hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
d05b754b90486210da606a98a2611876a9431e38 | 16,937 | ipynb | Jupyter Notebook | B2-NLP/Ajay_NLP_TopicModelling.ipynb | Shreyansh-Gupta/Open-contributions | e72a9ce2b0aa6a48081921bf8138b91ad259c422 | [
"MIT"
] | 61 | 2020-09-10T05:16:19.000Z | 2021-11-07T00:22:46.000Z | B2-NLP/Ajay_NLP_TopicModelling.ipynb | Shreyansh-Gupta/Open-contributions | e72a9ce2b0aa6a48081921bf8138b91ad259c422 | [
"MIT"
] | 72 | 2020-09-12T09:34:19.000Z | 2021-08-01T17:48:46.000Z | B2-NLP/Ajay_NLP_TopicModelling.ipynb | Shreyansh-Gupta/Open-contributions | e72a9ce2b0aa6a48081921bf8138b91ad259c422 | [
"MIT"
] | 571 | 2020-09-10T01:52:56.000Z | 2022-03-26T17:26:23.000Z | 29.871252 | 163 | 0.462656 | [
[
[
"## LDA (Latent Dirichlet Allocation)\n\nIn this notebook, I'll be showing you the practical example of topic modelling using LDA.\n\nFor this I'll be using ABC news headlines dataset from kaggle - https://www.kaggle.com/therohk/million-headlines",
"_____no_output_____"
]
],
[
[
"# Let's first read the dataset\nimport pandas as pd\n\ndf = pd.read_csv(\"abcnews-date-text.csv\")\n",
"_____no_output_____"
],
[
"# Let's check the head of the dataframe\n\ndf.head()",
"_____no_output_____"
]
],
[
[
"#### Here our main focus is the headline_text column because we will be using these headlines to extract the topics.",
"_____no_output_____"
]
],
[
[
"df1 = df[:50000].drop(\"publish_date\", axis = 1)",
"_____no_output_____"
]
],
[
[
"#### Here I am taking only 50000 records.",
"_____no_output_____"
]
],
[
[
"df1.head()",
"_____no_output_____"
],
[
"# Length of the data\n\nlen(df1)",
"_____no_output_____"
]
],
[
[
"### Preprocessing",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import CountVectorizer\n\ncv = CountVectorizer(max_df = 0.95, min_df = 3, stop_words = 'english')",
"_____no_output_____"
],
[
"# Create a document term matrix\n\ndtm = cv.fit_transform(df1[0:50000]['headline_text'])",
"_____no_output_____"
],
[
"dtm",
"_____no_output_____"
]
],
[
[
"### Let's perfrom LDA\n\n***Here I'll be assuming that there are 20 topics present in this document***",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition import LatentDirichletAllocation",
"_____no_output_____"
],
[
"lda = LatentDirichletAllocation(n_components = 20, random_state = 79)",
"_____no_output_____"
],
[
"# This will take some time to execute\n\nlda.fit(dtm)",
"_____no_output_____"
],
[
"topics = lda.transform(dtm)",
"_____no_output_____"
]
],
[
[
"### Let's print 15 most common words for all the 20 topics",
"_____no_output_____"
]
],
[
[
"for index,topic in enumerate(lda.components_):\n print(f'THE TOP 15 WORDS FOR TOPIC #{index}')\n print([cv.get_feature_names()[i] for i in topic.argsort()[-15:]])\n print('\\n')",
"THE TOP 15 WORDS FOR TOPIC #0\n['row', 'sale', 'telstra', 'indigenous', 'bid', 'campaign', 'budget', 'tax', 'airport', 'bomb', 'community', 'blast', 'funding', 'boost', 'security']\n\n\nTHE TOP 15 WORDS FOR TOPIC #1\n['says', 'saddam', 'dump', 'qaeda', 'broken', 'gm', 'city', 'waste', 'israel', 'gets', 'industry', 'al', 'warns', 'hill', 'future']\n\n\nTHE TOP 15 WORDS FOR TOPIC #2\n['debate', 'merger', 'real', 'local', 'centre', 'stop', 'woes', 'seeks', 'force', 'new', 'air', 'plan', 'chief', 'work', 'council']\n\n\nTHE TOP 15 WORDS FOR TOPIC #3\n['airs', 'opposition', 'staff', 'nsw', 'support', 'east', 'rate', 'teachers', 'pay', 'gold', 'west', 'strike', 'coast', 'south', 'concerns']\n\n\nTHE TOP 15 WORDS FOR TOPIC #4\n['soldiers', 'british', 'bali', 'forces', 'victims', 'iraqi', 'israeli', 'case', 'search', 'attack', 'appeal', 'missing', 'iraq', 'killed', 'baghdad']\n\n\nTHE TOP 15 WORDS FOR TOPIC #5\n['aims', 'plant', 'children', 'downer', 'nuclear', 'begin', 'says', 'sign', 'gas', 'deal', 'urges', 'north', 'korea', 'talks', 'new']\n\n\nTHE TOP 15 WORDS FOR TOPIC #6\n['china', 'fears', 'post', 'plan', 'discuss', 'jobs', 'leaders', 'meet', 'meeting', 'job', 'workers', 'bush', 'sars', 'iraq', 'war']\n\n\nTHE TOP 15 WORDS FOR TOPIC #7\n['praises', 'coach', 'summit', 'jones', 'suicide', 'battle', 'wallabies', 'thousands', 'terrorism', 'family', 'free', 'head', 'calls', 'test', 'tour']\n\n\nTHE TOP 15 WORDS FOR TOPIC #8\n['firefighters', 'league', 'way', 'education', 'red', 'beattie', 'issues', 'blaze', 'adelaide', 'title', 'race', 'lead', 'action', 'continues', 'takes']\n\n\nTHE TOP 15 WORDS FOR TOPIC #9\n['good', 'laws', 'union', 'insurance', 'fight', 'business', 'aid', 'doctors', 'new', 'group', 'help', 'rain', 'drought', 'farmers', 'qld']\n\n\nTHE TOP 15 WORDS FOR TOPIC #10\n['poll', 'lose', 'virus', 'parliament', 'labor', 'leave', 'changes', 'sheep', 'howard', 'lions', 'residents', 'service', 'election', 'iraq', 'pm']\n\n\nTHE TOP 15 WORDS FOR TOPIC #11\n['club', 'away', 'sets', 'figures', 'title', 'farm', 'says', 'cancer', 'hopes', 'win', 'big', 'open', 'minister', 'record', 'power']\n\n\nTHE TOP 15 WORDS FOR TOPIC #12\n['indian', 'arrest', 'attack', 'alleged', 'death', 'team', 'shooting', 'arrested', 'body', 'investigate', 'murder', 'man', 'trial', 'probe', 'police']\n\n\nTHE TOP 15 WORDS FOR TOPIC #13\n['fed', 'offer', 'regional', 'restrictions', 'wa', 'rail', 'nsw', 'act', 'rejects', 'plan', 'sa', 'vic', 'urged', 'water', 'govt']\n\n\nTHE TOP 15 WORDS FOR TOPIC #14\n['illegal', 'warned', 'protesters', 'threat', 'fishing', 'services', 'abuse', 'sars', 'care', 'war', 'study', 'anti', 'protest', 'inquiry', 'home']\n\n\nTHE TOP 15 WORDS FOR TOPIC #15\n['prices', 'film', 'long', 'week', 'dollar', 'fined', 'share', 'hits', 'company', 'makes', 'year', 'hit', 'backs', 'high', 'wins']\n\n\nTHE TOP 15 WORDS FOR TOPIC #16\n['perth', 'road', 'killed', 'dead', 'jailed', 'toll', 'accident', 'woman', 'injured', 'dies', 'charged', 'death', 'car', 'crash', 'man']\n\n\nTHE TOP 15 WORDS FOR TOPIC #17\n['old', 'black', 'win', 'market', 'aussie', 'australia', 'england', 'play', 'india', 'warning', 'victory', 'pakistan', 'final', 'world', 'cup']\n\n\nTHE TOP 15 WORDS FOR TOPIC #18\n['kill', 'aust', 'train', 'hears', 'indonesian', 'says', 'aceh', 'time', 'charge', 'man', 'charges', 'faces', 'troops', 'face', 'court']\n\n\nTHE TOP 15 WORDS FOR TOPIC #19\n['need', 'launch', 'spotlight', 'report', 'highway', 'new', 'mp', 'nats', 'target', 'plan', 'building', 'weapons', 'sought', 'highlights', 'students']\n\n\n"
]
],
[
[
"### Let's combine these topics with our original headlines",
"_____no_output_____"
]
],
[
[
"df1['Headline Topic'] = topics.argmax(axis = 1)",
"_____no_output_____"
],
[
"df1.head()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05b7bf9a8344685003a827ddd19ab8141fe445c | 146,619 | ipynb | Jupyter Notebook | Visualisation_codes.ipynb | punyajoy/biosbias | bedca0b8605e3e99d2a2b56c78a5b98c9839a77b | [
"MIT"
] | null | null | null | Visualisation_codes.ipynb | punyajoy/biosbias | bedca0b8605e3e99d2a2b56c78a5b98c9839a77b | [
"MIT"
] | null | null | null | Visualisation_codes.ipynb | punyajoy/biosbias | bedca0b8605e3e99d2a2b56c78a5b98c9839a77b | [
"MIT"
] | null | null | null | 242.746689 | 129,668 | 0.916402 | [
[
[
"%load_ext autoreload\n%autoreload 2\n\nimport preprocess \n",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"preprocess.main('CC*.pkl','BIOS.pkl')",
"Loading 'CC-MAIN-2018-34-bios.pkl'\nLoading 'CC-MAIN-2017-43-bios.pkl'\nLoading 'CC-MAIN-2014-41-bios.pkl'\nLoading 'CC-MAIN-2018-13-bios.pkl'\nLoading 'CC-MAIN-2013-20-bios.pkl'\nLoading 'CC-MAIN-2016-44-bios.pkl'\nLoading 'CC-MAIN-2018-05-bios.pkl'\n82,701/119,125 'different' name+titles (69.4%)\nProcessing bios...\nWrote 82,701 bios to 'BIOS.pkl'\n"
],
[
"!ls",
"aiethics\t\t CC-MAIN-2016-44-bios.pkl\tCC-MAIN-2018-34-log.txt\r\naiethics.yml\t\t CC-MAIN-2016-44-log.txt\tdownload_bios.py\r\nazure-pipelines.yml\t CC-MAIN-2017-43-bios.pkl\tfreq_titles.json\r\nBias_in_Bios_implement.ipynb CC-MAIN-2017-43-log.txt\tLICENSE\r\nBIOS.pkl\t\t CC-MAIN-2018-05-bios.pkl\tpreprocess.py\r\nCC-MAIN-2013-20-bios.pkl CC-MAIN-2018-05-log.txt\t__pycache__\r\nCC-MAIN-2013-20-log.txt CC-MAIN-2018-13-bios.pkl\tREADME.md\r\nCC-MAIN-2014-41-bios.pkl CC-MAIN-2018-13-log.txt\trecreate.sh\r\nCC-MAIN-2014-41-log.txt CC-MAIN-2018-34-bios.pkl\tUntitled.ipynb\r\n"
],
[
"import pickle\nall_bios = pickle.load( open( \"BIOS.pkl\", \"rb\" ) )",
"_____no_output_____"
]
],
[
[
"## Dictionary Details \n\n1. r[\"title\"] tells you the noramlized title\n2. r[\"gender\"] tells you the gender (binary for simplicity, determined from the pronouns)3. \n3. r[\"start_pos\"] indicates the length of the first sentence.\n4. r[\"raw\"] has the entire bio\n5. The field r[\"bio\"] contains a scrubbed version of the bio (with the person's name and obvious gender words (like she/he removed)\n\n\n## Problem Statement \n\nSo the classification task is to predict r[\"title\"] from r[\"raw\"][r[\"start_pos\"]:]\n",
"_____no_output_____"
],
[
"#### Example Dictionary Element",
"_____no_output_____"
]
],
[
[
"test_bio = all_bios[0]\ntest_bio['bio']",
"_____no_output_____"
],
[
"test_bio['raw']",
"_____no_output_____"
]
],
[
[
"### Distribution of occupation",
"_____no_output_____"
]
],
[
[
"occupation_dict={}\nfor bio in all_bios:\n occupation=bio['title']\n try:\n occupation_dict[occupation] = 1\n except KeyError:\n occupation_dict[occupation] += 1\n",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np\n\n\nkeys = x.keys()\nvals = x.values()\n\nplt.bar(keys, np.divide(list(vals), sum(vals)), label=\"Real distribution\")\n\nplt.ylim(0,1)\nplt.ylabel ('Percentage')\nplt.xlabel ('Significant number')\nplt.xticks(list(keys))\nplt.legend (bbox_to_anchor=(1, 1), loc=\"upper right\", borderaxespad=0.)\n\nplt.show()",
"_____no_output_____"
],
[
"import pandas as pd\nfrom matplotlib import pyplot as plt\nimport matplotlib as mpl\nimport seaborn as sns\n%matplotlib inline\n\n#Read in data & create total column\n\nimport pandas as pd\n\ntrain_data=pd.read_csv('Data/Train.csv')\nval_data =pd.read_csv('Data/Val.csv')\ntest_data =pd.read_csv('Data/Test.csv')\n\ntotal_data = pd.concat([train_data,test_data,val_data],axis=0)\n\n# #stacked_bar_data[\"total\"] = stacked_bar_data.Series1 + stacked_bar_data.Series2\n\n# # #Set general plot properties\n# sns.set_style(\"white\")\n# sns.set_context({\"figure.figsize\": (24, 10)})\n\n# # #Plot 1 - background - \"total\" (top) series\n# sns.barplot(x = stacked_bar_data.title, y = stacked_bar_data., color = \"red\")\n\n# # #Plot 2 - overlay - \"bottom\" series\n# # bottom_plot = sns.barplot(x = stacked_bar_data.Group, y = stacked_bar_data.Series1, color = \"#0000A3\")\n\n\n# # topbar = plt.Rectangle((0,0),1,1,fc=\"red\", edgecolor = 'none')\n# # bottombar = plt.Rectangle((0,0),1,1,fc='#0000A3', edgecolor = 'none')\n# # l = plt.legend([bottombar, topbar], ['Bottom Bar', 'Top Bar'], loc=1, ncol = 2, prop={'size':16})\n# # l.draw_frame(False)\n\n# # #Optional code - Make plot look nicer\n# # sns.despine(left=True)\n# # bottom_plot.set_ylabel(\"Y-axis label\")\n# # bottom_plot.set_xlabel(\"X-axis label\")\n\n# # #Set fonts to consistent 16pt size\n# # for item in ([bottom_plot.xaxis.label, bottom_plot.yaxis.label] +\n# # bottom_plot.get_xticklabels() + bottom_plot.get_yticklabels()):\n# # item.set_fontsize(16)\n",
"_____no_output_____"
],
[
"df=total_data.groupby(['title','gender'])['path'].count()\n",
"_____no_output_____"
],
[
"total_data['title'].unique()",
"_____no_output_____"
],
[
"df_to_plot=pd.DataFrame(columns=['title','M','F'])\nlist1=[]\n\nfor title in list(total_data['title'].unique()):\n try:\n list1.append((title, df[title,'M'],df[title,'F']))\n except:\n pass\ndf_to_plot=pd.DataFrame(list1,columns=['title','M','F'])\n\n",
"_____no_output_____"
],
[
"#total_data = pd.concat([train_data,test_data,val_data],axis=0)\n\ndf_to_plot[\"total\"] = df_to_plot['M'] + df_to_plot['F']\n\ndf_to_plot=df_to_plot.sort_values(['total'],ascending=False)\n# #Set general plot properties\nsns.set_style(\"white\")\nsns.set_context({\"figure.figsize\": (24, 10)})\n\n# #Plot 1 - background - \"total\" (top) series\nsns.barplot(x = df_to_plot.title, y = df_to_plot.total, color = \"green\")\n\n# #Plot 2 - overlay - \"bottom\" series\nbottom_plot = sns.barplot(x = df_to_plot.title, y = df_to_plot['M'], color = \"blue\")\n\n\ntopbar = plt.Rectangle((0,0),1,1,fc=\"green\", edgecolor = 'none')\nbottombar = plt.Rectangle((0,0),1,1,fc='blue', edgecolor = 'none')\nl = plt.legend([bottombar, topbar], ['Male', 'Female'], loc=1, ncol = 2, prop={'size':16})\nl.draw_frame(False)\n\n#Optional code - Make plot look nicer\nsns.despine(left=True)\nbottom_plot.set_ylabel(\"Log frequency\")\nplt.yscale('log')\n#Set fonts to consistent 16pt size\nfor item in ([bottom_plot.xaxis.label, bottom_plot.yaxis.label] +\n bottom_plot.get_xticklabels() + bottom_plot.get_yticklabels()):\n item.set_fontsize(28)\n item.set_rotation('vertical')\n#bottom_plot.set_xlabel(\"Occupation\")\nplt.tight_layout()\nbottom_plot.set_xlabel('')\nplt.savefig('data_distribution.png')",
"_____no_output_____"
]
],
[
[
"### Mithun add your codes here ",
"_____no_output_____"
],
[
"### Model 1 : Bag of words\n",
"_____no_output_____"
]
],
[
[
"word_dict={}\nfor bio in all_bios:\n index_to_start=bio['start_pos']\n tokens=bio['raw'][index_to_start:].split()\n for tok in tokens:\n tok = tok.strip().lower()\n try:\n word_dict[tok] += 1\n except:\n word_dict[tok] = 1\n \n\n",
"_____no_output_____"
],
[
"len(list(word_dict))",
"_____no_output_____"
],
[
"import nltk",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"from scipy.sparse import vstack, csr_matrix, save_npz, load_npz",
"_____no_output_____"
],
[
"!pip install scipy",
"Collecting scipy\n Using cached scipy-1.4.1-cp37-cp37m-manylinux1_x86_64.whl (26.1 MB)\nRequirement already satisfied: numpy>=1.13.3 in /home/punyajoy/.conda/envs/aiethics/lib/python3.7/site-packages (from scipy) (1.18.1)\n\u001b[31mERROR: keras 2.3.1 requires h5py, which is not installed.\u001b[0m\n\u001b[31mERROR: keras 2.3.1 requires keras-applications>=1.0.6, which is not installed.\u001b[0m\n\u001b[31mERROR: keras 2.3.1 requires keras-preprocessing>=1.0.5, which is not installed.\u001b[0m\n\u001b[31mERROR: keras 2.3.1 requires pyyaml, which is not installed.\u001b[0m\nInstalling collected packages: scipy\nSuccessfully installed scipy-1.4.1\n"
],
[
"df = pd.DataFrame(all_bios, columns =list(all_bios[0].keys()))",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\n",
"_____no_output_____"
],
[
"df_train,df_test_val=train_test_split(df, test_size=0.35, random_state=42,stratify=df['title'])\ndf_test,df_val=train_test_split(df_test_val, test_size=0.28, random_state=42,stratify=df_test_val['title'])",
"_____no_output_____"
],
[
"df_train.to_csv('Train.csv',index=False)\ndf_test.to_csv('Test.csv',index=False)\ndf_val.to_csv('Val.csv',index=False)",
"_____no_output_____"
],
[
"import heapq\nmost_freq = heapq.nlargest(50000, word_dict, key=word_dict.get)\n",
"_____no_output_____"
],
[
"dataset = []\n\n\nfor bio in all_bios:\n index_to_start=bio['start_pos']\n tokens=bio['raw'][index_to_start:].split()\n for tok in most_freq:\n if token in sentence_tokens:\n sent_vec.append(1)\n else:\n sent_vec.append(0)\n sentence_vectors.append(sent_vec)\n\nfor sentence in corpus:\n sentence_tokens = nltk.word_tokenize(sentence)\n sent_vec = []\n for token in most_freq:\n if token in sentence_tokens:\n sent_vec.append(1)\n else:\n sent_vec.append(0)\n sentence_vectors.append(sent_vec)\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05b85a11e9fcde5e1a84279f46c409316e0c2a0 | 76,430 | ipynb | Jupyter Notebook | pandas.ipynb | Nikhila-padmanabhan/Python-project | 69d740a1a79bd1927040848b829a6e9e7ac33f15 | [
"Apache-2.0"
] | null | null | null | pandas.ipynb | Nikhila-padmanabhan/Python-project | 69d740a1a79bd1927040848b829a6e9e7ac33f15 | [
"Apache-2.0"
] | null | null | null | pandas.ipynb | Nikhila-padmanabhan/Python-project | 69d740a1a79bd1927040848b829a6e9e7ac33f15 | [
"Apache-2.0"
] | null | null | null | 22.787716 | 1,441 | 0.443373 | [
[
[
"#pandas",
"_____no_output_____"
],
[
"#indexes are visible\n#2 types of data structure\n#1. sereis - vector - 1d\n#2.data framee - 2d\n#3. index - index is visible",
"_____no_output_____"
],
[
"import numpy as np\n\nimport pandas as pd",
"_____no_output_____"
],
[
"#descriptive statistics",
"_____no_output_____"
],
[
"data = pd.Series([0.25,0.5,0.75,1])\ndata",
"_____no_output_____"
],
[
"data.values",
"_____no_output_____"
],
[
"data.index",
"_____no_output_____"
],
[
"data.shape",
"_____no_output_____"
],
[
"data.describe",
"_____no_output_____"
],
[
"data.describe()",
"_____no_output_____"
],
[
"#explicit indexing - assigned manually, difficult for huge data\n#if it is given as an intiger it will replace the default indexing",
"_____no_output_____"
],
[
"data = pd.Series([0.25,0.5,0.75,1],\n index=['a','b','c','d'])\ndata\n ",
"_____no_output_____"
],
[
"data['b']",
"_____no_output_____"
],
[
"data[1]",
"_____no_output_____"
],
[
"population_dict={'California':351264283,\n 'Texas':54123648,\n 'New York':5545865245,\n 'Florida':745565323,\n 'Illinois':1243579}\npopulation = pd.Series(population_dict)\npopulation",
"_____no_output_____"
],
[
"population.values",
"_____no_output_____"
],
[
"population.index",
"_____no_output_____"
],
[
"population.describe",
"_____no_output_____"
],
[
"population.describe()",
"_____no_output_____"
],
[
"population[1]",
"_____no_output_____"
],
[
"population['Texas']",
"_____no_output_____"
],
[
"pd.Series([2,5,7,8])",
"_____no_output_____"
],
[
"pd.Series(5,index=[100,200,300,400,500])",
"_____no_output_____"
],
[
"pd.Series({2:'a',3:'b',8:'c'}, index=[2,8])",
"_____no_output_____"
],
[
"##dataframe objects",
"_____no_output_____"
],
[
"area_dict={'California':1264283,'Texas':5413648,'New York':565245,'Florida':745523,'Illinois':3454569}\narea = pd.Series(area_dict)\narea",
"_____no_output_____"
],
[
"states = pd.DataFrame({'population': population,\n 'area':area})\nstates",
"_____no_output_____"
],
[
"states.keys()",
"_____no_output_____"
],
[
"states.ndim",
"_____no_output_____"
],
[
"states.info()",
"<class 'pandas.core.frame.DataFrame'>\nIndex: 5 entries, California to Illinois\nData columns (total 2 columns):\npopulation 5 non-null int64\narea 5 non-null int64\ndtypes: int64(2)\nmemory usage: 280.0+ bytes\n"
],
[
"data = np.arange(12,24).reshape(4,3)\ndf = pd.DataFrame(data)\ndf",
"_____no_output_____"
],
[
"data = np.arange(12,24).reshape(4,3)\ndf = pd.DataFrame(data,columns=['a','b','c'],index=[5,8,9,4])\ndf",
"_____no_output_____"
],
[
"x= np.random.random(20).reshape(4,5)\ndf= pd.DataFrame(x, columns=[1,2,3,4,5],index=['a','b','c','d'])\ndf\n",
"_____no_output_____"
],
[
"y = [{'a':i,'b':2*i,'c':2**i+2}\n for i in range(1,4)]\npd.DataFrame(y)",
"_____no_output_____"
],
[
"a= np.zeros(3, dtype=[('A','i8'),('B','f8')])\na",
"_____no_output_____"
],
[
"pd.DataFrame(a)",
"_____no_output_____"
],
[
"#indexig are immutable arrays\n#indexes are orderd sets",
"_____no_output_____"
],
[
"indA = pd.Index([1,2,3,4,])\nindB = pd.Index([2,4,6,8,10])",
"_____no_output_____"
],
[
"indA&indB",
"_____no_output_____"
],
[
"indA | indB",
"_____no_output_____"
],
[
"indA ^ indB",
"_____no_output_____"
],
[
"data = pd.read_csv(\"births.csv\")\ndata",
"_____no_output_____"
],
[
"data.head() #top5 raws",
"_____no_output_____"
],
[
"data.tail() #last 5 raws",
"_____no_output_____"
],
[
"data.describe",
"_____no_output_____"
],
[
"data.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 15547 entries, 0 to 15546\nData columns (total 5 columns):\nyear 15547 non-null int64\nmonth 15547 non-null int64\nday 15067 non-null float64\ngender 15547 non-null object\nbirths 15547 non-null int64\ndtypes: float64(1), int64(3), object(1)\nmemory usage: 607.4+ KB\n"
],
[
"data.columns",
"_____no_output_____"
],
[
"data.index",
"_____no_output_____"
],
[
"data.shape",
"_____no_output_____"
],
[
"data.ndim",
"_____no_output_____"
],
[
"data.values",
"_____no_output_____"
],
[
"data['year']",
"_____no_output_____"
],
[
"import seaborn as sns \nsns.pairplot(data)",
"c:\\users\\student\\appdata\\local\\programs\\python\\python36\\lib\\site-packages\\numpy\\lib\\histograms.py:829: RuntimeWarning: invalid value encountered in greater_equal\n keep = (tmp_a >= first_edge)\nc:\\users\\student\\appdata\\local\\programs\\python\\python36\\lib\\site-packages\\numpy\\lib\\histograms.py:830: RuntimeWarning: invalid value encountered in less_equal\n keep &= (tmp_a <= last_edge)\n"
],
[
"datas = pd.read_csv(\"../Desktop/data/file.csv\")\ndatas",
"_____no_output_____"
],
[
"sns.pairplot(datas)",
"_____no_output_____"
],
[
"Data = pd.read_excel('sales.xlsx')\nData",
"_____no_output_____"
],
[
"sns.pairplot(Data)",
"_____no_output_____"
],
[
"path = \"../Desktop/1.txt\"\nwith open(path,'r') as file: #\"r\" indicates read \n print (file.read())\n Data = pd.DataFrame(file)\nData",
"_____no_output_____"
],
[
"Data = pd.read_csv('iris.data')\nData.head()",
"_____no_output_____"
],
[
"##opertations in Pandas",
"_____no_output_____"
],
[
"rng = np.random.RandomState()\nser = pd.Series(rng.random(10))\nser",
"_____no_output_____"
],
[
"id(rng)",
"_____no_output_____"
],
[
"type(ser)",
"_____no_output_____"
],
[
"df = pd.DataFrame(rng.randint(0,10,(4,4)),\n columns=['A','B','C','D'])\ndf",
"_____no_output_____"
],
[
"df.mean()",
"_____no_output_____"
],
[
"df.mean(1)",
"_____no_output_____"
],
[
"df.median()",
"_____no_output_____"
],
[
"np.exp(df)",
"_____no_output_____"
],
[
"np.sin(df*np.pi/4)",
"_____no_output_____"
],
[
"area = pd.Series({'Alaska':465356,'Texas':89535,'California':89553},\n name='Area')\npopulation = pd.Series({'Newyork':8658656,'Texas':583569535,'California':87856553},\n name='Population')\npopulation",
"_____no_output_____"
],
[
"area",
"_____no_output_____"
],
[
"population/area",
"_____no_output_____"
],
[
"area.index|population.index",
"_____no_output_____"
],
[
"A = pd.Series([2,6,3],index =[3,2,1])\nB = pd.Series([1,5,3],index =[0,3,2])\nA+B",
"_____no_output_____"
],
[
"A.add(B,fill_value=(0))",
"_____no_output_____"
],
[
"X= pd.DataFrame(rng.randint(0,20,(3,5)),\n columns=list('ABCDE'))\nX",
"_____no_output_____"
],
[
"Y = pd.DataFrame(rng.randint(0,20,(3,3)),\n columns=list('ABC'))\nY",
"_____no_output_____"
],
[
"Z =X.stack().mean()\nZ",
"_____no_output_____"
],
[
"X.add(Y,fill_value =Z)",
"_____no_output_____"
],
[
"p= rng.randint(10,size=(3,4))\np",
"_____no_output_____"
],
[
"p[2]",
"_____no_output_____"
],
[
"p-p[2]",
"_____no_output_____"
],
[
"s = pd.DataFrame(p, columns = list('QRST'))\ns",
"_____no_output_____"
],
[
"s.iloc[0] #iloc is used to select a particular row",
"_____no_output_____"
],
[
"s-s.iloc[0]",
"_____no_output_____"
],
[
"s.subtract(s['R'],axis=0)",
"_____no_output_____"
],
[
"halfrow = s.iloc[0,::2]\nhalfrow",
"_____no_output_____"
],
[
"s-halfrow",
"_____no_output_____"
],
[
"#Handliing the missing data",
"_____no_output_____"
],
[
"#nan\n#na\n#none\n#missing values has to be filled\n#1. finding NA\n#1Aa. identify NA\n#1ai. is null\n#1aii. not a null\n#2. fill NA \n#2a. fill_value\n#2b.fill_NA\n#2bi.bfill\n#2bii.ffill\n#3. drop - entire row will be deleted",
"_____no_output_____"
],
[
"a= np.array([1,None,2,3])\na",
"_____no_output_____"
],
[
"a.dtype #object datatype is for None values",
"_____no_output_____"
],
[
"b= np.array([1,4,2,3])\nb",
"_____no_output_____"
],
[
"b.dtype",
"_____no_output_____"
],
[
"a[0]",
"_____no_output_____"
],
[
"a[2]\n",
"_____no_output_____"
],
[
"for dtype in['object','int','float']:\n print(\"dtype=\", dtype)\n %timeit np.arange(1E6,dtype=dtype).sum()\n print()",
"_____no_output_____"
],
[
"for dtype in['object','int','float']:\n print(\"dtype=\", dtype)\n %timeit np.arange(1E6,dtype=dtype).mean()\n print()",
"_____no_output_____"
],
[
"#NAN",
"_____no_output_____"
],
[
"vals2= np.array([1,np.nan,3,4])\nvals2.dtype #nan data type is float",
"_____no_output_____"
],
[
"1+np.nan",
"_____no_output_____"
],
[
"vals2.sum(),vals2.min(),vals2.max(),vals2.mean()",
"_____no_output_____"
],
[
"np.nansum(vals2),np.nanmin(vals2),np.nanmax(vals2),np.nanmean(vals2) #can avoid nan values and perform operation",
"_____no_output_____"
],
[
"#nan and None",
"_____no_output_____"
],
[
"pd.Series([1,np.nan,2,None])",
"_____no_output_____"
],
[
"x= pd.Series(range(4),dtype=int)\nx",
"_____no_output_____"
],
[
"x[3]=None\nx",
"_____no_output_____"
],
[
"x.add(2)",
"_____no_output_____"
],
[
"#DATA TYPE CHANGES\n#1. FLOAT -no change \n#2. object - No change\n#3. intiger - cast to float - np.nan\n#4.Boolean - cast to objetc - None or np.nan",
"_____no_output_____"
],
[
"y = np.array([True,False,10,20,None,np.nan])\ny.dtype",
"_____no_output_____"
],
[
"y = pd.Series([True,False,10,20,None,np.nan])\ny.dtype",
"_____no_output_____"
],
[
"#null values",
"_____no_output_____"
],
[
"#isnull - find the null values in the data set\n#notnull - opposite of is null\n#dropna - filter the data with or without null values\n#fillna()-fill the values for NAN",
"_____no_output_____"
],
[
"data = pd.Series([1,np.nan,3,None,'hello'])\ndata",
"_____no_output_____"
],
[
"data.isnull()",
"_____no_output_____"
],
[
"Df= pd.DataFrame(data)\nDf",
"_____no_output_____"
],
[
"Df.isnull()",
"_____no_output_____"
],
[
"Df['age']=np.array([24,23,45,56,35])\nDf",
"_____no_output_____"
],
[
"Df.notnull()",
"_____no_output_____"
],
[
"data[data.notnull()]",
"_____no_output_____"
],
[
"Df[Df.notnull()]",
"_____no_output_____"
],
[
"#dropna() delete the null values without affecting the original data\n#fillna() will affect the original data",
"_____no_output_____"
],
[
"data.dropna()",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"data.fillna(1)",
"_____no_output_____"
],
[
"data.fillna(value = 10)",
"_____no_output_____"
],
[
"mean = np.nanmean(data)\nprint (mean)\ndata.fillna(value =mean) #data is manipulated",
"_____no_output_____"
],
[
"df = pd.DataFrame([[1, np.nan,2],\n [2,3,5],\n [np.nan,4,None]],columns=['A','B','C'])\ndf",
"_____no_output_____"
],
[
"df.dropna()",
"_____no_output_____"
],
[
"df.dropna(axis='columns')",
"_____no_output_____"
],
[
"df.dropna(axis='rows')",
"_____no_output_____"
],
[
"df['D']=np.nan\ndf",
"_____no_output_____"
],
[
"df.dropna(axis = 'columns',how = 'all')",
"_____no_output_____"
],
[
"df.dropna(axis = 'columns',how = 'any')",
"_____no_output_____"
],
[
"df.dropna(axis = 'rows',how = 'all')",
"_____no_output_____"
],
[
"df.dropna(axis = 'rows',how = 'any')",
"_____no_output_____"
],
[
"df.dropna(axis = 'rows',thresh =2) #number of available non null values =2\n",
"_____no_output_____"
],
[
"df.dropna(axis = 'columns', thresh =2)",
"_____no_output_____"
],
[
"df2 = pd.DataFrame({'Data':[10,20,30,np.nan,50,60],\n 'float':[1.5,2.5,3.2,4.5,5.5,np.nan],\n 'complex':[np.nan,2j+3,np.nan,23j+2,5j+2,np.nan]})\ndf2",
"_____no_output_____"
],
[
"df2.isnull()",
"_____no_output_____"
],
[
"df2.notnull()",
"_____no_output_____"
],
[
"df2.notna()",
"_____no_output_____"
],
[
"df2.dropna()",
"_____no_output_____"
],
[
"fill= np.nanmean(df2)\nfill",
"_____no_output_____"
],
[
"Fill_Missing_Value = df2.fillna(fill)\nFill_Missing_Value",
"_____no_output_____"
],
[
"Fill_Missing_Value = df2.fillna(fill,axis=1)\nFill_Missing_Value",
"_____no_output_____"
],
[
"data = pd.Series([1,np.nan,2,None,3],index=list('abcde'))\ndata",
"_____no_output_____"
],
[
"x=data.fillna(0)\nx",
"_____no_output_____"
]
],
[
[
"#forwardfill - ffill - none value will be filled with the previous data",
"_____no_output_____"
],
[
"#backwardfill= null value will be filled with the next value",
"_____no_output_____"
]
],
[
[
"data.fillna(method='ffill')",
"_____no_output_____"
],
[
"df3 = pd.DataFrame({'Data':[10,20,30,np.nan,50,60],\n 'float':[1.5,2.5,3.2,4.5,5.5,np.nan],\n })\ndf3",
"_____no_output_____"
],
[
"data.fillna(method='bfill')",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"Data = pd.read_csv('california_cities.csv')\nData",
"_____no_output_____"
],
[
"Data.head()",
"_____no_output_____"
],
[
"Data.tail()",
"_____no_output_____"
],
[
"Data.describe()",
"_____no_output_____"
],
[
"Data.info()",
"_____no_output_____"
],
[
"Data.columns",
"_____no_output_____"
],
[
"Data.index",
"_____no_output_____"
],
[
"Data.isnull().info()",
"_____no_output_____"
],
[
"Data1 = Data.drop(['Unnamed: 0'],axis =1) \nData1 ",
"_____no_output_____"
],
[
"Data1.isnull()",
"_____no_output_____"
],
[
"Data1.info()",
"_____no_output_____"
],
[
"#Data1['elevation_m']=np.nanmean(Data1['elevation_m'])\n#Data1 #original data will be affetced so not a recomanded method",
"_____no_output_____"
],
[
"Fill = np.nanmean(Data1['elevation_m'])\nData1['elevation_m']= Data1['elevation_m'].fillna(Fill)\nData1",
"_____no_output_____"
],
[
"Data1.info()",
"_____no_output_____"
],
[
"#hierarchical indexing",
"_____no_output_____"
],
[
"pd.__version__",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"index = [('California', 2000), ('California', 2010),\n ('New York', 2000), ('New York', 2010),\n ('Texas', 2000), ('Texas', 2010)]\npopulations = [33871648, 37253956,\n 18976457, 19378102,\n 20851820, 25145561]",
"_____no_output_____"
],
[
"populations",
"_____no_output_____"
],
[
"pop = pd.Series(populations, index=index)\npop",
"_____no_output_____"
],
[
"#pop [1:4]\npop['California',2000]",
"_____no_output_____"
],
[
"pop[[i for i in pop.index if i[1]==2010]]",
"_____no_output_____"
],
[
"for i in pop.index:\n if i[1]==2010:\n print(i,pop[i])",
"_____no_output_____"
],
[
"index = pd.MultiIndex.from_tuples(index)\nindex",
"_____no_output_____"
],
[
"pop =pop.reindex(index)\npop",
"_____no_output_____"
],
[
"pop['California']",
"_____no_output_____"
],
[
"pop[:,2010]",
"_____no_output_____"
],
[
"pop_df = pop.unstack()\npop_df",
"_____no_output_____"
],
[
"pop_df.stack()",
"_____no_output_____"
],
[
"pop",
"_____no_output_____"
],
[
"pop_df = pd.DataFrame ({'Total': pop,\n 'under18': [8865325656,35689545,\n 656898,458545545,\n 4455687,965856]})\npop_df",
"_____no_output_____"
],
[
"df = pd.DataFrame(np.random.rand(4,2),\n index =[['a','a','b','b'],[1,2,1,2]],\n columns=['data1','data2'])\ndf",
"_____no_output_____"
],
[
"data ={('california',2000):5589865365,\n ('california',2010):89888556,\n ('Texas',2000):78454533,\n ('Texas',2010):58963568,\n ('Newyork',2000):57989656,\n ('Newyork',2010):555655878}\npd.Series(data)",
"_____no_output_____"
],
[
"pd.MultiIndex.from_arrays([['a','a','b','b'],[1,2,1,2]])",
"_____no_output_____"
],
[
"pd.MultiIndex.from_tuples([('a',1),('a',2),('b',1),('b',2)])",
"_____no_output_____"
],
[
"pd.MultiIndex.from_product([['a','b'],[1,2]])",
"_____no_output_____"
],
[
"pd.MultiIndex(levels = [['a','b'],[1,2]],\n codes = [[0,0,1,1],[0,1,0,1]])",
"_____no_output_____"
],
[
"pop.index.names=['state','year']\npop",
"_____no_output_____"
],
[
"index = pd.MultiIndex.from_product([[2013,2014],[1,2,3]],\n names=['year','visit'])\ncolumns =pd.MultiIndex.from_product([['Rani','Raju','Sam'],['BMI','TEMP','WGHT']],\n names =['subject','type']) \n ",
"_____no_output_____"
],
[
"data=np.round(np.random.rand(6,9),2)\ndata+=37",
"_____no_output_____"
],
[
"health_data = pd.DataFrame(data,index=index,columns=columns)\nhealth_data",
"_____no_output_____"
],
[
"health_data['Rani']",
"_____no_output_____"
],
[
"import numpy as np\n\nimport pandas as pd",
"_____no_output_____"
],
[
"health_data.iloc[:3,:-3] # starts with oth row and ends with 2nd row, from right side till -3",
"_____no_output_____"
],
[
"health_data.iloc[:3]",
"_____no_output_____"
],
[
"idx = pd.IndexSlice\nhealth_data.loc[idx[:,1],idx[:,'TEMP']] #performing integer and string together",
"_____no_output_____"
],
[
"#sorted and unsorted indices\nindex = pd.MultiIndex.from_product([['a','c','b','d'],[1,2]])\ndata = pd.Series(np.random.rand(8),index=index)\ndata.index.names = ['char','int']\ndata",
"_____no_output_____"
],
[
"try:\n data[ 'a':'b']\nexcept KeyError as e:\n print(type(e))\n print(e)",
"_____no_output_____"
],
[
"data = data.sort_index()\ndata",
"_____no_output_____"
],
[
"pop.unstack(level =0)",
"_____no_output_____"
],
[
"pop.unstack().stack()",
"_____no_output_____"
],
[
"pop_flat = pop.reset_index(name='population')\npop_flat\n ",
"_____no_output_____"
],
[
"health_data",
"_____no_output_____"
],
[
"data_mean1 = health_data.mean(level='year')\ndata_mean1",
"_____no_output_____"
],
[
"data_mean2 = health_data.mean(level='visit')\ndata_mean2",
"_____no_output_____"
],
[
"data_mean1.mean(axis =1 ,level='type')",
"_____no_output_____"
]
]
] | [
"code",
"raw",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"raw",
"raw"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05b8eb3ff6642ee6e4c47f58b0ace3debde0c1e | 237,771 | ipynb | Jupyter Notebook | notebooks/CheckOutCT.ipynb | chang306/microstructure | 1e06c258d682682693f37e1a074b5a11a0c800d9 | [
"BSD-3-Clause"
] | null | null | null | notebooks/CheckOutCT.ipynb | chang306/microstructure | 1e06c258d682682693f37e1a074b5a11a0c800d9 | [
"BSD-3-Clause"
] | null | null | null | notebooks/CheckOutCT.ipynb | chang306/microstructure | 1e06c258d682682693f37e1a074b5a11a0c800d9 | [
"BSD-3-Clause"
] | null | null | null | 819.9 | 21,556 | 0.954847 | [
[
[
"from read_CT_txt_files import read_CT_txt_files\nimport matplotlib.pyplot as plt\n\nsite='2S16'\n\ndata_dir='../data/microCT/' + site + '/' \n\n[SSA_CT,height_min,height_max]=read_CT_txt_files(data_dir)\n\nfig,ax = plt.subplots()\nax.plot(6/917/SSA_CT*1000,height_min,label='microCT') #CT data\nax.set_xlabel('Equivalent diameter, mm')\nax.set_ylabel('Height above snow-soil interface [cm]')\n\nax.set_title(f'Site {site.upper()}')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Process all microCT and save the output.\n\n\nThis code begins by reading the CT data from every directory in ../data/microCT and processing it into a dataframe. \n\nIt then stores these dataframes in a dictionary (key: site code, value: dataframe).\n\nThe dictionary is then saved as a pickle file in data/microCT.",
"_____no_output_____"
]
],
[
[
"import os\nimport pandas as pd\nimport pickle\n\noutput_frames = {}\n\n\nfor site in os.listdir('../data/microCT'):\n \n if '.' not in site:\n\n data_dir='../data/microCT/' + site + '/' \n\n [SSA_CT,height_min,height_max]=read_CT_txt_files(data_dir)\n\n fig,ax = plt.subplots()\n ax.plot(6/917/SSA_CT*1000,height_min,label='microCT') #CT data\n ax.set_xlabel('Equivalent diameter, mm')\n ax.set_ylabel('Height above snow-soil interface [cm]')\n\n ax.set_title(f'Site {site.upper()}')\n plt.show()\n \n data_df = pd.DataFrame(\n {'height (cm)':height_min,\n 'SSA (m2/kg)':SSA_CT,\n 'Equiv. Diam (mm)':6/917/SSA_CT*1000,\n }\n )\n \n output_frames[site] = data_df\n \npickle.dump(output_frames, open('../data/microCT/processed_mCT.p', 'wb'))",
"_____no_output_____"
]
],
[
[
"# Test that the saved data can be read out and plotted again\n\n### The plots below should match the plots above!",
"_____no_output_____"
]
],
[
[
"# read data from pickle file\n\nframes = pickle.load(open('../data/microCT/processed_mCT.p', 'rb'))\n\nfor site in frames.keys():\n \n # extract dataframe from dict\n \n df = frames[site]\n \n # plot\n \n fig,ax = plt.subplots()\n ax.plot(df['Equiv. Diam (mm)'],\n df['height (cm)'])\n \n ax.set_xlabel('Equiv. Diam (mm)')\n ax.set_ylabel('Height above snow-soil interface [cm]')\n\n ax.set_title(f'Site {site.upper()}')\n \n plt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d05ba404a709113d8d2005d1a691553948cd37d6 | 339,539 | ipynb | Jupyter Notebook | notebooks/behavior_cloning_tutorial-Copy1.ipynb | Jetafull/CarND-Behavioral-Cloning-P3 | 09027e755d0b31053fcd99fa21f02c92856bf7b0 | [
"MIT"
] | null | null | null | notebooks/behavior_cloning_tutorial-Copy1.ipynb | Jetafull/CarND-Behavioral-Cloning-P3 | 09027e755d0b31053fcd99fa21f02c92856bf7b0 | [
"MIT"
] | null | null | null | notebooks/behavior_cloning_tutorial-Copy1.ipynb | Jetafull/CarND-Behavioral-Cloning-P3 | 09027e755d0b31053fcd99fa21f02c92856bf7b0 | [
"MIT"
] | null | null | null | 348.245128 | 111,256 | 0.92076 | [
[
[
"## Environment",
"_____no_output_____"
]
],
[
[
"%env CUDA_DEVICE_ORDER=PCI_BUS_ID\n%env CUDA_VISIBLE_DEVICES=0\nfrom pathlib import Path\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n%autosave 20\n\nimport csv\nimport pandas as pd\nfrom keras.backend import tf as ktf\nimport sys\nimport cv2\nimport six\n\n# keras\nimport keras\nfrom keras.models import Model\nfrom keras.models import Sequential\nfrom keras.regularizers import l2\nfrom keras.layers.core import Lambda\nfrom keras.optimizers import Adam\nfrom keras.layers.normalization import BatchNormalization\nfrom keras.callbacks import LearningRateScheduler\n\nfrom keras.models import Model\nfrom keras.layers import (\n Input,\n Activation,\n Dense,\n Flatten,\n Dropout\n)\nfrom keras.layers.convolutional import (\n Conv2D,\n MaxPooling2D,\n AveragePooling2D\n)\nfrom keras.layers.merge import add\nfrom keras import backend as K\nimport math\n\nimport tensorflow as tf\nconfig = tf.ConfigProto()\nconfig.gpu_options.allow_growth = True\nsession = tf.Session(config=config)\n\nROOT_PATH = Path('/home/downloads/CarND-Behavioral-Cloning-P3/')\n#ROOT_PATH=Path('/src')\n\nfrom tensorflow.python.client import device_lib\nprint(device_lib.list_local_devices())\n\n#SAMPLE_DATA_PATH = ROOT_PATH/'data/sample_data'\nSAMPLE_DATA_PATH = ROOT_PATH/'data/all'\nprint('tensorflow version: ', tf.__version__)\nprint('keras version: ', keras.__version__)\nprint('python version: ', sys.version_info)",
"env: CUDA_DEVICE_ORDER=PCI_BUS_ID\nenv: CUDA_VISIBLE_DEVICES=0\n"
]
],
[
[
"## Load images",
"_____no_output_____"
]
],
[
[
"#[str(x) for x in list(SAMPLE_DATA_PATH.iterdir())]\nlogs = pd.DataFrame()\nnum_tracks = [0, 0]\ninclude_folders = [\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/IMG',\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track1_recovery.csv',\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_drive4.csv',\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_curve.csv',\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track1_sampledata.csv',\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_drive3.csv',\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/backup',\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_drive5.csv',\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_reverse.csv',\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_drive2.csv',\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_drive1.csv',\n '/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track1_drive.csv'\n ]\nfor log_file in SAMPLE_DATA_PATH.glob('*.csv'):\n if str(log_file) not in include_folders:\n continue\n one_log = pd.read_csv(log_file)\n num_rows = one_log.shape[0]\n print(log_file, '\\t', num_rows)\n if str(log_file).find('track1') != -1:\n num_tracks[0] += num_rows\n else:\n num_tracks[1] += num_rows\n logs = pd.concat([logs, one_log], axis=0)\nprint('\\ntrack 1: ', num_tracks[0])\nprint('track 2: ', num_tracks[1])\nlogs.tail()",
"/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track1_recovery.csv \t 1458\n/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_drive4.csv \t 10252\n/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_curve.csv \t 6617\n/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track1_sampledata.csv \t 8036\n/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_drive3.csv \t 2039\n/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_drive5.csv \t 3098\n/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_reverse.csv \t 8873\n/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_drive2.csv \t 5465\n/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track2_drive1.csv \t 6120\n/home/downloads/CarND-Behavioral-Cloning-P3/data/all/driving_log_track1_drive.csv \t 7874\n\ntrack 1: 17368\ntrack 2: 42464\n"
]
],
[
[
"## Preprocessing and Augmentation",
"_____no_output_____"
]
],
[
[
"IMG_FOLDER_PATH = SAMPLE_DATA_PATH/'IMG'\n\ndef get_img_files(img_folder_path):\n image_files = []\n labels = dict()\n correction = 0.2\n\n for log in logs.iterrows():\n center, left, right, y = log[1][:4]\n for i, img_path in enumerate([center, left, right]):\n img_path = img_path.split('/')[-1].strip()\n abs_img_path = str(img_folder_path/img_path)\n if i == 1:\n y_corrected = y + correction # left\n elif i == 2:\n y_corrected = y - correction # right\n else:\n y_corrected = y\n\n image_files.append(abs_img_path)\n labels[abs_img_path] = y_corrected\n\n np.random.shuffle(image_files)\n trn_end_idx = int(len(image_files)*0.8)\n \n train_img_files = image_files[:trn_end_idx]\n val_img_files = image_files[trn_end_idx:]\n \n return train_img_files, val_img_files, labels",
"_____no_output_____"
],
[
"TRAIN_IMG_FILES, VAL_IMG_FILES, LABELS = get_img_files(IMG_FOLDER_PATH)",
"_____no_output_____"
],
[
"len(TRAIN_IMG_FILES), len(VAL_IMG_FILES), len(LABELS.keys())",
"_____no_output_____"
],
[
"def augment_data(img, y, probs=0.5):\n # flip\n if np.random.rand() > probs:\n img = np.fliplr(img)\n y = -y\n return img, y",
"_____no_output_____"
]
],
[
[
"## Create data generator for Keras model training",
"_____no_output_____"
]
],
[
[
"# adpated from https://stanford.edu/~shervine/blog/keras-how-to-generate-data-on-the-fly.html\nclass GeneratorFromFiles(keras.utils.Sequence):\n '''Generate data from list of image files.'''\n \n def __init__(self, list_files, labels, batch_size=64,\n dim=(160, 320, 3),\n post_dim=(66, 200, 3),\n shuffle=True, \n data_aug=None,\n resize=False):\n '''\n Paramters\n ----------\n list_files : a list of absolute path to image files\n labels : a dictionary mapping image files to labels (classes/continous value)\n batch_size : size for each batch\n dim : dimension for input image, height x width x number of channel\n shuffle : whether to shuffle data at each epoch\n '''\n self.dim = dim\n self.post_dim = post_dim if resize else dim\n self.batch_size = batch_size\n self.list_files = list_files\n self.labels = labels\n self.shuffle = shuffle\n self.data_aug = data_aug\n self.resize=resize\n \n self.on_epoch_end()\n \n def __len__(self):\n return int(len(self.list_files) / self.batch_size)\n \n def __getitem__(self, index):\n # generate indexes of the batch\n indexes = self.indexes[index*self.batch_size:(index+1)*self.batch_size]\n # find list of files\n list_files_batch = [self.list_files[k] for k in indexes]\n X, ys = self._generate(list_files_batch, self.data_aug)\n \n return X, ys\n \n def on_epoch_end(self):\n self.indexes = np.arange(len(self.list_files))\n if self.shuffle:\n np.random.shuffle(self.indexes)\n \n def _generate(self, list_files_batch, data_aug=None):\n X = np.zeros((self.batch_size, ) + self.post_dim)\n ys = np.zeros((self.batch_size))\n \n for i, img_file in enumerate(list_files_batch):\n x = plt.imread(img_file)\n if self.resize:\n x = cv2.resize(x, (self.post_dim[1], self.post_dim[0]))\n y = self.labels[img_file]\n if data_aug is not None:\n x, y = data_aug(x, y)\n X[i, ] = x\n ys[i] = y\n \n return X, ys",
"_____no_output_____"
]
],
[
[
"Visualize flipping the image",
"_____no_output_____"
]
],
[
[
"data_generator = GeneratorFromFiles(TRAIN_IMG_FILES, LABELS)\nres = next(iter(data_generator))\nplt.imshow(res[0][56].astype(int))",
"_____no_output_____"
],
[
"plt.imshow(augment_data(res[0][56], res[1][60], 0.0)[0].astype(int))",
"_____no_output_____"
],
[
"plt.imshow(cv2.resize(res[0][56], (200, 66)).astype(int))",
"_____no_output_____"
],
[
"plt.imshow(cv2.resize(augment_data(res[0][56], res[1][60], 0.0)[0], (200, 66)).astype(int))",
"_____no_output_____"
]
],
[
[
"## Model Architecture and Parameter",
"_____no_output_____"
],
[
"### Nvidia model",
"_____no_output_____"
]
],
[
[
"def _bn_act_dropout(input, dropout_rate):\n \"\"\"Helper to build a BN -> activation block\n \"\"\"\n norm = BatchNormalization(axis=2)(input)\n relu = Activation('elu')(norm)\n return Dropout(dropout_rate)(relu)\n \n\ndef _conv_bn_act_dropout(**conv_params):\n '''Helper to build a conv -> BN -> activation block -> dropout\n '''\n filters = conv_params['filters']\n kernel_size = conv_params['kernel_size']\n strides = conv_params.setdefault('strides', (1, 1))\n kernel_initializer = conv_params.setdefault('kernel_initializer', 'he_normal')\n padding = conv_params.setdefault('padding', 'valid')\n kernel_regularizer = conv_params.setdefault('kernel_regularizer', l2(1.e-4))\n dropout_rate = conv_params.setdefault('dropout_rate', 0.1)\n\n def f(input):\n conv = Conv2D(filters=filters, kernel_size=kernel_size,\n strides=strides, padding=padding,\n kernel_initializer=kernel_initializer,\n kernel_regularizer=kernel_regularizer)(input)\n return _bn_act_dropout(conv, dropout_rate)\n\n return f \n\ndef _dense_dropout(input, n, dropout_rate, dropout_multi=1):\n return Dropout(dropout_rate*dropout_multi)(Dense(n, activation='elu')(input))\n\ndef build_nvidia(in_shape, num_outputs, dropout_rate, dropout_multi=1):\n input = Input(shape=in_shape)\n in_layer = Lambda(lambda x: (x / 255.0) - 0.5, input_shape=(in_shape))(input)\n in_layer = _conv_bn_act_dropout(filters=24, kernel_size=(5, 5), strides=(2, 2), dropout_rate=dropout_rate)(in_layer)\n in_layer = _conv_bn_act_dropout(filters=36, kernel_size=(5, 5), strides=(2, 2), dropout_rate=dropout_rate)(in_layer)\n in_layer = _conv_bn_act_dropout(filters=48, kernel_size=(5, 5), strides=(2, 2), dropout_rate=dropout_rate)(in_layer)\n in_layer = _conv_bn_act_dropout(filters=64, kernel_size=(3, 3), strides=(1, 1), dropout_rate=dropout_rate)(in_layer)\n in_layer = _conv_bn_act_dropout(filters=64, kernel_size=(3, 3), strides=(1, 1), dropout_rate=dropout_rate)(in_layer)\n\n flatten = Flatten()(in_layer)\n flatten = _dense_dropout(flatten, 1000, dropout_rate, dropout_multi)\n flatten = _dense_dropout(flatten, 100, dropout_rate, dropout_multi)\n #flatten = _dense_dropout(flatten, 50, dropout_rate)\n flatten = Dense(50)(flatten)\n dense = Dense(units=num_outputs)(flatten)\n\n model = Model(inputs=input, outputs=dense)\n return model\n\n# learning rate schedule\ndef step_decay(epoch):\n initial_lrate = 1e-3\n drop = 0.5\n epochs_drop = 3\n lrate = initial_lrate * math.pow(drop, math.floor((1+epoch)/epochs_drop))\n return lrate",
"_____no_output_____"
],
[
"in_shape = (66, 200, 3)\n#in_shape = (160, 320, 3)\ndropout_rate = 0.2\nmodel = build_nvidia(in_shape, 1, dropout_rate, dropout_multi=2)\nopt = Adam(lr=1e-4)\nmodel.compile(loss='mse', optimizer=opt)\nlrate = LearningRateScheduler(step_decay)\ncallbacks_list = [lrate]\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) (None, 66, 200, 3) 0 \n_________________________________________________________________\nlambda_2 (Lambda) (None, 66, 200, 3) 0 \n_________________________________________________________________\nconv2d_3 (Conv2D) (None, 31, 98, 24) 1824 \n_________________________________________________________________\nbatch_normalization_1 (Batch (None, 31, 98, 24) 392 \n_________________________________________________________________\nactivation_1 (Activation) (None, 31, 98, 24) 0 \n_________________________________________________________________\ndropout_3 (Dropout) (None, 31, 98, 24) 0 \n_________________________________________________________________\nconv2d_4 (Conv2D) (None, 14, 47, 36) 21636 \n_________________________________________________________________\nbatch_normalization_2 (Batch (None, 14, 47, 36) 188 \n_________________________________________________________________\nactivation_2 (Activation) (None, 14, 47, 36) 0 \n_________________________________________________________________\ndropout_4 (Dropout) (None, 14, 47, 36) 0 \n_________________________________________________________________\nconv2d_5 (Conv2D) (None, 5, 22, 48) 43248 \n_________________________________________________________________\nbatch_normalization_3 (Batch (None, 5, 22, 48) 88 \n_________________________________________________________________\nactivation_3 (Activation) (None, 5, 22, 48) 0 \n_________________________________________________________________\ndropout_5 (Dropout) (None, 5, 22, 48) 0 \n_________________________________________________________________\nconv2d_6 (Conv2D) (None, 3, 20, 64) 27712 \n_________________________________________________________________\nbatch_normalization_4 (Batch (None, 3, 20, 64) 80 \n_________________________________________________________________\nactivation_4 (Activation) (None, 3, 20, 64) 0 \n_________________________________________________________________\ndropout_6 (Dropout) (None, 3, 20, 64) 0 \n_________________________________________________________________\nconv2d_7 (Conv2D) (None, 1, 18, 64) 36928 \n_________________________________________________________________\nbatch_normalization_5 (Batch (None, 1, 18, 64) 72 \n_________________________________________________________________\nactivation_5 (Activation) (None, 1, 18, 64) 0 \n_________________________________________________________________\ndropout_7 (Dropout) (None, 1, 18, 64) 0 \n_________________________________________________________________\nflatten_2 (Flatten) (None, 1152) 0 \n_________________________________________________________________\ndense_3 (Dense) (None, 1000) 1153000 \n_________________________________________________________________\ndropout_8 (Dropout) (None, 1000) 0 \n_________________________________________________________________\ndense_4 (Dense) (None, 100) 100100 \n_________________________________________________________________\ndropout_9 (Dropout) (None, 100) 0 \n_________________________________________________________________\ndense_5 (Dense) (None, 50) 5050 \n_________________________________________________________________\ndense_6 (Dense) (None, 1) 51 \n=================================================================\nTotal params: 1,390,369\nTrainable params: 1,389,959\nNon-trainable params: 410\n_________________________________________________________________\n"
]
],
[
[
"## Training and Validation",
"_____no_output_____"
]
],
[
[
"%%time\ntrn_data_generator = GeneratorFromFiles(TRAIN_IMG_FILES, LABELS, resize=True)\nval_data_generator = GeneratorFromFiles(VAL_IMG_FILES, LABELS, resize=True)\n\nmodel.fit_generator(trn_data_generator, \n validation_data=val_data_generator, \n epochs=12, \n workers=2, \n callbacks=callbacks_list,\n use_multiprocessing=False,\n verbose=1)",
"Epoch 1/12\n2243/2243 [==============================] - 241s 107ms/step - loss: 0.1739 - val_loss: 0.1210\nEpoch 2/12\n2243/2243 [==============================] - 238s 106ms/step - loss: 0.1171 - val_loss: 0.0942\nEpoch 3/12\n2243/2243 [==============================] - 225s 100ms/step - loss: 0.0940 - val_loss: 0.0839\nEpoch 4/12\n2243/2243 [==============================] - 225s 100ms/step - loss: 0.0851 - val_loss: 0.0869\nEpoch 5/12\n2243/2243 [==============================] - 226s 101ms/step - loss: 0.0795 - val_loss: 0.0709\nEpoch 6/12\n2243/2243 [==============================] - 225s 101ms/step - loss: 0.0727 - val_loss: 0.0668\nEpoch 7/12\n2243/2243 [==============================] - 226s 101ms/step - loss: 0.0707 - val_loss: 0.0686\nEpoch 8/12\n2243/2243 [==============================] - 225s 100ms/step - loss: 0.0697 - val_loss: 0.0644\nEpoch 9/12\n2243/2243 [==============================] - 226s 101ms/step - loss: 0.0661 - val_loss: 0.0619\nEpoch 10/12\n2243/2243 [==============================] - 225s 100ms/step - loss: 0.0650 - val_loss: 0.0607\nEpoch 11/12\n2243/2243 [==============================] - 225s 100ms/step - loss: 0.0644 - val_loss: 0.0607\nEpoch 12/12\n2243/2243 [==============================] - 226s 101ms/step - loss: 0.0621 - val_loss: 0.0584\nCPU times: user 1h 48min 21s, sys: 7min 32s, total: 1h 55min 53s\nWall time: 45min 34s\n"
],
[
"# model.load_weights(ROOT_PATH/'models/model-nvidia-base-2.h5')\n# trn_data_generator = GeneratorFromFiles(TRAIN_IMG_FILES, LABELS, resize=True)\n# val_data_generator = GeneratorFromFiles(VAL_IMG_FILES, LABELS, resize=True)",
"_____no_output_____"
]
],
[
[
"## Fine-Tuning the Model",
"_____no_output_____"
]
],
[
[
"%%time\nopt = Adam(lr=1e-5)\nmodel.compile(loss='mse', optimizer=opt)\nmodel.fit_generator(trn_data_generator, \n validation_data=val_data_generator, \n epochs=5, \n workers=3, \n use_multiprocessing=True,\n verbose=1)",
"Epoch 1/5\n2243/2243 [==============================] - 180s 80ms/step - loss: 0.0575 - val_loss: 0.0523\nEpoch 2/5\n2243/2243 [==============================] - 166s 74ms/step - loss: 0.0572 - val_loss: 0.0522\nEpoch 3/5\n2243/2243 [==============================] - 180s 80ms/step - loss: 0.0571 - val_loss: 0.0524\nEpoch 4/5\n2243/2243 [==============================] - 179s 80ms/step - loss: 0.0570 - val_loss: 0.0524\nEpoch 5/5\n2243/2243 [==============================] - 185s 82ms/step - loss: 0.0570 - val_loss: 0.0525\nCPU times: user 12min 31s, sys: 5min 37s, total: 18min 9s\nWall time: 14min 50s\n"
],
[
"%%time\nopt = Adam(lr=8e-6)\nmodel.compile(loss='mse', optimizer=opt)\nmodel.fit_generator(trn_data_generator, \n validation_data=val_data_generator, \n epochs=5, \n workers=3, \n use_multiprocessing=True,\n verbose=1)",
"Epoch 1/5\n2243/2243 [==============================] - 180s 80ms/step - loss: 0.0568 - val_loss: 0.0523\nEpoch 2/5\n2243/2243 [==============================] - 180s 80ms/step - loss: 0.0566 - val_loss: 0.0522\nEpoch 3/5\n2243/2243 [==============================] - 190s 85ms/step - loss: 0.0563 - val_loss: 0.0522\nEpoch 4/5\n1743/2243 [======================>.......] - ETA: 32s - loss: 0.0564"
]
],
[
[
"## Saving Model",
"_____no_output_____"
]
],
[
[
"model.save(ROOT_PATH/'models/model-nvidia-base-3.h5', include_optimizer=False)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d05ba4aa7473f99b996e1c91e29a331d025b15e1 | 32,313 | ipynb | Jupyter Notebook | AUTHOR_CLASSIFIER_NLTK.ipynb | smartysam9821/ML | 66422f0ca4b9e329ac16fb23ce3273fc6be93a7a | [
"MIT"
] | null | null | null | AUTHOR_CLASSIFIER_NLTK.ipynb | smartysam9821/ML | 66422f0ca4b9e329ac16fb23ce3273fc6be93a7a | [
"MIT"
] | null | null | null | AUTHOR_CLASSIFIER_NLTK.ipynb | smartysam9821/ML | 66422f0ca4b9e329ac16fb23ce3273fc6be93a7a | [
"MIT"
] | null | null | null | 34.049526 | 281 | 0.431684 | [
[
[
"import numpy as np\nimport os\nimport pandas as pd\nfrom sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer\nfrom sklearn.metrics.classification import classification_report, accuracy_score\nfrom sklearn.model_selection import cross_val_predict\nfrom nltk.corpus import stopwords\n",
"_____no_output_____"
],
[
"stop_words=stopwords.words('english')",
"_____no_output_____"
],
[
"trainX =np.array([])\nlabels = []\npath = '../Downloads/C50train/'\nauthors = os.listdir(path)[:10]; \nauthors = [i for i in authors if '.DS_Store' not in i]\nfor auth in authors:\n if authors != '.DS_Store':\n files = os.listdir(path + auth + '/');\n tmpX, tmpY = np.array([]), []\n for file in files:\n f = open(os.path.join(path, auth, file), 'r')\n data = f.read().replace('\\n', ' ')\n tmpX = np.append(tmpX,data)\n tmpY = tmpY + [auth]\n f.close()\n trainX = np.append(trainX, tmpX)\n labels = labels + tmpY",
"_____no_output_____"
],
[
"# initializing Count Vector\nvect = CountVectorizer()\nvect",
"_____no_output_____"
],
[
"# Split in to train and test data\nfrom sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(trainX, labels, train_size = 0.8)\n",
"_____no_output_____"
],
[
"# converting into pandas series\nX_train = pd.Series(X_train)\nX_test = pd.Series(X_test)\nX_train.head()\n",
"_____no_output_____"
],
[
"# learn the 'vocabulary' of the training data\nvect.fit(X_train)\n",
"_____no_output_____"
],
[
"# examine the fitted vocabulary\nvect.get_feature_names()[:50]\ntrain_vectors = vect.transform(X_train)\ntrain_vectors\n",
"_____no_output_____"
],
[
"train_vectors\ntest_vectors = vect.transform(X_test)\n",
"_____no_output_____"
],
[
"test_vectors\npd.DataFrame(train_vectors.toarray(), columns=vect.get_feature_names()).head(4)",
"_____no_output_____"
],
[
"from sklearn.naive_bayes import GaussianNB, MultinomialNB, BernoulliNB\nnb = GaussianNB()\nnb.fit(train_vectors.toarray(), y_train)",
"_____no_output_____"
],
[
"# generate your cross-validation prediction with 10 fold \n#Stratified sampling\ny_pred = cross_val_predict(nb, test_vectors.toarray(), y_test, cv=10)\nprint(classification_report(y_test, y_pred))\nprint(\"ACCURACY::\", accuracy_score(y_pred, y_test))",
"C:\\Users\\Admin\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_split.py:657: Warning: The least populated class in y has only 6 members, which is too few. The minimum number of members in any class cannot be less than n_splits=10.\n % (min_groups, self.n_splits)), Warning)\n"
],
[
"def classifier():\n X_train, X_test, y_train, y_test = train_test_split(trainX, labels, train_size = 0.8)\n \n # converting into pandas series\n X_train = pd.Series(X_train)\n X_test = pd.Series(X_test)\n \n vect.fit(X_train)\n # examine the fitted vocabulary\n \n vect.get_feature_names()[:50]\n train_vectors = vect.transform(X_train)\n train_vectors\n test_vectors = vect.transform(X_test)\n test_vectors\n pd.DataFrame(train_vectors.toarray(), columns=vect.get_feature_names()).head(4)\n nb.fit(train_vectors.toarray(), y_train)\n # generate your cross-validation prediction with 10 fold \n #Stratified sampling\n y_pred = cross_val_predict(nb, test_vectors.toarray(), y_test, cv=10)\n print(classification_report(y_test, y_pred))\n print(\"ACCURACY::\", accuracy_score(y_pred, y_test))",
"_____no_output_____"
],
[
"# initializing Count Vector\nvect = CountVectorizer(ngram_range=(1, 2))\n",
"_____no_output_____"
],
[
"classifier()",
"C:\\Users\\Admin\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_split.py:657: Warning: The least populated class in y has only 6 members, which is too few. The minimum number of members in any class cannot be less than n_splits=10.\n % (min_groups, self.n_splits)), Warning)\n"
],
[
"# initializing Count Vector\nvect = CountVectorizer(ngram_range=(1, 3))\n",
"_____no_output_____"
],
[
"classifier()",
"/Users/sumit/opt/anaconda3/lib/python3.7/site-packages/sklearn/model_selection/_split.py:657: Warning: The least populated class in y has only 7 members, which is too few. The minimum number of members in any class cannot be less than n_splits=10.\n % (min_groups, self.n_splits)), Warning)\n"
],
[
"# initializing Count Vector\nvect = CountVectorizer(ngram_range=(1, 2),stop_words=stop_words)",
"_____no_output_____"
],
[
"classifier()",
"C:\\Users\\Admin\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_split.py:657: Warning: The least populated class in y has only 7 members, which is too few. The minimum number of members in any class cannot be less than n_splits=10.\n % (min_groups, self.n_splits)), Warning)\n"
],
[
"vect = CountVectorizer(ngram_range=(1, 2),stop_words=stop_words, max_df=0.7)\nclassifer()",
"/Users/sumit/opt/anaconda3/lib/python3.7/site-packages/sklearn/model_selection/_split.py:657: Warning: The least populated class in y has only 7 members, which is too few. The minimum number of members in any class cannot be less than n_splits=10.\n % (min_groups, self.n_splits)), Warning)\n"
],
[
"vect = CountVectorizer(ngram_range=(1, 2),stop_words=stop_words, max_df=0.75)\nclassifier()",
"C:\\Users\\Admin\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_split.py:657: Warning: The least populated class in y has only 5 members, which is too few. The minimum number of members in any class cannot be less than n_splits=10.\n % (min_groups, self.n_splits)), Warning)\n"
],
[
"vect = CountVectorizer(ngram_range=(1, 2),stop_words=stop_words, max_df=0.8)\nclassifier()",
"C:\\Users\\Admin\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_split.py:657: Warning: The least populated class in y has only 5 members, which is too few. The minimum number of members in any class cannot be less than n_splits=10.\n % (min_groups, self.n_splits)), Warning)\n"
],
[
"vect = CountVectorizer(ngram_range=(1, 3),stop_words=stop_words, max_df=0.7)\nclassifier()",
"C:\\Users\\Admin\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_split.py:657: Warning: The least populated class in y has only 7 members, which is too few. The minimum number of members in any class cannot be less than n_splits=10.\n % (min_groups, self.n_splits)), Warning)\n"
],
[
"vect = CountVectorizer(ngram_range=(1, 3),stop_words=stop_words, max_df=0.75)\nclassifier()",
"C:\\Users\\Admin\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_split.py:657: Warning: The least populated class in y has only 4 members, which is too few. The minimum number of members in any class cannot be less than n_splits=10.\n % (min_groups, self.n_splits)), Warning)\n"
],
[
"vect = CountVectorizer(ngram_range=(1, 3),stop_words=stop_words, max_df=0.8)\nclassifier()",
"C:\\Users\\Admin\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_split.py:657: Warning: The least populated class in y has only 3 members, which is too few. The minimum number of members in any class cannot be less than n_splits=10.\n % (min_groups, self.n_splits)), Warning)\n"
],
[
"tf = TfidfVectorizer()\nclassifier()",
"C:\\Users\\Admin\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_split.py:657: Warning: The least populated class in y has only 7 members, which is too few. The minimum number of members in any class cannot be less than n_splits=10.\n % (min_groups, self.n_splits)), Warning)\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05ba5187c5bf4c99fc54d65dba9ecf2cbb3bcf5 | 190,091 | ipynb | Jupyter Notebook | notebooks/Fit-PLZ.ipynb | adrn/dr2-plz | 59c9cd536da4fe5d93a4a5bb744e36359e118878 | [
"MIT"
] | null | null | null | notebooks/Fit-PLZ.ipynb | adrn/dr2-plz | 59c9cd536da4fe5d93a4a5bb744e36359e118878 | [
"MIT"
] | null | null | null | notebooks/Fit-PLZ.ipynb | adrn/dr2-plz | 59c9cd536da4fe5d93a4a5bb744e36359e118878 | [
"MIT"
] | null | null | null | 330.019097 | 77,128 | 0.909938 | [
[
[
"import os\nos.environ['MACOSX_DEPLOYMENT_TARGET'] = '10.9' # HACK: needed for stan...\n\nimport astropy.coordinates as coord\nfrom astropy.table import Table, join, hstack\nimport astropy.units as u\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport numpy as np\nimport pystan\n\nfrom pyia import GaiaData",
"_____no_output_____"
],
[
"sm = pystan.StanModel('../stan/plz.stan')",
"INFO:pystan:COMPILING THE C++ CODE FOR MODEL anon_model_5fa9bbb8083cdec384c8bd3691721879 NOW.\n"
],
[
"tbl1 = Table.read('/Users/adrian/data/GaiaDR2/rrlyrae.fits')\ntbl2 = Table.read('/Users/adrian/data/GaiaDR2/vari_rrlyrae.fits')\ntbl1.meta = None\ntbl2.meta = None\ntbl2 = tbl2[[x for x in tbl2.colnames if x not in tbl1.colnames or x == 'source_id']]\ntbl = join(tbl1, tbl2, keys='source_id')",
"_____no_output_____"
],
[
"g = GaiaData(tbl)\nc = g.get_skycoord(distance=False)",
"_____no_output_____"
],
[
"k2014 = Table.read('/Users/adrian/data/Misc/Klein2014_rrlyrae.fit')\nkc = coord.SkyCoord(k2014['_RA'], k2014['_DE'], unit=u.deg)",
"_____no_output_____"
],
[
"idx, sep, _ = kc.match_to_catalog_sky(c)\njoin_tbl = hstack((tbl[idx[sep < 5*u.arcsec]], k2014[sep < 5*u.arcsec]))\n# join_tbl = tbl\n\ng = GaiaData(join_tbl)\ng.data['ebv'] = g.get_ebv()",
"_____no_output_____"
],
[
"plx_snr_mask = (g.parallax / g.parallax_error) > 10\n# plx_snr_mask = np.ones(len(g), dtype=bool)\n\nrp_nobs_mask = g.phot_rp_n_obs >= 20\n# rp_good_mask = np.isfinite(g.int_average_rp)\nrp_good_mask = np.isfinite(g.W1mag.value)\n\nfeh_mask = np.isfinite(g.metallicity) & (g.metallicity.value > -3) & (g.metallicity.value < 0)\nab_mask = g.best_classification == 'RRab'\nebv_mask = g.ebv < 0.5\npf_mask = (g.pf > 0.1*u.day) & (g.pf < 1*u.day)\n\nM = g.int_average_rp.value - g.get_distance(allow_negative=True).distmod.value\nabs_rp_mask = M < 1.\n\nall_mask = plx_snr_mask & rp_nobs_mask & rp_good_mask & feh_mask & ab_mask & ebv_mask & pf_mask & abs_rp_mask\nsub_g = g[all_mask]\n\n# HACK:\nsub_g = sub_g[sub_g.parallax > 0.5*u.mas]\nlen(sub_g)",
"/Users/adrian/anaconda/lib/python3.6/site-packages/ipykernel/__main__.py:8: RuntimeWarning: invalid value encountered in greater\n/Users/adrian/anaconda/lib/python3.6/site-packages/ipykernel/__main__.py:8: RuntimeWarning: invalid value encountered in less\n/Users/adrian/anaconda/lib/python3.6/site-packages/astropy/units/quantity.py:463: RuntimeWarning: invalid value encountered in greater\n result = super().__array_ufunc__(function, method, *arrays, **kwargs)\n/Users/adrian/anaconda/lib/python3.6/site-packages/astropy/units/quantity.py:463: RuntimeWarning: invalid value encountered in less\n result = super().__array_ufunc__(function, method, *arrays, **kwargs)\n/Users/adrian/anaconda/lib/python3.6/site-packages/ipykernel/__main__.py:14: RuntimeWarning: invalid value encountered in less\n"
],
[
"data = dict()\n\ndata['n_stars'] = len(sub_g)\ndata['Alambda'] = 0.1\n# kRP = 0.6104 # from https://www.aanda.org/articles/aa/pdf/2018/08/aa32843-18.pdf - taking just c1\n# data['Alambda'] = kRP * 3.1\n\ndata['plx'] = sub_g.parallax.value\ndata['plx_err'] = sub_g.parallax_error.value\n\n# data['mag'] = sub_g.int_average_rp.value\n# data['mag_err'] = sub_g.int_average_rp_error.value\ndata['mag'] = sub_g.W1mag.value\ndata['mag_err'] = sub_g.e_W1mag.value\n\ndata['EBV'] = sub_g.get_ebv()\ndata['EBV_err'] = 0.1 * data['EBV']\n\ndata['FeH'] = sub_g.metallicity.value\ndata['FeH_err'] = sub_g.metallicity_error.value\n\ndata['log10P'] = np.log10(sub_g.pf.value)\ndata['log10P_err'] = np.log10(np.e) * (sub_g.pf_error.value / sub_g.pf.value)\n\n# Sesar+2017\ndata['log10P_ref'] = np.log10(0.52854)\ndata['FeH_ref'] = -1.4",
"_____no_output_____"
],
[
"plx_samples = np.random.normal(sub_g.parallax.value, sub_g.parallax_error.value,\n size=(1024, len(sub_g)))\nDM_samples = coord.Distance(parallax=plx_samples * sub_g.parallax.unit).distmod.value\nDM_err = np.std(DM_samples, axis=0)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(6, 6))\n\nax.errorbar(sub_g.pf.value, data['mag'] - sub_g.distmod.value, \n yerr=np.sqrt(data['mag_err']**2 + DM_err**2), \n marker='o', ls='none', ecolor='#555555', color='k')\n\nax.set_xlabel('period [day]')\nax.set_ylabel('$M$ [day]')\n\nax.set_xlim(0.35, 0.8)\nax.set_ylim(0.1, -1.3)\n# ax.set_ylim(1.5, -0.5)\n\nax.set_xscale('log')",
"_____no_output_____"
],
[
"init = dict()\n\ninit['ln_s_M'] = np.log(0.1)\ninit['plx0'] = 0.\ninit['ln_sig_plx_add'] = np.log(1e-8)\ninit['f_plx'] = 1.\n\ninit['r'] = 1000 / sub_g.parallax.value\ninit['L'] = 250.\ninit['FeH_int'] = data['FeH']\n# init['EBV_int'] = data['EBV']\n# init['log10P_int'] = data['log10P']\n\ninit['a'] = -2.\ninit['b'] = 0.15\ninit['M_ref'] = -0.5",
"_____no_output_____"
],
[
"fit = sm.optimizing(data=data, init=init, iter=4096)\nsamples = sm.sampling(data=data, chains=1, init=[fit], control=dict(adapt_delta=1))",
"WARNING:pystan:n_eff / iter below 0.001 indicates that the effective sample size has likely been overestimated\nWARNING:pystan:Rhat above 1.1 or below 0.9 indicates that the chains very likely have not mixed\nWARNING:pystan:33 of 1000 iterations ended with a divergence (3.3 %).\nWARNING:pystan:Try running with adapt_delta larger than 1 to remove the divergences.\n"
],
[
"fit",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(6, 6))\n\nax.errorbar(sub_g.pf.value, data['mag'] - sub_g.distmod.value, \n yerr=np.sqrt(data['mag_err']**2 + DM_err**2), \n marker='o', ls='none', ecolor='#555555', color='k')\n\nxx = np.linspace(0., 1, 128)\n# for feh in np.arange(-2.5, -0.5+1e-3, 0.5):\n# M = (fit['a'] * (np.log10(xx) - data['log10P_ref']) +\n# # fit['b'] * (feh - data['FeH_ref']) +\n# fit['M_ref'])\n# ax.plot(xx, M, marker='')\n \nM = fit['a'] * (np.log10(xx) - data['log10P_ref']) + fit['M_ref']\nax.plot(xx, M, marker='')\n \nM = np.mean(samples['a']) * (np.log10(xx) - data['log10P_ref']) + np.mean(samples['M_ref'])\nax.plot(xx, M, marker='')\n\nax.set_xlabel('period [day]')\nax.set_ylabel('$M$ [day]')\n\nax.set_xlim(0.35, 0.8)\n# ax.set_ylim(0.1, -1.3)\nax.set_ylim(1.5, -0.5)\n\nax.set_xscale('log')\n\nax.axvline(10 ** data['log10P_ref'], zorder=-100, color='tab:blue', alpha=0.5)\nax.axhline(fit['M_ref'], zorder=-100, color='tab:blue', alpha=0.5)",
"/Users/adrian/anaconda/lib/python3.6/site-packages/ipykernel/__main__.py:14: RuntimeWarning: divide by zero encountered in log10\n/Users/adrian/anaconda/lib/python3.6/site-packages/ipykernel/__main__.py:17: RuntimeWarning: divide by zero encountered in log10\n"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(6, 6))\n\n# ax.errorbar(data['log10P'], data['plx'] - 1000 / fit['r'], data['plx_err'], ls='none')\nax.errorbar(data['log10P'], data['plx'] - 1000 / np.mean(samples['r'], axis=0), \n data['plx_err'], \n marker='o', ls='none', ecolor='#555555', color='k')\n\nax.axhline(np.mean(samples['plx0']), color='tab:red', zorder=-10)\n\nax.axhline(0, color='tab:blue', zorder=-100)\nax.set_ylim(-0.1, 0.1)",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05bae079152adb87bcb825619bd1b790d5ae26d | 9,780 | ipynb | Jupyter Notebook | jupyter/README.ipynb | MUSC-TBIC/etude-engine | 943608ae3458bfcecc5e1c0b24fb3aa5c8bc0cad | [
"Apache-2.0"
] | 9 | 2018-11-03T20:49:41.000Z | 2021-10-30T23:11:28.000Z | jupyter/README.ipynb | MUSC-TBIC/etude-engine | 943608ae3458bfcecc5e1c0b24fb3aa5c8bc0cad | [
"Apache-2.0"
] | 1 | 2019-06-04T17:17:41.000Z | 2019-06-04T17:17:41.000Z | jupyter/README.ipynb | MUSC-TBIC/etude-engine | 943608ae3458bfcecc5e1c0b24fb3aa5c8bc0cad | [
"Apache-2.0"
] | null | null | null | 29.281437 | 131 | 0.516258 | [
[
[
"from __future__ import print_function\n\nimport sys\nif sys.version_info[0] < 3: \n from StringIO import StringIO\nelse:\n from io import StringIO\n\nfrom os import chdir\nchdir( '../' )",
"_____no_output_____"
]
],
[
[
"Sample Runs\n=========\n\nBasic Run\n--------\nThe simplest test run requires that we specify a reference directory and a\ntest directory. The default file matching assumes that our reference and\ntest files match names exactly and both end in '.xml'. With just the\ntwo directory arguments, we get micro-average scores for the default\nmetrics across the full directory. ",
"_____no_output_____"
]
],
[
[
"!python etude.py \\\n --reference-input tests/data/i2b2_2016_track-1_reference \\\n --test-input tests/data/i2b2_2016_track-1_test\n",
"100% (10 of 10) |##########################| Elapsed Time: 0:00:01 Time: 0:00:01\n\nexact\tTP\tFP\tTN\tFN\nmicro-average\t340.0\t8.0\t0.0\t105.0\n"
]
],
[
[
"In the next sample runs, you can see how to include a per-file score breakdown and a per-annotation-type score breakdown.",
"_____no_output_____"
]
],
[
[
"!python etude.py \\\n --reference-input tests/data/i2b2_2016_track-1_reference \\\n --test-input tests/data/i2b2_2016_track-1_test \\\n --by-file",
"100% (10 of 10) |##########################| Elapsed Time: 0:00:01 Time: 0:00:01\n\nexact\tTP\tFP\tTN\tFN\nmicro-average\t340.0\t8.0\t0.0\t105.0\n0005_gs.xml\t31.0\t0.0\t0.0\t0.0\n0016_gs.xml\t21.0\t0.0\t0.0\t30.0\n0267_gs.xml\t27.0\t0.0\t0.0\t32.0\n0273_gs.xml\t0.0\t0.0\t0.0\t35.0\n0389_gs.xml\t26.0\t8.0\t0.0\t8.0\n0475_gs.xml\t45.0\t0.0\t0.0\t0.0\n0617_gs.xml\t32.0\t0.0\t0.0\t0.0\n0709_gs.xml\t41.0\t0.0\t0.0\t0.0\n0982_gs.xml\t95.0\t0.0\t0.0\t0.0\n0992_gs.xml\t22.0\t0.0\t0.0\t0.0\n"
],
[
"!python etude.py \\\n --reference-input tests/data/i2b2_2016_track-1_reference \\\n --test-input tests/data/i2b2_2016_track-1_test \\\n --by-type",
"100% (10 of 10) |##########################| Elapsed Time: 0:00:01 Time: 0:00:01\n\nexact\tTP\tFP\tTN\tFN\nmicro-average\t340.0\t8.0\t0.0\t105.0\nAge\t63.0\t2.0\t0.0\t29.0\nDateTime\t91.0\t2.0\t0.0\t33.0\nHCUnit\t61.0\t4.0\t0.0\t15.0\nOtherID\t7.0\t0.0\t0.0\t0.0\nOtherLoc\t1.0\t0.0\t0.0\t4.0\nOtherOrg\t18.0\t0.0\t0.0\t3.0\nPatient\t16.0\t0.0\t0.0\t3.0\nPhoneFax\t5.0\t0.0\t0.0\t1.0\nProvider\t54.0\t0.0\t0.0\t10.0\nStateCountry\t14.0\t0.0\t0.0\t7.0\nStreetCity\t4.0\t0.0\t0.0\t0.0\nZip\t4.0\t0.0\t0.0\t0.0\neAddress\t2.0\t0.0\t0.0\t0.0\n"
]
],
[
[
"Scoring on Different Fields\n-----------------------\n\nThe above examples show scoring based on the default key in the\nconfiguration file used for matching the reference to the test\nconfiguration. You may wish to group annotations on different fields,\nsuch as the parent class or long description.\n",
"_____no_output_____"
]
],
[
[
"!python etude.py \\\n --reference-input tests/data/i2b2_2016_track-1_reference \\\n --test-input tests/data/i2b2_2016_track-1_test \\\n --by-type\n",
"100% (10 of 10) |##########################| Elapsed Time: 0:00:01 Time: 0:00:01\n\nexact\tTP\tFP\tTN\tFN\nmicro-average\t340.0\t8.0\t0.0\t105.0\nAge\t63.0\t2.0\t0.0\t29.0\nDateTime\t91.0\t2.0\t0.0\t33.0\nHCUnit\t61.0\t4.0\t0.0\t15.0\nOtherID\t7.0\t0.0\t0.0\t0.0\nOtherLoc\t1.0\t0.0\t0.0\t4.0\nOtherOrg\t18.0\t0.0\t0.0\t3.0\nPatient\t16.0\t0.0\t0.0\t3.0\nPhoneFax\t5.0\t0.0\t0.0\t1.0\nProvider\t54.0\t0.0\t0.0\t10.0\nStateCountry\t14.0\t0.0\t0.0\t7.0\nStreetCity\t4.0\t0.0\t0.0\t0.0\nZip\t4.0\t0.0\t0.0\t0.0\neAddress\t2.0\t0.0\t0.0\t0.0\n"
],
[
"!python etude.py \\\n --reference-input tests/data/i2b2_2016_track-1_reference \\\n --test-input tests/data/i2b2_2016_track-1_test \\\n --by-type \\\n --score-key \"Parent\"",
"100% (10 of 10) |##########################| Elapsed Time: 0:00:01 Time: 0:00:01\n\nexact\tTP\tFP\tTN\tFN\nmicro-average\t341.0\t7.0\t0.0\t104.0\nAddress\t22.0\t0.0\t0.0\t7.0\nContact Information\t7.0\t0.0\t0.0\t1.0\nIdentifiers\t7.0\t0.0\t0.0\t0.0\nLocations\t80.0\t4.0\t0.0\t22.0\nNames\t70.0\t0.0\t0.0\t13.0\nTime\t155.0\t3.0\t0.0\t61.0\n"
],
[
"!python etude.py \\\n --reference-input tests/data/i2b2_2016_track-1_reference \\\n --test-input tests/data/i2b2_2016_track-1_test \\\n --by-type \\\n --score-key \"Long Name\"",
"100% (10 of 10) |##########################| Elapsed Time: 0:00:01 Time: 0:00:01\n\nexact\tTP\tFP\tTN\tFN\nmicro-average\t340.0\t8.0\t0.0\t105.0\nAge Greater than 89\t63.0\t2.0\t0.0\t29.0\nDate and Time Information\t91.0\t2.0\t0.0\t33.0\nElectronic Address Information\t2.0\t0.0\t0.0\t0.0\nHealth Care Provider Name\t54.0\t0.0\t0.0\t10.0\nHealth Care Unit Name\t61.0\t4.0\t0.0\t15.0\nOther ID Numbers\t7.0\t0.0\t0.0\t0.0\nOther Locations\t1.0\t0.0\t0.0\t4.0\nOther Organization Name\t18.0\t0.0\t0.0\t3.0\nPatient Name\t16.0\t0.0\t0.0\t3.0\nPhone, Fax, or Pager Number\t5.0\t0.0\t0.0\t1.0\nState or Country\t14.0\t0.0\t0.0\t7.0\nStreet City Name\t4.0\t0.0\t0.0\t0.0\nZIP Code\t4.0\t0.0\t0.0\t0.0\n"
]
],
[
[
"Testing\n=====\n\nUnit testing is done with the pytest module.\nBecause of a bug in how tests are processed in Python 2.7, you should run pytest indirectly rather than directly.\nAn [HTML-formatted coverage guide](../htmlcov/index.html) will be generated locally under the directory containing this code.",
"_____no_output_____"
]
],
[
[
"!python -m pytest --cov-report html --cov=./ tests",
"\u001b[1m============================= test session starts ==============================\u001b[0m\nplatform darwin -- Python 2.7.13, pytest-3.1.1, py-1.4.34, pluggy-0.4.0\nrootdir: /Users/pmh/git/etude, inifile:\nplugins: cov-2.5.1\ncollected 107 items \u001b[0m\u001b[1m1m\u001b[1m\n\u001b[0m\ntests/test_args_and_configs.py ..................\ntests/test_etude.py .......\ntests/test_scoring_metrics.py ...............................................................\ntests/test_text_extraction.py ...................\n\n---------- coverage: platform darwin, python 2.7.13-final-0 ----------\nCoverage HTML written to dir htmlcov\n\n\n\u001b[32m\u001b[1m========================== 107 passed in 3.50 seconds ==========================\u001b[0m\n"
]
],
[
[
"<html/index.html>",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d05bb351ac1fe922c2a0c7daad503c951143f51e | 27,170 | ipynb | Jupyter Notebook | xtr_tune_drop_lmse.ipynb | alexsyrom/datascience-ml-2 | 7f776560d06943f4aed853919d8d4aa6c32e6cc0 | [
"MIT"
] | null | null | null | xtr_tune_drop_lmse.ipynb | alexsyrom/datascience-ml-2 | 7f776560d06943f4aed853919d8d4aa6c32e6cc0 | [
"MIT"
] | null | null | null | xtr_tune_drop_lmse.ipynb | alexsyrom/datascience-ml-2 | 7f776560d06943f4aed853919d8d4aa6c32e6cc0 | [
"MIT"
] | null | null | null | 23.462867 | 127 | 0.466286 | [
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nfrom tqdm import tnrange, tqdm_notebook\nimport gc\nimport operator",
"_____no_output_____"
],
[
"sns.set_context('talk')",
"_____no_output_____"
],
[
"pd.set_option('display.max_columns', 500)",
"_____no_output_____"
],
[
"import warnings\nwarnings.filterwarnings('ignore', message='Changing the shape of non-C contiguous array')",
"_____no_output_____"
]
],
[
[
"# Read the data",
"_____no_output_____"
]
],
[
[
"dfXtrain = pd.read_csv('preprocessed_csv/train_tree.csv', index_col='id', sep=';')\ndfXtest = pd.read_csv('preprocessed_csv/test_tree.csv', index_col='id', sep=';')\ndfYtrain = pd.read_csv('preprocessed_csv/y_train_tree.csv', header=None, names=['ID', 'COTIS'], sep=';')",
"_____no_output_____"
],
[
"dfYtrain = dfYtrain.set_index('ID')",
"_____no_output_____"
]
],
[
[
"# Preprocessing",
"_____no_output_____"
],
[
"Вынесем var14, department и subreg.",
"_____no_output_____"
]
],
[
[
"dropped_col_names = ['var14', 'department', 'subreg'] \n\ndef drop_cols(df):\n return df.drop(dropped_col_names, axis=1), df[dropped_col_names]",
"_____no_output_____"
],
[
"train, dropped_train = drop_cols(dfXtrain)\ntest, dropped_test = drop_cols(dfXtest)",
"_____no_output_____"
]
],
[
[
"Добавим инфу о величине города из subreg'a",
"_____no_output_____"
]
],
[
[
"def add_big_city_cols(df, dropped_df):\n df['big'] = np.where(dropped_df['subreg'] % 100 == 0, 1, 0)\n df['average'] = np.where(dropped_df['subreg'] % 10 == 0, 1, 0)\n df['average'] = df['average'] - df['big']\n df['small'] = 1 - df['big'] - df['average']\n return df",
"_____no_output_____"
],
[
"train = add_big_city_cols(train, dropped_train)\ntest = add_big_city_cols(test, dropped_test)",
"_____no_output_____"
]
],
[
[
"Декодируем оставшиеся категориальные признаки",
"_____no_output_____"
]
],
[
[
"categorical = list(train.select_dtypes(exclude=[np.number]).columns)\ncategorical",
"_____no_output_____"
],
[
"list(test.select_dtypes(exclude=[np.number]).columns)",
"_____no_output_____"
],
[
"for col in categorical:\n print(col, train[col].nunique())",
"marque 154\nenergie_veh 5\nprofession 17\nvar6 5\nvar8 23\n"
]
],
[
[
"energie_veh и var6 с помощью get_dummies",
"_____no_output_____"
]
],
[
[
"small_cat = ['energie_veh', 'var6']",
"_____no_output_____"
],
[
"train = pd.get_dummies(train, columns=small_cat)\ntest = pd.get_dummies(test, columns=small_cat)",
"_____no_output_____"
]
],
[
[
"Для остальных посчитаем сглаженные средние таргета",
"_____no_output_____"
]
],
[
[
"big_cat = ['marque', 'profession', 'var8']",
"_____no_output_____"
]
],
[
[
"Описание для начала",
"_____no_output_____"
]
],
[
[
"df = pd.concat([dfYtrain.describe()] + [train[col].value_counts().describe() for col in big_cat], axis=1)\ndf",
"_____no_output_____"
]
],
[
[
"Сглаживать будем с 500",
"_____no_output_____"
],
[
"Будем использовать среднее, 25%, 50% и 75%",
"_____no_output_____"
],
[
"Декодирование",
"_____no_output_____"
]
],
[
[
"class EncodeWithAggregates():\n\n def __init__(self, cols, y_train, train, *tests):\n self.cols = cols\n self.y_train = y_train\n self.train = train\n self.tests = tests\n self.Xs = (self.train,) + self.tests\n \n self.smooth_coef = 500\n self.miss_val = 'NAN'\n self.percentiles = [25, 50, 75]\n self.names = ['Mean'] + [str(q) for q in self.percentiles]\n self.aggs = [np.mean] + [self.percentile_fix(q) for q in self.percentiles]\n self.miss_val_fills = [agg(y_train) for agg in self.aggs]\n self.train_aggs = [agg(y_train) for agg in self.aggs]\n\n def percentile_fix(self, q):\n def wrapped(a):\n return np.percentile(a, q)\n\n return wrapped\n \n \n def transform(self):\n for col in self.cols:\n self.encode(col)\n gc.collect()\n return self.Xs\n \n \n def encode(self, col):\n df = pd.concat([self.y_train, self.train[col]], axis=1)\n dfgb = df.groupby(col)\n dfsize = dfgb.size()\n dfsize.ix[self.miss_val] = 0\n \n for name, agg, miss_val_fill, train_agg in zip(self.names, self.aggs, self.miss_val_fills, self.train_aggs):\n dfm = dfgb.agg(agg)\n dfm.ix[self.miss_val] = miss_val_fill\n for X in self.Xs: \n agg_df = dfm.ix[X[col].fillna(self.miss_val)].set_index(X.index)[self.y_train.name]\n agg_size = dfsize.ix[X[col].fillna(self.miss_val)]\n agg_size = pd.DataFrame({'size': agg_size}).set_index(X.index)['size']\n agg_name = \"{}_{}\".format(col, name)\n X[agg_name] = (agg_df * agg_size + self.smooth_coef * train_agg) / (self.smooth_coef + agg_size)\n \n self.Xs = [X.drop(col, axis=1) for X in self.Xs]\n",
"_____no_output_____"
],
[
"train, test = EncodeWithAggregates(big_cat, dfYtrain['COTIS'], train, test).transform()",
"_____no_output_____"
],
[
"test.shape",
"_____no_output_____"
],
[
"train.shape",
"_____no_output_____"
],
[
"train.fillna(-9999, inplace=True)\ntest.fillna(-9999, inplace=True)",
"_____no_output_____"
],
[
"y_train = np.array(dfYtrain)\nx_train = np.array(train)\nx_test = np.array(test)",
"_____no_output_____"
]
],
[
[
"# Save routines",
"_____no_output_____"
]
],
[
[
"dfYtest = pd.DataFrame({'ID': dfXtest.index, 'COTIS': np.zeros(test.shape[0])})\ndfYtest = dfYtest[['ID', 'COTIS']]\ndfYtest.head()",
"_____no_output_____"
],
[
"def save_to_file(y, file_name):\n dfYtest['COTIS'] = y\n dfYtest.to_csv('results/{}'.format(file_name), index=False, sep=';')",
"_____no_output_____"
],
[
"model_name = 'lmse_without_size_xtr'\ndfYtest_stacking = pd.DataFrame({'ID': dfXtrain.index, model_name: np.zeros(train.shape[0])})\ndfYtest_stacking = dfYtest_stacking[['ID', model_name]]\ndfYtest_stacking.head()",
"_____no_output_____"
],
[
"def save_to_file_stacking(y, file_name):\n dfYtest_stacking[model_name] = y\n dfYtest_stacking.to_csv('stacking/{}'.format(file_name), index=False, sep=';')",
"_____no_output_____"
]
],
[
[
"# Train XGB",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import ExtraTreesRegressor",
"_____no_output_____"
],
[
"def plot_quality(grid_searcher, param_name):\n means = []\n stds = []\n for elem in grid_searcher.grid_scores_:\n means.append(np.mean(elem.cv_validation_scores))\n stds.append(np.sqrt(np.var(elem.cv_validation_scores)))\n means = np.array(means)\n stds = np.array(stds)\n \n params = grid_searcher.param_grid\n \n plt.figure(figsize=(10, 6))\n plt.plot(params[param_name], means)\n plt.fill_between(params[param_name], \\\n means + stds, means - stds, alpha = 0.3, facecolor='blue')\n plt.xlabel(param_name)\n plt.ylabel('MAPE')",
"_____no_output_____"
],
[
"def mape(y_true, y_pred): \n return -np.mean(np.abs((y_true - y_pred) / y_true)) * 100",
"_____no_output_____"
],
[
"def mape_scorer(est, X, y):\n gc.collect()\n return mape(y, est.predict(X))",
"_____no_output_____"
],
[
"class MyGS():\n class Element():\n def __init__(self):\n self.cv_validation_scores = []\n \n \n def add(self, score):\n self.cv_validation_scores.append(score)\n \n \n def __init__(self, param_grid, name, n_folds):\n self.param_grid = {name: param_grid}\n self.grid_scores_ = [MyGS.Element() for item in param_grid]\n \n \n def add(self, score, param_num):\n self.grid_scores_[param_num].add(score)\n ",
"_____no_output_____"
],
[
"validation_index = (dropped_train.department == 1) | (dropped_train.department > 90)\ntrain_index = ~validation_index",
"_____no_output_____"
],
[
"subtrain, validation = train[train_index], train[validation_index]\nx_subtrain = np.array(subtrain)\nx_validation = np.array(validation)\n\nysubtrain, yvalidation = dfYtrain[train_index], dfYtrain[validation_index]\ny_subtrain = np.array(ysubtrain).flatten()\ny_validation = np.array(yvalidation).flatten()",
"_____no_output_____"
],
[
"%%time\nest = ExtraTreesRegressor(n_estimators=10, max_features=51, \n max_depth=None, n_jobs=-1, random_state=42).fit(X=x_subtrain, y=np.log(y_subtrain))",
"CPU times: user 1min 26s, sys: 619 ms, total: 1min 26s\nWall time: 26 s\n"
],
[
"y_pred = est.predict(x_validation)\nmape(y_validation, np.exp(y_pred))",
"_____no_output_____"
],
[
"est",
"_____no_output_____"
],
[
"sample_weight_subtrain = np.power(y_subtrain, -1)",
"_____no_output_____"
],
[
"from sklearn.tree import DecisionTreeRegressor",
"_____no_output_____"
],
[
"%%time\ncount = 10000\nest = DecisionTreeRegressor(criterion='mae', max_depth=2, \n max_features=None, random_state=42).fit(\n X=x_subtrain[:count], y=y_subtrain[:count], sample_weight=sample_weight_subtrain[:count])",
"_____no_output_____"
],
[
"gc.collect()",
"_____no_output_____"
],
[
"y_pred = est.predict(x_validation)\nmape(y_validation, y_pred)",
"_____no_output_____"
]
],
[
[
"# Save",
"_____no_output_____"
]
],
[
[
"save_to_file_stacking(y_lmse_pred * 0.995, 'xbg_tune_eta015_num300_dropped_lmse.csv')",
"_____no_output_____"
],
[
"%%time\n\nparam = {'base_score':0.5, 'colsample_bylevel':1, 'colsample_bytree':1, 'gamma':0,\n 'eta':0.15, 'max_delta_step':0, 'max_depth':9,\n 'min_child_weight':1, 'nthread':-1,\n 'objective':'reg:linear', 'alpha':0, 'lambda':1,\n 'scale_pos_weight':1, 'seed':56, 'silent':True, 'subsample':1}\n\nnum_round = 180\n\ndtrain = xgb.DMatrix(x_train, \n label=np.log(y_train), \n missing=-9999,) \n #weight=weight_coef * np.power(y_train[train_index], -2) )\ndtest = xgb.DMatrix(x_test, missing=-9999)\nparam['base_score'] = np.percentile(np.log(y_train), 25)\nbst = xgb.train(param, dtrain, num_round)\ny_pred = np.exp(bst.predict(dtest))\ngc.collect()",
"CPU times: user 4min 17s, sys: 367 ms, total: 4min 17s\nWall time: 4min 18s\n"
],
[
"save_to_file(y_pred * 0.995, 'xbg_tune_eta015_num300_dropped_lmse.csv')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d05bc2f9107b0eb3d68eb76ee6d5355ef2cd767d | 109,425 | ipynb | Jupyter Notebook | beginners/04.ML_Modelling.ipynb | MKulfan/redbull-analytics-hol | 79cd6c2646350f84e5aaabe074e03d020a301781 | [
"UPL-1.0"
] | 40 | 2021-07-11T16:28:25.000Z | 2022-03-14T06:51:07.000Z | beginners/04.ML_Modelling.ipynb | MKulfan/redbull-analytics-hol | 79cd6c2646350f84e5aaabe074e03d020a301781 | [
"UPL-1.0"
] | 38 | 2021-06-21T14:39:35.000Z | 2022-02-17T12:14:07.000Z | beginners/04.ML_Modelling.ipynb | MKulfan/redbull-analytics-hol | 79cd6c2646350f84e5aaabe074e03d020a301781 | [
"UPL-1.0"
] | 46 | 2021-06-21T11:09:56.000Z | 2022-02-02T04:31:15.000Z | 78.497131 | 30,796 | 0.762769 | [
[
[
"# Test For The Best Machine Learning Algorithm For Prediction",
"_____no_output_____"
],
[
"This notebook takes about 40 minutes to run, but we've already run it and saved the data for you. Please read through it, though, so that you understand how we came to the conclusions we'll use moving forward.\n\n## Six Algorithms\n\nWe're going to compare six different algorithms to determine the best one to produce an accurate model for our predictions.\n\n### Logistic Regression\n\nLogistic Regression (LR) is a technique borrowed from the field of statistics. It is the go-to method for binary classification problems (problems with two class values). \n\n\n\nLogistic Regression is named for the function used at the core of the method: the logistic function. The logistic function is a probablistic method used to determine whether or not the driver will be the winner. Logistic Regression predicts probabilities.\n\n### Decision Tree\n\nA tree has many analogies in real life, and it turns out that it has influenced a wide area of machine learning, covering both classification and regression. In decision analysis, a decision tree can be used to visually and explicitly represent decisions and decision making.\n\n\n\nThis methodology is more commonly known as a \"learning decision tree\" from data, and the above tree is called a Classification tree because the goal is to classify a driver as the winner or not.\n\n### Random Forest\n\nRandom forest is a supervised learning algorithm. The \"forest\" it builds is an **ensemble of decision trees**, usually trained with the “bagging” method, a combination of learning models which increases the accuracy of the result.\n\nA random forest eradicates the limitations of a decision tree algorithm. It reduces the overfitting of datasets and increases precision. It generates predictions without requiring many configurations.\n\n\n\nHere's the difference between the Decision Tree and Random Forest methods:\n\n\n\n### Support Vector Machine Algorithm (SVC)\n\nSupport Vector Machines (SVMs) are a set of supervised learning methods used for classification, regression and detection of outliers.\n\nThe advantages of support vector machines are:\n\n- Effective in high dimensional spaces\n- Still effective in cases where number of dimensions is greater than the number of samples\n- Uses a subset of training points in the decision function (called support vectors), so it is also memory efficient\n- Versatile: different kernel functions can be specified for the decision function. Common kernels are provided, but it is also possible to specify custom kernels\n\nThe objective of a SVC (Support Vector Classifier) is to fit to the data you provide, returning a \"best fit\" hyperplane that divides, or categorizes, your data.\n\n### Gaussian Naive Bayes Algorithm\n\nNaive Bayes is a classification algorithm for binary (two-class) and multi-class classification problems. The technique is easiest to understand when described using binary or categorical input values. The representation used for naive Bayes is probabilities.\n\nA list of probabilities is stored to a file for a learned Naive Bayes model. This includes:\n\n- **Class Probabilities:** The probabilities of each class in the training dataset.\n- **Conditional Probabilities:** The conditional probabilities of each input value given each class value.\n\nNaive Bayes can be extended to real-value attributes, most commonly by assuming a Gaussian distribution. This extension of Naive Bayes is called Gaussian Naive Bayes. Other functions can be used to estimate the distribution of the data, but the Gaussian (or normal distribution) is the easiest to work with because you only need to estimate the mean and the standard deviation from your training data.\n\n### k Nearest Neighbor Algorithm (kNN)\n\nThe k-Nearest Neighbors (KNN) algorithm is a simple, supervised machine learning algorithm that can be used to solve both classification and regression problems.\n\nkNN works by finding the distances between a query and all of the examples in the data, selecting the specified number examples (k) closest to the query, then voting for the most frequent label (in the case of classification) or averages the labels (in the case of regression).\n\nThe kNN algorithm assumes the similarity between the new case/data and available cases, and puts the new case into the category that is most similar to the available categories.\n\n\n\n## Analyzing the Data\n\n### Feature Importance\n\nAnother great quality of the random forest algorithm is that it's easy to measure the relative importance of each feature to the prediction.\n\nThe Scikit-learn Python Library provides a great tool for this which measures a feature's importance by looking at how much the tree nodes that use that feature reduce impurity across all trees in the forest. It computes this score automatically for each feature after training, and scales the results so the sum of all importance is equal to one.\n\n### Data Visualization When Building a Model\n\nHow do you visualize the influence of the data? How do you frame the problem?\n\nAn important tool in the data scientist's toolkit is the power to visualize data using several excellent libraries such as Seaborn or MatPlotLib. Representing your data visually might allow you to uncover hidden correlations that you can leverage. Your visualizations might also help you to uncover bias or unbalanced data.\n\n\n\n### Splitting the Dataset\n\nPrior to training, you need to split your dataset into two or more parts of unequal size that still represent the data well. \n\n1. Training. This part of the dataset is fit to your model to train it. This set constitutes the majority of the original dataset.\n2. Testing. A test dataset is an independent group of data, often a subset of the original data, that you use to confirm the performance of the model you built.\n3. Validating. A validation set is a smaller independent group of examples that you use to tune the model's hyperparameters, or architecture, to improve the model. Depending on your data's size and the question you are asking, you might not need to build this third set.\n\n## Building the Model\n\nUsing your training data, your goal is to build a model, or a statistical representation of your data, using various algorithms to train it. Training a model exposes it to data and allows it to make assumptions about perceived patterns it discovers, validates, and accepts or rejects.\n\n\n### Decide on a Training Method\n\nDepending on your question and the nature of your data, you will choose a method to train it. Stepping through Scikit-learn's documentation, you can explore many ways to train a model. Depending on the results you get, you might have to try several different methods to build the best model. You are likely to go through a process whereby data scientists evaluate the performance of a model by feeding it unseen data, checking for accuracy, bias, and other quality-degrading issues, and selecting the most appropriate training method for the task at hand.\n\n### Train a Model\n\nArmed with your training data, you are ready to \"fit\" it to create a model. In many ML libraries you will find the code 'model.fit' - it is at this time that you send in your data as an array of values (usually 'X') and a feature variable (usually 'y').\n\n### Evaluate the Model\n\nOnce the training process is complete, you will be able to evaluate the model's quality by using test data to gauge its performance. This data is a subset of the original data that the model has not previously analyzed. You can print out a table of metrics about your model's quality.\n\n#### Model Fitting\n\nIn the Machine Learning context, model fitting refers to the accuracy of the model's underlying function as it attempts to analyze data with which it is not familiar.\n\n#### Underfitting and Overfitting\n\nUnderfitting and overfitting are common problems that degrade the quality of the model, as the model either doesn't fit well enough, or it fits too well. This causes the model to make predictions either too closely aligned or too loosely aligned with its training data. An overfit model predicts training data too well because it has learned the data's details and noise too well. An underfit model is not accurate as it can neither accurately analyze its training data nor data it has not yet 'seen'.\n\n\n\nLet's test out some algorithms to choose our path for modelling our predictions.\n\n",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"import time\nstart = time.time()",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport pickle\nfrom sklearn.metrics import confusion_matrix, precision_score\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.preprocessing import StandardScaler,LabelEncoder,OneHotEncoder\nfrom sklearn.model_selection import cross_val_score,StratifiedKFold,RandomizedSearchCV\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier, RandomForestRegressor\nfrom sklearn.svm import SVC\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.metrics import confusion_matrix,precision_score,f1_score,recall_score\nfrom sklearn.neural_network import MLPClassifier, MLPRegressor\nplt.style.use('seaborn')\n\nnp.set_printoptions(precision=4)",
"_____no_output_____"
],
[
"data = pd.read_csv('./data_f1/data_filtered.csv')",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"len(data)",
"_____no_output_____"
],
[
"dnf_by_driver = data.groupby('driver').sum()['driver_dnf']\ndriver_race_entered = data.groupby('driver').count()['driver_dnf']\ndriver_dnf_ratio = (dnf_by_driver/driver_race_entered)\ndriver_confidence = 1-driver_dnf_ratio\ndriver_confidence_dict = dict(zip(driver_confidence.index,driver_confidence))",
"_____no_output_____"
],
[
"driver_confidence_dict",
"_____no_output_____"
],
[
"dnf_by_constructor = data.groupby('constructor').sum()['constructor_dnf']\nconstructor_race_entered = data.groupby('constructor').count()['constructor_dnf']\nconstructor_dnf_ratio = (dnf_by_constructor/constructor_race_entered)\nconstructor_reliability = 1-constructor_dnf_ratio\nconstructor_reliability_dict = dict(zip(constructor_reliability.index,constructor_reliability))",
"_____no_output_____"
],
[
"constructor_reliability_dict",
"_____no_output_____"
],
[
"data['driver_confidence'] = data['driver'].apply(lambda x:driver_confidence_dict[x])\ndata['constructor_reliability'] = data['constructor'].apply(lambda x:constructor_reliability_dict[x])\n#removing retired drivers and constructors\nactive_constructors = ['Alpine F1', 'Williams', 'McLaren', 'Ferrari', 'Mercedes',\n 'AlphaTauri', 'Aston Martin', 'Alfa Romeo', 'Red Bull',\n 'Haas F1 Team']\nactive_drivers = ['Daniel Ricciardo', 'Mick Schumacher', 'Carlos Sainz',\n 'Valtteri Bottas', 'Lance Stroll', 'George Russell',\n 'Lando Norris', 'Sebastian Vettel', 'Kimi Räikkönen',\n 'Charles Leclerc', 'Lewis Hamilton', 'Yuki Tsunoda',\n 'Max Verstappen', 'Pierre Gasly', 'Fernando Alonso',\n 'Sergio Pérez', 'Esteban Ocon', 'Antonio Giovinazzi',\n 'Nikita Mazepin','Nicholas Latifi']\ndata['active_driver'] = data['driver'].apply(lambda x: int(x in active_drivers))\ndata['active_constructor'] = data['constructor'].apply(lambda x: int(x in active_constructors))",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"data.columns",
"_____no_output_____"
]
],
[
[
"## Directory to store Models",
"_____no_output_____"
]
],
[
[
"import os\nif not os.path.exists('./models'):\n os.mkdir('./models')",
"_____no_output_____"
],
[
"def position_index(x):\n if x<4:\n return 1\n if x>10:\n return 3\n else :\n return 2",
"_____no_output_____"
]
],
[
[
"## Model considering only Drivers",
"_____no_output_____"
]
],
[
[
"x_d= data[['GP_name','quali_pos','driver','age_at_gp_in_days','position','driver_confidence','active_driver']]",
"_____no_output_____"
],
[
"x_d = x_d[x_d['active_driver']==1]",
"_____no_output_____"
],
[
"sc = StandardScaler()\nle = LabelEncoder()\nx_d['GP_name'] = le.fit_transform(x_d['GP_name'])\nx_d['driver'] = le.fit_transform(x_d['driver'])\nx_d['GP_name'] = le.fit_transform(x_d['GP_name'])\nx_d['age_at_gp_in_days'] = sc.fit_transform(x_d[['age_at_gp_in_days']])\nX_d = x_d.drop(['position','active_driver'],1)\ny_d = x_d['position'].apply(lambda x: position_index(x))",
"_____no_output_____"
],
[
"#cross validation for diffrent models\nmodels = [LogisticRegression(),DecisionTreeClassifier(),RandomForestClassifier(),SVC(),GaussianNB(),KNeighborsClassifier()]\nnames = ['LogisticRegression','DecisionTreeClassifier','RandomForestClassifier','SVC','GaussianNB','KNeighborsClassifier']\nmodel_dict = dict(zip(models,names))\nmean_results_dri = []\nresults_dri = []\nname = []\nfor model in models:\n cv = StratifiedKFold(n_splits=10,random_state=1,shuffle=True)\n result = cross_val_score(model,X_d,y_d,cv=cv,scoring='accuracy')\n mean_results_dri.append(result.mean())\n results_dri.append(result)\n name.append(model_dict[model])\n print(f'{model_dict[model]} : {result.mean()}')",
"LogisticRegression : 0.9326015531660692\nDecisionTreeClassifier : 0.9178166069295101\nRandomForestClassifier : 0.9422811059907834\nSVC : 0.9270566649598907\nGaussianNB : 0.8591803208738693\nKNeighborsClassifier : 0.909511008704557\n"
],
[
"plt.figure(figsize=(15,10))\nplt.boxplot(x=results_dri,labels=name)\nplt.xlabel('Models')\nplt.ylabel('Accuracy')\nplt.title('Model performance comparision (drivers only)')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Model considering only Constructors",
"_____no_output_____"
]
],
[
[
"x_c = data[['GP_name','quali_pos','constructor','position','constructor_reliability','active_constructor']]",
"_____no_output_____"
],
[
"x_c = x_c[x_c['active_constructor']==1]",
"_____no_output_____"
],
[
"sc = StandardScaler()\nle = LabelEncoder()\nx_c['GP_name'] = le.fit_transform(x_c['GP_name'])\nx_c['constructor'] = le.fit_transform(x_c['constructor'])\nX_c = x_c.drop(['position','active_constructor'],1)\ny_c = x_c['position'].apply(lambda x: position_index(x))",
"_____no_output_____"
],
[
"#cross validation for diffrent models\nmodels = [LogisticRegression(),DecisionTreeClassifier(),RandomForestClassifier(),SVC(),GaussianNB(),KNeighborsClassifier()]\nnames = ['LogisticRegression','DecisionTreeClassifier','RandomForestClassifier','SVC','GaussianNB','KNeighborsClassifier']\nmodel_dict = dict(zip(models,names))\nmean_results_const = []\nresults_const = []\nname = []\nfor model in models:\n cv = StratifiedKFold(n_splits=10,random_state=1,shuffle=True)\n result = cross_val_score(model,X_c,y_c,cv=cv,scoring='accuracy')\n mean_results_const.append(result.mean())\n results_const.append(result)\n name.append(model_dict[model])\n print(f'{model_dict[model]} : {result.mean()}')",
