hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ecf024b4ca3f7b63e662c20153ac462d10ba3420 | 27,751 | ipynb | Jupyter Notebook | courses/machine_learning/deepdive/10_recommend/content_based_by_hand.ipynb | alivcor/training-data-analyst | 3ea2e955be0cdd52a391cef73cce439c1a99be0c | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive/10_recommend/content_based_by_hand.ipynb | alivcor/training-data-analyst | 3ea2e955be0cdd52a391cef73cce439c1a99be0c | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive/10_recommend/content_based_by_hand.ipynb | alivcor/training-data-analyst | 3ea2e955be0cdd52a391cef73cce439c1a99be0c | [
"Apache-2.0"
] | null | null | null | 36.610818 | 486 | 0.55598 | [
[
[
"## Content Based Filtering by hand\n\nThis lab illustrates how to implement a content based filter using low level Tensorflow operations. \nThe code here follows the technique explained in Module 2 of Recommendation Engines: Content Based Filtering.\n\n",
"_____no_output_____"
]
],
[
[
"!pip install tensorflow==2.1",
"Collecting tensorflow==2.1\n Downloading tensorflow-2.1.0-cp37-cp37m-manylinux2010_x86_64.whl (421.8 MB)\n\u001b[K |████████████████████████████████| 421.8 MB 14 kB/s s eta 0:00:01 |████████████████████████▊ | 326.6 MB 77.8 MB/s eta 0:00:02\n\u001b[?25hRequirement already satisfied: termcolor>=1.1.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow==2.1) (1.1.0)\nRequirement already satisfied: wrapt>=1.11.1 in /opt/conda/lib/python3.7/site-packages (from tensorflow==2.1) (1.12.1)\nCollecting tensorboard<2.2.0,>=2.1.0\n Downloading tensorboard-2.1.1-py3-none-any.whl (3.8 MB)\n\u001b[K |████████████████████████████████| 3.8 MB 82.3 MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied: keras-preprocessing>=1.1.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow==2.1) (1.1.2)\nRequirement already satisfied: opt-einsum>=2.3.2 in /opt/conda/lib/python3.7/site-packages (from tensorflow==2.1) (3.3.0)\nCollecting tensorflow-estimator<2.2.0,>=2.1.0rc0\n Downloading tensorflow_estimator-2.1.0-py2.py3-none-any.whl (448 kB)\n\u001b[K |████████████████████████████████| 448 kB 55.5 MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied: google-pasta>=0.1.6 in /opt/conda/lib/python3.7/site-packages (from tensorflow==2.1) (0.2.0)\nRequirement already satisfied: absl-py>=0.7.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow==2.1) (0.8.1)\nRequirement already satisfied: grpcio>=1.8.6 in /opt/conda/lib/python3.7/site-packages (from tensorflow==2.1) (1.32.0)\nRequirement already satisfied: numpy<2.0,>=1.16.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow==2.1) (1.18.5)\nCollecting scipy==1.4.1; python_version >= \"3\"\n Downloading scipy-1.4.1-cp37-cp37m-manylinux1_x86_64.whl (26.1 MB)\n\u001b[K |████████████████████████████████| 26.1 MB 50.1 MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied: six>=1.12.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow==2.1) (1.15.0)\nRequirement already satisfied: protobuf>=3.8.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow==2.1) (3.13.0)\nCollecting keras-applications>=1.0.8\n Downloading Keras_Applications-1.0.8-py3-none-any.whl (50 kB)\n\u001b[K |████████████████████████████████| 50 kB 8.1 MB/s eta 0:00:01\n\u001b[?25hRequirement already satisfied: wheel>=0.26; python_version >= \"3\" in /opt/conda/lib/python3.7/site-packages (from tensorflow==2.1) (0.35.1)\nCollecting astor>=0.6.0\n Downloading astor-0.8.1-py2.py3-none-any.whl (27 kB)\nCollecting gast==0.2.2\n Downloading gast-0.2.2.tar.gz (10 kB)\nRequirement already satisfied: setuptools>=41.0.0 in /opt/conda/lib/python3.7/site-packages (from tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (50.3.0)\nRequirement already satisfied: requests<3,>=2.21.0 in /opt/conda/lib/python3.7/site-packages (from tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (2.24.0)\nRequirement already satisfied: markdown>=2.6.8 in /opt/conda/lib/python3.7/site-packages (from tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (3.3.1)\nRequirement already satisfied: google-auth-oauthlib<0.5,>=0.4.1 in /opt/conda/lib/python3.7/site-packages (from tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (0.4.1)\nRequirement already satisfied: werkzeug>=0.11.15 in /opt/conda/lib/python3.7/site-packages (from tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (1.0.1)\nRequirement already satisfied: google-auth<2,>=1.6.3 in /opt/conda/lib/python3.7/site-packages (from tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (1.22.1)\nRequirement already satisfied: h5py in /opt/conda/lib/python3.7/site-packages (from keras-applications>=1.0.8->tensorflow==2.1) (2.10.0)\nRequirement already satisfied: idna<3,>=2.5 in /opt/conda/lib/python3.7/site-packages (from requests<3,>=2.21.0->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (2.10)\nRequirement already satisfied: chardet<4,>=3.0.2 in /opt/conda/lib/python3.7/site-packages (from requests<3,>=2.21.0->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (3.0.4)\nRequirement already satisfied: certifi>=2017.4.17 in /opt/conda/lib/python3.7/site-packages (from requests<3,>=2.21.0->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (2020.6.20)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /opt/conda/lib/python3.7/site-packages (from requests<3,>=2.21.0->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (1.25.10)\nRequirement already satisfied: importlib-metadata; python_version < \"3.8\" in /opt/conda/lib/python3.7/site-packages (from markdown>=2.6.8->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (2.0.0)\nRequirement already satisfied: requests-oauthlib>=0.7.0 in /opt/conda/lib/python3.7/site-packages (from google-auth-oauthlib<0.5,>=0.4.1->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (1.3.0)\nRequirement already satisfied: cachetools<5.0,>=2.0.0 in /opt/conda/lib/python3.7/site-packages (from google-auth<2,>=1.6.3->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (4.1.1)\nRequirement already satisfied: rsa<5,>=3.1.4; python_version >= \"3.5\" in /opt/conda/lib/python3.7/site-packages (from google-auth<2,>=1.6.3->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (4.6)\nRequirement already satisfied: pyasn1-modules>=0.2.1 in /opt/conda/lib/python3.7/site-packages (from google-auth<2,>=1.6.3->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (0.2.8)\nRequirement already satisfied: zipp>=0.5 in /opt/conda/lib/python3.7/site-packages (from importlib-metadata; python_version < \"3.8\"->markdown>=2.6.8->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (3.3.0)\nRequirement already satisfied: oauthlib>=3.0.0 in /opt/conda/lib/python3.7/site-packages (from requests-oauthlib>=0.7.0->google-auth-oauthlib<0.5,>=0.4.1->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (3.1.0)\nRequirement already satisfied: pyasn1>=0.1.3 in /opt/conda/lib/python3.7/site-packages (from rsa<5,>=3.1.4; python_version >= \"3.5\"->google-auth<2,>=1.6.3->tensorboard<2.2.0,>=2.1.0->tensorflow==2.1) (0.4.8)\nBuilding wheels for collected packages: gast\n Building wheel for gast (setup.py) ... \u001b[?25ldone\n\u001b[?25h Created wheel for gast: filename=gast-0.2.2-py3-none-any.whl size=7540 sha256=9081df2f424dd228de82394670104c60a92848ba81981934dd88f7ccbc0e8b0d\n Stored in directory: /home/jupyter/.cache/pip/wheels/21/7f/02/420f32a803f7d0967b48dd823da3f558c5166991bfd204eef3\nSuccessfully built gast\nInstalling collected packages: tensorboard, tensorflow-estimator, scipy, keras-applications, astor, gast, tensorflow\n Attempting uninstall: tensorboard\n Found existing installation: tensorboard 2.3.0\n Uninstalling tensorboard-2.3.0:\n Successfully uninstalled tensorboard-2.3.0\n Attempting uninstall: tensorflow-estimator\n Found existing installation: tensorflow-estimator 2.3.0\n Uninstalling tensorflow-estimator-2.3.0:\n Successfully uninstalled tensorflow-estimator-2.3.0\n Attempting uninstall: scipy\n Found existing installation: scipy 1.5.2\n Uninstalling scipy-1.5.2:\n Successfully uninstalled scipy-1.5.2\n Attempting uninstall: gast\n Found existing installation: gast 0.3.3\n Uninstalling gast-0.3.3:\n Successfully uninstalled gast-0.3.3\n Attempting uninstall: tensorflow\n Found existing installation: tensorflow 2.3.1\n Uninstalling tensorflow-2.3.1:\n Successfully uninstalled tensorflow-2.3.1\n\u001b[31mERROR: After October 2020 you may experience errors when installing or updating packages. This is because pip will change the way that it resolves dependency conflicts.\n\nWe recommend you use --use-feature=2020-resolver to test your packages with the new resolver before it becomes the default.\n\ntfx 0.23.0 requires attrs<20,>=19.3.0, but you'll have attrs 20.2.0 which is incompatible.\ntfx 0.23.0 requires google-resumable-media<0.7.0,>=0.6.0, but you'll have google-resumable-media 1.1.0 which is incompatible.\ntfx 0.23.0 requires pyarrow<0.18,>=0.17, but you'll have pyarrow 1.0.1 which is incompatible.\ntfx 0.23.0 requires tensorflow!=2.0.*,!=2.1.*,!=2.2.*,<3,>=1.15.2, but you'll have tensorflow 2.1.0 which is incompatible.\ntfx-bsl 0.23.0 requires pyarrow<0.18,>=0.17, but you'll have pyarrow 1.0.1 which is incompatible.\ntfx-bsl 0.23.0 requires tensorflow!=2.0.*,!=2.1.*,!=2.2.*,<3,>=1.15.2, but you'll have tensorflow 2.1.0 which is incompatible.\ntensorflow-transform 0.23.0 requires tensorflow!=2.0.*,!=2.1.*,!=2.2.*,<2.4,>=1.15.2, but you'll have tensorflow 2.1.0 which is incompatible.\ntensorflow-serving-api 2.3.0 requires tensorflow<3,>=2.3, but you'll have tensorflow 2.1.0 which is incompatible.\ntensorflow-probability 0.11.0 requires cloudpickle==1.3, but you'll have cloudpickle 1.6.0 which is incompatible.\ntensorflow-probability 0.11.0 requires gast>=0.3.2, but you'll have gast 0.2.2 which is incompatible.\ntensorflow-model-analysis 0.23.0 requires pyarrow<0.18,>=0.17, but you'll have pyarrow 1.0.1 which is incompatible.\ntensorflow-model-analysis 0.23.0 requires tensorflow!=2.0.*,!=2.1.*,!=2.2.*,<3,>=1.15.2, but you'll have tensorflow 2.1.0 which is incompatible.\ntensorflow-io 0.15.0 requires tensorflow<2.4.0,>=2.3.0, but you'll have tensorflow 2.1.0 which is incompatible.\ntensorflow-data-validation 0.23.1 requires joblib<0.15,>=0.12, but you'll have joblib 0.17.0 which is incompatible.\ntensorflow-data-validation 0.23.1 requires pyarrow<0.18,>=0.17, but you'll have pyarrow 1.0.1 which is incompatible.\ntensorflow-data-validation 0.23.1 requires tensorflow!=2.0.*,!=2.1.*,!=2.2.*,<3,>=1.15.2, but you'll have tensorflow 2.1.0 which is incompatible.\nkeras 2.4.0 requires tensorflow>=2.2.0, but you'll have tensorflow 2.1.0 which is incompatible.\u001b[0m\nSuccessfully installed astor-0.8.1 gast-0.2.2 keras-applications-1.0.8 scipy-1.4.1 tensorboard-2.1.1 tensorflow-2.1.0 tensorflow-estimator-2.1.0\n"
]
],
[
[
"Make sure to restart your kernel to ensure this change has taken place.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport tensorflow as tf\n\nprint(tf.__version__)",
"2.1.0\n"
]
],
[
[
"To start, we'll create our list of users, movies and features. While the users and movies represent elements in our database, for a content-based filtering method the features of the movies are likely hand-engineered and rely on domain knowledge to provide the best embedding space. Here we use the categories of Action, Sci-Fi, Comedy, Cartoon, and Drama to describe our movies (and thus our users).\n\nIn this example, we will assume our database consists of four users and six movies, listed below. ",
"_____no_output_____"
]
],
[
[
"users = ['Ryan', 'Danielle', 'Vijay', 'Chris']\nmovies = ['Star Wars', 'The Dark Knight', 'Shrek', 'The Incredibles', 'Bleu', 'Memento']\nfeatures = ['Action', 'Sci-Fi', 'Comedy', 'Cartoon', 'Drama']\n\nnum_users = len(users)\nnum_movies = len(movies)\nnum_feats = len(features)\nnum_recommendations = 2",
"_____no_output_____"
]
],
[
[
"### Initialize our users, movie ratings and features\n\nWe'll need to enter the user's movie ratings and the k-hot encoded movie features matrix. Each row of the users_movies matrix represents a single user's rating (from 1 to 10) for each movie. A zero indicates that the user has not seen/rated that movie. The movies_feats matrix contains the features for each of the given movies. Each row represents one of the six movies, the columns represent the five categories. A one indicates that a movie fits within a given genre/category. ",
"_____no_output_____"
]
],
[
[
"# each row represents a user's rating for the different movies\nusers_movies = tf.constant([\n [4, 6, 8, 0, 0, 0],\n [0, 0, 10, 0, 8, 3],\n [0, 6, 0, 0, 3, 7],\n [10, 9, 0, 5, 0, 2]],dtype=tf.float32)\n\n# features of the movies one-hot encoded\n# e.g. columns could represent ['Action', 'Sci-Fi', 'Comedy', 'Cartoon', 'Drama']\nmovies_feats = tf.constant([\n [1, 1, 0, 0, 1],\n [1, 1, 0, 0, 0],\n [0, 0, 1, 1, 0],\n [1, 0, 1, 1, 0],\n [0, 0, 0, 0, 1],\n [1, 0, 0, 0, 1]],dtype=tf.float32)",
"_____no_output_____"
]
],
[
[
"### Computing the user feature matrix\n\nWe will compute the user feature matrix; that is, a matrix containing each user's embedding in the five-dimensional feature space. ",
"_____no_output_____"
]
],
[
[
"## Computing user0's feature vector",
"_____no_output_____"
],
[
"a = tf.constant([[1,2,3],[4,5,6]])\nb = tf.expand_dims(a, axis=1)\nb",
"_____no_output_____"
],
[
"user0_movies = users_movies[0]\nuser0_movies = tf.expand_dims(user0_movies, axis=1)\nuser0_movies",
"_____no_output_____"
],
[
"movies_feats",
"_____no_output_____"
],
[
"user0_expanded_feats = user0_movies*movies_feats\nuser0_expanded_feats",
"_____no_output_____"
],
[
"user0_feats = tf.reduce_sum(user0_expanded_feats, axis=0)\nuser0_feats",
"_____no_output_____"
],
[
"#original",
"_____no_output_____"
],
[
"users_feats = tf.matmul(users_movies, movies_feats)\nusers_feats",
"_____no_output_____"
]
],
[
[
"Next we normalize each user feature vector to sum to 1. Normalizing isn't strictly neccesary, but it makes it so that rating magnitudes will be comparable between users.",
"_____no_output_____"
]
],
[
[
"users_feats = \nusers_feats",
"_____no_output_____"
]
],
[
[
"#### Ranking feature relevance for each user\n\nWe can use the users_feats computed above to represent the relative importance of each movie category for each user. ",
"_____no_output_____"
]
],
[
[
"top_users_features = \ntop_users_features",
"_____no_output_____"
],
[
" ",
"Ryan: ['Action', 'Sci-Fi', 'Comedy', 'Cartoon', 'Drama']\nDanielle: ['Drama', 'Comedy', 'Cartoon', 'Action', 'Sci-Fi']\nVijay: ['Action', 'Drama', 'Sci-Fi', 'Comedy', 'Cartoon']\nChris: ['Action', 'Sci-Fi', 'Drama', 'Comedy', 'Cartoon']\n"
]
],
[
[
"### Determining movie recommendations. \n\nWe'll now use the `users_feats` tensor we computed above to determine the movie ratings and recommendations for each user.\n\nTo compute the projected ratings for each movie, we compute the similarity measure between the user's feature vector and the corresponding movie feature vector. \n\nWe will use the dot product as our similarity measure. In essence, this is a weighted movie average for each user.",
"_____no_output_____"
]
],
[
[
"users_ratings = \nusers_ratings",
"_____no_output_____"
]
],
[
[
"The computation above finds the similarity measure between each user and each movie in our database. To focus only on the ratings for new movies, we apply a mask to the all_users_ratings matrix. \n\nIf a user has already rated a movie, we ignore that rating. This way, we only focus on ratings for previously unseen/unrated movies.",
"_____no_output_____"
]
],
[
[
"users_ratings_new = \nusers_ratings_new",
"_____no_output_____"
]
],
[
[
"Finally let's grab and print out the top 2 rated movies for each user",
"_____no_output_____"
]
],
[
[
"top_movies = \ntop_movies",
"_____no_output_____"
],
[
"for i in range(num_users):\n movie_names = [movies[index] for index in top_movies[i]]\n print('{}: {}'.format(users[i],movie_names))",
"Ryan: ['The Incredibles', 'Memento']\nDanielle: ['The Incredibles', 'Star Wars']\nVijay: ['Star Wars', 'The Incredibles']\nChris: ['Bleu', 'Shrek']\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecf02fb4386ae5c994d8f9b56eb886e1ff81fe7c | 68,593 | ipynb | Jupyter Notebook | examples/lazy_shared_calc.ipynb | blshao84/tributary | 20e41f782d46fa5d8c340629ed60719741702e7e | [
"Apache-2.0"
] | 357 | 2018-09-13T19:58:46.000Z | 2022-03-31T17:22:20.000Z | examples/lazy_shared_calc.ipynb | blshao84/tributary | 20e41f782d46fa5d8c340629ed60719741702e7e | [
"Apache-2.0"
] | 109 | 2018-09-13T18:37:00.000Z | 2022-03-27T00:59:49.000Z | examples/lazy_shared_calc.ipynb | blshao84/tributary | 20e41f782d46fa5d8c340629ed60719741702e7e | [
"Apache-2.0"
] | 36 | 2018-09-17T21:01:05.000Z | 2022-03-26T02:41:37.000Z | 66.081888 | 2,356 | 0.525054 | [
[
[
"import tributary.symbolic as ts\nimport tributary.lazy as tl",
"_____no_output_____"
],
[
"expr = ts.parse_expression(\"10sin**2 x**2 + 3xyz + tan theta\")",
"_____no_output_____"
],
[
"clz = ts.construct_lazy(expr)\n\nclass Derived(clz):\n def __init__(self, **kwargs):\n super(Derived, self).__init__(**kwargs)\n\n @tl.node\n def out2(self):\n return self.evaluate() + 1\n\n @tl.node\n def out3(self):\n return self.evaluate() + self.out2()\n\nx = Derived(x=1, y=2, z=3, theta=4)",
"_____no_output_____"
],
[
"x.evaluate()()",
"_____no_output_____"
],
[
"x.evaluate().graphviz()",
"_____no_output_____"
],
[
"x.out2()()",
"_____no_output_____"
],
[
"x.out2().graphviz()",
"_____no_output_____"
],
[
"x.out3()()",
"_____no_output_____"
],
[
"x.out3().graphviz()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf04ad703d77d1ed92788ca0ae51a72e6d77387 | 89,596 | ipynb | Jupyter Notebook | notebooks/interval.ipynb | elba-docker/analysis | 65c3bf36932bce35777ba4d2a2578f689e14b32c | [
"MIT"
] | 1 | 2020-04-23T06:19:49.000Z | 2020-04-23T06:19:49.000Z | notebooks/interval.ipynb | elba-docker/analysis | 65c3bf36932bce35777ba4d2a2578f689e14b32c | [
"MIT"
] | 1 | 2020-04-27T06:58:46.000Z | 2020-04-30T07:04:19.000Z | notebooks/interval.ipynb | elba-kubernetes/analysis | 65c3bf36932bce35777ba4d2a2578f689e14b32c | [
"MIT"
] | null | null | null | 25.686927 | 419 | 0.579289 | [
[
[
"# Add analysis to the path\nimport sys\nimport pathlib\nparent_dir = os.path.join(os.path.abspath(''), \"..\")\nsys.path.append(parent_dir)\n\nimport parsers\nimport matplotlib.pyplot as plt\nimport portion\nimport functools\nimport traceback\nimport math\nfrom aggregation import *\nfrom matplotlib import rc\nfrom collections import namedtuple\nfrom dataclasses import dataclass\nfrom pprint import pprint, pformat\n\nrc('font',**{'family': 'serif', 'size': 19})\nrc('text', usetex=True)\n\npath_to_results = os.path.normpath(os.path.join(parent_dir, \"archive\"))\nprint(path_to_results)\nworking_dir = os.path.normpath(os.path.join(parent_dir, \"working\"))\ndata = parsers.main(path_to_results, working_dir=working_dir)\nprint(\"Finished\")",
"/home/jazev/dev/cs4365/analysis/archive\nExtracting 30 top level archives on 4 workers\nFinished\n"
],
[
"CpuEntry = namedtuple('CpuEntry', 'time cpu interval')\n \ndef extract_intervals(iterable, predicate):\n first_matched = None\n last = None\n for (idx, item) in enumerate(iterable):\n if predicate(item):\n last = idx\n if first_matched is None:\n first_matched = idx\n else:\n if first_matched is not None:\n interval = portion.closed(first_matched, idx)\n first_matched = None\n last = None\n yield interval\n if first_matched is not None and last is not None:\n yield portion.closed(first_matched, last)\n\ndef get_cpu_entries(container: parsers.Container, sampling_period=0.1) -> List[CpuEntry]:\n if container is None:\n return []\n\n # Normalize entries from radvisor and moby\n entries = []\n if container.radvisor:\n entries.extend(CpuEntry(time=entry.read, cpu=entry.cpu.total, interval=None) for entry in container.radvisor[0].values())\n elif container.moby:\n entries.extend(CpuEntry(time=entry.read, cpu=entry.cpu.total, interval=None) for entry in container.moby.values())\n else:\n return []\n\n # Calculate the CPU percentages from the times by using the CPU time/timestamp deltas\n interval_zipped_lter = zip(\n find_deltas([entry.cpu for entry in entries]),\n find_deltas([entry.time for entry in entries]),\n entries[1:])\n utilization_entries = [CpuEntry(time=entry.time / 1E6, cpu=float(c) / t, interval=t)\n for (c, t, entry) in interval_zipped_lter if (float(c) / t) > 0] \n\n if sampling_period is not None:\n # Sample the CPU utilization\n times = [entry.time for entry in utilization_entries]\n cpus = [entry.cpu for entry in utilization_entries]\n intervals = [entry.interval for entry in utilization_entries]\n min_time = min(times)\n max_time = max(times)\n sampling_period_ms = sampling_period * 1E3\n sampling_intervals = pd.interval_range(\n start=min_time, end=max_time, freq=sampling_period_ms)\n cpu_df = pd.DataFrame({'cpu': cpus, 'time': times, 'interval': intervals})\n cpu_df['sampling_intervals'] = pd.cut(\n x=cpu_df['time'], bins=sampling_intervals, include_lowest=True)\n cpu_df = cpu_df.groupby('sampling_intervals').mean()\n return [CpuEntry(cpu=row[\"cpu\"], time=row[\"time\"], interval=row[\"interval\"]) for idx, row in cpu_df.iterrows() if row[\"cpu\"] > 0]\n else:\n return utilization_entries\n\n\ndef get_load_interval(host: parsers.TestHost, sampling_period=0.1, min_length: int=5, load_lower_bound=0.6) -> portion.Interval:\n load_interval_lists = []\n\n containers = host.containers()\n if not containers:\n return None\n\n for container in host.containers():\n sampled_entries = get_cpu_entries(container, sampling_period=sampling_period)\n \n # Perform aggregation on measurements\n def make_time_intervals(predicate):\n idle_index_intervals = list(extract_intervals(sampled_entries, predicate))\n return [portion.closed(sampled_entries[i.lower].time, sampled_entries[i.upper].time)\n for i in idle_index_intervals]\n \n load_intervals = [interval for interval in make_time_intervals(\n lambda e: e.cpu >= load_lower_bound) if interval.upper - interval.lower >= min_length - 1]\n load_interval_lists.append(load_intervals)\n\n # Find universal intersections of intervals\n def combine_interval_lists(a, b):\n combined_product_intervals = (i & j for i in a for j in b)\n return [interval for interval in combined_product_intervals if not interval.empty]\n\n load_intervals = functools.reduce(lambda a, b: combine_interval_lists(a, b),\n load_interval_lists, portion.open(-portion.inf, portion.inf))\n\n # Take union of all universally intersected intervals\n return functools.reduce(lambda i1, i2: i1 | i2, load_intervals, portion.empty())\n",
"_____no_output_____"
],
[
"\"\"\"\nInvestigate container CPU utilization distribution for a single test\n\"\"\"\n\ndef get_cpu_distribution(test):\n best_lower_threshold = None\n all_cpu_reads = []\n for replica in data[test].replicas:\n for host in replica.hosts.values():\n containers = host.containers()\n if containers:\n for container in containers:\n cpu_entries = get_cpu_entries(container, sampling_period=0.1)\n for entry in cpu_entries:\n all_cpu_reads.append(entry.cpu)\n cpu_df = pd.DataFrame({'cpu': all_cpu_reads})\n print(cpu_df.describe(include=\"all\"))",
"_____no_output_____"
],
[
"def describe_intervals(test: parsers.Test, top=0.10) -> List[str]:\n output = []\n try:\n output.append(f\"==== {test.id} ====\")\n # First, describe all intervals for the test\n hosts = flatten(replica.hosts.values() for replica in test.replicas)\n all_host_intervals = [host_collection_intervals(host) for host in hosts]\n all_container_intervals = flatten(all_host_intervals)\n all_intervals = flatten(all_container_intervals)\n intervals, _ = zip(*all_intervals)\n intervals_df = pd.DataFrame({'Read deltas (ms)': intervals})\n output.append(str(intervals_df.describe(include='all')))\n output.append(\"\")\n\n # Second, describe top percentage of intervals\n top_percent = []\n top_thresholds = []\n for container_list in all_container_intervals:\n container_intervals, _ = zip(*container_list)\n limit = np.quantile(container_intervals, 1 - top)\n top_thresholds.append(limit)\n top_percent.extend(i for i in container_intervals if i > limit)\n top_percent_df = pd.DataFrame({f'Top {top*100:.1f}% container read deltas (ms)': top_percent})\n output.append(str(top_percent_df.describe(include='all')))\n top_threshold_avg = np.mean(top_thresholds)\n output.append(f\"top_threshold_mu: {top_threshold_avg:.8f}\")\n output.append(\"\")\n\n # Third, describe all intervals during load and not during load\n load_interval_values = []\n not_load_interval_values = []\n thresholds = []\n for host, host_intervals in zip(hosts, all_host_intervals):\n containers = host.containers()\n if not containers:\n continue\n \n cpu_entries = []\n for container in containers:\n cpu_entries.extend(get_cpu_entries(container, sampling_period=0.1))\n\n if not cpu_entries:\n output.append(f\"[WARN] No CPU entries found for {host.replica_id} in test {host.id}\")\n continue\n\n # cpu_entries = flatten([get_cpu_entries(container, sampling_period=0.1) for container in containers])\n load_threshold = 0.5 * np.percentile([entry.cpu for entry in cpu_entries], 0.75)\n load_interval = get_load_interval(host, load_lower_bound=load_threshold)\n thresholds.append(load_threshold)\n for interval, timestamp in flatten(host_intervals):\n if timestamp in load_interval:\n load_interval_values.append(interval)\n else:\n not_load_interval_values.append(interval)\n # average threshold\n threshold_avg = np.mean(thresholds)\n # distributions\n during_load_df = pd.DataFrame({f'Read deltas during load (> {threshold_avg:.1f}% CPU) (ms)': load_interval_values})\n output.append(str(during_load_df.describe(include='all')))\n not_during_load_df = pd.DataFrame({f'Read deltas not during load (> {threshold_avg:.1f}% CPU) (ms)': not_load_interval_values})\n output.append(str(not_during_load_df.describe(include='all')))\n output.append(f\"threshold_mu: {threshold_avg*100:.8f}\")\n # total time/proportion\n total_during_load = np.sum(load_interval_values) / 1E3\n total_not_during_load = np.sum(not_load_interval_values) / 1E3\n load = total_during_load / (total_during_load + total_not_during_load)\n output.append(f\"total during load: {total_during_load:.8f}; total not during load: {total_not_during_load:.8f}; load %: {load*100:.8f}\")\n output.append(f\"=================\")\n except Exception as ex:\n output.append(\"~~~~~~ failed ~~~~~~\")\n output.append(str(ex))\n output.append(\"~~~~~~ exc ~~~~~~\")\n output.append(traceback.format_exc())\n output.append(\"~~~~~~ stack ~~~~~~\")\n output.append(traceback.format_stack())\n output.append(f\"=================\")\n return output\n\ndef list_to_str(l):\n if isinstance(l, str):\n return l\n elif isinstance(l, list):\n return \"\\n\".join([list_to_str(i) for i in l])\n else:\n return pprint.pformat(l)\n ",
"_____no_output_____"
],
[
"# warning: takes a long time, outputs all lines to all_output\n\ntests = [\n \"d-rc-50\", \"d-rc-100\", \"d-mc-50\", \"d-mc-100\",\n \"i-rc-50\", \"i-rc-100\", \"i-mc-50\", \"i-mc-100\",\n \"ii-rc-s\", \"ii-rc-b\", \"ii-mc-s\", \"ii-mc-b\",\n \"d-r-50\", \"d-r-100\", \"d-m-50\", \"d-m-100\",\n \"i-r-50\", \"i-r-100\", \"i-m-50\", \"i-m-100\",\n \"ii-r-s\", \"ii-r-b\", \"ii-m-s\", \"ii-m-b\"]\nnum = len(tests)\ndone = 0\nall_output = []\nwith Pool(cpu_count()) as pool:\n for output in pool.imap_unordered(describe_intervals, (data[test] for test in tests)):\n all_output.append(output)\n print(f\"{done+1}/{num} done\")\n done += 1\n",
"_____no_output_____"
],
[
"# output to file\nwith open(\"interval_out.txt\", \"w\") as file_out:\n file_out.write(list_to_str(all_output))",
"_____no_output_____"
],
[
"# print to output\nprint(list_to_str(all_output))",
"_____no_output_____"
],
[
"def get_cpu_df(container: parsers.Container) -> pd.DataFrame:\n cpu_entries = get_cpu_entries(container, sampling_period=None)\n cpu, timestamp, interval = zip(*((entry.cpu, entry.time, entry.interval) for entry in cpu_entries))\n return pd.DataFrame({'cpu': cpu, 'timestamp': normalize(timestamp), 'interval': [i / 1E6 for i in interval]})\n\ndef get_container_cpu_versus_interval_df(container: parsers.Container, window=5) -> pd.DataFrame:\n cpu_df = get_cpu_df(container)\n cpu_df[\"cpu_mean\"] = cpu_df[\"cpu\"].rolling(window=window).mean()\n return cpu_df[cpu_df['cpu_mean'].notnull()]\n\ndef aggregate_cpu_versus_interval(replica: parsers.TestReplica, window=5):\n aggregate_df = None\n for host in replica.hosts.values():\n containers = host.containers()\n if not containers:\n continue\n else:\n container_dfs = [get_container_cpu_versus_interval_df(container, window=window) for container in containers]\n for container_df in container_dfs:\n if aggregate_df is not None:\n aggregate_df = aggregate_df.append(container_df)\n else:\n aggregate_df = container_df\n return aggregate_df",
"_____no_output_____"
],
[
"# x - cpu utilization over a 0.5 second rolling interval\n# y - measured interval\n\nfrom matplotlib import colors\nimport matplotlib.ticker as mticker\n\nplt.rcParams['lines.solid_capstyle'] = 'round'\nspecial_positions = [50, 70, 100, 200, 500, 1000]\n\n\ndef expand(x, y, gap=1e-4):\n add = np.tile([0, gap, np.nan], len(x))\n x1 = np.repeat(x, 3) + add\n y1 = np.repeat(y, 3) + add\n return x1, y1\n\n\ndef remap(alpha, a, b, c, d):\n return c + ((alpha - a) / (b - a)) * (d - c)\n\n\ndef nrange(min, max, num=20, mode='linear'):\n points = None\n if mode == 'log':\n points = list(np.logspace(math.log10(min), math.log10(max), num=num + 1, endpoint=True))\n else:\n points = list(np.linspace(min, max, num=num + 1, endpoint=True))\n if not points:\n return []\n else:\n return zip(points[0:-1], points[1:])\n\n\ndef np_grid_density_reduction(data, x_mode='linear', y_mode='linear', slices=10, density_factor=0.4):\n min_x = np.min(data[:,0])\n max_x = np.max(data[:,0])\n min_y = np.min(data[:,1])\n max_y = np.max(data[:,1])\n\n x_slice_size = (max_x - min_x) / slices\n y_slice_size = (max_y - min_y) / slices\n\n reduced_segments = []\n max_cell_density = (1 / float(slices ** 2)) * density_factor\n max_cell_size = int(max_cell_density * data.size)\n rng = np.random.default_rng()\n\n for (x1, x2) in nrange(min_x, max_x, num=slices, mode=x_mode):\n x_filtered = data[(x1 <= data[:,0]) & (data[:,0] < x2)]\n # if the entire slice is less than the cell size, no need to break it up\n if x_filtered.size // 2 > max_cell_size:\n for (y1, y2) in nrange(min_y, max_y, num=slices, mode=y_mode):\n y_filtered = x_filtered[(y1 <= x_filtered[:,1]) & (x_filtered[:,1] < y2)]\n if y_filtered.size // 2 > max_cell_size:\n reduced_segments.append(rng.choice(y_filtered, max_cell_size, replace=False))\n else:\n reduced_segments.append(y_filtered)\n else:\n reduced_segments.append(x_filtered)\n \n return np.concatenate(reduced_segments)\n\n\ndef scatter_cpu_versus_interval(ax, df, color, label=None, size=5, alpha=0.5, overlap=True, slices=10, sample=1, min_size=1, max_size=1):\n # Perform binned density reduction\n intervals = df['interval']\n cpu = [c * 100 for c in df['cpu_mean']]\n\n # Convert array to numpy array and perform visual grid-based reduction\n zipped_data = list((c, i) for (c, i) in zip(cpu, intervals) if i > 37.5 and 0 <= c <= 100)\n data_np_arr = np.array(zipped_data)\n data_np_arr = np.random.default_rng().choice(data_np_arr, int(sample * (data_np_arr.size // 2)), replace=False)\n cpu = [i[0] for i in data_np_arr]\n intervals = [i[1] for i in data_np_arr]\n \n min_size_interval= 50\n max_size_interval = 200\n sizes = [w * size for w in (\n min_size if i < min_size_interval else\n max_size if i > max_size_interval else\n remap(i, min_size_interval, max_size_interval, min_size, max_size)\n for i in intervals)]\n\n if overlap:\n alphas = [min(1, alpha * w) for w in (\n 0.75 if i < special_positions[1] else\n 1.0 if i < special_positions[2] else\n 1.35 for i in intervals)]\n dotcolors=[colors.to_rgba(color, a) for a in alphas]\n ax.scatter(cpu, intervals, s=sizes, color=dotcolors, edgecolors='none', label=label)\n else:\n ax.plot(*expand(cpu, intervals), lw=size, color=color, alpha=alpha)\n\n\ndef multiprocess_aggregate_cpu_versus_interval(tup):\n replica, window = tup\n return aggregate_cpu_versus_interval(replica, window=window)\n\n\ndef test_aggregate_cpu_versus_interval(test, window=5):\n # replicas = [data[test].replicas[0]]\n replicas = data[test].replicas\n aggregate_df = None\n with Pool(cpu_count()) as pool:\n for df in pool.imap_unordered(multiprocess_aggregate_cpu_versus_interval, zip(replicas, [window] * len(replicas))):\n if aggregate_df is None:\n aggregate_df = df\n else:\n aggregate_df.append(df)\n return aggregate_df\n\n\ndef plot_test_comparison(test_a, test_b, test_a_legend, test_b_legend, axes_labels, title, y_max=None, window=5, ax=None):\n alpha = 0.75\n size = 10\n density_factor = 1\n sample = 0.2\n slices = 50\n color_a = (0.2, 0.4, 0.6)\n color_b = (1, 0.3, 0.3)\n \n test_a_df = test_aggregate_cpu_versus_interval(test_a, window=window).sample(frac=sample, replace=False, random_state=1)\n test_b_df = test_aggregate_cpu_versus_interval(test_b, window=window).sample(frac=sample, replace=False, random_state=1)\n\n if y_max is not None:\n test_a_df = test_a_df[test_a_df['interval'] < y_max]\n test_b_df = test_b_df[test_b_df['interval'] < y_max]\n\n ax = ax or plt.gca()\n if y_max is None:\n ticks = [*special_positions]\n else:\n ticks = [p for p in special_positions if p <= y_max]\n for t in ticks:\n ax.axhline(y=t, color='black', linestyle='dashed', alpha=0.4, lw=0.8, dashes=[4, 4])\n\n ax.set_title(title, pad=10)\n scatter_cpu_versus_interval(ax, test_a_df, color=color_a, slices=slices, size=size, sample=1, alpha=alpha, overlap=True, label=test_a_legend)\n scatter_cpu_versus_interval(ax, test_b_df, color=color_b, slices=slices, size=size, sample=1, alpha=alpha, overlap=True, label=test_b_legend)\n ax.set_yscale('log')\n if axes_labels[0]:\n ax.set_ylabel(\"Collection interval (ms)\")\n if axes_labels[1]:\n ax.set_xlabel(\"CPU Utilization rolling average (\\%)\")\n ax.xaxis.labelpad = 5\n if test_a_legend and test_b_legend:\n lgnd = ax.legend(loc=\"upper right\")\n lgnd.legendHandles[0]._sizes = [70]\n lgnd.legendHandles[1]._sizes = [70]\n ax.set_yticks(ticks)\n ax.get_yaxis().set_major_formatter(mticker.ScalarFormatter())\n ax.get_yaxis().set_minor_formatter(mticker.NullFormatter())\n\n\nfig, ((ax1, ax2), (ax3, ax4), (ax5, ax6)) = plt.subplots(nrows=3, ncols=2, figsize=(12,16))\nplt.subplots_adjust(left=None, bottom=None, right=None, top=None, wspace=0.19, hspace=0.26)\nplot_test_comparison(\"d-m-50\", \"d-r-50\", \"Moby\", \"rAdvisor\", (True, False), \"Direct [\\\\texttt{d-}], 50\\\\% max CPU\", 200, ax=ax1)\nplot_test_comparison(\"d-m-100\", \"d-r-100\", None, None, (False, False), \"Direct [\\\\texttt{d-}], 100\\\\% max CPU\", 200, ax=ax2)\nplot_test_comparison(\"i-m-50\", \"i-r-50\", None, None, (True, False), \"Synthetic [\\\\texttt{i-}], 50\\\\% max CPU\", 200, ax=ax3)\nplot_test_comparison(\"i-m-100\", \"i-r-100\", None, None, (False, False), \"Synthetic [\\\\texttt{i-}], 100\\\\% max CPU\", 200, ax=ax4)\nplot_test_comparison(\"ii-m-b\", \"ii-r-b\", None, None, (True, True), \"Application [\\\\texttt{ii-}], Bursty\", 70, ax=ax5)\nplot_test_comparison(\"ii-m-s\", \"ii-r-s\", None, None, (False, True), \"Application [\\\\texttt{ii-}], Sustained\", 70, ax=ax6)\nplt.savefig('cpu_interval_matrix.pdf', bbox_inches='tight')\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf056f5694406b7993e3cb0451170c426bdebac | 11,309 | ipynb | Jupyter Notebook | model-arima-aggregate.ipynb | esnmrd/phd | 8454ee54a4202bdbe846c17707c4a49fd8489721 | [
"MIT"
] | null | null | null | model-arima-aggregate.ipynb | esnmrd/phd | 8454ee54a4202bdbe846c17707c4a49fd8489721 | [
"MIT"
] | null | null | null | model-arima-aggregate.ipynb | esnmrd/phd | 8454ee54a4202bdbe846c17707c4a49fd8489721 | [
"MIT"
] | null | null | null | 38.862543 | 160 | 0.549739 | [
[
[
"#### Training temporally aggregated ARIMA models for FCR estimation to compare its performance with our vehicle-specific metamodels at low-res scales\r\n#### Ehsan Moradi, Ph.D. Candidate",
"_____no_output_____"
]
],
[
[
"# Load required libraries\r\nimport numpy as np\r\nimport pandas as pd\r\nimport matplotlib.pyplot as plt\r\nimport seaborn as sns\r\nfrom statsmodels.tsa.arima.model import ARIMA\r\nfrom sklearn.preprocessing import StandardScaler\r\nfrom sklearn.metrics import r2_score\r\n",
"_____no_output_____"
],
[
"# General settings\r\nVEHICLES = (\r\n \"019 Hyundai Elantra GT 2019 (2.0L Auto)\",\r\n \"025 Chevrolet Captiva 2010 (2.4L Auto)\",\r\n \"027 Chevrolet Cruze 2011 (1.8L Manual)\",\r\n)\r\nFEATURES = [\"SPD_KH\", \"ACC_MS2\", \"ALT_M\"]\r\nDEPENDENT = \"FCR_LH\"\r\nSETTINGS = {\r\n \"INPUT_TYPE\": \"ENSEMBLE\",\r\n \"INPUT_INDEX\": \"06\",\r\n \"OUTPUT_TYPE\": \"ARIMA\",\r\n \"OUTPUT_INDEX\": \"AGGREGATE\",\r\n}\r\nAGG_CFG = {\"DATETIME\": \"last\", \"ALT_M\": \"mean\", \"SPD_KH\": \"mean\",\r\n \"FCR_LH\": \"mean\", \"ACC_MS2\": \"mean\", \"FCR_LH_PRED_METAMODEL\": \"mean\"}\r\n",
"_____no_output_____"
],
[
"# Load sample data from Excel to a pandas dataframe\r\ndef load_from_Excel(vehicle, sheet, settings):\r\n directory = (\r\n \"../../Academia/PhD/Field Experiments/Veepeak/\"\r\n + vehicle\r\n + \"/Processed/\"\r\n + settings[\"INPUT_TYPE\"]\r\n + \"/\"\r\n )\r\n input_file = vehicle + \" - {0} - {1}.xlsx\".format(\r\n settings[\"INPUT_TYPE\"], settings[\"INPUT_INDEX\"]\r\n )\r\n input_path = directory + input_file\r\n df = pd.read_excel(input_path, sheet_name=sheet, header=0)\r\n return df\r\n",
"_____no_output_____"
],
[
"# Save the predicted field back to Excel file\r\ndef save_to_excel(df, vehicle, scale, settings):\r\n directory = (\r\n \"../../Academia/PhD/Field Experiments/Veepeak/\"\r\n + vehicle\r\n + \"/Processed/\"\r\n + settings[\"OUTPUT_TYPE\"]\r\n + \"/\"\r\n )\r\n output_file = vehicle + \" - {0} - {1} - {2}-SEC.xlsx\".format(\r\n settings[\"OUTPUT_TYPE\"], settings[\"OUTPUT_INDEX\"], scale\r\n )\r\n output_path = directory + output_file\r\n with pd.ExcelWriter(output_path, engine=\"openpyxl\", mode=\"w\") as writer:\r\n df.to_excel(writer, header=True, index=None)\r\n print(\"{} -> Data is saved to Excel successfully!\".format(vehicle))\r\n return None\r\n",
"_____no_output_____"
],
[
"# Training the ARIMA model for three temporal scales (1-sec, 5-sec, and 10-sec)\r\n# and generating out-of-sample predictions\r\npredictions, observations = {}, {}\r\nfor vehicle in VEHICLES:\r\n predictions[vehicle], observations[vehicle] = {}, {}\r\n df = load_from_Excel(vehicle, \"Sheet1\", SETTINGS)\r\n df = df[[\"DATETIME\", \"ALT_M\", \"SPD_KH\", \"FCR_LH\",\r\n \"ACC_MS2\", \"FCR_LH_PRED_METAMODEL\"]]\r\n for scale in [1, 5, 10]:\r\n dfs = df.groupby(df.index // scale).agg(AGG_CFG)\r\n # Apply feature scaling\r\n scaler_features = StandardScaler().fit(dfs[FEATURES])\r\n scaler_dependent = StandardScaler().fit(dfs[[DEPENDENT]])\r\n dfs[FEATURES] = scaler_features.transform(dfs[FEATURES])\r\n dfs[[DEPENDENT]] = scaler_dependent.transform(dfs[[DEPENDENT]])\r\n # Train-Test splitting (70%-30%)\r\n split_point = int(.7 * len(dfs))\r\n train = dfs[:split_point].copy(deep=True)\r\n # Train the ARIMA model\r\n # The AR order is chosen as 6 (in accordance with our RNN modeling lag order)\r\n # As the variables could be considered stationary (they are bounded and trendless), \"difference\" is set to 0.\r\n # Moving-average order of 3 is applied.\r\n model_l6 = ARIMA(train[DEPENDENT],\r\n exog=train[FEATURES], order=(6, 0, 3))\r\n fit_l6 = model_l6.fit(method_kwargs={\"warn_convergence\": False})\r\n # Out-of-sample prediction\r\n predictions[vehicle][scale] = fit_l6.predict(\r\n start=len(train), end=len(dfs) - 1, exog=dfs[FEATURES][split_point:]).values\r\n # Apply inverse scaling\r\n dfs[FEATURES] = scaler_features.inverse_transform(dfs[FEATURES])\r\n predictions[vehicle][scale] = scaler_dependent.inverse_transform(\r\n predictions[vehicle][scale])\r\n dfs[[DEPENDENT]] = scaler_dependent.inverse_transform(\r\n dfs[[DEPENDENT]])\r\n observations[vehicle][scale] = dfs[DEPENDENT][split_point:]\r\n dfs.loc[split_point:, \"FCR_LH_PRED_ARIMA_{0}_SEC\".format(\r\n scale)] = predictions[vehicle][scale]\r\n save_to_excel(dfs, vehicle, scale, SETTINGS)\r\n",
"_____no_output_____"
],
[
"# Time-series plot of ARIMA predictions vs. true observations for a selected time-window\r\nfor vehicle in VEHICLES:\r\n for scale in [1, 5, 10]:\r\n fig, ax = plt.subplots(figsize=(12, 4))\r\n sns.lineplot(x=range(750), y=predictions[vehicle][scale][0:750], color=\"blue\")\r\n sns.lineplot(x=range(750), y=observations[vehicle][scale][0:750], color=\"red\")\r\n plt.legend(labels=[\"Predictions (AR Order = 6)\", \"True Observations\"])\r\n plt.show()\r\n",
"_____no_output_____"
],
[
"# Scatter plot to compare ARIMA predictions and true observations\r\nfor vehicle in VEHICLES:\r\n for scale in [1, 5, 10]:\r\n fig, ax = plt.subplots(figsize=(4, 4))\r\n sns.scatterplot(x=observations[vehicle][scale], y=predictions[vehicle][scale])\r\n upper_bound = np.max([np.max(observations[vehicle][scale]),\r\n np.max(predictions[vehicle][scale])])\r\n plt.xlim(0, upper_bound)\r\n plt.ylim(0, upper_bound)\r\n plt.xlabel(\"True Observations\")\r\n plt.ylabel(\"ARIMA Predictions (AR Order = 6)\")\r\n plt.show()\r\n",
"_____no_output_____"
],
[
"# Calculate R-squared score of scaled ARIMA models\r\nfor vehicle in VEHICLES:\r\n for scale in [1, 5, 10]:\r\n print(\"{0}, {1}: {2}\".format(vehicle, scale, r2_score(\r\n observations[vehicle][scale], predictions[vehicle][scale])))\r\n",
"019 Hyundai Elantra GT 2019 (2.0L Auto), 1: 0.5256432994136848\n019 Hyundai Elantra GT 2019 (2.0L Auto), 5: 0.6599507495708109\n019 Hyundai Elantra GT 2019 (2.0L Auto), 10: 0.7143308152151611\n025 Chevrolet Captiva 2010 (2.4L Auto), 1: 0.10790025568393047\n025 Chevrolet Captiva 2010 (2.4L Auto), 5: 0.24965064042552265\n025 Chevrolet Captiva 2010 (2.4L Auto), 10: 0.22913369857489152\n027 Chevrolet Cruze 2011 (1.8L Manual), 1: 0.5548563597332341\n027 Chevrolet Cruze 2011 (1.8L Manual), 5: 0.6666841247201505\n027 Chevrolet Cruze 2011 (1.8L Manual), 10: 0.703432450942992\n"
],
[
"# Load sample data from Excel to a pandas dataframe (for scaled predictions)\r\ndef load_from_Excel_scaled(vehicle, scale, sheet):\r\n directory = (\r\n \"../../Academia/PhD/Field Experiments/Veepeak/\"\r\n + vehicle\r\n + \"/Processed/ARIMA/\"\r\n )\r\n input_file = vehicle + \" - ARIMA - AGGREGATE - {0}-SEC.xlsx\".format(scale)\r\n input_path = directory + input_file\r\n df = pd.read_excel(input_path, sheet_name=sheet, header=0)\r\n return df\r\n",
"_____no_output_____"
],
[
"# Calculate R-square score for scaled metamodel predictions\r\nfor vehicle in VEHICLES:\r\n for scale in [1, 5, 10]:\r\n df = load_from_Excel_scaled(vehicle, scale, \"Sheet1\")\r\n print(\"{0}, {1}: {2}\".format(vehicle, scale, r2_score(\r\n df[\"FCR_LH\"], df[\"FCR_LH_PRED_METAMODEL\"])))\r\n",
"019 Hyundai Elantra GT 2019 (2.0L Auto), 1: 0.7179144438996059\n019 Hyundai Elantra GT 2019 (2.0L Auto), 5: 0.827947997358645\n019 Hyundai Elantra GT 2019 (2.0L Auto), 10: 0.8397226533437434\n025 Chevrolet Captiva 2010 (2.4L Auto), 1: 0.8648514862384631\n025 Chevrolet Captiva 2010 (2.4L Auto), 5: 0.9226837940291782\n025 Chevrolet Captiva 2010 (2.4L Auto), 10: 0.9404964902260587\n027 Chevrolet Cruze 2011 (1.8L Manual), 1: 0.7647982784034113\n027 Chevrolet Cruze 2011 (1.8L Manual), 5: 0.8331106614564179\n027 Chevrolet Cruze 2011 (1.8L Manual), 10: 0.8440208423245168\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf06f19f9200e2eb3cfa69892b73bad13149025 | 3,760 | ipynb | Jupyter Notebook | Python/Dynamic_Programming_Optimal_Solution.ipynb | yingkaisha/notebook_code_vault | 08706d1b169f2d1ceced02d2920fe85bd6bc5541 | [
"Unlicense"
] | null | null | null | Python/Dynamic_Programming_Optimal_Solution.ipynb | yingkaisha/notebook_code_vault | 08706d1b169f2d1ceced02d2920fe85bd6bc5541 | [
"Unlicense"
] | null | null | null | Python/Dynamic_Programming_Optimal_Solution.ipynb | yingkaisha/notebook_code_vault | 08706d1b169f2d1ceced02d2920fe85bd6bc5541 | [
"Unlicense"
] | null | null | null | 22.380952 | 209 | 0.469149 | [
[
[
"# Dynamic Programming - Optimal Solution",
"_____no_output_____"
],
[
"### Perfect Squares\n\nGiven an integer n, return the least number of perfect square numbers that sum to n.\n\nA perfect square is an integer that is the square of an integer; in other words, it is the product of some integer with itself. For example, 1, 4, 9, and 16 are perfect squares while 3 and 11 are not.\n\nSource: https://leetcode.com/problems/perfect-squares/\n\nExample 1:\n\n```\nInput: n = 12\nOutput: 3\nExplanation: 12 = 4 + 4 + 4.\n```\n\nExample 2:\n\n```\nInput: n = 13\nOutput: 2\nExplanation: 13 = 4 + 9.\n```",
"_____no_output_____"
]
],
[
[
"class Solution:\n def numSquares(self, n):\n '''\n '''\n \n max_num = n\n max_val = n**2\n \n count_square = [9999,]*(max_val+1)\n count_square[0] = 0\n \n for num in range(max_num):\n for val in range(max_val+1):\n if num**2 <= val: \n count_square[val] = min(count_square[val], count_square[val-(num**2)]+1)\n \n return count_square[-1]",
"_____no_output_____"
],
[
"solver = Solution()\nsolver.numSquares(12)",
"_____no_output_____"
]
],
[
[
"### Largest Divisible Subset\n\nGiven a set of distinct positive integers nums, return the largest subset answer such that every pair (answer[i], answer[j]) of elements in this subset satisfies:\n\n```\nanswer[i] % answer[j] == 0, or\nanswer[j] % answer[i] == 0\n```\n\nIf there are multiple solutions, return any of them.\n\nSource: https://leetcode.com/problems/largest-divisible-subset/ \n\nExample 1:\n\n```\nInput: nums = [1,2,3]\nOutput: [1,2]\nExplanation: [1,3] is also accepted.\n```\n\nExample 2:\n\n```\nInput: nums = [1,2,4,8]\nOutput: [1,2,4,8]\n```",
"_____no_output_____"
]
],
[
[
"class Solution:\n def largestDivisibleSubset(self, nums):\n '''\n '''\n ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf09463073acc2013f62f3ad9d650e41101853a | 1,544 | ipynb | Jupyter Notebook | notebooks/py_1_nb/.ipynb_checkpoints/Untitled1-checkpoint.ipynb | xzzhang2/201811_budmc_Invitation2Py | 288178323151a07933d46ff4a2495e3e6793d866 | [
"MIT"
] | null | null | null | notebooks/py_1_nb/.ipynb_checkpoints/Untitled1-checkpoint.ipynb | xzzhang2/201811_budmc_Invitation2Py | 288178323151a07933d46ff4a2495e3e6793d866 | [
"MIT"
] | null | null | null | notebooks/py_1_nb/.ipynb_checkpoints/Untitled1-checkpoint.ipynb | xzzhang2/201811_budmc_Invitation2Py | 288178323151a07933d46ff4a2495e3e6793d866 | [
"MIT"
] | null | null | null | 16.602151 | 37 | 0.497409 | [
[
[
"1+1",
"_____no_output_____"
]
],
[
[
"# header\n*gh*",
"_____no_output_____"
]
]
] | [
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
]
] |
ecf0976add70dd73b9cdf3006138df6d363df06e | 24,743 | ipynb | Jupyter Notebook | examples/Interactive Widgets/Date Picker Widget.ipynb | 22sangho/ipython-in-depth | 500b91bdf932e6d03f4d87bcf89076ea1cfa0165 | [
"BSD-3-Clause"
] | 748 | 2015-01-05T05:48:49.000Z | 2022-02-27T01:05:42.000Z | examples/Interactive Widgets/Date Picker Widget.ipynb | 22sangho/ipython-in-depth | 500b91bdf932e6d03f4d87bcf89076ea1cfa0165 | [
"BSD-3-Clause"
] | 32 | 2015-04-02T22:25:41.000Z | 2022-01-18T05:31:46.000Z | examples/Interactive Widgets/Date Picker Widget.ipynb | 22sangho/ipython-in-depth | 500b91bdf932e6d03f4d87bcf89076ea1cfa0165 | [
"BSD-3-Clause"
] | 816 | 2015-01-04T04:19:15.000Z | 2022-03-17T20:57:19.000Z | 29.491061 | 224 | 0.547347 | [
[
[
"Before reading, make sure to review\n\n- [MVC prgramming](http://en.wikipedia.org/wiki/Model%E2%80%93view%E2%80%93controller)\n- [Backbone.js](https://www.codeschool.com/courses/anatomy-of-backbonejs)\n- [The widget IPEP](https://github.com/ipython/ipython/wiki/IPEP-23%3A-Backbone.js-Widgets)\n- [The original widget PR discussion](https://github.com/ipython/ipython/pull/4374)",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function # For py 2.7 compat\n\nimport ipywidgets as widgets # Widget definitions\nfrom IPython.display import display # Used to display widgets in the notebook\nfrom traitlets import Unicode # Used to declare attributes of our widget",
"_____no_output_____"
]
],
[
[
"# Abstract",
"_____no_output_____"
],
[
"This notebook implements a custom date picker widget,\nin order to demonstrate the widget creation process.\n\nTo create a custom widget, both Python and JavaScript code is required.",
"_____no_output_____"
],
[
"# Section 1 - Basics",
"_____no_output_____"
],
[
"## Python",
"_____no_output_____"
],
[
"When starting a project like this, it is often easiest to make a simple base implementation,\nto verify that the underlying framework is working as expected.\nTo start, we will create an empty widget and make sure that it can be rendered.\nThe first step is to define the widget in Python.",
"_____no_output_____"
]
],
[
[
"class DateWidget(widgets.DOMWidget):\n _view_name = Unicode('DatePickerView', sync=True)",
"_____no_output_____"
]
],
[
[
"Our widget inherits from `widgets.DOMWidget` since it is intended that it will be displayed in the notebook directly.\nThe `_view_name` trait is special; the widget framework will read the `_view_name` trait to determine what Backbone view the widget is associated with.\n**Using `sync=True` is very important** because it tells the widget framework that that specific traitlet should be synced between the front- and back-ends.",
"_____no_output_____"
],
[
"## JavaScript",
"_____no_output_____"
],
[
"In the IPython notebook [require.js](http://requirejs.org/) is used to load JavaScript dependencies.\nAll IPython widget code depends on `widgets/js/widget.js`,\nwhere the base widget model and base view are defined.\nWe use require.js to load this file:",
"_____no_output_____"
]
],
[
[
"%%javascript\n\nrequire([\"widgets/js/widget\"], function(WidgetManager){\n\n});",
"_____no_output_____"
]
],
[
[
"Now we need to define a view that can be used to represent the model.\nTo do this, the `IPython.DOMWidgetView` is extended.\n**A render function must be defined**.\nThe render function is used to render a widget view instance to the DOM.\nFor now, the render function renders a div that contains the text *Hello World!*\nLastly, the view needs to be registered with the widget manager, for which we need to load another module.\n\n**Final JavaScript code below:**",
"_____no_output_____"
]
],
[
[
"%%javascript\n\nrequire([\"widgets/js/widget\", \"widgets/js/manager\"], function(widget, manager){\n \n // Define the DatePickerView\n var DatePickerView = widget.DOMWidgetView.extend({\n render: function(){ this.$el.text('Hello World!'); },\n });\n \n // Register the DatePickerView with the widget manager.\n manager.WidgetManager.register_widget_view('DatePickerView', DatePickerView);\n});",
"_____no_output_____"
]
],
[
[
"## Test",
"_____no_output_____"
],
[
"To test what we have so far, create the widget, just like you would the builtin widgets:",
"_____no_output_____"
]
],
[
[
"DateWidget()",
"_____no_output_____"
]
],
[
[
"# Section 2 - Something useful",
"_____no_output_____"
],
[
"## Python",
"_____no_output_____"
],
[
"In the last section we created a simple widget that displayed *Hello World!*\nTo make an actual date widget, we need to add a property that will be synced between the Python model and the JavaScript model.\nThe new attribute must be a traitlet, so the widget machinery can handle it.\nThe traitlet must be constructed with a `sync=True` keyword argument, to tell the widget machinery knows to synchronize it with the front-end.\nAdding this to the code from the last section:",
"_____no_output_____"
]
],
[
[
"class DateWidget(widgets.DOMWidget):\n _view_name = Unicode('DatePickerView', sync=True)\n value = Unicode(sync=True)",
"_____no_output_____"
]
],
[
[
"## JavaScript",
"_____no_output_____"
],
[
"In the JavaScript, there is no need to define counterparts to the traitlets.\nWhen the JavaScript model is created for the first time,\nit copies all of the traitlet `sync=True` attributes from the Python model.\nWe need to replace *Hello World!* with an actual HTML date picker widget.",
"_____no_output_____"
]
],
[
[
"%%javascript\n\nrequire([\"widgets/js/widget\", \"widgets/js/manager\"], function(widget, manager){\n \n // Define the DatePickerView\n var DatePickerView = widget.DOMWidgetView.extend({\n render: function(){\n \n // Create the date picker control.\n this.$date = $('<input />')\n .attr('type', 'date')\n .appendTo(this.$el);\n },\n });\n \n // Register the DatePickerView with the widget manager.\n manager.WidgetManager.register_widget_view('DatePickerView', DatePickerView);\n});",
"_____no_output_____"
]
],
[
[
"In order to get the HTML date picker to update itself with the value set in the back-end, we need to implement an `update()` method.",
"_____no_output_____"
]
],
[
[
"%%javascript\n\nrequire([\"widgets/js/widget\", \"widgets/js/manager\"], function(widget, manager){\n \n // Define the DatePickerView\n var DatePickerView = widget.DOMWidgetView.extend({\n render: function(){\n \n // Create the date picker control.\n this.$date = $('<input />')\n .attr('type', 'date')\n .appendTo(this.$el);\n },\n \n update: function() {\n \n // Set the value of the date control and then call base.\n this.$date.val(this.model.get('value')); // ISO format \"YYYY-MM-DDTHH:mm:ss.sssZ\" is required\n return DatePickerView.__super__.update.apply(this);\n },\n });\n \n // Register the DatePickerView with the widget manager.\n manager.WidgetManager.register_widget_view('DatePickerView', DatePickerView);\n});",
"_____no_output_____"
]
],
[
[
"To get the changed value from the frontend to publish itself to the backend,\nwe need to listen to the change event triggered by the HTM date control and set the value in the model.\nAfter the date change event fires and the new value is set in the model,\nit is very important that we call `this.touch()` to let the widget machinery know which view changed the model.\nThis is important because the widget machinery needs to know which cell to route the message callbacks to.\n\n**Final JavaScript code below:**",
"_____no_output_____"
]
],
[
[
"%%javascript\n\n\nrequire([\"widgets/js/widget\", \"widgets/js/manager\"], function(widget, manager){\n \n // Define the DatePickerView\n var DatePickerView = widget.DOMWidgetView.extend({\n render: function(){\n \n // Create the date picker control.\n this.$date = $('<input />')\n .attr('type', 'date')\n .appendTo(this.$el);\n },\n \n update: function() {\n \n // Set the value of the date control and then call base.\n this.$date.val(this.model.get('value')); // ISO format \"YYYY-MM-DDTHH:mm:ss.sssZ\" is required\n return DatePickerView.__super__.update.apply(this);\n },\n \n // Tell Backbone to listen to the change event of input controls (which the HTML date picker is)\n events: {\"change\": \"handle_date_change\"},\n \n // Callback for when the date is changed.\n handle_date_change: function(event) {\n this.model.set('value', this.$date.val());\n this.touch();\n },\n });\n \n // Register the DatePickerView with the widget manager.\n manager.WidgetManager.register_widget_view('DatePickerView', DatePickerView);\n});",
"_____no_output_____"
]
],
[
[
"## Test",
"_____no_output_____"
],
[
"To test, create the widget the same way that the other widgets are created.",
"_____no_output_____"
]
],
[
[
"my_widget = DateWidget()\ndisplay(my_widget)",
"_____no_output_____"
]
],
[
[
"Display the widget again to make sure that both views remain in sync.",
"_____no_output_____"
]
],
[
[
"my_widget",
"_____no_output_____"
]
],
[
[
"Read the date from Python",
"_____no_output_____"
]
],
[
[
"my_widget.value",
"_____no_output_____"
]
],
[
[
"Set the date from Python",
"_____no_output_____"
]
],
[
[
"my_widget.value = \"1998-12-01\" # December 1st, 1998",
"_____no_output_____"
]
],
[
[
"# Section 3 - Extra credit",
"_____no_output_____"
],
[
"The 3rd party `dateutil` library is required to continue. https://pypi.python.org/pypi/python-dateutil",
"_____no_output_____"
]
],
[
[
"# Import the dateutil library to parse date strings.\nfrom dateutil import parser",
"_____no_output_____"
]
],
[
[
"In the last section we created a fully working date picker widget.\nNow we will add custom validation and support for labels.\nSo far, only the ISO date format \"YYYY-MM-DD\" is supported.\nNow, we will add support for all of the date formats recognized by the Python dateutil library.",
"_____no_output_____"
],
[
"## Python",
"_____no_output_____"
],
[
"The standard property name used for widget labels is `description`.\nIn the code block below, `description` has been added to the Python widget.",
"_____no_output_____"
]
],
[
[
"class DateWidget(widgets.DOMWidget):\n _view_name = Unicode('DatePickerView', sync=True)\n value = Unicode(sync=True)\n description = Unicode(sync=True)",
"_____no_output_____"
]
],
[
[
"The traitlet machinery searches the class that the trait is defined in for methods with \"`_changed`\" suffixed onto their names. Any method with the format \"`_X_changed`\" will be called when \"`X`\" is modified.\nWe can take advantage of this to perform validation and parsing of different date string formats.\nBelow, a method that listens to value has been added to the DateWidget.",
"_____no_output_____"
]
],
[
[
"class DateWidget(widgets.DOMWidget):\n _view_name = Unicode('DatePickerView', sync=True)\n value = Unicode(sync=True)\n description = Unicode(sync=True)\n\n # This function automatically gets called by the traitlet machinery when\n # value is modified because of this function's name.\n def _value_changed(self, name, old_value, new_value):\n pass",
"_____no_output_____"
]
],
[
[
"Now the function parses the date string,\nand only sets the value in the correct format.",
"_____no_output_____"
]
],
[
[
"class DateWidget(widgets.DOMWidget):\n _view_name = Unicode('DatePickerView', sync=True)\n value = Unicode(sync=True)\n description = Unicode(sync=True)\n \n # This function automatically gets called by the traitlet machinery when\n # value is modified because of this function's name.\n def _value_changed(self, name, old_value, new_value):\n \n # Parse the date time value.\n try:\n parsed_date = parser.parse(new_value)\n parsed_date_string = parsed_date.strftime(\"%Y-%m-%d\")\n except:\n parsed_date_string = ''\n \n # Set the parsed date string if the current date string is different.\n if self.value != parsed_date_string:\n self.value = parsed_date_string",
"_____no_output_____"
]
],
[
[
"Finally, a `CallbackDispatcher` is added so the user can perform custom validation.\nIf any one of the callbacks registered with the dispatcher returns False,\nthe new date is not set.\n\n**Final Python code below:**",
"_____no_output_____"
]
],
[
[
"class DateWidget(widgets.DOMWidget):\n _view_name = Unicode('DatePickerView', sync=True)\n value = Unicode(sync=True)\n description = Unicode(sync=True)\n \n def __init__(self, **kwargs):\n super(DateWidget, self).__init__(**kwargs)\n \n self.validate = widgets.CallbackDispatcher()\n \n # This function automatically gets called by the traitlet machinery when\n # value is modified because of this function's name.\n def _value_changed(self, name, old_value, new_value):\n \n # Parse the date time value.\n try:\n parsed_date = parser.parse(new_value)\n parsed_date_string = parsed_date.strftime(\"%Y-%m-%d\")\n except:\n parsed_date_string = ''\n \n # Set the parsed date string if the current date string is different.\n if old_value != new_value:\n valid = self.validate(parsed_date)\n if valid in (None, True):\n self.value = parsed_date_string\n else:\n self.value = old_value\n self.send_state() # The traitlet event won't fire since the value isn't changing.\n # We need to force the back-end to send the front-end the state\n # to make sure that the date control date doesn't change.",
"_____no_output_____"
]
],
[
[
"## JavaScript",
"_____no_output_____"
],
[
"Using the Javascript code from the last section,\nwe add a label to the date time object.\nThe label is a div with the `widget-hlabel` class applied to it.\n`widget-hlabel` is a class provided by the widget framework that applies special styling to a div to make it look like the rest of the horizontal labels used with the built-in widgets.\nSimilar to the `widget-hlabel` class is the `widget-hbox-single` class.\nThe `widget-hbox-single` class applies special styling to widget containers that store a single line horizontal widget.\n\nWe hide the label if the description value is blank.",
"_____no_output_____"
]
],
[
[
"%%javascript\n\nrequire([\"widgets/js/widget\", \"widgets/js/manager\"], function(widget, manager){\n \n // Define the DatePickerView\n var DatePickerView = widget.DOMWidgetView.extend({\n render: function(){\n this.$el.addClass('widget-hbox-single'); /* Apply this class to the widget container to make\n it fit with the other built in widgets.*/\n // Create a label.\n this.$label = $('<div />')\n .addClass('widget-hlabel')\n .appendTo(this.$el)\n .hide(); // Hide the label by default.\n \n // Create the date picker control.\n this.$date = $('<input />')\n .attr('type', 'date')\n .appendTo(this.$el);\n },\n \n update: function() {\n \n // Set the value of the date control and then call base.\n this.$date.val(this.model.get('value')); // ISO format \"YYYY-MM-DDTHH:mm:ss.sssZ\" is required\n \n // Hide or show the label depending on the existance of a description.\n var description = this.model.get('description');\n if (description == undefined || description == '') {\n this.$label.hide();\n } else {\n this.$label.show();\n this.$label.text(description);\n }\n \n return DatePickerView.__super__.update.apply(this);\n },\n \n // Tell Backbone to listen to the change event of input controls (which the HTML date picker is)\n events: {\"change\": \"handle_date_change\"},\n \n // Callback for when the date is changed.\n handle_date_change: function(event) {\n this.model.set('value', this.$date.val());\n this.touch();\n },\n });\n \n // Register the DatePickerView with the widget manager.\n manager.WidgetManager.register_widget_view('DatePickerView', DatePickerView);\n});",
"_____no_output_____"
]
],
[
[
"## Test",
"_____no_output_____"
],
[
"To test the drawing of the label we create the widget like normal but supply the additional description property a value.",
"_____no_output_____"
]
],
[
[
"# Add some additional widgets for aesthetic purpose\ndisplay(widgets.Text(description=\"First:\"))\ndisplay(widgets.Text(description=\"Last:\"))\n\nmy_widget = DateWidget()\ndisplay(my_widget)\nmy_widget.description=\"DOB:\"",
"_____no_output_____"
]
],
[
[
"Now we will try to create a widget that only accepts dates in the year 2014. We render the widget without a description to verify that it can still render without a label.",
"_____no_output_____"
]
],
[
[
"my_widget = DateWidget()\ndisplay(my_widget)\n\ndef require_2014(date):\n return not date is None and date.year == 2014\nmy_widget.validate.register_callback(require_2014)",
"_____no_output_____"
],
[
"# Try setting a valid date\nmy_widget.value = \"December 2, 2014\"",
"_____no_output_____"
],
[
"# Try setting an invalid date\nmy_widget.value = \"June 12, 1999\"",
"_____no_output_____"
],
[
"my_widget.value",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
ecf0c11f81650878d280dc49405324caac22562a | 4,114 | ipynb | Jupyter Notebook | Machine Learning/Snapchat Filter/Cartoon Filter.ipynb | StandingMike/data-science | 37cf6266a68c639bbba61f10d215ccd30f3b8f47 | [
"MIT"
] | null | null | null | Machine Learning/Snapchat Filter/Cartoon Filter.ipynb | StandingMike/data-science | 37cf6266a68c639bbba61f10d215ccd30f3b8f47 | [
"MIT"
] | null | null | null | Machine Learning/Snapchat Filter/Cartoon Filter.ipynb | StandingMike/data-science | 37cf6266a68c639bbba61f10d215ccd30f3b8f47 | [
"MIT"
] | null | null | null | 26.371795 | 122 | 0.524307 | [
[
[
"# Basic Snapchat Filter (Cartoon Face Mask)\n### Completed by Mike Willis in November 2021",
"_____no_output_____"
],
[
"## Goal: Cartoonify faces in video feed from live webcam\n\n### Steps\n1. **Capture video** feed from webcam\n2. **Recognize faces** in the video\n3. **Replace/Mask the face** region with your favorite cartoon character\n4. **Save the video** feed into a video file",
"_____no_output_____"
],
[
"#### Code to recognize faces",
"_____no_output_____"
]
],
[
[
"import cv2\nimport os",
"_____no_output_____"
]
],
[
[
"#### Import Cartoon Face",
"_____no_output_____"
]
],
[
[
"image = cv2.imread('data/halo_cartoon.jpg')",
"_____no_output_____"
]
],
[
[
"#### Create HAAR Cascade Filters",
"_____no_output_____"
]
],
[
[
"cascPathface = os.path.dirname(\n cv2.__file__) + \"/data/haarcascade_frontalface_alt2.xml\"\ncascPatheyes = os.path.dirname(\n cv2.__file__) + \"/data/haarcascade_eye_tree_eyeglasses.xml\"\n\nfaceCascade = cv2.CascadeClassifier(cascPathface)\neyeCascade = cv2.CascadeClassifier(cascPatheyes)",
"_____no_output_____"
]
],
[
[
"#### Putting it all together",
"_____no_output_____"
]
],
[
[
"# Open video capture\ncapture = cv2.VideoCapture(0)\n\n# Get height and width of Frame\nwidth = capture.get(cv2.CAP_PROP_FRAME_WIDTH)\nheight = capture.get(cv2.CAP_PROP_FRAME_HEIGHT)\n\n# Define video codec and file to write capture\nfourcc = cv2.VideoWriter_fourcc(*\"XVID\")\noutput_capture = cv2.VideoWriter('Mike_Task6_Cartoon_Filter_Output.avi', fourcc, 30.0, (int(width), int(height)))\n\nwhile True:\n # Capture frame-by-frame\n ret, frame = capture.read()\n gray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)\n faces = faceCascade.detectMultiScale(gray,\n scaleFactor=1.1,\n minNeighbors=5,\n minSize=(60, 60),\n flags=cv2.CASCADE_SCALE_IMAGE)\n for (x,y,w,h) in faces:\n cv2.rectangle(frame, (x, y), (x + w, y + h),(0,255,0), 2)\n faceROI = frame[y:y+h,x:x+w]\n image_resized = cv2.resize(image, (h, w), interpolation=cv2.INTER_AREA)\n # Display the resulting frame\n frame[y:y+h,x:x+w] = image_resized\n frame = cv2.flip(frame, 90)\n cv2.imshow('Face Video', frame)\n\n # Write the file\n output_capture.write(frame)\n if cv2.waitKey(1) & 0xFF == ord('q'):\n break\n\ncapture.release()\ncv2.destroyAllWindows()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf0ca4646ed27e9493a4cb7a0fa775a3ff3f5d9 | 11,798 | ipynb | Jupyter Notebook | docs/tutorials/average_optimizers_callback.ipynb | jeongukjae/addons | fe309184917f0c20b0b4b2caf34981877cdeea40 | [
"Apache-2.0"
] | 1,560 | 2018-11-26T23:57:34.000Z | 2022-03-27T10:37:34.000Z | docs/tutorials/average_optimizers_callback.ipynb | jeongukjae/addons | fe309184917f0c20b0b4b2caf34981877cdeea40 | [
"Apache-2.0"
] | 2,067 | 2018-11-28T04:40:23.000Z | 2022-03-31T11:36:50.000Z | docs/tutorials/average_optimizers_callback.ipynb | jeongukjae/addons | fe309184917f0c20b0b4b2caf34981877cdeea40 | [
"Apache-2.0"
] | 679 | 2018-11-27T14:39:25.000Z | 2022-03-31T10:09:22.000Z | 29.792929 | 299 | 0.506781 | [
[
[
"##### Copyright 2020 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Model Averaging\n\n<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/addons/tutorials/average_optimizers_callback\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/addons/blob/master/docs/tutorials/average_optimizers_callback.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/addons/blob/master/docs/tutorials/average_optimizers_callback.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/addons/docs/tutorials/average_optimizers_callback.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>\n",
"_____no_output_____"
],
[
"## Overview\n\nThis notebook demonstrates how to use Moving Average Optimizer along with the Model Average Checkpoint from tensorflow addons package.\n",
"_____no_output_____"
],
[
"## Moving Averaging \n\n> The advantage of Moving Averaging is that they are less prone to rampant loss shifts or irregular data representation in the latest batch. It gives a smooothened and a more genral idea of the model training until some point.\n\n## Stochastic Averaging\n\n> Stochastic Weight Averaging converges to wider optima. By doing so, it resembles geometric ensembeling. SWA is a simple method to improve model performance when used as a wrapper around other optimizers and averaging results from different points of trajectory of the inner optimizer.\n\n## Model Average Checkpoint \n\n> `callbacks.ModelCheckpoint` doesn't give you the option to save moving average weights in the middle of training, which is why Model Average Optimizers required a custom callback. Using the ```update_weights``` parameter, ```ModelAverageCheckpoint``` allows you to:\n1. Assign the moving average weights to the model, and save them.\n2. Keep the old non-averaged weights, but the saved model uses the average weights.",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"!pip install -U tensorflow-addons",
"_____no_output_____"
],
[
"import tensorflow as tf\nimport tensorflow_addons as tfa",
"_____no_output_____"
],
[
"import numpy as np\nimport os",
"_____no_output_____"
]
],
[
[
"## Build Model ",
"_____no_output_____"
]
],
[
[
"def create_model(opt):\n model = tf.keras.models.Sequential([\n tf.keras.layers.Flatten(), \n tf.keras.layers.Dense(64, activation='relu'),\n tf.keras.layers.Dense(64, activation='relu'),\n tf.keras.layers.Dense(10, activation='softmax')\n ])\n\n model.compile(optimizer=opt,\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])\n\n return model",
"_____no_output_____"
]
],
[
[
"## Prepare Dataset",
"_____no_output_____"
]
],
[
[
"#Load Fashion MNIST dataset\ntrain, test = tf.keras.datasets.fashion_mnist.load_data()\n\nimages, labels = train\nimages = images/255.0\nlabels = labels.astype(np.int32)\n\nfmnist_train_ds = tf.data.Dataset.from_tensor_slices((images, labels))\nfmnist_train_ds = fmnist_train_ds.shuffle(5000).batch(32)\n\ntest_images, test_labels = test",
"_____no_output_____"
]
],
[
[
"We will be comparing three optimizers here:\n\n* Unwrapped SGD\n* SGD with Moving Average\n* SGD with Stochastic Weight Averaging\n\nAnd see how they perform with the same model.",
"_____no_output_____"
]
],
[
[
"#Optimizers \nsgd = tf.keras.optimizers.SGD(0.01)\nmoving_avg_sgd = tfa.optimizers.MovingAverage(sgd)\nstocastic_avg_sgd = tfa.optimizers.SWA(sgd)",
"_____no_output_____"
]
],
[
[
"Both ```MovingAverage``` and ```StocasticAverage``` optimers use ```ModelAverageCheckpoint```.",
"_____no_output_____"
]
],
[
[
"#Callback \ncheckpoint_path = \"./training/cp-{epoch:04d}.ckpt\"\ncheckpoint_dir = os.path.dirname(checkpoint_path)\n\ncp_callback = tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_dir,\n save_weights_only=True,\n verbose=1)\navg_callback = tfa.callbacks.AverageModelCheckpoint(filepath=checkpoint_dir, \n update_weights=True)",
"_____no_output_____"
]
],
[
[
"## Train Model\n",
"_____no_output_____"
],
[
"### Vanilla SGD Optimizer ",
"_____no_output_____"
]
],
[
[
"#Build Model\nmodel = create_model(sgd)\n\n#Train the network\nmodel.fit(fmnist_train_ds, epochs=5, callbacks=[cp_callback])",
"_____no_output_____"
],
[
"#Evalute results\nmodel.load_weights(checkpoint_dir)\nloss, accuracy = model.evaluate(test_images, test_labels, batch_size=32, verbose=2)\nprint(\"Loss :\", loss)\nprint(\"Accuracy :\", accuracy)",
"_____no_output_____"
]
],
[
[
"### Moving Average SGD",
"_____no_output_____"
]
],
[
[
"#Build Model\nmodel = create_model(moving_avg_sgd)\n\n#Train the network\nmodel.fit(fmnist_train_ds, epochs=5, callbacks=[avg_callback])",
"_____no_output_____"
],
[
"#Evalute results\nmodel.load_weights(checkpoint_dir)\nloss, accuracy = model.evaluate(test_images, test_labels, batch_size=32, verbose=2)\nprint(\"Loss :\", loss)\nprint(\"Accuracy :\", accuracy)",
"_____no_output_____"
]
],
[
[
"### Stocastic Weight Average SGD ",
"_____no_output_____"
]
],
[
[
"#Build Model\nmodel = create_model(stocastic_avg_sgd)\n\n#Train the network\nmodel.fit(fmnist_train_ds, epochs=5, callbacks=[avg_callback])",
"_____no_output_____"
],
[
"#Evalute results\nmodel.load_weights(checkpoint_dir)\nloss, accuracy = model.evaluate(test_images, test_labels, batch_size=32, verbose=2)\nprint(\"Loss :\", loss)\nprint(\"Accuracy :\", accuracy)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecf0cb8058f8abbfe46c0bdb54048c624b3dd88f | 122,395 | ipynb | Jupyter Notebook | week1/kshitij/Q2 - Q/Attempt1_filesubmission_UnicycleModel.ipynb | naveenmoto/lablet102 | 24de9daa4ae75cbde93567a3239ede43c735cf03 | [
"MIT"
] | 1 | 2021-07-09T16:48:44.000Z | 2021-07-09T16:48:44.000Z | week1/kshitij/Q2 - Q/Attempt1_filesubmission_UnicycleModel.ipynb | naveenmoto/lablet102 | 24de9daa4ae75cbde93567a3239ede43c735cf03 | [
"MIT"
] | null | null | null | week1/kshitij/Q2 - Q/Attempt1_filesubmission_UnicycleModel.ipynb | naveenmoto/lablet102 | 24de9daa4ae75cbde93567a3239ede43c735cf03 | [
"MIT"
] | null | null | null | 122,395 | 122,395 | 0.796536 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib notebook\n%matplotlib inline\nimport doctest\nimport copy",
"_____no_output_____"
],
[
"#https://stackoverflow.com/questions/40137950/possible-to-run-python-doctest-on-a-jupyter-cell-function\ndef test(func):\n '''\n Use test as a decorator to a function with doctests in Jupyter notebook. \n Run the cell to see the results of the doctests.\n '''\n globs = copy.copy(globals())\n globs.update({func.__name__:func})\n doctest.run_docstring_examples(func, globs, verbose=True, name=func.__name__)\n return func",
"_____no_output_____"
]
],
[
[
"### Simulate straight line and circular movements with Unicycle model\n\nRobot is at the origin (0, 0) and facing North, i.e, $\\theta = \\pi/2$\n\nUse the Kinematics equations we had developed in class for the Unicycle model",
"_____no_output_____"
]
],
[
[
"#uncomment this decorator to test your code\n@test \ndef unicycle_model(curr_pose, v, w, dt=1.0):\n '''\n >>> unicycle_model((0.0,0.0,0.0), 1.0, 0.0)\n (1.0, 0.0, 0.0)\n >>> unicycle_model((0.0,0.0,0.0), 0.0, 1.0)\n (0.0, 0.0, 1.0)\n >>> unicycle_model((0.0, 0.0, 0.0), 1.0, 1.0)\n (1.0, 0.0, 1.0)\n '''\n ## write code to calculate next_pose\n # refer to the kinematic equations of a unicycle model\n # x = \n # y = \n # theta = \n x = curr_pose[0] + v*np.cos(curr_pose[2])*dt\n y = curr_pose[1] + v*np.sin(curr_pose[2])*dt\n theta = curr_pose[2] + w*dt\n \n # Keep theta bounded between [-pi, pi]\n theta = np.arctan2(np.sin(theta), np.cos(theta))\n # return calculated (x, y, theta)\n return (x,y,theta)",
"\nPYDEV DEBUGGER WARNING:\nsys.settrace() should not be used when the debugger is being used.\nThis may cause the debugger to stop working correctly.\nIf this is needed, please check: \nhttp://pydev.blogspot.com/2007/06/why-cant-pydev-debugger-work-with.html\nto see how to restore the debug tracing back correctly.\nCall Location:\n File \"/usr/lib/python3.7/doctest.py\", line 1487, in run\n sys.settrace(save_trace)\n\n"
]
],
[
[
"Now let us try to simulate motion along a straight line and circle\n\nLook at the kinematics equations you wrote - what should $v,\\omega$ if\n+ There is no change in $\\theta$\n+ $\\theta$ has to change from 0 to 360 degrees",
"_____no_output_____"
]
],
[
[
"#straight line\nstraight_trajectory = []\npose = (0, 0, np.pi/2)\nsteps = 10\n#fill in v and omega values\n#v = \nv = np.ones(steps)\n#w = \nw = np.zeros(steps)\n\nfor _ in range(steps):\n #instruction to take v, w and compute new pose\n straight_trajectory.append(pose)\n pose = unicycle_model(pose, v[_], w[_]) \n # store new pose\nstraight_trajectory = np.array(straight_trajectory) \n ",
"_____no_output_____"
],
[
"### Plot straight and circular trajectories\nplt.figure()\nplt.axes().set_aspect(\"equal\",\"datalim\")\nplt.plot(straight_trajectory[:,0],straight_trajectory[:,1])\n",
"_____no_output_____"
],
[
"#circle\ncircle_trajectory = []\npose = (0, 0, np.pi/2)\nsteps = 100\n#fill in v and omega values\n#v = \nv = 0.01*np.ones(steps)\n#w = \nw = np.ones(steps)\nfor _ in range(steps):\n #instruction to take v, w and compute new pose \n circle_trajectory.append(pose)\n pose = unicycle_model(pose, v[_], w[_],0.1) \n # store new pose\ncircle_trajectory = np.array(circle_trajectory) \n \n \n \n",
"_____no_output_____"
],
[
"### Plot straight and circular trajectories\nplt.figure()\nplt.axes().set_aspect(\"equal\",\"datalim\")\nplt.plot(circle_trajectory[:,0],circle_trajectory[:,1])\n",
"_____no_output_____"
]
],
[
[
"### Simulate Unicycle model with Open Loop control\n\nWe want the robot to follow these instructions\n\n**straight 10m, right turn, straight 5m, left turn, straight 8m, right turn**\n\nIt is in open loop; control commands have to be calculated upfront. How do we do it?\n\nTo keep things simple in the first iteration, we can fix $v = v_c$ and change only $\\omega$. To make it even simpler, $\\omega$ can take only 2 values \n+ 0 when the vehicle is going straight \n+ $\\omega = \\omega_c$ when turning\n\nThis leaves only 2 questions to be answered\n* What should be $v_c$ and $\\omega_c$?\n* When should $\\omega$ change from 0 and back?",
"_____no_output_____"
]
],
[
[
"v_c = 1 # m/s\nw_c = np.pi/6 # rad/s\n\n\n#calculate time taken to finish a quarter turn (pi/2)\nt_turn = 3 \n\n#calculate the time taken to finish straight segments\n# omega array is to be padded with equivalent zeros\n\n#t_straight1, t_straight_2, t_straight3 = \nt_straight_1, t_straight_2, t_straight_3 = 10,5,8\n\nall_w = [0]*t_straight_1 + [w_c]*t_turn + \\\n [0]*t_straight_2 + [w_c]*t_turn + \\\n [0]*t_straight_3 + [-w_c]*t_turn\nall_v = v_c*np.ones_like(all_w)\n\nprint(all_w)\nprint(all_v)",
"[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0.5235987755982988, 0.5235987755982988, 0.5235987755982988, 0, 0, 0, 0, 0, 0.5235987755982988, 0.5235987755982988, 0.5235987755982988, 0, 0, 0, 0, 0, 0, 0, 0, -0.5235987755982988, -0.5235987755982988, -0.5235987755982988]\n[1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.\n 1. 1. 1. 1. 1. 1. 1. 1.]\n"
]
],
[
[
"Let us make a cool function out of this!\n\nTake in as input a generic route and convert it into open-loop commands\n\nInput format: [(\"straight\", 5), (\"right\", 90), (\"straight\", 6), (\"left\", 85)]\n\nOutput: all_v, all_w",
"_____no_output_____"
]
],
[
[
"#route input format [(\"straight\", 5), (\"right\", 90), (\"straight\", 6), (\"left\", 85)]\ndef get_open_loop_commands(route, v_c, w_c):\n t_turn = int(np.pi/2/w_c) #seconds\n all_w = []\n #calculate time taken to finish a quarter turn (pi/2)\n for segment in route:\n if segment[0] == \"straight\":\n all_w += [0.0]*int(segment[1]/v_c)\n elif segment[0] == \"right\":\n all_w += [-w_c]*int(np.deg2rad(segment[1])/w_c)\n else:\n all_w += [w_c]*int(np.deg2rad(segment[1])/w_c)\n all_v = v_c*np.ones_like(all_w)\n #calculate the time taken to finish straight segments\n # omega array is to be padded with equivalent zeros\n return all_v, all_w",
"_____no_output_____"
],
[
"#route input format [(\"straight\", 5), (\"right\", 90), (\"straight\", 6), (\"left\", 85)]\n# using dictionaries\ndef get_open_loop_commands2(route, v_c, w_c):\n t_turn = int(np.pi/2/w_c) #seconds\n all_w = []\n omegas = {\"straight\":0, \"left\": w_c, \"right\":-w_c}\n #calculate time taken to finish a quarter turn (pi/2)\n for manoeuvre,command in route:\n u = np.ceil(command/v_c).astype('int')\n v = np.ceil(np.deg2rad(command)/w_c).astype('int')\n t_cmd = u if manoeuvre == 'straight' else v\n all_w += [omegas[manoeuvre]]*t_cmd\n all_v = v_c*np.ones_like(all_w)\n #calculate the time taken to finish straight segments\n # omega array is to be padded with equivalent zeros\n return all_v, all_w",
"_____no_output_____"
]
],
[
[
"### Unit test your function with the following inputs\n\n+ [(\"straight\", 5), (\"right\", 90), (\"straight\", 6), (\"left\", 85)]\n+ $v_c = 1$\n+ $w_c = \\pi/12$",
"_____no_output_____"
]
],
[
[
"get_open_loop_commands([(\"straight\", 5), (\"right\", 90), (\"straight\", 6), (\"left\", 85)],1,np.pi/12)\n\n ",
"_____no_output_____"
],
[
"robot_trajectory = []\npose = np.array([0, 0, np.pi/2])\nall_v, all_w = get_open_loop_commands([(\"straight\", 5), (\"right\", 90), (\"straight\", 6), (\"left\", 85)],1,np.pi/12)\n#print(all_w)\n#print(all_v)\nfor v, w in zip(all_v, all_w):\n #instruction to take v, w and compute new pose\n robot_trajectory.append(pose) \n pose = unicycle_model(pose,v,w,1.0) \n # store new pose\nrobot_trajectory = np.array(robot_trajectory) \n\nplt.figure()\nplt.axes().set_aspect(\"equal\",\"datalim\")\nplt.plot(robot_trajectory[:,0],robot_trajectory[:,1])\n#show first and last robot positions with + markers\nxi,yi, _ = robot_trajectory[0]\nxf,yf, _ = robot_trajectory[-1]\nplt.plot(xi, yi, 'g+', ms=10)\nplt.plot(xf, yf, 'r+', ms=10)",
"_____no_output_____"
],
[
"# plot robot trajectory\nplt.figure()\nplt.grid()\n#plt.plot( )\n\n#show first and last robot positions with + markers\n# example: plt.plot(0, 0, 'r+', ms=10)\n",
"_____no_output_____"
]
],
[
[
"### Improved capabilities!\n\n+ Slow the robot while turning ($v_c$ for turn and straight needed)\n\n+ How to accommodate a sampling time < 1.0s (hint: think of sampling instances instead of time)",
"_____no_output_____"
]
],
[
[
"#route input format [(\"straight\", 5), (\"right\", 90), (\"straight\", 6), (\"left\", 85)]\n# improved\ndef get_open_loop_commands3(route, v_slow, v_c, w_c):\n t_turn = int(np.pi/2/w_c) #seconds\n all_w = []\n omegas = {\"straight\":0, \"left\": w_c, \"right\":-w_c}\n #calculate time taken to finish a quarter turn (pi/2)\n for manoeuvre,command in route:\n u = np.ceil(command/v_c).astype('int')\n v = np.ceil(np.deg2rad(command)/w_c).astype('int')\n t_cmd = u if manoeuvre == 'straight' else v\n all_w += [omegas[manoeuvre]]*t_cmd\n all_v = v_c*np.ones_like(all_w)\n for i in range(len(all_w)):\n if not all_w[i] == 0:\n all_v[i] = v_slow \n\n #calculate the time taken to finish straight segments\n # omega array is to be padded with equivalent zeros\n return all_v, all_w",
"_____no_output_____"
],
[
"get_open_loop_commands3([(\"straight\", 5), (\"right\", 90), (\"straight\", 6), (\"left\", 85)],0.5,1,np.pi/12)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecf0cf66d25f0505563efb326d895ba24aa8ffc7 | 489,149 | ipynb | Jupyter Notebook | notebooks/FastAI-Balanced-Scenario-A-Experiments.ipynb | rambasnet/DeepLearning-TorTraffic | 03d0efcc2e9e6b0775610d0b3716887f1d493dee | [
"MIT"
] | 7 | 2020-06-22T20:35:44.000Z | 2022-03-03T04:15:52.000Z | notebooks/FastAI-Balanced-Scenario-A-Experiments.ipynb | rambasnet/DeepLearning-TorTrafficDetection | 03d0efcc2e9e6b0775610d0b3716887f1d493dee | [
"MIT"
] | 3 | 2019-11-17T05:34:10.000Z | 2021-10-08T15:12:42.000Z | notebooks/FastAI-Balanced-Scenario-A-Experiments.ipynb | rambasnet/DeepLearning-TorTrafficDetection | 03d0efcc2e9e6b0775610d0b3716887f1d493dee | [
"MIT"
] | 4 | 2020-11-21T13:14:40.000Z | 2022-02-13T09:44:21.000Z | 139.756857 | 19,708 | 0.856506 | [
[
[
"# Experiments on Scenario A with FastAI\n\nThis series of experiments was performed on a rebalanced subset of the Data from Scenario A where the new dataset is created by reducing a random sample of Non-Tor data to the equivalent size of the Tor data. The new dataset is comprised of exactly 50% Tor connection data and 50% Non-Tor connection data",
"_____no_output_____"
]
],
[
[
"import os, sys, glob, pprint\nfrom fastai.tabular import *\nfrom sklearn.utils import shuffle\nfrom sklearn.model_selection import train_test_split, StratifiedShuffleSplit\nimport pandas as pd\nimport numpy as np\n\n# set up pretty printer for easier data evaluation\np = pprint.PrettyPrinter(indent=4, width=30)\npretty = p.pprint\n\nprint('libraries loaded')",
"libraries loaded\n"
]
],
[
[
"## Prepare the data\n\nThe balanced datasets have already been balanced and cleaned for use, but we will do some re-cleaning to make sure the experiments without throwing an error. The files and their path are listed here\n\nNOTE: We manually went into the csv files after converting them from arff to remove Infinity cells, NaN cells, and to fill the empty initial column cell with the title Index. The reason we filled the initial column cell is because the fastAI library was throwing errors due to the empty cell title",
"_____no_output_____"
]
],
[
[
"dataPath: str = './balancedSetA'\ndata: list = [\n 'downsampled_merged_5s.csv',\n 'downsampled_SelectedFeatures-10s-TOR-NonTor.csv',\n 'downsampled_SelectedFeatures-15s-TOR-NonTor.csv',\n 'downsampled_TimeBasedFeatures-15s-TOR-NonTOR.csv',\n 'downsampled_TimeBasedFeatures-15s-TOR-NonTOR-15.csv',\n 'downsampled_TimeBasedFeatures-15s-TOR-NonTOR-85.csv',\n 'downsampled_TimeBasedFeatures-30s-TORNonTor.csv',\n 'downsampled_TimeBasedFeatures-30s-TORNonTOR-15.csv',\n 'downsampled_TimeBasedFeatures-30s-TORNonTOR-85.csv',\n 'downsampled_TimeBasedFeatures-60s-TOR-NonTor.csv',\n 'downsampled_TimeBasedFeatures-60s-TOR-NonTOR-15.csv',\n 'downsampled_TimeBasedFeatures-60s-TOR-NonTOR-85.csv',\n 'downsampled_TimeBasedFeatures-120s-TOR-NonTor.csv',\n 'downsampled_TimeBasedFeatures-120s-TOR-NonTOR-15.csv',\n 'downsampled_TimeBasedFeatures-120s-TOR-NonTOR-85.csv'\n]\n\n\ndef get_file_path(file):\n return os.path.join(dataPath, file)\n\ncsvFiles: list = list(map(get_file_path, data))\n\nprint(f'We will be running: {len(csvFiles)} experiments\\n')\npretty(csvFiles)",
"We will be running: 15 experiments\n\n[ './balancedSetA/downsampled_merged_5s.csv',\n './balancedSetA/downsampled_SelectedFeatures-10s-TOR-NonTor.csv',\n './balancedSetA/downsampled_SelectedFeatures-15s-TOR-NonTor.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-15s-TOR-NonTOR.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-15s-TOR-NonTOR-15.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-15s-TOR-NonTOR-85.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-30s-TORNonTor.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-30s-TORNonTOR-15.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-30s-TORNonTOR-85.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-60s-TOR-NonTor.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-60s-TOR-NonTOR-15.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-60s-TOR-NonTOR-85.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-120s-TOR-NonTor.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-120s-TOR-NonTOR-15.csv',\n './balancedSetA/downsampled_TimeBasedFeatures-120s-TOR-NonTOR-85.csv']\n"
]
],
[
[
"Here we create a series of helper functions to load, re-clean, and cache the data",
"_____no_output_____"
]
],
[
[
"def clean(df):\n # let's strip the whitspaces from column names\n df = df.rename(str.strip, axis='columns')\n\n # drop missing values/NaN etc.\n df.dropna(inplace=True)\n\n # drop Infinity rows and NaN string from each column\n for col in df.columns:\n \n indexNames = df[df[col]=='Infinity'].index\n if not indexNames.empty:\n print('deleting {} rows with Infinity in column {}'.format(len(indexNames), col))\n df.drop(indexNames, inplace=True)\n\n indexNames = df[df[col]=='inf'].index\n if not indexNames.empty:\n print('deleting {} rows with inf in column {}'.format(len(indexNames), col))\n df.drop(indexNames, inplace=True)\n\n indexNames = df[df[col]=='NaN'].index\n if not indexNames.empty:\n print('deleting {} rows with NaN in column {}'.format(len(indexNames), col))\n df.drop(indexNames, inplace=True)\n\n indexNames = df[df[col]=='nan'].index\n if not indexNames.empty:\n print('deleting {} rows with nan in column {}'.format(len(indexNames), col))\n df.drop(indexNames, inplace=True)\n\n # drop Source IP and Destination IP columns if they are present \n if 'Source IP' in df.columns:\n df.drop(columns=['Source IP'], inplace=True)\n\n if 'Destination IP' in df.columns:\n df.drop(columns=['Destination IP'], inplace=True)\n \n # convert Flow Bytes/s object & Flow Packets/s object into float type if they are present\n if 'Flow Bytes/s' in df.columns:\n df['Flow Bytes/s'] = df['Flow Bytes/s'].astype('float64')\n\n if 'Flow Packets/s' in df.columns:\n df['Flow Packets/s'] = df['Flow Packets/s'].astype('float64')\n\n return df\n \n\ndef load_data(filePath):\n \n # slice off the ./ from the filePath\n if filePath[0] == '.' and filePath[1] == '/':\n filePathClean: str = filePath[2::]\n pickleDump: str = f'./processed/{filePathClean}.pickle'\n else:\n pickleDump: str = f'./processed/{filePath}.pickle'\n \n print(f'Loading Dataset: {filePath}')\n print(f'\\tTo Dataset Cache: {pickleDump}\\n')\n \n # check if data already exists within cache\n if os.path.exists(pickleDump):\n df = pd.read_pickle(pickleDump)\n \n # if not, load data and clean it before caching it\n else:\n df = pd.read_csv(filePath, low_memory=False)\n df = clean(df)\n \n df.to_pickle(pickleDump)\n \n return df",
"_____no_output_____"
]
],
[
[
"Here we use the helper functions to map over the experiment files and load the csv files into memory",
"_____no_output_____"
]
],
[
[
"dataframes: list = list(map(load_data, csvFiles))",
"Loading Dataset: ./balancedSetA/downsampled_merged_5s.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_merged_5s.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_SelectedFeatures-10s-TOR-NonTor.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_SelectedFeatures-10s-TOR-NonTor.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_SelectedFeatures-15s-TOR-NonTor.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_SelectedFeatures-15s-TOR-NonTor.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-15s-TOR-NonTOR.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-15s-TOR-NonTOR.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-15s-TOR-NonTOR-15.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-15s-TOR-NonTOR-15.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-15s-TOR-NonTOR-85.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-15s-TOR-NonTOR-85.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-30s-TORNonTor.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-30s-TORNonTor.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-30s-TORNonTOR-15.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-30s-TORNonTOR-15.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-30s-TORNonTOR-85.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-30s-TORNonTOR-85.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-60s-TOR-NonTor.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-60s-TOR-NonTor.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-60s-TOR-NonTOR-15.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-60s-TOR-NonTOR-15.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-60s-TOR-NonTOR-85.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-60s-TOR-NonTOR-85.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-120s-TOR-NonTor.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-120s-TOR-NonTor.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-120s-TOR-NonTOR-15.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-120s-TOR-NonTOR-15.csv.pickle\n\nLoading Dataset: ./balancedSetA/downsampled_TimeBasedFeatures-120s-TOR-NonTOR-85.csv\n\tTo Dataset Cache: ./processed/balancedSetA/downsampled_TimeBasedFeatures-120s-TOR-NonTOR-85.csv.pickle\n\n"
]
],
[
[
"With the data cleaned, packaged, and loaded into memory, we now can create the functions to run the experiments",
"_____no_output_____"
]
],
[
[
"def run_experiment(experiment_number, df):\n try:\n dep_var = 'label'\n x = df[df[dep_var]=='TOR']\n except:\n dep_var = 'class1'\n x = df[df[dep_var]=='TOR']\n\n unused_categories: list = []\n \n if 'Source Port' in df.columns:\n unused_categories.append('Source Port')\n if 'Destination Port' in df.columns:\n unused_categories.append('Destination Port')\n if 'Protocol' in df.columns:\n unused_categories.append('Protocol')\n if 'Index' in df.columns:\n unused_categories.append('Index')\n if 'Unnamed: 0' in df.columns:\n unused_categories.append('Unnamed: 0')\n \n selected_features = list(set(df) - set(unused_categories) - set([dep_var]))\n print('Selected Features:')\n pretty(selected_features)\n print('\\nDependent Variable:', dep_var)\n \n procs = [FillMissing, Categorify, Normalize]\n sss = StratifiedShuffleSplit(n_splits = 1, test_size=0.2, random_state=0)\n \n for train_idx, test_idx in sss.split(df.index, df[dep_var]):\n data_fold = (TabularList.from_df(df, path=dataPath, cat_names=unused_categories, cont_names=selected_features, procs=procs)\n .split_by_idxs(train_idx, test_idx)\n .label_from_df(cols=dep_var)\n .databunch())\n \n \n # create model and learn\n model = tabular_learner(data_fold, layers=[50, 28], metrics=accuracy, callback_fns=ShowGraph)\n model.fit_one_cycle(cyc_len=10) #\n model.save('{}.model'.format(os.path.basename(csvFiles[experiment_number])))\n \n loss, acc = model.validate()\n print('loss {}: accuracy: {:.2f}%'.format(loss, acc*100))\n \n preds, y, losses = model.get_preds(with_loss=True)\n interp = ClassificationInterpretation(model, preds, y, losses)\n interp.plot_confusion_matrix()\n\n\ndef experiment_metadata(experiment_number, df):\n print(f'Experiment #{experiment_number + 1}\\n\\ndataset:\\t\\t{data[experiment_number]}\\nshape:\\t\\t\\t{df.shape}')\n try:\n id_column = 'label'\n tor_data = df[df[id_column]=='TOR']\n non_tor_data = df[df[id_column]=='nonTOR']\n if len(non_tor_data.index) == 0:\n non_tor_data = df[df[id_column] == 'NONTOR']\n except:\n id_column = 'class1'\n tor_data = df[df[id_column]=='TOR']\n non_tor_data = df[df[id_column]=='nonTOR']\n if len(non_tor_data.index) == 0:\n non_tor_data = df[df[id_column] == 'NONTOR']\n print(f'total TOR data:\\t\\t{len(tor_data.index)}\\ntotal Non-TOR data:\\t{len(non_tor_data.index)}\\n\\n')\n",
"_____no_output_____"
]
],
[
[
"Lets test that the experiments run when given an arbitrary set of data",
"_____no_output_____"
]
],
[
[
"experiment_metadata(12, dataframes[12])",
"Experiment #13\n\ndataset:\t\tdownsampled_TimeBasedFeatures-120s-TOR-NonTor.csv\nshape:\t\t\t(940, 25)\ntotal TOR data:\t\t470\ntotal Non-TOR data:\t470\n\n\n"
],
[
"run_experiment(12, dataframes[12])",
"Selected Features:\n[ 'max_biat',\n 'total_biat',\n 'mean_fiat',\n 'max_idle',\n 'total_fiat',\n 'min_idle',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
]
],
[
[
"Since our experiments are all neatly packaged in lists, we have created a simple generator to run each experiment",
"_____no_output_____"
]
],
[
[
"def run_experiments(dataframes):\n for experiment_number, df in enumerate(dataframes):\n experiment_metadata(experiment_number, df)\n yield run_experiment(experiment_number, df)",
"_____no_output_____"
],
[
"experiment = run_experiments(dataframes)",
"_____no_output_____"
]
],
[
[
"Here we run all of the experiments now that they have been set up",
"_____no_output_____"
]
],
[
[
"next(experiment)",
"Experiment #1\n\ndataset:\t\tdownsampled_merged_5s.csv\nshape:\t\t\t(29014, 28)\ntotal TOR data:\t\t14507\ntotal Non-TOR data:\t14507\n\n\nSelected Features:\n[ 'Bwd IAT Max',\n 'Idle Max',\n 'Active Std',\n 'Fwd IAT Mean',\n 'Fwd IAT Max',\n 'Active Mean',\n 'Idle Min',\n 'Active Max',\n 'Fwd IAT Min',\n 'Idle Mean',\n 'Bwd IAT Mean',\n 'Flow IAT Mean',\n 'Flow Bytes/s',\n 'Active Min',\n 'Flow IAT Max',\n 'Flow Duration',\n 'Flow Packets/s',\n 'Flow IAT Std',\n 'Flow IAT Min',\n 'Fwd IAT Std',\n 'Bwd IAT Std',\n 'Idle Std',\n 'Bwd IAT Min']\n\nDependent Variable: label\n"
],
[
"next(experiment)",
"Experiment #2\n\ndataset:\t\tdownsampled_SelectedFeatures-10s-TOR-NonTor.csv\nshape:\t\t\t(16088, 28)\ntotal TOR data:\t\t8044\ntotal Non-TOR data:\t8044\n\n\nSelected Features:\n[ 'Bwd IAT Max',\n 'Idle Max',\n 'Active Std',\n 'Fwd IAT Mean',\n 'Fwd IAT Max',\n 'Active Mean',\n 'Idle Min',\n 'Active Max',\n 'Fwd IAT Min',\n 'Idle Mean',\n 'Bwd IAT Mean',\n 'Flow IAT Mean',\n 'Flow Bytes/s',\n 'Active Min',\n 'Flow IAT Max',\n 'Flow Duration',\n 'Flow Packets/s',\n 'Flow IAT Std',\n 'Flow IAT Min',\n 'Fwd IAT Std',\n 'Bwd IAT Std',\n 'Idle Std',\n 'Bwd IAT Min']\n\nDependent Variable: label\n"
],
[
"next(experiment)",
"Experiment #3\n\ndataset:\t\tdownsampled_SelectedFeatures-15s-TOR-NonTor.csv\nshape:\t\t\t(6628, 6)\ntotal TOR data:\t\t3314\ntotal Non-TOR data:\t3314\n\n\nSelected Features:\n[ 'min_biat',\n 'max_biat',\n 'max_fiat',\n 'min_fiat']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #4\n\ndataset:\t\tdownsampled_TimeBasedFeatures-15s-TOR-NonTOR.csv\nshape:\t\t\t(6628, 25)\ntotal TOR data:\t\t3314\ntotal Non-TOR data:\t3314\n\n\nSelected Features:\n[ 'max_biat',\n 'total_biat',\n 'mean_fiat',\n 'max_idle',\n 'total_fiat',\n 'min_idle',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #5\n\ndataset:\t\tdownsampled_TimeBasedFeatures-15s-TOR-NonTOR-15.csv\nshape:\t\t\t(1030, 25)\ntotal TOR data:\t\t515\ntotal Non-TOR data:\t515\n\n\nSelected Features:\n[ 'max_biat',\n 'total_biat',\n 'mean_fiat',\n 'max_idle',\n 'total_fiat',\n 'min_idle',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #6\n\ndataset:\t\tdownsampled_TimeBasedFeatures-15s-TOR-NonTOR-85.csv\nshape:\t\t\t(5598, 25)\ntotal TOR data:\t\t2799\ntotal Non-TOR data:\t2799\n\n\nSelected Features:\n[ 'max_biat',\n 'total_biat',\n 'mean_fiat',\n 'max_idle',\n 'total_fiat',\n 'min_idle',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #7\n\ndataset:\t\tdownsampled_TimeBasedFeatures-30s-TORNonTor.csv\nshape:\t\t\t(3542, 25)\ntotal TOR data:\t\t1771\ntotal Non-TOR data:\t1771\n\n\nSelected Features:\n[ 'max_biat',\n 'total_biat',\n 'mean_fiat',\n 'max_idle',\n 'total_fiat',\n 'min_idle',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #8\n\ndataset:\t\tdownsampled_TimeBasedFeatures-30s-TORNonTOR-15.csv\nshape:\t\t\t(530, 25)\ntotal TOR data:\t\t265\ntotal Non-TOR data:\t265\n\n\nSelected Features:\n[ 'max_biat',\n 'total_biat',\n 'mean_fiat',\n 'max_idle',\n 'total_fiat',\n 'min_idle',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #9\n\ndataset:\t\tdownsampled_TimeBasedFeatures-30s-TORNonTOR-85.csv\nshape:\t\t\t(3012, 25)\ntotal TOR data:\t\t1506\ntotal Non-TOR data:\t1506\n\n\nSelected Features:\n[ 'max_biat',\n 'total_biat',\n 'mean_fiat',\n 'max_idle',\n 'total_fiat',\n 'min_idle',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #10\n\ndataset:\t\tdownsampled_TimeBasedFeatures-60s-TOR-NonTor.csv\nshape:\t\t\t(1828, 25)\ntotal TOR data:\t\t914\ntotal Non-TOR data:\t914\n\n\nSelected Features:\n[ 'max_biat',\n 'max_idle',\n 'mean_fiat',\n 'min_idle',\n 'std_biat',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_fiat',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #11\n\ndataset:\t\tdownsampled_TimeBasedFeatures-60s-TOR-NonTOR-15.csv\nshape:\t\t\t(272, 25)\ntotal TOR data:\t\t136\ntotal Non-TOR data:\t136\n\n\nSelected Features:\n[ 'max_biat',\n 'max_idle',\n 'mean_fiat',\n 'min_idle',\n 'std_biat',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_fiat',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #12\n\ndataset:\t\tdownsampled_TimeBasedFeatures-60s-TOR-NonTOR-85.csv\nshape:\t\t\t(1556, 25)\ntotal TOR data:\t\t778\ntotal Non-TOR data:\t778\n\n\nSelected Features:\n[ 'max_biat',\n 'max_idle',\n 'mean_fiat',\n 'min_idle',\n 'std_biat',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_fiat',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #13\n\ndataset:\t\tdownsampled_TimeBasedFeatures-120s-TOR-NonTor.csv\nshape:\t\t\t(940, 25)\ntotal TOR data:\t\t470\ntotal Non-TOR data:\t470\n\n\nSelected Features:\n[ 'max_biat',\n 'total_biat',\n 'mean_fiat',\n 'max_idle',\n 'total_fiat',\n 'min_idle',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #14\n\ndataset:\t\tdownsampled_TimeBasedFeatures-120s-TOR-NonTOR-15.csv\nshape:\t\t\t(132, 25)\ntotal TOR data:\t\t66\ntotal Non-TOR data:\t66\n\n\nSelected Features:\n[ 'max_biat',\n 'total_biat',\n 'mean_fiat',\n 'max_idle',\n 'total_fiat',\n 'min_idle',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
],
[
"next(experiment)",
"Experiment #15\n\ndataset:\t\tdownsampled_TimeBasedFeatures-120s-TOR-NonTOR-85.csv\nshape:\t\t\t(808, 25)\ntotal TOR data:\t\t404\ntotal Non-TOR data:\t404\n\n\nSelected Features:\n[ 'max_biat',\n 'total_biat',\n 'mean_fiat',\n 'max_idle',\n 'total_fiat',\n 'min_idle',\n 'std_flowiat',\n 'flowPktsPerSecond',\n 'min_biat',\n 'mean_biat',\n 'mean_idle',\n 'flowBytesPerSecond',\n 'min_flowiat',\n 'duration',\n 'std_active',\n 'max_fiat',\n 'max_active',\n 'min_fiat',\n 'std_idle',\n 'mean_flowiat',\n 'max_flowiat',\n 'mean_active',\n 'min_active']\n\nDependent Variable: class1\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf0d933a3477a141a930af1974f95781d5d645c | 946,592 | ipynb | Jupyter Notebook | deepthought/experiments/audiomostly2014/trialorder confusion.ipynb | maosenGao/deepthought | 4bd7e06463368920e55048276af20488becabf6a | [
"BSD-3-Clause"
] | 43 | 2015-02-01T15:37:38.000Z | 2022-01-08T05:34:04.000Z | deepthought/experiments/audiomostly2014/trialorder confusion.ipynb | maosenGao/deepthought | 4bd7e06463368920e55048276af20488becabf6a | [
"BSD-3-Clause"
] | 4 | 2015-02-12T11:28:38.000Z | 2016-10-08T17:28:49.000Z | deepthought/experiments/audiomostly2014/trialorder confusion.ipynb | maosenGao/deepthought | 4bd7e06463368920e55048276af20488becabf6a | [
"BSD-3-Clause"
] | 25 | 2015-10-04T17:49:53.000Z | 2021-06-02T15:35:47.000Z | 305.352258 | 53,461 | 0.914553 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecf0e0c43f092621cde1d56f84a1aef6b25f82a1 | 7,002 | ipynb | Jupyter Notebook | Week-06/2_LSTM.ipynb | Davidyz/ML-Tutorials-Season-2 | e9f0cbaf46e3207c0172d9cf2b6f527f9ff2198e | [
"MIT"
] | 14 | 2021-10-09T15:53:09.000Z | 2022-03-29T12:09:13.000Z | Week-06/2_LSTM.ipynb | Davidyz/ML-Tutorials-Season-2 | e9f0cbaf46e3207c0172d9cf2b6f527f9ff2198e | [
"MIT"
] | null | null | null | Week-06/2_LSTM.ipynb | Davidyz/ML-Tutorials-Season-2 | e9f0cbaf46e3207c0172d9cf2b6f527f9ff2198e | [
"MIT"
] | 10 | 2021-10-09T23:50:51.000Z | 2022-02-02T11:24:37.000Z | 34.835821 | 383 | 0.60954 | [
[
[
"# Long Short-term Memory model (LSTM)\n\nAs it has been mentioned in the previous notebook, the neurons of recurrent neural networks (RNNs) have an ability to retain cell memory that also influences the perception of new inputs. It all works fine, when we are dealing with problems with large separation between the predicted value and input, we face so-called ***vanishing gradient*** problem.\n\n#### Vanishing gradient\n\n\n\nIn simple terms, during the process of training a neural network, we are multiplying weights of each layers. However, if these weights (less than 1) are multiplied many times, we reach an extremely small values. Therefore, the weights of those earlier layers will not be changed signifficantly and our network will not be able to learn long-term dependencies.\n\nThis brings brings us to **LSTM** that introduces changes RNN in larger series analysis.\n\n\n## LSTM Structure\n\n\n\nThe main difference between the LSTM and RNN structure is that LSTM has special kind of units (so-called, **LSTM cells**) instead of regular neural network layers. In the following sections, we will have a look at each structural unit of LSTM cell.\n\n\n### Input\n\nAs it can be seen in the figure above, the input value is first concatenated to the output of the previous cell. This new input is then squashed between -1 and 1 using $tanh$ activation function:\n\n$g = tanh(x_tU^g + h_tV^g + b^g)$\n\nHere $U$, $V$ and $b$ are weights and bias respectively (*note that exponents g does not imply raising in power, but rather show that these are weights and biases at the input gate*).\n\nThis squashed input is then multiplied element-wise by the input layer of sigmoid activated nodes. The existance of sigmoid function (output in the range of 0-1) allows to \"eliminate\" insignifficant . The output of the sigmoid nodes can be expressed as:\n\n$i = \\sigma(x_tU^i + h_tV^i + b^i)$\n\nAfter element-wise multiplication, we get the output of our input section:\n\n$g*i$\n\n### Forget gate\n\nThe output of the input section is then passed through the forget gate loop. LSTM at this section has an internal state variable $s_t$ which is added to the lagged state ($s_{t-1}$). This addition operation is used instead of multiplication allows to reduce the risk of vanishing gradients while the forget gate itself helps model to learn only signifficant state variables.\n\nMathematically, the forget gate output can be expressed as:\n\n$f = \\sigma(x_tU^f + h_tV^f + b^f)$\n\nMultiplying this by the previous state value and adding the input gate output, we get the new state value:\n\n$s_t = s_{t-1}*f+g*i$\n\n### Output gate\n\nAt the output gate, we have to multiply the squashed ($tanh$ activation function) current state value by the forget output passed through the sigmoid function. In mathematical terms:\n\n$h_t = tanh(s_t)*\\sigma(x_tU^o + h_tV^o + b^o)$\n\n## Keras implementation\n\nAs we have now covered all required theory, we are going to look how all of it can be implemented in Keras (*we are not going to be covering implementation from scratch*).\n\nLet's create a simple sine function dataset:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport tensorflow as tf\nimport math",
"_____no_output_____"
],
[
"X_train = np.arange(0,100,0.5) \ny_train = np.sin(X_train)\n\nX_test = np.arange(100,200,0.5) \ny_test = np.sin(X_test)\n\ntrain_series = y_train.reshape((len(y_train), 1))\ntest_series = y_test.reshape((len(y_test), 1))\n\nplt.plot(X_train, y_train)\nplt.plot(X_test, y_test)",
"_____no_output_____"
]
],
[
[
"On the other hand, as you might remember from the RNN notebook, the sequential data models are trained by passing array of lagged data as an input and future data as the output. Let's create function, that translates our dataset into such batches (for the start, we will use look-back period of 1).",
"_____no_output_____"
]
],
[
[
"def dataset(data, look_back):\n X, y = [], []\n \n for i in range(len(data) - look_back):\n a = data[i:(i + look_back), ]\n X.append(a)\n y.append(data[i + look_back, ])\n return np.array(X), np.array(y)\n\nlook_back = 1\nX_train, y_train = dataset(train_series, look_back)\nX_test, y_test = dataset(test_series, look_back)",
"_____no_output_____"
]
],
[
[
"After splitting data into batches, we can now create our simple LSTM model",
"_____no_output_____"
]
],
[
[
"model = tf.keras.models.Sequential([\n #Create model\n ___\n])\n\n#Compile your model\n___\n\n#Fit model\n___",
"_____no_output_____"
]
],
[
[
"Finally, let's test our model with the validation dataset.",
"_____no_output_____"
]
],
[
[
"test_predictions = model.predict(X_test).reshape(199, 1)\nplt.plot(test_predictions)\nplt.plot(y_test)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf0e1376cf25ba244d766a85df681f92f25d65f | 3,410 | ipynb | Jupyter Notebook | iplPrediction.ipynb | vishu9219/ipl_prediction | 3b93d660019238be757ba4e89c3b26e1b6b48cc9 | [
"BSD-3-Clause"
] | null | null | null | iplPrediction.ipynb | vishu9219/ipl_prediction | 3b93d660019238be757ba4e89c3b26e1b6b48cc9 | [
"BSD-3-Clause"
] | null | null | null | iplPrediction.ipynb | vishu9219/ipl_prediction | 3b93d660019238be757ba4e89c3b26e1b6b48cc9 | [
"BSD-3-Clause"
] | null | null | null | 36.666667 | 234 | 0.56393 | [
[
[
"<a href=\"https://colab.research.google.com/github/vishu9219/ipl_prediction/blob/main/iplPrediction.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"Installing Python packages\npandas\nnumpy\nsklearn",
"_____no_output_____"
]
],
[
[
"! pip install pandas\n! pip install numpy\n! pip install sklearn\n! pip install random",
"Requirement already satisfied: pandas in /usr/local/lib/python3.7/dist-packages (1.1.5)\nRequirement already satisfied: numpy>=1.15.4 in /usr/local/lib/python3.7/dist-packages (from pandas) (1.19.5)\nRequirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.7/dist-packages (from pandas) (2.8.1)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.7/dist-packages (from pandas) (2018.9)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.7/dist-packages (from python-dateutil>=2.7.3->pandas) (1.15.0)\nRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (1.19.5)\nRequirement already satisfied: sklearn in /usr/local/lib/python3.7/dist-packages (0.0)\nRequirement already satisfied: scikit-learn in /usr/local/lib/python3.7/dist-packages (from sklearn) (0.22.2.post1)\nRequirement already satisfied: scipy>=0.17.0 in /usr/local/lib/python3.7/dist-packages (from scikit-learn->sklearn) (1.4.1)\nRequirement already satisfied: numpy>=1.11.0 in /usr/local/lib/python3.7/dist-packages (from scikit-learn->sklearn) (1.19.5)\nRequirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.7/dist-packages (from scikit-learn->sklearn) (1.0.1)\n\u001b[31mERROR: Could not find a version that satisfies the requirement random (from versions: none)\u001b[0m\n\u001b[31mERROR: No matching distribution found for random\u001b[0m\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf0e43c2be3dadf5948f1ed3b4f5c212ee26c99 | 22,338 | ipynb | Jupyter Notebook | r_examples/r_batch_transform/r_xgboost_batch_transform.ipynb | wmlba/amazon-sagemaker-examples | 340118df142cf76ecb2f2ab407fe5ffb257ed848 | [
"Apache-2.0"
] | 4 | 2020-06-15T14:07:43.000Z | 2020-06-28T12:34:13.000Z | r_examples/r_batch_transform/r_xgboost_batch_transform.ipynb | wmlba/amazon-sagemaker-examples | 340118df142cf76ecb2f2ab407fe5ffb257ed848 | [
"Apache-2.0"
] | null | null | null | r_examples/r_batch_transform/r_xgboost_batch_transform.ipynb | wmlba/amazon-sagemaker-examples | 340118df142cf76ecb2f2ab407fe5ffb257ed848 | [
"Apache-2.0"
] | 5 | 2020-06-27T12:15:51.000Z | 2020-06-28T12:34:14.000Z | 37.292154 | 419 | 0.603859 | [
[
[
"<h1>Batch Transform Using R with Amazon SageMaker</h1>\n\n**Note:** You will need to use R kernel in SageMaker for this notebook.\n\nThis sample Notebook describes how to do batch transform to make predictions for abalone age as measured by the number of rings in the shell. The notebook will use the public [abalone dataset](https://archive.ics.uci.edu/ml/datasets/abalone) hosted by [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/index.php).\n\nYou can find more details about SageMaker's Batch Trsnform here: \n- [Batch Transform](https://docs.aws.amazon.com/sagemaker/latest/dg/batch-transform.html) using a Transformer\n\nWe will use `reticulate` library to interact with SageMaker:\n- [`Reticulate` library](https://rstudio.github.io/reticulate/): provides an R interface to make API calls [Amazon SageMaker Python SDK](https://sagemaker.readthedocs.io/en/latest/index.html) to make API calls to Amazon SageMaker. The `reticulate` package translates between R and Python objects, and Amazon SageMaker provides a serverless data science environment to train and deploy ML models at scale.\n\nTable of Contents:\n- [Reticulating the Amazon SageMaker Python SDK](#Reticulating-the-Amazon-SageMaker-Python-SDK)\n- [Creating and Accessing the Data Storage](#Creating-and-accessing-the-data-storage)\n- [Downloading and Processing the Dataset](#Downloading-and-processing-the-dataset)\n- [Preparing the Dataset for Model Training](#Preparing-the-dataset-for-model-training)\n- [Creating a SageMaker Estimator](#Creating-a-SageMaker-Estimator)\n- [Batch Transform using SageMaker Transformer](#Batch-Transform-using-SageMaker-Transformer)\n- [Download the Batch Transform Output](#Download-the-Batch-Transform-Output)\n\n\n**Note:** The first portion of this notebook focused on data ingestion and preparing the data for model training is inspired by the data preparation part outlined in [\"Using R with Amazon SageMaker\"](https://github.com/awslabs/amazon-sagemaker-examples/blob/master/advanced_functionality/r_kernel/using_r_with_amazon_sagemaker.ipynb) notebook on AWS SageMaker Examples Github repository with some modifications.",
"_____no_output_____"
],
[
"<h3>Reticulating the Amazon SageMaker Python SDK</h3>\n\nFirst, load the `reticulate` library and import the `sagemaker` Python module. Once the module is loaded, use the `$` notation in R instead of the `.` notation in Python to use available classes. ",
"_____no_output_____"
]
],
[
[
"# Turn warnings off globally\noptions(warn=-1)",
"_____no_output_____"
],
[
"# Install reticulate library and import sagemaker\nlibrary(reticulate)\nsagemaker <- import('sagemaker')",
"_____no_output_____"
]
],
[
[
"<h3>Creating and Accessing the Data Storage</h3>\n\nThe `Session` class provides operations for working with the following [boto3](https://boto3.amazonaws.com/v1/documentation/api/latest/index.html) resources with Amazon SageMaker:\n\n* [S3](https://boto3.readthedocs.io/en/latest/reference/services/s3.html)\n* [SageMaker](https://boto3.readthedocs.io/en/latest/reference/services/sagemaker.html)\n\nLet's create an [Amazon Simple Storage Service](https://aws.amazon.com/s3/) bucket for your data. ",
"_____no_output_____"
]
],
[
[
"session <- sagemaker$Session()\nbucket <- session$default_bucket()\nprefix <- 'r-batch-transform'",
"_____no_output_____"
]
],
[
[
"**Note** - The `default_bucket` function creates a unique Amazon S3 bucket with the following name: \n\n`sagemaker-<aws-region-name>-<aws account number>`\n\nSpecify the IAM role's [ARN](https://docs.aws.amazon.com/general/latest/gr/aws-arns-and-namespaces.html) to allow Amazon SageMaker to access the Amazon S3 bucket. You can use the same IAM role used to create this Notebook:",
"_____no_output_____"
]
],
[
[
"role_arn <- sagemaker$get_execution_role()",
"_____no_output_____"
]
],
[
[
"<h3>Downloading and Processing the Dataset</h3>\n\nThe model uses the [abalone dataset](https://archive.ics.uci.edu/ml/datasets/abalone) from the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/index.php). First, download the data and start the [exploratory data analysis](https://en.wikipedia.org/wiki/Exploratory_data_analysis). Use tidyverse packages to read the data, plot the data, and transform the data into ML format for Amazon SageMaker:",
"_____no_output_____"
]
],
[
[
"library(readr)\ndata_file <- 'http://archive.ics.uci.edu/ml/machine-learning-databases/abalone/abalone.data'\nabalone <- read_csv(file = data_file, col_names = FALSE)\nnames(abalone) <- c('sex', 'length', 'diameter', 'height', 'whole_weight', 'shucked_weight', 'viscera_weight', 'shell_weight', 'rings')\nhead(abalone)",
"_____no_output_____"
]
],
[
[
"The output above shows that `sex` is a factor data type but is currently a character data type (F is Female, M is male, and I is infant). Change `sex` to a factor and view the statistical summary of the dataset:",
"_____no_output_____"
]
],
[
[
"abalone$sex <- as.factor(abalone$sex)\nsummary(abalone)",
"_____no_output_____"
]
],
[
[
"The summary above shows that the minimum value for `height` is 0.\n\nVisually explore which abalones have height equal to 0 by plotting the relationship between `rings` and `height` for each value of `sex`:",
"_____no_output_____"
]
],
[
[
"library(ggplot2)\noptions(repr.plot.width = 5, repr.plot.height = 4) \nggplot(abalone, aes(x = height, y = rings, color = sex)) + geom_point() + geom_jitter()",
"_____no_output_____"
]
],
[
[
"The plot shows multiple outliers: two infant abalones with a height of 0 and a few female and male abalones with greater heights than the rest. Let's filter out the two infant abalones with a height of 0.",
"_____no_output_____"
]
],
[
[
"library(dplyr)\nabalone <- abalone %>%\n filter(height != 0)",
"_____no_output_____"
]
],
[
[
"<h3>Preparing the Dataset for Model Training</h3>\n\nThe model needs three datasets: one each for training, testing, and validation. First, convert `sex` into a [dummy variable](https://en.wikipedia.org/wiki/Dummy_variable_(statistics)) and move the target, `rings`, to the first column. Amazon SageMaker algorithm require the target to be in the first column of the dataset.",
"_____no_output_____"
]
],
[
[
"abalone <- abalone %>%\n mutate(female = as.integer(ifelse(sex == 'F', 1, 0)),\n male = as.integer(ifelse(sex == 'M', 1, 0)),\n infant = as.integer(ifelse(sex == 'I', 1, 0))) %>%\n select(-sex)\nabalone <- abalone %>%\n select(rings:infant, length:shell_weight)\nhead(abalone)",
"_____no_output_____"
]
],
[
[
"Next, sample 70% of the data for training the ML algorithm. Split the remaining 30% into two halves, one for testing and one for validation:",
"_____no_output_____"
]
],
[
[
"abalone_train <- abalone %>%\n sample_frac(size = 0.7)\nabalone <- anti_join(abalone, abalone_train)\nabalone_test <- abalone %>%\n sample_frac(size = 0.5)\nabalone_valid <- anti_join(abalone, abalone_test)",
"_____no_output_____"
]
],
[
[
"Later in the notebook, we are going to use Batch Transform and Endpoint to make inference in two different ways and we will compare the results. The maximum number of rows that we can send to an endpoint for inference in one batch is 500 rows. We are going to reduce the number of rows for the test dataset to 500 and use this for batch and online inference for comparison. ",
"_____no_output_____"
]
],
[
[
"num_predict_rows <- 500\nabalone_test <- abalone_test[1:num_predict_rows, ]",
"_____no_output_____"
]
],
[
[
"Upload the training and validation data to Amazon S3 so that you can train the model. First, write the training and validation datasets to the local filesystem in .csv format:",
"_____no_output_____"
]
],
[
[
"write_csv(abalone_train, 'abalone_train.csv', col_names = FALSE)\nwrite_csv(abalone_valid, 'abalone_valid.csv', col_names = FALSE)\n\n# Remove target from test\nwrite_csv(abalone_test[-1], 'abalone_test.csv', col_names = FALSE)",
"_____no_output_____"
]
],
[
[
"Second, upload the two datasets to the Amazon S3 bucket into the `data` key:",
"_____no_output_____"
]
],
[
[
"s3_train <- session$upload_data(path = 'abalone_train.csv', \n bucket = bucket, \n key_prefix = paste(prefix,'data', sep = '/'))\ns3_valid <- session$upload_data(path = 'abalone_valid.csv', \n bucket = bucket, \n key_prefix = paste(prefix,'data', sep = '/'))\n\ns3_test <- session$upload_data(path = 'abalone_test.csv', \n bucket = bucket, \n key_prefix = paste(prefix,'data', sep = '/'))",
"_____no_output_____"
]
],
[
[
"Finally, define the Amazon S3 input types for the Amazon SageMaker algorithm:",
"_____no_output_____"
]
],
[
[
"s3_train_input <- sagemaker$s3_input(s3_data = s3_train,\n content_type = 'csv')\ns3_valid_input <- sagemaker$s3_input(s3_data = s3_valid,\n content_type = 'csv')",
"_____no_output_____"
]
],
[
[
"<hr>\n<h3>Creating a SageMaker Estimator</h3>\n\nAmazon SageMaker algorithm are available via a [Docker](https://www.docker.com/) container. To train an [XGBoost](https://en.wikipedia.org/wiki/Xgboost) model, specify the training containers in [Amazon Elastic Container Registry](https://aws.amazon.com/ecr/) (Amazon ECR) for the AWS Region.",
"_____no_output_____"
]
],
[
[
"registry <- sagemaker$amazon$amazon_estimator$registry(session$boto_region_name, algorithm='xgboost')\ncontainer <- paste(registry, '/xgboost:latest', sep='')\ncat('XGBoost Container Image URL: ', container)",
"_____no_output_____"
]
],
[
[
"Define an Amazon SageMaker [Estimator](http://sagemaker.readthedocs.io/en/latest/estimators.html), which can train any supplied algorithm that has been containerized with Docker. When creating the Estimator, use the following arguments:\n* **image_name** - The container image to use for training\n* **role** - The Amazon SageMaker service role\n* **train_instance_count** - The number of Amazon EC2 instances to use for training\n* **train_instance_type** - The type of Amazon EC2 instance to use for training\n* **train_volume_size** - The size in GB of the [Amazon Elastic Block Store](https://aws.amazon.com/ebs/) (Amazon EBS) volume to use for storing input data during training\n* **train_max_run** - The timeout in seconds for training\n* **input_mode** - The input mode that the algorithm supports\n* **output_path** - The Amazon S3 location for saving the training results (model artifacts and output files)\n* **output_kms_key** - The [AWS Key Management Service](https://aws.amazon.com/kms/) (AWS KMS) key for encrypting the training output\n* **base_job_name** - The prefix for the name of the training job\n* **sagemaker_session** - The Session object that manages interactions with Amazon SageMaker API",
"_____no_output_____"
]
],
[
[
"# Model artifacts and batch output\ns3_output <- paste('s3:/', bucket, prefix,'output', sep = '/')",
"_____no_output_____"
],
[
"# Estimator\nestimator <- sagemaker$estimator$Estimator(image_name = container,\n role = role_arn,\n train_instance_count = 1L,\n train_instance_type = 'ml.m5.4xlarge',\n train_volume_size = 30L,\n train_max_run = 3600L,\n input_mode = 'File',\n output_path = s3_output,\n output_kms_key = NULL,\n base_job_name = NULL,\n sagemaker_session = NULL)",
"_____no_output_____"
]
],
[
[
"**Note** - The equivalent to `None` in Python is `NULL` in R.\n\nNext, we Specify the [XGBoost hyperparameters](https://docs.aws.amazon.com/sagemaker/latest/dg/xgboost_hyperparameters.html) for the estimator. \n\nOnce the Estimator and its hyperparamters are specified, you can train (or fit) the estimator.",
"_____no_output_____"
]
],
[
[
"# Set Hyperparameters\nestimator$set_hyperparameters(eval_metric='rmse',\n objective='reg:linear',\n num_round=100L,\n rate_drop=0.3,\n tweedie_variance_power=1.4)",
"_____no_output_____"
],
[
"# Create a training job name\njob_name <- paste('sagemaker-r-xgboost', format(Sys.time(), '%H-%M-%S'), sep = '-')\n\n# Define the data channels for train and validation datasets\ninput_data <- list('train' = s3_train_input,\n 'validation' = s3_valid_input)\n\n# train the estimator\nestimator$fit(inputs = input_data, job_name = job_name)",
"_____no_output_____"
]
],
[
[
"<hr>\n\n<h3> Batch Transform using SageMaker Transformer </h3>",
"_____no_output_____"
],
[
"For more details on SageMaker Batch Transform, you can visit this example notebook on [Amazon SageMaker Batch Transform](https://github.com/awslabs/amazon-sagemaker-examples/blob/master/sagemaker_batch_transform/introduction_to_batch_transform/batch_transform_pca_dbscan_movie_clusters.ipynb).\n\nIn many situations, using a deployed model for making inference is not the best option, especially when the goal is not to make online real-time inference but to generate predictions from a trained model on a large dataset. In these situations, using Batch Transform may be more efficient and appropriate.\n\nThis section of the notebook explain how to set up the Batch Transform Job, and generate predictions.\n\nTo do this, we need to define the batch input data path on S3, and also where to save the generated predictions on S3.",
"_____no_output_____"
]
],
[
[
"# Define S3 path for Test data \ns3_test_url <- paste('s3:/', bucket, prefix, 'data','abalone_test.csv', sep = '/')",
"_____no_output_____"
]
],
[
[
"Then we create a `Transformer`. [Transformers](https://sagemaker.readthedocs.io/en/stable/transformer.html#transformer) take multiple paramters, including the following. For more details and the complete list visit the [documentation page](https://sagemaker.readthedocs.io/en/stable/transformer.html#transformer).\n\n- **model_name** (str) – Name of the SageMaker model being used for the transform job.\n- **instance_count** (int) – Number of EC2 instances to use.\n- **instance_type** (str) – Type of EC2 instance to use, for example, ‘ml.c4.xlarge’.\n\n- **output_path** (str) – S3 location for saving the transform result. If not specified, results are stored to a default bucket.\n\n- **base_transform_job_name** (str) – Prefix for the transform job when the transform() method launches. If not specified, a default prefix will be generated based on the training image name that was used to train the model associated with the transform job.\n\n- **sagemaker_session** (sagemaker.session.Session) – Session object which manages interactions with Amazon SageMaker APIs and any other AWS services needed. If not specified, the estimator creates one using the default AWS configuration chain.\n\nOnce we create a `Transformer` we can transform the batch input.",
"_____no_output_____"
]
],
[
[
"# Define a transformer\ntransformer <- estimator$transformer(instance_count=1L, \n instance_type='ml.m4.xlarge',\n output_path = s3_output)",
"_____no_output_____"
],
[
"# Do the batch transform\ntransformer$transform(s3_test_url,\n wait = TRUE)",
"_____no_output_____"
]
],
[
[
"<hr>\n<h3> Download the Batch Transform Output </h3>",
"_____no_output_____"
]
],
[
[
"# Download the file from S3 using S3Downloader to local SageMaker instance 'batch_output' folder\nsagemaker$s3$S3Downloader$download(paste(s3_output,\"abalone_test.csv.out\",sep = '/'),\n \"batch_output\")",
"_____no_output_____"
],
[
"# Read the batch csv from sagemaker local files\nlibrary(readr)\npredictions <- read_csv(file = 'batch_output/abalone_test.csv.out', col_names = 'predicted_rings')\nhead(predictions)",
"_____no_output_____"
]
],
[
[
"Column-bind the predicted rings to the test data:",
"_____no_output_____"
]
],
[
[
"# Concatenate predictions and test for comparison\nabalone_predictions <- cbind(predicted_rings = predictions, \n abalone_test)\n# Convert predictions to Integer\nabalone_predictions$predicted_rings = as.integer(abalone_predictions$predicted_rings);\nhead(abalone_predictions)",
"_____no_output_____"
],
[
"# Define a function to calculate RMSE\nrmse <- function(m, o){\n sqrt(mean((m - o)^2))\n}",
"_____no_output_____"
],
[
"# Calucalte RMSE\nabalone_rmse <- rmse(abalone_predictions$rings, abalone_predictions$predicted_rings)\ncat('RMSE for Batch Transform: ', round(abalone_rmse, digits = 2))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecf0fbfb92e97d22238323635f0c32a5fec23e9c | 22,253 | ipynb | Jupyter Notebook | data_codes/Fix and Save tatoeba data.ipynb | dudtjakdl/OpenNMT-Korean-To-English | 32fcdb860906f40f84375ec17a23ae32cb90baa0 | [
"Apache-2.0"
] | 11 | 2020-01-27T02:17:07.000Z | 2021-06-29T08:58:08.000Z | data_codes/Fix and Save tatoeba data.ipynb | dudtjakdl/OpenNMT-Korean-To-English | 32fcdb860906f40f84375ec17a23ae32cb90baa0 | [
"Apache-2.0"
] | null | null | null | data_codes/Fix and Save tatoeba data.ipynb | dudtjakdl/OpenNMT-Korean-To-English | 32fcdb860906f40f84375ec17a23ae32cb90baa0 | [
"Apache-2.0"
] | 4 | 2020-02-10T05:32:22.000Z | 2022-02-04T13:14:11.000Z | 33.870624 | 174 | 0.35775 | [
[
[
"import pandas as pd\nimport random",
"_____no_output_____"
],
[
"data = pd.read_csv(\"C:/Users/Soyoung Cho/Desktop/additional_data/tatoeba-kor-eng/kor.txt\", sep = \"\\t\", names = [\"English\", \"Korean\", \"etc\"])",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"del data['etc']",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"data = data[['Korean','English']]",
"_____no_output_____"
],
[
"Kor_list = list(data['Korean']) #모든 한글 문장이 담긴 리스트\nEng_list = list(data['English']) #모든 영어 문장이 담긴 리스트\n\nprint(Kor_list[:5])\nprint(\"\\n\")\nprint(Eng_list[:5])",
"['가.', '안녕.', '뛰어!', '뛰어.', '누구?']\n\n\n['Go.', 'Hi.', 'Run!', 'Run.', 'Who?']\n"
],
[
"result = list(zip(Kor_list,Eng_list))\n\nrandom.shuffle(result)\nresult \n\nKor_list, Eng_list = zip(*result)\n\nprint(Kor_list[:5])\nprint(\"\\n\")\nprint(Eng_list[:5])",
"('톰 컴퓨터는 바이러스에 감염되었어.', '이렇게나 짜증나다니!', '톰과 싸우는 건 무의미했어.', '특별히 좋아하는 노래같은 건 딱히 없어.', '톰의 아이디어를 썼어.')\n\n\n(\"A virus infected Tom's computer.\", 'How annoying!', 'There was no point in arguing with Tom.', \"I don't really have a favorite song.\", \"I used Tom's idea.\")\n"
],
[
"dict_ = {\"Korean\": [], \"English\" : []}\ndict_[\"Korean\"] = Kor_list\ndict_[\"English\"] = Eng_list",
"_____no_output_____"
],
[
"data = pd.DataFrame(dict_)\ndata",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"#data.to_csv(\"tatoeba_data.csv\", encoding = 'utf-8-sig', index = False, mode = \"w\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf10c78e6a96a58722b375855dfc54fa109125f | 62,879 | ipynb | Jupyter Notebook | notebooks/pollution/pollution.ipynb | CityScape-Datasets/labspt15-cityspire-g-ds | 41f85b5a662d25f6626942def150518b6d0c8d27 | [
"MIT"
] | null | null | null | notebooks/pollution/pollution.ipynb | CityScape-Datasets/labspt15-cityspire-g-ds | 41f85b5a662d25f6626942def150518b6d0c8d27 | [
"MIT"
] | null | null | null | notebooks/pollution/pollution.ipynb | CityScape-Datasets/labspt15-cityspire-g-ds | 41f85b5a662d25f6626942def150518b6d0c8d27 | [
"MIT"
] | 1 | 2021-03-05T03:32:18.000Z | 2021-03-05T03:32:18.000Z | 37.472586 | 162 | 0.292005 | [
[
[
"# EDA",
"_____no_output_____"
],
[
"### Import Data",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport statistics as stat",
"_____no_output_____"
],
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Mounted at /content/drive\n"
],
[
"pollution_df = pd.read_csv(\"/content/pollution_us_2000_2016.csv\")",
"_____no_output_____"
]
],
[
[
"### Explore Data",
"_____no_output_____"
]
],
[
[
"pollution_df.head(10)",
"_____no_output_____"
],
[
"pollution_df.shape",
"_____no_output_____"
],
[
"pollution_df.columns",
"_____no_output_____"
],
[
"pollution_df.describe(include = 'all')",
"_____no_output_____"
],
[
"pollution_df.dtypes",
"_____no_output_____"
],
[
"pollution_df.isna().sum()",
"_____no_output_____"
],
[
"pollution_df.count()",
"_____no_output_____"
]
],
[
[
"### Clean Data",
"_____no_output_____"
]
],
[
[
"pollution_df = pollution_df.drop(['Unnamed: 0','State Code','County Code','Site Num','Address','NO2 Units','O3 Units','SO2 Units','CO Units'],axis=1)\npollution_df.head()",
"_____no_output_____"
],
[
"# For this times series analysis, I am just going to stick with the air quality indicators\npollution = pollution_df[['State', 'County', 'City', 'Date Local', 'NO2 AQI', 'O3 AQI', 'SO2 AQI', 'CO AQI']]",
"_____no_output_____"
],
[
"# Dropping District of Columbia and Country of Mexico\npollution = pollution[pollution['State'] != 'Country of Mexico']\npollution = pollution[pollution['State'] != 'District of Columbia']",
"_____no_output_____"
],
[
"# For the time series analysis, changing Date Local from a string to a date format\npollution['Date Local'] = pd.to_datetime(pollution['Date Local'], format='%Y-%m-%d')",
"_____no_output_____"
],
[
"# Since there are a lot of duplicates in the data, I will group the data by state, county, and date\npollution = pollution.groupby(['State', 'City', 'County', 'Date Local']).agg(np.mean).reset_index()",
"_____no_output_____"
],
[
"summary_pollution = pollution.groupby(level=0)",
"_____no_output_____"
],
[
"summary_pollution.head()",
"_____no_output_____"
],
[
"pollution.to_csv('pollution.csv')",
"_____no_output_____"
],
[
"!cp pollution.csv \"drive/My Drive/\"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf11b16ae008105d89dad1d71c52335c48e197e | 11,354 | ipynb | Jupyter Notebook | Flag_Classification.ipynb | alok13/Algorithms-and-dataStructures | 5d31f50e5ecdf81c82372012ca882b90c6c286db | [
"Apache-2.0"
] | null | null | null | Flag_Classification.ipynb | alok13/Algorithms-and-dataStructures | 5d31f50e5ecdf81c82372012ca882b90c6c286db | [
"Apache-2.0"
] | null | null | null | Flag_Classification.ipynb | alok13/Algorithms-and-dataStructures | 5d31f50e5ecdf81c82372012ca882b90c6c286db | [
"Apache-2.0"
] | null | null | null | 32.255682 | 254 | 0.380483 | [
[
[
"<a href=\"https://colab.research.google.com/github/alok13/Algorithms-and-dataStructures/blob/master/Flag_Classification.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"Import tensorflow ",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function, unicode_literals\n\nimport os\nimport numpy as np\nimport glob\nimport shutil\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"try:\n # Use the %tensorflow_version magic if in colab.\n %tensorflow_version 2.x\nexcept Exception:\n pass\n\nimport tensorflow as tf",
"TensorFlow 2.x selected.\n"
]
],
[
[
"Challenges: Collect more data.\nUnderstand how number of classification makes model difficult.",
"_____no_output_____"
]
],
[
[
"\nos.listdir(\"Flags Data\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf120081c4124c404ac0e5c6ec2bf3eb178a40b | 7,113 | ipynb | Jupyter Notebook | MNIST_Python_TF/MNIST-Convet-SGD.ipynb | nv1620/DL-Frameworks-comparison | ed535cf3cd8783f2f4c59514f1084ccf4d829ce8 | [
"MIT"
] | 1 | 2020-02-10T07:55:00.000Z | 2020-02-10T07:55:00.000Z | MNIST_Python_TF/MNIST-Convet-SGD.ipynb | nv1620/DL-Frameworks-comparison | ed535cf3cd8783f2f4c59514f1084ccf4d829ce8 | [
"MIT"
] | null | null | null | MNIST_Python_TF/MNIST-Convet-SGD.ipynb | nv1620/DL-Frameworks-comparison | ed535cf3cd8783f2f4c59514f1084ccf4d829ce8 | [
"MIT"
] | null | null | null | 32.930556 | 149 | 0.521861 | [
[
[
"from __future__ import absolute_import, division, print_function, unicode_literals\n\nimport tensorflow as tf\n\nfrom tensorflow.keras import datasets, layers, models\nfrom tensorflow.python.keras.callbacks import TensorBoard\nimport matplotlib.pyplot as plt\nimport datetime\n\n# tensorflow version\nprint('tensorflow version:')\nprint(tf.__version__)",
"tensorflow version:\n2.0.0-alpha0\n"
],
[
"log_dir=\"logs\\\\fit\\\\SGDM\"\ntensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir=log_dir, histogram_freq=1)",
"_____no_output_____"
],
[
"(train_images, train_labels), (test_images, test_labels) = datasets.mnist.load_data()\n\ntrain_images = train_images.reshape((60000, 28, 28, 1))\ntest_images = test_images.reshape((10000, 28, 28, 1))\n\ntrain_images = train_images[:10000, :, :, :]\ntest_images = test_images[:3000, :, :, :]\ntrain_labels = train_labels[:10000]\ntest_labels = test_labels[:3000]\n\n# Normalize pixel values to be between 0 and 1\ntrain_images, test_images = train_images / 255.0, test_images / 255.0",
"_____no_output_____"
],
[
"model = models.Sequential()\nmodel.add(layers.Conv2D(3, (3, 3), activation='relu', input_shape=(28, 28, 1)))\nmodel.add(layers.MaxPooling2D((2, 2)))\nmodel.add(layers.Conv2D(64, (3, 3), activation='relu'))\nmodel.add(layers.MaxPooling2D((2, 2)))\nmodel.add(layers.Conv2D(64, (3, 3), activation='relu'))\nmodel.add(layers.Flatten())\nmodel.add(layers.Dense(64, activation='relu'))\nmodel.add(layers.Dense(10, activation='softmax'))",
"_____no_output_____"
],
[
"model.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d (Conv2D) (None, 26, 26, 3) 30 \n_________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 13, 13, 3) 0 \n_________________________________________________________________\nconv2d_1 (Conv2D) (None, 11, 11, 64) 1792 \n_________________________________________________________________\nmax_pooling2d_1 (MaxPooling2 (None, 5, 5, 64) 0 \n_________________________________________________________________\nconv2d_2 (Conv2D) (None, 3, 3, 64) 36928 \n_________________________________________________________________\nflatten (Flatten) (None, 576) 0 \n_________________________________________________________________\ndense (Dense) (None, 64) 36928 \n_________________________________________________________________\ndense_1 (Dense) (None, 10) 650 \n=================================================================\nTotal params: 76,328\nTrainable params: 76,328\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"model.compile(optimizer=tf.keras.optimizers.SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True),\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])\n\nhistory = model.fit(train_images, train_labels, epochs=10, callbacks=[tensorboard_callback])",
"Epoch 1/10\n10000/10000 [==============================] - 3s 347us/sample - loss: 0.6262 - accuracy: 0.8041\nEpoch 2/10\n10000/10000 [==============================] - 2s 168us/sample - loss: 0.1558 - accuracy: 0.9538\nEpoch 3/10\n10000/10000 [==============================] - 2s 183us/sample - loss: 0.1058 - accuracy: 0.9685\nEpoch 4/10\n10000/10000 [==============================] - 2s 192us/sample - loss: 0.0770 - accuracy: 0.9767\nEpoch 5/10\n10000/10000 [==============================] - 2s 192us/sample - loss: 0.0629 - accuracy: 0.9814\nEpoch 6/10\n10000/10000 [==============================] - 2s 176us/sample - loss: 0.0427 - accuracy: 0.9862\nEpoch 7/10\n10000/10000 [==============================] - 2s 204us/sample - loss: 0.0354 - accuracy: 0.9882\nEpoch 8/10\n10000/10000 [==============================] - ETA: 0s - loss: 0.0281 - accuracy: 0.99 - 2s 200us/sample - loss: 0.0278 - accuracy: 0.9914\nEpoch 9/10\n10000/10000 [==============================] - 2s 216us/sample - loss: 0.0207 - accuracy: 0.9933\nEpoch 10/10\n10000/10000 [==============================] - 2s 211us/sample - loss: 0.0257 - accuracy: 0.9908\n"
],
[
"test_loss, test_acc = model.evaluate(test_images, test_labels)",
"3000/3000 [==============================] - 0s 136us/sample - loss: 0.1084 - accuracy: 0.9703\n"
],
[
"print(test_acc)",
"0.97033334\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf144d942e7c4945709add11771a98b35324081 | 92,857 | ipynb | Jupyter Notebook | docs/examples/driver_examples/Qcodes example with HP8753D.ipynb | aaroncslau/Qcodes | ba1920198614e0923fbc046efcec2effb36db8f2 | [
"MIT"
] | 2 | 2019-02-14T00:07:06.000Z | 2021-03-30T03:38:06.000Z | docs/examples/driver_examples/Qcodes example with HP8753D.ipynb | qdev-dk/Qcodes | f587f0c4dd74271b3b9156167fbff3dceb2d185d | [
"MIT"
] | 22 | 2017-02-08T08:37:23.000Z | 2017-11-24T14:18:20.000Z | docs/examples/driver_examples/Qcodes example with HP8753D.ipynb | aaroncslau/Qcodes | ba1920198614e0923fbc046efcec2effb36db8f2 | [
"MIT"
] | 6 | 2017-03-31T21:01:08.000Z | 2019-08-20T09:25:22.000Z | 88.604008 | 29,502 | 0.744187 | [
[
[
"# Qcodes example with HP8753D\n\nThis is the example notebook illustrating how to use the QCoDeS driver\nfor the HP 8753D Network Analyzer.\n\nThroughout the notebook, we assume that a Mini-Circuits SLP-550+ Low Pass\nfilter is connected as the DUT.",
"_____no_output_____"
]
],
[
[
"# allow in-notebook matplotlib plots\n%matplotlib notebook\n\n# import QCoDeS\n\nimport qcodes as qc\nfrom qcodes.instrument_drivers.HP.HP8753D import HP8753D\n\n# we'll need this later\nfrom functools import partial\n\n# import logging\n# logging.basicConfig(level=logging.DEBUG)",
"_____no_output_____"
],
[
"# Instantiate the instrument\nvna = HP8753D('vna', 'GPIB0::6::INSTR')",
"Connected to: HEWLETT PACKARD 8753D (serial:0, firmware:6.14) in 0.23s\n"
],
[
"# for the sake of this tutorial, we reset the instrument\nvna.reset()\n\n# The following functions gives a nice self-explanatory\n# overview of the available instrument settings\n\nvna.print_readable_snapshot(update=True)",
"vna:\n\tparameter value\n--------------------------------------------------------------------------------\nIDN :\t{'vendor': 'HEWLETT PACKARD', 'model': '8753D', 'serial'...\naveraging :\tOFF \ndisplay_format :\tLog mag \ndisplay_reference :\t0 (dim. less)\ndisplay_scale :\t10 (dim. less)\nnumber_of_averages :\t16 \noutput_power :\t0 (dBm)\ns_parameter :\tS11 \nstart_freq :\t30000 (Hz)\nstop_freq :\t6e+09 (Hz)\nsweep_time :\t0.175 (s)\ntimeout :\t10 (s)\ntrace :\tNot available \ntrace_points :\t201 \n"
]
],
[
[
"## Single trace measurement",
"_____no_output_____"
]
],
[
[
"# Let's get a single trace of S21 with 10 averages\n# with a frequency range from 100 kHz to 1 GHz\n# on a linear scale\nvna.display_format('Lin mag')\nvna.s_parameter('S21')\nvna.start_freq(100e3)\nvna.stop_freq(1e9)\n\n# and let's adjust the y-scale to fit\nvna.display_scale(0.12)\nvna.display_reference(-0.1)\n\n# and finally enable averaging\nvna.averaging('ON')\nvna.number_of_averages(10)",
"_____no_output_____"
],
[
"# Now aquire a trace\nvna.trace.prepare_trace() # this must be called prior to a Measurement or Loop\ntracedata = qc.Measure(vna.trace).run() # this returns a dataset\nplot = qc.MatPlot(tracedata.arrays['vna_trace'])",
"DataSet:\n location = 'data/2017-11-02/#011_{name}_14-33-04'\n <Type> | <array_id> | <array.name> | <array.shape>\n Measured | vna_trace | trace | (201,)\nacquired at 2017-11-02 14:33:04\n"
