'Payment Method description'] = 'Direct Debits'\ndf_org.loc[df_org['Payment Method description'] != 'Direct Debits','Payment Method description'] = 'Non Direct Debits'\n\n### 12: Add target variable Over_Due_Category\n# 0: Not delay \n# 1: delay large than 0 and less than 30\n# 2: delay large than 30 and less than 60\n# 3: delay larger than 60\nholder = []\n\nfor value in df_org['Days Overdue (Delay)'].values:\n if value <= 0: holder.append(0)\n elif (value > 0) & (value <= 30): holder.append(1)\n elif (value > 30) & (value <= 60): holder.append(2)\n else: holder.append(3)\n\ndf_org['Over_Due_Category'] = holder\n\n### 13. Drop Variables\nto_drop=['Days Overdue (Delay)','Is_Pay_Delay']\ndf_org.drop(columns=to_drop,inplace=True)\n\n### 12. Quick Descriptive Analysis for the cleaned dataframe\nsweetviz.analyze(df_org).show_html()\n\n### 13. Save the dataset\ndf_org.to_csv('/content/drive/My Drive/Colab Notebooks/Data set/cleaned_data.csv',index=False)",
"_____no_output_____"
],
[
"df_org.head()",
"_____no_output_____"
],
[
"df_org.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 45841 entries, 0 to 45840\nData columns (total 11 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Cust_Num 45841 non-null int64 \n 1 ZIPCODE 45841 non-null object \n 2 Region 45841 non-null object \n 3 City 45841 non-null object \n 4 Age Of Customer(Months) 45841 non-null float64\n 5 DocumentNo 45841 non-null int64 \n 6 Payment Method description 45841 non-null object \n 7 Amount 45839 non-null float64\n 8 Payment Term 45841 non-null int64 \n 9 Customer_Total_Delay 45841 non-null int64 \n 10 Over_Due_Category 45841 non-null int64 \ndtypes: float64(2), int64(5), object(4)\nmemory usage: 3.8+ MB\n"
]
],
[
[
"# Second Exploritaty Data Analysis",
"_____no_output_____"
],
[
"**Exploritary Analysis**",
"_____no_output_____"
]
],
[
[
"df_org['Cust_Num'] = df_org['Cust_Num'].astype('string')\ndf_temp = df_org.sort_values(by='Customer_Total_Delay',ascending=False).copy()\n\nfig, ax1 = plt.subplots(figsize=(10,5))\nsns.set_style('darkgrid')\nsns.barplot(x='Customer_Total_Delay',y='Cust_Num',data=df_temp,palette='hls',ax=ax1)\nax1.set_ylim(bottom=20,top=0)\nplt.yticks(fontsize=9)\n\nplt.title('Top 20 Customers Have Highest Total Number of Delayed Payment',fontsize=15)\nplt.xlabel('Number of Delayed Payment',fontsize=12)\nplt.ylabel('Customer Series Number',fontsize=12)\n\nplt.savefig('/content/drive/My Drive/Colab Notebooks/Data set/Customer_Delay.png')\nplt.show()",
"_____no_output_____"
]
],
[
[
"**ECDF Chart**\n\nCustomer Business Life Circle Chart (upper left conor): \n> - 15% (0.15) of the customer have been doing business with us less than 140 months. \n> - 100% of the customer in this dataset have beening doing business with us less than 160 months. \n\nCustomer Total Number of Delayed Payment Chart (upper right conor): \n> - 25% of the customers has Total number of delayed payment less than 2000 invoices. \n> - 80% of the customers has total number of delayed payment less than 5000 invoices. \n\nInvoice Payment Term by Day Chart (Lower left conor): \n> - 83% of the invoices has invoice payment term less than 15 days. \n> - 17% of the invoices has invoice payment term between 50 and 150 days. ",
"_____no_output_____"
]
],
[
[
"x_age = np.sort(df_org['Age Of Customer(Months)'])\nx_amount = np.sort(df_org['Amount'])\nx_term = np.sort(df_org['Payment Term'])\nx_total_delay = np.sort(df_org['Customer_Total_Delay'])\n\ny_age = np.arange(1,len(x_age)+1) / len(x_age)\ny_amount = np.arange(1,len(x_amount)+1) / len(x_amount)\ny_term = np.arange(1,len(x_term)+1) / len(x_term)\ny_total_delay = np.arange(1,len(x_total_delay)+1) / len(x_total_delay)\n\nfig,[(ax1,ax2),(ax3,ax4)] = plt.subplots(nrows=2,ncols=2,figsize=(12,12))\nax1.plot(x_age,y_age,marker='.',linestyle='none',color='orange')\nax4.plot(x_amount,y_amount,marker='.',linestyle='none',color='green')\nax3.plot(x_term,y_term,marker='.',linestyle='none',color='green')\nax2.plot(x_total_delay,y_total_delay,marker='.',linestyle='none',color='orange')\n\nax1.set_xlabel('Customer Business Life Circle by Month')\nax1.set_ylabel('Percentage - ECDF')\n\nax4.set_xlabel('Invoice Amount')\nax4.set_ylabel('Percentage - ECDF')\n\nax3.set_xlabel('Invoice Payment Term by Day')\nax3.set_ylabel('Percentage - ECDF')\n\nax2.set_xlabel('Customer Total Number of Delay Payment')\nax2.set_ylabel('Percentage - ECDF')\n\nplt.title('ECDF Chart for Age of Cutomer, Total Delayed Payment, Payment Term, Invoice Amount',y=2.215,x=0,fontsize=15)\n\nplt.savefig('/content/drive/My Drive/Colab Notebooks/Data set/ECDF.png')\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Pie Chart for Geographic Region & Payment Method - Very Interesting**\n\n> - Among all six Top Occurancy Zipcode / Region / City, Non of them are using methods other than Direct Payment. \n\n> - It seems like payment methods such as Wire and Regulatory payments are not popular among our client base. ",
"_____no_output_____"
]
],
[
[
"fig, [ax1,ax2,ax3] = plt.subplots(nrows=3,ncols=1,figsize=(10,13))\nsns.countplot(x='ZIPCODE',data=df_org,hue='Payment Method description',order=df_org['ZIPCODE'].value_counts().index,ax=ax1)\nsns.countplot(x='Region',data=df_org,hue='Payment Method description',order=df_org['Region'].value_counts().index,ax=ax2)\nsns.countplot(x='City',data=df_org,hue='Payment Method description',order=df_org['City'].value_counts().index,ax=ax3)\n\nplt.title('Payment Method in Different Geographic Region',y=3.5,fontsize=15)\nplt.ylabel('Number of Count')\nax1.set_ylabel('Number of Count')\nax2.set_ylabel('Number of Count')\n\nplt.savefig('/content/drive/My Drive/Colab Notebooks/Data set/Geo_Payment_Type.png')\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Relationship Between Payment Amount & Customer Business Life Circle**\n\n> - It seems like the longer the customer doing business with company, the more likely they will granted higher loan Amount. \n> - It is interesting that for some large loan amount, the repayment method are non direct Debits such as wire transfer / regulatory payment. ",
"_____no_output_____"
]
],
[
[
"fig,ax1 = plt.subplots(figsize=(8,8))\nsns.scatterplot(x='Age Of Customer(Months)',y='Amount',data=df_org,hue='Payment Method description')\nax1.ticklabel_format(style='plain')\nplt.title('Relationship Between Payment Amount & Customer Business Life Circle',fontsize=15)\nplt.xlabel('Customer Business Life Circle in Month',fontsize=13)\nplt.ylabel('Payment Amount $',fontsize=13)\n\nplt.savefig('/content/drive/My Drive/Colab Notebooks/Data set/Payment_Life_Circle.png')\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Number of Total Delay for Each Customer & Payment Amount**\n> - It seems like for the more customer didn't pay the repayment on time, the less the loan they will granted.\n> - It interesting that those customer who has higher number of delayed payment, all of them are using Direct Payment. For those who not using the direct payment such as wire transfer, they did not have the late due payment. May be due to the wire transfer is more accessable than direct payment. ",
"_____no_output_____"
]
],
[
[
"fig,ax1 = plt.subplots(figsize=(8,8))\nsns.scatterplot(x='Customer_Total_Delay',y='Amount',data=df_org,hue='Payment Method description',ax=ax1)\nax1.ticklabel_format(style='plain')\nplt.title('Number of Total Delay for Each Customer & Payment Amount',fontsize=15)\nplt.xlabel('Number of Time Customer Delayed Payment',fontsize=12)\nplt.ylabel('Payment Amount $',fontsize=12)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Featuere Selection & Second Data Cleaning for Modeling",
"_____no_output_____"
]
],
[
[
"### 1: Encode the payment method\nzipcode_dict = {'Other':0,'AX0032':1,'AX0014':2,'AX0028':3,'AX0048':4,'AX0025':5,'AX0030':6}\nregion_dict = {'Other':0,'AA125':1,'AA113':2,'AA121':3,'AA127':4,'AA120':5,'AA123':6}\ncity_dict = {'Other':0,'AA42':1,'AA24':2,'AA38':3,'AA57':4,'AA35':5,'AA40':6}\npayment_dict = {'Direct Debits':1,'Non Direct Debits':0}\n\ndf_org['Payment Method description'] = df_org['Payment Method description'].map(payment_dict)\ndf_org['ZIPCODE'] = df_org['ZIPCODE'].map(zipcode_dict)\ndf_org['Region'] = df_org['Region'].map(region_dict)\ndf_org['City'] = df_org['City'].map(city_dict)\n\n### 2: Get the is due day weekend column\ndf_org['Net_Due_Date'] = pd.to_datetime(df_org['Net_Due_Date'])\ndf_org['Invoice_due_weekend'] = df_org['Net_Due_Date'].dt.dayofweek\ndf_org['Invoice_due_weekend'] = df_org['Invoice_due_weekend'] + 1 \ndf_org.loc[df_org['Invoice_due_weekend'].isin([1,2,3,4,5]),'Invoice_due_weekend'] = 'Weekday'\ndf_org.loc[df_org['Invoice_due_weekend'].isin([6,7]),'Invoice_due_weekend'] = 'Weekend'\ndf_org['Invoice_due_weekend'] = df_org['Invoice_due_weekend'].map({'Weekday':0,'Weekend':1})\n\n### 3: Drop the due day column\ndf_org.drop(columns='Net_Due_Date',inplace=True)\n\n### 4: Continouos variables normalization\ndf_org['Age Of Customer(Months)'] = (df_org['Age Of Customer(Months)'] - df_org['Age Of Customer(Months)'].mean()) / df_org['Age Of Customer(Months)'].std()\ndf_org['Amount'] = (df_org['Amount'] - df_org['Amount'].mean()) / df_org['Amount'].std()\ndf_org['Payment Term'] = (df_org['Payment Term'] - df_org['Payment Term'].mean()) / df_org['Payment Term'].std()\ndf_org['Customer_Total_Delay'] = (df_org['Customer_Total_Delay'] - df_org['Customer_Total_Delay'].mean()) / df_org['Customer_Total_Delay'].std()\n\n### 5: Re-order columns \ncol_order = ['Cust_Num','DocumentNo','ZIPCODE','Region','City','Payment Method description','Invoice_due_weekend',\n 'Age Of Customer(Months)','Amount','Payment Term','Customer_Total_Delay','Over_Due_Category']\n\ndf_org = df_org.reindex(columns=col_order)\n\n### 5-1: Remove 2 missing value from amount \ndf_org.drop(index=[45839,45840],inplace=True)\n\n### 6: Save the modeling data set\ndf_org.to_csv('/content/drive/My Drive/Colab Notebooks/Data set/modeling_data.csv',index=False)\n\n### 7: Get the Modeling dataset \nmodel_data = df_org.drop(['Cust_Num','DocumentNo'],axis=1).copy()",
"_____no_output_____"
],
[
"### 4: Chi_Squared Test for categorical variables \n# H0: two variables are independent\ndef chi_square_test(x_var):\n table = pd.crosstab(df_org['Over_Due_Category'],df_org[x_var])\n chi, pvalue, dof, ex = chi2_contingency(table.values)\n if pvalue > 0.05: print(x_var,'is independent and not include in model')\n else: print(x_var,'is related with y var and should include in model')\n\nfor col in ['ZIPCODE','Region','City','Payment Method description','Invoice_due_weekend']: \n chi_square_test(col)",
"ZIPCODE is related with y var and should include in model\nRegion is related with y var and should include in model\nCity is related with y var and should include in model\nPayment Method description is related with y var and should include in model\nInvoice_due_weekend is related with y var and should include in model\n"
],
[
"# 5: Feature Selection by Decision Tree \n# Since Customer_Total_Delay & Payment Term are highly correlated, and Payment Term is more important, so remove Customer_Total_Delay\n# Since the Zipcode, Region, and City essentially indicate a very similar object, so I only keep the zipcode\n# Payment Term & Customer_Total_Delay is strongly negative correlated\nmodel_data.loc[:,['Age Of Customer(Months)','Amount','Payment Term','Customer_Total_Delay']].corr()\n\nclassifer = ExtraTreesClassifier()\nresult = classifer.fit(indenpend_var,target_var)\n\nresult_df = pd.DataFrame({'Feature':indenpend_var.columns.values,'Importance':result.feature_importances_})\nresult_df.sort_values('Importance',ascending=False,inplace=True)\n\nsns.barplot(x='Importance',y='Feature',data=result_df)\nplt.title('Feature Importance')\nplt.show()\n\n# 6: Drop those columns mentioned above\nmodel_data.drop(['Region','City','Customer_Total_Delay'],axis=1,inplace=True)",
"_____no_output_____"
]
],
[
[
"**Multinomial Logistic Regresion Result**\n> - [How to Explain the result](https://https://stats.idre.ucla.edu/stata/output/multinomial-logistic-regression-2/)\n\n> - for Continouos variables (`Age Of Customer(Months)`, `Amount`,`Payment Term`), explain those as 1 std increase becasue I standarlized the data before regression. \n\n1. **std for Age of customer**: 21.1\n2. **std for Amount**: 68777.0\n3. **std for Payment Term**: 26.3\n",
"_____no_output_____"
]
],
[
[
"target_var = model_data['Over_Due_Category'].copy()\nindenpend_var = model_data.drop(['Over_Due_Category','Region','City','Customer_Total_Delay'],axis=1).copy()\nmod = sm.MNLogit(target_var,indenpend_var).fit()\nprint(mod.summary())",
" MNLogit Regression Results \n==============================================================================\nDep. Variable: Over_Due_Category No. Observations: 45839\nModel: MNLogit Df Residuals: 45821\nMethod: MLE Df Model: 15\nDate: Thu, 12 Nov 2020 Pseudo R-squ.: 0.08651\nTime: 04:47:07 Log-Likelihood: -32597.\nconverged: True LL-Null: -35684.\nCovariance Type: nonrobust LLR p-value: 0.000\n==============================================================================================\n Over_Due_Category=1 coef std err z P>|z| [0.025 0.975]\n----------------------------------------------------------------------------------------------\nZIPCODE 0.0678 0.007 10.092 0.000 0.055 0.081\nPayment Method description -0.6863 0.023 -29.747 0.000 -0.731 -0.641\nInvoice_due_weekend 1.3924 0.021 66.811 0.000 1.352 1.433\nAge Of Customer(Months) 0.2733 0.013 20.729 0.000 0.247 0.299\nAmount -0.0031 0.011 -0.295 0.768 -0.024 0.018\nPayment Term -0.0827 0.011 -7.318 0.000 -0.105 -0.061\n----------------------------------------------------------------------------------------------\n Over_Due_Category=2 coef std err z P>|z| [0.025 0.975]\n----------------------------------------------------------------------------------------------\nZIPCODE 0.8602 0.033 26.082 0.000 0.796 0.925\nPayment Method description -6.9125 0.158 -43.822 0.000 -7.222 -6.603\nInvoice_due_weekend 0.5270 0.076 6.940 0.000 0.378 0.676\nAge Of Customer(Months) 0.1722 0.017 9.852 0.000 0.138 0.207\nAmount -0.0975 0.044 -2.195 0.028 -0.185 -0.010\nPayment Term -0.3411 0.020 -17.122 0.000 -0.380 -0.302\n----------------------------------------------------------------------------------------------\n Over_Due_Category=3 coef std err z P>|z| [0.025 0.975]\n----------------------------------------------------------------------------------------------\nZIPCODE -0.8512 0.358 -2.377 0.017 -1.553 -0.149\nPayment Method description -6.1751 0.584 -10.566 0.000 -7.321 -5.030\nInvoice_due_weekend -0.6619 0.249 -2.659 0.008 -1.150 -0.174\nAge Of Customer(Months) 0.7050 0.074 9.472 0.000 0.559 0.851\nAmount 0.0131 0.016 0.826 0.409 -0.018 0.044\nPayment Term -0.3588 0.022 -16.245 0.000 -0.402 -0.316\n==============================================================================================\n"
]
],
[
[
"# Modeling",
"_____no_output_____"
]
],
[
[
"### Split the Train Test datasets\ntarget_var = model_data['Over_Due_Category'].copy()\nindenpend_var = model_data.drop('Over_Due_Category',axis=1).copy()\nx_train,x_test,y_train,y_test = train_test_split(indenpend_var,target_var,test_size=0.2,random_state=666)",
"_____no_output_____"
]
],
[
[
"**Logistic Regression Multinominal**",
"_____no_output_____"
]
],
[
[
"parameter_dict_logistic ={\n 'penalty':['l1','l2','elasticnet'],\n 'C':[0.1,0.3,0.5,0.7,0.9,1.0],\n 'solver':['newton-cg','sag','saga','lbfgs'],\n}\n\nlogistic_model = LogisticRegression()\n\ngrid_logistic = GridSearchCV(estimator=logistic_model,param_grid=parameter_dict_logistic,scoring='roc_auc_ovr_weighted',cv=5,n_jobs=-1)\n\ngrid_logistic.fit(x_train,y_train)\n\nprint(grid_logistic.best_params_)\nprint('the best roc_auc_weighted score is', grid_logistic.best_score_)",
"{'C': 0.7, 'penalty': 'l2', 'solver': 'lbfgs'}\nthe best roc_auc_weighted score is 0.706559351107521\n"
]
],
[
[
"**KNN Classifier**",
"_____no_output_____"
]
],
[
[
"parameter_KNN = {\n 'n_neighbors':[5,10,15,20,25,30,35,40,45,50,55,60],\n 'weights':['uniform','distance'],\n 'p':[1,2]\n}\n\nKNN_Classifier = KNeighborsClassifier()\n\ngrid_KNN = GridSearchCV(estimator=KNN_Classifier,param_grid=parameter_KNN,scoring='roc_auc_ovr_weighted',cv=5,n_jobs=-1)\n\ngrid_KNN.fit(x_train,y_train)\n\nprint(grid_KNN.best_params_)\nprint('the best roc_auc_weighted score is', grid_KNN.best_score_)",
"{'n_neighbors': 60, 'p': 1, 'weights': 'uniform'}\nthe best roc_auc_weighted score is 0.801720095159556\n"
]
],
[
[
"**Decision Tree**",
"_____no_output_____"
]
],
[
[
"parameter_tree = {\n 'criterion':['gini','entropy'],\n 'splitter':['best','random'],\n 'max_depth':[None,10,20,30,40,50,60],\n 'min_samples_split':[2,4,6,8,10,20,30]\n}\n\nTree_Classifier = DecisionTreeClassifier()\n\ngrid_tree = GridSearchCV(estimator=Tree_Classifier,param_grid=parameter_tree,scoring='roc_auc_ovr_weighted',cv=5,n_jobs=-1)\n\ngrid_tree.fit(x_train,y_train)\n\nprint(grid_tree.best_params_)\nprint('the best roc_auc_weighted score is', grid_tree.best_score_)",
"{'criterion': 'entropy', 'max_depth': 10, 'min_samples_split': 30, 'splitter': 'random'}\nthe best roc_auc_weighted score is 0.8007284133645829\n"
]
],
[
[
"**AUC_ROC Score**\n\n> - If you wanna plot the auc roc curve, it gonna be one for each level each model. I think score is good enough. ",
"_____no_output_____"
]
],
[
[
"classifier = [\n LogisticRegression(penalty='l2',solver='lbfgs',C=0.7),\n KNeighborsClassifier(n_neighbors=60,p=1,weights='uniform'),\n DecisionTreeClassifier(criterion='entropy',max_depth=10,min_samples_split=30,splitter='random'),\n GradientBoostingClassifier()\n]\n\nresult_table = pd.DataFrame(columns=['classifiers', 'AUC_ROC_SCORE'])\n\nfor cls in classifier:\n model = cls.fit(x_train,y_train)\n yprob = model.predict_proba(x_test)\n roc_auc = roc_auc_score(y_test,yprob,average='weighted',multi_class='ovr')\n result_table = result_table.append({\n 'classifiers':cls.__class__.__name__,\n 'AUC_ROC_SCORE':roc_auc\n },ignore_index=True)\n\n\nresult_table.sort_values('AUC_ROC_SCORE',inplace=True,ascending=False)\nresult_table.reset_index(inplace=True)\nresult_table.drop('index',axis=1,inplace=True)\n\nresult_table",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecee68a0186108364add8cdac4c81b9bb20bb3c4 | 2,100 | ipynb | Jupyter Notebook | OpenJudge/Zipper.ipynb | PKUfudawei/Data-Structure-Algorithm | 55d6d1764e46197f661163beffd0cec51c1aecd2 | [
"MIT"
] | null | null | null | OpenJudge/Zipper.ipynb | PKUfudawei/Data-Structure-Algorithm | 55d6d1764e46197f661163beffd0cec51c1aecd2 | [
"MIT"
] | null | null | null | OpenJudge/Zipper.ipynb | PKUfudawei/Data-Structure-Algorithm | 55d6d1764e46197f661163beffd0cec51c1aecd2 | [
"MIT"
] | null | null | null | 23.863636 | 82 | 0.420476 | [
[
[
"def zipper(s1,s2,s3):\n visited=[[False for _ in range(len(s2)+1)] for _ in range(len(s1)+1)]\n def dfs(a,b,c):\n if c>=len(s3):\n return True\n elif visited[a][b]==True:\n return False\n visited[a][b]=True\n if a<len(s1):\n if s3[c]==s1[a]:\n if dfs(a+1,b,c+1):\n return True\n if b<len(s2):\n if s3[c]==s2[b]:\n if dfs(a,b+1,c+1):\n return True\n \n return False\n \n return dfs(0,0,0)\n \n \ndef driver():\n n=int(input())\n for i in range(n):\n s1,s2,s3=input().split()\n print(f\"Data set {i+1}: \", end='')\n if zipper(s1,s2,s3):\n print('yes')\n else:\n print('no')\n \ndriver()",
"Data set 1: [[True, False], [False, False]]\n[[True, False], [True, False]]\nyes\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
ecee68c2b6ed15d10afcacb763cbf4f451051f6a | 4,967 | ipynb | Jupyter Notebook | programs/Understanding-Keras/model_or_weights/load_model_pre.ipynb | NCC-AI/Study | 44c6dcf62be4b91fd3b6b07b156b111f6f17402b | [
"Apache-2.0"
] | 1 | 2019-06-09T00:02:37.000Z | 2019-06-09T00:02:37.000Z | programs/Understanding-Keras/model_or_weights/load_model_pre.ipynb | NCC-AI/Study | 44c6dcf62be4b91fd3b6b07b156b111f6f17402b | [
"Apache-2.0"
] | null | null | null | programs/Understanding-Keras/model_or_weights/load_model_pre.ipynb | NCC-AI/Study | 44c6dcf62be4b91fd3b6b07b156b111f6f17402b | [
"Apache-2.0"
] | null | null | null | 31.04375 | 148 | 0.547614 | [
[
[
"import keras\nfrom keras.callbacks import ModelCheckpoint\nfrom keras.datasets import mnist\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Flatten\nfrom keras.layers import Conv2D, MaxPooling2D\nfrom keras import backend as K",
"Using TensorFlow backend.\n"
],
[
"batch_size = 128\nnum_classes = 10\nepochs = 5\n\n# the data, split between train and test sets\n(x_train, y_train), (x_test, y_test) = mnist.load_data()\n\nx_train = x_train.reshape(60000, 784)\nx_test = x_test.reshape(10000, 784)\nx_train = x_train.astype('float16')\nx_test = x_test.astype('float16')\nx_train /= 255\nx_test /= 255\n\n# convert class vectors to binary class matrices\ny_train = keras.utils.to_categorical(y_train, num_classes)\ny_test = keras.utils.to_categorical(y_test, num_classes)",
"_____no_output_____"
],
[
"def MyLoss(y_true, y_pred):\n return K.categorical_crossentropy(y_true, y_pred)\n\ndef MyMetrics(y_true, y_pred):\n return K.cast(K.equal(K.argmax(y_true, axis=-1),\n K.argmax(y_pred, axis=-1)),\n K.floatx())",
"_____no_output_____"
],
[
"model = Sequential()\nmodel.add(Dense(128, activation='relu', input_shape=(784,)))\nmodel.add(Dense(128, activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(Dense(num_classes, activation='softmax'))\n\nmodel.compile(loss=MyLoss,\n optimizer=keras.optimizers.Adadelta(),\n metrics=[MyMetrics])\n\ncallbacks = [ModelCheckpoint('b128_weights_f16.h5', monitor='val_loss', save_best_only=True, verbose=1, save_weights_only=True)]\n\nmodel.fit(x_train, y_train,\n batch_size=batch_size,\n epochs=epochs,\n verbose=1,\n validation_data=(x_test, y_test),\n callbacks=callbacks)",
"Train on 60000 samples, validate on 10000 samples\nEpoch 1/5\n60000/60000 [==============================] - 5s 84us/step - loss: 0.4161 - MyMetrics: 0.8760 - val_loss: 0.1618 - val_MyMetrics: 0.9511\n\nEpoch 00001: val_loss improved from inf to 0.16180, saving model to b128_weights_f16.h5\nEpoch 2/5\n60000/60000 [==============================] - 3s 44us/step - loss: 0.1857 - MyMetrics: 0.9459 - val_loss: 0.1180 - val_MyMetrics: 0.9630\n\nEpoch 00002: val_loss improved from 0.16180 to 0.11802, saving model to b128_weights_f16.h5\nEpoch 3/5\n60000/60000 [==============================] - 3s 44us/step - loss: 0.1366 - MyMetrics: 0.9601 - val_loss: 0.0995 - val_MyMetrics: 0.9684\n\nEpoch 00003: val_loss improved from 0.11802 to 0.09949, saving model to b128_weights_f16.h5\nEpoch 4/5\n60000/60000 [==============================] - 3s 43us/step - loss: 0.1103 - MyMetrics: 0.9676 - val_loss: 0.0863 - val_MyMetrics: 0.9728\n\nEpoch 00004: val_loss improved from 0.09949 to 0.08628, saving model to b128_weights_f16.h5\nEpoch 5/5\n60000/60000 [==============================] - 3s 44us/step - loss: 0.0906 - MyMetrics: 0.9737 - val_loss: 0.0792 - val_MyMetrics: 0.9757\n\nEpoch 00005: val_loss improved from 0.08628 to 0.07917, saving model to b128_weights_f16.h5\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
ecee744db36396044a446f99ba623c161f27e5a3 | 26,317 | ipynb | Jupyter Notebook | week3_model_free/seminar_alternative/seminar.ipynb | legendawes/Practical_RL | a8fc62f93a7ad271b3243cf50166552b74542962 | [
"MIT"
] | null | null | null | week3_model_free/seminar_alternative/seminar.ipynb | legendawes/Practical_RL | a8fc62f93a7ad271b3243cf50166552b74542962 | [
"MIT"
] | null | null | null | week3_model_free/seminar_alternative/seminar.ipynb | legendawes/Practical_RL | a8fc62f93a7ad271b3243cf50166552b74542962 | [
"MIT"
] | null | null | null | 93.322695 | 12,524 | 0.839875 | [
[
[
"## Alternative assignment\n\nHere you can find an alternative assignment notebook which does not require py2 or a physical screen.\n\nFor starters, please go to __qlearning.py__ file in the current folder and implement q-learning agent by following instructions in the file.",
"_____no_output_____"
]
],
[
[
"#XVFB will be launched if you run on a server\nimport os\nif type(os.environ.get(\"DISPLAY\")) is not str or len(os.environ.get(\"DISPLAY\"))==0:\n !bash ../xvfb start\n %env DISPLAY=:1\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom IPython.display import clear_output",
"_____no_output_____"
]
],
[
[
"### Demo on taxi\n\nHere we use the qlearning agent from before on taxi env from openai gym.\nYou will need to insert a few agent functions here.",
"_____no_output_____"
]
],
[
[
"import gym\nenv = gym.make(\"Taxi-v2\")\n\nn_actions = env.action_space.n",
"_____no_output_____"
],
[
"from qlearning import QLearningAgent\n\nagent = QLearningAgent(alpha=0.5,epsilon=0.1,discount=0.99,\n getLegalActions = lambda s: range(n_actions))",
"_____no_output_____"
],
[
"def play_and_train(env,agent,t_max=10**4):\n \"\"\"This function should \n - run a full game, actions given by agent.getAction(s)\n - train agent using agent.update(...) whenever possible\n - return total reward\"\"\"\n total_reward = 0.0\n s = env.reset()\n \n for t in range(t_max):\n a = <get agent to pick action given state s>\n \n next_s,r,done,_ = env.step(a)\n \n <train(update) agent for state s>\n \n s = next_s\n total_reward +=r\n if done:break\n \n return total_reward\n \n \n ",
"_____no_output_____"
],
[
"rewards = []\nfor i in range(1000):\n rewards.append(play_and_train(env,agent))\n \n agent.epsilon *= 0.999\n \n if i %100 ==0:\n clear_output(True)\n print agent.epsilon\n plt.plot(rewards)\n plt.show()\n ",
"0.0405980235923\n"
]
],
[
[
"# Main assignment\n\nUse agent to train efficiently on CartPole-v0\n\nThis environment has a continuous number of states, so you will have to group them into bins somehow.\n\nThe simplest way is to use `round(x,n_digits)` (or numpy round) to round real number to a given amount of digits.\n\nThe tricky part is to get the n_digits right for each state to train effectively.\n\nNote that you don't need to convert state to integers, but to __tuples__ of any kind of values.",
"_____no_output_____"
]
],
[
[
"#run xvfb and set %env DISPLAY=:1 if in binder or on a server\nenv = gym.make(\"CartPole-v0\")\nn_actions = env.action_space.n\n\nprint(\"first state:%s\"%(env.reset()))\nplt.imshow(env.render('rgb_array'))",
"[2017-02-06 23:09:05,557] Making new env: CartPole-v0\n"
],
[
"<your code below>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecee8d926903237493132b55716994600f194b18 | 51,766 | ipynb | Jupyter Notebook | sentiment-rnn/Sentiment_RNN_Exercise.ipynb | ahs8w/udacity_pytorch | 0f68bbfb0057005dc9535e16d7a1c4b86b580b12 | [
"MIT"
] | null | null | null | sentiment-rnn/Sentiment_RNN_Exercise.ipynb | ahs8w/udacity_pytorch | 0f68bbfb0057005dc9535e16d7a1c4b86b580b12 | [
"MIT"
] | null | null | null | sentiment-rnn/Sentiment_RNN_Exercise.ipynb | ahs8w/udacity_pytorch | 0f68bbfb0057005dc9535e16d7a1c4b86b580b12 | [
"MIT"
] | null | null | null | 37.376173 | 843 | 0.550265 | [
[
[
"# Sentiment Analysis with an RNN\n\nIn this notebook, you'll implement a recurrent neural network that performs sentiment analysis. \n>Using an RNN rather than a strictly feedforward network is more accurate since we can include information about the *sequence* of words. \n\nHere we'll use a dataset of movie reviews, accompanied by sentiment labels: positive or negative.\n\n<img src=\"assets/reviews_ex.png\" width=40%>\n\n### Network Architecture\n\nThe architecture for this network is shown below.\n\n<img src=\"assets/network_diagram.png\" width=40%>\n\n>**First, we'll pass in words to an embedding layer.** We need an embedding layer because we have tens of thousands of words, so we'll need a more efficient representation for our input data than one-hot encoded vectors. You should have seen this before from the Word2Vec lesson. You can actually train an embedding with the Skip-gram Word2Vec model and use those embeddings as input, here. However, it's good enough to just have an embedding layer and let the network learn a different embedding table on its own. *In this case, the embedding layer is for dimensionality reduction, rather than for learning semantic representations.*\n\n>**After input words are passed to an embedding layer, the new embeddings will be passed to LSTM cells.** The LSTM cells will add *recurrent* connections to the network and give us the ability to include information about the *sequence* of words in the movie review data. \n\n>**Finally, the LSTM outputs will go to a sigmoid output layer.** We're using a sigmoid function because positive and negative = 1 and 0, respectively, and a sigmoid will output predicted, sentiment values between 0-1. \n\nWe don't care about the sigmoid outputs except for the **very last one**; we can ignore the rest. We'll calculate the loss by comparing the output at the last time step and the training label (pos or neg).",
"_____no_output_____"
],
[
"---\n### Load in and visualize the data",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# read data from text files\nwith open('data/reviews.txt', 'r') as f:\n reviews = f.read()\nwith open('data/labels.txt', 'r') as f:\n labels = f.read()",
"_____no_output_____"
],
[
"print(reviews[:2000])\nprint(\"\\n\")\nprint(labels[:20])",
"bromwell high is a cartoon comedy . it ran at the same time as some other programs about school life such as teachers . my years in the teaching profession lead me to believe that bromwell high s satire is much closer to reality than is teachers . the scramble to survive financially the insightful students who can see right through their pathetic teachers pomp the pettiness of the whole situation all remind me of the schools i knew and their students . when i saw the episode in which a student repeatedly tried to burn down the school i immediately recalled . . . . . . . . . at . . . . . . . . . . high . a classic line inspector i m here to sack one of your teachers . student welcome to bromwell high . i expect that many adults of my age think that bromwell high is far fetched . what a pity that it isn t \nstory of a man who has unnatural feelings for a pig . starts out with a opening scene that is a terrific example of absurd comedy . a formal orchestra audience is turned into an insane violent mob by the crazy chantings of it s singers . unfortunately it stays absurd the whole time with no general narrative eventually making it just too off putting . even those from the era should be turned off . the cryptic dialogue would make shakespeare seem easy to a third grader . on a technical level it s better than you might think with some good cinematography by future great vilmos zsigmond . future stars sally kirkland and frederic forrest can be seen briefly . \nhomelessness or houselessness as george carlin stated has been an issue for years but never a plan to help those on the street that were once considered human who did everything from going to school work or vote for the matter . most people think of the homeless as just a lost cause while worrying about things such as racism the war on iraq pressuring kids to succeed technology the elections inflation or worrying if they ll be next to end up on the streets . br br but what if y\n\n\npositive\nnegative\npo\n"
]
],
[
[
"## Data pre-processing\n\nThe first step when building a neural network model is getting your data into the proper form to feed into the network. Since we're using embedding layers, we'll need to encode each word with an integer. We'll also want to clean it up a bit.\n\nYou can see an example of the reviews data above. Here are the processing steps, we'll want to take:\n>* We'll want to get rid of periods and extraneous punctuation.\n* Also, you might notice that the reviews are delimited with newline characters `\\n`. To deal with those, I'm going to split the text into each review using `\\n` as the delimiter. \n* Then I can combined all the reviews back together into one big string.\n\nFirst, let's remove all punctuation. Then get all the text without the newlines and split it into individual words.",
"_____no_output_____"
]
],
[
[
"from string import punctuation\n\nprint(punctuation)\n\n# get rid of punctuation\nreviews = reviews.lower() # lowercase, standardize\nall_text = ''.join([c for c in reviews if c not in punctuation])",
"!\"#$%&'()*+,-./:;<=>?@[\\]^_`{|}~\n"
],
[
"# split by new lines and spaces\nreviews_split = all_text.split('\\n')\nall_text = ' '.join(reviews_split)\n\n# create a list of words\nwords = all_text.split()",
"_____no_output_____"
],
[
"words[:30]",
"_____no_output_____"
]
],
[
[
"### Encoding the words\n\nThe embedding lookup requires that we pass in integers to our network. The easiest way to do this is to create dictionaries that map the words in the vocabulary to integers. Then we can convert each of our reviews into integers so they can be passed into the network.\n\n> **Exercise:** Now you're going to encode the words with integers. Build a dictionary that maps words to integers. Later we're going to pad our input vectors with zeros, so make sure the integers **start at 1, not 0**.\n> Also, convert the reviews to integers and store the reviews in a new list called `reviews_ints`. ",
"_____no_output_____"
]
],
[
[
"count = Counter(words)\nvocab = sorted(count, key=count.get, reverse=True)\nvocab_to_int = {word: i for i,word in enumerate(vocab, 1)}",
"_____no_output_____"
],
[
"# feel free to use this import \nfrom collections import Counter\n\nuniq_words = Counter(words).most_common()\n## Build a dictionary that maps words to integers\nvocab_to_int = {word[0]: i for i,word in enumerate(uniq_words, 1)}",
"_____no_output_____"
],
[
"## use the dict to tokenize each review in reviews_split\n## store the tokenized reviews in reviews_ints\nreviews_ints = [[vocab_to_int[word] for word in rev.split()] for rev in reviews_split]",
"_____no_output_____"
]
],
[
[
"**Test your code**\n\nAs a text that you've implemented the dictionary correctly, print out the number of unique words in your vocabulary and the contents of the first, tokenized review.",
"_____no_output_____"
]
],
[
[
"# stats about vocabulary\nprint('Unique words: ', len((vocab_to_int))) # should ~ 74000+\nprint(\"\\n\")\n\n# print tokens in first review\nprint(\"Tokenized review: \\n\", reviews_ints[:1])",
"Unique words: 74072\n\n\nTokenized review: \n [[21025, 308, 6, 3, 1050, 207, 8, 2138, 32, 1, 171, 57, 15, 49, 81, 5785, 44, 382, 110, 140, 15, 5194, 60, 154, 9, 1, 4975, 5852, 475, 71, 5, 260, 12, 21025, 308, 13, 1978, 6, 74, 2395, 5, 613, 73, 6, 5194, 1, 24103, 5, 1983, 10166, 1, 5786, 1499, 36, 51, 66, 204, 145, 67, 1199, 5194, 19869, 1, 37442, 4, 1, 221, 883, 31, 2988, 71, 4, 1, 5787, 10, 686, 2, 67, 1499, 54, 10, 216, 1, 383, 9, 62, 3, 1406, 3686, 783, 5, 3483, 180, 1, 382, 10, 1212, 13583, 32, 308, 3, 349, 341, 2913, 10, 143, 127, 5, 7690, 30, 4, 129, 5194, 1406, 2326, 5, 21025, 308, 10, 528, 12, 109, 1448, 4, 60, 543, 102, 12, 21025, 308, 6, 227, 4146, 48, 3, 2211, 12, 8, 215, 23]]\n"
]
],
[
[
"### Encoding the labels\n\nOur labels are \"positive\" or \"negative\". To use these labels in our network, we need to convert them to 0 and 1.\n\n> **Exercise:** Convert labels from `positive` and `negative` to 1 and 0, respectively, and place those in a new list, `encoded_labels`.",
"_____no_output_____"
]
],
[
[
"labels_split = labels.split('\\n')\n\n# 1=positive, 0=negative label conversion\nencoded_labels = np.array([1 if label=='positive' else 0 for label in labels_split])\nencoded_labels[:5]",
"_____no_output_____"
]
],
[
[
"### Removing Outliers\n\nAs an additional pre-processing step, we want to make sure that our reviews are in good shape for standard processing. That is, our network will expect a standard input text size, and so, we'll want to shape our reviews into a specific length. We'll approach this task in two main steps:\n\n1. Getting rid of extremely long or short reviews; the outliers\n2. Padding/truncating the remaining data so that we have reviews of the same length.\n\n<img src=\"assets/outliers_padding_ex.png\" width=40%>\n\nBefore we pad our review text, we should check for reviews of extremely short or long lengths; outliers that may mess with our training.",
"_____no_output_____"
]
],
[
[
"# outlier review stats\nreview_lens = Counter([len(x) for x in reviews_ints])\nprint(\"Zero-length reviews: {}\".format(review_lens[0]))\nprint(\"Maximum review length: {}\".format(max(review_lens)))",
"Zero-length reviews: 1\nMaximum review length: 2514\n"
]
],
[
[
"Okay, a couple issues here. We seem to have one review with zero length. And, the maximum review length is way too many steps for our RNN. We'll have to remove any super short reviews and truncate super long reviews. This removes outliers and should allow our model to train more efficiently.\n\n> **Exercise:** First, remove *any* reviews with zero length from the `reviews_ints` list and their corresponding label in `encoded_labels`.",
"_____no_output_____"
]
],
[
[
"idx = [ii for ii,review in enumerate(reviews_ints) if len(review) == 0]",
"_____no_output_____"
],
[
"print('Number of reviews before removing outliers: ', len(reviews_ints))\n\n## remove any reviews/labels with zero length from the reviews_ints list.\nidx = [ii for ii,review in enumerate(reviews_ints) if len(review) == 0][0]\ndel reviews_ints[idx]\nencoded_labels = np.delete(encoded_labels, idx)\n\nprint('Number of reviews after removing outliers: ', len(reviews_ints))",
"Number of reviews before removing outliers: 25001\nNumber of reviews after removing outliers: 25000\n"
]
],
[
[
"---\n## Padding sequences\n\nTo deal with both short and very long reviews, we'll pad or truncate all our reviews to a specific length. For reviews shorter than some `seq_length`, we'll pad with 0s. For reviews longer than `seq_length`, we can truncate them to the first `seq_length` words. A good `seq_length`, in this case, is 200.\n\n> **Exercise:** Define a function that returns an array `features` that contains the padded data, of a standard size, that we'll pass to the network. \n* The data should come from `review_ints`, since we want to feed integers to the network. \n* Each row should be `seq_length` elements long. \n* For reviews shorter than `seq_length` words, **left pad** with 0s. That is, if the review is `['best', 'movie', 'ever']`, `[117, 18, 128]` as integers, the row will look like `[0, 0, 0, ..., 0, 117, 18, 128]`. \n* For reviews longer than `seq_length`, use only the first `seq_length` words as the feature vector.\n\nAs a small example, if the `seq_length=10` and an input review is: \n```\n[117, 18, 128]\n```\nThe resultant, padded sequence should be: \n\n```\n[0, 0, 0, 0, 0, 0, 0, 117, 18, 128]\n```\n\n**Your final `features` array should be a 2D array, with as many rows as there are reviews, and as many columns as the specified `seq_length`.**\n\nThis isn't trivial and there are a bunch of ways to do this. But, if you're going to be building your own deep learning networks, you're going to have to get used to preparing your data.",
"_____no_output_____"
]
],
[
[
"r = reviews_ints[0]\nl = len(r)\nr = np.pad(r, (200-l,0), 'constant', constant_values=0)\nr",
"_____no_output_____"
],
[
"def pad_features(reviews_ints, seq_length):\n ''' Return features of review_ints, where each review is padded with 0's \n or truncated to the input seq_length.\n '''\n ## implement function\n \n features=[]\n for review in reviews_ints:\n l = len(review)\n if l < seq_length:\n review = np.pad(review, (seq_length-l,0), 'constant', constant_values=0) \n features.append(review[:seq_length])\n \n return np.stack(features)",
"_____no_output_____"
],
[
"# Test your implementation!\n\nseq_length = 200\n\nfeatures = pad_features(reviews_ints, seq_length=seq_length)\n\n## test statements - do not change - ##\nassert len(features)==len(reviews_ints), \"Your features should have as many rows as reviews.\"\nassert len(features[0])==seq_length, \"Each feature row should contain seq_length values.\"\n\n# print first 10 values of the first 30 batches \nprint(features[:30,:10])",
"[[ 0 0 0 0 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]\n [22382 42 46418 15 706 17139 3389 47 77 35]\n [ 4505 505 15 3 3342 162 8312 1652 6 4819]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 54 10 14 116 60 798 552 71 364 5]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 1 330 578 34 3 162 748 2731 9 325]\n [ 9 11 10171 5305 1946 689 444 22 280 673]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 1 307 10399 2069 1565 6202 6528 3288 17946 10628]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 21 122 2069 1565 515 8181 88 6 1325 1182]\n [ 1 20 6 76 40 6 58 81 95 5]\n [ 54 10 84 329 26230 46427 63 10 14 614]\n [ 11 20 6 30 1436 32317 3769 690 15100 6]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 40 26 109 17952 1422 9 1 327 4 125]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 10 499 1 307 10399 55 74 8 13 30]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]]\n"
]
],
[
[
"## Training, Validation, Test\n\nWith our data in nice shape, we'll split it into training, validation, and test sets.\n\n> **Exercise:** Create the training, validation, and test sets. \n* You'll need to create sets for the features and the labels, `train_x` and `train_y`, for example. \n* Define a split fraction, `split_frac` as the fraction of data to **keep** in the training set. Usually this is set to 0.8 or 0.9. \n* Whatever data is left will be split in half to create the validation and *testing* data.",
"_____no_output_____"
]
],
[
[
"split_frac = 0.8\nsplit_idx = int(len(features)*split_frac)\ntrain_x, remaining_x = features[:split_idx], features[split_idx:]\ntrain_y, remaining_y = encoded_labels[:split_idx], encoded_labels[split_idx:]\n\ntest_idx = int(len(remaining_x)*0.5)\nval_x, test_x = remaining_x[:test_idx], remaining_x[test_idx:]\nval_y, test_y = remaining_y[:test_idx], remaining_y[test_idx:]\n## split data into training, validation, and test data (features and labels, x and y)\n# mask = np.random.rand(len(features))>split_frac\n\n## print out the shapes of your resultant feature data\nlen(train_x), len(val_x), len(test_x)",
"_____no_output_____"
]
],
[
[
"**Check your work**\n\nWith train, validation, and test fractions equal to 0.8, 0.1, 0.1, respectively, the final, feature data shapes should look like:\n```\n Feature Shapes:\nTrain set: \t\t (20000, 200) \nValidation set: \t(2500, 200) \nTest set: \t\t (2500, 200)\n```",
"_____no_output_____"
],
[
"---\n## DataLoaders and Batching\n\nAfter creating training, test, and validation data, we can create DataLoaders for this data by following two steps:\n1. Create a known format for accessing our data, using [TensorDataset](https://pytorch.org/docs/stable/data.html#) which takes in an input set of data and a target set of data with the same first dimension, and creates a dataset.\n2. Create DataLoaders and batch our training, validation, and test Tensor datasets.\n\n```\ntrain_data = TensorDataset(torch.from_numpy(train_x), torch.from_numpy(train_y))\ntrain_loader = DataLoader(train_data, batch_size=batch_size)\n```\n\nThis is an alternative to creating a generator function for batching our data into full batches.",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torch.utils.data import TensorDataset, DataLoader\n\n# create Tensor datasets\ntrain_data = TensorDataset(torch.from_numpy(train_x), torch.from_numpy(train_y))\nvalid_data = TensorDataset(torch.from_numpy(val_x), torch.from_numpy(val_y))\ntest_data = TensorDataset(torch.from_numpy(test_x), torch.from_numpy(test_y))\n\n# dataloaders\nbatch_size = 50\n\n# make sure to SHUFFLE your data\ntrain_loader = DataLoader(train_data, shuffle=True, batch_size=batch_size)\nvalid_loader = DataLoader(valid_data, shuffle=True, batch_size=batch_size)\ntest_loader = DataLoader(test_data, shuffle=True, batch_size=batch_size)",
"_____no_output_____"
],
[
"# obtain one batch of training data\ndataiter = iter(train_loader)\nsample_x, sample_y = dataiter.next()\n\nprint('Sample input size: ', sample_x.size()) # batch_size, seq_length\nprint('Sample input: \\n', sample_x)\nprint()\nprint('Sample label size: ', sample_y.size()) # batch_size\nprint('Sample label: \\n', sample_y)",
"Sample input size: torch.Size([50, 200])\nSample input: \n tensor([[ 11, 6, 35, ..., 783, 5, 29],\n [ 1, 697, 3875, ..., 5, 3882, 7],\n [ 0, 0, 0, ..., 234, 148, 11],\n ...,\n [ 0, 0, 0, ..., 3, 45, 4],\n [ 1, 382, 4885, ..., 5, 28, 376],\n [ 101, 31, 21, ..., 3, 469, 24188]])\n\nSample label size: torch.Size([50])\nSample label: \n tensor([0, 0, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 1,\n 1, 0, 1, 0, 1, 1, 0, 0, 1, 0, 0, 1, 0, 1, 1, 1, 0, 1, 0, 0, 1, 0, 1, 1,\n 0, 1])\n"
]
],
[
[
"---\n# Sentiment Network with PyTorch\n\nBelow is where you'll define the network.\n\n<img src=\"assets/network_diagram.png\" width=40%>\n\nThe layers are as follows:\n1. An [embedding layer](https://pytorch.org/docs/stable/nn.html#embedding) that converts our word tokens (integers) into embeddings of a specific size.\n2. An [LSTM layer](https://pytorch.org/docs/stable/nn.html#lstm) defined by a hidden_state size and number of layers\n3. A fully-connected output layer that maps the LSTM layer outputs to a desired output_size\n4. A sigmoid activation layer which turns all outputs into a value 0-1; return **only the last sigmoid output** as the output of this network.\n\n### The Embedding Layer\n\nWe need to add an [embedding layer](https://pytorch.org/docs/stable/nn.html#embedding) because there are 74000+ words in our vocabulary. It is massively inefficient to one-hot encode that many classes. So, instead of one-hot encoding, we can have an embedding layer and use that layer as a lookup table. You could train an embedding layer using Word2Vec, then load it here. But, it's fine to just make a new layer, using it for only dimensionality reduction, and let the network learn the weights.\n\n\n### The LSTM Layer(s)\n\nWe'll create an [LSTM](https://pytorch.org/docs/stable/nn.html#lstm) to use in our recurrent network, which takes in an input_size, a hidden_dim, a number of layers, a dropout probability (for dropout between multiple layers), and a batch_first parameter.\n\nMost of the time, you're network will have better performance with more layers; between 2-3. Adding more layers allows the network to learn really complex relationships. \n\n> **Exercise:** Complete the `__init__`, `forward`, and `init_hidden` functions for the SentimentRNN model class.\n\nNote: `init_hidden` should initialize the hidden and cell state of an lstm layer to all zeros, and move those state to GPU, if available.",
"_____no_output_____"
]
],
[
[
"# First checking if GPU is available\ntrain_on_gpu=torch.cuda.is_available()\n\nif(train_on_gpu):\n print('Training on GPU.')\nelse:\n print('No GPU available, training on CPU.')",
"Training on GPU.\n"
],
[
"import torch.nn as nn",
"_____no_output_____"
],
[
"class SentimentRNN(nn.Module):\n \"\"\"\n The RNN model that will be used to perform Sentiment analysis.\n \"\"\"\n\n def __init__(self, vocab_size, output_size, embedding_dim, hidden_dim, n_layers, drop_prob=0.5):\n \"\"\"\n Initialize the model by setting up the layers.\n \"\"\"\n super(SentimentRNN, self).__init__()\n\n self.output_size = output_size\n self.n_layers = n_layers\n self.hidden_dim = hidden_dim\n \n # define all layers\n self.embed = nn.Embedding(vocab_size, embedding_dim)\n self.lstm = nn.LSTM(embedding_dim, hidden_dim, n_layers, dropout=drop_prob, batch_first=True)\n self.drop = nn.Dropout(drop_prob)\n self.fc = nn.Linear(hidden_dim, output_size)\n\n def forward(self, x, hidden):\n \"\"\"\n Perform a forward pass of our model on some input and hidden state.\n \"\"\"\n out,hidden = self.lstm(self.embed(x), hidden)\n sig_out = torch.sigmoid(self.fc(self.drop(out)))\n # return last sigmoid output and hidden state\n return sig_out[:,-1], hidden\n \n \n def init_hidden(self, batch_size):\n ''' Initializes hidden state '''\n # Create two new tensors with sizes n_layers x batch_size x hidden_dim,\n # initialized to zero, for hidden state and cell state of LSTM\n weight = next(self.parameters()).data \n if (train_on_gpu):\n hidden = (weight.new(self.n_layers, batch_size, self.hidden_dim).zero_().cuda(),\n weight.new(self.n_layers, batch_size, self.hidden_dim).zero_().cuda())\n else:\n hidden = (weight.new(self.n_layers, batch_size, self.hidden_dim).zero_(),\n weight.new(self.n_layers, batch_size, self.hidden_dim).zero_())\n\n return hidden",
"_____no_output_____"
]
],
[
[
"## Instantiate the network\n\nHere, we'll instantiate the network. First up, defining the hyperparameters.\n\n* `vocab_size`: Size of our vocabulary or the range of values for our input, word tokens.\n* `output_size`: Size of our desired output; the number of class scores we want to output (pos/neg).\n* `embedding_dim`: Number of columns in the embedding lookup table; size of our embeddings.\n* `hidden_dim`: Number of units in the hidden layers of our LSTM cells. Usually larger is better performance wise. Common values are 128, 256, 512, etc.\n* `n_layers`: Number of LSTM layers in the network. Typically between 1-3\n\n> **Exercise:** Define the model hyperparameters.\n",
"_____no_output_____"
]
],
[
[
"# Instantiate the model w/ hyperparams\nvocab_size = len(vocab_to_int)+1 # padding idx\noutput_size = 1\nembedding_dim = 128\nhidden_dim = 256\nn_layers = 2\n\nnet = SentimentRNN(vocab_size, output_size, embedding_dim, hidden_dim, n_layers)\n\nprint(net)",
"SentimentRNN(\n (embed): Embedding(74073, 400)\n (lstm): LSTM(400, 256, num_layers=2, batch_first=True, dropout=0.5)\n (drop): Dropout(p=0.5)\n (fc): Linear(in_features=256, out_features=1, bias=True)\n)\n"
]
],
[
[
"---\n## Training\n\nBelow is the typical training code. If you want to do this yourself, feel free to delete all this code and implement it yourself. You can also add code to save a model by name.\n\n>We'll also be using a new kind of cross entropy loss, which is designed to work with a single Sigmoid output. [BCELoss](https://pytorch.org/docs/stable/nn.html#bceloss), or **Binary Cross Entropy Loss**, applies cross entropy loss to a single value between 0 and 1.\n\nWe also have some data and training hyparameters:\n\n* `lr`: Learning rate for our optimizer.\n* `epochs`: Number of times to iterate through the training dataset.\n* `clip`: The maximum gradient value to clip at (to prevent exploding gradients).",
"_____no_output_____"
]
],
[
[
"# loss and optimization functions\nlr=0.001\n\ncriterion = nn.BCELoss()\noptimizer = torch.optim.Adam(net.parameters(), lr=lr)\n",
"_____no_output_____"
],
[
"# training params\n\nepochs = 4 # 3-4 is approx where I noticed the validation loss stop decreasing\n\ncounter = 0\nprint_every = 100\nclip=5 # gradient clipping\n\n# move model to GPU, if available\nif(train_on_gpu):\n net.cuda()\n\nnet.train()\n# train for some number of epochs\nfor e in range(epochs):\n # initialize hidden state\n h = net.init_hidden(batch_size)\n\n # batch loop\n for inputs, labels in train_loader:\n counter += 1\n\n if(train_on_gpu):\n inputs, labels = inputs.cuda(), labels.cuda()\n\n # Creating new variables for the hidden state, otherwise\n # we'd backprop through the entire training history\n h = tuple([each.data for each in h])\n\n # zero accumulated gradients\n net.zero_grad()\n\n # get the output from the model\n output, h = net(inputs, h)\n\n # calculate the loss and perform backprop\n loss = criterion(output.squeeze(), labels.float())\n loss.backward()\n # `clip_grad_norm` helps prevent the exploding gradient problem in RNNs / LSTMs.\n nn.utils.clip_grad_norm_(net.parameters(), clip)\n optimizer.step()\n\n # loss stats\n if counter % print_every == 0:\n # Get validation loss\n val_h = net.init_hidden(batch_size)\n val_losses = []\n net.eval()\n for inputs, labels in valid_loader:\n\n # Creating new variables for the hidden state, otherwise\n # we'd backprop through the entire training history\n val_h = tuple([each.data for each in val_h])\n\n if(train_on_gpu):\n inputs, labels = inputs.cuda(), labels.cuda()\n\n output, val_h = net(inputs, val_h)\n val_loss = criterion(output.squeeze(), labels.float())\n\n val_losses.append(val_loss.item())\n\n net.train()\n print(\"Epoch: {}/{}...\".format(e+1, epochs),\n \"Step: {}...\".format(counter),\n \"Loss: {:.6f}...\".format(loss.item()),\n \"Val Loss: {:.6f}\".format(np.mean(val_losses)))\n \n \n# Loss: 0.479497... Val Loss: 0.425451 embedding: 128, hidden: 256\n# Loss: 0.351858... Val Loss: 0.494416 \"\", reshape before/after fc\n# Loss: 0.257153... Val Loss: 0.504479 embedding: 400",
"Epoch: 1/4... Step: 100... Loss: 0.649368... Val Loss: 0.652598\nEpoch: 1/4... Step: 200... Loss: 0.592028... Val Loss: 0.644787\nEpoch: 1/4... Step: 300... Loss: 0.638039... Val Loss: 0.633251\nEpoch: 1/4... Step: 400... Loss: 0.550851... Val Loss: 0.604788\nEpoch: 2/4... Step: 500... Loss: 0.493080... Val Loss: 0.543076\nEpoch: 2/4... Step: 600... Loss: 0.438792... Val Loss: 0.537041\nEpoch: 2/4... Step: 700... Loss: 0.605888... Val Loss: 0.480132\nEpoch: 2/4... Step: 800... Loss: 0.490987... Val Loss: 0.456182\nEpoch: 3/4... Step: 900... Loss: 0.193564... Val Loss: 0.492796\nEpoch: 3/4... Step: 1000... Loss: 0.251907... Val Loss: 0.466576\nEpoch: 3/4... Step: 1100... Loss: 0.158107... Val Loss: 0.574199\nEpoch: 3/4... Step: 1200... Loss: 0.349664... Val Loss: 0.452875\nEpoch: 4/4... Step: 1300... Loss: 0.218790... Val Loss: 0.474196\nEpoch: 4/4... Step: 1400... Loss: 0.284669... Val Loss: 0.511473\nEpoch: 4/4... Step: 1500... Loss: 0.120573... Val Loss: 0.511209\nEpoch: 4/4... Step: 1600... Loss: 0.257153... Val Loss: 0.504479\n"
]
],
[
[
"---\n## Testing\n\nThere are a few ways to test your network.\n\n* **Test data performance:** First, we'll see how our trained model performs on all of our defined test_data, above. We'll calculate the average loss and accuracy over the test data.\n\n* **Inference on user-generated data:** Second, we'll see if we can input just one example review at a time (without a label), and see what the trained model predicts. Looking at new, user input data like this, and predicting an output label, is called **inference**.",
"_____no_output_____"
]
],
[
[
"# Get test data loss and accuracy\n\ntest_losses = [] # track loss\nnum_correct = 0\n\n# init hidden state\nh = net.init_hidden(batch_size)\n\nnet.eval()\n# iterate over test data\nfor inputs, labels in test_loader:\n\n # Creating new variables for the hidden state, otherwise\n # we'd backprop through the entire training history\n h = tuple([each.data for each in h])\n\n if(train_on_gpu):\n inputs, labels = inputs.cuda(), labels.cuda()\n \n # get predicted outputs\n output, h = net(inputs, h)\n \n # calculate loss\n test_loss = criterion(output.squeeze(), labels.float())\n test_losses.append(test_loss.item())\n \n # convert output probabilities to predicted class (0 or 1)\n pred = torch.round(output.squeeze()) # rounds to the nearest integer\n \n # compare predictions to true label\n correct_tensor = pred.eq(labels.float().view_as(pred))\n correct = np.squeeze(correct_tensor.numpy()) if not train_on_gpu else np.squeeze(correct_tensor.cpu().numpy())\n num_correct += np.sum(correct)\n\n\n# -- stats! -- ##\n# avg test loss\nprint(\"Test loss: {:.3f}\".format(np.mean(test_losses)))\n\n# accuracy over all test data\ntest_acc = num_correct/len(test_loader.dataset)\nprint(\"Test accuracy: {:.3f}\".format(test_acc))",
"Test loss: 0.520\nTest accuracy: 0.804\n"
]
],
[
[
"### Inference on a test review\n\nYou can change this test_review to any text that you want. Read it and think: is it pos or neg? Then see if your model predicts correctly!\n \n> **Exercise:** Write a `predict` function that takes in a trained net, a plain text_review, and a sequence length, and prints out a custom statement for a positive or negative review!\n* You can use any functions that you've already defined or define any helper functions you want to complete `predict`, but it should just take in a trained net, a text review, and a sequence length.\n",
"_____no_output_____"
]
],
[
[
"# negative test review\ntest_review_neg = 'The worst movie I have seen; acting was terrible and I want my money back. This movie had bad acting and the dialogue was slow.'\n",
"_____no_output_____"
],
[
"def tokenize_test(review):\n review = ''.join([c for c in review.lower() if c not in punctuation]) # lowercase, remove punctuation\n review_tok = [vocab_to_int[word] for word in review.split()]\n return review_tok",
"_____no_output_____"
],
[
"def predict(net, test_review, sequence_length=200):\n ''' Prints out whether a give review is predicted to be \n positive or negative in sentiment, using a trained model.\n \n params:\n net - A trained net \n test_review - a review made of normal text and punctuation\n sequence_length - the padded length of a review\n '''\n \n net.eval()\n \n h = net.init_hidden(1)\n tokens = tokenize_test(test_review)\n x = pad_features([tokens], sequence_length)\n x = torch.from_numpy(x)\n \n if(train_on_gpu): x = x.cuda()\n \n # get predicted outputs\n output, h = net(x, h)\n # print custom response based on whether test_review is pos/neg\n pred = torch.round(output.squeeze())\n res = 'positive' if pred else 'negative'\n print(f\"This review is {res}!\")",
"_____no_output_____"
],
[
"# positive test review\ntest_review_pos = 'This movie had the best acting and the dialogue was so good. I loved it.'\n",
"_____no_output_____"
],
[
"# call function\n# try negative and positive reviews!\nseq_length=200\npredict(net, test_review_pos, seq_length)",
"This review is positive!\n"
]
],
[
[
"### Try out test_reviews of your own!\n\nNow that you have a trained model and a predict function, you can pass in _any_ kind of text and this model will predict whether the text has a positive or negative sentiment. Push this model to its limits and try to find what words it associates with positive or negative.\n\nLater, you'll learn how to deploy a model like this to a production environment so that it can respond to any kind of user data put into a web app!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
eceeabba29bf144d717c5bb8fa1e590c5f0c34b3 | 280,754 | ipynb | Jupyter Notebook | RL/Monte Carlo/BlackJack_RL.ipynb | CodeLogist/ML-NewBie-s-Heaven | 01aa31cd31ba3b365d5152cee0514cabefca4784 | [
"MIT"
] | 1 | 2020-06-25T21:43:38.000Z | 2020-06-25T21:43:38.000Z | RL/Monte Carlo/BlackJack_RL.ipynb | CodeLogist/ML-NewBie-s-Heaven | 01aa31cd31ba3b365d5152cee0514cabefca4784 | [
"MIT"
] | null | null | null | RL/Monte Carlo/BlackJack_RL.ipynb | CodeLogist/ML-NewBie-s-Heaven | 01aa31cd31ba3b365d5152cee0514cabefca4784 | [
"MIT"
] | 1 | 2020-06-06T13:27:45.000Z | 2020-06-06T13:27:45.000Z | 1,132.072581 | 274,002 | 0.941771 | [
[
[
"import sys\nimport gym\nimport numpy as np\nfrom collections import defaultdict\n\nfrom plot_util import plot_blackjack_values,plot_policy",
"_____no_output_____"
],
[
"env=gym.make('Blackjack-v0')",
"_____no_output_____"
],
[
"print(env.observation_space)\nprint(env.action_space)",
"Tuple(Discrete(32), Discrete(11), Discrete(2))\nDiscrete(2)\n"
],
[
"for i_episode in range(3):\n state=env.reset()\n while True:\n print(state)\n action=env.action_space.sample()\n state,reward,done,info=env.step(action)\n if done:\n print('End Game! Reward: ',reward)\n print('You won :) \\n') if reward>0 else print('You lost:( \\n')\n break",
"(20, 10, False)\nEnd Game! Reward: -1.0\nYou lost:( \n\n(15, 10, False)\n(20, 10, False)\nEnd Game! Reward: -1\nYou lost:( \n\n(15, 4, False)\nEnd Game! Reward: 1.0\nYou won :) \n\n"
]
],
[
[
"##Monte Carlo Prediction\n",
"_____no_output_____"
]
],
[
[
"def generate_episode_from_limit_stochastic(env):\n episode=[]\n state=env.reset()\n while True:\n probs=[0.8, 0.2] if state[0]>18 else [0.2, 0.8]\n action=np.random.choice(np.arange(2),p=probs)\n next_state,reward,done,info=env.step(action)\n episode.append((state,action,reward))\n state=next_state\n if done:\n break\n return episode",
"_____no_output_____"
],
[
"for i in range(3):\n print(generate_episode_from_limit_stochastic(env))",
"[((15, 5, False), 1, 0), ((20, 5, False), 0, -1.0)]\n[((20, 10, False), 1, -1)]\n[((12, 10, True), 0, 1.0)]\n"
],
[
"def mc_prediction_q(env,num_episodes,generate_episode,gamma=1.0):\n returns_sum=defaultdict(lambda: np.zeros(env.action_space.n))\n N=defaultdict(lambda: np.zeros(env.action_space.n))\n Q=defaultdict(lambda: np.zeros(env.action_space.n))\n\n for i_episode in range(num_episodes):\n episode=generate_episode_from_limit_stochastic(env)\n states,actions,rewards=zip(*episode)\n discounts = np.array([gamma**i for i in range(len(rewards)+1)])\n for i,state in enumerate(states):\n returns_sum[state][actions[i]]+=sum(rewards[i:]*discounts[:-(1+i)])\n N[state][actions[i]]+=1.0\n Q[state][actions[i]]=returns_sum[state][actions[i]]/N[state][actions[i]]\n return Q",
"_____no_output_____"
],
[
"# obtain the action-value function\nQ = mc_prediction_q(env, 500000, generate_episode_from_limit_stochastic)\n\n# obtain the corresponding state-value function\nV_to_plot = dict((k,(k[0]>18)*(np.dot([0.8, 0.2],v)) + (k[0]<=18)*(np.dot([0.2, 0.8],v))) \\\n for k, v in Q.items())\n\n# plot the state-value function\nplot_blackjack_values(V_to_plot)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
eceead26dd2c525b3b4cccad7592b8e5909c39ea | 8,495 | ipynb | Jupyter Notebook | data-wrangling/.ipynb_checkpoints/extract_gene_sequences-checkpoint.ipynb | nextstrain/seasonal-cov | 9dee4e145d003b839729b86ff9c7b74bb8483e55 | [
"MIT"
] | 4 | 2020-03-24T21:54:12.000Z | 2020-03-26T19:12:05.000Z | data-wrangling/.ipynb_checkpoints/extract_gene_sequences-checkpoint.ipynb | blab/seasonal-cov-adaptive-evolution | 9dee4e145d003b839729b86ff9c7b74bb8483e55 | [
"MIT"
] | null | null | null | data-wrangling/.ipynb_checkpoints/extract_gene_sequences-checkpoint.ipynb | blab/seasonal-cov-adaptive-evolution | 9dee4e145d003b839729b86ff9c7b74bb8483e55 | [
"MIT"
] | null | null | null | 39.511628 | 156 | 0.524662 | [
[
[
"from Bio import SeqIO\nfrom Bio.SeqRecord import SeqRecord\nfrom Bio.SeqFeature import SeqFeature, FeatureLocation\nfrom Bio import AlignIO\nfrom collections import Counter ",
"_____no_output_____"
],
[
"#Gene positions for each virus\ndef get_virus_genes(virus):\n if virus == 'nl63':\n genes_dict = {'replicase1ab':\"replicase polyprotein 1ab\", 'spike':\"spike protein\", 'protein3':\"protein 3\", \n 'envelope':\"envelope protein\", 'membrane':\"membrane protein\", 'nucleocapsid':\"nucleocapsid protein\"}\n elif virus == '229e':\n genes_dict = {'replicase1ab':\"replicase polyprotein 1ab\", 'replicase1a': \"replicase polyprotein 1a\", 'spike':\"surface glycoprotein\", \n 'protein4a':\"4a protein\", 'protein4b':\"4b protein\",\n 'envelope':\"envelope protein\", 'membrane':\"membrane protein\", 'nucleocapsid':\"nucleocapsid protein\"}\n elif virus == 'hku1':\n genes_dict = {'replicase1ab':\"orf1ab polyprotein\", 'he':\"hemagglutinin-esterase glycoprotein\", \n 'spike':\"spike glycoprotein\", 'nonstructural4':\"non-structural protein\",\n 'envelope':\"small membrane protein\", 'membrane':\"membrane glycoprotein\", \n 'nucleocapsid':\"nucleocapsid phosphoprotein\", 'nucleocapsid2':\"nucleocapsid phosphoprotein 2\"}\n elif virus == 'oc43':\n genes_dict = {'replicase1ab':\"replicase polyprotein\", 'nonstructural2a':\"NS2a protein\",\n 'he':\"HE protein\", 'spike':\"S protein\", 'nonstructural2':\"NS2 protein\",\n 'envelope':\"NS3 protein\", 'membrane':\"M protein\", \n 'nucleocapsid':\"N protein\", 'n2protein':\"N2 protein\"}\n return genes_dict\n",
"_____no_output_____"
],
[
"#Gene positions for each virus\ndef get_gene_position(virus, gene, sequence):\n genes_dict = get_virus_genes(virus)\n \n for seq_record in SeqIO.parse(\"../../\"+str(virus)+\"/config/\"+str(virus)+\"_full_reference.gb\", \"genbank\"):\n for feature in seq_record.features:\n if feature.type == 'CDS':\n if feature.qualifiers['product'] == [genes_dict[gene]]:\n gene_nt = feature.location.extract(sequence)\n\n return gene_nt\n ",
"_____no_output_____"
],
[
"#From aligned .fasta, extract just the portion of the genome encoding each gene\ndef extract_genes(virus):\n \n aligned_fasta = \"../../\"+str(virus)+\"/results/aligned_\"+str(virus)+\"_full.fasta\"\n original_fasta = \"../../\"+str(virus)+\"/data/\"+str(virus)+\"_full.fasta\"\n \n genes_dict = get_virus_genes(virus)\n genes = [k for k,v in genes_dict.items()]\n \n for gene in genes:\n output_fasta = \"../../\"+str(virus)+\"/data/\"+str(virus)+\"_\"+str(gene)+\".fasta\"\n gene_sequences = {}\n with open(aligned_fasta, \"r\") as handle:\n alignment = SeqIO.parse(handle, \"fasta\")\n for aligned_record in alignment:\n gene_nt = get_gene_position(virus, gene, aligned_record.seq)\n gene_nt_str = str(gene_nt)\n #Throw out sequences that don't cover gene\n num_unaligned_gene = Counter(gene_nt_str)['N']\n if num_unaligned_gene < (len(gene_nt)/2):\n gene_sequences[aligned_record.id] = gene_nt\n \n gene_entries = []\n\n with open(original_fasta, \"r\") as handle_2:\n metadata = SeqIO.parse(handle_2, \"fasta\")\n for meta_record in metadata:\n strain_name = meta_record.id.split('|')[1]\n if str(strain_name) in gene_sequences.keys():\n gene_record = SeqRecord(gene_sequences[strain_name], id=meta_record.id, description=meta_record.id)\n gene_entries.append(gene_record)\n\n SeqIO.write(gene_entries, output_fasta, \"fasta\")\n",