"LogisticRegression : 0.9330210772833724\nDecisionTreeClassifier : 0.9290398126463701\nRandomForestClassifier : 0.9449648711943794\nSVC : 0.9515222482435597\nGaussianNB : 0.8519906323185011\nKNeighborsClassifier : 0.9370023419203747\n"
],
[
"plt.figure(figsize=(15,10))\nplt.boxplot(x=results_const,labels=name)\nplt.xlabel('Models')\nplt.ylabel('Accuracy')\nplt.title('Model performance comparision (Teams only)')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Model considering both Drivers and Constructors",
"_____no_output_____"
]
],
[
[
"cleaned_data = data[['GP_name','quali_pos','constructor','driver','position','driver_confidence','constructor_reliability','active_driver','active_constructor']]\ncleaned_data = cleaned_data[(cleaned_data['active_driver']==1)&(cleaned_data['active_constructor']==1)]\ncleaned_data.to_csv('./data_f1/cleaned_data.csv',index=False)",
"_____no_output_____"
]
],
[
[
"### Build your X dataset with next columns:\n- GP_name\n- quali_pos to predict the classification cluster (1,2,3) \n- constructor\n- driver\n- position\n- driver confidence\n- constructor_reliability\n- active_driver\n- active_constructor",
"_____no_output_____"
],
[
"### Filter the dataset for this Model \"Driver + Constructor\" all active drivers and constructors",
"_____no_output_____"
],
[
"### Create Standard Scaler and Label Encoder for the different features in order to have a similar scale for all features",
"_____no_output_____"
],
[
"### Prepare the X (Features dataset) and y for predicted value. \nIn our case, we want to calculate the cluster of final position for ech driver using the \"position_index\" function",
"_____no_output_____"
]
],
[
[
"# Implement X, y",
"_____no_output_____"
]
],
[
[
"### Applied the same list of ML Algorithms for cross validation of different models\n\nAnd Store the accuracy Mean Value in order to compare with previous ML Models",
"_____no_output_____"
]
],
[
[
"mean_results = []\nresults = []\nname = []",
"_____no_output_____"
],
[
"# cross validation for different models\n",
"_____no_output_____"
]
],
[
[
"### Use the same boxplot plotter used in the previous Models",
"_____no_output_____"
]
],
[
[
"# Implement boxplot\n",
"_____no_output_____"
]
],
[
[
"# Comparing The 3 ML Models\n\nLet's see mean score of our three assumptions.",
"_____no_output_____"
]
],
[
[
"lr = [mean_results[0],mean_results_dri[0],mean_results_const[0]]\ndtc = [mean_results[1],mean_results_dri[1],mean_results_const[1]]\nrfc = [mean_results[2],mean_results_dri[2],mean_results_const[2]]\nsvc = [mean_results[3],mean_results_dri[3],mean_results_const[3]]\ngnb = [mean_results[4],mean_results_dri[4],mean_results_const[4]]\nknn = [mean_results[5],mean_results_dri[5],mean_results_const[5]]\nfont1 = {\n 'family':'serif',\n 'color':'black',\n 'weight':'normal',\n 'size':18\n}\nfont2 = {\n 'family':'serif',\n 'color':'black',\n 'weight':'bold',\n 'size':12\n}\nx_ax = np.arange(3)\nplt.figure(figsize=(30,15))\nbar1 = plt.bar(x_ax,lr,width=0.1,align='center', label=\"Logistic Regression\")\nbar2 = plt.bar(x_ax+0.1,dtc,width=0.1,align='center', label=\"DecisionTree\")\nbar3 = plt.bar(x_ax+0.2,rfc,width=0.1,align='center', label=\"RandomForest\")\nbar4 = plt.bar(x_ax+0.3,svc,width=0.1,align='center', label=\"SVC\")\nbar5 = plt.bar(x_ax+0.4,gnb,width=0.1,align='center', label=\"GaussianNB\")\nbar6 = plt.bar(x_ax+0.5,knn,width=0.1,align='center', label=\"KNN\")\nplt.text(0.05,1,'CV score for combined data',fontdict=font1)\nplt.text(1.04,1,'CV score only driver data',fontdict=font1)\nplt.text(2,1,'CV score only team data',fontdict=font1)\nfor bar in bar1.patches:\n yval = bar.get_height()\n plt.text(bar.get_x()+0.01,yval+0.01,f'{round(yval*100,2)}%',fontdict=font2)\nfor bar in bar2.patches:\n yval = bar.get_height()\n plt.text(bar.get_x()+0.01,yval+0.01,f'{round(yval*100,2)}%',fontdict=font2)\nfor bar in bar3.patches:\n yval = bar.get_height()\n plt.text(bar.get_x()+0.01,yval+0.01,f'{round(yval*100,2)}%',fontdict=font2)\nfor bar in bar4.patches:\n yval = bar.get_height()\n plt.text(bar.get_x()+0.01,yval+0.01,f'{round(yval*100,2)}%',fontdict=font2)\nfor bar in bar5.patches:\n yval = bar.get_height()\n plt.text(bar.get_x()+0.01,yval+0.01,f'{round(yval*100,2)}%',fontdict=font2)\nfor bar in bar6.patches:\n yval = bar.get_height()\n plt.text(bar.get_x()+0.01,yval+0.01,f'{round(yval*100,2)}%',fontdict=font2)\nplt.legend(loc='center', bbox_to_anchor=(0.5, -0.10), shadow=False, ncol=6)\n\nplt.show()",
"_____no_output_____"
],
[
"end = time.time()",
"_____no_output_____"
],
[
"import datetime\nstr(datetime.timedelta(seconds=(end - start)))",
"_____no_output_____"
],
[
"print(str(end - start)+\" seconds\")",
"62.024924516677856 seconds\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d05bca764315644e0da3c721f1e097288777bd6a | 62,163 | ipynb | Jupyter Notebook | notebooks/Model Building.ipynb | Reasmey/adsi_beer_app | 345bab07d6fe579c019a06660cffa5d13718e03c | [
"FTL"
] | null | null | null | notebooks/Model Building.ipynb | Reasmey/adsi_beer_app | 345bab07d6fe579c019a06660cffa5d13718e03c | [
"FTL"
] | null | null | null | notebooks/Model Building.ipynb | Reasmey/adsi_beer_app | 345bab07d6fe579c019a06660cffa5d13718e03c | [
"FTL"
] | null | null | null | 30.728127 | 195 | 0.371523 | [
[
[
"# Model building\n\nhttps://www.kaggle.com/vadbeg/pytorch-nn-with-embeddings-and-catboost/notebook#PyTorch\n\nmostly based off this example, plus parts of code form tutorial 5 lab 3",
"_____no_output_____"
]
],
[
[
"# import load_data function from \n%load_ext autoreload\n%autoreload 2\n\n# fix system path\nimport sys\nsys.path.append(\"/home/jovyan/work\")",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nimport torch\nfrom torch.utils.data import Dataset, DataLoader",
"_____no_output_____"
],
[
"import random\n\ndef set_seed(seed):\n random.seed(seed)\n np.random.seed(seed)\n \n torch.manual_seed(seed)\n torch.backends.cudnn.deterministick = True\n torch.backends.cudnn.benchmark = False \n \nset_seed(27)",
"_____no_output_____"
],
[
"from src.data.sets import load_sets\n\nX_train, y_train, X_val, y_val, X_test, y_test = load_sets()",
"_____no_output_____"
],
[
"X_test.shape",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"X_val.shape",
"_____no_output_____"
],
[
"# need to convert to tensors\nfrom src.models.pytorch import EmbeddingDataset",
"_____no_output_____"
],
[
"train_dataset = EmbeddingDataset(X_train, \n targets=y_train,\n cat_cols_idx=[0],\n cont_cols_idx=[1,2,3,4])\n\nval_dataset = EmbeddingDataset(X_val, \n targets=y_val,\n cat_cols_idx=[0],\n cont_cols_idx=[1,2,3,4])\n\n\ntest_dataset = EmbeddingDataset(X_test,\n cat_cols_idx=[0],\n cont_cols_idx=[1,2,3,4],\n is_train=False)",
"_____no_output_____"
],
[
"print(f'First element of train_dataset: {train_dataset[1]}',\n f'First element of val_dataset: {val_dataset[1]}',\n f'First element of test_dataset: {test_dataset[1]}',sep='\\n')",
"First element of train_dataset: {'data': [tensor([4918.]), tensor([-3.2047, -2.1777, -0.3572, -0.4001])], 'target': tensor(13)}\nFirst element of val_dataset: {'data': [tensor([163.]), tensor([-1.0545, -0.5545, -1.0901, -1.0832])], 'target': tensor(31)}\nFirst element of test_dataset: {'data': [tensor([701.]), tensor([ 0.3790, 0.2570, 0.3757, -0.4001])]}\n"
],
[
"# embedding example\nclass ClassificationEmbdNN(torch.nn.Module):\n \n def __init__(self, emb_dims, no_of_cont=None):\n super(ClassificationEmbdNN, self).__init__()\n \n self.emb_layers = torch.nn.ModuleList([torch.nn.Embedding(x, y)\n for x, y in emb_dims])\n \n no_of_embs = sum([y for x, y in emb_dims])\n self.no_of_embs = no_of_embs\n self.emb_dropout = torch.nn.Dropout(0.2)\n \n self.no_of_cont = 0\n if no_of_cont:\n self.no_of_cont = no_of_cont\n self.bn_cont = torch.nn.BatchNorm1d(no_of_cont)\n \n self.fc1 = torch.nn.Linear(in_features=self.no_of_embs + self.no_of_cont, \n out_features=208)\n self.dropout1 = torch.nn.Dropout(0.2)\n self.bn1 = torch.nn.BatchNorm1d(208)\n self.act1 = torch.nn.ReLU()\n \n self.fc2 = torch.nn.Linear(in_features=208, \n out_features=208)\n self.dropout2 = torch.nn.Dropout(0.2)\n self.bn2 = torch.nn.BatchNorm1d(208)\n self.act2 = torch.nn.ReLU()\n \n# self.fc3 = torch.nn.Linear(in_features=256, \n# out_features=64)\n# self.dropout3 = torch.nn.Dropout(0.2)\n# self.bn3 = torch.nn.BatchNorm1d(64)\n# self.act3 = torch.nn.ReLU()\n \n self.fc3 = torch.nn.Linear(in_features=208, \n out_features=104)\n self.act3 = torch.nn.Softmax()\n \n def forward(self, x_cat, x_cont=None):\n if self.no_of_embs != 0:\n x = [emb_layer(x_cat[:, i])\n for i, emb_layer in enumerate(self.emb_layers)]\n \n x = torch.cat(x, 1)\n x = self.emb_dropout(x)\n \n if self.no_of_cont != 0:\n x_cont = self.bn_cont(x_cont)\n \n if self.no_of_embs != 0:\n x = torch.cat([x, x_cont], 1)\n else:\n x = x_cont\n \n x = self.fc1(x)\n x = self.dropout1(x)\n x = self.bn1(x)\n x = self.act1(x)\n \n x = self.fc2(x)\n x = self.dropout2(x)\n x = self.bn2(x)\n x = self.act2(x)\n \n# x = self.fc3(x)\n# x = self.dropout3(x)\n# x = self.bn3(x)\n# x = self.act3(x)\n \n x = self.fc3(x)\n x = self.act3(x)\n \n return x",
"_____no_output_____"
],
[
"model = ClassificationEmbdNN(emb_dims=[[5742, 252]], \n no_of_cont=4)",
"_____no_output_____"
],
[
"from src.models.pytorch import get_device\n\ndevice = get_device()\nmodel.to(device)",
"_____no_output_____"
],
[
"print(model)",
"ClassificationEmbdNN(\n (emb_layers): ModuleList(\n (0): Embedding(5724, 1000)\n )\n (emb_dropout): Dropout(p=0.2, inplace=False)\n (bn_cont): BatchNorm1d(4, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (fc1): Linear(in_features=1004, out_features=208, bias=True)\n (dropout1): Dropout(p=0.2, inplace=False)\n (bn1): BatchNorm1d(208, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (act1): ReLU()\n (fc2): Linear(in_features=208, out_features=208, bias=True)\n (dropout2): Dropout(p=0.2, inplace=False)\n (bn2): BatchNorm1d(208, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (act2): ReLU()\n (fc3): Linear(in_features=208, out_features=104, bias=True)\n (act3): Softmax(dim=None)\n)\n"
],
[
"criterion = torch.nn.CrossEntropyLoss()",
"_____no_output_____"
],
[
"optimizer = torch.optim.Adam(model.parameters(), lr=0.1)",
"_____no_output_____"
],
[
"BATCH_SIZE = 300\nN_EPOCHS = 10",
"_____no_output_____"
],
[
"train_loader = DataLoader(train_dataset,batch_size=BATCH_SIZE)\n\nvalid_loader = DataLoader(val_dataset,batch_size=BATCH_SIZE)",
"_____no_output_____"
],
[
"next(iter(train_loader))",
"_____no_output_____"
],
[
"next(iter(valid_loader))",
"_____no_output_____"
],
[
"from tqdm import tqdm_notebook as tqdm",
"_____no_output_____"
],
[
"def train_network(model, train_loader, valid_loader,\n loss_func, optimizer, n_epochs=20,\n saved_model='model.pt'):\n \n device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')\n model.to(device)\n \n train_losses = list()\n valid_losses = list()\n \n valid_loss_min = np.Inf\n \n for epoch in range(n_epochs):\n train_loss = 0.0\n valid_loss = 0.0\n \n# train_auc = 0.0\n# valid_auc = 0.0\n \n train_acc = 0.0\n valid_acc = 0.0\n \n model.train()\n for batch in tqdm(train_loader):\n optimizer.zero_grad()\n \n output = model(batch['data'][0].to(device, \n dtype=torch.long),\n batch['data'][1].to(device, \n dtype=torch.float))\n \n \n loss = loss_func(output, batch['target'].to(device, \n dtype=torch.long))\n \n loss.backward()\n optimizer.step()\n # Calculate global accuracy\n train_acc += (output.argmax(1) == batch['target']).sum().item()\n# train_auc += roc_auc_score(batch['target'].cpu().numpy(),\n# output.detach().cpu().numpy(),\n# multi_class = \"ovo\")\n\n train_loss += loss.item() * batch['data'][0].size(0) #!!!\n \n\n model.eval()\n for batch in tqdm(valid_loader):\n output = model(batch['data'][0].to(device, \n dtype=torch.long),\n batch['data'][1].to(device, \n dtype=torch.float))\n \n \n loss = loss_func(output, batch['target'].to(device, \n dtype=torch.long))\n \n# valid_auc += roc_auc_score(batch['target'].cpu().numpy(),\n# output.detach().cpu().numpy(),\n# multi_class = \"ovo\")\n valid_loss += loss.item() * batch['data'][0].size(0) #!!!\n # Calculate global accuracy\n valid_acc += (output.argmax(1) == batch['target']).sum().item()\n \n# train_loss = np.sqrt(train_loss / len(train_loader.sampler.indices))\n# valid_loss = np.sqrt(valid_loss / len(valid_loader.sampler.indices))\n\n# train_auc = train_auc / len(train_loader)\n# valid_auc = valid_auc / len(valid_loader)\n \n# train_losses.append(train_loss)\n# valid_losses.append(valid_loss)\n\n print('Epoch: {}. Training loss: {:.6f}. Validation loss: {:.6f}'\n .format(epoch, train_loss, valid_loss))\n print('Training AUC: {:.6f}. Validation AUC: {:.6f}'\n .format(train_acc, valid_acc))\n \n if valid_loss < valid_loss_min: # let's save the best weights to use them in prediction\n print('Validation loss decreased ({:.6f} --> {:.6f}). Saving model...'\n .format(valid_loss_min, valid_loss))\n \n torch.save(model.state_dict(), saved_model)\n valid_loss_min = valid_loss\n \n \n return train_losses, valid_losses",
"_____no_output_____"
],
[
"train_losses, valid_losses = train_network(model=model, \n train_loader=train_loader, \n valid_loader=valid_loader, \n loss_func=criterion, \n optimizer=optimizer,\n n_epochs=N_EPOCHS, \n saved_model='../models/embed_3layers.pt')",
"/opt/conda/lib/python3.7/site-packages/ipykernel_launcher.py:24: TqdmDeprecationWarning: This function will be removed in tqdm==5.0.0\nPlease use `tqdm.notebook.tqdm` instead of `tqdm.tqdm_notebook`\n"
]
],
[
[
"#### forgot to divide the loss and accuracy by length of data set",
"_____no_output_____"
]
],
[
[
"print('Training Accuracy: {:.2f}%'.format(5926.0/300.0))\nprint('Validation Accuracy: {:.2f}%'.format(2361.0/300.0))",
"Training Accuracy: 19.75%\nValidation Accuracy: 7.87%\n"
]
],
[
[
"# Predict with test set",
"_____no_output_____"
]
],
[
[
"def predict(data_loader, model):\n model.eval()\n device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')\n \n model.to(device)\n \n with torch.no_grad():\n predictions = None\n \n for i, batch in enumerate(tqdm(data_loader)): \n \n output = model(batch['data'][0].to(device, \n dtype=torch.long), \n batch['data'][1].to(device, \n dtype=torch.float)).cpu().numpy()\n \n if i == 0:\n predictions = output\n \n else: \n \n predictions = np.vstack((predictions, output))\n \n return predictions",
"_____no_output_____"
],
[
"model.load_state_dict(torch.load('../models/embed_3layers.pt'))\n\ntest_loader = DataLoader(test_dataset, \n batch_size=BATCH_SIZE)\n\nnn_predictions = predict(test_loader, model)",
"/opt/conda/lib/python3.7/site-packages/ipykernel_launcher.py:10: TqdmDeprecationWarning: This function will be removed in tqdm==5.0.0\nPlease use `tqdm.notebook.tqdm` instead of `tqdm.tqdm_notebook`\n # Remove the CWD from sys.path while we load stuff.\n"
],
[
"nn_predictions",
"_____no_output_____"
],
[
"test_acc = (nn_predictions.argmax(1) == y_test).sum().item()",
"_____no_output_____"
],
[
"test_acc/300",
"_____no_output_____"
],
[
"from sklearn.metrics import roc_auc_score, classification_report",
"_____no_output_____"
],
[
"# compute other metrics\nroc_auc_score(y_test,nn_predictions, multi_class='ovr', average='macro')",
"_____no_output_____"
],
[
"print(y_test)\nprint(nn_predictions.argmax(1))",
"[ 83 102 11 ... 43 9 14]\n[73 50 80 ... 12 32 99]\n"
],
[
"def convert_cr_to_dataframe(report_dict:dict) -> pd.DataFrame:\n \"\"\"\n Converts the dictionary format of the Classification Report (CR) to a\n dataframe for easy of sorting\n :param report_dict: The dictionary returned by \n sklearn.metrics.classification_report.\n :return: Returns a dataframe of the same information.\n \"\"\"\n beer_style = list(report_dict.keys())\n beer_style.remove('accuracy')\n beer_style.remove('macro avg')\n beer_style.remove('weighted avg')\n precision = []\n recall = []\n f1 = []\n support = []\n for key, value in report_dict.items():\n if key not in ['accuracy', 'macro avg', 'weighted avg']:\n precision.append(value['precision'])\n recall.append(value['recall'])\n f1.append(value['f1-score'])\n support.append(value['support'])\n result = pd.DataFrame({'beer_style': beer_style,\n 'precision': precision,\n 'recall': recall,\n 'f1': f1,\n 'support': support})\n return result",
"_____no_output_____"
],
[
"from joblib import load\n\nlbel_encoders = load('../models/label_encoders.joblib')",
"_____no_output_____"
],
[
"report_dict = classification_report(label_encoders['beer_style'].inverse_transform(y_test),\n label_encoders['beer_style'].inverse_transform(nn_predictions.argmax(1)),\n output_dict=True)\nreport_df = convert_cr_to_dataframe(report_dict)\nprint(report_df)\n#classification_report(y_test, nn_predictions.argmax(1))",
" beer_style precision recall f1 support\n0 Altbier 0.004481 0.013342 0.006709 1574\n1 American Adjunct Lager 0.000000 0.000000 0.000000 6047\n2 American Amber / Red Ale 0.031808 0.013229 0.018686 9298\n3 American Amber / Red Lager 0.141541 0.091302 0.111002 1851\n4 American Barleywine 0.001211 0.000184 0.000319 5439\n.. ... ... ... ... ...\n99 Vienna Lager 0.000386 0.000548 0.000453 1825\n100 Weizenbock 0.000000 0.000000 0.000000 1900\n101 Wheatwine 0.000000 0.000000 0.000000 769\n102 Winter Warmer 0.000000 0.000000 0.000000 4140\n103 Witbier 0.018517 0.052798 0.027419 6023\n\n[104 rows x 5 columns]\n"
],
[
"torch.save(model, \"../models/model.pt\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05be888f52aaaf39bf00cb3d50fde4c1cbf77ab | 40,952 | ipynb | Jupyter Notebook | notebooks/Example 7 - Portfolio Optimizer Performance.ipynb | wangjiehui11235/alpha-mind | eecddc8d2bb1ae8e9ebde980a02b6bea61c77e26 | [
"MIT"
] | 1 | 2020-05-18T20:57:25.000Z | 2020-05-18T20:57:25.000Z | notebooks/Example 7 - Portfolio Optimizer Performance.ipynb | atefar2/alpha-mind | 66d839affb5d81d31d5cac7e5e224278e3f99a8b | [
"MIT"
] | null | null | null | notebooks/Example 7 - Portfolio Optimizer Performance.ipynb | atefar2/alpha-mind | 66d839affb5d81d31d5cac7e5e224278e3f99a8b | [
"MIT"
] | null | null | null | 35.425606 | 178 | 0.424912 | [
[
[
"* 比较不同组合组合优化器在不同规模问题上的性能;\n\n* 下面的结果主要比较``alphamind``和``python``中其他优化器的性能差别,我们将尽可能使用``cvxopt``中的优化器,其次选择``scipy``;\n\n* 由于``scipy``在``ashare_ex``上面性能太差,所以一般忽略``scipy``在这个股票池上的表现;\n\n* 时间单位都是毫秒。\n\n* 请在环境变量中设置`DB_URI`指向数据库",
"_____no_output_____"
]
],
[
[
"import os\nimport timeit\nimport numpy as np\nimport pandas as pd\nimport cvxpy\nfrom alphamind.api import *\nfrom alphamind.portfolio.linearbuilder import linear_builder\nfrom alphamind.portfolio.meanvariancebuilder import mean_variance_builder\nfrom alphamind.portfolio.meanvariancebuilder import target_vol_builder\n\npd.options.display.float_format = '{:,.2f}'.format",
"_____no_output_____"
]
],
[
[
"## 0. 数据准备\n------------------",
"_____no_output_____"
]
],
[
[
"ref_date = '2018-02-08'\nu_names = ['sh50', 'hs300', 'zz500', 'zz800', 'zz1000', 'ashare_ex']\nb_codes = [16, 300, 905, 906, 852, None]\nrisk_model = 'short'\nfactor = 'EPS'\nlb = 0.0\nub = 0.1\ndata_source = os.environ['DB_URI']\nengine = SqlEngine(data_source)\n\nuniverses = [Universe(u_name) for u_name in u_names]\ncodes_set = [engine.fetch_codes(ref_date, universe=universe) for universe in universes]\ndata_set = [engine.fetch_data(ref_date, factor, codes, benchmark=b_code, risk_model=risk_model) for codes, b_code in zip(codes_set, b_codes)]",
"_____no_output_____"
]
],
[
[
"## 1. 线性优化(带线性限制条件)\n---------------------------------",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(columns=u_names, index=['cvxpy', 'alphamind'])\n\nnumber = 1\n\nfor u_name, sample_data in zip(u_names, data_set):\n factor_data = sample_data['factor']\n er = factor_data[factor].values\n n = len(er)\n lbound = np.ones(n) * lb\n ubound = np.ones(n) * ub\n \n risk_constraints = np.ones((n, 1))\n risk_target = (np.array([1.]), np.array([1.]))\n\n status, y, x1 = linear_builder(er, lbound, ubound, risk_constraints, risk_target)\n elasped_time1 = timeit.timeit(\"linear_builder(er, lbound, ubound, risk_constraints, risk_target)\", number=number, globals=globals()) / number * 1000\n\n A_eq = risk_constraints.T\n b_eq = np.array([1.])\n \n w = cvxpy.Variable(n)\n curr_risk_exposure = w * risk_constraints\n \n constraints = [w >= lbound,\n w <= ubound,\n curr_risk_exposure == risk_target[0]]\n \n objective = cvxpy.Minimize(-w.T * er)\n prob = cvxpy.Problem(objective, constraints)\n \n prob.solve(solver='ECOS')\n elasped_time2 = timeit.timeit(\"prob.solve(solver='ECOS')\",\n number=number, globals=globals()) / number * 1000\n\n np.testing.assert_almost_equal(x1 @ er, np.array(w.value).flatten() @ er, 4)\n\n df.loc['alphamind', u_name] = elasped_time1\n df.loc['cvxpy', u_name] = elasped_time2\n alpha_logger.info(f\"{u_name} is finished\")",
"2019-02-10 00:39:31,620 - ALPHA_MIND - INFO - sh50 is finished\n2019-02-10 00:39:31,648 - ALPHA_MIND - INFO - hs300 is finished\n2019-02-10 00:39:31,682 - ALPHA_MIND - INFO - zz500 is finished\n2019-02-10 00:39:31,738 - ALPHA_MIND - INFO - zz800 is finished\n2019-02-10 00:39:31,789 - ALPHA_MIND - INFO - zz1000 is finished\nD:\\ProgramData\\anaconda3\\lib\\site-packages\\cvxpy-1.0.10-py3.6-win-amd64.egg\\cvxpy\\problems\\problem.py:614: RuntimeWarning: overflow encountered in long_scalars\n if self.max_big_small_squared < big*small**2:\nD:\\ProgramData\\anaconda3\\lib\\site-packages\\cvxpy-1.0.10-py3.6-win-amd64.egg\\cvxpy\\problems\\problem.py:615: RuntimeWarning: overflow encountered in long_scalars\n self.max_big_small_squared = big*small**2\n2019-02-10 00:39:31,962 - ALPHA_MIND - INFO - ashare_ex is finished\n"
],
[
"df",
"_____no_output_____"
],
[
"prob.value",
"_____no_output_____"
]
],
[
[
"## 2. 线性优化(带L1限制条件)\n-----------------------",
"_____no_output_____"
]
],
[
[
"from cvxpy import pnorm\n\ndf = pd.DataFrame(columns=u_names, index=['cvxpy', 'alphamind (clp simplex)', 'alphamind (clp interior)', 'alphamind (ecos)'])\nturn_over_target = 0.5\nnumber = 1\n\nfor u_name, sample_data in zip(u_names, data_set):\n factor_data = sample_data['factor']\n er = factor_data[factor].values\n n = len(er)\n lbound = np.ones(n) * lb\n ubound = np.ones(n) * ub\n \n if 'weight' in factor_data:\n current_position = factor_data.weight.values\n else:\n current_position = np.ones_like(er) / len(er)\n\n risk_constraints = np.ones((len(er), 1))\n risk_target = (np.array([1.]), np.array([1.]))\n\n status, y, x1 = linear_builder(er,\n lbound,\n ubound,\n risk_constraints,\n risk_target,\n turn_over_target=turn_over_target,\n current_position=current_position,\n method='interior')\n elasped_time1 = timeit.timeit(\"\"\"linear_builder(er,\n lbound,\n ubound,\n risk_constraints,\n risk_target,\n turn_over_target=turn_over_target,\n current_position=current_position,\n method='interior')\"\"\", number=number, globals=globals()) / number * 1000\n \n w = cvxpy.Variable(n)\n curr_risk_exposure = risk_constraints.T @ w\n \n constraints = [w >= lbound,\n w <= ubound,\n curr_risk_exposure == risk_target[0],\n pnorm(w - current_position, 1) <= turn_over_target]\n \n objective = cvxpy.Minimize(-w.T * er)\n prob = cvxpy.Problem(objective, constraints)\n \n prob.solve(solver='ECOS')\n elasped_time2 = timeit.timeit(\"prob.solve(solver='ECOS')\",\n number=number, globals=globals()) / number * 1000\n \n status, y, x2 = linear_builder(er,\n lbound,\n ubound,\n risk_constraints,\n risk_target,\n turn_over_target=turn_over_target,\n current_position=current_position,\n method='simplex')\n elasped_time3 = timeit.timeit(\"\"\"linear_builder(er,\n lbound,\n ubound,\n risk_constraints,\n risk_target,\n turn_over_target=turn_over_target,\n current_position=current_position,\n method='simplex')\"\"\", number=number, globals=globals()) / number * 1000\n \n status, y, x3 = linear_builder(er,\n lbound,\n ubound,\n risk_constraints,\n risk_target,\n turn_over_target=turn_over_target,\n current_position=current_position,\n method='ecos')\n elasped_time4 = timeit.timeit(\"\"\"linear_builder(er,\n lbound,\n ubound,\n risk_constraints,\n risk_target,\n turn_over_target=turn_over_target,\n current_position=current_position,\n method='ecos')\"\"\", number=number, globals=globals()) / number * 1000\n \n \n np.testing.assert_almost_equal(x1 @ er, np.array(w.value).flatten() @ er, 4)\n np.testing.assert_almost_equal(x2 @ er, np.array(w.value).flatten() @ er, 4)\n np.testing.assert_almost_equal(x3 @ er, np.array(w.value).flatten() @ er, 4)\n\n df.loc['alphamind (clp interior)', u_name] = elasped_time1\n df.loc['alphamind (clp simplex)', u_name] = elasped_time3\n df.loc['alphamind (ecos)', u_name] = elasped_time4\n df.loc['cvxpy', u_name] = elasped_time2\n alpha_logger.info(f\"{u_name} is finished\")",
"2019-02-10 00:39:32,075 - ALPHA_MIND - INFO - sh50 is finished\n2019-02-10 00:39:32,209 - ALPHA_MIND - INFO - hs300 is finished\n2019-02-10 00:39:32,469 - ALPHA_MIND - INFO - zz500 is finished\n2019-02-10 00:39:32,998 - ALPHA_MIND - INFO - zz800 is finished\n2019-02-10 00:39:33,755 - ALPHA_MIND - INFO - zz1000 is finished\n2019-02-10 00:39:40,815 - ALPHA_MIND - INFO - ashare_ex is finished\n"
],
[
"df",
"_____no_output_____"
]
],
[
[
"## 3. Mean - Variance 优化 (无约束)\n-----------------------",
"_____no_output_____"
]
],
[
[
"from cvxpy import *\n\ndf = pd.DataFrame(columns=u_names, index=['cvxpy', 'alphamind'])\nnumber = 1\n\nfor u_name, sample_data in zip(u_names, data_set):\n all_styles = risk_styles + industry_styles + ['COUNTRY']\n factor_data = sample_data['factor']\n risk_cov = sample_data['risk_cov'][all_styles].values\n risk_exposure = factor_data[all_styles].values\n special_risk = factor_data.srisk.values\n sec_cov = risk_exposure @ risk_cov @ risk_exposure.T / 10000 + np.diag(special_risk ** 2) / 10000\n er = factor_data[factor].values\n n = len(er)\n\n bm = np.zeros(n)\n lbound = -np.ones(n) * np.inf\n ubound = np.ones(n) * np.inf\n risk_model = dict(cov=None, factor_cov=risk_cov/10000., factor_loading=risk_exposure, idsync=(special_risk**2)/10000.)\n\n status, y, x1 = mean_variance_builder(er,\n risk_model,\n bm,\n lbound,\n ubound,\n None,\n None,\n lam=1)\n elasped_time1 = timeit.timeit(\"\"\"mean_variance_builder(er,\n risk_model,\n bm,\n lbound,\n ubound,\n None,\n None,\n lam=1)\"\"\",\n number=number, globals=globals()) / number * 1000\n \n w = cvxpy.Variable(n)\n risk = sum_squares(multiply(special_risk / 100., w)) + quad_form((w.T * risk_exposure).T, risk_cov / 10000.)\n objective = cvxpy.Minimize(-w.T * er + 0.5 * risk)\n prob = cvxpy.Problem(objective)\n prob.solve(solver='ECOS')\n elasped_time2 = timeit.timeit(\"prob.solve(solver='ECOS')\",\n number=number, globals=globals()) / number * 1000\n \n u1 = -x1 @ er + 0.5 * x1 @ sec_cov @ x1\n x2 = np.array(w.value).flatten()\n u2 = -x2 @ er + 0.5 * x2 @ sec_cov @ x2\n \n np.testing.assert_array_almost_equal(u1, u2, 4)\n\n df.loc['alphamind', u_name] = elasped_time1\n df.loc['cvxpy', u_name] = elasped_time2\n alpha_logger.info(f\"{u_name} is finished\")",
"2019-02-10 00:39:40,951 - ALPHA_MIND - INFO - sh50 is finished\n2019-02-10 00:39:41,217 - ALPHA_MIND - INFO - hs300 is finished\n2019-02-10 00:39:41,592 - ALPHA_MIND - INFO - zz500 is finished\n2019-02-10 00:39:42,175 - ALPHA_MIND - INFO - zz800 is finished\n2019-02-10 00:39:42,796 - ALPHA_MIND - INFO - zz1000 is finished\n2019-02-10 00:39:45,458 - ALPHA_MIND - INFO - ashare_ex is finished\n"
],
[
"df",
"_____no_output_____"
]
],
[
[
"## 4. Mean - Variance 优化 (Box约束)\n---------------",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(columns=u_names, index=['cvxpy', 'alphamind'])\nnumber = 1\n\nfor u_name, sample_data in zip(u_names, data_set):\n all_styles = risk_styles + industry_styles + ['COUNTRY']\n factor_data = sample_data['factor']\n risk_cov = sample_data['risk_cov'][all_styles].values\n risk_exposure = factor_data[all_styles].values\n special_risk = factor_data.srisk.values\n sec_cov = risk_exposure @ risk_cov @ risk_exposure.T / 10000 + np.diag(special_risk ** 2) / 10000\n er = factor_data[factor].values\n n = len(er)\n\n bm = np.zeros(n)\n lbound = np.zeros(n)\n ubound = np.ones(n) * 0.1\n \n risk_model = dict(cov=None, factor_cov=risk_cov/10000., factor_loading=risk_exposure, idsync=(special_risk**2)/10000.)\n\n status, y, x1 = mean_variance_builder(er,\n risk_model,\n bm,\n lbound,\n ubound,\n None,\n None)\n elasped_time1 = timeit.timeit(\"\"\"mean_variance_builder(er,\n risk_model,\n bm,\n lbound,\n ubound,\n None,\n None)\"\"\",\n number=number, globals=globals()) / number * 1000\n \n w = cvxpy.Variable(n)\n risk = sum_squares(multiply(special_risk / 100., w)) + quad_form((w.T * risk_exposure).T, risk_cov / 10000.)\n objective = cvxpy.Minimize(-w.T * er + 0.5 * risk)\n constraints = [w >= lbound,\n w <= ubound]\n prob = cvxpy.Problem(objective, constraints)\n prob.solve(solver='ECOS')\n elasped_time2 = timeit.timeit(\"prob.solve(solver='ECOS')\",\n number=number, globals=globals()) / number * 1000\n \n u1 = -x1 @ er + 0.5 * x1 @ sec_cov @ x1\n x2 = np.array(w.value).flatten()\n u2 = -x2 @ er + 0.5 * x2 @ sec_cov @ x2\n \n np.testing.assert_array_almost_equal(u1, u2, 4)\n\n df.loc['alphamind', u_name] = elasped_time1\n df.loc['cvxpy', u_name] = elasped_time2\n alpha_logger.info(f\"{u_name} is finished\")",
"2019-02-10 00:39:45,561 - ALPHA_MIND - INFO - sh50 is finished\n2019-02-10 00:39:45,704 - ALPHA_MIND - INFO - hs300 is finished\n2019-02-10 00:39:45,943 - ALPHA_MIND - INFO - zz500 is finished\n2019-02-10 00:39:46,729 - ALPHA_MIND - INFO - zz800 is finished\n2019-02-10 00:39:47,099 - ALPHA_MIND - INFO - zz1000 is finished\n2019-02-10 00:39:49,245 - ALPHA_MIND - INFO - ashare_ex is finished\n"
],
[
"df",
"_____no_output_____"
]
],
[
[
"## 5. Mean - Variance 优化 (Box约束以及线性约束)\n----------------",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(columns=u_names, index=['cvxpy', 'alphamind'])\nnumber = 1\n\nfor u_name, sample_data in zip(u_names, data_set):\n all_styles = risk_styles + industry_styles + ['COUNTRY']\n factor_data = sample_data['factor']\n risk_cov = sample_data['risk_cov'][all_styles].values\n risk_exposure = factor_data[all_styles].values\n special_risk = factor_data.srisk.values\n sec_cov = risk_exposure @ risk_cov @ risk_exposure.T / 10000 + np.diag(special_risk ** 2) / 10000\n er = factor_data[factor].values\n n = len(er)\n \n bm = np.zeros(n)\n lbound = np.zeros(n)\n ubound = np.ones(n) * 0.1\n \n risk_constraints = np.ones((len(er), 1))\n risk_target = (np.array([1.]), np.array([1.]))\n \n risk_model = dict(cov=None, factor_cov=risk_cov/10000., factor_loading=risk_exposure, idsync=(special_risk**2)/10000.)\n\n status, y, x1 = mean_variance_builder(er,\n risk_model,\n bm,\n lbound,\n ubound,\n risk_constraints,\n risk_target)\n elasped_time1 = timeit.timeit(\"\"\"mean_variance_builder(er,\n risk_model,\n bm,\n lbound,\n ubound,\n risk_constraints,\n risk_target)\"\"\",\n number=number, globals=globals()) / number * 1000\n \n w = cvxpy.Variable(n)\n risk = sum_squares(multiply(special_risk / 100., w)) + quad_form((w.T * risk_exposure).T, risk_cov / 10000.)\n objective = cvxpy.Minimize(-w.T * er + 0.5 * risk)\n curr_risk_exposure = risk_constraints.T @ w\n constraints = [w >= lbound,\n w <= ubound,\n curr_risk_exposure == risk_target[0]]\n prob = cvxpy.Problem(objective, constraints)\n prob.solve(solver='ECOS')\n elasped_time2 = timeit.timeit(\"prob.solve(solver='ECOS')\",\n number=number, globals=globals()) / number * 1000\n\n u1 = -x1 @ er + 0.5 * x1 @ sec_cov @ x1\n x2 = np.array(w.value).flatten()\n u2 = -x2 @ er + 0.5 * x2 @ sec_cov @ x2\n \n np.testing.assert_array_almost_equal(u1, u2, 4)\n\n df.loc['alphamind', u_name] = elasped_time1\n df.loc['cvxpy', u_name] = elasped_time2\n alpha_logger.info(f\"{u_name} is finished\")",
"2019-02-10 00:39:49,364 - ALPHA_MIND - INFO - sh50 is finished\n2019-02-10 00:39:49,480 - ALPHA_MIND - INFO - hs300 is finished\n2019-02-10 00:39:49,680 - ALPHA_MIND - INFO - zz500 is finished\n2019-02-10 00:39:50,021 - ALPHA_MIND - INFO - zz800 is finished\n2019-02-10 00:39:50,458 - ALPHA_MIND - INFO - zz1000 is finished\n2019-02-10 00:39:52,248 - ALPHA_MIND - INFO - ashare_ex is finished\n"
],
[
"df",
"_____no_output_____"
]
],
[
[
"## 6. 线性优化(带二次限制条件)\n-------------------------",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(columns=u_names, index=['cvxpy', 'alphamind'])\nnumber = 1\ntarget_vol = 0.5\n\n\nfor u_name, sample_data in zip(u_names, data_set):\n all_styles = risk_styles + industry_styles + ['COUNTRY']\n factor_data = sample_data['factor']\n risk_cov = sample_data['risk_cov'][all_styles].values\n risk_exposure = factor_data[all_styles].values\n special_risk = factor_data.srisk.values\n sec_cov = risk_exposure @ risk_cov @ risk_exposure.T / 10000 + np.diag(special_risk ** 2) / 10000\n er = factor_data[factor].values\n n = len(er)\n \n if 'weight' in factor_data:\n bm = factor_data.weight.values\n else:\n bm = np.ones_like(er) / n\n lbound = np.zeros(n)\n ubound = np.ones(n) * 0.1\n \n risk_constraints = np.ones((n, 1))\n risk_target = (np.array([bm.sum()]), np.array([bm.sum()]))\n risk_model = dict(cov=None, factor_cov=risk_cov/10000., factor_loading=risk_exposure, idsync=(special_risk**2)/10000.)\n\n status, y, x1 = target_vol_builder(er,\n risk_model,\n bm,\n lbound,\n ubound,\n risk_constraints,\n risk_target,\n vol_target=target_vol)\n elasped_time1 = timeit.timeit(\"\"\"target_vol_builder(er,\n risk_model,\n bm,\n lbound,\n ubound,\n risk_constraints,\n risk_target,\n vol_target=target_vol)\"\"\",\n number=number, globals=globals()) / number * 1000\n \n w = cvxpy.Variable(n)\n risk = sum_squares(multiply(special_risk / 100., w)) + quad_form((w.T * risk_exposure).T, risk_cov / 10000.)\n objective = cvxpy.Minimize(-w.T * er)\n curr_risk_exposure = risk_constraints.T @ w\n constraints = [w >= lbound,\n w <= ubound,\n curr_risk_exposure == risk_target[0],\n risk <= target_vol * target_vol]\n prob = cvxpy.Problem(objective, constraints)\n prob.solve(solver='ECOS')\n elasped_time2 = timeit.timeit(\"prob.solve(solver='ECOS')\",\n number=number, globals=globals()) / number * 1000\n\n u1 = -x1 @ er\n x2 = np.array(w.value).flatten()\n u2 = -x2 @ er\n \n np.testing.assert_array_almost_equal(u1, u2, 4)\n\n df.loc['alphamind', u_name] = elasped_time1\n df.loc['cvxpy', u_name] = elasped_time2\n alpha_logger.info(f\"{u_name} is finished\")",
"2019-02-10 00:39:52,370 - ALPHA_MIND - INFO - sh50 is finished\n2019-02-10 00:39:52,532 - ALPHA_MIND - INFO - hs300 is finished\n2019-02-10 00:39:52,798 - ALPHA_MIND - INFO - zz500 is finished\n2019-02-10 00:39:53,099 - ALPHA_MIND - INFO - zz800 is finished\n2019-02-10 00:39:53,589 - ALPHA_MIND - INFO - zz1000 is finished\n2019-02-10 00:39:55,894 - ALPHA_MIND - INFO - ashare_ex is finished\n"
],
[
"df",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05beb9bf0a25ff4f7213586f3182fbd9eaf68c7 | 2,401 | ipynb | Jupyter Notebook | 01_(Paula)TrainAE.ipynb | hernanlira/hl_stargaze | 930042e7c004ff4b7a57f91a324de36a74c59579 | [
"Apache-2.0"
] | null | null | null | 01_(Paula)TrainAE.ipynb | hernanlira/hl_stargaze | 930042e7c004ff4b7a57f91a324de36a74c59579 | [
"Apache-2.0"
] | null | null | null | 01_(Paula)TrainAE.ipynb | hernanlira/hl_stargaze | 930042e7c004ff4b7a57f91a324de36a74c59579 | [
"Apache-2.0"
] | null | null | null | 25.010417 | 99 | 0.556018 | [
[
[
"Based on **Train-AEmodel-GRU2x32-encoding16-AEmodel-DR5-ps-SDSS-QSO-balanced-wandb.ipynb**\n\n## To-do",
"_____no_output_____"
]
],
[
[
"gpu_info = !nvidia-smi\ngpu_info = '\\n'.join(gpu_info)\nif gpu_info.find('failed') >= 0:\n print('Select the Runtime > \"Change runtime type\" menu to enable a GPU accelerator, ')\n print('and then re-execute this cell.')\nelse:\n print(gpu_info)",
"zsh:1: command not found: nvidia-smi\n"
],
[
"from psutil import virtual_memory\nram_gb = virtual_memory().total / 1e9\nprint('Your runtime has {:.1f} gigabytes of available RAM\\n'.format(ram_gb))\n\nif ram_gb < 20:\n print('To enable a high-RAM runtime, select the Runtime > \"Change runtime type\"')\n print('menu, and then select High-RAM in the Runtime shape dropdown. Then, ')\n print('re-execute this cell.')\nelse:\n print('You are using a high-RAM runtime!')",
"Your runtime has 8.6 gigabytes of available RAM\n\nTo enable a high-RAM runtime, select the Runtime > \"Change runtime type\"\nmenu, and then select High-RAM in the Runtime shape dropdown. Then, \nre-execute this cell.\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
]
] |
d05bf51d7491496471d00674677ad4ada28509ed | 174,687 | ipynb | Jupyter Notebook | notebooks/cifar-10/pixelShuffle.ipynb | henriwoodcock/Applying-Modern-Best-Practices-to-Autoencoders | ed2b27a4c6ea3a787fc222b5debdd96cbbafddf9 | [
"MIT"
] | null | null | null | notebooks/cifar-10/pixelShuffle.ipynb | henriwoodcock/Applying-Modern-Best-Practices-to-Autoencoders | ed2b27a4c6ea3a787fc222b5debdd96cbbafddf9 | [
"MIT"
] | null | null | null | notebooks/cifar-10/pixelShuffle.ipynb | henriwoodcock/Applying-Modern-Best-Practices-to-Autoencoders | ed2b27a4c6ea3a787fc222b5debdd96cbbafddf9 | [
"MIT"
] | null | null | null | 272.522621 | 74,482 | 0.891429 | [
[
[
"# Pixel Shuffle\n\nThis notebook is a comparison between two best practices. Pixel shuffle and upsampling followed by a convolution.",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"from fastai import *\nfrom fastai.tabular import *\nimport pandas as pd\nfrom torchsummary import summary\nimport torch\nfrom torch import nn\nimport imageio\nimport torch\nimport glob\nfrom fastai.vision import *\nimport os\nfrom torch import nn\nimport torch.nn.functional as F",
"_____no_output_____"
]
],
[
[
"## Data",
"_____no_output_____"
]
],
[
[
"colab = True\nif colab:\n from google.colab import drive\n drive.mount('/content/drive', force_remount = True)\n %cp \"/content/drive/My Drive/autoencoder-training/data.zip\" .\n !unzip -q data.zip\n image_path = \"data\"\n %cp \"/content/drive/My Drive/autoencoder-training/model_layers.py\" .\n %cp \"/content/drive/My Drive/autoencoder-training/baseline_model.py\" .\n %cp \"/content/drive/My Drive/autoencoder-training/pixelShuffle_model.py\" .\n import pixelShuffle_model\nelse: \n os.chdir(\"../\")\n image_path = os.getcwd() + \"/data\"",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&response_type=code&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly\n\nEnter your authorization code:\n··········\nMounted at /content/drive\n"
],
[
"np.random.seed(3333)\ntorch.manual_seed(3333)\n\nsize = 32\nbatchsize = 128\n\n#tfms = get_transforms(do_flip = True)\ntfms = get_transforms(do_flip=True, flip_vert=True, max_rotate=10, max_zoom=1.1, max_lighting=0.2, max_warp=0.2, p_affine=0, \n p_lighting=0.75)\nsrc = (ImageImageList.from_folder(image_path).split_by_folder().label_from_func(lambda x: x))\ndata = (src.transform(tfms, size=size, tfm_y=True)\n .databunch(bs=batchsize)\n .normalize(imagenet_stats, do_y = False))",
"_____no_output_____"
]
],
[
[
"## Model",
"_____no_output_____"
]
],
[
[
"autoencoder = pixelShuffle_model.autoencoder()",
"_____no_output_____"
],
[
"learn = Learner(data, autoencoder, loss_func = F.mse_loss)",
"_____no_output_____"
],
[
"learn.fit_one_cycle(5)",
"_____no_output_____"
],
[
"learn.lr_find()",
"_____no_output_____"
],
[
"learn.recorder.plot(suggestion=True)",
"Min numerical gradient: 7.59E-07\nMin loss divided by 10: 2.75E-07\n"
],
[
"learn.metrics = [mean_squared_error, mean_absolute_error, r2_score, explained_variance]",
"_____no_output_____"
],
[
"learn.fit_one_cycle(10, max_lr = 1e-03)",
"_____no_output_____"
]
],
[
[
"## Results",
"_____no_output_____"
],
[
"### Training",
"_____no_output_____"
]
],
[
[
"learn.show_results(ds_type=DatasetType.Train)",
"_____no_output_____"
]
],
[
[
"### Validation",
"_____no_output_____"
]
],
[
[
"learn.show_results(ds_type=DatasetType.Valid)",
"_____no_output_____"
],
[
"torch.save(autoencoder, \"/content/drive/My Drive/autoencoder-training/pixelShuffle-Cifar10.pt\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05bfe14600632d800260dae253236c0f8211a14 | 4,680 | ipynb | Jupyter Notebook | make_172_imagenet_6_class_data-Copy1.ipynb | BbChip0103/research_2d_bspl | b4d1305bd911ac0dc96b2c5d9064aa82315d11f0 | [
"MIT"
] | 1 | 2021-04-30T10:28:15.000Z | 2021-04-30T10:28:15.000Z | make_172_imagenet_6_class_data-Copy1.ipynb | BbChip0103/research_2d_bspl | b4d1305bd911ac0dc96b2c5d9064aa82315d11f0 | [
"MIT"
] | null | null | null | make_172_imagenet_6_class_data-Copy1.ipynb | BbChip0103/research_2d_bspl | b4d1305bd911ac0dc96b2c5d9064aa82315d11f0 | [
"MIT"
] | null | null | null | 24.375 | 96 | 0.522436 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport os\nimport os.path as path\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"index_list = [\n [0,2],\n [3,58],\n [59,63],\n [64,182],\n [183,185],\n [186,190]\n]\ny_list = ['bed', 'bird', 'cat', 'dog', 'house', 'tree']",
"_____no_output_____"
],
[
"base_path = 'data'",
"_____no_output_____"
],
[
"# Load npz\nsavepath = path.join(base_path, '172_bbc.npz')\nl = np.load(savepath)\nbbc = l['bbc']\nbbclabel = l['bbclabel']\n\nsavepath = path.join(base_path, '172_dht.npz')\nl = np.load(savepath)\ndht = l['dht']\ndhtlabel = l['dhtlabel']",
"_____no_output_____"
],
[
"savepath = path.join(base_path, '172_val.npz')\nl = np.load(savepath)\nvalimg = l['valimg']\nvallabel = l['vallabel']",
"_____no_output_____"
],
[
"img_shape = (172, 172, 3)",
"_____no_output_____"
],
[
"x_train = np.zeros((len(bbc)+len(dht), *img_shape))\n\nfor i in range(len(bbc)):\n x_train[i] = bbc[i].reshape(img_shape)\nfor i in range(len(dht)):\n x_train[len(bbc)+i] = dht[i].reshape(img_shape)",
"_____no_output_____"
],
[
"a = np.argmax(bbclabel, axis=1)\nb = np.argmax(dhtlabel, axis=1)\ny_train = np.hstack((a,b))\n\nfor i, [begin, end] in enumerate(index_list):\n y_train[(y_train >= begin) & (y_train <= end)] = i ",
"_____no_output_____"
],
[
"valimg = l['valimg']\nvallabel = l['vallabel']\n\nx_val = np.zeros((len(valimg), *img_shape))\nfor i in range(len(valimg)):\n x_val[i] = valimg[i].reshape(img_shape)\n \ny_val = np.argmax(vallabel, axis=1)\nfor i, [begin, end] in enumerate(index_list):\n y_val[(y_val >= begin) & (y_val <= end)] = i ",
"_____no_output_____"
]
],
[
[
"y_label = np.argmax(y_data, axis=1)\ny_text = ['bed', 'bird', 'cat', 'dog', 'house', 'tree']\ny_table = {i:text for i, text in enumerate(y_text)}\ny_table_array = np.array([(i, text) for i, text in enumerate(y_text)])",
"_____no_output_____"
],
[
"x_train_temp, x_test, y_train_temp, y_test = train_test_split(\n x_2d_data, y_label, test_size=0.2, random_state=42, stratify=y_label)\n\nx_train, x_val, y_train, y_val = train_test_split(\n x_train_temp, y_train_temp, test_size=0.25, random_state=42, stratify=y_train_temp)\n\nx_train.shape, y_train.shape, x_val.shape, y_val.shape, x_test.shape, y_test.shape",
"_____no_output_____"
]
],
[
[
"np.savez_compressed(path.join(base_path, 'imagenet_6_class_172_train_data_1.npz'), \n x_data=x_train, y_data=y_train, y_list=y_list)\nnp.savez_compressed(path.join(base_path, 'imagenet_6_class_172_val_data_1.npz'), \n x_data=x_val, y_data=y_val, y_list=y_list)",
"_____no_output_____"
]
]
] | [
"code",
"raw",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"raw",
"raw"
],
[
"code"
]
] |
d05c0b3ab127a3e16ee8169dc4d133dba38bccbb | 31,131 | ipynb | Jupyter Notebook | 03...learn_python.ipynb | ram574/Python-Learning | 1029978679a81674cd19d52f4c9ce022721657ec | [
"MIT"
] | null | null | null | 03...learn_python.ipynb | ram574/Python-Learning | 1029978679a81674cd19d52f4c9ce022721657ec | [
"MIT"
] | null | null | null | 03...learn_python.ipynb | ram574/Python-Learning | 1029978679a81674cd19d52f4c9ce022721657ec | [
"MIT"
] | null | null | null | 22.157295 | 1,127 | 0.434422 | [
[
[
"# Control Flow",
"_____no_output_____"
],
[
"### Python if else",
"_____no_output_____"
]
],
[
[
"def multiply(a, b):\n \"\"\"Function to multiply\"\"\"\n print(a * b)\nprint(multiply.__doc__)\nmultiply(5,2)",
"Function to multiply\n10\n"
],
[
"def func():\n \"\"\"Function to check i is greater or smaller\"\"\"\n i=10\n if i>5:\n print(\"i is greater than 5\")\n else:\n print(\"i is less than 15\")\nprint(func.__doc__)\nfunc()",
"Function to check i is greater or smaller\ni is greater than 5\n"
]
],
[
[
"### Nested if",
"_____no_output_____"
]
],
[
[
"if i==20:\n print(\"i is 10\")\nif i<15:\n print(\"i is less than 15\")\n if i>15:\n print(\"i is greater than 15\")\n else:\n print(\"Not present\") ",
"_____no_output_____"
]
],
[
[
"### if-elif-else ladder",
"_____no_output_____"
]
],
[
[
"def func():\n i=10\n if i==10:\n print(\"i is equal to 10\")\n elif i==15:\n print(\"Not present\")\n elif i==20:\n print('i am there')\n else:\n print(\"none\")\nfunc()",
"_____no_output_____"
]
],
[
[
"### Python for loop ",
"_____no_output_____"
]
],
[
[
"def func():\n var = input(\"enter number:\")\n x = int(var)\n for i in range(x):\n print(i)\nfunc()",
"enter number:10\n0\n1\n2\n3\n4\n5\n6\n7\n8\n9\n"
],
[
"## Lists iteration\ndef func():\n print(\"List Iteration\")\n l = [\"tulasi\", \"ram\", \"ponaganti\"]\n for i in l:\n print(i)\nfunc()\n\n# Iterating over a tuple (immutable)\ndef func():\n print(\"\\nTuple Iteration\")\n t = (\"tulasi\", \"ram\", \"ponaganti\")\n for i in t:\n print(i)\nfunc()\n \n# Iterating over a String\ndef func():\n print(\"\\nString Iteration\")\n s = \"tulasi\"\n for i in s:\n print(i)\nfunc()\n\n# Iterating over dictionary\ndef func():\n print(\"\\nDictionary Iteration\")\n d = dict()\n d['xyz'] = 123\n d['abc'] = 345\n for i in d:\n print(\"% s % d\" % (i, d[i]))\nfunc()",
"List Iteration\ntulasi\nram\nponaganti\n\nTuple Iteration\ntulasi\nram\nponaganti\n\nString Iteration\nt\nu\nl\na\ns\ni\n\nDictionary Iteration\nxyz 123\nabc 345\n"
]
],
[
[
"### Python for Loop with Continue Statement",
"_____no_output_____"
]
],
[
[
"def func():\n for letter in 'tulasiram':\n if letter == 'a':\n continue\n print(letter)\nfunc()",
"t\nu\nl\ns\ni\nr\nm\n"
]
],
[
[
"### Python For Loop with Break Statement",
"_____no_output_____"
]
],
[
[
"def func():\n for letter in 'tulasiram':\n if letter == 'a':\n break\n print('Current Letter :', letter)\nfunc()",
"Current Letter : t\nCurrent Letter : u\nCurrent Letter : l\n"
]
],
[
[
"### Python For Loop with Pass Statement",
"_____no_output_____"
]
],
[
[
"list = ['tulasi','ram','ponaganti']\n\ndef func():\n #An empty loop\n for list in 'ponaganti':\n pass\n print('Last Letter :', list)\nfunc()",
"Last Letter : i\n"
]
],
[
[
"### Python range",
"_____no_output_____"
]
],
[
[
"def func():\n sum=0\n for i in range(1,5):\n sum = sum + i\n print(sum)\n\nfunc()",
"10\n"
],
[
"def func():\n i=5\n for x in range(i):\n i = i+x\n print(i)\nfunc()",
"5\n6\n8\n11\n15\n"
]
],
[
[
"### Python for loop with else",
"_____no_output_____"
]
],
[
[
"for i in range(1, 4):\n print(i)\nelse: # Executed because no break in for\n print(\"No Break\\n\")\nfor i in range(1, 4):\n print(i)\n break\nelse: # Not executed as there is a break\n print(\"No Break\")",
"1\n2\n3\nNo Break\n\n1\n"
],
[
"### Using all for loop statements in small program\ndef func():\n var = input(\"enter number:\")\n x = int(var)\n for i in range(x):\n option = input(\"print, skip, or exit\")\n if option==\"print\":\n print(i)\n elif option=='skip':\n continue\n elif option=='exit':\n break\n print(\"Good bye....!\")\n\nfunc()",
"enter number:10\nprint, skip, or exit20\nprint, skip, or exit30\nprint, skip, or exit40\nprint, skip, or exit50\nprint, skip, or exit60\nprint, skip, or exit70\nprint, skip, or exit80\nprint, skip, or exit90\nprint, skip, or exit20\nprint, skip, or exit40\nGood bye....!\n"
],
[
"### Working with lists\ndef func():\n product_prices = []\n for i in range(5):\n product = input(\"How much the product cost ?\")\n product = float(product)\n product_prices.append(product)\n print(product_prices)\n print(\"Total price : \" , sum(product_prices))\n print(\"High cost of product :\" , max(product_prices))\n print(\"average price of products\", sum(product_prices)/len(product_prices))\nfunc()",
"How much the product cost ?10\nHow much the product cost ?20\nHow much the product cost ?30\nHow much the product cost ?40\nHow much the product cost ?50\n[10.0, 20.0, 30.0, 40.0, 50.0]\nTotal price : 150.0\nHigh cost of product : 50.0\naverage price of products 30.0\n"
],
[
"### Nested for loop\n### one to Twelve time tables using for loop\ndef func():\n for num1 in range(1,13):\n for num2 in range(1,13):\n print(num1, \"*\", num2, \"=\", num1*num2)\nfunc()",
"1 * 1 = 1\n1 * 2 = 2\n1 * 3 = 3\n1 * 4 = 4\n1 * 5 = 5\n1 * 6 = 6\n1 * 7 = 7\n1 * 8 = 8\n1 * 9 = 9\n1 * 10 = 10\n1 * 11 = 11\n1 * 12 = 12\n2 * 1 = 2\n2 * 2 = 4\n2 * 3 = 6\n2 * 4 = 8\n2 * 5 = 10\n2 * 6 = 12\n2 * 7 = 14\n2 * 8 = 16\n2 * 9 = 18\n2 * 10 = 20\n2 * 11 = 22\n2 * 12 = 24\n3 * 1 = 3\n3 * 2 = 6\n3 * 3 = 9\n3 * 4 = 12\n3 * 5 = 15\n3 * 6 = 18\n3 * 7 = 21\n3 * 8 = 24\n3 * 9 = 27\n3 * 10 = 30\n3 * 11 = 33\n3 * 12 = 36\n4 * 1 = 4\n4 * 2 = 8\n4 * 3 = 12\n4 * 4 = 16\n4 * 5 = 20\n4 * 6 = 24\n4 * 7 = 28\n4 * 8 = 32\n4 * 9 = 36\n4 * 10 = 40\n4 * 11 = 44\n4 * 12 = 48\n5 * 1 = 5\n5 * 2 = 10\n5 * 3 = 15\n5 * 4 = 20\n5 * 5 = 25\n5 * 6 = 30\n5 * 7 = 35\n5 * 8 = 40\n5 * 9 = 45\n5 * 10 = 50\n5 * 11 = 55\n5 * 12 = 60\n6 * 1 = 6\n6 * 2 = 12\n6 * 3 = 18\n6 * 4 = 24\n6 * 5 = 30\n6 * 6 = 36\n6 * 7 = 42\n6 * 8 = 48\n6 * 9 = 54\n6 * 10 = 60\n6 * 11 = 66\n6 * 12 = 72\n7 * 1 = 7\n7 * 2 = 14\n7 * 3 = 21\n7 * 4 = 28\n7 * 5 = 35\n7 * 6 = 42\n7 * 7 = 49\n7 * 8 = 56\n7 * 9 = 63\n7 * 10 = 70\n7 * 11 = 77\n7 * 12 = 84\n8 * 1 = 8\n8 * 2 = 16\n8 * 3 = 24\n8 * 4 = 32\n8 * 5 = 40\n8 * 6 = 48\n8 * 7 = 56\n8 * 8 = 64\n8 * 9 = 72\n8 * 10 = 80\n8 * 11 = 88\n8 * 12 = 96\n9 * 1 = 9\n9 * 2 = 18\n9 * 3 = 27\n9 * 4 = 36\n9 * 5 = 45\n9 * 6 = 54\n9 * 7 = 63\n9 * 8 = 72\n9 * 9 = 81\n9 * 10 = 90\n9 * 11 = 99\n9 * 12 = 108\n10 * 1 = 10\n10 * 2 = 20\n10 * 3 = 30\n10 * 4 = 40\n10 * 5 = 50\n10 * 6 = 60\n10 * 7 = 70\n10 * 8 = 80\n10 * 9 = 90\n10 * 10 = 100\n10 * 11 = 110\n10 * 12 = 120\n11 * 1 = 11\n11 * 2 = 22\n11 * 3 = 33\n11 * 4 = 44\n11 * 5 = 55\n11 * 6 = 66\n11 * 7 = 77\n11 * 8 = 88\n11 * 9 = 99\n11 * 10 = 110\n11 * 11 = 121\n11 * 12 = 132\n12 * 1 = 12\n12 * 2 = 24\n12 * 3 = 36\n12 * 4 = 48\n12 * 5 = 60\n12 * 6 = 72\n12 * 7 = 84\n12 * 8 = 96\n12 * 9 = 108\n12 * 10 = 120\n12 * 11 = 132\n12 * 12 = 144\n"
]
],
[
[
"### Python while loop",
"_____no_output_____"
]
],
[
[
"## Single line statement\n\ndef func():\n '''first one'''\n count = 0\n while (count < 5): count = count + 1; print(\"Tulasi Ram\")\nprint(func.__doc__)\nfunc()\n\n### or\n\ndef func():\n '''Second one'''\n count = 0\n while (count < 5): \n count = count + 1 \n print(\"Tulasi Ram\")\nprint(func.__doc__)\nfunc()",
"first one\nTulasi Ram\nTulasi Ram\nTulasi Ram\nTulasi Ram\nTulasi Ram\nSecond one\nTulasi Ram\nTulasi Ram\nTulasi Ram\nTulasi Ram\nTulasi Ram\n"
],
[
"def func():\n list = [\"ram\",\"tulasi\",\"ponaganti\"]\n while list:\n print(list.pop())\nfunc()",
"ponaganti\ntulasi\nram\n"
],
[
"def func():\n i=0\n for i in range(10):\n i+=1\n return i\nfunc()",
"_____no_output_____"
],
[
"def func():\n i = 0\n a = ['tulasi','ram','ponaganti']\n while i < len(a):\n if a[i] == 'tulasi' or a[i] == 'ram':\n i += 1\n continue\n print('Current word :', a[i])\n i+=1\nfunc()",
"Current word : ponaganti\n"
],
[
"def func():\n i = 0\n a = ['tulasi','ram','ponaganti']\n while i < len(a):\n if a[i] == 'ponaganti':\n i += 1\n break\n print('Current word :', a[i])\n i+=1\nfunc()",
"Current word : tulasi\nCurrent word : ram\n"
],
[
"def func():\n i = 0\n a = ['tulasi','ram','ponaganti']\n while i < len(a):\n if a[i] == 'tulasi':\n i += 1\n pass\n print('Current word :', a[i])\n i+=1\nfunc()",
"Current word : ram\nCurrent word : ponaganti\n"
],
[
"def whileElseFunc():\n i=0\n while i<10:\n i+=1\n print(i)\n else:\n print('no break')\n\nwhileElseFunc()\n",
"1\n2\n3\n4\n5\n6\n7\n8\n9\n10\nno break\n"
]
],
[
[
"### using break in loops",
"_____no_output_____"
]
],
[
[
"def func():\n i=0\n for i in range(10):\n i+=1\n print(i)\n break\n else:\n print('no break')\nfunc() ",
"1\n"
]
],
[
[
"### using continue in loops",
"_____no_output_____"
]
],
[
[
"def func():\n i=0\n for i in range(10):\n i+=1\n print(i)\n continue\n else:\n for i in range(5):\n i+=1\n print(i)\n break\n \nfunc()",
"1\n2\n3\n4\n5\n6\n7\n8\n9\n10\n1\n"
],
[
"def func():\n i=0\n for i in range(10):\n i+=1\n print(i)\n pass\n else:\n for i in range(5):\n i+=1\n print(i) \nfunc()",
"1\n2\n3\n4\n5\n6\n7\n8\n9\n10\n1\n2\n3\n4\n5\n"
]
],
[
[
"#### Looping techniques using enumerate()",
"_____no_output_____"
]
],
[
[
"def enumearteFunc():\n list =['tulasi','ram','ponaganti']\n for key in enumerate(list):\n print(key)\nenumearteFunc()\n\ndef enumearteFunc():\n list =['tulasi','ram','ponaganti']\n for key, value in enumerate(list):\n print(value)\nenumearteFunc()",
"(0, 'tulasi')\n(1, 'ram')\n(2, 'ponaganti')\ntulasi\nram\nponaganti\n"
],
[
"def zipFunc():\n list1 = ['name', 'firstname', 'lastname']\n list2 = ['ram', 'tulasi', 'ponaganti']\n# using zip() to combine two containers\n# and print values\n for list1, list2 in zip(list1, list2):\n print('What is your {0}? I am {1}.'.format(list1, list2))\nzipFunc()",
"What is your name? I am ram.\nWhat is your firstname? I am tulasi.\nWhat is your lastname? I am ponaganti.\n"
]
],
[
[
"\"\"\" Using iteritem(): iteritems() is used to loop through the \n dictionary printing the dictionary key-value pair sequentially \n which is used before Python 3 version\n \n Using items(): items() performs the similar task on dictionary as \n iteritems() but have certain disadvantages when compared with iteritems() \"\"\"",
"_____no_output_____"
]
],
[
[
"def itemFunc():\n name = {\"name\": \"tulasi\", \"firstname\": \"ram\"}\n print(\"The key value pair using items is : \")\n for key, value in name.items():\n print(key, value)\nitemFunc()",
"The key value pair using items is : \nname tulasi\nfirstname ram\n"
]
],
[
[
"sorting the list items using loop",
"_____no_output_____"
]
],
[
[
"def sortedFunc():\n list = ['ram','tulasi','ponaganti']\n for i in list:\n print(sorted(i))\n continue\n for i in reversed(list):\n print(i, end=\" \")\nsortedFunc()",
"['a', 'm', 'r']\n['a', 'i', 'l', 's', 't', 'u']\n['a', 'a', 'g', 'i', 'n', 'n', 'o', 'p', 't']\nponaganti tulasi ram "
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d05c0c2205299a5697cc5be818ca1277bef2f1b7 | 621,188 | ipynb | Jupyter Notebook | caffe/examples/detection.ipynb | dymil/dec | e551e01f90a3d67d2ef9c90e968c8faf5d8f857d | [
"MIT"
] | 408 | 2015-11-19T21:50:16.000Z | 2022-03-22T08:17:26.000Z | caffe/examples/detection.ipynb | XuJiaMing1997/dec | e551e01f90a3d67d2ef9c90e968c8faf5d8f857d | [
"MIT"
] | 29 | 2016-05-18T10:24:00.000Z | 2021-09-26T21:43:46.000Z | caffe/examples/detection.ipynb | XuJiaMing1997/dec | e551e01f90a3d67d2ef9c90e968c8faf5d8f857d | [
"MIT"
] | 152 | 2015-11-24T17:30:36.000Z | 2021-11-11T07:17:03.000Z | 734.264775 | 223,295 | 0.930886 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d05c1cf45da52b27cb65afadef774464c696a87e | 19,999 | ipynb | Jupyter Notebook | CNNTracker1-2.ipynb | clatfd/Coronary-Artery-Tracking-via-3D-CNN-Classification | ce26bed713552773436a2f8d90d83f274e932ebb | [
"MIT"
] | null | null | null | CNNTracker1-2.ipynb | clatfd/Coronary-Artery-Tracking-via-3D-CNN-Classification | ce26bed713552773436a2f8d90d83f274e932ebb | [
"MIT"
] | null | null | null | CNNTracker1-2.ipynb | clatfd/Coronary-Artery-Tracking-via-3D-CNN-Classification | ce26bed713552773436a2f8d90d83f274e932ebb | [
"MIT"
] | 2 | 2021-12-04T13:13:56.000Z | 2022-02-15T21:09:36.000Z | 31.29734 | 226 | 0.551228 | [
[
[
"# Code for artery tracking \n#simplified from 1-1",
"_____no_output_____"
],
[
"%load_ext autoreload\n%autoreload 2\n\nimport os\nos.environ[\"CUDA_VISIBLE_DEVICES\"]=\"0\"\nimport torch\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\n\nimport json\nimport cv2\nimport os\nimport matplotlib.pyplot as plt\nimport copy\nimport numpy as np\nimport pickle\nimport glob\nimport datetime\nimport pickle\n",
"_____no_output_____"
],
[
"%%javascript\n$('<div id=\"toc\"></div>').css({position: 'fixed', top: '120px', left: 0}).appendTo(document.body);\n$.getScript('https://kmahelona.github.io/ipython_notebook_goodies/ipython_notebook_toc.js');",
"_____no_output_____"
],
[
"# Load CNN Tracker\nimport sys\nsys.path.append(r'U:\\LiChen\\AICafe\\CNNTracker')\nfrom models.centerline_net import CenterlineNet\nfrom centerline_train_tools.data_provider_argu import DataGenerater\nfrom centerline_train_tools.centerline_trainner import Trainer\nimport torch",
"_____no_output_____"
],
[
"#import iCafe Python\nimport numpy as np\nimport sys\n#sys.path.append(r'\\\\DESKTOP2\\Ftensorflow\\LiChen\\iCafe')\nsys.path.insert(0,r'\\\\DESKTOP4\\Dtensorflow\\LiChen\\iCafePython')\nfrom iCafePython import iCafe\nfrom iCafePython import SnakeList,Snake,SWCNode,Point3D",
"_____no_output_____"
]
],
[
[
"# Load CNNTracker",
"_____no_output_____"
]
],
[
[
"#only need to select one model\n#Model 1 CNN tracker for ICA TOF MRA\nswc_name = 'cnn_snake'\n\nimport sys\nsys.path.append(r'U:\\LiChen\\AICafe\\CNNTracker')\nfrom models.centerline_net import CenterlineNet\n\nmax_points = 500\nprob_thr = 0.85\n\ninfer_model = CenterlineNet(n_classes=max_points)\ncheckpoint_path_infer = r\"D:\\tensorflow\\LiChen\\AICafe\\CNNTracker\\CNNTracker1-1\\classification_checkpoints\\centerline_net_model_Epoch_29.pkl\"\n\ncheckpoint = torch.load(checkpoint_path_infer)\nnet_dict = checkpoint['net_dict']\ninfer_model.load_state_dict(net_dict)\ninfer_model.to(device)\ninfer_model.eval()",
"_____no_output_____"
],
[
"#Model 2 CNN tracker for Coronary CTA\nswc_name = 'cnn_snake'\nmax_points = 500\nprob_thr = 0.85\n\ninfer_model = CenterlineNet(n_classes=max_points)\n\ncheckpoint_path_infer = r\"D:\\tensorflow\\LiChen\\AICafe\\CNNTracker\\CNNTracker2-1\\classification_checkpoints\\centerline_net_model_Epoch_81.pkl\"\n\ncheckpoint = torch.load(checkpoint_path_infer)\nnet_dict = checkpoint['net_dict']\ninfer_model.load_state_dict(net_dict)\ninfer_model.to(device)\ninfer_model.eval()",
"_____no_output_____"
],
[
"#Model 3 CNN tracker for LATTE\nswc_name = 'cnn_snake'\nmax_points = 500\nprob_thr = 0.85\n\ninfer_model = CenterlineNet(n_classes=max_points)\ncheckpoint_path_infer = r\"D:\\tensorflow\\LiChen\\AICafe\\CNNTracker\\CNNTracker4-1\\classification_checkpoints\\centerline_net_model_Epoch_99.pkl\"\n\n\n\ncheckpoint = torch.load(checkpoint_path_infer)\nnet_dict = checkpoint['net_dict']\ninfer_model.load_state_dict(net_dict)\ninfer_model.to(device)\ninfer_model.eval()",
"_____no_output_____"
]
],
[
[
"# Load datasets",
"_____no_output_____"
]
],
[
[
"dbname = 'BRAVEAI'\nicafe_dir = r'\\\\DESKTOP2\\GiCafe\\result/'\nseg_model_name = 'LumenSeg2-3'\n\nwith open(icafe_dir+'/'+dbname+'/db.list','rb') as fp:\n dblist = pickle.load(fp)\ntrain_list = dblist['train']\nval_list = dblist['val']\ntest_list = dblist['test']\npilist = [pi.split('/')[1] for pi in dblist['test']]\nlen(pilist)",
"_____no_output_____"
],
[
"dbname = 'RotterdanCoronary'\nicafe_dir = r'\\\\DESKTOP2\\GiCafe\\result/'\npilist = ['0_dataset05_U']\nseg_model_name = 'CoronarySeg1-8-5'",
"_____no_output_____"
],
[
"dbname = 'UNC'\nicafe_dir = r'\\\\DESKTOP2\\GiCafe\\result/'\nseg_model_name = 'LumenSeg5-1'\nwith open(icafe_dir+'/'+dbname+'/db.list','rb') as fp:\n dblist = pickle.load(fp)\npilist = [pi.split('/')[1] for pi in dblist['test']]\nlen(pilist)",
"_____no_output_____"
],
[
"dbname = 'HarborViewT1Pre'\nicafe_dir = r'\\\\DESKTOP2\\GiCafe\\result/'\npilist = ['0_ID%d_U'%i for i in [2,9,10,11,12]]\nlen(pilist)",
"_____no_output_____"
],
[
"# MERGE \ndbname = 'CAREIIMERGEGT'\nicafe_dir = r'\\\\DESKTOP2\\GiCafe\\result/'\nseg_model_name = 'LumenSeg6-1'\nwith open(icafe_dir+'/'+dbname+'/db.list','rb') as fp:\n dblist = pickle.load(fp)\npilist = [pi.split('/')[1] for pi in dblist['test']]\nlen(pilist)",
"_____no_output_____"
],
[
"dbname = 'IPH-Sup-TOF-FullCoverage'\nicafe_dir = r'\\\\DESKTOP2\\GiCafe\\result/'\nseg_model_name = 'LumenSeg7-1'\ndblist_name = icafe_dir+'/'+dbname+'/db.list'\n\nwith open(dblist_name,'rb') as fp:\n dblist = pickle.load(fp)\n \npilist = [pi.split('/')[1] for pi in dblist['test']]\nlen(pilist)",
"_____no_output_____"
],
[
"dbname = 'WALLIAI'\nicafe_dir = r'\\\\DESKTOP2\\GiCafe\\result/'\nseg_model_name = 'LumenSeg8-1'\ndblist_name = icafe_dir+'/'+dbname+'/db.list'\n\nwith open(dblist_name,'rb') as fp:\n dblist = pickle.load(fp)\n \npilist = [pi.split('/')[1] for pi in dblist['test']]\nlen(pilist),pilist\n",
"_____no_output_____"
]
],
[
[
"# Tracking",
"_____no_output_____"
]
],
[
[
"# from s.whole.modelname to swc traces\nfrom iCafePython.connect.ext import extSnake\nimport SimpleITK as sitk\n\n#redo artery tracing\nRETRACE = 1\n#redo artery tree contraint\nRETREE = 1\n\n#segmentation src\nseg_src = 's.whole.'+seg_model_name\n\n#Lumen segmentation threshold. \n# Lower value will cause too many noise branches, and neighboring branches will merge as one\n# Higher value will reduce the traces detectable\nSEGTHRES = 0.5\n\n#max search range in merge/branch, unit in mm\n# Higher value will allow larger gap and merge parts of broken arteries, \n# but will also force noise branches to be merged in the tree\nsearch_range_thres = 10\n\n#which ves to build graph for artery labeling\ngraph_ves = 'seg_ves_ext_tree2'\n\nDEBUG = 0\n\n\nfor pi in pilist[20:19:-1]:\n print('='*10,'Start processing',pilist.index(pi),'/',len(pilist),pi,'='*10)\n if not os.path.exists(icafe_dir+'/'+dbname+'/'+pi):\n os.mkdir(icafe_dir+'/'+dbname+'/'+pi)\n \n icafem = iCafe(icafe_dir+'/'+dbname+'/'+pi)\n \n #select correct version of s.whole from potentially multiple segmentation versions and save as s.whole\n icafem.loadImg(seg_src)\n icafem.saveImg('s.whole',icafem.I[seg_src],np.float16)\n icafem.loadImg('s.whole')\n\n #export v.tif for 3d visualization if icafe project does not have one already\n if 'v' not in icafem.listAvailImgs():\n vimg = copy.copy(icafem.I['s.whole'])\n vimg[vimg<0] = 0\n vimg = (vimg*255).astype(np.uint16)\n icafem.saveImg('v',vimg,np.int16)\n \n #Tracing\n if RETRACE or not icafem.existPath('seg_ves_ext.swc'):\n if 's.whole' not in icafem.I:\n icafem.loadImg('s.whole')\n seg_ves_snakelist = icafem.constructSkeleton(icafem.I['s.whole']>SEGTHRES)\n \n #load image\n file_name = icafem.getPath('o')\n re_spacing_img = sitk.GetArrayFromImage(sitk.ReadImage(file_name))\n\n seg_ves_snakelist = icafem.readSnake('seg_ves')\n seg_ves_ext_snakelist = extSnake(seg_ves_snakelist,infer_model,re_spacing_img,DEBUG=DEBUG)\n icafem.writeSWC('seg_ves_ext',seg_ves_ext_snakelist)\n else:\n seg_ves_ext_snakelist = icafem.readSnake('seg_ves_ext')\n print('read from existing seg ves ext')\n if seg_ves_ext_snakelist.NSnakes==0:\n print('no snake found in seg ves, abort',pi)\n continue\n \n if RETREE or not icafem.existPath('seg_ves_ext_tree.swc'):\n if 's.whole' not in icafem.I:\n icafem.loadImg('s.whole')\n if icafem.xml.res is None:\n icafem.xml.setResolution(0.296875)\n icafem.xml.writexml()\n tree_snakelist = seg_ves_ext_snakelist.tree(icafem,search_range=search_range_thres/icafem.xml.res,int_src='o',DEBUG=DEBUG)\n icafem.writeSWC('seg_ves_ext_tree', tree_snakelist)\n tree_snakelist = tree_snakelist.tree(icafem,search_range=search_range_thres/3/icafem.xml.res,int_src='s.whole',DEBUG=DEBUG)\n icafem.writeSWC('seg_ves_ext_tree2', tree_snakelist)\n \n tree_main_snakelist = tree_snakelist.mainArtTree(dist_thres=10)\n icafem.writeSWC('seg_ves_ext_tree2_main',tree_main_snakelist)\n \n ",
"_____no_output_____"
]
],
[
[
"# Artery labeling",
"_____no_output_____"
]
],
[
[
"from iCafePython.artlabel.artlabel import ArtLabel\nart_label_predictor = ArtLabel()",
"_____no_output_____"
],
[
"for pi in pilist[:]:\n print('='*10,'Start processing',pilist.index(pi),'/',len(pilist),pi,'='*10)\n if not os.path.exists(icafe_dir+'/'+dbname+'/'+pi):\n os.mkdir(icafe_dir+'/'+dbname+'/'+pi)\n \n icafem = iCafe(icafe_dir+'/'+dbname+'/'+pi)\n \n #generate (simplified node!=2) graph for GNN art labeling\n G = icafem.generateGraph(graph_ves,None,graphtype='graphsim', mode='test', trim=1)\n if len(G.nodes())<5:\n print('too few snakes for artlabeling')\n continue\n icafem.writeGraph(G,graphtype='graphsim')\n\n #predict landmarks\n pred_landmark, ves_end_pts = art_label_predictor.pred(icafem.getPath('graphsim'),icafem.xml.res)\n #complete graph Gcom for finding the pts in the path\n Gcom = icafem.generateGraph(graph_ves, None, graphtype='graphcom')\n ves_snakelist = findSnakeFromPts(Gcom,G,ves_end_pts)\n print('@@@predict',len(pred_landmark),'landmarks',ves_snakelist)\n #save landmark and ves\n icafem.xml.landmark = pred_landmark\n icafem.xml.writexml()\n icafem.writeSWC('ves_pred', ves_snakelist)",
"_____no_output_____"
],
[
"vimg = vimg[:,:,::-1]",
"_____no_output_____"
],
[
"np.max(vimg)",
"_____no_output_____"
],
[
"icafem.saveImg('v',vimg,np.float16)",
"_____no_output_____"
],
[
"import tifffile\na = tifffile.imread(r\"\\\\DESKTOP2\\GiCafe\\result\\WALLI\\47_WALLI-V-09-1-B_M\\TH_47_WALLI-V-09-1-B_Mv.tif\")",
"_____no_output_____"
],
[
"np.max(a[118])",
"_____no_output_____"
]
],
[
[
"# Eval",
"_____no_output_____"
]
],
[
[
"def eval_simple(snakelist):\n snakelist = copy.deepcopy(snakelist)\n _ = snakelist.resampleSnakes(1)\n #ground truth snakelist from icafem.veslist\n all_metic = snakelist.motMetric(icafem.veslist)\n metric_dict = all_metic.metrics(['MOTA','IDF1','MOTP','IDS'])\n #ref_snakelist = icafem.readSnake('ves')\n snakelist.compRefSnakelist(icafem.vessnakelist)\n metric_dict['OV'], metric_dict['OF'], metric_dict['OT'], metric_dict['AI'], metric_dict['UM'], metric_dict['UMS'], metric_dict['ref_UM'], metric_dict['ref_UMS'], metric_dict['mean_diff'] = snakelist.evalCompDist()\n str = ''\n metric_dict_simple = ['MOTA','IDF1','MOTP','IDS','OV']\n for key in metric_dict_simple:\n str += key+'\\t'\n str += '\\n'\n for key in metric_dict_simple:\n if type(metric_dict[key]) == int:\n str += '%d\\t'%metric_dict[key]\n else:\n str += '%.3f\\t'%metric_dict[key]\n print(str)\n return metric_dict",
"_____no_output_____"
],
[
"# calculate metric and save in each pi folder\nREFEAT = 0\nfor pi in pilist[:1]:\n print('='*10,'Start processing',pilist.index(pi),'/',len(pilist),pi,'='*10)\n \n icafem = iCafe(icafe_dir+'/'+pi)\n \n if REFEAT or not icafem.existPath('metric.pickle'):\n print('init metric')\n all_metric_dict = {}\n else:\n print('load metric')\n with open(icafem.getPath('metric.pickle'),'rb') as fp:\n all_metric_dict = pickle.load(fp)\n \n for vesname in ['seg_ves_ext_tree2_main']:\n #for vesname in ['seg_raw','seg_ves_ext_main','seg_ves_ext_tree2']:\n #comparison methods\n #for vesname in ['frangi_ves','seg_unet','seg_raw','raw_sep','cnn_snake','dcat_snake','seg_ves_ext_tree2_main']:\n if vesname in all_metric_dict:\n continue\n print('-'*10,vesname,'-'*10)\n pred_snakelist = icafem.readSnake(vesname)\n if pred_snakelist.NSnakes==0:\n print('no snake',pi,vesname)\n continue\n all_metric_dict[vesname] = eval_simple(pred_snakelist.resampleSnakes(1))\n \n with open(icafem.getPath('metric.pickle'),'wb') as fp:\n pickle.dump(all_metric_dict,fp)\n ",
"_____no_output_____"
],
[
"#check feat\npi = pilist[0]\nicafem = iCafe(icafe_dir+'/'+pi)\nwith open(icafem.getPath('metric.pickle'),'rb') as fp:\n all_metric_dict = pickle.load(fp)\nall_metric_dict",
"_____no_output_____"
],
[
"#collect feats from pickle\neval_vesname = {'frangi_ves':'Frangi','seg_unet':'U-Net','seg_raw':'DDT',\n 'raw_sep':'iCafe','cnn_snake':'CNN Tracker','dcat_snake':'DCAT','seg_ves_ext_tree2_main':'DOST (ours)',\n 'seg_ves':'DOST (initial curve)','seg_ves_ext_main':'DOST (deep snake)','seg_ves_ext_tree2':'DOST tree'}\nfeats = {}\nfor vesname in eval_vesname:\n feats[vesname] = {}\n \nfor pi in pilist[:]:\n icafem = iCafe(icafe_dir+'/'+dbname+'/'+pi)\n if not icafem.existPath('metric.pickle'):\n continue\n \n with open(icafem.getPath('metric.pickle'),'rb') as fp:\n all_metric_dict = pickle.load(fp)\n \n #for vesname in all_metric_dict:\n for vesname in eval_vesname:\n if vesname not in all_metric_dict:\n print('no',vesname,'in',pi)\n continue\n for metric in all_metric_dict[vesname]:\n if metric not in feats[vesname]:\n feats[vesname][metric] = []\n feats[vesname][metric].append(all_metric_dict[vesname][metric]) \n \n\nsel_metrics = ['OV','AI', 'MOTA', 'IDF1', 'IDS']\nprint('\\t'.join(['']+sel_metrics))\nfor vesname in feats:\n featstr = eval_vesname[vesname]+'\\t'\n for metric in sel_metrics:\n if metric in ['IDS']:\n featstr += '%.1f\\t'%np.mean(feats[vesname][metric]) \n else:\n featstr += '%.3f\\t'%np.mean(feats[vesname][metric])\n print(featstr)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d05c39c9f8da5156b61c1d1ec784a45007d667ef | 10,183 | ipynb | Jupyter Notebook | Assignment day5.ipynb | Raghavstyleking/LetsUpgrade-Python-Essentials | a8931ac417a228ebbde070d3cf267a0d25aba658 | [
"Apache-2.0"
] | null | null | null | Assignment day5.ipynb | Raghavstyleking/LetsUpgrade-Python-Essentials | a8931ac417a228ebbde070d3cf267a0d25aba658 | [
"Apache-2.0"
] | null | null | null | Assignment day5.ipynb | Raghavstyleking/LetsUpgrade-Python-Essentials | a8931ac417a228ebbde070d3cf267a0d25aba658 | [
"Apache-2.0"
] | null | null | null | 62.858025 | 6,952 | 0.567613 | [
[
[
"# Assignment of Day 5",
"_____no_output_____"
]
],
[
[
"lst1 = [1,5,6,4,1,2,3,5]\nlst2 = [1,5,6,5,1,2,3,6]\n\nlst = [1,1,5]\ncount = 0\nr=0\nfor x in lst:\n for y in lst1[r:]:\n r+=1\n if (x==y):\n count+=1\n break;\n else:\n pass\n \n\nif(count==3):\n print(\"it’s a Match\")\nelse:\n print(\"it’s Gone\")",
"it’s a Match\n"
],
[
"count = 0\nr=0\nfor x in lst:\n for y in lst2[r:]:\n r+=1\n if (x==y):\n count+=1\n break;\n else:\n pass\n \n\nif(count==3):\n print(\"it’s a Match\")\nelse:\n print(\"it’s Gone\")",
"it’s Gone\n"
],
[
"#Make a Function for prime numbers and use Filter to filter out all the prime numbers from 1-2500\ncheck = 0\ndef prime(num):\n if num > 1:\n for i in range(2, num):\n if (num % i) == 0:\n break\n else:\n return num\nnumber = filter(prime,range(1,2500))\n\n\nprint(list(number))",
"[3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101, 103, 105, 107, 109, 111, 113, 115, 117, 119, 121, 123, 125, 127, 129, 131, 133, 135, 137, 139, 141, 143, 145, 147, 149, 151, 153, 155, 157, 159, 161, 163, 165, 167, 169, 171, 173, 175, 177, 179, 181, 183, 185, 187, 189, 191, 193, 195, 197, 199, 201, 203, 205, 207, 209, 211, 213, 215, 217, 219, 221, 223, 225, 227, 229, 231, 233, 235, 237, 239, 241, 243, 245, 247, 249, 251, 253, 255, 257, 259, 261, 263, 265, 267, 269, 271, 273, 275, 277, 279, 281, 283, 285, 287, 289, 291, 293, 295, 297, 299, 301, 303, 305, 307, 309, 311, 313, 315, 317, 319, 321, 323, 325, 327, 329, 331, 333, 335, 337, 339, 341, 343, 345, 347, 349, 351, 353, 355, 357, 359, 361, 363, 365, 367, 369, 371, 373, 375, 377, 379, 381, 383, 385, 387, 389, 391, 393, 395, 397, 399, 401, 403, 405, 407, 409, 411, 413, 415, 417, 419, 421, 423, 425, 427, 429, 431, 433, 435, 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463, 465, 467, 469, 471, 473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501, 503, 505, 507, 509, 511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539, 541, 543, 545, 547, 549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577, 579, 581, 583, 585, 587, 589, 591, 593, 595, 597, 599, 601, 603, 605, 607, 609, 611, 613, 615, 617, 619, 621, 623, 625, 627, 629, 631, 633, 635, 637, 639, 641, 643, 645, 647, 649, 651, 653, 655, 657, 659, 661, 663, 665, 667, 669, 671, 673, 675, 677, 679, 681, 683, 685, 687, 689, 691, 693, 695, 697, 699, 701, 703, 705, 707, 709, 711, 713, 715, 717, 719, 721, 723, 725, 727, 729, 731, 733, 735, 737, 739, 741, 743, 745, 747, 749, 751, 753, 755, 757, 759, 761, 763, 765, 767, 769, 771, 773, 775, 777, 779, 781, 783, 785, 787, 789, 791, 793, 795, 797, 799, 801, 803, 805, 807, 809, 811, 813, 815, 817, 819, 821, 823, 825, 827, 829, 831, 833, 835, 837, 839, 841, 843, 845, 847, 849, 851, 853, 855, 857, 859, 861, 863, 865, 867, 869, 871, 873, 875, 877, 879, 881, 883, 885, 887, 889, 891, 893, 895, 897, 899, 901, 903, 905, 907, 909, 911, 913, 915, 917, 919, 921, 923, 925, 927, 929, 931, 933, 935, 937, 939, 941, 943, 945, 947, 949, 951, 953, 955, 957, 959, 961, 963, 965, 967, 969, 971, 973, 975, 977, 979, 981, 983, 985, 987, 989, 991, 993, 995, 997, 999, 1001, 1003, 1005, 1007, 1009, 1011, 1013, 1015, 1017, 1019, 1021, 1023, 1025, 1027, 1029, 1031, 1033, 1035, 1037, 1039, 1041, 1043, 1045, 1047, 1049, 1051, 1053, 1055, 1057, 1059, 1061, 1063, 1065, 1067, 1069, 1071, 1073, 1075, 1077, 1079, 1081, 1083, 1085, 1087, 1089, 1091, 1093, 1095, 1097, 1099, 1101, 1103, 1105, 1107, 1109, 1111, 1113, 1115, 1117, 1119, 1121, 1123, 1125, 1127, 1129, 1131, 1133, 1135, 1137, 1139, 1141, 1143, 1145, 1147, 1149, 1151, 1153, 1155, 1157, 1159, 1161, 1163, 1165, 1167, 1169, 1171, 1173, 1175, 1177, 1179, 1181, 1183, 1185, 1187, 1189, 1191, 1193, 1195, 1197, 1199, 1201, 1203, 1205, 1207, 1209, 1211, 1213, 1215, 1217, 1219, 1221, 1223, 1225, 1227, 1229, 1231, 1233, 1235, 1237, 1239, 1241, 1243, 1245, 1247, 1249, 1251, 1253, 1255, 1257, 1259, 1261, 1263, 1265, 1267, 1269, 1271, 1273, 1275, 1277, 1279, 1281, 1283, 1285, 1287, 1289, 1291, 1293, 1295, 1297, 1299, 1301, 1303, 1305, 1307, 1309, 1311, 1313, 1315, 1317, 1319, 1321, 1323, 1325, 1327, 1329, 1331, 1333, 1335, 1337, 1339, 1341, 1343, 1345, 1347, 1349, 1351, 1353, 1355, 1357, 1359, 1361, 1363, 1365, 1367, 1369, 1371, 1373, 1375, 1377, 1379, 1381, 1383, 1385, 1387, 1389, 1391, 1393, 1395, 1397, 1399, 1401, 1403, 1405, 1407, 1409, 1411, 1413, 1415, 1417, 1419, 1421, 1423, 1425, 1427, 1429, 1431, 1433, 1435, 1437, 1439, 1441, 1443, 1445, 1447, 1449, 1451, 1453, 1455, 1457, 1459, 1461, 1463, 1465, 1467, 1469, 1471, 1473, 1475, 1477, 1479, 1481, 1483, 1485, 1487, 1489, 1491, 1493, 1495, 1497, 1499, 1501, 1503, 1505, 1507, 1509, 1511, 1513, 1515, 1517, 1519, 1521, 1523, 1525, 1527, 1529, 1531, 1533, 1535, 1537, 1539, 1541, 1543, 1545, 1547, 1549, 1551, 1553, 1555, 1557, 1559, 1561, 1563, 1565, 1567, 1569, 1571, 1573, 1575, 1577, 1579, 1581, 1583, 1585, 1587, 1589, 1591, 1593, 1595, 1597, 1599, 1601, 1603, 1605, 1607, 1609, 1611, 1613, 1615, 1617, 1619, 1621, 1623, 1625, 1627, 1629, 1631, 1633, 1635, 1637, 1639, 1641, 1643, 1645, 1647, 1649, 1651, 1653, 1655, 1657, 1659, 1661, 1663, 1665, 1667, 1669, 1671, 1673, 1675, 1677, 1679, 1681, 1683, 1685, 1687, 1689, 1691, 1693, 1695, 1697, 1699, 1701, 1703, 1705, 1707, 1709, 1711, 1713, 1715, 1717, 1719, 1721, 1723, 1725, 1727, 1729, 1731, 1733, 1735, 1737, 1739, 1741, 1743, 1745, 1747, 1749, 1751, 1753, 1755, 1757, 1759, 1761, 1763, 1765, 1767, 1769, 1771, 1773, 1775, 1777, 1779, 1781, 1783, 1785, 1787, 1789, 1791, 1793, 1795, 1797, 1799, 1801, 1803, 1805, 1807, 1809, 1811, 1813, 1815, 1817, 1819, 1821, 1823, 1825, 1827, 1829, 1831, 1833, 1835, 1837, 1839, 1841, 1843, 1845, 1847, 1849, 1851, 1853, 1855, 1857, 1859, 1861, 1863, 1865, 1867, 1869, 1871, 1873, 1875, 1877, 1879, 1881, 1883, 1885, 1887, 1889, 1891, 1893, 1895, 1897, 1899, 1901, 1903, 1905, 1907, 1909, 1911, 1913, 1915, 1917, 1919, 1921, 1923, 1925, 1927, 1929, 1931, 1933, 1935, 1937, 1939, 1941, 1943, 1945, 1947, 1949, 1951, 1953, 1955, 1957, 1959, 1961, 1963, 1965, 1967, 1969, 1971, 1973, 1975, 1977, 1979, 1981, 1983, 1985, 1987, 1989, 1991, 1993, 1995, 1997, 1999, 2001, 2003, 2005, 2007, 2009, 2011, 2013, 2015, 2017, 2019, 2021, 2023, 2025, 2027, 2029, 2031, 2033, 2035, 2037, 2039, 2041, 2043, 2045, 2047, 2049, 2051, 2053, 2055, 2057, 2059, 2061, 2063, 2065, 2067, 2069, 2071, 2073, 2075, 2077, 2079, 2081, 2083, 2085, 2087, 2089, 2091, 2093, 2095, 2097, 2099, 2101, 2103, 2105, 2107, 2109, 2111, 2113, 2115, 2117, 2119, 2121, 2123, 2125, 2127, 2129, 2131, 2133, 2135, 2137, 2139, 2141, 2143, 2145, 2147, 2149, 2151, 2153, 2155, 2157, 2159, 2161, 2163, 2165, 2167, 2169, 2171, 2173, 2175, 2177, 2179, 2181, 2183, 2185, 2187, 2189, 2191, 2193, 2195, 2197, 2199, 2201, 2203, 2205, 2207, 2209, 2211, 2213, 2215, 2217, 2219, 2221, 2223, 2225, 2227, 2229, 2231, 2233, 2235, 2237, 2239, 2241, 2243, 2245, 2247, 2249, 2251, 2253, 2255, 2257, 2259, 2261, 2263, 2265, 2267, 2269, 2271, 2273, 2275, 2277, 2279, 2281, 2283, 2285, 2287, 2289, 2291, 2293, 2295, 2297, 2299, 2301, 2303, 2305, 2307, 2309, 2311, 2313, 2315, 2317, 2319, 2321, 2323, 2325, 2327, 2329, 2331, 2333, 2335, 2337, 2339, 2341, 2343, 2345, 2347, 2349, 2351, 2353, 2355, 2357, 2359, 2361, 2363, 2365, 2367, 2369, 2371, 2373, 2375, 2377, 2379, 2381, 2383, 2385, 2387, 2389, 2391, 2393, 2395, 2397, 2399, 2401, 2403, 2405, 2407, 2409, 2411, 2413, 2415, 2417, 2419, 2421, 2423, 2425, 2427, 2429, 2431, 2433, 2435, 2437, 2439, 2441, 2443, 2445, 2447, 2449, 2451, 2453, 2455, 2457, 2459, 2461, 2463, 2465, 2467, 2469, 2471, 2473, 2475, 2477, 2479, 2481, 2483, 2485, 2487, 2489, 2491, 2493, 2495, 2497, 2499]\n"
],
[
"#Make a Lambda function for capitalizing the whole sentence passed using arguments.\n#And map all the sentences in the List, with the lambda functions\n\narr = []\nst = [\"hey this is sai\",\"I am in mumbai\",\"....\"]\nfor x in st:\n arr.append(x.upper())\n \nprint(arr)",
"['HEY THIS IS SAI', 'I AM IN MUMBAI', '....']\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d05c3a800dd36df7ddfd7378b8f1e85cd44c2e5a | 371,505 | ipynb | Jupyter Notebook | 1_1_Image_Representation/6_3. Average Brightness.ipynb | georgiagn/CVND_Exercises | 4de186c80d14ed7d1e61c6bc51098ad0d9b4c54b | [
"MIT"
] | 1 | 2020-11-16T20:18:21.000Z | 2020-11-16T20:18:21.000Z | 1_1_Image_Representation/6_3. Average Brightness.ipynb | georgiagn/CVND_Exercises | 4de186c80d14ed7d1e61c6bc51098ad0d9b4c54b | [
"MIT"
] | null | null | null | 1_1_Image_Representation/6_3. Average Brightness.ipynb | georgiagn/CVND_Exercises | 4de186c80d14ed7d1e61c6bc51098ad0d9b4c54b | [
"MIT"
] | null | null | null | 1,040.630252 | 140,208 | 0.95624 | [
[
[
"# Day and Night Image Classifier\n---\n\nThe day/night image dataset consists of 200 RGB color images in two categories: day and night. There are equal numbers of each example: 100 day images and 100 night images.\n\nWe'd like to build a classifier that can accurately label these images as day or night, and that relies on finding distinguishing features between the two types of images!\n\n*Note: All images come from the [AMOS dataset](http://cs.uky.edu/~jacobs/datasets/amos/) (Archive of Many Outdoor Scenes).*\n",
"_____no_output_____"
],
[
"### Import resources\n\nBefore you get started on the project code, import the libraries and resources that you'll need.",
"_____no_output_____"
]
],
[
[
"import cv2 # computer vision library\nimport helpers\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Training and Testing Data\nThe 200 day/night images are separated into training and testing datasets. \n\n* 60% of these images are training images, for you to use as you create a classifier.\n* 40% are test images, which will be used to test the accuracy of your classifier.\n\nFirst, we set some variables to keep track of some where our images are stored:\n\n image_dir_training: the directory where our training image data is stored\n image_dir_test: the directory where our test image data is stored",
"_____no_output_____"
]
],
[
[
"# Image data directories\nimage_dir_training = \"day_night_images/training/\"\nimage_dir_test = \"day_night_images/test/\"",
"_____no_output_____"
]
],
[
[
"## Load the datasets\n\nThese first few lines of code will load the training day/night images and store all of them in a variable, `IMAGE_LIST`. This list contains the images and their associated label (\"day\" or \"night\"). \n\nFor example, the first image-label pair in `IMAGE_LIST` can be accessed by index: \n``` IMAGE_LIST[0][:]```.\n",
"_____no_output_____"
]
],
[
[
"# Using the load_dataset function in helpers.py\n# Load training data\nIMAGE_LIST = helpers.load_dataset(image_dir_training)\n",
"_____no_output_____"
]
],
[
[
"## Construct a `STANDARDIZED_LIST` of input images and output labels.\n\nThis function takes in a list of image-label pairs and outputs a **standardized** list of resized images and numerical labels.",
"_____no_output_____"
]
],
[
[
"# Standardize all training images\nSTANDARDIZED_LIST = helpers.standardize(IMAGE_LIST)",
"_____no_output_____"
]
],
[
[
"## Visualize the standardized data\n\nDisplay a standardized image from STANDARDIZED_LIST.",
"_____no_output_____"
]
],
[
[
"# Display a standardized image and its label\n\n# Select an image by index\nimage_num = 0\nselected_image = STANDARDIZED_LIST[image_num][0]\nselected_label = STANDARDIZED_LIST[image_num][1]\n\n# Display image and data about it\nplt.imshow(selected_image)\nprint(\"Shape: \"+str(selected_image.shape))\nprint(\"Label [1 = day, 0 = night]: \" + str(selected_label))\n",
"Shape: (600, 1100, 3)\nLabel [1 = day, 0 = night]: 1\n"
]
],
[
[
"# Feature Extraction\n\nCreate a feature that represents the brightness in an image. We'll be extracting the **average brightness** using HSV colorspace. Specifically, we'll use the V channel (a measure of brightness), add up the pixel values in the V channel, then divide that sum by the area of the image to get the average Value of the image.\n",
"_____no_output_____"
],
[
"## RGB to HSV conversion\n\nBelow, a test image is converted from RGB to HSV colorspace and each component is displayed in an image.",
"_____no_output_____"
]
],
[
[
"# Convert and image to HSV colorspace\n# Visualize the individual color channels\n\nimage_num = 0\ntest_im = STANDARDIZED_LIST[image_num][0]\ntest_label = STANDARDIZED_LIST[image_num][1]\n\n# Convert to HSV\nhsv = cv2.cvtColor(test_im, cv2.COLOR_RGB2HSV)\n\n# Print image label\nprint('Label: ' + str(test_label))\n\n# HSV channels\nh = hsv[:,:,0]\ns = hsv[:,:,1]\nv = hsv[:,:,2]\n\n# Plot the original image and the three channels\nf, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(20,10))\nax1.set_title('Standardized image')\nax1.imshow(test_im)\nax2.set_title('H channel')\nax2.imshow(h, cmap='gray')\nax3.set_title('S channel')\nax3.imshow(s, cmap='gray')\nax4.set_title('V channel')\nax4.imshow(v, cmap='gray')\n",
"Label: 1\n"
]
],
[
[
"---\n### Find the average brightness using the V channel\n\nThis function takes in a **standardized** RGB image and returns a feature (a single value) that represent the average level of brightness in the image. We'll use this value to classify the image as day or night.",
"_____no_output_____"
]
],
[
[
"# Find the average Value or brightness of an image\ndef avg_brightness(rgb_image):\n \n # Convert image to HSV\n hsv = cv2.cvtColor(rgb_image, cv2.COLOR_RGB2HSV)\n\n # Add up all the pixel values in the V channel\n sum_brightness = np.sum(hsv[:,:,2])\n \n ## TODO: Calculate the average brightness using the area of the image\n # and the sum calculated above\n avg = 0\n avg = sum_brightness/rgb_image.shape[0]/rgb_image.shape[1]\n \n return avg",
"_____no_output_____"
],
[
"# Testing average brightness levels\n# Look at a number of different day and night images and think about \n# what average brightness value separates the two types of images\n\n# As an example, a \"night\" image is loaded in and its avg brightness is displayed\nimage_num = 190\ntest_im = STANDARDIZED_LIST[image_num][0]\n\navg = avg_brightness(test_im)\nprint('Avg brightness: ' + str(avg))\nplt.imshow(test_im)",
"Avg brightness: 35.217\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05c3b154d279ab6af776e4e8b6de5c45205477d | 4,121 | ipynb | Jupyter Notebook | Presentation.ipynb | schabertrobbinger/jupyter-notebook-slides | 6d6aecb8c6096a178e8c444e38762fed1620cca3 | [
"MIT"
] | null | null | null | Presentation.ipynb | schabertrobbinger/jupyter-notebook-slides | 6d6aecb8c6096a178e8c444e38762fed1620cca3 | [
"MIT"
] | null | null | null | Presentation.ipynb | schabertrobbinger/jupyter-notebook-slides | 6d6aecb8c6096a178e8c444e38762fed1620cca3 | [
"MIT"
] | null | null | null | 23.548571 | 253 | 0.576316 | [
[
[
"**Fact: Amazon.com is rife with deceptive product marketing.**",
"_____no_output_____"
],
[
"<img src=\"reviews.png\">\n\nIf you squint hard enough, you can see that Warren Buffett is **not** actually the author of this book...",
"_____no_output_____"
],
[
"It is also easy to guess why this book has so many five star reviews:\n\n<img src=\"suspiciousreview.png\">",
"_____no_output_____"
],
[
"**Question: Can we improve on Amazon.com's ratings and review ranking algorithm?**",
"_____no_output_____"
],
[
"It is not clear, for example, that Amazon's ratings are meaningful at all.\nBy analyzing a little over two million video game reviews from Amazon.com, I concluded the positivity bias seen in the case above is far from a rare occurence:\n\n<img src=\"ratingshistogram1.png\">",
"_____no_output_____"
],
[
"Perhaps unsurprisingly given the above, the number of review upvotes actually appears to be anticorrelated with rating:\n\n<img src=\"ratingsupvotehistogram.png\">",
"_____no_output_____"
],
[
"**1) What should correlate well with the number of review upvotes?**",
"_____no_output_____"
],
[
"Since the number of upvotes is a measure of how helpful a given review was to consumers, I guessed that the length the review text should correlate with helpfulness because more information (\"Substantive content\") is provided by the reviewer:\n\n<img src=\"upvoteplot1.png\">",
"_____no_output_____"
],
[
"**2) Does it make sense for Amazon.com to preferentially rank newer reviews and reviews by its annointed Vine contributors?**",
"_____no_output_____"
],
[
"Is there concern that older reviews will necessarily garner a greater number of upvotes than newer reviews, solely because they have been around for longer? If so, this does not seem to be strongly reflected in the data:\n\n<img src=\"timestamphistogram.png\">",
"_____no_output_____"
],
[
"<img src=\"topreviewer.png\">\n<img src=\"bestreview.png\">",
"_____no_output_____"
],
[
"**We seem well on our way to a complementary alternative to Amazon.com's current system, even before doing any machine learning!**\n\n**Next step: Identify key words and phrases which indicate products should be avoided.**",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d05c5d392de37956214b187c3cc03d17197f317c | 5,709 | ipynb | Jupyter Notebook | JupyterNotebooks/Labs/Lab 3.ipynb | CometSmudge/CMPT-220L-903-21S | 635b97f54bb1f6e2acf9262e1daa891fd8d8c69d | [
"MIT"
] | null | null | null | JupyterNotebooks/Labs/Lab 3.ipynb | CometSmudge/CMPT-220L-903-21S | 635b97f54bb1f6e2acf9262e1daa891fd8d8c69d | [
"MIT"
] | null | null | null | JupyterNotebooks/Labs/Lab 3.ipynb | CometSmudge/CMPT-220L-903-21S | 635b97f54bb1f6e2acf9262e1daa891fd8d8c69d | [
"MIT"
] | null | null | null | 24.502146 | 152 | 0.496409 | [
[
[
"# Lab Three\n---\n\nFor this lab we're going to be making and using a bunch of functions. \n\nOur Goals are:\n- Switch Case\n- Looping\n- Making our own functions\n- Combining functions\n- Structuring solutions",
"_____no_output_____"
]
],
[
[
"// Give me an example of you using switch case.\nString house = \"BlueLions\";\n\nswitch(house){\n case \"BlueLions\":\n System.out.println(\"Dimitri\");\n case \"BlackEagles\":\n System.put.println(\"Edelgard\");\n case \"GoldenDeer\":\n System.out.println(\"Claude\");\n}",
"_____no_output_____"
],
[
"// Give me an example of you using a for loop\nfor int x = 10; x > -1; x--) {\n System.out.println(x);\n}",
"_____no_output_____"
],
[
"// Give me an example of you using a for each loop\nint[] numbers = {1, 2, 3, 4, 5};\n\nfor (int number: numbers) {\n System.out.println(number);\n}",
"_____no_output_____"
],
[
"// Give me an example of you using a while loop\nint x = 0;\nint stop = 11;\nwhile (x < stop) {\n System.out.println(\"not 10\");\n x++;\n}\nSystem.out.println(\"is 10\");",
"not 10\nnot 10\nnot 10\nnot 10\nnot 10\nnot 10\nnot 10\nnot 10\nnot 10\nnot 10\nnot 10\nis 10\n"
],
[
"// I want you to write a function that will take in a number and raise it to the power given. \n\n// For example if given the numbers 2 and 3. The math that the function should do is 2^3 and should print out or return 8. Print the output.\n\nint base = 2;\nint power = 3;\n\nint newbase = base;\nfor (int x = 1; x < power; x++){\n newbase = newbase * base;\n \n}\nSystem.out.println(newbase);",
"8\n"
],
[
"// I want you to write a function that will take in a list and see how many times a given number is in the list. \n\n// For example if the array given is [2,3,5,2,3,6,7,8,2] and the number given is 2 the function should print out or return 3. Print the output.\nint[] numbers = {2,3,5,2,3,6,7,8,2};\nint givenNumber = 2;\nint counter = 0;\nfor (int number: numbers) {\n if (number == givenNumber){\n counter++;\n }\n}\nSystem.out.println(counter);",
"3\n"
],
[
"// Give me a function that gives the answer to the pythagorean theorem. \n// I'd like you to reuse the exponent function from above as well as the functions below to make your function.\n\n// If you don't remember the pythagorean theorem the formula is (a^2 + b^2 = c^2). Given a and b as parameters i'd like you to return c. \n// If this doesn't make sense look up `Pythagorean Theorem Formula` on google.\n\nint addition(int a, int b) {\n int answer = a + b;\n return answer;\n}\n\nint division(int a, int b) {\n int answer = a / b;\n return answer;\n}\nint a = 3;\nint b = 4;\nint a = a * a;\nint b = b * b;\nint addition(int a, int b) {\n int answer = a + b;\nreturn answer;\n\n\n\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05c689efc697b2e1c0f548039489399616e2c40 | 24,650 | ipynb | Jupyter Notebook | practice_project.ipynb | Abhishekauti21/dsmp-pre-work | 718b1c4149306c84d22763df9dcc5959015267f8 | [
"MIT"
] | 2 | 2020-03-29T18:42:36.000Z | 2020-05-17T10:45:08.000Z | practice_project.ipynb | Abhishekauti21/dsmp-pre-work | 718b1c4149306c84d22763df9dcc5959015267f8 | [
"MIT"
] | null | null | null | practice_project.ipynb | Abhishekauti21/dsmp-pre-work | 718b1c4149306c84d22763df9dcc5959015267f8 | [
"MIT"
] | null | null | null | 36.736215 | 1,141 | 0.484503 | [
[
[
"<a href=\"https://colab.research.google.com/github/Abhishekauti21/dsmp-pre-work/blob/master/practice_project.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"class test:\n def __init__(self,a):\n self.a=a\n\n def display(self):\n print(self.a)\nobj=test()\nobj.display()",
"_____no_output_____"
],
[
"def f1():\n x=100\n print(x)\nx=+1\nf1()",
"100\n"
],
[
"area = { 'living' : [400, 450], 'living' : [650, 800], 'kitchen' : [300, 250], 'garage' : [250, 0]}\nprint (area['living'])",
"[650, 800]\n"
],
[
"List_1=[2,6,7,8]\nList_2=[2,6,7,8]\nprint(List_1[-2] + List_2[2])",
"14\n"
],
[
"d = {0: 'a', 1: 'b', 2: 'c'}\nfor x, y in d.items():\n print(x, y)",
"0 a\n1 b\n2 c\n"
],
[
"Numbers=[10,5,7,8,9,5]\nprint(max(Numbers)-min(Numbers))",
"5\n"
],
[
"fo = open(\"foo.txt\", \"read+\")\nprint(\"Name of the file: \", fo.name)\n\n# Assuming file has following 5 lines\n# This is 1st line\n# This is 2nd line\n# This is 3rd line\n# This is 4th line\n# This is 5th line\n\nfor index in range(5):\n line = fo.readline()\n print(\"Line No {} - {}\".format(index, line))\n #Close opened file\nfo.close()",
"_____no_output_____"
],
[
"x = \"abcdef\"\nwhile i in x:\n print(i, end=\" \")\n",
"_____no_output_____"
],
[
"def cube(x):\n return x * x * x \nx = cube(3) \nprint (x)\n",
"27\n"
],
[
" print(((True) or (False) and (False) or (False)))\n",
"True\n"
],
[
"x1=int('16')\nx2=8 + 8\nx3= (4**2)\n\nprint(x1 is x2 is x3)\n",
"True\n"
],
[
"Word='warrior knights' ,A=Word[9:14],B=Word[-13:-16:-1]\nB+A",
"_____no_output_____"
],
[
"def to_upper(k):\n return k.upper()\nx = ['ab', 'cd']\nprint(list(map(to_upper, x)))",
"['AB', 'CD']\n"
],
[
"my_string = \"hello world\"\nk = [(i.upper(), len(i)) for i in my_string]\nprint(k)",
"[('H', 1), ('E', 1), ('L', 1), ('L', 1), ('O', 1), (' ', 1), ('W', 1), ('O', 1), ('R', 1), ('L', 1), ('D', 1)]\n"
],
[
"from csv import reader\n\ndef explore_data(dataset, start, end, rows_and_columns=False):\n \"\"\"Explore the elements of a list.\n \n Print the elements of a list starting from the index 'start'(included) upto the index 'end' (excluded).\n \n Keyword arguments:\n dataset -- list of which we want to see the elements\n start -- index of the first element we want to see, this is included\n end -- index of the stopping element, this is excluded \n rows_and_columns -- this parameter is optional while calling the function. It takes binary values, either True or False. If true, print the dimension of the list, else dont.\n \"\"\"\n \n \n dataset_slice = dataset[start:end] \n for row in dataset_slice:\n print(row)\n print('\\n') # adds a new (empty) line between rows\n \n if rows_and_columns:\n print('Number of rows:', len(dataset))\n print('Number of columns:', len(dataset[0]))\n \n \n\n\ndef duplicate_and_unique_movies(dataset, index_):\n \"\"\"Check the duplicate and unique entries.\n \n We have nested list. This function checks if the rows in the list is unique or duplicated based on the element at index 'index_'.\n It prints the Number of duplicate entries, along with some examples of duplicated entry.\n \n Keyword arguments:\n dataset -- two dimensional list which we want to explore\n index_ -- column index at which the element in each row would be checked for duplicacy \n \n \"\"\"\n \n duplicate = []\n unique = []\n\n for movie in dataset:\n name = movie[index_]\n if name in unique:\n duplicate.append(name)\n else:\n unique.append(name)\n\n print('Number of duplicate Movies:', len(duplicate))\n print('\\n')\n print('Examples of duplicate Movies:', duplicate[:15])\n \n\n\n\ndef movies_lang(dataset, index_, lang_):\n \"\"\"Extract the movies of a particular language.\n \n Of all the movies available in all languages, this function extracts all the movies in a particular laguage.\n Once you ahve extracted the movies, call the explore_data() to print first few rows.\n \n Keyword arguments:\n dataset -- list containing the details of the movie\n index_ -- index which is to be compared for langauges\n lang_ -- desired language for which we want to filter out the movies\n \n Returns:\n movies_ -- list with details of the movies in selected language\n \n \"\"\"\n movies_ = []\n\n for movie in movies:\n lang = movie[index_]\n if lang == lang_:\n movies_.append(movie)\n\n print(\"Examples of Movies in English Language:\") \n explore_data(movies_, 0, 3, True)\n return movies_\n \n\n\ndef rate_bucket(dataset, rate_low, rate_high):\n \"\"\"Extract the movies within the specified ratings.\n \n This function extracts all the movies that has rating between rate_low and high_rate.\n Once you ahve extracted the movies, call the explore_data() to print first few rows.\n \n Keyword arguments:\n dataset -- list containing the details of the movie\n rate_low -- lower range of rating\n rate_high -- higher range of rating\n \n Returns:\n rated_movies -- list of the details of the movies with required ratings\n \"\"\"\n\n rated_movies = []\n\n for movie in dataset:\n vote_avg = float(movie[-4])\n if ((vote_avg >= rate_low) & (vote_avg <= rate_high)):\n rated_movies.append(movie)\n\n print(\"Examples of Movies in required rating bucket:\") \n explore_data(rated_movies, 0, 3, True)\n return rated_movies\n\n\n# Read the data file and store it as a list 'movies'\nopened_file = open(path, encoding=\"utf8\")\nread_file = reader(opened_file)\nmovies = list(read_file)\n\n# The first row is header. Extract and store it in 'movies_header'.\nmovies_header = movies[0]\nprint(\"Movies Header:\\n\", movies_header)\n\n# Subset the movies dataset such that the header is removed from the list and store it back in movies\nmovies = movies[1:]\n\n\n\n# Delete wrong data\n# Explore the row #4553. You will see that as apart from the id, description, status and title, no other information is available.\n# Hence drop this row.\n\nprint(\"Entry at index 4553:\")\nexplore_data(movies, 4553, 4554)\n\ndel movies[4553]\n\n\n\n\n# Using explore_data() with appropriate parameters, view the details of the first 5 movies.\nprint(\"First 5 Entries:\")\nexplore_data(movies, 0, 5, True)\n\n\n\n# Our dataset might have more than one entry for a movie. Call duplicate_and_unique_movies() with index of the name to check the same.\n\nduplicate_and_unique_movies(movies, 13)\n\n\n# We saw that there are 3 movies for which the there are multiple entries. \n# Create a dictionary, 'reviews_max' that will have the name of the movie as key, and the maximum number of reviews as values.\n\nreviews_max = {}\n\nfor movie in movies:\n name = movie[13]\n n_reviews = float(movie[12])\n \n if name in reviews_max and reviews_max[name] < n_reviews:\n reviews_max[name] = n_reviews\n \n elif name not in reviews_max:\n reviews_max[name] = n_reviews\n \nlen(reviews_max)\n\n# Create a list 'movies_clean', which will filter out the duplicate movies and contain the rows with maximum number of reviews for duplicate movies, as stored in 'review_max'. \n\nmovies_clean = []\nalready_added = []\n\nfor movie in movies:\n name = movie[13]\n n_reviews = float(movie[12])\n \n if (reviews_max[name] == n_reviews) and (name not in already_added):\n movies_clean.append(movie)\n already_added.append(name)\n \nlen(movies_clean)\n\n\n\n# Calling movies_lang(), extract all the english movies and store it in movies_en.\n\nmovies_en = movies_lang(movies_clean, 3, 'en')\n\n\n\n# Call the rate_bucket function to see the movies with rating higher than 8.\n\nhigh_rated_movies = rate_bucket(movies_en, 8, 10)\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05c68c3c3e20091eca3d342e88f07c3fc67366f | 490,081 | ipynb | Jupyter Notebook | Model/Resnet_18.ipynb | reyvnth/COVIDX | de3ece96f5b39d61b3c4ef381529807c257d5f49 | [
"Apache-2.0"
] | 2 | 2020-12-09T16:20:32.000Z | 2021-09-30T15:40:16.000Z | Model/Resnet_18.ipynb | reyvnth/COVIDX | de3ece96f5b39d61b3c4ef381529807c257d5f49 | [
"Apache-2.0"
] | null | null | null | Model/Resnet_18.ipynb | reyvnth/COVIDX | de3ece96f5b39d61b3c4ef381529807c257d5f49 | [
"Apache-2.0"
] | 2 | 2020-12-09T16:19:44.000Z | 2021-02-01T05:20:30.000Z | 490,081 | 490,081 | 0.942338 | [
[
[
"# Detecting COVID-19 with Chest X Ray using PyTorch\n\nImage classification of Chest X Rays in one of three classes: Normal, Viral Pneumonia, COVID-19\n\nDataset from [COVID-19 Radiography Dataset](https://www.kaggle.com/tawsifurrahman/covid19-radiography-database) on Kaggle",
"_____no_output_____"
],
[
"# Importing Libraries",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/gdrive')",
"Drive already mounted at /gdrive; to attempt to forcibly remount, call drive.mount(\"/gdrive\", force_remount=True).\n"
],
[
"%matplotlib inline\n\nimport os\nimport shutil\nimport copy\nimport random\nimport torch\nimport torch.nn as nn\nimport torchvision\nimport torch.optim as optim\nfrom torch.optim import lr_scheduler\nimport numpy as np\nimport seaborn as sns\nimport time\nfrom sklearn.metrics import confusion_matrix\n\nfrom PIL import Image\nimport matplotlib.pyplot as plt\n\ntorch.manual_seed(0)\n\nprint('Using PyTorch version', torch.__version__)",
"Using PyTorch version 1.7.0+cu101\n"
]
],
[
[
"# Preparing Training and Test Sets",
"_____no_output_____"
]
],
[
[
"class_names = ['Non-Covid', 'Covid']\nroot_dir = '/gdrive/My Drive/Research_Documents_completed/Data/Data/'\nsource_dirs = ['non', 'covid']",
"_____no_output_____"
]
],
[
[
"# Creating Custom Dataset",
"_____no_output_____"
]
],
[
[
"class ChestXRayDataset(torch.utils.data.Dataset):\n def __init__(self, image_dirs, transform):\n def get_images(class_name):\n images = [x for x in os.listdir(image_dirs[class_name]) if x.lower().endswith('png') or x.lower().endswith('jpg')]\n print(f'Found {len(images)} {class_name} examples')\n return images\n \n self.images = {}\n self.class_names = ['Non-Covid', 'Covid']\n \n for class_name in self.class_names:\n self.images[class_name] = get_images(class_name)\n \n self.image_dirs = image_dirs\n self.transform = transform\n \n \n def __len__(self):\n return sum([len(self.images[class_name]) for class_name in self.class_names])\n \n \n def __getitem__(self, index):\n class_name = random.choice(self.class_names)\n index = index % len(self.images[class_name])\n image_name = self.images[class_name][index]\n image_path = os.path.join(self.image_dirs[class_name], image_name)\n image = Image.open(image_path).convert('RGB')\n return self.transform(image), self.class_names.index(class_name)",
"_____no_output_____"
]
],
[
[
"# Image Transformations",
"_____no_output_____"
]
],
[
[
"train_transform = torchvision.transforms.Compose([\n torchvision.transforms.Resize(size=(224, 224)),\n torchvision.transforms.RandomHorizontalFlip(),\n torchvision.transforms.ToTensor(),\n torchvision.transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])\n])\n\ntest_transform = torchvision.transforms.Compose([\n torchvision.transforms.Resize(size=(224, 224)),\n torchvision.transforms.ToTensor(),\n torchvision.transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])\n])",
"_____no_output_____"
]
],
[
[
"# Prepare DataLoader",
"_____no_output_____"
]
],
[
[