]
],
[
[
"## Acquiring traces while sweeping",
"_____no_output_____"
]
],
[
[
"# Now we'll vary the output power and acquire a trace for each power\n# to examine how more power reduces the measurement noise\n\n# We have to ensure that the VNA finishes averaging before moving\n# to the next power\n\nn_avgs = 10\nvna.number_of_averages(n_avgs)\n# make a Task that runs N sweeps and waits for them to finish\nrun_sweeper = qc.Task(partial(vna.run_N_times, n_avgs))\n\n\n# prepare the trace\nvna.trace.prepare_trace()\n\n# set up a Loop\nloop = qc.Loop(vna.output_power.sweep(-84, -40, num=20)).each(run_sweeper, vna.trace)\ndata = loop.get_data_set(name='HP8753D_tutorial')\n\n# set up plotting\nplot = qc.QtPlot() # create a plot\nplot.add(data.vna_trace) # add a graph to the plot\n# run the loop\n_ = loop.with_bg_task(plot.update, plot.save).run() ",
"Started at 2017-11-02 14:33:27\nDataSet:\n location = 'data/2017-11-02/#012_HP8753D_tutorial_14-33-15'\n <Type> | <array_id> | <array.name> | <array.shape>\n Setpoint | vna_output_power_set | output_power | (20,)\n Measured | vna_trace | trace | (20, 201)\nFinished at 2017-11-02 14:34:40\n"
],
[
"# The resulting plot is indeed pretty\nplot",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecf1493add0d3d9bc00cc4a0c820086ae5ca516b | 12,619 | ipynb | Jupyter Notebook | StackExchange/MyAttempt.ipynb | RaulRPrado/LearningDataScience | 0a7d6cfffadd7cdd657be0609b7f088a19f9deb5 | [
"MIT"
] | 1 | 2022-01-05T13:49:27.000Z | 2022-01-05T13:49:27.000Z | StackExchange/MyAttempt.ipynb | RaulRPrado/LearningDataScience | 0a7d6cfffadd7cdd657be0609b7f088a19f9deb5 | [
"MIT"
] | null | null | null | StackExchange/MyAttempt.ipynb | RaulRPrado/LearningDataScience | 0a7d6cfffadd7cdd657be0609b7f088a19f9deb5 | [
"MIT"
] | null | null | null | 26.344468 | 340 | 0.508202 | [
[
[
"# Introduction\n\nThis is a problem from HackerRank.\n\nStack Exchange is an information powerhouse, built on the power of crowdsourcing. It has 105 different topics and each topic has a library of questions which have been asked and answered by knowledgeable members of the StackExchange community. The topics are as diverse as travel, cooking, programming, engineering and photography.\n\nWe have hand-picked ten different topics (such as Electronics, Mathematics, Photography etc.) from Stack Exchange, and we provide you with a set of questions from these topics.\n\nGiven a question and an excerpt, your task is to identify which among the 10 topics it belongs to.\n\nGetting started with text classification\n\nFor those getting started with this fascinating domain of text classification, here's a wonderful Youtube video of Professor Dan Jurafsky from Stanford, explaining the Naive Bayes classification algorithm, which you could consider using as a starting point\n\nInput Format\nThe first line will be an integer N. N lines follow each line being a valid JSON object. The following fields of raw data are given in json\n\nquestion (string) : The text in the title of the question.\nexcerpt (string) : Excerpt of the question body.\ntopic (string) : The topic under which the question was posted.\nThe input for the program has all the fields but topic which you have to predict as the answer.\n\nConstraints\n1 <= N <= 22000\ntopic is of ascii format\nquestion is of UTF-8 format\nexcerpt is of UTF-8 format\n\nOutput Format\nFor each question that is given as a JSON object, output the topic of the question as predicted by your model separated by newlines.\n\nThe training file is available here. It is also present in the current directory in which your code is executed.\n\nSample Input\n12345\njson_object\njson_object\njson_object\n.\n.\n.\njson_object\nSample Output\n\nelectronics\nsecurity\nphoto\n.\n.\n.\nmathematica\nSample testcases can be downloaded here for offline training. When you submit your solution to us, you can assume that the training file can be accessed by reading \"training.json\" which will be placed in the same folder as the one in which your program is being executed.\n\nScoring\n\nWhile the contest is going on, the score shown to you will be on the basis of the Sample Test file. The final score will be based on the Hidden Testcase only and there will be no weightage for your score on the Sample Test.\n\nScore = MaxScore for the test case * (C/T)\nWhere C = Number of topics identified correctly and\nT = total number of test JSONs in the input file.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport json",
"_____no_output_____"
]
],
[
[
"# Data Preparation",
"_____no_output_____"
]
],
[
[
"data = {'topic': list(), 'question': list()}\nwith open('training.json', 'r') as file:\n \n n_data = None\n for line in file:\n\n if n_data is None:\n n_data = int(line)\n else:\n d = json.loads(line)\n data['topic'].append(d['topic'])\n data['question'].append(d['question'])",
"_____no_output_____"
],
[
"labels, counts = np.unique(data['topic'], return_counts=True)",
"_____no_output_____"
],
[
"print(dict(zip(labels, counts)))",
"{'android': 2239, 'apple': 2064, 'electronics': 2079, 'gis': 2383, 'mathematica': 1369, 'photo': 1945, 'scifi': 2333, 'security': 1899, 'unix': 1965, 'wordpress': 1943}\n"
],
[
"from sklearn.preprocessing import OrdinalEncoder",
"_____no_output_____"
],
[
"topic_enc = OrdinalEncoder().fit(np.array(data['topic']).reshape(-1, 1))\ny = topic_enc.transform(np.array(data['topic']).reshape(-1, 1)).ravel()",
"_____no_output_____"
],
[
"X = np.array(data['question'])",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import TfidfVectorizer",
"_____no_output_____"
],
[
"vectorizer = TfidfVectorizer(stop_words='english', max_features=3500, decode_error='ignore')",
"_____no_output_____"
],
[
"X_train_vec = vectorizer.fit_transform(X_train)\nX_test_vec = vectorizer.transform(X_test)",
"_____no_output_____"
]
],
[
[
"# Modeling",
"_____no_output_____"
],
[
"## MLP",
"_____no_output_____"
]
],
[
[
"from sklearn.neural_network import MLPClassifier",
"_____no_output_____"
],
[
"clf = MLPClassifier(\n hidden_layer_sizes=(200, 20),\n batch_size=20,\n learning_rate='constant',\n learning_rate_init=0.001,\n early_stopping=True\n).fit(X_train_vec, y_train)",
"_____no_output_____"
],
[
"y_pred = clf.predict(X_test_vec)",
"_____no_output_____"
],
[
"from sklearn.metrics import classification_report",
"_____no_output_____"
],
[
"print(classification_report(y_test, y_pred))",
" precision recall f1-score support\n\n 0.0 0.72 0.82 0.77 445\n 1.0 0.78 0.63 0.70 426\n 2.0 0.63 0.72 0.67 399\n 3.0 0.85 0.80 0.83 474\n 4.0 0.72 0.56 0.63 278\n 5.0 0.78 0.81 0.80 380\n 6.0 0.69 0.87 0.77 500\n 7.0 0.71 0.65 0.68 384\n 8.0 0.56 0.54 0.55 357\n 9.0 0.85 0.77 0.81 401\n\n accuracy 0.73 4044\n macro avg 0.73 0.72 0.72 4044\nweighted avg 0.73 0.73 0.73 4044\n\n"
]
],
[
[
"## Naive Bayes",
"_____no_output_____"
]
],
[
[
"from sklearn.naive_bayes import MultinomialNB",
"_____no_output_____"
],
[
"clf_nb = MultinomialNB().fit(X_train_vec, y_train)",
"_____no_output_____"
],
[
"y_pred = clf_nb.predict(X_test_vec)",
"_____no_output_____"
],
[
"print(classification_report(y_test, y_pred))",
" precision recall f1-score support\n\n 0.0 0.74 0.84 0.79 445\n 1.0 0.78 0.72 0.75 426\n 2.0 0.81 0.80 0.81 399\n 3.0 0.74 0.91 0.81 474\n 4.0 0.91 0.54 0.68 278\n 5.0 0.82 0.89 0.86 380\n 6.0 0.94 0.85 0.89 500\n 7.0 0.80 0.74 0.77 384\n 8.0 0.65 0.66 0.66 357\n 9.0 0.83 0.86 0.85 401\n\n accuracy 0.80 4044\n macro avg 0.80 0.78 0.79 4044\nweighted avg 0.80 0.80 0.79 4044\n\n"
],
[
"y_pred_str = topic_enc.inverse_transform(y_pred.reshape(-1, 1))",
"_____no_output_____"
],
[
"y_pred_str",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf14a099159690d055b0d53f0e61fbad304b19d | 124,668 | ipynb | Jupyter Notebook | reu2021/specific_bots/RonBot.ipynb | levon003/wiki-ores-feedback | 29e7f1a41b16a7c57448d5bbc5801653debbc115 | [
"MIT"
] | 2 | 2022-03-27T19:24:30.000Z | 2022-03-29T16:15:31.000Z | reu2021/specific_bots/RonBot.ipynb | levon003/wiki-ores-feedback | 29e7f1a41b16a7c57448d5bbc5801653debbc115 | [
"MIT"
] | 1 | 2021-04-23T21:03:45.000Z | 2021-04-23T21:03:45.000Z | reu2021/specific_bots/RonBot.ipynb | levon003/wiki-ores-feedback | 29e7f1a41b16a7c57448d5bbc5801653debbc115 | [
"MIT"
] | null | null | null | 69.802912 | 23,788 | 0.671239 | [
[
[
"# RonBot\n\nhttps://en.wikipedia.org/wiki/User:RonBot\n\nBot is no longer active, operator has since passed away\n\ntried to look for discussion of malfunctionmost recent talk on user's page is from 2020 and can't find anything there\n\ndiscussion about brokenimage task possibly being broken in March 2019\nhttps://en.wikipedia.org/wiki/Wikipedia:Bots/Noticeboard/Archive_13#User:RonBot_trouble_possibly_in_need_of_intervention\n\ndiscussion about footballer category task in July 2018 https://en.wikipedia.org/wiki/Wikipedia:Bot_requests/Archive_76#Association_footballers_not_categorized_by_position\n",
"_____no_output_____"
]
],
[
[
"%reload_ext autoreload\n%autoreload 2\n%matplotlib inline",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"from datetime import datetime",
"_____no_output_____"
],
[
"df = pd.read_csv('../revs_scored_jan.tsv', sep='\\t', header=0)",
"_____no_output_____"
],
[
"days = df.rev_timestamp.map(lambda ts: datetime.utcfromtimestamp(ts).day)\ndf['day'] = days",
"_____no_output_____"
],
[
"sdf_ronbot = df[df.day<22]\nsdf_ronbot = sdf_ronbot[sdf_ronbot.user_text == \"RonBot\"]\n\nsdf_reverted = sdf_ronbot[sdf_ronbot.is_reverted] \nsdf_reverted = sdf_reverted[sdf_reverted.seconds_to_revert.astype('str').astype('int')<86400]",
"_____no_output_____"
],
[
"sdf1 = sdf_reverted\nsdf1.revert_id = sdf1.revert_id.astype('int')\n\nsdf2 = df[df.user_is_bot == False]\nsdf2 = sdf2[sdf2.is_revert]",
"_____no_output_____"
],
[
"reverts_by_human = pd.merge(sdf1, sdf2, \n how='inner', \n left_on='revert_id', \n right_on='rev_id', \n suffixes=('', '_reverter')\n)",
"_____no_output_____"
],
[
"reverts_by_human0 = reverts_by_human[reverts_by_human.page_namespace == 0]",
"_____no_output_____"
],
[
"# summary of variables\n# sdf_ronbot = all edits by ronbot w/in time frame\n# sdf_reverted = all reverted edits by ronbot w/in time frame\n# reverts_by_human = all edits of Ronbot that were reverted by a human\n# reverts_by_human0 = all edits by Ronbot in namespace 0 reverted by a human",
"_____no_output_____"
]
],
[
[
"## sample of edits reverted by humans in namespace 0",
"_____no_output_____"
]
],
[
[
"len(reverts_by_human0)",
"_____no_output_____"
],
[
"reverts_by_human0.sample(n=20, random_state=1).reset_index()",
"_____no_output_____"
]
],
[
[
"0. 877208766 on Adil Mezgour page, for task 7. ronbot adds category (footballers not categorized by position). user inter%anthro undoes it, says position category already present. indeed, article does list category \"association football forwards\" already\n1. 877208664 on Adham El Idrissi page, for task 7. ronbot adds category. user inter&anthro deletes it, says position category already present. indeed, category \"association football forwards\" is already present.\n2. 877208212 on Adamo Coulibaly page, for task 7. ronbot adds category (footballers not categorized by position). user Inter%anthro undoes it, says position category already present. indeed, article does list category \"association football forwards\" already\n3. 877209086 on dif page, for task 7. ronbot adds category (footballers not categorized by position). user Inter%anthro undoes it, says position category already present. indeed, article does list category \"association football midfielders\" already\n4. 877202703 same as above, user GiantSnowman reverts, in midfielders category\n5. 877208346 same as above, user inter&anthro reverts, in defenders category, edits are 12 hours apart on Jan 7\n6. 877277070 same as above, user Jmorrison230582 reverts, in category forwards, edits are 2 hours apart on Jan 7\n7. 876292953 on Eastern world page, RonBot adds broken image template. another user vsmith reverts to edit before alleged vandalism that broke the image ***not a malfunction***. edits about 30 mins apart, on Jan 1.\n8. 877208884 same footballer edits, reverting user is Inter&anthro, forwards category already present, edits about 12 hours apart on Jan 7\n9. 877279563 same footballer edits, reverting user is walter gorlitz, in category defenders, about 10 mins apart 7 Jan\n10. 877208746 same footballer edits, reverting user is Inter&anthro, in category defenders, edits about 11 hours apart on Jan 7\n11. 877277582 same footballer edits, reverting user Mattythewhite, in category midfielders, edits about 7 hours apart on Jan 7\n12. 876293084 on Johnny Sins page, RonBot adds a BrokenImage template. user Gotitbro reverts, says no broken image. indeed the image seems to be correct.\n13. 879469682 on Daksha page, RonBot adds BrokenImage template (task 12). user KylieTastic reverts to a different edit to fix the name of the image. ***not a malfunction***\n14. 877208422 same football edits, reverting user is GiantSnowman, in category midfielders, edits are about 3 hours apart Jan 7\n15. 877208840 same football edits, reverting user is inter&anthro, in category defenders, 12 hours apart on Jan 7\n16. 877209113 same football edits, reverting user is Inter&anthro, in category defenders, edits about 1 hour apart on Jan 7\n17. 877208707 same football edits, reverting user Inter&anthro, in category forwards, edits about 11 hours apart on Jan 7\n18. 877201219 same football edits, reverting user Mattythewhite, in category forwards, edits about 20 hours apart. first edit is 30 mins before midnight on 6 Jan, second 18:00 on Jan 7\n19. 877207378 same football edits, reverting user Daemonickangaroo2018, in category midfielders, edits about 13 hours apart on Jan 7",
"_____no_output_____"
]
],
[
[
"# edits reverted by human\nreverts_by_human.groupby(\"day\", as_index=False).count()[[\"day\", \"rev_id\"]]",
"_____no_output_____"
],
[
"# these numbers are different from Sokona's below because I did w/in 24 hours, hers are reverted in general\nsdf_reverted.groupby(\"day\", as_index=False).count()[[\"day\", \"rev_id\"]]",
"_____no_output_____"
],
[
"# total edits\nsdf_ronbot.groupby(\"day\", as_index=False).count()[[\"day\",\"rev_id\"]]",
"_____no_output_____"
],
[
"#made a dataframe of all of Ronbots reverted revisions and then of all the humans that are reverting\ndf_ronbot = sdf_ronbot\nreverted_ronbot = df_ronbot[df_ronbot.is_reverted == True]\n\ndf_bots = df[df.day < 22]\ndf_humans = df_bots[df_bots.user_is_bot == False]\ndf_human_reverters = df_humans[df_humans.is_revert == True]\n#ronbot_self_reverts = reverted_ronbot[reverted_ronbot.is_self_revert == True]",
"_____no_output_____"
],
[
"#then made a dataframe of all the revisions reverted within 24 hours\n\ndf_reverted = df[df.is_reverted]\ndf_reverted = df_reverted[df_reverted.seconds_to_revert.astype('str').astype('int')<86400]\ndf_reverted.revert_id = df_reverted.revert_id.astype('int')",
"_____no_output_____"
],
[
"#merged to get all the human reverters and who it was reverting (can be bot or human)\n\ndf_human_active_reverts = pd.merge(df_reverted, df_human_reverters, \n how='inner', \n left_on='revert_id', \n right_on='rev_id', \n suffixes=('', '_reverter')\n)",
"_____no_output_____"
],
[
"#deleted columns I didn't need and then subsetted the dataframe to any revisions of RonBot reverted by a human\ndf_human_active_reverts = df_human_active_reverts[['rev_id','user_text', 'user_is_bot','revert_id','user_text_reverter', 'is_self_revert_reverter', 'page_namespace','day']]\nreverted_ronbot_by_h = df_human_active_reverts[df_human_active_reverts.user_text == \"RonBot\"]\nreverted_ronbot_by_h",
"_____no_output_____"
],
[
"#graph of all of ronbots revisions by day, can clearly see the most was on January 7th\ncounts, bin_edges = np.histogram(df_ronbot.day, bins=len(set(df_ronbot.day)))\nplt.plot(bin_edges[:-1], counts)\nplt.ylabel(\"Revision count\")\nplt.xlabel(\"Date\")\nplt.title(\"All of Ronbot's Revisions by Day\")\nplt.xticks(ticks=[1, 7, 14, 21], labels=['Jan 1', 'Jan 7', 'Jan 14', 'Jan 21'])\nplt.show()",
"_____no_output_____"
],
[
"\"\"\"graph of all of ronbots reverted revisions by day, can clearly see the most was on January 7th, out of ~2000 total edits, \n~1750 were reverted on that day (~87%)\"\"\"\ncounts, bin_edges = np.histogram(reverted_ronbot.day, bins=len(set(reverted_ronbot.day)))\nplt.plot(bin_edges[:-1], counts)\nplt.ylabel(\"Reverted Revision count\")\nplt.xlabel(\"Date\")\nplt.title(\"All of Ronbot's Reverted Revisions by Day\")\nplt.xticks(ticks=[1, 7, 14, 21], labels=['Jan 1', 'Jan 7', 'Jan 14', 'Jan 21'])\nplt.show()",
"_____no_output_____"
],
[
"\"\"\"graph of all of ronbots reverted revisions by a human by day, can clearly see the most was on January 7th, out of ~2000 total edits,\n~1750 were reverted on that day (~87%), and out of all reverted revisions, 400 (~20%) were reverteed by humans\"\"\"\ncounts, bin_edges = np.histogram(reverted_ronbot_by_h.day, bins=len(set(reverted_ronbot_by_h.day)))\nplt.plot(bin_edges[:-1], counts)\nplt.ylabel(\"Revision count\")\nplt.xlabel(\"Date\")\nplt.title(\"All of Ronbot's Reverted Revisions (by a Human) by Day\")\nplt.xticks(ticks=[1, 7, 14, 21], labels=['Jan 1', 'Jan 7', 'Jan 14', 'Jan 21'])\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf16479f17fbd4edf993006f28d7d6a89815c53 | 41,064 | ipynb | Jupyter Notebook | Fake_news_LSTM_Example.ipynb | dd0me/COVID-19 | 6f20e47bc05de3c77cc195fbd05c9337471a192b | [
"CC-BY-4.0"
] | null | null | null | Fake_news_LSTM_Example.ipynb | dd0me/COVID-19 | 6f20e47bc05de3c77cc195fbd05c9337471a192b | [
"CC-BY-4.0"
] | null | null | null | Fake_news_LSTM_Example.ipynb | dd0me/COVID-19 | 6f20e47bc05de3c77cc195fbd05c9337471a192b | [
"CC-BY-4.0"
] | null | null | null | 42.246914 | 632 | 0.536285 | [
[
[
"<a href=\"https://colab.research.google.com/github/dd0me/COVID-19/blob/master/Fake_news_LSTM_Example.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"# Application: fake news and deceptive language detection\n\nIn this notebook, we will look at how we can use hybrid embeddings in the context of NLP tasks. In particular, we will see how to use and adapt deep learning architectures to take into account hybrid knowledge sources to classify documents. ",
"_____no_output_____"
],
[
"## Basic document classification using deep learning\nFirst, we will introduce a basic pipeline for training a deep learning model to perform text classification.\n\n### Dataset: deceptive language (fake hotel reviews)\nAs a first dataset, we will use the [deceptive opnion spam](http://myleott.com/op-spam.html) dataset (See the exercises below for a couple of more challenging datasets on fake news detection).\n\nThis corpus contains:\n * 400 truthful positive reviews from TripAdvisor\n * 400 deceptive positive reviews from Mechanical Turk\n * 400 truthful negative reviews from Expedia, Hotels.com, Orbitz, Priceline, TripAdvisor and Yelp\n * 400 deceptive negative reviews from Mechanical Turk\n \nThe dataset is described in more detail in the following papers:\n \n [M. Ott, Y. Choi, C. Cardie, and J.T. Hancock. 2011. Finding Deceptive Opinion Spam by Any Stretch of the Imagination. In Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies.](http://arxiv.org/abs/1107.4557)\n \n [M. Ott, C. Cardie, and J.T. Hancock. 2013. Negative Deceptive Opinion Spam. In Proceedings of the 2013 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies.](http://www.aclweb.org/anthology/N13-1053)\n \n For convenience, we have included the dataset as part of our GitHub tutorial repository.",
"_____no_output_____"
]
],
[
[
"%ls",
"_____no_output_____"
],
[
"%cd /content\n!git clone https://github.com/hybridnlp/tutorial\n!head -n2 /content/tutorial/datasamples/deceptive-opinion.csv",
"_____no_output_____"
]
],
[
[
"The last two lines show that the dataset is distributed as a comma-separated-value file with various fields. For our purposes, we are only interested in fields:\n * `deceptive`: this can be either *truthful* or *deceptive*\n * `text`: the plain text of the review\n \nThe other fields: `hotel` (name), `polarity` (positive or negative) and `source` (where the review comes from) are not relevant for us in this notebook.\n\nLet's first load the dataset in a format that is easier to feed into a text classification model. What we need is an object with fields:\n * `texts`: an array of texts\n * `categories`: an array of textual tags (e.g. *truthful* or *deceptive*) \n * `tags`: an array of integer tags (the categories)\n * `id2tag`: a map from the integer identifier to the textual identifier for the tag\n \nThe following cell produces such an object:",
"_____no_output_____"
]
],
[
[
"import pandas as pd # for handling tables a DataFrames\nimport tutorial.scripts.classification as clsion # library for text classification",
"_____no_output_____"
],
[
"hotel_df = pd.read_csv('/content/tutorial/datasamples/deceptive-opinion.csv',\n names=[\"deceptive\", \"hotel\", \"polarity\", \"source\", \"text\"])\nhotel_df = hotel_df[1:].reset_index() # first row is the header, so remove\nhotel_wnscd_df = pd.read_csv('/content/tutorial/datasamples/deceptive-opinion.tlgs_wnscd',\n names=['text_tlgs_wnscd'])\nhotel_df = pd.concat([hotel_df, hotel_wnscd_df], axis=1)\nraw_hotel_ds = clsion.read_classification_corpus(hotel_df, text_fields=['text'], tag_field='deceptive')\nraw_hotel_wnscd_ds = clsion.read_classification_corpus(hotel_df, text_fields=['text_tlgs_wnscd'], tag_field='deceptive')",
"_____no_output_____"
]
],
[
[
"The previous cell has actually loaded two versions of the dataset: \n * `raw_hotel_ds` contains the actual texts as originally published\n * `raw_hotel_wnscd_ds` provides the WordNet disambiguated `tlgs` tokenization (see notebooke 03 on Vecsigrafo for more details about this format). This is needed because we don't have a python method to automatically disambiguate a text using WordNet, so we provide this disambiguated version as part of the GitHub repo for this tutorial.",
"_____no_output_____"
]
],
[
[
"hotel_df[:5]",
"_____no_output_____"
]
],
[
[
"We can print a couple of examples from both datasets. ",
"_____no_output_____"
]
],
[
[
"clsion.sanity_check(raw_hotel_ds)",
"_____no_output_____"
],
[
"clsion.sanity_check(raw_hotel_wnscd_ds)",
"_____no_output_____"
]
],
[
[
"Cleaning the raw text often produces better results; we can do this as follows:",
"_____no_output_____"
]
],
[
[
"cl_hotel_ds = clsion.clean_ds_texts(raw_hotel_ds)\nclsion.sanity_check(cl_hotel_ds)",
"_____no_output_____"
]
],
[
[
"### Tokenize and index the dataset\n\nAs we said above, the raw datasets consist of `texts`, `categories` and `tags`. There are different ways to process the texts before passing it to a deep learning architecture, but typically they involve:\n * **tokenization**: how to split each document into basic forms which can be represented as vectors. In this notebook we will use tokenizations which result in words and synsets, but there are also architectures that accept character-level or n-grams of characters.\n * **indexing** of the text: in this step, the tokenized text is compared to a **vocabulary** (or, if no vocabulary is provided, it can be used to create a vocabulary), a list of words, so that you can assign a unique integer identifier to each token. You need this so that tokens will then be represented as embedding or vectors in a matrix. So having an identifier will enable you to know which row in the matrix corresponds to which token in the vocabulary.\n \nThe `clsion` library, included in the tutorial GitHub repo, already provides various indexing methods for text classification datasets. In the next cell we apply *simple indexing*, which uses white-space tokenization and creates a vocabulary based on the input dataset.",
"_____no_output_____"
]
],
[
[
"csim_hotel_ds = clsion.simple_index_ds(cl_hotel_ds)",
"_____no_output_____"
]
],
[
[
"Since the vocabulary was created based on the dataset, all tokens in the dataset are also in the vocabulary. In the next sections, we will see examples where embeddings are provided during indexing. \n\nThe following cell prints a couple of characteristics of the indexed dataset.",
"_____no_output_____"
]
],
[
[
"print(\n 'vocab size:', len(csim_hotel_ds['vocab_embedding']['w2i']),\n 'dim:', csim_hotel_ds['vocab_embedding']['dim'],\n 'vectors:', csim_hotel_ds['vocab_embedding']['vecs'])",
"_____no_output_____"
]
],
[
[
"As we can see, the vocabulary is quite small (about 11K words). By default, it specifies that the vocabulary embeddings should be of dimention 150, but no vectors are specified. This means the model can assign random embeddings to the 11K words.\n\n### Define the experiment to run\nThe `clsion` allows us to specify experiments to run: given an indexed dataset, we can execute a text classification experiment by specifying various hyper-parameters as follows:\n\n\n ",
"_____no_output_____"
]
],
[
[
"experiment1 = {\n 'hotel_csim': {\n 'indexed_dataset': csim_hotel_ds,\n 'executor': clsion.execute_experiment,\n 'hparams': clsion.merge_hparams([\n clsion.common_hparams, clsion.biLSTM_hparams, \n clsion.calc_hparams(csim_hotel_ds), \n { \n 'epochs': 20\n }\n ])\n }\n}",
"_____no_output_____"
]
],
[
[
"Under the hood, the library creates a Bidirectional LSTM model as requested (the library also can create other model architectures such as convolutional NNs).\n\nSince our dataset is fairly small, we don't need a very deep model. A fairly simple bidirectional LSTM should be sufficient. The generated model will consist of the following layers:\n * The **input layer**: is a tensor of shape $(l, )$, where $l$ is the number of tokens for each document. The empty second parameter will let us pass the model different number of input documents, as long as they all have the same number of tokens.\n * The **embedding layer** converts the each input document (a sequence of word ids) into a sequence of embeddings. Since we are not yet using pre-computed embeddings, these will be generated at random and trained with the rest of parameters in the model.\n * The **lstm layer**s: one or more bidirectional LSTMs. Explaining these in detail is out of the scope of this tutorial. Suffice it to say, each layer goes through each embedding in the sequence and produces a new embedding taking into account previous and posterior embeddings. The final layer only produces a single embedding, which represents the full document.\n * The **dense layer**: is a fully connected neural network that maps the output embedding of the final layer to a vector of 2 dimensions which can be compared to the manual labelled tag. \n \nFinally, we can run our experiment using the `n_cross_val` method. Depending on whether you have an environment with a GPU this can be a bit slow, so we only train a model once. (In practice, model resuls may vary due to random initializations, so it's usually a good idea to run the same model several times to get an average evaluation metric and an idea of how stable the model is.)",
"_____no_output_____"
]
],
[
[
"ex1_df, ex1_best_run = clsion.n_cross_val(experiment1, n=1)",
"_____no_output_____"
]
],
[
[
"The first element of the result is a DataFrame containing test results and a record of the used parameters.",
"_____no_output_____"
]
],
[
[
"ex1_df",
"_____no_output_____"
]
],
[
[
"### Discussion\n\nBidirectional LSTMs are really good at learning patters in text. However, this way of training a model will tend to overfit the training dataset. Since our dataset is fairly small and narrow: it only contains texts about hotel reviews, we should not expect this model to be able to detect fake reviews about other products or services. Similarly, we should not expect this model to be applicable to detecting other types of deceptive texts such as fake news.\n\nThe reason why such a model is very tied to the training dataset is that even the vocabulary is derived from the dataset: it will be biased towards words (and senses of those words) related to hotel reviews. Vocabulary about other products, services and topics cannot be learned from the input dataset.\n\nFurthermore, since no pre-trained embeddings were used, the model had to learn the embedding weights from scratch based on the signal provided by the 'deceptive' tags. It did not have an opportunity to learn more generic relations between words from a wider corpus.\n\nFor these reasons it is a good idea to use pre-trained embeddings as we show in the following sections.",
"_____no_output_____"
],
[
"## Using HolE embeddings\n\nIn this section we use embeddings learned using `HolE` and trained on WordNet 3.0. As we have seen in previous notebooks, such embeddings capture the relations specified in the WordNet knowledge graph. As such, synset embeddings tend to encode useful knowledge. However, lemma embeddings tend to be of poorer quality.",
"_____no_output_____"
],
[
"### Download the embeddings\n\nExecute the following cell to download and unpack the embeddings. If you recently executed previous notebooks as part of this tutorial, you may still have these in your environment.",
"_____no_output_____"
]
],
[
[
"!mkdir /content/vec/\n%cd /content/vec/\n!wget https://zenodo.org/record/1446214/files/wn-en-3.0-HolE-500e-150d.tar.gz\n!tar -xzf wn-en-3.0-HolE-500e-150d.tar.gz",
"_____no_output_____"
],
[
"%ls /content/vec/",
"_____no_output_____"
]
],
[
[
"### Load the embeddings and convert to the format expected by `clsion`\n\nThe provided embeddings are in `swivel`'s binary + vocab format. However, the `clsion` library expects a different python datastructure. Furtheremore, it will be easier to match the lemmas in the dataset to plain text rather than the `lem_<lemma_word>` format used to encode the HolE vocabulary, hence we need to do some cleaning of the vocabulary. This occurs in the following cells:",
"_____no_output_____"
]
],
[
[
"import tutorial.scripts.swivel.vecs as vecs\nvocab_file = '/content/vec/wn-en-3.1-HolE-500e.vocab.txt'\nholE_voc_file = '/content/vec/wn-en-3.1-HolE-500e.clean.vocab.txt'\nwith open(holE_voc_file, 'w', encoding='utf_8') as wf:\n with open(vocab_file, 'r', encoding='utf_8') as f:\n for word in f.readlines():\n word = word.strip()\n if not word:\n continue\n if word.startswith('lem_'):\n word = word.replace('lem_', '').replace('_', ' ')\n print(word, file=wf)\nvecbin = '/content/vec/wn-en-3.1-HolE-500e.tsv.bin'\nwnHolE = vecs.Vecs(holE_voc_file, vecbin)",
"_____no_output_____"
],
[
"import array\nimport tutorial.scripts.swivel.vecs as vecs\n\ndef load_swivel_bin_vocab_embeddings(bin_file, vocab_file):\n vectors = vecs.Vecs(vocab_file, bin_file)\n vecarr = array.array(str('d'))\n for idx in range(len(vectors.vocab)):\n vec = vectors.vecs[idx].tolist()[0]\n vecarr.extend(float(x) for x in vec)\n return {'itos': vectors.vocab,\n 'stoi': vectors.word_to_idx,\n 'vecs': vecarr,\n 'source': 'swivel' + bin_file,\n 'dim': vectors.vecs.shape[1]}\nwnHolE_emb=load_swivel_bin_vocab_embeddings(vecbin, holE_voc_file)",
"_____no_output_____"
]
],
[
[
"Now that we have the WordNet HolE embedding in the right format, we can explore some of the 'words' in the vocabulary:",
"_____no_output_____"
]
],
[
[
"wnHolE_emb['itos'][150000] # integer to string",
"_____no_output_____"
]
],
[
[
"### Tokenize and index the dataset\nAs in the previous case, we need to tokenize the raw dataset. However, since we now have access to the WordNet HolE embeddings, it make sense to use the WordNet disambiguated version of the text (i.e. `raw_hotel_wnscd_ds`). The `clsion` library already provides a method `index_ds_wnet` to perform tokenization and indexing using the expected WordNet encoding for synsets. ",
"_____no_output_____"
]
],
[
[
"wn_hotel_ds = clsion.index_ds_wnet(raw_hotel_wnscd_ds, wnHolE_emb)",
"_____no_output_____"
],
[
"print(\n 'vocab size:', len(wn_hotel_ds['vocab_embedding']['w2i']),\n 'dim:', wn_hotel_ds['vocab_embedding']['dim'])",
"_____no_output_____"
]
],
[
[
"The above produces an `ls` tokenization of the input text, which means that each original token is mapped to both a lemma and a synset. The model will then use both of these to map each token to the concatenation of the lemma and synset embedding. Since the WordNet HolE has 150 dimensions, each token will be represented by a 300 dimensional embedding (the concatenation of the lemma and synset embedding).\n\n### Define the experiment and run\nWe define the experiment using this new dataset as follows, the main change is that we do not want the embedding layer to be trainable, since we want to maintain the knowledge learned via HolE from WordNet. The model should only train the LSTM and dense layers to predict whether the input text is deceptive or not.",
"_____no_output_____"
]
],
[
[
"experiment2 = {\n 'hotel_wn_holE': {\n 'indexed_dataset': wn_hotel_ds,\n 'executor': clsion.execute_experiment,\n 'hparams': clsion.merge_hparams([\n clsion.common_hparams, clsion.biLSTM_hparams, \n clsion.calc_hparams(wn_hotel_ds), \n { \n 'epochs': 20,\n 'emb_trainable': False\n }\n ])\n }\n}",
"_____no_output_____"
],
[
"ex2_df, ex2_best_run = clsion.n_cross_val(experiment2, n=1)",
"_____no_output_____"
],
[
"ex2_df",
"_____no_output_____"
]
],
[
[
"### Discussion\nAlthough the model performs worse than the `csim` version, we can expect the model to be applicable to closely related domains. The hope is that, even if words did not appear in the training dataset, the model will be able to exploit embedding similarities learned from WordNet to generalise the 'deceptive' classification.",
"_____no_output_____"
],
[
"## Using Vecsigrafo UMBC WNet embeddings\n",
"_____no_output_____"
],
[
"### Download the embeddings\n\nIf you executed previous notebooks, you may already have the embedding in your environment.",
"_____no_output_____"
]
],
[
[
"%mkdir /content/umbc\n%mkdir /content/umbc/vec\nfull_precomp_url = 'https://zenodo.org/record/1446214/files/vecsigrafo_umbc_tlgs_ls_f_6e_160d_row_embedding.tar.gz'\nfull_precomp_targz = '/content/umbc/vec/tlgs_wnscd_ls_f_6e_160d_row_embedding.tar.gz'\n!wget {full_precomp_url} -O {full_precomp_targz}",
"_____no_output_____"
],
[
"!tar -xzf {full_precomp_targz} -C /content/umbc/vec/\nfull_precomp_vec_path = '/content/umbc/vec/vecsi_tlgs_wnscd_ls_f_6e_160d'",
"_____no_output_____"
],
[
"%ls /content/umbc/vec/vecsi_tlgs_wnscd_ls_f_6e_160d/",
"_____no_output_____"
]
],
[
[
"Since the embeddings were distributed as `tsv` files, we can use the `load_tsv_embeddings` method. Training models with all 1.4M vocab elements requires a lot of RAM, so we limit ourselves to only the first 250K vocab elements (these are the most frequent lemmas and synsets in UMBC).",
"_____no_output_____"
]
],
[
[
"def simple_lemmas(word):\n if word.startswith('lem_'):\n return word.replace('lem_', '').replace('_', ' ')\n else:\n return word\n \nwn_vecsi_umbc_emb = clsion.load_tsv_embeddings(full_precomp_vec_path + '/row_embedding.tsv', \n max_words=250000,\n word_map_fn=simple_lemmas\n )",
"_____no_output_____"
]
],
[
[
"### Tokenize and index dataset",
"_____no_output_____"
]
],
[
[
"wn_v_umbc_hotel_ds = clsion.index_ds_wnet(raw_hotel_wnscd_ds, wn_vecsi_umbc_emb)",
"_____no_output_____"
],
[
"print(\n 'vocab size:', len(wn_v_umbc_hotel_ds['vocab_embedding']['w2i']),\n 'dim:', wn_v_umbc_hotel_ds['vocab_embedding']['dim'])",
"_____no_output_____"
]
],
[
[
"### Define the experiment and run",
"_____no_output_____"
]
],
[
[
"experiment3 = {\n 'hotel_wn_vecsi_umbc': {\n 'indexed_dataset': wn_v_umbc_hotel_ds,\n 'executor': clsion.execute_experiment,\n 'hparams': clsion.merge_hparams([\n clsion.common_hparams, clsion.biLSTM_hparams, \n clsion.calc_hparams(wn_v_umbc_hotel_ds), \n { \n 'epochs': 20,\n 'emb_trainable': False\n }\n ])\n }\n}",
"_____no_output_____"
],
[
"ex3_df, ex3_best_run = clsion.n_cross_val(experiment3, n=1)",
"_____no_output_____"
],
[
"ex3_df",
"_____no_output_____"
]
],
[
[
"## Combine HolE and UMBC embeddings\nOne of the advantages of embeddings as a knowledge representation device is that they are trivial to combine. In the previous experiments we have tried to use lemma and synset embeddings derived from:\n * WordNet via HolE: these embeddings *encode* the knowledge derived from the structure of the WordNet Knowledge Graph\n * the Shallow Connectivity disambiguation of the UMBC corpus: these embeddings *encode* the knowledge derived from trying to predict the lemmas and synsets from their contexts.\n \nSince the embeddings encode different types of knowledge, it can be useful to use both embeddings at the same time when passing them to the deep learning model, as shown in this section.",
"_____no_output_____"
],
[
"### Combine the embeddings\nWe use the `concat_embs` method, which will go through the vocabularies of both input embeddings and concatenate them. Missing embeddings from one vocabulary will be mapped to the zero vector. Note that since `wnHolE_emb` has dimension 150 and `wn_vecsi_umbc_emb` has dimension 160, the resulting embedding will have dimension 310. (Besides concatenation, you could also experiment with other merging operations such as summation, substraction or averaging of the embeddings).",
"_____no_output_____"
]
],
[
[
"wn_vh_emb = clsion.concat_embs(wn_vecsi_umbc_emb, wnHolE_emb)",
"_____no_output_____"
],
[
"synsets = [w for w in wn_vh_emb['itos'] if w.startswith('wn31_')]\nprint('vocab has ', len(wn_vh_emb['itos']), '\"words\"', len(synsets), 'of which are synsets')",
"_____no_output_____"
],
[
"wn_vh_hotel_ds = clsion.index_ds_wnet(raw_hotel_wnscd_ds, wn_vh_emb)",
"_____no_output_____"
],
[
"experiment4 = {\n 'hotel_wn_vecsi_umbc': {\n 'indexed_dataset': wn_vh_hotel_ds,\n 'executor': clsion.execute_experiment,\n 'hparams': clsion.merge_hparams([\n clsion.common_hparams, clsion.biLSTM_hparams, \n clsion.calc_hparams(wn_vh_hotel_ds), \n { \n 'epochs': 20,\n 'emb_trainable': False\n }\n ])\n }\n}",
"_____no_output_____"
],
[
"ex4_df, _ = clsion.n_cross_val(experiment4, n=1)",
"_____no_output_____"
]
],
[
[
"## Discussion and results\nIn this notebook we have shown how to use use different types of embeddings as part of a deep learning text classification pipeline. We have not performed detailed experiments on the WordNet-based embeddings used in this notebook and, because the dataset is fairly small, the results can have quite a bit of variance depending on the initialization parameters. However, we have performed studies based on Cogito-based embeddings. The tables below shows some of our results:\n\nThe first set of results correspond to experiment 1 above. We trained the embeddings but explored various tokenizations strategies. \n\n\n | code | $\\mu$ acc | $\\sigma$ acc | tok | vocab | emb | trainable |\n | ------- | --------- | ------------ | ----------- | ----- | ------------------- | --------------------- |\n | sim | 0.8200 | 0.023 | ws | ds | random | y | \n | tok | 0.8325 | 0.029 | keras | ds | random | y | \n | csim | 0.8513 | 0.014 | clean ws | ds | random | y | \n | ctok | 0.8475 | 0.026 | clean keras | ds | random | y | \n\nAs discussed above, this approach produces the best test results, but the trained models are very specific to the training dataset. The current practice is therefore to use pre-trained word-embeddings. FastText embeddings tend to yield the best performance. We got the following results.\n\n | code | $\\mu$ acc | $\\sigma$ acc | tok | vocab | emb | trainable |\n | ------- | --------- | ------------ | ----------- | ----- | ------------------- | --------------------- |\n| ft-wiki | 0.7356 | 0.042 | ws | 250K | `wiki-en.vec` | n |\n | ft-wiki | 0.7775 | 0.044 | clean ws | 250K | `wiki-en.vec` | n |\n \n Next, we tried using HolE embedding trained on sensigrafo 14.2, which had very poor results:\n \n | code | $\\mu$ acc | $\\sigma$ acc | tok | vocab | emb | trainable |\n | ------- | --------- | ------------ | ----------- | ----- | ------------------- | --------------------- | \n | HolE_sensi | 0.6512 | 0.044 | cogito `s` | 250K | `HolE-en.14.2_500e` | n |\n\nNext we tried vecsigrafo trained on both wikipedia and umbc, either using only lemmas, only syncons or both lemmas and syncons. Using both lemmas and syncons always is better.\n\n | code | $\\mu$ acc | $\\sigma$ acc | tok | vocab | emb | trainable |\n | ------- | --------- | ------------ | ----------- | ----- | ------------------- | --------------------- | \n| v_wiki_l | 0.7450 | 0.050 | cogito `l` | 250K | `tlgs_ls_f_6e_160d` | n |\n | v_wiki_s | 0.7363 | 0.039 | cogito `s` | 250K | `tlgs_ls_f_6e_160d` | n |\n | v_wiki_ls | 0.7450 | 0.032 | cogito `ls` | 250K | `tlgs_ls_f_6e_160d` | n |\n | v_umbc_ls | 0.7413 | 0.038 | cogito `ls` | 250K | `tlgs_ls_6e_160d` | n |\n | v_umbc_l | 0.7350 | 0.041 | cogito `l` | 250K | `tlgs_ls_6e_160d` | n |\n | v_umbc_s | 0.7606 | 0.032 | cogito `s` | 250K | `tlgs_ls_6e_160d` | n |\n\n\nFinally, like in the experiment 4 above, we concatenated vecsigrafos (both lemmas and syncons) with HolE embeddings (only syncons, since lemmas tend to be poor quality). This produced the best results with a mean test accuracy of 79.31%. This is still lower than `csim`, but we expect this model to be more generic and applicable to other domains besides hotel reviews.\n\n | code | $\\mu$ acc | $\\sigma$ acc | tok | vocab | emb | trainable |\n | ------- | --------- | ------------ | ----------- | ----- | ------------------- | --------------------- | \n| vw_H_s | 0.7413 | 0.033 | cogito `s` | 304K | `tlgs_lsf`, `HolE` | n |\n | vw_H_ls | 0.7213 | 0.067 | cogito `ls` | 250K | `tlgs_lsf`, `HolE` | n |\n | vw_ls_H_s | 0.7275 | 0.041 | cogito `ls` | 250K | `tlgs_lsf`, `HolE` | n |\n | vu_H_s | 0.7669 | 0.043 | cogito `s` | 309K | `tlgs_ls`, `HolE` | n |\n | vu_ls_H_s | 0.7188 | 0.043 | cogito `ls` | 250K | `tlgs_ls`, `HolE` | n |\n | vu_ls_H_s | 0.7225 | 0.033 | cogito `l` | 250K | `tlgs_ls`, `HolE` | n |\n | vu_ls_H_s | 0.7788 | 0.033 | cogito `s` | 250K | `tlgs_ls`, `HolE` | n |\n | vu_ls_H_s | 0.7800 | 0.035 | cl cog `s` | 250K | `tlgs_ls`, `HolE` | n |\n | vu_ls_H_s | 0.7644 | 0.044 | cl cog `l` | 250K | `tlgs_ls`, `HolE` | n |\n | vu_ls_H_s |**0.7931** | 0.045 | cl cog `ls` | 250K | `tlgs_ls`, `HolE` | n |\n | vu_ls_H_s | 0.7838 | 0.028 | cl cog `s` | 500K | `tlgs_ls`, `HolE` | n |\n | vu_ls_H_s | ? | ? | cl cog `l` | 500K | `tlgs_ls`, `HolE` | n |\n | vu_ls_H_s | 0.7819 | 0.035 | cl cog `ls` | 500K | `tlgs_ls`, `HolE` | n |\n \n Finally, we have also experimented with a new type of embeddings, called contextual embeddings. Described in [Peters, M. E., Neumann, M., Iyyer, M., Gardner, M., Clark, C., Lee, K., & Zettlemoyer, L. (2018). Deep contextualized word representations. ](http://arxiv.org/abs/1802.05365). However, we did not manage to reproduce good results with this approach. \n \n| code | $\\mu$ acc | $\\sigma$ acc | tok | vocab | emb | trainable |\n | ------- | --------- | ------------ | ----------- | ----- | ------------------- | --------------------- | \n | elmo | 0.7250 | 0.039 | nltk sent | $\\infty$ | `elmo-5.5B` | n (0.1 dropout) |\n | elmo | 0.7269 | 0.038 | nltk sent | $\\infty$ | `elmo-5.5B` | n (0.5 dropout, 20ep) |\n \nWe have not yet tried applying more recent contextual embeddings, like those produced by BERT, but based on results reported elsewhere we assume these should produce very good results. We encourage you to use the [hugginface transformer library](https://huggingface.co/transformers/) introduced and used in [previous](https://colab.research.google.com/github/hybridnlp/tutorial/blob/master/01a_nlm_and_contextual_embeddings.ipynb) [notebooks](https://colab.research.google.com/github/hybridnlp/tutorial/blob/master/03a_LMMS_Transigrafo.ipynb) to fine-tune BERT for this task and we welcome pull-requests with your results.",
"_____no_output_____"
],
[
"# Further Exercises\n\n## Use the `fake_news` dataset from UMichigan\n",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
ecf164a3475cf4b6ddd37efedcf1f0a2e86601c9 | 428,047 | ipynb | Jupyter Notebook | my_nbs/dl1/00_notebook_tutorial-Copy1.ipynb | sanzgadea/fastai-v3 | d0131c1639e63d7a8a7901542d17afb805a91e56 | [
"Apache-2.0"
] | null | null | null | my_nbs/dl1/00_notebook_tutorial-Copy1.ipynb | sanzgadea/fastai-v3 | d0131c1639e63d7a8a7901542d17afb805a91e56 | [
"Apache-2.0"
] | 3 | 2021-05-20T15:34:22.000Z | 2022-02-26T07:04:35.000Z | my_nbs/dl1/00_notebook_tutorial-Copy1.ipynb | sanzgadea/fastai-v3 | d0131c1639e63d7a8a7901542d17afb805a91e56 | [
"Apache-2.0"
] | null | null | null | 353.465731 | 387,208 | 0.937993 | [
[
[
"**Important note:** You should always work on a duplicate of the course notebook. On the page you used to open this, tick the box next to the name of the notebook and click duplicate to easily create a new version of this notebook.\n\nYou will get errors each time you try to update your course repository if you don't do this, and your changes will end up being erased by the original course version.",
"_____no_output_____"
],
[
"# Welcome to Jupyter Notebooks!",
"_____no_output_____"
],
[
"If you want to learn how to use this tool you've come to the right place. This article will teach you all you need to know to use Jupyter Notebooks effectively. You only need to go through Section 1 to learn the basics and you can go into Section 2 if you want to further increase your productivity.",
"_____no_output_____"
],
[
"You might be reading this tutorial in a web page (maybe Github or the course's webpage). We strongly suggest to read this tutorial in a (yes, you guessed it) Jupyter Notebook. This way you will be able to actually *try* the different commands we will introduce here.",
"_____no_output_____"
],
[
"## Section 1: Need to Know",
"_____no_output_____"
],
[
"### Introduction",
"_____no_output_____"
],
[
"Let's build up from the basics, what is a Jupyter Notebook? Well, you are reading one. It is a document made of cells. You can write like I am writing now (markdown cells) or you can perform calculations in Python (code cells) and run them like this:",
"_____no_output_____"
]
],
[
[
"1+5**2",
"_____no_output_____"
]
],
[
[
"Cool huh? This combination of prose and code makes Jupyter Notebook ideal for experimentation: we can see the rationale for each experiment, the code and the results in one comprehensive document. In fast.ai, each lesson is documented in a notebook and you can later use that notebook to experiment yourself. \n\nOther renowned institutions in academy and industry use Jupyter Notebook: Google, Microsoft, IBM, Bloomberg, Berkeley and NASA among others. Even Nobel-winning economists [use Jupyter Notebooks](https://paulromer.net/jupyter-mathematica-and-the-future-of-the-research-paper/) for their experiments and some suggest that Jupyter Notebooks will be the [new format for research papers](https://www.theatlantic.com/science/archive/2018/04/the-scientific-paper-is-obsolete/556676/).",
"_____no_output_____"
],
[
"### Writing",
"_____no_output_____"
],
[
"A type of cell in which you can write like this is called _Markdown_. [_Markdown_](https://en.wikipedia.org/wiki/Markdown) is a very popular markup language. To specify that a cell is _Markdown_ you need to click in the drop-down menu in the toolbar and select _Markdown_.",
"_____no_output_____"
],
[
"Click on the the '+' button on the left and select _Markdown_ from the toolbar.",
"_____no_output_____"
],
[
"I just did it :)",
"_____no_output_____"
],
[
"Now you can type your first _Markdown_ cell. Write 'My first markdown cell' and press run.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"You should see something like this:",
"_____no_output_____"
],
[
"My first markdown cell",
"_____no_output_____"
],
[
"Now try making your first _Code_ cell: follow the same steps as before but don't change the cell type (when you add a cell its default type is _Code_). Type something like 3/2. You should see '1.5' as output.",
"_____no_output_____"
]
],
[
[
"3/2",
"_____no_output_____"
],
[
"9**9",
"_____no_output_____"
]
],
[
[
"### Modes",
"_____no_output_____"
],
[
"If you made a mistake in your *Markdown* cell and you have already ran it, you will notice that you cannot edit it just by clicking on it. This is because you are in **Command Mode**. Jupyter Notebooks have two distinct modes:\n\n1. **Edit Mode**: Allows you to edit a cell's content.\n\n2. **Command Mode**: Allows you to edit the notebook as a whole and use keyboard shortcuts but not edit a cell's content. \n\nYou can toggle between these two by either pressing <kbd>ESC</kbd> and <kbd>Enter</kbd> or clicking outside a cell or inside it (you need to double click if its a Markdown cell). You can always know which mode you're on since the current cell has a green border if in **Edit Mode** and a blue border in **Command Mode**. Try it!",
"_____no_output_____"
],
[
"### Other Important Considerations",
"_____no_output_____"
],
[
"1. Your notebook is autosaved every 120 seconds. If you want to manually save it you can just press the save button on the upper left corner or press <kbd>s</kbd> in **Command Mode**.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"2. To know if your kernel is computing or not you can check the dot in your upper right corner. If the dot is full, it means that the kernel is working. If not, it is idle. You can place the mouse on it and see the state of the kernel be displayed.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"3. There are a couple of shortcuts you must know about which we use **all** the time (always in **Command Mode**). These are:\n\n<kbd>Shift</kbd>+<kbd>Enter</kbd>: Runs the code or markdown on a cell\n\n<kbd>Up Arrow</kbd>+<kbd>Down Arrow</kbd>: Toggle across cells\n\n<kbd>b</kbd>: Create new cell\n\n<kbd>0</kbd>+<kbd>0</kbd>: Reset Kernel\n\nYou can find more shortcuts in the Shortcuts section below.",
"_____no_output_____"
],
[
"4. You may need to use a terminal in a Jupyter Notebook environment (for example to git pull on a repository). That is very easy to do, just press 'New' in your Home directory and 'Terminal'. Don't know how to use the Terminal? We made a tutorial for that as well. You can find it [here](https://course.fast.ai/terminal_tutorial.html).",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"That's it. This is all you need to know to use Jupyter Notebooks. That said, we have more tips and tricks below ↓↓↓",
"_____no_output_____"
],
[
"## Section 2: Going deeper",
"_____no_output_____"
],
[
"### Markdown formatting",
"_____no_output_____"
],
[
"#### Italics, Bold, Strikethrough, Inline, Blockquotes and Links",
"_____no_output_____"
],
[
"The five most important concepts to format your code appropriately when using markdown are:\n \n1. *Italics*: Surround your text with '\\_' or '\\*'\n2. **Bold**: Surround your text with '\\__' or '\\**'\n3. `inline`: Surround your text `with` '\\`'\n4. > blockquote: Place '\\>' before your text.\n5. [Links](https://course.fast.ai/): Surround the text you want to link with '\\[\\]' and place the link adjacent to the text, surrounded with '()'\n",
"_____no_output_____"
],
[
"#### Headings",
"_____no_output_____"
],
[
"Notice that including a hashtag before the text in a markdown cell makes the text a heading. The number of hashtags you include will determine the priority of the header ('#' is level one, '##' is level two, '###' is level three and '####' is level four). We will add three new cells with the '+' button on the left to see how every level of heading looks.",
"_____no_output_____"
],
[
"Double click on some headings and find out what level they are!",
"_____no_output_____"
],
[
"#### Hello World\n`Surprised to see me?`",
"_____no_output_____"
],
[
"> Yes!",
"_____no_output_____"
],
[
"**Yuhuu** [Link](www.wikipedia.com)",
"_____no_output_____"
],
[
"#### Lists",
"_____no_output_____"
],
[
"There are three types of lists in markdown.",
"_____no_output_____"
],
[
"Ordered list:\n\n1. Step 1\n 2. Step 1B\n3. Step 3",
"_____no_output_____"
],
[
"Unordered list\n\n* learning rate\n* cycle length\n* weight decay",
"_____no_output_____"
],
[
"Task list\n\n- [x] Learn Jupyter Notebooks\n - [x] Writing\n - [x] Modes\n - [x] Other Considerations\n- [ ] Change the world",
"_____no_output_____"
],
[
"Double click on each to see how they are built! ",
"_____no_output_____"
],
[
"### Code Capabilities",
"_____no_output_____"
],
[
"**Code** cells are different than **Markdown** cells in that they have an output cell. This means that we can _keep_ the results of our code within the notebook and share them. Let's say we want to show a graph that explains the result of an experiment. We can just run the necessary cells and save the notebook. The output will be there when we open it again! Try it out by running the next four cells.",
"_____no_output_____"
]
],
[
[
"# Import necessary libraries\nfrom fastai.vision import * \nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"from PIL import Image",
"_____no_output_____"
],
[
"a = 1\nb = a + 1\nc = b + a + 1\nd = c + b + a + 1\na, b, c ,d",
"_____no_output_____"
],
[
"plt.plot([a,b,c,d])\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can also print images while experimenting. I am watching you.",
"_____no_output_____"
]
],
[
[
"Image.open('images/notebook_tutorial/cat_example.jpg')",
"_____no_output_____"
]
],
[
[
"### Running the app locally",
"_____no_output_____"
],
[
"You may be running Jupyter Notebook from an interactive coding environment like Gradient, Sagemaker or Salamander. You can also run a Jupyter Notebook server from your local computer. What's more, if you have installed Anaconda you don't even need to install Jupyter (if not, just `pip install jupyter`).\n\nYou just need to run `jupyter notebook` in your terminal. Remember to run it from a folder that contains all the folders/files you will want to access. You will be able to open, view and edit files located within the directory in which you run this command but not files in parent directories.\n\nIf a browser tab does not open automatically once you run the command, you should CTRL+CLICK the link starting with 'https://localhost:' and this will open a new tab in your default browser.",
"_____no_output_____"
],
[
"### Creating a notebook",
"_____no_output_____"
],
[
"Click on 'New' in the upper right corner and 'Python 3' in the drop-down list (we are going to use a [Python kernel](https://github.com/ipython/ipython) for all our experiments).\n\n\n\nNote: You will sometimes hear people talking about the Notebook 'kernel'. The 'kernel' is just the Python engine that performs the computations for you. ",
"_____no_output_____"
],
[
"### Shortcuts and tricks",
"_____no_output_____"
],
[
"#### Command Mode Shortcuts",
"_____no_output_____"
],
[
"There are a couple of useful keyboard shortcuts in `Command Mode` that you can leverage to make Jupyter Notebook faster to use. Remember that to switch back and forth between `Command Mode` and `Edit Mode` with <kbd>Esc</kbd> and <kbd>Enter</kbd>.",
"_____no_output_____"
],
[
"<kbd>m</kbd>: Convert cell to Markdown",
"_____no_output_____"
],
[
"<kbd>y</kbd>: Convert cell to Code",
"_____no_output_____"
],
[
"<kbd>D</kbd>+<kbd>D</kbd>: Delete the cell(if it's not the only cell) or delete the content of the cell and reset cell to Code(if only one cell left)",
"_____no_output_____"
],
[
"<kbd>o</kbd>: Toggle between hide or show output",
"_____no_output_____"
]
],
[
[
"%magic\n\n",
"_____no_output_____"
]
],
[
[
"<kbd>Shift</kbd>+<kbd>Arrow up/Arrow down</kbd>: Selects multiple cells. Once you have selected them you can operate on them like a batch (run, copy, paste etc).",
"_____no_output_____"
],
[
"<kbd>Shift</kbd>+<kbd>M</kbd>: Merge selected cells.",
"_____no_output_____"
],
[
"<kbd>Shift</kbd>+<kbd>Tab</kbd>: [press these two buttons at the same time, once] Tells you which parameters to pass on a function\n\n<kbd>Shift</kbd>+<kbd>Tab</kbd>: [press these two buttons at the same time, three times] Gives additional information on the method",
"_____no_output_____"
],
[
"#### Cell Tricks",
"_____no_output_____"
]
],
[
[
"from fastai import*\nfrom fastai.vision import *",
"_____no_output_____"
]
],
[
[
"There are also some tricks that you can code into a cell.",
"_____no_output_____"
],
[
"`?function-name`: Shows the definition and docstring for that function",
"_____no_output_____"
]
],
[
[
"?ImageDataBunch",
"_____no_output_____"
]
],
[
[
"`??function-name`: Shows the source code for that function",
"_____no_output_____"
]
],
[
[
"??ImageDataBunch",
"_____no_output_____"
]
],
[
[
"`doc(function-name)`: Shows the definition, docstring **and links to the documentation** of the function\n(only works with fastai library imported)",
"_____no_output_____"
]
],
[
[
"doc(ImageDataBunch)",
"_____no_output_____"
]
],
[
[
"#### Line Magics",
"_____no_output_____"
],
[
"Line magics are functions that you can run on cells and take as an argument the rest of the line from where they are called. You call them by placing a '%' sign before the command. The most useful ones are:",
"_____no_output_____"
],
[
"`%matplotlib inline`: This command ensures that all matplotlib plots will be plotted in the output cell within the notebook and will be kept in the notebook when saved.",
"_____no_output_____"
],
[
"`%reload_ext autoreload`, `%autoreload 2`: Reload all modules before executing a new line. If a module is edited, it is not necessary to rerun the import commands, the modules will be reloaded automatically.",
"_____no_output_____"
],
[
"These three commands are always called together at the beginning of every notebook.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n%reload_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"`%timeit`: Runs a line a ten thousand times and displays the average time it took to run it.",
"_____no_output_____"
]
],
[
[
"%timeit [i+1 for i in range(1000)]",
"54.4 µs ± 1.37 µs per loop (mean ± std. dev. of 7 runs, 10000 loops each)\n"
]
],
[
[
"`%debug`: Allows to inspect a function which is showing an error using the [Python debugger](https://docs.python.org/3/library/pdb.html).",
"_____no_output_____"
]
],
[
[
"for i in range(1000):\n a = i+1\n b = 'string'\n c = b+1",
"_____no_output_____"
],
[
"%debug",
"> <ipython-input-14-8d78ff778454>(4)<module>()\n 1 for i in range(1000):\n 2 a = i+1\n 3 b = 'string'\n----> 4 c = b+1\n\nipdb> c\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecf16cfee34af9936be5e088dfbf4314eae9fe4d | 8,272 | ipynb | Jupyter Notebook | jupyter_notebooks/solving/LWE/solveLWE_withoutError.ipynb | TomMasterThesis/sca_on_newhope | 3c72da72076119fb22a64e965c21d340f597cee1 | [
"Apache-2.0"
] | null | null | null | jupyter_notebooks/solving/LWE/solveLWE_withoutError.ipynb | TomMasterThesis/sca_on_newhope | 3c72da72076119fb22a64e965c21d340f597cee1 | [
"Apache-2.0"
] | null | null | null | jupyter_notebooks/solving/LWE/solveLWE_withoutError.ipynb | TomMasterThesis/sca_on_newhope | 3c72da72076119fb22a64e965c21d340f597cee1 | [
"Apache-2.0"
] | null | null | null | 25.142857 | 109 | 0.459381 | [
[
[
"# Learning With Errors (LWE)",
"_____no_output_____"
],
[
"## import libs",
"_____no_output_____"
]
],
[
[
"#!conda install pycrypto",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nimport scipy.stats\nimport math\nimport itertools\nimport random\nfrom Crypto.Util import number\n\nimport matplotlib.pyplot as plt\nfrom matplotlib import collections as matcoll",
"_____no_output_____"
]
],
[
[
"## Set vector s (secret)",
"_____no_output_____"
],
[
"Choose $s \\in \\mathbb{Z}^m_p$ with arbitrary $m \\in \\mathbb{N}$",
"_____no_output_____"
]
],
[
[
"#s = np.array([2, 3]) \n#s = np.array([10, 13, 9]) \n#s = np.array([10, 13, 9, 11]) \n#s = np.array([10, 13, 9, 11, 3]) \ns = np.array([10, 13, 9, 11, 3, 10, 13, 9, 11, 3, 10]) \nn = len(s)",
"_____no_output_____"
]
],
[
[
"## parameters",
"_____no_output_____"
]
],
[
[
"random.seed(42) #set seed\n\n# modulus \n#p = 17 # only prime numbers (it has to be a finite field)\np = number.getPrime(n.bit_length()**2, randfunc=np.random.bytes) # using pycrypto lib (p = O(n^2))\nprint(\"Prime:\", p)\n\n#size parameter\nm = n\nprint('Count of equations:', m)",
"Prime: 47293\nCount of equations: 11\n"
]
],
[
[
"## Construct the LWE problem without error",
"_____no_output_____"
],
[
"#### Construct A, b",
"_____no_output_____"
]
],
[
[
"A = np.random.randint(0, p, size=(m, n))\n\nb = (np.matmul(A, s))%p # system of linear equations without perturbation",
"_____no_output_____"
]
],
[
[
"## Solving",
"_____no_output_____"
],
[
"### Modified Gaussian Elimination",
"_____no_output_____"
]
],
[
[
"# Iterative Algorithm (xgcd)\ndef iterative_egcd(a, b):\n x,y, u,v = 0,1, 1,0\n while a != 0:\n q,r = b//a,b%a; m,n = x-u*q,y-v*q # use x//y for floor \"floor division\"\n b,a, x,y, u,v = a,r, u,v, m,n\n return b, x, y\n\ndef modinv(a, m):\n g, x, y = iterative_egcd(a, m) \n if g != 1:\n return None\n else:\n return x % m\n \ndef solve_linear_congruence(a, b, m):\n \"\"\" Describe all solutions to ax = b (mod m), or raise ValueError. \"\"\"\n g = math.gcd(a, m)\n if b % g:\n raise ValueError(\"No solutions\")\n a, b, m = a//g, b//g, m//g\n return modinv(a, m) * b % m, m\n\ndef print_solutions(a, b, m):\n print(f\"Solving the congruence: {a}x = {b} (mod {m})\")\n x, mx = solve_linear_congruence(a, b, m)\n \n print(f\"Particular solution: x = {x}\")\n print(f\"General solution: x = {x} (mod {mx})\")\n \n# for debug\nprint_solutions(272, 256, 1009)",
"Solving the congruence: 272x = 256 (mod 1009)\nParticular solution: x = 179\nGeneral solution: x = 179 (mod 1009)\n"
],
[
"def gaussianEliminationForward(A, b, modulus):\n (m, n) = A.shape\n \n A = np.copy(A[:n][:])\n b = np.copy(b[:n])\n \n \n for j in range(n): # quadratic matrix\n i = j\n while(i<n-1):\n rowUpper = A[i, :]\n rowUpperLeader = rowUpper[j]\n leftUpper = b[i]\n rowLower = A[i+1, :]\n rowLowerLeader = rowLower[j]\n leftLower = b[i+1]\n\n if rowLowerLeader==0:\n pass\n elif rowUpperLeader==0 and rowLowerLeader!=0:\n # swap rows\n A[[i, i+1]] = A[[i+1, i]]\n b[[i, i+1]] = b[[i+1, i]]\n i=j-1 # redo column\n \n elif rowUpperLeader!=0 and rowLowerLeader!=0:\n lcm = np.lcm(rowUpperLeader, rowLowerLeader)\n rowLowerNew = (lcm/rowLowerLeader)*rowLower - (lcm/rowUpperLeader)*rowUpper\n leftLowerNew = (lcm/rowLowerLeader)*leftLower - (lcm/rowUpperLeader)*leftUpper\n \n A[i+1, :] = rowLowerNew%modulus\n b[i+1] = leftLowerNew%modulus\n \n i+=1\n\n return A, b\n\n\n\n \ndef gaussianEliminationBackward(A, b, modulus):\n (m, n) = A.shape\n x = np.zeros(m)\n \n for i in range(n-1, -1, -1):\n equLeft = A[i, :]\n equLeftCoef = equLeft[i]\n equRight = b[i]\n equRightCoef = equRight - np.dot(x, equLeft)\n \n solution, mx = solve_linear_congruence(equLeftCoef, equRightCoef, modulus)\n x[i] = solution\n \n return x\n\n# for debug\n#print(A[:n])\nA_new, b_new = gaussianEliminationForward(A, b, p)\n#print(A_new)\n#print()\n#print(b[:n].astype(int))\n#print(b_new.astype(int))\n#print()\n#print(scipy.linalg.solve(A[:m], b[:m]))\n#print(scipy.linalg.solve(A_new, b_new))",
"_____no_output_____"
],
[
"try:\n A_new, b_new = gaussianEliminationForward(A, b, p)\n x = gaussianEliminationBackward(A_new%p, b_new%p, p)\n print(\"Guess:\", x.astype(int)%p, \"\\t\", \"Right Solution:\", s)\nexcept ValueError: # occurs by linear dependency in the matrix subsetA\n print(\"linear dependency\")",
"Guess: [10 13 9 11 3 10 13 9 11 3 10] \t Right Solution: [10 13 9 11 3 10 13 9 11 3 10]\n"
]
],
[
[
"sometimes I got wrong solution because of integer overflow. Particularly with big p",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecf17cd10b1f8f8e39226786244f0385ad6164b1 | 105,587 | ipynb | Jupyter Notebook | demo/tissues/tissue_neighborhood_quantification.ipynb | cavenel/pyclesperanto_prototype | 701f8b6741833d1943b4b78a28b1f0b06a5519b4 | [
"BSD-3-Clause"
] | 2 | 2020-07-01T06:20:44.000Z | 2020-07-01T09:36:48.000Z | demo/tissues/tissue_neighborhood_quantification.ipynb | cavenel/pyclesperanto_prototype | 701f8b6741833d1943b4b78a28b1f0b06a5519b4 | [
"BSD-3-Clause"
] | null | null | null | demo/tissues/tissue_neighborhood_quantification.ipynb | cavenel/pyclesperanto_prototype | 701f8b6741833d1943b4b78a28b1f0b06a5519b4 | [
"BSD-3-Clause"
] | 1 | 2020-06-29T18:40:54.000Z | 2020-06-29T18:40:54.000Z | 342.814935 | 17,932 | 0.946632 | [
[
[
"# Tissue neighborhood quantification\n\nIn this notebook, we will analyse neighborhood-relationships between cells. \nWe count the number of neighbors for each cell and take a look at this number in a parametric image.\nAfterwards, we average this number locally between neighbors.\n\n## Test data\nLet's generate some cells and take a look at the borders between them",
"_____no_output_____"
]
],
[
[
"import pyclesperanto_prototype as cle\nimport numpy as np\nfrom skimage.io import imshow\nimport matplotlib",
"_____no_output_____"
],
[
"# Generate artificial cells as test data\ntissue = cle.artificial_tissue_2d()\n\ncle.imshow(tissue, labels=True)",
"_____no_output_____"
],
[
"membranes = cle.detect_label_edges(tissue)\ncle.imshow(membranes)",
"_____no_output_____"
]
],
[
[
"# Analysis and visualization of neighbor count",
"_____no_output_____"
]
],
[
[
"touch_matrix = cle.generate_touch_matrix(tissue)\nneighbor_count = cle.count_touching_neighbors(touch_matrix)\n\nparametric_image = cle.replace_intensities(tissue, neighbor_count)\ncle.imshow(parametric_image, min_display_intensity=0, max_display_intensity=10, color_map='jet')\n",
"_____no_output_____"
]
],
[
[
"## Average the measurement between cells to reduce noise\nMean of touching neighbors",
"_____no_output_____"
]
],
[
[
"local_mean_neighbor_count = cle.mean_of_touching_neighbors(neighbor_count, touch_matrix)\n\nparametric_image = cle.replace_intensities(tissue, local_mean_neighbor_count)\ncle.imshow(parametric_image, min_display_intensity=0, max_display_intensity=10, color_map='jet')",
"_____no_output_____"
]
],
[
[
"Median of touching neighbors",
"_____no_output_____"
]
],
[
[
"local_median_neighbor_count = cle.median_of_touching_neighbors(neighbor_count, touch_matrix)\n\nparametric_image = cle.replace_intensities(tissue, local_median_neighbor_count)\ncle.imshow(parametric_image, min_display_intensity=0, max_display_intensity=10, color_map='jet')",
"_____no_output_____"
],
[
"local_minimum_neighbor_count = cle.minimum_of_touching_neighbors(neighbor_count, touch_matrix)\n\nparametric_image = cle.replace_intensities(tissue, local_minimum_neighbor_count)\ncle.imshow(parametric_image, min_display_intensity=0, max_display_intensity=10, color_map='jet')",
"_____no_output_____"
],
[
"local_maximum_neighbor_count = cle.maximum_of_touching_neighbors(neighbor_count, touch_matrix)\n\nparametric_image = cle.replace_intensities(tissue, local_maximum_neighbor_count)\ncle.imshow(parametric_image, min_display_intensity=0, max_display_intensity=10, color_map='jet')",
"_____no_output_____"
],
[
"local_standard_deviation_neighbor_count = cle.standard_deviation_of_touching_neighbors(neighbor_count, touch_matrix)\n\nparametric_image = cle.replace_intensities(tissue, local_standard_deviation_neighbor_count)\ncle.imshow(parametric_image, min_display_intensity=0, max_display_intensity=10, color_map='jet')",
"_____no_output_____"
],
[
"# most popular number of neighbors locally\nlocal_mode_neighbor_count = cle.mode_of_touching_neighbors(neighbor_count, touch_matrix)\n\nparametric_image = cle.replace_intensities(tissue, local_mode_neighbor_count)\ncle.imshow(parametric_image, min_display_intensity=0, max_display_intensity=10, color_map='jet')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecf17f4ac8248b837336dd8f5b309fa9019dd01c | 757,592 | ipynb | Jupyter Notebook | 16_reinforcement_learning.ipynb | JapneetSingh/handson-ml | cabe792b2376b330c1bd764681045733853899aa | [
"Apache-2.0"
] | null | null | null | 16_reinforcement_learning.ipynb | JapneetSingh/handson-ml | cabe792b2376b330c1bd764681045733853899aa | [
"Apache-2.0"
] | null | null | null | 16_reinforcement_learning.ipynb | JapneetSingh/handson-ml | cabe792b2376b330c1bd764681045733853899aa | [
"Apache-2.0"
] | 2 | 2017-07-03T12:30:55.000Z | 2019-07-05T20:38:07.000Z | 65.096408 | 84,965 | 0.662445 | [
[
[
"**Chapter 16 – Reinforcement Learning**",
"_____no_output_____"
],
[
"This notebook contains all the sample code and solutions to the exercices in chapter 16.",
"_____no_output_____"
],
[
"# Setup",
"_____no_output_____"
],
[
"First, let's make sure this notebook works well in both python 2 and 3, import a few common modules, ensure MatplotLib plots figures inline and prepare a function to save the figures:",
"_____no_output_____"
]
],
[
[
"# To support both python 2 and python 3\nfrom __future__ import division, print_function, unicode_literals\n\n# Common imports\nimport numpy as np\nimport numpy.random as rnd\nimport os\n\n# to make this notebook's output stable across runs\nrnd.seed(42)\n\n# To plot pretty figures and animations\n%matplotlib nbagg\nimport matplotlib\nimport matplotlib.animation as animation\nimport matplotlib.pyplot as plt\nplt.rcParams['axes.labelsize'] = 14\nplt.rcParams['xtick.labelsize'] = 12\nplt.rcParams['ytick.labelsize'] = 12\n\n# Where to save the figures\nPROJECT_ROOT_DIR = \".\"\nCHAPTER_ID = \"rl\"\n\ndef save_fig(fig_id, tight_layout=True):\n path = os.path.join(PROJECT_ROOT_DIR, \"images\", CHAPTER_ID, fig_id + \".png\")\n print(\"Saving figure\", fig_id)\n if tight_layout:\n plt.tight_layout()\n plt.savefig(path, format='png', dpi=300)",
"_____no_output_____"
]
],
[
[
"# Introduction to OpenAI gym",
"_____no_output_____"
],
[
"In this notebook we will be using [OpenAI gym](https://gym.openai.com/), a great toolkit for developing and comparing Reinforcement Learning algorithms. It provides many environments for your learning *agents* to interact with. Let's start by importing `gym`:",
"_____no_output_____"
]
],
[
[
"import gym",
"_____no_output_____"
]
],
[
[
"Next we will load the MsPacman environment, version 0.",
"_____no_output_____"
]
],
[
[
"env = gym.make('MsPacman-v0')",
"INFO:gym.envs.registration:Making new env: MsPacman-v0\n[2016-10-23 14:19:54,435] Making new env: MsPacman-v0\n"
]
],
[
[
"Let's initialize the environment by calling is `reset()` method. This returns an observation:",
"_____no_output_____"
]
],
[
[
"obs = env.reset()",
"_____no_output_____"
]
],
[
[
"Observations vary depending on the environment. In this case it is an RGB image represented as a 3D NumPy array of shape [width, height, channels] (with 3 channels: Red, Green and Blue). In other environments it may return different objects, as we will see later.",
"_____no_output_____"
]
],
[
[
"obs.shape",
"_____no_output_____"
]
],
[
[
"An environment can be visualized by calling its `render()` method, and you can pick the rendering mode (the rendering options depend on the environment). In this example we will set `mode=\"rgb_array\"` to get an image of the environment as a NumPy array:",
"_____no_output_____"
]
],
[
[
"img = env.render(mode=\"rgb_array\")",
"_____no_output_____"
]
],
[
[
"Let's plot this image:",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(5,4))\nplt.imshow(img)\nplt.axis(\"off\")\nsave_fig(\"MsPacman\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"Welcome back to the 1980s! :)",
"_____no_output_____"
],
[
"In this environment, the rendered image is simply equal to the observation (but in many environments this is not the case):",
"_____no_output_____"
]
],
[
[
"(img == obs).all()",
"_____no_output_____"
]
],
[
[
"Let's create a little helper function to plot an environment:",
"_____no_output_____"
]
],
[
[
"def plot_environment(env, figsize=(5,4)):\n plt.close() # or else nbagg sometimes plots in the previous cell\n plt.figure(figsize=figsize)\n img = env.render(mode=\"rgb_array\")\n plt.imshow(img)\n plt.axis(\"off\")\n plt.show()",
"_____no_output_____"
]
],
[
[
"Let's see how to interact with an environment. Your agent will need to select an action from an \"action space\" (the set of possible actions). Let's see what this environment's action space looks like:",
"_____no_output_____"
]
],
[
[
"env.action_space",
"_____no_output_____"
]
],
[
[
"`Discrete(9)` means that the possible actions are integers 0 through 8, which represents the 9 possible positions of the joystick (0=center, 1=up, 2=right, 3=left, 4=down, 5=upper-right, 6=upper-left, 7=lower-right, 8=lower-left).",
"_____no_output_____"
],
[
"Next we need to tell the environment which action to play, and it will compute the next step of the game. Let's go left for 110 steps, then lower left for 40 steps:",
"_____no_output_____"
]
],
[
[
"env.reset()\nfor step in range(110):\n env.step(3) #left\nfor step in range(40):\n env.step(8) #lower-left",
"_____no_output_____"
]
],
[
[
"Where are we now?",
"_____no_output_____"
]
],
[
[
"plot_environment(env)",
"_____no_output_____"
]
],
[
[
"The `step()` function actually returns several important objects:",
"_____no_output_____"
]
],
[
[
"obs, reward, done, info = env.step(0)",
"_____no_output_____"
]
],
[
[
"The observation tells the agent what the environment looks like, as discussed earlier. This is a 210x160 RGB image:",
"_____no_output_____"
]
],
[
[
"obs.shape",
"_____no_output_____"
]
],
[
[
"The environment also tells the agent how much reward it got during the last step:",
"_____no_output_____"
]
],
[
[
"reward",
"_____no_output_____"
]
],
[
[
"When the game is over, the environment returns `done=True`:",
"_____no_output_____"
]
],
[
[
"done",
"_____no_output_____"
]
],
[
[
"Finally, `info` is an environment-specific dictionary that can provide some extra information about the internal state of the environment. This is useful for debugging, but your agent should not use this information for learning (it would be cheating).",
"_____no_output_____"
]
],
[
[
"info",
"_____no_output_____"
]
],
[
[
"Let's play one full game (with 3 lives), by moving in random directions for 10 steps at a time, recording each frame:",
"_____no_output_____"
]
],
[
[
"frames = []\n\nn_max_steps = 1000\nn_change_steps = 10\n\nobs = env.reset()\nfor step in range(n_max_steps):\n img = env.render(mode=\"rgb_array\")\n frames.append(img)\n if step % n_change_steps == 0:\n action = env.action_space.sample() # play randomly\n obs, reward, done, info = env.step(action)\n if done:\n break",
"_____no_output_____"
]
],
[
[
"Now show the animation (it's a bit jittery within Jupyter):",
"_____no_output_____"
]
],
[
[
"def update_scene(num, frames, patch):\n patch.set_data(frames[num])\n return patch,\n\ndef plot_animation(frames, repeat=False, interval=40):\n plt.close() # or else nbagg sometimes plots in the previous cell\n fig = plt.figure()\n patch = plt.imshow(frames[0])\n plt.axis('off')\n return animation.FuncAnimation(fig, update_scene, fargs=(frames, patch), frames=len(frames), repeat=repeat, interval=interval)",
"_____no_output_____"
],
[
"video = plot_animation(frames)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Once you have finished playing with an environment, you should close it to free up resources:",
"_____no_output_____"
]
],
[
[
"env.close()",
"_____no_output_____"
]
],
[
[
"To code our first learning agent, we will be using a simpler environment: the Cart-Pole. ",
"_____no_output_____"
],
[
"# A simple environment: the Cart-Pole",
"_____no_output_____"
],
[
"The Cart-Pole is a very simple environment composed of a cart that can move left or right, and pole placed vertically on top of it. The agent must move the cart left or right to keep the pole upright.",
"_____no_output_____"
]
],
[
[
"env = gym.make(\"CartPole-v0\")",
"INFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:20:27,624] Making new env: CartPole-v0\n"
],
[
"obs = env.reset()",
"_____no_output_____"
],
[
"obs",
"_____no_output_____"
]
],
[
[
"The observation is a 1D NumPy array composed of 4 floats: they represent the cart's horizontal position, its velocity, the angle of the pole (0 = vertical), and the angular velocity. Let's render the environment... unfortunately we need to fix an annoying rendering issue first.",
"_____no_output_____"
],
[
"## Fixing the rendering issue",
"_____no_output_____"
],
[
"Some environments (including the Cart-Pole) require access to your display, which opens up a separate window, even if you specify the `rgb_array` mode. In general you can safely ignore that window. However, if Jupyter is running on a headless server (ie. without a screen) it will raise an exception. One way to avoid this is to install a fake X server like Xvfb. You can start Jupyter using the `xvfb-run` command:\n\n $ xvfb-run -s \"-screen 0 1400x900x24\" jupyter notebook\n\nIf you are running this notebook using Binder, then this has been taken care of for you. If not, and you don't want to worry about Xvfb, then you can just use the following rendering function for the Cart-Pole:",
"_____no_output_____"
]
],
[
[
"from PIL import Image, ImageDraw\n\ntry:\n from pyglet.gl import gl_info\n openai_cart_pole_rendering = True # no problem, let's use OpenAI gym's rendering function\nexcept Exception:\n openai_cart_pole_rendering = False # probably no X server available, let's use our own rendering function\n\ndef render_cart_pole(env, obs):\n if openai_cart_pole_rendering:\n # use OpenAI gym's rendering function\n return env.render(mode=\"rgb_array\")\n else:\n # rendering for the cart pole environment (in case OpenAI gym can't do it)\n img_w = 600\n img_h = 400\n cart_w = img_w // 12\n cart_h = img_h // 15\n pole_len = img_h // 3.5\n pole_w = img_w // 80 + 1\n x_width = 2\n max_ang = 0.2\n bg_col = (255, 255, 255)\n cart_col = 0x000000 # Blue Green Red\n pole_col = 0x669acc # Blue Green Red\n\n pos, vel, ang, ang_vel = obs\n img = Image.new('RGB', (img_w, img_h), bg_col)\n draw = ImageDraw.Draw(img)\n cart_x = pos * img_w // x_width + img_w // x_width\n cart_y = img_h * 95 // 100\n top_pole_x = cart_x + pole_len * np.sin(ang)\n top_pole_y = cart_y - cart_h // 2 - pole_len * np.cos(ang)\n draw.line((0, cart_y, img_w, cart_y), fill=0)\n draw.rectangle((cart_x - cart_w // 2, cart_y - cart_h // 2, cart_x + cart_w // 2, cart_y + cart_h // 2), fill=cart_col) # draw cart\n draw.line((cart_x, cart_y - cart_h // 2, top_pole_x, top_pole_y), fill=pole_col, width=pole_w) # draw pole\n return np.array(img)\n\ndef plot_cart_pole(env, obs):\n plt.close() # or else nbagg sometimes plots in the previous cell\n img = render_cart_pole(env, obs)\n plt.imshow(img)\n plt.axis(\"off\")\n plt.show()",
"_____no_output_____"
],
[
"plot_cart_pole(env, obs)",
"_____no_output_____"
]
],
[
[
"Now let's look at the action space:",
"_____no_output_____"
]
],
[
[
"env.action_space",
"_____no_output_____"
]
],
[
[
"Yep, just two possible actions: accelerate towards the left or towards the right. Let's push the cart left until the pole falls:",
"_____no_output_____"
]
],
[
[
"obs = env.reset()\nwhile True:\n obs, reward, done, info = env.step(0)\n if done:\n break",
"_____no_output_____"
],
[
"plt.close() # or else nbagg sometimes plots in the previous cell\nimg = render_cart_pole(env, obs)\nplt.imshow(img)\nplt.axis(\"off\")\nsave_fig(\"cart_pole_plot\")",
"_____no_output_____"
]
],
[
[
"Notice that the game is over when the pole tilts too much, not when it actually falls. Now let's reset the environment and push the cart to right instead:",
"_____no_output_____"
]
],
[
[
"obs = env.reset()\nwhile True:\n obs, reward, done, info = env.step(1)\n if done:\n break",
"_____no_output_____"
],
[
"plot_cart_pole(env, obs)",
"_____no_output_____"
]
],
[
[
"Looks like it's doing what we're telling it to do. Now how can we make the poll remain upright? We will need to define a _policy_ for that. This is the strategy that the agent will use to select an action at each step. It can use all the past actions and observations to decide what to do.",
"_____no_output_____"
],
[
"# A simple hard-coded policy",
"_____no_output_____"
],
[
"Let's hard code a simple strategy: if the pole is tilting to the left, then push the cart to the left, and _vice versa_. Let's see if that works:",
"_____no_output_____"
]
],
[
[
"frames = []\n\nn_max_steps = 1000\nn_change_steps = 10\n\nobs = env.reset()\nfor step in range(n_max_steps):\n img = render_cart_pole(env, obs)\n frames.append(img)\n\n # hard-coded policy\n position, velocity, angle, angular_velocity = obs\n if angle < 0:\n action = 0\n else:\n action = 1\n\n obs, reward, done, info = env.step(action)\n if done:\n break",
"_____no_output_____"
],
[
"video = plot_animation(frames)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Nope, the system is unstable and after just a few wobbles, the pole ends up too tilted: game over. We will need to be smarter than that!",
"_____no_output_____"
],
[
"# Neural Network Policies",
"_____no_output_____"
],
[
"Let's create a neural network that will take observations as inputs, and output the action to take for each observation. To choose an action, the network will first estimate a probability for each action, then select an action randomly according to the estimated probabilities. In the case of the Cart-Pole environment, there are just two possible actions (left or right), so we only need one output neuron: it will output the probability `p` of the action 0 (left), and of course the probability of action 1 (right) will be `1 - p`.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom tensorflow.contrib.layers import fully_connected\n\n# 1. Specify the network architecture\nn_inputs = 4 # == env.observation_space.shape[0]\nn_hidden = 4 # it's a simple task, we don't need more than this\nn_outputs = 1 # only outputs the probability of accelerating left\ninitializer = tf.contrib.layers.variance_scaling_initializer()\n\n# 2. Build the neural network\nX = tf.placeholder(tf.float32, shape=[None, n_inputs])\nhidden = fully_connected(X, n_hidden, activation_fn=tf.nn.elu,\n weights_initializer=initializer)\noutputs = fully_connected(hidden, n_outputs, activation_fn=tf.nn.sigmoid,\n weights_initializer=initializer)\n\n# 3. Select a random action based on the estimated probabilities\np_left_and_right = tf.concat(concat_dim=1, values=[outputs, 1 - outputs])\naction = tf.multinomial(tf.log(p_left_and_right), num_samples=1)\n\ninit = tf.initialize_all_variables()",
"_____no_output_____"
]
],
[
[
"In this particular environment, the past actions and observations can safely be ignored, since each observation contains the environment's full state. If there were some hidden state then you may need to consider past actions and observations in order to try to infer the hidden state of the environment. For example, if the environment only revealed the position of the cart but not its velocity, you would have to consider not only the current observation but also the previous observation in order to estimate the current velocity. Another example is if the observations are noisy: you may want to use the past few observations to estimate the most likely current state. Our problem is thus as simple as can be: the current observation is noise-free and contains the environment's full state.",
"_____no_output_____"
],
[
"You may wonder why we are picking a random action based on the probability given by the policy network, rather than just picking the action with the highest probability. This approach lets the agent find the right balance between _exploring_ new actions and _exploiting_ the actions that are known to work well. Here's an analogy: suppose you go to a restaurant for the first time, and all the dishes look equally appealing so you randomly pick one. If it turns out to be good, you can increase the probability to order it next time, but you shouldn't increase that probability to 100%, or else you will never try out the other dishes, some of which may be even better than the one you tried.",
"_____no_output_____"
],
[
"Let's randomly initialize this policy neural network and use it to play one game:",
"_____no_output_____"
]
],
[
[
"n_max_steps = 1000\nframes = []\n\nwith tf.Session() as sess:\n init.run()\n obs = env.reset()\n for step in range(n_max_steps):\n img = render_cart_pole(env, obs)\n frames.append(img)\n action_val = action.eval(feed_dict={X: obs.reshape(1, n_inputs)})\n obs, reward, done, info = env.step(action_val[0][0])\n if done:\n break\n\nenv.close()",
"_____no_output_____"
]
],
[
[
"Now let's look at how well this randomly initialized policy network performed:",
"_____no_output_____"
]
],
[
[
"video = plot_animation(frames)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Yeah... pretty bad. The neural network will have to learn to do better. First let's see if it is capable of learning the basic policy we used earlier: go left if the pole is tilting left, and go right if it is tilting right. The following code defines the same neural network but we add the target probabilities `y`, and the training operations (`cross_entropy`, `optimizer` and `training_op`):",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom tensorflow.contrib.layers import fully_connected\n\ntf.reset_default_graph()\n\nn_inputs = 4\nn_hidden = 4\nn_outputs = 1\n\nlearning_rate = 0.01\n\ninitializer = tf.contrib.layers.variance_scaling_initializer()\n\nX = tf.placeholder(tf.float32, shape=[None, n_inputs])\ny = tf.placeholder(tf.float32, shape=[None, n_outputs])\n\nhidden = fully_connected(X, n_hidden, activation_fn=tf.nn.elu, weights_initializer=initializer)\nlogits = fully_connected(hidden, n_outputs, activation_fn=None)\noutputs = tf.nn.sigmoid(logits) # probability of action 0 (left)\np_left_and_right = tf.concat(concat_dim=1, values=[outputs, 1 - outputs])\naction = tf.multinomial(tf.log(p_left_and_right), num_samples=1)\n\ncross_entropy = tf.nn.sigmoid_cross_entropy_with_logits(logits, y)\noptimizer = tf.train.AdamOptimizer(learning_rate)\ntraining_op = optimizer.minimize(cross_entropy)\n\ninit = tf.initialize_all_variables()\nsaver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"We can make the same net play in 10 different environments in parallel, and train for 1000 iterations. We also reset environments when they are done.",
"_____no_output_____"
]
],
[
[
"n_environments = 10\nn_iterations = 1000\n\nenvs = [gym.make(\"CartPole-v0\") for _ in range(n_environments)]\nobservations = [env.reset() for env in envs]\n\nwith tf.Session() as sess:\n init.run()\n for iteration in range(n_iterations):\n target_probas = np.array([([1.] if obs[2] < 0 else [0.]) for obs in observations]) # if angle<0 we want proba(left)=1., or else proba(left)=0.\n action_val, _ = sess.run([action, training_op], feed_dict={X: np.array(observations), y: target_probas})\n for env_index, env in enumerate(envs):\n obs, reward, done, info = env.step(action_val[env_index][0])\n observations[env_index] = obs if not done else env.reset()\n saver.save(sess, \"my_policy_net_basic.ckpt\")\n\nfor env in envs:\n env.close()",
"INFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:07,450] Making new env: CartPole-v0\nINFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:07,459] Making new env: CartPole-v0\nINFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:07,469] Making new env: CartPole-v0\nINFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:07,480] Making new env: CartPole-v0\nINFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:07,489] Making new env: CartPole-v0\nINFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:07,499] Making new env: CartPole-v0\nINFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:07,508] Making new env: CartPole-v0\nINFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:07,518] Making new env: CartPole-v0\nINFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:07,527] Making new env: CartPole-v0\nINFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:07,537] Making new env: CartPole-v0\n"
],
[
"def render_policy_net(model_path, action, X, n_max_steps = 1000):\n frames = []\n env = gym.make(\"CartPole-v0\")\n obs = env.reset()\n with tf.Session() as sess:\n saver.restore(sess, model_path)\n for step in range(n_max_steps):\n img = render_cart_pole(env, obs)\n frames.append(img)\n action_val = action.eval(feed_dict={X: obs.reshape(1, n_inputs)})\n obs, reward, done, info = env.step(action_val[0][0])\n if done:\n break\n env.close()\n return frames ",
"_____no_output_____"
],
[
"frames = render_policy_net(\"my_policy_net_basic.ckpt\", action, X)\nvideo = plot_animation(frames)\nplt.show()",
"INFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:09,941] Making new env: CartPole-v0\n"
]
],
[
[
"Looks like it learned the policy correctly. Now let's see if it can learn a better policy on its own.",
"_____no_output_____"
],
[
"# Policy Gradients",
"_____no_output_____"
],
[
"To train this neural network we will need to define the target probabilities `y`. If an action is good we should increase its probability, and conversely if it is bad we should reduce it. But how do we know whether an action is good or bad? The problem is that most actions have delayed effects, so when you win or lose points in a game, it is not clear which actions contributed to this result: was it just the last action? Or the last 10? Or just one action 50 steps earlier? This is called the _credit assignment problem_.\n\nThe _Policy Gradients_ algorithm tackles this problem by first playing multiple games, then making the actions in good games slightly more likely, while actions in bad games are made slightly less likely. First we play, then we go back and think about what we did.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom tensorflow.contrib.layers import fully_connected\n\ntf.reset_default_graph()\n\nn_inputs = 4\nn_hidden = 4\nn_outputs = 1\n\nlearning_rate = 0.01\n\ninitializer = tf.contrib.layers.variance_scaling_initializer()\n\nX = tf.placeholder(tf.float32, shape=[None, n_inputs])\n\nhidden = fully_connected(X, n_hidden, activation_fn=tf.nn.elu, weights_initializer=initializer)\nlogits = fully_connected(hidden, n_outputs, activation_fn=None)\noutputs = tf.nn.sigmoid(logits) # probability of action 0 (left)\np_left_and_right = tf.concat(concat_dim=1, values=[outputs, 1 - outputs])\naction = tf.multinomial(tf.log(p_left_and_right), num_samples=1)\n\ny = 1. - tf.to_float(action)\ncross_entropy = tf.nn.sigmoid_cross_entropy_with_logits(logits, y)\noptimizer = tf.train.AdamOptimizer(learning_rate)\ngrads_and_vars = optimizer.compute_gradients(cross_entropy)\ngradients = [grad for grad, variable in grads_and_vars]\ngradient_placeholders = []\ngrads_and_vars_feed = []\nfor grad, variable in grads_and_vars:\n gradient_placeholder = tf.placeholder(tf.float32, shape=grad.get_shape())\n gradient_placeholders.append(gradient_placeholder)\n grads_and_vars_feed.append((gradient_placeholder, variable))\ntraining_op = optimizer.apply_gradients(grads_and_vars_feed)\n\ninit = tf.initialize_all_variables()\nsaver = tf.train.Saver()",
"_____no_output_____"
],
[
"def discount_rewards(rewards, discount_rate):\n discounted_rewards = np.zeros(len(rewards))\n cumulative_rewards = 0\n for step in reversed(range(len(rewards))):\n cumulative_rewards = rewards[step] + cumulative_rewards * discount_rate\n discounted_rewards[step] = cumulative_rewards\n return discounted_rewards\n\ndef discount_and_normalize_rewards(all_rewards, discount_rate):\n all_discounted_rewards = [discount_rewards(rewards, discount_rate) for rewards in all_rewards]\n flat_rewards = np.concatenate(all_discounted_rewards)\n reward_mean = flat_rewards.mean()\n reward_std = flat_rewards.std()\n return [(discounted_rewards - reward_mean)/reward_std for discounted_rewards in all_discounted_rewards]",
"_____no_output_____"
],
[
"discount_rewards([10, 0, -50], discount_rate=0.8)",
"_____no_output_____"
],
[
"discount_and_normalize_rewards([[10, 0, -50], [10, 20]], discount_rate=0.8)",
"_____no_output_____"
],
[
"env = gym.make(\"CartPole-v0\")\n\nn_games_per_update = 10\nn_max_steps = 1000\nn_iterations = 250\nsave_iterations = 10\ndiscount_rate = 0.95\n\nwith tf.Session() as sess:\n init.run()\n for iteration in range(n_iterations):\n print(\"\\r{}\\tTotal rewards: \".format(iteration), end=\"\")\n all_rewards = []\n all_gradients = []\n for game in range(n_games_per_update):\n current_rewards = []\n current_gradients = []\n obs = env.reset()\n for step in range(n_max_steps):\n action_val, gradients_val = sess.run([action, gradients], feed_dict={X: obs.reshape(1, n_inputs)})\n obs, reward, done, info = env.step(action_val[0][0])\n current_rewards.append(reward)\n current_gradients.append(gradients_val)\n if done:\n break\n all_rewards.append(current_rewards)\n all_gradients.append(current_gradients)\n print(np.sum(current_rewards), end=\" \")\n\n all_rewards = discount_and_normalize_rewards(all_rewards, discount_rate=discount_rate)\n feed_dict = {}\n for var_index, gradient_placeholder in enumerate(gradient_placeholders):\n mean_gradients = np.mean([reward * all_gradients[game_index][step][var_index]\n for game_index, rewards in enumerate(all_rewards)\n for step, reward in enumerate(rewards)], axis=0)\n feed_dict[gradient_placeholder] = mean_gradients\n sess.run(training_op, feed_dict=feed_dict)\n if iteration % save_iterations == 0:\n saver.save(sess, \"my_policy_net_pg.ckpt\")",
"INFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:21:27,628] Making new env: CartPole-v0\n"
],
[
"env.close()",
"_____no_output_____"
],
[
"frames = render_policy_net(\"my_policy_net_pg.ckpt\", action, X, n_max_steps=1000)\nvideo = plot_animation(frames)\nplt.show()",
"INFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 14:32:17,325] Making new env: CartPole-v0\n"
]
],
[
[
"# Markov Chains",
"_____no_output_____"
]
],
[
[
"transition_probabilities = [\n [0.7, 0.2, 0.0, 0.1], # from s0 to s0, s1, s2, s3\n [0.0, 0.0, 0.9, 0.1], # from s1 to ...\n [0.0, 1.0, 0.0, 0.0], # from s2 to ...\n [0.0, 0.0, 0.0, 1.0], # from s3 to ...\n ]\n\nn_max_steps = 50\n\ndef print_sequence(start_state=0):\n current_state = start_state\n print(\"States:\", end=\" \")\n for step in range(n_max_steps):\n print(current_state, end=\" \")\n if current_state == 3:\n break\n current_state = rnd.choice(range(4), p=transition_probabilities[current_state])\n else:\n print(\"...\", end=\"\")\n print()\n\nfor _ in range(10):\n print_sequence()",
"States: 0 0 3 \nStates: 0 1 2 1 2 1 2 1 2 1 3 \nStates: 0 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 3 \nStates: 0 3 \nStates: 0 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 3 \nStates: 0 1 3 \nStates: 0 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 ...\nStates: 0 0 3 \nStates: 0 0 0 1 2 1 2 1 3 \nStates: 0 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 3 \n"
]
],
[
[
"# Markov Decision Process",
"_____no_output_____"
]
],
[
[
"transition_probabilities = [\n [[0.7, 0.3, 0.0], [1.0, 0.0, 0.0], [0.8, 0.2, 0.0]], # in s0, if action a0 then proba 0.7 to state s0 and 0.3 to state s1, etc.\n [[0.0, 1.0, 0.0], None, [0.0, 0.0, 1.0]],\n [None, [0.8, 0.1, 0.1], None],\n ]\n\nrewards = [\n [[+10, 0, 0], [0, 0, 0], [0, 0, 0]],\n [[0, 0, 0], [0, 0, 0], [0, 0, -50]],\n [[0, 0, 0], [+40, 0, 0], [0, 0, 0]],\n ]\n\npossible_actions = [[0, 1, 2], [0, 2], [1]]\n\ndef policy_fire(state):\n return [0, 2, 1][state]\n\ndef policy_random(state):\n return rnd.choice(possible_actions[state])\n\ndef policy_safe(state):\n return [0, 0, 1][state]\n\nclass MDPEnvironment(object):\n def __init__(self, start_state=0):\n self.start_state=start_state\n self.reset()\n def reset(self):\n self.total_rewards = 0\n self.state = self.start_state\n def step(self, action):\n next_state = rnd.choice(range(3), p=transition_probabilities[self.state][action])\n reward = rewards[self.state][action][next_state]\n self.state = next_state\n self.total_rewards += reward\n return self.state, reward\n\ndef run_episode(policy, n_steps, start_state=0, display=True):\n env = MDPEnvironment()\n if display:\n print(\"States (+rewards):\", end=\" \")\n for step in range(n_steps):\n if display:\n if step == 10:\n print(\"...\", end=\" \")\n elif step < 10:\n print(env.state, end=\" \")\n action = policy(env.state)\n state, reward = env.step(action)\n if display and step < 10:\n if reward:\n print(\"({})\".format(reward), end=\" \")\n if display:\n print(\"Total rewards =\", env.total_rewards)\n return env.total_rewards\n\nfor policy in (policy_fire, policy_random, policy_safe):\n all_totals = []\n print(policy.__name__)\n for episode in range(1000):\n all_totals.append(run_episode(policy, n_steps=100, display=(episode<5)))\n print(\"Summary: mean={:.1f}, std={:1f}, min={}, max={}\".format(np.mean(all_totals), np.std(all_totals), np.min(all_totals), np.max(all_totals)))\n print()",
"policy_fire\nStates (+rewards): 0 (10) 0 (10) 0 1 (-50) 2 2 2 (40) 0 (10) 0 (10) 0 (10) ... Total rewards = 210\nStates (+rewards): 0 1 (-50) 2 (40) 0 (10) 0 (10) 0 1 (-50) 2 2 (40) 0 (10) ... Total rewards = 70\nStates (+rewards): 0 (10) 0 1 (-50) 2 (40) 0 (10) 0 (10) 0 (10) 0 (10) 0 (10) 0 (10) ... Total rewards = 70\nStates (+rewards): 0 1 (-50) 2 1 (-50) 2 (40) 0 (10) 0 1 (-50) 2 (40) 0 ... Total rewards = -10\nStates (+rewards): 0 1 (-50) 2 (40) 0 (10) 0 (10) 0 1 (-50) 2 (40) 0 (10) 0 (10) ... Total rewards = 290\nSummary: mean=121.1, std=129.333766, min=-330, max=470\n\npolicy_random\nStates (+rewards): 0 1 (-50) 2 1 (-50) 2 (40) 0 1 (-50) 2 2 (40) 0 ... Total rewards = -60\nStates (+rewards): 0 (10) 0 0 0 0 0 (10) 0 0 0 (10) 0 ... Total rewards = -30\nStates (+rewards): 0 1 1 (-50) 2 (40) 0 0 1 1 1 1 ... Total rewards = 10\nStates (+rewards): 0 (10) 0 (10) 0 0 0 0 1 (-50) 2 (40) 0 0 ... Total rewards = 0\nStates (+rewards): 0 0 (10) 0 1 (-50) 2 (40) 0 0 0 0 (10) 0 (10) ... Total rewards = 40\nSummary: mean=-22.1, std=88.152740, min=-380, max=200\n\npolicy_safe\nStates (+rewards): 0 1 1 1 1 1 1 1 1 1 ... Total rewards = 0\nStates (+rewards): 0 1 1 1 1 1 1 1 1 1 ... Total rewards = 0\nStates (+rewards): 0 (10) 0 (10) 0 (10) 0 1 1 1 1 1 1 ... Total rewards = 30\nStates (+rewards): 0 (10) 0 1 1 1 1 1 1 1 1 ... Total rewards = 10\nStates (+rewards): 0 1 1 1 1 1 1 1 1 1 ... Total rewards = 0\nSummary: mean=22.3, std=26.244312, min=0, max=170\n\n"
]
],
[
[
"# Q-Learning",
"_____no_output_____"
],
[
"Q-Learning will learn the optimal policy by watching the random policy play.",
"_____no_output_____"
]
],
[
[
"n_states = 3\nn_actions = 3\nn_steps = 20000\nalpha = 0.01\ngamma = 0.99\nexploration_policy = policy_random\nq_values = np.full((n_states, n_actions), -np.inf)\nfor state, actions in enumerate(possible_actions):\n q_values[state][actions]=0\n\nenv = MDPEnvironment()\nfor step in range(n_steps):\n action = exploration_policy(env.state)\n state = env.state\n next_state, reward = env.step(action)\n next_value = np.max(q_values[next_state]) # greedy policy\n q_values[state, action] = (1-alpha)*q_values[state, action] + alpha*(reward + gamma * next_value)",
"_____no_output_____"
],
[
"def optimal_policy(state):\n return np.argmax(q_values[state])",
"_____no_output_____"
],
[
"q_values",
"_____no_output_____"
],
[
"all_totals = []\nfor episode in range(1000):\n all_totals.append(run_episode(optimal_policy, n_steps=100, display=(episode<5)))\nprint(\"Summary: mean={:.1f}, std={:1f}, min={}, max={}\".format(np.mean(all_totals), np.std(all_totals), np.min(all_totals), np.max(all_totals)))\nprint()",
"States (+rewards): 0 (10) 0 (10) 0 1 (-50) 2 (40) 0 (10) 0 1 (-50) 2 (40) 0 (10) ... Total rewards = 230\nStates (+rewards): 0 (10) 0 (10) 0 (10) 0 1 (-50) 2 2 1 (-50) 2 (40) 0 (10) ... Total rewards = 90\nStates (+rewards): 0 1 (-50) 2 (40) 0 (10) 0 (10) 0 (10) 0 (10) 0 (10) 0 (10) 0 (10) ... Total rewards = 170\nStates (+rewards): 0 1 (-50) 2 (40) 0 (10) 0 (10) 0 (10) 0 (10) 0 (10) 0 (10) 0 (10) ... Total rewards = 220\nStates (+rewards): 0 1 (-50) 2 (40) 0 (10) 0 1 (-50) 2 (40) 0 (10) 0 (10) 0 (10) ... Total rewards = -50\nSummary: mean=125.6, std=127.363464, min=-290, max=500\n\n"
]
],
[
[
"# Learning to play MsPacman using Deep Q-Learning",
"_____no_output_____"
]
],
[
[
"env = gym.make(\"MsPacman-v0\")\nobs = env.reset()",
"INFO:gym.envs.registration:Making new env: MsPacman-v0\n[2016-10-23 14:33:23,403] Making new env: MsPacman-v0\n"
],
[
"obs.shape",
"_____no_output_____"
],
[
"env.action_space",
"_____no_output_____"
]
],
[
[
"## Preprocessing",
"_____no_output_____"
],
[
"Preprocessing the images is optional but greatly speeds up training.",
"_____no_output_____"
]
],
[
[
"mspacman_color = np.array([210, 164, 74]).mean()\n\ndef preprocess_observation(obs):\n img = obs[1:176:2, ::2] # crop and downsize\n img = img.mean(axis=2) # to greyscale\n img[img==mspacman_color] = 0 # Improve contrast\n img = (img - 128) / 128 - 1 # normalize from -1. to 1.\n return img.reshape(88, 80, 1)\n\nimg = preprocess_observation(obs)",
"_____no_output_____"
],
[
"plt.figure(figsize=(11, 7))\nplt.subplot(121)\nplt.title(\"Original observation (160×210 RGB)\")\nplt.imshow(obs)\nplt.axis(\"off\")\nplt.subplot(122)\nplt.title(\"Preprocessed observation (88×80 greyscale)\")\nplt.imshow(img.reshape(88, 80), interpolation=\"nearest\", cmap=\"gray\")\nplt.axis(\"off\")\nsave_fig(\"preprocessing_plot\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Build DQN",
"_____no_output_____"
]
],
[
[
"tf.reset_default_graph()\n\nfrom tensorflow.contrib.layers import convolution2d, fully_connected\n\ninput_height = 88\ninput_width = 80\ninput_channels = 1\nconv_n_maps = [32, 64, 64]\nconv_kernel_sizes = [(8,8), (4,4), (3,3)]\nconv_strides = [4, 2, 1]\nconv_paddings = [\"SAME\"]*3 \nconv_activation = [tf.nn.relu]*3\nn_hidden_inputs = 64 * 11 * 10 # conv3 has 64 maps of 11x10 each\nn_hidden = 512\nhidden_activation = tf.nn.relu\nn_outputs = env.action_space.n\ninitializer = tf.contrib.layers.variance_scaling_initializer()\n\nlearning_rate = 0.01\n\ndef q_network(X_state, scope):\n prev_layer = X_state\n conv_layers = []\n with tf.variable_scope(scope) as scope:\n for n_maps, kernel_size, stride, padding, activation in zip(conv_n_maps, conv_kernel_sizes, conv_strides, conv_paddings, conv_activation):\n prev_layer = convolution2d(prev_layer, num_outputs=n_maps, kernel_size=kernel_size, stride=stride, padding=padding, activation_fn=activation, weights_initializer=initializer)\n conv_layers.append(prev_layer)\n last_conv_layer_flat = tf.reshape(prev_layer, shape=[-1, n_hidden_inputs])\n hidden = fully_connected(last_conv_layer_flat, n_hidden, activation_fn=hidden_activation, weights_initializer=initializer)\n outputs = fully_connected(hidden, n_outputs, activation_fn=None)\n trainable_vars = {var.name[len(scope.name):]: var for var in tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope=scope.name)}\n return outputs, trainable_vars\n\nX_state = tf.placeholder(tf.float32, shape=[None, input_height, input_width, input_channels])\nactor_q_values, actor_vars = q_network(X_state, scope=\"q_networks/actor\") # acts\ncritic_q_values, critic_vars = q_network(X_state, scope=\"q_networks/critic\") # learns\n\ncopy_ops = [actor_var.assign(critic_vars[var_name])\n for var_name, actor_var in actor_vars.items()]\ncopy_critic_to_actor = tf.group(*copy_ops)\n\nwith tf.variable_scope(\"train\"):\n X_action = tf.placeholder(tf.int32, shape=[None])\n y = tf.placeholder(tf.float32, shape=[None, 1])\n q_value = tf.reduce_sum(critic_q_values * tf.one_hot(X_action, n_outputs),\n reduction_indices=1, keep_dims=True)\n cost = tf.reduce_mean(tf.square(y - q_value))\n global_step = tf.Variable(0, trainable=False, name='global_step')\n optimizer = tf.train.AdamOptimizer(learning_rate)\n training_op = optimizer.minimize(cost, global_step=global_step)\n \ninit = tf.initialize_all_variables()\nsaver = tf.train.Saver()",
"_____no_output_____"
],
[
"actor_vars",
"_____no_output_____"
],
[
"from collections import deque\n\nreplay_memory_size = 10000\nreplay_memory = deque([], maxlen=replay_memory_size)\n\ndef sample_memories(batch_size):\n indices = rnd.permutation(len(replay_memory))[:batch_size]\n cols = [[], [], [], [], []] # state, action, reward, next_state, continue\n for idx in indices:\n memory = replay_memory[idx]\n for col, value in zip(cols, memory):\n col.append(value)\n cols = [np.array(col) for col in cols]\n return cols[0], cols[1], cols[2].reshape(-1, 1), cols[3], cols[4].reshape(-1, 1)",
"_____no_output_____"
],
[
"eps_min = 0.05\neps_max = 1.0\neps_decay_steps = 50000\nimport sys\n\ndef epsilon_greedy(q_values, step):\n epsilon = max(eps_min, eps_max - (eps_max-eps_min) * step/eps_decay_steps)\n print(\" epsilon {}\".format(epsilon), end=\"\")\n sys.stdout.flush()\n if rnd.rand() < epsilon:\n return rnd.randint(n_outputs) # random action\n else:\n return np.argmax(q_values) # optimal action",
"_____no_output_____"
],
[
"n_steps = 100000 # total number of training steps\ntraining_start = 1000 # start training after 1,000 game iterations\ntraining_interval = 3 # run a training step every 3 game iterations\nsave_steps = 50 # save the model every 50 training steps\ncopy_steps = 25 # copy the critic to the actor every 25 training steps\ndiscount_rate = 0.95\nskip_start = 90 # Skip the start of every game (it's just waiting time).\nbatch_size = 50\niteration = 0 # game iterations\ncheckpoint_path = \"my_dqn.ckpt\"\ndone = True # env needs to be reset\n\nwith tf.Session() as sess:\n if os.path.isfile(checkpoint_path):\n saver.restore(sess, checkpoint_path)\n else:\n init.run()\n while True:\n step = global_step.eval()\n if step >= n_steps:\n break\n iteration += 1\n print(\"\\rIteration {}\\tTraining step {}/{} ({:.1f}%)\".format(iteration, step, n_steps, step * 100 / n_steps), end=\"\")\n if done: # game over, start again\n obs = env.reset()\n for skip in range(skip_start): # skip boring game iterations at the start of each game\n obs, reward, done, info = env.step(0)\n state = preprocess_observation(obs)\n\n # Actor evaluates what to do\n q_values = actor_q_values.eval(feed_dict={X_state: [state]})\n action = epsilon_greedy(q_values, step)\n\n # Actor plays\n obs, reward, done, info = env.step(action)\n next_state = preprocess_observation(obs)\n\n # Let's memorize what happened\n replay_memory.append((state, action, reward, next_state, 1.0 - done))\n state = next_state\n\n if iteration < training_start or iteration % training_interval != 0:\n continue\n \n # Critic learns\n X_state_val, X_action_val, rewards, X_next_state_val, continues = sample_memories(batch_size)\n next_q_values = actor_q_values.eval(feed_dict={X_state: X_next_state_val})\n y_val = rewards + continues * discount_rate * np.max(next_q_values, axis=1, keepdims=True)\n training_op.run(feed_dict={X_state: X_state_val, X_action: X_action_val, y: y_val})\n\n # Regularly copy critic to actor\n if step % copy_steps == 0:\n copy_critic_to_actor.run()\n\n # And save regularly\n if step % save_steps == 0:\n saver.save(sess, checkpoint_path)\n",
"Iteration 7695\tTraining step 3499/3500 (100.0%) epsilon 0.933519"
]
],
[
[
"## DQN for the Cart-Pole",
"_____no_output_____"
]
],
[
[
"eps_min = 0.1\neps_max = 1.0\neps_decay_steps = 20000\nimport sys\n\ndef epsilon_greedy(q_values, step):\n epsilon = max(eps_min, eps_max - (eps_max-eps_min) * step/eps_decay_steps)\n print(\" epsilon {}\".format(epsilon), end=\"\")\n sys.stdout.flush()\n if rnd.rand() < epsilon:\n return rnd.randint(n_outputs) # random action\n else:\n return np.argmax(q_values) # optimal action",
"_____no_output_____"
],
[
"import tensorflow as tf\nfrom tensorflow.contrib.layers import fully_connected\n\ntf.reset_default_graph()\n\nn_inputs = 4\nn_hidden = 4\nn_outputs = 2\n\nlearning_rate = 0.01\n\ninitializer = tf.contrib.layers.variance_scaling_initializer()\n\ndef q_network(X_state, scope):\n with tf.variable_scope(scope) as scope:\n hidden = fully_connected(X_state, n_hidden, activation_fn=tf.nn.elu, weights_initializer=initializer)\n outputs = fully_connected(hidden, n_outputs, activation_fn=None)\n trainable_vars = {var.name[len(scope.name):]: var for var in tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope=scope.name)}\n return outputs, trainable_vars \n\nX_state = tf.placeholder(tf.float32, shape=[None, n_inputs])\nactor_q_values, actor_vars = q_network(X_state, scope=\"q_networks/actor\") # acts\ncritic_q_values, critic_vars = q_network(X_state, scope=\"q_networks/critic\") # learns\n\ncopy_ops = [actor_var.assign(critic_vars[var_name])\n for var_name, actor_var in actor_vars.items()]\ncopy_critic_to_actor = tf.group(*copy_ops)\n\nwith tf.variable_scope(\"train\"):\n X_action = tf.placeholder(tf.int32, shape=[None])\n y = tf.placeholder(tf.float32, shape=[None, 1])\n q_value = tf.reduce_sum(critic_q_values * tf.one_hot(X_action, n_outputs),\n reduction_indices=1, keep_dims=True)\n cost = tf.reduce_mean(tf.square(y - q_value))\n global_step = tf.Variable(0, trainable=False, name='global_step')\n optimizer = tf.train.AdamOptimizer(learning_rate)\n training_op = optimizer.minimize(cost, global_step=global_step)\n \ninit = tf.initialize_all_variables()\nsaver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_steps = 50000 # total number of training steps\ntraining_start = 1000 # start training after 1,000 game iterations\ntraining_interval = 3 # run a training step every 3 game iterations\nsave_steps = 50 # save the model every 50 training steps\ncopy_steps = 25 # copy the critic to the actor every 25 training steps\ndiscount_rate = 0.95\nbatch_size = 50\niteration = 0 # game iterations\ncheckpoint_path = \"my_dqn.ckpt\"\ndone = True # env needs to be reset\n\nenv = gym.make(\"CartPole-v0\")\n\nreplay_memory.clear()\n\nwith tf.Session() as sess:\n if os.path.isfile(checkpoint_path):\n saver.restore(sess, checkpoint_path)\n else:\n init.run()\n while True:\n step = global_step.eval()\n if step >= n_steps:\n break\n iteration += 1\n print(\"\\rIteration {}\\tTraining step {}/{} ({:.1f}%)\".format(iteration, step, n_steps, step * 100 / n_steps), end=\"\")\n if done: # game over, start again\n obs = env.reset()\n state = obs\n\n # Actor evaluates what to do\n q_values = actor_q_values.eval(feed_dict={X_state: [state]})\n action = epsilon_greedy(q_values, step)\n\n # Actor plays\n obs, reward, done, info = env.step(action)\n next_state = obs\n\n # Let's memorize what happened\n replay_memory.append((state, action, reward, next_state, 1.0 - done))\n state = next_state\n\n if iteration < training_start or iteration % training_interval != 0:\n continue\n \n # Critic learns\n X_state_val, X_action_val, rewards, X_next_state_val, continues = sample_memories(batch_size)\n next_q_values = actor_q_values.eval(feed_dict={X_state: X_next_state_val})\n y_val = rewards + continues * discount_rate * np.max(next_q_values, axis=1, keepdims=True)\n training_op.run(feed_dict={X_state: X_state_val, X_action: X_action_val, y: y_val})\n\n # Regularly copy critic to actor\n if step % copy_steps == 0:\n copy_critic_to_actor.run()\n\n # And save regularly\n if step % save_steps == 0:\n saver.save(sess, checkpoint_path)\n",
"INFO:gym.envs.registration:Making new env: CartPole-v0\n[2016-10-23 15:08:49,678] Making new env: CartPole-v0\n"
],
[
"n_max_steps = 1000\n\nframes = []\nobs = env.reset()\nwith tf.Session() as sess:\n saver.restore(sess, checkpoint_path)\n for step in range(n_max_steps):\n img = render_cart_pole(env, obs)\n frames.append(img)\n actor_q_values_val = actor_q_values.eval(feed_dict={X_state: obs.reshape(1, n_inputs)})\n action_val = np.argmax(actor_q_values_val)\n obs, reward, done, info = env.step(action_val)\n if done:\n break",
"_____no_output_____"
],
[
"len(frames)",
"_____no_output_____"
],
[
"video = plot_animation(frames)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Exercise solutions",
"_____no_output_____"
],
[
"Coming soon...",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ecf1a46179ff598c75ef8d945d6764c3558f0d21 | 7,107 | ipynb | Jupyter Notebook | Jupyter Books/IngestTickers.ipynb | rriley99/TechnicalAnalysis | 61b178d338bcbb809fb3001f6e80ac26933be0cf | [
"MIT"
] | 1 | 2021-01-04T00:16:10.000Z | 2021-01-04T00:16:10.000Z | Jupyter Books/IngestTickers.ipynb | rriley99/TechnicalAnalysis | 61b178d338bcbb809fb3001f6e80ac26933be0cf | [
"MIT"
] | null | null | null | Jupyter Books/IngestTickers.ipynb | rriley99/TechnicalAnalysis | 61b178d338bcbb809fb3001f6e80ac26933be0cf | [
"MIT"
] | 1 | 2021-01-29T12:08:43.000Z | 2021-01-29T12:08:43.000Z | 36.634021 | 1,219 | 0.562966 | [
[
[
"\"\"\" Purpose of this program is to load ticker/quote data into a local postgres database, eventually this may\nbe expanded to something like CosmosDB. Added bool columns for S&P, NASDAQ and DOW.\n\"\"\"\nimport psycopg2\nimport pandas as pd\nimport yahoo_fin.stock_info as si\nimport time",
"_____no_output_____"
],
[
"conn = psycopg2.connect(\"dbname=StonksGoUp user=postgres host=localhost password=admin\")\ncur = conn.cursor()\ndata = si.get_data('msft')\nSQL_stockdata = \"\"\" \n CREATE TABLE stockdata (\n ticker varchar(5) NOT NULL,\n quotedate date NOT NULL,\n open numeric NOT NULL,\n high numeric NOT NULL,\n low numeric NOT NULL,\n close numeric NOT NULL,\n adjclose numeric NOT NULL,\n volume bigint,\n CONSTRAINT pk_stockdata PRIMARY KEY (ticker, quotedate)\n);\n\"\"\"\nSQL_tickers = \"\"\"\n CREATE TABLE tickers (\n ticker varchar(10) NOT NULL,\n isdow boolean NOT NULL DEFAULT false,\n isnasdaq boolean NOT NULL DEFAULT false,\n issp500 boolean NOT NULL DEFAULT false,\n createddate timestamp with time zone NOT NULL DEFAULT now(),\n CONSTRAINT pk_tickers PRIMARY KEY (ticker)\n );\n\"\"\"\n# cur.execute(SQL_stockdata, conn)\n# cur.execute(SQL_tickers, conn)\n# conn.commit()\n\n\n",
"_____no_output_____"
],
[
"\"\"\"INSERT data into the tickers reference table\"\"\"\n\ndf_ticker = pd.DataFrame()\nnasdaq = si.tickers_nasdaq()\nsp500 = si.tickers_sp500()\ndow = si.tickers_dow()\nother = si.tickers_other()\n\ntickers = list(set([*nasdaq, *sp500, *dow, *other]))\n\ndf_ticker['ticker'] = tickers\ndf_ticker.replace(\"\", float(\"NaN\"), inplace=True)\ndf_ticker.dropna(subset = [\"ticker\"], inplace=True)\n\ndf_ticker['isdow'] = df_ticker['ticker'].isin(dow)\ndf_ticker['issp500'] = df_ticker['ticker'].isin(sp500)\ndf_ticker['isnasdaq'] = df_ticker['ticker'].isin(nasdaq)\n\ninsert = [list(row) for row in df_ticker.itertuples(index=False)]\n\nSQL_Ticker_insert= \"\"\" INSERT INTO public.tickers(ticker,isdow, isnasdaq, issp500) VALUES (%s,%s,%s,%s) ON CONFLICT DO NOTHING\"\"\"\ncur.executemany(SQL_Ticker_insert, insert)\nconn.commit()\n\nprint(f'{cur.rowcount} rows inserted.')",
"_____no_output_____"
],
[
"\"\"\"INSERT quote data into stockdata table\"\"\"\n\nSQL_stockdata_insert = \"\"\"INSERT INTO public.stockdata (quotedate, open, high, low, close, adjclose, volume, ticker)\n VALUES(%s,%s,%s,%s,%s,%s,%s,%s) ON CONFLICT DO NOTHING\"\"\"\nfor ticker in df_ticker['ticker']:\n time.sleep(3)\n print(ticker)\n try: \n data = si.get_data(ticker)\n insert_data = [list(row) for row in data.itertuples()]\n cur.executemany(SQL_stockdata_insert, insert_data)\n except: pass\n conn.commit()",
"_____no_output_____"
],
[
"cur.close()\nconn.close()",
"_____no_output_____"
],
[
"tables = ('tickers', 'stocks')\nSQL_check = f\"\"\"\n SELECT tablename\n FROM pg_catalog.pg_tables\n WHERE schemaname != 'pg_catalog' \n AND schemaname != 'information_schema'\n AND tablename IN {tables};\n \"\"\"\n\ncur.execute(SQL_check, conn)\n#tables_returned = cur.fetchall()\ntables_returned = [x[0] for x in cur.fetchall()]\nprint(tables_returned)\n#tables_returned - tables\nlist_diff = [i for i in tables + tables_returned if i not in tables or i not in tables_returned]\nprint(list_diff)",
"['tickers']\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf1b9e6d04fb41d0cdeb5bb9bb4649a9c16a019 | 142,726 | ipynb | Jupyter Notebook | 08-machine_learning_jupyter/.ipynb_checkpoints/plot_iris_dataset-checkpoint.ipynb | iproduct/coulse-ml | 65577fd4202630d3d5cb6333ddc51cede750fb5a | [
"Apache-2.0"
] | 1 | 2020-10-02T15:48:42.000Z | 2020-10-02T15:48:42.000Z | 08-machine_learning_jupyter/.ipynb_checkpoints/plot_iris_dataset-checkpoint.ipynb | iproduct/coulse-ml | 65577fd4202630d3d5cb6333ddc51cede750fb5a | [
"Apache-2.0"
] | null | null | null | 08-machine_learning_jupyter/.ipynb_checkpoints/plot_iris_dataset-checkpoint.ipynb | iproduct/coulse-ml | 65577fd4202630d3d5cb6333ddc51cede750fb5a | [
"Apache-2.0"
] | null | null | null | 1,005.112676 | 104,024 | 0.958031 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n# The Iris Dataset\n\nThis data sets consists of 3 different types of irises'\n(Setosa, Versicolour, and Virginica) petal and sepal\nlength, stored in a 150x4 numpy.ndarray\n\nThe rows being the samples and the columns being:\nSepal Length, Sepal Width, Petal Length and Petal Width.\n\nThe below plot uses the first two features.\nSee `here <https://en.wikipedia.org/wiki/Iris_flower_data_set>`_ for more\ninformation on this dataset.\n",
"_____no_output_____"
]
],
[
[
"print(__doc__)\n\n\n# Code source: Gaël Varoquaux\n# Modified for documentation by Jaques Grobler\n# License: BSD 3 clause\n\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.mplot3d import Axes3D\nfrom sklearn import datasets\nfrom sklearn.decomposition import PCA\n\n# import some data to play with\niris = datasets.load_iris()\nX = iris.data[:, :2] # we only take the first two features.\ny = iris.target\n\nx_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5\ny_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5\n\nplt.figure(2, figsize=(8, 6))\nplt.clf()\n\n# Plot the training points\nplt.scatter(X[:, 0], X[:, 1], c=y, cmap=plt.cm.Set1,\n edgecolor='k')\nplt.xlabel('Sepal length')\nplt.ylabel('Sepal width')\n\nplt.xlim(x_min, x_max)\nplt.ylim(y_min, y_max)\nplt.xticks(())\nplt.yticks(())\n\n# To getter a better understanding of interaction of the dimensions\n# plot the first three PCA dimensions\nfig = plt.figure(1, figsize=(8, 6))\nax = Axes3D(fig, elev=-150, azim=110)\nX_reduced = PCA(n_components=3).fit_transform(iris.data)\nax.scatter(X_reduced[:, 0], X_reduced[:, 1], X_reduced[:, 2], c=y,\n cmap=plt.cm.Set1, edgecolor='k', s=40)\nax.set_title(\"First three PCA directions\")\nax.set_xlabel(\"1st eigenvector\")\nax.w_xaxis.set_ticklabels([])\nax.set_ylabel(\"2nd eigenvector\")\nax.w_yaxis.set_ticklabels([])\nax.set_zlabel(\"3rd eigenvector\")\nax.w_zaxis.set_ticklabels([])\n\nplt.show()",
"Automatically created module for IPython interactive environment\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf1bbc1e84e40dffcc9ad949b847113e16ba2a9 | 2,543 | ipynb | Jupyter Notebook | Teals/2019 Projects/Spring 2019 TEALS Project Student Guide/Week 3/ProjectConclusions.ipynb | Bhaskers-Blu-Org2/k12 | 9a30499107e65760376a9a13ec8e72b0451ec538 | [
"MIT"
] | 3 | 2020-05-09T10:10:01.000Z | 2020-09-13T02:21:55.000Z | Teals/2019 Projects/Spring 2019 TEALS Project Student Guide/Week 3/ProjectConclusions.ipynb | microsoft/k12 | d84b9d57444fa91238c44720891811aa2e0c7ed0 | [
"MIT"
] | null | null | null | Teals/2019 Projects/Spring 2019 TEALS Project Student Guide/Week 3/ProjectConclusions.ipynb | microsoft/k12 | d84b9d57444fa91238c44720891811aa2e0c7ed0 | [
"MIT"
] | 4 | 2020-06-30T16:16:36.000Z | 2020-11-26T02:38:28.000Z | 79.46875 | 1,947 | 0.71569 | [
[
[
"# Cognitive Hackathon: Project Conclusions\n\n## Overview\nProject summaries are created for team presentations.\n\n## Objectives\nWith your team, create a summary of your project, including data inputs and JSON outputs and interpretation, as well as your conclusions. Put results into your notebooks to use for presentations next week. \n\nSummaries should answer these questions:\n\n* Project Goals\n* Cognitive Services\n* Data Sets\n* Project Output and Result\n* Conclusions\n* New Directions\n\nConclusions shouls reflect the feedback form the team. As a guide, here are detailed descriptions of suggested questions to answer in your project conclusions:\n\n### Project Goals\nWhat is the goal of the project?\nWho would benefit from this idea? How?\nWhat are the challenges?\n\n### Cognitive Services\nHow will cognitive services help make the project a reality?\nAre the provided cognitive services sufficient to meet the projects requirements? If so then how? In not then what more is needed?\nWhat are the important aspects of the cognitive analysis that must work in order for the project to succeed?\nWhat are some of the risks in using cognitive services for this project?\n\n### Data Sets\nWhat data sets (images, text) would be necessary to make this project a success?\nWhat will be the sources of that data and how will it be gathered?\n\n### Project Output and Result\nWhat are the outputs of this project? Decisions, analysis, data, etc.\nWho will use this output? How?\nWhat would constitute useful output?\n\n### Conclusions\nWhat were the successes in this project? What were the challenges?\nWhat would be the best case scenario for the success of this project? How would it affect the world?\n\n### New Directions\nWhat were some of the best ideas that came up for other projects?\nWould those also use cognitive services or something else?\n\n## Summary\nProject conclusions were formulated by each team.",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
ecf1c0b7f985a4ec9446c0dd9e19f7f869011cac | 94,703 | ipynb | Jupyter Notebook | post_submission_Mon_Mar_30_PT5_DS_Sprint5_SC_linear_models.ipynb | LambdaTheda/DS-Unit-2-Linear-Models | cc910e08b45bab7ac4a5cd06c59646203422d190 | [
"MIT"
] | null | null | null | post_submission_Mon_Mar_30_PT5_DS_Sprint5_SC_linear_models.ipynb | LambdaTheda/DS-Unit-2-Linear-Models | cc910e08b45bab7ac4a5cd06c59646203422d190 | [
"MIT"
] | null | null | null | post_submission_Mon_Mar_30_PT5_DS_Sprint5_SC_linear_models.ipynb | LambdaTheda/DS-Unit-2-Linear-Models | cc910e08b45bab7ac4a5cd06c59646203422d190 | [
"MIT"
] | null | null | null | 35.913159 | 630 | 0.388911 | [
[
[
"<a href=\"https://colab.research.google.com/github/LambdaTheda/DS-Unit-2-Linear-Models/blob/master/post_submission_Mon_Mar_30_PT5_DS_Sprint5_SC_linear_models.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"_Lambda School Data Science, Unit 2_\n \n# Linear Models Sprint Challenge\n\nTo demonstrate mastery on your Sprint Challenge, do all the required, numbered instructions in this notebook.\n\nTo earn a score of \"3\", also do all the stretch goals.\n\nYou are permitted and encouraged to do as much data exploration as you want.",
"_____no_output_____"
],
[
"### Part 1, Classification\n- 1.1. Do train/test split. Arrange data into X features matrix and y target vector\n- 1.2. Use scikit-learn to fit a logistic regression model\n- 1.3. Report classification metric: accuracy\n\n### Part 2, Regression\n- 2.1. Begin with baselines for regression\n- 2.2. Do train/validate/test split\n- 2.3. Arrange data into X features matrix and y target vector\n- 2.4. Do one-hot encoding\n- 2.5. Use scikit-learn to fit a linear regression or ridge regression model\n- 2.6. Report validation MAE and $R^2$\n\n### Stretch Goals, Regression\n- Make at least 2 visualizations to explore relationships between features and target. You may use any visualization library\n- Try at least 3 feature combinations. You may select features manually, or automatically\n- Report validation MAE and $R^2$ for each feature combination you try\n- Report test MAE and $R^2$ for your final model\n- Print or plot the coefficients for the features in your model",
"_____no_output_____"
]
],
[
[
"# If you're in Colab...\nimport sys\nif 'google.colab' in sys.modules:\n !pip install category_encoders==2.*\n !pip install pandas-profiling==2.*\n !pip install plotly==4.*",
"_____no_output_____"
]
],
[
[
"# Part 1, Classification: Predict Blood Donations 🚑\nOur dataset is from a mobile blood donation vehicle in Taiwan. The Blood Transfusion Service Center drives to different universities and collects blood as part of a blood drive.\n\nThe goal is to predict whether the donor made a donation in March 2007, using information about each donor's history.\n\nGood data-driven systems for tracking and predicting donations and supply needs can improve the entire supply chain, making sure that more patients get the blood transfusions they need.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ndonors = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/blood-transfusion/transfusion.data')\nassert donors.shape == (748,5)\n\ndonors = donors.rename(columns={\n 'Recency (months)': 'months_since_last_donation', \n 'Frequency (times)': 'number_of_donations', \n 'Monetary (c.c. blood)': 'total_volume_donated', \n 'Time (months)': 'months_since_first_donation', \n 'whether he/she donated blood in March 2007': 'made_donation_in_march_2007'\n})",
"_____no_output_____"
]
],
[
[
"Notice that the majority class (did not donate blood in March 2007) occurs about 3/4 of the time. \n\nThis is the accuracy score for the \"majority class baseline\" (the accuracy score we'd get by just guessing the majority class every time).",
"_____no_output_____"
]
],
[
[
"# Accuracy score for the \"majority class baseline\"\ndonors['made_donation_in_march_2007'].value_counts(normalize=True)",
"_____no_output_____"
]
],
[
[
"## 1.1. Do train/test split. Arrange data into X features matrix and y target vector\n\nDo these steps in either order.\n\nUse scikit-learn's train/test split function to split randomly. (You can include 75% of the data in the train set, and hold out 25% for the test set, which is the default.)",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nX = donors.drop(columns = 'made_donation_in_march_2007') # features matrix; drop target column\ny = donors.made_donation_in_march_2007 # target array\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state = 43)\n\n#(You can include 75% of the data in the train set, and hold out 25% for the test set, which is the default.)\ntrain_test_split(X, y, random_state = 43, train_size = 0.75)",
"_____no_output_____"
]
],
[
[
"### 1.1a) Exploratory Data Analysis",
"_____no_output_____"
]
],
[
[
"donors.head()",
"_____no_output_____"
],
[
"donors.describe()",
"_____no_output_____"
]
],
[
[
"## 1.2. Use scikit-learn to fit a logistic regression model\n\nYou may use any number of features",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade category_encoders\nimport category_encoders as ce\n\nfrom sklearn.linear_model import LogisticRegressionCV\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.impute import SimpleImputer\n\ntarget = 'made_donation_in_march_2007'\nfeatures = donors.columns.drop([target]) # can just be donors.columns? Since dropped in 1.1?\n\n# Set validation X and y values\nX_val = X_test\ny_val = y_test\n\nprint(X_train.shape, X_val.shape)\n#-----------------------\n\n# One Hot Encode categorical data into numerical for model\nencoder = ce.OneHotEncoder(use_cat_names=True)\n\nX_train_encoded = encoder.fit_transform(X_train)\nX_val_encoded = encoder.transform(X_val)\n\nprint(X_train_encoded.shape, X_val_encoded.shape)\n#-----------------------\n\n# Impute missing values using mean\nimputer = SimpleImputer(strategy = 'mean')\nX_train_imputed = imputer.fit_transform(X_train_encoded)\nX_val_imputed = imputer.transform(X_val_encoded)\n#-----------------------\n\n# Standardize units\nscaler = StandardScaler() # instantiate\nX_train_scaled = scaler.fit_transform(X_train_imputed)\nX_val_scaled = scaler.transform(X_val_imputed)\n#-----------------------\n\n# Fit model\nmodel = LogisticRegressionCV(cv = 5, n_jobs = -1, random_state = 42)\nmodel.fit(X_train_scaled, y_train)\n\n",
"_____no_output_____"
]
],
[
[
"## 1.3. Report classification metric: accuracy\n\nWhat is your model's accuracy on the test set?\n\nDon't worry if your model doesn't beat the majority class baseline. That's okay!\n\n_\"The combination of some data and an aching desire for an answer does not ensure that a reasonable answer can be extracted from a given body of data.\"_ —[John Tukey](https://en.wikiquote.org/wiki/John_Tukey)\n\n(Also, if we used recall score instead of accuracy score, then your model would almost certainly beat the baseline. We'll discuss how to choose and interpret evaluation metrics throughout this unit.)\n",
"_____no_output_____"
]
],
[
[
"print('Validation Accuracy: ', model.score(X_val_scaled, y_val))",
"Validation Accuracy: 0.8021390374331551\n"
]
],
[
[
"# Part 2, Regression: Predict home prices in Ames, Iowa 🏠\n\nYou'll use historical housing data. ***There's a data dictionary at the bottom of the notebook.*** \n\nRun this code cell to load the dataset:\n\n\n\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nURL = 'https://drive.google.com/uc?export=download&id=1522WlEW6HFss36roD_Cd9nybqSuiVcCK'\nhomes = pd.read_csv(URL)\nassert homes.shape == (2904, 47)",
"_____no_output_____"
]
],
[
[
"## 2.1. Begin with baselines\n\nWhat is the Mean Absolute Error and R^2 score for a mean baseline? (You can get these estimated scores using all your data, before splitting it.)",
"_____no_output_____"
],
[
"### 2.1a) Exploratory Data Analysis",
"_____no_output_____"
]
],
[
[
"homes.describe()",
"_____no_output_____"
],
[
"homes.head()",
"_____no_output_____"
]
],
[
[
"### 2.1b) Common Regression baseline model: guessing the mean for every sample\n",
"_____no_output_____"
]
],
[
[
"guess = homes['SalePrice'].mean()\nerrors = guess - homes['SalePrice']\nerrors",
"_____no_output_____"
],
[
"from sklearn.metrics import mean_absolute_error, r2_score\n\n# MAE\nmean_absolute_error = errors.abs().mean()\nmean_absolute_error",
"_____no_output_____"
],
[
"# R^2 Score\nr2_score(homes['SalePrice'], errors)",
"_____no_output_____"
]
],
[
[
"## 2.2. Do train/validate/test split\n\nTrain on houses sold in the years 2006 - 2008. (1,920 rows)\n\nValidate on house sold in 2009. (644 rows)\n\nTest on houses sold in 2010. (340 rows)",
"_____no_output_____"
],
[
"### 2.2a) Split Train dataset",
"_____no_output_____"
]
],
[
[
"# TRAIN on houses sold in the years 2006 - 2008. (1,920 rows)\n# Locate data between those dates, inclusive\nhomes.loc['2006-01-01' : '2008-31-12']",
"_____no_output_____"
],
[
"# Filter train data\ntrain = homes[homes['Yr_Sold'].isin([2006, 2007, 2008])]\ntrain\ntrain.shape",
"_____no_output_____"
]
],
[
[
"### 2.2b) Split Validate dataset",
"_____no_output_____"
]
],
[
[
"#VALIDATE on house sold in 2009. (644 rows)\n#Filter test data\nval = homes[homes['Yr_Sold'] == 2009]\nval\nval.shape",
"_____no_output_____"
]
],
[
[
"### 2.2c) Split Test dataset",
"_____no_output_____"
]
],
[
[
"#TEST on houses sold in 2010. (340 rows)\n#Filter and set aside test data\ntest = homes[homes['Yr_Sold'] == 2010]\n#\ntest.shape",
"_____no_output_____"
]
],
[
[
"## 2.3. Arrange data into X features matrix and y target vector\n\nSelect at least one numeric feature and at least one categorical feature.\n\nOtherwise, you may choose whichever features and however many you want.",
"_____no_output_____"
]
],
[
[
"#y target vector\ntarget = 'SalePrice'\n\nprint(f'Our y target vector is the home\\'s {target}.')",
"Our y target vector is the home's SalePrice.\n"
],
[
"# Explore datatypes in homes dataframe\nhomes.dtypes",
"_____no_output_____"
],
[
"# Set X features matrix; select features columns to predict SalePrice with\nfeatures = ['Bldg_Type', 'Overall_Cond']\n\nprint(f' Our X features matrix consists of {len(features)} categorical and numerical features, aka columns, respectively: {features}.')",
" Our X features matrix consists of 2 categorical and numerical features, aka columns, respectively: ['Bldg_Type', 'Overall_Cond'].\n"
],
[
"# Set Xs and ys; split into train/validate/test datasets\nX_train = train[features]\ny_test = train[target]\n\nX_test = test[features]\ny_test = test[target]\n\nX_val = val[features]\ny_val = val[target]\n\nprint(f'X_train is {X_train.shape}') ",
"X_train is (1920, 2)\n"
],
[
"# print X_test.shape and y_test.shape\n'''\nin mar 30 ref Copy of Unit 2 TEST 1 - 5pm retake jan28 DS_Sprint_Challenge_Linear_Models.ipynb\nhttps://colab.research.google.com/drive/1fQhP8oRhEScOPWn2wr0-8SrEIuUsTgle#scrollTo=kvtcovmVpsQn\ngot X_test is (340, 2) and y_test is (340,); HERE, get X_test is (340, 6) and y_test is (340,)-\nDIFF # cols for X_test\n''' \nprint(f'X_test is {X_test.shape} and y_test is {y_test.shape}')",
"X_test is (340, 6) and y_test is (340,)\n"
]
],
[
[
"## 2.4. Do one-hot encoding\n\nEncode your categorical feature(s).",
"_____no_output_____"
]
],
[
[
"import category_encoders as ce\n\nencoder = ce.OneHotEncoder(use_cat_names=True)\n\nX_train_encoded = encoder.fit_transform(X_train)\nX_val_encoded = encoder.transform(X_val)\nX_test_encoded = encoder.transform(X_test)\n\nprint(X_train_encoded.shape, X_val_encoded.shape, X_test_encoded.shape)",
"_____no_output_____"
],
[
"# Standardize\nscaler = StandardScaler() # instantiate\nX_train_scaled = scaler.fit_transform(X_train_imputed)\nX_val_scaled = scaler.transform(X_val_imputed)",
"_____no_output_____"
],
[
"y_train.shape",
"_____no_output_____"
],
[
"X_train_scaled.shape",
"_____no_output_____"
],
[
"X_train_scaled.dtype",
"_____no_output_____"
]
],
[
[
"## 2.5. Use scikit-learn to fit a linear regression or ridge regression model\nFit your model.",
"_____no_output_____"
]
],
[
[
"#ATTEMPT # 1 USING RIDGE REGRESSION\nfrom sklearn.linear_model import RidgeCV\nfrom sklearn.preprocessing import StandardScaler\n\nscaler = StandardScaler()\n\n#X_train_scaled = scaler.fit_transform(X_train_encoded)\nX_train_scaled.shape\n\n#X_val_scaled = scaler.transform(X_val_encoded) # NotFittedError: This StandardScaler instance is not fitted yet. Call 'fit' with appropriate arguments before using this estimator.\ny_train.shape\n\n#ATTEMPTED FIX for ridge.fit(X_train_scaled, y_train) #ValueError: Found input variables with inconsistent numbers of samples: [1920, 561]\n#y_train = homes[target] # using https://stackoverflow.com/questions/54146387/sklearn-valueerrorfound-input-variables-with-inconsistent-numbers-of-samples\n#y_train.head\n\nridge = RidgeCV()\nridge.fit(X_train_scaled, y_train) #ValueError: Found input variables with inconsistent numbers of samples: [1920, 561]",
"_____no_output_____"
],
[
"\n# ATTEMPT 1: Using LINEAR REGRESSION; Following the 5 step process \n\nimport numpy as np\n\n# 1) Import the appropriate estimator class from Scikit-Learn library\nfrom sklearn.linear_model import LinearRegression\n\n# 2) Instantiate this class\nmodel = LinearRegression()\n\n'''\n# 3) Arrange X features Matrix & y target vector\nX = X_train_encoded\ny = y_train\n\n# 3a) try to fix below: model.fit(X_train_encoded, y_train) #ValueError: Found input variables with inconsistent numbers of samples: [1920, 561]; because of transforming ?\n\n#X = np.asarray([X_train_encoded])\n#y = np.asarray([y_train])\n'''\n\n# 4) Fit the model\nmodel.fit(X_train_encoded, y_train) #ValueError: Found input variables with inconsistent numbers of samples: [1920, 561]; because of transforming ? DOUBT IT\n#model.fit(X_train_scaled, y_train) #ValueError: cannot copy sequence with size 1920 to array axis with dimension 6\n\n#5) Apply the model to new data\nX_test = X_test_encoded\ny_pred = model.predict(X_test) #ValueError: matmul: Input operand 1 has a mismatch in its core dimension 0, with gufunc signature (n?,k),(k,m?)->(n?,m?) (size 4 is different from 6)\n\n",
"_____no_output_____"
],
[
"\n# ATTEMPT 2: Using LINEAR REGRESSION\n\n# 1) Import the appropriate estimator class from Scikit-Learn library\nfrom sklearn.linear_model import LinearRegression\n\n# 2) Instantiate this class \nmodel = LinearRegression()\n",
"_____no_output_____"
],
[
"#2a) Reshape X to match y shape? from # from https://stackoverflow.com/questions/44181664/sklearn-valueerror-found-input-variables-with-inconsistent-numbers-of-samples\n#model.fit(X.reshape(561,), y_train) # ATTEMPT # 1 TO FIX using numpy arrays; AttributeError: 'DataFrame' object has no attribute 'reshape'\n\n#2b) ATTEMPT # 2 TO FIX using numpy arrays https://datascience.stackexchange.com/questions/20199/train-test-split-error-found-input-variables-with-inconsistent-numbers-of-sam\n# and https://docs.scipy.org/doc/numpy/reference/arrays.indexing.html\n\n'''To fix this error:\n\n1. Remove the extra list from inside of np.array() when defining X or remove the extra dimension afterwards with the following command: X = X.reshape(X.shape[1:]). Now, the shape of X will be (6, 29).\n2. Transpose X by running X = X.transpose() to get equal number of samples in X and Y. Now, the shape of X will be (29, 6) and the shape of Y will be (29,).\n\nX = X.reshape(X.reshape(X.shape[1:561])) #shape of X should be (1920, 6)? from Encoding Section above\n\nX = X.transpose() # Now the shape of X should be (6, 1920), and y will be 561? (shape of y_train from above cell)\n'''\n# 3) Fit the model\n# model.fit(X_train_encoded, y_train) \n# model.fit(X_train_scaled, y_train)\n\n",
"_____no_output_____"
]
],
[
[
"## 2.6. Report validation MAE and $R^2$\n\nWhat is your model's Mean Absolute Error and $R^2$ score on the validation set? (You are not graded on how high or low your validation scores are.)",
"_____no_output_____"
]
],
[
[
"# Linear Regression validation metrics for VALIDATION data: \n# NotFittedError: This LinearRegression instance is not fitted yet. Call 'fit' with appropriate arguments before using this estimator.\nfrom sklearn.metrics import mean_absolute_error, mean_squared_error, r2_score\n\ny_pred = model.predict(X_val_encoded)\n\nmse = mean_squared_error(y_val, y_pred) #Mean squared error\n\nrmse = np.sqrt(mse) #root mean squared Error\n\nmae = mean_absolute_error(y_val, y_pred) #mean absolute Error\n\nr2 = r2_score(y_val, y_pred) #r^2\n\nprint('Mean Squared Error:', mse)\nprint('Root Mean Squared Error:', rmse)\nprint('Mean Absolute Error:', mae)\nprint('R^2:', r2)",
"_____no_output_____"
]
],
[
[