"_____no_output_____"
],
[
"extract_genes('nl63')",
"_____no_output_____"
],
[
"###Old below",
"_____no_output_____"
],
[
"#From aligned .fasta, extract just the portion of the genome encoding Spike\ndef extract_spike(aligned_fasta, original_fasta, output_spike_fasta, output_he_fasta):\n \n spike_sequences = {}\n he_sequences = {}\n with open(aligned_fasta, \"r\") as handle:\n alignment = SeqIO.parse(handle, \"fasta\")\n for record in alignment:\n spike_nt = record.seq[23643:27729]\n spike_nt_str = str(spike_nt)\n #Throw out sequences that don't cover Spike\n num_unaligned_spike = Counter(spike_nt_str)['N']\n if num_unaligned_spike < (len(spike_nt)/2):\n spike_sequences[record.id] = spike_nt\n \n he_nt = record.seq[22354:23629]\n he_nt_str = str(he_nt)\n #Throw out sequences that don't cover HE\n num_unaligned_he = Counter(he_nt_str)['N']\n if num_unaligned_he < (len(he_nt)/2):\n he_sequences[record.id] = he_nt\n \n print(len(spike_sequences))\n print(len(he_sequences))\n \n spike_entries = []\n he_entries = []\n with open(original_fasta, \"r\") as handle_2:\n metadata = SeqIO.parse(handle_2, \"fasta\")\n for record in metadata:\n gb_id = record.id.split('|')[0]\n if gb_id in spike_sequences.keys():\n spike_record = SeqRecord(spike_sequences[gb_id], id=record.id, description=record.id)\n spike_entries.append(spike_record)\n if gb_id in he_sequences.keys():\n he_record = SeqRecord(he_sequences[gb_id], id=record.id, description=record.id)\n he_entries.append(he_record)\n \n SeqIO.write(spike_entries, output_spike_fasta, \"fasta\")\n SeqIO.write(he_entries, output_he_fasta, \"fasta\")",
"_____no_output_____"
],
[
"extract_spike(\"../nextstrain/seasonal-corona-genome/results/aligned_hku1.fasta\", \n \"../nextstrain/seasonal-corona-genome/data/hku1_datefix.fasta\", \n \"../nextstrain/seasonal-corona-beta/data/hku1_spike_genomealign.fasta\", \n \"../nextstrain/seasonal-corona-beta/data/hku1_he_genomealign.fasta\")\n",
"39\n29\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eceec2ac7bf6bbf7f308be002e209612bfe61030 | 83,985 | ipynb | Jupyter Notebook | Pull Points Example.ipynb | dljthomas1/Get-Pull-Points | a25580e94df74925039daf96dce567c0233404ab | [
"MIT"
] | null | null | null | Pull Points Example.ipynb | dljthomas1/Get-Pull-Points | a25580e94df74925039daf96dce567c0233404ab | [
"MIT"
] | null | null | null | Pull Points Example.ipynb | dljthomas1/Get-Pull-Points | a25580e94df74925039daf96dce567c0233404ab | [
"MIT"
] | null | null | null | 178.691489 | 50,864 | 0.871275 | [
[
[
"# Pull Points Script Example\nThis Jupyter Notebook demonstrates my pull points script in action. I create this script principally to get around some of the query limits when pulling Geographic data. For example, when using the Google Places API, Google only allows you to pull for first 60 results. If you want data for a whole city, for example, you need to find some way of carving that city up into small sections that you can pull data on. This script does exactly that.\n\nIn the first step we import our city boundary shapefile (you can also skip this stage and import a polygon directly if you have one).",
"_____no_output_____"
]
],
[
[
"# Import your shapefile polygon\nimport geopandas as gpd\ngdf = gpd.read_file('test_data/test_data.shp')\ngdf.plot()",
"_____no_output_____"
]
],
[
[
"If you're working with actual geographic data then you should probably convert the Coordinate Reference System to a local metric one. In the example below, I use OSMNX's bbox from point function. The function is ordinarily used to return a bounding box of a particular size around a point. I don't actually want a bounding box, but the same function includes the ability to project the bouding box into a local local metric projection and returns the local CRS in the process. We just want to extract this CRS.",
"_____no_output_____"
]
],
[
[
"import osmnx as ox\n\n# Get the centre point of our polygon which we will feed into OSMNX\ncentroid = gdf.iloc[0]['geometry'].centroid.y, gdf.iloc[0]['geometry'].centroid.x\n\n# Get the local CRS from OSMXN\ncrs = ox.utils_geo.bbox_from_point(centroid, project_utm = True, return_crs = True)[-1]\ncrs",
"_____no_output_____"
],
[
"# Covnert our GDF into a local system\ngdf = gdf.to_crs(crs)\n\n# Extract the polgyon from the GDF\npolygon = gdf.iloc[0]['geometry']\npolygon",
"_____no_output_____"
],
[
"# Import my pull points script\nfrom pull_points import get_pull_points\nhelp(get_pull_points)",
"Help on function get_pull_points in module pull_points:\n\nget_pull_points(poly, n, plot_map=False, plot_legend=False)\n This function will take a polygon, and try to segment it into an n approximately equally\n sized sections, returning the centre points of each section.\n \n If your polygon is bounday of a pyshical place (eg a city, town, etc) you should first \n convert the coordinate reference system (CRS) to a local metric projection. See the Jupyter\n notebook on my Github profile as an example.\n \n poly Polgon on which you wish to generate 'pull points'.\n n (int) The number of sections you wish to split the polygon up into\n plot_map True/False. Do you want to plot a map of the results\n plot legend Do you want to plot the legend of the map?\n\n"
],
[
"df = get_pull_points(polygon, 10, plot_map = True, plot_legend = True)\ndf",
"_____no_output_____"
],
[
"# Convert our points back into longitude and latitudes\ndf.crs = crs\ndf = df.to_crs(\"epsg:4326\")\ndf['x'] = df.geometry.apply(lambda x: x.x)\ndf['y'] = df.geometry.apply(lambda x: x.y)\ndf",
"_____no_output_____"
]
],
[
[
"You can now iterate through these points to pull data from your desired API.\n\nAs an example, Initially, I explained you can only pull 60 results through Google Places API. Now you can pull 600 results (60 results from 10 locations around the city).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
eceecefe78d77cf26dc48cd0eda66a63b9925ded | 2,829 | ipynb | Jupyter Notebook | src/vqa/.ipynb_checkpoints/QuestionImage_Visualization-checkpoint.ipynb | esteng/vagueness | 6c9627575188b325cfb72181931c95b774ca18b0 | [
"MIT"
] | null | null | null | src/vqa/.ipynb_checkpoints/QuestionImage_Visualization-checkpoint.ipynb | esteng/vagueness | 6c9627575188b325cfb72181931c95b774ca18b0 | [
"MIT"
] | null | null | null | src/vqa/.ipynb_checkpoints/QuestionImage_Visualization-checkpoint.ipynb | esteng/vagueness | 6c9627575188b325cfb72181931c95b774ca18b0 | [
"MIT"
] | null | null | null | 29.46875 | 128 | 0.568752 | [
[
[
"import json\nimport re\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nfrom PIL import Image\n\nwith open('/home/jgualla1/vagueness/src/jimena_work/output.json') as f:\n total_question_dict = json.load(f)\n \nwith open('/export/a14/jgualla1/v2_mscoco_val2014_complementary_pairs.json') as h:\n total_pairs_dict = json.load(h)\n \n#print(total_question_dict)\nprint(total_pairs_dict)",
"_____no_output_____"
],
[
"#img=mpimg.imread('/export/a14/jgualla1/val2014/COCO_val2014_000000000001.jpg')\n#imgplot = plt.imshow(img)\n#plt.show()ff\n#filename = '/export/a14/jgualla1/val2014/COCO_val2014_000000190753.jpg'\n#image = Image.open(filename)\n#image.show()\n#print(len(total_question_dict))\nfor key in total_question_dict:\n question_dict = total_question_dict[key]\n #print(question_dict)\n #print(question_dict['imageId'])\n question_id = question_dict['question_id']\n image_id = question_dict['imageId']\n image_length = len(str(image_id))\n needed_length = 12 - image_length\n needed_zeros = str(0)\n for x in range(0, needed_length-1):\n needed_zeros = needed_zeros + str(0)\n image = mpimg.imread(\"/export/a14/jgualla1/val2014/COCO_val2014_\" + str(needed_zeros) + str(image_id) + \".jpg\")\n print(question_dict['question'])\n print(question_dict['question_id'])\n for pair in total_pairs_dict:\n if pair[0] == question_id:\n print(\"Question pair id: \" + str(pair[1]))\n print(question_dict['question_answers'][0])\n plt.imshow(image)\n plt.axis(\"off\")\n plt.imshow(image)\n plt.show()\n print(\"________________________________________\")\n print(\" \")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
eceed04494d28532322a33d2c577c1882d514400 | 26,408 | ipynb | Jupyter Notebook | 02_Filtering_&_Sorting/Chipotle/Exercises.ipynb | WayneMao/pandas_exercises | 5627955485d10bb8484e7b6bf9104f3c19da39b0 | [
"BSD-3-Clause"
] | null | null | null | 02_Filtering_&_Sorting/Chipotle/Exercises.ipynb | WayneMao/pandas_exercises | 5627955485d10bb8484e7b6bf9104f3c19da39b0 | [
"BSD-3-Clause"
] | null | null | null | 02_Filtering_&_Sorting/Chipotle/Exercises.ipynb | WayneMao/pandas_exercises | 5627955485d10bb8484e7b6bf9104f3c19da39b0 | [
"BSD-3-Clause"
] | null | null | null | 30.600232 | 135 | 0.349174 | [
[
[
"# Ex1 - Filtering and Sorting Data",
"_____no_output_____"
],
[
"This time we are going to pull data directly from the internet.\nSpecial thanks to: https://github.com/justmarkham for sharing the dataset and materials.\n\n### Step 1. Import the necessary libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"### Step 2. Import the dataset from this [address](https://raw.githubusercontent.com/justmarkham/DAT8/master/data/chipotle.tsv). ",
"_____no_output_____"
],
[
"### Step 3. Assign it to a variable called chipo.",
"_____no_output_____"
]
],
[
[
"url = 'https://raw.githubusercontent.com/justmarkham/DAT8/master/data/chipotle.tsv'\n\nchipo = pd.read_csv(url,sep='\\t')\nprint(chipo.shape)\nchipo.head()\n",
"(4622, 5)\n"
]
],
[
[
"### Step 4. How many products cost more than $10.00?",
"_____no_output_____"
]
],
[
[
"chipo.item_price = [float(item[1:-1]) for item in chipo.item_price]\n#print(price)\nchipo.head()",
"_____no_output_____"
],
[
"# delete the duplicates in item_name and quantity\nchipo_filtered = chipo.drop_duplicates(['item_name','quantity'])\n\n# select only the products with quantity equals to 1\nchipo_one_prod = chipo_filtered[chipo_filtered.quantity == 1]\n\nchipo_one_prod[chipo_one_prod['item_price']>10].item_name.nunique()\nchipo_one_prod",
"(103, 5)\n"
]
],
[
[
"### Step 5. What is the price of each item? \n###### print a data frame with only two columns item_name and item_price",
"_____no_output_____"
]
],
[
[
"#chipo[(chipo['item_name'] == 'Chicken Bowl') & (chipo['quantity'] == 1)]\n\ntemp_chipo = pd.concat([chipo.quantity,chipo.item_name],axis=1)\ntemp_chipo",
"_____no_output_____"
]
],
[
[
"### Step 6. Sort by the name of the item",
"_____no_output_____"
]
],
[
[
"chipo.sort_values('item_name')",
"_____no_output_____"
]
],
[
[
"### Step 7. What was the quantity of the most expensive item ordered?",
"_____no_output_____"
]
],
[
[
"chipo.sort_values('item_price',ascending=False).head(1)",
"_____no_output_____"
]
],
[
[
"### Step 8. How many times were a Veggie Salad Bowl ordered?",
"_____no_output_____"
]
],
[
[
"chipo.item_name[chipo.item_name == 'Veggie Salad Bowl'].value_counts()",
"_____no_output_____"
]
],
[
[
"### Step 9. How many times people orderd more than one Canned Soda?",
"_____no_output_____"
]
],
[
[
"temp = chipo[(chipo.item_name == 'Canned Soda') & (chipo.quantity > 1)]\nlen(temp)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
eceedd1e71a3a39cadb4cbb42e13dec0322fbc89 | 433,646 | ipynb | Jupyter Notebook | WeatherPy/WeatherPy.ipynb | Connelito/Python-API-challenge | 46a24eb21513fc8ae73b8ca40b192e5ec2e7a35a | [
"ADSL"
] | null | null | null | WeatherPy/WeatherPy.ipynb | Connelito/Python-API-challenge | 46a24eb21513fc8ae73b8ca40b192e5ec2e7a35a | [
"ADSL"
] | null | null | null | WeatherPy/WeatherPy.ipynb | Connelito/Python-API-challenge | 46a24eb21513fc8ae73b8ca40b192e5ec2e7a35a | [
"ADSL"
] | null | null | null | 224.920124 | 43,540 | 0.897594 | [
[
[
"# WeatherPy\n----\n\n#### Note\n* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.",
"_____no_output_____"
]
],
[
[
"# Dependencies\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport json\nimport requests\nimport scipy.stats as st\nfrom scipy.stats import linregress\nfrom datetime import date\nfrom api_keys import weather_api_key, g_key\nfrom citipy import citipy as cp",
"_____no_output_____"
],
[
"# Output File (CSV)\noutput_data_file = \"output_data/cities.csv\"",
"_____no_output_____"
]
],
[
[
"## Generate Cities List",
"_____no_output_____"
]
],
[
[
"# List for holding lat_lngs and cities\nlat_lngs = []\ncities = []\n\n# Create a set of random lat and lng combinations\nlats = np.random.uniform(low=-90.000, high=90.000, size=1500)\nlngs = np.random.uniform(low=-180.000, high=180.000, size=1500)\nlat_lngs = zip(lats, lngs)\n\n# Identify nearest city for each lat, lng combination\nfor lat_lng in lat_lngs:\n city = cp.nearest_city(lat_lng[0], lat_lng[1]).city_name\n \n # If the city is unique, then add it to a our cities list\n if city not in cities:\n cities.append(city)\n\n# Print the city count to confirm sufficient count\nlen(cities)",
"_____no_output_____"
]
],
[
[
"### Perform API Calls\n* Perform a weather check on each city using a series of successive API calls.\n* Include a print log of each city as it'sbeing processed (with the city number and city name).\n",
"_____no_output_____"
]
],
[
[
"city_name = []\ncloudiness = []\ncountry = []\ndate = []\nhumidity = []\nlat = []\nlng = []\nmax_temp = []\nwind_speed = []\nindex_counter = 0\nset_counter = 1",
"_____no_output_____"
],
[
"print(\"Beginning Data Retrieval \")\nprint(\"-----------------------------\")\n\nbase_url = \"http://api.openweathermap.org/data/2.5/weather?\"\nunits = \"imperial\"\nquery_url = f\"{base_url}appid={weather_api_key}&units={units}&q=\"\n\n\n# Grab data for each city name in cities list\nfor index, city in enumerate(cities, start = 1):\n try:\n response = requests.get(query_url + city).json()\n city_name.append(response[\"name\"])\n cloudiness.append(response[\"clouds\"][\"all\"])\n country.append(response[\"sys\"][\"country\"])\n date.append(response[\"dt\"])\n humidity.append(response[\"main\"][\"humidity\"])\n lat.append(response[\"coord\"][\"lat\"])\n lng.append(response[\"coord\"][\"lon\"])\n max_temp.append(response['main']['temp_max'])\n wind_speed.append(response[\"wind\"][\"speed\"])\n \n if index_counter > 49:\n index_counter = 0\n set_counter = set_counter + 1\n \n else:\n index_counter = index_counter + 1\n \n print(f\"Processing Record {index_counter} of Set {set_counter} : {city}\") \n \n except(KeyError, IndexError):\n print(\"City not found. Skipping...\")\n\nprint(\"-----------------------------\")\nprint(\"Data Retrieval Complete\")\nprint(\"-----------------------------\")",
"Beginning Data Retrieval \n-----------------------------\nProcessing Record 1 of Set 1 : puerto ayora\nProcessing Record 2 of Set 1 : qena\nProcessing Record 3 of Set 1 : mar del plata\nProcessing Record 4 of Set 1 : baykit\nProcessing Record 5 of Set 1 : santa engracia\nProcessing Record 6 of Set 1 : houston\nProcessing Record 7 of Set 1 : mataura\nProcessing Record 8 of Set 1 : butaritari\nProcessing Record 9 of Set 1 : hilo\nProcessing Record 10 of Set 1 : daru\nProcessing Record 11 of Set 1 : jalu\nProcessing Record 12 of Set 1 : thompson\nProcessing Record 13 of Set 1 : rikitea\nProcessing Record 14 of Set 1 : nikolskoye\nProcessing Record 15 of Set 1 : qaanaaq\nProcessing Record 16 of Set 1 : petrolina\nProcessing Record 17 of Set 1 : arraial do cabo\nProcessing Record 18 of Set 1 : ilulissat\nProcessing Record 19 of Set 1 : kilgore\nProcessing Record 20 of Set 1 : provideniya\nProcessing Record 21 of Set 1 : houma\nProcessing Record 22 of Set 1 : poum\nProcessing Record 23 of Set 1 : novyy urgal\nProcessing Record 24 of Set 1 : atuona\nProcessing Record 25 of Set 1 : jamestown\nProcessing Record 26 of Set 1 : bredasdorp\nProcessing Record 27 of Set 1 : albany\nProcessing Record 28 of Set 1 : yellowknife\nProcessing Record 29 of Set 1 : hermanus\nProcessing Record 30 of Set 1 : marsa matruh\nProcessing Record 31 of Set 1 : yefremov\nProcessing Record 32 of Set 1 : carlisle\nProcessing Record 33 of Set 1 : sitka\nCity not found. Skipping...\nProcessing Record 34 of Set 1 : huarmey\nProcessing Record 35 of Set 1 : raudeberg\nProcessing Record 36 of Set 1 : irituia\nProcessing Record 37 of Set 1 : bluff\nProcessing Record 38 of Set 1 : atasu\nProcessing Record 39 of Set 1 : grand gaube\nProcessing Record 40 of Set 1 : hobart\nProcessing Record 41 of Set 1 : boali\nProcessing Record 42 of Set 1 : boguchany\nProcessing Record 43 of Set 1 : juneau\nProcessing Record 44 of Set 1 : mezen\nProcessing Record 45 of Set 1 : tuktoyaktuk\nProcessing Record 46 of Set 1 : longyearbyen\nCity not found. Skipping...\nProcessing Record 47 of Set 1 : coahuayana\nProcessing Record 48 of Set 1 : punta arenas\nProcessing Record 49 of Set 1 : veraval\nProcessing Record 50 of Set 1 : tasiilaq\nProcessing Record 0 of Set 2 : kavieng\nCity not found. Skipping...\nProcessing Record 1 of Set 2 : avarua\nCity not found. Skipping...\nProcessing Record 2 of Set 2 : hithadhoo\nProcessing Record 3 of Set 2 : kargasok\nProcessing Record 4 of Set 2 : gannan\nCity not found. Skipping...\nProcessing Record 5 of Set 2 : kichera\nProcessing Record 6 of Set 2 : owando\nCity not found. Skipping...\nProcessing Record 7 of Set 2 : san policarpo\nProcessing Record 8 of Set 2 : rocha\nProcessing Record 9 of Set 2 : christchurch\nProcessing Record 10 of Set 2 : mahebourg\nProcessing Record 11 of Set 2 : port elizabeth\nCity not found. Skipping...\nProcessing Record 12 of Set 2 : kanashevo\nProcessing Record 13 of Set 2 : natal\nProcessing Record 14 of Set 2 : saint-philippe\nProcessing Record 15 of Set 2 : khatanga\nProcessing Record 16 of Set 2 : saskylakh\nProcessing Record 17 of Set 2 : ternate\nProcessing Record 18 of Set 2 : ushuaia\nProcessing Record 19 of Set 2 : verkhnyaya inta\nProcessing Record 20 of Set 2 : candolim\nProcessing Record 21 of Set 2 : port hardy\nProcessing Record 22 of Set 2 : leningradskiy\nProcessing Record 23 of Set 2 : ribeira grande\nProcessing Record 24 of Set 2 : georgetown\nProcessing Record 25 of Set 2 : katherine\nProcessing Record 26 of Set 2 : kapaa\nProcessing Record 27 of Set 2 : sorong\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 28 of Set 2 : bonavista\nProcessing Record 29 of Set 2 : kaitangata\nProcessing Record 30 of Set 2 : gerash\nProcessing Record 31 of Set 2 : barrow\nProcessing Record 32 of Set 2 : arlit\nProcessing Record 33 of Set 2 : smalininkai\nProcessing Record 34 of Set 2 : port blair\nProcessing Record 35 of Set 2 : werda\nProcessing Record 36 of Set 2 : hasaki\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 37 of Set 2 : cuamba\nProcessing Record 38 of Set 2 : busselton\nProcessing Record 39 of Set 2 : vangaindrano\nProcessing Record 40 of Set 2 : kimbe\nCity not found. Skipping...\nProcessing Record 41 of Set 2 : kungurtug\nProcessing Record 42 of Set 2 : surgut\nCity not found. Skipping...\nProcessing Record 43 of Set 2 : palmer\nCity not found. Skipping...\nProcessing Record 44 of Set 2 : sungaipenuh\nProcessing Record 45 of Set 2 : chuy\nCity not found. Skipping...\nProcessing Record 46 of Set 2 : kruisfontein\nProcessing Record 47 of Set 2 : port lincoln\nProcessing Record 48 of Set 2 : meulaboh\nProcessing Record 49 of Set 2 : castro\nProcessing Record 50 of Set 2 : turukhansk\nCity not found. Skipping...\nProcessing Record 0 of Set 3 : aswan\nProcessing Record 1 of Set 3 : alexandria\nProcessing Record 2 of Set 3 : isangel\nProcessing Record 3 of Set 3 : homer\nProcessing Record 4 of Set 3 : kumluca\nProcessing Record 5 of Set 3 : upernavik\nProcessing Record 6 of Set 3 : iqaluit\nProcessing Record 7 of Set 3 : wah\nProcessing Record 8 of Set 3 : save\nProcessing Record 9 of Set 3 : campbell river\nProcessing Record 10 of Set 3 : dikson\nProcessing Record 11 of Set 3 : kalat\nProcessing Record 12 of Set 3 : codrington\nProcessing Record 13 of Set 3 : santiago del estero\nProcessing Record 14 of Set 3 : yafran\nProcessing Record 15 of Set 3 : raton\nProcessing Record 16 of Set 3 : pogranichnyy\nProcessing Record 17 of Set 3 : ozernovskiy\nProcessing Record 18 of Set 3 : nuuk\nProcessing Record 19 of Set 3 : hovd\nProcessing Record 20 of Set 3 : onokhoy\nProcessing Record 21 of Set 3 : iquitos\nProcessing Record 22 of Set 3 : cape town\nProcessing Record 23 of Set 3 : hearst\nProcessing Record 24 of Set 3 : buraydah\nProcessing Record 25 of Set 3 : lavrentiya\nProcessing Record 26 of Set 3 : asau\nProcessing Record 27 of Set 3 : new norfolk\nCity not found. Skipping...\nProcessing Record 28 of Set 3 : vila velha\nProcessing Record 29 of Set 3 : ulladulla\nProcessing Record 30 of Set 3 : clyde river\nProcessing Record 31 of Set 3 : margate\nProcessing Record 32 of Set 3 : santa rosa\nProcessing Record 33 of Set 3 : faya\nProcessing Record 34 of Set 3 : vaini\nProcessing Record 35 of Set 3 : port macquarie\nProcessing Record 36 of Set 3 : chumphon\nProcessing Record 37 of Set 3 : sao joao da barra\nCity not found. Skipping...\nProcessing Record 38 of Set 3 : saint-louis\nProcessing Record 39 of Set 3 : chokurdakh\nProcessing Record 40 of Set 3 : aklavik\nCity not found. Skipping...\nProcessing Record 41 of Set 3 : yeppoon\nProcessing Record 42 of Set 3 : bambous virieux\nProcessing Record 43 of Set 3 : machico\nProcessing Record 44 of Set 3 : constitucion\nProcessing Record 45 of Set 3 : pochutla\nProcessing Record 46 of Set 3 : kiunga\nProcessing Record 47 of Set 3 : atar\nProcessing Record 48 of Set 3 : otaru\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 49 of Set 3 : ambilobe\nProcessing Record 50 of Set 3 : pacific grove\nProcessing Record 0 of Set 4 : sobolevo\nProcessing Record 1 of Set 4 : ekibastuz\nProcessing Record 2 of Set 4 : malanje\nProcessing Record 3 of Set 4 : mitsamiouli\nProcessing Record 4 of Set 4 : gambela\nCity not found. Skipping...\nProcessing Record 5 of Set 4 : fortuna\nProcessing Record 6 of Set 4 : kupino\nProcessing Record 7 of Set 4 : nalut\nProcessing Record 8 of Set 4 : arica\nProcessing Record 9 of Set 4 : birjand\nProcessing Record 10 of Set 4 : kapenguria\nProcessing Record 11 of Set 4 : makakilo city\nProcessing Record 12 of Set 4 : nioro\nProcessing Record 13 of Set 4 : coquimbo\nProcessing Record 14 of Set 4 : tecpan\nProcessing Record 15 of Set 4 : leo\nCity not found. Skipping...\nProcessing Record 16 of Set 4 : dingle\nProcessing Record 17 of Set 4 : ustikolina\nProcessing Record 18 of Set 4 : bratsk\nProcessing Record 19 of Set 4 : ambanja\nProcessing Record 20 of Set 4 : tepelene\nCity not found. Skipping...\nProcessing Record 21 of Set 4 : bone\nProcessing Record 22 of Set 4 : beringovskiy\nProcessing Record 23 of Set 4 : luderitz\nProcessing Record 24 of Set 4 : yumen\nProcessing Record 25 of Set 4 : yulara\nProcessing Record 26 of Set 4 : bekhtery\nProcessing Record 27 of Set 4 : mountain home\nProcessing Record 28 of Set 4 : kualakapuas\nProcessing Record 29 of Set 4 : itoman\nProcessing Record 30 of Set 4 : port alfred\n"
]
],
[
[
"### Convert Raw Data to DataFrame\n* Export the city data into a .csv.\n* Display the DataFrame",
"_____no_output_____"
]
],
[
[
"# Create dataframe with data from API calls\nweather_df = pd.DataFrame({\n \"Lng\": lng,\n \"Lat\": lat, \n \"Country\": country,\n \"City\": city_name, \n \"Max Temp\": max_temp, \n \"Humidity\": humidity,\n \"Cloudiness\": cloudiness,\n \"Wind Speed\": wind_speed,\n \"Date\": date\n })",
"_____no_output_____"
],
[
"# Display dataframe\nweather_df.head()",
"_____no_output_____"
],
[
"# Output dataframe to CSV\nweather_df.to_csv(\"output_data/cities.csv\", index=False, header=True)",
"_____no_output_____"
]
],
[
[
"## Inspect the data and remove the cities where the humidity > 100%.\n----\nSkip this step if there are no cities that have humidity > 100%. ",
"_____no_output_____"
]
],
[
[
"# inspect the basic statistical values of the dataframe, paying attention to make sure no max humidity over 100\nweather_df.describe()",
"_____no_output_____"
],
[
"# Get the indices of cities that have humidity over 100%.\nbad_data = []\n\n# if any cities have humidity over 100%, print the index values of those lines\nif weather_df[\"Humidity\"].max() > 100:\n bad_data = weather_df.loc[weather_df[\"Humidity\"]>100, :].index.values\n print(bad_data)\n",
"_____no_output_____"
],
[
"# Make a new DataFrame equal to the city data to drop all humidity outliers by index.\n# Passing \"inplace=False\" will make a copy of the city_data DataFrame, which we call \"clean_city_data\".\nclean_city_df = weather_df.drop(bad_data, inplace=False)",
"_____no_output_____"
],
[
"clean_city_df.describe()",
"_____no_output_____"
]
],
[
[
"## Plotting the Data\n* Use proper labeling of the plots using plot titles (including date of analysis) and axes labels.\n* Save the plotted figures as .pngs.",
"_____no_output_____"
]
],
[
[
"# Scatter plot function\ndef scatter(yvar, title, ylabel, fig_name):\n plt.scatter(clean_city_df[\"Lat\"],clean_city_df[yvar],color=\"steelblue\", edgecolors=\"black\")\n plt.title(\"City Latitude vs \" + title + \" (\" + '07/25/21' + \")\")\n plt.ylabel(ylabel)\n plt.xlabel(\"Latitude\")\n plt.grid()\n \n # save figure as png\n plt.savefig(\"output_data/Lat_v_\" + fig_name + \".png\")\n \n # display scatter\n return plt.show()",
"_____no_output_____"
]
],
[
[
"## Latitude vs. Temperature Plot",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Temp scatter plot\nscatter(\"Max Temp\", \"Max Temperature\", \"Max Temperature (F)\", \"Temp\")",
"_____no_output_____"
]
],
[
[
"There appears to be a bellcurve relationship between Latitude and Max Temp at the time I recorded the data.",
"_____no_output_____"
],
[
"## Latitude vs. Humidity Plot",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Humidity scatter plot\nscatter(\"Humidity\", \"Humidity\", \"Humidity (%)\", \"Humid\")",
"_____no_output_____"
]
],
[
[
"There does not appear to be any obvious relationship between latitude and humdity at the time I recorded the data. However, most humidity levels were above 60%.",
"_____no_output_____"
],
[
"## Latitude vs. Cloudiness Plot",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Cloudiness scatter plot\nscatter(\"Cloudiness\", \"Cloudiness\", \"Cloudiness (%)\", \"Cloud\")",
"_____no_output_____"
]
],
[
[
"There doesn't appear to be any obvious relationship between latitude and cloudiness at the time I recorded the data.",
"_____no_output_____"
],
[
"## Latitude vs. Wind Speed Plot",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Wind Speed scatter plot\nscatter(\"Wind Speed\", \"Wind Speed\", \"Wind Speed (mph)\", \"Wind\")",
"_____no_output_____"
]
],
[
[
"There doesn't appear to be any obvious relationship between latitude and wind speed at the time I recorded the data. However, most wind speeds were below 10 mph.",
"_____no_output_____"
],
[
"## Linear Regression",
"_____no_output_____"
]
],
[
[
"# create new DataFrames that only contain northern and southern hemisphere data, respectively\n\nnorth_hem = clean_city_df.loc[clean_city_df[\"Lat\"]>=0,:]\nsouth_hem = clean_city_df.loc[clean_city_df[\"Lat\"]<0,:]",
"_____no_output_____"
],
[
"# Generate hemisphere scatter plots with linear regressions\ndef scatter_and_regression(x_axis, y_axis, title, ylabel, fig_name, hemi, eq_loc):\n \n # Generate linear regression\n (slope, intercept, rvalue, pvalue, stderr) = linregress(x_axis, y_axis)\n regress_values = x_axis * slope + intercept\n line_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\n \n # Plot linear regression\n plt.plot(x_axis, regress_values, color=\"red\")\n plt.annotate(line_eq, eq_loc, color=\"red\", size=15)\n \n # Create scatter\n plt.scatter(x_axis, y_axis, color=\"steelblue\")\n plt.title(hemi + \" Hemisphere: City Latitude vs \" + title + \" (\" + '07/25/21' + \")\")\n plt.ylabel(ylabel)\n plt.xlabel(\"Latitude\")\n \n # save figure as png \n plt.savefig(\"output_data/Lat_v_\" + fig_name + \".png\")\n \n # print r-value for analysis\n print(f\"The r-value is: {round(rvalue,2)}\")\n \n # display scatter & linear regression \n return plt.show()",
"_____no_output_____"
]
],
[
[
"#### Northern Hemisphere - Max Temp vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Temp scatter & linear regression for Northern Hemisphere\nscatter_and_regression(north_hem[\"Lat\"], north_hem[\"Max Temp\"], \n \"Max Temperature\", \"Max Temperature (F)\", \n \"Temp_North\", \"Northern\", (0,40))",
"The r-value is: -0.74\n"
]
],
[
[
"#### Southern Hemisphere - Max Temp vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Temp scatter & linear regression for Southern Hemisphere\nscatter_and_regression(south_hem[\"Lat\"], south_hem[\"Max Temp\"], \n \"Max Temperature\", \"Max Temperature (F)\", \n \"Temp_South\", \"Southern\", (-55,80))",
"The r-value is: 0.7\n"
]
],
[
[
"* There is a strong negative correlation between latitude and max temperature for the northern hemisphere at the time I recorded the data.\n* There is a strong positive correlation between latitude and max temperature for the southern hemisphere at the time I recorded the data.\n* Both charts are very similar relationships for the northern and southern hemisphere's.",
"_____no_output_____"
],
[
"#### Northern Hemisphere - Humidity (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Humidity scatter & linear regression for Northern Hemisphere\nscatter_and_regression(north_hem[\"Lat\"], north_hem[\"Humidity\"], \n \"Humidity\", \"Humidity (%)\", \n \"Humid_North\", \"Northern\", (0,10))",
"The r-value is: 0.1\n"
]
],
[
[
"#### Southern Hemisphere - Humidity (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Humidity scatter & linear regression for Southern Hemisphere\nscatter_and_regression(south_hem[\"Lat\"], south_hem[\"Humidity\"], \n \"Humidity\", \"Humidity (%)\", \n \"Humid_South\", \"Southern\", (-55,20))",
"The r-value is: 0.09\n"
]
],
[
[
"* There is a very weak positive correlation between latitude and humidity for the northern hemisphere at the time I recorded the data.\n* There is a very weak positive correlation between latitude and humidity for the southern hemisphere at the time I recorded the data.",
"_____no_output_____"
],
[
"#### Northern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Cloudiness scatter & linear regression for Northern Hemisphere\nscatter_and_regression(north_hem[\"Lat\"], north_hem[\"Cloudiness\"], \n \"Cloudiness\", \"Cloudiness (%)\", \n \"Cloud_North\", \"Northern\", (0,0))",
"The r-value is: 0.07\n"
]
],
[
[
"#### Southern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Cloudiness scatter & linear regression for Southern Hemisphere\nscatter_and_regression(south_hem[\"Lat\"], south_hem[\"Cloudiness\"], \n \"Cloudiness\", \"Cloudiness (%)\", \n \"Cloud_South\", \"Southern\", (-55,25))",
"The r-value is: 0.26\n"
]
],
[
[
"* There is a very weak positive correlation between latitude and cloudiness for the northern hemisphere at the time I recorded the data.\n* There is a weak positive correlation between latitude and cloudiness for the southern hemisphere at the time I recorded the data.",
"_____no_output_____"
],
[
"#### Northern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Wind Speed scatter & linear regression for Northern Hemisphere\nscatter_and_regression(north_hem[\"Lat\"], north_hem[\"Wind Speed\"], \n \"Wind Speed\", \"Wind Speed (mph)\", \n \"Wind_North\", \"Northern\", (40,25))",
"The r-value is: 0.16\n"
]
],
[
[
"#### Southern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Create Lat vs Wind Speed scatter & linear regression for Southern Hemisphere\nscatter_and_regression(south_hem[\"Lat\"], south_hem[\"Wind Speed\"], \n \"Wind Speed\", \"Wind Speed (mph)\", \n \"Wind_South\", \"Southern\", (-25,25))",
"The r-value is: -0.18\n"
]
],
[
[
"* There is a very weak positive correlation between latitude and wind speed for the northern hemisphere at the time I recorded the data.\n* There is a very weak positive correlation between latitude and wind speed for the southern hemisphere at the time I recorded the data.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eceede2c7ae6d9cc0b9da4520d8662c452901252 | 92,269 | ipynb | Jupyter Notebook | zip_size/size_224.ipynb | allnes/age_classifier | b2390ab05ef070c09a11dfc93b3f097340e6366d | [
"BSD-3-Clause"
] | null | null | null | zip_size/size_224.ipynb | allnes/age_classifier | b2390ab05ef070c09a11dfc93b3f097340e6366d | [
"BSD-3-Clause"
] | null | null | null | zip_size/size_224.ipynb | allnes/age_classifier | b2390ab05ef070c09a11dfc93b3f097340e6366d | [
"BSD-3-Clause"
] | 1 | 2020-02-18T16:01:06.000Z | 2020-02-18T16:01:06.000Z | 266.67341 | 36,390 | 0.887102 | [
[
[
"<a href=\"https://colab.research.google.com/github/allnes/age_classifier/blob/master/zip_size/size_224.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"%tensorflow_version 1.x\n\nfrom google.colab import drive\nimport os\n\ndrive.mount('/content/drive')\nos.chdir('/content/drive/My Drive/DL_DATA_GRAPH/BUILD/cnn_graph/')\n!ls\n\n# !git clone https://github.com/mdeff/cnn_graph\n# !git status\n# !git fetch\n# !git checkout graph_train\n!git pull origin graph_train\n\n# classes = { 2, 3, 4, 5, 6, 7, 9 }\n# zip_size = { 64, 80, 96, 112, 128, 160, 192, 224 }\n\n%run age_classifier/age_classes_fcn \\\n --path_project='/content/drive/My Drive/DL_DATA_GRAPH/' \\\n --path_data='NEW/resize/converted_data_resize_224.npz' \\\n --zip_size=224 \\\n --class_count=5 \\\n --fcn_count=700 \\\n\n%matplotlib inline",
"TensorFlow 1.x selected.\nGo to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&response_type=code&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly\n\nEnter your authorization code:\n··········\nMounted at /content/drive\nage_classifier\tLICENSE.txt rcv1.ipynb summaries\ncheckpoints\tmakefile README.md\t trials\nlib\t\tnips2016 requirements.txt usage.ipynb\nFrom https://github.com/allnes/cnn_graph\n * branch graph_train -> FETCH_HEAD\nAlready up to date.\n['arr_0', 'arr_1']\n(729, 50176)\n(729,)\n--> Reshape data\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
eceee13e05eb795b87435c4e109ef01b7256bde3 | 18,264 | ipynb | Jupyter Notebook | docs/_downloads/a5fa2446bb57fcfef260001e46171916/transfer_learning_tutorial.ipynb | jessemin/PyTorch-tutorials-kr | bcb015e5b4eb4013f3ee03374c2669733bfd09ca | [
"BSD-3-Clause"
] | 1 | 2019-12-05T05:16:44.000Z | 2019-12-05T05:16:44.000Z | docs/_downloads/a5fa2446bb57fcfef260001e46171916/transfer_learning_tutorial.ipynb | jessemin/PyTorch-tutorials-kr | bcb015e5b4eb4013f3ee03374c2669733bfd09ca | [
"BSD-3-Clause"
] | null | null | null | docs/_downloads/a5fa2446bb57fcfef260001e46171916/transfer_learning_tutorial.ipynb | jessemin/PyTorch-tutorials-kr | bcb015e5b4eb4013f3ee03374c2669733bfd09ca | [
"BSD-3-Clause"
] | null | null | null | 83.018182 | 3,132 | 0.628449 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n전이학습(Transfer Learning) 튜토리얼\n====================================\n**Author**: `Sasank Chilamkurthy <https://chsasank.github.io>`_\n **번역**: `박정환 <http://github.com/9bow>`_\n\n이 튜토리얼에서는 전이학습(Transfer Learning)을 이용하여 신경망을 어떻게 학습시키는지\n배워보겠습니다. 전이학습에 대해서 더 알아보시려면\n`CS231n 노트 <http://cs231n.github.io/transfer-learning/>`__ 를 읽어보시면 좋습니다.\n\n위 노트를 인용해보면,\n\n 실제로 충분한 크기의 데이터셋을 갖추기는 상대적으로 드물기 때문에,\n (무작위 초기화를 통해) 바닥부터(from scratch) 전체 합성곱 신경망(Convolutional\n Network)를 학습하는 사람은 거의 없습니다. 대신, 매우 큰 데이터셋(예.\n 100가지 분류(Category)에 대해 120만개의 이미지가 포함된 ImageNet)에서 합성곱\n 신경망(ConvNet)을 미리 학습(Pretrain)한 후, 이 합성곱 신경망을 관심있는 작업\n (task of interest)을 위한 초기화(initialization) 또는 고정 특징 추출기(fixed\n feature extractor)로 사용합니다.\n\n이러한 2가지의 주요한 전이학습 시나리오는 다음과 같습니다:\n\n- **합성곱 신경망의 미세조정(Finetuning)**: 무작위 초기화 대신, 신경망을\n ImageNet 1000 데이터셋 등으로 미리 학습한 신경망으로 초기화합니다. 학습의 나머지\n 과정들은 평상시와 같습니다.\n- **고정 특정 추출기로써의 합성곱 신경망**: 여기서는 마지막의 완전히 연결\n (Fully-connected)된 계층을 제외한 모든 신경망의 가중치를 고정합니다. 이\n 마지막의 완전히 연결된 계층은 새로운 무작위의 가중치를 갖는 계층으로 대체되어\n 이 계층만 학습합니다.\n\n\n",
"_____no_output_____"
]
],
[
[
"# License: BSD\n# Author: Sasank Chilamkurthy\n\nfrom __future__ import print_function, division\n\nimport torch\nimport torch.nn as nn\nimport torch.optim as optim\nfrom torch.optim import lr_scheduler\nimport numpy as np\nimport torchvision\nfrom torchvision import datasets, models, transforms\nimport matplotlib.pyplot as plt\nimport time\nimport os\nimport copy\n\nplt.ion() # interactive mode",
"_____no_output_____"
]
],
[
[
"데이터 불러오기\n---------------\n\n데이터를 불러오기 위해 torchvision과 torch.utils.data 패키지를 사용하겠습니다.\n\n오늘 풀고자 하는 문제는 **개미** 와 **벌** 을 분류하는 모델을 학습하는 것입니다.\n각각의 분류에는 75개의 검증용 이미지(validation image)가 있습니다. 일반적으로,\n만약 바닥부터 학습을 한다면, 이는 일반화하기에는 아주 작은 데이터셋입니다.\n하지만 전이학습을 사용할 것이므로, 합리적으로 잘 일반화해 할 수 있습니다.\n\n이 데이터셋은 ImageNet의 아주 작은 부분(Subset)입니다.\n\n.. Note ::\n 데이터를 `여기 <https://download.pytorch.org/tutorial/hymenoptera_data.zip>`_\n 에서 다운로드 받아 현재 디렉토리에 압축을 푸십시오.\n\n",
"_____no_output_____"
]
],
[
[
"# 학습을 위한 데이터 증가(Augmentation)와 일반화하기\n# 단지 검증을 위한 일반화하기\ndata_transforms = {\n 'train': transforms.Compose([\n transforms.RandomResizedCrop(224),\n transforms.RandomHorizontalFlip(),\n transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])\n ]),\n 'val': transforms.Compose([\n transforms.Resize(256),\n transforms.CenterCrop(224),\n transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])\n ]),\n}\n\ndata_dir = 'hymenoptera_data'\nimage_datasets = {x: datasets.ImageFolder(os.path.join(data_dir, x),\n data_transforms[x])\n for x in ['train', 'val']}\ndataloaders = {x: torch.utils.data.DataLoader(image_datasets[x], batch_size=4,\n shuffle=True, num_workers=4)\n for x in ['train', 'val']}\ndataset_sizes = {x: len(image_datasets[x]) for x in ['train', 'val']}\nclass_names = image_datasets['train'].classes\n\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")",
"_____no_output_____"
]
],
[
[
"일부 이미지 시각화하기\n^^^^^^^^^^^^^^^^^^^^^^^^^\n데이터 증가를 이해하기 위해 일부 학습용 이미지를 시각화해보겠습니다.\n\n",
"_____no_output_____"
]
],
[
[
"def imshow(inp, title=None):\n \"\"\"Imshow for Tensor.\"\"\"\n inp = inp.numpy().transpose((1, 2, 0))\n mean = np.array([0.485, 0.456, 0.406])\n std = np.array([0.229, 0.224, 0.225])\n inp = std * inp + mean\n inp = np.clip(inp, 0, 1)\n plt.imshow(inp)\n if title is not None:\n plt.title(title)\n plt.pause(0.001) # pause a bit so that plots are updated\n\n\n# Get a batch of training data\ninputs, classes = next(iter(dataloaders['train']))\n\n# Make a grid from batch\nout = torchvision.utils.make_grid(inputs)\n\nimshow(out, title=[class_names[x] for x in classes])",
"_____no_output_____"
]
],
[
[
"모델 학습하기\n--------------\n\n이제 모델을 학습하기 위한 일반 함수를 작성해보겠습니다. 여기서는 다음 내용들을\n설명합니다:\n\n- Learning Rate 관리(Scheduling)\n- 최적의 모델 구하기\n\n아래에서 ``scheduler`` 매개변수는 ``torch.optim.lr_scheduler`` 의 LR Scheduler\n객체(Object)입니다.\n\n",
"_____no_output_____"
]
],
[
[
"def train_model(model, criterion, optimizer, scheduler, num_epochs=25):\n since = time.time()\n\n best_model_wts = copy.deepcopy(model.state_dict())\n best_acc = 0.0\n\n for epoch in range(num_epochs):\n print('Epoch {}/{}'.format(epoch, num_epochs - 1))\n print('-' * 10)\n\n # Each epoch has a training and validation phase\n for phase in ['train', 'val']:\n if phase == 'train':\n scheduler.step()\n model.train() # Set model to training mode\n else:\n model.eval() # Set model to evaluate mode\n\n running_loss = 0.0\n running_corrects = 0\n\n # Iterate over data.\n for inputs, labels in dataloaders[phase]:\n inputs = inputs.to(device)\n labels = labels.to(device)\n\n # zero the parameter gradients\n optimizer.zero_grad()\n\n # forward\n # track history if only in train\n with torch.set_grad_enabled(phase == 'train'):\n outputs = model(inputs)\n _, preds = torch.max(outputs, 1)\n loss = criterion(outputs, labels)\n\n # backward + optimize only if in training phase\n if phase == 'train':\n loss.backward()\n optimizer.step()\n\n # statistics\n running_loss += loss.item() * inputs.size(0)\n running_corrects += torch.sum(preds == labels.data)\n\n epoch_loss = running_loss / dataset_sizes[phase]\n epoch_acc = running_corrects.double() / dataset_sizes[phase]\n\n print('{} Loss: {:.4f} Acc: {:.4f}'.format(\n phase, epoch_loss, epoch_acc))\n\n # deep copy the model\n if phase == 'val' and epoch_acc > best_acc:\n best_acc = epoch_acc\n best_model_wts = copy.deepcopy(model.state_dict())\n\n print()\n\n time_elapsed = time.time() - since\n print('Training complete in {:.0f}m {:.0f}s'.format(\n time_elapsed // 60, time_elapsed % 60))\n print('Best val Acc: {:4f}'.format(best_acc))\n\n # load best model weights\n model.load_state_dict(best_model_wts)\n return model",
"_____no_output_____"
]
],
[
[
"모델 예측값 시각화하기\n^^^^^^^^^^^^^^^^^^^^^^^\n\n일부 이미지에 대한 예측값을 보여주는 일반화된(Generic) 함수입니다.\n\n\n",
"_____no_output_____"
]
],
[
[
"def visualize_model(model, num_images=6):\n was_training = model.training\n model.eval()\n images_so_far = 0\n fig = plt.figure()\n\n with torch.no_grad():\n for i, (inputs, labels) in enumerate(dataloaders['val']):\n inputs = inputs.to(device)\n labels = labels.to(device)\n\n outputs = model(inputs)\n _, preds = torch.max(outputs, 1)\n\n for j in range(inputs.size()[0]):\n images_so_far += 1\n ax = plt.subplot(num_images//2, 2, images_so_far)\n ax.axis('off')\n ax.set_title('predicted: {}'.format(class_names[preds[j]]))\n imshow(inputs.cpu().data[j])\n\n if images_so_far == num_images:\n model.train(mode=was_training)\n return\n model.train(mode=was_training)",
"_____no_output_____"
]
],
[
[
"합성곱 신경망 미세조정(Finetuning)\n----------------------------------\n\n미리 학습한 모델을 불러온 후 마지막의 완전히 연결된 계층을 재설정(reset)합니다.\n\n\n",
"_____no_output_____"
]
],
[
[
"model_ft = models.resnet18(pretrained=True)\nnum_ftrs = model_ft.fc.in_features\nmodel_ft.fc = nn.Linear(num_ftrs, 2)\n\nmodel_ft = model_ft.to(device)\n\ncriterion = nn.CrossEntropyLoss()\n\n# Observe that all parameters are being optimized\noptimizer_ft = optim.SGD(model_ft.parameters(), lr=0.001, momentum=0.9)\n\n# Decay LR by a factor of 0.1 every 7 epochs\nexp_lr_scheduler = lr_scheduler.StepLR(optimizer_ft, step_size=7, gamma=0.1)",
"_____no_output_____"
]
],
[
[
"학습 및 평가하기\n^^^^^^^^^^^^^^^^^^\n\nCPU에서 15-25분 가량 소요될 것입니다. 그래도 GPU에서는 1분도 걸리지 않습니다.\n\n\n",
"_____no_output_____"
]
],
[
[
"model_ft = train_model(model_ft, criterion, optimizer_ft, exp_lr_scheduler,\n num_epochs=25)",
"_____no_output_____"
],
[
"visualize_model(model_ft)",
"_____no_output_____"
]
],
[
[
"고정 특정 추출기로써의 합성곱 신경망\n-------------------------------------\n\n이제, 마지막 계층을 제외한 모든 신경망을 고정(freeze)할 필요가 있습니다.\n``requires_grad == False`` 를 설정하여 매개변수를 고정하여 ``backward()`` 에서\n경사도(gradient)가 계산되지 않도록 해야합니다.\n\n이 부분에 대한 문서는\n`여기 <http://pytorch.org/docs/notes/autograd.html#excluding-subgraphs-from-backward>`__\n에서 확인할 수 있습니다.\n\n\n",
"_____no_output_____"
]
],
[
[
"model_conv = torchvision.models.resnet18(pretrained=True)\nfor param in model_conv.parameters():\n param.requires_grad = False\n\n# Parameters of newly constructed modules have requires_grad=True by default\nnum_ftrs = model_conv.fc.in_features\nmodel_conv.fc = nn.Linear(num_ftrs, 2)\n\nmodel_conv = model_conv.to(device)\n\ncriterion = nn.CrossEntropyLoss()\n\n# Observe that only parameters of final layer are being optimized as\n# opoosed to before.\noptimizer_conv = optim.SGD(model_conv.fc.parameters(), lr=0.001, momentum=0.9)\n\n# Decay LR by a factor of 0.1 every 7 epochs\nexp_lr_scheduler = lr_scheduler.StepLR(optimizer_conv, step_size=7, gamma=0.1)",
"_____no_output_____"
]
],
[
[
"학습 및 평가하기\n^^^^^^^^^^^^^^^^^\n\nCPU에서 실행하는 경우 이전 시나리오와 비교했을 때 약 절반 가량의 시간이 소요됩니다.\n이는 대부분의 신경망에서 경사도를 계산할 필요가 없을 것으로 기대합니다. 하지만,\n순전파(forward)는 계산해야 할 필요가 있습니다.\n\n\n",
"_____no_output_____"
]
],
[
[
"model_conv = train_model(model_conv, criterion, optimizer_conv,\n exp_lr_scheduler, num_epochs=25)",
"_____no_output_____"
],
[
"visualize_model(model_conv)\n\nplt.ioff()\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
eceee35a95449d579b36e0d68541296256e9dcc9 | 46,088 | ipynb | Jupyter Notebook | farm_planning/farm_planning.ipynb | jacksonwalters/modeling-examples | 549fb263a3aa469ceaf9cbaf98a4d55ff98a13f5 | [
"Apache-2.0"
] | null | null | null | farm_planning/farm_planning.ipynb | jacksonwalters/modeling-examples | 549fb263a3aa469ceaf9cbaf98a4d55ff98a13f5 | [
"Apache-2.0"
] | null | null | null | farm_planning/farm_planning.ipynb | jacksonwalters/modeling-examples | 549fb263a3aa469ceaf9cbaf98a4d55ff98a13f5 | [
"Apache-2.0"
] | null | null | null | 38.567364 | 688 | 0.525668 | [
[
[
"# Farm Planning\n\n## Objective and Prerequisites\n\nThis model is an example of a production planning problem. In such problems — which are common across a wide range of industries — choices must be made regarding which products to produce and which resources to use to produce these products.\n\nIn this example, we will model and solve a multi-period production planning problem. In this case, the purpose of the application is to optimize the operation of a farm over five years. The farmer has a certain number of farmable acres and cows. The cows can be sold or used to produce milk, which can also be sold for profit. Food for the cows is grown on the farmable acres. The aim is to create an optimal operational plan for the next five years to maximize the total profit of the farm. \nThis includes determining optimal strategies for cultivation, for managing the cows, and for the overall operation of the farm\n\nMore information on this type of model can be found in example #8 of the fifth edition of Model Building in Mathematical Programming, by H. Paul Williams on pages 262-263 and 312-315.\n\nThis modeling example is at the advanced level, where we assume that you know Python and the Gurobi Python API and that you have advanced knowledge of building mathematical optimization models. Typically, the objective function and/or constraints of these examples are complex or require advanced features of the Gurobi Python API.\n\n**Download the Repository** <br /> You can download the repository containing this and other examples by clicking [here](https://github.com/Gurobi/modeling-examples/archive/master.zip). \n\n**Gurobi License** <br />\nIn order to run this Jupyter Notebook properly, you must have a Gurobi license. If you do not have one, you can request an [evaluation license](https://www.gurobi.com/downloads/request-an-evaluation-license/?utm_source=3PW&utm_medium=OT&utm_campaign=WW-MU-AGR-OR-O_LEA-PR_NO-Q3_FY20_WW_JPME_farm-planning_COM_EVAL_GITHUB_&utm_term=farm-planning-problem&utm_content=C_JPM) as a *commercial user*, or download a [free license](https://www.gurobi.com/academia/academic-program-and-licenses/?utm_source=3PW&utm_medium=OT&utm_campaign=WW-MU-AGR-OR-O_LEA-PR_NO-Q3_FY20_WW_JPME_farm-planning_ACADEMIC_EVAL_GITHUB_&utm_term=farm-planning-problem&utm_content=C_JPM) as an *academic user*.\n\n---\n## Problem Description\n\nA farmer with a 200-acre farm wishes to create a five-year production plan.\n\nCurrently, he has a herd of 120 cows comprising 20 heifers (young female cows) and 100 adult dairy cows. Each heifer requires 2/3 of an acre to support it and each dairy cow requires one acre.\n\nOn average, a dairy cow produces 1.1 calves per year. Half of these calves will be bullocks that are sold shortly after birth for an average of $\\$30$ each. The remaining calves, heifers, can either be sold for $\\$40$ or raised until age two when they will become dairy cows. For the current year, all heifers identified for sale have already been sold.\n\nThe general practice is to sell all dairy cows at the age of 12 for an average of $\\$120$ each. However, each year an average of $5\\%$ of heifers and $2\\%$ of dairy cows die. At present, the farmer’s herd of 120 cows is evenly distributed with 10 cows per age from newborn to 11 years old.\n\nThe milk from a dairy cow can be sold for $\\$370$ annually. The farmer can currently house up to 130 cows, but this capacity limit can be increased for $\\$200$ per additional cow.\n\nEach dairy cow requires 0.6 tons of grain and 0.7 tons of sugar beet per year. Both of these can be grown on the farm. Each acre can yield 1.5 tons of sugar beet. However, only 80 acres are suitable for growing grain, and those acres have different levels of productivity as follows:\n\n| Land Group | Area (Acres) | Grain Production (Tons/Acre) |\n| --- | --- | --- |\n| Group 1 | 20 | 1.10 |\n| Group 2 | 30 | 0.90 |\n| Group 3 | 20 | 0.80 |\n| Group 4 | 10 | 0.65 |\n\nSugar beet can be bought for $\\$70$ a ton and sold for $\\$58$ a ton. Grain can be bought for $\\$90$ a ton and sold for $\\$75$ a ton.\n\nThe annual labor requirements for cows as well as grain and sugar beet production are as follows:\n\n| <i></i> | Labor Required (Hr/Year) |\n| --- | --- |\n| Heifer | 10 |\n| Milk-producing Cow | 42 |\n| Acre Devoted to Grain | 4 |\n| Acre Devoted to Sugar Beet| 14 |\n\nOther annual costs are as follows:\n\n| <i></i> | Cost (USD/Year) |\n| --- | --- |\n| Heifer | 50 |\n| Milk-producing Cow | 100 |\n| Acre Devoted to Grain | 15 |\n| Acre Devoted to Sugar Beet| 10 |\n\nLabor currently costs the farmer $\\$4000$ per year and that cost provides 5,500 hours of labor. Additional labor can be paid for at the rate of $\\$1.20$ per hour.\n\nAny capital expenditure can be financed with a 10-year loan at $15\\%$ interest annually. The interest and principal are paid back in 10 equal annual payments. It is prohibited for the annual cash flow to be negative.\n\nThe farmer does not want to reduce the total number of dairy cows at the end of the five-year period by more than $50\\%$, nor does he want to increase their number by more than $75\\%$.\n\nWhat plan should the farmer follow over the next five years in order to maximize profit?\n\n---\n## Model Formulation\n\n### Sets and Indices\n\n$t \\in \\text{Years}=\\{1,2,\\dots,5\\}$: Set of years.\n\n$l \\in \\text{Lands}=\\{1,2,3,4\\}$: Set of land groups.\n\n$k \\in \\text{Ages}=\\{1,2,\\dots,12\\}$: Set of cow ages.\n\n### Parameters\n\n$\\text{Installment} \\in \\mathbb{R}^+$: Annual payment for each $\\$200$ loan. It can be computed as follows.\n\n$$\n\\text{Installment}= \\text{Loan}*r*\\frac{(1+r)^n}{(1+r)^n-1},\n$$\n\nwhere $\\text{Loan}$ is the loan amount of $\\$200$, $r$ is the interest rate of $15\\%$, and $n$ represents the ten-year period. Note that the book reports a value of $\\$39.71$ instead of $\\$39.85$.\n\n$\\text{Housing_cap} \\in \\mathbb{N}$: Number of cows that can be housed.\n\n$\\text{Land_cap} \\in \\mathbb{R}^+$: Land available (in Acres).\n\n$\\text{Labor_cap} \\in \\mathbb{R}^+$: Regular labor available (in hours) in a year.\n\n$\\text{GR_intake} \\in \\mathbb{R}^+$: Tons of grain consumed by a dairy cow in a year.\n\n$\\text{SB_intake} \\in \\mathbb{R}^+$: Tons of sugar beet consumed by a dairy cow in a year.\n\n$\\text{HF_land} \\in \\mathbb{R}^+$: Acres required for sustaining each heifer.\n\n$\\text{HF_labor} \\in \\mathbb{R}^+$: Hours of labor required in a year by each heifer.\n\n$\\text{Cow_labor} \\in \\mathbb{R}^+$: Hours of labor required in a year by each dairy cow.\n\n$\\text{GR_labor} \\in \\mathbb{R}^+$: Hours of labor required in a year by each acre of land devoted to grains.\n\n$\\text{SB_labor} \\in \\mathbb{R}^+$: Hours of labor required in a year by each acre of land devoted to sugar beet.\n\n$\\text{HF_decay} \\in [0,1] \\subset \\mathbb{R}^+$: Average proportion of heifers that die in a year.\n\n$\\text{Cow_decay} \\in [0,1] \\subset \\mathbb{R}^+$: Average proportion of dairy cows that die in a year.\n\n$\\text{Birthrate} \\in \\mathbb{R}^+$: Expected number of calves produced by a dairy cow in a year.\n\n$\\text{Min_final_cows} \\in \\mathbb{N}$: Minimum number of dairy cows at the end of the planning horizon.\n\n$\\text{Max_final_cows} \\in \\mathbb{N}$: Maximum number of dairy cows at the end of the planning horizon.\n\n$\\text{Initial_HF} \\in \\mathbb{R}^+$: Number of heifers of each age at the beginning of the planning horizon.\n\n$\\text{Initial_cows} \\in \\mathbb{R}^+$: Number of dairy cows of each age at the beginning of the planning horizon.\n\n$\\text{BL_price} \\in \\mathbb{R}^+$: Price for selling one bullock.\n\n$\\text{HF_price} \\in \\mathbb{R}^+$: Price for selling one heifer.\n\n$\\text{Cow_price} \\in \\mathbb{R}^+$: Price for selling one dairy cow.\n\n$\\text{Milk_price} \\in \\mathbb{R}^+$: Price for selling the milk produced by a dairy cow in a year.\n\n$\\text{GR_price} \\in \\mathbb{R}^+$: Price for selling a ton of grain.\n\n$\\text{SB_price} \\in \\mathbb{R}^+$: Price for selling a ton of sugar beet.\n\n$\\text{GR_cost} \\in \\mathbb{R}^+$: Cost for purchasing a ton of grain.\n\n$\\text{SB_cost} \\in \\mathbb{R}^+$: Cost for purchasing a ton of sugar beet.\n\n$\\text{Overtime_cost} \\in \\mathbb{R}^+$: Cost for getting an hour of overtime.\n\n$\\text{Regular_time_cost} \\in \\mathbb{R}^+$: Cost for having 5,500 hours of labor in regular time.\n\n$\\text{HF_cost} \\in \\mathbb{R}^+$: Yearly cost for supporting a heifer.\n\n$\\text{Cow_cost} \\in \\mathbb{R}^+$: Yearly cost for supporting a dairy cow.\n\n$\\text{GR_land_cost} \\in \\mathbb{R}^+$: Yearly cost for supporting an acre of land devoted to grain.\n\n$\\text{SB_land_cost} \\in \\mathbb{R}^+$: Yearly cost for supporting an acre of land devoted to sugar beet.\n\n$\\text{SB_yield} \\in \\mathbb{R}^+$: Sugar beet yield.\n\n$\\text{GR_yield}_l \\in \\mathbb{R}^+$: Grain yield at land group $l$.\n\n$\\text{GR_area}_l \\in \\mathbb{R}^+$: Number of acres suitable for growing grain in land group $l$.\n\n### Decision Variables\n\n$\\text{Outlay}_t \\in \\mathbb{R}^+$: Amount of money (in units of $\\$200$) spent on renting houses in year $t$.\n\n$\\text{Overtime}_t \\in \\mathbb{R}^+$: Number of extra labor hours needed in year $t$.\n\n$\\text{Newborn}_t \\in \\mathbb{R}^+$: Number of newborn heifers left in year $t$ to be raised.\n\n$\\text{HF_sell}_t \\in \\mathbb{R}^+$: Number of newborn heifers to sell in year $t$.\n\n$\\text{Profit}_t \\in \\mathbb{R}^+$: Profit attained in year $t$.\n\n$\\text{SB_buy}_t \\in \\mathbb{R}^+$: Number of tons of sugar beet to buy in year $t$.\n\n$\\text{SB_sell}_t \\in \\mathbb{R}^+$: Number of tons of sugar beet to sell in year $t$.\n\n$\\text{GR_buy}_t \\in \\mathbb{R}^+$: Number of tons of grain to buy in year $t$.\n\n$\\text{GR_sell}_t \\in \\mathbb{R}^+$: Number of tons of grain to sell in year $t$.\n\n$\\text{SB}_t \\in \\mathbb{R}^+$: Number of tons of sugar beet to grow in year $t$.\n\n$\\text{GR}_{t,l} \\in \\mathbb{R}^+$: Number of tons of grain to grow in year $t$ at land group $l$.\n\n$\\text{Cows}_{t,k} \\in \\mathbb{R}^+$: Number of cows of age $k$ available in year $t$.\n\n\n### Objective Function\n\n- **Profit**: Maximize the total profit (in USD) of the planning horizon. Notice that, to make capital expenditure as costly in latter years as in former ones, it is necessary to subtract pending loan payments.\n\n\\begin{equation}\n\\text{Maximize} \\quad Z = \\sum_{t \\in \\text{Years}}{\\text{Profit}_t - installment*(t+4)*\\text{Outlay}_t}\n\\end{equation}\n\n### Constraints\n\n- **Housing Capacity**: Livestock cannot exceed the installed capacity plus house rental in year $t$.\n\n\\begin{equation}\n\\text{Newborn}_t + \\sum_{k \\in \\text{Ages} \\setminus \\{12\\}}{\\text{Cows}_{t,k}} \\leq \\text{Housing_cap} + \\sum_{d \\in \\text{Years}: d \\leq t}{\\text{Outlay}_d} \\quad \\forall t \\in \\text{Years}\n\\end{equation}\n\n- **Food Consumption**: There must be enough food to feed the livestock in year $t$.\n\n- Grain.\n\n\\begin{equation}\n\\sum_{k \\in \\text{Ages} \\setminus \\{1,12\\}}{\\text{GR_intake}*\\text{Cows}_{t,k}} \\leq \\text{GR_buy}_t - \\text{GR_sell}_t + \\sum_{l \\in \\text{Lands}}{\\text{GR}_{t,l}} \\quad \\forall t \\in \\text{Years}\n\\end{equation}\n\n- Sugar beet.\n\n\\begin{equation}\n\\sum_{k \\in \\text{Ages} \\setminus \\{1,12\\}}{\\text{SB_intake}*\\text{Cows}_{t,k}} \\leq \\text{SB_buy}_t - \\text{SB_sell}_t + \\text{SB}_t \\quad \\forall t \\in \\text{Years}\n\\end{equation}\n\n- **Grain Growing**: Grain produced on land $l$ cannot exceed the production capacity of year $t$.\n\n\\begin{equation}\n\\text{GR}_{t,l} \\leq \\text{GR_yield}_l*\\text{GR_area}_l \\quad \\forall (t,l) \\in \\text{Years} \\times \\text{Lands}\n\\end{equation}\n\n- **Land Capacity**: Use of space in year $t$ cannot exceed available land.\n\n\\begin{equation}\n\\frac{1}{\\text{SB_yield}}*\\text{SB}_t + \\text{HF_land}*(\\text{Newborn}_t + \\text{Cow}_{t,1}) + \n\\end{equation}\n\n\\begin{equation}\n\\sum_{k \\in \\text{Ages} \\setminus \\{1,12\\}}{\\text{Cows}_{t,k}} + \n\\end{equation}\n\n\\begin{equation}\n\\sum_{l \\in \\text{Lands}}{\\frac{\\text{GR}_{t,l}}{\\text{GR_yield}_l}} \\leq \\text{Land_cap} \\quad \\forall t \\in \\text{Years}\n\\end{equation}\n\n- **Labor**: Labor required to run the farm in year $t$ cannot exceed contracted time plus overtime.\n\n\\begin{equation}\n\\text{HF_labor}*(\\text{Newborn}_t + \\text{Cow}_{t,1}) + \n\\end{equation}\n\n\\begin{equation}\n\\sum_{k \\in \\text{Ages} \\setminus \\{1,12\\}}{\\text{Cow_labor}*\\text{Cow}_{t,k}} + \n\\end{equation}\n\n\\begin{equation}\n\\sum_{l \\in \\text{Lands}}{\\frac{\\text{GR_labor}*\\text{GR}_{t,l}}{\\text{GR_yield}_l}} + \n\\end{equation}\n\n\\begin{equation}\n\\frac{\\text{SB_labor}*\\text{SB}_t}{\\text{SB_yield}} \\leq \\text{Labor_cap} + \\text{Overtime}_t \\qquad \\forall t \\in \\text{Years}\n\\end{equation}\n\n\n- **Continuity**: Livestock in year $t$ have to survive the previous year.\n\n\\begin{equation}\n\\text{Cows}_{t,1} = (1-\\text{HF_decay})*\\text{Newborn}_{t-1} \\quad \\forall t \\in \\text{Years} \\setminus \\{1\\}\n\\end{equation}\n\n\\begin{equation}\n\\text{Cows}_{t,2} = (1-\\text{HF_decay})*\\text{Cows}_{t-1,1} \\quad \\forall t \\in \\text{Years} \\setminus \\{1\\}\n\\end{equation}\n\n\\begin{equation}\n\\text{Cows}_{t,k+1} = (1-\\text{Cow_decay})*\\text{Cows}_{t-1,k} \\quad \\forall (t,k) \\in \\text{Years} \\setminus \\{1\\} \\times \\text{Ages} \\setminus \\{1,12\\}\n\\end{equation}\n\n- **Heifers Birth**: Heifers born in year $t$ depend on the number of dairy cows.\n\n\\begin{equation}\n\\text{Newborn}_t + \\text{HF_sell}_t = \\sum_{k \\in \\text{Ages} \\setminus \\{1,12\\}}{\\frac{\\text{Birthrate}}{2}*\\text{Cows}_{t,k}} \\quad \\forall t \\in \\text{Years}\n\\end{equation}\n\n- **Final Dairy Cows**: The number of dairy cows at the end of the planning horizon must be within tolerances.\n\n\\begin{equation}\n\\text{Min_final_cows} \\leq \\sum_{k \\in \\text{Ages} \\setminus \\{1,12\\}}{\\text{Cows}_{5,k}} \\leq \\text{Max_final_cows}\n\\end{equation}\n\n- **Initial Conditions**: Set the number of livestock available at the beginning of the planning horizon.\n\n\\begin{equation}\n\\text{Cows}_{1,1} = \\text{Initial_HF}\n\\end{equation}\n\n\\begin{equation}\n\\text{Cows}_{1,2} = \\text{Initial_HF}\n\\end{equation}\n\n\\begin{equation}\n\\text{Cows}_{1,k} = \\text{Initial_cows} \\quad \\forall k \\in \\text{Ages} \\setminus \\{1,2\\}\n\\end{equation}\n\n- **Yearly Profit**: Profit in year $t$ is driven by operations from crops and livestock, after accounting for labor, land and financial costs.\n\n\\begin{equation}\n\\text{Profit}_t = \\frac{\\text{BL_price}*\\text{Birthrate}}{2}*\\sum_{k \\in \\text{Ages} \\setminus \\{1,12\\}}{\\text{Cows}_{t,k}}\n+ \\text{HF_price}*\\text{HF_sell}_t\n+ \\text{Cow_price}*\\text{Cows}_{t,12}\n\\end{equation}\n\n\\begin{equation}\n+ \\text{Milk_price}*\\sum_{k \\in \\text{Ages} \\setminus \\{1,12\\}}{\\text{Cows}_{t,k}}\n\\end{equation}\n\n\\begin{equation}\n+ \\text{GR_price}*\\text{GR_sell}_t\n+ \\text{SB_price}*\\text{SB_sell}_t\n\\end{equation}\n\n\\begin{equation}\n- \\text{GR_cost}*\\text{GR_buy}_t\n- \\text{SB_cost}*\\text{SB_buy}_t\n\\end{equation}\n\n\\begin{equation}\n- \\text{Overtime_cost}*\\text{Overtime}_t\n- \\text{Regular_time_cost}\n\\end{equation}\n\n\\begin{equation}\n- \\text{HF_cost}*(\\text{Newborn}_t + \\text{Cows}_{t,1})\n- \\text{Cow_cost}*\\sum_{k \\in \\text{Ages} \\setminus \\{1,12\\}}{\\text{Cows}_{t,k}}\n\\end{equation}\n\n\\begin{equation}\n- \\text{GR_land_cost}*\\sum_{l \\in \\text{Lands}}{\\frac{\\text{GR}_{t,l}}{\\text{GR_yield}_l}}\n- \\text{SB_land_cost}*\\frac{\\text{SB}_t}{\\text{SB_yield}} \\\\\n- \\text{Installment}*\\sum_{d \\in \\text{Years}:d \\leq t}{\\text{Outlay}_d}\n\\quad \\forall t \\in \\text{Years}\n\\end{equation}\n\n---\n## Python Implementation\n\nWe import the Gurobi Python Module and other Python libraries.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\n\nimport gurobipy as gp\nfrom gurobipy import GRB\n\n# tested with Python 3.7.0 & Gurobi 9.0",
"_____no_output_____"
]
],
[
[
"## Input Data\nWe define all the input data of the model.",
"_____no_output_____"
]
],
[
[
"# Parameters\n\nyears = [1,2,3,4,5]\nlands = [1,2,3,4]\nages = [1,2,3,4,5,6,7,8,9,10,11,12]\ncow_ages = [2,3,4,5,6,7,8,9,10,11]\n\ngr_area = {1: 20.0, 2: 30.0, 3: 20.0, 4: 10.0}\ngr_yield = {1: 1.1, 2: 0.9, 3: 0.8, 4:0.65}\nsb_yield = 1.5\nhousing_cap = 130\ngr_intake = 0.6\nsb_intake = 0.7\nhf_land = 2/3.0\nland_cap = 200\nhf_labor = 10/100.0\ncow_labor = 42/100.0\ngr_labor = 4/100.0\nsb_labor = 14/100.0\nlabor_cap = 5500/100.0\ncow_decay = 0.02\nhf_decay = 0.05\ninitial_hf = 9.5\ninitial_cows = 9.8\nbirthrate = 1.1\nmin_final_cows = 50\nmax_final_cows = 175\nbl_price = 30\nhf_price = 40\ncow_price = 120\nmilk_price = 370\ngr_price = 75\nsb_price = 58\ngr_cost = 90\nsb_cost = 70\novertime_cost = 120\nregular_time_cost = 4000\nhf_cost = 50\ncow_cost = 100\ngr_land_cost = 15\nsb_land_cost = 10\ninstallment = 39.71",
"_____no_output_____"
]
],
[
[
"## Model Deployment\n\nWe create a model and the variables. For each, year we have continuous variables, which tell us how much sugar beet is grown (in tons), how much grain is bought (in tons), how much grain is sold (in tons), how much sugar beet is bought (in tons), how much sugar beet is sold (in tons), how much extra labor is recruited, how much capital outlay there is, how many heifers are sold at birth, how much profit there is, and how many cows are newborns (age 0).\n\nFor each year and each land group, there is a continuous variable, which tells us how much grain is grown on that land group. For each year and each cow age, there is a continuous variable that tells us how many cows exists in the current year of that age.",
"_____no_output_____"
]
],
[
[
"model = gp.Model('Farming')\nsb = model.addVars(years, vtype=GRB.CONTINUOUS, name=\"SB\")\ngr_buy = model.addVars(years, vtype=GRB.CONTINUOUS, name=\"GR_buy\")\ngr_sell = model.addVars(years, vtype=GRB.CONTINUOUS, name=\"GR_sell\")\nsb_buy = model.addVars(years, vtype=GRB.CONTINUOUS, name=\"SB_buy\")\nsb_sell = model.addVars(years, vtype=GRB.CONTINUOUS, name=\"SB_sell\")\novertime = model.addVars(years, vtype=GRB.CONTINUOUS, name=\"Overtime\")\noutlay = model.addVars(years, vtype=GRB.CONTINUOUS, name=\"Outlay\")\nhf_sell = model.addVars(years, vtype=GRB.CONTINUOUS, name=\"HF_sell\")\nnewborn = model.addVars(years, vtype=GRB.CONTINUOUS, name=\"Newborn\")\nprofit = model.addVars(years, vtype=GRB.CONTINUOUS, name=\"Profit\")\ngr = model.addVars(years, lands, vtype=GRB.CONTINUOUS, name=\"GR\")\ncows = model.addVars(years, ages, vtype=GRB.CONTINUOUS, name=\"Cows\")",
"Using license file c:\\gurobi\\gurobi.lic\nSet parameter TokenServer to value SANTOS-SURFACE-\n"
]
],
[
[
"Next we insert the constraints:\n\nThere is only a housing capacity for 130 cows per year. It is possible that there are more than 130 cows, but housing these additional cows will require additional costs related to renting houses, which is covered with the `Outlay` variables.",
"_____no_output_____"
]
],
[
[
"# 1. Housing capacity\n\nHousingCap = model.addConstrs((newborn[year] +\n cows[year,1] +\n gp.quicksum(cows[year,age] for age in cow_ages) -\n gp.quicksum(outlay[d] for d in years if d <= year)\n <= housing_cap for year in years), name=\"Housing_cap\")",
"_____no_output_____"
]
],
[
[
"After selling, buying, and producing grains and sugar beet, there needs to be enough of them in storage to feed all the cows.",
"_____no_output_____"
]
],
[
[
"# 2.1 Food consumption (Grain)\n\nGrainConsumption = model.addConstrs((gp.quicksum(gr_intake*cows[year, age] for age in cow_ages)\n <= gr_buy[year] - gr_sell[year] + gr.sum(year, '*')\n for year in years), name=\"Grain_consumption\")\n\n# 2.1 Food consumption (Sugar beet)\nSugarbeetConsumption = model.addConstrs((gp.quicksum(sb_intake*cows[year, age] for age in cow_ages)\n <= sb_buy[year] - sb_sell[year] + sb[year]\n for year in years), name=\"Sugar_beet_consumption\")",
"_____no_output_____"
]
],
[
[
"The grain produced on a land group cannot exceed the specified production capacity of that land group.",
"_____no_output_____"
]
],
[
[
"# 3. Grain growing\n\nGrainGrowing = model.addConstrs((gr[year, land] <= gr_yield[land]*gr_area[land]\n for year in years for land in lands), name=\"Grain_growing\")",
"_____no_output_____"
]
],
[
[
"Each cow needs a certain number of acres to support it. The amount depends on the age of the cow. There are 200 acres available at most.",
"_____no_output_____"
]
],
[
[