"train_dirs = {\n 'Non-Covid': '/gdrive/My Drive/Research_Documents_completed/Data/Data/non/',\n 'Covid': '/gdrive/My Drive/Research_Documents_completed/Data/Data/covid/'\n}\n#train_dirs = {\n# 'Non-Covid': '/gdrive/My Drive/Data/Data/non/',\n# 'Covid': '/gdrive/My Drive/Data/Data/covid/'\n#}\n\ntrain_dataset = ChestXRayDataset(train_dirs, train_transform)",
"Found 3265 Non-Covid examples\nFound 203 Covid examples\n"
],
[
"test_dirs = {\n 'Non-Covid': '/gdrive/My Drive/Research_Documents_completed/Data/Data/test/non/',\n 'Covid': '/gdrive/My Drive/Research_Documents_completed/Data/Data/test/covid/'\n}\n\ntest_dataset = ChestXRayDataset(test_dirs, test_transform)",
"Found 3000 Non-Covid examples\nFound 200 Covid examples\n"
],
[
"batch_size = 25\n\ndl_train = torch.utils.data.DataLoader(train_dataset, batch_size=batch_size, shuffle=True)\ndl_test = torch.utils.data.DataLoader(test_dataset, batch_size=batch_size, shuffle=True)\nprint(dl_train)\nprint('Number of training batches', len(dl_train))\nprint('Number of test batches', len(dl_test))",
"<torch.utils.data.dataloader.DataLoader object at 0x7f3c11961048>\nNumber of training batches 139\nNumber of test batches 128\n"
]
],
[
[
"# Data Visualization",
"_____no_output_____"
]
],
[
[
"\nclass_names = train_dataset.class_names\n\n\ndef show_images(images, labels, preds):\n plt.figure(figsize=(30, 20))\n for i, image in enumerate(images):\n plt.subplot(1, 25, i + 1, xticks=[], yticks=[])\n image = image.numpy().transpose((1, 2, 0))\n mean = np.array([0.485, 0.456, 0.406])\n std = np.array([0.229, 0.224, 0.225])\n image = image * std + mean\n image = np.clip(image, 0., 1.)\n plt.imshow(image)\n col = 'green'\n if preds[i] != labels[i]:\n col = 'red'\n \n plt.xlabel(f'{class_names[int(labels[i].numpy())]}')\n plt.ylabel(f'{class_names[int(preds[i].numpy())]}', color=col)\n plt.tight_layout()\n plt.show()",
"_____no_output_____"
],
[
"images, labels = next(iter(dl_train))\nshow_images(images, labels, labels)",
"_____no_output_____"
],
[
"images, labels = next(iter(dl_test))\nshow_images(images, labels, labels)",
"_____no_output_____"
]
],
[
[
"# Creating the Model",
"_____no_output_____"
]
],
[
[
"resnet18 = torchvision.models.resnet18(pretrained=True)\n\nprint(resnet18)",
"ResNet(\n (conv1): Conv2d(3, 64, kernel_size=(7, 7), stride=(2, 2), padding=(3, 3), bias=False)\n (bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (maxpool): MaxPool2d(kernel_size=3, stride=2, padding=1, dilation=1, ceil_mode=False)\n (layer1): Sequential(\n (0): BasicBlock(\n (conv1): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (1): BasicBlock(\n (conv1): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (layer2): Sequential(\n (0): BasicBlock(\n (conv1): Conv2d(64, 128, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (downsample): Sequential(\n (0): Conv2d(64, 128, kernel_size=(1, 1), stride=(2, 2), bias=False)\n (1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (1): BasicBlock(\n (conv1): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (layer3): Sequential(\n (0): BasicBlock(\n (conv1): Conv2d(128, 256, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (downsample): Sequential(\n (0): Conv2d(128, 256, kernel_size=(1, 1), stride=(2, 2), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (1): BasicBlock(\n (conv1): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (layer4): Sequential(\n (0): BasicBlock(\n (conv1): Conv2d(256, 512, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (downsample): Sequential(\n (0): Conv2d(256, 512, kernel_size=(1, 1), stride=(2, 2), bias=False)\n (1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (1): BasicBlock(\n (conv1): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (avgpool): AdaptiveAvgPool2d(output_size=(1, 1))\n (fc): Linear(in_features=512, out_features=1000, bias=True)\n)\n"
],
[
"resnet18.fc = torch.nn.Linear(in_features=512, out_features=2)\nloss_fn = torch.nn.CrossEntropyLoss()\noptimizer = torch.optim.Adam(resnet18.parameters(), lr=3e-5)",
"_____no_output_____"
],
[
"print(resnet18)",
"ResNet(\n (conv1): Conv2d(3, 64, kernel_size=(7, 7), stride=(2, 2), padding=(3, 3), bias=False)\n (bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (maxpool): MaxPool2d(kernel_size=3, stride=2, padding=1, dilation=1, ceil_mode=False)\n (layer1): Sequential(\n (0): BasicBlock(\n (conv1): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (1): BasicBlock(\n (conv1): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (layer2): Sequential(\n (0): BasicBlock(\n (conv1): Conv2d(64, 128, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (downsample): Sequential(\n (0): Conv2d(64, 128, kernel_size=(1, 1), stride=(2, 2), bias=False)\n (1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (1): BasicBlock(\n (conv1): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (layer3): Sequential(\n (0): BasicBlock(\n (conv1): Conv2d(128, 256, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (downsample): Sequential(\n (0): Conv2d(128, 256, kernel_size=(1, 1), stride=(2, 2), bias=False)\n (1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (1): BasicBlock(\n (conv1): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (layer4): Sequential(\n (0): BasicBlock(\n (conv1): Conv2d(256, 512, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (downsample): Sequential(\n (0): Conv2d(256, 512, kernel_size=(1, 1), stride=(2, 2), bias=False)\n (1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (1): BasicBlock(\n (conv1): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace=True)\n (conv2): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn2): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n )\n (avgpool): AdaptiveAvgPool2d(output_size=(1, 1))\n (fc): Linear(in_features=512, out_features=2, bias=True)\n)\n"
],
[
"def show_preds():\n resnet18.eval()\n images, labels = next(iter(dl_test))\n outputs = resnet18(images)\n _, preds = torch.max(outputs, 1)\n show_images(images, labels, preds)",
"_____no_output_____"
],
[
"show_preds()",
"_____no_output_____"
]
],
[
[
"# Training the Model",
"_____no_output_____"
]
],
[
[
"def train(epochs):\n best_model_wts = copy.deepcopy(resnet18.state_dict())\n b_acc = 0.0\n t_loss = []\n t_acc = []\n avg_t_loss=[]\n avg_t_acc=[]\n v_loss = []\n v_acc=[]\n avg_v_loss = []\n avg_v_acc = []\n ep = []\n print('Starting training..')\n for e in range(0, epochs):\n ep.append(e+1)\n print('='*20)\n print(f'Starting epoch {e + 1}/{epochs}')\n print('='*20)\n\n train_loss = 0.\n val_loss = 0.\n train_accuracy = 0\n total_train = 0\n correct_train = 0\n\n resnet18.train() # set model to training phase\n\n for train_step, (images, labels) in enumerate(dl_train):\n optimizer.zero_grad()\n outputs = resnet18(images)\n _, pred = torch.max(outputs, 1)\n loss = loss_fn(outputs, labels)\n loss.backward()\n optimizer.step()\n train_loss += loss.item()\n train_loss /= (train_step + 1)\n _, predicted = torch.max(outputs, 1)\n total_train += labels.nelement()\n correct_train += sum((predicted == labels).numpy())\n train_accuracy = correct_train / total_train\n t_loss.append(train_loss)\n t_acc.append(train_accuracy)\n \n if train_step % 20 == 0:\n print('Evaluating at step', train_step)\n print(f'Training Loss: {train_loss:.4f}, Training Accuracy: {train_accuracy:.4f}')\n\n accuracy = 0. \n resnet18.eval() # set model to eval phase\n\n for val_step, (images, labels) in enumerate(dl_test):\n outputs = resnet18(images)\n loss = loss_fn(outputs, labels)\n val_loss += loss.item()\n\n _, preds = torch.max(outputs, 1)\n accuracy += sum((preds == labels).numpy())\n\n val_loss /= (val_step + 1)\n accuracy = accuracy/len(test_dataset)\n print(f'Validation Loss: {val_loss:.4f}, Validation Accuracy: {accuracy:.4f}')\n v_loss.append(val_loss)\n v_acc.append(accuracy)\n show_preds()\n\n resnet18.train()\n\n if accuracy > b_acc:\n b_acc = accuracy\n \n \n avg_t_loss.append(sum(t_loss)/len(t_loss))\n avg_v_loss.append(sum(v_loss)/len(v_loss))\n avg_t_acc.append(sum(t_acc)/len(t_acc))\n avg_v_acc.append(sum(v_acc)/len(v_acc))\n \n\n best_model_wts = copy.deepcopy(resnet18.state_dict())\n print('Best validation Accuracy: {:4f}'.format(b_acc)) \n print('Training complete..')\n \n plt.plot(ep, avg_t_loss, 'g', label='Training loss')\n plt.plot(ep, avg_v_loss, 'b', label='validation loss')\n plt.title('Training and Validation loss for each epoch')\n plt.xlabel('Epochs')\n plt.ylabel('Loss')\n plt.legend()\n plt.savefig('/gdrive/My Drive/Research_Documents_completed/Resnet18_completed/resnet18_loss.png')\n plt.show()\n\n plt.plot(ep, avg_t_acc, 'g', label='Training accuracy')\n plt.plot(ep, avg_v_acc, 'b', label='validation accuracy')\n plt.title('Training and Validation Accuracy for each epoch')\n plt.xlabel('Epochs')\n plt.ylabel('Accuracy')\n plt.legend()\n plt.savefig('/gdrive/My Drive/Research_Documents_completed/Resnet18_completed/resnet18_accuarcy.png')\n plt.show()\n\n torch.save(resnet18.state_dict(),'/gdrive/My Drive/Research_Documents_completed/Resnet18_completed/resnet18.pt') ",
"_____no_output_____"
],
[
"%%time\n\ntrain(epochs=5)",
"Starting training..\n====================\nStarting epoch 1/5\n====================\nEvaluating at step 0\nTraining Loss: 0.8522, Training Accuracy: 0.4800\n"
]
],
[
[
"# Final Results",
"_____no_output_____"
],
[
"VALIDATION LOSS AND TRAINING LOSS VS EPOCH\nVALIDATION ACCURACY AND TRAINING ACCURACY VS EPOCH\nBEST ACCURACY ERROR..\n",
"_____no_output_____"
]
],
[
[
"show_preds()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d05c7ff06fb2d6c08d620f353884cf397338097e | 29,167 | ipynb | Jupyter Notebook | docs/source/tutorials/1-getting-started/plotting-target-pixel-files.ipynb | alex-w/lightkurve | 6c687e1027ce59b08fabeba69f2358ab00ffefa9 | [
"MIT"
] | null | null | null | docs/source/tutorials/1-getting-started/plotting-target-pixel-files.ipynb | alex-w/lightkurve | 6c687e1027ce59b08fabeba69f2358ab00ffefa9 | [
"MIT"
] | null | null | null | docs/source/tutorials/1-getting-started/plotting-target-pixel-files.ipynb | alex-w/lightkurve | 6c687e1027ce59b08fabeba69f2358ab00ffefa9 | [
"MIT"
] | null | null | null | 30.256224 | 719 | 0.61405 | [
[
[
"# Plotting Target Pixel Files with Lightkurve",
"_____no_output_____"
],
[
"## Learning Goals\n\nBy the end of this tutorial, you will:\n\n- Learn how to download and plot target pixel files from the data archive using [Lightkurve](https://docs.lightkurve.org).\n- Be able to plot the target pixel file background.\n- Be able to extract and plot flux from a target pixel file.\n",
"_____no_output_____"
],
[
"## Introduction",
"_____no_output_____"
],
[
"The [*Kepler*](https://www.nasa.gov/mission_pages/kepler/main/index.html), [*K2*](https://www.nasa.gov/mission_pages/kepler/main/index.html), and [*TESS*](https://tess.mit.edu/) telescopes observe stars for long periods of time, from just under a month to four years. By doing so they observe how the brightnesses of stars change over time.\n\nPixels around targeted stars are cut out and stored as *target pixel files* at each observing cadence. In this tutorial, we will learn how to use Lightkurve to download and understand the different photometric data stored in a target pixel file, and how to extract flux using basic aperture photometry.\n\nIt is useful to read the accompanying tutorial discussing how to use target pixel file products with Lightkurve before starting this tutorial. It is recommended that you also read the tutorial on using *Kepler* light curve products with Lightkurve, which will introduce you to some specifics on how *Kepler*, *K2*, and *TESS* make observations, and how these are displayed as light curves. It also introduces some important terms and concepts that are referred to in this tutorial.\n\n*Kepler* observed a single field in the sky, although not all stars in this field were recorded. Instead, pixels were selected around certain targeted stars. These cutout images are called target pixel files, or TPFs. By combining the amount of flux in the pixels where the star appears, you can make a measurement of the amount of light from a star in that observation. The pixels chosen to include in this measurement are referred to as an *aperture*.\n\nTPFs are typically the first port of call when studying a star with *Kepler*, *K2*, or *TESS*. They allow us to see where our data is coming from, and identify potential sources of noise or systematic trends. In this tutorial, we will use the *Kepler* mission as the main example, but these tools equally apply to *TESS* and *K2* as well.",
"_____no_output_____"
],
[
"## Imports\n\nThis tutorial requires:\n- **[Lightkurve](https://docs.lightkurve.org)** to work with TPF files.\n- [**Matplotlib**](https://matplotlib.org/) for plotting.",
"_____no_output_____"
]
],
[
[
"import lightkurve as lk\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## 1. Downloading a TPF",
"_____no_output_____"
],
[
"A TPF contains the original imaging data from which a light curve is derived. Besides the brightness data measured by the charge-coupled device (CCD) camera, a TPF also includes post-processing information such as an estimate of the astronomical background, and a recommended pixel aperture for extracting a light curve. \n\nFirst, we download a target pixel file. We will use one quarter's worth of *Kepler* data for the star named [Kepler-8](http://www.openexoplanetcatalogue.com/planet/Kepler-8%20b/), a star somewhat larger than the Sun, and the host of a [hot Jupiter planet](https://en.wikipedia.org/wiki/Hot_Jupiter).",
"_____no_output_____"
]
],
[
[
"search_result = lk.search_targetpixelfile(\"Kepler-8\", author=\"Kepler\", quarter=4, cadence=\"long\")\nsearch_result",
"_____no_output_____"
],
[
"tpf = search_result.download()",
"_____no_output_____"
]
],
[
[
"This TPF contains data for every cadence in the quarter we downloaded. Let's focus on the first cadence for now, which we can select using zero-based indexing as follows:",
"_____no_output_____"
]
],
[
[
"first_cadence = tpf[0]\nfirst_cadence",
"_____no_output_____"
]
],
[
[
"## 2. Flux and Background",
"_____no_output_____"
],
[
"At each cadence the TPF has a number of photometry data properties. These are:\n\n- `flux_bkg`: the astronomical background of the image.\n- `flux_bkg_err`: the statistical uncertainty on the background flux.\n- `flux`: the stellar flux after the background is removed.\n- `flux_err`: the statistical uncertainty on the stellar flux after background removal.\n\nThese properties can be accessed via a TPF object as follows:",
"_____no_output_____"
]
],
[
[
"first_cadence.flux.value",
"_____no_output_____"
]
],
[
[
"And you can plot the data as follows:",
"_____no_output_____"
]
],
[
[
"first_cadence.plot(column='flux');",
"_____no_output_____"
]
],
[
[
"Alternatively, if you are working directly with a FITS file, you can access the data in extension 1 (for example, `first_cadence.hdu[1].data['FLUX']`). Note that you can find all of the details on the structure and contents of TPF files in Section 2.3.2 of the [*Kepler* Archive Manual](http://archive.stsci.edu/files/live/sites/mast/files/home/missions-and-data/kepler/_documents/archive_manual.pdf).",
"_____no_output_____"
],
[
"When plotting data using the `plot()` function, what you are seeing in the TPF is the flux *after* the background has been removed. This background flux typically consists of [zodiacal light](https://en.wikipedia.org/wiki/Zodiacal_light) or earthshine (especially in *TESS* observations). The background is typically smooth and changes on scales much larger than a single TPF. In *Kepler*, the background is estimated for the CCD as a whole, before being extracted from each TPF in that CCD. You can learn more about background removal in Section 4.2 of the [*Kepler* Data Processing Handbook](http://archive.stsci.edu/files/live/sites/mast/files/home/missions-and-data/kepler/_documents/KSCI-19081-002-KDPH.pdf).",
"_____no_output_____"
],
[
"Now, let's compare the background to the background-subtracted flux to get a sense of scale. We can do this using the `plot()` function's `column` keyword. By default the function plots the flux, but we can change this to plot the background, as well as other data such as the error on each pixel's flux.",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(2,2, figsize=(16,16))\nfirst_cadence.plot(ax=axes[0,0], column='FLUX')\nfirst_cadence.plot(ax=axes[0,1], column='FLUX_BKG')\nfirst_cadence.plot(ax=axes[1,0], column='FLUX_ERR')\nfirst_cadence.plot(ax=axes[1,1], column='FLUX_BKG_ERR');",
"_____no_output_____"
]
],
[
[
"From looking at the color scale on both plots, you may see that the background flux is very low compared to the total flux emitted by a star. This is expected — stars are bright! But these small background corrections become important when looking at the very small scale changes caused by planets or stellar oscillations. Understanding the background is an important part of astronomy with *Kepler*, *K2*, and *TESS*.",
"_____no_output_____"
],
[
"If the background is particularly bright and you want to see what the TPF looks like with it included, passing the `bkg=True` argument to the `plot()` method will show the TPF with the flux added on top of the background, representing the total flux recorded by the spacecraft.",
"_____no_output_____"
]
],
[
[
"first_cadence.plot(bkg=True);",
"_____no_output_____"
]
],
[
[
"In this case, the background is low and the star is bright, so it doesn't appear to make much of a difference.",
"_____no_output_____"
],
[
"## 3. Apertures",
"_____no_output_____"
],
[
"As part of the data processing done by the *Kepler* pipeline, each TPF includes a recommended *optimal aperture mask*. This aperture mask is optimized to ensure that the stellar signal has a high signal-to-noise ratio, with minimal contamination from the background.",
"_____no_output_____"
],
[
"The optimal aperture is stored in the TPF as the `pipeline_mask` property. We can have a look at it by calling it here:",
"_____no_output_____"
]
],
[
[
"first_cadence.pipeline_mask",
"_____no_output_____"
]
],
[
[
"As you can see, it is a Boolean array detailing which pixels are included. We can plot this aperture over the top of our TPF using the `plot()` function, and passing in the mask to the `aperture_mask` keyword. This will highlight the pixels included in the aperture mask using red hatched lines.",
"_____no_output_____"
]
],
[
[
"first_cadence.plot(aperture_mask=first_cadence.pipeline_mask);",
"_____no_output_____"
]
],
[
[
"You don't necessarily have to pass in the `pipeline_mask` to the `plot()` function; it can be any mask you create yourself, provided it is the right shape. An accompanying tutorial explains how to create such custom apertures, and goes into aperture photometry in more detail. For specifics on the selection of *Kepler*'s optimal apertures, read the [*Kepler* Data Processing Handbook](https://archive.stsci.edu/files/live/sites/mast/files/home/missions-and-data/kepler/_documents/KSCI-19081-002-KDPH.pdf), Section 7, *Finding Optimal Apertures in Kepler Data*.",
"_____no_output_____"
],
[
"## 4. Simple Aperture Photometry",
"_____no_output_____"
],
[
"Finally, let's learn how to perform simple aperture photometry (SAP) using the provided optimal aperture in `pipeline_mask` and the TPF.",
"_____no_output_____"
],
[
"Using the full TPF for all cadences in the quarter, we can perform aperture photometry using the `to_lightcurve()` method as follows:",
"_____no_output_____"
]
],
[
[
"lc = tpf.to_lightcurve()",
"_____no_output_____"
]
],
[
[
"This method returns a `LightCurve` object which details the flux and flux centroid position at each cadence:",
"_____no_output_____"
]
],
[
[
"lc",
"_____no_output_____"
]
],
[
[
"Note that this [`KeplerLightCurve`](https://docs.lightkurve.org/api/lightkurve.lightcurve.KeplerLightCurve.html) object has fewer data columns than in light curves downloaded directly from MAST. This is because we are extracting our light curve directly from the TPF using minimal processing, whereas light curves created using the official pipeline include more processing and more columns.\n\nWe can visualize the light curve as follows:",
"_____no_output_____"
]
],
[
[
"lc.plot();",
"_____no_output_____"
]
],
[
[
"This light curve is similar to the SAP light curve we previously encountered in the light curve tutorial.",
"_____no_output_____"
],
[
"### Note\n\nThe background flux can be plotted in a similar way, using the [`get_bkg_lightcurve()`](https://docs.lightkurve.org/api/lightkurve.targetpixelfile.KeplerTargetPixelFile.html#lightkurve.targetpixelfile.KeplerTargetPixelFile.get_bkg_lightcurve) method. This does not require an aperture, but instead sums the flux in the TPF's `FLUX_BKG` column at each timestamp. ",
"_____no_output_____"
]
],
[
[
"bkg = tpf.get_bkg_lightcurve()\nbkg.plot();",
"_____no_output_____"
]
],
[
[
"Inspecting the background in this way is useful to identify signals which appear to be present in the background rather than in the astronomical object under study.",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"## Exercises",
"_____no_output_____"
],
[
"Some stars, such as the planet-hosting star Kepler-10, have been observed both with *Kepler* and *TESS*. In this exercise, download and plot both the *TESS* and *Kepler* TPFs, along with the optimal apertures. You can do this by either selecting the TPFs from the list returned by [`search_targetpixelfile()`](https://docs.lightkurve.org/api/lightkurve.search.search_targetpixelfile.html), or by using the `mission` keyword argument when searching.\n\nBoth *Kepler* and *TESS* produce target pixel file data products, but these can look different across the two missions. *TESS* is focused on brighter stars and has larger pixels, so a star that might occupy many pixels in *Kepler* may only occupy a few in *TESS*.\n\nHow do light curves extracted from both of them compare?",
"_____no_output_____"
]
],
[
[
"#datalist = lk.search_targetpixelfile(...)\n",
"_____no_output_____"
],
[
"#soln:\ndatalist = lk.search_targetpixelfile(\"Kepler-10\")\ndatalist",
"_____no_output_____"
],
[
"kep = datalist[6].download()\ntes = datalist[15].download()",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(1, 2, figsize=(14,6))\nkep.plot(ax=axes[0], aperture_mask=kep.pipeline_mask, scale='log')\ntes.plot(ax=axes[1], aperture_mask=tes.pipeline_mask)\nfig.tight_layout();",
"_____no_output_____"
],
[
"lc_kep = kep.to_lightcurve()\nlc_tes = tes.to_lightcurve()",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(1, 2, figsize=(14,6), sharey=True)\nlc_kep.flatten().plot(ax=axes[0], c='k', alpha=.8)\nlc_tes.flatten().plot(ax=axes[1], c='k', alpha=.8);",
"_____no_output_____"
]
],
[
[
"If you plot the light curves for both missions side by side, you will see a stark difference. The *Kepler* data has a much smaller scatter, and repeating transits are visible. This is because *Kepler*'s pixels were smaller, and so could achieve a higher precision on fainter stars. *TESS* has larger pixels and therefore focuses on brighter stars. For stars like Kepler-10, it would be hard to detect a planet using *TESS* data alone.",
"_____no_output_____"
],
[
"## About this Notebook",
"_____no_output_____"
],
[
"**Authors:** Oliver Hall ([email protected]), Geert Barentsen\n\n**Updated On**: 2020-09-15",
"_____no_output_____"
],
[
"## Citing Lightkurve and Astropy\n\nIf you use `lightkurve` or `astropy` for published research, please cite the authors. Click the buttons below to copy BibTeX entries to your clipboard.",
"_____no_output_____"
],
[
"lk.show_citation_instructions()",
"_____no_output_____"
],
[
"<img style=\"float: right;\" src=\"https://raw.githubusercontent.com/spacetelescope/notebooks/master/assets/stsci_pri_combo_mark_horizonal_white_bkgd.png\" alt=\"Space Telescope Logo\" width=\"200px\"/>\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d05c9b3fc5040d4f6d86a26f6a18f92079cd3160 | 13,370 | ipynb | Jupyter Notebook | docs/site/tutorials/python_interoperability.ipynb | texasmichelle/swift | 37821ab7a57bc3013be20565a9a52f94145e2225 | [
"Apache-2.0"
] | 6,260 | 2018-04-26T17:54:25.000Z | 2022-03-31T02:14:24.000Z | docs/site/tutorials/python_interoperability.ipynb | texasmichelle/swift | 37821ab7a57bc3013be20565a9a52f94145e2225 | [
"Apache-2.0"
] | 427 | 2018-04-26T18:04:28.000Z | 2022-03-29T21:40:15.000Z | docs/site/tutorials/python_interoperability.ipynb | texasmichelle/swift | 37821ab7a57bc3013be20565a9a52f94145e2225 | [
"Apache-2.0"
] | 748 | 2018-04-26T17:58:10.000Z | 2022-03-13T00:52:21.000Z | 28.206751 | 290 | 0.583396 | [
[
[
"##### Copyright 2018 The TensorFlow Authors. [Licensed under the Apache License, Version 2.0](#scrollTo=ByZjmtFgB_Y5).",
"_____no_output_____"
]
],
[
[
"// #@title Licensed under the Apache License, Version 2.0 (the \"License\"); { display-mode: \"form\" }\n// Licensed under the Apache License, Version 2.0 (the \"License\");\n// you may not use this file except in compliance with the License.\n// You may obtain a copy of the License at\n//\n// https://www.apache.org/licenses/LICENSE-2.0\n//\n// Unless required by applicable law or agreed to in writing, software\n// distributed under the License is distributed on an \"AS IS\" BASIS,\n// WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n// See the License for the specific language governing permissions and\n// limitations under the License.",
"_____no_output_____"
]
],
[
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/swift/tutorials/python_interoperability\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/swift/blob/main/docs/site/tutorials/python_interoperability.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/swift/blob/main/docs/site/tutorials/python_interoperability.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"# Python interoperability\n\nSwift For TensorFlow supports Python interoperability.\n\nYou can import Python modules from Swift, call Python functions, and convert values between Swift and Python.",
"_____no_output_____"
]
],
[
[
"import PythonKit\nprint(Python.version)",
"_____no_output_____"
]
],
[
[
"## Setting the Python version",
"_____no_output_____"
],
[
"By default, when you `import Python`, Swift searches system library paths for the newest version of Python installed. \nTo use a specific Python installation, set the `PYTHON_LIBRARY` environment variable to the `libpython` shared library provided by the installation. For example: \n\n`export PYTHON_LIBRARY=\"~/anaconda3/lib/libpython3.7m.so\"`\n\nThe exact filename will differ across Python environments and platforms.",
"_____no_output_____"
],
[
"Alternatively, you can set the `PYTHON_VERSION` environment variable, which instructs Swift to search system library paths for a matching Python version. Note that `PYTHON_LIBRARY` takes precedence over `PYTHON_VERSION`.\n\nIn code, you can also call the `PythonLibrary.useVersion` function, which is equivalent to setting `PYTHON_VERSION`.",
"_____no_output_____"
]
],
[
[
"// PythonLibrary.useVersion(2)\n// PythonLibrary.useVersion(3, 7)",
"_____no_output_____"
]
],
[
[
"__Note: you should run `PythonLibrary.useVersion` right after `import Python`, before calling any Python code. It cannot be used to dynamically switch Python versions.__",
"_____no_output_____"
],
[
"Set `PYTHON_LOADER_LOGGING=1` to see [debug output for Python library loading](https://github.com/apple/swift/pull/20674#discussion_r235207008). ",
"_____no_output_____"
],
[
"## Basics\n\nIn Swift, `PythonObject` represents an object from Python.\nAll Python APIs use and return `PythonObject` instances.\n\nBasic types in Swift (like numbers and arrays) are convertible to `PythonObject`. In some cases (for literals and functions taking `PythonConvertible` arguments), conversion happens implicitly. To explicitly cast a Swift value to `PythonObject`, use the `PythonObject` initializer.\n\n`PythonObject` defines many standard operations, including numeric operations, indexing, and iteration.",
"_____no_output_____"
]
],
[
[
"// Convert standard Swift types to Python.\nlet pythonInt: PythonObject = 1\nlet pythonFloat: PythonObject = 3.0\nlet pythonString: PythonObject = \"Hello Python!\"\nlet pythonRange: PythonObject = PythonObject(5..<10)\nlet pythonArray: PythonObject = [1, 2, 3, 4]\nlet pythonDict: PythonObject = [\"foo\": [0], \"bar\": [1, 2, 3]]\n\n// Perform standard operations on Python objects.\nprint(pythonInt + pythonFloat)\nprint(pythonString[0..<6])\nprint(pythonRange)\nprint(pythonArray[2])\nprint(pythonDict[\"bar\"])",
"_____no_output_____"
],
[
"// Convert Python objects back to Swift.\nlet int = Int(pythonInt)!\nlet float = Float(pythonFloat)!\nlet string = String(pythonString)!\nlet range = Range<Int>(pythonRange)!\nlet array: [Int] = Array(pythonArray)!\nlet dict: [String: [Int]] = Dictionary(pythonDict)!\n\n// Perform standard operations.\n// Outputs are the same as Python!\nprint(Float(int) + float)\nprint(string.prefix(6))\nprint(range)\nprint(array[2])\nprint(dict[\"bar\"]!)",
"_____no_output_____"
]
],
[
[
"`PythonObject` defines conformances to many standard Swift protocols:\n* `Equatable`\n* `Comparable`\n* `Hashable`\n* `SignedNumeric`\n* `Strideable`\n* `MutableCollection`\n* All of the `ExpressibleBy_Literal` protocols\n\nNote that these conformances are not type-safe: crashes will occur if you attempt to use protocol functionality from an incompatible `PythonObject` instance.",
"_____no_output_____"
]
],
[
[
"let one: PythonObject = 1\nprint(one == one)\nprint(one < one)\nprint(one + one)\n\nlet array: PythonObject = [1, 2, 3]\nfor (i, x) in array.enumerated() {\n print(i, x)\n}",
"_____no_output_____"
]
],
[
[
"To convert tuples from Python to Swift, you must statically know the arity of the tuple.\n\nCall one of the following instance methods:\n- `PythonObject.tuple2`\n- `PythonObject.tuple3`\n- `PythonObject.tuple4`",
"_____no_output_____"
]
],
[
[
"let pythonTuple = Python.tuple([1, 2, 3])\nprint(pythonTuple, Python.len(pythonTuple))\n\n// Convert to Swift.\nlet tuple = pythonTuple.tuple3\nprint(tuple)",
"_____no_output_____"
]
],
[
[
"## Python builtins\n\nAccess Python builtins via the global `Python` interface.",
"_____no_output_____"
]
],
[
[
"// `Python.builtins` is a dictionary of all Python builtins.\n_ = Python.builtins\n\n// Try some Python builtins.\nprint(Python.type(1))\nprint(Python.len([1, 2, 3]))\nprint(Python.sum([1, 2, 3]))",
"_____no_output_____"
]
],
[
[
"## Importing Python modules\n\nUse `Python.import` to import a Python module. It works like the `import` keyword in `Python`.",
"_____no_output_____"
]
],
[
[
"let np = Python.import(\"numpy\")\nprint(np)\nlet zeros = np.ones([2, 3])\nprint(zeros)",
"_____no_output_____"
]
],
[
[
"Use the throwing function `Python.attemptImport` to perform safe importing.",
"_____no_output_____"
]
],
[
[
"let maybeModule = try? Python.attemptImport(\"nonexistent_module\")\nprint(maybeModule)",
"_____no_output_____"
]
],
[
[
"## Conversion with `numpy.ndarray`\n\nThe following Swift types can be converted to and from `numpy.ndarray`:\n- `Array<Element>`\n- `ShapedArray<Scalar>`\n- `Tensor<Scalar>`\n\nConversion succeeds only if the `dtype` of the `numpy.ndarray` is compatible with the `Element` or `Scalar` generic parameter type.\n\nFor `Array`, conversion from `numpy` succeeds only if the `numpy.ndarray` is 1-D.",
"_____no_output_____"
]
],
[
[
"import TensorFlow\n\nlet numpyArray = np.ones([4], dtype: np.float32)\nprint(\"Swift type:\", type(of: numpyArray))\nprint(\"Python type:\", Python.type(numpyArray))\nprint(numpyArray.shape)",
"_____no_output_____"
],
[
"// Examples of converting `numpy.ndarray` to Swift types.\nlet array: [Float] = Array(numpy: numpyArray)!\nlet shapedArray = ShapedArray<Float>(numpy: numpyArray)!\nlet tensor = Tensor<Float>(numpy: numpyArray)!\n\n// Examples of converting Swift types to `numpy.ndarray`.\nprint(array.makeNumpyArray())\nprint(shapedArray.makeNumpyArray())\nprint(tensor.makeNumpyArray())\n\n// Examples with different dtypes.\nlet doubleArray: [Double] = Array(numpy: np.ones([3], dtype: np.float))!\nlet intTensor = Tensor<Int32>(numpy: np.ones([2, 3], dtype: np.int32))!",
"_____no_output_____"
]
],
[
[
"## Displaying images\n\nYou can display images in-line using `matplotlib`, just like in Python notebooks.",
"_____no_output_____"
]
],
[
[
"// This cell is here to display plots inside a Jupyter Notebook.\n// Do not copy it into another environment.\n%include \"EnableIPythonDisplay.swift\"\nprint(IPythonDisplay.shell.enable_matplotlib(\"inline\"))",
"_____no_output_____"
],
[
"let np = Python.import(\"numpy\")\nlet plt = Python.import(\"matplotlib.pyplot\")\n\nlet time = np.arange(0, 10, 0.01)\nlet amplitude = np.exp(-0.1 * time)\nlet position = amplitude * np.sin(3 * time)\n\nplt.figure(figsize: [15, 10])\n\nplt.plot(time, position)\nplt.plot(time, amplitude)\nplt.plot(time, -amplitude)\n\nplt.xlabel(\"Time (s)\")\nplt.ylabel(\"Position (m)\")\nplt.title(\"Oscillations\")\n\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05cac15429018f239a45c235dee8c7812d06136 | 18,887 | ipynb | Jupyter Notebook | old/IMDb+reviews+Sentiment+Analysis-ML.ipynb | dzlab/deepprojects | c8a0327217f1517a008e76da51b77c80f5392d68 | [
"Unlicense"
] | 28 | 2018-12-02T09:52:01.000Z | 2021-11-06T20:23:50.000Z | old/IMDb+reviews+Sentiment+Analysis-ML.ipynb | dzlab/deepprojects | c8a0327217f1517a008e76da51b77c80f5392d68 | [
"Unlicense"
] | 1 | 2021-12-01T15:42:44.000Z | 2022-01-23T20:11:42.000Z | old/IMDb+reviews+Sentiment+Analysis-ML.ipynb | dzlab/deepprojects | c8a0327217f1517a008e76da51b77c80f5392d68 | [
"Unlicense"
] | 7 | 2018-12-04T18:52:11.000Z | 2021-04-08T17:06:42.000Z | 29.979365 | 1,411 | 0.554297 | [
[
[
"import numpy as np\nimport pandas as pd\nimport scipy as sp\n\nfrom scipy import sparse\n\nimport nltk\nfrom nltk.corpus import stopwords\nfrom nltk.stem.porter import PorterStemmer\n\nimport string\nimport re\nimport glob\n\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.feature_extraction.text import CountVectorizer, FeatureHasher\n\nimport keras\nfrom keras.optimizers import Adam\nfrom keras.callbacks import ModelCheckpoint\nfrom keras.layers import Dense, Embedding, LSTM, Dropout\nfrom keras.models import Sequential\n\nimport matplotlib.pyplot as plt\n\nprint('Keras version: %s' % keras.__version__)\n\nPATH = \"data/aclImdb\"",
"Keras version: 2.1.5\n"
],
[
"# or use nltk or spacy\nhtmltag = re.compile(r'<.*?>')\nnumbers = re.compile(r'[0-9]')\nquotes = re.compile(r'\\\"|`')\npunctuation = re.compile(r'([%s])'% string.punctuation)\nenglish_stopwords =set(stopwords.words('english'))\nstemmer = PorterStemmer()",
"_____no_output_____"
],
[
"# read files in the given tree, using subfolders as the target classes\ndef read_files(folder, subfolders):\n corpus, labels = [], []\n for index, label in enumerate(subfolders):\n path = '/'.join([folder, label, '*.txt'])\n for filename in glob.glob(path):\n corpus.append(open(filename, 'r').read())\n labels.append(index)\n return corpus, np.array(labels).astype(np.int)\n\n# pre-processor\ndef preprocess(s):\n # lowercase\n s = s.lower()\n # remove html tags\n s = htmltag.sub(' ', s)\n # remove numbers\n s = numbers.sub(' ', s)\n # remove quotes\n s = quotes.sub(' ', s)\n # replace puctuation\n s = punctuation.sub(' ', s)\n return s\n \n# tokenization\ndef tokenize(s):\n # use a serious tokenizer\n tokens = nltk.word_tokenize(s)\n # remove stopwords\n tokens = filter(lambda w: not w in english_stopwords, tokens)\n # stem words\n tokens = [stemmer.stem(token) for token in tokens]\n return tokens",
"_____no_output_____"
],
[
"#coprus_train_pos = [open(filename, 'r').read() for filename in glob.glob(PATH + '/train/pos/*.txt')]\n#coprus_train_neg = [open(filename, 'r').read() for filename in glob.glob(PATH + '/train/neg/*.txt')]\ncorpus_train, y_train = read_files(PATH + '/train', ['neg', 'pos'])\ncorpus_test, y_test = read_files(PATH + '/test', ['neg', 'pos'])",
"_____no_output_____"
],
[
"len(corpus_train), len(y_train), corpus_train[0], y_train[0], corpus_train[24999], y_train[24999]",
"_____no_output_____"
],
[
"len(corpus_test), len(y_test), corpus_test[0], y_test[0]",
"_____no_output_____"
],
[
"vectorizer = CountVectorizer(preprocessor=preprocess, tokenizer=tokenize)\nterm_doc_train = vectorizer.fit_transform(corpus_train)\nterm_doc_test = vectorizer.transform(corpus_test)",
"_____no_output_____"
],
[
"vocab = vectorizer.get_feature_names()\nvocab[100:102]",
"_____no_output_____"
],
[
"vocab_size = len(vocab)\nh = FeatureHasher(n_features=10, input_type='string')\nf = h.fit_transform(['q', 'w'])\nf.shape, f.toarray()",
"_____no_output_____"
],
[
"term_doc_train[0]",
"_____no_output_____"
],
[
"term_doc_train[100].toarray()",
"_____no_output_____"
],
[
"vectorizer.vocabulary_['cool']",
"_____no_output_____"
],
[
"# Multinomial Naive Bayes\nalpha = 0.1 # smoothing parameter\nclass MultinomialNaiveBayes():\n \"\"\"\n Arguments:\n alpha: smoothing parameter\n \"\"\"\n def __init__(self, alpha=0.1):\n self.b = 0\n self.r = 0\n self.alpha = alpha\n\n def fit(self, X, y):\n # bias\n N_pos = (y==1).shape[0]\n N_neg = (y==0).shape[0]\n self.b = np.log(N_pos / N_neg)\n # count of occurences for every token in vocabulary as they appear in positive samples\n p = alpha + X[y==1].sum(axis=0)\n p_l1 = np.linalg.norm(p, ord=1) # L1 norm\n # count of occurences for every token in vocabulary as they appear in negative samples\n q = alpha + X[y==0].sum(axis=0)\n q_l1 = np.linalg.norm(q, ord=1) # L1 norm\n # log count ratio\n self.r = np.log((p/p_l1) / (q/q_l1))\n #self.r = sp.sparse.csr_matrix(self.r.T)\n return self.r, self.b\n\n def predict(self, X):\n y_pred = np.sign(sp.sparse.csr_matrix.dot(X, self.r.T) + self.b)\n y_pred[y_pred==-1] = 0\n return y_pred\n \n def score(self, X, y):\n y_predict = self.predict(X)\n y_reshaped = np.reshape(y, y_predict.shape)\n return (y_reshaped == y_predict).mean()",
"_____no_output_____"
],
[
"model = MultinomialNaiveBayes()\nr, b = model.fit(term_doc_train, y_train)\nb, r.shape, term_doc_train.shape",
"_____no_output_____"
],
[
"term_doc_train.shape, r.shape, term_doc_train[0], r",
"_____no_output_____"
],
[
"# accuracy on training set\ny_pred = model.predict(term_doc_train)\n#y_train = np.reshape(y_train, (25000, 1))\n(np.reshape(y_train, (25000, 1)) == y_pred).mean()",
"_____no_output_____"
],
[
"# accuracy on validation set\ny_pred2 = model.predict(term_doc_test)\n#y_test = np.reshape(y_test, (25000, 1))\n(np.reshape(y_test, (25000, 1)) == y_pred2).mean()",
"_____no_output_____"
],
[
"# now let's binary term document\nterm_doc_train = term_doc_train.sign() # turn everything into 1 or 0\nterm_doc_test = term_doc_test.sign() # turn everything into 1 or 0\nterm_doc_train.shape, term_doc_test.shape",
"_____no_output_____"
],
[
"model = MultinomialNaiveBayes()\nmodel.fit(term_doc_train, y_train)\naccuracy_train = model.score(term_doc_train, y_train)\naccuracy_test = model.score(term_doc_test, y_test)\naccuracy_train, accuracy_test",
"_____no_output_____"
],
[
"term_doc_train.shape, y_train.shape, term_doc_train[y_train==0].sum(axis=0).shape, term_doc_train[y_train==1].sum(axis=0).shape",
"_____no_output_____"
],
[
"(y_train==0).shape, (y_train==1).shape, y_pred.shape",
"_____no_output_____"
],
[
"# now with plain logistic regression\nmodel = LogisticRegression()\nmodel.fit(term_doc_train, y_train)\n# accuracy on training\ny_pred = model.predict(term_doc_train)\naccuracy_train = (y_train == y_pred).mean()\n# accuracy on validation\ny_pred = model.predict(term_doc_test)\naccuracy_test = (y_test == y_pred).mean()\naccuracy_train, accuracy_test",
"_____no_output_____"
],
[
"# now with regularized logistic regression\nmodel = LogisticRegression(C=0.01, dual=True)\nmodel.fit(term_doc_train, y_train)\n# accuracy on training\ny_pred = model.predict(term_doc_train)\naccuracy_train = (y_train == y_pred).mean()\n# accuracy on validation\ny_pred = model.predict(term_doc_test)\naccuracy_test = (y_test == y_pred).mean()\naccuracy_train, accuracy_test",
"_____no_output_____"
],
[
"# now combining Naive Base and Logistic Regression\n\"\"\"\nclass NBLR(keras.Model):\n def __init__(self):\n super(NBLR, self).__init__(name='NBLR')\n self.softmax = keras.layers.Activation('softmax')\n\n def call(self, inputs):\n out = self.softmax(inputs)\n return out\n\nmodel = NBLR()\nmodel.compile(loss='mean_squared_error', optimizer='sgd', metrics=['accuracy'])\nlosses = model.fit(x=term_doc_train, y=y_train)\n\"\"\"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05cae1cb358a6f8cd8af81887cbfa4f4b95a01c | 7,172 | ipynb | Jupyter Notebook | generative-models/LDA.ipynb | yusueliu/murphy-book | 71d62cc083a683fb861be1e5acb8eeb948b00c54 | [
"Apache-2.0"
] | 2 | 2019-03-25T22:22:23.000Z | 2019-09-29T20:46:58.000Z | generative-models/LDA.ipynb | yusueliu/murphy-book | 71d62cc083a683fb861be1e5acb8eeb948b00c54 | [
"Apache-2.0"
] | null | null | null | generative-models/LDA.ipynb | yusueliu/murphy-book | 71d62cc083a683fb861be1e5acb8eeb948b00c54 | [
"Apache-2.0"
] | 1 | 2021-12-24T01:14:12.000Z | 2021-12-24T01:14:12.000Z | 55.596899 | 399 | 0.647936 | [
[
[
"## LDA\n\nThe graphical model representation of LDA is given blow:\n<img src=\"figures/LDA.png\">\n\nThe basic idea of LDA is that documents are represented as random mixtures over latent topics, where each topic is characterized by a distribution over words.\n\nLDA assumes the following generative process for each document $\\mathbf{w}$ in a corpus $\\mathcal{D}$:\n1. Choose $N\\sim$ Poisson($\\xi$).\n2. Choose $\\theta \\sim $Dir($\\alpha$).\n3. For each of the $N$ words $w_n$:\na) Choose a topic $z_n\\sim$ Multinomial($\\theta$)\nb) Choose a word $w_n$ from $p(w_n|z_n, \\beta)$, a multinomial probability conditioned on the topic $z_n$.\n\nSeveral simplifying assumptions are made in this basic model:\n1. The dimensionality $k$ of the Dirichlet distribution (and thus the dimensionality of the topic variable $z$) is assumed known and fixed.\n2. The word probabilities are parameterized by a $k\\times V$ matrix $\\beta$ where $\\beta_{ij} = p(w^j=1 | z^i=1)$, which for now we treat as a fixed quantity that is to be estimated.\n3. The Poisson assumption is not critical to anything that follows and more realistic document length distributions can be used as needed.\n\nA $k$-dimensional Dirichlet random variable $\\theta$ can take values in the $(k-1)$-simplex (a $k$-vector $\\theta$ in the $(k-1)$-simplex if $\\theta_i\\ge 0, \\sum_{i=1}^k\\theta_i=1$), and has the following probability density on this simplex:\n\n$$\np(\\theta|\\alpha) = \\frac{\\Gamma(\\sum_{i=1}^k\\alpha_i)}{\\prod_{i=1}^k\\Gamma(\\alpha_i)}\\theta_1^{\\alpha_1-1}\\cdots\\theta_k^{a_k-1},\n$$\nwhere the parameter $\\alpha$ is a $k$-vector with components $\\alpha_i > 0$, and where $\\Gamma(x)$ is the Gamma function.\n\nGiven the parameters $\\alpha$ and $\\beta$, the joint distribution of a topic mixture $\\theta$, a set of $N$ topics $\\mathbf{z}$, and a set of $N$ words $\\mathbf{w}$ is given by:\n$$\np(\\theta, \\mathbf{z}|\\alpha, \\beta) = p(\\theta | \\alpha) \\prod_{n=1}^N p(z_n|\\theta)p(w_n|z_n,\\beta),\n$$\nwhere $p(z_n|\\theta)$ is simply $\\theta_i$ for the unique $i$ such that $z_n^i = 1$. Integrating over $\\theta$ and summing over $z$, we obtain the marginal distribution of a document:\n$$\np(\\mathbf{w}|\\alpha,\\beta) = \\int p(\\theta|\\alpha)\\left(\\prod_{n=1}^N\\sum_{z_n}p(z_n|\\theta) p(w_n|z_n,\\beta)\\right)d\\theta.\n$$\nFinally, taking the product of the marginal probabilities of single documents, we obtain the probability of a corpus:\n$$\np(\\mathcal{D}|\\alpha,\\beta) = \\prod_{d=1}^M\\int p(\\theta_d|\\alpha)\\left(\\prod_{n=1}^{N_d}\\sum_{z_{d_n}}p(z_{d_n}|\\theta_d)p(w_{d_n}| z_{d_n},\\beta)\\right)d\\theta_d.\n$$",
"_____no_output_____"
],
[
"There are **three** levels to the LDA representation. The parameters $\\alpha$ and $\\beta$ are corpus level parameters, assumed to be sampled once in the process of generating a corpus.The variables $\\theta_d$ are document-level variables, sampled once per document. Finally the variables $z_{dn}$ and $w_{dn}$ are word-level variables and are sampled once for each word in each document.\n\nNote the topic node is sampled *repeatedly* within the document. Under LDA, documents can be associated with multiple topics.",
"_____no_output_____"
],
[
"### Inference\nThe key inference problem that we need to solve in order to use LDA is that of computing the posterior distribution of the hidden variables given a document:\n$$\np(\\theta, \\mathbf{z}|\\mathbf{w}, \\alpha, \\beta) = \\frac{p(\\theta, \\mathbf{z}, \\mathbf{w}| \\alpha, \\beta)}{p(\\mathbf{w}|\\alpha, \\beta)}.\n$$\nUnfortunately, this distribution is intractable to compute in general. We can however use a variety of variety of approximate inference algorithms.\n\n### Variational Inference\nConvexity based variational algorithm for inference in LDA.\n\nBasic idea:\n- Use Jensen's inequality to obtain an adjustable lower bound on the log likelihood ",
"_____no_output_____"
],
[
"## Probabilistic latent semantic indexing\n\nThis model posits that a document label $d$ and a word $w_n$ are conditionally independent given an unobserved topic $z$:\n\n$$\np(d, w_n) = p(d)\\sum_{z}p(w_n|z)p(z|d).\n$$\n\nThe pLSI model attempts to relax the simplifying assumption made in the mixture of unigrams model that each document is generated from only one topic. In a sense, it does capture the possibility that a document may contain multiple topics since $p(z|d)$ serves as the mixture weights of the topics for a particular document $d$. \n\nHowever, we need to note several problems:\n\n1. $d$ is a dummy index into the list of documents in the *training set*. Thus, $d$ is a multinomial random variable with as many possible values as there are training documents and the model learns the topic mixtures $p(z|d)$ only for those documents on which it is trained.\n\n2. Also stems from the use of a distribution index by training documents, is that the number of parameters which must be estimated grows linearly with the number of training documents. The parameters for a $k$-topic PLSI model are $k$ multinomial distributions of size $V$ and $M$ mixtures over the $k$ hidden topics. This gives $kV + kM$ parameters and therefore linear growth in $M$. \n\n$\\therefore$ pLSI is not a well-defined generative model of documents; there is no natural way to use it to assign probability to a previously seen document. Also, this linear growth in parameters suggests that the model is prone to overfitting. ",
"_____no_output_____"
],
[
"The principal advantages of generative models such as LDA include their modularity and their extensibility. As a probabilistic module, LDA can be readily embedded in a more complex model\n\nLDA overcomes both problems of pLSI by treating the topic mixture weights as a $k$-parameter hidden *random variables* rather than a large set of individual parameters which are explicitly linked to the training set. ",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d05cb43157fe3f519bbbc69778104ebaf5d7f215 | 301,396 | ipynb | Jupyter Notebook | Prototype Notebook/Example_1_Sandstone.ipynb | nre-aachen/gempy | 7e3a613545e62c2311a0350bcc8b955d84809fc6 | [
"MIT"
] | null | null | null | Prototype Notebook/Example_1_Sandstone.ipynb | nre-aachen/gempy | 7e3a613545e62c2311a0350bcc8b955d84809fc6 | [
"MIT"
] | null | null | null | Prototype Notebook/Example_1_Sandstone.ipynb | nre-aachen/gempy | 7e3a613545e62c2311a0350bcc8b955d84809fc6 | [
"MIT"
] | null | null | null | 117.137971 | 55,752 | 0.79159 | [
[
[
"# Example 1: Sandstone Model",
"_____no_output_____"
]
],
[
[
"# Importing\nimport theano.tensor as T\nimport theano\nimport sys, os\nsys.path.append(\"../GeMpy\")\nsys.path.append(\"../\")\n# Importing GeMpy modules\nimport gempy as GeMpy\n\n# Reloading (only for development purposes)\nimport importlib\nimportlib.reload(GeMpy)\n\n# Usuful packages\nimport numpy as np\nimport pandas as pn\n\nimport matplotlib.pyplot as plt\n\n# This was to choose the gpu\nos.environ['CUDA_LAUNCH_BLOCKING'] = '1'\n\n# Default options of printin\nnp.set_printoptions(precision = 6, linewidth= 130, suppress = True)\n\n#%matplotlib inline\n%matplotlib inline",
"_____no_output_____"
],
[
"# Importing the data from csv files and settign extent and resolution\ngeo_data = GeMpy.create_data([696000,747000,6863000,6950000,-20000, 200],[ 50, 50, 50],\n path_f = os.pardir+\"/input_data/a_Foliations.csv\",\n path_i = os.pardir+\"/input_data/a_Points.csv\")\n\n# Assigning series to formations as well as their order (timewise)\nGeMpy.set_data_series(geo_data, {\"EarlyGranite_Series\":geo_data.formations[-1], \n \"BIF_Series\":(geo_data.formations[0], geo_data.formations[1]),\n \"SimpleMafic_Series\":geo_data.formations[2]}, \n order_series = [\"EarlyGranite_Series\",\n \"BIF_Series\",\n \"SimpleMafic_Series\"], verbose=0)",
"_____no_output_____"
],
[
"GeMpy.data_to_pickle(geo_data, 'sandstone')",
"_____no_output_____"
],
[
"inter = GeMpy.InterpolatorInput(geo_data)",
"I am in the setting\nfloat32\nI am here\n[2, 2]\n"
],
[
"inter.interpolator.tg.n_formation.get_value()",
"_____no_output_____"
],
[
"import numpy as np\nnp.zeros((100,0))",
"_____no_output_____"
],
[
"100000/1000",
"_____no_output_____"
],
[
"GeMpy.plot_data(geo_data)",
"_____no_output_____"
],
[
"geo_data.formations",
"_____no_output_____"
],
[
"di = GeMpy.InterpolatorInput(geo_data)",
"I am in the setting\nfloat32\nI am here\n[2, 2]\n"
],
[
"di.data.get_formation_number()",
"_____no_output_____"
],
[
"geo_data_s = GeMpy.select_series(geo_data, ['EarlyGranite_Series'])",
"_____no_output_____"
],
[
"# Preprocessing data to interpolate: This rescales the coordinates between 0 and 1 for stability issues.\n# Here we can choose also the drift degree (in new updates I will change it to be possible to change the\n# grade after compilation). From here we can set also the data type of the operations in case you want to\n# use the GPU. Verbose is huge. There is a large list of strings that select what you want to print while\n# the computation.\ndata_interp = GeMpy.set_interpolator(geo_data,\n dtype=\"float32\",\n verbose=[])",
"_____no_output_____"
],
[
"# This cell will go to the backend\n\n# Set all the theano shared parameters and return the symbolic variables (the input of the theano function)\ninput_data_T = data_interp.interpolator.tg.input_parameters_list()\n\n# Prepare the input data (interfaces, foliations data) to call the theano function.\n#Also set a few theano shared variables with the len of formations series and so on\ninput_data_P = data_interp.interpolator.data_prep() \n\n# Compile the theano function.\ndebugging = theano.function(input_data_T, data_interp.interpolator.tg.whole_block_model(), on_unused_input='ignore',\n allow_input_downcast=True, profile=True)",
"[9 9 9]\n"
],
[
"%%timeit\n# Solve model calling the theano function\nsol = debugging(input_data_P[0], input_data_P[1], input_data_P[2], input_data_P[3],input_data_P[4], input_data_P[5])\n",
"1 loop, best of 3: 5.34 s per loop\n"
],
[
"lith = sol[-1,0,:]",
"_____no_output_____"
],
[
"np.save('sandstone_lith', lith)",
"_____no_output_____"
],
[
"a = geo_data.grid.grid[:,0].astype(bool)",
"_____no_output_____"
],
[
"a2 = a.reshape(50,50,50)",
"_____no_output_____"
],
[
"a2[:,:,0]",
"_____no_output_____"
],
[
"geo_data.grid.grid",
"_____no_output_____"
],
[
"50*50",
"_____no_output_____"
],
[
"geo_data.data_to_pickle('sandstone')",
"_____no_output_____"
],
[
"a2 = a1[:2500]\n\n",
"_____no_output_____"
],
[
"g = geo_data.grid.grid\nh = geo_data.grid.grid[:2500]",
"_____no_output_____"
],
[
"%%timeit\neu(g,h)",
"1 loop, best of 3: 1.73 s per loop\n"
],
[
"def squared_euclidean_distances(x_1, x_2):\n \"\"\"\n Compute the euclidian distances in 3D between all the points in x_1 and x_2\n Args:\n x_1 (theano.tensor.matrix): shape n_points x number dimension\n x_2 (theano.tensor.matrix): shape n_points x number dimension\n\n Returns:\n theano.tensor.matrix: Distancse matrix. shape n_points x n_points\n \"\"\"\n\n # T.maximum avoid negative numbers increasing stability\n\n\n return sqd",
"_____no_output_____"
],
[
"x_1 = T.matrix()\nx_2 = T.matrix()\n\nsqd = T.sqrt(T.maximum(\n (x_1**2).sum(1).reshape((x_1.shape[0], 1)) +\n (x_2**2).sum(1).reshape((1, x_2.shape[0])) -\n 2 * x_1.dot(x_2.T), 0\n))\neu = theano.function([x_1, x_2], sqd)",
"_____no_output_____"
],
[
"from evtk.hl import gridToVTK \n\nimport numpy as np \n\n# Dimensions \n\n\nnx, ny, nz = 50, 50, 50 \n\nlx = geo_data.extent[0]-geo_data.extent[1]\nly = geo_data.extent[2]-geo_data.extent[3]\nlz = geo_data.extent[4]-geo_data.extent[5]\n\ndx, dy, dz = lx/nx, ly/ny, lz/nz \n\nncells = nx * ny * nz \n\nnpoints = (nx + 1) * (ny + 1) * (nz + 1) \n\n# Coordinates \n\nx = np.arange(0, lx + 0.1*dx, dx, dtype='float64') \n\ny = np.arange(0, ly + 0.1*dy, dy, dtype='float64') \n\nz = np.arange(0, lz + 0.1*dz, dz, dtype='float64') \n\nx += geo_data.extent[0]\ny +=geo_data.extent[2]\nz +=geo_data.extent[5]\n\n# Variables \n \nlitho = sol[-1,0,:].reshape( (nx, ny, nz))\n\n\n\ngridToVTK(\"./sandstone\", x, y, z, cellData = {\"lithology\" : litho},) ",
"_____no_output_____"
],
[
"geo_data.extent[4]",
"_____no_output_____"
],
[
"# Plot the block model. \nGeMpy.plot_section(geo_data, 13, block = sol[-1,0,:], direction='x', plot_data = True)",
"_____no_output_____"
],
[
"geo_res = pn.read_csv('olaqases.vox')",
"_____no_output_____"
],
[
"geo_res = geo_res.iloc[9:]\n",
"_____no_output_____"
],
[
"geo_res['nx 50'].unique(), geo_data.formations",
"_____no_output_____"
],
[
"ip_addresses = geo_data.interfaces[\"formation\"].unique()\nip_dict = dict(zip(ip_addresses, range(1, len(ip_addresses) + 1)))\nip_dict['Murchison'] = 0\nip_dict['out'] = 0\nip_dict['SimpleMafic'] = 4\ngeo_res_num = geo_res['nx 50'].replace(ip_dict)\n",
"_____no_output_____"
],
[
"geo_res_num",
"_____no_output_____"
],
[
"ip_dict",
"_____no_output_____"
],
[
"(geo_res_num.shape[0]), sol[-1,0,:].shape[0]",
"_____no_output_____"
],
[
"sol[-1,0, :][7]",
"_____no_output_____"
],
[
"geo_res_num:",
"_____no_output_____"
],
[
"geo_res_num.as_matrix().astype(int)",
"_____no_output_____"
],
[
"plt.imshow( geo_res_num.as_matrix().reshape(50, 50, 50)[:, 23, :], origin=\"bottom\", cmap=\"viridis\" )",
"_____no_output_____"
],
[
"plt.imshow( sol[-1,0,:].reshape(50, 50, 50)[:, 23, :].T, origin=\"bottom\", cmap=\"viridis\" )",
"_____no_output_____"
],
[
"# Plot the block model. \nGeMpy.plot_section(geo_data, 13, block = geo_res_num.as_matrix(), direction='y', plot_data = True)",
"_____no_output_____"
],
[
"50*50*50",
"_____no_output_____"
],
[
"np.unique(sol[-1,0,:])",
"_____no_output_____"
],
[
"# Formation number and formation\ndata_interp.interfaces.groupby('formation number').formation.unique()",
"_____no_output_____"
],
[
"data_interp.interpolator.tg.u_grade_T.get_value()",
"_____no_output_____"
],
[
"np.unique(sol)",
"_____no_output_____"
],
[
"#np.save('SandstoneSol', sol)\nnp.count_nonzero(np.load('SandstoneSol.npy') == sol)",
"_____no_output_____"
],
[
"sol.shape",
"_____no_output_____"
],
[
"GeMpy.PlotData(geo_data).plot3D_steno(sol[-1,0,:], 'Sandstone', description='The sandstone model')",
"\n>> Welcome to the Python client library for Steno3D!\n\nCredentials file found: /home/miguel/.steno3d_client/credentials\nAccessing API developer key for @leguark\nWelcome to Steno3D! You are logged in as @leguark\nVerifying your quota for public projects...\nThis PUBLIC project will be viewable by everyone.\nTotal progress: 100% - Uploading: project Sandstone\nComplete!\nhttps://steno3d.com/resource/volume/BWCZxkJUmmdHurZg3mGs\n"
],
[
"np.linspace(geo_data.extent[0], geo_data.extent[1], geo_data.resolution[0], retstep=True)",
"_____no_output_____"
],
[
"np.diff(np.linspace(geo_data.extent[0], geo_data.extent[1], geo_data.resolution[0], retstep=False)).shape",
"_____no_output_____"
],
[
"(geo_data.extent[1]- geo_data.extent[0])/ geo_data.resolution[0]-4",
"_____no_output_____"
],
[
"(geo_data.extent[1]- geo_data.extent[0])/39",
"_____no_output_____"
],
[
"# So far this is a simple 3D visualization. I have to adapt it into GeMpy \n\nlith0 = sol == 0\nlith1 = sol == 1\nlith2 = sol == 2\nlith3 = sol == 3\nlith4 = sol == 4\nnp.unique(sol)\n\nimport ipyvolume.pylab as p3\n\np3.figure(width=800)\n\np3.scatter(geo_data.grid.grid[:,0][lith0],\n geo_data.grid.grid[:,1][lith0],\n geo_data.grid.grid[:,2][lith0], marker='box', color = 'blue', size = 0.1 )\n\np3.scatter(geo_data.grid.grid[:,0][lith1],\n geo_data.grid.grid[:,1][lith1],\n geo_data.grid.grid[:,2][lith1], marker='box', color = 'yellow', size = 1 )\n\np3.scatter(geo_data.grid.grid[:,0][lith2],\n geo_data.grid.grid[:,1][lith2],\n geo_data.grid.grid[:,2][lith2], marker='box', color = 'green', size = 1 )\n\np3.scatter(geo_data.grid.grid[:,0][lith3],\n geo_data.grid.grid[:,1][lith3],\n geo_data.grid.grid[:,2][lith3], marker='box', color = 'pink', size = 1 )\n\np3.scatter(geo_data.grid.grid[:,0][lith4],\n geo_data.grid.grid[:,1][lith4],\n geo_data.grid.grid[:,2][lith4], marker='box', color = 'red', size = 1 )\n\np3.xlim(np.min(geo_data.grid.grid[:,0]),np.min(geo_data.grid.grid[:,0])+2175.0*40)\np3.ylim(np.min(geo_data.grid.grid[:,1]),np.max(geo_data.grid.grid[:,1]))\np3.zlim(np.min(geo_data.grid.grid[:,2]),np.min(geo_data.grid.grid[:,2])+2175.0*40)#np.max(geo_data.grid.grid[:,2]))\n\np3.show()",
"_____no_output_____"
],
[
"# The profile at the moment sucks because all what is whithin a scan is not subdivided\ndebugging.profile.summary()",
"Function profiling\n==================\n Message: <ipython-input-6-22dcf15bad61>:3\n Time in 5 calls to Function.__call__: 1.357155e+01s\n Time in Function.fn.__call__: 1.357096e+01s (99.996%)\n Time in thunks: 1.357014e+01s (99.990%)\n Total compile time: 2.592983e+01s\n Number of Apply nodes: 95\n Theano Optimizer time: 1.642699e+01s\n Theano validate time: 3.617525e-02s\n Theano Linker time (includes C, CUDA code generation/compiling): 9.462233e+00s\n Import time 1.913705e-01s\n Node make_thunk time 9.450990e+00s\n Node forall_inplace,cpu,scan_fn}(Elemwise{Maximum}[(0, 0)].0, Subtensor{int64:int64:int8}.0, Subtensor{int64:int64:int8}.0, Subtensor{int64:int64:int8}.0, Subtensor{int64:int64:int8}.0, Subtensor{int64:int64:int8}.0, Subtensor{int64:int64:int8}.0, IncSubtensor{InplaceSet;:int64:}.0, grade of the universal drift, <TensorType(float64, matrix)>, <TensorType(float64, vector)>, Value of the formation, Position of the dips, Rest of the points of the layers, Reference points for every layer, Angle of every dip, Azimuth, Polarity, InplaceDimShuffle{x,x}.0, InplaceDimShuffle{x,x}.0, Elemwise{Composite{((sqr(sqr(i0)) * sqr(i0)) * i0)}}.0, Elemwise{Composite{(sqr(sqr(i0)) * i0)}}.0, Elemwise{Composite{(sqr(i0) * i0)}}.0, Elemwise{mul,no_inplace}.0, Elemwise{neg,no_inplace}.0, Elemwise{mul,no_inplace}.0, Elemwise{true_div,no_inplace}.0, Elemwise{Mul}[(0, 1)].0, Elemwise{mul,no_inplace}.0, Elemwise{Composite{(i0 * Composite{sqr(sqr(i0))}(i1))}}.0, Elemwise{Composite{(((i0 * i1) / sqr(i2)) + i3)}}.0, Reshape{2}.0) time 9.275085e+00s\n Node Elemwise{Composite{Switch(LT((i0 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2)), i3), (i4 - i0), Switch(GE((i0 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2)), (i2 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2))), (i5 + i0), Switch(LE((i2 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2)), i3), (i5 + i0), i0)))}}(Elemwise{Composite{minimum(minimum(minimum(minimum(minimum(i0, i1), i2), i3), i4), i5)}}.0, TensorConstant{1}, Elemwise{add,no_inplace}.0, TensorConstant{0}, TensorConstant{-2}, TensorConstant{2}) time 3.851414e-03s\n Node Elemwise{Composite{Switch(i0, Switch(LT((i1 + i2), i3), i3, (i1 + i2)), Switch(LT(i1, i2), i1, i2))}}(Elemwise{lt,no_inplace}.0, Elemwise{Composite{minimum(minimum(minimum(minimum(minimum(i0, i1), i2), i3), i4), i5)}}.0, Elemwise{Composite{Switch(LT((i0 + i1), i2), i2, (i0 + i1))}}.0, TensorConstant{0}) time 3.796577e-03s\n Node Elemwise{Composite{minimum(minimum(minimum(minimum(minimum(i0, i1), i2), i3), i4), i5)}}(Elemwise{Composite{Switch(LT((i0 + i1), i2), i2, (i0 + i1))}}.0, Elemwise{sub,no_inplace}.0, Elemwise{Composite{Switch(LT((i0 + i1), i2), i2, (i0 + i1))}}.0, Elemwise{sub,no_inplace}.0, Elemwise{Composite{Switch(LT((i0 + i1), i2), i2, (i0 + i1))}}.0, Elemwise{sub,no_inplace}.0) time 3.589630e-03s\n Node Elemwise{Composite{(((i0 - maximum(i1, i2)) - i3) + maximum(i4, i5))}}[(0, 0)](Elemwise{Composite{Switch(LT(i0, i1), (i0 + i2), i0)}}.0, Elemwise{Composite{minimum(((i0 + i1) - i2), i3)}}.0, TensorConstant{1}, TensorConstant{1}, Elemwise{Composite{((maximum(i0, i1) - Switch(LT(i2, i3), (i2 + i4), i2)) + i1)}}[(0, 2)].0, TensorConstant{2}) time 3.567696e-03s\n\nTime in all call to theano.grad() 0.000000e+00s\nTime since theano import 74.908s\nClass\n---\n<% time> <sum %> <apply time> <time per call> <type> <#call> <#apply> <Class name>\n 99.6% 99.6% 13.517s 2.70e+00s Py 5 1 theano.scan_module.scan_op.Scan\n 0.2% 99.9% 0.034s 6.76e-03s Py 5 1 theano.tensor.basic.Nonzero\n 0.1% 100.0% 0.012s 4.31e-05s C 285 57 theano.tensor.elemwise.Elemwise\n 0.0% 100.0% 0.005s 9.77e-04s C 5 1 theano.tensor.subtensor.AdvancedSubtensor1\n 0.0% 100.0% 0.001s 2.78e-04s C 5 1 theano.tensor.subtensor.IncSubtensor\n 0.0% 100.0% 0.000s 2.13e-06s C 40 8 theano.tensor.subtensor.Subtensor\n 0.0% 100.0% 0.000s 3.59e-06s C 15 3 theano.tensor.basic.Reshape\n 0.0% 100.0% 0.000s 7.26e-07s C 65 13 theano.tensor.basic.ScalarFromTensor\n 0.0% 100.0% 0.000s 9.44e-06s C 5 1 theano.tensor.basic.AllocEmpty\n 0.0% 100.0% 0.000s 1.88e-06s C 20 4 theano.compile.ops.Shape_i\n 0.0% 100.0% 0.000s 1.86e-06s C 20 4 theano.tensor.elemwise.DimShuffle\n 0.0% 100.0% 0.000s 8.58e-07s C 5 1 theano.compile.ops.Rebroadcast\n ... (remaining 0 Classes account for 0.00%(0.00s) of the runtime)\n\nOps\n---\n<% time> <sum %> <apply time> <time per call> <type> <#call> <#apply> <Op name>\n 99.6% 99.6% 13.517s 2.70e+00s Py 5 1 forall_inplace,cpu,scan_fn}\n 0.2% 99.9% 0.034s 6.76e-03s Py 5 1 Nonzero\n 0.1% 99.9% 0.011s 5.50e-04s C 20 4 Elemwise{mul,no_inplace}\n 0.0% 100.0% 0.005s 9.77e-04s C 5 1 AdvancedSubtensor1\n 0.0% 100.0% 0.001s 2.78e-04s C 5 1 IncSubtensor{InplaceSet;:int64:}\n 0.0% 100.0% 0.001s 1.77e-04s C 5 1 Elemwise{eq,no_inplace}\n 0.0% 100.0% 0.000s 7.26e-07s C 65 13 ScalarFromTensor\n 0.0% 100.0% 0.000s 9.44e-06s C 5 1 AllocEmpty{dtype='float64'}\n 0.0% 100.0% 0.000s 4.32e-06s C 10 2 Subtensor{int64}\n 0.0% 100.0% 0.000s 1.40e-06s C 30 6 Subtensor{int64:int64:int8}\n 0.0% 100.0% 0.000s 1.88e-06s C 20 4 Shape_i{0}\n 0.0% 100.0% 0.000s 3.39e-06s C 10 2 Reshape{1}\n 0.0% 100.0% 0.000s 2.31e-06s C 10 2 InplaceDimShuffle{x,x}\n 0.0% 100.0% 0.000s 7.63e-07s C 30 6 Elemwise{le,no_inplace}\n 0.0% 100.0% 0.000s 1.45e-06s C 15 3 Elemwise{Composite{Switch(LT((i0 + i1), i2), i2, (i0 + i1))}}\n 0.0% 100.0% 0.000s 4.01e-06s C 5 1 Elemwise{true_div,no_inplace}\n 0.0% 100.0% 0.000s 4.01e-06s C 5 1 Reshape{2}\n 0.0% 100.0% 0.000s 3.81e-06s C 5 1 Elemwise{Composite{Switch(LT((i0 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2)), i3), (i4 - i0), Switch(GE((i0 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2)), (i2 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2))), (i5 + i0), Switch(LE((i2 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2)), i3), (i5 + i0), i0)))}}\n 0.0% 100.0% 0.000s 1.21e-06s C 15 3 Elemwise{sub,no_inplace}\n 0.0% 100.0% 0.000s 3.62e-06s C 5 1 Elemwise{Composite{(i0 * Composite{sqr(sqr(i0))}(i1))}}\n ... (remaining 27 Ops account for 0.00%(0.00s) of the runtime)\n\nApply\n------\n<% time> <sum %> <apply time> <time per call> <#call> <id> <Apply name>\n 99.6% 99.6% 13.517s 2.70e+00s 5 93 forall_inplace,cpu,scan_fn}(Elemwise{Maximum}[(0, 0)].0, Subtensor{int64:int64:int8}.0, Subtensor{int64:int64:int8}.0, Subtensor{int64:int64:int8}.0, Subtensor{int64:int64:int8}.0, Subtensor{int64:int64:int8}.0, Subtensor{int64:int64:int8}.0, IncSubtensor{InplaceSet;:int64:}.0, grade of the universal drift, <TensorType(float64, matrix)>, <TensorType(float64, vector)>, Value of the formation, Position of the dips, Rest of the points of the layers, Re\n input 0: dtype=int64, shape=no shape, strides=no strides \n input 1: dtype=int64, shape=no shape, strides=no strides \n input 2: dtype=int64, shape=no shape, strides=no strides \n input 3: dtype=int64, shape=no shape, strides=no strides \n input 4: dtype=int64, shape=no shape, strides=no strides \n input 5: dtype=int64, shape=no shape, strides=no strides \n input 6: dtype=int64, shape=no shape, strides=no strides \n input 7: dtype=float64, shape=no shape, strides=no strides \n input 8: dtype=int64, shape=no shape, strides=no strides \n input 9: dtype=float64, shape=no shape, strides=no strides \n input 10: dtype=float64, shape=no shape, strides=no strides \n input 11: dtype=float64, shape=no shape, strides=no strides \n input 12: dtype=float32, shape=no shape, strides=no strides \n input 13: dtype=float32, shape=no shape, strides=no strides \n input 14: dtype=float32, shape=no shape, strides=no strides \n input 15: dtype=float32, shape=no shape, strides=no strides \n input 16: dtype=float32, shape=no shape, strides=no strides \n input 17: dtype=float32, shape=no shape, strides=no strides \n input 18: dtype=float64, shape=no shape, strides=no strides \n input 19: dtype=float64, shape=no shape, strides=no strides \n input 20: dtype=float64, shape=no shape, strides=no strides \n input 21: dtype=float64, shape=no shape, strides=no strides \n input 22: dtype=float64, shape=no shape, strides=no strides \n input 23: dtype=float64, shape=no shape, strides=no strides \n input 24: dtype=float64, shape=no shape, strides=no strides \n input 25: dtype=float64, shape=no shape, strides=no strides \n input 26: dtype=float64, shape=no shape, strides=no strides \n input 27: dtype=float64, shape=no shape, strides=no strides \n input 28: dtype=float64, shape=no shape, strides=no strides \n input 29: dtype=float64, shape=no shape, strides=no strides \n input 30: dtype=float64, shape=no shape, strides=no strides \n input 31: dtype=float64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.2% 99.9% 0.034s 6.76e-03s 5 37 Nonzero(Reshape{1}.0)\n input 0: dtype=float64, shape=no shape, strides=no strides \n output 0: dtype=int64, shape=no shape, strides=no strides \n 0.1% 99.9% 0.011s 2.19e-03s 5 27 Elemwise{mul,no_inplace}(<TensorType(float64, matrix)>, Elemwise{eq,no_inplace}.0)\n input 0: dtype=float64, shape=no shape, strides=no strides \n input 1: dtype=bool, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.005s 9.77e-04s 5 51 AdvancedSubtensor1(Reshape{1}.0, Subtensor{int64}.0)\n input 0: dtype=float64, shape=no shape, strides=no strides \n input 1: dtype=int64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.001s 2.78e-04s 5 91 IncSubtensor{InplaceSet;:int64:}(AllocEmpty{dtype='float64'}.0, Rebroadcast{0}.0, Constant{1})\n input 0: dtype=float64, shape=no shape, strides=no strides \n input 1: dtype=float64, shape=no shape, strides=no strides \n input 2: dtype=int64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.001s 1.77e-04s 5 15 Elemwise{eq,no_inplace}(InplaceDimShuffle{0,x}.0, TensorConstant{(1, 1) of 0})\n input 0: dtype=float64, shape=no shape, strides=no strides \n input 1: dtype=int8, shape=no shape, strides=no strides \n output 0: dtype=bool, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 9.44e-06s 5 88 AllocEmpty{dtype='float64'}(Elemwise{Composite{(Switch(LT(maximum(i0, i1), i2), (maximum(i0, i1) + i3), (maximum(i0, i1) - i2)) + i4)}}[(0, 0)].0, Shape_i{0}.0)\n input 0: dtype=int64, shape=no shape, strides=no strides \n input 1: dtype=int64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 5.25e-06s 5 34 Reshape{1}(Elemwise{mul,no_inplace}.0, TensorConstant{(1,) of -1})\n input 0: dtype=float64, shape=no shape, strides=no strides \n input 1: dtype=int64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 4.63e-06s 5 44 Subtensor{int64}(Nonzero.0, Constant{0})\n input 0: dtype=int64, shape=no shape, strides=no strides \n input 1: dtype=int64, shape=no shape, strides=no strides \n output 0: dtype=int64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 4.29e-06s 5 23 Elemwise{mul,no_inplace}(TensorConstant{(1, 1) of 4.0}, InplaceDimShuffle{x,x}.0)\n input 0: dtype=float64, shape=no shape, strides=no strides \n input 1: dtype=float64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 4.01e-06s 5 94 Subtensor{int64}(forall_inplace,cpu,scan_fn}.0, ScalarFromTensor.0)\n input 0: dtype=float64, shape=no shape, strides=no strides \n input 1: dtype=int64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 4.01e-06s 5 58 Reshape{2}(AdvancedSubtensor1.0, TensorConstant{[-1 3]})\n input 0: dtype=float64, shape=no shape, strides=no strides \n input 1: dtype=int64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 4.01e-06s 5 29 Elemwise{true_div,no_inplace}(TensorConstant{(1, 1) of -14.0}, Elemwise{sqr,no_inplace}.0)\n input 0: dtype=float64, shape=no shape, strides=no strides \n input 1: dtype=float64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 3.81e-06s 5 38 Elemwise{Composite{Switch(LT((i0 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2)), i3), (i4 - i0), Switch(GE((i0 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2)), (i2 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2))), (i5 + i0), Switch(LE((i2 - Composite{Switch(LT(i0, i1), i0, i1)}(i1, i2)), i3), (i5 + i0), i0)))}}(Elemwise{Composite{minimum(minimum(minimum(minimum(minimum(i0, i1), i2), i3), i4), i5)}}.0, TensorConstant{1}, Elemwise{add,no_inplace}.\n input 0: dtype=int64, shape=no shape, strides=no strides \n input 1: dtype=int64, shape=no shape, strides=no strides \n input 2: dtype=int64, shape=no shape, strides=no strides \n input 3: dtype=int8, shape=no shape, strides=no strides \n input 4: dtype=int64, shape=no shape, strides=no strides \n input 5: dtype=int64, shape=no shape, strides=no strides \n output 0: dtype=int64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 3.62e-06s 5 17 Elemwise{Composite{(i0 * Composite{sqr(sqr(i0))}(i1))}}(TensorConstant{(1, 1) of 15.0}, InplaceDimShuffle{x,x}.0)\n input 0: dtype=float64, shape=no shape, strides=no strides \n input 1: dtype=float64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 3.62e-06s 5 2 Shape_i{0}(Length of interfaces in every series)\n input 0: dtype=int64, shape=no shape, strides=no strides \n output 0: dtype=int64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 3.58e-06s 5 20 Elemwise{Composite{((sqr(sqr(i0)) * sqr(i0)) * i0)}}(InplaceDimShuffle{x,x}.0)\n input 0: dtype=float64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 3.39e-06s 5 81 Subtensor{int64:int64:int8}(Length of interfaces in every series, ScalarFromTensor.0, ScalarFromTensor.0, Constant{1})\n input 0: dtype=int64, shape=no shape, strides=no strides \n input 1: dtype=int64, shape=no shape, strides=no strides \n input 2: dtype=int64, shape=no shape, strides=no strides \n input 3: dtype=int8, shape=no shape, strides=no strides \n output 0: dtype=int64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 3.19e-06s 5 8 Elemwise{Composite{(((i0 * i1) / sqr(i2)) + i3)}}(TensorConstant{14.0}, <TensorType(float64, scalar)>, <TensorType(float64, scalar)>, <TensorType(float64, scalar)>)\n input 0: dtype=float64, shape=no shape, strides=no strides \n input 1: dtype=float64, shape=no shape, strides=no strides \n input 2: dtype=float64, shape=no shape, strides=no strides \n input 3: dtype=float64, shape=no shape, strides=no strides \n output 0: dtype=float64, shape=no shape, strides=no strides \n 0.0% 100.0% 0.000s 3.00e-06s 5 77 Elemwise{Composite{(((i0 - maximum(i1, i2)) - i3) + maximum(i4, i5))}}[(0, 0)](Elemwise{Composite{Switch(LT(i0, i1), (i0 + i2), i0)}}.0, Elemwise{Composite{minimum(((i0 + i1) - i2), i3)}}.0, TensorConstant{1}, TensorConstant{1}, Elemwise{Composite{((maximum(i0, i1) - Switch(LT(i2, i3), (i2 + i4), i2)) + i1)}}[(0, 2)].0, TensorConstant{2})\n input 0: dtype=int64, shape=no shape, strides=no strides \n input 1: dtype=int64, shape=no shape, strides=no strides \n input 2: dtype=int8, shape=no shape, strides=no strides \n input 3: dtype=int8, shape=no shape, strides=no strides \n input 4: dtype=int64, shape=no shape, strides=no strides \n input 5: dtype=int8, shape=no shape, strides=no strides \n output 0: dtype=int64, shape=no shape, strides=no strides \n ... (remaining 75 Apply instances account for 0.00%(0.00s) of the runtime)\n\nHere are tips to potentially make your code run faster\n (if you think of new ones, suggest them on the mailing list).\n Test them first, as they are not guaranteed to always provide a speedup.\n Sorry, no tip for today.\n"
]
],
[
[
"#### Below here so far is deprecated",
"_____no_output_____"
],
[
"First we make a GeMpy instance with most of the parameters default (except range that is given by the project). Then we also fix the extension and the resolution of the domain we want to interpolate. Finally we compile the function, only needed once every time we open the project (the guys of theano they are working on letting loading compiled files, even though in our case it is not a big deal).\n\n*General note. So far the reescaling factor is calculated for all series at the same time. GeoModeller does it individually for every potential field. I have to look better what this parameter exactly means*",
"_____no_output_____"
],
[
"All input data is stored in pandas dataframes under, ```self.Data.Interances``` and ```self.Data.Foliations```:",
"_____no_output_____"
],
[
"In case of disconformities, we can define which formation belong to which series using a dictionary. Until python 3.6 is important to specify the order of the series otherwise is random",
"_____no_output_____"
],
[
"Now in the data frame we should have the series column too",
"_____no_output_____"
],
[
"Next step is the creating of a grid. So far only regular. By default it takes the extent and the resolution given in the `import_data` method.",
"_____no_output_____"
]
],
[
[
"# Create a class Grid so far just regular grid\n#GeMpy.set_grid(geo_data)\n#GeMpy.get_grid(geo_data)",
"_____no_output_____"
]
],
[
[
"## Plotting raw data",
"_____no_output_____"
],
[
"The object Plot is created automatically as we call the methods above. This object contains some methods to plot the data and the results.\n\nIt is possible to plot a 2D projection of the data in a specific direction using the following method. Also is possible to choose the series you want to plot. Additionally all the key arguments of seaborn lmplot can be used.",
"_____no_output_____"
]
],
[
[
"#GeMpy.plot_data(geo_data, 'y', geo_data.series.columns.values[1])",
"_____no_output_____"
]
],
[
[
"## Class Interpolator",
"_____no_output_____"
],
[
"This class will take the data from the class Data and calculate potential fields and block. We can pass as key arguments all the variables of the interpolation. I recommend not to touch them if you do not know what are you doing. The default values should be good enough. Also the first time we execute the method, we will compile the theano function so it can take a bit of time.",
"_____no_output_____"
]
],
[
[
"%debug",
"> \u001b[1;32m/home/bl3/PycharmProjects/GeMpy/GeMpy/GeMpy.py\u001b[0m(46)\u001b[0;36mrescale_data\u001b[1;34m()\u001b[0m\n\u001b[1;32m 44 \u001b[1;33m\u001b[1;33m\u001b[0m\u001b[0m\n\u001b[0m\u001b[1;32m 45 \u001b[1;33m \u001b[0mnew_coord_extent\u001b[0m \u001b[1;33m=\u001b[0m \u001b[0m_np\u001b[0m\u001b[1;33m.\u001b[0m\u001b[0mzeros_like\u001b[0m\u001b[1;33m(\u001b[0m\u001b[0mgeo_data\u001b[0m\u001b[1;33m.\u001b[0m\u001b[0mextent\u001b[0m\u001b[1;33m)\u001b[0m\u001b[1;33m\u001b[0m\u001b[0m\n\u001b[0m\u001b[1;32m---> 46 \u001b[1;33m \u001b[0mnew_coord_extent\u001b[0m\u001b[1;33m[\u001b[0m\u001b[1;33m:\u001b[0m\u001b[1;36m2\u001b[0m\u001b[1;33m]\u001b[0m \u001b[1;33m=\u001b[0m \u001b[0mgeo_data\u001b[0m\u001b[1;33m.\u001b[0m\u001b[0mextent\u001b[0m\u001b[1;33m[\u001b[0m\u001b[1;33m:\u001b[0m\u001b[1;36m2\u001b[0m\u001b[1;33m]\u001b[0m \u001b[1;33m-\u001b[0m \u001b[0mcenters\u001b[0m\u001b[1;33m[\u001b[0m\u001b[1;36m0\u001b[0m\u001b[1;33m]\u001b[0m \u001b[1;33m/\u001b[0m \u001b[0mrescaling_factor\u001b[0m \u001b[1;33m+\u001b[0m \u001b[1;36m0.5001\u001b[0m\u001b[1;33m\u001b[0m\u001b[0m\n\u001b[0m\u001b[1;32m 47 \u001b[1;33m \u001b[0mnew_coord_extent\u001b[0m\u001b[1;33m[\u001b[0m\u001b[1;36m2\u001b[0m\u001b[1;33m:\u001b[0m\u001b[1;36m4\u001b[0m\u001b[1;33m]\u001b[0m \u001b[1;33m=\u001b[0m \u001b[0mgeo_data\u001b[0m\u001b[1;33m.\u001b[0m\u001b[0mextent\u001b[0m\u001b[1;33m[\u001b[0m\u001b[1;36m2\u001b[0m\u001b[1;33m:\u001b[0m\u001b[1;36m4\u001b[0m\u001b[1;33m]\u001b[0m \u001b[1;33m-\u001b[0m \u001b[0mcenters\u001b[0m\u001b[1;33m[\u001b[0m\u001b[1;36m1\u001b[0m\u001b[1;33m]\u001b[0m \u001b[1;33m/\u001b[0m \u001b[0mrescaling_factor\u001b[0m \u001b[1;33m+\u001b[0m \u001b[1;36m0.5001\u001b[0m\u001b[1;33m\u001b[0m\u001b[0m\n\u001b[0m\u001b[1;32m 48 \u001b[1;33m \u001b[0mnew_coord_extent\u001b[0m\u001b[1;33m[\u001b[0m\u001b[1;36m4\u001b[0m\u001b[1;33m:\u001b[0m\u001b[1;36m6\u001b[0m\u001b[1;33m]\u001b[0m \u001b[1;33m=\u001b[0m \u001b[0mgeo_data\u001b[0m\u001b[1;33m.\u001b[0m\u001b[0mextent\u001b[0m\u001b[1;33m[\u001b[0m\u001b[1;36m4\u001b[0m\u001b[1;33m:\u001b[0m\u001b[1;36m6\u001b[0m\u001b[1;33m]\u001b[0m \u001b[1;33m-\u001b[0m \u001b[0mcenters\u001b[0m\u001b[1;33m[\u001b[0m\u001b[1;36m2\u001b[0m\u001b[1;33m]\u001b[0m \u001b[1;33m/\u001b[0m \u001b[0mrescaling_factor\u001b[0m \u001b[1;33m+\u001b[0m \u001b[1;36m0.5001\u001b[0m\u001b[1;33m\u001b[0m\u001b[0m\n\u001b[0m\n\u0001\u0002ipdb> \u0001\u0002geo_data.extent[:2]\n[696000, 747000]\n\u0001\u0002ipdb> \u0001\u0002centers[0]\n396789.0625\n\u0001\u0002ipdb> \u0001\u0002new_coord_extent[:2]\narray([0, 0])\n\u0001\u0002ipdb> \u0001\u0002geo_data.extent[:2] - centers[0] / rescaling_factor + 0.5001\n*** TypeError: unsupported operand type(s) for -: 'list' and 'float'\n\u0001\u0002ipdb> \u0001\u0002geo_data.extent[:2] - centers[0] / rescaling_factor\n*** TypeError: unsupported operand type(s) for -: 'list' and 'float'\n\u0001\u0002ipdb> \u0001\u0002geo_data.extent[:2] - centers[0]\n*** TypeError: unsupported operand type(s) for -: 'list' and 'float'\n\u0001\u0002ipdb> \u0001\u0002centers[0]\n396789.0625\n\u0001\u0002ipdb> \u0001\u0002geo_data.extent[:2] - centers[0].as_matrix(\n*** SyntaxError: unexpected EOF while parsing\n\u0001\u0002ipdb> \u0001\u0002geo_data.extent[:2] - centers[0].as_matrix()\n*** AttributeError: 'float' object has no attribute 'as_matrix'\n\u0001\u0002ipdb> \u0001\u0002geo_data.extent[:2] - centers.as_matrix()[0]\n*** TypeError: unsupported operand type(s) for -: 'list' and 'float'\n\u0001\u0002ipdb> \u0001\u0002exit\n"
],
[
"geo_data.interpolator.results",
"_____no_output_____"
],
[
"geo_data.interpolator.tg.c_o_T.get_value(), geo_data.interpolator.tg.a_T.get_value()",
"_____no_output_____"
],
[
"geo_data.interpolator.compile_potential_field_function()",
"_____no_output_____"
],
[
"geo_data.interpolator.compute_potential_fields('BIF_Series',verbose = 3)",
"The serie formations are SimpleMafic2|SimpleBIF\nThe formations are: \nLayers \n X Y Z formation series\n0 735484.817806 6.891936e+06 -1819.319309 SimpleMafic2 BIF_Series\n1 729854.915982 6.891938e+06 -1432.263309 SimpleMafic2 BIF_Series\n2 724084.267161 6.891939e+06 -4739.830309 SimpleMafic2 BIF_Series\n3 733521.625000 6.895282e+06 521.555240 SimpleMafic2 BIF_Series\n4 721933.375000 6.884592e+06 496.669295 SimpleMafic2 BIF_Series\n5 724251.000000 6.886909e+06 484.550926 SimpleMafic2 BIF_Series\n6 727316.313000 6.886460e+06 478.254423 SimpleMafic2 BIF_Series\n7 729858.250000 6.887134e+06 484.259574 SimpleMafic2 BIF_Series\n8 732699.250000 6.885040e+06 494.526481 SimpleMafic2 BIF_Series\n9 716849.500000 6.887358e+06 508.981894 SimpleMafic2 BIF_Series\n10 719017.625000 6.892218e+06 508.179387 SimpleMafic2 BIF_Series\n11 739179.440691 6.891936e+06 -552.591309 SimpleBIF BIF_Series\n12 735564.599804 6.891936e+06 -2652.196309 SimpleBIF BIF_Series\n13 730009.009977 6.891938e+06 -2088.409309 SimpleBIF BIF_Series\n14 718795.791326 6.891941e+06 -2773.169309 SimpleBIF BIF_Series\n15 724143.386160 6.891939e+06 -5569.907309 SimpleBIF BIF_Series\n16 723877.188000 6.899768e+06 529.152169 SimpleBIF BIF_Series\n17 732998.313000 6.898049e+06 521.619609 SimpleBIF BIF_Series\n18 743689.438000 6.891769e+06 512.811278 SimpleBIF BIF_Series\n19 712961.813000 6.882722e+06 547.826016 SimpleBIF BIF_Series\n20 716284.875000 6.891346e+06 515.586860 SimpleBIF BIF_Series\n21 718942.875000 6.897600e+06 538.490136 SimpleBIF BIF_Series\n22 722157.625000 6.882947e+06 481.747055 SimpleBIF BIF_Series\n23 723952.000000 6.885488e+06 480.122832 SimpleBIF BIF_Series\n24 728736.813000 6.885488e+06 477.929009 SimpleBIF BIF_Series\n25 738829.813000 6.878087e+06 470.081431 SimpleBIF BIF_Series \n foliations \n X Y Z azimuth dip polarity \\\n0 739426.627684 6.891935e+06 75.422691 220.0 70.0 1 \n1 717311.112372 6.891941e+06 -1497.488309 90.0 60.0 1 \n\n formation series G_x G_y G_z \n0 SimpleBIF BIF_Series -0.604023 -7.198463e-01 0.34202 \n1 SimpleBIF BIF_Series 0.866025 5.302876e-17 0.50000 \nDual Kriging weights: [ nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan nan\n nan nan nan nan]\n"
],
[
"geo_data.interpolator.potential_fields",
"_____no_output_____"
],
[
"geo_data.interpolator.results",
"_____no_output_____"
],
[
"geo_data.interpolator.tg.c_resc.get_value()",
"_____no_output_____"
]
],
[
[
"Now we could visualize the individual potential fields as follow:",
"_____no_output_____"
],
[
"### Early granite",
"_____no_output_____"
]
],
[
[
"GeMpy.plot_potential_field(geo_data,10, n_pf=0)",
"_____no_output_____"
]
],
[
[
"### BIF Series",
"_____no_output_____"
]
],
[
[
"GeMpy.plot_potential_field(geo_data,13, n_pf=1, cmap = \"magma\", plot_data = True,\n verbose = 5)",
"_____no_output_____"
]
],
[
[
"### SImple mafic",
"_____no_output_____"
]
],
[
[
"GeMpy.plot_potential_field(geo_data, 10, n_pf=2)",
"_____no_output_____"
]
],
[
[
"## Optimizing the export of lithologies\n\nBut usually the final result we want to get is the final block. The method `compute_block_model` will compute the block model, updating the attribute `block`. This attribute is a theano shared function that can return a 3D array (raveled) using the method `get_value()`.",
"_____no_output_____"
]
],
[
[
"GeMpy.compute_block_model(geo_data)",
"../GeMpy/GeMpy.py:38: UserWarning: Using default interpolation values\n warnings.warn('Using default interpolation values')\n"
],
[
"#GeMpy.set_interpolator(geo_data, u_grade = 0, compute_potential_field=True)",
"_____no_output_____"
]
],
[
[
"And again after computing the model in the Plot object we can use the method `plot_block_section` to see a 2D section of the model",
"_____no_output_____"
]
],
[
[
"GeMpy.plot_section(geo_data, 13, direction='y')",
"_____no_output_____"
]
],
[
[
"## Export to vtk. (*Under development*)",
"_____no_output_____"
]
],
[
[
"\"\"\"Export model to VTK\n\nExport the geology blocks to VTK for visualisation of the entire 3-D model in an\nexternal VTK viewer, e.g. Paraview.\n\n..Note:: Requires pyevtk, available for free on: https://github.com/firedrakeproject/firedrake/tree/master/python/evtk\n\n**Optional keywords**:\n - *vtk_filename* = string : filename of VTK file (default: output_name)\n - *data* = np.array : data array to export to VKT (default: entire block model)\n\"\"\"\nvtk_filename = \"noddyFunct2\"\n\nextent_x = 10\nextent_y = 10\nextent_z = 10\n\ndelx = 0.2\ndely = 0.2\ndelz = 0.2\nfrom pyevtk.hl import gridToVTK\n# Coordinates\nx = np.arange(0, extent_x + 0.1*delx, delx, dtype='float64')\ny = np.arange(0, extent_y + 0.1*dely, dely, dtype='float64')\nz = np.arange(0, extent_z + 0.1*delz, delz, dtype='float64')\n\n# self.block = np.swapaxes(self.block, 0, 2)\ngridToVTK(vtk_filename, x, y, z, cellData = {\"geology\" : sol})",
"_____no_output_____"
]
],
[
[
"## Performance Analysis",
"_____no_output_____"
],
[
"One of the advantages of theano is the posibility to create a full profile of the function. This has to be included in at the time of the creation of the function. At the moment it should be active (the downside is larger compilation time and I think also a bit in the computation so be careful if you need a fast call)",
"_____no_output_____"
],
[
"### CPU",
"_____no_output_____"
],
[
"The following profile is with a 2 core laptop. Nothing spectacular.",
"_____no_output_____"
],
[
"Looking at the profile we can see that most of time is in pow operation (exponential). This probably is that the extent is huge and we are doing it with too much precision. I am working on it",
"_____no_output_____"
],
[
"### GPU",
"_____no_output_____"
]
],
[
[
"%%timeit\n\n# Compute the block\nGeMpy.compute_block_model(geo_data, [0,1,2], verbose = 0)",
"1 loop, best of 3: 1.74 s per loop\n"
],
[
"geo_data.interpolator._interpolate.profile.summary()",
"Function profiling\n==================\n Message: ../GeMpy/DataManagement.py:994\n Time in 3 calls to Function.__call__: 8.400567e-01s\n Time in Function.fn.__call__: 8.395956e-01s (99.945%)\n Time in thunks: 8.275988e-01s (98.517%)\n Total compile time: 3.540267e+00s\n Number of Apply nodes: 342\n Theano Optimizer time: 2.592782e+00s\n Theano validate time: 1.640296e-01s\n Theano Linker time (includes C, CUDA code generation/compiling): 8.665011e-01s\n Import time 1.915064e-01s\n\nTime in all call to theano.grad() 0.000000e+00s\nTime since theano import 72.847s\nClass\n---\n<% time> <sum %> <apply time> <time per call> <type> <#call> <#apply> <Class name>\n 57.3% 57.3% 0.474s 2.87e-03s C 165 55 theano.tensor.elemwise.Elemwise\n 10.1% 67.4% 0.084s 2.79e-03s C 30 10 theano.tensor.blas.Dot22Scalar\n 9.6% 77.0% 0.079s 9.81e-04s C 81 27 theano.sandbox.cuda.basic_ops.HostFromGpu\n 6.4% 83.4% 0.053s 8.89e-03s Py 6 2 theano.tensor.basic.Nonzero\n 6.4% 89.8% 0.053s 1.77e-02s Py 3 1 theano.tensor.nlinalg.MatrixInverse\n 5.1% 95.0% 0.042s 2.01e-03s C 21 7 theano.tensor.elemwise.Sum\n 2.3% 97.2% 0.019s 3.13e-03s C 6 2 theano.sandbox.cuda.basic_ops.GpuAdvancedSubtensor1\n 0.9% 98.1% 0.007s 5.00e-04s C 15 5 theano.tensor.basic.Alloc\n 0.5% 98.6% 0.004s 2.34e-04s C 18 6 theano.sandbox.cuda.basic_ops.GpuAlloc\n 0.5% 99.1% 0.004s 1.43e-04s C 27 9 theano.sandbox.cuda.basic_ops.GpuJoin\n 0.4% 99.5% 0.004s 3.59e-05s C 102 34 theano.sandbox.cuda.basic_ops.GpuElemwise\n 0.2% 99.7% 0.001s 5.48e-05s C 27 9 theano.sandbox.cuda.basic_ops.GpuFromHost\n 0.1% 99.8% 0.001s 1.49e-05s C 66 22 theano.sandbox.cuda.basic_ops.GpuReshape\n 0.0% 99.9% 0.000s 4.41e-05s C 6 2 theano.compile.ops.DeepCopyOp\n 0.0% 99.9% 0.000s 2.63e-06s C 72 24 theano.tensor.subtensor.IncSubtensor\n 0.0% 99.9% 0.000s 2.80e-06s C 48 16 theano.sandbox.cuda.basic_ops.GpuSubtensor\n 0.0% 99.9% 0.000s 1.13e-06s C 114 38 theano.sandbox.cuda.basic_ops.GpuDimShuffle\n 0.0% 99.9% 0.000s 3.96e-05s C 3 1 theano.sandbox.cuda.basic_ops.GpuAllocEmpty\n 0.0% 100.0% 0.000s 3.20e-05s Py 3 1 theano.tensor.extra_ops.FillDiagonal\n 0.0% 100.0% 0.000s 1.23e-06s C 69 23 theano.tensor.elemwise.DimShuffle\n ... (remaining 9 Classes account for 0.03%(0.00s) of the runtime)\n\nOps\n---\n<% time> <sum %> <apply time> <time per call> <type> <#call> <#apply> <Op name>\n 36.2% 36.2% 0.300s 9.98e-02s C 3 1 Elemwise{Composite{(i0 * i1 * LT(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4), i5) * (((i6 + ((i7 * Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4)) / i8)) - ((i9 * Composite{(sqr(i0) * i0)}(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4))) / i10)) + ((i11 * Composite{(sqr(sqr(i0)) * i0)}(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4))) / i12)))}}[(0, 4)]\n 19.2% 55.4% 0.159s 5.30e-02s C 3 1 Elemwise{Composite{(i0 * ((LT(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3), i4) * ((i5 + (i6 * Composite{(sqr(i0) * i0)}((Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3) / i4))) + (i7 * Composite{((sqr(sqr(i0)) * sqr(i0)) * i0)}((Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3) / i4)))) - ((i8 * sqr((Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3) / i4))) + (i9 * Composite{(sqr(sqr(i0)\n 10.1% 65.5% 0.084s 2.79e-03s C 30 10 Dot22Scalar\n 9.6% 75.1% 0.079s 9.81e-04s C 81 27 HostFromGpu\n 6.4% 81.5% 0.053s 8.89e-03s Py 6 2 Nonzero\n 6.4% 88.0% 0.053s 1.77e-02s Py 3 1 MatrixInverse\n 5.0% 92.9% 0.041s 4.58e-03s C 9 3 Sum{axis=[0], acc_dtype=float64}\n 2.3% 95.2% 0.019s 3.13e-03s C 6 2 GpuAdvancedSubtensor1\n 0.9% 96.1% 0.008s 1.27e-03s C 6 2 Elemwise{Mul}[(0, 1)]\n 0.9% 97.0% 0.007s 5.00e-04s C 15 5 Alloc\n 0.6% 97.6% 0.005s 1.72e-03s C 3 1 Elemwise{Composite{((i0 / i1) + ((i2 * i3) / i1) + ((i4 * i2 * i5) / i6))}}[(0, 0)]\n 0.5% 98.1% 0.004s 2.76e-04s C 15 5 GpuAlloc\n 0.5% 98.6% 0.004s 1.43e-04s C 27 9 GpuJoin\n 0.3% 98.9% 0.002s 4.95e-05s C 45 15 GpuElemwise{sub,no_inplace}\n 0.2% 99.1% 0.001s 5.48e-05s C 27 9 GpuFromHost\n 0.2% 99.2% 0.001s 1.08e-04s C 12 4 Elemwise{Cast{float64}}\n 0.1% 99.3% 0.001s 9.13e-05s C 12 4 Sum{axis=[1], acc_dtype=float64}\n 0.1% 99.5% 0.001s 6.88e-05s C 15 5 GpuElemwise{mul,no_inplace}\n 0.1% 99.6% 0.001s 6.47e-05s C 15 5 Elemwise{Sqr}[(0, 0)]\n 0.1% 99.7% 0.001s 1.61e-05s C 60 20 GpuReshape{2}\n ... (remaining 75 Ops account for 0.29%(0.00s) of the runtime)\n\nApply\n------\n<% time> <sum %> <apply time> <time per call> <#call> <id> <Apply name>\n 36.2% 36.2% 0.300s 9.98e-02s 3 332 Elemwise{Composite{(i0 * i1 * LT(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4), i5) * (((i6 + ((i7 * Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4)) / i8)) - ((i9 * Composite{(sqr(i0) * i0)}(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4))) / i10)) + ((i11 * Composite{(sqr(sqr(i0)) * i0)}(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4))) / i12)))}}[(0, 4)](HostFromGpu.0, HostFromGpu.0, Reshape{2\n 19.2% 55.4% 0.159s 5.30e-02s 3 331 Elemwise{Composite{(i0 * ((LT(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3), i4) * ((i5 + (i6 * Composite{(sqr(i0) * i0)}((Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3) / i4))) + (i7 * Composite{((sqr(sqr(i0)) * sqr(i0)) * i0)}((Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3) / i4)))) - ((i8 * sqr((Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3) / i4))) + (i9 * Composite{(sqr(sqr(i0)) * i0)}((C\n 6.4% 61.8% 0.053s 1.77e-02s 3 318 MatrixInverse(IncSubtensor{InplaceSet;int64::, int64:int64:}.0)\n 5.3% 67.1% 0.044s 1.46e-02s 3 180 Nonzero(HostFromGpu.0)\n 5.0% 72.1% 0.042s 1.39e-02s 3 269 Dot22Scalar(Elemwise{Cast{float64}}.0, InplaceDimShuffle{1,0}.0, TensorConstant{2.0})\n 3.8% 75.9% 0.031s 1.04e-02s 3 335 Sum{axis=[0], acc_dtype=float64}(Elemwise{Composite{(i0 * i1 * LT(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4), i5) * (((i6 + ((i7 * Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4)) / i8)) - ((i9 * Composite{(sqr(i0) * i0)}(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4))) / i10)) + ((i11 * Composite{(sqr(sqr(i0)) * i0)}(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i2, i3, i4))) / i12)))}}[(0, 4)].0)\n 3.5% 79.4% 0.029s 9.55e-03s 3 286 HostFromGpu(GpuJoin.0)\n 3.0% 82.3% 0.025s 8.18e-03s 3 329 HostFromGpu(GpuSubtensor{:int64:}.0)\n 2.5% 84.8% 0.021s 6.96e-03s 3 268 Dot22Scalar(Elemwise{Cast{float64}}.0, InplaceDimShuffle{1,0}.0, TensorConstant{2.0})\n 2.5% 87.4% 0.021s 6.93e-03s 3 267 Dot22Scalar(Elemwise{Cast{float64}}.0, InplaceDimShuffle{1,0}.0, TensorConstant{2.0})\n 1.6% 89.0% 0.013s 4.46e-03s 3 216 GpuAdvancedSubtensor1(GpuReshape{1}.0, Subtensor{int64}.0)\n 1.3% 90.3% 0.011s 3.56e-03s 3 328 HostFromGpu(GpuSubtensor{int64:int64:}.0)\n 1.2% 91.4% 0.010s 3.22e-03s 3 200 Nonzero(HostFromGpu.0)\n 0.9% 92.4% 0.008s 2.53e-03s 3 333 Elemwise{Mul}[(0, 1)](HostFromGpu.0, InplaceDimShuffle{1,0}.0, HostFromGpu.0)\n 0.9% 93.3% 0.007s 2.47e-03s 3 235 Alloc(Subtensor{:int64:}.0, Elemwise{Composite{((i0 // i1) + i2)}}[(0, 0)].0, TensorConstant{1}, TensorConstant{1}, Elemwise{Composite{Switch(LT(i0, i1), Switch(LT((i2 + i0), i1), i1, (i2 + i0)), Switch(LT(i0, i2), i0, i2))}}.0)\n 0.9% 94.1% 0.007s 2.36e-03s 3 334 Sum{axis=[0], acc_dtype=float64}(Elemwise{Composite{(i0 * ((LT(Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3), i4) * ((i5 + (i6 * Composite{(sqr(i0) * i0)}((Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3) / i4))) + (i7 * Composite{((sqr(sqr(i0)) * sqr(i0)) * i0)}((Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3) / i4)))) - ((i8 * sqr((Composite{Cast{float32}(sqrt(((i0 + i1) - i2)))}(i1, i2, i3) / i4))) + (i9 * \n 0.6% 94.8% 0.005s 1.79e-03s 3 226 GpuAdvancedSubtensor1(GpuReshape{1}.0, Subtensor{int64}.0)\n 0.6% 95.4% 0.005s 1.72e-03s 3 337 Elemwise{Composite{((i0 / i1) + ((i2 * i3) / i1) + ((i4 * i2 * i5) / i6))}}[(0, 0)](Sum{axis=[0], acc_dtype=float64}.0, InplaceDimShuffle{x}.0, InplaceDimShuffle{x}.0, Sum{axis=[0], acc_dtype=float64}.0, TensorConstant{(1,) of -1.0}, Sum{axis=[0], acc_dtype=float64}.0, InplaceDimShuffle{x}.0)\n 0.6% 96.0% 0.005s 1.69e-03s 3 330 HostFromGpu(GpuSubtensor{int64::}.0)\n 0.5% 96.5% 0.004s 1.37e-03s 3 153 HostFromGpu(GpuReshape{1}.0)\n ... (remaining 322 Apply instances account for 3.51%(0.03s) of the runtime)\n\nHere are tips to potentially make your code run faster\n (if you think of new ones, suggest them on the mailing list).\n Test them first, as they are not guaranteed to always provide a speedup.\n - Try installing amdlibm and set the Theano flag lib.amdlibm=True. This speeds up only some Elemwise operation.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
d05cc06f96fcab5650572e08ff4c30e1330d3c5e | 7,148 | ipynb | Jupyter Notebook | C1W3_L3_CustomLayerWithActivation.ipynb | 100rab-S/TensorFlow-Advanced-Techniques | dad049ba457d928386713b95149af3fccd6d3524 | [
"MIT"
] | 1 | 2021-06-01T07:47:20.000Z | 2021-06-01T07:47:20.000Z | C1W3_L3_CustomLayerWithActivation.ipynb | 100rab-S/TensorFlow-Advanced-Techniques | dad049ba457d928386713b95149af3fccd6d3524 | [
"MIT"
] | null | null | null | C1W3_L3_CustomLayerWithActivation.ipynb | 100rab-S/TensorFlow-Advanced-Techniques | dad049ba457d928386713b95149af3fccd6d3524 | [
"MIT"
] | null | null | null | 35.562189 | 485 | 0.490487 | [
[
[
"<a href=\"https://colab.research.google.com/github/100rab-S/TensorFlow-Advanced-Techniques/blob/main/C1W3_L3_CustomLayerWithActivation.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Ungraded Lab: Activation in Custom Layers\n\nIn this lab, we extend our knowledge of building custom layers by adding an activation parameter. The implementation is pretty straightforward as you'll see below.",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"try:\n # %tensorflow_version only exists in Colab.\n %tensorflow_version 2.x\nexcept Exception:\n pass\n\nimport tensorflow as tf\nfrom tensorflow.keras.layers import Layer",
"_____no_output_____"
]
],
[
[
"## Adding an activation layer\n\nTo use the built-in activations in Keras, we can specify an `activation` parameter in the `__init__()` method of our custom layer class. From there, we can initialize it by using the `tf.keras.activations.get()` method. This takes in a string identifier that corresponds to one of the [available activations](https://keras.io/api/layers/activations/#available-activations) in Keras. Next, you can now pass in the forward computation to this activation in the `call()` method.",
"_____no_output_____"
]
],
[
[
"class SimpleDense(Layer):\n\n # add an activation parameter\n def __init__(self, units=32, activation=None):\n super(SimpleDense, self).__init__()\n self.units = units\n \n # define the activation to get from the built-in activation layers in Keras\n self.activation = tf.keras.activations.get(activation)\n\n\n def build(self, input_shape): # we don't need to change anything in this method to add activation to our custom layer\n w_init = tf.random_normal_initializer()\n self.w = tf.Variable(name=\"kernel\",\n initial_value=w_init(shape=(input_shape[-1], self.units),\n dtype='float32'),\n trainable=True)\n b_init = tf.zeros_initializer()\n self.b = tf.Variable(name=\"bias\",\n initial_value=b_init(shape=(self.units,), dtype='float32'),\n trainable=True)\n #super().build(input_shape)\n\n\n def call(self, inputs):\n \n # pass the computation to the activation layer\n return self.activation(tf.matmul(inputs, self.w) + self.b)",
"_____no_output_____"
]
],
[
[
"We can now pass in an activation parameter to our custom layer. The string identifier is mostly the same as the function name so 'relu' below will get `tf.keras.activations.relu`.",
"_____no_output_____"
]
],
[
[
"mnist = tf.keras.datasets.mnist\n\n(x_train, y_train),(x_test, y_test) = mnist.load_data()\nx_train, x_test = x_train / 255.0, x_test / 255.0\n\nmodel = tf.keras.models.Sequential([\n tf.keras.layers.Flatten(input_shape=(28, 28)),\n SimpleDense(128, activation='relu'),\n tf.keras.layers.Dropout(0.2),\n tf.keras.layers.Dense(10, activation='softmax')\n])\n\nmodel.compile(optimizer='adam',\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])\n\nmodel.fit(x_train, y_train, epochs=5)\nmodel.evaluate(x_test, y_test)",
"Downloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/mnist.npz\n11493376/11490434 [==============================] - 0s 0us/step\nEpoch 1/5\n1875/1875 [==============================] - 5s 2ms/step - loss: 0.4861 - accuracy: 0.8560\nEpoch 2/5\n1875/1875 [==============================] - 4s 2ms/step - loss: 0.1496 - accuracy: 0.9553\nEpoch 3/5\n1875/1875 [==============================] - 4s 2ms/step - loss: 0.1051 - accuracy: 0.9682\nEpoch 4/5\n1875/1875 [==============================] - 4s 2ms/step - loss: 0.0864 - accuracy: 0.9735\nEpoch 5/5\n1875/1875 [==============================] - 4s 2ms/step - loss: 0.0727 - accuracy: 0.9771\n313/313 [==============================] - 1s 1ms/step - loss: 0.0716 - accuracy: 0.9778\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d05cc26d361f44a4fe57555318e6d0feaa44087b | 15,595 | ipynb | Jupyter Notebook | _notebooks/2020-05-12-hosted_kfp_gcf.ipynb | amygdala/fastpages | cc113e9c35f25b0c6a55392346f72cd0a520d45d | [
"Apache-2.0"
] | null | null | null | _notebooks/2020-05-12-hosted_kfp_gcf.ipynb | amygdala/fastpages | cc113e9c35f25b0c6a55392346f72cd0a520d45d | [
"Apache-2.0"
] | 1 | 2021-07-06T18:36:10.000Z | 2021-07-06T18:36:10.000Z | _notebooks/2020-05-12-hosted_kfp_gcf.ipynb | amygdala/fastpages | cc113e9c35f25b0c6a55392346f72cd0a520d45d | [
"Apache-2.0"
] | null | null | null | 41.476064 | 412 | 0.631164 | [
[
[
"# Using Google Cloud Functions to support event-based triggering of Cloud AI Platform Pipelines\n> This post shows how you can run a Cloud AI Platform Pipeline from a Google Cloud Function, providing a way for Pipeline runs to be triggered by events.\n\n- toc: true \n- badges: true\n- comments: true\n- categories: [ml, pipelines, mlops, kfp, gcf]\n",
"_____no_output_____"
],
[
"This example shows how you can run a [Cloud AI Platform Pipeline](https://cloud.google.com/blog/products/ai-machine-learning/introducing-cloud-ai-platform-pipelines) from a [Google Cloud Function](https://cloud.google.com/functions/docs/), thus providing a way for Pipeline runs to be triggered by events (in the interim before this is supported by Pipelines itself). \n\nIn this example, the function is triggered by the addition of or update to a file in a [Google Cloud Storage](https://cloud.google.com/storage/) (GCS) bucket, but Cloud Functions can have other triggers too (including [Pub/Sub](https://cloud.google.com/pubsub/docs/)-based triggers).\n\nThe example is Google Cloud Platform (GCP)-specific, and requires a [Cloud AI Platform Pipelines](https://cloud.google.com/ai-platform/pipelines/docs) installation using Pipelines version >= 0.4. To run this example as a notebook, click on one of the badges at the top of the page or see [here](https://github.com/amygdala/code-snippets/blob/master/ml/notebook_examples/functions/hosted_kfp_gcf.ipynb).\n\n(If you are instead interested in how to do this with a Kubeflow-based pipelines installation, see [this notebook](https://github.com/amygdala/kubeflow-examples/blob/cookbook/cookbook/pipelines/notebooks/gcf_kfp_trigger.ipynb)).\n",
"_____no_output_____"
],
[
"## Setup\n\n### Create a Cloud AI Platform Pipelines installation\n\nFollow the instructions in the [documentation](https://cloud.google.com/ai-platform/pipelines/docs) to create a Cloud AI Platform Pipelines installation. ",
"_____no_output_____"
],
[
"### Identify (or create) a Cloud Storage bucket to use for the example",
"_____no_output_____"
],
[
"**Before executing the next cell**, edit it to **set the `TRIGGER_BUCKET` environment variable** to a Google Cloud Storage bucket ([create a bucket first](https://console.cloud.google.com/storage/browser) if necessary). Do *not* include the `gs://` prefix in the bucket name.\n\nWe'll deploy the GCF function so that it will trigger on new and updated files (blobs) in this bucket.",