"# Stretch Goals, Regression\n- Make at least 2 visualizations to explore relationships between features and target. You may use any visualization library\n- Try at least 3 feature combinations. You may select features manually, or automatically\n- Report validation MAE and $R^2$ for each feature combination you try\n- Report test MAE and $R^2$ for your final model\n- Print or plot the coefficients for the features in your model",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"# Data Dictionary \n\nHere's a description of the data fields:\n\n```\n1st_Flr_SF: First Floor square feet\n\nBedroom_AbvGr: Bedrooms above grade (does NOT include basement bedrooms)\n\nBldg_Type: Type of dwelling\n\t\t\n 1Fam\tSingle-family Detached\t\n 2FmCon\tTwo-family Conversion; originally built as one-family dwelling\n Duplx\tDuplex\n TwnhsE\tTownhouse End Unit\n TwnhsI\tTownhouse Inside Unit\n \nBsmt_Half_Bath: Basement half bathrooms\n\nBsmt_Full_Bath: Basement full bathrooms\n\nCentral_Air: Central air conditioning\n\n N\tNo\n Y\tYes\n\t\t\nCondition_1: Proximity to various conditions\n\t\n Artery\tAdjacent to arterial street\n Feedr\tAdjacent to feeder street\t\n Norm\tNormal\t\n RRNn\tWithin 200' of North-South Railroad\n RRAn\tAdjacent to North-South Railroad\n PosN\tNear positive off-site feature--park, greenbelt, etc.\n PosA\tAdjacent to postive off-site feature\n RRNe\tWithin 200' of East-West Railroad\n RRAe\tAdjacent to East-West Railroad\n\t\nCondition_2: Proximity to various conditions (if more than one is present)\n\t\t\n Artery\tAdjacent to arterial street\n Feedr\tAdjacent to feeder street\t\n Norm\tNormal\t\n RRNn\tWithin 200' of North-South Railroad\n RRAn\tAdjacent to North-South Railroad\n PosN\tNear positive off-site feature--park, greenbelt, etc.\n PosA\tAdjacent to postive off-site feature\n RRNe\tWithin 200' of East-West Railroad\n RRAe\tAdjacent to East-West Railroad\n \n Electrical: Electrical system\n\n SBrkr\tStandard Circuit Breakers & Romex\n FuseA\tFuse Box over 60 AMP and all Romex wiring (Average)\t\n FuseF\t60 AMP Fuse Box and mostly Romex wiring (Fair)\n FuseP\t60 AMP Fuse Box and mostly knob & tube wiring (poor)\n Mix\tMixed\n \n Exter_Cond: Evaluates the present condition of the material on the exterior\n\t\t\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor\n \n Exter_Qual: Evaluates the quality of the material on the exterior \n\t\t\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor\n\t\t\nExterior_1st: Exterior covering on house\n\n AsbShng\tAsbestos Shingles\n AsphShn\tAsphalt Shingles\n BrkComm\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n CemntBd\tCement Board\n HdBoard\tHard Board\n ImStucc\tImitation Stucco\n MetalSd\tMetal Siding\n Other\tOther\n Plywood\tPlywood\n PreCast\tPreCast\t\n Stone\tStone\n Stucco\tStucco\n VinylSd\tVinyl Siding\n Wd Sdng\tWood Siding\n WdShing\tWood Shingles\n\t\nExterior_2nd: Exterior covering on house (if more than one material)\n\n AsbShng\tAsbestos Shingles\n AsphShn\tAsphalt Shingles\n BrkComm\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n CemntBd\tCement Board\n HdBoard\tHard Board\n ImStucc\tImitation Stucco\n MetalSd\tMetal Siding\n Other\tOther\n Plywood\tPlywood\n PreCast\tPreCast\n Stone\tStone\n Stucco\tStucco\n VinylSd\tVinyl Siding\n Wd Sdng\tWood Siding\n WdShing\tWood Shingles\n \nFoundation: Type of foundation\n\t\t\n BrkTil\tBrick & Tile\n CBlock\tCinder Block\n PConc\tPoured Contrete\t\n Slab\tSlab\n Stone\tStone\n Wood\tWood\n\t\t\nFull_Bath: Full bathrooms above grade\n\nFunctional: Home functionality (Assume typical unless deductions are warranted)\n\n Typ\tTypical Functionality\n Min1\tMinor Deductions 1\n Min2\tMinor Deductions 2\n Mod\tModerate Deductions\n Maj1\tMajor Deductions 1\n Maj2\tMajor Deductions 2\n Sev\tSeverely Damaged\n Sal\tSalvage only\n\t\t\nGr_Liv_Area: Above grade (ground) living area square feet\n \nHalf_Bath: Half baths above grade\n\nHeating: Type of heating\n\t\t\n Floor\tFloor Furnace\n GasA\tGas forced warm air furnace\n GasW\tGas hot water or steam heat\n Grav\tGravity furnace\t\n OthW\tHot water or steam heat other than gas\n Wall\tWall furnace\n\t\t\nHeating_QC: Heating quality and condition\n\n Ex\tExcellent\n Gd\tGood\n TA\tAverage/Typical\n Fa\tFair\n Po\tPoor\n\nHouse_Style: Style of dwelling\n\t\n 1Story\tOne story\n 1.5Fin\tOne and one-half story: 2nd level finished\n 1.5Unf\tOne and one-half story: 2nd level unfinished\n 2Story\tTwo story\n 2.5Fin\tTwo and one-half story: 2nd level finished\n 2.5Unf\tTwo and one-half story: 2nd level unfinished\n SFoyer\tSplit Foyer\n SLvl\tSplit Level\n\nKitchen_AbvGr: Kitchens above grade\n\nKitchen_Qual: Kitchen quality\n\n Ex\tExcellent\n Gd\tGood\n TA\tTypical/Average\n Fa\tFair\n Po\tPoor\n\nLandContour: Flatness of the property\n\n Lvl\tNear Flat/Level\t\n Bnk\tBanked - Quick and significant rise from street grade to building\n HLS\tHillside - Significant slope from side to side\n Low\tDepression\n\t\t\nLand_Slope: Slope of property\n\t\t\n Gtl\tGentle slope\n Mod\tModerate Slope\t\n Sev\tSevere Slope\n\nLot_Area: Lot size in square feet\n\nLot_Config: Lot configuration\n\n Inside\tInside lot\n Corner\tCorner lot\n CulDSac\tCul-de-sac\n FR2\tFrontage on 2 sides of property\n FR3\tFrontage on 3 sides of property\n\nLot_Shape: General shape of property\n\n Reg\tRegular\t\n IR1\tSlightly irregular\n IR2\tModerately Irregular\n IR3\tIrregular\n\nMS_SubClass: Identifies the type of dwelling involved in the sale.\t\n\n 20\t1-STORY 1946 & NEWER ALL STYLES\n 30\t1-STORY 1945 & OLDER\n 40\t1-STORY W/FINISHED ATTIC ALL AGES\n 45\t1-1/2 STORY - UNFINISHED ALL AGES\n 50\t1-1/2 STORY FINISHED ALL AGES\n 60\t2-STORY 1946 & NEWER\n 70\t2-STORY 1945 & OLDER\n 75\t2-1/2 STORY ALL AGES\n 80\tSPLIT OR MULTI-LEVEL\n 85\tSPLIT FOYER\n 90\tDUPLEX - ALL STYLES AND AGES\n 120\t1-STORY PUD (Planned Unit Development) - 1946 & NEWER\n 150\t1-1/2 STORY PUD - ALL AGES\n 160\t2-STORY PUD - 1946 & NEWER\n 180\tPUD - MULTILEVEL - INCL SPLIT LEV/FOYER\n 190\t2 FAMILY CONVERSION - ALL STYLES AND AGES\n\nMS_Zoning: Identifies the general zoning classification of the sale.\n\t\t\n A\tAgriculture\n C\tCommercial\n FV\tFloating Village Residential\n I\tIndustrial\n RH\tResidential High Density\n RL\tResidential Low Density\n RP\tResidential Low Density Park \n RM\tResidential Medium Density\n\nMas_Vnr_Type: Masonry veneer type\n\n BrkCmn\tBrick Common\n BrkFace\tBrick Face\n CBlock\tCinder Block\n None\tNone\n Stone\tStone\n\nMo_Sold: Month Sold (MM)\n\nNeighborhood: Physical locations within Ames city limits\n\n Blmngtn\tBloomington Heights\n Blueste\tBluestem\n BrDale\tBriardale\n BrkSide\tBrookside\n ClearCr\tClear Creek\n CollgCr\tCollege Creek\n Crawfor\tCrawford\n Edwards\tEdwards\n Gilbert\tGilbert\n IDOTRR\tIowa DOT and Rail Road\n MeadowV\tMeadow Village\n Mitchel\tMitchell\n Names\tNorth Ames\n NoRidge\tNorthridge\n NPkVill\tNorthpark Villa\n NridgHt\tNorthridge Heights\n NWAmes\tNorthwest Ames\n OldTown\tOld Town\n SWISU\tSouth & West of Iowa State University\n Sawyer\tSawyer\n SawyerW\tSawyer West\n Somerst\tSomerset\n StoneBr\tStone Brook\n Timber\tTimberland\n Veenker\tVeenker\n\t\t\t\nOverall_Cond: Rates the overall condition of the house\n\n 10\tVery Excellent\n 9\tExcellent\n 8\tVery Good\n 7\tGood\n 6\tAbove Average\t\n 5\tAverage\n 4\tBelow Average\t\n 3\tFair\n 2\tPoor\n 1\tVery Poor\n\nOverall_Qual: Rates the overall material and finish of the house\n\n 10\tVery Excellent\n 9\tExcellent\n 8\tVery Good\n 7\tGood\n 6\tAbove Average\n 5\tAverage\n 4\tBelow Average\n 3\tFair\n 2\tPoor\n 1\tVery Poor\n\nPaved_Drive: Paved driveway\n\n Y\tPaved \n P\tPartial Pavement\n N\tDirt/Gravel\n\nRoof_Matl: Roof material\n\n ClyTile\tClay or Tile\n CompShg\tStandard (Composite) Shingle\n Membran\tMembrane\n Metal\tMetal\n Roll\tRoll\n Tar&Grv\tGravel & Tar\n WdShake\tWood Shakes\n WdShngl\tWood Shingles\n\nRoof_Style: Type of roof\n\n Flat\tFlat\n Gable\tGable\n Gambrel\tGabrel (Barn)\n Hip\tHip\n Mansard\tMansard\n Shed\tShed\n\nSalePrice: the sales price for each house\n\nSale_Condition: Condition of sale\n\n Normal\tNormal Sale\n Abnorml\tAbnormal Sale - trade, foreclosure, short sale\n AdjLand\tAdjoining Land Purchase\n Alloca\tAllocation - two linked properties with separate deeds, typically condo with a garage unit\t\n Family\tSale between family members\n Partial\tHome was not completed when last assessed (associated with New Homes)\n\nSale_Type: Type of sale\n\t\t\n WD \tWarranty Deed - Conventional\n CWD\tWarranty Deed - Cash\n VWD\tWarranty Deed - VA Loan\n New\tHome just constructed and sold\n COD\tCourt Officer Deed/Estate\n Con\tContract 15% Down payment regular terms\n ConLw\tContract Low Down payment and low interest\n ConLI\tContract Low Interest\n ConLD\tContract Low Down\n Oth\tOther\n\t\nStreet: Type of road access to property\n\n Grvl\tGravel\t\n Pave\tPaved\n \t\nTotRms_AbvGrd: Total rooms above grade (does not include bathrooms)\n\nUtilities: Type of utilities available\n\t\t\n AllPub\tAll public Utilities (E,G,W,& S)\t\n NoSewr\tElectricity, Gas, and Water (Septic Tank)\n NoSeWa\tElectricity and Gas Only\n ELO\tElectricity only\t\n\t\nYear_Built: Original construction date\n\nYear_Remod/Add: Remodel date (same as construction date if no remodeling or additions)\n\t\t\t\t\t\t\nYr_Sold: Year Sold (YYYY)\t\n\n```",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecf1c2dc4999b23851ca50ad59563a4b384ee5e3 | 40,695 | ipynb | Jupyter Notebook | 05. Deep Neural Networks.ipynb | lmarti/ml-sklearn-sernageomin | 955a9cc080ee1f0b6c17d8d5548462a95d242ff1 | [
"MIT"
] | null | null | null | 05. Deep Neural Networks.ipynb | lmarti/ml-sklearn-sernageomin | 955a9cc080ee1f0b6c17d8d5548462a95d242ff1 | [
"MIT"
] | null | null | null | 05. Deep Neural Networks.ipynb | lmarti/ml-sklearn-sernageomin | 955a9cc080ee1f0b6c17d8d5548462a95d242ff1 | [
"MIT"
] | null | null | null | 30.346756 | 247 | 0.549601 | [
[
[
"<div align='left' style=\"width:38%;overflow:hidden;\">\n<a href='http://inria.fr'>\n<img src='https://github.com/lmarti/jupyter_custom/raw/master/imgs/inr_logo_rouge.png' alt='Inria logo' title='Inria'/>\n</a>\n</div>",
"_____no_output_____"
],
[
"# Machine Learning with `scikit-learn`\n\n# Deep Neural Networks\n\n## by [Nayat Sánchez Pi](http://www.nayatsanchezpi.com) and [Luis Martí](http://lmarti.com)\n\n$\\renewcommand{\\vec}[1]{\\boldsymbol{#1}}$",
"_____no_output_____"
]
],
[
[
"import random\n\nimport numpy as np\nimport pandas as pd\n\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport matplotlib.cm as cm\nfrom mpl_toolkits.mplot3d import Axes3D",
"_____no_output_____"
],
[
"plt.rc('font', family='serif')\n\n# numpy - pretty matrix \nnp.set_printoptions(precision=3, threshold=1000, edgeitems=5, linewidth=80, suppress=True)\n\nimport seaborn\nseaborn.set(style='whitegrid'); seaborn.set_context('talk')\n\n%matplotlib inline\n%config InlineBackend.figure_format = 'retina'",
"_____no_output_____"
],
[
"from ipywidgets import interact, interactive, fixed\nimport ipywidgets as widgets",
"_____no_output_____"
],
[
"# tikzmagic extesion for figures - https://github.com/mkrphys/ipython-tikzmagic\n%load_ext tikzmagic",
"_____no_output_____"
],
[
"# fixing a seed for reproducibility, do not do this in real life. \nrandom.seed(a=42) ",
"_____no_output_____"
]
],
[
[
"### About the notebook/slides\n\n* The slides are _programmed_ as a [Jupyter](http://jupyter.org)/[IPython](https://ipython.org/) notebook.\n* **Feel free to try them and experiment on your own by launching the notebooks.**",
"_____no_output_____"
],
[
"If you are using [nbviewer](http://nbviewer.jupyter.org) you can change to slides mode by clicking on the icon:\n\n<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-3\"><span/></div>\n <div class=\"col-md-6\">\n <img alt='view as slides' src='https://github.com/lmarti/jupyter_custom/raw/master/imgs/view-as-slides.png'/>\n </div>\n <div class=\"col-md-3\" align='center'><span/></div>\n </div>\n</div>",
"_____no_output_____"
],
[
"## Big data\n\n* *Buzzword* implying that you are able to handle large amounts of data.\n* Algorithmical and technical challenges.\n* Dimensions:\n * Number of records or entries (this is mostly a technical challenge).\n * Number of variables to take into account.\n * Complex data representation i.e. images.",
"_____no_output_____"
],
[
"## Handling images",
"_____no_output_____"
],
[
"<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-1\" align='center'>\n </div>\n <div class=\"col-md-10\">\n <img src='imgs/06/challenges.jpeg'/>\n </div>\n <div class=\"col-md-1\" align='center'>\n </div>\n </div>\n</div>",
"_____no_output_____"
],
[
"# Dealing with this complex and high-dimensional data\n\n* Intuitively we'd expect networks with many more hidden layers to be more powerful.\n* Such networks could use the intermediate layers to build up multiple levels of abstraction.\n* Extract progressively more abstract features.\n* A deep network could have a better performance than a shallow one with the same number of neurons?\n\nDeep neural networks at last!",
"_____no_output_____"
],
[
"For example, doing visual pattern recognition:\n* neurons in the first hidden layer might learn to recognize edges,\n* neurons in the second layer could learn to recognize more complex shapes, say triangle or rectangles, built up from edges. \n* The third layer would then recognize still more complex shapes. \n* And so on.",
"_____no_output_____"
],
[
"<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-3\" align='center'>\n </div>\n <div class=\"col-md-6\">\n <img src='imgs/06/AI_system_parts.png'/>\n <small> From [Theoretical Motivations for Deep Learning](http://rinuboney.github.io/2015/10/18/theoretical-motivations-deep-learning.html)</small>\n </div>\n <div class=\"col-md-3\" align='center'>\n </div>\n </div>\n</div>\n",
"_____no_output_____"
],
[
"# Learning deep MLP\n\n* Intuitively, nothing stop us from training deep MLPs with the backpropagation algorithm.\n* However, results are worst than *shallow* architectures.\n\n### Why?\n\n* This contradicts our intuition.\n* In the worst case we should have layers \"doing nothing\" but not worsening the results.",
"_____no_output_____"
],
[
"## Investigating the issue\n\n* We need a measure of of progress of learning -> our gradients.\n* I have already told you that is important to check the gradients.\n* We have the vectors $\\vec{\\delta}^1,\\ldots,\\vec{\\delta}^l,\\ldots$ of the deltas corresponding to each layer.\n* We can use the norm of the vector $\\left|\\vec{\\delta}^l\\right|$ as an indicator of how much learning is taking place in each layer. ",
"_____no_output_____"
],
[
"Gradient descent with just 1,000 training images, trained over 500 epochs on the MNIST dataset.\n\n<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-3\" align='center'>\n </div>\n <div class=\"col-md-6\">\n <img src='imgs/06/training_speed_2_layers.png'/>\n </div>\n <div class=\"col-md-3\" align='center'>\n </div>\n </div>\n</div>\n<small>More examples on http://neuralnetworksanddeeplearning.com/chap5.html.</small>",
"_____no_output_____"
],
[
"* The two layers start out learning at very different speeds\n* The speed in both layers then drops very quickly, before rebounding. \n* *The first hidden layer learns much more slowly than the second hidden layer*.",
"_____no_output_____"
],
[
"## What about a three hidden layers network?\n\n<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-3\" align='center'>\n </div>\n <div class=\"col-md-6\">\n <img src='imgs/06/training_speed_3_layers.png'/>\n </div>\n <div class=\"col-md-3\" align='center'>\n </div>\n </div>\n</div>",
"_____no_output_____"
],
[
"## And with four?\n\n<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-3\" align='center'>\n </div>\n <div class=\"col-md-6\">\n <img src='imgs/06/training_speed_4_layers.png'/>\n </div>\n <div class=\"col-md-3\" align='center'>\n </div>\n </div>\n</div>",
"_____no_output_____"
],
[
"### The phenomenon is known as the \n# Vanishing/Exploding Gradient Problem\n\nWas reported by:\n* Gradient flow in recurrent nets: the difficulty of learning long-term dependencies, by Sepp Hochreiter, Yoshua Bengio, Paolo Frasconi, and Jürgen Schmidhuber (2001).\n* Sepp Hochreiter's earlier Diploma Thesis, Untersuchungen zu dynamischen neuronalen Netzen (1991, in German).\n\nBut, probably every body that worked in neural networks had eventually made an experiment like the previous one.",
"_____no_output_____"
],
[
"* It turns out that the gradient in deep neural networks is **unstable**, \n* tending to either **explode** or **vanish** in earlier layers. \n* This instability is a fundamental problem for gradient-based learning in deep neural networks. \n* It's something we need to understand and address.",
"_____no_output_____"
],
[
"# What's causing the vanishing gradient problem?\n\nLets picture a simple deep network:\n\n<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-2\" align='center'>\n </div>\n <div class=\"col-md-8\">\n <div class='well well-sm'>\n <img src='imgs/06/tikz37.png'/>\n </div>\n </div>\n <div class=\"col-md-2\" align='center'>\n </div>\n </div>\n</div>\nHere, $w_1, w_2,\\ldots$, are the weights, $b_1,b_2,\\ldots$ are the biases, and $C$ is some cost function. \n$$\n\\frac{\\partial C}{\\partial b_1} = \nf'(\\text{net}_1)\\times w_2 \\times f'(\\text{net}_2) \\times w_3 \\times f'(\\text{net}_3) \\times w_4 \\times f'(\\text{net}_5) \\times\n\\frac{\\partial C}{\\partial \\hat{y}}.\n$$",
"_____no_output_____"
],
[
"\"What makes deep networks hard to train?\" is a complex question:\n* instabilities associated to gradient-based learning in deep networks.\n* Evidence suggest that there is also a role played by the choice of activation function, \n* the way weights are initialized, and \n* how learning by gradient descent is implemented. \n\nMany factors play a role in making deep networks hard to train, and understanding all those factors is still a subject of ongoing research.",
"_____no_output_____"
],
[
"# What of we train each layer separately?",
"_____no_output_____"
],
[
" ## Autoencoders\n\n* An autoencoder is typically a MLP neural network which aims to learn a compressed, distributed representation (encoding) of a dataset.\n* They learn to predict their inputs.",
"_____no_output_____"
],
[
"<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-2\"><span/></div>\n <div class=\"col-md-8\">\n <img class='img-thumbnail' alt='Perceptron' src='imgs/autoencoder.png'/>\n </div>\n <div class=\"col-md-2\" align='center'><span/></div>\n </div>\n</div>",
"_____no_output_____"
],
[
"## Stacked autoencoders",
"_____no_output_____"
],
[
"<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-2\"><span/></div>\n <div class=\"col-md-8\">\n <img class='img-thumbnail' alt='Perceptron' src='imgs/stacked-autoencoder.png'/>\n </div>\n <div class=\"col-md-2\" align='center'><span/></div>\n </div>\n</div>",
"_____no_output_____"
],
[
"\n* The hidden layer of autoencoder $t$ acts as an input layer to autoencoder $t+1$.\n* The input layer of the first autoencoder is the input layer for the whole network.\n\nThe greedy layer-wise training procedure works like this:\n* Train the hidden layers as autoencoders in succession.\n* Train final (output layer) to predict targets.",
"_____no_output_____"
],
[
"# Can we look back (again) to nature for inspiration?",
"_____no_output_____"
],
[
"# Convolutional Neural Networks (CNNs / ConvNets)\n\nConvolutional Neural Networks are very similar to ordinary neural networks:\n\n* They are made up of neurons that have learnable weights and biases. \n* Each neuron receives some inputs, performs a dot product and optionally follows it with a non-linear activation. \n* The whole network still expresses a single differentiable score function: from the raw image pixels on one end to class scores at the other.\n* They still have a loss function on the last (fully-connected) layer and all the tips/tricks we debated for learning regular NN still apply.\n\nDifference: \n* ConvNets make the explicit assumption that the inputs are images, this allows to encode certain properties into the architecture. \n* Forward function more efficient to implement and vastly reduce the amount of parameters in the network.\n* Main application: image classification.",
"_____no_output_____"
],
[
"## Yet another problem with regular NNs\n\n* Regular Neural Nets don’t scale well to full images.\n* Images only of size $32\\times32\\times3$ (32 wide, 32 high, 3 color channels), imply that the first hidden layer will have $32\\times 32\\times3 = 3072$ weights.\n* This fully-connected structure does not scale to larger images. \n* For example, an image of more \"normal\" size, e.g. $200\\times 200\\times3$, would lead to neurons that have $200\\times 200\\times3$ = $120,000$ weights!\n* Moreover, we would almost certainly want to have several such neurons, so the parameters would add up quickly! \n* Clearly, this full connectivity is wasteful.\n* The huge number of parameters would quickly lead to overfitting.",
"_____no_output_____"
],
[
"Convolutional Neural Networks take advantage of the fact that the input consists of images\n\n* constrain the architecture in a more sensible way. \n* unlike a regular Neural Network, the layers of a ConvNet have neurons arranged in 3 dimensions: width, height, depth.\n* neurons in a layer will only be connected to a small region of the layer before it, instead of all of the neurons in a fully-connected manner.\n* The ConvNet architecture we will reduce the full image into a single vector of class scores, arranged along the depth dimension.",
"_____no_output_____"
],
[
"<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-5\">\n Regular network <img src='imgs/06/conv.jpeg'/>\n <div class=\"col-md-2\" align='center'>\n \n </div>\n </div>\n <div class=\"col-md-5\">\n ConvNet <img src='imgs/06/cnn.jpeg'/>\n </div>\n </div>\n</div>",
"_____no_output_____"
],
[
"## Key concepts\n\n* **Local receptive fields**: We won't connect every input pixel to every hidden neuron. Instead, we only make connections in small, localized regions of the input image.\n* **Shared weights and biases**.\n* **Pooling**: usually used immediately after convolutional layers. They simplify the information in the output from the convolutional layer.",
"_____no_output_____"
],
[
"## ConvNets layer types\n\n* As we described above, a ConvNet is a sequence of layers, and\n* every layer of a ConvNet transforms one volume of activations to another through a differentiable function. \n* Three main types of layers to build ConvNet architectures: \n * Convolutional Layer, \n * Rectified linear units (RELU) Layer,\n * Pooling Layer, and \n * Fully-Connected Layer (exactly as seen in regular Neural Networks). \n\nWe will stack these layers to form a full ConvNet architecture.",
"_____no_output_____"
],
[
"<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-1\" align='center'>\n </div>\n <div class=\"col-md-10\">\n <div class='well well-sm' align='center'>\n <img src='imgs/06/convnet.jpeg'/>\n </div>\n </div>\n <div class=\"col-md-1\" align='center'>\n </div>\n </div>\n</div>",
"_____no_output_____"
],
[
"## Convolutional Layer\n\nThe convolutional layer is the core building block of a convolutional network that does most of the computational heavy lifting.\n* consist of a set of learnable filters. \n* Every filter is small spatially (along width and height), but extends through the full depth of the input volume. ",
"_____no_output_____"
],
[
"For example,\n* a typical filter on a first layer of a ConvNet might have size 5x5x3 (i.e. 5 pixels width and height, and 3 color channels).\n* During the forward pass, we slide (*convolve*) each filter across the width and height of the input volume and compute dot products between the entries of the filter and the input at any position. \n* We will produce a 2-dimensional activation map that gives the responses of that filter at every spatial position.\n\n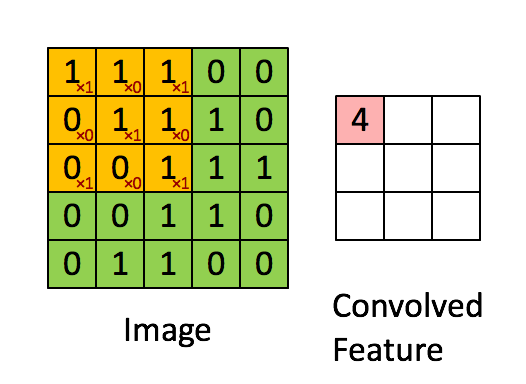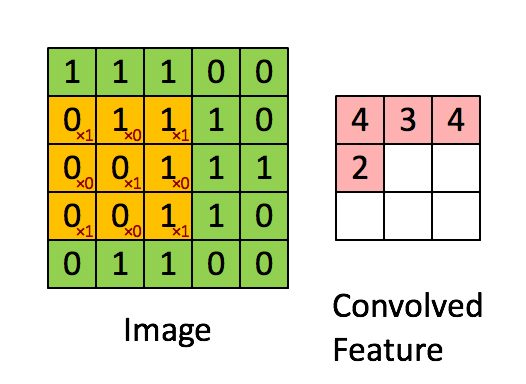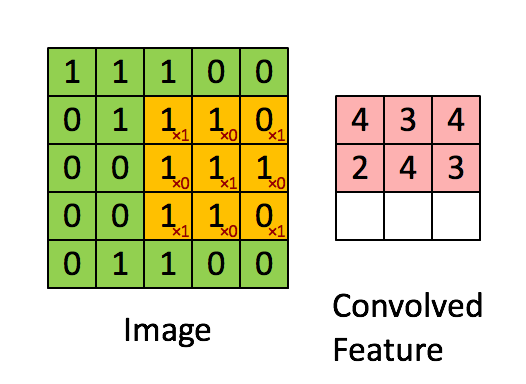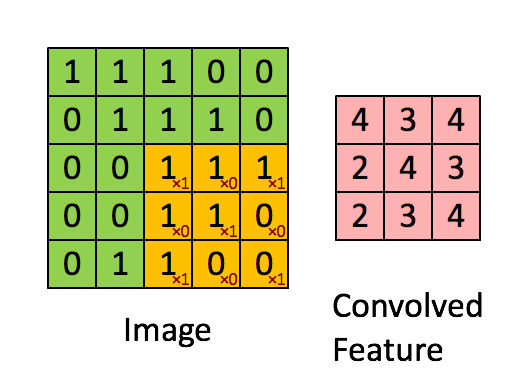",
"_____no_output_____"
],
[
"* The network will learn filters that activate when they see some type of visual feature:\n * an edge of some orientation or a blotch of some color on the first layer, or \n * entire honeycomb or wheel-like patterns on higher layers of the network.\n* We will have an entire set of filters in each CONV layer (e.g. 12 filters), and each of them will produce a separate 2-dimensional activation map. We will stack these activation maps along the depth dimension and produce the output volume.",
"_____no_output_____"
],
[
"## Spatial arrangement\n\nHow many neurons there are in the output volume and how they are arranged?\nThree hyperparameters control the size of the output volume: \n* depth, \n* stride and \n* zero-padding.",
"_____no_output_____"
],
[
"### Depth of the output volume \n\nCorresponds to the number of filters we would like to use, \n* each learning to look for something different in the input. \n\nFor example, if the first Convolutional Layer takes as input the raw image, then different neurons along the depth dimension may activate in presence of various oriented edged, or blobs of color.",
"_____no_output_____"
],
[
"### Stride\n\n* We must specify the stride with which we slide the filter. \n* When the stride is 1 then we move the filters one pixel at a time.\n* When the stride is 2 (or uncommonly 3 or more, though this is rare in practice) then the filters jump 2 pixels at a time as we slide them around. \n* This will produce smaller output volumes spatially.",
"_____no_output_____"
],
[
"### Zero padding\n\n* Sometimes it is convenient to pad the input volume with zeros around the border.\n* The nice feature of zero padding is that it will allow us to control the spatial size of the output volumes.",
"_____no_output_____"
],
[
"### Shared weight and biases\n\n* All neurons in the layer detect the same *feature*.\n* We need to add layers to encode more features.\n\n<div align='center'>\n<img src='http://neuralnetworksanddeeplearning.com/images/tikz46.png' width='47%'/>\n</div>",
"_____no_output_____"
],
[
"<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-1\" align='center'>\n </div>\n <div class=\"col-md-10\">\n <div class='well well-sm' align='center'>\n <img src='imgs/06/weights.jpeg'/>\n </div>\n </div>\n <div class=\"col-md-1\" align='center'>\n </div>\n </div>\n</div>\nKrizhevsky et al. Each of the 96 filters shown here is of size $[11\\times11\\times3]$, and each one is shared by the $55\\times55$ neurons in one depth slice",
"_____no_output_____"
],
[
"## Summarizing the Convolutional layer\n* Accepts a volume of size $W_1\\times H_1\\times D_1$\n* Requires four hyperparameters:\n * Number of filters $K$,\n * their spatial extent $F$,\n * the stride $S$,\n * the amount of zero padding $P$.\nProduces a volume of size $W_2\\times H_2\\times D_2$ where:\n$$\nW_2 = \\frac{W_1−F+2P}{S+1},\\ H_2 = \\frac{H_1−F+2P}{S+1},\\ D_2=K\n$$",
"_____no_output_____"
],
[
"## Pooling\n\n* Subsampling layers reduce the size of the input. \n\n* There are multiple ways to subsample, but the most popular are:\n - max pooling (most popular), \n - average pooling, and\n - stochastic pooling.",
"_____no_output_____"
],
[
"In max-pooling, a pooling unit simply outputs the maximum activation in the 2×22×2 input region:\n<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-1\" align='center'>\n </div>\n <div class=\"col-md-10\">\n <div class='well well-sm' align='center'>\n <img src='http://neuralnetworksanddeeplearning.com/images/tikz47.png'/>\n </div>\n </div>\n <div class=\"col-md-1\" align='center'>\n </div>\n </div>\n</div>",
"_____no_output_____"
],
[
"We can see convolution as the application of a filter or a dimensionality reduction.\n\n* Convolutional layers apply a number of filters to the input. \n* The result of one filter applied across the image is called feature map.\n* If the previous layer is also convolutional, the filters are applied across all of it’s FMs with different weights, so each input FM is connected to each output FM. \n> The intuition behind the shared weights across the image is that the features will be detected regardless of their location, while the multiplicity of filters allows each of them to detect different set of features.",
"_____no_output_____"
],
[
"* The convolutional architecture is quite different to the architecture of traditional neural network.\n* But the overall picture is similar: \n * a network made of many simple units, \n * whose behaviors are determined by their weights and biases. \nThe overall goal is still the same: to use training data to train the network's weights and biases so that the network does a good job classifying input.",
"_____no_output_____"
],
[
"## Restricted Boltzmann machines\n\nRestricted Boltzmann machines (RBM) are generative stochastic neural network that can learn a probability distribution over its set of inputs.\n\n<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-3\" align='center'>\n </div>\n <div class=\"col-md-6\">\n <div class='well well-sm' align='center'>\n <img src='imgs/06/rbm.png'/>\n </div>\n </div>\n <div class=\"col-md-3\" align='center'>\n </div>\n </div>\n</div>",
"_____no_output_____"
],
[
"## Training RBMs: Contrastive Divergence\n\n* **Positive phase:**\n - An input sample $\\vec{x}$ is presented to the input layer. \n - $\\vec{x}$ is propagated to the hidden layer in a similar manner to the feedforward networks. \n - The result is the hidden layer activations, $\\vec{h}$.\n* **Negative phase:**\n - Propagate $\\vec{h}$ back to the visible layer with result resulting in a $\\vec{x}'$.\n - $\\vec{x}'$ back to the hidden layer.\n* **Weight update**:\n$$ \\vec{w}(t+1) = \\vec{w}(t) + \\alpha (\\vec{x}\\vec{h}^{\\intercal} -\\vec{x'}\\vec{h'}^{\\intercal}).$$\n\n* The positive phase reflects the network internal representation of the data.\n* The negative phase represents an attempt to recreate the data based on this internal representation. ",
"_____no_output_____"
],
[
"* The goal is that the \"generated\" data to be as close as possible to the \"real\" one.\n* This is reflected in the weight update formula.\n\n> In other words, the net has a perception of how the input data must be represented, so it tries to reproduce the data based on this perception. If its reproduction isn’t close enough to reality, it makes an adjustment and tries again.",
"_____no_output_____"
],
[
"## Deep Belief Networks\n\nRestricted Boltzmann machines can be stacked to create a class of neural networks known as deep belief networks (DBNs).\n<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-3\" align='center'>\n </div>\n <div class=\"col-md-6\">\n <div class='well well-sm' align='center'>\n <img src='imgs/06/deep-belief.png'/>\n </div>\n </div>\n <div class=\"col-md-3\" align='center'>\n </div>\n </div>\n</div>",
"_____no_output_____"
],
[
"## Programming Deep Learning\n\n* Torch - An open source software library for machine learning based on the Lua programming language.\n* Caffe - A deep learning framework.\n* Apache SINGA - A deep learning platform developed for scalability, usability and extensibility.\n\nMy current preference:\n* Tensorflow",
"_____no_output_____"
],
[
"# General Principles\n\n* Supervise the learning process (did I mentioned that you should check your gradients?).\n* Use a correct experimental methodology.\n* Contrast your results with a baseline method.",
"_____no_output_____"
],
[
"# [Applications of Deep Learning](https://en.wikipedia.org/wiki/Deep_learning#Applications)\n\nThere are many successfull applications, for example:\n\n* Computer vision and image recognition;\n* Speech recognition;\n* Natural language processing $\\rightarrow$ probabilistic context free grammars;\n* Anomaly detection on many variables;\n* ... and many more. ",
"_____no_output_____"
],
[
"# Final remarks\n\n* Deep learning as a step towards realising *strong AI*;\n* thus many organizations have become interested in its use for particular applications.\n * see https://en.wikipedia.org/wiki/Deep_learning#Commercial_activities\n* Better understanding of mental processes.\n* Deep learning $\\iff$ big data.",
"_____no_output_____"
],
[
"<hr/>\n<div class=\"container-fluid\">\n <div class=\"row\">\n <div class=\"col-md-3\" align='center'>\n <img align='center' alt=\"Creative Commons License\" style=\"border-width:0\" src=\"https://i.creativecommons.org/l/by-nc-sa/4.0/88x31.png\"/>\n </div>\n <div class=\"col-md-9\">\n This work is licensed under a [Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License](http://creativecommons.org/licenses/by-nc-sa/4.0/).\n </div>\n </div>\n</div>",
"_____no_output_____"
]
],
[
[
"# this code is here for cosmetic reasons\nfrom IPython.core.display import HTML\nfrom urllib.request import urlopen\nHTML(urlopen('https://raw.githubusercontent.com/lmarti/jupyter_custom/master/custom.include').read().decode('utf-8'))",
"_____no_output_____"
]
],
[
[
" ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecf1cd9d419a8b6ef51b5d1dda7d8526e07f2478 | 39,026 | ipynb | Jupyter Notebook | new_beginning/cluster_generator_human.ipynb | kiri93/graco | fea1a82df51c2d3afe36dc73229936ce4ddf11cd | [
"Apache-2.0"
] | null | null | null | new_beginning/cluster_generator_human.ipynb | kiri93/graco | fea1a82df51c2d3afe36dc73229936ce4ddf11cd | [
"Apache-2.0"
] | null | null | null | new_beginning/cluster_generator_human.ipynb | kiri93/graco | fea1a82df51c2d3afe36dc73229936ce4ddf11cd | [
"Apache-2.0"
] | null | null | null | 44.448747 | 2,012 | 0.586327 | [
[
[
"from pyclustering.cluster.kmedoids import kmedoids\nfrom functools import partial\nfrom random import sample\n\nimport os\nimport time\nimport numpy as np\nimport pandas as pd\nimport networkx as nx",
"_____no_output_____"
],
[
"DATA_DIRECTORY = \"/media/clusterduck123/joe/data\"\nHUMAN_DIRECTORY = f\"{DATA_DIRECTORY}/processed-data/human\"\nNETWORK_DIRECTORY = f\"{HUMAN_DIRECTORY}/networks\"\nMATRIX_DIRECTORY = f\"{HUMAN_DIRECTORY}/distance-matrices\"\nANNOTATION_DIRECTORY = f\"{HUMAN_DIRECTORY}/annotations\"",
"_____no_output_____"
]
],
[
[
"# Clustering",
"_____no_output_____"
]
],
[
[
"def get_number_of_pre_runs(CLUSTER_DIRECTORY, distance, n_clusters = 99):\n splitted_file_names = [name.split('_') for name in os.listdir(CLUSTER_DIRECTORY)]\n pre_runs = [int(run) for run, ncluster, db_txt in splitted_file_names if ncluster == str(n_clusters)]\n if pre_runs:\n return max(pre_runs)\n else:\n return -1",
"_____no_output_____"
]
],
[
[
"## GDV",
"_____no_output_____"
]
],
[
[
"MIN_CLUSTERS = 2\nMAX_CLUSTERS = 100\n\nall_distances = [filename.split('_')[0] for filename in os.listdir(f\"{MATRIX_DIRECTORY}/GDV\")]",
"_____no_output_____"
],
[
"method = 'kmedoid'\n\nfor run in range(4):\n for distance in {'canberra'}:\n print(distance)\n \n CLUSTER_DIRECTORY = f\"{HUMAN_DIRECTORY}/clusterings/GDV/{distance}/{method}\"\n if not os.path.exists(CLUSTER_DIRECTORY):\n os.makedirs(CLUSTER_DIRECTORY)\n \n df = pd.read_csv(f\"{MATRIX_DIRECTORY}/GDV/{distance}_BioGRID.txt\", delimiter=' ')\n D = df.values.astype(float) \n\n t1 = time.time()\n for n_clusters in range(MIN_CLUSTERS, MAX_CLUSTERS+1):\n initial_medoids = sample(range(len(D)), n_clusters)\n kmedoids_instance = kmedoids(D, initial_medoids, data_type='distance_matrix')\n kmedoids_instance.process()\n \n nr = get_number_of_pre_runs(CLUSTER_DIRECTORY, distance, MAX_CLUSTERS)\n\n with open(f\"{CLUSTER_DIRECTORY}/{nr+1}_{n_clusters}_BioGRID.txt\", 'w') as f:\n for cluster in kmedoids_instance.get_clusters():\n f.write(' '.join(df.columns[cluster]) + '\\n')\n t2 = time.time()\n print(f'{n_clusters}: {t2-t1:.2f}sec', end='\\r')\n print()",
"canberra\n100: 5300.11sec\ncanberra\n8: 421.59sec\r"
]
],
[
[
"# GCV-A",
"_____no_output_____"
]
],
[
[
"feature = 'GCV-A'\n\nMIN_CLUSTERS = 2\nMAX_CLUSTERS = 100\n\nall_distances = sorted('_'.join(filename.split('_')[:-1]) \n for filename in os.listdir(f\"{MATRIX_DIRECTORY}/{feature}\"))",
"_____no_output_____"
],
[
"method = 'kmedoid'\n\nfor run in range(5):\n for distance in {''}:\n print(distance)\n \n CLUSTER_DIRECTORY = f\"{HUMAN_DIRECTORY}/clusterings/{feature}/{distance}/{method}\"\n if not os.path.exists(CLUSTER_DIRECTORY):\n os.makedirs(CLUSTER_DIRECTORY)\n \n df = pd.read_csv(f\"{MATRIX_DIRECTORY}/{feature}/{distance}_BioGRID.txt\", delimiter=' ')\n D = df.values.astype(float) \n\n t1 = time.time()\n for n_clusters in range(MIN_CLUSTERS, MAX_CLUSTERS+1):\n initial_medoids = sample(range(len(D)), n_clusters)\n kmedoids_instance = kmedoids(D, initial_medoids, data_type='distance_matrix')\n kmedoids_instance.process()\n \n nr = get_number_of_pre_runs(CLUSTER_DIRECTORY, distance, MAX_CLUSTERS)\n\n with open(f\"{CLUSTER_DIRECTORY}/{nr+1}_{n_clusters}_BioGRID.txt\", 'w') as f:\n for cluster in kmedoids_instance.get_clusters():\n f.write(' '.join(df.columns[cluster]) + '\\n')\n t2 = time.time()\n print(f'{n_clusters}: {t2-t1:.2f}sec', end='\\r')\n print()",
"_____no_output_____"
]
],
[
[
"## GCV-A",
"_____no_output_____"
]
],
[
[
"MIN_CLUSTERS = 2\nMAX_CLUSTERS = 100\n\nall_distances = [filename.split('_')[0] for filename in os.listdir(f\"{MATRIX_DIRECTORY}/GCV-A\")]",
"_____no_output_____"
],
[
"method = 'kmedoid'\n\nfor run in range(40):\n for distance in ['normalized1-linf',\n 'normalized1-l2',\n 'normalized1-l1']:\n print(distance)\n \n CLUSTER_DIRECTORY = f\"{YEAST_DIRECTORY}/clusterings/GCV-A/{distance}/{method}\"\n if not os.path.exists(CLUSTER_DIRECTORY):\n os.makedirs(CLUSTER_DIRECTORY)\n \n df = pd.read_csv(f\"{MATRIX_DIRECTORY}/GCV-A/{distance}_BioGRID.txt\", delimiter=' ')\n D = df.values.astype(float) \n\n t1 = time.time()\n for n_clusters in range(MIN_CLUSTERS, MAX_CLUSTERS+1):\n initial_medoids = sample(range(len(D)), n_clusters)\n kmedoids_instance = kmedoids(D, initial_medoids, data_type='distance_matrix')\n kmedoids_instance.process()\n \n nr = get_number_of_pre_runs(CLUSTER_DIRECTORY, distance, MAX_CLUSTERS)\n\n with open(f\"{CLUSTER_DIRECTORY}/{nr+1}_{n_clusters}_BioGRID.txt\", 'w') as f:\n for cluster in kmedoids_instance.get_clusters():\n f.write(' '.join(df.columns[cluster]) + '\\n')\n t2 = time.time()\n print(f'{n_clusters}: {t2-t1:.2f}sec', end='\\r')\n print()",
"normalized1-linf\n100: 567.56sec\nnormalized1-l2\n100: 553.66sec\nnormalized1-l1\n100: 568.33sec\nnormalized1-linf\n100: 566.88sec\nnormalized1-l2\n100: 543.78sec\nnormalized1-l1\n100: 542.83sec\nnormalized1-linf\n100: 562.03sec\nnormalized1-l2\n100: 561.84sec\nnormalized1-l1\n100: 565.90sec\nnormalized1-linf\n100: 585.38sec\nnormalized1-l2\n100: 553.77sec\nnormalized1-l1\n100: 569.14sec\nnormalized1-linf\n100: 544.02sec\nnormalized1-l2\n100: 557.46sec\nnormalized1-l1\n100: 550.29sec\nnormalized1-linf\n100: 561.53sec\nnormalized1-l2\n100: 546.45sec\nnormalized1-l1\n100: 565.12sec\nnormalized1-linf\n100: 555.60sec\nnormalized1-l2\n100: 548.69sec\nnormalized1-l1\n100: 579.96sec\nnormalized1-linf\n100: 568.23sec\nnormalized1-l2\n100: 558.88sec\nnormalized1-l1\n100: 575.31sec\nnormalized1-linf\n100: 552.40sec\nnormalized1-l2\n100: 550.22sec\nnormalized1-l1\n100: 570.18sec\nnormalized1-linf\n100: 589.95sec\nnormalized1-l2\n100: 594.89sec\nnormalized1-l1\n100: 600.24sec\nnormalized1-linf\n100: 591.80sec\nnormalized1-l2\n100: 576.68sec\nnormalized1-l1\n100: 600.33sec\nnormalized1-linf\n100: 579.74sec\nnormalized1-l2\n100: 589.04sec\nnormalized1-l1\n100: 605.94sec\nnormalized1-linf\n100: 576.16sec\nnormalized1-l2\n100: 582.09sec\nnormalized1-l1\n100: 623.37sec\nnormalized1-linf\n100: 605.80sec\nnormalized1-l2\n100: 601.76sec\nnormalized1-l1\n100: 621.67sec\nnormalized1-linf\n100: 605.21sec\nnormalized1-l2\n100: 603.54sec\nnormalized1-l1\n100: 607.22sec\nnormalized1-linf\n100: 603.71sec\nnormalized1-l2\n100: 601.16sec\nnormalized1-l1\n100: 613.27sec\nnormalized1-linf\n100: 610.51sec\nnormalized1-l2\n100: 600.49sec\nnormalized1-l1\n100: 603.19sec\nnormalized1-linf\n100: 609.79sec\nnormalized1-l2\n100: 598.49sec\nnormalized1-l1\n100: 606.13sec\nnormalized1-linf\n100: 611.47sec\nnormalized1-l2\n100: 604.88sec\nnormalized1-l1\n100: 602.60sec\nnormalized1-linf\n100: 609.47sec\nnormalized1-l2\n100: 599.80sec\nnormalized1-l1\n100: 613.32sec\nnormalized1-linf\n100: 604.82sec\nnormalized1-l2\n100: 600.13sec\nnormalized1-l1\n100: 619.02sec\nnormalized1-linf\n100: 599.87sec\nnormalized1-l2\n100: 598.86sec\nnormalized1-l1\n100: 622.18sec\nnormalized1-linf\n100: 594.47sec\nnormalized1-l2\n100: 602.70sec\nnormalized1-l1\n100: 612.16sec\nnormalized1-linf\n100: 610.14sec\nnormalized1-l2\n100: 600.80sec\nnormalized1-l1\n100: 625.80sec\nnormalized1-linf\n100: 610.94sec\nnormalized1-l2\n100: 602.27sec\nnormalized1-l1\n100: 614.08sec\nnormalized1-linf\n100: 602.53sec\nnormalized1-l2\n100: 598.38sec\nnormalized1-l1\n100: 607.15sec\nnormalized1-linf\n100: 609.22sec\nnormalized1-l2\n100: 599.97sec\nnormalized1-l1\n100: 613.66sec\nnormalized1-linf\n100: 609.92sec\nnormalized1-l2\n100: 598.88sec\nnormalized1-l1\n100: 604.45sec\nnormalized1-linf\n100: 611.63sec\nnormalized1-l2\n100: 603.19sec\nnormalized1-l1\n100: 624.35sec\nnormalized1-linf\n100: 605.05sec\nnormalized1-l2\n100: 601.79sec\nnormalized1-l1\n100: 625.59sec\nnormalized1-linf\n100: 600.77sec\nnormalized1-l2\n100: 596.41sec\nnormalized1-l1\n100: 611.99sec\nnormalized1-linf\n100: 612.23sec\nnormalized1-l2\n100: 596.56sec\nnormalized1-l1\n100: 614.23sec\nnormalized1-linf\n100: 609.06sec\nnormalized1-l2\n100: 604.03sec\nnormalized1-l1\n100: 560.15sec\nnormalized1-linf\n100: 557.21sec\nnormalized1-l2\n100: 552.48sec\nnormalized1-l1\n100: 559.74sec\nnormalized1-linf\n100: 561.37sec\nnormalized1-l2\n100: 552.93sec\nnormalized1-l1\n100: 560.18sec\nnormalized1-linf\n100: 558.31sec\nnormalized1-l2\n100: 546.69sec\nnormalized1-l1\n100: 567.09sec\nnormalized1-linf\n100: 562.88sec\nnormalized1-l2\n100: 548.73sec\nnormalized1-l1\n100: 565.21sec\nnormalized1-linf\n100: 559.56sec\nnormalized1-l2\n100: 551.87sec\nnormalized1-l1\n100: 559.50sec\nnormalized1-linf\n100: 566.21sec\nnormalized1-l2\n100: 589.19sec\nnormalized1-l1\n100: 608.29sec\nnormalized1-linf\n100: 581.74sec\nnormalized1-l2\n100: 591.30sec\nnormalized1-l1\n100: 586.87sec\n"
]
],
[
[
"## GCV-G",
"_____no_output_____"
]
],
[
[
"MIN_CLUSTERS = 2\nMAX_CLUSTERS = 100\n\nall_distances = [filename.split('_')[0] for filename in os.listdir(f\"{MATRIX_DIRECTORY}/GCV-G\")]",
"_____no_output_____"
],
[
"method = 'kmedoid'\n\nfor run in range(10):\n for distance in ['normalized1-linf',\n 'normalized1-l2',\n 'normalized1-l1']:\n print(distance)\n \n CLUSTER_DIRECTORY = f\"{YEAST_DIRECTORY}/clusterings/GCV-G/{distance}/{method}\"\n if not os.path.exists(CLUSTER_DIRECTORY):\n os.makedirs(CLUSTER_DIRECTORY)\n \n df = pd.read_csv(f\"{MATRIX_DIRECTORY}/GCV-G/{distance}_BioGRID.txt\", delimiter=' ')\n D = df.values.astype(float) \n\n t1 = time.time()\n for n_clusters in range(MIN_CLUSTERS, MAX_CLUSTERS+1):\n initial_medoids = sample(range(len(D)), n_clusters)\n kmedoids_instance = kmedoids(D, initial_medoids, data_type='distance_matrix')\n kmedoids_instance.process()\n \n nr = get_number_of_pre_runs(CLUSTER_DIRECTORY, distance, MAX_CLUSTERS)\n\n with open(f\"{CLUSTER_DIRECTORY}/{nr+1}_{n_clusters}_BioGRID.txt\", 'w') as f:\n for cluster in kmedoids_instance.get_clusters():\n f.write(' '.join(df.columns[cluster]) + '\\n')\n t2 = time.time()\n print(f'{n_clusters}: {t2-t1:.2f}sec', end='\\r')\n print()",
"_____no_output_____"
]
],
[
[
"# GCV-AD",
"_____no_output_____"
]
],
[
[
"MIN_CLUSTERS = 2\nMAX_CLUSTERS = 100\n\nall_distances = [filename.split('_')[0] for filename in os.listdir(f\"{MATRIX_DIRECTORY}/GCV-AD\")]",
"_____no_output_____"
],
[
"method = 'kmedoid'\n\nfor run in range(50):\n for distance in ['normalized1-linf',\n 'normalized1-l2',\n 'normalized1-l1']:\n print(distance)\n \n CLUSTER_DIRECTORY = f\"{YEAST_DIRECTORY}/clusterings/GCV-AD/{distance}/{method}\"\n if not os.path.exists(CLUSTER_DIRECTORY):\n os.makedirs(CLUSTER_DIRECTORY)\n \n df = pd.read_csv(f\"{MATRIX_DIRECTORY}/GCV-AD/{distance}_BioGRID.txt\", delimiter=' ')\n D = df.values.astype(float) \n\n t1 = time.time()\n for n_clusters in range(MIN_CLUSTERS, MAX_CLUSTERS+1):\n initial_medoids = sample(range(len(D)), n_clusters)\n kmedoids_instance = kmedoids(D, initial_medoids, data_type='distance_matrix')\n kmedoids_instance.process()\n \n nr = get_number_of_pre_runs(CLUSTER_DIRECTORY, distance, MAX_CLUSTERS)\n\n with open(f\"{CLUSTER_DIRECTORY}/{nr+1}_{n_clusters}_BioGRID.txt\", 'w') as f:\n for cluster in kmedoids_instance.get_clusters():\n f.write(' '.join(df.columns[cluster]) + '\\n')\n t2 = time.time()\n print(f'{n_clusters}: {t2-t1:.2f}sec', end='\\r')\n print()",
"normalized1-linf\n100: 552.05sec\nnormalized1-l2\n100: 582.32sec\nnormalized1-l1\n100: 591.60sec\nnormalized1-linf\n100: 592.12sec\nnormalized1-l2\n100: 615.60sec\nnormalized1-l1\n100: 606.99sec\nnormalized1-linf\n100: 596.02sec\nnormalized1-l2\n100: 609.16sec\nnormalized1-l1\n100: 601.76sec\nnormalized1-linf\n100: 588.87sec\nnormalized1-l2\n100: 603.07sec\nnormalized1-l1\n100: 581.43sec\nnormalized1-linf\n100: 561.65sec\nnormalized1-l2\n100: 579.20sec\nnormalized1-l1\n100: 568.50sec\nnormalized1-linf\n100: 557.98sec\nnormalized1-l2\n100: 570.04sec\nnormalized1-l1\n100: 566.63sec\nnormalized1-linf\n100: 557.91sec\nnormalized1-l2\n100: 567.28sec\nnormalized1-l1\n100: 564.90sec\nnormalized1-linf\n100: 561.06sec\nnormalized1-l2\n100: 569.79sec\nnormalized1-l1\n100: 566.39sec\nnormalized1-linf\n100: 557.00sec\nnormalized1-l2\n100: 571.23sec\nnormalized1-l1\n100: 571.42sec\nnormalized1-linf\n100: 562.81sec\nnormalized1-l2\n100: 569.62sec\nnormalized1-l1\n100: 568.80sec\nnormalized1-linf\n100: 557.26sec\nnormalized1-l2\n100: 568.03sec\nnormalized1-l1\n100: 567.45sec\nnormalized1-linf\n100: 557.30sec\nnormalized1-l2\n100: 569.75sec\nnormalized1-l1\n100: 572.71sec\nnormalized1-linf\n100: 557.41sec\nnormalized1-l2\n100: 569.04sec\nnormalized1-l1\n100: 571.37sec\nnormalized1-linf\n100: 556.92sec\nnormalized1-l2\n100: 579.15sec\nnormalized1-l1\n100: 567.35sec\nnormalized1-linf\n100: 564.81sec\nnormalized1-l2\n100: 576.87sec\nnormalized1-l1\n100: 566.93sec\nnormalized1-linf\n100: 560.95sec\nnormalized1-l2\n100: 576.49sec\nnormalized1-l1\n100: 567.48sec\nnormalized1-linf\n100: 561.86sec\nnormalized1-l2\n100: 570.67sec\nnormalized1-l1\n100: 556.66sec\nnormalized1-linf\n100: 559.21sec\nnormalized1-l2\n100: 579.34sec\nnormalized1-l1\n100: 571.09sec\nnormalized1-linf\n100: 559.17sec\nnormalized1-l2\n100: 580.28sec\nnormalized1-l1\n100: 566.17sec\nnormalized1-linf\n100: 559.00sec\nnormalized1-l2\n100: 578.81sec\nnormalized1-l1\n100: 567.59sec\nnormalized1-linf\n100: 558.27sec\nnormalized1-l2\n100: 578.41sec\nnormalized1-l1\n100: 569.74sec\nnormalized1-linf\n100: 561.82sec\nnormalized1-l2\n100: 570.08sec\nnormalized1-l1\n100: 565.61sec\nnormalized1-linf\n100: 562.61sec\nnormalized1-l2\n100: 570.04sec\nnormalized1-l1\n100: 570.10sec\nnormalized1-linf\n100: 558.96sec\nnormalized1-l2\n100: 579.97sec\nnormalized1-l1\n100: 561.90sec\nnormalized1-linf\n100: 559.38sec\nnormalized1-l2\n100: 580.08sec\nnormalized1-l1\n100: 567.54sec\nnormalized1-linf\n100: 563.70sec\nnormalized1-l2\n100: 571.60sec\nnormalized1-l1\n100: 568.59sec\nnormalized1-linf\n100: 557.68sec\nnormalized1-l2\n100: 576.82sec\nnormalized1-l1\n100: 569.30sec\nnormalized1-linf\n100: 563.13sec\nnormalized1-l2\n100: 616.81sec\nnormalized1-l1\n80: 506.50sec\r"
]
],
[
[
"# GCV-DG",
"_____no_output_____"
]
],
[
[
"MIN_CLUSTERS = 2\nMAX_CLUSTERS = 100\n\nall_distances = [filename.split('_')[0] for filename in os.listdir(f\"{MATRIX_DIRECTORY}/GCV-DG\")]",
"_____no_output_____"
],
[
"method = 'kmedoid'\n\nfor run in range(50):\n for distance in ['normalized1-linf',\n 'normalized1-l2',\n 'normalized1-l1']:\n print(distance)\n \n CLUSTER_DIRECTORY = f\"{YEAST_DIRECTORY}/clusterings/GCV-DG/{distance}/{method}\"\n if not os.path.exists(CLUSTER_DIRECTORY):\n os.makedirs(CLUSTER_DIRECTORY)\n \n df = pd.read_csv(f\"{MATRIX_DIRECTORY}/GCV-DG/{distance}_BioGRID.txt\", delimiter=' ')\n D = df.values.astype(float) \n\n t1 = time.time()\n for n_clusters in range(MIN_CLUSTERS, MAX_CLUSTERS+1):\n initial_medoids = sample(range(len(D)), n_clusters)\n kmedoids_instance = kmedoids(D, initial_medoids, data_type='distance_matrix')\n kmedoids_instance.process()\n \n nr = get_number_of_pre_runs(CLUSTER_DIRECTORY, distance, MAX_CLUSTERS)\n\n with open(f\"{CLUSTER_DIRECTORY}/{nr+1}_{n_clusters}_BioGRID.txt\", 'w') as f:\n for cluster in kmedoids_instance.get_clusters():\n f.write(' '.join(df.columns[cluster]) + '\\n')\n t2 = time.time()\n print(f'{n_clusters}: {t2-t1:.2f}sec', end='\\r')\n print()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecf1d48bfde3563ba57abb6976b13a5812c1e6ba | 41,098 | ipynb | Jupyter Notebook | jupyter/topic05_bagging_rf/topic5_part1_bagging.ipynb | ivan-magda/mlcourse_open_homeworks | bc67fe6b872655e8e5628ec14b01fde407c5eb3c | [
"MIT"
] | 1 | 2018-10-24T08:35:29.000Z | 2018-10-24T08:35:29.000Z | jupyter/topic05_bagging_rf/topic5_part1_bagging.ipynb | ivan-magda/mlcourse_open_homeworks | bc67fe6b872655e8e5628ec14b01fde407c5eb3c | [
"MIT"
] | null | null | null | jupyter/topic05_bagging_rf/topic5_part1_bagging.ipynb | ivan-magda/mlcourse_open_homeworks | bc67fe6b872655e8e5628ec14b01fde407c5eb3c | [
"MIT"
] | 3 | 2019-10-03T22:32:24.000Z | 2021-01-13T10:09:22.000Z | 167.065041 | 24,772 | 0.852377 | [
[
[
"<center>\n<img src=\"../../img/ods_stickers.jpg\">\n## Открытый курс по машинному обучению\n</center>\nАвторы материала: Data Science интерн Ciklum, студент магистерской программы CSDS UCU Виталий Радченко, программист-исследователь Mail.ru Group, старший преподаватель Факультета Компьютерных Наук ВШЭ Юрий Кашницкий. Материал распространяется на условиях лицензии [Creative Commons CC BY-NC-SA 4.0](https://creativecommons.org/licenses/by-nc-sa/4.0/). Можно использовать в любых целях (редактировать, поправлять и брать за основу), кроме коммерческих, но с обязательным упоминанием автора материала.",
"_____no_output_____"
],
[
"# <center> Тема 5. Композиции алгоритмов, случайный лес</center>\n## <center> Часть 1. Bagging</center>",
"_____no_output_____"
],
[
"Из прошлых лекций вы уже узнали про разные алгоритмы классификации, а также научились правильно валидироваться и оценивать качество модели. Но что делать, если вы уже нашли лучшую модель и повысить точность модели больше не можете? В таком случае нужно применить более продвинутые техники машинного обучения, которые можно объединить словом «ансамбли». Ансамбль — это некая совокупность, части которой образуют единое целое. Из повседневной жизни вы знаете музыкальные ансамбли, где объединены несколько музыкальных инструментов, архитектурные ансамбли с разными зданиями и т.д. \n\n### Ансамбли\n\nХорошим примером ансамблей считается теорема Кондорсе «о жюри присяжных» (1784). Если каждый член жюри присяжных имеет независимое мнение, и если вероятность правильного решения члена жюри больше 0.5, то тогда вероятность правильного решения присяжных в целом возрастает с увеличением количества членов жюри и стремится к единице. Если же вероятность быть правым у каждого из членов жюри меньше 0.5, то вероятность принятия правильного решения присяжными в целом монотонно уменьшается и стремится к нулю с увеличением количества присяжных. \n- $\\large N $ — количество присяжных\n- $\\large p $ — вероятность правильного решения присяжного\n- $\\large \\mu $ — вероятность правильного решения всего жюри\n- $\\large m $ — минимальное большинство членов жюри, $ m = floor(N/2) + 1 $\n- $\\large C_N^i$ — число [сочетаний](https://ru.wikipedia.org/wiki/%D0%A1%D0%BE%D1%87%D0%B5%D1%82%D0%B0%D0%BD%D0%B8%D0%B5) из $N$ по $i$\n$$ \\large \\mu = \\sum_{i=m}^{N}C_N^ip^i(1-p)^{N-i} $$\nЕсли $\\large p > 0 $, то $\\large \\mu > p $\nЕсли $\\large N \\rightarrow \\infty $, то $\\large \\mu \\rightarrow 1 $\n<img src=\"../../img/bull.png\" align=\"right\" width=15% height=15%>\nДавайте рассмотрим ещё один пример ансамблей — \"Мудрость толпы\". Фрэнсис Гальтон в 1906 году посетил рынок, где проводилась некая лотерея для крестьян. \nИх собралось около 800 человек и они пытались угадать вес быка, который стоял перед ними. Его вес составлял 1198 фунтов. Ни один крестьянин не угадал точный вес быка, но если посчитать среднее от их предсказаний, то получим 1197 фунтов.\nЭту идею уменьшения ошибки применили и в машинном обучении.\n\n\n### Бутстрэп\n\nBagging (от Bootstrap aggregation) — это один из первых и самых простых видов ансамблей. Он был придуман [Ле́о Бре́йманом](https://ru.wikipedia.org/wiki/Брейман,_Лео) в 1994 году. Бэггинг основан на статистическом методе бутстрэппинга, который позволяет оценивать многие статистики сложных моделей.\n\nМетод бутстрэпа заключается в следующем. Пусть имеется выборка $\\large X$ размера $\\large N$. Равномерно возьмем из выборки $\\large N$ объектов с возвращением. Это означает, что мы будем $\\large N$ раз выбирать произвольный объект выборки (считаем, что каждый объект «достается» с одинаковой вероятностью $\\large \\frac{1}{N}$), причем каждый раз мы выбираем из всех исходных $\\large N$ объектов. Можно представить себе мешок, из которого достают шарики: выбранный на каком-то шаге шарик возвращается обратно в мешок, и следующий выбор опять делается равновероятно из того же числа шариков. Отметим, что из-за возвращения среди них окажутся повторы. Обозначим новую выборку через $\\large X_1$. Повторяя процедуру $\\large M$ раз, сгенерируем $\\large M$ подвыборок $\\large X_1, \\dots, X_M$. Теперь мы имеем достаточно большое число выборок и можем оценивать различные статистики исходного распределения.\n\n\n\nДавайте для примера возьмем вам уже известный датасет `telecom_churn` из прошлых уроков нашего курса. Напомним, что это задача бинарной классификации оттока клиентов. Одним из самых важных признаков в этом датасете является количество звонков в сервисный центр, которые были сделаны клиентом. Давайте попробуем визулизировать данные и посмотреть на распределение данного признака.\n\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom matplotlib import pyplot as plt\nplt.style.use('ggplot')\nplt.rcParams['figure.figsize'] = 10, 6\nimport seaborn as sns\n%matplotlib inline\n\ntelecom_data = pd.read_csv('../../data/telecom_churn.csv')\n\nfig = sns.kdeplot(telecom_data[telecom_data['Churn'] == False]['Customer service calls'], label = 'Loyal')\nfig = sns.kdeplot(telecom_data[telecom_data['Churn'] == True]['Customer service calls'], label = 'Churn') \nfig.set(xlabel='Количество звонков', ylabel='Плотность') \nplt.show()",
"_____no_output_____"
]
],
[
[
"Как вы уже могли заметить, количество звонков в сервисный центр у лояльных клиентов меньше, чем у наших бывших клиентов. Теперь было бы хорошо оценить сколько в среднем делает звонков каждая из групп. Так как данных в нашем датасете мало, то искать среднее не совсем правильно, лучше применить наши новые знания бутстрэпа. Давайте сгенерируем 1000 новых подвыборок из нашей генеральной совокупности и сделаем интервальную оценку среднего. ",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndef get_bootstrap_samples(data, n_samples):\n # функция для генерации подвыборок с помощью бутстрэпа\n indices = np.random.randint(0, len(data), (n_samples, len(data)))\n samples = data[indices]\n \n return samples\n\ndef stat_intervals(stat, alpha):\n # функция для интервальной оценки\n boundaries = np.percentile(stat, [100 * alpha / 2., 100 * (1 - alpha / 2.)])\n \n return boundaries\n\n# сохранение в отдельные numpy массивы данных по лояльным и уже бывшим клиентам\nloyal_calls = telecom_data[telecom_data['Churn'] == False]['Customer service calls'].values\nchurn_calls= telecom_data[telecom_data['Churn'] == True]['Customer service calls'].values\n\n# ставим seed для воспроизводимости результатов\nnp.random.seed(0)\n\n# генерируем выборки с помощью бутстрэра и сразу считаем по каждой из них среднее\nloyal_mean_scores = [np.mean(sample) \n for sample in get_bootstrap_samples(loyal_calls, 1000)]\nchurn_mean_scores = [np.mean(sample) \n for sample in get_bootstrap_samples(churn_calls, 1000)]\n\n# выводим интервальную оценку среднего\nprint(\"Service calls from loyal: mean interval\", stat_intervals(loyal_mean_scores, 0.05))\nprint(\"Service calls from churn: mean interval\", stat_intervals(churn_mean_scores, 0.05))",
"Service calls from loyal: mean interval [1.4077193 1.49473684]\nService calls from churn: mean interval [2.0621118 2.39761905]\n"
]
],
[
[
"\nВ итоге мы получили, что с 95% вероятностью среднее число звонков от лояльных клиентов будет лежать в промежутке между 1.40 и 1.50, в то время как наши бывшие клиенты звонили в среднем от 2.06 до 2.40 раз. Также ещё можно обратить внимание, что интервал для лояльных клиентов уже, что довольно логично, так как они звонят редко (в основном 0, 1 или 2 раза), а недовольные клиенты будут звонить намного чаще, но со временем их терпение закончится, и они поменяют оператора.\n\n### Бэггинг\n\n\nТеперь вы имеете представление о бустрэпе, и мы можем перейти непосредственно к бэггингу. Пусть имеется обучающая выборка $\\large X$. С помощью бутстрэпа сгенерируем из неё выборки $\\large X_1, \\dots, X_M$. Теперь на каждой выборке обучим свой классификатор $\\large a_i(x)$. Итоговый классификатор будет усреднять ответы всех этих алгоритмов (в случае классификации это соответствует голосованию): $\\large a(x) = \\frac{1}{M}\\sum_{i = 1}^M a_i(x)$. Эту схему можно представить картинкой ниже.\n\n<img src=\"../../img/bagging.png\" alt=\"image\"/>\n\nРассмотрим задачу регрессии с базовыми алгоритмами $\\large b_1(x), \\dots , b_n(x)$. Предположим, что существует истинная функция ответа для всех объектов $\\large y(x)$, а также задано распределение на объектах $\\large p(x)$. В этом случае мы можем записать ошибку каждой функции регрессии $$ \\large \\varepsilon_i(x) = b_i(x) − y(x), i = 1, \\dots, n$$\nи записать матожидание среднеквадратичной ошибки $$ \\large E_x(b_i(x) − y(x))^{2} = E_x \\varepsilon_i (x). $$\n\nСредняя ошибка построенных функций регрессии имеет вид $$ \\large E_1 = \\frac{1}{n}E_x \\sum_{i=1}^n \\varepsilon_i^{2}(x) $$\n\nПредположим, что ошибки несмещены и некоррелированы: \n\n$$ \\large \\begin{array}{rcl} E_x\\varepsilon_i(x) &=& 0, \\\\\nE_x\\varepsilon_i(x)\\varepsilon_j(x) &=& 0, i \\neq j. \\end{array}$$\n\nПостроим теперь новую функцию регрессии, которая будет усреднять ответы построенных нами функций:\n$$ \\large a(x) = \\frac{1}{n}\\sum_{i=1}^{n}b_i(x) $$\n\nНайдем ее среднеквадратичную ошибку:\n\n$$ \\large \\begin{array}{rcl}E_n &=& E_x\\Big(\\frac{1}{n}\\sum_{i=1}^{n}b_i(x)-y(x)\\Big)^2 \\\\\n&=& E_x\\Big(\\frac{1}{n}\\sum_{i=1}^{n}\\varepsilon_i\\Big)^2 \\\\\n&=& \\frac{1}{n^2}E_x\\Big(\\sum_{i=1}^{n}\\varepsilon_i^2(x) + \\sum_{i \\neq j}\\varepsilon_i(x)\\varepsilon_j(x)\\Big) \\\\\n&=& \\frac{1}{n}E_1\\end{array}$$\n\nТаким образом, усреднение ответов позволило уменьшить средний квадрат ошибки в n раз!\n\nНапомним вам из нашего предыдущего [урока](https://habrahabr.ru/company/ods/blog/323890/#razlozhenie-oshibki-na-smeschenie-i-razbros-bias-variance-decomposition), как раскладывается общая ошибка:\n$$\\large \\begin{array}{rcl} \n\\text{Err}\\left(\\vec{x}\\right) &=& \\mathbb{E}\\left[\\left(y - \\hat{f}\\left(\\vec{x}\\right)\\right)^2\\right] \\\\\n&=& \\sigma^2 + f^2 + \\text{Var}\\left(\\hat{f}\\right) + \\mathbb{E}\\left[\\hat{f}\\right]^2 - 2f\\mathbb{E}\\left[\\hat{f}\\right] \\\\\n&=& \\left(f - \\mathbb{E}\\left[\\hat{f}\\right]\\right)^2 + \\text{Var}\\left(\\hat{f}\\right) + \\sigma^2 \\\\\n&=& \\text{Bias}\\left(\\hat{f}\\right)^2 + \\text{Var}\\left(\\hat{f}\\right) + \\sigma^2\n\\end{array}$$\n\nБэггинг позволяет снизить дисперсию (variance) обучаемого классификатора, уменьшая величину, на сколько ошибка будет отличаться, если обучать модель на разных наборах данных, или другими словами, предотвращает переобучение. Эффективность бэггинга достигается благодаря тому, что базовые алгоритмы, обученные по различным подвыборкам, получаются достаточно различными, и их ошибки взаимно компенсируются при голосовании, а также за счёт того, что объекты-выбросы могут не попадать в некоторые обучающие подвыборки.\n\nВ библиотеке `scikit-learn` есть реализация `BaggingRegressor` и `BaggingClassifier`, которая позволяет использовать большинство других алгоритмов \"внутри\". Рассмотрим на практике как работает бэггинг и сравним его с деревом решений, воспользуясь примером из [документации](http://scikit-learn.org/stable/auto_examples/ensemble/plot_bias_variance.html#sphx-glr-auto-examples-ensemble-plot-bias-variance-py).\n\n\n\nОшибка дерева решений\n$$ \\large 0.0255 (Err) = 0.0003 (Bias^2) + 0.0152 (Var) + 0.0098 (\\sigma^2) $$\nОшибка бэггинга\n$$ \\large 0.0196 (Err) = 0.0004 (Bias^2) + 0.0092 (Var) + 0.0098 (\\sigma^2) $$\n\nПо графику и результатам выше видно, что ошибка дисперсии намного меньше при бэггинге, как мы и доказали теоретически выше. \n\nБэггинг эффективен на малых выборках, когда исключение даже малой части обучающих объектов приводит к построению существенно различных базовых классификаторов. В случае больших выборок обычно генерируют подвыборки существенно меньшей длины.\n\nСледует отметить, что рассмотренный нами пример не очень применим на практике, поскольку мы сделали предположение о некоррелированности ошибок, что редко выполняется. Если это предположение неверно, то уменьшение ошибки оказывается не таким значительным. В следующих лекциях мы рассмотрим более сложные методы объединения алгоритмов в композицию, которые позволяют добиться высокого качества в реальных задачах.\n\n### Out-of-bag error\n\nЗабегая вперед, отметим, что при использовании случайных лесов нет необходимости в кросс-валидации или в отдельном тестовом наборе, чтобы получить несмещенную оценку ошибки набора тестов. Внутренняя оценка во время работы получается следующим образом:\n\nКаждое дерево строится с использованием разных образцов бутстрэпа из исходных данных. Примерно 37% примеров остаются вне выборки бутстрэпа и не используется при построении k-го дерева.\n\nЭто можно легко доказать: пусть в выборке $\\large \\ell$ объектов. На каждом шаге все объекты попадают в подвыборку с возвращением равновероятно, т.е отдельный объект — с вероятностью $\\large\\frac{1}{\\ell}.$ Вероятность того, что объект НЕ попадет в подвыборку (т.е. его не взяли $\\large \\ell$ раз): $\\large (1 - \\frac{1}{\\ell})^\\ell$. При $\\large \\ell \\rightarrow +\\infty$ получаем один из \"замечательных\" пределов $\\large \\frac{1}{e}$. Тогда вероятность попадания конкретного объекта в подвыборку $\\large \\approx 1 - \\frac{1}{e} \\approx 63\\%$.\n\nДавайте рассмотрим, как это работает на практике:\n\n\nНа рисунке видно, что наш классификатор ошибся в 4 наблюдениях, которые мы не использовали для тренировки. Значит точность нашего классификатора: $\\large \\frac{11}{15}*100\\% = 73.33\\%$\n\nПолучается, что каждый базовый алгоритм обучается на ~63% исходных объектов. Значит, на оставшихся ~37% его можно сразу проверять. Out-of-Bag оценка — это усредненная оценка базовых алгоритмов на тех ~37% данных, на которых они не обучались.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecf1d91ffb3c3d711bbdaf4d44e92fa30af05861 | 5,544 | ipynb | Jupyter Notebook | 2.1-AFirstLookAtANeuralNet.ipynb | marcemq/DL-with-python | 6ac2e780cfe5a6ef93fbc59fded4d5eb02ac6ee8 | [
"CC0-1.0"
] | null | null | null | 2.1-AFirstLookAtANeuralNet.ipynb | marcemq/DL-with-python | 6ac2e780cfe5a6ef93fbc59fded4d5eb02ac6ee8 | [
"CC0-1.0"
] | null | null | null | 2.1-AFirstLookAtANeuralNet.ipynb | marcemq/DL-with-python | 6ac2e780cfe5a6ef93fbc59fded4d5eb02ac6ee8 | [
"CC0-1.0"
] | null | null | null | 23.1 | 107 | 0.511905 | [
[
[
"import keras\nkeras.__version__",
"_____no_output_____"
]
],
[
[
"# A first look at a neural network\n\nInitial neural net to for MNIST dataset to verify that we have all the software requirements in place",
"_____no_output_____"
]
],
[
[
"from keras.datasets import mnist\n(train_images, train_labels),(test_images,test_labels) = mnist.load_data()",
"_____no_output_____"
],
[
"# checking train data shape/len\nprint(train_images.shape)\nprint(len(train_labels))\ntrain_labels",
"(60000, 28, 28)\n60000\n"
],
[
"# checking test data shape/len\nprint(test_images.shape)\nprint(len(test_labels))\ntest_labels",
"(10000, 28, 28)\n10000\n"
],
[
"# Network architecture\nfrom keras import models\nfrom keras import layers\nfrom tensorflow.keras.utils import to_categorical\n\nnetwork = models.Sequential()\nnetwork.add(layers.Dense(512, activation='relu', input_shape=(28*28,)))\nnetwork.add(layers.Dense(10, activation='softmax'))",
"_____no_output_____"
],
[
"# Compilation step\nnetwork.compile(optimizer='rmsprop', loss='categorical_crossentropy', metrics=['accuracy'])",
"_____no_output_____"
],
[
"# preparing the image data\ntrain_images = train_images.reshape((60000, 28 * 28))\ntrain_images = train_images.astype('float32') / 255\n\ntest_images = test_images.reshape((10000, 28 * 28))\ntest_images = test_images.astype('float32') / 255",
"_____no_output_____"
],
[
"# preparing the labels\ntrain_labels = to_categorical(train_labels)\ntest_labels = to_categorical(test_labels)",
"_____no_output_____"
],
[
"# traing neural net\nnetwork.fit(train_images, train_labels, epochs=5, batch_size=128)",
"Epoch 1/5\n469/469 [==============================] - 2s 3ms/step - loss: 0.2586 - accuracy: 0.9255\nEpoch 2/5\n469/469 [==============================] - 1s 3ms/step - loss: 0.1032 - accuracy: 0.9696\nEpoch 3/5\n469/469 [==============================] - 1s 3ms/step - loss: 0.0682 - accuracy: 0.9794\nEpoch 4/5\n469/469 [==============================] - 1s 3ms/step - loss: 0.0503 - accuracy: 0.9847\nEpoch 5/5\n469/469 [==============================] - 1s 3ms/step - loss: 0.0385 - accuracy: 0.9886\n"
],
[
"# testing model performance with test_images, test_label\ntest_loss, test_acc = network.evaluate(test_images, test_labels)\nprint('test_acc:', test_acc)",
"313/313 [==============================] - 0s 638us/step - loss: 0.0725 - accuracy: 0.9763\ntest_acc: 0.9763000011444092\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf1d9a2ac55811a8fb66ecd6a14fa78e5d8d188 | 8,765 | ipynb | Jupyter Notebook | notebooks/State.ipynb | sheldon-cheah/cppkernel | 212c81f34c2f144d605fc0be4a90327989ab7625 | [
"BSD-3-Clause"
] | null | null | null | notebooks/State.ipynb | sheldon-cheah/cppkernel | 212c81f34c2f144d605fc0be4a90327989ab7625 | [
"BSD-3-Clause"
] | null | null | null | notebooks/State.ipynb | sheldon-cheah/cppkernel | 212c81f34c2f144d605fc0be4a90327989ab7625 | [
"BSD-3-Clause"
] | null | null | null | 8,765 | 8,765 | 0.681689 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecf1df73e0bc91fd934e887b44d4883c85236e24 | 2,183 | ipynb | Jupyter Notebook | notebooks/chap7-tb.ipynb | maruel/deepdream | 39e1883d7ee0c4393ba0b35bd7cc9ac2066e4e72 | [
"Apache-2.0"
] | 1 | 2022-02-01T21:27:52.000Z | 2022-02-01T21:27:52.000Z | notebooks/chap7-tb.ipynb | maruel/deepdream | 39e1883d7ee0c4393ba0b35bd7cc9ac2066e4e72 | [
"Apache-2.0"
] | null | null | null | notebooks/chap7-tb.ipynb | maruel/deepdream | 39e1883d7ee0c4393ba0b35bd7cc9ac2066e4e72 | [
"Apache-2.0"
] | null | null | null | 30.746479 | 109 | 0.596427 | [
[
[
"%matplotlib widget\n%load_ext tensorboard\n%tensorboard --logdir ./logdir/tensorboard\nimport time\nfrom matplotlib import pyplot as plt\nimport numpy as np\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.keras import layers",
"_____no_output_____"
],
[
"def get_mnist_model():\n inputs = keras.Input(shape=(28 * 28,))\n features = layers.Dense(512, activation=\"relu\")(inputs)\n features = layers.Dropout(0.5)(features)\n outputs = layers.Dense(10, activation=\"softmax\")(features)\n model = keras.Model(inputs, outputs)\n return model\n\n(images, labels), (test_images, test_labels) = keras.datasets.mnist.load_data()\nimages = images.reshape((60000, 28 * 28)).astype(\"float32\") / 255\ntest_images = test_images.reshape((10000, 28 * 28)).astype(\"float32\") / 255\ntrain_img, val_img = images[10000:], images[:10000]\ntrain_lbl, val_lbl = labels[10000:], labels[:10000]\n\nmdl = get_mnist_model()\nmdl.compile(optimizer=\"rmsprop\", loss=\"sparse_categorical_crossentropy\", metrics=[\"accuracy\"])\ntb = keras.callbacks.TensorBoard(log_dir=\"./logdir/tensorboard\")\n_ = mdl.fit(train_img, train_lbl, epochs=10, validation_data=(val_img, val_lbl), callbacks=[tb])",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
ecf1e864282e0ae1804c8dab1e8aa220b1d073c3 | 94,264 | ipynb | Jupyter Notebook | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | azimgoogle/pytorch-challenge | eed0b5c7433832a39ac4bc037e32bec2f91c73ae | [
"MIT"
] | null | null | null | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | azimgoogle/pytorch-challenge | eed0b5c7433832a39ac4bc037e32bec2f91c73ae | [
"MIT"
] | null | null | null | intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Exercises).ipynb | azimgoogle/pytorch-challenge | eed0b5c7433832a39ac4bc037e32bec2f91c73ae | [
"MIT"
] | null | null | null | 79.146935 | 16,724 | 0.706601 | [
[
[
"# Neural networks with PyTorch\n\nDeep learning networks tend to be massive with dozens or hundreds of layers, that's where the term \"deep\" comes from. You can build one of these deep networks using only weight matrices as we did in the previous notebook, but in general it's very cumbersome and difficult to implement. PyTorch has a nice module `nn` that provides a nice way to efficiently build large neural networks.",
"_____no_output_____"
]
],
[
[
"# Import necessary packages\n\n%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nimport numpy as np\nimport torch\n\nimport helper\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"\nNow we're going to build a larger network that can solve a (formerly) difficult problem, identifying text in an image. Here we'll use the MNIST dataset which consists of greyscale handwritten digits. Each image is 28x28 pixels, you can see a sample below\n\n<img src='assets/mnist.png'>\n\nOur goal is to build a neural network that can take one of these images and predict the digit in the image.\n\nFirst up, we need to get our dataset. This is provided through the `torchvision` package. The code below will download the MNIST dataset, then create training and test datasets for us. Don't worry too much about the details here, you'll learn more about this later.",
"_____no_output_____"
]
],
[
[
"### Run this cell\n\nfrom torchvision import datasets, transforms\n\n# Define a transform to normalize the data\ntransform = transforms.Compose([transforms.ToTensor(),\n transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),\n ])\n\n# Download and load the training data\ntrainset = datasets.MNIST('MNIST_data/', download=True, train=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)",
"_____no_output_____"
]
],
[
[
"We have the training data loaded into `trainloader` and we make that an iterator with `iter(trainloader)`. Later, we'll use this to loop through the dataset for training, like\n\n```python\nfor image, label in trainloader:\n ## do things with images and labels\n```\n\nYou'll notice I created the `trainloader` with a batch size of 64, and `shuffle=True`. The batch size is the number of images we get in one iteration from the data loader and pass through our network, often called a *batch*. And `shuffle=True` tells it to shuffle the dataset every time we start going through the data loader again. But here I'm just grabbing the first batch so we can check out the data. We can see below that `images` is just a tensor with size `(64, 1, 28, 28)`. So, 64 images per batch, 1 color channel, and 28x28 images.",
"_____no_output_____"
]
],
[
[
"dataiter = iter(trainloader)\nimages, labels = dataiter.next()\nprint(type(images))\nprint(images.shape)\nprint(labels.shape)",
"<class 'torch.Tensor'>\ntorch.Size([64, 1, 28, 28])\ntorch.Size([64])\n"
]
],
[
[
"This is what one of the images looks like. ",
"_____no_output_____"
]
],
[
[
"plt.imshow(images[1].numpy().squeeze(), cmap='Greys_r');",
"_____no_output_____"
]
],
[
[
"First, let's try to build a simple network for this dataset using weight matrices and matrix multiplications. Then, we'll see how to do it using PyTorch's `nn` module which provides a much more convenient and powerful method for defining network architectures.\n\nThe networks you've seen so far are called *fully-connected* or *dense* networks. Each unit in one layer is connected to each unit in the next layer. In fully-connected networks, the input to each layer must be a one-dimensional vector (which can be stacked into a 2D tensor as a batch of multiple examples). However, our images are 28x28 2D tensors, so we need to convert them into 1D vectors. Thinking about sizes, we need to convert the batch of images with shape `(64, 1, 28, 28)` to a have a shape of `(64, 784)`, 784 is 28 times 28. This is typically called *flattening*, we flattened the 2D images into 1D vectors.\n\nPreviously you built a network with one output unit. Here we need 10 output units, one for each digit. We want our network to predict the digit shown in an image, so what we'll do is calculate probabilities that the image is of any one digit or class. This ends up being a discrete probability distribution over the classes (digits) that tells us the most likely class for the image. That means we need 10 output units for the 10 classes (digits). We'll see how to convert the network output into a probability distribution next.\n\n> **Exercise:** Flatten the batch of images `images`. Then build a multi-layer network with 784 input units, 256 hidden units, and 10 output units using random tensors for the weights and biases. For now, use a sigmoid activation for the hidden layer. Leave the output layer without an activation, we'll add one that gives us a probability distribution next.",
"_____no_output_____"
]
],
[
[
"## Your solution\ndef activation(x):\n \"\"\" Sigmoid activation function \n \n Arguments\n ---------\n x: torch.Tensor\n \"\"\"\n return 1/(1+torch.exp(-x))\n\nim = images.view(64, 784)\nW1 = torch.randn(784, 256)\nW2 = torch.randn(256, 10)\n\nB1 = torch.randn(1, 256)\nB2 = torch.randn(1, 10)\n\nh = activation(torch.mm(im,W1) + B1)\nY = torch.mm(h, W2) + B2\nY\n\n# out = # output of your network, should have shape (64,10)",
"_____no_output_____"
],
[
"images.shape",
"_____no_output_____"
],
[
"## Solution\ndef activation(x):\n return 1/(1+torch.exp(-x))\n\n# Flatten the input images\ninputs = images.view(images.shape[0], -1)\n\n# Create parameters\nw1 = torch.randn(784, 256)\nb1 = torch.randn(256)\n\nw2 = torch.randn(256, 10)\nb2 = torch.randn(10)\n\nh = activation(torch.mm(inputs, w1) + b1)\n\nout = torch.mm(h, w2) + b2\nout",
"_____no_output_____"
]
],
[
[
"Now we have 10 outputs for our network. We want to pass in an image to our network and get out a probability distribution over the classes that tells us the likely class(es) the image belongs to. Something that looks like this:\n<img src='assets/image_distribution.png' width=500px>\n\nHere we see that the probability for each class is roughly the same. This is representing an untrained network, it hasn't seen any data yet so it just returns a uniform distribution with equal probabilities for each class.\n\nTo calculate this probability distribution, we often use the [**softmax** function](https://en.wikipedia.org/wiki/Softmax_function). Mathematically this looks like\n\n$$\n\\Large \\sigma(x_i) = \\cfrac{e^{x_i}}{\\sum_k^K{e^{x_k}}}\n$$\n\nWhat this does is squish each input $x_i$ between 0 and 1 and normalizes the values to give you a proper probability distribution where the probabilites sum up to one.\n\n> **Exercise:** Implement a function `softmax` that performs the softmax calculation and returns probability distributions for each example in the batch. Note that you'll need to pay attention to the shapes when doing this. If you have a tensor `a` with shape `(64, 10)` and a tensor `b` with shape `(64,)`, doing `a/b` will give you an error because PyTorch will try to do the division across the columns (called broadcasting) but you'll get a size mismatch. The way to think about this is for each of the 64 examples, you only want to divide by one value, the sum in the denominator. So you need `b` to have a shape of `(64, 1)`. This way PyTorch will divide the 10 values in each row of `a` by the one value in each row of `b`. Pay attention to how you take the sum as well. You'll need to define the `dim` keyword in `torch.sum`. Setting `dim=0` takes the sum across the rows while `dim=1` takes the sum across the columns.",
"_____no_output_____"
]
],
[
[
"def softmax(x):\n ## TODO: Implement the softmax function here\n y = torch.exp(x)\n denominator = torch.sum(y, dim = 1).view(y.shape[0],1)##out.shape finds row number of the inputted matrix\n y /= denominator\n return y\n\n# Here, out should be the output of the network in the previous excercise with shape (64,10)\nprobabilities = softmax(out)\n\n# Does it have the right shape? Should be (64, 10)\nprint(probabilities.shape)\n# Does it sum to 1?\nprint(probabilities.sum(dim=1))",
"torch.Size([64, 10])\ntensor([1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000])\n"
]
],
[
[
"## Building networks with PyTorch\n\nPyTorch provides a module `nn` that makes building networks much simpler. Here I'll show you how to build the same one as above with 784 inputs, 256 hidden units, 10 output units and a softmax output.",
"_____no_output_____"
]
],
[
[
"from torch import nn",
"_____no_output_____"
],
[
"class Network(nn.Module):\n def __init__(self):\n super().__init__()\n \n # Inputs to hidden layer linear transformation\n self.hidden = nn.Linear(784, 256)\n # Output layer, 10 units - one for each digit\n self.output = nn.Linear(256, 10)\n \n # Define sigmoid activation and softmax output \n self.sigmoid = nn.Sigmoid()\n self.softmax = nn.Softmax(dim=1)\n print(type(self))\n \n def forward(self, x):\n # Pass the input tensor through each of our operations\n x = self.hidden(x)\n x = self.sigmoid(x)\n x = self.output(x)\n x = self.softmax(x)\n print(type(self))\n \n return x",
"_____no_output_____"
]
],
[
[
"Let's go through this bit by bit.\n\n```python\nclass Network(nn.Module):\n```\n\nHere we're inheriting from `nn.Module`. Combined with `super().__init__()` this creates a class that tracks the architecture and provides a lot of useful methods and attributes. It is mandatory to inherit from `nn.Module` when you're creating a class for your network. The name of the class itself can be anything.\n\n```python\nself.hidden = nn.Linear(784, 256)\n```\n\nThis line creates a module for a linear transformation, $x\\mathbf{W} + b$, with 784 inputs and 256 outputs and assigns it to `self.hidden`. The module automatically creates the weight and bias tensors which we'll use in the `forward` method. You can access the weight and bias tensors once the network once it's create at `net.hidden.weight` and `net.hidden.bias`.\n\n```python\nself.output = nn.Linear(256, 10)\n```\n\nSimilarly, this creates another linear transformation with 256 inputs and 10 outputs.\n\n```python\nself.sigmoid = nn.Sigmoid()\nself.softmax = nn.Softmax(dim=1)\n```\n\nHere I defined operations for the sigmoid activation and softmax output. Setting `dim=1` in `nn.Softmax(dim=1)` calculates softmax across the columns.\n\n```python\ndef forward(self, x):\n```\n\nPyTorch networks created with `nn.Module` must have a `forward` method defined. It takes in a tensor `x` and passes it through the operations you defined in the `__init__` method.\n\n```python\nx = self.hidden(x)\nx = self.sigmoid(x)\nx = self.output(x)\nx = self.softmax(x)\n```\n\nHere the input tensor `x` is passed through each operation a reassigned to `x`. We can see that the input tensor goes through the hidden layer, then a sigmoid function, then the output layer, and finally the softmax function. It doesn't matter what you name the variables here, as long as the inputs and outputs of the operations match the network architecture you want to build. The order in which you define things in the `__init__` method doesn't matter, but you'll need to sequence the operations correctly in the `forward` method.\n\nNow we can create a `Network` object.",
"_____no_output_____"
]
],
[
[
"# Create the network and look at it's text representation\nmodel = Network()\nmodel",
"<class '__main__.Network'>\n"
]
],
[
[
"You can define the network somewhat more concisely and clearly using the `torch.nn.functional` module. This is the most common way you'll see networks defined as many operations are simple element-wise functions. We normally import this module as `F`, `import torch.nn.functional as F`.",
"_____no_output_____"
]
],
[
[
"import torch.nn.functional as F\n\nclass Network(nn.Module):\n def __init__(self):\n super().__init__()\n # Inputs to hidden layer linear transformation\n self.hidden = nn.Linear(784, 256)\n # Output layer, 10 units - one for each digit\n self.output = nn.Linear(256, 10)\n \n def forward(self, x):\n # Hidden layer with sigmoid activation\n x = F.sigmoid(self.hidden(x))\n # Output layer with softmax activation\n x = F.softmax(self.output(x), dim=1)\n \n return x",
"_____no_output_____"
]
],
[
[
"### Activation functions\n\nSo far we've only been looking at the softmax activation, but in general any function can be used as an activation function. The only requirement is that for a network to approximate a non-linear function, the activation functions must be non-linear. Here are a few more examples of common activation functions: Tanh (hyperbolic tangent), and ReLU (rectified linear unit).\n\n<img src=\"assets/activation.png\" width=700px>\n\nIn practice, the ReLU function is used almost exclusively as the activation function for hidden layers.",
"_____no_output_____"
],
[
"### Your Turn to Build a Network\n\n<img src=\"assets/mlp_mnist.png\" width=600px>\n\n> **Exercise:** Create a network with 784 input units, a hidden layer with 128 units and a ReLU activation, then a hidden layer with 64 units and a ReLU activation, and finally an output layer with a softmax activation as shown above. You can use a ReLU activation with the `nn.ReLU` module or `F.relu` function.",
"_____no_output_____"
]
],
[
[
"## Your solution here\n\nclass Network(nn.Module):\n def __init__(self):\n super().__init__()\n # Inputs to hidden layer linear transformation\n self.fc1 = nn.Linear(784, 128)\n self.fc2 = nn.Linear(128, 64)\n # Output layer, 10 units - one for each digit\n self.fc3 = nn.Linear(64, 10)\n \n def forward(self, x):\n # Hidden layer with sigmoid activation\n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n # Output layer with softmax activation\n x = F.softmax(self.fc3(x), dim=1)\n \n return x\n\nmodel = Network()\nmodel",
"_____no_output_____"
]
],
[
[
"### Initializing weights and biases\n\nThe weights and such are automatically initialized for you, but it's possible to customize how they are initialized. The weights and biases are tensors attached to the layer you defined, you can get them with `model.fc1.weight` for instance.",
"_____no_output_____"
]
],
[
[
"print(type(model.fc1.weight))\nprint(model.fc1.weight)\nprint(type(model.fc1.bias))\nprint(model.fc1.bias)",
"<class 'torch.nn.parameter.Parameter'>\nParameter containing:\ntensor([[-0.0357, 0.0023, -0.0331, ..., 0.0278, 0.0328, -0.0201],\n [ 0.0231, 0.0033, -0.0249, ..., 0.0258, 0.0173, -0.0040],\n [ 0.0017, -0.0085, -0.0272, ..., -0.0024, -0.0154, -0.0193],\n ...,\n [ 0.0342, 0.0175, -0.0161, ..., 0.0127, 0.0075, 0.0004],\n [-0.0004, -0.0073, 0.0293, ..., 0.0148, 0.0315, -0.0043],\n [-0.0299, 0.0035, 0.0045, ..., -0.0073, -0.0212, 0.0146]],\n requires_grad=True)\n<class 'torch.nn.parameter.Parameter'>\nParameter containing:\ntensor([-0.0267, 0.0324, 0.0240, 0.0246, 0.0117, 0.0229, -0.0239, -0.0216,\n -0.0209, 0.0216, 0.0083, -0.0091, -0.0240, 0.0325, -0.0159, 0.0167,\n -0.0336, -0.0240, 0.0243, -0.0355, 0.0044, 0.0259, -0.0307, 0.0188,\n -0.0317, 0.0052, 0.0117, 0.0289, -0.0256, -0.0089, -0.0116, -0.0286,\n -0.0313, 0.0156, 0.0196, -0.0150, -0.0009, -0.0186, -0.0222, -0.0274,\n 0.0188, 0.0264, 0.0303, 0.0145, -0.0020, 0.0317, 0.0056, 0.0073,\n -0.0104, 0.0347, -0.0086, -0.0192, 0.0155, 0.0119, 0.0197, 0.0070,\n -0.0347, 0.0294, -0.0102, 0.0094, 0.0247, -0.0165, 0.0060, -0.0085,\n -0.0345, 0.0096, 0.0125, -0.0275, 0.0108, 0.0016, -0.0314, -0.0219,\n 0.0058, 0.0075, -0.0204, -0.0030, -0.0233, -0.0151, -0.0282, -0.0348,\n 0.0179, -0.0228, 0.0261, -0.0169, -0.0219, -0.0138, -0.0076, -0.0147,\n -0.0251, 0.0129, -0.0254, -0.0066, -0.0211, 0.0181, -0.0298, -0.0273,\n -0.0001, -0.0333, -0.0202, 0.0209, 0.0044, -0.0280, 0.0023, 0.0147,\n -0.0065, 0.0108, 0.0086, -0.0057, 0.0240, -0.0055, 0.0196, -0.0087,\n 0.0217, -0.0346, 0.0013, 0.0004, 0.0344, 0.0066, -0.0323, 0.0322,\n -0.0062, 0.0242, 0.0189, -0.0007, 0.0201, 0.0031, -0.0088, 0.0111],\n requires_grad=True)\n"
]
],
[
[
"For custom initialization, we want to modify these tensors in place. These are actually autograd *Variables*, so we need to get back the actual tensors with `model.fc1.weight.data`. Once we have the tensors, we can fill them with zeros (for biases) or random normal values.",
"_____no_output_____"
]
],
[
[
"type(model.fc1.bias.data)\ntype(model.fc1.bias)",
"_____no_output_____"
],
[
"# Set biases to all zeros\nmodel.fc1.bias.data.fill_(0)",
"_____no_output_____"
],
[
"# sample from random normal with standard dev = 0.01\nmodel.fc1.weight.data.normal_(std=0.01)",
"_____no_output_____"
]
],
[
[
"### Forward pass\n\nNow that we have a network, let's see what happens when we pass in an image.",
"_____no_output_____"
]
],
[
[
"# Grab some data \ndataiter = iter(trainloader)\nimages, labels = dataiter.next()\n\n# Resize images into a 1D vector, new shape is (batch size, color channels, image pixels) \nimages.resize_(64, 1, 784)\n# or images.resize_(images.shape[0], 1, 784) to automatically get batch size\n\n# Forward pass through the network\nimg_idx = 0\nps = model.forward(images[img_idx,:])\n\nimg = images[img_idx]\nhelper.view_classify(img.view(1, 28, 28), ps)",
"_____no_output_____"
]
],
[
[
"As you can see above, our network has basically no idea what this digit is. It's because we haven't trained it yet, all the weights are random!\n\n### Using `nn.Sequential`\n\nPyTorch provides a convenient way to build networks like this where a tensor is passed sequentially through operations, `nn.Sequential` ([documentation](https://pytorch.org/docs/master/nn.html#torch.nn.Sequential)). Using this to build the equivalent network:",
"_____no_output_____"
]
],
[
[
"# Hyperparameters for our network\ninput_size = 784\nhidden_sizes = [128, 64]\noutput_size = 10\n\n# Build a feed-forward network\nmodel = nn.Sequential(nn.Linear(input_size, hidden_sizes[0]),\n nn.ReLU(),\n nn.Linear(hidden_sizes[0], hidden_sizes[1]),\n nn.ReLU(),\n nn.Linear(hidden_sizes[1], output_size),\n nn.Softmax(dim=1))\nprint(model)\n\n# Forward pass through the network and display output\nimages, labels = next(iter(trainloader))\nimages.resize_(images.shape[0], 1, 784)\nps = model.forward(images[0,:])\nhelper.view_classify(images[0].view(1, 28, 28), ps)",
"Sequential(\n (0): Linear(in_features=784, out_features=128, bias=True)\n (1): ReLU()\n (2): Linear(in_features=128, out_features=64, bias=True)\n (3): ReLU()\n (4): Linear(in_features=64, out_features=10, bias=True)\n (5): Softmax()\n)\n"
]
],
[
[
"The operations are availble by passing in the appropriate index. For example, if you want to get first Linear operation and look at the weights, you'd use `model[0]`.",
"_____no_output_____"
]
],
[
[
"print(model[0])\nmodel[0].weight",
"Linear(in_features=784, out_features=128, bias=True)\n"
]
],
[
[
"You can also pass in an `OrderedDict` to name the individual layers and operations, instead of using incremental integers. Note that dictionary keys must be unique, so _each operation must have a different name_.",
"_____no_output_____"
]
],
[
[
"from collections import OrderedDict\nmodel = nn.Sequential(OrderedDict([\n ('fc1', nn.Linear(input_size, hidden_sizes[0])),\n ('relu1', nn.ReLU()),\n ('fc2', nn.Linear(hidden_sizes[0], hidden_sizes[1])),\n ('relu2', nn.ReLU()),\n ('output', nn.Linear(hidden_sizes[1], output_size)),\n ('softmax', nn.Softmax(dim=1))]))\nmodel",
"_____no_output_____"
]
],
[
[
"Now you can access layers either by integer or the name",
"_____no_output_____"
]
],
[
[
"print(model[0])\nprint(model.fc1)",
"Linear(in_features=784, out_features=128, bias=True)\nLinear(in_features=784, out_features=128, bias=True)\n"
]
],
[
[
"In the next notebook, we'll see how we can train a neural network to accuractly predict the numbers appearing in the MNIST images.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecf1e89cc45e3e7444b1cefe3d22a2ece81bf0ee | 231,759 | ipynb | Jupyter Notebook | analysis/HighDensity/collisions.ipynb | SpeedJack/pecsn | 40c757cddec978e06de766c9dff00abf57ccd6b3 | [
"MIT"
] | null | null | null | analysis/HighDensity/collisions.ipynb | SpeedJack/pecsn | 40c757cddec978e06de766c9dff00abf57ccd6b3 | [
"MIT"
] | null | null | null | analysis/HighDensity/collisions.ipynb | SpeedJack/pecsn | 40c757cddec978e06de766c9dff00abf57ccd6b3 | [
"MIT"
] | null | null | null | 199.792241 | 96,216 | 0.853404 | [
[
[
"# Number of Collisions Factorial Analysis (High Density Scenario)",
"_____no_output_____"
]
],
[
[
"import os\nimport math\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom itertools import product, chain, combinations\nfrom scipy import stats\nfrom IPython.display import display, HTML\n%matplotlib inline\n\ndef parse_if_number(s):\n try: return float(s)\n except: return True if s==\"true\" else False if s==\"false\" else s if s else None\n\ndef parse_ndarray(s):\n return np.fromstring(s, sep=' ') if s else None\n\ndef get_file_name(name):\n return name.replace(':', '-')",
"_____no_output_____"
]
],
[
[
"## Config",
"_____no_output_____"
]
],
[
[
"inputFile = 'collisions.csv'\nrepetitionsCount = -1 # -1 = auto-detect\nfactors = ['m', 'D']\n\ntIntervalAlpha = 0.95\n\nplotSize = (10, 10)\nplotStyle = 'seaborn-whitegrid'\nsaveFigures = False\n\n# Filter scalars\nscalarsFilter = ['Floorplan.collisions:sum']\n# Filter vectors\nvectorsFilter = []\n# Percentiles\npercentiles = [0.25, 0.5, 0.75, 0.9, 0.95]\n\n# Performance indexes\nperfIndexes = [\n ('Floorplan.collisions:sum', 'total number of collisions'),\n]\n\n# Transformations\ntransformations = [\n]\n\nintPercentiles = [int(i*100) for i in percentiles]\nvecPerfIndexes = []\n#for intPercentile in intPercentiles:\n# vecPerfIndexes.append(('broadcastTime' + str(intPercentile), 'Broadcast time needed to reach the ' + str(intPercentile) + 'th percentile of the coverage'))\nfor v in vecPerfIndexes:\n perfIndexes.append(v)\n #transformations.append((v[0], lambda x: math.log(x)))",
"_____no_output_____"
]
],
[
[
"## Load scalars",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('exported_data/' + inputFile, converters = {\n 'attrvalue': parse_if_number,\n 'binedges': parse_ndarray,\n 'binvalues': parse_ndarray,\n 'vectime': parse_ndarray,\n 'vecvalue': parse_ndarray,\n})",
"_____no_output_____"
],
[
"if repetitionsCount <= 0: # auto-detect\n repetitionsCount = int(df[df.attrname == 'repetition']['attrvalue'].max()) + 1\nprint('Repetitions:', repetitionsCount)\n\n# Computed\nfactorsCount = len(factors)\n\nif saveFigures:\n os.makedirs('figures', exist_ok=True)",
"Repetitions: 10\n"
],
[
"scalars = df[(df.type == 'scalar') | ((df.type == 'itervar') & (df.attrname != 'TO')) | ((df.type == 'param') & (df.attrname == 'Floorplan.userCount')) | ((df.type == 'runattr') & (df.attrname == 'repetition'))]\nscalars = scalars.assign(qname = scalars.attrname.combine_first(scalars.module + '.' + scalars.name))\nfor index, row in scalars[scalars.type == 'itervar'].iterrows():\n val = scalars.loc[index, 'attrvalue']\n if isinstance(val, str) and not all(c.isdigit() for c in val):\n scalars.loc[index, 'attrvalue'] = eval(val)\nscalars.value = scalars.value.combine_first(scalars.attrvalue.astype('float64'))\nscalars_wide = scalars.pivot_table(index=['run'], columns='qname', values='value')\nscalars_wide.sort_values([*factors, 'repetition'], inplace=True)\ncount = 0\nfor index in scalars_wide.index:\n config = count // repetitionsCount\n scalars_wide.loc[index, 'config'] = config\n count += 1\nscalars_wide = scalars_wide[['config', 'repetition', *factors, *scalarsFilter]]\n\nconfigsCount = int(scalars_wide['config'].max()) + 1\ntotalSims = configsCount*repetitionsCount\ndisplay(HTML(\"<style>div.output_scroll { height: auto; max-height: 48em; }</style>\"))\npd.set_option('display.max_rows', totalSims)\npd.set_option('display.max_columns', 100)\n\n# coverage\n#scalars_wide['coveredUsersPercent'] = scalars_wide['Floorplan.coveredUsers:sum'] / (scalars_wide['Floorplan.userCount'] - 1)",
"_____no_output_____"
]
],
[
[
"## Load vectors",
"_____no_output_____"
]
],
[
[
"vectors = df[df.type == 'vector']\nvectors = vectors.assign(qname = vectors.module + '.' + vectors.name)\nfor index in scalars_wide.index:\n r = index\n cfg = scalars_wide.loc[index, 'config']\n rep = scalars_wide.loc[index, 'repetition']\n vectors.loc[vectors.run == r, 'config'] = cfg\n vectors.loc[vectors.run == r, 'repetition'] = rep\nvectors = vectors[vectors.qname.isin(vectorsFilter)]\nvectors.sort_values(['config', 'repetition', 'qname'], inplace=True)\nvectors = vectors[['config', 'repetition', 'qname', 'vectime', 'vecvalue']]",
"_____no_output_____"
]
],
[
[
"## Compute scalars from vectors",
"_____no_output_____"
]
],
[
[
"def get_percentile(percentile, vectime, vecvalue, totalvalue):\n tofind = percentile * totalvalue\n idx = 0\n csum = vecvalue.cumsum()\n for value in csum:\n if value >= tofind:\n return vectime[idx]\n idx += 1\n return math.inf\n\nfor index, row in vectors.iterrows():\n for vecPerf, percentile in zip(vecPerfIndexes, percentiles):\n vecPerfIndex = vecPerf[0]\n cfg = row['config']\n rep = row['repetition']\n if vecPerfIndex.startswith('broadcastTime'):\n total = scalars_wide[(scalars_wide['config'] == cfg) & (scalars_wide['repetition'] == rep)]['Floorplan.userCount'].values[0] - 1\n else:\n raise Exception('Need to specify total for ' + vecPerfIndex + '. (coding required)')\n value = get_percentile(percentile, row['vectime'], row['vecvalue'], total)\n scalars_wide.loc[(scalars_wide['config'] == cfg) & (scalars_wide['repetition'] == rep), vecPerfIndex] = value",
"_____no_output_____"
]
],
[
[
"## Apply transformations",
"_____no_output_____"
]
],
[
[
"for col, transform in transformations:\n scalars_wide[col] = scalars_wide[col].map(transform, 'ignore')",
"_____no_output_____"
]
],
[
[
"## Full factorial",
"_____no_output_____"
]
],
[
[
"for cfg in range(0, configsCount):\n for perfIndex, _ in perfIndexes:\n mean = scalars_wide[scalars_wide['config'] == cfg][perfIndex].mean()\n variance = scalars_wide[scalars_wide['config'] == cfg][perfIndex].var()\n _, positiveInterval = tuple(v*math.sqrt(variance/repetitionsCount) for v in stats.t.interval(tIntervalAlpha, repetitionsCount - 1))\n negerr = positiveInterval\n poserr = positiveInterval\n if perfIndex == 'coveredUsersPercent':\n poserr = min(1 - mean, positiveInterval)\n scalars_wide.loc[scalars_wide['config'] == cfg, perfIndex + 'Mean'] = mean\n scalars_wide.loc[scalars_wide['config'] == cfg, perfIndex + 'Variance'] = variance\n scalars_wide.loc[scalars_wide['config'] == cfg, perfIndex + 'Negerr'] = negerr\n scalars_wide.loc[scalars_wide['config'] == cfg, perfIndex + 'Poserr'] = poserr\nscalars_wide = scalars_wide[scalars_wide['repetition'] == 0]\n\nfor perfIndex, _ in perfIndexes:\n del scalars_wide[perfIndex]\ndel scalars_wide['repetition']\n#del scalars_wide['Floorplan.userCount']\n#del scalars_wide['Floorplan.coveredUsers:sum']\ndel scalars_wide['config']\n\nscalars_wide ",
"_____no_output_____"
],
[
"for xFactor in factors:\n print('Plotting with', xFactor, 'on the x axis...')\n otherFactors = [fac for fac in factors if fac != xFactor]\n current = scalars_wide.sort_values([xFactor, *otherFactors])\n count = 0\n lastVal = None\n for index,row in current.iterrows():\n if lastVal != None and lastVal != row[xFactor]:\n count = 0\n current.loc[index, 'config'] = count\n count += 1\n lastVal = row[xFactor]\n x = current[xFactor].unique().tolist()\n for perfIndex, perfIndexDesc in perfIndexes:\n plt.figure(figsize=plotSize)\n plt.style.use(plotStyle)\n for cfg in range(0, int(current['config'].max()) + 1):\n y = current[current['config'] == cfg][perfIndex + 'Mean'].tolist()\n poserr = current[current['config'] == cfg][perfIndex + 'Poserr'].tolist()\n negerr = current[current['config'] == cfg][perfIndex + 'Negerr'].tolist()\n realy = []\n realx = []\n realne = []\n realpe = []\n curIdx = 0\n for val in y:\n if not math.isinf(val):\n realy.append(val)\n realx.append(x[curIdx])\n realne.append(negerr[curIdx])\n realpe.append(poserr[curIdx])\n curIdx += 1\n y = realy\n negerr = realne\n poserr = realpe\n err = [negerr, poserr]\n lbl = \"\"\n for fac in otherFactors:\n lbl += fac + '=' + str(current[current['config'] == cfg][fac].tolist()[0]) + ', '\n lbl = lbl[:-2]\n plt.errorbar(x=np.array(realx), y=np.array(y), yerr=np.array(err), capsize=3, linestyle='-', marker='.', markersize=10, label=lbl)\n plt.title('Full factorial plot for ' + perfIndexDesc)\n plt.ylabel(perfIndex)\n plt.xlabel(xFactor)\n plt.legend()\n if saveFigures:\n fig = plt.gcf()\n fig.savefig('figures/' + get_file_name(perfIndex) + '-' + xFactor + '-ffplot.png')\n plt.show()\n print('########################################')\n print()",
"Plotting with m on the x axis...\n"
]
],
[
[
"## Observations\n\nThe number of collisions linearly depends on `D`.\n\nAlso, trickle relaying is very effective in reducing the number of collisions: low values of `m` allows for a low number of collisions.",
"_____no_output_____"
],
[
"### Rerun this notebook\n\nTo rerun this notebook, you can:\n- just rerun the simulations with the corresponding configuration: `./simulate.sh -s HighDensity -c HighDensityCollisions -o collisions` (you will get slighly different results)\n- download our datasets from `https://drive.google.com/file/d/1ZFRV2DecoTvax9lngEsuPPw8Cz1DXvLc/view?usp=sharing` (login with UNIPI institutional account)\n- use our seed to rerun the simulations. Add `seed-set = ${runnumber}22727` to the configuration",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ecf1f159c646aab34c344480e388b930ea8937eb | 31,510 | ipynb | Jupyter Notebook | ds_book/docs/Lesson1a_annex.ipynb | kathrynberger/tensorflow-eo-training | 1ac7d4e26ddab9b64876b526efe4780796a62ad6 | [
"Apache-2.0"
] | 15 | 2022-01-07T18:24:22.000Z | 2022-01-27T05:11:02.000Z | ds_book/docs/Lesson1a_annex.ipynb | kathrynberger/tensorflow-eo-training | 1ac7d4e26ddab9b64876b526efe4780796a62ad6 | [
"Apache-2.0"
] | 56 | 2021-11-19T08:13:28.000Z | 2021-12-09T18:30:23.000Z | ds_book/docs/Lesson1a_annex.ipynb | developmentseed/servir-amazonia-ml | 2d8382d93e77fcd026c2107d84c2e7a107986f94 | [
"Apache-2.0"
] | 7 | 2022-01-10T09:37:54.000Z | 2022-01-21T17:04:47.000Z | 84.477212 | 1,152 | 0.736052 | [
[
[
"# Annex to introduction content",
"_____no_output_____"
],
[
"## Objectives",
"_____no_output_____"
],
[
"The goal of this notebook is to teach some basics of machine learning, deep learning and the TensorFlow framework. Here you will find both the explanations of key concepts and the illustrative programs. ",
"_____no_output_____"
],
[
"### What is Machine Learning?\n\nMachine learning (ML) is a subset of artificial intelligence (AI), which in broad terms, is defined as the ability of a machine to simulate intelligent human behavior. \n\nThe intention of AI and by relation, ML, is to enable machines to learn patterns and subsequently automate certain tasks using sequestered knowledge. These tasks, which are otherwise nominally performed by humans, typically emote complex characteristics that a human similarly learns through pattern recognition.\n\nMachine learning was coined in the 1950s by AI pioneer Arthur Samuel as the \"field of study that gives computers the ability to learn without explicitly being programmed.\" \n\nTo illustrate the importance of ML, we may compare traditional programming and ML. Whereas the former requires humans to create the program with detailed instructions for the computer to follow, ML allows the computer to program itself and learn the instructions through self-guided interaction and analysis. This difference confers benefits in many ways, namely: \n1) time savings on behalf of the human programmer, \n2) time savings on behalf of a human manual interpreter,\n3) overhead involved in describing step-wise instructions for a complex task such as, for example, how to recognize natural oil seeps versus anthropogenically-derived oil splills in satellite imagery\n\nAt its core, machine learning is founded on the consumption of data, and ideally lots of it. ML learns from data provided to it, and generally speaking, the more data the smarter the model. The model trains itself to recognize patterns and features in the data, which then enables it to make predictions about related subject matter. \n\nHumans still have a role in this process. The appropriate ML algorithm has to be selected and supplied with useful information. Furthermore, human programmers can bootstrap an ML model and help reduce its learning curve by tuning certain parameters. \n\nThere are several subcategories of machine learning:\n\n1) **Supervised machine learning** involves training a model with labeled data sets that explicitly give examples of predictive features and their target attribute(s). Most geospatial ML applications are of this type, such as the task of supplying ground truth labels of deforestation events with corresponding satellite imagery to a model during training so that it can predict deforestation events in satellite imagery absent of labels.\n\n2) **Unsupervised machine learning** involves tasking a model to search for patterns in data without the guidance of labels. This is often used to explore data and find patterns that human programmers aren’t explicitly looking for, or when ground truth labels don't exist. As another geospatial example, unsupervised ML might be used to classify land cover without expert knowledge of a specific terrain and its land use categories. For classification, one would manually assign labels to clusters after an unsupervised algorithm is run to find clusters in a dataset.\n\n3) **Self-supervised machine learning** is very new and growing in its application. It sits at the intersection of the former two ML types, and is revolutionary in its ability to perform like a supervised approach albeit with far less labeled data. It does this by learning common sense, which many consider the \"dark matter of artificial intelligence\" ([Facebook AI](https://ai.facebook.com/blog/self-supervised-learning-the-dark-matter-of-intelligence/)). Common sense enables people to learn new concepts or skills without requiring massive amounts of guidance or teaching for every single objective. Rather, common sense is founded by a wealth of background knowledge gathered through observations and experience over time. Self-supervised ML leverages an understanding of the structure of the data by learning supervisory signals from the data itself, such that it can infer any withheld portion of the data from the remaining data. A geospatial example of this might be predicting urban structures that are diverse across different geographies, yet descriptive enough for a human interpreter to infer their identity using common sense.\n\n4) **Reinforcement machine learning**, at last, is a form of ML in which machines learn by way of trial and error to perform optimized actions by being rewarded or penalized. Reinforcement learning can produce models capable of autonomous decision-making and action by iteratively giving feedback on the relative correctness of its decisions and actions.\n\n",
"_____no_output_____"
],
[
"```{important}\nThere are also some problems where machine learning is uniquely equipped to learn insights and make decisions when a human might not, such as drawing relationships from combined spectral indices in a complex terrain. \n```",
"_____no_output_____"
],
[
"### What are Neural Networks?",
"_____no_output_____"
],
[
"Artificial neural networks (ANNs) are a specific, biologically-inspired class of machine learning algorithms. They are modeled after the structure and function of the human brain, in which tens of billions of nodes called neurons are connected through synapses. One can think of the neuron as an elementary processing unit, which processes incoming data and passes along a derived message if the data is weighed to be useful. The many synapses, or message pathways, in a neural network are not uniform in strength, and can become weaker or stronger as more data is consumed and more feedback is received over time. That characteristic is in part why neurons are programmable and responsive to granular and/or system level changes so impressively.\n\n:::{figure-md} neuron-fig\n<img src=\"images/neuron-structure.jpg\" width=\"450px\">\n\nBiological neuron (from [https://training.seer.cancer.gov/anatomy/nervous/tissue.html](https://training.seer.cancer.gov/anatomy/nervous/tissue.html)).\n:::\n\nANNs are essentially program that makes decisions by weighing the evidence and responding to feedback. By varying the input data, types of parameters and their values, we can get different models of decision-making.\n\n:::{figure-md} neuralnet_basic-fig\n<img src=\"https://miro.medium.com/max/1100/1*x6KWjKTOBhUYL0MRX4M3oQ.png\" width=\"450px\">\n\nBasic neural network from [https://towardsdatascience.com/machine-learning-for-beginners-an-introduction-to-neural-networks-d49f22d238f9](https://towardsdatascience.com/machine-learning-for-beginners-an-introduction-to-neural-networks-d49f22d238f9).\n:::\n\nIn network architectures, neurons are grouped in layers, with synapses traversing the interstitial space between neurons in one layer and the next. As data passes through successive layers of the network, features are derived, combined and interpreted in a low-level to high-level trajectory. For example, in the intial layers of a network, you might see a model begin to detect crude lines and edges, and then in the intermediate layers you see the lines combined to form a building, and then the surrounding context or building color or texture might be involved in the latter layers to predict the the type of building.",
"_____no_output_____"
],
[
"#### What are Convolutional Neural Networks?\n\nA Convolutional Neural Network (ConvNet/CNN) is a form of deep learning inspired by the organization of the human visual cortex, in which individual neurons respond to stimuli within a constrained region of the visual field known as the receptive field. Several receptive fields overlap to account for the entire visual area. \n\nIn artificial CNNs, an input matrix such as an image is given importance per various aspects and objects in the image through a moving, convoling receptive field. Very little pre-processing is required for CNNs relative to other classification methods as the need for upfront feature-engineering is removed. Rather, CNNs learn the correct filters and consequent features on their own, provided enough training time and examples. ",
"_____no_output_____"
],
[
":::{figure-md} convolution-fig\n<img src=\"https://miro.medium.com/max/1400/1*Fw-ehcNBR9byHtho-Rxbtw.gif\" width=\"450px\">\n\nConvolution of a kernal over an input matrix from [https://towardsdatascience.com/intuitively-understanding-convolutions-for-deep-learning-1f6f42faee1](https://towardsdatascience.com/intuitively-understanding-convolutions-for-deep-learning-1f6f42faee1).\n:::\n\n#### What is a kernel/filter?\nA kernel is matrix smaller than the input. It acts as a receptive field that moves over the input matrix from left to right and top to bottom and filters for features in the image.\n\n#### What is stride?\nStride refers to the number of pixels that the kernel shifts at each step in its navigation of the input matrix. \n\n#### What is a convolution operation?\nThe convolution operation is the combination of two functions to produce a third function as a result. In effect, it is a merging of two sets of information, the the kernel and the input matrix. The dot products produced by the kernel and the input matrix at each stride are the new values in the resulting matrix, also known as a feature map. \n\n:::{figure-md} convolution-arithmetic-fig\n<img src=\"https://theano-pymc.readthedocs.io/en/latest/_images/numerical_no_padding_no_strides.gif\" width=\"450px\">\n\nConvolution of a kernal over an input matrix from [https://theano-pymc.readthedocs.io/en/latest/tutorial/conv_arithmetic.html](https://theano-pymc.readthedocs.io/en/latest/tutorial/conv_arithmetic.html).\n:::\n\n#### Convolution operation using 3D filter\nAn input image is often represented as a 3D matrix with a dimension for width (pixels), height (pixels), and depth (channels). In the case of an optical image with red, green and blue channels, the kernel/filter matrix is shaped with the same channel depth as the input and the weighted sum of dot products is computed across all 3 dimensions. \n\n#### What is padding?\nAfter a convolution operation, the feature map is by defualt smaller than the original input matrix. To maintain the same spatial dimensions between input matrix and output feature map, we may pad the input matrix with a border of zeroes or ones. There are two types of padding:\n1. Same padding: a border of zeroes or ones is added to match the input/output dimensions\n2. Valid padding: no border is added and the output dimensions are not matched to the input\n\nWe use same padding often because it allows us to construct deeper networks. Without it, the progressive downsizing of the feature maps would constrain how many convolutional layers could be used before the feature map becomes too small.\n\n:::{figure-md} padding-fig\n<img src=\"https://miro.medium.com/max/666/1*noYcUAa_P8nRilg3Lt_nuA.png\" width=\"450px\">\n\n[Padding an input matrix with zeroes](https://ayeshmanthaperera.medium.com/what-is-padding-in-cnns-71b21fb0dd7).\n:::",
"_____no_output_____"
],
[
"### What is Deep Learning?\n",
"_____no_output_____"
],
[
"Deep learning is defined by neural networks with depth, i.e. many layers and connections. The reason for why deep learning is so highly performant lies in the degree of abstraction made possible by feature extraction across so many layers in which each neuron, or processing unit, is interacting with input from neurons in previous layers and making decisions accordingly. The deepest layers of a network once trained can be capabale inferring highly abstract concepts, such as what differentiates a school from a house in satellite imagery.",
"_____no_output_____"
],
[
"```{admonition} **Cost of deep learning**\nDeep learning requires a lot of data to learn from and usually a significant amount of computing power, so it can be expensive depending on the scope of the problem. \n```",
"_____no_output_____"
],
[
"#### Training and Testing Data\n\nThe dataset (e.g. all images and their labels) are split into training, validation and testing sets. A common ratio is 70:20:10 percent, train:validation:test. If randomly split, it is important to check that all class labels exist in all sets and are well represented.\n\n```{important} Why do we need validation and test data? Are they redundant?\nWe need separate test data to evaluate the performance of the model because the validation data is used during training to measure error and therefore inform updates to the model parameters. Therefore, validation data is not unbiased ot the model. A need for new, wholly unseen data to test with is required.\n```",
"_____no_output_____"
],
[
"#### Activation Function, Weights and Biases\n\nIn a neural network, neurons in one layer are connected to neurons in the next layer. As information passes from one neuron to the next, the information is conditioned by the weight of the synapse and is subjected to a bias. As it turns out, these variables, the weights and biases, play a significant role in determining if the information passes further beyond the current neuron.\n\nThe activation function decides whether or not the output from one neuron is useful or not based on a threshold value, and therefore, whether it will be carried from one layer to the next.\n\nWeights, as a reminder, control the signal (or the strength of the connection) between two neurons in two consecutive layers. In other words, a weight decides how much influence the information coming from one neuron will have on the decision made by the next neuron. Smaller weights correlate with less influence from one neuron to the next. \n\nBiases are values which help determine whether or not the activation output from a neuron is going to be passed forward through the network. Each Neuron has a bias, and it is the combination of the weighted sum from the input layer (where weighted sum = weights * input matrix) + the bias that decides the activation of a neuron. In the absence of a bias value, the neuron may not be activated by considering only the weighted sum from input layer. For example, if the weighted sum from the input layer is negative, and the activation function only fires when the weighted sum is greater than zero, the neuron won’t fire. If the neuron doesn’t fire / is not activated, the information from this neuron is not passed through rest of neural network. Adding a bias term of 1, for example, to the weighted sum would make the output of the neuron positive, in doing so allowing the neuron to fire and creating more range with respect to weights which will activate and hence be used throughout the network. Stated simply, bias increases the flexibility of the model by giving credence to a larger range of weights.\n\n:::{figure-md} activation-fig\n<img src=\"https://cdn-images-1.medium.com/max/651/1*UA30b0mJUPYoPvN8yJr2iQ.jpeg\" width=\"450px\">\n\n[Weights, bias, activation](https://laptrinhx.com/statistics-is-freaking-hard-wtf-is-activation-function-207913705/).\n:::\n\nDuring training, the weights and biases are learned and updated using the training and validation dataset to fit the data and reduce error of prediction values relative to target values.",
"_____no_output_____"
],
[
"```{important}\n- **Activation function**: decides whether or not the output from one neuron is useful or not\n- **Weights**: control the signal between neurons in consecutive layers\n- **Biases**: a threshold value that determines the activation of each neuron \n- Weights and biases are the learnable parameters of a deep learning model\n```",
"_____no_output_____"
],
[
"#### Hyper-parameters\n\nImportantly, neural networks train in cycles, where the input data passes through the network, a relationship between input data and target values is learned, a prediction is made, the prediction value is measured for error relative to its true value, and the errors are used to inform updates to parameters in the network, feeding into the next cycle of learning and prediction using the updated information. Bear in mind, unless your dataset is very small, it has to be fed to the model in smaller parts (known as **batches**) in order to avoid memory overload or resource exhaustion.\n\nThe **learning rate** controls how much we want the model to change in response to the estimated error after each training cycle\nThe **batch size** determines the portion of our training dataset that can be fed to the model during each cycle. Stated otherwise, batch size controls the number of training samples to work through before the model’s internal parameters are updated.\n\nThe learning rate is a hyperparameter that controls how much the model may change in response to the estimated error each time the model weights are updated. Choosing the learning rate is challenging as a value too small may result in a long training process that could take a long time to converge on an optimal set of parameters, whereas a value too large may result in learning a sub-optimal set of weights too fast, getting stuck at a local optimum and consequently missing the global optimum. \n\nThink of a batch as a for-loop iterating over one or more samples and making predictions. At the end of the batch's forward pass through the network, the predictions are compared to the expected output variables and an error is calculated. The error is back propogated through the network to adjust the parameters with respect to the error. A training dataset can be divided into one or more batches.\n\nAn **epoch** is defined as the point when all training samples, aka the entire dataset, has passed through the neural network once. The number of epochs controls how many times the entire dataset is cycled through and analyzed by the neural network. We should expect to see the error progressively reduce throughout the course of successive epochs.\n\nThe **optimization function** is really important. It’s what we use to change the attributes of your neural network such as weights and biases in order to reduce the losses. The goal of an optimization function is to minimize the error produced by the model.\n\nThe **loss function**, also known as the cost function, measures how much the model needs to improve based on the prediction errors relative to the true values during training. \n\nThe **accuracy metric** measures the performance of a model. For example, a pixel to pixel comparison for agreement on class.\n\nNote: the **activation function** is also a hyper-parameter.\n\n:::{figure-md} loss_curve-fig\n<img src=\"https://miro.medium.com/max/810/1*UUHvSixG7rX2EfNFTtqBDA.gif\" width=\"450px\">\n\n[Loss curve](https://towardsdatascience.com/machine-learning-fundamentals-via-linear-regression-41a5d11f5220).\n::::::{figure-md} loss_curve-fig\n<img src=\"https://miro.medium.com/max/810/1*UUHvSixG7rX2EfNFTtqBDA.gif\" width=\"450px\">\n\n[Loss curve](https://towardsdatascience.com/machine-learning-fundamentals-via-linear-regression-41a5d11f5220).\n:::",
"_____no_output_____"
],
[
"#### Common Deep Learning Algorithms for Computer Vision",
"_____no_output_____"
],
[
"- Image classification: classifying whole images, e.g. image with clouds, image without clouds\n- Object detection: identifying locations of objects in an image and classifying them, e.g. identify bounding boxes of cars and planes in satellite imagery\n- Semantic segmentation: classifying individual pixels in an image, e.g. land cover classification\n- Instance segmentation: classifying individual pixels in an image in terms of both class and individual membership, e.g. detecting unique agricultural field polygons and classifying them\n- Generative Adversarial: a type of image generation where synthetic images are created from real ones, e.g. creating synthetic landscapes from real landscape images",
"_____no_output_____"
],
[
"#### Semantic Segmentation\nTo pair with the content of these tutorials, we will demonstrate semantic segmentation (supervised) to map land use categories and illegal gold mining activity. \n- Semantic = of or relating to meaning (class)\n- Segmentation = division (of image) into separate parts",
"_____no_output_____"
],
[
"#### U-Net Segmentation Architecture\n\nSemantic segmentation is often distilled into the combination of an encoder and a decoder. An encoder generates logic or feedback from input data, and a decoder takes that feedback and translates it to output data in the same form as the input.\n\nThe U-Net model, which is one of many deep learning segmentation algorithms, has a great illustration of this structure. In Fig. 8, the encoder is on the left side of the model. It consists of consecutive convolutional layers, each followed by ReLU and a max pooling operation to encode feature representations at multiple scales. The encoder can be represented by most feature extraction networks designed for classification. In the initial convolutional layers, the filters learn low level features in an image such as lines or edges. Progressing through further layers, the filters learn more abstract features such as combinations of lines and colors. The encoder downsamples as it moves from extracting low-level, granular features to high level abstract features. \n\nThe decoder, on the right side of the Fig. 8 diagram, is tasked to semantically project the discriminative features learned by the encoder onto the original pixel space to render a dense classification. The decoder consists of deconvolution and concatenation with corresponding features from the encoder followed by regular convolution operations. \n\nDeconvolution in a CNN is used to restore the dimensions of feature maps to the original size of the input image. This operation is also referred to as transposed convolution, upconvolution or upsampling. With deonvolution, the goal is to progressively upsample feature maps to pair with the size of the corresponding concatenation blocks from the encoder. You may see the gray and green arrows, where we concatenate two feature maps together. The main contribution of U-Net in this sense is that while upsampling in the network we are also concatenating the higher resolution feature maps from the encoder network with the upsampled features in order to better learn representations with following convolutions. Since upsampling is a sparse operation we need a good prior from earlier stages to better represent the localization.\n\nFollowing the decoder is the final classification layer, which computes the pixel-wise classification for each cell in the final feature map.\n\nThese models are often applied to computer vision problems where regions of pixel space are representative of a unique class. A semantic segmentation model enables direct localization and quantification of predicted classes.\n\nAlso to note, batch normalization is used as a way of accelerating training and many studies have found it to be important to use to obtain state-of-the-art results on benchmark problems. With batch normalization, each element of a layer in a neural network is normalized to zero mean and unit variance, based on its statistics within a mini-batch. \n\nReLU is an operation, an activation function to be specific, that induces non-linearity. This function intakes the feature map from a convolution operation and remaps it such that any positive value stays exactly the same, and any negative value becomes zero.\n\n:::{figure-md} relu-graph-fig\n<img src=\"https://miro.medium.com/max/3200/1*w48zY6o9_5W9iesSsNabmQ.gif\" width=\"450px\">\n\n[ReLU activation function](https://medium.com/ai%C2%B3-theory-practice-business/magic-behind-activation-function-c6fbc5e36a92).\n:::\n\nMax pooling is used to summarize a feature map and only retain the important structural elements, foregoing the more granular detail that may not be significant to the modeling task. This helps to denoise the signal and helps with computational efficiency. It works similar to convolution in that a kernel with a stride is applied to the feature map and only the maximum value within each patch is reserved.\n\n:::{figure-md} maxpooling-fig\n<img src=\"https://thumbs.gfycat.com/FirstMediumDalmatian-size_restricted.gif\" width=\"450px\">\n\n[Max pooling with a kernal over an input matrix](https://gfycat.com/firstmediumdalmatian).\n:::\n\n:::{figure-md} relu-maxpooling-fig\n<img src=\"https://miro.medium.com/max/1000/1*cmGESKfSZLH2ksqF_kBgfQ.gif\" width=\"450px\">\n\n[ReLU applied to an input matrix](https://towardsdatascience.com/a-laymans-guide-to-building-your-first-image-classification-model-in-r-using-keras-b285deac6572).\n:::\n\n:::{figure-md} relu-maxpooling-fig\n<img src=\"https://miro.medium.com/max/1000/1*cmGESKfSZLH2ksqF_kBgfQ.gif\" width=\"450px\">\n\n[ReLU applied to an input matrix](https://towardsdatascience.com/a-laymans-guide-to-building-your-first-image-classification-model-in-r-using-keras-b285deac6572).\n:::",
"_____no_output_____"
],
[
"#### Auxiliary Notes\n\nGenerally speaking, after training the model is complete, we can use the same evaluation metrics for deep learning as we do for classical ML. So, as an example, for segmentation, confusion matrix and f1-score are applicable in both classical ML (i.e. random forest) and in deep learning (i.e. semantic segmentation).\n\nYet, still there is the question of when we should use deep learning instead of classical ML. One of the great strengths of deep learning is its built in feature extraction, which can handle very complex and /or abstract data. A good rule of thumb: when either 1) the data is very complex and we do not want to do a lot of processing to define the features manually, or 2) when the task suits well to automation and scaling beyond human processing speed, deep learning is a good choice. In contrast, when the data is relatively uniform and/or the feature space is not too noisy, or, when our input data is not exceedingly vast, then classical ML is probably all that is necessary. \n\nTypically, when selecting a model architecture, you start with what are you trying to model (is it to classify an image, detect an object, so on so forth) and then, within that associated family of algorithms, you check for the state of the art model at the present time. Developers all over build and test their models against standard benchmark datasets so as to compare performances fairly. You can reference the performance scores of the different architectures to decide which is the best, but you’ll want to keep in mind the notion of architecture complexity. Some might perform extremely well, but the architectures under the hood are enormous and computationally expensive to use. A simple model architecture will comparatively cost less than a complex one, so identifying the right balance of complexity and performance in relation to training time and executional cost is important to factor in.\n\nYour choice of a loss function is a function of the problem itself. If it’s a binary classification problem, you might choose binary cross-entropy. If it’s a multi-class classification problem, you might choose multi-class cross entropy. In both, cross entropy entails comparing the predicted class probabilities for, let’s say a given pixel, and choosing the highest probability class. We will use an adaption on this loss function for a class-imbalanced dataset. It’s called focal loss and uses class weighting based on frequency in the dataset to tonify the losses associated with each class.\n\nLastly, when we instantiate the model, we can initialize the weights to random values or all zeroes and let the model learn the correct values. Or, we can adopt the trained weights from another model to expedite the learning progress. Remember, in the initial convolutional layers, it’s just simple features that are learned - like combinations of lines and colors - and since that is generally extensible to many image-related applications, we can adopt the weights trained to learn those low-level features and start with that foundational knowledge. This makes it such that the network only really needs to learn the high level features unique to your training data. More on this later on.\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecf20069f288f44b9f3a6afebce97635418d4595 | 121,155 | ipynb | Jupyter Notebook | src/HCI_Project.ipynb | PRISHIta123/DSER_Movie_Subtitles | b1e1cc0f957ba2a61663ab370fa330dad1b331ce | [
"MIT"
] | null | null | null | src/HCI_Project.ipynb | PRISHIta123/DSER_Movie_Subtitles | b1e1cc0f957ba2a61663ab370fa330dad1b331ce | [
"MIT"
] | null | null | null | src/HCI_Project.ipynb | PRISHIta123/DSER_Movie_Subtitles | b1e1cc0f957ba2a61663ab370fa330dad1b331ce | [
"MIT"
] | null | null | null | 39.567276 | 3,670 | 0.514704 | [
[
[
"**Import Libraries**",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport nltk\nimport chardet\nfrom importlib import reload\nimport re",
"_____no_output_____"
],
[
"import sys\nreload(sys)\nsys.getdefaultencoding()",
"_____no_output_____"
]
],
[
[
"**Read Movies Metadata, Lines and Characters**",
"_____no_output_____"
]
],
[
[
"movies_metadata=pd.read_csv(\"../cornell movie-dialogs corpus/movies_metadata.csv\")",
"_____no_output_____"
],
[
"f1 = open(\"../cornell movie-dialogs corpus/movie_lines.csv\", \"rb\")",
"_____no_output_____"
],
[
"movie_lines= f1.read().decode(errors='replace').splitlines()\nprint(type(movie_lines))",
"<class 'list'>\n"
],
[
"f2 =open(\"C://Users/PrishitaRay/Desktop/DSER_Movie_Subtitles/cornell movie-dialogs corpus/movie_characters_metadata.csv\",\"rb\")",
"_____no_output_____"
],
[
"movie_characters_metadata = f2.read().decode(errors='replace').splitlines()",
"_____no_output_____"
],
[
"movies_metadata=np.array(movies_metadata)\nmovies_metadata",
"_____no_output_____"
],
[
"movie_lines=np.array(movie_lines)\nmovie_lines",
"_____no_output_____"
],
[
"L=[]\nfor i in range(0,len(movie_lines)):\n x=movie_lines[i].split(',')\n L.append(x)",
"_____no_output_____"
],
[
"lines=[]",
"_____no_output_____"
],
[
"for i in range(0,len(L)):\n l=[]\n for j in range(0,5):\n l.append(L[i][j])\n lines.append(l) ",
"_____no_output_____"
],
[
"lines[0]",
"_____no_output_____"
],
[
"movie_characters=np.array(movie_characters_metadata)\nmovie_characters ",
"_____no_output_____"
],
[
"L=[]\nfor i in range(0,len(movie_characters)):\n x=movie_characters[i].split(',')\n L.append(x)",
"_____no_output_____"
],
[
"characters=[]",
"_____no_output_____"
],
[
"for i in range(0,len(L)):\n l=[]\n for j in range(0,6):\n l.append(L[i][j])\n characters.append(l)",
"_____no_output_____"
],
[
"characters[0]",
"_____no_output_____"
],
[
"L=[]\nfor i in range(0,len(lines)):\n L.append(lines[i][2])",
"_____no_output_____"
],
[
"ids=set(L)\nmovie_ids=list(ids)\nmovie_ids[0:10]",
"_____no_output_____"
]
],
[
[
"**Map Movie Lines to respective Movie IDs**",
"_____no_output_____"
]
],
[
[
"lines_dict=dict.fromkeys(movie_ids)",
"_____no_output_____"
],
[
"for x in movie_ids:\n l=[]\n for i in range(0,len(lines)):\n if lines[i][2]==x:\n l.append(lines[i][4])\n lines_dict[x]=l",
"_____no_output_____"
],
[
"lines_dict[' m1 '][0:10]",
"_____no_output_____"
]
],
[
[
"**Map Movie Characters to respective Movie IDs**",
"_____no_output_____"
]
],
[
[
"characters_dict=dict.fromkeys(movie_ids)",
"_____no_output_____"
],
[
"for x in movie_ids:\n l=[]\n for i in range(0,len(characters)):\n if characters[i][2]==x:\n l.append(characters[i][1])\n characters_dict[x]=l",
"_____no_output_____"
],
[
"characters_dict[' m2 ']",
"_____no_output_____"
],
[
"movies_dict=dict.fromkeys(movie_ids)\nfor x in movie_ids:\n l=[]\n for i in range(0,len(characters)):\n if characters[i][2]==x:\n l.append(characters[i][3])\n break\n movies_dict[x]=l",
"_____no_output_____"
],
[
"movies_dict[' m3 ']",
"_____no_output_____"
]
],
[
[
"**Map lines spoken in the movie to their actual speakers**",
"_____no_output_____"
]
],
[
[
"actual_speakers=dict.fromkeys(movie_ids)\nfor x in movie_ids:\n l=[]\n for i in range(0,len(lines)):\n if lines[i][2]==x:\n l.append(lines[i][3])\n actual_speakers[x]=l",
"_____no_output_____"
],
[
"actual_speakers[' m4 '][0:50]",
"_____no_output_____"
],
[
"import tensorflow as tf\nfrom tensorflow.contrib.layers import fully_connected",
"_____no_output_____"
]
],
[
[
"**Perform word tokenization and POS Tagging on the movie lines**",
"_____no_output_____"
]
],
[
[
"from nltk import pos_tag,word_tokenize\n\nl=lines_dict[' m510 ']\np=[]\np.append(pos_tag(word_tokenize(l[3])))\np",
"_____no_output_____"
],
[
"POS_Tags=[]\nfor key in lines_dict.keys():\n p=[]\n for x in lines_dict[key]:\n p.append(pos_tag(word_tokenize(x)))\n POS_Tags.append(p)",
"_____no_output_____"
],
[
"POS_Tags[100][0:10]",
"_____no_output_____"
]
],
[
[
"**Lemmatize the POS Tagged Lines using NLTK's inbuilt WordNetLemmatizer**",
"_____no_output_____"
]
],
[
[
"from nltk.stem import WordNetLemmatizer\nlemmatizer=WordNetLemmatizer()",
"_____no_output_____"
],
[
"for i in range(0,len(POS_Tags)):\n for j in range(0,len(POS_Tags[i])):\n for k in range(0,len(POS_Tags[i][j])):\n l=list(POS_Tags[i][j][k])\n l[0]=lemmatizer.lemmatize(l[0])\n t=tuple(l)\n POS_Tags[i][j][k]=t ",
"_____no_output_____"
],
[
"lemmatized_with_POS=POS_Tags",
"_____no_output_____"
],
[
"lemmatized_with_POS[600][0:10]",
"_____no_output_____"
]
],
[
[
"**Remove Stop Words from Lemmatized Lines**",
"_____no_output_____"
]
],
[
[
"remove_stop_words=[]",
"_____no_output_____"
],
[
"from nltk.corpus import stopwords\nimport nltk",
"_____no_output_____"
],
[
"stop_words = stopwords.words('english')",
"_____no_output_____"
],
[
"stop_words[:10]",
"_____no_output_____"
],
[
"for i in range(0,len(POS_Tags)):\n for j in range(0,len(POS_Tags[i])):\n n=len(POS_Tags[i][j])\n for k in range(0,n):\n l=list(POS_Tags[i][j][k])\n l[0]=l[0].lower()\n t=tuple(l)\n POS_Tags[i][j][k]=t",
"_____no_output_____"
],
[
"len(POS_Tags)",
"_____no_output_____"
],
[
"for i in range(0,len(POS_Tags)):\n l2=[]\n for j in range(0,len(POS_Tags[i])):\n l1=[]\n for k in range(0,len(POS_Tags[i][j])):\n l=list(POS_Tags[i][j][k])\n if l[0] not in stop_words:\n l1.append(POS_Tags[i][j][k])\n l2.append(l1)\n remove_stop_words.append(l2)",
"_____no_output_____"
]
],
[
[
"**Create pairs of consecutive dialogs from the processed movie lines**",
"_____no_output_____"
]
],
[
[
"pp_lines_dict=dict.fromkeys(movie_ids)\n\ncnt=0\nfor key in pp_lines_dict.keys():\n l=[]\n for j in range(0,len(POS_Tags[cnt])):\n s=\"\"\n for pair in POS_Tags[cnt][j]:\n if s==\"\":\n s=s+pair[0]\n else:\n s=s+\" \"+pair[0]\n l.append(s)\n pp_lines_dict[key]=l\n cnt= cnt+1",
"_____no_output_____"
]
],
[
[
"**Read the Switchboard Dialog Act Corpus for training**",
"_____no_output_____"
]
],
[
[
"import os\nfiles=os.listdir(\"C://Users/PrishitaRay/Desktop/DSER_Movie_Subtitles/Switchboard Dialog Act Corpus/swda\")\nsw_train=[]\n\nfor i in range(1,len(files)):\n sw_train.append(files[i])\n \nprint(len(sw_train))",
"1140\n"
]
],
[
[
"**Create lists of Dialogs and their corresponding Dialog Acts from the Corpus**",
"_____no_output_____"
]
],
[
[
"#Training on SwitchBoard Dialogs Act Corpus\nimport re\nos.chdir(\"C://Users/PrishitaRay/Desktop/DSER_Movie_Subtitles/Switchboard Dialog Act Corpus/swda\")\ntokenized_dialogs=[]\ndialog_acts=[]",
"_____no_output_____"
],
[
"dialogs=[]\nfor f in sw_train:\n file= pd.read_csv(f)\n d_act= list(file['act_tag'])\n text_with_pos= list(file['pos'])\n \n for d in d_act:\n dialog_acts.append(d)\n\n for i in range(0,len(d_act)):\n #Removing unneccessary delimiters from text\n pos= str(text_with_pos[i]).strip()\n word_tag = map((lambda x : tuple(x.split(\"/\"))), re.split(r\"\\s+\", pos))\n word_tag = filter((lambda x : len(x) == 2), word_tag)\n\n w= [x[0] for x in word_tag]\n tokenized_dialogs.append(w)\n separator=' '\n d= separator.join(w)\n dialogs.append(d)",
"_____no_output_____"
],
[
"print(len(dialogs))\nprint(len(dialog_acts))\nprint(dialogs[0:5])\nprint(dialog_acts[0:5])",
"221512\n221512\n['Okay .', 'So ,', 'I guess ,', 'What kind of experience do you , do you have , then with child care ?', 'I think , uh , I wonder if that worked .']\n['o', 'qw', 'qy^d', '+', '+']\n"
],
[
"#Testing for first movie only\ntest_data=[]\nfor key in pp_lines_dict.keys():\n #List of sentences with word embeddings for each movie\n swe=[]\n for line in pp_lines_dict[key]:\n tokenized = re.sub('[,?.]','', line).lower().split(' ')\n separator=' '\n line=separator.join(tokenized)\n test_data.append(line)\n \n break",
"_____no_output_____"
]
],
[
[
"**Create vocabulary of words in the Corpus based on their occurence frequencies**",
"_____no_output_____"
]
],
[
[
"#List of words\nlow=[]\n\nfor dialog in tokenized_dialogs:\n for token in dialog:\n low.append(token) \n\nlow_cnt={k:v for v,k in enumerate(np.unique(low))}\n\nsorted_lc=sorted(low_cnt.items(), key = lambda kv:(kv[1], kv[0]),reverse=True)[0:10000]\nvocab=dict(sorted_lc)\nprint(len(vocab))\n#print(vocab)",
"10000\n"
]
],
[
[
"**Encode words in a dialog to be trained using One Hot Encoding over the Vocabulary Length along with padding**",
"_____no_output_____"
],
[
"**Generate a sparse matrix of the dialog acts after One Hot Encoding**",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import LabelEncoder,OneHotEncoder\nfrom keras.preprocessing.text import one_hot\nfrom keras.preprocessing.sequence import pad_sequences\nVOCAB_LEN=len(vocab.keys())\nn_timesteps=20\nk=5\nencoded_docs = [one_hot(d, VOCAB_LEN) for d in dialogs]\nmax_length = 20\npadded_docs = pad_sequences(encoded_docs, maxlen=max_length, padding='post')\n\nX= padded_docs[0:100000]\ndialog_acts=np.array(dialog_acts)\nlabel_encoder = LabelEncoder()\nvec = label_encoder.fit_transform(dialog_acts)\nenc= OneHotEncoder(sparse=False)\nY = enc.fit_transform(vec.reshape((vec.shape[0]),1))\nY= Y[0:100000]",
"c:\\users\\prishitaray\\appdata\\local\\programs\\python\\python35\\lib\\site-packages\\sklearn\\preprocessing\\_encoders.py:363: FutureWarning: The handling of integer data will change in version 0.22. Currently, the categories are determined based on the range [0, max(values)], while in the future they will be determined based on the unique values.\nIf you want the future behaviour and silence this warning, you can specify \"categories='auto'\".\nIn case you used a LabelEncoder before this OneHotEncoder to convert the categories to integers, then you can now use the OneHotEncoder directly.\n warnings.warn(msg, FutureWarning)\n"
]
],
[
[
"**Create the Keras Bidirectional LSTMs Model (check the paper in the README file for reference)** ",
"_____no_output_____"
]
],
[
[
"from keras.models import Sequential\nfrom keras.layers import LSTM\nfrom keras.layers import Dropout\nfrom keras.layers import Flatten\nfrom keras.layers import Bidirectional\nfrom keras.layers import TimeDistributed\nfrom keras.layers import Dense\nfrom keras import optimizers\nfrom keras.layers.embeddings import Embedding\n\nmodel= Sequential()\nmodel.add(Embedding(VOCAB_LEN, 200, input_length=20))\nmodel.add(Dropout(0.5))\nmodel.add(Bidirectional(LSTM(100,return_sequences=True)))\n#k=5 for previous 5 timesteps\nmodel.add(LSTM(100))\n#One hot encoding of Dialog Act class\nmodel.add(Dense(303, activation='softmax'))\nsgd = optimizers.SGD(lr=1, decay=0.8, momentum=0.9, nesterov=True)\nmodel.compile(loss='categorical_crossentropy', optimizer=sgd, metrics=['accuracy'])\nprint(model.summary())\nmodel.fit(X, Y, epochs=50,batch_size=32,verbose=2)\n# evaluate the model\nloss, accuracy = model.evaluate(X, Y, verbose=2)\nprint('Accuracy: %f' % (accuracy*100))",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding_1 (Embedding) (None, 20, 200) 2000000 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 20, 200) 0 \n_________________________________________________________________\nbidirectional_1 (Bidirection (None, 20, 200) 240800 \n_________________________________________________________________\nlstm_2 (LSTM) (None, 100) 120400 \n_________________________________________________________________\ndense_1 (Dense) (None, 303) 30603 \n=================================================================\nTotal params: 2,391,803\nTrainable params: 2,391,803\nNon-trainable params: 0\n_________________________________________________________________\nNone\nEpoch 1/50\n - 555s - loss: 2.1822 - acc: 0.4574\nEpoch 2/50\n - 510s - loss: 2.0779 - acc: 0.4743\nEpoch 3/50\n - 573s - loss: 2.0627 - acc: 0.4756\nEpoch 4/50\n - 477s - loss: 2.0321 - acc: 0.4805\nEpoch 5/50\n - 441s - loss: 2.0065 - acc: 0.4874\nEpoch 6/50\n - 448s - loss: 1.9935 - acc: 0.4908\nEpoch 7/50\n - 437s - loss: 1.9865 - acc: 0.4921\nEpoch 8/50\n - 454s - loss: 1.9815 - acc: 0.4926\nEpoch 9/50\n - 439s - loss: 1.9772 - acc: 0.4932\nEpoch 10/50\n - 530s - loss: 1.9747 - acc: 0.4932\nEpoch 11/50\n - 600s - loss: 1.9712 - acc: 0.4938\nEpoch 12/50\n - 615s - loss: 1.9694 - acc: 0.4942\nEpoch 13/50\n - 465s - loss: 1.9672 - acc: 0.4940\nEpoch 14/50\n - 463s - loss: 1.9655 - acc: 0.4947\nEpoch 15/50\n - 465s - loss: 1.9650 - acc: 0.4943\nEpoch 16/50\n - 465s - loss: 1.9625 - acc: 0.4943\nEpoch 17/50\n - 470s - loss: 1.9615 - acc: 0.4945\nEpoch 18/50\n - 470s - loss: 1.9600 - acc: 0.4952\nEpoch 19/50\n - 464s - loss: 1.9585 - acc: 0.4950\nEpoch 20/50\n - 457s - loss: 1.9583 - acc: 0.4950\nEpoch 21/50\n - 465s - loss: 1.9574 - acc: 0.4950\nEpoch 22/50\n - 458s - loss: 1.9560 - acc: 0.4955\nEpoch 23/50\n - 465s - loss: 1.9548 - acc: 0.4954\nEpoch 24/50\n - 452s - loss: 1.9539 - acc: 0.4955\nEpoch 25/50\n - 466s - loss: 1.9535 - acc: 0.4954\nEpoch 26/50\n - 516s - loss: 1.9529 - acc: 0.4957\nEpoch 27/50\n - 481s - loss: 1.9523 - acc: 0.4956\nEpoch 28/50\n - 479s - loss: 1.9516 - acc: 0.4955\nEpoch 29/50\n - 475s - loss: 1.9508 - acc: 0.4957\nEpoch 30/50\n - 526s - loss: 1.9503 - acc: 0.4955\nEpoch 31/50\n - 519s - loss: 1.9491 - acc: 0.4957\nEpoch 32/50\n - 516s - loss: 1.9485 - acc: 0.4961\nEpoch 33/50\n - 545s - loss: 1.9480 - acc: 0.4958\nEpoch 34/50\n - 500s - loss: 1.9481 - acc: 0.4956\nEpoch 35/50\n - 471s - loss: 1.9472 - acc: 0.4956\nEpoch 36/50\n - 492s - loss: 1.9470 - acc: 0.4958\nEpoch 37/50\n - 465s - loss: 1.9465 - acc: 0.4958\nEpoch 38/50\n - 496s - loss: 1.9461 - acc: 0.4959\nEpoch 39/50\n - 506s - loss: 1.9453 - acc: 0.4954\nEpoch 40/50\n - 555s - loss: 1.9448 - acc: 0.4965\nEpoch 41/50\n - 494s - loss: 1.9447 - acc: 0.4956\nEpoch 42/50\n - 441s - loss: 1.9443 - acc: 0.4959\nEpoch 43/50\n - 454s - loss: 1.9434 - acc: 0.4958\nEpoch 44/50\n - 444s - loss: 1.9430 - acc: 0.4960\nEpoch 45/50\n - 457s - loss: 1.9428 - acc: 0.4963\nEpoch 46/50\n - 444s - loss: 1.9423 - acc: 0.4959\nEpoch 47/50\n - 465s - loss: 1.9418 - acc: 0.4962\nEpoch 48/50\n - 440s - loss: 1.9414 - acc: 0.4963\nEpoch 49/50\n - 448s - loss: 1.9414 - acc: 0.4960\nEpoch 50/50\n - 437s - loss: 1.9412 - acc: 0.4960\nAccuracy: 49.806000\n"