"# 4. Land capacity\n\nLandCap = model.addConstrs((sb[year]/sb_yield + hf_land*(newborn[year] + cows[year,1])\n + gp.quicksum((1/gr_yield[land])*gr[year, land] for land in lands)\n + gp.quicksum(cows[year, age] for age in cow_ages)\n <= land_cap for year in years), name=\"Land_capacity\")",
"_____no_output_____"
]
],
[
[
"Each cow and each acre requires a certain amount of worker time to maintain. The farm is currently able to provide a fixed number of worker hours in a year. Any additional work that needs to be done requires external workers, for which there will be an additional cost.",
"_____no_output_____"
]
],
[
[
"# 5. Labor\n\nLabor = model.addConstrs((hf_labor*(newborn[year] + cows[year,1])\n + gp.quicksum(cow_labor*cows[year, age] for age in cow_ages)\n + gp.quicksum(gr_labor/gr_yield[land]*gr[year,land] for land in lands)\n + sb_labor/sb_yield*sb[year] \n <= labor_cap + overtime[year] for year in years), name=\"Labor\")",
"_____no_output_____"
]
],
[
[
"Each year a certain percentage of the cows die, depending on their age.",
"_____no_output_____"
]
],
[
[
"# 6.1 Continuity\n\nContinuity1 = model.addConstrs((cows[year,1] == (1-hf_decay)*newborn[year-1] \n for year in years if year > min(years)),\n name=\"Continuity_a\")\n\n# 6.2 Continuity\n\nContinuity2 = model.addConstrs((cows[year,2] == (1-hf_decay)*cows[year-1,1] \n for year in years if year > min(years)),\n name=\"Continuity_b\")\n\n# 6.3 Continuity\n\nContinuity3 = model.addConstrs((cows[year,age+1] == (1-cow_decay)*cows[year-1,age] \n for year in years for age in cow_ages if year > min(years)),\n name=\"Continuity_c\")",
"_____no_output_____"
]
],
[
[
"Keep track of the number of cows; cows can come into/out of the model by being bought or sold or through birth.",
"_____no_output_____"
]
],
[
[
"# 7. Heifers birth\n\nHeifersBirth = model.addConstrs((newborn[year] + hf_sell[year] \n == gp.quicksum(birthrate/2*cows[year,age] for age in cow_ages) for year in years)\n , name=\"Heifers_birth\")",
"_____no_output_____"
]
],
[
[
"At the end of the five years, the farmer wants at least 50 and at most 175 diary cows.",
"_____no_output_____"
]
],
[
[
"# 8. Final dairy cows\nFinalDairyCows = model.addRange(gp.quicksum(cows[max(years), age] for age in cow_ages), min_final_cows, max_final_cows, name=\"Final_dairy_cows\" )",
"_____no_output_____"
]
],
[
[
"In the first year, there are 9.5 one-year-old cows and 9.5 two-year-old cows. In addition, there are 9.8 cows for each age from three to 12. Note that we are solving this as an LP model to make it easier to solve. This can lead to fractional values for variables, which are in reality integers. ",
"_____no_output_____"
]
],
[
[
"# 9.1-9.2 Initial conditions\n\nInitialHeifers = model.addConstrs((initial_hf == cows[1, age] for age in ages if age < 3),\n name=\"Initial_conditions\")\n\n# 9.3 Initial conditions\n\nInitialCows = model.addConstrs((initial_cows == cows[1, age] for age in ages if age >= 3),\n name=\"Initial_condition_cows\")",
"_____no_output_____"
]
],
[
[
"The following constraints determine yearly profits. The costs for labor currently total $\\$4,000$. The profit is influenced by the selling of bullocks and heifers, selling of 12-year-old-cows, selling of milk, selling of grain, selling of sugar beet, buying of grain, buying of sugar beet, labor costs, heifer costs, dairy cow costs, grain costs, sugar beet costs, and capital costs.",
"_____no_output_____"
]
],
[
[
"# 10. Yearly profit\n\nYearlyProfit = model.addConstrs((profit[year]\n == bl_price*birthrate/2*gp.quicksum(cows[year, age] for age in cow_ages)\n + hf_price*hf_sell[year] + cow_price*cows[year, 12]\n + milk_price*gp.quicksum(cows[year, age] for age in cow_ages)\n + gr_price*gr_sell[year] + sb_price*sb_sell[year]\n - gr_cost*gr_buy[year] - sb_cost*sb_buy[year]\n - overtime_cost*overtime[year] - regular_time_cost\n - hf_cost*(newborn[year] + cows[year,1])\n - cow_cost*gp.quicksum(cows[year, age] for age in cow_ages)\n - gr_land_cost*gp.quicksum(gr[year, land]/gr_yield[land] for land in lands)\n - sb_land_cost*sb[year]/sb_yield\n - installment*gp.quicksum(outlay[d] for d in years if d <= year)\n for year in years), name=\"Yearly_profit\")",
"_____no_output_____"
]
],
[
[
"The total profit of the planning horizon consists of the calculated profits minus the loan payments that remain pending:",
"_____no_output_____"
]
],
[
[
"# 0. Total profit\n\nmodel.setObjective(gp.quicksum(profit[year] - installment*(year+4)*outlay[year] for year in years), GRB.MAXIMIZE)",
"_____no_output_____"
]
],
[
[
"Next, we start the optimization and Gurobi finds the optimal solution.",
"_____no_output_____"
]
],
[
[
"model.optimize()",
"Gurobi Optimizer version 9.0.0 build v9.0.0rc2 (win64)\nOptimize a model with 116 rows, 131 columns and 734 nonzeros\nModel fingerprint: 0x1612a027\nCoefficient statistics:\n Matrix range [4e-02, 3e+02]\n Objective range [1e+00, 4e+02]\n Bounds range [1e+02, 1e+02]\n RHS range [7e+00, 4e+03]\nPresolve removed 84 rows and 67 columns\nPresolve time: 0.01s\nPresolved: 32 rows, 64 columns, 252 nonzeros\n\nIteration Objective Primal Inf. Dual Inf. Time\n 0 5.1200000e+32 5.000000e+30 5.120000e+02 0s\n 24 1.2171917e+05 0.000000e+00 0.000000e+00 0s\n\nSolved in 24 iterations and 0.01 seconds\nOptimal objective 1.217191729e+05\n"
]
],
[
[
"---\n## Analysis\n\nThe optimal plan results in a total profit of $\\$121,719.17$ over the five-year period the model covers. The detailed plan for each year is as follows.\n\n### Financial Plan\n\nThis plan determines the profit and outlay at each period of the planning horizon. For example, the profit in year 1 is $\\$21,906.1$ and the outlay is zero dollars.",
"_____no_output_____"
]
],
[
[
"rows = [\"Profit\", \"Outlay\"]\ncolumns = years.copy()\nfinance_plan = pd.DataFrame(columns=columns, index=rows, data=0.0)\n\nfor year in years:\n if (abs(profit[year].x) > 1e-6):\n finance_plan.loc[\"Profit\", year] = np.round(profit[year].x, 1)\n if (abs(outlay[year].x) > 1e-6):\n finance_plan.loc[\"Outlay\", year] = np.round(outlay[year].x, 1)\nfinance_plan",
"_____no_output_____"
]
],
[
[
"### Plan for Grains\nThis plan defines the number of tons of grain to grow for each type of land group (rows) at each year of the planning horizon (columns). It also defines the number of tons of grain to buy and sell at each year of the planning horizon. For example, we are going to grow 22 tons of grain at land group 1 during year 2 . In addition, we are going to buy 35.1 tons of grain during year 2.",
"_____no_output_____"
]
],
[
[
"rows = lands.copy() + [\"Buy\", \"Sell\"]\ncolumns = years.copy()\ngr_plan = pd.DataFrame(columns=columns, index=rows, data=0.0)\n\nfor year, land in gr.keys():\n if (abs(gr[year, land].x) > 1e-6):\n gr_plan.loc[land, year] = np.round(gr[year, land].x, 1)\nfor year in years:\n if (abs(gr_buy[year].x) > 1e-6):\n gr_plan.loc[\"Buy\", year] = np.round(gr_buy[year].x, 1)\n if (abs(gr_sell[year].x) > 1e-6):\n gr_plan.loc[\"Sell\", year] = np.round(gr_sell[year].x, 1)\ngr_plan",
"_____no_output_____"
]
],
[
[
"### Plan for Sugar Beet\nThis plan defines the number of tons of sugar beet to grow at each year of the planning horizon (columns). It also defines the number of tons of sugar beet to buy and sell at each year of the planning horizon. For example, we are going to grow 94 tons of sugar beet during year 2 . In addition, we are going to sell 27.4 tons of sugar beet during year 2.",
"_____no_output_____"
]
],
[
[
"rows = [\"Grow\", \"Buy\", \"Sell\"]\ncolumns = years.copy()\nsb_plan = pd.DataFrame(columns=columns, index=rows, data=0.0)\n\nfor year in years:\n if (abs(sb[year].x) > 1e-6):\n sb_plan.loc[\"Grow\", year] = np.round(sb[year].x, 1)\n if (abs(sb_buy[year].x) > 1e-6):\n sb_plan.loc[\"Buy\", year] = np.round(sb_buy[year].x, 1)\n if (abs(sb_sell[year].x) > 1e-6):\n sb_plan.loc[\"Sell\", year] = np.round(sb_sell[year].x, 1)\nsb_plan",
"_____no_output_____"
]
],
[
[
"### Plan for Heifers\n\nThis plan shows number of heifers to sell and raise at each period of the planning horizon. For example, we are going to sell 40.8 heifers and raise 11.6 heifers during year 2.",
"_____no_output_____"
]
],
[
[
"rows = [\"Sell\", \"Raise\"]\ncolumns = years.copy()\nlivestock_plan = pd.DataFrame(columns=columns, index=rows, data=0.0)\n\nfor year in years:\n if (abs(hf_sell[year].x) > 1e-6):\n livestock_plan.loc[\"Sell\", year] = np.round(hf_sell[year].x, 1)\n if (abs(newborn[year].x) > 1e-6):\n livestock_plan.loc[\"Raise\", year] = np.round(newborn[year].x, 1)\nlivestock_plan",
"_____no_output_____"
]
],
[
[
"---\n## References\n\nH. Paul Williams, Model Building in Mathematical Programming, fifth edition.\n\nCopyright © 2020 Gurobi Optimization, LLC",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eceee5c969ffc33fd8314e62a3890534a16ece0c | 7,992 | ipynb | Jupyter Notebook | docs/lectures/lecture30/notebook/s8-a1-challenge.ipynb | r34g4n/2020-CS109A | 665100fec24309edb818a51bc8c29db2912d370f | [
"MIT"
] | 81 | 2020-08-17T10:18:50.000Z | 2022-03-14T00:10:17.000Z | docs/lectures/lecture30/notebook/s8-a1-challenge.ipynb | SBalas/2020-CS109A | 3eb01ac57adbef09c7dbb10eda7408dd4545b3f7 | [
"MIT"
] | 1 | 2022-02-09T06:15:51.000Z | 2022-02-09T12:42:44.000Z | docs/lectures/lecture30/notebook/s8-a1-challenge.ipynb | SBalas/2020-CS109A | 3eb01ac57adbef09c7dbb10eda7408dd4545b3f7 | [
"MIT"
] | 95 | 2020-08-29T22:49:34.000Z | 2022-03-25T18:36:13.000Z | 30.387833 | 269 | 0.588463 | [
[
[
"# Title\n\n**Exercise: A.1- MLP using Keras**\n\n# Description\n\nThe aim of this exercise is to come up with a simple Multi-layer perceptron classifier using tensorflow.",
"_____no_output_____"
],
[
"<img src=\"../img/image.png\" style=\"width: 500px;\">",
"_____no_output_____"
],
[
"The dataset used here is the Iris dataset, same as the one used for logistic regression classification. This dataset has several features such as sepal length, sepal width and so on, to predict which of the iris species that particular flower belongs to here. \n\n# Instructions:\n1. Read the csv file as a pandas dataframe.\n2. Assign the dependent and independent variables. The species is your dependent variable. All the other columns are your predictors.\n3. Split the dataset into train and validation sets.\n4. Define the network parameters for the MLP.\n5. Initialise the weights and biases of the network.\n6. Define the MLP model with input, hidden and output layers.\n7. Fit the model on the training data.\n8. Compute and print the train and validation accuracy.\n9. Compare the output of the MLP classifier with that of the logistic model you had earlier fit in `Session 5 Exercise B.1`. Find out which model performs better and why it does so?\n\n# Hints:\n\n\n<a href=\"https://keras.io/api/layers/merging_layers/add/\" target=\"_blank\">keras.add()</a> : To add a layer to the model\n\n\n<a href=\"https://keras.io/api/models/sequential/\" target=\"_blank\">keras.fit()</a> : Fit the model for the data\n\n\n<a href=\"https://www.tensorflow.org/api_docs/python/tf/keras/Model\" target=\"_blank\">model.evaluate()</a> : Evaluate model performance on predictors vs true values\n\nNote: This exercise is **auto-graded and you can try multiple attempts.**",
"_____no_output_____"
]
],
[
[
"# Import the necessary libraries\nimport tensorflow as tf\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.model_selection import train_test_split\nimport pandas as pd\n%matplotlib inline\n\ntf.keras.backend.clear_session() # For easy reset of notebook state.",
"_____no_output_____"
],
[
"# Read the file 'IRIS.csv'\n\ndf = pd.read_csv('IRIS.csv')",
"_____no_output_____"
],
[
"# Take a quick look at the dataset\ndf.head()",
"_____no_output_____"
],
[
"# We use one-hot-encoding to encode the 'species' labels using pd.get_dummies\n\none_hot_df = pd.get_dummies(df[___], prefix='species')",
"_____no_output_____"
],
[
"# The predictor variables are all columns except species\nX = df.drop([___],axis=1).values\n\n# The response variable is the one-hot-encoded species values\ny = one_hot_df.values",
"_____no_output_____"
],
[
"# We divide our data into test and train sets with 80% training size\n\nX_train, X_test, y_train, y_test = train_test_split(___,___,train_size=___)",
"_____no_output_____"
],
[
"# To build the MLP, we will use the keras library\n\nmodel = tf.keras.models.Sequential(name='MLP')\n\n# To initialise our model we set some parameters \n# commonly defined in an MLP design\n\n# The number of nodes in a hidden layer\nn_hidden = ___\n\n# The number of nodes in the input layer (features)\nn_input = ___\n\n# The number of nodes in the output layer\nn_output = ___",
"_____no_output_____"
],
[
"# We add the first hidden layer with `n_hidden` number of neurons \n# and 'relu' activation\nmodel.add((tf.keras.layers.Dense(n_hidden,input_dim=___, activation = ___,name='hidden')))\n\n# The second layer is the final layer in our case, using 'softmax' on the output labels\nmodel.add(tf.keras.layers.Dense(n_output, activation = ___,name='output'))",
"_____no_output_____"
],
[
"# Now we compile the model using 'categorical_crossentropy' loss, \n# optimizer as 'sgd' and 'accuracy' as a metric\nmodel.compile(optimizer=___,\n loss=___,\n metrics=[___])",
"_____no_output_____"
],
[
"# You can see an overview of the model you built using .summary()\nmodel.summary()",
"_____no_output_____"
],
[
"# We fit the model, and save it to a variable 'history' that can be \n# accessed later to analyze the training profile\n# We also set validation_split=0.2 for 20% of training data to be \n# used for validation\n# verbose=0 means you will not see the output after every epoch. \n# Set verbose=1 to see it\n\nhistory = model.fit(___,___, epochs = ___, batch_size = 16,verbose=0,validation_split=___)",
"_____no_output_____"
],
[
"# Here we plot the training and validation loss and accuracy\n\nfig, ax = plt.subplots(1,2,figsize = (16,4))\nax[0].plot(history.history['loss'],'r',label = 'Training Loss')\nax[0].plot(history.history['val_loss'],'b',label = 'Validation Loss')\nax[1].plot(history.history['accuracy'],'r',label = 'Training Accuracy')\nax[1].plot(history.history['val_accuracy'],'b',label = 'Validation Accuracy')\nax[0].legend()\nax[1].legend()\nax[0].set_xlabel('Epochs')\nax[1].set_xlabel('Epochs');\nax[0].set_ylabel('Loss')\nax[1].set_ylabel('Accuracy %');\nfig.suptitle('MLP Training', fontsize = 24)",
"_____no_output_____"
],
[
"### edTest(test_accuracy) ###\n# Once you have near-perfect validation accuracy, time to evaluate model performance on test set \n\ntrain_accuracy = model.evaluate(___,___)[1]\ntest_accuracy = model.evaluate(___,___)[1]\nprint(f'The training set accuracy for the model is {train_accuracy}\\\n \\n The test set accuracy for the model is {test_accuracy}')",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eceee8407dbc06d6f85a67f7bb1bd490c3ed3305 | 15,848 | ipynb | Jupyter Notebook | notebooks/JHU_COVID-19.ipynb | JoaoRodrigues/COVID-19-data | 9f79602f5523ab76400c324ea4d09a393f0f2fa8 | [
"BSD-3-Clause"
] | 1 | 2020-03-25T18:09:22.000Z | 2020-03-25T18:09:22.000Z | notebooks/JHU_COVID-19.ipynb | JoaoRodrigues/COVID-19-data | 9f79602f5523ab76400c324ea4d09a393f0f2fa8 | [
"BSD-3-Clause"
] | null | null | null | notebooks/JHU_COVID-19.ipynb | JoaoRodrigues/COVID-19-data | 9f79602f5523ab76400c324ea4d09a393f0f2fa8 | [
"BSD-3-Clause"
] | null | null | null | 32.276986 | 326 | 0.493879 | [
[
[
"# 2019 Novel Coronavirus (SARS-CoV-2) and COVID-19 Unpivoted Data\n\nThe following script takes data from the repository of the 2019 Novel Coronavirus Visual Dashboard operated by Johns Hopkins University's Center for Systems Science and Engineering (JHU CSSE). It will apply necessary cleansing/reformatting to make it use in traditional relational databases and data visualization tools.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport os\nfrom datetime import datetime, date, timedelta\nimport pycountry\nfrom copy import deepcopy",
"_____no_output_____"
],
[
"# papermill parameters\noutput_folder = \"../output/\"",
"_____no_output_____"
]
],
[
[
"Data until 22MAR2020 is stored in a cache. This is collated and reshaped data from previous days.",
"_____no_output_____"
]
],
[
[
"pre_2203_data = pd.read_csv(\"https://s3-us-west-1.amazonaws.com/starschema.covid/CSSEGISandData_COVID-19_until_0322.csv\",keep_default_na=False)",
"_____no_output_____"
]
],
[
[
"Daily reports from and including 23MAR2020 are downloaded from the JHU CSSE GIS and Data Github repository.",
"_____no_output_____"
]
],
[
[
"def urls():\n return [template.format(month=dt.month, day=dt.day, year=dt.year) for dt in dates]",
"_____no_output_____"
],
[
"def retrieve_and_merge():\n dates = [date(year=2020, month=3, day=23) + timedelta(n) for n in range(int((datetime.now().date() - datetime(year=2020, month=3, day=23).date()).days))]\n template = \"https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_daily_reports/{month:02d}-{day:02d}-{year}.csv\"\n \n res = pd.DataFrame()\n \n for dt in dates:\n df = pd.read_csv(template.format(year=dt.year,\n month=dt.month,\n day=dt.day),keep_default_na=False)\n df[\"Date\"] = dt\n res = res.append(df, ignore_index=True)\n return res.melt(id_vars=[col for col in res.columns if col not in [\"Confirmed\", \"Deaths\", \"Recovered\", \"Active\"]],\n var_name=\"Case_Type\",\n value_name=\"Cases\").drop([\"Last_Update\"], axis=1).rename(columns={\"Long_\": \"Long\",\n \"Country_Region\": \"Country/Region\",\n \"Province_State\": \"Province/State\"})",
"_____no_output_____"
],
[
"df = retrieve_and_merge()",
"_____no_output_____"
]
],
[
[
"The original dataset stores the number of `Cases` for a given day in columns. \nThis is not useful for reporting, thus we move these date columns to rows:",
"_____no_output_____"
],
[
"## Data Quality",
"_____no_output_____"
],
[
"We use `pycountry` to resolve geographies.",
"_____no_output_____"
],
[
"A number of states have inconsistent naming or special characters, such as `Taiwan*`. These are normalised through a replacement `dict` with ISO3166-1 compliant names. Data is then aggregated for each division by date and case type.",
"_____no_output_____"
]
],
[
[
"changed_names = {\n \"Holy See\": \"Holy See (Vatican City State)\",\n \"Vatican City\": \"Holy See (Vatican City State)\",\n \"Hong Kong SAR\": \"Hong Kong\",\n \"Iran (Islamic Republic of)\": \"Iran, Islamic Republic of\",\n \"Iran\": \"Iran, Islamic Republic of\",\n \"Macao SAR\": \"Macao\",\n \"Macau\": \"Macao\",\n \"Republic of Korea\": \"Korea, Republic of\",\n \"South Korea\": \"Korea, Republic of\",\n \"Korea, South\": \"Korea, Republic of\",\n \"Republic of Moldova\": \"Moldova, Republic of\",\n \"Russia\": \"Russian Federation\",\n \"Saint Martin\": \"Sint Maarten (Dutch part)\",\n \"St. Martin\": \"Sint Maarten (Dutch part)\",\n \"Taipei and environs\": \"Taiwan, Province of China\",\n \"Vietnam\": \"Viet Nam\",\n \"occupied Palestinian territory\": \"Palestine, State of\",\n \"Taiwan*\": \"Taiwan, Province of China\",\n \"Congo (Brazzaville)\": \"Congo\",\n \"Congo (Kinshasa)\": \"Congo, The Democratic Republic of the\",\n \"Gambia, The\": \"Gambia\",\n \"The Gambia\": \"Gambia\",\n \"Tanzania\": \"Tanzania, United Republic of\",\n \"US\": \"United States\",\n \"Curacao\": \"Curaçao\",\n \"Brunei\": \"Brunei Darussalam\",\n \"Cote d'Ivoire\": \"Côte d'Ivoire\",\n \"Moldova\": \"Moldova, Republic of\",\n \"The Bahamas\": \"Bahamas\",\n \"Venezuela\": \"Venezuela, Bolivarian Republic of\",\n \"Bolivia\": \"Bolivia, Plurinational State of\",\n \"East Timor\": \"Timor-Leste\",\n \"Cape Verde\": \"Cabo Verde\"\n}\n\ndef normalize_names(df):\n df[\"Country/Region\"] = df[\"Country/Region\"].replace(changed_names)\n df[\"Cases\"] = df[\"Cases\"].replace('',0).astype(int)\n \n return(df.groupby(by=[\"Country/Region\",\"Province/State\", \"Date\", \"Case_Type\"], as_index=False).agg({\"Cases\": \"sum\", \"Long\": \"first\", \"Lat\": \"first\"}))",
"_____no_output_____"
],
[
"df = normalize_names(df)",
"_____no_output_____"
]
],
[
[
"## Adding ISO3166-1 and ISO3166-2 identifiers",
"_____no_output_____"
],
[
"To facilitate easy recognition, ISO3166-1 identifiers are added to all countries and ISO3166-2 identifiers are added where appropriate. This is the case where subregional data exists:\n\n* Australia\n* Canada\n* France (`France` for metropolitan France, separate regions for DOM/TOMs\n* PRC\n* US\n* UK (the `UK` province identifier encompasses only Great Britain and Northern Ireland, other dependencies reporting to the UK authorities are separate subdivisions)\n* The Kingdom of the Netherlands (`Netherlands` encompasses the constituent country of the Netherlands, and the other constituent countries register cases as separate provinces of the Kingdom of the Netherlands)",
"_____no_output_____"
]
],
[
[
"def resolve_iso3166_1(row):\n if row[\"Country/Region\"] is not \"Cruise Ship\":\n if pycountry.countries.get(name=row[\"Country/Region\"]):\n row[\"ISO3166-1\"] = pycountry.countries.get(name=row[\"Country/Region\"]).alpha_2\n else:\n row[\"ISO3166-1\"] = \"\"\n return row",
"_____no_output_____"
],
[
"df = df.apply(resolve_iso3166_1, axis=1)",
"_____no_output_____"
]
],
[
[
"We then encode level 2 IDs:",
"_____no_output_____"
]
],
[
[
"fr_subdivisions = {\"France\": \"FR\",\n \"French Guiana\": \"GF\",\n \"French Polynesia\": \"PF\",\n \"Guadeloupe\": \"GUA\",\n \"Mayotte\": \"YT\",\n \"Reunion\": \"RE\",\n \"Saint Barthelemy\": \"BL\",\n \"St Martin\": \"MF\"}\n\nnl_subdivisions = {\"Netherlands\": \"NL\",\n \"Aruba\": \"AW\",\n \"Curacao\": \"CW\"}\n\ncn_subdivisions = {'Jilin': 'CN-JL',\n 'Xizang': 'CN-XZ',\n 'Anhui': 'CN-AH',\n 'Jiangsu': 'CN-JS',\n 'Yunnan': 'CN-YN',\n 'Beijing': 'CN-BJ',\n 'Jiangxi': 'CN-JX',\n 'Zhejiang': 'CN-ZJ',\n 'Chongqing': 'CN-CQ',\n 'Liaoning': 'CN-LN',\n 'Fujian': 'CN-FJ',\n 'Guangdong': 'CN-GD',\n 'Inner Mongolia': 'CN-NM',\n 'Gansu': 'CN-GS',\n 'Ningxia': 'CN-NX',\n 'Guangxi': 'CN-GX',\n 'Qinghai': 'CN-QH',\n 'Guizhou': 'CN-GZ',\n 'Sichuan': 'CN-SC',\n 'Henan': 'CN-HA',\n 'Shandong': 'CN-SD',\n 'Hubei': 'CN-HB',\n 'Shanghai': 'CN-SH',\n 'Hebei': 'CN-HE',\n 'Shaanxi': 'CN-SN',\n 'Hainan': 'CN-HI',\n 'Shanxi': 'CN-SX',\n 'Tianjin': 'CN-TJ',\n 'Heilongjiang': 'CN-HL',\n 'Hunan': 'CN-HN',\n 'Xinjiang': 'CN-XJ',\n 'Tibet': \"CN-XZ\"}\n\nuk_subdivisions = {\"United Kingdom\": \"UK\",\n \"Cayman Islands\": \"KY\",\n \"Channel Islands\": \"CHA\",\n \"Gibraltar\": \"GI\",\n \"Montserrat\": \"MS\"}\n\nsubdivisions = {\n \"AU\": {subdivision.name: subdivision.code.replace(\"AU-\", \"\") for subdivision in pycountry.subdivisions.get(country_code=\"AU\")},\n \"CA\": {subdivision.name: subdivision.code.replace(\"CA-\", \"\") for subdivision in pycountry.subdivisions.get(country_code=\"CA\")},\n \"US\": {subdivision.name: subdivision.code.replace(\"US-\", \"\") for subdivision in pycountry.subdivisions.get(country_code=\"US\")},\n \"GB\": uk_subdivisions,\n \"CN\": cn_subdivisions,\n \"NL\": nl_subdivisions,\n \"FR\": fr_subdivisions\n}",
"_____no_output_____"
],
[
"countries_with_subdivisions = list(subdivisions.keys())\n\ndef resolve_iso3166_2(row):\n if row[\"ISO3166-1\"] in countries_with_subdivisions:\n row[\"ISO3166-2\"] = subdivisions[row[\"ISO3166-1\"]].get(row[\"Province/State\"])\n else:\n row[\"ISO3166-2\"] = \"\"\n return row",
"_____no_output_____"
],
[
"df = df.apply(resolve_iso3166_2, axis=1)",
"_____no_output_____"
]
],
[
[
"## Calculating case changes",
"_____no_output_____"
],
[
"We concatenate the new import data frame, `df`, with the pre-23MAR2020 data to facilitate calculating differences, and sort it by primary keys and `Date`.",
"_____no_output_____"
]
],
[
[
"result = pre_2203_data.append(df, sort=\"False\")\nresult[\"Date\"] = pd.to_datetime(result[\"Date\"])",
"_____no_output_____"
]
],
[
[
"Next, we sort the data by primary keys and `Date` to ensure we can add a `Difference` column as a window function.",
"_____no_output_____"
]
],
[
[
"result = result.sort_values(by=[\"Country/Region\", \"Province/State\", \"Case_Type\", \"Date\"], ascending=True)",
"_____no_output_____"
],
[
"result[\"Difference\"] = result[\"Cases\"] - result.groupby([\"Country/Region\", \"Province/State\", \"Case_Type\"])[\"Cases\"].shift(periods=1)",
"_____no_output_____"
],
[
"result[\"Difference\"] = result[\"Difference\"].fillna(0)",
"_____no_output_____"
]
],
[
[
"## Adding timestamp",
"_____no_output_____"
],
[
"Before we save the file locally, we add the `Last_Update_Date` in `UTC` time zone.",
"_____no_output_____"
]
],
[
[
"result[\"Last_Update_Date\"] = datetime.utcnow()",
"_____no_output_____"
]
],
[
[
"## Output",
"_____no_output_____"
],
[
"Finally, we store the output in the `output` folder as `JHU_COVID-19.csv` as an unindexed CSV file.",
"_____no_output_____"
]
],
[
[
"result.to_csv(output_folder + \"JHU_COVID-19.csv\", index=False, columns=[\"Country/Region\",\n \"Province/State\",\n \"Date\",\n \"Case_Type\",\n \"Cases\",\n \"Long\",\n \"Lat\",\n \"ISO3166-1\",\n \"ISO3166-2\",\n \"Difference\",\n \"Last_Update_Date\"])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
eceef4d9b3dfc6e7cc8b1392a2bb46496ab70150 | 223,787 | ipynb | Jupyter Notebook | examples/Tutorial 13.ipynb | AzipSauhabah/Riskfolio-Lib | d02b03e665cfa8b12e41cb441c75c6fbcf1a4365 | [
"BSD-3-Clause"
] | null | null | null | examples/Tutorial 13.ipynb | AzipSauhabah/Riskfolio-Lib | d02b03e665cfa8b12e41cb441c75c6fbcf1a4365 | [
"BSD-3-Clause"
] | null | null | null | examples/Tutorial 13.ipynb | AzipSauhabah/Riskfolio-Lib | d02b03e665cfa8b12e41cb441c75c6fbcf1a4365 | [
"BSD-3-Clause"
] | null | null | null | 250.882287 | 77,916 | 0.889913 | [
[
[
"# Riskfolio-Lib Tutorial: \n<br>__[Financionerioncios](https://financioneroncios.wordpress.com)__\n<br>__[Orenji](https://www.orenj-i.net)__\n<br>__[Riskfolio-Lib](https://riskfolio-lib.readthedocs.io/en/latest/)__\n<br>__[Dany Cajas](https://www.linkedin.com/in/dany-cajas/)__\n<a href='https://ko-fi.com/B0B833SXD' target='_blank'><img height='36' style='border:0px;height:36px;' src='https://cdn.ko-fi.com/cdn/kofi1.png?v=2' border='0' alt='Buy Me a Coffee at ko-fi.com' /></a> \n\n## Part XIII: Riskfolio-Lib and Xlwings\n\n## 1. Downloading the data:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport yfinance as yf\nimport warnings\n\nwarnings.filterwarnings(\"ignore\")\n\nyf.pdr_override()\npd.options.display.float_format = '{:.4%}'.format\n\n# Date range\nstart = '2016-01-01'\nend = '2019-12-30'\n\n# Tickers of assets\nassets = ['JCI', 'TGT', 'CMCSA', 'CPB', 'MO', 'NBL', 'APA', 'MMC', 'JPM',\n 'ZION', 'PSA', 'BAX', 'BMY', 'LUV', 'PCAR', 'TXT', 'DHR',\n 'DE', 'MSFT', 'HPQ', 'SEE', 'VZ', 'CNP', 'NI']\nassets.sort()\n\n# Downloading data\ndata = yf.download(assets, start = start, end = end)\ndata = data.loc[:,('Adj Close', slice(None))]\ndata.columns = assets",
"[*********************100%***********************] 24 of 24 completed\n"
],
[
"# Calculating returns\n\nY = data[assets].pct_change().dropna()\n\ndisplay(Y.head())",
"_____no_output_____"
]
],
[
[
"## 2. Estimating Mean Variance Portfolios\n\n### 2.1 Calculating the portfolio that maximizes Sharpe ratio.",
"_____no_output_____"
]
],
[
[
"import riskfolio.Portfolio as pf\n\n# Building the portfolio object\nport = pf.Portfolio(returns=Y)\n# Calculating optimum portfolio\n\n# Select method and estimate input parameters:\n\nmethod_mu='hist' # Method to estimate expected returns based on historical data.\nmethod_cov='hist' # Method to estimate covariance matrix based on historical data.\n\nport.assets_stats(method_mu=method_mu, method_cov=method_cov, d=0.94)\n\n# Estimate optimal portfolio:\n\nmodel='Classic' # Could be Classic (historical), BL (Black Litterman) or FM (Factor Model)\nrm = 'MV' # Risk measure used, this time will be variance\nobj = 'Sharpe' # Objective function, could be MinRisk, MaxRet, Utility or Sharpe\nhist = True # Use historical scenarios for risk measures that depend on scenarios\nrf = 0 # Risk free rate\nl = 0 # Risk aversion factor, only useful when obj is 'Utility'\n\nw = port.optimization(model=model, rm=rm, obj=obj, rf=rf, l=l, hist=hist)\n\ndisplay(w.T)",
"_____no_output_____"
]
],
[
[
"### 2.2 Plotting portfolio composition",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport riskfolio.PlotFunctions as plf\n\n# Plotting the composition of the portfolio\n\nfig_1, ax_1 = plt.subplots(figsize=(10,6))\n\nax_1 = plf.plot_pie(w=w, title='Sharpe Mean Variance', others=0.05, nrow=25, cmap = \"tab20\",\n height=6, width=10, ax=ax_1)",
"_____no_output_____"
]
],
[
[
"### 2.3 Calculate efficient frontier",
"_____no_output_____"
]
],
[
[
"points = 50 # Number of points of the frontier\n\nfrontier = port.efficient_frontier(model=model, rm=rm, points=points, rf=rf, hist=hist)\n\ndisplay(frontier.T.head())",
"_____no_output_____"
],
[
"# Plotting the efficient frontier\n\nlabel = 'Max Risk Adjusted Return Portfolio' # Title of point\nmu = port.mu # Expected returns\ncov = port.cov # Covariance matrix\nreturns = port.returns # Returns of the assets\n\nfig_2, ax_2 = plt.subplots(figsize=(10,6))\n\nplf.plot_frontier(w_frontier=frontier, mu=mu, cov=cov, returns=returns, rm=rm,\n rf=rf, alpha=0.01, cmap='viridis', w=w, label=label,\n marker='*', s=16, c='r', height=6, width=10, ax=ax_2)",
"_____no_output_____"
],
[
"# Plotting efficient frontier composition\n\nfig_3, ax_3 = plt.subplots(figsize=(10,6))\n\nplf.plot_frontier_area(w_frontier=frontier, cmap=\"tab20\", height=6, width=10, ax=ax_3)",
"_____no_output_____"
]
],
[
[
"## 3. Combining Riskfolio-Lib and Xlwings\n\n### 3.1 Creating an Empty Excel Workbook",
"_____no_output_____"
]
],
[
[
"import xlwings as xw\n\n# Creating an empty Excel Workbook\n\nwb = xw.Book() \nsheet1 = wb.sheets[0]\nsheet1.name = 'Charts'\nsheet2 = wb.sheets.add('Frontier')\nsheet3 = wb.sheets.add('Optimal Weights')",
"_____no_output_____"
]
],
[
[
"### 3.2 Adding Pictures to Sheet 1",
"_____no_output_____"
]
],
[
[
"sheet1.pictures.add(fig_1, name = \"Weights\",\n update = True, \n top = sheet1.range(\"A1\").top,\n left = sheet1.range(\"A1\").left)\n\nsheet1.pictures.add(fig_2, name = \"Frontier\",\n update = True, \n top = sheet1.range(\"M1\").top,\n left = sheet1.range(\"M1\").left)\n\nsheet1.pictures.add(fig_3, name = \"Composition\",\n update = True, \n top = sheet1.range(\"A30\").top,\n left = sheet1.range(\"A30\").left)",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/dcajasn/Riskfolio-Lib/master/examples/Fig1.png\">",
"_____no_output_____"
],
[
"### 3.2 Adding Data to Sheet 2 and Sheet 3",
"_____no_output_____"
]
],
[
[
"# Writing the weights of the frontier in the Excel Workbook\n\nsheet2.range('A1').value = frontier.applymap('{:.6%}'.format)\n\n# Writing the optimal weights in the Excel Workbook\n\nsheet3.range('A1').value = w.applymap('{:.6%}'.format)",
"_____no_output_____"
]
],
[
[
"<img src=\"https://raw.githubusercontent.com/dcajasn/Riskfolio-Lib/master/examples/Fig2.png\">\n<img src=\"https://raw.githubusercontent.com/dcajasn/Riskfolio-Lib/master/examples/Fig3.png\">",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eceefaa86965b483b4bfe84c296c7277db167973 | 35,768 | ipynb | Jupyter Notebook | intro-to-pytorch/Part 3 - Training Neural Networks (Exercises).ipynb | manucalop/deep-learning-v2-pytorch | 5f67a46abf465c2b8a025bd823a41bec10e55368 | [
"MIT"
] | null | null | null | intro-to-pytorch/Part 3 - Training Neural Networks (Exercises).ipynb | manucalop/deep-learning-v2-pytorch | 5f67a46abf465c2b8a025bd823a41bec10e55368 | [
"MIT"
] | null | null | null | intro-to-pytorch/Part 3 - Training Neural Networks (Exercises).ipynb | manucalop/deep-learning-v2-pytorch | 5f67a46abf465c2b8a025bd823a41bec10e55368 | [
"MIT"
] | null | null | null | 51.390805 | 9,112 | 0.68237 | [
[
[
"# Training Neural Networks\n\n[Video](https://youtu.be/9ILiZwbi9dA)\n\nThe network we built in the previous part isn't so smart, it doesn't know anything about our handwritten digits. Neural networks with non-linear activations work like universal function approximators. There is some function that maps your input to the output. For example, images of handwritten digits to class probabilities. The power of neural networks is that we can train them to approximate this function, and basically any function given enough data and compute time.\n\n<img src=\"assets/function_approx.png\" width=500px>\n\nAt first the network is naive, it doesn't know the function mapping the inputs to the outputs. We train the network by showing it examples of real data, then adjusting the network parameters such that it approximates this function.\n\nTo find these parameters, we need to know how poorly the network is predicting the real outputs. For this we calculate a **loss function** (also called the cost), a measure of our prediction error. For example, the mean squared loss is often used in regression and binary classification problems\n\n$$\n\\large \\ell = \\frac{1}{2n}\\sum_i^n{\\left(y_i - \\hat{y}_i\\right)^2}\n$$\n\nwhere $n$ is the number of training examples, $y_i$ are the true labels, and $\\hat{y}_i$ are the predicted labels.\n\nBy minimizing this loss with respect to the network parameters, we can find configurations where the loss is at a minimum and the network is able to predict the correct labels with high accuracy. We find this minimum using a process called **gradient descent**. The gradient is the slope of the loss function and points in the direction of fastest change. To get to the minimum in the least amount of time, we then want to follow the gradient (downwards). You can think of this like descending a mountain by following the steepest slope to the base.\n\n<img src='assets/gradient_descent.png' width=350px>",
"_____no_output_____"
],
[
"## Backpropagation\n\nFor single layer networks, gradient descent is straightforward to implement. However, it's more complicated for deeper, multilayer neural networks like the one we've built. Complicated enough that it took about 30 years before researchers figured out how to train multilayer networks.\n\nTraining multilayer networks is done through **backpropagation** which is really just an application of the chain rule from calculus. It's easiest to understand if we convert a two layer network into a graph representation.\n\n<img src='assets/backprop_diagram.png' width=550px>\n\nIn the forward pass through the network, our data and operations go from bottom to top here. We pass the input $x$ through a linear transformation $L_1$ with weights $W_1$ and biases $b_1$. The output then goes through the sigmoid operation $S$ and another linear transformation $L_2$. Finally we calculate the loss $\\ell$. We use the loss as a measure of how bad the network's predictions are. The goal then is to adjust the weights and biases to minimize the loss.\n\nTo train the weights with gradient descent, we propagate the gradient of the loss backwards through the network. Each operation has some gradient between the inputs and outputs. As we send the gradients backwards, we multiply the incoming gradient with the gradient for the operation. Mathematically, this is really just calculating the gradient of the loss with respect to the weights using the chain rule.\n\n$$\n\\large \\frac{\\partial \\ell}{\\partial W_1} = \\frac{\\partial L_1}{\\partial W_1} \\frac{\\partial S}{\\partial L_1} \\frac{\\partial L_2}{\\partial S} \\frac{\\partial \\ell}{\\partial L_2}\n$$\n\n**Note:** I'm glossing over a few details here that require some knowledge of vector calculus, but they aren't necessary to understand what's going on.\n\nWe update our weights using this gradient with some learning rate $\\alpha$. \n\n$$\n\\large W^\\prime_1 = W_1 - \\alpha \\frac{\\partial \\ell}{\\partial W_1}\n$$\n\nThe learning rate $\\alpha$ is set such that the weight update steps are small enough that the iterative method settles in a minimum.",
"_____no_output_____"
],
[
"## Losses in PyTorch\n\nLet's start by seeing how we calculate the loss with PyTorch. Through the `nn` module, PyTorch provides losses such as the cross-entropy loss (`nn.CrossEntropyLoss`). You'll usually see the loss assigned to `criterion`. As noted in the last part, with a classification problem such as MNIST, we're using the softmax function to predict class probabilities. With a softmax output, you want to use cross-entropy as the loss. To actually calculate the loss, you first define the criterion then pass in the output of your network and the correct labels.\n\nSomething really important to note here. Looking at [the documentation for `nn.CrossEntropyLoss`](https://pytorch.org/docs/stable/nn.html#torch.nn.CrossEntropyLoss),\n\n> This criterion combines `nn.LogSoftmax()` and `nn.NLLLoss()` in one single class.\n>\n> The input is expected to contain scores for each class.\n\nThis means we need to pass in the raw output of our network into the loss, not the output of the softmax function. This raw output is usually called the *logits* or *scores*. We use the logits because softmax gives you probabilities which will often be very close to zero or one but floating-point numbers can't accurately represent values near zero or one ([read more here](https://docs.python.org/3/tutorial/floatingpoint.html)). It's usually best to avoid doing calculations with probabilities, typically we use log-probabilities.",
"_____no_output_____"
]
],
[
[
"# The MNIST datasets are hosted on yann.lecun.com that has moved under CloudFlare protection\n# Run this script to enable the datasets download\n# Reference: https://github.com/pytorch/vision/issues/1938\n\nfrom six.moves import urllib\nopener = urllib.request.build_opener()\nopener.addheaders = [('User-agent', 'Mozilla/5.0')]\nurllib.request.install_opener(opener)",
"_____no_output_____"
],
[
"import torch\nfrom torch import nn\nimport torch.nn.functional as F\nfrom torchvision import datasets, transforms\n\n# Define a transform to normalize the data\ntransform = transforms.Compose([transforms.ToTensor(),\n transforms.Normalize((0.5,), (0.5,)),\n ])\n# Download and load the training data\ntrainset = datasets.MNIST('~/.pytorch/MNIST_data/', download=True, train=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)",
"_____no_output_____"
]
],
[
[
"### Note\nIf you haven't seen `nn.Sequential` yet, please finish the end of the Part 2 notebook.",
"_____no_output_____"
]
],
[
[
"# Build a feed-forward network\nmodel = nn.Sequential(nn.Linear(784, 128),\n nn.ReLU(),\n nn.Linear(128, 64),\n nn.ReLU(),\n nn.Linear(64, 10))\n\n# Define the loss\ncriterion = nn.CrossEntropyLoss()\n\n# Get our data\ndataiter = iter(trainloader)\n\nimages, labels = next(dataiter)\n\n# Flatten images\nimages = images.view(images.shape[0], -1)\n\n# Forward pass, get our logits\nlogits = model(images)\n# Calculate the loss with the logits and the labels\nloss = criterion(logits, labels)\n\nprint(loss)",
"tensor(2.3151, grad_fn=<NllLossBackward0>)\n"
]
],
[
[
"In my experience it's more convenient to build the model with a log-softmax output using `nn.LogSoftmax` or `F.log_softmax` ([documentation](https://pytorch.org/docs/stable/nn.html#torch.nn.LogSoftmax)). Then you can get the actual probabilities by taking the exponential `torch.exp(output)`. With a log-softmax output, you want to use the negative log likelihood loss, `nn.NLLLoss` ([documentation](https://pytorch.org/docs/stable/nn.html#torch.nn.NLLLoss)).\n\n>**Exercise:** Build a model that returns the log-softmax of the output and calculate the loss using the negative log likelihood loss. Note that for `nn.LogSoftmax` and `F.log_softmax` you'll need to set the `dim` keyword argument appropriately. `dim=0` calculates softmax across the rows, so each column sums to 1, while `dim=1` calculates across the columns so each row sums to 1. Think about what you want the output to be and choose `dim` appropriately.",
"_____no_output_____"
]
],
[
[
"# TODO: Build a feed-forward network\nmodel = nn.Sequential(nn.Linear(784, 128),\n nn.ReLU(),\n nn.Linear(128,64),\n nn.ReLU(),\n nn.Linear(64,10),\n nn.LogSoftmax(dim=1))\n\n# TODO: Define the loss\ncriterion = nn.NLLLoss()\n\n### Run this to check your work\n# Get our data\ndataiter = iter(trainloader)\n\nimages, labels = next(dataiter)\n\n# Flatten images\nimages = images.view(images.shape[0], -1)\n\n# Forward pass, get our logits\nlogits = model(images)\n# Calculate the loss with the logits and the labels\nloss = criterion(logits, labels)\n\nprint(loss)",
"tensor(2.2924, grad_fn=<NllLossBackward0>)\n"
]
],
[
[
"[Video](https://youtu.be/zBWlOeX2sQM)\n\n## Autograd\n\nNow that we know how to calculate a loss, how do we use it to perform backpropagation? Torch provides a module, `autograd`, for automatically calculating the gradients of tensors. We can use it to calculate the gradients of all our parameters with respect to the loss. Autograd works by keeping track of operations performed on tensors, then going backwards through those operations, calculating gradients along the way. To make sure PyTorch keeps track of operations on a tensor and calculates the gradients, you need to set `requires_grad = True` on a tensor. You can do this at creation with the `requires_grad` keyword, or at any time with `x.requires_grad_(True)`.\n\nYou can turn off gradients for a block of code with the `torch.no_grad()` content:\n```python\nx = torch.zeros(1, requires_grad=True)\n>>> with torch.no_grad():\n... y = x * 2\n>>> y.requires_grad\nFalse\n```\n\nAlso, you can turn on or off gradients altogether with `torch.set_grad_enabled(True|False)`.\n\nThe gradients are computed with respect to some variable `z` with `z.backward()`. This does a backward pass through the operations that created `z`.",
"_____no_output_____"
]
],
[
[
"x = torch.randn(2,2, requires_grad=True)\nprint(x)",
"tensor([[ 0.9777, 0.0922],\n [-1.1834, -0.2655]], requires_grad=True)\n"
],
[
"y = x**2\nprint(y)",
"tensor([[0.9559, 0.0085],\n [1.4004, 0.0705]], grad_fn=<PowBackward0>)\n"
]
],
[
[
"Below we can see the operation that created `y`, a power operation `PowBackward0`.",
"_____no_output_____"
]
],
[
[
"## grad_fn shows the function that generated this variable\nprint(y.grad_fn)",
"<PowBackward0 object at 0x7f55d5220760>\n"
]
],
[
[
"The autograd module keeps track of these operations and knows how to calculate the gradient for each one. In this way, it's able to calculate the gradients for a chain of operations, with respect to any one tensor. Let's reduce the tensor `y` to a scalar value, the mean.",
"_____no_output_____"
]
],
[
[
"z = y.mean()\nprint(z)",
"tensor(0.6088, grad_fn=<MeanBackward0>)\n"
]
],
[
[
"You can check the gradients for `x` and `y` but they are empty currently.",
"_____no_output_____"
]
],
[
[
"print(x.grad)",
"None\n"
]
],
[
[
"To calculate the gradients, you need to run the `.backward` method on a Variable, `z` for example. This will calculate the gradient for `z` with respect to `x`\n\n$$\n\\frac{\\partial z}{\\partial x} = \\frac{\\partial}{\\partial x}\\left[\\frac{1}{n}\\sum_i^n x_i^2\\right] = \\frac{x}{2}\n$$",
"_____no_output_____"
]
],
[
[
"z.backward()\nprint(x.grad)\nprint(x/2)",
"tensor([[ 0.4888, 0.0461],\n [-0.5917, -0.1327]])\ntensor([[ 0.4888, 0.0461],\n [-0.5917, -0.1327]], grad_fn=<DivBackward0>)\n"
]
],
[
[
"These gradients calculations are particularly useful for neural networks. For training we need the gradients of the cost with respect to the weights. With PyTorch, we run data forward through the network to calculate the loss, then, go backwards to calculate the gradients with respect to the loss. Once we have the gradients we can make a gradient descent step. ",
"_____no_output_____"
],
[
"## Loss and Autograd together\n\nWhen we create a network with PyTorch, all of the parameters are initialized with `requires_grad = True`. This means that when we calculate the loss and call `loss.backward()`, the gradients for the parameters are calculated. These gradients are used to update the weights with gradient descent. Below you can see an example of calculating the gradients using a backwards pass.",
"_____no_output_____"
]
],
[
[
"# Build a feed-forward network\nmodel = nn.Sequential(nn.Linear(784, 128),\n nn.ReLU(),\n nn.Linear(128, 64),\n nn.ReLU(),\n nn.Linear(64, 10),\n nn.LogSoftmax(dim=1))\n\ncriterion = nn.NLLLoss()\ndataiter = iter(trainloader)\nimages, labels = next(dataiter)\nimages = images.view(images.shape[0], -1)\n\nlogits = model(images)\nloss = criterion(logits, labels)",
"_____no_output_____"
],
[
"print('Before backward pass: \\n', model[0].weight.grad)\n\nloss.backward()\n\nprint('After backward pass: \\n', model[0].weight.grad)",
"Before backward pass: \n None\nAfter backward pass: \n tensor([[-0.0012, -0.0012, -0.0012, ..., -0.0012, -0.0012, -0.0012],\n [ 0.0015, 0.0015, 0.0015, ..., 0.0015, 0.0015, 0.0015],\n [-0.0031, -0.0031, -0.0031, ..., -0.0031, -0.0031, -0.0031],\n ...,\n [-0.0024, -0.0024, -0.0024, ..., -0.0024, -0.0024, -0.0024],\n [-0.0049, -0.0049, -0.0049, ..., -0.0049, -0.0049, -0.0049],\n [-0.0025, -0.0025, -0.0025, ..., -0.0025, -0.0025, -0.0025]])\n"
]
],
[
[
"## Training the network!\n\nThere's one last piece we need to start training, an optimizer that we'll use to update the weights with the gradients. We get these from PyTorch's [`optim` package](https://pytorch.org/docs/stable/optim.html). For example we can use stochastic gradient descent with `optim.SGD`. You can see how to define an optimizer below.",
"_____no_output_____"
]
],
[
[
"from torch import optim\n\n# Optimizers require the parameters to optimize and a learning rate\noptimizer = optim.SGD(model.parameters(), lr=0.01)",
"_____no_output_____"
]
],
[
[
"Now we know how to use all the individual parts so it's time to see how they work together. Let's consider just one learning step before looping through all the data. The general process with PyTorch:\n\n* Make a forward pass through the network \n* Use the network output to calculate the loss\n* Perform a backward pass through the network with `loss.backward()` to calculate the gradients\n* Take a step with the optimizer to update the weights\n\nBelow I'll go through one training step and print out the weights and gradients so you can see how it changes. Note that I have a line of code `optimizer.zero_grad()`. When you do multiple backwards passes with the same parameters, the gradients are accumulated. This means that you need to zero the gradients on each training pass or you'll retain gradients from previous training batches.",
"_____no_output_____"
]
],
[
[
"print('Initial weights - ', model[0].weight)\n\ndataiter = iter(trainloader)\nimages, labels = next(dataiter)\nimages.resize_(64, 784)\n\n# Clear the gradients, do this because gradients are accumulated\noptimizer.zero_grad()\n\n# Forward pass, then backward pass, then update weights\noutput = model(images)\nloss = criterion(output, labels)\nloss.backward()\nprint('Gradient -', model[0].weight.grad)",
"Initial weights - Parameter containing:\ntensor([[ 0.0327, 0.0021, -0.0077, ..., -0.0030, 0.0341, -0.0254],\n [ 0.0085, 0.0311, -0.0216, ..., 0.0152, -0.0310, 0.0097],\n [ 0.0255, 0.0038, -0.0059, ..., 0.0010, -0.0310, 0.0254],\n ...,\n [ 0.0010, 0.0231, 0.0232, ..., 0.0003, 0.0177, -0.0173],\n [-0.0078, 0.0011, 0.0071, ..., 0.0246, -0.0118, -0.0281],\n [ 0.0352, -0.0065, -0.0087, ..., 0.0159, 0.0112, 0.0087]],\n requires_grad=True)\nGradient - tensor([[-0.0005, -0.0005, -0.0005, ..., -0.0005, -0.0005, -0.0005],\n [ 0.0003, 0.0003, 0.0003, ..., 0.0003, 0.0003, 0.0003],\n [-0.0055, -0.0055, -0.0055, ..., -0.0055, -0.0055, -0.0055],\n ...,\n [ 0.0004, 0.0004, 0.0004, ..., 0.0004, 0.0004, 0.0004],\n [ 0.0056, 0.0056, 0.0056, ..., 0.0056, 0.0056, 0.0056],\n [ 0.0020, 0.0020, 0.0020, ..., 0.0020, 0.0020, 0.0020]])\n"
],
[
"# Take an update step and view the new weights\noptimizer.step()\nprint('Updated weights - ', model[0].weight)",
"Updated weights - Parameter containing:\ntensor([[ 0.0327, 0.0021, -0.0077, ..., -0.0030, 0.0341, -0.0254],\n [ 0.0085, 0.0311, -0.0216, ..., 0.0152, -0.0310, 0.0097],\n [ 0.0256, 0.0039, -0.0058, ..., 0.0010, -0.0309, 0.0254],\n ...,\n [ 0.0010, 0.0230, 0.0232, ..., 0.0003, 0.0177, -0.0173],\n [-0.0079, 0.0010, 0.0070, ..., 0.0245, -0.0119, -0.0282],\n [ 0.0352, -0.0065, -0.0087, ..., 0.0159, 0.0111, 0.0087]],\n requires_grad=True)\n"
]
],
[
[
"### Training for real\n\nNow we'll put this algorithm into a loop so we can go through all the images. Some nomenclature, one pass through the entire dataset is called an *epoch*. So here we're going to loop through `trainloader` to get our training batches. For each batch, we'll be doing a training pass where we calculate the loss, do a backwards pass, and update the weights.\n\n>**Exercise:** Implement the training pass for our network. If you implemented it correctly, you should see the training loss drop with each epoch.",
"_____no_output_____"
]
],
[
[
"## Your solution here\n\nmodel = nn.Sequential(nn.Linear(784, 128),\n nn.ReLU(),\n nn.Linear(128, 64),\n nn.ReLU(),\n nn.Linear(64, 10),\n nn.LogSoftmax(dim=1))\n\ncriterion = nn.NLLLoss()\noptimizer = optim.SGD(model.parameters(), lr=0.003)\n\nepochs = 5\nfor e in range(epochs):\n running_loss = 0 #To get the loss of the epoch\n for images, labels in trainloader:\n # Flatten MNIST images into a 784 long vector\n images = images.view(images.shape[0], -1)\n \n # TODO: Training pass\n optimizer.zero_grad()\n \n output = model(images)\n loss = criterion(output, labels)\n loss.backward()\n optimizer.step()\n \n running_loss += loss.item()\n else:\n print(f\"Training loss: {running_loss/len(trainloader)}\")",
"Training loss: 1.950085681638738\nTraining loss: 0.9164738921976802\nTraining loss: 0.5511519496184168\nTraining loss: 0.4412556580865561\nTraining loss: 0.3900658352526902\nTraining loss: 0.3605045829214521\nTraining loss: 0.3406065811655288\nTraining loss: 0.3256806004355584\nTraining loss: 0.3132319454508804\nTraining loss: 0.3025838366068248\n"
]
],
[
[
"With the network trained, we can check out it's predictions.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport helper\n\ndataiter = iter(trainloader)\nimages, labels = next(dataiter)\n\nimg = images[0].view(1, 784)\n# Turn off gradients to speed up this part\nwith torch.no_grad():\n logps = model(img)\n\n# Output of the network are log-probabilities, need to take exponential for probabilities\nps = torch.exp(logps)\nhelper.view_classify(img.view(1, 28, 28), ps)",
"_____no_output_____"
]
],
[
[
"Now our network is brilliant. It can accurately predict the digits in our images. Next up you'll write the code for training a neural network on a more complex dataset.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
eceeffadf899d338127115cb0b641cfd5d99bdef | 14,058 | ipynb | Jupyter Notebook | datasets/allen_institute_mouse_brain_cell_diversity.ipynb | reconstrue/transcriptomics_on_colab | d30c5b049b0022bc10d5d585dce89e83c9ddd7b6 | [
"Apache-2.0"
] | 1 | 2020-02-06T18:46:06.000Z | 2020-02-06T18:46:06.000Z | datasets/allen_institute_mouse_brain_cell_diversity.ipynb | reconstrue/single_cell_on_colab | d30c5b049b0022bc10d5d585dce89e83c9ddd7b6 | [
"Apache-2.0"
] | 83 | 2019-11-22T22:56:22.000Z | 2020-04-08T23:06:12.000Z | datasets/allen_institute_mouse_brain_cell_diversity.ipynb | reconstrue/transcriptomics_on_colab | d30c5b049b0022bc10d5d585dce89e83c9ddd7b6 | [
"Apache-2.0"
] | null | null | null | 29.72093 | 297 | 0.49751 | [
[
[
"# Allen Institute' Mouse Brain Cell Diversity dataset\n\n~75K cells. ~6GB of data.\n\nFound via alleninstitute.org: [Cell Types Database: RNA-Seq Data](https://portal.brain-map.org/atlases-and-data/rnaseq#Mouse_Cortex_and_Hip)",
"_____no_output_____"
],
[
"## Legal\n\nApache 2.0 licensed by Reconstrue 2019\n\nStarted 2019-12-13 by cut and paste from repo's datashader_on_colab.ipynb.",
"_____no_output_____"
],
[
"## Set up",
"_____no_output_____"
]
],
[
[
"# Datashader is all that needs to be installed. Takes a few seconds.\n!pip install --quiet datashader",
"_____no_output_____"
],
[
"import umap\nimport numpy as np\nimport pandas as pd\nimport requests\nimport os\nimport datashader as ds\nimport datashader.utils as utils\nimport datashader.transfer_functions as tf\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport zipfile\nimport urllib.request\n\nsns.set(font_scale=2.0, style=\"white\")",
"_____no_output_____"
]
],
[
[
"#### Data load\n\nScroll on down to the above page's section, [Cell Diversity in the Mouse Cortex and Hippocampus\nRNA-Seq Data Summary](https://portal.brain-map.org/atlases-and-data/rnaseq#Mouse_Cortex_and_Hip). There are 8 files in the Cell Diversity dataset. Two are needed just to scatter plot:\n- 2d-coordinates.zip: columns = `sample_name`, `tsne_1`, `tsne_2`\n- sample-annotations.zip: `sample_name`",
"_____no_output_____"
],
[
"##### 2d-coordinates.zip\nThey have a pre-learned embedding, [2D coordinates](https://transcriptomic-viewer-downloads.s3-us-west-2.amazonaws.com/mouse/2d-coordinates.zip) Just a CSV with X and Y coordinates for all the cells. It's a ZIP file 1.5 MB, containg a CSV file.\tIt's describeds as:\n> TSNE coordinates for each cell, as shown in the Transcriptomics Explorer\n",
"_____no_output_____"
]
],
[
[
"# In not already cached, download and unzip the data to a CSV file on local file system\n\n# Basically: zip_url => csv_filename\nzip_url = 'https://transcriptomic-viewer-downloads.s3-us-west-2.amazonaws.com/mouse/2d-coordinates.zip'\nzip_filename = '2d-coordinates.zip'\ncsv_filename = '2d_coordinates.csv'",
"_____no_output_____"
],
[
"if not os.path.isfile(zip_filename):\n print(f'Downloading to {zip_filename} from {zip_url}')\n # Download the file from `url` and save it locally under `file_name`:\n urllib.request.urlretrieve(zip_url, zip_filename)\nelse:\n print(f'Using pre-cached {zip_filename}')\n\nif not os.path.isfile(csv_filename):\n !unzip {zip_filename}\n\n!ls -lh",
"_____no_output_____"
]
],
[
[
"\n[Wikipedia: Zip_(file_format)](https://en.wikipedia.org/wiki/Zip_(file_format)):\n>Most of the signatures end with the short integer 0x4b50, which is stored in little-endian ordering. Viewed as an ASCII string this reads \"PK\", the initials of the inventor Phil Katz. Thus, when a ZIP file is viewed in a text editor the first two bytes of the file are usually \"PK\".",
"_____no_output_____"
]
],
[
[
"with open(zip_filename, 'rb') as zippee:\n print('zip header:' + str(zippee.read(4)))",
"_____no_output_____"
],
[
"import tarfile\nprint(f'Is it a tarfile: {tarfile.is_tarfile(zip_filename)}')\nprint(f'Is it a zipfile: {zipfile.is_zipfile(zip_filename)}')\n\nimport gzip\nimport csv\n\n#if not os.path.isfile(csv_filename):\n# with zipfile.ZipFile(zip_filename) as zip:\n# #zip.infolist()\n# #with zip.open('file.csv') as myZip:\n# df = pd.read_csv(zip) \n#else:\n# print(f'Using pre-cached {csv_filename}')\n \nimport google.colab as colab\n\nsource_df = pd.read_csv(csv_filename)\ncolab.data_table.DataTable(source_df)\n#source_df.describe",
"_____no_output_____"
]
],
[
[
"##### sample-annotations.zip\n\nThis file has the info needed to color the points\n\n",
"_____no_output_____"
]
],
[
[
"annot_zip_url = 'https://transcriptomic-viewer-downloads.s3-us-west-2.amazonaws.com/mouse/sample-annotations.zip'\nannot_zip_filename = 'sample-annotations.zip'\nannot_csv_filename = 'sample_annotations.csv'\n",
"_____no_output_____"
],
[
"import urllib.request\n\nif not os.path.isfile(annot_zip_filename):\n print(f'Downloading to {annot_zip_filename} from {annot_zip_url}\\n')\n # Download the file from `url` and save it locally under `file_name`:\n urllib.request.urlretrieve(annot_zip_url, annot_zip_filename)\nelse:\n print(f'Using pre-cached {annot_zip_filename}\\n')\n\n# Python zipfile was choking on this so let bash take care of it\nif not os.path.isfile(annot_csv_filename):\n !unzip {annot_zip_filename}\n\n!ls -lh\n",
"_____no_output_____"
],
[
"annot_df = pd.read_csv(annot_csv_filename, dtype={'class_label':'category', 'subclass_label':'category'})\nannot_df.dtypes",
"_____no_output_____"
],
[
"colab.data_table.DataTable(annot_df)",
"_____no_output_____"
]
],
[
[
"## Datashader plot",
"_____no_output_____"
]
],
[
[
"%%time\n\n# Plot via Datashader\nfrom colorcet import fire\n\n#df = pd.DataFrame(embedding, columns=('x', 'y'))\n#df['class'] = pd.Series([str(x) for x in target], dtype=\"category\")\n\ncvs = ds.Canvas(plot_width=500, plot_height=500)\nagg = cvs.points(source_df, 'tsne_1', 'tsne_2')#, 'blue') #, ds.count_cat('color'))\nimg = tf.shade(agg, color_key=fire, how='eq_hist')\n\nutils.export_image(img, filename='cell_diversity', background='#f8f8f8')\n\nimage = plt.imread('cell_diversity.png')\nfig, ax = plt.subplots(figsize=(13, 13))\n\nligth_gray = '#f8f8f8'\nfor spine in ax.spines.values():\n spine.set_edgecolor(ligth_gray)\n\nplt.imshow(image)\nplt.setp(ax, xticks=[], yticks=[])\nplt.title(\"Allen Institute's Cell Diversity pre-built tSNE, \\n\"\n \"rendered via Datashader\",\n fontsize=12)\n\nplt.show()",
"_____no_output_____"
],
[
"\n# JFT-TODO: what I need is to get cluster color categorical?\n#source_df['color'] = source_df['tsne_1']\n#source_df['color2'] = pd.Series(source_df['tsne_2'][:74985])\n\n#class_labels = pd.DataFrame(sample_annos_df['class_label'][:74985])\n#class_labels.describe()\n\n\n#pd.Series(sample_annos_df['class_label'][:74985])\n#merged = pd.concat([source_df,class_labels], axis=1)\n#list(merged.columns)\n#merged.describe()\n",
"_____no_output_____"
],
[
"#source_df.describe()\ncolab.data_table.DataTable(source_df)",
"_____no_output_____"
],
[
"#print(len(source_df.index))\n#print(len(sample_annos_df.index))\n#list(sample_annos_df.columns)",
"_____no_output_____"
],
[
"samples_df = pd.merge(source_df, annot_df, on='sample_name')\ncolab.data_table.DataTable(samples_df)",
"_____no_output_____"
],
[
"samples_df.dtypes",
"_____no_output_____"
],
[
"#%%time\n\n# Plot via Datashader\nimport colorcet\n\n#df = pd.DataFrame(embedding, columns=('x', 'y'))\n#df['class'] = pd.Series([str(x) for x in target], dtype=\"category\")\n\ncvs = ds.Canvas(plot_width=500, plot_height=500)\nagg = cvs.points(samples_df, 'tsne_1', 'tsne_2', ds.count_cat('subclass_label'))\nimg = tf.shade(agg, color_key=colorcet.glasbey_dark, how='eq_hist')\n\nutils.export_image(img, filename='cell_diversity', background='#f8f8f8')\n\nimage = plt.imread('cell_diversity.png')\nfig, ax = plt.subplots(figsize=(13, 13))\n\nligth_gray = '#f8f8f8'\nfor spine in ax.spines.values():\n spine.set_edgecolor(ligth_gray)\n\nplt.imshow(image)\nplt.setp(ax, xticks=[], yticks=[])\nplt.title(\"Allen Institute's Cell Diversity pre-built tSNE, \\n\"\n \"rendered via Datashader\",\n fontsize=12)\n\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecef0a06a524734434e42dc2a02f19d11c1e882d | 10,219 | ipynb | Jupyter Notebook | markdown_generator/PubsFromBib.ipynb | fsame/fsame.github.io | 38c4581d3e8637cb6c8c2562ac70d3862ebb7e53 | [
"MIT"
] | null | null | null | markdown_generator/PubsFromBib.ipynb | fsame/fsame.github.io | 38c4581d3e8637cb6c8c2562ac70d3862ebb7e53 | [
"MIT"
] | null | null | null | markdown_generator/PubsFromBib.ipynb | fsame/fsame.github.io | 38c4581d3e8637cb6c8c2562ac70d3862ebb7e53 | [
"MIT"
] | null | null | null | 43.300847 | 1,018 | 0.468245 | [
[
[
"# Publications markdown generator for academicpages\n\nTakes a set of bibtex of publications and converts them for use with [academicpages.github.io](academicpages.github.io). This is an interactive Jupyter notebook ([see more info here](http://jupyter-notebook-beginner-guide.readthedocs.io/en/latest/what_is_jupyter.html)). \n\nThe core python code is also in `pubsFromBibs.py`. \nRun either from the `markdown_generator` folder after replacing updating the publist dictionary with:\n* bib file names\n* specific venue keys based on your bib file preferences\n* any specific pre-text for specific files\n* Collection Name (future feature)\n\nTODO: Make this work with other databases of citations, \nTODO: Merge this with the existing TSV parsing solution",
"_____no_output_____"
]
],
[
[
"from pybtex.database.input import bibtex\nimport pybtex.database.input.bibtex \nfrom time import strptime\nimport string\nimport html\nimport os\nimport re",
"_____no_output_____"
],
[
"#todo: incorporate different collection types rather than a catch all publications, requires other changes to template\npublist = {\n \"proceeding\": {\n \"file\" : \"proceedings.bib\",\n \"venuekey\": \"booktitle\",\n \"venue-pretext\": \"In the proceedings of \",\n \"collection\" : {\"name\":\"publications\",\n \"permalink\":\"/publication/\"}\n \n },\n \"journal\":{\n \"file\": \"pubs.bib\",\n \"venuekey\" : \"journal\",\n \"venue-pretext\" : \"\",\n \"collection\" : {\"name\":\"publications\",\n \"permalink\":\"/publication/\"}\n } \n}",
"_____no_output_____"
],
[
"html_escape_table = {\n \"&\": \"&\",\n '\"': \""\",\n \"'\": \"'\"\n }\n\ndef html_escape(text):\n \"\"\"Produce entities within text.\"\"\"\n return \"\".join(html_escape_table.get(c,c) for c in text)",
"_____no_output_____"
],
[
"for pubsource in publist:\n parser = bibtex.Parser()\n bibdata = parser.parse_file(publist[pubsource][\"file\"])\n\n #loop through the individual references in a given bibtex file\n for bib_id in bibdata.entries:\n #reset default date\n pub_year = \"1900\"\n pub_month = \"01\"\n pub_day = \"01\"\n \n b = bibdata.entries[bib_id].fields\n \n try:\n pub_year = f'{b[\"year\"]}'\n\n #todo: this hack for month and day needs some cleanup\n if \"month\" in b.keys(): \n if(len(b[\"month\"])<3):\n pub_month = \"0\"+b[\"month\"]\n pub_month = pub_month[-2:]\n elif(b[\"month\"] not in range(12)):\n tmnth = strptime(b[\"month\"][:3],'%b').tm_mon \n pub_month = \"{:02d}\".format(tmnth) \n else:\n pub_month = str(b[\"month\"])\n if \"day\" in b.keys(): \n pub_day = str(b[\"day\"])\n\n \n pub_date = pub_year+\"-\"+pub_month+\"-\"+pub_day\n \n #strip out {} as needed (some bibtex entries that maintain formatting)\n clean_title = b[\"title\"].replace(\"{\", \"\").replace(\"}\",\"\").replace(\"\\\\\",\"\").replace(\" \",\"-\") \n\n url_slug = re.sub(\"\\\\[.*\\\\]|[^a-zA-Z0-9_-]\", \"\", clean_title)\n url_slug = url_slug.replace(\"--\",\"-\")\n\n md_filename = (str(pub_date) + \"-\" + url_slug + \".md\").replace(\"--\",\"-\")\n html_filename = (str(pub_date) + \"-\" + url_slug).replace(\"--\",\"-\")\n\n #Build Citation from text\n citation = \"\"\n\n #citation authors - todo - add highlighting for primary author?\n for author in bibdata.entries[bib_id].persons[\"author\"]:\n citation = citation+\" \"+author.first_names[0]+\" \"+author.last_names[0]+\", \"\n\n #citation title\n citation = citation + \"\\\"\" + html_escape(b[\"title\"].replace(\"{\", \"\").replace(\"}\",\"\").replace(\"\\\\\",\"\")) + \".\\\"\"\n\n #add venue logic depending on citation type\n venue = publist[pubsource][\"venue-pretext\"]+b[publist[pubsource][\"venuekey\"]].replace(\"{\", \"\").replace(\"}\",\"\").replace(\"\\\\\",\"\")\n\n citation = citation + \" \" + html_escape(venue)\n citation = citation + \", \" + pub_year + \".\"\n\n \n ## YAML variables\n md = \"---\\ntitle: \\\"\" + html_escape(b[\"title\"].replace(\"{\", \"\").replace(\"}\",\"\").replace(\"\\\\\",\"\")) + '\"\\n'\n \n md += \"\"\"collection: \"\"\" + publist[pubsource][\"collection\"][\"name\"]\n\n md += \"\"\"\\npermalink: \"\"\" + publist[pubsource][\"collection\"][\"permalink\"] + html_filename\n \n note = False\n if \"note\" in b.keys():\n if len(str(b[\"note\"])) > 5:\n md += \"\\nexcerpt: '\" + html_escape(b[\"note\"]) + \"'\"\n note = True\n\n md += \"\\ndate: \" + str(pub_date) \n\n md += \"\\nvenue: '\" + html_escape(venue) + \"'\"\n \n url = False\n if \"url\" in b.keys():\n if len(str(b[\"url\"])) > 5:\n md += \"\\npaperurl: '\" + b[\"url\"] + \"'\"\n url = True\n\n md += \"\\ncitation: '\" + html_escape(citation) + \"'\"\n\n md += \"\\n---\"\n\n \n ## Markdown description for individual page\n if note:\n md += \"\\n\" + html_escape(b[\"note\"]) + \"\\n\"\n\n if url:\n md += \"\\n[Access paper here](\" + b[\"url\"] + \"){:target=\\\"_blank\\\"}\\n\" \n else:\n md += \"\\nUse [Google Scholar](https://scholar.google.com/scholar?q=\"+html.escape(clean_title.replace(\"-\",\"+\"))+\"){:target=\\\"_blank\\\"} for full citation\"\n\n md_filename = os.path.basename(md_filename)\n\n with open(\"../_publications/\" + md_filename, 'w') as f:\n f.write(md)\n print(f'SUCESSFULLY PARSED {bib_id}: \\\"', b[\"title\"][:60],\"...\"*(len(b['title'])>60),\"\\\"\")\n # field may not exist for a reference\n except KeyError as e:\n print(f'WARNING Missing Expected Field {e} from entry {bib_id}: \\\"', b[\"title\"][:30],\"...\"*(len(b['title'])>30),\"\\\"\")\n continue\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecef17c2f135c21a218076c655e48c88bb6aa9f2 | 55 | ipynb | Jupyter Notebook | Jupyter Notebook/prod.ipynb | Raikan10/Hello-world-1 | 1029686f751e85216c05d7a9d60c64a829600f96 | [
"MIT"
] | 1,428 | 2018-10-03T15:15:17.000Z | 2019-03-31T18:38:36.000Z | Jupyter Notebook/prod.ipynb | Raikan10/Hello-world-1 | 1029686f751e85216c05d7a9d60c64a829600f96 | [
"MIT"
] | 1,162 | 2018-10-03T15:05:49.000Z | 2018-10-18T14:17:52.000Z | Jupyter Notebook/prod.ipynb | Raikan10/Hello-world-1 | 1029686f751e85216c05d7a9d60c64a829600f96 | [
"MIT"