"_____no_output_____"
]
],
[
[
"%env TRIGGER_BUCKET=REPLACE_WITH_YOUR_GCS_BUCKET_NAME",
"_____no_output_____"
]
],
[
[
"### Give Cloud Function's service account the necessary access\n\nFirst, make sure the Cloud Function API [is enabled](https://console.cloud.google.com/apis/library/cloudfunctions.googleapis.com?q=functions).\n\nCloud Functions uses the project's 'appspot' acccount for its service account. It will have the form: \n`[email protected]`. (This is also the project's App Engine service account).\n\n- Go to your project's [IAM - Service Account page](https://console.cloud.google.com/iam-admin/serviceaccounts).\n- Find the ` [email protected]` account and copy its email address.\n- Find the project's Compute Engine (GCE) default service account (this is the default account used for the Pipelines installation). It will have a form like this: `[email protected]`.\n Click the checkbox next to the GCE service account, and in the 'INFO PANEL' to the right, click **ADD MEMBER**. Add the Functions service account (`[email protected]`) as a **Project Viewer** of the GCE service account. \n \n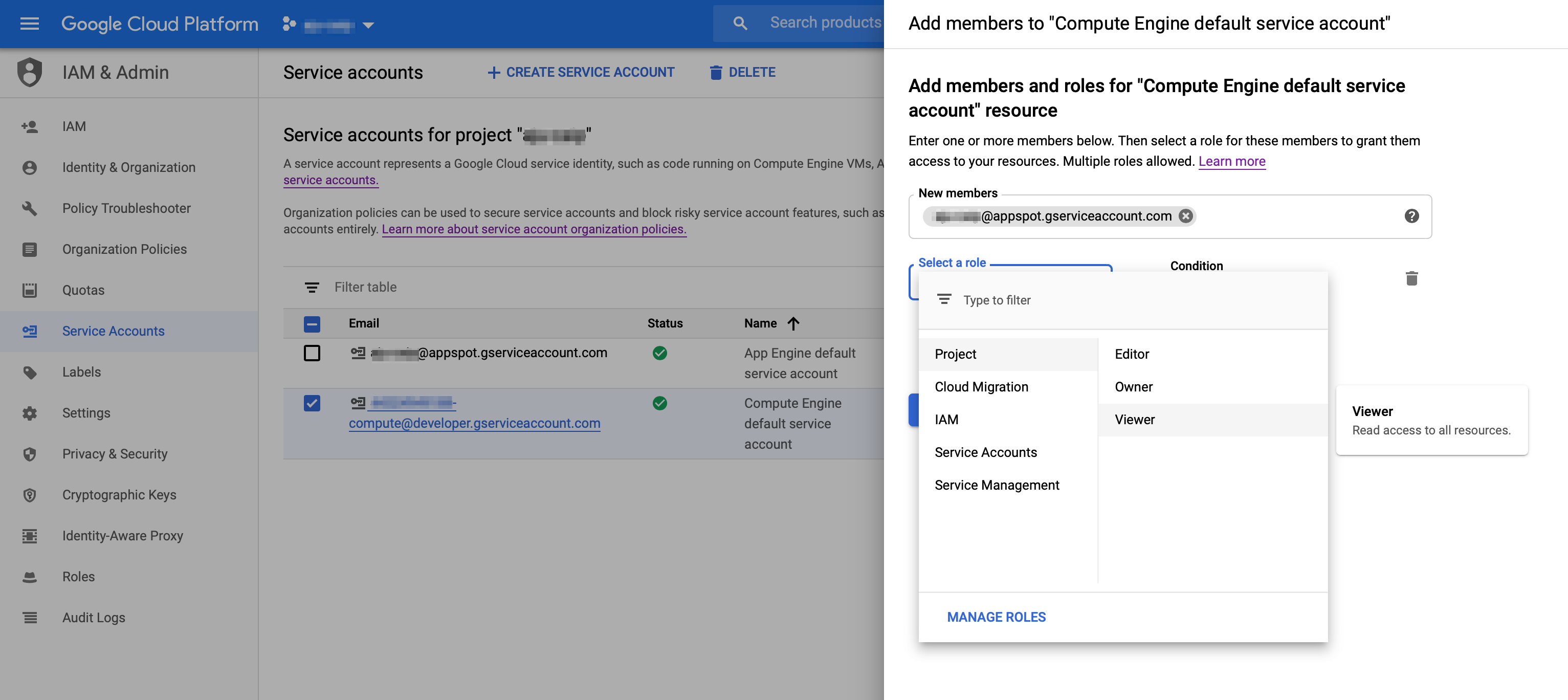 ",
"_____no_output_____"
],
[
"Next, configure your `TRIGGER_BUCKET` to allow the Functions service account access to that bucket. \n\n- Navigate in the console to your list of buckets in the [Storage Browser](https://console.cloud.google.com/storage/browser).\n- Click the checkbox next to the `TRIGGER_BUCKET`. In the 'INFO PANEL' to the right, click **ADD MEMBER**. Add the service account (`[email protected]`) with `Storage Object Admin` permissions. (While not tested, giving both Object view and create permissions should also suffice).\n\n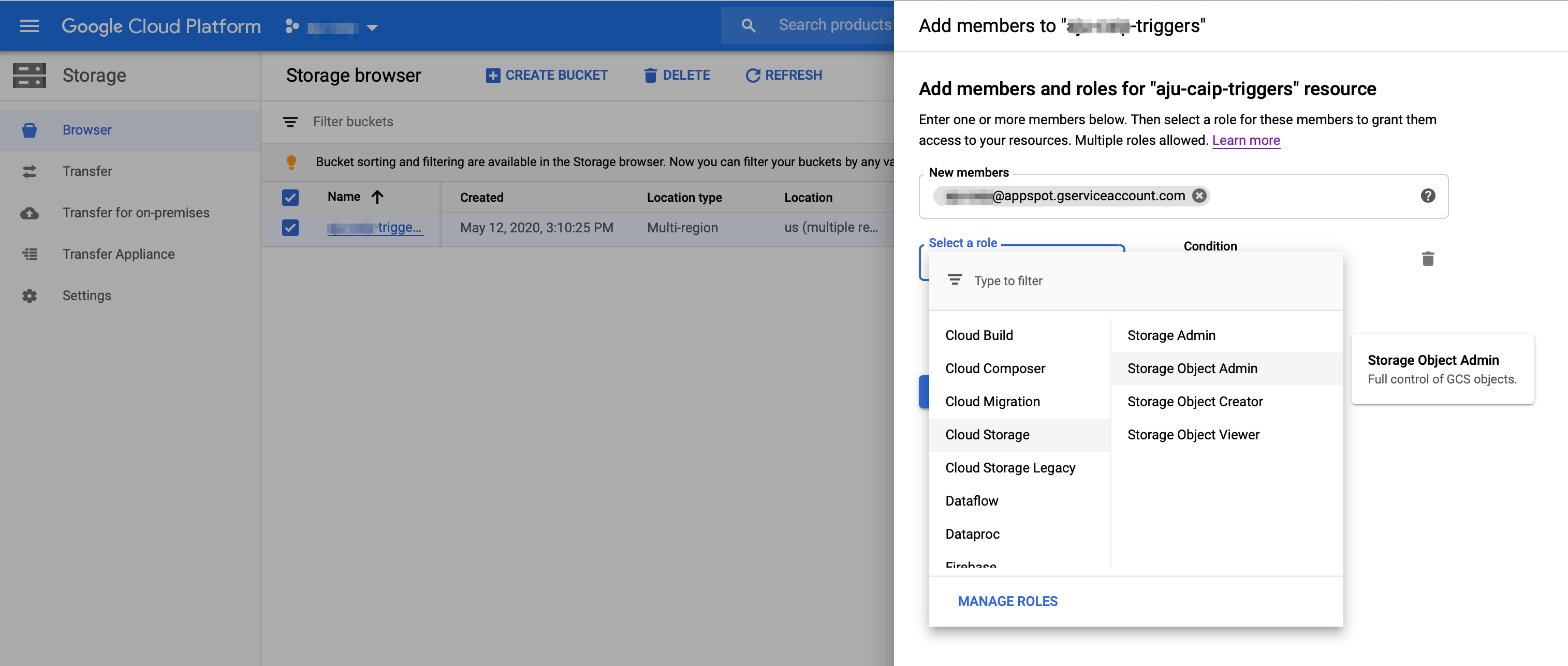",
"_____no_output_____"
],
[
"## Create a simple GCF function to test your configuration\n\nFirst we'll generate and deploy a simple GCF function, to test that the basics are properly configured. ",
"_____no_output_____"
]
],
[
[
"%%bash\nmkdir -p functions",
"_____no_output_____"
]
],
[
[
"We'll first create a `requirements.txt` file, to indicate what packages the GCF code requires to be installed. (We won't actually need `kfp` for this first 'sanity check' version of a GCF function, but we'll need it below for the second function we'll create, that deploys a pipeline).",
"_____no_output_____"
]
],
[
[
"%%writefile functions/requirements.txt\nkfp",
"_____no_output_____"
]
],
[
[
"Next, we'll create a simple GCF function in the `functions/main.py` file:",
"_____no_output_____"
]
],
[
[
"%%writefile functions/main.py\nimport logging\n\ndef gcs_test(data, context):\n \"\"\"Background Cloud Function to be triggered by Cloud Storage.\n This generic function logs relevant data when a file is changed.\n\n Args:\n data (dict): The Cloud Functions event payload.\n context (google.cloud.functions.Context): Metadata of triggering event.\n Returns:\n None; the output is written to Stackdriver Logging\n \"\"\"\n\n logging.info('Event ID: {}'.format(context.event_id))\n logging.info('Event type: {}'.format(context.event_type))\n logging.info('Data: {}'.format(data))\n logging.info('Bucket: {}'.format(data['bucket']))\n logging.info('File: {}'.format(data['name']))\n file_uri = 'gs://%s/%s' % (data['bucket'], data['name'])\n logging.info('Using file uri: %s', file_uri)\n\n logging.info('Metageneration: {}'.format(data['metageneration']))\n logging.info('Created: {}'.format(data['timeCreated']))\n logging.info('Updated: {}'.format(data['updated']))",
"_____no_output_____"
]
],
[
[
"Deploy the GCF function as follows. (You'll need to **wait a moment or two for output of the deployment to display in the notebook**). You can also run this command from a notebook terminal window in the `functions` subdirectory.",
"_____no_output_____"
]
],
[
[
"%%bash\ncd functions\ngcloud functions deploy gcs_test --runtime python37 --trigger-resource ${TRIGGER_BUCKET} --trigger-event google.storage.object.finalize",
"_____no_output_____"
]
],
[
[
"After you've deployed, test your deployment by adding a file to the specified `TRIGGER_BUCKET`. You can do this easily by visiting the **Storage** panel in the Cloud Console, clicking on the bucket in the list, and then clicking on **Upload files** in the bucket details view.\n\nThen, check in the logs viewer panel (https://console.cloud.google.com/logs/viewer) to confirm that the GCF function was triggered and ran correctly. You can select 'Cloud Function' in the first pulldown menu to filter on just those log entries.",
"_____no_output_____"
],
[
"## Deploy a Pipeline from a GCF function\n\nNext, we'll create a GCF function that deploys an AI Platform Pipeline when triggered. First, preserve your existing main.py in a backup file:",
"_____no_output_____"
]
],
[
[
"%%bash\ncd functions\nmv main.py main.py.bak",
"_____no_output_____"
]
],
[
[
"Then, **before executing the next cell**, **edit the `HOST` variable** in the code below. You'll replace `<your_endpoint>` with the correct value for your installation.\n\nTo find this URL, visit the [Pipelines panel](https://console.cloud.google.com/ai-platform/pipelines/) in the Cloud Console. \nFrom here, you can find the URL by clicking on the **SETTINGS** link for the Pipelines installation you want to use, and copying the 'host' string displayed in the client example code (prepend `https://` to that string in the code below). \nYou can alternately click on **OPEN PIPELINES DASHBOARD** for the Pipelines installation, and copy that URL, removing the `/#/pipelines` suffix.",
"_____no_output_____"
]
],
[
[
"%%writefile functions/main.py\nimport logging\nimport datetime\nimport logging\nimport time\n \nimport kfp\nimport kfp.compiler as compiler\nimport kfp.dsl as dsl\n \nimport requests\n \n# TODO: replace with your Pipelines endpoint URL\nHOST = 'https://<your_endpoint>.pipelines.googleusercontent.com'\n\[email protected](\n name='Sequential',\n description='A pipeline with two sequential steps.'\n)\ndef sequential_pipeline(filename='gs://ml-pipeline-playground/shakespeare1.txt'):\n \"\"\"A pipeline with two sequential steps.\"\"\"\n op1 = dsl.ContainerOp(\n name='filechange',\n image='library/bash:4.4.23',\n command=['sh', '-c'],\n arguments=['echo \"%s\" > /tmp/results.txt' % filename],\n file_outputs={'newfile': '/tmp/results.txt'})\n op2 = dsl.ContainerOp(\n name='echo',\n image='library/bash:4.4.23',\n command=['sh', '-c'],\n arguments=['echo \"%s\"' % op1.outputs['newfile']]\n )\n \ndef get_access_token():\n url = 'http://metadata.google.internal/computeMetadata/v1/instance/service-accounts/default/token'\n r = requests.get(url, headers={'Metadata-Flavor': 'Google'})\n r.raise_for_status()\n access_token = r.json()['access_token']\n return access_token\n \ndef hosted_kfp_test(data, context):\n logging.info('Event ID: {}'.format(context.event_id))\n logging.info('Event type: {}'.format(context.event_type))\n logging.info('Data: {}'.format(data))\n logging.info('Bucket: {}'.format(data['bucket']))\n logging.info('File: {}'.format(data['name']))\n file_uri = 'gs://%s/%s' % (data['bucket'], data['name'])\n logging.info('Using file uri: %s', file_uri)\n \n logging.info('Metageneration: {}'.format(data['metageneration']))\n logging.info('Created: {}'.format(data['timeCreated']))\n logging.info('Updated: {}'.format(data['updated']))\n \n token = get_access_token() \n logging.info('attempting to launch pipeline run.')\n ts = int(datetime.datetime.utcnow().timestamp() * 100000)\n client = kfp.Client(host=HOST, existing_token=token)\n compiler.Compiler().compile(sequential_pipeline, '/tmp/sequential.tar.gz')\n exp = client.create_experiment(name='gcstriggered') # this is a 'get or create' op\n res = client.run_pipeline(exp.id, 'sequential_' + str(ts), '/tmp/sequential.tar.gz',\n params={'filename': file_uri})\n logging.info(res)\n\n",
"_____no_output_____"
]
],
[
[
"Next, deploy the new GCF function. As before, **it will take a moment or two for the results of the deployment to display in the notebook**.",
"_____no_output_____"
]
],
[
[
"%%bash\ncd functions\ngcloud functions deploy hosted_kfp_test --runtime python37 --trigger-resource ${TRIGGER_BUCKET} --trigger-event google.storage.object.finalize",
"_____no_output_____"
]
],
[
[
"Add another file to your `TRIGGER_BUCKET`. This time you should see both GCF functions triggered. The `hosted_kfp_test` function will deploy the pipeline. You'll be able to see it running at your Pipeline installation's endpoint, `https://<your_endpoint>.pipelines.googleusercontent.com/#/pipelines`, under the given Pipelines Experiment (`gcstriggered` as default).",
"_____no_output_____"
],
[
"------------------------------------------\nCopyright 2020, Google, LLC.\nLicensed under the Apache License, Version 2.0 (the \"License\");\nyou may not use this file except in compliance with the License.\nYou may obtain a copy of the License at\n\n http://www.apache.org/licenses/LICENSE-2.0\n\nUnless required by applicable law or agreed to in writing, software\ndistributed under the License is distributed on an \"AS IS\" BASIS,\nWITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\nSee the License for the specific language governing permissions and\nlimitations under the License.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d05cc9c0bf5a981c0ddd26a48511ea4bc0b84da7 | 117,637 | ipynb | Jupyter Notebook | Notebooks/python 9 - Herramientas_Estadisticas.ipynb | diegour1/HerramientasComputacionales | 6a2bb36fd7756854d151467e7b4dcc93c0949fc5 | [
"MIT"
] | 2 | 2020-08-31T20:24:15.000Z | 2021-03-26T04:09:18.000Z | Notebooks/python 9 - Herramientas_Estadisticas.ipynb | diegour1/HerramientasComputacionales | 6a2bb36fd7756854d151467e7b4dcc93c0949fc5 | [
"MIT"
] | null | null | null | Notebooks/python 9 - Herramientas_Estadisticas.ipynb | diegour1/HerramientasComputacionales | 6a2bb36fd7756854d151467e7b4dcc93c0949fc5 | [
"MIT"
] | 1 | 2020-11-03T17:47:03.000Z | 2020-11-03T17:47:03.000Z | 150.238825 | 37,236 | 0.893401 | [
[
[
"# Herramientas Estadisticas",
"_____no_output_____"
],
[
"# Contenido:\n\n1.Estadistica:\n\n - Valor medio.\n - Mediana.\n - Desviacion estandar.\n \n\n2.Histogramas:\n \n - Histrogramas con python.\n - Histogramas con numpy.\n - Como normalizar un histograma.\n \n \n3.Distribuciones:\n \n - Como obtener una distribucion a partir de un histograma.\n - Distribucion Normal\n - Distribucion de Poisson\n - Distribucion Binomial",
"_____no_output_____"
],
[
"# 1. Estadistica\n\n## Promedio \n\nEl promedio de una variable $x$ esta definado como:\n\n$\\bar{x} = \\dfrac{\\sum{x_i}}{N} $\n\n## Mediana \n\nLa mediana de un conjunto de datos, es el valor al cual el conjunto de datos\nse divide en dos: \n\nEjemplo: \n\nsea $x$ = [1, 4, 7, 7, 3, 3, 1] la mediana de $median(x) = 3$\n\nFormalmente la mediana se define como el valor $x_m$ que divide la funcion de probabilidad $F(x)$ en partes iguales.\n\n$F(x_m) = \\dfrac{1}{2}$\n\n## El valor mas probable\n\nEs el valor con mayor probabilidad $x_p$.\n\nEjemplo: \n\nsea $x$ = [1, 4, 7, 7, 3, 2, 1] el valor mas probable es $x_p = 7$\n\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"# %pylab inline \n\ndef mi_mediana(lista):\n x = sorted(lista)\n d = int(len(x)/2)\n if(len(x)%2==0):\n return (x[d-1] + x[d])*0.5\n else:\n return x[d-1]\n \nx_input = [1,3,4,5,5,7,7,6,8,6]\nmi_mediana(x_input)\n\nprint(mi_mediana(x_input) == np.median(x_input))",
"True\n"
]
],
[
[
"## Problemas de no saber estadística\n\nEste tipo de conceptos parecen sencillos. Pero no siempre son claros para todo el mundo.",
"_____no_output_____"
]
],
[
[
"x = np.arange(1, 12)\ny = np.random.random(11)*10\nplt.figure(figsize=(12, 5))\nfig = plt.subplot(1, 2, 1)\nplt.scatter(x, y, c='purple', alpha=0.8, s=60)\ny_mean = np.mean(y)\ny_median = np.median(y)\nplt.axhline(y_mean, c='g', lw=3, label=r\"$\\rm{Mean}$\")\nplt.axhline(y_median, c='r', lw=3, label=r\"$\\rm{Median}$\")\nplt.legend(fontsize=20)\nfig = plt.subplot(1, 2, 2)\nh = plt.hist(x, alpha=0.6, histtype='bar', ec='black')\nprint(y)",
"[9.33745032 0.46206052 3.07349261 8.65709198 6.44733954 2.5552359\n 8.93987727 8.24695437 5.62111292 4.64621772 0.05366015]\n"
]
],
[
[
"# Desviacion estandar\n\nEs el promedio de las incertidumbres de las mediciones $x_i$\n\n$\\sigma = \\sqrt{\\dfrac{1}{n-1} \\sum(x_{i} - \\bar{x})^2}$\n\nDonde $n$ es el número de la muestra\n\nAdicionalmente la ${\\bf{varianza}}$ se define como:\n\n$\\bar{x^2} - \\bar{x}^{2}$\n\n$\\sigma^2 = \\dfrac{1}{N} \\sum(x_{i} - \\bar{x})^2$\n\nY es una medida similar a la desviacion estandar que da cuenta de la \ndispersion de los datos alrededor del promedio.\nDonde $N$ es la población total.\n\n# Función de Correlación\n\n$cor(x, y) = \\dfrac{<({(x-\\bar{x})(y-\\bar{y})})>}{\\sigma_x \\sigma_{y}} $\n\n# Ejercicio:\n\nCompruebe si se cumplen las siguientes propiedades:\n\n1. Cor(X,Y) = Cor(Y, X)\n2. Cor(X,X) = 1\n3. Cor(X,-X) = -1\n4. Cor(aX+b, cY + d) = Cor(X, Y), si a y c != 0",
"_____no_output_____"
]
],
[
[
"x = np.arange(1, 12)\ny = np.random.random(11)*10\nplt.figure(figsize=(9, 5))\ny_mean = np.mean(y)\ny_median = np.median(y)\nplt.axhline(y_mean, c='g', lw=3, label=r\"$\\rm{Mean}$\")\nplt.axhline(y_median, c='r', lw=3, label=r\"$\\rm{Median}$\")\nsigma_y = np.std(y)\nplt.axhspan(y_mean-sigma_y, y_mean + sigma_y, facecolor='g', alpha=0.5, label=r\"$\\rm{\\sigma}$\")\nplt.legend(fontsize=20)\nplt.scatter(x, y, c='purple', alpha=0.8, s=60)\nplt.ylim(-2, 14)",
"_____no_output_____"
],
[
"print (\"Variancia = \", np.var(y))\nprint (\"Desviacion estandar = \", np.std(y))",
"Variancia = 7.888849132964844\nDesviacion estandar = 2.8087095138096507\n"
]
],
[
[
"## Referencias: \n \nPara mas funciones estadisticas que se pueden usar en python ver:\n \n- NumPy: http://docs.scipy.org/doc/numpy/reference/routines.statistics.html\n- SciPy: http://docs.scipy.org/doc/scipy/reference/stats.html\n",
"_____no_output_____"
],
[
"# Histogramas",
"_____no_output_____"
],
[
"## 1. hist\n\nhist es una funcion de python que genera un histograma a partir de un array de datos.",
"_____no_output_____"
]
],
[
[
"x = np.random.random(200)\nplt.subplot(2,2,1)\nplt.title(\"A simple hist\")\nh = plt.hist(x)\nplt.subplot(2,2,2)\nplt.title(\"bins\")\nh = plt.hist(x, bins=20)\nplt.subplot(2,2,3)\nplt.title(\"alpha\")\nh = plt.hist(x, bins=20, alpha=0.6)\nplt.subplot(2,2,4)\nplt.title(\"histtype\")\nh = plt.hist(x, bins=20, alpha=0.6, histtype='stepfilled')",
"_____no_output_____"
]
],
[
[
"## 2. Numpy-histogram",
"_____no_output_____"
]
],
[
[
"N, bins = np.histogram(caras, bins=15)",
"_____no_output_____"
],
[
"plt.plot(bins[0:-1], N)",
"_____no_output_____"
]
],
[
[
"# Histogramas 2D",
"_____no_output_____"
]
],
[
[
"x = np.random.random(500)\ny = np.random.random(500)\n\nplt.subplot(4, 2, 1)\nplt.hexbin(x, y, gridsize=15, cmap=\"gray\")\nplt.colorbar()\nplt.subplot(4, 2, 2)\ndata = plt.hist2d(x, y, bins=15, cmap=\"binary\")\nplt.colorbar()\nplt.subplot(4, 2, 3)\nplt.hexbin(x, y, gridsize=15)\nplt.colorbar()\nplt.subplot(4, 2, 4)\ndata = plt.hist2d(x, y, bins=15)\nplt.colorbar()",
"_____no_output_____"
]
],
[
[
"# Como normalizar un histograma. \n\nNormalizar un histograma significa que la integral del histograma sea 1. ",
"_____no_output_____"
]
],
[
[
"x = np.random.random(10)*4\nplt.title(\"Como no normalizar un histograma\", fontsize=25)\nh = plt.hist(x, normed=\"True\")",
"_____no_output_____"
],
[
"print (\"El numero tamaño del bin debe de ser de la unidad\")\nplt.title(\"Como normalizar un histograma\", fontsize=25)\nh = hist(x, normed=\"True\", bins=4)\n",
"_____no_output_____"
]
],
[
[
"Cual es la probabilidad de sacar 9 veces cara en 10 lanzamientos?",
"_____no_output_____"
],
[
"# Distribución de Probabilidad:\n\nLas distribuciones de probabilidad dan información de cual es la probabilidad de que una variable aleatoria $x$ aprezca en un intervalo dado. ¿Si tenemos un conjunto de datos como podemos conocer la distribucion de probabilidad?",
"_____no_output_____"
]
],
[
[
"x = np.random.random(100)*10\nplt.subplot(1, 2, 1)\nh = plt.hist(x)\nplt.subplot(1, 2, 2)\nhisto, bin_edges = np.histogram(x, density=True)\nplt.bar(bin_edges[:-1], histo, width=1)\nplt.xlim(min(bin_edges), max(bin_edges))",
"_____no_output_____"
]
],
[
[
"# Distribución Normal: Descripcion Matemática.\n\n$f(x, \\mu, \\sigma) = \\dfrac{1}{\\sigma \\sqrt(2\\pi)} e^{-\\dfrac{(x-\\mu)^2}{2\\sigma^2}} $\n\ndonde $\\sigma$ es la desviacion estandar y $\\mu$ la media de los datos $x$\n\nEs una función de distribucion de probabilidad que esta totalmente determinada por los parametros $\\mu$ y $\\sigma$. \n\nLa funcion es simetrica alrededor de $\\mu$.\n\nEn python podemos usar scipy para hacer uso de la función normal.",
"_____no_output_____"
]
],
[
[
"import scipy.stats",
"_____no_output_____"
],
[
"x = np.linspace(0, 1, 100)\nn_dist = scipy.stats.norm(0.5, 0.1)\nplt.plot(x, n_dist.pdf(x))",
"_____no_output_____"
]
],
[
[
"## Podemos generar numeros aleatorios con una distribucion normal:",
"_____no_output_____"
]
],
[
[
"x = np.random.normal(0.0, 1.0, 1000)\ny = np.random.normal(0.0, 2.0, 1000)\nw = np.random.normal(0.0, 3.0, 1000)\nz = np.random.normal(0.0, 4.0, 1000)",
"_____no_output_____"
],
[
"histo = plt.hist(z, alpha=0.2, histtype=\"stepfilled\", color='r')\nhisto = plt.hist(w, alpha=0.4, histtype=\"stepfilled\", color='b')\nhisto = plt.hist(y, alpha=0.6, histtype=\"stepfilled\", color='k')\nhisto = plt.hist(x, alpha=0.8, histtype=\"stepfilled\", color='g')\n\nplt.title(r\"$\\rm{Distribuciones\\ normales\\ con\\ diferente\\ \\sigma}$\", fontsize=20)",
"_____no_output_____"
]
],
[
[
"**Intervalo de confianza**\n\n$\\sigma_1$ = 68% de los datos van a estar dentro de 1$\\sigma$\n\n$\\sigma_2$ = 95% de los datos van a estar dentro de 2$\\sigma$\n\n$\\sigma_3$ = 99.7% de los datos van a estar dentro de 3$\\sigma$\n\n\n\n",
"_____no_output_____"
],
[
"### Ejercicio: Generen distribuciones normales con:\n- $\\mu = 5$ y $\\sigma = 2$ \n- $\\mu = -3$ y $\\sigma = -2$\n- $\\mu = 4$ y $\\sigma = 5$\n\n#### Grafiquen las PDF,CDF sobre los mismos ejes, con distintos colores y leyendas. Qué observan? (Una gráfica con PDF y otra con CDF).",
"_____no_output_____"
],
[
"# Ejercicio:\n1. Realize graficas de:\n 1. Diferencia de Caras - Sellos para 40 y 20 mediciones cada una con mayor numero de lanzamientos que la anterior. (abs(cara-sello)vs Numero de lanzamientos)\n 2. La razon (sara/sello) en funcion del Numero de lanzamientos.\n Comente los resultados.\n2. Repita los graficos anteriores pero ahora hagalos en escala logaritmica.\n Comente los resultados.\n3. Haga graficos de el promedio de abs(cara - sello) en funcion del numero de lanzamientos en escala logaritmica.\ny otro con el promedio de (cara/sello).\n Comente los reultados.\n4. Repita el punto anterior pero esta vez con la desviación estandar.\n comente los resultados.",
"_____no_output_____"
],
[
"Imaginemos por un momento el siguiente experimento: \n \nQueremos estudiar la probabilidad de que al lanzar una moneda obtengamos cara o sello, de antamento sabemos que esta es del 50%.\nPero analizemos un poco mas a fondo, ¿Cual será la probabilidad de sacar 10 caras consecutivas?\n\nPara responder proponemos el siguiente método:\n\n1. Lanzamos una moneda 10 veces y miramos si sale cara o sello y guardamos estos datos. \n2. Repetimos este procedimiento y 1000 veces.",
"_____no_output_____"
],
[
"## Funcion que lanza la moneda N veces.",
"_____no_output_____"
]
],
[
[
"def coinflip(N):\n cara = 0\n sello = 0\n i=0\n while i < N:\n x = np.random.randint(0, 10)/5.0\n if x >= 1.0:\n cara+=1\n elif x<1.0:\n sello+=1\n i+=1\n return cara/N, sello/N",
"_____no_output_____"
]
],
[
[
"## Función que hace M veces N lanzamientos.",
"_____no_output_____"
]
],
[
[
"def realizaciones(M, N):\n caras=[]\n for i in range(M):\n x, y = coinflip(N)\n caras.append(x)\n return caras",
"_____no_output_____"
],
[
"hist(caras, normed=True, bins=20)",
"_____no_output_____"
],
[
"caras = realizaciones(100000, 30.)",
"_____no_output_____"
]
],
[
[
"# PDF ",
"_____no_output_____"
]
],
[
[
"N, bins = np.histogram(x, density=True)\nplt.plot(bins[0:-1], N)",
"_____no_output_____"
]
],
[
[
"# CDF",
"_____no_output_____"
]
],
[
[
"h = plt.hist(x, cumulative=True, bins=20)",
"_____no_output_____"
]
],
[
[
"# References: \n- Ejemplo de la Moneda: Introduction to computation and programming using Python. , John Guttag. Pagina 179.\n- Ejemplos de estadistica en python: http://nbviewer.ipython.org/github/dhuppenkothen/ClassicalStatsPython/blob/master/classicalstatsexamples.ipynb\n- Para ver una derivación matematica: A Modern course in Statistical Physics, Reichl, Pagina 191.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d05ce49fbcce982c88436e6ff76cd0ce72993045 | 13,235 | ipynb | Jupyter Notebook | .ipynb_checkpoints/cities_to_html-checkpoint.ipynb | lmm9/Web-Design-Challenge | aa061debd23724ae50f3f2c6fdcf2ab16d2135b1 | [
"ADSL"
] | null | null | null | .ipynb_checkpoints/cities_to_html-checkpoint.ipynb | lmm9/Web-Design-Challenge | aa061debd23724ae50f3f2c6fdcf2ab16d2135b1 | [
"ADSL"
] | null | null | null | .ipynb_checkpoints/cities_to_html-checkpoint.ipynb | lmm9/Web-Design-Challenge | aa061debd23724ae50f3f2c6fdcf2ab16d2135b1 | [
"ADSL"
] | null | null | null | 29.280973 | 89 | 0.345372 | [
[
[
"# Dependencies\nimport pandas as pd",
"_____no_output_____"
],
[
"# Store filepath in a variable\ncity_raw = \"Resources/cities.csv\"",
"_____no_output_____"
],
[
"# Read our Data file with the pandas library\n# Not every CSV requires an encoding, but be aware this can come up\ncity_data = pd.read_csv(city_raw, encoding=\"UTF-8\")",
"_____no_output_____"
],
[
"# Show just the header\ncity_data.head()",
"_____no_output_____"
],
[
"city_data.set_index('City_ID', inplace = True)\ncity_data.head()",
"_____no_output_____"
],
[
"city_data.to_html('city_data.html')",
"_____no_output_____"
],
[
"# Head does not change the DataFrame--it only displays it\nfile_one_df.head()",
"_____no_output_____"
],
[
"# Export file as a CSV, without the Pandas index, but with the header\nfile_one_df.to_csv(\"Output/fileOne.csv\", index=False, header=True)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05d17b2216244d428143732bc63ef70d7f52793 | 314,481 | ipynb | Jupyter Notebook | 04-Aspect_Based_Opinion_Mining/code/01-Build_Model.ipynb | ayan1995/DS_projects | 1ced9cf21c6e41c442470111c6d81e8ecd71eaa0 | [
"Apache-2.0"
] | null | null | null | 04-Aspect_Based_Opinion_Mining/code/01-Build_Model.ipynb | ayan1995/DS_projects | 1ced9cf21c6e41c442470111c6d81e8ecd71eaa0 | [
"Apache-2.0"
] | null | null | null | 04-Aspect_Based_Opinion_Mining/code/01-Build_Model.ipynb | ayan1995/DS_projects | 1ced9cf21c6e41c442470111c6d81e8ecd71eaa0 | [
"Apache-2.0"
] | null | null | null | 88.015953 | 373 | 0.301821 | [
[
[
"# 01 - Sentence Classification Model Building\n",
"_____no_output_____"
],
[
"# Parse & clearn labeled training data",
"_____no_output_____"
]
],
[
[
"import xml.etree.ElementTree as ET\ntree = ET.parse('../data/Restaurants_Train.xml')\nroot = tree.getroot()",
"_____no_output_____"
],
[
"root",
"_____no_output_____"
],
[
"# Use this dataframe for multilabel classification\n# Must use scikitlearn's multilabel binarizer\n\nlabeled_reviews = []\nfor sentence in root.findall(\"sentence\"):\n entry = {}\n aterms = []\n aspects = []\n if sentence.find(\"aspectTerms\"):\n for aterm in sentence.find(\"aspectTerms\").findall(\"aspectTerm\"):\n aterms.append(aterm.get(\"term\"))\n if sentence.find(\"aspectCategories\"):\n for aspect in sentence.find(\"aspectCategories\").findall(\"aspectCategory\"):\n aspects.append(aspect.get(\"category\"))\n entry[\"text\"], entry[\"terms\"], entry[\"aspects\"]= sentence[0].text, aterms, aspects\n labeled_reviews.append(entry)\nlabeled_df = pd.DataFrame(labeled_reviews)\nprint(\"there are\",len(labeled_reviews),\"reviews in this training set\")\n# print(sentence.find(\"aspectCategories\").findall(\"aspectCategory\").get(\"category\"))",
"_____no_output_____"
],
[
"# Save annotated reviews\nlabeled_df.to_pickle(\"annotated_reviews_df.pkl\")\nlabeled_df.head()",
"_____no_output_____"
]
],
[
[
"# Training the model with Naive Bayes\n1. replace pronouns with neural coref\n2. train the model with naive bayes",
"_____no_output_____"
]
],
[
[
"from neuralcoref import Coref\nimport en_core_web_lg\nspacy = en_core_web_lg.load()\ncoref = Coref(nlp=spacy)\n\n# Define function for replacing pronouns using neuralcoref\ndef replace_pronouns(text):\n coref.one_shot_coref(text)\n return coref.get_resolved_utterances()[0]",
"Loading neuralcoref model from /home/ubuntu/anaconda3/lib/python3.6/site-packages/neuralcoref/weights/\nLoading embeddings from /home/ubuntu/anaconda3/lib/python3.6/site-packages/neuralcoref/weights/static_word\nLoading embeddings from /home/ubuntu/anaconda3/lib/python3.6/site-packages/neuralcoref/weights/tuned_word\n"
],
[
"# Read annotated reviews df, which is the labeled dataset for training\n# This is located in the pickled files folder\nannotated_reviews_df = pd.read_pickle(\"../pickled_files/annotated_reviews_df.pkl\")\nannotated_reviews_df.head(3)",
"_____no_output_____"
],
[
"# Create a new column for text whose pronouns have been replaced\nannotated_reviews_df[\"text_pro\"] = annotated_reviews_df.text.map(lambda x: replace_pronouns(x))",
"_____no_output_____"
],
[
"# uncomment below to pickle the new df\n# annotated_reviews_df.to_pickle(\"annotated_reviews_df2.pkl\")\n\n# Read pickled file with replaced pronouns if it exists already\nannotated_reviews_df = pd.read_pickle(\"annotated_reviews_df2.pkl\")",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import MultiLabelBinarizer\n\n# Convert the multi-labels into arrays\nmlb = MultiLabelBinarizer()\ny = mlb.fit_transform(annotated_reviews_df.aspects)\nX = annotated_reviews_df.text_pro\n\n# Split data into train and test set\nX_train, X_test, y_train, y_test = train_test_split(\n X, y, test_size=0.25, random_state=0)\n\n# save the the fitted binarizer labels\n# This is important: it contains the how the multi-label was binarized, so you need to\n# load this in the next folder in order to undo the transformation for the correct labels.\nfilename = 'mlb.pkl'\npickle.dump(mlb, open(filename, 'wb'))",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.feature_extraction.text import TfidfTransformer\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.naive_bayes import MultinomialNB\nfrom skmultilearn.problem_transform import LabelPowerset\nimport numpy as np\n\n# LabelPowerset allows for multi-label classification\n# Build a pipeline for multinomial naive bayes classification\ntext_clf = Pipeline([('vect', CountVectorizer(stop_words = \"english\",ngram_range=(1, 1))),\n ('tfidf', TfidfTransformer(use_idf=False)),\n ('clf', LabelPowerset(MultinomialNB(alpha=1e-1))),])\ntext_clf = text_clf.fit(X_train, y_train)\npredicted = text_clf.predict(X_test)\n\n# Calculate accuracy\nnp.mean(predicted == y_test)",
"_____no_output_____"
],
[
"# Test if SVM performs better\nfrom sklearn.linear_model import SGDClassifier\ntext_clf_svm = Pipeline([('vect', CountVectorizer()),\n ('tfidf', TfidfTransformer()),\n ('clf-svm', LabelPowerset(\n SGDClassifier(loss='hinge', penalty='l2',\n alpha=1e-3, max_iter=6, random_state=42)))])\n_ = text_clf_svm.fit(X_train, y_train)\npredicted_svm = text_clf_svm.predict(X_test)\n\n#Calculate accuracy\nnp.mean(predicted_svm == y_test)",
"_____no_output_____"
],
[
"import pickle\n# Train naive bayes on full dataset and save model\ntext_clf = Pipeline([('vect', CountVectorizer(stop_words = \"english\",ngram_range=(1, 1))),\n ('tfidf', TfidfTransformer(use_idf=False)),\n ('clf', LabelPowerset(MultinomialNB(alpha=1e-1))),])\ntext_clf = text_clf.fit(X, y)\n\n# save the model to disk\nfilename = 'naive_model1.pkl'\npickle.dump(text_clf, open(filename, 'wb'))",
"_____no_output_____"
]
],
[
[
"At this point, we can move on to 02-Sentiment analysis notebook, which will load the fitted Naive bayes model.",
"_____no_output_____"
]
],
[
[
"#mlb.inverse_transform(predicted)\npred_df = pd.DataFrame(\n {'text_pro': X_test,\n 'pred_category': mlb.inverse_transform(predicted)\n })",
"_____no_output_____"
],
[
"pd.set_option('display.max_colwidth', -1)\npred_df.head()",
"_____no_output_____"
]
],
[
[
"## Some scrap code below which wasn't used",
"_____no_output_____"
]
],
[
[
"# Save annotated reviews\nlabeled_df.to_pickle(\"annotated_reviews_df.pkl\")\nlabeled_df.head()",
"_____no_output_____"
],
[
"# This code was for parsing out terms & their relations to aspects\n# However, the terms were not always hyponyms of the aspects, so they were unusable\naspects = {\"food\":[],\"service\":[],\"anecdotes/miscellaneous\":[], \"ambience\":[], \"price\":[]}\nfor i in range(len(labeled_df)):\n if len(labeled_df.aspects[i]) == 1:\n if labeled_df.terms[i] != []:\n for terms in labeled_df.terms[i]:\n aspects[labeled_df.aspects[i][0]].append(terms.lower())\nfor key in aspects:\n aspects[key] = list(set(aspects[key]))",
"_____no_output_____"
],
[
"terms = []\nfor i in labeled_df.terms:\n for j in i:\n if j not in terms:\n terms.append(j)\nprint(\"there are\", len(terms),\"unique terms\")",
"_____no_output_____"
],
[
"# Use this dataframe if doing the classifications separately as binary classifications\nlabeled_reviews2 = []\nfor sentence in root.findall(\"sentence\"):\n entry = {\"food\":0,\"service\":0,\"anecdotes/miscellaneous\":0, \"ambience\":0, \"price\":0}\n aterms = []\n aspects = []\n if sentence.find(\"aspectTerms\"):\n for aterm in sentence.find(\"aspectTerms\").findall(\"aspectTerm\"):\n aterms.append(aterm.get(\"term\"))\n if sentence.find(\"aspectCategories\"):\n for aspect in sentence.find(\"aspectCategories\").findall(\"aspectCategory\"):\n if aspect.get(\"category\") in entry.keys():\n entry[aspect.get(\"category\")] = 1\n entry[\"text\"], entry[\"terms\"] = sentence[0].text, aterms\n labeled_reviews2.append(entry)\nlabeled_df2 = pd.DataFrame(labeled_reviews2)\n# print(sentence.find(\"aspectCategories\").findall(\"aspectCategory\").get(\"category\"))",
"_____no_output_____"
],
[
"labeled_df2.iloc[:,:5].sum()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d05d18e02bd3cd86ee12ef63254b8d638dd43393 | 57,392 | ipynb | Jupyter Notebook | old_versions/1main-v4-MCMC-symmetry.ipynb | danhtaihoang/categorical-variables | 6993315bc9664496896883262ff5bf6125e8d0d0 | [
"MIT"
] | null | null | null | old_versions/1main-v4-MCMC-symmetry.ipynb | danhtaihoang/categorical-variables | 6993315bc9664496896883262ff5bf6125e8d0d0 | [
"MIT"
] | null | null | null | old_versions/1main-v4-MCMC-symmetry.ipynb | danhtaihoang/categorical-variables | 6993315bc9664496896883262ff5bf6125e8d0d0 | [
"MIT"
] | null | null | null | 96.619529 | 20,652 | 0.804485 | [
[
[
"# Network inference of categorical variables: non-sequential data",
"_____no_output_____"
]
],
[
[
"import sys\nimport numpy as np\nfrom scipy import linalg\nfrom sklearn.preprocessing import OneHotEncoder\n\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nimport inference\nimport fem",
"_____no_output_____"
],
[
"# setting parameter:\nnp.random.seed(1)\n\nn = 20 # number of positions\nm = 5 # number of values at each position\nl = int(((n*m)**2)) # number of samples\ng = 2.\n\nnm = n*m",
"_____no_output_____"
],
[
"def itab(n,m): \n i1 = np.zeros(n)\n i2 = np.zeros(n)\n for i in range(n):\n i1[i] = i*m\n i2[i] = (i+1)*m\n\n return i1.astype(int),i2.astype(int)",
"_____no_output_____"
],
[
"# generate coupling matrix w0:\ndef generate_interactions(n,m,g):\n nm = n*m\n w = np.random.normal(0.0,g/np.sqrt(nm),size=(nm,nm))\n i1tab,i2tab = itab(n,m)\n\n for i in range(n): \n i1,i2 = i1tab[i],i2tab[i] \n w[i1:i2,:] -= w[i1:i2,:].mean(axis=0) \n\n for i in range(n):\n i1,i2 = i1tab[i],i2tab[i]\n w[i1:i2,i1:i2] = 0. # no self-interactions\n\n for i in range(nm):\n for j in range(nm):\n if j > i: w[i,j] = w[j,i] \n \n return w",
"_____no_output_____"
],
[
"i1tab,i2tab = itab(n,m)",
"_____no_output_____"
],
[
"w0 = inference.generate_interactions(n,m,g)",
"_____no_output_____"
],
[
"#plt.imshow(w0,cmap='rainbow',origin='lower')\n#plt.clim(-0.5,0.5)\n#plt.colorbar(fraction=0.045, pad=0.05,ticks=[-0.5,0,0.5])\n#plt.show()\n#print(w0)",
"_____no_output_____"
],
[
"def generate_sequences2(w,n,m,l):\n i1tab,i2tab = itab(n,m)\n\n # initial s (categorical variables)\n s_ini = np.random.randint(0,m,size=(l,n)) # integer values\n #print(s_ini)\n\n # onehot encoder \n enc = OneHotEncoder(n_values=m)\n s = enc.fit_transform(s_ini).toarray()\n print(s)\n\n nrepeat = 500\n for irepeat in range(nrepeat):\n for i in range(n):\n i1,i2 = i1tab[i],i2tab[i]\n\n h = s.dot(w[i1:i2,:].T) # h[t,i1:i2]\n h_old = (s[:,i1:i2]*h).sum(axis=1) # h[t,i0]\n k = np.random.randint(0,m,size=l)\n\n for t in range(l):\n if np.exp(h[t,k[t]] - h_old[t]) > np.random.rand():\n s[t,i1:i2] = 0.\n s[t,i1+k[t]] = 1.\n return s ",
"_____no_output_____"
],
[
"# 2018.11.07: Tai\ndef nrgy(s,w):\n l = s.shape[0]\n n,m = 20,3\n \n i1tab,i2tab = itab(n,m)\n p = np.zeros((l,n))\n for i in range(n):\n i1,i2 = i1tab[i],i2tab[i]\n \n h = s.dot(w[i1:i2,:].T)\n #e = (s[:,i1:i2]*h).sum(axis=1)\n \n #p[:,i] = np.exp(e) \n #p_sum = np.sum(np.exp(h),axis=1) \n #p[:,i] /= p_sum\n \n p[:,i] = np.exp((s[:,i1:i2]*h).sum(axis=1))/(np.exp(h).sum(axis=1))\n \n #like = p.sum(axis=1)\n \n return np.sum(np.log(p),axis=1)",
"_____no_output_____"
],
[
"# Vipul:\ndef nrgy_vp(onehot,w):\n nrgy = onehot*(onehot.dot(w.T))\n# print(nrgy - np.log(2*np.cosh(nrgy)))\n return np.sum(nrgy - np.log(2*np.cosh(nrgy)),axis=1) #ln prob",
"_____no_output_____"
],
[
"def generate_sequences_vp(w,n_positions,n_residues,n_seq):\n n_size = n_residues*n_positions\n n_trial = 100*(n_size) #monte carlo steps to find the right sequences\n \n b = np.zeros((n_size))\n\n trial_seq = np.tile(np.random.randint(0,n_residues,size=(n_positions)),(n_seq,1))\n print(trial_seq[0])\n enc = OneHotEncoder(n_values=n_residues)\n onehot = enc.fit_transform(trial_seq).toarray()\n old_nrgy = nrgy(onehot,w) #+ n_positions*(n_residues-1)*np.log(2)\n \n for trial in range(n_trial):\n# print('before',np.mean(old_nrgy))\n index_array = np.random.choice(range(n_positions),size=2,replace=False)\n index,index1 = index_array[0],index_array[1]\n r_trial = np.random.randint(0,n_residues,size=(n_seq))\n\n r_trial1 = np.random.randint(0,n_residues,size=(n_seq))\n \n mod_seq = np.copy(trial_seq)\n mod_seq[:,index] = r_trial\n\n mod_seq[:,index1] = r_trial1\n\n mod_nrgy = nrgy(enc.fit_transform(mod_seq).toarray(),w) #+ n_positions*(n_residues-1)*np.log(2)\n \n seq_change = mod_nrgy-old_nrgy > np.log(np.random.rand(n_seq))\n #seq_change = mod_nrgy/(old_nrgy+mod_nrgy) > np.random.rand(n_seq)\n if trial>n_size:\n trial_seq[seq_change,index] = r_trial[seq_change]\n trial_seq[seq_change,index1] = r_trial1[seq_change]\n old_nrgy[seq_change] = mod_nrgy[seq_change]\n else:\n best_seq = np.argmax(mod_nrgy-old_nrgy)\n trial_seq = np.tile(mod_seq[best_seq],(n_seq,1))\n old_nrgy = np.tile(mod_nrgy[best_seq],(n_seq))\n \n if trial%(10*n_size) == 0: print('after',np.mean(old_nrgy))#,trial_seq[0:5])\n print(trial_seq[:10,:10])\n #return trial_seq\n return enc.fit_transform(trial_seq).toarray()",
"_____no_output_____"
],
[
"s = generate_sequences_vp(w0,n,m,l)",
"[2 4 4 2 1 3 3 3 1 3 0 1 4 0 3 4 4 1 4 1]\n('after', -22.591166261502227)\n('after', -20.250554625306833)\n('after', -20.267284933491265)\n('after', -20.243528271612313)\n('after', -20.223310673851287)\n('after', -20.26470319297705)\n('after', -20.24805766353499)\n('after', -20.25960534241031)\n('after', -20.255155719254432)\n('after', -20.261671663076385)\n[[4 0 3 2 3 3 1 1 0 1]\n [0 3 4 3 0 0 1 4 0 1]\n [0 1 0 4 4 3 2 4 0 2]\n [3 2 0 2 0 1 2 1 1 3]\n [4 4 3 2 1 3 0 1 2 1]\n [4 2 3 0 0 0 4 4 3 2]\n [1 1 2 0 3 1 3 1 3 4]\n [4 4 1 0 1 0 4 4 3 2]\n [3 1 2 2 3 4 4 3 4 4]\n [1 4 4 0 0 4 0 3 2 0]]\n"
],
[
"def generate_sequences_time_series(s_ini,w,n,m): \n i1tab,i2tab = itab(n,m) \n\n l = s_ini.shape[0]\n \n # initial s (categorical variables)\n #s_ini = np.random.randint(0,m,size=(l,n)) # integer values\n #print(s_ini)\n\n # onehot encoder \n enc = OneHotEncoder(n_values=m)\n s = enc.fit_transform(s_ini).toarray()\n #print(s) \n\n ntrial = 20*m\n\n for t in range(l-1):\n h = np.sum(s[t,:]*w[:,:],axis=1)\n for i in range(n):\n i1,i2 = i1tab[i],i2tab[i]\n \n k = np.random.randint(0,m) \n for itrial in range(ntrial): \n k2 = np.random.randint(0,m) \n while k2 == k:\n k2 = np.random.randint(0,m)\n \n if np.exp(h[i1+k2]- h[i1+k]) > np.random.rand():\n k = k2\n \n s[t+1,i1:i2] = 0.\n s[t+1,i1+k] = 1.\n \n return s",
"_____no_output_____"
],
[
"# generate non-sequences from time series\n\n#l1 = 100\n#s_ini = np.random.randint(0,m,size=(l1,n)) # integer values\n\n#s = np.zeros((l,nm))\n#for t in range(l):\n# np.random.seed(t+10)\n# s[t,:] = generate_sequences_time_series(s_ini,w0,n,m)[-1,:] ",
"_____no_output_____"
],
[
"print(s.shape)\nprint(s[:10,:10])",
"(10000, 100)\n[[0. 0. 0. 0. 1. 1. 0. 0. 0. 0.]\n [1. 0. 0. 0. 0. 0. 0. 0. 1. 0.]\n [1. 0. 0. 0. 0. 0. 1. 0. 0. 0.]\n [0. 0. 0. 1. 0. 0. 0. 1. 0. 0.]\n [0. 0. 0. 0. 1. 0. 0. 0. 0. 1.]\n [0. 0. 0. 0. 1. 0. 0. 1. 0. 0.]\n [0. 1. 0. 0. 0. 0. 1. 0. 0. 0.]\n [0. 0. 0. 0. 1. 0. 0. 0. 0. 1.]\n [0. 0. 0. 1. 0. 0. 1. 0. 0. 0.]\n [0. 1. 0. 0. 0. 0. 0. 0. 0. 1.]]\n"
],
[
"## 2018.11.07: for non sequencial data\ndef fit_additive(s,n,m):\n nloop = 10\n i1tab,i2tab = itab(n,m)\n\n nm = n*m\n nm1 = nm - m\n\n w_infer = np.zeros((nm,nm))\n\n for i in range(n):\n i1,i2 = i1tab[i],i2tab[i]\n\n # remove column i\n x = np.hstack([s[:,:i1],s[:,i2:]])\n \n x_av = np.mean(x,axis=0)\n dx = x - x_av\n c = np.cov(dx,rowvar=False,bias=True)\n c_inv = linalg.pinv(c,rcond=1e-15)\n\n #print(c_inv.shape)\n\n h = s[:,i1:i2].copy()\n for iloop in range(nloop):\n h_av = h.mean(axis=0)\n dh = h - h_av\n\n dhdx = dh[:,:,np.newaxis]*dx[:,np.newaxis,:]\n dhdx_av = dhdx.mean(axis=0)\n\n w = np.dot(dhdx_av,c_inv)\n \n #w = w - w.mean(axis=0) \n\n h = np.dot(x,w.T)\n\n p = np.exp(h)\n p_sum = p.sum(axis=1)\n \n #p /= p_sum[:,np.newaxis] \n for k in range(m):\n p[:,k] = p[:,k]/p_sum[:]\n \n h += s[:,i1:i2] - p\n \n w_infer[i1:i2,:i1] = w[:,:i1]\n w_infer[i1:i2,i2:] = w[:,i1:]\n \n return w_infer\n\nw2 = fit_additive(s,n,m)\n\nplt.plot([-1,1],[-1,1],'r--')\nplt.scatter(w0,w2)",
"_____no_output_____"
],
[
"i1tab,i2tab = itab(n,m) \n\nnloop = 5\nnm1 = nm - m\n\nw_infer = np.zeros((nm,nm))\nwini = np.random.normal(0.0,1./np.sqrt(nm),size=(nm,nm1))\n\nfor i in range(n):\n\n i1,i2 = i1tab[i],i2tab[i]\n\n x = np.hstack([s[:,:i1],s[:,i2:]])\n y = s.copy()\n\n # covariance[ia,ib]\n cab_inv = np.empty((m,m,nm1,nm1))\n eps = np.empty((m,m,l))\n for ia in range(m):\n for ib in range(m):\n if ib != ia:\n eps[ia,ib,:] = y[:,i1+ia] - y[:,i1+ib]\n\n which_ab = eps[ia,ib,:] !=0. \n xab = x[which_ab] \n\n # ----------------------------\n xab_av = np.mean(xab,axis=0)\n dxab = xab - xab_av\n cab = np.cov(dxab,rowvar=False,bias=True)\n cab_inv[ia,ib,:,:] = linalg.pinv(cab,rcond=1e-15)\n\n\n w = wini[i1:i2,:].copy() \n for iloop in range(nloop):\n h = np.dot(x,w.T)\n\n for ia in range(m):\n wa = np.zeros(nm1)\n for ib in range(m):\n if ib != ia:\n\n which_ab = eps[ia,ib,:] !=0.\n\n eps_ab = eps[ia,ib,which_ab]\n xab = x[which_ab]\n\n # ----------------------------\n xab_av = np.mean(xab,axis=0)\n dxab = xab - xab_av\n\n h_ab = h[which_ab,ia] - h[which_ab,ib] \n ha = np.divide(eps_ab*h_ab,np.tanh(h_ab/2.), out=np.zeros_like(h_ab), where=h_ab!=0) \n\n dhdx = (ha - ha.mean())[:,np.newaxis]*dxab\n dhdx_av = dhdx.mean(axis=0)\n\n wab = cab_inv[ia,ib,:,:].dot(dhdx_av) # wa - wb\n\n wa += wab\n w[ia,:] = wa/m\n\n w_infer[i1:i2,:i1] = w[:,:i1]\n w_infer[i1:i2,i2:] = w[:,i1:]\n \n #return w_infer",
"_____no_output_____"
],
[
"plt.plot([-1,1],[-1,1],'r--')\nplt.scatter(w0,w_infer)\n#plt.scatter(w0[0:3,3:],w[0:3,:])",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05d1cec27967ea6cde428fa0e21971da403e912 | 565,757 | ipynb | Jupyter Notebook | question3.ipynb | kanishk779/SMAI-2 | 05c332995148a6e0c10ca96018c28a65b04005c7 | [
"MIT"
] | null | null | null | question3.ipynb | kanishk779/SMAI-2 | 05c332995148a6e0c10ca96018c28a65b04005c7 | [
"MIT"
] | null | null | null | question3.ipynb | kanishk779/SMAI-2 | 05c332995148a6e0c10ca96018c28a65b04005c7 | [
"MIT"
] | null | null | null | 487.301464 | 463,144 | 0.929295 | [
[
[
"\n1. You are provided the titanic dataset. Load the dataset and perform splitting into training and test sets with 70:30 ratio randomly using test train split.\n2. Use the Logistic regression created from scratch (from the prev question) in this question as well.\n3. Data cleaning plays a major role in this question. Report all the methods used by you in the ipynb.\n\n--> \n\ni. Check for missing values\n\nii. Drop Columns & Handle missing values\n\niii. Create dummies for categorical features\n\nyou are free to perform other data cleaning to improve your results.\n\n\n\n4. Report accuracy score, Confusion matrix, heat map, classifiaction report and any other metrics you feel useful.",
"_____no_output_____"
],
[
"dataset link : \nhttps://iiitaphyd-my.sharepoint.com/:f:/g/personal/apurva_jadhav_students_iiit_ac_in/Eictt5_qmoxNqezgQQiMWeIBph4sxlfA6jWAJNPnV2SF9Q?e=mQmYN0 \n\n(titanic.csv)\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import accuracy_score, f1_score,confusion_matrix,r2_score\nsns.set(style=\"darkgrid\")",
"_____no_output_____"
],
[
"df = pd.read_csv('titanic.csv')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"print('Missing Values in the columns : \\n')\nprint(df.isnull().sum())",
"Missing Values in the columns : \n\nPassengerId 0\nSurvived 0\nPclass 0\nName 0\nSex 0\nAge 177\nSibSp 0\nParch 0\nTicket 0\nFare 0\nCabin 687\nEmbarked 2\ndtype: int64\n"
],
[
"df.describe(include='all')",
"_____no_output_____"
]
],
[
[
"## Data cleaning\n1. **Removal** :-\n- Remove *Name* column as this attribute does not affect the *Survived* status of the passenger. And moreover we can see that each person has a unique name hence there is no point considering this column.\n- Remove *Ticket* because there are 681 unique values of ticket and moreover if there is some correlation between the ticket and *Survived* status that can be captured by *Fare*.\n- Remove *Cabin* as there are lot of missing values",
"_____no_output_____"
]
],
[
[
"df = df.drop(columns=['Name', 'Ticket', 'Cabin', 'PassengerId'])\ns1 = sns.barplot(data = df, y='Survived' , hue='Sex' , x='Sex')\ns1.set_title('Male-Female Survival')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Females had a better survival rate than male.",
"_____no_output_____"
]
],
[
[
"sns.pairplot(df, hue='Survived')",
"_____no_output_____"
]
],
[
[
"### Categorical data\nFor categorical variables where no ordinal relationship exists, the integer encoding may not be enough, at best, or misleading to the model at worst.\n\nForcing an ordinal relationship via an ordinal encoding and allowing the model to assume a natural ordering between categories may result in poor performance or unexpected results (predictions halfway between categories).\n\nIn this case, a one-hot encoding can be applied to the ordinal representation. This is where the integer encoded variable is removed and one new binary variable is added for each unique integer value in the variable.",
"_____no_output_____"
],
[
"### Dummy Variables\nThe one-hot encoding creates one binary variable for each category.\n\nThe problem is that this representation includes redundancy. For example, if we know that [1, 0, 0] represents “blue” and [0, 1, 0] represents “green” we don’t need another binary variable to represent “red“, instead we could use 0 values for both “blue” and “green” alone, e.g. [0, 0].\n\nThis is called a dummy variable encoding, and always represents C categories with C-1 binary variables.",
"_____no_output_____"
]
],
[
[
"from numpy import mean\ns1 = sns.barplot(data = df, y='Survived' , hue='Embarked' , x='Embarked', estimator=mean)\ns1.set_title('Survival vs Boarding place')\nplt.show()",
"_____no_output_____"
],
[
"carrier_count = df['Embarked'].value_counts()\nsns.barplot(x=carrier_count.index, y=carrier_count.values, alpha=0.9)\nplt.title('Frequency Distribution of Boarding place')\nplt.ylabel('Number of Occurrences', fontsize=12)\nplt.xlabel('Places', fontsize=12)\nplt.show()",
"_____no_output_____"
],
[
"df = pd.get_dummies(df, columns=['Sex', 'Embarked'], prefix=['Sex', 'Embarked'])\ndf.head()",
"_____no_output_____"
],
[
"print('Missing Values in the columns : \\n')\nprint(df.isnull().sum())",
"Missing Values in the columns : \n\nSurvived 0\nPclass 0\nAge 177\nSibSp 0\nParch 0\nFare 0\nSex_female 0\nSex_male 0\nEmbarked_C 0\nEmbarked_Q 0\nEmbarked_S 0\ndtype: int64\n"
],
[
"df = df.fillna(df['Age'].mean())",
"_____no_output_____"
],
[
"print('Missing Values in the columns : \\n')\nprint(df.isnull().sum())",
"Missing Values in the columns : \n\nSurvived 0\nPclass 0\nAge 0\nSibSp 0\nParch 0\nFare 0\nSex_female 0\nSex_male 0\nEmbarked_C 0\nEmbarked_Q 0\nEmbarked_S 0\ndtype: int64\n"
],
[
"df = df.astype(np.float64)",
"_____no_output_____"
],
[
"Y = df['Survived']\nY = np.array(Y)\ndf.drop(columns=['Survived'], inplace=True)",
"_____no_output_____"
],
[
"def standardise(df, col):\n df[col] = (df[col] - df[col].mean())/df[col].std()\n return df\n\nfor col in df.columns:\n df = standardise(df, col)",
"_____no_output_____"
],
[
"import copy\nX = copy.deepcopy(df.to_numpy())\nX.shape",
"_____no_output_____"
],
[
"x_train, x_test, y_train, y_test = train_test_split(X, Y, test_size=0.3, shuffle=True)\nx_train.shape",
"_____no_output_____"
],
[
"class MyLogisticRegression:\n def __init__(self, train_data, Y):\n self.data = train_data # It is assumed that data is normalized and shuffled (rows, cols)\n self.Y = Y[:, np.newaxis]\n self.b = np.random.randn()\n self.cols = self.data.shape[1]\n self.rows = self.data.shape[0]\n self.weights = np.random.randn(self.cols, 1) # Initialising weights to 1, shape (cols, 1)\n self.num_iterations = 600\n self.learning_rate = 0.0001\n self.batch_size = 20\n self.errors = []\n \n @staticmethod\n def sigmoid(x):\n return 1/(1 + np.exp(-x))\n \n def calc_mini_batches(self):\n new_data = np.hstack((self.data, self.Y))\n np.random.shuffle(new_data)\n \n rem = self.rows % self.batch_size\n num = self.rows // self.batch_size\n till = self.batch_size * num\n if num > 0:\n dd = np.array(np.vsplit(new_data[ :till, :], num))\n X_batch = dd[:, :, :-1]\n Y_batch = dd[:, :, -1]\n \n return X_batch, Y_batch\n\n def update_weights(self, X, Y):\n Y_predicted = self.predict(X) # Remember that X has data stored along the row for one sample\n gradient = np.dot(np.transpose(X), Y_predicted - Y)\n self.b = self.b - np.sum(Y_predicted - Y)\n self.weights = self.weights - (self.learning_rate * gradient) # vector subtraction\n \n def print_error(self):\n Y_Predicted = self.predict(self.data)\n class_one = self.Y == 1\n class_two = np.invert(class_one)\n val = np.sum(np.log(Y_Predicted[class_one]))\n val += np.sum(np.log(1 - Y_Predicted[class_two]))\n self.errors.append(-val)\n print(-val)\n \n def gradient_descent(self):\n for j in range(self.num_iterations):\n X, Y = self.calc_mini_batches()\n num_batches = X.shape[0]\n for i in range(num_batches):\n self.update_weights(X[i, :, :], Y[i, :][:, np.newaxis]) # update the weights\n if (j+1)%100 == 0:\n self.print_error()\n plt.plot(self.errors)\n plt.style.use('ggplot')\n plt.xlabel('iteration')\n plt.ylabel('')\n plt.title('Error Vs iteration')\n plt.show()\n \n def predict(self, X):\n # X is 2 dimensional array, samples along the rows\n return self.sigmoid(np.dot(X, self.weights) + self.b)",
"_____no_output_____"
],
[
"reg = MyLogisticRegression(x_train, y_train)\nreg.gradient_descent()\ny_pred = reg.predict(x_test)\npred = y_pred >= 0.5\npred = pred.astype(int)\nprint('accuracy : {a}'.format(a=accuracy_score(y_test, pred)))\nprint('f1 score : {a}'.format(a = f1_score(y_test, pred)))",
"392.59193782039017\n342.7883291939533\n1023.2092246075381\n471.4541619889718\n354.30022553279963\n310.7894615918599\n"
],
[
"confusion_matrix(y_test, pred)",
"_____no_output_____"
],
[
"sns.heatmap(confusion_matrix(y_test, pred))",
"_____no_output_____"
],
[
"from sklearn.metrics import classification_report\nprint(classification_report(y_test, pred))",
" precision recall f1-score support\n\n 0.0 0.82 0.86 0.84 153\n 1.0 0.80 0.75 0.77 115\n\n accuracy 0.81 268\n macro avg 0.81 0.80 0.80 268\nweighted avg 0.81 0.81 0.81 268\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05d1e11bcba0f649830c8095e83921457ae973a | 46,296 | ipynb | Jupyter Notebook | Materials_to_be_Graded/Jupyter_files/datacrimes33556-5y.ipynb | SommerKim/school_grades | 97770e518c9ff5c248687f8cc4cf65879a23edf6 | [
"MIT"
] | 2 | 2021-03-14T19:47:21.000Z | 2021-05-15T14:00:47.000Z | Materials_to_be_Graded/Jupyter_files/datacrimes33556-5y.ipynb | SommerKim/school_grades | 97770e518c9ff5c248687f8cc4cf65879a23edf6 | [
"MIT"
] | null | null | null | Materials_to_be_Graded/Jupyter_files/datacrimes33556-5y.ipynb | SommerKim/school_grades | 97770e518c9ff5c248687f8cc4cf65879a23edf6 | [
"MIT"
] | null | null | null | 54.788166 | 17,944 | 0.625 | [
[
[
"#dependencies\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport datetime\n#file path\ncsv_path = \"../Resources/SA/crimedata-33556-5yr.csv\"\ncrime_data_df = pd.read_csv(csv_path)\ncrime_data_df.head(10)",
"_____no_output_____"
],
[
"#identify columns names used in the data\ncrime_data_df.columns",
"_____no_output_____"
],
[
"#remove some columns from the data \nreduced_crime_data_df = crime_data_df.loc[:, ['Reported Date', 'Case Number', 'Crime Type', 'Crime Type Extension', 'Address'\n , 'zip']]\nreduced_crime_data_df.head(10)",
"_____no_output_____"
],
[
"#columns to use in the table below \ncolumns = [\n \"Reported Date\", \"Case Number\", \"Crime Type\", \"Crime Type Extension\",\"Address\", \"zip\"\n]\n#create a new DF for non Traffic related crimes\ndata_crime_no_traffic_df = reduced_crime_data_df.loc[reduced_crime_data_df[\"Crime Type\"] != \"TRAFFIC\", columns]\ndata_crime_no_traffic_df_33556 = data_crime_no_traffic_df.loc[data_crime_no_traffic_df[\"zip\"] == 33556, columns]\ndata_crime_no_traffic_df_33556.head(10)\n",
"_____no_output_____"
],
[
"#count all non traffic related crimes\ndata_crime_no_traffic_df_33556[\"Case Number\"].count()",
"_____no_output_____"
],
[
"#list of crimes and count\ncrime_type = data_crime_no_traffic_df_33556.groupby(\"Crime Type\")\ncrime_type[\"Case Number\"].count()",
"_____no_output_____"
],
[
"#dates when a crime had been recorded\ncrime_date = data_crime_no_traffic_df_33556.groupby(\"Reported Date\")\ncrime_date[\"Case Number\"].count()",
"_____no_output_____"
],
[
"#build a year columns to count annual crimes on the following cells\ndata_crime_no_traffic_df_33556['year'] = pd.DatetimeIndex(data_crime_no_traffic_df_33556['Reported Date']).year\ndata_crime_no_traffic_df_33556.head()",
"_____no_output_____"
],
[
"#annual count of crimes recorded\ncrime_date_annual = data_crime_no_traffic_df_33556.groupby(\"year\")\ncrime_date_annual[\"Case Number\"].count()",
"_____no_output_____"
],
[
"year =[2016,2017,2018,2019,2020]\ncrimes_count =[228,221,175,149,144]\n\nplt.plot(year,crimes_count)\nplt.title('5 years Crime Trend in the zip code 33556 ')\nplt.xlabel('Years')\nplt.ylabel('Crimes count')\nplt.xticks(rotation=90, horizontalalignment='right', fontweight='light', fontsize='medium',)\nplt.yticks(horizontalalignment='right')\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05d1e95aff566744a12e2251d0b989f72761abe | 1,054 | ipynb | Jupyter Notebook | 2021 Осенний семестр/Практическое задание 4_5/Якимов_Задание 4_5_1.ipynb | mosalov/Notebook_For_AI_Main | a693d29bf0bdcf824cb4f1eca86ff54b67ba7428 | [
"MIT"
] | 6 | 2021-09-20T10:28:18.000Z | 2022-03-14T18:39:17.000Z | 2021 Осенний семестр/Практическое задание 4_5/Якимов_Задание 4_5_1.ipynb | mosalov/Notebook_For_AI_Main | a693d29bf0bdcf824cb4f1eca86ff54b67ba7428 | [
"MIT"
] | 122 | 2020-09-07T11:57:57.000Z | 2022-03-22T06:47:03.000Z | 2021 Осенний семестр/Практическое задание 4_5/Якимов_Задание 4_5_1.ipynb | mosalov/Notebook_For_AI_Main | a693d29bf0bdcf824cb4f1eca86ff54b67ba7428 | [
"MIT"
] | 97 | 2020-09-07T11:32:19.000Z | 2022-03-31T10:27:38.000Z | 17.864407 | 42 | 0.476281 | [
[
[
"import numpy\nimport pandas\nimport matplotlib.pyplot\na=(4,4)\narr=numpy.ones(a)\narr=arr*5\nb=(2,8)\narr=arr.reshape(b)\nprint(arr)",
"[[5. 5. 5. 5. 5. 5. 5. 5.]\n [5. 5. 5. 5. 5. 5. 5. 5.]]\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d05d3954edd476a9554e9f637da60ce035170abf | 21,744 | ipynb | Jupyter Notebook | test/test-aligntts.ipynb | ishine/malaya-speech | fd34afc7107af1656dff4b3201fa51dda54fde18 | [
"MIT"
] | 111 | 2020-08-31T04:58:54.000Z | 2022-03-29T15:44:18.000Z | test/test-aligntts.ipynb | ishine/malaya-speech | fd34afc7107af1656dff4b3201fa51dda54fde18 | [
"MIT"
] | 14 | 2020-12-16T07:27:22.000Z | 2022-03-15T17:39:01.000Z | test/test-aligntts.ipynb | ishine/malaya-speech | fd34afc7107af1656dff4b3201fa51dda54fde18 | [
"MIT"
] | 29 | 2021-02-09T08:57:15.000Z | 2022-03-12T14:09:19.000Z | 113.25 | 1,874 | 0.684511 | [
[
[
"import os\n\nos.environ['CUDA_VISIBLE_DEVICES'] = '1'",
"_____no_output_____"
],
[
"import sys\n\nSOURCE_DIR = os.path.dirname(os.path.dirname(os.path.abspath(__name__)))\nsys.path.insert(0, SOURCE_DIR)",
"_____no_output_____"
],
[
"import tensorflow as tf\n\ntf.compat.v1.enable_eager_execution()",
"_____no_output_____"
],
[
"x = tf.random.normal((2, 20, 80))\nx",
"_____no_output_____"
],
[
"from malaya_speech.train.model.aligntts.layer import FFTransformer",
"WARNING:tensorflow:From /Users/huseinzolkepli/Documents/malaya-speech/malaya_speech/train/optimizer/__init__.py:39: The name tf.train.AdagradOptimizer is deprecated. Please use tf.compat.v1.train.AdagradOptimizer instead.\n\nWARNING:tensorflow:From /Users/huseinzolkepli/Documents/malaya-speech/malaya_speech/train/optimizer/__init__.py:40: The name tf.train.AdamOptimizer is deprecated. Please use tf.compat.v1.train.AdamOptimizer instead.\n\nWARNING:tensorflow:From /Users/huseinzolkepli/Documents/malaya-speech/malaya_speech/train/optimizer/__init__.py:41: The name tf.train.FtrlOptimizer is deprecated. Please use tf.compat.v1.train.FtrlOptimizer instead.\n\nWARNING:tensorflow:From /Users/huseinzolkepli/Documents/malaya-speech/malaya_speech/train/optimizer/__init__.py:43: The name tf.train.RMSPropOptimizer is deprecated. Please use tf.compat.v1.train.RMSPropOptimizer instead.\n\nWARNING:tensorflow:From /Users/huseinzolkepli/Documents/malaya-speech/malaya_speech/train/optimizer/__init__.py:44: The name tf.train.GradientDescentOptimizer is deprecated. Please use tf.compat.v1.train.GradientDescentOptimizer instead.\n\nWARNING:tensorflow:\nThe TensorFlow contrib module will not be included in TensorFlow 2.0.\nFor more information, please see:\n * https://github.com/tensorflow/community/blob/master/rfcs/20180907-contrib-sunset.md\n * https://github.com/tensorflow/addons\n * https://github.com/tensorflow/io (for I/O related ops)\nIf you depend on functionality not listed there, please file an issue.\n\n"
],
[
"fftransformer = FFTransformer(in_out_channels = 80, num_heads = 6)",
"_____no_output_____"
],
[
"fftransformer(x)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05d3f69e294e13b0a5f795cfa5a2a1ef36957b5 | 30,039 | ipynb | Jupyter Notebook | Week05/From NumPy to Logistic Regression.ipynb | HowardNTUST/HackNTU_Data_2017 | ad8e753a16719b6f9396d88b313a5757f5ed4794 | [
"MIT"
] | null | null | null | Week05/From NumPy to Logistic Regression.ipynb | HowardNTUST/HackNTU_Data_2017 | ad8e753a16719b6f9396d88b313a5757f5ed4794 | [
"MIT"
] | null | null | null | Week05/From NumPy to Logistic Regression.ipynb | HowardNTUST/HackNTU_Data_2017 | ad8e753a16719b6f9396d88b313a5757f5ed4794 | [
"MIT"
] | 1 | 2019-02-24T17:41:45.000Z | 2019-02-24T17:41:45.000Z | 32.439525 | 4,724 | 0.635907 | [
[
[
"起手式,導入 numpy, matplotlib",
"_____no_output_____"
]
],
[
[
"from PIL import Image\nimport numpy as np\n%matplotlib inline\nimport matplotlib\nimport matplotlib.pyplot as plt\nmatplotlib.style.use('bmh')\nmatplotlib.rcParams['figure.figsize']=(8,5)",
"_____no_output_____"
]
],
[
[
"使用之前下載的 mnist 資料,載入訓練資料 `train_set` 和測試資料 `test_set`",
"_____no_output_____"
]
],
[
[
"import gzip\nimport pickle\nwith gzip.open('../Week02/mnist.pkl.gz', 'rb') as f:\n train_set, validation_set, test_set = pickle.load(f, encoding='latin1')\n \ntrain_X, train_y = train_set\nvalidation_X, validation_y = validation_set\ntest_X, test_y = test_set",
"_____no_output_____"
]
],
[
[
"之前的看圖片函數",
"_____no_output_____"
]
],
[
[
"from IPython.display import display\ndef showX(X):\n int_X = (X*255).clip(0,255).astype('uint8')\n # N*784 -> N*28*28 -> 28*N*28 -> 28 * 28N\n int_X_reshape = int_X.reshape(-1,28,28).swapaxes(0,1).reshape(28,-1)\n display(Image.fromarray(int_X_reshape))\n# 訓練資料, X 的前 20 筆\nshowX(train_X[:20])",
"_____no_output_____"
]
],
[
[
"train_set 是用來訓練我們的模型用的\n\n我們的模型是很簡單的 logistic regression 模型,用到的參數只有一個 784x10 的矩陣 W 和一個長度 10 的向量 b。\n\n我們先用均勻隨機亂數來設定 W 和 b 。 ",
"_____no_output_____"
]
],
[
[
"W = np.random.uniform(low=-1, high=1, size=(28*28,10))\nb = np.random.uniform(low=-1, high=1, size=10)\n",
"_____no_output_____"
]
],
[
[
"完整的模型如下\n將圖片看成是長度 784 的向量 x\n\n計算 $Wx+b$, 然後再取 $exp$。 最後得到的十個數值。將這些數值除以他們的總和。\n我們希望出來的數字會符合這張圖片是這個數字的機率。\n\n### $ \\Pr(Y=i|x, W, b) = \\frac {e^{W_i x + b_i}} {\\sum_j e^{W_j x + b_j}}$\n",
"_____no_output_____"
],
[
"先拿第一筆資料試試看, x 是輸入。 y 是這張圖片對應到的數字(以這個例子來說 y=5)。",
"_____no_output_____"
]
],
[
[
"x = train_X[0]\ny = train_y[0]\nshowX(x)\ny",
"_____no_output_____"
]
],
[
[
"先計算 $e^{Wx+b} $",
"_____no_output_____"
]
],
[
[
"Pr = np.exp(x @ W + b)\nPr.shape",
"_____no_output_____"
]
],
[
[
"然後 normalize,讓總和變成 1 (符合機率的意義)",
"_____no_output_____"
]
],
[
[
"Pr = Pr/Pr.sum()\nPr",
"_____no_output_____"
]
],
[
[
"由於 $W$ 和 $b$ 都是隨機設定的,所以上面我們算出的機率也是隨機的。\n\n正確解是 $y=5$, 運氣好有可能猜中\n\n為了要評斷我們的預測的品質,要設計一個評斷誤差的方式,我們用的方法如下(不是常見的方差,而是用熵的方式來算,好處是容易微分,效果好)",
"_____no_output_____"
],
[
"\n\n## $ loss = - \\log(\\Pr(Y=y|x, W,b)) $\n",
"_____no_output_____"
],
[
"上述的誤差評分方式,常常稱作 error 或者 loss,數學式可能有點費解。實際計算其實很簡單,就是下面的式子",
"_____no_output_____"
]
],
[
[
"loss = -np.log(Pr[y])\nloss",
"_____no_output_____"
]
],
[
[
"### 想辦法改進。 \n我們用一種被稱作是 gradient descent 的方式來改善我們的誤差。\n\n因為我們知道 gradient 是讓函數上升最快的方向。所以我們如果朝 gradient 的反方向走一點點(也就是下降最快的方向),那麼得到的函數值應該會小一點。\n\n記得我們的變數是 $W$ 和 $b$ (裡面總共有 28*20+10 個變數),所以我們要把 $loss$ 對 $W$ 和 $b$ 裡面的每一個參數來偏微分。\n\n還好這個偏微分是可以用手算出他的形式,而最後偏微分的式子也不會很複雜。",
"_____no_output_____"
],
[
"$loss$ 展開後可以寫成\n$loss = \\log(\\sum_j e^{W_j x + b_j}) - W_i x - b_i$",
"_____no_output_____"
],
[
" 對 $k \\neq i$ 時, $loss$ 對 $b_k$ 的偏微分是 \n $$ \\frac{e^{W_k x + b_k}}{\\sum_j e^{W_j x + b_j}} = \\Pr(Y=k | x, W, b)$$\n對 $k = i$ 時, $loss$ 對 $b_k$ 的偏微分是 \n$$ \\Pr(Y=k | x, W, b) - 1$$\n\n",
"_____no_output_____"
]
],
[
[
"gradb = Pr.copy()\ngradb[y] -= 1\nprint(gradb)",
"[ 1.11201478e-03 2.32129668e-06 3.47186834e-03 3.64416088e-03\n 9.89922844e-01 -9.99616538e-01 4.67890738e-09 3.02581069e-04\n 1.11720864e-07 1.16063080e-03]\n"
]
],
[
[
"對 $W$ 的偏微分也不難\n\n 對 $k \\neq i$ 時, $loss$ 對 $W_{k,t}$ 的偏微分是 \n $$ \\frac{e^{W_k x + b_k} W_{k,t} x_t}{\\sum_j e^{W_j x + b_j}} = \\Pr(Y=k | x, W, b) x_t$$\n對 $k = i$ 時, $loss$ 對 $W_{k,t}$ 的偏微分是 \n$$ \\Pr(Y=k | x, W, b) x_t - x_t$$\n\n",
"_____no_output_____"
]
],
[
[
"print(Pr.shape, x.shape, W.shape)\ngradW = x.reshape(784,1) @ Pr.reshape(1,10)\ngradW[:, y] -= x",
"(10,) (784,) (784, 10)\n"
]
],
[
[
"算好 gradient 後,讓 W 和 b 分別往 gradient 反方向走一點點,得到新的 W 和 b",
"_____no_output_____"
]
],
[
[
"W -= 0.1 * gradW\nb -= 0.1 * gradb",
"_____no_output_____"
]
],
[
[
"再一次計算 $\\Pr$ 以及 $loss$",
"_____no_output_____"
]
],
[
[
"Pr = np.exp(x @ W + b)\nPr = Pr/Pr.sum()\nloss = -np.log(Pr[y])\nloss",
"_____no_output_____"
]
],
[
[
"### Q\n* 看看 Pr , 然後找出機率最大者, predict y 值\n* 再跑一遍上面程序,看看誤差是否變小?\n* 拿其他的測試資料來看看,我們的 W, b 學到了什麼?",
"_____no_output_____"
],
[
"我們將同樣的方式輪流對五萬筆訓練資料來做,看看情形會如何",
"_____no_output_____"
]
],
[
[
"W = np.random.uniform(low=-1, high=1, size=(28*28,10))\nb = np.random.uniform(low=-1, high=1, size=10)\nscore = 0\nN=50000*20\nd = 0.001\nlearning_rate = 1e-2\nfor i in range(N):\n if i%50000==0:\n print(i, \"%5.3f%%\"%(score*100))\n x = train_X[i%50000]\n y = train_y[i%50000]\n Pr = np.exp( x @ W +b)\n Pr = Pr/Pr.sum()\n loss = -np.log(Pr[y])\n score *=(1-d)\n if Pr.argmax() == y:\n score += d\n gradb = Pr.copy()\n gradb[y] -= 1\n gradW = x.reshape(784,1) @ Pr.reshape(1,10)\n gradW[:, y] -= x\n W -= learning_rate * gradW\n b -= learning_rate * gradb\n ",
"0 0.000%\n50000 87.490%\n100000 89.497%\n150000 90.022%\n200000 90.377%\n250000 90.599%\n300000 91.002%\n350000 91.298%\n400000 91.551%\n450000 91.613%\n500000 91.678%\n550000 91.785%\n600000 91.792%\n650000 91.889%\n700000 91.918%\n750000 91.946%\n800000 91.885%\n850000 91.955%\n900000 91.954%\n950000 92.044%\n"
]
],
[
[
"結果發現正確率大約是 92%, 但這是對訓練資料而不是對測試資料\n\n而且,一筆一筆的訓練資也有點慢,線性代數的特點就是能夠向量運算。如果把很多筆 $x$ 當成列向量組合成一個矩陣(然後叫做 $X$),由於矩陣乘法的原理,我們還是一樣計算 $WX+b$ , 就可以同時得到多筆結果。\n\n下面的函數,可以一次輸入多筆 $x$, 同時一次計算多筆 $x$ 的結果和準確率。",
"_____no_output_____"
]
],
[
[
"def compute_Pr(X):\n Pr = np.exp(X @ W + b)\n return Pr/Pr.sum(axis=1, keepdims=True)\ndef compute_accuracy(Pr, y):\n return (Pr.argmax(axis=1)==y).mean()",
"_____no_output_____"
]
],
[
[
"下面是更新過得訓練過程, 當 i%100000 時,順便計算一下 test accuracy 和 valid accuracy。",
"_____no_output_____"
]
],
[
[
"%%timeit -r 1 -n 1\ndef compute_Pr(X):\n Pr = np.exp(X @ W + b)\n return Pr/Pr.sum(axis=1, keepdims=True)\ndef compute_accuracy(Pr, y):\n return (Pr.argmax(axis=1)==y).mean()\n\nW = np.random.uniform(low=-1, high=1, size=(28*28,10))\nb = np.random.uniform(low=-1, high=1, size=10)\nscore = 0\nN=20000\nbatch_size = 128\nlearning_rate = 0.5\nfor i in range(0, N):\n if (i+1)%2000==0: \n test_score = compute_accuracy(compute_Pr(test_X), test_y)*100 \n train_score = compute_accuracy(compute_Pr(train_X), train_y)*100\n print(i+1, \"%5.2f%%\"%test_score, \"%5.2f%%\"%train_score)\n # 隨機選出一些訓練資料出來\n rndidx = np.random.choice(train_X.shape[0], batch_size, replace=False)\n X, y = train_X[rndidx], train_y[rndidx]\n # 一次計算所有的 Pr\n Pr = compute_Pr(X)\n # 計算平均 gradient \n Pr_one_y = Pr-np.eye(10)[y]\n gradb = Pr_one_y.mean(axis=0)\n gradW = X.T @ (Pr_one_y) / batch_size\n # 更新 W 和 ba\n W -= learning_rate * gradW\n b -= learning_rate * gradb",
"2000 90.50% 90.47%\n4000 91.17% 91.56%\n6000 91.72% 92.03%\n8000 91.86% 92.25%\n10000 92.03% 92.52%\n12000 92.14% 92.88%\n14000 92.34% 92.81%\n16000 92.29% 92.99%\n18000 92.18% 93.13%\n20000 92.06% 93.12%\n1 loop, best of 1: 1min 8s per loop\n"
]
],
[
[
"最後得到的準確率是 92%-93%\n\n不算完美,不過畢竟這只有一個矩陣而已。",
"_____no_output_____"
],
[
"光看數據沒感覺,我們來看看前十筆測試資料跑起來的情形\n\n可以看到前十筆只有錯一個",
"_____no_output_____"
]
],
[
[
"Pr = compute_Pr(test_X[:10])\npred_y =Pr.argmax(axis=1)\nfor i in range(10):\n print(pred_y[i], test_y[i])\n showX(test_X[i])",
"7 7\n"
]
],
[
[
"看看前一百筆資料中,是哪些情況算錯",
"_____no_output_____"
]
],
[
[
"Pr = compute_Pr(test_X[:100])\npred_y = Pr.argmax(axis=1)\nfor i in range(100):\n if pred_y[i] != test_y[i]:\n print(pred_y[i], test_y[i])\n showX(test_X[i])",
"6 5\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d05d40dbfbd13186a9d265dcfb001ffd18e9c9ee | 13,440 | ipynb | Jupyter Notebook | widgets.ipynb | satish-annigeri/Notebooks | 92a7dc1d4cf4aebf73bba159d735a2e912fc88bb | [
"CC0-1.0"
] | null | null | null | widgets.ipynb | satish-annigeri/Notebooks | 92a7dc1d4cf4aebf73bba159d735a2e912fc88bb | [
"CC0-1.0"
] | null | null | null | widgets.ipynb | satish-annigeri/Notebooks | 92a7dc1d4cf4aebf73bba159d735a2e912fc88bb | [
"CC0-1.0"
] | null | null | null | 69.278351 | 7,056 | 0.75506 | [
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt\n\ndef plot(amplitude, color):\n fig, ax = plt.subplots(figsize=(4,3 ), subplot_kw={'axisbg': '#EEEEEE', 'axisbelow': True})\n ax.grid(color='w', linewidth=2, linestyle='solid')\n x = np.linspace(0, 10, 1000)\n ax.plot(x, amplitude * np.sin(x), color=color, lw=5, alpha=0.4)\n ax.set_xlim(0, 10)\n ax.set_ylim(-1.1, 1.1)\n return fig\n\nfig = plot(0.1, 'blue')\nplt.show()\n\nfrom IPython.html.widgets import interact, FloatSliderWidget, RadioButtonsWidget\n\n#interact(plot, amplitude=FloatSliderWidget(0.1, 1.0, 0.1), color=RadioButtonsWidget(['blue', 'green', 'red']))",
"_____no_output_____"
],
[
"from IPython.html.widgets import interact, interactive, fixed\nfrom IPython.html import widgets\nfrom IPython.display import clear_output, display, HTML\n\ndef show_args(**kwargs):\n s = '<h3>Arguments:</h3><table>\\n'\n for k,v in kwargs.items():\n s += '<tr><td>{0}</td><td>{1}</td></tr>\\n'.format(k,v)\n s += '</table>'\n display(HTML(s))\n return\n\nshow_args(a=10, b='Hi There', c=True)",
"_____no_output_____"
],
[
"i = interact(show_args,\n Temp=(0,10),\n Current=(0.,10.,0.01),\n z=True,\n Text=u'Type here!',\n #Algorithm=['This','That','Other'],\n a=widgets.FloatSliderWidget(min=-10.0, max=10.0, step=0.1, value=5.0)\n )",
"_____no_output_____"
],
[
"i.widget",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d05d5137720080c10b6473a0594d04cafd439fa1 | 7,905 | ipynb | Jupyter Notebook | Notebooks/00.ipynb | eunicenjuguna/Python4Bioinformatics2020 | 90dc206e05a285f783103ba9add48930bb2dbcde | [
"CC-BY-4.0"
] | null | null | null | Notebooks/00.ipynb | eunicenjuguna/Python4Bioinformatics2020 | 90dc206e05a285f783103ba9add48930bb2dbcde | [
"CC-BY-4.0"
] | null | null | null | Notebooks/00.ipynb | eunicenjuguna/Python4Bioinformatics2020 | 90dc206e05a285f783103ba9add48930bb2dbcde | [
"CC-BY-4.0"
] | null | null | null | 47.335329 | 434 | 0.65642 | [
[
[
"# Python For Bioinformatics\n\nIntroduction to Python for Bioinformatics - available at https://github.com/kipkurui/Python4Bioinformatics.\n\n<small><small><i>\n\n## Attribution\nThese tutorials are an adaptation of the Introduction to Python for Maths by [Andreas Ernst](http://users.monash.edu.au/~andreas), available from https://gitlab.erc.monash.edu.au/andrease/Python4Maths.git. The original version was written by Rajath Kumar and is available at https://github.com/rajathkumarmp/Python-Lectures.\n\nThese notes have been greatly amended and updated for the MSC Bioinformatics and Molecular Biology at Pwani university, sponsored by EANBiT by [Caleb Kibet](https://twitter.com/calkibet)\n</small></small></i>\n\n# Quick Introduction to Jupyter Notebooks\n\nThroughout this course, we will be using Jupyter Notebooks. Although the HPC you will be using will have Jupyter setup, these notes are provided for you want to set it up in your Computer. \n\n## Introduction\nThe Jupyter Notebook is an interactive computing environment that enables users to author notebooks, which contain a complete and self-contained record of a computation. These notebooks can be shared more efficiently. The notebooks may contain:\n* Live code\n* Interactive widgets\n* Plots\n* Narrative text\n* Equations\n* Images\n* Video\n\nIt is good to note that \"Jupyter\" is a loose acronym meaning Julia, Python, and R; the primary languages supported by Jupyter. \n\nThe notebook can allow a computational researcher to create reproducible documentation of their research. As Bioinformatics is datacentric, use of Jupyter Notebooks increases research transparency, hence promoting open science. \n\n## First Steps\n\n### Installation\n\n1. [Download Miniconda](https://www.anaconda.com/download/) for your specific OS to your home directory\n - Linux: `wget https://repo.continuum.io/miniconda/Miniconda3-latest-Linux-x86_64.sh`\n - Mac: `curl https://repo.continuum.io/miniconda/Miniconda3-latest-MacOSX-x86_64.sh`\n2. Run:\n - `bash Miniconda3-latest-Linux-x86_64.sh`\n - `bash Miniconda3-latest-MacOSX-x86_64.sh`\n3. Follow all the prompts: if unsure, accept defaults\n4. Close and re-open your terminal\n5. If the installation is successful, you should see a list of installed packages with\n - `conda list`\nIf the command cannot be found, you can add Anaconda bin to the path using:\n ` export PATH=~/anaconda3/bin:$PATH`\n\nFor reproducible analysis, you can [create a conda environment](https://conda.io/docs/user-guide/tasks/manage-environments.html) with all the Python packages you used.\n\n `conda create --name bioinf python jupyter`\n \nTo activate the conda environment:\n `source activate bioinf`\n\nHaving set-up conda environment, you can install any package you need using pip. \n\n`conda install jupyter`\n`conda install -c conda-forge jupyterlab`\n\nor by using pip\n\n`pip3 install jupyter`\n\nThen you can quickly launch it using:\n\n`jupyter notebook` or `jupyter lab`\n\nNB: We will use a jupyter lab for training. \n\n\nA Jupyter notebook is made up of many cells. Each cell can contain Python code. You can execute a cell by clicking on it and pressing `Shift-Enter` or `Ctrl-Enter` (run without moving to the next line). \n\n### Further help\n\nTo learn more about Jupyter notebooks, check [the official introduction](http://nbviewer.jupyter.org/github/jupyter/notebook/blob/master/docs/source/examples/Notebook/Notebook%20Basics.ipynb) and [some useful Jupyter Tricks](https://www.dataquest.io/blog/jupyter-notebook-tips-tricks-shortcuts/). \n\nBook: http://www.ict.ru.ac.za/Resources/cspw/thinkcspy3/thinkcspy3.pdf\n\n# Python for Bioinformatics\n\n## Introduction\n\nPython is a modern, robust, high-level programming language. It is straightforward to pick up even if you are entirely new to programming. \n\nPython, similar to other languages like Matlab or R, is interpreted hence runs slowly compared to C++, Fortran or Java. However, writing programs in Python is very quick. Python has an extensive collection of libraries for everything from scientific computing to web services. It caters for object-oriented and functional programming with a module system that allows large and complex applications to be developed in Python. \n\nThese lectures are using Jupyter notebooks which mix Python code with documentation. The python notebooks can be run on a web server or stand-alone on a computer.\n\n\n## Contents\n\nThis course is broken up into a number of notebooks (lectures).\n### Session 1\n* [01](01.ipynb) Basic data types and operations (numbers, strings) \n* [02](02.ipynb) String manipulation \n\n### Session 2\n* [03](03.ipynb) Data structures: Lists and Tuples\n* [04](04.ipynb) Data structures (continued): dictionaries\n\n### Session 3\n* [05](05.ipynb) Control statements: if, for, while, try statements\n* [06](06.ipynb) Functions\n* [07](07.ipynb) Files, Scripting and Modules\n### Session 4\n* [08](08.ipynb) Data Analysis and plotting with Pandas\n* [09](09.ipynb) Reproducible Bioinformatics Research\n* [10](10.ipynb) Introduction to Biopython\n\nThis is a tutorial style introduction to Python. For a quick reminder/summary of Python syntax, the following [Quick Reference Card](http://www.cs.put.poznan.pl/csobaniec/software/python/py-qrc.html) may be useful. A longer and more detailed tutorial style introduction to python is available from the python site at: https://docs.python.org/3/tutorial/.\n\n\n## How to learn from this resource?\n\nDownload all the notebooks from [Python4Bioinformatics](https://github.com/kipkurui/Python4Bioinformatics2019). The easiest way to do that is to clone the GitHub repository to your working directory using any of the following commands:\n\n git clone https://github.com/kipkurui/Python4Bioinformatics2019.git\n\nor\n\n wget https://github.com/kipkurui/Python4Bioinformatics2019/archive/master.zip\n \n unzip master.zip\n \n rm master.zip\n \n\n## How to Contribute\n\nTo contribute, fork the repository, make some updates and send me a pull request. \n\nAlternatively, you can open an issue. \n\n## License\nThis work is licensed under the Creative Commons Attribution 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/.\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
d05d53e23b5bc4989a0dcc089a01a5f02055a4f2 | 482,825 | ipynb | Jupyter Notebook | Untitled2.ipynb | kialio/FlareFinder | 503db208df35a5a1f0958b875d5f55c7105b5abf | [
"MIT"
] | null | null | null | Untitled2.ipynb | kialio/FlareFinder | 503db208df35a5a1f0958b875d5f55c7105b5abf | [
"MIT"
] | null | null | null | Untitled2.ipynb | kialio/FlareFinder | 503db208df35a5a1f0958b875d5f55c7105b5abf | [
"MIT"
] | null | null | null | 136.893961 | 1,695 | 0.475787 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d05d5c04ab1559acd58045c253ae36255efa6073 | 35,459 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Skorodumov.Lab2-checkpoint.ipynb | SkorodumovAlex/SIAODLabs | 13617860163e065b221ab59ad774ee4df706e6b8 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Skorodumov.Lab2-checkpoint.ipynb | SkorodumovAlex/SIAODLabs | 13617860163e065b221ab59ad774ee4df706e6b8 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Skorodumov.Lab2-checkpoint.ipynb | SkorodumovAlex/SIAODLabs | 13617860163e065b221ab59ad774ee4df706e6b8 | [
"MIT"
] | null | null | null | 66.278505 | 8,376 | 0.576102 | [
[
[
"Скородумов Александр",
"_____no_output_____"
],
[
"БВТ1904",
"_____no_output_____"
],
[
"Лабораторная работа №2 Методы поиска\n",
"_____no_output_____"
],
[
"№1",
"_____no_output_____"
]
],
[
[
"#Импорты\nfrom IPython.display import HTML, display\nfrom tabulate import tabulate\nimport random\nimport time",
"_____no_output_____"
],
[
"#Рандомная генерация\ndef random_matrix(m = 50, n = 50, min_limit = -250, max_limit = 1016):\n return [[random.randint(min_limit, max_limit) for _ in range(n)] for _ in range(m)]",
"_____no_output_____"
],
[
"#Бинарный поиск\nclass BinarySearchMap:\n def __init__(self):\n self.data = [] # хранилище (key, value) значений\n\n def search(self, key):\n \"\"\" Поиск индекса (во всех случаях лучше левосторонний,\n чтоб insert вставлял по убыванию) \"\"\"\n l = 0\n r = len(self.data)\n while l < r:\n m = (l + r) // 2\n if self.data[m][0] < key:\n l = m + 1\n else:\n r = m\n return l \n \n def __setitem__(self, key, value):\n \"\"\" Добавить элемент \"\"\"\n index = self.search(key)\n # если ключ уже есть в таблице, то надо заменить значение\n if index < len(self.data) and self.data[index][0] == key:\n self.data[index] = (key, value)\n else:\n # иначе добавляем новую запись\n self.data.insert(index, (key, value))\n \n def __delitem__(self, key):\n \"\"\" Удалить элемент \"\"\"\n index = self.search(key)\n self.data.pop(index)\n \n def __getitem__(self, key):\n \"\"\" Получить элемент \"\"\"\n index = self.search(key)\n found_key, val = self.data[index]\n # если найденный индекс выдает запрашиваемый ключ\n if found_key == key:\n return val\n raise KeyError()\n",
"_____no_output_____"
],
[
"#Фибоначчиев поиск\nfib_c = [0, 1]\ndef fib(n):\n if len(fib_c) - 1 < n:\n fib_c.append(fib(n - 1) + fib(n - 2))\n return fib_c[n]\n\nclass FibonacciMap(BinarySearchMap):\n def search(self, key):\n m = 0 \n while fib(m) < len(self.data): \n m += 1\n offset = 0\n while fib(m) > 1:\n i = min(offset + fib(m - 1), len(self.data) - 1)\n if key > self.data[i][0]:\n offset = i\n elif key == self.data[i][0]:\n return i\n m -= 1\n if len(self.data) and self.data[offset][0] < key:\n return offset + 1\n return 0",
"_____no_output_____"
],
[
"#Интерполяционный поиск\ndef nearest_mid(input_list, lower_bound_index, upper_bound_index, search_value):\n return lower_bound_index + \\\n (upper_bound_index - lower_bound_index) * \\\n (search_value - input_list[lower_bound_index]) // \\\n (input_list[upper_bound_index][0] - input_list[lower_bound_index][0])\n\nclass InterpolateMap(BinarySearchMap):\n def interpolation_search(self, term):\n size_of_list = len(self.data) - 1\n\n index_of_first_element = 0\n index_of_last_element = size_of_list\n\n while index_of_first_element <= index_of_last_element:\n mid_point = nearest_mid(self.data, index_of_first_element, index_of_last_element, term)\n\n if mid_point > index_of_last_element or mid_point < index_of_first_element:\n return None\n\n if self.data[mid_point][0] == term:\n return mid_point\n\n if term > self.data[mid_point][0]:\n index_of_first_element = mid_point + 1\n else:\n index_of_last_element = mid_point - 1\n\n if index_of_first_element > index_of_last_element:\n return None",
"_____no_output_____"
],
[
"#Бинарное дерево\nclass Tree:\n def __init__(self, key, value):\n self.key = key\n self.value = value\n self.left = self.right = None\n \nclass BinaryTreeMap:\n root = None\n \n def insert(self, tree, key, value):\n if tree is None:\n return Tree(key, value)\n if tree.key > key:\n tree.left = self.insert(tree.left, key, value)\n elif tree.key < key:\n tree.right = self.insert(tree.right, key, value)\n else:\n tree.value = value\n return tree\n \n def search(self, tree, key):\n if tree is None or tree.key == key:\n return tree\n if tree.key > key:\n return self.search(tree.left, key)\n return self.search(tree.right, key)\n \n def __getitem__(self, key):\n tree = self.search(self.root, key)\n if tree is not None:\n return tree.value\n raise KeyError()\n \n def __setitem__(self, key, value):\n if self.root is None:\n self.root = self.insert(self.root, key, value)\n else: self.insert(self.root, key, value)",
"_____no_output_____"
]
],
[
[
"№2",
"_____no_output_____"
]
],
[
[
"#Простое рехеширование\nclass HashMap:\n def __init__(self):\n self.size = 0\n self.data = []\n self._resize()\n \n def _hash(self, key, i):\n return (hash(key) + i) % len(self.data)\n \n def _find(self, key):\n i = 0;\n index = self._hash(key, i);\n while self.data[index] is not None and self.data[index][0] != key:\n i += 1\n index = self._hash(key, i);\n return index;\n \n def _resize(self):\n temp = self.data\n self.data = [None] * (2*len(self.data) + 1)\n for item in temp:\n if item is not None:\n self.data[self._find(item[0])] = item\n \n def __setitem__(self, key, value):\n if self.size + 1 > len(self.data) // 2:\n self._resize()\n index = self._find(key)\n if self.data[index] is None: \n self.size += 1\n self.data[index] = (key, value)\n \n def __getitem__(self, key):\n index = self._find(key)\n if self.data[index] is not None:\n return self.data[index][1]\n raise KeyError()",
"_____no_output_____"
],
[
"#Рехеширование с помощью псевдослучайных чисел\nclass RandomHashMap(HashMap):\n _rand_c = [5323]\n \n def _rand(self, i):\n if len(self._rand_c) - 1 < i:\n self._rand_c.append(self._rand(i - 1))\n return (123456789 * self._rand_c[i] + 987654321) % 65546\n \n def _hash(self, key, i):\n return (hash(key) + self._rand(i)) % len(self.data)",
"_____no_output_____"
],
[
"#Метод Цепочек\nclass ChainMap:\n def __init__(self):\n self.size = 0\n self.data = []\n self._resize()\n \n def _hash(self, key):\n return hash(key) % len(self.data)\n \n def _insert(self, index, item):\n if self.data[index] is None:\n self.data[index] = [item]\n return True\n else:\n for i, item_ in enumerate(self.data[index]):\n if item_[0] == item[0]:\n self.data[index][i] = item\n return False\n self.data[index].append(item)\n return True\n \n def _resize(self):\n temp = self.data\n self.data = [None] * (2*len(self.data) + 1)\n for bucket in temp:\n if bucket is not None:\n for key, value in bucket:\n self._insert(self._hash(key), (key, value))\n \n def __setitem__(self, key, value):\n if self.size + 1 > len(self.data) // 1.5:\n self._resize()\n if self._insert(self._hash(key), (key, value)): \n self.size += 1\n \n def __getitem__(self, key):\n index = self._hash(key)\n if self.data[index] is not None:\n for key_, value in self.data[index]:\n if key_ == key:\n return value\n raise KeyError()",
"_____no_output_____"
]
],
[
[
"Сравнение алгоритмов",
"_____no_output_____"
]
],
[
[
"алгоритмы = {\n 'Бинарный поиск': BinarySearchMap,\n 'Фибоначчиева поиск': FibonacciMap,\n 'Интерполяционный поиск': InterpolateMap,\n 'Бинарное дерево': BinaryTreeMap,\n 'Простое рехэширование': HashMap,\n 'Рехэширование с помощью псевдослучайных чисел': RandomHashMap,\n 'Метод цепочек': ChainMap,\n 'Стандартная функция поиска': dict\n}\n\n\nзатраченное_время = {}\nтестовые_набор = random_matrix(50, 1000)\nfor имя_алгоритма, Таблица in алгоритмы.items():\n копия_наборов = тестовые_набор.copy()\n время_начало = time.perf_counter()\n for набор in копия_наборов:\n таблица = Таблица()\n for значение, ключ in enumerate(набор):\n таблица[ключ] = значение\n assert таблица[ключ] == значение, f'Найденный элемент не соответствует записанному'\n время_конца = time.perf_counter()\n затраченное_время[имя_алгоритма] = (время_конца - время_начало) / len(тестовые_набор)\n\nотсортированная_таблица_затраченного_времени = sorted(затраченное_время.items(), key=lambda kv: kv[1])\ntabulate(отсортированная_таблица_затраченного_времени, headers=['Алгоритм','Время'], tablefmt='html', showindex=\"always\")",
"_____no_output_____"
]
],
[
[
"№3",
"_____no_output_____"
]
],
[
[
"#Вывод результата\ndef tag(x, color='white'):\n return f'<td style=\"width:24px;height:24px;text-align:center;\" bgcolor=\"{color}\">{x}</td>'\nth = ''.join(map(tag, ' abcdefgh '))\ndef chessboard(data):\n row = lambda i: ''.join([\n tag('<span style=\"font-size:24px\">*</span>' * v,\n color='white' if (i+j+1)%2 else 'silver')\n for j, v in enumerate(data[i])])\n tb = ''.join([f'<tr>{tag(8-i)}{row(i)}{tag(8-i)}</tr>' for i in range(len(data))])\n return HTML(f'<table>{th}{tb}{th}</table>')",
"_____no_output_____"
],
[
"#Создание доски\narr = [[0] * 8 for i in range(8)]\narr[1][2] = 1\nchessboard(arr)",
"_____no_output_____"
],
[
"#Алгоритм\ndef check_place(rows, row, column):\n \"\"\" Проверяет, если board[column][row] под атакой других ферзей \"\"\"\n for i in range(row):\n if rows[i] == column or \\\n rows[i] - i == column - row or \\\n rows[i] + i == column + row:\n return False\n return True\n\ntotal_shown = 0\ndef put_queen(rows=[0]*8, row=0):\n \"\"\" Пытается подобрать место для ферзя, которое не находится под атакой других \"\"\"\n if row == 8: # мы уместили всех 8 ферзей и можем показать доску\n arr = [[0] * 8 for i in range(8)]\n for row, column in enumerate(rows):\n arr[row][column] = 1\n return chessboard(arr)\n else:\n for column in range(8):\n if check_place(rows, row, column):\n rows[row] = column\n board = put_queen(rows, row + 1)\n if board: return board\n \nput_queen()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d05d5ecb85d3bd62468cd9d2a1e419df37668a99 | 233,193 | ipynb | Jupyter Notebook | sentiment_analysis_experiment/Sentiment_analysis_experiment_1.ipynb | sreramk1/sentiment-analysis | 6b0866d6631a7edf1ccf344b727d33758917d88b | [
"Apache-2.0"
] | null | null | null | sentiment_analysis_experiment/Sentiment_analysis_experiment_1.ipynb | sreramk1/sentiment-analysis | 6b0866d6631a7edf1ccf344b727d33758917d88b | [
"Apache-2.0"
] | null | null | null | sentiment_analysis_experiment/Sentiment_analysis_experiment_1.ipynb | sreramk1/sentiment-analysis | 6b0866d6631a7edf1ccf344b727d33758917d88b | [
"Apache-2.0"
] | null | null | null | 60.885901 | 26,278 | 0.600837 | [
[
[
"import numpy as np\n\nimport tensorflow_datasets as tfds\nimport tensorflow as tf\ntf.config.run_functions_eagerly(False) \n#tfds.disable_progress_bar()",
"_____no_output_____"
],
[
"tf.version.VERSION",
"_____no_output_____"
],
[
"import pandas as pd ",
"_____no_output_____"
],
[
"dataset = pd.read_csv(\"/content/drive/MyDrive/sentiment-dataset/airline_sentiment_analysis.csv\")",
"_____no_output_____"
],
[
"print (dataset[:10])\nprint (dataset[len(dataset) - 10:])",
" Unnamed: 0 ... text\n0 1 ... @VirginAmerica plus you've added commercials t...\n1 3 ... @VirginAmerica it's really aggressive to blast...\n2 4 ... @VirginAmerica and it's a really big bad thing...\n3 5 ... @VirginAmerica seriously would pay $30 a fligh...\n4 6 ... @VirginAmerica yes, nearly every time I fly VX...\n5 8 ... @virginamerica Well, I didn't…but NOW I DO! :-D\n6 9 ... @VirginAmerica it was amazing, and arrived an ...\n7 11 ... @VirginAmerica I <3 pretty graphics. so muc...\n8 12 ... @VirginAmerica This is such a great deal! Alre...\n9 13 ... @VirginAmerica @virginmedia I'm flying your #f...\n\n[10 rows x 3 columns]\n Unnamed: 0 ... text\n11531 14627 ... @AmericanAir Flight Cancelled Flightled, can't...\n11532 14628 ... Thank you. “@AmericanAir: @jlhalldc Customer R...\n11533 14629 ... @AmericanAir How do I change my flight if the ...\n11534 14630 ... @AmericanAir Thanks! He is.\n11535 14631 ... @AmericanAir thx for nothing on getting us out...\n11536 14633 ... @AmericanAir my flight was Cancelled Flightled...\n11537 14634 ... @AmericanAir right on cue with the delays👌\n11538 14635 ... @AmericanAir thank you we got on a different f...\n11539 14636 ... @AmericanAir leaving over 20 minutes Late Flig...\n11540 14638 ... @AmericanAir you have my money, you change my ...\n\n[10 rows x 3 columns]\n"
],
[
"def process(txt):\n return ' '.join(word for word in txt.split(' ') if not word.startswith('@'))\n\nprocess(\" word1 word2 word3 @word4 word5 word6\")",
"_____no_output_____"
],
[
"dataset_processed = pd.DataFrame.copy(dataset, deep=True)\n\ndataset_processed['text'] = dataset['text'].apply(process)\nprint(dataset_processed[:3])\nprint(dataset_processed[len(dataset_processed) - 3:])\n",
" Unnamed: 0 ... text\n0 1 ... plus you've added commercials to the experienc...\n1 3 ... it's really aggressive to blast obnoxious \"ent...\n2 4 ... and it's a really big bad thing about it\n\n[3 rows x 3 columns]\n Unnamed: 0 ... text\n11538 14635 ... thank you we got on a different flight to Chic...\n11539 14636 ... leaving over 20 minutes Late Flight. No warnin...\n11540 14638 ... you have my money, you change my flight, and d...\n\n[3 rows x 3 columns]\n"
],
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"def process_label(label):\n if label == \"negative\":\n return 0\n elif label == \"positive\":\n return 1\n raise Exception(\"unrecognized label\")",
"_____no_output_____"
],
[
"dataset_processed['airline_sentiment'] = dataset_processed['airline_sentiment'].apply(process_label)",
"_____no_output_____"
],
[
"dataset_train, dataset_test = train_test_split(dataset_processed, test_size = 0.2)",
"_____no_output_____"
],
[
"dataset_train[100:125]",
"_____no_output_____"
],
[
"len(dataset_train)",
"_____no_output_____"
],
[
"BUFFER_SIZE = 10000\nBATCH_SIZE = 64",
"_____no_output_____"
],
[
"dataset_train_text_tf = tf.convert_to_tensor(dataset_train['text'], dtype=tf.string)\ndataset_train_label_tf = tf.convert_to_tensor(dataset_train['airline_sentiment'], dtype=tf.float32)\n\ndataset_test_text_tf = tf.convert_to_tensor(dataset_test['text'], dtype=tf.string)\ndataset_test_lable_tf = tf.convert_to_tensor(dataset_test['airline_sentiment'], dtype=tf.float32)\n\ndataset_train_tf = tf.data.Dataset.from_tensor_slices((dataset_train_text_tf, dataset_train_label_tf))\ndataset_test_tf = tf.data.Dataset.from_tensor_slices((dataset_test_text_tf, dataset_test_lable_tf))\n",
"_____no_output_____"
],
[
"count = 10\ni = 0\nfor ele in dataset_train_tf.as_numpy_iterator():\n if i >= count: \n break\n print (ele)\n i += 1",
"(b'why must you always delay my Late Flight night Orlando flights? \\xf0\\x9f\\x92\\x94', 0.0)\n(b'So appreciated!', 1.0)\n(b'thanks, keep up the good work', 1.0)\n(b\"we never received that $15 credit for inoperable tv's on our SFO > JFK flight 2 weeks ago. never got an email...\", 0.0)\n(b'what response? Is our flight out of Montrose Cancelled Flightled or not?', 0.0)\n(b\"so you don't have a pilot now for #clt \\xe2\\x9c\\x88 #ord for at least another hour. Why on earth would you board the plane? Makes no sense!\", 0.0)\n(b\"LUV Ya Too!!!! I will sing a song for y'all when I finally get on that plane back to Nashville!!! #LOVESOUTHWESTAIR\", 1.0)\n(b\"she's the type of person that can make a customers day! I fly 100+ times a year & she's one of the top flight attendants I've had!\", 1.0)\n(b\"really not acceptable. Just informed plane won't start. Chartering bus to take passengers to jfk.\", 0.0)\n(b'Also, been on hold for 30 minutes with your \"customer service\" to find out when my new flight is scheduled bc your site SUCKS', 0.0)\n"
],
[
"train_dataset_batched_tf = dataset_train_tf.shuffle(BUFFER_SIZE).batch(BATCH_SIZE).prefetch(tf.data.AUTOTUNE)\ntest_dataset_batched_tf = dataset_test_tf.batch(BATCH_SIZE).prefetch(tf.data.AUTOTUNE)",
"_____no_output_____"
],
[
"count = 1\ni = 0\nfor ele in train_dataset_batched_tf.as_numpy_iterator():\n if i >= count: \n break\n print (ele)\n i += 1",
"(array([b\"Fine. Would you have them call me? I left a message, was told it would be 2 hours for a call. Haven't heard anything yet.\",\n b\"thanks! Y'all have some of the best customer service left in the industry.\",\n b\"it's been 2 hrs of wait on the phone a) worst customer services b) trying to know where my suitcase Is and way MORE \\xf0\\x9f\\x98\\xa4#ANGRY\",\n b\"I cheated on you, and I'm sorry. I'll never do it again. has given my wife and I the worst start to a honeymoon ever\",\n b'a $100 - totaled. Not happy. Not at all.',\n b'is gettin fancy! #Mint #LieFlat Nice work on the menu #LobsterMac #BloodyMary #JetSetter http://t.co/zf5wjgtXzT',\n b\"when trying to check-in online, it says to call...now I've been on hold for 2 hours...what to do?\",\n b'.@USAirways trying to get a partner PNR, and have spent more than 1 hour on hold. I know its snowing somewhere, but this is awful',\n b'not an issue but I think training & information would help. Great ppl but service needs to switch from individual to group better',\n b'worst experience with you. Cancelled Flightled flight, no voucher and no luggage because \"ramp was broken.\" No other ramps in Charlotte??',\n b\"I fly normally. This doesn't happen to me with them. I'll let your flyers provide their own feedback. Thank you.\",\n b'I guess the Kit Kat looks tasty... not going near that \"sandwich.\"',\n b\"Thank y'all for being an amazing airline who knows how to treat their customers. you guys rock!\",\n b\"yet again you disappoint. Sitting at IAD for UA3728 for 3.5hrs and you can't seem to know why the plane hasn't left Albany. #Fail\",\n b'once again flying AA4285, once again 60+ min delay because of mechanical issues. Perhaps you should consider maintenance?',\n b'great. Looking forward to your response to my DM then',\n b'...be found when he checks in. Now no info. Please DM me and help me fix this NOW. (3/3)',\n b\"Hey, first time flyer next week - excited! But I'm having a hard time getting my flights added to my Elevate account. Help?\",\n b'gate agents are now working with everyone to resolve connecting flight issues which is my concern',\n b'Rebooked for tomorrow morning. Never been here - not sure what I can see before tomorrow morning!',\n b'this is not a fair set up. I payed for a full seat. I should get access to a full seat. http://t.co/SbA0ARicyq',\n b\"customers aren't dumb. These revenue based programs will hurt everyone. Not gonna save money like you think\",\n b'thank you for always have the most amazing customer service! Bring on The Disney Princess Half Marathon',\n b'I did see that! Working on picking up a trip or two as we type.',\n b'no my concerns were not addressed',\n b'please help! No bags, no way to get through to customer service since 8AM this morning! Help!!',\n b' Yes. Dale at Baggage office was wonderful. But not everyone is on the same page down there... We had a 6 hour wait!',\n b'appreciate it!!',\n b\"Issue is JFK. Pilot explained once JFK reopens we can get scheduled back there, but why can't we divert to LGA? Closer than ACY!\",\n b\"by far the worst airline in history. I'll never ever fly your garbage again\",\n b\"ok it's now been 7 months waiting to hear from airline. I gave them quite a bit more than the 30 days requested! Terrible service\",\n b\"passengers seated, crew ready #WheresThePilot? Flt1088 from ORD. Hope he isn't at the bar.\",\n b'worst airline ever! Staff is nasty, wifi down bags are delayed due to weather?? And now the belt is broken. Selling UAL stock in AM',\n b\"that's what you have said for years, you are losing customers!!!!!!\",\n b\"nope I gave up - maybe they'll deliver it\",\n b'And now the flight Flight Booking Problems site is totally down. Folks, what is the problem?',\n b\"only thing confusing me is why I lost priority boarding? I'm a mileage plus card member \\xf0\\x9f\\x98\\x94\",\n b'all right, but can you give me an email to write to ?',\n b'thanks for the show! \\xf0\\x9f\\x91\\x8d',\n b'it took ages for one snapchat story to load. one. ONE. I will demolish you',\n b\"we're home, you guys recovered, now we can laugh about it and the extra day in barbados. Will you open Cuba soon?\",\n b'negative. Done wasting time with amateurs at customer service. Thanks for at least offering.',\n b'- thanks. She submitted a damaged bag complaint online...is there anything else we can do? #goodcustomerservice',\n b'The delay is nothing but the personnel being so combative up to the point of saying \"what\\'s the hury, the plane is not leaving',\n b'Thanks, she did her best. Staying the night in Dallas, new trial to Detroit via Atlanta tomorrow, assuming no Cancelled Flightlations.',\n b'how can I get travel question answered quickly... Online and calling not helping with this busy day',\n b'Not even on the bag status...will take actions against this company is incredible how irresponsible are with the costumer',\n b'now maintenance issues with flight 5639 and more issues with passengers that will miss connections needing to get off',\n b'Aww Thanks AA..DFW was on GMA up here this AM..so i understand ..Btw A.A is my Airline when im able to trv..Love you guys.:)',\n b'is non existent and I will take this as far as needed.Why hide behind a corporate logo? Provide a number #tcf #useless #amateur',\n b\"Thanks for the reminder of a few older flights I'd taken and the easy access to add points to my new JB account! Awesome service.\",\n b'I Cancelled Flighted my flight. I really don\\xe2\\x80\\x99t need this much trouble.',\n b\"it wasn't a delay so much as a straight Cancelled Flightlation. Weather wasn't an issue either.\",\n b\"flt. 4567 departure time has changed five times in the last 20 minutes. Why don't you figure out a solution and announce once?\",\n b'I am signed up for notifications. This is the first trip I was not updated on. Not sure why this happened.',\n b\"Hi there, looks like my connection is delayed too so I'll make it. Thanks!\",\n b\"How best to talk with an agent to reschedule Cancelled Flighted flight? No one answers at AA. Know it's busy, but need help.Thanks\",\n b\"Thanks, both airlines said that it is located at AA Detroit. Also was informed that it flew with AA, which shouldn't matter.\",\n b'I have a flight from omaha to chicago (en route to NYC) and they are seating me and my partner separate, please fix this res# ILC0HP',\n b\"We're having 2 grandbabies in 2 weeks -- will travel to DC for the births. Thank you for the reasonable fares! See you Saturday!\",\n b'what maintenance? The flight landed from Jamaica, has to go through security then get to term 3 then cleaned then board',\n b'Hidden City forces me into crappy seat even though exit row is available on the first leg. Your support cannot fix. :-(',\n b'#BQONPA flight #1641 delayed from POS-MIA, missed #2214 MIA- ATL need seat on last flight to ATL.',\n b\"customer service failure aside, one would think you guys would care about inaccurate manifests. I'm sure TSA would.\"],\n dtype=object), array([0., 1., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 1., 0., 0., 1., 0.,\n 0., 0., 0., 0., 0., 1., 1., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0.,\n 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 1.,\n 0., 0., 0., 0., 1., 0., 1., 0., 1., 0., 0., 0., 0.], dtype=float32))\n"
],
[
"print(dataset_train_tf)\nprint(dataset_test_tf)",
"<TensorSliceDataset shapes: ((), ()), types: (tf.string, tf.float32)>\n<TensorSliceDataset shapes: ((), ()), types: (tf.string, tf.float32)>\n"
],
[
"#VOCAB_SIZE = 1000\nencoder = tf.keras.layers.TextVectorization()\n #max_tokens=VOCAB_SIZE)\nencoder.adapt(train_dataset_batched_tf.map(lambda text, label: text))",
"_____no_output_____"
],
[
"count_0 = len(train_dataset_batched_tf)\ncount = 0\nfor ds in train_dataset_batched_tf:\n count += len(ds[0])\n print(len(ds[0]))\ncount",
"_____no_output_____"
],
[
"encoder(\"hello world HELLO WORLD\")[:].numpy()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\n\ndef plot_graphs(history, metric):\n plt.plot(history.history[metric])\n plt.plot(history.history['val_'+metric], '')\n plt.xlabel(\"Epochs\")\n plt.ylabel(metric)\n plt.legend([metric, 'val_'+metric])",
"_____no_output_____"
],
[
"vocab = np.array(encoder.get_vocabulary())\nvocab[100:150]",
"_____no_output_____"
],
[
"for example, label in dataset_train_tf.take(1):\n print('texts: ', example.numpy())\n print()\n print('labels: ', label.numpy())",
"texts: b'why must you always delay my Late Flight night Orlando flights? \\xf0\\x9f\\x92\\x94'\n\nlabels: 0.0\n"
],
[
"model = tf.keras.Sequential([\n encoder,\n tf.keras.layers.Embedding(\n input_dim=len(encoder.get_vocabulary()),\n output_dim=64,\n # Use masking to handle the variable sequence lengths\n mask_zero=True),\n tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(64)),\n\n tf.keras.layers.Dense(64, activation='relu'),\n tf.keras.layers.Dense(32, activation='relu'),\n tf.keras.layers.Dense(16, activation='relu'),\n tf.keras.layers.Dense(8, activation='relu'),\n tf.keras.layers.Dense(1) #, activation='sigmoid')\n ])",
"_____no_output_____"
],
[
"model.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ntext_vectorization (TextVect (None, None) 0 \n_________________________________________________________________\nembedding (Embedding) (None, None, 64) 770176 \n_________________________________________________________________\nbidirectional (Bidirectional (None, 128) 66048 \n_________________________________________________________________\ndense (Dense) (None, 64) 8256 \n_________________________________________________________________\ndense_1 (Dense) (None, 32) 2080 \n_________________________________________________________________\ndense_2 (Dense) (None, 16) 528 \n_________________________________________________________________\ndense_3 (Dense) (None, 8) 136 \n_________________________________________________________________\ndense_4 (Dense) (None, 1) 9 \n=================================================================\nTotal params: 847,233\nTrainable params: 847,233\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"encoder(\"hello world. This is great\").numpy()",
"_____no_output_____"
],
[
"model.compile(loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),\n optimizer=tf.keras.optimizers.Adam(1e-4),\n metrics=['accuracy']) #run_eagerly=True)",
"_____no_output_____"
],
[
"history = model.fit(train_dataset_batched_tf, epochs=10,\n validation_data=test_dataset_batched_tf,\n validation_steps=30)",
"Epoch 1/10\n145/145 [==============================] - 20s 87ms/step - loss: 0.6000 - accuracy: 0.7973 - val_loss: 0.4479 - val_accuracy: 0.7917\nEpoch 2/10\n145/145 [==============================] - 10s 71ms/step - loss: 0.4006 - accuracy: 0.7973 - val_loss: 0.3862 - val_accuracy: 0.7917\nEpoch 3/10\n145/145 [==============================] - 10s 71ms/step - loss: 0.3055 - accuracy: 0.7973 - val_loss: 0.3105 - val_accuracy: 0.7917\nEpoch 4/10\n145/145 [==============================] - 10s 72ms/step - loss: 0.2383 - accuracy: 0.7973 - val_loss: 0.2937 - val_accuracy: 0.7917\nEpoch 5/10\n145/145 [==============================] - 10s 72ms/step - loss: 0.1840 - accuracy: 0.8438 - val_loss: 0.2728 - val_accuracy: 0.8823\nEpoch 6/10\n145/145 [==============================] - 10s 71ms/step - loss: 0.1276 - accuracy: 0.9469 - val_loss: 0.2823 - val_accuracy: 0.9141\nEpoch 7/10\n145/145 [==============================] - 10s 71ms/step - loss: 0.0915 - accuracy: 0.9742 - val_loss: 0.3264 - val_accuracy: 0.9115\nEpoch 8/10\n145/145 [==============================] - 10s 69ms/step - loss: 0.0703 - accuracy: 0.9816 - val_loss: 0.3343 - val_accuracy: 0.9130\nEpoch 9/10\n145/145 [==============================] - 10s 70ms/step - loss: 0.0537 - accuracy: 0.9874 - val_loss: 0.3435 - val_accuracy: 0.9146\nEpoch 10/10\n145/145 [==============================] - 10s 70ms/step - loss: 0.0408 - accuracy: 0.9900 - val_loss: 0.3936 - val_accuracy: 0.9172\n"
],
[
"test_loss, test_acc = model.evaluate(test_dataset_batched_tf)\n\nprint('Test Loss:', test_loss)\nprint('Test Accuracy:', test_acc)",
"37/37 [==============================] - 1s 16ms/step - loss: 0.3950 - accuracy: 0.9181\nTest Loss: 0.3949778378009796\nTest Accuracy: 0.9181463718414307\n"
],
[
"",
"_____no_output_____"
],
[