]
],
[
[
"**Predict Dialog acts over movie lines**",
"_____no_output_____"
]
],
[
[
"encoded_docs = [one_hot(d, VOCAB_LEN) for d in test_data]\nmax_length = 20\nX_test = pad_sequences(encoded_docs, maxlen=max_length, padding='post')\nohe_Y_pred=model.predict(X_test)",
"_____no_output_____"
],
[
"da=list(set(dialog_acts))\npred_DA=[]\nfor i in range(0,len(ohe_Y_pred)):\n pred=list(ohe_Y_pred[i])\n maxpos=pred.index(max(pred))\n pred_DA.append(da[maxpos])\n\nprint(pred_DA)",
"['ba', 'ba', 'ar^r', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'ar^r', 'qw', 'ba', 'ar^r', 'ba', 'ar^r', 'ar^r', 'ba', 'qw', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'qw', 'ba', 'ba', 'ar^r', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'qw', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'qw', 'ba', 'ar^r', 'ba', 'ba', 'qw', 'ba', 'ar^r', 'ar^r', 'ar^r', 'ar^r', 'ar^r', 'qw', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'qw', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'qw', 'ar^r', 'ba', 'ba', 'qw', 'ar^r', 'ar^r', 'qw', 'ba', 'ba', 'ar^r', 'ba', 'ar^r', 'ba', 'qw', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ar^r', 'ar^r', 'ar^r', 'ar^r', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'qw', 'ba', 'ar^r', 'qw', 'ar^r', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'qw', 'ba', 'ba', 'qw', 'ba', 'ba', 'qw', 'ba', 'ba', 'qw', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ar^r', 'ba', 'ar^r', 'ar^r', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ar^r', 'qw', 'ba', 'ar^r', 'ar^r', 'ar^r', 'ar^r', 'qw', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'qw', 'ba', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ar^r', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'qw', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'qw', 'ba', 'ar^r', 'ba', 'ar^r', 'ba', 'qw', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ar^r', 'ar^r', 'ar^r', 'ba', 'qw', 'qw', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'qw', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'qw', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'qw', 'ar^r', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ba', 'ba', 'ba', 'qw', 'ar^r', 'ar^r', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'qw', 'ar^r', 'ar^r', 'ar^r', 'qw', 'ar^r', 'ba', 'ar^r', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'qw', 'ba', 'ba', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ba', 'ar^r', 'ar^r', 'ba', 'ar^r', 'ar^r', 'ar^r', 'ba', 'ar^r', 'ar^r', 'qw', 'ar^r', 'ba', 'ar^r', 'ba', 'ar^r', 'ba', 'ar^r', 'ar^r', 'ar^r', 'ar^r']\n"
]
],
[
[
"**Rules to determine speaker turns based on rules for a pair of dialog acts**",
"_____no_output_____"
]
],
[
[
"turn_indexes=[]\nfor i in range(0,len(pred_DA)-1):\n #d is for the current dialog, nd is for the next dialog in the pair\n d=pred_DA[i]\n nd=pred_DA[i+1]\n if nd=='b' or nd=='sv' or nd=='aa' or nd=='ba' or nd=='bk' or nd=='fo_o_fw_by_bc' or nd=='bf' or nd=='ft' or nd=='bd':\n turn_indexes.append(i+1)\n elif d=='qo' or d=='qy^d' or d=='qy' or d=='qw' or d=='^g' or d=='qrr':\n if nd=='ny' or nd=='nn' or nd=='na' or nd=='ng' or nd=='no' or nd=='sd' or nd=='sv'or nd=='arp_nd' or nd=='aap_am' or nd=='ng' or nd=='oo_co_cc' or nd=='ar^r':\n turn_indexes.append(i+1)\n elif d=='ad' and nd=='no':\n turn_indexes.append(i+1)\n elif d=='fp' and nd=='sd':\n turn_indexes.append(i+1)\n elif d=='qw^d':\n if nd=='sd' or nd=='ng':\n turn_indexes.append(i+1)\n elif d=='fc' and nd=='aa':\n turn_indexes.append(i+1)\n elif d=='%' or d=='ba' or d=='sv' or d=='sd':\n if nd=='^q':\n turn_indexes.append(i+1)\n elif d=='qh':\n turn_indexes.append(i+1)\n elif d=='fa' or d=='ft' or d=='oo_co_cc' or d=='fc':\n if nd=='bd':\n turn_indexes.append(i+1)\n elif d=='bf' and nd=='aap_am':\n turn_indexes.append(i+1)\n elif d=='bh' or d=='^g':\n if nd=='aa':\n turn_indexes.append(i+1)\n elif d=='t3' and nd=='x':\n turn_indexes.append(i+1)\n elif d=='^q' and nd=='br':\n turn_indexes.append(i+1)\n elif d=='^h':\n if nd=='qw^d' or nd=='br' or nd=='bd' or nd=='^q':\n turn_indexes.append(i+1)\n elif d=='br' and nd=='ad':\n turn_indexes.append(i+1)\n elif d=='arp_nd':\n if nd=='t3' or nd=='oo_co_cc' or nd=='bd' or nd=='ft' or nd=='qw^d':\n turn_indexes.append(i+1)\n elif d=='^2':\n turn_indexes.append(i+1)\n elif d=='h':\n if nd=='bd' or nd=='^q':\n turn_indexes.append(i+1)\n elif d=='t1':\n if nd=='qw^d' or nd=='t3' or nd=='arp_nd' or nd=='no' or nd=='b^m':\n turn_indexes.append(i+1)\n elif d=='ad' and nd=='ft':\n turn_indexes.append(i+1)\n elif d=='%':\n turn_indexes.append(i+1)\n elif d=='fo_o_fw_by_bc' and nd=='bk':\n turn_indexes.append(i+1)",
"_____no_output_____"
]
],
[
[
"**Sample Speaker IDs at predicted speaker turns in the movie and display to the user**",
"_____no_output_____"
]
],
[
[
"print(\"Speaker turns for movie: \",movies_dict[' m0 '])\nfor i in range(0,len(lines_dict[' m0 '])):\n if i in turn_indexes:\n print(lines_dict[' m0 '][i],\"(speaker turn): \",actual_speakers[' m0 '][i])\n else:\n print(lines_dict[' m0 '][i])",
"Speaker turns for movie: [' 10 things i hate about you ']\n They do not!\n They do to! (speaker turn): CAMERON \n I hope so.\n She okay? (speaker turn): CAMERON \n Let's go.\n Wow\n Okay -- you're gonna need to learn how to lie. (speaker turn): BIANCA \n No (speaker turn): CAMERON \n\" I'm kidding. You know how sometimes you just become this \"\"persona\"\"? And you don't know how to quit?\" (speaker turn): BIANCA \n Like my fear of wearing pastels?\n\" The \"\"real you\"\".\"\n What good stuff? (speaker turn): BIANCA \n I figured you'd get to the good stuff eventually.\n Thank God! If I had to hear one more story about your coiffure... (speaker turn): CAMERON \n\" Me. This endless ...blonde babble. I'm like\n What crap?\n do you listen to this crap? (speaker turn): BIANCA \n No...\n\" Then Guillermo says (speaker turn): BIANCA \n You always been this selfish? (speaker turn): CAMERON \n But\n Then that's all you had to say.\n\" Well (speaker turn): BIANCA \n\" You never wanted to go out with 'me (speaker turn): CAMERON \n I was? (speaker turn): BIANCA \n\" I looked for you back at the party (speaker turn): CAMERON \n Tons\n Have fun tonight? (speaker turn): CAMERON \n I believe we share an art instructor (speaker turn): CAMERON \n You know Chastity?\n\" Looks like things worked out tonight (speaker turn): CAMERON \n Hi. (speaker turn): BIANCA \n Who knows? All I've ever heard her say is that she'd dip before dating a guy that smokes. (speaker turn): BIANCA \n So that's the kind of guy she likes? Pretty ones? (speaker turn): CAMERON \n\" Lesbian? No. I found a picture of Jared Leto in one of her drawers (speaker turn): BIANCA \n She's not a... (speaker turn): CAMERON \n I'm workin' on it. But she doesn't seem to be goin' for him. (speaker turn): CAMERON \n\" I really\n Sure have. (speaker turn): CAMERON \n\" Eber's Deep Conditioner every two days. And I never (speaker turn): BIANCA \n How do you get your hair to look like that? (speaker turn): CAMERON \n You're sweet.\n You have my word. As a gentleman\n I counted on you to help my cause. You and that thug are obviously failing. Aren't we ever going on our date? (speaker turn): BIANCA \n You got something on your mind?\n Where? (speaker turn): BIANCA \n There. (speaker turn): CAMERON \n\" Well (speaker turn): CAMERON \n How is our little Find the Wench A Date plan progressing? (speaker turn): BIANCA \n Forget French. (speaker turn): BIANCA \n That's because it's such a nice one. (speaker turn): CAMERON \n I don't want to know how to say that though. I want to know useful things. Like where the good stores are. How much does champagne cost? Stuff like Chat. I have never in my life had to point out my head to someone. (speaker turn): BIANCA \n Right. See? You're ready for the quiz. (speaker turn): CAMERON \n C'esc ma tete. This is my head\n Let me see what I can do. (speaker turn): CAMERON \n\" Gosh\n That's a shame. (speaker turn): CAMERON \n\" Unsolved mystery. She used to be really popular when she started high school (speaker turn): BIANCA \n Why?\n Seems like she could get a date easy enough... (speaker turn): CAMERON \n\" The thing is (speaker turn): BIANCA \n Cameron. (speaker turn): CAMERON \n\" No (speaker turn): BIANCA \n Forget it. (speaker turn): CAMERON \n You're asking me out. That's so cute. What's your name again? (speaker turn): BIANCA \n Okay... then how 'bout we try out some French cuisine. Saturday? Night?\n Not the hacking and gagging and spitting part. Please. (speaker turn): BIANCA \n\" Well (speaker turn): CAMERON \n Can we make this quick? Roxanne Korrine and Andrew Barrett are having an incredibly horrendous public break- up on the quad. Again.\n I did.\n You think you ' re the only sophomore at the prom?\n I don't have to be home 'til two. (speaker turn): CHASTITY \n I have to be home in twenty minutes. (speaker turn): BIANCA \n All I know is -- I'd give up my private line to go out with a guy like Joey. (speaker turn): CHASTITY \n\" Sometimes I wonder if the guys we're supposed to want to go out with are the ones we actually want to go out with\n\" Bianca (speaker turn): CHASTITY \n Combination. I don't know -- I thought he'd be different. More of a gentleman... (speaker turn): BIANCA \n Is he oily or dry? (speaker turn): CHASTITY \n\" He practically proposed when he found out we had the same dermatologist. I mean. Dr. Bonchowski is great an all\n\" Would you mind getting me a drink\n Great\n Joey. (speaker turn): BIANCA \n Who? (speaker turn): CHASTITY \n Where did he go? He was just here. (speaker turn): BIANCA \n You might wanna think about it\n No. (speaker turn): CHASTITY \n Did you change your hair?\n You know the deal. I can ' t go if Kat doesn't go -- (speaker turn): BIANCA \n\" Listen (speaker turn): JOEY \n You're concentrating awfully hard considering it's gym class.\n\" Hi (speaker turn): BIANCA \n\" Hey\n My agent says I've got a good shot at being the Prada guy next year.\n Neat...\n\" It's a gay cruise line\n Queen Harry?\n\" So yeah\n Hopefully. (speaker turn): BIANCA \n\" Exactly So (speaker turn): JOEY \n Expensive? (speaker turn): BIANCA \n It's more (speaker turn): JOEY \n Perm? (speaker turn): KAT \n Patrick -- is that- a. (speaker turn): BIANCA \n It's just you. (speaker turn): KAT \n Is that woman a complete fruit-loop or is it just me?\n No! I just wanted (speaker turn): BIANCA \n What? To completely damage me? To send me to therapy forever? What?\n I just wanted -- (speaker turn): BIANCA \n You set me up. (speaker turn): KAT \n Let go! (speaker turn): BIANCA \n So did you (speaker turn): KAT \n\" You looked beautiful last night (speaker turn): BIANCA \n\" I guess I'll never know (speaker turn): BIANCA \n\" Not all experiences are good (speaker turn): KAT \n\" God\n I guess I thought I was protecting you. (speaker turn): KAT \n I'm not stupid enough to repeat your mistakes.\n That's not (speaker turn): KAT \n\" No. you didn't! If you really thought I could make my own decisions (speaker turn): BIANCA \n I wanted to let you make up your own mind about him.\n Why didn't you tell me?\n\" After that (speaker turn): KAT \n But (speaker turn): BIANCA \n\" Just once. Afterwards (speaker turn): KAT \n You did what?\n He said everyone was doing it. So I did it.\n As in... (speaker turn): BIANCA \n\" Now I do. Back then (speaker turn): KAT \n But you hate Joey (speaker turn): BIANCA \n\" He was (speaker turn): KAT \n Why?\n In 9th. For a month (speaker turn): KAT \n What? (speaker turn): BIANCA \n\" Joey never told you we went out (speaker turn): KAT \n\" I wish I had that luxury. I'm the only sophomore that got asked to the prom and I can't go (speaker turn): BIANCA \n\" I do care. But I'm a firm believer in doing something for your own reasons (speaker turn): KAT \n Like you care.\n\" Listen\n You're welcome. (speaker turn): KAT \n\" I don't get you. You act like you're too good for any of this (speaker turn): BIANCA \n I really don't think I need any social advice from you right now. (speaker turn): BIANCA \n\" Bianca (speaker turn): KAT \n Can we go now?\n You are so completely unbalanced. (speaker turn): BIANCA \n\" Yeah (speaker turn): BIANCA \n It's Shakespeare. Maybe you've heard of him? (speaker turn): KAT \n Like I'm supposed to know what that even means. (speaker turn): BIANCA \n At least I'm not a clouted fen- sucked hedge-pig.\n Can't you forget for just one night that you're completely wretched?\n\" Bogey Lowenstein's party is normal (speaker turn): BIANCA \n What's normal?\n\" You're ruining my life' Because you won't be normal (speaker turn): BIANCA \n I think you're a freak. I think you do this to torture me. And I think you suck. (speaker turn): BIANCA \n What do you think? (speaker turn): KAT \n\" Oh\n It means that Gigglepuss is playing at Club Skunk and we're going. (speaker turn): KAT \n\" Oh my God\n Can you at least start wearing a bra?\n I have the potential to smack the crap out of you if you don't get out of my way. (speaker turn): KAT \n\" Nowhere... Hi (speaker turn): BIANCA \n Where've you been?\n\" I have a date (speaker turn): BIANCA \n I'm missing something.\n Fine. I see that I'm a prisoner in my own house. I'm not a daughter. I'm a possession! (speaker turn): BIANCA \n You're not going unless your sister goes. End of story.\n\" He's not a \"\"hot rod\"\". Whatever that is.\" (speaker turn): BIANCA \n\" It's that hot rod Joey\n\" No (speaker turn): BIANCA \n The prom? Kat has a date? (speaker turn): WALTER \n\" Daddy (speaker turn): BIANCA \n Because she'll scare them away. (speaker turn): WALTER \n Why?\n Promise me you won't talk to any boys unless your sister is present.\n Just for a minute (speaker turn): WALTER \n\" Daddy\n Wear the belly before you go. (speaker turn): WALTER \n It's just a party. Daddy. (speaker turn): BIANCA \n\" Oh\n\" If Kat's not going\n\" Daddy (speaker turn): BIANCA \n\" It's just a party. Daddy (speaker turn): BIANCA \n Otherwise known as an orgy? (speaker turn): WALTER \n\" If you must know\n And where're you going? (speaker turn): WALTER \n\" Daddy (speaker turn): BIANCA \n Exactly my point\n But she doesn't want to date. (speaker turn): BIANCA \n\" But it's not fair -- she's a mutant\n Then neither will you. And I'll get to sleep at night.\n What if she never starts dating?\n No! You're not dating until your sister starts dating. End of discussion.\n\" Now don't get upset. Daddy\n Just sent 'em through. (speaker turn): BRUCE \n\" Padua girls. One tall\n Never (speaker turn): BRUCE \n Fan of a fan. You see a couple of minors come in? (speaker turn): PATRICK \n Didn't have you pegged for a Gigglepuss fan. Aren't they a little too pre-teen belly-button ring for you?\n\" Always a pleasure (speaker turn): PATRICK \n\" Best case scenario (speaker turn): MICHAEL \n You humiliated the woman! Sacrifice yourself on the altar of dignity and even the score. (speaker turn): CAMERON \n\" No (speaker turn): CAMERON \n\" Buttholus extremus. But hey (speaker turn): MICHAEL \n The hell is that? What kind of 'guy just picks up a girl and carries her away while you're talking to her? (speaker turn): CAMERON \n Extremely unfortunate maneuver.\n\" Hell (speaker turn): CAMERON \n You told me that part already.\n It's her favorite band.\n Assail your ears for one night. (speaker turn): MICHAEL \n Okay! I wasn't sure\n He's pretty! (speaker turn): MICHAEL \n Dead at forty-one. (speaker turn): MICHAEL \n Her favorite uncle\n It's a lung cancer issue (speaker turn): MICHAEL \n Number one. She hates smokers\n Are you kidding? He'll piss himself with joy. He's the ultimate kiss ass. (speaker turn): MICHAEL \n Will Bogey get bent?\n\" In that case (speaker turn): MICHAEL \n This is it. A golden opportunity. Patrick can ask Katarina to the party. (speaker turn): CAMERON \n\" Like we had a choice? Besides -- when you let the enemy think he's orchestrating the battle (speaker turn): MICHAEL \n You got him involved? (speaker turn): CAMERON \n Hey -- I've gotta have a few clients when I get to Wall Street.\n I thought you hated those people.\n\" You know (speaker turn): MICHAEL \n That's what I just said\n Did she actually say she'd go out with you? (speaker turn): MICHAEL \n Forget his reputation. Do you think we've got a plan or not? (speaker turn): CAMERON \n\" I'm serious\n They always let felons sit in on Honors Biology? (speaker turn): CAMERON \n No kidding. He's a criminal. I heard he lit a state trooper on fire. He just got out of Alcatraz... (speaker turn): MICHAEL \n He seems like he thrives on danger\n What makes you think he'll do it? (speaker turn): MICHAEL \n You wanna go out with him? (speaker turn): MICHAEL \n What about him?\n\" Unlikely (speaker turn): MICHAEL \n\" I teach her French (speaker turn): CAMERON \n\" The mewling (speaker turn): MICHAEL \n That's her? Bianca's sister?\n\" Yeah (speaker turn): MICHAEL \n\" You could consecrate with her (speaker turn): MICHAEL \n You mean I'd get a chance to talk to her? (speaker turn): CAMERON \n Guess who just signed up for a tutor? (speaker turn): MICHAEL \n Sure do ... my Mom's from Canada\n You know French? (speaker turn): MICHAEL \n\" Joey Dorsey? Perma-shit-grin. I wish I could say he's a moron\n He always have that shit-eating grin? (speaker turn): CAMERON \n\" Because they're bred to. Their mothers liked guys like that\n Why do girls like that always like guys like that?\n\" I could start with your haircut (speaker turn): MICHAEL \n Why not?\n Bianca Stratford. Sophomore. Don't even think about it\n Who is she? (speaker turn): CAMERON \n\" You burn (speaker turn): MICHAEL \n That girl -- I -- (speaker turn): CAMERON \n\" Yeah (speaker turn): MICHAEL \n That I'm used to. (speaker turn): CAMERON \n Couple thousand. Most of them evil (speaker turn): MICHAEL \n How many people go here? (speaker turn): CAMERON \n Get out! (speaker turn): MICHAEL \n Thirty-two. (speaker turn): CAMERON \n How many people were in your old school?\n\" Yeah. A couple. We're outnumbered by the cows (speaker turn): CAMERON \n I was kidding. People actually live there? (speaker turn): MICHAEL \n\" North\n So -- which Dakota you from? (speaker turn): MICHAEL \n C'mon. I'm supposed to give you the tour. (speaker turn): MICHAEL \n So they tell me...\n You the new guy?\n You get the girl. (speaker turn): CAMERON \n What's the worst?\n Where? (speaker turn): PATRICK \n She kissed me. (speaker turn): CAMERON \n You makin' any headway? (speaker turn): PATRICK \n She just needs time to cool off I'll give it a day. (speaker turn): PATRICK \n She hates you with the fire of a thousand suns . That's a direct quote\n I don ' t know. I decided not to nail her when she was too drunk to remember it. (speaker turn): PATRICK \n What'd you do to her?\n\" Then\n Sure (speaker turn): CAMERON \n Cameron -- do you like the girl?\n\" She's partial to Joey\n What 're you talking about?\n It's off. The whole thing.\n\" Cameron\n\" Don't make me do it (speaker turn): PATRICK \n Gigglepuss is playing there tomorrow night. (speaker turn): CAMERON \n So what does that give me? I'm supposed to buy her some noodles and a book and sit around listening to chicks who can't play their instruments? (speaker turn): PATRICK \n\" Okay -- Likes: Thai food (speaker turn): CAMERON \n I've retrieved certain pieces of information on Miss Katarina Stratford I think you'll find helpful. (speaker turn): CAMERON \n What've you got for me? (speaker turn): PATRICK \n Yeah -- we'll see. (speaker turn): PATRICK \n And he means that strictly in a non- prison-movie type of way.\n And why would I do that?\n Leave my sister alone. (speaker turn): KAT \n Your sister here?\n Away. (speaker turn): KAT \n Where ya goin?\n Not at all (speaker turn): JOEY \n Hey -- do you mind? (speaker turn): KAT \n They're running the rest of me next month. (speaker turn): JOEY \n\" Yeah (speaker turn): KAT \n\" The vintage look is over\n Enough with the Barbie n' Ken shit. I know. (speaker turn): JOEY \n\" I don't know (speaker turn): PATRICK \n Get her to act like a human\n Do what? (speaker turn): PATRICK \n How'd you do it? (speaker turn): JOEY \n A deal's a deal. (speaker turn): JOEY \n It's about time. (speaker turn): PATRICK \n\" Forget her sister (speaker turn): PATRICK \n Forget it. (speaker turn): JOEY \n A hundred bucks a date. (speaker turn): PATRICK \n What?\n I just upped my price (speaker turn): PATRICK \n I got her under control. She just acts crazed in public to keep up the image. (speaker turn): PATRICK \n Watching the bitch trash my car doesn't count as a date.\n I'm on it (speaker turn): PATRICK \n\" When I shell out fifty\n\" Fifty (speaker turn): PATRICK \n Take it or leave it. This isn't a negotiation. (speaker turn): JOEY \n\" Fine\n I can't take a girl like that out on twenty bucks. (speaker turn): PATRICK \n How much? (speaker turn): PATRICK \n I can't date her sister until that one gets a boyfriend. And that's the catch. She doesn't want a boyfriend. (speaker turn): JOEY \n You're gonna pay me to take out some girl? (speaker turn): PATRICK \n\" You got it (speaker turn): JOEY \n But you'd go out with her if you had the cake? (speaker turn): JOEY \n You need money to take a girl out (speaker turn): PATRICK \n You just said (speaker turn): JOEY \n\" Sure (speaker turn): PATRICK \n\" Yeah (speaker turn): JOEY \n\" Two legs (speaker turn): PATRICK \n What do you think? (speaker turn): JOEY \n Yeah\n Hey -- it's all for the higher good right? (speaker turn): MICHAEL \n You better not fuck this up. I'm heavily invested. (speaker turn): JOEY \n What? We took bathes together when we were kids.\n You and Verona? (speaker turn): JOEY \n\" Uh (speaker turn): MICHAEL \n I hear you're helpin' Verona. (speaker turn): JOEY \n So what you need to do is recruit a guy who'll go out with her. Someone who's up for the job. (speaker turn): MICHAEL \n Does this conversation have a purpose? (speaker turn): JOEY \n But she can't go out with you because her sister is this insane head case and no one will go out with her. right? (speaker turn): MICHAEL \n We're not. (speaker turn): JOEY \n\" Well (speaker turn): MICHAEL \n We don't chat. (speaker turn): JOEY \n Nope - just came by to chat (speaker turn): MICHAEL \n Are you lost?\n Hey. (speaker turn): MICHAEL \n\" Oh (speaker turn): KAT \n William - he asked me to meet him here.\n Who?\n Have you seen him? (speaker turn): MANDELLA \n\" Oh\n You ' re looking at this from the wrong perspective. We're making a statement. (speaker turn): KAT \n\" Okay (speaker turn): MANDELLA \n\" Listen to you! You sound like Betty (speaker turn): KAT \n\" Well (speaker turn): MANDELLA \n Can you even imagine? Who the hell would go to this a bastion of commercial excess? (speaker turn): KAT \n I got drunk. I puked. I got rejected. It was big fun. (speaker turn): KAT \n You didn't (speaker turn): MANDELLA \n I did Bianca a favor and it backfired.\n You didn't have a choice? Where's Kat and what have you done with her? (speaker turn): MANDELLA \n I didn't have a choice. (speaker turn): KAT \n You went to the party? I thought we were officially opposed to suburban social activity. (speaker turn): MANDELLA \n Who cares? (speaker turn): KAT \n What'd he say?\n No fear. (speaker turn): KAT \n You think this'll work?\n\" If I was Bianca (speaker turn): KAT \n Does it matter?\n\" I appreciate your efforts toward a speedy death\n Neither has his heterosexuality. (speaker turn): KAT \n That's never been proven (speaker turn): MANDELLA \n William didn't even go to high school (speaker turn): KAT \n William would never have gone to a state school.\n\" So he has this huge raging fit about Sarah Lawrence and insists that I go to his male-dominated\n You could always go with me. I'm sure William has some friends. (speaker turn): MANDELLA \n The people at this school are so incredibly foul. (speaker turn): KAT \n But imagine the things he'd say during sex.\n\" I realize that the men of this fine institution are severely lacking (speaker turn): KAT \n An attempted slit.\n What's this? (speaker turn): KAT \n Just a little. (speaker turn): MANDELLA \n\" Mandella (speaker turn): KAT \n Block E? (speaker turn): KAT \n He always look so (speaker turn): MANDELLA \n I'm sure he's completely incapable of doing anything that interesting. (speaker turn): KAT \n That's Pat Verona? The one who was gone for a year? I heard he was doing porn movies.\n Patrick Verona Random skid. (speaker turn): KAT \n Who's that? (speaker turn): MANDELLA \n Don ' t you even dare. . . (speaker turn): PATRICK \n\" Oh (speaker turn): KAT \n Because I like to torture you.\n Why is my veggie burger the only burnt object on this grill?\n\" Yeah (speaker turn): PATRICK \n Is that right? (speaker turn): KAT \n\" Besides\n I thought you could use it. When you start your band. (speaker turn): PATRICK \n A Fender Strat. You bought this? (speaker turn): KAT \n I didn't care about the money. (speaker turn): PATRICK \n\" Really? What was it like? A down payment now (speaker turn): KAT \n It wasn't like that. (speaker turn): PATRICK \n You were paid to take me out! By -- the one person I truly hate. I knew it was a set-up! (speaker turn): KAT \n Wait I... (speaker turn): PATRICK \n It gets worse -- you still have your freshman yearbook? (speaker turn): PATRICK \n That ' s completely adorable! (speaker turn): KAT \n\" That's where I was last year. She'd never lived alone -- my grandfather died -- I stayed with her. I wasn't in jail\n What? (speaker turn): KAT \n My grandmother's . (speaker turn): PATRICK \n\" Look (speaker turn): KAT \n Oh huh\n It's just something I had. You know\n It's Scurvy's. His date got convicted. Where'd you get the dress?\n How'd you get a tux at the last minute? (speaker turn): KAT \n Nothing! There's nothing in it for me. Just the pleasure of your company.\n\" Answer the question (speaker turn): KAT \n You need therapy. Has anyone ever told you that?\n You tell me. (speaker turn): KAT \n So I have to have a motive to be with you?\n Create a little drama? Start a new rumor? What? (speaker turn): KAT \n Because I don't want to. It's a stupid tradition. (speaker turn): KAT \n Why not? (speaker turn): PATRICK \n\" No (speaker turn): KAT \n No what? (speaker turn): PATRICK \n No. (speaker turn): KAT \n You know what I mean (speaker turn): PATRICK \n Is that a request or a command? (speaker turn): KAT \n Go to the prom with me\n You're amazingly self-assured. Has anyone ever told you that?\n No one else knows\n What?\n You're sweet. And sexy. And completely hot for me.\n No -- something real. Something no one else knows. (speaker turn): KAT \n I hate peas.\n Tell me something true.\n I know the porn career's a lie. (speaker turn): KAT \n Hearsay. (speaker turn): PATRICK \n The duck?\n Fallacy. (speaker turn): PATRICK \n State trooper? (speaker turn): KAT \n For. . . ? (speaker turn): KAT \n You up for it? (speaker turn): PATRICK \n You never disappointed me.\n How?\n Then you screwed up (speaker turn): PATRICK \n Something like that (speaker turn): KAT \n\" So if you disappoint them from the start (speaker turn): PATRICK \n I don't like to do what people expect. Then they expect it all the time and they get disappointed when you change. (speaker turn): KAT \n Yes (speaker turn): PATRICK \n Acting the way we do. (speaker turn): KAT \n So what's your excuse?\n\" Yeah\n A soft side? Who knew? (speaker turn): PATRICK \n I dazzled him with my wit (speaker turn): KAT \n So how'd you get Chapin to look the other way? (speaker turn): PATRICK \n Good call.\n I figured it had to be something ridiculous to win your respect. And piss you off. (speaker turn): PATRICK \n The Partridge Family? (speaker turn): KAT \n Maybe.\n You want me to climb up and show you how to get down? (speaker turn): KAT \n Forget it. I'm stayin'. (speaker turn): PATRICK \n Put your right foot there -- (speaker turn): KAT \n Try lookin' at it from this angle\n C'mon. It's not that bad\n I guess I never told you I'm afraid of heights.\n\" Look up (speaker turn): PATRICK \n He left! I sprung the dickhead and he cruised on me.\n Other than my upchuck reflex? Nothing. (speaker turn): KAT \n So what did I have an effect on ? (speaker turn): PATRICK \n Don't for one minute think that you had any effect whatsoever on my panties. (speaker turn): KAT \n Unwelcome? I guess someone still has her panties in a twist. (speaker turn): PATRICK \n Unwelcome. (speaker turn): KAT \n Wholesome. (speaker turn): PATRICK \n Pleasant?\n You 're so -- (speaker turn): KAT \n I heard there was a poetry reading.\n What are you doing here? (speaker turn): KAT \n\" Excuse me (speaker turn): PATRICK \n\" No offense (speaker turn): PATRICK \n BIANCA (speaker turn): KAT \n Who? (speaker turn): PATRICK \n He just wants me to be someone I'm not.\n So what ' s up with your dad? He a pain in the ass? (speaker turn): PATRICK \n I'm gettin' there (speaker turn): PATRICK \n\" Oh (speaker turn): KAT \n You don't strike me as the type that would ask permission. (speaker turn): PATRICK \n My father wouldn't approve of that that (speaker turn): KAT \n Start a band?\n This.\n Do what? (speaker turn): PATRICK \n"
]
],
[
[
"**Train Naive Bayes Classifier over ISEAR Dataset to predict emotions**",
"_____no_output_____"
]
],
[
[
"import string\nfrom textblob.classifiers import NaiveBayesClassifier\n\nemotion_df=pd.read_csv(\"C://Users/PrishitaRay/Desktop/DSER_Movie_Subtitles/ISEAR.csv\",header=None)\n\nexclude=set(string.punctuation)\nlemma=WordNetLemmatizer()\n\nneg=['not', 'neither', 'nor', 'but', 'however', 'although', 'nonetheless', 'despite', 'except','even though', 'yet']\n\nem_list = list(emotion_df[0][0:1000])\ntext_list = list(emotion_df[1][0:1000])\n\ntrain = []\nsum_text_list = []\ne_score_dict = {}\n\nfor i in range(0,len(text_list)):\n stop_free = \" \".join([x for x in text_list[i].lower().split() if x not in stop_words if x not in neg])\n punc_free = \"\".join([ch for ch in stop_free if ch not in exclude])\n text_list[i] = \" \".join([lemmatizer.lemmatize(word) for word in punc_free.split()])\n\nfor i in range(0,len(em_list)):\n l=[]\n l.append(text_list[i])\n \n l.append(em_list[i])\n train.append(l)\n\ncl = NaiveBayesClassifier(train)\n\nemotion_data=dict.fromkeys(movie_ids)\n\ncnt=0\nfor key in emotion_data.keys():\n l=[]\n for j in range(0,len(POS_Tags[cnt])):\n s=\"\"\n for pair in POS_Tags[cnt][j]:\n if s==\"\":\n s=s+pair[0]\n else:\n s=s+\" \"+pair[0]\n l.append(s)\n emotion_data[key]=l\n cnt= cnt+1\n \ntest_em_mov0=emotion_data[' m0 ']\n\nfor line in test_em_mov0:\n em=cl.classify(line)\n print(line+\" :\"+em)\n",
"they do not ! :guilt\nthey do to ! :shame\ni hope so . :sadness\nshe okay ? :joy\nlet 's go . :guilt\nwow :joy\nokay -- you 're gon na need to learn how to lie . :disgust\nno :joy\n`` i 'm kidding . you know how sometimes you just become this `` '' persona '' '' ? and you do n't know how to quit ? '' :disgust\nlike my fear of wearing pastel ? :joy\n`` the `` '' real you '' '' . '' :joy\nwhat good stuff ? :joy\ni figured you 'd get to the good stuff eventually . :joy\nthank god ! if i had to hear one more story about your coiffure ... :joy\n`` me . this endless ... blonde babble . i 'm like :sadness\nwhat crap ? :joy\ndo you listen to this crap ? :anger\nno ... :joy\n`` then guillermo say :joy\nyou always been this selfish ? :joy\nbut :joy\nthen that 's all you had to say . :disgust\n`` well :joy\n`` you never wanted to go out with 'me :guilt\ni wa ? :joy\n`` i looked for you back at the party :joy\ntons :joy\nhave fun tonight ? :joy\ni believe we share an art instructor :sadness\nyou know chastity ? :joy\n`` looks like thing worked out tonight :guilt\nhi . :joy\nwho know ? all i 've ever heard her say is that she 'd dip before dating a guy that smoke . :anger\nso that 's the kind of guy she like ? pretty one ? :sadness\n`` lesbian ? no . i found a picture of jared leto in one of her drawer :disgust\nshe 's not a ... :joy\ni 'm workin ' on it . but she doe n't seem to be goin ' for him . :anger\n`` i really :joy\nsure have . :joy\n`` eber 's deep conditioner every two day . and i never :shame\nhow do you get your hair to look like that ? :shame\nyou 're sweet . :joy\nyou have my word . as a gentleman :joy\ni counted on you to help my cause . you and that thug are obviously failing . are n't we ever going on our date ? :disgust\nyou got something on your mind ? :anger\nwhere ? :joy\nthere . :joy\n`` well :joy\nhow is our little find the wench a date plan progressing ? :sadness\nforget french . :sadness\nthat 's because it 's such a nice one . :sadness\ni do n't want to know how to say that though . i want to know useful thing . like where the good store are . how much doe champagne cost ? stuff like chat . i have never in my life had to point out my head to someone . :guilt\nright . see ? you 're ready for the quiz . :joy\nc'esc ma tete . this is my head :sadness\nlet me see what i can do . :anger\n`` gosh :joy\nthat 's a shame . :guilt\n`` unsolved mystery . she used to be really popular when she started high school :joy\nwhy ? :joy\nseems like she could get a date easy enough ... :guilt\n`` the thing is :sadness\ncameron . :joy\n`` no :joy\nforget it . :sadness\nyou 're asking me out . that 's so cute . what 's your name again ? :sadness\nokay ... then how 'bout we try out some french cuisine . saturday ? night ? :fear\nnot the hacking and gagging and spitting part . please . :joy\n`` well :joy\ncan we make this quick ? roxanne korrine and andrew barrett are having an incredibly horrendous public break- up on the quad . again . :disgust\ni did . :joy\nyou think you ' re the only sophomore at the prom ? :joy\ni do n't have to be home 'til two . :shame\ni have to be home in twenty minute . :fear\nall i know is -- i 'd give up my private line to go out with a guy like joey . :guilt\n`` sometimes i wonder if the guy we 're supposed to want to go out with are the one we actually want to go out with :guilt\n`` bianca :joy\ncombination . i do n't know -- i thought he 'd be different . more of a gentleman ... :shame\nis he oily or dry ? :sadness\n`` he practically proposed when he found out we had the same dermatologist . i mean . dr. bonchowski is great an all :joy\n`` would you mind getting me a drink :sadness\ngreat :joy\njoey . :joy\nwho ? :joy\nwhere did he go ? he wa just here . :guilt\nyou might wan na think about it :sadness\nno . :joy\ndid you change your hair ? :joy\nyou know the deal . i can ' t go if kat doe n't go -- :joy\n`` listen :sadness\nyou 're concentrating awfully hard considering it 's gym class . :sadness\n`` hi :joy\n`` hey :joy\nmy agent say i 've got a good shot at being the prada guy next year . :joy\nneat ... :joy\n`` it 's a gay cruise line :sadness\nqueen harry ? :joy\n`` so yeah :sadness\nhopefully . :joy\n`` exactly so :sadness\nexpensive ? :joy\nit 's more :sadness\nperm ? :joy\npatrick -- is that- a . :sadness\nit 's just you . :joy\nis that woman a complete fruit-loop or is it just me ? :sadness\nno ! i just wanted :joy\nwhat ? to completely damage me ? to send me to therapy forever ? what ? :disgust\ni just wanted -- :joy\nyou set me up . :disgust\nlet go ! :guilt\nso did you :joy\n`` you looked beautiful last night :joy\n`` i guess i 'll never know :sadness\n`` not all experience are good :joy\n`` god :joy\ni guess i thought i wa protecting you . :joy\ni 'm not stupid enough to repeat your mistake . :guilt\nthat 's not :sadness\n`` no . you did n't ! if you really thought i could make my own decision :sadness\ni wanted to let you make up your own mind about him . :guilt\nwhy did n't you tell me ? :anger\n`` after that :sadness\nbut :joy\n`` just once . afterwards :joy\nyou did what ? :joy\nhe said everyone wa doing it . so i did it . :guilt\nas in ... :sadness\n`` now i do . back then :shame\nbut you hate joey :joy\n`` he wa :joy\nwhy ? :joy\nin 9th . for a month :joy\nwhat ? :joy\n`` joey never told you we went out :joy\n`` i wish i had that luxury . i 'm the only sophomore that got asked to the prom and i ca n't go :guilt\n`` i do care . but i 'm a firm believer in doing something for your own reason :guilt\nlike you care . :joy\n`` listen :sadness\nyou 're welcome . :joy\n`` i do n't get you . you act like you 're too good for any of this :shame\ni really do n't think i need any social advice from you right now . :shame\n`` bianca :joy\ncan we go now ? :guilt\nyou are so completely unbalanced . :joy\n`` yeah :joy\nit 's shakespeare . maybe you 've heard of him ? :sadness\nlike i 'm supposed to know what that even mean . :shame\nat least i 'm not a clouted fen- sucked hedge-pig . :sadness\nca n't you forget for just one night that you 're completely wretched ? :disgust\n`` bogey lowenstein 's party is normal :joy\nwhat 's normal ? :joy\n`` you 're ruining my life ' because you wo n't be normal :joy\ni think you 're a freak . i think you do this to torture me . and i think you suck . :disgust\nwhat do you think ? :joy\n`` oh :sadness\nit mean that gigglepuss is playing at club skunk and we 're going . :shame\n`` oh my god :sadness\ncan you at least start wearing a bra ? :sadness\ni have the potential to smack the crap out of you if you do n't get out of my way . :shame\n`` nowhere ... hi :joy\nwhere 've you been ? :joy\n`` i have a date :guilt\ni 'm missing something . :sadness\nfine . i see that i 'm a prisoner in my own house . i 'm not a daughter . i 'm a possession ! :guilt\nyou 're not going unless your sister go . end of story . :guilt\n`` he 's not a `` '' hot rod '' '' . whatever that is . '' :sadness\n`` it 's that hot rod joey :sadness\n`` no :joy\nthe prom ? kat ha a date ? :guilt\n`` daddy :joy\nbecause she 'll scare them away . :sadness\nwhy ? :joy\npromise me you wo n't talk to any boy unless your sister is present . :shame\njust for a minute :joy\n`` daddy :joy\nwear the belly before you go . :joy\nit 's just a party . daddy . :shame\n`` oh :sadness\n`` if kat 's not going :sadness\n`` daddy :joy\n`` it 's just a party . daddy :shame\notherwise known a an orgy ? :joy\n`` if you must know :joy\nand where 're you going ? :disgust\n`` daddy :joy\nexactly my point :sadness\nbut she doe n't want to date . :disgust\n`` but it 's not fair -- she 's a mutant :sadness\nthen neither will you . and i 'll get to sleep at night . :fear\nwhat if she never start dating ? :sadness\nno ! you 're not dating until your sister start dating . end of discussion . :joy\n`` now do n't get upset . daddy :anger\njust sent 'em through . :joy\n`` padua girl . one tall :sadness\nnever :sadness\nfan of a fan . you see a couple of minor come in ? :disgust\ndid n't have you pegged for a gigglepuss fan . are n't they a little too pre-teen belly-button ring for you ? :guilt\n`` always a pleasure :joy\n`` best case scenario :sadness\nyou humiliated the woman ! sacrifice yourself on the altar of dignity and even the score . :disgust\n`` no :joy\n`` buttholus extremus . but hey :joy\nthe hell is that ? what kind of 'guy just pick up a girl and carry her away while you 're talking to her ? :disgust\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf21267e5bc84ed1ba54b1cdc0e999e619ea106 | 32,420 | ipynb | Jupyter Notebook | 08-machine_learning_jupyter/matplotlib_demo.ipynb | iproduct/coulse-ml | 65577fd4202630d3d5cb6333ddc51cede750fb5a | [
"Apache-2.0"
] | 1 | 2020-10-02T15:48:42.000Z | 2020-10-02T15:48:42.000Z | 08-machine_learning_jupyter/matplotlib_demo.ipynb | iproduct/coulse-ml | 65577fd4202630d3d5cb6333ddc51cede750fb5a | [
"Apache-2.0"
] | null | null | null | 08-machine_learning_jupyter/matplotlib_demo.ipynb | iproduct/coulse-ml | 65577fd4202630d3d5cb6333ddc51cede750fb5a | [
"Apache-2.0"
] | null | null | null | 558.965517 | 31,232 | 0.947502 | [
[
[
"import numpy as np\nfrom matplotlib import pyplot as plt\n\nif __name__ == \"__main__\":\n ys = 200 + np.random.randn(100)\n x = [x for x in range(len(ys))]\n\n plt.plot(x, ys, '-')\n plt.fill_between(x, ys, 195, where=(ys > 200), facecolor='g', alpha=0.6)\n\n plt.title(\"Sample Visualization\")\n plt.show()\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
ecf22c8ce6e9f585ad6a9cb129d2548984572da4 | 53,941 | ipynb | Jupyter Notebook | examples/sklearn/demo_exponentiated_gradient_reduction_sklearn.ipynb | giandos200/AIF360 | 71b0ed1894ab3c2d45cf32d2b049c9ec43792e4a | [
"Apache-2.0"
] | 982 | 2018-09-12T17:19:11.000Z | 2020-07-13T21:26:24.000Z | examples/sklearn/demo_exponentiated_gradient_reduction_sklearn.ipynb | JeffinSiby/AIF360 | 746e763191ef46ba3ab5c601b96ce3f6dcb772fd | [
"Apache-2.0"
] | 109 | 2018-09-12T20:39:43.000Z | 2020-07-09T20:12:00.000Z | examples/sklearn/demo_exponentiated_gradient_reduction_sklearn.ipynb | JeffinSiby/AIF360 | 746e763191ef46ba3ab5c601b96ce3f6dcb772fd | [
"Apache-2.0"
] | 335 | 2018-09-13T15:35:09.000Z | 2020-07-06T10:56:12.000Z | 39.229818 | 360 | 0.504384 | [
[
[
"# Sklearn compatible Exponentiated Gradient Reduction\n\nExponentiated gradient reduction is an in-processing technique that reduces fair classification to a sequence of cost-sensitive classification problems, returning a randomized classifier with the lowest empirical error subject to \nfair classification constraints. The code for exponentiated gradient reduction wraps the source class \n`fairlearn.reductions.ExponentiatedGradient` available in the https://github.com/fairlearn/fairlearn library,\nlicensed under the MIT Licencse, Copyright Microsoft Corporation.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\n\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.model_selection import GridSearchCV, train_test_split\nfrom sklearn.preprocessing import OneHotEncoder\n\nfrom aif360.sklearn.inprocessing import ExponentiatedGradientReduction\n\nfrom aif360.sklearn.datasets import fetch_adult\nfrom aif360.sklearn.metrics import disparate_impact_ratio, average_odds_error, generalized_fpr\nfrom aif360.sklearn.metrics import generalized_fnr, difference",
"/Users/sohiniupadhyay/Desktop/AIF360/aif360/sklearn/inprocessing/grid_search_reduction.py:85: SyntaxWarning: \"is\" with a literal. Did you mean \"==\"?\n if constraints is \"GroupLoss\":\n/Users/sohiniupadhyay/Desktop/AIF360/aif360/sklearn/inprocessing/grid_search_reduction.py:94: SyntaxWarning: \"is\" with a literal. Did you mean \"==\"?\n if loss is \"ZeroOne\":\n/Users/sohiniupadhyay/Desktop/AIF360/aif360/sklearn/datasets/tempeh_datasets.py:38: SyntaxWarning: \"is\" with a literal. Did you mean \"==\"?\n if subset is \"train\":\n/Users/sohiniupadhyay/Desktop/AIF360/aif360/sklearn/datasets/tempeh_datasets.py:40: SyntaxWarning: \"is\" with a literal. Did you mean \"==\"?\n elif subset is \"test\":\n"
]
],
[
[
"### Loading data",
"_____no_output_____"
],
[
"Datasets are formatted as separate `X` (# samples x # features) and `y` (# samples x # labels) DataFrames. The index of each DataFrame contains protected attribute values per sample. Datasets may also load a `sample_weight` object to be used with certain algorithms/metrics. All of this makes it so that aif360 is compatible with scikit-learn objects.\n\nFor example, we can easily load the Adult dataset from UCI with the following line:",
"_____no_output_____"
]
],
[
[
"X, y, sample_weight = fetch_adult()\nX.head()",
"_____no_output_____"
],
[
"# there is one unused category ('Never-worked') that was dropped during dropna\nX.workclass.cat.remove_unused_categories(inplace=True)",
"_____no_output_____"
]
],
[
[
"We can then map the protected attributes to integers,",
"_____no_output_____"
]
],
[
[
"X.index = pd.MultiIndex.from_arrays(X.index.codes, names=X.index.names)\ny.index = pd.MultiIndex.from_arrays(y.index.codes, names=y.index.names)",
"_____no_output_____"
]
],
[
[
"and the target classes to 0/1,",
"_____no_output_____"
]
],
[
[
"y = pd.Series(y.factorize(sort=True)[0], index=y.index)",
"_____no_output_____"
]
],
[
[
"split the dataset,",
"_____no_output_____"
]
],
[
[
"(X_train, X_test,\n y_train, y_test) = train_test_split(X, y, train_size=0.7, random_state=1234567)",
"_____no_output_____"
]
],
[
[
"We use Pandas for one-hot encoding for easy reference to columns associated with protected attributes, information necessary for Exponentiated Gradient Reduction",
"_____no_output_____"
]
],
[
[
"X_train, X_test = pd.get_dummies(X_train), pd.get_dummies(X_test)\nX_train.head()",
"_____no_output_____"
]
],
[
[
"The protected attribute information is also replicated in the labels:",
"_____no_output_____"
]
],
[
[
"y_train.head()",
"_____no_output_____"
]
],
[
[
"### Running metrics",
"_____no_output_____"
],
[
"With the data in this format, we can easily train a scikit-learn model and get predictions for the test data:",
"_____no_output_____"
]
],
[
[
"y_pred = LogisticRegression(solver='lbfgs').fit(X_train, y_train).predict(X_test)\nlr_acc = accuracy_score(y_test, y_pred)\nprint(lr_acc)",
"0.8373995724920764\n"
]
],
[
[
"We can assess how close the predictions are to equality of odds.\n\n`average_odds_error()` computes the (unweighted) average of the absolute values of the true positive rate (TPR) difference and false positive rate (FPR) difference, i.e.:\n\n$$ \\tfrac{1}{2}\\left(|FPR_{D = \\text{unprivileged}} - FPR_{D = \\text{privileged}}| + |TPR_{D = \\text{unprivileged}} - TPR_{D = \\text{privileged}}|\\right) $$",
"_____no_output_____"
]
],
[
[
"lr_aoe_sex = average_odds_error(y_test, y_pred, prot_attr='sex')\nprint(lr_aoe_sex)",
"0.09897521109915139\n"
],
[
"lr_aoe_race = average_odds_error(y_test, y_pred, prot_attr='race')\nprint(lr_aoe_race)",
"0.00867568807624941\n"
]
],
[
[
"### Exponentiated Gradient Reduction",
"_____no_output_____"
],
[
"Choose a base model for the randomized classifier",
"_____no_output_____"
]
],
[
[
"estimator = LogisticRegression(solver='lbfgs')",
"_____no_output_____"
]
],
[
[
"Determine the columns associated with the protected attribute(s)",
"_____no_output_____"
]
],
[
[
"prot_attr_cols = [colname for colname in X_train if \"sex\" in colname or \"race\" in colname]",
"_____no_output_____"
]
],
[
[
"Train the randomized classifier and observe test accuracy. Other options for `constraints` include \"DemographicParity\", \"TruePositiveRateDifference,\" and \"ErrorRateRatio.\"",
"_____no_output_____"
]
],
[
[
"np.random.seed(0) #for reproducibility\nexp_grad_red = ExponentiatedGradientReduction(prot_attr=prot_attr_cols, \n estimator=estimator, \n constraints=\"EqualizedOdds\",\n drop_prot_attr=False)\nexp_grad_red.fit(X_train, y_train)\negr_acc = exp_grad_red.score(X_test, y_test)\nprint(egr_acc)\n\n# Check for that accuracy is comparable\nassert abs(lr_acc-egr_acc)<=0.03",
"lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n"
],
[
"egr_aoe_sex = average_odds_error(y_test, exp_grad_red.predict(X_test), prot_attr='sex')\nprint(egr_aoe_sex)\n\n# Check for improvement in average odds error for sex\nassert egr_aoe_sex<lr_aoe_sex",
"0.018426256067917424\n"
],
[
"egr_aoe_race = average_odds_error(y_test, exp_grad_red.predict(X_test), prot_attr='race')\nprint(egr_aoe_race)\n\n# Check for improvement in average odds error for race\n# assert egr_aoe_race<lr_aoe_race",
"0.005848503310276698\n"
]
],
[
[
"Number of calls made to base model algorithm",
"_____no_output_____"
]
],
[
[
"exp_grad_red.model._n_oracle_calls",
"_____no_output_____"
]
],
[
[
"Maximum calls permitted",
"_____no_output_____"
]
],
[
[
"exp_grad_red.T",
"_____no_output_____"
]
],
[
[
"Instead of passing in a value for `constraints`, we can also pass a `fairlearn.reductions.moment` object in for `constraints_moment`. You could use a predefined moment as we do below or create a custom moment using the fairlearn library.",
"_____no_output_____"
]
],
[
[
"import fairlearn.reductions as red \n\nnp.random.seed(0) #need for reproducibility\nexp_grad_red2 = ExponentiatedGradientReduction(prot_attr=prot_attr_cols, \n estimator=estimator, \n constraints=red.EqualizedOdds(),\n drop_prot_attr=False)\nexp_grad_red2.fit(X_train, y_train)\nexp_grad_red2.score(X_test, y_test)",
"lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\nlbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n"
],
[
"average_odds_error(y_test, exp_grad_red2.predict(X_test), prot_attr='sex')",
"_____no_output_____"
],
[
"average_odds_error(y_test, exp_grad_red2.predict(X_test), prot_attr='race')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecf23579eda423d7805e2f26ad8717b359ee7dfc | 4,467 | ipynb | Jupyter Notebook | _pages/AI/TensorFlow/src/NCIA-CNN/Day_03_01_auto_encoder.ipynb | shpimit/shpimit.github.io | 83d1f920f75c2871f8e62f045db29a9b2d93f87b | [
"MIT"
] | 1 | 2018-05-13T12:57:32.000Z | 2018-05-13T12:57:32.000Z | _pages/AI/TensorFlow/src/NCIA-CNN/Day_03_01_auto_encoder.ipynb | shpimit/shpimit.github.io | 83d1f920f75c2871f8e62f045db29a9b2d93f87b | [
"MIT"
] | null | null | null | _pages/AI/TensorFlow/src/NCIA-CNN/Day_03_01_auto_encoder.ipynb | shpimit/shpimit.github.io | 83d1f920f75c2871f8e62f045db29a9b2d93f87b | [
"MIT"
] | null | null | null | 28.819355 | 109 | 0.505709 | [
[
[
"# Day_03_01_auto_encoder.py\nimport tensorflow as tf\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom tensorflow.examples.tutorials.mnist import input_data",
"_____no_output_____"
],
[
"def model_1(ph_x):\n enc_w = tf.get_variable('enc_w1', shape=[784, 256], initializer=tf.initializers.glorot_normal())\n enc_b = tf.Variable(tf.zeros([256]))\n\n enc = tf.matmul(ph_x, enc_w) + enc_b\n relu = tf.nn.relu(enc)\n\n dec_w = tf.get_variable('dec_w1', shape=[256, 784], initializer=tf.initializers.glorot_normal())\n dec_b = tf.Variable(tf.zeros([784]))\n\n dec = tf.matmul(relu, dec_w) + dec_b\n # 0~1을 벗어나면 noise가 나와서....\n # return tf.nn.relu(dec)\n return tf.nn.sigmoid(dec)",
"_____no_output_____"
]
],
[
[
"# 문제\n## 1번 함수를 tf.layers로 수정하세요",
"_____no_output_____"
]
],
[
[
"def model_2(ph_x):\n\n output = tf.layers.dense(ph_x, 256, activation=tf.nn.relu)\n return tf.layers.dense(output, 784, activation=tf.nn.sigmoid)",
"_____no_output_____"
],
[
"def auto_encoder():\n mnist = input_data.read_data_sets('mnist')\n\n # ph_x = tf.placeholder(tf.float32)\n # tf.layer를 사용할때는 shape을 지정 해준다.\n # -1은 명확하게 존잰할때, None은 모를때 사용\n ph_x = tf.placeholder(tf.float32, shape=[None, 784])\n\n output = model_2(ph_x)\n\n # loss_i = tf.nn.sparse_softmax_cross_entropy_with_logits(logits=z, labels=ph_y)\n loss = tf.reduce_mean((ph_x - output) ** 2)\n\n # 모델 정확도 높이기 위해 아님 -> 빨리 수렴한다.\n # optimizer = tf.train.GradientDescentOptimizer(learning_rate=0.1)\n optimizer = tf.train.AdamOptimizer(learning_rate=0.01)\n train = optimizer.minimize(loss=loss)\n\n # sess로 변수의 값을 알수 있다.\n sess = tf.Session()\n sess.run(tf.global_variables_initializer())\n\n epochs = 10\n batch_size = 100 # 출구를 몇개씩 조사할건지\n n_iters = mnist.train.num_examples // batch_size # 550\n\n # epcochs를 1번 돌고 념\n for i in range(epochs):\n c = 0\n for j in range(n_iters):\n xx, _ = mnist.train.next_batch(batch_size)\n\n sess.run(train, feed_dict={ph_x: xx})\n c += sess.run(loss, {ph_x: xx})\n\n print(i, c/ n_iters)\n\n # ---------------------------------------------------------------- #\n sample_count = 10\n samples = sess.run(output, {ph_x: mnist.test.images[:sample_count]})\n\n _, ax = plt.subplots(2, sample_count, figsize=(sample_count, 2))\n\n for i in range(sample_count):\n ax[0, i].set_axis_off()\n ax[1, i].set_axis_off()\n\n ax[0][i].imshow(np.reshape(mnist.test.images[i], [28, 28]), cmap='gray')\n ax[1][i].imshow(np.reshape(samples[i], [28, 28]), cmap='gray')\n\n # ax[0][i].imshow(np.reshape(mnist.test.images[i], [28, 28]))\n # ax[1][i].imshow(np.reshape(samples[i], [28, 28]))\n\n plt.tight_layout()\n plt.show()\n\n sess.close()",
"_____no_output_____"
],
[
"auto_encoder()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecf241f083e1e9a900168268f344d192fc417ecb | 2,326 | ipynb | Jupyter Notebook | notebooks/book1/18/regtreeSurfaceDemo.ipynb | karm-patel/pyprobml | af8230a0bc0d01bb0f779582d87e5856d25e6211 | [
"MIT"
] | null | null | null | notebooks/book1/18/regtreeSurfaceDemo.ipynb | karm-patel/pyprobml | af8230a0bc0d01bb0f779582d87e5856d25e6211 | [
"MIT"
] | null | null | null | notebooks/book1/18/regtreeSurfaceDemo.ipynb | karm-patel/pyprobml | af8230a0bc0d01bb0f779582d87e5856d25e6211 | [
"MIT"
] | null | null | null | 28.365854 | 132 | 0.438951 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecf246da2ca3239cc93217b2690f7fd09692cffe | 13,263 | ipynb | Jupyter Notebook | examples/tutorials/translations/hindi/Part 04 - Federated Learning via Trusted Aggregator.ipynb | NicoSerranoP/PySyft | 87fcd566c46fce4c16d363c94396dd26bd82a016 | [
"Apache-2.0"
] | 3 | 2020-11-24T05:15:57.000Z | 2020-12-07T09:52:45.000Z | examples/tutorials/translations/hindi/Part 04 - Federated Learning via Trusted Aggregator.ipynb | NicoSerranoP/PySyft | 87fcd566c46fce4c16d363c94396dd26bd82a016 | [
"Apache-2.0"
] | 2 | 2020-03-09T09:17:06.000Z | 2020-04-09T13:33:12.000Z | examples/tutorials/translations/hindi/Part 04 - Federated Learning via Trusted Aggregator.ipynb | NicoSerranoP/PySyft | 87fcd566c46fce4c16d363c94396dd26bd82a016 | [
"Apache-2.0"
] | 1 | 2022-03-25T00:07:51.000Z | 2022-03-25T00:07:51.000Z | 32.34878 | 486 | 0.494081 | [
[
[
"# भाग 4: मॉडल एवरेजिंग के साथ फेडरेटेड लर्निंग\n\n**रिकैप:** इस ट्यूटोरियल के भाग 2 में, हमने फेडरेटेड लर्निंग के एक बहुत ही सरल संस्करण का उपयोग करके एक मॉडल को प्रशिक्षित किया। इसके लिए प्रत्येक डेटा अधिकारी को अपने ग्रेडिएंट को देखने में सक्षम होने के लिए मॉडल अधिकारी पर भरोसा करना आवश्यक था।\n\n**विवरण:** इस ट्यूटोरियल में, हम दिखाएंगे कि अंतिम परिणामी मॉडल, मॉडल के अधिकारी को (हमें), वापस भेजे जाने से पहले एक विश्वसनीय \"सुरक्षित कर्मचारी\" द्वारा भार को अनुमति देने के लिए भाग 3 से उन्नत एकत्रीकरण उपकरण का उपयोग कैसे करें।\n\nइस तरह, केवल सुरक्षित कर्मचारी ही देख सकते हैं कि किसका वजन किसके पास से आया है। हम यह बताने में सक्षम हो सकते हैं कि मॉडल के कौन से हिस्से बदले गए, लेकिन हम यह नहीं जानते कि कौन सा कार्यकर्ता (बॉब या ऐलिस) कौन सा परिवर्तन करता है, जो गोपनीयता की एक परत बनाता है।\n\nलेखक:\n - Andrew Trask - Twitter: [@iamtrask](https://twitter.com/iamtrask)\n - Jason Mancuso - Twitter: [@jvmancuso](https://twitter.com/jvmancuso)\n \n nbTranslate का उपयोग करके अनुवादित\n\nसंपादक:\n\n - Urvashi Raheja - Github: [@raheja](https://github.com/raheja)\n \n",
"_____no_output_____"
]
],
[
[
"import torch\nimport syft as sy\nimport copy\nhook = sy.TorchHook(torch)\nfrom torch import nn, optim",
"_____no_output_____"
]
],
[
[
"# चरण 1: डेटा अधिकारी बनाएँ\n\nसबसे पहले, हम दो डेटा अधिकारी (बॉब और ऐलिस) बनाने जा रहे हैं, जिनमें से प्रत्येक के पास कम डेटा है। हम \"secure_worker\" नामक एक सुरक्षित मशीन को इनिशियलाइज़ करने जा रहे हैं। व्यवहार में यह सुरक्षित हार्डवेयर हो सकता है (जैसे इंटेल का SGX) या केवल एक विश्वसनीय मध्यस्थ।",
"_____no_output_____"
]
],
[
[
"# create a couple workers\n\nbob = sy.VirtualWorker(hook, id=\"bob\")\nalice = sy.VirtualWorker(hook, id=\"alice\")\nsecure_worker = sy.VirtualWorker(hook, id=\"secure_worker\")\n\n\n# A Toy Dataset\ndata = torch.tensor([[0,0],[0,1],[1,0],[1,1.]], requires_grad=True)\ntarget = torch.tensor([[0],[0],[1],[1.]], requires_grad=True)\n\n# get pointers to training data on each worker by\n# sending some training data to bob and alice\nbobs_data = data[0:2].send(bob)\nbobs_target = target[0:2].send(bob)\n\nalices_data = data[2:].send(alice)\nalices_target = target[2:].send(alice)",
"_____no_output_____"
]
],
[
[
"# चरण 2: हमारा मॉडल बनाएं\n\nइस उदाहरण के लिए, हम एक सरल रैखिक (Linear) मॉडल के साथ प्रशिक्षित करने जा रहे हैं। हम इसे प्रारंभिक रूप से PyTorch के nn.Linear कंस्ट्रक्टर का उपयोग करके शुरू कर सकते हैं।",
"_____no_output_____"
]
],
[
[
"# Iniitalize A Toy Model\nmodel = nn.Linear(2,1)",
"_____no_output_____"
]
],
[
[
"# चरण 3: ऐलिस और बॉब को मॉडल की एक प्रति भेजें\n\nअगला, हमें एलिस और बॉब को वर्तमान मॉडल की एक प्रति भेजने की आवश्यकता है ताकि वे अपने स्वयं के डेटासेट पर सीखने के चरणों का प्रदर्शन कर सकें।",
"_____no_output_____"
]
],
[
[
"bobs_model = model.copy().send(bob)\nalices_model = model.copy().send(alice)\n\nbobs_opt = optim.SGD(params=bobs_model.parameters(),lr=0.1)\nalices_opt = optim.SGD(params=alices_model.parameters(),lr=0.1)",
"_____no_output_____"
]
],
[
[
"# चरण 4: ट्रेन बॉब और ऐलिस के मॉडल (समानांतर में)\n\nजैसा कि सिक्योर एवरेजिंग के माध्यम से फेडरेटेड लर्निंग के साथ पारंपरिक है, प्रत्येक डेटा अधिकारी पहले मॉडल को एक साथ औसतन होने से पहले स्थानीय रूप से कई पुनरावृत्तियों के लिए अपने मॉडल को प्रशिक्षित करता है।",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n\n # Train Bob's Model\n bobs_opt.zero_grad()\n bobs_pred = bobs_model(bobs_data)\n bobs_loss = ((bobs_pred - bobs_target)**2).sum()\n bobs_loss.backward()\n\n bobs_opt.step()\n bobs_loss = bobs_loss.get().data\n\n # Train Alice's Model\n alices_opt.zero_grad()\n alices_pred = alices_model(alices_data)\n alices_loss = ((alices_pred - alices_target)**2).sum()\n alices_loss.backward()\n\n alices_opt.step()\n alices_loss = alices_loss.get().data\n \n print(\"Bob:\" + str(bobs_loss) + \" Alice:\" + str(alices_loss))",
"_____no_output_____"
]
],
[
[
"# चरण 5: एक सुरक्षित कार्यकर्ता को दोनों अद्यतन मॉडल भेजें\n\nअब जब प्रत्येक डेटा अधिकारी के पास आंशिक रूप से प्रशिक्षित मॉडल है, तो उन्हें सुरक्षित तरीके से एक साथ औसत करने का समय है। हम ऐलिस और बॉब को उनके मॉडल को सुरक्षित (विश्वसनीय) सर्वर पर भेजने के निर्देश देकर इसे प्राप्त करते हैं।\n\nध्यान दें कि हमारे एपीआई के इस उपयोग का अर्थ है कि प्रत्येक मॉडल को सुरक्षित रूप से सुरक्षित_वर्कर को भेजा जाता है। हम इसे कभी नहीं देखते हैं।",
"_____no_output_____"
]
],
[
[
"alices_model.move(secure_worker)",
"_____no_output_____"
],
[
"bobs_model.move(secure_worker)",
"_____no_output_____"
]
],
[
[
"# चरण 6: औसत मॉडल",
"_____no_output_____"
],
[
"अंत में, इस प्रशिक्षण युग के लिए अंतिम चरण बॉब और एलिस के प्रशिक्षित मॉडलों को एक साथ औसत करना है और फिर हमारे वैश्विक \"मॉडल\" के लिए मूल्यों को निर्धारित करने के लिए इसका उपयोग करना है।",
"_____no_output_____"
]
],
[
[
"with torch.no_grad():\n model.weight.set_(((alices_model.weight.data + bobs_model.weight.data) / 2).get())\n model.bias.set_(((alices_model.bias.data + bobs_model.bias.data) / 2).get())\n",
"_____no_output_____"
]
],
[
[
"# दोहराएं\n\nऔर अब हमें बस इस कई बार पुनरावृति करने की आवश्यकता है!",
"_____no_output_____"
]
],
[
[
"iterations = 10\nworker_iters = 5\n\nfor a_iter in range(iterations):\n \n bobs_model = model.copy().send(bob)\n alices_model = model.copy().send(alice)\n\n bobs_opt = optim.SGD(params=bobs_model.parameters(),lr=0.1)\n alices_opt = optim.SGD(params=alices_model.parameters(),lr=0.1)\n\n for wi in range(worker_iters):\n\n # Train Bob's Model\n bobs_opt.zero_grad()\n bobs_pred = bobs_model(bobs_data)\n bobs_loss = ((bobs_pred - bobs_target)**2).sum()\n bobs_loss.backward()\n\n bobs_opt.step()\n bobs_loss = bobs_loss.get().data\n\n # Train Alice's Model\n alices_opt.zero_grad()\n alices_pred = alices_model(alices_data)\n alices_loss = ((alices_pred - alices_target)**2).sum()\n alices_loss.backward()\n\n alices_opt.step()\n alices_loss = alices_loss.get().data\n \n alices_model.move(secure_worker)\n bobs_model.move(secure_worker)\n with torch.no_grad():\n model.weight.set_(((alices_model.weight.data + bobs_model.weight.data) / 2).get())\n model.bias.set_(((alices_model.bias.data + bobs_model.bias.data) / 2).get())\n \n print(\"Bob:\" + str(bobs_loss) + \" Alice:\" + str(alices_loss))",
"_____no_output_____"
]
],
[
[
"अंत में, हम यह सुनिश्चित करना चाहते हैं कि हमारा परिणामी मॉडल सही तरीके से सीखे, इसलिए हम इसका मूल्यांकन एक परीक्षण डेटासेट पर करेंगे। इस छोटी सी समस्या में, हम मूल डेटा का उपयोग करेंगे, लेकिन व्यवहार में हम यह समझने के लिए नए डेटा का उपयोग करना चाहेंगे कि मॉडल अनदेखी उदाहरणों के लिए कितना सामान्य है।",
"_____no_output_____"
]
],
[
[
"preds = model(data)\nloss = ((preds - target) ** 2).sum()",
"_____no_output_____"
],
[
"print(preds)\nprint(target)\nprint(loss.data)",
"_____no_output_____"
]
],
[
[
"इस छोटे उदाहरण में, औसत मॉडल स्थानीय स्तर पर प्रशिक्षित एक सादे मॉडल के सापेक्ष व्यवहार कर रहा है, हालांकि हम प्रत्येक कार्यकर्ता के प्रशिक्षण डेटा को उजागर किए बिना इसे प्रशिक्षित करने में सक्षम थे। हम मॉडल अधिकारी को डेटा रिसाव को रोकने के लिए एक विश्वसनीय एग्रीगेटर पर प्रत्येक कार्यकर्ता से अपडेट किए गए मॉडल को एकत्र करने में सक्षम थे।\n\nभविष्य के ट्यूटोरियल में, हम अपने विश्वसनीय एकत्रीकरण को सीधे ग्रेडिएंट के साथ करने का लक्ष्य रखेंगे, ताकि हम मॉडल को बेहतर ग्रेडिएंट अनुमानों के साथ अपडेट कर सकें और एक मजबूत मॉडल पर पहुंच सकें।",
"_____no_output_____"
],
[
"# बधाई हो!!! - समुदाय में शामिल होने का समय!\n\nइस नोटबुक ट्यूटोरियल को पूरा करने पर बधाई! यदि आपने इसका आनंद लिया है और एआई और एआई सप्लाई चेन (डेटा) के विकेन्द्रीकृत स्वामित्व के संरक्षण की ओर आंदोलन में शामिल होना चाहते हैं, तो आप निम्न तरीकों से ऐसा कर सकते हैं!\n\n### GitHub पर स्टार PySyft\n\nहमारे समुदाय की मदद करने का सबसे आसान तरीका सिर्फ रिपॉजिटरी को अभिनीत करना है! यह हमारे द्वारा बनाए जा रहे कूल टूल्स के बारे में जागरूकता बढ़ाने में मदद करता है।\n\n- [स्टार PySyft](https://github.com/OpenMined/PySyft)\n\n### हमारे Slack में शामिल हों!\n\nनवीनतम प्रगति पर अद्यतित रहने का सबसे अच्छा तरीका हमारे समुदाय में शामिल होना है! [http://slack.openmined.org](http://slack.openmined.org) पर फॉर्म भरकर आप ऐसा कर सकते हैं\n\n### एक कोड परियोजना में शामिल हों!\n\nहमारे समुदाय में योगदान करने का सबसे अच्छा तरीका एक कोड योगदानकर्ता बनना है! किसी भी समय आप PySyft GitHub जारी करने वाले पृष्ठ पर जा सकते हैं और \"Projects\" के लिए फ़िल्टर कर सकते हैं। यह आपको सभी शीर्ष स्तर के टिकट दिखाएगा कि आप किन परियोजनाओं में शामिल हो सकते हैं! यदि आप किसी परियोजना में शामिल नहीं होना चाहते हैं, लेकिन आप थोड़ी सी कोडिंग करना चाहते हैं, तो आप \"good first issue\" चिह्नित गीथहब मुद्दों की खोज करके अधिक \"one off\" मिनी-प्रोजेक्ट्स की तलाश कर सकते हैं।\n\n- [PySyft Projects](https://github.com/OpenMined/PySyft/issues?q=is%3Aopen+is%3Aissue+label%3AProject)\n- [Good First Issue Tickets](https://github.com/OpenMined/PySyft/issues?q=is%3Aopen+is%3Aissue+label%3A%22good+first+issue%22)\n\n### दान करना\n\nयदि आपके पास हमारे कोडबेस में योगदान करने का समय नहीं है, लेकिन फिर भी समर्थन उधार देना चाहते हैं, तो आप हमारे ओपन कलेक्टिव में भी एक बैकर बन सकते हैं। सभी दान हमारी वेब होस्टिंग और अन्य सामुदायिक खर्च जैसे कि हैकाथॉन और मीटअप की ओर जाते हैं!\n\n[OpenMined का ओपन कलेक्टिव पेज](https://opencollective.com/openmined)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
ecf248ddf0084a3c6f8fe41738f99e366c06aaa8 | 36,357 | ipynb | Jupyter Notebook | src/check_cond_env_pose.ipynb | gamerDecathlete/neural_jacobian_estimation | 44deed91f0650830dd2da1796e67d084f0918995 | [
"Apache-2.0"
] | 4 | 2021-03-26T23:58:44.000Z | 2022-01-17T18:06:49.000Z | src/check_cond_env_pose.ipynb | gamerDecathlete/neural_jacobian_estimation | 44deed91f0650830dd2da1796e67d084f0918995 | [
"Apache-2.0"
] | null | null | null | src/check_cond_env_pose.ipynb | gamerDecathlete/neural_jacobian_estimation | 44deed91f0650830dd2da1796e67d084f0918995 | [
"Apache-2.0"
] | null | null | null | 443.378049 | 34,924 | 0.950794 | [
[
[
"from environments import SimulatorKinovaGripper, MultiPointReacher\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"dt = 0.05\n\n\n\n",
"_____no_output_____"
],
[
"gym = MultiPointReacher(dt= dt)\nfor i in range(100):\n gym.render()\n gym.reset()\nplt.show()\ngym.render()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecf24e4f27a71d6215fab1b0898f7737382f4489 | 753,795 | ipynb | Jupyter Notebook | Chapter-6.ipynb | chomskim/Text-Analytics-with-Python | 256fbfa142c7b05de4381d4520a1f32fa1a4972d | [
"Apache-2.0"
] | 2 | 2020-06-28T11:15:01.000Z | 2021-01-05T02:21:13.000Z | Chapter-6.ipynb | chomskim/Text-Analytics-with-Python | 256fbfa142c7b05de4381d4520a1f32fa1a4972d | [
"Apache-2.0"
] | null | null | null | Chapter-6.ipynb | chomskim/Text-Analytics-with-Python | 256fbfa142c7b05de4381d4520a1f32fa1a4972d | [
"Apache-2.0"
] | 3 | 2018-12-23T11:31:21.000Z | 2022-01-25T23:01:25.000Z | 400.528693 | 266,446 | 0.908845 | [
[
[
"# CHAPTER 6 Text Similarity and Clustering\n",
"_____no_output_____"
],
[
"## Text Normalization\n",
"_____no_output_____"
]
],
[
[
"from contractions import CONTRACTION_MAP\nimport re\nimport nltk\nimport string\nfrom nltk.stem import WordNetLemmatizer\nfrom HTMLParser import HTMLParser\nimport unicodedata\n\nstopword_list = nltk.corpus.stopwords.words('english')\nstopword_list = stopword_list + ['mr', 'mrs', 'come', 'go', 'get',\n 'tell', 'listen', 'one', 'two', 'three',\n 'four', 'five', 'six', 'seven', 'eight',\n 'nine', 'zero', 'join', 'find', 'make',\n 'say', 'ask', 'tell', 'see', 'try', 'back',\n 'also']\nwnl = WordNetLemmatizer()\nhtml_parser = HTMLParser()\n\ndef tokenize_text(text):\n tokens = nltk.word_tokenize(text) \n tokens = [token.strip() for token in tokens]\n return tokens\n\ndef expand_contractions(text, contraction_mapping):\n \n contractions_pattern = re.compile('({})'.format('|'.join(contraction_mapping.keys())), \n flags=re.IGNORECASE|re.DOTALL)\n def expand_match(contraction):\n match = contraction.group(0)\n first_char = match[0]\n expanded_contraction = contraction_mapping.get(match)\\\n if contraction_mapping.get(match)\\\n else contraction_mapping.get(match.lower()) \n expanded_contraction = first_char+expanded_contraction[1:]\n return expanded_contraction\n \n expanded_text = contractions_pattern.sub(expand_match, text)\n expanded_text = re.sub(\"'\", \"\", expanded_text)\n return expanded_text\n \n \nfrom pattern.en import tag\nfrom nltk.corpus import wordnet as wn\n\n# Annotate text tokens with POS tags\ndef pos_tag_text(text):\n \n def penn_to_wn_tags(pos_tag):\n if pos_tag.startswith('J'):\n return wn.ADJ\n elif pos_tag.startswith('V'):\n return wn.VERB\n elif pos_tag.startswith('N'):\n return wn.NOUN\n elif pos_tag.startswith('R'):\n return wn.ADV\n else:\n return None\n \n tagged_text = tag(text)\n tagged_lower_text = [(word.lower(), penn_to_wn_tags(pos_tag))\n for word, pos_tag in\n tagged_text]\n return tagged_lower_text\n \n# lemmatize text based on POS tags \ndef lemmatize_text(text):\n \n pos_tagged_text = pos_tag_text(text)\n lemmatized_tokens = [wnl.lemmatize(word, pos_tag) if pos_tag\n else word \n for word, pos_tag in pos_tagged_text]\n lemmatized_text = ' '.join(lemmatized_tokens)\n return lemmatized_text\n \n\ndef remove_special_characters(text):\n tokens = tokenize_text(text)\n pattern = re.compile('[{}]'.format(re.escape(string.punctuation)))\n filtered_tokens = filter(None, [pattern.sub(' ', token) for token in tokens])\n filtered_text = ' '.join(filtered_tokens)\n return filtered_text\n \n \ndef remove_stopwords(text):\n tokens = tokenize_text(text)\n filtered_tokens = [token for token in tokens if token not in stopword_list]\n filtered_text = ' '.join(filtered_tokens) \n return filtered_text\n\ndef keep_text_characters(text):\n filtered_tokens = []\n tokens = tokenize_text(text)\n for token in tokens:\n if re.search('[a-zA-Z]', token):\n filtered_tokens.append(token)\n filtered_text = ' '.join(filtered_tokens)\n return filtered_text\n\ndef unescape_html(parser, text):\n \n return parser.unescape(text)\n\ndef normalize_corpus(corpus, lemmatize=True, \n only_text_chars=False,\n tokenize=False):\n \n normalized_corpus = [] \n for text in corpus:\n text = html_parser.unescape(text)\n text = expand_contractions(text, CONTRACTION_MAP)\n if lemmatize:\n text = lemmatize_text(text)\n else:\n text = text.lower()\n text = remove_special_characters(text)\n text = remove_stopwords(text)\n if only_text_chars:\n text = keep_text_characters(text)\n \n if tokenize:\n text = tokenize_text(text)\n normalized_corpus.append(text)\n else:\n normalized_corpus.append(text)\n \n return normalized_corpus\n\n\ndef parse_document(document):\n document = re.sub('\\n', ' ', document)\n if isinstance(document, str):\n document = document\n elif isinstance(document, unicode):\n return unicodedata.normalize('NFKD', document).encode('ascii', 'ignore')\n else:\n raise ValueError('Document is not string or unicode!')\n document = document.strip()\n sentences = nltk.sent_tokenize(document)\n sentences = [sentence.strip() for sentence in sentences]\n return sentences\n",
"_____no_output_____"
]
],
[
[
"## Feature Extraction\n",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer\n\ndef build_feature_matrix(documents, feature_type='frequency',\n ngram_range=(1, 1), min_df=0.0, max_df=1.0):\n\n feature_type = feature_type.lower().strip() \n \n if feature_type == 'binary':\n vectorizer = CountVectorizer(binary=True, min_df=min_df,\n max_df=max_df, ngram_range=ngram_range)\n elif feature_type == 'frequency':\n vectorizer = CountVectorizer(binary=False, min_df=min_df,\n max_df=max_df, ngram_range=ngram_range)\n elif feature_type == 'tfidf':\n vectorizer = TfidfVectorizer(min_df=min_df, max_df=max_df, \n ngram_range=ngram_range)\n else:\n raise Exception(\"Wrong feature type entered. Possible values: 'binary', 'frequency', 'tfidf'\")\n\n feature_matrix = vectorizer.fit_transform(documents).astype(float)\n \n return vectorizer, feature_matrix",
"_____no_output_____"
]
],
[
[
"## Analyzing Term Similarity\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom scipy.stats import itemfreq\n\ndef vectorize_terms(terms):\n terms = [term.lower() for term in terms]\n terms = [np.array(list(term)) for term in terms]\n terms = [np.array([ord(char) for char in term]) \n for term in terms]\n return terms\n \ndef boc_term_vectors(word_list):\n word_list = [word.lower() for word in word_list]\n unique_chars = np.unique(\n np.hstack([list(word) \n for word in word_list]))\n word_list_term_counts = [{char: count for char, count in itemfreq(list(word))}\n for word in word_list]\n \n boc_vectors = [np.array([int(word_term_counts.get(char, 0)) \n for char in unique_chars])\n for word_term_counts in word_list_term_counts]\n return list(unique_chars), boc_vectors\n",
"_____no_output_____"
],
[
"root = 'Believe'\nterm1 = 'beleive'\nterm2 = 'bargain'\nterm3 = 'Elephant' \n\nterms = [root, term1, term2, term3]\n\nvec_root, vec_term1, vec_term2, vec_term3 = vectorize_terms(terms)\nprint '''\nroot: {}\nterm1: {}\nterm2: {}\nterm3: {}\n'''.format(vec_root, vec_term1, vec_term2, vec_term3)\n",
"\nroot: [ 98 101 108 105 101 118 101]\nterm1: [ 98 101 108 101 105 118 101]\nterm2: [ 98 97 114 103 97 105 110]\nterm3: [101 108 101 112 104 97 110 116]\n\n"
],
[
"features, (boc_root, boc_term1, boc_term2, boc_term3) = boc_term_vectors(terms)\nprint 'Features:', features\nprint '''\nroot: {}\nterm1: {}\nterm2: {}\nterm3: {}\n'''.format(boc_root, boc_term1, boc_term2, boc_term3)\n",
"Features: ['a', 'b', 'e', 'g', 'h', 'i', 'l', 'n', 'p', 'r', 't', 'v']\n\nroot: [0 1 3 0 0 1 1 0 0 0 0 1]\nterm1: [0 1 3 0 0 1 1 0 0 0 0 1]\nterm2: [2 1 0 1 0 1 0 1 0 1 0 0]\nterm3: [1 0 2 0 1 0 1 1 1 0 1 0]\n\n"
]
],
[
[
"### Hamming Distance\n",
"_____no_output_____"
]
],
[
[
"def hamming_distance(u, v, norm=False):\n if u.shape != v.shape:\n raise ValueError('The vectors must have equal lengths.')\n return (u != v).sum() if not norm else (u != v).mean()\n",
"_____no_output_____"
],
[
"# DEMOS!\n\n# build the term vectors here \nroot_term = root\nroot_vector = vec_root\nroot_boc_vector = boc_root\n\nterms = [term1, term2, term3]\nvector_terms = [vec_term1, vec_term2, vec_term3]\nboc_vector_terms = [boc_term1, boc_term2, boc_term3]\n",
"_____no_output_____"
],
[
"# HAMMING DISTANCE DEMO\nfor term, vector_term in zip(terms, vector_terms):\n print 'Hamming distance between root: {} and term: {} is {}'.format(root_term,\n term,\n hamming_distance(root_vector, vector_term, norm=False))\n",
"Hamming distance between root: Believe and term: beleive is 2\nHamming distance between root: Believe and term: bargain is 6\n"
],
[
"for term, vector_term in zip(terms, vector_terms):\n print 'Normalized Hamming distance between root: {} and term: {} is {}'.format(root_term,\n term,\n round(hamming_distance(root_vector, vector_term, norm=True), 2))\n",
"Normalized Hamming distance between root: Believe and term: beleive is 0.29\nNormalized Hamming distance between root: Believe and term: bargain is 0.86\n"
]
],
[
[
"### Manhattan Distance\n",
"_____no_output_____"
]
],
[
[
"def manhattan_distance(u, v, norm=False):\n if u.shape != v.shape:\n raise ValueError('The vectors must have equal lengths.')\n return abs(u - v).sum() if not norm else abs(u - v).mean()\n\n",
"_____no_output_____"
],
[
"for term, vector_term in zip(terms, vector_terms):\n print 'Manhattan distance between root: {} and term: {} is {}'.format(root_term,\n term,\n manhattan_distance(root_vector, vector_term, norm=False))\n",
"Manhattan distance between root: Believe and term: beleive is 8\nManhattan distance between root: Believe and term: bargain is 38\n"
],
[
"for term, vector_term in zip(terms, vector_terms):\n print 'Normalized Manhattan distance between root: {} and term: {} is {}'.format(root_term,\n term,\n round(manhattan_distance(root_vector, vector_term, norm=True),2))\n",
"Normalized Manhattan distance between root: Believe and term: beleive is 1.14\nNormalized Manhattan distance between root: Believe and term: bargain is 5.43\n"
]
],
[
[
"### Euclidean Distance\n",
"_____no_output_____"
]
],
[
[
"def euclidean_distance(u,v):\n if u.shape != v.shape:\n raise ValueError('The vectors must have equal lengths.')\n distance = np.sqrt(np.sum(np.square(u - v)))\n return distance\n",
"_____no_output_____"
],
[
"for term, vector_term in zip(terms, vector_terms):\n print 'Euclidean distance between root: {} and term: {} is {}'.format(root_term,\n term,\n round(euclidean_distance(root_vector, vector_term),2))\n\n",
"Euclidean distance between root: Believe and term: beleive is 5.66\nEuclidean distance between root: Believe and term: bargain is 17.94\n"
]
],
[
[
"### Levenshtein Edit Distance\n",
"_____no_output_____"
]
],
[
[
"import copy\nimport pandas as pd\n\ndef levenshtein_edit_distance(u, v):\n # convert to lower case\n u = u.lower()\n v = v.lower()\n # base cases\n if u == v: return 0\n elif len(u) == 0: return len(v)\n elif len(v) == 0: return len(u)\n # initialize edit distance matrix\n edit_matrix = []\n # initialize two distance matrices \n du = [0] * (len(v) + 1)\n dv = [0] * (len(v) + 1)\n # du: the previous row of edit distances\n for i in range(len(du)):\n du[i] = i\n # dv : the current row of edit distances \n for i in range(len(u)):\n dv[0] = i + 1\n # compute cost as per algorithm\n for j in range(len(v)):\n cost = 0 if u[i] == v[j] else 1\n dv[j + 1] = min(dv[j] + 1, du[j + 1] + 1, du[j] + cost)\n # assign dv to du for next iteration\n for j in range(len(du)):\n du[j] = dv[j]\n # copy dv to the edit matrix\n edit_matrix.append(copy.copy(dv))\n # compute the final edit distance and edit matrix \n distance = dv[len(v)]\n edit_matrix = np.array(edit_matrix)\n edit_matrix = edit_matrix.T\n edit_matrix = edit_matrix[1:,]\n edit_matrix = pd.DataFrame(data=edit_matrix,\n index=list(v),\n columns=list(u))\n return distance, edit_matrix\n \n",
"_____no_output_____"
],
[
"for term in terms:\n edit_d, edit_m = levenshtein_edit_distance(root_term, term)\n print 'Computing distance between root: {} and term: {}'.format(root_term,\n term)\n print 'Levenshtein edit distance is {}'.format(edit_d)\n print 'The complete edit distance matrix is depicted below'\n print edit_m\n print '-'*30 \n",
"Computing distance between root: Believe and term: beleive\nLevenshtein edit distance is 2\nThe complete edit distance matrix is depicted below\n b e l i e v e\nb 0 1 2 3 4 5 6\ne 1 0 1 2 3 4 5\nl 2 1 0 1 2 3 4\ne 3 2 1 1 1 2 3\ni 4 3 2 1 2 2 3\nv 5 4 3 2 2 2 3\ne 6 5 4 3 2 3 2\n------------------------------\nComputing distance between root: Believe and term: bargain\nLevenshtein edit distance is 6\nThe complete edit distance matrix is depicted below\n b e l i e v e\nb 0 1 2 3 4 5 6\na 1 1 2 3 4 5 6\nr 2 2 2 3 4 5 6\ng 3 3 3 3 4 5 6\na 4 4 4 4 4 5 6\ni 5 5 5 4 5 5 6\nn 6 6 6 5 5 6 6\n------------------------------\nComputing distance between root: Believe and term: Elephant\nLevenshtein edit distance is 7\nThe complete edit distance matrix is depicted below\n b e l i e v e\ne 1 1 2 3 4 5 6\nl 2 2 1 2 3 4 5\ne 3 2 2 2 2 3 4\np 4 3 3 3 3 3 4\nh 5 4 4 4 4 4 4\na 6 5 5 5 5 5 5\nn 7 6 6 6 6 6 6\nt 8 7 7 7 7 7 7\n------------------------------\n"
]
],
[
[
"### Cosine Distance and Similarity\n",
"_____no_output_____"
]
],
[
[
"def cosine_distance(u, v):\n distance = 1.0 - (np.dot(u, v) / \n (np.sqrt(sum(np.square(u))) * np.sqrt(sum(np.square(v))))\n )\n return distance\n",
"_____no_output_____"
],
[
"for term, boc_term in zip(terms, boc_vector_terms):\n print 'Analyzing similarity between root: {} and term: {}'.format(root_term,\n term)\n distance = round(cosine_distance(root_boc_vector, boc_term),2)\n similarity = 1 - distance \n print 'Cosine distance is {}'.format(distance)\n print 'Cosine similarity is {}'.format(similarity)\n print '-'*40\n",
"Analyzing similarity between root: Believe and term: beleive\nCosine distance is -0.0\nCosine similarity is 1.0\n----------------------------------------\nAnalyzing similarity between root: Believe and term: bargain\nCosine distance is 0.82\nCosine similarity is 0.18\n----------------------------------------\nAnalyzing similarity between root: Believe and term: Elephant\nCosine distance is 0.39\nCosine similarity is 0.61\n----------------------------------------\n"
]
],
[
[
"## Analyzing Document Similarity\n",
"_____no_output_____"
]
],
[
[
"#from normalization import normalize_corpus\n#from utils import build_feature_matrix\nimport numpy as np\n",
"_____no_output_____"
],
[
"toy_corpus = ['The sky is blue',\n'The sky is blue and beautiful',\n'Look at the bright blue sky!',\n'Python is a great Programming language',\n'Python and Java are popular Programming languages',\n'Among Programming languages, both Python and Java are the most used in Analytics',\n'The fox is quicker than the lazy dog',\n'The dog is smarter than the fox',\n'The dog, fox and cat are good friends']\n",
"_____no_output_____"
],
[
"query_docs = ['The fox is definitely smarter than the dog',\n 'Java is a static typed programming language unlike Python',\n 'I love to relax under the beautiful blue sky!'] \n\n",
"_____no_output_____"
],
[
"# normalize and extract features from the toy corpus\nnorm_corpus = normalize_corpus(toy_corpus, lemmatize=True)\ntfidf_vectorizer, tfidf_features = build_feature_matrix(norm_corpus,\n feature_type='tfidf',\n ngram_range=(1, 1), \n min_df=0.0, max_df=1.0)\n \n",
"_____no_output_____"
],
[
"# normalize and extract features from the query corpus\nnorm_query_docs = normalize_corpus(query_docs, lemmatize=True) \nquery_docs_tfidf = tfidf_vectorizer.transform(norm_query_docs)\n",
"_____no_output_____"
]
],
[
[
"### Cosine Similarity\n",
"_____no_output_____"
]
],
[
[
"def compute_cosine_similarity(doc_features, corpus_features,\n top_n=3):\n # get document vectors\n doc_features = doc_features[0]\n # compute similarities\n #similarity = np.dot(doc_features, corpus_features.T)\n similarity = doc_features * corpus_features.T\n similarity = similarity.toarray()[0]\n # get docs with highest similarity scores\n top_docs = similarity.argsort()[::-1][:top_n]\n top_docs_with_score = [(index, round(similarity[index], 3))\n for index in top_docs]\n return top_docs_with_score\n",
"_____no_output_____"
],
[
"print 'Document Similarity Analysis using Cosine Similarity'\nprint '='*60\nfor index, doc in enumerate(query_docs):\n \n doc_tfidf = query_docs_tfidf[index]\n top_similar_docs = compute_cosine_similarity(doc_tfidf,\n tfidf_features,\n top_n=2)\n print 'Document',index+1 ,':', doc\n print 'Top', len(top_similar_docs), 'similar docs:'\n print '-'*40 \n for doc_index, sim_score in top_similar_docs:\n print 'Doc num: {} Similarity Score: {}\\nDoc: {}'.format(doc_index+1,\n sim_score,\n toy_corpus[doc_index]) \n print '-'*40 \n print \n",
"Document Similarity Analysis using Cosine Similarity\n============================================================\nDocument 1 : The fox is definitely smarter than the dog\nTop 2 similar docs:\n----------------------------------------\nDoc num: 8 Similarity Score: 1.0\nDoc: The dog is smarter than the fox\n----------------------------------------\nDoc num: 7 Similarity Score: 0.426\nDoc: The fox is quicker than the lazy dog\n----------------------------------------\n\nDocument 2 : Java is a static typed programming language unlike Python\nTop 2 similar docs:\n----------------------------------------\nDoc num: 5 Similarity Score: 0.837\nDoc: Python and Java are popular Programming languages\n----------------------------------------\nDoc num: 6 Similarity Score: 0.661\nDoc: Among Programming languages, both Python and Java are the most used in Analytics\n----------------------------------------\n\nDocument 3 : I love to relax under the beautiful blue sky!\nTop 2 similar docs:\n----------------------------------------\nDoc num: 2 Similarity Score: 1.0\nDoc: The sky is blue and beautiful\n----------------------------------------\nDoc num: 1 Similarity Score: 0.72\nDoc: The sky is blue\n----------------------------------------\n\n"
]
],
[
[
"### Hellinger-Bhattacharya Distance\n",
"_____no_output_____"
]
],
[
[
"def compute_hellinger_bhattacharya_distance(doc_features, corpus_features,\n top_n=3):\n # get document vectors \n doc_features = doc_features.toarray()[0]\n corpus_features = corpus_features.toarray()\n # compute hb distances\n distance = np.hstack(\n np.sqrt(0.5 *\n np.sum(\n np.square(np.sqrt(doc_features) - \n np.sqrt(corpus_features)), \n axis=1)))\n # get docs with lowest distance scores \n top_docs = distance.argsort()[:top_n]\n top_docs_with_score = [(index, round(distance[index], 3))\n for index in top_docs]\n return top_docs_with_score \n",
"_____no_output_____"
],
[
"print 'Document Similarity Analysis using Hellinger-Bhattacharya distance'\nprint '='*60\nfor index, doc in enumerate(query_docs):\n \n doc_tfidf = query_docs_tfidf[index]\n top_similar_docs = compute_hellinger_bhattacharya_distance(doc_tfidf,\n tfidf_features,\n top_n=2)\n print 'Document',index+1 ,':', doc\n print 'Top', len(top_similar_docs), 'similar docs:'\n print '-'*40 \n for doc_index, sim_score in top_similar_docs:\n print 'Doc num: {} Distance Score: {}\\nDoc: {}'.format(doc_index+1,\n sim_score,\n toy_corpus[doc_index]) \n print '-'*40 \n print \n",
"Document Similarity Analysis using Hellinger-Bhattacharya distance\n============================================================\nDocument 1 : The fox is definitely smarter than the dog\nTop 2 similar docs:\n----------------------------------------\nDoc num: 8 Distance Score: 0.0\nDoc: The dog is smarter than the fox\n----------------------------------------\nDoc num: 7 Distance Score: 0.96\nDoc: The fox is quicker than the lazy dog\n----------------------------------------\n\nDocument 2 : Java is a static typed programming language unlike Python\nTop 2 similar docs:\n----------------------------------------\nDoc num: 5 Distance Score: 0.53\nDoc: Python and Java are popular Programming languages\n----------------------------------------\nDoc num: 4 Distance Score: 0.766\nDoc: Python is a great Programming language\n----------------------------------------\n\nDocument 3 : I love to relax under the beautiful blue sky!\nTop 2 similar docs:\n----------------------------------------\nDoc num: 2 Distance Score: 0.0\nDoc: The sky is blue and beautiful\n----------------------------------------\nDoc num: 1 Distance Score: 0.602\nDoc: The sky is blue\n----------------------------------------\n\n"
]
],
[
[
"### Okapi BM25 Ranking\n",
"_____no_output_____"
]
],
[
[
"import scipy.sparse as sp \n\ndef compute_corpus_term_idfs(corpus_features, norm_corpus):\n \n dfs = np.diff(sp.csc_matrix(corpus_features, copy=True).indptr)\n dfs = 1 + dfs # to smoothen idf later\n total_docs = 1 + len(norm_corpus)\n idfs = 1.0 + np.log(float(total_docs) / dfs)\n return idfs\n\n",
"_____no_output_____"
],
[
"def compute_bm25_similarity(doc_features, corpus_features,\n corpus_doc_lengths, avg_doc_length,\n term_idfs, k1=1.5, b=0.75, top_n=3):\n # get corpus bag of words features\n corpus_features = corpus_features.toarray()\n # convert query document features to binary features\n # this is to keep a note of which terms exist per document\n doc_features = doc_features.toarray()[0]\n doc_features[doc_features >= 1] = 1\n \n # compute the document idf scores for present terms\n doc_idfs = doc_features * term_idfs\n # compute numerator expression in BM25 equation\n numerator_coeff = corpus_features * (k1 + 1)\n numerator = np.multiply(doc_idfs, numerator_coeff)\n # compute denominator expression in BM25 equation\n denominator_coeff = k1 * (1 - b + \n (b * (corpus_doc_lengths / \n avg_doc_length)))\n denominator_coeff = np.vstack(denominator_coeff)\n denominator = corpus_features + denominator_coeff\n # compute the BM25 score combining the above equations\n bm25_scores = np.sum(np.divide(numerator,\n denominator),\n axis=1)\n # get top n relevant docs with highest BM25 score \n top_docs = bm25_scores.argsort()[::-1][:top_n]\n top_docs_with_score = [(index, round(bm25_scores[index], 3))\n for index in top_docs]\n return top_docs_with_score\n",
"_____no_output_____"
],
[
"vectorizer, corpus_features = build_feature_matrix(norm_corpus,\n feature_type='frequency')\nquery_docs_features = vectorizer.transform(norm_query_docs)\n\ndoc_lengths = [len(doc.split()) for doc in norm_corpus] \navg_dl = np.average(doc_lengths) \ncorpus_term_idfs = compute_corpus_term_idfs(corpus_features,\n norm_corpus)\n",
"_____no_output_____"
],
[
"print 'Document Similarity Analysis using BM25'\nprint '='*60\nfor index, doc in enumerate(query_docs):\n \n doc_features = query_docs_features[index]\n top_similar_docs = compute_bm25_similarity(doc_features,\n corpus_features,\n doc_lengths,\n avg_dl,\n corpus_term_idfs,\n k1=1.5, b=0.75,\n top_n=2)\n print 'Document',index+1 ,':', doc\n print 'Top', len(top_similar_docs), 'similar docs:'\n print '-'*40 \n for doc_index, sim_score in top_similar_docs:\n print 'Doc num: {} BM25 Score: {}\\nDoc: {}'.format(doc_index+1,\n sim_score,\n toy_corpus[doc_index]) \n print '-'*40 \n print",
"Document Similarity Analysis using BM25\n============================================================\nDocument 1 : The fox is definitely smarter than the dog\nTop 2 similar docs:\n----------------------------------------\nDoc num: 8 BM25 Score: 7.334\nDoc: The dog is smarter than the fox\n----------------------------------------\nDoc num: 7 BM25 Score: 3.88\nDoc: The fox is quicker than the lazy dog\n----------------------------------------\n\nDocument 2 : Java is a static typed programming language unlike Python\nTop 2 similar docs:\n----------------------------------------\nDoc num: 5 BM25 Score: 7.248\nDoc: Python and Java are popular Programming languages\n----------------------------------------\nDoc num: 6 BM25 Score: 6.042\nDoc: Among Programming languages, both Python and Java are the most used in Analytics\n----------------------------------------\n\nDocument 3 : I love to relax under the beautiful blue sky!\nTop 2 similar docs:\n----------------------------------------\nDoc num: 2 BM25 Score: 7.334\nDoc: The sky is blue and beautiful\n----------------------------------------\nDoc num: 1 BM25 Score: 4.984\nDoc: The sky is blue\n----------------------------------------\n\n"
]
],
[
[
"## Document Clustering\n",
"_____no_output_____"
],
[
"## Clustering Greatest Movies of All Time\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nmovie_data = pd.read_csv('movie_data.csv')\n\nprint movie_data.head()\n",
" Title Synopsis\n0 The Godfather In late summer 1945, guests are gathered for t...\n1 The Shawshank Redemption In 1947, Andy Dufresne (Tim Robbins), a banker...\n2 Schindler's List The relocation of Polish Jews from surrounding...\n3 Raging Bull The film opens in 1964, where an older and fat...\n4 Casablanca In the early years of World War II, December 1...\n"
],
[
"movie_titles = movie_data['Title'].tolist()\nmovie_synopses = movie_data['Synopsis'].tolist()\n\nprint 'Movie:', movie_titles[0]\nprint 'Movie Synopsis:', movie_synopses[0][:1000]\n",
"Movie: The Godfather\nMovie Synopsis: In late summer 1945, guests are gathered for the wedding reception of Don Vito Corleone's daughter Connie (Talia Shire) and Carlo Rizzi (Gianni Russo). Vito (Marlon Brando), the head of the Corleone Mafia family, is known to friends and associates as \"Godfather.\" He and Tom Hagen (Robert Duvall), the Corleone family lawyer, are hearing requests for favors because, according to Italian tradition, \"no Sicilian can refuse a request on his daughter's wedding day.\" One of the men who asks the Don for a favor is Amerigo Bonasera, a successful mortician and acquaintance of the Don, whose daughter was brutally beaten by two young men because she refused their advances; the men received minimal punishment. The Don is disappointed in Bonasera, who'd avoided most contact with the Don due to Corleone's nefarious business dealings. The Don's wife is godmother to Bonasera's shamed daughter, a relationship the Don uses to extract new loyalty from the undertaker. The Don agrees to have his men punish \n"
],
[
"#from normalization import normalize_corpus\n#from utils import build_feature_matrix\n\n# normalize corpus\nnorm_movie_synopses = normalize_corpus(movie_synopses,\n lemmatize=True,\n only_text_chars=True)\n\n# extract tf-idf features\nvectorizer, feature_matrix = build_feature_matrix(norm_movie_synopses,\n feature_type='tfidf',\n min_df=0.24, max_df=0.85,\n ngram_range=(1, 2))\n# view number of features\nprint feature_matrix.shape \n",
"(100, 307)\n"
],
[
"# get feature names\nfeature_names = vectorizer.get_feature_names()\n\n# print sample features\nprint feature_names[:20] \n",
"[u'able', u'accept', u'across', u'act', u'agree', u'alive', u'allow', u'alone', u'along', u'already', u'although', u'always', u'another', u'anything', u'apartment', u'appear', u'approach', u'arm', u'army', u'around']\n"
]
],
[
[
"### K-means Clustering\n",
"_____no_output_____"
]
],
[
[
"from sklearn.cluster import KMeans\n\ndef k_means(feature_matrix, num_clusters=5):\n km = KMeans(n_clusters=num_clusters,\n max_iter=10000)\n km.fit(feature_matrix)\n clusters = km.labels_\n return km, clusters\n",
"_____no_output_____"
],
[
"num_clusters = 5 \nkm_obj, clusters = k_means(feature_matrix=feature_matrix,\n num_clusters=num_clusters)\nmovie_data['Cluster'] = clusters\n\n\nfrom collections import Counter\n# get the total number of movies per cluster\nc = Counter(clusters)\nprint c.items()\n",
"[(0, 46), (1, 12), (2, 19), (3, 17), (4, 6)]\n"
],
[
"def get_cluster_data(clustering_obj, movie_data, \n feature_names, num_clusters,\n topn_features=10):\n\n cluster_details = {} \n # get cluster centroids\n ordered_centroids = clustering_obj.cluster_centers_.argsort()[:, ::-1]\n # get key features for each cluster\n # get movies belonging to each cluster\n for cluster_num in range(num_clusters):\n cluster_details[cluster_num] = {}\n cluster_details[cluster_num]['cluster_num'] = cluster_num\n key_features = [feature_names[index] \n for index \n in ordered_centroids[cluster_num, :topn_features]]\n cluster_details[cluster_num]['key_features'] = key_features\n \n movies = movie_data[movie_data['Cluster'] == cluster_num]['Title'].values.tolist()\n cluster_details[cluster_num]['movies'] = movies\n \n return cluster_details\n \n \n \ndef print_cluster_data(cluster_data):\n # print cluster details\n for cluster_num, cluster_details in cluster_data.items():\n print 'Cluster {} details:'.format(cluster_num)\n print '-'*20\n print 'Key features:', cluster_details['key_features']\n print 'Movies in this cluster:'\n print ', '.join(cluster_details['movies'])\n print '='*40\n",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nfrom sklearn.manifold import MDS\nfrom sklearn.metrics.pairwise import cosine_similarity\nimport random\nfrom matplotlib.font_manager import FontProperties\n",
"_____no_output_____"
],
[
"def plot_clusters(num_clusters, feature_matrix,\n cluster_data, movie_data,\n plot_size=(16,8)):\n # generate random color for clusters \n def generate_random_color():\n color = '#%06x' % random.randint(0, 0xFFFFFF)\n return color\n # define markers for clusters \n markers = ['o', 'v', '^', '<', '>', '8', 's', 'p', '*', 'h', 'H', 'D', 'd']\n # build cosine distance matrix\n cosine_distance = 1 - cosine_similarity(feature_matrix) \n # dimensionality reduction using MDS\n mds = MDS(n_components=2, dissimilarity=\"precomputed\", \n random_state=1)\n # get coordinates of clusters in new low-dimensional space\n plot_positions = mds.fit_transform(cosine_distance) \n x_pos, y_pos = plot_positions[:, 0], plot_positions[:, 1]\n # build cluster plotting data\n cluster_color_map = {}\n cluster_name_map = {}\n for cluster_num, cluster_details in cluster_data.items():\n # assign cluster features to unique label\n cluster_color_map[cluster_num] = generate_random_color()\n cluster_name_map[cluster_num] = ', '.join(cluster_details['key_features'][:5]).strip()\n # map each unique cluster label with its coordinates and movies\n cluster_plot_frame = pd.DataFrame({'x': x_pos,\n 'y': y_pos,\n 'label': movie_data['Cluster'].values.tolist(),\n 'title': movie_data['Title'].values.tolist()\n })\n grouped_plot_frame = cluster_plot_frame.groupby('label')\n # set plot figure size and axes\n fig, ax = plt.subplots(figsize=plot_size) \n ax.margins(0.05)\n # plot each cluster using co-ordinates and movie titles\n for cluster_num, cluster_frame in grouped_plot_frame:\n marker = markers[cluster_num] if cluster_num < len(markers) \\\n else np.random.choice(markers, size=1)[0]\n ax.plot(cluster_frame['x'], cluster_frame['y'], \n marker=marker, linestyle='', ms=12,\n label=cluster_name_map[cluster_num], \n color=cluster_color_map[cluster_num], mec='none')\n ax.set_aspect('auto')\n ax.tick_params(axis= 'x', which='both', bottom='off', top='off', \n labelbottom='off')\n ax.tick_params(axis= 'y', which='both', left='off', top='off', \n labelleft='off')\n fontP = FontProperties()\n fontP.set_size('small') \n ax.legend(loc='upper center', bbox_to_anchor=(0.5, -0.01), fancybox=True, \n shadow=True, ncol=5, numpoints=1, prop=fontP) \n #add labels as the film titles\n for index in range(len(cluster_plot_frame)):\n ax.text(cluster_plot_frame.ix[index]['x'], \n cluster_plot_frame.ix[index]['y'], \n cluster_plot_frame.ix[index]['title'], size=8) \n # show the plot \n plt.show() \n",
"_____no_output_____"
],
[
"cluster_data = get_cluster_data(clustering_obj=km_obj,\n movie_data=movie_data,\n feature_names=feature_names,\n num_clusters=num_clusters,\n topn_features=5) \n\nprint_cluster_data(cluster_data) \n\nplot_clusters(num_clusters=num_clusters, \n feature_matrix=feature_matrix,\n cluster_data=cluster_data, \n movie_data=movie_data,\n plot_size=(16,8)) \n",
"Cluster 0 details:\n--------------------\nKey features: [u'police', u'apartment', u'man', u'car', u'woman']\nMovies in this cluster:\nThe Shawshank Redemption, Schindler's List, Casablanca, One Flew Over the Cuckoo's Nest, The Wizard of Oz, Psycho, Sunset Blvd., Vertigo, On the Waterfront, West Side Story, Star Wars, E.T. the Extra-Terrestrial, 2001: A Space Odyssey, The Silence of the Lambs, It's a Wonderful Life, Some Like It Hot, 12 Angry Men, Gandhi, Unforgiven, Rocky, A Streetcar Named Desire, To Kill a Mockingbird, The Best Years of Our Lives, My Fair Lady, Ben-Hur, The Treasure of the Sierra Madre, The Apartment, High Noon, The Pianist, The French Connection, A Place in the Sun, Midnight Cowboy, Mr. Smith Goes to Washington, Annie Hall, Good Will Hunting, Tootsie, Fargo, Pulp Fiction, The Maltese Falcon, A Clockwork Orange, Taxi Driver, Double Indemnity, Rebel Without a Cause, Rear Window, The Third Man, North by Northwest\n========================================\nCluster 1 details:\n--------------------\nKey features: [u'family', u'brother', u'father', u'year', u'fight']\nMovies in this cluster:\nThe Godfather, Raging Bull, Titanic, The Godfather: Part II, Doctor Zhivago, Goodfellas, The King's Speech, Rain Man, Out of Africa, Giant, The Grapes of Wrath, Stagecoach\n========================================\nCluster 2 details:\n--------------------\nKey features: [u'soldier', u'kill', u'army', u'men', u'war']\nMovies in this cluster:\nGone with the Wind, Lawrence of Arabia, The Bridge on the River Kwai, Dr. Strangelove or: How I Learned to Stop Worrying and Love the Bomb, Apocalypse Now, The Lord of the Rings: The Return of the King, Gladiator, From Here to Eternity, Saving Private Ryan, Raiders of the Lost Ark, Patton, Braveheart, The Good, the Bad and the Ugly, Butch Cassidy and the Sundance Kid, Platoon, Dances with Wolves, The Deer Hunter, All Quiet on the Western Front, Shane\n========================================\nCluster 3 details:\n--------------------\nKey features: [u'love', u'john', u'father', u'marry', u'film']\nMovies in this cluster:\nCitizen Kane, Forrest Gump, The Sound of Music, Singin' in the Rain, Amadeus, The Philadelphia Story, An American in Paris, It Happened One Night, Terms of Endearment, The Green Mile, Close Encounters of the Third Kind, Network, Nashville, The Graduate, American Graffiti, Wuthering Heights, Yankee Doodle Dandy\n========================================\nCluster 4 details:\n--------------------\nKey features: [u'water', u'girl', u'attempt', u'cross', u'father']\nMovies in this cluster:\nChinatown, Jaws, The Exorcist, City Lights, The African Queen, Mutiny on the Bounty\n========================================\n"
]
],
[
[
"### Affinity Propagation\n",
"_____no_output_____"
]
],
[
[
"from sklearn.cluster import AffinityPropagation\n \ndef affinity_propagation(feature_matrix):\n \n sim = feature_matrix * feature_matrix.T\n sim = sim.todense()\n ap = AffinityPropagation()\n ap.fit(sim)\n clusters = ap.labels_ \n return ap, clusters\n",
"_____no_output_____"
],
[
"# get clusters using affinity propagation\nap_obj, clusters = affinity_propagation(feature_matrix=feature_matrix)\nmovie_data['Cluster'] = clusters\n\n# get the total number of movies per cluster\nc = Counter(clusters) \nprint c.items() \n\n# get total clusters\ntotal_clusters = len(c)\nprint 'Total Clusters:', total_clusters\n",
"[(0, 5), (1, 6), (2, 12), (3, 6), (4, 2), (5, 7), (6, 10), (7, 7), (8, 4), (9, 8), (10, 3), (11, 4), (12, 5), (13, 7), (14, 4), (15, 3), (16, 7)]\nTotal Clusters: 17\n"
],
[
"cluster_data = get_cluster_data(clustering_obj=ap_obj,\n movie_data=movie_data,\n feature_names=feature_names,\n num_clusters=total_clusters,\n topn_features=5) \n\nprint_cluster_data(cluster_data) \n",
"Cluster 0 details:\n--------------------\nKey features: [u'able', u'always', u'cover', u'end', u'charge']\nMovies in this cluster:\nThe Godfather, The Godfather: Part II, Doctor Zhivago, The Pianist, Goodfellas\n========================================\nCluster 1 details:\n--------------------\nKey features: [u'alive', u'accept', u'around', u'agree', u'attack']\nMovies in this cluster:\nCasablanca, One Flew Over the Cuckoo's Nest, Titanic, 2001: A Space Odyssey, The Silence of the Lambs, Good Will Hunting\n========================================\nCluster 2 details:\n--------------------\nKey features: [u'apartment', u'film', u'final', u'fall', u'due']\nMovies in this cluster:\nThe Shawshank Redemption, Vertigo, West Side Story, Rocky, Tootsie, Nashville, The Graduate, The Maltese Falcon, A Clockwork Orange, Taxi Driver, Rear Window, The Third Man\n========================================\nCluster 3 details:\n--------------------\nKey features: [u'arrest', u'film', u'evening', u'final', u'fall']\nMovies in this cluster:\nThe Wizard of Oz, Psycho, E.T. the Extra-Terrestrial, My Fair Lady, Ben-Hur, Close Encounters of the Third Kind\n========================================\nCluster 4 details:\n--------------------\nKey features: [u'become', u'film', u'city', u'army', u'die']\nMovies in this cluster:\n12 Angry Men, Mr. Smith Goes to Washington\n========================================\nCluster 5 details:\n--------------------\nKey features: [u'behind', u'city', u'father', u'appear', u'allow']\nMovies in this cluster:\nForrest Gump, Amadeus, Gladiator, Braveheart, The Exorcist, A Place in the Sun, Double Indemnity\n========================================\nCluster 6 details:\n--------------------\nKey features: [u'body', u'allow', u'although', u'city', u'break']\nMovies in this cluster:\nSchindler's List, Gone with the Wind, Lawrence of Arabia, Star Wars, The Lord of the Rings: The Return of the King, From Here to Eternity, Raiders of the Lost Ark, The Best Years of Our Lives, The Deer Hunter, Stagecoach\n========================================\nCluster 7 details:\n--------------------\nKey features: [u'brother', u'bring', u'close', u'although', u'car']\nMovies in this cluster:\nGandhi, Unforgiven, To Kill a Mockingbird, The Good, the Bad and the Ugly, Butch Cassidy and the Sundance Kid, High Noon, Shane\n========================================\nCluster 8 details:\n--------------------\nKey features: [u'child', u'everyone', u'attempt', u'fall', u'face']\nMovies in this cluster:\nChinatown, Jaws, The African Queen, Mutiny on the Bounty\n========================================\nCluster 9 details:\n--------------------\nKey features: [u'continue', u'bring', u'daughter', u'break', u'allow']\nMovies in this cluster:\nThe Bridge on the River Kwai, Dr. Strangelove or: How I Learned to Stop Worrying and Love the Bomb, Apocalypse Now, Saving Private Ryan, Patton, Platoon, Dances with Wolves, All Quiet on the Western Front\n========================================\nCluster 10 details:\n--------------------\nKey features: [u'despite', u'drop', u'family', u'confront', u'drive']\nMovies in this cluster:\nThe Treasure of the Sierra Madre, City Lights, Midnight Cowboy\n========================================\nCluster 11 details:\n--------------------\nKey features: [u'discover', u'always', u'feel', u'city', u'act']\nMovies in this cluster:\nRaging Bull, It Happened One Night, Rain Man, Rebel Without a Cause\n========================================\nCluster 12 details:\n--------------------\nKey features: [u'discuss', u'alone', u'drop', u'business', u'consider']\nMovies in this cluster:\nSingin' in the Rain, An American in Paris, The Apartment, Annie Hall, Network\n========================================\nCluster 13 details:\n--------------------\nKey features: [u'due', u'final', u'day', u'ever', u'eventually']\nMovies in this cluster:\nOn the Waterfront, It's a Wonderful Life, Some Like It Hot, The French Connection, Fargo, Pulp Fiction, North by Northwest\n========================================\nCluster 14 details:\n--------------------\nKey features: [u'early', u'able', u'end', u'charge', u'allow']\nMovies in this cluster:\nA Streetcar Named Desire, The King's Speech, Giant, The Grapes of Wrath\n========================================\nCluster 15 details:\n--------------------\nKey features: [u'enter', u'eventually', u'cut', u'accept', u'even']\nMovies in this cluster:\nThe Philadelphia Story, The Green Mile, American Graffiti\n========================================\nCluster 16 details:\n--------------------\nKey features: [u'far', u'allow', u'apartment', u'anything', u'car']\nMovies in this cluster:\nCitizen Kane, Sunset Blvd., The Sound of Music, Out of Africa, Terms of Endearment, Wuthering Heights, Yankee Doodle Dandy\n========================================\n"
],
[
"plot_clusters(num_clusters=num_clusters, \n feature_matrix=feature_matrix,\n cluster_data=cluster_data, \n movie_data=movie_data,\n plot_size=(16,8)) \n",
"_____no_output_____"
]
],
[
[
"### Ward’s Agglomerative Hierarchical Clustering\n",
"_____no_output_____"
]
],
[
[
"from scipy.cluster.hierarchy import ward, dendrogram\n\ndef ward_hierarchical_clustering(feature_matrix):\n \n cosine_distance = 1 - cosine_similarity(feature_matrix)\n linkage_matrix = ward(cosine_distance)\n return linkage_matrix\n \ndef plot_hierarchical_clusters(linkage_matrix, movie_data, figure_size=(8,12)):\n # set size\n fig, ax = plt.subplots(figsize=figure_size) \n movie_titles = movie_data['Title'].values.tolist()\n # plot dendrogram\n ax = dendrogram(linkage_matrix, orientation=\"left\", labels=movie_titles)\n plt.tick_params(axis= 'x', \n which='both', \n bottom='off',\n top='off',\n labelbottom='off')\n plt.tight_layout()\n #plt.savefig('ward_hierachical_clusters.png', dpi=200)\n plt.show()\n",
"_____no_output_____"
],
[
"# build ward's linkage matrix \nlinkage_matrix = ward_hierarchical_clustering(feature_matrix)\n# plot the dendrogram\nplot_hierarchical_clusters(linkage_matrix=linkage_matrix,\n movie_data=movie_data,\n figure_size=(8,10))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecf25a856543fa12fe39e661d42626deec77a753 | 94,308 | ipynb | Jupyter Notebook | Course4/Week1/Convolution_model_Application_v1a.ipynb | pranavkantgaur/CourseraDLSpecialization | 6e76df71ab40cccb9762282f95531ef9d541a27f | [
"MIT"
] | null | null | null | Course4/Week1/Convolution_model_Application_v1a.ipynb | pranavkantgaur/CourseraDLSpecialization | 6e76df71ab40cccb9762282f95531ef9d541a27f | [
"MIT"
] | null | null | null | Course4/Week1/Convolution_model_Application_v1a.ipynb | pranavkantgaur/CourseraDLSpecialization | 6e76df71ab40cccb9762282f95531ef9d541a27f | [
"MIT"
] | null | null | null | 96.134557 | 18,850 | 0.793506 | [
[
[
"# Convolutional Neural Networks: Application\n\nWelcome to Course 4's second assignment! In this notebook, you will:\n\n- Implement helper functions that you will use when implementing a TensorFlow model\n- Implement a fully functioning ConvNet using TensorFlow \n\n**After this assignment you will be able to:**\n\n- Build and train a ConvNet in TensorFlow for a classification problem \n\nWe assume here that you are already familiar with TensorFlow. If you are not, please refer the *TensorFlow Tutorial* of the third week of Course 2 (\"*Improving deep neural networks*\").",
"_____no_output_____"
],
[
"### <font color='darkblue'> Updates to Assignment <font>\n\n#### If you were working on a previous version\n* The current notebook filename is version \"1a\". \n* You can find your work in the file directory as version \"1\".\n* To view the file directory, go to the menu \"File->Open\", and this will open a new tab that shows the file directory.\n\n#### List of Updates\n* `initialize_parameters`: added details about tf.get_variable, `eval`. Clarified test case.\n* Added explanations for the kernel (filter) stride values, max pooling, and flatten functions.\n* Added details about softmax cross entropy with logits.\n* Added instructions for creating the Adam Optimizer.\n* Added explanation of how to evaluate tensors (optimizer and cost).\n* `forward_propagation`: clarified instructions, use \"F\" to store \"flatten\" layer.\n* Updated print statements and 'expected output' for easier visual comparisons.\n* Many thanks to Kevin P. Brown (mentor for the deep learning specialization) for his suggestions on the assignments in this course!",
"_____no_output_____"
],
[
"## 1.0 - TensorFlow model\n\nIn the previous assignment, you built helper functions using numpy to understand the mechanics behind convolutional neural networks. Most practical applications of deep learning today are built using programming frameworks, which have many built-in functions you can simply call. \n\nAs usual, we will start by loading in the packages. ",
"_____no_output_____"
]
],
[
[
"import math\nimport numpy as np\nimport h5py\nimport matplotlib.pyplot as plt\nimport scipy\nfrom PIL import Image\nfrom scipy import ndimage\nimport tensorflow as tf\nfrom tensorflow.python.framework import ops\nfrom cnn_utils import *\n\n%matplotlib inline\nnp.random.seed(1)",
"_____no_output_____"
]
],
[
[
"Run the next cell to load the \"SIGNS\" dataset you are going to use.",
"_____no_output_____"
]
],
[
[
"# Loading the data (signs)\nX_train_orig, Y_train_orig, X_test_orig, Y_test_orig, classes = load_dataset()",
"_____no_output_____"
]
],
[
[
"As a reminder, the SIGNS dataset is a collection of 6 signs representing numbers from 0 to 5.\n\n<img src=\"images/SIGNS.png\" style=\"width:800px;height:300px;\">\n\nThe next cell will show you an example of a labelled image in the dataset. Feel free to change the value of `index` below and re-run to see different examples. ",
"_____no_output_____"
]
],
[
[
"# Example of a picture\nindex = 6\nplt.imshow(X_train_orig[index])\nprint (\"y = \" + str(np.squeeze(Y_train_orig[:, index])))",
"y = 2\n"
]
],
[
[
"In Course 2, you had built a fully-connected network for this dataset. But since this is an image dataset, it is more natural to apply a ConvNet to it.\n\nTo get started, let's examine the shapes of your data. ",
"_____no_output_____"
]
],
[
[
"X_train = X_train_orig/255.\nX_test = X_test_orig/255.\nY_train = convert_to_one_hot(Y_train_orig, 6).T\nY_test = convert_to_one_hot(Y_test_orig, 6).T\nprint (\"number of training examples = \" + str(X_train.shape[0]))\nprint (\"number of test examples = \" + str(X_test.shape[0]))\nprint (\"X_train shape: \" + str(X_train.shape))\nprint (\"Y_train shape: \" + str(Y_train.shape))\nprint (\"X_test shape: \" + str(X_test.shape))\nprint (\"Y_test shape: \" + str(Y_test.shape))\nconv_layers = {}",
"number of training examples = 1080\nnumber of test examples = 120\nX_train shape: (1080, 64, 64, 3)\nY_train shape: (1080, 6)\nX_test shape: (120, 64, 64, 3)\nY_test shape: (120, 6)\n"
]
],
[
[
"### 1.1 - Create placeholders\n\nTensorFlow requires that you create placeholders for the input data that will be fed into the model when running the session.\n\n**Exercise**: Implement the function below to create placeholders for the input image X and the output Y. You should not define the number of training examples for the moment. To do so, you could use \"None\" as the batch size, it will give you the flexibility to choose it later. Hence X should be of dimension **[None, n_H0, n_W0, n_C0]** and Y should be of dimension **[None, n_y]**. [Hint: search for the tf.placeholder documentation\"](https://www.tensorflow.org/api_docs/python/tf/placeholder).",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: create_placeholders\n\ndef create_placeholders(n_H0, n_W0, n_C0, n_y):\n \"\"\"\n Creates the placeholders for the tensorflow session.\n \n Arguments:\n n_H0 -- scalar, height of an input image\n n_W0 -- scalar, width of an input image\n n_C0 -- scalar, number of channels of the input\n n_y -- scalar, number of classes\n \n Returns:\n X -- placeholder for the data input, of shape [None, n_H0, n_W0, n_C0] and dtype \"float\"\n Y -- placeholder for the input labels, of shape [None, n_y] and dtype \"float\"\n \"\"\"\n\n ### START CODE HERE ### (≈2 lines)\n X = tf.placeholder(tf.float32, shape=(None, n_H0, n_W0, n_C0))\n Y = tf.placeholder(tf.float32, shape=(None, n_y))\n ### END CODE HERE ###\n \n return X, Y",
"_____no_output_____"
],
[
"X, Y = create_placeholders(64, 64, 3, 6)\nprint (\"X = \" + str(X))\nprint (\"Y = \" + str(Y))",
"X = Tensor(\"Placeholder:0\", shape=(?, 64, 64, 3), dtype=float32)\nY = Tensor(\"Placeholder_1:0\", shape=(?, 6), dtype=float32)\n"
]
],
[
[
"**Expected Output**\n\n<table> \n<tr>\n<td>\n X = Tensor(\"Placeholder:0\", shape=(?, 64, 64, 3), dtype=float32)\n\n</td>\n</tr>\n<tr>\n<td>\n Y = Tensor(\"Placeholder_1:0\", shape=(?, 6), dtype=float32)\n\n</td>\n</tr>\n</table>",
"_____no_output_____"
],
[
"### 1.2 - Initialize parameters\n\nYou will initialize weights/filters $W1$ and $W2$ using `tf.contrib.layers.xavier_initializer(seed = 0)`. You don't need to worry about bias variables as you will soon see that TensorFlow functions take care of the bias. Note also that you will only initialize the weights/filters for the conv2d functions. TensorFlow initializes the layers for the fully connected part automatically. We will talk more about that later in this assignment.\n\n**Exercise:** Implement initialize_parameters(). The dimensions for each group of filters are provided below. Reminder - to initialize a parameter $W$ of shape [1,2,3,4] in Tensorflow, use:\n```python\nW = tf.get_variable(\"W\", [1,2,3,4], initializer = ...)\n```\n#### tf.get_variable()\n[Search for the tf.get_variable documentation](https://www.tensorflow.org/api_docs/python/tf/get_variable). Notice that the documentation says:\n```\nGets an existing variable with these parameters or create a new one.\n```\nSo we can use this function to create a tensorflow variable with the specified name, but if the variables already exist, it will get the existing variable with that same name.\n",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: initialize_parameters\n\ndef initialize_parameters():\n \"\"\"\n Initializes weight parameters to build a neural network with tensorflow. The shapes are:\n W1 : [4, 4, 3, 8]\n W2 : [2, 2, 8, 16]\n Note that we will hard code the shape values in the function to make the grading simpler.\n Normally, functions should take values as inputs rather than hard coding.\n Returns:\n parameters -- a dictionary of tensors containing W1, W2\n \"\"\"\n \n tf.set_random_seed(1) # so that your \"random\" numbers match ours\n \n ### START CODE HERE ### (approx. 2 lines of code)\n W1 = tf.get_variable(\"W1\", [4,4,3, 8], initializer=tf.contrib.layers.xavier_initializer(seed=0))\n W2 = tf.get_variable(\"W2\", [2,2,8, 16], initializer=tf.contrib.layers.xavier_initializer(seed = 0))\n ### END CODE HERE ###\n\n parameters = {\"W1\": W1,\n \"W2\": W2}\n \n return parameters",
"_____no_output_____"
],
[
"tf.reset_default_graph()\nwith tf.Session() as sess_test:\n parameters = initialize_parameters()\n init = tf.global_variables_initializer()\n sess_test.run(init)\n print(\"W1[1,1,1] = \\n\" + str(parameters[\"W1\"].eval()[1,1,1]))\n print(\"W1.shape: \" + str(parameters[\"W1\"].shape))\n print(\"\\n\")\n print(\"W2[1,1,1] = \\n\" + str(parameters[\"W2\"].eval()[1,1,1]))\n print(\"W2.shape: \" + str(parameters[\"W2\"].shape))",
"W1[1,1,1] = \n[ 0.00131723 0.14176141 -0.04434952 0.09197326 0.14984085 -0.03514394\n -0.06847463 0.05245192]\nW1.shape: (4, 4, 3, 8)\n\n\nW2[1,1,1] = \n[-0.08566415 0.17750949 0.11974221 0.16773748 -0.0830943 -0.08058\n -0.00577033 -0.14643836 0.24162132 -0.05857408 -0.19055021 0.1345228\n -0.22779644 -0.1601823 -0.16117483 -0.10286498]\nW2.shape: (2, 2, 8, 16)\n"
]
],
[
[
"** Expected Output:**\n\n```\nW1[1,1,1] = \n[ 0.00131723 0.14176141 -0.04434952 0.09197326 0.14984085 -0.03514394\n -0.06847463 0.05245192]\nW1.shape: (4, 4, 3, 8)\n\n\nW2[1,1,1] = \n[-0.08566415 0.17750949 0.11974221 0.16773748 -0.0830943 -0.08058\n -0.00577033 -0.14643836 0.24162132 -0.05857408 -0.19055021 0.1345228\n -0.22779644 -0.1601823 -0.16117483 -0.10286498]\nW2.shape: (2, 2, 8, 16)\n```",
"_____no_output_____"
],
[
"### 1.3 - Forward propagation\n\nIn TensorFlow, there are built-in functions that implement the convolution steps for you.\n\n- **tf.nn.conv2d(X,W, strides = [1,s,s,1], padding = 'SAME'):** given an input $X$ and a group of filters $W$, this function convolves $W$'s filters on X. The third parameter ([1,s,s,1]) represents the strides for each dimension of the input (m, n_H_prev, n_W_prev, n_C_prev). Normally, you'll choose a stride of 1 for the number of examples (the first value) and for the channels (the fourth value), which is why we wrote the value as `[1,s,s,1]`. You can read the full documentation on [conv2d](https://www.tensorflow.org/api_docs/python/tf/nn/conv2d).\n\n- **tf.nn.max_pool(A, ksize = [1,f,f,1], strides = [1,s,s,1], padding = 'SAME'):** given an input A, this function uses a window of size (f, f) and strides of size (s, s) to carry out max pooling over each window. For max pooling, we usually operate on a single example at a time and a single channel at a time. So the first and fourth value in `[1,f,f,1]` are both 1. You can read the full documentation on [max_pool](https://www.tensorflow.org/api_docs/python/tf/nn/max_pool).\n\n- **tf.nn.relu(Z):** computes the elementwise ReLU of Z (which can be any shape). You can read the full documentation on [relu](https://www.tensorflow.org/api_docs/python/tf/nn/relu).\n\n- **tf.contrib.layers.flatten(P)**: given a tensor \"P\", this function takes each training (or test) example in the batch and flattens it into a 1D vector. \n * If a tensor P has the shape (m,h,w,c), where m is the number of examples (the batch size), it returns a flattened tensor with shape (batch_size, k), where $k=h \\times w \\times c$. \"k\" equals the product of all the dimension sizes other than the first dimension.\n * For example, given a tensor with dimensions [100,2,3,4], it flattens the tensor to be of shape [100, 24], where 24 = 2 * 3 * 4. You can read the full documentation on [flatten](https://www.tensorflow.org/api_docs/python/tf/contrib/layers/flatten).\n\n- **tf.contrib.layers.fully_connected(F, num_outputs):** given the flattened input F, it returns the output computed using a fully connected layer. You can read the full documentation on [full_connected](https://www.tensorflow.org/api_docs/python/tf/contrib/layers/fully_connected).\n\nIn the last function above (`tf.contrib.layers.fully_connected`), the fully connected layer automatically initializes weights in the graph and keeps on training them as you train the model. Hence, you did not need to initialize those weights when initializing the parameters.\n\n\n#### Window, kernel, filter\nThe words \"window\", \"kernel\", and \"filter\" are used to refer to the same thing. This is why the parameter `ksize` refers to \"kernel size\", and we use `(f,f)` to refer to the filter size. Both \"kernel\" and \"filter\" refer to the \"window.\"",
"_____no_output_____"
],
[
"**Exercise**\n\nImplement the `forward_propagation` function below to build the following model: `CONV2D -> RELU -> MAXPOOL -> CONV2D -> RELU -> MAXPOOL -> FLATTEN -> FULLYCONNECTED`. You should use the functions above. \n\nIn detail, we will use the following parameters for all the steps:\n - Conv2D: stride 1, padding is \"SAME\"\n - ReLU\n - Max pool: Use an 8 by 8 filter size and an 8 by 8 stride, padding is \"SAME\"\n - Conv2D: stride 1, padding is \"SAME\"\n - ReLU\n - Max pool: Use a 4 by 4 filter size and a 4 by 4 stride, padding is \"SAME\"\n - Flatten the previous output.\n - FULLYCONNECTED (FC) layer: Apply a fully connected layer without an non-linear activation function. Do not call the softmax here. This will result in 6 neurons in the output layer, which then get passed later to a softmax. In TensorFlow, the softmax and cost function are lumped together into a single function, which you'll call in a different function when computing the cost. ",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: forward_propagation\n\ndef forward_propagation(X, parameters):\n \"\"\"\n Implements the forward propagation for the model:\n CONV2D -> RELU -> MAXPOOL -> CONV2D -> RELU -> MAXPOOL -> FLATTEN -> FULLYCONNECTED\n \n Note that for simplicity and grading purposes, we'll hard-code some values\n such as the stride and kernel (filter) sizes. \n Normally, functions should take these values as function parameters.\n \n Arguments:\n X -- input dataset placeholder, of shape (input size, number of examples)\n parameters -- python dictionary containing your parameters \"W1\", \"W2\"\n the shapes are given in initialize_parameters\n\n Returns:\n Z3 -- the output of the last LINEAR unit\n \"\"\"\n \n # Retrieve the parameters from the dictionary \"parameters\" \n W1 = parameters['W1']\n W2 = parameters['W2']\n \n ### START CODE HERE ###\n # CONV2D: stride of 1, padding 'SAME'\n Z1 = tf.nn.conv2d(X ,W1, strides = [1,1,1,1], padding = 'SAME')\n # RELU\n A1 = tf.nn.relu(Z1)\n # MAXPOOL: window 8x8, stride 8, padding 'SAME'\n P1 = tf.nn.max_pool(A1, ksize = [1,8,8,1], strides = [1,8,8,1], padding = 'SAME')\n # CONV2D: filters W2, stride 1, padding 'SAME'\n Z2 = tf.nn.conv2d(P1, W2, strides=[1,1,1,1], padding='SAME')\n # RELU\n A2 = tf.nn.relu(Z2)\n # MAXPOOL: window 4x4, stride 4, padding 'SAME'\n P2 = tf.nn.max_pool(A2, ksize=[1, 4,4, 1], strides=[1,4,4,1], padding='SAME')\n # FLATTEN\n F = tf.contrib.layers.flatten(P2)\n # FULLY-CONNECTED without non-linear activation function (not not call softmax).\n # 6 neurons in output layer. Hint: one of the arguments should be \"activation_fn=None\" \n Z3 = tf.contrib.layers.fully_connected(F, 6, activation_fn=None)\n ### END CODE HERE ###\n\n return Z3",
"_____no_output_____"
],
[
"tf.reset_default_graph()\n\nwith tf.Session() as sess:\n np.random.seed(1)\n X, Y = create_placeholders(64, 64, 3, 6)\n parameters = initialize_parameters()\n Z3 = forward_propagation(X, parameters)\n init = tf.global_variables_initializer()\n sess.run(init)\n a = sess.run(Z3, {X: np.random.randn(2,64,64,3), Y: np.random.randn(2,6)})\n print(\"Z3 = \\n\" + str(a))",
"Z3 = \n[[-0.44670227 -1.57208765 -1.53049231 -2.31013036 -1.29104376 0.46852064]\n [-0.17601591 -1.57972014 -1.4737016 -2.61672091 -1.00810647 0.5747785 ]]\n"
]
],
[
[
"**Expected Output**:\n\n```\nZ3 = \n[[-0.44670227 -1.57208765 -1.53049231 -2.31013036 -1.29104376 0.46852064]\n [-0.17601591 -1.57972014 -1.4737016 -2.61672091 -1.00810647 0.5747785 ]]\n```",
"_____no_output_____"
],
[
"### 1.4 - Compute cost\n\nImplement the compute cost function below. Remember that the cost function helps the neural network see how much the model's predictions differ from the correct labels. By adjusting the weights of the network to reduce the cost, the neural network can improve its predictions.\n\nYou might find these two functions helpful: \n\n- **tf.nn.softmax_cross_entropy_with_logits(logits = Z, labels = Y):** computes the softmax entropy loss. This function both computes the softmax activation function as well as the resulting loss. You can check the full documentation [softmax_cross_entropy_with_logits](https://www.tensorflow.org/api_docs/python/tf/nn/softmax_cross_entropy_with_logits).\n- **tf.reduce_mean:** computes the mean of elements across dimensions of a tensor. Use this to calculate the sum of the losses over all the examples to get the overall cost. You can check the full documentation [reduce_mean](https://www.tensorflow.org/api_docs/python/tf/reduce_mean).\n\n#### Details on softmax_cross_entropy_with_logits (optional reading)\n* Softmax is used to format outputs so that they can be used for classification. It assigns a value between 0 and 1 for each category, where the sum of all prediction values (across all possible categories) equals 1.\n* Cross Entropy is compares the model's predicted classifications with the actual labels and results in a numerical value representing the \"loss\" of the model's predictions.\n* \"Logits\" are the result of multiplying the weights and adding the biases. Logits are passed through an activation function (such as a relu), and the result is called the \"activation.\"\n* The function is named `softmax_cross_entropy_with_logits` takes logits as input (and not activations); then uses the model to predict using softmax, and then compares the predictions with the true labels using cross entropy. These are done with a single function to optimize the calculations.\n\n** Exercise**: Compute the cost below using the function above.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: compute_cost \n\ndef compute_cost(Z3, Y):\n \"\"\"\n Computes the cost\n \n Arguments:\n Z3 -- output of forward propagation (output of the last LINEAR unit), of shape (number of examples, 6)\n Y -- \"true\" labels vector placeholder, same shape as Z3\n \n Returns:\n cost - Tensor of the cost function\n \"\"\"\n \n ### START CODE HERE ### (1 line of code)\n cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits = Z3, labels = Y))\n ### END CODE HERE ###\n \n return cost",
"_____no_output_____"
],
[
"tf.reset_default_graph()\n\nwith tf.Session() as sess:\n np.random.seed(1)\n X, Y = create_placeholders(64, 64, 3, 6)\n parameters = initialize_parameters()\n Z3 = forward_propagation(X, parameters)\n cost = compute_cost(Z3, Y)\n init = tf.global_variables_initializer()\n sess.run(init)\n a = sess.run(cost, {X: np.random.randn(4,64,64,3), Y: np.random.randn(4,6)})\n print(\"cost = \" + str(a))",
"cost = 2.91034\n"
]
],
[
[
"**Expected Output**: \n```\ncost = 2.91034\n```",
"_____no_output_____"
],
[
"## 1.5 Model \n\nFinally you will merge the helper functions you implemented above to build a model. You will train it on the SIGNS dataset. \n\n**Exercise**: Complete the function below. \n\nThe model below should:\n\n- create placeholders\n- initialize parameters\n- forward propagate\n- compute the cost\n- create an optimizer\n\nFinally you will create a session and run a for loop for num_epochs, get the mini-batches, and then for each mini-batch you will optimize the function. [Hint for initializing the variables](https://www.tensorflow.org/api_docs/python/tf/global_variables_initializer)",
"_____no_output_____"
],
[
"#### Adam Optimizer\nYou can use `tf.train.AdamOptimizer(learning_rate = ...)` to create the optimizer. The optimizer has a `minimize(loss=...)` function that you'll call to set the cost function that the optimizer will minimize.\n\nFor details, check out the documentation for [Adam Optimizer](https://www.tensorflow.org/api_docs/python/tf/train/AdamOptimizer)",
"_____no_output_____"
],
[
"#### Random mini batches\nIf you took course 2 of the deep learning specialization, you implemented `random_mini_batches()` in the \"Optimization\" programming assignment. This function returns a list of mini-batches. It is already implemented in the `cnn_utils.py` file and imported here, so you can call it like this:\n```Python\nminibatches = random_mini_batches(X, Y, mini_batch_size = 64, seed = 0)\n```\n(You will want to choose the correct variable names when you use it in your code).",
"_____no_output_____"
],
[
"#### Evaluating the optimizer and cost\n\nWithin a loop, for each mini-batch, you'll use the `tf.Session` object (named `sess`) to feed a mini-batch of inputs and labels into the neural network and evaluate the tensors for the optimizer as well as the cost. Remember that we built a graph data structure and need to feed it inputs and labels and use `sess.run()` in order to get values for the optimizer and cost.\n\nYou'll use this kind of syntax:\n```\noutput_for_var1, output_for_var2 = sess.run(\n fetches=[var1, var2],\n feed_dict={var_inputs: the_batch_of_inputs,\n var_labels: the_batch_of_labels}\n )\n```\n* Notice that `sess.run` takes its first argument `fetches` as a list of objects that you want it to evaluate (in this case, we want to evaluate the optimizer and the cost). \n* It also takes a dictionary for the `feed_dict` parameter. \n* The keys are the `tf.placeholder` variables that we created in the `create_placeholders` function above. \n* The values are the variables holding the actual numpy arrays for each mini-batch. \n* The sess.run outputs a tuple of the evaluated tensors, in the same order as the list given to `fetches`. \n\nFor more information on how to use sess.run, see the documentation [tf.Sesssion#run](https://www.tensorflow.org/api_docs/python/tf/Session#run) documentation.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: model\n\ndef model(X_train, Y_train, X_test, Y_test, learning_rate = 0.009,\n num_epochs = 100, minibatch_size = 64, print_cost = True):\n \"\"\"\n Implements a three-layer ConvNet in Tensorflow:\n CONV2D -> RELU -> MAXPOOL -> CONV2D -> RELU -> MAXPOOL -> FLATTEN -> FULLYCONNECTED\n \n Arguments:\n X_train -- training set, of shape (None, 64, 64, 3)\n Y_train -- test set, of shape (None, n_y = 6)\n X_test -- training set, of shape (None, 64, 64, 3)\n Y_test -- test set, of shape (None, n_y = 6)\n learning_rate -- learning rate of the optimization\n num_epochs -- number of epochs of the optimization loop\n minibatch_size -- size of a minibatch\n print_cost -- True to print the cost every 100 epochs\n \n Returns:\n train_accuracy -- real number, accuracy on the train set (X_train)\n test_accuracy -- real number, testing accuracy on the test set (X_test)\n parameters -- parameters learnt by the model. They can then be used to predict.\n \"\"\"\n \n ops.reset_default_graph() # to be able to rerun the model without overwriting tf variables\n tf.set_random_seed(1) # to keep results consistent (tensorflow seed)\n seed = 3 # to keep results consistent (numpy seed)\n (m, n_H0, n_W0, n_C0) = X_train.shape \n n_y = Y_train.shape[1] \n costs = [] # To keep track of the cost\n \n # Create Placeholders of the correct shape\n ### START CODE HERE ### (1 line)\n X, Y = tf.placeholder(tf.float32, shape=(None, n_H0, n_W0, n_C0)), tf.placeholder(tf.float32, shape=(None, n_y))\n ### END CODE HERE ###\n\n # Initialize parameters\n ### START CODE HERE ### (1 line)\n parameters = initialize_parameters()\n ### END CODE HERE ###\n \n # Forward propagation: Build the forward propagation in the tensorflow graph\n ### START CODE HERE ### (1 line)\n Z3 = forward_propagation(X, parameters)\n ### END CODE HERE ###\n \n # Cost function: Add cost function to tensorflow graph\n ### START CODE HERE ### (1 line)\n cost = compute_cost(Z3, Y)\n ### END CODE HERE ###\n \n # Backpropagation: Define the tensorflow optimizer. Use an AdamOptimizer that minimizes the cost.\n ### START CODE HERE ### (1 line)\n optimizer = tf.train.AdamOptimizer(learning_rate = learning_rate).minimize(loss=cost)\n ### END CODE HERE ###\n \n # Initialize all the variables globally\n init = tf.global_variables_initializer()\n \n # Start the session to compute the tensorflow graph\n with tf.Session() as sess:\n \n # Run the initialization\n sess.run(init)\n \n # Do the training loop\n for epoch in range(num_epochs):\n\n minibatch_cost = 0.\n num_minibatches = int(m / minibatch_size) # number of minibatches of size minibatch_size in the train set\n seed = seed + 1\n minibatches = random_mini_batches(X_train, Y_train, minibatch_size, seed)\n\n for minibatch in minibatches:\n\n # Select a minibatch\n (minibatch_X, minibatch_Y) = minibatch\n \"\"\"\n # IMPORTANT: The line that runs the graph on a minibatch.\n # Run the session to execute the optimizer and the cost.\n # The feedict should contain a minibatch for (X,Y).\n \"\"\"\n ### START CODE HERE ### (1 line)\n _ , temp_cost = sess.run(fetches=[optimizer, cost], feed_dict={X: minibatch_X,\n Y: minibatch_Y})\n ### END CODE HERE ###\n \n minibatch_cost += temp_cost / num_minibatches\n \n\n # Print the cost every epoch\n if print_cost == True and epoch % 5 == 0:\n print (\"Cost after epoch %i: %f\" % (epoch, minibatch_cost))\n if print_cost == True and epoch % 1 == 0:\n costs.append(minibatch_cost)\n \n \n # plot the cost\n plt.plot(np.squeeze(costs))\n plt.ylabel('cost')\n plt.xlabel('iterations (per tens)')\n plt.title(\"Learning rate =\" + str(learning_rate))\n plt.show()\n\n # Calculate the correct predictions\n predict_op = tf.argmax(Z3, 1)\n correct_prediction = tf.equal(predict_op, tf.argmax(Y, 1))\n \n # Calculate accuracy on the test set\n accuracy = tf.reduce_mean(tf.cast(correct_prediction, \"float\"))\n print(accuracy)\n train_accuracy = accuracy.eval({X: X_train, Y: Y_train})\n test_accuracy = accuracy.eval({X: X_test, Y: Y_test})\n print(\"Train Accuracy:\", train_accuracy)\n print(\"Test Accuracy:\", test_accuracy)\n \n return train_accuracy, test_accuracy, parameters",
"_____no_output_____"
]
],
[
[
"Run the following cell to train your model for 100 epochs. Check if your cost after epoch 0 and 5 matches our output. If not, stop the cell and go back to your code!",
"_____no_output_____"
]
],
[
[
"_, _, parameters = model(X_train, Y_train, X_test, Y_test)",
"Cost after epoch 0: 1.917929\nCost after epoch 5: 1.506757\nCost after epoch 10: 0.955359\nCost after epoch 15: 0.845802\nCost after epoch 20: 0.701174\nCost after epoch 25: 0.571977\nCost after epoch 30: 0.518435\nCost after epoch 35: 0.495806\nCost after epoch 40: 0.429827\nCost after epoch 45: 0.407291\nCost after epoch 50: 0.366394\nCost after epoch 55: 0.376922\nCost after epoch 60: 0.299491\nCost after epoch 65: 0.338870\nCost after epoch 70: 0.316400\nCost after epoch 75: 0.310413\nCost after epoch 80: 0.249549\nCost after epoch 85: 0.243457\nCost after epoch 90: 0.200031\nCost after epoch 95: 0.175452\n"
]
],
[
[
"**Expected output**: although it may not match perfectly, your expected output should be close to ours and your cost value should decrease.\n\n<table> \n<tr>\n <td> \n **Cost after epoch 0 =**\n </td>\n\n <td> \n 1.917929\n </td> \n</tr>\n<tr>\n <td> \n **Cost after epoch 5 =**\n </td>\n\n <td> \n 1.506757\n </td> \n</tr>\n<tr>\n <td> \n **Train Accuracy =**\n </td>\n\n <td> \n 0.940741\n </td> \n</tr> \n\n<tr>\n <td> \n **Test Accuracy =**\n </td>\n\n <td> \n 0.783333\n </td> \n</tr> \n</table>",
"_____no_output_____"
],
[
"Congratulations! You have finished the assignment and built a model that recognizes SIGN language with almost 80% accuracy on the test set. If you wish, feel free to play around with this dataset further. You can actually improve its accuracy by spending more time tuning the hyperparameters, or using regularization (as this model clearly has a high variance). \n\nOnce again, here's a thumbs up for your work! ",
"_____no_output_____"
]
],
[
[
"fname = \"images/thumbs_up.jpg\"\nimage = np.array(ndimage.imread(fname, flatten=False))\nmy_image = scipy.misc.imresize(image, size=(64,64))\nplt.imshow(my_image)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
ecf270a3cc0cab748c567027c62df7a0cb129f78 | 18,473 | ipynb | Jupyter Notebook | DirectProgramming/DPC++/Jupyter/oneapi-essentials-training/04_DPCPP_Sub_Groups/Sub_Groups.ipynb | daverous/oneAPI-samples | bafdcc8c6cade9eed74fd946bc93d8be4f5a4a00 | [
"MIT"
] | null | null | null | DirectProgramming/DPC++/Jupyter/oneapi-essentials-training/04_DPCPP_Sub_Groups/Sub_Groups.ipynb | daverous/oneAPI-samples | bafdcc8c6cade9eed74fd946bc93d8be4f5a4a00 | [
"MIT"
] | null | null | null | DirectProgramming/DPC++/Jupyter/oneapi-essentials-training/04_DPCPP_Sub_Groups/Sub_Groups.ipynb | daverous/oneAPI-samples | bafdcc8c6cade9eed74fd946bc93d8be4f5a4a00 | [
"MIT"
] | null | null | null | 32.126957 | 326 | 0.55822 | [
[
[
"# Subgroups",
"_____no_output_____"
],
[
"##### Sections\n- [What are Subgroups?](#What-are-Subgroups?)\n- [How a Subgroup Maps to Graphics Hardware](#How-a-Subgroup-Maps-to-Graphics-Hardware)\n- _Code:_ [Subgroup info](#Subgroup-info)\n- _Code:_ [Subgroup shuffle operations](#Subgroup-shuffle-operations)\n- _Code:_ [Subgroup Collectives](#Subgroup-Collectives)",
"_____no_output_____"
],
[
"## Learning Objectives",
"_____no_output_____"
],
[
"- Understand advantages of using Subgroups in Data Parallel C++ (DPC++)\n- Take advantage of Subgroup collectives in ND-Range kernel implementation\n- Use Subgroup Shuffle operations to avoid explicit memory operations",
"_____no_output_____"
],
[
"## What are Subgroups?",
"_____no_output_____"
],
[
"On many modern hardware platforms, __a subset of the work-items in a work-group__ are executed simultaneously or with additional scheduling guarantees. These subset of work-items are called subgroups. Leveraging subgroups will help to __map execution to low-level hardware__ and may help in achieving higher performance.",
"_____no_output_____"
],
[
"## Subgroups in ND-Range Kernel Execution",
"_____no_output_____"
],
[
"Parallel execution with the ND_RANGE Kernel helps to group work items that map to hardware resources. This helps to __tune applications for performance__.\n\nThe execution range of an ND-range kernel is divided into __work-groups__, __subgroups__ and __work-items__ as shown in picture below.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"## How a Subgroup Maps to Graphics Hardware",
"_____no_output_____"
],
[
"| | |\n|:---:|:---|\n| __Work-item__ | Represents the individual instances of a kernel function. | \n| __Work-group__ | The entire iteration space is divided into smaller groups called work-groups, work-items within a work-group are scheduled on a single compute unit on hardware. | \n| __Subgroup__ | A subset of work-items within a work-group that are executed simultaneously, may be mapped to vector hardware. (DPC++) | \n",
"_____no_output_____"
],
[
"The picture below shows how work-groups and subgroups map to __Intel® Gen11 Graphics Hardware__.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"## Why use Subgroups?",
"_____no_output_____"
],
[
"- Work-items in a sub-group can __communicate directly using shuffle operations__, without explicit memory operations.\n- Work-items in a sub-group can synchronize using sub-group barriers and __guarantee memory consistency__ using sub-group memory fences.\n- Work-items in a sub-group have access to __sub-group collectives__, providing fast implementations of common parallel patterns.",
"_____no_output_____"
],
[
"## sub_group class",
"_____no_output_____"
],
[
"The subgroup handle can be obtained from the nd_item using the __get_sub_group()__",
"_____no_output_____"
],
[
"```cpp\n intel::sub_group sg = item.get_sub_group();\n```",
"_____no_output_____"
],
[
"Once you have the subgroup handle, you can query for more information about the subgroup, do shuffle operations or use collective functions.",
"_____no_output_____"
],
[
"## Subgroup info",
"_____no_output_____"
],
[
"The subgroup handle can be queried to get other information like number of work-items in subgroup, or number of subgroups in a work-group which will be needed for developers to implement kernel code using subgroups:\n- __get_local_id()__ returns the index of the work-item within its subgroup\n- __get_local_range()__ returns the size of sub_group \n- __get_group_id()__ returns the index of the subgroup\n- __get_group_range()__ returns the number of subgroups within the parent work-group\n\n\n```cpp\n h.parallel_for(nd_range<1>(64,64), [=](nd_item<1> item){\n /* get sub_group handle */\n intel::sub_group sg = item.get_sub_group();\n /* query sub_group and print sub_group info once per sub_group */\n if(sg.get_local_id()[0] == 0){\n out << \"sub_group id: \" << sg.get_group_id()[0]\n << \" of \" << sg.get_group_range()\n << \", size=\" << sg.get_local_range()[0] \n << endl;\n }\n });\n```",
"_____no_output_____"
],
[
"### Lab Exercise: Subgroup Info",
"_____no_output_____"
],
[
"The DPC++ code below demonstrates subgroup query methods to print sub-group info: Inspect code, there are no modifications necessary:\n1. Inspect the code cell below and click run ▶ to save the code to file\n2. Next run ▶ the cell in the __Build and Run__ section below the code to compile and execute the code.",
"_____no_output_____"
]
],
[
[