] | 3,909 | 2018-10-03T15:07:19.000Z | 2019-03-31T18:39:08.000Z | 11 | 19 | 0.6 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecef1d2e89453db8ff3ac3a4ef9f4c75077ff2e9 | 10,258 | ipynb | Jupyter Notebook | Mission_to_Mars.ipynb | Bettinadavis11/Mission_to_Mars | caa26290437b692d02d3defa77dcf7174e984b5e | [
"MIT"
] | null | null | null | Mission_to_Mars.ipynb | Bettinadavis11/Mission_to_Mars | caa26290437b692d02d3defa77dcf7174e984b5e | [
"MIT"
] | null | null | null | Mission_to_Mars.ipynb | Bettinadavis11/Mission_to_Mars | caa26290437b692d02d3defa77dcf7174e984b5e | [
"MIT"
] | null | null | null | 26.50646 | 1,076 | 0.457692 | [
[
[
"#import splinter beautiful soup\nfrom splinter import Browser\nfrom bs4 import BeautifulSoup as soup\nfrom webdriver_manager.chrome import ChromeDriverManager\nimport pandas as pd",
"_____no_output_____"
],
[
"executable_path = {'executable_path': 'chromedriver.exe'}\nbrowser = Browser('chrome', **executable_path, headless=False)",
"_____no_output_____"
],
[
"# Visit the mars nasa news site\nurl = 'https://redplanetscience.com'\nbrowser.visit(url)\n# Optional delay for loading the page\nbrowser.is_element_present_by_css('div.list_text', wait_time=1)",
"_____no_output_____"
],
[
"html = browser.html\nnews_soup = soup(html, 'html.parser')\nslide_elem = news_soup.select_one('div.list_text')",
"_____no_output_____"
],
[
"slide_elem.find('div', class_='content_title')",
"_____no_output_____"
],
[
"# Use the parent element to find the first `a` tag and save it as `news_title`\nnews_title = slide_elem.find('div', class_='content_title').get_text()\nnews_title",
"_____no_output_____"
],
[
"# Use the parent element to find the paragraph text\nnews_p = slide_elem.find('div', class_='article_teaser_body').get_text()\nnews_p",
"_____no_output_____"
]
],
[
[
"### Featured images",
"_____no_output_____"
]
],
[
[
"# Visit URL\nurl = 'https://spaceimages-mars.com'\nbrowser.visit(url)",
"_____no_output_____"
],
[
"# Find and click the full image button\nfull_image_elem = browser.find_by_tag('button')[1]\nfull_image_elem.click()",
"_____no_output_____"
],
[
"# Parse the resulting html with soup\nhtml = browser.html\nimg_soup = soup(html, 'html.parser')",
"_____no_output_____"
],
[
"# Find the relative image url\nimg_url_rel = img_soup.find('img', class_='fancybox-image').get('src')\nimg_url_rel",
"_____no_output_____"
],
[
"# Use the base URL to create an absolute URL\nimg_url = f'https://spaceimages-mars.com/{img_url_rel}'\nimg_url",
"_____no_output_____"
],
[
"df = pd.read_html('https://galaxyfacts-mars.com')[0]\ndf.columns=['description', 'Mars', 'Earth']\ndf.set_index('description', inplace=True)\ndf",
"_____no_output_____"
],
[
"df = pd.read_html('https://galaxyfacts-mars.com')[0]\ndf.columns=['description', 'Mars', 'Earth']\ndf.set_index('description', inplace=True)\ndf.to_html()",
"_____no_output_____"
],
[
"browser.quit()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecef28c3174cce94046758648c107eec5a64bc4a | 29,112 | ipynb | Jupyter Notebook | 6. Temporal Difference Learning/notebooks/Double-Q-Learning.ipynb | A-Pai/Reinforcement-Learning | ff460e9df09f508f7bf8eddc99943b617a0915ba | [
"MIT"
] | 101 | 2017-12-07T14:14:30.000Z | 2022-02-24T09:41:05.000Z | 6. Temporal Difference Learning/notebooks/Double-Q-Learning.ipynb | Jack-926/Reinforcement-Learning | bcc7b1521c922c7092a61cb0bfabe6c8de34084f | [
"MIT"
] | 4 | 2019-05-18T05:05:29.000Z | 2020-06-25T06:49:56.000Z | 6. Temporal Difference Learning/notebooks/Double-Q-Learning.ipynb | Jack-926/Reinforcement-Learning | bcc7b1521c922c7092a61cb0bfabe6c8de34084f | [
"MIT"
] | 33 | 2018-05-20T15:52:05.000Z | 2022-03-29T08:42:50.000Z | 242.6 | 26,924 | 0.917079 | [
[
[
"import gym, sys\n\n%matplotlib inline\nsys.path.append('../..')\nsys.path.append('..')\n\nimport algorithms, visualize, utils",
"_____no_output_____"
]
],
[
[
"## Cliff Walking",
"_____no_output_____"
]
],
[
[
"env = gym.make('CliffWalking-v0')",
"_____no_output_____"
],
[
"Q, sum_rewards = algorithms.double_qlearning(env, alpha=.5, epsilon=0.1, num_episodes=500)",
"_____no_output_____"
],
[
"visualize.rewards(sum_rewards)",
"_____no_output_____"
]
],
[
[
"### Run Environment Interactively",
"_____no_output_____"
]
],
[
[
"utils.run_environment_greedy(env, Q)",
"Current State: 47\no o o o o o o o o o o o\no o o o o o o o o o o o\no o o o o o o o o o o o\no C C C C C C C C C C x\n\nFinished\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecef2feb789d5f434a255cd94d496d48c39df1c0 | 6,875 | ipynb | Jupyter Notebook | Join/spatial_joins.ipynb | jdgomezmo/gee | 7016c47ee902dbf60b1aeb6319424c61c1107345 | [
"MIT"
] | 1 | 2020-11-16T22:07:42.000Z | 2020-11-16T22:07:42.000Z | Join/spatial_joins.ipynb | tingli3/earthengine-py-notebooks | 7016c47ee902dbf60b1aeb6319424c61c1107345 | [
"MIT"
] | null | null | null | Join/spatial_joins.ipynb | tingli3/earthengine-py-notebooks | 7016c47ee902dbf60b1aeb6319424c61c1107345 | [
"MIT"
] | null | null | null | 43.238994 | 1,031 | 0.585455 | [
[
[
"<table class=\"ee-notebook-buttons\" align=\"left\">\n <td><a target=\"_blank\" href=\"https://github.com/giswqs/earthengine-py-notebooks/tree/master/Join/spatial_joins.ipynb\"><img width=32px src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" /> View source on GitHub</a></td>\n <td><a target=\"_blank\" href=\"https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Join/spatial_joins.ipynb\"><img width=26px src=\"https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png\" />Notebook Viewer</a></td>\n <td><a target=\"_blank\" href=\"https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Join/spatial_joins.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" /> Run in Google Colab</a></td>\n</table>",
"_____no_output_____"
],
[
"## Install Earth Engine API and geemap\nInstall the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.\nThe following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.\n\n**Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).",
"_____no_output_____"
]
],
[
[
"# Installs geemap package\nimport subprocess\n\ntry:\n import geemap\nexcept ImportError:\n print('geemap package not installed. Installing ...')\n subprocess.check_call([\"python\", '-m', 'pip', 'install', 'geemap'])\n\n# Checks whether this notebook is running on Google Colab\ntry:\n import google.colab\n import geemap.eefolium as geemap\nexcept:\n import geemap\n\n# Authenticates and initializes Earth Engine\nimport ee\n\ntry:\n ee.Initialize()\nexcept Exception as e:\n ee.Authenticate()\n ee.Initialize() ",
"_____no_output_____"
]
],
[
[
"## Create an interactive map \nThe default basemap is `Google Maps`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/basemaps.py) can be added using the `Map.add_basemap()` function. ",
"_____no_output_____"
]
],
[
[
"Map = geemap.Map(center=[40,-100], zoom=4)\nMap",
"_____no_output_____"
]
],
[
[
"## Add Earth Engine Python script ",
"_____no_output_____"
]
],
[
[
"# Add Earth Engine dataset\n# Load a primary 'collection': protected areas (Yosemite National Park).\nprimary = ee.FeatureCollection(\"WCMC/WDPA/current/polygons\") \\\n .filter(ee.Filter.eq('NAME', 'Yosemite National Park'))\n\n# Load a secondary 'collection': power plants.\npowerPlants = ee.FeatureCollection('WRI/GPPD/power_plants')\n\n# Define a spatial filter, with distance 100 km.\ndistFilter = ee.Filter.withinDistance(**{\n 'distance': 100000,\n 'leftField': '.geo',\n 'rightField': '.geo',\n 'maxError': 10\n})\n\n# Define a saveAll join.\ndistSaveAll = ee.Join.saveAll(**{\n 'matchesKey': 'points',\n 'measureKey': 'distance'\n})\n\n# Apply the join.\nspatialJoined = distSaveAll.apply(primary, powerPlants, distFilter)\n\n# Print the result.\n# print(spatialJoined.getInfo())\nMap.centerObject(spatialJoined, 10)\nMap.addLayer(ee.Image().paint(spatialJoined, 1, 3), {}, 'Spatial Joined')\n",
"_____no_output_____"
]
],
[
[
"## Display Earth Engine data layers ",
"_____no_output_____"
]
],
[
[
"Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.\nMap",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecef31bbe51d7d315548e1cf0200e7ad1169477e | 6,836 | ipynb | Jupyter Notebook | PLM_example.ipynb | charliedondapati/PLM | b6e005b032d052546e2f604806594a529f1df151 | [
"BSD-2-Clause-FreeBSD"
] | 1 | 2021-08-06T15:49:19.000Z | 2021-08-06T15:49:19.000Z | PLM_example.ipynb | charliedondapati/PLM | b6e005b032d052546e2f604806594a529f1df151 | [
"BSD-2-Clause-FreeBSD"
] | null | null | null | PLM_example.ipynb | charliedondapati/PLM | b6e005b032d052546e2f604806594a529f1df151 | [
"BSD-2-Clause-FreeBSD"
] | 2 | 2021-04-09T18:55:05.000Z | 2021-08-06T15:49:23.000Z | 25.225092 | 117 | 0.447338 | [
[
[
"### **Installing PLM package**",
"_____no_output_____"
]
],
[
[
"!pip install PLM\n",
"_____no_output_____"
]
],
[
[
"### **Process Dataset**\n\n\n* Target should be output column followed by the label\n* Target is preceeded by Input features\n\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\ndf = pd.read_csv('equ23_data.csv', header=None)\ntarget = np.array(df[df.columns[[-2, -1]]])\ndata = np.array(df.iloc[:, :-2])",
"_____no_output_____"
]
],
[
[
"\n\n---\n\n\n\n* pelm() is the wrapper function with calls train and test methods internally.\n* The number of epochs can be specified as a parameter, the default is epochs=20.\n\n",
"_____no_output_____"
]
],
[
[
"from PLM.pelm import pelm\n\nn = 10;\nparameter1 = 10;\nparameter2 = 10;\nmodel_number = 3;\n\npelm(data, target, model_number, n=n, p=parameter1, s=parameter2)",
"Execution time: 43.6878502368927 secs\nMin error: 0.013624696718351184\nMean error: 0.14071896365248865\n"
]
],
[
[
"\n\n\n---\n\n\n* plm_train() & plm_train() methods can be called seperately from the module\n* Dataset should rightly processed for calling the specific functions\n* plm_train() & plm_test() methods used BELM(Bi-directional ELM) package internally\n\n",
"_____no_output_____"
]
],
[
[
"X_train, X_test, Y_train, Y_test = train_test_split(data, target, test_size=0.3)\nL_train = Y_train[:, -1].reshape(-1, 1)\nL_test = Y_test[:, -1].reshape(-1, 1)\n\nY_test = Y_test[:, 0].reshape(-1, 1)\nY_train = Y_train[:, 0].reshape(-1, 1)",
"_____no_output_____"
],
[
"from PLM.pelm import pelm, plm_train, plm_test\n\nn = 10;\nparameter1 = 10;\nparameter2 = 10;\nmodel_number = 3;\n\nd, t, l, rl, net = plm_train(X_train, Y_train, L_train, n=n, s1=parameter1, s2=parameter2, c=model_number);",
"_____no_output_____"
],
[
"e, svm_acc = plm_test(d, l, X_test, Y_test, L_test, net, c=model_number)\nprint(\"Error(rmse): \",e)",
"Error(rmse): 0.06363327438862462\n"
]
],
[
[
"### **Bi-directional Extreme Learning Machine**\n\n\n* Last column should be the output column \n* Output column is preceeded by the input features\n\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.model_selection import train_test_split\n\ndf = pd.read_csv('sample_data.csv', header=None)\noutput = np.array(df[df.columns[-1]])\ndata = np.array(df.iloc[:, :-1])\n\nX_train, X_test, Y_train, Y_test = train_test_split(data, output, test_size=0.3)\nY_train = Y_train.reshape(-1, 1)\nY_test = Y_test.reshape(-1, 1)",
"_____no_output_____"
],
[
"from BELM.belm import BELM\n\nbelm = BELM(X_train.shape[1], Y_train.shape[1], precision=\"single\")\nbelm.add_neurons(2, 'sigm')\nbelm.train(X_train, Y_train)\nyhat = belm.predict(X_test)\ne = belm.error(Y_test, yhat)\nprint(\"Error(rmse):\",e)",
"Error(rmse) = 0.13198138049742777\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecef3d5c4b0c3842d0180e85cf18503cff639b24 | 11,023 | ipynb | Jupyter Notebook | partitioner_examples.ipynb | no-identd/partitioner | b3024a1e63e4a547125772a3af45a1ea74330295 | [
"Apache-2.0"
] | 1 | 2019-08-08T00:47:44.000Z | 2019-08-08T00:47:44.000Z | partitioner_examples.ipynb | NeelShah18/partitioner | d52920d1272f8d7f5d3eae508e4b0a35ea04b825 | [
"Apache-2.0"
] | null | null | null | partitioner_examples.ipynb | NeelShah18/partitioner | d52920d1272f8d7f5d3eae508e4b0a35ea04b825 | [
"Apache-2.0"
] | null | null | null | 27.626566 | 573 | 0.530709 | [
[
[
"# Example usages of the partitioner module \n### JRW, 4/19/2017\nTo load the module, run:",
"_____no_output_____"
]
],
[
[
"from partitioner.tools import partitioner",
"_____no_output_____"
]
],
[
[
"### Set up\nNote that partitioner utilized relatively-large training data files. Thus, this module will likely not be downloaded with any data (e.g., if downloaded from `pip`). If this is the case, training data may be downloaded through the `.download()` method. Note that this will initiate a prompt, to which a response is required.",
"_____no_output_____"
],
[
"```\npa = partitioner()\npa.download()\n```",
"_____no_output_____"
],
[
"### High-performance English model\nOnce the training data has been downloaded, the following will load all English data sets. This requires significant memory resources, but results in a high-performance (see https://arxiv.org/pdf/1608.02025.pdf for details) model:",
"_____no_output_____"
]
],
[
[
"pa = partitioner(language = \"en\", doPOS = True, doLFD = True, q = {\"type\": 0.77, \"POS\": 0.71})\nprint(\"\\n\".join(pa.partition(\"How could something like this simply pop up out of the blue?\")))",
"How\n \ncould\n \nsomething\n \nlike\n \nthis\n \nsimply\n \npop up\n \nout of the blue\n?\n"
]
],
[
[
"Note that this utilizes the parameterization determined in the above article. To change the threshold partition probabilities for both wordforms (type) and part-of-speech (POS), try the following. Note, lower values of q makes it more difficult for words to join together, and values outside of [0,1] will result in random partitions, which are discussed below.",
"_____no_output_____"
]
],
[
[
"print(pa.q)\npa.q['type'] = 0.5\nprint(\"\\n\".join(pa.partition(\"How could something like this simply pop up out of the blue?\")))",
"{'type': 0.76, 'POS': 0.46}\nHow\n \ncould\n \nsomething\n \nlike\n \nthis\n \nsimply\n \npop up\n \nout of\n \nthe\n \nblue\n?\n"
]
],
[
[
"### Reduced memory overhead\nFirst, clear the data and then load all but the largest (Wikipedia) MWE dataset. Note: partitioner will not be able to resolve as many named entities without Wikipedia.",
"_____no_output_____"
]
],
[
[
"pa.q['type'] = 0.76\npa.clear()\npa.language = \"en\"\nfor source in [\"wordnet\", \"tweebank\", \"trustpilot\", \"ted\", \"streusle\", \"ritter\", \"lowlands\"]:\n pa.source = source\n pa.load()\nprint(\"\\n\".join(pa.partition(\"How could something like this simply pop up out of the blue?\")))",
"How\n \ncould\n \nsomething\n \nlike\n \nthis\n \nsimply\n \npop up\n \nout of\n \nthe\n \nblue\n?\n"
]
],
[
[
"### Run partitioner in a different language\npartitioner comes with starter data from Wikipedia for nine other languages besides English: Dutch (nl), Finnish (fi), German (de), Greek (el), Italian (it), Polish (pl), Portuguese (pt), Russian (ru), and Spanish (es). Note that this is only starter data for these languages, which being from Wikipedia will mostly only cover nouns, as opposed to more conversational language. To learn more about how data are annotated for MWE segmentation, see https://www.cs.cmu.edu/~nschneid/mwecorpus.pdf for more information on comprehensive MWE annotations.",
"_____no_output_____"
]
],
[
[
"pa.clear()\npa.language = \"de\"\npa.source = \"\"\npa.load()\nprint(\"\\n\".join(pa.partition(\"Die binäre Suche ist ein Algorithmus.\")))",
"Warning: no known contractions for the de language.\nDie\n \nbinäre Suche\n \nist\n \nein\n \nAlgorithmus\n.\n"
]
],
[
[
"### Partition a whole text file\nIn addition to segmenting lines of text, partitioner can be applied to whole files to produce aggregated counts. This results in a rank-frequency distribution, which can be assessed for a bag-of-phrases goodness of fit ($R^2$). ",
"_____no_output_____"
]
],
[
[
"pa.clear()\npa.language = \"en\"\npa.source = \"streusle\"\npa.load()\npa.partitionText(textfile=\"README.md\")\npa.testFit()\nprint(\"R-squared: \"+str(round(pa.rsq,2)))\nprint(\"\")\nphrases = sorted(pa.frequencies, key = lambda x: pa.frequencies[x], reverse = True)\nfor j in range(25):\n phrase = phrases[j]\n print(\"\\\"\"+phrase+\"\\\": \"+str(pa.frequencies[phrase]))",
"R-squared: 0.47\n\n\" \": 289.0\n\"\n\": 52.0\n\",\": 29.0\n\">\": 24.0\n\"'\": 16.0\n\"the\": 16.0\n\".\": 15.0\n\"#\": 12.0\n\"\"\": 12.0\n\"partitioner\": 10.0\n\":\": 8.0\n\"(\": 7.0\n\"of\": 7.0\n\"=\": 7.0\n\"data\": 7.0\n\")\": 7.0\n\"a\": 6.0\n\"from\": 5.0\n\"for\": 4.0\n\"pa\": 4.0\n\"with\": 4.0\n\"The\": 4.0\n\"to\": 3.0\n\"source\": 3.0\n\"segmentation\": 3.0\n"
]
],
[
[
"### Run non-deterministic partitions\nThe partitioner project and module grew out of a more simplistic, probabilistic framework. Instead of using the MWE partitions, we can maintain the training data and just partition at random, acording to the loaded probabilities. Random partitions ensue when the threshold parameters are outside of [0,1]. To really see the effects, clear out all partition data and use the uniform random partition probability.\n\nAlso, to run random partitions it is best to turn off part-of-speech tagging, the longest first defined (LFD) algorithm (which ensures that all partitioned MWEs are in fact defined), in addition to limiting the gap size to zero. Note that different runs on the same sentence will produce different partitions.",
"_____no_output_____"
]
],
[
[
"pa.clear()\nprint(pa.qunif)\npa.q['type'] = -1; pa.q['POS'] = -1\npa.doLFD = False\npa.doPOS = False\npa.maxgap = 0\nprint(\"\\n\".join(pa.partition(\"Randomness is hard to manage.\")))\nprint(\"\\n\\n\")\nprint(\"\\n\".join(pa.partition(\"Randomness is hard to manage.\")))",
"0.5\nRandomness\n \nis hard to\n \nmanage.\n\n\n\nRandomness is hard to manage.\n"
]
],
[
[
"### Compute non-deterministic partition expectations\nRather can computing one-off non-deterministic partitions, which are the result of a random process, we can also compute the expectation. For a given phrase, the computed amount is the \n\n* \"expected frequency that a phrase is partitioned from a text, given the partition probabilities\"\n\nEssentially, these may be treated like counts, generalizing the n-grams framework.",
"_____no_output_____"
]
],
[
[
"print(pa.expectation(\"On average, randomness is dull.\"))",
"{'average, randomness': 0.125, 'is': 0.25, 'On average': 0.25, 'On average, randomness': 0.125, 'average, randomness is dull': 0.03125, ' ': 2.0, 'randomness is dull.': 0.0625, 'On average, randomness is dull.': 0.03125, ',': 0.5, '.': 0.5, 'is dull.': 0.125, 'average, randomness is dull.': 0.03125, 'On': 0.5, 'On average, randomness is': 0.0625, 'randomness is': 0.125, 'randomness is dull': 0.0625, 'dull': 0.25, 'dull.': 0.25, 'randomness': 0.25, 'average': 0.25, 'On average, randomness is dull': 0.03125, 'is dull': 0.125, 'average, randomness is': 0.0625}\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecef4bd1fd850eb57ef627cf00d7198876f72c2d | 156,916 | ipynb | Jupyter Notebook | Applying AI to 2D Medical Imaging Data/Models for Classification of 2D Medical Images/Exercise - Differentiate Between Models/Exercise.ipynb | mayank1101/AI-for-Healthcare | 90a70863d8731cbac0662067030340c68374af15 | [
"MIT"
] | 2 | 2020-12-15T19:35:39.000Z | 2021-09-21T19:47:28.000Z | Applying AI to 2D Medical Imaging Data/Models for Classification of 2D Medical Images/Exercise - Differentiate Between Models/Exercise.ipynb | mayank1101/AI-for-Healthcare | 90a70863d8731cbac0662067030340c68374af15 | [
"MIT"
] | null | null | null | Applying AI to 2D Medical Imaging Data/Models for Classification of 2D Medical Images/Exercise - Differentiate Between Models/Exercise.ipynb | mayank1101/AI-for-Healthcare | 90a70863d8731cbac0662067030340c68374af15 | [
"MIT"
] | 2 | 2021-12-27T15:13:32.000Z | 2022-03-27T22:43:25.000Z | 336.729614 | 51,276 | 0.939037 | [
[
[
"%matplotlib inline\n\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport scipy.stats\n\nimport skimage\nfrom skimage import io\nimport glob\n\nimport sklearn\nfrom scipy.ndimage import gaussian_filter",
"_____no_output_____"
]
],
[
[
"## First we'll do background segmentation:",
"_____no_output_____"
]
],
[
[
"## Read in two mammo images: \ndense = io.imread('dense/mdb003.pgm')\nfatty = io.imread('fatty/mdb005.pgm')",
"_____no_output_____"
],
[
"plt.imshow(dense)",
"_____no_output_____"
],
[
"plt.imshow(fatty)",
"_____no_output_____"
],
[
"x = plt.hist(dense.ravel(),bins=256)",
"_____no_output_____"
],
[
"x = plt.hist(fatty.ravel(),bins=256)",
"_____no_output_____"
],
[
"## Next, experiment with different cut-off intensity thresholds to try to separate the background of the image\n## Uncomment the code below and play with the value of 'thresh' to create two new binarized images\n\nthresh = 30\n\ndense_smooth = gaussian_filter(dense, sigma = 5)\ndense_bin = (dense_smooth > thresh) * 255\n\nfatty_smooth = gaussian_filter(fatty, sigma = 5)\nfatty_bin = (fatty_smooth > thresh) * 255",
"_____no_output_____"
],
[
"## Visualize the binarized images to see if the threshold you chose separates the breast tissue from the background\nplt.imshow(dense_bin)",
"_____no_output_____"
],
[
"plt.imshow(fatty_bin)",
"_____no_output_____"
]
],
[
[
"Experiment with different values of 'thresh' above until you are satisfied that you are able to create a reasonable separation of tissue from background.\n\nOne image pre-processing trick you might try before binarizing is _smoothing_ which you perform with a gaussian filter. Try adding the following step before binarization: \n\nimg_smooth = gaussian_filter(img, sigma = 5)\n\nWhere changing the value of _sigma_ will change the amount of smoothing. ",
"_____no_output_____"
],
[
"## Once you have chosen your value of thresh, let's use it to see if we can classify dense v. fatty breast tissue: ",
"_____no_output_____"
]
],
[
[
"## Let's first get all of the intensity values of the breast tissue for our fatty breast images using the\n## segmentation method above\nfatty_imgs = glob.glob(\"fatty/*\")\ndense_imgs = glob.glob(\"dense/*\")\n\nfatty_intensities = []\n\nfor i in fatty_imgs: \n \n img = plt.imread(i)\n img_mask = (img > thresh)\n fatty_intensities.extend(img[img_mask].tolist())\n\nx = plt.hist(fatty_intensities,bins=256)",
"_____no_output_____"
],
[
"## Same for dense breast images \ndense_intensities = []\n\nfor i in dense_imgs: \n \n img = plt.imread(i)\n img_mask = (img > thresh)\n dense_intensities.extend(img[img_mask].tolist())\n \nx = plt.hist(dense_intensities,bins=256)",
"_____no_output_____"
],
[
"## Use scipy.stats.mode to get the mode of the two distributions above\n## scipy.stats.mode returns mode and bin-count\n\ndense_mode = scipy.stats.mode(dense_intensities)[0][0]\nfatty_mode = scipy.stats.mode(fatty_intensities)[0][0]\nprint('dense_mode: ',dense_mode,', fatty_mode: ',fatty_mode)",
"dense_mode: 176 , fatty_mode: 140\n"
],
[
"## Loop through all of the fatty images, binarize them using your threshold, and compare the peaks of the \n## distributions of the *tissue only* to the peaks of the distributions of all fatty and all dense breast images: \n\nfor i in fatty_imgs: \n \n img = plt.imread(i)\n img_mask = (img > thresh)\n \n ## Use scipy.stats.mode to get the mode of the tissue in the image: \n img_mode = scipy.stats.mode(img[img_mask])[0][0] \n \n fatty_delta = img_mode - fatty_mode\n dense_delta = img_mode - dense_mode\n \n if (np.abs(fatty_delta) < np.abs(dense_delta)):\n print(\"Fatty\")\n else:\n print(\"Dense\")",
"Fatty\nFatty\nFatty\nFatty\nDense\nDense\nFatty\nDense\nFatty\nFatty\n"
],
[
"## Loop through all of the dense images, binarize them using your threshold, and compare the peaks of the \n## distributions of the *tissue only* to the peaks of the distributions of all fatty and all dense breast images: \n\nfor i in dense_imgs: \n \n img = plt.imread(i)\n img_mask = (img > thresh)\n \n ## Use scipy.stats.mode to get the mode of the tissue in the image: \n img_mode = scipy.stats.mode(img[img_mask])[0][0]\n \n fatty_delta = img_mode - scipy.stats.mode(fatty_intensities)[0][0]\n dense_delta = img_mode - scipy.stats.mode(dense_intensities)[0][0]\n \n if (np.abs(fatty_delta) < np.abs(dense_delta)):\n print(\"Fatty\")\n else:\n print(\"Dense\")",
"Dense\nDense\nDense\nDense\nDense\nDense\nDense\nDense\nDense\nDense\n"
]
],
[
[
"### Reference Dense Brest vs Fatty Brest\nhttps://www.cdc.gov/cancer/breast/basic_info/dense-breasts.htm#:~:text=Breast%20density%20reflects%20the%20amount,as%20seen%20on%20a%20mammogram.&text=The%20breasts%20are%20almost%20entirely,about%2040%25%20of%20women).",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecef6fed414d715c4c583feaa665d59dbbb5e3ee | 196,736 | ipynb | Jupyter Notebook | singlelayer-perceptron-how-to-implement.ipynb | vitorglemos/kaggle-kernel-algorithm | 73aa4a55b3e0ad8d884e952dfa77d550937e5ab3 | [
"MIT"
] | 1 | 2020-08-15T05:10:42.000Z | 2020-08-15T05:10:42.000Z | singlelayer-perceptron-how-to-implement.ipynb | vitorglemos/kaggle-kernel-algorithm | 73aa4a55b3e0ad8d884e952dfa77d550937e5ab3 | [
"MIT"
] | null | null | null | singlelayer-perceptron-how-to-implement.ipynb | vitorglemos/kaggle-kernel-algorithm | 73aa4a55b3e0ad8d884e952dfa77d550937e5ab3 | [
"MIT"
] | 1 | 2020-10-14T14:46:11.000Z | 2020-10-14T14:46:11.000Z | 554.185915 | 148,276 | 0.941795 | [
[
[
"# Artificial Neural Networks \n## Introduction \n\n<img src=\"https://media.springernature.com/original/springer-static/image/art%3A10.1007%2Fs40846-016-0191-3/MediaObjects/40846_2016_191_Fig1_HTML.gif\">\n\n<p style=\"text-align: justify;\">Artificial Neural Networks are mathematical models inspired by the human brain, specifically the ability to learn, process, and perform tasks. The Artificial Neural Networks are powerful tools that assist in solving complex problems linked mainly in the area of combinatorial optimization and machine learning. In this context, artificial neural networks have the most varied applications possible, as such models can adapt to the situations presented, ensuring a gradual increase in performance without any human interference. We can say that the Artificial Neural Networks are potent methods can give computers a new possibility, that is, a machine does not get stuck to preprogrammed rules and opens up various options to learn from its own mistakes.</p>",
"_____no_output_____"
],
[
"## Biologic Model\n\n<img src=\"https://www.neuroskills.com/images/photo-500x500-neuron.png\">\n<p style=\"text-align: justify;\">Artificial neurons are designed to mimic aspects of their biological counterparts. The neuron is one of the fundamental units that make up the entire brain structure of the central nervous system; such cells are responsible for transmitting information through the electrical potential difference in their membrane. In this context, a biological neuron can be divided as follows.</p>\n\n**Dendrites** – are thin branches located in the nerve cell. These cells act on receiving nerve input from other parts of our body.\n\n**Soma** – acts as a summation function. As positive and negative signals (exciting and inhibiting, respectively) arrive in the soma from the dendrites they are added together.\n\n**Axon** – gets its signal from the summation behavior which occurs inside the soma. It is formed by a single extended filament located throughout the neuron. The axon is responsible for sending nerve impulses to the external environment of a cell.",
"_____no_output_____"
],
[
"## Artificial Neuron as Mathematic Notation\nIn general terms, an input X is multiplied by a weight W and added a bias b producing the net activation. \n<img style=\"max-width:60%;max-height:60%;\" src=\"https://miro.medium.com/max/1290/1*-JtN9TWuoZMz7z9QKbT85A.png\">\n\nWe can summarize an artificial neuron with the following mathematical expression:\n$$\n\\hat{y} = f\\left(\\text{net}\\right)= f\\left(\\vec{w}\\cdot\\vec{x}+b\\right) = f\\left(\\sum_{i=1}^{n}{w_i x_i + b}\\right)\n$$",
"_____no_output_____"
],
[
"## The SigleLayer Perceptron\n\n<p style=\"text-align: justify;\">The Perceptron and its learning algorithm pioneered the research in neurocomputing. the perceptron is an algorithm for supervised learning of binary classifiers [1]. A binary classifier is a function which can decide whether or not an input, represented by a vector of numbers, belongs to some specific class. It is a type of linear classifier, i.e. a classification algorithm that makes its predictions based on a linear predictor function combining a set of weights with the feature vector.<p>\n \n<img src=\"https://www.edureka.co/blog/wp-content/uploads/2017/12/Perceptron-Learning-Algorithm_03.gif\">\n \n#### References\n \n- Freund, Y.; Schapire, R. E. (1999). \"Large margin classification using the perceptron algorithm\" (PDF). Machine Learning\n\n- Aizerman, M. A.; Braverman, E. M.; Rozonoer, L. I. (1964). \"Theoretical foundations of the potential function method in pattern recognition learning\". Automation and Remote Control. 25: 821–837.\n \n- Mohri, Mehryar and Rostamizadeh, Afshin (2013). Perceptron Mistake Bounds.",
"_____no_output_____"
],
[
"## The SingleLayer Perceptron Learning\nLearning goes by calculating the prediction of the perceptron:\n\n### Basic Neuron \n$$\n\\hat{y} = f\\left(\\vec{w}\\cdot\\vec{x} + b) = f( w_{1}x_{1} + w_2x_{2} + \\cdots + w_nx_{n}+b\\right)\\,\n$$\n\nAfter that, we update the weights and the bias using as:\n\n$$\n\\hat{w_i} = w_i + \\alpha (y - \\hat{y}) x_{i} \\,,\\ i=1,\\ldots,n\\,;\\\\\n$$\n$$\n\\hat{b} = b + \\alpha (y - \\hat{y})\\,.\n$$",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nclass SingleLayerPerceptron:\n def __init__(self, my_weights, my_bias, learningRate=0.05):\n self.weights = my_weights\n self.bias = my_bias\n self.learningRate = learningRate\n \n def activation(self, net):\n answer = 1 if net > 0 else 0\n return answer\n \n def neuron(self, inputs):\n neuronArchitecture = np.dot(self.weights, inputs) + self.bias\n return neuronArchitecture\n \n def neuron_propagate(self, inputs):\n processing = self.neuron(inputs)\n return self.activation(processing) \n \n def training(self, inputs, output):\n output_prev = self.neuron_propagate(inputs)\n self.weights = [W + X * self.learningRate * (output - output_prev)\n for (W, X) in zip(self.weights, inputs)]\n self.bias += self.learningRate * (output - output_prev)\n error_calculation = np.abs(output_prev - output)\n return error_calculation",
"_____no_output_____"
],
[
"data = pd.DataFrame(columns=('x1', 'x2'), data=np.random.uniform(size=(600,2)))\ndata.head()",
"_____no_output_____"
],
[
"def show_dataset(data, ax):\n data[data.y==1].plot(kind='scatter', ax=ax, x='x1', y='x2', color='blue')\n data[data.y==0].plot(kind='scatter', ax=ax, x='x1', y='x2', color='red')\n plt.grid()\n plt.title(' My Dataset')\n ax.set_xlim(-0.1,1.1)\n ax.set_ylim(-0.1,1.1)\n \ndef testing(inputs):\n answer = int(np.sum(inputs) > 1)\n return answer\n\ndata['y'] = data.apply(testing, axis=1)",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(10,10))\nshow_dataset(data, fig.gca())",
"_____no_output_____"
],
[
"InitialWeights = [0.1, 0.1]\nInitialBias = 0.01\nLearningRate = 0.1\nSLperceptron = SingleLayerPerceptron(InitialWeights, \n InitialBias,\n LearningRate)",
"_____no_output_____"
],
[
"import random, itertools\n\ndef showAll(perceptron, data, threshold, ax=None):\n if ax==None:\n fig = plt.figure(figsize=(5,4))\n ax = fig.gca()\n \n show_dataset(data, ax)\n show_threshold(perceptron, ax)\n title = 'training={}'.format(threshold + 1)\n ax.set_title(title)\n \ndef trainingData(SinglePerceptron, inputs):\n count = 0 \n for i, line in inputs.iterrows():\n count = count + SinglePerceptron.training(line[0:2], \n line[2])\n \n return count\n\ndef limit(neuron, inputs):\n weights_0 = neuron.weights[0]\n weights_1 = neuron.weights[1]\n bias = neuron.bias\n threshold = -weights_0 * inputs - bias\n threshold = threshold / weights_1\n return threshold\n\ndef show_threshold(SinglePerceptron, ax):\n xlim = plt.gca().get_xlim()\n ylim = plt.gca().get_ylim()\n \n x2 = [limit(SinglePerceptron, x1) for x1 in xlim]\n \n ax.plot(xlim, x2, color=\"yellow\")\n ax.set_xlim(-0.1,1.1)\n ax.set_ylim(-0.1,1.1)\n\nf, axarr = plt.subplots(3, 4, sharex=True, sharey=True, figsize=(12,12))\naxs = list(itertools.chain.from_iterable(axarr))\nuntil = 12\nfor interaction in range(until):\n showAll(SLperceptron, data, interaction, ax=axs[interaction])\n trainingData(SLperceptron, data)",
"_____no_output_____"
]
],
[
[
"Example using Multilayer Perceptron (no libraries): https://www.kaggle.com/vitorgamalemos/iris-flower-using-multilayer-perceptron/",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecef75da4cf7f44d028b967f6040de4efd2e25f9 | 2,469 | ipynb | Jupyter Notebook | Algorithms/Sorting_Algorithms/Cycle_Sort/Python/Cycle_Sort.ipynb | darshanjain2000/OneDayOneAlgo | 0e286927aadb618f780de04ace83907aaf80bfa9 | [
"MIT"
] | 32 | 2020-05-23T07:40:31.000Z | 2021-02-02T18:14:30.000Z | Algorithms/Sorting_Algorithms/Cycle_Sort/Python/Cycle_Sort.ipynb | darshanjain2000/OneDayOneAlgo | 0e286927aadb618f780de04ace83907aaf80bfa9 | [
"MIT"
] | 45 | 2020-05-22T10:30:51.000Z | 2020-12-28T08:17:13.000Z | Algorithms/Sorting_Algorithms/Cycle_Sort/Python/Cycle_Sort.ipynb | darshanjain2000/OneDayOneAlgo | 0e286927aadb618f780de04ace83907aaf80bfa9 | [
"MIT"
] | 31 | 2020-05-22T10:18:16.000Z | 2020-10-23T07:52:35.000Z | 24.939394 | 113 | 0.456865 | [
[
[
"def cycleSort(array): \n \n for cycleStart in range(0, len(array) - 1): # Looping through the array to find the correct postition\n item = array[cycleStart] \n pos = cycleStart \n \n for i in range(cycleStart + 1, len(array)): \n if array[i] < item: \n pos += 1\n \n if pos == cycleStart: # Ignore if item is same item is found. \n continue\n \n while item == array[pos]: # Insert the item there or right after any duplicates. \n pos += 1\n array[pos], item = item, array[pos] \n \n while pos != cycleStart: # Rotate the rest of the cycle. \n \n pos = cycleStart # Position to put the item\n for i in range(cycleStart + 1, len(array)): \n if array[i] < item: \n pos += 1\n \n while item == array[pos]: # Put the item there or right after any duplicates. \n pos += 1\n array[pos], item = item, array[pos] ",
"_____no_output_____"
],
[
"li = [1, 34, 55, 22, 24, 78, 96, 8, 81, 64, 77, 65]",
"_____no_output_____"
],
[
"cycleSort(li)",
"_____no_output_____"
],
[
"print(\"After sort : \")\nfor _ in range(0, len(li)):\n print(li[_], end = \" \")",
"After sort : \n1 8 22 24 34 55 64 65 77 78 81 96 "
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
ecef764d32f3bbff89cb30039235be74b96d7f69 | 501,749 | ipynb | Jupyter Notebook | debug/url_func.ipynb | gabelev/MISMI | b3d34632e491d9c96824f9a58cabc093755d8b83 | [
"MIT"
] | null | null | null | debug/url_func.ipynb | gabelev/MISMI | b3d34632e491d9c96824f9a58cabc093755d8b83 | [
"MIT"
] | null | null | null | debug/url_func.ipynb | gabelev/MISMI | b3d34632e491d9c96824f9a58cabc093755d8b83 | [
"MIT"
] | null | null | null | 71.250923 | 140 | 0.598528 | [
[
[
"def url_builder(page_range, terms):\n base_url = 'http://www.amazon.com/s/rh=%2Ck%3A'\n page = '&page='\n plus = '+'\n search_terms = plus.join(terms.split())\n return [base_url + search_terms + page + str(i) for i in range(1, page_range+1)]",
"_____no_output_____"
],
[
"url_builder(4, 'after happiness')",
"_____no_output_____"
],
[
"def url_dict_builder(page_range, list_of_terms):\n result = {}\n for term in list_of_terms:\n urls_term = {}\n urls = url_builder(page_range, term)\n result[term] = urls\n return result",
"_____no_output_____"
],
[
"terms_list = ['after happiness',\n'career advancement',\n'fullest potential',\n'success formula',\n'professional growth',\n'formula for success',\n'finding happiness',\n'change your thinking',\n'positive psychology',\n'self-improvement',\n'motivational books',\n'personal success',\n'professional success',\n'personal growth',\n'self improvement',\n'self-help',\n'positive thinking',\n'positive genius',\n'achieving happiness',\n'science of happiness',\n'positive inception',\n'achieving success',\n'positive inspiration',\n'success accelerants',\n'spreading happiness',\n'positive change',\n'navigating the workplace',\n'first job after college',\n'career advancement',\n'AEI',\n'getting what you want',\n'college graduation books',\n'American Enterprise Institute',\n'professional growth',\n'soft skills',\n'college graduation gifts',\n'career advice',\n'college graduation gift',\n'adult life',\n'bad boss',\n'professional success',\n'personal success',\n'graduation books',\n'career success',\n'living well',\n'workplace culture',\n'communication skills',\n'personal development',\n'office manners',\n'getting ahead',\n'professional writing',\n'living a good life',\n'corporate ladder',\n'professional growth',\n'professional success',\n'getting ahead',\n'business classics',\n'collaborative management',\n'power of relationships',\n'getting what you want',\n'making friends',\n'making connections',\n'connecting to people',\n'personal success',\n'personal growth',\n'relationship building',\n'business classic',\n'connecting',\n'business success',\n'social media networking',\n'communication skills',\n'meaningful relationships',\n'meaningful connections',\n'connecting to the world',\n'networking',\n'business relationships',\n'IHHP',\n'handling stress',\n'nerves of steel',\n'interview stress',\n'business psychology',\n'overcoming obstacles',\n'personal improvement',\n'power of pressure',\n'pressure solutions',\n'decision-making',\n'personal success',\n'decision making',\n'self-help',\n'stress management',\n'self improvement',\n'self help',\n'professional success',\n'business success',\n'COTE',\n'nature of pressure',\n'face pressure',\n'pressure traps',\n'pressure management',\n'performance under pressure',\n'decisions under pressure',\n'success under pressure',\n'handling pressure',\n'health and human potential',\n'performance anxiety',\n'startup founders',\n'job for tomorrow',\n'intelligent risk',\n'add value',\n'personal brand',\n'career growth',\n'build a career',\n'business inspiration',\n'personal branding',\n'adding value',\n'professional growth',\n'professional inspiration',\n'building a career',\n'career plans',\n'entrepreneurship',\n'professional development',\n'your brand',\n'personal development',\n'success stories',\n'career guide',\n'career guides',\n'personal success',\n'personal growth',\n'founders',\n'startup success',\n'startup',\n'adapt to the future',\n'transform your career',\n'invest in yourself',\n'lifeline relationships',\n'inner circle',\n'building deep relationships',\n'deep relationships',\n'building trusting relationships',\n'dream team',\n'trusting relationships',\n'existential intelligence',\n'self help',\n'business',\n'relationships',\n'networking',\n'self-help',\n'building relationships',\n'building trust',\n'self improvement',\n'self-improvement',\n'leadership',\n'career development',\n'making connections',\n'goal-setting',\n'sparring',\n'corporate sparring',\n'building relationships at work',\n'professional relationships',\n'trusted team',\n'meaningful connections',\n'creativity',\n'creative genius',\n'disruptive innovation',\n'rediscover creativity',\n'innovation',\n'breakthrough creativity',\n'rethink',\n're-think',\n'creative',\n'personal improvement',\n'personal success',\n'achieve potential',\n'achieving potential',\n'full potential',\n'unlock potential',\n'unlocking potential',\n'self growth',\n'art',\n'self discovery',\n'self-discovery',\n'know yourself',\n'unlock your potential',\n'unlocking your creativity',\n'unlock your creativity',\n'unleash creativity',\n'unleash genius',\n'philosophy of creativity',\n'creative value',\n'outside the box thinking',\n'37 signals',\n'callaboration',\n'co-working space',\n'business',\n'work from home',\n'virtual workspace',\n'telecommuting',\n'remote work',\n'remote jobs',\n'real estate footprint',\n'work at home jobs',\n'work from home opportunities',\n'work at home',\n'work from home moms',\n'working from home',\n'work from home careers',\n'telecommuting jobs',\n'telecommute',\n'virtual office space',\n'under one roof',\n'remote business',\n'work remotely',\n'working remotely',\n'work remotely from home',\n'commute',\n'work from home jobs',\n'business psychology',\n'essentialist',\n'essential',\n'minimalism',\n'minimalist',\n'essential living',\n'how to be more produtive',\n'how to be more productive at work',\n'being more productive',\n'increase productivity',\n'increasing productivity',\n'increased productivity',\n'productivity tools',\n'achieve more',\n'mental clairty',\n'mental focus',\n'design your life',\n'time management',\n'time management skills',\n'time management tools',\n'effective time management',\n'power of choice',\n'what really matters',\n'vital few',\n'trivial many',\n'less but better',\n'less is more',\n'psychology',\n'digital reputation',\n'reputation management',\n'digital footprint',\n'digital future',\n'online reputation management',\n'brand reputation',\n'online reputation',\n'personal data',\n'personalized data',\n'privacy management',\n'reputation.com',\n'data privacy',\n'online privacy',\n'online presense',\n'privacy settings',\n'privacy issues',\n'digital privacy',\n'digital privacy issues',\n'online privacy issues',\n'social media privacy',\n'millenial privacy',\n'reputation economics',\n'privacy advocate',\n'social media mistakes',\n'personal data collection',\n'personal data protection',\n'reputation rich',\n'power of reputation',\n'Maine',\n'orchard',\n'apple',\n'apple orchard',\n'love triangle',\n'psychic',\n'premonitions',\n'Amish cooking',\n'Amish recipes',\n'Amish Romance',\n'Christian Fiction',\n'Christian Literature',\n'Christian Romance',\n'Contemporary Christian Romance',\n'Contemporary Religious Fiction',\n'Religious Fiction',\n'Religious Romance',\n'Women’s Christian Fiction',\n'Women’s Religious Fiction',\n'Contemporary Christian Fiction',\n'amish fiction',\n'clean fiction',\n'clean reads',\n'clean romance',\n'inspirational romance',\n'inspirational fiction',\n'bonnet',\n'plain',\n'buggy',\n'Kapp',\n'Englischer',\n'Old Order Amish',\n'Old Order Mennonite',\n'Maine',\n'orchard',\n'apple',\n'apple orchard',\n'gardening',\n'garden',\n'psychic',\n'premonitions',\n'Amish cooking',\n'Amish recipes',\n'Amish Romance',\n'Christian Fiction',\n'Christian Literature',\n'Christian Romance',\n'Contemporary Christian Romance',\n'Contemporary Religious Fiction',\n'Religious Fiction',\n'Religious Romance',\n'Women%27s Christian Fiction',\n'Women%27s Religious Fiction',\n'Contemporary Christian Fiction',\n'amish fiction',\n'clean fiction',\n'clean reads',\n'clean romance',\n'inspirational romance',\n'inspirational fiction',\n'bonnet',\n'plain',\n'buggy',\n'Kapp',\n'Englischer',\n'Old Order Amish',\n'Old Order Mennonite',\n'prayer',\n'rape',\n'shunning',\n'Mennonite',\n'Amish quilt',\n'Amish Romance',\n'Christian Fiction',\n'Christian Literature',\n'Christian Romance',\n'Contemporary Christian Romance',\n'Contemporary Religious Fiction',\n'Religious Fiction',\n'Religious Romance',\n'Women’s Christian Fiction',\n'Women’s Religious Fiction',\n'Contemporary Christian Fiction',\n'amish fiction',\n'clean fiction',\n'clean reads',\n'clean romance',\n'inspirational romance',\n'inspirational fiction',\n'bonnet',\n'plain',\n'buggy',\n'Kapp',\n'Englischer',\n'Old Order Amish',\n'Old Order Mennonite',\n'culture clash',\n'single parent',\n'parenthood',\n'widow',\n'widowhood',\n'conversion',\n'Amish Romance',\n'Christian Fiction',\n'Christian Literature',\n'Christian Romance',\n'Contemporary Christian Romance',\n'Contemporary Religious Fiction',\n'Religious Fiction',\n'Religious Romance',\n'Women’s Christian Fiction',\n'Women’s Religious Fiction',\n'Contemporary Christian Fiction',\n'amish fiction',\n'clean fiction',\n'clean reads',\n'clean romance',\n'inspirational romance',\n'inspirational fiction',\n'bonnet',\n'plain',\n'buggy',\n'Kapp',\n'Englischer',\n'Old Order Amish',\n'Old Order Mennonite',\n'Amish',\n'christian fiction',\n'amish fiction',\n'pennsylvania',\n'Florida',\n'Spring',\n'brothers',\n'restaurant',\n'diner',\n'novella',\n'Mennonite',\n'Amish cooking',\n'Amish recipes',\n'orchard',\n'Amish Romance',\n'Christian Fiction',\n'Christian Literature',\n'Christian Romance',\n'Contemporary Christian Romance',\n'Contemporary Religious Fiction',\n'Religious Fiction',\n'Religious Romance',\n'Women’s Christian Fiction',\n'Women’s Religious Fiction',\n'Contemporary Christian Fiction',\n'amish fiction',\n'clean fiction',\n'clean reads',\n'clean romance',\n'inspirational romance',\n'inspirational fiction',\n'bonnet',\n'plain',\n'buggy',\n'Kapp',\n'Englischer',\n'Old Order Amish',\n'Old Order Mennonite',\n'holiday',\n'Christmas',\n'Advent',\n'novella',\n'mistaken identity',\n'arts and crafts',\n'arts & crafts',\n'woodworking',\n'woodworker',\n'Amish Romance',\n'Christian Fiction',\n'Christian Literature',\n'Christian Romance',\n'Contemporary Christian Romance',\n'Contemporary Religious Fiction',\n'Religious Fiction',\n'Religious Romance',\n'Women’s Christian Fiction',\n'Women’s Religious Fiction',\n'Contemporary Christian Fiction',\n'amish fiction',\n'clean fiction',\n'clean reads',\n'clean romance',\n'inspirational romance',\n'inspirational fiction',\n'bonnet',\n'plain',\n'buggy',\n'Kapp',\n'Englischer',\n'Old Order Amish',\n'Old Order Mennonite',\n'amish fiction',\n'christian fiction',\n'twins',\n'twin',\n'switched at birth',\n'Amish Romance',\n'Christian Fiction',\n'Christian Literature',\n'Christian Romance',\n'Contemporary Christian Romance',\n'Contemporary Religious Fiction',\n'Religious Fiction',\n'Religious Romance',\n'Women’s Christian Fiction',\n'Women’s Religious Fiction',\n'Contemporary Christian Fiction',\n'amish fiction',\n'clean fiction',\n'clean reads',\n'clean romance',\n'inspirational romance',\n'inspirational fiction',\n'bonnet',\n'plain',\n'buggy',\n'Kapp',\n'Englischer',\n'Old Order Amish',\n'Old Order Mennonite',\n'B&B',\n'BNB',\n'bed and breakfast',\n'bed & breakfast',\n'Kansas',\n'mistaken identity',\n'Mennonite fiction',\n'Amish Romance',\n'Christian Fiction',\n'Christian Literature',\n'Christian Romance',\n'Contemporary Christian Romance',\n'Contemporary Religious Fiction',\n'Religious Fiction',\n'Religious Romance',\n'Women’s Christian Fiction',\n'Women’s Religious Fiction',\n'Contemporary Christian Fiction',\n'amish fiction',\n'clean fiction',\n'clean reads',\n'clean romance',\n'inspirational romance',\n'inspirational fiction',\n'bonnet',\n'plain',\n'buggy',\n'Kapp',\n'Englischer',\n'Old Order Amish',\n'Old Order Mennonite',\n'Mennonite Romance']",
"_____no_output_____"
],
[
"# url_dict_builder(20, terms_list)",
"_____no_output_____"
],
[
"len(terms_list)",
"_____no_output_____"
],
[
"term_set = set(terms_list)",
"_____no_output_____"
],
[
"len(term_set)",
"_____no_output_____"
],
[
"url_dict_builder(20, term_set)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecef88779c82e3bc299863777821bd4f1342694a | 807,242 | ipynb | Jupyter Notebook | 2020-11-24-age_pure_pytorch.ipynb | jvanelteren/ds | 45bab16fda74bc92180a6f6576f08fdd9aa96528 | [
"MIT"
] | null | null | null | 2020-11-24-age_pure_pytorch.ipynb | jvanelteren/ds | 45bab16fda74bc92180a6f6576f08fdd9aa96528 | [
"MIT"
] | null | null | null | 2020-11-24-age_pure_pytorch.ipynb | jvanelteren/ds | 45bab16fda74bc92180a6f6576f08fdd9aa96528 | [
"MIT"
] | null | null | null | 1,019.244949 | 373,635 | 0.951135 | [
[
[
"# Face2Age\n> Predicting someone's age from their face\n\n- toc: false\n- branch: master\n- badges: true\n- comments: true\n- author: Jesse van Elteren\n- image: images/age_recognition.png\n- categories: []",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"This blog is about predicting someone’s age by their face. I've also made an webpage where you [can try to beat the computer](http://35.206.67.46/).\n\nDeep learning has seen incredible results the last couple of year. As long as you have a large enough dataset, it you can transform any input to any output. A couple of examples:\n\n* Image classification, where you map an image to a label (input an image of a frog, computer outputs ‘frog’)\n* Sentence generation (if I start a sentence with ‘Today, the weather is’, computer outputs ‘great, let's take a walk’.\n* Recommender system (which is the next video that will be most likely to keep the user watching)\nAs an aside, deep learning is not without it’s ethical problems, for example these social media recommender systems have a [tendency to radicalize people](https://www.nytimes.com/2018/03/10/opinion/sunday/youtube-politics-radical.html), since that maximizes their 'engagement' to the platform. \n \nIn this project, I’ve taken on a image regression problem. It’s a nice problem, because the answer is not always obvious: some people look older or younger than they really are. By showing the computer model examples and giving feedback how wrong it is on its prediction, the model is improving. We don’t have to explain anything about how an old or young person looks like. Can you imagine how difficult it would be to program how to recognize wrinkles? This is essentially the wonder of neural networks.\n\nWhen training a model, an important part is the performance metric. For this task of guessing someone’s age, I’ve chosen Mean Absolute Error, basically how many years you’ve guessed wrong. A prediction of 12 on an actual age of 10 means the MAE is 2, just as a prediction of 8 also has an MAE of 2.\n\nThe dataset contains about 10.000 images, I’ve trained the model on 70% of the dataset. That leaves about 3k images which the model has not seen. We use this to test it’s performance. In theory, this performance will generalize to other unseen images. In practice that remains to be seen, since real life images can be much messier, e.g. in quality, zoom level and background.\n\nBelow you can see it getting better over time. The horizontal axis displays how many times we feed the training set to the model. The left graph shows performance on the training set, the right graph on the test set. Initially, it’s off by about 11 years, and slowly converging to a MAE of 2 years. But taking the performance on the training dataset is cheating, we are interested in it’s performance on unseen images! The performance there converges to around 4.2.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"# hide\nimport time\nimport functools\nimport random\nfrom pathlib import Path\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport copy\nfrom PIL import Image\n\nimport torch\nfrom torch.utils.data import Dataset, DataLoader, TensorDataset\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torch.utils.tensorboard import SummaryWriter\nfrom torch.optim import lr_scheduler, swa_utils\nimport torchvision\nfrom torchvision.datasets import ImageFolder, DatasetFolder\nfrom torchvision import transforms\nfrom diskcache import Cache",
"_____no_output_____"
],
[
"# hide\ncache = Cache('.cache/diskcache')\nDEVICE = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\nPATH = 'data/face_age'\n\[email protected]()\ndef imgpath_to_normalized_tensor(imgpath):\n # makes a tensor, scales range to 0-1 and normalizes to same as imagenet\n normalize = transforms.Normalize(mean=[0.485, 0.456, 0.406],\n std=[0.229, 0.224, 0.225])\n img = normalize(transforms.PILToTensor()(Image.open(imgpath)).float()/255)\n return img\n\n\nclass Ageset(Dataset):\n def __init__(self, path, transforms = None, valid=False, split_pct = 0.3):\n self.image_paths = list(Path(path).rglob(\"*.png\"))\n random.seed(42)\n random.shuffle(self.image_paths)\n split_point = int(len(self)*0.3)\n if valid:\n self.image_paths = self.image_paths[:split_point]\n # print('len validation dataset', len(self.image_paths))\n else:\n self.image_paths = self.image_paths[split_point:]\n # print('len train dataset', len(self.image_paths))\n def __len__(self):\n return len(self.image_paths)\n \n def show_image(self,i):\n return Image.open(self.image_paths[i])\n\n @classmethod # somehow this is needed for diskcache to work properly. Or define the function outside of the class\n @functools.lru_cache(maxsize=None)\n @cache.memoize()\n def imgpath_to_normalized_tensor(cls,imgpath):\n # makes a tensor, scales range to 0-1 and normalizes to same as imagenet\n normalize = transforms.Normalize(mean=[0.485, 0.456, 0.406],\n std=[0.229, 0.224, 0.225])\n img = normalize(transforms.PILToTensor()(Image.open(imgpath)).float()/255)\n return img\n \n def __getitem__(self,i):\n if isinstance(i, slice):\n # return [self[n] for n,_ in enumerate(self.image_paths[i])]\n # maybe better to return a new ageset to preserve the functions\n ret = copy.deepcopy(self)\n ret.image_paths = ret.image_paths[i]\n return ret\n\n return (self.imgpath_to_normalized_tensor(self.image_paths[i]),\n int(self.image_paths[i].parent.name))\n",
"_____no_output_____"
],
[
"# hide\n# this was a small experiment where I wanted to load the full dataset onto GPU. The dataset was too large though\ndef construct_tensor_dataset(path, max_len=None):\n # this took 20 GB RAM max\n image_paths = list(Path(path).rglob(\"*.png\"))\n random.shuffle(image_paths)\n if max_len:\n image_paths = image_paths[:max_len]\n xs = torch.empty(len(image_paths), 3,200,200)\n print('empty created')\n for i, loc in enumerate(image_paths):\n if i%1000==0: print(i)\n xs[i] = imgpath_to_normalized_tensor(loc)\n torch.save(xs,'data/input/xs')\n ys = torch.stack([torch.Tensor([int(Path(loc).parent.name)]) for loc in image_paths])\n torch.save(ys,'data/input/ys')\n return None\n\nclass AgeTensorDataset(TensorDataset):\n def __init__(self, xs, ys, valid=False, split_pct=0.3):\n length = len(xs)\n split = int(xs.shape[0]*split_pct)\n if valid:\n super().__init__(xs[:split],ys[:split])\n else:\n super().__init__(xs[split:],ys[split:])\n\n def __getitem__(self,x):\n return super().__getitem__(x)",
"_____no_output_____"
],
[
"# hide\nclass AgeResnet(nn.Module):\n # a resnet with a fresh fully connected layer on top\n def __init__(self, size='18', feat_extract=False):\n super().__init__()\n resnet = 'torchvision.models.resnet'+size+'(pretrained=True)'\n resnet = eval(resnet)\n modules=list(resnet.children())[:-1]\n self.resnet =nn.Sequential(*modules)\n\n if feat_extract:\n # with feature extraction we only train the linear layer and keep the resnet parameters fixed \n for m in self.modules():\n m.requires_grad_(False)\n\n self.fc = nn.Linear(in_features=512, out_features=1, bias=True)\n nn.init.kaiming_normal_(self.fc.weight)\n\n def forward(self,x):\n out = self.resnet(x)\n x = torch.flatten(out, 1)\n return self.fc(x)",
"_____no_output_____"
],
[
"# hide\ndef determine_size(dataset):\n # a helper function to determine the size of the dataset\n num_items = len(dataset)\n img_dimensions = list(dataset[0][0].shape)\n bytes_per_fp32 = 4\n bytes_per_gb = 1024**3\n size_in_gb = num_items * int(np.product(img_dimensions)) * bytes_per_fp32 / bytes_per_gb\n print('items in dataset', num_items, 'img_dimensions', img_dimensions, 'size of ds in memory in gb:', size_in_gb)",
"_____no_output_____"
],
[
"# hide\ndef train():\n # the training function, which allows for different types of training\n # writes tensorboard stats and saves best model\n # early stopping can be implemented with NOT_IMPROVE_COUNT\n best_loss = 1000000000\n best_model = None\n not_improve_count = 0\n loss = {'train':[], 'val':[]}\n\n for epoch in range(NUM_EPOCH):\n print(f'Starting epoch {epoch}')\n start_time = time.time()\n for phase in ['train', 'val']:\n if phase == 'train':\n model.train()\n else:\n model.eval()\n\n total_loss = 0\n for data in dls[phase]:\n x, y = data[0].to(DEVICE), data[1].to(DEVICE)\n with torch.set_grad_enabled(phase == 'train'):\n pred = model(x)\n loss = loss_fn(y, pred)\n total_loss += loss * len(y)\n if phase == 'train':\n loss.backward()\n opt.step()\n opt.zero_grad()\n if SWA_ENABLED and epoch > SWA_START:\n # stochastic weight averaging\n swa_model.update_parameters(model)\n swa_sched.step()\n elif SCHED_ENABLED:\n # learning rate scheduling, not really useful icm Adam\n sched.step(loss)\n writer.add_scalar('lr/scheduler', sched.get_last_lr()[0], epoch)\n writer.add_scalar('lr/optparamgroup0', opt.param_groups[0]['lr'], epoch)\n writer.add_scalar('batchloss/'+phase, loss, epoch)\n \n writer.add_scalar('loss/'+phase, total_loss/len(dls[phase].dataset), epoch)\n \n if total_loss < best_loss:\n best_loss = total_loss\n best_model = copy.deepcopy(model.state_dict())\n not_improve_count = 0\n else:\n not_improve_count += 1\n if not_improve_count > EARLY_STOP:\n print('early stopping!')\n break\n \n print(f\"loss after epoch {epoch} : {total_loss / len(dls['val'].dataset)}\")\n writer.add_scalar('time', (time.time()-start_time)/60, epoch)\n\n if SWA_ENABLED:\n swa_model.to('cpu')\n swa_utils.update_bn(train_dl, swa_model)\n swa_model.to(DEVICE)\n total_loss_train = 0\n total_loss_val = 0\n\n with torch.no_grad():\n for data in train_dl:\n x, y = data[0].to(DEVICE), data[1].to(DEVICE)\n total_loss_train += loss_fn(y, model(x)) * len(y)\n writer.add_scalar('loss/train', total_loss_train/len(train_set), epoch+1)\n swa_model.eval()\n\n for data in val_dl:\n x, y = data[0].to(DEVICE), data[1].to(DEVICE)\n total_loss_val += loss_fn(y, model(x)) * len(y)\n writer.add_scalar('loss/val', total_loss_train/len(val_set), epoch+1)\n swa_model.avg_fn=None\n torch.save(swa_model.state_dict() ,'data/models/'+'swa_model'+str((total_loss_val/len(val_set)).item()))\n torch.save(best_model,'data/models/'+'model'+str((best_loss/len(val_set)).item()))\n writer.flush()\n writer.close()",
"_____no_output_____"
],
[
"# hide\n# adam works best with lr of 0.001 (tested 0.1 and 0.01)\n# adam without scheduler works best\n# first it took 6 minutes to load all the datasets. With lru cache it was immediate (6GB memory use). With disk cache it took about 1-2 minutes. Great result. Finally what worked even better is not do any preloading, but just lru_cache getitem. This shaved off the initial preloading \n# feature extraction led to MSE of 12 after 40 epochs. Didnt really work. Maybe unfreeze more\n# adam was outperformed in feature extraction, but for finetuning it worked better\n# larger bs converges better and runs sligthly faster (about 10%) [512, 256,64,8]\n# SWA gives an extreme improvement, from loss 3.7 to 2.7!!, ran 40 epochs, then 15 SWA epochs\n\n\ndef mae_loss(y, pred):\n return (torch.abs(y-pred.T)).mean()\nloss_fn = mae_loss\n\nNUM_EPOCH = 40\nSWA_START = 40\nLR = 0.001\nBATCH_SIZE = 512\nDEVICE = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\nSWA_ENABLED = False\nSCHED_ENABLED = False\nEARLY_STOP = 5\n\n\ntorch.cuda.empty_cache()\n\ntrain_set = Ageset(\"data/face_age\")\ndetermine_size(train_set)\ntrain_dl = DataLoader(train_set, batch_size=BATCH_SIZE, shuffle=True)\n\nval_set = Ageset(\"data/face_age\", valid=True)\ndetermine_size(val_set) \nval_dl = DataLoader(val_set, batch_size=BATCH_SIZE, shuffle=True)\ndls = {'train': train_dl, 'val': val_dl}\n\nfeat={True:'feat_ext', False:'finetune'}\nopts = {0:'adam',1:'adabound'}\n\n\n# the below is a somewhat hacked loop to iteratively train different models and gauge the performance with tensorboard\nfor i in [False]:\n for j in range(1):\n for BATCH_SIZE in [256]:\n for res in [18]:\n torch.cuda.empty_cache()\n\n writer = SummaryWriter(comment=f'{feat[i]} opt {opts[j]} epoch {NUM_EPOCH} SWA_START {SWA_START} LR BATCH_SIZE {LR}')\n model = AgeResnet(size=str(res), feat_extract=i)\n model = model.to(DEVICE)\n\n if j ==0:\n opt = torch.optim.Adam(model.parameters(), LR)\n if j ==1:\n opt = adabound.AdaBound(model.parameters(), lr=1e-3, final_lr=0.1)\n\n if SCHED_ENABLED:\n # if i ==0:\n # sched = torch.optim.lr_scheduler.ReduceLROnPlateau(opt)\n # sched.get_last_lr = lambda: [1]\n # if i == 1:\n # sched = torch.optim.lr_scheduler.OneCycleLR(opt, LR, steps_per_epoch=len(train_dl), epochs=NUM_EPOCH)\n # if i == 2:\n sched = torch.optim.lr_scheduler.MultiplicativeLR(opt, lr_lambda=lambda x: 1)\n if SWA_ENABLED:\n swa_model = swa_utils.AveragedModel(model)\n swa_sched = swa_utils.SWALR(opt, swa_lr = 0.0005)\n train()",
"items in dataset 6837 img_dimensions [3, 200, 200] size of ds in memory in gb: 3.0563771724700928\nitems in dataset 2930 img_dimensions [3, 200, 200] size of ds in memory in gb: 1.309812068939209\nStarting epoch 0\nloss after epoch 0 : 12.120803833007812\nStarting epoch 1\nloss after epoch 1 : 8.390729904174805\nStarting epoch 2\nloss after epoch 2 : 10.963115692138672\nStarting epoch 3\nloss after epoch 3 : 8.751303672790527\nStarting epoch 4\nloss after epoch 4 : 6.533456802368164\nStarting epoch 5\nloss after epoch 5 : 5.988646030426025\nStarting epoch 6\nloss after epoch 6 : 5.376057147979736\nStarting epoch 7\nloss after epoch 7 : 4.809545040130615\nStarting epoch 8\nloss after epoch 8 : 5.354997634887695\nStarting epoch 9\nloss after epoch 9 : 4.8012542724609375\nStarting epoch 10\nloss after epoch 10 : 5.545410633087158\nStarting epoch 11\nloss after epoch 11 : 5.049632549285889\nStarting epoch 12\nloss after epoch 12 : 4.769824504852295\nStarting epoch 13\nloss after epoch 13 : 4.643012046813965\nStarting epoch 14\nloss after epoch 14 : 4.807333946228027\nStarting epoch 15\nloss after epoch 15 : 5.639796257019043\nStarting epoch 16\nloss after epoch 16 : 5.593840599060059\nStarting epoch 17\nloss after epoch 17 : 4.487029075622559\nStarting epoch 18\nloss after epoch 18 : 4.736180782318115\nStarting epoch 19\nloss after epoch 19 : 4.9413652420043945\nStarting epoch 20\nloss after epoch 20 : 5.095068454742432\nStarting epoch 21\nloss after epoch 21 : 4.473381519317627\nStarting epoch 22\nloss after epoch 22 : 4.699722766876221\nStarting epoch 23\nloss after epoch 23 : 4.602048397064209\nStarting epoch 24\nloss after epoch 24 : 4.7109293937683105\nStarting epoch 25\nloss after epoch 25 : 4.4954328536987305\nStarting epoch 26\nloss after epoch 26 : 4.651307106018066\nStarting epoch 27\nloss after epoch 27 : 4.680775165557861\nStarting epoch 28\nloss after epoch 28 : 4.300748825073242\nStarting epoch 29\nloss after epoch 29 : 4.537985801696777\nStarting epoch 30\nloss after epoch 30 : 5.001606464385986\nStarting epoch 31\nloss after epoch 31 : 4.299375534057617\nStarting epoch 32\nloss after epoch 32 : 4.338717937469482\nStarting epoch 33\nloss after epoch 33 : 4.439096450805664\nStarting epoch 34\nloss after epoch 34 : 4.383476257324219\nStarting epoch 35\nloss after epoch 35 : 5.083619594573975\nStarting epoch 36\nloss after epoch 36 : 4.240642070770264\nStarting epoch 37\nloss after epoch 37 : 4.3507981300354\nStarting epoch 38\nloss after epoch 38 : 4.774952411651611\nStarting epoch 39\nloss after epoch 39 : 4.568401336669922\n"
],
[
"# hide\ndef gen_pred_df(model, val_items=None, valid=True):\n # generates pd dataframe with all the predictions\n model.eval()\n model.to(DEVICE)\n \n val_set = Ageset(\"data/face_age\", valid=valid)[:val_items] if val_items else Ageset(\"data/face_age\", valid=valid)\n data =[(model(x[None].to(DEVICE)).item(),y) for x,y in val_set]\n df = pd.DataFrame(data, columns=['pred', 'actual'])\n df['loss'] = abs(df['pred']-df['actual'])\n df['path'] = val_set.image_paths\n return df",
"_____no_output_____"
],
[
"# hide\nfrom IPython.display import set_matplotlib_formats\ndef plot_top_losses(df, valid=True, top_loss=True):\n #plots top 9 of the dataframe\n # if top_loss, will sort the dataframe to display the worst performing items\n if top_loss:\n df = df.sort_values('loss', ascending=False)\n else:\n df = df.sample(frac=1)\n set_matplotlib_formats('svg')\n fig,ax = plt.subplots(3,3,figsize=(7,7))\n val_set = Ageset('data/face_age', valid=valid)\n for i, a in enumerate(ax.flat):\n pred = int(df.iloc[i]['pred'])\n actual = df.iloc[i]['actual']\n a.set_ylabel('44')\n a.imshow(val_set.show_image(df.iloc[i].name))\n a.set_title('Actual: ' + str(actual) + ' Model: ' + str(pred), size=10)\n a.axis('off')\n fig.tight_layout() \n set_matplotlib_formats('png')\n plt.subplots_adjust(wspace=0.1, hspace=0.15)\n return plt",
"_____no_output_____"
],
[
"# hide\nmodel = AgeResnet()\nmodel.load_state_dict(torch.load('/ds/data/models/model4.18')) \n# when using CPU: ,map_location=torch.device('cpu')\ndf = gen_pred_df(model, valid=True)",
"_____no_output_____"
]
],
[
[
"Let's check out predictions on some random images in the test set. In the title the actual age and the predicted age by the model.",
"_____no_output_____"
]
],
[
[
"# hide_input\nplot = plot_top_losses(df, top_loss=False)\nplot.show()",
"_____no_output_____"
]
],
[
[
"That looks pretty good! To take a more general approach, let's plot the all the images from the test set in a graph.",
"_____no_output_____"
]
],
[
[
"# hide_input\nplt.scatter(df['pred'],df['actual'], s=0.5)\nplt.figsize=(10,10)\nplt.axis([0,120, 0, 120])\nplt.gca().set_aspect('equal', adjustable='box')\nplt.title('Predictions are ballpark accurate')\nplt.xlabel('Predicted age')\nplt.ylabel('Actual age');",
"_____no_output_____"
]
],
[
[
"There are definitely some errors, but overall it seems reasonable. It's also interesting to plot the faces where the error was largest.",
"_____no_output_____"
]
],
[
[
"# hide_input\nplot_top_losses(df,valid=True).show()",
"_____no_output_____"