"\nsample_text = ('good it\\'s great')\npredictions = model.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('bad. It\\'s very bad. Worse')\npredictions = model.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('This airlines is the best')\npredictions = model.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('I will never fly with you')\npredictions = model.predict(np.array([sample_text]))\nprint(predictions)\n\n\nsample_text = ('I will never recommend you')\npredictions = model.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('I will always recommend you')\npredictions = model.predict(np.array([sample_text]))\nprint(predictions)\n\n\nsample_text = ('Will be a long time before I recommend you to anyone.')\npredictions = model.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('I liked the way you guys organize yourself')\npredictions = model.predict(np.array([sample_text]))\nprint(predictions)",
"[[2.4241579]]\n[[-3.679005]]\n[[1.5192201]]\n[[-1.4085932]]\n[[-0.5680525]]\n[[1.4155738]]\n[[-3.8368037]]\n[[0.36423916]]\n"
],
[
"",
"_____no_output_____"
],
[
"model.set_weights",
"_____no_output_____"
],
[
"encoder_new= None\nencoder_new = tf.keras.layers.TextVectorization()",
"_____no_output_____"
],
[
"encoder_new.get_config()",
"_____no_output_____"
],
[
"encoder_new.adapt(np.array([['hell']], dtype=np.object), batch_size=None)",
"_____no_output_____"
],
[
"encoder_new.set_weights(encoder.get_weights())",
"_____no_output_____"
],
[
"encoder(\"hello world\").numpy()",
"_____no_output_____"
],
[
"encoder_new(\"hello world\").numpy()",
"_____no_output_____"
],
[
"model2 = tf.keras.Sequential([\n encoder,\n tf.keras.layers.Embedding(\n input_dim=len(encoder.get_vocabulary()),\n output_dim=64,\n # Use masking to handle the variable sequence lengths\n mask_zero=True),\n tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(64)),\n\n tf.keras.layers.Dense(64, activation='relu'),\n tf.keras.layers.Dense(32, activation='relu'),\n tf.keras.layers.Dense(16, activation='relu'),\n tf.keras.layers.Dense(8, activation='relu'),\n tf.keras.layers.Dense(1) #, activation='sigmoid')\n ])",
"_____no_output_____"
],
[
"model2.compile(loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),\n optimizer=tf.keras.optimizers.Adam(1e-4),\n metrics=['accuracy']) #run_eagerly=True)",
"_____no_output_____"
],
[
"layers = []\nfor layer in model.layers:\n layers.append(layer.get_weights())",
"_____no_output_____"
],
[
"i = 0\nfor layer in model2.layers:\n # if i == 0:\n # i += 1\n # continue\n print(layer.get_weights()[0].dtype)\n layer.set_weights(layers[i])\n i += 1",
"object\nfloat32\nfloat32\nfloat32\nfloat32\nfloat32\nfloat32\nfloat32\n"
],
[
"sample_text = ('good it\\'s great')\npredictions = model2.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('bad. It\\'s very bad. Worse')\npredictions = model2.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('This airlines is the best')\npredictions = model2.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('I will never fly with you')\npredictions = model2.predict(np.array([sample_text]))\nprint(predictions)\n\n\nsample_text = ('I will never recommend you')\npredictions = model2.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('I will always recommend you')\npredictions = model2.predict(np.array([sample_text]))\nprint(predictions)\n\n\nsample_text = ('Will be a long time before I recommend you to anyone.')\npredictions = model2.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('I liked the way you guys organize yourself')\npredictions = model2.predict(np.array([sample_text]))\nprint(predictions)",
"[[2.4241579]]\n[[-3.679005]]\n[[1.5192201]]\n[[-1.4085932]]\n[[-0.5680525]]\n[[1.4155738]]\n[[-3.8368037]]\n[[0.36423916]]\n"
],
[
"import json\n",
"_____no_output_____"
],
[
"class NdarrayEncoder(json.JSONEncoder):\n def default(self, obj):\n if isinstance(obj, np.ndarray):\n return obj.tolist()\n if isinstance(obj, bytes):\n return obj.decode('utf-8')\n print(obj)\n return json.JSONEncoder().default(self, obj)",
"_____no_output_____"
],
[
"layersInList = []\n\nfor layer in model.layers:\n layersInList.append(layer.get_weights())",
"_____no_output_____"
],
[
"weightsInJson = json.dumps(layersInList, cls=NdarrayEncoder)\n",
"_____no_output_____"
],
[
"with open(\"weights.json\", \"w\") as json_file:\n json_file.write(weightsInJson)",
"_____no_output_____"
],
[
"with open(\"weights.json\", \"r\") as json_file_r:\n weightsInListRead = json_file_r.read()",
"_____no_output_____"
],
[
"weightsReadData = json.loads(weightsInListRead)",
"_____no_output_____"
],
[
"# def isIterable(obj):\n# if hasattr(obj, '__iter__') and hasattr(obj, '__next__') and hasattr('__getitem__'):\n# return True\n# return False\n\ndef convertStringToBytesInObject(convertableObj):\n if isinstance(convertableObj, list):\n i = 0\n for item in convertableObj:\n if isinstance(item, str):\n convertableObj[i] = item.encode()\n elif isinstance(item, list): \n convertStringToBytesInObject(item)\n i += 1\n else:\n print(convertableObj)\n raise Exception(\" expected to be iterable \")\n\n",
"_____no_output_____"
],
[
"isIterable([])",
"_____no_output_____"
],
[
"# convertStringToBytesInObject(weightsReadData)",
"_____no_output_____"
],
[
"encoder_new= None\nencoder_new = tf.keras.layers.TextVectorization()",
"_____no_output_____"
],
[
"encoder_new.get_config()",
"_____no_output_____"
],
[
"encoder_new.adapt(np.array([['hell']], dtype=np.object), batch_size=None)",
"_____no_output_____"
],
[
"encoder_new.set_weights(weightsReadData[0])",
"_____no_output_____"
],
[
"encoder_new(\"hello world\").numpy()",
"_____no_output_____"
],
[
"encoder(\"hello world\").numpy()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"model3 = tf.keras.Sequential([\n encoder_new,\n tf.keras.layers.Embedding(\n input_dim=len(encoder_new.get_vocabulary()),\n output_dim=64,\n # Use masking to handle the variable sequence lengths\n mask_zero=True),\n tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(64)),\n\n tf.keras.layers.Dense(64, activation='relu'),\n tf.keras.layers.Dense(32, activation='relu'),\n tf.keras.layers.Dense(16, activation='relu'),\n tf.keras.layers.Dense(8, activation='relu'),\n tf.keras.layers.Dense(1) #, activation='sigmoid')\n ])",
"_____no_output_____"
],
[
"model3.compile(loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),\n optimizer=tf.keras.optimizers.Adam(1e-4),\n metrics=['accuracy']) #run_eagerly=True)",
"_____no_output_____"
],
[
"layers2 = []\nfor layerWeights in weightsReadData:\n layers2.append(layerWeights)\n print(len(layerWeights))",
"1\n1\n6\n2\n2\n2\n2\n2\n"
],
[
"def convertToNdarray(obj):\n if isinstance(obj, list):\n return np.asarray([convertToNdarray(o) for o in obj])\n else:\n return obj\n",
"_____no_output_____"
],
[
"layers2 = convertToNdarray(layers2)",
"/usr/local/lib/python3.7/dist-packages/numpy/core/_asarray.py:83: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray\n return array(a, dtype, copy=False, order=order)\n"
],
[
"layers2 = [np.array(layer, dtype=object) for layer in layers2]",
"_____no_output_____"
],
[
"i = 0\nfor layer in model3.layers:\n # if i == 0:\n # i += 1\n # continue\n print(layer.get_weights()[0].dtype)\n layer.set_weights(layers2[i])\n i += 1",
"object\nfloat32\nfloat32\nfloat32\nfloat32\nfloat32\nfloat32\nfloat32\n"
],
[
"sample_text = ('good it\\'s great')\npredictions = model3.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('bad. It\\'s very bad. Worse')\npredictions = model3.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('This airlines is the best')\npredictions = model3.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('I will never fly with you')\npredictions = model3.predict(np.array([sample_text]))\nprint(predictions)\n\n\nsample_text = ('I will never recommend you')\npredictions = model3.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('I will always recommend you')\npredictions = model3.predict(np.array([sample_text]))\nprint(predictions)\n\n\nsample_text = ('Will be a long time before I recommend you to anyone.')\npredictions = model3.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('I liked the way you guys organize yourself')\npredictions = model3.predict(np.array([sample_text]))\nprint(predictions)",
"[[2.4241579]]\n[[-3.679005]]\n[[1.5192201]]\n[[-1.4085932]]\n[[-0.5680525]]\n[[1.4155738]]\n[[-3.8368037]]\n[[0.36423916]]\n"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"len(weightsReadData)",
"_____no_output_____"
],
[
"len(model3.layers)",
"_____no_output_____"
],
[
"len(weightsReadData[2])",
"_____no_output_____"
],
[
"len(model3.layers[2].get_weights())",
"_____no_output_____"
],
[
"len(model2.layers[2].get_weights())",
"_____no_output_____"
],
[
"hw = b'hello world'",
"_____no_output_____"
],
[
"json.dumps(hw.decode('utf-8'))",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"tf.keras.layers.serialize(encoder)",
"_____no_output_____"
],
[
"encoder.get_weights()",
"_____no_output_____"
],
[
"encoder_new= None\nencoder_new = tf.keras.layers.TextVectorization()\n",
"_____no_output_____"
],
[
"encoder_new.get_config()",
"_____no_output_____"
],
[
"encoder_new.adapt([['hell']], batch_size=None)",
"_____no_output_____"
],
[
"encoder_new.set_weights(encoder.get_weights())",
"_____no_output_____"
],
[
"# encoder_new.set_vocabulary(encoder.get_vocabulary())",
"_____no_output_____"
],
[
"encoder(\"hello world\").numpy()",
"_____no_output_____"
],
[
"encoder_new(\"hello world\").numpy()",
"_____no_output_____"
],
[
"dataset_train_batched_text = np.array_split(dataset_train['text'],len(dataset_train['text'])/BATCH_SIZE)\ndataset_train_batched_class = np.array_split(dataset_train['airline_sentiment'], len(dataset_train['airline_sentiment'])/BATCH_SIZE)\n\ndataset_test_batched_text = np.array_split(dataset_test['text'],len(dataset_test['text'])/BATCH_SIZE)\ndataset_test_batched_class = np.array_split(dataset_test['airline_sentiment'], len(dataset_test['airline_sentiment'])/BATCH_SIZE)\n",
"_____no_output_____"
],
[
"print (len(dataset_train))\nprint (len(dataset_test))\nprint (\" ------------------------ \")\nprint (len(dataset_train_batched_text))\nprint (len(dataset_train_batched_class))\nprint (len(dataset_train_batched_text[len(dataset_train_batched_text)- 1]))\nprint (len(dataset_train_batched_class[len(dataset_train_batched_text)- 1]))\nprint (\" ------------------------ \")\nprint (len(dataset_test_batched_text))\nprint (len(dataset_test_batched_class))\nprint (len(dataset_test_batched_text[len(dataset_test_batched_text)- 1]))\nprint (len(dataset_test_batched_class[len(dataset_test_batched_class)- 1]))",
"9232\n2309\n ------------------------ \n144\n144\n64\n64\n ------------------------ \n36\n36\n64\n64\n"
],
[
"dataset_test_batched_text_tmp = np.asarray(dataset_test_batched_text, dtype=object)\ndataset_test_batched_class_tmp = np.asarray(dataset_test_batched_class, dtype=object)\n\ndataset_train_batched_text_tmp = np.asarray(dataset_train_batched_text, dtype=object)\ndataset_train_batched_class_tmp = np.asarray(dataset_train_batched_class, dtype=object)\n\nnp_dataset_test_batched_text = []\nnp_dataset_test_batched_class = []\nnp_dataset_train_batched_text = []\nnp_dataset_train_batched_class = []\n\nfor itr in dataset_test_batched_text_tmp:\n np_dataset_test_batched_text.append(itr.to_numpy())\n\nfor itr in dataset_test_batched_class_tmp:\n np_dataset_test_batched_class.append(itr.to_numpy())\n\n\nfor itr in dataset_train_batched_text_tmp:\n np_dataset_train_batched_text.append(itr.to_numpy())\n\nfor itr in dataset_train_batched_class_tmp:\n np_dataset_train_batched_class.append(itr.to_numpy())\n\n\nnp_dataset_test_batched_text = np.asarray(np_dataset_test_batched_text, dtype=object)\nnp_dataset_test_batched_class = np.asarray(np_dataset_test_batched_class, dtype=object)\nnp_dataset_train_batched_text = np.asarray(np_dataset_train_batched_text, dtype=object)\nnp_dataset_train_batched_class = np.asarray(np_dataset_train_batched_class, dtype=object)",
"_____no_output_____"
],
[
"np_dataset_test_batched_text[len(np_dataset_test_batched_text)- 1][0]",
"_____no_output_____"
],
[
"tf_dataset_test_batched_text = tf.data.Dataset.from_tensor_slices(np_dataset_test_batched_text)\ntf_dataset_test_batched_text\n",
"_____no_output_____"
],
[
"VOCAB_SIZE = 1000\nencoder = tf.keras.layers.TextVectorization()\n #max_tokens=VOCAB_SIZE)\nencoder.adapt(np_dataset_train_batched_text)",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\n\ndef plot_graphs(history, metric):\n plt.plot(history.history[metric])\n plt.plot(history.history['val_'+metric], '')\n plt.xlabel(\"Epochs\")\n plt.ylabel(metric)\n plt.legend([metric, 'val_'+metric])\n",
"_____no_output_____"
],
[
"dataset_2, info = tfds.load('imdb_reviews', with_info=True,\n as_supervised=True)\ntrain_dataset, test_dataset = dataset_2['train'], dataset_2['test']\n\ntrain_dataset.element_spec",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"for example, label in train_dataset.take(1):\n print('text: ', example.numpy())\n print('label: ', label.numpy())\n",
"text: b\"This was an absolutely terrible movie. Don't be lured in by Christopher Walken or Michael Ironside. Both are great actors, but this must simply be their worst role in history. Even their great acting could not redeem this movie's ridiculous storyline. This movie is an early nineties US propaganda piece. The most pathetic scenes were those when the Columbian rebels were making their cases for revolutions. Maria Conchita Alonso appeared phony, and her pseudo-love affair with Walken was nothing but a pathetic emotional plug in a movie that was devoid of any real meaning. I am disappointed that there are movies like this, ruining actor's like Christopher Walken's good name. I could barely sit through it.\"\nlabel: 0\n"
],
[
"BUFFER_SIZE = 10000\nBATCH_SIZE = 64\n",
"_____no_output_____"
],
[
"train_dataset = train_dataset.shuffle(BUFFER_SIZE).batch(BATCH_SIZE).prefetch(tf.data.AUTOTUNE)\ntest_dataset = test_dataset.batch(BATCH_SIZE).prefetch(tf.data.AUTOTUNE)\n",
"_____no_output_____"
],
[
"train_dataset.as_numpy_iterator()",
"_____no_output_____"
],
[
"for example, label in train_dataset.take(1):\n print('texts: ', example.numpy()[:3])\n print()\n print('labels: ', label.numpy()[:])\n",
"texts: [b\"Eleven different Film Makers from different parts of the world are assembled in this film to present their views and ideas about the WTC attack. This is one of the best effort you will see in any Film. Films like this are rarely made and appreciated. This film tries to touch every possible core of WTC. Here are some of the most important stories from the film that makes this film so unique.<br /><br />There is the story from Samira Makhmalbaf (Iran) where somewhere in Iran people are preparing for the attacks from America. There a teacher is trying to educate her students by informing them about Innocent People being killed in WTC massacre. Then comes a story from Youssef Chahine (Egypt) where a Film Maker comes across face-to-face conversation with a Dead Soldier in the WTC attack and a Dead Hard Core Terrorist who was involved in WTC attack. Then we see a story from Idrissa Ouedraogo (Burkina Faso) where a group of Five Innocent children's sees Osama Bin Laden and plans to kidnap him and win the reward money from America. Then we see the story from Alejandro Gozalez Inarritu (Mexico) where you see a Black Screen and slowly you see the real footage of WTC buildings coming down. And the people who are stuck in the building are jumping out of it to save their lives. The other most important story is from Mira Nair (India) where a mother is struggling to get respect for her Dead Son whose name is falsely trapped in WTC massacre! After September 11 attack, Our heart beat automatically starts pumping if we hear two names anywhere in the world.. First is World Trade Centre and the second is Osama! This film totally changes our perception and makes a strong point by claiming something more to it.<br /><br />I will definitely recommend this movie to everyone who loves to have such kinds of Home DVD Collection. Definitely worth every penny you spend. But please don't expect anything more apart from Films in this DVD. There is of course Filmographies of the Film Makers but No Extra Features.\"\n b\"Interesting to read comments by viewers regarding Omega Code... many of the overwhelmingly positive comments were lifted almost word for word from TBN broadcasts... the movie looks as if it were made to go directly to video, to be stocked besides the three-part rapture series that was done by some other religious group in the 70s.. dont remember it? You wont remember this one either in a year or two. This is the first movie I have ever seen where it was implied that it was your religious duty to go to it and buy as many tickets as possible to save souls... very shameful... this just goes to show that if you are a televangelist's son, you too can play high-roller Hollywood producer with lil ole ladies tithe money...\"\n b\"While this film certainly does possess the stench of a bad film, it's surprisingly watchable on several levels. First, for old movie fans, it's interesting to see the leading role played by Dean Jagger (no relation to Mick). While Jagger later went on to a very respectable role as a supporting actor (even garnering the Oscar in this category for 12 O'CLOCK HIGH), here his performance is truly unique since he actually has a full head of hair (I never saw him this way before) and because he was by far the worst actor in the film. This film just goes to show that if an actor cannot act in his earlier films doesn't mean he can't eventually learn to be a great actor. Another good example of this phenomenon is Paul Newman, whose first movie (THE SILVER CHALICE) is considered one of the worst films of the 1950s.<br /><br />A second reason to watch the film is the shear cheesiness of it all. The writing is bad, the acting is bad and the special effects are bad. For example, when Jagger and an unnamed Cambodian are wading through the water, it's obvious they are really just walking in place and the background is poorly projected behind them. Plus, once they leave the water, their costumes are 100% dry!!! Horrid continuity and mindlessly bad dialog abounds throughout the film--so much so that it's hard to imagine why they didn't ask Bela Lugosi or George Zucco to star in the film--since both of them starred in many grade-z horror films. In many ways, this would be a perfect example for a film class on how NOT to make a film.<br /><br />So, while giving it a 3 is probably a bit over-generous, it's fun to laugh at and short so it's worth a look for bad film fans.\"]\n\nlabels: [1 0 0 0 0 1 1 0 1 0 0 0 0 1 1 1 1 1 0 1 0 1 1 1 1 0 1 1 1 0 1 0 1 0 1 0 1\n 1 1 0 0 1 0 0 1 0 1 0 1 0 1 1 0 1 1 0 1 1 1 0 0 1 1 1]\n"
],
[
"VOCAB_SIZE = 1000\nencoder = tf.keras.layers.experimental.preprocessing.TextVectorization(\n max_tokens=VOCAB_SIZE)\nencoder.adapt(train_dataset.map(lambda text, label: text))",
"_____no_output_____"
],
[
"vocab = np.array(encoder.get_vocabulary())\nvocab[:]",
"_____no_output_____"
],
[
"encoded_example = encoder(example)[:3].numpy()\nencoded_example",
"_____no_output_____"
],
[
"for n in range(3):\n print(\"Original: \", example[n].numpy())\n print(\"Round-trip: \", \" \".join(vocab[encoded_example[n]]))\n print()\n",
"Original: b\"Eleven different Film Makers from different parts of the world are assembled in this film to present their views and ideas about the WTC attack. This is one of the best effort you will see in any Film. Films like this are rarely made and appreciated. This film tries to touch every possible core of WTC. Here are some of the most important stories from the film that makes this film so unique.<br /><br />There is the story from Samira Makhmalbaf (Iran) where somewhere in Iran people are preparing for the attacks from America. There a teacher is trying to educate her students by informing them about Innocent People being killed in WTC massacre. Then comes a story from Youssef Chahine (Egypt) where a Film Maker comes across face-to-face conversation with a Dead Soldier in the WTC attack and a Dead Hard Core Terrorist who was involved in WTC attack. Then we see a story from Idrissa Ouedraogo (Burkina Faso) where a group of Five Innocent children's sees Osama Bin Laden and plans to kidnap him and win the reward money from America. Then we see the story from Alejandro Gozalez Inarritu (Mexico) where you see a Black Screen and slowly you see the real footage of WTC buildings coming down. And the people who are stuck in the building are jumping out of it to save their lives. The other most important story is from Mira Nair (India) where a mother is struggling to get respect for her Dead Son whose name is falsely trapped in WTC massacre! After September 11 attack, Our heart beat automatically starts pumping if we hear two names anywhere in the world.. First is World Trade Centre and the second is Osama! This film totally changes our perception and makes a strong point by claiming something more to it.<br /><br />I will definitely recommend this movie to everyone who loves to have such kinds of Home DVD Collection. Definitely worth every penny you spend. But please don't expect anything more apart from Films in this DVD. There is of course Filmographies of the Film Makers but No Extra Features.\"\nRound-trip: [UNK] different film [UNK] from different parts of the world are [UNK] in this film to present their [UNK] and ideas about the [UNK] [UNK] this is one of the best effort you will see in any film films like this are [UNK] made and [UNK] this film tries to [UNK] every possible [UNK] of [UNK] here are some of the most important stories from the film that makes this film so [UNK] br there is the story from [UNK] [UNK] [UNK] where [UNK] in [UNK] people are [UNK] for the [UNK] from america there a [UNK] is trying to [UNK] her [UNK] by [UNK] them about [UNK] people being killed in [UNK] [UNK] then comes a story from [UNK] [UNK] [UNK] where a film [UNK] comes across [UNK] [UNK] with a dead [UNK] in the [UNK] [UNK] and a dead hard [UNK] [UNK] who was involved in [UNK] [UNK] then we see a story from [UNK] [UNK] [UNK] [UNK] where a group of five [UNK] [UNK] [UNK] [UNK] [UNK] [UNK] and [UNK] to [UNK] him and [UNK] the [UNK] money from america then we see the story from [UNK] [UNK] [UNK] [UNK] where you see a black screen and [UNK] you see the real footage of [UNK] [UNK] coming down and the people who are [UNK] in the [UNK] are [UNK] out of it to save their lives the other most important story is from [UNK] [UNK] [UNK] where a mother is [UNK] to get [UNK] for her dead son whose name is [UNK] [UNK] in [UNK] [UNK] after [UNK] [UNK] [UNK] our heart [UNK] [UNK] starts [UNK] if we hear two [UNK] [UNK] in the world first is world [UNK] [UNK] and the second is [UNK] this film totally [UNK] our [UNK] and makes a strong point by [UNK] something more to itbr br i will definitely recommend this movie to everyone who [UNK] to have such [UNK] of home dvd [UNK] definitely worth every [UNK] you [UNK] but please dont expect anything more apart from films in this dvd there is of course [UNK] of the film [UNK] but no [UNK] features \n\nOriginal: b\"Interesting to read comments by viewers regarding Omega Code... many of the overwhelmingly positive comments were lifted almost word for word from TBN broadcasts... the movie looks as if it were made to go directly to video, to be stocked besides the three-part rapture series that was done by some other religious group in the 70s.. dont remember it? You wont remember this one either in a year or two. This is the first movie I have ever seen where it was implied that it was your religious duty to go to it and buy as many tickets as possible to save souls... very shameful... this just goes to show that if you are a televangelist's son, you too can play high-roller Hollywood producer with lil ole ladies tithe money...\"\nRound-trip: interesting to read comments by viewers [UNK] [UNK] [UNK] many of the [UNK] [UNK] comments were [UNK] almost word for word from [UNK] [UNK] the movie looks as if it were made to go [UNK] to video to be [UNK] [UNK] the [UNK] [UNK] series that was done by some other [UNK] group in the 70s dont remember it you wont remember this one either in a year or two this is the first movie i have ever seen where it was [UNK] that it was your [UNK] [UNK] to go to it and buy as many [UNK] as possible to save [UNK] very [UNK] this just goes to show that if you are a [UNK] son you too can play [UNK] hollywood [UNK] with [UNK] [UNK] [UNK] [UNK] money \n\nOriginal: b\"While this film certainly does possess the stench of a bad film, it's surprisingly watchable on several levels. First, for old movie fans, it's interesting to see the leading role played by Dean Jagger (no relation to Mick). While Jagger later went on to a very respectable role as a supporting actor (even garnering the Oscar in this category for 12 O'CLOCK HIGH), here his performance is truly unique since he actually has a full head of hair (I never saw him this way before) and because he was by far the worst actor in the film. This film just goes to show that if an actor cannot act in his earlier films doesn't mean he can't eventually learn to be a great actor. Another good example of this phenomenon is Paul Newman, whose first movie (THE SILVER CHALICE) is considered one of the worst films of the 1950s.<br /><br />A second reason to watch the film is the shear cheesiness of it all. The writing is bad, the acting is bad and the special effects are bad. For example, when Jagger and an unnamed Cambodian are wading through the water, it's obvious they are really just walking in place and the background is poorly projected behind them. Plus, once they leave the water, their costumes are 100% dry!!! Horrid continuity and mindlessly bad dialog abounds throughout the film--so much so that it's hard to imagine why they didn't ask Bela Lugosi or George Zucco to star in the film--since both of them starred in many grade-z horror films. In many ways, this would be a perfect example for a film class on how NOT to make a film.<br /><br />So, while giving it a 3 is probably a bit over-generous, it's fun to laugh at and short so it's worth a look for bad film fans.\"\nRound-trip: while this film certainly does [UNK] the [UNK] of a bad film its [UNK] [UNK] on several [UNK] first for old movie fans its interesting to see the leading role played by [UNK] [UNK] no [UNK] to [UNK] while [UNK] later went on to a very [UNK] role as a supporting actor even [UNK] the oscar in this [UNK] for [UNK] [UNK] high here his performance is truly unique since he actually has a full head of [UNK] i never saw him this way before and because he was by far the worst actor in the film this film just goes to show that if an actor cannot act in his earlier films doesnt mean he cant eventually learn to be a great actor another good example of this [UNK] is paul [UNK] whose first movie the [UNK] [UNK] is [UNK] one of the worst films of the [UNK] br a second reason to watch the film is the [UNK] [UNK] of it all the writing is bad the acting is bad and the special effects are bad for example when [UNK] and an [UNK] [UNK] are [UNK] through the [UNK] its obvious they are really just [UNK] in place and the background is poorly [UNK] behind them plus once they leave the [UNK] their [UNK] are [UNK] [UNK] [UNK] [UNK] and [UNK] bad dialog [UNK] throughout the [UNK] much so that its hard to imagine why they didnt ask [UNK] [UNK] or george [UNK] to star in the [UNK] both of them [UNK] in many [UNK] horror films in many ways this would be a perfect example for a film class on how not to make a filmbr br so while giving it a 3 is probably a bit [UNK] its fun to laugh at and short so its worth a look for bad film fans \n\n"
],
[
"\n\n",
"_____no_output_____"
],
[
"import os\nmodel2 = None\nprint(os.listdir('/content/drive/MyDrive/sentiment/'))\nif len(os.listdir('/content/drive/MyDrive/sentiment/')) == 0:\n model = tf.keras.Sequential([\n encoder,\n tf.keras.layers.Embedding(\n input_dim=len(encoder.get_vocabulary()),\n output_dim=64,\n # Use masking to handle the variable sequence lengths\n mask_zero=True),\n tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(64)),\n tf.keras.layers.Dense(64, activation='relu'),\n tf.keras.layers.Dense(1, activation='sigmoid')\n ])\n print (\"created model\")\nelse:\n model2 = tf.keras.models.load_model (\"/content/drive/MyDrive/sentiment/\")\n print (\"loaded model\")",
"['variables', 'assets', 'saved_model.pb', 'keras_metadata.pb']\n"
],
[
"if model2 is not None:\n model = model2",
"_____no_output_____"
],
[
"print([layer.supports_masking for layer in model.layers])\n",
"[False, True, True, True, True]\n"
],
[
"# predict on a sample text without padding.\n\nsample_text = ('The movie was cool. The animation and the graphics '\n 'were out of this world. I would recommend this movie.')\npredictions = model.predict(np.array([sample_text]))\nprint(predictions[0])\n",
"WARNING:tensorflow:6 out of the last 10 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7fa5d69c5320> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.\n"
],
[
"# predict on a sample text with padding\n\npadding = \"the \" * 2000\npredictions = model.predict(np.array([sample_text, padding]))\nprint(predictions[0])\n",
"[0.5011921]\n"
],
[
"model.compile(loss=tf.keras.losses.BinaryCrossentropy(from_logits=False),\n optimizer=tf.keras.optimizers.Adam(1e-4),\n metrics=['accuracy']) #run_eagerly=True)\n",
"_____no_output_____"
],
[
"history = model.fit(train_dataset, epochs=5,\n validation_data=test_dataset,\n validation_steps=30)\n",
"Epoch 1/5\n391/391 [==============================] - 112s 285ms/step - loss: 0.6542 - accuracy: 0.6130 - val_loss: 0.5631 - val_accuracy: 0.7479\nEpoch 2/5\n391/391 [==============================] - 110s 279ms/step - loss: 0.4339 - accuracy: 0.8212 - val_loss: 0.4028 - val_accuracy: 0.8385\nEpoch 3/5\n391/391 [==============================] - 110s 280ms/step - loss: 0.3584 - accuracy: 0.8520 - val_loss: 0.3456 - val_accuracy: 0.8615\nEpoch 4/5\n391/391 [==============================] - 108s 274ms/step - loss: 0.3338 - accuracy: 0.8620 - val_loss: 0.3362 - val_accuracy: 0.8615\nEpoch 5/5\n391/391 [==============================] - 107s 271ms/step - loss: 0.3217 - accuracy: 0.8693 - val_loss: 0.3322 - val_accuracy: 0.8604\n"
],
[
"test_loss, test_acc = model.evaluate(test_dataset)\n\nprint('Test Loss:', test_loss)\nprint('Test Accuracy:', test_acc)\n",
"391/391 [==============================] - 56s 143ms/step - loss: 0.3299 - accuracy: 0.8616\nTest Loss: 0.3298826515674591\nTest Accuracy: 0.8615999817848206\n"
],
[
"# predict on a sample text without padding.\n\nsample_text = ('good is great')\npredictions = model.predict(np.array([sample_text]))\nprint(predictions)\n\nsample_text = ('bad equals very bad. Worse')\npredictions = model.predict(np.array([sample_text]))\nprint(predictions)\n",
"[[1.9038882]]\n[[-1.7162021]]\n"
],
[
"x = tfds.as_numpy(test_dataset)",
"_____no_output_____"
],
[
"for ele in train_dataset.as_numpy_iterator():\n print (ele)\n print (\"---------------------\")",
"_____no_output_____"
],
[
"?plt.figure(figsize=(16, 6))\nplt.subplot(1, 2, 1)\nplot_graphs(history, 'accuracy')\nplt.subplot(1, 2, 2)\nplot_graphs(history, 'loss')\n",
"_____no_output_____"
],
[
"m = tf.keras.metrics.Accuracy()\nm.update_state([[0], [2], [3], [4]], [[0], [2], [3], [4]])\nm.result().numpy()",
"_____no_output_____"
],
[
"import copy\nvicab2 = copy.deepcopy(vocab)",
"_____no_output_____"
],
[
"vicab2.sort()",
"_____no_output_____"
],
[
"vicab2",
"_____no_output_____"
],
[
"\nlst = []\ndef func(text, label):\n lst.append([text, label])\n return text, label\n\n\ntest_dataset.map(func)",
"_____no_output_____"
],
[
"lst",
"_____no_output_____"
],
[
"for ele in test_dataset.as_numpy_iterator():\n print (ele)",
"_____no_output_____"
],
[
"tf.keras.models.save_model(model=model, filepath=\"/content/drive/MyDrive/sentiment/\")",
"WARNING:absl:Found untraced functions such as lstm_cell_4_layer_call_fn, lstm_cell_4_layer_call_and_return_conditional_losses, lstm_cell_5_layer_call_fn, lstm_cell_5_layer_call_and_return_conditional_losses, lstm_cell_4_layer_call_fn while saving (showing 5 of 10). These functions will not be directly callable after loading.\n"
],
[
"tf.saved_model.save(obj=model, export_dir=\"/content/drive/MyDrive/sentiment\")",
"WARNING:absl:Found untraced functions such as lstm_cell_19_layer_call_fn, lstm_cell_19_layer_call_and_return_conditional_losses, lstm_cell_20_layer_call_fn, lstm_cell_20_layer_call_and_return_conditional_losses, lstm_cell_19_layer_call_fn while saving (showing 5 of 10). These functions will not be directly callable after loading.\n"
],
[
"model.save(\"/content/drive/MyDrive/sentiment/model\", save_format=\"tf\")",
"WARNING:absl:Found untraced functions such as lstm_cell_19_layer_call_fn, lstm_cell_19_layer_call_and_return_conditional_losses, lstm_cell_20_layer_call_fn, lstm_cell_20_layer_call_and_return_conditional_losses, lstm_cell_19_layer_call_fn while saving (showing 5 of 10). These functions will not be directly callable after loading.\n"
],
[
"for layer in model.layers: print(layer.get_config(), layer.get_weights())",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"input_array = np.random.randint(len(encoder.get_vocabulary()), size=(3, 1))\n\nmodel_temp = tf.keras.Sequential()\nmodel_temp.add(encoder)\nmodel_temp.add(tf.keras.layers.Embedding( \n input_dim=len(encoder.get_vocabulary()),\n output_dim=64,\n # Use masking to handle the variable sequence lengths\n mask_zero=True))\nmodel_temp.compile('rmsprop', 'mse')\n# output_array = model_temp.predict(\"hello world this is great!\")\n# print(output_array.shape)\n\n\nsample_text = ('The movie was cool. The animation and the graphics '\n 'were out of this world. I would recommend this movie.')\npredictions = model_temp.predict(np.array([sample_text]))\nprint(len(predictions))\nprint(len(predictions[0]))\nprint(len(predictions[0][0]))\n\n\n\n\n\n# The model will take as input an integer matrix of size (batch,\n# input_length), and the largest integer (i.e. word index) in the input\n# should be no larger than 999 (vocabulary size).\n# Now model.output_shape is (None, 10, 64), where `None` is the batch\n# dimension.\n# input_array = np.random.randint(900, size=(3, 10))\n# model_temp.compile('rmsprop', 'mse')\n# output_array = model_temp.predict(input_array)\n# print(output_array.shape)\n\n",
"1\n19\n64\n"
],
[
"print (output_array[0][0])",
"[ 0.0051492 -0.01514421 0.0233087 -0.03882884 0.01593846]\n"
],
[
"input_array[0][0]",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05d67e78b52fa5913e8f985d914ca20a6ab2e35 | 27,963 | ipynb | Jupyter Notebook | Examples/simple_U-Net.ipynb | thgnaedi/DeepRain | 4cf9323901f38898a3b119faf07e2869630046c3 | [
"BSD-3-Clause"
] | 29 | 2018-10-15T09:55:45.000Z | 2022-03-04T07:23:32.000Z | Examples/simple_U-Net.ipynb | thgnaedi/DeepRain | 4cf9323901f38898a3b119faf07e2869630046c3 | [
"BSD-3-Clause"
] | 30 | 2018-10-23T09:06:15.000Z | 2020-01-08T04:39:24.000Z | Examples/simple_U-Net.ipynb | thgnaedi/DeepRain | 4cf9323901f38898a3b119faf07e2869630046c3 | [
"BSD-3-Clause"
] | 9 | 2019-08-29T10:00:21.000Z | 2022-01-09T02:48:44.000Z | 60.395248 | 137 | 0.625362 | [
[
[
"import tensorflow as tf\nfrom tensorflow.python.keras.utils import HDF5Matrix\nfrom tensorflow.python.keras.models import Model\nfrom tensorflow.python.keras.layers import (Input, Lambda, Conv2D, MaxPooling2D, Flatten, Dense, Dropout, \n Lambda, Activation, BatchNormalization, concatenate, UpSampling2D, \n ZeroPadding2D)\n\n\nfrom sklearn.metrics import mean_squared_error, mean_absolute_error, confusion_matrix\n\n\nfrom matplotlib import pyplot as plt\n%matplotlib inline\n\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"```Python\nx_train = HDF5Matrix(\"data.h5\", \"x_train\")\nx_valid = HDF5Matrix(\"data.h5\", \"x_valid\")\n```\nshapes should be:\n* (1355578, 432, 560, 1)\n* (420552, 432, 560, 1)",
"_____no_output_____"
]
],
[
[
"def gen_data(shape=0, name=\"input\"):\n data = np.random.rand(512, 512, 4)\n label = data[:,:,-1]\n \n return tf.constant(data.reshape(1,512,512,4).astype(np.float32)), tf.constant(label.reshape(1,512,512,1).astype(np.float32))\n\n## NOTE:\n## Tensor 4D -> Batch,X,Y,Z\n## Tesnor max. float32!\n\nd, l = gen_data(0,0)\nprint(d.shape, l.shape)",
"(1, 512, 512, 4) (1, 512, 512, 1)\n"
],
[
"def unet():\n inputs, label = gen_data()\n input_shape = inputs.shape\n #down0a = Conv2D(16, (3, 3), padding='same')(inputs)\n down0a = Conv2D(16, kernel_size=(3, 3), padding='same', input_shape=input_shape)(inputs)\n down0a_pool = MaxPooling2D((2, 2), strides=(2, 2))(down0a)\n print(\"down0a.shape:\",down0a.shape,\"\\ndwnpool.shap:\", down0a_pool.shape)#?!? letztes != Batch?\n #dim0 = Batch\n #dim1,dim2 = X,Y\n #dim3 = Kanaele\n \n up1 = UpSampling2D((3, 3))(down0a)\n print(\"upsamp.shape:\",up1.shape) #UpSampling ändert dim1, dim2... somit (?,X,Y,?) evtl. Batch auf dim0 ?\n \nunet()",
"down0a.shape: (1, 512, 512, 16) \ndwnpool.shap: (1, 256, 256, 16)\nupsamp.shape: (1, 1536, 1536, 16)\n"
],
[
"def unet2(input_shape, output_length):\n inputs = Input(shape=input_shape, name=\"input\")\n \n # 512\n down0a = Conv2D(16, (3, 3), padding='same')(inputs)\n down0a = BatchNormalization()(down0a)\n down0a = Activation('relu')(down0a)\n down0a = Conv2D(16, (3, 3), padding='same')(down0a)\n down0a = BatchNormalization()(down0a)\n down0a = Activation('relu')(down0a)\n down0a_pool = MaxPooling2D((2, 2), strides=(2, 2))(down0a)\n # 256\n down0 = Conv2D(32, (3, 3), padding='same')(down0a_pool)\n down0 = BatchNormalization()(down0)\n down0 = Activation('relu')(down0)\n down0 = Conv2D(32, (3, 3), padding='same')(down0)\n down0 = BatchNormalization()(down0)\n down0 = Activation('relu')(down0)\n down0_pool = MaxPooling2D((2, 2), strides=(2, 2))(down0)\n # 128\n down1 = Conv2D(64, (3, 3), padding='same')(down0_pool)\n down1 = BatchNormalization()(down1)\n down1 = Activation('relu')(down1)\n down1 = Conv2D(64, (3, 3), padding='same')(down1)\n down1 = BatchNormalization()(down1)\n down1 = Activation('relu')(down1)\n down1_pool = MaxPooling2D((2, 2), strides=(2, 2))(down1)\n # 64\n down2 = Conv2D(128, (3, 3), padding='same')(down1_pool)\n down2 = BatchNormalization()(down2)\n down2 = Activation('relu')(down2)\n down2 = Conv2D(128, (3, 3), padding='same')(down2)\n down2 = BatchNormalization()(down2)\n down2 = Activation('relu')(down2)\n down2_pool = MaxPooling2D((2, 2), strides=(2, 2))(down2)\n # 8\n center = Conv2D(1024, (3, 3), padding='same')(down2_pool)\n center = BatchNormalization()(center)\n center = Activation('relu')(center)\n center = Conv2D(1024, (3, 3), padding='same')(center)\n center = BatchNormalization()(center)\n center = Activation('relu')(center)\n # center\n up2 = UpSampling2D((2, 2))(center)\n up2 = concatenate([down2, up2], axis=3)\n up2 = Conv2D(128, (3, 3), padding='same')(up2)\n up2 = BatchNormalization()(up2)\n up2 = Activation('relu')(up2)\n up2 = Conv2D(128, (3, 3), padding='same')(up2)\n up2 = BatchNormalization()(up2)\n up2 = Activation('relu')(up2)\n up2 = Conv2D(128, (3, 3), padding='same')(up2)\n up2 = BatchNormalization()(up2)\n up2 = Activation('relu')(up2)\n # 64\n up1 = UpSampling2D((2, 2))(up2)\n up1 = concatenate([down1, up1], axis=3)\n up1 = Conv2D(64, (3, 3), padding='same')(up1)\n up1 = BatchNormalization()(up1)\n up1 = Activation('relu')(up1)\n up1 = Conv2D(64, (3, 3), padding='same')(up1)\n up1 = BatchNormalization()(up1)\n up1 = Activation('relu')(up1)\n up1 = Conv2D(64, (3, 3), padding='same')(up1)\n up1 = BatchNormalization()(up1)\n up1 = Activation('relu')(up1)\n # 128\n up0 = UpSampling2D((2, 2))(up1)\n up0 = concatenate([down0, up0], axis=3)\n up0 = Conv2D(32, (3, 3), padding='same')(up0)\n up0 = BatchNormalization()(up0)\n up0 = Activation('relu')(up0)\n up0 = Conv2D(32, (3, 3), padding='same')(up0)\n up0 = BatchNormalization()(up0)\n up0 = Activation('relu')(up0)\n up0 = Conv2D(32, (3, 3), padding='same')(up0)\n up0 = BatchNormalization()(up0)\n up0 = Activation('relu')(up0)\n # 256\n up0a = UpSampling2D((2, 2))(up0)\n up0a = concatenate([down0a, up0a], axis=3)\n up0a = Conv2D(16, (3, 3), padding='same')(up0a)\n up0a = BatchNormalization()(up0a)\n up0a = Activation('relu')(up0a)\n up0a = Conv2D(16, (3, 3), padding='same')(up0a)\n up0a = BatchNormalization()(up0a)\n up0a = Activation('relu')(up0a)\n up0a = Conv2D(16, (3, 3), padding='same')(up0a)\n up0a = BatchNormalization()(up0a)\n up0a = Activation('relu')(up0a)\n # 512\n output = Conv2D(1, (1, 1), activation='relu')(up0a)\n \n model = Model(inputs=inputs, outputs=output)\n model.compile(loss=\"mean_squared_error\", optimizer='adam')\n return model\n\nd = unet2((512,512,4),(512,512,1))\n",
"_____no_output_____"
]
],
[
[
"Anschließend:\n```Python\noutput_length = 1\ninput_length = output_length + 1\ninput_shape=(432, 560, input_length)\n\nmodel_1 = unet(input_shape, output_length)\nmodel_1.fit(x_train_1, y_train_1, batch_size = 16, epochs = 25, \n validation_data=(x_valid_1, y_valid_1))\n```",
"_____no_output_____"
]
],
[
[
"d.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput (InputLayer) (None, 512, 512, 4) 0 \n__________________________________________________________________________________________________\nconv2d_116 (Conv2D) (None, 512, 512, 16) 592 input[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_111 (BatchN (None, 512, 512, 16) 64 conv2d_116[0][0] \n__________________________________________________________________________________________________\nactivation_111 (Activation) (None, 512, 512, 16) 0 batch_normalization_111[0][0] \n__________________________________________________________________________________________________\nconv2d_117 (Conv2D) (None, 512, 512, 16) 2320 activation_111[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_112 (BatchN (None, 512, 512, 16) 64 conv2d_117[0][0] \n__________________________________________________________________________________________________\nactivation_112 (Activation) (None, 512, 512, 16) 0 batch_normalization_112[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_22 (MaxPooling2D) (None, 256, 256, 16) 0 activation_112[0][0] \n__________________________________________________________________________________________________\nconv2d_118 (Conv2D) (None, 256, 256, 32) 4640 max_pooling2d_22[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_113 (BatchN (None, 256, 256, 32) 128 conv2d_118[0][0] \n__________________________________________________________________________________________________\nactivation_113 (Activation) (None, 256, 256, 32) 0 batch_normalization_113[0][0] \n__________________________________________________________________________________________________\nconv2d_119 (Conv2D) (None, 256, 256, 32) 9248 activation_113[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_114 (BatchN (None, 256, 256, 32) 128 conv2d_119[0][0] \n__________________________________________________________________________________________________\nactivation_114 (Activation) (None, 256, 256, 32) 0 batch_normalization_114[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_23 (MaxPooling2D) (None, 128, 128, 32) 0 activation_114[0][0] \n__________________________________________________________________________________________________\nconv2d_120 (Conv2D) (None, 128, 128, 64) 18496 max_pooling2d_23[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_115 (BatchN (None, 128, 128, 64) 256 conv2d_120[0][0] \n__________________________________________________________________________________________________\nactivation_115 (Activation) (None, 128, 128, 64) 0 batch_normalization_115[0][0] \n__________________________________________________________________________________________________\nconv2d_121 (Conv2D) (None, 128, 128, 64) 36928 activation_115[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_116 (BatchN (None, 128, 128, 64) 256 conv2d_121[0][0] \n__________________________________________________________________________________________________\nactivation_116 (Activation) (None, 128, 128, 64) 0 batch_normalization_116[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_24 (MaxPooling2D) (None, 64, 64, 64) 0 activation_116[0][0] \n__________________________________________________________________________________________________\nconv2d_122 (Conv2D) (None, 64, 64, 128) 73856 max_pooling2d_24[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_117 (BatchN (None, 64, 64, 128) 512 conv2d_122[0][0] \n__________________________________________________________________________________________________\nactivation_117 (Activation) (None, 64, 64, 128) 0 batch_normalization_117[0][0] \n__________________________________________________________________________________________________\nconv2d_123 (Conv2D) (None, 64, 64, 128) 147584 activation_117[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_118 (BatchN (None, 64, 64, 128) 512 conv2d_123[0][0] \n__________________________________________________________________________________________________\nactivation_118 (Activation) (None, 64, 64, 128) 0 batch_normalization_118[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_25 (MaxPooling2D) (None, 32, 32, 128) 0 activation_118[0][0] \n__________________________________________________________________________________________________\nconv2d_124 (Conv2D) (None, 32, 32, 1024) 1180672 max_pooling2d_25[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_119 (BatchN (None, 32, 32, 1024) 4096 conv2d_124[0][0] \n__________________________________________________________________________________________________\nactivation_119 (Activation) (None, 32, 32, 1024) 0 batch_normalization_119[0][0] \n__________________________________________________________________________________________________\nconv2d_125 (Conv2D) (None, 32, 32, 1024) 9438208 activation_119[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_120 (BatchN (None, 32, 32, 1024) 4096 conv2d_125[0][0] \n__________________________________________________________________________________________________\nactivation_120 (Activation) (None, 32, 32, 1024) 0 batch_normalization_120[0][0] \n__________________________________________________________________________________________________\nup_sampling2d_22 (UpSampling2D) (None, 64, 64, 1024) 0 activation_120[0][0] \n__________________________________________________________________________________________________\nconcatenate_21 (Concatenate) (None, 64, 64, 1152) 0 activation_118[0][0] \n up_sampling2d_22[0][0] \n__________________________________________________________________________________________________\nconv2d_126 (Conv2D) (None, 64, 64, 128) 1327232 concatenate_21[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_121 (BatchN (None, 64, 64, 128) 512 conv2d_126[0][0] \n__________________________________________________________________________________________________\nactivation_121 (Activation) (None, 64, 64, 128) 0 batch_normalization_121[0][0] \n__________________________________________________________________________________________________\nconv2d_127 (Conv2D) (None, 64, 64, 128) 147584 activation_121[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_122 (BatchN (None, 64, 64, 128) 512 conv2d_127[0][0] \n__________________________________________________________________________________________________\nactivation_122 (Activation) (None, 64, 64, 128) 0 batch_normalization_122[0][0] \n__________________________________________________________________________________________________\nconv2d_128 (Conv2D) (None, 64, 64, 128) 147584 activation_122[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_123 (BatchN (None, 64, 64, 128) 512 conv2d_128[0][0] \n__________________________________________________________________________________________________\nactivation_123 (Activation) (None, 64, 64, 128) 0 batch_normalization_123[0][0] \n__________________________________________________________________________________________________\nup_sampling2d_23 (UpSampling2D) (None, 128, 128, 128 0 activation_123[0][0] \n__________________________________________________________________________________________________\nconcatenate_22 (Concatenate) (None, 128, 128, 192 0 activation_116[0][0] \n up_sampling2d_23[0][0] \n__________________________________________________________________________________________________\nconv2d_129 (Conv2D) (None, 128, 128, 64) 110656 concatenate_22[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_124 (BatchN (None, 128, 128, 64) 256 conv2d_129[0][0] \n__________________________________________________________________________________________________\nactivation_124 (Activation) (None, 128, 128, 64) 0 batch_normalization_124[0][0] \n__________________________________________________________________________________________________\nconv2d_130 (Conv2D) (None, 128, 128, 64) 36928 activation_124[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_125 (BatchN (None, 128, 128, 64) 256 conv2d_130[0][0] \n__________________________________________________________________________________________________\nactivation_125 (Activation) (None, 128, 128, 64) 0 batch_normalization_125[0][0] \n__________________________________________________________________________________________________\nconv2d_131 (Conv2D) (None, 128, 128, 64) 36928 activation_125[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_126 (BatchN (None, 128, 128, 64) 256 conv2d_131[0][0] \n__________________________________________________________________________________________________\nactivation_126 (Activation) (None, 128, 128, 64) 0 batch_normalization_126[0][0] \n__________________________________________________________________________________________________\nup_sampling2d_24 (UpSampling2D) (None, 256, 256, 64) 0 activation_126[0][0] \n__________________________________________________________________________________________________\nconcatenate_23 (Concatenate) (None, 256, 256, 96) 0 activation_114[0][0] \n up_sampling2d_24[0][0] \n__________________________________________________________________________________________________\nconv2d_132 (Conv2D) (None, 256, 256, 32) 27680 concatenate_23[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_127 (BatchN (None, 256, 256, 32) 128 conv2d_132[0][0] \n__________________________________________________________________________________________________\nactivation_127 (Activation) (None, 256, 256, 32) 0 batch_normalization_127[0][0] \n__________________________________________________________________________________________________\nconv2d_133 (Conv2D) (None, 256, 256, 32) 9248 activation_127[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_128 (BatchN (None, 256, 256, 32) 128 conv2d_133[0][0] \n__________________________________________________________________________________________________\nactivation_128 (Activation) (None, 256, 256, 32) 0 batch_normalization_128[0][0] \n__________________________________________________________________________________________________\nconv2d_134 (Conv2D) (None, 256, 256, 32) 9248 activation_128[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_129 (BatchN (None, 256, 256, 32) 128 conv2d_134[0][0] \n__________________________________________________________________________________________________\nactivation_129 (Activation) (None, 256, 256, 32) 0 batch_normalization_129[0][0] \n__________________________________________________________________________________________________\nup_sampling2d_25 (UpSampling2D) (None, 512, 512, 32) 0 activation_129[0][0] \n__________________________________________________________________________________________________\nconcatenate_24 (Concatenate) (None, 512, 512, 48) 0 activation_112[0][0] \n up_sampling2d_25[0][0] \n__________________________________________________________________________________________________\nconv2d_135 (Conv2D) (None, 512, 512, 16) 6928 concatenate_24[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_130 (BatchN (None, 512, 512, 16) 64 conv2d_135[0][0] \n__________________________________________________________________________________________________\nactivation_130 (Activation) (None, 512, 512, 16) 0 batch_normalization_130[0][0] \n__________________________________________________________________________________________________\nconv2d_136 (Conv2D) (None, 512, 512, 16) 2320 activation_130[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_131 (BatchN (None, 512, 512, 16) 64 conv2d_136[0][0] \n__________________________________________________________________________________________________\nactivation_131 (Activation) (None, 512, 512, 16) 0 batch_normalization_131[0][0] \n__________________________________________________________________________________________________\nconv2d_137 (Conv2D) (None, 512, 512, 16) 2320 activation_131[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_132 (BatchN (None, 512, 512, 16) 64 conv2d_137[0][0] \n__________________________________________________________________________________________________\nactivation_132 (Activation) (None, 512, 512, 16) 0 batch_normalization_132[0][0] \n__________________________________________________________________________________________________\nconv2d_138 (Conv2D) (None, 512, 512, 1) 17 activation_132[0][0] \n==================================================================================================\nTotal params: 12,790,209\nTrainable params: 12,783,713\nNon-trainable params: 6,496\n__________________________________________________________________________________________________\n"
],
[
"#ToDo: now learn something!",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05d6ec784f0b65ee60163a0572a3aad913877ee | 24,667 | ipynb | Jupyter Notebook | imooc/PCA/PCA.ipynb | imgoodman/play-with-machine-learning | 377da326aba699e76295cb9020f9fa392febf1a4 | [
"Apache-2.0"
] | null | null | null | imooc/PCA/PCA.ipynb | imgoodman/play-with-machine-learning | 377da326aba699e76295cb9020f9fa392febf1a4 | [
"Apache-2.0"
] | null | null | null | imooc/PCA/PCA.ipynb | imgoodman/play-with-machine-learning | 377da326aba699e76295cb9020f9fa392febf1a4 | [
"Apache-2.0"
] | null | null | null | 68.519444 | 17,032 | 0.790368 | [
[
[
"import numpy as np",
"_____no_output_____"
],
[
"class PCA:\n def __init__(self, n_components):\n \"\"\"\n 初始化PCA\n \"\"\"\n assert n_components>=1, \"n_components 必须大于1\"\n \n self.n_components=n_components\n self.components_=None\n \n def fit(self, X, eta=0.01,n_iters=1e4):\n \"\"\"\n 获得数据集X的n_components主成分\n \"\"\"\n assert self.n_components <= X.shape[1], \"主成分数量不能大于数据的维度\"\n \n def demean(X):\n \"\"\"\n 将数据集的均值变为0\n \"\"\"\n return X - np.mean(X, axis=0)\n def f(w, X):\n \"\"\"\n 目标函数\n \"\"\"\n return np.sum(X.dot(w)**2)/len(X)\n def df(w, X):\n \"\"\"\n 目标函数的梯度\n \"\"\"\n return X.T.dot(X.dot(w))*2/len(X)\n def direction(w):\n \"\"\"\n 将向量转化为标准向量\n \"\"\"\n return w/np.linalg.norm(w)\n def first_components(X, initial_w, eta=0.01, n_iters=1e4, epsilon=1e-8):\n w=direction(initial_w)\n cur_iter=0\n \n while cur_iter<n_iters:\n gradient=df(w, X)\n last_w=w\n w=w+eta*gradient\n w=direction(w)\n if abs(f(w,X) - f(last_w, X))<epsilon:\n break\n cur_iter+=1\n return w\n X_pca=demean(X)\n self.components_=np.empty((self.n_components, X.shape[1]))\n for i in range(self.n_components):\n initial_w=np.random.random(X_pca.shape[1])\n w=first_components(X_pca, initial_w, eta, n_iters)\n self.components_[i,:]=w\n \n X_pca=X_pca - X_pca.dot(w).reshape(-1,1)*w\n return self\n \n def transform(self, X):\n \"\"\"\n 将X,映射到各个主成分分量中\n \"\"\"\n assert X.shape[1]==self.components_.shape[1], \"维度要一致\"\n \n return X.dot(self.components_.T)\n \n def inverse_transform(self,X):\n \"\"\"\n 将X,反向映射到原来的特征空间\n \"\"\"\n assert X.shape[1]==self.components_.shape[0]\n \n return X.dot(self.components_)\n \n def __repr__(self):\n return \"PCA(n_components=%d)\" % self.n_components",
"_____no_output_____"
],
[
"X=np.empty((100,2))\nX[:,0]=np.random.uniform(0.0,100.0,size=100)\nX[:,1]=0.75*X[:,0]+3.0+np.random.normal(0.0,10.0,size=100)",
"_____no_output_____"
],
[
"X.shape",
"_____no_output_____"
],
[
"pca=PCA(n_components=2)\npca.fit(X)",
"_____no_output_____"
],
[
"pca.components_",
"_____no_output_____"
],
[
"pca=PCA(n_components=1)\npca.fit(X)",
"_____no_output_____"
],
[
"pca.components_",
"_____no_output_____"
],
[
"x_reduction=pca.transform(X)",
"_____no_output_____"
],
[
"x_reduction.shape",
"_____no_output_____"
],
[
"x_restore=pca.inverse_transform(x_reduction)",
"_____no_output_____"
],
[
"x_restore.shape",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"plt.scatter(X[:,0],X[:,1], color='b',alpha=0.5)\nplt.scatter(x_restore[:,0], x_restore[:,1], color='r', alpha=0.5)\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05d7596c569ba380e5ee9f2c1fef5fd1af06f56 | 18,639 | ipynb | Jupyter Notebook | assignment1/two_layer_net.ipynb | ayush29feb/cs231n | 155f411ee353eff3d46ecfff5fa9c6c37aa2afb0 | [
"MIT"
] | 1 | 2016-12-20T05:45:00.000Z | 2016-12-20T05:45:00.000Z | assignment1/two_layer_net.ipynb | ayush29feb/cs231n | 155f411ee353eff3d46ecfff5fa9c6c37aa2afb0 | [
"MIT"
] | null | null | null | assignment1/two_layer_net.ipynb | ayush29feb/cs231n | 155f411ee353eff3d46ecfff5fa9c6c37aa2afb0 | [
"MIT"
] | null | null | null | 36.618861 | 524 | 0.588175 | [
[
[
"# Implementing a Neural Network\nIn this exercise we will develop a neural network with fully-connected layers to perform classification, and test it out on the CIFAR-10 dataset.",
"_____no_output_____"
]
],
[
[
"# A bit of setup\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom cs231n.classifiers.neural_net import TwoLayerNet\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n# for auto-reloading external modules\n# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython\n%load_ext autoreload\n%autoreload 2\n\ndef rel_error(x, y):\n \"\"\" returns relative error \"\"\"\n return np.max(np.abs(x - y) / (np.maximum(1e-8, np.abs(x) + np.abs(y))))",
"/Users/ayush/anaconda2/lib/python2.7/site-packages/matplotlib/font_manager.py:273: UserWarning: Matplotlib is building the font cache using fc-list. This may take a moment.\n warnings.warn('Matplotlib is building the font cache using fc-list. This may take a moment.')\n"
]
],
[
[
"We will use the class `TwoLayerNet` in the file `cs231n/classifiers/neural_net.py` to represent instances of our network. The network parameters are stored in the instance variable `self.params` where keys are string parameter names and values are numpy arrays. Below, we initialize toy data and a toy model that we will use to develop your implementation.",
"_____no_output_____"
]
],
[
[
"# Create a small net and some toy data to check your implementations.\n# Note that we set the random seed for repeatable experiments.\n\ninput_size = 4\nhidden_size = 10\nnum_classes = 3\nnum_inputs = 5\n\ndef init_toy_model():\n np.random.seed(0)\n return TwoLayerNet(input_size, hidden_size, num_classes, std=1e-1)\n\ndef init_toy_data():\n np.random.seed(1)\n X = 10 * np.random.randn(num_inputs, input_size)\n y = np.array([0, 1, 2, 2, 1])\n return X, y\n\nnet = init_toy_model()\nX, y = init_toy_data()",
"_____no_output_____"
]
],
[
[
"# Forward pass: compute scores\nOpen the file `cs231n/classifiers/neural_net.py` and look at the method `TwoLayerNet.loss`. This function is very similar to the loss functions you have written for the SVM and Softmax exercises: It takes the data and weights and computes the class scores, the loss, and the gradients on the parameters. \n\nImplement the first part of the forward pass which uses the weights and biases to compute the scores for all inputs.",
"_____no_output_____"
]
],
[
[
"scores = net.loss(X)\nprint 'Your scores:'\nprint scores\nprint\nprint 'correct scores:'\ncorrect_scores = np.asarray([\n [-0.81233741, -1.27654624, -0.70335995],\n [-0.17129677, -1.18803311, -0.47310444],\n [-0.51590475, -1.01354314, -0.8504215 ],\n [-0.15419291, -0.48629638, -0.52901952],\n [-0.00618733, -0.12435261, -0.15226949]])\nprint correct_scores\nprint\n\n# The difference should be very small. We get < 1e-7\nprint 'Difference between your scores and correct scores:'\nprint np.sum(np.abs(scores - correct_scores))",
"Your scores:\n[[-0.81233741 -1.27654624 -0.70335995]\n [-0.17129677 -1.18803311 -0.47310444]\n [-0.51590475 -1.01354314 -0.8504215 ]\n [-0.15419291 -0.48629638 -0.52901952]\n [-0.00618733 -0.12435261 -0.15226949]]\n\ncorrect scores:\n[[-0.81233741 -1.27654624 -0.70335995]\n [-0.17129677 -1.18803311 -0.47310444]\n [-0.51590475 -1.01354314 -0.8504215 ]\n [-0.15419291 -0.48629638 -0.52901952]\n [-0.00618733 -0.12435261 -0.15226949]]\n\nDifference between your scores and correct scores:\n3.68027209255e-08\n"
]
],
[
[
"# Forward pass: compute loss\nIn the same function, implement the second part that computes the data and regularizaion loss.",
"_____no_output_____"
]
],
[
[
"loss, _ = net.loss(X, y, reg=0.1)\ncorrect_loss = 1.30378789133\n\n# should be very small, we get < 1e-12\nprint 'Difference between your loss and correct loss:'\nprint np.sum(np.abs(loss - correct_loss))",
"Difference between your loss and correct loss:\n1.79856129989e-13\n"
]
],
[
[
"# Backward pass\nImplement the rest of the function. This will compute the gradient of the loss with respect to the variables `W1`, `b1`, `W2`, and `b2`. Now that you (hopefully!) have a correctly implemented forward pass, you can debug your backward pass using a numeric gradient check:",
"_____no_output_____"
]
],
[
[
"from cs231n.gradient_check import eval_numerical_gradient\n\n# Use numeric gradient checking to check your implementation of the backward pass.\n# If your implementation is correct, the difference between the numeric and\n# analytic gradients should be less than 1e-8 for each of W1, W2, b1, and b2.\n\nloss, grads = net.loss(X, y, reg=0.1)\n\n# these should all be less than 1e-8 or so\nfor param_name in grads:\n f = lambda W: net.loss(X, y, reg=0.1)[0]\n param_grad_num = eval_numerical_gradient(f, net.params[param_name], verbose=False)\n print param_grad_num.shape\n print '%s max relative error: %e' % (param_name, rel_error(param_grad_num, grads[param_name]))",
"(4, 10)\nW1 max relative error: 1.000000e+00\n"
]
],
[
[
"# Train the network\nTo train the network we will use stochastic gradient descent (SGD), similar to the SVM and Softmax classifiers. Look at the function `TwoLayerNet.train` and fill in the missing sections to implement the training procedure. This should be very similar to the training procedure you used for the SVM and Softmax classifiers. You will also have to implement `TwoLayerNet.predict`, as the training process periodically performs prediction to keep track of accuracy over time while the network trains.\n\nOnce you have implemented the method, run the code below to train a two-layer network on toy data. You should achieve a training loss less than 0.2.",
"_____no_output_____"
]
],
[
[
"net = init_toy_model()\nstats = net.train(X, y, X, y,\n learning_rate=1e-1, reg=1e-5,\n num_iters=100, verbose=False)\n\nprint 'Final training loss: ', stats['loss_history'][-1]\n\n# plot the loss history\nplt.plot(stats['loss_history'])\nplt.xlabel('iteration')\nplt.ylabel('training loss')\nplt.title('Training Loss history')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Load the data\nNow that you have implemented a two-layer network that passes gradient checks and works on toy data, it's time to load up our favorite CIFAR-10 data so we can use it to train a classifier on a real dataset.",
"_____no_output_____"
]
],
[
[
"from cs231n.data_utils import load_CIFAR10\n\ndef get_CIFAR10_data(num_training=49000, num_validation=1000, num_test=1000):\n \"\"\"\n Load the CIFAR-10 dataset from disk and perform preprocessing to prepare\n it for the two-layer neural net classifier. These are the same steps as\n we used for the SVM, but condensed to a single function. \n \"\"\"\n # Load the raw CIFAR-10 data\n cifar10_dir = 'cs231n/datasets/cifar-10-batches-py'\n X_train, y_train, X_test, y_test = load_CIFAR10(cifar10_dir)\n \n # Subsample the data\n mask = range(num_training, num_training + num_validation)\n X_val = X_train[mask]\n y_val = y_train[mask]\n mask = range(num_training)\n X_train = X_train[mask]\n y_train = y_train[mask]\n mask = range(num_test)\n X_test = X_test[mask]\n y_test = y_test[mask]\n\n # Normalize the data: subtract the mean image\n mean_image = np.mean(X_train, axis=0)\n X_train -= mean_image\n X_val -= mean_image\n X_test -= mean_image\n\n # Reshape data to rows\n X_train = X_train.reshape(num_training, -1)\n X_val = X_val.reshape(num_validation, -1)\n X_test = X_test.reshape(num_test, -1)\n\n return X_train, y_train, X_val, y_val, X_test, y_test\n\n\n# Invoke the above function to get our data.\nX_train, y_train, X_val, y_val, X_test, y_test = get_CIFAR10_data()\nprint 'Train data shape: ', X_train.shape\nprint 'Train labels shape: ', y_train.shape\nprint 'Validation data shape: ', X_val.shape\nprint 'Validation labels shape: ', y_val.shape\nprint 'Test data shape: ', X_test.shape\nprint 'Test labels shape: ', y_test.shape",
"_____no_output_____"
]
],
[
[
"# Train a network\nTo train our network we will use SGD with momentum. In addition, we will adjust the learning rate with an exponential learning rate schedule as optimization proceeds; after each epoch, we will reduce the learning rate by multiplying it by a decay rate.",
"_____no_output_____"
]
],
[
[
"input_size = 32 * 32 * 3\nhidden_size = 50\nnum_classes = 10\nnet = TwoLayerNet(input_size, hidden_size, num_classes)\n\n# Train the network\nstats = net.train(X_train, y_train, X_val, y_val,\n num_iters=1000, batch_size=200,\n learning_rate=1e-4, learning_rate_decay=0.95,\n reg=0.5, verbose=True)\n\n# Predict on the validation set\nval_acc = (net.predict(X_val) == y_val).mean()\nprint 'Validation accuracy: ', val_acc\n\n",
"_____no_output_____"
]
],
[
[
"# Debug the training\nWith the default parameters we provided above, you should get a validation accuracy of about 0.29 on the validation set. This isn't very good.\n\nOne strategy for getting insight into what's wrong is to plot the loss function and the accuracies on the training and validation sets during optimization.\n\nAnother strategy is to visualize the weights that were learned in the first layer of the network. In most neural networks trained on visual data, the first layer weights typically show some visible structure when visualized.",
"_____no_output_____"
]
],
[
[
"# Plot the loss function and train / validation accuracies\nplt.subplot(2, 1, 1)\nplt.plot(stats['loss_history'])\nplt.title('Loss history')\nplt.xlabel('Iteration')\nplt.ylabel('Loss')\n\nplt.subplot(2, 1, 2)\nplt.plot(stats['train_acc_history'], label='train')\nplt.plot(stats['val_acc_history'], label='val')\nplt.title('Classification accuracy history')\nplt.xlabel('Epoch')\nplt.ylabel('Clasification accuracy')\nplt.show()",
"_____no_output_____"
],
[
"from cs231n.vis_utils import visualize_grid\n\n# Visualize the weights of the network\n\ndef show_net_weights(net):\n W1 = net.params['W1']\n W1 = W1.reshape(32, 32, 3, -1).transpose(3, 0, 1, 2)\n plt.imshow(visualize_grid(W1, padding=3).astype('uint8'))\n plt.gca().axis('off')\n plt.show()\n\nshow_net_weights(net)",
"_____no_output_____"
]
],
[
[
"# Tune your hyperparameters\n\n**What's wrong?**. Looking at the visualizations above, we see that the loss is decreasing more or less linearly, which seems to suggest that the learning rate may be too low. Moreover, there is no gap between the training and validation accuracy, suggesting that the model we used has low capacity, and that we should increase its size. On the other hand, with a very large model we would expect to see more overfitting, which would manifest itself as a very large gap between the training and validation accuracy.\n\n**Tuning**. Tuning the hyperparameters and developing intuition for how they affect the final performance is a large part of using Neural Networks, so we want you to get a lot of practice. Below, you should experiment with different values of the various hyperparameters, including hidden layer size, learning rate, numer of training epochs, and regularization strength. You might also consider tuning the learning rate decay, but you should be able to get good performance using the default value.\n\n**Approximate results**. You should be aim to achieve a classification accuracy of greater than 48% on the validation set. Our best network gets over 52% on the validation set.\n\n**Experiment**: You goal in this exercise is to get as good of a result on CIFAR-10 as you can, with a fully-connected Neural Network. For every 1% above 52% on the Test set we will award you with one extra bonus point. Feel free implement your own techniques (e.g. PCA to reduce dimensionality, or adding dropout, or adding features to the solver, etc.).",
"_____no_output_____"
]
],
[
[
"best_net = None # store the best model into this \n\n#################################################################################\n# TODO: Tune hyperparameters using the validation set. Store your best trained #\n# model in best_net. #\n# #\n# To help debug your network, it may help to use visualizations similar to the #\n# ones we used above; these visualizations will have significant qualitative #\n# differences from the ones we saw above for the poorly tuned network. #\n# #\n# Tweaking hyperparameters by hand can be fun, but you might find it useful to #\n# write code to sweep through possible combinations of hyperparameters #\n# automatically like we did on the previous exercises. #\n#################################################################################\npass\n#################################################################################\n# END OF YOUR CODE #\n#################################################################################",
"_____no_output_____"
],
[
"# visualize the weights of the best network\nshow_net_weights(best_net)",
"_____no_output_____"
]
],
[
[
"# Run on the test set\nWhen you are done experimenting, you should evaluate your final trained network on the test set; you should get above 48%.\n\n**We will give you extra bonus point for every 1% of accuracy above 52%.**",
"_____no_output_____"
]
],
[
[
"test_acc = (best_net.predict(X_test) == y_test).mean()\nprint 'Test accuracy: ', test_acc",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d05d88b1114f7fdd1c97490c839464700e6b5c5e | 75,566 | ipynb | Jupyter Notebook | ranking_stocks_by_category.ipynb | pritishyuvraj/profit-from-stock | b66d1988c4fd209687900f04015b0ddef0f4b35b | [
"MIT"
] | null | null | null | ranking_stocks_by_category.ipynb | pritishyuvraj/profit-from-stock | b66d1988c4fd209687900f04015b0ddef0f4b35b | [
"MIT"
] | null | null | null | ranking_stocks_by_category.ipynb | pritishyuvraj/profit-from-stock | b66d1988c4fd209687900f04015b0ddef0f4b35b | [
"MIT"
] | null | null | null | 28.472494 | 2,016 | 0.51191 | [
[
[
"import yfinance as yf \nimport pandas as pd\nimport csv",
"_____no_output_____"
],
[
"# Address to folders\nstock_info_directory = \"/Users/pyuvraj/CCPP/data_for_profit_from_stock/all_stocks_historical_prices/stocks\"\nranked_growth_stocks = stock_info_directory + \"/ranked_stock_prices.csv\"",
"_____no_output_____"
],
[
"msft = yf.Ticker(\"MSFT\")",
"_____no_output_____"
],
[
"print(msft)",
"yfinance.Ticker object <MSFT>\n"
],
[
"dir(msft)",
"_____no_output_____"
],
[
"msft.recommendations",
"_____no_output_____"
],
[
"msft.info",
"_____no_output_____"
],
[
"msft.recommendations[:-5]",
"_____no_output_____"
],
[
"all_growth_stocks = pd.read_csv(ranked_growth_stocks)",
"_____no_output_____"
],
[
"print(all_growth_stocks)",
" stock_name stock_growth\n0 CEI.csv -3.779877e+02\n1 INPX.csv -3.609980e+02\n2 CHFS.csv -3.583176e+02\n3 TRNX.csv -3.518619e+02\n4 SLS.csv -3.515691e+02\n... ... ...\n4464 TT.csv 7.229298e+04\n4465 WEI.csv 1.167866e+05\n4466 OSW.csv 1.195446e+05\n4467 JPT.csv 2.849168e+05\n4468 RCP.csv 1.263946e+06\n\n[4469 rows x 2 columns]\n"
],
[
"only_positive_growth_stocks = all_growth_stocks.drop(all_growth_stocks[all_growth_stocks.stock_growth < 1].index)",
"_____no_output_____"
],
[
"print(only_positive_growth_stocks)",
" stock_name stock_growth\n2176 LARK.csv 1.035300e+00\n2177 EHI.csv 1.042683e+00\n2178 TVE.csv 1.047389e+00\n2179 INBKL.csv 1.099508e+00\n2180 GYC.csv 1.166316e+00\n... ... ...\n4464 TT.csv 7.229298e+04\n4465 WEI.csv 1.167866e+05\n4466 OSW.csv 1.195446e+05\n4467 JPT.csv 2.849168e+05\n4468 RCP.csv 1.263946e+06\n\n[2293 rows x 2 columns]\n"
],
[
"stock_category = []\nfor index, row in only_positive_growth_stocks.iterrows():\n# if index > 2181: break\n print(row.stock_name, row.stock_growth)\n stock_name = row.stock_name[:-4]\n print(stock_name)\n try:\n stock_object = yf.Ticker(stock_name)\n print(stock_object.info['sector'])\n stock_category.append([row.stock_name, row.stock_growth, stock_object.info['sector']])\n except Exception:\n pass\nprint(stock_category)\n\nwith open(stock_info_directory + \"/category_wise_ranked_growth_stocks.csv\", \"w\", newline=\"\") as f:\n writer = csv.writer(f)\n writer.writerows(stock_category)",
"LARK.csv 1.0352997848863992\nLARK\nFinancial Services\nEHI.csv 1.0426829974356742\nEHI\nFinancial Services\nTVE.csv 1.0473885985452114\nTVE\nINBKL.csv 1.099507602004044\nINBKL\nGYC.csv 1.1663162969851673\nGYC\nCVX.csv 1.1675958503527184\nCVX\nEnergy\nTGH.csv 1.1684064211146443\nTGH\nIndustrials\nSYF.csv 1.1927655935143535\nSYF\nFinancial Services\nMSD.csv 1.1978174803030346\nMSD\nFinancial Services\nUSIO.csv 1.2177710564793074\nUSIO\nTechnology\nKMI.csv 1.221811767553656\nKMI\nEnergy\nPFO.csv 1.2277742529973477\nPFO\nFinancial Services\nFLEX.csv 1.2381421209592425\nFLEX\nTechnology\nADM.csv 1.257204445971973\nADM\nConsumer Defensive\nPMO.csv 1.2598701507659789\nPMO\nFinancial Services\nBGX.csv 1.275758242318708\nBGX\nFinancial Services\nHTA.csv 1.279409338649769\nHTA\nReal Estate\nMXL.csv 1.2933283435100371\nMXL\nTechnology\nSGU.csv 1.328476855761373\nSGU\nEnergy\nCTAA.csv 1.3313899856090712\nCTAA\nAJX.csv 1.3482907745138863\nAJX\nReal Estate\nGLU.csv 1.3588154428615962\nGLU\nFinancial Services\nEAB.csv 1.3678261511041927\nEAB\nSMFG.csv 1.3738055696216591\nSMFG\nFinancial Services\nPBB.csv 1.4117822623141905\nPBB\nHYI.csv 1.4702286475225967\nHYI\nFinancial Services\nBWA.csv 1.4709704827327528\nBWA\nConsumer Cyclical\nNWS.csv 1.5317006141021183\nNWS\nOLEM.csv 1.5809145672754248\nOLEM\nNXGN.csv 1.5965166619039481\nNXGN\nHealthcare\nJPS.csv 1.6098280899558604\nJPS\nFinancial Services\nKIQ.csv 1.6163687112554788\nKIQ\nIndustrials\nUSAS.csv 1.6601623396012766\nUSAS\nBasic Materials\nJAZZ.csv 1.6618074974285353\nJAZZ\nHealthcare\nHTH.csv 1.6733352255538887\nHTH\nFinancial Services\nASRVP.csv 1.6944173128846307\nASRVP\nTGP.csv 1.7004465933874788\nTGP\nEnergy\nAEGN.csv 1.705463819824796\nAEGN\nHYT.csv 1.7276540136824692\nHYT\nFinancial Services\nWSBC.csv 1.7375871952388842\nWSBC\nFinancial Services\nFSB.csv 1.7440115580680278\nFSB\nDSX.csv 1.7718203315905114\nDSX\nIndustrials\nBOKFL.csv 1.7762719274390166\nBOKFL\nMUA.csv 1.8006499954243322\nMUA\nFinancial Services\nDTJ.csv 1.8174229651440128\nDTJ\nDTUS.csv 1.8500431302977864\nDTUS\nZNH.csv 1.8504595287165413\nZNH\nIndustrials\nSYPR.csv 1.8517480568668088\nSYPR\nConsumer Cyclical\nINBK.csv 1.905453818745552\nINBK\nFinancial Services\nABM.csv 1.905716340376496\nABM\nIndustrials\nBZM.csv 1.93808411495979\nBZM\nWIW.csv 1.9448591210455528\nWIW\nFinancial Services\nBP.csv 1.9752660604813173\nBP\nEnergy\nELU.csv 1.9937522349980312\nELU\nCPIX.csv 2.003631820325481\nCPIX\nHealthcare\nWLKP.csv 2.0809908930822503\nWLKP\nBasic Materials\nFLC.csv 2.081027812214068\nFLC\nFinancial Services\nNID.csv 2.1275706656805373\nNID\nFinancial Services\nCDXC.csv 2.1516285732767604\nCDXC\nHealthcare\nGGT.csv 2.1664004003042443\nGGT\nFinancial Services\nELSE.csv 2.1819045677503235\nELSE\nTechnology\nBHC.csv 2.219508999154135\nBHC\nHealthcare\nAG.csv 2.223349479072735\nAG\nBasic Materials\nNUV.csv 2.2598609753968413\nNUV\nFinancial Services\nGFY.csv 2.2805118391649177\nGFY\nSBGI.csv 2.300359269549091\nSBGI\nCommunication Services\nJHS.csv 2.3107076895852727\nJHS\nFinancial Services\nSIMO.csv 2.325167562614656\nSIMO\nTechnology\nORRF.csv 2.386917857287916\nORRF\nFinancial Services\nDEX.csv 2.3893860138119694\nDEX\nFinancial Services\nJRI.csv 2.4332198261799918\nJRI\nFinancial Services\nTVC.csv 2.4825240069897307\nTVC\nGMTA.csv 2.493279896726143\nGMTA\nPMM.csv 2.520446361006881\nPMM\nFinancial Services\nWNC.csv 2.589482651015356\nWNC\nIndustrials\nPAM.csv 2.6032464579540147\nPAM\nUtilities\nSMCI.csv 2.6193105114686333\nSMCI\nTechnology\nVMI.csv 2.6357695076086616\nVMI\nIndustrials\nTPCO.csv 2.6388057480580045\nTPCO\nOXM.csv 2.643381322347416\nOXM\nConsumer Cyclical\nSUNS.csv 2.6518871600156957\nSUNS\nFinancial Services\nRFP.csv 2.6734368779166573\nRFP\nBasic Materials\nPINC.csv 2.7176534039384372\nPINC\nHealthcare\nAAT.csv 2.7278768399630757\nAAT\nReal Estate\nMNE.csv 2.7798844136521232\nMNE\nJHI.csv 2.799214683909629\nJHI\nFinancial Services\nPFD.csv 2.8167885424502384\nPFD\nFinancial Services\nPICO.csv 2.824212262037337\nPICO\nSENEB.csv 2.848365553902454\nSENEB\nConsumer Defensive\nPARR.csv 2.8581830098622985\nPARR\nEnergy\nPKX.csv 2.9225857585648285\nPKX\nBasic Materials\nLMHB.csv 2.928692836923898\nLMHB\nFRD.csv 2.9297101215122154\nFRD\nBasic Materials\nFMBI.csv 3.0002742139367893\nFMBI\nFinancial Services\nHTLD.csv 3.015795144351089\nHTLD\nIndustrials\nNWSA.csv 3.0649344301948864\nNWSA\nCommunication Services\nUNB.csv 3.1251745031191795\nUNB\nFinancial Services\nGSK.csv 3.1301353764570816\nGSK\nHealthcare\nBTA.csv 3.2099803584940303\nBTA\nFinancial Services\nDCOM.csv 3.2237295899710268\nDCOM\nFinancial Services\nUMPQ.csv 3.2249928030922788\nUMPQ\nFinancial Services\nRJN.csv 3.2572768315335696\nRJN\nPIM.csv 3.257343585563268\nPIM\nFinancial Services\nUCI.csv 3.264420397702024\nUCI\nSVT.csv 3.3102342993436142\nSVT\nIndustrials\nSJI.csv 3.3332323854005548\nSJI\nUtilities\nGBAB.csv 3.3789640532545424\nGBAB\nFinancial Services\nBCM.csv 3.4027291794944503\nBCM\nLFC.csv 3.4713902432956534\nLFC\nFinancial Services\nDISCA.csv 3.484430064210372\nDISCA\nCommunication Services\nTZOO.csv 3.4891760824864235\nTZOO\nConsumer Cyclical\nWVVIP.csv 3.506149754844746\nWVVIP\nConsumer Defensive\nCHY.csv 3.5230147962566014\nCHY\nFinancial Services\nFSLR.csv 3.542697720413223\nFSLR\nTechnology\nDSL.csv 3.589690885992276\nDSL\nFinancial Services\nRQI.csv 3.6567923691928734\nRQI\nFinancial Services\nINSI.csv 3.6836479351954567\nINSI\nFinancial Services\nCYCCP.csv 3.72564150168985\nCYCCP\nHealthcare\nLDP.csv 3.736844012379184\nLDP\nFinancial Services\nARDC.csv 3.750583191007493\nARDC\nFinancial Services\nIMKTA.csv 3.755412243985468\nIMKTA\nConsumer Defensive\nKOP.csv 3.7579318768424006\nKOP\nBasic Materials\nEXFO.csv 3.761356406345893\nEXFO\nTechnology\nPRCP.csv 3.8110632389703927\nPRCP\nHTGC.csv 3.817950584657721\nHTGC\nFinancial Services\nPOWL.csv 3.8279555961117686\nPOWL\nIndustrials\nDLHC.csv 3.881554836279896\nDLHC\nIndustrials\nBTT.csv 3.9396466467651168\nBTT\nFinancial Services\nPBCTP.csv 3.9553775650213066\nPBCTP\nFinancial Services\nPBF.csv 3.987280890262863\nPBF\nEnergy\nNTCT.csv 3.989045084141134\nNTCT\nTechnology\nCRK.csv 4.020263865120649\nCRK\nEnergy\nMPV.csv 4.043496164283688\nMPV\nFinancial Services\nGRX.csv 4.074433015805406\nGRX\nFinancial Services\nVLY.csv 4.155023321829038\nVLY\nFinancial Services\nNIQ.csv 4.170793275495002\nNIQ\nFinancial Services\nUEC.csv 4.227227377074058\nUEC\nEnergy\nUHS.csv 4.235639812762925\nUHS\nHealthcare\nCIK.csv 4.248757853722679\nCIK\nFinancial Services\nNZF.csv 4.260581639283637\nNZF\nFinancial Services\nIBKC.csv 4.277852751432004\nIBKC\nAMS.csv 4.341598322954482\nAMS\nHealthcare\nNEN.csv 4.348670444120109\nNEN\nReal Estate\nCNTY.csv 4.358350626043077\nCNTY\nConsumer Cyclical\nOFC.csv 4.381531672870423\nOFC\nReal Estate\nAVK.csv 4.40429861884991\nAVK\nFinancial Services\nAPAM.csv 4.421733313833292\nAPAM\nFinancial Services\nVIV.csv 4.436193526238763\nVIV\nCommunication Services\nNHS.csv 4.44754161308774\nNHS\nFinancial Services\nETX.csv 4.551747543187697\nETX\nFinancial Services\nPDT.csv 4.5768939064377605\nPDT\nFinancial Services\nTNAV.csv 4.60436602049295\nTNAV\nSCD.csv 4.608498683622844\nSCD\nFinancial Services\nDMB.csv 4.618282454904291\nDMB\nFinancial Services\nGBLI.csv 4.631373576815909\nGBLI\nFinancial Services\nGGZ.csv 4.635417561082144\nGGZ\nFinancial Services\nBNS.csv 4.672158808163892\nBNS\nFinancial Services\nTR.csv 4.701266584625041\nTR\nConsumer Defensive\nCHRW.csv 4.710972341545434\nCHRW\nIndustrials\nVRAY.csv 4.737820976391141\nVRAY\nHealthcare\nDLTR.csv 4.854123450467885\nDLTR\nConsumer Defensive\nPSX.csv 4.884613933903744\nPSX\nEnergy\nPNFP.csv 4.960472077803022\nPNFP\nFinancial Services\nNXQ.csv 4.971206684444355\nNXQ\nFinancial Services\nTFII.csv 5.0033925861350514\nTFII\nIndustrials\nSNFCA.csv 5.015829969299882\nSNFCA\nFinancial Services\nHTBK.csv 5.021812840671141\nHTBK\nFinancial Services\nDRUA.csv 5.026280058285726\nDRUA\nACH.csv 5.05030381768302\nACH\nBasic Materials\nWIA.csv 5.129418188397389\nWIA\nFinancial Services\nFORR.csv 5.162912587998352\nFORR\nIndustrials\nSSB.csv 5.171189757704322\nSSB\nFinancial Services\nJGH.csv 5.183511222388411\nJGH\nFinancial Services\nHSTM.csv 5.2073039758847335\nHSTM\nHealthcare\nOLP.csv 5.220889978212906\nOLP\nReal Estate\nUCIB.csv 5.239036023341546\nUCIB\nFORK.csv 5.243069361461348\nFORK\nCTHR.csv 5.247231019513375\nCTHR\nConsumer Cyclical\nVBF.csv 5.269588126870099\nVBF\nFinancial Services\nCUBE.csv 5.318697276139549\nCUBE\nReal Estate\nPRGX.csv 5.349438767679182\nPRGX\nERF.csv 5.35886004711972\nERF\nEnergy\nFUND.csv 5.406978584165662\nFUND\nFinancial Services\nWEA.csv 5.422683195311691\nWEA\nFinancial Services\nRTIX.csv 5.425732501685809\nRTIX\nNVG.csv 5.4436192603139695\nNVG\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05da6412df94bb4b831cbd91e872034cd921061 | 34,128 | ipynb | Jupyter Notebook | Design_Figure.ipynb | WagnerLabPapers/Waskom_JNeurosci_2014 | e5dd8ef62e6bbbb0e1991f896fa26d45da48752a | [
"BSD-3-Clause"
] | 13 | 2015-02-04T23:04:40.000Z | 2019-06-20T01:48:17.000Z | Design_Figure.ipynb | mwaskom/Waskom_JNeurosci_2014 | e5dd8ef62e6bbbb0e1991f896fa26d45da48752a | [
"BSD-3-Clause"
] | null | null | null | Design_Figure.ipynb | mwaskom/Waskom_JNeurosci_2014 | e5dd8ef62e6bbbb0e1991f896fa26d45da48752a | [
"BSD-3-Clause"
] | 3 | 2016-05-02T19:19:34.000Z | 2022-02-15T08:18:58.000Z | 169.791045 | 27,511 | 0.847193 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d05dc138290659a8a74892803aeeea9fd27283a8 | 464,975 | ipynb | Jupyter Notebook | Final_Colab.ipynb | jared-garalde/sme_deploy_heroku | 941657a77ba3d7fa2528ccf2b91dd77f7819b5e0 | [
"MIT"
] | null | null | null | Final_Colab.ipynb | jared-garalde/sme_deploy_heroku | 941657a77ba3d7fa2528ccf2b91dd77f7819b5e0 | [
"MIT"
] | null | null | null | Final_Colab.ipynb | jared-garalde/sme_deploy_heroku | 941657a77ba3d7fa2528ccf2b91dd77f7819b5e0 | [
"MIT"
] | null | null | null | 669.028777 | 391,272 | 0.93273 | [
[
[
"import pandas as pd\nimport numpy as np\n# import pymssql\n# from fuzzywuzzy import fuzz\nimport json\nimport tweepy\nfrom collections import defaultdict\nfrom datetime import datetime\nimport re\n# import pyodbc\nfrom wordcloud import WordCloud\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nfrom wordcloud import WordCloud\nimport string, nltk, re, json, tweepy, gensim, scipy.sparse, pickle, pyLDAvis, pyLDAvis.gensim\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom nltk.stem import WordNetLemmatizer\nfrom nltk.corpus import stopwords\nfrom gensim import matutils, models, corpora\nimport warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
]
],
[
[
"# Social Media Analysis",
"_____no_output_____"
],
[
"## EDA",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('./meme_cleaning.csv')",
"_____no_output_____"
],
[
"df_sentiment = pd.read_csv('563_df_sentiments.csv')",
"_____no_output_____"
],
[
"df_sentiment = df_sentiment.drop(columns=['Unnamed: 0', 'Unnamed: 0.1', 'Unnamed: 0.1.1'])",
"_____no_output_____"
],
[
"df_sentiment.head()",
"_____no_output_____"
],
[
"#Extract all words that begin with # and turn the results into a dataframe\r\ntemp = df_sentiment['Tweet'].str.lower().str.extractall(r\"(#\\w+)\")\r\ntemp.columns = ['unnamed']\r\n# Convert the multiple hashtag values into a list\r\ntemp = temp.groupby(level = 0)['unnamed'].apply(list)\r\n# Save the result as a feature in the original dataset\r\ndf_sentiment['hashtags'] = temp",
"_____no_output_____"
],
[
"for i in range(len(df_sentiment)):\r\n if df_sentiment.loc[i, 'No_of_Retweets'] >= 4:\r\n df_sentiment.loc[i, 'No_of_Retweets'] = 4\r\n\r\nfor i in range(len(df_sentiment)):\r\n if df_sentiment.loc[i, 'No_of_Likes'] >= 10:\r\n df_sentiment.loc[i, 'No_of_Likes'] = 10",
"_____no_output_____"
],
[
"retweet_df = df_sentiment.groupby(['No_of_Retweets', 'vaderSentiment']).vaderSentimentScores.agg(count='count').reset_index()\r\nlike_df = df_sentiment.groupby(['No_of_Likes', 'vaderSentiment']).vaderSentimentScores.agg(count='count').reset_index()\r\nclassify_df = df_sentiment.vaderSentiment.value_counts().reset_index()\r\ndf_sentiment.Labels = df_sentiment.Labels.fillna('')",
"_____no_output_____"
],
[
"df_likes_dict = df_sentiment.groupby('No_of_Likes').vaderSentimentScores.agg(count='count').to_dict()['count']\r\ndf_retweet_dict = df_sentiment.groupby('No_of_Retweets').vaderSentimentScores.agg(count='count').to_dict()['count']",
"_____no_output_____"
],
[
"for i in range(len(like_df)):\r\n like_df.loc[i, 'Normalized_count'] = like_df.loc[i, 'count'] / df_likes_dict[like_df.loc[i, 'No_of_Likes']]\r\n\r\nfor i in range(len(retweet_df)):\r\n retweet_df.loc[i, 'Normalized_count'] = retweet_df.loc[i, 'count'] / df_retweet_dict[retweet_df.loc[i, 'No_of_Retweets']]",
"_____no_output_____"
]
],
[
[
"## Sentiment",
"_____no_output_____"
]
],
[
[
"g = sns.catplot(x = \"No_of_Likes\", y = \"Normalized_count\", hue = \"vaderSentiment\", data = like_df, kind = \"bar\")",
"_____no_output_____"
],
[
"g = sns.catplot(x = \"No_of_Retweets\", y = \"Normalized_count\", hue = \"vaderSentiment\", data = retweet_df, kind = \"bar\")",
"_____no_output_____"
],
[
"plt.pie(classify_df['vaderSentiment'], labels=classify_df['index']);",
"_____no_output_____"
],
[
"l = []\r\nfor i in range(len(df_sentiment)):\r\n for element in df_sentiment.loc[i, 'Labels'].split():\r\n if element != 'Font':\r\n l.append(element)",
"_____no_output_____"
]
],
[
[
"## Word Cloud",
"_____no_output_____"
]
],
[
[
"wordcloud = WordCloud(width = 800, height = 800, \r\n background_color ='white', \r\n min_font_size = 10).generate(str(l))\r\n\r\nplt.figure(figsize = (8, 8), facecolor = None) \r\nplt.imshow(wordcloud, interpolation='bilinear')\r\nplt.axis(\"off\")\r\nplt.tight_layout(pad = 0) \r\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Topic Modeling",
"_____no_output_____"
]
],
[
[
"cv = CountVectorizer(stop_words='english')\r\ndata_cv = cv.fit_transform(df.Tweet)\r\nwords = cv.get_feature_names()\r\ndata_dtm = pd.DataFrame(data_cv.toarray(), columns=cv.get_feature_names())\r\npickle.dump(cv, open(\"cv_stop.pkl\", \"wb\"))",
"_____no_output_____"
],
[
"data_dtm_transpose = data_dtm.transpose()",
"_____no_output_____"
],
[
"sparse_counts = scipy.sparse.csr_matrix(data_dtm_transpose)\r\ncorpus = matutils.Sparse2Corpus(sparse_counts)",
"_____no_output_____"
],
[
"cv = pickle.load(open(\"cv_stop.pkl\", \"rb\"))\r\nid2word = dict((v, k) for k, v in cv.vocabulary_.items())\r\nword2id = dict((k, v) for k, v in cv.vocabulary_.items())\r\nd = corpora.Dictionary()\r\nd.id2token = id2word\r\nd.token2id = word2id",
"_____no_output_____"
],
[
"lda = gensim.models.ldamodel.LdaModel(corpus=corpus, id2word=id2word, num_topics=3, passes=10)\r\nlda.print_topics()",
"_____no_output_____"
],
[
"pyLDAvis.enable_notebook()\r\nvis = pyLDAvis.gensim.prepare(lda, corpus, d)\r\nvis",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05de1f65cc0416666679f766c6e31f2878a66c1 | 10,259 | ipynb | Jupyter Notebook | Cls5-Dimentionality Reduction/DimensionalityReduction-CaseStudy2-solution.ipynb | tuhinssam/MLResources | 5410ce33bc6f3a951b0f94c32bf82748a8c5bd6c | [
"MIT"
] | 1 | 2020-01-31T06:18:30.000Z | 2020-01-31T06:18:30.000Z | Cls5-Dimentionality Reduction/DimensionalityReduction-CaseStudy2-solution.ipynb | tuhinssam/MLResources | 5410ce33bc6f3a951b0f94c32bf82748a8c5bd6c | [
"MIT"
] | null | null | null | Cls5-Dimentionality Reduction/DimensionalityReduction-CaseStudy2-solution.ipynb | tuhinssam/MLResources | 5410ce33bc6f3a951b0f94c32bf82748a8c5bd6c | [
"MIT"
] | null | null | null | 23.263039 | 224 | 0.452383 | [
[
[
"### 1. We shall use the same dataset used in previous assignment - digits. Make a 80-20 train/test split.\n[Hint: Explore datasets module from scikit learn]\n### 2. Using scikit learn perform a LDA on the dataset. Find out the number of components in the projected subspace.\n[Hint: Refer to discriminant analysis module of scikit learn]\n### 3. Transform the dataset and fit a logistic regression and observe the accuracy. Compare it with the previous model based on PCA in terms of accuracy and model complexity.\n[Hint: Project both the train and test samples to the new subspace]",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"from sklearn.datasets import load_digits",
"_____no_output_____"
],
[
"digits = load_digits()",
"_____no_output_____"
],
[
"digits.data",
"_____no_output_____"
],
[
"digits.data.shape",
"_____no_output_____"
],
[
"digits.target.shape",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(digits.data,digits.target, test_size=0.2, random_state = 5)",
"_____no_output_____"
],
[
"from sklearn.discriminant_analysis import LinearDiscriminantAnalysis",
"_____no_output_____"
],
[
"lda = LinearDiscriminantAnalysis()",
"_____no_output_____"
],
[
"lda.fit(X_train,y_train)",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\sklearn\\discriminant_analysis.py:388: UserWarning: Variables are collinear.\n warnings.warn(\"Variables are collinear.\")\n"
],
[
"predictedlda = lda.predict(X_test)",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score",
"_____no_output_____"
],
[
"accuracy_score(y_test,predicted)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression",
"_____no_output_____"
],
[
"logreg = LogisticRegression(random_state=5)",
"_____no_output_____"
],
[
"logreg.fit(X_train,y_train)",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\sklearn\\linear_model\\logistic.py:433: FutureWarning: Default solver will be changed to 'lbfgs' in 0.22. Specify a solver to silence this warning.\n FutureWarning)\nC:\\ProgramData\\Anaconda3\\lib\\site-packages\\sklearn\\linear_model\\logistic.py:460: FutureWarning: Default multi_class will be changed to 'auto' in 0.22. Specify the multi_class option to silence this warning.\n \"this warning.\", FutureWarning)\n"
],
[
"logregpredict = logreg.predict(X_test)",
"_____no_output_____"
],
[
"accuracy_score(y_test, logregpredict)",
"_____no_output_____"
],
[
"dfcompare = pd.DataFrame({\"Actual\":y_test,\"Predicted\":logregpredict})",
"_____no_output_____"
],
[
"dfcompare.sample(10)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05de82b13c461ca610fe7c2f4ef06b06deaa692 | 259,107 | ipynb | Jupyter Notebook | Neural Network Assignment.ipynb | DeepLearningVision-2019/a3-neural-network-class-munozgce | 011e82a7b72400a1d236334d91fdad21cb9adb6b | [
"MIT"
] | null | null | null | Neural Network Assignment.ipynb | DeepLearningVision-2019/a3-neural-network-class-munozgce | 011e82a7b72400a1d236334d91fdad21cb9adb6b | [
"MIT"
] | null | null | null | Neural Network Assignment.ipynb | DeepLearningVision-2019/a3-neural-network-class-munozgce | 011e82a7b72400a1d236334d91fdad21cb9adb6b | [
"MIT"
] | null | null | null | 547.794926 | 220,256 | 0.941144 | [
[
[
"### Project: Create a neural network class\n\n---\n\nBased on previous code examples, develop a neural network class that is able to classify any dataset provided. The class should create objects based on the desired network architecture:\n\n1. Number of inputs\n2. Number of hidden layers\n3. Number of neurons per layer\n4. Number of outputs\n5. Learning rate\n\nThe class must have the train, and predict functions.\n\nTest the neural network class on the datasets provided below: Use the input data to train the network, and then pass new inputs to predict on. Print the expected label and the predicted label for the input you used. Print the accuracy of the training after predicting on different inputs.\n\nUse matplotlib to plot the error that the train method generates.\n\n**Don't forget to install Keras and tensorflow in your environment!**\n\n---",
"_____no_output_____"
],
[
"### Import the needed Packages",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\n# Needed for the mnist data\nfrom keras.datasets import mnist\nfrom keras.utils import to_categorical ",
"_____no_output_____"
]
],
[
[
"### Define the class",
"_____no_output_____"
]
],
[
[
"class NeuralNetwork:\n\n def __init__(self, architecture, alpha):\n \n \n '''\n layers: List of integers which represents the architecture of the network.\n alpha: Learning rate.\n '''\n # TODO: Initialize the list of weights matrices, then store\n # the network architecture and learning rate\n self.layers = architecture \n self.alpha = alpha \n np.random.seed(13)\n self.FW = np.random.randn(architecture[0], architecture[2])\n self.MW = np.empty((architecture[0] - 1, architecture[2], architecture[2]))\n \n for x in range(architecture[0] - 2):\n self.MW[x] = np.random.randn(architecture[2], architecture[2])\n \n self.LW = np.random.randn(architecture[2], architecture[3])\n self.FB = np.random.randn(architecture[2])\n self.MB = np.random.randn(architecture[1] - 1, architecture[2])\n self.LB = np.random.randn(architecture[3])\n \n pass\n \n \n def __repr__(self): \n \n return \"NeuralNetwork: {}\".format( \"-\".join(str(l) for l in self.layers))\n\n def softmax(self, X): \n \n expX = np.exp(X)\n return expX / expX.sum(axis=1, keepdims=True)\n \n def sigmoid(self, x):\n # the sigmoid for a given input value\n \n return 1.0 / (1.0 + np.exp(-x))\n \n def sigmoid_deriv(self, x):\n # the derivative of the sigmoid\n \n return x * (1 - x)\n \n def predict(self, inputs):\n # TODO: Define the predict function\n self.newWeights = np.empty((self.layers[1], inputs.shape[0], self.layers[2]))\n self.newWeights[0] = self.sigmoid(np.dot(inputs, self.FW) + self.FB)\n for x in range(self.layers[0] - 2):\n self.newWeights[x+1] = self.sigmoid(np.dot(self.newWeights[x], self.MW[x]) + self.MB[x])\n finalLevel = self.softmax( np.dot(self.newWeights[len(self.newWeights)-1], self.LW) + self.LB)\n \n return finalLevel\n \n def train(self, inputs, labels, epochs = 1000, displayUpdate = 100):\n \n fail = []\n for i in range(epochs):\n \n hop = self.predict(inputs)\n error = labels - hope\n error1 = np.dot(error * self.sigmoid_deriv(hope), self.LW.T)\n delta = np.dot(error * self.sigmoid_deriv(hope), self.LW.T) * self.sigmoid_deriv(self.newWeights[len(self.newWeights) - 1])\n\n self.LB += * self.alpha\n self.LW += np.dot(self.newWeights[len(self.newWeights) - 1].T, error * self.sigmoid_deriv(hope)) * self.alpha\n \n for x in range(self.layers[1] - 1):\n self.MW[(len(self.MW) - 1) - x] += np.dot(self.newWeights[(len(self.newWeights) - 2) - x].T, delta) * self.alpha\n delta = np.sum(delta)\n self.MB[x] += delta * self.alpha\n delta = np.dot(delta, self.MW[(len(self.MW) - 1) - x]) * self.sigmoid_deriv(self.newWeights[(len(self.newWeights) - 2) - x])\n \n self.FW += np.dot(inputs.T, delta) * self.alpha\n delta2 = np.sum(delta)\n self.FB += delta2 * self.alpha\n \n fail.append(np.mean(np.abs(level_error)))\n \n return fail\n \n ",
"_____no_output_____"
]
],
[
[
"### Test datasets",
"_____no_output_____"
],
[
"#### XOR",
"_____no_output_____"
]
],
[
[
"# input dataset\nXOR_inputs = np.array([ \n [0,0],\n [0,1],\n [1,0],\n [1,1]\n ])\n\n# labels dataset \nXOR_labels = np.array([[0,1,1,0]]).T",
"_____no_output_____"
],
[
"#TODO: Test the class with the XOR data\n\n\n",
"_____no_output_____"
]
],
[
[
"#### Multiple classes",
"_____no_output_____"
]
],
[
[
"# Creates the data points for each class\nclass_1 = np.random.randn(700, 2) + np.array([0, -3]) \nclass_2 = np.random.randn(700, 2) + np.array([3, 3]) \nclass_3 = np.random.randn(700, 2) + np.array([-3, 3])\nfeature_set = np.vstack([class_1, class_2, class_3])\nlabels = np.array([0]*700 + [1]*700 + [2]*700)\none_hot_labels = np.zeros((2100, 3))\n\nfor i in range(2100): \n one_hot_labels[i, labels[i]] = 1\n\nplt.figure(figsize=(10,10)) \nplt.scatter(feature_set[:,0], feature_set[:,1], c=labels, s=30, alpha=0.5) \nplt.show()",
"_____no_output_____"
],
[
"#TODO: Test the class with the multiple classes data\n\nr = NeuralNetwork([2,2,5,3], 0.01)\nfails = r.train(feature_set, one_hot_labels, 10000, 1000)\nfig, ax = plt.subplots(1,1)\nax.set_ylabel('Error')\nax.plot(fails)\ntest = np.array([[0,-4]])\nprint(str(test) + str(network.predict(test)))",
"Error: 0.47806009636665425\nError: 0.007121601211763323\nError: 0.005938405234795827\nError: 0.005920131593441376\nError: 0.00585185558757003\nError: 0.0049490985751735606\nError: 0.004301948969147726\nError: 0.0038221899933325782\nError: 0.003507891190406313\nError: 0.003280260509683804\n"
]
],
[
[
"#### On the mnist data set\n\n---\nTrain the network to classify hand drawn digits.\n\nFor this data set, if the training step is taking too long, you can try to adjust the architecture of the network to have fewer layers, or you could try to train it with fewer input. The data has already been loaded and preprocesed so that it can be used with the network.\n\n---",
"_____no_output_____"
]
],
[
[
"# Load the train and test data from the mnist data set\n(train_images, train_labels), (test_images, test_labels) = mnist.load_data()\n\n# Plot a sample data point\nplt.title(\"Label: \" + str(train_labels[0]))\nplt.imshow(train_images[0], cmap=\"gray\")",
"_____no_output_____"
],
[
"# Standardize the data\n\n# Flatten the images\ntrain_images = train_images.reshape((60000, 28 * 28))\n# turn values from 0-255 to 0-1\ntrain_images = train_images.astype('float32') / 255 \n\ntest_images = test_images.reshape((10000, 28 * 28)) \ntest_images = test_images.astype('float32') / 255\n\n# Create one hot encoding for the labels\ntrain_labels = to_categorical(train_labels) \ntest_labels = to_categorical(test_labels)",
"_____no_output_____"
],
[
"# TODO: Test the class with the mnist data. Test the training of the network with the test_images data, and \n# record the accuracy of the classification.\n\nr = NeuralNetwork([2,2,5,3], 0.01)\nfails = r.train(feature_set, one_hot_labels, 10000, 1000)\nfig, ax = plt.subplots(1,1)\nax.set_ylabel('Error')\nax.plot(fails)\ntest = np.array([[0,-3]])\nprf = network.predict(test_images[0:1000])\none_hot_test_labels = to_categorical(test_labels[0:1000])\nnp.set_printoptions(precision=10, suppress= True, linewidth=75)\nguess = np.copy(prf)\nguess[guess > 0.5] = 1\nguess[guess < 0.5] = 0\nfails = []\nfor index, (guess, label) in enumerate(zip(predictions[0:10], one_hot_test_labels[0:10])):\n if not np.array_equal(prediction,label):\n fails.append((index, prediction, label))\n\nfor img, plot in zip(fails, plots):\n plot.imshow(test_images[img[0]].reshape(28,28), cmap = \"gray\")\n plot.set_title(str(img[1]))",
"Error: 0.47806009636665425\nError: 0.007121601211763323\nError: 0.005938405234795827\nError: 0.005920131593441376\nError: 0.00585185558757003\nError: 0.0049490985751735606\nError: 0.004301948969147726\nError: 0.0038221899933325782\nError: 0.003507891190406313\nError: 0.003280260509683804\n"
]
],
[
[
"After predicting on the *test_images*, use matplotlib to display some of the images that were not correctly classified. Then, answer the following questions: \n\n1. **Why do you think those were incorrectly classified?**\n2. **What could you try doing to improve the classification accuracy?**\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
d05df1186886201a19ad4a2b38b2713a82fd56a4 | 8,280 | ipynb | Jupyter Notebook | Pandas Assignment.ipynb | oguro515/ADS-Assignment-1 | 735a408bd2d26ca703372c36bd515fcbc65997d4 | [
"MIT"
] | null | null | null | Pandas Assignment.ipynb | oguro515/ADS-Assignment-1 | 735a408bd2d26ca703372c36bd515fcbc65997d4 | [
"MIT"
] | null | null | null | Pandas Assignment.ipynb | oguro515/ADS-Assignment-1 | 735a408bd2d26ca703372c36bd515fcbc65997d4 | [
"MIT"
] | null | null | null | 19.856115 | 197 | 0.506401 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d05e17b8af0282aac2ecda0347ffbdd6c1edb5bb | 399,046 | ipynb | Jupyter Notebook | _notebooks/2021-12-26-titanic.ipynb | rhkrehtjd/kaggle | ff24969da923991573fdccd1f8b1f5bd7920004d | [