"%%writefile lab/sub_group_info.cpp\n//==============================================================\n// Copyright © 2020 Intel Corporation\n//\n// SPDX-License-Identifier: MIT\n// =============================================================\n#include <CL/sycl.hpp>\nusing namespace sycl;\n\nstatic const size_t N = 64; // global size\nstatic const size_t B = 64; // work-group size\n\nint main() {\n queue q;\n std::cout << \"Device : \" << q.get_device().get_info<info::device::name>() << std::endl;\n\n q.submit([&](handler &h) {\n //# setup sycl stream class to print standard output from device code\n auto out = stream(1024, 768, h);\n\n //# nd-range kernel\n h.parallel_for(nd_range<1>(N, B), [=](nd_item<1> item) {\n //# get sub_group handle\n intel::sub_group sg = item.get_sub_group();\n\n //# query sub_group and print sub_group info once per sub_group\n if (sg.get_local_id()[0] == 0) {\n out << \"sub_group id: \" << sg.get_group_id()[0] << \" of \"\n << sg.get_group_range() << \", size=\" << sg.get_local_range()[0]\n << endl;\n }\n });\n }).wait();\n}",
"_____no_output_____"
]
],
[
[
"#### Build and Run\nSelect the cell below and click run ▶ to compile and execute the code:",
"_____no_output_____"
]
],
[
[
"! chmod 755 q; chmod 755 run_sub_group_info.sh; if [ -x \"$(command -v qsub)\" ]; then ./q run_sub_group_info.sh; else ./run_sub_group_info.sh; fi",
"_____no_output_____"
]
],
[
[
"_If the Jupyter cells are not responsive or if they error out when you compile the code samples, please restart the Jupyter Kernel: \n\"Kernel->Restart Kernel and Clear All Outputs\" and compile the code samples again_.",
"_____no_output_____"
],
[
"## Sub-group shuffle operations",
"_____no_output_____"
],
[
"One of the most useful features of subgroups is the ability to __communicate directly between individual work-items__ without explicit memory operations.\n\nShuffle operations enable us to remove work-group local memory usage from our kernels and/or to __avoid unnecessary repeated accesses to global memory__.\n\nThe code below uses `shuffle_xor` to swap the values of two work-items:\n\n```cpp\n h.parallel_for(nd_range<1>(N,B), [=](nd_item<1> item){\n intel::sub_group sg = item.get_sub_group();\n size_t i = item.get_global_id(0);\n /* Shuffles */\n //data[i] = sg.shuffle(data[i], 2);\n //data[i] = sg.shuffle_up(0, data[i], 1);\n //data[i] = sg.shuffle_down(data[i], 0, 1);\n data[i] = sg.shuffle_xor(data[i], 1);\n });\n\n```\n\n<img src=\"assets/shuffle_xor.png\" alt=\"shuffle_xor\" width=\"300\"/>",
"_____no_output_____"
],
[
"### Lab Exercise: Subgroup Shuffle",
"_____no_output_____"
],
[
"The code below uses subgroup shuffle to swap items in a subgroup. You can try other shuffle operations or change the fixed constant in the shuffle function.\n\nThe DPC++ code below demonstrates sub-group shuffle operations: Inspect code, there are no modifications necessary:\n\n1. Inspect the code cell below and click run ▶ to save the code to file.\n\n2. Next run ▶ the cell in the __Build and Run__ section below the code to compile and execute the code.",
"_____no_output_____"
]
],
[
[
"%%writefile lab/sub_group_shuffle.cpp\n//==============================================================\n// Copyright © 2020 Intel Corporation\n//\n// SPDX-License-Identifier: MIT\n// =============================================================\n#include <CL/sycl.hpp>\nusing namespace sycl;\n\nstatic const size_t N = 256; // global size\nstatic const size_t B = 64; // work-group size\n\nint main() {\n queue q;\n std::cout << \"Device : \" << q.get_device().get_info<info::device::name>() << std::endl;\n\n //# initialize data array using usm\n int *data = static_cast<int *>(malloc_shared(N * sizeof(int), q));\n for (int i = 0; i < N; i++) data[i] = i;\n for (int i = 0; i < N; i++) std::cout << data[i] << \" \";\n std::cout << std::endl << std::endl;\n\n q.parallel_for(nd_range<1>(N, B), [=](nd_item<1> item) {\n intel::sub_group sg = item.get_sub_group();\n size_t i = item.get_global_id(0);\n\n //# swap adjasent items in array using sub_group shuffle_xor\n data[i] = sg.shuffle_xor(data[i], 1);\n }).wait();\n\n for (int i = 0; i < N; i++) std::cout << data[i] << \" \";\n free(data, q);\n return 0;\n}",
"_____no_output_____"
]
],
[
[
"#### Build and Run\nSelect the cell below and click run ▶ to compile and execute the code:",
"_____no_output_____"
]
],
[
[
"! chmod 755 q; chmod 755 run_sub_group_shuffle.sh; if [ -x \"$(command -v qsub)\" ]; then ./q run_sub_group_shuffle.sh; else ./run_sub_group_shuffle.sh; fi",
"_____no_output_____"
]
],
[
[
"_If the Jupyter cells are not responsive or if they error out when you compile the code samples, please restart the Jupyter Kernel: \n\"Kernel->Restart Kernel and Clear All Outputs\" and compile the code samples again_.",
"_____no_output_____"
],
[
"## Subgroup Collectives",
"_____no_output_____"
],
[
"The collective functions provide implementations of closely-related common parallel patterns. \n\nProviding these implementations as library functions instead __increases developer productivity__ and gives implementations the ability to __generate highly optimized code__ for individual target devices.\n\n```cpp\n h.parallel_for(nd_range<1>(N,B), [=](nd_item<1> item){\n intel::sub_group sg = item.get_sub_group();\n size_t i = item.get_global_id(0);\n /* Collectives */\n data[i] = reduce(sg, data[i], intel::plus<>());\n //data[i] = reduce(sg, data[i], intel::maximum<>());\n //data[i] = reduce(sg, data[i], intel::minimum<>());\n });\n\n```",
"_____no_output_____"
],
[
"### Lab Exercise: Subgroup Collectives",
"_____no_output_____"
],
[
"The code below uses subgroup collectives to add all items in a subgroup. You can change \"_plus_\" to \"_maximum_\" and check output.\n\nThe DPC++ code below demonstrates sub-group collectives: Inspect code, there are no modifications necessary:\n\n1. Inspect the code cell below and click run ▶ to save the code to file.\n\n2. Next run ▶ the cell in the __Build and Run__ section below the code to compile and execute the code.",
"_____no_output_____"
]
],
[
[
"%%writefile lab/sub_group_collective.cpp\n//==============================================================\n// Copyright © 2020 Intel Corporation\n//\n// SPDX-License-Identifier: MIT\n// =============================================================\n#include <CL/sycl.hpp>\nusing namespace sycl;\n\nstatic const size_t N = 256; // global size\nstatic const size_t B = 64; // work-group size\n\nint main() {\n queue q;\n std::cout << \"Device : \" << q.get_device().get_info<info::device::name>() << std::endl;\n\n //# initialize data array using usm\n int *data = static_cast<int *>(malloc_shared(N * sizeof(int), q));\n for (int i = 0; i < N; i++) data[i] = 1 + i;\n for (int i = 0; i < N; i++) std::cout << data[i] << \" \";\n std::cout << std::endl << std::endl;\n\n q.parallel_for(nd_range<1>(N, B), [=](nd_item<1> item) {\n intel::sub_group sg = item.get_sub_group();\n size_t i = item.get_global_id(0);\n\n //# Adds all elements in sub_group using sub_group collectives\n int sum = reduce(sg, data[i], intel::plus<>());\n\n //# write sub_group sum in first location for each sub_group\n if (sg.get_local_id()[0] == 0) {\n data[i] = sum;\n } else {\n data[i] = 0;\n }\n }).wait();\n\n for (int i = 0; i < N; i++) std::cout << data[i] << \" \";\n free(data, q);\n return 0;\n}",
"_____no_output_____"
]
],
[
[
"#### Build and Run\nSelect the cell below and click run ▶ to compile and execute the code:",
"_____no_output_____"
]
],
[
[
"! chmod 755 q; chmod 755 run_sub_group_collective.sh; if [ -x \"$(command -v qsub)\" ]; then ./q run_sub_group_collective.sh; else ./run_sub_group_collective.sh; fi",
"_____no_output_____"
]
],
[
[
"_If the Jupyter cells are not responsive or if they error out when you compile the code samples, please restart the Jupyter Kernel: \n\"Kernel->Restart Kernel and Clear All Outputs\" and compile the code samples again_.",
"_____no_output_____"
],
[
"## Summary",
"_____no_output_____"
],
[
"Subgroups allow kernel programming that maps executions at low-level hardware and may help in achieving higher levels of performance.",
"_____no_output_____"
],
[
"<html><body><span style=\"color:green\"><h1>Survey</h1></span></body></html>\n\n[We would appreciate any feedback you’d care to give, so that we can improve the overall training quality and experience. Thanks! ](https://intel.az1.qualtrics.com/jfe/form/SV_574qnSw6eggbn1z)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecf2865bbd853cc42af7fc08f8821b3407a8d2c9 | 18,504 | ipynb | Jupyter Notebook | PAT MeOH Syn_adiabat.ipynb | santiago-salas-v/walas | 65c1a86f9f2aaf6757d139aa6ce658ab1c750f0d | [
"MIT"
] | null | null | null | PAT MeOH Syn_adiabat.ipynb | santiago-salas-v/walas | 65c1a86f9f2aaf6757d139aa6ce658ab1c750f0d | [
"MIT"
] | null | null | null | PAT MeOH Syn_adiabat.ipynb | santiago-salas-v/walas | 65c1a86f9f2aaf6757d139aa6ce658ab1c750f0d | [
"MIT"
] | null | null | null | 36.934132 | 273 | 0.46136 | [
[
[
"## Methanol-Synthese\n\n$T_{ein}=493,15K$\n\np=50 bar\n\n### Bilanzen\nStoffbilanzen\n\n$\\dot n_i = \\dot n_{i, 0} + \\sum_{j}{\\nu_ij \\xi_j}$ \n\nEnergiebilanz\n\n$\\begin{array}{ll}\n0 &= \\dot Q + (\\dot n (\\Delta H^\\circ_{(T)}-\\Delta H^\\circ_0))_{ein}-\n(\\dot n (\\Delta H^\\circ_{(T)}-\\Delta H^\\circ_0))_{aus} + \n\\sum\\limits_{j}{\\xi_j (-\\Delta Hr_j(T))}\\\\\n&= 0 + (\\dot n (\\Delta H^\\circ_{(T)}-\\Delta H^\\circ_0))_{ein}-\n(\\dot n (\\Delta H^\\circ_{(T)}-\\Delta H^\\circ_0))_{aus} + \n\\sum\\limits_{j}{\\xi_j (-\\Delta Hr_j(T))}\\\\\n\\end{array}\\\\\n$\n\n### Gleichgewichtskonstanten\n\n$\\begin{array}{ll}\nexp \\left(- \\frac{\\Delta G_i}{R T} \\right) &= K_p K_{\\phi^{eq}} = K_x \\prod\\limits_{i} \\left( \\frac{p}{p^0}\\right)^{\\nu_i} K_{\\phi^{eq}} \\\\\n&=\\prod\\limits_{i} (n_i)^{\\nu_i}\\left( \\frac{p}{p^0}\\right)^{\\sum\\limits_{i} \\nu_i}(n)^{-\\sum\\limits_{i} \\nu_i} K_{\\phi^{eq}}\\end{array}$\n\n$p^0 = 1 bar$\n\nIdealer Gas, $K_{\\phi^{eq}}=1$ \n\n**Methode A)** Geringe Veränderung der Reaktionsenthalpie mit der Temperatur\n\nVan't Hoff, $\\frac{d ln K}{dT} = -\\frac{\\Delta H}{R T^2} \\sim \\Rightarrow ln \\left(\\frac{K}{K'} \\right) = -\\frac{\\Delta H^0}{R}\\left(\\frac{1}{T} - \\frac{1}{T'} \\right)$\n\n$\\begin{array}{ll}\nK_{(493,15K)} &= K_{(298,15K)} \\times exp \\left[-\\frac{\\Delta H^0}{R}\\left(\\frac{1}{493,15 K} - \\frac{1}{298,15 K} \\right)\\right] \\\\\n&= \\prod_i (n_i)^{\\nu_i}\\left( \\frac{p}{p^0}\\right)^{\\sum_i \\nu_i}(n)^{-\\sum_i \\nu_i}\\end{array}$ \n\n**Methode B)** Wechselwirkung der Reaktionsenthalpie mit der Temperatur [SVNA]\n\n$\\Delta H^\\circ = \\Delta H_0^\\circ + R \\int\\limits_{T_0}^{T}{\\frac{\\Delta Cp^\\circ}{R}dT}$\n\n$\\Delta S^\\circ = \\Delta S_0^\\circ + R \\int\\limits_{T_0}^{T}{\\frac{\\Delta Cp^\\circ}{R}\\frac{dT}{T}}$\n\n$\\Delta G^\\circ = \\Delta H^\\circ - T \\Delta S^\\circ = \\Delta H_0^\\circ + R \\int\\limits_{T_0}^{T}{\\frac{\\Delta Cp^\\circ}{R}dT} - T \\Delta S_0^\\circ - R T \\int\\limits_{T_0}^{T}{\\frac{\\Delta Cp^\\circ}{R}\\frac{dT}{T}}$\n\n$\\Delta S_0^\\circ = \\frac{\\Delta H_0^\\circ - \\Delta G_0^\\circ}{T_0}$\n\n$\\Delta G^\\circ = \\Delta H_0^\\circ - \\frac{T}{T_0}(\\Delta H_0^\\circ -\\Delta G_0^\\circ) + R \\int\\limits_{T_0}^{T}{\\frac{\\Delta Cp^\\circ}{R}dT} - R T \\int\\limits_{T_0}^{T}{\\frac{\\Delta Cp^\\circ}{R}\\frac{dT}{T}}$\n\n$\\begin{array}{ll}\nK_{(T)} &= exp \\left(-\\frac{\\Delta H_0^\\circ}{R T} + \\frac{(\\Delta H_0^\\circ -\\Delta G_0^\\circ)}{R T_0} - \\frac{1}{T}\\int\\limits_{T_0}^{T}{\\frac{\\Delta Cp^\\circ}{R}dT} + \\int\\limits_{T_0}^{T}{\\frac{\\Delta Cp^\\circ}{R}\\frac{dT}{T}}\\right) \\\\\n&= \\prod_i (n_i)^{\\nu_i}\\left( \\frac{p}{p^0}\\right)^{\\sum_i \\nu_i}(n)^{-\\sum_i \\nu_i}\\end{array}$ \n\nSomit läßt sich K(T) bestimmen, insofern man über einen Ausdruck für $Cp_i(T)$ verfügt. Bei geringer Veränderung der Wärmekapazität Cp im Temperatur-Bereich kann man auch einen bestimmten Mittelwert als ~konstant einsetzen.\n\n**Methode C)** Gibbs'sche Energie-Funktion - Gef(T) - aus Thermochemischen Tabellen [BP]\n\n$-Gef(T) = -[G(T)-H(298,15)]/T$\n\n$-R ln(K) = \\sum\\nu_i Gef_i - \\sum \\nu_i H_i(298,15K)/T$\n\nIn thermochemischen Tabellen [BP] sind die Werte -Gef(T) verfügbar. \n\n### Literaturhinweise\n* [SVNA] Smith J.M., Van Ness H.C., Abbott M.M.; Introduction to chemical engineering thermodynamics; 6th ed.; McGraw-Hill; New York; 2001; S. 458-462.\n* [BP] Barin Isan, Platzki Gregor; Thermochemical data of pure substances; 3. ed.; VCH; New York; 1995.",
"_____no_output_____"
],
[
"# Adiabatischer Fall",
"_____no_output_____"
]
],
[
[
"from scipy import optimize\nimport numpy as np\n\np = 50. # bar\ntemp = 273.15 + 220. # K\nt0_ref = 298.15 # K\nr = 8.314 # J/(mol K)\n\nnamen = ['CO', 'H2', 'CO2', 'H2O', 'CH3OH']\n\nn0co = 50.\nn0h2 = 100.\nn0co2 = 0.\nn0h2o = 0.\nn0ch3oh = 0.\n\nne = np.array([n0co, n0h2, n0co2, n0h2o, n0ch3oh])\n\nnuij = np.array([[-1, -2, 0, 0, +1] ,\n [0, -3, -1, +1, +1], \n [-1, +1, +1, -1, 0]]).T\n\nh_298 = np.array(\n [-110.541, 0., -393.505, -241.826,-201.167]) * 1000 # J/mol\n\ng_298 = np.array(\n [-169.474, -38.962, -457.240, -298.164, -272.667]) * 1000 # J/mol\n\n# Berechne delta Cp(T) mit Temperaturfunktionen für ideale Gase (SVN).\n\n# Koeffizienten für Cp(T)/R = A + B*T + C*T^2 + D*T^-2, T[=]K\n# Nach rechts hin: A, B, C, D\n# Nach unten hin: CO, H2, CO2, H2O, CH3OH\ncp_coefs = np.array([\n [\n y.replace(',', '.') for y in x.split('\\t')\n ] for x in \"\"\"\n3,3760E+00\t5,5700E-04\t0,0000E+00\t-3,1000E+03\n3,2490E+00\t4,2200E-04\t0,0000E+00\t8,3000E+03\n5,4570E+00\t1,0450E-03\t0,0000E+00\t-1,1570E+05\n3,4700E+00\t1,4500E-03\t0,0000E+00\t1,2100E+04\n2,2110E+00\t1,2216E-02\t-3,4500E-06\t0,0000E+00\n\"\"\".split('\\n') if len(x)>0], dtype=float)\n\ndef cp(t):\n return r * (\n cp_coefs[:,0] + \n cp_coefs[:,1] * t + \n cp_coefs[:,2] * t**2 + \n cp_coefs[:,3] * t**-2\n ) # J/(mol K)\n\n# Berechne H(T), G(T) und K(T) mit Cp(T)\n\ndef h(t):\n return (\n h_298 + \n r * cp_coefs[:,0]*(t-t0_ref) + \n r * cp_coefs[:,1]/2.*(t**2-t0_ref**2) + \n r * cp_coefs[:,2]/3.*(t**3-t0_ref**3) -\n r * cp_coefs[:,3]*(1/t-1/t0_ref)\n ) # J/mol\n\ndef g(t, h_t):\n return (\n h_t - t/t0_ref*(h_298 - g_298) -\n r * cp_coefs[:,0]*t*np.log(t/t0_ref) -\n r * cp_coefs[:,1]*t**2*(1-t0_ref/t) - \n r * cp_coefs[:,2]/2.*t**3*(1-(t0_ref/t)**2) +\n r * cp_coefs[:,3]/2.*1/t*(1-(t/t0_ref)**2)\n ) # J/mol\n\ndef k(t, g_t):\n delta_g_t = nuij.T.dot(g_t)\n return np.exp(-delta_g_t/(r * t))\n\n\ndelta_gr_298 = nuij.T.dot(g_298)\n\ndelta_hr_298 = nuij.T.dot(h_298)\n\ncp_493 = cp(493.15) # J/(mol K)\nh_493 = h(493.15) # J/mol\ng_493 = g(493.15, h_493) # J/mol\nk_493 = k(493.15, g_493) # []\n\nfor i, f in enumerate(delta_hr_298):\n print('Delta H_' + str(i+1) + '(298.15K)=' + str(f/1000.) + 'kJ/mol')\n\nprint('\\n')\nfor i, f in enumerate(k_493):\n print('K' + str(i+1) + '(493K)=' + str(f))\nprint('\\n')\n\nn0 = np.array([n0co, n0h2, n0ch3oh])\n \ndef fun(x_vec): \n nco = x_vec[0]\n nh2 = x_vec[1]\n nch3oh = x_vec[2]\n xi1 = x_vec[3]\n t = x_vec[4]\n \n n = np.array([nco, nh2, nch3oh])\n \n cp_t = cp(t)\n h_t = h(t)\n g_t = g(t, h_t)\n k_t = k(t, g_t)\n \n h_ein = h_493[[1, 2, -1]]\n cp_ein = cp_493[[1, 2, -1]]\n cp_t = cp_t[[1, 2, -1]]\n h_t = h_t[[1, 2, -1]]\n g_t = g_t[[1, 2, -1]]\n \n delta_h_t = nuij[[1, 2, -1]].T.dot(h_t) # J/mol\n \n f1 = -nco + n0co - xi1\n f2 = -nh2 + n0h2 -2*xi1\n f3 = -nch3oh + n0ch3oh +xi1\n f4 = -k_t[0] * (nco * nh2**2) + \\\n nch3oh * (p/1.)**-2 * (nco + nh2 + nch3oh)**-(-2)\n #f5 = np.sum(\n # np.multiply(n0, h_ein) - \n # np.multiply(n, h_t)\n #) + xi1 * (-delta_h_t[0])\n f5 = np.sum(\n np.multiply(n0, cp_ein)*493.15 - \n np.multiply(n, cp_t)*t \n ) + xi1 * (-delta_h_t[0])\n \n return [f1, f2, f3, f4, f5]\n\nx0 = np.append(n0, [0., 493.15])\n\nsol = optimize.root(fun, x0)\nf_final = - sol.x[:3].reshape([3,1]) + ne[[0,1,4]].reshape([3,1]) + nuij[:,0][[0,1,4]].reshape([3,1])*sol.x[-2]\n\nprint(sol)\nprint('\\n\\n')\nprint('Zustand der Optimisierungs-Funktionen\\n')\nprint(f_final)\n\nprint('\\n\\n')\nprint('T_ein=493.15K, p=50 bar, in adiabatischem Reaktor')\nprint('Lösung für nur einzige Reaktion (ohne CO2):\\n')\nfor i, f in enumerate(sol.x[:2]):\n print('n_' + namen[i] + '= ' + str(f) + ' mol')\nprint('n_' + namen[-1] + '= ' + str(sol.x[2]) + ' mol')\nprint('T= ' + str(sol.x[-1]) + ' K')\n\nn0 = np.array([n0co, n0h2, n0co2, n0h2o, n0ch3oh])\nn0 = ne\n\n# Lösung des einfacheren Falls in schwierigerem Fall einwenden.\ndef fun(x_vec): \n nco = x_vec[0]\n nh2 = x_vec[1]\n nco2 = x_vec[2]\n nh2o = x_vec[3]\n nch3oh = x_vec[4]\n xi1 = x_vec[5]\n xi2 = x_vec[6]\n xi3 = x_vec[7]\n t = x_vec[8]\n \n n = np.array([nco, nh2, nco2, nh2o, nch3oh])\n xi = np.array([xi1, xi2, xi3])\n \n h_ein = h_493\n cp_ein = cp_493\n cp_t = cp(t)\n h_t = h(t)\n g_t = g(t, h_t)\n k_t = k(t, g_t)\n \n delta_h_t = nuij.T.dot(h_t) # J/mol\n \n f1 = -nco + n0co - xi1 +0 -xi3\n f2 = -nh2 + n0h2 -2*xi1 -3*xi2 +xi3\n f3 = -nco2 + n0co2 +0 -xi2 +xi3\n f4 = -nh2o + n0h2o +0 +xi2 -xi3\n f5 = -nch3oh + n0ch3oh +xi1 +xi2 -0\n f6 = -k_t[0] * (nco * nh2**2) + \\\n nch3oh * (p/1.)**-2 * (nco + nh2 + nco2 + nh2o + nch3oh)**-(-2)\n f7 = -k_t[1] * (nco2 * nh2**3) + \\\n nch3oh * nh2o * (p/1.)**-2 * (nco + nh2 + nco2 + nh2o + nch3oh)**-(-2)\n f8 = -k_t[2] * (nco * nh2o) + \\\n nco2 * nh2 * (p/1.)**0 * (nco + nh2 + nco2 + nh2o + nch3oh)**-0\n f9 = np.sum(\n np.multiply(n0, (h_ein-h_298)) - \n np.multiply(n, (h_t-h_298))) + np.dot(xi, -delta_h_t)\n #f9 = np.sum(\n # np.multiply(n0, cp_ein)*493.15 - \n # np.multiply(n, cp_t)*t) + np.dot(xi, -delta_h_t)\n \n return [f1, f2, f3, f4, f5, f6, f7, f8, f9]\n\nx0 = np.append(n0, [0., 0., 0., sol.x[-1]])\n\nsol = optimize.root(fun, x0)\n\nf_final = - sol.x[:5].reshape([5,1]) + ne.reshape([5,1]) + nuij.dot(sol.x[5:-1].reshape([3,1]))\n\nprint('\\n\\n')\nprint('T_ein=493.15K, p=50 bar, in adiabatischem Reaktor.')\nprint('Lösung für alle drei Reaktionen, mit CO2:\\n')\nfor i, f in enumerate(sol.x[:5]):\n print('n_' + namen[i] + '= ' + str(f) + ' mol')\n\nprint('\\n')\n\nfor i, f in enumerate(sol.x[5:-1]):\n print('xi_' + str(i) + '= ' + str(f) + ' mol')\n \nprint('\\n')\n \nprint('T=' + str(sol.x[-1]) + ' K, oder...')\nprint('T=' + str(sol.x[-1]-273.15) + ' °C')\n\nprint('\\n')\nprint('0 = Q + Sum(Delta H)_ein - Sum(Delta H)_aus')\nbilanz = np.sum(\n np.multiply(n0, (h_493-h_298)) -\n np.multiply(sol.x[:5], (h(sol.x[-1])-h_298))\n) + np.dot(sol.x[5:-1], -nuij.T.dot(h(sol.x[-1]))) \nannaeherung = np.sum(\n np.multiply(n0, cp_493)*493.15 -\n np.multiply(sol.x[:5], cp(sol.x[-1]))*sol.x[-1]\n) + np.dot(sol.x[5:-1], -nuij.T.dot(h(sol.x[-1])))\nprint('-Q = (n.(H_t-H_298))_ein -(n.(H_t-H_298))_aus + Sum(xi_j * (-Delta Hr_j)) = ' + \n str(bilanz) + 'J/h')\nprint('-Q = (n Cp T)_ein - (n Cp T)_aus + Sum(xi_j * (-Delta H_j)) = ' + \n str(annaeherung) + 'J/h' + \n '(nur Annäherung. Fehler: ' + '{:.2f}'.format(\n (annaeherung-bilanz)/bilanz) + ')' )\nprint('\\n\\n')\nprint('Zustand der Optimisierungs-Funktionen\\n')\nprint(f_final)",
"Delta H_1(298.15K)=-90.626kJ/mol\nDelta H_2(298.15K)=-49.488kJ/mol\nDelta H_3(298.15K)=-41.138kJ/mol\n\n\nK1(493K)=0.0088102868389\nK2(493K)=5.7133657404e-05\nK3(493K)=154.204845956\n\n\n fjac: array([[ -6.98220403e-05, -2.13470694e-16, -9.52744512e-21,\n -5.12921178e-03, -9.99986843e-01],\n [ 1.57166023e-02, -9.97133966e-03, -4.60048510e-18,\n 9.99813608e-01, -5.12942058e-03],\n [ -3.73103145e-01, -6.50699281e-01, 6.61350208e-01,\n -6.24396114e-04, 2.92538676e-05],\n [ -8.20521478e-01, 5.63874061e-01, 9.19102575e-02,\n 1.85216484e-02, -3.77114697e-05],\n [ 4.32771916e-01, 5.08465399e-01, 7.44424883e-01,\n -1.73205518e-03, -2.13332210e-05]])\n fun: array([ -4.44089210e-16, -8.88178420e-16, 0.00000000e+00,\n -5.52020651e-10, 6.98491931e-10])\n message: 'The solution converged.'\n nfev: 20\n qtf: array([ 1.44182179e-06, -1.56560613e-06, 9.40392430e-10,\n -2.90851842e-08, 2.75554739e-09])\n r: array([ 1.43221252e+04, 2.25742130e+04, 3.02905388e+04,\n -2.03411166e+05, 7.23437740e+03, 1.00287427e+02,\n 9.86723160e+01, -3.95568925e+03, 1.27484710e+02,\n -1.51205819e+00, 1.01056495e+01, -2.68089360e-01,\n -6.18361166e+01, 1.94702164e+00, -2.21688703e-03])\n status: 1\n success: True\n x: array([ 46.22169757, 92.44339514, 3.77830243, 3.77830243,\n 614.06573205])\n\n\n\nZustand der Optimisierungs-Funktionen\n\n[[ -4.44089210e-16]\n [ -8.88178420e-16]\n [ 0.00000000e+00]]\n\n\n\nT_ein=493.15K, p=50 bar, in adiabatischem Reaktor\nLösung für nur einzige Reaktion (ohne CO2):\n\nn_CO= 46.2216975716 mol\nn_H2= 92.4433951433 mol\nn_CH3OH= 3.77830242836 mol\nT= 614.065732048 K\n\n\n\nT_ein=493.15K, p=50 bar, in adiabatischem Reaktor.\nLösung für alle drei Reaktionen, mit CO2:\n\nn_CO= 45.3678348003 mol\nn_H2= 90.7356696005 mol\nn_CO2= 1.85405940675e-16 mol\nn_H2O= 1.4430304831e-17 mol\nn_CH3OH= 4.63216519974 mol\n\n\nxi_0= -2.64802822417 mol\nxi_1= 7.28019342391 mol\nxi_2= 7.28019342391 mol\n\n\nT=606.759730614 K, oder...\nT=333.609730614 °C\n\n\n0 = Q + Sum(Delta H)_ein - Sum(Delta H)_aus\n-Q = (n.(H_t-H_298))_ein -(n.(H_t-H_298))_aus + Sum(xi_j * (-Delta Hr_j)) = -1.21246557683e-07J/h\n-Q = (n Cp T)_ein - (n Cp T)_aus + Sum(xi_j * (-Delta H_j)) = -18389.5024134J/h(nur Annäherung. Fehler: 151670305242.10)\n\n\n\nZustand der Optimisierungs-Funktionen\n\n[[ 0.00000000e+00]\n [ 0.00000000e+00]\n [ -1.85405941e-16]\n [ -1.44303048e-17]\n [ 0.00000000e+00]]\n"
],
[
"print('Lösung, in 30 Dezimalzahlen')\nprint('')\nfor part in sol.x:\n print('{:.30g}'.format(part).replace('.',','))",
"Lösung, in 30 Dezimalzahlen\n\n45,367834800264532191249600146\n90,7356696005290643824992002919\n1,85405940674631041373621166986e-16\n1,44303048310322819158516687663e-17\n4,63216519973546780875039985403\n-2,64802822417277949895719757478\n7,28019342390824775179680727888\n7,28019342390824775179680727888\n606,759730614496675116242840886\n"
]
],
[
[
"# Min(G)\n\n$\\Delta G_{f i}^{0} + R T ln(y_i \\hat{\\phi_i} P/P^0) + \\sum\\limits_{k}{ \\lambda_k a_{ik}}=0 \\hspace{10mm} (i=1,2,...,N)$\n\n$\\sum\\limits_{i}{n_i a_{ik}}=A_k \\hspace{10mm} (k = 1,2,...,\\omega)$",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
ecf2a0f3a41daf7b9b9888fcbf2656727e3f7199 | 3,952 | ipynb | Jupyter Notebook | demo/projects/tutorials/ex1_database.ipynb | cloudcalvin/spira | 2dcaef188f2bc8c3839e1b5ff0be027e0cd4908c | [
"MIT"
] | null | null | null | demo/projects/tutorials/ex1_database.ipynb | cloudcalvin/spira | 2dcaef188f2bc8c3839e1b5ff0be027e0cd4908c | [
"MIT"
] | 1 | 2021-10-17T10:18:04.000Z | 2021-10-17T10:18:04.000Z | demo/projects/tutorials/ex1_database.ipynb | cloudcalvin/spira | 2dcaef188f2bc8c3839e1b5ff0be027e0cd4908c | [
"MIT"
] | null | null | null | 22.078212 | 102 | 0.516194 | [
[
[
"# Connect to Database\n\nOne the most powerful functionalities of SPiRA is effectively connecting data to \ncell instances. This examples shows how data from the defined RDD are connected\nto a class using parameters. By connecting parameters to a class through a \nfield allows the given data to be intercepted and manipulated before fully \ncommitting it to the class instance.",
"_____no_output_____"
],
[
"## Demonstrates\n\n1. How to link process data from the RDD to default parameter values.\n2. How to change parameters when creating an instance.\n3. How to switch to a different RDD by simply importing a new database file.\n4. Add documentation to a specific parameter.",
"_____no_output_____"
]
],
[
[
"import spira\nfrom spira import param",
"\n---------------------------------------------\n[RDD] SPiRA-default\n\n[SPiRA] Version 0.0.2-Auron - MIT License\n---------------------------------------------\n"
]
],
[
[
"The Rule Deck Database has to be imported before use. Importing a specific \nRDD script will initialize and create the data tree. ",
"_____no_output_____"
]
],
[
[
"RDD = spira.get_rule_deck()",
"_____no_output_____"
]
],
[
[
"Create a pcell using data from the currently set fabrication process.",
"_____no_output_____"
]
],
[
[
"class PCell(spira.Cell):\n\n layer = param.LayerField(number=RDD.BAS.LAYER.number, doc='Layer for the first polygon.')\n width = param.FloatField(default=RDD.BAS.WIDTH, doc='Box shape width.')\n\npcell = PCell()\nprint(pcell.layer)\nprint(pcell.width)",
"[SPiRA: Layer] ('', layer 4, datatype 0)\n1.5\n"
]
],
[
[
"Switch to a different database.",
"_____no_output_____"
]
],
[
[
"print(RDD)\nfrom demo.pdks.process.aist_pdk import database\nprint(RDD)",
"< RDD SPiRA-default>\n\n---------------------------------------------\n[RDD] AiST\n< RDD AiST>\n"
]
],
[
[
"Display parameter documentation.",
"_____no_output_____"
]
],
[
[
"print(PCell.layer.__doc__)\nprint(PCell.width.__doc__)",
"Layer for the first polygon.\nBox shape width.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf2a6b90b61d27d13acd5c21cb201b3ca9c2cae | 166,185 | ipynb | Jupyter Notebook | tutorials/create_advanced.ipynb | gisce/pandapower | 12e68eff41c1bb71436901d96867ce9571102600 | [
"BSD-3-Clause"
] | 1 | 2019-03-14T05:27:43.000Z | 2019-03-14T05:27:43.000Z | tutorials/create_advanced.ipynb | HaoranDennis/pandapower | 22c8680d3373879e792fe7478bd2dde4ea8cb018 | [
"BSD-3-Clause"
] | null | null | null | tutorials/create_advanced.ipynb | HaoranDennis/pandapower | 22c8680d3373879e792fe7478bd2dde4ea8cb018 | [
"BSD-3-Clause"
] | null | null | null | 32.464349 | 295 | 0.336583 | [
[
[
"# Create Networks - Advanced",
"_____no_output_____"
],
[
"This tutorial shows how to create a more complex pandapower network step by step. The network includes every element which is availiable in the pandapower framework.\n\nThe final network looks like this:\n\n<img src=\"pics/example_network.png\" width=\"50%\">",
"_____no_output_____"
],
[
"The structural information about this network are stored in csv tables in the example_advanced folder.\n\nFor a better overview the creation of the individual components is divided in three steps. Each step handles one of the three voltage levels: high, medium and low voltage. We star by initializing an empty pandapower network:",
"_____no_output_____"
]
],
[
[
"#import the pandapower module\nimport pandapower as pp\nimport pandas as pd\n\n#create an empty network \nnet = pp.create_empty_network()",
"_____no_output_____"
]
],
[
[
"## High voltage level",
"_____no_output_____"
],
[
"### Buses\n\n<img src=\"pics/example_network_buses_hv_detail.png\" width=\"50%\">",
"_____no_output_____"
],
[
"\nThere are two 380 kV and five 110 kV busbars (type=\"b\"). The 380/110 kV substation is modeled in detail with all nodes and switches, which is why we need additional nodes (type=\"b\") to connect the switches.",
"_____no_output_____"
]
],
[
[
"# Double busbar\npp.create_bus(net, name='Double Busbar 1', vn_kv=380, type='b')\npp.create_bus(net, name='Double Busbar 2', vn_kv=380, type='b')\nfor i in range(10):\n pp.create_bus(net, name='Bus DB T%s' % i, vn_kv=380, type='n')\nfor i in range(1, 5):\n pp.create_bus(net, name='Bus DB %s' % i, vn_kv=380, type='n')\n\n# Single busbar\npp.create_bus(net, name='Single Busbar', vn_kv=110, type='b')\nfor i in range(1, 6):\n pp.create_bus(net, name='Bus SB %s' % i, vn_kv=110, type='n')\nfor i in range(1, 6):\n for j in [1, 2]:\n pp.create_bus(net, name='Bus SB T%s.%s' % (i, j), vn_kv=110, type='n')\n\n# Remaining buses\nfor i in range(1, 5):\n pp.create_bus(net, name='Bus HV%s' % i, vn_kv=110, type='n')\n\n# show bustable\nnet.bus",
"_____no_output_____"
]
],
[
[
"### Lines\n\n<img src=\"pics/example_network_lines_hv.png\" width=\"40%\">",
"_____no_output_____"
],
[
"The information about the 6 HV lines are stored in a csv file that we load from the hard drive:",
"_____no_output_____"
]
],
[
[
"hv_lines = pd.read_csv('example_advanced/hv_lines.csv', sep=';', header=0, decimal=',')\nhv_lines",
"_____no_output_____"
]
],
[
[
"and use to create all lines:",
"_____no_output_____"
]
],
[
[
"# create lines\nfor _, hv_line in hv_lines.iterrows():\n from_bus = pp.get_element_index(net, \"bus\", hv_line.from_bus)\n to_bus = pp.get_element_index(net, \"bus\", hv_line.to_bus)\n pp.create_line(net, from_bus, to_bus, length_km=hv_line.length,std_type=hv_line.std_type, name=hv_line.line_name, parallel=hv_line.parallel)\n\n# show line table\nnet.line",
"_____no_output_____"
]
],
[
[
"### Transformer\n\n<img src=\"pics/example_network_trafos_hv.png\" width=\"40%\">",
"_____no_output_____"
],
[
"The 380/110 kV transformer connects the buses \"Bus DB 1\" and \"Bus DB 2\". We use the get_element_index function from the pandapower toolbox to find the bus indices of the buses with these names and create a transformer by directly specifying the parameters:",
"_____no_output_____"
]
],
[
[
"hv_bus = pp.get_element_index(net, \"bus\", \"Bus DB 2\")\nlv_bus = pp.get_element_index(net, \"bus\", \"Bus SB 1\")\npp.create_transformer_from_parameters(net, hv_bus, lv_bus, sn_mva=300, vn_hv_kv=380, vn_lv_kv=110, vkr_percent=0.06,\n vk_percent=8, pfe_kw=0, i0_percent=0, tp_pos=0, shift_degree=0, name='EHV-HV-Trafo')\n\nnet.trafo # show trafo table",
"_____no_output_____"
]
],
[
[
"### Switches\n\n<img src=\"pics/example_network_switches_hv.png\" width=\"60%\">",
"_____no_output_____"
],
[
"Now we create the switches to connect the buses in the transformer station. The switch configuration is stored in the following csv table:",
"_____no_output_____"
]
],
[
[
"hv_bus_sw = pd.read_csv('example_advanced/hv_bus_sw.csv', sep=';', header=0, decimal=',')\nhv_bus_sw",
"_____no_output_____"
],
[
"# Bus-bus switches\nfor _, switch in hv_bus_sw.iterrows():\n from_bus = pp.get_element_index(net, \"bus\", switch.from_bus)\n to_bus = pp.get_element_index(net, \"bus\", switch.to_bus)\n pp.create_switch(net, from_bus, to_bus, et=switch.et, closed=switch.closed, type=switch.type, name=switch.bus_name)\n\n# Bus-line switches\nhv_buses = net.bus[(net.bus.vn_kv == 380) | (net.bus.vn_kv == 110)].index\nhv_ls = net.line[(net.line.from_bus.isin(hv_buses)) & (net.line.to_bus.isin(hv_buses))]\nfor _, line in hv_ls.iterrows():\n pp.create_switch(net, line.from_bus, line.name, et='l', closed=True, type='LBS', name='Switch %s - %s' % (net.bus.name.at[line.from_bus], line['name']))\n pp.create_switch(net, line.to_bus, line.name, et='l', closed=True, type='LBS', name='Switch %s - %s' % (net.bus.name.at[line.to_bus], line['name']))\n\n# Trafo-line switches\npp.create_switch(net, pp.get_element_index(net, \"bus\", 'Bus DB 2'), pp.get_element_index(net, \"trafo\", 'EHV-HV-Trafo'), et='t', closed=True, type='LBS', name='Switch DB2 - EHV-HV-Trafo')\npp.create_switch(net, pp.get_element_index(net, \"bus\", 'Bus SB 1'), pp.get_element_index(net, \"trafo\", 'EHV-HV-Trafo'), et='t', closed=True, type='LBS', name='Switch SB1 - EHV-HV-Trafo')\n\n# show switch table\nnet.switch",
"_____no_output_____"
]
],
[
[
"### External Grid\n\n<img src=\"pics/example_network_ext_grids_hv.png\" width=\"40%\">",
"_____no_output_____"
],
[
"We equip the high voltage side of the transformer with an external grid connection:",
"_____no_output_____"
]
],
[
[
"pp.create_ext_grid(net, pp.get_element_index(net, \"bus\", 'Double Busbar 1'), vm_pu=1.03, va_degree=0, name='External grid',\n s_sc_max_mva=10000, rx_max=0.1, rx_min=0.1)\n\nnet.ext_grid # show external grid table",
"_____no_output_____"
]
],
[
[
"### Loads\n\n<img src=\"pics/example_network_loads_hv.png\" width=\"40%\">",
"_____no_output_____"
],
[
"The five loads in the HV network are defined in the following csv file:",
"_____no_output_____"
]
],
[
[
"hv_loads = pd.read_csv('example_advanced/hv_loads.csv', sep=';', header=0, decimal=',')\nhv_loads",
"_____no_output_____"
],
[
"for _, load in hv_loads.iterrows():\n bus_idx = pp.get_element_index(net, \"bus\", load.bus)\n pp.create_load(net, bus_idx, p_mw=load.p, q_mvar=load.q, name=load.load_name)\n\n# show load table\nnet.load",
"_____no_output_____"
]
],
[
[
"### Generator\n\n<img src=\"pics/example_network_gens_hv.png\" width=\"40%\">",
"_____no_output_____"
],
[
"The voltage controlled generator is created with an active power of 100 MW (negative for generation) and a voltage set point of 1.03 per unit:",
"_____no_output_____"
]
],
[
[
"pp.create_gen(net, pp.get_element_index(net, \"bus\", 'Bus HV4'), vm_pu=1.03, p_mw=100, name='Gas turbine')\n\n# show generator table\nnet.gen",
"_____no_output_____"
]
],
[
[
"### Static generators\n\n<img src=\"pics/example_network_sgens_hv.png\" width=\"40%\">",
"_____no_output_____"
],
[
"We create this wind park with an active power of 20 MW (negative for generation) and a reactive power of -4 Mvar. To classify the generation as a wind park, we set type to \"WP\":",
"_____no_output_____"
]
],
[
[
"pp.create_sgen(net, pp.get_element_index(net, \"bus\", 'Bus SB 5'), p_mw=20, q_mvar=4, sn_mva=45, \n type='WP', name='Wind Park')\n\n# show static generator table\nnet.sgen",
"_____no_output_____"
]
],
[
[
"### Shunt\n\n<img src=\"pics/example_network_shunts_hv.png\" width=\"40%\">",
"_____no_output_____"
]
],
[
[
"pp.create_shunt(net, pp.get_element_index(net, \"bus\", 'Bus HV1'), p_mw=0, q_mvar=0.960, name='Shunt')\n\n# show shunt table\nnet.shunt",
"_____no_output_____"
]
],
[
[
"### External network equivalents\n\n\n<img src=\"pics/example_network_ext_equi_hv.png\" width=\"40%\">",
"_____no_output_____"
],
[
"The two remaining elements are impedances and extended ward equivalents:",
"_____no_output_____"
]
],
[
[
"# Impedance\npp.create_impedance(net, pp.get_element_index(net, \"bus\", 'Bus HV3'), pp.get_element_index(net, \"bus\", 'Bus HV1'), \n rft_pu=0.074873, xft_pu=0.198872, sn_mva=100, name='Impedance')\n\n# show impedance table\nnet.impedance",
"_____no_output_____"
],
[
"# xwards\npp.create_xward(net, pp.get_element_index(net, \"bus\", 'Bus HV3'), ps_mw=23.942, qs_mvar=-12.24187, pz_mw=2.814571, \n qz_mvar=0, r_ohm=0, x_ohm=12.18951, vm_pu=1.02616, name='XWard 1')\npp.create_xward(net, pp.get_element_index(net, \"bus\", 'Bus HV1'), ps_mw=3.776, qs_mvar=-7.769979, pz_mw=9.174917, \n qz_mvar=0, r_ohm=0, x_ohm=50.56217, vm_pu=1.024001, name='XWard 2')\n\n# show xward table\nnet.xward",
"_____no_output_____"
]
],
[
[
"## Medium voltage level",
"_____no_output_____"
],
[
"### Buses\n\n<img src=\"pics/example_network_buses_mv.png\" width=\"50%\">",
"_____no_output_____"
]
],
[
[
"pp.create_bus(net, name='Bus MV0 20kV', vn_kv=20, type='n')\nfor i in range(8):\n pp.create_bus(net, name='Bus MV%s' % i, vn_kv=10, type='n')\n\n#show only medium voltage bus table\nmv_buses = net.bus[(net.bus.vn_kv == 10) | (net.bus.vn_kv == 20)]\nmv_buses",
"_____no_output_____"
]
],
[
[
"### Lines\n\n<img src=\"pics/example_network_lines_mv.png\" width=\"50%\">",
"_____no_output_____"
]
],
[
[
"mv_lines = pd.read_csv('example_advanced/mv_lines.csv', sep=';', header=0, decimal=',')\nfor _, mv_line in mv_lines.iterrows():\n from_bus = pp.get_element_index(net, \"bus\", mv_line.from_bus)\n to_bus = pp.get_element_index(net, \"bus\", mv_line.to_bus)\n pp.create_line(net, from_bus, to_bus, length_km=mv_line.length, std_type=mv_line.std_type, name=mv_line.line_name)\n\n# show only medium voltage lines\nnet.line[net.line.from_bus.isin(mv_buses.index)]",
"_____no_output_____"
]
],
[
[
"### 3 Winding Transformer\n\n<img src=\"pics/example_network_trafos_mv.png\" width=\"50%\">",
"_____no_output_____"
],
[
"The three winding transformer transforms its high voltage level to two different lower voltage levels, in this case from 110 kV to 20 kV and 10 kV.",
"_____no_output_____"
]
],
[
[
"hv_bus = pp.get_element_index(net, \"bus\", \"Bus HV2\")\nmv_bus = pp.get_element_index(net, \"bus\", \"Bus MV0 20kV\")\nlv_bus = pp.get_element_index(net, \"bus\", \"Bus MV0\")\npp.create_transformer3w_from_parameters(net, hv_bus, mv_bus, lv_bus, vn_hv_kv=110, vn_mv_kv=20, vn_lv_kv=10, \n sn_hv_mva=40, sn_mv_mva=15, sn_lv_mva=25, vk_hv_percent=10.1, \n vk_mv_percent=10.1, vk_lv_percent=10.1, vkr_hv_percent=0.266667, \n vkr_mv_percent=0.033333, vkr_lv_percent=0.04, pfe_kw=0, i0_percent=0, \n shift_mv_degree=30, shift_lv_degree=30, tap_side=\"hv\", tap_neutral=0, tap_min=-8, \n tap_max=8, tap_step_percent=1.25, tap_pos=0, name='HV-MV-MV-Trafo')\n\n# show transformer3w table\nnet.trafo3w",
"_____no_output_____"
]
],
[
[
"### Switches",
"_____no_output_____"
]
],
[
[
"# Bus-line switches\nmv_buses = net.bus[(net.bus.vn_kv == 10) | (net.bus.vn_kv == 20)].index\nmv_ls = net.line[(net.line.from_bus.isin(mv_buses)) & (net.line.to_bus.isin(mv_buses))]\nfor _, line in mv_ls.iterrows():\n pp.create_switch(net, line.from_bus, line.name, et='l', closed=True, type='LBS', name='Switch %s - %s' % (net.bus.name.at[line.from_bus], line['name']))\n pp.create_switch(net, line.to_bus, line.name, et='l', closed=True, type='LBS', name='Switch %s - %s' % (net.bus.name.at[line.to_bus], line['name']))\n\n# open switch\nopen_switch_id = net.switch[(net.switch.name == 'Switch Bus MV5 - MV Line5')].index\nnet.switch.closed.loc[open_switch_id] = False\n\n#show only medium voltage switch table\nnet.switch[net.switch.bus.isin(mv_buses)]",
"_____no_output_____"
]
],
[
[
"### Loads\n\n<img src=\"pics/example_network_loads_mv.png\" width=\"50%\">",
"_____no_output_____"
]
],
[
[
"mv_loads = pd.read_csv('example_advanced/mv_loads.csv', sep=';', header=0, decimal=',')\nfor _, load in mv_loads.iterrows():\n bus_idx = pp.get_element_index(net, \"bus\", load.bus)\n pp.create_load(net, bus_idx, p_mw=load.p, q_mvar=load.q, name=load.load_name)\n\n# show only medium voltage loads\nnet.load[net.load.bus.isin(mv_buses)]",
"_____no_output_____"
]
],
[
[
"### Static generators\n\n<img src=\"pics/example_network_sgens_mv.png\" width=\"50%\">",
"_____no_output_____"
]
],
[
[
"mv_sgens = pd.read_csv('example_advanced/mv_sgens.csv', sep=';', header=0, decimal=',')\nfor _, sgen in mv_sgens.iterrows():\n bus_idx = pp.get_element_index(net, \"bus\", sgen.bus)\n pp.create_sgen(net, bus_idx, p_mw=sgen.p, q_mvar=sgen.q, sn_mva=sgen.sn, type=sgen.type, name=sgen.sgen_name)\n\n# show only medium voltage static generators\nnet.sgen[net.sgen.bus.isin(mv_buses)]",
"_____no_output_____"
]
],
[
[
"## Low voltage level",
"_____no_output_____"
],
[
"### Busses\n\n<img src=\"pics/example_network_buses_lv.png\" width=\"50%\">",
"_____no_output_____"
]
],
[
[
"pp.create_bus(net, name='Bus LV0', vn_kv=0.4, type='n')\nfor i in range(1, 6):\n pp.create_bus(net, name='Bus LV1.%s' % i, vn_kv=0.4, type='m')\nfor i in range(1, 5):\n pp.create_bus(net, name='Bus LV2.%s' % i, vn_kv=0.4, type='m')\npp.create_bus(net, name='Bus LV2.2.1', vn_kv=0.4, type='m')\npp.create_bus(net, name='Bus LV2.2.2', vn_kv=0.4, type='m')\n\n# show only low voltage buses\nnet.bus[net.bus.vn_kv == 0.4]",
"_____no_output_____"
]
],
[
[
"### Lines\n\n<img src=\"pics/example_network_lines_lv.png\" width=\"50%\">",
"_____no_output_____"
]
],
[
[
"# create lines\nlv_lines = pd.read_csv('example_advanced/lv_lines.csv', sep=';', header=0, decimal=',')\nfor _, lv_line in lv_lines.iterrows():\n from_bus = pp.get_element_index(net, \"bus\", lv_line.from_bus)\n to_bus = pp.get_element_index(net, \"bus\", lv_line.to_bus)\n pp.create_line(net, from_bus, to_bus, length_km=lv_line.length, std_type=lv_line.std_type, name=lv_line.line_name)\n\n# show only low voltage lines\nnet.line[net.line.from_bus.isin(lv_buses.index)]",
"_____no_output_____"
]
],
[
[
"### Transformer\n\n<img src=\"pics/example_network_trafos_lv.png\" width=\"50%\">",
"_____no_output_____"
]
],
[
[
"hv_bus = pp.get_element_index(net, \"bus\", \"Bus MV4\")\nlv_bus = pp.get_element_index(net, \"bus\",\"Bus LV0\")\npp.create_transformer_from_parameters(net, hv_bus, lv_bus, sn_mva=.4, vn_hv_kv=10, vn_lv_kv=0.4, vkr_percent=1.325, vk_percent=4, pfe_kw=0.95, i0_percent=0.2375, tap_side=\"hv\", tap_neutral=0, tap_min=-2, tap_max=2, tap_step_percent=2.5, tp_pos=0, shift_degree=150, name='MV-LV-Trafo')\n\n#show only low voltage transformer\nnet.trafo[net.trafo.lv_bus.isin(lv_buses.index)]",
"_____no_output_____"
]
],
[
[
"### Switches",
"_____no_output_____"
]
],
[
[
"lv_buses\n# Bus-line switches\nlv_ls = net.line[(net.line.from_bus.isin(lv_buses.index)) & (net.line.to_bus.isin(lv_buses.index))]\nfor _, line in lv_ls.iterrows():\n pp.create_switch(net, line.from_bus, line.name, et='l', closed=True, type='LBS', name='Switch %s - %s' % (net.bus.name.at[line.from_bus], line['name']))\n pp.create_switch(net, line.to_bus, line.name, et='l', closed=True, type='LBS', name='Switch %s - %s' % (net.bus.name.at[line.to_bus], line['name']))\n\n# Trafo-line switches\npp.create_switch(net, pp.get_element_index(net, \"bus\", 'Bus MV4'), pp.get_element_index(net, \"trafo\", 'MV-LV-Trafo'), et='t', closed=True, type='LBS', name='Switch MV4 - MV-LV-Trafo')\npp.create_switch(net, pp.get_element_index(net, \"bus\", 'Bus LV0'), pp.get_element_index(net, \"trafo\", 'MV-LV-Trafo'), et='t', closed=True, type='LBS', name='Switch LV0 - MV-LV-Trafo')\n\n# show only low vvoltage switches\nnet.switch[net.switch.bus.isin(lv_buses.index)]\n",
"_____no_output_____"
]
],
[
[
"### Loads\n\n<img src=\"pics/example_network_loads_lv.png\" width=\"50%\">",
"_____no_output_____"
]
],
[
[
"lv_loads = pd.read_csv('example_advanced/lv_loads.csv', sep=';', header=0, decimal=',')\nfor _, load in lv_loads.iterrows():\n bus_idx = pp.get_element_index(net, \"bus\", load.bus)\n pp.create_load(net, bus_idx, p_mw=load.p, q_mvar=load.q, name=load.load_name)\n \n# show only low voltage loads\nnet.load[net.load.bus.isin(lv_buses.index)]",
"_____no_output_____"
]
],
[
[
"### Static generators\n\n<img src=\"pics/example_network_sgens_lv.png\" width=\"50%\">",
"_____no_output_____"
]
],
[
[
"lv_sgens = pd.read_csv('example_advanced/lv_sgens.csv', sep=';', header=0, decimal=',')\nfor _, sgen in lv_sgens.iterrows():\n bus_idx = pp.get_element_index(net, \"bus\", sgen.bus)\n pp.create_sgen(net, bus_idx, p_mw=sgen.p, q_mvar=sgen.q, sn_mva=sgen.sn, type=sgen.type, name=sgen.sgen_name)\n\n# show only low voltage static generators\nnet.sgen[net.sgen.bus.isin(lv_buses.index)]",
"_____no_output_____"
]
],
[
[
"## Run a Power Flow",
"_____no_output_____"
]
],
[
[
"pp.runpp(net, calculate_voltage_angles=True, init=\"dc\")\nnet",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf2cc31194b48871ed54fed5e1f433978889d51 | 2,675 | ipynb | Jupyter Notebook | RungeKuttaMethod.ipynb | sekilas13/WebGL-Orbiter | 14d80d654486d02ad35cbb1fc52e32287cff557c | [
"MIT-0",
"MIT"
] | 27 | 2016-03-22T12:19:58.000Z | 2022-03-20T07:47:03.000Z | RungeKuttaMethod.ipynb | sekilas13/WebGL-Orbiter | 14d80d654486d02ad35cbb1fc52e32287cff557c | [
"MIT-0",
"MIT"
] | 17 | 2018-07-22T05:09:14.000Z | 2022-02-27T06:38:30.000Z | RungeKuttaMethod.ipynb | sekilas13/WebGL-Orbiter | 14d80d654486d02ad35cbb1fc52e32287cff557c | [
"MIT-0",
"MIT"
] | 12 | 2016-10-16T17:32:14.000Z | 2022-03-16T03:24:45.000Z | 24.318182 | 113 | 0.419065 | [
[
[
"\n# Gravitational acceleration\n\n$x$ is a position vector here.\n\n$$\nf(x) = -\\frac{x}{|x|^3}\n$$\n\nNewton equation\n\n$$\n\\frac{d^2 x}{dt^2} = f(x) = -\\frac{x}{|x|^3}\n$$\n\nDefine an intermediate variable to make it first-order differetial equation $ v \\equiv \\frac{dx}{dt} $\n\n$$\n\\frac{dv}{dt} = -\\frac{x}{|x|^3} \\\\\n\\frac{dx}{dt} = v\n$$\n\nMake the differential deltas\n\n$$\n\\Delta v = \\frac{x}{|x|^3} \\Delta t \\\\\n\\Delta x = v \\Delta t\n$$\n\n# Euler method\n\nFormal definition:\n\n$$\nx_{n+1} = x_n + hf'(t_n, x_n)\n$$\n\nIn our case:\n\n$$\nv_{n+1} = v_n + h \\frac{x}{|x|^3} \\\\\nx_{t+1} = x_n + h v(t)\n$$\n\n\n# Modified Euler method (Midpoint method or Second-order Runge Kutta method)\n\nFormal definition:\n\n\\begin{align*}\nk_1 &= h f'(t_n, x_n) \\\\\nk_2 &= h f'(t_n + \\frac 12 h, x_n + \\frac 12 k_1) \\\\\nx_{n+1} &= x_n + k_2\n\\end{align*}\n\nIn our case:\n\n\\begin{align*}\n\\Delta v_1 &= h f(x) \\\\\n\\Delta x_1 &= h v(t) \\\\\n\\Delta v_2 &= h f(x + \\Delta x_1 / 2) \\\\\n\\Delta x_2 &= h (v + \\Delta v_1 / 2) \\\\\nv(t+h) &= v(t) + \\Delta v_2 \\\\\nx(t+h) &= x(t) + \\Delta x_2\n\\end{align*}\n\n## Fourth-order Runge-Kutta method\n\nFormal definition:\n\n\\begin{align*}\nk_1 &= h f'(t_n, x_n) \\\\\nk_2 &= h f'(t_n + \\frac 12 h, x_n + \\frac 12 k_1) \\\\\nk_3 &= h f'(t_n + \\frac 12 h, x_n + \\frac 12 k_2) \\\\\nk_4 &= h f'(t_n + h, x_n + k_3) \\\\\nx_{n+1} &= x_n + \\frac{k_1}{6} + \\frac{k_2}{3} + \\frac{k_3}{3} + \\frac{k_4}{6}\n\\end{align*}\n\nIn our case: ?\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
ecf2e30a7b0cee413583b11d7365d73b6025b2ff | 28,399 | ipynb | Jupyter Notebook | neural-networks-deep-learning/Week 4/Deep Neural Network Application.ipynb | Samra10/Deep-Learning-Coursera | ca3689e14d58f516690ac3e2b05dc62334b61150 | [
"Unlicense"
] | null | null | null | neural-networks-deep-learning/Week 4/Deep Neural Network Application.ipynb | Samra10/Deep-Learning-Coursera | ca3689e14d58f516690ac3e2b05dc62334b61150 | [
"Unlicense"
] | null | null | null | neural-networks-deep-learning/Week 4/Deep Neural Network Application.ipynb | Samra10/Deep-Learning-Coursera | ca3689e14d58f516690ac3e2b05dc62334b61150 | [
"Unlicense"
] | null | null | null | 34.717604 | 341 | 0.546639 | [
[
[
"# Deep Neural Network for Image Classification: Application\n\nWhen you finish this, you will have finished the last programming assignment of Week 4, and also the last programming assignment of this course! \n\nYou will use use the functions you'd implemented in the previous assignment to build a deep network, and apply it to cat vs non-cat classification. Hopefully, you will see an improvement in accuracy relative to your previous logistic regression implementation. \n\n**After this assignment you will be able to:**\n- Build and apply a deep neural network to supervised learning. \n\nLet's get started!",
"_____no_output_____"
],
[
"## 1 - Packages",
"_____no_output_____"
],
[
"Let's first import all the packages that you will need during this assignment. \n- [numpy](www.numpy.org) is the fundamental package for scientific computing with Python.\n- [matplotlib](http://matplotlib.org) is a library to plot graphs in Python.\n- [h5py](http://www.h5py.org) is a common package to interact with a dataset that is stored on an H5 file.\n- [PIL](http://www.pythonware.com/products/pil/) and [scipy](https://www.scipy.org/) are used here to test your model with your own picture at the end.\n- dnn_app_utils provides the functions implemented in the \"Building your Deep Neural Network: Step by Step\" assignment to this notebook.\n- np.random.seed(1) is used to keep all the random function calls consistent. It will help us grade your work.",
"_____no_output_____"
]
],
[
[
"import time\nimport numpy as np\nimport h5py\nimport matplotlib.pyplot as plt\nimport scipy\nfrom PIL import Image\nfrom scipy import ndimage\nfrom dnn_app_utils_v3 import *\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (5.0, 4.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n%load_ext autoreload\n%autoreload 2\n\nnp.random.seed(1)",
"_____no_output_____"
]
],
[
[
"## 2 - Dataset\n\nYou will use the same \"Cat vs non-Cat\" dataset as in \"Logistic Regression as a Neural Network\" (Assignment 2). The model you had built had 70% test accuracy on classifying cats vs non-cats images. Hopefully, your new model will perform a better!\n\n**Problem Statement**: You are given a dataset (\"data.h5\") containing:\n - a training set of m_train images labelled as cat (1) or non-cat (0)\n - a test set of m_test images labelled as cat and non-cat\n - each image is of shape (num_px, num_px, 3) where 3 is for the 3 channels (RGB).\n\nLet's get more familiar with the dataset. Load the data by running the cell below.",
"_____no_output_____"
]
],
[
[
"train_x_orig, train_y, test_x_orig, test_y, classes = load_data()",
"_____no_output_____"
]
],
[
[
"The following code will show you an image in the dataset. Feel free to change the index and re-run the cell multiple times to see other images. ",
"_____no_output_____"
]
],
[
[
"# Example of a picture\nindex = 10\nplt.imshow(train_x_orig[index])\nprint (\"y = \" + str(train_y[0,index]) + \". It's a \" + classes[train_y[0,index]].decode(\"utf-8\") + \" picture.\")",
"_____no_output_____"
],
[
"# Explore your dataset \nm_train = train_x_orig.shape[0]\nnum_px = train_x_orig.shape[1]\nm_test = test_x_orig.shape[0]\n\nprint (\"Number of training examples: \" + str(m_train))\nprint (\"Number of testing examples: \" + str(m_test))\nprint (\"Each image is of size: (\" + str(num_px) + \", \" + str(num_px) + \", 3)\")\nprint (\"train_x_orig shape: \" + str(train_x_orig.shape))\nprint (\"train_y shape: \" + str(train_y.shape))\nprint (\"test_x_orig shape: \" + str(test_x_orig.shape))\nprint (\"test_y shape: \" + str(test_y.shape))",
"_____no_output_____"
]
],
[
[
"As usual, you reshape and standardize the images before feeding them to the network. The code is given in the cell below.\n\n<img src=\"images/imvectorkiank.png\" style=\"width:450px;height:300px;\">\n\n<caption><center> <u>Figure 1</u>: Image to vector conversion. <br> </center></caption>",
"_____no_output_____"
]
],
[
[
"# Reshape the training and test examples \ntrain_x_flatten = train_x_orig.reshape(train_x_orig.shape[0], -1).T # The \"-1\" makes reshape flatten the remaining dimensions\ntest_x_flatten = test_x_orig.reshape(test_x_orig.shape[0], -1).T\n\n# Standardize data to have feature values between 0 and 1.\ntrain_x = train_x_flatten/255.\ntest_x = test_x_flatten/255.\n\nprint (\"train_x's shape: \" + str(train_x.shape))\nprint (\"test_x's shape: \" + str(test_x.shape))\n",
"_____no_output_____"
]
],
[
[
"$12,288$ equals $64 \\times 64 \\times 3$ which is the size of one reshaped image vector.",
"_____no_output_____"
],
[
"## 3 - Architecture of your model",
"_____no_output_____"
],
[
"Now that you are familiar with the dataset, it is time to build a deep neural network to distinguish cat images from non-cat images.\n\nYou will build two different models:\n- A 2-layer neural network\n- An L-layer deep neural network\n\nYou will then compare the performance of these models, and also try out different values for $L$. \n\nLet's look at the two architectures.\n\n### 3.1 - 2-layer neural network\n\n<img src=\"images/2layerNN_kiank.png\" style=\"width:650px;height:400px;\">\n<caption><center> <u>Figure 2</u>: 2-layer neural network. <br> The model can be summarized as: ***INPUT -> LINEAR -> RELU -> LINEAR -> SIGMOID -> OUTPUT***. </center></caption>\n\n<u>Detailed Architecture of figure 2</u>:\n- The input is a (64,64,3) image which is flattened to a vector of size $(12288,1)$. \n- The corresponding vector: $[x_0,x_1,...,x_{12287}]^T$ is then multiplied by the weight matrix $W^{[1]}$ of size $(n^{[1]}, 12288)$.\n- You then add a bias term and take its relu to get the following vector: $[a_0^{[1]}, a_1^{[1]},..., a_{n^{[1]}-1}^{[1]}]^T$.\n- You then repeat the same process.\n- You multiply the resulting vector by $W^{[2]}$ and add your intercept (bias). \n- Finally, you take the sigmoid of the result. If it is greater than 0.5, you classify it to be a cat.\n\n### 3.2 - L-layer deep neural network\n\nIt is hard to represent an L-layer deep neural network with the above representation. However, here is a simplified network representation:\n\n<img src=\"images/LlayerNN_kiank.png\" style=\"width:650px;height:400px;\">\n<caption><center> <u>Figure 3</u>: L-layer neural network. <br> The model can be summarized as: ***[LINEAR -> RELU] $\\times$ (L-1) -> LINEAR -> SIGMOID***</center></caption>\n\n<u>Detailed Architecture of figure 3</u>:\n- The input is a (64,64,3) image which is flattened to a vector of size (12288,1).\n- The corresponding vector: $[x_0,x_1,...,x_{12287}]^T$ is then multiplied by the weight matrix $W^{[1]}$ and then you add the intercept $b^{[1]}$. The result is called the linear unit.\n- Next, you take the relu of the linear unit. This process could be repeated several times for each $(W^{[l]}, b^{[l]})$ depending on the model architecture.\n- Finally, you take the sigmoid of the final linear unit. If it is greater than 0.5, you classify it to be a cat.\n\n### 3.3 - General methodology\n\nAs usual you will follow the Deep Learning methodology to build the model:\n 1. Initialize parameters / Define hyperparameters\n 2. Loop for num_iterations:\n a. Forward propagation\n b. Compute cost function\n c. Backward propagation\n d. Update parameters (using parameters, and grads from backprop) \n 4. Use trained parameters to predict labels\n\nLet's now implement those two models!",
"_____no_output_____"
],
[
"## 4 - Two-layer neural network\n\n**Question**: Use the helper functions you have implemented in the previous assignment to build a 2-layer neural network with the following structure: *LINEAR -> RELU -> LINEAR -> SIGMOID*. The functions you may need and their inputs are:\n```python\ndef initialize_parameters(n_x, n_h, n_y):\n ...\n return parameters \ndef linear_activation_forward(A_prev, W, b, activation):\n ...\n return A, cache\ndef compute_cost(AL, Y):\n ...\n return cost\ndef linear_activation_backward(dA, cache, activation):\n ...\n return dA_prev, dW, db\ndef update_parameters(parameters, grads, learning_rate):\n ...\n return parameters\n```",
"_____no_output_____"
]
],
[
[
"### CONSTANTS DEFINING THE MODEL ####\nn_x = 12288 # num_px * num_px * 3\nn_h = 7\nn_y = 1\nlayers_dims = (n_x, n_h, n_y)",
"_____no_output_____"
],
[
"# GRADED FUNCTION: two_layer_model\n\ndef two_layer_model(X, Y, layers_dims, learning_rate = 0.0075, num_iterations = 3000, print_cost=False):\n \"\"\"\n Implements a two-layer neural network: LINEAR->RELU->LINEAR->SIGMOID.\n \n Arguments:\n X -- input data, of shape (n_x, number of examples)\n Y -- true \"label\" vector (containing 0 if cat, 1 if non-cat), of shape (1, number of examples)\n layers_dims -- dimensions of the layers (n_x, n_h, n_y)\n num_iterations -- number of iterations of the optimization loop\n learning_rate -- learning rate of the gradient descent update rule\n print_cost -- If set to True, this will print the cost every 100 iterations \n \n Returns:\n parameters -- a dictionary containing W1, W2, b1, and b2\n \"\"\"\n \n np.random.seed(1)\n grads = {}\n costs = [] # to keep track of the cost\n m = X.shape[1] # number of examples\n (n_x, n_h, n_y) = layers_dims\n \n # Initialize parameters dictionary, by calling one of the functions you'd previously implemented\n ### START CODE HERE ### (≈ 1 line of code)\n parameters = None\n ### END CODE HERE ###\n \n # Get W1, b1, W2 and b2 from the dictionary parameters.\n W1 = parameters[\"W1\"]\n b1 = parameters[\"b1\"]\n W2 = parameters[\"W2\"]\n b2 = parameters[\"b2\"]\n \n # Loop (gradient descent)\n\n for i in range(0, num_iterations):\n\n # Forward propagation: LINEAR -> RELU -> LINEAR -> SIGMOID. Inputs: \"X, W1, b1, W2, b2\". Output: \"A1, cache1, A2, cache2\".\n ### START CODE HERE ### (≈ 2 lines of code)\n A1, cache1 = None\n A2, cache2 = None\n ### END CODE HERE ###\n \n # Compute cost\n ### START CODE HERE ### (≈ 1 line of code)\n cost = None\n ### END CODE HERE ###\n \n # Initializing backward propagation\n dA2 = - (np.divide(Y, A2) - np.divide(1 - Y, 1 - A2))\n \n # Backward propagation. Inputs: \"dA2, cache2, cache1\". Outputs: \"dA1, dW2, db2; also dA0 (not used), dW1, db1\".\n ### START CODE HERE ### (≈ 2 lines of code)\n dA1, dW2, db2 = None\n dA0, dW1, db1 = None\n ### END CODE HERE ###\n \n # Set grads['dWl'] to dW1, grads['db1'] to db1, grads['dW2'] to dW2, grads['db2'] to db2\n grads['dW1'] = dW1\n grads['db1'] = db1\n grads['dW2'] = dW2\n grads['db2'] = db2\n \n # Update parameters.\n ### START CODE HERE ### (approx. 1 line of code)\n parameters = None\n ### END CODE HERE ###\n\n # Retrieve W1, b1, W2, b2 from parameters\n W1 = parameters[\"W1\"]\n b1 = parameters[\"b1\"]\n W2 = parameters[\"W2\"]\n b2 = parameters[\"b2\"]\n \n # Print the cost every 100 training example\n if print_cost and i % 100 == 0:\n print(\"Cost after iteration {}: {}\".format(i, np.squeeze(cost)))\n if print_cost and i % 100 == 0:\n costs.append(cost)\n \n # plot the cost\n\n plt.plot(np.squeeze(costs))\n plt.ylabel('cost')\n plt.xlabel('iterations (per tens)')\n plt.title(\"Learning rate =\" + str(learning_rate))\n plt.show()\n \n return parameters",
"_____no_output_____"
]
],
[
[
"Run the cell below to train your parameters. See if your model runs. The cost should be decreasing. It may take up to 5 minutes to run 2500 iterations. Check if the \"Cost after iteration 0\" matches the expected output below, if not click on the square (⬛) on the upper bar of the notebook to stop the cell and try to find your error.",
"_____no_output_____"
]
],
[
[
"parameters = two_layer_model(train_x, train_y, layers_dims = (n_x, n_h, n_y), num_iterations = 2500, print_cost=True)",
"_____no_output_____"
]
],
[
[
"**Expected Output**:\n<table> \n <tr>\n <td> **Cost after iteration 0**</td>\n <td> 0.6930497356599888 </td>\n </tr>\n <tr>\n <td> **Cost after iteration 100**</td>\n <td> 0.6464320953428849 </td>\n </tr>\n <tr>\n <td> **...**</td>\n <td> ... </td>\n </tr>\n <tr>\n <td> **Cost after iteration 2400**</td>\n <td> 0.048554785628770206 </td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"Good thing you built a vectorized implementation! Otherwise it might have taken 10 times longer to train this.\n\nNow, you can use the trained parameters to classify images from the dataset. To see your predictions on the training and test sets, run the cell below.",
"_____no_output_____"
]
],
[
[
"predictions_train = predict(train_x, train_y, parameters)",
"_____no_output_____"
]
],
[
[
"**Expected Output**:\n<table> \n <tr>\n <td> **Accuracy**</td>\n <td> 1.0 </td>\n </tr>\n</table>",
"_____no_output_____"
]
],
[
[
"predictions_test = predict(test_x, test_y, parameters)",
"_____no_output_____"
]
],
[
[
"**Expected Output**:\n\n<table> \n <tr>\n <td> **Accuracy**</td>\n <td> 0.72 </td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"**Note**: You may notice that running the model on fewer iterations (say 1500) gives better accuracy on the test set. This is called \"early stopping\" and we will talk about it in the next course. Early stopping is a way to prevent overfitting. \n\nCongratulations! It seems that your 2-layer neural network has better performance (72%) than the logistic regression implementation (70%, assignment week 2). Let's see if you can do even better with an $L$-layer model.",
"_____no_output_____"
],
[
"## 5 - L-layer Neural Network\n\n**Question**: Use the helper functions you have implemented previously to build an $L$-layer neural network with the following structure: *[LINEAR -> RELU]$\\times$(L-1) -> LINEAR -> SIGMOID*. The functions you may need and their inputs are:\n```python\ndef initialize_parameters_deep(layers_dims):\n ...\n return parameters \ndef L_model_forward(X, parameters):\n ...\n return AL, caches\ndef compute_cost(AL, Y):\n ...\n return cost\ndef L_model_backward(AL, Y, caches):\n ...\n return grads\ndef update_parameters(parameters, grads, learning_rate):\n ...\n return parameters\n```",
"_____no_output_____"
]
],
[
[
"### CONSTANTS ###\nlayers_dims = [12288, 20, 7, 5, 1] # 4-layer model",
"_____no_output_____"
],
[
"# GRADED FUNCTION: L_layer_model\n\ndef L_layer_model(X, Y, layers_dims, learning_rate = 0.0075, num_iterations = 3000, print_cost=False):#lr was 0.009\n \"\"\"\n Implements a L-layer neural network: [LINEAR->RELU]*(L-1)->LINEAR->SIGMOID.\n \n Arguments:\n X -- data, numpy array of shape (number of examples, num_px * num_px * 3)\n Y -- true \"label\" vector (containing 0 if cat, 1 if non-cat), of shape (1, number of examples)\n layers_dims -- list containing the input size and each layer size, of length (number of layers + 1).\n learning_rate -- learning rate of the gradient descent update rule\n num_iterations -- number of iterations of the optimization loop\n print_cost -- if True, it prints the cost every 100 steps\n \n Returns:\n parameters -- parameters learnt by the model. They can then be used to predict.\n \"\"\"\n\n np.random.seed(1)\n costs = [] # keep track of cost\n \n # Parameters initialization. (≈ 1 line of code)\n ### START CODE HERE ###\n parameters = None\n ### END CODE HERE ###\n \n # Loop (gradient descent)\n for i in range(0, num_iterations):\n\n # Forward propagation: [LINEAR -> RELU]*(L-1) -> LINEAR -> SIGMOID.\n ### START CODE HERE ### (≈ 1 line of code)\n AL, caches = None\n ### END CODE HERE ###\n \n # Compute cost.\n ### START CODE HERE ### (≈ 1 line of code)\n cost = None\n ### END CODE HERE ###\n \n # Backward propagation.\n ### START CODE HERE ### (≈ 1 line of code)\n grads = None\n ### END CODE HERE ###\n \n # Update parameters.\n ### START CODE HERE ### (≈ 1 line of code)\n parameters = None\n ### END CODE HERE ###\n \n # Print the cost every 100 training example\n if print_cost and i % 100 == 0:\n print (\"Cost after iteration %i: %f\" %(i, cost))\n if print_cost and i % 100 == 0:\n costs.append(cost)\n \n # plot the cost\n plt.plot(np.squeeze(costs))\n plt.ylabel('cost')\n plt.xlabel('iterations (per tens)')\n plt.title(\"Learning rate =\" + str(learning_rate))\n plt.show()\n \n return parameters",
"_____no_output_____"
]
],
[
[
"You will now train the model as a 4-layer neural network. \n\nRun the cell below to train your model. The cost should decrease on every iteration. It may take up to 5 minutes to run 2500 iterations. Check if the \"Cost after iteration 0\" matches the expected output below, if not click on the square (⬛) on the upper bar of the notebook to stop the cell and try to find your error.",
"_____no_output_____"
]
],
[
[
"parameters = L_layer_model(train_x, train_y, layers_dims, num_iterations = 2500, print_cost = True)",
"_____no_output_____"
]
],
[
[
"**Expected Output**:\n<table> \n <tr>\n <td> **Cost after iteration 0**</td>\n <td> 0.771749 </td>\n </tr>\n <tr>\n <td> **Cost after iteration 100**</td>\n <td> 0.672053 </td>\n </tr>\n <tr>\n <td> **...**</td>\n <td> ... </td>\n </tr>\n <tr>\n <td> **Cost after iteration 2400**</td>\n <td> 0.092878 </td>\n </tr>\n</table>",
"_____no_output_____"
]
],
[
[
"pred_train = predict(train_x, train_y, parameters)",
"_____no_output_____"
]
],
[
[
"<table>\n <tr>\n <td>\n **Train Accuracy**\n </td>\n <td>\n 0.985645933014\n </td>\n </tr>\n</table>",
"_____no_output_____"
]
],
[
[
"pred_test = predict(test_x, test_y, parameters)",
"_____no_output_____"
]
],
[
[
"**Expected Output**:\n\n<table> \n <tr>\n <td> **Test Accuracy**</td>\n <td> 0.8 </td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"Congrats! It seems that your 4-layer neural network has better performance (80%) than your 2-layer neural network (72%) on the same test set. \n\nThis is good performance for this task. Nice job! \n\nThough in the next course on \"Improving deep neural networks\" you will learn how to obtain even higher accuracy by systematically searching for better hyperparameters (learning_rate, layers_dims, num_iterations, and others you'll also learn in the next course). ",
"_____no_output_____"
],
[
"## 6) Results Analysis\n\nFirst, let's take a look at some images the L-layer model labeled incorrectly. This will show a few mislabeled images. ",
"_____no_output_____"
]
],
[
[
"print_mislabeled_images(classes, test_x, test_y, pred_test)",
"_____no_output_____"
]
],
[
[
"**A few types of images the model tends to do poorly on include:** \n- Cat body in an unusual position\n- Cat appears against a background of a similar color\n- Unusual cat color and species\n- Camera Angle\n- Brightness of the picture\n- Scale variation (cat is very large or small in image) ",
"_____no_output_____"
],
[
"## 7) Test with your own image (optional/ungraded exercise) ##\n\nCongratulations on finishing this assignment. You can use your own image and see the output of your model. To do that:\n 1. Click on \"File\" in the upper bar of this notebook, then click \"Open\" to go on your Coursera Hub.\n 2. Add your image to this Jupyter Notebook's directory, in the \"images\" folder\n 3. Change your image's name in the following code\n 4. Run the code and check if the algorithm is right (1 = cat, 0 = non-cat)!",
"_____no_output_____"
]
],
[
[
"## START CODE HERE ##\nmy_image = \"my_image.jpg\" # change this to the name of your image file \nmy_label_y = [1] # the true class of your image (1 -> cat, 0 -> non-cat)\n## END CODE HERE ##\n\nfname = \"images/\" + my_image\nimage = np.array(ndimage.imread(fname, flatten=False))\nmy_image = scipy.misc.imresize(image, size=(num_px,num_px)).reshape((num_px*num_px*3,1))\nmy_image = my_image/255.\nmy_predicted_image = predict(my_image, my_label_y, parameters)\n\nplt.imshow(image)\nprint (\"y = \" + str(np.squeeze(my_predicted_image)) + \", your L-layer model predicts a \\\"\" + classes[int(np.squeeze(my_predicted_image)),].decode(\"utf-8\") + \"\\\" picture.\")",
"_____no_output_____"
]
],
[
[
"**References**:\n\n- for auto-reloading external module: http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecf30d657623a18fc6bb0c37bb7cb8faa9ab7cfb | 33,149 | ipynb | Jupyter Notebook | Supriya Kumari 2.ipynb | s77upriy/Sweetmemo | eb37dd00bd1c8e86eb6ce421679b3e3c89507906 | [
"BSD-2-Clause"
] | null | null | null | Supriya Kumari 2.ipynb | s77upriy/Sweetmemo | eb37dd00bd1c8e86eb6ce421679b3e3c89507906 | [
"BSD-2-Clause"
] | null | null | null | Supriya Kumari 2.ipynb | s77upriy/Sweetmemo | eb37dd00bd1c8e86eb6ce421679b3e3c89507906 | [
"BSD-2-Clause"
] | null | null | null | 23.148743 | 921 | 0.526471 | [
[
[
"<center>\n <img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/IDSNlogo.png\" width=\"300\" alt=\"cognitiveclass.ai logo\" />\n</center>\n\n# String Operations\n\nEstimated time needed: **15** minutes\n\n## Objectives\n\nAfter completing this lab you will be able to:\n\n* Work with Strings\n* Perform operations on String\n* Manipulate Strings using indexing and escape sequences\n",
"_____no_output_____"
],
[
"<h2>Table of Contents</h2>\n<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <ul>\n <li>\n <a href=\"https://#strings\">What are Strings?</a>\n </li>\n <li>\n <a href=\"https://#index\">Indexing</a>\n <ul>\n <li><a href=\"https://neg/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\">Negative Indexing</a></li>\n <li><a href=\"https://slice/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\">Slicing</a></li>\n <li><a href=\"https://stride/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\">Stride</a></li>\n <li><a href=\"https://concat/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\">Concatenate Strings</a></li>\n </ul>\n </li>\n <li>\n <a href=\"https://#escape\">Escape Sequences</a>\n </li>\n <li>\n <a href=\"https://#operations\">String Operations</a>\n </li>\n <li>\n <a href=\"https://#quiz\">Quiz on Strings</a>\n </li>\n </ul>\n\n</div>\n\n<hr>\n",
"_____no_output_____"
],
[
"<h2 id=\"strings\">What are Strings?</h2>\n",
"_____no_output_____"
],
[
"The following example shows a string contained within 2 quotation marks:\n",
"_____no_output_____"
]
],
[
[
"# Use quotation marks for defining string\n\n\"Michael Jackson\"",
"_____no_output_____"
]
],
[
[
"We can also use single quotation marks:\n",
"_____no_output_____"
]
],
[
[
"# Use single quotation marks for defining string\n\n'Michael Jackson'",
"_____no_output_____"
]
],
[
[
"A string can be a combination of spaces and digits:\n",
"_____no_output_____"
]
],
[
[
"# Digitals and spaces in string\n\n'1 2 3 4 5 6 '",
"_____no_output_____"
]
],
[
[
"A string can also be a combination of special characters :\n",
"_____no_output_____"
]
],
[
[
"# Special characters in string\n\n'@#2_#]&*^%$'",
"_____no_output_____"
]
],
[
[
"We can print our string using the print statement:\n",
"_____no_output_____"
]
],
[
[
"# Print the string\n\nprint(\"hello!\")",
"_____no_output_____"
]
],
[
[
"We can bind or assign a string to another variable:\n",
"_____no_output_____"
]
],
[
[
"# Assign string to variable\n\nname = \"Michael Jackson\"\nname",
"_____no_output_____"
]
],
[
[
"<hr>\n",
"_____no_output_____"
],
[
"<h2 id=\"index\">Indexing</h2>\n",
"_____no_output_____"
],
[
"It is helpful to think of a string as an ordered sequence. Each element in the sequence can be accessed using an index represented by the array of numbers:\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%201/images/StringsIndex.png\" width=\"600\" align=\"center\" />\n",
"_____no_output_____"
],
[
"The first index can be accessed as follows:\n",
"_____no_output_____"
],
[
"<hr/>\n<div class=\"alert alert-success alertsuccess\" style=\"margin-top: 20px\">\n[Tip]: Because indexing starts at 0, it means the first index is on the index 0.\n</div>\n<hr/>\n",
"_____no_output_____"
]
],
[
[
"# Print the first element in the string\n\nprint(name[0])",
"_____no_output_____"
]
],
[
[
"We can access index 6:\n",
"_____no_output_____"
]
],
[
[
"# Print the element on index 6 in the string\n\nprint(name[6])",
"_____no_output_____"
]
],
[
[
"Moreover, we can access the 13th index:\n",
"_____no_output_____"
]
],
[
[
"# Print the element on the 13th index in the string\n\nprint(name[13])",
"_____no_output_____"
]
],
[
[
"<h3 id=\"neg\">Negative Indexing</h3>\n",
"_____no_output_____"
],
[
"We can also use negative indexing with strings:\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%201/images/StringsNeg.png\" width=\"600\" align=\"center\" />\n",
"_____no_output_____"
],
[
"Negative index can help us to count the element from the end of the string.\n",
"_____no_output_____"
],
[
"The last element is given by the index -1:\n",
"_____no_output_____"
]
],
[
[
"# Print the last element in the string\nName=\"Mickael Jackson\"\nprint(Name[-1])",
"n\n"
]
],
[
[
"The first element can be obtained by index -15:\n",
"_____no_output_____"
]
],
[
[
"# Print the first element in the string\nName=\"Mickael Jackson\"\nprint(Name[-15])",
"M\n"
]
],
[
[
"We can find the number of characters in a string by using <code>len</code>, short for length:\n",
"_____no_output_____"
]
],
[
[
"# Find the length of string\n\nlen(\"Michael Jackson\")",
"_____no_output_____"
]
],
[
[
"<h3 id=\"slice\">Slicing</h3>\n",
"_____no_output_____"
],
[
"We can obtain multiple characters from a string using slicing, we can obtain the 0 to 4th and 8th to the 12th element:\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%201/images/StringsSlice.png\" width=\"600\" align=\"center\" />\n",
"_____no_output_____"
],
[
"<hr/>\n<div class=\"alert alert-success alertsuccess\" style=\"margin-top: 20px\">\n[Tip]: When taking the slice, the first number means the index (start at 0), and the second number means the length from the index to the last element you want (start at 1)\n</div>\n<hr/>\n",
"_____no_output_____"
]
],
[
[
"# Take the slice on variable name with only index 0 to index 3\nname=\"Mickael Jackson\"\nname[0:4]",
"_____no_output_____"
],
[
"# Take the slice on variable name with only index 8 to index 11\nname=\"Mickael Jackson\"\nname[8:12]",
"_____no_output_____"
]
],
[
[
"<h3 id=\"stride\">Stride</h3>\n",
"_____no_output_____"
],
[
"We can also input a stride value as follows, with the '2' indicating that we are selecting every second variable:\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%201/images/StringsStride.png\" width=\"600\" align=\"center\" />\n",
"_____no_output_____"
]
],
[
[
"# Get every second element. The elments on index 1, 3, 5 ...\nname=\"Mickael Jackson\"\nname[::2]",
"_____no_output_____"
]
],
[
[
"We can also incorporate slicing with the stride. In this case, we select the first five elements and then use the stride:\n",
"_____no_output_____"
]
],
[
[
"# Get every second element in the range from index 0 to index 4\nname=\"Mickael Jackson\"\nname[0:5:2]",
"_____no_output_____"
]
],
[
[
"<h3 id=\"concat\">Concatenate Strings</h3>\n",
"_____no_output_____"
],
[
"We can concatenate or combine strings by using the addition symbols, and the result is a new string that is a combination of both:\n",
"_____no_output_____"
]
],
[
[
"# Concatenate two strings\nname=\"Mickael Jackson\"\nstatement = name + \" is the best\"\nstatement",
"_____no_output_____"
]
],
[
[
"To replicate values of a string we simply multiply the string by the number of times we would like to replicate it. In this case, the number is three. The result is a new string, and this new string consists of three copies of the original string:\n",
"_____no_output_____"
]
],
[
[
"# Print the string for 3 times\n\n3 * \"Michael Jackson \"",
"_____no_output_____"
]
],
[
[
"You can create a new string by setting it to the original variable. Concatenated with a new string, the result is a new string that changes from Michael Jackson to “Michael Jackson is the best\".\n",
"_____no_output_____"
]
],
[
[
"# Concatenate strings\n\nname = \"Michael Jackson\"\nname = name + \" is the best\"\nname",
"_____no_output_____"
]
],
[
[
"<hr>\n",
"_____no_output_____"
],
[
"<h2 id=\"escape\">Escape Sequences</h2>\n",
"_____no_output_____"
],
[
"Back slashes represent the beginning of escape sequences. Escape sequences represent strings that may be difficult to input. For example, back slash \"n\" represents a new line. The output is given by a new line after the back slash \"n\" is encountered:\n",
"_____no_output_____"
]
],
[
[
"# New line escape sequence\n\nprint(\" Michael Jackson \\n is the best\" )",
" Michael Jackson \n is the best\n"
]
],
[
[
"Similarly, back slash \"t\" represents a tab:\n",
"_____no_output_____"
]
],
[
[
"# Tab escape sequence\n\nprint(\" Michael Jackson \\t is the best\" )",
" Michael Jackson \t is the best\n"
]
],
[
[
"If you want to place a back slash in your string, use a double back slash:\n",
"_____no_output_____"
]
],
[
[
"# Include back slash in string\n\nprint(\" Michael Jackson \\\\ is the best\" )",
" Michael Jackson \\ is the best\n"
]
],
[
[
"We can also place an \"r\" before the string to display the backslash:\n",
"_____no_output_____"
]
],
[
[
"# r will tell python that string will be display as raw string\n\nprint(r\" Michael Jackson \\ is the best\" )",
" Michael Jackson \\ is the best\n"
]
],
[
[
"<hr>\n",
"_____no_output_____"
],
[
"<h2 id=\"operations\">String Operations</h2>\n",
"_____no_output_____"
],
[
"There are many string operation methods in Python that can be used to manipulate the data. We are going to use some basic string operations on the data.\n",
"_____no_output_____"
],
[
"Let's try with the method <code>upper</code>; this method converts lower case characters to upper case characters:\n",
"_____no_output_____"
]
],
[
[
"# Convert all the characters in string to upper case\n\na = \"Thriller is the sixth studio album\"\nprint(\"before upper:\", a)\nb = a.upper()\nprint(\"After upper:\", b)",
"before upper: Thriller is the sixth studio album\nAfter upper: THRILLER IS THE SIXTH STUDIO ALBUM\n"
]
],
[
[
"The method <code>replace</code> replaces a segment of the string, i.e. a substring with a new string. We input the part of the string we would like to change. The second argument is what we would like to exchange the segment with, and the result is a new string with the segment changed:\n",
"_____no_output_____"
]
],
[
[
"# Replace the old substring with the new target substring is the segment has been found in the string\n\na = \"Michael Jackson is the best\"\nb = a.replace('Michael', 'Janet')\nb",
"_____no_output_____"
]
],
[
[
"The method <code>find</code> finds a sub-string. The argument is the substring you would like to find, and the output is the first index of the sequence. We can find the sub-string <code>jack</code> or <code>el<code>.\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%201/images/StringsFind.png\" width=\"600\" align=\"center\" />\n",
"_____no_output_____"
]
],
[
[
"# Find the substring in the string. Only the index of the first elment of substring in string will be the output\n\nname = \"Michael Jackson\"\nname.find('el')",
"_____no_output_____"
],
[
"# Find the substring in the string.\n\nname.find('Jack')",
"_____no_output_____"
]
],
[
[
"If the sub-string is not in the string then the output is a negative one. For example, the string 'Jasdfasdasdf' is not a substring:\n",
"_____no_output_____"
]
],
[
[
"# If cannot find the substring in the string\n\nname.find('Jasdfasdasdf')",
"_____no_output_____"
]
],
[
[
"<hr>\n",
"_____no_output_____"
],
[
"<h2 id=\"quiz\">Quiz on Strings</h2>\n",
"_____no_output_____"
],
[
"What is the value of the variable <code>a</code> after the following code is executed?\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute \n\na = \"1\"\nprint(a)",
"1\n"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\n\"1\"\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"What is the value of the variable <code>b</code> after the following code is executed?\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n\nb = \"2\"\nb",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\n\"2\"\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"What is the value of the variable <code>c</code> after the following code is executed?\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute \n\nc = a + b\nc",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\n\"12\"\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"<hr>\n",
"_____no_output_____"
],
[
"Consider the variable <code>d</code> use slicing to print out the first three elements:\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n\nd = \"ABCDEFG\"\nd[0:3]",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nprint(d[:3]) \n\n# or \n\nprint(d[0:3])\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"<hr>\n",
"_____no_output_____"
],
[
"Use a stride value of 2 to print out every second character of the string <code>e</code>:\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n\ne = 'clocrkr1e1c1t'\ne[::2]",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nprint(e[::2])\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"<hr>\n",
"_____no_output_____"
],
[
"Print out a backslash:\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nprint(\"\\\\\\\\\")\nprint(r\"\\ \")",
"\\\\\n\\ \n"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nprint(\"\\\\\\\\\")\n\nor\n\nprint(r\"\\ \")\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"<hr>\n",
"_____no_output_____"
],
[
"Convert the variable <code>f</code> to uppercase:\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n\nf = \"You are wrong\"\nf.upper()",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nf.upper()\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"<hr>\n",
"_____no_output_____"
],
[
"Consider the variable <code>g</code>, and find the first index of the sub-string <code>snow</code>:\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n\ng = \"Mary had a little lamb Little lamb, little lamb Mary had a little lamb \\\nIts fleece was white as snow And everywhere that Mary went Mary went, Mary went \\\nEverywhere that Mary went The lamb was sure to go\"\ng.find(\"snow\")\n",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\ng.find(\"snow\")\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"In the variable <code>g</code>, replace the sub-string <code>Mary</code> with <code>Bob</code>:\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\ng.replace(\"Mary\",\"Bob\")",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\ng.replace(\"Mary\", \"Bob\")\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"<hr>\n<h2>The last exercise!</h2>\n<p>Congratulations, you have completed your first lesson and hands-on lab in Python. However, there is one more thing you need to do. The Data Science community encourages sharing work. The best way to share and showcase your work is to share it on GitHub. By sharing your notebook on GitHub you are not only building your reputation with fellow data scientists, but you can also show it off when applying for a job. Even though this was your first piece of work, it is never too early to start building good habits. So, please read and follow <a href=\"https://cognitiveclass.ai/blog/data-scientists-stand-out-by-sharing-your-notebooks/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\" target=\"_blank\">this article</a> to learn how to share your work.\n<hr>\n",
"_____no_output_____"
],
[
"## Author\n\n<a href=\"https://www.linkedin.com/in/joseph-s-50398b136/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\" target=\"_blank\">Joseph Santarcangelo</a>\n\n## Change Log\n\n| Date (YYYY-MM-DD) | Version | Changed By | Change Description |\n| ----------------- | ------- | ---------- | ----------------------------------- |\n| 2020-11-11 | 2.1 | Aije | Updated variable names to lowercase |\n| 2020-08-26 | 2.0 | Lavanya | Moved lab to course repo in GitLab |\n\n## <h3 align=\"center\"> © IBM Corporation 2020. All rights reserved. <h3/>\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ecf3684a0c62fe2dea47423bea7bd87528fb9fb9 | 196,691 | ipynb | Jupyter Notebook | pulsar_search/period_finding/shor.ipynb | NLESC-quantum/quantum_comp | 84cf681bb7d64e00bcd89aa489bffc2271df0e3b | [
"Apache-2.0"
] | null | null | null | pulsar_search/period_finding/shor.ipynb | NLESC-quantum/quantum_comp | 84cf681bb7d64e00bcd89aa489bffc2271df0e3b | [
"Apache-2.0"
] | 18 | 2021-06-24T06:38:57.000Z | 2022-03-30T14:34:11.000Z | pulsar_search/period_finding/shor.ipynb | NLeSC/quantum_comp | 84cf681bb7d64e00bcd89aa489bffc2271df0e3b | [
"Apache-2.0"
] | null | null | null | 111.566081 | 49,800 | 0.843511 | [
[
[
"# Shor's Algorithm",
"_____no_output_____"
],
[
"Shor’s algorithm is famous for factoring integers in polynomial time. Since the best-known classical algorithm requires superpolynomial time to factor the product of two primes, the widely used cryptosystem, RSA, relies on factoring being impossible for large enough integers.\n\nIn this chapter we will focus on the quantum part of Shor’s algorithm, which actually solves the problem of _period finding_. Since a factoring problem can be turned into a period finding problem in polynomial time, an efficient period finding algorithm can be used to factor integers efficiently too. For now its enough to show that if we can compute the period of $a^x\\bmod N$ efficiently, then we can also efficiently factor. Since period finding is a worthy problem in its own right, we will first solve this, then discuss how this can be used to factor in section 5.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nfrom qiskit import QuantumCircuit, Aer, transpile, assemble\nfrom qiskit.visualization import plot_histogram\nfrom math import gcd\nfrom numpy.random import randint\nimport pandas as pd\nfrom fractions import Fraction\nprint(\"Imports Successful\")",
"Imports Successful\n"
]
],
[
[
"## 1. The Problem: Period Finding\n\nLet’s look at the periodic function:\n\n$$ f(x) = a^x \\bmod{N}$$\n\n<details>\n <summary>Reminder: Modulo & Modular Arithmetic (Click here to expand)</summary>\n\nThe modulo operation (abbreviated to 'mod') simply means to find the remainder when dividing one number by another. For example:\n\n$$ 17 \\bmod 5 = 2 $$\n\nSince $17 \\div 5 = 3$ with remainder $2$. (i.e. $17 = (3\\times 5) + 2$). In Python, the modulo operation is denoted through the <code>%</code> symbol.\n\nThis behaviour is used in <a href=\"https://en.wikipedia.org/wiki/Modular_arithmetic\">modular arithmetic</a>, where numbers 'wrap round' after reaching a certain value (the modulus). Using modular arithmetic, we could write:\n\n$$ 17 = 2 \\pmod 5$$\n\nNote that here the $\\pmod 5$ applies to the entire equation (since it is in parenthesis), unlike the equation above where it only applied to the left-hand side of the equation.\n</details>\n\nwhere $a$ and $N$ are positive integers, $a$ is less than $N$, and they have no common factors. The period, or order ($r$), is the smallest (non-zero) integer such that:\n\n$$a^r \\bmod N = 1 $$ \n\nWe can see an example of this function plotted on the graph below. Note that the lines between points are to help see the periodicity and do not represent the intermediate values between the x-markers.",
"_____no_output_____"
]
],
[
[
"N = 35\na = 3\n\n# Calculate the plotting data\nxvals = np.arange(35)\nyvals = [np.mod(a**x, N) for x in xvals]\n\n# Use matplotlib to display it nicely\nfig, ax = plt.subplots()\nax.plot(xvals, yvals, linewidth=1, linestyle='dotted', marker='x')\nax.set(xlabel='$x$', ylabel='$%i^x$ mod $%i$' % (a, N),\n title=\"Example of Periodic Function in Shor's Algorithm\")\ntry: # plot r on the graph\n r = yvals[1:].index(1) +1 \n plt.annotate('', xy=(0,1), xytext=(r,1), arrowprops=dict(arrowstyle='<->'))\n plt.annotate('$r=%i$' % r, xy=(r/3,1.5))\nexcept ValueError:\n print('Could not find period, check a < N and have no common factors.')",
"_____no_output_____"
]
],
[
[
"## 2. The Solution\n\nShor’s solution was to use [quantum phase estimation](./quantum-phase-estimation.html) on the unitary operator:\n\n$$ U|y\\rangle \\equiv |ay \\bmod N \\rangle $$\n\nTo see how this is helpful, let’s work out what an eigenstate of U might look like. If we started in the state $|1\\rangle$, we can see that each successive application of U will multiply the state of our register by $a \\pmod N$, and after $r$ applications we will arrive at the state $|1\\rangle$ again. For example with $a = 3$ and $N = 35$:\n\n$$\\begin{aligned}\nU|1\\rangle &= |3\\rangle & \\\\\nU^2|1\\rangle &= |9\\rangle \\\\\nU^3|1\\rangle &= |27\\rangle \\\\\n& \\vdots \\\\\nU^{(r-1)}|1\\rangle &= |12\\rangle \\\\\nU^r|1\\rangle &= |1\\rangle \n\\end{aligned}$$",
"_____no_output_____"
]
],
[
[
"ax.set(xlabel='Number of applications of U', ylabel='End state of register',\n title=\"Effect of Successive Applications of U\")\nfig",
"_____no_output_____"
]
],
[
[
"So a superposition of the states in this cycle ($|u_0\\rangle$) would be an eigenstate of $U$:\n\n$$|u_0\\rangle = \\tfrac{1}{\\sqrt{r}}\\sum_{k=0}^{r-1}{|a^k \\bmod N\\rangle} $$\n\n\n<details>\n <summary>Click to Expand: Example with $a = 3$ and $N=35$</summary>\n\n$$\\begin{aligned}\n|u_0\\rangle &= \\tfrac{1}{\\sqrt{12}}(|1\\rangle + |3\\rangle + |9\\rangle \\dots + |4\\rangle + |12\\rangle) \\\\[10pt]\nU|u_0\\rangle &= \\tfrac{1}{\\sqrt{12}}(U|1\\rangle + U|3\\rangle + U|9\\rangle \\dots + U|4\\rangle + U|12\\rangle) \\\\[10pt]\n &= \\tfrac{1}{\\sqrt{12}}(|3\\rangle + |9\\rangle + |27\\rangle \\dots + |12\\rangle + |1\\rangle) \\\\[10pt]\n &= |u_0\\rangle\n\\end{aligned}$$\n</details>\n\n\nThis eigenstate has an eigenvalue of 1, which isn’t very interesting. A more interesting eigenstate could be one in which the phase is different for each of these computational basis states. Specifically, let’s look at the case in which the phase of the $k$th state is proportional to $k$:\n\n$$\\begin{aligned}\n|u_1\\rangle &= \\tfrac{1}{\\sqrt{r}}\\sum_{k=0}^{r-1}{e^{-\\tfrac{2\\pi i k}{r}}|a^k \\bmod N\\rangle}\\\\[10pt]\nU|u_1\\rangle &= e^{\\tfrac{2\\pi i}{r}}|u_1\\rangle \n\\end{aligned}\n$$\n\n<details>\n <summary>Click to Expand: Example with $a = 3$ and $N=35$</summary>\n\n$$\\begin{aligned}\n|u_1\\rangle &= \\tfrac{1}{\\sqrt{12}}(|1\\rangle + e^{-\\tfrac{2\\pi i}{12}}|3\\rangle + e^{-\\tfrac{4\\pi i}{12}}|9\\rangle \\dots + e^{-\\tfrac{20\\pi i}{12}}|4\\rangle + e^{-\\tfrac{22\\pi i}{12}}|12\\rangle) \\\\[10pt]\nU|u_1\\rangle &= \\tfrac{1}{\\sqrt{12}}(|3\\rangle + e^{-\\tfrac{2\\pi i}{12}}|9\\rangle + e^{-\\tfrac{4\\pi i}{12}}|27\\rangle \\dots + e^{-\\tfrac{20\\pi i}{12}}|12\\rangle + e^{-\\tfrac{22\\pi i}{12}}|1\\rangle) \\\\[10pt]\nU|u_1\\rangle &= e^{\\tfrac{2\\pi i}{12}}\\cdot\\tfrac{1}{\\sqrt{12}}(e^{\\tfrac{-2\\pi i}{12}}|3\\rangle + e^{-\\tfrac{4\\pi i}{12}}|9\\rangle + e^{-\\tfrac{6\\pi i}{12}}|27\\rangle \\dots + e^{-\\tfrac{22\\pi i}{12}}|12\\rangle + e^{-\\tfrac{24\\pi i}{12}}|1\\rangle) \\\\[10pt]\nU|u_1\\rangle &= e^{\\tfrac{2\\pi i}{12}}|u_1\\rangle\n\\end{aligned}$$\n\n(We can see $r = 12$ appears in the denominator of the phase.)\n</details>\n\nThis is a particularly interesting eigenvalue as it contains $r$. In fact, $r$ has to be included to make sure the phase differences between the $r$ computational basis states are equal. This is not the only eigenstate with this behaviour; to generalise this further, we can multiply an integer, $s$, to this phase difference, which will show up in our eigenvalue:\n\n$$\\begin{aligned}\n|u_s\\rangle &= \\tfrac{1}{\\sqrt{r}}\\sum_{k=0}^{r-1}{e^{-\\tfrac{2\\pi i s k}{r}}|a^k \\bmod N\\rangle}\\\\[10pt]\nU|u_s\\rangle &= e^{\\tfrac{2\\pi i s}{r}}|u_s\\rangle \n\\end{aligned}\n$$\n\n<details>\n <summary>Click to Expand: Example with $a = 3$ and $N=35$</summary>\n\n$$\\begin{aligned}\n|u_s\\rangle &= \\tfrac{1}{\\sqrt{12}}(|1\\rangle + e^{-\\tfrac{2\\pi i s}{12}}|3\\rangle + e^{-\\tfrac{4\\pi i s}{12}}|9\\rangle \\dots + e^{-\\tfrac{20\\pi i s}{12}}|4\\rangle + e^{-\\tfrac{22\\pi i s}{12}}|12\\rangle) \\\\[10pt]\nU|u_s\\rangle &= \\tfrac{1}{\\sqrt{12}}(|3\\rangle + e^{-\\tfrac{2\\pi i s}{12}}|9\\rangle + e^{-\\tfrac{4\\pi i s}{12}}|27\\rangle \\dots + e^{-\\tfrac{20\\pi i s}{12}}|12\\rangle + e^{-\\tfrac{22\\pi i s}{12}}|1\\rangle) \\\\[10pt]\nU|u_s\\rangle &= e^{\\tfrac{2\\pi i s}{12}}\\cdot\\tfrac{1}{\\sqrt{12}}(e^{-\\tfrac{2\\pi i s}{12}}|3\\rangle + e^{-\\tfrac{4\\pi i s}{12}}|9\\rangle + e^{-\\tfrac{6\\pi i s}{12}}|27\\rangle \\dots + e^{-\\tfrac{22\\pi i s}{12}}|12\\rangle + e^{-\\tfrac{24\\pi i s}{12}}|1\\rangle) \\\\[10pt]\nU|u_s\\rangle &= e^{\\tfrac{2\\pi i s}{12}}|u_s\\rangle\n\\end{aligned}$$\n\n</details>\n\nWe now have a unique eigenstate for each integer value of $s$ where $$0 \\leq s \\leq r-1.$$ Very conveniently, if we sum up all these eigenstates, the different phases cancel out all computational basis states except $|1\\rangle$:\n\n$$ \\tfrac{1}{\\sqrt{r}}\\sum_{s=0}^{r-1} |u_s\\rangle = |1\\rangle$$\n\n<details>\n <summary>Click to Expand: Example with $a = 7$ and $N=15$</summary>\n\nFor this, we will look at a smaller example where $a = 7$ and $N=15$. In this case $r=4$:\n\n$$\\begin{aligned}\n\\tfrac{1}{2}(\\quad|u_0\\rangle &= \\tfrac{1}{2}(|1\\rangle \\hphantom{e^{-\\tfrac{2\\pi i}{12}}}+ |7\\rangle \\hphantom{e^{-\\tfrac{12\\pi i}{12}}} + |4\\rangle \\hphantom{e^{-\\tfrac{12\\pi i}{12}}} + |13\\rangle)\\dots \\\\[10pt]\n+ |u_1\\rangle &= \\tfrac{1}{2}(|1\\rangle + e^{-\\tfrac{2\\pi i}{4}}|7\\rangle + e^{-\\tfrac{\\hphantom{1}4\\pi i}{4}}|4\\rangle + e^{-\\tfrac{\\hphantom{1}6\\pi i}{4}}|13\\rangle)\\dots \\\\[10pt]\n+ |u_2\\rangle &= \\tfrac{1}{2}(|1\\rangle + e^{-\\tfrac{4\\pi i}{4}}|7\\rangle + e^{-\\tfrac{\\hphantom{1}8\\pi i}{4}}|4\\rangle + e^{-\\tfrac{12\\pi i}{4}}|13\\rangle)\\dots \\\\[10pt]\n+ |u_3\\rangle &= \\tfrac{1}{2}(|1\\rangle + e^{-\\tfrac{6\\pi i}{4}}|7\\rangle + e^{-\\tfrac{12\\pi i}{4}}|4\\rangle + e^{-\\tfrac{18\\pi i}{4}}|13\\rangle)\\quad) = |1\\rangle \\\\[10pt]\n\\end{aligned}$$\n\n</details>\n\nSince the computational basis state $|1\\rangle$ is a superposition of these eigenstates, which means if we do QPE on $U$ using the state $|1\\rangle$, we will measure a phase:\n\n$$\\phi = \\frac{s}{r}$$\n\nWhere $s$ is a random integer between $0$ and $r-1$. We finally use the [continued fractions](https://en.wikipedia.org/wiki/Continued_fraction) algorithm on $\\phi$ to find $r$. The circuit diagram looks like this (note that this diagram uses Qiskit's qubit ordering convention):\n\n<img src=\"images/shor_circuit_1.svg\">\n\nWe will next demonstrate Shor’s algorithm using Qiskit’s simulators. For this demonstration we will provide the circuits for $U$ without explanation, but in section 4 we will discuss how circuits for $U^{2^j}$ can be constructed efficiently.",
"_____no_output_____"
],
[
"## 3. Qiskit Implementation\n\nIn this example we will solve the period finding problem for $a=7$ and $N=15$. We provide the circuits for $U$ where:\n\n$$U|y\\rangle = |ay\\bmod 15\\rangle $$\n\nwithout explanation. To create $U^x$, we will simply repeat the circuit $x$ times. In the next section we will discuss a general method for creating these circuits efficiently. The function `c_amod15` returns the controlled-U gate for `a`, repeated `power` times.",
"_____no_output_____"
]
],
[
[
"def c_amod15(a, power):\n \"\"\"Controlled multiplication by a mod 15\"\"\"\n if a not in [2,7,8,11,13]:\n raise ValueError(\"'a' must be 2,7,8,11 or 13\")\n U = QuantumCircuit(4) \n for iteration in range(power):\n if a in [2,13]:\n U.swap(0,1)\n U.swap(1,2)\n U.swap(2,3)\n if a in [7,8]:\n U.swap(2,3)\n U.swap(1,2)\n U.swap(0,1)\n if a == 11:\n U.swap(1,3)\n U.swap(0,2)\n if a in [7,11,13]:\n for q in range(4):\n U.x(q)\n U = U.to_gate()\n U.name = \"%i^%i mod 15\" % (a, power)\n c_U = U.control()\n return c_U",
"_____no_output_____"
]
],
[
[
"We will use 8 counting qubits:",
"_____no_output_____"
]
],
[
[
"# Specify variables\nn_count = 8 # number of counting qubits\na = 7",
"_____no_output_____"
]
],
[
[
"We also import the circuit for the QFT (you can read more about the QFT in the [quantum Fourier transform chapter](./quantum-fourier-transform.html#generalqft)):",
"_____no_output_____"
]
],
[
[
"def qft_dagger(n):\n \"\"\"n-qubit QFTdagger the first n qubits in circ\"\"\"\n qc = QuantumCircuit(n)\n # Don't forget the Swaps!\n for qubit in range(n//2):\n qc.swap(qubit, n-qubit-1)\n for j in range(n):\n for m in range(j):\n qc.cp(-np.pi/float(2**(j-m)), m, j)\n qc.h(j)\n qc.name = \"QFT†\"\n return qc",
"_____no_output_____"
]
],
[
[
"With these building blocks we can easily construct the circuit for Shor's algorithm:",
"_____no_output_____"
]
],
[
[
"# Create QuantumCircuit with n_count counting qubits\n# plus 4 qubits for U to act on\nqc = QuantumCircuit(n_count + 4, n_count)\n\n# Initialize counting qubits\n# in state |+>\nfor q in range(n_count):\n qc.h(q)\n \n# And auxiliary register in state |1>\nqc.x(3+n_count)\n\n# Do controlled-U operations\nfor q in range(n_count):\n qc.append(c_amod15(a, 2**q), \n [q] + [i+n_count for i in range(4)])\n\n# Do inverse-QFT\nqc.append(qft_dagger(n_count), range(n_count))\n\n# Measure circuit\nqc.measure(range(n_count), range(n_count))\nqc.draw(\"mpl\", fold=-1) # -1 means 'do not fold' ",
"_____no_output_____"
]
],
[
[
"Let's see what results we measure:",
"_____no_output_____"
]
],
[
[
"aer_sim = Aer.get_backend('aer_simulator')\nt_qc = transpile(qc, aer_sim)\nqobj = assemble(t_qc)\nresults = aer_sim.run(qobj).result()\ncounts = results.get_counts()\nplot_histogram(counts)",
"_____no_output_____"
]
],
[
[
"Since we have 8 qubits, these results correspond to measured phases of:",
"_____no_output_____"
]
],
[
[
"rows, measured_phases = [], []\nfor output in counts:\n decimal = int(output, 2) # Convert (base 2) string to decimal\n phase = decimal/(2**n_count) # Find corresponding eigenvalue\n measured_phases.append(phase)\n # Add these values to the rows in our table:\n rows.append([f\"{output}(bin) = {decimal:>3}(dec)\", \n f\"{decimal}/{2**n_count} = {phase:.2f}\"])\n# Print the rows in a table\nheaders=[\"Register Output\", \"Phase\"]\ndf = pd.DataFrame(rows, columns=headers)\nprint(df)",
" Register Output Phase\n0 01000000(bin) = 64(dec) 64/256 = 0.25\n1 10000000(bin) = 128(dec) 128/256 = 0.50\n2 00000000(bin) = 0(dec) 0/256 = 0.00\n3 11000000(bin) = 192(dec) 192/256 = 0.75\n"
]
],
[
[
"We can now use the continued fractions algorithm to attempt to find $s$ and $r$. Python has this functionality built in: We can use the `fractions` module to turn a float into a `Fraction` object, for example:",
"_____no_output_____"
]
],
[
[
"Fraction(0.666)",
"_____no_output_____"
]
],
[
[
"Because this gives fractions that return the result exactly (in this case, `0.6660000...`), this can give gnarly results like the one above. We can use the `.limit_denominator()` method to get the fraction that most closely resembles our float, with denominator below a certain value:",
"_____no_output_____"
]
],
[
[
"# Get fraction that most closely resembles 0.666\n# with denominator < 15\nFraction(0.666).limit_denominator(15)",
"_____no_output_____"
]
],
[
[
"Much nicer! The order (r) must be less than N, so we will set the maximum denominator to be `15`:",
"_____no_output_____"
]
],
[
[
"rows = []\nfor phase in measured_phases:\n frac = Fraction(phase).limit_denominator(15)\n rows.append([phase, f\"{frac.numerator}/{frac.denominator}\", frac.denominator])\n# Print as a table\nheaders=[\"Phase\", \"Fraction\", \"Guess for r\"]\ndf = pd.DataFrame(rows, columns=headers)\nprint(df)",
" Phase Fraction Guess for r\n0 0.25 1/4 4\n1 0.50 1/2 2\n2 0.00 0/1 1\n3 0.75 3/4 4\n"
]
],
[
[
"We can see that two of the measured eigenvalues provided us with the correct result: $r=4$, and we can see that Shor’s algorithm has a chance of failing. These bad results are because $s = 0$, or because $s$ and $r$ are not coprime and instead of $r$ we are given a factor of $r$. The easiest solution to this is to simply repeat the experiment until we get a satisfying result for $r$.\n\n### Quick Exercise\n\n- Modify the circuit above for values of $a = 2, 8, 11$ and $13$. What results do you get and why?",
"_____no_output_____"
],
[
"## 4. Modular Exponentiation\n\nYou may have noticed that the method of creating the $U^{2^j}$ gates by repeating $U$ grows exponentially with $j$ and will not result in a polynomial time algorithm. We want a way to create the operator:\n\n$$ U^{2^j}|y\\rangle = |a^{2^j}y \\bmod N \\rangle $$\n\nthat grows polynomially with $j$. Fortunately, calculating:\n\n$$ a^{2^j} \\bmod N$$\n\nefficiently is possible. Classical computers can use an algorithm known as _repeated squaring_ to calculate an exponential. In our case, since we are only dealing with exponentials of the form $2^j$, the repeated squaring algorithm becomes very simple:",
"_____no_output_____"
]
],
[
[
"def a2jmodN(a, j, N):\n \"\"\"Compute a^{2^j} (mod N) by repeated squaring\"\"\"\n for i in range(j):\n a = np.mod(a**2, N)\n return a",
"_____no_output_____"
],
[
"a2jmodN(7, 2049, 53)",
"_____no_output_____"
]
],
[
[
"If an efficient algorithm is possible in Python, then we can use the same algorithm on a quantum computer. Unfortunately, despite scaling polynomially with $j$, modular exponentiation circuits are not straightforward and are the bottleneck in Shor’s algorithm. A beginner-friendly implementation can be found in reference [1].\n\n## 5. Factoring from Period Finding\n\nNot all factoring problems are difficult; we can spot an even number instantly and know that one of its factors is 2. In fact, there are [specific criteria](https://nvlpubs.nist.gov/nistpubs/FIPS/NIST.FIPS.186-4.pdf#%5B%7B%22num%22%3A127%2C%22gen%22%3A0%7D%2C%7B%22name%22%3A%22XYZ%22%7D%2C70%2C223%2C0%5D) for choosing numbers that are difficult to factor, but the basic idea is to choose the product of two large prime numbers.\n\nA general factoring algorithm will first check to see if there is a shortcut to factoring the integer (is the number even? Is the number of the form $N = a^b$?), before using Shor’s period finding for the worst-case scenario. Since we aim to focus on the quantum part of the algorithm, we will jump straight to the case in which N is the product of two primes.\n\n### Example: Factoring 15\n\nTo see an example of factoring on a small number of qubits, we will factor 15, which we all know is the product of the not-so-large prime numbers 3 and 5.",
"_____no_output_____"
]
],
[
[
"N = 15",
"_____no_output_____"
]
],
[
[
"The first step is to choose a random number, $a$, between $1$ and $N-1$:",
"_____no_output_____"
]
],
[
[
"np.random.seed(1) # This is to make sure we get reproduceable results\na = randint(2, 15)\nprint(a)",
"7\n"
]
],
[
[
"Next we quickly check it isn't already a non-trivial factor of $N$:",
"_____no_output_____"
]
],
[
[
"from math import gcd # greatest common divisor\ngcd(a, N)",
"_____no_output_____"
]
],
[
[
"Great. Next, we do Shor's order finding algorithm for `a = 7` and `N = 15`. Remember that the phase we measure will be $s/r$ where:\n\n$$ a^r \\bmod N = 1 $$\n\nand $s$ is a random integer between 0 and $r-1$.",
"_____no_output_____"
]
],
[
[
"def qpe_amod15(a):\n n_count = 8\n qc = QuantumCircuit(4+n_count, n_count)\n for q in range(n_count):\n qc.h(q) # Initialize counting qubits in state |+>\n qc.x(3+n_count) # And auxiliary register in state |1>\n for q in range(n_count): # Do controlled-U operations\n qc.append(c_amod15(a, 2**q), \n [q] + [i+n_count for i in range(4)])\n qc.append(qft_dagger(n_count), range(n_count)) # Do inverse-QFT\n qc.measure(range(n_count), range(n_count))\n # Simulate Results\n aer_sim = Aer.get_backend('aer_simulator')\n # Setting memory=True below allows us to see a list of each sequential reading\n t_qc = transpile(qc, aer_sim)\n qobj = assemble(t_qc, shots=1)\n result = aer_sim.run(qobj, memory=True).result()\n readings = result.get_memory()\n print(\"Register Reading: \" + readings[0])\n phase = int(readings[0],2)/(2**n_count)\n print(\"Corresponding Phase: %f\" % phase)\n return phase",
"_____no_output_____"
]
],
[
[
"From this phase, we can easily find a guess for $r$:",
"_____no_output_____"
]
],
[
[
"phase = qpe_amod15(a) # Phase = s/r\nFraction(phase).limit_denominator(15) # Denominator should (hopefully!) tell us r",
"Register Reading: 01000000\nCorresponding Phase: 0.250000\n"
],
[
"frac = Fraction(phase).limit_denominator(15)\ns, r = frac.numerator, frac.denominator\nprint(r)",
"4\n"
]
],
[
[
"Now we have $r$, we might be able to use this to find a factor of $N$. Since:\n\n$$a^r \\bmod N = 1 $$\n\nthen:\n\n$$(a^r - 1) \\bmod N = 0 $$\n\nwhich means $N$ must divide $a^r-1$. And if $r$ is also even, then we can write:\n\n$$a^r -1 = (a^{r/2}-1)(a^{r/2}+1)$$\n\n(if $r$ is not even, we cannot go further and must try again with a different value for $a$). There is then a high probability that the greatest common divisor of $N$ and either $a^{r/2}-1$, or $a^{r/2}+1$ is a proper factor of $N$ [2]:",
"_____no_output_____"
]
],
[
[
"guesses = [gcd(a**(r//2)-1, N), gcd(a**(r//2)+1, N)]\nprint(guesses)",
"[3, 5]\n"
]
],
[
[
"The cell below repeats the algorithm until at least one factor of 15 is found. You should try re-running the cell a few times to see how it behaves.",
"_____no_output_____"
]
],
[
[
"a = 7\nfactor_found = False\nattempt = 0\nwhile not factor_found:\n attempt += 1\n print(\"\\nAttempt %i:\" % attempt)\n phase = qpe_amod15(a) # Phase = s/r\n frac = Fraction(phase).limit_denominator(N) # Denominator should (hopefully!) tell us r\n r = frac.denominator\n print(\"Result: r = %i\" % r)\n if phase != 0:\n # Guesses for factors are gcd(x^{r/2} ±1 , 15)\n guesses = [gcd(a**(r//2)-1, N), gcd(a**(r//2)+1, N)]\n print(\"Guessed Factors: %i and %i\" % (guesses[0], guesses[1]))\n for guess in guesses:\n if guess not in [1,N] and (N % guess) == 0: # Check to see if guess is a factor\n print(\"*** Non-trivial factor found: %i ***\" % guess)\n factor_found = True",
"\nAttempt 1:\nRegister Reading: 00000000\nCorresponding Phase: 0.000000\nResult: r = 1\n\nAttempt 2:\nRegister Reading: 11000000\nCorresponding Phase: 0.750000\nResult: r = 4\nGuessed Factors: 3 and 5\n*** Non-trivial factor found: 3 ***\n*** Non-trivial factor found: 5 ***\n"
]
],
[
[
"## 6. References\n\n1. Stephane Beauregard, _Circuit for Shor's algorithm using 2n+3 qubits,_ [arXiv:quant-ph/0205095](https://arxiv.org/abs/quant-ph/0205095)\n\n2. M. Nielsen and I. Chuang, _Quantum Computation and Quantum Information,_ Cambridge Series on Information and the Natural Sciences (Cambridge University Press, Cambridge, 2000). (Page 633)",
"_____no_output_____"
]
],
[
[
"import qiskit.tools.jupyter\n%qiskit_version_table",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf369f29e7b465e73d4716cb719653b2c940f3f | 275,817 | ipynb | Jupyter Notebook | BigData.ipynb | yohanesnuwara/public-geoscience-data | 061e3587be9c1820f9c1359282eb0e037640984e | [
"MIT"
] | 7 | 2020-08-26T04:49:52.000Z | 2021-11-08T14:39:46.000Z | BigData.ipynb | yohanesnuwara/public-geoscience-data | 061e3587be9c1820f9c1359282eb0e037640984e | [
"MIT"
] | null | null | null | BigData.ipynb | yohanesnuwara/public-geoscience-data | 061e3587be9c1820f9c1359282eb0e037640984e | [
"MIT"
] | 1 | 2020-10-19T08:24:47.000Z | 2020-10-19T08:24:47.000Z | 234.140068 | 41,012 | 0.880301 | [
[
[
"<a href=\"https://colab.research.google.com/github/yohanesnuwara/reservoir-qi/blob/master/BigData.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Handling Big Volume of Well Data",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"Access Google Drive to get data",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&response_type=code&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly\n\nEnter your authorization code:\n··········\nMounted at /content/drive\n"
],
[
"import sys\nsys.path.append('/content/drive/My Drive/Colab Notebooks')",
"_____no_output_____"
]
],
[
[
"Read multiple files. In the folder, there are 46 LAS files from Norne Field.",
"_____no_output_____"
]
],
[
[
"import glob\nimport os\n\nfile_path = \"/content/drive/My Drive/Colab Notebooks/well_logs\"\nread_files = glob.glob(os.path.join(file_path, \"*.las\"))\nread_files",
"_____no_output_____"
]
],
[
[
"Get the name of the well from the file.",
"_____no_output_____"
]
],
[
[
"well_names = []\n\nfor files in read_files:\n files = os.path.splitext(os.path.basename(files))[0]\n well_names.append(files)",
"_____no_output_____"
],
[
"well_names",
"_____no_output_____"
]
],
[
[
"If the well name too long, shorten it by renaming it.",
"_____no_output_____"
]
],
[
[
"well_names = np.array(well_names)\nwellnames = np.array(['B1BH', 'E2AH', 'B1AHT2', 'K1HT2',\n 'B1H', 'B2H', 'B3H', 'B4AH',\n 'B4BH', 'B4CH', 'B4DHT2', 'B4H',\n 'C1H', 'C2H', 'C3H', 'C4AH',\n 'C4H', 'D1AH', 'D1BH', 'D1CH',\n 'D1H', 'D2HT2', 'D3AH', 'D3BY1HT2',\n 'D3BY2H', 'D3H', 'D4AH', 'D4AHT2',\n 'D4H', 'E1H', 'E2H', 'E3AH',\n 'E3AHT2', 'E3BH', 'E3CHT2','E3H',\n 'E4AH', 'E4AHT2', 'E4H','E4HT2',\n 'F1H', 'F2H', 'F3H', \n 'F4H','K1H', 'K3H'])",
"_____no_output_____"
]
],
[
[
"Import `lasio` library to import LAS data",
"_____no_output_____"
]
],
[
[
"!pip install lasio",
"Collecting lasio\n Downloading https://files.pythonhosted.org/packages/94/fa/b06aa199d8eaf85a8e6d417021d2aeff9b5c59c92c9a669046408b211bc9/lasio-0.24.1-py3-none-any.whl\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from lasio) (1.17.5)\nInstalling collected packages: lasio\nSuccessfully installed lasio-0.24.1\n"
],
[
"import lasio",
"_____no_output_____"
]
],
[
[
"Read LAS file (if command: `Header section Parameter regexp=~P was not found.`, it's OK)",
"_____no_output_____"
]
],
[
[
"lases = []\n\nfor files in read_files:\n las = lasio.read(files)\n lases.append(las)",
"Header section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\nHeader section Parameter regexp=~P was not found.\n"
]
],
[
[
"Input the name of the well you want to view in the `find` and check what data is present.",
"_____no_output_____"
]
],
[
[
"find = 'B2H'\nid_ = np.int64(np.where(wellnames==find)).item()\nlases[id_].keys()",
"_____no_output_____"
]
],
[
[
"Check more details.",
"_____no_output_____"
]
],
[
[
"lases[id_].curves",
"_____no_output_____"
]
],
[
[
"# ***",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"append = np.append(lases[1].data['NPHI'], lases[2].data['NPHI'])\nappend2 = np.append(append, lases[3].data['NPHI'])\nappend3 = np.append(append2, lases[5].data['NPHI'])\nappend4 = np.append(append3, lases[6].data['NPHI'])\nappend4 = append4[np.logical_not(np.isnan(append4))] # delete NaN values, otherwise sns distplot can't plot\nplt.figure(figsize=(20,8))\nsns.distplot(append4, bins=50, color='green', hist_kws=dict(edgecolor=\"black\", linewidth=1))\nplt.show()",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\n#import data\n#conists of one column of datapoints as 2.231, -0.1516, 1.564, etc\n# data=np.loadtxt(\"https://github.com/HomeworkHelpOnline/KDE-Task/raw/master/dataset1.txt\")\ndata=append4\n\n#normalized histogram of loaded datase\nhist, bins = np.histogram(data,bins=100,range=(np.min(data),np.max(data)) ,density=True)\nwidth = 0.7 * (bins[1] - bins[0])\ncenter = (bins[:-1] + bins[1:]) / 2\n\n\n#generate data with double random()\ngeneratedData=np.zeros(1000)\nmaxData=np.max(data)\nminData=np.min(data)\ni=0\nwhile i<1000:\n randNo=np.random.rand(1)*(maxData-minData)-np.absolute(minData)\n if np.random.rand(1)<=hist[np.argmax(randNo<(center+(bins[1] - bins[0])/2))-1]:\n generatedData[i]=randNo\n i+=1\n\n#normalized histogram of generatedData\nhist1, bins2 = np.histogram(generatedData,bins=100,range=(np.min(data),np.max(data)), density=True)\nwidth2 = 0.7 * (bins2[1] - bins2[0])\ncenter2 = (bins2[:-1] + bins2[1:]) / 2\n\n#plot both histograms\nplt.figure(figsize=(20,8))\nplt.subplot(1,2,1)\nplt.title(\"Original Data\")\nsns.distplot(data, bins=50, color ='blue', hist_kws=dict(edgecolor=\"black\", linewidth=1))\nplt.subplot(1,2,2)\nplt.title(\"Generated Data\")\nsns.distplot(generatedData, bins=50, color ='red', hist_kws=dict(edgecolor=\"black\", linewidth=1))",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\n#https://en.wikipedia.org/wiki/Gaussian_function\ndef gaussian(x,b=1):\n return np.exp(-x**2/(2*b**2))/(b*np.sqrt(2*np.pi))\n \n#import data\n#conists of one column of datapoints as 2.231, -0.1516, 1.564, etc\n# data=np.loadtxt(\"https://github.com/HomeworkHelpOnline/KDE-Task/raw/master/dataset1.txt\")\ndata=append4\n\nN=100 #Number of bins\nlenDataset = len(data)\n#normalized histogram of loaded datase\nhist, bins = np.histogram(data, bins=N, range=(np.min(data), np.max(data)), density=True)\nwidth = 0.7 * (bins[1] - bins[0])\ndx=(bins[1] - bins[0])\ncenter = (bins[:-1] + bins[1:]) / 2\n\n##Generate data\n#There are few options here - save values of KDE for every small dx \n#OR save all the dataset and generate probability for every x we will test.\n#We choose the second option here.\n\nsumPdfSilverman=np.zeros(len(center))\n#Silverman's Rule to find optimal bandwidth\n#https://en.wikipedia.org/wiki/Kernel_density_estimation#Practical_estimation_of_the_bandwidth\nh=1.06*np.std(data)*lenDataset**(-1/5.0)\n\nfor i in range(0, lenDataset):\n sumPdfSilverman+=((gaussian(center[:, None]-data[i],h))/lenDataset)[:,0]\n \n#So here we have to sum 1000 gaussians at generated random x to evaluate probability that this x exists in new generated dataset.\ni=0\ngeneratedDataPdfSilverman=np.zeros(1000)\nwhile i<1000:\n randNo=np.random.rand(1)*(np.max(data)-np.min(data))-np.absolute(np.min(data))\n if np.random.rand(1)<=np.sum((gaussian(randNo-data,h))/lenDataset):\n generatedDataPdfSilverman[i]=randNo\n i+=1\n\n#Our second approach to calculate optimal bandwidth h using least-squares cross validation\n#This looks a bit tricky, take a look at the theory explanation in the related article if you need to.\nh_test = np.linspace(0.01, 1, 100) #h values to iterate for testing\nL = np.zeros(len(h_test))\nfhati = np.zeros(len(data)) #fhati\ncenter\niteration=0\nfor h_iter in h_test:\n #find first part of equation\n for i in range(0, lenDataset):\n fhat = 0\n fhat+=((gaussian(center[:, None]-data[i],h_iter))/lenDataset)[:,0]\n \n #find second part of equation for sum fhati\n for i in range (0, lenDataset):\n mask=np.ones(lenDataset,dtype=bool)\n mask[i]=0\n fhati[i]=np.sum(gaussian(data[mask]-data[i],h_iter))/(lenDataset-1)\n \n L[iteration]=np.sum(fhat**2)*dx-2*np.mean(fhati)\n iteration=iteration+1\n\nh2=h_test[np.argmin(L)]\n#we can look how L looks like, depending on h\nfig0, ax0 = plt.subplots(1,1, figsize=(14,8))\nax0.plot(h_test,L)\nfig0.savefig(\"Function_to_minimize[h,L_value].jpg\")\n\n#resulting PDF with found h2\nsumPdfLSCV=np.zeros(len(center))\nfor i in range(0, lenDataset):\n sumPdfLSCV+=((gaussian(center[:, None]-data[i],h2))/lenDataset)[:,0]\n\n#So here we have to sum 1000 gaussians at generated random x to evaluate probability that this x exists in new generated dataset.\ni=0\ngeneratedDataPdfCV=np.zeros(1000)\nwhile i<1000:\n randNo=np.random.rand(1)*(np.max(data)-np.min(data))-np.absolute(np.min(data))\n if np.random.rand(1)<=np.sum((gaussian(randNo-data,h2))/lenDataset):\n generatedDataPdfCV[i]=randNo\n i+=1\n\n\n##Plotting\nfig, ax = plt.subplots(2,2, figsize=(14,8), sharey=True )\n#Estimated PDF using Silverman's calculation for h\nax[0,0].plot(center, sumPdfSilverman, '-k', linestyle=\"dashed\")\nax[0,0].set_title('KDE, Silvermans bandwidth h=%.2f' % h)\n\n#Histogram for generated data using KDE and h found using Silverman's method\nhist2, bins2 = np.histogram(generatedDataPdfSilverman, bins=N, range=(np.min(data), np.max(data)), density=True)\nax[1,0].bar(center, hist2, align='center', width=width, fc='#AAAAFF')\nax[1,0].set_title('Generated, Silvermans bandwidth h=%.2f' % h)\n\n#Estimated PDF using Least-squares cross-validation for h\nax[0,1].plot(center, sumPdfLSCV, '-k', linestyle=\"dashed\")\nax[0,1].set_title('KDE, LSCV bandwidth h=%.2f' % h2)\n\n#Histogram for generated data using KDE and h found using LSCV\nhist3, bins3 = np.histogram(generatedDataPdfCV, bins=N, range=(np.min(data), np.max(data)), density=True)\nax[1,1].bar(center, hist3, align='center', width=width, fc='#AAAAFF')\nax[1,1].set_title('Generated, LSCV bandwidth h=%.2f' % h2)\n\n#note that PDF found using KDE does not sum up exactly to one, because we ignore the side-spread",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\n#https://en.wikipedia.org/wiki/Gaussian_function\ndef gaussian(x,b=1):\n return np.exp(-x**2/(2*b**2))/(b*np.sqrt(2*np.pi))\n \n#import data\n#conists of one column of datapoints as 2.231, -0.1516, 1.564, etc\n# data=np.loadtxt(\"https://github.com/HomeworkHelpOnline/KDE-Task/raw/master/dataset1.txt\")\ndata=append4\n\nN=100 #Number of bins\nlenDataset = len(data)\n#normalized histogram of loaded datase\nhist, bins = np.histogram(data, bins=N, range=(np.min(data), np.max(data)), density=True)\nwidth = 0.7 * (bins[1] - bins[0])\ndx=(bins[1] - bins[0])\ncenter = (bins[:-1] + bins[1:]) / 2\n\n##Generate data\n#There are few options here - save values of KDE for every small dx \n#OR save all the dataset and generate probability for every x we will test.\n#We choose the second option here.\n\nsumPdfSilverman=np.zeros(len(center))\n#Silverman's Rule to find optimal bandwidth\n#https://en.wikipedia.org/wiki/Kernel_density_estimation#Practical_estimation_of_the_bandwidth\nh=1.06*np.std(data)*lenDataset**(-1/5.0)\n\nfor i in range(0, lenDataset):\n sumPdfSilverman+=((gaussian(center[:, None]-data[i],h))/lenDataset)[:,0]\n \n#So here we have to sum 1000 gaussians at generated random x to evaluate probability that this x exists in new generated dataset.\ni=0\ngeneratedDataPdfSilverman=np.zeros(1000)\nwhile i<1000:\n randNo=np.random.rand(1)*(np.max(data)-np.min(data))-np.absolute(np.min(data))\n if np.random.rand(1)<=np.sum((gaussian(randNo-data,h))/lenDataset):\n generatedDataPdfSilverman[i]=randNo\n i+=1\n\n# #Our second approach to calculate optimal bandwidth h using least-squares cross validation\n# #This looks a bit tricky, take a look at the theory explanation in the related article if you need to.\n# h_test = np.linspace(0.01, 1, 100) #h values to iterate for testing\n# L = np.zeros(len(h_test))\n# fhati = np.zeros(len(data)) #fhati\n# center\n# iteration=0\n# for h_iter in h_test:\n# #find first part of equation\n# for i in range(0, lenDataset):\n# fhat = 0\n# fhat+=((gaussian(center[:, None]-data[i],h_iter))/lenDataset)[:,0]\n \n# #find second part of equation for sum fhati\n# for i in range (0, lenDataset):\n# mask=np.ones(lenDataset,dtype=bool)\n# mask[i]=0\n# fhati[i]=np.sum(gaussian(data[mask]-data[i],h_iter))/(lenDataset-1)\n \n# L[iteration]=np.sum(fhat**2)*dx-2*np.mean(fhati)\n# iteration=iteration+1\n\n# h2=h_test[np.argmin(L)]\n# #we can look how L looks like, depending on h\n# fig0, ax0 = plt.subplots(1,1, figsize=(14,8))\n# ax0.plot(h_test,L)\n# fig0.savefig(\"Function_to_minimize[h,L_value].jpg\")\n\nh2 = 0.01\n\n#resulting PDF with found h2\nsumPdfLSCV=np.zeros(len(center))\nfor i in range(0, lenDataset):\n sumPdfLSCV+=((gaussian(center[:, None]-data[i],h2))/lenDataset)[:,0]\n\n#So here we have to sum 1000 gaussians at generated random x to evaluate probability that this x exists in new generated dataset.\ni=0\ngeneratedDataPdfCV=np.zeros(1000)\nwhile i<1000:\n randNo=np.random.rand(1)*(np.max(data)-np.min(data))-np.absolute(np.min(data))\n if np.random.rand(1)<=np.sum((gaussian(randNo-data,h2))/lenDataset):\n generatedDataPdfCV[i]=randNo\n i+=1\n\n\n##Plotting\nfig, ax = plt.subplots(2,2, figsize=(14,8), sharey=True )\n#Estimated PDF using Silverman's calculation for h\nax[0,0].plot(center, sumPdfSilverman, '-k', linestyle=\"dashed\")\nax[0,0].set_title('KDE, Silvermans bandwidth h=%.2f' % h)\n\n#Histogram for generated data using KDE and h found using Silverman's method\nhist2, bins2 = np.histogram(generatedDataPdfSilverman, bins=N, range=(np.min(data), np.max(data)), density=True)\nax[1,0].bar(center, hist2, align='center', width=width, fc='#AAAAFF')\nax[1,0].set_title('Generated, Silvermans bandwidth h=%.2f' % h)\n\n#Estimated PDF using Least-squares cross-validation for h\nax[0,1].plot(center, sumPdfLSCV, '-k', linestyle=\"dashed\")\nax[0,1].set_title('KDE, LSCV bandwidth h=%.2f' % h2)\n\n#Histogram for generated data using KDE and h found using LSCV\nhist3, bins3 = np.histogram(generatedDataPdfCV, bins=N, range=(np.min(data), np.max(data)), density=True)\nax[1,1].bar(center, hist3, align='center', width=width, fc='#AAAAFF')\nax[1,1].set_title('Generated, LSCV bandwidth h=%.2f' % h2)\n\n#note that PDF found using KDE does not sum up exactly to one, because we ignore the side-spread",
"_____no_output_____"
],
[
"h",
"_____no_output_____"
],
[
"cumulative = np.cumsum(hist)*dx #original dataset\ncumulativeHist = np.cumsum(hist1)*dx #histogram generated\ncumulativeKDE_Silverman = np.cumsum(hist2)*dx #KDE Silverman's h generated\ncumulativeKDE_LSCV = np.cumsum(hist3)*dx #KDE LSCV generated\n\nDHist=np.max(np.absolute(cumulative-cumulativeHist))\nDKDE_Silverman=np.max(np.absolute(cumulative-cumulativeKDE_Silverman))\nDKDE_LSCV=np.max(np.absolute(cumulative-cumulativeKDE_LSCV))\n\nfig, ax = plt.subplots(1,3, figsize=(16,4), sharey=True)\nax[0].set_ylim([0,1.4])\nax[0].plot(cumulative, label=\"Original data\")\nax[0].plot(cumulativeHist, label=\"Histogram generated\")\nax[0].legend()\nax[0].set_title('Dmax = %.3f' % DHist, y=0.05, x=0.7)\n\nax[1].plot(cumulative, label=\"Original data\")\nax[1].plot(cumulativeKDE_Silverman, label=\"KDE_Silverman generated\")\nax[1].legend()\nax[1].set_title('Dmax = %.3f' % DKDE_Silverman, y=0.05, x=0.7)\n\nax[2].plot(cumulative, label=\"Original data\")\nax[2].plot(cumulativeKDE_LSCV, label=\"KDE_LSCV generated\")\nax[2].legend()\nax[2].set_title('Dmax = %.3f' % DKDE_LSCV, y=0.05, x=0.7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Gaussian, Epanechnikov, Tophat KDE",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom scipy.stats import norm\nfrom sklearn.neighbors import KernelDensity\n\n# Plot a 1D density example\n\n# N = 100\n# np.random.seed(1)\n# X = np.concatenate((np.random.normal(0, 1, int(0.3 * N)),\n# np.random.normal(5, 1, int(0.7 * N))))[:, np.newaxis]\n\nX = np.c_[append4]\n\nX_plot = np.linspace(0, max(append4), 1000)[:, np.newaxis]\n\ntrue_dens = (0.3 * norm(0, 1).pdf(X_plot[:, 0]) + 0.7 * norm(5, 1).pdf(X_plot[:, 0]))\n\nfig, ax = plt.subplots()\nax.fill(X_plot[:, 0], true_dens, fc='black', alpha=0.2, label='input distribution')\n\nfor kernel in ['epanechnikov', 'tophat', 'gaussian']:\n kde = KernelDensity(kernel=kernel, bandwidth=0.5).fit(X)\n log_dens = kde.score_samples(X_plot)\n ax.plot(X_plot[:, 0], np.exp(log_dens), '-',\n label=\"kernel = '{0}'\".format(kernel))\n\nax.text(6, 0.38, \"N={0} points\".format(N))\n\nax.legend(loc='upper left')\nax.plot(X[:, 0], -0.005 - 0.01 * np.random.random(X.shape[0]), '+k')\n\nax.set_xlim(0, 1)\nax.set_ylim(-0.02, 0.4)\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf37221da5cab59c96dcb43941230b828e9d1e4 | 42,429 | ipynb | Jupyter Notebook | dli-learning-path/accelerate-ml-with-gpu/notebook/XGBoost-on-skLearn-cuML-snapML.ipynb | robin2008/wmla-assets | 93617cade8deddbd94d2a5bc8c5d19e5d1d0b516 | [
"Apache-2.0"
] | 5 | 2019-09-04T02:26:07.000Z | 2022-01-25T13:39:30.000Z | dli-learning-path/accelerate-ml-with-gpu/notebook/XGBoost-on-skLearn-cuML-snapML.ipynb | robin2008/wmla-assets | 93617cade8deddbd94d2a5bc8c5d19e5d1d0b516 | [
"Apache-2.0"
] | 11 | 2019-10-01T03:12:12.000Z | 2022-02-10T14:06:34.000Z | dli-learning-path/accelerate-ml-with-gpu/notebook/XGBoost-on-skLearn-cuML-snapML.ipynb | robin2008/wmla-assets | 93617cade8deddbd94d2a5bc8c5d19e5d1d0b516 | [
"Apache-2.0"
] | 30 | 2019-08-23T18:05:05.000Z | 2022-03-02T15:16:14.000Z | 33.861931 | 487 | 0.507318 | [
[
[
"# Train a Xgboost Model with Watson Machine Learning \n\nNotebook created by Zeming Zhao on June, 2021\n\nXGBoost is an implementation of gradient boosted decision trees designed for speed and performance. which is an algorithm that has recently been dominating applied machine learning and Kaggle competitions for structured or tabular data.\n\nSnapBoost implements a boosting machine that can be used to construct an ensemble of decision trees. It can be used for both clasification and regression tasks. In constrast to other boosting frameworks, Snap ML’s boosting machine dose not utilize a fixed maximal tree depth at each boosting iteration. Instead, the tree depth is sampled at each boosting iteration according to a discrete uniform distribution. The fit and predict functions accept numpy.ndarray data structures.\n\nThis notebook covers the following sections:\n\n1. [Setup Xgboost Model using xgboost lib](#xgboost-model)<br>\n\n1. [Training the model on CPU with Watson Machine Learning Accelerator](#xgboost-cpu)<br>\n\n1. [Training the model on GPU with Watson Machine Learning Accelerator](#xgboost-gpu)<br>\n\n1. [Setup SnapBoost Model using snapML lib on CPU](#snapml-model)<br>\n\n1. [Training the model on CPU with Watson Machine Learning Accelerator](#snapml-cpu)<br>\n\n1. [Setup SnapBoost Model using snapML lib on GPU](#snapml-model-gpu)<br>\n\n1. [Training the model on GPU with Watson Machine Learning Accelerator](#snapml-gpu)<br>",
"_____no_output_____"
],
[
"## Preparations\n### Prepare directory and file for writing Xgboost engine.",
"_____no_output_____"
]
],
[
[
"from pathlib import Path\nmodel_dir = f'/project_data/data_asset/models' \nmodel_base_name = f'main.py'\nPath(model_dir).mkdir(exist_ok=True)\nprint(\"create model directory done.\")",
"create model directory done.\n"
]
],
[
[
"<a id = \"xgboost-model\"></a>\n## Step 1 : Setup Xgboost model\n### create a Xgboost Model based on Xgboost lib ",
"_____no_output_____"
]
],
[
[
"model_main='xgboost-'+model_base_name",
"_____no_output_____"
],
[
"%%writefile {model_dir}/{model_main}\n\nimport os, datetime\nimport numpy as np\nimport pandas as pd\nfrom sklearn.datasets import make_classification\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import precision_score, recall_score, accuracy_score\nimport xgboost as xgb\n\nos.environ['KMP_DUPLICATE_LIB_OK']='True'\n\n# Define Parameters for a large regression\nn_samples = 2**13 \nn_features = 899 \nn_info = 600 \ndata_type = np.float32\n\n# Generate Data using scikit-learn\nX,y = make_classification(n_samples=n_samples,\n n_features=n_features,\n n_informative=n_info,\n random_state=123, n_classes=2)\n\nX = pd.DataFrame(X.astype(data_type))\ny = pd.Series(y.astype(np.int32))\n\nX_train, X_test, y_train, y_test = train_test_split(X, y,\n test_size = 0.2,\n random_state=0)\n\nprint(\"Number of examples: %d\" % (X_train.shape[0]))\nprint(\"Number of features: %d\" % (X_train.shape[1]))\nprint(\"Number of classes: %d\" % (len(np.unique(y_train))))\n\nD_train = xgb.DMatrix(X_train, label=y_train)\nD_test = xgb.DMatrix(X_test, label=y_test)\n\n# set parameters\nparam = {\n 'eta': 0.3, \n 'max_depth': 6, \n 'objective': 'multi:softprob', \n 'num_class': 3} \n\nsteps = 20 # The number of training iterations\n\n# setup model and train\nstart = datetime.datetime.now()\nmodel = xgb.train(param, D_train, steps)\nend = datetime.datetime.now()\nprint (\"Xgboost train timecost: %.2gs\" % ((end-start).total_seconds()))\n\n# predict\nstart = datetime.datetime.now()\npreds = model.predict(D_test)\nend = datetime.datetime.now()\nprint (\"Xgboost predict timecost: %.2gs\" % ((end-start).total_seconds()))\n\n# check result\nbest_preds = np.asarray([np.argmax(line) for line in preds])\nprint(\"Precision = {}\".format(precision_score(y_test, best_preds, average='macro')))\nprint(\"Recall = {}\".format(recall_score(y_test, best_preds, average='macro')))\nprint(\"Accuracy = {}\".format(accuracy_score(y_test, best_preds)))\n\n# save the xgboost model into a file\nimport pickle\nfilename = './xgboost_model.pkl'\npickle.dump(model, open(filename, 'wb')) \nprint(\"Xgboost model saved successfully.\")",
"Overwriting /project_data/data_asset/models/xgboost-main.py\n"
]
],
[
[
"<a id = \"xgboost-cpu\"></a>\n## Step 2 : Training the Xgboost model on CPU with Watson Machine Learning Accelerator\n### Prepare the libs for job submission",
"_____no_output_____"
]
],
[
[
"import requests\nfrom requests.packages.urllib3.exceptions import InsecureRequestWarning\nrequests.packages.urllib3.disable_warnings(InsecureRequestWarning)\n\nfrom matplotlib import pyplot as plt\n%pylab inline\n\nimport base64\nimport json\nimport time\nimport urllib",
"Populating the interactive namespace from numpy and matplotlib\n"
]
],
[
[
"### Configuring your environment and project details\nTo set up your project details, provide your credentials in this cell. You must include your cluster URL, username, and password.",
"_____no_output_____"
]
],
[
[
"# please enter Watson Machine Learning Accelerator host name\nhostname='wmla-console-wmla.apps.dse-perf.cpolab.ibm.com'\n# login='username:password' # please enter the login and password\nlogin='mluser1:mluser1'\nes = base64.b64encode(login.encode('utf-8')).decode(\"utf-8\")\n# print(es)\ncommonHeaders={'Authorization': 'Basic '+es}\nreq = requests.Session()\nauth_url = 'https://{}/auth/v1/logon'.format(hostname)\nprint(auth_url)\na=requests.get(auth_url,headers=commonHeaders, verify=False)\naccess_token=a.json()['accessToken']\n# print(\"Access_token: \", access_token)\ndl_rest_url = 'https://{}/platform/rest/deeplearning/v1'.format(hostname)\ncommonHeaders={'accept': 'application/json', 'X-Auth-Token': access_token}\nreq = requests.Session()\n# Health check\nconfUrl = 'https://{}/platform/rest/deeplearning/v1/conf'.format(hostname)\nr = req.get(confUrl, headers=commonHeaders, verify=False)",
"https://wmla-console-wmla.apps.dse-perf.cpolab.ibm.com/auth/v1/logon\n"
]
],
[
[
"### Define the status checking fuction",
"_____no_output_____"
]
],
[
[
"import tarfile\nimport tempfile\nimport os\nimport json\nimport pprint\nimport pandas as pd\nfrom IPython.display import clear_output\n\ndef query_job_status(job_id,refresh_rate=3) :\n\n execURL = dl_rest_url +'/execs/'+ job_id['id']\n pp = pprint.PrettyPrinter(indent=2)\n\n keep_running=True\n res=None\n while(keep_running):\n res = req.get(execURL, headers=commonHeaders, verify=False)\n monitoring = pd.DataFrame(res.json(), index=[0])\n pd.set_option('max_colwidth', 120)\n clear_output()\n print(\"Refreshing every {} seconds\".format(refresh_rate))\n display(monitoring)\n pp.pprint(res.json())\n if(res.json()['state'] not in ['PENDING_CRD_SCHEDULER', 'SUBMITTED','RUNNING']) :\n keep_running=False\n time.sleep(refresh_rate)\n return res",
"_____no_output_____"
]
],
[
[
"### Define the job submission fuction",
"_____no_output_____"
]
],
[
[
"def submit_job_to_wmla (args, files) :\n starttime = datetime.datetime.now()\n r = requests.post(dl_rest_url+'/execs?args='+args, files=files,\n headers=commonHeaders, verify=False)\n if not r.ok:\n print('submit job failed: code=%s, %s'%(r.status_code, r.content))\n job_status = query_job_status(r.json(),refresh_rate=5)\n endtime = datetime.datetime.now()\n print(\"\\nTotallly training cost: \", (endtime - starttime).seconds, \" seconds.\")",
"_____no_output_____"
]
],
[
[
"### Define the submittion parameters",
"_____no_output_____"
]
],
[
[
"# specify the model file, conda env, device type and device number\nargs = '--exec-start tensorflow --cs-datastore-meta type=fs \\\n--workerDeviceNum 1 \\\n--workerMemory 32G \\\n--workerDeviceType cpu \\\n--conda-env-name rapids-21.06-new \\\n--model-main ' + model_main\nprint(args)",
"--exec-start tensorflow --cs-datastore-meta type=fs --workerDeviceNum 1 --workerMemory 32G --workerDeviceType cpu --conda-env-name rapids-21.06-new --model-main xgboost-main.py\n"
],
[
"files = {'file': open(\"{0}/{1}\".format(model_dir,model_main),'rb')}\nsubmit_job_to_wmla (args, files)",
"Refreshing every 5 seconds\n"
]
],
[
[
"<a id = \"xgboost-gpu\"></a>\n## Step 3 : Training the Xgboost model on GPU with Watson Machine Learning Accelerator\n### Define the submittion parameters using conda env with GPU version XGBoost",
"_____no_output_____"
]
],
[
[
"# specify the conda env of xgboost and worker device type\nargs = '--exec-start tensorflow --cs-datastore-meta type=fs \\\n--workerDeviceNum 1 \\\n--workerMemory 32G \\\n--workerDeviceType gpu \\\n--conda-env-name dlipy3 \\\n--model-main ' + model_main\nprint(args)",
"--exec-start tensorflow --cs-datastore-meta type=fs --workerDeviceNum 1 --workerMemory 32G --workerDeviceType gpu --conda-env-name dlipy3 --model-main xgboost-main.py\n"
],
[
"files = {'file': open(\"{0}/{1}\".format(model_dir,model_main),'rb')}\nsubmit_job_to_wmla (args, files)",
"Refreshing every 5 seconds\n"
]
],
[
[
"<a id = \"snapml-model\"></a>\n## Step 4 : Setup Snap Boosting model using snapML\n### Create a Snap Boosting model based on snapML ",
"_____no_output_____"
]
],
[
[
"model_main='snapboost-'+model_base_name",
"_____no_output_____"
],
[
"%%writefile {model_dir}/{model_main}\n\nimport os, datetime\nimport numpy as np\nimport pandas as pd\nfrom sklearn.datasets import make_classification\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import precision_score, recall_score, accuracy_score\nfrom snapml import BoostingMachineClassifier as SnapBoostingMachineClassifier\n\nos.environ['KMP_DUPLICATE_LIB_OK']='True'\n\n# Define Parameters for a large regression\nn_samples = 2**13 \nn_features = 899 \nn_info = 600 \ndata_type = np.float32\n\n# Generate Data using scikit-learn\nX,y = make_classification(n_samples=n_samples,\n n_features=n_features,\n n_informative=n_info,\n random_state=123, n_classes=2)\n\nX = pd.DataFrame(X.astype(data_type))\ny = pd.Series(y.astype(np.int32))\n\nX_train, X_test, y_train, y_test = train_test_split(X, y,\n test_size = 0.2,\n random_state=0)\n\nprint(\"Number of examples: %d\" % (X_train.shape[0]))\nprint(\"Number of features: %d\" % (X_train.shape[1]))\nprint(\"Number of classes: %d\" % (len(np.unique(y_train))))\n\nsnap_model = SnapBoostingMachineClassifier(random_state=42, \n n_jobs=1,\n hist_nbins=256,\n num_round=20) # same interations number with xgboost\n\n# setup model and train\nstart = datetime.datetime.now()\nsnap_model.fit(X_train.values, y_train.values) #.train(param, D_train, steps)\nend = datetime.datetime.now()\nprint (\"Xgboost train timecost: %.2gs\" % ((end-start).total_seconds()))\n\n# predict\nstart = datetime.datetime.now()\npreds = snap_model.predict(X_test.values)\nend = datetime.datetime.now()\nprint (\"Xgboost predict timecost: %.2gs\" % ((end-start).total_seconds()))\n\n# check result\nbest_preds = np.asarray([np.argmax(line) for line in preds])\nprint(\"Precision = {}\".format(precision_score(y_test, best_preds, average='macro')))\nprint(\"Recall = {}\".format(recall_score(y_test, best_preds, average='macro')))\nprint(\"Accuracy = {}\".format(accuracy_score(y_test, best_preds)))\n\n# save the xgboost model into a file\nimport pickle\nfilename = './snapboost_model.pkl'\npickle.dump(snap_model, open(filename, 'wb')) \nprint(\"Snapboost model saved successfully.\")",
"Overwriting /project_data/data_asset/models/snapboost-gpu-main.py\n"
]
],
[
[
"<a id = \"snapml-cpu\"></a>\n## Step 5 : Training the SnapML model on CPU with Watson Machine Learning Accelerator\n### Re-define the submission parameters",
"_____no_output_____"
]
],
[
[
"# specify the model file, conda env, device type and device number\nargs = '--exec-start tensorflow --cs-datastore-meta type=fs \\\n--workerDeviceNum 1 \\\n--workerMemory 32G \\\n--workerDeviceType cpu \\\n--conda-env-name snapml-177rc \\\n--model-main ' + model_main\nprint(args)",
"--exec-start tensorflow --cs-datastore-meta type=fs --workerDeviceNum 1 --workerMemory 32G --workerDeviceType cpu --conda-env-name snapml-177rc --model-main snapboost-gpu-main.py\n"
],
[
"files = {'file': open(\"{0}/{1}\".format(model_dir,model_main),'rb')}\nsubmit_job_to_wmla (args, files)",
"Refreshing every 5 seconds\n"
]
],
[
[
"<a id = \"snapml-model-gpu\"></a>\n## Step 6 : Setup SnapBoost model using snapML on GPU\n### Create a SnapBoost model based on snapML ",
"_____no_output_____"
]
],
[
[
"model_main='snapboost-gpu-'+model_base_name",
"_____no_output_____"
],
[
"%%writefile {model_dir}/{model_main}\n\nimport os, datetime\nimport numpy as np\nimport pandas as pd\nfrom sklearn.datasets import make_classification\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import precision_score, recall_score, accuracy_score\nfrom snapml import BoostingMachineClassifier as SnapBoostingMachineClassifier\n\nos.environ['KMP_DUPLICATE_LIB_OK']='True'\n\n# Define Parameters for a large regression\nn_samples = 2**13 \nn_features = 899 \nn_info = 600 \ndata_type = np.float32\n\n# Generate Data using scikit-learn\nX,y = make_classification(n_samples=n_samples,\n n_features=n_features,\n n_informative=n_info,\n random_state=123, n_classes=2)\n\nX = pd.DataFrame(X.astype(data_type))\ny = pd.Series(y.astype(np.int32))\n\nX_train, X_test, y_train, y_test = train_test_split(X, y,\n test_size = 0.2,\n random_state=0)\n\nprint(\"Number of examples: %d\" % (X_train.shape[0]))\nprint(\"Number of features: %d\" % (X_train.shape[1]))\nprint(\"Number of classes: %d\" % (len(np.unique(y_train))))\n\nsnap_model = SnapBoostingMachineClassifier(use_gpu=True, # use gpu\n random_state=42, \n n_jobs=1,\n hist_nbins=256,\n num_round=20) # same interations number with xgboost\n\n# setup model and train\nstart = datetime.datetime.now()\nsnap_model.fit(X_train.values, y_train.values) #.train(param, D_train, steps)\nend = datetime.datetime.now()\nprint (\"Xgboost train timecost: %.2gs\" % ((end-start).total_seconds()))\n\n# predict\nstart = datetime.datetime.now()\npreds = snap_model.predict(X_test.values)\nend = datetime.datetime.now()\nprint (\"Xgboost predict timecost: %.2gs\" % ((end-start).total_seconds()))\n\n# check result\nbest_preds = np.asarray([np.argmax(line) for line in preds])\nprint(\"Precision = {}\".format(precision_score(y_test, best_preds, average='macro')))\nprint(\"Recall = {}\".format(recall_score(y_test, best_preds, average='macro')))\nprint(\"Accuracy = {}\".format(accuracy_score(y_test, best_preds)))\n\n# save the xgboost model into a file\nimport pickle\nfilename = './snapboost_model.pkl'\npickle.dump(snap_model, open(filename, 'wb')) \nprint(\"Snapboost model saved successfully.\")",
"Overwriting /project_data/data_asset/models/snapboost-gpu-main.py\n"
]
],
[
[
"<a id = \"snapml-gpu\"></a>\n## Step 7 : Training the SnapML model on GPU with Watson Machine Learning Accelerator\n### Re-define the submission parameters",
"_____no_output_____"
]
],
[
[
"# specify the model file, conda env, device type and device number\nargs = '--exec-start tensorflow --cs-datastore-meta type=fs \\\n--workerDeviceNum 1 \\\n--workerMemory 32G \\\n--workerDeviceType gpu \\\n--conda-env-name snapml-177rc \\\n--model-main ' + model_main\n# --msd-env CUDA_FORCE_PTX_JIT=1 \\\nprint(args)",
"--exec-start tensorflow --cs-datastore-meta type=fs --workerDeviceNum 1 --workerMemory 32G --workerDeviceType gpu --conda-env-name snapml-177rc --model-main snapboost-gpu-main.py\n"
],
[
"#files = {'file': open('/project_data/data_asset/models/snapboost-gpu-main.py', 'rb')}\nfiles = {'file': open(\"{0}/{1}\".format(model_dir,model_main),'rb')}\nsubmit_job_to_wmla (args, files)",
"Refreshing every 5 seconds\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecf37e6a7075430a23e5a7f7077397e5fbfbff0f | 543,089 | ipynb | Jupyter Notebook | notebook.ipynb | MarvelAmazon/titanic-data-analyis | cc8ed2e2ac5b393c05c627cfd259908c354273a1 | [
"MIT"
] | null | null | null | notebook.ipynb | MarvelAmazon/titanic-data-analyis | cc8ed2e2ac5b393c05c627cfd259908c354273a1 | [
"MIT"
] | null | null | null | notebook.ipynb | MarvelAmazon/titanic-data-analyis | cc8ed2e2ac5b393c05c627cfd259908c354273a1 | [
"MIT"
] | 1 | 2020-06-22T20:25:34.000Z | 2020-06-22T20:25:34.000Z | 165.273585 | 56,492 | 0.878307 | [
[
[
"## Variable Notes\n<ul>\n<li>\n<ol>\npclass: A proxy for socio-economic status (SES)\n<li>1st = Upper </li>\n<li>2nd = Middle </li>\n<li>3rd = Lower </li>\n</ol>\n</li>\n\n<li>age: Age is fractional if less than 1. If the age is estimated, is it in the form of xx.5</li>\n\n<li>sibsp: The dataset defines family relations in this way...\n<ul>\n<li>Sibling = brother, sister, stepbrother, stepsister</li>\n<li>Spouse = husband, wife (mistresses and fiancés were ignored)</li>\n</ul>\n</li>\n\n<li>parch: The dataset defines family relations in this way...\n<ul>\n<li>Parent = mother, father</li>\n<li>Child = daughter, son, stepdaughter, stepson</li>\n<li>Some children travelled only with a nanny, therefore parch=0 for them.</li>\n</ul>\n</li>\n</ul>",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n!pip install matplotlib",
"Requirement already satisfied: matplotlib in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (3.1.3)\nRequirement already satisfied: python-dateutil>=2.1 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from matplotlib) (2.8.1)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from matplotlib) (2.4.6)\nRequirement already satisfied: cycler>=0.10 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from matplotlib) (0.10.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from matplotlib) (1.1.0)\nRequirement already satisfied: numpy>=1.11 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from matplotlib) (1.17.2)\nRequirement already satisfied: six>=1.5 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from python-dateutil>=2.1->matplotlib) (1.15.0)\nRequirement already satisfied: setuptools in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from kiwisolver>=1.0.1->matplotlib) (45.2.0.post20200210)\n"
],
[
"!pip install Flask-Bootstrap4",
"Requirement already satisfied: Flask-Bootstrap4 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (4.0.2)\nRequirement already satisfied: visitor in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from Flask-Bootstrap4) (0.1.3)\nRequirement already satisfied: Flask>=0.8 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from Flask-Bootstrap4) (1.1.2)\nRequirement already satisfied: dominate in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from Flask-Bootstrap4) (2.5.1)\nRequirement already satisfied: Jinja2>=2.10.1 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from Flask>=0.8->Flask-Bootstrap4) (2.11.2)\nRequirement already satisfied: itsdangerous>=0.24 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from Flask>=0.8->Flask-Bootstrap4) (1.1.0)\nRequirement already satisfied: click>=5.1 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from Flask>=0.8->Flask-Bootstrap4) (7.1.2)\nRequirement already satisfied: Werkzeug>=0.15 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from Flask>=0.8->Flask-Bootstrap4) (1.0.1)\nRequirement already satisfied: MarkupSafe>=0.23 in c:\\users\\yvel marcelin\\anaconda3\\lib\\site-packages (from Jinja2>=2.10.1->Flask>=0.8->Flask-Bootstrap4) (1.1.1)\n"
]
],
[
[