],
[
"# hide\n# some faces that were wrongly labelled were removed from dataset\nrm = ['data/face_age/005/2547.png',\n'data/face_age/001/3829.png',\n'data/face_age/069/8150.png',\n'data/face_age/001/5497.png',\n'data/face_age/001/4313.png',\n'data/face_age/021/8242.png',\n'data/face_age/070/1536.png',\n'data/face_age/001/7326.png',\n'data/face_age/021/1634.png',\n'data/face_age/008/7254.png']\nfor f in rm:\n print(f)\n os.unlink(Path(f))",
"data/face_age/005/2547.png\n"
]
],
[
[
"In most of the cases the model was simply off, but there are also faces which look much older or younger, or very blurry ones. Which highlights the importance of understanding the dataset and potentially removing outliers from it.\n\nAnother approach is to visualize the average error by age.",
"_____no_output_____"
]
],
[
[
"# hide_input\nset_matplotlib_formats('png')\ngroup = df.groupby(['actual']).agg(['count','mean'])\nfig,ax = plt.subplots(2,1,figsize=(4,8))\nax[0].scatter(group.index, group['loss']['mean'], s=1)\nax[0].set_title('Higher average error for higher ages')\nax[1].scatter(group.index, group['loss']['count'], s=1)\nax[0].axis([0,120, 0, 12])\nax[1].set_title('Fewer images in dataset for higher ages')\nax[0].set_xlabel(\"Actual age\")\nax[0].set_ylabel(\"Error in years\")\nax[1].set_ylabel(\"Number of items in dataset\")\nax[1].set_xlabel(\"Actual age\");\nax[1].axis([0,120, 0, 150])\nplt.subplots_adjust(wspace=0.2, hspace=0.3)\n",
"_____no_output_____"
],
[
"# hide\n# to launch tensorboard, run the following command in terminal\npython -m tensorboard.main --logdir=runs --host=0.0.0.0 --port=6006",
"_____no_output_____"
],
[
"# hide\n# dumps predictions into format that backend can use\ndef save_preds_for_app(df):\n df['pred']= df['pred'].astype(int)\n df['path']= df['path'].astype(str)\n # saving something as pickle file\n import pickle\n with open('app/models/predictions.pickle', 'wb') as handle:\n pickle.dump(df, handle, protocol=pickle.HIGHEST_PROTOCOL)\nsave_preds_for_app(df) ",
"_____no_output_____"
]
],
[
[
"The error gets larger as someone gets older, which makes perfect sense: when you see a baby you are not going to guess wrong by more then 5 years, but for someone age 50 that is more difficult. \n\nYou could argue for a slight improvement in the predictions with ages > 60, but it could also be an anomaly. There are few really old faces in the dataset, which could be of influence. There are also 300 images of babies age 0 in the dataset, which I removed from the second chart to have a better visualisation.\n\nI’ve build a [webpage](http://35.206.67.46/) where you can try out if you can beat the computer, or even upload a selfie for fun to see how old the computer thinks you are. Please take the results with a grain of salt. I’ve taken some selfies and the computer estimated me around 27 to 42, which is ballpark accurate. However, a condolence card of my grandmother age 92 was classified as 62, and although she did look young, 62 was an underestimation.\n\nI’ve had much fun with this project. I hope you like it as well and will [try out the application](http://35.206.67.46/). No guarantees it will be online indefinately by the wayw.",
"_____no_output_____"
],
[
"> Note: Technical details on how it was build: Pytorch, Pretrained resnet-18 model. Application runs on Google Cloud Platform, with a Nginx and Gunicorn running in Docker container. Python backend in Fastapi. Code available on [Github](https://github.com/jvanelteren/age_prediction), dataset on [Kaggle](https://www.kaggle.com/frabbisw/facial-age)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ecef8afdfe46e783cb6d1bf030f3c060ea11aaf8 | 34,607 | ipynb | Jupyter Notebook | ImplicitSpectral1DDiff.ipynb | tapashreepradhan/phase-field-modelling-nptel | 8da1b8271481c3032b9e486620ab86d5eff3a20c | [
"Apache-2.0"
] | null | null | null | ImplicitSpectral1DDiff.ipynb | tapashreepradhan/phase-field-modelling-nptel | 8da1b8271481c3032b9e486620ab86d5eff3a20c | [
"Apache-2.0"
] | null | null | null | ImplicitSpectral1DDiff.ipynb | tapashreepradhan/phase-field-modelling-nptel | 8da1b8271481c3032b9e486620ab86d5eff3a20c | [
"Apache-2.0"
] | null | null | null | 34,607 | 34,607 | 0.925622 | [
[
[
"# Spectral Techniques \n\nAgain we are going to be solving \n\n> ### $ \\frac{\\partial c}{\\partial t} = D \\frac{\\partial ^ 2c}{\\partial x^2} $\n\nThis time we are going to be using Fourier Transformations while doing it. \n\nHere basically we transform x vector in the expression for c(x,t) into k domain, which is also known as wave vector of reciprocal space. \n\nWe are going to the Discrete Fourier Transformation (DFT) Algorithm for the most efficient computation here. \n\nThe finally expression that we would need to implement in our code is :\n\n> ### $ cft^{ t + \\Delta t} = \\frac {cft ^{t}} { 1 + Dk^2 \\Delta t} $ \n\nwhere\n> cft = fourier tranform of c\n\n> D = Diffusivity\n\nThis equation is known as the Spectral Technique. \n\nAlso Fourier Transformations imply Periodic Boundary Conditions which will be in terms of k. \n",
"_____no_output_____"
]
],
[
[
"# importing libraries\nimport numpy as np\nfrom matplotlib import pyplot as plt",
"_____no_output_____"
],
[
"# ----- Defining Parameters ----- #\n\nN = 128 # grid spaces\nD = 1.0\ndt = 1.0\nx = np.zeros(N) # composition vector\nfor i in range (0, N):\n x[i] = i+1\n\nM = 2.0 # no. of wavelengths included in the domain\n\nc = np.zeros(N) # concentration vector\n\nfor j in range(0, N):\n c[j] = 0.5 * (1 + np.sin(2 * np.pi * M * (j+1) / N)) # sinusoidal concentration profile\n\nplt.plot(x,c, color = \"red\") # initial plot \n\n\nhalfN = N/2 # half the no. of grid spaces\n\ndelk = 2 * np.pi / N # wave vector\n\nctilde = np.zeros(N) # F.T of concentration vector\n\n# ------------- Loops -------------- #\n\nfor k in range (0,40): # no. of firgures to be plotted\n for m in range (0,500): # no. of timesteps\n \n ctilde = np.fft.fft(c) # invoking F.T\n \n # Implementation of PBC \n for i in range (0, N):\n \n if ( i < halfN) :\n k = i * delk\n if ( i >= halfN) :\n k = (i-N+1) * delk\n # implementation of spectral technique\n ctilde[i]= ctilde[i] / ( 1 + D * (k**2) * dt)\n \n c = np.real(np.fft.ifft(ctilde)) # taking inverse F.T\n plt.plot(x,c, color = \"blue\", linestyle = \"dotted\")\n plt.xlabel (\"Composition, x\")\n plt.ylabel (\"Concentration, c\")\n plt.title (\"C vs X\")",
"_____no_output_____"
]
],
[
[
"As we can see in the plot, wherever the curvature is positive, that region is going to increase and wherever the curvature is negative, that region is going to decrease.\n\nThus, the system will go towards homogenization. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
ecef93db3726159b55788dd9c38538f5fe8b4970 | 963 | ipynb | Jupyter Notebook | Table_Of_Contents.ipynb | jehalladay/RustAssemblyNotebooks | 5da19ae1c1d61ddae57ce69ab27b63c56c022df9 | [
"MIT"
] | null | null | null | Table_Of_Contents.ipynb | jehalladay/RustAssemblyNotebooks | 5da19ae1c1d61ddae57ce69ab27b63c56c022df9 | [
"MIT"
] | null | null | null | Table_Of_Contents.ipynb | jehalladay/RustAssemblyNotebooks | 5da19ae1c1d61ddae57ce69ab27b63c56c022df9 | [
"MIT"
] | null | null | null | 19.26 | 68 | 0.485981 | [
[
[
"## Beginners Rust and Web Assembly Notebook\n\n### Table of Contents\n \n#### [1 - Compiling using rustc and cargo](Chapter_1.ipynb)\n\n#### [2 - Syntax, Data Types, and Output](Chapter_2.ipynb)\n\n#### [3](Chapter_3.ipynb)\n\n#### [4](Chapter_4.ipynb)\n\n#### [5](Chapter_5.ipynb)\n\n#### [6](Chapter_6.ipynb)",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
ecef9d233b163cd155a08006838dc820f1561fc2 | 11,954 | ipynb | Jupyter Notebook | patch_creation/CreatePatches.ipynb | UVA-DSI-2019-Capstones/CHRC | 3b89fb6039e435f383754f933537201402391a07 | [
"MIT"
] | null | null | null | patch_creation/CreatePatches.ipynb | UVA-DSI-2019-Capstones/CHRC | 3b89fb6039e435f383754f933537201402391a07 | [
"MIT"
] | null | null | null | patch_creation/CreatePatches.ipynb | UVA-DSI-2019-Capstones/CHRC | 3b89fb6039e435f383754f933537201402391a07 | [
"MIT"
] | 1 | 2019-09-07T14:01:14.000Z | 2019-09-07T14:01:14.000Z | 27.671296 | 296 | 0.533545 | [
[
[
"import math\nimport os\n\nimport numpy as np\nimport openslide\nfrom PIL import Image\nfrom openslide import OpenSlideError\nfrom openslide.deepzoom import DeepZoomGenerator\nimport pandas as pd\n\nfrom pyspark.ml.linalg import Vectors\nimport pyspark.sql.functions as F\nfrom scipy.ndimage.morphology import binary_fill_holes\nfrom skimage.color import rgb2gray\nfrom skimage.feature import canny\nfrom skimage.morphology import binary_closing, binary_dilation, disk",
"_____no_output_____"
],
[
"tile_size = 3000\nsample_size = 3000\ngrayscale = False\nnum_partitions = 200\ntraining = True\nsave_jpegs = True\nconvert2DF = False\nrow_indices = False\ntrain_frac = 0.8\nsample_frac=0.01\nseed = 42\noverlap = 0",
"_____no_output_____"
],
[
"# open slide image\nslide = openslide.open_slide('/home/ss4yd/test/data/C17-83_04.svs')\n\ngenerator = DeepZoomGenerator(slide, tile_size=tile_size, overlap=overlap, limit_bounds=True)\n\nhighest_zoom_level = generator.level_count - 1\n# mag = int(slide.properties[openslide.PROPERTY_NAME_OBJECTIVE_POWER])\nzoom_level = highest_zoom_level\n\nslide_num = os.path.join('./data', 'C17-83_04.svs').split('.')[-2].split('/')[-1]",
"_____no_output_____"
],
[
"cols, rows = generator.level_tiles[zoom_level]\ntile_indices = [(slide_num, tile_size, overlap, zoom_level, col, row)\n for col in range(cols) for row in range(rows)]",
"_____no_output_____"
],
[
"tiles = []\nfor index in tile_indices:\n slide_num, tile_size, overlap, zoom_level, col, row = index\n tile = np.asarray(generator.get_tile(zoom_level, (col, row)))\n tiles.append(tile)",
"_____no_output_____"
],
[
"tiles[0].shape",
"_____no_output_____"
],
[
"def optical_density(tile):\n tile = tile.astype(np.float64)\n od = -np.log((tile+1)/240)\n return od",
"_____no_output_____"
],
[
"def keep_tile(tile, tile_size, tissue_threshold = 0.50):\n if tile.shape[0:2] == (tile_size, tile_size):\n print(\"inside if\")\n tile_orig = tile\n tile = rgb2gray(tile)\n tile = 1 - tile\n \n tile = canny(tile)\n \n tile = binary_closing(tile, disk(10))\n tile = binary_dilation(tile, disk(10))\n tile = binary_fill_holes(tile)\n percentage = tile.mean()\n \n check1 = percentage >= tissue_threshold\n\n # Check 2\n # Convert to optical density values\n tile = optical_density(tile_orig)\n # Threshold at beta\n beta = 0.15\n tile = np.min(tile, axis=2) >= beta\n # Apply morphology for same reasons as above.\n tile = binary_closing(tile, disk(2))\n tile = binary_dilation(tile, disk(2))\n tile = binary_fill_holes(tile)\n percentage = tile.mean()\n check2 = percentage >= tissue_threshold\n print(check1, check2)\n return check1 and check2\n else:\n return False\n",
"_____no_output_____"
],
[
"keep_tile(tiles[0], 3000, tissue_threshold=50)",
"inside if\n"
],
[
"def process_tile(tile, sample_size, grayscale):\n \n \"\"\"\n Process a tile into a group of smaller samples.\n Cut up a tile into smaller blocks of sample_size x sample_size pixels,\n change the shape of each sample from (H, W, channels) to\n (channels, H, W), then flatten each into a vector of length\n channels*H*W.\n Args:\n tile_tuple: A (slide_num, tile) tuple, where slide_num is an\n integer, and tile is a 3D NumPy array of shape\n (tile_size, tile_size, channels).\n sample_size: The new width and height of the square samples to be\n generated.\n grayscale: Whether or not to generate grayscale samples, rather\n than RGB.\n Returns:\n A list of (slide_num, sample) tuples representing cut up tiles,\n where each sample is a 3D NumPy array of shape\n (sample_size_x, sample_size_y, channels).\n \"\"\"\n if grayscale:\n tile = rgb2gray(tile)[:, :, np.newaxis] # Grayscale\n # Save disk space and future IO time by converting from [0,1] to [0,255],\n # at the expense of some minor loss of information.\n tile = np.round(tile * 255).astype(\"uint8\")\n x, y, ch = tile.shape\n # 1. Reshape into a 5D array of (num_x, sample_size_x, num_y, sample_size_y, ch), where\n # num_x and num_y are the number of chopped tiles on the x and y axes, respectively.\n # 2. Swap sample_size_x and num_y axes to create\n # (num_x, num_y, sample_size_x, sample_size_y, ch).\n # 3. Combine num_x and num_y into single axis, returning\n # (num_samples, sample_size_x, sample_size_y, ch).\n samples = (tile.reshape((x // sample_size, sample_size, y // sample_size, sample_size, ch))\n .swapaxes(1, 2)\n .reshape((-1, sample_size, sample_size, ch)))\n samples = [(slide_num, sample) for sample in list(samples)]\n return samples\n",
"_____no_output_____"
],
[
"filter_tiles = [tile for tile in tiles if keep_tile(tile,tile_size, 0.50)]",
"inside if\n"
],
[
"keep_tile(tiles[3], 4000, tissue_threshold=50)",
"_____no_output_____"
],
[
"samples = [process_tile(tile, sample_size, False) for tile in filter_tiles]",
"_____no_output_____"
],
[
"def save_nonlabelled_sample_2_jpeg(sample, save_dir):\n \"\"\"\n Save the sample without labels into JPEG\n Args:\n sample_element: a sample tuple without labels, e.g. (slide_num, sample)\n save_dir: the file directory at which to save JPEGs\n \"\"\"\n slide_num, img_value = sample\n filename = '{slide_num}_{hash}.jpeg'.format(\n slide_num=slide_num, hash=np.random.randint(1e4))\n filepath = os.path.join(save_dir, filename)\n save_jpeg_help(img_value, filepath)\n\n\ndef save_jpeg_help(img_value, filepath):\n \"\"\"\n Save data into JPEG\n Args:\n img_value: the image value with the size (img_size_x, img_size_y, channels)\n file path: the file path at which to save JPEGs\n \"\"\"\n dir = os.path.dirname(filepath)\n os.makedirs(dir, exist_ok=True)\n img = Image.fromarray(img_value.astype(np.uint8), 'RGB')\n img.save(filepath)\n",
"_____no_output_____"
],
[
"for sample in samples:\n save_nonlabelled_sample_2_jpeg(sample[0], './')",
"_____no_output_____"
],
[
"samples",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecefaead4500793247a9ec69aab3746cf053ae46 | 879,996 | ipynb | Jupyter Notebook | Code/Model 1 (regression trees)/AAT-DT_v13.ipynb | SamIlic/Forecasting-Stock-Returns-via-Supervised-Learning | 1947c311397676434d8224aefb837dda481e1bc5 | [
"MIT"
] | 2 | 2019-08-13T20:07:26.000Z | 2021-04-15T02:31:26.000Z | Code/Model 1 (regression trees)/AAT-DT_v13.ipynb | SamIlic/Forecasting-Stock-Returns-via-Stacking | 1947c311397676434d8224aefb837dda481e1bc5 | [
"MIT"
] | null | null | null | Code/Model 1 (regression trees)/AAT-DT_v13.ipynb | SamIlic/Forecasting-Stock-Returns-via-Stacking | 1947c311397676434d8224aefb837dda481e1bc5 | [
"MIT"
] | null | null | null | 263.550764 | 103,840 | 0.904353 | [
[
[
"## Advanced Algorithmic Trading DT - V13\n\n## Updates from Last Version\nTrial of shift today's price so we dont cheat\n\n\n\n## Contents\n- Basic Setup\n- Clean Data & Create Technical Indicator Variables\n- ",
"_____no_output_____"
],
[
"#### Import Packages",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport math\n\nimport datetime\nimport gc\n\nfrom sklearn.ensemble import (BaggingRegressor, RandomForestRegressor, AdaBoostRegressor)\nfrom sklearn.model_selection import ParameterGrid\nfrom sklearn.tree import DecisionTreeRegressor\nfrom sklearn import linear_model\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.linear_model import BayesianRidge\nfrom sklearn.metrics import mean_squared_error\n\nfrom technical_indicators import * # import all function\n\nfrom sklearn.model_selection import TimeSeriesSplit\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import r2_score\nfrom sklearn.preprocessing import scale\n\nfrom sklearn.decomposition import PCA\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.cross_decomposition import PLSRegression\n",
"_____no_output_____"
]
],
[
[
"#### Set Parameters",
"_____no_output_____"
]
],
[
[
"# Set the random seed, number of estimators and the \"step factor\" used to plot the graph of MSE for each method\nrandom_state = 42 # Seed\nn_jobs = -1 # -1 --> all Processors # Parallelisation factor for bagging, random forests (controls the number of processor cores used)\nn_estimators = 200 # total number of estimators ot use in the MSE graph\nstep_factor = 10 # controls granularity of calculation by stepping through the number of estimators\naxis_step = int(n_estimators / step_factor) # 1000/10 = 100 separate calculations will be performed for each of the 3 ensebmle methods\n",
"_____no_output_____"
]
],
[
[
"#### Read in Data via GitHub URL",
"_____no_output_____"
]
],
[
[
"url = \"https://raw.githubusercontent.com/meenmo/Stat479_Project/master/Data/IBM.csv\"\ndf_ORIGINAL = pd.read_csv(url)",
"_____no_output_____"
],
[
"plot_path = 'C:/Users/meenm/OneDrive - UW-Madison/Github/Stat479_project2/Stat479_Project/Plots/'",
"_____no_output_____"
],
[
"display(df_ORIGINAL.head())",
"_____no_output_____"
]
],
[
[
"***\n## Clean Data & Create Technical Indicator Variables\n\n- Create Deep copy of dataframe\n- Use Adjusted Close Data\n- Drop Close \n- Rename \"Adj. Close\" as \"Close\"\n- Create Lagged Features\n- Drop NaN\n- Create Technical Indicator Variables\n- Drop NaN\n- Re-set index as Date",
"_____no_output_____"
]
],
[
[
"df_features = df_ORIGINAL.copy(deep=True) # Create Deep\ndf_features.drop(['Close'], axis = 1, inplace = True) # drop close column\ndf_features.columns = ['Date', 'High', 'Low', 'Open', 'Volume', 'Close'] # Close is actually Adj. Close\n\ndf_features['Date'] = pd.to_datetime(df_features['Date'])\n#df_features.head() # sanity check\n\n\n\"\"\"\nCreates Lagged Returns \n- given OHLCV dataframe\n- numer of lagged days\n\"\"\"\ndef create_lag_features(df, lag_days):\n df_ret = df.copy()\n \n # iterate through the lag days to generate lag values up to lag_days + 1\n for i in range(1,lag_days + 2):\n df_lag = df_ret[['Date', 'Close']].copy()\n # generate dataframe to shift index by i day.\n df_lag['Date'] = df_lag['Date'].shift(-i)\n df_lag.columns = ['Date', 'value_lag' + str(i)]\n # combine the valuelag\n df_ret = pd.merge(df_ret, df_lag, how = 'left', left_on = ['Date'], right_on = ['Date'])\n \n #frees memory\n del df_lag\n \n # calculate today's percentage lag\n df_ret['Today'] = (df_ret['Close'] - df_ret['value_lag1'])/(df_ret['value_lag1']) * 100.0 \n \n # calculate percentage lag\n for i in range(1, lag_days + 1):\n df_ret['lag' + str(i)] = (df_ret['value_lag'+ str(i)] - df_ret['value_lag'+ str(i+1)])/(df_ret['value_lag'+str(i+1)]) * 100.0\n \n # drop unneeded columns which are value_lags\n for i in range(1, lag_days + 2):\n df_ret.drop(['value_lag' + str(i)], axis = 1, inplace = True)\n \n return df_ret\n\n\n### Run Function\ndf_features = create_lag_features(df_features, 5) # 5 lag features\n#df_features.head(7)\n\n# drop earlier data with missing lag features\ndf_features.dropna(inplace=True)\n# reset index\ndf_features.reset_index(drop = True, inplace = True)\n\n\n#### GENERATE TECHNICAL INDICATORS FEATURES\ndf_features = standard_deviation(df_features, 14)\n\ndf_features = relative_strength_index(df_features, 14) # periods\ndf_features = average_directional_movement_index(df_features, 14, 13) # n, n_ADX\ndf_features = moving_average(df_features, 21) # periods\ndf_features = exponential_moving_average(df_features, 21) # periods\ndf_features = momentum(df_features, 14) # \n\ndf_features = average_true_range(df_features, 14)\ndf_features = bollinger_bands(df_features, 21)\ndf_features = ppsr(df_features)\ndf_features = stochastic_oscillator_k(df_features)\ndf_features = stochastic_oscillator_d(df_features, 14)\ndf_features = trix(df_features, 14)\ndf_features = macd(df_features, 26, 12)\ndf_features = mass_index(df_features)\ndf_features = vortex_indicator(df_features, 14)\n\ndf_features = kst_oscillator(df_features, 10, 10, 10, 15, 10, 15, 20, 30)\ndf_features = true_strength_index(df_features, 25, 13)\n\n#df_features = accumulation_distribution(df_features, 14) # Causes Problems, apparently\ndf_features = chaikin_oscillator(df_features)\ndf_features = money_flow_index(df_features, 14)\ndf_features = on_balance_volume(df_features, 14)\ndf_features = force_index(df_features, 14)\ndf_features = ease_of_movement(df_features, 14)\ndf_features = commodity_channel_index(df_features, 14)\ndf_features = keltner_channel(df_features, 14)\ndf_features = ultimate_oscillator(df_features)\ndf_features = donchian_channel(df_features, 14)\n \n#drop earlier data with missing lag features\ndf_features.dropna(inplace=True)\ndf_features = df_features.reset_index(drop = True)\n\n\n\n###########################################################################################\n# Store Variables now for plots later\ndaily_index = df_features.index\ndaily_returns = df_features[\"Today\"]\ndaily_price = df_features[\"Close\"]\n\n# Re-set \"Date\" as the index\ndf_features = df_features.set_index('Date')\n\n### Sanity Check\ndf_features.head(10)\n",
"_____no_output_____"
]
],
[
[
"## Standardize Data & Create X & y\n\n- Drop all data used to create technical indicators (this is done in the book)\n- Then Standardize, necessary for PLS\n- Run PLS\n- Select Appropriate number of components\n- Create X & y\n\nNOTE: some technical indicators use Present data, but for simplicity, just ignore this",
"_____no_output_____"
]
],
[
[
"df_features['FutureReturns'] = df_features['Today'].shift(-1)\ndf_features.dropna(inplace=True)",
"_____no_output_____"
],
[
"### Standardize Data\n##########################################################################################\n# Drop Columns\nlist_of_columns_to_exclude = [\"High\", \"Low\", \"Open\", \"Volume\",\"Close\", \"FutureReturns\"]\nX_temp_standardized = df_features.copy(deep=True)\nX_temp_standardized.drop(list_of_columns_to_exclude, axis = 1, inplace = True) # drop columns\n\n# Standardize\nX_temp_standardized\ndates = X_temp_standardized.index # get dates to set as index after data is standardized\nnames = X_temp_standardized.columns # Get column names first\nX_temp_standardized = StandardScaler().fit_transform(X_temp_standardized)\n\n# Convert to DataFrame\nX_temp_standardized = pd.DataFrame(X_temp_standardized, columns=names, index=dates)\nX = X_temp_standardized\n\n\n### Get y\n##########################################################################################\ny_temp = pd.DataFrame(df_features[\"FutureReturns\"], index=X.index) # can only standardize a dataframe\nsc = StandardScaler()\ny = sc.fit_transform(y_temp) # Standardize, cause we did it for our original variables\ny = pd.DataFrame(y, index=X.index, columns=[\"FutureReturns\"]) # convert back to dataframe\ny = y[\"FutureReturns\"] # now re-get y as a Pandas Series\n\n### Sanity Check\nprint(\"Shape of X: \", X.shape)\nprint(\"Shape of y: \", y.shape)\n\n# Check Types\nprint(type(X)) # Needs to be <class 'pandas.core.frame.DataFrame'>\nprint(type(y)) # Needs ro be <class 'pandas.core.series.Series'>",
"Shape of X: (4189, 43)\nShape of y: (4189,)\n<class 'pandas.core.frame.DataFrame'>\n<class 'pandas.core.series.Series'>\n"
]
],
[
[
"#### Split: Train & Validatte / Test\n\n- Train & Validate: < '2018-01-01'\n- Test: >= '2018-01-01'",
"_____no_output_____"
]
],
[
[
"X_train_all = X.loc[(X.index < '2018-01-01')]\ny_train_all = y[X_train_all.index]\n\n# # creates all test data which is all after January 2018\nX_test = X.loc[(X.index >= '2018-01-01'),:]\ny_test = y[X_test.index]\n\n### Sanity Check\nprint(\"Shape of X_train_all: \", X_train_all.shape)\nprint(\"Shape of y_train_all: \", y_train_all.shape)\nprint(\"Shape of X_test: \", X_test.shape)\nprint(\"Shape of y_test: \", y_test.shape)",
"Shape of X_train_all: (3979, 43)\nShape of y_train_all: (3979,)\nShape of X_test: (210, 43)\nShape of y_test: (210,)\n"
]
],
[
[
"## Time Series Train Test Split ---- \n",
"_____no_output_____"
],
[
"### Random Forest",
"_____no_output_____"
]
],
[
[
"\"\"\"\nExecute Random Forest for differnt number of Time Series Splits\n\"\"\"\ndef Call_Random_Forest(numSplits):\n ### Prepare Random Forest\n ##############################################################################\n # Initialize Random Forest Instance\n rf = RandomForestRegressor(n_estimators=150, n_jobs=-1, random_state=123, max_features=\"sqrt\", min_samples_split=4, max_depth=30, min_samples_leaf=1)\n\n rf_mse = [] # MSE\n rf_r2 = [] # R2\n\n \n ### Time Series Split\n ##############################################################################\n splits = TimeSeriesSplit(n_splits=numSplits) # 3 splits\n\n splitCount = 0 # dummy count var to track current split num in print statements\n for train_index, test_index in splits.split(X_train_all):\n splitCount += 1 \n\n # Train Split\n X_train = X_train_all.iloc[train_index,:]\n y_train = y[X_train.index]\n\n # Validate Split\n X_val = X_train_all.iloc[test_index,:]\n y_val = y[X_val.index]\n\n# # Print Statements\n# print(\"~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\")\n# print(\"Split: \", splitCount)\n# print('Observations: ', (X_train.shape[0] + X_val.shape[0]))\n# #print('Cutoff date, or first date in validation data: ', X_val.iloc[0,0])\n# print('Training Observations: ', (X_train.shape[0]))\n# print('Testing Observations: ', (X_val.shape[0]))\n# print(\"~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\")\n\n ### Run Random Forest\n rf.fit(X_train, y_train)\n prediction = rf.predict(X_val)\n\n mse = mean_squared_error(y_val, prediction)\n r2 = r2_score(y_val, prediction)\n\n rf_mse.append(mse)\n rf_r2.append(r2)\n\n # print(\"rf_mse: \", rf_mse)\n # print(\"rf_r2: \", rf_r2)\n\n\n ### Time Series Split\n ##############################################################################\n # Plot the chart of MSE versus number of estimators\n plt.figure(figsize=(12, 7))\n plt.title('Random Forest - MSE & R-Squared')\n\n ### MSE\n plt.plot(list(range(1,splitCount+1)), rf_mse, 'b-', color=\"blue\", label='MSE')\n plt.plot(list(range(1,splitCount+1)), rf_r2, 'b-', color=\"green\", label='R-Squared')\n plt.plot(list(range(1,splitCount+1)), np.array([0] * splitCount), 'b-', color=\"red\", label='Zero')\n\n\n plt.legend(loc='upper right')\n plt.xlabel('Train/Test Split Number')\n plt.ylabel('Mean Squared Error & R-Squared')\n plt.show()\n print(\"rf_r2: \", rf_r2)\n #print(rf.feature_importances_)\n print(\"Mean r2: \", np.mean(rf_r2))",
"_____no_output_____"
]
],
[
[
"### Bagging",
"_____no_output_____"
]
],
[
[
"\"\"\"\nExecute Bagging for differnt number of Time Series Splits\n\"\"\"\ndef Call_Bagging(numSplits):\n ### Prepare Bagging\n ##############################################################################\n # Initialize Bagging Instance\n bagging = BaggingRegressor(n_estimators=150, n_jobs=-1, random_state=123)\n\n bagging_mse = [] # MSE\n bagging_r2 = [] # R2\n\n\n ### Time Series Split\n ##############################################################################\n splits = TimeSeriesSplit(n_splits=numSplits) # 3 splits\n\n splitCount = 0 # dummy count var to track current split num in print statements\n for train_index, test_index in splits.split(X_train_all):\n splitCount += 1 \n\n # Train Split\n X_train = X_train_all.iloc[train_index,:]\n y_train = y[X_train.index]\n\n # Validate Split\n X_val = X_train_all.iloc[test_index,:]\n y_val = y[X_val.index]\n\n# # Print Statements\n# print(\"~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\")\n# print(\"Split: \", splitCount)\n# print('Observations: ', (X_train.shape[0] + X_test.shape[0]))\n# #print('Cutoff date, or first date in validation data: ', X_val.iloc[0,0])\n# print('Training Observations: ', (X_train.shape[0]))\n# print('Testing Observations: ', (X_test.shape[0]))\n# print(\"~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\")\n\n ### Run Random Forest\n bagging.fit(X_train, y_train)\n prediction = bagging.predict(X_val)\n\n mse = mean_squared_error(y_val, prediction)\n r2 = r2_score(y_val, prediction)\n \n\n bagging_mse.append(mse)\n bagging_r2.append(r2)\n\n\n ### Time Series Split\n ##############################################################################\n # Plot the chart of MSE versus number of estimators\n plt.figure(figsize=(12, 7))\n plt.title('Bagging - MSE & R-Squared')\n\n ### MSE\n plt.plot(list(range(1,splitCount+1)), bagging_mse, 'b-', color=\"blue\", label='MSE')\n plt.plot(list(range(1,splitCount+1)), bagging_r2, 'b-', color=\"green\", label='R-Squared')\n plt.plot(list(range(1,splitCount+1)), np.array([0] * splitCount), 'b-', color=\"red\", label='Zero')\n\n\n plt.legend(loc='upper right')\n plt.xlabel('Train/Test Split Number')\n plt.ylabel('Mean Squared Error & R-Squared')\n plt.show()\n print(\"bagging_r2: \", bagging_r2)\n print(np.mean(bagging_r2))",
"_____no_output_____"
]
],
[
[
"### Boosting",
"_____no_output_____"
]
],
[
[
"\"\"\"\nExecute Random Forest for differnt number of Time Series Splits\n\"\"\"\ndef Call_Boosting(numSplits):\n ### Prepare Boosting\n ##############################################################################\n # Initialize Boosting Instance \n boosting = AdaBoostRegressor(DecisionTreeRegressor(),\n n_estimators=150, random_state=123,learning_rate=0.01)\n\n boosting_mse = [] # MSE\n boosting_r2 = [] # R2\n\n\n ### Time Series Split\n ##############################################################################\n splits = TimeSeriesSplit(n_splits=numSplits) # 3 splits\n\n splitCount = 0 # dummy count var to track current split num in print statements\n for train_index, test_index in splits.split(X_train_all):\n splitCount += 1 \n\n # Train Split\n X_train = X_train_all.iloc[train_index,:]\n y_train = y[X_train.index]\n\n # Validate Split\n X_val = X_train_all.iloc[test_index,:]\n y_val = y[X_val.index]\n\n# # Print Statements\n# print(\"~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\")\n# print(\"Split: \", splitCount)\n# print('Observations: ', (X_train.shape[0] + X_test.shape[0]))\n# #print('Cutoff date, or first date in validation data: ', X_val.iloc[0,0])\n# print('Training Observations: ', (X_train.shape[0]))\n# print('Testing Observations: ', (X_test.shape[0]))\n# print(\"~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\")\n\n ### Run Random Forest\n boosting.fit(X_train, y_train)\n prediction = boosting.predict(X_val)\n\n mse = mean_squared_error(y_val, prediction)\n r2 = r2_score(y_val, prediction)\n\n boosting_mse.append(mse)\n boosting_r2.append(r2)\n\n\n ### Time Series Split\n ##############################################################################\n # Plot the chart of MSE versus number of estimators\n plt.figure(figsize=(12, 7))\n plt.title('Boosting - MSE & R-Squared')\n\n ### MSE\n plt.plot(list(range(1,splitCount+1)), boosting_mse, 'b-', color=\"blue\", label='MSE')\n plt.plot(list(range(1,splitCount+1)), boosting_r2, 'b-', color=\"green\", label='R-Squared')\n plt.plot(list(range(1,splitCount+1)), np.array([0] * splitCount), 'b-', color=\"red\", label='Zero')\n\n\n plt.legend(loc='upper right')\n plt.xlabel('Train/Test Split Number')\n plt.ylabel('Mean Squared Error & R-Squared')\n plt.show()\n print(\"boosting_r2: \", boosting_r2)",
"_____no_output_____"
]
],
[
[
"### Linear Regression\n",
"_____no_output_____"
]
],
[
[
"\"\"\"\nExecute Linear Regression for different number of Time Series Splits\n\"\"\"\ndef Call_Linear(numSplits):\n ### Prepare Random Forest\n ##############################################################################\n # Initialize Random Forest Instance\n linear = LinearRegression(n_jobs=-1, normalize=True, fit_intercept=False) # if we don't fit the intercept we get a better prediction\n\n linear_mse = [] # MSE\n linear_r2 = [] # R2\n\n\n ### Time Series Split\n ##############################################################################\n splits = TimeSeriesSplit(n_splits=numSplits) # 3 splits\n\n splitCount = 0 # dummy count var to track current split num in print statements\n for train_index, test_index in splits.split(X_train_all):\n splitCount += 1 \n\n # Train Split\n X_train = X_train_all.iloc[train_index,:]\n y_train = y[X_train.index]\n\n # Validate Split\n X_val = X_train_all.iloc[test_index,:]\n y_val = y[X_val.index]\n\n# # Print Statements\n# print(\"~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\")\n# print(\"Split: \", splitCount)\n# print('Observations: ', (X_train.shape[0] + X_test.shape[0]))\n# #print('Cutoff date, or first date in validation data: ', X_val.iloc[0,0])\n# print('Training Observations: ', (X_train.shape[0]))\n# print('Testing Observations: ', (X_test.shape[0]))\n# print(\"~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\")\n\n ### Run Random Forest\n linear.fit(X_train, y_train)\n prediction = linear.predict(X_val)\n\n mse = mean_squared_error(y_val, prediction)\n r2 = r2_score(y_val, prediction)\n r2 = np.corrcoef(y_val, prediction)[0, 1] \n r2 = r2*r2 # square of correlation coefficient --> R-squared\n\n linear_mse.append(mse)\n linear_r2.append(r2)\n\n\n ### Time Series Split\n ##############################################################################\n # Plot the chart of MSE versus number of estimators\n plt.figure(figsize=(12, 7))\n plt.title('Linear Regression - MSE & R-Squared')\n\n ### MSE\n plt.plot(list(range(1,splitCount+1)), linear_mse, 'b-', color=\"blue\", label='MSE')\n plt.plot(list(range(1,splitCount+1)), linear_r2, 'b-', color=\"green\", label='R-Squared')\n plt.plot(list(range(1,splitCount+1)), np.array([0] * splitCount), 'b-', color=\"red\", label='Zero')\n\n\n plt.legend(loc='upper right')\n plt.xlabel('Train/Test Split Number')\n plt.ylabel('Mean Squared Error & R-Squared')\n plt.show()\n print(\"linear_r2: \", linear_r2)",
"_____no_output_____"
]
],
[
[
"### Misc. Graphs ---- Price, Returns & Cumulative Returns",
"_____no_output_____"
]
],
[
[
"# figure dimenstions\nlength = 15\nheight = 5\n\n### Prices\nplt.figure(figsize=(length, height))\nplt.title('IBM Adj Close Price Graph')\nplt.plot(daily_index, daily_price, 'b-', color=\"blue\", label='Prices')\nplt.legend(loc='upper right')\nplt.xlabel('Days')\nplt.ylabel('Prices')\n#plt.savefig(plot_path+'IBM Adj Close Price Graph')\nplt.show()\n\n### Returns\nplt.figure(figsize=(length, height))\nplt.title('IBM Daily Returns')\nplt.plot(daily_index, daily_returns, 'b-', color=\"blue\", label='Returns')\nplt.legend(loc='upper right')\nplt.xlabel('Days')\nplt.ylabel('Returns')\n#plt.savefig(plot_path+'IBM Daily Returns')\nplt.show()\n\n### Cumulative Returns\nplt.figure(figsize=(length, height))\nplt.title('IBM Cumulative Returns')\ncumulative_returns = daily_returns.cumsum()\nplt.plot(daily_index, cumulative_returns, 'b-', color=\"green\", label='Cumulative Returns')\nplt.legend(loc='upper right')\nplt.xlabel('Days')\nplt.ylabel('Cumulative Return')\n#plt.savefig(plot_path+'IBM Cumulative Returns')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### First - A Note on R-Squared\n\n##### What Does A Negative R Squared Value Mean?\n\n- What does R-squared tell us?\n - It tells us whether a horizonal line through the vertical mean of the data is a better predictor\n- For a Linear Regression\n - R-squared is just the coreelation coefficient squared\n - R-squared can't be negative, becasue at 0, it becomes the horizontal line\n- For All other Model\n - For practical purposes, the lowest R2 you can get is zero, but only because the assumption is that if your regression line is not better than using the mean, then you will just use the mean value. \n - However if your regression line is worse than using the mean value, the r squared value that you calculate will be negative.\n - Note that the reason R2 can't be negative in the linear regression case is just due to chance and how linear regression is contructed\n ",
"_____no_output_____"
],
[
"*** \n*** \n*** \n\n## Grid Search for Best Model",
"_____no_output_____"
],
[
"## Revised RandomForest",
"_____no_output_____"
]
],
[
[
"\"\"\"\nExecute Random Forest for differnt number of Time Series Splits\nParameters\n - numSplits: Number of Train/Validation splits to run in order to compute our final model (bigger --> more accurate & more complex)\n - maxDepth: Max Depth of the DT\n - minSamplesSplit: min samples to split (basically pruning)\n \nReturns\n - Average R-Squared of the DT model across all Train/Validation splits\n\"\"\"\ndef Call_Random_Forest_Grid_Search(numSplits, minSamplesSplit, maxDepth):\n ### Prepare Random Forest\n ##############################################################################\n # Initialize Random Forest Instance\n rf_mse = [] # MSE\n rf_r2 = [] # R2\n rf = RandomForestRegressor(n_estimators=150, n_jobs=-1, random_state=123, max_features=\"sqrt\", \n max_depth=maxDepth, \n min_samples_split=minSamplesSplit)\n\n ### Time Series Split\n ##############################################################################\n splits = TimeSeriesSplit(n_splits=numSplits) # 3 splits\n\n splitCount = 0 # dummy count var to track current split num in print statements\n for train_index, test_index in splits.split(X_train_all):\n splitCount += 1 \n\n # Train Split\n X_train = X_train_all.iloc[train_index,:]\n y_train = y[X_train.index]\n\n # Validate Split\n X_val = X_train_all.iloc[test_index,:]\n y_val = y[X_val.index]\n\n ### Run Random Forest\n rf.fit(X_train, y_train)\n prediction = rf.predict(X_val)\n\n mse = mean_squared_error(y_val, prediction)\n r2 = r2_score(y_val, prediction)\n\n rf_mse.append(mse)\n rf_r2.append(r2)\n\n return np.mean(rf_mse), np.mean(rf_r2)",
"_____no_output_____"
],
[
"# Call_Random_Forest_Grid_Search(numSplits, minSamplessplit, maxDepth)\n\nminSamplesSplit_list = [2,5,10]\nmaxDepth_list = [15,20,25]\n\nbest_model_parameters = [0,0]\nbest_model_param_mse = [0,0]\nmax_r2 = -100\nmin_mse = 100\ncount = 0\n\nrf_df = pd.DataFrame()\n# Loop over all possible parameters\nfor minSamplesSplit in minSamplesSplit_list:\n for maxDepth in maxDepth_list:\n count += 1\n temp_mean_mse, temp_mean_r2 = Call_Random_Forest_Grid_Search(10, minSamplesSplit, maxDepth) # Call Random Forest Train/Validation\n print(\"temp_mean \", count, \": \", temp_mean_r2)\n if temp_mean_r2 > max_r2:\n max_r2 = temp_mean_r2 # store new max\n best_model_parameters[0] = minSamplesSplit\n best_model_parameters[1] = maxDepth\n if temp_mean_mse < min_mse:\n min_mse = temp_mean_mse\n best_model_param_mse[0] = minSamplesSplit\n best_model_param_mse[1] = maxDepth\n \n rf_df.loc[count,'min_sample_split'] = minSamplesSplit\n rf_df.loc[count,'max_depth'] = maxDepth\n rf_df.loc[count,'mean_r2'] = temp_mean_r2\n rf_df.loc[count, 'mearn_mse'] = temp_mean_mse\n\nprint(\"Best R2: \", max_r2) \nprint(best_model_parameters)\nprint(\"Best MSE: \", min_mse) \nprint(best_model_param_mse)",
"temp_mean 1 : -0.3260546656037615\ntemp_mean 2 : -0.40359007779873135\ntemp_mean 3 : -0.3961887900276575\ntemp_mean 4 : -0.2863225006865322\ntemp_mean 5 : -0.3624610537281746\ntemp_mean 6 : -0.36383610018587376\ntemp_mean 7 : -0.2858894498687869\ntemp_mean 8 : -0.3467666836288161\ntemp_mean 9 : -0.3551411348399384\nBest R2: -0.2858894498687869\n[10, 15]\nBest MSE: 1.0461347284116116\n[5, 15]\n"
]
],
[
[
"## Best Model RF\n - min_samples_split = 10\n - max_depth = 15",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\n\n# plot nicely\nx_index = np.unique(rf_df.loc[:,'min_sample_split'])\ny_index = np.unique((rf_df.loc[:,'max_depth']))\na = pd.DataFrame(index = x_index, columns = y_index)\n\nfor i in x_index:\n for j in y_index:\n a.loc[i,j] = list(rf_df.loc[(rf_df['min_sample_split'] == i) & (rf_df['max_depth'] == j),'mean_r2'])[0]#.astype(float)\na = a.apply(pd.to_numeric) \nsns.heatmap(a)\nplt.xlabel('max_depth')\nplt.ylabel('min_samples_split')\nplt.title('r2')\nplt.savefig(plot_path+'Best_Model_RF')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Bagging",
"_____no_output_____"
]
],
[
[
"\n\"\"\"\nExecute Bagging for differnt number of Time Series Splits\nParameters\n - numSplits: Number of Train/Validation splits to run in order to compute our final model (bigger --> more accurate & more complex)\n - min_samples_split: minimum samples to split\n - max_depth: depth of tree\n \nReturns\n - Average R-Squared of the DT model across all Train/Validation splits\"\"\"\ndef Call_Bagging_Grid_Search(numSplits, min_samples_split, max_depth):\n ### Prepare Bagging\n ##############################################################################\n # Initialize Bagging Instance\n bagging_mse = [] # MSE\n bagging_r2 = [] # R2\n bagging = BaggingRegressor(DecisionTreeRegressor(min_samples_split = min_samples_split, max_depth = max_depth),\n n_estimators=150, random_state=123,\n #n_jobs = -1,\n max_samples=100,\n max_features=20)\n# bagging = BaggingRegressor(n_estimators=150, n_jobs=-1, random_state=123)\n\n ### Time Series Split\n ##############################################################################\n splits = TimeSeriesSplit(n_splits=numSplits) # 3 splits\n\n splitCount = 0 # dummy count var to track current split num in print statements\n for train_index, test_index in splits.split(X_train_all):\n splitCount += 1 \n\n # Train Split\n X_train = X_train_all.iloc[train_index,:]\n y_train = y[X_train.index]\n\n # Validate Split\n X_val = X_train_all.iloc[test_index,:]\n y_val = y[X_val.index]\n\n ### Run Bagging\n bagging.fit(X_train, y_train)\n prediction = bagging.predict(X_val)\n\n mse = mean_squared_error(y_val, prediction)\n r2 = r2_score(y_val, prediction)\n \n\n bagging_mse.append(mse)\n bagging_r2.append(r2)\n\n return np.mean(bagging_mse), np.mean(bagging_r2)",
"_____no_output_____"
],
[
"min_sample_split = [5,10,15]\nmax_depth = [10,15,20]\n\nbest_model_parameters_bag = [0,0]\nbest_model_param_bag_mse = [0,0]\nmax_r2_bag = -100\nmin_mse_bag = 100\ncount = 0\n\nbag_df = pd.DataFrame()\n# Loop over all possible parameters\nfor minSampleSplit in min_sample_split:\n for maxDepth in max_depth:\n count += 1\n temp_mean_mse, temp_mean_r2 = Call_Bagging_Grid_Search(10, minSampleSplit, maxDepth) # Call Boosting Train/Validation\n print(\"temp_mean \", count, \": \", temp_mean_r2)\n if temp_mean_r2 > max_r2_bag:\n max_r2_bag = temp_mean_r2 # store new max\n best_model_parameters_bag[0] = minSampleSplit\n best_model_parameters_bag[1] = maxDepth\n if temp_mean_mse < min_mse_bag:\n min_mse_bag = temp_mean_mse # store new max\n best_model_param_bag_mse[0] = minSampleSplit\n best_model_param_bag_mse[1] = maxDepth\n \n bag_df.loc[count,'min_sample_split'] = minSampleSplit\n bag_df.loc[count,'max_depth'] = maxDepth\n bag_df.loc[count,'mean_r2'] = temp_mean_r2\n bag_df.loc[count, 'mean_mse'] = temp_mean_mse\n \n\nprint(\"Best R2: \", max_r2_bag) \nprint(best_model_parameters_bag)\nprint(\"Best MSE: \", min_mse_bag) \nprint(best_model_param_bag_mse) ",
"temp_mean 1 : -0.0613786377557111\ntemp_mean 2 : -0.059978445053838515\ntemp_mean 3 : -0.060473558284150665\ntemp_mean 4 : -0.05652461616362579\ntemp_mean 5 : -0.054507688411346754\ntemp_mean 6 : -0.05455572302221251\ntemp_mean 7 : -0.05924282770450871\ntemp_mean 8 : -0.05980189878909546\ntemp_mean 9 : -0.0594608974854162\nBest R2: -0.054507688411346754\n[10, 15]\nBest MSE: 0.8556329443265843\n[10, 15]\n"
],
[
"import seaborn as sns\n\n# plot nicely\nx_index = np.unique(bag_df.loc[:,'min_sample_split'])\ny_index = np.unique((bag_df.loc[:,'max_depth']))\na = pd.DataFrame(index = x_index, columns = y_index)\n\nfor i in x_index:\n for j in y_index:\n a.loc[i,j] = list(bag_df.loc[(bag_df['min_sample_split'] == i) & (bag_df['max_depth'] == j),'mean_r2'])[0]#.astype(float)\na = a.apply(pd.to_numeric) \nsns.heatmap(a)\nplt.xlabel('max_depth')\nplt.ylabel('min_samples_split')\nplt.title('r2')\nplt.savefig(plot_path+'Best_Model_Bagging')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Best Model: Bagging\n\n##### According to Grid Search, our best model is:\n- minsampleleaf = 10\n- maxdepth = 15\n- I had to put max-samples as 100, because some of the split has max samples of only slightly over 100",
"_____no_output_____"
],
[
"# AdaBoost",
"_____no_output_____"
]
],
[
[
"\"\"\"\nExecute Boosting for differnt number of Time Series Splits\nParameters\n - numSplits: Number of Train/Validation splits to run in order to compute our final model (bigger --> more accurate & more complex)\n - min_samples_split: Mininum number of samples to split. if a leaf node has greater than this parameter, we can still split\n - max_depth: depth of tree\n - learning_rate: gradient descent alpha\n \nReturns\n - Average R-Squared of the DT model across all Train/Validation splits\"\"\"\ndef Call_Boosting_Grid_Search(numSplits, min_samples_split, max_depth, learning_rate):\n ### Prepare Boosting\n ##############################################################################\n # Initialize Bagging Instance\n boost_mse = [] # MSE\n boost_r2 = [] # R2\n boost = AdaBoostRegressor(DecisionTreeRegressor(max_features = 'sqrt', random_state = 123, min_samples_split = min_samples_split,\n max_depth = max_depth),\n n_estimators=150, random_state=123,learning_rate=learning_rate)\n\n\n ### Time Series Split\n ##############################################################################\n splits = TimeSeriesSplit(n_splits=numSplits) # 3 splits\n\n splitCount = 0 # dummy count var to track current split num in print statements\n for train_index, test_index in splits.split(X_train_all):\n splitCount += 1 \n\n # Train Split\n X_train = X_train_all.iloc[train_index,:]\n y_train = y[X_train.index]\n\n # Validate Split\n X_val = X_train_all.iloc[test_index,:]\n y_val = y[X_val.index]\n\n\n ### Run Boosting\n boost.fit(X_train, y_train)\n prediction = boost.predict(X_val)\n\n mse = mean_squared_error(y_val, prediction)\n r2 = r2_score(y_val, prediction)\n \n\n boost_mse.append(mse)\n boost_r2.append(r2)\n\n return np.mean(boost_mse),np.mean(boost_r2)",
"_____no_output_____"
],
[
"### WARNING TAKES VERY LONG TO RUN\nmin_sample_split = [5,10,15]\nmax_depth = [5,10,15]\nlearning_rate = [0.01,0.1,1]\n\nbest_model_parameters_boost = [0,0,0]\nbest_model_param_boost_mse = [0,0,0]\nmax_r2_boosting = -100\nmin_mse_boosting = 100\ncount = 0\n\nboosting_df = pd.DataFrame()\n# Loop over all possible parameters\nfor minSampleSplit in min_sample_split:\n for maxDepth in max_depth:\n for learn in learning_rate:\n count += 1\n temp_mean_mse, temp_mean_r2 = Call_Boosting_Grid_Search(10, minSampleSplit, maxDepth, learn) # Call Boosting Train/Validation\n print(\"temp_mean \", count, \": \", temp_mean_r2)\n if temp_mean_r2 > max_r2_boosting:\n max_r2_boosting = temp_mean_r2 # store new max\n best_model_parameters_boost[0] = minSampleSplit\n best_model_parameters_boost[1] = maxDepth\n best_model_parameters_boost[2] = learn\n if temp_mean_mse < min_mse_boosting:\n min_mse_boosting = temp_mean_mse # store new max\n best_model_param_boost_mse[0] = minSampleSplit\n best_model_param_boost_mse[1] = maxDepth\n best_model_param_boost_mse[2] = learn\n boosting_df.loc[count,'min_sample_split'] = minSampleSplit\n boosting_df.loc[count,'max_depth'] = maxDepth\n boosting_df.loc[count, 'Learning_rate'] = learn\n boosting_df.loc[count,'mean_r2'] = temp_mean_r2\n \nprint(\"Best R2: \", max_r2_boosting) \nprint(best_model_parameters_boost)\nprint(\"Best MSE: \", min_mse_boosting) \nprint(best_model_param_boost_mse)",
"temp_mean 1 : -0.013864130499985828\ntemp_mean 2 : -0.043979240423140095\ntemp_mean 3 : -0.2923320589309241\ntemp_mean 4 : -0.03789874217575312\ntemp_mean 5 : -0.06520359579258461\ntemp_mean 6 : -0.16001909224645977\ntemp_mean 7 : -0.09828306848676785\ntemp_mean 8 : -0.1327629256271357\ntemp_mean 9 : -0.18611915166131782\ntemp_mean 10 : -0.015055898998584182\ntemp_mean 11 : -0.060781524580330884\ntemp_mean 12 : -0.23680920328538738\ntemp_mean 13 : -0.04548028040570747\ntemp_mean 14 : -0.10129823518028927\ntemp_mean 15 : -0.192120662955232\ntemp_mean 16 : -0.0947961490171861\ntemp_mean 17 : -0.13027457712751897\ntemp_mean 18 : -0.16466857696325013\ntemp_mean 19 : -0.014009367732869349\ntemp_mean 20 : -0.059311555956203764\ntemp_mean 21 : -0.26940682547141737\ntemp_mean 22 : -0.04267404899820879\ntemp_mean 23 : -0.08757112147348804\ntemp_mean 24 : -0.22587775029679608\ntemp_mean 25 : -0.09200502371076831\ntemp_mean 26 : -0.1697707202265931\ntemp_mean 27 : -0.22275256224458548\nBest R2: -0.013864130499985828\n[5, 5, 0.01]\nBest MSE: 0.8286563346243154\n[5, 5, 0.01]\n"
],
[
"display(boosting_df)",
"_____no_output_____"
],
[
"import seaborn as sns\nimport numpy as np\n\n# plot nicely\n\n\np = 0\nfor learn in (np.unique(boosting_df.loc[:,'Learning_rate'])):\n\n x_index = np.unique(boosting_df.loc[:,'min_sample_split'])\n y_index = np.unique((boosting_df.loc[:,'max_depth']))\n a = pd.DataFrame(index = x_index, columns = y_index)\n \n for i in x_index:\n for j in y_index:\n a.loc[i,j] = list(boosting_df.loc[(boosting_df['min_sample_split'] == i) & (boosting_df['max_depth'] == j) & (boosting_df['Learning_rate'] == learn),'mean_r2'])[0]#.astype(float)\n a = a.apply(pd.to_numeric) \n sns.heatmap(a)\n plt.savefig(plot_path+'Best_Model_Boosting_'+str(p))\n p+=1\n # f, (ax1,ax2,ax3) = matplotlib.pyplot.subplots(1, 3)\n \n \n plt.title('learning_rate' + str(learn))\n plt.xlabel('max_depth')\n plt.ylabel('min_samples_leaf')\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Best Model:\n\n#### According to Grid Search, our best model is:\n- min_samples_leaf = 5\n- max_depth = 5\n- learning_rate = 1",
"_____no_output_____"
],
[
"# Linear Regression",
"_____no_output_____"
]
],
[
[
"\"\"\"\nExecute LR for differnt number of Time Series Splits\nParameters\n - numSplits: Number of Train/Validation splits to run in order to compute our final model (bigger --> more accurate & more complex)\n \nReturns\n - Average R-Squared of the DT model across all Train/Validation splits\"\"\"\ndef Call_LR_Grid_Search(numSplits):\n ### Prepare LR\n ##############################################################################\n # Initialize Bagging Instance\n lr_mse = [] # MSE\n lr_r2 = [] # R2\n lr = LinearRegression()\n\n\n ### Time Series Split\n ##############################################################################\n splits = TimeSeriesSplit(n_splits=numSplits) # 3 splits\n\n splitCount = 0 # dummy count var to track current split num in print statements\n for train_index, test_index in splits.split(X_train_all):\n splitCount += 1 \n\n # Train Split\n X_train = X_train_all.iloc[train_index,:]\n y_train = y[X_train.index]\n\n # Validate Split\n X_val = X_train_all.iloc[test_index,:]\n y_val = y[X_val.index]\n\n\n ### Run Boosting\n lr.fit(X_train, y_train)\n prediction = lr.predict(X_val)\n\n mse = mean_squared_error(y_val, prediction)\n r2 = r2_score(y_val, prediction)\n \n\n lr_mse.append(mse)\n lr_r2.append(r2)\n\n return np.mean(lr_mse),np.mean(lr_r2)",
"_____no_output_____"
],
[
"lr_mse, lr_r2 = Call_LR_Grid_Search(10)\nprint(\"MSE: \", lr_mse)\nprint('r2:', lr_r2)",
"MSE: 1.1566064724561058\nr2: -0.5404404557165396\n"
]
],
[
[
"***\n# FINAL PREDICTION - Fixed from V7",
"_____no_output_____"
]
],
[
[
"rf_train_pred = []\nbag_train_pred = []\nboost_train_pred = []\nlr_train_pred = []\nrf_prediction = []\nbag_prediction = []\nboost_prediction = []\nlr_prediction = []",
"_____no_output_____"
]
],
[
[
"#### Set Final Model Parameters",
"_____no_output_____"
]
],
[
[
"### Number of Testing Splits/Times we Re-train the model on latest data\nN = 10\n\n### Random Forest\nmax_depth_rf = 30\nmin_samples_split_rf = 2\n\n### Bagging\nmax_depth_bagging = 15\nmin_samples_split_bagging = 5\nmax_samples_bagging = 1500\nmax_features = 20\n\n### Boosting\nmax_depth_boosting = 5\nmin_samples_split_boosting = 5\nlearning_rate = 1",
"_____no_output_____"
]
],
[
[
"#### Run Final Models (Random Forest, Bagging, Boosting)",
"_____no_output_____"
]
],
[
[
"## WARNING Takes long to run\nsplits = TimeSeriesSplit(n_splits=N)\n\nrf = RandomForestRegressor(n_estimators=150, \n #n_jobs=-1, \n random_state=123, max_features=\"sqrt\", \n max_depth=max_depth_rf, \n min_samples_split=min_samples_split_rf)\n\nbag = BaggingRegressor(DecisionTreeRegressor(max_features = 'sqrt', random_state = 123, \n max_depth = max_depth_bagging, \n min_samples_split = min_samples_split_bagging),\n n_estimators=150, \n #n_jobs=-1, \n random_state=123,\n max_samples=max_samples_bagging,\n max_features=max_features)\n\nboost = AdaBoostRegressor(DecisionTreeRegressor(max_features = 'sqrt', random_state = 123, \n max_depth = max_depth_boosting,\n min_samples_split = min_samples_split_boosting),\n n_estimators=150, \n random_state=123,\n learning_rate=learning_rate)\n\nlr = LinearRegression()\n\n\n# predict first fold of test set, need to change 21 if using different than 10 cv\n### Run Random Forest\nrf.fit(X_train_all, y_train_all)\nprediction = rf.predict(X_test.iloc[0:20,:])\nrf_prediction.append(prediction)\nrf_train_pred.append(rf.predict(X_train_all))\n\n# y_val = y[X_test.iloc[0:21,:].index]\n\n# mse = mean_squared_error(y_val, prediction)\n# r2 = r2_score(y_val, prediction)\n\n# ### Run bagging\nbag.fit(X_train_all, y_train_all)\nprediction = bag.predict(X_test.iloc[0:20,:])\nbag_prediction.append(prediction)\nbag_train_pred.append(bag.predict(X_train_all))\n\n\n# y_val = y[X_test.iloc[0:21,:].index]\n\n# mse = mean_squared_error(y_val, prediction)\n# r2 = r2_score(y_val, prediction)\n\n## Run boosting\nboost.fit(X_train_all, y_train_all)\nprediction = boost.predict(X_test.iloc[0:20,:])\nboost_prediction.append(prediction)\nboost_train_pred.append(boost.predict(X_train_all))\n\n# y_val = y[X_test.iloc[0:21,:].index]\n\n# mse = mean_squared_error(y_val, prediction)\n# r2 = r2_score(y_val, prediction)\n\n## Run lr\nlr.fit(X_train_all, y_train_all)\nprediction = lr.predict(X_test.iloc[0:20,:])\nlr_prediction.append(prediction)\nlr_train_pred.append(lr.predict(X_train_all))\n\n\n# y_val = y[X_test.iloc[0:21,:].index]\n\n\nsplitCount = 0\nfor train_index, test_index in splits.split(X_test):\n splitCount += 1\n \n # Calculate NEW Train/Test indices\n train_index = X_train_all.shape[0] + len(train_index)\n \n # Train Split\n X_train = X.iloc[0:train_index,:]\n y_train = y[X_train.index]\n \n # Validate Split\n X_val = X_test.iloc[test_index[0]:(test_index[-1]+1),:]\n y_val = y[X_val.index]\n \n ### Run Random Forest\n rf.fit(X_train, y_train)\n prediction = rf.predict(X_val)\n rf_prediction.append(prediction)\n rf_train_pred.append(rf.predict(X_train))\n\n \n# mse = mean_squared_error(y_val, prediction)\n# r2 = r2_score(y_val, prediction)\n\n# rf_metrics.append([mse,r2])\n \n ### Run bagging\n bag.fit(X_train, y_train)\n prediction = bag.predict(X_val)\n bag_prediction.append(prediction)\n bag_train_pred.append(bag.predict(X_train))\n\n\n \n# mse = mean_squared_error(y_val, prediction)\n# r2 = r2_score(y_val, prediction)\n\n# bag_metrics.append([mse,r2])\n \n ## Run boosting\n boost.fit(X_train, y_train)\n prediction = boost.predict(X_val)\n boost_prediction.append(prediction)\n boost_train_pred.append(boost.predict(X_train))\n\n\n# mse = mean_squared_error(y_val, prediction)\n# r2 = r2_score(y_val, prediction)\n\n# boost_metrics.append([mse,r2])\n\n ## Linear Regression\n lr.fit(X_train, y_train)\n prediction = lr.predict(X_val)\n lr_prediction.append(prediction)\n lr_train_pred.append(lr.predict(X_train))\n\n \n print(splitCount)",
"1\n2\n3\n4\n5\n6\n7\n8\n9\n10\n"
],
[
"#make into one array instead of 10x10 matrix\nrf_prediction = np.array(rf_prediction)\nrf_prediction = np.concatenate(rf_prediction, axis=0)\nbag_prediction = np.array(bag_prediction)\nbag_prediction = np.concatenate(bag_prediction, axis=0)\nboost_prediction = np.array(boost_prediction)\nboost_prediction = np.concatenate(boost_prediction, axis=0)\nlr_prediction = np.array(lr_prediction)\nlr_prediction = np.concatenate(lr_prediction, axis=0)",
"_____no_output_____"
],
[
"# evaluate \nrf_mse = mean_squared_error(y_test[0:210], rf_prediction)\nrf_r2 = r2_score(y_test[0:210], rf_prediction)\nbag_mse = mean_squared_error(y_test[0:210], bag_prediction)\nbag_r2 = r2_score(y_test[0:210], bag_prediction)\nboost_mse = mean_squared_error(y_test[0:210], boost_prediction)\nboost_r2 = r2_score(y_test[0:210], boost_prediction)\nlr_mse = mean_squared_error(y_test[0:210], lr_prediction)\nlr_r2 = r2_score(y_test[0:210], lr_prediction)\n\nprint('rf_mse:', rf_mse)\nprint('rf_r2:', rf_r2)\nprint('bag_mse:', bag_mse)\nprint('bag_r2:', bag_r2)\nprint('boost_mse:', boost_mse)\nprint('boost_r2:', boost_r2)\nprint('lr_mse:', lr_mse)\nprint('lr_r2:', lr_r2)",
"rf_mse: 1.1723327247240511\nrf_r2: -0.10031700554311529\nbag_mse: 1.1095992557800227\nbag_r2: -0.041437217203099896\nboost_mse: 1.064271811183617\nboost_r2: 0.0011057887673068256\nlr_mse: 1.3414309195982936\nlr_r2: -0.2590275964042281\n"
],
[
"plt.plot(['RF','Bagging','Boosting', 'LR'],[rf_mse, bag_mse, boost_mse, lr_mse],label='MSE')\nplt.plot([rf_r2, bag_r2, boost_r2, lr_r2],label='R^2')\nplt.legend()\nplt.title('MSE & R^2 of final test prediction')\n#plt.savefig(plot_path+'MSE_R2_linear_pred')\nplt.show()",
"_____no_output_____"
],
[
"# plot best model which is Random Forest\nplt.figure(figsize=(15, 7))\nplt.plot(sc.inverse_transform(np.array(y_test)))\nplt.plot(sc.inverse_transform(rf_prediction))\nplt.plot(list(range(1,210+1)), np.array([0] * 210), 'b-', color=\"red\", label='Zero')\nplt.legend([\"Realized Returns\",\"Forecasted Returns\",\"Zero\"])\n\nplt.ylabel('% Returns')\nplt.xlabel('days')\nplt.title('Random Forest: Predicted Returns vs Actual Returns')\n#plt.savefig(plot_path+'RF_Predicted_Returns_Actual_Returns')\nplt.show()",
"_____no_output_____"
],
[
"# plot best model which is Bagging\nplt.figure(figsize=(15, 7))\nplt.plot(sc.inverse_transform(np.array(y_test)))\nplt.plot(sc.inverse_transform(bag_prediction))\nplt.plot(list(range(1,210+1)), np.array([0] * 210), 'b-', color=\"red\", label='Zero')\nplt.legend([\"Realized Returns\",\"Forecasted Returns\",\"Zero\"])\n\nplt.ylabel('% Returns')\nplt.xlabel('days')\nplt.title('Bagging: Predicted Returns vs Actual Returns')\n#plt.savefig(plot_path+'Bagging_Predicted_Returns_Actual_Returns')\nplt.show()",
"_____no_output_____"
],
[
"# plot best model which is boosting\nplt.figure(figsize=(15, 7))\nplt.plot(sc.inverse_transform(np.array(y_test)))\nplt.plot(sc.inverse_transform(boost_prediction))\nplt.plot(list(range(1,210+1)), np.array([0] * 210), 'b-', color=\"red\", label='Zero')\nplt.legend([\"Realized Returns\",\"Forecasted Returns\",\"Zero\"])\n\n\nplt.ylabel('% Returns')\nplt.xlabel('days')\nplt.title('Boosting: Predicted Returns vs Actual Returns')\n#plt.savefig(plot_path+'Boosting_Predicted_Returns_Actual_Returns')\nplt.show()",
"_____no_output_____"
],
[
"# plot best model which is LR\nplt.figure(figsize=(15, 7))\nplt.plot(sc.inverse_transform(np.array(y_test)))\nplt.plot(sc.inverse_transform(lr_prediction))\nplt.plot(list(range(1,210+1)), np.array([0] * 210), 'b-', color=\"red\", label='Zero')\nplt.legend([\"Realized Returns\",\"Forecasted Returns\",\"Zero\"])\n\n\nplt.ylabel('% Returns')\nplt.xlabel('days')\nplt.title('LR: Predicted Returns vs Actual Returns')\n#plt.savefig(plot_path+'LR_Predicted_Returns_Actual_Returns')\nplt.show()",
"_____no_output_____"
],
[
"X_train.columns",
"_____no_output_____"
],
[
"importances = boost.feature_importances_\nindices = np.argsort(importances)[::-1]\nplot_path = 'C:/Users/meenm/OneDrive - UW-Madison/Github/Stat479_project2/Stat479_Project/Plots/'\n\n# Plot the feature importances of the forest\nplt.figure(figsize=(18,5))\nplt.title(\"Boosting Feature importances\")\nplt.bar(range(X_train.shape[1]), importances[indices])\n# If you want to define your own labels,\n# change indices to a list of labels on the following line.\nplt.xticks(range(X_train.shape[1]), indices)\nplt.xlim([-1, X_train.shape[1]])\nplt.ylabel('Feature Importances')\nplt.xlabel('Feature Number')\nplt.savefig(plot_path+'Boosting_Feature_Importance.png')\nplt.show()",
"_____no_output_____"
],
[
"boost.feature_importances_",
"_____no_output_____"
],
[
"# RF Prediction\ncorrect = 0\n\nconf_mat_rf = np.array([[0,0],[0,0]])\n\nfor ind, i in enumerate(rf_prediction):\n if rf_prediction[ind] <= 0 and y_test[ind] <= 0:\n correct = correct + 1\n conf_mat_rf[0,0] = conf_mat_rf[0,0] + 1\n elif rf_prediction[ind] >= 0 and y_test[ind] >= 0:\n correct = correct + 1\n conf_mat_rf[1,1] = conf_mat_rf[1,1] + 1\n elif rf_prediction[ind] >= 0 and y_test[ind] < 0:\n conf_mat_rf[1,0] = conf_mat_rf[1,0] + 1\n elif rf_prediction[ind] <=0 and y_test[ind] > 0:\n conf_mat_rf[0,1] = conf_mat_rf[0,1] + 1\n \nprint('Accuracy', correct/210)\nprint(conf_mat_rf)",
"Accuracy 0.5523809523809524\n[[44 33]\n [61 72]]\n"
],
[
"# Bag Prediction\ncorrect = 0\n\nconf_mat_bag = np.array([[0,0],[0,0]])\n\nfor ind, i in enumerate(bag_prediction):\n if bag_prediction[ind] <= 0 and y_test[ind] <= 0:\n correct = correct + 1\n conf_mat_bag[0,0] = conf_mat_bag[0,0] + 1\n elif bag_prediction[ind] >= 0 and y_test[ind] >= 0:\n correct = correct + 1\n conf_mat_bag[1,1] = conf_mat_bag[1,1] + 1\n elif bag_prediction[ind] >= 0 and y_test[ind] < 0:\n conf_mat_bag[1,0] = conf_mat_bag[1,0] + 1\n elif bag_prediction[ind] <=0 and y_test[ind] > 0:\n conf_mat_bag[0,1] = conf_mat_bag[0,1] + 1\n \nprint('Accuracy', correct/210)\nprint(conf_mat_bag)",
"Accuracy 0.5428571428571428\n[[35 26]\n [70 79]]\n"
],
[
"# BINARY PREDICTION ACCURATE 53.3% of time for boosting \ncorrect = 0\n\nconf_mat_boost = np.array([[0,0],[0,0]])\n\nfor ind, i in enumerate(boost_prediction):\n if boost_prediction[ind] >= 0 and y_test[ind] >= 0:\n correct = correct + 1\n conf_mat_boost[0,0] = conf_mat_boost[0,0] + 1\n elif boost_prediction[ind] <= 0 and y_test[ind] <= 0:\n correct = correct + 1\n conf_mat_boost[1,1] = conf_mat_boost[1,1] + 1\n elif boost_prediction[ind] >= 0 and y_test[ind] < 0:\n conf_mat_boost[1,0] = conf_mat_boost[1,0] + 1\n elif boost_prediction[ind] <=0 and y_test[ind] > 0:\n conf_mat_boost[0,1] = conf_mat_boost[0,1] + 1\n \nprint('Accuracy', correct/210)\nprint(conf_mat_boost)",
"Accuracy 0.5333333333333333\n[[50 55]\n [43 62]]\n"
],
[
"# BINARY PREDICTION ACCURATE 53.3% of time for LR \ncorrect = 0\n\nconf_mat_lr = np.array([[0,0],[0,0]])\n\nfor ind, i in enumerate(lr_prediction):\n if lr_prediction[ind] >= 0 and y_test[ind] >= 0:\n correct = correct + 1\n conf_mat_lr[0,0] = conf_mat_lr[0,0] + 1\n elif lr_prediction[ind] <= 0 and y_test[ind] <= 0:\n correct = correct + 1\n conf_mat_lr[1,1] = conf_mat_lr[1,1] + 1\n elif lr_prediction[ind] >= 0 and y_test[ind] < 0:\n conf_mat_lr[1,0] = conf_mat_lr[1,0] + 1\n elif lr_prediction[ind] <=0 and y_test[ind] > 0:\n conf_mat_lr[0,1] = conf_mat_lr[0,1] + 1\n \nprint('Accuracy', correct/210)\nprint(conf_mat_lr)",
"Accuracy 0.5238095238095238\n[[82 23]\n [77 28]]\n"
]
],
[
[
"## Confusion Matrix",
"_____no_output_____"
]
],
[
[
"rf_prediction_binary = np.where(rf_prediction>=0,1,0)\nbag_prediction_binary = np.where(bag_prediction>=0,1,0)\nboost_prediction_binary = np.where(boost_prediction>=0,1,0)\nlr_prediction_binary = np.where(lr_prediction>=0,1,0)\ny_test_binary = np.where(y_test>=0,1,0)",
"_____no_output_____"
],
[
"from mlxtend.evaluate import confusion_matrix\n\ncm_rf = confusion_matrix(rf_prediction_binary,y_test_binary)\ncm_bagging = confusion_matrix(bag_prediction_binary,y_test_binary)\ncm_boosting = confusion_matrix(boost_prediction_binary,y_test_binary)\ncm_lr = confusion_matrix(lr_prediction_binary,y_test_binary)\n\ndisplay(cm_rf)\ndisplay(cm_bagging)\ndisplay(cm_boosting)\ndisplay(cm_lr)",
"_____no_output_____"
],
[
"from mlxtend.plotting import plot_confusion_matrix\nimport matplotlib.pyplot as plt\n\nfig, ax = plot_confusion_matrix(conf_mat=cm_rf)\nplt.title('Random Forest: Confusion Matrix')\nfig, ax = plot_confusion_matrix(conf_mat=cm_bagging)\nplt.title('Bagging: Confusion Matrix')\nfig, ax = plot_confusion_matrix(conf_mat=cm_boosting)\nplt.title('Boosting: Confusion Matrix')\nfig, ax = plot_confusion_matrix(conf_mat=cm_lr)\nplt.title('Linear Regression: Confusion Matrix')",
"_____no_output_____"
],
[
"from sklearn.preprocessing import MinMaxScaler\n\nrf_prediction_prob = MinMaxScaler().fit_transform(rf_prediction.reshape(-1,1))\nrf_prediction_prob = rf_prediction_prob.flatten('C')\nbag_prediction_prob = MinMaxScaler().fit_transform(bag_prediction.reshape(-1,1))\nbag_prediction_prob = bag_prediction_prob.flatten('C')\nboost_prediction_prob = MinMaxScaler().fit_transform(boost_prediction.reshape(-1,1))\nboost_prediction_prob = boost_prediction_prob.flatten('C')\nlr_prediction_prob = MinMaxScaler().fit_transform(lr_prediction.reshape(-1,1))\nlr_prediction_prob = lr_prediction_prob.flatten('C')",
"_____no_output_____"
],
[
"def confusion_matrix_binary(y_true, y_predicted):\n\n tp, fn, fp, tn = 0, 0, 0, 0\n \n for i, j in zip(y_true, y_predicted):\n if i == j:\n if i == 0:\n tp += 1\n else:\n tn += 1\n else:\n if i == 0:\n fn += 1\n else:\n fp += 1\n \n conf_matrix = np.zeros(4).reshape(2, 2).astype(int)\n conf_matrix[0, 0] = tp\n conf_matrix[0, 1] = fn\n conf_matrix[1, 0] = fp\n conf_matrix[1, 1] = tn \n \n return conf_matrix",
"_____no_output_____"
],
[
"def binary_cm_from_multiclass(y_true, y_predicted, positive_label):\n \n y_true_ary = np.array(y_true)\n y_predicted_ary = np.array(y_predicted)\n \n y_true_mod = np.where(y_true_ary != positive_label, 1, 0) # YOUR CODE\n y_predicted_mod = np.where(y_predicted_ary != positive_label, 1, 0) # YOUR CODE\n \n cm = confusion_matrix_binary(y_true_mod, y_predicted_mod)\n return cm",