"Apache-2.0"
] | null | null | null | _notebooks/2021-12-26-titanic.ipynb | rhkrehtjd/kaggle | ff24969da923991573fdccd1f8b1f5bd7920004d | [
"Apache-2.0"
] | null | null | null | _notebooks/2021-12-26-titanic.ipynb | rhkrehtjd/kaggle | ff24969da923991573fdccd1f8b1f5bd7920004d | [
"Apache-2.0"
] | null | null | null | 917.347126 | 110,844 | 0.95502 | [
[
[
"# Titanic 4",
"_____no_output_____"
],
[
"> ### `Pclass, Sex, Age`",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nplt.style.use('seaborn')\nsns.set(font_scale=2.5)\n\nimport missingno as msno \nimport warnings\nwarnings.filterwarnings('ignore')\n%matplotlib inline\ndf_train=pd.read_csv('C:/Users/ehfus/Downloads/titanic/train.csv')\ndf_test=pd.read_csv('C:/Users/ehfus/Downloads/titanic/test.csv')",
"_____no_output_____"
],
[
"# violin plot\nfig,ax=plt.subplots(1,2,figsize=(18,8))\nsns.violinplot('Pclass','Age',hue='Survived',data=df_train,sacle='count',split=True,ax=ax[0])\nax[0].set_title('Pclass and Age vs Survived')\nax[0].set_yticks(range(0,110,10))\n\nsns.violinplot('Sex','Age',hue='Survived',data=df_train,scale='count',split=True, ax=ax[1])\nax[1].set_title('Sexx and Age vs Survived')\nax[1].set_yticks(range(0,110,10))\nplt.show()",
"_____no_output_____"
]
],
[
[
"- scale에도 option이 여러가지 있음, google에서 확인해볼 것",
"_____no_output_____"
],
[
"> ### `Embarked : 탑승한 항구`",
"_____no_output_____"
]
],
[
[
"f,ax=plt.subplots(1,1,figsize=(7,7))\ndf_train[['Embarked','Survived']]\\\n.groupby(['Embarked'], as_index=True).mean()\\\n.sort_values(by='Survived', ascending=False)\\\n.plot.bar(ax=ax)",
"_____no_output_____"
]
],
[
[
"- `sort_values` 또는 `sort_index`도 사용 가능",
"_____no_output_____"
]
],
[
[
"f,ax=plt.subplots(2,2,figsize=(20,15)) #2차원임/ 1,2는 1차원\nsns.countplot('Embarked',data=df_train, ax=ax[0,0])\nax[0,0].set_title('(1) No. Of Passengers Boared')\n\nsns.countplot('Embarked',hue='Sex',data=df_train, ax=ax[0,1])\nax[0,1].set_title('(2) Male-Female split for embarked')\n\nsns.countplot('Embarked', hue='Survived', data=df_train, ax=ax[1,0])\nax[1,0].set_title('(3) Embarked vs Survived')\n\nsns.countplot('Embarked', hue='Pclass', data=df_train, ax=ax[1,1])\nax[1,1].set_title('(4) Embarked vs Survived')\n\nplt.subplots_adjust(wspace=0.2, hspace=0.5) # 상하좌우간격 맞춰줌\nplt.show()",
"_____no_output_____"
]
],
[
[
"> ### `Family - SibSp + ParCh`",
"_____no_output_____"
]
],
[
[
"df_train['FamilySize']=df_train['SibSp'] + df_train['Parch'] + 1",
"_____no_output_____"
],
[
"print('Maximum size of Family : ',df_train['FamilySize'].max())\nprint('Minimum size of Family : ',df_train['FamilySize'].min())",
"Maximum size of Family : 11\nMinimum size of Family : 1\n"
]
],
[
[
"- Pandas series는 연산이 가능",
"_____no_output_____"
]
],
[
[
"f,ax=plt.subplots(1,3,figsize=(40,10))\nsns.countplot('FamilySize', data=df_train, ax=ax[0])\nax[0].set_title('(1) No. Of Passenger Boarded', y=1.02)\n\nsns.countplot('FamilySize', hue='Survived',data=df_train, ax=ax[1])\nax[1].set_title('(2) Survived countplot depending on FamilySize', y=1.02)\n\ndf_train[['FamilySize','Survived']].groupby(['FamilySize'],as_index=True).mean().sort_values(by='Survived',ascending=False).plot.bar(ax=ax[2])\nax[2].set_title('(3) Survived rate depending on FamilySize',y=1.02)\n\nplt.subplots_adjust(wspace=0.2,hspace=0.5)\nplt.show()",
"_____no_output_____"
]
],
[
[
"> ### `Fare : 요금, 연속형 변수`",
"_____no_output_____"
],
[
"- distplot ?? 시리즈에 히스토그램을 그려줌,Skewness? 왜도임 + 첨도도 있음\n\n- 왜도? 첨도?\n\n- python에서 나타내는 함수는?",
"_____no_output_____"
]
],
[
[
"fig,ax=plt.subplots(1,1,figsize=(8,8))\ng=sns.distplot(df_train['Fare'], color='b',label='Skewness{:.2f}'.format(df_train['Fare'].skew()),ax=ax)\ng=g.legend(loc='best')",
"_____no_output_____"
]
],
[
[
"- skewness가 5정도로 꽤 큼 -> 좌로 많이 치우쳐져 있음 -> 그대로 모델에 학습시키면 성능이 낮아질 수 있음",
"_____no_output_____"
]
],
[
[
"df_train['Fare']=df_train['Fare'].map(lambda i: np.log(i) if i>0 else 0)",
"_____no_output_____"
]
],
[
[
"df_train['Fare']의 값을 적절하게 변형 중",
"_____no_output_____"
]
],
[
[
"fig,ax=plt.subplots(1,1,figsize=(8,8))\ng=sns.distplot(df_train['Fare'], color='b',label='Skewness{:.2f}'.format(df_train['Fare'].skew()),ax=ax)\ng=g.legend(loc='best')",
"_____no_output_____"
]
],
[
[
"이런 작업(log로 변환)을 통해 skewness가 0으로 근접하게 해주었음",
"_____no_output_____"
]
],
[
[
"df_train['Ticket'].value_counts()",
"_____no_output_____"
]
],
[
[
"정말 다양한 자료형태가 결합되어 있음 -> 적절한 변형이 필요해 보임",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d05e2a16a80fc74f0762aec1be22db352a188235 | 26,543 | ipynb | Jupyter Notebook | Machine Learning 2_Using Advanced Machine Learning Models/Reference Material/190053-Reactors-DS-Tr2-Sec1-2-PCA.ipynb | raspyweather/Reactors | a8e05ec1e585958f932c4c304a595f03bd15db39 | [
"MIT"
] | 385 | 2019-10-21T14:36:08.000Z | 2022-03-31T16:35:53.000Z | Machine_Learning_2/Reference Material/190053-Reactors-DS-Tr2-Sec1-2-PCA.ipynb | debsen077/Reactors | 638227d015937920a58489694db22bce05357ff2 | [
"MIT"
] | 115 | 2019-10-19T02:41:58.000Z | 2022-03-04T23:00:41.000Z | Machine_Learning_2/Reference Material/190053-Reactors-DS-Tr2-Sec1-2-PCA.ipynb | debsen077/Reactors | 638227d015937920a58489694db22bce05357ff2 | [
"MIT"
] | 303 | 2019-10-18T07:27:40.000Z | 2022-03-29T12:44:01.000Z | 32.212379 | 745 | 0.614927 | [
[
[
"# Section 1.2: Dimension reduction and principal component analysis (PCA)\n\nOne of the iron laws of data science is know as the \"curse of dimensionality\": as the number of considered features (dimensions) of a feature space increases, the number of data configurations can grow exponentially and thus the number observations (data points) needed to account for these configurations must also increase. Because this fact of life has huge ramifications for the time, computational effort, and memory required it is often desirable to reduce the number of dimensions we have to work with.\n\nOne way to accomplish this is by reducing the number of features considered in an analysis. After all, not all features are created equal, and some yield more insight for a given analysis than others. While this type of feature engineering is necessary in any data-science project, we can really only take it so far; up to a point, considering more features can often increase the accuracy of a classifier. (For example, consder how many features could increase the accuracy of classifying images as cats or dogs.)\n\n## PCA in theory\n\nAnother way to reduce the number of dimensions that we have to work with is by projecting our feature space into a lower dimensional space. The reason why we can do this is that in most real-world problems, data points are not spread uniformly across all dimensions. Some features might be near constant, while others are highly correlated, which means that those data points lie close to a lower-dimensional subspace.\n\nIn the image below, the data points are not spread across the entire plane, but are nicely clumped, roughly in an oval. Because the cluster (or, indeed, any cluster) is roughly elliptical, it can be mathematically described by two values: its major (long) axis and its minor (short) axis. These axes form the *principal components* of the cluster. \n\n<img align=\"center\" style=\"padding-right:10px;\" src=\"Images/PCA.png\">\n\nIn fact, we can construct a whole new feature space around this cluster, defined by two *eigenvectors* (the vectors that define the linear transformation to this new feature space), $c_{1}$ and $c_{2}$. Better still, we don't have to consider all of the dimensions of this new space. Intuitively, we can see that most of the points lie on or close to the line that runs through $c_{1}$. So, if we project the cluster down from two dimensions to that single dimension, we capture most of the information about this data sense while simplifying our analysis. This ability to extract most of the information from a dataset by considering only a fraction of its definitive eigenvectors forms the heart of principal component analysis (PCA).\n\n## Import modules and dataset\n\nYou will need to clean and prepare the data in order to conduct PCA on it, so pandas will be essential. You will also need NumPy, a bit of Scikit Learn, and pyplot.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.decomposition import PCA\nfrom sklearn.preprocessing import StandardScaler\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"The dataset we’ll use here is the same one drawn from the [U.S. Department of Agriculture National Nutrient Database for Standard Reference](https://www.ars.usda.gov/northeast-area/beltsville-md-bhnrc/beltsville-human-nutrition-research-center/nutrient-data-laboratory/docs/usda-national-nutrient-database-for-standard-reference/) that you prepared in Section 1.1. Remember to set the encoding to `latin_1` (for those darn µg).",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('Data/USDA-nndb-combined.csv', encoding='latin_1')",
"_____no_output_____"
]
],
[
[
"We can check the number of columns and rows using the `info()` method for the `DataFrame`.",
"_____no_output_____"
]
],
[
[
"df.info()",
"_____no_output_____"
]
],
[
[
"> **Exercise**\n>\n> Can you think of a more concise way to check the number of rows and columns in a `DataFrame`? (***Hint:*** Use one of the [attributes](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html) of the `DataFrame`.)",
"_____no_output_____"
],
[
"## Handle `null` values\n\nBecause this is a real-world dataset, it is a safe bet that it has `null` values in it. We could first check to see if this is true. However, later on in this section, we will have to transform our data using a function that cannot use `NaN` values, so we might as well drop rows containing those values.",
"_____no_output_____"
],
[
"> **Exercise**\n>\n> Drop rows from the `DataFrame` that contain `NaN` values. (If you need help remembering which method to use, see [this page](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.dropna.html).)",
"_____no_output_____"
],
[
"> **Exercise solution**\n>\n> The correct code to use is `df = df.dropna()`.",
"_____no_output_____"
],
[
"Now let’s see how many rows we have left.",
"_____no_output_____"
]
],
[
[
"df.shape",
"_____no_output_____"
]
],
[
[
"Dropping those rows eliminated 76 percent of our data (8989 entries to 2190). An imperfect state of affairs, but we still have enough for our purposes in this section.\n\n> **Key takeaway:** Another solution to removing `null` values is to impute values for them, but this can be tricky. Should we handle missing values as equal to 0? What about a fatty food with `NaN` for `Lipid_Tot_(g)`? We could try taking the averages of values surrounding a `NaN`, but what about foods that are right next to rows containing foods from radically different food groups? It is possible to make justifiable imputations for missing values, but it can be important to involve subject-matter experts (SMEs) in that process.",
"_____no_output_____"
],
[
"## Split off descriptive columns\n\nOut descriptive columns (such as `FoodGroup` and `Shrt_Desc`) pose challenges for us when it comes time to perform PCA because they are categorical rather than numerical features, so we will split our `DataFrame` in to one containing the descriptive information and one containing the nutritional information.",
"_____no_output_____"
]
],
[
[
"desc_df = df.iloc[:, [0, 1, 2]+[i for i in range(50,54)]]\ndesc_df.set_index('NDB_No', inplace=True)\ndesc_df.head()",
"_____no_output_____"
]
],
[
[
"> **Question**\n>\n> Why was it necessary to structure the `iloc` method call the way we did in the code cell above? What did it accomplish? Why was it necessary set the `desc_df` index to `NDB_No`?",
"_____no_output_____"
]
],
[
[
"nutr_df = df.iloc[:, :-5]\nnutr_df.head()",
"_____no_output_____"
]
],
[
[
"> **Question**\n>\n> What did the `iloc` syntax do in the code cell above?",
"_____no_output_____"
]
],
[
[
"nutr_df = nutr_df.drop(['FoodGroup', 'Shrt_Desc'], axis=1)",
"_____no_output_____"
]
],
[
[
"> **Exercise**\n>\n> Now set the index of `nutr_df` to use `NDB_No`.",
"_____no_output_____"
],
[
"> **Exercise solution**\n>\n> The correct code for students to use here is `nutr_df.set_index('NDB_No', inplace=True)`.",
"_____no_output_____"
],
[
"Now let’s take a look at `nutr_df`.",
"_____no_output_____"
]
],
[
[
"nutr_df.head()",
"_____no_output_____"
]
],
[
[
"## Check for correlation among features\n\nOne thing that can skew our classification results is correlation among our features. Recall that the whole reason that PCA works is that it exploits the correlation among data points to project our feature-space into a lower-dimensional space. However, if some of our features are highly correleted to begin with, these relationships might create spurious clusters of data in our PCA.\n\nThe code to check for correlations in our data isn't long, but it takes too long (up to 10 to 20 minutes) to run for a course like this. Instead, the table below shows the output from that code:\n\n| | column | row | corr |\n|--:|------------------:|------------------:|-----:|\n| 0 | Folate\\_Tot\\_(µg) | Folate\\_DFE\\_(µg) | 0.98 |\n| 1 | Folic\\_Acid\\_(µg) | Folate\\_DFE\\_(µg) | 0.95 |\n| 2 | Folate\\_DFE\\_(µg) | Folate\\_Tot\\_(µg) | 0.98 |\n| 3 | Vit\\_A\\_RAE | Retinol\\_(µg) | 0.99 |\n| 4 | Retinol\\_(µg) | Vit\\_A\\_RAE | 0.99 |\n| 5 | Vit\\_D\\_µg | Vit\\_D\\_IU | 1 |\n| 6 | Vit\\_D\\_IU | Vit\\_D\\_µg | 1 |\n\nAs it turns out, dropping `Folate_DFE_(µg)`, `Vit_A_RAE`, and `Vit_D_IU` will eliminate the correlations enumerated in the table above.",
"_____no_output_____"
]
],
[
[
"nutr_df.drop(['Folate_DFE_(µg)', 'Vit_A_RAE', 'Vit_D_IU'], \n inplace=True, axis=1)\nnutr_df.head()",
"_____no_output_____"
]
],
[
[
"## Normalize and center the data\n\nOur numeric data comes in a variety of mass units (grams, milligrams, and micrograms) and one energy unit (kilocalories). In order to make an apples-to-apples comparison (pun intended) of the nutritional data, we need to first *normalize* the data and make it more normally distributed (that is, make the distribution of the data look more like a familiar bell curve).\n\nTo help see why we need to normalize the data, let's look at a histogram of all of the columns.",
"_____no_output_____"
]
],
[
[
"ax = nutr_df.hist(bins=50, xlabelsize=-1, ylabelsize=-1, figsize=(11,11))",
"_____no_output_____"
]
],
[
[
"Not a bell curve in sight. Worse, a lot of the data is clumped at or around 0. We will use the Box-Cox Transformation on the data, but it requires strictly positive input, so we will add 1 to every value in each column.",
"_____no_output_____"
]
],
[
[
"nutr_df = nutr_df + 1",
"_____no_output_____"
]
],
[
[
"Now for the transformation. The [Box-Cox Transformation](https://www.statisticshowto.datasciencecentral.com/box-cox-transformation/) performs the transformation $y(\\lambda) = \\dfrac{y^{\\lambda}-1}{\\lambda}$ for $\\lambda \\neq 0$ and $y(\\lambda) = log y$ for $\\lambda = 0$ for all values $y$ in a given column. SciPy has a particularly useful `boxcox()` function that can automatically calculate the $\\lambda$ for each column that best normalizes the data in that column. (However, it is does not support `NaN` values; scikit-learn has a comparable `boxcox()` function that is `NaN`-safe, but it is not available on the version of scikit-learn that comes with Azure notebooks.)",
"_____no_output_____"
]
],
[
[
"from scipy.stats import boxcox\n\nnutr_df_TF = pd.DataFrame(index=nutr_df.index)\nfor col in nutr_df.columns.values:\n nutr_df_TF['{}_TF'.format(col)] = boxcox(nutr_df.loc[:, col])[0]",
"_____no_output_____"
]
],
[
[
"Let's now take a look at the `DataFrame` containing the transformed data.",
"_____no_output_____"
]
],
[
[
"ax = nutr_df_TF.hist(bins=50, xlabelsize=-1, ylabelsize=-1, figsize=(11,11))",
"_____no_output_____"
]
],
[
[
"Few of these columns looks properly normal, but it is enough to now center the data.\n\nOur data units were incompatible to begin with, and the transformations have not improved that. But we can address that by centering the data around 0; that is, we will again transform the data, this time so that every column has a mean of 0 and a standard deviation of 1. Scikit-learn has a convenient function for this.",
"_____no_output_____"
]
],
[
[
"nutr_df_TF = StandardScaler().fit_transform(nutr_df_TF)",
"_____no_output_____"
]
],
[
[
"You can satisfy your self that the data is now centered by using the `mean()` method on the `DataFrame`.",
"_____no_output_____"
]
],
[
[
"print(\"mean: \", np.round(nutr_df_TF.mean(), 2))",
"_____no_output_____"
]
],
[
[
"> **Exercise**\n>\n> Find the standard deviation for the `nutr_df_TF`. (If you need a hint as to which method to use, see [this page](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.std.html).)",
"_____no_output_____"
],
[
"> **Exercise solution**\n>\n> The correct code to use here is `print(\"s.d.: \", np.round(nutr_df_TF.std(), 2))`.",
"_____no_output_____"
],
[
"## PCA in practice\n\nIt is finally time to perform the PCA on our data. (As stated before, even with pretty clean data, a lot of effort has to go into preparing the data for analysis.)",
"_____no_output_____"
]
],
[
[
"fit = PCA()\npca = fit.fit_transform(nutr_df_TF)",
"_____no_output_____"
]
],
[
[
"So, now that we have peformed the PCA on our data, what do we actually have? Remember that PCA is foremost about finding the eigenvectors for our data. We then want to select some subset of those vectors to form the lower-dimensional subspace in which to analyze our data.\n\nNot all of the eigenvectors are created equal. Just a few of them will account for the majority of the variance in the data. (Put another way, a subspace composed of just a few of the eigenvectors will retain the majority of the information from our data.) We want to focus on those vectors.\n\nTo help us get a sense of how many vectors we should use, consider this scree graph of the variance for the PCA components, which plots the variance explained by the components from greatest to least.",
"_____no_output_____"
]
],
[
[
"plt.plot(fit.explained_variance_ratio_)",
"_____no_output_____"
]
],
[
[
"This is where data science can become an art. As a rule of thumb, we want to look for \"elbow\" in the graph, which is the point at which the few components have captured the majority of the variance in the data (after that point, we are only adding complexity to the analysis for increasingly diminishing returns). In this particular case, that appears to be at about five components.\n\nWe can take the cumulative sum of the first five components to see how much variance they capture in total.",
"_____no_output_____"
]
],
[
[
"print(fit.explained_variance_ratio_[:5].sum())",
"_____no_output_____"
]
],
[
[
"So our five components capture about 70 percent of the variance. We can see what fewer or additional components would yield by looking at the cumulative variance for all of the components.",
"_____no_output_____"
]
],
[
[
"print(fit.explained_variance_ratio_.cumsum())",
"_____no_output_____"
]
],
[
[
"We can also examine this visually.",
"_____no_output_____"
]
],
[
[
"plt.plot(np.cumsum(fit.explained_variance_ratio_))\nplt.title(\"Cumulative Explained Variance Graph\")",
"_____no_output_____"
]
],
[
[
"Ultimately, it is a matter of judgment as to how many components to use, but five vectors (and 70 percent of the variance) will suffice for our purposes in this section.\n\nTo aid further analysis, let's now put those five components into a DataFrame.",
"_____no_output_____"
]
],
[
[
"pca_df = pd.DataFrame(pca[:, :5], index=df.index)\npca_df.head()",
"_____no_output_____"
]
],
[
[
"Each column represents one of the eigenvectors, and each row is one of the coordinates that defines that vector in five-dimensional space.\n\nWe will want to add the FoodGroup column back in to aid with our interpretation of the data later on. Let's also rename the component-columns $c_{1}$ through $c_{5}$ so that we know what we are looking at.",
"_____no_output_____"
]
],
[
[
"pca_df = pca_df.join(desc_df)\npca_df.drop(['Shrt_Desc', 'GmWt_Desc1', 'GmWt_2', 'GmWt_Desc2', 'Refuse_Pct'], \n axis=1, inplace=True)\npca_df.rename(columns={0:'c1', 1:'c2', 2:'c3', 3:'c4', 4:'c5'}, \n inplace=True)\npca_df.head()",
"_____no_output_____"
]
],
[
[
"Don't worry that the FoodGroup column has all `NaN` values: it is not a vector, so it has no vector coordinates.\n\nOne last thing we should demonstrate is that each of the components is mutually perpendicular (or orthogonal in math-speak). One way of expressing that condition is that each component-vector should perfectly correspond with itself and not correlate at all (positively or negatively) with any other vector.",
"_____no_output_____"
]
],
[
[
"np.round(pca_df.corr(), 5)",
"_____no_output_____"
]
],
[
[
"## Interpreting the results\n\nWhat do our vectors mean? Put another way, what kinds of foods populate the differnt clusters we have discovered among the data?\n\nTo see these results, we will create pandas Series for each of the components, index them by feature, and then sort them in descreasing order (so that a higher number represents a feature that is positively correlated with that vector and negative numbers represent low correlation).",
"_____no_output_____"
]
],
[
[
"vects = fit.components_[:5]",
"_____no_output_____"
],
[
"c1 = pd.Series(vects[0], index=nutr_df.columns)\nc1.sort_values(ascending=False)",
"_____no_output_____"
]
],
[
[
"Our first cluster is defined by foods that are high in protein and minerals like selenium and zinc while also being low in sugars and vitamin C. Even to a non-specialist, these sound like foods such as meat, poultry, or legumes.\n\n> **Key takeaway:** Particularly when it comes to interpretation, subject-matter expertise can prove essential to producing high-quality analysis. For this reason, you should also try to include SMEs in your data -cience projects.",
"_____no_output_____"
]
],
[
[
"c2 = pd.Series(vects[1], index=nutr_df.columns)\nc2.sort_values(ascending=False)",
"_____no_output_____"
]
],
[
[
"Our second group is foods that are high in fiber and folic acid and low in cholesterol.\n\n> **Exercise**\n>\n> Find the sorted output for $c_{3}$, $c_{4}$, and $c_{5}$.\n>\n> ***Hint:*** Remember that Python uses zero-indexing.",
"_____no_output_____"
],
[
"Even without subject-matter expertise, it is possible to get a more accurate sense of the kinds of foods are defined by each component? Yes! This is the reason we merged the `FoodGroup` column back into `pca_df`. We will sort that `DataFrame` by the components and count the values from `FoodGroup` for the top items.",
"_____no_output_____"
]
],
[
[
"pca_df.sort_values(by='c1')['FoodGroup'][:500].value_counts()",
"_____no_output_____"
]
],
[
[
"We can do the same thing for $c_{2}$.",
"_____no_output_____"
]
],
[
[
"pca_df.sort_values(by='c2')['FoodGroup'][:500].value_counts()",
"_____no_output_____"
]
],
[
[
"> **Exercise**\n>\n> Repeat this process for $c_{3}$, $c_{4}$, and $c_{5}$.",
"_____no_output_____"
],
[
"> **A parting note:** `Baby Foods` and some other categories might seem to dominate several of the categories. This is a product of all of the rows we had to drop that had `NaN` values. If we look at all of the value counts for `FoodGroup`, we will see that they are not evenly distributed, with some categories far more represented than others.",
"_____no_output_____"
]
],
[
[
"df['FoodGroup'].value_counts()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d05e2ab6a67253b72e95417444a98caae7e978a1 | 9,674 | ipynb | Jupyter Notebook | examples/notebooks/geemap_and_ipyleaflet.ipynb | hugoledoux/geemap | 10c665d06976cf9ac2d732309334be1044df71b8 | [
"MIT"
] | 3 | 2020-09-26T16:58:40.000Z | 2021-11-08T09:31:06.000Z | examples/notebooks/geemap_and_ipyleaflet.ipynb | hugoledoux/geemap | 10c665d06976cf9ac2d732309334be1044df71b8 | [
"MIT"
] | null | null | null | examples/notebooks/geemap_and_ipyleaflet.ipynb | hugoledoux/geemap | 10c665d06976cf9ac2d732309334be1044df71b8 | [
"MIT"
] | 1 | 2020-04-16T19:54:13.000Z | 2020-04-16T19:54:13.000Z | 30.613924 | 1,027 | 0.598408 | [
[
[
"<table class=\"ee-notebook-buttons\" align=\"left\">\n <td><a target=\"_blank\" href=\"https://github.com/giswqs/geemap/tree/master/examples/notebooks/geemap_and_ipyleaflet.ipynb\"><img width=32px src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" /> View source on GitHub</a></td>\n <td><a target=\"_blank\" href=\"https://nbviewer.jupyter.org/github/giswqs/geemap/blob/master/examples/notebooks/geemap_and_ipyleaflet.ipynb\"><img width=26px src=\"https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png\" />Notebook Viewer</a></td>\n <td><a target=\"_blank\" href=\"https://colab.research.google.com/github/giswqs/geemap/blob/master/examples/notebooks/geemap_and_ipyleaflet.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" /> Run in Google Colab</a></td>\n</table>",
"_____no_output_____"
],
[
"## Install Earth Engine API and geemap\nInstall the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.\nThe following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.\n\n**Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).",
"_____no_output_____"
]
],
[
[
"# Installs geemap package\nimport subprocess\n\ntry:\n import geemap\nexcept ImportError:\n print('geemap package not installed. Installing ...')\n subprocess.check_call([\"python\", '-m', 'pip', 'install', 'geemap'])\n\n# Checks whether this notebook is running on Google Colab\ntry:\n import google.colab\n import geemap.eefolium as emap\nexcept:\n import geemap as emap\n\n# Authenticates and initializes Earth Engine\nimport ee\n\ntry:\n ee.Initialize()\nexcept Exception as e:\n ee.Authenticate()\n ee.Initialize() ",
"_____no_output_____"
]
],
[
[
"## Create an interactive map",
"_____no_output_____"
]
],
[
[
"import geemap\nMap = geemap.Map(center=(40, -100), zoom=4)\nMap.add_minimap(position='bottomright')\nMap",
"_____no_output_____"
]
],
[
[
"## Add tile layers\n\nFor example, you can Google Map tile layer:",
"_____no_output_____"
]
],
[
[
"url = 'https://mt1.google.com/vt/lyrs=m&x={x}&y={y}&z={z}'\nMap.add_tile_layer(url, name='Google Map', attribution='Google')",
"_____no_output_____"
]
],
[
[
"Add Google Terrain tile layer:",
"_____no_output_____"
]
],
[
[
"url = 'https://mt1.google.com/vt/lyrs=p&x={x}&y={y}&z={z}'\nMap.add_tile_layer(url, name='Google Terrain', attribution='Google')",
"_____no_output_____"
]
],
[
[
"## Add WMS layers\nMore WMS layers can be found at <https://viewer.nationalmap.gov/services/>.",
"_____no_output_____"
],
[
"For example, you can add NAIP imagery.",
"_____no_output_____"
]
],
[
[
"url = 'https://services.nationalmap.gov/arcgis/services/USGSNAIPImagery/ImageServer/WMSServer?'\nMap.add_wms_layer(url=url, layers='0', name='NAIP Imagery', format='image/png')",
"_____no_output_____"
]
],
[
[
"Add USGS 3DEP Elevation Dataset",
"_____no_output_____"
]
],
[
[
"url = 'https://elevation.nationalmap.gov/arcgis/services/3DEPElevation/ImageServer/WMSServer?'\nMap.add_wms_layer(url=url, layers='3DEPElevation:None', name='3DEP Elevation', format='image/png')",
"_____no_output_____"
]
],
[
[
"## Capture user inputs",
"_____no_output_____"
]
],
[
[
"import geemap\nfrom ipywidgets import Label\nfrom ipyleaflet import Marker\n\nMap = geemap.Map(center=(40, -100), zoom=4)\n\nlabel = Label()\ndisplay(label)\n\ncoordinates = []\n\ndef handle_interaction(**kwargs):\n latlon = kwargs.get('coordinates')\n if kwargs.get('type') == 'mousemove':\n label.value = str(latlon)\n elif kwargs.get('type') == 'click':\n coordinates.append(latlon)\n Map.add_layer(Marker(location=latlon))\n\nMap.on_interaction(handle_interaction)\n\nMap",
"_____no_output_____"
],
[
"print(coordinates)",
"_____no_output_____"
]
],
[
[
"## A simpler way for capturing user inputs",
"_____no_output_____"
]
],
[
[
"import geemap\nMap = geemap.Map(center=(40, -100), zoom=4)\ncluster = Map.listening(event='click', add_marker=True)\nMap",
"_____no_output_____"
],
[
"# Get the last mouse clicked coordinates\nMap.last_click",
"_____no_output_____"
],
[
"# Get all the mouse clicked coordinates\nMap.all_clicks",
"_____no_output_____"
]
],
[
[
"## SplitMap control",
"_____no_output_____"
]
],
[
[
"import geemap\nfrom ipyleaflet import *\n\nMap = geemap.Map(center=(47.50, -101), zoom=7)\n\nright_layer = WMSLayer(\n url = 'https://ndgishub.nd.gov/arcgis/services/Imagery/AerialImage_ND_2017_CIR/ImageServer/WMSServer?',\n layers = 'AerialImage_ND_2017_CIR',\n name = 'AerialImage_ND_2017_CIR',\n format = 'image/png'\n)\n\nleft_layer = WMSLayer(\n url = 'https://ndgishub.nd.gov/arcgis/services/Imagery/AerialImage_ND_2018_CIR/ImageServer/WMSServer?',\n layers = 'AerialImage_ND_2018_CIR',\n name = 'AerialImage_ND_2018_CIR',\n format = 'image/png'\n)\n\ncontrol = SplitMapControl(left_layer=left_layer, right_layer=right_layer)\nMap.add_control(control)\nMap.add_control(LayersControl(position='topright'))\nMap.add_control(FullScreenControl())\n\nMap",
"_____no_output_____"
],
[
"import geemap\nMap = geemap.Map()\nMap.split_map(left_layer='HYBRID', right_layer='ESRI')\nMap",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05e3b166822bb133067795e0cc5b3306722ec86 | 13,652 | ipynb | Jupyter Notebook | Training.ipynb | nitinsrswt/age_gender_predictions | ee3110d9519bf9ab63b5d5cd44ca4722dc6be632 | [
"MIT"
] | null | null | null | Training.ipynb | nitinsrswt/age_gender_predictions | ee3110d9519bf9ab63b5d5cd44ca4722dc6be632 | [
"MIT"
] | null | null | null | Training.ipynb | nitinsrswt/age_gender_predictions | ee3110d9519bf9ab63b5d5cd44ca4722dc6be632 | [
"MIT"
] | null | null | null | 39.229885 | 263 | 0.59017 | [
[
[
"# Gender and Age Detection ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport os\nimport matplotlib.pyplot as plt\nimport cv2\nfrom tensorflow.keras.models import Sequential, load_model, Model\nfrom tensorflow.keras.layers import Conv2D, MaxPool2D, Dense, Dropout, BatchNormalization, Flatten, Input\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"# Defining the path .\ndatasetFolder = r\"C:\\Users\\ACER\\Documents\\Gender Detection\\DataSets\\UTKFace\"",
"_____no_output_____"
],
[
"# Creating empty list.\n\npixels = []\nage = []\ngender = []\nfor img in os.listdir(datasetFolder) : # os.listdir opens the directory \"datasetFolder\"\n # Label of each image is splitted on \"_\" and required information is stored in required variable.\n ages = img.split(\"_\")[0] \n genders = img.split(\"_\")[1]\n img = cv2.imread(str(datasetFolder) + \"/\" + str(img)) # Reading each image from the path of folder provided.\n img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB) # Converting the input image from BGR to RGB as computer by default sees an image in BGR.\n \n # Appending necessary data in respective created lists.\n pixels.append(np.array(img))\n age.append(np.array(ages))\n gender.append(np.array(genders))\n\n# Converting list to array\nage = np.array(age, dtype = np.int64) \npixels = np.array(pixels)\ngender = np.array(gender, np.uint64)",
"_____no_output_____"
],
[
"# Printing the length of the pixel .\np = len(pixels)\nprint(f\"No. of images working upon {p}\")",
"No. of images working upon 23708\n"
],
[
"# Splitting the images in train and test dataset.\nx_train, x_test, y_train, y_test = train_test_split(pixels, age, random_state = 100)\n\n# Splitting the dataset in train and test dataset as gender as.\nx_train_2, x_test_2, y_train_2, y_test_2 = train_test_split(pixels, gender, random_state = 100)",
"_____no_output_____"
],
[
"# Checking the shape of the images set. Here (200, 200, 3) are height, width and channel of the images respectively.\nx_train.shape, x_train_2.shape, x_test.shape, x_test_2.shape, ",
"_____no_output_____"
],
[
"# Checking the shape of the target variable.\ny_train.shape, y_train_2.shape, y_test.shape, y_test_2.shape",
"_____no_output_____"
]
],
[
[
"###### Below cell of code is used to create layers of a convolution neural network model. The layers in a CNN model are : \n* Input Layer\n* Convolution Layer\n* ReLu Layer\n* Pooling Layer\n* Fully Connected Network",
"_____no_output_____"
]
],
[
[
"inputLayer = Input(shape = (200, 200, 3)) # From the Input Model called from keras.models. Again (200, 200, 3) are height, width and channel of the images respectively.\nconvLayer1 = Conv2D(140,(3,3), activation = 'relu')(inputLayer) \n'''An activation function is basically just a simple function that transforms its inputs into outputs that have a certain range.\nAlso the ReLu activation transforms the -ve vaulues into 0 and positive remains the same, hence it is known as half rectifier as\nwell.'''\nconvLayer2 = Conv2D(130,(3,3), activation = 'relu')(convLayer1) # Creating seccond layer of CNN.\nbatch1 = BatchNormalization()(convLayer2) # Normalizing the data.\npoolLayer3 = MaxPool2D((2,2))(batch1) # Creating third, Pool Layer of the CNN.\nconvLayer3 = Conv2D(120,(3,3), activation = 'relu')(poolLayer3) # Adding the third Layer.\nbatch2 = BatchNormalization()(convLayer3) # Normalizing the layer.\npoolLayer4 = MaxPool2D((2,2))(batch2) #Adding fourth layer of CNN. \nflt = Flatten()(poolLayer4) # Flattening the data.",
"_____no_output_____"
],
[
"age_model = Dense(128,activation=\"relu\")(flt) # Here 128 is the no. of neurons connected with the flatten data layer.\nage_model = Dense(64,activation=\"relu\")(age_model) #Now as we move down, no. of neurons are reducing with previous neurons connected to them.\nage_model = Dense(32,activation=\"relu\")(age_model) \nage_model = Dense(1,activation=\"relu\")(age_model)",
"_____no_output_____"
],
[
"gender_model = Dense(128,activation=\"relu\")(flt) # The same work as above with 128 neurons is done for gender predictive model.\ngender_model = Dense(80,activation=\"relu\")(gender_model)\ngender_model = Dense(64,activation=\"relu\")(gender_model)\ngender_model = Dense(32,activation=\"relu\")(gender_model)\ngender_model = Dropout(0.5)(gender_model) # Drop-out layer is added to dodge the overfitting of the model.\n'''Softmax is a mathematical function that converts a vector of numbers into a vector of probabilities, where the probabilities \nof each value are proportional to the relative scale of each value in the vector. Here it is used as an activation function.'''\ngender_model = Dense(2,activation=\"softmax\")(gender_model) ",
"_____no_output_____"
]
],
[
[
"###### Below cell of code is to make an object of the Model from keras.models.",
"_____no_output_____"
]
],
[
[
"model = Model(inputs=inputLayer,outputs=[age_model,gender_model]) # Adding the input layer and the output layer in our model and making the object.\nmodel.compile(optimizer=\"adam\",loss=[\"mse\",\"sparse_categorical_crossentropy\"],metrics=['mae','accuracy']) \nmodel.summary() # To get the summary of our model.",
"Model: \"functional_1\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) [(None, 200, 200, 3) 0 \n__________________________________________________________________________________________________\nconv2d (Conv2D) (None, 198, 198, 140 3920 input_1[0][0] \n__________________________________________________________________________________________________\nconv2d_1 (Conv2D) (None, 196, 196, 130 163930 conv2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization (BatchNorma (None, 196, 196, 130 520 conv2d_1[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 98, 98, 130) 0 batch_normalization[0][0] \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 96, 96, 120) 140520 max_pooling2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_1 (BatchNor (None, 96, 96, 120) 480 conv2d_2[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_1 (MaxPooling2D) (None, 48, 48, 120) 0 batch_normalization_1[0][0] \n__________________________________________________________________________________________________\nflatten (Flatten) (None, 276480) 0 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\ndense_3 (Dense) (None, 80) 22118480 flatten[0][0] \n__________________________________________________________________________________________________\ndense_4 (Dense) (None, 64) 5184 dense_3[0][0] \n__________________________________________________________________________________________________\ndense (Dense) (None, 64) 17694784 flatten[0][0] \n__________________________________________________________________________________________________\ndense_5 (Dense) (None, 32) 2080 dense_4[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 32) 2080 dense[0][0] \n__________________________________________________________________________________________________\ndropout (Dropout) (None, 32) 0 dense_5[0][0] \n__________________________________________________________________________________________________\ndense_2 (Dense) (None, 1) 33 dense_1[0][0] \n__________________________________________________________________________________________________\ndense_6 (Dense) (None, 2) 66 dropout[0][0] \n==================================================================================================\nTotal params: 40,132,077\nTrainable params: 40,131,577\nNon-trainable params: 500\n__________________________________________________________________________________________________\n"
],
[
"save = model.fit(x_train,[y_train,y_train_2], validation_data=(x_test,[y_test,y_test_2]),epochs=50)\nmodel.save(\"model.h5\")",
"Epoch 1/50\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05e3d692a0b24b4c1d80428e294bfe3cd1eb53d | 4,377 | ipynb | Jupyter Notebook | notebooks/generate_node_features/from old pc/corpus_2018_level_0_1_2_3.ipynb | PacoPacov/site_similarity | db21c2e7b6a6a8cdaccb55703017990fa72d2593 | [
"MIT"
] | null | null | null | notebooks/generate_node_features/from old pc/corpus_2018_level_0_1_2_3.ipynb | PacoPacov/site_similarity | db21c2e7b6a6a8cdaccb55703017990fa72d2593 | [
"MIT"
] | null | null | null | notebooks/generate_node_features/from old pc/corpus_2018_level_0_1_2_3.ipynb | PacoPacov/site_similarity | db21c2e7b6a6a8cdaccb55703017990fa72d2593 | [
"MIT"
] | 1 | 2020-08-03T10:22:33.000Z | 2020-08-03T10:22:33.000Z | 22.106061 | 116 | 0.549692 | [
[
[
"import sys,os\nsys.path.append(\"/home/panayot/Documents/site_simility\")",
"_____no_output_____"
],
[
"from utils",
"_____no_output_____"
],
[
"from utils.notebook_utils import combined_nodes_referral_sites_audience_overlap",
"_____no_output_____"
],
[
"d = combined_nodes_referral_sites_audience_overlap(data_year='2018', level=3)",
"For file \"modified_corpus_2018_referral_sites.json\" -> load 1062 records\nFor file \"modified_corpus_2018_referral_sites_level_1.json\" -> load 2835 records\nFor file \"modified_corpus_2018_referral_sites_level_2.json\" -> load 7099 records\nFor file \"modified_corpus_2018_referral_sites_level_3.json\" -> load 15767 records\nLoaded 30889 nodes with records level <= 3 and child size:149110\nreferral_sites node size: 68157 audience_overlap node size: 149178\n"
],
[
"import redis\n\nred = redis.Redis()",
"_____no_output_____"
],
[
"unique_nodes = set()\n\nfor l, r in d:\n unique_nodes.add(l)\n unique_nodes.add(r)\n\nprint('Sites that needs to extract alexa section info: ', len([el for el in unique_nodes if not red.get(el)]))",
"Sites that needs to extract alexa section info: 38835\n"
],
[
"from utils.notebook_utils import get_alexa_information_sections",
"_____no_output_____"
],
[
"alexa_data_for_nodes_level_0_and_1 = get_alexa_information_sections(unique_nodes)",
"_____no_output_____"
],
[
"from datetime import datetime\n\nnow = datetime.now()\n\ncurrent_time = now.strftime(\"%H:%M:%S\")\nprint(\"Current Time =\", current_time)\n\nresult = list(zip(unique_nodes, alexa_data_for_nodes_level_0_and_1))",
"Current Time = 01:20:07\n"
],
[
"from datetime import datetime\n\nnow = datetime.now()\n\ncurrent_time = now.strftime(\"%H:%M:%S\")\nprint(\"Current Time =\", current_time) # started 5:02 - 05:27:49 for 2500",
"Current Time = 06:42:44\n"
],
[
"import json\n\n\nwith open('corpus_2018_levels_0_1_2_3_data.json', 'w') as f:\n json.dump(dict(result), f)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05e4bf1eef5b8b573948187b0995f1c338154bd | 24,192 | ipynb | Jupyter Notebook | introduction_to_amazon_algorithms/lda_topic_modeling/LDA-Introduction.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
] | 2 | 2021-03-31T21:10:44.000Z | 2021-04-03T04:27:26.000Z | introduction_to_amazon_algorithms/lda_topic_modeling/LDA-Introduction.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
] | 1 | 2021-03-25T18:31:29.000Z | 2021-03-25T18:31:29.000Z | introduction_to_amazon_algorithms/lda_topic_modeling/LDA-Introduction.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
] | 2 | 2021-02-24T02:44:02.000Z | 2021-08-19T06:49:50.000Z | 38.769231 | 888 | 0.64294 | [
[
[
"# An Introduction to SageMaker LDA\n\n***Finding topics in synthetic document data using Spectral LDA algorithms.***\n\n---\n\n1. [Introduction](#Introduction)\n1. [Setup](#Setup)\n1. [Training](#Training)\n1. [Inference](#Inference)\n1. [Epilogue](#Epilogue)",
"_____no_output_____"
],
[
"# Introduction\n***\n\nAmazon SageMaker LDA is an unsupervised learning algorithm that attempts to describe a set of observations as a mixture of distinct categories. Latent Dirichlet Allocation (LDA) is most commonly used to discover a user-specified number of topics shared by documents within a text corpus. Here each observation is a document, the features are the presence (or occurrence count) of each word, and the categories are the topics. Since the method is unsupervised, the topics are not specified up front, and are not guaranteed to align with how a human may naturally categorize documents. The topics are learned as a probability distribution over the words that occur in each document. Each document, in turn, is described as a mixture of topics.\n\nIn this notebook we will use the Amazon SageMaker LDA algorithm to train an LDA model on some example synthetic data. We will then use this model to classify (perform inference on) the data. The main goals of this notebook are to,\n\n* learn how to obtain and store data for use in Amazon SageMaker,\n* create an AWS SageMaker training job on a data set to produce an LDA model,\n* use the LDA model to perform inference with an Amazon SageMaker endpoint.\n\nThe following are ***not*** goals of this notebook:\n\n* understand the LDA model,\n* understand how the Amazon SageMaker LDA algorithm works,\n* interpret the meaning of the inference output\n\nIf you would like to know more about these things take a minute to run this notebook and then check out the SageMaker LDA Documentation and the **LDA-Science.ipynb** notebook.",
"_____no_output_____"
]
],
[
[
"!conda install -y scipy",
"_____no_output_____"
],
[
"%matplotlib inline\n\nimport os, re\n\nimport boto3\nimport matplotlib.pyplot as plt\nimport numpy as np\nnp.set_printoptions(precision=3, suppress=True)\n\n# some helpful utility functions are defined in the Python module\n# \"generate_example_data\" located in the same directory as this\n# notebook\nfrom generate_example_data import generate_griffiths_data, plot_lda, match_estimated_topics\n\n# accessing the SageMaker Python SDK\nimport sagemaker\nfrom sagemaker.amazon.common import RecordSerializer\nfrom sagemaker.serializers import CSVSerializer\nfrom sagemaker.deserializers import JSONDeserializer",
"_____no_output_____"
]
],
[
[
"# Setup\n\n***\n\n*This notebook was created and tested on an ml.m4.xlarge notebook instance.*\n\nBefore we do anything at all, we need data! We also need to setup our AWS credentials so that AWS SageMaker can store and access data. In this section we will do four things:\n\n1. [Setup AWS Credentials](#SetupAWSCredentials)\n1. [Obtain Example Dataset](#ObtainExampleDataset)\n1. [Inspect Example Data](#InspectExampleData)\n1. [Store Data on S3](#StoreDataonS3)",
"_____no_output_____"
],
[
"## Setup AWS Credentials\n\nWe first need to specify some AWS credentials; specifically data locations and access roles. This is the only cell of this notebook that you will need to edit. In particular, we need the following data:\n\n* `bucket` - An S3 bucket accessible by this account.\n * Used to store input training data and model data output.\n * Should be within the same region as this notebook instance, training, and hosting.\n* `prefix` - The location in the bucket where this notebook's input and and output data will be stored. (The default value is sufficient.)\n* `role` - The IAM Role ARN used to give training and hosting access to your data.\n * See documentation on how to create these.\n * The script below will try to determine an appropriate Role ARN.",
"_____no_output_____"
]
],
[
[
"from sagemaker import get_execution_role\n\nsession = sagemaker.Session()\nrole = get_execution_role()\nbucket = session.default_bucket()\nprefix = 'sagemaker/DEMO-lda-introduction'\n\nprint('Training input/output will be stored in {}/{}'.format(bucket, prefix))\nprint('\\nIAM Role: {}'.format(role))",
"_____no_output_____"
]
],
[
[
"## Obtain Example Data\n\n\nWe generate some example synthetic document data. For the purposes of this notebook we will omit the details of this process. All we need to know is that each piece of data, commonly called a *\"document\"*, is a vector of integers representing *\"word counts\"* within the document. In this particular example there are a total of 25 words in the *\"vocabulary\"*.\n\n$$\n\\underbrace{w}_{\\text{document}} = \\overbrace{\\big[ w_1, w_2, \\ldots, w_V \\big] }^{\\text{word counts}},\n\\quad\nV = \\text{vocabulary size}\n$$\n\nThese data are based on that used by Griffiths and Steyvers in their paper [Finding Scientific Topics](http://psiexp.ss.uci.edu/research/papers/sciencetopics.pdf). For more information, see the **LDA-Science.ipynb** notebook.",
"_____no_output_____"
]
],
[
[
"print('Generating example data...')\nnum_documents = 6000\nnum_topics = 5\nknown_alpha, known_beta, documents, topic_mixtures = generate_griffiths_data(\n num_documents=num_documents, num_topics=num_topics)\nvocabulary_size = len(documents[0])\n\n# separate the generated data into training and tests subsets\nnum_documents_training = int(0.9*num_documents)\nnum_documents_test = num_documents - num_documents_training\n\ndocuments_training = documents[:num_documents_training]\ndocuments_test = documents[num_documents_training:]\n\ntopic_mixtures_training = topic_mixtures[:num_documents_training]\ntopic_mixtures_test = topic_mixtures[num_documents_training:]\n\nprint('documents_training.shape = {}'.format(documents_training.shape))\nprint('documents_test.shape = {}'.format(documents_test.shape))",
"_____no_output_____"
]
],
[
[
"## Inspect Example Data\n\n*What does the example data actually look like?* Below we print an example document as well as its corresponding known *topic-mixture*. A topic-mixture serves as the \"label\" in the LDA model. It describes the ratio of topics from which the words in the document are found.\n\nFor example, if the topic mixture of an input document $\\mathbf{w}$ is,\n\n$$\\theta = \\left[ 0.3, 0.2, 0, 0.5, 0 \\right]$$\n\nthen $\\mathbf{w}$ is 30% generated from the first topic, 20% from the second topic, and 50% from the fourth topic. For more information see **How LDA Works** in the SageMaker documentation as well as the **LDA-Science.ipynb** notebook.\n\nBelow, we compute the topic mixtures for the first few training documents. As we can see, each document is a vector of word counts from the 25-word vocabulary and its topic-mixture is a probability distribution across the five topics used to generate the sample dataset.",
"_____no_output_____"
]
],
[
[
"print('First training document =\\n{}'.format(documents[0]))\nprint('\\nVocabulary size = {}'.format(vocabulary_size))",
"_____no_output_____"
],
[
"print('Known topic mixture of first document =\\n{}'.format(topic_mixtures_training[0]))\nprint('\\nNumber of topics = {}'.format(num_topics))\nprint('Sum of elements = {}'.format(topic_mixtures_training[0].sum()))",
"_____no_output_____"
]
],
[
[
"Later, when we perform inference on the training data set we will compare the inferred topic mixture to this known one.\n\n---\n\nHuman beings are visual creatures, so it might be helpful to come up with a visual representation of these documents. In the below plots, each pixel of a document represents a word. The greyscale intensity is a measure of how frequently that word occurs. Below we plot the first few documents of the training set reshaped into 5x5 pixel grids.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nfig = plot_lda(documents_training, nrows=3, ncols=4, cmap='gray_r', with_colorbar=True)\nfig.suptitle('Example Document Word Counts')\nfig.set_dpi(160)",
"_____no_output_____"
]
],
[
[
"## Store Data on S3\n\nA SageMaker training job needs access to training data stored in an S3 bucket. Although training can accept data of various formats we convert the documents MXNet RecordIO Protobuf format before uploading to the S3 bucket defined at the beginning of this notebook. We do so by making use of the SageMaker Python SDK utility `RecordSerializer`.",
"_____no_output_____"
]
],
[
[
"# convert documents_training to Protobuf RecordIO format\nrecordio_protobuf_serializer = RecordSerializer()\nfbuffer = recordio_protobuf_serializer.serialize(documents_training)\n\n# upload to S3 in bucket/prefix/train\nfname = 'lda.data'\ns3_object = os.path.join(prefix, 'train', fname)\nboto3.Session().resource('s3').Bucket(bucket).Object(s3_object).upload_fileobj(fbuffer)\n\ns3_train_data = 's3://{}/{}'.format(bucket, s3_object)\nprint('Uploaded data to S3: {}'.format(s3_train_data))",
"_____no_output_____"
]
],
[
[
"# Training\n\n***\n\nOnce the data is preprocessed and available in a recommended format the next step is to train our model on the data. There are number of parameters required by SageMaker LDA configuring the model and defining the computational environment in which training will take place.\n\nFirst, we specify a Docker container containing the SageMaker LDA algorithm. For your convenience, a region-specific container is automatically chosen for you to minimize cross-region data communication. Information about the locations of each SageMaker algorithm is available in the documentation.",
"_____no_output_____"
]
],
[
[
"from sagemaker.amazon.amazon_estimator import get_image_uri\n# select the algorithm container based on this notebook's current location\n\nregion_name = boto3.Session().region_name\ncontainer = get_image_uri(region_name, 'lda')\n\nprint('Using SageMaker LDA container: {} ({})'.format(container, region_name))",
"_____no_output_____"
]
],
[
[
"Particular to a SageMaker LDA training job are the following hyperparameters:\n\n* **`num_topics`** - The number of topics or categories in the LDA model.\n * Usually, this is not known a priori.\n * In this example, howevever, we know that the data is generated by five topics.\n\n* **`feature_dim`** - The size of the *\"vocabulary\"*, in LDA parlance.\n * In this example, this is equal 25.\n\n* **`mini_batch_size`** - The number of input training documents.\n\n* **`alpha0`** - *(optional)* a measurement of how \"mixed\" are the topic-mixtures.\n * When `alpha0` is small the data tends to be represented by one or few topics.\n * When `alpha0` is large the data tends to be an even combination of several or many topics.\n * The default value is `alpha0 = 1.0`.\n\nIn addition to these LDA model hyperparameters, we provide additional parameters defining things like the EC2 instance type on which training will run, the S3 bucket containing the data, and the AWS access role. Note that,\n\n* Recommended instance type: `ml.c4`\n* Current limitations:\n * SageMaker LDA *training* can only run on a single instance.\n * SageMaker LDA does not take advantage of GPU hardware.\n * (The Amazon AI Algorithms team is working hard to provide these capabilities in a future release!)",
"_____no_output_____"
]
],
[
[
"# specify general training job information\nlda = sagemaker.estimator.Estimator(\n container,\n role,\n output_path='s3://{}/{}/output'.format(bucket, prefix),\n train_instance_count=1,\n train_instance_type='ml.c4.2xlarge',\n sagemaker_session=session,\n)\n\n# set algorithm-specific hyperparameters\nlda.set_hyperparameters(\n num_topics=num_topics,\n feature_dim=vocabulary_size,\n mini_batch_size=num_documents_training,\n alpha0=1.0,\n)\n\n# run the training job on input data stored in S3\nlda.fit({'train': s3_train_data})",
"_____no_output_____"
]
],
[
[
"If you see the message\n\n> `===== Job Complete =====`\n\nat the bottom of the output logs then that means training sucessfully completed and the output LDA model was stored in the specified output path. You can also view information about and the status of a training job using the AWS SageMaker console. Just click on the \"Jobs\" tab and select training job matching the training job name, below:",
"_____no_output_____"
]
],
[
[
"print('Training job name: {}'.format(lda.latest_training_job.job_name))",
"_____no_output_____"
]
],
[
[
"# Inference\n\n***\n\nA trained model does nothing on its own. We now want to use the model we computed to perform inference on data. For this example, that means predicting the topic mixture representing a given document.\n\nWe create an inference endpoint using the SageMaker Python SDK `deploy()` function from the job we defined above. We specify the instance type where inference is computed as well as an initial number of instances to spin up.",
"_____no_output_____"
]
],
[
[
"lda_inference = lda.deploy(\n initial_instance_count=1,\n instance_type='ml.m4.xlarge', # LDA inference may work better at scale on ml.c4 instances\n)",
"_____no_output_____"
]
],
[
[
"Congratulations! You now have a functioning SageMaker LDA inference endpoint. You can confirm the endpoint configuration and status by navigating to the \"Endpoints\" tab in the AWS SageMaker console and selecting the endpoint matching the endpoint name, below: ",
"_____no_output_____"
]
],
[
[
"print('Endpoint name: {}'.format(lda_inference.endpoint_name))",
"_____no_output_____"
]
],
[
[
"With this realtime endpoint at our fingertips we can finally perform inference on our training and test data.\n\nWe can pass a variety of data formats to our inference endpoint. In this example we will demonstrate passing CSV-formatted data. Other available formats are JSON-formatted, JSON-sparse-formatter, and RecordIO Protobuf. We make use of the SageMaker Python SDK utilities `CSVSerializer` and `JSONDeserializer` when configuring the inference endpoint.",
"_____no_output_____"
]
],
[
[
"lda_inference.serializer = CSVSerializer()\nlda_inference.deserializer = JSONDeserializer()",
"_____no_output_____"
]
],
[
[
"We pass some test documents to the inference endpoint. Note that the serializer and deserializer will atuomatically take care of the datatype conversion from Numpy NDArrays.",
"_____no_output_____"
]
],
[
[
"results = lda_inference.predict(documents_test[:12])\n\nprint(results)",
"_____no_output_____"
]
],
[
[
"It may be hard to see but the output format of SageMaker LDA inference endpoint is a Python dictionary with the following format.\n\n```\n{\n 'predictions': [\n {'topic_mixture': [ ... ] },\n {'topic_mixture': [ ... ] },\n {'topic_mixture': [ ... ] },\n ...\n ]\n}\n```\n\nWe extract the topic mixtures, themselves, corresponding to each of the input documents.",
"_____no_output_____"
]
],
[
[
"computed_topic_mixtures = np.array([prediction['topic_mixture'] for prediction in results['predictions']])\n\nprint(computed_topic_mixtures)",
"_____no_output_____"
]
],
[
[
"If you decide to compare these results to the known topic mixtures generated in the [Obtain Example Data](#ObtainExampleData) Section keep in mind that SageMaker LDA discovers topics in no particular order. That is, the approximate topic mixtures computed above may be permutations of the known topic mixtures corresponding to the same documents.",
"_____no_output_____"
]
],
[
[
"print(topic_mixtures_test[0]) # known test topic mixture\nprint(computed_topic_mixtures[0]) # computed topic mixture (topics permuted)",
"_____no_output_____"
]
],
[
[
"## Stop / Close the Endpoint\n\nFinally, we should delete the endpoint before we close the notebook.\n\nTo do so execute the cell below. Alternately, you can navigate to the \"Endpoints\" tab in the SageMaker console, select the endpoint with the name stored in the variable `endpoint_name`, and select \"Delete\" from the \"Actions\" dropdown menu. ",
"_____no_output_____"
]
],
[
[
"sagemaker.Session().delete_endpoint(lda_inference.endpoint_name)",
"_____no_output_____"
]
],
[
[
"# Epilogue\n\n---\n\nIn this notebook we,\n\n* generated some example LDA documents and their corresponding topic-mixtures,\n* trained a SageMaker LDA model on a training set of documents,\n* created an inference endpoint,\n* used the endpoint to infer the topic mixtures of a test input.\n\nThere are several things to keep in mind when applying SageMaker LDA to real-word data such as a corpus of text documents. Note that input documents to the algorithm, both in training and inference, need to be vectors of integers representing word counts. Each index corresponds to a word in the corpus vocabulary. Therefore, one will need to \"tokenize\" their corpus vocabulary.\n\n$$\n\\text{\"cat\"} \\mapsto 0, \\; \\text{\"dog\"} \\mapsto 1 \\; \\text{\"bird\"} \\mapsto 2, \\ldots\n$$\n\nEach text document then needs to be converted to a \"bag-of-words\" format document.\n\n$$\nw = \\text{\"cat bird bird bird cat\"} \\quad \\longmapsto \\quad w = [2, 0, 3, 0, \\ldots, 0]\n$$\n\nAlso note that many real-word applications have large vocabulary sizes. It may be necessary to represent the input documents in sparse format. Finally, the use of stemming and lemmatization in data preprocessing provides several benefits. Doing so can improve training and inference compute time since it reduces the effective vocabulary size. More importantly, though, it can improve the quality of learned topic-word probability matrices and inferred topic mixtures. For example, the words *\"parliament\"*, *\"parliaments\"*, *\"parliamentary\"*, *\"parliament's\"*, and *\"parliamentarians\"* are all essentially the same word, *\"parliament\"*, but with different conjugations. For the purposes of detecting topics, such as a *\"politics\"* or *governments\"* topic, the inclusion of all five does not add much additional value as they all essentiall describe the same feature.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d05e4fa7087cdaa39c5b3f8e8382ce105391eee5 | 1,896 | ipynb | Jupyter Notebook | audience.ipynb | berthubert/audience-minutes | c91fff267409fb3d1703b82f3f103c3cddfc3f3f | [
"MIT"
] | 30 | 2021-06-27T02:25:02.000Z | 2022-03-06T15:04:25.000Z | audience.ipynb | berthubert/audience-minutes | c91fff267409fb3d1703b82f3f103c3cddfc3f3f | [
"MIT"
] | 2 | 2021-06-27T20:58:14.000Z | 2022-02-27T12:47:16.000Z | audience.ipynb | berthubert/audience-minutes | c91fff267409fb3d1703b82f3f103c3cddfc3f3f | [
"MIT"
] | null | null | null | 23.121951 | 97 | 0.545359 | [
[
[
"%matplotlib notebook\n%precision 2\nimport matplotlib\nimport matplotlib.pyplot as plt\nplt.rcParams['figure.figsize'] = [9.5, 6]\nimport pandas",
"_____no_output_____"
],
[
"percs=pandas.read_csv(\"percs.csv\")\nbusy=percs.groupby([\"url\"]).count().sort_values([\"count\"], ascending=False).head(10)\nbusy",
"_____no_output_____"
],
[
"# given 10% sampling and once a minute measurements, one sample is 10 minutes\n# so this gives a table with reading hours\nbusy.perc*10/60",
"_____no_output_____"
],
[
"# you can raise or lower the number of bins depending on how much data you have\nfor url in busy.index:\n plt.figure()\n plt.hist(percs[(percs.url==url)].perc, bins=10, density=True)\n plt.grid()\n plt.ylabel(\"Density\")\n plt.xlabel(\"Location as percentage of page length\")\n plt.title(\"Sampled density of readership for\\n\"+url)\n\n print(url)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d05e68e1fc32c8ea5619dd0f36550706eb3f59c0 | 154,612 | ipynb | Jupyter Notebook | courses/machine_learning/deepdive2/text_classification/solutions/word2vec.ipynb | juancaob/training-data-analyst | 85ad8c70849466bb87a6c6eb01cd0db883277d51 | [
"Apache-2.0"
] | 2 | 2021-12-29T10:49:00.000Z | 2021-12-31T13:42:35.000Z | courses/machine_learning/deepdive2/text_classification/solutions/word2vec.ipynb | juancaob/training-data-analyst | 85ad8c70849466bb87a6c6eb01cd0db883277d51 | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive2/text_classification/solutions/word2vec.ipynb | juancaob/training-data-analyst | 85ad8c70849466bb87a6c6eb01cd0db883277d51 | [
"Apache-2.0"
] | null | null | null | 29.66462 | 521 | 0.467583 | [
[
[
"# Word2Vec\n\n**Learning Objectives**\n\n1. Compile all steps into one function\n2. Prepare training data for Word2Vec\n3. Model and Training\n4. Embedding lookup and analysis\n\n\n\n\n## Introduction \nWord2Vec is not a singular algorithm, rather, it is a family of model architectures and optimizations that can be used to learn word embeddings from large datasets. Embeddings learned through Word2Vec have proven to be successful on a variety of downstream natural language processing tasks.\n\nNote: This notebook is based on [Efficient Estimation of Word Representations in Vector Space](https://arxiv.org/pdf/1301.3781.pdf) and\n[Distributed\nRepresentations of Words and Phrases and their Compositionality](https://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf). It is not an exact implementation of the papers. Rather, it is intended to illustrate the key ideas.\n\nThese papers proposed two methods for learning representations of words: \n\n* **Continuous Bag-of-Words Model** which predicts the middle word based on surrounding context words. The context consists of a few words before and after the current (middle) word. This architecture is called a bag-of-words model as the order of words in the context is not important.\n* **Continuous Skip-gram Model** which predict words within a certain range before and after the current word in the same sentence. A worked example of this is given below.\n\n\nYou'll use the skip-gram approach in this notebook. First, you'll explore skip-grams and other concepts using a single sentence for illustration. Next, you'll train your own Word2Vec model on a small dataset. This notebook also contains code to export the trained embeddings and visualize them in the [TensorFlow Embedding Projector](http://projector.tensorflow.org/).\n\n\nEach learning objective will correspond to a __#TODO__ in the [student lab notebook](../labs/word2vec.ipynb) -- try to complete that notebook first before reviewing this solution notebook.",
"_____no_output_____"
],
[
"## Skip-gram and Negative Sampling ",
"_____no_output_____"
],
[
"While a bag-of-words model predicts a word given the neighboring context, a skip-gram model predicts the context (or neighbors) of a word, given the word itself. The model is trained on skip-grams, which are n-grams that allow tokens to be skipped (see the diagram below for an example). The context of a word can be represented through a set of skip-gram pairs of `(target_word, context_word)` where `context_word` appears in the neighboring context of `target_word`. ",
"_____no_output_____"
],
[
"Consider the following sentence of 8 words.\n> The wide road shimmered in the hot sun. \n\nThe context words for each of the 8 words of this sentence are defined by a window size. The window size determines the span of words on either side of a `target_word` that can be considered `context word`. Take a look at this table of skip-grams for target words based on different window sizes.",
"_____no_output_____"
],
[
"Note: For this tutorial, a window size of *n* implies n words on each side with a total window span of 2*n+1 words across a word.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"The training objective of the skip-gram model is to maximize the probability of predicting context words given the target word. For a sequence of words *w<sub>1</sub>, w<sub>2</sub>, ... w<sub>T</sub>*, the objective can be written as the average log probability",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"where `c` is the size of the training context. The basic skip-gram formulation defines this probability using the softmax function.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"where *v* and *v<sup>'<sup>* are target and context vector representations of words and *W* is vocabulary size. ",
"_____no_output_____"
],
[
"Computing the denominator of this formulation involves performing a full softmax over the entire vocabulary words which is often large (10<sup>5</sup>-10<sup>7</sup>) terms. ",
"_____no_output_____"
],
[
"The [Noise Contrastive Estimation](https://www.tensorflow.org/api_docs/python/tf/nn/nce_loss) loss function is an efficient approximation for a full softmax. With an objective to learn word embeddings instead of modelling the word distribution, NCE loss can be [simplified](https://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf) to use negative sampling. ",
"_____no_output_____"
],
[
"The simplified negative sampling objective for a target word is to distinguish the context word from *num_ns* negative samples drawn from noise distribution *P<sub>n</sub>(w)* of words. More precisely, an efficient approximation of full softmax over the vocabulary is, for a skip-gram pair, to pose the loss for a target word as a classification problem between the context word and *num_ns* negative samples. ",
"_____no_output_____"
],
[
"A negative sample is defined as a (target_word, context_word) pair such that the context_word does not appear in the `window_size` neighborhood of the target_word. For the example sentence, these are few potential negative samples (when `window_size` is 2).\n\n```\n(hot, shimmered)\n(wide, hot)\n(wide, sun)\n```",
"_____no_output_____"
],
[
"In the next section, you'll generate skip-grams and negative samples for a single sentence. You'll also learn about subsampling techniques and train a classification model for positive and negative training examples later in the tutorial.",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"# Use the chown command to change the ownership of repository to user.\n!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst",
"_____no_output_____"
],
[
"!pip install -q tqdm",
"_____no_output_____"
],
[
"# You can use any Python source file as a module by executing an import statement in some other Python source file.\n# The import statement combines two operations; it searches for the named module, then it binds the\n# results of that search to a name in the local scope.\nimport io\nimport itertools\nimport numpy as np\nimport os\nimport re\nimport string\nimport tensorflow as tf\nimport tqdm\n\nfrom tensorflow.keras import Model, Sequential\nfrom tensorflow.keras.layers import Activation, Dense, Dot, Embedding, Flatten, GlobalAveragePooling1D, Reshape\nfrom tensorflow.keras.layers.experimental.preprocessing import TextVectorization",
"_____no_output_____"
]
],
[
[
"Please check your tensorflow version using the cell below.",
"_____no_output_____"
]
],
[
[
"# Show the currently installed version of TensorFlow\nprint(\"TensorFlow version: \",tf.version.VERSION)",
"TensorFlow version: 2.6.0\n"
],
[
"SEED = 42 \nAUTOTUNE = tf.data.experimental.AUTOTUNE",
"_____no_output_____"
]
],
[
[
"### Vectorize an example sentence",
"_____no_output_____"
],
[
"Consider the following sentence: \n`The wide road shimmered in the hot sun.`\n\nTokenize the sentence:",
"_____no_output_____"
]
],
[
[
"sentence = \"The wide road shimmered in the hot sun\"\ntokens = list(sentence.lower().split())\nprint(len(tokens))",
"8\n"
]
],
[
[
"Create a vocabulary to save mappings from tokens to integer indices.",
"_____no_output_____"
]
],
[
[
"vocab, index = {}, 1 # start indexing from 1\nvocab['<pad>'] = 0 # add a padding token \nfor token in tokens:\n if token not in vocab: \n vocab[token] = index\n index += 1\nvocab_size = len(vocab)\nprint(vocab)",
"{'<pad>': 0, 'the': 1, 'wide': 2, 'road': 3, 'shimmered': 4, 'in': 5, 'hot': 6, 'sun': 7}\n"
]
],
[
[
"Create an inverse vocabulary to save mappings from integer indices to tokens.",
"_____no_output_____"
]
],
[
[
"inverse_vocab = {index: token for token, index in vocab.items()}\nprint(inverse_vocab)",
"{0: '<pad>', 1: 'the', 2: 'wide', 3: 'road', 4: 'shimmered', 5: 'in', 6: 'hot', 7: 'sun'}\n"
]
],
[
[
"Vectorize your sentence.\n",
"_____no_output_____"
]
],
[
[
"example_sequence = [vocab[word] for word in tokens]\nprint(example_sequence)",
"[1, 2, 3, 4, 5, 1, 6, 7]\n"
]
],
[
[
"### Generate skip-grams from one sentence",
"_____no_output_____"
],
[
"The `tf.keras.preprocessing.sequence` module provides useful functions that simplify data preparation for Word2Vec. You can use the `tf.keras.preprocessing.sequence.skipgrams` to generate skip-gram pairs from the `example_sequence` with a given `window_size` from tokens in the range `[0, vocab_size)`.\n\nNote: `negative_samples` is set to `0` here as batching negative samples generated by this function requires a bit of code. You will use another function to perform negative sampling in the next section.\n",
"_____no_output_____"
]
],
[
[
"window_size = 2\npositive_skip_grams, _ = tf.keras.preprocessing.sequence.skipgrams(\n example_sequence, \n vocabulary_size=vocab_size,\n window_size=window_size,\n negative_samples=0)\nprint(len(positive_skip_grams))",
"26\n"
]
],
[
[
"Take a look at few positive skip-grams.",
"_____no_output_____"
]
],
[
[
"for target, context in positive_skip_grams[:5]:\n print(f\"({target}, {context}): ({inverse_vocab[target]}, {inverse_vocab[context]})\")",
"(1, 3): (the, road)\n(4, 1): (shimmered, the)\n(5, 6): (in, hot)\n(4, 2): (shimmered, wide)\n(3, 2): (road, wide)\n"
]
],
[
[
"### Negative sampling for one skip-gram ",
"_____no_output_____"
],
[
"The `skipgrams` function returns all positive skip-gram pairs by sliding over a given window span. To produce additional skip-gram pairs that would serve as negative samples for training, you need to sample random words from the vocabulary. Use the `tf.random.log_uniform_candidate_sampler` function to sample `num_ns` number of negative samples for a given target word in a window. You can call the funtion on one skip-grams's target word and pass the context word as true class to exclude it from being sampled.\n",
"_____no_output_____"
],
[
"Key point: *num_ns* (number of negative samples per positive context word) between [5, 20] is [shown to work](https://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf) best for smaller datasets, while *num_ns* between [2,5] suffices for larger datasets. ",
"_____no_output_____"
]
],
[
[
"# Get target and context words for one positive skip-gram.\ntarget_word, context_word = positive_skip_grams[0]\n\n# Set the number of negative samples per positive context. \nnum_ns = 4\n\ncontext_class = tf.reshape(tf.constant(context_word, dtype=\"int64\"), (1, 1))\nnegative_sampling_candidates, _, _ = tf.random.log_uniform_candidate_sampler(\n true_classes=context_class, # class that should be sampled as 'positive'\n num_true=1, # each positive skip-gram has 1 positive context class\n num_sampled=num_ns, # number of negative context words to sample\n unique=True, # all the negative samples should be unique\n range_max=vocab_size, # pick index of the samples from [0, vocab_size]\n seed=SEED, # seed for reproducibility\n name=\"negative_sampling\" # name of this operation\n)\nprint(negative_sampling_candidates)\nprint([inverse_vocab[index.numpy()] for index in negative_sampling_candidates])",
"tf.Tensor([2 1 4 3], shape=(4,), dtype=int64)\n['wide', 'the', 'shimmered', 'road']\n"
]
],
[
[
"### Construct one training example",
"_____no_output_____"
],
[
"For a given positive `(target_word, context_word)` skip-gram, you now also have `num_ns` negative sampled context words that do not appear in the window size neighborhood of `target_word`. Batch the `1` positive `context_word` and `num_ns` negative context words into one tensor. This produces a set of positive skip-grams (labelled as `1`) and negative samples (labelled as `0`) for each target word.",
"_____no_output_____"
]
],
[
[
"# Add a dimension so you can use concatenation (on the next step).\nnegative_sampling_candidates = tf.expand_dims(negative_sampling_candidates, 1)\n\n# Concat positive context word with negative sampled words.\ncontext = tf.concat([context_class, negative_sampling_candidates], 0)\n\n# Label first context word as 1 (positive) followed by num_ns 0s (negative).\nlabel = tf.constant([1] + [0]*num_ns, dtype=\"int64\") \n\n# Reshape target to shape (1,) and context and label to (num_ns+1,).\ntarget = tf.squeeze(target_word)\ncontext = tf.squeeze(context)\nlabel = tf.squeeze(label)",
"_____no_output_____"
]
],
[
[
"Take a look at the context and the corresponding labels for the target word from the skip-gram example above. ",
"_____no_output_____"
]
],
[
[
"print(f\"target_index : {target}\")\nprint(f\"target_word : {inverse_vocab[target_word]}\")\nprint(f\"context_indices : {context}\")\nprint(f\"context_words : {[inverse_vocab[c.numpy()] for c in context]}\")\nprint(f\"label : {label}\")",
"target_index : 1\ntarget_word : the\ncontext_indices : [3 2 1 4 3]\ncontext_words : ['road', 'wide', 'the', 'shimmered', 'road']\nlabel : [1 0 0 0 0]\n"
]
],
[
[
"A tuple of `(target, context, label)` tensors constitutes one training example for training your skip-gram negative sampling Word2Vec model. Notice that the target is of shape `(1,)` while the context and label are of shape `(1+num_ns,)`",
"_____no_output_____"
]
],
[
[
"print(f\"target :\", target)\nprint(f\"context :\", context )\nprint(f\"label :\", label )",
"target : tf.Tensor(1, shape=(), dtype=int32)\ncontext : tf.Tensor([3 2 1 4 3], shape=(5,), dtype=int64)\nlabel : tf.Tensor([1 0 0 0 0], shape=(5,), dtype=int64)\n"
]
],
[
[
"### Summary",
"_____no_output_____"
],
[
"This picture summarizes the procedure of generating training example from a sentence. \n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"## Lab Task 1: Compile all steps into one function\n",
"_____no_output_____"
],
[
"### Skip-gram Sampling table ",
"_____no_output_____"
],
[
"A large dataset means larger vocabulary with higher number of more frequent words such as stopwords. Training examples obtained from sampling commonly occuring words (such as `the`, `is`, `on`) don't add much useful information for the model to learn from. [Mikolov et al.](https://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf) suggest subsampling of frequent words as a helpful practice to improve embedding quality. ",
"_____no_output_____"
],
[
"The `tf.keras.preprocessing.sequence.skipgrams` function accepts a sampling table argument to encode probabilities of sampling any token. You can use the `tf.keras.preprocessing.sequence.make_sampling_table` to generate a word-frequency rank based probabilistic sampling table and pass it to `skipgrams` function. Take a look at the sampling probabilities for a `vocab_size` of 10.",
"_____no_output_____"
]
],
[
[
"sampling_table = tf.keras.preprocessing.sequence.make_sampling_table(size=10)\nprint(sampling_table)",
"[0.00315225 0.00315225 0.00547597 0.00741556 0.00912817 0.01068435\n 0.01212381 0.01347162 0.01474487 0.0159558 ]\n"
]
],
[
[
"`sampling_table[i]` denotes the probability of sampling the i-th most common word in a dataset. The function assumes a [Zipf's distribution](https://en.wikipedia.org/wiki/Zipf%27s_law) of the word frequencies for sampling.",
"_____no_output_____"
],
[
"Key point: The `tf.random.log_uniform_candidate_sampler` already assumes that the vocabulary frequency follows a log-uniform (Zipf's) distribution. Using these distribution weighted sampling also helps approximate the Noise Contrastive Estimation (NCE) loss with simpler loss functions for training a negative sampling objective.",
"_____no_output_____"
],
[
"### Generate training data",
"_____no_output_____"
],
[
"Compile all the steps described above into a function that can be called on a list of vectorized sentences obtained from any text dataset. Notice that the sampling table is built before sampling skip-gram word pairs. You will use this function in the later sections.",
"_____no_output_____"
]
],
[
[
"# Generates skip-gram pairs with negative sampling for a list of sequences\n# (int-encoded sentences) based on window size, number of negative samples\n# and vocabulary size.\ndef generate_training_data(sequences, window_size, num_ns, vocab_size, seed):\n # Elements of each training example are appended to these lists.\n targets, contexts, labels = [], [], []\n\n # Build the sampling table for vocab_size tokens.\n # TODO 1a\n sampling_table = tf.keras.preprocessing.sequence.make_sampling_table(vocab_size)\n\n # Iterate over all sequences (sentences) in dataset.\n for sequence in tqdm.tqdm(sequences):\n\n # Generate positive skip-gram pairs for a sequence (sentence).\n positive_skip_grams, _ = tf.keras.preprocessing.sequence.skipgrams(\n sequence, \n vocabulary_size=vocab_size,\n sampling_table=sampling_table,\n window_size=window_size,\n negative_samples=0)\n \n # Iterate over each positive skip-gram pair to produce training examples \n # with positive context word and negative samples.\n # TODO 1b\n for target_word, context_word in positive_skip_grams:\n context_class = tf.expand_dims(\n tf.constant([context_word], dtype=\"int64\"), 1)\n negative_sampling_candidates, _, _ = tf.random.log_uniform_candidate_sampler(\n true_classes=context_class,\n num_true=1, \n num_sampled=num_ns, \n unique=True, \n range_max=vocab_size, \n seed=SEED, \n name=\"negative_sampling\")\n \n # Build context and label vectors (for one target word)\n negative_sampling_candidates = tf.expand_dims(\n negative_sampling_candidates, 1)\n\n context = tf.concat([context_class, negative_sampling_candidates], 0)\n label = tf.constant([1] + [0]*num_ns, dtype=\"int64\")\n\n # Append each element from the training example to global lists.\n targets.append(target_word)\n contexts.append(context)\n labels.append(label)\n\n return targets, contexts, labels",
"_____no_output_____"
]
],
[
[
"## Lab Task 2: Prepare training data for Word2Vec",
"_____no_output_____"
],
[
"With an understanding of how to work with one sentence for a skip-gram negative sampling based Word2Vec model, you can proceed to generate training examples from a larger list of sentences!",
"_____no_output_____"
],
[
"### Download text corpus\n",
"_____no_output_____"
],
[
"You will use a text file of Shakespeare's writing for this tutorial. Change the following line to run this code on your own data.",
"_____no_output_____"
]
],
[
[
"path_to_file = tf.keras.utils.get_file('shakespeare.txt', 'https://storage.googleapis.com/download.tensorflow.org/data/shakespeare.txt')",
"Downloading data from https://storage.googleapis.com/download.tensorflow.org/data/shakespeare.txt\n"
]
],
[
[
"Read text from the file and take a look at the first few lines. ",
"_____no_output_____"
]
],
[
[
"with open(path_to_file) as f: \n lines = f.read().splitlines()\nfor line in lines[:20]:\n print(line)",
"First Citizen:\nBefore we proceed any further, hear me speak.\n\nAll:\nSpeak, speak.\n\nFirst Citizen:\nYou are all resolved rather to die than to famish?\n\nAll:\nResolved. resolved.\n\nFirst Citizen:\nFirst, you know Caius Marcius is chief enemy to the people.\n\nAll:\nWe know't, we know't.\n\nFirst Citizen:\nLet us kill him, and we'll have corn at our own price.\n"
]
],
[
[
"Use the non empty lines to construct a `tf.data.TextLineDataset` object for next steps.",
"_____no_output_____"
]
],
[
[
"# TODO 2a\ntext_ds = tf.data.TextLineDataset(path_to_file).filter(lambda x: tf.cast(tf.strings.length(x), bool))",
"_____no_output_____"
]
],
[
[
"### Vectorize sentences from the corpus",
"_____no_output_____"
],
[
"You can use the `TextVectorization` layer to vectorize sentences from the corpus. Learn more about using this layer in this [Text Classification](https://www.tensorflow.org/tutorials/keras/text_classification) tutorial. Notice from the first few sentences above that the text needs to be in one case and punctuation needs to be removed. To do this, define a `custom_standardization function` that can be used in the TextVectorization layer.",
"_____no_output_____"
]
],
[
[
"# We create a custom standardization function to lowercase the text and \n# remove punctuation.\ndef custom_standardization(input_data):\n lowercase = tf.strings.lower(input_data)\n return tf.strings.regex_replace(lowercase,\n '[%s]' % re.escape(string.punctuation), '')\n\n# Define the vocabulary size and number of words in a sequence.\nvocab_size = 4096\nsequence_length = 10\n\n# Use the text vectorization layer to normalize, split, and map strings to\n# integers. Set output_sequence_length length to pad all samples to same length.\nvectorize_layer = TextVectorization(\n standardize=custom_standardization,\n max_tokens=vocab_size,\n output_mode='int',\n output_sequence_length=sequence_length)",
"_____no_output_____"
]
],
[
[
"Call `adapt` on the text dataset to create vocabulary.\n",
"_____no_output_____"
]
],
[
[
"vectorize_layer.adapt(text_ds.batch(1024))",
"_____no_output_____"
]
],
[
[
"Once the state of the layer has been adapted to represent the text corpus, the vocabulary can be accessed with `get_vocabulary()`. This function returns a list of all vocabulary tokens sorted (descending) by their frequency. ",
"_____no_output_____"
]
],
[
[
"# Save the created vocabulary for reference.\ninverse_vocab = vectorize_layer.get_vocabulary()\nprint(inverse_vocab[:20])",
"['', '[UNK]', 'the', 'and', 'to', 'i', 'of', 'you', 'my', 'a', 'that', 'in', 'is', 'not', 'for', 'with', 'me', 'it', 'be', 'your']\n"
]
],
[
[
"The vectorize_layer can now be used to generate vectors for each element in the `text_ds`.",
"_____no_output_____"
]
],
[
[
"def vectorize_text(text):\n text = tf.expand_dims(text, -1)\n return tf.squeeze(vectorize_layer(text))\n\n# Vectorize the data in text_ds.\ntext_vector_ds = text_ds.batch(1024).prefetch(AUTOTUNE).map(vectorize_layer).unbatch()",
"_____no_output_____"
]
],
[
[
"### Obtain sequences from the dataset",
"_____no_output_____"
],
[
"You now have a `tf.data.Dataset` of integer encoded sentences. To prepare the dataset for training a Word2Vec model, flatten the dataset into a list of sentence vector sequences. This step is required as you would iterate over each sentence in the dataset to produce positive and negative examples. \n\nNote: Since the `generate_training_data()` defined earlier uses non-TF python/numpy functions, you could also use a `tf.py_function` or `tf.numpy_function` with `tf.data.Dataset.map()`.",
"_____no_output_____"
]
],
[
[
"sequences = list(text_vector_ds.as_numpy_iterator())\nprint(len(sequences))",
"32777\n"
]
],
[
[
"Take a look at few examples from `sequences`.\n",
"_____no_output_____"
]
],
[
[
"for seq in sequences[:5]:\n print(f\"{seq} => {[inverse_vocab[i] for i in seq]}\")",
"[ 89 270 0 0 0 0 0 0 0 0] => ['first', 'citizen', '', '', '', '', '', '', '', '']\n[138 36 982 144 673 125 16 106 0 0] => ['before', 'we', 'proceed', 'any', 'further', 'hear', 'me', 'speak', '', '']\n[34 0 0 0 0 0 0 0 0 0] => ['all', '', '', '', '', '', '', '', '', '']\n[106 106 0 0 0 0 0 0 0 0] => ['speak', 'speak', '', '', '', '', '', '', '', '']\n[ 89 270 0 0 0 0 0 0 0 0] => ['first', 'citizen', '', '', '', '', '', '', '', '']\n"
]
],
[
[
"### Generate training examples from sequences",
"_____no_output_____"
],
[
"`sequences` is now a list of int encoded sentences. Just call the `generate_training_data()` function defined earlier to generate training examples for the Word2Vec model. To recap, the function iterates over each word from each sequence to collect positive and negative context words. Length of target, contexts and labels should be same, representing the total number of training examples.",
"_____no_output_____"
]
],
[
[
"targets, contexts, labels = generate_training_data(\n sequences=sequences, \n window_size=2, \n num_ns=4, \n vocab_size=vocab_size, \n seed=SEED)\nprint(len(targets), len(contexts), len(labels))",
"\r 0%| | 0/32777 [00:00<?, ?it/s]"
]
],
[
[
"### Configure the dataset for performance",
"_____no_output_____"
],
[
"To perform efficient batching for the potentially large number of training examples, use the `tf.data.Dataset` API. After this step, you would have a `tf.data.Dataset` object of `(target_word, context_word), (label)` elements to train your Word2Vec model!",
"_____no_output_____"
]
],
[
[
"BATCH_SIZE = 1024\nBUFFER_SIZE = 10000\ndataset = tf.data.Dataset.from_tensor_slices(((targets, contexts), labels))\ndataset = dataset.shuffle(BUFFER_SIZE).batch(BATCH_SIZE, drop_remainder=True)\nprint(dataset)",
"<BatchDataset shapes: (((1024,), (1024, 5, 1)), (1024, 5)), types: ((tf.int32, tf.int64), tf.int64)>\n"
]
],
[
[
"Add `cache()` and `prefetch()` to improve performance.",
"_____no_output_____"
]
],
[
[
"dataset = dataset.cache().prefetch(buffer_size=AUTOTUNE)\nprint(dataset)",
"<PrefetchDataset shapes: (((1024,), (1024, 5, 1)), (1024, 5)), types: ((tf.int32, tf.int64), tf.int64)>\n"
]
],
[
[
"## Lab Task 3: Model and Training",
"_____no_output_____"
],
[
"The Word2Vec model can be implemented as a classifier to distinguish between true context words from skip-grams and false context words obtained through negative sampling. You can perform a dot product between the embeddings of target and context words to obtain predictions for labels and compute loss against true labels in the dataset.",
"_____no_output_____"
],
[
"### Subclassed Word2Vec Model",
"_____no_output_____"
],
[
"Use the [Keras Subclassing API](https://www.tensorflow.org/guide/keras/custom_layers_and_models) to define your Word2Vec model with the following layers:\n\n\n* `target_embedding`: A `tf.keras.layers.Embedding` layer which looks up the embedding of a word when it appears as a target word. The number of parameters in this layer are `(vocab_size * embedding_dim)`.\n* `context_embedding`: Another `tf.keras.layers.Embedding` layer which looks up the embedding of a word when it appears as a context word. The number of parameters in this layer are the same as those in `target_embedding`, i.e. `(vocab_size * embedding_dim)`.\n* `dots`: A `tf.keras.layers.Dot` layer that computes the dot product of target and context embeddings from a training pair.\n* `flatten`: A `tf.keras.layers.Flatten` layer to flatten the results of `dots` layer into logits.\n\nWith the sublassed model, you can define the `call()` function that accepts `(target, context)` pairs which can then be passed into their corresponding embedding layer. Reshape the `context_embedding` to perform a dot product with `target_embedding` and return the flattened result.",
"_____no_output_____"
],
[
"Key point: The `target_embedding` and `context_embedding` layers can be shared as well. You could also use a concatenation of both embeddings as the final Word2Vec embedding.",
"_____no_output_____"
]
],
[
[
"class Word2Vec(Model):\n def __init__(self, vocab_size, embedding_dim):\n super(Word2Vec, self).__init__()\n self.target_embedding = Embedding(vocab_size, \n embedding_dim,\n input_length=1,\n name=\"w2v_embedding\", )\n self.context_embedding = Embedding(vocab_size, \n embedding_dim, \n input_length=num_ns+1)\n self.dots = Dot(axes=(3,2))\n self.flatten = Flatten()\n\n def call(self, pair):\n target, context = pair\n we = self.target_embedding(target)\n ce = self.context_embedding(context)\n dots = self.dots([ce, we])\n return self.flatten(dots)",
"_____no_output_____"
]
],
[
[
"### Define loss function and compile model\n",
"_____no_output_____"
],
[
"For simplicity, you can use `tf.keras.losses.CategoricalCrossEntropy` as an alternative to the negative sampling loss. If you would like to write your own custom loss function, you can also do so as follows:\n\n``` python\ndef custom_loss(x_logit, y_true):\n return tf.nn.sigmoid_cross_entropy_with_logits(logits=x_logit, labels=y_true)\n```\n\nIt's time to build your model! Instantiate your Word2Vec class with an embedding dimension of 128 (you could experiment with different values). Compile the model with the `tf.keras.optimizers.Adam` optimizer. ",
"_____no_output_____"
]
],
[
[
"# TODO 3a\nembedding_dim = 128\nword2vec = Word2Vec(vocab_size, embedding_dim)\nword2vec.compile(optimizer='adam',\n loss=tf.keras.losses.CategoricalCrossentropy(from_logits=True),\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"Also define a callback to log training statistics for tensorboard.",
"_____no_output_____"
]
],
[
[
"tensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir=\"logs\")",
"_____no_output_____"
]
],
[
[
"Train the model with `dataset` prepared above for some number of epochs.",
"_____no_output_____"
]
],
[
[
"word2vec.fit(dataset, epochs=20, callbacks=[tensorboard_callback])",
"Epoch 1/20\n"
]
],
[
[
"Tensorboard now shows the Word2Vec model's accuracy and loss.",
"_____no_output_____"
]
],
[
[
"!tensorboard --bind_all --port=8081 --load_fast=false --logdir logs",
"_____no_output_____"
]
],
[
[
"Run the following command in **Cloud Shell:**\n\n<code>gcloud beta compute ssh --zone <instance-zone> <notebook-instance-name> --project <project-id> -- -L 8081:localhost:8081</code> \n\nMake sure to replace <instance-zone>, <notebook-instance-name> and <project-id>.\n\nIn Cloud Shell, click *Web Preview* > *Change Port* and insert port number *8081*. Click *Change and Preview* to open the TensorBoard.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"**To quit the TensorBoard, click Kernel > Interrupt kernel**.",
"_____no_output_____"
],
[
"## Lab Task 4: Embedding lookup and analysis",
"_____no_output_____"
],
[
"Obtain the weights from the model using `get_layer()` and `get_weights()`. The `get_vocabulary()` function provides the vocabulary to build a metadata file with one token per line. ",
"_____no_output_____"
]
],
[
[
"# TODO 4a\nweights = word2vec.get_layer('w2v_embedding').get_weights()[0]\nvocab = vectorize_layer.get_vocabulary()",
"_____no_output_____"
]
],
[
[
"Create and save the vectors and metadata file. ",
"_____no_output_____"
]
],
[
[
"out_v = io.open('vectors.tsv', 'w', encoding='utf-8')\nout_m = io.open('metadata.tsv', 'w', encoding='utf-8')\n\nfor index, word in enumerate(vocab):\n if index == 0: continue # skip 0, it's padding.\n vec = weights[index] \n out_v.write('\\t'.join([str(x) for x in vec]) + \"\\n\")\n out_m.write(word + \"\\n\")\nout_v.close()\nout_m.close()",
"_____no_output_____"
]
],
[
[
"Download the `vectors.tsv` and `metadata.tsv` to analyze the obtained embeddings in the [Embedding Projector](https://projector.tensorflow.org/).",
"_____no_output_____"
]
],
[
[
"try:\n from google.colab import files\n files.download('vectors.tsv')\n files.download('metadata.tsv')\nexcept Exception as e:\n pass",
"_____no_output_____"
]
],
[
[
"## Next steps\n",
"_____no_output_____"
],
[
"This tutorial has shown you how to implement a skip-gram Word2Vec model with negative sampling from scratch and visualize the obtained word embeddings.\n\n* To learn more about word vectors and their mathematical representations, refer to these [notes](https://web.stanford.edu/class/cs224n/readings/cs224n-2019-notes01-wordvecs1.pdf).\n\n* To learn more about advanced text processing, read the [Transformer model for language understanding](https://www.tensorflow.org/tutorials/text/transformer) tutorial.\n\n* If you’re interested in pre-trained embedding models, you may also be interested in [Exploring the TF-Hub CORD-19 Swivel Embeddings](https://www.tensorflow.org/hub/tutorials/cord_19_embeddings_keras), or the [Multilingual Universal Sentence Encoder](https://www.tensorflow.org/hub/tutorials/cross_lingual_similarity_with_tf_hub_multilingual_universal_encoder)\n\n* You may also like to train the model on a new dataset (there are many available in [TensorFlow Datasets](https://www.tensorflow.org/datasets)).\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d05e78b008b5290f3641ce4ac617dc4e21c624b2 | 8,739 | ipynb | Jupyter Notebook | csharp-101/04-Numbers and Integer Math.ipynb | ScriptBox99/dotnet-csharp-notebooks | 2d943dca4db5d19751ba35efc1b3a90451aa6afd | [
"MIT"
] | 1 | 2022-03-31T02:41:47.000Z | 2022-03-31T02:41:47.000Z | csharp-101/04-Numbers and Integer Math.ipynb | ScriptBox99/dotnet-csharp-notebooks | 2d943dca4db5d19751ba35efc1b3a90451aa6afd | [
"MIT"
] | null | null | null | csharp-101/04-Numbers and Integer Math.ipynb | ScriptBox99/dotnet-csharp-notebooks | 2d943dca4db5d19751ba35efc1b3a90451aa6afd | [
"MIT"
] | null | null | null | 24.478992 | 182 | 0.446275 | [
[
[
"# Numbers and Integer Math\n\nWatch the full [C# 101 video](https://www.youtube.com/watch?v=jEE0pWTq54U&list=PLdo4fOcmZ0oVxKLQCHpiUWun7vlJJvUiN&index=5) for this module.",
"_____no_output_____"
],
[
"## Integer Math\n\nYou have a few `integers` defined below. An `integer` is a positive or negative whole number.\n> Before you run the code, what should c be?",
"_____no_output_____"
],
[
"## Addition",
"_____no_output_____"
]
],
[
[
"int a = 18;\nint b = 6;\nint c = a + b;\nConsole.WriteLine(c);",
"24\r\n"
]
],
[
[
"## Subtraction",
"_____no_output_____"
]
],
[
[
"int c = a - b;\nConsole.WriteLine(c);",
"12\r\n"
]
],
[
[
"## Multiplication",
"_____no_output_____"
]
],
[
[
"int c = a * b;\nConsole.WriteLine(c);",
"108\r\n"
]
],
[
[
"## Division",
"_____no_output_____"
]
],
[
[
"int c = a / b;\nConsole.WriteLine(c);",
"3\r\n"
]
],
[
[
"# Order of operations\n\nC# follows the order of operation when it comes to math. That is, it does multiplication and division first, then addition and subtraction.\n> What would the math be if C# didn't follow the order of operation, and instead just did math left to right?",
"_____no_output_____"
]
],
[
[
"int a = 5;\nint b = 4;\nint c = 2;\nint d = a + b * c;\nConsole.WriteLine(d);",
"13\r\n"
]
],
[
[
"## Using parenthesis\n\nYou can also force different orders by putting parentheses around whatever you want done first\n> Try it out",
"_____no_output_____"
]
],
[
[
"int d = (a + b) * c;\nConsole.WriteLine(d);",
"18\r\n"
]
],
[
[
"You can make math as long and complicated as you want.\n> Can you make this line even more complicated?",
"_____no_output_____"
]
],
[
[
"int d = (a + b) - 6 * c + (12 * 4) / 3 + 12;\nConsole.WriteLine(d);",
"25\r\n"
]
],
[
[
"## Integers: Whole numbers no matter what\n\nInteger math will always produce integers. What that means is that even when math should result in a decimal or fraction, the answer will be truncated to a whole number.\n> Check it out. WHat should the answer truly be?",
"_____no_output_____"
]
],
[
[
"int a = 7;\nint b = 4;\nint c = 3;\nint d = (a + b) / c;\nConsole.WriteLine(d);",
"3\r\n"
]
],
[
[
"# Playground\n\nPlay around with what you've learned! Here's some starting ideas:\n> Do you have any homework or projects that need math? Try using code in place of a calculator!\n>\n> How do integers round? Do they always round up? down? to the nearest integer?\n>\n> How do the Order of Operations work? Play around with parentheses.",
"_____no_output_____"
]
],
[
[
"Console.WriteLine(\"Playground\");",
"Playground\r\n"
]
],
[
[
"# Continue learning\n\nThere are plenty more resources out there to learn!\n> [⏩ Next Module - Numbers and Integer Precision](http://tinyurl.com/csharp-notebook05)\n>\n> [⏪ Last Module - Searching Strings](http://tinyurl.com/csharp-notebook03)\n>\n> [Watch the video](https://www.youtube.com/watch?v=jEE0pWTq54U&list=PLdo4fOcmZ0oVxKLQCHpiUWun7vlJJvUiN&index=5)\n>\n> [Documentation: Numbers in C#](https://docs.microsoft.com/dotnet/csharp/tour-of-csharp/tutorials/numbers-in-csharp?WT.mc_id=Educationalcsharp-c9-scottha)\n>\n> [Start at the beginning: What is C#?](https://www.youtube.com/watch?v=BM4CHBmAPh4&list=PLdo4fOcmZ0oVxKLQCHpiUWun7vlJJvUiN&index=1)",
"_____no_output_____"
],
[
"# Other resources\n\nHere's some more places to explore:\n> [Other 101 Videos](https://dotnet.microsoft.com/learn/videos?WT.mc_id=csharpnotebook-35129-website)\n>\n> [Microsoft Learn](https://docs.microsoft.com/learn/dotnet/?WT.mc_id=csharpnotebook-35129-website)\n>\n> [C# Documentation](https://docs.microsoft.com/dotnet/csharp/?WT.mc_id=csharpnotebook-35129-website)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d05e800fef0653bf1467051e2fbb3a64371ae795 | 31,079 | ipynb | Jupyter Notebook | 5-Clustering/1-Visualize/solution/R/lesson_14-R.ipynb | LyhourChhen/ML-For-Beginners | 1cc73ef3438bb6c6f84096c7b40597695a7de79b | [
"MIT"
] | 2 | 2021-09-14T08:38:22.000Z | 2021-12-17T04:18:27.000Z | 5-Clustering/1-Visualize/solution/R/lesson_14-R.ipynb | duduyu11/ML-For-Beginners | fe357399521b7bbf7fbb7f15d70be5bbd68f4b05 | [
"MIT"
] | null | null | null | 5-Clustering/1-Visualize/solution/R/lesson_14-R.ipynb | duduyu11/ML-For-Beginners | fe357399521b7bbf7fbb7f15d70be5bbd68f4b05 | [
"MIT"
] | 1 | 2021-11-30T16:55:17.000Z | 2021-11-30T16:55:17.000Z | 63.556237 | 798 | 0.552817 | [
[
[