"<img src=\"ressources/image1.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image2.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image3.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image16.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image17.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image18.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image21.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image4.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image5.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image6.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image7.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image8.png\">",
"_____no_output_____"
]
],
[
[
"import numpy as np\nX = np.array([1,2,4,3,4,2,4,3,4,3])\nY1 = 3*X\nY = np.array([2,5,4,7.5,6,2,1,2,1,6])\nY2 =np.array([2.1,4,8.5,6.1,8.2,4,6,6,6,4])\nlen(Y2) == len(X)",
"_____no_output_____"
],
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nmy_dict = dict(x =X,y=Y)\ndf = pd.DataFrame(my_dict)\ndf.head()\ndf.corr()\n\n",
"_____no_output_____"
],
[
"plt.scatter(X,Y)\nplt.plot(X,3*X)",
"_____no_output_____"
],
[
"import seaborn as sns; sns.set(color_codes=True)\ntips = sns.load_dataset(\"tips\")\nax = sns.regplot(x=X, y=Y1, data=tips)",
"_____no_output_____"
],
[
"import seaborn as sns; sns.set(color_codes=True)\ntips = sns.load_dataset(\"tips\")\nax = sns.regplot(x=X, y=Y, data=tips)",
"_____no_output_____"
],
[
"import seaborn as sns; sns.set(color_codes=True)\ntips = sns.load_dataset(\"tips\")\nax = sns.regplot(x=X, y=-Y2, data=tips)\nmy_dict = dict(x =X,y=-Y2,y1=Y)\n\n",
"_____no_output_____"
],
[
"X = np.array([1,2,4,5,3,5,3,4,2,3,2])\nlen(X)",
"_____no_output_____"
],
[
"X1= -2*X",
"_____no_output_____"
],
[
"X2 = [1,4.2,2,3,5,2,4,5,2,4,2]\nlen(X2)",
"_____no_output_____"
],
[
"Y = 2*X1 +X2\nmap_dict= dict(x=X,x1=X1,x2=X2,y=Y)\ndf = pd.DataFrame(map_dict)",
"_____no_output_____"
],
[
"import seaborn as sns; sns.set(color_codes=True)\ntips = sns.load_dataset(\"tips\")\nax = sns.regplot(x=X, y=Y, data=tips)\n",
"_____no_output_____"
],
[
"import seaborn as sns; sns.set(color_codes=True)\ntips = sns.load_dataset(\"tips\")\nax = sns.regplot(x=X1, y=Y, data=tips)\n",
"_____no_output_____"
],
[
"import seaborn as sns; sns.set(color_codes=True)\ntips = sns.load_dataset(\"tips\")\nax = sns.regplot(x=X2, y=Y, data=tips)",
"_____no_output_____"
],
[
"df.corr()",
"_____no_output_____"
],
[
"#Step5: Annotate each cell with the numeric value\nfig, ax = plt.subplots(figsize=(10,6))\nsns.heatmap(df.corr(), center=0, cmap='BrBG', annot=True) #adjusting the color and adding annotation (actual correlation values) makes it easier to form a conclusion ",
"_____no_output_____"
]
],
[
[
"<img src=\"ressources/image9.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image10.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image11.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image12.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image13.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image14.png\">",
"_____no_output_____"
],
[
"<img src=\"ressources/image15.png\">",
"_____no_output_____"
],
[
"# 1. Import all librairies and datasets",
"_____no_output_____"
]
],
[
[
"# import all librairies needed for this model\n\nimport pandas as pd # for dataframe computaion\nimport numpy as np # vector computation\nimport matplotlib.pyplot as plt # for plot\nimport seaborn as sns # for plot\nfrom sklearn.preprocessing import LabelEncoder, StandardScaler # for data preprocessing\nfrom sklearn.linear_model import LogisticRegression # for logistic regression\nfrom sklearn.model_selection import train_test_split # for splitting and train and test datastet randomly\nfrom sklearn.metrics import classification_report # for metrics and model evaluation\nfrom sklearn.impute import SimpleImputer # for data preprocessing",
"_____no_output_____"
],
[
"# import dataset \nurl_test = \"datasets/test.csv\"\nurl_train= \"datasets/train.csv\"",
"_____no_output_____"
],
[
"train_df = pd.read_csv(url_train)\ntest_df = pd.read_csv(url_test)",
"_____no_output_____"
]
],
[
[
"# 2. Missing Values",
"_____no_output_____"
]
],
[
[
"train_df.isnull().sum()",
"_____no_output_____"
],
[
"np.round(train_df.isna().sum()/train_df.shape[0],2)",
"_____no_output_____"
],
[
"cols = [col for col in train_df.columns if col not in ['PassengerId','Cabin']]\ntrain_df = train_df[cols]",
"_____no_output_____"
],
[
"train_df.isna().sum()",
"_____no_output_____"
],
[
"# SimpleImputer\n\n# Embarked column\nsimple_1 = SimpleImputer(missing_values=np.nan, strategy='most_frequent')\nvalues = simple_1.fit_transform(train_df[['Embarked']].values)\ntrain_df['Embarked'] = values",
"_____no_output_____"
],
[
"# Embarked column\nsimple_1 = SimpleImputer(missing_values=np.nan, strategy='median')\nvalues = simple_1.fit_transform(train_df[['Age']].values)\ntrain_df['Age'] = values",
"_____no_output_____"
],
[
"train_df.isna().sum()",
"_____no_output_____"
],
[
"col_cat = ['Survived','Pclass','Sex','SibSp','Parch','Embarked']\nfor col in col_cat:\n print(col, train_df[col].unique())",
"Survived [0 1]\nPclass [3 1 2]\nSex ['male' 'female']\nSibSp [1 0 3 4 2 5 8]\nParch [0 1 2 5 3 4 6]\nEmbarked ['S' 'C' 'Q']\n"
],
[
"map_sex = {'male':1,'female':0}\ntrain_df['Sex'] = train_df['Sex'].replace(map_sex)",
"_____no_output_____"
],
[
"embarked_map = {'S':0,'C':1,'Q':2}\ntrain_df['Embarked'] = train_df['Embarked'].replace(embarked_map)",
"_____no_output_____"
],
[
"col_cat = ['Survived','Pclass','Sex','SibSp','Parch','Embarked']\nfor col in col_cat:\n print(col, train_df[col].unique())",
"Survived [0 1]\nPclass [3 1 2]\nSex [1 0]\nSibSp [1 0 3 4 2 5 8]\nParch [0 1 2 5 3 4 6]\nEmbarked [0 1 2]\n"
],
[
"train_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 891 entries, 0 to 890\nData columns (total 10 columns):\nSurvived 891 non-null int64\nPclass 891 non-null int64\nName 891 non-null object\nSex 891 non-null int64\nAge 891 non-null float64\nSibSp 891 non-null int64\nParch 891 non-null int64\nTicket 891 non-null object\nFare 891 non-null float64\nEmbarked 891 non-null int64\ndtypes: float64(2), int64(6), object(2)\nmemory usage: 69.7+ KB\n"
]
],
[
[
"# 3. Verify well data types",
"_____no_output_____"
]
],
[
[
"train_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 891 entries, 0 to 890\nData columns (total 10 columns):\nSurvived 891 non-null int64\nPclass 891 non-null int64\nName 891 non-null object\nSex 891 non-null int64\nAge 891 non-null float64\nSibSp 891 non-null int64\nParch 891 non-null int64\nTicket 891 non-null object\nFare 891 non-null float64\nEmbarked 891 non-null int64\ndtypes: float64(2), int64(6), object(2)\nmemory usage: 69.7+ KB\n"
],
[
"test_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 418 entries, 0 to 417\nData columns (total 11 columns):\nPassengerId 418 non-null int64\nPclass 418 non-null int64\nName 418 non-null object\nSex 418 non-null object\nAge 332 non-null float64\nSibSp 418 non-null int64\nParch 418 non-null int64\nTicket 418 non-null object\nFare 417 non-null float64\nCabin 91 non-null object\nEmbarked 418 non-null object\ndtypes: float64(2), int64(4), object(5)\nmemory usage: 36.0+ KB\n"
]
],
[
[
"# 4. Find oultiers",
"_____no_output_____"
],
[
"## 1 Example",
"_____no_output_____"
]
],
[
[
"Q1 = train_df.describe().T['25%']\nQ3 = train_df.describe().T['75%']\ntrain_IQR = Q3-Q1\ntrain_IQR",
"_____no_output_____"
],
[
"Q1 = test_df.describe().T['25%']\nQ3 = test_df.describe().T['75%']\ntest_IQR = Q3-Q1\ntest_IQR",
"_____no_output_____"
]
],
[
[
"## Example 2",
"_____no_output_____"
]
],
[
[
"#Step1: Calculate the IQR\ntrain_df_iqr = train_df\nQ1 = train_df_iqr.quantile(0.25)\nQ3 = train_df_iqr.quantile(0.75)\nIQR = Q3 - Q1\nMIN_RANGE = Q1 - 1.5 * IQR\nMAX_RANGE = Q3+ 1.5 *IQR\n(train_df_iqr < (Q1 - 1.5 * IQR))",
"_____no_output_____"
],
[
"\ndf_outliers = (train_df_iqr < (Q1 - 1.5 * IQR)) |(train_df_iqr > (Q3 + 1.5 * IQR))\ndf_outliers.head()\ntrain_out = train_df_iqr[~((train_df_iqr < (Q1 - 1.5 * IQR)) |(train_df_iqr > (Q3 + 1.5 * IQR))).any(axis=1)]\ntrain_out.shape\ntrain_out.tail()",
"_____no_output_____"
],
[
"percent_ =round( (train_out.shape[0] / train_df.shape[0]) *100,2)\npercent_",
"_____no_output_____"
],
[
"train_df.boxplot()",
"_____no_output_____"
]
],
[
[
"# 5. Removes duplicates",
"_____no_output_____"
]
],
[
[
"train_out.shape",
"_____no_output_____"
],
[
"train_out.drop_duplicates(inplace=True)\ntrain_out.shape",
"C:\\Users\\Yvel Marcelin\\anaconda3\\lib\\site-packages\\ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
]
],
[
[
"# 6. Exploratory Data Analysis",
"_____no_output_____"
],
[
"## UNIVARIATE EDA",
"_____no_output_____"
]
],
[
[
"col_cat",
"_____no_output_____"
],
[
"for col in col_cat:\n display(train_df.groupby(col).size().to_frame().rename(columns={0:'count'}))",
"_____no_output_____"
],
[
"# Survivded\ndf =train_df.groupby(['Survived']).size().to_frame().rename(columns={0:'count'}).reset_index()\nlabels= ['dead','alive']\nvalues= df['count']\ncolors = ['#FA8F0A','#F0DA32']\nexplode = (0.1, 0)\nplt.pie(values, labels=labels, colors=colors, autopct='%1.1f%%', explode=explode, shadow=True)\nplt.show()",
"_____no_output_____"
],
[
"# Survivded\ndf =train_df[train_df.Survived == 0].groupby(['Sex']).size().to_frame().rename(columns={0:'count'}).reset_index()\nlabels= ['female','male']\nvalues= df['count']\ncolors = ['#FA8F0A','#F0DA32']\nexplode = (0.1, 0)\nplt.pie(values, labels=labels, colors=colors, autopct='%1.1f%%', explode=explode, shadow=True)",
"_____no_output_____"
],
[
"# Survivded\ndf =train_df[train_df.Survived == 0].groupby(['Pclass']).size().to_frame().rename(columns={0:'count'}).reset_index()\nlabels=df['Pclass'] \nvalues= df['count']\ncolors = ['#FA8F0A','#F0DA32','red']\nexplode = (0.1, 0,0)\nplt.pie(values, labels=labels, colors=colors, autopct='%1.1f%%', explode=explode, shadow=True)",
"_____no_output_____"
],
[
"df.plot(kind='barh',x='Pclass',y = 'count')",
"_____no_output_____"
]
],
[
[
"## Numerical values",
"_____no_output_____"
]
],
[
[
"col_num = [x for x in train_df.columns if x not in col_cat+['Name','Ticket']]\ncol_num",
"_____no_output_____"
],
[
"df_num = train_df[col_num]\ndf_num[col_num[0]].plot(kind='hist')",
"_____no_output_____"
],
[
"sns.distplot(df_num[col_num[0]], bins=10, kde=True, rug=True);",
"_____no_output_____"
],
[
"df_num = train_df[col_num]\ndf_num[col_num[1]].plot(kind='hist')",
"_____no_output_____"
],
[
"sns.distplot(df_num[col_num[1]], bins=10, kde=True, rug=True);",
"_____no_output_____"
],
[
"import seaborn as sns; sns.set(color_codes=True)\nax = sns.regplot(x=col_num[0], y=col_num[1], data=df_num)",
"_____no_output_____"
],
[
"train_df.corr()",
"_____no_output_____"
],
[
"#Step5: Annotate each cell with the numeric value\nfig, ax = plt.subplots(figsize=(10,6))\nsns.heatmap(train_df.corr(), center=0, cmap='BrBG', annot=True) ",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(10,6))\nsns.heatmap(train_out.corr(), center=0, cmap='BrBG', annot=True) ",
"_____no_output_____"
],
[
"def change_age_rage(x):\n if x >= 20 and x<= 40:\n return 1\n return 0",
"_____no_output_____"
],
[
"def change_fare_rage(x):\n if x >= 0 and x<= 100:\n return 1\n return 0",
"_____no_output_____"
],
[
"train_df['Fare'] = train_df['Fare'].apply(lambda x : change_fare_rage(x))\n#Step5: Annotate each cell with the numeric value\nfig, ax = plt.subplots(figsize=(10,6))\nsns.heatmap(train_df.corr(), center=0, annot=True) ",
"_____no_output_____"
],
[
"# Null values \ndisplay(train_df.isnull().sum())\n\n",
"_____no_output_____"
],
[
"display(train_df.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 891 entries, 0 to 890\nData columns (total 10 columns):\nSurvived 891 non-null int64\nPclass 891 non-null int64\nName 891 non-null object\nSex 891 non-null int64\nAge 891 non-null float64\nSibSp 891 non-null int64\nParch 891 non-null int64\nTicket 891 non-null object\nFare 891 non-null int64\nEmbarked 891 non-null int64\ndtypes: float64(1), int64(7), object(2)\nmemory usage: 69.7+ KB\n"
],
[
"np.round(train_df.isna().sum()/train_df.shape[0],2)",
"_____no_output_____"
],
[
"train_df.describe()\n",
"_____no_output_____"
],
[
"train_df.boxplot()",
"_____no_output_____"
],
[
"columns = [x for x in train_df.columns if x not in ['PassengerId','Cabin']]",
"_____no_output_____"
],
[
"for col in columns:\n print(col,len(train_df[col].unique()) )",
"Survived 2\nPclass 3\nName 891\nSex 2\nAge 88\nSibSp 7\nParch 7\nTicket 681\nFare 2\nEmbarked 3\n"
],
[
"train_df[columns].dtypes",
"_____no_output_____"
],
[
"col_cat = ['Survived','Pclass','Sex','SibSp','Parch','Embarked']\nfor col in col_cat:\n print(col, train_df[col].unique())",
"Survived [0 1]\nPclass [3 1 2]\nSex [1 0]\nSibSp [1 0 3 4 2 5 8]\nParch [0 1 2 5 3 4 6]\nEmbarked [0 1 2]\n"
],
[
"map_sex = {'female': 0,'male': 1}\ntrain_df['Sex_num'] = train_df['Sex'].replace(map_sex)",
"_____no_output_____"
],
[
"col_cat = ['Survived','Pclass','Sex','SibSp','Parch','Embarked']\nfor col in col_cat:\n print(col, train_df[col].unique())",
"_____no_output_____"
],
[
"map_embarked = {'S': 0,'C': 1, 'Q':3}",
"_____no_output_____"
],
[
"train_df['Embarked_Num'] = train_df['Embarked'].replace(map_embarked)",
"_____no_output_____"
],
[
"train_df[['Sex_num','Sex']]",
"_____no_output_____"
],
[
"# SimpleImputer\n\n# Embarked column\nsimple_1 = SimpleImputer(missing_values=np.nan, strategy='most_frequent')\nvalues = simple_1.fit_transform(train_df[['Embarked']].values)\ntrain_df['Embarked'] = values",
"_____no_output_____"
],
[
"train_df['Embarked'].isna().sum()",
"_____no_output_____"
],
[
"train_df",
"_____no_output_____"
],
[
"col =['Survived','Pclass','Name']\n\ndf1 = train_df[col]\n\nfilter_ = df1['Survived'] == 0",
"_____no_output_____"
],
[
"df1[filter_]",
"_____no_output_____"
],
[
"pd.crosstab(df1.Survived, df1.Pclass).apply(lambda r: r/r.sum(), axis=1)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf38cf09cb37fe855cea91b9f7ac0f33fa0bbd7 | 65,942 | ipynb | Jupyter Notebook | Iterations/integrated_model/integrated_model.ipynb | BasemSuleiman/Intelligent_Failure_Prediction | 772318d9242f7c3f476a1c8f5c1aaeb3459b39f6 | [
"MIT"
] | null | null | null | Iterations/integrated_model/integrated_model.ipynb | BasemSuleiman/Intelligent_Failure_Prediction | 772318d9242f7c3f476a1c8f5c1aaeb3459b39f6 | [
"MIT"
] | null | null | null | Iterations/integrated_model/integrated_model.ipynb | BasemSuleiman/Intelligent_Failure_Prediction | 772318d9242f7c3f476a1c8f5c1aaeb3459b39f6 | [
"MIT"
] | null | null | null | 54.542597 | 1,603 | 0.571335 | [
[
[
"## Load pretrained weak models",
"_____no_output_____"
]
],
[
[
"import xgboost as xgb\nfrom sklearn.linear_model import LogisticRegression\nimport pickle\nimport numpy as np\nimport sys\nimport sklearn.svm.classes\nimport pandas as pd\nimport os\n\n\n#path constants\ntrain_path = '../data/final/train/'\ntest_path = '../data/final/test/'\n\n#type constants\nvehicle_types = ['ZVe44', 'ZV573', 'ZV63d', 'ZVfd4', 'ZVa9c', 'ZVa78', 'ZV252']\n\n#label dataframe\nlabel_df = pd.read_csv('../data/final/label.csv', delimiter = ',', encoding = 'utf-8')\n\n#sys.path.append(r'D:/ProgramData/Anaconda3/Lib/site-packages/sklearn/svm/')\n\ncluster_n = 36\n\nok = 2\npos = 1\nnot_found = 0.5\nneg = 0\n\n#load all the cluster oriented models and map them with the corresponding vehicle type\nsaved_model_path = '../models/final/'\nGBDT_map = dict()\nfor vehicle_type in vehicle_types:\n GBDT_list = list()\n df=pd.read_csv(saved_model_path+vehicle_type+'/status.csv', sep=',',header=None)\n df = df.iloc[:,0].to_numpy()\n for i in range(len(df)):\n if df[i] == ok:\n fn = saved_model_path +vehicle_type+'/trainedXGB'+str(i)+'.pkl'\n print(fn)\n with open(fn, 'rb') as file:\n pickle_model = pickle.load(file)\n GBDT_list.append(pickle_model)\n elif df[i] == neg:\n print('neg')\n GBDT_list.append(neg)\n elif df[i] == pos:\n print('pos')\n GBDT_list.append(pos)\n else:\n print('not found')\n GBDT_list.append(not_found)\n GBDT_map[vehicle_type] = GBDT_list",
"../models/final/ZVe44/trainedXGB0.pkl\n../models/final/ZVe44/trainedXGB1.pkl\n../models/final/ZVe44/trainedXGB2.pkl\n../models/final/ZVe44/trainedXGB3.pkl\n../models/final/ZVe44/trainedXGB4.pkl\n../models/final/ZVe44/trainedXGB5.pkl\npos\nnot found\npos\npos\nnot found\nnot found\n../models/final/ZVe44/trainedXGB12.pkl\n../models/final/ZVe44/trainedXGB13.pkl\n../models/final/ZVe44/trainedXGB14.pkl\n../models/final/ZVe44/trainedXGB15.pkl\n../models/final/ZVe44/trainedXGB16.pkl\n../models/final/ZVe44/trainedXGB17.pkl\n../models/final/ZVe44/trainedXGB18.pkl\n../models/final/ZVe44/trainedXGB19.pkl\n../models/final/ZVe44/trainedXGB20.pkl\n../models/final/ZVe44/trainedXGB21.pkl\n../models/final/ZVe44/trainedXGB22.pkl\n../models/final/ZVe44/trainedXGB23.pkl\n../models/final/ZVe44/trainedXGB24.pkl\n../models/final/ZVe44/trainedXGB25.pkl\n../models/final/ZVe44/trainedXGB26.pkl\npos\n../models/final/ZVe44/trainedXGB28.pkl\n../models/final/ZVe44/trainedXGB29.pkl\n../models/final/ZVe44/trainedXGB30.pkl\n../models/final/ZVe44/trainedXGB31.pkl\n../models/final/ZVe44/trainedXGB32.pkl\n../models/final/ZVe44/trainedXGB33.pkl\n../models/final/ZVe44/trainedXGB34.pkl\n../models/final/ZVe44/trainedXGB35.pkl\n../models/final/ZV573/trainedXGB0.pkl\n../models/final/ZV573/trainedXGB1.pkl\n../models/final/ZV573/trainedXGB2.pkl\n../models/final/ZV573/trainedXGB3.pkl\n../models/final/ZV573/trainedXGB4.pkl\n../models/final/ZV573/trainedXGB5.pkl\n../models/final/ZV573/trainedXGB6.pkl\n../models/final/ZV573/trainedXGB7.pkl\nneg\nnot found\nnot found\nnot found\n../models/final/ZV573/trainedXGB12.pkl\n../models/final/ZV573/trainedXGB13.pkl\n../models/final/ZV573/trainedXGB14.pkl\n../models/final/ZV573/trainedXGB15.pkl\n../models/final/ZV573/trainedXGB16.pkl\n../models/final/ZV573/trainedXGB17.pkl\n../models/final/ZV573/trainedXGB18.pkl\n../models/final/ZV573/trainedXGB19.pkl\n../models/final/ZV573/trainedXGB20.pkl\n../models/final/ZV573/trainedXGB21.pkl\n../models/final/ZV573/trainedXGB22.pkl\n../models/final/ZV573/trainedXGB23.pkl\n../models/final/ZV573/trainedXGB24.pkl\n../models/final/ZV573/trainedXGB25.pkl\n../models/final/ZV573/trainedXGB26.pkl\nneg\n../models/final/ZV573/trainedXGB28.pkl\n../models/final/ZV573/trainedXGB29.pkl\n../models/final/ZV573/trainedXGB30.pkl\n../models/final/ZV573/trainedXGB31.pkl\n../models/final/ZV573/trainedXGB32.pkl\n../models/final/ZV573/trainedXGB33.pkl\n../models/final/ZV573/trainedXGB34.pkl\n../models/final/ZV573/trainedXGB35.pkl\n../models/final/ZV63d/trainedXGB0.pkl\n../models/final/ZV63d/trainedXGB1.pkl\n../models/final/ZV63d/trainedXGB2.pkl\n../models/final/ZV63d/trainedXGB3.pkl\n../models/final/ZV63d/trainedXGB4.pkl\n../models/final/ZV63d/trainedXGB5.pkl\n../models/final/ZV63d/trainedXGB6.pkl\n../models/final/ZV63d/trainedXGB7.pkl\n../models/final/ZV63d/trainedXGB8.pkl\nneg\nnot found\nnot found\n../models/final/ZV63d/trainedXGB12.pkl\n../models/final/ZV63d/trainedXGB13.pkl\n../models/final/ZV63d/trainedXGB14.pkl\n../models/final/ZV63d/trainedXGB15.pkl\n../models/final/ZV63d/trainedXGB16.pkl\n../models/final/ZV63d/trainedXGB17.pkl\n../models/final/ZV63d/trainedXGB18.pkl\n../models/final/ZV63d/trainedXGB19.pkl\n../models/final/ZV63d/trainedXGB20.pkl\n../models/final/ZV63d/trainedXGB21.pkl\n../models/final/ZV63d/trainedXGB22.pkl\n../models/final/ZV63d/trainedXGB23.pkl\n../models/final/ZV63d/trainedXGB24.pkl\npos\nnot found\npos\n../models/final/ZV63d/trainedXGB28.pkl\n../models/final/ZV63d/trainedXGB29.pkl\n../models/final/ZV63d/trainedXGB30.pkl\n../models/final/ZV63d/trainedXGB31.pkl\n../models/final/ZV63d/trainedXGB32.pkl\n../models/final/ZV63d/trainedXGB33.pkl\n../models/final/ZV63d/trainedXGB34.pkl\n../models/final/ZV63d/trainedXGB35.pkl\n../models/final/ZVfd4/trainedXGB0.pkl\n../models/final/ZVfd4/trainedXGB1.pkl\n../models/final/ZVfd4/trainedXGB2.pkl\n../models/final/ZVfd4/trainedXGB3.pkl\npos\nnot found\n../models/final/ZVfd4/trainedXGB6.pkl\npos\nnot found\nnot found\nnot found\nnot found\n../models/final/ZVfd4/trainedXGB12.pkl\n../models/final/ZVfd4/trainedXGB13.pkl\n../models/final/ZVfd4/trainedXGB14.pkl\n../models/final/ZVfd4/trainedXGB15.pkl\n../models/final/ZVfd4/trainedXGB16.pkl\n../models/final/ZVfd4/trainedXGB17.pkl\n../models/final/ZVfd4/trainedXGB18.pkl\n../models/final/ZVfd4/trainedXGB19.pkl\n../models/final/ZVfd4/trainedXGB20.pkl\n../models/final/ZVfd4/trainedXGB21.pkl\n../models/final/ZVfd4/trainedXGB22.pkl\n../models/final/ZVfd4/trainedXGB23.pkl\nnot found\nnot found\nnot found\nnot found\n../models/final/ZVfd4/trainedXGB28.pkl\n../models/final/ZVfd4/trainedXGB29.pkl\n../models/final/ZVfd4/trainedXGB30.pkl\n../models/final/ZVfd4/trainedXGB31.pkl\n../models/final/ZVfd4/trainedXGB32.pkl\n../models/final/ZVfd4/trainedXGB33.pkl\n../models/final/ZVfd4/trainedXGB34.pkl\n../models/final/ZVfd4/trainedXGB35.pkl\n../models/final/ZVa9c/trainedXGB0.pkl\n../models/final/ZVa9c/trainedXGB1.pkl\n../models/final/ZVa9c/trainedXGB2.pkl\n../models/final/ZVa9c/trainedXGB3.pkl\n../models/final/ZVa9c/trainedXGB4.pkl\n../models/final/ZVa9c/trainedXGB5.pkl\n../models/final/ZVa9c/trainedXGB6.pkl\nnot found\nnot found\nnot found\nnot found\nnot found\n../models/final/ZVa9c/trainedXGB12.pkl\n../models/final/ZVa9c/trainedXGB13.pkl\n../models/final/ZVa9c/trainedXGB14.pkl\n../models/final/ZVa9c/trainedXGB15.pkl\n../models/final/ZVa9c/trainedXGB16.pkl\n../models/final/ZVa9c/trainedXGB17.pkl\n../models/final/ZVa9c/trainedXGB18.pkl\n../models/final/ZVa9c/trainedXGB19.pkl\n../models/final/ZVa9c/trainedXGB20.pkl\n../models/final/ZVa9c/trainedXGB21.pkl\n../models/final/ZVa9c/trainedXGB22.pkl\n../models/final/ZVa9c/trainedXGB23.pkl\nneg\nnot found\npos\nneg\n../models/final/ZVa9c/trainedXGB28.pkl\n../models/final/ZVa9c/trainedXGB29.pkl\n../models/final/ZVa9c/trainedXGB30.pkl\n../models/final/ZVa9c/trainedXGB31.pkl\n../models/final/ZVa9c/trainedXGB32.pkl\n../models/final/ZVa9c/trainedXGB33.pkl\n../models/final/ZVa9c/trainedXGB34.pkl\n../models/final/ZVa9c/trainedXGB35.pkl\n../models/final/ZVa78/trainedXGB0.pkl\n../models/final/ZVa78/trainedXGB1.pkl\n../models/final/ZVa78/trainedXGB2.pkl\nnot found\n../models/final/ZVa78/trainedXGB4.pkl\n../models/final/ZVa78/trainedXGB5.pkl\npos\nnot found\nnot found\nnot found\nnot found\nnot found\n../models/final/ZVa78/trainedXGB12.pkl\n../models/final/ZVa78/trainedXGB13.pkl\n../models/final/ZVa78/trainedXGB14.pkl\nnot found\n../models/final/ZVa78/trainedXGB16.pkl\n../models/final/ZVa78/trainedXGB17.pkl\n../models/final/ZVa78/trainedXGB18.pkl\nnot found\n../models/final/ZVa78/trainedXGB20.pkl\n../models/final/ZVa78/trainedXGB21.pkl\n../models/final/ZVa78/trainedXGB22.pkl\nnot found\n../models/final/ZVa78/trainedXGB24.pkl\n../models/final/ZVa78/trainedXGB25.pkl\nnot found\nnot found\n../models/final/ZVa78/trainedXGB28.pkl\n../models/final/ZVa78/trainedXGB29.pkl\n../models/final/ZVa78/trainedXGB30.pkl\nnot found\n../models/final/ZVa78/trainedXGB32.pkl\n../models/final/ZVa78/trainedXGB33.pkl\n../models/final/ZVa78/trainedXGB34.pkl\nnot found\n../models/final/ZV252/trainedXGB0.pkl\n../models/final/ZV252/trainedXGB1.pkl\n../models/final/ZV252/trainedXGB2.pkl\n../models/final/ZV252/trainedXGB3.pkl\nnot found\nnot found\npos\nnot found\nnot found\nnot found\nnot found\nnot found\n../models/final/ZV252/trainedXGB12.pkl\n../models/final/ZV252/trainedXGB13.pkl\n../models/final/ZV252/trainedXGB14.pkl\n../models/final/ZV252/trainedXGB15.pkl\n../models/final/ZV252/trainedXGB16.pkl\n../models/final/ZV252/trainedXGB17.pkl\n../models/final/ZV252/trainedXGB18.pkl\n../models/final/ZV252/trainedXGB19.pkl\nneg\n../models/final/ZV252/trainedXGB21.pkl\n../models/final/ZV252/trainedXGB22.pkl\n../models/final/ZV252/trainedXGB23.pkl\nnot found\nnot found\nnot found\nnot found\nnot found\nnot found\nnot found\nneg\n../models/final/ZV252/trainedXGB32.pkl\n../models/final/ZV252/trainedXGB33.pkl\n../models/final/ZV252/trainedXGB34.pkl\n../models/final/ZV252/trainedXGB35.pkl\n"
],
[
"GBDT_map['ZVe44']",
"_____no_output_____"
]
],
[
[
"## Load data to generate score tensor",
"_____no_output_____"
]
],
[
[
"def getLabel(filename, label_df):\n idx = label_df.loc[label_df['sample_file_name'] == filename]\n return idx.iloc[0]['label']\n\nfeature_thresholds = dict()\nfeature_thresholds[1] = [3000,5000] #engine rpm\nfeature_thresholds[2] = [4500,7000] #oil pump rpm\nfeature_thresholds[7] = [700, 1650, 2500] #displacement current\n#3x3x4 = 36 clusters\n\ncluster_dict = dict()\n\ndef clear_dict():\n global cluster_dict\n cluster_dict = dict()\n for i in range(36):\n cluster_dict[i] = None\n \ndef clustering(df, feature_thresholds, keys, cluster_n, this_num):\n if len(keys) == 0:\n #print('cluster '+str(this_num)+':'+note)\n global cluster_dict\n if len(df) == 0:\n cluster_dict[this_num] = None\n else:\n cluster_dict[this_num] = df\n else:\n keys_ = keys.copy()\n key = keys_.pop(0)\n thresholds = feature_thresholds[key]\n prev = 0\n cluster_n = int(cluster_n / (len(thresholds)+1))\n i = 0\n for val in thresholds:\n new_df = df[(df.iloc[:,key] > prev) & (df.iloc[:,key] <= val)]\n clustering(new_df, feature_thresholds, keys_, cluster_n, this_num + cluster_n*i)\n prev = val\n i+=1\n \n new_df = df[df.iloc[:,key] > prev]\n clustering(new_df, feature_thresholds, keys_, cluster_n, this_num + cluster_n*i)\n i+=1\n \n\ndef feature_tensor_gen(path, label_df, model_list):\n#path: train_path or test_path\n#vehicle_type: one string element under vehicle_types = ['ZVe44', 'ZV573', 'ZV63d', 'ZVfd4', 'ZVa9c', 'ZVa78', 'ZV252']\n n_cluster = 36\n #these are variables to calculate traversing progress (DO NOT CHANGE)\n counts_per_percent = int(len(os.listdir(path)) / 100)\n percentage_completion = 0\n counter = 0\n \n #pooling result from 50 weak learners then concatenated with the label\n feature_tensor = np.empty((0, cluster_n+1))\n \n #thresholds to categorize data points\n feature_thresholds = dict()\n feature_thresholds[1] = [3000,5000] #engine rpm\n feature_thresholds[2] = [4500,7000] #oil pump rpm\n feature_thresholds[7] = [700, 1650, 2500] #displacement current\n \n global cluster_dict\n \n for file in os.listdir(path):\n \n sample_df = pd.read_csv(path + '/' + file, delimiter = ',', encoding = 'utf-8')\n n = len(sample_df)\n label = getLabel(file, label_df)\n feature_vector = list()\n clear_dict()\n clustering(sample_df, feature_thresholds, list(feature_thresholds.keys()), cluster_n, 0)\n \n for i in range(cluster_n):\n df = cluster_dict[i]\n \n if df is None or len(df) == 0:\n feature_vector.append(np.nan)\n continue\n \n model = model_list[i]\n pooling_score = 0\n if model == 0:\n pooling_score = 0\n elif model == 0.5:\n pooling_score = np.nan\n elif model == 1:\n pooling_score = 1\n else:\n result = model.predict(df.iloc[:,:-1], validate_features=False)\n pooling_score = np.average(result)\n feature_vector.append(pooling_score)\n \n feature_vector.append(label)\n feature_vector = np.array(feature_vector) \n feature_tensor = np.append(feature_tensor, [feature_vector], axis=0)\n # --------------------------------------------------------------------------\n # NO NEED TO CHANGE ANYTHING BELOW\n \n #belows are to show traversing progress (DO NOT CHANGE)\n counter += 1\n if counter == counts_per_percent:\n counter = 0\n percentage_completion += 1\n print('traversing files under', path, ':', percentage_completion, \"%\", end=\"\\r\", flush=True)\n return feature_tensor",
"_____no_output_____"
],
[
"train_tensor = dict()\ntest_tensor = dict()\n\ntrain_path = '../data/final/train/'\ntest_path = '../data/final/test/'\n\nfor vehicle_type in vehicle_types:\n\n train_tensor[vehicle_type] = feature_tensor_gen(train_path+vehicle_type, label_df, GBDT_map[vehicle_type])\n test_tensor[vehicle_type] = feature_tensor_gen(test_path+vehicle_type, label_df, GBDT_map[vehicle_type])",
"traversing files under ../data/final/train/ZVe44 : 6 %\r"
]
],
[
[
"## Output score tensor",
"_____no_output_____"
]
],
[
[
"#print(feature_tensor.shape)\ntensor_path = '../data/final/feature_tensors'\nif not os.path.exists(tensor_path):\n os.makedirs(tensor_path)\nfor vehicle_type in vehicle_types:\n trainset = train_tensor[vehicle_type]\n testset = test_tensor[vehicle_type]\n np.savetxt(tensor_path+'/'+vehicle_type+\"_train.csv\", trainset, delimiter=\",\")\n np.savetxt(tensor_path+'/'+vehicle_type+\"_test.csv\", testset, delimiter=\",\")\n ",
"_____no_output_____"
],
[
"path = '../data/final/feature_tensors/'\nprint(path)\ntrain_tensor = dict()\ntest_tensor = dict()\nfor vehicle_type in vehicle_types:\n train_tensor[vehicle_type] =pd.read_csv(path+vehicle_type+'_train.csv',sep=',',header=None).to_numpy()\n print(train_tensor[vehicle_type].shape)\n test_tensor[vehicle_type] = pd.read_csv(path+vehicle_type+'_test.csv', sep=',',header=None).to_numpy()\n print(test_tensor[vehicle_type].shape)",
"../data/final/feature_tensors/\n(13883, 37)\n(3471, 37)\n(49193, 37)\n(12299, 37)\n(3869, 37)\n(968, 37)\n(938, 37)\n(235, 37)\n(4178, 37)\n(1045, 37)\n(8208, 37)\n(2052, 37)\n(345, 37)\n(87, 37)\n"
]
],
[
[
"# Test report of GBDT kernel",
"_____no_output_____"
]
],
[
[
"from sklearn import metrics\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.metrics import classification_report\ndef performance_summary(vehicle_type, train, test):\n print('Model performance report for vehicle type:', vehicle_type)\n params = {'booster': 'gbtree', 'eta': 1, 'max_depth': 16, 'gamma' : 1.5}\n bst = xgb.XGBClassifier(**params)\n bst.fit(train[:,:36],train[:,36])\n \n y_hat = bst.predict(train[:,:36])\n acc = accuracy_score(train[:,36], y_hat)\n print('train acc:',acc)\n y_hat = bst.predict(test[:,:36])\n acc = accuracy_score(test[:,36], y_hat)\n print(classification_report(test[:,36], y_hat,digits=4))\n print('test acc:',acc)\n fpr, tpr, thresholds = metrics.roc_curve(test[:,36], y_hat, pos_label=1)\n print('AUC:',metrics.auc(fpr, tpr))\n with open('../models/final/'+vehicle_type+'_integrated_model.pkl', 'wb') as f:\n pickle.dump(bst,f)\n return acc * test.shape[0]\n\ncorrect = 0\nnums = 0\nfor vehicle_type in vehicle_types:\n nums += test_tensor[vehicle_type].shape[0]\n correct += performance_summary(vehicle_type, train_tensor[vehicle_type], test_tensor[vehicle_type])\nprint('Overal test acc:', correct / nums)",
"Model performance report for vehicle type: ZVe44\ntrain acc: 0.987682777497659\n precision recall f1-score support\n\n 0.0 0.6750 0.6672 0.6711 1731\n 1.0 0.6727 0.6805 0.6766 1740\n\n accuracy 0.6739 3471\n macro avg 0.6739 0.6739 0.6738 3471\nweighted avg 0.6739 0.6739 0.6739 3471\n\ntest acc: 0.6738692019590896\nAUC: 0.67385206876631\nModel performance report for vehicle type: ZV573\ntrain acc: 0.9791637021527453\n precision recall f1-score support\n\n 0.0 0.6441 0.6450 0.6446 6253\n 1.0 0.6323 0.6315 0.6319 6046\n\n accuracy 0.6383 12299\n macro avg 0.6382 0.6382 0.6382 12299\nweighted avg 0.6383 0.6383 0.6383 12299\n\ntest acc: 0.6383445808602325\nAUC: 0.6382311548346307\nModel performance report for vehicle type: ZV63d\ntrain acc: 0.9966399586456449\n precision recall f1-score support\n\n 0.0 0.7018 0.7435 0.7220 421\n 1.0 0.7931 0.7569 0.7746 547\n\n accuracy 0.7510 968\n macro avg 0.7474 0.7502 0.7483 968\nweighted avg 0.7534 0.7510 0.7517 968\n\ntest acc: 0.7510330578512396\nAUC: 0.7501617546800298\nModel performance report for vehicle type: ZVfd4\ntrain acc: 1.0\n precision recall f1-score support\n\n 0.0 0.8507 0.8261 0.8382 138\n 1.0 0.7624 0.7938 0.7778 97\n\n accuracy 0.8128 235\n macro avg 0.8066 0.8100 0.8080 235\nweighted avg 0.8143 0.8128 0.8133 235\n\ntest acc: 0.8127659574468085\nAUC: 0.8099506947557149\nModel performance report for vehicle type: ZVa9c\ntrain acc: 0.9988032551460029\n precision recall f1-score support\n\n 0.0 0.7872 0.8057 0.7964 597\n 1.0 0.7327 0.7098 0.7211 448\n\n accuracy 0.7646 1045\n macro avg 0.7600 0.7578 0.7587 1045\nweighted avg 0.7639 0.7646 0.7641 1045\n\ntest acc: 0.7645933014354067\nAUC: 0.757758285474994\nModel performance report for vehicle type: ZVa78\ntrain acc: 0.9939083820662769\n precision recall f1-score support\n\n 0.0 0.6720 0.7033 0.6873 1011\n 1.0 0.6982 0.6667 0.6821 1041\n\n accuracy 0.6847 2052\n macro avg 0.6851 0.6850 0.6847 2052\nweighted avg 0.6853 0.6847 0.6846 2052\n\ntest acc: 0.6846978557504874\nAUC: 0.684965380811078\nModel performance report for vehicle type: ZV252\ntrain acc: 0.9971014492753624\n precision recall f1-score support\n\n 0.0 0.8077 0.8571 0.8317 49\n 1.0 0.8000 0.7368 0.7671 38\n\n accuracy 0.8046 87\n macro avg 0.8038 0.7970 0.7994 87\nweighted avg 0.8043 0.8046 0.8035 87\n\ntest acc: 0.8045977011494253\nAUC: 0.7969924812030076\nOveral test acc: 0.6638884754675795\n"
]
],
[
[
"# Test report of AVG Pooling Kernel",
"_____no_output_____"
]
],
[
[
"from sklearn import metrics\nfrom sklearn.metrics import accuracy_score\n\ndef test_report(vehicle_type, train, test):\n print('summary of test accuracy for vehicle type:', vehicle_type)\n arr = np.copy(train)\n where_are_NaNs = np.isnan(arr)\n arr[where_are_NaNs] = 0.5\n scores = np.mean(arr[:,0:36],axis=1)\n scores = [1 if num >= 0.5 else 0 for num in scores]\n scores = np.array(scores)\n acc = accuracy_score(train[:,36], scores)\n print('Train acc:', acc)\n \n arr = np.copy(test)\n where_are_NaNs = np.isnan(arr)\n arr[where_are_NaNs] = 0.5\n scores = np.mean(arr[:,0:36],axis=1)\n scores = [1 if num >= 0.5 else 0 for num in scores]\n scores = np.array(scores)\n acc = accuracy_score(test[:,36], scores)\n print(classification_report(test[:,36], scores, digits=4))\n print('Test acc:', acc)\n fpr, tpr, thresholds = metrics.roc_curve(test[:,36], scores, pos_label=1)\n print('AUC:',metrics.auc(fpr, tpr))\n correct = int(acc * test.shape[0])\n #print(correct,'/',test.shape[0])\n return correct \navg_acc = 0\nfor i in range(1):\n path = '../data/final/feature_tensors/'\n print(path)\n train_tensor = dict()\n test_tensor = dict()\n for vehicle_type in vehicle_types:\n train_tensor[vehicle_type] =pd.read_csv(path+vehicle_type+'_train.csv',sep=',',header=None).to_numpy()\n test_tensor[vehicle_type] = pd.read_csv(path+vehicle_type+'_test.csv', sep=',',header=None).to_numpy()\n\n\n\n correct = 0\n nums = 0\n for vehicle_type in vehicle_types:\n nums += test_tensor[vehicle_type].shape[0]\n correct += test_report(vehicle_type, train_tensor[vehicle_type], test_tensor[vehicle_type])\n avg_acc += (correct / nums)\n print('Test acc:', correct / nums)\nprint('average accuracy:', avg_acc)",
"../data/final/feature_tensors/\nsummary of test accuracy for vehicle type: ZVe44\nTrain acc: 0.8461427645321616\n precision recall f1-score support\n\n 0.0 0.6979 0.6979 0.6979 1731\n 1.0 0.6994 0.6994 0.6994 1740\n\n accuracy 0.6986 3471\n macro avg 0.6986 0.6986 0.6986 3471\nweighted avg 0.6986 0.6986 0.6986 3471\n\nTest acc: 0.6986459233650245\nAUC: 0.6986438972887907\nsummary of test accuracy for vehicle type: ZV573\nTrain acc: 0.8176163275262741\n precision recall f1-score support\n\n 0.0 0.6738 0.6387 0.6558 6253\n 1.0 0.6454 0.6801 0.6623 6046\n\n accuracy 0.6591 12299\n macro avg 0.6596 0.6594 0.6590 12299\nweighted avg 0.6598 0.6591 0.6590 12299\n\nTest acc: 0.6590779738190097\nAUC: 0.6594262474819232\nsummary of test accuracy for vehicle type: ZV63d\nTrain acc: 0.9441716205737917\n precision recall f1-score support\n\n 0.0 0.7767 0.5534 0.6463 421\n 1.0 0.7186 0.8775 0.7901 547\n\n accuracy 0.7366 968\n macro avg 0.7476 0.7155 0.7182 968\nweighted avg 0.7438 0.7366 0.7276 968\n\nTest acc: 0.7365702479338843\nAUC: 0.715478945837151\nsummary of test accuracy for vehicle type: ZVfd4\nTrain acc: 1.0\n precision recall f1-score support\n\n 0.0 0.8759 0.8696 0.8727 138\n 1.0 0.8163 0.8247 0.8205 97\n\n accuracy 0.8511 235\n macro avg 0.8461 0.8472 0.8466 235\nweighted avg 0.8513 0.8511 0.8512 235\n\nTest acc: 0.851063829787234\nAUC: 0.8471537427162708\nsummary of test accuracy for vehicle type: ZVa9c\nTrain acc: 0.9892292963140259\n precision recall f1-score support\n\n 0.0 0.7586 0.8526 0.8028 597\n 1.0 0.7647 0.6384 0.6959 448\n\n accuracy 0.7608 1045\n macro avg 0.7616 0.7455 0.7494 1045\nweighted avg 0.7612 0.7608 0.7570 1045\n\nTest acc: 0.7607655502392344\nAUC: 0.745494586025365\nsummary of test accuracy for vehicle type: ZVa78\nTrain acc: 0.8745126705653021\n precision recall f1-score support\n\n 0.0 0.7088 0.6044 0.6524 1011\n 1.0 0.6639 0.7589 0.7082 1041\n\n accuracy 0.6827 2052\n macro avg 0.6863 0.6816 0.6803 2052\nweighted avg 0.6860 0.6827 0.6807 2052\n\nTest acc: 0.6827485380116959\nAUC: 0.6816189067234485\nsummary of test accuracy for vehicle type: ZV252\nTrain acc: 0.9971014492753624\n precision recall f1-score support\n\n 0.0 0.7593 0.8367 0.7961 49\n 1.0 0.7576 0.6579 0.7042 38\n\n accuracy 0.7586 87\n macro avg 0.7584 0.7473 0.7502 87\nweighted avg 0.7585 0.7586 0.7560 87\n\nTest acc: 0.7586206896551724\nAUC: 0.7473147153598282\nTest acc: 0.6799622959765839\naverage accuracy: 0.6799622959765839\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf3b7d51c9c2bfc0a658fe024a3c09041b896c2 | 20,348 | ipynb | Jupyter Notebook | MagicSquareGame/MagicSquareGame.ipynb | zhang-lucy/QuantumKatas | 9761d057602d0fdbda4fa6761d45440ee141d97b | [
"MIT"
] | 1 | 2020-07-09T16:15:48.000Z | 2020-07-09T16:15:48.000Z | MagicSquareGame/MagicSquareGame.ipynb | zhang-lucy/QuantumKatas | 9761d057602d0fdbda4fa6761d45440ee141d97b | [
"MIT"
] | null | null | null | MagicSquareGame/MagicSquareGame.ipynb | zhang-lucy/QuantumKatas | 9761d057602d0fdbda4fa6761d45440ee141d97b | [
"MIT"
] | 1 | 2020-07-26T16:19:22.000Z | 2020-07-26T16:19:22.000Z | 33.357377 | 343 | 0.583989 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecf3ba8b9cb421889a4dbf15d7f31ff44fafa64a | 1,864 | ipynb | Jupyter Notebook | collab.ipynb | mrdavidgagnon/ArduinoThrustSensor | c1434949a2559e5aada39b348252f884c7035998 | [
"MIT"
] | 1 | 2016-10-26T22:11:44.000Z | 2016-10-26T22:11:44.000Z | collab.ipynb | mrdavidgagnon/ArduinoThrustSensor | c1434949a2559e5aada39b348252f884c7035998 | [
"MIT"
] | null | null | null | collab.ipynb | mrdavidgagnon/ArduinoThrustSensor | c1434949a2559e5aada39b348252f884c7035998 | [
"MIT"
] | null | null | null | 31.066667 | 238 | 0.493026 | [
[
[
"<a href=\"https://colab.research.google.com/github/mrdavidgagnon/ArduinoThrustSensor/blob/master/collab.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"Change 2\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
ecf3c2a9db0bc628f98e5bdffe579572ec30f210 | 421,556 | ipynb | Jupyter Notebook | CRUISE_SHIP_ACCIDENT_MODEL_PREDICTION.ipynb | vngeno/KNN-AND-NAIVE-BAYES-CLASSIFICATION-PREDICTION-MODEL | c7456fb2746b4077086b140a94daaaae926b4e0e | [
"MIT"
] | null | null | null | CRUISE_SHIP_ACCIDENT_MODEL_PREDICTION.ipynb | vngeno/KNN-AND-NAIVE-BAYES-CLASSIFICATION-PREDICTION-MODEL | c7456fb2746b4077086b140a94daaaae926b4e0e | [
"MIT"
] | null | null | null | CRUISE_SHIP_ACCIDENT_MODEL_PREDICTION.ipynb | vngeno/KNN-AND-NAIVE-BAYES-CLASSIFICATION-PREDICTION-MODEL | c7456fb2746b4077086b140a94daaaae926b4e0e | [
"MIT"
] | null | null | null | 160.22653 | 141,790 | 0.852539 | [
[
[
"<a href=\"https://colab.research.google.com/github/vngeno/KNN-AND-NAIVE-BAYES-CLASSIFICATION-MODEL/blob/main/CRUISE_SHIP_ACCIDENT_MODEL_PREDICTION.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"## 1. Defining the Question",
"_____no_output_____"
],
[
"### a) Specifying the Data Analytic Question\n\nWe will work as data scientists for a cruise ship company to build a model that predicts whether or not a passenger survives an accident in the cruise ship based on the features provided",
"_____no_output_____"
],
[
"#Understanding the Concept\n\nThe study is based on a true life event of the Royal Mail Ship (RMS) which was a British luxury passenger liner that sank on April 14–15, 1912, during its maiden voyage, en route to New York City from Southampton, England, killing about 1,500 (see Researcher’s Note: Titanic) passengers and ship personnel. One of the most famous tragedies in modern history, it inspired numerous stories, several films, and a musical and has been the subject of much scholarship and scientific speculation.",
"_____no_output_____"
],
[
"### b) Defining the Metric for Success\n\n\nWe will create a model that will allow the company to accurately predict wether a person will survive in a cruise ship accident or not.",
"_____no_output_____"
],
[
"### d) Recording the Experimental Design\n\n\nOur analysis will take the below flow:\n\n1.)Defining the question\n\n2.)Data Understanding\n\n3.)Exploratory Data Analysis\n\n4.)Implementing the Solution\n\n6.)Challenging the Solution",
"_____no_output_____"
],
[
"## 2. Reading the Data",
"_____no_output_____"
]
],
[
[
"# Loading the Data from the source i.e. csv\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns; sns.set(font_scale=1.2)\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")\nfrom sklearn.svm import SVC\nfrom sklearn.decomposition import PCA\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import confusion_matrix,accuracy_score\n",
"_____no_output_____"
],
[
"data_train = pd.read_csv('/content/train (5).csv')\ndata_train.head()",
"_____no_output_____"
],
[
"data_test = pd.read_csv('/content/test (1).csv')\ndata_test.head()",
"_____no_output_____"
],
[
"#Checking the columns\nprint(data_test.columns)\nprint('***********************************************************************************')\nprint(data_test.columns)\n",
"Index(['PassengerId', 'Pclass', 'Name', 'Sex', 'Age', 'SibSp', 'Parch',\n 'Ticket', 'Fare', 'Cabin', 'Embarked'],\n dtype='object')\n***********************************************************************************\nIndex(['PassengerId', 'Pclass', 'Name', 'Sex', 'Age', 'SibSp', 'Parch',\n 'Ticket', 'Fare', 'Cabin', 'Embarked'],\n dtype='object')\n"
],
[
"# Determining the no. of records in our datasets\n#\nprint(data_train.shape)\nprint(data_test.shape)",
"(891, 12)\n(418, 11)\n"
],
[
"# Checking whether each column has an appropriate datatype\n#\nprint(data_test.info())\nprint('******************************************************************************************')\nprint(data_train.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 418 entries, 0 to 417\nData columns (total 11 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 PassengerId 418 non-null int64 \n 1 Pclass 418 non-null int64 \n 2 Name 418 non-null object \n 3 Sex 418 non-null object \n 4 Age 332 non-null float64\n 5 SibSp 418 non-null int64 \n 6 Parch 418 non-null int64 \n 7 Ticket 418 non-null object \n 8 Fare 417 non-null float64\n 9 Cabin 91 non-null object \n 10 Embarked 418 non-null object \ndtypes: float64(2), int64(4), object(5)\nmemory usage: 36.0+ KB\nNone\n******************************************************************************************\n<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 891 entries, 0 to 890\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 PassengerId 891 non-null int64 \n 1 Survived 891 non-null int64 \n 2 Pclass 891 non-null int64 \n 3 Name 891 non-null object \n 4 Sex 891 non-null object \n 5 Age 714 non-null float64\n 6 SibSp 891 non-null int64 \n 7 Parch 891 non-null int64 \n 8 Ticket 891 non-null object \n 9 Fare 891 non-null float64\n 10 Cabin 204 non-null object \n 11 Embarked 889 non-null object \ndtypes: float64(2), int64(5), object(5)\nmemory usage: 83.7+ KB\nNone\n"
]
],
[
[
"## 5. Tidying the Dataset",
"_____no_output_____"
]
],
[
[
"#Checking for outliers\ndata_test.boxplot()",
"_____no_output_____"
],
[
"data_train.boxplot()",
"_____no_output_____"
],
[
"#Loop method to check for outliers visually using a boxplot\n#Test dataset\ncol_names = ['Age','SibSp', 'Parch', 'Fare']\n\nfig, ax = plt.subplots(len(col_names), figsize= (12,40))\n\nfor i, col_val in enumerate(col_names):\n sns.boxplot(y=data_test[col_val], ax= ax[i])\n ax[i].set_title('Box plot - {}'.format(col_val), fontsize= 10)\n ax[i].set_xlabel(col_val, fontsize= 8)\nplt.show()",
"_____no_output_____"
],
[
"col_names = ['Age','SibSp', 'Parch', 'Fare']\n\nfig, ax = plt.subplots(len(col_names), figsize= (12,40))\n\nfor i, col_val in enumerate(col_names):\n sns.boxplot(y=data_train[col_val], ax= ax[i])\n ax[i].set_title('Box plot - {}'.format(col_val), fontsize= 10)\n ax[i].set_xlabel(col_val, fontsize= 8)\nplt.show()",
"_____no_output_____"
]
],
[
[
"I decided to keep the outliers as they were in the age, fare, Parch and SibSp columns and removing or changing their values would give false results during modelling",
"_____no_output_____"
]
],
[
[
"# Identifying the Missing Data\n#\ndata_test.isnull().sum()",
"_____no_output_____"
],
[
"data_train.isnull().sum()",
"_____no_output_____"
],
[
"#Dropping the missing values in the datasets\ndata_test.dropna(inplace=True)\ndata_train.dropna(inplace=True)",
"_____no_output_____"
],
[
"print(data_test.duplicated().nunique())\nprint(data_train.duplicated().nunique())",
"1\n1\n"
],
[
"#Dropping columns\ndata_train.drop(['Name','Embarked', 'Fare','Ticket','Cabin'], axis = 1, inplace=True)\ndata_train",
"_____no_output_____"
]
],
[
[
"# 6. Exploratory Analysis",
"_____no_output_____"
],
[
"##Univariate Analysis",
"_____no_output_____"
]
],
[
[
"#The summary statistics are as follows\nprint(data_train.describe())",
" PassengerId Survived Pclass Age SibSp Parch\ncount 183.000000 183.000000 183.000000 183.000000 183.000000 183.000000\nmean 455.366120 0.672131 1.191257 35.674426 0.464481 0.475410\nstd 247.052476 0.470725 0.515187 15.643866 0.644159 0.754617\nmin 2.000000 0.000000 1.000000 0.920000 0.000000 0.000000\n25% 263.500000 0.000000 1.000000 24.000000 0.000000 0.000000\n50% 457.000000 1.000000 1.000000 36.000000 0.000000 0.000000\n75% 676.000000 1.000000 1.000000 47.500000 1.000000 1.000000\nmax 890.000000 1.000000 3.000000 80.000000 3.000000 4.000000\n"
],
[
"data_train.head(2)",
"_____no_output_____"
],
[
"data_train['Sex'].value_counts().plot(kind='barh')\nplt.title('Gender');",
"_____no_output_____"
],
[
"#Skewness\n#Positive values indicate that the tail of the data is right-skewed.\nprint('Skewness of Age: ' +str(data_train['Age'].skew()))",
"Skewness of Age: 0.00967583943600615\n"
],
[
"#Negative values of kurtosis as seen in the Age column indicate that a distribution is flat and has thin tails. Platykurtic distributions have negative kurtosis values.\n#A platykurtic distribution is flatter (less peaked) when compared with the normal distribution, with fewer values in its shorter (i.e. lighter and thinner) tails.\nprint('Kurtosis of Age: ' +str(data_train['Age'].kurt()))",
"Kurtosis of Age: -0.22618354101694615\n"
]
],
[
[
"**Univariate Interpretation**\n\nThere were more males than female passengers hence there could be higher number of males who will not survive or will survive the accident.",
"_____no_output_____"
],
[
"##Bivariate Analysis",
"_____no_output_____"
]
],
[
[
"#The sns pairplot below shows lack of correlation between the features\nsns.pairplot(data_train)\n",
"_____no_output_____"
],
[
"#Pearson's heatmap to check correlation between the variables\ncorr = data_train.corr() \nplt.figure(figsize = (20,10))\nsns.heatmap(corr, xticklabels=corr.columns, yticklabels=corr.columns, annot=True)\nplt.show()",
"_____no_output_____"
],
[
"data_train.groupby('Sex')['Survived'].value_counts().unstack().plot.bar(stacked=True);",
"_____no_output_____"
],
[
"data_train.groupby('SibSp')['Survived'].value_counts().unstack().plot.bar(stacked=True);",
"_____no_output_____"
],
[
"data_train.groupby('Parch')['Survived'].value_counts().unstack().plot.bar(stacked=True);",
"_____no_output_____"
],
[
"data_train.groupby('Pclass')['Survived'].value_counts().unstack().plot.bar(stacked=True);",
"_____no_output_____"
]
],
[
[
"**Bivariate Interpretation**\n\nHere Sibsp means Number of Siblings/Spouses Aboard\n\nParch - Number of Parents/Children\n\nPclass - Passenger class.\n\nThere's negligible correlation in the variables\n\nThere were more females that survived compared to males.\n\nThere were more parents/children that survived\n\nPassenger class 1 had most surivors and also had the most deaths.",
"_____no_output_____"
],
[
"##Linear Discriminant Analysis",
"_____no_output_____"
]
],
[
[
"# Data Reduction\n#\n#WE'LL APPLY LDA HERE\n\n#Preparing the data for encoding\n#We'll drop some columns that are not necessary for the study then convert the remaining columns to numericals\ntrain = data_train.copy(deep=True)\ntrain['Sex'] = data_train['Sex'].astype('category')\ntrain['Age'] = data_train['Age'].astype('category')\ntrain['SibSp'] = data_train['SibSp'].astype('category')\ntrain['PassengerId'] = data_train['PassengerId'].astype('category')\ntrain['Parch'] = data_train['Parch'].astype('category')\ntrain['Pclass'] = data_train['Pclass'].astype('category')\ntrain.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 183 entries, 1 to 889\nData columns (total 7 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 PassengerId 183 non-null category\n 1 Survived 183 non-null int64 \n 2 Pclass 183 non-null category\n 3 Sex 183 non-null category\n 4 Age 183 non-null category\n 5 SibSp 183 non-null category\n 6 Parch 183 non-null category\ndtypes: category(6), int64(1)\nmemory usage: 14.1 KB\n"
],
[
"#Using label encoding\nfrom sklearn.preprocessing import LabelEncoder\nlabelencoder = LabelEncoder()\ntrain = train.copy(deep=True)\ntrain['Sex']=labelencoder.fit_transform(train['Sex'])\ntrain['Sex']",
"_____no_output_____"
],
[
"train.columns",
"_____no_output_____"
],
[
"# #Rearranging the columns to have Survived as the first column\ntrain = train[['Survived', 'Parch', 'Pclass', 'Sex', 'Age', 'SibSp', 'PassengerId']]\ntrain",
"_____no_output_____"
],
[
"#Separating the target label (price) from the other features\n\nfeat = train.iloc[:,1:6]\nlabel = train['Survived']",
"_____no_output_____"
],
[
"\n#Subject our frame to LDA model\n\n#import LDA method from sklearn library\nfrom sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA\n\n#creating an instance of LDA\n\nlda = LDA()\n\nlda_ = lda.fit(feat, label)\nlda_feat = lda_.transform(feat)\n\n#Displaying coefficients of the reducted columns\nlda_.coef_",
"_____no_output_____"
],
[
"\nnew_df = pd.DataFrame(index=feat.columns.values, data=lda_.coef_[0].T)\nnew_df.sort_values(0, ascending=False)\n",
"_____no_output_____"
]
],
[
[
"**LDA INTERPRETATION**\n\nThe major features that determine survival of a passenger is Number of siblings or spouses on board, followed by age then number of Parents/Children.\n\nSex and Passenger class have minimal determination of passenger survival",
"_____no_output_____"
],
[
"#Implementing the Solution",
"_____no_output_____"
],
[
"##Modelling",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"###KNN Classifier Model",
"_____no_output_____"
]
],
[
[
"# Splitting our dataset into its attributes and labels\nX = train.iloc[:,1:6].values\ny = train['Survived'].values",
"_____no_output_____"
],
[
"# Train Test Split\n# To avoid over-fitting, we will divide our dataset into training and test splits, \n# which gives us a better idea as to how our algorithm performed during the testing phase. \n# This way our algorithm is tested on un-seen data\nfrom sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.20)",
"_____no_output_____"
],
[
"# Feature Scaling\n# Before making any actual predictions, it is always a good practice to scale the features \n# so that all of them can be uniformly evaluated.\nfrom sklearn.preprocessing import StandardScaler\nscaler = StandardScaler()\nscaler.fit(X_train)\n\nX_train = scaler.transform(X_train)\nX_test = scaler.transform(X_test)",
"_____no_output_____"
],
[
"# Training and Predictions\n# In the second line, this class is initialized with one parameter, i.e. n_neigbours.\n# This is basically the value for the K. There is no ideal value for K and it is selected after testing and evaluation, \n# however to start out, 5 seems to be the most commonly used value for KNN algorithm.\nfrom sklearn.neighbors import KNeighborsClassifier\nclassifier = KNeighborsClassifier(n_neighbors=5)\nclassifier.fit(X_train, y_train)",
"_____no_output_____"
],
[
"#We'll now make predictions on our test data\ny_pred = classifier.predict(X_test)",
"_____no_output_____"
],
[
"#Evaluating the Algorithm\n# In evaluating an algorithm, confusion matrix, precision, F1 score are the commonly used metrics\n# The confusion_matrix and classification_report methods of the sklearn.metrics can be used to calculate these metrics.\nfrom sklearn.metrics import classification_report, confusion_matrix\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.metrics import precision_score, recall_score\nprint(confusion_matrix(y_test, y_pred))\nprint(classification_report(y_test, y_pred))\nprint(accuracy_score(y_test, y_pred))\nprecision, recall = precision_score(y_test, y_pred), recall_score(y_test, y_pred)\nprint(precision, recall)\n",
"[[ 6 2]\n [ 6 23]]\n precision recall f1-score support\n\n 0 0.50 0.75 0.60 8\n 1 0.92 0.79 0.85 29\n\n accuracy 0.78 37\n macro avg 0.71 0.77 0.73 37\nweighted avg 0.83 0.78 0.80 37\n\n0.7837837837837838\n0.92 0.7931034482758621\n"
],
[
"# Train Test Split with 70/30\nfrom sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30)",
"_____no_output_____"
],
[
"#Feature Scaling\nscaler = StandardScaler()\nscaler.fit(X_train)\nX_train = scaler.transform(X_train)\nX_test = scaler.transform(X_test)",
"_____no_output_____"
],
[
"#Fitting the KNN model\nclassifier = KNeighborsClassifier(n_neighbors=5)\nclassifier.fit(X_train, y_train)",
"_____no_output_____"
],
[
"#We'll now make predictions on our test data\ny_pred = classifier.predict(X_test)",
"_____no_output_____"
],
[
"#Evaluating the algorithm\nprint(confusion_matrix(y_test, y_pred))\nprint(classification_report(y_test, y_pred))\nprint(accuracy_score(y_test, y_pred))\nprecision, recall = precision_score(y_test, y_pred), recall_score(y_test, y_pred)\nprint(precision, recall)",
"[[ 9 4]\n [ 9 33]]\n precision recall f1-score support\n\n 0 0.50 0.69 0.58 13\n 1 0.89 0.79 0.84 42\n\n accuracy 0.76 55\n macro avg 0.70 0.74 0.71 55\nweighted avg 0.80 0.76 0.78 55\n\n0.7636363636363637\n0.8918918918918919 0.7857142857142857\n"
],
[
"# Train Test Split with 60/40\nfrom sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.40)",
"_____no_output_____"
],
[
"#Feature Scaling\nscaler = StandardScaler()\nscaler.fit(X_train)\nX_train = scaler.transform(X_train)\nX_test = scaler.transform(X_test)",
"_____no_output_____"
],
[
"#Fitting the KNN model\nclassifier = KNeighborsClassifier(n_neighbors=5)\nclassifier.fit(X_train, y_train)",
"_____no_output_____"
],
[
"#We'll now make predictions on our test data\ny_pred = classifier.predict(X_test)",
"_____no_output_____"
],
[
"#Evaluating the algorithm\nprint(confusion_matrix(y_test, y_pred))\nprint(classification_report(y_test, y_pred))\nprint(accuracy_score(y_test, y_pred))\nprecision, recall = precision_score(y_test, y_pred), recall_score(y_test, y_pred)\nprint(precision, recall)",
"[[10 17]\n [ 4 43]]\n precision recall f1-score support\n\n 0 0.71 0.37 0.49 27\n 1 0.72 0.91 0.80 47\n\n accuracy 0.72 74\n macro avg 0.72 0.64 0.65 74\nweighted avg 0.72 0.72 0.69 74\n\n0.7162162162162162\n0.7166666666666667 0.9148936170212766\n"
]
],
[
[
"**KNN CLASSIFIER MODEL INTERPRETATION**\n\nFrom the above, we notice that our model gets better having increased the test size. \nGenerally the model is a good fit with a F1 score of 0.49 and precision of 71%, accuracy of 71% using the largest test size of 40.\n\nThe confusion matrix also keeps getting better with 10 & 43 accurate values on the 3rd model",
"_____no_output_____"
],
[
"Regression\n",
"_____no_output_____"
]
],
[
[
"from sklearn.neighbors import KNeighborsRegressor\nfrom sklearn.metrics import mean_squared_error",
"_____no_output_____"
],
[
"\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=.3, random_state=0)",
"_____no_output_____"
],
[
"clf = KNeighborsRegressor(11)\nclf.fit(X_train, y_train)",
"_____no_output_____"
],
[
"#Making our prediction\nfrom sklearn.metrics import mean_absolute_error\nfrom sklearn.metrics import r2_score\nfrom sklearn.metrics import accuracy_score\ny_pred = clf.predict(X_test)\nprint(mean_squared_error(y_test, y_pred))\nprint(mean_absolute_error(y_test, y_pred))",
"0.18707738542449287\n0.3785123966942147\n"
]
],
[
[
"**KNN REGRESSOR MODEL INTERPRETATION**\n\n\nThe RMSE score is quite low hence the regressor model is also a good fit",
"_____no_output_____"
],
[
"###NAIVE BAYES CLASSIFIER ",
"_____no_output_____"
],
[
"####Gaussian Naive Bayes Classifier",
"_____no_output_____"
]
],
[
[
"#Splitting the data to train size of 70/30\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=6) ",
"_____no_output_____"
],
[
"# Training our model\nfrom sklearn.naive_bayes import GaussianNB\nclf = GaussianNB() \nmodel = clf.fit(X_train, y_train) ",
"_____no_output_____"
],
[
"#Predicting our test predictors\npredicted = model.predict(X_test)\nprint(np.mean(predicted == y_test))",
"0.7454545454545455\n"
]
],
[
[
"**GAUSSIAN NAIVE BAYES CLASSIFIER INTERPRETATION**\n\nThe above result shows that, there's a 75% chance of passenger survival during the accident. ",
"_____no_output_____"
],
[
"#Challenging the solution\n\nGenerally the data we worked with was accurate, however we had to drop a number of missing values hence that should be investigated further. \n\n",
"_____no_output_____"
],
[
"#Follow up questions",
"_____no_output_____"
],
[
"### a). Did we have the right data?\n\nThe data we had is right and has produced quite accurate results even without getting rid of outliers",
"_____no_output_____"
],
[
"### b). Do we need other data to answer our question?\n\nOur question is well answered",
"_____no_output_____"
],
[
"### c). Did we have the right question?\n\nYes we did have the correct question",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecf3c4c3fa60e17b15f84b3f7b7db2ff7a147256 | 7,063 | ipynb | Jupyter Notebook | Tensorflow101.ipynb | codexponent/tensorflow101 | 82dc32f8dedbee1c92a41631752fffd45123be04 | [
"MIT"
] | null | null | null | Tensorflow101.ipynb | codexponent/tensorflow101 | 82dc32f8dedbee1c92a41631752fffd45123be04 | [
"MIT"
] | null | null | null | Tensorflow101.ipynb | codexponent/tensorflow101 | 82dc32f8dedbee1c92a41631752fffd45123be04 | [
"MIT"
] | null | null | null | 23.233553 | 553 | 0.555571 | [
[
[
"## <center>Tensorflow.</center>",
"_____no_output_____"
],
[
"##### TensorFlow™ is an open source software library for high performance numerical computation. Its flexible architecture allows easy deployment of computation across a variety of platforms (CPUs, GPUs, TPUs), and from desktops to clusters of servers to mobile and edge devices. Originally developed by researchers and engineers from the Google Brain team within Google’s AI organization, it comes with strong support for machine learning and deep learning and the flexible numerical computation core is used across many other scientific domains.",
"_____no_output_____"
],
[
"### <center>Importing Tensorflow</center>",
"_____no_output_____"
],
[
"##### To use TensorFlow, we need to import the library called tensorflow. We imported it with the name \"tf\", so the modules can be accessed by tf.moduleName",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf",
"_____no_output_____"
]
],
[
[
"### <center>Sessions</center>",
"_____no_output_____"
],
[
"##### Sessions are contex for creating a graph inside tensorflow. The graphs need session for the computaion of the values",
"_____no_output_____"
]
],
[
[
"a = tf.constant(12)\nb = tf.constant(13)\nc = tf.multiply(12, 13)",
"_____no_output_____"
],
[
"with tf.Session() as session:\n print(session.run(c))",
"156\n"
]
],
[
[
"### <center>Matrix Multiplications</center>",
"_____no_output_____"
],
[
"##### As we all know, most of the images are just matrix tables, matrix tables of pixel values. So, most of the computer vision task relies on matrix multiplications of the matrices.",
"_____no_output_____"
]
],
[
[
"matrixA = tf.constant([[3, 4], [4, 5]])\nmatrixB = tf.constant([[5, 6], [2, 3]])\nmatrixC = tf.matmul(matrixA, matrixB)\n\n# Don't get confused with tf.multiply and tf.matmul as the first one does element wise multiplications \n# and the latter one gives the dot product of the two matrices",
"_____no_output_____"
],
[
"with tf.Session() as session:\n print(session.run(matrixC))",
"[[23 30]\n [30 39]]\n"
]
],
[
[
"### <center>Variables</center>",
"_____no_output_____"
],
[
"##### A TensorFlow variable is the best way to represent shared, persistent state manipulated by your program. Variables are manipulated via the tf.Variable class. A tf.Variable represents a tensor whose value can be changed by running ops on it.",
"_____no_output_____"
]
],
[
[
"variableA = tf.Variable(0)\nvariableB = tf.constant(5)\nactivity1 = tf.assign(variableA, variableB)",
"_____no_output_____"
],
[
"with tf.Session() as session:\n # To be able to use variables in a computation graph it is necessary to initialize them before \n # running the graph in a session.\n session.run(tf.global_variables_initializer())\n session.run(activity1)\n print(session.run(variableA))",
"5\n"
]
],
[
[
"### <center>Placeholders</center>",
"_____no_output_____"
],
[
"##### A placeholder is simply a variable that we will assign data to at a later date. Unlike variables, the placeholders get's their data from outside of the computational graph.",
"_____no_output_____"
]
],
[
[
"placeholder1 = tf.placeholder(dtype=tf.float32)",
"_____no_output_____"
],
[
"with tf.Session() as session:\n print(session.run(placeholder1, feed_dict={placeholder1: 5}))",
"5.0\n"
],
[
"placeholder2 = tf.placeholder(dtype=tf.float32)\nplaceholder3 = tf.placeholder(dtype=tf.float32)\nactivity2 = tf.pow(placeholder2, placeholder3)",
"_____no_output_____"
],
[
"with tf.Session() as session:\n activity3 = session.run(activity2, feed_dict={placeholder2: 2, placeholder3: 3})\n print(activity3)",
"8.0\n"
]
],
[
[
"### <center>Thanks for completing this lesson!</center>",
"_____no_output_____"
],
[
"<hr>\n##### Notebook created by: <a href=\"https://www.linkedin.com/in/sulabhshrestha/\"> Sulabh Shrestha </a></h4> \n##### Copyright © 2018 [CodeExponent]",
"_____no_output_____"
],
[
"<hr>\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ecf3c98ef7e7f4d983fddb41772d9a61e85e334f | 79,433 | ipynb | Jupyter Notebook | aquaponics-master/notebooks/Test Heterotrophic.ipynb | JunwenLIU/Aquaponics_system_Simulink | 5048012730815f3e8bb5ccb254c7e1d4be8a84d4 | [
"Apache-2.0"
] | null | null | null | aquaponics-master/notebooks/Test Heterotrophic.ipynb | JunwenLIU/Aquaponics_system_Simulink | 5048012730815f3e8bb5ccb254c7e1d4be8a84d4 | [
"Apache-2.0"
] | null | null | null | aquaponics-master/notebooks/Test Heterotrophic.ipynb | JunwenLIU/Aquaponics_system_Simulink | 5048012730815f3e8bb5ccb254c7e1d4be8a84d4 | [
"Apache-2.0"
] | null | null | null | 420.280423 | 42,792 | 0.939874 | [
[
[
"# Test Fish Heterotrophic Nutrient Sector #\n\nPrimarily used for debugging and making sure module runs fine on its own.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom gekko import GEKKO\nimport matplotlib.pyplot as plt\n\nfrom aquaponics import Aquaponics",
"_____no_output_____"
],
[
"a = Aquaponics('heterotrophic')\nm = a.get_model()\n\ntf = 30\nsteps = tf * 10 + 1\nm.time = np.linspace(0,tf,steps)\nm.options.IMODE = 4\nm.options.SOLVER = 3\n\nm.solve(disp=False)",
"_____no_output_____"
],
[
"%matplotlib inline\nplt.figure(figsize=(12,6))\nplt.title('Heterotrophic Food Quantity')\nax = plt.subplot(211)\nplt.plot(m.time, a.HFe)\nplt.grid()\nplt.ylabel('Energy (kcal / pond)')\n\nax = plt.subplot(212, sharex=ax)\nplt.plot(m.time, a.HFp)\nplt.grid()\nplt.ylabel('Protein (g protein / pond))')\n\nplt.xlim(0, tf)\nplt.xlabel('Time (days)')",
"_____no_output_____"
]
],
[
[
"## Heterotrophic and Autotrophic ##",
"_____no_output_____"
]
],
[
[
"a = Aquaponics('autotrophic', 'heterotrophic')\nm = a.get_model()\n\ntf = 30\nsteps = tf * 10 + 1\nm.time = np.linspace(0,tf,steps)\nm.options.IMODE = 4\nm.options.SOLVER = 3\n\nm.solve(disp=False)",
"_____no_output_____"
],
[
"%matplotlib inline\nplt.figure(figsize=(12,6))\nax = plt.subplot(211)\nplt.plot(m.time, a.AFe, label='Energy (kcal / pond)')\nplt.plot(m.time, a.AFp, label='Protein (g protein / pond)')\nplt.grid()\nplt.ylabel('Autotrophic')\nplt.legend()\n\nax = plt.subplot(212, sharex=ax)\nplt.plot(m.time, a.HFe, label='Energy (kcal / pond)')\nplt.plot(m.time, a.HFp, label='Protein (g protein / pond)')\nplt.grid()\nplt.ylabel('Heterotrophic')\nplt.legend()\n\nplt.xlim(0, tf)\nplt.xlabel('Time (days)')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecf3d0af175fe200061df1cce8d67afa4de42706 | 858 | ipynb | Jupyter Notebook | 5-Monitoring/solutions/a-solution.ipynb | marciomocellin/amazon-sagemaker-workshop | b2142a17404f9390afd1b29149f69fdafca97e72 | [
"MIT-0"
] | 8 | 2021-06-02T12:19:56.000Z | 2021-08-04T12:30:20.000Z | 5-Monitoring/solutions/a-solution.ipynb | marciomocellin/amazon-sagemaker-workshop | b2142a17404f9390afd1b29149f69fdafca97e72 | [
"MIT-0"
] | 1 | 2021-08-03T21:29:01.000Z | 2021-08-03T21:29:01.000Z | 5-Monitoring/solutions/a-solution.ipynb | marciomocellin/amazon-sagemaker-workshop | b2142a17404f9390afd1b29149f69fdafca97e72 | [
"MIT-0"
] | 5 | 2021-05-26T14:16:43.000Z | 2021-08-24T16:56:05.000Z | 26 | 220 | 0.599068 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecf3d0d94e0c5cbcd3d7c645bfa47ca0b24755b9 | 2,283 | ipynb | Jupyter Notebook | Sample_OneCell.ipynb | kohlisimranjit/cs224u | 7a1813a78c268e50dc9cb6855f5fe8e13a0d883d | [
"Apache-2.0"
] | null | null | null | Sample_OneCell.ipynb | kohlisimranjit/cs224u | 7a1813a78c268e50dc9cb6855f5fe8e13a0d883d | [
"Apache-2.0"
] | null | null | null | Sample_OneCell.ipynb | kohlisimranjit/cs224u | 7a1813a78c268e50dc9cb6855f5fe8e13a0d883d | [
"Apache-2.0"
] | null | null | null | 34.074627 | 309 | 0.6417 | [
[
[
"### Your original system [3 points]\n\nThis question asks you to design your own model. You can of course include steps made above (ideally, the above questions informed your system design!), but your model should not be literally identical to any of the above models. Other ideas: retrofitting, autoencoders, GloVe, subword modeling, ... \n\nRequirements:\n\n1. Your code must operate on one of the count matrices in `data/vsmdata`. You can choose which one. __Other pretrained vectors cannot be introduced__.\n\n1. Your code must be self-contained, so that we can work with your model directly in your homework submission notebook. If your model depends on external data or other resources, please submit a ZIP archive containing these resources along with your submission.\n\nIn the cell below, please provide a brief technical description of your original system, so that the teaching team can gain an understanding of what it does. This will help us to understand your code and analyze all the submissions to identify patterns and strategies.",
"_____no_output_____"
]
],
[
[
"# Enter your system description in this cell.\n# This is my code description attached here as requested\n# My peak score was: #\n# This is my code\n# You can see that my system is based on ....\nif 'IS_GRADESCOPE_ENV' not in os.environ:\n display(full_word_similarity_evaluation(giga5))\n\n# Please do not remove this comment.",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
ecf3d23606e178258d478e1bdcef2f41a2e41bf8 | 28,666 | ipynb | Jupyter Notebook | my_portfolio_analysis/life_calc.ipynb | laye0619/my_portfolio_analysis | d3989036bd890341f5e25b0b65ca31f1c089919d | [
"MIT"
] | null | null | null | my_portfolio_analysis/life_calc.ipynb | laye0619/my_portfolio_analysis | d3989036bd890341f5e25b0b65ca31f1c089919d | [
"MIT"
] | null | null | null | my_portfolio_analysis/life_calc.ipynb | laye0619/my_portfolio_analysis | d3989036bd890341f5e25b0b65ca31f1c089919d | [
"MIT"
] | null | null | null | 80.522472 | 18,102 | 0.75689 | [
[
[
"import pandas as pd\npd.options.display.max_rows = 1000\n%matplotlib inline",
"_____no_output_____"
],
[
"def return_period_calc(on_job:bool, report_df: pd.DataFrame, year: int, start_money: float, cpi: float, ann_cost: float, ann_return: float, return_period: int) -> set:\n current_money = start_money\n current_ann_cost = ann_cost\n current_year = year\n for i in range(return_period):\n current_ann_cost = current_ann_cost*(1+cpi)\n report_df = pd.concat([report_df, pd.Series(\n {'year': current_year, 'start_money': current_money, 'current_cost': current_ann_cost})], axis=1)\n current_money -= current_ann_cost\n if on_job:\n current_money += 100\n if i == (return_period-1):\n current_money = current_money*(1+ann_return)**return_period\n i += 1\n current_year += 1\n return current_money, current_ann_cost, report_df",
"_____no_output_____"
],
[
"# set params\ninit_money = 1000.0 # 初始资金\ncpi = 0.03 # 每年CPI涨幅\nann_cost = 50.0 # 每年生活费用 - 单位:年\nann_return = 0.1 # 年化回报率\nreturn_period = 5 # 年化回报周期 - 单位:年\n\nmoney = init_money\ncost = ann_cost\nyear = 2022\nreport_df = pd.DataFrame()\nfired = True\nwhile True:\n (money, cost, report_df) = return_period_calc(\n on_job=fired,\n report_df=report_df,\n year=year,\n start_money=money,\n cpi=cpi,\n ann_cost=cost,\n ann_return=ann_return,\n return_period=return_period\n )\n fired = False\n year += return_period\n if money <= 0 or money >= 10000:\n break\nreport_df = report_df.T.set_index('year')\ndisplay(report_df)\nreport_df.plot()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecf3d9367d5be460269ae2d612f1806479c77a36 | 321,898 | ipynb | Jupyter Notebook | notebooks/06_information_geometry.ipynb | ninamiolane/geomstats | b68cf068eceba2c3c1855ccbaa08ad42c69bbde1 | [
"MIT"
] | 10 | 2018-01-28T17:16:44.000Z | 2022-02-27T02:42:41.000Z | notebooks/06_information_geometry.ipynb | ninamiolane/geomstats | b68cf068eceba2c3c1855ccbaa08ad42c69bbde1 | [
"MIT"
] | 67 | 2018-01-05T17:15:32.000Z | 2018-05-11T18:50:30.000Z | notebooks/06_information_geometry.ipynb | ninamiolane/geomstats | b68cf068eceba2c3c1855ccbaa08ad42c69bbde1 | [
"MIT"
] | 3 | 2017-12-15T23:42:07.000Z | 2020-07-08T16:50:28.000Z | 643.796 | 135,150 | 0.947117 | [
[
[
"# Tutorial: Information geometry",
"_____no_output_____"
],
[
"Lead author: Alice Le Brigant.\n\nDisclaimer: this notebook requires the use of the ```numpy``` backend.",
"_____no_output_____"
],
[
"## Introduction\n\nInformation geometry is a branch of mathematics at the crossroads of statistics and differential geometry, focused on the study of probability distributions from a geometric point of view. One of the tools of information geometry is the Fisher information distance, which allows to compare probability distributions inside a given parametric family. In that sense, information geometry is an alternative approach to optimal transport. \n\nThe Fisher information metric or Fisher-Rao metric - although the latter usually denotes its non parametric counterpart - is a Riemannian metric defined on the space of parameters of a family of distributions using the Fisher information matrix. This metric is invariant under change of parameterization. Moreover it is the only Riemannian metric compatible with the notion of information contained by the model on the parameter, in the sense that it is the only metric that preserves the geometry of a parametric model after transformation by a sufficient statistic (Cencov's theorem). For an overview, see [[A2016]](#References).",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
],
[
"Before starting this tutorial, we set the working directory to be the root of the geomstats repository. In order to have the code working on your machine, you need to change this path to the path of your geomstats repository.",
"_____no_output_____"
]
],
[
[
"import os\nimport subprocess\n\ngeomstats_gitroot_path = subprocess.check_output(\n ['git', 'rev-parse', '--show-toplevel'], \n universal_newlines=True)\n\nos.chdir(geomstats_gitroot_path[:-1])\n\nprint('Working directory: ', os.getcwd())",
"Working directory: /Users/alicelebrigant/ownCloud/Python/SpyderProjects/geomstats\n"
],
[
"import matplotlib\nimport matplotlib.image as mpimg\nimport matplotlib.pyplot as plt\nfrom matplotlib.lines import Line2D\n\nimport geomstats.backend as gs\nimport geomstats.visualization as visualization",
"INFO: Using numpy backend\n"
]
],
[
[
"## Normal distributions",
"_____no_output_____"
],
[
"The Fisher information geometry of the family of normal distributions is arguably the most well-known. The space of parameters is the upper half-plane where the x-coordinate encodes the mean and the y-coordinate the standard deviation. Quite remarkably, the Fisher information metric induces the hyperbolic geometry of the Poincare half plane [[AM1981]](#References). To start, we need an instance of the class ```NormalDistributions``` and its Fisher information metric.",
"_____no_output_____"
]
],
[
[
"from geomstats.information_geometry.normal import NormalDistributions\n\nnormal = NormalDistributions()\nfisher_metric = normal.metric",
"_____no_output_____"
]
],
[
[
"Using the ```visualization``` module, we can plot the geodesic between two points, each defining the parameters (mean and standard deviation) for a normal distribution. We recognise the shape of a geodesic of the Poincare half-plane, namely a half-circle orthogonal to the x-axis.",
"_____no_output_____"
]
],
[
[
"point_a = gs.array([1., 1.])\npoint_b = gs.array([3., 1.])\n\ngeodesic_ab_fisher = fisher_metric.geodesic(point_a, point_b)\n\nn_points = 20\nt = gs.linspace(0, 1, n_points)",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(10, 5))\nax = fig.add_subplot(111)\ncc = gs.zeros((n_points, 3))\ncc[:, 2] = gs.linspace(0, 1, n_points)\n\nvisualization.plot(\n geodesic_ab_fisher(t), ax=ax, space='H2_poincare_half_plane', label='point on geodesic', color=cc)\n\nax.set_xlim(0., 4.)\nax.set_ylim(0., 2.)\nax.set_title('Geodesic between two normal distributions for the Fisher-Rao metric')\nax.legend();",
"_____no_output_____"
]
],
[
[
"Each point of the geodesic defines a normal distribution, and so we obtain an optimal interpolation between the distributions corresponding to ```point_a``` and ```point_b```, which we can visualize in terms of probability density functions.",
"_____no_output_____"
]
],
[
[
"pdfs = normal.point_to_pdf(geodesic_ab_fisher(t))\nx = gs.linspace(-3., 7., 100)\n\nfig = plt.figure(figsize=(10, 5))\nfor i in range(n_points):\n plt.plot(x, pdfs(x)[:, i], color=cc[i, :])\nplt.title('Corresponding interpolation between pdfs');",
"_____no_output_____"
]
],
[
[
"Another possibility to compare probability distributions is given by the $L^2$-Wasserstein metric, central in optimal transport. In the case of normal distributions, the $L^2$-Wasserstein metric induces the Euclidean geometry on the upper half plane [[BGKL2017]](#References). Therefore, the Wasserstein distance between two normal distributions with different means and same variance (```point_a``` and ```point_b```) will not change when this common variance is increased (```point_c``` and ```point_d```), while the corresponding Fisher information distance will decrease, as can be deduced from the shape of the geodesic. This can be interpreted as a consequence of the increasing overlap of the corresponding probability densities.",
"_____no_output_____"
]
],
[
[
"from geomstats.geometry.euclidean import Euclidean\n\nplane = Euclidean(2)\nwasserstein_metric = plane.metric\n\npoint_c = gs.array([1., 3.])\npoint_d = gs.array([3., 3.])\n\ngeodesic_cd_fisher = fisher_metric.geodesic(point_c, point_d)\ngeodesic_ab_wasserstein = wasserstein_metric.geodesic(point_a, point_b)\ngeodesic_cd_wasserstein = wasserstein_metric.geodesic(point_c, point_d)\n\npoints = gs.stack((point_a, point_b, point_c, point_d))\npdfs = normal.point_to_pdf(points)",
"_____no_output_____"
],
[
"%matplotlib inline\n\nfig = plt.figure(figsize=(12, 5))\nax1 = fig.add_subplot(121)\n\nvisualization.plot(\n gs.vstack((geodesic_ab_fisher(t), geodesic_cd_fisher(t))), \n ax=ax1, space='H2_poincare_half_plane', label='Fisher information geodesic',\n color='black')\nvisualization.plot(\n gs.vstack((geodesic_ab_wasserstein(t), geodesic_cd_wasserstein(t))),\n ax=ax1, space='H2_poincare_half_plane', label='Wasserstein geodesic',\n color='black', alpha=0.5)\nvisualization.plot(\n gs.stack((point_a, point_b)), ax=ax1, space='H2_poincare_half_plane', \n label='points a and b', s=100)\nvisualization.plot(\n gs.stack((point_c, point_d)), ax=ax1, space='H2_poincare_half_plane', \n label='points c and d', s=100)\n\nax1.set_xlim(0., 4.)\nax1.set_ylim(0., 4.)\nax1.legend();\n\nax2 = fig.add_subplot(122)\nx = gs.linspace(-3., 7., 100)\nlines = [Line2D([0], [0], color='C0'),\n Line2D([0], [0], color='C1')]\nax2.plot(x, pdfs(x)[:, :2], c='C0')\nax2.plot(x, pdfs(x)[:, 2:], c='C1')\nax2.legend(lines, ['pdfs a and b', 'pdfs c and d']);",
"_____no_output_____"
]
],
[
[
"## Beta distributions",
"_____no_output_____"
],
[
"Let us now consider the example of beta distributions, where the space of parameters is the first quadrant. In this case, the geodesics for the Fisher-Rao metric do not have a closed form, but can be found numerically [[LGRP2020]](#References). Here we plot an example of geodesic ball.",
"_____no_output_____"
]
],
[
[
"from geomstats.information_geometry.beta import BetaDistributions\n\nbeta = BetaDistributions()",
"_____no_output_____"
],
[
"n_rays = 50\ncenter = gs.array([2., 2.])\ntheta = gs.linspace(-gs.pi, gs.pi, n_rays)\ndirections = gs.transpose(\n gs.stack((gs.cos(theta), gs.sin(theta))))\n\nfig = plt.figure(figsize=(5, 5))\nax = fig.add_subplot(111)\nray_length = 0.25\ndirection_norms = beta.metric.squared_norm(directions, center)**(1/2)\nunit_vectors = directions/gs.expand_dims(direction_norms, 1)\ninitial_vectors = ray_length * unit_vectors\n\nn_points = 10\nt = gs.linspace(0., 1., n_points)\nfor j in range(n_rays):\n geod = beta.metric.geodesic(\n initial_point=center, initial_tangent_vec=initial_vectors[j, :])\n ax.plot(*gs.transpose(gs.array([geod(k) for k in t])))\nax.set_xlim(1, 3)\nax.set_ylim(1, 3)\nax.set_title('Geodesic ball of the space of beta distributions');",
"_____no_output_____"
]
],
[
[
"Now we consider an application to the study of the leaf inclination angle distribution of plants. The leaf angle distribution among a common plant species can be appropriately represented by a beta distribution ([CPR2018](#References)). The dataset `leaves` ([CPR2018](#References)) contains pairs of beta distribution parameters, each describing the distribution of the inclination angles of leaves inside a given plant species. These species are divided into 5 categories according to inclination angle distribution type: spherical, erectophile, uniform, planophile and plagiophile.",
"_____no_output_____"
]
],
[
[
"import geomstats.datasets.utils as data_utils\n\nbeta_param, distrib_type = data_utils.load_leaves()",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(5, 5))\nfor distrib in set(distrib_type):\n points = beta_param[distrib_type==distrib, :]\n plt.plot(points[:, 0], points[:, 1], 'o', label=distrib)\nplt.title('Beta parameters of the leaf inclination angle distributions of 172 different species')\nplt.legend();",
"_____no_output_____"
]
],
[
[
"Using the ```FrechetMean``` learning class, we can compute the leaf inclination angle mean distribution among the species of type 'planophile'.",
"_____no_output_____"
]
],
[
[
"from geomstats.learning.frechet_mean import FrechetMean\n\npoints_plan = beta_param[distrib_type=='planophile', :]\n\nmean = FrechetMean(metric=beta.metric)\nmean.fit(points_plan)\n\nmean_estimate = mean.estimate_",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(5, 5))\nplt.plot(points_plan[:, 0], points_plan[:, 1], 'o', label='planophile')\nplt.plot(*mean_estimate, 'o', markersize=10, label='mean planophile')\nplt.title('Beta parameters of the leaf inclination angle mean distribution '\n 'of species of planophile type')\nplt.legend();",
"_____no_output_____"
]
],
[
[
"## References\n\n.. [A2016] S. Amari. Information geometry and its applications. Vol. 194. Springer, 2016.\n\n.. [AM1981] C. Atkinson and A. FS Mitchell. Rao’s distance measure. Sankhya: The Indian Journal of Statistics. Series A, pp. 345–365, 1981.\n\n.. [BGKL2017] J. Bigot, R. Gouet, T. Klein and A. López. Geodesic PCA in the Wasserstein space by convex PCA. In Annales de l'Institut Henri Poincaré, Probabilités et Statistiques. Vol. 53. No. 1. Institut Henri Poincaré, 2017.\n\n.. [CPR2018] F. Chianucci, J. Pisek, K. Raabe et al. A dataset of leaf inclination angles for temperate and boreal broadleaf woody species. Annals of Forest Science Vol. 75, No. 50, 2018. https://doi.org/10.17632/4rmc7r8zvy.2.\n\n.. [LGRP2020] A. Le Brigant, N. Guigui, S. Rebbah and S. Puechmorel, Classifying histograms of medical data using information geometry of beta distributions. IFAC-PapersOnLine, Vol. 54, No. 9, 514-520, 2021.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
ecf3dce574038c7533697296db25525681f8dea7 | 345,666 | ipynb | Jupyter Notebook | intro.ipynb | mmckerns/tuthpc | bf5b1a6b5e31ce4703fe344a51514f85a8e93394 | [
"BSD-3-Clause"
] | 116 | 2015-07-07T12:58:37.000Z | 2022-03-04T08:04:19.000Z | intro.ipynb | meitric/tuthpc | 178f06202a087137859969fb8bbb2b0dd4b37414 | [
"BSD-3-Clause"
] | 6 | 2015-07-07T15:32:38.000Z | 2016-07-20T10:04:48.000Z | intro.ipynb | meitric/tuthpc | 178f06202a087137859969fb8bbb2b0dd4b37414 | [
"BSD-3-Clause"
] | 62 | 2015-07-07T13:12:41.000Z | 2021-02-22T21:15:03.000Z | 78.811218 | 106 | 0.817558 | [
[
[
"# Motivating example: betting strategies in monte carlo games",
"_____no_output_____"
]
],
[
[
"%%file roll.py\n\"\"\"\nrolls a slightly biased (100-sided) die\n\"\"\"\n\nimport random\n\n\ndef win(x):\n '''\n a simple win/loss: where win if (100 > x > 50)\n '''\n return int(bool(100 > x > 50))\n\n\ndef die():\n ''' \n a simple win/loss: where win if (100 > random(1,100) > 50)\n ''' \n return win(random.randint(1,100))\n\n\n# EOF",
"Overwriting roll.py\n"
],
[
"%%file use_roll.py\n\"\"\"\nusage testing of roll\n\"\"\"\n\nimport roll\n\ndef roll_dice(N):\n \"roll a biased 100-sided die N times\"\n rolls = []\n for i in range(N):\n rolls.append(roll.die())\n return rolls\n\n\nif __name__ == '__main__':\n N = 25\n print(roll_dice(N))\n\n\n# EOF",
"Overwriting use_roll.py\n"
],
[
"!python use_roll.py",
"[1, 1, 0, 1, 0, 0, 1, 0, 1, 1, 0, 0, 1, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0]\r\n"
],
[
"%%file strategy.py\n\"\"\"\nbetting strategies\n\nforked from: http://pythonprogramming.net/dashboard/#tab_montecarloyo\n\"\"\"\n\nimport roll\n\n\ndef simple(funds, initial_wager, wager_count, *args):\n '''\n Simple bettor, betting the same amount each time.\n '''\n value = funds\n wager = initial_wager\n history = [funds] + [0]*(wager_count)\n\n currentWager = 1\n\n while currentWager <= wager_count:\n if roll.die():\n value += wager\n history[currentWager] = value\n else:\n value -= wager\n if value <= 0:\n break\n history[currentWager] = value\n currentWager += 1\n\n return history\n\n\ndef martingale(funds, initial_wager, wager_count, wscale=1, lscale=1):\n '''martingale bettor, \"doubling-down\" (actually, \"*scaling-down\")'''\n value = funds\n wager = initial_wager\n history = [funds] + [0]*(wager_count)\n\n currentWager = 1\n\n # since we'll be betting based on previous bet outcome #\n previousWager = 'win'\n\n # since we'll be doubling #\n previousWagerAmount = initial_wager\n\n while currentWager <= wager_count:\n if previousWager == 'win':\n if roll.die():\n value += wager\n history[currentWager] = value\n else:\n value -= wager\n if value <= 0:\n break\n history[currentWager] = value\n previousWager = 'loss'\n previousWagerAmount = wager\n elif previousWager == 'loss':\n if roll.die():\n wager = previousWagerAmount * wscale\n if (value - wager) < 0:\n wager = value\n value += wager\n history[currentWager] = value\n wager = initial_wager\n previousWager = 'win'\n else:\n wager = previousWagerAmount * lscale\n if (value - wager) < 0:\n wager = value\n value -= wager\n if value <= 0:\n break\n history[currentWager] = value\n previousWager = 'loss'\n previousWagerAmount = wager\n\n currentWager += 1\n\n return history\n\n\ndef dAlembert(funds, initial_wager, wager_count, *args):\n '''d'Alembert bettor'''\n value = funds\n wager = initial_wager\n history = [funds] + [0]*(wager_count)\n\n currentWager = 1\n\n # since we'll be betting based on previous bet outcome #\n previousWager = 'win'\n\n # since we'll be doubling #\n previousWagerAmount = initial_wager\n\n while currentWager <= wager_count:\n if previousWager == 'win':\n if wager == initial_wager:\n pass\n else:\n wager -= initial_wager\n if roll.die():\n value += wager\n history[currentWager] = value\n else:\n value -= wager\n if value <= 0:\n break\n history[currentWager] = value\n previousWager = 'loss'\n previousWagerAmount = wager\n elif previousWager == 'loss':\n wager = previousWagerAmount + initial_wager\n if (value - wager) < 0:\n wager = value\n if roll.die():\n value += wager\n history[currentWager] = value\n previousWager = 'win'\n else:\n value -= wager\n if value <= 0:\n break\n history[currentWager] = value\n previousWager = 'loss'\n previousWagerAmount = wager\n\n currentWager += 1\n\n return history\n\n\n# EOF",
"Overwriting strategy.py\n"
],
[
"%%file use_strategy.py\n\"\"\"\nusage testing of strategy\n\"\"\"\n\nimport numpy as np\nimport matplotlib.pylab as mpl\nimport strategy\n\n\ndef plot_trajectory(initial_funds, initial_bets, number_bets, *args, **kwds):\n \"plot the rolls of a 100-sided die for all strategies\"\n bettors = (strategy.simple, strategy.martingale, strategy.dAlembert)\n for bettor in bettors:\n mpl.plot(bettor(initial_funds, initial_bets, number_bets, *args, **kwds))\n # baseline\n mpl.plot(initial_funds * np.ones(number_bets+1), lw=2)\n return\n\n\nif __name__ == '__main__':\n initial_funds = 10000\n initial_bet = 100\n number_bets = 1000\n W = L = 2\n\n plot_trajectory(initial_funds, initial_bet, number_bets, W, L)\n mpl.show()\n\n\n# EOF",
"Overwriting use_strategy.py\n"
],
[
"!python2.7 use_strategy.py",
"_____no_output_____"
],
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import matplotlib.pylab as mpl\nimport use_strategy as use\ninitial_funds = 10000\ninitial_bet = 100\nnumber_bets = 1000\nW = L = 2\n\nuse.plot_trajectory(initial_funds, initial_bet, number_bets, W, L)\nmpl.show()",
"_____no_output_____"
],
[
"%%file trials.py\n\"\"\"\nmonte carlo trials for betting strategies, and measures of success\n\"\"\"\n\nimport numpy as np\nfrom strategy import simple, martingale, dAlembert\n\n\ndef monte(bettor, initial_funds, initial_bet, number_bets, number_players, W,L):\n \"monte carlo run for a betting strategy\"\n history = []\n while len(history) < number_players:\n history.append(bettor(initial_funds, initial_bet, number_bets, W,L))\n return np.array(history)\n\n\ndef alive(history, number_players):\n \"find the percentage of players that are not broke\"\n return 100. * sum(np.asarray(history, bool).T[-1])/number_players\n\n\ndef gains(history, number_players, initial_funds):\n \"find the percentage of players that have profited\"\n return 100. * sum(history.T[-1] > initial_funds)/number_players\n\n\ndef profit(history, number_players, initial_funds):\n \"find the total profit\"\n return np.max(history.T[-1]) - initial_funds\n\n\ndef margin(history, number_players, initial_funds):\n \"find the percentage the return on investment is over the initial funds\"\n initial = number_players * initial_funds\n return 100.* (sum(history.T[-1]) - initial)/initial\n\n\n# EOF",
"Overwriting trials.py\n"
],
[
"%%file use_trials.py\n\"\"\"\nusage testing of trials\n\"\"\"\n\nimport numpy as np\nimport matplotlib.pylab as mpl\nimport trials\n\n\ndef plot_trajectories(bettor, initial_funds, initial_bet, number_bets, number_players, W,L):\n \"plot the Monte Carlo trajectories of a 100-sided die for the selected strategy\"\n history = trials.monte(bettor, initial_funds, initial_bet, number_bets, number_players, W,L)\n mpl.plot(history.T)\n mpl.plot(initial_funds * np.ones(number_bets+1), lw=2)\n\n print \"survived: {}%\".format(trials.alive(history, number_players))\n print \"profited: {}%\".format(trials.gains(history, number_players, initial_funds))\n funds = trials.margin(history, number_players, initial_funds)\n funds = np.mean(funds)*initial_funds\n win = funds >= 0\n print \"ave profit: {}${:.2f}\".format('' if win else '-', funds if win else -funds)\n funds = trials.profit(history, number_players, initial_funds)\n win = funds >= 0\n print \"max profit: {}${:.2f}\".format('' if win else '-', funds if win else -funds)\n return \n\n\nif __name__ == '__main__':\n import strategy\n initial_funds = 10000\n initial_bet = 100\n number_bets = 1000\n number_players = 100\n bettor = strategy.martingale\n W = L = 2.0\n\n plot_trajectories(bettor, initial_funds, initial_bet, number_bets, number_players, W,L)\n mpl.show()\n\n\n# EOF",
"Overwriting use_trials.py\n"
],
[
"import matplotlib.pylab as mpl\nimport use_trials as use\nimport strategy\ninitial_funds = 10000\ninitial_bet = 100\nnumber_bets = 1000\nnumber_players = 100\nbettor = strategy.martingale\nW = L = 2.0\n\nuse.plot_trajectories(bettor, initial_funds, initial_bet, number_bets, number_players, W,L)\nmpl.show()",
"survived: 15.0%\nprofited: 15.0%\nave profit: -$221700.00\nmax profit: $53000.00\n"
],
[
"import matplotlib.pylab as mpl\nimport use_trials as use\nimport strategy\ninitial_funds = 10000\ninitial_bet = 100\nnumber_bets = 1000\nnumber_players = 100\nbettor = strategy.simple\nW = L = 2.0\n\nuse.plot_trajectories(bettor, initial_funds, initial_bet, number_bets, number_players, W,L)\nmpl.show()",
"survived: 99.0%\nprofited: 27.0%\nave profit: -$179800.00\nmax profit: $5200.00\n"
],
[
"%%file optimize.py\n'''\nmonte carlo optimization of betting strategies for a given quantitative measure\n'''\n\nimport numpy as np\nfrom itertools import imap\nimport strategy\nfrom trials import monte, alive, gains, profit, margin\n\n\ndef monte_stats(bettor, initial_funds, initial_bet, number_bets, number_players, W,L):\n \"get the desired stats from a monte carlo run\"\n settings = np.seterr(over='ignore', invalid='ignore')\n history = monte(bettor, initial_funds, initial_bet, number_bets, number_players, W,L)\n survived = alive(history, number_players)\n profited = gains(history, number_players, initial_funds)\n max_profit = profit(history, number_players, initial_funds)\n ave_profit = margin(history, number_players, initial_funds)\n np.seterr(**settings)\n return survived, profited, max_profit, ave_profit\n\n\ndef safety_metric(initial_funds, survived, profited, max_profit, ave_profit):\n \"we define optimality with (4*profited + survived)/5\"\n return (4*profited + survived)/5\n\n\ndef profit_metric(initial_funds, survived, profited, max_profit, ave_profit):\n \"we define optimality with (max_profit - ave_profit)\"\n return max_profit - (np.mean(ave_profit) * initial_funds)\n\n\ndef optimize_WL(metric, players=100, funds=None, bet=None, number_bets=None, symmetric=False):\n \"soewhat hacky Monte Carlo optimization of betting parameters for martingale strategy\"\n samples = int(np.random.uniform(100., 1000.)) if players is None else int(players)\n if funds is None: funds = np.random.uniform(1000. ,1000000.)\n if bet is None: bet = np.random.uniform(1. ,1000.)\n number_bets = int(np.random.uniform(10.,10000.)) if number_bets is None else int(number_bets)\n bettor = strategy.martingale\n \n W = np.random.uniform(0.1, 10., samples)\n L = W if symmetric else np.random.uniform(0.1, 10., samples)\n \n def measure(W,L):\n \"let's hide this, because it's ugly (*groan*)\" # good indicator a class should be built\n return metric(funds, *monte_stats(bettor, funds, bet, number_bets, samples, W,L))\n \n # use imap to run the monte carlo, because it's cool\n results = imap(measure, W,L)\n\n i = 0\n best_result = 0.0\n best_value = W[0],L[0]\n for result in results:\n if result > best_result:\n best_result = result\n best_value = W[i],L[i]\n print \"best: %s @ %s\" % (best_result, best_value)\n i += 1\n if not i%np.floor_divide(samples, 10):\n print \"{:.2f}% done\".format(100. * i / samples)\n\n return best_value\n\n\nif __name__ == '__main__':\n initial_funds = 10000\n initial_bet = 100\n number_bets = 100\n number_players = 300 # XXX: THIS SHOULD BE MUCH LARGER\n metric = safety_metric\n symmetric = True\n\n print \"optimizing W,L for symmetric=%s\\n%s\\n\" % (symmetric, metric.__doc__)\n optimize_WL(metric, number_players, initial_funds, initial_bet, number_bets, symmetric)\n\n\n# EOF",
"Overwriting optimize.py\n"
],
[
"!python2.7 optimize.py",
"optimizing W,L for symmetric=True\nwe define optimality with (4*profited + survived)/5\n\nbest: 2.33333333333 @ (8.5757103167409081, 8.5757103167409081)\nbest: 4.66666666667 @ (8.3045062569027834, 8.3045062569027834)\nbest: 51.4666666667 @ (0.30982064128937659, 0.30982064128937659)\nbest: 56.8 @ (0.30217794763987715, 0.30217794763987715)\nbest: 67.0666666667 @ (1.5010348603055301, 1.5010348603055301)\n10.00% done\n20.00% done\nbest: 73.7333333333 @ (1.6427232291502374, 1.6427232291502374)\n30.00% done\n40.00% done\n50.00% done\n60.00% done\nbest: 73.9333333333 @ (1.7790024945283196, 1.7790024945283196)\n70.00% done\n80.00% done\n90.00% done\n100.00% done\n"
]
],
[
[
"Now on to [the tutorial](efficient.ipynb)...",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecf3e7bcb76b8729b04f2e92a73deb2a8d05383c | 589,404 | ipynb | Jupyter Notebook | Assignment-7.ipynb | shobes572/DSI_Module-25 | 686bcd48f5ef2919a7df71a9c7e033e6f36006f5 | [
"MIT"
] | null | null | null | Assignment-7.ipynb | shobes572/DSI_Module-25 | 686bcd48f5ef2919a7df71a9c7e033e6f36006f5 | [
"MIT"
] | null | null | null | Assignment-7.ipynb | shobes572/DSI_Module-25 | 686bcd48f5ef2919a7df71a9c7e033e6f36006f5 | [
"MIT"
] | null | null | null | 1,056.27957 | 573,100 | 0.948387 | [
[
[
"import numpy as np\nimport pandas as pd\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nfrom sklearn.datasets import fetch_openml\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.decomposition import PCA",
"_____no_output_____"
]
],
[
[
"### 1. Load the dataset and make your preprocessing like normalizing the data.",
"_____no_output_____"
]
],
[
[
"mnist = fetch_openml('Fashion-MNIST', version=1, cache=True)",
"_____no_output_____"
],
[
"target_dict = {\n 0: 'T-shirt/top',\n 1: 'Trouser',\n 2: 'Pullover',\n 3: 'Dress',\n 4: 'Coat',\n 5: 'Sandal',\n 6: 'Shirt',\n 7: 'Sneaker',\n 8: 'Bag',\n 9: 'Ankle Boot',\n}",
"_____no_output_____"
],
[
"X = pd.DataFrame(mnist['data'])\ny = pd.Series(mnist['target'])\nX = X.sample(10000, random_state=36)\ny = y.iloc[X.index]\nX = X.reset_index(drop=True)\ny = y.astype(int).reset_index(drop=True)\ny = y.replace(target_dict)\ndisplay(\n X,\n y\n)",
"_____no_output_____"
],
[
"scaler = StandardScaler()\nscaled_X = scaler.fit_transform(X)\ndisplay(scaled_X)",
"_____no_output_____"
],
[
"pca_model = PCA(n_components=2)\npca_X = pca_model.fit_transform(scaled_X)\npca_X",
"_____no_output_____"
],
[
"plt.figure(figsize=(12, 11))\nsns.scatterplot(pca_X[:, 0], pca_X[:, 1], hue=y, legend='full')\nplt.show()",
"_____no_output_____"
]
],
[
[
"No, there is still a significant amout of overlap despite seeing multiple slightly segregated clusters",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
ecf3f656ba77f9d3ed43e9271833d14baee49aef | 3,222 | ipynb | Jupyter Notebook | MHD/FEniCS/MHD/CG/PicardIter_Direct/.ipynb_checkpoints/Untitled0-checkpoint.ipynb | wathen/PHD | 35524f40028541a4d611d8c78574e4cf9ddc3278 | [
"MIT"
] | 3 | 2020-10-25T13:30:20.000Z | 2021-08-10T21:27:30.000Z | MHD/FEniCS/MHD/CG/PicardIter_Direct/.ipynb_checkpoints/Untitled0-checkpoint.ipynb | wathen/PHD | 35524f40028541a4d611d8c78574e4cf9ddc3278 | [
"MIT"
] | null | null | null | MHD/FEniCS/MHD/CG/PicardIter_Direct/.ipynb_checkpoints/Untitled0-checkpoint.ipynb | wathen/PHD | 35524f40028541a4d611d8c78574e4cf9ddc3278 | [
"MIT"
] | 3 | 2019-10-28T16:12:13.000Z | 2020-01-13T13:59:44.000Z | 40.275 | 556 | 0.578212 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecf3f8f332cfc5b68a88689e698dfa7afb2a1eb3 | 196,015 | ipynb | Jupyter Notebook | P1.ipynb | Philippe-Capdepuy/CarND-LaneLines-P1 | 624e70ee461a466793a48f5b6da0259d7135414f | [
"MIT"
] | null | null | null | P1.ipynb | Philippe-Capdepuy/CarND-LaneLines-P1 | 624e70ee461a466793a48f5b6da0259d7135414f | [
"MIT"
] | null | null | null | P1.ipynb | Philippe-Capdepuy/CarND-LaneLines-P1 | 624e70ee461a466793a48f5b6da0259d7135414f | [
"MIT"
] | null | null | null | 78.879276 | 121,953 | 0.773135 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecf400ee999cd4db987d94b8bd395d8e9f9fbe74 | 1,408 | ipynb | Jupyter Notebook | Lecture Material/08_File_IO/08.8_Summary.ipynb | knherrera/pcc-cis-012-intro-to-programming-python | f2fb8ec5b242fc6deb5c0e2abda60bb91171aad4 | [
"MIT"
] | 23 | 2020-02-19T22:07:17.000Z | 2021-08-19T20:43:21.000Z | Lecture Material/08_File_IO/08.8_Summary.ipynb | knherrera/pcc-cis-012-intro-to-programming-python | f2fb8ec5b242fc6deb5c0e2abda60bb91171aad4 | [
"MIT"
] | 12 | 2020-03-04T04:34:38.000Z | 2021-02-23T04:28:31.000Z | Lecture Material/08_File_IO/08.8_Summary.ipynb | knherrera/pcc-cis-012-intro-to-programming-python | f2fb8ec5b242fc6deb5c0e2abda60bb91171aad4 | [
"MIT"
] | 18 | 2020-03-05T05:21:11.000Z | 2022-03-05T05:57:12.000Z | 27.076923 | 87 | 0.574574 | [
[
[
"# 9.8 Summary\n\nCongrats! You've completed this module. In this module you learned:\n\n- What a file is.\n- Where files are stored on the computing device.\n- What are file paths and the differences between absolute and relative paths.\n- Line ending and why they matter.\n- The `pathlib` library and its basic usage.\n- How to open and close a file properly.\n- How to read and write opened files.\n- Some common scenarios for file IO such as:\n - Iteratting over lines in a file.\n - Searching within a file.\n - Appending to a file.\n - Working with two files at the same time.\n- What a binary file is and how to work with them.\n- Reading the CSV and JSON file formats.\n\nKeep up the good work and see you soon.",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
ecf409bf2e9beb2c5bc19c0c0eed7b11c02c339e | 10,063 | ipynb | Jupyter Notebook | extract_df.ipynb | ferdinandwp/dataprep | c5fe22e6011610908c275e8f6591ba92529e8127 | [
"MIT"
] | null | null | null | extract_df.ipynb | ferdinandwp/dataprep | c5fe22e6011610908c275e8f6591ba92529e8127 | [
"MIT"
] | null | null | null | extract_df.ipynb | ferdinandwp/dataprep | c5fe22e6011610908c275e8f6591ba92529e8127 | [
"MIT"
] | null | null | null | 34.940972 | 1,046 | 0.503826 | [
[
[
"# Import Library\nimport numpy as np\nimport pandas as pd\nimport os\nimport sqlalchemy as db\nimport mysql.connector\nfrom mysql.connector import errorcode\nfrom sqlalchemy import create_engine\nimport pymysql\nimport xlrd\nimport decimal\nfrom datetime import date, datetime\n\nD = decimal.Decimal\n",
"_____no_output_____"
],
[
"data_path1 = 'C:\\\\Users\\\\fwidjojoputra\\\\Desktop\\\\data\\\\'\ndata_path2 = 'E:\\\\EASi\\\\5-Operations\\\\50-Management\\\\500-DataCentral\\\\3-Super User\\\\Development\\\\data_repository\\\\msp_data\\\\'\n\nfile_name1 = os.path.join(data_path1,'resources.csv') \nfile_name2 = os.path.join(data_path1,'projects.csv') \nfile_name3 = os.path.join(data_path1,'tasks.csv') \nfile_name4 = os.path.join(data_path1,'assignments.csv') \nfile_name5 = os.path.join(data_path1,'tasktime_phased.csv')\nfile_name6 = os.path.join(data_path2,'psoft_data.csv')\nfile_name7 = os.path.join(data_path2,'psoft_prj_data.csv')",
"_____no_output_____"
],
[
"df1 = pd.read_csv(file_name1) # res\ndf2 = pd.read_csv(file_name2) # prj\ndf3 = pd.read_csv(file_name3) # task\ndf4 = pd.read_csv(file_name4) # ass\ndf5 = pd.read_csv(file_name5) # task_tp\ndf6 = pd.read_csv(file_name6) # psoft_data\ndf7 = pd.read_csv(file_name7) # psoft__prj_data",
"_____no_output_____"
],
[
"#-------------------- Data prep RESOURCES --------------------#\nres_df = df1.loc[:,['DataCentralID',\n 'ResourceId']]\n\nprj_df = df2.loc[:,['ProjectId',\n 'SOWNb',\n 'ProjectActualFinishDate',\n 'year']]\n\ntsk_df = df3.loc[:,['TaskId',\n 'ParentTaskId',\n 'ProjectId',\n 'TaskName']]\n\nass_df = df4.loc[:,['ProjectId',\n 'TaskId',\n 'AssignmentId',\n 'ResourceId']]\n\ntsktp_df = df5.loc[:,['TaskId',\n 'ProjectId',\n 'TaskWork',\n 'TaskCost',\n 'TaskName']] \n\npsoft_df = df6.loc[:,['Customer',\n 'Category',\n 'Order No',\n 'Job Req #',\n 'ID',\n 'Name',\n 'Earn Code',\n 'Blended Rate',\n 'Job Code']]\n\npsoft_prj_df = df7.loc[:,['Project',\n 'Psoft Project ID',\n 'EASi ID',\n 'Job Req',\n 'Bill Rate']]\n\nres_df.rename({'DataCentralID':'easi_id',\n 'ResourceId':'res_id'},axis=1, inplace=True)\n\nprj_df.rename({'SOWNb':'sow_no',\n 'ProjectId':'prj_id',\n 'ProjectActualFinishDate':'end_date'},axis=1, inplace=True)\n\ntsk_df.rename({'TaskId':'task_id',\n 'ParentTaskId':'parent_id',\n 'ProjectId':'prj_id',\n 'TaskName':'task_name'},axis=1, inplace=True)\n\nass_df.rename({'ProjectId':'prj_id',\n 'TaskId':'task_id',\n 'AssignmentId':'ass_id',\n 'ResourceId':'res_id'},axis=1, inplace=True)\n\ntsktp_df.rename({'TaskId':'task_id',\n 'ProjectId':'prj_id',\n 'TaskWork':'task_work',\n 'TaskCost':'task_cost',\n 'TaskName':'task_name'},axis=1, inplace=True)\n\npsoft_df.rename({'Customer':'client',\n 'Category':'cat',\n 'Order No':'ord_no',\n 'Job Req #':'job_req',\n 'ID':'easi_id',\n 'Name':'res_name',\n 'Earn Code':'earn_code',\n 'Blended Rate':'br',\n 'Job Code':'job_code'},axis=1, inplace=True)\n\npsoft_prj_df.rename({'Project':'sow_no',\n 'Psoft Project ID':'psoft_prj_id',\n 'EASi ID':'easi_id',\n 'Job Req':'job_req',\n 'Bill Rate':'br'},axis=1, inplace=True)",
"_____no_output_____"
],
[
"prj_tsk_df = tsk_df.merge(prj_df, on='prj_id', how='left')\nprj_task_ass_df = prj_tsk_df.merge(ass_df, on='task_id', how='inner')\nprj_task_ass_res_df = prj_task_ass_df.merge(res_df, on='res_id', how='left')\n\n# group dataset by task_id\ntask_timephased_df = tsktp_df.groupby('task_id')['task_work','task_cost'].sum()\ntsktp_df1 = task_timephased_df.reset_index()\ntsktp_df1['br'] = tsktp_df1['task_cost']/tsktp_df1['task_work']\n\nall_df = prj_task_ass_res_df.merge(tsktp_df1, on='task_id', how='left')",
"_____no_output_____"
],
[
"# convert easi id to int\nall_df = all_df.fillna(value='0')\ntest_df = all_df[all_df.easi_id != 'xxx']\ntest_df['easi_id'] = test_df['easi_id'].astype('int64')\ntest_df['br'] = test_df['br'].astype('float64')",
"_____no_output_____"
],
[
"# remove id 0 and 1\ntest_df2 = test_df[test_df.easi_id != 0]\ntest_df3 = test_df2[test_df2.easi_id != 1]\ntest_df = test_df3.round(2)\n# Collect required data for merging\ninit_df = test_df.loc[:,['sow_no',\n 'task_name',\n 'easi_id',\n 'br']]",
"_____no_output_____"
],
[
"init_df['easi_id'].nunique(), init_df.shape\ninit_df['br'].nunique()",
"_____no_output_____"
],
[
"# Handle psoft data\n# psoft_df['easi_id'].nunique(), psoft_df.shape\npsoft_df['br'].nunique()",
"_____no_output_____"
],
[
"# Combined set\nfinal_df = init_df.merge(psoft_df, left_on=['easi_id','br'], right_on=['easi_id','br'], how='inner')\nfinal_df.shape",
"_____no_output_____"
],
[
"final_df.shape, psoft_prj_df.shape",
"_____no_output_____"
],
[
"extract_df = final_df.merge(psoft_prj_df, left_on=['sow_no','easi_id','br','job_req'], right_on=['sow_no','easi_id','br','job_req'], how='inner')\nextract_df.head(50) ",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf41856145f111b20c14d3c92efff7c69b64175 | 281,359 | ipynb | Jupyter Notebook | notebooks/GLM-HMM for blockworld.ipynb | nhat-le/ssm | 2f386c04bf7540b0075f40b5d0ae3923296d8bfd | [
"MIT"
] | null | null | null | notebooks/GLM-HMM for blockworld.ipynb | nhat-le/ssm | 2f386c04bf7540b0075f40b5d0ae3923296d8bfd | [
"MIT"
] | null | null | null | notebooks/GLM-HMM for blockworld.ipynb | nhat-le/ssm | 2f386c04bf7540b0075f40b5d0ae3923296d8bfd | [
"MIT"
] | null | null | null | 250.319395 | 39,540 | 0.913573 | [
[
[
"# Input Driven Observations (\"GLM-HMM\")\n\nNotebook prepared by Zoe Ashwood: feel free to email me with feedback or questions (zashwood at cs dot princeton dot edu).\n\nThis notebook demonstrates the \"InputDrivenObservations\" class, and illustrates its use in the context of modeling decision-making data as in Ashwood et al. (2020) ([Mice alternate between discrete strategies during perceptual\ndecision-making](https://www.biorxiv.org/content/10.1101/2020.10.19.346353v1.full.pdf)).\n\nCompared to the model considered in the notebook [\"2 Input Driven HMM\"](https://github.com/lindermanlab/ssm/blob/master/notebooks/2%20Input%20Driven%20HMM.ipynb), Ashwood et al. (2020) assumes a stationary transition matrix where transition probabilities *do not* depend on external inputs. However, observation probabilities now *do* depend on external covariates according to:\n\n$$\n\\begin{align}\n\\Pr(y_t = c \\mid z_{t} = k, u_t, w_{kc}) = \n\\frac{\\exp\\{w_{kc}^\\mathsf{T} u_t\\}}\n{\\sum_{c'=1}^C \\exp\\{w_{kc'}^\\mathsf{T} u_t\\}}\n\\end{align}\n$$\n\nwhere $c \\in \\{1, ..., C\\}$ indicates the categorical class for the observation, $u_{t} \\in \\mathbb{R}^{M}$ is the set of input covariates, and $w_{kc} \\in \\mathbb{R}^{M}$ is the set of input weights associated with state $k$ and class $c$. These weights, along with the transition matrix and initial state probabilities, will be learned.\n\nIn Ashwood et al. (2020), $C = 2$ as $y_{t}$ represents the binary choice made by an animal during a 2AFC (2-Alternative Forced Choice) task. The above equation then reduces to:\n\n$$\n\\begin{align}\n\\Pr(y_t = 1 \\mid z_{t} = k, u_t, w_{k}) = \n\\frac{1}\n{1 + \\exp\\{-w_{k}^\\mathsf{T} u_t\\}}.\n\\end{align}\n$$\n\nand only a single set of weights is associated with each state.",
"_____no_output_____"
],
[
"## 1. Setup\nThe line `import ssm` imports the package for use. Here, we have also imported a few other packages for plotting.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport numpy.random as npr\nimport matplotlib.pyplot as plt\nimport ssm\nfrom ssm.util import one_hot, find_permutation\n\n%matplotlib inline\n\nnpr.seed(0)",
"_____no_output_____"
]
],
[
[
"## 2. Input Driven Observations\nWe create a HMM with input-driven observations and 'standard' (stationary) transitions with the following line: \n```python\n ssm.HMM(num_states, obs_dim, input_dim, observations=\"input_driven_obs\", observation_kwargs=dict(C=num_categories), transitions=\"standard\")\n```\n\nAs in Ashwood et al. (2020), we are going to model an animal's binary choice data during a decision-making task, so we will set `num_categories=2` because the animal only has two options available to it. We will also set `obs_dim = 1` because the dimensionality of the observation data is 1 (if we were also modeling, for example, the binned reaction time of the animal, we could set `obs_dim = 2`). For the sake of simplicity, we will assume that an animal's choice in a particular state is only affected by the external stimulus associated with that particular trial, and its innate choice bias. Thus, we will set `input_dim = 2` and we will simulate input data that resembles sequences of stimuli in what follows. In Ashwood et al. (2020), they found that many mice used 3 decision-making states when performing 2AFC tasks. We will, thus, set `num_states = 3`.",
"_____no_output_____"
],
[
"### 2a. Initialize GLM-HMM",
"_____no_output_____"
]
],
[
[
"# Set the parameters of the GLM-HMM\nnum_states = 3 # number of discrete states\nobs_dim = 1 # number of observed dimensions\nnum_categories = 2 # number of categories for output\ninput_dim = 2 # input dimensions\n\n# Make a GLM-HMM\ntrue_glmhmm = ssm.HMM(num_states, obs_dim, input_dim, observations=\"input_driven_obs\", \n observation_kwargs=dict(C=num_categories), transitions=\"standard\")",
"_____no_output_____"
]
],
[
[
"## Generate test data",
"_____no_output_____"
]
],
[
[
"def categorical(p):\n return (p.cumsum(-1) >= np.random.uniform(size=p.shape[:-1])[..., None]).argmax(-1)",
"_____no_output_____"
],
[
"zcurr = 0\nscurr = 0\n\ndef next_state(zcurr):\n zlst = [0, 1, 2]\n if np.random.uniform() < 0.1: #switch\n zlst.pop(zcurr)\n return zlst[0] if np.random.uniform() > 0.5 else zlst[1]\n return zcurr\n\ndef next_target(scurr):\n return scurr if np.random.uniform() > 0.1 else 1 - scurr\n\ndef sigmoid(x):\n return 1 / (1 + np.exp(-x))\n\nzlst = [0]\nylst = [0]\nslst = [0]\nrlst = [0]\nw = np.array([[5, 0], [0, 5], [0, -5]])\nnp.random.seed(0)\nfor i in range(1000):\n zcurr = next_state(zcurr)\n zlst.append(zcurr)\n \n scurr = next_target(scurr)\n slst.append(scurr)\n \n yprob = w[zcurr,0] * (int(ylst[i]) * 2 - 1) * (int(rlst[i]) * 2 - 1) + w[zcurr, 1]\n y = np.random.uniform() < sigmoid(yprob)\n ylst.append(y)\n \n rcurr = y == scurr\n rlst.append(rcurr)\n",
"_____no_output_____"
],
[
"inpts1 = (np.array(ylst, dtype='int') * 2 - 1) * (np.array(rlst, dtype='int') * 2 - 1)\ninpts2 = np.ones(len(ylst))\ninpts = np.array([inpts1, inpts2]).T\ntrue_choices = np.array([ylst], dtype='int').T\ninpts = inpts[:-1,:]\ntrue_choices = true_choices[1:]\nprint('inpts shape:', inpts.shape, '. true choices shape:', true_choices.shape)",
"inpts shape: (1000, 2) . true choices shape: (1000, 1)\n"
],
[
"plt.plot(rlst,label='r')\nplt.plot(zlst, label='z')\nplt.plot(ylst, label='y')\nplt.plot(slst, label='s')\nplt.xlim(0, 100)\nplt.legend()",
"_____no_output_____"
]
],
[
[
"## 3. Fit GLM-HMM and perform recovery analysis",
"_____no_output_____"
],
[
"### 3a. Maximum Likelihood Estimation",
"_____no_output_____"
],
[
"Now we instantiate a new GLM-HMM and check that we can recover the generative parameters in simulated data:",
"_____no_output_____"
]
],
[
[
"new_glmhmm = ssm.HMM(num_states, obs_dim, input_dim, observations=\"input_driven_obs\", \n observation_kwargs=dict(C=num_categories), transitions=\"standard\")\n\nN_iters = 200 # maximum number of EM iterations. Fitting with stop earlier if increase in LL is below tolerance specified by tolerance parameter\nfit_ll = new_glmhmm.fit(true_choices, inputs=inpts, method=\"em\", num_iters=N_iters, tolerance=10**-4)",
"_____no_output_____"
],
[
"# Plot the log probabilities of the true and fit models. Fit model final LL should be greater \n# than or equal to true LL.\nfig = plt.figure(figsize=(4, 3), dpi=80, facecolor='w', edgecolor='k')\nplt.plot(fit_ll, label=\"EM\")\n#plt.plot([0, len(fit_ll)], true_ll * np.ones(2), ':k', label=\"True\")\nplt.legend(loc=\"lower right\")\nplt.xlabel(\"EM Iteration\")\nplt.xlim(0, len(fit_ll))\nplt.ylabel(\"Log Probability\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 3b. Retrieved parameters",
"_____no_output_____"
],
[
"Compare retrieved weights and transition matrices to generative parameters. To do this, we may first need to permute the states of the fit GLM-HMM relative to the\ngenerative model. One way to do this uses the `find_permutation` function from `ssm`:",
"_____no_output_____"
]
],
[
[
"new_glmhmm.permute(find_permutation(true_latents[0], new_glmhmm.most_likely_states(true_choices[0], input=inpts[0])))",
"_____no_output_____"
]
],
[
[
"Now plot generative and retrieved weights for GLMs (analogous plot to Figure S1c in \nAshwood et al. (2020)):",
"_____no_output_____"
]
],
[
[
"new_glmhmm.ob",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(4, 3), dpi=80, facecolor='w', edgecolor='k')\ncols = ['#ff7f00', '#4daf4a', '#377eb8']\nrecovered_weights = new_glmhmm.observations.params\nfor k in range(num_states):\n if k ==0:\n# plt.plot(range(input_dim), gen_weights[k][0], marker='o',\n# color=cols[k], linestyle='-',\n# lw=1.5, label=\"generative\")\n plt.plot(range(input_dim), recovered_weights[k][0], color=cols[k],\n lw=1.5, label = \"recovered\", linestyle = '--')\n else:\n# plt.plot(range(input_dim), gen_weights[k][0], marker='o',\n# color=cols[k], linestyle='-',\n# lw=1.5, label=\"\")\n plt.plot(range(input_dim), recovered_weights[k][0], color=cols[k],\n lw=1.5, label = '', linestyle = '--')\nplt.yticks(fontsize=10)\nplt.ylabel(\"GLM weight\", fontsize=15)\nplt.xlabel(\"covariate\", fontsize=15)\nplt.xticks([0, 1], ['stimulus', 'bias'], fontsize=12, rotation=45)\nplt.axhline(y=0, color=\"k\", alpha=0.5, ls=\"--\")\nplt.legend()\nplt.title(\"Weight recovery\", fontsize=15)",
"_____no_output_____"
]
],
[
[
"Now plot generative and retrieved transition matrices (analogous plot to Figure S1c in \nAshwood et al. (2020)):",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(5, 2.5), dpi=80, facecolor='w', edgecolor='k')\nplt.subplot(1, 2, 1)\n# gen_trans_mat = np.exp(gen_log_trans_mat)[0]\n# plt.imshow(gen_trans_mat, vmin=-0.8, vmax=1, cmap='bone')\n# for i in range(gen_trans_mat.shape[0]):\n# for j in range(gen_trans_mat.shape[1]):\n# text = plt.text(j, i, str(np.around(gen_trans_mat[i, j], decimals=2)), ha=\"center\", va=\"center\",\n# color=\"k\", fontsize=12)\n# plt.xlim(-0.5, num_states - 0.5)\n# plt.xticks(range(0, num_states), ('1', '2', '3'), fontsize=10)\n# plt.yticks(range(0, num_states), ('1', '2', '3'), fontsize=10)\n# plt.ylim(num_states - 0.5, -0.5)\n# plt.ylabel(\"state t\", fontsize = 15)\n# plt.xlabel(\"state t+1\", fontsize = 15)\n# plt.title(\"generative\", fontsize = 15)\n\n\nplt.subplot(1, 2, 2)\nrecovered_trans_mat = np.exp(new_glmhmm.transitions.log_Ps)\nplt.imshow(recovered_trans_mat, vmin=-0.8, vmax=1, cmap='bone')\nfor i in range(recovered_trans_mat.shape[0]):\n for j in range(recovered_trans_mat.shape[1]):\n text = plt.text(j, i, str(np.around(recovered_trans_mat[i, j], decimals=2)), ha=\"center\", va=\"center\",\n color=\"k\", fontsize=12)\nplt.xlim(-0.5, num_states - 0.5)\nplt.xticks(range(0, num_states), ('1', '2', '3'), fontsize=10)\nplt.yticks(range(0, num_states), ('1', '2', '3'), fontsize=10)\nplt.ylim(num_states - 0.5, -0.5)\nplt.title(\"recovered\", fontsize = 15)\nplt.subplots_adjust(0, 0, 1, 1)\n",
"_____no_output_____"
]
],
[
[
"### 3c. Posterior State Probabilities",
"_____no_output_____"
],
[
"Let's now plot $p(z_{t} = k|\\mathbf{y}, \\{u_{t}\\}_{t=1}^{T})$, the posterior state probabilities, which give the probability of the animal being in state k at trial t.",
"_____no_output_____"
]
],
[
[
"# Get expected states:\nposterior_probs = [new_glmhmm.expected_states(data=data, input=inpt)[0]\n for data, inpt\n in zip([true_choices], [inpts])]",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(5, 2.5), dpi=80, facecolor='w', edgecolor='k')\nsess_id = 0 #session id; can choose any index between 0 and num_sess-1\nfor k in range(num_states):\n plt.plot(posterior_probs[sess_id][:, k], label=\"State \" + str(k + 1), lw=2,\n color=cols[k])\nplt.ylim((-0.01, 1.01))\nplt.yticks([0, 0.5, 1], fontsize = 10)\nplt.xlabel(\"trial #\", fontsize = 15)\nplt.ylabel(\"p(state)\", fontsize = 15)\nplt.xlim(0, 100)",
"_____no_output_____"
],
[
"decoded_states = np.argmax(posterior_probs[0], axis=1)\ncopy_states = decoded_states.copy()\ncopy_states[decoded_states == 1] = 0\ncopy_states[decoded_states == 0] = 1\n\n\n# plt.plot(rlst,label='r')\nplt.plot(zlst, label='z')\nplt.plot(copy_states)\n# plt.plot(ylst, label='y')\n# plt.plot(slst, label='s')\nplt.xlim(0, 500)\nplt.legend()",
"_____no_output_____"
],
[
"np.sum(np.array(copy_states, dtype='int') == np.array(zlst, dtype='int'))",
"<ipython-input-68-1bd8fa2af5f7>:1: DeprecationWarning: elementwise comparison failed; this will raise an error in the future.\n np.sum(np.array(copy_states, dtype='int') == np.array(zlst, dtype='int'))\n"
],
[
"np.array(zlst) == np.array(copy_states)",
"<ipython-input-74-98e8faa0aabf>:1: DeprecationWarning: elementwise comparison failed; this will raise an error in the future.\n np.array(zlst) == np.array(copy_states)\n"
],
[
"x1 = np.array(zlst)[1:]\nx2 = np.array(copy_states)\nnp.sum(x1==x2)",
"_____no_output_____"
],
[
"x1.shape",
"_____no_output_____"
]
],
[
[
"With these posterior state probabilities, we can assign trials to states and then plot the fractional occupancy of each state:",
"_____no_output_____"
]
],
[
[
"# concatenate posterior probabilities across sessions\nposterior_probs_concat = np.concatenate(posterior_probs)\n# get state with maximum posterior probability at particular trial:\nstate_max_posterior = np.argmax(posterior_probs_concat, axis = 1)\n# now obtain state fractional occupancies:\n_, state_occupancies = np.unique(state_max_posterior, return_counts=True)\nstate_occupancies = state_occupancies/np.sum(state_occupancies)",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(2, 2.5), dpi=80, facecolor='w', edgecolor='k')\nfor z, occ in enumerate(state_occupancies):\n plt.bar(z, occ, width = 0.8, color = cols[z])\nplt.ylim((0, 1))\nplt.xticks([0, 1, 2], ['1', '2', '3'], fontsize = 10)\nplt.yticks([0, 0.5, 1], ['0', '0.5', '1'], fontsize=10)\nplt.xlabel('state', fontsize = 15)\nplt.ylabel('frac. occupancy', fontsize=15)",
"_____no_output_____"
]
],
[
[
"## 4. Fit GLM-HMM and perform recovery analysis: Maximum A Priori Estimation",
"_____no_output_____"
],
[
"Above, we performed Maximum Likelihood Estimation to retrieve the generative parameters of the GLM-HMM in simulated data. In the small data regime, where we do not have many trials available to us, we may instead want to perform Maximum A Priori (MAP) Estimation in order to incorporate a prior term and restrict the range for the best fitting parameters. Unfortunately, what is meant by 'small data regime' is problem dependent and will be affected by the number of states in the generative GLM-HMM, and the specific parameters of the generative model, amongst other things. In practice, we may perform both Maximum Likelihood Estimation and MAP estimation and compare the ability of the fit models to make predictions on held-out data (see Section 5 on Cross-Validation below). \n\nThe prior we consider for the GLM-HMM is the product of a Gaussian prior on the GLM weights, $W$, and a Dirichlet prior on the transition matrix, $A$:\n\n$$\n\\begin{align}\n\\Pr(W, A) &= \\mathcal{N}(W|0, \\Sigma) \\Pr(A|\\alpha) \\\\&= \\mathcal{N}(W|0, diag(\\sigma^{2}, \\cdots, \\sigma^{2})) \\prod_{j=1}^{K} \\dfrac{1}{B(\\alpha)} \\prod_{k=1}^{K} A_{jk}^{\\alpha -1}\n\\end{align}\n$$\n\nThere are two hyperparameters controlling the strength of the prior: $\\sigma$ and $\\alpha$. The larger the value of $\\sigma$ and if $\\alpha = 1$, the more similar MAP estimation will become to Maximum Likelihood Estimation, and the prior term will become an additive offset to the objective function of the GLM-HMM that is independent of the values of $W$ and $A$. In comparison, setting $\\sigma = 2$ and $\\alpha = 2$ will result in the prior no longer being independent of $W$ and $\\alpha$. \n\nIn order to perform MAP estimation for the GLM-HMM with `ssm`, the new syntax is:\n\n```python\nssm.HMM(num_states, obs_dim, input_dim, observations=\"input_driven_obs\", \n observation_kwargs=dict(C=num_categories,prior_sigma=prior_sigma),\n transitions=\"sticky\", transition_kwargs=dict(alpha=prior_alpha,kappa=0))```\n\nwhere `prior_sigma` is the $\\sigma$ parameter from above, and `prior_alpha` is the $\\alpha$ parameter.",
"_____no_output_____"
]
],
[
[
"# Instantiate GLM-HMM and set prior hyperparameters\nprior_sigma = 2\nprior_alpha = 2\nmap_glmhmm = ssm.HMM(num_states, obs_dim, input_dim, observations=\"input_driven_obs\", \n observation_kwargs=dict(C=num_categories,prior_sigma=prior_sigma),\n transitions=\"sticky\", transition_kwargs=dict(alpha=prior_alpha,kappa=0))",
"_____no_output_____"
],
[
"# Fit GLM-HMM with MAP estimation:\n_ = map_glmhmm.fit(true_choices, inputs=inpts, method=\"em\", num_iters=N_iters, tolerance=10**-4)",
"_____no_output_____"
]
],
[
[
"Compare final likelihood of data with MAP estimation and MLE to likelihood under generative model (note: we cannot use log_probability that is output of `fit` function as this incorporates prior term, which is not comparable between generative and MAP models). We want to check that MAP and MLE likelihood values are higher than true likelihood; if they are not, this may indicate a poor initialization and that we should refit these models.",
"_____no_output_____"
]
],
[
[
"true_likelihood = true_glmhmm.log_likelihood(true_choices, inputs=inpts)\nmle_final_ll = new_glmhmm.log_likelihood(true_choices, inputs=inpts) \nmap_final_ll = map_glmhmm.log_likelihood(true_choices, inputs=inpts) ",
"_____no_output_____"
],
[
"# Plot these values\nfig = plt.figure(figsize=(2, 2.5), dpi=80, facecolor='w', edgecolor='k')\nloglikelihood_vals = [true_likelihood, mle_final_ll, map_final_ll]\ncolors = ['Red', 'Navy', 'Purple']\nfor z, occ in enumerate(loglikelihood_vals):\n plt.bar(z, occ, width = 0.8, color = colors[z])\nplt.ylim((true_likelihood-5, true_likelihood+15))\nplt.xticks([0, 1, 2], ['true', 'mle', 'map'], fontsize = 10)\nplt.xlabel('model', fontsize = 15)\nplt.ylabel('loglikelihood', fontsize=15)",
"_____no_output_____"
]
],
[
[
"## 5. Cross Validation",
"_____no_output_____"
],
[
"To assess which model is better - the model fit via Maximum Likelihood Estimation, or the model fit via MAP estimation - we can investigate the predictive power of these fit models on held-out test data sets.",
"_____no_output_____"
]
],
[
[
"# Create additional input sequences to be used as held-out test data\nnum_test_sess = 10\ntest_inpts = np.ones((num_test_sess, num_trials_per_sess, input_dim)) \ntest_inpts[:,:,0] = np.random.choice(stim_vals, (num_test_sess, num_trials_per_sess)) \ntest_inpts = list(test_inpts)",
"_____no_output_____"
],
[
"# Create set of test latents and choices to accompany input sequences:\ntest_latents, test_choices = [], []\nfor sess in range(num_test_sess):\n test_z, test_y = true_glmhmm.sample(num_trials_per_sess, input=test_inpts[sess])\n test_latents.append(test_z)\n test_choices.append(test_y)",
"_____no_output_____"
],
[
"# Compare likelihood of test_choices for model fit with MLE and MAP:\nmle_test_ll = new_glmhmm.log_likelihood(test_choices, inputs=test_inpts) \nmap_test_ll = map_glmhmm.log_likelihood(test_choices, inputs=test_inpts) ",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(2, 2.5), dpi=80, facecolor='w', edgecolor='k')\nloglikelihood_vals = [mle_test_ll, map_test_ll]\ncolors = ['Navy', 'Purple']\nfor z, occ in enumerate(loglikelihood_vals):\n plt.bar(z, occ, width = 0.8, color = colors[z])\nplt.ylim((mle_test_ll-2, mle_test_ll+5))\nplt.xticks([0, 1], ['mle', 'map'], fontsize = 10)\nplt.xlabel('model', fontsize = 15)\nplt.ylabel('loglikelihood', fontsize=15)",
"_____no_output_____"
]
],
[
[
"Here we see that the model fit with MAP estimation achieves higher likelihood on the held-out dataset than the model fit with MLE, so we would choose this model as the best model of animal decision-making behavior (although we'd probably want to perform multiple fold cross-validation to be sure that this is the case in all instantiations of test data). \n\nLet's finish by comparing the retrieved weights and transition matrices from MAP estimation to the generative parameters.",
"_____no_output_____"
]
],
[
[
"map_glmhmm.permute(find_permutation(true_latents[0], map_glmhmm.most_likely_states(true_choices[0], input=inpts[0])))",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(6, 3), dpi=80, facecolor='w', edgecolor='k')\ncols = ['#ff7f00', '#4daf4a', '#377eb8']\nplt.subplot(1,2,1)\nrecovered_weights = new_glmhmm.observations.params\nfor k in range(num_states):\n if k ==0: # show labels only for first state\n plt.plot(range(input_dim), gen_weights[k][0], marker='o',\n color=cols[k],\n lw=1.5, label=\"generative\")\n plt.plot(range(input_dim), recovered_weights[k][0], color=cols[k],\n lw=1.5, label = 'recovered', linestyle='--') \n else:\n plt.plot(range(input_dim), gen_weights[k][0], marker='o',\n color=cols[k], \n lw=1.5, label=\"\")\n plt.plot(range(input_dim), recovered_weights[k][0], color=cols[k],\n lw=1.5, label = '', linestyle='--')\nplt.yticks(fontsize=10)\nplt.ylabel(\"GLM weight\", fontsize=15)\nplt.xlabel(\"covariate\", fontsize=15)\nplt.xticks([0, 1], ['stimulus', 'bias'], fontsize=12, rotation=45)\nplt.axhline(y=0, color=\"k\", alpha=0.5, ls=\"--\")\nplt.title(\"MLE\", fontsize = 15)\nplt.legend()\n\nplt.subplot(1,2,2)\nrecovered_weights = map_glmhmm.observations.params\nfor k in range(num_states):\n plt.plot(range(input_dim), gen_weights[k][0], marker='o',\n color=cols[k],\n lw=1.5, label=\"\", linestyle = '-')\n plt.plot(range(input_dim), recovered_weights[k][0], color=cols[k],\n lw=1.5, label = '', linestyle='--')\nplt.yticks(fontsize=10)\nplt.xticks([0, 1], ['', ''], fontsize=12, rotation=45)\nplt.axhline(y=0, color=\"k\", alpha=0.5, ls=\"--\")\nplt.title(\"MAP\", fontsize = 15)",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(7, 2.5), dpi=80, facecolor='w', edgecolor='k')\nplt.subplot(1, 3, 1)\ngen_trans_mat = np.exp(gen_log_trans_mat)[0]\nplt.imshow(gen_trans_mat, vmin=-0.8, vmax=1, cmap='bone')\nfor i in range(gen_trans_mat.shape[0]):\n for j in range(gen_trans_mat.shape[1]):\n text = plt.text(j, i, str(np.around(gen_trans_mat[i, j], decimals=2)), ha=\"center\", va=\"center\",\n color=\"k\", fontsize=12)\nplt.xlim(-0.5, num_states - 0.5)\nplt.xticks(range(0, num_states), ('1', '2', '3'), fontsize=10)\nplt.yticks(range(0, num_states), ('1', '2', '3'), fontsize=10)\nplt.ylim(num_states - 0.5, -0.5)\nplt.ylabel(\"state t\", fontsize = 15)\nplt.xlabel(\"state t+1\", fontsize = 15)\nplt.title(\"generative\", fontsize = 15)\n\n\nplt.subplot(1, 3, 2)\nrecovered_trans_mat = np.exp(new_glmhmm.transitions.log_Ps)\nplt.imshow(recovered_trans_mat, vmin=-0.8, vmax=1, cmap='bone')\nfor i in range(recovered_trans_mat.shape[0]):\n for j in range(recovered_trans_mat.shape[1]):\n text = plt.text(j, i, str(np.around(recovered_trans_mat[i, j], decimals=2)), ha=\"center\", va=\"center\",\n color=\"k\", fontsize=12)\nplt.xlim(-0.5, num_states - 0.5)\nplt.xticks(range(0, num_states), ('1', '2', '3'), fontsize=10)\nplt.yticks(range(0, num_states), ('1', '2', '3'), fontsize=10)\nplt.ylim(num_states - 0.5, -0.5)\nplt.title(\"recovered - MLE\", fontsize = 15)\nplt.subplots_adjust(0, 0, 1, 1)\n\n\nplt.subplot(1, 3, 3)\nrecovered_trans_mat = np.exp(map_glmhmm.transitions.log_Ps)\nplt.imshow(recovered_trans_mat, vmin=-0.8, vmax=1, cmap='bone')\nfor i in range(recovered_trans_mat.shape[0]):\n for j in range(recovered_trans_mat.shape[1]):\n text = plt.text(j, i, str(np.around(recovered_trans_mat[i, j], decimals=2)), ha=\"center\", va=\"center\",\n color=\"k\", fontsize=12)\nplt.xlim(-0.5, num_states - 0.5)\nplt.xticks(range(0, num_states), ('1', '2', '3'), fontsize=10)\nplt.yticks(range(0, num_states), ('1', '2', '3'), fontsize=10)\nplt.ylim(num_states - 0.5, -0.5)\nplt.title(\"recovered - MAP\", fontsize = 15)\nplt.subplots_adjust(0, 0, 1, 1)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
ecf41ec1a0bbe63c3b2b1cf89aab4bfe9b12c13d | 4,151 | ipynb | Jupyter Notebook | Comments Verifier.ipynb | lanking520/NVBugsLib | 6b5b08d575946cf5d1a4fb1a097d4830a730fc42 | [
"MIT"
] | 2 | 2017-09-23T21:35:42.000Z | 2019-02-12T06:51:49.000Z | Comments Verifier.ipynb | lanking520/NVBugsLib | 6b5b08d575946cf5d1a4fb1a097d4830a730fc42 | [
"MIT"
] | 1 | 2018-09-27T23:42:00.000Z | 2018-09-28T16:50:35.000Z | Comments Verifier.ipynb | lanking520/NVBugsLib | 6b5b08d575946cf5d1a4fb1a097d4830a730fc42 | [
"MIT"
] | null | null | null | 34.305785 | 874 | 0.608287 | [
[
[
"# coding=utf8\nfrom SentenceParserPython3 import SentenceParser\nimport pandas as pd\nimport numpy as np\nfrom bs4 import BeautifulSoup\nimport sys\nimport re\ndef printProgressBar (iteration, total, prefix = '', suffix = '', decimals = 1, length = 100, fill = '='):\n percent = (\"{0:.\" + str(decimals) + \"f}\").format(100 * (iteration / float(total)))\n filledLength = int(length * iteration // total)\n bar = fill * filledLength + '.' * (length - filledLength)\n sys.stdout.write('\\r%s |%s| %s%% %s' % (prefix, bar, percent, suffix))\n sys.stdout.flush()",
"_____no_output_____"
],
[
"def matchgit(test_str):\n teststr = re.sub(r'[\\n\\r\\t\\a\\b]+', ' ', test_str)\n pattern = r'\\[Git Change .*\\](.*)Bug ([0-9]+)'\n templist = []\n for item in re.findall(pattern, test_str):\n templist.append(item[0])\n return templist, re.sub(pattern, ' ', test_str)\n\nmatchgit(mystr)",
"_____no_output_____"
],
[
"mystr = \"\"\"\n\n[Git Change 2081e0570c0c63a6c90c15194d4667098502eea0 by Michael Frydrych \n(Branch: dev-gfx Repo: tegra/tests-graphics/android)] \n\nhwcomposer: use correct egl context\n\nEgl context needs to be reactivated after the control returns from\nhwcomposer set back to the test. At least the compression tests\ndefined in test_gl_compression.cpp have had this fault. Failure to\nreactivate its own egl context may have led to destruction of objects\nfrom hwcomposer context which has been active at that time.\n\nBug 1899629\n\nChange-Id: I98b6f75d61d9f5b1514f051f9beafcf0732a92aa\nReviewed-on: http://git-master/r/1490341\n(cherry picked from commit 0935ae6f713d05497e7ab832e6ddd48ea567a0d6)\nReviewed-on: http://git-master/r/1493015\nReviewed-by: Michael Frydrych <[email protected]>\nTested-by: Michael Frydrych <[email protected]>\n\"\"\"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
ecf42ea079659d1d66294e1830195528d6a4e470 | 228,855 | ipynb | Jupyter Notebook | kg-alphadiv.ipynb | amnona/paper-daycare | 40b07ae61f2f72e40dd38b74bc57c0ac9dcaa15c | [
"MIT"
] | null | null | null | kg-alphadiv.ipynb | amnona/paper-daycare | 40b07ae61f2f72e40dd38b74bc57c0ac9dcaa15c | [
"MIT"
] | 2 | 2020-09-06T15:26:23.000Z | 2020-09-06T16:23:04.000Z | kg-alphadiv.ipynb | amnona/paper-daycare | 40b07ae61f2f72e40dd38b74bc57c0ac9dcaa15c | [
"MIT"
] | null | null | null | 161.620763 | 33,636 | 0.892639 | [
[
[
"import calour as ca\nimport calour_utils as cu",
"failed to load logging config file\n"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\nimport scipy\nimport numpy as np\nimport matplotlib as mpl\nimport pandas as pd\nimport scipy as sp;\n",
"_____no_output_____"
],
[
"pwd",
"_____no_output_____"
],
[
"ca.set_log_level(11)",
"_____no_output_____"
],
[
"%matplotlib inline",
"_____no_output_____"
],
[
"db=ca.database._get_database_class('dbbact')",
"creating logger\n"
]
],
[
[
"# Load the (rarified) data",
"_____no_output_____"
]
],
[
[
"datn = ca.read_amplicon('data/gan-subsampled.biom', 'data/gan-subsampled_sample.txt', min_reads=1000, normalize=None)",
"2021-10-05 17:57:03 INFO loaded 268 samples, 14052 features\n2021-10-05 17:57:03 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:57:03 INFO After filtering, 268 remain.\n"
]
],
[
[
"## Now load the 10 additional rarified experiments",
"_____no_output_____"
]
],
[
[
"datr = datn.copy()",
"_____no_output_____"
],
[
"for i in range(10):\n exp = ca.read_amplicon('./data/rarefactions/gan-subsampled-%s.biom' % i, 'data/gan-subsampled_sample.txt', min_reads=1000, normalize=None)\n datr=datr.join_experiments(exp, 'exp-%s' % i, prefixes=['o','j'])",
"2021-10-05 17:57:04 INFO loaded 268 samples, 14052 features\n2021-10-05 17:57:05 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:57:05 INFO After filtering, 268 remain.\n2021-10-05 17:57:05 INFO Both experiments contain same sample IDs - adding prefixes\n2021-10-05 17:57:09 INFO loaded 268 samples, 14052 features\n2021-10-05 17:57:10 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:57:10 INFO After filtering, 268 remain.\n2021-10-05 17:57:14 INFO loaded 268 samples, 14052 features\n2021-10-05 17:57:15 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:57:15 INFO After filtering, 268 remain.\n2021-10-05 17:57:15 INFO Both experiments contain same sample IDs - adding prefixes\n2021-10-05 17:57:20 INFO loaded 268 samples, 14052 features\n2021-10-05 17:57:20 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:57:20 INFO After filtering, 268 remain.\n2021-10-05 17:57:26 INFO loaded 268 samples, 14052 features\n2021-10-05 17:57:26 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:57:26 INFO After filtering, 268 remain.\n2021-10-05 17:57:26 INFO Both experiments contain same sample IDs - adding prefixes\n2021-10-05 17:57:32 INFO loaded 268 samples, 14052 features\n2021-10-05 17:57:32 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:57:32 INFO After filtering, 268 remain.\n2021-10-05 17:57:38 INFO loaded 268 samples, 14052 features\n2021-10-05 17:57:38 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:57:38 INFO After filtering, 268 remain.\n2021-10-05 17:57:38 INFO Both experiments contain same sample IDs - adding prefixes\n2021-10-05 17:57:44 INFO loaded 268 samples, 14052 features\n2021-10-05 17:57:44 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:57:44 INFO After filtering, 268 remain.\n2021-10-05 17:57:50 INFO loaded 268 samples, 14052 features\n2021-10-05 17:57:50 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:57:50 INFO After filtering, 268 remain.\n2021-10-05 17:57:50 INFO Both experiments contain same sample IDs - adding prefixes\n2021-10-05 17:58:03 INFO loaded 268 samples, 14052 features\n2021-10-05 17:58:03 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:58:03 INFO After filtering, 268 remain.\n"
]
],
[
[
"### Get rid of features with 0 reads",
"_____no_output_____"
]
],
[
[
"datn=datn.filter_sum_abundance(0.000001)\ndatr=datr.filter_sum_abundance(0.000001)",
"2021-10-05 17:58:10 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:58:10 INFO After filtering, 1274 remain.\n2021-10-05 17:58:10 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:58:10 INFO After filtering, 1452 remain.\n"
],
[
"datn.plot(sample_field='kindergarten',barx_fields=['Time'])",
"/Users/amnon/git/calour/calour/heatmap/heatmap.py:308: MatplotlibDeprecationWarning: You are modifying the state of a globally registered colormap. In future versions, you will not be able to modify a registered colormap in-place. To remove this warning, you can make a copy of the colormap first. cmap = copy.copy(mpl.cm.get_cmap(\"viridis\"))\n cmap.set_bad(bad_color)\n"
]
],
[
[
"# Plot the alpha-diversity plot",
"_____no_output_____"
]
],
[
[
"datr.sparse=False",
"_____no_output_____"
],
[
"def get_sample_alpha(exp):\n alpha = {}\n alphastd = {}\n for cid, cexp in exp.iterate('sample_ID'):\n cnum = (cexp.data>0).sum(axis = 1)\n alpha[cid] = np.mean(cnum)\n alphastd[cid] = np.std(cnum)\n return alpha, alphastd",
"_____no_output_____"
],
[
"allmean,allstd=get_sample_alpha(datr)",
"_____no_output_____"
],
[
"dats=datn.copy()\nfor cid in allmean.keys():\n dats.sample_metadata.loc[dats.sample_metadata['sample_ID']==cid,'avgspecies'] = allmean[cid]\n dats.sample_metadata.loc[dats.sample_metadata['sample_ID']==cid,'stdspecies'] = allstd[cid]\n",
"_____no_output_____"
],
[
"datn.sample_metadata['avgspecies'] = np.sum(datn.data>0, axis=1)",
"_____no_output_____"
],
[
"f=plt.figure()\ntt=dats.filter_samples('kindergarten',['Home','Family'],negate=True)\ntt=tt.filter_samples('Time',1)\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[0.8,0.8,1,1],ecolor='k',barsabove=True)\ntt=dats.filter_samples('Time',2)\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[0.6,0.6,1,1],ecolor='k',barsabove=True)\ntt=dats.filter_samples('Time',3)\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[0.2,0.2,1,1],ecolor='k',barsabove=True)\ntt=dats.filter_samples('kindergarten',['Home','Family'],negate=True)\ntt=dats.filter_samples('Time',4)\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[0,0,1,1],ecolor='k',barsabove=True)\ntt=dats.filter_samples('Time',5)\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[0,0,0.5,1],ecolor='k',barsabove=True)\ntt=dats.filter_samples('kindergarten','Home')\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[1,0,0,1],ecolor='k',barsabove=True)\nplt.xlim([0,35])\nplt.ylim([0,220])\nplt.xlabel('Age (months)')\nplt.ylabel('Number of species')\nplt.legend(['Day care sample 1','Day care sample 2','Day care sample 3','Day care sample 4','Day care sample 5','Home care'])",
"AmpliconExperiment with 41 samples, 1274 features\nAmpliconExperiment with 54 samples, 1274 features\nAmpliconExperiment with 52 samples, 1274 features\nAmpliconExperiment with 57 samples, 1274 features\nAmpliconExperiment with 40 samples, 1274 features\nAmpliconExperiment with 24 samples, 1274 features\n"
],
[
"f.savefig('figures/fig-alpha-age-scatter.pdf')",
"_____no_output_____"
]
],
[
[
"# Do age binning and p-value for alpha diversity",
"_____no_output_____"
]
],
[
[
"def draw_age_alpha_onebar(threshold=0.5):\n '''Draw the per age-bin alpha diversity from home and kindergarten kids, and print the significance\n \n Parameters\n ----------\n threshold: float, optional\n the minimal number of reads for a feature to be defined as present\n \n \n Returns\n f: matplotlib.figure\n the figure with the bar plot\n '''\n f=plt.figure()\n colors=['r']\n colors.extend(list(plt.cm.Blues(np.linspace(0.5,1,5))))\n res=pd.DataFrame(columns=['age','hmean','gmean', 'hstd', 'gstd','agegrp'])\n# tt.sample_metadata['avgspecies']=np.sum(tt.data>=threshold,axis=1)\n for cage_cat, cexp in tt.iterate('age_cat', axis=0):\n cage_cat_num = cexp.sample_metadata.iloc[0]['age_cat_num']\n tthome = cexp.filter_samples('kindergarten','Home')\n ttgan = cexp.filter_samples('kindergarten','Home',negate=True)\n alpha_home=np.nanmean(tthome.sample_metadata['avgspecies'])\n alpha_gan=np.nanmean(ttgan.sample_metadata['avgspecies'])\n alpha_home_std=np.nanstd(tthome.sample_metadata['avgspecies']) / np.sqrt(len(tthome.sample_metadata))\n alpha_gan_std=np.nanstd(ttgan.sample_metadata['avgspecies']) / np.sqrt(len(ttgan.sample_metadata))\n print('age %s, home: %f, gan: %f' % (cage_cat, alpha_home, alpha_gan))\n print(sp.stats.mannwhitneyu(tthome.sample_metadata['avgspecies'],ttgan.sample_metadata['avgspecies']))\n print('gan %d samples, home %d samples' % (len(ttgan.sample_metadata), len(tthome.sample_metadata)))\n plt.bar(cage_cat_num, alpha_home, yerr=alpha_home_std, width=1.5, color=colors[0])\n plt.bar(cage_cat_num+1.5, alpha_gan, yerr=alpha_gan_std, width=1.5, color=colors[1])\n plt.ylim([0,120])\n plt.xlim([8,34])\n plt.legend(['Home care','Day care'],loc='upper left')\n plt.xticks(res['age']+1,res['agegrp'])\n plt.ylabel('number of species')\n return f",
"_____no_output_____"
]
],
[
[
"### Add age bin (0-5, 5-10 months etc.) to each sample",
"_____no_output_____"
]
],
[
[
"tt=cu.numeric_to_categories(dats,'age_months','age_cat',np.arange(0,35,5),inplace=False)",
"_____no_output_____"
],
[
"tt.sample_metadata.age_cat.value_counts()",
"_____no_output_____"
]
],
[
[
"### Get rid of timepoint1 samples (since did not have time to mingle in the kindergarten)",
"_____no_output_____"
]
],
[
[
"tt=tt.filter_samples('Time',[1],negate=True)",
"_____no_output_____"
],
[
"tt.sample_metadata.age_cat.value_counts()",
"_____no_output_____"
],
[
"f = draw_age_alpha_onebar()",
"age 10-15, home: 53.171717, gan: 66.560606\nMannwhitneyuResult(statistic=197.5, pvalue=0.05324726879944413)\ngan 66 samples, home 9 samples\nage 5-10, home: 39.363636, gan: 44.973485\nMannwhitneyuResult(statistic=78.5, pvalue=0.12052758369985261)\ngan 24 samples, home 9 samples\nage >30, home: 58.818182, gan: 109.038961\nMannwhitneyuResult(statistic=1.0, pvalue=0.19136654444261297)\ngan 7 samples, home 1 samples\nage 25-30, home: 89.454545, gan: 111.416268\nMannwhitneyuResult(statistic=3.0, pvalue=0.031653363113564224)\ngan 19 samples, home 2 samples\nage 20-25, home: 85.090909, gan: 105.716883\nMannwhitneyuResult(statistic=25.0, pvalue=0.07192593482742682)\ngan 35 samples, home 3 samples\nage 15-20, home: nan, gan: 88.495544\nMannwhitneyuResult(statistic=0.0, pvalue=0.0)\ngan 51 samples, home 0 samples\nage 0-5, home: nan, gan: 41.090909\nMannwhitneyuResult(statistic=0.0, pvalue=0.0)\ngan 1 samples, home 0 samples\n"