"_____no_output_____"
],
[
"from collections import OrderedDict\n\ndef plot_roc_curve_plus_auc(y_true, y_score, col, lb,lim, pos_label=1, num_thresholds=100):\n\n # INSERT YOUR CODE FROM THE PREVIOUS EXERCISE HERE\n # BUT MODIFY IT SUCH THAT IT ALSO RETURNS THE\n # ROC Area Under the Curve\n \n # MODIFY THIS CELL\n y_true_ary = np.array(y_true)\n y_score_ary = np.array(y_score)\n x_axis_values = []\n y_axis_values = []\n thresholds = np.linspace(0., 1., num_thresholds)\n \n num_positives = len(np.where(y_true_ary == pos_label)[0]) # YOUR CODE\n num_negatives = len(np.where(y_true_ary != pos_label)[0]) # YOUR CODE\n \n plt.plot([0,1],[0,1],linestyle='--',color='black',label='Random Guess')\n\n for i, thr in enumerate(thresholds):\n \n binarized_scores = np.where(y_score >= thr, pos_label, int(not pos_label))\n \n positive_predictions = len(np.where(binarized_scores == pos_label)[0]) # YOUR CODE\n \n cm = binary_cm_from_multiclass(y_true_ary, binarized_scores, pos_label)\n num_true_positives = cm[0][0] #YOUR CODE\n num_false_positives = cm[1][0]# YOUR CODE\n \n fpr = cm[1][0]/(cm[1][0] + cm[1][1])\n tpr = cm[0][0]/(cm[0][1] + cm[0][0])\n \n x_axis_values.append(fpr) # YOUR CODE\n y_axis_values.append(tpr) # YOUR CODE\n \n plt.step(x_axis_values, y_axis_values, where='post', color=col, label=lb)\n plt.xlim([0., lim])\n plt.ylim([0., lim])\n plt.ylabel('True Positive Rate')\n plt.xlabel('False Positive Rate')\n \n #Preventing Repeating Labeles on the plot\n handles, labels = plt.gca().get_legend_handles_labels()\n by_label = OrderedDict(zip(labels, handles))\n plt.legend(by_label.values(), by_label.keys())\n \n plt.title('ROC curve')\n \n # sort x and y values to prevent getting negative values\n x_axis_values_sorted = x_axis_values.sort()\n y_axis_values_sorted = y_axis_values.sort()\n roc_auc = np.trapz(y_axis_values, x_axis_values)\n\n return roc_auc",
"_____no_output_____"
],
[
"auc_rf = plot_roc_curve_plus_auc(y_test_binary, rf_prediction_prob, 'blue', 'Random Forest', 1,pos_label=1)\nauc_bagging = plot_roc_curve_plus_auc(y_test_binary, bag_prediction_prob,'green', 'Bagging', 1,pos_label=1)\nauc_boosting = plot_roc_curve_plus_auc(y_test_binary, boost_prediction_prob,'brown', 'Boosting',1, pos_label=1)\nauc_lr = plot_roc_curve_plus_auc(y_test_binary, lr_prediction_prob,'orange', 'Linear Regression',1, pos_label=1)\n\nplt.savefig(plot_path+\"ROC Curve_1\")",
"_____no_output_____"
],
[
"#ROC Curve with 0.4 of xlim/ylim\nauc_rf = plot_roc_curve_plus_auc(y_test_binary, rf_prediction_prob, 'blue', 'Random Forest', 0.4,pos_label=1)\nauc_bagging = plot_roc_curve_plus_auc(y_test_binary, bag_prediction_prob,'green', 'Bagging', 0.4,pos_label=1)\nauc_boosting = plot_roc_curve_plus_auc(y_test_binary, boost_prediction_prob,'brown', 'Boosting',0.4, pos_label=1)\nauc_lr = plot_roc_curve_plus_auc(y_test_binary, lr_prediction_prob,'orange', 'Linear Regression',0.4, pos_label=1)\n\nplt.savefig(plot_path+\"ROC Curve_2\")",
"_____no_output_____"
],
[
"auc_dic = {\"auc_rf\":auc_rf,\"auc_bagging\":auc_bagging,\"auc_boosting\":auc_boosting, \"auc_lr\":auc_lr}\n\nfor i in auc_dic.keys():\n print('ROC AUC',i, ': %.4f' % auc_dic[i])",
"ROC AUC auc_rf : 0.5516\nROC AUC auc_bagging : 0.5713\nROC AUC auc_boosting : 0.5683\nROC AUC auc_lr : 0.5449\n"
],
[
"# plot best model which is boosting\nplt.figure(figsize=(15, 7))\nplt.plot(sc.inverse_transform(np.array(y_test[0:210])).cumsum())\nplt.plot(sc.inverse_transform(boost_prediction).cumsum())\nplt.plot(list(range(1,210+1)), np.array([0] * 210), 'b-', color=\"red\", label='Zero')\nplt.legend([\"Cumulative Realized Returns\",\"Cumulative Forecasted Returns\",\"Zero\"])\n\nplt.ylabel('% Returns')\nplt.xlabel('days')\nplt.title('Boosting: Predicted Returns vs Actual Returns')\nplt.savefig(plot_path+'Boosting_Pred_Returns_vs_Actual_Returns')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecefc9ae83812637ca4eaa91a50a66578c7d8abc | 103,749 | ipynb | Jupyter Notebook | MySQL_exercises/.ipynb_checkpoints/MySQL_Exercise_05_Summaries_of_Groups_of_Data-checkpoint.ipynb | Renatochaz/SQL-for-Big-Data | 7e92fa525d8caabb6f10a1c3d208d5582e272269 | [
"FSFAP"
] | null | null | null | MySQL_exercises/.ipynb_checkpoints/MySQL_Exercise_05_Summaries_of_Groups_of_Data-checkpoint.ipynb | Renatochaz/SQL-for-Big-Data | 7e92fa525d8caabb6f10a1c3d208d5582e272269 | [
"FSFAP"
] | null | null | null | MySQL_exercises/.ipynb_checkpoints/MySQL_Exercise_05_Summaries_of_Groups_of_Data-checkpoint.ipynb | Renatochaz/SQL-for-Big-Data | 7e92fa525d8caabb6f10a1c3d208d5582e272269 | [
"FSFAP"
] | null | null | null | 30.469604 | 570 | 0.312552 | [
[
[
"Copyright Jana Schaich Borg/Attribution-NonCommercial 4.0 International (CC BY-NC 4.0)",
"_____no_output_____"
],
[
"# MySQL Exercise 5: Summaries of Groups of Data\n\nSo far you've learned how to select, reformat, manipulate, order, and summarize data from a single table in database. In this lesson, you are going to learn how to summarize multiple subsets of your data in the same query. The method for doing this is to include a \"GROUP BY\" clause in your SQL query. \n \n \n\n## The GROUP BY clause\n\nThe GROUP BY clause comes after the WHERE clause, but before ORDER BY or LIMIT:\n\n<img src=\"https://duke.box.com/shared/static/lc0yjfz1339q3mc04ne13m2q5tkzy3x7.jpg\" width=400 alt=\"SELECT FROM WHERE ORDER BY\" />\n\nThe GROUP BY clause is easy to incorporate into your queries. In fact, it might be a little too easy to incorporate into MySQL queries, because it can be used incorrectly in MySQL queries even when no error message is displayed. As a consequence, I suggest you adopt a healthy dose of caution every time you use the GROUP BY clause. By the end of this lesson, you will understand why. When used correctly, though, GROUP BY is one of the most useful and efficient parts of an SQL query, and once you are comfortable using it, you will use it very frequently.\n\n**To get started, load the SQL library and the Dognition database, and set the dognition database as the default:**\n",
"_____no_output_____"
]
],
[
[
"%load_ext sql\n%sql mysql://studentuser:studentpw@localhost/dognitiondb\n%sql USE dognitiondb",
" * mysql://studentuser:***@localhost/dognitiondb\n0 rows affected.\n"
]
],
[
[
"Let's return to a question from MySQL Exercise 4. How would you query the average rating for each of the 40 tests in the Reviews table? As we discussed, one very inefficient method to do that would be to write 40 separate queries with each one having a different test name in the WHERE conditional clause. Then you could copy or transcribe the results from all 40 queries into one place. But that wouldn't be very pleasant. Here's how you could do the same thing using one query that has a GROUP BY clause:\n\n```mySQL\nSELECT test_name, AVG(rating) AS AVG_Rating\nFROM reviews\nGROUP BY test_name\n```\n\nThis query will output the average rating for each test. More technically, this query will instruct MySQL to average all the rows that have the same value in the test_name column.\n\nNotice that I included test_name in the SELECT statement. As a strong rule of thumb, if you are grouping by a column, you should also include that column in the SELECT statement. If you don't do this, you won't know to which group each row of your output corresponds. \n \n**To see what I mean, try the query above without test_name included in the SELECT statement:**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT AVG(rating) AS avg_rating\nFROM reviews\nGROUP BY test_name LIMIT 3;",
" * mysql://studentuser:***@localhost/dognitiondb\n3 rows affected.\n"
]
],
[
[
"You can form groups using derived values as well as original columns. To illustrate this, let's address another question: how many tests were completed during each month of the year?\n\nTo answer this question, we need to take advantage of another datetime function described in the website below:\n\nhttp://www.w3resource.com/mysql/date-and-time-functions/date-and-time-functions.php\n\nMONTH() will return a number representing the month of a date entry. To get the total number of tests completed each month, you could put the MONTH function into the GROUP BY clause, in this case through an alias:\n\n```mySQL\nSELECT test_name, MONTH(created_at) AS Month, COUNT(created_at) AS Num_Completed_Tests\nFROM complete_tests\nGROUP BY Month;\n```\n\nYou can also group by multiple columns or derived fields. If we wanted to determine the total number of each type of test completed each month, you could include both \"test_name\" and the derived \"Month\" field in the GROUP BY clause, separated by a comma.\n\n```mySQL\nSELECT test_name, MONTH(created_at) AS Month, COUNT(created_at) AS Num_Completed_Tests\nFROM complete_tests\nGROUP BY test_name, Month;\n```\n\nMySQL allows you to use aliases in a GROUP BY clause, but some database systems do not. If you are using a database system that does NOT accept aliases in GROUP BY clauses, you can still group by derived fields, but you have to duplicate the calculation for the derived field in the GROUP BY clause in addition to including the derived field in the SELECT clause:\n\n```mySQL\nSELECT test_name, MONTH(created_at) AS Month, COUNT(created_at) AS Num_Completed_Tests\nFROM complete_tests\nGROUP BY test_name, MONTH(created_at);\n``` \n \nTry the query once with test_name first in the GROUP BY list, and once with Month first in the GROUP BY list below. Inspect the outputs:",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT test_name, MONTH(created_at) AS Month, COUNT(created_at) AS Num_Completed_Tests\nFROM complete_tests\nGROUP BY Month, test_name LIMIT 5;",
" * mysql://studentuser:***@localhost/dognitiondb\n5 rows affected.\n"
],
[
"%%sql\nSELECT test_name, MONTH(created_at) AS Month, COUNT(created_at) AS Num_Completed_Tests\nFROM complete_tests\nGROUP BY test_name, Month LIMIT 5;",
" * mysql://studentuser:***@localhost/dognitiondb\n5 rows affected.\n"
]
],
[
[
"Notice that in the first case, the first block of rows share the same test_name, but are broken up into separate months (for those of you who took the \"Data Visualization and Communication with Tableau\" course of this specialization, this is similar to what would happen if you put test_name first and created_at second on the rows or columns shelf in Tableau). \n\nIn the second case, the first block of rows share the same month, but are broken up into separate tests (this is similar to what would happen if you put created_at first and test_name second on the rows or columns shelf in Tableau). If you were to visualize these outputs, they would look like the charts below.\n\n<img src=\"https://duke.box.com/shared/static/y6l3ldxg289brid5mmn886qweglt65qv.jpg\" width=800 alt=\"ORDER\" />\n\nDifferent database servers might default to ordering the outputs in a certain way, but you shouldn't rely on that being the case. To ensure the output is ordered in a way you intend, add an ORDER BY clause to your grouped query using the syntax you already know and have practiced:\n\n```mySQL\nSELECT test_name, MONTH(created_at) AS Month, COUNT(created_at) AS Num_Completed_Tests\nFROM complete_tests\nGROUP BY test_name, Month\nORDER BY test_name ASC, Month ASC;\n```\n\n**Question 1: Output a table that calculates the number of distinct female and male dogs in each breed group of the Dogs table, sorted by the total number of dogs in descending order (the sex/breed_group pair with the greatest number of dogs should have 8466 unique Dog_Guids):**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT COUNT(DISTINCT dog_guid) as num_dogs, breed_group, SUM(gender = 'female') as Female, SUM(gender = 'male') as Male \nFROM dogs\ngroup by breed_group\nORDER BY num_dogs DESC;\n\nSELECT COUNT(DISTINCT dog_guid) as num_dogs, breed_group, gender\nFROM dogs\ngroup by gender, breed_group\nORDER BY num_dogs DESC;",
" * mysql://studentuser:***@localhost/dognitiondb\n9 rows affected.\n18 rows affected.\n"
]
],
[
[
"Some database servers, including MySQL, allow you to use numbers in place of field names in the GROUP BY or ORDER BY fields to reduce the overall length of the queries. I tend to avoid this abbreviated method of writing queries because I find it challenging to troubleshoot when you are writing complicated queries with many fields, but it does allow you to write queries faster. To use this method, assign each field in your SELECT statement a number acording to the order the field appears in the SELECT statement. In the following statement:\n\n```mySQL\nSELECT test_name, MONTH(created_at) AS Month, COUNT(created_at) AS Num_Completed_Tests\n```\n\ntest_name would be #1, Month would be #2, and Num_Completed_Tests would be #3. You could then rewrite the query above to read:\n\n```mySQL\nSELECT test_name, MONTH(created_at) AS Month, COUNT(created_at) AS Num_Completed_Tests\nFROM complete_tests\nGROUP BY 1, 2\nORDER BY 1 ASC, 2 ASC;\n```\n\n**Question 2: Revise the query your wrote in Question 1 so that it uses only numbers in the GROUP BY and ORDER BY fields.**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT COUNT(DISTINCT dog_guid) as num_dogs, breed_group, gender\nFROM dogs\ngroup by 2, 3\nORDER BY 1 DESC;",
" * mysql://studentuser:***@localhost/dognitiondb\n18 rows affected.\n"
]
],
[
[
"## The HAVING clause\n\nJust like you can query subsets of rows using the WHERE clause, you can query subsets of aggregated groups using the HAVING clause. However, wheras the expression that follows a WHERE clause has to be applicable to each row of data in a column, the expression that follows a HAVING clause has to be applicable or computable using a group of data. \n\nIf you wanted to examine the number of tests completed only during the winter holiday months of November and December, you would need to use a WHERE clause, because the month a test was completed in is recorded in each row. Your query might look like this:\n\n```mySQL\nSELECT test_name, MONTH(created_at) AS Month, COUNT(created_at) AS Num_Completed_Tests\nFROM complete_tests\nWHERE MONTH(created_at)=11 OR MONTH(created_at)=12\nGROUP BY 1, 2\nORDER BY 3 DESC;\n```\nIf you then wanted to output only the test-month pairs that had at least 20 records in them, you would add a HAVING clause, because the stipulation of at least 20 records only makes sense and is only computable at the aggregated group level:\n\n```mySQL\nSELECT test_name, MONTH(created_at) AS Month, COUNT(created_at) AS Num_Completed_Tests\nFROM complete_tests\nWHERE MONTH(created_at)=11 OR MONTH(created_at)=12\nGROUP BY 1, 2\nHAVING COUNT(created_at)>=20\nORDER BY 3 DESC;\n```\n\n**Question 3: Revise the query your wrote in Question 2 so that it (1) excludes the NULL and empty string entries in the breed_group field, and (2) excludes any groups that don't have at least 1,000 distinct Dog_Guids in them. Your result should contain 8 rows. (HINT: sometimes empty strings are registered as non-NULL values. You might want to include the following line somewhere in your query to exclude these values as well):**\n\n```mySQL\nbreed_group!=\"\"\n```\n",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT COUNT(DISTINCT dog_guid) as num_dogs, breed_group, gender\nFROM dogs\nGROUP BY 2, 3\nHAVING breed_group!=\"\" AND num_dogs >= 1000\nORDER BY 1 DESC;",
" * mysql://studentuser:***@localhost/dognitiondb\n8 rows affected.\n"
]
],
[
[
"We will review several issues that can be tricky about using GROUP BY in your queries in the next lesson, but those issues will make more sense once you are sure you are comfortable with the basic functionality of the GROUP BY and HAVING clauses. \n \n \n\n## Practice incorporating GROUP BY and HAVING into your own queries.\n\n**Question 4: Write a query that outputs the average number of tests completed and average mean inter-test-interval for every breed type, sorted by the average number of completed tests in descending order (popular hybrid should be the first row in your output).**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT breed_type, AVG(total_tests_completed) as avg_tests, AVG(mean_iti_days)\nFROM dogs \nGROUP BY breed_type \nORDER BY 2 DESC LIMIT 10;",
" * mysql://studentuser:***@localhost/dognitiondb\n4 rows affected.\n"
]
],
[
[
"**Question 5: Write a query that outputs the average amount of time it took customers to complete each type of test where any individual reaction times over 6000 hours are excluded and only average reaction times that are greater than 0 seconds are included (your output should end up with 67 rows).**\n",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT test_name, AVG(TIMESTAMPDIFF(HOUR,start_time,end_time)) as avg_duration\nFROM exam_answers\nWHERE TIMESTAMPDIFF(MINUTE,start_time,end_time) <= 6000\nGROUP BY test_name\nHAVING AVG(TIMESTAMPDIFF(MINUTE,start_time,end_time)) > 0\nORDER BY 2 DESC LIMIT 10;",
" * mysql://studentuser:***@localhost/dognitiondb\n10 rows affected.\n"
]
],
[
[
"**Question 6: Write a query that outputs the total number of unique User_Guids in each combination of State and ZIP code (postal code) in the United States, sorted first by state name in ascending alphabetical order, and second by total number of unique User_Guids in descending order (your first state should be AE and there should be 5043 rows in total in your output).**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT user_guid, state, zip as postal_code\nFROM users\nWHERE country = 'US'\nGROUP BY state, zip\nORDER BY state ASC LIMIT 10;",
" * mysql://studentuser:***@localhost/dognitiondb\n10 rows affected.\n"
]
],
[
[
"**Question 7: Write a query that outputs the total number of unique User_Guids in each combination of State and ZIP code in the United States *that have at least 5 users*, sorted first by state name in ascending alphabetical order, and second by total number of unique User_Guids in descending order (your first state/ZIP code combination should be AZ/86303).**",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT COUNT(user_guid) as total_users, state, zip\nFROM users\nWHERE country = 'US'\nGROUP BY state, zip\nHAVING total_users >= 5\nORDER BY state ASC, total_users DESC;",
" * mysql://studentuser:***@localhost/dognitiondb\n377 rows affected.\n"
]
],
[
[
"<mark>**Be sure to watch the next video before beginning Exercise 6, the next set of MySQL exercises</mark>,** **and feel free to practice any other queries you wish below!**",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecefcc6757bcc55c8dfbe012f994b6664a91275b | 372,011 | ipynb | Jupyter Notebook | section_graph_slam/graphbasedslam4.ipynb | kentaroy47/LNPR_BOOK_CODES | f0d1bef336423ebdf04539ce833f0ce4cffc51f5 | [
"MIT"
] | 148 | 2019-03-27T00:20:16.000Z | 2022-03-30T22:34:11.000Z | section_graph_slam/graphbasedslam4.ipynb | kentaroy47/LNPR_BOOK_CODES | f0d1bef336423ebdf04539ce833f0ce4cffc51f5 | [
"MIT"
] | 3 | 2018-11-07T04:33:13.000Z | 2018-12-31T01:35:16.000Z | section_graph_slam/graphbasedslam4.ipynb | kentaroy47/LNPR_BOOK_CODES | f0d1bef336423ebdf04539ce833f0ce4cffc51f5 | [
"MIT"
] | 116 | 2019-04-18T08:35:53.000Z | 2022-03-24T05:17:46.000Z | 119.887528 | 223,523 | 0.782256 | [
[
[
"import sys \nsys.path.append('../scripts/')\nfrom kf import * #誤差楕円を描くのに利用",
"_____no_output_____"
],
[
"def make_ax(): #axisの準備\n fig = plt.figure(figsize=(4,4))\n ax = fig.add_subplot(111)\n ax.set_aspect('equal')\n ax.set_xlim(-5,5) \n ax.set_ylim(-5,5) \n ax.set_xlabel(\"X\",fontsize=10) \n ax.set_ylabel(\"Y\",fontsize=10) \n return ax\n\ndef draw_trajectory(xs, ax): #軌跡の描画\n poses = [xs[s] for s in range(len(xs))]\n ax.scatter([e[0] for e in poses], [e[1] for e in poses], s=5, marker=\".\", color=\"black\")\n ax.plot([e[0] for e in poses], [e[1] for e in poses], linewidth=0.5, color=\"black\")\n \ndef draw_observations(xs, zlist, ax): #センサ値の描画\n for s in range(len(xs)):\n if s not in zlist:\n continue\n \n for obs in zlist[s]:\n x, y, theta = xs[s]\n ell, phi = obs[1][0], obs[1][1]\n mx = x + ell*math.cos(theta + phi)\n my = y + ell*math.sin(theta + phi)\n ax.plot([x,mx], [y,my], color=\"pink\", alpha=0.5)\n \ndef draw_edges(edges, ax): ###graphbasedslam2draw\n for e in edges:\n ax.plot([e.x1[0], e.x2[0]], [e.x1[1] ,e.x2[1]], color=\"red\", alpha=0.5)\n\ndef draw(xs, zlist, edges): #引数追加\n ax = make_ax()\n draw_observations(xs, zlist, ax)\n draw_trajectory(xs, ax)\n draw_edges(edges, ax) #追加\n plt.show()",
"_____no_output_____"
],
[
"def read_data():#データの読み込み\n hat_xs = {} #軌跡のデータ(ステップ数をキーにして姿勢を保存)\n zlist = {} #センサ値のデータ(ステップ数をキーにして、さらにその中にランドマークのIDとセンサ値をタプルで保存)\n\n with open(\"log.txt\") as f:\n for line in f.readlines():\n tmp = line.rstrip().split()\n\n step = int(tmp[1])\n if tmp[0] == \"x\": #姿勢のレコードの場合\n hat_xs[step] = np.array([float(tmp[2]), float(tmp[3]), float(tmp[4])]).T\n elif tmp[0] == \"z\": #センサ値のレコードの場合\n if step not in zlist: #まだ辞書が空の時は空の辞書を作る\n zlist[step] = []\n zlist[step].append((int(tmp[2]), np.array([float(tmp[3]), float(tmp[4]), float(tmp[5])]).T))\n \n return hat_xs, zlist",
"_____no_output_____"
],
[
"class ObsEdge: ###graphbasedslam4matrix\n def __init__(self, t1, t2, z1, z2, xs, sensor_noise_rate=[0.14, 0.05, 0.05]): #sensor_noise_rate追加\n assert z1[0] == z2[0] \n\n self.t1, self.t2 = t1, t2 \n self.x1, self.x2 = xs[t1], xs[t2]\n self.z1, self.z2 = z1[1], z2[1]\n \n s1 = math.sin(self.x1[2] + self.z1[1]) \n c1 = math.cos(self.x1[2] + self.z1[1])\n s2 = math.sin(self.x2[2] + self.z2[1])\n c2 = math.cos(self.x2[2] + self.z2[1])\n \n ##誤差の計算##\n hat_e = self.x2 - self.x1 + np.array([\n self.z2[0]*c2 - self.z1[0]*c1, \n self.z2[0]*s2 - self.z1[0]*s1,\n self.z2[1] - self.z2[2] - self.z1[1] + self.z1[2]\n ])\n while hat_e[2] >= math.pi: hat_e[2] -= math.pi*2\n while hat_e[2] < -math.pi: hat_e[2] += math.pi*2\n \n ##精度行列の作成## \n Q1 = np.diag([(self.z1[0]*sensor_noise_rate[0])**2, sensor_noise_rate[1]**2, sensor_noise_rate[2]**2])\n R1 = - np.array([[c1, -self.z1[0]*s1, 0],\n [s1, self.z1[0]*c1, 0],\n [ 0, 1, -1]])\n \n Q2 = np.diag([(self.z2[0]*sensor_noise_rate[0])**2, sensor_noise_rate[1]**2, sensor_noise_rate[2]**2])\n R2 = np.array([[c2, -self.z2[0]*s2, 0],\n [s2, self.z2[0]*c2, 0],\n [ 0, 1, -1]])\n \n Sigma = R1.dot(Q1).dot(R1.T) + R2.dot(Q2).dot(R2.T)\n Omega = np.linalg.inv(Sigma)\n \n print(Sigma)",
"_____no_output_____"
],
[
"import itertools\ndef make_edges(hat_xs, zlist):\n landmark_keys_zlist = {} #ランドマークのIDをキーにして観測された時刻とセンサ値を記録\n\n for step in zlist: #キーを時刻からランドマークのIDへ\n for z in zlist[step]:\n landmark_id = z[0]\n if landmark_id not in landmark_keys_zlist: \n landmark_keys_zlist[landmark_id] = []\n\n landmark_keys_zlist[landmark_id].append((step, z))\n \n edges = []\n for landmark_id in landmark_keys_zlist:\n step_pairs = list(itertools.combinations(landmark_keys_zlist[landmark_id], 2)) #時刻のペアを作成\n edges += [ObsEdge(xz1[0], xz2[0], xz1[1], xz2[1], hat_xs) for xz1, xz2 in step_pairs]\n \n return edges",
"_____no_output_____"
],
[
"hat_xs, zlist = read_data() ###graphbasedslam4exec\nedges = make_edges(hat_xs, zlist)\ndraw(hat_xs, zlist, edges)",
"[[ 0.12541578 -0.00496595 0.00054303]\n [-0.00496595 0.0174828 0.00882297]\n [ 0.00054303 0.00882297 0.01 ]]\n[[ 0.21193083 -0.14192213 0.00957162]\n [-0.14192213 0.31788804 0.01053672]\n [ 0.00957162 0.01053672 0.01 ]]\n[[ 0.19548551 -0.12110289 0.00863156]\n [-0.12110289 0.26220417 0.01024947]\n [ 0.00863156 0.01024947 0.01 ]]\n[[ 0.16496596 -0.07748165 0.00614591]\n [-0.07748165 0.14288966 0.00974061]\n [ 0.00614591 0.00974061 0.01 ]]\n[[ 0.16023894 -0.06377287 0.0050494 ]\n [-0.06377287 0.1034876 0.00975108]\n [ 0.0050494 0.00975108 0.01 ]]\n[[ 0.11660836 -0.02344136 0.00281867]\n [-0.02344136 0.04188691 0.00825788]\n [ 0.00281867 0.00825788 0.01 ]]\n[[ 0.09651109 -0.00930661 0.00191347]\n [-0.00930661 0.02534225 0.00712307]\n [ 0.00191347 0.00712307 0.01 ]]\n[[ 0.08630317 -0.00238696 0.00147377]\n [-0.00238696 0.01907312 0.00609948]\n [ 0.00147377 0.00609948 0.01 ]]\n[[ 0.22344506 0.14226925 0.00846139]\n [ 0.14226925 0.2566526 -0.00084402]\n [ 0.00846139 -0.00084402 0.01 ]]\n[[0.14416869 0.07153365 0.00673083]\n [0.07153365 0.16396783 0.00141714]\n [0.00673083 0.00141714 0.01 ]]\n[[0.12941508 0.05757632 0.00647949]\n [0.05757632 0.15163042 0.00204741]\n [0.00647949 0.00204741 0.01 ]]\n[[0.10662666 0.03092457 0.00472484]\n [0.03092457 0.08658787 0.00294148]\n [0.00472484 0.00294148 0.01 ]]\n[[0.08896496 0.01008 0.0028692 ]\n [0.01008 0.03929036 0.00412997]\n [0.0028692 0.00412997 0.01 ]]\n[[0.08719467 0.00773667 0.00199805]\n [0.00773667 0.02524617 0.00412654]\n [0.00199805 0.00412654 0.01 ]]\n[[ 3.16438529e-01 1.22371319e-03 -1.66869535e-04]\n [ 1.22371319e-03 4.04228908e-02 -3.51975221e-03]\n [-1.66869535e-04 -3.51975221e-03 1.00000000e-02]]\n[[ 0.40653983 0.03473101 0.00104295]\n [ 0.03473101 0.05620982 -0.00503846]\n [ 0.00104295 -0.00503846 0.01 ]]\n[[ 0.23407362 0.03669221 0.00169736]\n [ 0.03669221 0.04029807 -0.00180511]\n [ 0.00169736 -0.00180511 0.01 ]]\n[[ 1.69933970e-01 3.13456864e-02 1.91647109e-03]\n [ 3.13456864e-02 3.47453763e-02 -1.14600372e-04]\n [ 1.91647109e-03 -1.14600372e-04 1.00000000e-02]]\n[[0.13967791 0.03046262 0.00235514]\n [0.03046262 0.03703836 0.00093646]\n [0.00235514 0.00093646 0.01 ]]\n[[0.11048971 0.01830573 0.00192141]\n [0.01830573 0.02722583 0.00221307]\n [0.00192141 0.00221307 0.01 ]]\n[[ 0.17281467 -0.15073511 0.01038919]\n [-0.15073511 0.31426837 0.00911657]\n [ 0.01038919 0.00911657 0.01 ]]\n[[ 0.15636935 -0.12991587 0.00944914]\n [-0.12991587 0.25858449 0.00882932]\n [ 0.00944914 0.00882932 0.01 ]]\n[[ 0.1258498 -0.08629463 0.00696348]\n [-0.08629463 0.13926998 0.00832047]\n [ 0.00696348 0.00832047 0.01 ]]\n[[ 0.12112278 -0.07258586 0.00586698]\n [-0.07258586 0.09986793 0.00833094]\n [ 0.00586698 0.00833094 0.01 ]]\n[[ 0.0774922 -0.03225434 0.00363624]\n [-0.03225434 0.03826724 0.00683774]\n [ 0.00363624 0.00683774 0.01 ]]\n[[ 0.05739493 -0.01811959 0.00273105]\n [-0.01811959 0.02172257 0.00570292]\n [ 0.00273105 0.00570292 0.01 ]]\n[[ 0.04718701 -0.01119994 0.00229134]\n [-0.01119994 0.01545345 0.00467933]\n [ 0.00229134 0.00467933 0.01 ]]\n[[ 0.1843289 0.13345626 0.00927896]\n [ 0.13345626 0.25303293 -0.00226416]\n [ 0.00927896 -0.00226416 0.01 ]]\n[[ 1.05052534e-01 6.27206683e-02 7.54839934e-03]\n [ 6.27206683e-02 1.60348152e-01 -3.00264721e-06]\n [ 7.54839934e-03 -3.00264721e-06 1.00000000e-02]]\n[[0.09029892 0.04876334 0.00729707]\n [0.04876334 0.14801075 0.00062726]\n [0.00729707 0.00062726 0.01 ]]\n[[0.0675105 0.02211159 0.00554242]\n [0.02211159 0.0829682 0.00152133]\n [0.00554242 0.00152133 0.01 ]]\n[[0.0498488 0.00126701 0.00368677]\n [0.00126701 0.03567068 0.00270983]\n [0.00368677 0.00270983 0.01 ]]\n[[ 0.04807851 -0.00107631 0.00281562]\n [-0.00107631 0.0216265 0.00270639]\n [ 0.00281562 0.00270639 0.01 ]]\n[[ 0.27732237 -0.00758927 0.0006507 ]\n [-0.00758927 0.03680322 -0.0049399 ]\n [ 0.0006507 -0.0049399 0.01 ]]\n[[ 0.36742367 0.02591802 0.00186052]\n [ 0.02591802 0.05259014 -0.0064586 ]\n [ 0.00186052 -0.0064586 0.01 ]]\n[[ 0.19495747 0.02787923 0.00251493]\n [ 0.02787923 0.03667839 -0.00322525]\n [ 0.00251493 -0.00322525 0.01 ]]\n[[ 0.13081781 0.0225327 0.00273404]\n [ 0.0225327 0.0311257 -0.00153474]\n [ 0.00273404 -0.00153474 0.01 ]]\n[[ 0.10056175 0.02164963 0.00317271]\n [ 0.02164963 0.03341869 -0.00048368]\n [ 0.00317271 -0.00048368 0.01 ]]\n[[0.07137355 0.00949275 0.00273898]\n [0.00949275 0.02360616 0.00079292]\n [0.00273898 0.00079292 0.01 ]]\n[[ 0.2428844 -0.26687205 0.01847772]\n [-0.26687205 0.55898973 0.01054307]\n [ 0.01847772 0.01054307 0.01 ]]\n[[ 0.21236485 -0.22325081 0.01599207]\n [-0.22325081 0.43967522 0.01003421]\n [ 0.01599207 0.01003421 0.01 ]]\n[[ 0.20763783 -0.20954203 0.01489556]\n [-0.20954203 0.40027317 0.01004468]\n [ 0.01489556 0.01004468 0.01 ]]\n[[ 0.16400725 -0.16921051 0.01266483]\n [-0.16921051 0.33867247 0.00855148]\n [ 0.01266483 0.00855148 0.01 ]]\n[[ 0.14390999 -0.15507576 0.01175963]\n [-0.15507576 0.32212781 0.00741666]\n [ 0.01175963 0.00741666 0.01 ]]\n[[ 0.13370206 -0.14815611 0.01131993]\n [-0.14815611 0.31585868 0.00639307]\n [ 0.01131993 0.00639307 0.01 ]]\n[[ 2.70843953e-01 -3.49990947e-03 1.83075450e-02]\n [-3.49990947e-03 5.53438167e-01 -5.50419780e-04]\n [ 1.83075450e-02 -5.50419780e-04 1.00000000e-02]]\n[[ 0.19156759 -0.07423551 0.01657699]\n [-0.07423551 0.46075339 0.00171074]\n [ 0.01657699 0.00171074 0.01 ]]\n[[ 0.17681397 -0.08819284 0.01632565]\n [-0.08819284 0.44841599 0.00234101]\n [ 0.01632565 0.00234101 0.01 ]]\n[[ 0.15402555 -0.11484459 0.014571 ]\n [-0.11484459 0.38337344 0.00323508]\n [ 0.014571 0.00323508 0.01 ]]\n[[ 0.13636385 -0.13568916 0.01271536]\n [-0.13568916 0.33607592 0.00442357]\n [ 0.01271536 0.00442357 0.01 ]]\n[[ 0.13459356 -0.13803248 0.01184421]\n [-0.13803248 0.32203174 0.00442014]\n [ 0.01184421 0.00442014 0.01 ]]\n[[ 0.36383742 -0.14454544 0.00967929]\n [-0.14454544 0.33720845 -0.00322615]\n [ 0.00967929 -0.00322615 0.01 ]]\n[[ 0.45393872 -0.11103815 0.01088911]\n [-0.11103815 0.35299538 -0.00474486]\n [ 0.01088911 -0.00474486 0.01 ]]\n[[ 0.28147252 -0.10907694 0.01154352]\n [-0.10907694 0.33708363 -0.00151151]\n [ 0.01154352 -0.00151151 0.01 ]]\n[[ 2.17332862e-01 -1.14423469e-01 1.17626297e-02]\n [-1.14423469e-01 3.31530939e-01 1.78998377e-04]\n [ 1.17626297e-02 1.78998377e-04 1.00000000e-02]]\n[[ 0.1870768 -0.11530654 0.01220129]\n [-0.11530654 0.33382393 0.00123006]\n [ 0.01220129 0.00123006 0.01 ]]\n[[ 0.1578886 -0.12746342 0.01176757]\n [-0.12746342 0.3240114 0.00250666]\n [ 0.01176757 0.00250666 0.01 ]]\n[[ 0.19591953 -0.20243157 0.01505202]\n [-0.20243157 0.38399135 0.00974696]\n [ 0.01505202 0.00974696 0.01 ]]\n[[ 0.19119251 -0.18872279 0.01395551]\n [-0.18872279 0.34458929 0.00975744]\n [ 0.01395551 0.00975744 0.01 ]]\n[[ 0.14756193 -0.14839128 0.01172477]\n [-0.14839128 0.2829886 0.00826423]\n [ 0.01172477 0.00826423 0.01 ]]\n[[ 0.12746466 -0.13425652 0.01081958]\n [-0.13425652 0.26644394 0.00712942]\n [ 0.01081958 0.00712942 0.01 ]]\n[[ 0.11725674 -0.12733688 0.01037988]\n [-0.12733688 0.26017481 0.00610583]\n [ 0.01037988 0.00610583 0.01 ]]\n[[ 0.25439863 0.01731933 0.01736749]\n [ 0.01731933 0.4977543 -0.00083767]\n [ 0.01736749 -0.00083767 0.01 ]]\n[[ 0.17512226 -0.05341627 0.01563693]\n [-0.05341627 0.40506952 0.00142349]\n [ 0.01563693 0.00142349 0.01 ]]\n[[ 0.16036865 -0.0673736 0.0153856 ]\n [-0.0673736 0.39273211 0.00205376]\n [ 0.0153856 0.00205376 0.01 ]]\n[[ 0.13758023 -0.09402535 0.01363095]\n [-0.09402535 0.32768957 0.00294783]\n [ 0.01363095 0.00294783 0.01 ]]\n[[ 0.11991853 -0.11486992 0.0117753 ]\n [-0.11486992 0.28039205 0.00413632]\n [ 0.0117753 0.00413632 0.01 ]]\n[[ 0.11814824 -0.11721324 0.01090416]\n [-0.11721324 0.26634786 0.00413289]\n [ 0.01090416 0.00413289 0.01 ]]\n[[ 0.3473921 -0.12372621 0.00873924]\n [-0.12372621 0.28152458 -0.0035134 ]\n [ 0.00873924 -0.0035134 0.01 ]]\n[[ 0.4374934 -0.09021891 0.00994905]\n [-0.09021891 0.29731151 -0.00503211]\n [ 0.00994905 -0.00503211 0.01 ]]\n[[ 0.2650272 -0.0882577 0.01060346]\n [-0.0882577 0.28139976 -0.00179876]\n [ 0.01060346 -0.00179876 0.01 ]]\n[[ 2.00887540e-01 -9.36042324e-02 1.08225760e-02]\n [-9.36042324e-02 2.75847067e-01 -1.08249727e-04]\n [ 1.08225760e-02 -1.08249727e-04 1.00000000e-02]]\n[[ 0.17063148 -0.0944873 0.01126124]\n [-0.0944873 0.27814005 0.00094281]\n [ 0.01126124 0.00094281 0.01 ]]\n[[ 0.14144328 -0.10664419 0.01082751]\n [-0.10664419 0.26832752 0.00221942]\n [ 0.01082751 0.00221942 0.01 ]]\n[[ 0.16067296 -0.14510155 0.01146985]\n [-0.14510155 0.22527478 0.00924858]\n [ 0.01146985 0.00924858 0.01 ]]\n[[ 0.11704238 -0.10477004 0.00923912]\n [-0.10477004 0.16367409 0.00775538]\n [ 0.00923912 0.00775538 0.01 ]]\n[[ 0.09694511 -0.09063529 0.00833392]\n [-0.09063529 0.14712943 0.00662056]\n [ 0.00833392 0.00662056 0.01 ]]\n[[ 0.08673719 -0.08371564 0.00789422]\n [-0.08371564 0.1408603 0.00559697]\n [ 0.00789422 0.00559697 0.01 ]]\n[[ 0.22387908 0.06094057 0.01488184]\n [ 0.06094057 0.37843979 -0.00134652]\n [ 0.01488184 -0.00134652 0.01 ]]\n[[ 0.14460271 -0.00979503 0.01315128]\n [-0.00979503 0.28575501 0.00091464]\n [ 0.01315128 0.00091464 0.01 ]]\n[[ 0.1298491 -0.02375236 0.01289995]\n [-0.02375236 0.2734176 0.0015449 ]\n [ 0.01289995 0.0015449 0.01 ]]\n[[ 0.10706068 -0.05040411 0.0111453 ]\n [-0.05040411 0.20837506 0.00243897]\n [ 0.0111453 0.00243897 0.01 ]]\n[[ 0.08939898 -0.07124868 0.00928965]\n [-0.07124868 0.16107754 0.00362747]\n [ 0.00928965 0.00362747 0.01 ]]\n[[ 0.08762869 -0.07359201 0.0084185 ]\n [-0.07359201 0.14703335 0.00362403]\n [ 0.0084185 0.00362403 0.01 ]]\n[[ 0.31687255 -0.08010497 0.00625358]\n [-0.08010497 0.16221007 -0.00402226]\n [ 0.00625358 -0.00402226 0.01 ]]\n[[ 0.40697385 -0.04659767 0.0074634 ]\n [-0.04659767 0.177997 -0.00554096]\n [ 0.0074634 -0.00554096 0.01 ]]\n[[ 0.23450765 -0.04463647 0.00811781]\n [-0.04463647 0.16208525 -0.00230761]\n [ 0.00811781 -0.00230761 0.01 ]]\n[[ 0.17036799 -0.04998299 0.00833692]\n [-0.04998299 0.15653256 -0.0006171 ]\n [ 0.00833692 -0.0006171 0.01 ]]\n[[ 0.14011193 -0.05086606 0.00877559]\n [-0.05086606 0.15882554 0.00043396]\n [ 0.00877559 0.00043396 0.01 ]]\n[[ 0.11092373 -0.06302295 0.00834186]\n [-0.06302295 0.14901301 0.00171056]\n [ 0.00834186 0.00171056 0.01 ]]\n[[ 0.11231536 -0.09106126 0.00814261]\n [-0.09106126 0.12427204 0.00776585]\n [ 0.00814261 0.00776585 0.01 ]]\n[[ 0.0922181 -0.07692651 0.00723742]\n [-0.07692651 0.10772737 0.00663103]\n [ 0.00723742 0.00663103 0.01 ]]\n[[ 0.08201018 -0.07000686 0.00679772]\n [-0.07000686 0.10145825 0.00560744]\n [ 0.00679772 0.00560744 0.01 ]]\n[[ 0.21915207 0.07464935 0.01378533]\n [ 0.07464935 0.33903773 -0.00133605]\n [ 0.01378533 -0.00133605 0.01 ]]\n[[0.1398757 0.00391375 0.01205477]\n [0.00391375 0.24635295 0.00092511]\n [0.01205477 0.00092511 0.01 ]]\n[[ 0.12512208 -0.01004358 0.01180344]\n [-0.01004358 0.23401555 0.00155537]\n [ 0.01180344 0.00155537 0.01 ]]\n[[ 0.10233366 -0.03669533 0.01004879]\n [-0.03669533 0.168973 0.00244945]\n [ 0.01004879 0.00244945 0.01 ]]\n[[ 0.08467196 -0.0575399 0.00819314]\n [-0.0575399 0.12167548 0.00363794]\n [ 0.00819314 0.00363794 0.01 ]]\n[[ 0.08290167 -0.05988323 0.007322 ]\n [-0.05988323 0.1076313 0.0036345 ]\n [ 0.007322 0.0036345 0.01 ]]\n[[ 0.31214553 -0.06639619 0.00515707]\n [-0.06639619 0.12280802 -0.00401179]\n [ 0.00515707 -0.00401179 0.01 ]]\n[[ 0.40224684 -0.03288889 0.00636689]\n [-0.03288889 0.13859494 -0.00553049]\n [ 0.00636689 -0.00553049 0.01 ]]\n[[ 0.22978063 -0.03092769 0.0070213 ]\n [-0.03092769 0.12268319 -0.00229714]\n [ 0.0070213 -0.00229714 0.01 ]]\n[[ 0.16564097 -0.03627421 0.00724042]\n [-0.03627421 0.1171305 -0.00060663]\n [ 0.00724042 -0.00060663 0.01 ]]\n[[ 0.13538491 -0.03715729 0.00767908]\n [-0.03715729 0.11942349 0.00044443]\n [ 0.00767908 0.00044443 0.01 ]]\n[[ 0.10619671 -0.04931417 0.00724535]\n [-0.04931417 0.10961096 0.00172103]\n [ 0.00724535 0.00172103 0.01 ]]\n[[ 0.04858751 -0.03659499 0.00500668]\n [-0.03659499 0.04612668 0.00513783]\n [ 0.00500668 0.00513783 0.01 ]]\n[[ 0.03837959 -0.02967534 0.00456698]\n [-0.02967534 0.03985755 0.00411424]\n [ 0.00456698 0.00411424 0.01 ]]\n[[ 0.17552148 0.11498086 0.0115546 ]\n [ 0.11498086 0.27743704 -0.00282925]\n [ 0.0115546 -0.00282925 0.01 ]]\n[[ 0.09624511 0.04424526 0.00982404]\n [ 0.04424526 0.18475226 -0.00056809]\n [ 0.00982404 -0.00056809 0.01 ]]\n[[8.14914947e-02 3.02879323e-02 9.57270499e-03]\n [3.02879323e-02 1.72414855e-01 6.21725582e-05]\n [9.57270499e-03 6.21725582e-05 1.00000000e-02]]\n[[0.05870308 0.00363618 0.00781805]\n [0.00363618 0.10737231 0.00095624]\n [0.00781805 0.00095624 0.01 ]]\n[[ 0.04104137 -0.01720839 0.00596241]\n [-0.01720839 0.06007479 0.00214474]\n [ 0.00596241 0.00214474 0.01 ]]\n[[ 0.03927109 -0.01955171 0.00509126]\n [-0.01955171 0.04603061 0.0021413 ]\n [ 0.00509126 0.0021413 0.01 ]]\n[[ 0.26851495 -0.02606467 0.00292634]\n [-0.02606467 0.06120732 -0.00550499]\n [ 0.00292634 -0.00550499 0.01 ]]\n[[ 0.35861625 0.00744262 0.00413616]\n [ 0.00744262 0.07699425 -0.00702369]\n [ 0.00413616 -0.00702369 0.01 ]]\n[[ 0.18615004 0.00940383 0.00479057]\n [ 0.00940383 0.0610825 -0.00379034]\n [ 0.00479057 -0.00379034 0.01 ]]\n[[ 0.12201039 0.0040573 0.00500968]\n [ 0.0040573 0.05552981 -0.00209984]\n [ 0.00500968 -0.00209984 0.01 ]]\n[[ 0.09175433 0.00317423 0.00544835]\n [ 0.00317423 0.05782279 -0.00104877]\n [ 0.00544835 -0.00104877 0.01 ]]\n[[ 0.06256612 -0.00898265 0.00501462]\n [-0.00898265 0.04801027 0.00022783]\n [ 0.00501462 0.00022783 0.01 ]]\n[[ 0.01828233 -0.01554059 0.00366179]\n [-0.01554059 0.02331289 0.00297942]\n [ 0.00366179 0.00297942 0.01 ]]\n[[ 0.15542422 0.12911561 0.0106494 ]\n [ 0.12911561 0.26089237 -0.00396407]\n [ 0.0106494 -0.00396407 0.01 ]]\n[[ 0.07614785 0.05838002 0.00891884]\n [ 0.05838002 0.1682076 -0.00170291]\n [ 0.00891884 -0.00170291 0.01 ]]\n[[ 0.06139423 0.04442268 0.00866751]\n [ 0.04442268 0.15587019 -0.00107264]\n [ 0.00866751 -0.00107264 0.01 ]]\n[[ 0.03860581 0.01777094 0.00691286]\n [ 0.01777094 0.09082764 -0.00017857]\n [ 0.00691286 -0.00017857 0.01 ]]\n[[ 0.02094411 -0.00307364 0.00505721]\n [-0.00307364 0.04353013 0.00100992]\n [ 0.00505721 0.00100992 0.01 ]]\n[[ 0.01917382 -0.00541696 0.00418607]\n [-0.00541696 0.02948594 0.00100648]\n [ 0.00418607 0.00100648 0.01 ]]\n[[ 0.24841768 -0.01192992 0.00202114]\n [-0.01192992 0.04466266 -0.0066398 ]\n [ 0.00202114 -0.0066398 0.01 ]]\n[[ 0.33851899 0.02157737 0.00323096]\n [ 0.02157737 0.06044959 -0.00815851]\n [ 0.00323096 -0.00815851 0.01 ]]\n[[ 0.16605278 0.02353858 0.00388537]\n [ 0.02353858 0.04453784 -0.00492516]\n [ 0.00388537 -0.00492516 0.01 ]]\n[[ 0.10191313 0.01819205 0.00410448]\n [ 0.01819205 0.03898514 -0.00323465]\n [ 0.00410448 -0.00323465 0.01 ]]\n[[ 0.07165707 0.01730898 0.00454315]\n [ 0.01730898 0.04127813 -0.00218359]\n [ 0.00454315 -0.00218359 0.01 ]]\n[[ 0.04246886 0.0051521 0.00410942]\n [ 0.0051521 0.0314656 -0.00090699]\n [ 0.00410942 -0.00090699 0.01 ]]\n[[ 0.14521629 0.13603526 0.0102097 ]\n [ 0.13603526 0.25462325 -0.00498766]\n [ 0.0102097 -0.00498766 0.01 ]]\n[[ 0.06593993 0.06529967 0.00847914]\n [ 0.06529967 0.16193847 -0.0027265 ]\n [ 0.00847914 -0.0027265 0.01 ]]\n[[ 0.05118631 0.05134233 0.00822781]\n [ 0.05134233 0.14960107 -0.00209624]\n [ 0.00822781 -0.00209624 0.01 ]]\n[[ 0.02839789 0.02469059 0.00647316]\n [ 0.02469059 0.08455852 -0.00120216]\n [ 0.00647316 -0.00120216 0.01 ]]\n[[ 1.07361893e-02 3.84601262e-03 4.61750964e-03]\n [ 3.84601262e-03 3.72609996e-02 -1.36697938e-05]\n [ 4.61750964e-03 -1.36697938e-05 1.00000000e-02]]\n[[ 8.96590052e-03 1.50269068e-03 3.74636437e-03]\n [ 1.50269068e-03 2.32168164e-02 -1.71063042e-05]\n [ 3.74636437e-03 -1.71063042e-05 1.00000000e-02]]\n[[ 0.23820976 -0.00501027 0.00158144]\n [-0.00501027 0.03839353 -0.0076634 ]\n [ 0.00158144 -0.0076634 0.01 ]]\n[[ 0.32831106 0.02849702 0.00279126]\n [ 0.02849702 0.05418046 -0.0091821 ]\n [ 0.00279126 -0.0091821 0.01 ]]\n[[ 0.15584486 0.03045823 0.00344567]\n [ 0.03045823 0.03826871 -0.00594875]\n [ 0.00344567 -0.00594875 0.01 ]]\n[[ 0.0917052 0.0251117 0.00366478]\n [ 0.0251117 0.03271602 -0.00425824]\n [ 0.00366478 -0.00425824 0.01 ]]\n[[ 0.06144914 0.02422863 0.00410345]\n [ 0.02422863 0.03500901 -0.00320718]\n [ 0.00410345 -0.00320718 0.01 ]]\n[[ 0.03226094 0.01207175 0.00366972]\n [ 0.01207175 0.02519648 -0.00193058]\n [ 0.00366972 -0.00193058 0.01 ]]\n[[ 0.20308182 0.20995587 0.01546675]\n [ 0.20995587 0.39951795 -0.00967 ]\n [ 0.01546675 -0.00967 0.01 ]]\n[[ 0.1883282 0.19599854 0.01521542]\n [ 0.19599854 0.38718055 -0.00903973]\n [ 0.01521542 -0.00903973 0.01 ]]\n[[ 0.16553978 0.16934679 0.01346077]\n [ 0.16934679 0.322138 -0.00814566]\n [ 0.01346077 -0.00814566 0.01 ]]\n[[ 0.14787808 0.14850222 0.01160512]\n [ 0.14850222 0.27484048 -0.00695716]\n [ 0.01160512 -0.00695716 0.01 ]]\n[[ 0.14610779 0.14615889 0.01073398]\n [ 0.14615889 0.2607963 -0.0069606 ]\n [ 0.01073398 -0.0069606 0.01 ]]\n[[ 0.37535165 0.13964593 0.00856906]\n [ 0.13964593 0.27597302 -0.01460689]\n [ 0.00856906 -0.01460689 0.01 ]]\n[[ 0.46545295 0.17315322 0.00977888]\n [ 0.17315322 0.29175994 -0.0161256 ]\n [ 0.00977888 -0.0161256 0.01 ]]\n[[ 0.29298675 0.17511443 0.01043328]\n [ 0.17511443 0.27584819 -0.01289225]\n [ 0.01043328 -0.01289225 0.01 ]]\n[[ 0.22884709 0.16976791 0.0106524 ]\n [ 0.16976791 0.2702955 -0.01120174]\n [ 0.0106524 -0.01120174 0.01 ]]\n[[ 0.19859103 0.16888483 0.01109106]\n [ 0.16888483 0.27258849 -0.01015067]\n [ 0.01109106 -0.01015067 0.01 ]]\n[[ 0.16940283 0.15672795 0.01065734]\n [ 0.15672795 0.26277596 -0.00887407]\n [ 0.01065734 -0.00887407 0.01 ]]\n[[ 0.10905183 0.12526294 0.01348486]\n [ 0.12526294 0.29449577 -0.00677857]\n [ 0.01348486 -0.00677857 0.01 ]]\n[[ 0.08626341 0.09861119 0.01173021]\n [ 0.09861119 0.22945322 -0.0058845 ]\n [ 0.01173021 -0.0058845 0.01 ]]\n[[ 0.06860171 0.07776662 0.00987456]\n [ 0.07776662 0.1821557 -0.004696 ]\n [ 0.00987456 -0.004696 0.01 ]]\n[[ 0.06683142 0.0754233 0.00900342]\n [ 0.0754233 0.16811152 -0.00469944]\n [ 0.00900342 -0.00469944 0.01 ]]\n[[ 0.29607528 0.06891034 0.0068385 ]\n [ 0.06891034 0.18328824 -0.01234573]\n [ 0.0068385 -0.01234573 0.01 ]]\n[[ 0.38617659 0.10241763 0.00804832]\n [ 0.10241763 0.19907517 -0.01386444]\n [ 0.00804832 -0.01386444 0.01 ]]\n[[ 0.21371038 0.10437884 0.00870273]\n [ 0.10437884 0.18316342 -0.01063109]\n [ 0.00870273 -0.01063109 0.01 ]]\n[[ 0.14957073 0.09903231 0.00892184]\n [ 0.09903231 0.17761072 -0.00894058]\n [ 0.00892184 -0.00894058 0.01 ]]\n[[ 0.11931467 0.09814924 0.0093605 ]\n [ 0.09814924 0.17990371 -0.00788951]\n [ 0.0093605 -0.00788951 0.01 ]]\n[[ 0.09012646 0.08599236 0.00892678]\n [ 0.08599236 0.17009118 -0.00661291]\n [ 0.00892678 -0.00661291 0.01 ]]\n[[ 0.0715098 0.08465386 0.01147888]\n [ 0.08465386 0.21711582 -0.00525423]\n [ 0.01147888 -0.00525423 0.01 ]]\n[[ 0.0538481 0.06380929 0.00962323]\n [ 0.06380929 0.1698183 -0.00406574]\n [ 0.00962323 -0.00406574 0.01 ]]\n[[ 0.05207781 0.06146597 0.00875209]\n [ 0.06146597 0.15577412 -0.00406917]\n [ 0.00875209 -0.00406917 0.01 ]]\n[[ 0.28132167 0.054953 0.00658717]\n [ 0.054953 0.17095084 -0.01171546]\n [ 0.00658717 -0.01171546 0.01 ]]\n[[ 0.37142297 0.0884603 0.00779698]\n [ 0.0884603 0.18673776 -0.01323417]\n [ 0.00779698 -0.01323417 0.01 ]]\n[[ 0.19895676 0.0904215 0.00845139]\n [ 0.0904215 0.17082601 -0.01000082]\n [ 0.00845139 -0.01000082 0.01 ]]\n[[ 0.13481711 0.08507498 0.00867051]\n [ 0.08507498 0.16527332 -0.00831031]\n [ 0.00867051 -0.00831031 0.01 ]]\n[[ 0.10456105 0.08419191 0.00910917]\n [ 0.08419191 0.16756631 -0.00725925]\n [ 0.00910917 -0.00725925 0.01 ]]\n[[ 0.07537285 0.07203502 0.00867544]\n [ 0.07203502 0.15775378 -0.00598265]\n [ 0.00867544 -0.00598265 0.01 ]]\n[[ 0.03105968 0.03715754 0.00786858]\n [ 0.03715754 0.10477575 -0.00317167]\n [ 0.00786858 -0.00317167 0.01 ]]\n[[ 0.02928939 0.03481422 0.00699744]\n [ 0.03481422 0.09073157 -0.0031751 ]\n [ 0.00699744 -0.0031751 0.01 ]]\n[[ 0.25853325 0.02830126 0.00483252]\n [ 0.02830126 0.10590829 -0.01082139]\n [ 0.00483252 -0.01082139 0.01 ]]\n[[ 0.34863455 0.06180855 0.00604233]\n [ 0.06180855 0.12169521 -0.0123401 ]\n [ 0.00604233 -0.0123401 0.01 ]]\n[[ 0.17616835 0.06376976 0.00669674]\n [ 0.06376976 0.10578346 -0.00910675]\n [ 0.00669674 -0.00910675 0.01 ]]\n[[ 0.11202869 0.05842323 0.00691586]\n [ 0.05842323 0.10023077 -0.00741624]\n [ 0.00691586 -0.00741624 0.01 ]]\n[[ 0.08177263 0.05754016 0.00735452]\n [ 0.05754016 0.10252376 -0.00636518]\n [ 0.00735452 -0.00636518 0.01 ]]\n[[ 0.05258443 0.04538327 0.00692079]\n [ 0.04538327 0.09271123 -0.00508857]\n [ 0.00692079 -0.00508857 0.01 ]]\n[[ 0.01162769 0.01396964 0.00514179]\n [ 0.01396964 0.04343405 -0.00198661]\n [ 0.00514179 -0.00198661 0.01 ]]\n[[ 0.24087155 0.00745668 0.00297687]\n [ 0.00745668 0.05861077 -0.0096329 ]\n [ 0.00297687 -0.0096329 0.01 ]]\n[[ 0.33097285 0.04096398 0.00418669]\n [ 0.04096398 0.0743977 -0.0111516 ]\n [ 0.00418669 -0.0111516 0.01 ]]\n[[ 0.15850664 0.04292518 0.0048411 ]\n [ 0.04292518 0.05848595 -0.00791825]\n [ 0.0048411 -0.00791825 0.01 ]]\n[[ 0.09436699 0.03757866 0.00506021]\n [ 0.03757866 0.05293325 -0.00622775]\n [ 0.00506021 -0.00622775 0.01 ]]\n[[ 0.06411093 0.03669558 0.00549887]\n [ 0.03669558 0.05522624 -0.00517668]\n [ 0.00549887 -0.00517668 0.01 ]]\n[[ 0.03492272 0.0245387 0.00506515]\n [ 0.0245387 0.04541371 -0.00390008]\n [ 0.00506515 -0.00390008 0.01 ]]\n[[ 0.23910126 0.00511336 0.00210572]\n [ 0.00511336 0.04456659 -0.00963633]\n [ 0.00210572 -0.00963633 0.01 ]]\n[[ 0.32920256 0.03862065 0.00331554]\n [ 0.03862065 0.06035351 -0.01115504]\n [ 0.00331554 -0.01115504 0.01 ]]\n[[ 0.15673635 0.04058186 0.00396995]\n [ 0.04058186 0.04444176 -0.00792169]\n [ 0.00396995 -0.00792169 0.01 ]]\n[[ 0.0925967 0.03523533 0.00418906]\n [ 0.03523533 0.03888907 -0.00623118]\n [ 0.00418906 -0.00623118 0.01 ]]\n[[ 0.06234064 0.03435226 0.00462773]\n [ 0.03435226 0.04118206 -0.00518012]\n [ 0.00462773 -0.00518012 0.01 ]]\n[[ 0.03315244 0.02219538 0.004194 ]\n [ 0.02219538 0.03136953 -0.00390352]\n [ 0.004194 -0.00390352 0.01 ]]\n[[ 0.55844642 0.03210769 0.00115062]\n [ 0.03210769 0.07553023 -0.01880133]\n [ 0.00115062 -0.01880133 0.01 ]]\n[[ 0.38598022 0.0340689 0.00180503]\n [ 0.0340689 0.05961848 -0.01556798]\n [ 0.00180503 -0.01556798 0.01 ]]\n[[ 0.32184056 0.02872237 0.00202414]\n [ 0.02872237 0.05406579 -0.01387747]\n [ 0.00202414 -0.01387747 0.01 ]]\n[[ 0.2915845 0.0278393 0.00246281]\n [ 0.0278393 0.05635878 -0.01282641]\n [ 0.00246281 -0.01282641 0.01 ]]\n[[ 0.2623963 0.01568242 0.00202908]\n [ 0.01568242 0.04654625 -0.0115498 ]\n [ 0.00202908 -0.0115498 0.01 ]]\n[[ 0.47608152 0.06757619 0.00301485]\n [ 0.06757619 0.07540541 -0.01708669]\n [ 0.00301485 -0.01708669 0.01 ]]\n[[ 0.41194186 0.06222967 0.00323396]\n [ 0.06222967 0.06985272 -0.01539618]\n [ 0.00323396 -0.01539618 0.01 ]]\n[[ 0.3816858 0.06134659 0.00367263]\n [ 0.06134659 0.0721457 -0.01434511]\n [ 0.00367263 -0.01434511 0.01 ]]\n[[ 0.3524976 0.04918971 0.0032389 ]\n [ 0.04918971 0.06233317 -0.01306851]\n [ 0.0032389 -0.01306851 0.01 ]]\n[[ 0.23947566 0.06419087 0.00388837]\n [ 0.06419087 0.05394097 -0.01216283]\n [ 0.00388837 -0.01216283 0.01 ]]\n[[ 0.2092196 0.0633078 0.00432703]\n [ 0.0633078 0.05623395 -0.01111176]\n [ 0.00432703 -0.01111176 0.01 ]]\n[[ 0.18003139 0.05115092 0.00389331]\n [ 0.05115092 0.04642142 -0.00983516]\n [ 0.00389331 -0.00983516 0.01 ]]\n[[ 0.14507994 0.05796127 0.00454615]\n [ 0.05796127 0.05068126 -0.00942125]\n [ 0.00454615 -0.00942125 0.01 ]]\n[[ 0.11589174 0.04580439 0.00411242]\n [ 0.04580439 0.04086873 -0.00814465]\n [ 0.00411242 -0.00814465 0.01 ]]\n[[ 0.08563568 0.04492132 0.00455108]\n [ 0.04492132 0.04316172 -0.00709359]\n [ 0.00455108 -0.00709359 0.01 ]]\n[[ 0.05805537 0.02773175 -0.01627905]\n [ 0.02773175 0.42189053 0.00109834]\n [-0.01627905 0.00109834 0.01 ]]\n[[ 0.05468359 0.02302165 -0.01559736]\n [ 0.02302165 0.39270667 0.00082573]\n [-0.01559736 0.00082573 0.01 ]]\n[[ 0.0466368 0.02488281 -0.01380778]\n [ 0.02488281 0.32983958 0.00083917]\n [-0.01380778 0.00083917 0.01 ]]\n[[ 0.04215856 0.02606358 -0.01240273]\n [ 0.02606358 0.2945626 0.00082888]\n [-0.01240273 0.00082888 0.01 ]]\n[[ 0.03871478 0.03042721 -0.01069028]\n [ 0.03042721 0.26829388 0.00146464]\n [-0.01069028 0.00146464 0.01 ]]\n[[ 0.32829986 0.16188244 -0.01423302]\n [ 0.16188244 0.37867065 0.01062526]\n [-0.01423302 0.01062526 0.01 ]]\n[[ 0.24624085 0.14998306 -0.01468337]\n [ 0.14998306 0.38308745 0.00908317]\n [-0.01468337 0.00908317 0.01 ]]\n[[ 0.19895684 0.14358747 -0.01522051]\n [ 0.14358747 0.39644416 0.0080065 ]\n [-0.01522051 0.0080065 0.01 ]]\n[[ 0.0893779 0.07090833 -0.01310779]\n [ 0.07090833 0.31647845 0.0049806 ]\n [-0.01310779 0.0049806 0.01 ]]\n[[ 0.07936754 0.06243936 -0.01264898]\n [ 0.06243936 0.30451091 0.00460078]\n [-0.01264898 0.00460078 0.01 ]]\n[[ 0.05980853 0.04599794 -0.01154072]\n [ 0.04599794 0.28155024 0.00369558]\n [-0.01154072 0.00369558 0.01 ]]\n[[ 0.04660046 0.03335115 -0.01001712]\n [ 0.03335115 0.26410044 0.00284862]\n [-0.01001712 0.00284862 0.01 ]]\n[[ 0.23697061 -0.11101096 -0.00236603]\n [-0.11101096 0.42576843 0.00877581]\n [-0.00236603 0.00877581 0.01 ]]\n[[ 0.21579816 -0.08473502 -0.00336707]\n [-0.08473502 0.38455605 0.00842871]\n [-0.00336707 0.00842871 0.01 ]]\n[[ 0.19014171 -0.05638352 -0.00449176]\n [-0.05638352 0.34539039 0.00795167]\n [-0.00449176 0.00795167 0.01 ]]\n[[ 0.09525463 0.00192916 -0.00673883]\n [ 0.00192916 0.28494849 0.00538066]\n [-0.00673883 0.00538066 0.01 ]]\n[[ 0.07487333 0.00830172 -0.00683338]\n [ 0.00830172 0.28099795 0.0045294 ]\n [-0.00683338 0.0045294 0.01 ]]\n[[ 0.04674788 0.02148447 -0.0073586 ]\n [ 0.02148447 0.27091029 0.00277746]\n [-0.0073586 0.00277746 0.01 ]]\n[[ 0.03787043 -0.00754453 -0.01367267]\n [-0.00754453 0.2939031 -0.00041695]\n [-0.01367267 -0.00041695 0.01 ]]\n[[ 0.02982364 -0.00568337 -0.01188309]\n [-0.00568337 0.23103601 -0.00040351]\n [-0.01188309 -0.00040351 0.01 ]]\n[[ 0.0253454 -0.00450259 -0.01047804]\n [-0.00450259 0.19575903 -0.00041381]\n [-0.01047804 -0.00041381 0.01 ]]\n[[ 2.19016169e-02 -1.38967041e-04 -8.76558634e-03]\n [-1.38967041e-04 1.69490313e-01 2.21956830e-04]\n [-8.76558634e-03 2.21956830e-04 1.00000000e-02]]\n[[ 0.3114867 0.13131627 -0.01230833]\n [ 0.13131627 0.27986708 0.00938258]\n [-0.01230833 0.00938258 0.01 ]]\n[[ 0.22942769 0.11941688 -0.01275868]\n [ 0.11941688 0.28428388 0.00784048]\n [-0.01275868 0.00784048 0.01 ]]\n[[ 0.18214368 0.11302129 -0.01329582]\n [ 0.11302129 0.29764059 0.00676381]\n [-0.01329582 0.00676381 0.01 ]]\n[[ 0.07256474 0.04034216 -0.0111831 ]\n [ 0.04034216 0.21767488 0.00373792]\n [-0.0111831 0.00373792 0.01 ]]\n[[ 0.06255438 0.03187318 -0.01072429]\n [ 0.03187318 0.20570734 0.0033581 ]\n [-0.01072429 0.0033581 0.01 ]]\n[[ 0.04299537 0.01543177 -0.00961602]\n [ 0.01543177 0.18274667 0.0024529 ]\n [-0.00961602 0.0024529 0.01 ]]\n[[ 0.0297873 0.00278498 -0.00809243]\n [ 0.00278498 0.16529687 0.00160594]\n [-0.00809243 0.00160594 0.01 ]]\n[[ 0.22015745 -0.14157713 -0.00044134]\n [-0.14157713 0.32696486 0.00753312]\n [-0.00044134 0.00753312 0.01 ]]\n[[ 0.198985 -0.1153012 -0.00144238]\n [-0.1153012 0.28575248 0.00718602]\n [-0.00144238 0.00718602 0.01 ]]\n[[ 0.17332855 -0.08694969 -0.00256707]\n [-0.08694969 0.24658682 0.00670899]\n [-0.00256707 0.00670899 0.01 ]]\n[[ 0.07844147 -0.02863701 -0.00481414]\n [-0.02863701 0.18614492 0.00413798]\n [-0.00481414 0.00413798 0.01 ]]\n[[ 0.05806017 -0.02226445 -0.00490869]\n [-0.02226445 0.18219438 0.00328671]\n [-0.00490869 0.00328671 0.01 ]]\n[[ 0.02993472 -0.0090817 -0.00543391]\n [-0.0090817 0.17210672 0.00153477]\n [-0.00543391 0.00153477 0.01 ]]\n[[ 0.02645186 -0.01039347 -0.0112014 ]\n [-0.01039347 0.20185215 -0.00067612]\n [-0.0112014 -0.00067612 0.01 ]]\n[[ 0.02197362 -0.00921269 -0.00979635]\n [-0.00921269 0.16657517 -0.00068642]\n [-0.00979635 -0.00068642 0.01 ]]\n[[ 1.85298390e-02 -4.84906836e-03 -8.08389233e-03]\n [-4.84906836e-03 1.40306453e-01 -5.06519965e-05]\n [-8.08389233e-03 -5.06519965e-05 1.00000000e-02]]\n[[ 0.30811492 0.12660617 -0.01162663]\n [ 0.12660617 0.25068322 0.00910997]\n [-0.01162663 0.00910997 0.01 ]]\n[[ 0.22605591 0.11470678 -0.01207699]\n [ 0.11470678 0.25510002 0.00756788]\n [-0.01207699 0.00756788 0.01 ]]\n[[ 0.1787719 0.10831119 -0.01261413]\n [ 0.10831119 0.26845673 0.00649121]\n [-0.01261413 0.00649121 0.01 ]]\n[[ 0.06919296 0.03563206 -0.01050141]\n [ 0.03563206 0.18849102 0.00346531]\n [-0.01050141 0.00346531 0.01 ]]\n[[ 0.0591826 0.02716308 -0.0100426 ]\n [ 0.02716308 0.17652348 0.00308549]\n [-0.0100426 0.00308549 0.01 ]]\n[[ 0.03962359 0.01072167 -0.00893433]\n [ 0.01072167 0.15356281 0.00218029]\n [-0.00893433 0.00218029 0.01 ]]\n[[ 0.02641552 -0.00192513 -0.00741074]\n [-0.00192513 0.13611301 0.00133333]\n [-0.00741074 0.00133333 0.01 ]]\n[[ 2.16785676e-01 -1.46287233e-01 2.40354108e-04]\n [-1.46287233e-01 2.97781000e-01 7.26051234e-03]\n [ 2.40354108e-04 7.26051234e-03 1.00000000e-02]]\n[[ 0.19561322 -0.1200113 -0.00076069]\n [-0.1200113 0.25656862 0.00691341]\n [-0.00076069 0.00691341 0.01 ]]\n[[ 0.16995677 -0.0916598 -0.00188538]\n [-0.0916598 0.21740296 0.00643638]\n [-0.00188538 0.00643638 0.01 ]]\n[[ 0.07506969 -0.03334711 -0.00413245]\n [-0.03334711 0.15696106 0.00386537]\n [-0.00413245 0.00386537 0.01 ]]\n[[ 0.05468839 -0.02697455 -0.00422699]\n [-0.02697455 0.15301052 0.0030141 ]\n [-0.00422699 0.0030141 0.01 ]]\n[[ 0.02656294 -0.0137918 -0.00475221]\n [-0.0137918 0.14292286 0.00126217]\n [-0.00475221 0.00126217 0.01 ]]\n[[ 0.01392683 -0.00735153 -0.00800677]\n [-0.00735153 0.10370808 -0.00067298]\n [-0.00800677 -0.00067298 0.01 ]]\n[[ 1.04830482e-02 -2.98791043e-03 -6.29431709e-03]\n [-2.98791043e-03 7.74393616e-02 -3.72140689e-05]\n [-6.29431709e-03 -3.72140689e-05 1.00000000e-02]]\n[[ 0.30006813 0.12846732 -0.00983706]\n [ 0.12846732 0.18781613 0.00912341]\n [-0.00983706 0.00912341 0.01 ]]\n[[ 0.21800912 0.11656794 -0.01028741]\n [ 0.11656794 0.19223293 0.00758131]\n [-0.01028741 0.00758131 0.01 ]]\n[[ 0.17072511 0.11017235 -0.01082455]\n [ 0.11017235 0.20558964 0.00650464]\n [-0.01082455 0.00650464 0.01 ]]\n[[ 0.06114617 0.03749322 -0.00871183]\n [ 0.03749322 0.12562393 0.00347875]\n [-0.00871183 0.00347875 0.01 ]]\n[[ 0.05113581 0.02902424 -0.00825302]\n [ 0.02902424 0.11365639 0.00309892]\n [-0.00825302 0.00309892 0.01 ]]\n[[ 0.0315768 0.01258283 -0.00714476]\n [ 0.01258283 0.09069572 0.00219373]\n [-0.00714476 0.00219373 0.01 ]]\n[[ 1.83687324e-02 -6.39676327e-05 -5.62116087e-03]\n [-6.39676327e-05 7.32459158e-02 1.34676598e-03]\n [-5.62116087e-03 1.34676598e-03 1.00000000e-02]]\n[[ 0.20873888 -0.14442607 0.00202993]\n [-0.14442607 0.23491391 0.00727395]\n [ 0.00202993 0.00727395 0.01 ]]\n[[ 0.18756643 -0.11815014 0.00102889]\n [-0.11815014 0.19370153 0.00692685]\n [ 0.00102889 0.00692685 0.01 ]]\n[[ 1.61909980e-01 -8.97986381e-02 -9.58042973e-05]\n [-8.97986381e-02 1.54535865e-01 6.44981714e-03]\n [-9.58042973e-05 6.44981714e-03 1.00000000e-02]]\n[[ 0.0670229 -0.03148595 -0.00234287]\n [-0.03148595 0.09409397 0.00387881]\n [-0.00234287 0.00387881 0.01 ]]\n[[ 0.0466416 -0.02511339 -0.00243742]\n [-0.02511339 0.09014343 0.00302754]\n [-0.00243742 0.00302754 0.01 ]]\n[[ 0.01851615 -0.01193064 -0.00296264]\n [-0.01193064 0.08005577 0.0012756 ]\n [-0.00296264 0.0012756 0.01 ]]\n[[ 6.00480703e-03 -1.80713426e-03 -4.88926639e-03]\n [-1.80713426e-03 4.21623800e-02 -4.75090893e-05]\n [-4.88926639e-03 -4.75090893e-05 1.00000000e-02]]\n[[ 0.29558989 0.1296481 -0.00843201]\n [ 0.1296481 0.15253915 0.00911311]\n [-0.00843201 0.00911311 0.01 ]]\n[[ 0.21353088 0.11774871 -0.00888236]\n [ 0.11774871 0.15695595 0.00757102]\n [-0.00888236 0.00757102 0.01 ]]\n[[ 0.16624687 0.11135313 -0.0094195 ]\n [ 0.11135313 0.17031265 0.00649435]\n [-0.0094195 0.00649435 0.01 ]]\n[[ 0.05666793 0.03867399 -0.00730678]\n [ 0.03867399 0.09034694 0.00346845]\n [-0.00730678 0.00346845 0.01 ]]\n[[ 0.04665757 0.03020502 -0.00684797]\n [ 0.03020502 0.07837941 0.00308863]\n [-0.00684797 0.00308863 0.01 ]]\n[[ 0.02709856 0.0137636 -0.0057397 ]\n [ 0.0137636 0.05541874 0.00218343]\n [-0.0057397 0.00218343 0.01 ]]\n[[ 0.01389049 0.00111681 -0.00421611]\n [ 0.00111681 0.03796893 0.00133647]\n [-0.00421611 0.00133647 0.01 ]]\n[[ 0.20426064 -0.1432453 0.00343498]\n [-0.1432453 0.19963693 0.00726366]\n [ 0.00343498 0.00726366 0.01 ]]\n[[ 0.18308819 -0.11696936 0.00243394]\n [-0.11696936 0.15842455 0.00691656]\n [ 0.00243394 0.00691656 0.01 ]]\n[[ 0.15743174 -0.08861786 0.00130925]\n [-0.08861786 0.11925888 0.00643952]\n [ 0.00130925 0.00643952 0.01 ]]\n[[ 0.06254466 -0.03030518 -0.00093782]\n [-0.03030518 0.05881699 0.00386851]\n [-0.00093782 0.00386851 0.01 ]]\n[[ 0.04216336 -0.02393262 -0.00103237]\n [-0.02393262 0.05486644 0.00301725]\n [-0.00103237 0.00301725 0.01 ]]\n[[ 0.01403791 -0.01074987 -0.00155759]\n [-0.01074987 0.04477879 0.00126531]\n [-0.00155759 0.00126531 0.01 ]]\n[[ 0.29214611 0.13401172 -0.00671955]\n [ 0.13401172 0.12627043 0.00974888]\n [-0.00671955 0.00974888 0.01 ]]\n[[ 0.2100871 0.12211234 -0.00716991]\n [ 0.12211234 0.13068723 0.00820678]\n [-0.00716991 0.00820678 0.01 ]]\n[[ 0.16280308 0.11571675 -0.00770705]\n [ 0.11571675 0.14404394 0.00713011]\n [-0.00770705 0.00713011 0.01 ]]\n[[ 0.05322415 0.04303762 -0.00559433]\n [ 0.04303762 0.06407823 0.00410422]\n [-0.00559433 0.00410422 0.01 ]]\n[[ 0.04321379 0.03456864 -0.00513552]\n [ 0.03456864 0.05211069 0.0037244 ]\n [-0.00513552 0.0037244 0.01 ]]\n[[ 0.02365478 0.01812723 -0.00402725]\n [ 0.01812723 0.02915003 0.0028192 ]\n [-0.00402725 0.0028192 0.01 ]]\n[[ 0.01044671 0.00548043 -0.00250365]\n [ 0.00548043 0.01170022 0.00197224]\n [-0.00250365 0.00197224 0.01 ]]\n[[ 0.20081686 -0.13888167 0.00514744]\n [-0.13888167 0.17336821 0.00789942]\n [ 0.00514744 0.00789942 0.01 ]]\n[[ 0.17964441 -0.11260574 0.00414639]\n [-0.11260574 0.13215584 0.00755232]\n [ 0.00414639 0.00755232 0.01 ]]\n[[ 0.15398796 -0.08425424 0.0030217 ]\n [-0.08425424 0.09299017 0.00707529]\n [ 0.0030217 0.00707529 0.01 ]]\n[[ 0.05910088 -0.02594155 0.00077463]\n [-0.02594155 0.03254827 0.00450428]\n [ 0.00077463 0.00450428 0.01 ]]\n[[ 0.03871958 -0.01956899 0.00068009]\n [-0.01956899 0.02859773 0.00365301]\n [ 0.00068009 0.00365301 0.01 ]]\n[[ 0.01059413 -0.00638624 0.00015487]\n [-0.00638624 0.01851007 0.00190107]\n [ 0.00015487 0.00190107 0.01 ]]\n[[ 0.49967218 0.25356757 -0.01071265]\n [ 0.25356757 0.241064 0.0173674 ]\n [-0.01071265 0.0173674 0.01 ]]\n[[ 0.45238816 0.24717198 -0.01124979]\n [ 0.24717198 0.25442071 0.01629073]\n [-0.01124979 0.01629073 0.01 ]]\n[[ 0.34280923 0.17449285 -0.00913707]\n [ 0.17449285 0.174455 0.01326484]\n [-0.00913707 0.01326484 0.01 ]]\n[[ 0.33279887 0.16602387 -0.00867826]\n [ 0.16602387 0.16248746 0.01288502]\n [-0.00867826 0.01288502 0.01 ]]\n[[ 0.31323986 0.14958246 -0.00756999]\n [ 0.14958246 0.13952679 0.01197982]\n [-0.00756999 0.01197982 0.01 ]]\n[[ 0.30003179 0.13693567 -0.0060464 ]\n [ 0.13693567 0.12207699 0.01113286]\n [-0.0060464 0.01113286 0.01 ]]\n[[ 0.49040194 -0.00742644 0.00160469]\n [-0.00742644 0.28374498 0.01706004]\n [ 0.00160469 0.01706004 0.01 ]]\n[[0.46922949 0.0188495 0.00060365]\n [0.0188495 0.2425326 0.01671294]\n [0.00060365 0.01671294 0.01 ]]\n[[ 0.44357304 0.047201 -0.00052104]\n [ 0.047201 0.20336694 0.01623591]\n [-0.00052104 0.01623591 0.01 ]]\n[[ 0.34868596 0.10551368 -0.00276811]\n [ 0.10551368 0.14292504 0.0136649 ]\n [-0.00276811 0.0136649 0.01 ]]\n[[ 0.32830466 0.11188624 -0.00286265]\n [ 0.11188624 0.1389745 0.01281363]\n [-0.00286265 0.01281363 0.01 ]]\n[[ 0.30017921 0.12506899 -0.00338787]\n [ 0.12506899 0.12888684 0.01106169]\n [-0.00338787 0.01106169 0.01 ]]\n[[ 0.37032916 0.2352726 -0.01170014]\n [ 0.2352726 0.25883751 0.01474864]\n [-0.01170014 0.01474864 0.01 ]]\n[[ 0.26075022 0.16259347 -0.00958742]\n [ 0.16259347 0.1788718 0.01172275]\n [-0.00958742 0.01172275 0.01 ]]\n[[ 0.25073986 0.15412449 -0.00912862]\n [ 0.15412449 0.16690426 0.01134292]\n [-0.00912862 0.01134292 0.01 ]]\n[[ 0.23118085 0.13768308 -0.00802035]\n [ 0.13768308 0.14394359 0.01043773]\n [-0.00802035 0.01043773 0.01 ]]\n[[ 0.21797278 0.12503628 -0.00649675]\n [ 0.12503628 0.12649379 0.00959076]\n [-0.00649675 0.00959076 0.01 ]]\n[[ 0.40834293 -0.01932583 0.00115434]\n [-0.01932583 0.28816178 0.01551795]\n [ 0.00115434 0.01551795 0.01 ]]\n[[3.87170478e-01 6.95011078e-03 1.53294562e-04]\n [6.95011078e-03 2.46949402e-01 1.51708476e-02]\n [1.53294562e-04 1.51708476e-02 1.00000000e-02]]\n[[ 0.36151403 0.03530161 -0.0009714 ]\n [ 0.03530161 0.20778374 0.01469381]\n [-0.0009714 0.01469381 0.01 ]]\n[[ 0.26662695 0.0936143 -0.00321846]\n [ 0.0936143 0.14734184 0.01212281]\n [-0.00321846 0.01212281 0.01 ]]\n[[ 0.24624565 0.09998686 -0.00331301]\n [ 0.09998686 0.1433913 0.01127154]\n [-0.00331301 0.01127154 0.01 ]]\n[[ 0.2181202 0.1131696 -0.00383823]\n [ 0.1131696 0.13330364 0.0095196 ]\n [-0.00383823 0.0095196 0.01 ]]\n[[ 0.21346621 0.15619788 -0.01012456]\n [ 0.15619788 0.1922285 0.01064608]\n [-0.01012456 0.01064608 0.01 ]]\n[[ 0.20345585 0.1477289 -0.00966575]\n [ 0.1477289 0.18026097 0.01026625]\n [-0.00966575 0.01026625 0.01 ]]\n[[ 0.18389684 0.13128749 -0.00855748]\n [ 0.13128749 0.1573003 0.00936106]\n [-0.00855748 0.00936106 0.01 ]]\n[[ 0.17068877 0.11864069 -0.00703389]\n [ 0.11864069 0.13985049 0.00851409]\n [-0.00703389 0.00851409 0.01 ]]\n[[ 0.36105892 -0.02572141 0.0006172 ]\n [-0.02572141 0.30151849 0.01444128]\n [ 0.0006172 0.01444128 0.01 ]]\n[[ 0.33988646 0.00055452 -0.00038384]\n [ 0.00055452 0.26030611 0.01409418]\n [-0.00038384 0.01409418 0.01 ]]\n[[ 0.31423002 0.02890602 -0.00150853]\n [ 0.02890602 0.22114044 0.01361714]\n [-0.00150853 0.01361714 0.01 ]]\n[[ 0.21934294 0.08721871 -0.0037556 ]\n [ 0.08721871 0.16069855 0.01104614]\n [-0.0037556 0.01104614 0.01 ]]\n[[ 0.19896163 0.09359127 -0.00385015]\n [ 0.09359127 0.156748 0.01019487]\n [-0.00385015 0.01019487 0.01 ]]\n[[ 0.17083619 0.10677402 -0.00437537]\n [ 0.10677402 0.14666035 0.00844293]\n [-0.00437537 0.00844293 0.01 ]]\n[[ 0.09387691 0.07504977 -0.00755303]\n [ 0.07504977 0.10029526 0.00724036]\n [-0.00755303 0.00724036 0.01 ]]\n[[ 0.0743179 0.05860835 -0.00644477]\n [ 0.05860835 0.07733459 0.00633516]\n [-0.00644477 0.00633516 0.01 ]]\n[[ 0.06110983 0.04596156 -0.00492117]\n [ 0.04596156 0.05988478 0.0054882 ]\n [-0.00492117 0.0054882 0.01 ]]\n[[ 0.25147998 -0.09840055 0.00272992]\n [-0.09840055 0.22155278 0.01141538]\n [ 0.00272992 0.01141538 0.01 ]]\n[[ 0.23030753 -0.07212461 0.00172888]\n [-0.07212461 0.1803404 0.01106828]\n [ 0.00172888 0.01106828 0.01 ]]\n[[ 0.20465108 -0.04377311 0.00060419]\n [-0.04377311 0.14117473 0.01059125]\n [ 0.00060419 0.01059125 0.01 ]]\n[[ 0.109764 0.01453957 -0.00164288]\n [ 0.01453957 0.08073284 0.00802024]\n [-0.00164288 0.00802024 0.01 ]]\n[[ 0.0893827 0.02091213 -0.00173743]\n [ 0.02091213 0.07678229 0.00716897]\n [-0.00173743 0.00716897 0.01 ]]\n[[ 0.06125725 0.03409488 -0.00226265]\n [ 0.03409488 0.06669464 0.00541704]\n [-0.00226265 0.00541704 0.01 ]]\n[[ 0.06430754 0.05013938 -0.00598596]\n [ 0.05013938 0.06536705 0.00595534]\n [-0.00598596 0.00595534 0.01 ]]\n[[ 0.05109947 0.03749258 -0.00446236]\n [ 0.03749258 0.04791725 0.00510838]\n [-0.00446236 0.00510838 0.01 ]]\n[[ 0.24146962 -0.10686952 0.00318873]\n [-0.10686952 0.20958524 0.01103556]\n [ 0.00318873 0.01103556 0.01 ]]\n[[ 0.22029717 -0.08059359 0.00218769]\n [-0.08059359 0.16837286 0.01068846]\n [ 0.00218769 0.01068846 0.01 ]]\n[[ 0.19464072 -0.05224209 0.001063 ]\n [-0.05224209 0.1292072 0.01021143]\n [ 0.001063 0.01021143 0.01 ]]\n[[ 0.09975364 0.0060706 -0.00118407]\n [ 0.0060706 0.0687653 0.00764042]\n [-0.00118407 0.00764042 0.01 ]]\n[[ 0.07937234 0.01244316 -0.00127862]\n [ 0.01244316 0.06481476 0.00678915]\n [-0.00127862 0.00678915 0.01 ]]\n[[ 0.05124689 0.02562591 -0.00180384]\n [ 0.02562591 0.0547271 0.00503721]\n [-0.00180384 0.00503721 0.01 ]]\n[[ 0.03154046 0.02105117 -0.00335409]\n [ 0.02105117 0.02495658 0.00420318]\n [-0.00335409 0.00420318 0.01 ]]\n[[ 0.22191062 -0.12331094 0.004297 ]\n [-0.12331094 0.18662457 0.01013036]\n [ 0.004297 0.01013036 0.01 ]]\n[[ 0.20073816 -0.097035 0.00329595]\n [-0.097035 0.1454122 0.00978326]\n [ 0.00329595 0.00978326 0.01 ]]\n[[ 0.17508171 -0.0686835 0.00217126]\n [-0.0686835 0.10624653 0.00930623]\n [ 0.00217126 0.00930623 0.01 ]]\n[[ 8.01946332e-02 -1.03708141e-02 -7.58040071e-05]\n [-1.03708141e-02 4.58046351e-02 6.73522265e-03]\n [-7.58040071e-05 6.73522265e-03 1.00000000e-02]]\n[[ 0.05981333 -0.00399826 -0.00017035]\n [-0.00399826 0.04185409 0.00588395]\n [-0.00017035 0.00588395 0.01 ]]\n[[ 0.03168788 0.00918449 -0.00069557]\n [ 0.00918449 0.03176644 0.00413202]\n [-0.00069557 0.00413202 0.01 ]]\n[[ 0.20870255 -0.13595773 0.00582059]\n [-0.13595773 0.16917477 0.0092834 ]\n [ 0.00582059 0.0092834 0.01 ]]\n[[ 0.18753009 -0.1096818 0.00481955]\n [-0.1096818 0.12796239 0.0089363 ]\n [ 0.00481955 0.0089363 0.01 ]]\n[[ 0.16187364 -0.08133029 0.00369486]\n [-0.08133029 0.08879672 0.00845927]\n [ 0.00369486 0.00845927 0.01 ]]\n[[ 0.06698656 -0.02301761 0.00144779]\n [-0.02301761 0.02835483 0.00588826]\n [ 0.00144779 0.00588826 0.01 ]]\n[[ 0.04660526 -0.01664505 0.00135324]\n [-0.01664505 0.02440428 0.00503699]\n [ 0.00135324 0.00503699 0.01 ]]\n[[ 0.01847981 -0.0034623 0.00082803]\n [-0.0034623 0.01431663 0.00328505]\n [ 0.00082803 0.00328505 0.01 ]]\n[[ 0.37790024 -0.2540439 0.01247064]\n [-0.2540439 0.28963038 0.01486348]\n [ 0.01247064 0.01486348 0.01 ]]\n[[ 0.35224379 -0.2256924 0.01134595]\n [-0.2256924 0.25046472 0.01438645]\n [ 0.01134595 0.01438645 0.01 ]]\n[[ 0.25735671 -0.16737972 0.00909888]\n [-0.16737972 0.19002282 0.01181544]\n [ 0.00909888 0.01181544 0.01 ]]\n[[ 0.23697541 -0.16100716 0.00900434]\n [-0.16100716 0.18607228 0.01096417]\n [ 0.00900434 0.01096417 0.01 ]]\n[[ 0.20884996 -0.14782441 0.00847912]\n [-0.14782441 0.17598462 0.00921224]\n [ 0.00847912 0.00921224 0.01 ]]\n[[ 0.33107134 -0.19941647 0.0103449 ]\n [-0.19941647 0.20925234 0.01403935]\n [ 0.0103449 0.01403935 0.01 ]]\n[[ 0.23618426 -0.14110378 0.00809784]\n [-0.14110378 0.14881044 0.01146834]\n [ 0.00809784 0.01146834 0.01 ]]\n[[ 0.21580296 -0.13473122 0.00800329]\n [-0.13473122 0.1448599 0.01061707]\n [ 0.00800329 0.01061707 0.01 ]]\n[[ 0.18767751 -0.12154847 0.00747807]\n [-0.12154847 0.13477224 0.00886514]\n [ 0.00747807 0.00886514 0.01 ]]\n[[ 0.21052781 -0.11275228 0.00697315]\n [-0.11275228 0.10964478 0.01099131]\n [ 0.00697315 0.01099131 0.01 ]]\n[[ 0.19014651 -0.10637972 0.0068786 ]\n [-0.10637972 0.10569423 0.01014004]\n [ 0.0068786 0.01014004 0.01 ]]\n[[ 0.16202106 -0.09319697 0.00635338]\n [-0.09319697 0.09560658 0.0083881 ]\n [ 0.00635338 0.0083881 0.01 ]]\n[[ 0.09525943 -0.04806704 0.00463153]\n [-0.04806704 0.04525234 0.00756903]\n [ 0.00463153 0.00756903 0.01 ]]\n[[ 0.06713398 -0.03488429 0.00410632]\n [-0.03488429 0.03516468 0.0058171 ]\n [ 0.00410632 0.0058171 0.01 ]]\n[[ 0.04675268 -0.02851173 0.00401177]\n [-0.02851173 0.03121414 0.00496583]\n [ 0.00401177 0.00496583 0.01 ]]\n[[ 0.32756366 0.35252286 -0.02073293]\n [ 0.35252286 0.71162202 0.012381 ]\n [-0.02073293 0.012381 0.01 ]]\n[[ 0.3730754 0.36925209 -0.01998325]\n [ 0.36925209 0.6760648 0.01361559]\n [-0.01998325 0.01361559 0.01 ]]\n[[ 0.34501542 0.30138752 -0.01669516]\n [ 0.30138752 0.53202944 0.0133937 ]\n [-0.01669516 0.0133937 0.01 ]]\n[[ 0.80296464 0.33073848 -0.01480326]\n [ 0.33073848 0.53985847 0.020451 ]\n [-0.01480326 0.020451 0.01 ]]\n[[ 0.72033265 0.32918638 -0.01501442]\n [ 0.32918638 0.53386958 0.01947049]\n [-0.01501442 0.01947049 0.01 ]]\n[[ 0.83686029 0.26514998 -0.01304502]\n [ 0.26514998 0.51697165 0.02087024]\n [-0.01304502 0.02087024 0.01 ]]\n[[ 0.50359585 0.17890015 -0.01062635]\n [ 0.17890015 0.46819854 0.01655545]\n [-0.01062635 0.01655545 0.01 ]]\n[[ 0.43495928 0.15449563 -0.00952819]\n [ 0.15449563 0.46851366 0.01538921]\n [-0.00952819 0.01538921 0.01 ]]\n[[0.33601058 0.01542221 0.00115325]\n [0.01542221 0.9322739 0.01181809]\n [0.00115325 0.01181809 0.01 ]]\n[[ 2.82483011e-01 7.82212955e-02 -7.24444679e-04]\n [ 7.82212955e-02 7.90706112e-01 1.06178510e-02]\n [-7.24444679e-04 1.06178510e-02 1.00000000e-02]]\n[[ 0.25898638 0.10668025 -0.00121164]\n [ 0.10668025 0.75630323 0.00979854]\n [-0.00121164 0.00979854 0.01 ]]\n[[ 0.33721312 0.32022174 -0.01788443]\n [ 0.32022174 0.5372488 0.01314024]\n [-0.01788443 0.01314024 0.01 ]]\n[[ 0.30915314 0.25235717 -0.01459634]\n [ 0.25235717 0.39321344 0.01291835]\n [-0.01459634 0.01291835 0.01 ]]\n[[ 0.76710236 0.28170812 -0.01270444]\n [ 0.28170812 0.40104248 0.01997565]\n [-0.01270444 0.01997565 0.01 ]]\n[[ 0.68447037 0.28015602 -0.01291561]\n [ 0.28015602 0.39505359 0.01899514]\n [-0.01291561 0.01899514 0.01 ]]\n[[ 0.80099801 0.21611963 -0.01094621]\n [ 0.21611963 0.37815565 0.02039489]\n [-0.01094621 0.02039489 0.01 ]]\n[[ 0.46773357 0.1298698 -0.00852753]\n [ 0.1298698 0.32938254 0.0160801 ]\n [-0.00852753 0.0160801 0.01 ]]\n[[ 0.399097 0.10546527 -0.00742938]\n [ 0.10546527 0.32969766 0.01491385]\n [-0.00742938 0.01491385 0.01 ]]\n[[ 0.3001483 -0.03360815 0.00325207]\n [-0.03360815 0.79345791 0.01134274]\n [ 0.00325207 0.01134274 0.01 ]]\n[[0.24662073 0.02919094 0.00137437]\n [0.02919094 0.65189012 0.0101425 ]\n [0.00137437 0.0101425 0.01 ]]\n[[0.2231241 0.05764989 0.00088718]\n [0.05764989 0.61748724 0.00932318]\n [0.00088718 0.00932318 0.01 ]]\n[[ 0.35466488 0.2690864 -0.01384666]\n [ 0.2690864 0.35765622 0.01415294]\n [-0.01384666 0.01415294 0.01 ]]\n[[ 0.81261409 0.29843735 -0.01195476]\n [ 0.29843735 0.36548525 0.02121024]\n [-0.01195476 0.02121024 0.01 ]]\n[[ 0.72998211 0.29688525 -0.01216592]\n [ 0.29688525 0.35949636 0.02022973]\n [-0.01216592 0.02022973 0.01 ]]\n[[ 0.84650975 0.23284886 -0.01019653]\n [ 0.23284886 0.34259843 0.02162948]\n [-0.01019653 0.02162948 0.01 ]]\n[[ 0.51324531 0.14659903 -0.00777785]\n [ 0.14659903 0.29382532 0.01731469]\n [-0.00777785 0.01731469 0.01 ]]\n[[ 0.44460873 0.12219451 -0.0066797 ]\n [ 0.12219451 0.29414044 0.01614845]\n [-0.0066797 0.01614845 0.01 ]]\n[[ 0.34566004 -0.01687892 0.00400175]\n [-0.01687892 0.75790068 0.01257733]\n [ 0.00400175 0.01257733 0.01 ]]\n[[0.29213247 0.04592017 0.00212405]\n [0.04592017 0.61633289 0.01137709]\n [0.00212405 0.01137709 0.01 ]]\n[[0.26863584 0.07437912 0.00163686]\n [0.07437912 0.58193001 0.01055778]\n [0.00163686 0.01055778 0.01 ]]\n[[ 0.78455412 0.23057278 -0.00866667]\n [ 0.23057278 0.22144989 0.02098835]\n [-0.00866667 0.02098835 0.01 ]]\n[[ 0.70192213 0.22902069 -0.00887783]\n [ 0.22902069 0.21546101 0.02000784]\n [-0.00887783 0.02000784 0.01 ]]\n[[ 0.81844977 0.16498429 -0.00690844]\n [ 0.16498429 0.19856307 0.02140759]\n [-0.00690844 0.02140759 0.01 ]]\n[[ 0.48518533 0.07873446 -0.00448976]\n [ 0.07873446 0.14978996 0.0170928 ]\n [-0.00448976 0.0170928 0.01 ]]\n[[ 0.41654876 0.05432994 -0.00339161]\n [ 0.05432994 0.15010508 0.01592655]\n [-0.00339161 0.01592655 0.01 ]]\n[[ 0.31760006 -0.08474349 0.00728984]\n [-0.08474349 0.61386532 0.01235544]\n [ 0.00728984 0.01235544 0.01 ]]\n[[ 0.26407249 -0.0219444 0.00541214]\n [-0.0219444 0.47229753 0.0111552 ]\n [ 0.00541214 0.0111552 0.01 ]]\n[[0.24057586 0.00651455 0.00492495]\n [0.00651455 0.43789466 0.01033588]\n [0.00492495 0.01033588 0.01 ]]\n[[ 1.15987134 0.25837164 -0.00698593]\n [ 0.25837164 0.22329004 0.02706514]\n [-0.00698593 0.02706514 0.01 ]]\n[[ 1.27639899 0.19433524 -0.00501653]\n [ 0.19433524 0.2063921 0.02846489]\n [-0.00501653 0.02846489 0.01 ]]\n[[ 0.94313454 0.10808541 -0.00259785]\n [ 0.10808541 0.157619 0.0241501 ]\n [-0.00259785 0.0241501 0.01 ]]\n[[ 0.87449797 0.08368089 -0.0014997 ]\n [ 0.08368089 0.15793411 0.02298386]\n [-0.0014997 0.02298386 0.01 ]]\n[[ 0.77554927 -0.05539253 0.00918174]\n [-0.05539253 0.62169436 0.01941274]\n [ 0.00918174 0.01941274 0.01 ]]\n[[0.7220217 0.00740656 0.00730405]\n [0.00740656 0.48012657 0.0182125 ]\n [0.00730405 0.0182125 0.01 ]]\n[[0.69852508 0.03586551 0.00681685]\n [0.03586551 0.44572369 0.01739319]\n [0.00681685 0.01739319 0.01 ]]\n[[ 1.193767 0.19278315 -0.0052277 ]\n [ 0.19278315 0.20040321 0.02748438]\n [-0.0052277 0.02748438 0.01 ]]\n[[ 0.86050256 0.10653332 -0.00280902]\n [ 0.10653332 0.15163011 0.02316959]\n [-0.00280902 0.02316959 0.01 ]]\n[[ 0.79186598 0.08212879 -0.00171087]\n [ 0.08212879 0.15194523 0.02200335]\n [-0.00171087 0.02200335 0.01 ]]\n[[ 0.69291729 -0.05694463 0.00897058]\n [-0.05694463 0.61570547 0.01843223]\n [ 0.00897058 0.01843223 0.01 ]]\n[[0.63938972 0.00585446 0.00709288]\n [0.00585446 0.47413768 0.01723199]\n [0.00709288 0.01723199 0.01 ]]\n[[0.61589309 0.03431341 0.00660569]\n [0.03431341 0.4397348 0.01641268]\n [0.00660569 0.01641268 0.01 ]]\n[[ 9.77030201e-01 4.24969182e-02 -8.39620039e-04]\n [ 4.24969182e-02 1.34732170e-01 2.45693406e-02]\n [-8.39620039e-04 2.45693406e-02 1.00000000e-02]]\n[[9.08393626e-01 1.80923966e-02 2.58531285e-04]\n [1.80923966e-02 1.35047288e-01 2.34030946e-02]\n [2.58531285e-04 2.34030946e-02 1.00000000e-02]]\n[[ 0.80944493 -0.12098103 0.01093998]\n [-0.12098103 0.59880753 0.01983198]\n [ 0.01093998 0.01983198 0.01 ]]\n[[ 0.75591736 -0.05818194 0.00906228]\n [-0.05818194 0.45723974 0.01863174]\n [ 0.00906228 0.01863174 0.01 ]]\n[[ 0.73242073 -0.02972298 0.00857509]\n [-0.02972298 0.42283686 0.01781242]\n [ 0.00857509 0.01781242 0.01 ]]\n[[ 0.57512918 -0.06815743 0.00267721]\n [-0.06815743 0.08627418 0.01908831]\n [ 0.00267721 0.01908831 0.01 ]]\n[[ 0.47618049 -0.20723086 0.01335865]\n [-0.20723086 0.55003443 0.01551719]\n [ 0.01335865 0.01551719 0.01 ]]\n[[ 0.42265292 -0.14443177 0.01148096]\n [-0.14443177 0.40846663 0.01431695]\n [ 0.01148096 0.01431695 0.01 ]]\n[[ 0.39915629 -0.11597282 0.01099377]\n [-0.11597282 0.37406376 0.01349764]\n [ 0.01099377 0.01349764 0.01 ]]\n[[ 0.40754391 -0.23163538 0.01445681]\n [-0.23163538 0.55034954 0.01435094]\n [ 0.01445681 0.01435094 0.01 ]]\n[[ 0.35401634 -0.16883629 0.01257911]\n [-0.16883629 0.40878175 0.0131507 ]\n [ 0.01257911 0.0131507 0.01 ]]\n[[ 0.33051972 -0.14037734 0.01209192]\n [-0.14037734 0.37437888 0.01233139]\n [ 0.01209192 0.01233139 0.01 ]]\n[[ 0.25506765 -0.30790972 0.02326056]\n [-0.30790972 0.872542 0.00957959]\n [ 0.02326056 0.00957959 0.01 ]]\n[[ 0.23157102 -0.27945076 0.02277336]\n [-0.27945076 0.83813912 0.00876027]\n [ 0.02277336 0.00876027 0.01 ]]\n[[ 0.17804345 -0.21665167 0.02089567]\n [-0.21665167 0.69657133 0.00756003]\n [ 0.02089567 0.00756003 0.01 ]]\n[[ 0.15481891 0.07588792 -0.02711606]\n [ 0.07588792 1.15790472 0.00207834]\n [-0.02711606 0.00207834 0.01 ]]\n[[ 0.12940855 0.08143642 -0.02310995]\n [ 0.08143642 0.88894066 0.00291172]\n [-0.02310995 0.00291172 0.01 ]]\n[[ 0.13152098 0.08765123 -0.02214961]\n [ 0.08765123 0.84022484 0.00346818]\n [-0.02214961 0.00346818 0.01 ]]\n[[ 0.52791337 0.2786762 -0.02222558]\n [ 0.2786762 0.89445292 0.01232156]\n [-0.02222558 0.01232156 0.01 ]]\n[[ 0.49558019 0.23271545 -0.02097299]\n [ 0.23271545 0.83440061 0.01197019]\n [-0.02097299 0.01197019 0.01 ]]\n[[ 0.35797846 0.12751136 -0.01821796]\n [ 0.12751136 0.72789937 0.01000385]\n [-0.01821796 0.01000385 0.01 ]]\n[[ 0.59968987 -0.26023066 -0.00584249]\n [-0.26023066 0.9368225 0.01321765]\n [-0.00584249 0.01321765 0.01 ]]\n[[ 0.5667442 -0.25322343 -0.005745 ]\n [-0.25322343 0.93777488 0.01277957]\n [-0.005745 0.01277957 0.01 ]]\n[[ 0.37171111 -0.15117477 -0.00718019]\n [-0.15117477 0.84294299 0.00998732]\n [-0.00718019 0.00998732 0.01 ]]\n[[ 0.11553015 0.09721082 -0.02150077]\n [ 0.09721082 0.75255828 0.0034603 ]\n [-0.02150077 0.0034603 0.01 ]]\n[[ 0.11764259 0.10342563 -0.02054043]\n [ 0.10342563 0.70384247 0.00401677]\n [-0.02054043 0.00401677 0.01 ]]\n[[ 0.51403497 0.2944506 -0.0206164 ]\n [ 0.2944506 0.75807055 0.01287014]\n [-0.0206164 0.01287014 0.01 ]]\n[[ 0.48170179 0.24848985 -0.0193638 ]\n [ 0.24848985 0.69801824 0.01251877]\n [-0.0193638 0.01251877 0.01 ]]\n[[ 0.34410007 0.14328576 -0.01660878]\n [ 0.14328576 0.591517 0.01055243]\n [-0.01660878 0.01055243 0.01 ]]\n[[ 0.58581148 -0.24445626 -0.00423331]\n [-0.24445626 0.80044013 0.01376623]\n [-0.00423331 0.01376623 0.01 ]]\n[[ 0.5528658 -0.23744903 -0.00413581]\n [-0.23744903 0.80139251 0.01332815]\n [-0.00413581 0.01332815 0.01 ]]\n[[ 0.35783272 -0.13540037 -0.005571 ]\n [-0.13540037 0.70656062 0.0105359 ]\n [-0.005571 0.0105359 0.01 ]]\n[[ 0.09223222 0.10897413 -0.01653432]\n [ 0.10897413 0.4348784 0.00485015]\n [-0.01653432 0.00485015 0.01 ]]\n[[ 0.48862461 0.2999991 -0.01661029]\n [ 0.2999991 0.48910649 0.01370352]\n [-0.01661029 0.01370352 0.01 ]]\n[[ 0.45629143 0.25403835 -0.01535769]\n [ 0.25403835 0.42905417 0.01335215]\n [-0.01535769 0.01335215 0.01 ]]\n[[ 0.3186897 0.14883426 -0.01260267]\n [ 0.14883426 0.32255293 0.01138581]\n [-0.01260267 0.01138581 0.01 ]]\n[[ 5.60401112e-01 -2.38907758e-01 -2.27202352e-04]\n [-2.38907758e-01 5.31476066e-01 1.45996098e-02]\n [-2.27202352e-04 1.45996098e-02 1.00000000e-02]]\n[[ 5.27455437e-01 -2.31900531e-01 -1.29705385e-04]\n [-2.31900531e-01 5.32428439e-01 1.41615276e-02]\n [-1.29705385e-04 1.41615276e-02 1.00000000e-02]]\n[[ 0.33242235 -0.12985188 -0.00156489]\n [-0.12985188 0.43759655 0.01136928]\n [-0.00156489 0.01136928 0.01 ]]\n[[ 0.49073704 0.30621391 -0.01564994]\n [ 0.30621391 0.44039067 0.01425999]\n [-0.01564994 0.01425999 0.01 ]]\n[[ 0.45840386 0.26025316 -0.01439735]\n [ 0.26025316 0.38033836 0.01390861]\n [-0.01439735 0.01390861 0.01 ]]\n[[ 0.32080214 0.15504907 -0.01164233]\n [ 0.15504907 0.27383712 0.01194227]\n [-0.01164233 0.01194227 0.01 ]]\n[[ 0.56251355 -0.23269295 0.00073314]\n [-0.23269295 0.48276025 0.01515608]\n [ 0.00073314 0.01515608 0.01 ]]\n[[ 0.52956788 -0.22568572 0.00083064]\n [-0.22568572 0.48371262 0.01471799]\n [ 0.00083064 0.01471799 0.01 ]]\n[[ 0.33453479 -0.12363706 -0.00060455]\n [-0.12363706 0.38888074 0.01192575]\n [-0.00060455 0.01192575 0.01 ]]\n[[ 0.85479625 0.45127813 -0.01447332]\n [ 0.45127813 0.43456644 0.02276199]\n [-0.01447332 0.02276199 0.01 ]]\n[[ 0.71719452 0.34607404 -0.0117183 ]\n [ 0.34607404 0.3280652 0.02079565]\n [-0.0117183 0.02079565 0.01 ]]\n[[ 9.58905933e-01 -4.16679785e-02 6.57171307e-04]\n [-4.16679785e-02 5.36988334e-01 2.40094501e-02]\n [ 6.57171307e-04 2.40094501e-02 1.00000000e-02]]\n[[ 9.25960258e-01 -3.46607509e-02 7.54668275e-04]\n [-3.46607509e-02 5.37940707e-01 2.35713678e-02]\n [ 7.54668275e-04 2.35713678e-02 1.00000000e-02]]\n[[ 7.30927176e-01 6.73879045e-02 -6.80520518e-04]\n [ 6.73879045e-02 4.43108818e-01 2.07791205e-02]\n [-6.80520518e-04 2.07791205e-02 1.00000000e-02]]\n[[ 0.68486134 0.30011328 -0.0104657 ]\n [ 0.30011328 0.26801289 0.02044428]\n [-0.0104657 0.02044428 0.01 ]]\n[[ 0.92657275 -0.08762873 0.00190976]\n [-0.08762873 0.47693602 0.02365808]\n [ 0.00190976 0.02365808 0.01 ]]\n[[ 0.89362708 -0.08062151 0.00200726]\n [-0.08062151 0.4778884 0.02322 ]\n [ 0.00200726 0.02322 0.01 ]]\n[[6.98593995e-01 2.14271493e-02 5.72073149e-04]\n [2.14271493e-02 3.83056507e-01 2.04277484e-02]\n [5.72073149e-04 2.04277484e-02 1.00000000e-02]]\n[[ 0.78897103 -0.19283282 0.00466479]\n [-0.19283282 0.37043478 0.02169174]\n [ 0.00466479 0.02169174 0.01 ]]\n[[ 0.75602535 -0.1858256 0.00476228]\n [-0.1858256 0.37138716 0.02125366]\n [ 0.00476228 0.02125366 0.01 ]]\n[[ 0.56099227 -0.08377694 0.0033271 ]\n [-0.08377694 0.27655527 0.01846141]\n [ 0.0033271 0.01846141 0.01 ]]\n[[ 0.99773676 -0.57356761 0.01713775]\n [-0.57356761 0.58031029 0.02446746]\n [ 0.01713775 0.02446746 0.01 ]]\n[[ 0.80270368 -0.47151896 0.01570257]\n [-0.47151896 0.4854784 0.02167521]\n [ 0.01570257 0.02167521 0.01 ]]\n[[ 0.76975801 -0.46451173 0.01580006]\n [-0.46451173 0.48643077 0.02123713]\n [ 0.01580006 0.02123713 0.01 ]]\n[[ 0.75838149 -0.15215087 -0.00493686]\n [-0.15215087 0.14045595 -0.02184427]\n [-0.00493686 -0.02184427 0.01 ]]\n[[ 0.67118258 -0.12884489 -0.00419902]\n [-0.12884489 0.12431305 -0.02036614]\n [-0.00419902 -0.02036614 0.01 ]]\n[[ 0.64710515 -0.12279476 -0.00396852]\n [-0.12279476 0.12036212 -0.01990956]\n [-0.00396852 -0.01990956 0.01 ]]\n[[ 0.55010435 -0.11646133 -0.00376498]\n [-0.11646133 0.10748518 -0.017715 ]\n [-0.00376498 -0.017715 0.01 ]]\n[[ 0.4943492 -0.11698555 -0.0039364 ]\n [-0.11698555 0.10078137 -0.01595903]\n [-0.0039364 -0.01595903 0.01 ]]\n[[ 0.46880683 -0.11587527 -0.00397672]\n [-0.11587527 0.09764566 -0.01483572]\n [-0.00397672 -0.01483572 0.01 ]]\n[[ 0.58737847 -0.29017332 -0.01592748]\n [-0.29017332 0.59109162 -0.01700744]\n [-0.01592748 -0.01700744 0.01 ]]\n[[ 0.52413578 -0.21205176 -0.01239772]\n [-0.21205176 0.35005062 -0.01588061]\n [-0.01239772 -0.01588061 0.01 ]]\n[[ 0.51630358 -0.20067862 -0.01154031]\n [-0.20067862 0.30406405 -0.01580233]\n [-0.01154031 -0.01580233 0.01 ]]\n[[ 0.51327081 -0.19309956 -0.01070808]\n [-0.19309956 0.26434931 -0.01588323]\n [-0.01070808 -0.01588323 0.01 ]]\n[[ 0.46241509 -0.13950218 -0.00837252]\n [-0.13950218 0.17031723 -0.01384908]\n [-0.00837252 -0.01384908 0.01 ]]\n[[ 0.44890924 -0.12269945 -0.00749663]\n [-0.12269945 0.14444085 -0.0127775 ]\n [-0.00749663 -0.0127775 0.01 ]]\n[[ 0.69649162 0.05714068 -0.01055657]\n [ 0.05714068 0.28101951 -0.00307361]\n [-0.01055657 -0.00307361 0.01 ]]\n[[ 0.60492039 -0.00648663 -0.00895287]\n [-0.00648663 0.20710527 -0.00477004]\n [-0.00895287 -0.00477004 0.01 ]]\n[[ 0.56968635 -0.02984823 -0.00826997]\n [-0.02984823 0.18092724 -0.00555227]\n [-0.00826997 -0.00555227 0.01 ]]\n[[ 0.53566021 -0.05269341 -0.00748346]\n [-0.05269341 0.15517885 -0.00641325]\n [-0.00748346 -0.00641325 0.01 ]]\n[[ 0.47713111 -0.09375807 -0.00537623]\n [-0.09375807 0.10916741 -0.00836062]\n [-0.00537623 -0.00836062 0.01 ]]\n[[ 0.55111772 -0.05782833 -0.00220565]\n [-0.05782833 0.07863944 -0.01867092]\n [-0.00220565 -0.01867092 0.01 ]]\n[[ 0.52704028 -0.0517782 -0.00197515]\n [-0.0517782 0.07468851 -0.01821434]\n [-0.00197515 -0.01821434 0.01 ]]\n[[ 0.43003949 -0.04544477 -0.00177161]\n [-0.04544477 0.06181157 -0.01601978]\n [-0.00177161 -0.01601978 0.01 ]]\n[[ 0.37428434 -0.04596899 -0.00194304]\n [-0.04596899 0.05510776 -0.01426382]\n [-0.00194304 -0.01426382 0.01 ]]\n[[ 0.34874197 -0.04485871 -0.00198335]\n [-0.04485871 0.05197204 -0.0131405 ]\n [-0.00198335 -0.0131405 0.01 ]]\n[[ 0.46731361 -0.21915676 -0.01393411]\n [-0.21915676 0.54541801 -0.01531222]\n [-0.01393411 -0.01531222 0.01 ]]\n[[ 0.40407092 -0.1410352 -0.01040436]\n [-0.1410352 0.304377 -0.0141854 ]\n [-0.01040436 -0.0141854 0.01 ]]\n[[ 0.39623872 -0.12966206 -0.00954694]\n [-0.12966206 0.25839044 -0.01410712]\n [-0.00954694 -0.01410712 0.01 ]]\n[[ 0.39320595 -0.12208301 -0.00871471]\n [-0.12208301 0.2186757 -0.01418801]\n [-0.00871471 -0.01418801 0.01 ]]\n[[ 0.34235022 -0.06848562 -0.00637915]\n [-0.06848562 0.12464362 -0.01215386]\n [-0.00637915 -0.01215386 0.01 ]]\n[[ 0.32884438 -0.05168289 -0.00550327]\n [-0.05168289 0.09876724 -0.01108228]\n [-0.00550327 -0.01108228 0.01 ]]\n[[ 0.57642675 0.12815724 -0.00856321]\n [ 0.12815724 0.23534589 -0.0013784 ]\n [-0.00856321 -0.0013784 0.01 ]]\n[[ 0.48485553 0.06452993 -0.00695951]\n [ 0.06452993 0.16143166 -0.00307482]\n [-0.00695951 -0.00307482 0.01 ]]\n[[ 0.44962149 0.04116833 -0.0062766 ]\n [ 0.04116833 0.13525362 -0.00385705]\n [-0.0062766 -0.00385705 0.01 ]]\n[[ 0.41559535 0.01832315 -0.00549009]\n [ 0.01832315 0.10950524 -0.00471804]\n [-0.00549009 -0.00471804 0.01 ]]\n[[ 0.35706625 -0.02274151 -0.00338286]\n [-0.02274151 0.0634938 -0.00666541]\n [-0.00338286 -0.00666541 0.01 ]]\n[[ 0.43984138 -0.02847222 -0.0012373 ]\n [-0.02847222 0.0585456 -0.01673621]\n [-0.0012373 -0.01673621 0.01 ]]\n[[ 0.34284059 -0.02213879 -0.00103376]\n [-0.02213879 0.04566867 -0.01454165]\n [-0.00103376 -0.01454165 0.01 ]]\n[[ 0.28708544 -0.02266301 -0.00120519]\n [-0.02266301 0.03896486 -0.01278568]\n [-0.00120519 -0.01278568 0.01 ]]\n[[ 0.26154306 -0.02155273 -0.0012455 ]\n [-0.02155273 0.03582914 -0.01166236]\n [-0.0012455 -0.01166236 0.01 ]]\n[[ 0.38011471 -0.19585078 -0.01319626]\n [-0.19585078 0.52927511 -0.01383408]\n [-0.01319626 -0.01383408 0.01 ]]\n[[ 0.31687202 -0.11772922 -0.00966651]\n [-0.11772922 0.2882341 -0.01270726]\n [-0.00966651 -0.01270726 0.01 ]]\n[[ 0.30903981 -0.10635608 -0.00880909]\n [-0.10635608 0.24224753 -0.01262898]\n [-0.00880909 -0.01262898 0.01 ]]\n[[ 0.30600705 -0.09877703 -0.00797687]\n [-0.09877703 0.2025328 -0.01270987]\n [-0.00797687 -0.01270987 0.01 ]]\n[[ 0.25515132 -0.04517964 -0.0056413 ]\n [-0.04517964 0.10850071 -0.01067572]\n [-0.0056413 -0.01067572 0.01 ]]\n[[ 0.24164547 -0.02837691 -0.00476542]\n [-0.02837691 0.08262433 -0.00960414]\n [-0.00476542 -0.00960414 0.01 ]]\n[[ 4.89227851e-01 1.51463218e-01 -7.82536052e-03]\n [ 1.51463218e-01 2.19202990e-01 9.97423549e-05]\n [-7.82536052e-03 9.97423549e-05 1.00000000e-02]]\n[[ 0.39765662 0.08783591 -0.00622166]\n [ 0.08783591 0.14528875 -0.00159669]\n [-0.00622166 -0.00159669 0.01 ]]\n[[ 0.36242259 0.06447431 -0.00553876]\n [ 0.06447431 0.11911072 -0.00237891]\n [-0.00553876 -0.00237891 0.01 ]]\n[[ 0.32839645 0.04162913 -0.00475225]\n [ 0.04162913 0.09336234 -0.0032399 ]\n [-0.00475225 -0.0032399 0.01 ]]\n[[ 0.26986734 0.00056447 -0.00264501]\n [ 0.00056447 0.04735089 -0.00518727]\n [-0.00264501 -0.00518727 0.01 ]]\n[[ 0.31876315 -0.01608866 -0.00080326]\n [-0.01608866 0.04171773 -0.01408507]\n [-0.00080326 -0.01408507 0.01 ]]\n[[ 0.263008 -0.01661288 -0.00097469]\n [-0.01661288 0.03501393 -0.0123291 ]\n [-0.00097469 -0.0123291 0.01 ]]\n[[ 0.23746562 -0.0155026 -0.001015 ]\n [-0.0155026 0.03187821 -0.01120578]\n [-0.001015 -0.01120578 0.01 ]]\n[[ 0.35603727 -0.18980065 -0.01296576]\n [-0.18980065 0.52532418 -0.0133775 ]\n [-0.01296576 -0.0133775 0.01 ]]\n[[ 0.29279458 -0.11167909 -0.00943601]\n [-0.11167909 0.28428317 -0.01225068]\n [-0.00943601 -0.01225068 0.01 ]]\n[[ 0.28496237 -0.10030595 -0.00857859]\n [-0.10030595 0.2382966 -0.0121724 ]\n [-0.00857859 -0.0121724 0.01 ]]\n[[ 0.28192961 -0.0927269 -0.00774637]\n [-0.0927269 0.19858187 -0.0122533 ]\n [-0.00774637 -0.0122533 0.01 ]]\n[[ 0.23107388 -0.03912951 -0.0054108 ]\n [-0.03912951 0.10454978 -0.01021915]\n [-0.0054108 -0.01021915 0.01 ]]\n[[ 0.21756803 -0.02232678 -0.00453492]\n [-0.02232678 0.0786734 -0.00914757]\n [-0.00453492 -0.00914757 0.01 ]]\n[[ 0.46515041 0.15751335 -0.00759486]\n [ 0.15751335 0.21525206 0.00055632]\n [-0.00759486 0.00055632 0.01 ]]\n[[ 0.37357918 0.09388604 -0.00599116]\n [ 0.09388604 0.14133782 -0.00114011]\n [-0.00599116 -0.00114011 0.01 ]]\n[[ 0.33834515 0.07052444 -0.00530826]\n [ 0.07052444 0.11515979 -0.00192233]\n [-0.00530826 -0.00192233 0.01 ]]\n[[ 0.30431901 0.04767926 -0.00452175]\n [ 0.04767926 0.0894114 -0.00278332]\n [-0.00452175 -0.00278332 0.01 ]]\n[[ 0.2457899 0.0066146 -0.00241452]\n [ 0.0066146 0.04339996 -0.00473069]\n [-0.00241452 -0.00473069 0.01 ]]\n[[ 0.16600721 -0.01027945 -0.00077115]\n [-0.01027945 0.02213699 -0.01013454]\n [-0.00077115 -0.01013454 0.01 ]]\n[[ 0.14046483 -0.00916917 -0.00081146]\n [-0.00916917 0.01900127 -0.00901123]\n [-0.00081146 -0.00901123 0.01 ]]\n[[ 0.25903648 -0.18346722 -0.01276222]\n [-0.18346722 0.51244724 -0.01118294]\n [-0.01276222 -0.01118294 0.01 ]]\n[[ 0.19579379 -0.10534566 -0.00923247]\n [-0.10534566 0.27140623 -0.01005612]\n [-0.00923247 -0.01005612 0.01 ]]\n[[ 0.18796158 -0.09397252 -0.00837505]\n [-0.09397252 0.22541967 -0.00997784]\n [-0.00837505 -0.00997784 0.01 ]]\n[[ 0.18492882 -0.08639346 -0.00754283]\n [-0.08639346 0.18570493 -0.01005874]\n [-0.00754283 -0.01005874 0.01 ]]\n[[ 0.13407309 -0.03279608 -0.00520726]\n [-0.03279608 0.09167285 -0.00802459]\n [-0.00520726 -0.00802459 0.01 ]]\n[[ 0.12056724 -0.01599335 -0.00433138]\n [-0.01599335 0.06579646 -0.00695301]\n [-0.00433138 -0.00695301 0.01 ]]\n[[ 0.36814962 0.16384678 -0.00739132]\n [ 0.16384678 0.20237512 0.00275088]\n [-0.00739132 0.00275088 0.01 ]]\n[[ 0.27657839 0.10021947 -0.00578762]\n [ 0.10021947 0.12846088 0.00105445]\n [-0.00578762 0.00105445 0.01 ]]\n[[ 0.24134436 0.07685787 -0.00510472]\n [ 0.07685787 0.10228285 0.00027223]\n [-0.00510472 0.00027223 0.01 ]]\n[[ 0.20731822 0.05401269 -0.00431821]\n [ 0.05401269 0.07653447 -0.00058876]\n [-0.00431821 -0.00058876 0.01 ]]\n[[ 0.14878911 0.01294803 -0.00221097]\n [ 0.01294803 0.03052302 -0.00253613]\n [-0.00221097 -0.00253613 0.01 ]]\n[[ 0.08470968 -0.0096934 -0.00098289]\n [-0.0096934 0.01229746 -0.00725526]\n [-0.00098289 -0.00725526 0.01 ]]\n[[ 0.20328133 -0.18399145 -0.01293365]\n [-0.18399145 0.50574343 -0.00942698]\n [-0.01293365 -0.00942698 0.01 ]]\n[[ 0.14003864 -0.10586988 -0.0094039 ]\n [-0.10586988 0.26470242 -0.00830015]\n [-0.0094039 -0.00830015 0.01 ]]\n[[ 0.13220643 -0.09449674 -0.00854648]\n [-0.09449674 0.21871586 -0.00822188]\n [-0.00854648 -0.00822188 0.01 ]]\n[[ 0.12917367 -0.08691769 -0.00771425]\n [-0.08691769 0.17900112 -0.00830277]\n [-0.00771425 -0.00830277 0.01 ]]\n[[ 0.07831794 -0.0333203 -0.00537869]\n [-0.0333203 0.08496904 -0.00626862]\n [-0.00537869 -0.00626862 0.01 ]]\n[[ 0.06481209 -0.01651758 -0.0045028 ]\n [-0.01651758 0.05909265 -0.00519704]\n [-0.0045028 -0.00519704 0.01 ]]\n[[ 0.31239447 0.16332255 -0.00756275]\n [ 0.16332255 0.19567131 0.00450685]\n [-0.00756275 0.00450685 0.01 ]]\n[[ 0.22082324 0.09969525 -0.00595904]\n [ 0.09969525 0.12175707 0.00281042]\n [-0.00595904 0.00281042 0.01 ]]\n[[ 0.18558921 0.07633364 -0.00527614]\n [ 0.07633364 0.09557904 0.00202819]\n [-0.00527614 0.00202819 0.01 ]]\n[[ 0.15156307 0.05348846 -0.00448963]\n [ 0.05348846 0.06983066 0.00116721]\n [-0.00448963 0.00116721 0.01 ]]\n[[ 0.09303396 0.0124238 -0.0023824 ]\n [ 0.0124238 0.02381922 -0.00078016]\n [-0.0023824 -0.00078016 0.01 ]]\n[[ 0.17773895 -0.18288117 -0.01297396]\n [-0.18288117 0.50260771 -0.00830366]\n [-0.01297396 -0.00830366 0.01 ]]\n[[ 0.11449626 -0.1047596 -0.00944421]\n [-0.1047596 0.26156671 -0.00717684]\n [-0.00944421 -0.00717684 0.01 ]]\n[[ 0.10666406 -0.09338646 -0.00858679]\n [-0.09338646 0.21558014 -0.00709856]\n [-0.00858679 -0.00709856 0.01 ]]\n[[ 0.10363129 -0.08580741 -0.00775456]\n [-0.08580741 0.1758654 -0.00717945]\n [-0.00775456 -0.00717945 0.01 ]]\n[[ 0.05277557 -0.03221002 -0.005419 ]\n [-0.03221002 0.08183332 -0.0051453 ]\n [-0.005419 -0.0051453 0.01 ]]\n[[ 0.03926972 -0.0154073 -0.00454312]\n [-0.0154073 0.05595694 -0.00407372]\n [-0.00454312 -0.00407372 0.01 ]]\n[[ 0.2868521 0.16443283 -0.00760306]\n [ 0.16443283 0.1925356 0.00563016]\n [-0.00760306 0.00563016 0.01 ]]\n[[ 0.19528087 0.10080553 -0.00599936]\n [ 0.10080553 0.11862136 0.00393373]\n [-0.00599936 0.00393373 0.01 ]]\n[[ 0.16004683 0.07744392 -0.00531645]\n [ 0.07744392 0.09244333 0.00315151]\n [-0.00531645 0.00315151 0.01 ]]\n[[ 0.12602069 0.05459874 -0.00452994]\n [ 0.05459874 0.06669494 0.00229052]\n [-0.00452994 0.00229052 0.01 ]]\n[[ 0.06749159 0.01353408 -0.00242271]\n [ 0.01353408 0.0206835 0.00034315]\n [-0.00242271 0.00034315 0.01 ]]\n[[ 0.23306791 -0.27905765 -0.02139497]\n [-0.27905765 0.75501267 -0.00934856]\n [-0.02139497 -0.00934856 0.01 ]]\n[[ 0.2252357 -0.26768451 -0.02053755]\n [-0.26768451 0.70902611 -0.00927028]\n [-0.02053755 -0.00927028 0.01 ]]\n[[ 0.22220294 -0.26010546 -0.01970532]\n [-0.26010546 0.66931137 -0.00935117]\n [-0.01970532 -0.00935117 0.01 ]]\n[[ 0.17134721 -0.20650807 -0.01736976]\n [-0.20650807 0.57527929 -0.00731702]\n [-0.01736976 -0.00731702 0.01 ]]\n[[ 0.15784136 -0.18970535 -0.01649388]\n [-0.18970535 0.54940291 -0.00624544]\n [-0.01649388 -0.00624544 0.01 ]]\n[[ 0.40542374 -0.00986521 -0.01955382]\n [-0.00986521 0.68598156 0.00345844]\n [-0.01955382 0.00345844 0.01 ]]\n[[ 0.31385251 -0.07349252 -0.01795012]\n [-0.07349252 0.61206733 0.00176201]\n [-0.01795012 0.00176201 0.01 ]]\n[[ 0.27861848 -0.09685412 -0.01726721]\n [-0.09685412 0.58588929 0.00097979]\n [-0.01726721 0.00097979 0.01 ]]\n[[ 2.44592340e-01 -1.19699304e-01 -1.64807037e-02]\n [-1.19699304e-01 5.60140911e-01 1.18804224e-04]\n [-1.64807037e-02 1.18804224e-04 1.00000000e-02]]\n[[ 0.18606323 -0.16076396 -0.01437347]\n [-0.16076396 0.51412947 -0.00182857]\n [-0.01437347 -0.00182857 0.01 ]]\n[[ 0.16199301 -0.18956295 -0.0170078 ]\n [-0.18956295 0.4679851 -0.00814346]\n [-0.0170078 -0.00814346 0.01 ]]\n[[ 0.15896025 -0.18198389 -0.01617557]\n [-0.18198389 0.42827036 -0.00822435]\n [-0.01617557 -0.00822435 0.01 ]]\n[[ 0.10810452 -0.12838651 -0.01384001]\n [-0.12838651 0.33423828 -0.0061902 ]\n [-0.01384001 -0.0061902 0.01 ]]\n[[ 0.09459867 -0.11158378 -0.01296413]\n [-0.11158378 0.3083619 -0.00511862]\n [-0.01296413 -0.00511862 0.01 ]]\n[[ 0.34218105 0.06825635 -0.01602407]\n [ 0.06825635 0.44494056 0.00458527]\n [-0.01602407 0.00458527 0.01 ]]\n[[ 0.25060982 0.00462904 -0.01442037]\n [ 0.00462904 0.37102632 0.00288884]\n [-0.01442037 0.00288884 0.01 ]]\n[[ 0.21537578 -0.01873256 -0.01373746]\n [-0.01873256 0.34484829 0.00210661]\n [-0.01373746 0.00210661 0.01 ]]\n[[ 0.18134965 -0.04157774 -0.01295095]\n [-0.04157774 0.3190999 0.00124563]\n [-0.01295095 0.00124563 0.01 ]]\n[[ 0.12282054 -0.0826424 -0.01084372]\n [-0.0826424 0.27308846 -0.00070174]\n [-0.01084372 -0.00070174 0.01 ]]\n[[ 0.15112804 -0.17061075 -0.01531816]\n [-0.17061075 0.3822838 -0.00814607]\n [-0.01531816 -0.00814607 0.01 ]]\n[[ 0.10027232 -0.11701337 -0.01298259]\n [-0.11701337 0.28825171 -0.00611192]\n [-0.01298259 -0.00611192 0.01 ]]\n[[ 0.08676647 -0.10021064 -0.01210671]\n [-0.10021064 0.26237533 -0.00504034]\n [-0.01210671 -0.00504034 0.01 ]]\n[[ 0.33434885 0.07962949 -0.01516665]\n [ 0.07962949 0.39895399 0.00466355]\n [-0.01516665 0.00466355 0.01 ]]\n[[ 0.24277762 0.01600218 -0.01356295]\n [ 0.01600218 0.32503975 0.00296712]\n [-0.01356295 0.00296712 0.01 ]]\n[[ 0.20754358 -0.00735942 -0.01288005]\n [-0.00735942 0.29886172 0.00218489]\n [-0.01288005 0.00218489 0.01 ]]\n[[ 0.17351744 -0.0302046 -0.01209354]\n [-0.0302046 0.27311334 0.00132391]\n [-0.01209354 0.00132391 0.01 ]]\n[[ 0.11498834 -0.07126926 -0.0099863 ]\n [-0.07126926 0.22710189 -0.00062346]\n [-0.0099863 -0.00062346 0.01 ]]\n[[ 0.09723955 -0.10943431 -0.01215037]\n [-0.10943431 0.24853698 -0.00619281]\n [-0.01215037 -0.00619281 0.01 ]]\n[[ 0.0837337 -0.09263159 -0.01127448]\n [-0.09263159 0.22266059 -0.00512123]\n [-0.01127448 -0.00512123 0.01 ]]\n[[ 0.33131608 0.08720854 -0.01433442]\n [ 0.08720854 0.35923925 0.00458265]\n [-0.01433442 0.00458265 0.01 ]]\n[[ 0.23974485 0.02358124 -0.01273072]\n [ 0.02358124 0.28532501 0.00288622]\n [-0.01273072 0.00288622 0.01 ]]\n[[ 2.04510815e-01 2.19633366e-04 -1.20478180e-02]\n [ 2.19633366e-04 2.59146984e-01 2.10399785e-03]\n [-1.20478180e-02 2.10399785e-03 1.00000000e-02]]\n[[ 0.17048468 -0.02262555 -0.01126131]\n [-0.02262555 0.2333986 0.00124301]\n [-0.01126131 0.00124301 0.01 ]]\n[[ 0.11195557 -0.06369021 -0.00915408]\n [-0.06369021 0.18738716 -0.00070436]\n [-0.00915408 -0.00070436 0.01 ]]\n[[ 0.03287797 -0.0390342 -0.00893892]\n [-0.0390342 0.12862851 -0.00308708]\n [-0.00893892 -0.00308708 0.01 ]]\n[[ 0.28046035 0.14080593 -0.01199886]\n [ 0.14080593 0.26520717 0.0066168 ]\n [-0.01199886 0.0066168 0.01 ]]\n[[ 0.18888913 0.07717862 -0.01039516]\n [ 0.07717862 0.19129293 0.00492037]\n [-0.01039516 0.00492037 0.01 ]]\n[[ 0.15365509 0.05381702 -0.00971225]\n [ 0.05381702 0.1651149 0.00413815]\n [-0.00971225 0.00413815 0.01 ]]\n[[ 0.11962895 0.03097184 -0.00892574]\n [ 0.03097184 0.13936652 0.00327716]\n [-0.00892574 0.00327716 0.01 ]]\n[[ 0.06109985 -0.01009282 -0.00681851]\n [-0.01009282 0.09335507 0.00132979]\n [-0.00681851 0.00132979 0.01 ]]\n[[ 0.2669545 0.15760866 -0.01112298]\n [ 0.15760866 0.23933079 0.00768838]\n [-0.01112298 0.00768838 0.01 ]]\n[[ 0.17538328 0.09398135 -0.00951928]\n [ 0.09398135 0.16541655 0.00599195]\n [-0.00951928 0.00599195 0.01 ]]\n[[ 0.14014924 0.07061974 -0.00883637]\n [ 0.07061974 0.13923852 0.00520973]\n [-0.00883637 0.00520973 0.01 ]]\n[[ 0.1061231 0.04777457 -0.00804986]\n [ 0.04777457 0.11349013 0.00434874]\n [-0.00804986 0.00434874 0.01 ]]\n[[ 0.047594 0.0067099 -0.00594263]\n [ 0.0067099 0.06747869 0.00240137]\n [-0.00594263 0.00240137 0.01 ]]\n[[ 0.42296566 0.27382148 -0.01257922]\n [ 0.27382148 0.30199521 0.01569584]\n [-0.01257922 0.01569584 0.01 ]]\n[[ 0.38773162 0.25045988 -0.01189631]\n [ 0.25045988 0.27581718 0.01491361]\n [-0.01189631 0.01491361 0.01 ]]\n[[ 0.35370548 0.2276147 -0.0111098 ]\n [ 0.2276147 0.25006879 0.01405263]\n [-0.0111098 0.01405263 0.01 ]]\n[[ 0.29517638 0.18655004 -0.00900257]\n [ 0.18655004 0.20405735 0.01210526]\n [-0.00900257 0.01210526 0.01 ]]\n[[ 0.29616039 0.18683257 -0.01029261]\n [ 0.18683257 0.20190294 0.01321718]\n [-0.01029261 0.01321718 0.01 ]]\n[[ 0.26213425 0.16398739 -0.0095061 ]\n [ 0.16398739 0.17615455 0.0123562 ]\n [-0.0095061 0.0123562 0.01 ]]\n[[ 0.20360515 0.12292273 -0.00739887]\n [ 0.12292273 0.13014311 0.01040883]\n [-0.00739887 0.01040883 0.01 ]]\n[[ 0.22690022 0.14062579 -0.0088232 ]\n [ 0.14062579 0.14997652 0.01157397]\n [-0.0088232 0.01157397 0.01 ]]\n[[ 0.16837111 0.09956113 -0.00671597]\n [ 0.09956113 0.10396508 0.0096266 ]\n [-0.00671597 0.0096266 0.01 ]]\n[[ 0.13434497 0.07671595 -0.00592946]\n [ 0.07671595 0.07821669 0.00876562]\n [-0.00592946 0.00876562 0.01 ]]\n[[ 0.2627867 -0.25051937 -0.01585057]\n [-0.25051937 0.42602457 -0.01161294]\n [-0.01585057 -0.01161294 0.01 ]]\n[[ 0.17274991 -0.16184332 -0.01441662]\n [-0.16184332 0.34282478 -0.00830232]\n [-0.01441662 -0.00830232 0.01 ]]\n[[ 0.22456301 -0.01845399 -0.01967937]\n [-0.01845399 0.67479891 -0.00323099]\n [-0.01967937 -0.00323099 0.01 ]]\n[[ 0.20263872 -0.04316484 -0.01807229]\n [-0.04316484 0.55452167 -0.00363575]\n [-0.01807229 -0.00363575 0.01 ]]\n[[ 0.18664641 -0.0539778 -0.01493019]\n [-0.0539778 0.36882424 -0.00316825]\n [-0.01493019 -0.00316825 0.01 ]]\n[[ 0.21703643 -0.03353162 -0.01378258]\n [-0.03353162 0.32080884 -0.00156905]\n [-0.01378258 -0.00156905 0.01 ]]\n[[ 0.84859482 -0.08315561 -0.00784091]\n [-0.08315561 0.26264623 0.00900975]\n [-0.00784091 0.00900975 0.01 ]]\n[[ 0.6617281 -0.11873941 -0.00697031]\n [-0.11873941 0.23678183 0.00688732]\n [-0.00697031 0.00688732 0.01 ]]\n[[ 0.67496444 -0.18251048 -0.00519198]\n [-0.18251048 0.24885714 0.00703281]\n [-0.00519198 0.00703281 0.01 ]]\n[[ 0.41182693 -0.20905735 -0.00339847]\n [-0.20905735 0.24553215 0.00323158]\n [-0.00339847 0.00323158 0.01 ]]\n[[ 0.16498719 -0.17896089 -0.01621157]\n [-0.17896089 0.42960543 -0.00777667]\n [-0.01621157 -0.00777667 0.01 ]]\n[[ 0.2168003 -0.03557156 -0.02147433]\n [-0.03557156 0.76157956 -0.00270534]\n [-0.02147433 -0.00270534 0.01 ]]\n[[ 0.194876 -0.06028241 -0.01986724]\n [-0.06028241 0.64130232 -0.0031101 ]\n [-0.01986724 -0.0031101 0.01 ]]\n[[ 0.17888369 -0.07109537 -0.01672515]\n [-0.07109537 0.45560489 -0.00264261]\n [-0.01672515 -0.00264261 0.01 ]]\n[[ 0.20927372 -0.05064919 -0.01557753]\n [-0.05064919 0.40758949 -0.0010434 ]\n [-0.01557753 -0.0010434 0.01 ]]\n[[ 0.8408321 -0.10027318 -0.00963586]\n [-0.10027318 0.34942688 0.0095354 ]\n [-0.00963586 0.0095354 0.01 ]]\n[[ 0.65396538 -0.13585698 -0.00876526]\n [-0.13585698 0.32356248 0.00741297]\n [-0.00876526 0.00741297 0.01 ]]\n[[ 0.66720173 -0.19962805 -0.00698693]\n [-0.19962805 0.33563779 0.00755846]\n [-0.00698693 0.00755846 0.01 ]]\n[[ 0.40406422 -0.22617492 -0.00519342]\n [-0.22617492 0.3323128 0.00375723]\n [-0.00519342 0.00375723 0.01 ]]\n[[ 1.26763502e-01 5.31044952e-02 -2.00403698e-02]\n [ 5.31044952e-02 6.78379769e-01 6.05273094e-04]\n [-2.00403698e-02 6.05273094e-04 1.00000000e-02]]\n[[ 1.04839205e-01 2.83936454e-02 -1.84332853e-02]\n [ 2.83936454e-02 5.58102526e-01 2.00514106e-04]\n [-1.84332853e-02 2.00514106e-04 1.00000000e-02]]\n[[ 0.0888469 0.01758069 -0.01529119]\n [ 0.01758069 0.37240509 0.00066801]\n [-0.01529119 0.00066801 0.01 ]]\n[[ 0.11923692 0.03802686 -0.01414358]\n [ 0.03802686 0.3243897 0.00226722]\n [-0.01414358 0.00226722 0.01 ]]\n[[ 0.7507953 -0.01159712 -0.0082019 ]\n [-0.01159712 0.26622708 0.01284601]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecefd8f39a3ad3a8eb267389856871c396709579 | 1,794 | ipynb | Jupyter Notebook | Untitled.ipynb | robinhundt/cns-journal-club-u-net-cnn | d0754c6db18c6840e3e2838e05f7a3527c4a9606 | [
"MIT"
] | null | null | null | Untitled.ipynb | robinhundt/cns-journal-club-u-net-cnn | d0754c6db18c6840e3e2838e05f7a3527c4a9606 | [
"MIT"
] | null | null | null | Untitled.ipynb | robinhundt/cns-journal-club-u-net-cnn | d0754c6db18c6840e3e2838e05f7a3527c4a9606 | [
"MIT"
] | null | null | null | 18.121212 | 91 | 0.512263 | [
[
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"from keras.models import Sequential\n\nmodel = Sequential()",
"Using TensorFlow backend.\n"
],
[
"from keras.layers import Dense\nmodel.add(Dense(units=64, activation='relu', input_dim=100))\nmodel.add(Dense(units=10, activation='softmax'))",
"_____no_output_____"
],
[
"model.compile(loss='categorical_crossentropy', optimizer='sgd', metrics=['accuracy'])",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"a = np.eye(4)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecefe9eeac255ab02151ef8bd207ea7022098fc7 | 173,498 | ipynb | Jupyter Notebook | MIT-States playground.ipynb | patrickphatnguyen/TIRG-Vietnamese | 4df1558e578415db6102bc05a116fb5a78cbb7d1 | [
"Apache-2.0"
] | null | null | null | MIT-States playground.ipynb | patrickphatnguyen/TIRG-Vietnamese | 4df1558e578415db6102bc05a116fb5a78cbb7d1 | [
"Apache-2.0"
] | 1 | 2021-03-30T08:58:17.000Z | 2021-03-30T08:58:17.000Z | MIT-States playground.ipynb | patrickphat/TIRG-Vietnamese | 4df1558e578415db6102bc05a116fb5a78cbb7d1 | [
"Apache-2.0"
] | null | null | null | 113.545812 | 129,188 | 0.829024 | [
[
[
"%load_ext autoreload\n%autoreload 2",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"from main import load_dataset\nimport os \nclass opt_config:\n def __init__(self):\n self.dataset = \"mitstates_vn\"\n self.dataset_path = \"../data/MITStates/release_dataset\" \n self.model = \"tirg\" \n self.loss = \"soft_triplet\" \n self.comment = \"css3d_tirg\"\n self.embed_dim = 512\n self.learning_rate = 1e-2\n self.weight_decay = 1e-6 \n self.f = \"\"\n self.learning_rate_decay_frequency = 99999999\n self.batch_size = 32\n self.num_epochs = 100\n self.n_epochs_valudations = 5\n self.loader_num_workers = 4\n self.pretrained_weights = None\n \nopt = opt_config()\n\ntrainset, testset = load_dataset(opt)",
"Reading dataset mitstates_vn\n82732 test queries\ntrainset size: 43207\ntestset size: 10546\n"
],
[
"trainset.generate_random_query_target()\n",
"_____no_output_____"
],
[
"path = opt.dataset_path\n\nnouns = set()\nadjs = set()\n\nfor f in os.listdir(path + '/images'):\n if ' ' not in f:\n continue\n adj, noun = f.split()\n nouns.add(noun)\n adjs.add(adj)",
"_____no_output_____"
],
[
"len(nouns)",
"_____no_output_____"
],
[
"adjs",
"_____no_output_____"
],
[
"for adj in adjs:\n if adj not in vocab_dict.keys():\n print(adj)",
"brushed\nadj\n"
],
[
"vocab_dict = {\n \"ancient\":\"cổ xưa\",\n \"barren\":\"cằn cỗi\",\n \"bent\":\"cong vẹo\",\n \"blunt\":\"cùn\",\n \"bright\":\"sáng sủa\",\n \"broken\":\"hư hỏng\",\n \"browned\": \"chiên vàng\",\n \"brushed\": \"chải chuốt\",\n \"burnt\": \"thiêu đốt\",\n \"caramelized\":\"caramen hóa\",\n \"chipped\":\"sứt mẻ\",\n \"clean\":\"sạch sẽ\",\n \"clear\":\"thông thoáng\",\n \"closed\":\"đóng lại\",\n \"cloudy\":\"nhiều mây\",\n \"cluttered\":\"lộn xộn\",\n \"coiled\":\"uốn khúc\",\n \"cooked\":\"nấu chín\",\n \"cored\":\"bỏ hạt\",\n \"cracked\":\"nứt nẻ\",\n \"creased\": \"nhàu nát\",\n \"crinkled\": \"nhàu nát\",\n \"crumpled\": \"nhàu nát\",\n \"crushed\": \"nghiền nát\",\n \"curved\": \"uốn cong\",\n 'cut': \"cắt\",\n 'damp': \"ẩm ướt\",\n 'dark': \"tối\",\n 'deflated': \"xẹp xuống\",\n 'dented': \"móp\",\n 'diced': \"cắt vuông\",\n 'dirty': \"dơ bẩn\",\n 'draped': \"phủ lên\",\n 'dry': \"khô\",\n 'dull': \"ảm đạm\",\n 'empty': \"trống vắng\",\n 'engraved': \"điêu khắc\",\n 'eroded': \"xói mòn\",\n 'fallen': \"rơi\",\n 'filled': \"rót đầy\",\n 'foggy': \"sương mù\",\n 'folded': \"gấp lại\",\n 'frayed': \"sờn rách\",\n 'fresh': \"tươi mới\",\n 'frozen': \"đông lạnh\",\n 'full': \"lấp đầy\",\n 'grimy': \"dơ bẩn\",\n 'heavy': \"nặng nề\",\n 'huge': \"to lớn\",\n 'inflated': \"thổi phồng\",\n 'large': \"to lớn\",\n 'lightweight': \"nhẹ nhàng\",\n 'loose': \"lỏng lẻo\",\n 'mashed': \"nghiền nát\",\n 'melted': \"tan chảy\",\n 'modern': \"hiện đại\",\n 'moldy': \"mốc meo\",\n 'molten': \"nấu chảy\",\n 'mossy': \"rêu phong\",\n 'muddy': \"bùn lầy\",\n 'murky': \"âm u\",\n 'narrow': \"chất hẹp\",\n 'new': \"mới mẻ\",\n 'old': \"cũ kĩ\",\n 'open': \"mở\",\n 'painted': \"sơn màu\",\n 'peeled': \"bóc vỏ\",\n 'pierced': \"xỏ lỗ\",\n 'pressed': \"ép lại\",\n 'pureed': \"xảy nguyễn\",\n 'raw': \"còn sống\",\n 'ripe': \"chín muồi\",\n 'ripped': \"xé\",\n 'rough': \"sần sùi\",\n 'ruffled': \"nhăn nhúm\",\n 'runny': \"chảy nước\",\n 'rusty': \"gì sét\",\n 'scratched': \"trầy\",\n 'sharp': \"sắt nhọn\",\n 'shattered': \"bể tan\",\n 'shiny': \"sáng bóng\",\n 'short': \"thấp ngắn\",\n 'sliced': \"cắt lát\",\n 'small': \"nhỏ bé\",\n 'smooth': \"mượt mà\",\n 'spilled': \"chảy nước\",\n 'splintered': \"mảnh vụn\",\n 'squished': \"nhăn nheo\",\n 'standing': \"đứng thẳng\",\n 'steaming': \"hấp hơi\",\n 'straight': \"thẳng\",\n 'sunny': \"ánh nắng\",\n 'tall': \"cao\",\n 'thawed': \"rã đông\",\n 'thick': \"dày\",\n 'thin': \"mỏng\",\n 'tight': \"bó sát\",\n 'tiny': \"nhỏ bé\",\n 'toppled': \"làm ngã\",\n 'torn':\"rách nát\",\n 'unpainted': \"chưa sơn màu\",\n 'unripe': \"chưa chín\",\n 'upright': \"thẳng đứng\",\n 'verdant': \"xanh tươi\",\n 'viscous': \"nhầy nhớt\",\n 'weathered': \"phong hóa\",\n 'wet': \"ẩm ướt\",\n 'whipped': \"\",\n 'wide': \"rộng\",\n 'wilted': \"héo tàn\",\n 'windblown': \"gió thổi\",\n 'winding': \"quanh co\",\n 'worn': \"hao mòn\",\n 'wrinkled': \"nếp nhăn\",\n 'young': \"trẻ trung\"\n}",
"_____no_output_____"
],
[
"len(vocab_dict)",
"_____no_output_____"
],
[
"from torch_functions import UnNormalize\nfrom matplotlib import pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"rand = np.random.randint(10000)\nitem = trainset.__getitem__(rand)\nunorm = UnNormalize(mean=(0.485, 0.456, 0.406), std=(0.229, 0.224, 0.225))\n\nsource_img_data = unorm(item[\"source_img_data\"]).detach().numpy()\nsource_img_data = np.einsum(\"ijk->jki\",source_img_data)\ntarget_img_data = unorm(item[\"target_img_data\"]).detach().numpy()\ntarget_img_data = np.einsum(\"ijk->jki\",target_img_data)\nmod = item[\"mod\"][\"str\"]\nplt.subplot(1,2,1)\nplt.imshow(source_img_data)\nplt.subplot(1,2,2)\nplt.imshow(target_img_data)\nprint(mod)",
"xảy_nguyễn\n"
],
[
"trainset.get_all_texts()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
eceff5328bfe20d2a8c3354cea02440dc11c0ec0 | 146,347 | ipynb | Jupyter Notebook | Lesson04/.ipynb_checkpoints/Exercise31-checkpoint.ipynb | TrainingByPackt/Interactive-Data-Visualization-with-Python | 3113fb7cf7fd5b12cc79c448c2017a2f151bedd3 | [
"MIT"
] | 73 | 2019-08-22T01:43:11.000Z | 2022-03-25T03:12:02.000Z | Lesson04/.ipynb_checkpoints/Exercise31-checkpoint.ipynb | TrainingByPackt/Interactive-Data-Visualization-with-Python | 3113fb7cf7fd5b12cc79c448c2017a2f151bedd3 | [
"MIT"
] | 3 | 2019-11-02T18:02:59.000Z | 2019-12-05T04:18:42.000Z | Lesson04/.ipynb_checkpoints/Exercise31-checkpoint.ipynb | TrainingByPackt/Interactive-Data-Visualization-with-Python | 3113fb7cf7fd5b12cc79c448c2017a2f151bedd3 | [
"MIT"
] | 79 | 2019-08-16T10:46:50.000Z | 2022-03-22T04:04:41.000Z | 32.27768 | 154 | 0.453299 | [
[
[
"import altair as alt\nimport pandas as pd",
"_____no_output_____"
],
[
"hpi_url = \"https://raw.githubusercontent.com/TrainingByPackt/Interactive-Data-Visualization-with-Python/master/datasets/hpi_data_countries.tsv\"\n#read it into a DataFrame using pandas\nhpi_df = pd.read_csv(hpi_url, sep='\\t')",
"_____no_output_____"
],
[
"# multiple altair charts placed one after the other\nchart = alt.Chart(hpi_df).mark_circle().encode(\ny='Happy Planet Index',\ncolor='Region:N'\n)\nchart1 = chart.encode(x='Wellbeing (0-10)')\nchart2 = chart.encode(x='Life Expectancy (years)')\nalt.vconcat(chart1, chart2)",
"_____no_output_____"
],
[
"# multiple altair charts placed horizontally next to each other\nchart = alt.Chart(hpi_df).mark_circle().encode(\ny='Happy Planet Index',\ncolor='Region:N'\n)\nchart1 = chart.encode(x='Wellbeing (0-10)')\nchart2 = chart.encode(x='Life Expectancy (years)')\nalt.hconcat(chart1, chart2)",
"_____no_output_____"
],
[
"# hover and tooltip across multiple charts\nselected_area = alt.selection_interval()\nchart = alt.Chart(hpi_df).mark_circle().encode(\ny='Happy Planet Index',\ncolor=alt.condition(selected_area, 'Region', alt.\nvalue('lightgray'))\n).add_selection(\nselected_area\n)\nchart1 = chart.encode(x='Wellbeing (0-10)')\nchart2 = chart.encode(x='Life Expectancy (years)')\nchart1 | chart2",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecf005a348b70aa8eebfe8e5cc58ae8e009092f2 | 203,548 | ipynb | Jupyter Notebook | CONTENT/PYTHON/_Jupyter-notebooks/4-Visualization-with-Matplotlib.ipynb | impastasyndrome/DS-ALGO-OFFICIAL | c85ec9cf0af0009f038b7a571a7ac1fb466b7f3a | [
"Apache-2.0"
] | 11 | 2021-02-18T04:53:44.000Z | 2022-01-16T10:57:39.000Z | CONTENT/PYTHON/_Jupyter-notebooks/4-Visualization-with-Matplotlib.ipynb | impastasyndrome/DS-ALGO-OFFICIAL | c85ec9cf0af0009f038b7a571a7ac1fb466b7f3a | [
"Apache-2.0"
] | 162 | 2021-03-09T01:52:11.000Z | 2022-03-12T01:09:07.000Z | CONTENT/PYTHON/_Jupyter-notebooks/4-Visualization-with-Matplotlib.ipynb | impastasyndrome/DS-ALGO-OFFICIAL | c85ec9cf0af0009f038b7a571a7ac1fb466b7f3a | [
"Apache-2.0"
] | 8 | 2021-02-18T05:12:34.000Z | 2022-03-06T19:02:14.000Z | 163.361156 | 32,638 | 0.899552 | [
[
[
"ISRC Python Workshop: Visualization with Matplotlib\n\n___Visualization with Matplotlib and Seaborn___",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"@author: Zhiya Zuo\n\n@email: [email protected]",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"The following imports the main module in package `matplotlib`. Note that `%matplotlib inline` means we want the plot to be inline in our notebook instead of getting a pop-up windows.",
"_____no_output_____"
]
],
[
[
"from matplotlib import pyplot as plt\r\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"<hr>",
"_____no_output_____"
],
[
"#### Introduction",
"_____no_output_____"
],
[
"Visualization is one of the most important things of data analysis. Besides just producing ___readable___ plots, we should make an effort to improve the overall attractiveness of the plots. `matplotlib` is a powerful package for ___Python___ users. Let's start with an example.",
"_____no_output_____"
],
[
"First we generate sample data",
"_____no_output_____"
]
],
[
[
"X_arr = np.arange(10)\r\nY_arr = 3*X_arr + 2 + np.random.random(size=X_arr.size) # linear with some noise\r\nprint(X_arr)\r\nprint(Y_arr)",
"[0 1 2 3 4 5 6 7 8 9]\n[ 2.01486134 5.25099205 8.51321917 11.03441301 14.57193933 17.29545064\n 20.35492655 23.4540255 26.15927674 29.59403265]\n"
]
],
[
[
"To plot a simple scatter plot, we can use `plt.scatter()` function",
"_____no_output_____"
]
],
[
[
"plt.scatter(X_arr, Y_arr)",
"_____no_output_____"
]
],
[
[
"See how easy this is? However, this is still too simple - we do not even have X or Y axis labels. Further we may want to change the scatter markers. Let's get a better plot!",
"_____no_output_____"
]
],
[
[
"# Use `+` as marker; color set as `g` (green); size proportion to Y values\nplt.scatter(X_arr, Y_arr, marker='+', c='g', s=Y_arr*10) \n# How about adding a line to it? Let's use `plt.plot()`\n# set line style to dashed; color as `k` (black) \nplt.plot(X_arr, Y_arr, linestyle='dashed', color='k')\n# set x/y axis limits: first two are xlow and xhigh; last two are ylow and yhigh\nplt.axis([0, 10, 0, 35])\n# set x/y labels\nplt.xlabel('My X Axis')\nplt.ylabel('My Y Axis')\n# set title\nplt.title('My First Plot')",
"_____no_output_____"
]
],
[
[
"##### Anatomy of a Figure",
"_____no_output_____"
],
[
"Before we go deeper, let's take a look at the structure of a figure in `matplotlib`:\n\n<img width=700 src=\"https://matplotlib.org/_images/anatomy1.png\">",
"_____no_output_____"
],
[
"As you can see, we have done things for many of the elements. There's no need to memorize them, though. We can always Google and find information on specific parts that we want to update.",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"#### Coding style (personal recommendation)",
"_____no_output_____"
],
[
"I myself prefer the following way of getting a figure with `matplotlib`:",
"_____no_output_____"
]
],
[
[
"# `plt.subplots()` returns a figure object (which is the whole thing as shown above)\n# and `axes` that control specific plots in the figure.\n# Here our \"subplots\" layout is by default 1 row and 1 col and therefore 1 plot\nfig, ax = plt.subplots()\n# Setting figure size\nfig.set_figwidth(6)\nfig.set_figheight(5)\n# plot should be done on the `axis`: ax\nax.plot(X_arr, Y_arr)",
"_____no_output_____"
]
],
[
[
"Applying what we did earlier:",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots()\nfig.set_figwidth(6)\nfig.set_figheight(5)\n# What we just did, applying to `ax`\nax.scatter(X_arr, Y_arr, marker='+', c='g', s=Y_arr*10) \nax.plot(X_arr, Y_arr, linestyle='dashed', color='k')\nax.axis([0, 10, 0, 35])\nax.set_xlabel('My X Axis')\nax.set_ylabel('My Y Axis')\nax.set_title('My First Plot')",
"_____no_output_____"
]
],
[
[
"This is especially useful when handling multiple plots in one figure. Let's use an example of 4 plots: 2x2 layout",
"_____no_output_____"
]
],
[
[
"# Now the returned `ax` would be array with a shape a 2x2\nfig, ax_arr = plt.subplots(nrows=2, ncols=2)\nax_arr.shape",
"_____no_output_____"
]
],
[
[
"We can plot in each of them by adding `plot` to each of the `axes` using a `for` loop. (In reality we should plot different things in different subplots)",
"_____no_output_____"
]
],
[
[
"# Now the returned `ax` would be array with a shape a 2x2\nfig, ax_arr = plt.subplots(nrows=2, ncols=2)\nfor ax_row in ax_arr:\n for ax in ax_row:\n ax.plot(X_arr, Y_arr)",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"#### Different plots",
"_____no_output_____"
],
[
"Instead of the simpletst scatter/line plots, there are of course other options.",
"_____no_output_____"
],
[
"##### Histogram",
"_____no_output_____"
],
[
"Let's use a Gaussian distribution for illustration",
"_____no_output_____"
]
],
[
[
"mu, sigma = 15, 1\ngaussian_arr = np.random.normal(mu, sigma, size=10000)\nnp.mean(gaussian_arr), np.std(gaussian_arr, ddof=1)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\n# `hist()` will return something but we usually do not need.\nfreq_arr, bin_arr, _ = ax.hist(gaussian_arr)",
"_____no_output_____"
]
],
[
[
"We can actually customize and make it prettier",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots()\n# Facecolor set to green; transparency (`alpha`) level: 30%\nfreq_arr, bin_arr, _ = ax.hist(gaussian_arr, facecolor='g', alpha=0.3)\n# Add grid\nax.grid()",
"_____no_output_____"
]
],
[
[
"##### Boxplot",
"_____no_output_____"
],
[
"With the guaussian data, we can use a different form: box plot",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots()\nax.boxplot(gaussian_arr, \n vert=False, # verticle\n showfliers=False, # do not show outliers\n showmeans=True, # show the mean\n labels=['Gaussian'] # group name (label)\n )",
"_____no_output_____"
]
],
[
[
"##### Bar chart",
"_____no_output_____"
],
[
"Random data for bar chart input:",
"_____no_output_____"
]
],
[
[
"bar_arr = np.array(['Spring', 'Summer', 'Fall', 'Winder'])\nfreq_arr = np.random.randint(0, 100, 4) # generate 4 random integers for bar chart\nfreq_arr",
"_____no_output_____"
]
],
[
[
"We can also add error bars:",
"_____no_output_____"
]
],
[
[
"yerr_arr = np.random.randint(5, 10, 4)\nyerr_arr",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nax.bar(bar_arr, freq_arr, # X and Y\n yerr = yerr_arr, # error bars\n color='gold',\n )",
"_____no_output_____"
]
],
[
[
"##### Group these plots together!",
"_____no_output_____"
]
],
[
[
"fig, ax_arr = plt.subplots(nrows=2, ncols=2, \n # let their scales be different\n sharex=False, sharey=False)\nfig.set_figwidth(12)\nfig.set_figheight(8)\n# set global title\nfig.suptitle(\"My first subplots\")\n\n## first\nax_arr[0, 0].scatter(X_arr, Y_arr, marker='+', c='g', s=Y_arr*10) \nax_arr[0, 0].plot(X_arr, Y_arr, linestyle='dashed', color='k')\nax_arr[0, 0].axis([0, 10, 0, 35])\nax_arr[0, 0].set_title('My First Plot')\n## second \nax_arr[0, 1].hist(gaussian_arr, facecolor='g', alpha=0.3)\nax_arr[0, 1].set_title('Histogram')\n## third\nax_arr[1, 0].boxplot(gaussian_arr, vert=False, showfliers=False,\n showmeans=True, labels=['Gaussian'])\nax_arr[1, 0].set_title('Box plot')\n## last one\nax_arr[1,1].bar(bar_arr, freq_arr, \n yerr = yerr_arr, color='gold')\nax_arr[1, 1].set_title('Bar chart')",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"#### Seaborn: Made life easier",
"_____no_output_____"
],
[
"Finally, I would like to introduce [`seaborn`](https://seaborn.pydata.org/). This package really helps me get some good-looking plots for journal/conference submissions. Specifically, I would like to show you two examples.",
"_____no_output_____"
]
],
[
[
"import seaborn as sns",
"_____no_output_____"
]
],
[
[
"##### Example 1: Heatmap",
"_____no_output_____"
],
[
"I would use the example of correlataion matrix. First I load a sample data `iris`",
"_____no_output_____"
]
],
[
[
"df = sns.load_dataset('iris')\ndf.head()",
"_____no_output_____"
]
],
[
[
"Let's get the correlation between the first 3 columns:",
"_____no_output_____"
]
],
[
[
"corr_df = df[df.columns[:3]].corr()\ncorr_df",
"_____no_output_____"
]
],
[
[
"Getting a heatmap in `seaborn` is very straightfoward:",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots()\nfig.set_figwidth(8)\nfig.set_figheight(7)\nsns.heatmap(corr_df, \n annot=True, # show the values\n fmt=\".2f\", # format of the value\n cmap=sns.light_palette(\"#F4D03F\", as_cmap=True), # color map to be used \n ax=ax # plot on the `ax` we just instaniated\n )",
"_____no_output_____"
]
],
[
[
"##### Example 2: Linear regression",
"_____no_output_____"
],
[
"Using the same dataset, let's plot the bivariate regression between sepal length and width for different species:",
"_____no_output_____"
]
],
[
[
"sns.lmplot(x=\"sepal_length\", y=\"sepal_width\", \n data=df, # the data to be used\n col=\"species\", # seperate data into column based on `species`\n hue=\"species\", # color for each subplot (facet)\n col_wrap=3, # wrap into a new row for every 2 columns\n ci=None, # do not show confidence interval\n palette=\"muted\", # color palette to use\n size=4, # size of each subplot (facet)\n )",
"_____no_output_____"
]
],
[
[
"Note that `lmplot()` is a wrapper of ['FacetGrid'](https://seaborn.pydata.org/generated/seaborn.FacetGrid.html) so we cannot set the `axes` for this figure.",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"#### Some helpful tutorials...\n\nFinally, I list some tutorials that I think will greatly benefit the understanding of the packages mentioend above:\n\n- https://matplotlib.org/users/pyplot_tutorial.html\n- https://matplotlib.org/faq/usage_faq.html#parts-of-a-figure\n- https://seaborn.pydata.org/examples/index.html",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ecf00630b54598c4c00f6a5fe0ae9befbaf5b1b7 | 420,853 | ipynb | Jupyter Notebook | PyCitySchools_Challenge.ipynb | ayyray/School_District_Analysis | f90e2cdd86d2dad7be5dc49958748a93df013453 | [
"MIT"
] | null | null | null | PyCitySchools_Challenge.ipynb | ayyray/School_District_Analysis | f90e2cdd86d2dad7be5dc49958748a93df013453 | [
"MIT"
] | null | null | null | PyCitySchools_Challenge.ipynb | ayyray/School_District_Analysis | f90e2cdd86d2dad7be5dc49958748a93df013453 | [
"MIT"
] | null | null | null | 33.920609 | 201 | 0.397471 | [
[
[
"## Importing Data ",
"_____no_output_____"
]
],
[
[
"# Dependencies and Setup\nimport pandas as pd\n\n# File to Load (Remember to change the path if needed.)\nschool_data_to_load = \"Resources/schools_complete.csv\"\nstudent_data_to_load = \"Resources/students_complete.csv\"\n\n# Read the School Data and Student Data and store into a Pandas DataFrame\nschool_data_df = pd.read_csv(school_data_to_load)\nstudent_data_df = pd.read_csv(student_data_to_load)\n\n# Cleaning Student Names and Replacing Substrings in a Python String\n# Add each prefix and suffix to remove to a list.\nprefixes_suffixes = [\"Dr. \", \"Mr. \",\"Ms. \", \"Mrs. \", \"Miss \", \" MD\", \" DDS\", \" DVM\", \" PhD\"]\n\n# Iterate through the words in the \"prefixes_suffixes\" list and replace them with an empty space, \"\".\nfor word in prefixes_suffixes:\n student_data_df[\"student_name\"] = student_data_df[\"student_name\"].str.replace(word,\"\")\n\n# Check names.\nstudent_data_df.head(10)",
"/var/folders/8s/_5tkv_c11cxd85qkch7sw7hm0000gn/T/ipykernel_3262/3904321406.py:18: FutureWarning: The default value of regex will change from True to False in a future version.\n student_data_df[\"student_name\"] = student_data_df[\"student_name\"].str.replace(word,\"\")\n"
]
],
[
[
"## Deliverable 1: Replace the reading and math scores.\n\n### Replace the 9th grade reading and math scores at Thomas High School with NaN",
"_____no_output_____"
]
],
[
[
"# Install numpy using conda install numpy or pip install numpy. \n# Step 1. Import numpy as np.\nimport numpy as np",
"_____no_output_____"
],
[
"# Step 2. Use the loc method on the student_data_df to select all the reading scores from the 9th grade at Thomas High School and replace them with NaN.\nstudent_data_df.loc[ (student_data_df[\"school_name\"] == \"Thomas High School\") \n & (student_data_df[\"grade\"]== \"9th\") \n & (student_data_df[\"reading_score\"] > 0), \"reading_score\"] = np.nan ",
"_____no_output_____"
],
[
"# Step 3. Refactor the code in Step 2 to replace the math scores with NaN.\nstudent_data_df.loc[ (student_data_df[\"school_name\"] == \"Thomas High School\") \n & (student_data_df[\"grade\"]== \"9th\") \n & (student_data_df[\"math_score\"] > 0), \"math_score\"] = np.nan ",
"_____no_output_____"
],
[
"# Step 4. Check the student data for NaN's. ",
"_____no_output_____"
],
[
"student_data_df.tail(10)",
"_____no_output_____"
]
],
[
[
"## Deliverable 2 : Repeat the school district analysis\n\n\n### District Summary",
"_____no_output_____"
]
],
[
[
"# Combine the data into a single dataset\nschool_data_complete_df = pd.merge(student_data_df, school_data_df, how=\"right\", on=[\"school_name\", \"school_name\"])\nschool_data_complete_df.head()",
"_____no_output_____"
],
[
"# Calculate the Totals (Schools and Students)\nschool_count = len(school_data_complete_df[\"school_name\"].unique())\nstudent_count = school_data_complete_df[\"Student ID\"].count()\n\n# Calculate the Total Budget\ntotal_budget = school_data_df[\"budget\"].sum()",
"_____no_output_____"
],
[
"# Calculate the Average Scores using the \"clean_student_data\".\naverage_reading_score = school_data_complete_df[\"reading_score\"].mean()\naverage_math_score = school_data_complete_df[\"math_score\"].mean()",
"_____no_output_____"
],
[
"# Step 1. Get the number of students that are in ninth grade at Thomas High School.\n# These students have no grades. \n\nninth_THS_count = school_data_complete_df.loc[ (school_data_complete_df[\"school_name\"] == \"Thomas High School\") \n & (school_data_complete_df[\"grade\"] == \"9th\"), \"Student ID\"].count() \n# Get the total student count \nstudent_count = school_data_complete_df[\"Student ID\"].count()\n\n\n# Step 2. Subtract the number of students that are in ninth grade at \n# Thomas High School from the total student count to get the new total student count.\nnew_school_count = student_count - ninth_THS_count\nnew_school_count",
"_____no_output_____"
],
[
"# Calculate the passing rates using the \"clean_student_data\".\npassing_math_count = school_data_complete_df[(school_data_complete_df[\"math_score\"] >= 70)].count()[\"student_name\"]\npassing_reading_count = school_data_complete_df[(school_data_complete_df[\"reading_score\"] >= 70)].count()[\"student_name\"]",
"_____no_output_____"
],
[
"# Step 3. Calculate the passing percentages with the new total student count.\nnew_passing_math_percentage = passing_math_count / float(new_school_count) * 100\nnew_passing_reading_percentage = passing_reading_count / float(new_school_count) * 100\nprint(new_passing_reading_percentage)\nprint(new_passing_math_percentage)",
"85.6596657108166\n74.76039164018704\n"
],
[
"# Calculate the students who passed both reading and math.\npassing_math_reading = school_data_complete_df[(school_data_complete_df[\"math_score\"] >= 70)\n & (school_data_complete_df[\"reading_score\"] >= 70)]\n\n# Calculate the number of students that passed both reading and math.\noverall_passing_math_reading_count = passing_math_reading[\"student_name\"].count()\n\n\n# Step 4.Calculate the overall passing percentage with new total student count.\nnew_overall_passing_percentage = overall_passing_math_reading_count / new_school_count * 100\nprint(new_overall_passing_percentage)",
"64.85571830840374\n"
],
[
"# Create a DataFrame\ndistrict_summary_df = pd.DataFrame(\n [{\"Total Schools\": school_count, \n \"Total Students\": student_count, \n \"Total Budget\": total_budget,\n \"Average Math Score\": average_math_score, \n \"Average Reading Score\": average_reading_score,\n \"% Passing Math\": new_passing_math_percentage,\n \"% Passing Reading\": new_passing_reading_percentage,\n \"% Overall Passing\": new_overall_passing_percentage}])\n\n\n\n# Format the \"Total Students\" to have the comma for a thousands separator.\ndistrict_summary_df[\"Total Students\"] = district_summary_df[\"Total Students\"].map(\"{:,}\".format)\n# Format the \"Total Budget\" to have the comma for a thousands separator, a decimal separator and a \"$\".\ndistrict_summary_df[\"Total Budget\"] = district_summary_df[\"Total Budget\"].map(\"${:,.2f}\".format)\n# Format the columns.\ndistrict_summary_df[\"Average Math Score\"] = district_summary_df[\"Average Math Score\"].map(\"{:.1f}\".format)\ndistrict_summary_df[\"Average Reading Score\"] = district_summary_df[\"Average Reading Score\"].map(\"{:.1f}\".format)\ndistrict_summary_df[\"% Passing Math\"] = district_summary_df[\"% Passing Math\"].map(\"{:.1f}\".format)\ndistrict_summary_df[\"% Passing Reading\"] = district_summary_df[\"% Passing Reading\"].map(\"{:.1f}\".format)\ndistrict_summary_df[\"% Overall Passing\"] = district_summary_df[\"% Overall Passing\"].map(\"{:.1f}\".format)\n\n# Display the data frame\ndistrict_summary_df.head()",
"_____no_output_____"
]
],
[
[
"### School Summary",
"_____no_output_____"
]
],
[
[
"# Determine the School Type\nper_school_types = school_data_df.set_index([\"school_name\"])[\"type\"]\n\n# Calculate the total student count.\nper_school_counts = school_data_complete_df[\"school_name\"].value_counts()\n\n# Calculate the total school budget and per capita spending\nper_school_budget = school_data_complete_df.groupby([\"school_name\"]).mean()[\"budget\"]\n# Calculate the per capita spending.\nper_school_capita = per_school_budget / per_school_counts\n\n# Calculate the average test scores.\nper_school_math = school_data_complete_df.groupby([\"school_name\"]).mean()[\"math_score\"]\nper_school_reading = school_data_complete_df.groupby([\"school_name\"]).mean()[\"reading_score\"]\n\n# Calculate the passing scores by creating a filtered DataFrame.\nper_school_passing_math = school_data_complete_df[(school_data_complete_df[\"math_score\"] >= 70)]\nper_school_passing_reading = school_data_complete_df[(school_data_complete_df[\"reading_score\"] >= 70)]\n\n# Calculate the number of students passing math and passing reading by school.\nper_school_passing_math = per_school_passing_math.groupby([\"school_name\"]).count()[\"student_name\"]\nper_school_passing_reading = per_school_passing_reading.groupby([\"school_name\"]).count()[\"student_name\"]\n\n# Calculate the percentage of passing math and reading scores per school.\nper_school_passing_math = per_school_passing_math / per_school_counts * 100\nper_school_passing_reading = per_school_passing_reading / per_school_counts * 100\n\n# Calculate the students who passed both reading and math.\nper_passing_math_reading = school_data_complete_df[(school_data_complete_df[\"reading_score\"] >= 70)\n & (school_data_complete_df[\"math_score\"] >= 70)]\n\n# Calculate the number of students passing math and passing reading by school.\nper_passing_math_reading = per_passing_math_reading.groupby([\"school_name\"]).count()[\"student_name\"]\n\n# Calculate the percentage of passing math and reading scores per school.\nper_overall_passing_percentage = per_passing_math_reading / per_school_counts * 100",
"_____no_output_____"
],
[
"# Create the DataFrame\nper_school_summary_df = pd.DataFrame({\n \"School Type\": per_school_types,\n \"Total Students\": per_school_counts,\n \"Total School Budget\": per_school_budget,\n \"Per Student Budget\": per_school_capita,\n \"Average Math Score\": per_school_math,\n \"Average Reading Score\": per_school_reading,\n \"% Passing Math\": per_school_passing_math,\n \"% Passing Reading\": per_school_passing_reading,\n \"% Overall Passing\": per_overall_passing_percentage})\n\n\nper_school_summary_df",
"_____no_output_____"
],
[
"# Format the Total School Budget and the Per Student Budget\nper_school_summary_df[\"Total School Budget\"] = per_school_summary_df[\"Total School Budget\"].map(\"${:,.2f}\".format)\nper_school_summary_df[\"Per Student Budget\"] = per_school_summary_df[\"Per Student Budget\"].map(\"${:,.2f}\".format)\n\n# Display the data frame\n\n",
"_____no_output_____"
],
[
"# Step 5. Get the number of 10th-12th graders from Thomas High School (THS).\nTHS_10_12_graders = student_data_df.loc[(student_data_df[\"school_name\"] == \"Thomas High School\")\n & ((student_data_df[\"grade\"] == \"10th\") | (student_data_df[\"grade\"] == \"11th\") | (student_data_df[\"grade\"] == \"12th\"))][\"grade\"].count()\nTHS_10_12_graders\n\n",
"_____no_output_____"
],
[
"# Step 6. Get all the students passing math from THS\nTHS_passing_math_df = student_data_df.loc[(student_data_df[\"math_score\"] >= 70)\n & (student_data_df[\"school_name\"] == \"Thomas High School\") \n & ((student_data_df[\"grade\"] == \"10th\") | (student_data_df[\"grade\"] == \"11th\") | (student_data_df[\"grade\"] == \"12th\"))].count()\nTHS_passing_math_df",
"_____no_output_____"
],
[
"# Step 7. Get all the students passing reading from THS\nTHS_passing_reading_df = student_data_df.loc[(student_data_df[\"reading_score\"] >= 70)\n & (student_data_df[\"school_name\"] == \"Thomas High School\") \n & ((student_data_df[\"grade\"] == \"10th\") | (student_data_df[\"grade\"] == \"11th\") | (student_data_df[\"grade\"] == \"12th\"))].count()\nTHS_passing_reading_df",
"_____no_output_____"
],
[
"# Step 8. Get all the students passing math and reading from THS\nTHS_math_reading_passing_df = student_data_df.loc[(student_data_df[\"math_score\"] >= 70)\n & (student_data_df[\"reading_score\"] >= 70)\n & (student_data_df[\"school_name\"] == \"Thomas High School\") \n & ((student_data_df[\"grade\"] == \"10th\") | (student_data_df[\"grade\"] == \"11th\") | (student_data_df[\"grade\"] == \"12th\"))].count()\nTHS_math_reading_passing_df",
"_____no_output_____"
],
[
"# Step 9. Calculate the percentage of 10th-12th grade students passing math from Thomas High School. \nTHS_passing_math_percentage = THS_passing_math_df[\"math_score\"]/THS_10_12_graders *100\nTHS_passing_math_percentage",
"_____no_output_____"
],
[