"## **Nigerian Music scraped from Spotify - an analysis**\r\n\r\nClustering is a type of [Unsupervised Learning](https://wikipedia.org/wiki/Unsupervised_learning) that presumes that a dataset is unlabelled or that its inputs are not matched with predefined outputs. It uses various algorithms to sort through unlabeled data and provide groupings according to patterns it discerns in the data.\r\n\r\n[**Pre-lecture quiz**](https://white-water-09ec41f0f.azurestaticapps.net/quiz/27/)\r\n\r\n### **Introduction**\r\n\r\n[Clustering](https://link.springer.com/referenceworkentry/10.1007%2F978-0-387-30164-8_124) is very useful for data exploration. Let's see if it can help discover trends and patterns in the way Nigerian audiences consume music.\r\n\r\n> ✅ Take a minute to think about the uses of clustering. In real life, clustering happens whenever you have a pile of laundry and need to sort out your family members' clothes 🧦👕👖🩲. In data science, clustering happens when trying to analyze a user's preferences, or determine the characteristics of any unlabeled dataset. Clustering, in a way, helps make sense of chaos, like a sock drawer.\r\n\r\nIn a professional setting, clustering can be used to determine things like market segmentation, determining what age groups buy what items, for example. Another use would be anomaly detection, perhaps to detect fraud from a dataset of credit card transactions. Or you might use clustering to determine tumors in a batch of medical scans.\r\n\r\n✅ Think a minute about how you might have encountered clustering 'in the wild', in a banking, e-commerce, or business setting.\r\n\r\n> 🎓 Interestingly, cluster analysis originated in the fields of Anthropology and Psychology in the 1930s. Can you imagine how it might have been used?\r\n\r\nAlternately, you could use it for grouping search results - by shopping links, images, or reviews, for example. Clustering is useful when you have a large dataset that you want to reduce and on which you want to perform more granular analysis, so the technique can be used to learn about data before other models are constructed.\r\n\r\n✅ Once your data is organized in clusters, you assign it a cluster Id, and this technique can be useful when preserving a dataset's privacy; you can instead refer to a data point by its cluster id, rather than by more revealing identifiable data. Can you think of other reasons why you'd refer to a cluster Id rather than other elements of the cluster to identify it?\r\n\r\n### Getting started with clustering\r\n\r\n> 🎓 How we create clusters has a lot to do with how we gather up the data points into groups. Let's unpack some vocabulary:\r\n>\r\n> 🎓 ['Transductive' vs. 'inductive'](https://wikipedia.org/wiki/Transduction_(machine_learning))\r\n>\r\n> Transductive inference is derived from observed training cases that map to specific test cases. Inductive inference is derived from training cases that map to general rules which are only then applied to test cases.\r\n>\r\n> An example: Imagine you have a dataset that is only partially labelled. Some things are 'records', some 'cds', and some are blank. Your job is to provide labels for the blanks. If you choose an inductive approach, you'd train a model looking for 'records' and 'cds', and apply those labels to your unlabeled data. This approach will have trouble classifying things that are actually 'cassettes'. A transductive approach, on the other hand, handles this unknown data more effectively as it works to group similar items together and then applies a label to a group. In this case, clusters might reflect 'round musical things' and 'square musical things'.\r\n>\r\n> 🎓 ['Non-flat' vs. 'flat' geometry](https://datascience.stackexchange.com/questions/52260/terminology-flat-geometry-in-the-context-of-clustering)\r\n>\r\n> Derived from mathematical terminology, non-flat vs. flat geometry refers to the measure of distances between points by either 'flat' ([Euclidean](https://wikipedia.org/wiki/Euclidean_geometry)) or 'non-flat' (non-Euclidean) geometrical methods.\r\n>\r\n> 'Flat' in this context refers to Euclidean geometry (parts of which are taught as 'plane' geometry), and non-flat refers to non-Euclidean geometry. What does geometry have to do with machine learning? Well, as two fields that are rooted in mathematics, there must be a common way to measure distances between points in clusters, and that can be done in a 'flat' or 'non-flat' way, depending on the nature of the data. [Euclidean distances](https://wikipedia.org/wiki/Euclidean_distance) are measured as the length of a line segment between two points. [Non-Euclidean distances](https://wikipedia.org/wiki/Non-Euclidean_geometry) are measured along a curve. If your data, visualized, seems to not exist on a plane, you might need to use a specialized algorithm to handle it.\r\n\r\n<p >\r\n <img src=\"../../images/flat-nonflat.png\"\r\n width=\"600\"/>\r\n <figcaption>Infographic by Dasani Madipalli</figcaption>\r\n\r\n\r\n\r\n> 🎓 ['Distances'](https://web.stanford.edu/class/cs345a/slides/12-clustering.pdf)\r\n>\r\n> Clusters are defined by their distance matrix, e.g. the distances between points. This distance can be measured a few ways. Euclidean clusters are defined by the average of the point values, and contain a 'centroid' or center point. Distances are thus measured by the distance to that centroid. Non-Euclidean distances refer to 'clustroids', the point closest to other points. Clustroids in turn can be defined in various ways.\r\n>\r\n> 🎓 ['Constrained'](https://wikipedia.org/wiki/Constrained_clustering)\r\n>\r\n> [Constrained Clustering](https://web.cs.ucdavis.edu/~davidson/Publications/ICDMTutorial.pdf) introduces 'semi-supervised' learning into this unsupervised method. The relationships between points are flagged as 'cannot link' or 'must-link' so some rules are forced on the dataset.\r\n>\r\n> An example: If an algorithm is set free on a batch of unlabelled or semi-labelled data, the clusters it produces may be of poor quality. In the example above, the clusters might group 'round music things' and 'square music things' and 'triangular things' and 'cookies'. If given some constraints, or rules to follow (\"the item must be made of plastic\", \"the item needs to be able to produce music\") this can help 'constrain' the algorithm to make better choices.\r\n>\r\n> 🎓 'Density'\r\n>\r\n> Data that is 'noisy' is considered to be 'dense'. The distances between points in each of its clusters may prove, on examination, to be more or less dense, or 'crowded' and thus this data needs to be analyzed with the appropriate clustering method. [This article](https://www.kdnuggets.com/2020/02/understanding-density-based-clustering.html) demonstrates the difference between using K-Means clustering vs. HDBSCAN algorithms to explore a noisy dataset with uneven cluster density.\r\n\r\nDeepen your understanding of clustering techniques in this [Learn module](https://docs.microsoft.com/learn/modules/train-evaluate-cluster-models?WT.mc_id=academic-15963-cxa)\r\n\r\n### **Clustering algorithms**\r\n\r\nThere are over 100 clustering algorithms, and their use depends on the nature of the data at hand. Let's discuss some of the major ones:\r\n\r\n- **Hierarchical clustering**. If an object is classified by its proximity to a nearby object, rather than to one farther away, clusters are formed based on their members' distance to and from other objects. Hierarchical clustering is characterized by repeatedly combining two clusters.\r\n\r\n\r\n<p >\r\n <img src=\"../../images/hierarchical.png\"\r\n width=\"600\"/>\r\n <figcaption>Infographic by Dasani Madipalli</figcaption>\r\n\r\n\r\n\r\n- **Centroid clustering**. This popular algorithm requires the choice of 'k', or the number of clusters to form, after which the algorithm determines the center point of a cluster and gathers data around that point. [K-means clustering](https://wikipedia.org/wiki/K-means_clustering) is a popular version of centroid clustering which separates a data set into pre-defined K groups. The center is determined by the nearest mean, thus the name. The squared distance from the cluster is minimized.\r\n\r\n<p >\r\n <img src=\"../../images/centroid.png\"\r\n width=\"600\"/>\r\n <figcaption>Infographic by Dasani Madipalli</figcaption>\r\n\r\n\r\n\r\n- **Distribution-based clustering**. Based in statistical modeling, distribution-based clustering centers on determining the probability that a data point belongs to a cluster, and assigning it accordingly. Gaussian mixture methods belong to this type.\r\n\r\n- **Density-based clustering**. Data points are assigned to clusters based on their density, or their grouping around each other. Data points far from the group are considered outliers or noise. DBSCAN, Mean-shift and OPTICS belong to this type of clustering.\r\n\r\n- **Grid-based clustering**. For multi-dimensional datasets, a grid is created and the data is divided amongst the grid's cells, thereby creating clusters.\r\n\r\nThe best way to learn about clustering is to try it for yourself, so that's what you'll do in this exercise.\r\n\r\nWe'll require some packages to knock-off this module. You can have them installed as: `install.packages(c('tidyverse', 'tidymodels', 'DataExplorer', 'summarytools', 'plotly', 'paletteer', 'corrplot', 'patchwork'))`\r\n\r\nAlternatively, the script below checks whether you have the packages required to complete this module and installs them for you in case some are missing.\r\n",
"_____no_output_____"
]
],
[
[
"suppressWarnings(if(!require(\"pacman\")) install.packages(\"pacman\"))\r\n\r\npacman::p_load('tidyverse', 'tidymodels', 'DataExplorer', 'summarytools', 'plotly', 'paletteer', 'corrplot', 'patchwork')\r\n",
"_____no_output_____"
]
],
[
[
"## Exercise - cluster your data\n\nClustering as a technique is greatly aided by proper visualization, so let's get started by visualizing our music data. This exercise will help us decide which of the methods of clustering we should most effectively use for the nature of this data.\n\nLet's hit the ground running by importing the data.\n",
"_____no_output_____"
]
],
[
[
"# Load the core tidyverse and make it available in your current R session\r\nlibrary(tidyverse)\r\n\r\n# Import the data into a tibble\r\ndf <- read_csv(file = \"https://raw.githubusercontent.com/microsoft/ML-For-Beginners/main/5-Clustering/data/nigerian-songs.csv\")\r\n\r\n# View the first 5 rows of the data set\r\ndf %>% \r\n slice_head(n = 5)\r\n",
"_____no_output_____"
]
],
[
[
"Sometimes, we may want some little more information on our data. We can have a look at the `data` and `its structure` by using the [*glimpse()*](https://pillar.r-lib.org/reference/glimpse.html) function:\n\n",
"_____no_output_____"
]
],
[
[
"# Glimpse into the data set\r\ndf %>% \r\n glimpse()\r\n",
"_____no_output_____"
]
],
[
[
"Good job!💪\n\nWe can observe that `glimpse()` will give you the total number of rows (observations) and columns (variables), then, the first few entries of each variable in a row after the variable name. In addition, the *data type* of the variable is given immediately after each variable's name inside `< >`.\n\n`DataExplorer::introduce()` can summarize this information neatly:\n",
"_____no_output_____"
]
],
[
[
"# Describe basic information for our data\r\ndf %>% \r\n introduce()\r\n\r\n# A visual display of the same\r\ndf %>% \r\n plot_intro()\r\n",
"_____no_output_____"
]
],
[
[
"Awesome! We have just learnt that our data has no missing values.\n\nWhile we are at it, we can explore common central tendency statistics (e.g [mean](https://en.wikipedia.org/wiki/Arithmetic_mean) and [median](https://en.wikipedia.org/wiki/Median)) and measures of dispersion (e.g [standard deviation](https://en.wikipedia.org/wiki/Standard_deviation)) using `summarytools::descr()`\n",
"_____no_output_____"
]
],
[
[
"# Describe common statistics\r\ndf %>% \r\n descr(stats = \"common\")\r\n",
"_____no_output_____"
]
],
[
[
"Let's look at the general values of the data. Note that popularity can be `0`, which show songs that have no ranking. We'll remove those shortly.\n\n> 🤔 If we are working with clustering, an unsupervised method that does not require labeled data, why are we showing this data with labels? In the data exploration phase, they come in handy, but they are not necessary for the clustering algorithms to work.\n\n### 1. Explore popular genres\n\nLet's go ahead and find out the most popular genres 🎶 by making a count of the instances it appears.\n",
"_____no_output_____"
]
],
[
[
"# Popular genres\r\ntop_genres <- df %>% \r\n count(artist_top_genre, sort = TRUE) %>% \r\n# Encode to categorical and reorder the according to count\r\n mutate(artist_top_genre = factor(artist_top_genre) %>% fct_inorder())\r\n\r\n# Print the top genres\r\ntop_genres\r\n",
"_____no_output_____"
]
],
[
[
"That went well! They say a picture is worth a thousand rows of a data frame (actually nobody ever says that 😅). But you get the gist of it, right?\n\nOne way to visualize categorical data (character or factor variables) is using barplots. Let's make a barplot of the top 10 genres:\n",
"_____no_output_____"
]
],
[
[
"# Change the default gray theme\r\ntheme_set(theme_light())\r\n\r\n# Visualize popular genres\r\ntop_genres %>%\r\n slice(1:10) %>% \r\n ggplot(mapping = aes(x = artist_top_genre, y = n,\r\n fill = artist_top_genre)) +\r\n geom_col(alpha = 0.8) +\r\n paletteer::scale_fill_paletteer_d(\"rcartocolor::Vivid\") +\r\n ggtitle(\"Top genres\") +\r\n theme(plot.title = element_text(hjust = 0.5),\r\n # Rotates the X markers (so we can read them)\r\n axis.text.x = element_text(angle = 90))\r\n",
"_____no_output_____"
]
],
[
[
"Now it's way easier to identify that we have `missing` genres 🧐!\n\n> A good visualisation will show you things that you did not expect, or raise new questions about the data - Hadley Wickham and Garrett Grolemund, [R For Data Science](https://r4ds.had.co.nz/introduction.html)\n\nNote, when the top genre is described as `Missing`, that means that Spotify did not classify it, so let's get rid of it.\n",
"_____no_output_____"
]
],
[
[
"# Visualize popular genres\r\ntop_genres %>%\r\n filter(artist_top_genre != \"Missing\") %>% \r\n slice(1:10) %>% \r\n ggplot(mapping = aes(x = artist_top_genre, y = n,\r\n fill = artist_top_genre)) +\r\n geom_col(alpha = 0.8) +\r\n paletteer::scale_fill_paletteer_d(\"rcartocolor::Vivid\") +\r\n ggtitle(\"Top genres\") +\r\n theme(plot.title = element_text(hjust = 0.5),\r\n # Rotates the X markers (so we can read them)\r\n axis.text.x = element_text(angle = 90))\r\n",
"_____no_output_____"
]
],
[
[
"From the little data exploration, we learn that the top three genres dominate this dataset. Let's concentrate on `afro dancehall`, `afropop`, and `nigerian pop`, additionally filter the dataset to remove anything with a 0 popularity value (meaning it was not classified with a popularity in the dataset and can be considered noise for our purposes):\n\n",
"_____no_output_____"
]
],
[
[
"nigerian_songs <- df %>% \r\n # Concentrate on top 3 genres\r\n filter(artist_top_genre %in% c(\"afro dancehall\", \"afropop\",\"nigerian pop\")) %>% \r\n # Remove unclassified observations\r\n filter(popularity != 0)\r\n\r\n\r\n\r\n# Visualize popular genres\r\nnigerian_songs %>%\r\n count(artist_top_genre) %>%\r\n ggplot(mapping = aes(x = artist_top_genre, y = n,\r\n fill = artist_top_genre)) +\r\n geom_col(alpha = 0.8) +\r\n paletteer::scale_fill_paletteer_d(\"ggsci::category10_d3\") +\r\n ggtitle(\"Top genres\") +\r\n theme(plot.title = element_text(hjust = 0.5))\r\n",
"_____no_output_____"
]
],
[
[
"Let's see whether there is any apparent linear relationship among the numerical variables in our data set. This relationship is quantified mathematically by the [correlation statistic](https://en.wikipedia.org/wiki/Correlation).\n\nThe correlation statistic is a value between -1 and 1 that indicates the strength of a relationship. Values above 0 indicate a *positive* correlation (high values of one variable tend to coincide with high values of the other), while values below 0 indicate a *negative* correlation (high values of one variable tend to coincide with low values of the other).\n",
"_____no_output_____"
]
],
[
[
"# Narrow down to numeric variables and fid correlation\r\ncorr_mat <- nigerian_songs %>% \r\n select(where(is.numeric)) %>% \r\n cor()\r\n\r\n# Visualize correlation matrix\r\ncorrplot(corr_mat, order = 'AOE', col = c('white', 'black'), bg = 'gold2') \r\n",
"_____no_output_____"
]
],
[
[
"The data is not strongly correlated except between `energy` and `loudness`, which makes sense, given that loud music is usually pretty energetic. `Popularity` has a correspondence to `release date`, which also makes sense, as more recent songs are probably more popular. Length and energy seem to have a correlation too.\n\nIt will be interesting to see what a clustering algorithm can make of this data!\n\n> 🎓 Note that correlation does not imply causation! We have proof of correlation but no proof of causation. An [amusing web site](https://tylervigen.com/spurious-correlations) has some visuals that emphasize this point.\n\n### 2. Explore data distribution\n\nLet's ask some more subtle questions. Are the genres significantly different in the perception of their danceability, based on their popularity? Let's examine our top three genres data distribution for popularity and danceability along a given x and y axis using [density plots](https://www.khanacademy.org/math/ap-statistics/density-curves-normal-distribution-ap/density-curves/v/density-curves).\n",
"_____no_output_____"
]
],
[
[
"# Perform 2D kernel density estimation\r\ndensity_estimate_2d <- nigerian_songs %>% \r\n ggplot(mapping = aes(x = popularity, y = danceability, color = artist_top_genre)) +\r\n geom_density_2d(bins = 5, size = 1) +\r\n paletteer::scale_color_paletteer_d(\"RSkittleBrewer::wildberry\") +\r\n xlim(-20, 80) +\r\n ylim(0, 1.2)\r\n\r\n# Density plot based on the popularity\r\ndensity_estimate_pop <- nigerian_songs %>% \r\n ggplot(mapping = aes(x = popularity, fill = artist_top_genre, color = artist_top_genre)) +\r\n geom_density(size = 1, alpha = 0.5) +\r\n paletteer::scale_fill_paletteer_d(\"RSkittleBrewer::wildberry\") +\r\n paletteer::scale_color_paletteer_d(\"RSkittleBrewer::wildberry\") +\r\n theme(legend.position = \"none\")\r\n\r\n# Density plot based on the danceability\r\ndensity_estimate_dance <- nigerian_songs %>% \r\n ggplot(mapping = aes(x = danceability, fill = artist_top_genre, color = artist_top_genre)) +\r\n geom_density(size = 1, alpha = 0.5) +\r\n paletteer::scale_fill_paletteer_d(\"RSkittleBrewer::wildberry\") +\r\n paletteer::scale_color_paletteer_d(\"RSkittleBrewer::wildberry\")\r\n\r\n\r\n# Patch everything together\r\nlibrary(patchwork)\r\ndensity_estimate_2d / (density_estimate_pop + density_estimate_dance)\r\n",
"_____no_output_____"
]
],
[
[
"We see that there are concentric circles that line up, regardless of genre. Could it be that Nigerian tastes converge at a certain level of danceability for this genre?\n\nIn general, the three genres align in terms of their popularity and danceability. Determining clusters in this loosely-aligned data will be a challenge. Let's see whether a scatter plot can support this.\n",
"_____no_output_____"
]
],
[
[
"# A scatter plot of popularity and danceability\r\nscatter_plot <- nigerian_songs %>% \r\n ggplot(mapping = aes(x = popularity, y = danceability, color = artist_top_genre, shape = artist_top_genre)) +\r\n geom_point(size = 2, alpha = 0.8) +\r\n paletteer::scale_color_paletteer_d(\"futurevisions::mars\")\r\n\r\n# Add a touch of interactivity\r\nggplotly(scatter_plot)\r\n",
"_____no_output_____"
]
],
[
[
"A scatterplot of the same axes shows a similar pattern of convergence.\n\nIn general, for clustering, you can use scatterplots to show clusters of data, so mastering this type of visualization is very useful. In the next lesson, we will take this filtered data and use k-means clustering to discover groups in this data that see to overlap in interesting ways.\n\n## **🚀 Challenge**\n\nIn preparation for the next lesson, make a chart about the various clustering algorithms you might discover and use in a production environment. What kinds of problems is the clustering trying to address?\n\n## [**Post-lecture quiz**](https://white-water-09ec41f0f.azurestaticapps.net/quiz/28/)\n\n## **Review & Self Study**\n\nBefore you apply clustering algorithms, as we have learned, it's a good idea to understand the nature of your dataset. Read more on this topic [here](https://www.kdnuggets.com/2019/10/right-clustering-algorithm.html)\n\nDeepen your understanding of clustering techniques:\n\n- [Train and Evaluate Clustering Models using Tidymodels and friends](https://rpubs.com/eR_ic/clustering)\n\n- Bradley Boehmke & Brandon Greenwell, [*Hands-On Machine Learning with R*](https://bradleyboehmke.github.io/HOML/)*.*\n\n## **Assignment**\n\n[Research other visualizations for clustering](https://github.com/microsoft/ML-For-Beginners/blob/main/5-Clustering/1-Visualize/assignment.md)\n\n## THANK YOU TO:\n\n[Jen Looper](https://www.twitter.com/jenlooper) for creating the original Python version of this module ♥️\n\n[`Dasani Madipalli`](https://twitter.com/dasani_decoded) for creating the amazing illustrations that make machine learning concepts more interpretable and easier to understand.\n\nHappy Learning,\n\n[Eric](https://twitter.com/ericntay), Gold Microsoft Learn Student Ambassador.\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d05e8e648aec8d62bfe41e2bf53eb5da96d3e176 | 18,547 | ipynb | Jupyter Notebook | camilo_torres_botero/.ipynb_checkpoints/Ejercicios 1.3 Random Networks Vs. Real Networks-checkpoint.ipynb | spulido99/NetworksAnalysis | 3f6f585305f5825e25488bae8c6b427bc18436c6 | [
"MIT"
] | null | null | null | camilo_torres_botero/.ipynb_checkpoints/Ejercicios 1.3 Random Networks Vs. Real Networks-checkpoint.ipynb | spulido99/NetworksAnalysis | 3f6f585305f5825e25488bae8c6b427bc18436c6 | [
"MIT"
] | null | null | null | camilo_torres_botero/.ipynb_checkpoints/Ejercicios 1.3 Random Networks Vs. Real Networks-checkpoint.ipynb | spulido99/NetworksAnalysis | 3f6f585305f5825e25488bae8c6b427bc18436c6 | [
"MIT"
] | null | null | null | 110.39881 | 14,670 | 0.866879 | [
[
[
"# Ejercicios Random Networks vs Real Networks",
"_____no_output_____"
],
[
"## Ejercicios Diferencia en Distribución de Grados\n\nCompare la distribución de grados de una red real contra una red aleatoria.\n\n- Baje un red real de SNAP\n- Cree una red aleatoria con el mismo número de links y nodos\n- Compare la distribución de grados",
"_____no_output_____"
]
],
[
[
"import networkx as nx\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline\n\nedges = []\nfor line in open('CA-HepTh.txt'):\n if line[0] != '#':\n edge = line.replace('\\n','').split('\\t')\n edges.append((edge[0],edge[1]))\n \nG=nx.Graph()\nG.add_edges_from(edges)\n\nd = G.degree()\n#degrees = [degree for _, d.items()]\n#print(d)\n\nN = len(G.nodes())\np = (2*len(edges))/(N*(N-1))\nG_rand = nx.gnp_random_graph(N,p)\n\nsns.distplot(list(G.degree().values()))\nsns.distplot(list(G_rand.degree().values()))",
"C:\\Users\\Camil\\Anaconda3\\lib\\site-packages\\statsmodels\\nonparametric\\kdetools.py:20: VisibleDeprecationWarning: using a non-integer number instead of an integer will result in an error in the future\n y = X[:m/2+1] + np.r_[0,X[m/2+1:],0]*1j\n"
]
],
[
[
"## Ejercicios Comparación Tamaño del componente Gigante\n\nGenere varias realizaciones de la red aleatoria y compare el tamaño del componente gigante contra el de la red real",
"_____no_output_____"
],
[
"## Ejercicio Comparación Número de componentes\n\nGenera varias realizaciones de la red aleatoria y compare la cantidad de componentes",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d05e93b81313b48fbcd32f56da5aa286f6f63cbe | 40,532 | ipynb | Jupyter Notebook | tutorials/optimization/5_admm_optimizer.ipynb | prasad-kumkar/qiskit-tutorials | 3b87a1fc0a9cc7c558cbae3ea90a9e5b005eeb72 | [
"Apache-2.0"
] | null | null | null | tutorials/optimization/5_admm_optimizer.ipynb | prasad-kumkar/qiskit-tutorials | 3b87a1fc0a9cc7c558cbae3ea90a9e5b005eeb72 | [
"Apache-2.0"
] | null | null | null | tutorials/optimization/5_admm_optimizer.ipynb | prasad-kumkar/qiskit-tutorials | 3b87a1fc0a9cc7c558cbae3ea90a9e5b005eeb72 | [
"Apache-2.0"
] | 1 | 2021-02-20T04:34:25.000Z | 2021-02-20T04:34:25.000Z | 77.057034 | 10,952 | 0.798505 | [
[
[
"# ADMM Optimizer",
"_____no_output_____"
],
[
"## Introduction",
"_____no_output_____"
],
[
"The ADMM Optimizer can solve classes of mixed-binary constrained optimization problems, hereafter (MBCO), which often appear in logistic, finance, and operation research. In particular, the ADMM Optimizer here designed can tackle the following optimization problem $(P)$:\n\n$$\n\\min_{x \\in \\mathcal{X},u\\in\\mathcal{U} \\subseteq \\mathbb{R}^l } \\quad q(x) + \\varphi(u),\n$$\n\nsubject to the constraints:\n\n$$\n\\mathrm{s.t.:~} \\quad G x = b, \\quad g(x) \\leq 0, \\quad \\ell(x, u) \\leq 0, \n$$\n\nwith the corresponding functional assumptions.\n\n1. Function $q: \\mathbb{R}^n \\to \\mathbb{R}$ is quadratic, i.e., $q(x) = x^{\\intercal} Q x + a^{\\intercal} x$ for a given symmetric squared matrix $Q \\in \\mathbb{R}^n \\times \\mathbb{R}^n, Q = Q^{\\intercal}$, and vector $a \\in \\mathbb{R}^n$;\n2. The set $\\mathcal{X} = \\{0,1\\}^n = \\{x_{(i)} (1-x_{(i)}) = 0, \\forall i\\}$ enforces the binary constraints;\n3. Matrix $G\\in\\mathbb{R}^n \\times \\mathbb{R}^{n'}$, vector $b \\in \\mathbb{R}^{n'}$, and function $g: \\mathbb{R}^n \\to \\mathbb{R}$ is convex;\n4. Function $\\varphi: \\mathbb{R}^l \\to \\mathbb{R}$ is convex and $\\mathcal{U}$ is a convex set;\n5. Function $\\ell: \\mathbb{R}^n\\times \\mathbb{R}^l \\to \\mathbb{R}$ is *jointly* convex in $x, u$.",
"_____no_output_____"
],
[
"In order to solve MBO problems, [1] proposed heuristics for $(P)$ based on the Alternating Direction Method of Multipliers (ADMM) [2]. ADMM is an operator splitting algorithm with a long history in convex optimization, and it is known to have residual, objective and dual variable convergence properties, provided that convexity assumptions are holding.\n\nThe method of [1] (referred to as 3-ADMM-H) leverages the ADMM operator-splitting procedure to devise a decomposition for certain classes of MBOs into:\n- a QUBO subproblem to be solved by on the quantum device via variational algorithms, such as VQE or QAOA;\n- continuous convex constrained subproblem, which can be efficiently solved with classical optimization solvers.\n\nThe algorithm 3-ADMM-H works as follows:\n\n0. Initialization phase (set the parameters and the QUBO and convex solvers);\n1. For each ADMM iterations ($k = 1, 2, \\ldots, $) untill termination:\n - Solve a properly defined QUBO subproblem (with a classical or quantum solver);\n - Solve properly defined convex problems (with a classical solver);\n - Update the dual variables.\n2. Return optimizers and cost.\n\n \nA comprehensive discussion on the conditions for convergence, feasibility and optimality of the algorithm can be found in [1]. A variant with 2 ADMM blocks, namely a QUBO subproblem, and a continuous convex constrained subproblem, is also introduced in [1].\n\n## References\n\n[1] [C. Gambella and A. Simonetto, *Multi-block ADMM heuristics for mixed-binary optimization, on classical and quantum computers,* arXiv preprint arXiv:2001.02069 (2020).](https://arxiv.org/abs/2001.02069)\n\n[2] [S. Boyd, N. Parikh, E. Chu, B. Peleato, and J. Eckstein, *Distributed optimization and statistical learning via the alternating direction method of multipliers,* Foundations and Trends in Machine learning, 3, 1–122 (2011).](https://web.stanford.edu/~boyd/papers/pdf/admm_distr_stats.pdf)",
"_____no_output_____"
],
[
"## Initialization\nFirst of all we load all the packages that we need.",
"_____no_output_____"
]
],
[
[
"import time\nfrom typing import List, Optional, Any\nimport numpy as np\nimport matplotlib.pyplot as plt \n\nfrom docplex.mp.model import Model\n\nfrom qiskit import BasicAer\nfrom qiskit.aqua.algorithms import QAOA, NumPyMinimumEigensolver\nfrom qiskit.optimization.algorithms import CobylaOptimizer, MinimumEigenOptimizer\nfrom qiskit.optimization.problems import QuadraticProgram\nfrom qiskit.optimization.algorithms.admm_optimizer import ADMMParameters, ADMMOptimizer\n\n# If CPLEX is installed, you can uncomment this line to import the CplexOptimizer.\n# CPLEX can be used in this tutorial to solve the convex continuous problem, \n# but also as a reference to solve the QUBO, or even the full problem.\n# \n# from qiskit.optimization.algorithms import CplexOptimizer",
"_____no_output_____"
]
],
[
[
"We first initialize all the algorithms we plan to use later in this tutorial.\n\nTo solve the QUBO problems we can choose between \n- `MinimumEigenOptimizer` using different `MinimumEigensolver`, such as `VQE`, `QAOA` or `NumpyMinimumEigensolver` (classical)\n- `GroverOptimizer`\n- `CplexOptimizer` (classical, if CPLEX is installed)\n\nand to solve the convex continuous problems we can choose between the following classical solvers:\n- `CplexOptimizer` (if CPLEX is installed)\n- `CobylaOptimizer`\n\nIn case CPLEX is not available, the `CobylaOptimizer` (for convex continuous problems) and the `MinimumEigenOptimizer` using the `NumpyMinimumEigensolver` (for QUBOs) can be used as classical alternatives to CPLEX for testing, validation, and benchmarking.",
"_____no_output_____"
]
],
[
[
"# define COBYLA optimizer to handle convex continuous problems.\ncobyla = CobylaOptimizer()\n\n# define QAOA via the minimum eigen optimizer\nqaoa = MinimumEigenOptimizer(QAOA(quantum_instance=BasicAer.get_backend('statevector_simulator')))\n\n# exact QUBO solver as classical benchmark\nexact = MinimumEigenOptimizer(NumPyMinimumEigensolver()) # to solve QUBOs\n\n# in case CPLEX is installed it can also be used for the convex problems, the QUBO, \n# or as a benchmark for the full problem.\n#\n# cplex = CplexOptimizer()",
"_____no_output_____"
]
],
[
[
"## Example\n\nWe test 3-ADMM-H algorithm on a simple Mixed-Binary Quadratic Problem with equality and inequality constraints (Example 6 reported in [1]). We first construct a docplex problem and then load it into a `QuadraticProgram`.",
"_____no_output_____"
]
],
[
[
"# construct model using docplex\nmdl = Model('ex6')\n\nv = mdl.binary_var(name='v')\nw = mdl.binary_var(name='w')\nt = mdl.binary_var(name='t')\nu = mdl.continuous_var(name='u')\n\nmdl.minimize(v + w + t + 5 * (u-2)**2)\nmdl.add_constraint(v + 2 * w + t + u <= 3, \"cons1\")\nmdl.add_constraint(v + w + t >= 1, \"cons2\")\nmdl.add_constraint(v + w == 1, \"cons3\")\n\n# load quadratic program from docplex model\nqp = QuadraticProgram()\nqp.from_docplex(mdl)\nprint(qp.export_as_lp_string())",
"\\ This file has been generated by DOcplex\n\\ ENCODING=ISO-8859-1\n\\Problem name: ex6\n\nMinimize\n obj: v + w + t - 20 u + [ 10 u^2 ]/2 + 20\nSubject To\n cons1: v + 2 w + t + u <= 3\n cons2: v + w + t >= 1\n cons3: v + w = 1\n\nBounds\n 0 <= v <= 1\n 0 <= w <= 1\n 0 <= t <= 1\n\nBinaries\n v w t\nEnd\n\n"
]
],
[
[
"## Classical Solution\n\n3-ADMM-H needs a QUBO optimizer to solve the QUBO subproblem, and a continuous optimizer to solve the continuous convex constrained subproblem. We first solve the problem classically: we use the `MinimumEigenOptimizer` with the `NumPyMinimumEigenSolver` as a classical and exact QUBO solver and we use the `CobylaOptimizer` as a continuous convex solver. 3-ADMM-H supports any other suitable solver available in Qiskit. For instance, VQE, QAOA, and GroverOptimizer can be invoked as quantum solvers, as demonstrated later.\nIf CPLEX is installed, the `CplexOptimizer` can also be used as both, a QUBO and convex solver.",
"_____no_output_____"
],
[
"### Parameters\nThe 3-ADMM-H are wrapped in class `ADMMParameters`. Customized parameter values can be set as arguments of the class. In this example, parameters $\\rho, \\beta$ are initialized to $1001$ and $1000$, respectively. The penalization `factor_c` of equality constraints $Gx = b$ is set to $900$. The tolerance `tol` for primal residual convergence is set to `1.e-6`. \nIn this case, the 3-block implementation is guaranteed to converge for Theorem 4 of [1], because the inequality constraint with the continuous variable is always active. The 2-block implementation can be run by setting `three_block=False`, and practically converges to a feasible not optimal solution. \n",
"_____no_output_____"
]
],
[
[
"admm_params = ADMMParameters(\n rho_initial=1001, \n beta=1000, \n factor_c=900,\n max_iter=100,\n three_block=True, tol=1.e-6\n )",
"_____no_output_____"
]
],
[
[
"### Calling 3-ADMM-H algorithm\nTo invoke the 3-ADMM-H algorithm, an instance of the `ADMMOptimizer` class needs to be created. This takes ADMM-specific parameters and the subproblem optimizers separately into the constructor. The solution returned is an instance of `OptimizationResult` class.",
"_____no_output_____"
]
],
[
[
"# define QUBO optimizer\nqubo_optimizer = exact\n# qubo_optimizer = cplex # uncomment to use CPLEX instead\n\n# define classical optimizer\nconvex_optimizer = cobyla\n# convex_optimizer = cplex # uncomment to use CPLEX instead\n\n# initialize ADMM with classical QUBO and convex optimizer\nadmm = ADMMOptimizer(params=admm_params, \n qubo_optimizer=qubo_optimizer,\n continuous_optimizer=convex_optimizer)",
"_____no_output_____"
],
[
"# run ADMM to solve problem\nresult = admm.solve(qp)",
"_____no_output_____"
]
],
[
[
"### Classical Solver Result\nThe 3-ADMM-H solution can be then printed and visualized. The `x` attribute of the solution contains respectively, the\nvalues of the binary decision variables and the values of the continuous decision variables. The `fval` is the objective\nvalue of the solution.",
"_____no_output_____"
]
],
[
[
"print(\"x={}\".format(result.x))\nprint(\"fval={:.2f}\".format(result.fval))",
"x=[0.0, 1.0, 0.0, 1.0000000000000002]\nfval=6.00\n"
]
],
[
[
"Solution statistics can be accessed in the `state` field and visualized. We here display the convergence of 3-ADMM-H, in terms of primal residuals.",
"_____no_output_____"
]
],
[
[
"plt.plot(result.state.residuals)\nplt.xlabel(\"Iterations\")\nplt.ylabel(\"Residuals\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Quantum Solution\nWe now solve the same optimization problem with QAOA as QUBO optimizer, running on simulated quantum device. \nFirst, one need to select the classical optimizer of the eigensolver QAOA. Then, the simulation backened is set. Finally, \nthe eigensolver is wrapped into the `MinimumEigenOptimizer` class. A new instance of `ADMMOptimizer` is populated with QAOA as QUBO optimizer.",
"_____no_output_____"
]
],
[
[
"# define QUBO optimizer\nqubo_optimizer = qaoa \n\n# define classical optimizer\nconvex_optimizer = cobyla\n# convex_optimizer = cplex # uncomment to use CPLEX instead\n\n# initialize ADMM with quantum QUBO optimizer and classical convex optimizer\nadmm_q = ADMMOptimizer(params=admm_params, \n qubo_optimizer=qubo_optimizer,\n continuous_optimizer=convex_optimizer)",
"_____no_output_____"
],
[
"# run ADMM to solve problem\nresult_q = admm_q.solve(qp)",
"_____no_output_____"
]
],
[
[
"### Quantum Solver Results\nHere we present the results obtained from the quantum solver. As in the example above `x` stands for the solution, the `fval` is for objective value.",
"_____no_output_____"
]
],
[
[
"print(\"x={}\".format(result_q.x))\nprint(\"fval={:.2f}\".format(result_q.fval))",
"x=[0.0, 1.0, 0.0, 1.0000000000000002]\nfval=6.00\n"
],
[
"plt.clf()\nplt.plot(result_q.state.residuals)\nplt.xlabel(\"Iterations\")\nplt.ylabel(\"Residuals\")\nplt.show()",
"_____no_output_____"
],
[
"import qiskit.tools.jupyter\n%qiskit_version_table\n%qiskit_copyright",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d05e97890500c6a8162e22267e898819eeda0129 | 185,068 | ipynb | Jupyter Notebook | notebooks/Plot_results-parallel_run-cmaes.ipynb | hannanabdul55/seldonian-fairness | d02aaa3b62170df66f7a2962a32fa7d54028de78 | [
"MIT"
] | 2 | 2022-01-05T05:25:23.000Z | 2022-03-13T15:01:41.000Z | notebooks/Plot_results-parallel_run-cmaes.ipynb | hannanabdul55/seldonian-fairness | d02aaa3b62170df66f7a2962a32fa7d54028de78 | [
"MIT"
] | 1 | 2021-02-21T18:31:35.000Z | 2021-02-21T18:31:35.000Z | notebooks/Plot_results-parallel_run-cmaes.ipynb | hannanabdul55/seldonian-fairness | d02aaa3b62170df66f7a2962a32fa7d54028de78 | [
"MIT"
] | 1 | 2022-03-13T15:01:45.000Z | 2022-03-13T15:01:45.000Z | 979.195767 | 31,208 | 0.956043 | [
[
[
"%matplotlib inline\n%load_ext autoreload\n%autoreload 2\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport pickle\nimport glob\nfrom plot_results import *",
"_____no_output_____"
]
],
[
[
"# CMAES parallel run for 50 trials hard barrier",
"_____no_output_____"
]
],
[
[
"folder = 'result_cmaes_50tr_30n'\n\nconfig = pickle.load(open(folder + \"/config.p\", \"rb\"))\nexps = pickle.load(open(folder + \"/exps.p\", \"rb\"))\nres = pickle.load(open(list(glob.glob(folder + '/final_res*.p'))[0], 'rb'))\nplot_results(res, config['opt'])",
"_____no_output_____"
]
],
[
[
"# CMAES run for 50 trials with stratification",
"_____no_output_____"
]
],
[
[
"folder = 'result_cmaes_50tr_30n_stratify'\n\nconfig = pickle.load(open(folder + \"/config.p\", \"rb\"))\nexps = pickle.load(open(folder + \"/exps.p\", \"rb\"))\nres = pickle.load(open(list(glob.glob(folder + '/final_res*.p'))[0], 'rb'))\nplot_results(res, config['opt'])",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d05e9b0ccc8bb6ff91544d0dba761292db4da01b | 300,822 | ipynb | Jupyter Notebook | examples/notebooks/04 Initial SoC.ipynb | brosaplanella/liionpack | 4c3f61b6f28e1419974c8572669d70fc173a6959 | [
"MIT"
] | null | null | null | examples/notebooks/04 Initial SoC.ipynb | brosaplanella/liionpack | 4c3f61b6f28e1419974c8572669d70fc173a6959 | [
"MIT"
] | null | null | null | examples/notebooks/04 Initial SoC.ipynb | brosaplanella/liionpack | 4c3f61b6f28e1419974c8572669d70fc173a6959 | [
"MIT"
] | null | null | null | 817.451087 | 96,396 | 0.955858 | [
[
[
"# 4 Setting the initial SoC",
"_____no_output_____"
],
[
"Setting the initial SoC for your pack is performed with an argument passed to the solve algorithm. Currently the same value is applied to each battery but in future it will be possible to vary the SoC across the pack.",
"_____no_output_____"
]
],
[
[
"import liionpack as lp\nimport pybamm\nimport numpy as np\nimport matplotlib.pyplot as plt",
"c:\\users\\tom\\code\\pybamm\\pybamm\\expression_tree\\functions.py:204: RuntimeWarning: invalid value encountered in sign\n return self.function(*evaluated_children)\n"
]
],
[
[
"Lets set up the most simple pack possible with 1 battery and very low busbar resistance to compare to a pure PyBaMM simulation",
"_____no_output_____"
]
],
[
[
"Rsmall = 1e-6",
"_____no_output_____"
],
[
"netlist = lp.setup_circuit(Np=1, Ns=1, Rb=Rsmall, Rc=Rsmall, Ri=5e-2, V=4.0, I=1.0)",
"_____no_output_____"
],
[
"# Heat transfer coefficients\nhtc = np.ones(1) * 10\n# PyBaMM parameters\nchemistry = pybamm.parameter_sets.Chen2020\nparameter_values = pybamm.ParameterValues(chemistry=chemistry)",
"_____no_output_____"
],
[
"# Cycling experiment\nexperiment = pybamm.Experiment(\n [\n (\n \"Discharge at 1 A for 1000 s or until 3.3 V\",\n \"Rest for 1000 s\",\n \"Charge at 1 A for 1000 s or until 4.0 V\",\n \"Rest for 1000 s\",\n )\n ]\n * 3, period=\"10 s\"\n)",
"_____no_output_____"
],
[
"SoC = 0.5",
"_____no_output_____"
],
[
"# Solve pack\noutput = lp.solve(netlist=netlist,\n parameter_values=parameter_values,\n experiment=experiment,\n htc=htc, initial_soc=SoC)",
"c:\\users\\tom\\code\\pybamm\\pybamm\\expression_tree\\functions.py:204: RuntimeWarning: invalid value encountered in sign\n return self.function(*evaluated_children)\nSolving Pack: 100%|███████████████████████████████████████████████████████████████| 1200/1200 [00:06<00:00, 191.77it/s]\n"
]
],
[
[
"Let's compare to the PyBaMM simulation",
"_____no_output_____"
]
],
[
[
"parameter_values = pybamm.ParameterValues(chemistry=chemistry)\nparameter_values.update({\"Total heat transfer coefficient [W.m-2.K-1]\": 10.0})\nsim = lp.create_simulation(parameter_values, experiment, make_inputs=False)",
"_____no_output_____"
],
[
"sol = sim.solve(initial_soc=SoC)",
"_____no_output_____"
],
[
"def compare(sol, output):\n # Get pack level results\n time = sol[\"Time [s]\"].entries\n v_pack = output[\"Pack terminal voltage [V]\"]\n i_pack = output[\"Pack current [A]\"]\n v_batt = sol[\"Terminal voltage [V]\"].entries\n i_batt = sol[\"Current [A]\"].entries\n\n # Plot pack voltage and current\n _, (axl, axr) = plt.subplots(1, 2, tight_layout=True, figsize=(15, 10), sharex=True, sharey=True)\n axl.plot(time[1:], v_pack, color=\"green\", label=\"simulation\")\n axl.set_xlabel(\"Time [s]\")\n axl.set_ylabel(\"Pack terminal voltage [V]\", color=\"green\")\n axl2 = axl.twinx()\n axl2.plot(time[1:], i_pack, color=\"black\", label=\"simulation\")\n axl2.set_ylabel(\"Pack current [A]\", color=\"black\")\n axl2.set_title(\"Liionpack Simulation\")\n axr.plot(time, v_batt, color=\"red\", label=\"simulation\")\n axr.set_xlabel(\"Time [s]\")\n axr.set_ylabel(\"Battery terminal voltage [V]\", color=\"red\")\n axr2 = axr.twinx()\n axr2.plot(time, i_batt, color=\"blue\", label=\"simulation\")\n axr2.set_ylabel(\"Battery current [A]\", color=\"blue\")\n axr2.set_title(\"Single PyBaMM Simulation\")",
"_____no_output_____"
],
[
"compare(sol, output)",
"_____no_output_____"
]
],
[
[
"Now lets start the simulation from a different state of charge",
"_____no_output_____"
]
],
[
[
"SoC = 0.25",
"_____no_output_____"
],
[
"# Solve pack\noutput = lp.solve(netlist=netlist,\n parameter_values=parameter_values,\n experiment=experiment,\n htc=htc, initial_soc=SoC)",
"Solving Pack: 100%|███████████████████████████████████████████████████████████████| 1200/1200 [00:06<00:00, 188.69it/s]\n"
],
[
"compare(sol, output)",
"_____no_output_____"
]
],
[
[
"Here we are still comparing to the PyBaMM simulation at 0.5 SoC and we can see that liionpack started at a lower voltage corresponding to a lower SoC.",
"_____no_output_____"
]
],
[
[
"parameter_values = pybamm.ParameterValues(chemistry=chemistry)\nparameter_values.update({\"Total heat transfer coefficient [W.m-2.K-1]\": 10.0})\nsim = lp.create_simulation(parameter_values, experiment, make_inputs=False)\nsol = sim.solve(initial_soc=SoC)",
"_____no_output_____"
]
],
[
[
"Now we can re-run the PyBaMM simulation and compare again",
"_____no_output_____"
]
],
[
[
"compare(sol, output)",
"_____no_output_____"
],
[
"lp.draw_circuit(netlist)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05eb59bff3d453414e0d71cad21223440d3710b | 26,618 | ipynb | Jupyter Notebook | 01_donnees_en_tables/correction/03_1_format_csv_correction.ipynb | efloti/cours-nsi-premiere | 5b05bc81e5f8d7df47bf5785068b4bf4d1e357bb | [
"CC0-1.0"
] | null | null | null | 01_donnees_en_tables/correction/03_1_format_csv_correction.ipynb | efloti/cours-nsi-premiere | 5b05bc81e5f8d7df47bf5785068b4bf4d1e357bb | [
"CC0-1.0"
] | null | null | null | 01_donnees_en_tables/correction/03_1_format_csv_correction.ipynb | efloti/cours-nsi-premiere | 5b05bc81e5f8d7df47bf5785068b4bf4d1e357bb | [
"CC0-1.0"
] | null | null | null | 23.41073 | 201 | 0.50526 | [
[
[
"# Découverte du format CSV - *Comma-Separated values*",
"_____no_output_____"
],
[
"**Plan du document**\n- Le format **CSV**\n- Représenter des données CSV avec Python\n - Première solution: un tableau de tuples\n - **Deuxième solution**: un tableau de *tuples nommés* (dictionnaires)\n - l'*unpacking*,\n - l'opération *zip*\n - syntaxe en *compréhension des dictionnaires*\n - **synthèse**: CSV -> tableau de tuples nommés",
"_____no_output_____"
],
[
"## Le format CSV",
"_____no_output_____"
],
[
"*Comma*: virgule; *CSV*: valeurs séparées par des virgule.",
"_____no_output_____"
],
[
"CSV est un format **textuel** (par opposition à *binaire*) qui sert à représenter des **données en tables**; voici à quoi cela ressemble:\n\n```\nnom,prenom,date_naissance\nDurand,Jean-Pierre,23/05/1985\nDupont,Christophe,15/12/1967\nTerta,Henry,12/06/1978 \n```",
"_____no_output_____"
],
[
"On devine qu'il s'agit d'informations à propos d'individus: Jean pierre Durand né le 23 mai 1985, etc. En informatique on parle de **collection de données**.\n\nLa première ligne précise le sens des valeurs trouvées aux lignes suivantes; ses valeurs `nom`, `prenom`, `date_naissance` sont appelées **descripteurs** ou encore **attributs**.\n\nLes lignes suivantes correspondent à des individus différents; en informatique on parle souvent d'**objets** ou d'**entités**.\n\nChaque «*objet*» (ici individu) correspond à une ligne: les **valeurs** qu'on y trouve sont associées aux *descripteurs* de même position.",
"_____no_output_____"
],
[
"On peut (re)présenter la même information plus agréablement avec un rendu table:\n\n| nom | prenom | date_naissance |\n| ------------- |:-------------:| -----:|\n| Durand | Jean-Pierre | 23/05/1985 |\n| Dupont | Christophe | 15/12/1967 |\n| Tertra | Henry | 12/06/1978 |",
"_____no_output_____"
],
[
"## Représenter des données CSV avec Python",
"_____no_output_____"
],
[
"### Première solution: une liste de tuples",
"_____no_output_____"
],
[
"Cela donnerait `[('Durand', 'Jean-Pierre', '23/05/1985'), ('Dupont',..),(..)]`",
"_____no_output_____"
],
[
"On y parvient assez simplement à l'aide de `str.split(..)`:",
"_____no_output_____"
]
],
[
[
"donnees_CSV = \"\"\"nom,prenom,date_naissance\nDurand,Jean-Pierre,23/05/1985\nDupont,Christophe,15/12/1967\nTerta,Henry,12/06/1978\"\"\"",
"_____no_output_____"
],
[
"etape1 = donnees_CSV.split('\\n')\netape1",
"_____no_output_____"
],
[
"etape2 = [obj.split(',') for obj in etape1]\netape2 # une liste de liste",
"_____no_output_____"
],
[
"etape3 = [tuple(obj) for obj in etape2]\netape3 # une liste de tuple",
"_____no_output_____"
],
[
"fin = etape3[1:] # un petit slice\nfin # sans l'en-tête",
"_____no_output_____"
]
],
[
[
"#### À faire toi-même\n\nOn peut parvenir à `fin` à partir de `donnees_CSV` en **une seule fois** par *composition* ... essais!",
"_____no_output_____"
]
],
[
[
"# deux_en_un = [ obj.split(',') for obj in donnees_CSV.split('\\n') ]\n# trois_en_un = [ tuple( obj.split(',') ) for obj in donnees_CSV.split('\\n') ]\n# tu peux essayer de faire deux_en_un, puis trois_en_un avant.\nquatre_en_un = [ tuple( obj.split(',') ) for obj in donnees_CSV.split('\\n') ][1:]",
"_____no_output_____"
],
[
"# pour tester\nassert quatre_en_un == fin",
"_____no_output_____"
]
],
[
[
"___",
"_____no_output_____"
],
[
"L'inconvénient de cette représentation c'est qu'elle «oublie» les descripteurs.\n\nPourquoi ne pas les conserver comme à l'étape3? Pour éviter d'avoir un tableau *hétérogène*: le premier élément ne serait pas un «objet». De tels tableaux sont plus difficile à manipuler.",
"_____no_output_____"
],
[
"### Deuxième solution: un tableau de *tuples nommés*",
"_____no_output_____"
],
[
"**n-uplet (ou tuples) nommés**: tuple dont chaque valeur est associée à un descripteur.\n\nMalheureusement Python ne possède pas un tel type par défaut (il existe toutefois dans la bibliothèque standard).\n\nPour représenter ce type, nous utiliserons un dictionnaire dont les clés sont les descripteurs; voici un exemple:\n```python\n{'nom': 'Durand', 'prenom': 'Jean-Pierre', 'date_naissance': '23/05/1985'}\n```",
"_____no_output_____"
],
[
"Pour y parvenir, nous partons de:",
"_____no_output_____"
]
],
[
[
"donnees_CSV = \"\"\"nom,prenom,date_naissance\nDurand,Jean-Pierre,23/05/1985\nDupont,Christophe,15/12/1967\nTerta,Henry,12/06/1978\"\"\"",
"_____no_output_____"
]
],
[
[
"Les étapes qui suivent servent à séparer les descripteurs et les objets:",
"_____no_output_____"
]
],
[
[
"tmp = donnees_CSV.split('\\n')\ntmp",
"_____no_output_____"
],
[
"descripteurs_str = tmp[0]\ndescripteurs = tuple(descripteurs_str.split(','))\nprint(f\"le tuple des descripteurs: {descripteurs}\")",
"_____no_output_____"
],
[
"donnees_str = tmp[1:]\ndonnees_str",
"_____no_output_____"
],
[
"objets = [tuple(obj.split(',')) for obj in donnees_str]\nprint(f\"la liste des objets (des personnes ici):\\n {objets}\")",
"la liste des objets (des personnes ici):\n [('Durand', 'Jean-Pierre', '23/05/1985'), ('Dupont', 'Christophe', '15/12/1967'), ('Terta', 'Henry', '12/06/1978')]\n"
]
],
[
[
"#### À faire toi-même\n\nPeux-tu compléter les parties manquantes pour obtenir le même résultat plus rapidement?",
"_____no_output_____"
]
],
[
[
"descripteurs = tuple( donnees_CSV.split('\\n')[0].split(',') )\nobjets = [ tuple( ligne.split(',') ) for ligne in donnees_CSV.split('\\n')[1:] ]\nprint(f\"- les descripteurs:\\n\\t {descripteurs}\\n- les objets:\\n\\t {objets}\")",
"- les descripteurs:\n\t ('nom', 'prenom', 'date_naissance')\n- les objets:\n\t [('Durand', 'Jean-Pierre', '23/05/1985'), ('Dupont', 'Christophe', '15/12/1967'), ('Terta', 'Henry', '12/06/1978')]\n"
]
],
[
[
"______",
"_____no_output_____"
],
[
"#### À faire toi-même - *découverte de l'**unpacking** (déballage)*\n\nPeux-tu réaliser le traitement précédent en **vraiment** une seule ligne? Pour cela observe les trois exemples qui suivent:",
"_____no_output_____"
]
],
[
[
"# exemple1 d'unpacking\ntete, *queue = [1, 2, 3, 4]\nprint(f\"La tête: {tete} et la queue: {queue}\")",
"La tête: 1 et la queue: [2, 3, 4]\n"
],
[
"# exemple2 d'unpacking\nun, deux, *reste = [1, 2, 3, 4]\nprint(f\"un: {un}\\ndeux: {deux}\\nreste: {reste}\")",
"5 6\n"
],
[
"# exemple3 d'unpacking\ntete, *corps, pied = [1,2,3,4]\nprint(f\"tete: {tete}\\ncorps: {corps}\\npied: {pied}\")",
"3 4\n"
],
[
"# À toi de jouer!\ndescripteurs, *objets = [tuple(d.split(',')) for d in donnees_CSV.split('\\n')]\nprint(f\"les descripteurs:\\n\\t {descripteurs}\\nles objets:\\n\\t {objets}\")",
"les descripteurs:\n\t ('nom', 'prenom', 'date_naissance')\nles objets:\n\t [('Durand', 'Jean-Pierre', '23/05/1985'), ('Dupont', 'Christophe', '15/12/1967'), ('Terta', 'Henry', '12/06/1978')]\n"
]
],
[
[
"____",
"_____no_output_____"
],
[
"Arrivé à ce stade nous voudrions combiner:\n- `('descr1', 'descr2', ...)` et `('v1', 'v2', ...)` en ...\n- `{'descr1': 'v1', 'descr2': 'v2', ..}` (n-uplet nommé) ",
"_____no_output_____"
],
[
"#### Appareiller deux séquences - `zip`",
"_____no_output_____"
],
[
"On a souvent besoin de grouper par paires deux séquences de même longueur `len`.\n\n*Ex*: je **dispose** de `['a', 'b', 'c']` et `[3, 2, 1]`\n\nj'ai **besoin de** `[('a', 3), ('b', 2), ('c', 1)]`.",
"_____no_output_____"
],
[
"#### À faire toi-même",
"_____no_output_____"
],
[
"La fonction `appareiller(t1, t2)` prend deux tableaux de même taille en argument et renvoie un tableau obtenue en appararillant les éléments de `t1` et `t2` de même index.\n\nCompléter le code qui suit pour résoudre ce problème",
"_____no_output_____"
]
],
[
[
"def appareiller(t1, t2):\n assert len(t1) == len(t2)\n t = []\n for i in range(len(t1)):\n couple = (t1[i], t2[i])\n t.append( couple )\n return t\n\n# autre solution avec la syntaxe en compréhension\ndef appareiller2(t1, t2):\n assert len(t1) == len(t2)\n return [\n (t1[i], t2[i])\n for i in range(len(t1))\n ]",
"_____no_output_____"
],
[
"# vérifier votre solution\ntab1 = ['a', 'b', 'c']\ntab2 = [3, 2, 1]\nassert appareiller(tab1, tab2) == [('a', 3), ('b', 2), ('c', 1)]\nassert appareiller2(tab1, tab2) == [('a', 3), ('b', 2), ('c', 1)]",
"_____no_output_____"
]
],
[
[
"___",
"_____no_output_____"
],
[
"Un cas d'utilisation fréquent de l'apparaillement est la lecture dans une boucle des paires",
"_____no_output_____"
]
],
[
[
"# tester moi\ntab1 = ['a', 'b', 'c']\ntab2 = [3, 2, 1]\nfor a, b in appareiller(tab1, tab2):\n print(f'a vaut \"{a}\" et b vaut \"{b}\"')",
"a vaut \"a\" et b vaut \"3\"\na vaut \"b\" et b vaut \"2\"\na vaut \"c\" et b vaut \"1\"\n"
]
],
[
[
"en fait, Python dispose d'une fonction prédéfinie `zip(seq1, seq2, ...)` qui fait la même chose avec des «séquences» (`list` est un cas particulier de séquence).\n\n*note*: `zip`?? penser à la «fermeture-éclair» d'un blouson ...",
"_____no_output_____"
]
],
[
[
"z = zip(tab1, tab2)\nprint(z)\nprint(list(z))",
"_____no_output_____"
]
],
[
[
"*note*: elle renvoie un objet spécial de type `zip` car on l'utilise souvent dans une boucle directement c'est-à-dire sans mémoriser le zip (un peu comme avec `range`)",
"_____no_output_____"
]
],
[
[
"# tester moi\ntab1 = ['a', 'b', 'c']\ntab2 = [3, 2, 1]\nfor a, b in zip(tab1, tab):\n print(f'a vaut \"{a}\" et b vaut \"{b}\"')",
"a vaut \"a\" et b vaut \"3\"\na vaut \"b\" et b vaut \"2\"\n"
]
],
[
[
"#### Découverte: la syntaxe en compréhension est aussi valable pour les `dict`",
"_____no_output_____"
],
[
"Voici un exemple simple:",
"_____no_output_____"
]
],
[
[
"modele_tuple_nomme = {desc: None for desc in descripteurs}\nmodele_tuple_nomme",
"_____no_output_____"
]
],
[
[
"Bien Noter que la partie avant `for` est de la forme `<cle>: <val>`.\n\nOn utilise généralement cela avec `zip`:",
"_____no_output_____"
]
],
[
[
"cles = (\"cle1\", \"cle2\", \"cle3\")\nvaleurs = (\"ah\", \"oh\", \"hein\")\n{c: v for c, v in zip(cles, valeurs)} # zip fonctionne aussi avec des tuples de même longueur!",
"_____no_output_____"
]
],
[
[
"Voici encore un exemple bien utile pour réaliser un tableau à partir de données CSV.",
"_____no_output_____"
]
],
[
[
"cles = (\"cle1\", \"cle2\", \"cle3\")\nobjets = [(\"ah\", \"oh\", \"hein\"), ('riri', 'fifi', 'loulou')]\n# on veut un tableau de tuples nommés\n[ {desc: val for desc, val in zip(cles, objet)} for objet in objets ]",
"_____no_output_____"
]
],
[
[
"### Synthèse: retour au problème des données au format CSV",
"_____no_output_____"
],
[
"En combinant tout ce que tu as appris et les exemples précédents, tu devrais être capable d'obtenir notre liste de n-uplets nommés en quelques lignes ... Non?\n\n*rappel*: au départ, on **dispose de**\n```python\ndonnees_CSV = \"\"\"nom,prenom,date_naissance\nDurand,Jean-Pierre,23/05/1985\nDupont,Christophe,15/12/1967\nTerta,Henry,12/06/1978\"\"\"\n```\n\nau final, on veut **produire** une liste de *tuples nommés*:\n```python\n[\n {'nom': 'Durand', 'prenom': 'Jean-Pierre', 'date_naissance': '23/05/1985'},\n {'nom': 'Dupont', 'prenom': 'Christophe', 'date_naissance': '15/12/1967'},\n {'nom': 'Terta', 'prenom': 'Henry', 'date_naissance': '12/06/1978'}\n]\n```\n",
"_____no_output_____"
],
[
"Voici comment y parvenir en deux «compréhensions»",
"_____no_output_____"
]
],
[
[
"descripteurs, *objets = [tuple(ligne.split(',')) for ligne in donnees_CSV.split('\\n')]\n\nobjets = [ # sur plusieurs ligne pour plus de clarté.\n {\n desc: val for desc, val in zip(descripteurs, obj)\n }\n for obj in objets\n]\nobjets",
"_____no_output_____"
]
],
[
[
"#### À faire toi-même",
"_____no_output_____"
],
[
"La syntaxe en compréhension des listes et des dictionnaires est utile et puissante mais nécessite pas mal d'investissement pour être bien maîtrisée.\n\nPour cette raison, reprend le problème en écrivant une fonction `csv_vers_objets(csv_str)` qui prend en argument la chaîne au format csv et renvoie le tableau de n-uplets nommés correspondant.\n\nNous la réutiliserons dans le 05_applications...",
"_____no_output_____"
]
],
[
[
"def csv_vers_objets(csv_str):\n descripteurs, *objets = [tuple(ligne.split(',')) for ligne in csv_str.split('\\n')]\n\n objets = [ # sur plusieurs ligne pour plus de clarté.\n {\n desc: val for desc, val in zip(descripteurs, obj)\n }\n for obj in objets\n ]\n return objets\n\ncsv_vers_objets(donnees_CSV)",
"_____no_output_____"
],
[
"assert csv_vers_objets(donnees_CSV) == objets",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
d05ec26a292f426b66bfe08ffe1640689a8c372d | 54,368 | ipynb | Jupyter Notebook | ipy_notebooks/old/current_neural_net-normalized-culled.ipynb | mattjshannon/swsnet | ea27ab708b9e626074da11476bfa31f617339c68 | [
"BSD-3-Clause"
] | null | null | null | ipy_notebooks/old/current_neural_net-normalized-culled.ipynb | mattjshannon/swsnet | ea27ab708b9e626074da11476bfa31f617339c68 | [
"BSD-3-Clause"
] | 20 | 2018-07-17T23:30:21.000Z | 2018-12-13T21:52:45.000Z | ipy_notebooks/old/current_neural_net-normalized-culled.ipynb | mattjshannon/swsnet | ea27ab708b9e626074da11476bfa31f617339c68 | [
"BSD-3-Clause"
] | null | null | null | 84.030912 | 12,664 | 0.837478 | [
[
[
"# TF neural net with normalized ISO spectra",
"_____no_output_____"
]
],
[
[
"# TensorFlow and tf.keras\nimport tensorflow as tf\nfrom tensorflow import keras\n\n# Helper libraries\nimport glob\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nfrom concurrent.futures import ProcessPoolExecutor\nfrom IPython.core.debugger import set_trace as st\nfrom sklearn.model_selection import train_test_split\nfrom time import time\n\n# My modules\nfrom swsnet import helpers\n\nprint(tf.__version__)",
"1.10.0\n"
]
],
[
[
"## Dataset: ISO-SWS (normalized, culled)",
"_____no_output_____"
]
],
[
[
"# Needed directories\nbase_dir = '../data/isosws_atlas/'\n\n# Pickles containing our spectra in the form of pandas dataframes:\nspec_dir = base_dir + 'spectra_normalized/'\nspec_files = np.sort(glob.glob(spec_dir + '*.pkl'))\n\n# Metadata pickle (pd.dataframe). Note each entry contains a pointer to the corresponding spectrum pickle.\nmetadata = base_dir + 'metadata_sorted_normalized_culled.pkl'",
"_____no_output_____"
]
],
[
[
"#### Labels ('group'):\n\n1. Naked stars\n2. Stars with dust\n3. Warm, dusty objects\n4. Cool, dusty objects\n5. Very red objects\n6. Continuum-free objects but having emission lines\n7. Flux-free and/or fatally flawed spectra",
"_____no_output_____"
],
[
"### Subset 1: all data included",
"_____no_output_____"
]
],
[
[
"features, labels = helpers.load_data(base_dir=base_dir, metadata=metadata,\n only_ok_data=False, clean=False, verbose=False)",
"_____no_output_____"
],
[
"print(features.shape)\nprint(labels.shape)",
"(1235, 359)\n(1235,)\n"
]
],
[
[
"### Subset 2: exclude group 7",
"_____no_output_____"
]
],
[
[
"features_clean, labels_clean = \\\n helpers.load_data(base_dir=base_dir, metadata=metadata,\n only_ok_data=False, clean=True, verbose=False)",
"_____no_output_____"
],
[
"print(features_clean.shape)\nprint(labels_clean.shape)",
"(1058, 359)\n(1058,)\n"
]
],
[
[
"### Subset 3: exclude group 7, uncertain data",
"_____no_output_____"
]
],
[
[
"features_certain, labels_certain = \\\n helpers.load_data(base_dir=base_dir, metadata=metadata,\n only_ok_data=True, clean=False, verbose=False)",
"_____no_output_____"
],
[
"print(features_certain.shape)\nprint(labels_certain.shape)",
"(851, 359)\n(851,)\n"
]
],
[
[
"# Testing l2norms",
"_____no_output_____"
]
],
[
[
"def neural(features, labels, test_size=0.3, l2norm=0.01):\n\n X_train, X_test, y_train, y_test = \\\n train_test_split(features, labels, test_size=test_size, random_state = 42)\n\n # Sequential model, 7 classes of output.\n model = keras.Sequential()\n model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm), input_dim=359))\n model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))\n model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))\n model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))\n model.add(keras.layers.Dense(7, activation='softmax'))\n\n # Early stopping condition.\n callback = [tf.keras.callbacks.EarlyStopping(monitor='acc', patience=5, verbose=0)]\n\n # Recompile model and fit.\n model.compile(optimizer=keras.optimizers.Adam(0.0005),\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])\n # model.fit(X_train, y_train, epochs=50, batch_size=32, verbose=False)\n model.fit(X_train, y_train, epochs=100, batch_size=32, callbacks=callback, verbose=False)\n\n # Check accuracy.\n score = model.evaluate(X_test, y_test, verbose=0)\n accuracy = score[1]\n print(\"L2 norm, accuracy: \", l2norm, accuracy)\n \n return model, test_size, accuracy",
"_____no_output_____"
],
[
"# for l2norm in (0.1, 0.01, 0.001, 0.0001, 0.00001):\n# model, test_size, accuracy = neural(features, labels, l2norm=l2norm)",
"L2 norm, accuracy: 0.1 0.4660194177650711\nL2 norm, accuracy: 0.01 0.7313915869178895\nL2 norm, accuracy: 0.001 0.7766990302835853\nL2 norm, accuracy: 0.0001 0.7637540466577104\nL2 norm, accuracy: 1e-05 0.7669902922266124\n"
],
[
"# for l2norm in (0.1, 0.01, 0.001, 0.0001, 0.00001):\n# model, test_size, accuracy = neural(features_clean, labels_clean, l2norm=l2norm)",
"L2 norm, accuracy: 0.1 0.48113207622144205\nL2 norm, accuracy: 0.01 0.7452830199925404\nL2 norm, accuracy: 0.001 0.7798742123369901\nL2 norm, accuracy: 0.0001 0.7955974831521136\nL2 norm, accuracy: 1e-05 0.8018867935774461\n"
],
[
"# for l2norm in (0.1, 0.01, 0.001, 0.0001, 0.00001):\n# model, test_size, accuracy = neural(features_certain, labels_certain, l2norm=l2norm)",
"L2 norm, accuracy: 0.1 0.5859375\nL2 norm, accuracy: 0.01 0.828125\nL2 norm, accuracy: 0.001 0.8671875\nL2 norm, accuracy: 0.0001 0.89453125\nL2 norm, accuracy: 1e-05 0.87890625\n"
],
[
"# for l2norm in (0.001, 0.0001, 0.00001, 0.000001):\n# model, test_size, accuracy = neural(features_certain, labels_certain, l2norm=l2norm)",
"L2 norm, accuracy: 0.001 0.88671875\nL2 norm, accuracy: 0.0001 0.859375\nL2 norm, accuracy: 1e-05 0.86328125\nL2 norm, accuracy: 1e-06 0.859375\n"
]
],
[
[
"***",
"_____no_output_____"
],
[
"# Testing training size vs. accuracy",
"_____no_output_____"
],
[
"Model:",
"_____no_output_____"
]
],
[
[
"def run_NN(input_tuple):\n \"\"\"Run a Keras NN for the purpose of examining the effect of training set size.\n \n Args:\n features (ndarray): Array containing the spectra (fluxes).\n labels (ndarray): Array containing the group labels for the spectra.\n test_size (float): Fraction of test size relative to (test + training).\n \n Returns:\n test_size (float): Input test_size, just a sanity check!\n accuracy (float): Accuracy of this neural net when applied to the test set.\n \"\"\"\n \n features, labels, test_size = input_tuple\n l2norm = 0.001\n \n X_train, X_test, y_train, y_test = \\\n train_test_split(features, labels, test_size=test_size, random_state = 42)\n \n # Sequential model, 7 classes of output.\n model = keras.Sequential()\n model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm), input_dim=359))\n model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))\n model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))\n model.add(keras.layers.Dense(64, activation='relu', kernel_regularizer=keras.regularizers.l2(l2norm)))\n model.add(keras.layers.Dense(7, activation='softmax'))\n\n # Early stopping condition.\n callback = [tf.keras.callbacks.EarlyStopping(monitor='acc', patience=5, verbose=0)]\n\n # Recompile model and fit.\n model.compile(optimizer=keras.optimizers.Adam(0.0005),\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])\n # model.fit(X_train, y_train, epochs=50, batch_size=32, verbose=False)\n model.fit(X_train, y_train, epochs=100, batch_size=32, callbacks=callback, verbose=False)\n\n # Check accuracy.\n score = model.evaluate(X_test, y_test, verbose=0)\n accuracy = score[1]\n# print(\"Test size, accuracy: \", test_size, accuracy)\n\n return test_size, accuracy",
"_____no_output_____"
],
[
"def run_networks(search_map):\n # Run the networks in parallel.\n start = time()\n pool = ProcessPoolExecutor(max_workers=14)\n results = list(pool.map(run_NN, search_map))\n end = time()\n print('Took %.3f seconds' % (end - start))\n\n run_matrix = np.array(results)\n return run_matrix\n\ndef plot_results(run_matrix):\n # Examine results.\n plt.plot(run_matrix.T[0], run_matrix.T[1], 's', mfc='w', ms=5, mew=2, mec='r');\n plt.xlabel('Test size (fraction)');\n plt.ylabel('Test accuracy');\n plt.minorticks_on();\n# plt.xlim(left=0); \n return",
"_____no_output_____"
]
],
[
[
"Search space (training size):",
"_____no_output_____"
]
],
[
[
"# Values of test_size to probe.\nsearch_space = np.arange(0.14, 0.60, 0.02)\nprint('Size of test set considered: ', search_space)\n\n# Number of iterations for each test_size value.\nn_iterations = 20\n\n# Create a vector to iterate over.\nrx = np.array([search_space] * n_iterations).T\nsearch_space_full = rx.flatten()\n\nprint('Number of iterations per test_size: ', n_iterations)\nprint('Total number of NN iterations required: ', n_iterations * len(search_space))",
"Size of test set considered: [0.14 0.16 0.18 0.2 0.22 0.24 0.26 0.28 0.3 0.32 0.34 0.36 0.38 0.4\n 0.42 0.44 0.46 0.48 0.5 0.52 0.54 0.56 0.58]\nNumber of iterations per test_size: 20\nTotal number of NN iterations required: 460\n"
],
[
"# Wrap up tuple inputs for running in parallel.\nsearch_map = [(features, labels, x) for x in search_space_full]\nsearch_map_clean = [(features_clean, labels_clean, x) for x in search_space_full]\nsearch_map_certain = [(features_certain, labels_certain, x) for x in search_space_full]",
"_____no_output_____"
],
[
"run_matrix = run_networks(search_map)\nrun_matrix_clean = run_networks(search_map_clean)\nrun_matrix_certain = run_networks(search_map_certain)",
"Took 344.395 seconds\nTook 307.249 seconds\nTook 264.682 seconds\n"
]
],
[
[
"## Full set:",
"_____no_output_____"
]
],
[
[
"plot_results(run_matrix)",
"_____no_output_____"
]
],
[
[
"## Clean set:",
"_____no_output_____"
]
],
[
[
"plot_results(run_matrix_clean)",
"_____no_output_____"
]
],
[
[
"## Certain set:",
"_____no_output_____"
]
],
[
[
"plot_results(run_matrix_certain)",
"_____no_output_____"
]
],
[
[
"***",
"_____no_output_____"
],
[
"Based on the above, probably need to do more data preprocessing:\n- e.g., remove untrustworthy data",
"_____no_output_____"
]
],
[
[
"# save_path = '../models/nn_sorted_normalized_culled.h5'",
"_____no_output_____"
],
[
"# model.save(save_path)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
d05ecfff9054822f1949be80202f542ca122ad34 | 272,724 | ipynb | Jupyter Notebook | MNIST_data.ipynb | hotpocket/DigitRecognizer | e24b78fdbd19070108f7a33416549e4ab263b5d5 | [
"MIT"
] | null | null | null | MNIST_data.ipynb | hotpocket/DigitRecognizer | e24b78fdbd19070108f7a33416549e4ab263b5d5 | [
"MIT"
] | null | null | null | MNIST_data.ipynb | hotpocket/DigitRecognizer | e24b78fdbd19070108f7a33416549e4ab263b5d5 | [
"MIT"
] | null | null | null | 292.308682 | 212,076 | 0.886028 | [
[
[
"Filename: MNIST_data.ipynb\n\nFrom <a href=\"http://neuralnetworksanddeeplearning.com/chap1.html\"> this </a> book\n\nAbbreviation: MNIST = Modified (handwritten digits data set from the U.S.) National Institute of Standards and Technology\n\nPurpose: Explore the MNIST digits data to get familiar with the content and quality of the data.",
"_____no_output_____"
]
],
[
[
"import mnist_loader\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n\n# training_data, validation_data, test_data = mnist_loader.load_data_wrapper()\n#training_data, validation_data, test_data = mnist_loader.load_data()\ntraining, validation, test = mnist_loader.load_data()\nstruct = [{'name': 'training', 'data': training[0], 'label': training[1]},\n {'name': 'validation', 'data': validation[0], 'label': validation[1]},\n {'name': 'test', 'data': test[0], 'label': test[1]}]\n",
"_____no_output_____"
]
],
[
[
"Training, validation, and test data structures are 2 element tuples having the following structure:\n* <pre>[[p,i,x,e,l,s, , i,n, i,m,a,g,e, ,1], [...]]</pre>\n* <pre>[num_represented_by_image1, ...]",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(1, 3)\n\ntrain = pd.Series(training_data[1])\ntrain.hist(ax=axes[0])\naxes[0].set_title(\"Training Data\")\n#display(train.describe())\n\nvalidate = pd.Series(validation_data[1])\nvalidate.hist(ax=axes[1])\naxes[1].set_title(\"Validation Data\")\n#display(validate.describe())\n\ntest = pd.Series(test_data[1])\ntest.hist(ax=axes[2])\naxes[2].set_title(\"Test Data\")\n#display(test.describe())\n",
"_____no_output_____"
],
[
"display(\"Distribution of validation data values\")\nvalues = pd.Series(validation_data[1])\nvalues.hist()\nvalues.describe()",
"_____no_output_____"
],
[
"display(\"Distribution of validation data values\")\nvalues = pd.Series(test_data[1])\nvalues.hist()\nvalues.describe()",
"_____no_output_____"
]
],
[
[
"<h3>Training Data</h3>",
"_____no_output_____"
]
],
[
[
"pixels = pd.DataFrame(training_data[0])\ndisplay(\"Images: {} Pixels-per-image: {}\".format(*pixels.shape))\npixels.head()\n#pixels.T.describe() # takes FOREVER ... \n",
"_____no_output_____"
],
[
"print('\\033[1m'+\"validation_data:\"+'\\033[0m')\ndisplay(validation_data)\nprint('{1:32s}{0}'.format(type(validation_data),'\\033[1m'+\"validation_data type:\"+'\\033[0m'))\nprint('{1:32s}{0}'.format(len(validation_data),'\\033[1m'+\"num of components:\"+'\\033[0m'))\nprint('')\nprint('{1:32s}{0}'.format(type(validation_data[0]),'\\033[1m'+\"first component type:\"+'\\033[0m'))\nprint('{1:32s}{0}'.format(len(validation_data[0]),'\\033[1m'+\"num of sub-components:\"+'\\033[0m'))\nprint('')\nprint('{1:32s}{0}'.format(type(validation_data[1]),'\\033[1m'+\"second component type:\"+'\\033[0m'))\nprint('{1:32s}{0}'.format(len(validation_data[1]),'\\033[1m'+\"num of sub-components:\"+'\\033[0m'))",
"\u001b[1mvalidation_data:\u001b[0m\n"
],
[
"print('\\033[1m'+\"test_data:\"+'\\033[0m')\ndisplay(test_data)\nprint('{1:32s}{0}'.format(type(test_data),'\\033[1m'+\"test_data type:\"+'\\033[0m'))\nprint('{1:32s}{0}'.format(len(test_data),'\\033[1m'+\"num of components:\"+'\\033[0m'))\nprint('')\nprint('{1:32s}{0}'.format(type(test_data[0]),'\\033[1m'+\"first component type:\"+'\\033[0m'))\nprint('{1:32s}{0}'.format(len(test_data[0]),'\\033[1m'+\"num of sub-components:\"+'\\033[0m'))\nprint('')\nprint('{1:32s}{0}'.format(type(test_data[1]),'\\033[1m'+\"second component type:\"+'\\033[0m'))\nprint('{1:32s}{0}'.format(len(test_data[1]),'\\033[1m'+\"num of sub-components:\"+'\\033[0m'))",
"\u001b[1mtest_data:\u001b[0m\n"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\nprint(type(training_data[0][0]))\nprint(len(training_data[0][0]))\nprint(28*28)\n# break data into 28 x 28 square array (from 1 x 784 array)\nplottable_image = np.reshape(training_data[0][0], (28, 28))\n#display(plottable_image)\n# plot\nplt.imshow(plottable_image, cmap='gray_r')\nplt.show()",
"<class 'numpy.ndarray'>\n784\n784\n"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n\ntable_cols = 18\ntable_rows = 12\n\nrand_grid = np.random.rand(28, 28)\n# plottable_image = np.reshape(training_data[0][0], (28, 28))\n\nk = 0\nfor i in range(0,table_cols) :\n for j in range(0,table_rows) :\n if i==0 and j==0 :\n plottable_images = [rand_grid]\n else :\n plottable_images.append( np.reshape(training_data[0][k], (28, 28)) )\n k += 1\n\nprint(len(plottable_images))\n\nfig, axes = plt.subplots(table_rows, table_cols, figsize=(15, 12),\n subplot_kw={'xticks': [], 'yticks': []})\nfig.subplots_adjust(hspace=0.5, wspace=0)\n\nfor i, ax in enumerate(axes.flat):\n ax.imshow(plottable_images[i], cmap='gray_r')\n if i == 0 :\n ax.set_title(\"rand\")\n else :\n digit = str(training_data[1][i-1])\n index = str(i-1)\n ax.set_title(\"({}) {}\".format(index, digit))\n\nplt.show()",
"216\n"
],
[
"print(len(list(training_data)))\nprint(len(list(validation_data)))\nprint(len(list(test_data)))",
"2\n2\n2\n"
],
[
"training_data, validation_data, test_data = mnist_loader.load_data_wrapper()",
"_____no_output_____"
],
[
"training_data = list(training_data)\nvalidation_data = list(validation_data)\ntest_data = list(test_data)",
"_____no_output_____"
],
[
"print(len(training_data))\nprint(len(validation_data))\nprint(len(test_data))",
"2\n2\n2\n"
],
[
"print(len(training_data))\nprint(len(validation_data))\nprint(len(test_data))",
"50000\n10000\n10000\n"
],
[
"print(training_data)",
"(array([[0., 0., 0., ..., 0., 0., 0.],\n [0., 0., 0., ..., 0., 0., 0.],\n [0., 0., 0., ..., 0., 0., 0.],\n ...,\n [0., 0., 0., ..., 0., 0., 0.],\n [0., 0., 0., ..., 0., 0., 0.],\n [0., 0., 0., ..., 0., 0., 0.]], dtype=float32), array([5, 0, 4, ..., 8, 4, 8]))\n"
],
[
"print(training_data[0][1])",
"[0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0.19921875 0.62109375 0.98828125 0.62109375 0.1953125\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0.1875 0.9296875\n 0.984375 0.984375 0.984375 0.92578125 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0.2109375 0.88671875 0.98828125 0.984375 0.93359375\n 0.91015625 0.984375 0.22265625 0.0234375 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0.0390625 0.234375 0.875\n 0.984375 0.98828125 0.984375 0.7890625 0.328125 0.984375\n 0.98828125 0.4765625 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0.63671875 0.984375 0.984375 0.984375 0.98828125\n 0.984375 0.984375 0.375 0.73828125 0.98828125 0.65234375\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0.19921875 0.9296875\n 0.98828125 0.98828125 0.7421875 0.4453125 0.98828125 0.890625\n 0.18359375 0.30859375 0.99609375 0.65625 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0.1875 0.9296875 0.984375 0.984375 0.69921875\n 0.046875 0.29296875 0.47265625 0.08203125 0. 0.\n 0.98828125 0.94921875 0.1953125 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0.1484375 0.64453125\n 0.98828125 0.91015625 0.8125 0.328125 0. 0.\n 0. 0. 0. 0. 0.98828125 0.984375\n 0.64453125 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0.02734375 0.6953125 0.984375 0.9375 0.27734375\n 0.07421875 0.109375 0. 0. 0. 0.\n 0. 0. 0.98828125 0.984375 0.76171875 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.22265625\n 0.984375 0.984375 0.24609375 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0.98828125 0.984375 0.76171875 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0.7734375 0.98828125 0.7421875\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0.99609375 0.98828125\n 0.765625 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0.296875 0.9609375 0.984375 0.4375 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0.98828125 0.984375 0.578125 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0.33203125 0.984375\n 0.8984375 0.09765625 0. 0. 0. 0.\n 0. 0. 0. 0. 0.02734375 0.52734375\n 0.98828125 0.7265625 0.046875 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0.33203125 0.984375 0.87109375 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0.02734375 0.51171875 0.984375 0.87890625 0.27734375\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0.33203125 0.984375 0.56640625 0. 0. 0.\n 0. 0. 0. 0. 0.1875 0.64453125\n 0.984375 0.67578125 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0.3359375 0.98828125\n 0.87890625 0. 0. 0. 0. 0.\n 0. 0.4453125 0.9296875 0.98828125 0.6328125 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0.33203125 0.984375 0.97265625 0.5703125\n 0.1875 0.11328125 0.33203125 0.6953125 0.87890625 0.98828125\n 0.87109375 0.65234375 0.21875 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0.33203125 0.984375 0.984375 0.984375 0.89453125 0.83984375\n 0.984375 0.984375 0.984375 0.765625 0.5078125 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0.109375 0.77734375\n 0.984375 0.984375 0.98828125 0.984375 0.984375 0.91015625\n 0.56640625 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0.09765625 0.5 0.984375\n 0.98828125 0.984375 0.55078125 0.14453125 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. ]\n"
],
[
"training_data.shape",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05ed899c9a1edf96178def8c358f806ffb2e9b1 | 11,030 | ipynb | Jupyter Notebook | docs/refactoring/performance/asyncio-example.ipynb | veit/jupyter-tutorial-de | abe8cd576125d4c860bf17e9d4dc1e19614f79f1 | [
"BSD-3-Clause"
] | null | null | null | docs/refactoring/performance/asyncio-example.ipynb | veit/jupyter-tutorial-de | abe8cd576125d4c860bf17e9d4dc1e19614f79f1 | [
"BSD-3-Clause"
] | 3 | 2021-01-01T16:13:59.000Z | 2021-07-27T15:41:39.000Z | docs/refactoring/performance/asyncio-example.ipynb | veit/jupyter-tutorial-de | abe8cd576125d4c860bf17e9d4dc1e19614f79f1 | [
"BSD-3-Clause"
] | null | null | null | 26.771845 | 327 | 0.55893 | [
[
[
"# `asyncio` Beispiel\n\nAb IPython≥7.0 könnt ihr `asyncio` direkt in Jupyter Notebooks verwenden; seht auch [IPython 7.0, Async REPL](https://blog.jupyter.org/ipython-7-0-async-repl-a35ce050f7f7).",
"_____no_output_____"
],
[
"Wenn ihr die Fehlermeldung `RuntimeError: This event loop is already running` erhaltet, hilft euch vielleicht [nest-asyncio] weiter.\n\nIhr könnt das Paket in eurer Jupyter- oder JupyterHub-Umgebung installieren mit\n\n```bash\n$ pipenv install nest-asyncio\n```\n\nIhr könnt es dann in euer Notebook importieren und verwenden mit:",
"_____no_output_____"
]
],
[
[
"import nest_asyncio\n\nnest_asyncio.apply()",
"_____no_output_____"
]
],
[
[
"## Einfaches *Hello world*-Beispiel",
"_____no_output_____"
]
],
[
[
"import asyncio\n\nasync def hello():\n print('Hello')\n await asyncio.sleep(1)\n print('world')\n\nawait hello()",
"Hello\nworld\n"
]
],
[
[
"## Ein bisschen näher an einem realen Beispiel",
"_____no_output_____"
]
],
[
[
"import asyncio\nimport random\n\n\nasync def produce(queue, n):\n for x in range(1, n + 1):\n # produce an item\n print('producing {}/{}'.format(x, n))\n # simulate i/o operation using sleep\n await asyncio.sleep(random.random())\n item = str(x)\n # put the item in the queue\n await queue.put(item)\n\n # indicate the producer is done\n await queue.put(None)\n\n\nasync def consume(queue):\n while True:\n # wait for an item from the producer\n item = await queue.get()\n if item is None:\n # the producer emits None to indicate that it is done\n break\n\n # process the item\n print('consuming {}'.format(item))\n # simulate i/o operation using sleep\n await asyncio.sleep(random.random())\n\n\nloop = asyncio.get_event_loop()\nqueue = asyncio.Queue(loop=loop)\nasyncio.ensure_future(produce(queue, 10), loop=loop)\nloop.run_until_complete(consume(queue))",
"producing 1/10\nproducing 2/10\nconsuming 1\nproducing 3/10\nconsuming 2\nproducing 4/10\nconsuming 3\nproducing 5/10\nconsuming 4\nproducing 6/10\nconsuming 5\nproducing 7/10\nconsuming 6\nproducing 8/10\nconsuming 7\nproducing 9/10\nconsuming 8\nproducing 10/10\nconsuming 9\nconsuming 10\n"
]
],
[
[
"## Ausnahmebehandlung\n\n> **Siehe auch:** [set_exception_handler](https://docs.python.org/3/library/asyncio-eventloop.html#asyncio.loop.set_exception_handler)",
"_____no_output_____"
]
],
[
[
"def main():\n loop = asyncio.get_event_loop()\n # May want to catch other signals too\n signals = (signal.SIGHUP, signal.SIGTERM, signal.SIGINT)\n for s in signals:\n loop.add_signal_handler(\n s, lambda s=s: asyncio.create_task(shutdown(loop, signal=s)))\n loop.set_exception_handler(handle_exception)\n queue = asyncio.Queue()",
"_____no_output_____"
]
],
[
[
"## Testen mit `pytest`",
"_____no_output_____"
],
[
"### Beispiel:",
"_____no_output_____"
]
],
[
[
"import pytest\n\[email protected]\nasync def test_consume(mock_get, mock_queue, message, create_mock_coro):\n mock_get.side_effect = [message, Exception(\"break while loop\")]\n\n with pytest.raises(Exception, match=\"break while loop\"):\n await consume(mock_queue)",
"_____no_output_____"
]
],
[
[
"### Bibliotheken von Drittanbietern\n\n* [pytest-asyncio](https://github.com/pytest-dev/pytest-asyncio) hat hilfreiche Dinge wie Test-Fixtures für `event_loop`, `unused_tcp_port`, und `unused_tcp_port_factory`; und die Möglichkeit zum Erstellen eurer eigenen [asynchronen Fixtures](https://github.com/pytest-dev/pytest-asyncio/#async-fixtures).\n* [asynctest](https://asynctest.readthedocs.io/en/latest/index.html) verfügt über hilfreiche Werkzeuge, einschließlich Coroutine-Mocks und [exhaust_callbacks](https://asynctest.readthedocs.io/en/latest/asynctest.helpers.html#asynctest.helpers.exhaust_callbacks) so dass wir `await task` nicht manuell erstellen müssen.\n* [aiohttp](https://docs.aiohttp.org/en/stable/) hat ein paar wirklich nette eingebaute Test-Utilities.",
"_____no_output_____"
],
[
"## Debugging\n\n`asyncio` hat bereits einen [debug mode](https://docs.python.org/3.6/library/asyncio-dev.html#debug-mode-of-asyncio) in der Standardbibliothek. Ihr könnt ihn einfach mit der Umgebungsvariablen `PYTHONASYNCIODEBUG` oder im Code mit `loop.set_debug(True)` aktivieren.",
"_____no_output_____"
],
[
"### Verwendet den Debug-Modus zum Identifizieren langsamer asynchroner Aufrufe\n\nDer Debug-Modus von `asyncio` hat einen kleinen eingebauten Profiler. Wenn der Debug-Modus aktiviert ist, protokolliert `asyncio` alle asynchronen Aufrufe, die länger als 100 Millisekunden dauern.",
"_____no_output_____"
],
[
"### Debugging im Produktivbetrieb mit `aiodebug`\n\n[aiodebug](https://github.com/qntln/aiodebug) ist eine kleine Bibliothek zum Überwachen und Testen von Asyncio-Programmen.",
"_____no_output_____"
],
[
"#### Beispiel",
"_____no_output_____"
]
],
[
[
"from aiodebug import log_slow_callbacks\n\ndef main():\n loop = asyncio.get_event_loop()\n log_slow_callbacks.enable(0.05)",
"_____no_output_____"
]
],
[
[
"## Logging\n\n[aiologger](https://github.com/b2wdigital/aiologger) ermöglicht eine nicht-blockierendes Logging.",
"_____no_output_____"
],
[
"## Asynchrone Widgets\n\n> **Seht auch:** [Asynchronous Widgets](https://ipywidgets.readthedocs.io/en/stable/examples/Widget%20Asynchronous.html)",
"_____no_output_____"
]
],
[
[
"def wait_for_change(widget, value):\n future = asyncio.Future()\n def getvalue(change):\n # make the new value available\n future.set_result(change.new)\n widget.unobserve(getvalue, value)\n widget.observe(getvalue, value)\n return future",
"_____no_output_____"
],
[
"from ipywidgets import IntSlider\nslider = IntSlider()\n\nasync def f():\n for i in range(10):\n print('did work %s'%i)\n x = await wait_for_change(slider, 'value')\n print('async function continued with value %s'%x)\nasyncio.ensure_future(f())\n\nslider",
"_____no_output_____"
]
],
[
[
"## Weiterlesen\n\n* Lynn Root: [asyncio: We Did It Wrong](https://www.roguelynn.com/words/asyncio-we-did-it-wrong/)\n* Mike Driscoll: [An Intro to asyncio](https://www.blog.pythonlibrary.org/2016/07/26/python-3-an-intro-to-asyncio/)\n* Yeray Diaz: [Asyncio Coroutine Patterns: Beyond await](https://medium.com/python-pandemonium/asyncio-coroutine-patterns-beyond-await-a6121486656f)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d05ee64c8fedbaf5ff40af385d49ce781e1fda8b | 26,829 | ipynb | Jupyter Notebook | B - A Closer Look at Word Embeddings.ipynb | Andrews2017/pytorch-sentiment-analysis | bc5b2e0221abc289aee789c02812bb57495be7ff | [
"MIT"
] | 3,595 | 2018-03-26T19:44:39.000Z | 2022-03-31T08:45:12.000Z | B - A Closer Look at Word Embeddings.ipynb | v-mk-s/pytorch-sentiment-analysis | b4efbefa47672174394a8b6a27d4e7bc193bc224 | [
"MIT"
] | 105 | 2018-09-07T14:49:27.000Z | 2022-03-01T05:43:11.000Z | B - A Closer Look at Word Embeddings.ipynb | v-mk-s/pytorch-sentiment-analysis | b4efbefa47672174394a8b6a27d4e7bc193bc224 | [
"MIT"
] | 1,066 | 2018-06-12T00:58:01.000Z | 2022-03-27T09:03:54.000Z | 29.353392 | 612 | 0.573931 | [
[
[
"# B - A Closer Look at Word Embeddings\n\nWe have very briefly covered how word embeddings (also known as word vectors) are used in the tutorials. In this appendix we'll have a closer look at these embeddings and find some (hopefully) interesting results.\n\nEmbeddings transform a one-hot encoded vector (a vector that is 0 in elements except one, which is 1) into a much smaller dimension vector of real numbers. The one-hot encoded vector is also known as a *sparse vector*, whilst the real valued vector is known as a *dense vector*. \n\nThe key concept in these word embeddings is that words that appear in similar _contexts_ appear nearby in the vector space, i.e. the Euclidean distance between these two word vectors is small. By context here, we mean the surrounding words. For example in the sentences \"I purchased some items at the shop\" and \"I purchased some items at the store\" the words 'shop' and 'store' appear in the same context and thus should be close together in vector space.\n\nYou may have also heard about *word2vec*. *word2vec* is an algorithm (actually a bunch of algorithms) that calculates word vectors from a corpus. In this appendix we use *GloVe* vectors, *GloVe* being another algorithm to calculate word vectors. If you want to know how *word2vec* works, check out a two part series [here](http://mccormickml.com/2016/04/19/word2vec-tutorial-the-skip-gram-model/) and [here](http://mccormickml.com/2017/01/11/word2vec-tutorial-part-2-negative-sampling/), and if you want to find out more about *GloVe*, check the website [here](https://nlp.stanford.edu/projects/glove/).\n\nIn PyTorch, we use word vectors with the `nn.Embedding` layer, which takes a _**[sentence length, batch size]**_ tensor and transforms it into a _**[sentence length, batch size, embedding dimensions]**_ tensor.\n\nIn tutorial 2 onwards, we also used pre-trained word embeddings (specifically the GloVe vectors) provided by TorchText. These embeddings have been trained on a gigantic corpus. We can use these pre-trained vectors within any of our models, with the idea that as they have already learned the context of each word they will give us a better starting point for our word vectors. This usually leads to faster training time and/or improved accuracy.\n\nIn this appendix we won't be training any models, instead we'll be looking at the word embeddings and finding a few interesting things about them.\n\nA lot of the code from the first half of this appendix is taken from [here](https://github.com/spro/practical-pytorch/blob/master/glove-word-vectors/glove-word-vectors.ipynb). For more information about word embeddings, go [here](https://monkeylearn.com/blog/word-embeddings-transform-text-numbers/). \n\n## Loading the GloVe vectors\n\nFirst, we'll load the GloVe vectors. The `name` field specifies what the vectors have been trained on, here the `6B` means a corpus of 6 billion words. The `dim` argument specifies the dimensionality of the word vectors. GloVe vectors are available in 50, 100, 200 and 300 dimensions. There is also a `42B` and `840B` glove vectors, however they are only available at 300 dimensions.",
"_____no_output_____"
]
],
[
[
"import torchtext.vocab\n\nglove = torchtext.vocab.GloVe(name = '6B', dim = 100)\n\nprint(f'There are {len(glove.itos)} words in the vocabulary')",
"There are 400000 words in the vocabulary\n"
]
],
[
[
"As shown above, there are 400,000 unique words in the GloVe vocabulary. These are the most common words found in the corpus the vectors were trained on. **In these set of GloVe vectors, every single word is lower-case only.**\n\n`glove.vectors` is the actual tensor containing the values of the embeddings.",
"_____no_output_____"
]
],
[
[
"glove.vectors.shape",
"_____no_output_____"
]
],
[
[
"We can see what word is associated with each row by checking the `itos` (int to string) list. \n\nBelow implies that row 0 is the vector associated with the word 'the', row 1 for ',' (comma), row 2 for '.' (period), etc.",
"_____no_output_____"
]
],
[
[
"glove.itos[:10]",
"_____no_output_____"
]
],
[
[
"We can also use the `stoi` (string to int) dictionary, in which we input a word and receive the associated integer/index. If you try get the index of a word that is not in the vocabulary, you receive an error.",
"_____no_output_____"
]
],
[
[
"glove.stoi['the']",
"_____no_output_____"
]
],
[
[
"We can get the vector of a word by first getting the integer associated with it and then indexing into the word embedding tensor with that index.",
"_____no_output_____"
]
],
[
[
"glove.vectors[glove.stoi['the']].shape",
"_____no_output_____"
]
],
[
[
"We'll be doing this a lot, so we'll create a function that takes in word embeddings and a word then returns the associated vector. It'll also throw an error if the word doesn't exist in the vocabulary.",
"_____no_output_____"
]
],
[
[
"def get_vector(embeddings, word):\n assert word in embeddings.stoi, f'*{word}* is not in the vocab!'\n return embeddings.vectors[embeddings.stoi[word]]",
"_____no_output_____"
]
],
[
[
"As before, we use a word to get the associated vector.",
"_____no_output_____"
]
],
[
[
"get_vector(glove, 'the').shape",
"_____no_output_____"
]
],
[
[
"## Similar Contexts\n\nNow to start looking at the context of different words. \n\nIf we want to find the words similar to a certain input word, we first find the vector of this input word, then we scan through our vocabulary calculating the distance between the vector of each word and our input word vector. We then sort these from closest to furthest away.\n\nThe function below returns the closest 10 words to an input word vector:",
"_____no_output_____"
]
],
[
[
"import torch\n\ndef closest_words(embeddings, vector, n = 10):\n \n distances = [(word, torch.dist(vector, get_vector(embeddings, word)).item())\n for word in embeddings.itos]\n \n return sorted(distances, key = lambda w: w[1])[:n]",
"_____no_output_____"
]
],
[
[
"Let's try it out with 'korea'. The closest word is the word 'korea' itself (not very interesting), however all of the words are related in some way. Pyongyang is the capital of North Korea, DPRK is the official name of North Korea, etc.\n\nInterestingly, we also get 'Japan' and 'China', implies that Korea, Japan and China are frequently talked about together in similar contexts. This makes sense as they are geographically situated near each other. ",
"_____no_output_____"
]
],
[
[
"word_vector = get_vector(glove, 'korea')\n\nclosest_words(glove, word_vector)",
"_____no_output_____"
]
],
[
[
"Looking at another country, India, we also get nearby countries: Thailand, Malaysia and Sri Lanka (as two separate words). Australia is relatively close to India (geographically), but Thailand and Malaysia are closer. So why is Australia closer to India in vector space? This is most probably due to India and Australia appearing in the context of [cricket](https://en.wikipedia.org/wiki/Cricket) matches together.",
"_____no_output_____"
]
],
[
[
"word_vector = get_vector(glove, 'india')\n\nclosest_words(glove, word_vector)",
"_____no_output_____"
]
],
[
[
"We'll also create another function that will nicely print out the tuples returned by our `closest_words` function.",
"_____no_output_____"
]
],
[
[
"def print_tuples(tuples):\n for w, d in tuples:\n print(f'({d:02.04f}) {w}') ",
"_____no_output_____"
]
],
[
[
"A final word to look at, 'sports'. As we can see, the closest words are most of the sports themselves. ",
"_____no_output_____"
]
],
[
[
"word_vector = get_vector(glove, 'sports')\n\nprint_tuples(closest_words(glove, word_vector))",
"(0.0000) sports\n(3.5875) sport\n(4.4590) soccer\n(4.6508) basketball\n(4.6561) baseball\n(4.8028) sporting\n(4.8763) football\n(4.9624) professional\n(4.9824) entertainment\n(5.0975) media\n"
]
],
[
[
"## Analogies\n\nAnother property of word embeddings is that they can be operated on just as any standard vector and give interesting results.\n\nWe'll show an example of this first, and then explain it:",
"_____no_output_____"
]
],
[
[
"def analogy(embeddings, word1, word2, word3, n=5):\n \n #get vectors for each word\n word1_vector = get_vector(embeddings, word1)\n word2_vector = get_vector(embeddings, word2)\n word3_vector = get_vector(embeddings, word3)\n \n #calculate analogy vector\n analogy_vector = word2_vector - word1_vector + word3_vector\n \n #find closest words to analogy vector\n candidate_words = closest_words(embeddings, analogy_vector, n+3)\n \n #filter out words already in analogy\n candidate_words = [(word, dist) for (word, dist) in candidate_words \n if word not in [word1, word2, word3]][:n]\n \n print(f'{word1} is to {word2} as {word3} is to...')\n \n return candidate_words",
"_____no_output_____"
],
[
"print_tuples(analogy(glove, 'man', 'king', 'woman'))",
"man is to king as woman is to...\n(4.0811) queen\n(4.6429) monarch\n(4.9055) throne\n(4.9216) elizabeth\n(4.9811) prince\n"
]
],
[
[
"This is the canonical example which shows off this property of word embeddings. So why does it work? Why does the vector of 'woman' added to the vector of 'king' minus the vector of 'man' give us 'queen'?\n\nIf we think about it, the vector calculated from 'king' minus 'man' gives us a \"royalty vector\". This is the vector associated with traveling from a man to his royal counterpart, a king. If we add this \"royality vector\" to 'woman', this should travel to her royal equivalent, which is a queen!\n\nWe can do this with other analogies too. For example, this gets an \"acting career vector\":",
"_____no_output_____"
]
],
[
[
"print_tuples(analogy(glove, 'man', 'actor', 'woman'))",
"man is to actor as woman is to...\n(2.8133) actress\n(5.0039) comedian\n(5.1399) actresses\n(5.2773) starred\n(5.3085) screenwriter\n"
]
],
[
[
"For a \"baby animal vector\":",
"_____no_output_____"
]
],
[
[
"print_tuples(analogy(glove, 'cat', 'kitten', 'dog'))",
"cat is to kitten as dog is to...\n(3.8146) puppy\n(4.2944) rottweiler\n(4.5888) puppies\n(4.6086) pooch\n(4.6520) pug\n"
]
],
[
[
"A \"capital city vector\":",
"_____no_output_____"
]
],
[
[
"print_tuples(analogy(glove, 'france', 'paris', 'england'))",
"france is to paris as england is to...\n(4.1426) london\n(4.4938) melbourne\n(4.7087) sydney\n(4.7630) perth\n(4.7952) birmingham\n"
]
],
[
[
"A \"musician's genre vector\":",
"_____no_output_____"
]
],
[
[
"print_tuples(analogy(glove, 'elvis', 'rock', 'eminem'))",
"elvis is to rock as eminem is to...\n(5.6597) rap\n(6.2057) rappers\n(6.2161) rapper\n(6.2444) punk\n(6.2690) hop\n"
]
],
[
[
"And an \"ingredient vector\":",
"_____no_output_____"
]
],
[
[
"print_tuples(analogy(glove, 'beer', 'barley', 'wine'))",
"beer is to barley as wine is to...\n(5.6021) grape\n(5.6760) beans\n(5.8174) grapes\n(5.9035) lentils\n(5.9454) figs\n"
]
],
[
[
"## Correcting Spelling Mistakes\n\nAnother interesting property of word embeddings is that they can actually be used to correct spelling mistakes! \n\nWe'll put their findings into code and briefly explain them, but to read more about this, check out the [original thread](http://forums.fast.ai/t/nlp-any-libraries-dictionaries-out-there-for-fixing-common-spelling-errors/16411) and the associated [write-up](https://blog.usejournal.com/a-simple-spell-checker-built-from-word-vectors-9f28452b6f26).\n\nFirst, we need to load up the much larger vocabulary GloVe vectors, this is due to the spelling mistakes not appearing in the smaller vocabulary. \n\n**Note**: these vectors are very large (~2GB), so watch out if you have a limited internet connection.",
"_____no_output_____"
]
],
[
[
"glove = torchtext.vocab.GloVe(name = '840B', dim = 300)",
"_____no_output_____"
]
],
[
[
"Checking the vocabulary size of these embeddings, we can see we now have over 2 million unique words in our vocabulary!",
"_____no_output_____"
]
],
[
[
"glove.vectors.shape",
"_____no_output_____"
]
],
[
[
"As the vectors were trained with a much larger vocabulary on a larger corpus of text, the words that appear are a little different. Notice how the words 'north', 'south', 'pyongyang' and 'dprk' no longer appear in the most closest words to 'korea'.",
"_____no_output_____"
]
],
[
[
"word_vector = get_vector(glove, 'korea')\n\nprint_tuples(closest_words(glove, word_vector))",
"(0.0000) korea\n(3.9857) taiwan\n(4.4022) korean\n(4.9016) asia\n(4.9593) japan\n(5.0721) seoul\n(5.4058) thailand\n(5.6025) singapore\n(5.7010) russia\n(5.7240) hong\n"
]
],
[
[
"Our first step to correcting spelling mistakes is looking at the vector for a misspelling of the word 'reliable'.",
"_____no_output_____"
]
],
[
[
"word_vector = get_vector(glove, 'relieable')\n\nprint_tuples(closest_words(glove, word_vector))",
"(0.0000) relieable\n(5.0366) relyable\n(5.2610) realible\n(5.4719) realiable\n(5.5402) relable\n(5.5917) relaible\n(5.6412) reliabe\n(5.8802) relaiable\n(5.9593) stabel\n(5.9981) consitant\n"
]
],
[
[
"Notice how the correct spelling, \"reliable\", does not appear in the top 10 closest words. Surely the misspellings of a word should appear next to the correct spelling of the word as they appear in the same context, right? \n\nThe hypothesis is that misspellings of words are all equally shifted away from their correct spelling. This is because articles of text that contain spelling mistakes are usually written in an informal manner where correct spelling doesn't matter as much (such as tweets/blog posts), thus spelling errors will appear together as they appear in context of informal articles.\n\nSimilar to how we created analogies before, we can create a \"correct spelling\" vector. This time, instead of using a single example to create our vector, we'll use the average of multiple examples. This will hopefully give better accuracy!\n\nWe first create a vector for the correct spelling, 'reliable', then calculate the difference between the \"reliable vector\" and each of the 8 misspellings of 'reliable'. As we are going to concatenate these 8 misspelling tensors together we need to unsqueeze a \"batch\" dimension to them.",
"_____no_output_____"
]
],
[
[
"reliable_vector = get_vector(glove, 'reliable')\n\nreliable_misspellings = ['relieable', 'relyable', 'realible', 'realiable', \n 'relable', 'relaible', 'reliabe', 'relaiable']\n\ndiff_reliable = [(reliable_vector - get_vector(glove, s)).unsqueeze(0) \n for s in reliable_misspellings]",
"_____no_output_____"
]
],
[
[
"We take the average of these 8 'difference from reliable' vectors to get our \"misspelling vector\".",
"_____no_output_____"
]
],
[
[
"misspelling_vector = torch.cat(diff_reliable, dim = 0).mean(dim = 0)",
"_____no_output_____"
]
],
[
[
"We can now correct other spelling mistakes using this \"misspelling vector\" by finding the closest words to the sum of the vector of a misspelled word and the \"misspelling vector\".\n\nFor a misspelling of \"because\":",
"_____no_output_____"
]
],
[
[
"word_vector = get_vector(glove, 'becuase')\n\nprint_tuples(closest_words(glove, word_vector + misspelling_vector))",
"(6.1090) because\n(6.4250) even\n(6.4358) fact\n(6.4914) sure\n(6.5094) though\n(6.5601) obviously\n(6.5682) reason\n(6.5856) if\n(6.6099) but\n(6.6415) why\n"
]
],
[
[
"For a misspelling of \"definitely\":",
"_____no_output_____"
]
],
[
[
"word_vector = get_vector(glove, 'defintiely')\n\nprint_tuples(closest_words(glove, word_vector + misspelling_vector))",
"(5.4070) definitely\n(5.5643) certainly\n(5.7192) sure\n(5.8152) well\n(5.8588) always\n(5.8812) also\n(5.9557) simply\n(5.9667) consider\n(5.9821) probably\n(5.9948) definately\n"
]
],
[
[
"For a misspelling of \"consistent\":",
"_____no_output_____"
]
],
[
[
"word_vector = get_vector(glove, 'consistant')\n\nprint_tuples(closest_words(glove, word_vector + misspelling_vector))",
"(5.9641) consistent\n(6.3674) reliable\n(7.0195) consistant\n(7.0299) consistently\n(7.1605) accurate\n(7.2737) fairly\n(7.3037) good\n(7.3520) reasonable\n(7.3801) dependable\n(7.4027) ensure\n"
]
],
[
[
"For a misspelling of \"package\":",
"_____no_output_____"
]
],
[
[
"word_vector = get_vector(glove, 'pakage')\n\nprint_tuples(closest_words(glove, word_vector + misspelling_vector))",
"(6.6117) package\n(6.9315) packages\n(7.0195) pakage\n(7.0911) comes\n(7.1241) provide\n(7.1469) offer\n(7.1861) reliable\n(7.2431) well\n(7.2434) choice\n(7.2453) offering\n"
]
],
[
[
"For a more in-depth look at this, check out the [write-up](https://blog.usejournal.com/a-simple-spell-checker-built-from-word-vectors-9f28452b6f26).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d05eed03361b69b61566efd17f416510ff868cf1 | 133,258 | ipynb | Jupyter Notebook | 2. Best Time To Visit Munich.ipynb | rmnng/dsblogpost | 568fe2fc455be5119cdf78466831fcd2c47a57ab | [
"MIT"
] | null | null | null | 2. Best Time To Visit Munich.ipynb | rmnng/dsblogpost | 568fe2fc455be5119cdf78466831fcd2c47a57ab | [
"MIT"
] | null | null | null | 2. Best Time To Visit Munich.ipynb | rmnng/dsblogpost | 568fe2fc455be5119cdf78466831fcd2c47a57ab | [
"MIT"
] | null | null | null | 116.078397 | 84,508 | 0.837263 | [
[
[
"### This notebook explores the calendar of Munich listings to answer the question: \n## What is the most expensive and the cheapest time to visit Munich?",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom matplotlib import pyplot as plt\nimport seaborn as sns\nsns.set()",
"_____no_output_____"
],
[
"LOCATION = 'munich'\n\ndf_list = pd.read_csv(LOCATION + '/listings.csv.gz')\ndf_reviews = pd.read_csv(LOCATION + '/reviews.csv.gz')\ndf_cal = pd.read_csv(LOCATION + '/calendar.csv.gz')",
"_____no_output_____"
],
[
"pd.options.display.max_rows=10\npd.options.display.max_columns=None\npd.options.display.max_colwidth=30",
"_____no_output_____"
]
],
[
[
"___",
"_____no_output_____"
],
[
"### Calendar",
"_____no_output_____"
],
[
"#### First look into to data and types for each column:",
"_____no_output_____"
]
],
[
[
"df_cal",
"_____no_output_____"
],
[
"df_cal.dtypes",
"_____no_output_____"
]
],
[
[
"___",
"_____no_output_____"
],
[
"#### Some data types are wrong. In order to be able to work with the data, we need to change some datatypes.",
"_____no_output_____"
],
[
"First convert **date** to *datetime* type:",
"_____no_output_____"
]
],
[
[
"df_cal['date'] = pd.to_datetime(df_cal['date'])",
"_____no_output_____"
]
],
[
[
"**Price** needs to converted to *float* in order to be able to work with it.",
"_____no_output_____"
]
],
[
[
"df_cal['price']=df_cal['price'].replace(to_replace='[\\$,]', value='', regex=True).astype(float)\ndf_cal['adjusted_price']=df_cal['adjusted_price'].replace(to_replace='[\\$,]', value='', regex=True).astype(float)",
"_____no_output_____"
]
],
[
[
"This is how it looks now:",
"_____no_output_____"
]
],
[
[
"df_cal.head()",
"_____no_output_____"
]
],
[
[
"And this are the corrected data types:",
"_____no_output_____"
]
],
[
[
"df_cal.dtypes",
"_____no_output_____"
]
],
[
[
"___",
"_____no_output_____"
],
[
"### First question to be answered is, what is the price distribution over the year:",
"_____no_output_____"
],
[
"Let's calculate the mean price over all listings for each day of the year:",
"_____no_output_____"
],
[
"First check if we have *NULL* values in the data frame.",
"_____no_output_____"
]
],
[
[
"df_cal.isnull().sum()",
"_____no_output_____"
]
],
[
[
"*NULL* values have impact to the average (even if very small due to the small number of missing values), let's drop all rows with *NULL* **price**.",
"_____no_output_____"
]
],
[
[
"df_cal.dropna(subset=['price'], inplace=True)",
"_____no_output_____"
]
],
[
[
"Now let's group all listings by **date** and calculate the average **price** of all listings for each day:",
"_____no_output_____"
]
],
[
[
"mean_price = df_cal[['date', 'price']].groupby(by='date').mean().reset_index()",
"_____no_output_____"
],
[
"mean_price",
"_____no_output_____"
]
],
[
[
"And plot the result:",
"_____no_output_____"
]
],
[
[
"## use the plot method and scale the size based on ploted values\nscale_from = mean_price['price'][1:-2].min()*0.95\nscale_to = mean_price['price'][1:-2].max()*1.02\n\nmean_price.set_index('date')[1:-2].plot(kind='line', y='price', figsize=(20,10), grid=True).set_ylim(scale_from, scale_to);",
"_____no_output_____"
]
],
[
[
"### HERE WE ARE! There are two interesting observations:\n#### 1. There is a peak in the second half of September: **\"Welcome to the Octoberfest!**\"\n#### 2. The price apparently depends on the day of week. Let's have a closer look at it.",
"_____no_output_____"
],
[
"___",
"_____no_output_____"
],
[
"### Second question: What is the price distribution within a week?",
"_____no_output_____"
],
[
"To be able to have a close look at prices, let's introduce the **day_of_week** column.",
"_____no_output_____"
]
],
[
[
"df_cal['day_of_week'] = df_cal['date'].dt.dayofweek",
"_____no_output_____"
]
],
[
[
"Let's group the prices for each day of week and get the average price:",
"_____no_output_____"
]
],
[
[
"mean_price_dow = df_cal[['day_of_week', 'price']].groupby(by='day_of_week').mean().reset_index()",
"_____no_output_____"
],
[
"mean_price_dow",
"_____no_output_____"
]
],
[
[
"It's difficult to interpret index-based day of week. Let's convert it to strings from Monday to Sunday:",
"_____no_output_____"
]
],
[
[
"def convert_day_of_week(day_idx):\n '''\n This function convert index based day of week to string\n 0 - Monday\n 6 - Sunday\n \n if the day_idx oís out of this range, this index will be returned\n ''' \n \n if(day_idx>6 or day_idx<0):\n return day_idx\n \n lst = ['Monday', 'Tuesday', 'Wednesday', 'Thursday', 'Friday', 'Saturday', 'Sunday']\n return lst[day_idx]",