],
[
"f.savefig('./figures/alpha-barplot-age.pdf')",
"_____no_output_____"
]
],
[
[
"# Look at correlation between age and alpha-diversity",
"_____no_output_____"
],
[
"### Day-care",
"_____no_output_____"
]
],
[
[
"tt=dats.filter_samples('kindergarten',['Home'],negate=True)",
"_____no_output_____"
],
[
"sp.stats.pearsonr(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies)",
"_____no_output_____"
],
[
"sp.stats.spearmanr(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies)",
"_____no_output_____"
],
[
"sp.stats.linregress(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies)",
"_____no_output_____"
]
],
[
[
"### Home-care",
"_____no_output_____"
]
],
[
[
"tth=dats.filter_samples('kindergarten',['Home'])",
"_____no_output_____"
],
[
"sp.stats.pearsonr(tth.sample_metadata.age_months,tth.sample_metadata.avgspecies)",
"_____no_output_____"
],
[
"sp.stats.spearmanr(tth.sample_metadata.age_months,tth.sample_metadata.avgspecies)",
"_____no_output_____"
],
[
"sp.stats.linregress(tth.sample_metadata.age_months,tth.sample_metadata.avgspecies)",
"_____no_output_____"
]
],
[
[
"# Calculate the p-value for the slope difference\n## Using random permutations of the kindergarten/home labels",
"_____no_output_____"
]
],
[
[
"def get_slope_pval(exp, num_perm=1000):\n '''Get the slope p-value using random label permutations\n \n Parameters\n ----------\n exp: calour.Experiment\n The experiment to test\n num_perm: int, optional\n Number of permutations to test\n \n Returns:\n res: list of float\n The slope difference for each iteration.\n NOTE: res[0] is without permutation, res[1:-1] are the permuted results\n '''\n res = []\n np.random.seed(2020)\n cexp = exp.copy()\n for cperm in range(num_perm):\n # calculate the slopes\n tt=cexp.filter_samples('kindergarten',['Home'],negate=True)\n dc=sp.stats.linregress(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies)\n tth=cexp.filter_samples('kindergarten',['Home'])\n hc=sp.stats.linregress(tth.sample_metadata.age_months,tth.sample_metadata.avgspecies)\n res.append(dc.slope-hc.slope)\n # permute the kindergarten labels\n cexp.sample_metadata['kindergarten']=cexp.sample_metadata['kindergarten'].sample(frac=1).values\n print('orig_diff: %f' % res[0])\n num_big = np.sum(np.abs(res)>=res[0])\n print('pval: %d/%d=%f' % (num_big, len(res), num_big/len(res) ))\n num_ss = np.sum(res>=res[0])\n print('single sided pval: %d/%d=%f' % (num_ss, len(res), num_ss/len(res) ))\n return res",
"_____no_output_____"
],
[
"b=get_slope_pval(dats,5000)",
"orig_diff: 1.329668\npval: 768/5000=0.153600\nsingle sided pval: 333/5000=0.066600\n"
],
[
"plt.figure()\n_=plt.hist(b,50)",
"_____no_output_____"
]
],
[
[
"# Alpha diversity regression plot for day care and home care",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LinearRegression\nfrom scipy.linalg import lstsq\n\nf=plt.figure()\ntt=dats.filter_samples('kindergarten',['Home'],negate=True)\nplt.plot(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,'o',c=[0.8,0.8,1,1])\ntt=dats.filter_samples('kindergarten',['Home'],negate=False)\nplt.plot(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,'o',c=[1,0,0,1])\n\ntt=dats.filter_samples('kindergarten',['Home'],negate=True)\nxpos=np.array(tt.sample_metadata.age_months).reshape(-1, 1)\npearson = scipy.stats.pearsonr(tt.sample_metadata.age_months, tt.sample_metadata.avgspecies)\nprint('day care pearson correlation:')\nprint(pearson)\nreg = LinearRegression().fit(xpos,tt.sample_metadata.avgspecies)\n# reg = lstsq(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies)\nx_pred = np.array([[0],[35]])\ny_pred = reg.predict(x_pred)\nplt.plot(x_pred,y_pred,'-')\nx_text=np.array([[17.5]])\ny_text = reg.predict(x_text)\nplt.text(x_text-5,y_text+20,'y=%.2f*x+%.2f' % (reg.coef_, reg.intercept_),size=14,c='b')\nplt.xlim([0,35])\nplt.ylim([0,200])\nprint('daycare - slope %f, intercept %f, rsq %f' % (reg.coef_, reg.intercept_, reg.score(xpos, tt.sample_metadata.avgspecies)))\nrslope1 = reg.coef_\n\ntt=dats.filter_samples('kindergarten',['Home'],negate=False)\nxpos=np.array(tt.sample_metadata.age_months).reshape(-1, 1)\npearson = scipy.stats.pearsonr(tt.sample_metadata.age_months, tt.sample_metadata.avgspecies)\nprint('home care pearson correlation:')\nprint(pearson)\nreg = LinearRegression().fit(xpos,tt.sample_metadata.avgspecies)\nprint('homecare - slope %f, intercept %f, rsq %f' % (reg.coef_, reg.intercept_, reg.score(xpos, tt.sample_metadata.avgspecies)))\n# reg = lstsq(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies)\nx_pred = np.array([[0],[35]])\ny_pred = reg.predict(x_pred)\nplt.plot(x_pred,y_pred,'-r')\nx_text=np.array([[17.5]])\ny_text = reg.predict(x_text)\nplt.text(x_text,y_text-20,'y=%.2f*x+%.2f' % (reg.coef_, reg.intercept_),size=14,c='r')\nrslope2 = reg.coef_\nplt.xlim([3,35])\nplt.ylim([0,200])\n\n# calculate p-value using 1000 random label permutations\nvals = []\npdat=dats.copy()\nnp.random.seed(2021)\nfor cperm in range(1000):\n pdat.sample_metadata['kindergarten'] = np.random.permutation(pdat.sample_metadata['kindergarten'])\n tt=pdat.filter_samples('kindergarten',['Home'],negate=True)\n xpos=np.array(tt.sample_metadata.age_months).reshape(-1, 1)\n reg = LinearRegression().fit(xpos,tt.sample_metadata.avgspecies)\n slope1 = reg.coef_\n tt=pdat.filter_samples('kindergarten',['Home'],negate=False)\n xpos=np.array(tt.sample_metadata.age_months).reshape(-1, 1)\n reg = LinearRegression().fit(xpos,tt.sample_metadata.avgspecies)\n slope2 = reg.coef_\n vals.append(slope1-slope2)\n\nnum_big = np.sum(np.array(vals) >= rslope1-rslope2)\nprint('%d cases where got >= diff' % num_big)\npval = ((num_big+1)/(len(vals)+1))\nprint('pval %f' % pval)\nplt.title('pval %f' % pval)\nplt.xlabel('Age (months)')\nplt.ylabel('Number of species')\nplt.legend(['home care', 'day care'])\n\nf.savefig('supplementary/fig-alpha-regression.pdf')\n",
"day care pearson correlation:\n(0.6594310994311885, 7.943291488965394e-32)\ndaycare - slope 3.397911, intercept 22.884162, rsq 0.434849\nhome care pearson correlation:\n(0.67210610388437, 0.0003217119225773184)\nhomecare - slope 2.068243, intercept 25.488051, rsq 0.451727\n69 cases where got >= diff\npval 0.069930\n"
]
],
[
[
"# Look also at 10k reads/sample for review",
"_____no_output_____"
]
],
[
[
"dat10k = ca.read_amplicon('./data/gan-subsampled-10k.biom', 'data/gan-subsampled-10k_sample.txt', min_reads=1000, normalize=None)",
"2021-10-05 17:59:32 INFO loaded 232 samples, 14052 features\n2021-10-05 17:59:33 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:59:33 INFO After filtering, 232 remain.\n"
],
[
"dat10k=dat10k.filter_sum_abundance(0.000001)",
"2021-10-05 17:59:33 WARNING Do you forget to normalize your data? It is required before running this function\n2021-10-05 17:59:33 INFO After filtering, 1376 remain.\n"
],
[
"allmean,allstd=get_sample_alpha(dat10k)",
"_____no_output_____"
],
[
"dats=dat10k.copy()\nfor cid in allmean.keys():\n dats.sample_metadata.loc[dats.sample_metadata['sample_ID']==cid,'avgspecies'] = allmean[cid]\n dats.sample_metadata.loc[dats.sample_metadata['sample_ID']==cid,'stdspecies'] = allstd[cid]\n",
"_____no_output_____"
],
[
"f=plt.figure()\ntt=dats.filter_samples('kindergarten',['Home','Family'],negate=True)\ntt=tt.filter_samples('Time',1)\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[0.8,0.8,1,1],ecolor='k',barsabove=True)\ntt=dats.filter_samples('Time',2)\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[0.6,0.6,1,1],ecolor='k',barsabove=True)\ntt=dats.filter_samples('Time',3)\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[0.2,0.2,1,1],ecolor='k',barsabove=True)\ntt=dats.filter_samples('kindergarten',['Home','Family'],negate=True)\ntt=dats.filter_samples('Time',4)\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[0,0,1,1],ecolor='k',barsabove=True)\ntt=dats.filter_samples('Time',5)\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[0,0,0.5,1],ecolor='k',barsabove=True)\ntt=dats.filter_samples('kindergarten','Home')\nprint(tt)\nplt.errorbar(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,yerr=tt.sample_metadata.stdspecies,fmt='o',c=[1,0,0,1],ecolor='k',barsabove=True)\nplt.xlim([0,35])\nplt.ylim([0,220])\nplt.xlabel('Age (months)')\nplt.ylabel('Number of species')\nplt.legend(['Day care sample 1','Day care sample 2','Day care sample 3','Day care sample 4','Day care sample 5','Home care'])",
"AmpliconExperiment with 31 samples, 1376 features\nAmpliconExperiment with 51 samples, 1376 features\nAmpliconExperiment with 41 samples, 1376 features\nAmpliconExperiment with 51 samples, 1376 features\nAmpliconExperiment with 35 samples, 1376 features\nAmpliconExperiment with 23 samples, 1376 features\n"
],
[
"f.savefig('review-figs/fig-alpha-age-scatter-10k.pdf')",
"_____no_output_____"
]
],
[
[
"### and the age category graph",
"_____no_output_____"
]
],
[
[
"tt=cu.numeric_to_categories(dats,'age_months','age_cat',np.arange(0,35,5),inplace=False)",
"_____no_output_____"
],
[
"tt.sample_metadata.age_cat.value_counts()",
"_____no_output_____"
]
],
[
[
"### Get rid of timepoint1 samples (since did not have time to mingle in the kindergarten)",
"_____no_output_____"
]
],
[
[
"tt=tt.filter_samples('Time',[1],negate=True)",
"_____no_output_____"
],
[
"tt.sample_metadata.age_cat.value_counts()",
"_____no_output_____"
],
[
"f = draw_age_alpha_onebar()\nplt.ylim([0,150])",
"age 10-15, home: 62.000000, gan: 78.964912\nMannwhitneyuResult(statistic=170.0, pvalue=0.053995994600332095)\ngan 57 samples, home 9 samples\nage 5-10, home: 48.125000, gan: 54.842105\nMannwhitneyuResult(statistic=50.0, pvalue=0.08756563663571082)\ngan 19 samples, home 8 samples\nage >30, home: 63.000000, gan: 127.714286\nMannwhitneyuResult(statistic=0.0, pvalue=0.0938906360495762)\ngan 7 samples, home 1 samples\nage 25-30, home: 105.500000, gan: 132.526316\nMannwhitneyuResult(statistic=3.5, pvalue=0.03601884009533429)\ngan 19 samples, home 2 samples\nage 20-25, home: 98.666667, gan: 127.470588\nMannwhitneyuResult(statistic=17.0, pvalue=0.031076245526067255)\ngan 34 samples, home 3 samples\nage 15-20, home: nan, gan: 105.195122\nMannwhitneyuResult(statistic=0.0, pvalue=0.0)\ngan 41 samples, home 0 samples\nage 0-5, home: nan, gan: 48.000000\nMannwhitneyuResult(statistic=0.0, pvalue=0.0)\ngan 1 samples, home 0 samples\n"
],
[
"f.savefig('review-figs/alpha-diversity-bins-10k.pdf')",
"_____no_output_____"
]
],
[
[
"### and the regression plot",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LinearRegression\nfrom scipy.linalg import lstsq\n\nf=plt.figure()\ntt=dats.filter_samples('kindergarten',['Home'],negate=True)\nplt.plot(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,'o',c=[0.8,0.8,1,1])\ntt=dats.filter_samples('kindergarten',['Home'],negate=False)\nplt.plot(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies,'o',c=[1,0,0,1])\n\ntt=dats.filter_samples('kindergarten',['Home'],negate=True)\nxpos=np.array(tt.sample_metadata.age_months).reshape(-1, 1)\npearson = scipy.stats.pearsonr(tt.sample_metadata.age_months, tt.sample_metadata.avgspecies)\nprint('day care pearson correlation:')\nprint(pearson)\nreg = LinearRegression().fit(xpos,tt.sample_metadata.avgspecies)\n# reg = lstsq(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies)\nx_pred = np.array([[0],[35]])\ny_pred = reg.predict(x_pred)\nplt.plot(x_pred,y_pred,'-')\nx_text=np.array([[17.5]])\ny_text = reg.predict(x_text)\nplt.text(x_text-5,y_text+20,'y=%.2f*x+%.2f' % (reg.coef_, reg.intercept_),size=14,c='b')\nplt.xlim([0,35])\nplt.ylim([0,200])\nprint('daycare - slope %f, intercept %f, rsq %f' % (reg.coef_, reg.intercept_, reg.score(xpos, tt.sample_metadata.avgspecies)))\nrslope1 = reg.coef_\n\ntt=dats.filter_samples('kindergarten',['Home'],negate=False)\nxpos=np.array(tt.sample_metadata.age_months).reshape(-1, 1)\npearson = scipy.stats.pearsonr(tt.sample_metadata.age_months, tt.sample_metadata.avgspecies)\nprint('home care pearson correlation:')\nprint(pearson)\nreg = LinearRegression().fit(xpos,tt.sample_metadata.avgspecies)\nprint('homecare - slope %f, intercept %f, rsq %f' % (reg.coef_, reg.intercept_, reg.score(xpos, tt.sample_metadata.avgspecies)))\n# reg = lstsq(tt.sample_metadata.age_months,tt.sample_metadata.avgspecies)\nx_pred = np.array([[0],[35]])\ny_pred = reg.predict(x_pred)\nplt.plot(x_pred,y_pred,'-r')\nx_text=np.array([[17.5]])\ny_text = reg.predict(x_text)\nplt.text(x_text,y_text-20,'y=%.2f*x+%.2f' % (reg.coef_, reg.intercept_),size=14,c='r')\nrslope2 = reg.coef_\nplt.xlim([3,35])\nplt.ylim([0,200])\n\n# calculate p-value using 1000 random label permutations\nvals = []\npdat=dats.copy()\nnp.random.seed(2021)\nfor cperm in range(1000):\n pdat.sample_metadata['kindergarten'] = np.random.permutation(pdat.sample_metadata['kindergarten'])\n tt=pdat.filter_samples('kindergarten',['Home'],negate=True)\n xpos=np.array(tt.sample_metadata.age_months).reshape(-1, 1)\n reg = LinearRegression().fit(xpos,tt.sample_metadata.avgspecies)\n slope1 = reg.coef_\n tt=pdat.filter_samples('kindergarten',['Home'],negate=False)\n xpos=np.array(tt.sample_metadata.age_months).reshape(-1, 1)\n reg = LinearRegression().fit(xpos,tt.sample_metadata.avgspecies)\n slope2 = reg.coef_\n vals.append(slope1-slope2)\n\nnum_big = np.sum(np.array(vals) >= rslope1-rslope2)\nprint('%d cases where got >= diff' % num_big)\npval = ((num_big+1)/(len(vals)+1))\nprint('pval %f' % pval)\nplt.title('pval %f' % pval)\nplt.xlabel('Age (months)')\nplt.ylabel('Number of species')\nplt.legend(['home care', 'day care'])\n\nf.savefig('review-figs/fig-alpha-regression-10k.pdf')\n",
"day care pearson correlation:\n(0.6626270405628927, 8.53541167242263e-28)\ndaycare - slope 3.947950, intercept 29.328787, rsq 0.439075\nhome care pearson correlation:\n(0.6584032865273745, 0.0006364031066222026)\nhomecare - slope 2.251725, intercept 32.640004, rsq 0.433495\n49 cases where got >= diff\npval 0.049950\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf4362d9b52dd1d50f4d38e3cfdbe554d386c79 | 426,162 | ipynb | Jupyter Notebook | 02 TensorFlow/tutorial/07 Use function API to define layers.ipynb | asong1997/Time-Series-Analysis | 6fc55ebae2b7820241eb4cf7a5f26db933df79c8 | [
"Apache-2.0"
] | 242 | 2021-04-24T15:53:25.000Z | 2022-03-31T10:51:18.000Z | 02 TensorFlow/tutorial/07 Use function API to define layers.ipynb | asong1997/Time-Series-Analysis | 6fc55ebae2b7820241eb4cf7a5f26db933df79c8 | [
"Apache-2.0"
] | null | null | null | 02 TensorFlow/tutorial/07 Use function API to define layers.ipynb | asong1997/Time-Series-Analysis | 6fc55ebae2b7820241eb4cf7a5f26db933df79c8 | [
"Apache-2.0"
] | 86 | 2021-04-25T02:54:56.000Z | 2022-03-29T02:56:50.000Z | 431.775076 | 207,736 | 0.904489 | [
[
[
"import numpy as np\n\nimport tensorflow as tf\n\nfrom tensorflow import keras\nfrom tensorflow.keras import layers\n\ntf.keras.backend.clear_session() # 重置notebook中keras的状态",
"_____no_output_____"
],
[
"inputs = keras.Input(shape=(784,))",
"_____no_output_____"
],
[
"img_inputs = keras.Input(shape=(32, 32, 3))",
"_____no_output_____"
],
[
"inputs.shape",
"_____no_output_____"
],
[
"inputs.dtype",
"_____no_output_____"
],
[
"dense = layers.Dense(64, activation='relu')\nx = dense(inputs)",
"_____no_output_____"
],
[
"x = layers.Dense(64, activation='relu')(x)\noutputs = layers.Dense(10)(x)",
"_____no_output_____"
],
[
"model = keras.Model(inputs=inputs, outputs=outputs, name='mnist_model')",
"_____no_output_____"
],
[
"model.summary()",
"Model: \"mnist_model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 784)] 0 \n_________________________________________________________________\ndense (Dense) (None, 64) 50240 \n_________________________________________________________________\ndense_1 (Dense) (None, 64) 4160 \n_________________________________________________________________\ndense_2 (Dense) (None, 10) 650 \n=================================================================\nTotal params: 55,050\nTrainable params: 55,050\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"keras.utils.plot_model(model, 'my_first_model.png',dpi=150)",
"_____no_output_____"
],
[
"keras.utils.plot_model(model, 'my_first_model_with_shape_info.png', show_layer_names=True, show_shapes=True, dpi=150)",
"_____no_output_____"
],
[
"(x_train, y_train), (x_test, y_test) = keras.datasets.mnist.load_data()\n\nx_train = x_train.reshape(60000, 784).astype('float32') / 255\nx_test = x_test.reshape(10000, 784).astype('float32') / 255\n\nmodel.compile(loss=keras.losses.SparseCategoricalCrossentropy(from_logits=True),\n optimizer=keras.optimizers.RMSprop(),\n metrics=['accuracy'])\n\nhistory = model.fit(x_train, y_train,\n batch_size=64,\n epochs=5,\n validation_split=0.2)\n\ntest_scores = model.evaluate(x_test, y_test, verbose=2)\nprint('Test loss:', test_scores[0])\nprint('Test accuracy:', test_scores[1])",
"Train on 48000 samples, validate on 12000 samples\nEpoch 1/5\n48000/48000 [==============================] - 4s 93us/sample - loss: 0.3414 - accuracy: 0.9040 - val_loss: 0.1884 - val_accuracy: 0.9439\nEpoch 2/5\n48000/48000 [==============================] - 3s 67us/sample - loss: 0.1616 - accuracy: 0.9513 - val_loss: 0.1532 - val_accuracy: 0.9578\nEpoch 3/5\n48000/48000 [==============================] - 3s 65us/sample - loss: 0.1197 - accuracy: 0.9648 - val_loss: 0.1253 - val_accuracy: 0.9640\nEpoch 4/5\n48000/48000 [==============================] - 3s 65us/sample - loss: 0.0947 - accuracy: 0.9715 - val_loss: 0.1185 - val_accuracy: 0.9668\nEpoch 5/5\n48000/48000 [==============================] - 3s 64us/sample - loss: 0.0801 - accuracy: 0.9762 - val_loss: 0.1061 - val_accuracy: 0.9704\n10000/10000 - 1s - loss: 0.0919 - accuracy: 0.9745\nTest loss: 0.091879672590876\nTest accuracy: 0.9745\n"
],
[
"model.save('path_to_my_model')\ndel model\n# 从文件重新创建完全相同的模型:\nmodel = keras.models.load_model('path_to_my_model')",
"WARNING:tensorflow:From C:\\anaconda3\\envs\\keras\\lib\\site-packages\\tensorflow_core\\python\\ops\\resource_variable_ops.py:1786: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.\nInstructions for updating:\nIf using Keras pass *_constraint arguments to layers.\nINFO:tensorflow:Assets written to: path_to_my_model\\assets\n"
],
[
"encoder_input = keras.Input(shape=(28, 28, 1), name='img')\nx = layers.Conv2D(16, 3, activation='relu')(encoder_input)\nx = layers.Conv2D(32, 3, activation='relu')(x)\nx = layers.MaxPooling2D(3)(x)\nx = layers.Conv2D(32, 3, activation='relu')(x)\nx = layers.Conv2D(16, 3, activation='relu')(x)\nencoder_output = layers.GlobalMaxPooling2D()(x)\n\nencoder = keras.Model(encoder_input, encoder_output, name='encoder')\nencoder.summary()\n\nx = layers.Reshape((4, 4, 1))(encoder_output)\nx = layers.Conv2DTranspose(16, 3, activation='relu')(x)\nx = layers.Conv2DTranspose(32, 3, activation='relu')(x)\nx = layers.UpSampling2D(3)(x)\nx = layers.Conv2DTranspose(16, 3, activation='relu')(x)\ndecoder_output = layers.Conv2DTranspose(1, 3, activation='relu')(x)\n\nautoencoder = keras.Model(encoder_input, decoder_output, name='autoencoder')\nautoencoder.summary()",
"Model: \"encoder\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nimg (InputLayer) [(None, 28, 28, 1)] 0 \n_________________________________________________________________\nconv2d (Conv2D) (None, 26, 26, 16) 160 \n_________________________________________________________________\nconv2d_1 (Conv2D) (None, 24, 24, 32) 4640 \n_________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 8, 8, 32) 0 \n_________________________________________________________________\nconv2d_2 (Conv2D) (None, 6, 6, 32) 9248 \n_________________________________________________________________\nconv2d_3 (Conv2D) (None, 4, 4, 16) 4624 \n_________________________________________________________________\nglobal_max_pooling2d (Global (None, 16) 0 \n=================================================================\nTotal params: 18,672\nTrainable params: 18,672\nNon-trainable params: 0\n_________________________________________________________________\nModel: \"autoencoder\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nimg (InputLayer) [(None, 28, 28, 1)] 0 \n_________________________________________________________________\nconv2d (Conv2D) (None, 26, 26, 16) 160 \n_________________________________________________________________\nconv2d_1 (Conv2D) (None, 24, 24, 32) 4640 \n_________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 8, 8, 32) 0 \n_________________________________________________________________\nconv2d_2 (Conv2D) (None, 6, 6, 32) 9248 \n_________________________________________________________________\nconv2d_3 (Conv2D) (None, 4, 4, 16) 4624 \n_________________________________________________________________\nglobal_max_pooling2d (Global (None, 16) 0 \n_________________________________________________________________\nreshape (Reshape) (None, 4, 4, 1) 0 \n_________________________________________________________________\nconv2d_transpose (Conv2DTran (None, 6, 6, 16) 160 \n_________________________________________________________________\nconv2d_transpose_1 (Conv2DTr (None, 8, 8, 32) 4640 \n_________________________________________________________________\nup_sampling2d (UpSampling2D) (None, 24, 24, 32) 0 \n_________________________________________________________________\nconv2d_transpose_2 (Conv2DTr (None, 26, 26, 16) 4624 \n_________________________________________________________________\nconv2d_transpose_3 (Conv2DTr (None, 28, 28, 1) 145 \n=================================================================\nTotal params: 28,241\nTrainable params: 28,241\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"encoder_input = keras.Input(shape=(28, 28, 1), name='original_img')\nx = layers.Conv2D(16, 3, activation='relu')(encoder_input)\nx = layers.Conv2D(32, 3, activation='relu')(x)\nx = layers.MaxPooling2D(3)(x)\nx = layers.Conv2D(32, 3, activation='relu')(x)\nx = layers.Conv2D(16, 3, activation='relu')(x)\nencoder_output = layers.GlobalMaxPooling2D()(x)\n\nencoder = keras.Model(encoder_input, encoder_output, name='encoder')\nencoder.summary()\n\ndecoder_input = keras.Input(shape=(16,), name='encoded_img')\nx = layers.Reshape((4, 4, 1))(decoder_input)\nx = layers.Conv2DTranspose(16, 3, activation='relu')(x)\nx = layers.Conv2DTranspose(32, 3, activation='relu')(x)\nx = layers.UpSampling2D(3)(x)\nx = layers.Conv2DTranspose(16, 3, activation='relu')(x)\ndecoder_output = layers.Conv2DTranspose(1, 3, activation='relu')(x)\n\ndecoder = keras.Model(decoder_input, decoder_output, name='decoder')\ndecoder.summary()\n\nautoencoder_input = keras.Input(shape=(28, 28, 1), name='img')\nencoded_img = encoder(autoencoder_input)\ndecoded_img = decoder(encoded_img)\nautoencoder = keras.Model(autoencoder_input, decoded_img, name='autoencoder')\nautoencoder.summary()",
"Model: \"encoder\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\noriginal_img (InputLayer) [(None, 28, 28, 1)] 0 \n_________________________________________________________________\nconv2d_4 (Conv2D) (None, 26, 26, 16) 160 \n_________________________________________________________________\nconv2d_5 (Conv2D) (None, 24, 24, 32) 4640 \n_________________________________________________________________\nmax_pooling2d_1 (MaxPooling2 (None, 8, 8, 32) 0 \n_________________________________________________________________\nconv2d_6 (Conv2D) (None, 6, 6, 32) 9248 \n_________________________________________________________________\nconv2d_7 (Conv2D) (None, 4, 4, 16) 4624 \n_________________________________________________________________\nglobal_max_pooling2d_1 (Glob (None, 16) 0 \n=================================================================\nTotal params: 18,672\nTrainable params: 18,672\nNon-trainable params: 0\n_________________________________________________________________\nModel: \"decoder\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nencoded_img (InputLayer) [(None, 16)] 0 \n_________________________________________________________________\nreshape_1 (Reshape) (None, 4, 4, 1) 0 \n_________________________________________________________________\nconv2d_transpose_4 (Conv2DTr (None, 6, 6, 16) 160 \n_________________________________________________________________\nconv2d_transpose_5 (Conv2DTr (None, 8, 8, 32) 4640 \n_________________________________________________________________\nup_sampling2d_1 (UpSampling2 (None, 24, 24, 32) 0 \n_________________________________________________________________\nconv2d_transpose_6 (Conv2DTr (None, 26, 26, 16) 4624 \n_________________________________________________________________\nconv2d_transpose_7 (Conv2DTr (None, 28, 28, 1) 145 \n=================================================================\nTotal params: 9,569\nTrainable params: 9,569\nNon-trainable params: 0\n_________________________________________________________________\nModel: \"autoencoder\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nimg (InputLayer) [(None, 28, 28, 1)] 0 \n_________________________________________________________________\nencoder (Model) (None, 16) 18672 \n_________________________________________________________________\ndecoder (Model) (None, 28, 28, 1) 9569 \n=================================================================\nTotal params: 28,241\nTrainable params: 28,241\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"def get_model():\n inputs = keras.Input(shape=(128,))\n outputs = layers.Dense(1)(inputs)\n return keras.Model(inputs, outputs)\n\nmodel1 = get_model()\nmodel2 = get_model()\nmodel3 = get_model()\n\ninputs = keras.Input(shape=(128,))\ny1 = model1(inputs)\ny2 = model2(inputs)\ny3 = model3(inputs)\noutputs = layers.average([y1, y2, y3])\nensemble_model = keras.Model(inputs=inputs, outputs=outputs)\nensemble_model.summary()",
"Model: \"model_3\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_6 (InputLayer) [(None, 128)] 0 \n__________________________________________________________________________________________________\nmodel (Model) (None, 1) 129 input_6[0][0] \n__________________________________________________________________________________________________\nmodel_1 (Model) (None, 1) 129 input_6[0][0] \n__________________________________________________________________________________________________\nmodel_2 (Model) (None, 1) 129 input_6[0][0] \n__________________________________________________________________________________________________\naverage (Average) (None, 1) 0 model[1][0] \n model_1[1][0] \n model_2[1][0] \n==================================================================================================\nTotal params: 387\nTrainable params: 387\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
],
[
"num_tags = 12 # 问题标签的数量\nnum_words = 10000 # 预处理文本数据时获得的词汇量\nnum_departments = 4 # 预测部门数\n\ntitle_input = keras.Input(shape=(None,), name='title') # 可变长度的整数序列\nbody_input = keras.Input(shape=(None,), name='body') # 可变长度的整数序列\ntags_input = keras.Input(shape=(num_tags,), name='tags') # 大小为num_tags的二进制向量\n\n# 将标题中的每个单词嵌入到64维向量中\ntitle_features = layers.Embedding(num_words, 64)(title_input)\n# 将文本中的每个单词嵌入到64维向量中\nbody_features = layers.Embedding(num_words, 64)(body_input)\n\n# 将标题中嵌入单词的序列减少为单个128维向量\ntitle_features = layers.LSTM(128)(title_features)\n# 将body内嵌入词的序列化为单个32维向量\nbody_features = layers.LSTM(32)(body_features)\n\n# 通过concatenate(级联)将所有可用功能合并到单个向量中\n# 它以张量列表作为输入,除了级联轴外,它们均具有相同的形状,并返回单个张量,即所有输入的级联。\nx = layers.concatenate([title_features, body_features, tags_input])\n\n# 通过特征使用逻辑回归以进行优先级预测\npriority_pred = layers.Dense(1, name='priority')(x)\n# 通过特征对部分进行分类\ndepartment_pred = layers.Dense(num_departments, name='department')(x)\n\n# 实例化预测优先级和部门的端到端模型\nmodel = keras.Model(inputs=[title_input, body_input, tags_input],\n outputs=[priority_pred, department_pred])",
"_____no_output_____"
],
[
"keras.utils.plot_model(model, 'multi_input_and_output_model.png', show_shapes=True, dpi=150)",
"_____no_output_____"
],
[
"model.compile(optimizer=keras.optimizers.RMSprop(1e-3),\n loss=[keras.losses.BinaryCrossentropy(from_logits=True),\n keras.losses.CategoricalCrossentropy(from_logits=True)],\n loss_weights=[1., 0.2])",
"_____no_output_____"
],
[
"model.compile(optimizer=keras.optimizers.RMSprop(1e-3),\n loss={'priority':keras.losses.BinaryCrossentropy(from_logits=True),\n 'department': keras.losses.CategoricalCrossentropy(from_logits=True)},\n loss_weights=[1., 0.2])",
"_____no_output_____"
],
[
"title_data = np.random.randint(num_words, size=(1280, 10))\ntitle_data",
"_____no_output_____"
],
[
"priority_targets = np.random.random(size=(1280, 1))\npriority_targets",
"_____no_output_____"
],
[
"# 构造虚拟输入数据\ntitle_data = np.random.randint(num_words, size=(1280, 10))\nbody_data = np.random.randint(num_words, size=(1280, 100))\ntags_data = np.random.randint(2, size=(1280, num_tags)).astype('float32')\n\n# 构造期望输出数据\npriority_targets = np.random.random(size=(1280, 1))\ndept_targets = np.random.randint(2, size=(1280, num_departments))\n\n# 训练\nmodel.fit({'title': title_data, 'body': body_data, 'tags': tags_data},\n {'priority': priority_targets, 'department': dept_targets},\n epochs=2,\n batch_size=32)",
"Train on 1280 samples\nEpoch 1/2\n1280/1280 [==============================] - 9s 7ms/sample - loss: 1.3411 - priority_loss: 0.7009 - department_loss: 3.2008\nEpoch 2/2\n1280/1280 [==============================] - 1s 669us/sample - loss: 1.3339 - priority_loss: 0.6987 - department_loss: 3.1757\n"
],
[
"tf.keras.backend.clear_session()\n\ninputs = keras.Input(shape=(32, 32, 3), name='img')\nx = layers.Conv2D(32, 3, activation='relu')(inputs)\nx = layers.Conv2D(64, 3, activation='relu')(x)\nblock_1_output = layers.MaxPooling2D(3)(x) \n\nx = layers.Conv2D(64, 3, activation='relu', padding='same')(block_1_output)\nx = layers.Conv2D(64, 3, activation='relu', padding='same')(x)\nblock_2_output = layers.add([x, block_1_output]) # 输入张量列表,返回输入的总和。\n\nx = layers.Conv2D(64, 3, activation='relu', padding='same')(block_2_output)\nx = layers.Conv2D(64, 3, activation='relu', padding='same')(x)\nblock_3_output = layers.add([x, block_2_output])\n\nx = layers.Conv2D(64, 3, activation='relu')(block_3_output)\nx = layers.GlobalAveragePooling2D()(x)\nx = layers.Dense(256, activation='relu')(x)\nx = layers.Dropout(0.5)(x)\noutputs = layers.Dense(10)(x)\n\nmodel = keras.Model(inputs, outputs, name='toy_resnet')\nmodel.summary()",
"Model: \"toy_resnet\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\nimg (InputLayer) [(None, 32, 32, 3)] 0 \n__________________________________________________________________________________________________\nconv2d (Conv2D) (None, 30, 30, 32) 896 img[0][0] \n__________________________________________________________________________________________________\nconv2d_1 (Conv2D) (None, 28, 28, 64) 18496 conv2d[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 9, 9, 64) 0 conv2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 9, 9, 64) 36928 max_pooling2d[0][0] \n__________________________________________________________________________________________________\nconv2d_3 (Conv2D) (None, 9, 9, 64) 36928 conv2d_2[0][0] \n__________________________________________________________________________________________________\nadd (Add) (None, 9, 9, 64) 0 conv2d_3[0][0] \n max_pooling2d[0][0] \n__________________________________________________________________________________________________\nconv2d_4 (Conv2D) (None, 9, 9, 64) 36928 add[0][0] \n__________________________________________________________________________________________________\nconv2d_5 (Conv2D) (None, 9, 9, 64) 36928 conv2d_4[0][0] \n__________________________________________________________________________________________________\nadd_1 (Add) (None, 9, 9, 64) 0 conv2d_5[0][0] \n add[0][0] \n__________________________________________________________________________________________________\nconv2d_6 (Conv2D) (None, 7, 7, 64) 36928 add_1[0][0] \n__________________________________________________________________________________________________\nglobal_average_pooling2d (Globa (None, 64) 0 conv2d_6[0][0] \n__________________________________________________________________________________________________\ndense (Dense) (None, 256) 16640 global_average_pooling2d[0][0] \n__________________________________________________________________________________________________\ndropout (Dropout) (None, 256) 0 dense[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 10) 2570 dropout[0][0] \n==================================================================================================\nTotal params: 223,242\nTrainable params: 223,242\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
],
[
"keras.utils.plot_model(model, 'mini_resnet.png', show_shapes=True, dpi=150)",
"_____no_output_____"
],
[
"(x_train, y_train), (x_test, y_test) = keras.datasets.cifar10.load_data()\n\nx_train = x_train.astype('float32') / 255.\nx_test = x_test.astype('float32') / 255.\ny_train = keras.utils.to_categorical(y_train, 10)\ny_test = keras.utils.to_categorical(y_test, 10)\n\nmodel.compile(optimizer=keras.optimizers.RMSprop(1e-3),\n loss=keras.losses.CategoricalCrossentropy(from_logits=True),\n metrics=['acc'])\n\nmodel.fit(x_train, y_train,\n batch_size=64,\n epochs=1,\n validation_split=0.2)",
"Downloading data from https://www.cs.toronto.edu/~kriz/cifar-10-python.tar.gz\n 188416/170498071 [..............................] - ETA: 46:16"
],
[
"# 嵌入映射到128维向量的1000个单词\nshared_embedding = layers.Embedding(1000, 128)\n\ntext_input_a = keras.Input(shape=(None,), dtype='int32')\n\ntext_input_b = keras.Input(shape=(None,), dtype='int32')\n\n# 重用同一层来编码两个输入\nencoded_input_a = shared_embedding(text_input_a)\nencoded_input_b = shared_embedding(text_input_b)",
"_____no_output_____"
],
[
"tf.keras.backend.clear_session()\nvgg19 = tf.keras.applications.VGG19()",
"_____no_output_____"
],
[
"feat_extraction_model = keras.Model(inputs=vgg19.input, outputs=features_list)\n\nimg = np.random.random((1, 224, 224, 3)).astype('float32')\nextracted_features = feat_extraction_model(img)",
"_____no_output_____"
],
[
"class CustomDense(layers.Layer):\n def __init__(self, units=32):\n super(CustomDense, self).__init__()\n self.units = units\n\n def build(self, input_shape):\n self.w = self.add_weight(shape=(input_shape[-1], self.units),\n initializer='random_normal',\n trainable=True)\n self.b = self.add_weight(shape=(self.units,),\n initializer='random_normal',\n trainable=True)\n\n def call(self, inputs):\n return tf.matmul(inputs, self.w) + self.b\n\n\ninputs = keras.Input((4,))\noutputs = CustomDense(10)(inputs)\n\nmodel = keras.Model(inputs, outputs)",
"_____no_output_____"
],
[
"class CustomDense(layers.Layer):\n\n def __init__(self, units=32):\n super(CustomDense, self).__init__()\n self.units = units\n\n def build(self, input_shape):\n self.w = self.add_weight(shape=(input_shape[-1], self.units),\n initializer='random_normal',\n trainable=True)\n self.b = self.add_weight(shape=(self.units,),\n initializer='random_normal',\n trainable=True)\n\n def call(self, inputs):\n return tf.matmul(inputs, self.w) + self.b\n\n def get_config(self):\n return {'units': self.units}\n\n\ninputs = keras.Input((4,))\noutputs = CustomDense(10)(inputs)\n\nmodel = keras.Model(inputs, outputs)\nconfig = model.get_config()\n\nnew_model = keras.Model.from_config(config, custom_objects={'CustomDense': CustomDense})",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf445aef633fc248ddf3c1618489462e741fe25 | 8,138 | ipynb | Jupyter Notebook | tutorial-contents-notebooks/201_torch_numpy.ipynb | zikai1/PyTorch-Tutorial | 0988ec46f0190a526c09b072be0caf1f7e0a1e8b | [
"MIT"
] | 4 | 2018-11-19T11:37:27.000Z | 2020-03-25T15:25:14.000Z | tutorial-contents-notebooks/201_torch_numpy.ipynb | ziyue246/PyTorch-Tutorial | 46c516fc109809f4299e6bbcd045f8637aef4dd6 | [
"MIT"
] | null | null | null | tutorial-contents-notebooks/201_torch_numpy.ipynb | ziyue246/PyTorch-Tutorial | 46c516fc109809f4299e6bbcd045f8637aef4dd6 | [
"MIT"
] | 4 | 2019-02-02T13:44:00.000Z | 2021-09-02T03:43:42.000Z | 20.345 | 145 | 0.440157 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
ecf4473eb29fc2a392cfa3225d1ec7818eec0e77 | 190,948 | ipynb | Jupyter Notebook | custom.ipynb | wosigor/nft-image-generator | 22dc1f162dd0b8657126194cabee1419d50979ba | [
"MIT"
] | null | null | null | custom.ipynb | wosigor/nft-image-generator | 22dc1f162dd0b8657126194cabee1419d50979ba | [
"MIT"
] | null | null | null | custom.ipynb | wosigor/nft-image-generator | 22dc1f162dd0b8657126194cabee1419d50979ba | [
"MIT"
] | null | null | null | 178.456075 | 18,345 | 0.532412 | [
[
[
"from PIL import Image \nfrom IPython.display import display \nimport random\nimport json\nimport os\nimport pprint",
"_____no_output_____"
],
[
"# path = \"./trait-layers\"\npath = \"./high-res-traits\"\ntrait_directories = os.listdir(path)\ntrait_directories.remove(\".DS_Store\")\ntrait_directories.sort()\nprint(trait_directories)\ntrait_files = {}\nfor file in trait_directories:\n if file in trait_files:\n continue\n else:\n trait_files[file] = os.listdir(path + \"/\" + file)\n \nfor t in trait_files:\n trait_files[t].sort()\n print(t)\n print(trait_files[t])",
"['01_background', '02_base', '03_eyebrow', '04_eye', '05_lashes', '06_nose', '07_makeup', '08_jewelry', '09_clothes', '10_hair', '11_hat', '12_necklace', '13_rings', '14_sunglasses', '15_others']\n01_background\n['01.png', '01b.png', '01c.png', '02a.png', '02b.png', '02c.png', '03a.png', '03b.png', '03c.png', '04a.png', '04b.png', '04c.png', '04d.png', '05a.png', '05b.png', '05c.png', '05d.png', '06a.png', '06b.png', '06c.png', '06d.png']\n02_base\n['01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png']\n03_eyebrow\n['01.png', '02.png', '03.png', '04.png']\n04_eye\n['01.png', '02.png', '03.png', '04.png', '05.png']\n05_lashes\n['01a.png', '01b.png', '01c.png', '01d.png', '01e.png']\n06_nose\n['01.png']\n07_makeup\n['01.png', '02.png', '03.png', '04.png', '05.png', '06.png', '07.png', '08.png', '09.png', '10a.png', '10b.png', '10c.png', '10d.png', '11a.png']\n08_jewelry\n['01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03a.png', '03b.png', '03c.png', '03d.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '05b.png', '05c.png', '05d.png', '05e.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png']\n09_clothes\n['.DS_Store', '01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '05b.png', '05c.png', '05d.png', '05e.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png', '07a.png', '07b.png', '07c.png', '07d.png', '07e.png', '07f.png', '08a.png', '08b.png', '09a.png', '09b.png', '09c.png', '10a.png', '10b.png', '10c.png', '10d.png', '11a.png', '11b.png', '11c.png', '11d.png', '12a.png', '12b.png', '12c.png', '12d.png', '13a.png', '13b.png', '13c.png', '14a.png', '14b.png', '14c.png', '14d.png', '14e.png', '14f.png', '15a.png', '15b.png', '15c.png', '15d.png', '15e.png']\n10_hair\n['01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03a.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '06a.png', '06b.png', '06c.png', '06d.png']\n11_hat\n['01a.png', '01b.png', '01c.png', '01d.png', '02.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '05b.png', '05c.png', '05d.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png', '07a.png', '07b.png', '07c.png', '07d.png', '07e.png', '07f.png', '08a.png', '08b.png', '08c.png', '08d.png', '09a.png', '09b.png', '09c.png', '09d.png', '10a.png', '10b.png', '10c.png', '10d.png', '11a.png', '11b.png', '11c.png', '11d.png', '12a.png', '12b.png', '12c.png', '12d.png']\n12_necklace\n['.DS_Store', '01.png', '02.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '05a.png', '05b.png', '05c.png', '05d.png', '06a.png', '06b.png', '06c.png', '06d.png', '08d.png', '08e.png']\n13_rings\n['01a.png', '01b.png', '01c.png', '02a.png', '02b.png', '02c.png', '03a.png', '03b.png', '03c.png', '04a.png', '04b.png', '04c.png', '05a.png', '05b.png', '05c.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png']\n14_sunglasses\n['01.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03.png', '04.png', '05a.png', '05b.png', '05c.png', '05d.png', '05e.png', '06a.png', '06b.png', '06c.png', '06d.png', '07a.png', '07b.png', '07c.png', '08a.png', '08b.png', '08c.png', '08d.png', '08e.png', '09a.png', '09b.png', '09c.png', '09d.png', '09e.png', '10a.png', '10b.png', '10c.png', '10d.png', '10e.png', '11a.png', '11b.png', '11c.png', '11d.png', '12a.png', '12b.png', '12c.png', '12d.png', '13a.png', '13b.png', '13c.png', '13d.png', '14a.png', '14b.png', '14c.png', '14d.png']\n15_others\n['01a.png', '01b.png', '01c.png', '01d.png', '01e.png']\n"
],
[
"# Each image is made up a series of traits\n# The weightings for each trait drive the rarity and add up to 100%\n\ntraits = {}\n\nbackground = ['01.png', '01b.png', '01c.png', '02a.png', '02b.png', '02c.png', '03a.png', '03b.png', '03c.png', '04a.png', '04b.png', '04c.png', '04d.png', '05a.png', '05b.png', '05c.png', '05d.png', '06a.png', '06b.png', '06c.png', '06d.png']\nbackground_weights = [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 4, 4, 4, 4, 4]\n\nbase = ['01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png']\nbase_weights = [25, 5, 5, 5, 5, 5, 5, 5, 5, 5, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4]\n\neyebrow = ['01.png', '02.png', '03.png', '04.png']\neyebrow_weights = [25, 25, 25, 25]\n\neye = ['01.png', '02.png', '03.png', '04.png', '05.png']\neye_weights = [25, 20, 20, 20, 5]\n\nlashes = ['', '01a.png', '01b.png', '01c.png', '01d.png', '01e.png']\nlashes_weights = [30, 14, 14, 14, 14, 14]\n\nnose = ['', '01.png']\nnose_weights = [50, 50]\n\nmakeup = ['', '01.png', '02.png', '03.png', '04.png', '05.png', '06.png', '07.png', '08.png', '09.png', '10a.png', '10b.png', '10c.png', '10d.png', '11a.png']\nmakeup_weights = [40, 4, 4, 4, 4, 4, 5, 5, 5, 4, 4, 4, 4, 4, 4]\n\njewelry = ['', '01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03a.png', '03b.png', '03c.png', '03d.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '05b.png', '05c.png', '05d.png', '05e.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png']\njewelry_over = ['05a.png', '05b.png', '05c.png', '05d.png', '05e.png']\njewelry_weights = [10, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3]\n\nclothes = ['', '01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '05b.png', '05c.png', '05d.png', '05e.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png', '07a.png', '07b.png', '07c.png', '07d.png', '07e.png', '07f.png', '08a.png', '08b.png', '09a.png', '09b.png', '09c.png', '10a.png', '10b.png', '10c.png', '10d.png', '11a.png', '11b.png', '11c.png', '11d.png', '12a.png', '12b.png', '12c.png', '12d.png', '13a.png', '13b.png', '13c.png', '14a.png', '14b.png', '14c.png', '14d.png', '14e.png', '14f.png', '15a.png', '15b.png', '15c.png', '15d.png', '15e.png']\nclothes_no_hat = ['09a.png', '09b.png', '09c.png', '14a.png', '14b.png', '14c.png', '14d.png', '14e.png', '14f.png']\nclothes_no_sunglasses = ['09a.png', '09b.png', '09c.png']\nclothes_necklace = ['07a.png', '07b.png', '07c.png', '07d.png', '07e.png', '07f.png', '08a.png', '08b.png', '09a.png', '09b.png', '09c.png', '11a.png', '11b.png', '11c.png', '11d.png', '12a.png', '12b.png', '12c.png', '12d.png']\nclothes_weights = [1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1]\n\nnecklace = ['', '01.png', '02.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '05a.png', '05b.png', '05c.png', '05d.png', '06a.png', '06b.png', '06c.png', '06d.png', '08d.png', '08e.png']\nnecklace_no_clothes = ['01.png']\nnecklace_weights = [60, 0.2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2]\n\nhair = ['', '01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03a.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '06a.png', '06b.png', '06c.png', '06d.png']\nhair_no_hat = ['03a.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '06a.png', '06b.png', '06c.png', '06d.png']\nhair_no_necklace = ['05a.png']\nhair_weights = [65, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 2, 1.5, 1.5, 1.5, 1.5, 1.5, 4.5, 1.5, 1.5, 1.5, 1.5]\n\nhat = ['', '01a.png', '01b.png', '01c.png', '01d.png', '02.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '05b.png', '05c.png', '05d.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png', '07a.png', '07b.png', '07c.png', '07d.png', '07e.png', '07f.png', '08a.png', '08b.png', '08c.png', '08d.png', '09a.png', '09b.png', '09c.png', '09d.png', '10a.png', '10b.png', '10c.png', '10d.png', '11a.png', '11b.png', '11c.png', '11d.png', '12a.png', '12b.png', '12c.png', '12d.png']\nhat_no_glasses = ['08a.png', '08b.png', '08c.png', '08d.png', '10a.png', '10b.png', '10c.png', '10d.png', '12a.png', '12b.png', '12c.png', '12d.png']\nhat_no_har = []\nhat_weights = [40, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1.5, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1]\n\nrings = ['', '01a.png', '01b.png', '01c.png', '02a.png', '02b.png', '02c.png', '03a.png', '03b.png', '03c.png', '04a.png', '04b.png', '04c.png', '05a.png', '05b.png', '05c.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png']\nrings_weights = [40, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3]\n\nsunglasses = ['', '01.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03.png', '04.png', '05a.png', '05b.png', '05c.png', '05d.png', '05e.png', '06a.png', '06b.png', '06c.png', '06d.png', '07a.png', '07b.png', '07c.png', '08a.png', '08b.png', '08c.png', '08d.png', '08e.png', '09a.png', '09b.png', '09c.png', '09d.png', '09e.png', '10a.png', '10b.png', '10c.png', '10d.png', '10e.png', '11a.png', '11b.png', '11c.png', '11d.png', '12a.png', '12b.png', '12c.png', '12d.png', '13a.png', '13b.png', '13c.png', '13d.png', '14a.png', '14b.png', '14c.png', '14d.png']\nsunglasses_no_hat = ['07a.png', '07b.png', '07c.png', '11a.png', '11b.png', '11c.png', '11d.png']\nsunglasses_mask_no_hair = ['07a.png', '07b.png', '07c.png']\nsunglasses_weights = [52, 1, 1, 1, 1, 1, 1, 1, 0.5, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0.2, 0.2, 0.2, 0.2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1]\n\nothers = ['', '01a.png', '01b.png', '01c.png', '01d.png', '01e.png']\nothers_weights = [95, 1, 1, 1, 1, 1]\n\n\n# Dictionary variable for each trait. \n# Each trait corresponds to its file name\n\ntraits['background'] = background\ntraits['base'] = base\ntraits['eyebrow'] = eyebrow\ntraits['eye'] = eye\ntraits['lashes'] = lashes\ntraits['nose'] = nose\ntraits['makeup'] = makeup\ntraits['jewelry'] = jewelry\ntraits['clothes'] = clothes\ntraits['necklace'] = necklace\ntraits['hair'] = hair\ntraits['hat'] = hat\ntraits['rings'] = rings\ntraits['sunglasses'] = sunglasses\ntraits['others'] = others\nprint(traits)\n\n\ndirectory_names = ['01_background', '02_base', '03_eyebrow', '04_eye', '05_lashes', '06_nose', '07_makeup', '08_jewelry', '09_clothes', '10_hair', '11_hat', '12_necklace', '13_rings', '14_sunglasses', '15_others']\ndirectory_mappings = {}\nfor i, s in enumerate(directory_names):\n for t in traits.keys():\n if t in s:\n directory_mappings[t] = directory_names[i]\nprint(directory_mappings)\n \n",
"{'background': ['01.png', '01b.png', '01c.png', '02a.png', '02b.png', '02c.png', '03a.png', '03b.png', '03c.png', '04a.png', '04b.png', '04c.png', '04d.png', '05a.png', '05b.png', '05c.png', '05d.png', '06a.png', '06b.png', '06c.png', '06d.png'], 'base': ['01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png'], 'eyebrow': ['01.png', '02.png', '03.png', '04.png'], 'eye': ['01.png', '02.png', '03.png', '04.png', '05.png'], 'lashes': ['', '01a.png', '01b.png', '01c.png', '01d.png', '01e.png'], 'nose': ['', '01.png'], 'makeup': ['', '01.png', '02.png', '03.png', '04.png', '05.png', '06.png', '07.png', '08.png', '09.png', '10a.png', '10b.png', '10c.png', '10d.png', '11a.png'], 'jewelry': ['', '01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03a.png', '03b.png', '03c.png', '03d.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '05b.png', '05c.png', '05d.png', '05e.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png'], 'clothes': ['', '01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '05b.png', '05c.png', '05d.png', '05e.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png', '07a.png', '07b.png', '07c.png', '07d.png', '07e.png', '07f.png', '08a.png', '08b.png', '09a.png', '09b.png', '09c.png', '10a.png', '10b.png', '10c.png', '10d.png', '11a.png', '11b.png', '11c.png', '11d.png', '12a.png', '12b.png', '12c.png', '12d.png', '13a.png', '13b.png', '13c.png', '14a.png', '14b.png', '14c.png', '14d.png', '14e.png', '14f.png', '15a.png', '15b.png', '15c.png', '15d.png', '15e.png'], 'necklace': ['', '01.png', '02.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '05a.png', '05b.png', '05c.png', '05d.png', '06a.png', '06b.png', '06c.png', '06d.png', '08d.png', '08e.png'], 'hair': ['', '01a.png', '01b.png', '01c.png', '01d.png', '01e.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03a.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '06a.png', '06b.png', '06c.png', '06d.png'], 'hat': ['', '01a.png', '01b.png', '01c.png', '01d.png', '02.png', '03a.png', '03b.png', '03c.png', '03d.png', '03e.png', '04a.png', '04b.png', '04c.png', '04d.png', '04e.png', '05a.png', '05b.png', '05c.png', '05d.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png', '07a.png', '07b.png', '07c.png', '07d.png', '07e.png', '07f.png', '08a.png', '08b.png', '08c.png', '08d.png', '09a.png', '09b.png', '09c.png', '09d.png', '10a.png', '10b.png', '10c.png', '10d.png', '11a.png', '11b.png', '11c.png', '11d.png', '12a.png', '12b.png', '12c.png', '12d.png'], 'rings': ['', '01a.png', '01b.png', '01c.png', '02a.png', '02b.png', '02c.png', '03a.png', '03b.png', '03c.png', '04a.png', '04b.png', '04c.png', '05a.png', '05b.png', '05c.png', '06a.png', '06b.png', '06c.png', '06d.png', '06e.png'], 'sunglasses': ['', '01.png', '02a.png', '02b.png', '02c.png', '02d.png', '02e.png', '03.png', '04.png', '05a.png', '05b.png', '05c.png', '05d.png', '05e.png', '06a.png', '06b.png', '06c.png', '06d.png', '07a.png', '07b.png', '07c.png', '08a.png', '08b.png', '08c.png', '08d.png', '08e.png', '09a.png', '09b.png', '09c.png', '09d.png', '09e.png', '10a.png', '10b.png', '10c.png', '10d.png', '10e.png', '11a.png', '11b.png', '11c.png', '11d.png', '12a.png', '12b.png', '12c.png', '12d.png', '13a.png', '13b.png', '13c.png', '13d.png', '14a.png', '14b.png', '14c.png', '14d.png'], 'others': ['', '01a.png', '01b.png', '01c.png', '01d.png', '01e.png']}\n{'background': '01_background', 'base': '02_base', 'eyebrow': '03_eyebrow', 'eye': '04_eye', 'lashes': '05_lashes', 'nose': '06_nose', 'makeup': '07_makeup', 'jewelry': '08_jewelry', 'clothes': '09_clothes', 'hair': '10_hair', 'hat': '11_hat', 'necklace': '12_necklace', 'rings': '13_rings', 'sunglasses': '14_sunglasses', 'others': '15_others'}\n"
],
[
"# Custom image\ntraits = {}\nimage = {}\nimage['background'] = '01c.png'\nimage['base'] = '04e.png'\nimage['eyebrow'] = '03.png'\nimage['eye'] = '02.png'\nimage['lashes'] = '01c.png'\n# image['nose'] = '01.png'\nimage['makeup'] = '01.png'\nimage['jewelry'] = '01d.png'\n# image['necklace'] = '06b.png'\nimage['clothes'] = '07f.png'\nimage['hair'] = '04d.png'\n# image['hat'] = '04d.png'\n# image['rings'] = '05b.png'\nimage['sunglasses'] = '11c.png'\n# image['others'] = ''\n\n\n\nimgs = []\nfor trait in image:\n img = Image.open(f'./high-res-traits/{directory_mappings[trait]}/{image[trait]}').convert('RGBA')\n imgs.append(img)\n\n#Create each composite\nstart_image = Image.alpha_composite(imgs[0], imgs[1])\nnext_image = start_image\nfor idx, val in enumerate(imgs):\n if idx < 2:\n continue\n else:\n next_image = Image.alpha_composite(start_image, imgs[idx])\n start_image = next_image\n\n#Convert to RGB\nrgb_im = next_image.convert('RGB')\n# resize images for sampling. Remove below line for final images\nsmaller_img = rgb_im.resize((1024,1024),Image.ANTIALIAS)\nfile_name = 'thetechie.png'\nsmaller_img.save(\"./custom_images/\" + file_name)\n\n",
"_____no_output_____"
],
[
"## Generate Traits\n\nTOTAL_IMAGES = 100 # Number of random unique images we want to generate\n\nall_images = [] \n\n# A recursive function to generate unique image combinations\ndef create_new_image():\n \n new_image = {} #\n\n # For each trait category, select a random trait based on the weightings \n new_image [\"background\"] = random.choices(background, background_weights)[0]\n new_image [\"base\"] = random.choices(base, base_weights)[0]\n new_image [\"makeup\"] = random.choices(makeup, makeup_weights)[0]\n new_image [\"eye\"] = random.choices(eye, eye_weights)[0]\n new_image [\"hair\"] = random.choices(hair, hair_weights)[0]\n new_image [\"hat\"] = random.choices(hat, hat_weights)[0]\n if new_image[\"hat\"] in hat_special:\n new_image [\"necklace\"] = necklace[0]\n else:\n new_image [\"necklace\"] = random.choices(necklace, necklace_weights)[0]\n new_image [\"rings\"] = random.choices(rings, rings_weights)[0]\n new_image [\"sunglasses\"] = random.choices(sunglasses, sunglasses_weights)[0]\n\n \n if new_image in all_images:\n return create_new_image()\n else:\n return new_image\n \n \n# Generate the unique combinations based on trait weightings\nfor i in range(TOTAL_IMAGES): \n \n new_trait_image = create_new_image()\n \n all_images.append(new_trait_image)\n \n",
"_____no_output_____"
],
[
"# Returns true if all images are unique\ndef all_images_unique(all_images):\n seen = list()\n return not any(i in seen or seen.append(i) for i in all_images)\n\nprint(\"Are all images unique?\", all_images_unique(all_images))",
"Are all images unique? True\n"
],
[
"# Add token Id to each image\ni = 200\nfor item in all_images:\n item[\"tokenId\"] = i\n i = i + 1",
"_____no_output_____"
],
[
"print(all_images)",
"[{'background': '01.png', 'base': '02a.png', 'makeup': '02.png', 'eye': '01.png', 'hair': '', 'hat': '05a.png', 'necklace': '11d.png', 'rings': '', 'sunglasses': '06b.png', 'tokenId': 200}, {'background': '04c.png', 'base': '02c.png', 'makeup': '', 'eye': '04.png', 'hair': '', 'hat': '03b.png', 'necklace': '04d.png', 'rings': '04.png', 'sunglasses': '02c.png', 'tokenId': 201}, {'background': '05b.png', 'base': '01a.png', 'makeup': '', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '09a.png', 'rings': '', 'sunglasses': '', 'tokenId': 202}, {'background': '04d.png', 'base': '02e.png', 'makeup': '', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '06b.png', 'rings': '', 'sunglasses': '05d.png', 'tokenId': 203}, {'background': '05c.png', 'base': '01a.png', 'makeup': '', 'eye': '01.png', 'hair': '01a.png', 'hat': '01b.png', 'necklace': '02.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 204}, {'background': '03b.png', 'base': '03b.png', 'makeup': '', 'eye': '04.png', 'hair': '02e.png', 'hat': '', 'necklace': '06a.png', 'rings': '', 'sunglasses': '', 'tokenId': 205}, {'background': '05d.png', 'base': '04b.png', 'makeup': '02.png', 'eye': '01.png', 'hair': '', 'hat': '04e.png', 'necklace': '11e.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 206}, {'background': '04b.png', 'base': '01b.png', 'makeup': '02.png', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '02.png', 'rings': '04.png', 'sunglasses': '02d.png', 'tokenId': 207}, {'background': '01c.png', 'base': '04b.png', 'makeup': '02.png', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '', 'rings': '03.png', 'sunglasses': '', 'tokenId': 208}, {'background': '01b.png', 'base': '01a.png', 'makeup': '03.png', 'eye': '01.png', 'hair': '', 'hat': '04c.png', 'necklace': '05a.png', 'rings': '', 'sunglasses': '', 'tokenId': 209}, {'background': '02a.png', 'base': '03c.png', 'makeup': '01.png', 'eye': '01.png', 'hair': '', 'hat': '06b.png', 'necklace': '10d.png', 'rings': '', 'sunglasses': '', 'tokenId': 210}, {'background': '04b.png', 'base': '01a.png', 'makeup': '01.png', 'eye': '01.png', 'hair': '', 'hat': '01c.png', 'necklace': '', 'rings': '', 'sunglasses': '', 'tokenId': 211}, {'background': '01.png', 'base': '01a.png', 'makeup': '02.png', 'eye': '01.png', 'hair': '02d.png', 'hat': '07d.png', 'necklace': '', 'rings': '05.png', 'sunglasses': '', 'tokenId': 212}, {'background': '01.png', 'base': '02b.png', 'makeup': '', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '08a.png', 'rings': '', 'sunglasses': '', 'tokenId': 213}, {'background': '02a.png', 'base': '02b.png', 'makeup': '05.png', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '08a.png', 'rings': '', 'sunglasses': '02d.png', 'tokenId': 214}, {'background': '02b.png', 'base': '04c.png', 'makeup': '04.png', 'eye': '04.png', 'hair': '', 'hat': '04b.png', 'necklace': '08a.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 215}, {'background': '01b.png', 'base': '04d.png', 'makeup': '', 'eye': '01.png', 'hair': '02a.png', 'hat': '', 'necklace': '11d.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 216}, {'background': '05a.png', 'base': '03a.png', 'makeup': '01.png', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '03a.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 217}, {'background': '01c.png', 'base': '01d.png', 'makeup': '02.png', 'eye': '03.png', 'hair': '02c.png', 'hat': '06a.png', 'necklace': '03a.png', 'rings': '', 'sunglasses': '04.png', 'tokenId': 218}, {'background': '01.png', 'base': '03e.png', 'makeup': '', 'eye': '01.png', 'hair': '', 'hat': '04a.png', 'necklace': '06b.png', 'rings': '', 'sunglasses': '05e.png', 'tokenId': 219}, {'background': '04b.png', 'base': '04d.png', 'makeup': '', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '', 'rings': '', 'sunglasses': '05e.png', 'tokenId': 220}, {'background': '03c.png', 'base': '03d.png', 'makeup': '05.png', 'eye': '02.png', 'hair': '01c.png', 'hat': '01b.png', 'necklace': '10d.png', 'rings': '', 'sunglasses': '', 'tokenId': 221}, {'background': '01.png', 'base': '01a.png', 'makeup': '02.png', 'eye': '02.png', 'hair': '', 'hat': '06d.png', 'necklace': '', 'rings': '', 'sunglasses': '05b.png', 'tokenId': 222}, {'background': '05b.png', 'base': '03e.png', 'makeup': '', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '09c.png', 'rings': '', 'sunglasses': '', 'tokenId': 223}, {'background': '01c.png', 'base': '04b.png', 'makeup': '', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '10c.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 224}, {'background': '03c.png', 'base': '04e.png', 'makeup': '01.png', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '08c.png', 'rings': '04.png', 'sunglasses': '', 'tokenId': 225}, {'background': '02c.png', 'base': '02e.png', 'makeup': '', 'eye': '01.png', 'hair': '', 'hat': '04e.png', 'necklace': '11e.png', 'rings': '', 'sunglasses': '06c.png', 'tokenId': 226}, {'background': '01c.png', 'base': '03d.png', 'makeup': '05.png', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '04d.png', 'rings': '', 'sunglasses': '', 'tokenId': 227}, {'background': '04a.png', 'base': '04e.png', 'makeup': '01.png', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '12b.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 228}, {'background': '03b.png', 'base': '01a.png', 'makeup': '04.png', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '09e.png', 'rings': '05.png', 'sunglasses': '06a.png', 'tokenId': 229}, {'background': '01b.png', 'base': '03b.png', 'makeup': '04.png', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '07b.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 230}, {'background': '01.png', 'base': '03d.png', 'makeup': '01.png', 'eye': '02.png', 'hair': '', 'hat': '07d.png', 'necklace': '', 'rings': '01.png', 'sunglasses': '05a.png', 'tokenId': 231}, {'background': '01b.png', 'base': '01a.png', 'makeup': '04.png', 'eye': '05.png', 'hair': '', 'hat': '05a.png', 'necklace': '05c.png', 'rings': '04.png', 'sunglasses': '', 'tokenId': 232}, {'background': '02b.png', 'base': '01a.png', 'makeup': '01.png', 'eye': '03.png', 'hair': '01d.png', 'hat': '06c.png', 'necklace': '07c.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 233}, {'background': '01b.png', 'base': '01c.png', 'makeup': '04.png', 'eye': '01.png', 'hair': '02a.png', 'hat': '04a.png', 'necklace': '12e.png', 'rings': '', 'sunglasses': '04.png', 'tokenId': 234}, {'background': '03a.png', 'base': '01d.png', 'makeup': '', 'eye': '04.png', 'hair': '02c.png', 'hat': '', 'necklace': '07d.png', 'rings': '', 'sunglasses': '', 'tokenId': 235}, {'background': '03b.png', 'base': '01e.png', 'makeup': '01.png', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '08d.png', 'rings': '01.png', 'sunglasses': '04.png', 'tokenId': 236}, {'background': '01.png', 'base': '02a.png', 'makeup': '02.png', 'eye': '04.png', 'hair': '02a.png', 'hat': '', 'necklace': '07c.png', 'rings': '', 'sunglasses': '05c.png', 'tokenId': 237}, {'background': '01b.png', 'base': '01a.png', 'makeup': '04.png', 'eye': '04.png', 'hair': '02d.png', 'hat': '', 'necklace': '09a.png', 'rings': '02.png', 'sunglasses': '01.png', 'tokenId': 238}, {'background': '05d.png', 'base': '04c.png', 'makeup': '04.png', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '05b.png', 'rings': '', 'sunglasses': '', 'tokenId': 239}, {'background': '01c.png', 'base': '02b.png', 'makeup': '05.png', 'eye': '02.png', 'hair': '02a.png', 'hat': '', 'necklace': '07d.png', 'rings': '02.png', 'sunglasses': '', 'tokenId': 240}, {'background': '05d.png', 'base': '02a.png', 'makeup': '', 'eye': '03.png', 'hair': '02d.png', 'hat': '', 'necklace': '06a.png', 'rings': '01.png', 'sunglasses': '02d.png', 'tokenId': 241}, {'background': '04b.png', 'base': '02b.png', 'makeup': '04.png', 'eye': '05.png', 'hair': '01e.png', 'hat': '', 'necklace': '06a.png', 'rings': '', 'sunglasses': '', 'tokenId': 242}, {'background': '01c.png', 'base': '02c.png', 'makeup': '02.png', 'eye': '05.png', 'hair': '', 'hat': '04a.png', 'necklace': '07b.png', 'rings': '02.png', 'sunglasses': '', 'tokenId': 243}, {'background': '05a.png', 'base': '04a.png', 'makeup': '', 'eye': '02.png', 'hair': '02b.png', 'hat': '04b.png', 'necklace': '06c.png', 'rings': '01.png', 'sunglasses': '02e.png', 'tokenId': 244}, {'background': '03b.png', 'base': '01a.png', 'makeup': '04.png', 'eye': '01.png', 'hair': '', 'hat': '05c.png', 'necklace': '07d.png', 'rings': '', 'sunglasses': '05a.png', 'tokenId': 245}, {'background': '02a.png', 'base': '01b.png', 'makeup': '02.png', 'eye': '05.png', 'hair': '', 'hat': '', 'necklace': '07b.png', 'rings': '', 'sunglasses': '', 'tokenId': 246}, {'background': '04d.png', 'base': '03b.png', 'makeup': '03.png', 'eye': '05.png', 'hair': '', 'hat': '', 'necklace': '06d.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 247}, {'background': '01c.png', 'base': '01a.png', 'makeup': '', 'eye': '01.png', 'hair': '', 'hat': '05c.png', 'necklace': '08a.png', 'rings': '02.png', 'sunglasses': '', 'tokenId': 248}, {'background': '02b.png', 'base': '04c.png', 'makeup': '04.png', 'eye': '04.png', 'hair': '', 'hat': '06c.png', 'necklace': '09c.png', 'rings': '', 'sunglasses': '', 'tokenId': 249}, {'background': '05d.png', 'base': '01a.png', 'makeup': '', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '04a.png', 'rings': '', 'sunglasses': '05b.png', 'tokenId': 250}, {'background': '03c.png', 'base': '04b.png', 'makeup': '04.png', 'eye': '01.png', 'hair': '01c.png', 'hat': '05a.png', 'necklace': '05c.png', 'rings': '', 'sunglasses': '', 'tokenId': 251}, {'background': '03b.png', 'base': '03e.png', 'makeup': '03.png', 'eye': '04.png', 'hair': '01b.png', 'hat': '', 'necklace': '08a.png', 'rings': '', 'sunglasses': '02d.png', 'tokenId': 252}, {'background': '05b.png', 'base': '01e.png', 'makeup': '02.png', 'eye': '01.png', 'hair': '01c.png', 'hat': '07b.png', 'necklace': '', 'rings': '03.png', 'sunglasses': '', 'tokenId': 253}, {'background': '05b.png', 'base': '01c.png', 'makeup': '', 'eye': '05.png', 'hair': '02b.png', 'hat': '', 'necklace': '06a.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 254}, {'background': '02c.png', 'base': '02d.png', 'makeup': '', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '11d.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 255}, {'background': '04d.png', 'base': '01a.png', 'makeup': '01.png', 'eye': '01.png', 'hair': '02a.png', 'hat': '07c.png', 'necklace': '', 'rings': '', 'sunglasses': '', 'tokenId': 256}, {'background': '01.png', 'base': '03d.png', 'makeup': '01.png', 'eye': '05.png', 'hair': '01d.png', 'hat': '', 'necklace': '', 'rings': '02.png', 'sunglasses': '02b.png', 'tokenId': 257}, {'background': '03b.png', 'base': '01a.png', 'makeup': '05.png', 'eye': '05.png', 'hair': '', 'hat': '06e.png', 'necklace': '05a.png', 'rings': '', 'sunglasses': '', 'tokenId': 258}, {'background': '03b.png', 'base': '01e.png', 'makeup': '05.png', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '03c.png', 'rings': '04.png', 'sunglasses': '', 'tokenId': 259}, {'background': '04d.png', 'base': '03c.png', 'makeup': '04.png', 'eye': '03.png', 'hair': '01a.png', 'hat': '', 'necklace': '11d.png', 'rings': '', 'sunglasses': '', 'tokenId': 260}, {'background': '05b.png', 'base': '01c.png', 'makeup': '', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '10c.png', 'rings': '', 'sunglasses': '06a.png', 'tokenId': 261}, {'background': '03c.png', 'base': '04c.png', 'makeup': '02.png', 'eye': '01.png', 'hair': '', 'hat': '03e.png', 'necklace': '11a.png', 'rings': '', 'sunglasses': '', 'tokenId': 262}, {'background': '04d.png', 'base': '03a.png', 'makeup': '', 'eye': '01.png', 'hair': '', 'hat': '03b.png', 'necklace': '07d.png', 'rings': '', 'sunglasses': '', 'tokenId': 263}, {'background': '05a.png', 'base': '04c.png', 'makeup': '', 'eye': '01.png', 'hair': '', 'hat': '07a.png', 'necklace': '', 'rings': '', 'sunglasses': '', 'tokenId': 264}, {'background': '04d.png', 'base': '04a.png', 'makeup': '03.png', 'eye': '02.png', 'hair': '', 'hat': '03a.png', 'necklace': '07b.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 265}, {'background': '04d.png', 'base': '04c.png', 'makeup': '05.png', 'eye': '04.png', 'hair': '', 'hat': '01c.png', 'necklace': '04a.png', 'rings': '', 'sunglasses': '', 'tokenId': 266}, {'background': '01b.png', 'base': '03d.png', 'makeup': '', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '10a.png', 'rings': '', 'sunglasses': '03.png', 'tokenId': 267}, {'background': '01.png', 'base': '01a.png', 'makeup': '03.png', 'eye': '04.png', 'hair': '01c.png', 'hat': '03d.png', 'necklace': '04d.png', 'rings': '02.png', 'sunglasses': '02a.png', 'tokenId': 268}, {'background': '04c.png', 'base': '03e.png', 'makeup': '', 'eye': '02.png', 'hair': '', 'hat': '07e.png', 'necklace': '', 'rings': '04.png', 'sunglasses': '', 'tokenId': 269}, {'background': '04b.png', 'base': '01a.png', 'makeup': '', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '10a.png', 'rings': '', 'sunglasses': '', 'tokenId': 270}, {'background': '01.png', 'base': '02d.png', 'makeup': '04.png', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '09e.png', 'rings': '03.png', 'sunglasses': '03.png', 'tokenId': 271}, {'background': '03c.png', 'base': '01a.png', 'makeup': '05.png', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '07a.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 272}, {'background': '03b.png', 'base': '03a.png', 'makeup': '04.png', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '12d.png', 'rings': '01.png', 'sunglasses': '05a.png', 'tokenId': 273}, {'background': '04d.png', 'base': '03e.png', 'makeup': '05.png', 'eye': '01.png', 'hair': '', 'hat': '01c.png', 'necklace': '03b.png', 'rings': '02.png', 'sunglasses': '02a.png', 'tokenId': 274}, {'background': '01c.png', 'base': '03e.png', 'makeup': '03.png', 'eye': '03.png', 'hair': '', 'hat': '01b.png', 'necklace': '08a.png', 'rings': '04.png', 'sunglasses': '', 'tokenId': 275}, {'background': '03a.png', 'base': '01c.png', 'makeup': '01.png', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '10d.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 276}, {'background': '03c.png', 'base': '03b.png', 'makeup': '05.png', 'eye': '01.png', 'hair': '02a.png', 'hat': '07e.png', 'necklace': '', 'rings': '', 'sunglasses': '', 'tokenId': 277}, {'background': '01b.png', 'base': '04e.png', 'makeup': '05.png', 'eye': '03.png', 'hair': '01a.png', 'hat': '04c.png', 'necklace': '', 'rings': '', 'sunglasses': '', 'tokenId': 278}, {'background': '03b.png', 'base': '01a.png', 'makeup': '', 'eye': '02.png', 'hair': '', 'hat': '06b.png', 'necklace': '03c.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 279}, {'background': '02c.png', 'base': '03b.png', 'makeup': '01.png', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '08d.png', 'rings': '', 'sunglasses': '', 'tokenId': 280}, {'background': '04c.png', 'base': '01a.png', 'makeup': '02.png', 'eye': '04.png', 'hair': '', 'hat': '06e.png', 'necklace': '12c.png', 'rings': '', 'sunglasses': '05a.png', 'tokenId': 281}, {'background': '05c.png', 'base': '03e.png', 'makeup': '', 'eye': '03.png', 'hair': '', 'hat': '07e.png', 'necklace': '', 'rings': '', 'sunglasses': '', 'tokenId': 282}, {'background': '04d.png', 'base': '02e.png', 'makeup': '01.png', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '', 'rings': '03.png', 'sunglasses': '', 'tokenId': 283}, {'background': '02a.png', 'base': '04a.png', 'makeup': '', 'eye': '03.png', 'hair': '02b.png', 'hat': '', 'necklace': '08d.png', 'rings': '', 'sunglasses': '', 'tokenId': 284}, {'background': '05d.png', 'base': '01c.png', 'makeup': '01.png', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '08a.png', 'rings': '', 'sunglasses': '01.png', 'tokenId': 285}, {'background': '01b.png', 'base': '01d.png', 'makeup': '02.png', 'eye': '05.png', 'hair': '02a.png', 'hat': '', 'necklace': '04b.png', 'rings': '', 'sunglasses': '02e.png', 'tokenId': 286}, {'background': '02c.png', 'base': '03b.png', 'makeup': '01.png', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '06d.png', 'rings': '04.png', 'sunglasses': '01.png', 'tokenId': 287}, {'background': '03a.png', 'base': '01a.png', 'makeup': '', 'eye': '04.png', 'hair': '01c.png', 'hat': '', 'necklace': '12c.png', 'rings': '', 'sunglasses': '', 'tokenId': 288}, {'background': '05d.png', 'base': '01c.png', 'makeup': '05.png', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '09a.png', 'rings': '', 'sunglasses': '05a.png', 'tokenId': 289}, {'background': '05d.png', 'base': '02c.png', 'makeup': '', 'eye': '01.png', 'hair': '', 'hat': '05c.png', 'necklace': '08b.png', 'rings': '02.png', 'sunglasses': '', 'tokenId': 290}, {'background': '02c.png', 'base': '01b.png', 'makeup': '', 'eye': '03.png', 'hair': '', 'hat': '03a.png', 'necklace': '08b.png', 'rings': '', 'sunglasses': '', 'tokenId': 291}, {'background': '03c.png', 'base': '02c.png', 'makeup': '', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '07d.png', 'rings': '01.png', 'sunglasses': '02d.png', 'tokenId': 292}, {'background': '05d.png', 'base': '02a.png', 'makeup': '', 'eye': '02.png', 'hair': '', 'hat': '05c.png', 'necklace': '12d.png', 'rings': '01.png', 'sunglasses': '05d.png', 'tokenId': 293}, {'background': '04a.png', 'base': '04a.png', 'makeup': '02.png', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '02.png', 'rings': '03.png', 'sunglasses': '02a.png', 'tokenId': 294}, {'background': '01c.png', 'base': '02c.png', 'makeup': '03.png', 'eye': '04.png', 'hair': '01a.png', 'hat': '', 'necklace': '03b.png', 'rings': '', 'sunglasses': '', 'tokenId': 295}, {'background': '05b.png', 'base': '01e.png', 'makeup': '05.png', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '05a.png', 'rings': '04.png', 'sunglasses': '', 'tokenId': 296}, {'background': '01.png', 'base': '02c.png', 'makeup': '', 'eye': '01.png', 'hair': '', 'hat': '01d.png', 'necklace': '03e.png', 'rings': '', 'sunglasses': '', 'tokenId': 297}, {'background': '03a.png', 'base': '01a.png', 'makeup': '04.png', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '07e.png', 'rings': '', 'sunglasses': '05a.png', 'tokenId': 298}, {'background': '03a.png', 'base': '02e.png', 'makeup': '', 'eye': '03.png', 'hair': '', 'hat': '01d.png', 'necklace': '12c.png', 'rings': '04.png', 'sunglasses': '', 'tokenId': 299}]\n"
],
[
"# Calculate Trait Counts\ntrait_stats = {}\nsum_all_traits = 0\nfor trait in traits:\n sum_all_traits += len(traits[trait])\ntrait_stats['sum_all_traits'] = sum_all_traits\nprint(\"Total items in collection: \" + str(sum_all_traits))\n \ntrait_group_counts = {}\n \nfor trait in traits:\n trait_group_counts[trait] = 0\n trait_counts = {}\n for trait_value in traits[trait]:\n trait_counts[trait_value] = 0\n trait_group_counts[trait] = trait_counts\n\nfor image in all_images:\n for trait in trait_group_counts:\n trait_group_counts[trait][image[trait]] += 1\n \nprint(trait_group_counts)",
"Total items in collection: 163\n{'background': {'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}, 'base': {'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}, 'makeup': {'': 35, '01.png': 15, '02.png': 15, '03.png': 7, '04.png': 15, '05.png': 13}, 'eye': {'01.png': 31, '02.png': 19, '03.png': 19, '04.png': 22, '05.png': 9}, 'hair': {'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}, 'hat': {'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}, 'necklace': {'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}, 'rings': {'': 52, '01.png': 15, '02.png': 8, '03.png': 13, '04.png': 10, '05.png': 2}, 'sunglasses': {'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}}\n"
],
[
"## Calculate rarity scores\nrarity_scores = {}\nfor trait, values in trait_group_counts.items():\n rarity_scores_traits = {}\n for val in values:\n if values[val] == 0:\n next\n else:\n print(trait)\n print(values)\n rarity_score = 1 / (values[val] / TOTAL_IMAGES) \n rarity_scores_traits[val] = { 'rarity_score': rarity_score, 'count': values[val]}\n rarity_scores[trait] = rarity_scores_traits\nprint(rarity_scores)\n\n## Save meta stats\nwith open('./metadata/all-stats.json', 'w') as outfile:\n json.dump(rarity_scores, outfile, indent=4)",
"background\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbackground\n{'01.png': 11, '01b.png': 9, '01c.png': 9, '02a.png': 4, '02b.png': 3, '02c.png': 5, '03a.png': 5, '03b.png': 9, '03c.png': 7, '04a.png': 2, '04b.png': 5, '04c.png': 3, '04d.png': 9, '05a.png': 3, '05b.png': 6, '05c.png': 2, '05d.png': 8}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nbase\n{'01a.png': 22, '01b.png': 3, '01c.png': 6, '01d.png': 3, '01e.png': 4, '02a.png': 4, '02b.png': 4, '02c.png': 6, '02d.png': 2, '02e.png': 4, '03a.png': 3, '03b.png': 6, '03c.png': 2, '03d.png': 5, '03e.png': 7, '04a.png': 4, '04b.png': 4, '04c.png': 6, '04d.png': 2, '04e.png': 3}\nmakeup\n{'': 35, '01.png': 15, '02.png': 15, '03.png': 7, '04.png': 15, '05.png': 13}\nmakeup\n{'': 35, '01.png': 15, '02.png': 15, '03.png': 7, '04.png': 15, '05.png': 13}\nmakeup\n{'': 35, '01.png': 15, '02.png': 15, '03.png': 7, '04.png': 15, '05.png': 13}\nmakeup\n{'': 35, '01.png': 15, '02.png': 15, '03.png': 7, '04.png': 15, '05.png': 13}\nmakeup\n{'': 35, '01.png': 15, '02.png': 15, '03.png': 7, '04.png': 15, '05.png': 13}\nmakeup\n{'': 35, '01.png': 15, '02.png': 15, '03.png': 7, '04.png': 15, '05.png': 13}\neye\n{'01.png': 31, '02.png': 19, '03.png': 19, '04.png': 22, '05.png': 9}\neye\n{'01.png': 31, '02.png': 19, '03.png': 19, '04.png': 22, '05.png': 9}\neye\n{'01.png': 31, '02.png': 19, '03.png': 19, '04.png': 22, '05.png': 9}\neye\n{'01.png': 31, '02.png': 19, '03.png': 19, '04.png': 22, '05.png': 9}\neye\n{'01.png': 31, '02.png': 19, '03.png': 19, '04.png': 22, '05.png': 9}\nhair\n{'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}\nhair\n{'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}\nhair\n{'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}\nhair\n{'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}\nhair\n{'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}\nhair\n{'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}\nhair\n{'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}\nhair\n{'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}\nhair\n{'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}\nhair\n{'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}\nhair\n{'': 71, '01a.png': 4, '01b.png': 1, '01c.png': 5, '01d.png': 2, '01e.png': 1, '02a.png': 7, '02b.png': 3, '02c.png': 2, '02d.png': 3, '02e.png': 1}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nhat\n{'': 54, '01a.png': 0, '01b.png': 3, '01c.png': 3, '01d.png': 2, '02.png': 0, '03a.png': 2, '03b.png': 2, '03c.png': 0, '03d.png': 1, '03e.png': 1, '04a.png': 3, '04b.png': 2, '04c.png': 2, '04d.png': 0, '04e.png': 2, '05a.png': 3, '05b.png': 0, '05c.png': 4, '05d.png': 0, '06a.png': 1, '06b.png': 2, '06c.png': 2, '06d.png': 1, '06e.png': 2, '07a.png': 1, '07b.png': 1, '07c.png': 1, '07d.png': 2, '07e.png': 3, '07f.png': 0}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nnecklace\n{'': 15, '01.png': 0, '02.png': 3, '03a.png': 2, '03b.png': 2, '03c.png': 2, '03d.png': 0, '03e.png': 1, '04a.png': 2, '04b.png': 1, '04c.png': 0, '04d.png': 3, '05a.png': 3, '05b.png': 1, '05c.png': 2, '05d.png': 0, '06a.png': 4, '06b.png': 2, '06c.png': 1, '06d.png': 2, '07a.png': 1, '07b.png': 4, '07c.png': 2, '07d.png': 5, '07e.png': 1, '08a.png': 7, '08b.png': 2, '08c.png': 1, '08d.png': 3, '09a.png': 3, '09b.png': 0, '09c.png': 2, '09d.png': 0, '09e.png': 2, '10a.png': 2, '10b.png': 0, '10c.png': 2, '10d.png': 3, '10e.png': 0, '11a.png': 1, '11b.png': 0, '11c.png': 0, '11d.png': 4, '11e.png': 2, '12a.png': 0, '12b.png': 1, '12c.png': 3, '12d.png': 2, '12e.png': 1}\nrings\n{'': 52, '01.png': 15, '02.png': 8, '03.png': 13, '04.png': 10, '05.png': 2}\nrings\n{'': 52, '01.png': 15, '02.png': 8, '03.png': 13, '04.png': 10, '05.png': 2}\nrings\n{'': 52, '01.png': 15, '02.png': 8, '03.png': 13, '04.png': 10, '05.png': 2}\nrings\n{'': 52, '01.png': 15, '02.png': 8, '03.png': 13, '04.png': 10, '05.png': 2}\nrings\n{'': 52, '01.png': 15, '02.png': 8, '03.png': 13, '04.png': 10, '05.png': 2}\nrings\n{'': 52, '01.png': 15, '02.png': 8, '03.png': 13, '04.png': 10, '05.png': 2}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\nsunglasses\n{'': 63, '01.png': 3, '02a.png': 3, '02b.png': 1, '02c.png': 1, '02d.png': 5, '02e.png': 2, '03.png': 2, '04.png': 3, '05a.png': 6, '05b.png': 2, '05c.png': 1, '05d.png': 2, '05e.png': 2, '06a.png': 2, '06b.png': 1, '06c.png': 1, '06d.png': 0}\n{'background': {'01.png': {'rarity_score': 9.090909090909092, 'count': 11}, '01b.png': {'rarity_score': 11.11111111111111, 'count': 9}, '01c.png': {'rarity_score': 11.11111111111111, 'count': 9}, '02a.png': {'rarity_score': 25.0, 'count': 4}, '02b.png': {'rarity_score': 33.333333333333336, 'count': 3}, '02c.png': {'rarity_score': 20.0, 'count': 5}, '03a.png': {'rarity_score': 20.0, 'count': 5}, '03b.png': {'rarity_score': 11.11111111111111, 'count': 9}, '03c.png': {'rarity_score': 14.285714285714285, 'count': 7}, '04a.png': {'rarity_score': 50.0, 'count': 2}, '04b.png': {'rarity_score': 20.0, 'count': 5}, '04c.png': {'rarity_score': 33.333333333333336, 'count': 3}, '04d.png': {'rarity_score': 11.11111111111111, 'count': 9}, '05a.png': {'rarity_score': 33.333333333333336, 'count': 3}, '05b.png': {'rarity_score': 16.666666666666668, 'count': 6}, '05c.png': {'rarity_score': 50.0, 'count': 2}, '05d.png': {'rarity_score': 12.5, 'count': 8}}, 'base': {'01a.png': {'rarity_score': 4.545454545454546, 'count': 22}, '01b.png': {'rarity_score': 33.333333333333336, 'count': 3}, '01c.png': {'rarity_score': 16.666666666666668, 'count': 6}, '01d.png': {'rarity_score': 33.333333333333336, 'count': 3}, '01e.png': {'rarity_score': 25.0, 'count': 4}, '02a.png': {'rarity_score': 25.0, 'count': 4}, '02b.png': {'rarity_score': 25.0, 'count': 4}, '02c.png': {'rarity_score': 16.666666666666668, 'count': 6}, '02d.png': {'rarity_score': 50.0, 'count': 2}, '02e.png': {'rarity_score': 25.0, 'count': 4}, '03a.png': {'rarity_score': 33.333333333333336, 'count': 3}, '03b.png': {'rarity_score': 16.666666666666668, 'count': 6}, '03c.png': {'rarity_score': 50.0, 'count': 2}, '03d.png': {'rarity_score': 20.0, 'count': 5}, '03e.png': {'rarity_score': 14.285714285714285, 'count': 7}, '04a.png': {'rarity_score': 25.0, 'count': 4}, '04b.png': {'rarity_score': 25.0, 'count': 4}, '04c.png': {'rarity_score': 16.666666666666668, 'count': 6}, '04d.png': {'rarity_score': 50.0, 'count': 2}, '04e.png': {'rarity_score': 33.333333333333336, 'count': 3}}, 'makeup': {'': {'rarity_score': 2.857142857142857, 'count': 35}, '01.png': {'rarity_score': 6.666666666666667, 'count': 15}, '02.png': {'rarity_score': 6.666666666666667, 'count': 15}, '03.png': {'rarity_score': 14.285714285714285, 'count': 7}, '04.png': {'rarity_score': 6.666666666666667, 'count': 15}, '05.png': {'rarity_score': 7.692307692307692, 'count': 13}}, 'eye': {'01.png': {'rarity_score': 3.2258064516129035, 'count': 31}, '02.png': {'rarity_score': 5.2631578947368425, 'count': 19}, '03.png': {'rarity_score': 5.2631578947368425, 'count': 19}, '04.png': {'rarity_score': 4.545454545454546, 'count': 22}, '05.png': {'rarity_score': 11.11111111111111, 'count': 9}}, 'hair': {'': {'rarity_score': 1.4084507042253522, 'count': 71}, '01a.png': {'rarity_score': 25.0, 'count': 4}, '01b.png': {'rarity_score': 100.0, 'count': 1}, '01c.png': {'rarity_score': 20.0, 'count': 5}, '01d.png': {'rarity_score': 50.0, 'count': 2}, '01e.png': {'rarity_score': 100.0, 'count': 1}, '02a.png': {'rarity_score': 14.285714285714285, 'count': 7}, '02b.png': {'rarity_score': 33.333333333333336, 'count': 3}, '02c.png': {'rarity_score': 50.0, 'count': 2}, '02d.png': {'rarity_score': 33.333333333333336, 'count': 3}, '02e.png': {'rarity_score': 100.0, 'count': 1}}, 'hat': {'': {'rarity_score': 1.8518518518518516, 'count': 54}, '01b.png': {'rarity_score': 33.333333333333336, 'count': 3}, '01c.png': {'rarity_score': 33.333333333333336, 'count': 3}, '01d.png': {'rarity_score': 50.0, 'count': 2}, '03a.png': {'rarity_score': 50.0, 'count': 2}, '03b.png': {'rarity_score': 50.0, 'count': 2}, '03d.png': {'rarity_score': 100.0, 'count': 1}, '03e.png': {'rarity_score': 100.0, 'count': 1}, '04a.png': {'rarity_score': 33.333333333333336, 'count': 3}, '04b.png': {'rarity_score': 50.0, 'count': 2}, '04c.png': {'rarity_score': 50.0, 'count': 2}, '04e.png': {'rarity_score': 50.0, 'count': 2}, '05a.png': {'rarity_score': 33.333333333333336, 'count': 3}, '05c.png': {'rarity_score': 25.0, 'count': 4}, '06a.png': {'rarity_score': 100.0, 'count': 1}, '06b.png': {'rarity_score': 50.0, 'count': 2}, '06c.png': {'rarity_score': 50.0, 'count': 2}, '06d.png': {'rarity_score': 100.0, 'count': 1}, '06e.png': {'rarity_score': 50.0, 'count': 2}, '07a.png': {'rarity_score': 100.0, 'count': 1}, '07b.png': {'rarity_score': 100.0, 'count': 1}, '07c.png': {'rarity_score': 100.0, 'count': 1}, '07d.png': {'rarity_score': 50.0, 'count': 2}, '07e.png': {'rarity_score': 33.333333333333336, 'count': 3}}, 'necklace': {'': {'rarity_score': 6.666666666666667, 'count': 15}, '02.png': {'rarity_score': 33.333333333333336, 'count': 3}, '03a.png': {'rarity_score': 50.0, 'count': 2}, '03b.png': {'rarity_score': 50.0, 'count': 2}, '03c.png': {'rarity_score': 50.0, 'count': 2}, '03e.png': {'rarity_score': 100.0, 'count': 1}, '04a.png': {'rarity_score': 50.0, 'count': 2}, '04b.png': {'rarity_score': 100.0, 'count': 1}, '04d.png': {'rarity_score': 33.333333333333336, 'count': 3}, '05a.png': {'rarity_score': 33.333333333333336, 'count': 3}, '05b.png': {'rarity_score': 100.0, 'count': 1}, '05c.png': {'rarity_score': 50.0, 'count': 2}, '06a.png': {'rarity_score': 25.0, 'count': 4}, '06b.png': {'rarity_score': 50.0, 'count': 2}, '06c.png': {'rarity_score': 100.0, 'count': 1}, '06d.png': {'rarity_score': 50.0, 'count': 2}, '07a.png': {'rarity_score': 100.0, 'count': 1}, '07b.png': {'rarity_score': 25.0, 'count': 4}, '07c.png': {'rarity_score': 50.0, 'count': 2}, '07d.png': {'rarity_score': 20.0, 'count': 5}, '07e.png': {'rarity_score': 100.0, 'count': 1}, '08a.png': {'rarity_score': 14.285714285714285, 'count': 7}, '08b.png': {'rarity_score': 50.0, 'count': 2}, '08c.png': {'rarity_score': 100.0, 'count': 1}, '08d.png': {'rarity_score': 33.333333333333336, 'count': 3}, '09a.png': {'rarity_score': 33.333333333333336, 'count': 3}, '09c.png': {'rarity_score': 50.0, 'count': 2}, '09e.png': {'rarity_score': 50.0, 'count': 2}, '10a.png': {'rarity_score': 50.0, 'count': 2}, '10c.png': {'rarity_score': 50.0, 'count': 2}, '10d.png': {'rarity_score': 33.333333333333336, 'count': 3}, '11a.png': {'rarity_score': 100.0, 'count': 1}, '11d.png': {'rarity_score': 25.0, 'count': 4}, '11e.png': {'rarity_score': 50.0, 'count': 2}, '12b.png': {'rarity_score': 100.0, 'count': 1}, '12c.png': {'rarity_score': 33.333333333333336, 'count': 3}, '12d.png': {'rarity_score': 50.0, 'count': 2}, '12e.png': {'rarity_score': 100.0, 'count': 1}}, 'rings': {'': {'rarity_score': 1.923076923076923, 'count': 52}, '01.png': {'rarity_score': 6.666666666666667, 'count': 15}, '02.png': {'rarity_score': 12.5, 'count': 8}, '03.png': {'rarity_score': 7.692307692307692, 'count': 13}, '04.png': {'rarity_score': 10.0, 'count': 10}, '05.png': {'rarity_score': 50.0, 'count': 2}}, 'sunglasses': {'': {'rarity_score': 1.5873015873015872, 'count': 63}, '01.png': {'rarity_score': 33.333333333333336, 'count': 3}, '02a.png': {'rarity_score': 33.333333333333336, 'count': 3}, '02b.png': {'rarity_score': 100.0, 'count': 1}, '02c.png': {'rarity_score': 100.0, 'count': 1}, '02d.png': {'rarity_score': 20.0, 'count': 5}, '02e.png': {'rarity_score': 50.0, 'count': 2}, '03.png': {'rarity_score': 50.0, 'count': 2}, '04.png': {'rarity_score': 33.333333333333336, 'count': 3}, '05a.png': {'rarity_score': 16.666666666666668, 'count': 6}, '05b.png': {'rarity_score': 50.0, 'count': 2}, '05c.png': {'rarity_score': 100.0, 'count': 1}, '05d.png': {'rarity_score': 50.0, 'count': 2}, '05e.png': {'rarity_score': 50.0, 'count': 2}, '06a.png': {'rarity_score': 50.0, 'count': 2}, '06b.png': {'rarity_score': 100.0, 'count': 1}, '06c.png': {'rarity_score': 100.0, 'count': 1}}}\n"
],
[
"#### Generate Metadata for all Traits \nMETADATA_FILE_NAME = './metadata/all-traits.json'; \nwith open(METADATA_FILE_NAME, 'w') as outfile:\n json.dump(all_images, outfile, indent=4)",
"_____no_output_____"
],
[
"#### Generate Images \nfor item in all_images:\n print(item)\n imgs = []\n for trait in item:\n if trait == 'tokenId':\n continue \n # don't create images which have a blank option\n if item[trait] == '':\n continue\n# img = Image.open(f'./trait-layers/{directory_mappings[trait]}/{item[trait]}').convert('RGBA')\n img = Image.open(f'./high-res-traits/{directory_mappings[trait]}/{item[trait]}').convert('RGBA')\n# img = img.resize((400,400),Image.ANTIALIAS)\n imgs.append(img)\n\n #Create each composite\n start_image = Image.alpha_composite(imgs[0], imgs[1])\n next_image = start_image\n for idx, val in enumerate(imgs):\n if idx < 2:\n continue\n else:\n next_image = Image.alpha_composite(start_image, imgs[idx])\n start_image = next_image\n\n #Convert to RGB\n rgb_im = next_image.convert('RGB')\n # resize images for sampling. Remove below line for final images\n smaller_img = rgb_im.resize((1024,1024),Image.ANTIALIAS)\n file_name = str(item[\"tokenId\"]) + \".png\"\n smaller_img.save(\"./images/\" + file_name)\n \n ",
"{'background': '01.png', 'base': '02a.png', 'makeup': '02.png', 'eye': '01.png', 'hair': '', 'hat': '05a.png', 'necklace': '11d.png', 'rings': '', 'sunglasses': '06b.png', 'tokenId': 200}\n{'background': '04c.png', 'base': '02c.png', 'makeup': '', 'eye': '04.png', 'hair': '', 'hat': '03b.png', 'necklace': '04d.png', 'rings': '04.png', 'sunglasses': '02c.png', 'tokenId': 201}\n{'background': '05b.png', 'base': '01a.png', 'makeup': '', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '09a.png', 'rings': '', 'sunglasses': '', 'tokenId': 202}\n{'background': '04d.png', 'base': '02e.png', 'makeup': '', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '06b.png', 'rings': '', 'sunglasses': '05d.png', 'tokenId': 203}\n{'background': '05c.png', 'base': '01a.png', 'makeup': '', 'eye': '01.png', 'hair': '01a.png', 'hat': '01b.png', 'necklace': '02.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 204}\n{'background': '03b.png', 'base': '03b.png', 'makeup': '', 'eye': '04.png', 'hair': '02e.png', 'hat': '', 'necklace': '06a.png', 'rings': '', 'sunglasses': '', 'tokenId': 205}\n{'background': '05d.png', 'base': '04b.png', 'makeup': '02.png', 'eye': '01.png', 'hair': '', 'hat': '04e.png', 'necklace': '11e.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 206}\n{'background': '04b.png', 'base': '01b.png', 'makeup': '02.png', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '02.png', 'rings': '04.png', 'sunglasses': '02d.png', 'tokenId': 207}\n{'background': '01c.png', 'base': '04b.png', 'makeup': '02.png', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '', 'rings': '03.png', 'sunglasses': '', 'tokenId': 208}\n{'background': '01b.png', 'base': '01a.png', 'makeup': '03.png', 'eye': '01.png', 'hair': '', 'hat': '04c.png', 'necklace': '05a.png', 'rings': '', 'sunglasses': '', 'tokenId': 209}\n{'background': '02a.png', 'base': '03c.png', 'makeup': '01.png', 'eye': '01.png', 'hair': '', 'hat': '06b.png', 'necklace': '10d.png', 'rings': '', 'sunglasses': '', 'tokenId': 210}\n{'background': '04b.png', 'base': '01a.png', 'makeup': '01.png', 'eye': '01.png', 'hair': '', 'hat': '01c.png', 'necklace': '', 'rings': '', 'sunglasses': '', 'tokenId': 211}\n{'background': '01.png', 'base': '01a.png', 'makeup': '02.png', 'eye': '01.png', 'hair': '02d.png', 'hat': '07d.png', 'necklace': '', 'rings': '05.png', 'sunglasses': '', 'tokenId': 212}\n{'background': '01.png', 'base': '02b.png', 'makeup': '', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '08a.png', 'rings': '', 'sunglasses': '', 'tokenId': 213}\n{'background': '02a.png', 'base': '02b.png', 'makeup': '05.png', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '08a.png', 'rings': '', 'sunglasses': '02d.png', 'tokenId': 214}\n{'background': '02b.png', 'base': '04c.png', 'makeup': '04.png', 'eye': '04.png', 'hair': '', 'hat': '04b.png', 'necklace': '08a.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 215}\n{'background': '01b.png', 'base': '04d.png', 'makeup': '', 'eye': '01.png', 'hair': '02a.png', 'hat': '', 'necklace': '11d.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 216}\n{'background': '05a.png', 'base': '03a.png', 'makeup': '01.png', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '03a.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 217}\n{'background': '01c.png', 'base': '01d.png', 'makeup': '02.png', 'eye': '03.png', 'hair': '02c.png', 'hat': '06a.png', 'necklace': '03a.png', 'rings': '', 'sunglasses': '04.png', 'tokenId': 218}\n{'background': '01.png', 'base': '03e.png', 'makeup': '', 'eye': '01.png', 'hair': '', 'hat': '04a.png', 'necklace': '06b.png', 'rings': '', 'sunglasses': '05e.png', 'tokenId': 219}\n{'background': '04b.png', 'base': '04d.png', 'makeup': '', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '', 'rings': '', 'sunglasses': '05e.png', 'tokenId': 220}\n{'background': '03c.png', 'base': '03d.png', 'makeup': '05.png', 'eye': '02.png', 'hair': '01c.png', 'hat': '01b.png', 'necklace': '10d.png', 'rings': '', 'sunglasses': '', 'tokenId': 221}\n{'background': '01.png', 'base': '01a.png', 'makeup': '02.png', 'eye': '02.png', 'hair': '', 'hat': '06d.png', 'necklace': '', 'rings': '', 'sunglasses': '05b.png', 'tokenId': 222}\n{'background': '05b.png', 'base': '03e.png', 'makeup': '', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '09c.png', 'rings': '', 'sunglasses': '', 'tokenId': 223}\n{'background': '01c.png', 'base': '04b.png', 'makeup': '', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '10c.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 224}\n{'background': '03c.png', 'base': '04e.png', 'makeup': '01.png', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '08c.png', 'rings': '04.png', 'sunglasses': '', 'tokenId': 225}\n{'background': '02c.png', 'base': '02e.png', 'makeup': '', 'eye': '01.png', 'hair': '', 'hat': '04e.png', 'necklace': '11e.png', 'rings': '', 'sunglasses': '06c.png', 'tokenId': 226}\n{'background': '01c.png', 'base': '03d.png', 'makeup': '05.png', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '04d.png', 'rings': '', 'sunglasses': '', 'tokenId': 227}\n{'background': '04a.png', 'base': '04e.png', 'makeup': '01.png', 'eye': '02.png', 'hair': '', 'hat': '', 'necklace': '12b.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 228}\n{'background': '03b.png', 'base': '01a.png', 'makeup': '04.png', 'eye': '01.png', 'hair': '', 'hat': '', 'necklace': '09e.png', 'rings': '05.png', 'sunglasses': '06a.png', 'tokenId': 229}\n{'background': '01b.png', 'base': '03b.png', 'makeup': '04.png', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '07b.png', 'rings': '03.png', 'sunglasses': '', 'tokenId': 230}\n{'background': '01.png', 'base': '03d.png', 'makeup': '01.png', 'eye': '02.png', 'hair': '', 'hat': '07d.png', 'necklace': '', 'rings': '01.png', 'sunglasses': '05a.png', 'tokenId': 231}\n{'background': '01b.png', 'base': '01a.png', 'makeup': '04.png', 'eye': '05.png', 'hair': '', 'hat': '05a.png', 'necklace': '05c.png', 'rings': '04.png', 'sunglasses': '', 'tokenId': 232}\n{'background': '02b.png', 'base': '01a.png', 'makeup': '01.png', 'eye': '03.png', 'hair': '01d.png', 'hat': '06c.png', 'necklace': '07c.png', 'rings': '01.png', 'sunglasses': '', 'tokenId': 233}\n{'background': '01b.png', 'base': '01c.png', 'makeup': '04.png', 'eye': '01.png', 'hair': '02a.png', 'hat': '04a.png', 'necklace': '12e.png', 'rings': '', 'sunglasses': '04.png', 'tokenId': 234}\n{'background': '03a.png', 'base': '01d.png', 'makeup': '', 'eye': '04.png', 'hair': '02c.png', 'hat': '', 'necklace': '07d.png', 'rings': '', 'sunglasses': '', 'tokenId': 235}\n{'background': '03b.png', 'base': '01e.png', 'makeup': '01.png', 'eye': '03.png', 'hair': '', 'hat': '', 'necklace': '08d.png', 'rings': '01.png', 'sunglasses': '04.png', 'tokenId': 236}\n{'background': '01.png', 'base': '02a.png', 'makeup': '02.png', 'eye': '04.png', 'hair': '02a.png', 'hat': '', 'necklace': '07c.png', 'rings': '', 'sunglasses': '05c.png', 'tokenId': 237}\n{'background': '01b.png', 'base': '01a.png', 'makeup': '04.png', 'eye': '04.png', 'hair': '02d.png', 'hat': '', 'necklace': '09a.png', 'rings': '02.png', 'sunglasses': '01.png', 'tokenId': 238}\n{'background': '05d.png', 'base': '04c.png', 'makeup': '04.png', 'eye': '04.png', 'hair': '', 'hat': '', 'necklace': '05b.png', 'rings': '', 'sunglasses': '', 'tokenId': 239}\n{'background': '01c.png', 'base': '02b.png', 'makeup': '05.png', 'eye': '02.png', 'hair': '02a.png', 'hat': '', 'necklace': '07d.png', 'rings': '02.png', 'sunglasses': '', 'tokenId': 240}\n{'background': '05d.png', 'base': '02a.png', 'makeup': '', 'eye': '03.png', 'hair': '02d.png', 'hat': '', 'necklace': '06a.png', 'rings': '01.png', 'sunglasses': '02d.png', 'tokenId': 241}\n{'background': '04b.png', 'base': '02b.png', 'makeup': '04.png', 'eye': '05.png', 'hair': '01e.png', 'hat': '', 'necklace': '06a.png', 'rings': '', 'sunglasses': '', 'tokenId': 242}\n{'background': '01c.png', 'base': '02c.png', 'makeup': '02.png', 'eye': '05.png', 'hair': '', 'hat': '04a.png', 'necklace': '07b.png', 'rings': '02.png', 'sunglasses': '', 'tokenId': 243}\n{'background': '05a.png', 'base': '04a.png', 'makeup': '', 'eye': '02.png', 'hair': '02b.png', 'hat': '04b.png', 'necklace': '06c.png', 'rings': '01.png', 'sunglasses': '02e.png', 'tokenId': 244}\n"
],
[
"#### Generate Metadata for each Image \n\nf = open('./metadata/all-traits.json',) \ndata = json.load(f)\n\n\nIMAGES_BASE_URI = \"ADD_IMAGES_BASE_URI_HERE\"\nPROJECT_NAME = \"ADD_PROJECT_NAME_HERE\"\n\ndef getAttribute(key, value):\n return {\n \"trait_type\": key,\n \"value\": value\n }\nfor i in data:\n token_id = i['tokenId']\n token = {\n \"image\": IMAGES_BASE_URI + str(token_id) + '.png',\n \"tokenId\": token_id,\n \"name\": PROJECT_NAME + ' ' + str(token_id),\n \"attributes\": []\n }\n for attr in i:\n token[\"attributes\"].append(getAttribute(attr, i[attr]))\n\n print(token)\n with open('./metadata/' + str(token_id), 'w') as outfile:\n json.dump(token, outfile, indent=4)\nf.close()",
"{'image': 'ADD_IMAGES_BASE_URI_HERE0.png', 'tokenId': 0, 'name': 'ADD_PROJECT_NAME_HERE 0', 'attributes': [{'trait_type': 'background', 'value': '05b.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': '02d.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '05a.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 0}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE1.png', 'tokenId': 1, 'name': 'ADD_PROJECT_NAME_HERE 1', 'attributes': [{'trait_type': 'background', 'value': '05d.png'}, {'trait_type': 'base', 'value': '03c.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '04e.png'}, {'trait_type': 'necklace', 'value': '08d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '05e.png'}, {'trait_type': 'tokenId', 'value': 1}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE2.png', 'tokenId': 2, 'name': 'ADD_PROJECT_NAME_HERE 2', 'attributes': [{'trait_type': 'background', 'value': '05d.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '02e.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '06d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 2}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE3.png', 'tokenId': 3, 'name': 'ADD_PROJECT_NAME_HERE 3', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '02b.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '11c.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 3}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE4.png', 'tokenId': 4, 'name': 'ADD_PROJECT_NAME_HERE 4', 'attributes': [{'trait_type': 'background', 'value': '03c.png'}, {'trait_type': 'base', 'value': '01e.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '10d.png'}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 4}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE5.png', 'tokenId': 5, 'name': 'ADD_PROJECT_NAME_HERE 5', 'attributes': [{'trait_type': 'background', 'value': '04c.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '01b.png'}, {'trait_type': 'necklace', 'value': '03d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 5}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE6.png', 'tokenId': 6, 'name': 'ADD_PROJECT_NAME_HERE 6', 'attributes': [{'trait_type': 'background', 'value': '05a.png'}, {'trait_type': 'base', 'value': '04d.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '09e.png'}, {'trait_type': 'rings', 'value': '01.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 6}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE7.png', 'tokenId': 7, 'name': 'ADD_PROJECT_NAME_HERE 7', 'attributes': [{'trait_type': 'background', 'value': '02c.png'}, {'trait_type': 'base', 'value': '03c.png'}, {'trait_type': 'makeup', 'value': '02.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '11d.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 7}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE8.png', 'tokenId': 8, 'name': 'ADD_PROJECT_NAME_HERE 8', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '02a.png'}, {'trait_type': 'hat', 'value': '03b.png'}, {'trait_type': 'necklace', 'value': '11a.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 8}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE9.png', 'tokenId': 9, 'name': 'ADD_PROJECT_NAME_HERE 9', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '03b.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '06b.png'}, {'trait_type': 'necklace', 'value': '09b.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': '02c.png'}, {'trait_type': 'tokenId', 'value': 9}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE10.png', 'tokenId': 10, 'name': 'ADD_PROJECT_NAME_HERE 10', 'attributes': [{'trait_type': 'background', 'value': '05d.png'}, {'trait_type': 'base', 'value': '03e.png'}, {'trait_type': 'makeup', 'value': '03.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '03a.png'}, {'trait_type': 'necklace', 'value': '10b.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '06d.png'}, {'trait_type': 'tokenId', 'value': 10}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE11.png', 'tokenId': 11, 'name': 'ADD_PROJECT_NAME_HERE 11', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': '02a.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '09d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 11}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE12.png', 'tokenId': 12, 'name': 'ADD_PROJECT_NAME_HERE 12', 'attributes': [{'trait_type': 'background', 'value': '03b.png'}, {'trait_type': 'base', 'value': '04c.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '07d.png'}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': '03.png'}, {'trait_type': 'tokenId', 'value': 12}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE13.png', 'tokenId': 13, 'name': 'ADD_PROJECT_NAME_HERE 13', 'attributes': [{'trait_type': 'background', 'value': '05d.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '11c.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': '05b.png'}, {'trait_type': 'tokenId', 'value': 13}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE14.png', 'tokenId': 14, 'name': 'ADD_PROJECT_NAME_HERE 14', 'attributes': [{'trait_type': 'background', 'value': '03b.png'}, {'trait_type': 'base', 'value': '03a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '03d.png'}, {'trait_type': 'necklace', 'value': '04a.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 14}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE15.png', 'tokenId': 15, 'name': 'ADD_PROJECT_NAME_HERE 15', 'attributes': [{'trait_type': 'background', 'value': '03a.png'}, {'trait_type': 'base', 'value': '01c.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '05.png'}, {'trait_type': 'hair', 'value': '01d.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '07d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 15}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE16.png', 'tokenId': 16, 'name': 'ADD_PROJECT_NAME_HERE 16', 'attributes': [{'trait_type': 'background', 'value': '02c.png'}, {'trait_type': 'base', 'value': '03a.png'}, {'trait_type': 'makeup', 'value': '02.png'}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '09e.png'}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': '01.png'}, {'trait_type': 'tokenId', 'value': 16}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE17.png', 'tokenId': 17, 'name': 'ADD_PROJECT_NAME_HERE 17', 'attributes': [{'trait_type': 'background', 'value': '02c.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': '01e.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '04b.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 17}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE18.png', 'tokenId': 18, 'name': 'ADD_PROJECT_NAME_HERE 18', 'attributes': [{'trait_type': 'background', 'value': '03a.png'}, {'trait_type': 'base', 'value': '03a.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '02c.png'}, {'trait_type': 'hat', 'value': '01d.png'}, {'trait_type': 'necklace', 'value': '05c.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 18}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE19.png', 'tokenId': 19, 'name': 'ADD_PROJECT_NAME_HERE 19', 'attributes': [{'trait_type': 'background', 'value': '05a.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': '02d.png'}, {'trait_type': 'hat', 'value': '05d.png'}, {'trait_type': 'necklace', 'value': '08a.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '04.png'}, {'trait_type': 'tokenId', 'value': 19}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE20.png', 'tokenId': 20, 'name': 'ADD_PROJECT_NAME_HERE 20', 'attributes': [{'trait_type': 'background', 'value': '01c.png'}, {'trait_type': 'base', 'value': '02e.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '04b.png'}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '02b.png'}, {'trait_type': 'tokenId', 'value': 20}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE21.png', 'tokenId': 21, 'name': 'ADD_PROJECT_NAME_HERE 21', 'attributes': [{'trait_type': 'background', 'value': '05d.png'}, {'trait_type': 'base', 'value': '02d.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '01b.png'}, {'trait_type': 'hat', 'value': '07e.png'}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': '01.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 21}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE22.png', 'tokenId': 22, 'name': 'ADD_PROJECT_NAME_HERE 22', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '01b.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '10c.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '02c.png'}, {'trait_type': 'tokenId', 'value': 22}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE23.png', 'tokenId': 23, 'name': 'ADD_PROJECT_NAME_HERE 23', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '03c.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '04c.png'}, {'trait_type': 'necklace', 'value': '05d.png'}, {'trait_type': 'rings', 'value': '01.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 23}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE24.png', 'tokenId': 24, 'name': 'ADD_PROJECT_NAME_HERE 24', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '03.png'}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '06b.png'}, {'trait_type': 'rings', 'value': '01.png'}, {'trait_type': 'sunglasses', 'value': '05d.png'}, {'trait_type': 'tokenId', 'value': 24}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE25.png', 'tokenId': 25, 'name': 'ADD_PROJECT_NAME_HERE 25', 'attributes': [{'trait_type': 'background', 'value': '01c.png'}, {'trait_type': 'base', 'value': '02b.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '05d.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 25}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE26.png', 'tokenId': 26, 'name': 'ADD_PROJECT_NAME_HERE 26', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '04d.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '10c.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': '04.png'}, {'trait_type': 'tokenId', 'value': 26}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE27.png', 'tokenId': 27, 'name': 'ADD_PROJECT_NAME_HERE 27', 'attributes': [{'trait_type': 'background', 'value': '02b.png'}, {'trait_type': 'base', 'value': '02a.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '01d.png'}, {'trait_type': 'hat', 'value': '05a.png'}, {'trait_type': 'necklace', 'value': '09a.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 27}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE28.png', 'tokenId': 28, 'name': 'ADD_PROJECT_NAME_HERE 28', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '06b.png'}, {'trait_type': 'tokenId', 'value': 28}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE29.png', 'tokenId': 29, 'name': 'ADD_PROJECT_NAME_HERE 29', 'attributes': [{'trait_type': 'background', 'value': '02b.png'}, {'trait_type': 'base', 'value': '04a.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '06e.png'}, {'trait_type': 'necklace', 'value': '08c.png'}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': '04.png'}, {'trait_type': 'tokenId', 'value': 29}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE30.png', 'tokenId': 30, 'name': 'ADD_PROJECT_NAME_HERE 30', 'attributes': [{'trait_type': 'background', 'value': '03c.png'}, {'trait_type': 'base', 'value': '04a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': '01a.png'}, {'trait_type': 'hat', 'value': '05c.png'}, {'trait_type': 'necklace', 'value': '08c.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '05c.png'}, {'trait_type': 'tokenId', 'value': 30}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE31.png', 'tokenId': 31, 'name': 'ADD_PROJECT_NAME_HERE 31', 'attributes': [{'trait_type': 'background', 'value': '05c.png'}, {'trait_type': 'base', 'value': '04a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '03b.png'}, {'trait_type': 'necklace', 'value': '04a.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 31}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE32.png', 'tokenId': 32, 'name': 'ADD_PROJECT_NAME_HERE 32', 'attributes': [{'trait_type': 'background', 'value': '04c.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '07c.png'}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': '01.png'}, {'trait_type': 'sunglasses', 'value': '06a.png'}, {'trait_type': 'tokenId', 'value': 32}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE33.png', 'tokenId': 33, 'name': 'ADD_PROJECT_NAME_HERE 33', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '02.png'}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': '01c.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '06c.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 33}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE34.png', 'tokenId': 34, 'name': 'ADD_PROJECT_NAME_HERE 34', 'attributes': [{'trait_type': 'background', 'value': '04a.png'}, {'trait_type': 'base', 'value': '02d.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '01c.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '09e.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '01.png'}, {'trait_type': 'tokenId', 'value': 34}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE35.png', 'tokenId': 35, 'name': 'ADD_PROJECT_NAME_HERE 35', 'attributes': [{'trait_type': 'background', 'value': '03a.png'}, {'trait_type': 'base', 'value': '02b.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '05d.png'}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 35}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE36.png', 'tokenId': 36, 'name': 'ADD_PROJECT_NAME_HERE 36', 'attributes': [{'trait_type': 'background', 'value': '05a.png'}, {'trait_type': 'base', 'value': '03d.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '01a.png'}, {'trait_type': 'necklace', 'value': '08a.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 36}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE37.png', 'tokenId': 37, 'name': 'ADD_PROJECT_NAME_HERE 37', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '04b.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '09b.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 37}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE38.png', 'tokenId': 38, 'name': 'ADD_PROJECT_NAME_HERE 38', 'attributes': [{'trait_type': 'background', 'value': '05a.png'}, {'trait_type': 'base', 'value': '02d.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': '01e.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '08d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 38}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE39.png', 'tokenId': 39, 'name': 'ADD_PROJECT_NAME_HERE 39', 'attributes': [{'trait_type': 'background', 'value': '04c.png'}, {'trait_type': 'base', 'value': '02a.png'}, {'trait_type': 'makeup', 'value': '02.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': '02b.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '06b.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 39}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE40.png', 'tokenId': 40, 'name': 'ADD_PROJECT_NAME_HERE 40', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '03.png'}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '07b.png'}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 40}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE41.png', 'tokenId': 41, 'name': 'ADD_PROJECT_NAME_HERE 41', 'attributes': [{'trait_type': 'background', 'value': '03b.png'}, {'trait_type': 'base', 'value': '01d.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '10d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 41}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE42.png', 'tokenId': 42, 'name': 'ADD_PROJECT_NAME_HERE 42', 'attributes': [{'trait_type': 'background', 'value': '02a.png'}, {'trait_type': 'base', 'value': '01c.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '05a.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '06b.png'}, {'trait_type': 'tokenId', 'value': 42}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE43.png', 'tokenId': 43, 'name': 'ADD_PROJECT_NAME_HERE 43', 'attributes': [{'trait_type': 'background', 'value': '04d.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '05.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '07f.png'}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 43}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE44.png', 'tokenId': 44, 'name': 'ADD_PROJECT_NAME_HERE 44', 'attributes': [{'trait_type': 'background', 'value': '02a.png'}, {'trait_type': 'base', 'value': '02d.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '03a.png'}, {'trait_type': 'necklace', 'value': '11d.png'}, {'trait_type': 'rings', 'value': '04.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 44}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE45.png', 'tokenId': 45, 'name': 'ADD_PROJECT_NAME_HERE 45', 'attributes': [{'trait_type': 'background', 'value': '02b.png'}, {'trait_type': 'base', 'value': '03e.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '02e.png'}, {'trait_type': 'hat', 'value': '01b.png'}, {'trait_type': 'necklace', 'value': '10e.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': '05a.png'}, {'trait_type': 'tokenId', 'value': 45}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE46.png', 'tokenId': 46, 'name': 'ADD_PROJECT_NAME_HERE 46', 'attributes': [{'trait_type': 'background', 'value': '05c.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': '02c.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '07e.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 46}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE47.png', 'tokenId': 47, 'name': 'ADD_PROJECT_NAME_HERE 47', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '03d.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': '02b.png'}, {'trait_type': 'hat', 'value': '01c.png'}, {'trait_type': 'necklace', 'value': '05b.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '03.png'}, {'trait_type': 'tokenId', 'value': 47}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE48.png', 'tokenId': 48, 'name': 'ADD_PROJECT_NAME_HERE 48', 'attributes': [{'trait_type': 'background', 'value': '03a.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '08a.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 48}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE49.png', 'tokenId': 49, 'name': 'ADD_PROJECT_NAME_HERE 49', 'attributes': [{'trait_type': 'background', 'value': '01c.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '09d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '05c.png'}, {'trait_type': 'tokenId', 'value': 49}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE50.png', 'tokenId': 50, 'name': 'ADD_PROJECT_NAME_HERE 50', 'attributes': [{'trait_type': 'background', 'value': '05a.png'}, {'trait_type': 'base', 'value': '03b.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '07d.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': '02e.png'}, {'trait_type': 'tokenId', 'value': 50}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE51.png', 'tokenId': 51, 'name': 'ADD_PROJECT_NAME_HERE 51', 'attributes': [{'trait_type': 'background', 'value': '05b.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': '01e.png'}, {'trait_type': 'hat', 'value': '05a.png'}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': '01.png'}, {'trait_type': 'tokenId', 'value': 51}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE52.png', 'tokenId': 52, 'name': 'ADD_PROJECT_NAME_HERE 52', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '03a.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '06e.png'}, {'trait_type': 'necklace', 'value': '09d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 52}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE53.png', 'tokenId': 53, 'name': 'ADD_PROJECT_NAME_HERE 53', 'attributes': [{'trait_type': 'background', 'value': '01c.png'}, {'trait_type': 'base', 'value': '01b.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '03d.png'}, {'trait_type': 'necklace', 'value': '04b.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 53}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE54.png', 'tokenId': 54, 'name': 'ADD_PROJECT_NAME_HERE 54', 'attributes': [{'trait_type': 'background', 'value': '05a.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '03e.png'}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 54}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE55.png', 'tokenId': 55, 'name': 'ADD_PROJECT_NAME_HERE 55', 'attributes': [{'trait_type': 'background', 'value': '05c.png'}, {'trait_type': 'base', 'value': '03e.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '08a.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 55}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE56.png', 'tokenId': 56, 'name': 'ADD_PROJECT_NAME_HERE 56', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '05d.png'}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': '02e.png'}, {'trait_type': 'tokenId', 'value': 56}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE57.png', 'tokenId': 57, 'name': 'ADD_PROJECT_NAME_HERE 57', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '03a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '03c.png'}, {'trait_type': 'necklace', 'value': '08d.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': '06b.png'}, {'trait_type': 'tokenId', 'value': 57}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE58.png', 'tokenId': 58, 'name': 'ADD_PROJECT_NAME_HERE 58', 'attributes': [{'trait_type': 'background', 'value': '03c.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '03.png'}, {'trait_type': 'eye', 'value': '05.png'}, {'trait_type': 'hair', 'value': '02a.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '05c.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': '02e.png'}, {'trait_type': 'tokenId', 'value': 58}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE59.png', 'tokenId': 59, 'name': 'ADD_PROJECT_NAME_HERE 59', 'attributes': [{'trait_type': 'background', 'value': '02b.png'}, {'trait_type': 'base', 'value': '04d.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': '02e.png'}, {'trait_type': 'hat', 'value': '03d.png'}, {'trait_type': 'necklace', 'value': '05a.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': '06b.png'}, {'trait_type': 'tokenId', 'value': 59}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE60.png', 'tokenId': 60, 'name': 'ADD_PROJECT_NAME_HERE 60', 'attributes': [{'trait_type': 'background', 'value': '05d.png'}, {'trait_type': 'base', 'value': '02e.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '04a.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '02a.png'}, {'trait_type': 'tokenId', 'value': 60}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE61.png', 'tokenId': 61, 'name': 'ADD_PROJECT_NAME_HERE 61', 'attributes': [{'trait_type': 'background', 'value': '05a.png'}, {'trait_type': 'base', 'value': '02b.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '02.png'}, {'trait_type': 'necklace', 'value': '11b.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 61}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE62.png', 'tokenId': 62, 'name': 'ADD_PROJECT_NAME_HERE 62', 'attributes': [{'trait_type': 'background', 'value': '01c.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '02.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '02e.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '04d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '05a.png'}, {'trait_type': 'tokenId', 'value': 62}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE63.png', 'tokenId': 63, 'name': 'ADD_PROJECT_NAME_HERE 63', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '02e.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '11c.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '05b.png'}, {'trait_type': 'tokenId', 'value': 63}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE64.png', 'tokenId': 64, 'name': 'ADD_PROJECT_NAME_HERE 64', 'attributes': [{'trait_type': 'background', 'value': '05b.png'}, {'trait_type': 'base', 'value': '01c.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': '02c.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '05a.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 64}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE65.png', 'tokenId': 65, 'name': 'ADD_PROJECT_NAME_HERE 65', 'attributes': [{'trait_type': 'background', 'value': '05a.png'}, {'trait_type': 'base', 'value': '03d.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '04a.png'}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 65}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE66.png', 'tokenId': 66, 'name': 'ADD_PROJECT_NAME_HERE 66', 'attributes': [{'trait_type': 'background', 'value': '05b.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '12d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 66}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE67.png', 'tokenId': 67, 'name': 'ADD_PROJECT_NAME_HERE 67', 'attributes': [{'trait_type': 'background', 'value': '02c.png'}, {'trait_type': 'base', 'value': '04c.png'}, {'trait_type': 'makeup', 'value': '03.png'}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': '02b.png'}, {'trait_type': 'hat', 'value': '01c.png'}, {'trait_type': 'necklace', 'value': '04d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 67}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE68.png', 'tokenId': 68, 'name': 'ADD_PROJECT_NAME_HERE 68', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '02c.png'}, {'trait_type': 'makeup', 'value': '03.png'}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '07d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 68}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE69.png', 'tokenId': 69, 'name': 'ADD_PROJECT_NAME_HERE 69', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '06b.png'}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 69}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE70.png', 'tokenId': 70, 'name': 'ADD_PROJECT_NAME_HERE 70', 'attributes': [{'trait_type': 'background', 'value': '02b.png'}, {'trait_type': 'base', 'value': '01d.png'}, {'trait_type': 'makeup', 'value': '02.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '03e.png'}, {'trait_type': 'necklace', 'value': '05d.png'}, {'trait_type': 'rings', 'value': '01.png'}, {'trait_type': 'sunglasses', 'value': '06b.png'}, {'trait_type': 'tokenId', 'value': 70}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE71.png', 'tokenId': 71, 'name': 'ADD_PROJECT_NAME_HERE 71', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '03c.png'}, {'trait_type': 'necklace', 'value': '03e.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 71}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE72.png', 'tokenId': 72, 'name': 'ADD_PROJECT_NAME_HERE 72', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '01b.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '01a.png'}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 72}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE73.png', 'tokenId': 73, 'name': 'ADD_PROJECT_NAME_HERE 73', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '04d.png'}, {'trait_type': 'makeup', 'value': '03.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '01.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 73}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE74.png', 'tokenId': 74, 'name': 'ADD_PROJECT_NAME_HERE 74', 'attributes': [{'trait_type': 'background', 'value': '01c.png'}, {'trait_type': 'base', 'value': '01d.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': '01c.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '04d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 74}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE75.png', 'tokenId': 75, 'name': 'ADD_PROJECT_NAME_HERE 75', 'attributes': [{'trait_type': 'background', 'value': '03c.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': '01c.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '11a.png'}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 75}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE76.png', 'tokenId': 76, 'name': 'ADD_PROJECT_NAME_HERE 76', 'attributes': [{'trait_type': 'background', 'value': '04a.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 76}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE77.png', 'tokenId': 77, 'name': 'ADD_PROJECT_NAME_HERE 77', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '03a.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '06c.png'}, {'trait_type': 'rings', 'value': '01.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 77}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE78.png', 'tokenId': 78, 'name': 'ADD_PROJECT_NAME_HERE 78', 'attributes': [{'trait_type': 'background', 'value': '02c.png'}, {'trait_type': 'base', 'value': '03b.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': '01b.png'}, {'trait_type': 'hat', 'value': '01b.png'}, {'trait_type': 'necklace', 'value': '11e.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 78}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE79.png', 'tokenId': 79, 'name': 'ADD_PROJECT_NAME_HERE 79', 'attributes': [{'trait_type': 'background', 'value': '04b.png'}, {'trait_type': 'base', 'value': '03b.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': '02c.png'}, {'trait_type': 'hat', 'value': '04e.png'}, {'trait_type': 'necklace', 'value': '08d.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 79}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE80.png', 'tokenId': 80, 'name': 'ADD_PROJECT_NAME_HERE 80', 'attributes': [{'trait_type': 'background', 'value': '05c.png'}, {'trait_type': 'base', 'value': '04e.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '06d.png'}, {'trait_type': 'necklace', 'value': '06a.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '04.png'}, {'trait_type': 'tokenId', 'value': 80}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE81.png', 'tokenId': 81, 'name': 'ADD_PROJECT_NAME_HERE 81', 'attributes': [{'trait_type': 'background', 'value': '03b.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '07a.png'}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 81}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE82.png', 'tokenId': 82, 'name': 'ADD_PROJECT_NAME_HERE 82', 'attributes': [{'trait_type': 'background', 'value': '05c.png'}, {'trait_type': 'base', 'value': '04a.png'}, {'trait_type': 'makeup', 'value': '02.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '01e.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '07d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 82}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE83.png', 'tokenId': 83, 'name': 'ADD_PROJECT_NAME_HERE 83', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '02b.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '05.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '06e.png'}, {'trait_type': 'necklace', 'value': '05b.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 83}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE84.png', 'tokenId': 84, 'name': 'ADD_PROJECT_NAME_HERE 84', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '05.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '10d.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 84}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE85.png', 'tokenId': 85, 'name': 'ADD_PROJECT_NAME_HERE 85', 'attributes': [{'trait_type': 'background', 'value': '03b.png'}, {'trait_type': 'base', 'value': '02e.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '03.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '06d.png'}, {'trait_type': 'necklace', 'value': '04b.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': '02c.png'}, {'trait_type': 'tokenId', 'value': 85}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE86.png', 'tokenId': 86, 'name': 'ADD_PROJECT_NAME_HERE 86', 'attributes': [{'trait_type': 'background', 'value': '02a.png'}, {'trait_type': 'base', 'value': '04c.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '02c.png'}, {'trait_type': 'hat', 'value': '06a.png'}, {'trait_type': 'necklace', 'value': '05a.png'}, {'trait_type': 'rings', 'value': '04.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 86}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE87.png', 'tokenId': 87, 'name': 'ADD_PROJECT_NAME_HERE 87', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '03c.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': '02d.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '08b.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 87}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE88.png', 'tokenId': 88, 'name': 'ADD_PROJECT_NAME_HERE 88', 'attributes': [{'trait_type': 'background', 'value': '04b.png'}, {'trait_type': 'base', 'value': '03d.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '04d.png'}, {'trait_type': 'necklace', 'value': '04a.png'}, {'trait_type': 'rings', 'value': '01.png'}, {'trait_type': 'sunglasses', 'value': '06d.png'}, {'trait_type': 'tokenId', 'value': 88}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE89.png', 'tokenId': 89, 'name': 'ADD_PROJECT_NAME_HERE 89', 'attributes': [{'trait_type': 'background', 'value': '05c.png'}, {'trait_type': 'base', 'value': '04b.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '02c.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '07c.png'}, {'trait_type': 'rings', 'value': '01.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 89}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE90.png', 'tokenId': 90, 'name': 'ADD_PROJECT_NAME_HERE 90', 'attributes': [{'trait_type': 'background', 'value': '01b.png'}, {'trait_type': 'base', 'value': '02d.png'}, {'trait_type': 'makeup', 'value': '02.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '01e.png'}, {'trait_type': 'hat', 'value': '04d.png'}, {'trait_type': 'necklace', 'value': '04a.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': '06a.png'}, {'trait_type': 'tokenId', 'value': 90}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE91.png', 'tokenId': 91, 'name': 'ADD_PROJECT_NAME_HERE 91', 'attributes': [{'trait_type': 'background', 'value': '03c.png'}, {'trait_type': 'base', 'value': '02b.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '01d.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '08b.png'}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': '06d.png'}, {'trait_type': 'tokenId', 'value': 91}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE92.png', 'tokenId': 92, 'name': 'ADD_PROJECT_NAME_HERE 92', 'attributes': [{'trait_type': 'background', 'value': '05d.png'}, {'trait_type': 'base', 'value': '01d.png'}, {'trait_type': 'makeup', 'value': '05.png'}, {'trait_type': 'eye', 'value': '05.png'}, {'trait_type': 'hair', 'value': '01e.png'}, {'trait_type': 'hat', 'value': '01d.png'}, {'trait_type': 'necklace', 'value': '04b.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': '06d.png'}, {'trait_type': 'tokenId', 'value': 92}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE93.png', 'tokenId': 93, 'name': 'ADD_PROJECT_NAME_HERE 93', 'attributes': [{'trait_type': 'background', 'value': '01c.png'}, {'trait_type': 'base', 'value': '01a.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': '02a.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '11a.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 93}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE94.png', 'tokenId': 94, 'name': 'ADD_PROJECT_NAME_HERE 94', 'attributes': [{'trait_type': 'background', 'value': '01.png'}, {'trait_type': 'base', 'value': '04b.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '08a.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 94}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE95.png', 'tokenId': 95, 'name': 'ADD_PROJECT_NAME_HERE 95', 'attributes': [{'trait_type': 'background', 'value': '03a.png'}, {'trait_type': 'base', 'value': '01e.png'}, {'trait_type': 'makeup', 'value': '01.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': '04e.png'}, {'trait_type': 'necklace', 'value': '11c.png'}, {'trait_type': 'rings', 'value': '03.png'}, {'trait_type': 'sunglasses', 'value': '06c.png'}, {'trait_type': 'tokenId', 'value': 95}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE96.png', 'tokenId': 96, 'name': 'ADD_PROJECT_NAME_HERE 96', 'attributes': [{'trait_type': 'background', 'value': '01c.png'}, {'trait_type': 'base', 'value': '02b.png'}, {'trait_type': 'makeup', 'value': '04.png'}, {'trait_type': 'eye', 'value': '02.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '10c.png'}, {'trait_type': 'rings', 'value': '05.png'}, {'trait_type': 'sunglasses', 'value': '02c.png'}, {'trait_type': 'tokenId', 'value': 96}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE97.png', 'tokenId': 97, 'name': 'ADD_PROJECT_NAME_HERE 97', 'attributes': [{'trait_type': 'background', 'value': '04b.png'}, {'trait_type': 'base', 'value': '04e.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '02c.png'}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': '11c.png'}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 97}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE98.png', 'tokenId': 98, 'name': 'ADD_PROJECT_NAME_HERE 98', 'attributes': [{'trait_type': 'background', 'value': '04a.png'}, {'trait_type': 'base', 'value': '03e.png'}, {'trait_type': 'makeup', 'value': '03.png'}, {'trait_type': 'eye', 'value': '04.png'}, {'trait_type': 'hair', 'value': ''}, {'trait_type': 'hat', 'value': ''}, {'trait_type': 'necklace', 'value': ''}, {'trait_type': 'rings', 'value': ''}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 98}]}\n{'image': 'ADD_IMAGES_BASE_URI_HERE99.png', 'tokenId': 99, 'name': 'ADD_PROJECT_NAME_HERE 99', 'attributes': [{'trait_type': 'background', 'value': '02b.png'}, {'trait_type': 'base', 'value': '04c.png'}, {'trait_type': 'makeup', 'value': ''}, {'trait_type': 'eye', 'value': '01.png'}, {'trait_type': 'hair', 'value': '01e.png'}, {'trait_type': 'hat', 'value': '06b.png'}, {'trait_type': 'necklace', 'value': '05d.png'}, {'trait_type': 'rings', 'value': '02.png'}, {'trait_type': 'sunglasses', 'value': ''}, {'trait_type': 'tokenId', 'value': 99}]}\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf456c1c8b5799dba6a3f059cd550eed048e569 | 11,711 | ipynb | Jupyter Notebook | notebooks/AUP110_W14_Exercise.ipynb | htchu/AU110Programming | 6c22e3d202afb0ba90ef02378900270ab7ab657b | [
"MIT"
] | 1 | 2021-09-13T11:49:58.000Z | 2021-09-13T11:49:58.000Z | notebooks/AUP110_W14_Exercise.ipynb | htchu/AU110Programming | 6c22e3d202afb0ba90ef02378900270ab7ab657b | [
"MIT"
] | null | null | null | notebooks/AUP110_W14_Exercise.ipynb | htchu/AU110Programming | 6c22e3d202afb0ba90ef02378900270ab7ab657b | [
"MIT"
] | null | null | null | 20.654321 | 70 | 0.418581 | [
[
[
"# AU Fundamentals of Python Programming-Ex14\r\n* 主題1-M1 Q01-Q03\r\n* 主題2-M2 Q01-Q03\r\n* 主題3-M3 Q01-Q05\r\n",
"_____no_output_____"
],
[
"##(Input)輸入參數",
"_____no_output_____"
]
],
[
[
"instr = input() #輸入是一個字串,例如: 7\r\na = int(instr) #用int()轉成整數變數a\r\nprint(a)",
"_____no_output_____"
],
[
"a = int(input())\r\nprint(a)",
"_____no_output_____"
],
[
"b = float(input())\r\nprint(b)",
"_____no_output_____"
],
[
"instr = input() #輸入是一個字串,例如: 7 9 8\r\nparas = instr.split() #將輸入字串用split()切開\r\na = int(paras[0]) #用int()轉成整數變數a\r\nb = int(paras[1]) #用int()轉成整數變數b\r\nc = int(paras[2]) #用int()轉成整數變數c\r\nprint(a, b, c)",
"_____no_output_____"
],
[
"a, b, c = map(int, input().split()) #用map()將輸入轉成整數變數a, b, c\r\nprint(a, b, c)",
"_____no_output_____"
]
],
[
[
"## (Output)輸出結果",
"_____no_output_____"
],
[
"%-formatting 格式化列印",
"_____no_output_____"
]
],
[
[
"a = 1.234\r\nb = 2.345\r\nprint(\"{0}四捨五入的結果= {0:.2f}\".format(a))\r\nprint(\"{0}四捨五入的結果= {0:.2f}\".format(b))",
"_____no_output_____"
]
],
[
[
"## M1 Problems:Q1-Q3",
"_____no_output_____"
],
[
"### M1-Q01 單位換算:英哩轉公里\r\n\r\n問題描述:\r\n試撰寫一程式,可由鍵盤輸入英哩(float),程式的輸出為公里(float),其轉換公式如下\r\n: 1 英哩 = 1.6 公里\r\n\r\n輸入說明:\r\n輸入欲轉換之英哩數(float)。\r\n\r\n輸出說明:\r\n輸出公里(float),取到小數點以下第二位,最後必須有換行字元。\r\n\r\n範例\r\n\r\n| Sample Input: | Sample Output: |\r\n|:----------------|:-------------------------|\r\n| 90.8 | 145.28⏎ |\r\n| 95.4 | 152.64⏎|",
"_____no_output_____"
],
[
"### M1-Q02 攝氏轉華氏溫度\r\n問題描述:\r\n讓使用者輸入一攝氏溫度,輸出相對應的華氏溫度。\r\n$F = C × 9/5 + 32$\r\n\r\n輸入說明:\r\n輸入一攝氏溫度(float)。\r\n\r\n輸出說明:\r\n輸出相對應的華氏溫度(float),計算到小數第2位四捨五入,最後必須有換行字元。\r\n\r\n範例:\r\n\r\n| Sample Input: | Sample Output: |\r\n|:----------------|:-------------------------|\r\n| 30.8 | 87.44⏎|\r\n| 16.9 | 62.42⏎|",
"_____no_output_____"
],
[
"### M1-Q03 合、差及乘積\r\n問題描述:輸入兩個整數,計算其合、差及乘積。\r\n\r\n輸入說明:輸入兩個整數(int)。\r\n\r\n輸出說明:輸出兩個整數的合(int)、差(int)及乘積(int),最後必須有換行字元。\r\n\r\n| Sample Input: | Sample Output: |\r\n|:----------------|:-------------------------|\r\n| 7 | 7+3=10 |\r\n| 3 | 7-3=4 |\r\n| | 7*3=21 |\r\n| | 7/3=2(1)⏎|",
"_____no_output_____"
],
[
"## M2 Problems:Q1-Q3",
"_____no_output_____"
],
[
"### M2-Q01 最大值與最小值 \r\n問題描述:\r\n寫一個程式來找出輸入的 5 個數字的最大值和最小值,數值不限定為整數,且值可存放於 float 型態數值內。\r\n\r\n輸入說明:\r\n輸入5個數字\r\n\r\n輸出說明:\r\n輸出數列中的最大值與最小值,輸出時需附上小數點後兩位數字,最後必須有換行字元。\r\n\r\n\r\n| Sample Input: | Sample Output: |\r\n|:----------------|:-------------------------|\r\n|-2 -15.2 0 9.5 100 | max=100.00 |\r\n| |min=-15.20⏎ |\r\n| 0 3 52.7 998 135 | max=998.00 |\r\n| |min=0.00⏎ |",
"_____no_output_____"
],
[
"### M2-Q02 '*'三角形\r\n(時間限制:2 秒)\r\n\r\n問題描述:\r\n讓使用者輸入一正整數 n,利用迴圈以字元 '*' 輸出高度為 n 的三角形。\r\n\r\n輸入說明:\r\n輸入一正整數 n。\r\n\r\n輸出說明:\r\n利用迴圈以字元 '*' 輸出高度為 n 的三角形,最後必須有換行字元。\r\n\r\n| Sample Input: | Sample Output: |\r\n|:----------------|:-------------------------|\r\n|4 | \\* |\r\n| |\\*\\* |\r\n| | \\*\\*\\* |\r\n| |\\*\\*\\*\\*⏎ |\r\n",
"_____no_output_____"
],
[
"### M2-Q03 '*'反向三角形\r\n問題描述:\r\n讓使用者輸入一正整數 n,利用迴圈以字元 '*' 輸出高度為 n 的三角形。\r\n\r\n輸入說明:\r\n輸入一正整數 n。\r\n\r\n輸出說明:\r\n利用迴圈以字元 '*' 輸出高度為 n 的三角形,最後必須有換行字元。\r\n\r\n| Sample Input: | Sample Output: |\r\n|:----------------|:-------------------------|\r\n|4 | \\* |\r\n| | \\*\\* |\r\n| | \\*\\*\\* |\r\n| |\\*\\*\\*\\*⏎ |",
"_____no_output_____"
],
[
"## M3 Problems:Q1-Q5",
"_____no_output_____"
],
[
"### M3Q1. 反向字串\r\n\r\n問題描述:\r\n輸入一個字串,並把它反向輸出。\r\n\r\n輸入說明:\r\n輸入一個字串。\r\n\r\n輸出說明:\r\n將輸入字串反向輸出,最後必須有換行字元。\r\n\r\n| Sample Input: | Sample Output: |\r\n|:----------------|:-------------------------|\r\n|AsIACSIE | EISCAIsA |\r\n| |⏎|",
"_____no_output_____"
],
[
"### M3Q2. 亂數選號程式\r\n\r\n問題描述:\r\n請設計一樂透亂數選號程式,由1~42 中選出6 個不重覆的數字組合並輸出。\r\n請使用以下方式:\r\n```\r\nimport random\r\nrandom.seed(10) #seed() 就是設定亂數種子,可以使每次跑出的亂數序列,都會是一樣的。\r\nprint(random.randint(0, 42)) #randint(begin, end) 隨機選取一個數字\r\n```\r\n輸入說明:輸入seed。\r\n輸出說明:由1~42 中選出6 個不重覆的數字組合並輸出,數字間請以tab 作為間格,最後必須有\r\n換行字元。\r\n\r\n| Sample Input: | Sample Output: |\r\n|:----------------|:-------------------------|\r\n|23323456 | 34 40 41 18 24 33 |\r\n| |⏎|",
"_____no_output_____"
]
],
[
[
"nums=list(range(1, 43))\r\nprint(nums)",
"_____no_output_____"
]
],
[
[
"### M3Q3. 陣列行列互換\r\n\r\n問題描述:宣告一個5*7 的二維整數陣列,使用者輸入陣列元素(0~100),並將行列互換輸出。\r\n\r\n輸入說明:輸入陣列元素(0~100)。\r\n\r\n輸出說明:行列互換輸出,數字以tab 間格,最後必須有換行字元。\r\n\r\n| Sample Input: | Sample Output: |\r\n|:----------------|:-------------------------|\r\n|81 7 7 10 97 0 97 |81 90 9 28 5|\r\n|90 67 8 25 1 39 34|7 67 54 8 1 |\r\n|9 54 63 53 53 55 77|7 8 63 17 95|\r\n|28 8 17 50 41 99 89|10 25 53 50 99 |\r\n| 5 1 95 99 76 92 60|97 1 53 41 76|\r\n| | 0 39 55 99 92 |\r\n| |97 34 77 89 60 |\r\n| |⏎ |",
"_____no_output_____"
],
[