"# Step 10. Calculate the percentage of 10th-12th grade students passing reading from Thomas High School.\nTHS_passing_reading_percentage = THS_passing_reading_df[\"reading_score\"]/THS_10_12_graders *100\nTHS_passing_reading_percentage",
"_____no_output_____"
],
[
"# Step 11. Calculate the overall passing percentage of 10th-12th grade from Thomas High School. \nTHS_overall_passing_percentage = THS_math_reading_passing_df[\"student_name\"]/THS_10_12_graders *100\nTHS_overall_passing_percentage",
"_____no_output_____"
],
[
"# Step 12. Replace the passing math percent for Thomas High School in the per_school_summary_df.\nper_school_summary_df.loc[[\"Thomas High School\"],[\"% Passing Math\"]] = THS_passing_math_percentage\nTHS_passing_math_percentage\n",
"_____no_output_____"
],
[
"# Step 13. Replace the passing reading percentage for Thomas High School in the per_school_summary_df.\nper_school_summary_df.loc[[\"Thomas High School\"],[\"% Passing Reading\"]] = THS_passing_reading_percentage\nTHS_passing_reading_percentage",
"_____no_output_____"
],
[
"# Step 14. Replace the overall passing percentage for Thomas High School in the per_school_summary_df.\nper_school_summary_df.loc[[\"Thomas High School\"],[\"% Overall Passing\"]] = THS_overall_passing_percentage\nTHS_overall_passing_percentage",
"_____no_output_____"
],
[
"per_school_summary_df.tail(3)\n",
"_____no_output_____"
]
],
[
[
"### High and Low Performing Schools",
"_____no_output_____"
]
],
[
[
"# Sort and show top five schools.\ntop_5_schools = per_school_summary_df.sort_values([\"% Overall Passing\"], ascending=False)\ntop_5_schools.head(5)",
"_____no_output_____"
],
[
"# Sort and show bottom five schools.\nbottom_5_schools = per_school_summary_df.sort_values([\"% Overall Passing\"], ascending=True)\nbottom_5_schools.tail(5)",
"_____no_output_____"
]
],
[
[
"### Math and Reading Scores by Grade",
"_____no_output_____"
]
],
[
[
"# Create a Series of scores by grade levels using conditionals.\nnew_ninth_graders = school_data_complete_df[(school_data_complete_df[\"grade\"] == \"9th\")]\ntenth_graders = school_data_complete_df[(school_data_complete_df[\"grade\"] == \"10th\")]\neleventh_graders = school_data_complete_df[(school_data_complete_df[\"grade\"] == \"11th\")]\ntwelth_graders = school_data_complete_df[(school_data_complete_df[\"grade\"] == \"12th\")]\n\n# Group each school Series by the school name for the average math score.\nnew_ninth_graders_math_scores = new_ninth_graders.groupby([\"school_name\"]).mean()[\"math_score\"]\ntenth_graders_math_scores = tenth_graders.groupby([\"school_name\"]).mean()[\"math_score\"]\neleventh_graders_math_scores = eleventh_graders.groupby([\"school_name\"]).mean()[\"math_score\"]\ntwelth_graders_math_scores = twelth_graders.groupby([\"school_name\"]).mean()[\"math_score\"]\n\n# Group each school Series by the school name for the average reading score.\nnew_ninth_graders_reading_scores = new_ninth_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]\ntenth_graders_reading_scores = tenth_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]\neleventh_graders_reading_scores = eleventh_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]\ntwelth_graders_reading_scores = twelth_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]\n\n#NEW_ninth_graders_reading_scores\n#NEW_ninth_graders\n\n\n",
"_____no_output_____"
],
[
"# Combine each Series for average math scores by school into single data frame.\nnew_math_scores_by_grade = pd.DataFrame({\n \"9th\":new_ninth_graders_math_scores,\n \"10th\":tenth_graders_math_scores, \n \"11th\":eleventh_graders_math_scores,\n \"12th\":twelth_graders_math_scores\n})\nnew_math_scores_by_grade",
"_____no_output_____"
],
[
"# Combine each Series for average reading scores by school into single data frame.\nnew_reading_scores_by_grade = pd.DataFrame({\n \"9th\":new_ninth_graders_reading_scores,\n \"10th\":tenth_graders_reading_scores, \n \"11th\":eleventh_graders_reading_scores,\n \"12th\":twelth_graders_reading_scores\n})\nnew_reading_scores_by_grade",
"_____no_output_____"
],
[
"# Format each grade column.\nnew_math_scores_by_grade[\"9th\"] = new_math_scores_by_grade[\"9th\"].map(\"{:.1f}\".format)\nnew_math_scores_by_grade[\"10th\"] = new_math_scores_by_grade[\"10th\"].map(\"{:.1f}\".format)\nnew_math_scores_by_grade[\"11th\"] = new_math_scores_by_grade[\"11th\"].map(\"{:.1f}\".format)\nnew_math_scores_by_grade[\"12th\"] = new_math_scores_by_grade[\"12th\"].map(\"{:.1f}\".format)\nnew_math_Scores_by_grade = new_math_scores_by_grade[[\"9th\", \"10th\", \"11th\", \"12th\"]]\n#new_math_scores_by_grade = math_scores_by_grade[\n #[\"9th\", \"10th\", \"11th\", \"12th\"]]\n\n#new_math_scores_by_grade.index.name= None (below)\n\nnew_reading_scores_by_grade[\"9th\"] = new_reading_scores_by_grade[\"9th\"].map(\"{:.1f}\".format)\nnew_reading_scores_by_grade[\"10th\"] = new_reading_scores_by_grade[\"10th\"].map(\"{:.1f}\".format)\nnew_reading_scores_by_grade[\"11th\"] = new_reading_scores_by_grade[\"11th\"].map(\"{:.1f}\".format)\nnew_reading_scores_by_grade[\"12th\"] = new_reading_scores_by_grade[\"12th\"].map(\"{:.1f}\".format)\nnew_reading_Scores_by_grade = new_reading_scores_by_grade[[\"9th\", \"10th\", \"11th\", \"12th\"]]\n#new_reading_scores_by_grade = reading_scores_by_grade[\n #[\"9th\", \"10th\", \"11th\", \"12th\"]]\n\n#new_reading_scores_by_grade.index.name = None (below)\n\n\n",
"_____no_output_____"
],
[
"# Remove the index.\nnew_math_scores_by_grade = new_math_scores_by_grade[\n [\"9th\", \"10th\", \"11th\", \"12th\"]]\n\nnew_math_scores_by_grade.index.name= None\n\n# Display the data frame\nnew_math_scores_by_grade",
"_____no_output_____"
],
[
"## Remove the index.\nnew_reading_scores_by_grade = new_reading_scores_by_grade[\n [\"9th\", \"10th\", \"11th\", \"12th\"]]\n\nnew_reading_scores_by_grade.index.name = None\n\n\n# Display the data frame\nnew_reading_scores_by_grade",
"_____no_output_____"
]
],
[
[
"### Scores by School Spending",
"_____no_output_____"
]
],
[
[
"# Establish the spending bins and group names.\nspending_bins = [0, 585, 630, 645, 675]\ngroup_names = [\"<$584\", \"$585-$629\", \"$630-$644\", \"$645-$675\"]\n\n# Categorize spending based on the bins.\nper_school_summary_df[\"Spending Ranges (Per Student)\"] = pd.cut(per_school_capita, spending_bins, labels=group_names)\nper_school_summary_df",
"_____no_output_____"
],
[
"# Calculate averages for the desired columns. \nspending_math_scores = per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"Average Math Score\"]\nspending_reading_scores = per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"Average Reading Score\"]\nspending_passing_math_scores = per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"% Passing Math\"]\nspending_passing_reading_scores = per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"% Passing Reading\"]\noverall_passing_spending = per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"% Overall Passing\"]\noverall_passing_spending",
"_____no_output_____"
],
[
"# Create the DataFrame\nspending_summary_df = pd.DataFrame({\n \"Average Math Score\": spending_math_scores,\n \"Average Reading Score\": spending_reading_scores,\n \"% Passing Math\": spending_passing_math_scores,\n \"% Passing Reading\": spending_passing_reading_scores,\n \"% Overall Passing\": overall_passing_spending\n})\nspending_summary_df",
"_____no_output_____"
],
[
"# Format the DataFrame \nspending_summary_df[\"Average Math Score\"] = spending_summary_df[\"Average Math Score\"].map(\"{:.1f}\".format)\nspending_summary_df[\"Average Reading Score\"] = spending_summary_df[\"Average Reading Score\"].map(\"{:.1f}\".format)\nspending_summary_df[\"% Passing Math\"] = spending_summary_df[\"% Passing Math\"].map(\"{:.0f}\".format)\nspending_summary_df[\"% Passing Reading\"] = spending_summary_df[\"% Passing Reading\"].map(\"{:.0f}\".format)\nspending_summary_df[\"% Overall Passing\"] = spending_summary_df[\"% Overall Passing\"].map(\"{:.0f}\".format)\nspending_summary_df",
"_____no_output_____"
]
],
[
[
"### Scores by School Size",
"_____no_output_____"
]
],
[
[
"# Establish the bins.\nsize_bins = [0,1000,2000,5000]\ngroup_names = [\"Small (<1000)\", \"Medium (1000-2000)\", \"Large (2000-5000)\"]\n\n# Categorize spending based on the bins.\nper_school_summary_df[\"School Size\"] = pd.cut(per_school_summary_df[\"Total Students\"], size_bins, labels=group_names)\nper_school_summary_df",
"_____no_output_____"
],
[
"# Calculate averages for the desired columns. \nsize_math_scores = per_school_summary_df.groupby([\"School Size\"]).mean()[\"Average Math Score\"]\nsize_reading_scores = per_school_summary_df.groupby([\"School Size\"]).mean()[\"Average Reading Score\"]\nsize_passing_math = per_school_summary_df.groupby([\"School Size\"]).mean()[\"% Passing Math\"]\nsize_passing_reading = per_school_summary_df.groupby([\"School Size\"]).mean()[\"% Passing Reading\"]\nsize_overall_passing = per_school_summary_df.groupby([\"School Size\"]).mean()[\"% Overall Passing\"]\nsize_overall_passing",
"_____no_output_____"
],
[
"# Assemble into DataFrame. \nsize_summary_df = pd.DataFrame({\n \"Average Math Score\": size_math_scores,\n \"Average Reading Score\": size_reading_scores,\n \"% Passing Math\": size_passing_math,\n \"% Passing Reading\": size_passing_reading,\n \"% Overall Passing\": size_overall_passing\n})\nsize_summary_df",
"_____no_output_____"
],
[
"# Format the DataFrame \nsize_summary_df[\"Average Math Score\"] = size_summary_df[\"Average Math Score\"].map(\"{:.1f}\".format)\nsize_summary_df[\"Average Reading Score\"] = size_summary_df[\"Average Reading Score\"].map(\"{:.1f}\".format)\nsize_summary_df[\"% Passing Math\"] = size_summary_df[\"% Passing Math\"].map(\"{:.0f}\".format)\nsize_summary_df[\"% Passing Reading\"] = size_summary_df[\"% Passing Reading\"].map(\"{:.0f}\".format)\nsize_summary_df[\"% Overall Passing\"] = size_summary_df[\"% Overall Passing\"].map(\"{:.0f}\".format)\nsize_summary_df",
"_____no_output_____"
]
],
[
[
"### Scores by School Type",
"_____no_output_____"
]
],
[
[
"# Calculate averages for the desired columns. \ntype_math_scores = per_school_summary_df.groupby([\"School Type\"]).mean()[\"Average Math Score\"]\ntype_reading_scores = per_school_summary_df.groupby([\"School Type\"]).mean()[\"Average Reading Score\"]\ntype_passing_math = per_school_summary_df.groupby([\"School Type\"]).mean()[\"% Passing Math\"]\ntype_passing_reading = per_school_summary_df.groupby([\"School Type\"]).mean()[\"% Passing Reading\"]\ntype_overall_passing = per_school_summary_df.groupby([\"School Type\"]).mean()[\"% Overall Passing\"]\n",
"_____no_output_____"
],
[
"# Assemble into DataFrame.\ntype_summary_df = pd.DataFrame({\n \"Average Math Score\": type_math_scores,\n \"Average Reading Score\": type_reading_scores,\n \"% Passing Math\": type_passing_math,\n \"% Passing Reading\": type_passing_reading,\n \"% Overall Passing\": type_overall_passing\n})\ntype_summary_df",
"_____no_output_____"
],
[
"# Format the DataFrame \ntype_summary_df[\"Average Math Score\"] = type_summary_df[\"Average Math Score\"].map(\"{:.1f}\".format)\ntype_summary_df[\"Average Reading Score\"] = type_summary_df[\"Average Reading Score\"].map(\"{:.1f}\".format)\ntype_summary_df[\"% Passing Math\"] = type_summary_df[\"% Passing Math\"].map(\"{:.0f}\".format)\ntype_summary_df[\"% Passing Reading\"] = type_summary_df[\"% Passing Reading\"].map(\"{:.0f}\".format)\ntype_summary_df[\"% Overall Passing\"] = type_summary_df[\"% Overall Passing\"].map(\"{:.0f}\".format)\ntype_summary_df",
"_____no_output_____"
]
],
[
[
"# End of Challenge (Everything below is practice from module)",
"_____no_output_____"
]
],
[
[
"# Add the Pandas dependency.\nimport pandas as pd",
"_____no_output_____"
],
[
"# Files to load\nschool_data_to_load = \"Resources/schools_complete.csv\"\nstudent_data_to_load = \"Resources/students_complete.csv\"\n",
"_____no_output_____"
],
[
"# Read the school data file and store it in a Pandas DataFrame.\nschool_data_df = pd.read_csv(school_data_to_load)\nschool_data_df\n",
"_____no_output_____"
],
[
"# Read the student data file and store it in a Pandas DataFrame.\nstudent_data_df = pd.read_csv(student_data_to_load)\nstudent_data_df.head()",
"_____no_output_____"
],
[
"# Determine if there are any missing values in the school data.\nschool_data_df.count()",
"_____no_output_____"
],
[
"# Determine if there are any missing values in the student data.\nstudent_data_df.count()",
"_____no_output_____"
],
[
"# Determine if there are any missing values in the school data.\nschool_data_df.isnull()",
"_____no_output_____"
],
[
"# Determine if there are any missing values in the student data.\nstudent_data_df.isnull()",
"_____no_output_____"
],
[
"# Determine if there are any missing values in the student data.\nstudent_data_df.isnull().sum()",
"_____no_output_____"
],
[
"# Determine if there are not any missing values in the student data.\nstudent_data_df.notnull().sum()",
"_____no_output_____"
],
[
"# Determine data types for the school DataFrame.\nschool_data_df.dtypes",
"_____no_output_____"
],
[
"# Add each prefix and suffix to remove to a list.\nprefixes_suffixes = [\"Dr. \", \"Mr. \",\"Ms. \", \"Mrs. \", \"Miss \", \" MD\", \" DDS\", \" DVM\", \" PhD\"]\n\n# Iterate through the \"prefixes_suffixes\" list and replace them with an empty space, \"\" when it appears in the student's name.\nfor word in prefixes_suffixes:\n student_data_df[\"student_name\"] = student_data_df[\"student_name\"].str.replace(word,\"\")",
"/var/folders/8s/_5tkv_c11cxd85qkch7sw7hm0000gn/T/ipykernel_3262/924126586.py:6: FutureWarning: The default value of regex will change from True to False in a future version.\n student_data_df[\"student_name\"] = student_data_df[\"student_name\"].str.replace(word,\"\")\n"
],
[
"student_data_df",
"_____no_output_____"
],
[
"# Combine the data into a single dataset.\nschool_data_complete_df = pd.merge(student_data_df, school_data_df, on=[\"school_name\", \"school_name\"])\nschool_data_complete_df",
"_____no_output_____"
],
[
"# Get the total number of students.\nstudent_count = school_data_complete_df.count()\nstudent_count",
"_____no_output_____"
],
[
"school_data_complete_df[\"Student ID\"].count()",
"_____no_output_____"
],
[
"# Calculate the total number of schools.\nschool_count = school_data_df[\"school_name\"].count()\nschool_count",
"_____no_output_____"
],
[
"# Calculate the total number of schools\nschool_count_2 = school_data_complete_df[\"school_name\"].unique()\nschool_count_2",
"_____no_output_____"
],
[
"len(school_data_complete_df[\"school_name\"].unique())",
"_____no_output_____"
],
[
"# Calculate the total budget.\ntotal_budget = school_data_df[\"budget\"].sum()\ntotal_budget",
"_____no_output_____"
],
[
"# Calculate the average reading score.\naverage_reading_score = school_data_complete_df[\"reading_score\"].mean()\naverage_reading_score",
"_____no_output_____"
],
[
"# Calculate the average math score.\naverage_math_score = school_data_complete_df[\"math_score\"].mean()\naverage_math_score",
"_____no_output_____"
],
[
"passing_math = school_data_complete_df[\"math_score\"] >= 70\npassing_reading = school_data_complete_df[\"reading_score\"] >= 70\npassing_math",
"_____no_output_____"
],
[
"passing_reading",
"_____no_output_____"
],
[
"# Get all the students who are passing math in a new DataFrame.\npassing_math = school_data_complete_df[school_data_complete_df[\"math_score\"] >= 70]\npassing_math.head()",
"_____no_output_____"
],
[
"# Get all the students who are passing reading in a new DataFrame.\npassing_reading = school_data_complete_df[school_data_complete_df[\"reading_score\"] >= 70]\npassing_reading.head()",
"_____no_output_____"
],
[
"# Trying out len to calcualte number of students passing math.\nlen(passing_math)",
"_____no_output_____"
],
[
"# Correct way to determine students passing math using count () method.\npassing_math[\"student_name\"].count()",
"_____no_output_____"
],
[
"# Calculate the number of students passing math.\npassing_math_count = passing_math[\"student_name\"].count()\n\n# Calculate the number of students passing reading.\npassing_reading_count = passing_reading[\"student_name\"].count()\n",
"_____no_output_____"
],
[
"print(passing_math_count)\nprint(passing_reading_count)",
"29370\n33610\n"
],
[
"# Student count from earlier was calcualted using this code (the = part).\nstudent_count = school_data_complete_df[\"Student ID\"].count()",
"_____no_output_____"
],
[
"# Calculate the percent that passed math.\npassing_math_percentage = passing_math_count / float(student_count) * 100\n\n# Calculate the percent that passed reading.\npassing_reading_percentage = passing_reading_count / float(student_count) * 100\n",
"_____no_output_____"
],
[
"print(passing_math_percentage)\nprint(passing_reading_percentage)",
"74.9808526933878\n85.80546336482001\n"
],
[
"# Calculate the students who passed both math and reading.\npassing_math_reading = school_data_complete_df[(school_data_complete_df[\"math_score\"] >= 70) & (school_data_complete_df[\"reading_score\"] >= 70)]\npassing_math_reading",
"_____no_output_____"
],
[
"# Calculate the number of students who passed both math and reading.\noverall_passing_math_reading_count = passing_math_reading[\"student_name\"].count()\noverall_passing_math_reading_count",
"_____no_output_____"
],
[
"# Calculate the overall passing percentage.\n\noverall_passing_percentage = overall_passing_math_reading_count / student_count * 100\noverall_passing_percentage",
"_____no_output_____"
],
[
"# Adding a list of values with keys to create a new DataFrame.\ndistrict_summary_df = pd.DataFrame(\n [{\"Total Schools\": school_count,\n \"Total Students\": student_count,\n \"Total Budget\": total_budget,\n \"Average Math Score\": average_math_score,\n \"Average Reading Score\": average_reading_score,\n \"% Passing Math\": passing_math_percentage,\n \"% Passing Reading\": passing_reading_percentage,\n \"% Overall Passing\": overall_passing_percentage}])\ndistrict_summary_df\n",
"_____no_output_____"
],
[
"# Format the \"Total Students\" to have the comma for a thousands separator.\ndistrict_summary_df[\"Total Students\"] = district_summary_df[\"Total Students\"].map(\"{:,}\".format)\n\ndistrict_summary_df[\"Total Students\"]",
"_____no_output_____"
],
[
"district_summary_df",
"_____no_output_____"
],
[
"# Format \"Total Budget\" to have the comma for a thousands separator, a decimal separator, and a \"$\".\n\ndistrict_summary_df[\"Total Budget\"] = district_summary_df[\"Total Budget\"].map(\"${:,.2f}\".format)\n\ndistrict_summary_df[\"Total Budget\"]",
"_____no_output_____"
],
[
"district_summary_df",
"_____no_output_____"
],
[
"# Format the columns.\ndistrict_summary_df[\"Average Math Score\"] = district_summary_df[\"Average Math Score\"].map(\"{:.1f}\".format)\n\ndistrict_summary_df[\"Average Reading Score\"] = district_summary_df[\"Average Reading Score\"].map(\"{:.1f}\".format)\n\ndistrict_summary_df[\"% Passing Math\"] = district_summary_df[\"% Passing Math\"].map(\"{:.0f}\".format)\n\ndistrict_summary_df[\"% Passing Reading\"] = district_summary_df[\"% Passing Reading\"].map(\"{:.0f}\".format)\n\ndistrict_summary_df[\"% Overall Passing\"] = district_summary_df[\"% Overall Passing\"].map(\"{:.0f}\".format)\n\ndistrict_summary_df",
"_____no_output_____"
],
[
"# Reorder the columns in the order you want them to appear.\nnew_column_order = [\"Total Schools\", \"Total Students\", \"Total Budget\",\"Average Math Score\", \"Average Reading Score\", \"% Passing Math\", \"% Passing Reading\", \"% Overall Passing\"]\n\n# Assign district summary df the new column order.\ndistrict_summary_df = district_summary_df[new_column_order]\ndistrict_summary_df",
"_____no_output_____"
],
[
"# Determine the school type.\nper_school_types = school_data_df.set_index([\"school_name\"])[\"type\"]\nper_school_types",
"_____no_output_____"
],
[
"# Add the per_school_types into a DataFrame for testing.\ndf = pd.DataFrame(per_school_types)\ndf",
"_____no_output_____"
],
[
"# Calculate the total student count.\nper_school_counts = school_data_df[\"size\"]\nper_school_counts",
"_____no_output_____"
],
[
"# Calculate the total student count.\nper_school_counts = school_data_df.set_index([\"school_name\"])[\"size\"]\nper_school_counts",
"_____no_output_____"
],
[
"# Calculate the total student count.\nper_school_counts = school_data_complete_df[\"school_name\"].value_counts()\nper_school_counts",
"_____no_output_____"
],
[
"# Calculate the total school budget.\nper_school_budget = school_data_df.set_index([\"school_name\"])[\"budget\"]\nper_school_budget\n",
"_____no_output_____"
],
[
"# Calculate the per capita spending.\nper_school_capita = per_school_budget / per_school_counts\nper_school_capita",
"_____no_output_____"
],
[
"# Calculate the math scores.\nstudent_school_math = student_data_df.set_index([\"school_name\"])[\"math_score\"]\nstudent_school_math",
"_____no_output_____"
],
[
"# Calculate the average math scores.\nper_school_averages = school_data_complete_df.groupby([\"school_name\"]).mean()\nper_school_averages",
"_____no_output_____"
],
[
"# Calculate the average test scores.\nper_school_math = school_data_complete_df.groupby([\"school_name\"]).mean()[\"math_score\"]\n\nper_school_reading = school_data_complete_df.groupby([\"school_name\"]).mean()[\"reading_score\"]\n\nper_school_math",
"_____no_output_____"
],
[
"per_school_reading",
"_____no_output_____"
],
[
"# Calculate the passing scores by creating a filtered DataFrame.\nper_school_passing_math = school_data_complete_df[(school_data_complete_df[\"math_score\"] >= 70)]\n\nper_school_passing_reading = school_data_complete_df[(school_data_complete_df[\"reading_score\"] >= 70)]\nper_school_passing_math",
"_____no_output_____"
],
[
"per_school_passing_reading",
"_____no_output_____"
],
[
"# Calculate the number of students passing math and passing reading by school.\nper_school_passing_math = per_school_passing_math.groupby([\"school_name\"]).count()[\"student_name\"]\n\nper_school_passing_reading = per_school_passing_reading.groupby([\"school_name\"]).count()[\"student_name\"]\nper_school_passing_math",
"_____no_output_____"
],
[
"per_school_passing_reading",
"_____no_output_____"
],
[
"# Calculate the percentage of passing math and reading scores per school.\nper_school_passing_math = per_school_passing_math / per_school_counts * 100\n\nper_school_passing_reading = per_school_passing_reading / per_school_counts * 100\n",
"_____no_output_____"
],
[
"per_school_passing_math ",
"_____no_output_____"
],
[
"per_school_passing_reading ",
"_____no_output_____"
],
[
"# Calculate the students who passed both math and reading.\nper_passing_math_reading = school_data_complete_df[(school_data_complete_df[\"math_score\"] >= 70) & (school_data_complete_df[\"reading_score\"] >= 70)]\n\nper_passing_math_reading.head()",
"_____no_output_____"
],
[
"# Calculate the number of students who passed both math and reading.\nper_passing_math_reading = per_passing_math_reading.groupby([\"school_name\"]).count()[\"student_name\"]\n",
"_____no_output_____"
],
[
"per_passing_math_reading",
"_____no_output_____"
],
[
"# Calculate the overall passing percentage.\nper_overall_passing_percentage = per_passing_math_reading / per_school_counts * 100",
"_____no_output_____"
],
[
"per_overall_passing_percentage",
"_____no_output_____"
],
[
"# Adding a list of values with keys to create a new DataFrame.\n\nper_school_summary_df = pd.DataFrame({\n \"School Type\": per_school_types,\n \"Total Students\": per_school_counts,\n \"Total School Budget\": per_school_budget,\n \"Per Student Budget\": per_school_capita,\n \"Average Math Score\": per_school_math,\n \"Average Reading Score\": per_school_reading,\n \"% Passing Math\": per_school_passing_math,\n \"% Passing Reading\": per_school_passing_reading,\n \"% Overall Passing\": per_overall_passing_percentage})\nper_school_summary_df.head()\n",
"_____no_output_____"
],
[
"# Format the Total School Budget and the Per Student Budget columns.\nper_school_summary_df[\"Total School Budget\"] = per_school_summary_df[\"Total School Budget\"].map(\"${:,.2f}\".format)\n\nper_school_summary_df[\"Per Student Budget\"] = per_school_summary_df[\"Per Student Budget\"].map(\"${:,.2f}\".format)\n",
"_____no_output_____"
],
[
"# Display the data frame\nper_school_summary_df.head()",
"_____no_output_____"
],
[
"# Sort and show top five schools.\ntop_schools = per_school_summary_df.sort_values([\"% Overall Passing\"], ascending=False)\n\ntop_schools.head()",
"_____no_output_____"
],
[
"# Sort and show top five schools.\nbottom_schools = per_school_summary_df.sort_values([\"% Overall Passing\"], ascending=True)\n\nbottom_schools.head()",
"_____no_output_____"
],
[
"# Create a grade level DataFrames.\nninth_graders = school_data_complete_df[(school_data_complete_df[\"grade\"] == \"9th\")]\n\ntenth_graders = school_data_complete_df[(school_data_complete_df[\"grade\"] == \"10th\")]\n\neleventh_graders = school_data_complete_df[(school_data_complete_df[\"grade\"] == \"11th\")]\n\ntwelfth_graders = school_data_complete_df[(school_data_complete_df[\"grade\"] == \"12th\")]",
"_____no_output_____"
],
[
"ninth_graders",
"_____no_output_____"
],
[
"tenth_graders",
"_____no_output_____"
],
[
"eleventh_graders",
"_____no_output_____"
],
[
"twelfth_graders",
"_____no_output_____"
],
[
"# Group each grade level DataFrame by the school name for the average math score.\nninth_grade_math_scores = ninth_graders.groupby([\"school_name\"]).mean()[\"math_score\"]\n\ntenth_grade_math_scores = tenth_graders.groupby([\"school_name\"]).mean()[\"math_score\"]\n\neleventh_grade_math_scores = eleventh_graders.groupby([\"school_name\"]).mean()[\"math_score\"]\n\ntwelfth_grade_math_scores = twelfth_graders.groupby([\"school_name\"]).mean()[\"math_score\"]",
"_____no_output_____"
],
[
"ninth_grade_math_scores",
"_____no_output_____"
],
[
"tenth_grade_math_scores",
"_____no_output_____"
],
[
"eleventh_grade_math_scores",
"_____no_output_____"
],
[
"twelfth_grade_math_scores",
"_____no_output_____"
],
[
"# Group each grade level DataFrame by the school name for the average reading score.\nninth_grade_reading_scores = ninth_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]\n\ntenth_grade_reading_scores = tenth_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]\n\neleventh_grade_reading_scores = eleventh_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]\n\ntwelfth_grade_reading_scores = twelfth_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]",
"_____no_output_____"
],
[
"ninth_grade_reading_scores",
"_____no_output_____"
],
[
"tenth_grade_reading_scores",
"_____no_output_____"
],
[
"eleventh_grade_reading_scores",
"_____no_output_____"
],
[
"twelfth_grade_reading_scores",
"_____no_output_____"
],
[
"# Combine each grade level Series for average math scores by school into a single DataFrame.\nmath_scores_by_grade = pd.DataFrame({\n \"9th\": ninth_grade_math_scores,\n \"10th\": tenth_grade_math_scores,\n \"11th\": eleventh_grade_math_scores,\n \"12th\": twelfth_grade_math_scores})\n\nmath_scores_by_grade.head()\n",
"_____no_output_____"
],
[
"# Combine each grade level Series for average reading scores by school into a single DataFrame.\nreading_scores_by_grade = pd.DataFrame({\n \"9th\": ninth_grade_reading_scores,\n \"10th\": tenth_grade_reading_scores,\n \"11th\": eleventh_grade_reading_scores,\n \"12th\": twelfth_grade_reading_scores})\n\nreading_scores_by_grade.head()",
"_____no_output_____"
],
[
"# Format each grade column.\nmath_scores_by_grade[\"9th\"] = math_scores_by_grade[\"9th\"].map(\"{:.1f}\".format)\n\nmath_scores_by_grade[\"10th\"] = math_scores_by_grade[\"10th\"].map(\"{:.1f}\".format)\n\nmath_scores_by_grade[\"11th\"] = math_scores_by_grade[\"11th\"].map(\"{:.1f}\".format)\n\nmath_scores_by_grade[\"12th\"] = math_scores_by_grade[\"12th\"].map(\"{:.1f}\".format)\n\n# Make sure the columns are in the correct order.\nmath_scores_by_grade = math_scores_by_grade[\n [\"9th\", \"10th\", \"11th\", \"12th\"]]\n\n# Remove the index name.\nmath_scores_by_grade.index.name = None\n# Display the DataFrame.\nmath_scores_by_grade.head()",
"_____no_output_____"
],
[
"# Format each grade column.\nreading_scores_by_grade[\"9th\"] = reading_scores_by_grade[\"9th\"].map(\"{:,.1f}\".format)\n\nreading_scores_by_grade[\"10th\"] = reading_scores_by_grade[\"10th\"].map(\"{:,.1f}\".format)\n\nreading_scores_by_grade[\"11th\"] = reading_scores_by_grade[\"11th\"].map(\"{:,.1f}\".format)\n\nreading_scores_by_grade[\"12th\"] = reading_scores_by_grade[\"12th\"].map(\"{:,.1f}\".format)\n\n# Make sure the columns are in the correct order.\nreading_scores_by_grade = reading_scores_by_grade[\n [\"9th\", \"10th\", \"11th\", \"12th\"]]\n\n# Remove the index name.\nreading_scores_by_grade.index.name = None\n# Display the data frame.\nreading_scores_by_grade.head()",
"_____no_output_____"
],
[
"# Get the descriptive statistics for the per_school_capita.\nper_school_capita.describe()",
"_____no_output_____"
],
[
"per_school_capita",
"_____no_output_____"
],
[
"# Cut the per_school_capita into the spending ranges.\nspending_bins = [0, 585, 615, 645, 675]\npd.cut(per_school_capita, spending_bins)",
"_____no_output_____"
],
[
"# Cut the per_school_capita into the spending ranges.\nspending_bins = [0, 585, 615, 645, 675]\nper_school_capita.groupby(pd.cut(per_school_capita, spending_bins)).count()\n",
"_____no_output_____"
],
[
"# Cut the per_school_capita into the spending ranges.\nspending_bins = [0, 585, 630, 645, 675]\nper_school_capita.groupby(pd.cut(per_school_capita, spending_bins)).count()",
"_____no_output_____"
],
[
"# Establish the spending bins and group names.\nspending_bins = [0, 585, 630, 645, 675]\ngroup_names = [\"<$584\", \"$585-629\", \"$630-644\", \"$645-675\"]",
"_____no_output_____"
],
[
"# Categorize spending based on the bins.\nper_school_summary_df[\"Spending Ranges (Per Student)\"] = pd.cut(per_school_capita, spending_bins, labels=group_names)\n\nper_school_summary_df",
"_____no_output_____"
],
[
"# Calculate averages for the desired columns.\nspending_math_scores = per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"Average Math Score\"]\n\nspending_reading_scores = per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"Average Reading Score\"]\n\nspending_passing_math = per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"% Passing Math\"]\n\nspending_passing_reading = per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"% Passing Reading\"]\n\noverall_passing_spending = per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"% Overall Passing\"]",
"_____no_output_____"
],
[
"spending_math_scores",
"_____no_output_____"
],
[
"spending_reading_scores",
"_____no_output_____"
],
[
"spending_passing_math",
"_____no_output_____"
],
[
"spending_passing_reading",
"_____no_output_____"
],
[
"overall_passing_spending",
"_____no_output_____"
],
[
"# Assemble into DataFrame.\nspending_summary_df = pd.DataFrame({\n \"Average Math Score\" : spending_math_scores,\n \"Average Reading Score\": spending_reading_scores,\n \"% Passing Math\": spending_passing_math,\n \"% Passing Reading\": spending_passing_reading,\n \"% Overall Passing\": overall_passing_spending})\n\nspending_summary_df\n",
"_____no_output_____"
],
[
"# Formatting\nspending_summary_df[\"Average Math Score\"] = spending_summary_df[\"Average Math Score\"].map(\"{:.1f}\".format)\n\nspending_summary_df[\"Average Reading Score\"] = spending_summary_df[\"Average Reading Score\"].map(\"{:.1f}\".format)\n\nspending_summary_df[\"% Passing Math\"] = spending_summary_df[\"% Passing Math\"].map(\"{:.0f}\".format)\n\nspending_summary_df[\"% Passing Reading\"] = spending_summary_df[\"% Passing Reading\"].map(\"{:.0f}\".format)\n\nspending_summary_df[\"% Overall Passing\"] = spending_summary_df[\"% Overall Passing\"].map(\"{:.0f}\".format)\n\nspending_summary_df",
"_____no_output_____"
],
[
"per_school_counts",
"_____no_output_____"
],
[
"# Establish the bins.\nsize_bins = [0, 1000, 2000, 5000]\ngroup_names = [\"Small (<1000)\", \"Medium (1000-2000)\", \"Large (2000-5000)\"]",
"_____no_output_____"
],
[
"# Categorize spending based on the bins.\nper_school_summary_df[\"School Size\"] = pd.cut(per_school_summary_df[\"Total Students\"], size_bins, labels=group_names)\n\nper_school_summary_df.head()",
"_____no_output_____"
],
[
"# Calculate averages for the desired columns.\nsize_math_scores = per_school_summary_df.groupby([\"School Size\"]).mean()[\"Average Math Score\"]\n\nsize_reading_scores = per_school_summary_df.groupby([\"School Size\"]).mean()[\"Average Reading Score\"]\n\nsize_passing_math = per_school_summary_df.groupby([\"School Size\"]).mean()[\"% Passing Math\"]\n\nsize_passing_reading = per_school_summary_df.groupby([\"School Size\"]).mean()[\"% Passing Reading\"]\n\nsize_overall_passing = per_school_summary_df.groupby([\"School Size\"]).mean()[\"% Overall Passing\"]",
"_____no_output_____"
],
[
"size_math_scores",
"_____no_output_____"
],
[
"size_reading_scores",
"_____no_output_____"
],
[
"size_passing_math",
"_____no_output_____"
],
[
"size_passing_reading",
"_____no_output_____"
],
[
"size_overall_passing",
"_____no_output_____"
],
[
"# Calculate averages for the desired columns.\ntype_math_scores = per_school_summary_df.groupby([\"School Type\"]).mean()[\"Average Math Score\"]\n\ntype_reading_scores = per_school_summary_df.groupby([\"School Type\"]).mean()[\"Average Reading Score\"]\n\ntype_passing_math = per_school_summary_df.groupby([\"School Type\"]).mean()[\"% Passing Math\"]\n\ntype_passing_reading = per_school_summary_df.groupby([\"School Type\"]).mean()[\"% Passing Reading\"]\n\ntype_overall_passing = per_school_summary_df.groupby([\"School Type\"]).mean()[\"% Overall Passing\"]\n",
"_____no_output_____"
],
[
"type_math_scores",
"_____no_output_____"
],
[
"type_reading_scores",
"_____no_output_____"
],
[
"type_passing_math",
"_____no_output_____"
],
[
"type_passing_reading",
"_____no_output_____"
],
[
"type_overall_passing",
"_____no_output_____"
],
[
"# Assemble into DataFrame.\ntype_summary_df = pd.DataFrame({\n \"Average Math Score\" : type_math_scores,\n \"Average Reading Score\": type_reading_scores,\n \"% Passing Math\": type_passing_math,\n \"% Passing Reading\": type_passing_reading,\n \"% Overall Passing\": type_overall_passing})\n\ntype_summary_df",
"_____no_output_____"
],
[
"# Formatting\ntype_summary_df[\"Average Math Score\"] = type_summary_df[\"Average Math Score\"].map(\"{:.1f}\".format)\n\ntype_summary_df[\"Average Reading Score\"] = type_summary_df[\"Average Reading Score\"].map(\"{:.1f}\".format)\n\ntype_summary_df[\"% Passing Math\"] = type_summary_df[\"% Passing Math\"].map(\"{:.0f}\".format)\n\ntype_summary_df[\"% Passing Reading\"] = type_summary_df[\"% Passing Reading\"].map(\"{:.0f}\".format)\n\ntype_summary_df[\"% Overall Passing\"] = type_summary_df[\"% Overall Passing\"].map(\"{:.0f}\".format)\n\ntype_summary_df",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf0161b0010ec75246d7f920ae8f956950d9e0f | 34,777 | ipynb | Jupyter Notebook | notebooks/Explore.ipynb | learningequality/sushi-chef-aflatoun | 40b8db813779eb4664a44554a759b4cac982af5a | [
"MIT"
] | null | null | null | notebooks/Explore.ipynb | learningequality/sushi-chef-aflatoun | 40b8db813779eb4664a44554a759b4cac982af5a | [
"MIT"
] | 1 | 2019-09-20T19:55:27.000Z | 2019-09-20T19:55:27.000Z | notebooks/Explore.ipynb | learningequality/sushi-chef-aflatoun | 40b8db813779eb4664a44554a759b4cac982af5a | [
"MIT"
] | null | null | null | 66.878846 | 176 | 0.730512 | [
[
[
"import json\nimport logging\nimport os\n\n\nfrom aflatoun_chef import (\n AFLATOUN_LICENSE,\n AFLATOUN_CONTENT_BASE_DIR,\n LANGUAGE_FOLDER_LOOKUP,\n CONTENT_FOLDERS_FOR_LANG)\n\nfrom aflatoun_chef import get_metadata_file_path, get_metadata\nfrom aflatoun_chef import build_ricecooker_json_tree\n \nfrom aflatoun_chef import LOGGER\nLOGGER.addHandler(logging.StreamHandler())\n\nimport pprint\npp = pprint.PrettyPrinter(indent=4, width=80)",
"_____no_output_____"
],
[
"# lang = 'fr'\n# lang_dir = LANGUAGE_FOLDER_LOOKUP[lang]\n\n# channel_base_dir = os.path.join(AFLATOUN_CONTENT_BASE_DIR, lang_dir)\n# content_folders = list(os.walk(channel_base_dir))\n\n# content_folders",
"_____no_output_____"
],
[
"build_ricecooker_json_tree(None,{'lang':'fr'}, '/tmp/yo.json')\n",
"Starting to build the ricecooker_json_tree\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl41.1\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl41.2\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl41.3\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl41.4\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl41.5\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl41.6\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl42.1\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl42.2\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl42.3\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl42.4\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl43.1\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl43.2\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl43.3\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl43.4\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl43.5\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl44.1\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl44.2\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl44.3\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl44.4\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl44.5\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl44.6\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl44.7\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl51.1\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl51.2\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl51.3\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl51.4\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl51.5\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl51.6\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl52.1\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl52.2\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl52.3\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl52.4\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl53.1\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl53.2\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl53.3\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl53.4\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl53.5\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl54.1\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl54.2\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl54.3\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl54.4\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl54.5\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl54.6\nprocessing folder content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 5ème niveau/faafl54.7\nprocessing folder content/aflatoun_tree/aflatoun/French/Manuel de Formation\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/01fr_les_origines_de_aflatoun\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/02fr_les_clubs_dans_la_programme\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/03fr_les_activities_programme_principal\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/04fr_cde_la_qualite_de_leducation\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/05fr_legalite_des_genres\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/06fr_les_enfants_et_lespargne\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/07fr_entreprises_sociales_financieres_des_enfants\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/08fr_temps_de_parole_de_lenseignant\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/09fr_brainstorming\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/10fr_larbre_probleme\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/11fr_theatre_image\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/12fr_tableau_cva\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/13fr_cartes_memoire\nprocessing folder content/aflatoun_tree/aflatoun/French/Modules de support pour la formation continue/14fr_travail_de_groupe\n"
],
[
"ex_path = 'content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl44.4/03faafl44.4qr.exercise.zip'\nex_path = 'content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl41.1/03faafl41.1qr.exercise.zip'\nex_path = 'content/aflatoun_tree/aflatoun/French/Les Séries Aflatoun/Plan de leçon 4ème niveau/faafl43.5/01faafl43.5qp.exercise.zip'",
"_____no_output_____"
],
[
"import zipfile\narchive = zipfile.ZipFile(ex_path, 'r')\n#\njson_bytes = archive.read('exercise.json')\nexercise_json = json.loads(json_bytes)\nprint(exercise_json)\n#\njson_bytes = archive.read('assessment_items.json')\nquestions = json.loads(json_bytes)\nlen(questions)\nquestions[0]\n",
"{'title': '01faafl43.5qp', 'description': 'Quiz de préparation', 'all_assessment_items': [425, 426, 427, 428]}\n"
],
[
"ex_path",
"_____no_output_____"
],
[
"# exercise_json\nexercise_zip_to_dict(ex_path)['questions'][1].__dict__",
"{'title': '01faafl43.5qp', 'description': 'Quiz de préparation', 'all_assessment_items': [425, 426, 427, 428]}\n"
],
[
"q.__dict__",
"_____no_output_____"
],
[
"os.path.splitext('faafl43.5qp.exercise.zip')",
"_____no_output_____"
],
[
"SingleSelectQuestion?",
"_____no_output_____"
],
[
"from ricecooker.classes.nodes import ExerciseNode",
"_____no_output_____"
],
[
"ExerciseNode?",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf023e66a9dd55958d6e9e5eab54193441eabe4 | 183,922 | ipynb | Jupyter Notebook | lectures/lecture-12.ipynb | oseledets/nla2016 | 6cb20593d9c8bf8f979859f489366f11b926e696 | [
"MIT"
] | 22 | 2016-10-20T09:57:37.000Z | 2021-04-29T04:48:28.000Z | lectures/lecture-12.ipynb | oseledets/nla2016 | 6cb20593d9c8bf8f979859f489366f11b926e696 | [
"MIT"
] | 4 | 2017-09-07T08:43:59.000Z | 2017-09-25T09:02:37.000Z | lectures/lecture-12.ipynb | oseledets/nla2016 | 6cb20593d9c8bf8f979859f489366f11b926e696 | [
"MIT"
] | 29 | 2016-10-25T07:02:05.000Z | 2019-10-09T04:35:45.000Z | 148.443906 | 50,572 | 0.869173 | [
[
[
"# Lecture 12: Iterative methods-2",
"_____no_output_____"
],
[
"## Previous lecture\n- Arnoldi orthogonalization of Krylov subspaces\n- Lanczos for the symmetric case\n- Energy functional and conjugate gradient method\n- Convergence analysis\n- Non-symmetric case: idea of GMRES",
"_____no_output_____"
],
[
"## Today lecture\n- More iterative methods \n- The concept of preconditioners",
"_____no_output_____"
],
[
"## What methods to use\n\n- If a matrix is symmetric (Hermitian) positive definite, use CG method.\n- If a matrix is symmetric by indefinite, we can use MINRES method (GMRES applied to a symmetric system)\n- If a matrix is non-symmetric and not very big, use GMRES\n- Ia a matrix is non-symmetric and we can store limited amount of vectors, use either: GMRES with restarts, or BiCGStab (the latter of the product with $A^{\\top}$ is also available).",
"_____no_output_____"
],
[
"## MINRES\n\nThe MINRES method is GMRES applied to a symmetric system. We minimize\n\n$$\\Vert A Q_j x_j - f \\Vert = \\Vert Q_j \\widehat{x}_j + h_{j, j-1} q_j \\widehat{x}_j - f \\Vert = \\Vert Q_{j+1} \\widehat{H}_{j+1} \\widehat{x}_j - f \\Vert \\rightarrow \\min$$\n\nwhich is equivalent to a linear least squares with an **almost tridiagonal** matrix\n\n$$\\Vert \\widehat{H}_{j+1} x_{j} - \\gamma e_0 \\Vert \\rightarrow \\min.$$\n\nIn a similar fashion, we can derive short-term recurrences.\n\nA careful implementation of MINRES requires at most $5$ vectors to be stored.",
"_____no_output_____"
],
[
"## Difference between MINRES and CG\n- MINRES minimizes $\\Vert Ax_k - f \\Vert$ over the Krylov subspace\n- CG minimize $(Ax, x) - 2(f, x)$ over the Krylov subspace\n- MINRES works for indefinite (i.e., non-positive definite problems).\n\nCG stores less vectors ($3$ instead of $5$). \n\nNow, let us talk about non-symmetric systems.",
"_____no_output_____"
],
[
"## Non-symmetric systems\nThe main disadvantage of GMRES: we have to store all the vectors, so the memory cost grows with each step. \n\nWe can do **restarts** (i.e. get a new residual and a new Krylov subspace): we find some approximate solution $x$ \n\nand now solve the linear system for the correction:\n\n$$A(x + e) = f, \\quad Ae = f - Ax,$$\n\nand generate the new **Krylov subspace** from the residual vector. This spoils the convergence, as we will see from the demo.\n\nBiConjugate gradient (named **BiCg**, proposed by Fletcher) avoids that using \"short recurrences\" like in the CG method.\n\nIt is very unstable in the original form, thus a stabilized version has been proposed (named **BiCgStab**).",
"_____no_output_____"
],
[
"## Idea of biconjugate gradient\n\nIdea of BiCG method is to use the normal equations:\n\n$$A^* A x = A^* f,$$\n\nand apply the CG method to it.\n\nThe condition number has squared, thus we need **stabilization**.\n\nThe stabilization idea proposed by Van der Vorst et al. improves the stability.\n\nLet us do some demo for a simple non-symmetric matrix.",
"_____no_output_____"
]
],
[
[
"#### import numpy as np\nimport scipy.sparse.linalg\n%matplotlib inline\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport scipy as sp\nn = 300\nex = np.ones(n);\nlp1 = -sp.sparse.spdiags(np.vstack((ex, -(2 + 1./n)*ex, (1 + 1./n) * ex)), [-1, 0, 1], n, n, 'csr'); \nrhs = np.ones(n)\nee = sp.sparse.eye(n)\n\n#lp2 = sp.kron(lp1, ee) + sp.kron(ee, lp1)\n#rhs = np.ones(n * n)\nres_all = []\nres_all_no = []\nres_all_bicg = []\ndef my_print(r):\n res_all.append(r)\n \ndef my_print1(r):\n res_all_no.append(r)\n\ndef my_print2(x): #For BiCGStab they have another callback, please rewrite\n res_all_bicg.append(np.linalg.norm(lp1.dot(x) - rhs))\n \n#res_all_bicg = np.array(res_all_bicg)\nsol = scipy.sparse.linalg.gmres(lp1, rhs, restart=20, callback=my_print)\nsol1 = scipy.sparse.linalg.gmres(lp1, rhs, restart=n, callback=my_print1)\nsol2 = scipy.sparse.linalg.bicgstab(lp1, rhs, x0=np.zeros(n), callback=my_print2)\nres_all_bicg = np.array(res_all_bicg)/res_all_bicg[0]\n\nlim = 350\nplt.semilogy(res_all[:lim], marker='.',color='k', label='GMRES, restart=20')\nplt.semilogy(res_all_no[:lim], marker='x',color='r', label='GMRES, no restart')\nplt.semilogy(res_all_bicg[:lim], label='BiCGStab', marker='o')\n\nplt.xlabel('Iteration number')\nplt.ylabel('Residual')\nplt.legend(loc='best')`",
"_____no_output_____"
]
],
[
[
"## Notes about BiCG\n\nA practical implementation of BiCG uses **two-sided Lanczos process** ",
"_____no_output_____"
]
],
[
[
"#### import numpy as np\nimport scipy.sparse.linalg\n%matplotlib inline\nimport matplotlib.pyplot as plt\nfrom scipy.sparse.linalg import minres, gmres\nimport numpy as np\nimport scipy as sp\nn = 300\nex = np.ones(n);\nlp1 = -sp.sparse.spdiags(np.vstack((ex, -(2 + 1./n)*ex, (1 + 1./n) * ex)), [-1, 0, 1], n, n, 'csr'); \nrhs = np.ones(n)\nee = sp.sparse.eye(n)\n\n#lp2 = sp.kron(lp1, ee) + sp.kron(ee, lp1)\n#rhs = np.ones(n * n)\nres_all = []\nres_all_no = []\nres_all_bicg = []\ndef my_print(r):\n res_all.append(r)\n \ndef my_print1(r):\n res_all_no.append(r)\n\ndef my_print2(x): #For BiCGStab they have another callback, please rewrite\n res_all_bicg.append(np.linalg.norm(lp1.dot(x) - rhs))\n \n#res_all_bicg = np.array(res_all_bicg)\nsol = scipy.sparse.linalg.gmres(lp1, rhs, callback=my_print)\nsol1 = scipy.sparse.linalg.gmres(lp1, rhs, restart=n, callback=my_print1)\nsol2 = scipy.sparse.linalg.bicgstab(lp1, rhs, x0=np.zeros(n), callback=my_print2)\nres_all_bicg = np.array(res_all_bicg)/res_all_bicg[0]\n\nlim = 350\nplt.semilogy(res_all[:lim], marker='.',color='k', label='GMRES, restart=20')\nplt.semilogy(res_all_no[:lim], marker='x',color='r', label='GMRES, no restart')\nplt.semilogy(res_all_bicg[:lim], label='BiCGStab', marker='o')\n\nplt.xlabel('Iteration number')\nplt.ylabel('Residual')\nplt.legend(loc='best')",
"_____no_output_____"
]
],
[
[
"## BiConjugate Gradients\n\nBiCG theory is nasty. Since we solve \n\n$$( A^* A ) x = f, $$\n\n there are two options. \n\n1. Use $K(A^* A, A^* f)$ to generate the subspace. That leads to square of condition number\n2. Instead, use two Krylov subspaces $K(A)$ and $K(A^*)$ to generate two basises that are **biorthogonal** (so-called biorthogonal Lanczos).\n\nThe goal is to compute the Petrov-Galerkin projection \n\n$$W^* A V \\widehat{x} = W^* f$$\n\nwith columns $W$ from the Krylov subspace of $A^*$, $V$ from $A$. \n\nThat may lead to instabilities if we try to recompute the solutions in the efficient way. It is related to the **pivoting** (which we did not use in CG), and it is not naturally implemented here.",
"_____no_output_____"
],
[
"## BiCGStab\n\nBiCGStab is frequently used, and represent a **stabilized** version of BiCG. It has faster and smoother convergence than original BiCG method.\n\n\nThe formulas can be found, for example, here https://en.wikipedia.org/wiki/Biconjugate_gradient_stabilized_method\n\nIt is a combination of BiCG step followed by GMRES(1) step in order to smooth the convergence.\n\nFor more detailed reading, please consult the book \"Iterative Krylov Methods for Large Linear Systems\" by H. Van-der Voorst.\n\nA short demo to compare \"Stabilized\" vs \"Non-stabilized\" versions.",
"_____no_output_____"
]
],
[
[
"#### import numpy as np\nimport scipy.sparse.linalg\n%matplotlib inline\nimport matplotlib.pyplot as plt\nfrom scipy.sparse.linalg import minres, gmres\nimport numpy as np\nimport scipy as sp\nn = 300\nex = np.ones(n);\nlp1 = -sp.sparse.spdiags(np.vstack((ex, -(2 + 1./n)*ex, (1 + 1./n) * ex)), [-1, 0, 1], n, n, 'csr'); \nrhs = np.random.randn(n)\nee = sp.sparse.eye(n)\n\n#lp2 = sp.kron(lp1, ee) + sp.kron(ee, lp1)\n#rhs = np.ones(n * n)\nres_all_bicg = []\nres_all_bicgstab = []\n\ndef my_print3(x):\n res_all_bicg.append(np.linalg.norm(lp1.dot(x) - rhs))\n\ndef my_print2(x): #For BiCGStab they have another callback, please rewrite\n res_all_bicgstab.append(np.linalg.norm(lp1.dot(x) - rhs))\n \n#res_all_bicg = np.array(res_all_bicg)\nsol2 = scipy.sparse.linalg.bicg(lp1, rhs, x0=np.zeros(n), callback=my_print3)\nsol2 = scipy.sparse.linalg.bicgstab(lp1, rhs, x0=np.zeros(n), callback=my_print2)\nres_all_bicg = np.array(res_all_bicg)/res_all_bicg[0]\nres_all_bicgstab = np.array(res_all_bicgstab)/res_all_bicgstab[0]\n\nlim = 350\nplt.semilogy(res_all_bicgstab[:lim], marker='.',color='k', label='BiCGStab')\nplt.semilogy(res_all_bicg[:lim], marker='x',color='r', label='BiCG')\n\nplt.xlabel('Iteration number')\nplt.ylabel('Residual')\nplt.legend(loc='best')",
"_____no_output_____"
]
],
[
[
"## \"Nonlinear GMRES\" or Anderson acceleration\n\nWe can apply the GMRES-like idea to speed up the convergence of a given fixed-point iteration\n\n$$x_{k+1} = \\Phi(x_k).$$\n\nThis was actually older than the GMRES, and known as an Direct Inversion in Iterated Subspaces in Quantum Chemistry, or **Anderson Acceleration**.\n\nIdea: \n\n**Use history** for the update, \n\n$$x_{k+1} = \\Phi(x_k) + \\sum_{s=1}^m \\alpha_s (x_{k - s} - \\Phi(x_{k - s})), $$\n\nand the parameters $\\alpha_s$ are selected to minimize the norm of the residual.",
"_____no_output_____"
],
[
"## Battling the condition number\n\nThe condition number problem is **un-avoidable** if only the matrix-by-vector product is used.\n\nThus we need an **army of preconditioners** to solve it.\n\nThere are several **general purpose** preconditioners that we can use,\n\nbut often for a particular problem a special design is needed.",
"_____no_output_____"
],
[
"## Preconditioner: general concept\n\nThe general concept of the preconditioner is simple:\n\nGiven a linear system \n\n$$A x = f,$$\n\nwe want to find the matrix $P_R$ (or $P_L$) such that \n\n1. Condition number of $AP_R^{-1}$ (right preconditioner) or $P^{-1}_LA$ (right preconditioner) or $P^{-1}_L A P_R^{-1}$ is better than for $A$\n2. We can easily solve $P_Ly = g$ or $P_Ry = g$ for any $g$ (otherwise we could choose e.g. $P_L = A$)\n\nThen we solve for (right preconditioner)\n\n$$ AP_R^{-1} y = f \\quad \\Rightarrow \\quad P_R x = y$$ \n\nor (left preconditioner)\n\n$$ P_L^{-1} A x = P_L^{-1}f,$$ \nor even both\n$$ P_L^{-1} A P_R^{-1} y = P_L^{-1}f \\quad \\Rightarrow \\quad P_R x = y.$$ \n\nThe best choice is of course $P = A,$ but this does not make life easier.\n\nOne of the ideas is to use other iterative methods (beside Krylov) as preconditioners.",
"_____no_output_____"
],
[
"## Other iterative methods as preconditioners\nThere are other iterative methods that we have not mentioned. \n\n1. Jacobi method\n2. Gauss-Seidel\n3. SOR (Successive over-relaxation)",
"_____no_output_____"
],
[
"## Jacobi method (as preconditioner)\n\nConsider again the matrix with non-zero diagonal. To get the **Jacobi method** is when you express the diagonal element:\n\n$$a_{ii} x_i = -\\sum_{i \\ne j} a_{ij} x_j + f_i$$\n\nand use this to iteratively update $x_i$:\n$$ x_i^{(k+1)} = -\\frac{1}{a_{ii}}\\left( \\sum_{i \\ne j} a_{ij} x_j^{(k)} + f_i \\right),$$\nor in the matrix form\n$$\nx^{(k+1)} = D^{-1}\\left((D-A)x^{(k)} + f\\right) \n$$\nwhere $D = \\mathrm{diag}(A)$ and finally\n$$\nx^{(k+1)} = x^{(k)} - D^{-1}(Ax^{(k)} - f).\n$$\n\nSo, Jacobi method is nothing, but simple Richardson iteration with $\\tau=1$ and left preconditioner $P = D$ - diagonal of a matrix. Therefore we will refer to $P = \\mathrm{diag}(A)$ as **Jacobi preconditioner**. Note that it can be used for any other method like Chebyshev or Krylov-type methods.",
"_____no_output_____"
],
[
"## Properties of the Jacobi preconditioner\n\nJacobi preconditioner:\n\n1. Very easy to compute and apply\n2. Works well for diagonally dominant matrices (remember the Gershgorin circle theorem!)\n3. Useless if all diagonal entries are the same (proportional to the identity matrix)",
"_____no_output_____"
],
[
"## Gauss-Seidel (as preconditioner)\nAnother well-known method is **Gauss-Seidel method**. \n\nIts canonical form is very similar to the Jacobi method, with a small difference. When we update $x_i$ as\n\n$$x_i^{(k+1)} := -\\frac{1}{a_{ii}}\\left( \\sum_{j =1}^{i-1} a_{ij} x_j^{(k+1)} +\\sum_{j = i+1}^n a_{ij} x_j^{(k)} - f_i \\right)$$\n\nwe **use it in the later updates**. In the Jacobi method we use the full vector from the previous iteration.\n\nIts matrix form is more complicated.",
"_____no_output_____"
],
[
"## Gauss-Seidel: matrix version\n\nGiven $A = A^{*} > 0$ we have \n\n$$A = L + D + L^{*},$$\n\nwhere $D$ is the diagonal of $A$, $L$ is lower-triangular part with zero on the diagonal.\n\nOne iteration of the GS method reads\n$$\nx^{(k+1)} = x^{(k)} - (L + D)^{-1}(Ax^{(k)} - f).\n$$\nand we refer to the preconditioner $P = L+D$ as **Gauss-Seidel preconditioner**.\n\n**Good news: ** $\\rho(I - (L+D)^{-1} A) < 1, $\n\nwhere $\\rho$ is the spectral radius,\n\ni.e. for a positive definite matrix GS-method always converges.",
"_____no_output_____"
],
[
"## Gauss-Seidel and coordinate descent\n\nGS-method can be viewed as a coordinate descent method, applied to the energy functional\n\n$$F(x) = (Ax, x) - 2(f, x)$$\n\nwith the iteration \n\n$$x_i := \\arg \\min_z F(x_1, \\ldots, x_{i-1}, z, x_{i+1}, \\ldots, x_d).$$\n\nMoreover, the order in which we eliminate variables, is really important!",
"_____no_output_____"
],
[
"## Side note: Nonlinear Gauss-Seidel (a.k.a coordinate descent)\nIf $F$ is given, and we optimize one coordinate at a time, we have\n\n\n$$x_i := \\arg \\min_z F(x_1, \\ldots, x_{i-1}, z, x_{i+1}, \\ldots, x_d).$$\n\n**Note** the convergence result for block coordinate descent for the case of a general functional $F$: \n\nit converges locally with the speed of the GS-method applied to the **Hessian** $$H = \\nabla^2 F$$ of the functional.\n\nThus, if $F$ is twice differentiable and $x_*$ is the local minimum, then $H > 0$ can Gauss-Seidel converges.",
"_____no_output_____"
],
[
"## Successive overrelaxation (as preconditioner)\n\nWe can even introduce a parameter into the GS-method preconditioner, giving a **successive over-relaxation** (**SOR**) method:\n\n$$\nx^{(k+1)} = x^{(k)} - \\omega (D + \\omega L)^{-1}(Ax^{(k)} - f).\n$$\n\n$$P = \\frac{1}{\\omega}(D+\\omega L).$$\n\nConverges for $0<\\omega < 2$. Optimal selection of $\\omega$ is **not trivial**. Note that $\\omega = 1$ gives us a Gauss-Seidel preconditioner.\n",
"_____no_output_____"
],
[
"## Preconditioners for sparse matrices\n\nIf $A$ is sparse, one iteration of Jacobi, GS and SOR method is cheap. \n\nFor GS, we need to solve linear system with a sparse triangular matrix $L$, which costs $\\mathcal{O}(nnz)$.\n\nFor sparse matrices, however, there are more complicated algorithms, based on the idea of **approximate** LU-decomposition.\n\nRemember the motivation for CG: possibility of the early stopping, how to do approximate LU-decomposition for a sparse matrix? ",
"_____no_output_____"
],
[
"## Remember the Gaussian elimination\nLet us remember the basic method for solving linear systems:\n\nDecompose the matrix $A$ in the form \n\n$$A = P_1 L U P^{\\top}_2, $$\n\nwhere $P_1$ and $P_2$ are certain **permutation** matrices (which do the pivoting).\n\nThe most natural idea is to use **sparse** $L$ and $U$. \n\nIt is not possible without **fill-in** growth for example for matrices, coming from 2D/3D Partial Differential equations (PDEs).\n\nWhat to do?",
"_____no_output_____"
],
[
"## Incomplete LU\n\n\nSuppose you want to eliminate a variable $x_1$,\n\nand the equations have the form\n\n$$5 x_1 + x_4 + x_{10} = 1, \\quad 3 x_1 + x_4 + x_8 = 0, \\ldots,$$\n\nand in all other equations $x_1$ are not present. \n\nAfter the elimination, only $x_{10}$ will enter additionally to the second equation (new fill-in).\n\n$$x_4 + x_8 + 3(1 - x_4 - x_{10})/5 = 0$$\n\nIn the Incomplete $LU$ case (actually, ILU(0)) we just throw away the **new fill-in**.",
"_____no_output_____"
],
[
"## Incomplete-LU: formal definition\n\nWe run the usual LU-decomposition cycle, but avoid inserting non-zeros **other** than the initial non-zero pattern. \n\n``\n L = np.zeros((n, n))\n U = np.zeros((n, n))\n for k in range(n): #Eliminate one row \n L[k, k] = 1\n for i in range(k+1, n):\n L[i, k] = a[i, k] / a[k, k]\n for j in range(k+1, n):\n a[i, j] = a[i, j] - L[i, k] * a[k, j] #New fill-ins appear here\n for j in range(k, n):\n U[k, j] = a[k, j]\n``",
"_____no_output_____"
],
[
"## ILU(k)\n\nYousef Saad (who is the author of GMRES) also had a seminal paper on the **Incomplete LU** decomposition\n\nA good book on the topic is Saad, Yousef (1996), Iterative methods for sparse linear systems \n\nAnd he proposed **ILU(k)** method, which has a nice interpretation in terms of graphs.",
"_____no_output_____"
],
[
"## ILU(k): idea\n\nThe idea of ILU(k) is very instructive and is based on the connection between sparse matrices and graphs.\n\nSuppose you have an $n \\times n$ matrix $A$ and a corresponding adjacency graph.\n\nThen we eliminate one variable (vertex) and get a smaller system of size $(n-1) \\times (n-1)$.\n\nNew edges (=fill-in) appears between high-order neighbors.",
"_____no_output_____"
],
[
"## LU & graphs\n\nThe new edge can appear only between vertices that had common neighbour: it means, that they are second-order neigbours. This is also a sparsity pattern of the matrix \n\n$$A^2.$$\n\nThe **ILU(k)** idea is to leave only the elements in $L$ and $U$ that are $k$-order neighbours in the original graph.\n\nThe ILU(2) is very efficient, but for some reason completely abandoned (i.e. there is no implementation in MATLAB and scipy).\n\nThere is an original Sparsekit software by Saad, which works quite well.",
"_____no_output_____"
],
[
"## ILU Thresholded (ILUT)\nA much more popular approach is based on the so-called **thresholded LU**.\n\nYou do the standard Gaussian elimination with fill-ins, but either:\n\n- Throw away elements that are smaller than threshold, and/or control the amount of non-zeros you are allowed to store.\n\n- The smaller is the threshold, the better is the preconditioner, but more memory it takes.\n\nIt is denoted ILUT($\\tau$).",
"_____no_output_____"
],
[
"## Symmetric positive definite case\n\nIn the SPD case, instead of incomplete LU you should use Incomplete Cholesky, which is twice faster and consumes twice less memory.\n\nBoth **ILUT** and **Ichol** are implemented in scipy and you can try (nothing quite fancy, but it works).",
"_____no_output_____"
],
[
"## Second-order LU preconditioners\n\nThere is a more efficient (but much less popular due to the limit of open-source implementations) **second-order** LU factorization [proposed by I. Kaporin](http://www.researchgate.net/profile/I_Kaporin/publication/242940993_High_quality_preconditioning_of_a_general_symmetric_positive_definite_matrix_based_on_its_UTU__UTR__RTU-decomposition/links/53f72ad90cf2888a74976f54.pdf)\n\nThe idea is to approximate the matrix in the form\n\n$$A \\approx U_2 U^{\\top}_2 + U^{\\top}_2 R_2 + R^{\\top}_2 U_2,$$\n\nwhich is just the expansion of the $UU^{\\top}$ with respect to the perturbation of $U$. \n\nwhere $U_1$ and $U_2$ are low-triangular and sparse, whereare $R_2$ is small with respect to the drop tolerance parameter.\n\n",
"_____no_output_____"
],
[
"## What else?\nBesides the ILU/IC approaches, there is a very efficient approach, called **algebraic multigrid (AMG)**.\n\nIt was first proposed by Ruge and Stuben in 1987.\n\nIt is probably the only **black-box** method that is asymptotically optimal for 2D/3D problems \n\n(at least, for elliptic problems).\n\nThe idea comes from the **geometric multigrid**.\n\n\n",
"_____no_output_____"
],
[
"## Multigrid: just the basic idea\nThe basic idea of the multigrid is to consider a set of problems on **coarser meshes** (instead of the original problem).\n\nThen we solve a system on a coarse mesh, and interpolate it to a finer mesh.\n\nGeometric multigrid uses the information about the meshes, operators, discretization..\n\nAMG method tries to guess it from the **matrix**.\n\nMore in the **FastPDE course** :)",
"_____no_output_____"
],
[
"## Summary of this part\n\n- Jacobi, Gauss-Seidel, SSOR (as preconditioners)\n- Incomplete LU, three flavours: ILU(k), ILUT, ILU2",
"_____no_output_____"
],
[
"## Iterative methods for other NLA problems\n\nUp to now, we only talked about the iterative methods for **linear systems**.\n\nThere are other important large-scale problems:\n\n1. (Partial) Eigenvalue problem: $Ax_k = \\lambda_k x_k.$\n2. (Partial) SVD problem: $A v_k = \\sigma_k u_k, \\quad A^* u_k = \\sigma_k v_k$.\n\n**It is extremely difficult to compute all eigenvalues/singular values of a matrix**\n\n(singular vectors / eigenvectors are not sparse, so we need to store them all!).\n\nBut it is possible to solve **partial eigenvalue problems**. \n\nRecall, that the SVD follows from the symmetric eigenvalue problem.",
"_____no_output_____"
],
[
"## Iterative method for eigenvalue problems\n\nWe started the journey through iterative methods from the power method to compute the eigenvector, corresponding to the largest eigenvalue.\n\n$$x := \\frac{Ax}{\\Vert Ax \\Vert}.$$\n\nThe solution obviously lies in the Krylov space. \n\nIn the general symmetric case, this eigenvector maximizes the **Rayleigh quotient**\n\n$$ x_* = \\arg \\max_x \\frac{(Ax, x)}{(x, x)}.$$\n\nThus, we can try to look for the approximate solution in a subspace $Q$, and find the coefficients from the reduced eigenvalue problem\n\n$$(Q^* A Q) \\widehat{x} = \\widehat{\\lambda} \\widehat{x},$$\n\nwith the hope that the eigenvalue of $Q^* A Q$ will converge to the eigenvalue of $A$.\n\nThe machinery is similar to linear systems.",
"_____no_output_____"
],
[
"## General Krylov scheme for eigenvalues\n\nA general Krylov-Arnoldi-Lanczos scheme would be as follows: \n\n1. Generate the basis in the $j$-th Krylov subspace\n2. Compute the Galerkin projection $A_j = Q^*_j A Q_j$\n3. Compute (some of) the eigenvalues of $A_j$ (also called **Ritz values**) as an approximation to the eigenvalues of $A$.",
"_____no_output_____"
],
[
"## Difficulties\n\nThere are advantages/difficulties of this approach. \n- Approximation to several eigenvalues obtained simultaneouslu\n- At each step we need to recompute eigenvalues/eigenvectors of a tridiagonal (in the symmetric case) matrix $A$, and there is no simple update formula, and there is no way to efficiently recompute the eigenvalue/eigenvector in an efficient way (compared to linear system)\n- Thus, no robust stopping criteta is availabl\n",
"_____no_output_____"
],
[
"## Alternatives\n\nAs an alternative, there are several methods:\n\n- Jacobi-Davidson\n- Locally Optimal Block Conjugate Gradient \n- Inverse iteration\n\nThey focus on some of the eigenvalues and use different principles.",
"_____no_output_____"
],
[
"## Next lecture\n\n- Eigenvalue problems in more details.",
"_____no_output_____"
],
[
"# Questions?",
"_____no_output_____"
]
],
[
[
"from IPython.core.display import HTML\ndef css_styling():\n styles = open(\"./styles/custom.css\", \"r\").read()\n return HTML(styles)\ncss_styling()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.