"### M3Q4. 數字矩陣\r\n\r\n問題描述:設計一方法 $𝐹(ℎ,w)$ 印出寬𝑤、高ℎ 如下的數字矩陣。(以 𝑤 = 5,ℎ = 3為例)\r\n```\r\n1 2 3 4 5\r\n2 4 6 8 10\r\n3 6 9 12 15\r\n```\r\n其中第二行是第一行的兩倍、第三行是第一行的三倍、以此類推。\r\n\r\n輸入說明:\r\n分別輸入整數寬𝑤、高ℎ。\r\n\r\n輸出說明:\r\n輸出寬𝑤、高ℎ如下的數字矩陣,數字間請以tab 間格,最後必須有換行字元。\r\n\r\n| Sample Input: | Sample Output: |\r\n|:----------------|:-------------------------|\r\n|5 3 |1 2 3 4 5|\r\n| | 2 4 6 8 10 |\r\n| |3 6 9 12 15 |\r\n| |⏎ |",
"_____no_output_____"
],
[
"### M3Q5. $𝑪(m,n)$\r\n\r\n問題描述:\r\n輸入兩個整數,輸出$𝑪(m,n)$。\r\n$𝑪(m,n)=m!/n!(m-n)!$\r\n\r\n輸入說明:\r\n分別輸入整數 m、n。\r\n\r\n輸出說明:\r\n輸出$𝑪(m,n)$最後必須有換行字元。\r\n\r\n範例:\r\n\r\n| Sample Input: | Sample Output: |\r\n|:----------------|:-------------------------|\r\n|8 6 |28|\r\n| |⏎ |",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
ecf45d34d258538688650da52c4a66902b67ec77 | 1,622 | ipynb | Jupyter Notebook | nbs/06_other/rss_feeds.ipynb | andrewtruong/notes | 24e3a7d822f746ac4ab6c3186bc44dccb4f0e169 | [
"Apache-2.0"
] | null | null | null | nbs/06_other/rss_feeds.ipynb | andrewtruong/notes | 24e3a7d822f746ac4ab6c3186bc44dccb4f0e169 | [
"Apache-2.0"
] | null | null | null | nbs/06_other/rss_feeds.ipynb | andrewtruong/notes | 24e3a7d822f746ac4ab6c3186bc44dccb4f0e169 | [
"Apache-2.0"
] | null | null | null | 20.794872 | 156 | 0.541307 | [
[
[
"# RSS Feeds\n> Recipes for RSS Feeds",
"_____no_output_____"
]
],
[
[
"import feedparser\nimport pandas as pd",
"_____no_output_____"
],
[
"json = feedparser.parse('https://forums.redflagdeals.com/feed/forum/9')\ndf = pd.json_normalize(json['entries'])",
"_____no_output_____"
],
[
"search_terms = ['EVGA', 'G15', 'wifi', 'laptop']\nq = '|'.join(search_terms)",
"_____no_output_____"
],
[
"interesting_links = df.loc[(df.title.str.contains(q, regex=True, case=False)) | (df.summary.str.contains(q, regex=True, case=False)), 'link'].tolist()",
"_____no_output_____"
],
[
"df.loc[df.title.str.contains(q, regex=True), 'id'].tolist()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecf460489eca1c6b6eb6e53267514232bde693dc | 36,464 | ipynb | Jupyter Notebook | Gradient Boosting for molecular properties/pmp-data-test.ipynb | dhananjayraut/ML_projects | e7b5008c4039bfa057cc6f7d991224fd2d268eb6 | [
"MIT"
] | null | null | null | Gradient Boosting for molecular properties/pmp-data-test.ipynb | dhananjayraut/ML_projects | e7b5008c4039bfa057cc6f7d991224fd2d268eb6 | [
"MIT"
] | null | null | null | Gradient Boosting for molecular properties/pmp-data-test.ipynb | dhananjayraut/ML_projects | e7b5008c4039bfa057cc6f7d991224fd2d268eb6 | [
"MIT"
] | null | null | null | 31.059625 | 121 | 0.353061 | [
[
[
"# This Python 3 environment comes with many helpful analytics libraries installed\n# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python\n# For example, here's several helpful packages to load in \n\nimport numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\nimport matplotlib.pyplot as plt\nfrom os import listdir\nfrom os.path import isfile, join\nfrom multiprocessing import Pool\n# Input data files are available in the \"../input/\" directory.\n# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory\n\nimport os\nroot = '../input/champs-scalar-coupling/'\nQm = '../input/quantum-machine-9-aka-qm9/dsgdb9nsd.xyz/'\n# Any results you write to the current directory are saved as output.",
"_____no_output_____"
],
[
"train = pd.read_csv(root+'train.csv')\nstructures = pd.read_csv(root+'structures.csv')\ntype_list = train['type'].unique().tolist()",
"_____no_output_____"
],
[
"def get_atoms(name):\n path = root + 'structures/' + name + '.xyz'\n qmp = Qm + name + '.xyz'\n mm = pd.read_csv(qmp,sep='\\t',engine='python', skiprows=2, skipfooter=3, names=range(5))[4]\n if mm.dtype == 'O':\n mm = mm.str.replace('*^','e',regex=False).astype(float)\n lis = []\n file = open(path,'r')\n number = int(file.readline())\n file.readline()\n for i in range(number): \n line = file.readline().split()\n lis.append([line[0],np.array(line[1:]).astype(float),mm[i]])\n file.close()\n return lis\nprint(*get_atoms('dsgdb9nsd_000007'),sep=' \\n')",
"['C', array([-0.018704 , 1.52558201, 0.01043281]), -0.345672] \n['C', array([ 0.00210374, -0.00388191, 0.00199882]), -0.345672] \n['H', array([0.99487275, 1.93974324, 0.0029412 ]), 0.115222] \n['H', array([-0.54207611, 1.92361063, -0.86511735]), 0.115225] \n['H', array([-0.52524112, 1.91417308, 0.90002399]), 0.115225] \n['H', array([ 0.52548654, -0.40190784, 0.87754395]), 0.115225] \n['H', array([-1.01147651, -0.4180338 , 0.00950849]), 0.115222] \n['H', array([ 0.50862619, -0.3924704 , -0.88760117]), 0.115225]\n"
],
[
"def get_distance(coor1, coor2):\n return np.linalg.norm(coor2 - coor1)\ndef unit_vector(vector):\n return vector / np.linalg.norm(vector)\ndef angle_between(v1, v2):\n v1_u = unit_vector(v1)\n v2_u = unit_vector(v2)\n return np.degrees(np.arccos(np.clip(np.dot(v1_u, v2_u), -1.0, 1.0)))\ndef angle_bw_points(p1,p2,p3):\n return angle_between(p2-p1,p3-p1)",
"_____no_output_____"
],
[
"a = np.array([0,0,0])\nb = np.array([2,0,0])\nc = np.array([0,0,4])\nprint(angle_bw_points(a,b,c))",
"90.0\n"
],
[
"def get_next_points(name,index0,index1,k=5):\n lis,dis = get_atoms(name), []\n if index1 < index0:\n at0 = lis.pop(index0)\n at1 = lis.pop(index1)\n else:\n at1 = lis.pop(index1)\n at0 = lis.pop(index0)\n for i in lis: dis.append(min(get_distance(at0[1], i[1]),get_distance(at1[1], i[1])))\n a = np.argsort(np.array(dis))[:k]\n #a = np.argwhere(np.array(dis) < get_distance(at0[1], at1[1])).T[0]\n return [lis[i] for i in a.tolist()]",
"_____no_output_____"
],
[
"def get_point(name,index):\n lis = get_atoms(name)\n return lis[index]",
"_____no_output_____"
],
[
"get_next_points('dsgdb9nsd_000007',0,2)",
"_____no_output_____"
],
[
"def get_from_type(lis):\n ret = []\n for i in lis:\n if i == 'H': ret.append(1.0)\n elif i == 'C': ret.append(2.0)\n elif i == 'O': ret.append(3.0)\n elif i == 'N': ret.append(4.0)\n elif i == 'F': ret.append(5.0)\n else: ret.append(0.0)\n return ret",
"_____no_output_____"
],
[
"def get_feature_list(df,j):\n name = df['molecule_name'][j]\n index0 = df['atom_index_0'][j]\n index1 = df['atom_index_1'][j]\n lis = get_next_points(name,index0,index1,8)\n p0 = get_point(name,index0)\n p1 = get_point(name,index1)\n fea = get_from_type(p0[0])\n fea.append(p0[2])\n fea.extend(get_from_type(p1[0]))\n fea.append(p1[2])\n fea.append(get_distance(p0[1], p1[1]))\n for i in lis: \n fea.extend(get_from_type(i[0]))\n fea.append(i[2])\n fea.extend([get_distance(p0[1], i[1]),get_distance(p1[1], i[1]),\n angle_bw_points(p0[1],p1[1],i[1]), angle_bw_points(p1[1],p0[1],i[1])])\n for i in range(8 - len(lis)):\n fea.extend(get_from_type('I'))\n fea.append(0)\n fea.extend([50.0, 50.0, 45.0, 45.0])\n return [np.float32(k) for k in fea]",
"_____no_output_____"
],
[
"df_ini = train",
"_____no_output_____"
],
[
"featr = ['type0','charge0','type1','charge1','distance',\n'type_0','charge_0','dist0_0','dist1_0','angle0_0','angle1_0',\n'type_1','charge_1','dist0_1','dist1_1','angle0_1','angle1_1',\n'type_2','charge_2','dist0_2','dist1_2','angle0_2','angle1_2',\n'type_3','charge_3','dist0_3','dist1_3','angle0_3','angle1_3',\n'type_4','charge_4','dist0_4','dist1_4','angle0_4','angle1_4',\n'type_5','charge_5','dist0_5','dist1_5','angle0_5','angle1_5',\n'type_6','charge_6','dist0_6','dist1_6','angle0_6','angle1_6',\n'type_7','charge_7','dist0_7','dist1_7','angle0_7','angle1_7']",
"_____no_output_____"
],
[
"from tqdm import trange\ndef get_dataframe(df_ini):\n arr = np.zeros((df_ini.shape[0], 53))\n for j in range(df_ini.shape[0]):\n fea = get_feature_list(df_ini,j)\n arr[j,:] = np.array(fea)\n #print(fea)\n break\n return pd.DataFrame(arr)\nv = get_dataframe(df_ini)\nv.columns = featr\nv.head()",
"_____no_output_____"
],
[
"def tt(j):\n return get_feature_list(df_ini,j)\ndef get_dataframe(df_ini):\n with Pool(8) as p:\n a = p.map(tt,[i for i in range(df_ini.shape[0])])\n return pd.DataFrame(np.array(a))",
"_____no_output_____"
],
[
"# %%time\n# df_ini = train\n# df_t = get_dataframe(train)",
"_____no_output_____"
],
[
"# df_t.columns = featr\n# dd = pd.concat([train,df_t],axis=1)\n# dd.head()",
"_____no_output_____"
],
[
"# print(dd.shape)\n# print(train.shape)\n# dd.to_csv('train_.csv')",
"_____no_output_____"
],
[
"%%time\ntest = pd.read_csv(root+'test.csv')\ndf_ini = test\ndf_t = get_dataframe(test)",
"CPU times: user 2min 15s, sys: 21.3 s, total: 2min 37s\nWall time: 3h 8min 16s\n"
],
[
"df_t.columns = featr\ndd = pd.concat([test,df_t],axis=1)\ndd.head()",
"_____no_output_____"
],
[
"print(dd.shape)\nprint(test.shape)\ndd.to_csv('test_.csv')",
"(2505542, 58)\n(2505542, 5)\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf462bcbf34713f4150e25f0572893cdc475594 | 253,216 | ipynb | Jupyter Notebook | LM_analysis/LM_analysis_bootstrap_2.ipynb | Living-with-machines/lwm_ARTIDIGH_2020_OCR_impact_downstream_NLP_tasks | 1b2027f624f457bda734e547378ec725bb5a0881 | [
"CC-BY-4.0"
] | 5 | 2020-04-26T20:34:02.000Z | 2021-06-17T12:34:24.000Z | LM_analysis/LM_analysis_bootstrap_2.ipynb | alan-turing-institute/lwm_ARTIDIGH_2020_OCR_impact_downstream_NLP_tasks | 1b2027f624f457bda734e547378ec725bb5a0881 | [
"CC-BY-4.0"
] | null | null | null | LM_analysis/LM_analysis_bootstrap_2.ipynb | alan-turing-institute/lwm_ARTIDIGH_2020_OCR_impact_downstream_NLP_tasks | 1b2027f624f457bda734e547378ec725bb5a0881 | [
"CC-BY-4.0"
] | 1 | 2020-06-12T12:26:20.000Z | 2020-06-12T12:26:20.000Z | 405.1456 | 137,884 | 0.924562 | [
[
[
"# Language Model analysis",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport time\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nfrom gensim.models import Word2Vec\nfrom gensim import logging\nlogging.basicConfig(format='%(asctime)s : %(levelname)s : %(message)s', level=logging.INFO)",
"_____no_output_____"
],
[
"# embedding models, base model\n#model_path = \"/Users/khosseini/myJobs/ATI/Projects/2019/Living-with-Machines-code/language-lab-mro/lexicon_expansion/interactive_expansion/models/all_books/w2v_005/w2v_words.model\"\nmodel_path = \"./LMs/embedding_model_scratch_corrected.model\"\nw2v_corrected = Word2Vec.load(model_path)",
"2019-11-21 11:36:42,747 : INFO : loading Word2Vec object from ./LMs/embedding_model_scratch_corrected.model\n2019-11-21 11:36:43,185 : INFO : loading wv recursively from ./LMs/embedding_model_scratch_corrected.model.wv.* with mmap=None\n2019-11-21 11:36:43,186 : INFO : loading vectors from ./LMs/embedding_model_scratch_corrected.model.wv.vectors.npy with mmap=None\n2019-11-21 11:36:43,297 : INFO : setting ignored attribute vectors_norm to None\n2019-11-21 11:36:43,299 : INFO : loading vocabulary recursively from ./LMs/embedding_model_scratch_corrected.model.vocabulary.* with mmap=None\n2019-11-21 11:36:43,299 : INFO : loading trainables recursively from ./LMs/embedding_model_scratch_corrected.model.trainables.* with mmap=None\n2019-11-21 11:36:43,300 : INFO : loading syn1neg from ./LMs/embedding_model_scratch_corrected.model.trainables.syn1neg.npy with mmap=None\n2019-11-21 11:36:43,406 : INFO : setting ignored attribute cum_table to None\n2019-11-21 11:36:43,406 : INFO : loaded ./LMs/embedding_model_scratch_corrected.model\n"
],
[
"def found_neighbors(myrow, embedding, colname='vocab', topn=1):\n try:\n vocab_neigh = embedding.wv.most_similar([myrow['vocab']], topn=topn)\n return list(np.array(vocab_neigh)[:, 0])\n except KeyError:\n return []",
"_____no_output_____"
],
[
"def jaccard_similarity_df(myrow, colname_1, colname_2, num_items=False, make_lowercase=True):\n \"\"\"\n Jaccard similarity between two documents (e.g., OCR and Human) on flattened list of words\n \"\"\"\n if not num_items:\n list1 = myrow[colname_1]\n list2 = myrow[colname_2]\n else:\n list1 = myrow[colname_1][:num_items]\n list2 = myrow[colname_2][:num_items]\n if make_lowercase:\n list1 = [x.lower() for x in list1]\n list2 = [x.lower() for x in list2]\n intersection = len(list(set(list1).intersection(list2)))\n union = (len(list1) + len(list2)) - intersection\n return float(intersection) / union",
"_____no_output_____"
],
[
"words_corrected = []\nfor item in w2v_corrected.wv.vocab:\n words_corrected.append([item, int(w2v_corrected.wv.vocab[item].count)])",
"_____no_output_____"
],
[
"pd_words = pd.DataFrame(words_corrected, columns=['vocab', 'count'])",
"_____no_output_____"
],
[
"pd_words = pd_words.sort_values(by=['count'], ascending=False)\nprint(\"size: {}\".format(len(pd_words)))\npd_words.head()",
"size: 179735\n"
],
[
"pd2search = pd_words[0:1000]\npd2search",
"_____no_output_____"
]
],
[
[
"# Quality bands 1, 2",
"_____no_output_____"
]
],
[
[
"start_run = input(\"This will take a long time! Continue? (y/n)\")\n\nif start_run == 'y':\n neigh_jaccard_bands_1_2 = []\n\n for i_model in range(0, 50):\n\n w2v_em_corr_qual_1_2 = Word2Vec.load('./LMs/w2v_005_EM_corr_qual_1_2_%05i.model' % i_model)\n w2v_em_ocr_qual_1_2 = Word2Vec.load('./LMs/w2v_005_EM_ocr_qual_1_2_%05i.model' % i_model)\n\n #for topn in [1, 2, 5, 10, 50, 100, 500, 1000, 5000, 10000, 50000]:\n for topn in [50000]:\n print(\"topn: {}\".format(topn))\n t1 = time.time()\n\n pd2search = pd_words[0:1000]\n pd2search['w2v_em_corr_qual_1_2'] = pd2search.apply(found_neighbors, args=[w2v_em_corr_qual_1_2, \n 'vocab', \n topn], axis=1)\n print(\"corr: {}\".format(time.time() - t1))\n pd2search['w2v_em_ocr_qual_1_2'] = pd2search.apply(found_neighbors, args=[w2v_em_ocr_qual_1_2,\n 'vocab', \n topn], axis=1)\n print(\"ocr: {}\".format(time.time() - t1))\n\n mytopn_range = [1, 2, 5, \n 10, 20, 50, \n 100, 200, 500, \n 1000, 2000, 5000, \n 10000, 20000, 50000]\n for mytopn in mytopn_range:\n pd2search['jaccard_qual_1_2'] = \\\n pd2search.apply(jaccard_similarity_df, \n args=['w2v_em_corr_qual_1_2', \"w2v_em_ocr_qual_1_2\", mytopn], \n axis=1)\n\n neigh_jaccard_bands_1_2.append(\n [mytopn, \n pd2search['jaccard_qual_1_2'].mean(), \n pd2search['jaccard_qual_1_2'].std(),\n i_model\n ])\n print(\"total: {}\".format(time.time() - t1))\n np.save(\"neigh_jaccard_bands_1_2.npy\", np.array(neigh_jaccard_bands_1_2))\n\n neigh_jaccard_bands_1_2 = np.array(neigh_jaccard_bands_1_2)\n np.save(\"neigh_jaccard_bands_1_2.npy\", neigh_jaccard_bands_1_2)",
"This will take a long time! Continue? (y/n)n\n"
],
[
"# neigh_jaccard_bands_1_2 = []\n\n# for i_model in range(0, 50):\n\n# w2v_em_corr_qual_1_2 = Word2Vec.load('./LMs/w2v_005_EM_corr_qual_1_2_%05i.model' % i_model)\n# w2v_em_ocr_qual_1_2 = Word2Vec.load('./LMs/w2v_005_EM_ocr_qual_1_2_%05i.model' % i_model)\n\n# for topn in [1, 2, 5, 10, 50, 100, 500, 1000, 5000, 10000, 50000]:\n# print(\"topn: {}\".format(topn))\n# t1 = time.time()\n\n# pd2search = pd_words[0:1000]\n# pd2search['w2v_em_corr_qual_1_2'] = pd2search.apply(found_neighbors, args=[w2v_em_corr_qual_1_2, \n# 'vocab', \n# topn], axis=1)\n# print(\"corr: {}\".format(time.time() - t1))\n# pd2search['w2v_em_ocr_qual_1_2'] = pd2search.apply(found_neighbors, args=[w2v_em_ocr_qual_1_2, \n# 'vocab', \n# topn], axis=1)\n# pd2search['jaccard_qual_1_2'] = \\\n# pd2search.apply(jaccard_similarity_df, args=['w2v_em_corr_qual_1_2', \n# \"w2v_em_ocr_qual_1_2\", \n# True], \n# axis=1)\n\n# neigh_jaccard_bands_1_2.append(\n# [topn, \n# pd2search['jaccard_qual_1_2'].mean(), \n# pd2search['jaccard_qual_1_2'].std(),\n# i_model\n# ])\n\n# print(\"total: {}\".format(time.time() - t1))\n\n# neigh_jaccard_bands_1_2 = np.array(neigh_jaccard_bands_1_2)",
"_____no_output_____"
],
[
"neigh_jaccard_bands_1_2_all = np.load(\"./bootstrap_results/neigh_jaccard_bands_1_2.npy\")\nneigh_jaccard_bands_3_4 = np.load(\"./bootstrap_results/neigh_jaccard_bands_3_4.npy\")",
"_____no_output_____"
],
[
"neigh_jaccard_bands_1_2 = np.split(neigh_jaccard_bands_1_2_all, 50)",
"_____no_output_____"
],
[
"plt.figure(figsize=(20, 10))\n\nplt.plot([-10, -9], [10, 10],\n c='k', alpha=0.5,\n marker='o', markersize=10,\n lw=5,\n label='Quality bands=1,2')\n\nplt.plot(neigh_jaccard_bands_1_2[0][:, 0], neigh_jaccard_bands_1_2[0][:, 1],\n c='k', alpha=0.1,\n marker='o', markersize=10,\n lw=5)\n\nfor i in range(1, 50):\n plt.plot(neigh_jaccard_bands_1_2[i][:, 0], neigh_jaccard_bands_1_2[i][:, 1],\n #np.std(neigh_jaccard_bands_1_2, axis=0)[:, 1],\n c='k', alpha=0.1, #fmt='-o',\n marker='o', markersize=10,\n lw=5)\n #label='Quality bands=1,2')\n\nplt.plot(neigh_jaccard_bands_3_4[:, 0], neigh_jaccard_bands_3_4[:, 1], \n 'r-o', alpha=1.0, markersize=10,\n lw=5,\n label='Quality bands=3,4')\n\nplt.grid(linewidth=2)\nplt.xticks(size=32)\nplt.yticks(size=32)\nplt.xlabel(\"#neighbours\", size=42)\nplt.ylabel(\"Jaccard similarity\", size=42)\nplt.xscale(\"log\")\nplt.xlim(0.9, 60000)\nplt.ylim(0.05, 0.6)\n\nplt.legend(prop={'size': 36})\nplt.show()\n#plt.xlim(0, 20000)",
"_____no_output_____"
],
[
"plt.figure(figsize=(20, 10))\n\nplt.errorbar(neigh_jaccard_bands_1_2[i][:, 0], neigh_jaccard_bands_1_2[i][:, 1],\n np.std(neigh_jaccard_bands_1_2, axis=0)[:, 1],\n c='k', alpha=1.0,\n marker='o', markersize=10,\n lw=5,\n label='Quality bands=1,2')\n\nplt.plot(neigh_jaccard_bands_3_4[:, 0], neigh_jaccard_bands_3_4[:, 1], \n 'r-o', alpha=1.0, markersize=10,\n lw=5,\n label='Quality bands=3,4')\n\nplt.grid()\nplt.xticks(size=30)\nplt.yticks(size=30)\nplt.xlabel(\"#neighbours\", size=40)\nplt.ylabel(\"Jaccard similarity\", size=40)\nplt.xscale(\"log\")\nplt.xlim(0.9, 60000)\nplt.ylim(0.05, 0.6)\n\nplt.legend(prop={'size': 30})\nplt.show()\n#plt.xlim(0, 20000)",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 5))\n\nplt.errorbar(neigh_jaccard_bands_1_2[0][:, 0], np.mean(neigh_jaccard_bands_1_2, axis=0)[:, 1],\n np.std(neigh_jaccard_bands_1_2, axis=0)[:, 1],\n c='k', alpha=1.0, fmt='-o',\n #marker='o',\n lw=4,\n label='Quality bands=1,2')\n\nplt.plot(neigh_jaccard_bands_3_4[:, 0], neigh_jaccard_bands_3_4[:, 1], \n 'r-o', alpha=1.0, \n lw=4,\n label='Quality bands=3,4')\n\nplt.grid()\nplt.xticks(size=20)\nplt.yticks(size=20)\nplt.xlabel(\"#neighbours\", size=24)\nplt.ylabel(\"Jaccard similarity\", size=24)\nplt.xscale(\"log\")\nplt.xlim(0.9, 60000)\nplt.ylim(0.05, 0.6)\n\nplt.legend(prop={'size': 20})\nplt.show()\n#plt.xlim(0, 20000)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf468fb16279fc45e38d7196ea2a029ea057dc5 | 6,726 | ipynb | Jupyter Notebook | Chapter6_ObjectOriented/vector2d_exceptions.ipynb | tomex74/UdemyPythonPro | b4b83483fa2d3337a2860d53ff38e68eb38b3ac4 | [
"MIT"
] | null | null | null | Chapter6_ObjectOriented/vector2d_exceptions.ipynb | tomex74/UdemyPythonPro | b4b83483fa2d3337a2860d53ff38e68eb38b3ac4 | [
"MIT"
] | null | null | null | Chapter6_ObjectOriented/vector2d_exceptions.ipynb | tomex74/UdemyPythonPro | b4b83483fa2d3337a2860d53ff38e68eb38b3ac4 | [
"MIT"
] | null | null | null | 30.995392 | 885 | 0.518882 | [
[
[
"import numbers\nfrom math import sqrt\nfrom functools import total_ordering\n\n@total_ordering\nclass Vector2D:\n def __init__(self, x=0, y=0):\n if isinstance(x, numbers.Real) and isinstance(y, numbers.Real): \n self.x = x\n self.y = y\n else:\n raise TypeError('You must pass in int/float values for x and y!')\n\n def __call__(self):\n print(\"Calling the __call__ function!\")\n return self.__repr__()\n\n def __repr__(self):\n return 'vector.Vector2D({}, {})'.format(self.x, self.y)\n\n def __str__(self):\n return '({}, {})'.format(self.x, self.y)\n\n def __bool__(self):\n return bool(abs(self))\n\n def __abs__(self):\n return sqrt(pow(self.x, 2) + pow(self.y, 2))\n\n def check_vector_types(self, vector2):\n if not isinstance(self, Vector2D) or not isinstance(vector2, Vector2D):\n raise TypeError('You have to pass in two instances of the vector class!')\n\n def __eq__(self, other_vector):\n self.check_vector_types(other_vector)\n if self.x == other_vector.x and self.y == other_vector.y:\n return True\n else:\n return False\n\n def __lt__(self, other_vector):\n self.check_vector_types(other_vector)\n if abs(self) < abs(other_vector):\n return True\n else:\n return False\n \n def __add__(self, other_vector):\n self.check_vector_types(other_vector)\n x = self.x + other_vector.x\n y = self.y + other_vector.y\n return Vector2D(x, y)\n\n # try (== 1):\n # except (>= 1): \n # finally (optional): \n def __sub__(self, other_vector):\n try:\n x = self.x - other_vector.x\n y = self.y - other_vector.y\n return Vector2D(x, y)\n except AttributeError as e:\n print(\"AttributeError: {} was raised!\".format(e))\n #return self\n except Exception as e:\n print(\"Exception {}: {} was raised!\".format(type(e), e))\n\n def __mul__(self, other):\n if isinstance(other, Vector2D):\n return self.x * other.x + self.y * other.y\n elif isinstance(other, numbers.Real):\n return Vector2D(self.x * other, self.y * other)\n else:\n raise TypeError('You must pass in a vector instance or an int/float number!')\n\n def __truediv__(self, other):\n if isinstance(other, numbers.Real):\n if other != 0.0:\n return Vector2D(self.x / other, self.y / other)\n else:\n raise ValueError('You cannot divide by zero!')\n else:\n raise TypeError('You must pass in an int/float value!')",
"_____no_output_____"
],
[
"v1 = Vector2D(1, 2)\nv2 = Vector2D(1, 3)",
"_____no_output_____"
],
[
"v1 - 2\n\n#print(v1 / 0)",
"AttributeError: 'int' object has no attribute 'x' was raised!\n"
],
[
"import builtins\n\nbuiltin_list = [builtin for builtin in dir(builtins) if 'Error' in builtin]\n\nprint(builtin_list)",
"['ArithmeticError', 'AssertionError', 'AttributeError', 'BlockingIOError', 'BrokenPipeError', 'BufferError', 'ChildProcessError', 'ConnectionAbortedError', 'ConnectionError', 'ConnectionRefusedError', 'ConnectionResetError', 'EOFError', 'EnvironmentError', 'FileExistsError', 'FileNotFoundError', 'FloatingPointError', 'IOError', 'ImportError', 'IndentationError', 'IndexError', 'InterruptedError', 'IsADirectoryError', 'KeyError', 'LookupError', 'MemoryError', 'ModuleNotFoundError', 'NameError', 'NotADirectoryError', 'NotImplementedError', 'OSError', 'OverflowError', 'PermissionError', 'ProcessLookupError', 'RecursionError', 'ReferenceError', 'RuntimeError', 'SyntaxError', 'SystemError', 'TabError', 'TimeoutError', 'TypeError', 'UnboundLocalError', 'UnicodeDecodeError', 'UnicodeEncodeError', 'UnicodeError', 'UnicodeTranslateError', 'ValueError', 'ZeroDivisionError']\n"
],
[
"v1-2",
"AttributeError: 'int' object has no attribute 'x' was raised!\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
ecf47dbe83bdac57d916d5203df2daab9b01893a | 30,196 | ipynb | Jupyter Notebook | bar_chart.ipynb | sanabasangare/data-visualization | 09a03d0414941d28e312037ccaa0b283dbb2ec06 | [
"MIT"
] | null | null | null | bar_chart.ipynb | sanabasangare/data-visualization | 09a03d0414941d28e312037ccaa0b283dbb2ec06 | [
"MIT"
] | null | null | null | bar_chart.ipynb | sanabasangare/data-visualization | 09a03d0414941d28e312037ccaa0b283dbb2ec06 | [
"MIT"
] | null | null | null | 355.247059 | 27,510 | 0.904458 | [
[
[
"#### Bar chart plotting",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom collections import Counter\n\n\ndef bar_chart(plt):\n\n plt.figure(figsize=(20, 10))\n\n years = [2000, 2002, 2005, 2007, 2010, 2012, 2014, 2015]\n\n num_websites = [17, 38, 64, 121, 206, 697, 968, 863]\n\n xs = [i + 0.1 for i, _ in enumerate(years)]\n\n plt.bar(xs, num_websites, color='green')\n plt.ylabel(\"# of Websites (millions)\")\n plt.title(\"Total number of websites online\")\n\n plt.xticks([i + 0.5 for i, _ in enumerate(years)],\n years, color='blue')\n\n plt.show()\n\n\nif __name__ == \"__main__\":\n\n bar_chart(plt)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
ecf4805bd496ea7f35e0e47b82021a7600f505d1 | 4,477 | ipynb | Jupyter Notebook | Activity_2.ipynb | InoRM21/OOP-1-2 | 54eb63928689c2abad1d9a7ba655c9e3968630d3 | [
"Apache-2.0"
] | null | null | null | Activity_2.ipynb | InoRM21/OOP-1-2 | 54eb63928689c2abad1d9a7ba655c9e3968630d3 | [
"Apache-2.0"
] | null | null | null | Activity_2.ipynb | InoRM21/OOP-1-2 | 54eb63928689c2abad1d9a7ba655c9e3968630d3 | [
"Apache-2.0"
] | null | null | null | 28.698718 | 222 | 0.471521 | [
[
[
"<a href=\"https://colab.research.google.com/github/InoRM21/OOP-1-2/blob/main/Activity_2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"#Write a python program that converts the temperature Celsius to Fahrenheit. Create a class name Temperature.\n#Create Celsius as attribute name, Temp() as method, and temp1 as object name. F = 1.8xC + 32\n\nclass Temperature: \n def __init__(self,Celsius): \n self.Celsius = Celsius \n \n def Temp(self): \n return ((1.8*self.Celsius)+32)\n \ninput_temp = float(input(\"Input temperature in celsius: \"))\ntemp1 = Temperature(input_temp)\nprint(round(temp1.Temp(),2))",
"Input temperature in celsius: 30\n86.0\n"
]
],
[
[
"#Define an Area() method of the class that calculates the circle's area.",
"_____no_output_____"
]
],
[
[
"import math\n\nclass Circle: \n def __init__(self,radius): \n self.radius = radius \n \n def Area(self): \n return math.pi*(self.radius**2)\n\nradius = float(input(\"Enter the radius of the circle: \"))\ncircle1 = Circle(radius)\nprint (\"The Area of the circle of the radius\", radius, \"=\", round (circle1.Area(),4))",
"Enter the radius of the circle: 17\nThe Area of the circle of the radius 17.0 = 907.9203\n"
]
],
[
[
"#Define a Perimeter () method of the class which allows you to calculate the perimeter of the circle.",
"_____no_output_____"
]
],
[
[
"import math\n\nclass Circle: \n def __init__(self,radius): \n self.radius = radius \n \n def Perimeter(self): \n return 2*math.pi*(self.radius)\n\nradius = float(input(\"Enter the radius of the circle: \"))\ncircle1 = Circle(radius)\n\nprint (\"The Perimeter of the circle of the radius\", radius, \"=\", round(circle1.Perimeter(),4))",
"Enter the radius of the circle: 17\nThe Perimeter of the circle of the radius 17.0 = 106.8142\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
ecf4aa10d90fa2efef74378740ea19b19363439f | 491,293 | ipynb | Jupyter Notebook | Regression/Linear Models/GammaRegressor_MinMaxScalar.ipynb | shreepad-nade/ds-seed | 93ddd3b73541f436b6832b94ca09f50872dfaf10 | [
"Apache-2.0"
] | 53 | 2021-08-28T07:41:49.000Z | 2022-03-09T02:20:17.000Z | Regression/Linear Models/GammaRegressor_MinMaxScalar.ipynb | shreepad-nade/ds-seed | 93ddd3b73541f436b6832b94ca09f50872dfaf10 | [
"Apache-2.0"
] | 142 | 2021-07-27T07:23:10.000Z | 2021-08-25T14:57:24.000Z | Regression/Linear Models/GammaRegressor_MinMaxScalar.ipynb | shreepad-nade/ds-seed | 93ddd3b73541f436b6832b94ca09f50872dfaf10 | [
"Apache-2.0"
] | 38 | 2021-07-27T04:54:08.000Z | 2021-08-23T02:27:20.000Z | 647.289855 | 406,133 | 0.93983 | [
[
[
"# GammaRegressor with MinMaxScalar",
"_____no_output_____"
],
[
"This code template is for the regression analysis using Gamma Regression via data scaling technique MinMax Scalar",
"_____no_output_____"
],
[
"### Required Packages",
"_____no_output_____"
]
],
[
[
"import warnings\r\nimport numpy as np \r\nimport pandas as pd \r\nimport seaborn as se \r\nimport matplotlib.pyplot as plt \r\nfrom sklearn.preprocessing import LabelEncoder,StandardScaler,MinMaxScaler\r\nfrom sklearn.pipeline import make_pipeline\r\nfrom sklearn.model_selection import train_test_split \r\nfrom sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error \r\nfrom sklearn.linear_model import GammaRegressor\r\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"### Initialization\n\nFilepath of CSV file",
"_____no_output_____"
]
],
[
[
"#filepath\r\nfile_path= \"\"",
"_____no_output_____"
]
],
[
[
"List of features which are required for model training .",
"_____no_output_____"
]
],
[
[
"#x_values\r\nfeatures=[]",
"_____no_output_____"
]
],
[
[
"Target feature for prediction.",
"_____no_output_____"
]
],
[
[
"#y_value\r\ntarget=''",
"_____no_output_____"
]
],
[
[
"### Data Fetching\n\nPandas is an open-source, BSD-licensed library providing high-performance, easy-to-use data manipulation and data analysis tools.\n\nWe will use panda's library to read the CSV file using its storage path.And we use the head function to display the initial row or entry.",
"_____no_output_____"
]
],
[
[
"df=pd.read_csv(file_path)\r\ndf.head()",
"_____no_output_____"
]
],
[
[
"### Feature Selections\n\nIt is the process of reducing the number of input variables when developing a predictive model. Used to reduce the number of input variables to both reduce the computational cost of modelling and, in some cases, to improve the performance of the model.\n\nWe will assign all the required input features to X and target/outcome to Y.",
"_____no_output_____"
]
],
[
[
"X=df[features]\nY=df[target]",
"_____no_output_____"
]
],
[
[
"### Data Preprocessing\n\nSince the majority of the machine learning models in the Sklearn library doesn't handle string category data and Null value, we have to explicitly remove or replace null values. The below snippet have functions, which removes the null value if any exists. And convert the string classes data in the datasets by encoding them to integer classes.\n",
"_____no_output_____"
]
],
[
[
"def NullClearner(df):\n if(isinstance(df, pd.Series) and (df.dtype in [\"float64\",\"int64\"])):\n df.fillna(df.mean(),inplace=True)\n return df\n elif(isinstance(df, pd.Series)):\n df.fillna(df.mode()[0],inplace=True)\n return df\n else:return df\ndef EncodeX(df):\n return pd.get_dummies(df)",
"_____no_output_____"
]
],
[
[
"Calling preprocessing functions on the feature and target set.\n",
"_____no_output_____"
]
],
[
[
"x=X.columns.to_list()\nfor i in x:\n X[i]=NullClearner(X[i])\nX=EncodeX(X)\nY=NullClearner(Y)\nX.head()",
"_____no_output_____"
]
],
[
[
"#### Correlation Map\n\nIn order to check the correlation between the features, we will plot a correlation matrix. It is effective in summarizing a large amount of data where the goal is to see patterns.",
"_____no_output_____"
]
],
[
[
"f,ax = plt.subplots(figsize=(18, 18))\nmatrix = np.triu(X.corr())\nse.heatmap(X.corr(), annot=True, linewidths=.5, fmt= '.1f',ax=ax, mask=matrix)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Data Splitting\n\nThe train-test split is a procedure for evaluating the performance of an algorithm. The procedure involves taking a dataset and dividing it into two subsets. The first subset is utilized to fit/train the model. The second subset is used for prediction. The main motive is to estimate the performance of the model on new data.",
"_____no_output_____"
]
],
[
[
"x_train,x_test,y_train,y_test=train_test_split(X,Y,test_size=0.2,random_state=123)",
"_____no_output_____"
]
],
[
[
"### Model\n\nGeneralized Linear Model with a Gamma distribution.\n\nThis regressor uses the ‘log’ link function.\n\nFor Ref.\nhttps://scikit-learn.org/stable/modules/linear_model.html#generalized-linear-regression",
"_____no_output_____"
],
[
"#### MinMax Scalar\nTransform features by scaling each feature to a given range.\n\nThis estimator scales and translates each feature individually such that it is in the given range on the training set, e.g. between zero and one.\n\nFor more reference:-\nhttps://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.MinMaxScaler.html",
"_____no_output_____"
]
],
[
[
"model = make_pipeline(MinMaxScaler(),GammaRegressor())\nmodel.fit(x_train,y_train)",
"_____no_output_____"
]
],
[
[
"#### Model Accuracy\n\nWe will use the trained model to make a prediction on the test set.Then use the predicted value for measuring the accuracy of our model.\n\nscore: The score function returns the coefficient of determination R2 of the prediction.\n",
"_____no_output_____"
]
],
[
[
"print(\"Accuracy score {:.2f} %\\n\".format(model.score(x_test,y_test)*100))",
"Accuracy score 17.81 %\n\n"
]
],
[
[
"> **r2_score**: The **r2_score** function computes the percentage variablility explained by our model, either the fraction or the count of correct predictions. \n\n> **mae**: The **mean abosolute error** function calculates the amount of total error(absolute average distance between the real data and the predicted data) by our model. \n\n> **mse**: The **mean squared error** function squares the error(penalizes the model for large errors) by our model. ",
"_____no_output_____"
]
],
[
[
"y_pred=model.predict(x_test)\nprint(\"R2 Score: {:.2f} %\".format(r2_score(y_test,y_pred)*100))\nprint(\"Mean Absolute Error {:.2f}\".format(mean_absolute_error(y_test,y_pred)))\nprint(\"Mean Squared Error {:.2f}\".format(mean_squared_error(y_test,y_pred)))",
"R2 Score: 24.11 %\nMean Absolute Error 2.56\nMean Squared Error 14.04\n"
]
],
[
[
"#### Prediction Plot\n\nFirst, we make use of a plot to plot the actual observations, with x_train on the x-axis and y_train on the y-axis.\nFor the regression line, we will use x_train on the x-axis and then the predictions of the x_train observations on the y-axis.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(14,10))\nplt.plot(range(20),y_test[0:20], color = \"green\")\nplt.plot(range(20),model.predict(x_test[0:20]), color = \"red\")\nplt.legend([\"Actual\",\"prediction\"]) \nplt.title(\"Predicted vs True Value\")\nplt.xlabel(\"Record number\")\nplt.ylabel(target)\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Creator: Jay Shimpi , Github: [Profile](https://github.com/JayShimpi22)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecf4c1c022e283dc7b4641faa234d2316444d1d6 | 74,310 | ipynb | Jupyter Notebook | applications/jupyter-extension/nteract_on_jupyter/notebooks/implicit-pipelines-old.ipynb | jjhenkel/nteract | 088222484b59af14b1da22de4d0990d8925adf95 | [
"BSD-3-Clause"
] | null | null | null | applications/jupyter-extension/nteract_on_jupyter/notebooks/implicit-pipelines-old.ipynb | jjhenkel/nteract | 088222484b59af14b1da22de4d0990d8925adf95 | [
"BSD-3-Clause"
] | null | null | null | applications/jupyter-extension/nteract_on_jupyter/notebooks/implicit-pipelines-old.ipynb | jjhenkel/nteract | 088222484b59af14b1da22de4d0990d8925adf95 | [
"BSD-3-Clause"
] | null | null | null | 39.21372 | 129 | 0.447436 | [
[
[
"%load_ext autoreload\n%autoreload 2\n\nimport pandas as pd\nimport json\n\nfrom codebookold.python import *\nfrom codebookold.semantics import DSNotebooks as DSN\n\nEvaluator.use_ds_gh_2017()",
"_____no_output_____"
],
[
"flows = pd.read_csv('/data/gh-2017/results/flows-read-to-fit.csv')",
"_____no_output_____"
],
[
"flows",
"_____no_output_____"
],
[
"import time \n\nOPS = [\n ('filters', DSN.filters),\n ('single_compares', DSN.single_compares),\n ('projections', DSN.projections),\n ('pandas_reads', DSN.pandas_reads),\n ('likely_labels', DSN.likely_labels),\n ('likely_values', DSN.likely_values),\n ('column_additions', DSN.column_additions),\n ('sklearn_fits', DSN.sklearn_fits),\n ('train_test_splits', DSN.train_test_splits),\n ('sklearn_fit_transforms', DSN.sklearn_fit_transforms),\n ('applies', DSN.applies),\n ('copies', DSN.copies),\n ('fillna', DSN.fillna),\n ('as_matrix', DSN.as_matrix),\n ('array', DSN.array),\n ('reshape', DSN.reshape),\n ('drops', DSN.drops),\n ('astype', DSN.astype),\n ('dropna', DSN.dropna),\n ('replace', DSN.replace),\n ('new_data_frame', DSN.new_data_frame),\n ('join', DSN.join),\n ('merge', DSN.merge),\n ('reset_indices', DSN.reset_indices),\n ('astype', DSN.astype),\n ('get_dummies', DSN.get_dummies),\n ('maps', DSN.maps),\n ('sklearn_transforms', DSN.sklearn_transforms),\n]\n\nres_frames = []\nfor name, op in OPS:\n print('Working on op:={}'.format(name))\n start = time.perf_counter()\n res_frames.append(op())\n elapsed_time = time.perf_counter() - start\n print(f\"Total time: {elapsed_time:.4f}s\")\n \n\nall_ops = pd.concat([\n x[['gid', 'pretty']] for x in res_frames\n])",
"Working on op:=<functools._lru_cache_wrapper object at 0x7f2a985fe360>\n + Had only 38896 allowable files (pre-filter files)\n + File select time: 0.4777s\n + Found 38896 matching files\n + Query already compiled (cached) `/tmp/queries/873261015198eaef776dbb59abf6425f1b2dbdcfc2eb86f73f9f97f32435b199.dl`\n + Query time: 26.8220s\n + Collation time: 0.2895s\nTotal time: 27.6113s\nTotal time: 27.7510s\nWorking on op:=<functools._lru_cache_wrapper object at 0x7f2a985fe400>\n + Had only 38896 allowable files (pre-filter files)\n + File select time: 0.5104s\n + Found 38896 matching files\n + Query already compiled (cached) `/tmp/queries/2505957c1bec268e54ae0e6f148162270b5c3599e5bcb6b6cafd93588e9f7ab0.dl`\n + Query time: 52.3877s\n + Collation time: 0.7242s\nTotal time: 53.6462s\n + Had only 24066 allowable files (pre-filter files)\n + File select time: 0.5060s\n + Found 24066 matching files\n + Query already compiled (cached) `/tmp/queries/a06562c2592dbc319c9b74c14f7f99fdc5b1d98748317c263f3779d6db3060e0.dl`\n + Query time: 69.1848s\n + Collation time: 0.7770s\nTotal time: 71.0722s\nTotal time: 128.2815s\nWorking on op:=<functools._lru_cache_wrapper object at 0x7f2a985fe220>\n + Had only 38896 allowable files (pre-filter files)\n + File select time: 0.5099s\n + Found 38896 matching files\n + Query already compiled (cached) `/tmp/queries/1aec7d01205ffa455742566f930c9b35d082cf24891b58155e5518507cd0d3ab.dl`\n + Query time: 76.2684s\n + Collation time: 12.6764s\nTotal time: 89.4771s\n + Had only 38896 allowable files (pre-filter files)\n + File select time: 0.4956s\n + Found 38896 matching files\n + Query already compiled (cached) `/tmp/queries/dd28ea1b3f30d3c8c472919240a7f628ef1337f40447a4788c859fc99d3bf387.dl`\n + Query time: 58.0022s\n + Collation time: 2.6130s\nTotal time: 72.6907s\n + Had only 32582 allowable files (pre-filter files)\n + File select time: 0.5120s\n + Found 32582 matching files\n + Query already compiled (cached) `/tmp/queries/a9bdea0ab00941d5d30612f123e9e4bbe2610b42cef7ca15a9434f2d65c5c269.dl`\n + Query time: 26.5792s\n + Collation time: 0.5980s\nTotal time: 30.1213s\n + Had only 38896 allowable files (pre-filter files)\n + File select time: 0.5125s\n + Found 38896 matching files\n + Query already compiled (cached) `/tmp/queries/aa10ea9669be3d8ff9e6e758129fb5eaa06d173c9d2854567fe8e463b8c0f034.dl`\n + Query time: 26.1012s\n + Collation time: 0.7360s\nTotal time: 27.3723s\n + Had only 32485 allowable files (pre-filter files)\n + File select time: 0.5119s\n + Found 32485 matching files\n + Query already compiled (cached) `/tmp/queries/f1805307abdd39f2b484e4fda739c71cd951ab552be7a7e71844c50e86d2682b.dl`\n + Query time: 74.7209s\n + Collation time: 1.5789s\nTotal time: 77.5204s\n + Had only 32485 allowable files (pre-filter files)\n + File select time: 0.4933s\n + Found 32485 matching files\n + Query already compiled (cached) `/tmp/queries/dd28ea1b3f30d3c8c472919240a7f628ef1337f40447a4788c859fc99d3bf387.dl`\n + Query time: 26.3905s\n + Collation time: 0.3779s\nTotal time: 28.2475s\nTotal time: 555.7669s\nWorking on op:=<functools._lru_cache_wrapper object at 0x7f2a985e5f40>\n + File select time: 12.5868s\n + Found 273870 matching files\n + Query already compiled (cached) `/tmp/queries/2f7dfacf9a56ffca49ef8662fce6ca1437ce36387305c747bbf255c8d34bbaac.dl`\n + Query time: 152.8291s\n + Collation time: 0.8854s\nTotal time: 166.5292s\n + Had only 273280 allowable files (pre-filter files)\n + File select time: 0.5971s\n + Found 273280 matching files\n + Query already compiled (cached) `/tmp/queries/c923f271f8d26990e9ee62e9a7d194f13d1c32f2aeb33331a65fae8612a96fdd.dl`\n + Query time: 5547.3083s\n + Collation time: 2.6669s\nTotal time: 5552.0158s\n + Had only 244216 allowable files (pre-filter files)\n + File select time: 12.4811s\n + Found 162790 matching files\n + Query already compiled (cached) `/tmp/queries/17fef5c89ebf2eaf067ea57d734db8e920532ea57d78d63d27155afcf9c24a5c.dl`\n + Query time: 107.9243s\n + Collation time: 1.3148s\nTotal time: 126.2002s\n + Had only 244216 allowable files (pre-filter files)\n + File select time: 12.3082s\n + Found 9664 matching files\n + Query already compiled (cached) `/tmp/queries/dcb5685c5d82c4d52205550e8182eaefdc5b6fbbed3728320a905edcd82f0448.dl`\n + Query time: 14.5040s\n + Collation time: 0.0866s\nTotal time: 31.4280s\n + Had only 244216 allowable files (pre-filter files)\n + File select time: 12.1976s\n + Found 489 matching files\n + Query already compiled (cached) `/tmp/queries/945559f96711c9a2c805149410c2e41a955fb684da7ab87bbdd9a768e92da9a5.dl`\n + Query time: 8.3588s\n + Collation time: 0.0096s\nTotal time: 24.7826s\n + Had only 244216 allowable files (pre-filter files)\n + File select time: 12.5392s\n + Found 4025 matching files\n + Query already compiled (cached) `/tmp/queries/12956229f014097235ce90bfd04ab682e6cc3d305ffe1f7555438d9dbd0f310b.dl`\n + Query time: 10.1319s\n + Collation time: 0.0325s\nTotal time: 27.1218s\n + Had only 244216 allowable files (pre-filter files)\n + File select time: 12.1487s\n + Found 4587 matching files\n + Query already compiled (cached) `/tmp/queries/09841124799b2ec6401e4ab702a7ab246671a168f30b33e8f86c26cd05929b8c.dl`\n + Query time: 11.3413s\n + Collation time: 0.0364s\nTotal time: 27.9264s\n + Had only 244216 allowable files (pre-filter files)\n + File select time: 12.4378s\n + Found 3862 matching files\n + Query already compiled (cached) `/tmp/queries/8885ca5f01f482d0cc94c0c729d9956e6665cb327101992043971bb7677e5062.dl`\n + Query time: 9.9027s\n + Collation time: 0.0392s\nTotal time: 26.8095s\n + Had only 244216 allowable files (pre-filter files)\n + File select time: 12.1455s\n + Found 7401 matching files\n + Query already compiled (cached) `/tmp/queries/9e98334074ac3feff3aecc1004445bcc4e51b32e52756f62f2ec7caf5e3b54b0.dl`\n + Query time: 12.6135s\n + Collation time: 0.0621s\nTotal time: 28.9613s\nTotal time: 6015.0205s\nWorking on op:=<functools._lru_cache_wrapper object at 0x7f2a985e5ea0>\n + Had only 38896 allowable files (pre-filter files)\n + File select time: 12.1731s\n + Found 38896 matching files\n + Query already compiled (cached) `/tmp/queries/30e7c4a0de7b18515be9db9841827c6511f034e2b86980cff9ff16a322386d3b.dl`\n + Query time: 23.8720s\n + Collation time: 0.4805s\nTotal time: 36.5495s\n + Had only 38896 allowable files (pre-filter files)\n + File select time: 12.0379s\n + Found 38896 matching files\n + Query already compiled (cached) `/tmp/queries/cd7bda65d67c8769761afe27e2c4f8efe589b23bfb3392454144ce2d4bfc0f26.dl`\n + Query time: 27.9965s\n + Collation time: 0.6744s\nTotal time: 40.7333s\n + Had only 38896 allowable files (pre-filter files)\n + File select time: 0.5027s\n + Found 38896 matching files\n + Query already compiled (cached) `/tmp/queries/9391e0afc7e400595668b5c063f651fabdc8bf1835209059d0cf46a80bc944e2.dl`\n + Query time: 25.4670s\n + Collation time: 1.5254s\nTotal time: 28.3720s\n + Had only 38896 allowable files (pre-filter files)\n + File select time: 0.4583s\n + Found 38896 matching files\n + Query already compiled (cached) `/tmp/queries/c923f271f8d26990e9ee62e9a7d194f13d1c32f2aeb33331a65fae8612a96fdd.dl`\n + Query time: 164.4822s\n + Collation time: 0.9597s\nTotal time: 166.7421s\n + Had only 38896 allowable files (pre-filter files)\n + File select time: 0.4819s\n + Found 38896 matching files\n + Query already compiled (cached) `/tmp/queries/aa47e2acac1a6ccf6214100f541a1b2e328b166bc771b2839a7d390d64dec04a.dl`\n + Query time: 25.8079s\n + Collation time: 1.3886s\nTotal time: 29.1972s\n + Had only 38896 allowable files (pre-filter files)\n + File select time: 0.4845s\n + Found 38896 matching files\n + Query already compiled (cached) `/tmp/queries/82655e148666b64f2cd013040a248b13ed10a4ed2035e98e66850d1e34f780cb.dl`\n"
],
[
"# Add \"unmodeled\" calls\nall_calls = DSN.exec(\n call() % select('name') % 'call'\n)\n\n# Get the ones we don't have models for \nall_calls = all_calls[~all_calls.gid_call.isin(all_ops.gid)]\nall_calls['pretty'] = 'Unmodeled[' + all_calls.out_name_call + ']'\nall_calls['gid'] = all_calls.gid_call\n\n# Add these now as \"unmodeled\" operators\nall_ops = pd.concat([all_ops, all_calls])",
" + File select time: 0.0004s\n + Found 647 matching files\n + Query already compiled (cached) `/tmp/queries/9ebe5aef7224f64ac809216889d6b7a3e676366dcbbe66c21354d172ad66d14f.dl`\n + Query time: 0.5182s\n + Collation time: 0.1583s\nTotal time: 0.6820s\n"
],
[
"def abstract(gids):\n abstracted = []\n for gid in gids:\n as_op = all_ops[all_ops.gid == gid]\n if len(as_op) > 0:\n abstracted.append((gid, ' ; '.join(as_op.pretty.to_list())))\n continue\n abstracted.append((gid, '???'))\n return abstracted\n\nflows_df = DSN.flows_reads_to_fits()\n \nimplicit_flows = flows_df.sort_values(\n ['start_line_flow', 'start_col_flow']\n).groupby(\n ['fpath', 'gid_source', 'gid_sink']\n)[['gid_flow', 'out_to_flow', 'out_edge_flow']].agg(list)\n\nimplicit_flows.gid_flow = implicit_flows.gid_flow.apply(abstract)\nimplicit_flows.out_to_flow = implicit_flows.out_to_flow.apply(abstract)",
" + Had only 38 allowable files (pre-filter files)\n + File select time: 0.0005s\n + Found 38 matching files\n + Query already compiled (cached) `/tmp/queries/2e765f47f59bf7fd40e826c17ce7cbae50dfa93ded49da5b9b18138825e3e294.dl`\n + Query time: 0.4929s\n + Collation time: 0.0555s\nTotal time: 0.5563s\n"
],
[
"import networkx as nx\nfrom networkx.drawing.nx_pydot import read_dot\nfrom networkx.drawing.nx_agraph import to_agraph\n\ngraphs = []\n\nfor key, row in implicit_flows.iterrows():\n tmp_g = nx.DiGraph()\n for (ga, opa), (gb, opb), edge in zip(*row.to_list()):\n tmp_g.add_node(ga, label=opa)\n tmp_g.add_node(gb, label=opb)\n tmp_g.add_edge(ga, gb, label=edge)\n graphs.append(tmp_g)",
"_____no_output_____"
],
[
"def cleanup_graph(target):\n # Go through ??? nodes and remove them \n changes = True\n while changes:\n changes = False\n for node in list(nx.topological_sort(target)):\n if target.nodes[node]['label'] != '???':\n continue\n incoming = target.in_edges(node)\n outgoing = target.out_edges(node)\n for (s,_) in incoming:\n for (_,t) in outgoing:\n target.add_edge(s,t,label=\"collapsed\")\n target.remove_node(node)\n changes=True\n break\n \n # I want to remove \"def -> use\" edges if there's _any_ other path from a def to it's use\n # Same thing for \"non-pure\" edges --- if there's another path, that's more interesting to us\n removals = []\n for s,t,l in target.edges.data('label'):\n if l != 'direct-use' and l != 'non-pure':\n continue\n # Only query for (at most) two paths from generator...\n # if we get to two, we don't need to enumerate the rest\n gen = nx.all_simple_paths(target, s,t)\n if next(gen, None) is None:\n continue\n if next(gen, None) is None:\n continue\n removals.append((s,t))\n\n # Remove the marked edges\n for (s,t) in removals:\n target.remove_edge(s,t)\n \n # Chains of Likely[X] can be reduced to one Likely[X]\n \n return target\n\ngraphs = list(map(cleanup_graph, graphs))",
"_____no_output_____"
],
[
"with open('/app/flow.txt', 'w') as fh:\n for i,g in enumerate(list(graphs)):\n print('{}/{}'.format(i+1, len(graphs)))\n fh.write(str(to_agraph(g)) + '\\n---\\n\\n')\n",
"1/156\n2/156\n3/156\n4/156\n5/156\n6/156\n7/156\n8/156\n9/156\n10/156\n11/156\n12/156\n13/156\n14/156\n15/156\n16/156\n17/156\n18/156\n19/156\n20/156\n21/156\n22/156\n23/156\n24/156\n25/156\n26/156\n27/156\n28/156\n29/156\n30/156\n31/156\n32/156\n33/156\n34/156\n35/156\n36/156\n37/156\n38/156\n39/156\n40/156\n41/156\n42/156\n43/156\n44/156\n45/156\n46/156\n47/156\n48/156\n49/156\n50/156\n51/156\n52/156\n53/156\n54/156\n55/156\n56/156\n57/156\n58/156\n59/156\n60/156\n61/156\n62/156\n63/156\n64/156\n65/156\n66/156\n67/156\n68/156\n69/156\n70/156\n71/156\n72/156\n73/156\n74/156\n75/156\n76/156\n77/156\n78/156\n79/156\n80/156\n81/156\n82/156\n83/156\n84/156\n85/156\n86/156\n87/156\n88/156\n89/156\n90/156\n91/156\n92/156\n93/156\n94/156\n95/156\n96/156\n97/156\n98/156\n99/156\n100/156\n101/156\n102/156\n103/156\n104/156\n105/156\n106/156\n107/156\n108/156\n109/156\n110/156\n111/156\n112/156\n113/156\n114/156\n115/156\n116/156\n117/156\n118/156\n119/156\n120/156\n121/156\n122/156\n123/156\n124/156\n125/156\n126/156\n127/156\n128/156\n129/156\n130/156\n131/156\n132/156\n133/156\n134/156\n135/156\n136/156\n137/156\n138/156\n139/156\n140/156\n141/156\n142/156\n143/156\n144/156\n145/156\n146/156\n147/156\n148/156\n149/156\n150/156\n151/156\n152/156\n153/156\n154/156\n155/156\n156/156\n"
],
[
"print(to_agraph(tg))",
"strict digraph G {\n\tgraph [name=G];\n\tnode [label=\"\\N\"];\n\t4788157767577519914\t [label=\"\\\"Read[read_csv]\\\"\"];\n\t1757491248908626318\t [label=\"\\\"Project[Pclass,Sex,Age,Fare,Parch]\\\"\"];\n\t4788157767577519914 -> 1757491248908626318\t [color=gray,\n\t\tstyle=solid];\n\t-8387815119344862824\t [label=\"\\\"Unmodeled[append]\\\"\"];\n\t7190111499327850570\t [label=\"\\\"Unmodeled[fit_transform]\\\"\"];\n\t-8387815119344862824 -> 7190111499327850570\t [color=gray,\n\t\tstyle=solid];\n\t1757491248908626318 -> -8387815119344862824\t [color=black,\n\t\tstyle=solid];\n\t-1953037366365052216\t [label=\"\\\"???\\\"\"];\n\t-1671177202092878445\t [label=\"\\\"???\\\"\"];\n\t-1953037366365052216 -> -1671177202092878445\t [color=orange,\n\t\tstyle=dashed];\n\t-2246360499928749877\t [label=\"\\\"FitTransform[LabelEncoder]\\\"\"];\n\t-1953037366365052216 -> -2246360499928749877\t [color=gray,\n\t\tstyle=solid];\n\t526293004170640997\t [label=\"\\\"AsMatrix[]\\\"\"];\n\t-1671177202092878445 -> 526293004170640997\t [color=gray,\n\t\tstyle=solid];\n\t7190111499327850570 -> -1953037366365052216\t [color=gray,\n\t\tstyle=solid];\n\t-2246360499928749877 -> -1671177202092878445\t [color=gray,\n\t\tstyle=solid];\n\t-7406026133898430313\t [label=\"\\\"Likely[Values]\\\"\"];\n\t7668313745462718906\t [label=\"\\\"Fit[BernoulliNB]\\\"\"];\n\t-7406026133898430313 -> 7668313745462718906\t [color=black,\n\t\tstyle=solid];\n\t526293004170640997 -> -7406026133898430313\t [color=gray,\n\t\tstyle=solid];\n}\n\n"
],
[
"attrs = DSN.exec(\n attribute() % 'attr'\n | where | the_object_is(anything() % select_as('text', 'target'))\n | and_w | the_attribute_is(identifier() % select_as('text', 'field'))\n)\n\ncompares = DSN.exec(\n comparison(with_exactly_two_children()) % 'filter'\n | where | the_first_child_is(attribute(from_set(attrs, 'gid_attr')))\n | and_w | the_second_child_is(literal() % 'rhs')\n)\n\nuses_of_compares = DSN.exec(\n use_of(comparison(from_set(compares, 'gid_filter'))) % 'use'\n)\n\ntmp = DSN.exec(\n subscript(with_exactly_two_children()) % 'expr'\n | where | the_subscript_is(anything(from_set(uses_of_compares, 'gid_use')))\n | and_w | the_value_is(anything() % 'target')\n)",
" + File select time: 0.0004s\n + Found 647 matching files\n + Query already compiled (cached) `/tmp/queries/efcefa617ba78cfa5e04f73c381c6bf4ed711e8893ceaa7861506924acae00e5.dl`\n + Query time: 0.4972s\n + Collation time: 0.1220s\nTotal time: 0.6252s\n + Had only 562 allowable files (pre-filter files)\n + File select time: 0.0005s\n + Found 562 matching files\n + Query already compiled (cached) `/tmp/queries/c780e2968eb631f68891484cb58540307418990fd25f6a9d5185ec094e63b57d.dl`\n + Query time: 3.0908s\n + Collation time: 0.0067s\nTotal time: 3.2815s\n + Had only 64 allowable files (pre-filter files)\n + File select time: 0.0004s\n + Found 64 matching files\n + Query already compiled (cached) `/tmp/queries/42ea7c56f87191d3b8837bbd0c125adca5434fe009c92cb0f46b91494513ac04.dl`\n + Query time: 4.1820s\n + Collation time: 0.0053s\nTotal time: 4.1946s\n + Had only 64 allowable files (pre-filter files)\n + File select time: 0.0004s\n + Found 64 matching files\n + Profile time: 0.0911s\n + Compile time: 13.7693s\n + Query time: 0.3437s\n + Collation time: 0.0054s\nTotal time: 14.2177s\n"
],
[
"tmp = DSN.exec(attribute() % 'attr' |where| the_attribute() |isa| identifier() % 'field')",
" + File select time: 0.0004s\n + Found 647 matching files\n + Profile time: 0.0849s\n + Compile time: 12.1508s\n + Query time: 0.4709s\n + Collation time: 0.1671s\nTotal time: 12.8819s\n"
],
[
"DSN.projections()",
"INFO: Pandarallel will run on 20 workers.\nINFO: Pandarallel will use Memory file system to transfer data between the main process and workers.\n + File select time: 0.0004s\n + Found 647 matching files\n + Query already compiled (cached) `/tmp/queries/1aec7d01205ffa455742566f930c9b35d082cf24891b58155e5518507cd0d3ab.dl`\n + Query time: 1.0007s\n + Collation time: 0.1438s\nTotal time: 1.1507s\n + Had only 598 allowable files (pre-filter files)\n + File select time: 0.0005s\n + Found 598 matching files\n + Query already compiled (cached) `/tmp/queries/dd28ea1b3f30d3c8c472919240a7f628ef1337f40447a4788c859fc99d3bf387.dl`\n + Query time: 1.0451s\n + Collation time: 0.0280s\nTotal time: 1.1992s\n + Had only 302 allowable files (pre-filter files)\n + File select time: 0.0005s\n + Found 302 matching files\n + Query already compiled (cached) `/tmp/queries/c7afd45aaca01aedeb4493184eebddab4cf0708af1ddbc73f4cb5e3f0db00584.dl`\n + Query time: 0.4771s\n + Collation time: 0.0094s\nTotal time: 0.5145s\n + File select time: 0.0005s\n + Found 647 matching files\n + Query already compiled (cached) `/tmp/queries/aa10ea9669be3d8ff9e6e758129fb5eaa06d173c9d2854567fe8e463b8c0f034.dl`\n + Query time: 0.4977s\n + Collation time: 0.0121s\nTotal time: 0.5163s\n + Had only 251 allowable files (pre-filter files)\n + File select time: 0.0005s\n + Found 251 matching files\n + Query already compiled (cached) `/tmp/queries/da2823113bdb089852fe28d3f96d43c1862c86d79e9bb2a0acbd20cbf3356bca.dl`\n + Query time: 24.7471s\n + Collation time: 0.0162s\nTotal time: 24.7749s\n + Had only 251 allowable files (pre-filter files)\n + File select time: 0.0004s\n + Found 251 matching files\n + Profile time: 0.0916s\n + Compile time: 13.0674s\n + Query time: 0.4221s\n + Collation time: 0.0063s\nTotal time: 13.6001s\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf4e6276db1daf07075c08ce4146f75318fc061 | 1,190 | ipynb | Jupyter Notebook | Small pattern programming/s pattern.ipynb | mujahid2580/python-pattern-program | 8fc230f871b747abd70f819b2e6cfff72d93c2a7 | [
"MIT"
] | 1 | 2020-02-19T19:32:31.000Z | 2020-02-19T19:32:31.000Z | Small pattern programming/s pattern.ipynb | mujahid2580/pyhton-pattern-programming | 8fc230f871b747abd70f819b2e6cfff72d93c2a7 | [
"MIT"
] | null | null | null | Small pattern programming/s pattern.ipynb | mujahid2580/pyhton-pattern-programming | 8fc230f871b747abd70f819b2e6cfff72d93c2a7 | [
"MIT"
] | 2 | 2020-09-30T15:26:27.000Z | 2021-03-05T07:09:42.000Z | 19.193548 | 113 | 0.408403 | [
[
[
"for i in range(7):\n for j in range(7):\n if (((i==0 or i==6 or i==3) and j!=6) and j>0) or j==0 and i>0 and i<3 or j==6 and i>3 and i<6 :\n print(\"*\",end=\"\")\n else:\n print(end=\" \")\n print()",
" ***** \n* \n* \n ***** \n *\n *\n ***** \n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
ecf4e86067ba7f75826a641fb28e7d5b5be69af9 | 40,964 | ipynb | Jupyter Notebook | markdown_generator/publications.ipynb | eipapa/eipapa.github.io | f0ea04149e7b97320368639fc654162d86154e2f | [
"CC-BY-4.0",
"MIT"
] | 1 | 2020-02-11T00:06:56.000Z | 2020-02-11T00:06:56.000Z | markdown_generator/publications.ipynb | eipapa/eipapa.github.io | f0ea04149e7b97320368639fc654162d86154e2f | [
"CC-BY-4.0",
"MIT"
] | null | null | null | markdown_generator/publications.ipynb | eipapa/eipapa.github.io | f0ea04149e7b97320368639fc654162d86154e2f | [
"CC-BY-4.0",
"MIT"
] | null | null | null | 86.788136 | 2,904 | 0.664608 | [
[
[
"# Publications markdown generator for academicpages\n\nTakes a TSV of publications with metadata and converts them for use with [academicpages.github.io](academicpages.github.io). This is an interactive Jupyter notebook ([see more info here](http://jupyter-notebook-beginner-guide.readthedocs.io/en/latest/what_is_jupyter.html)). The core python code is also in `publications.py`. Run either from the `markdown_generator` folder after replacing `publications.tsv` with one containing your data.\n\nTODO: Make this work with BibTex and other databases of citations, rather than Stuart's non-standard TSV format and citation style.\n",
"_____no_output_____"
],
[
"## Data format\n\nThe TSV needs to have the following columns: pub_date, title, venue, excerpt, citation, site_url, and paper_url, with a header at the top. \n\n- `excerpt` and `paper_url` can be blank, but the others must have values. \n- `pub_date` must be formatted as YYYY-MM-DD.\n- `url_slug` will be the descriptive part of the .md file and the permalink URL for the page about the paper. The .md file will be `YYYY-MM-DD-[url_slug].md` and the permalink will be `https://[yourdomain]/publications/YYYY-MM-DD-[url_slug]`\n\nThis is how the raw file looks (it doesn't look pretty, use a spreadsheet or other program to edit and create).",
"_____no_output_____"
]
],
[
[
"!cat publications.tsv",
"pub_date\ttitle\tvenue\texcerpt\tcitation\turl_slug\tpaper_url\r\r\n2020-04-21\tCorporate Hackathons, How and Why? A multiple case study of motivation, project proposal and selection, goal setting, coordination, and project continuation.\tHuman-Computer Interaction Journal\t\"Time-bounded events such as hackathons, data dives, codefests, hack-days, sprints or edit-a-thons have increasingly gained attention from practitioners and researchers. Yet there is a paucity of research on corporate hackathons, which are nearly ubiquitous and present significant organizational, cultural, and managerial challenges. To provide a comprehensive understanding of team processes and broad array of outcomes of corporate hackathons, we conducted a mixed-methods, multiple case study of five teams that participated in a large scale corporate hackathon. Two teams were “pre-existing” teams (PETs) and three were newly-formed “flash” teams (FTs). Our analysis revealed that PETs coordinated almost as if it was just another day at the office while creating innovations within the boundary of their regular work, whereas FTs adopted role-based coordination adapted to the hackathon context while creating innovations beyond the boundary of their regular work. Project sustainability depended on how much effort the team put into finding a home for their projects and whether their project was a good fit with existing products in the organization’s product portfolio. Moreover, hackathon participation had perceived positive effects on participants’ skills, careers, and social networks.\"\tPe-Than, E.P.P., Nolte, A., Filippova, A., Bird, C., Scallen, S., and Herbsleb, J.D. (in press). Corporate Hackathons, How and Why? A Multiple Case Study of Motivation, Project Proposal and Selection, Goal Setting, Coordination, and Project Continuation. Human-Computer Interaction.\tPe-Than-hcij-2020\thttp://eipapa.github.io/files/Pe-Than-hcij-2020.pdf\r\r\n2019-06-13\tCollaborative Writing at Scale: A Case Study of Two Open-Text Projects Done on GitHub\tACM Collective Intelligence (CI'19)\t\"Work of all kinds is increasingly done in a networked digital environment comprised of multiple Internet-connected platforms offering varying affordances and serving communities with specific norms and values. Such an environment invites inclusive participation in collaborative production but, at the same time, challenges the roles and design of platforms traditionally used for specific kinds of work. Despite the earlier prevalence of shared editors, collaborative writing is now moving to online platforms with social networking functionality such as Wikipedia and GitHub. This study examines the evolution of digital text artifacts in a networked digital environment as revealed through a case study of two open text projects on GitHub.com – a popular social coding/software development platform. Our findings suggest that GitHub’s pull-based model effectively manages collaborative writing at scale through sophisticated version control and lightweight review as participation and visibility of the project increases. In this pull-based model, contributors either converge at a single project to perfect its artifacts, or adopt and tailor the original project to their needs. In sum, this study highlights a new mode of collaborative writing in which GitHub and other platforms are used, conventions are adopted, and roles are established.\"\tPe-Than, E.P.P., Dabbish, L., and Herbsleb, J.D. (2019). Collaborative Writing at Scale: A Case Study of Two Open-Text Projects Done on GitHub. Poster presented at the 7th ACM Conference on Collective Intelligence 2019 (CI'19). https://ci.acm.org/2019/assets/proceedings/CI_2019_paper_65.pdf\tCI-poster-2019\thttp://eipapa.github.io/files/CI-poster-2019.pdf\r\r\n2019-06-25\tCollaborative Writing at Scale: A Case Study of Two Open-Text Projects Done on GitHub\tThe Future of Work (HCIC'19) Workshop\t\"Collaborative Work of all kinds is increasingly done in a networked digital environment comprised of multiple Internet-connected platforms offering varying affordances and serving communities with specific norms and values. Such an environment invites inclusive participation in collaborative production but, at the same time, challenges the roles and design of platforms traditionally used for specific kinds of work. Despite the earlier prevalence of shared editors, collaborative writing is now moving to online platforms with social networking functionality such as Wikipedia and GitHub. This study examines the evolution of digital text artifacts in a networked digital environment as revealed through a case study of two open text projects on GitHub.com – a popular social coding/software development platform. Our findings suggest that working collaboratively in a networked digital environment enables production to occur across multiple platforms as the need for this is perceived. In particular, GitHub’s pull-based model effectively manages collaborative writing at scale through sophisticated version control and lightweight review as participation and visibility of the project increases. In this pull-based model, contributors either converge at a single project to perfect its artifacts, or adopt and tailor the original project to their needs. In sum, this study highlights a new mode of collaborative writing in which GitHub and other platforms are used, conventions are adopted, and roles are established.\"\t\"Pe-Than, E.P.P, Dabbish, L., and Herbsleb, J.D. (2019). Collaborative Writing at Scale: A Case Study of Two Open-Text Projects Done on GitHub. <i>Paper presented at The Future of Work, Human Computer Interaction Consortium (HCIC'19) Workshop</i> http://eipapa.github.io/files/HCIC-poster-2019.pdf.\"\tHCIC-2019\thttp://eipapa.github.io/files/HCIC-poster-2019.pdf\r\r\n2018-07-11\tDesigning Corporate Hackathons with a Purpose\tIEEE Software\t\"In hackathons, small teams work together over a specified period of time to complete a project of interest. Hackathons have become increasingly popular as a means to surface and prototype innovative and creative ideas for products, but their impact often goes beyond product innovation. Based on our empirical studies of 10 hackathons held by scientific communities, a corporation, and universities as well as the review of published literature, we discuss that hackathons can be organized around goals such as enriching social networks, facilitating collaborative learning, and workforce development. We also discuss design choices that can scaffold the organization of hackathons and their trade-offs. Design choices include identifying a suitable mixture of attendee skills, the selection process for projects and teams, and whether to hold a competitive or collaborative event. Hackathons can achieve multiple goals if designed carefully.\"\t\"Pe-Than, E.P.P., Nolte, A., Filippova, A., Bird, C., Scallen, S., and Herbsleb, J.D. (2018). Designing Corporate Hackathons with a Purpose. IEEE Software, 36 (1), pp. 15-22.\thttps://doi.org/10.1109/MS.2018.290110547\"\tIEEESW-2018\thttp://eipapa.github.io/files/IEEESW-2018.pdf\r\r\n2018-11-01\tYou Hacked and Now What?: - Exploring Outcomes of a Corporate Hackathon\tProceedings of the ACM on Human-Computer Interaction (PACM HCI, ACM CSCW'18)\t\"Time bounded events such as hackathons, data dives, codefests, hack-days, sprints or edit-a-thons have increasingly gained attention from practitioners and researchers. Existing research, however, has mainly focused on the event itself, while potential outcomes of hackathons have received limited attention. Furthermore, most research around hackathons focuses on collegiate or civic events. Research around hackathons internal to tech companies, which are nearly ubiquitous, and present significant organizational, cultural, and managerial challenges, remains scarce. In this paper we address this gap by presenting findings from a case study of five teams which participated in a large scale corporate hackathon. Most team members voiced their intentions to continue the projects their worked on during the hackathon, but those whose projects did get continued were characterized by meticulous preparation, a focus on executing a shared vision during the hackathon, extended dissemination activities afterwards and a fit to existing product lines. Such teams were led by individuals who perceived the hackathon as an opportunity to bring their idea to life and advance their careers, and who recruited teams who had a strong interest in the idea and in learning the skills necessary to contribute efficiently. Our analysis also revealed that individual team members perceived hackathon participation to have positive effects on their career parts, networks and skill development.\"\t\"Nolte, A., Pe-Than, E.P.P., Filippova, A., Bird, C., Scallen, S., and Herbsleb, J.D. (2018). You Hacked and Now What?: - Exploring Outcomes of a Corporate Hackathon. In Proceedings of the ACM on Human Computer Interaction, 2 (CSCW'18), Article 129, 23 pages. https://doi.org/10.1145/3274398\"\tCSCW-2018\thttp://eipapa.github.io/files/CSCW-2018.pdf\r\r\n2019-03-13\tUnderstanding Hackathons for Science: Collaboration, Affordances, and Outcomes\tiConference'19\t\"Nowadays, hackathons have become a popular way of bringing people together to engage in brief, intensive collaborative work. Despite being a brief activity, being collocated with team members and focused on a task—radical collocation—could improve collaboration of scientific software teams. Using a mixed-methods study of participants who attended two hackathons at Space Telescope Science Institute (STScI), we examined how hackathons can facilitate collaboration in scientific software teams which typically involve members from two different disciplines: science and software engineering. We found that hackathons created a focused interruption-free working environment in which team members were able to assess each other’s skills, focus together on a single project and leverage opportunities to exchange knowledge with other collocated participants, thereby allowing technical work to advance more efficiently. This study suggests “hacking” as a new and productive form of collaborative work in scientific software production.\"\t\"Pe-Than, E.P.P. and Herbsleb, J.D. (2019). Understanding Hackathons for Science: Collaboration, Affordances, and Outcomes. In Taylor N., Christian-Lamb C., Martin M., Nardi B. (eds) Information in Contemporary Society, iConference'19. Lecture Notes in Computer Science, vol 11420 (iConference'19), pp. 27-37. Springer, Cham. https://doi.org/10.1007/978-3-030-15742-5_3\"\tiConference-2019\thttp://eipapa.github.io/files/iConference-2019.pdf\r\r\n2018-10-01\tCollaborative Writing on GitHub: A Case Study of a Book Project\tCompanion of the 2018 ACM Conference on Computer Supported Cooperative Work and Social Computing (ACM CSCW'18)\t\"Social coding platforms such as GitHub are increasingly becoming a digital workspace for the production of non-software digital artifacts. Since GitHub offers unique features that are different from traditional ways of collaborative writing, it is interesting to investigate how GitHub features are used for writing. In this paper, we present the preliminary findings of a mixed-methods, case study of collaboration practices in a GitHub book project. We found that the use of GitHub depended on task interdependence and audience participation. GitHub's direct push method was used to coordinate both loosely- and tightly-coupled work, with the latter requiring collaborators to follow socially-accepted conventions. The pull-based method was adopted once the project was released to the public. While face-to-face and online meetings were prominent in the early phases, GitHub's issues became instrumental for communication and project management in later phases. Our findings have implications for the design of collaborative writing tools.\"\t\"Pe-Than, E.P.P., Dabbish, L., and Herbsleb, J.D. (2018). Collaborative Writing on GitHub: A Case Study of a Book Project. In Companion of the 2018 ACM Conference on Computer Supported Cooperative Work and Social Computing (CSCW’18), pp. 305-308. https://doi.org/10.1145/3272973.3274083\"\tCSCW-abstract-2018\thttp://eipapa.github.io/files/CSCW-abstract-2018.pdf\r\r\n2018-01-04\tThe 2nd Workshop on Hacking and Making at Time-Bounded Events: Current Trends and Next Steps in Research and Event Design\tExtended Abstracts of the 2018 ACM CHI Conference on Human Factors in Computing Systems (ACM CHI'18)\t\"Hackathons or Hackathon-style events, describe increasingly popular time-bounded intensive events across different fields and sectors. Often cited examples of hackathons include the demanding overnight competitive coding events, but there are many design variations for different audiences and with divergent aims. They offer a new form of collaboration by affording explicit, predictable, time-bounded spaces for interdependent work and engaging with new audiences. This one-day workshop will bring together researchers, experienced event organizers, and practitioners to share and discuss their practical experiences. Empirical insights from studying these events may help position the CHI community to better study, plan and design hackathon-style events and socio-technical systems that support new modes of production and collaboration.\"\t\"Pe-Than, E.P.P., Herbsleb, J.D., Nolte, A., Gerber, E., Fiore-Gartland, B., Chapman, B., Moser, A., and Wilkins-Diehr, N. (2018). The 2nd Workshop on Hacking and Making at Time-Bounded Events: Current Trends and Next Steps in Research and Event Design. <i>In Extended Abstracts of the 2018 CHI Conference on Human Factors in Computing Systems (CHI EA'18), </i>paper W35, pp. 1-8. https://doi.org/10.1145/3170427.3170615\"\tCHI-workshop-2018\thttp://eipapa.github.io/files/CHI-workshop-2018.pdf\r\r\n2019-01-01\tHackathons for Science, How and Why?\tThe 233rd Meeting of the American Astronomical Society (AAS Meeting #233)\t\"Based on our empirical studies of 14 hackathons held by a corporation (Microsoft OneWeek Hackathon), universities, and scientific communities including three hack days at Space Telescope Science Institute (STScI), we will present how hackathons can be designed to achieve specific goals in a semi-academic environment like STScI. Our recommendations are derived from the analysis of data collected through ethnographic observations, interviews, and questionnaires. Hackathons are not only a good way to foster innovation but also to provide learning and knowledge exchange opportunities, to create new and enhance existing social connections, to exercise new technical and leadership opportunities, and to get the needed work done or to make a quick progress on technical work. However, designing a hackathon involves careful upfront planning, project selection, team formation, goal setting, and follow-up activities. Before the hackathon, the organizers should ask potential participants to propose project ideas (e.g., highest priority needed work in the case of STScI), and elicit their skills, expertise, and project preference. The organizers should form teams by matching participants' skills to required skills for the projects. The resulting teams consist of a mix of members with varying levels of expertise. It is advisable for teams to perform preparatory work which includes appointing a team lead and having pre-event meetings where they discuss their plan for the event, break the projects into small individual tasks, assign tasks to team members, and familiarize themselves with the environment, project, and task. The organizers should advise teams to set realistic goals for the event and keep track of their progress toward these goals. At the end of the event, the organizers should advise teams to present their accomplishment and future plans, and encourage them to plan for future collaboration and designate a person to keep track of the progress. Examples include using the common free time to work side-by-side and self-organizing mini-hackathons (e.g., lunch hacks). With careful consideration of activities mentioned above, hackathons may provide a fruitful avenue of collaboration between astronomers and software experts.\"\t\"Pe-Than, E.P.P., Momcheva, I., Tollerud, E., and Herbsleb, J.D. (2019). Hackathons for Science, How and Why? Poster presented at the 233rd Meeting of the American Astronomical Society, AAS Meeting #233, id.459.11. https://ui.adsabs.harvard.edu/abs/2019AAS...23345911P/abstract\"\tAAS-poster-2019\thttp://eipapa.github.io/files/AAS-poster-2019.pdf\r\r\n2018-01-09\tAn Analysis of the PEARC 2018 Science Gateways Community Institute Hackathon: Lessons Learned\tPractice & Experience in Advanced Research Computing Conference Series (PEARC'18)\t\"Science Gateways are web portals on which data, software, instruments, and computing resources provide users, and scientists streamlined workflows through dynamically coded frontends. These web portals allow scientist to focus on research with little to no need to utilize programmatic skills. Hackathons are time-bounded events in which collaborators intensely focus on a given subject or problem in an effort to generate solutions. Hackathon events provide opportunities for participants with diverse backgrounds, mixed specialties, and broad skill sets to interact in a manner that promotes disruptive solutions. Hackathons come in a variety of different forms and serve many different purposes. These purposes include developing resources, infrastructure, practices, and culture for a community. The Science Gateways Community Institute team proposed a hackathon to introduce students attending the PEARC 2018 conference, to the concepts of Science Gateways. The aim of the hackathon was to stimulate interest in Science Gateways while developing projects that were of value to researchers from different disciplines. They believed that PEARC would be an ideal venue for this type of event, due to the community of researchers, students, and practitioners from various disciplines that were interested and skilled in the high-performance computing field attending the conference. The Science Gateways Community Institute utilized a hackathon formatted event to provide students with an opportunity to collaborate and generate targeted skills while creating needed features for production web portals. The student participants included a group of 17 graduate and undergraduate students with predominately underrepresented minorities. The hackathon followed the PEARC 2018 conference held in Pittsburgh, PA. The science gateways targeted during the event were MyGeoHub (Purdue University), Cosmic2 (San Diego Supercomputing Center), and SimCCS (Indiana University) with associated mentors serving as subject matter experts. This poster presents organizational methods and technologies used to coordinate the international planning team, lessons learned during the event, and deliverables presented by the student teams.\"\t\"Powell, J., Hayden, L., Nolte, A., Herbsleb, J.D., Pe-Than, E.P.P., Wong, M., Kalyanam, R., Ellet, K., Pamidighantam, S., Traxler, K., and Cannon, A. (2018). An Analysis of the PEARC 2018 Science Gateways Community Institute Hackathon: Lessons Learned. <i>Poster presented at Gateways 2018 organized by the Science Gateways Community Institute (SGCI)</i>. https://doi.org/10.6084/m9.figshare.7070309.v2\"\tSGCI-poster-2018\thttp://eipapa.github.io/files/SGCI-poster-2018.pdf\r\r\n2018-12-30\tThe 2nd Workshop on Hacking and Making at Time-Bounded Events.\tTechnical Report CMU-ISR-18-109\t\"In hackathons, small teams work together over a specified period of time to complete a project of interest. Such time-bounded hackathon-style events have become increasingly popular across different domains in recent years. Collegiate hackathons, just one of the many variants of hackathons, that are supported by the largest hackathon league (https://mlh.io/) alone attract over 65,000 participants among more than 200 events each year. Variously known as data dives, codefests, hack-days, sprints, edit-a-thons, mapathons, and so on, such events vary depending on different audiences and with divergent aims: for example, whether teams know each other beforehand, whether the event is structured as a competition with prizes, whether the event is open or requires membership or invitations, and whether the desired outcome is primarily a product innovation, learning a new skill, forming a community around a cause, solving a technical problem that requires intensive focus by a group, or just having fun. Taken together, hackathons offer new opportunities and challenges for collaboration by affording explicit, predictable, time-bounded spaces for collaborative work and engaging with new audiences. With the goal of discussing opportunities and challenges surrounding hackathons of different kinds, this one-day workshop brought together researchers, experienced event organizers, and practitioners to share and discuss their practical experiences. Empirical insights from studying these events may help position the CHI community to better study, plan and design hackathon-style events as socio-technical systems that support new modes of production and collaboration.\"\t\"Pe-Than, E.P.P., and Nolte, A. (Editors) (2018). The 2nd Workshop on Hacking and Making at Time-Bounded Events. <i>Technical Report CMU-ISR-18-109</i>, Carnegie Mellon University. http://eipapa.github.io/files/ISR-techreport-2018.pdf\"\tISR-techreport-2018\thttp://eipapa.github.io/files/ISR-techreport-2018.pdf\r\r\n"
]
],
[
[
"## Import pandas\n\nWe are using the very handy pandas library for dataframes.",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"## Import TSV\n\nPandas makes this easy with the read_csv function. We are using a TSV, so we specify the separator as a tab, or `\\t`.\n\nI found it important to put this data in a tab-separated values format, because there are a lot of commas in this kind of data and comma-separated values can get messed up. However, you can modify the import statement, as pandas also has read_excel(), read_json(), and others.",
"_____no_output_____"
]
],
[
[
"publications = pd.read_csv(\"publications.tsv\", sep=\"\\t\", header=0)\npublications\n",
"_____no_output_____"
]
],
[
[
"## Escape special characters\n\nYAML is very picky about how it takes a valid string, so we are replacing single and double quotes (and ampersands) with their HTML encoded equivilents. This makes them look not so readable in raw format, but they are parsed and rendered nicely.",
"_____no_output_____"
]
],
[
[
"html_escape_table = {\n \"&\": \"&\",\n '\"': \""\",\n \"'\": \"'\"\n }\n\ndef html_escape(text):\n \"\"\"Produce entities within text.\"\"\"\n return \"\".join(html_escape_table.get(c,c) for c in text)",
"_____no_output_____"
]
],
[
[
"## Creating the markdown files\n\nThis is where the heavy lifting is done. This loops through all the rows in the TSV dataframe, then starts to concatentate a big string (```md```) that contains the markdown for each type. It does the YAML metadata first, then does the description for the individual page.",
"_____no_output_____"
]
],
[
[
"import os\nfor row, item in publications.iterrows():\n \n md_filename = str(item.pub_date) + \"-\" + item.url_slug + \".md\"\n html_filename = str(item.pub_date) + \"-\" + item.url_slug\n year = item.pub_date[:4]\n \n ## YAML variables\n \n md = \"---\\ntitle: \\\"\" + item.title + '\"\\n'\n md += \"layout: archive\" + '\\n'\n md += \"\"\"collection: publications\"\"\"\n md += \"\"\"\\npermalink: /publication/\"\"\" + html_filename\n if len(str(item.excerpt)) > 5:\n md += \"\\nexcerpt: '\" + html_escape(item.excerpt) + \"'\"\n md += \"\\ndate: \" + str(item.pub_date) \n md += \"\\nvenue: '\" + html_escape(item.venue) + \"'\" \n# if len(str(item.paper_url)) > 5:\n# md += \"\\npaperurl: '\" + item.paper_url + \"'\"\n# md += \"\\ncitation: '\" + html_escape(item.citation) + \"'\"\n md += \"\\n---\"\n \n ## Markdown description for individual page\n \n if len(str(item.paper_url)) > 5:\n# md += \"\\n[<span style=\\\"color: #c41e3a\\\">Download PDF here.</span>](\" + item.paper_url + \")\\n\"\n md += \"\\n[Download PDF here.](\" + item.paper_url + \")\\n\"\n if len(str(item.excerpt)) > 5:\n md += \"\\n**Abstract**: \" + html_escape(item.excerpt) + \"\\n\" \n# md += \"\\nAbstract: \" + html_escape(item.description) + \"\\n\"\n md += \"\\n**Recommended citation**: \" + item.citation\n md_filename = os.path.basename(md_filename)\n with open(\"../_publications/\" + md_filename, 'w') as f:\n f.write(md)",
"_____no_output_____"
]
],
[
[
"These files are in the publications directory, one directory below where we're working from.",
"_____no_output_____"
]
],
[
[
"!ls ../_publications/",
"2018-01-04-CHI-workshop-2018.md 2019-01-01-AAS-poster-2019.md\r\n2018-01-09-SGCI-poster-2018.md 2019-03-13-iConference-2019.md\r\n2018-07-11-IEEESW-2018.md 2019-06-13-CI-poster-2019.md\r\n2018-10-01-CSCW-abstract-2018.md 2019-06-25-HCIC-2019.md\r\n2018-11-01-CSCW-2018.md 2020-04-21-Pe-Than-hcij-2020.md\r\n2018-12-30-ISR-techreport-2018.md \u001b[34mbackup\u001b[m\u001b[m\r\n"
],
[
"!cat ../_publications/2009-10-01-paper-title-number-1.md",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecf4f42098e14f8b2eebcd0014e62a7375c24ae3 | 4,429 | ipynb | Jupyter Notebook | _notebooks/2022-01-02-math_la_sym.ipynb | stevenchen521/blogging | d4905e1542aa2e3a06b3410ddde2b960d81884de | [
"Apache-2.0"
] | null | null | null | _notebooks/2022-01-02-math_la_sym.ipynb | stevenchen521/blogging | d4905e1542aa2e3a06b3410ddde2b960d81884de | [
"Apache-2.0"
] | null | null | null | _notebooks/2022-01-02-math_la_sym.ipynb | stevenchen521/blogging | d4905e1542aa2e3a06b3410ddde2b960d81884de | [
"Apache-2.0"
] | null | null | null | 35.717742 | 393 | 0.609167 | [
[
[
"# Properties of Symmetric Matrice\n> Recently I'm reviewing mathematic basis which I think would be useful for the the machine learning. In this post, I am going to review some important properties of symmetric matrix.\n\n- toc: true\n- comments: true\n- branch: master\n- badges: false\n- categories: [Math]\n- image: images/la_sym.png\n",
"_____no_output_____"
],
[
"# Definition\nBased on [wikipedia](https://en.wikipedia.org/wiki/Symmetric_matrix), in linear algebra, a symmetric matrix is a square matrix that is equal to its transpose. Formally, $A=A^T$ where $A$ is the matrix. The entries of the symmetric matrix $A$ are symmetric with respect to the main diagonal. More specificallt, suppose $a_{ij}$ denotes the emelent of $A$, then we have $a_{ij}=a_{ji}$.\n\nFor example the following is a symmetric matrix.\n\n\\begin{bmatrix}\n1 & 7 & 3\\\\7 & 4 & 5\\\\3 & 5 & 0\n\\end{bmatrix}\n",
"_____no_output_____"
],
[
"# Properties of Symmetric Matrice\n\n## 1. Eigenvevtors of the symmetric matrix are orthogonal to each other\n\nSuppose we have symmetric matrix $A$ with its eigenvectors $V=[v_1,v_2...v_n]$, we have $v_iv_j^T=\\vec0$ where $i,j\\in [1,n]$ \n**Proof**: the target is to prove $v_iv_j^T=\\vec0$ \n\n\n1. $Av_1=\\lambda_1v_1$ \n2. $Av_2=\\lambda_1v_2$ \n3. $A=A^T$ \n4. $v_1^TAv_2=v_1^T\\lambda_2v_2=(v_2^T\\lambda_2v_1)^T=(v_2^TAv_1)^T=(v_2^T\\lambda_1v_1)^T$\n\nFrom the equation 4 we have: \n$v_2^T\\lambda_1v_1 = v_2^T\\lambda_2v_1$ \n$(\\lambda_1-\\lambda_2)v_2^Tv_1= \\vec0$\n\nThere two situations here:\n- if $\\lambda_1<>\\lambda_2$, $v_2^Tv_1$ has be to zero.\n- if $\\lambda_1 == \\lambda_2$, since the eigenvectors are in the null space of $(A-\\lambda I)$, when one eigenvector corresponds to multiple eigenvectors, null space $N(A-\\lambda I)$ is not linear independent. In this case, we have infinite many choices for those eigenvectors and we can always choose them to be orthogonal.",
"_____no_output_____"
],
[
"## 2. Symmetric matrice are always diagonalizable if its eigenvalues are distinct\n\nSuppose symmetric matrix $A$ has distinct eigenvalues, it is linearly independent or invertable. Then we have $V^{-1}AV = \\Lambda$, where the matrix $V$ contains all the eigenvectors of $A$ and $\\Lambda$ is a diagonal matrix with $A$'s eigenvalues filled in the \ndiagonal. \n**Proof**: \n1. $AV=V\\Lambda$ \n2. $V^{-1}AV=V^{-1}V\\Lambda=\\Lambda$, $V$ is invertable because eigenvectors of symmetric are orthogonal to each other.\n",
"_____no_output_____"
],
[
"# Summary\n\nSymetric matrice are important in mahine learning. For instance the covariance matrice is symmetric. In this post I listed some properties of symmetric matric and also gave the proofs.",
"_____no_output_____"
],
[
"# Reference\n\nSho Nakagome. (2018). [Linear Algebra — Part 6: eigenvalues and eigenvectors](https://towardsdatascience.com/the-properties-and-application-of-symmetric-matrice-1dc3f183de5a)",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
ecf502276c0d3181e2e40635a7639b5254ddeaa7 | 4,645 | ipynb | Jupyter Notebook | 00_getting_started/0. before_you_get_started.ipynb | jorana/RP2020 | e189f4ccb7574431ea899e6731b8cb5beb0b2915 | [
"MIT"
] | null | null | null | 00_getting_started/0. before_you_get_started.ipynb | jorana/RP2020 | e189f4ccb7574431ea899e6731b8cb5beb0b2915 | [
"MIT"
] | null | null | null | 00_getting_started/0. before_you_get_started.ipynb | jorana/RP2020 | e189f4ccb7574431ea899e6731b8cb5beb0b2915 | [
"MIT"
] | null | null | null | 27.981928 | 377 | 0.571367 | [
[
[
"# Before you get started\nAuthor:\n\nJ. Angevaare // <[email protected]> // 2020-05-25",
"_____no_output_____"
],
[
"-----------\n\n# What is this project about anyway?\n\nGood question! We have recorded an introduction for you guys so you can start with that:\n - https://drive.google.com/file/d/140Y-svJHJQeuvn8JhJMkKJjveL0PqaPa/view?usp=sharing\n \nThere are some questions in there that you want to figure out before you continue, otherwise you'll have no idea what you are doing. You may find it very helpful that we have actually nicely written down what this 'Modulation' experiment actually is. This is the experiment where also the data that you are going to look at is coming from. The paper can be found here:\n - https://arxiv.org/abs/1804.02765\n \nAfter that, you can continue doing some (essential) checks below and start with the next tutorial where we take a look at a Co-60 spectrum.\n\nFinally if you run into a question related to radioactivity and you want a __very__ complete answer try the following (rather lengthy but excellent) reference:\n - Glenn Knoll - radiation detection and measurement\n\n-----------",
"_____no_output_____"
],
[
"-----------\n## Checking the packages\nBefore we can start with actually opening files and generating spectra we need to make sure you have a working python environment to work in. To this end we first need to have to cell below working otherwise we may need to install some packages.\n\nExecute the cell below:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport uproot\nimport numba",
"_____no_output_____"
]
],
[
[
"If you get somthing like:\n```\n---------------------------------------------------------------------------\nModuleNotFoundError Traceback (most recent call last)\n<ipython-input-2-b7b06ad0c640> in <module>\n----> 1 import numba\n\nModuleNotFoundError: No module named 'numba'\n```\nwe need to install that. You can do this by for example uncommenting the relevant line below (or if you are lazy just execute all of them or the last one).",
"_____no_output_____"
]
],
[
[
"# # Uncomment whatever you need\n# !pip install pandas\n# !pip install numpy\n# !pip install uproot\n# !pip install numba\n# !pip install matplotlib",
"_____no_output_____"
],
[
"# # Or, if you are lazy do this:\n# !pip install pandas numpy uproot numba matplotlib",
"_____no_output_____"
]
],
[
[
"# Checking python version\nWe assume you have python 3.6 or higher. Check that using the lines below:",
"_____no_output_____"
]
],
[
[
"import platform\nprint(platform.python_version())",
"3.8.3\n"
],
[
"print(f'This wonderderfull line will not work in old python versions. '\n f'If it does you have a good version: v{platform.python_version()}')",
"This wonderderfull line will not work in old python versions. If it does you have a good version: v3.8.3\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
ecf5094be441463931e12e120e34cf9c9115c914 | 195,247 | ipynb | Jupyter Notebook | solve.ipynb | maciejewiczow/psi-project-captcha | bcc7955e8bd4ac7791d5c381d6362a8cf14342e1 | [
"MIT"
] | null | null | null | solve.ipynb | maciejewiczow/psi-project-captcha | bcc7955e8bd4ac7791d5c381d6362a8cf14342e1 | [
"MIT"
] | null | null | null | solve.ipynb | maciejewiczow/psi-project-captcha | bcc7955e8bd4ac7791d5c381d6362a8cf14342e1 | [
"MIT"
] | null | null | null | 762.683594 | 74,074 | 0.849468 | [
[
[
"import cv2\nimport tensorflow as tf\nfrom os import path\nimport os\nimport random\nimport numpy as np\nfrom matplotlib import pyplot as plt\nimport model\nfrom model import make_model, get_checkpoint_dir\nfrom ModelArch import ModelArch\nfrom captchaSplitter import getRegionsFromImage, readImage\nimport importlib",
"_____no_output_____"
],
[
"importlib.reload(model)",
"_____no_output_____"
],
[
"arch = ModelArch.ARCH_3",
"_____no_output_____"
],
[
"checkpoint_path = path.join(\"checkpoints\", get_checkpoint_dir(arch), \"chkpt\")",
"_____no_output_____"
],
[
"model = make_model((50,50,3), arch)",
"_____no_output_____"
],
[
"model.load_weights(checkpoint_path)",
"_____no_output_____"
],
[
"datasetPath = \"lettersDataset_final\"",
"_____no_output_____"
],
[
"labels = [f.name for f in os.scandir(path.join(datasetPath, \"train\")) if f.is_dir()]",
"_____no_output_____"
],
[
"inputCaptchaPath = \"dataset\\\\caixa\\\\1m5ed.gif\"\n# folder = \"lettersDataset_final\\\\test\\\\n\\\\\"\n# inputCaptchaPath = path.join(folder, random.choice(os.listdir(folder)))",
"_____no_output_____"
],
[
"image = readImage(inputCaptchaPath)\nplt.imshow(image)",
"_____no_output_____"
],
[
"letters = getRegionsFromImage(image)[0]\nletters = [cv2.resize(letter, (50,50)) for letter in letters] \nletters = [cv2.cvtColor(gray, cv2.COLOR_GRAY2RGB) for gray in letters] ",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(nrows=1, ncols=len(letters))\nfig.set_figwidth(25)\n\nfor (i, letter) in enumerate(letters):\n ax[i].imshow(letter)",
"_____no_output_____"
],
[
"predictions = model.predict(np.stack(letters))",
"_____no_output_____"
],
[
"labeledPredictions = [zip(labels, prediction) for prediction in predictions]\npredictedLetters = [max(prediction, key=lambda x: x[1]) for prediction in labeledPredictions]",
"_____no_output_____"
],
[
"text = \"\".join([letter for (letter, confidence) in predictedLetters])\nconfidences = [confidence for (l, confidence) in predictedLetters]",
"_____no_output_____"
],
[
"print(text)\nprint(confidences)",
"1m5ed\n[0.98699456, 0.99670583, 0.999995, 0.9950848, 0.9995079]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
ecf51407d7daa9d9b1b158b5d4404bf01accefd5 | 19,099 | ipynb | Jupyter Notebook | experiments/testing/.ipynb_checkpoints/ROLO_network_test_all-checkpoint.ipynb | AloshkaD/tracker_for_autonomous_robots | 226fa82d8ee7bb9106b840c3438a300cba85918e | [
"Apache-2.0"
] | null | null | null | experiments/testing/.ipynb_checkpoints/ROLO_network_test_all-checkpoint.ipynb | AloshkaD/tracker_for_autonomous_robots | 226fa82d8ee7bb9106b840c3438a300cba85918e | [
"Apache-2.0"
] | null | null | null | experiments/testing/.ipynb_checkpoints/ROLO_network_test_all-checkpoint.ipynb | AloshkaD/tracker_for_autonomous_robots | 226fa82d8ee7bb9106b840c3438a300cba85918e | [
"Apache-2.0"
] | null | null | null | 47.7475 | 1,158 | 0.544426 | [
[
[
"# Copyright (c) <2016> <GUANGHAN NING>. All Rights Reserved.\n \n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n\n# http://www.apache.org/licenses/LICENSE-2.0\n\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License. \n\n'''\nScript File: ROLO_network_test_all.py\n\nDescription:\n\n\tROLO is short for Recurrent YOLO, aimed at simultaneous object detection and tracking\n\tPaper: http://arxiv.org/abs/1607.05781\n\tAuthor: Guanghan Ning\n\tWebpage: http://guanghan.info/\n'''\n\n# Imports\nimport sys\npath_to_utils = '/home/a/SDC/defence/ROLO/utils'\nsys.path.extend([path_to_utils])\nimport ROLO_utils as utils\n\nimport tensorflow as tf\n#from tensorflow.models.rnn import rnn, rnn_cell\nfrom tensorflow.contrib import rnn\nfrom tensorflow.python.ops.nn import rnn_cell\nfrom tensorflow.core.protobuf import saver_pb2\nimport cv2\n\nimport numpy as np\nimport os.path\nimport time\nimport random",
"_____no_output_____"
],
[
"# Copyright (c) <2016> <GUANGHAN NING>. All Rights Reserved.\n \n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n\n# http://www.apache.org/licenses/LICENSE-2.0\n\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License. \n\n'''\nScript File: ROLO_network_test_all.py\n\nDescription:\n\n\tROLO is short for Recurrent YOLO, aimed at simultaneous object detection and tracking\n\tPaper: http://arxiv.org/abs/1607.05781\n\tAuthor: Guanghan Ning\n\tWebpage: http://guanghan.info/\n'''\nimport sys\n# Imports\npath_to_utils = '/home/a/SDC/defence/ROLO/utils'\nsys.path.extend([path_to_utils])\nimport ROLO_utils as utils\n\nimport tensorflow as tf\n#from tensorflow.models.rnn import rnn, rnn_cell\nfrom tensorflow.contrib import rnn\nfrom tensorflow.python.ops.nn import rnn_cell\nfrom tensorflow.core.protobuf import saver_pb2\nimport cv2\n\nimport numpy as np\nimport os.path\nimport time\nimport random\n\n\nclass ROLO_TF:\n disp_console = True\n restore_weights = True#False\n\n # YOLO parameters\n fromfile = None\n tofile_img = 'test/output.jpg'\n tofile_txt = 'test/output.txt'\n imshow = True\n filewrite_img = False\n filewrite_txt = False\n disp_console = True\n yolo_weights_file = '/weights/YOLO_small.ckpt'\n alpha = 0.1\n threshold = 0.2\n iou_threshold = 0.5\n num_class = 20\n num_box = 2\n grid_size = 7\n classes = [\"aeroplane\", \"bicycle\", \"bird\", \"boat\", \"bottle\", \"bus\", \"car\", \"cat\", \"chair\", \"cow\", \"diningtable\", \"dog\", \"horse\", \"motorbike\", \"person\", \"pottedplant\", \"sheep\", \"sofa\", \"train\",\"tvmonitor\"]\n w_img, h_img = [352, 240]\n\n # ROLO Network Parameters\n rolo_weights_file = 'null'\n # rolo_weights_file = '/u03/Guanghan/dev/ROLO-dev/model_dropout_30.ckpt'\n lstm_depth = 3\n num_steps = 3 # number of frames as an input sequence\n num_feat = 4096\n num_predict = 6 # final output of LSTM 6 loc parameters\n num_gt = 4\n num_input = num_feat + num_predict # data input: 4096+6= 5002\n\n # ROLO Parameters\n batch_size = 1\n display_step = 1\n\n # tf Graph input\n x = tf.placeholder(\"float32\", [None, num_steps, num_input])\n istate = tf.placeholder(\"float32\", [None, 2*num_input]) #state & cell => 2x num_input\n y = tf.placeholder(\"float32\", [None, num_gt])\n\n # Define weights\n weights = {\n 'out': tf.Variable(tf.random_normal([num_input, num_predict]))\n }\n biases = {\n 'out': tf.Variable(tf.random_normal([num_predict]))\n }\n\n\n def __init__(self,argvs = []):\n print(\"ROLO init\")\n self.ROLO(argvs)\n def LSTM_single(self, name, _X, _istate, _weights, _biases):\n with tf.device('/gpu:0'):\n # input shape: (batch_size, n_steps, n_input)\n _X = tf.transpose(_X, [1, 0, 2]) # permute num_steps and batch_size\n # Reshape to prepare input to hidden activation\n _X = tf.reshape(_X, [self.num_steps * self.batch_size, self.num_input]) # (num_steps*batch_size, num_input)\n # Split data because rnn cell needs a list of inputs for the RNN inner loop\n #_X = tf.split(0, self.num_steps, _X) # n_steps * (batch_size, num_input)\n _X = tf.split(_X, self.num_steps , 0) # n_steps * (batch_size, num_input)\n \n cell = tf.nn.rnn_cell.LSTMCell(self.num_input, self.num_input)\n #cell = tf.contrib.rnn.LSTMCell(self.num_input, state_is_tuple=False)\n #print(_X.shape)\n state = _istate\n\n \n for step in range(self.num_steps):\n outputs, state = tf.nn.rnn(cell, [_X[step]], state)\n #outputs, state = tf.contrib.rnn.static_rnn(cell, [_X [step] ], state, dtype=tf.float32)\n tf.get_variable_scope().reuse_variables()\n return outputs\n # Experiment with dropout\n def dropout_features(self, feature, prob):\n num_drop = int(prob * 4096)\n drop_index = random.sample(xrange(4096), num_drop)\n for i in range(len(drop_index)):\n index = drop_index[i]\n feature[index] = 0\n return feature\n '''---------------------------------------------------------------------------------------'''\n def build_networks(self):\n if self.disp_console : print (\"Building ROLO graph...\")\n # Build rolo layers\n self.lstm_module = self.LSTM_single('lstm_test', self.x, self.istate, self.weights, self.biases)\n self.ious= tf.Variable(tf.zeros([self.batch_size]), name=\"ious\")\n self.sess = tf.Session()\n self.sess.run(tf.initialize_all_variables())\n #self.saver = tf.train.Saver()\n self.saver = tf.train.Saver(write_version = saver_pb2.SaverDef.V1)\n #self.saver.restore(self.sess, self.rolo_weights_file)\n if self.disp_console : print (\"Loading complete!\" + '\\n')\n\n\n def testing(self, x_path, y_path):\n total_loss = 0\n # Use rolo_input for LSTM training\n pred = self.LSTM_single('lstm_train', self.x, self.istate, self.weights, self.biases)\n #print(\"pred: \", pred)\n self.pred_location = pred[0][:, 4097:4101]\n #print(\"pred_location: \", self.pred_location)\n #print(\"self.y: \", self.y)\n self.correct_prediction = tf.square(self.pred_location - self.y)\n #print(\"self.correct_prediction: \", self.correct_prediction)\n self.accuracy = tf.reduce_mean(self.correct_prediction) * 100\n #print(\"self.accuracy: \", self.accuracy)\n #optimizer = tf.train.AdamOptimizer(learning_rate=self.learning_rate).minimize(self.accuracy) # Adam Optimizer\n\n # Initializing the variables\n init = tf.initialize_all_variables()\n # Launch the graph\n with tf.Session() as sess:\n\n if (self.restore_weights == True):\n sess.run(init)\n self.saver.restore(sess, self.rolo_weights_file)\n print (\"Loading complete!\" + '\\n')\n else:\n sess.run(init)\n\n id = 0 #don't change this\n total_time = 0.0\n #id= 1\n\n # Keep training until reach max iterations\n while id < self.testing_iters - self.num_steps:\n # Load training data & ground truth\n batch_xs = self.rolo_utils.load_yolo_output_test(x_path, self.batch_size, self.num_steps, id) # [num_of_examples, num_input] (depth == 1)\n\n # Apply dropout to batch_xs\n #for item in range(len(batch_xs)):\n # batch_xs[item] = self.dropout_features(batch_xs[item], 0.4)\n\n batch_ys = self.rolo_utils.load_rolo_gt_test(y_path, self.batch_size, self.num_steps, id)\n batch_ys = utils.locations_from_0_to_1(self.w_img, self.h_img, batch_ys)\n\n # Reshape data to get 3 seq of 5002 elements\n batch_xs = np.reshape(batch_xs, [self.batch_size, self.num_steps, self.num_input])\n batch_ys = np.reshape(batch_ys, [self.batch_size, 4])\n #print(\"Batch_ys: \", batch_ys)\n\n start_time = time.time()\n pred_location= sess.run(self.pred_location,feed_dict={self.x: batch_xs, self.y: batch_ys, self.istate: np.zeros((self.batch_size, 2*self.num_input))})\n cycle_time = time.time() - start_time\n total_time += cycle_time\n\n #print(\"ROLO Pred: \", pred_location)\n #print(\"len(pred) = \", len(pred_location))\n #print(\"ROLO Pred in pixel: \", pred_location[0][0]*self.w_img, pred_location[0][1]*self.h_img, pred_location[0][2]*self.w_img, pred_location[0][3]*self.h_img)\n #print(\"correct_prediction int: \", (pred_location + 0.1).astype(int))\n\n # Save pred_location to file\n utils.save_rolo_output_test(self.output_path, pred_location, id, self.num_steps, self.batch_size)\n\n #sess.run(optimizer, feed_dict={self.x: batch_xs, self.y: batch_ys, self.istate: np.zeros((self.batch_size, 2*self.num_input))})\n\n if id % self.display_step == 0:\n # Calculate batch loss\n loss = sess.run(self.accuracy, feed_dict={self.x: batch_xs, self.y: batch_ys, self.istate: np.zeros((self.batch_size, 2*self.num_input))})\n #print \"Iter \" + str(id*self.batch_size) + \", Minibatch Loss= \" + \"{:.6f}\".format(loss) #+ \"{:.5f}\".format(self.accuracy)\n total_loss += loss\n id += 1\n #print(id)\n\n #print \"Testing Finished!\"\n avg_loss = total_loss/id\n print (\"Avg loss: \" + str(avg_loss))\n print (\"Time Spent on Tracking: \" + str(total_time))\n print (\"fps: \" + str(id/total_time))\n #save_path = self.saver.save(sess, self.rolo_weights_file)\n #print(\"Model saved in file: %s\" % save_path)\n\n return None\n\n\n def ROLO(self, argvs):\n\n self.rolo_utils= utils.ROLO_utils()\n self.rolo_utils.loadCfg()\n self.params = self.rolo_utils.params\n\n arguments = self.rolo_utils.argv_parser(argvs)\n\n if self.rolo_utils.flag_train is True:\n self.training(utils.x_path, utils.y_path)\n elif self.rolo_utils.flag_track is True:\n self.build_networks()\n self.track_from_file(utils.file_in_path)\n elif self.rolo_utils.flag_detect is True:\n self.build_networks()\n self.detect_from_file(utils.file_in_path)\n else:\n print (\"Default: running ROLO test.\")\n self.build_networks()\n\n evaluate_st = 0\n evaluate_ed = 29\n\n for test in range(evaluate_st, evaluate_ed + 1):\n\n [self.w_img, self.h_img, sequence_name, dummy_1, self.testing_iters] = utils.choose_video_sequence(test)\n\n x_path = os.path.join('benchmark/DATA', sequence_name, 'yolo_out/')\n y_path = os.path.join('benchmark/DATA', sequence_name, 'groundtruth_rect.txt')\n self.output_path = os.path.join('benchmark/DATA', sequence_name, 'rolo_out_test/')\n utils.createFolder(self.output_path)\n\n #self.rolo_weights_file = '/u03/Guanghan/dev/ROLO-dev/output/ROLO_model/model_nodrop_30_2.ckpt' #no dropout\n #self.rolo_weights_file = '/u03/Guanghan/dev/ROLO-dev/output/ROLO_model/model_online.ckpt'\n #self.rolo_weights_file = '/u03/Guanghan/dev/ROLO-dev/output/MOLO/model_MOT.ckpt'\n #self.rolo_weights_file = '/u03/Guanghan/dev/ROLO-dev/output/MOLO/model_MOT_0.2.ckpt'\n\n #self.rolo_weights_file= '/u03/Guanghan/dev/ROLO-dev/output/ROLO_model/model_step6_exp0.ckpt'\n #self.rolo_weights_file= '/u03/Guanghan/dev/ROLO-dev/output/ROLO_model/model_step3_exp1.ckpt'\n #self.rolo_weights_file= '/u03/Guanghan/dev/ROLO-dev/output/ROLO_model/model_step6_exp2.ckpt'\n\n #self.rolo_weights_file= '/u03/Guanghan/dev/ROLO-dev/output/ROLO_model/model_step3_exp2.ckpt'\n #self.rolo_weights_file= '/u03/Guanghan/dev/ROLO-dev/output/ROLO_model/model_step9_exp2.ckpt'\n #self.rolo_weights_file= '/u03/Guanghan/dev/ROLO-dev/output/ROLO_model/model_step1_exp2.ckpt'\n\n self.rolo_weights_file= '/u03/Guanghan/dev/ROLO-dev/output/ROLO_model/model_step3_exp1_old.ckpt'\n\n self.num_steps = 3 # number of frames as an input sequence\n print(\"TESTING ROLO on video sequence: \", sequence_name)\n self.testing(x_path, y_path)\n\n\n '''----------------------------------------main-----------------------------------------------------'''\ndef main(argvs):\n ROLO_TF(argvs)\n\n\nif __name__=='__main__':\n main(' ')\n\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
ecf51f91547630468b501e11f660e570d40524e1 | 11,287 | ipynb | Jupyter Notebook | components/gcp/ml_engine/deploy/sample.ipynb | civitaspo/pipelines | 7276711700ee93ec0d7490523313bdad840acb83 | [
"Apache-2.0"
] | null | null | null | components/gcp/ml_engine/deploy/sample.ipynb | civitaspo/pipelines | 7276711700ee93ec0d7490523313bdad840acb83 | [
"Apache-2.0"
] | 484 | 2021-01-21T06:49:17.000Z | 2022-03-23T01:21:24.000Z | components/gcp/ml_engine/deploy/sample.ipynb | civitaspo/pipelines | 7276711700ee93ec0d7490523313bdad840acb83 | [
"Apache-2.0"
] | null | null | null | 39.883392 | 373 | 0.598476 | [
[
[
"# Name\n\nDeploying a trained model to Cloud Machine Learning Engine \n\n\n# Label\n\nCloud Storage, Cloud ML Engine, Kubeflow, Pipeline\n\n\n# Summary\n\nA Kubeflow Pipeline component to deploy a trained model from a Cloud Storage location to Cloud ML Engine.\n\n\n# Details\n\n\n## Intended use\n\nUse the component to deploy a trained model to Cloud ML Engine. The deployed model can serve online or batch predictions in a Kubeflow Pipeline.\n\n\n## Runtime arguments\n\n| Argument | Description | Optional | Data type | Accepted values | Default |\n|--------------------------|---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|----------|--------------|-----------------|---------|\n| model_uri | The URI of a Cloud Storage directory that contains a trained model file.<br/> Or <br/> An [Estimator export base directory](https://www.tensorflow.org/guide/saved_model#perform_the_export) that contains a list of subdirectories named by timestamp. The directory with the latest timestamp is used to load the trained model file. | No | GCSPath | | |\n| project_id | The ID of the Google Cloud Platform (GCP) project of the serving model. | No | GCPProjectID | | |\n| model_id | The name of the trained model. | Yes | String | | None |\n| version_id | The name of the version of the model. If it is not provided, the operation uses a random name. | Yes | String | | None |\n| runtime_version | The Cloud ML Engine runtime version to use for this deployment. If it is not provided, the default stable version, 1.0, is used. | Yes | String | | None |\n| python_version | The version of Python used in the prediction. If it is not provided, version 2.7 is used. You can use Python 3.5 if runtime_version is set to 1.4 or above. Python 2.7 works with all supported runtime versions. | Yes | String | | 2.7 |\n| model | The JSON payload of the new [model](https://cloud.google.com/ml-engine/reference/rest/v1/projects.models). | Yes | Dict | | None |\n| version | The new [version](https://cloud.google.com/ml-engine/reference/rest/v1/projects.models.versions) of the trained model. | Yes | Dict | | None |\n| replace_existing_version | Indicates whether to replace the existing version in case of a conflict (if the same version number is found.) | Yes | Boolean | | FALSE |\n| set_default | Indicates whether to set the new version as the default version in the model. | Yes | Boolean | | FALSE |\n| wait_interval | The number of seconds to wait in case the operation has a long run time. | Yes | Integer | | 30 |\n\n\n\n## Input data schema\n\nThe component looks for a trained model in the location specified by the `model_uri` runtime argument. The accepted trained models are:\n\n\n* [Tensorflow SavedModel](https://cloud.google.com/ml-engine/docs/tensorflow/exporting-for-prediction) \n* [Scikit-learn & XGBoost model](https://cloud.google.com/ml-engine/docs/scikit/exporting-for-prediction)\n\nThe accepted file formats are:\n\n* *.pb\n* *.pbtext\n* model.bst\n* model.joblib\n* model.pkl\n\n`model_uri` can also be an [Estimator export base directory, ](https://www.tensorflow.org/guide/saved_model#perform_the_export)which contains a list of subdirectories named by timestamp. The directory with the latest timestamp is used to load the trained model file.\n\n## Output\n| Name | Description | Type |\n|:------- |:---- | :--- |\n| job_id | The ID of the created job. | String |\n| job_dir | The Cloud Storage path that contains the trained model output files. | GCSPath |\n\n\n## Cautions & requirements\n\nTo use the component, you must:\n\n* [Set up the cloud environment](https://cloud.google.com/ml-engine/docs/tensorflow/getting-started-training-prediction#setup).\n* The component can authenticate to GCP. Refer to [Authenticating Pipelines to GCP](https://www.kubeflow.org/docs/gke/authentication-pipelines/) for details.\n* Grant read access to the Cloud Storage bucket that contains the trained model to the Kubeflow user service account.\n\n## Detailed description\n\nUse the component to: \n* Locate the trained model at the Cloud Storage location you specify.\n* Create a new model if a model provided by you doesn’t exist.\n* Delete the existing model version if `replace_existing_version` is enabled.\n* Create a new version of the model from the trained model.\n* Set the new version as the default version of the model if `set_default` is enabled.\n\nFollow these steps to use the component in a pipeline:\n\n1. Install the Kubeflow Pipeline SDK:\n\n",
"_____no_output_____"
]
],
[
[
"%%capture --no-stderr\n\n!pip3 install kfp --upgrade",
"_____no_output_____"
]
],
[
[
"2. Load the component using KFP SDK",
"_____no_output_____"
]
],
[
[
"import kfp.components as comp\n\nmlengine_deploy_op = comp.load_component_from_url(\n 'https://raw.githubusercontent.com/kubeflow/pipelines/1.4.0-rc.1/components/gcp/ml_engine/deploy/component.yaml')\nhelp(mlengine_deploy_op)",
"_____no_output_____"
]
],
[
[
"### Sample\nNote: The following sample code works in IPython notebook or directly in Python code.\n\nIn this sample, you deploy a pre-built trained model from `gs://ml-pipeline-playground/samples/ml_engine/census/trained_model/` to Cloud ML Engine. The deployed model is `kfp_sample_model`. A new version is created every time the sample is run, and the latest version is set as the default version of the deployed model.\n\n#### Set sample parameters",
"_____no_output_____"
]
],
[
[
"# Required Parameters\nPROJECT_ID = '<Please put your project ID here>'\n\n# Optional Parameters\nEXPERIMENT_NAME = 'CLOUDML - Deploy'\nTRAINED_MODEL_PATH = 'gs://ml-pipeline-playground/samples/ml_engine/census/trained_model/'",
"_____no_output_____"
]
],
[
[
"#### Example pipeline that uses the component",
"_____no_output_____"
]
],
[
[
"import kfp.dsl as dsl\nimport json\[email protected](\n name='CloudML deploy pipeline',\n description='CloudML deploy pipeline'\n)\ndef pipeline(\n model_uri = 'gs://ml-pipeline-playground/samples/ml_engine/census/trained_model/',\n project_id = PROJECT_ID,\n model_id = 'kfp_sample_model',\n version_id = '',\n runtime_version = '1.10',\n python_version = '',\n version = {},\n replace_existing_version = 'False',\n set_default = 'True',\n wait_interval = '30'):\n task = mlengine_deploy_op(\n model_uri=model_uri, \n project_id=project_id, \n model_id=model_id, \n version_id=version_id, \n runtime_version=runtime_version, \n python_version=python_version,\n version=version, \n replace_existing_version=replace_existing_version, \n set_default=set_default, \n wait_interval=wait_interval)",
"_____no_output_____"
]
],
[
[
"#### Compile the pipeline",
"_____no_output_____"
]
],
[
[
"pipeline_func = pipeline\npipeline_filename = pipeline_func.__name__ + '.zip'\nimport kfp.compiler as compiler\ncompiler.Compiler().compile(pipeline_func, pipeline_filename)",
"_____no_output_____"
]
],
[
[
"#### Submit the pipeline for execution",
"_____no_output_____"
]
],
[
[
"#Specify pipeline argument values\narguments = {}\n\n#Get or create an experiment and submit a pipeline run\nimport kfp\nclient = kfp.Client()\nexperiment = client.create_experiment(EXPERIMENT_NAME)\n\n#Submit a pipeline run\nrun_name = pipeline_func.__name__ + ' run'\nrun_result = client.run_pipeline(experiment.id, run_name, pipeline_filename, arguments)",
"_____no_output_____"
]
],
[
[
"## References\n* [Component python code](https://github.com/kubeflow/pipelines/blob/master/components/gcp/container/component_sdk/python/kfp_component/google/ml_engine/_deploy.py)\n* [Component docker file](https://github.com/kubeflow/pipelines/blob/master/components/gcp/container/Dockerfile)\n* [Sample notebook](https://github.com/kubeflow/pipelines/blob/master/components/gcp/ml_engine/deploy/sample.ipynb)\n* [Cloud Machine Learning Engine Model REST API](https://cloud.google.com/ml-engine/reference/rest/v1/projects.models)\n* [Cloud Machine Learning Engine Version REST API](https://cloud.google.com/ml-engine/reference/rest/v1/projects.versions)\n\n## License\nBy deploying or using this software you agree to comply with the [AI Hub Terms of Service](https://aihub.cloud.google.com/u/0/aihub-tos) and the [Google APIs Terms of Service](https://developers.google.com/terms/). To the extent of a direct conflict of terms, the AI Hub Terms of Service will control.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
ecf5268ef4749a752580dce76d2b65ce5df831b1 | 204,243 | ipynb | Jupyter Notebook | d2l-en/mxnet/chapter_appendix-mathematics-for-deep-learning/single-variable-calculus.ipynb | gr8khan/d2lai | 7c10432f38c80e86978cd075d0024902b47842a0 | [
"MIT"
] | null | null | null | d2l-en/mxnet/chapter_appendix-mathematics-for-deep-learning/single-variable-calculus.ipynb | gr8khan/d2lai | 7c10432f38c80e86978cd075d0024902b47842a0 | [
"MIT"
] | null | null | null | d2l-en/mxnet/chapter_appendix-mathematics-for-deep-learning/single-variable-calculus.ipynb | gr8khan/d2lai | 7c10432f38c80e86978cd075d0024902b47842a0 | [
"MIT"
] | null | null | null | 44.046366 | 888 | 0.490759 | [
[
[
"# Single Variable Calculus\n:label:`sec_single_variable_calculus`\n\nIn :numref:`sec_calculus`, we saw the basic elements of differential calculus. This section takes a deeper dive into the fundamentals of calculus and how we can understand and apply it in the context of machine learning.\n\n## Differential Calculus\nDifferential calculus is fundamentally the study of how functions behave under small changes. To see why this is so core to deep learning, let us consider an example.\n\nSuppose that we have a deep neural network where the weights are, for convenience, concatenated into a single vector $\\mathbf{w} = (w_1, \\ldots, w_n)$. Given a training dataset, we consider the loss of our neural network on this dataset, which we will write as $\\mathcal{L}(\\mathbf{w})$. \n\nThis function is extraordinarily complex, encoding the performance of all possible models of the given architecture on this dataset, so it is nearly impossible to tell what set of weights $\\mathbf{w}$ will minimize the loss. Thus, in practice, we often start by initializing our weights *randomly*, and then iteratively take small steps in the direction which makes the loss decrease as rapidly as possible.\n\nThe question then becomes something that on the surface is no easier: how do we find the direction which makes the weights decrease as quickly as possible? To dig into this, let us first examine the case with only a single weight: $L(\\mathbf{w}) = L(x)$ for a single real value $x$. \n\nLet us take $x$ and try to understand what happens when we change it by a small amount to $x + \\epsilon$. If you wish to be concrete, think a number like $\\epsilon = 0.0000001$. To help us visualize what happens, let us graph an example function, $f(x) = \\sin(x^x)$, over the $[0, 3]$.\n",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nfrom d2l import mxnet as d2l\nfrom IPython import display\nfrom mxnet import np, npx\nnpx.set_np()\n\n# Plot a function in a normal range\nx_big = np.arange(0.01, 3.01, 0.01)\nys = np.sin(x_big**x_big)\nd2l.plot(x_big, ys, 'x', 'f(x)')",
"_____no_output_____"
]
],
[
[
"At this large scale, the function's behavior is not simple. However, if we reduce our range to something smaller like $[1.75,2.25]$, we see that the graph becomes much simpler.\n",
"_____no_output_____"
]
],
[
[
"# Plot a the same function in a tiny range\nx_med = np.arange(1.75, 2.25, 0.001)\nys = np.sin(x_med**x_med)\nd2l.plot(x_med, ys, 'x', 'f(x)')",
"_____no_output_____"
]
],
[
[
"Taking this to an extreme, if we zoom into a tiny segment, the behavior becomes far simpler: it is just a straight line.\n",
"_____no_output_____"
]
],
[
[
"# Plot a the same function in a tiny range\nx_small = np.arange(2.0, 2.01, 0.0001)\nys = np.sin(x_small**x_small)\nd2l.plot(x_small, ys, 'x', 'f(x)')",
"_____no_output_____"
]
],
[
[
"This is the key observation of single variable calculus: the behavior of familiar functions can be modeled by a line in a small enough range. This means that for most functions, it is reasonable to expect that as we shift the $x$ value of the function by a little bit, the output $f(x)$ will also be shifted by a little bit. The only question we need to answer is, \"How large is the change in the output compared to the change in the input? Is it half as large? Twice as large?\"\n\nThus, we can consider the ratio of the change in the output of a function for a small change in the input of the function. We can write this formally as\n\n$$\n\\frac{L(x+\\epsilon) - L(x)}{(x+\\epsilon) - x} = \\frac{L(x+\\epsilon) - L(x)}{\\epsilon}.\n$$\n\nThis is already enough to start to play around with in code. For instance, suppose that we know that $L(x) = x^{2} + 1701(x-4)^3$, then we can see how large this value is at the point $x = 4$ as follows.\n",
"_____no_output_____"
]
],
[
[
"# Define our function\ndef L(x):\n return x**2 + 1701*(x-4)**3\n\n# Print the difference divided by epsilon for several epsilon\nfor epsilon in [0.1, 0.001, 0.0001, 0.00001]:\n print(f'epsilon = {epsilon:.5f} -> {(L(4+epsilon) - L(4)) / epsilon:.5f}')",
"epsilon = 0.10000 -> 25.11000\nepsilon = 0.00100 -> 8.00270\nepsilon = 0.00010 -> 8.00012\nepsilon = 0.00001 -> 8.00001\n"
]
],
[
[
"Now, if we are observant, we will notice that the output of this number is suspiciously close to $8$. Indeed, if we decrease $\\epsilon$, we will see value becomes progressively closer to $8$. Thus we may conclude, correctly, that the value we seek (the degree a change in the input changes the output) should be $8$ at the point $x=4$. The way that a mathematician encodes this fact is\n\n$$\n\\lim_{\\epsilon \\rightarrow 0}\\frac{L(4+\\epsilon) - L(4)}{\\epsilon} = 8.\n$$\n\nAs a bit of a historical digression: in the first few decades of neural network research, scientists used this algorithm (the *method of finite differences*) to evaluate how a loss function changed under small perturbation: just change the weights and see how the loss changed. This is computationally inefficient, requiring two evaluations of the loss function to see how a single change of one variable influenced the loss. If we tried to do this with even a paltry few thousand parameters, it would require several thousand evaluations of the network over the entire dataset! It was not solved until 1986 that the *backpropagation algorithm* introduced in :cite:`Rumelhart.Hinton.Williams.ea.1988` provided a way to calculate how *any* change of the weights together would change the loss in the same computation time as a single prediction of the network over the dataset.\n\nBack in our example, this value $8$ is different for different values of $x$, so it makes sense to define it as a function of $x$. More formally, this value dependent rate of change is referred to as the *derivative* which is written as\n\n$$\\frac{df}{dx}(x) = \\lim_{\\epsilon \\rightarrow 0}\\frac{f(x+\\epsilon) - f(x)}{\\epsilon}.$$\n:eqlabel:`eq_der_def`\n\nDifferent texts will use different notations for the derivative. For instance, all of the below notations indicate the same thing:\n\n$$\n\\frac{df}{dx} = \\frac{d}{dx}f = f' = \\nabla_xf = D_xf = f_x.\n$$\n\nMost authors will pick a single notation and stick with it, however even that is not guaranteed. It is best to be familiar with all of these. We will use the notation $\\frac{df}{dx}$ throughout this text, unless we want to take the derivative of a complex expression, in which case we will use $\\frac{d}{dx}f$ to write expressions like\n$$\n\\frac{d}{dx}\\left[x^4+\\cos\\left(\\frac{x^2+1}{2x-1}\\right)\\right].\n$$\n\nOftentimes, it is intuitively useful to unravel the definition of derivative :eqref:`eq_der_def` again to see how a function changes when we make a small change of $x$:\n\n$$\\begin{aligned} \\frac{df}{dx}(x) = \\lim_{\\epsilon \\rightarrow 0}\\frac{f(x+\\epsilon) - f(x)}{\\epsilon} & \\implies \\frac{df}{dx}(x) \\approx \\frac{f(x+\\epsilon) - f(x)}{\\epsilon} \\\\ & \\implies \\epsilon \\frac{df}{dx}(x) \\approx f(x+\\epsilon) - f(x) \\\\ & \\implies f(x+\\epsilon) \\approx f(x) + \\epsilon \\frac{df}{dx}(x). \\end{aligned}$$\n:eqlabel:`eq_small_change`\n\nThe last equation is worth explicitly calling out. It tells us that if you take any function and change the input by a small amount, the output would change by that small amount scaled by the derivative.\n\nIn this way, we can understand the derivative as the scaling factor that tells us how large of change we get in the output from a change in the input.\n\n## Rules of Calculus\n:label:`sec_derivative_table`\n\nWe now turn to the task of understanding how to compute the derivative of an explicit function. A full formal treatment of calculus would derive everything from first principles. We will not indulge in this temptation here, but rather provide an understanding of the common rules encountered.\n\n### Common Derivatives\nAs was seen in :numref:`sec_calculus`, when computing derivatives one can oftentimes use a series of rules to reduce the computation to a few core functions. We repeat them here for ease of reference.\n\n* **Derivative of constants.** $\\frac{d}{dx}c = 0$.\n* **Derivative of linear functions.** $\\frac{d}{dx}(ax) = a$.\n* **Power rule.** $\\frac{d}{dx}x^n = nx^{n-1}$.\n* **Derivative of exponentials.** $\\frac{d}{dx}e^x = e^x$.\n* **Derivative of the logarithm.** $\\frac{d}{dx}\\log(x) = \\frac{1}{x}$.\n\n### Derivative Rules\nIf every derivative needed to be separately computed and stored in a table, differential calculus would be near impossible. It is a gift of mathematics that we can generalize the above derivatives and compute more complex derivatives like finding the derivative of $f(x) = \\log\\left(1+(x-1)^{10}\\right)$. As was mentioned in :numref:`sec_calculus`, the key to doing so is to codify what happens when we take functions and combine them in various ways, most importantly: sums, products, and compositions.\n\n* **Sum rule.** $\\frac{d}{dx}\\left(g(x) + h(x)\\right) = \\frac{dg}{dx}(x) + \\frac{dh}{dx}(x)$.\n* **Product rule.** $\\frac{d}{dx}\\left(g(x)\\cdot h(x)\\right) = g(x)\\frac{dh}{dx}(x) + \\frac{dg}{dx}(x)h(x)$.\n* **Chain rule.** $\\frac{d}{dx}g(h(x)) = \\frac{dg}{dh}(h(x))\\cdot \\frac{dh}{dx}(x)$.\n\nLet us see how we may use :eqref:`eq_small_change` to understand these rules. For the sum rule, consider following chain of reasoning:\n\n$$\n\\begin{aligned}\nf(x+\\epsilon) & = g(x+\\epsilon) + h(x+\\epsilon) \\\\\n& \\approx g(x) + \\epsilon \\frac{dg}{dx}(x) + h(x) + \\epsilon \\frac{dh}{dx}(x) \\\\\n& = g(x) + h(x) + \\epsilon\\left(\\frac{dg}{dx}(x) + \\frac{dh}{dx}(x)\\right) \\\\\n& = f(x) + \\epsilon\\left(\\frac{dg}{dx}(x) + \\frac{dh}{dx}(x)\\right).\n\\end{aligned}\n$$\n\nBy comparing this result with the fact that $f(x+\\epsilon) \\approx f(x) + \\epsilon \\frac{df}{dx}(x)$, we see that $\\frac{df}{dx}(x) = \\frac{dg}{dx}(x) + \\frac{dh}{dx}(x)$ as desired. The intuition here is: when we change the input $x$, $g$ and $h$ jointly contribute to the change of the output by $\\frac{dg}{dx}(x)$ and $\\frac{dh}{dx}(x)$.\n\n\nThe product is more subtle, and will require a new observation about how to work with these expressions. We will begin as before using :eqref:`eq_small_change`:\n\n$$\n\\begin{aligned}\nf(x+\\epsilon) & = g(x+\\epsilon)\\cdot h(x+\\epsilon) \\\\\n& \\approx \\left(g(x) + \\epsilon \\frac{dg}{dx}(x)\\right)\\cdot\\left(h(x) + \\epsilon \\frac{dh}{dx}(x)\\right) \\\\\n& = g(x)\\cdot h(x) + \\epsilon\\left(g(x)\\frac{dh}{dx}(x) + \\frac{dg}{dx}(x)h(x)\\right) + \\epsilon^2\\frac{dg}{dx}(x)\\frac{dh}{dx}(x) \\\\\n& = f(x) + \\epsilon\\left(g(x)\\frac{dh}{dx}(x) + \\frac{dg}{dx}(x)h(x)\\right) + \\epsilon^2\\frac{dg}{dx}(x)\\frac{dh}{dx}(x). \\\\\n\\end{aligned}\n$$\n\n\nThis resembles the computation done above, and indeed we see our answer ($\\frac{df}{dx}(x) = g(x)\\frac{dh}{dx}(x) + \\frac{dg}{dx}(x)h(x)$) sitting next to $\\epsilon$, but there is the issue of that term of size $\\epsilon^{2}$. We will refer to this as a *higher-order term*, since the power of $\\epsilon^2$ is higher than the power of $\\epsilon^1$. We will see in a later section that we will sometimes want to keep track of these, however for now observe that if $\\epsilon = 0.0000001$, then $\\epsilon^{2}= 0.0000000000001$, which is vastly smaller. As we send $\\epsilon \\rightarrow 0$, we may safely ignore the higher order terms. As a general convention in this appendix, we will use \"$\\approx$\" to denote that the two terms are equal up to higher order terms. However, if we wish to be more formal we may examine the difference quotient\n\n$$\n\\frac{f(x+\\epsilon) - f(x)}{\\epsilon} = g(x)\\frac{dh}{dx}(x) + \\frac{dg}{dx}(x)h(x) + \\epsilon \\frac{dg}{dx}(x)\\frac{dh}{dx}(x),\n$$\n\nand see that as we send $\\epsilon \\rightarrow 0$, the right hand term goes to zero as well.\n\nFinally, with the chain rule, we can again progress as before using :eqref:`eq_small_change` and see that\n\n$$\n\\begin{aligned}\nf(x+\\epsilon) & = g(h(x+\\epsilon)) \\\\\n& \\approx g\\left(h(x) + \\epsilon \\frac{dh}{dx}(x)\\right) \\\\\n& \\approx g(h(x)) + \\epsilon \\frac{dh}{dx}(x) \\frac{dg}{dh}(h(x))\\\\\n& = f(x) + \\epsilon \\frac{dg}{dh}(h(x))\\frac{dh}{dx}(x),\n\\end{aligned}\n$$\n\nwhere in the second line we view the function $g$ as having its input ($h(x)$) shifted by the tiny quantity $\\epsilon \\frac{dh}{dx}(x)$.\n\nThese rule provide us with a flexible set of tools to compute essentially any expression desired. For instance,\n\n$$\n\\begin{aligned}\n\\frac{d}{dx}\\left[\\log\\left(1+(x-1)^{10}\\right)\\right] & = \\left(1+(x-1)^{10}\\right)^{-1}\\frac{d}{dx}\\left[1+(x-1)^{10}\\right]\\\\\n& = \\left(1+(x-1)^{10}\\right)^{-1}\\left(\\frac{d}{dx}[1] + \\frac{d}{dx}[(x-1)^{10}]\\right) \\\\\n& = \\left(1+(x-1)^{10}\\right)^{-1}\\left(0 + 10(x-1)^9\\frac{d}{dx}[x-1]\\right) \\\\\n& = 10\\left(1+(x-1)^{10}\\right)^{-1}(x-1)^9 \\\\\n& = \\frac{10(x-1)^9}{1+(x-1)^{10}}.\n\\end{aligned}\n$$\n\nWhere each line has used the following rules:\n\n1. The chain rule and derivative of logarithm.\n2. The sum rule.\n3. The derivative of constants, chain rule, and power rule.\n4. The sum rule, derivative of linear functions, derivative of constants.\n\nTwo things should be clear after doing this example:\n\n1. Any function we can write down using sums, products, constants, powers, exponentials, and logarithms can have its derivate computed mechanically by following these rules.\n2. Having a human follow these rules can be tedious and error prone!\n\nThankfully, these two facts together hint towards a way forward: this is a perfect candidate for mechanization! Indeed backpropagation, which we will revisit later in this section, is exactly that.\n\n### Linear Approximation\nWhen working with derivatives, it is often useful to geometrically interpret the approximation used above. In particular, note that the equation \n\n$$\nf(x+\\epsilon) \\approx f(x) + \\epsilon \\frac{df}{dx}(x),\n$$\n\napproximates the value of $f$ by a line which passes through the point $(x, f(x))$ and has slope $\\frac{df}{dx}(x)$. In this way we say that the derivative gives a linear approximation to the function $f$, as illustrated below:\n",
"_____no_output_____"
]
],
[
[
"# Compute sin\nxs = np.arange(-np.pi, np.pi, 0.01)\nplots = [np.sin(xs)]\n\n# Compute some linear approximations. Use d(sin(x)) / dx = cos(x)\nfor x0 in [-1.5, 0, 2]:\n plots.append(np.sin(x0) + (xs - x0) * np.cos(x0))\n\nd2l.plot(xs, plots, 'x', 'f(x)', ylim=[-1.5, 1.5])",
"_____no_output_____"
]
],
[
[
"### Higher Order Derivatives\n\nLet us now do something that may on the surface seem strange. Take a function $f$ and compute the derivative $\\frac{df}{dx}$. This gives us the rate of change of $f$ at any point.\n\nHowever, the derivative, $\\frac{df}{dx}$, can be viewed as a function itself, so nothing stops us from computing the derivative of $\\frac{df}{dx}$ to get $\\frac{d^2f}{dx^2} = \\frac{df}{dx}\\left(\\frac{df}{dx}\\right)$. We will call this the second derivative of $f$. This function is the rate of change of the rate of change of $f$, or in other words, how the rate of change is changing. We may apply the derivative any number of times to obtain what is called the $n$-th derivative. To keep the notation clean, we will denote the $n$-th derivative as \n\n$$\nf^{(n)}(x) = \\frac{d^{n}f}{dx^{n}} = \\left(\\frac{d}{dx}\\right)^{n} f.\n$$\n\nLet us try to understand *why* this is a useful notion. Below, we visualize $f^{(2)}(x)$, $f^{(1)}(x)$, and $f(x)$. \n\nFirst, consider the case that the second derivative $f^{(2)}(x)$ is a positive constant. This means that the slope of the first derivative is positive. As a result, the first derivative $f^{(1)}(x)$ may start out negative, becomes zero at a point, and then becomes positive in the end. This tells us the slope of our original function $f$ and therefore, the function $f$ itself decreases, flattens out, then increases. In other words, the function $f$ curves up, and has a single minimum as is shown in :numref:`fig_positive-second`.\n\n\n:label:`fig_positive-second`\n\n\nSecond, if the second derivative is a negative constant, that means that the first derivative is decreasing. This implies the first derivative may start out positive, becomes zero at a point, and then becomes negative. Hence, the function $f$ itself increases, flattens out, then decreases. In other words, the function $f$ curves down, and has a single maximum as is shown in :numref:`fig_negative-second`.\n\n\n:label:`fig_negative-second`\n\n\nThird, if the second derivative is a always zero, then the first derivative will never change---it is constant! This means that $f$ increases (or decreases) at a fixed rate, and $f$ is itself a straight line as is shown in :numref:`fig_zero-second`.\n\n\n:label:`fig_zero-second`\n\nTo summarize, the second derivative can be interpreted as describing the way that the function $f$ curves. A positive second derivative leads to a upwards curve, while a negative second derivative means that $f$ curves downwards, and a zero second derivative means that $f$ does not curve at all.\n\nLet us take this one step further. Consider the function $g(x) = ax^{2}+ bx + c$. We can then compute that\n\n$$\n\\begin{aligned}\n\\frac{dg}{dx}(x) & = 2ax + b \\\\\n\\frac{d^2g}{dx^2}(x) & = 2a.\n\\end{aligned}\n$$\n\nIf we have some original function $f(x)$ in mind, we may compute the first two derivatives and find the values for $a, b$, and $c$ that make them match this computation. Similarly to the previous section where we saw that the first derivative gave the best approximation with a straight line, this construction provides the best approximation by a quadratic. Let us visualize this for $f(x) = \\sin(x)$.\n",
"_____no_output_____"
]
],
[
[
"# Compute sin\nxs = np.arange(-np.pi, np.pi, 0.01)\nplots = [np.sin(xs)]\n\n# Compute some quadratic approximations. Use d(sin(x)) / dx = cos(x)\nfor x0 in [-1.5, 0, 2]:\n plots.append(np.sin(x0) + (xs - x0) * np.cos(x0) -\n (xs - x0)**2 * np.sin(x0) / 2)\n\nd2l.plot(xs, plots, 'x', 'f(x)', ylim=[-1.5, 1.5])",
"_____no_output_____"
]
],
[
[
"We will extend this idea to the idea of a *Taylor series* in the next section. \n\n### Taylor Series\n\n\nThe *Taylor series* provides a method to approximate the function $f(x)$ if we are given values for the first $n$ derivatives at a point $x_0$, i.e., $\\left\\{ f(x_0), f^{(1)}(x_0), f^{(2)}(x_0), \\ldots, f^{(n)}(x_0) \\right\\}$. The idea will be to find a degree $n$ polynomial that matches all the given derivatives at $x_0$.\n\nWe saw the case of $n=2$ in the previous section and a little algebra shows this is\n\n$$\nf(x) \\approx \\frac{1}{2}\\frac{d^2f}{dx^2}(x_0)(x-x_0)^{2}+ \\frac{df}{dx}(x_0)(x-x_0) + f(x_0).\n$$\n\nAs we can see above, the denominator of $2$ is there to cancel out the $2$ we get when we take two derivatives of $x^2$, while the other terms are all zero. Same logic applies for the first derivative and the value itself.\n\nIf we push the logic further to $n=3$, we will conclude that\n\n$$\nf(x) \\approx \\frac{\\frac{d^3f}{dx^3}(x_0)}{6}(x-x_0)^3 + \\frac{\\frac{d^2f}{dx^2}(x_0)}{2}(x-x_0)^{2}+ \\frac{df}{dx}(x_0)(x-x_0) + f(x_0).\n$$\n\nwhere the $6 = 3 \\times 2 = 3!$ comes from the constant we get in front if we take three derivatives of $x^3$.\n\n\nFurthermore, we can get a degree $n$ polynomial by \n\n$$\nP_n(x) = \\sum_{i = 0}^{n} \\frac{f^{(i)}(x_0)}{i!}(x-x_0)^{i}.\n$$\n\nwhere the notation \n\n$$\nf^{(n)}(x) = \\frac{d^{n}f}{dx^{n}} = \\left(\\frac{d}{dx}\\right)^{n} f.\n$$\n\n\nIndeed, $P_n(x)$ can be viewed as the best $n$-th degree polynomial approximation to our function $f(x)$.\n\nWhile we are not going to dive all the way into the error of the above approximations, it is worth mentioning the infinite limit. In this case, for well behaved functions (known as real analytic functions) like $\\cos(x)$ or $e^{x}$, we can write out the infinite number of terms and approximate the exactly same function\n\n$$\nf(x) = \\sum_{n = 0}^\\infty \\frac{f^{(n)}(x_0)}{n!}(x-x_0)^{n}.\n$$\n\nTake $f(x) = e^{x}$ as am example. Since $e^{x}$ is its own derivative, we know that $f^{(n)}(x) = e^{x}$. Therefore, $e^{x}$ can be reconstructed by taking the Taylor series at $x_0 = 0$, i.e.,\n\n$$\ne^{x} = \\sum_{n = 0}^\\infty \\frac{x^{n}}{n!} = 1 + x + \\frac{x^2}{2} + \\frac{x^3}{6} + \\cdots.\n$$\n\nLet us see how this works in code and observe how increasing the degree of the Taylor approximation brings us closer to the desired function $e^x$.\n",
"_____no_output_____"
]
],
[
[
"# Compute the exponential function\nxs = np.arange(0, 3, 0.01)\nys = np.exp(xs)\n\n# Compute a few Taylor series approximations\nP1 = 1 + xs\nP2 = 1 + xs + xs**2 / 2\nP5 = 1 + xs + xs**2 / 2 + xs**3 / 6 + xs**4 / 24 + xs**5 / 120\n\nd2l.plot(xs, [ys, P1, P2, P5], 'x', 'f(x)', legend=[\n \"Exponential\", \"Degree 1 Taylor Series\", \"Degree 2 Taylor Series\",\n \"Degree 5 Taylor Series\"])",
"_____no_output_____"
]
],
[
[
"Taylor series have two primary applications:\n\n1. *Theoretical applications*: Often when we try to understand a too complex function, using Taylor series enables us to turn it into a polynomial that we can work with directly.\n\n2. *Numerical applications*: Some functions like $e^{x}$ or $\\cos(x)$ are difficult for machines to compute. They can store tables of values at a fixed precision (and this is often done), but it still leaves open questions like \"What is the 1000-th digit of $\\cos(1)$?\" Taylor series are often helpful to answer such questions. \n\n\n## Summary\n\n* Derivatives can be used to express how functions change when we change the input by a small amount.\n* Elementary derivatives can be combined using derivative rules to create arbitrarily complex derivatives.\n* Derivatives can be iterated to get second or higher order derivatives. Each increase in order provides more fine grained information on the behavior of the function.\n* Using information in the derivatives of a single data example, we can approximate well behaved functions by polynomials obtained from the Taylor series.\n\n\n## Exercises\n\n1. What is the derivative of $x^3-4x+1$?\n2. What is the derivative of $\\log(\\frac{1}{x})$?\n3. True or False: If $f'(x) = 0$ then $f$ has a maximum or minimum at $x$?\n4. Where is the minimum of $f(x) = x\\log(x)$ for $x\\ge0$ (where we assume that $f$ takes the limiting value of $0$ at $f(0)$)?\n",
"_____no_output_____"
],
[
"[Discussions](https://discuss.d2l.ai/t/412)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
ecf5297dacb4250205813438d7517eb2087646ad | 5,831 | ipynb | Jupyter Notebook | Coding-Ninjas-Data-Structure-and-Algorithm-in-Python-main/DP-2.ipynb | SaiPrasanth212/Coding-Ninjas-Data-Structures-and-Algorithms | 8d318e47e315a987b76abbb730bd06bfb4f8919c | [
"MIT"
] | null | null | null | Coding-Ninjas-Data-Structure-and-Algorithm-in-Python-main/DP-2.ipynb | SaiPrasanth212/Coding-Ninjas-Data-Structures-and-Algorithms | 8d318e47e315a987b76abbb730bd06bfb4f8919c | [
"MIT"
] | null | null | null | Coding-Ninjas-Data-Structure-and-Algorithm-in-Python-main/DP-2.ipynb | SaiPrasanth212/Coding-Ninjas-Data-Structures-and-Algorithms | 8d318e47e315a987b76abbb730bd06bfb4f8919c | [
"MIT"
] | null | null | null | 26.03125 | 136 | 0.474704 | [
[
[
"Min Cost Path Problem",
"_____no_output_____"
]
],
[
[
"from sys import stdin\nMAX_VALUE = 2147483647\n\ndef minCostPathHelper(input, mRows, nCols, currRow, currCol) :\n if (currRow >= mRows) or (currCol >= nCols) :\n return MAX_VALUE\n\n \n if currRow == (mRows - 1) and currCol == (nCols - 1) :\n return input[currRow][currCol]\n\n downCost = minCostPathHelper(input, mRows, nCols, (currRow + 1), currCol)\n diagonalCost = minCostPathHelper(input, mRows, nCols, (currRow + 1), (currCol + 1))\n leftCost = minCostPathHelper(input, mRows, nCols, currRow, (currCol + 1))\n\n return input[currRow][currCol] + min(diagonalCost, downCost, leftCost)\n\n\n\ndef minCostPath(input, mRows, nCols) :\n if mRows == 0 :\n return MAX_VALUE\n \n return minCostPathHelper(input, mRows, nCols, 0, 0)\n\n\n\n\ndef take2DInput() :\n li = stdin.readline().rstrip().split(\" \")\n mRows = int(li[0])\n nCols = int(li[1])\n \n if mRows == 0 :\n return list(), 0, 0\n \n mat = [list(map(int, input().strip().split(\" \"))) for row in range(mRows)]\n return mat, mRows, nCols\n\n\n#main\nmat, mRows, nCols = take2DInput()\nprint(minCostPath(mat, mRows, nCols))",
"_____no_output_____"
]
],
[
[
"LCS - Problem",
"_____no_output_____"
]
],
[
[
"from sys import stdin\n\ndef lcs(s, t) :\n m = len(s)\n n = len(t)\n \n subProblems = [[0] * (n + 1) for i in range((m + 1))]\n \n\n for currStart in range(1, (m + 1)) :\n for currEnd in range(1, (n + 1)) :\n if s[m - currStart] == t[n - currEnd] :\n subProblems[currStart][currEnd] = 1 + subProblems[currStart - 1][currEnd - 1]\n else :\n subProblems[currStart][currEnd] = max(subProblems[currStart - 1][currEnd], subProblems[currStart][currEnd - 1])\n \n return subProblems[m][n]\n \n\n\n#main\ns = str(stdin.readline().rstrip())\nt = str(stdin.readline().rstrip())\n\nprint(lcs(s, t))",
"_____no_output_____"
]
],
[
[
"0 1 Knapsack - Problem",
"_____no_output_____"
]
],
[
[
"from sys import stdin\n\ndef knapsack(weights, values, n, maxWeight) :\n if (n == 0) or (maxWeight == 0) :\n return 0\n\n if weights[n - 1] > maxWeight :\n return knapsack(weights, values, n - 1, maxWeight)\n\n includeItem = values[n - 1] + knapsack(weights, values, n - 1, maxWeight - weights[n - 1])\n\n excludeItem = knapsack(weights, values, n - 1, maxWeight)\n\n return max(includeItem, excludeItem)\n\n\n\n\ndef takeInput() :\n n = int(stdin.readline().rstrip())\n\n if n == 0 :\n return list(), list(), n, 0\n\n weights = list(map(int, stdin.readline().rstrip().split(\" \")))\n values = list(map(int, stdin.readline().rstrip().split(\" \")))\n maxWeight = int(stdin.readline().rstrip())\n\n return weights, values, n, maxWeight\n\n\n#main\nweights, values, n, maxWeight = takeInput()\n\nprint(knapsack(weights, values, n, maxWeight))",
"_____no_output_____"
]
],
[
[
"Matrix Chain Multiplication",
"_____no_output_____"
]
],
[
[
"import sys\nfrom sys import stdin\ndef mcm(p,n):\n\tn+=1\n\tm=[[sys.maxsize for i in range (0,n+1)] for j in range (0,n+1)]\n\tfor i in range (1,n):\n\t\tm[i][i]=0\n\tfor l in range (2,n):\n\t\tfor i in range (1,n-l+1):\n\t\t\tj=i+l-1\n\t\t\tfor k in range (i,j):\n\t\t\t\tq=m[i][k]+m[k+1][j]+p[i-1]*p[k]*p[j]\n\t\t\t\tif(q<m[i][j]):\n\t\t\t\t\tm[i][j]=q\n\treturn m[1][n-1]\n\nn=int(stdin.readline().strip())\np=[int(i) for i in stdin.readline().strip().split()]\nprint(mcm(p,n))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.