"_____no_output_____"
],
[
"mean_price_dow['day_of_week'] = mean_price_dow['day_of_week'].apply(convert_day_of_week)",
"_____no_output_____"
],
[
"mean_price_dow",
"_____no_output_____"
]
],
[
[
"#### Now we can plot the result:",
"_____no_output_____"
]
],
[
[
"scale_from = mean_price_dow['price'].min()*0.95\nscale_to = mean_price_dow['price'].max()*1.02\n\nsns.set(rc={'figure.figsize':(15,5)})\nfig = sns.barplot(data=mean_price_dow, x='day_of_week', y='price', color='#0080bb');\nfig.set_ylim(scale_from, scale_to);\nfig.set_title('Prices for day of the week');\nfig.set_xlabel('Day of week');\nfig.set_ylabel('Price');",
"_____no_output_____"
]
],
[
[
"#### No surprise, the most expensive days are Friday and Saturday. The weekend in Munich can start!",
"_____no_output_____"
],
[
"___",
"_____no_output_____"
],
[
"## What is the most expensive and the cheapest time to visit Munich?",
"_____no_output_____"
],
[
"#### If you want to save your money, don't visit Munich during the Octoberfest, which is end of September, beginning of October. Do it better in April, it's much cheaper and you can enjoy green Munich. ",
"_____no_output_____"
],
[
"#### If you plan a short trip and you are not constrained by day of week, in order to save the money, Sunday to Thursday is the best choice for you.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d05ef2bd66bed06806504e8b1a4ec20943c75d60 | 20,195 | ipynb | Jupyter Notebook | doc/matplotlib/quickstart.ipynb | huifer/python_doc | 4b98a84cda656e8f28bafcd3f1507f8da82f3c96 | [
"Apache-2.0"
] | 2 | 2019-11-01T16:32:05.000Z | 2020-06-06T10:05:46.000Z | doc/matplotlib/quickstart.ipynb | huifer/python_doc | 4b98a84cda656e8f28bafcd3f1507f8da82f3c96 | [
"Apache-2.0"
] | 1 | 2019-10-30T00:40:47.000Z | 2020-01-23T06:35:24.000Z | doc/matplotlib/quickstart.ipynb | huifer/python_doc | 4b98a84cda656e8f28bafcd3f1507f8da82f3c96 | [
"Apache-2.0"
] | null | null | null | 137.380952 | 16,695 | 0.87279 | [
[
[
"# Matplotlib 快速入门\n## Matplotlib 是什么\n- 官方文档: https://matplotlib.org\n> Matplotlib is a Python 2D plotting library which produces publication-quality figures in a variety of hardcopy formats and interactive environments across platforms. Matplotlib can be used in Python scripts, the Python and IPython shell (à la MATLAB or Mathematica), web application servers, and various graphical user interface toolkits.\n- Matplotlib 在 Python 中作为一个绘制 2D 图像的库\n\n## 为什么用 Matplotlib\n1. 支持图像种类多\n1. 绘图简单\n\n## 安装\n`pip install matplotlib`\n\n## 核心\n- `matplotlib.pyplot` 绘图核心\n- 布局\n- 颜色\n- 图例\n- 文本\n- 交互\n- 输出\n\n## 术语\n\n## Title\n> 标题\n## Major tick\n> 刻度\n## Legend\n> 图例\n## Major tick label\n> 刻度上的标记\n## Grid\n> 后置网格\n\n## Line \n> Plot的一种折线图\n## Markers\n> 标记(可用散点图绘制)\n\n## Y axis label\n> 纵坐标标注\n## X axis label \n> 横坐标标注\n## Spines \n> 绘制范围\n\n| 英文 | 中文 |\n| ----------------- | --------- |\n| Annotation | 标注 |\n| Artist | 艺术家 |\n| Axes | 轴域 |\n| Axis | 轴/坐标轴 |\n| Bézier | 贝塞尔 |\n| Coordinate | 坐标 |\n| Coordinate System | 坐标系 |\n| Figure | 图形 |\n| Handle | 句柄 |\n| Handler | 处理器 |\n| Image | 图像 |\n| Legend | 图例 |\n| Line | 线条 |\n| Patch | 补丁 |\n| Path | 路径 |\n| Pick | 拾取 |\n| Subplot | 子图 |\n| Text | 文本 |\n| Tick | 刻度 |\n| Tick Label | 刻度标签 |\n| Transformation | 变换 |\n\n\n## 第一个Plot\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nplt.plot([1,2,3,2])\nplt.ylabel('some numbers')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
d05f09f0e494948adb21d9d0e37d2c8070046e5b | 5,083 | ipynb | Jupyter Notebook | eigen_broadcast_cheatsheet_(cpp).ipynb | arthursoprano/notebooks | f83c662fecd23c913c40b256d198190fbdcda45e | [
"MIT"
] | 19 | 2017-05-09T16:25:16.000Z | 2020-06-30T23:22:38.000Z | eigen_broadcast_cheatsheet_(cpp).ipynb | arthursoprano/notebooks | f83c662fecd23c913c40b256d198190fbdcda45e | [
"MIT"
] | 1 | 2019-01-25T13:50:39.000Z | 2019-01-25T14:30:39.000Z | eigen_broadcast_cheatsheet_(cpp).ipynb | arthursoprano/notebooks | f83c662fecd23c913c40b256d198190fbdcda45e | [
"MIT"
] | 7 | 2017-06-21T12:30:32.000Z | 2019-01-06T23:48:04.000Z | 22.1 | 165 | 0.460752 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d05f0ec5cbebb54890729bce38219a638a756b5d | 51,466 | ipynb | Jupyter Notebook | Notebooks/data_cleanup.ipynb | HadenMoore/yelp-dataset-challenge-1-ds | 9e99e6010ee56ea82ff816c5335653a390a978ce | [
"MIT"
] | 2 | 2020-01-10T16:56:49.000Z | 2020-02-17T17:08:11.000Z | Notebooks/data_cleanup.ipynb | HadenMoore/yelp-dataset-challenge-1-ds | 9e99e6010ee56ea82ff816c5335653a390a978ce | [
"MIT"
] | 2 | 2019-12-20T17:15:33.000Z | 2020-02-17T17:41:22.000Z | Notebooks/data_cleanup.ipynb | HadenMoore/yelp-dataset-challenge-1-ds | 9e99e6010ee56ea82ff816c5335653a390a978ce | [
"MIT"
] | 4 | 2019-11-06T22:28:11.000Z | 2020-02-17T20:22:30.000Z | 37.759354 | 4,153 | 0.47509 | [
[
[
"# second notebook for Yelp1 Labs 18 Project\n# data cleanup",
"_____no_output_____"
],
[
"# imports\n\n# dataframe\nimport pandas as pd\nimport json\n\n# NLP\nimport gensim\nfrom gensim.utils import simple_preprocess\nfrom gensim.parsing.preprocessing import STOPWORDS\nfrom gensim import corpora",
"_____no_output_____"
],
[
"# import review.json file from https://www.yelp.com/dataset\nwith open('/Users/ianforrest/Desktop/coding/repos/yelp/yelp_dataset/review.json') as f:\n review = json.loads(\"[\" + \n f.read().replace(\"}\\n{\", \"},\\n{\") + \n \"]\")",
"_____no_output_____"
],
[
"# convert review.json files to pandas DataFrame 'df_review'\ndf_review = pd.DataFrame(review)",
"_____no_output_____"
],
[
"# check df_review to make sure it was created correctly\ndf_review.head()",
"_____no_output_____"
],
[
"# check column names of df_review\ndf_review.columns",
"_____no_output_____"
],
[
"# check value counts of 'stars' column\ndf_review['stars'].value_counts()",
"_____no_output_____"
],
[
"# check value counts of useful column\ndf_review['useful'].value_counts()",
"_____no_output_____"
],
[
"# check value counts of funny column\ndf_review['funny'].value_counts()",
"_____no_output_____"
],
[
"# check value counts of cool column\ndf_review['cool'].value_counts()",
"_____no_output_____"
],
[
"# check text of random reviews in dataset as part of initial exploration\ndf_review.iloc[3244,7]",
"_____no_output_____"
],
[
"# check text of random reviews in dataset as part of initial exploration\ndf_review.iloc[2342553,7]",
"_____no_output_____"
],
[
"# check text of random reviews in dataset as part of initial exploration\ndf_review.iloc[3,7]",
"_____no_output_____"
],
[
"# export df_review to .csv\n#df_review.to_csv(r'/Users/ianforrest/Desktop/coding/repos/yelp/yelp_dataset/df_review.csv')",
"_____no_output_____"
],
[
"# create copy of dataframe to manipulate for model\ndf = df_review.copy()",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"# add 'total_votes' column to dataframe; total of 'useful', 'funny', 'cool' columns\ndf['total_votes'] = df['useful'] + df['funny'] + df['cool']",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"# drop unused columns from dataframe\ndf = df.drop(columns=['user_id', 'business_id', 'review_id', 'useful', 'funny', 'cool'])",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"# convert 'date' column to datetime format\ndf['date'] = pd.to_datetime(df['date'])",
"_____no_output_____"
],
[
"df.dtypes",
"_____no_output_____"
],
[
"# check value counts of 'total_votes' column\ndf['total_votes'].value_counts()",
"_____no_output_____"
],
[
"# limit dataframe to reviews with 0 or more total votes\ndf = df.loc[df['total_votes'] >= 0]",
"_____no_output_____"
],
[
"# check value counts of 'total_votes' column\ndf['total_votes'].value_counts()",
"_____no_output_____"
],
[
"# remove html code from text column\ndf['text'] = df['text'].str.replace('(\\d{1,2}[/. ](?:\\d{1,2}|January|Jan)[/. ]\\d{2}(?:\\d{2})?)', '')\ndf['text'] = df['text'].str.replace('\\n\\n', '') \ndf['text'] = df['text'].str.replace('\\\\n', '')\ndf['text'] = df['text'].str.replace('\\n', '')",
"_____no_output_____"
],
[
"# check text of random reviews in dataset to make sure HTML code is removed correctly\n# backslashes before apostrophes are for display purposes only to indicate apostrophes are not quotation marks\ndf.iloc[2342553,1]",
"_____no_output_____"
],
[
"# initiate STOPWORDS for NLP Processing\nSTOPWORDS = set(STOPWORDS).union(set(['I', 'We', 'i', 'we', 'it', \"it's\", \n 'it', 'the', 'this', 'they', 'They',\n 'he', 'He', 'she', 'She', '\\n', '\\n\\n']))",
"_____no_output_____"
],
[
"# create tokenize function to tokenize review text\ndef tokenize(text):\n return [token for token in simple_preprocess(text, deacc=True, min_len=4, max_len=40) if token not in STOPWORDS]",
"_____no_output_____"
],
[
"# add tokens column to dataframe \ndf['tokens'] = df['text'].apply(tokenize)",
"_____no_output_____"
],
[
"# check to make sure tokens were added to dataframe correctly\ndf.head()",
"_____no_output_____"
],
[
"# export cleaned dataframe with tokenized text to .csv file\ndf.to_csv(r'/Users/ianforrest/Desktop/coding/repos/yelp/yelp_dataset/df.csv')",
"_____no_output_____"
],
[
"df.sort_values(['total_votes'], ascending=False)",
"_____no_output_____"
],
[
"df.iloc[1292098,1]",
"_____no_output_____"
],
[
"df.dtypes",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05f27d82deee2e23acdea402b06115d02d0410f | 145,359 | ipynb | Jupyter Notebook | 05-ocr/ocr.ipynb | yizt/opencv-exercises | 95f332f3da1c8ab147017fef4dbe0514fcea9af6 | [
"Apache-2.0"
] | null | null | null | 05-ocr/ocr.ipynb | yizt/opencv-exercises | 95f332f3da1c8ab147017fef4dbe0514fcea9af6 | [
"Apache-2.0"
] | null | null | null | 05-ocr/ocr.ipynb | yizt/opencv-exercises | 95f332f3da1c8ab147017fef4dbe0514fcea9af6 | [
"Apache-2.0"
] | null | null | null | 203.299301 | 124,200 | 0.909266 | [
[
[
"import cv2\nfrom matplotlib import pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"img=cv2.imread('../tmp/out/000.jpg')\n#img=img[5:-5,5:-5]",
"_____no_output_____"
],
[
"gray=cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)",
"_____no_output_____"
],
[
"gray.shape,np.sum(gray,axis=0).shape",
"_____no_output_____"
],
[
"np.where(np.sum(~gray,axis=0)<=1000)",
"_____no_output_____"
],
[
"np.sum(~gray,axis=0)",
"_____no_output_____"
],
[
"106/3",
"_____no_output_____"
],
[
"np.sum(~gray,axis=0)[65:75]",
"_____no_output_____"
],
[
"np.sum(~gray,axis=0)[48:58]",
"_____no_output_____"
],
[
"gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\nvis = img.copy()\n# mser 检测最大稳定极值区域\nmser = cv2.MSER_create()\nregions = mser.detectRegions(gray)",
"_____no_output_____"
],
[
"msers, bboxes=regions",
"_____no_output_____"
],
[
"for i in msers[0]:\n print(i)",
"[ 6 16]\n[ 6 17]\n[ 6 19]\n[ 6 21]\n[ 6 22]\n[ 6 20]\n[ 6 18]\n[ 6 24]\n[ 6 23]\n[ 6 25]\n[ 6 26]\n[ 6 27]\n[ 6 28]\n[ 5 30]\n[ 5 31]\n[ 5 29]\n[ 5 28]\n[ 5 27]\n[ 7 21]\n[ 6 29]\n[ 5 26]\n[ 7 16]\n[ 4 33]\n[ 4 32]\n[ 5 32]\n[ 5 25]\n[ 6 15]\n[ 8 21]\n[ 9 21]\n[10 21]\n[11 21]\n[12 21]\n[13 21]\n[14 21]\n[15 21]\n[ 5 17]\n[17 21]\n[18 21]\n[16 21]\n[ 3 35]\n[ 3 34]\n[ 4 34]\n[19 21]\n[ 5 24]\n[21 21]\n[23 20]\n[23 21]\n[24 20]\n[24 21]\n[22 21]\n[20 21]\n[ 5 18]\n[ 5 20]\n[ 5 19]\n[ 5 16]\n[ 5 23]\n[ 4 31]\n[ 5 22]\n[ 5 21]\n[ 8 16]\n[ 6 30]\n[ 2 36]\n[ 2 35]\n"
],
[
"bboxes",
"_____no_output_____"
],
[
"len(bboxes),len(msers)",
"_____no_output_____"
],
[
"hulls = [cv2.convexHull(p.reshape(-1, 1, 2)) for p in msers]",
"_____no_output_____"
],
[
"hulls[0]",
"_____no_output_____"
],
[
"cv2.polylines(img, hulls[:1], 1, (0, 255, 0))",
"_____no_output_____"
],
[
"plt.figure(figsize=(25,20))\nplt.imshow(img)",
"_____no_output_____"
],
[
"for box in bboxes:\n x, y, w, h = box\n cv2.rectangle(img, (x,y),(x+w, y+h), (255, 0, 0), 2)",
"_____no_output_____"
],
[
"arr = np.empty((4, 2, 3), dtype=np.float32)\nprint(arr)\n# 打印字节串\nb_arr = arr.tostring()\nprint(b_arr)\n# 字节串转ndarray数据\narr_2 = np.frombuffer(b_arr, dtype=np.float32)\narr_2.shape = (4, 2, 3)\nprint(arr_2)",
"[[[1.946e-42 1.169e-42 1.945e-42]\n [1.183e-42 1.944e-42 1.187e-42]]\n\n [[1.941e-42 1.188e-42 1.937e-42]\n [1.190e-42 1.925e-42 1.190e-42]]\n\n [[1.924e-42 1.188e-42 1.923e-42]\n [1.171e-42 1.925e-42 1.167e-42]]\n\n [[1.927e-42 1.166e-42 1.939e-42]\n [1.166e-42 1.946e-42 1.167e-42]]]\nb'm\\x05\\x00\\x00B\\x03\\x00\\x00l\\x05\\x00\\x00L\\x03\\x00\\x00k\\x05\\x00\\x00O\\x03\\x00\\x00i\\x05\\x00\\x00P\\x03\\x00\\x00f\\x05\\x00\\x00Q\\x03\\x00\\x00^\\x05\\x00\\x00Q\\x03\\x00\\x00]\\x05\\x00\\x00P\\x03\\x00\\x00\\\\\\x05\\x00\\x00D\\x03\\x00\\x00^\\x05\\x00\\x00A\\x03\\x00\\x00_\\x05\\x00\\x00@\\x03\\x00\\x00h\\x05\\x00\\x00@\\x03\\x00\\x00m\\x05\\x00\\x00A\\x03\\x00\\x00'\n[[[1.946e-42 1.169e-42 1.945e-42]\n [1.183e-42 1.944e-42 1.187e-42]]\n\n [[1.941e-42 1.188e-42 1.937e-42]\n [1.190e-42 1.925e-42 1.190e-42]]\n\n [[1.924e-42 1.188e-42 1.923e-42]\n [1.171e-42 1.925e-42 1.167e-42]]\n\n [[1.927e-42 1.166e-42 1.939e-42]\n [1.166e-42 1.946e-42 1.167e-42]]]\n"
],
[
"type(b_arr)",
"_____no_output_____"
],
[
"from PIL import Image, ImageDraw,ImageFont\n\ndef put_texts(h, w, words, font):\n # 白底黑字\n img = np.ones((h, w, 3), np.uint8) * 255\n color = (255, 0, 0)\n #\n im = Image.fromarray(img)\n draw = ImageDraw.Draw(im)\n x, y = 2, 2\n for word in words:\n draw.text((x, y), word, color, font=font)\n word_w, word_h = font.getsize(word)\n if h > w:\n y += word_h\n else:\n w += word_w\n return np.array(im)",
"_____no_output_____"
],
[
"#font = ImageFont.truetype('/System/Library/Fonts/STHeiti Light.ttc')\nim=put_texts(100,32,'上大声道',font)\n",
"_____no_output_____"
],
[
"img = np.ones((100, 32, 3), np.uint8) * 255\ncolor = (255, 0, 0)\n#\nim = Image.fromarray(img)\ndraw = ImageDraw.Draw(im)",
"_____no_output_____"
],
[
"draw.text((2, 2), '空', color, font=font)",
"_____no_output_____"
],
[
"font.getsize('案例三等奖')",
"_____no_output_____"
],
[
"plt.imshow(im)",
"_____no_output_____"
],
[
"font.size",
"_____no_output_____"
],
[
"font=font.font_variant(size=20)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05f298f17a476a8ab55a220841f9ea655c78946 | 3,236 | ipynb | Jupyter Notebook | content/lessons/09/End-To-End-Example/ETEE-Bad-Password-Checker.ipynb | MahopacHS/spring2019-mollea1213 | c42e0f49bd2d9965ea1a58db7f72784fea167110 | [
"MIT"
] | null | null | null | content/lessons/09/End-To-End-Example/ETEE-Bad-Password-Checker.ipynb | MahopacHS/spring2019-mollea1213 | c42e0f49bd2d9965ea1a58db7f72784fea167110 | [
"MIT"
] | null | null | null | content/lessons/09/End-To-End-Example/ETEE-Bad-Password-Checker.ipynb | MahopacHS/spring2019-mollea1213 | c42e0f49bd2d9965ea1a58db7f72784fea167110 | [
"MIT"
] | 1 | 2019-02-05T12:52:27.000Z | 2019-02-05T12:52:27.000Z | 58.836364 | 1,246 | 0.613412 | [
[
[
"# End-To-End Example: Bad Password Checker\n\n- Read in list of bad passwords from file `bad-passwords.txt`\n- Main program loop which:\n - inputs a password \n - checks whether the password is \"good\" or \"bad\" by checking against the list\n - repeats this until you enter no password. \n\n",
"_____no_output_____"
]
],
[
[
"# read passwords into list\n#todo:\n#input: none, output: list of bad passwords\n# for each line in bad-passwords.txt\n# add to list\n\ndef read_passwords():\n bad_password_list = []\n filename = \"ETEE-bad-passwords.txt\"\n with open(filename) as f:\n for line in f:\n bad_password_list.append(line.strip())\n return bad_password_list\n\n# password in list?\n#input: password list and a password to check, output: True or False\n#todo\n# get index of password in list\n# return true\n# when ValueError return false\n\ndef password_in_list(password, bad_password_list):\n try:\n index = bad_password_list.index(password)\n return True\n except ValueError:\n return False\n\n# main program \nbad_password_list = read_passwords()\nprint(\"This program will check for quality passwords against a list of known bad passwords.\")\nwhile True:\n password = input(\"Enter a password or ENTER to quit: \")\n if password == \"\":\n break\n if password_in_list(password, bad_password_list):\n print(\"%s is a bad password. It is on the list.\" % (password))\n else:\n print(\"%s seems like an ok password. It is not on the list.\" % (password))\n",
"This program will check for quality passwords against a list of known bad passwords.\nEnter a password or ENTER to quit: 123456\n123456 is a bad password. It is on the list.\nEnter a password or ENTER to quit: fjdskafjoda;shv\nfjdskafjoda;shv seems like an ok password. It is not on the list.\nEnter a password or ENTER to quit: test\ntest is a bad password. It is on the list.\nEnter a password or ENTER to quit: pasword\npasword seems like an ok password. It is not on the list.\nEnter a password or ENTER to quit: password\npassword is a bad password. It is on the list.\nEnter a password or ENTER to quit: \n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
d05f2b7dbeb6637dbb5fd8776240b648976b5351 | 28,485 | ipynb | Jupyter Notebook | site/en/tutorials/generative/dcgan.ipynb | PhilipMay/docs | 4d291590e6352f7fda6175e4f663cceb287589d5 | [
"Apache-2.0"
] | 3 | 2020-01-28T11:36:06.000Z | 2020-01-28T12:15:04.000Z | site/en/tutorials/generative/dcgan.ipynb | PhilipMay/docs | 4d291590e6352f7fda6175e4f663cceb287589d5 | [
"Apache-2.0"
] | 1 | 2020-02-20T14:49:33.000Z | 2020-02-20T14:49:33.000Z | site/en/tutorials/generative/dcgan.ipynb | PhilipMay/docs | 4d291590e6352f7fda6175e4f663cceb287589d5 | [
"Apache-2.0"
] | 1 | 2020-03-04T00:12:25.000Z | 2020-03-04T00:12:25.000Z | 31.579823 | 435 | 0.515008 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Deep Convolutional Generative Adversarial Network",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/tutorials/generative/dcgan\">\n <img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />\n View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/tutorials/generative/dcgan.ipynb\">\n <img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />\n Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/tutorials/generative/dcgan.ipynb\">\n <img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />\n View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs/site/en/tutorials/generative/dcgan.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"This tutorial demonstrates how to generate images of handwritten digits using a [Deep Convolutional Generative Adversarial Network](https://arxiv.org/pdf/1511.06434.pdf) (DCGAN). The code is written using the [Keras Sequential API](https://www.tensorflow.org/guide/keras) with a `tf.GradientTape` training loop.",
"_____no_output_____"
],
[
"## What are GANs?\n[Generative Adversarial Networks](https://arxiv.org/abs/1406.2661) (GANs) are one of the most interesting ideas in computer science today. Two models are trained simultaneously by an adversarial process. A *generator* (\"the artist\") learns to create images that look real, while a *discriminator* (\"the art critic\") learns to tell real images apart from fakes.\n\n\n\nDuring training, the *generator* progressively becomes better at creating images that look real, while the *discriminator* becomes better at telling them apart. The process reaches equilibrium when the *discriminator* can no longer distinguish real images from fakes.\n\n\n\nThis notebook demonstrates this process on the MNIST dataset. The following animation shows a series of images produced by the *generator* as it was trained for 50 epochs. The images begin as random noise, and increasingly resemble hand written digits over time.\n\n\n\nTo learn more about GANs, we recommend MIT's [Intro to Deep Learning](http://introtodeeplearning.com/) course.",
"_____no_output_____"
],
[
"### Import TensorFlow and other libraries",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function, unicode_literals",
"_____no_output_____"
],
[
"try:\n # %tensorflow_version only exists in Colab.\n %tensorflow_version 2.x\nexcept Exception:\n pass\n",
"_____no_output_____"
],
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"tf.__version__",
"_____no_output_____"
],
[
"# To generate GIFs\n!pip install imageio",
"_____no_output_____"
],
[
"import glob\nimport imageio\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport os\nimport PIL\nfrom tensorflow.keras import layers\nimport time\n\nfrom IPython import display",
"_____no_output_____"
]
],
[
[
"### Load and prepare the dataset\n\nYou will use the MNIST dataset to train the generator and the discriminator. The generator will generate handwritten digits resembling the MNIST data.",
"_____no_output_____"
]
],
[
[
"(train_images, train_labels), (_, _) = tf.keras.datasets.mnist.load_data()",
"_____no_output_____"
],
[
"train_images = train_images.reshape(train_images.shape[0], 28, 28, 1).astype('float32')\ntrain_images = (train_images - 127.5) / 127.5 # Normalize the images to [-1, 1]",
"_____no_output_____"
],
[
"BUFFER_SIZE = 60000\nBATCH_SIZE = 256",
"_____no_output_____"
],
[
"# Batch and shuffle the data\ntrain_dataset = tf.data.Dataset.from_tensor_slices(train_images).shuffle(BUFFER_SIZE).batch(BATCH_SIZE)",
"_____no_output_____"
]
],
[
[
"## Create the models\n\nBoth the generator and discriminator are defined using the [Keras Sequential API](https://www.tensorflow.org/guide/keras#sequential_model).",
"_____no_output_____"
],
[
"### The Generator\n\nThe generator uses `tf.keras.layers.Conv2DTranspose` (upsampling) layers to produce an image from a seed (random noise). Start with a `Dense` layer that takes this seed as input, then upsample several times until you reach the desired image size of 28x28x1. Notice the `tf.keras.layers.LeakyReLU` activation for each layer, except the output layer which uses tanh.",
"_____no_output_____"
]
],
[
[
"def make_generator_model():\n model = tf.keras.Sequential()\n model.add(layers.Dense(7*7*256, use_bias=False, input_shape=(100,)))\n model.add(layers.BatchNormalization())\n model.add(layers.LeakyReLU())\n\n model.add(layers.Reshape((7, 7, 256)))\n assert model.output_shape == (None, 7, 7, 256) # Note: None is the batch size\n\n model.add(layers.Conv2DTranspose(128, (5, 5), strides=(1, 1), padding='same', use_bias=False))\n assert model.output_shape == (None, 7, 7, 128)\n model.add(layers.BatchNormalization())\n model.add(layers.LeakyReLU())\n\n model.add(layers.Conv2DTranspose(64, (5, 5), strides=(2, 2), padding='same', use_bias=False))\n assert model.output_shape == (None, 14, 14, 64)\n model.add(layers.BatchNormalization())\n model.add(layers.LeakyReLU())\n\n model.add(layers.Conv2DTranspose(1, (5, 5), strides=(2, 2), padding='same', use_bias=False, activation='tanh'))\n assert model.output_shape == (None, 28, 28, 1)\n\n return model",
"_____no_output_____"
]
],
[
[
"Use the (as yet untrained) generator to create an image.",
"_____no_output_____"
]
],
[
[
"generator = make_generator_model()\n\nnoise = tf.random.normal([1, 100])\ngenerated_image = generator(noise, training=False)\n\nplt.imshow(generated_image[0, :, :, 0], cmap='gray')",
"_____no_output_____"
]
],
[
[
"### The Discriminator\n\nThe discriminator is a CNN-based image classifier.",
"_____no_output_____"
]
],
[
[
"def make_discriminator_model():\n model = tf.keras.Sequential()\n model.add(layers.Conv2D(64, (5, 5), strides=(2, 2), padding='same',\n input_shape=[28, 28, 1]))\n model.add(layers.LeakyReLU())\n model.add(layers.Dropout(0.3))\n\n model.add(layers.Conv2D(128, (5, 5), strides=(2, 2), padding='same'))\n model.add(layers.LeakyReLU())\n model.add(layers.Dropout(0.3))\n\n model.add(layers.Flatten())\n model.add(layers.Dense(1))\n\n return model",
"_____no_output_____"
]
],
[
[
"Use the (as yet untrained) discriminator to classify the generated images as real or fake. The model will be trained to output positive values for real images, and negative values for fake images.",
"_____no_output_____"
]
],
[
[
"discriminator = make_discriminator_model()\ndecision = discriminator(generated_image)\nprint (decision)",
"_____no_output_____"
]
],
[
[
"## Define the loss and optimizers\n\nDefine loss functions and optimizers for both models.\n",
"_____no_output_____"
]
],
[
[
"# This method returns a helper function to compute cross entropy loss\ncross_entropy = tf.keras.losses.BinaryCrossentropy(from_logits=True)",
"_____no_output_____"
]
],
[
[
"### Discriminator loss\n\nThis method quantifies how well the discriminator is able to distinguish real images from fakes. It compares the discriminator's predictions on real images to an array of 1s, and the discriminator's predictions on fake (generated) images to an array of 0s.",
"_____no_output_____"
]
],
[
[
"def discriminator_loss(real_output, fake_output):\n real_loss = cross_entropy(tf.ones_like(real_output), real_output)\n fake_loss = cross_entropy(tf.zeros_like(fake_output), fake_output)\n total_loss = real_loss + fake_loss\n return total_loss",
"_____no_output_____"
]
],
[
[
"### Generator loss\nThe generator's loss quantifies how well it was able to trick the discriminator. Intuitively, if the generator is performing well, the discriminator will classify the fake images as real (or 1). Here, we will compare the discriminators decisions on the generated images to an array of 1s.",
"_____no_output_____"
]
],
[
[
"def generator_loss(fake_output):\n return cross_entropy(tf.ones_like(fake_output), fake_output)",
"_____no_output_____"
]
],
[
[
"The discriminator and the generator optimizers are different since we will train two networks separately.",
"_____no_output_____"
]
],
[
[
"generator_optimizer = tf.keras.optimizers.Adam(1e-4)\ndiscriminator_optimizer = tf.keras.optimizers.Adam(1e-4)",
"_____no_output_____"
]
],
[
[
"### Save checkpoints\nThis notebook also demonstrates how to save and restore models, which can be helpful in case a long running training task is interrupted.",
"_____no_output_____"
]
],
[
[
"checkpoint_dir = './training_checkpoints'\ncheckpoint_prefix = os.path.join(checkpoint_dir, \"ckpt\")\ncheckpoint = tf.train.Checkpoint(generator_optimizer=generator_optimizer,\n discriminator_optimizer=discriminator_optimizer,\n generator=generator,\n discriminator=discriminator)",
"_____no_output_____"
]
],
[
[
"## Define the training loop\n\n",
"_____no_output_____"
]
],
[
[
"EPOCHS = 50\nnoise_dim = 100\nnum_examples_to_generate = 16\n\n# We will reuse this seed overtime (so it's easier)\n# to visualize progress in the animated GIF)\nseed = tf.random.normal([num_examples_to_generate, noise_dim])",
"_____no_output_____"
]
],
[
[
"The training loop begins with generator receiving a random seed as input. That seed is used to produce an image. The discriminator is then used to classify real images (drawn from the training set) and fakes images (produced by the generator). The loss is calculated for each of these models, and the gradients are used to update the generator and discriminator.",
"_____no_output_____"
]
],
[
[
"# Notice the use of `tf.function`\n# This annotation causes the function to be \"compiled\".\[email protected]\ndef train_step(images):\n noise = tf.random.normal([BATCH_SIZE, noise_dim])\n\n with tf.GradientTape() as gen_tape, tf.GradientTape() as disc_tape:\n generated_images = generator(noise, training=True)\n\n real_output = discriminator(images, training=True)\n fake_output = discriminator(generated_images, training=True)\n\n gen_loss = generator_loss(fake_output)\n disc_loss = discriminator_loss(real_output, fake_output)\n\n gradients_of_generator = gen_tape.gradient(gen_loss, generator.trainable_variables)\n gradients_of_discriminator = disc_tape.gradient(disc_loss, discriminator.trainable_variables)\n\n generator_optimizer.apply_gradients(zip(gradients_of_generator, generator.trainable_variables))\n discriminator_optimizer.apply_gradients(zip(gradients_of_discriminator, discriminator.trainable_variables))",
"_____no_output_____"
],
[
"def train(dataset, epochs):\n for epoch in range(epochs):\n start = time.time()\n\n for image_batch in dataset:\n train_step(image_batch)\n\n # Produce images for the GIF as we go\n display.clear_output(wait=True)\n generate_and_save_images(generator,\n epoch + 1,\n seed)\n\n # Save the model every 15 epochs\n if (epoch + 1) % 15 == 0:\n checkpoint.save(file_prefix = checkpoint_prefix)\n\n print ('Time for epoch {} is {} sec'.format(epoch + 1, time.time()-start))\n\n # Generate after the final epoch\n display.clear_output(wait=True)\n generate_and_save_images(generator,\n epochs,\n seed)",
"_____no_output_____"
]
],
[
[
"**Generate and save images**\n\n",
"_____no_output_____"
]
],
[
[
"def generate_and_save_images(model, epoch, test_input):\n # Notice `training` is set to False.\n # This is so all layers run in inference mode (batchnorm).\n predictions = model(test_input, training=False)\n\n fig = plt.figure(figsize=(4,4))\n\n for i in range(predictions.shape[0]):\n plt.subplot(4, 4, i+1)\n plt.imshow(predictions[i, :, :, 0] * 127.5 + 127.5, cmap='gray')\n plt.axis('off')\n\n plt.savefig('image_at_epoch_{:04d}.png'.format(epoch))\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Train the model\nCall the `train()` method defined above to train the generator and discriminator simultaneously. Note, training GANs can be tricky. It's important that the generator and discriminator do not overpower each other (e.g., that they train at a similar rate).\n\nAt the beginning of the training, the generated images look like random noise. As training progresses, the generated digits will look increasingly real. After about 50 epochs, they resemble MNIST digits. This may take about one minute / epoch with the default settings on Colab.",
"_____no_output_____"
]
],
[
[
"train(train_dataset, EPOCHS)",
"_____no_output_____"
]
],
[
[
"Restore the latest checkpoint.",
"_____no_output_____"
]
],
[
[
"checkpoint.restore(tf.train.latest_checkpoint(checkpoint_dir))",
"_____no_output_____"
]
],
[
[
"## Create a GIF\n",
"_____no_output_____"
]
],
[
[
"# Display a single image using the epoch number\ndef display_image(epoch_no):\n return PIL.Image.open('image_at_epoch_{:04d}.png'.format(epoch_no))",
"_____no_output_____"
],
[
"display_image(EPOCHS)",
"_____no_output_____"
]
],
[
[
"Use `imageio` to create an animated gif using the images saved during training.",
"_____no_output_____"
]
],
[
[
"anim_file = 'dcgan.gif'\n\nwith imageio.get_writer(anim_file, mode='I') as writer:\n filenames = glob.glob('image*.png')\n filenames = sorted(filenames)\n last = -1\n for i,filename in enumerate(filenames):\n frame = 2*(i**0.5)\n if round(frame) > round(last):\n last = frame\n else:\n continue\n image = imageio.imread(filename)\n writer.append_data(image)\n image = imageio.imread(filename)\n writer.append_data(image)\n\nimport IPython\nif IPython.version_info > (6,2,0,''):\n display.Image(filename=anim_file)",
"_____no_output_____"
]
],
[
[
"If you're working in Colab you can download the animation with the code below:",
"_____no_output_____"
]
],
[
[
"try:\n from google.colab import files\nexcept ImportError:\n pass\nelse:\n files.download(anim_file)",
"_____no_output_____"
]
],
[
[
"## Next steps\n",
"_____no_output_____"
],
[
"This tutorial has shown the complete code necessary to write and train a GAN. As a next step, you might like to experiment with a different dataset, for example the Large-scale Celeb Faces Attributes (CelebA) dataset [available on Kaggle](https://www.kaggle.com/jessicali9530/celeba-dataset). To learn more about GANs we recommend the [NIPS 2016 Tutorial: Generative Adversarial Networks](https://arxiv.org/abs/1701.00160).\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d05f35d563aa837ab0a57f6e00c6e9a70a1894dc | 25,261 | ipynb | Jupyter Notebook | prep_mnist_padded1.ipynb | PopGalacticHistory/imagewalker | dd151d0698472aebdde0487a364c56fed048bb61 | [
"MIT"
] | 2 | 2021-04-28T13:33:45.000Z | 2021-11-09T14:31:09.000Z | prep_mnist_padded1.ipynb | PopGalacticHistory/imagewalker | dd151d0698472aebdde0487a364c56fed048bb61 | [
"MIT"
] | null | null | null | prep_mnist_padded1.ipynb | PopGalacticHistory/imagewalker | dd151d0698472aebdde0487a364c56fed048bb61 | [
"MIT"
] | 1 | 2021-03-07T13:25:59.000Z | 2021-03-07T13:25:59.000Z | 43.704152 | 192 | 0.620799 | [
[
[
"import pickle\nfrom misc import *\nimport SYCLOP_env as syc\nfrom RL_brain_b import DeepQNetwork\n",
"_____no_output_____"
],
[
"import cv2\nimport time",
"_____no_output_____"
],
[
"from mnist import MNIST\n\nmnist = MNIST('/home/bnapp/datasets/mnist/')\nimages, labels = mnist.load_training()\n# some_mnistSM =[ cv2.resize(1.+np.reshape(uu,[28,28]), dsize=(256, 256)) for uu in images[:2]]#[:4096]] \nsome_samples_for_setup= prep_mnist_padded_images(2)\n \n",
"_____no_output_____"
],
[
"# run_dir = 'saved_runs/run_syclop_generic1.py_noname_1576060868_0/' #padded mnist beta 0.1 speed penalty 5\n# result_type = 'nwk2.nwk'\n\n# run_dir = 'saved_runs/run_syclop_generic1.py_noname_1576147784_0/' #padded mnist beta 0.1 speed penalty 20\n\n\nrun_dir = 'saved_runs/run_syclop_generic1.py_noname_1576403573_0/' #padded mnist beta 0.1 speed penalty 0\nresult_type = 'tempX_1.nwk'\n",
"_____no_output_____"
],
[
"hp = HP()\nhp.mem_depth=1\nhp.logmode=False\nbatch_size=256\naction_space_size=9\n# images = some_mnistSM\nnumber_of_images = len(images)\nreward = syc.Rewards()\nobservation_size = 256*4\nRL = DeepQNetwork(action_space_size, observation_size*hp.mem_depth,#sensor.frame_size+2,\n reward_decay=0.99,\n e_greedy=1-1e-9,\n e_greedy0=1-1e-9,\n replace_target_iter=10,\n memory_size=100000,\n e_greedy_increment=0.0001,\n learning_rate=0.0025,\n double_q=False,\n dqn_mode=True,\n state_table=np.zeros([1,observation_size*hp.mem_depth]),\n soft_q_type='boltzmann',\n beta=0.1\n )\n\n\n",
"debug n_features: 1024\ndebug: (?, 1024)\ndebug: (?, 400)\ndebug: (?, 200)\ndebug: (?, 200)\ndebug: (?, 200)\ndebug n_features: 1024\ndebug: (?, 1024)\ndebug: (?, 400)\ndebug: (?, 200)\ndebug: (?, 200)\ndebug: (?, 200)\n"
],
[
"\ndef local_observer(sensor,agent):\n if hp.logmode:\n normfactor=1.0\n else:\n normfactor = 1.0/256.0\n return normfactor*np.concatenate([relu_up_and_down(sensor.central_dvs_view),\n relu_up_and_down(cv2.resize(1.0*sensor.dvs_view, dsize=(16, 16), interpolation=cv2.INTER_AREA))])\n\nobservation = np.random.uniform(0,1,size=[hp.mem_depth, observation_size])\n",
"_____no_output_____"
],
[
"scene_bb = [None]*batch_size\nsensor_bb =[None]*batch_size\nagent_bb = [None]*batch_size\naction_bb = [None]*batch_size\naction_list_bb = [None]*batch_size\nq_list_bb = [None]*batch_size\nobservation_bb = [None]*batch_size\n\nwith open(run_dir+'/hp.pkl','rb') as f:\n this_hp = pickle.load(f)\n\nfor bb in range(batch_size):\n scene_bb[bb] = syc.Scene(frame_list=some_samples_for_setup[0:1])\n sensor_bb[bb] = syc.Sensor()\n agent_bb[bb] = syc.Agent(max_q = [scene_bb[bb].maxx-sensor_bb[bb].hp.winx,scene_bb[bb].maxy-sensor_bb[bb].hp.winy])\n agent_bb[bb].hp.action_space = this_hp.agent.action_space\n",
"_____no_output_____"
],
[
"\nRL.dqn.load_nwk_param(run_dir+'/'+ result_type) \nwith open(run_dir+'/hp.pkl','rb') as f:\n this_hp = pickle.load(f)\n hp.fading_mem = this_hp.fading_mem +0.0 #to avoid assignment by address\nsize=(28,28)\noffset=(0,0)\naction_records=[]\nq_records=[]\nobservation_feeder=np.zeros([batch_size,1024])\nfor image_num,image in enumerate(images):\n step = 0\n episode = 0\nfor batch_num in range(len(images)//batch_size):\n for bb in range(batch_size):\n action_list_bb[bb] = []\n# q_list_bb[bb] = []\n observation_bb[bb] = np.random.uniform(0,1,size=[hp.mem_depth, observation_size])\n observation_bb[bb] = np.random.uniform(0,1,size=[hp.mem_depth, observation_size])\n# scene_bb[bb].current_frame = image_num[bb]\n \n #### sizing story:\n image_resized=cv2.resize(0.0+np.reshape(images[batch_num*batch_size+bb],[28,28]), dsize=size)\n scene_bb[bb].image = build_mnist_padded([image_resized],y_size=size[1],x_size=size[0],offset=offset)\n# scene_bb[bb].image = build_mnist_padded([images[batch_num*batch_size+bb]])\n\n agent_bb[bb].reset()\n agent_bb[bb].q_ana[1]=128./2.-32\n agent_bb[bb].q_ana[0]=128./2-32\n agent_bb[bb].q = np.int32(np.floor(agent_bb[bb].q_ana))\n\n sensor_bb[bb].reset()\n sensor_bb[bb].update(scene_bb[bb], agent_bb[bb])\n sensor_bb[bb].update(scene_bb[bb], agent_bb[bb])\n \n time1=time.time()\n for step_prime in range(1000):\n deep_time1=time.time()\n# action = RL.choose_action(observation.reshape([-1]))\n for bb in range(batch_size):\n observation_feeder[bb,:]=observation_bb[bb].reshape([1,-1])\n oo = RL.dqn.eval_eval(observation_feeder) \n boltzmann_measure = np.exp(RL.beta * (oo-np.max(oo,axis=1).reshape([-1,1]))) #todo here substracted max to avoid exponent exploding. need to be taken into a separate function!\n boltzmann_measure = boltzmann_measure / np.sum(boltzmann_measure, axis=1).reshape([-1,1])\n for bb in range(batch_size):\n action_bb[bb] = np.random.choice(list(range(RL.n_actions)),1, p=boltzmann_measure[bb,:].reshape([-1]))[0]\n \n# action_bb= [a for a in np.argmax(oo,axis=1)]\n deep_time2=time.time()\n shallow_time1=time.time()\n \n for bb in range(batch_size):\n agent_bb[bb].act(action_bb[bb])\n action_list_bb[bb].append(action_bb[bb])\n# q_list_bb[bb].append(agent_bb[bb].q_ana)\n sensor_bb[bb].update(scene_bb[bb],agent_bb[bb])\n observation_bb[bb] *= hp.fading_mem\n observation_bb[bb] += local_observer(sensor_bb[bb], agent_bb[bb]) # todo: generalize\n shallow_time2=time.time()\n# print('deep:',deep_time2-deep_time1,'shallow:',shallow_time2-shallow_time1)\n time2=time.time()\n \n print('batch num:',batch_num,'wall time consumed:',time2-time1)\n for bb in range(batch_size):\n action_records.append(action_list_bb[bb])\n# q_records.append(q_list_bb[bb])\n",
"batch num: 0 wall time consumed: 39.24245738983154\nbatch num: 1 wall time consumed: 38.755786180496216\nbatch num: 2 wall time consumed: 38.64829134941101\nbatch num: 3 wall time consumed: 38.89785671234131\nbatch num: 4 wall time consumed: 38.728580713272095\nbatch num: 5 wall time consumed: 38.78235936164856\nbatch num: 6 wall time consumed: 38.581660747528076\nbatch num: 7 wall time consumed: 39.75066304206848\nbatch num: 8 wall time consumed: 39.06776213645935\nbatch num: 9 wall time consumed: 38.79161477088928\nbatch num: 10 wall time consumed: 38.849507093429565\nbatch num: 11 wall time consumed: 39.05150532722473\nbatch num: 12 wall time consumed: 39.012853384017944\nbatch num: 13 wall time consumed: 38.74488806724548\nbatch num: 14 wall time consumed: 38.79856014251709\nbatch num: 15 wall time consumed: 38.77446150779724\nbatch num: 16 wall time consumed: 38.767223834991455\nbatch num: 17 wall time consumed: 39.92315196990967\nbatch num: 18 wall time consumed: 38.765279054641724\nbatch num: 19 wall time consumed: 38.805659532547\nbatch num: 20 wall time consumed: 38.73130774497986\nbatch num: 21 wall time consumed: 39.27479529380798\nbatch num: 22 wall time consumed: 38.756571769714355\nbatch num: 23 wall time consumed: 38.45664358139038\nbatch num: 24 wall time consumed: 38.717315435409546\nbatch num: 25 wall time consumed: 39.61017894744873\nbatch num: 26 wall time consumed: 39.40128445625305\nbatch num: 27 wall time consumed: 39.47049260139465\nbatch num: 28 wall time consumed: 38.7383348941803\nbatch num: 29 wall time consumed: 39.735931634902954\nbatch num: 30 wall time consumed: 40.09104347229004\nbatch num: 31 wall time consumed: 41.66463255882263\nbatch num: 32 wall time consumed: 40.277501344680786\nbatch num: 33 wall time consumed: 40.56296110153198\nbatch num: 34 wall time consumed: 39.97669959068298\nbatch num: 35 wall time consumed: 39.914886236190796\nbatch num: 36 wall time consumed: 40.13045620918274\nbatch num: 37 wall time consumed: 40.38657307624817\nbatch num: 38 wall time consumed: 39.783586263656616\nbatch num: 39 wall time consumed: 40.028661489486694\nbatch num: 40 wall time consumed: 40.360772371292114\nbatch num: 41 wall time consumed: 41.30896306037903\nbatch num: 42 wall time consumed: 39.71547174453735\nbatch num: 43 wall time consumed: 40.25914645195007\nbatch num: 44 wall time consumed: 40.036412477493286\nbatch num: 45 wall time consumed: 40.37951850891113\nbatch num: 46 wall time consumed: 39.95870351791382\nbatch num: 47 wall time consumed: 39.82537055015564\nbatch num: 48 wall time consumed: 40.28987503051758\nbatch num: 49 wall time consumed: 40.06290245056152\nbatch num: 50 wall time consumed: 40.1447548866272\nbatch num: 51 wall time consumed: 40.22420334815979\nbatch num: 52 wall time consumed: 40.22356343269348\nbatch num: 53 wall time consumed: 40.17773962020874\nbatch num: 54 wall time consumed: 40.32162666320801\nbatch num: 55 wall time consumed: 40.12950134277344\nbatch num: 56 wall time consumed: 40.187488079071045\nbatch num: 57 wall time consumed: 39.988410234451294\nbatch num: 58 wall time consumed: 39.85330414772034\nbatch num: 59 wall time consumed: 39.88715052604675\nbatch num: 60 wall time consumed: 40.449333906173706\nbatch num: 61 wall time consumed: 39.92989110946655\nbatch num: 62 wall time consumed: 39.81858730316162\nbatch num: 63 wall time consumed: 39.919777631759644\nbatch num: 64 wall time consumed: 40.10007667541504\nbatch num: 65 wall time consumed: 40.12239623069763\nbatch num: 66 wall time consumed: 40.0331175327301\nbatch num: 67 wall time consumed: 39.88394594192505\nbatch num: 68 wall time consumed: 40.360435247421265\nbatch num: 69 wall time consumed: 39.98176693916321\nbatch num: 70 wall time consumed: 39.97550654411316\nbatch num: 71 wall time consumed: 40.17575478553772\nbatch num: 72 wall time consumed: 39.98882985115051\nbatch num: 73 wall time consumed: 40.37295055389404\nbatch num: 74 wall time consumed: 39.69810390472412\nbatch num: 75 wall time consumed: 40.19703269004822\nbatch num: 76 wall time consumed: 40.05048322677612\nbatch num: 77 wall time consumed: 40.29378080368042\nbatch num: 78 wall time consumed: 40.02045488357544\nbatch num: 79 wall time consumed: 41.157652139663696\nbatch num: 80 wall time consumed: 39.684431314468384\nbatch num: 81 wall time consumed: 39.717286348342896\nbatch num: 82 wall time consumed: 39.86164832115173\nbatch num: 83 wall time consumed: 40.21325612068176\nbatch num: 84 wall time consumed: 40.22037100791931\nbatch num: 85 wall time consumed: 39.68337345123291\nbatch num: 86 wall time consumed: 40.03053689002991\nbatch num: 87 wall time consumed: 40.17217969894409\nbatch num: 88 wall time consumed: 39.75106716156006\nbatch num: 89 wall time consumed: 39.81740999221802\nbatch num: 90 wall time consumed: 40.09240770339966\nbatch num: 91 wall time consumed: 39.87443923950195\nbatch num: 92 wall time consumed: 39.76974630355835\nbatch num: 93 wall time consumed: 39.91161632537842\nbatch num: 94 wall time consumed: 39.9528648853302\nbatch num: 95 wall time consumed: 39.96637034416199\nbatch num: 96 wall time consumed: 39.79718732833862\nbatch num: 97 wall time consumed: 40.227434158325195\nbatch num: 98 wall time consumed: 39.94765782356262\nbatch num: 99 wall time consumed: 40.258211851119995\nbatch num: 100 wall time consumed: 39.804479360580444\nbatch num: 101 wall time consumed: 39.79862070083618\nbatch num: 102 wall time consumed: 40.080618143081665\nbatch num: 103 wall time consumed: 40.11222243309021\nbatch num: 104 wall time consumed: 39.73807454109192\nbatch num: 105 wall time consumed: 39.79391837120056\nbatch num: 106 wall time consumed: 40.10137128829956\nbatch num: 107 wall time consumed: 40.08752751350403\nbatch num: 108 wall time consumed: 40.17952060699463\nbatch num: 109 wall time consumed: 39.9392671585083\nbatch num: 110 wall time consumed: 39.90235996246338\nbatch num: 111 wall time consumed: 40.118677377700806\nbatch num: 112 wall time consumed: 40.059932231903076\nbatch num: 113 wall time consumed: 40.03299021720886\nbatch num: 114 wall time consumed: 40.237268686294556\nbatch num: 115 wall time consumed: 40.262397050857544\nbatch num: 116 wall time consumed: 40.16004204750061\nbatch num: 117 wall time consumed: 39.99065709114075\nbatch num: 118 wall time consumed: 39.77810502052307\nbatch num: 119 wall time consumed: 40.03807067871094\nbatch num: 120 wall time consumed: 39.98832440376282\nbatch num: 121 wall time consumed: 39.28131556510925\nbatch num: 122 wall time consumed: 39.37433624267578\nbatch num: 123 wall time consumed: 38.951510190963745\nbatch num: 124 wall time consumed: 39.33266592025757\nbatch num: 125 wall time consumed: 38.86020088195801\nbatch num: 126 wall time consumed: 39.24878764152527\nbatch num: 127 wall time consumed: 39.35402011871338\nbatch num: 128 wall time consumed: 39.00271916389465\nbatch num: 129 wall time consumed: 39.24384522438049\nbatch num: 130 wall time consumed: 39.21375608444214\nbatch num: 131 wall time consumed: 38.98906326293945\nbatch num: 132 wall time consumed: 39.283729791641235\nbatch num: 133 wall time consumed: 39.33757424354553\nbatch num: 134 wall time consumed: 39.18048429489136\nbatch num: 135 wall time consumed: 39.28144407272339\nbatch num: 136 wall time consumed: 39.12085843086243\nbatch num: 137 wall time consumed: 39.13159155845642\nbatch num: 138 wall time consumed: 39.38188409805298\nbatch num: 139 wall time consumed: 39.20531463623047\nbatch num: 140 wall time consumed: 39.19406533241272\nbatch num: 141 wall time consumed: 39.300915479660034\nbatch num: 142 wall time consumed: 39.288023948669434\nbatch num: 143 wall time consumed: 39.414305210113525\nbatch num: 144 wall time consumed: 39.35805058479309\nbatch num: 145 wall time consumed: 39.50939178466797\nbatch num: 146 wall time consumed: 39.003331899642944\nbatch num: 147 wall time consumed: 39.156272411346436\nbatch num: 148 wall time consumed: 39.64461612701416\nbatch num: 149 wall time consumed: 39.037638664245605\nbatch num: 150 wall time consumed: 39.3753604888916\nbatch num: 151 wall time consumed: 39.142780780792236\nbatch num: 152 wall time consumed: 39.38616895675659\nbatch num: 153 wall time consumed: 39.36794400215149\nbatch num: 154 wall time consumed: 39.10295057296753\nbatch num: 155 wall time consumed: 39.260515451431274\nbatch num: 156 wall time consumed: 39.134814977645874\n"
],
[
"len(action_records)",
"_____no_output_____"
],
[
"with open('mnist_padded_b0p1_v0_X28_Tx0y0_act_full1.pkl','wb') as f:\n pickle.dump([action_records[:30000],labels[:30000]],f)\n \nwith open('mnist_padded_b0p1_v0_X28_Tx0y0_act_full2.pkl','wb') as f:\n pickle.dump([action_records[30000:],labels[30000:]],f)",
"_____no_output_____"
],
[
"np.shape(sensor_bb[0].frame_view)",
"_____no_output_____"
],
[
"agent_bb[0].q_ana",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05f65d398e307009d4f0ecaf046d7b09ecf3138 | 18,992 | ipynb | Jupyter Notebook | PyMC Part 1 Done.ipynb | Journeyman08/BayesianSurvivalAnalysis | 9b7149c74123506c6532aa0ba4049c555f3bc083 | [
"MIT"
] | 3 | 2016-07-24T03:17:38.000Z | 2018-01-13T19:35:58.000Z | PyMC Part 1 Done.ipynb | Journeyman08/BayesianSurvivalAnalysis | 9b7149c74123506c6532aa0ba4049c555f3bc083 | [
"MIT"
] | null | null | null | PyMC Part 1 Done.ipynb | Journeyman08/BayesianSurvivalAnalysis | 9b7149c74123506c6532aa0ba4049c555f3bc083 | [
"MIT"
] | 2 | 2016-05-05T08:25:04.000Z | 2021-12-08T14:03:00.000Z | 41.197397 | 7,486 | 0.686394 | [
[
[
"import lifelines\nimport pymc as pm\nfrom pyBMA.CoxPHFitter import CoxPHFitter\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom numpy import log\nfrom datetime import datetime\nimport pandas as pd\n%matplotlib inline ",
"_____no_output_____"
]
],
[
[
"The first step in any data analysis is acquiring and munging the data\n\nOur starting data set can be found here:\n http://jakecoltman.com in the pyData post\n\nIt is designed to be roughly similar to the output from DCM's path to conversion\n\nDownload the file and transform it into something with the columns:\n\n id,lifetime,age,male,event,search,brand\n \nwhere lifetime is the total time that we observed someone not convert for and event should be 1 if we see a conversion and 0 if we don't. Note that all values should be converted into ints\n\nIt is useful to note that end_date = datetime.datetime(2016, 5, 3, 20, 36, 8, 92165)\n\n",
"_____no_output_____"
]
],
[
[
"running_id = 0\noutput = [[0]]\nwith open(\"E:/output.txt\") as file_open:\n for row in file_open.read().split(\"\\n\"):\n cols = row.split(\",\")\n if cols[0] == output[-1][0]:\n output[-1].append(cols[1])\n output[-1].append(True)\n else:\n output.append(cols)\n output = output[1:]\n \nfor row in output:\n if len(row) == 6:\n row += [datetime(2016, 5, 3, 20, 36, 8, 92165), False]\noutput = output[1:-1]",
"_____no_output_____"
],
[
"def convert_to_days(dt):\n day_diff = dt / np.timedelta64(1, 'D')\n if day_diff == 0:\n return 23.0\n else: \n return day_diff\n\ndf = pd.DataFrame(output, columns=[\"id\", \"advert_time\", \"male\",\"age\",\"search\",\"brand\",\"conversion_time\",\"event\"])\ndf[\"lifetime\"] = pd.to_datetime(df[\"conversion_time\"]) - pd.to_datetime(df[\"advert_time\"])\ndf[\"lifetime\"] = df[\"lifetime\"].apply(convert_to_days)\ndf[\"male\"] = df[\"male\"].astype(int)\ndf[\"search\"] = df[\"search\"].astype(int)\ndf[\"brand\"] = df[\"brand\"].astype(int)\ndf[\"age\"] = df[\"age\"].astype(int)\ndf[\"event\"] = df[\"event\"].astype(int)\ndf = df.drop('advert_time', 1)\ndf = df.drop('conversion_time', 1)\ndf = df.set_index(\"id\")\ndf = df.dropna(thresh=2)\ndf.median()",
"_____no_output_____"
],
[
"###Parametric Bayes\n#Shout out to Cam Davidson-Pilon",
"_____no_output_____"
],
[
"## Example fully worked model using toy data\n## Adapted from http://blog.yhat.com/posts/estimating-user-lifetimes-with-pymc.html\n## Note that we've made some corrections \n\nN = 2500\n\n##Generate some random data \nlifetime = pm.rweibull( 2, 5, size = N )\nbirth = pm.runiform(0, 10, N)\ncensor = ((birth + lifetime) >= 10)\nlifetime_ = lifetime.copy()\nlifetime_[censor] = 10 - birth[censor]\n\n\nalpha = pm.Uniform('alpha', 0, 20)\nbeta = pm.Uniform('beta', 0, 20)\n\[email protected]\ndef survival(value=lifetime_, alpha = alpha, beta = beta ):\n return sum( (1-censor)*(log( alpha/beta) + (alpha-1)*log(value/beta)) - (value/beta)**(alpha))\n\nmcmc = pm.MCMC([alpha, beta, survival ] )\nmcmc.sample(50000, 30000)",
" [----------- 29% ] 14864 of 50000 complete in 21.5 secHalting at iteration 14874 of 50000\n"
],
[
"pm.Matplot.plot(mcmc)\nmcmc.trace(\"alpha\")[:]",
"_____no_output_____"
]
],
[
[
"Problems: \n\n 1 - Try to fit your data from section 1 \n 2 - Use the results to plot the distribution of the median\n \nNote that the media of a Weibull distribution is:\n$$β(log 2)^{1/α}$$ ",
"_____no_output_____"
]
],
[
[
"censor = np.array(df[\"event\"].apply(lambda x: 0 if x else 1).tolist())\nalpha = pm.Uniform(\"alpha\", 0,50) \nbeta = pm.Uniform(\"beta\", 0,50) \n\[email protected]\ndef survival(value=df[\"lifetime\"], alpha = alpha, beta = beta ):\n return sum( (1-censor)*(np.log( alpha/beta) + (alpha-1)*np.log(value/beta)) - (value/beta)**(alpha))\n\n\nmcmc = pm.MCMC([alpha, beta, survival ] )\nmcmc.sample(10000)",
" [-----------------100%-----------------] 10000 of 10000 complete in 16.9 sec"
],
[
"def weibull_median(alpha, beta):\n return beta * ((log(2)) ** ( 1 / alpha))\nplt.hist([weibull_median(x[0], x[1]) for x in zip(mcmc.trace(\"alpha\"), mcmc.trace(\"beta\"))])",
"_____no_output_____"
]
],
[
[
"Problems:\n \n 4 - Try adjusting the number of samples for burning and thinnning\n 5 - Try adjusting the prior and see how it affects the estimate ",
"_____no_output_____"
]
],
[
[
"#### Adjust burn and thin, both paramters of the mcmc sample function",
"_____no_output_____"
],
[
"#### Narrow and broaden prior",
"_____no_output_____"
]
],
[
[
"Problems:\n \n 7 - Try testing whether the median is greater than a different values",
"_____no_output_____"
]
],
[
[
"#### Hypothesis testing",
"_____no_output_____"
]
],
[
[
"If we want to look at covariates, we need a new approach. \n\nWe'll use Cox proprtional hazards, a very popular regression model.\n\nTo fit in python we use the module lifelines:\n\nhttp://lifelines.readthedocs.io/en/latest/",
"_____no_output_____"
]
],
[
[
"### Fit a cox proprtional hazards model",
"_____no_output_____"
]
],
[
[
"Once we've fit the data, we need to do something useful with it. Try to do the following things:\n\n 1 - Plot the baseline survival function\n\n 2 - Predict the functions for a particular set of features\n\n 3 - Plot the survival function for two different set of features\n\n 4 - For your results in part 3 caculate how much more likely a death event is for one than the other for a given period of time",
"_____no_output_____"
]
],
[
[
"#### Plot baseline hazard function",
"_____no_output_____"
],
[
"#### Predict",
"_____no_output_____"
],
[
"#### Plot survival functions for different covariates",
"_____no_output_____"
],
[
"#### Plot some odds",
"_____no_output_____"
]
],
[
[
"Model selection\n\nDifficult to do with classic tools (here)\n\nProblem:\n\n 1 - Calculate the BMA coefficient values\n \n 2 - Try running with different priors",
"_____no_output_____"
]
],
[
[
"#### BMA Coefficient values",
"_____no_output_____"
],
[
"#### Different priors",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05f74e4ef1aac51c49926d8af4d094f0abe3a77 | 39,183 | ipynb | Jupyter Notebook | linear_regression_from_scratch.ipynb | arjunjanamatti/tf_nptel | fd7b1a7f5f5cf904c4dc4904c23830bfc3f2e235 | [
"Apache-2.0"
] | null | null | null | linear_regression_from_scratch.ipynb | arjunjanamatti/tf_nptel | fd7b1a7f5f5cf904c4dc4904c23830bfc3f2e235 | [
"Apache-2.0"
] | null | null | null | linear_regression_from_scratch.ipynb | arjunjanamatti/tf_nptel | fd7b1a7f5f5cf904c4dc4904c23830bfc3f2e235 | [
"Apache-2.0"
] | null | null | null | 141.454874 | 12,056 | 0.893959 | [
[
[
"* Normalizing helps in getting better convergence in training data",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"import pandas as pd\n\nraw_data = pd.read_csv('data.txt', sep = ',', header = None)\nX = raw_data.iloc[:,0].values\ny = raw_data.iloc[:,1].values\nX_1 = X.reshape(len(X), 1)\nplt.scatter(X,y)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Using scikit library",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(X_1, y, test_size=0.2, random_state=42)\n\n\nfrom sklearn.linear_model import LinearRegression\n\nmodel = LinearRegression()\nmodel.fit(X = X_train,\n y = y_train)\n\nprint(\"Accuracy of model: \", round((model.score(X = X_1,\n y = y)),2))\nprint()\nprint('Intercept: ', model.intercept_)\nprint()\nprint('slope: ', model.coef_)\n\npredicted_value = model.predict(X = X_1)\n\nplt.scatter(X, y)\nplt.plot(X, predicted_value)\nplt.show()",
"Accuracy of model: 0.59\n\nIntercept: 15.07636055026471\n\nslope: [1.19463787]\n"
]
],
[
[
"### Using gradient descent algorithm",
"_____no_output_____"
]
],
[
[
"b = 0\nw_1 = 0\ncount = 0\nw_1_list = []\nb_list = []\nloss_function_list = []\nwhile count < 1000:\n predicted_y = b + (w_1 * X)\n loss_function = (np.sum(predicted_y - y)**2) * 0.5\n gradient_of_w_1 = np.sum(((predicted_y - y) * X))\n gradient_of_b = np.sum(((predicted_y - y) * 1))\n learning_rate = 0.000001\n w_1 = w_1 - (learning_rate * gradient_of_w_1)\n b = b - (learning_rate * gradient_of_b)\n w_1_list.append(w_1)\n b_list.append(b)\n loss_function_list.append(loss_function)\n count = count + 1\n \npred = b_list[np.argmin(loss_function_list)] + ((w_1_list[np.argmin(loss_function_list)])* X)\n\nprint(\"Accuracy of model: \", round(r_square(y, pred),2))\nprint()\nprint('Intercept: ', model.intercept_)\nprint()\nprint('slope: ', model.coef_)\n\nplt.scatter(X, y)\nplt.plot(X, pred)\nplt.show()",
"Accuracy of model: 0.59\n\nIntercept: 15.07636055026471\n\nslope: [1.19463787]\n"
],
[
"def r_square(y, pred):\n mean_value = np.mean(y)\n total = np.sum((y - mean_value)**2)\n residuals = np.sum((y - pred)**2)\n return 1 - (residuals / total)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05f85c0ee1130ea15f9a58a15e477cd42131711 | 145,952 | ipynb | Jupyter Notebook | lectures/Feb-25-probability_distributions/probability_distributions.ipynb | nishadalal120/NEU-365P-385L-Spring-2021 | eff075482913a6c72737c578f1c5fc42527c12bb | [
"Unlicense"
] | 12 | 2021-01-05T18:26:42.000Z | 2021-03-11T19:26:07.000Z | lectures/Feb-25-probability_distributions/probability_distributions.ipynb | nishadalal120/NEU-365P-385L-Spring-2021 | eff075482913a6c72737c578f1c5fc42527c12bb | [
"Unlicense"
] | 1 | 2021-04-21T00:57:10.000Z | 2021-04-21T00:57:10.000Z | lectures/Feb-25-probability_distributions/probability_distributions.ipynb | nishadalal120/NEU-365P-385L-Spring-2021 | eff075482913a6c72737c578f1c5fc42527c12bb | [
"Unlicense"
] | 22 | 2021-01-21T18:52:41.000Z | 2021-04-15T20:22:20.000Z | 161.988901 | 42,676 | 0.906647 | [
[
[
"# Probability Distributions",
"_____no_output_____"
],
[
"# Some typical stuff we'll likely use",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%config InlineBackend.figure_format = 'retina'",
"_____no_output_____"
]
],
[
[
"# [SciPy](https://scipy.org)\n\n### [scipy.stats](https://docs.scipy.org/doc/scipy-0.14.0/reference/stats.html)",
"_____no_output_____"
]
],
[
[
"import scipy as sp\nimport scipy.stats as st",
"_____no_output_____"
]
],
[
[
"# Binomial Distribution\n\n### <font color=darkred> **Example**: A couple, who are both carriers for a recessive disease, wish to have 5 children. They want to know the probability that they will have four healthy kids.</font>\n\nIn this case the random variable is the number of healthy kids.",
"_____no_output_____"
]
],
[
[
"# number of trials (kids)\nn = 5\n\n# probability of success on each trial\n# i.e. probability that each child will be healthy = 1 - 0.5 * 0.5 = 0.75\np = 0.75\n\n# a binomial distribution object\ndist = st.binom(n, p)\n\n# probability of four healthy kids\ndist.pmf(4)",
"_____no_output_____"
],
[
"print(f\"The probability of having four healthy kids is {dist.pmf(4):.3f}\")",
"The probability of having four healthy kids is 0.396\n"
]
],
[
[
"### <font color=darkred>Probability to have each of 0-5 healthy kids.</font>",
"_____no_output_____"
]
],
[
[
"# all possible # of successes out of n trials\n# i.e. all possible outcomes of the random variable\n# i.e. all possible number of healthy kids = 0-5\nnumHealthyKids = np.arange(n+1)\n\nnumHealthyKids",
"_____no_output_____"
],
[
"# probability of obtaining each possible number of successes\n# i.e. probability of having each possible number of healthy children\npmf = dist.pmf(numHealthyKids)\n\npmf",
"_____no_output_____"
]
],
[
[
"### <font color=darkred>Visualize the probability to have each of 0-5 healthy kids.</font>",
"_____no_output_____"
]
],
[
[
"plt.bar(numHealthyKids, pmf)\nplt.xlabel('# healthy children', fontsize=18)\nplt.ylabel('probability', fontsize=18);",
"_____no_output_____"
]
],
[
[
"### <font color=darkred>Probability to have at least 4 healthy kids.</font>",
"_____no_output_____"
]
],
[
[
"# sum of probabilities of 4 and 5 healthy kids\npmf[-2:].sum()",
"_____no_output_____"
],
[
"# remaining probability after subtracting CDF for 3 kids\n1 - dist.cdf(3)",
"_____no_output_____"
],
[
"# survival function for 3 kids\ndist.sf(3)",
"_____no_output_____"
]
],
[
[
"### <font color=darkred>What is the expected number of healthy kids?</font>",
"_____no_output_____"
]
],
[
[
"print(f\"The expected number of healthy kids is {dist.mean()}\")",
"The expected number of healthy kids is 3.75\n"
]
],
[
[
"### <font color=darkred>How sure are we about the above estimate?</font>",
"_____no_output_____"
]
],
[
[
"print(f\"The expected number of healthy kids is {dist.mean()} ± {dist.std():.2f}\")",
"The expected number of healthy kids is 3.75 ± 0.97\n"
]
],
[
[
"# <font color=red> Exercise</font>\n\nShould the couple consider having six children?\n\n1. Plot the *pmf* for the probability of each possible number of healthy children.\n2. What's the probability that they will all be healthy?",
"_____no_output_____"
],
[
"# Poisson Distribution\n\n### <font color=darkred> **Example**: Assume that the rate of deleterious mutations is ~1.2 per diploid genome. What is the probability that an individual has 8 or more spontaneous deleterious mutations?</font>\n\nIn this case the random variable is the number of deleterious mutations within an individuals genome.",
"_____no_output_____"
]
],
[
[
"# the rate of deleterious mutations is 1.2 per diploid genome\nrate = 1.2\n\n# poisson distribution describing the predicted number of spontaneous mutations\ndist = st.poisson(rate)",
"_____no_output_____"
],
[
"# let's look at the probability for 0-10 mutations\nnumMutations = np.arange(11)\n\nplt.bar(numMutations, dist.pmf(numMutations))\nplt.xlabel('# mutations', fontsize=18)\nplt.ylabel('probability', fontsize=18);",
"_____no_output_____"
],
[
"print(f\"Probability of less than 8 mutations = {dist.cdf(7)}\")\nprint(f\"Probability of 8 or more mutations = {dist.sf(7)}\")\ndist.cdf(7) + dist.sf(7)",
"Probability of less than 8 mutations = 0.9999630211320938\nProbability of 8 or more mutations = 3.6978867906171055e-05\n"
]
],
[
[
"# <font color=red> Exercise</font>\n\nFor the above example, what is the probability that an individual has three or fewer mutations?",
"_____no_output_____"
],
[
"# Exponential Distribution\n\n### <font color=darkred> **Example**: Assume that a neuron spikes 1.5 times per second on average. Plot the probability density function of interspike intervals from zero to five seconds with a resolution of 0.01 seconds.</font>\n\nIn this case the random variable is the interspike interval time.",
"_____no_output_____"
]
],
[
[
"# spike rate per second\nrate = 1.5\n\n# exponential distribution describing the neuron's predicted interspike intervals\ndist = st.expon(loc=0, scale=1/rate)",
"_____no_output_____"
],
[
"# plot interspike intervals from 0-5 seconds at 0.01 sec resolution\nintervalsSec = np.linspace(0, 5, 501)\n\n# probability density for each interval\npdf = dist.pdf(intervalsSec)\n\nplt.plot(intervalsSec, pdf)\nplt.xlabel('interspike interval (sec)', fontsize=18)\nplt.ylabel('pdf', fontsize=18);",
"_____no_output_____"
]
],
[
[
"### <font color=darkred>What is the average interval?</font>",
"_____no_output_____"
]
],
[
[
"print(f\"Average interspike interval = {dist.mean():.2f} seconds.\")",
"Average interspike interval = 0.67 seconds.\n"
]
],
[
[
"### <font color=darkred>time constant = 1 / rate = mean</font>",
"_____no_output_____"
]
],
[
[
"tau = 1 / rate\ntau",
"_____no_output_____"
]
],
[
[
"### <font color=darkred> What is the probability that an interval will be between 1 and 2 seconds?</font>",
"_____no_output_____"
]
],
[
[
"prob1to2 = dist.cdf(2) - dist.cdf(1);\n\nprint(f\"Probability of an interspike interval being between 1 and 2 seconds is {prob1to2:.2f}\")",
"Probability of an interspike interval being between 1 and 2 seconds is 0.17\n"
]
],
[
[
"### <font color=darkred> For what time *T* is the probability that an interval is shorter than *T* equal to 25%?</font>",
"_____no_output_____"
]
],
[
[
"timeAtFirst25PercentOfDist = dist.ppf(0.25) # percent point function\n\nprint(f\"There is a 25% chance that an interval is shorter than {timeAtFirst25PercentOfDist:.2f} seconds.\")",
"There is a 25% chance that an interval is shorter than 0.19 seconds.\n"
]
],
[
[
"# <font color=red> Exercise</font>\n\nFor the above example, what is the probability that 3 seconds will pass without any spikes?",
"_____no_output_____"
],
[
"# Normal Distribution\n\n### <font color=darkred> **Example**: Under basal conditions the resting membrane voltage of a neuron fluctuates around -70 mV with a variance of 10 mV.</font>\n\nIn this case the random variable is the neuron's resting membrane voltage.",
"_____no_output_____"
]
],
[
[
"# mean resting membrane voltage (mV)\nmu = -70\n\n# standard deviation about the mean\nsd = np.sqrt(10)\n\n# normal distribution describing the neuron's predicted resting membrane voltage\ndist = st.norm(mu, sd)",
"_____no_output_____"
],
[
"# membrane voltages from -85 to -55 mV\nmV = np.linspace(-85, -55, 301)\n\n# probability density for each membrane voltage in mV\npdf = dist.pdf(mV)\n\nplt.plot(mV, pdf)\nplt.xlabel('membrane voltage (mV)', fontsize=18)\nplt.ylabel('pdf', fontsize=18);",
"_____no_output_____"
]
],
[
[
"### <font color=darkred> What range of membrane voltages (centered on the mean) account for 95% of the probability.</font>",
"_____no_output_____"
]
],
[
[
"low = dist.ppf(0.025) # first 2.5% of distribution\nhigh = dist.ppf(0.975) # first 97.5% of distribution\n\nprint(f\"95% of membrane voltages are expected to fall within {low :.1f} and {high :.1f} mV.\")",
"95% of membrane voltages are expected to fall within -76.2 and -63.8 mV.\n"
]
],
[
[
"# <font color=red> Exercise</font>\n\nIn a resting neuron, what's the probability that you would measure a membrane voltage greater than -65 mV?\n\nIf you meaassure -65 mV, is the neuron at rest?",
"_____no_output_____"
],
[
"# <font color=red> Exercise</font>\n\nWhat probability distribution might best describe the number of synapses per millimeter of dendrite?\n\n A) Binomial\n B) Poisson\n C) Exponential\n D) Normal",
"_____no_output_____"
],
[
"# <font color=red> Exercise</font>\n\nWhat probability distribution might best describe the time a protein spends in its active conformation?\n\n A) Binomial\n B) Poisson\n C) Exponential\n D) Normal",
"_____no_output_____"
],
[
"# <font color=red> Exercise</font>\n\nWhat probability distribution might best describe the weights of adult mice in a colony?\n\n A) Binomial\n B) Poisson\n C) Exponential\n D) Normal",
"_____no_output_____"
],
[
"# <font color=red> Exercise</font>\n\nWhat probability distribution might best describe the number of times a subject is able to identify the correct target in a series of trials?\n\n A) Binomial\n B) Poisson\n C) Exponential\n D) Normal",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d05f8ad393ce4d28a239766c45012931fc3e776b | 70,843 | ipynb | Jupyter Notebook | handwashing_notebook.ipynb | shukkkur/Analyzing-The-Discovery-of-Handwashing | 320cb0b3a9fd620a3d3d523f4df346c97cb51938 | [
"CC-BY-4.0"
] | 5 | 2021-08-28T16:13:59.000Z | 2022-02-04T05:51:34.000Z | handwashing_notebook.ipynb | shukkkur/Analyzing-The-Discovery-of-Handwashing | 320cb0b3a9fd620a3d3d523f4df346c97cb51938 | [
"CC-BY-4.0"
] | null | null | null | handwashing_notebook.ipynb | shukkkur/Analyzing-The-Discovery-of-Handwashing | 320cb0b3a9fd620a3d3d523f4df346c97cb51938 | [
"CC-BY-4.0"
] | null | null | null | 121.723368 | 33,192 | 0.845701 | [
[
[
"### Dr. Ignaz Semmelweis",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nfrom IPython.display import display",
"_____no_output_____"
],
[
"# Read datasets/yearly_deaths_by_clinic.csv into yearly\nyearly = pd.read_csv('datasets/yearly_deaths_by_clinic.csv')\n\n# Print out yearly\ndisplay(yearly)",
"_____no_output_____"
]
],
[
[
"### The alarming number of deaths",
"_____no_output_____"
]
],
[
[
"# Calculate proportion of deaths per no. births\nyearly['proportion_deaths'] = yearly.deaths / yearly.births\n\n# Extract Clinic 1 data into clinic_1 and Clinic 2 data into clinic_2\nclinic_1 = yearly[yearly.clinic == 'clinic 1']\nclinic_2 = yearly[yearly.clinic == 'clinic 2']\n\n# Print out clinic_1\ndisplay(clinic_2)",
"_____no_output_____"
]
],
[
[
"### Death at the clinics",
"_____no_output_____"
]
],
[
[
"# Plot yearly proportion of deaths at the two clinics\nax = clinic_1.plot(x='year', y='proportion_deaths', label='Clinic 1')\nclinic_2.plot(x='year', y='proportion_deaths', label='Clinic 2', ax=ax)\n\nplt.ylabel(\"Proportion deaths\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"### The handwashing",
"_____no_output_____"
]
],
[
[
"# Read datasets/monthly_deaths.csv into monthly\nmonthly = pd.read_csv('datasets/monthly_deaths.csv', parse_dates=['date'])\n\n# Calculate proportion of deaths per no. births\nmonthly[\"proportion_deaths\"] = monthly.deaths/monthly.births\n\n# Print out the first rows in monthly\ndisplay(monthly.head())",
"_____no_output_____"
]
],
[
[
"### The effect of handwashing",
"_____no_output_____"
]
],
[
[
"# Date when handwashing was made mandatory\nhandwashing_start = pd.to_datetime('1847-06-01')\n\n# Split monthly into before and after handwashing_start\nbefore_washing = monthly[monthly.date < handwashing_start]\nafter_washing = monthly[monthly.date >= handwashing_start]\n\n# Plot monthly proportion of deaths before and after handwashing\nax = before_washing.plot(x='date', \n y='proportion_deaths', label='Before Washing')\nafter_washing.plot(x='date',y='proportion_deaths', label='After Washing', ax=ax)\n\nplt.ylabel(\"Proportion deaths\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"### More handwashing, fewer deaths?",
"_____no_output_____"
]
],
[
[
"# Difference in mean monthly proportion of deaths due to handwashing\nbefore_proportion = before_washing.proportion_deaths\nafter_proportion = after_washing.proportion_deaths\n\nmean_diff = after_proportion.mean() - before_proportion.mean()\nprint(mean_diff)",
"-0.0839566075118334\n"
]
],
[
[
"### Bootstrap analysis",
"_____no_output_____"
]
],
[
[
"# A bootstrap analysis of the reduction of deaths due to handwashing\nboot_mean_diff = []\n\nfor i in range(3000):\n boot_before = before_proportion.sample(replace=True,n=len(before_proportion))\n boot_after = after_proportion.sample(replace=True,n=len(after_proportion))\n boot_mean_diff.append(boot_after.mean()-boot_before.mean())\n\n# Calculating a 95% confidence interval from boot_mean_diff \nconfidence_interval = pd.Series(boot_mean_diff).quantile([0.025, 0.975] )\nprint(confidence_interval)",
"0.025 -0.101638\n0.975 -0.067481\ndtype: float64\n"
]
],
[
[
"### Conclusion",
"_____no_output_____"
]
],
[
[
"# The data Semmelweis collected points to that:\ndoctors_should_wash_their_hands = True\nprint(doctors_should_wash_their_hands)",
"True\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d05f8dd61a9531c7bcf92bd06a31266d23a59a18 | 412,271 | ipynb | Jupyter Notebook | Neelesh_Color-Mappings_opencv.ipynb | Shreyansh-Gupta/Open-contributions | e72a9ce2b0aa6a48081921bf8138b91ad259c422 | [
"MIT"
] | 61 | 2020-09-10T05:16:19.000Z | 2021-11-07T00:22:46.000Z | Neelesh_Color-Mappings_opencv.ipynb | Shreyansh-Gupta/Open-contributions | e72a9ce2b0aa6a48081921bf8138b91ad259c422 | [
"MIT"
] | 72 | 2020-09-12T09:34:19.000Z | 2021-08-01T17:48:46.000Z | Neelesh_Color-Mappings_opencv.ipynb | Shreyansh-Gupta/Open-contributions | e72a9ce2b0aa6a48081921bf8138b91ad259c422 | [
"MIT"
] | 571 | 2020-09-10T01:52:56.000Z | 2022-03-26T17:26:23.000Z | 2,396.924419 | 139,756 | 0.96153 | [
[
[
"# Colorspaces\n\nLet's have a brief introduction into converting to different colorspaces! The video goes into more detail about colorspaces.\n\n",
"_____no_output_____"
]
],
[
[
"import cv2\nimport matplotlib.pyplot as plt\nimport numpy as np\n%matplotlib inline",
"_____no_output_____"
],
[
"img = cv2.imread('../DATA/00-puppy.jpg')",
"_____no_output_____"
]
],
[
[
"### Converting to Different Colorspaces",
"_____no_output_____"
]
],
[
[
"img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)\nplt.imshow(img)",
"_____no_output_____"
]
],
[
[
"**Converting to HSV**\nhttps://en.wikipedia.org/wiki/HSL_and_HSV",
"_____no_output_____"
]
],
[
[
"img = cv2.cvtColor(img, cv2.COLOR_BGR2HSV)\nplt.imshow(img)",
"_____no_output_____"
],
[
"img = cv2.imread('../DATA/00-puppy.jpg')\nimg = cv2.cvtColor(img, cv2.COLOR_BGR2HLS)\nplt.imshow(img)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d05f987ba6a006804e11639ff66a6fc99377f621 | 748,767 | ipynb | Jupyter Notebook | Elo merchant/Elo_mixer1.ipynb | nguyenphuhien13/Kaggle | 4aa0438e7d0c73d51e111cf2b7026545155701fb | [
"MIT"
] | 2 | 2019-07-09T06:31:44.000Z | 2019-07-09T19:41:11.000Z | Elo merchant/Elo_mixer1.ipynb | nguyenphuhien13/Kaggle | 4aa0438e7d0c73d51e111cf2b7026545155701fb | [
"MIT"
] | null | null | null | Elo merchant/Elo_mixer1.ipynb | nguyenphuhien13/Kaggle | 4aa0438e7d0c73d51e111cf2b7026545155701fb | [
"MIT"
] | null | null | null | 349.23834 | 335,024 | 0.921086 | [
[
[
"# Import the necessary libraries\nimport numpy as np\nimport pandas as pd\nimport os\nimport time\nimport warnings\nimport gc\ngc.collect()\nimport os\nfrom six.moves import urllib\nimport matplotlib\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport datetime\nwarnings.filterwarnings('ignore')\n%matplotlib inline\nplt.style.use('seaborn')\nfrom scipy import stats\nfrom scipy.stats import norm, skew\nfrom sklearn.preprocessing import StandardScaler",
"_____no_output_____"
],
[
"#Add All the Models Libraries\n# preprocessing\nfrom sklearn.preprocessing import LabelEncoder\nlabel_enc = LabelEncoder()\n# Scalers\nfrom sklearn.utils import shuffle\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.pipeline import FeatureUnion\n\n# Models\n\nfrom sklearn.linear_model import Lasso\nfrom sklearn.metrics import mean_squared_log_error,mean_squared_error, r2_score,mean_absolute_error\n\nfrom sklearn.model_selection import train_test_split #training and testing data split\nfrom sklearn import metrics #accuracy measure\nfrom sklearn.metrics import confusion_matrix #for confusion matrix\nfrom scipy.stats import reciprocal, uniform\n\nfrom sklearn.model_selection import StratifiedKFold, RepeatedKFold\n\n# Cross-validation\nfrom sklearn.model_selection import KFold #for K-fold cross validation\nfrom sklearn.model_selection import cross_val_score #score evaluation\nfrom sklearn.model_selection import cross_val_predict #prediction\nfrom sklearn.model_selection import cross_validate\n\n# GridSearchCV\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.model_selection import RandomizedSearchCV\n\n#Common data processors\nfrom sklearn.preprocessing import OneHotEncoder, LabelEncoder\nfrom sklearn import feature_selection\nfrom sklearn import model_selection\nfrom sklearn import metrics\nfrom sklearn.base import BaseEstimator, TransformerMixin\nfrom sklearn.utils import check_array\nfrom scipy import sparse",
"_____no_output_____"
],
[
"# to make this notebook's output stable across runs\nnp.random.seed(123)\ngc.collect()\n# To plot pretty figures\n%matplotlib inline\nplt.rcParams['axes.labelsize'] = 14\nplt.rcParams['xtick.labelsize'] = 12\nplt.rcParams['ytick.labelsize'] = 12",
"_____no_output_____"
],
[
"#Reduce the memory usage - by Panchajanya Banerjee\ndef reduce_mem_usage(df, verbose=True):\n numerics = ['int16', 'int32', 'int64', 'float16', 'float32', 'float64']\n start_mem = df.memory_usage().sum() / 1024**2 \n for col in df.columns:\n col_type = df[col].dtypes\n if col_type in numerics:\n c_min = df[col].min()\n c_max = df[col].max()\n if str(col_type)[:3] == 'int':\n if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:\n df[col] = df[col].astype(np.int8)\n elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:\n df[col] = df[col].astype(np.int16)\n elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:\n df[col] = df[col].astype(np.int32)\n elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:\n df[col] = df[col].astype(np.int64) \n else:\n if c_min > np.finfo(np.float16).min and c_max < np.finfo(np.float16).max:\n df[col] = df[col].astype(np.float16)\n elif c_min > np.finfo(np.float32).min and c_max < np.finfo(np.float32).max:\n df[col] = df[col].astype(np.float32)\n else:\n df[col] = df[col].astype(np.float64) \n end_mem = df.memory_usage().sum() / 1024**2\n if verbose: print('Mem. usage decreased to {:5.2f} Mb ({:.1f}% reduction)'.format(end_mem, 100 * (start_mem - end_mem) / start_mem))\n return df",
"_____no_output_____"
],
[
"train = reduce_mem_usage(pd.read_csv('train.csv',parse_dates=[\"first_active_month\"]))\ntest = reduce_mem_usage(pd.read_csv('test.csv', parse_dates=[\"first_active_month\"]))",
"Mem. usage decreased to 4.04 Mb (56.2% reduction)\nMem. usage decreased to 2.24 Mb (52.5% reduction)\n"
],
[
"test.first_active_month = test.first_active_month.fillna(pd.to_datetime('2017-09-01'))\ntest.isnull().sum()",
"_____no_output_____"
],
[
"# Now extract the month, year, day, weekday\ntrain[\"month\"] = train[\"first_active_month\"].dt.month\ntrain[\"year\"] = train[\"first_active_month\"].dt.year\ntrain['week'] = train[\"first_active_month\"].dt.weekofyear\ntrain['dayofweek'] = train['first_active_month'].dt.dayofweek\ntrain['days'] = (datetime.date(2018, 2, 1) - train['first_active_month'].dt.date).dt.days\ntrain['quarter'] = train['first_active_month'].dt.quarter\n\ntest[\"month\"] = test[\"first_active_month\"].dt.month\ntest[\"year\"] = test[\"first_active_month\"].dt.year\ntest['week'] = test[\"first_active_month\"].dt.weekofyear\ntest['dayofweek'] = test['first_active_month'].dt.dayofweek\ntest['days'] = (datetime.date(2018, 2, 1) - test['first_active_month'].dt.date).dt.days\ntest['quarter'] = test['first_active_month'].dt.quarter",
"_____no_output_____"
],
[
"# Taking Reference from Other Kernels\ndef aggregate_transaction_hist(trans, prefix): \n \n agg_func = {\n 'purchase_date' : ['max','min'],\n 'month_diff' : ['mean', 'min', 'max', 'var'],\n 'month_diff_lag' : ['mean', 'min', 'max', 'var'],\n 'weekend' : ['sum', 'mean'],\n 'authorized_flag': ['sum', 'mean'],\n 'category_1': ['sum','mean', 'max','min'],\n 'purchase_amount': ['sum', 'mean', 'max', 'min', 'std'],\n 'installments': ['sum', 'mean', 'max', 'min', 'std'], \n 'month_lag': ['max','min','mean','var'],\n 'card_id' : ['size'],\n 'month': ['nunique'],\n 'hour': ['nunique'],\n 'weekofyear': ['nunique'],\n 'dayofweek': ['nunique'],\n 'year': ['nunique'],\n 'subsector_id': ['nunique'],\n 'merchant_category_id' : ['nunique', lambda x:stats.mode(x)[0]],\n 'merchant_id' : ['nunique', lambda x:stats.mode(x)[0]],\n 'state_id' : ['nunique', lambda x:stats.mode(x)[0]],\n }\n \n agg_trans = trans.groupby(['card_id']).agg(agg_func)\n agg_trans.columns = [prefix + '_'.join(col).strip() for col in agg_trans.columns.values]\n agg_trans.reset_index(inplace=True)\n \n df = (trans.groupby('card_id').size().reset_index(name='{}transactions_count'.format(prefix)))\n \n agg_trans = pd.merge(df, agg_trans, on='card_id', how='left')\n \n return agg_trans",
"_____no_output_____"
],
[
"transactions = reduce_mem_usage(pd.read_csv('historical_transactions_clean_outlier.csv'))\ntransactions = transactions.loc[transactions.purchase_amount < 50,]\ntransactions['authorized_flag'] = transactions['authorized_flag'].map({'Y': 1, 'N': 0})\ntransactions['category_1'] = transactions['category_1'].map({'Y': 0, 'N': 1})",
"Mem. usage decreased to 1638.06 Mb (50.8% reduction)\n"
],
[
"#Feature Engineering - Adding new features \ntransactions['purchase_date'] = pd.to_datetime(transactions['purchase_date'])\ntransactions['year'] = transactions['purchase_date'].dt.year\ntransactions['weekofyear'] = transactions['purchase_date'].dt.weekofyear\ntransactions['month'] = transactions['purchase_date'].dt.month\ntransactions['dayofweek'] = transactions['purchase_date'].dt.dayofweek\ntransactions['weekend'] = (transactions.purchase_date.dt.weekday >=5).astype(int)\ntransactions['hour'] = transactions['purchase_date'].dt.hour \ntransactions['quarter'] = transactions['purchase_date'].dt.quarter\ntransactions['month_diff'] = ((pd.to_datetime('01/03/2018') - transactions['purchase_date']).dt.days)//30\ntransactions['month_diff_lag'] = transactions['month_diff'] + transactions['month_lag']\n\ngc.collect()",
"_____no_output_____"
],
[
"def aggregate_bymonth(trans, prefix): \n \n agg_func = {\n 'purchase_amount': ['sum', 'mean'],\n 'card_id' : ['size'],\n 'merchant_category_id' : ['nunique', lambda x:stats.mode(x)[0]],\n# 'merchant_id' : ['nunique', lambda x:stats.mode(x)[0]],\n }\n \n agg_trans = trans.groupby(['card_id','month','year']).agg(agg_func)\n agg_trans.columns = [prefix + '_'.join(col).strip() for col in agg_trans.columns.values]\n agg_trans.reset_index(inplace=True)\n \n df = (trans.groupby('card_id').size().reset_index(name='{}transactions_count'.format(prefix)))\n \n agg_trans = pd.merge(df, agg_trans, on='card_id', how='left')\n \n return agg_trans\n\nmerge = aggregate_bymonth(transactions, prefix='hist_')\nmerge = merge.drop(['hist_transactions_count'], axis = 1)\nmerge['Date'] = pd.to_datetime(merge[['year', 'month']].assign(Day=1))",
"_____no_output_____"
],
[
"df1 = merge.groupby(['card_id', 'hist_merchant_category_id_<lambda>']).size().reset_index(name='Count')\ndf1 = df1.loc[df1.Count > 1]\ndf1 = df1.groupby(['card_id']).agg({'Count':['sum']})\ndf1.columns = ['category_repeated_month']\n\ntrain = pd.merge(train, df1, on='card_id',how='left')\ntest = pd.merge(test, df1, on='card_id',how='left')\ndf1\ngc.collect()",
"_____no_output_____"
],
[
"## Second last month\namerge = merge.sort_values('Date').groupby('card_id', \n as_index=False).apply(lambda x: x.iloc[-2])[['card_id','hist_card_id_size','hist_purchase_amount_sum','hist_purchase_amount_mean']]\nnew_names = [(i,i+'_last2') for i in amerge.iloc[:, 1:].columns.values]\namerge.rename(columns = dict(new_names), inplace=True)\n\ntrain = pd.merge(train, amerge, on='card_id',how='left')\ntest = pd.merge(test, amerge, on='card_id',how='left')\n\ngc.collect()",
"_____no_output_____"
],
[
"# last month and first month\nmerge1 = merge.loc[merge.groupby('card_id').Date.idxmax(),:][[ 'card_id','hist_card_id_size',\n 'hist_purchase_amount_sum','hist_purchase_amount_mean']]\nnew_names = [(i,i+'_last') for i in merge1.iloc[:, 1:].columns.values]\nmerge1.rename(columns = dict(new_names), inplace=True)\n\nmerge2 = merge.loc[merge.groupby('card_id').Date.idxmin(),:][['card_id','hist_card_id_size',\n 'hist_purchase_amount_sum','hist_purchase_amount_mean']]\nnew_names = [(i,i+'_first') for i in merge2.iloc[:, 1:].columns.values]\nmerge2.rename(columns = dict(new_names), inplace=True)\ncomb = pd.merge(merge1, merge2, on='card_id',how='left')\n\ntrain = pd.merge(train, comb, on='card_id',how='left')\ntest = pd.merge(test, comb, on='card_id',how='left')\n\ngc.collect()",
"_____no_output_____"
],
[
"## Same merchant purchase\ndf = (transactions.groupby(['card_id','merchant_id','purchase_amount']).size().reset_index(name='count_hist'))\ndf['purchase_amount_hist'] = df.groupby(['card_id','merchant_id'])['purchase_amount'].transform('sum')\ndf['count_hist'] = df.groupby(['card_id','merchant_id'])['count_hist'].transform('sum')\ndf = df.drop_duplicates()\ndf = df.loc[df['count_hist'] >= 2]\nagg_func = {\n 'count_hist' : ['count'],\n 'purchase_amount_hist':['sum','mean'],\n 'purchase_amount':['sum','mean'],\n}\ndf = df.groupby(['card_id']).agg(agg_func)\ndf.columns = [''.join(col).strip() for col in df.columns.values]\nnew_names = [(i,i+'_merhist') for i in df.iloc[:, 3:].columns.values]\ndf.rename(columns = dict(new_names), inplace=True)\n\ntrain = pd.merge(train, df, on='card_id',how='left')\ntest = pd.merge(test, df, on='card_id',how='left')",
"_____no_output_____"
],
[
"# Same category purchase\ndf = (transactions.groupby(['card_id','merchant_category_id','purchase_amount']).size().reset_index(name='hist_count'))\ndf['hist_purchase_amount'] = df.groupby(['card_id','merchant_category_id'])['purchase_amount'].transform('sum')\ndf['hist_count'] = df.groupby(['card_id','merchant_category_id'])['hist_count'].transform('sum')\ndf = df.drop_duplicates()\ndf = df.loc[df['hist_count'] >= 2]\ndf['hist_count_4'] = 0\ndf.loc[df['hist_count'] >= 4, 'hist_count_4'] = 1\ndf['hist_mean4'] = 0\ndf.loc[df['hist_count'] >= 4, 'hist_mean4'] = df['hist_purchase_amount']/df['hist_count']\nagg_fun = {\n 'hist_count' : ['count'],\n 'hist_count_4' : ['sum'],\n 'hist_purchase_amount':['sum','mean'],\n 'hist_mean4' : ['sum','mean'],\n 'purchase_amount':['sum','mean'],\n}\ndf = df.groupby(['card_id']).agg(agg_fun)\ndf.columns = [''.join(col).strip() for col in df.columns.values]\nnew_names = [(i,'hist'+i) for i in df.iloc[:, 6:].columns.values]\ndf.rename(columns = dict(new_names), inplace=True)\n\ntrain = pd.merge(train, df, on='card_id',how='left')\ntest = pd.merge(test, df, on='card_id',how='left')",
"_____no_output_____"
],
[
"# agg_func = {'mean': ['mean'],}\n# for col in ['category_2','category_3']:\n# transactions[col+'_mean'] = transactions['purchase_amount'].groupby(transactions[col]).agg('mean')\n# transactions[col+'_max'] = transactions['purchase_amount'].groupby(transactions[col]).agg('max')\n# transactions[col+'_min'] = transactions['purchase_amount'].groupby(transactions[col]).agg('min')\n# transactions[col+'_var'] = transactions['purchase_amount'].groupby(transactions[col]).agg('var')\n# agg_func[col+'_mean'] = ['mean']\n# gc.collect()",
"_____no_output_____"
],
[
"merchants = reduce_mem_usage(pd.read_csv('merchants_clean.csv'))\nmerchants = merchants.drop(['Unnamed: 0', 'merchant_group_id', 'merchant_category_id',\n 'subsector_id', 'numerical_1', 'numerical_2', \n 'active_months_lag3','active_months_lag6',\n 'city_id', 'state_id'\n ], axis = 1)\nd = dict(zip(merchants.columns[1:], ['histchant_{}'.format(x) for x in (merchants.columns[1:])]))\nd.update({\"merchant_id\": \"hist_merchant_id_<lambda>\"})\nmerchants = merchants.rename(index=str, columns= d)\n## convert the month in business to categorical\nmerchants.histchant_active_months_lag12 = pd.cut(merchants.histchant_active_months_lag12, 4)",
"Mem. usage decreased to 16.91 Mb (71.2% reduction)\n"
],
[
"merge_trans = aggregate_transaction_hist(transactions, prefix='hist_')\nmerge_trans = merge_trans.merge(merchants, on = 'hist_merchant_id_<lambda>', how = 'left')\n## hist transaction frequency\nmerge_trans['hist_freq'] = merge_trans.hist_transactions_count/(((merge_trans.hist_purchase_date_max - \n merge_trans.hist_purchase_date_min).dt.total_seconds())/86400)\nmerge_trans['hist_freq_amount'] = merge_trans['hist_freq'] * merge_trans['hist_purchase_amount_mean']\nmerge_trans['hist_freq_install'] = merge_trans['hist_freq'] * merge_trans['hist_installments_mean']",
"_____no_output_____"
],
[
"cols = ['histchant_avg_sales_lag3','histchant_avg_purchases_lag3',\n 'histchant_avg_sales_lag6','histchant_avg_purchases_lag6',\n 'histchant_avg_sales_lag12','histchant_avg_purchases_lag12','hist_freq']\nfor col in cols:\n merge_trans[col] = pd.qcut(merge_trans[col], 4)",
"_____no_output_____"
],
[
"for col in cols:\n merge_trans[col].fillna(merge_trans[col].mode()[0], inplace=True)\n label_enc.fit(list(merge_trans[col].values)) \n merge_trans[col] = label_enc.transform(list(merge_trans[col].values))",
"_____no_output_____"
],
[
"for col in ['histchant_category_1','histchant_most_recent_sales_range','histchant_most_recent_purchases_range',\n 'histchant_active_months_lag12','histchant_category_4','histchant_category_2']:\n merge_trans[col].fillna(merge_trans[col].mode()[0], inplace=True)",
"_____no_output_____"
],
[
"label_enc.fit(list(merge_trans['hist_merchant_id_<lambda>'].values)) \nmerge_trans['hist_merchant_id_<lambda>'] = label_enc.transform(list(merge_trans['hist_merchant_id_<lambda>'].values))\nlabel_enc.fit(list(merge_trans['histchant_active_months_lag12'].values)) \nmerge_trans['histchant_active_months_lag12'] = label_enc.transform(list(merge_trans['histchant_active_months_lag12'].values))",
"_____no_output_____"
],
[
"#del transactions\ngc.collect()\ntrain = pd.merge(train, merge_trans, on='card_id',how='left')\ntest = pd.merge(test, merge_trans, on='card_id',how='left')\n#del merge_trans\ngc.collect()",
"_____no_output_____"
],
[
"#Feature Engineering - Adding new features \ntrain['hist_purchase_date_max'] = pd.to_datetime(train['hist_purchase_date_max'])\ntrain['hist_purchase_date_min'] = pd.to_datetime(train['hist_purchase_date_min'])\ntrain['hist_purchase_date_diff'] = (train['hist_purchase_date_max'] - train['hist_purchase_date_min']).dt.days\ntrain['hist_purchase_date_average'] = train['hist_purchase_date_diff']/train['hist_card_id_size']\ntrain['hist_purchase_date_uptonow'] = (pd.to_datetime('01/03/2018') - train['hist_purchase_date_max']).dt.days\ntrain['hist_purchase_date_uptomin'] = (pd.to_datetime('01/03/2018') - train['hist_purchase_date_min']).dt.days\ntrain['hist_first_buy'] = (train['hist_purchase_date_min'] - train['first_active_month']).dt.days\nfor feature in ['hist_purchase_date_max','hist_purchase_date_min']:\n train[feature] = train[feature].astype(np.int64) * 1e-9\ngc.collect()",
"_____no_output_____"
],
[
"#Feature Engineering - Adding new features \ntest['hist_purchase_date_max'] = pd.to_datetime(test['hist_purchase_date_max'])\ntest['hist_purchase_date_min'] = pd.to_datetime(test['hist_purchase_date_min'])\ntest['hist_purchase_date_diff'] = (test['hist_purchase_date_max'] - test['hist_purchase_date_min']).dt.days\ntest['hist_purchase_date_average'] = test['hist_purchase_date_diff']/test['hist_card_id_size']\ntest['hist_purchase_date_uptonow'] = (pd.to_datetime('01/03/2018') - test['hist_purchase_date_max']).dt.days\ntest['hist_purchase_date_uptomin'] = (pd.to_datetime('01/03/2018') - test['hist_purchase_date_min']).dt.days\ntest['hist_first_buy'] = (test['hist_purchase_date_min'] - test['first_active_month']).dt.days\nfor feature in ['hist_purchase_date_max','hist_purchase_date_min']:\n test[feature] = test[feature].astype(np.int64) * 1e-9\n\ngc.collect()",
"_____no_output_____"
],
[
"# Taking Reference from Other Kernels\ndef aggregate_transaction_new(trans, prefix): \n \n agg_func = {\n 'purchase_date' : ['max','min'],\n 'month_diff' : ['mean', 'min', 'max'],\n 'month_diff_lag' : ['mean', 'min', 'max'],\n 'weekend' : ['sum', 'mean'],\n 'authorized_flag': ['sum'],\n 'category_1': ['sum','mean', 'max','min'],\n 'purchase_amount': ['sum', 'mean', 'max', 'min'],\n 'installments': ['sum', 'mean', 'max', 'min'], \n 'month_lag': ['max','min','mean'],\n 'card_id' : ['size'],\n 'month': ['nunique'],\n 'hour': ['nunique'],\n 'weekofyear': ['nunique'],\n 'dayofweek': ['nunique'],\n 'year': ['nunique'],\n 'subsector_id': ['nunique'],\n 'merchant_category_id' : ['nunique', lambda x:stats.mode(x)[0]],\n 'merchant_id' : ['nunique', lambda x:stats.mode(x)[0]],\n 'state_id' : ['nunique', lambda x:stats.mode(x)[0]],\n }\n \n agg_trans = trans.groupby(['card_id']).agg(agg_func)\n agg_trans.columns = [prefix + '_'.join(col).strip() for col in agg_trans.columns.values]\n agg_trans.reset_index(inplace=True)\n \n df = (trans.groupby('card_id').size().reset_index(name='{}transactions_count'.format(prefix)))\n \n agg_trans = pd.merge(df, agg_trans, on='card_id', how='left')\n \n return agg_trans",
"_____no_output_____"
],
[
"# Now extract the data from the new transactions\nnew_transactions = reduce_mem_usage(pd.read_csv('new_merchant_transactions_clean_outlier.csv'))\nnew_transactions = new_transactions.loc[new_transactions.purchase_amount < 50,]\nnew_transactions['authorized_flag'] = new_transactions['authorized_flag'].map({'Y': 1, 'N': 0})\nnew_transactions['category_1'] = new_transactions['category_1'].map({'Y': 0, 'N': 1})",
"Mem. usage decreased to 106.71 Mb (52.5% reduction)\n"
],
[
"#Feature Engineering - Adding new features inspired by Chau's first kernel\nnew_transactions['purchase_date'] = pd.to_datetime(new_transactions['purchase_date'])\nnew_transactions['year'] = new_transactions['purchase_date'].dt.year\nnew_transactions['weekofyear'] = new_transactions['purchase_date'].dt.weekofyear\nnew_transactions['month'] = new_transactions['purchase_date'].dt.month\nnew_transactions['dayofweek'] = new_transactions['purchase_date'].dt.dayofweek\nnew_transactions['weekend'] = (new_transactions.purchase_date.dt.weekday >=5).astype(int)\nnew_transactions['hour'] = new_transactions['purchase_date'].dt.hour \nnew_transactions['quarter'] = new_transactions['purchase_date'].dt.quarter\nnew_transactions['is_month_start'] = new_transactions['purchase_date'].dt.is_month_start\nnew_transactions['month_diff'] = ((pd.to_datetime('01/03/2018') - new_transactions['purchase_date']).dt.days)//30\nnew_transactions['month_diff_lag'] = new_transactions['month_diff'] + new_transactions['month_lag']\n\ngc.collect()\n\n# new_transactions['Christmas_Day_2017'] = (pd.to_datetime('2017-12-25') - \n# new_transactions['purchase_date']).dt.days.apply(lambda x: x if x > 0 and x <= 15 else 0)\n# new_transactions['Valentine_Day_2017'] = (pd.to_datetime('2017-06-13') - \n# new_transactions['purchase_date']).dt.days.apply(lambda x: x if x > 0 and x <= 7 else 0)\n# #Black Friday : 24th November 2017\n# new_transactions['Black_Friday_2017'] = (pd.to_datetime('2017-11-27') - \n# new_transactions['purchase_date']).dt.days.apply(lambda x: x if x > 0 and x <= 7 else 0)\n\n# aggs = {'mean': ['mean'],}\n\n# for col in ['category_2','category_3']:\n# new_transactions[col+'_mean'] = new_transactions['purchase_amount'].groupby(new_transactions[col]).agg('mean')\n# new_transactions[col+'_max'] = new_transactions['purchase_amount'].groupby(new_transactions[col]).agg('max')\n# new_transactions[col+'_min'] = new_transactions['purchase_amount'].groupby(new_transactions[col]).agg('min')\n# new_transactions[col+'_var'] = new_transactions['purchase_amount'].groupby(new_transactions[col]).agg('var')\n# aggs[col+'_mean'] = ['mean']",
"_____no_output_____"
],
[
"new_merge = aggregate_bymonth(new_transactions, prefix='new_')\nnew_merge = new_merge.drop(['new_transactions_count'], axis = 1)\nnew_merge['Date'] = pd.to_datetime(new_merge[['year', 'month']].assign(Day=1))\n\ngc.collect()",
"_____no_output_____"
],
[
"merge1 = new_merge.loc[new_merge.groupby('card_id').Date.idxmax(),:][[ 'card_id','new_card_id_size',\n 'new_purchase_amount_sum','new_purchase_amount_mean']]\nnew_names = [(i,i+'_last') for i in merge1.iloc[:, 1:].columns.values]\nmerge1.rename(columns = dict(new_names), inplace=True)\n\n# merge2 = merge.loc[merge.groupby('card_id').Date.idxmin(),:][['card_id','new_card_id_size',\n# 'new_purchase_amount_sum','new_purchase_amount_mean']]\n# new_names = [(i,i+'_first') for i in merge2.iloc[:, 1:].columns.values]\n# merge2.rename(columns = dict(new_names), inplace=True)\n# comb = pd.merge(merge1, merge2, on='card_id',how='left')\n\ntrain = pd.merge(train, merge1, on='card_id',how='left')\ntest = pd.merge(test, merge1, on='card_id',how='left')\n\ngc.collect()",
"_____no_output_____"
],
[
"## Same merchant purchase\ndf = (new_transactions.groupby(['card_id','merchant_id','purchase_amount']).size().reset_index(name='count_new'))\ndf['purchase_amount_new'] = df.groupby(['card_id','merchant_id'])['purchase_amount'].transform('sum')\ndf['count_new'] = df.groupby(['card_id','merchant_id'])['count_new'].transform('sum')\ndf = df.drop_duplicates()\ndf = df.loc[df['count_new'] >= 2]\nagg_func = {\n 'count_new' : ['count'],\n 'purchase_amount_new':['sum','mean'],\n 'purchase_amount':['sum','mean'],\n}\ndf = df.groupby(['card_id']).agg(agg_func)\ndf.columns = [''.join(col).strip() for col in df.columns.values]\nnew_names = [(i,'new'+i) for i in df.iloc[:, 3:].columns.values]\ndf.rename(columns = dict(new_names), inplace=True)\n\ntrain = pd.merge(train, df, on='card_id',how='left')\ntest = pd.merge(test, df, on='card_id',how='left')",
"_____no_output_____"
],
[
"# Same category purchase\ndf = (new_transactions.groupby(['card_id','merchant_category_id']).size().reset_index(name='new_count'))\ndf['new_count'] = df.groupby(['card_id','merchant_category_id'])['new_count'].transform('sum')\ndf = df.drop_duplicates()\ndf = df.loc[df['new_count'] >= 2]\ndf['new_count_4'] = 0\ndf.loc[df['new_count'] >= 4, 'new_count_4'] = 1\nagg_fun = {\n 'new_count' : ['count'],\n 'new_count_4' : ['sum'],\n}\ndf = df.groupby(['card_id']).agg(agg_fun)\ndf.columns = [''.join(col).strip() for col in df.columns.values]\n\ntrain = pd.merge(train, df, on='card_id',how='left')\ntest = pd.merge(test, df, on='card_id',how='left')",
"_____no_output_____"
],
[
"merchants = reduce_mem_usage(pd.read_csv('merchants_clean.csv'))\nmerchants = merchants.drop(['Unnamed: 0', 'merchant_group_id', 'merchant_category_id',\n 'subsector_id', 'numerical_1', 'numerical_2', \n 'active_months_lag3','active_months_lag6',\n 'city_id', 'state_id',\n ], axis = 1)\nd = dict(zip(merchants.columns[1:], ['newchant_{}'.format(x) for x in (merchants.columns[1:])]))\nd.update({\"merchant_id\": \"new_merchant_id_<lambda>\"})\nmerchants = merchants.rename(index=str, columns= d)\n## convert the month in business to categorical\nmerchants.newchant_active_months_lag12 = pd.cut(merchants.newchant_active_months_lag12, 4)",
"Mem. usage decreased to 16.91 Mb (71.2% reduction)\n"
],
[
"merge_new = aggregate_transaction_new(new_transactions, prefix='new_')\nmerge_new = merge_new.merge(merchants, on = 'new_merchant_id_<lambda>', how = 'left')\n## new transaction frequency\nmerge_new['new_freq'] = merge_new.new_transactions_count/(((merge_new.new_purchase_date_max - \n merge_new.new_purchase_date_min).dt.total_seconds())/86400)\nmerge_new['new_freq_amount'] = merge_new['new_freq'] * merge_new['new_purchase_amount_mean']\nmerge_new['new_freq_install'] = merge_new['new_freq'] * merge_new['new_installments_mean']",
"_____no_output_____"
],
[
"cols = ['newchant_avg_sales_lag3','newchant_avg_purchases_lag3',\n 'newchant_avg_sales_lag6','newchant_avg_purchases_lag6',\n 'newchant_avg_sales_lag12','newchant_avg_purchases_lag12','new_freq']\nfor col in cols:\n merge_new[col] = pd.qcut(merge_new[col], 4)",
"_____no_output_____"
],
[
"for col in cols:\n merge_new[col].fillna(merge_new[col].mode()[0], inplace=True)\n label_enc.fit(list(merge_new[col].values)) \n merge_new[col] = label_enc.transform(list(merge_new[col].values))",
"_____no_output_____"
],
[
"for col in ['newchant_category_1','newchant_most_recent_sales_range','newchant_most_recent_purchases_range',\n 'newchant_active_months_lag12','newchant_category_4','newchant_category_2']:\n merge_new[col].fillna(merge_new[col].mode()[0], inplace=True)",
"_____no_output_____"
],
[
"label_enc.fit(list(merge_new['new_merchant_id_<lambda>'].values)) \nmerge_new['new_merchant_id_<lambda>'] = label_enc.transform(list(merge_new['new_merchant_id_<lambda>'].values))\nlabel_enc.fit(list(merge_new['newchant_active_months_lag12'].values)) \nmerge_new['newchant_active_months_lag12'] = label_enc.transform(list(merge_new['newchant_active_months_lag12'].values))",
"_____no_output_____"
],
[
"#del new_transactions\ngc.collect()\n\ntrain = pd.merge(train, merge_new, on='card_id',how='left')\ntest = pd.merge(test, merge_new, on='card_id',how='left')\n#del merge_new\n\ngc.collect()",
"_____no_output_____"
],
[
"train_na = train.isnull().sum()\ntrain_na = train_na.drop(train_na[train_na == 0].index).sort_values(ascending=False)\nmissing_data = pd.DataFrame({'Missing Value' :train_na})\nmissing_data.head(5)",
"_____no_output_____"
],
[
"for col in ['new_freq','new_purchase_amount_min','new_purchase_amount_max','newchant_category_4','new_weekend_mean',\n 'new_purchase_amount_mean','newchant_active_months_lag12','new_weekend_sum','newchant_avg_purchases_lag12',\n 'newchant_avg_sales_lag12','newchant_avg_purchases_lag6','newchant_avg_sales_lag6','new_category_1_sum',\n 'newchant_avg_purchases_lag3','newchant_avg_sales_lag3','new_category_1_mean','new_category_1_max',\n 'new_category_1_min','newchant_most_recent_purchases_range','newchant_most_recent_sales_range',\n 'newchant_category_1'] : # -1\n train[col] = train[col].fillna(-1)\n test[col] = test[col].fillna(-1)\n \nfor col in ['new_installments_min','new_installments_max','new_installments_mean','new_installments_sum',\n 'new_purchase_amount_sum','new_state_id_<lambda>' ]: # -2\n train[col] = train[col].fillna(-2)\n test[col] = test[col].fillna(-2)\n \nfor col in ['newchant_category_2','new_authorized_flag_sum','new_month_lag_min','new_month_lag_max','new_card_id_size',\n 'new_month_lag_mean','new_weekofyear_nunique','new_year_nunique','new_state_id_nunique',\n 'new_merchant_id_<lambda>','new_merchant_id_nunique','new_merchant_category_id_nunique',\n 'new_subsector_id_nunique','new_dayofweek_nunique','new_hour_nunique','new_month_nunique',\n 'new_transactions_count','new_count_4sum','new_countcount','hist_count_4sum','hist_countcount',\n 'hist_purchase_amountmean','hist_purchase_amountsum','purchase_amount_newmean','purchase_amount_newsum',\n 'count_newcount','purchase_amount_histmean','purchase_amount_histsum','count_histcount','hist_mean4mean',\n 'hist_mean4sum','newpurchase_amountmean','newpurchase_amountsum','purchase_amountmean_merhist',\n 'purchase_amountsum_merhist','histpurchase_amountmean','histpurchase_amountsum',\n 'new_merchant_category_id_<lambda>','category_repeated_month','new_purchase_amount_mean_last',\n 'new_purchase_amount_sum_last','new_card_id_size_last']: # 0\n train[col] = train[col].fillna(0)\n test[col] = test[col].fillna(0)\n \ntrain.new_month_diff_mean = train.new_month_diff_mean.fillna(23)\ntrain.new_month_diff_min = train.new_month_diff_min.fillna(23)\ntrain.new_month_diff_max = train.new_month_diff_max.fillna(24)\ntrain.new_month_diff_lag_mean = train.new_month_diff_lag_mean.fillna(24)\ntrain.new_month_diff_lag_min = train.new_month_diff_lag_min.fillna(24)\ntrain.new_month_diff_lag_max = train.new_month_diff_lag_max.fillna(24)\n\ntest.new_month_diff_mean = test.new_month_diff_mean.fillna(23)\ntest.new_month_diff_min = test.new_month_diff_min.fillna(23)\ntest.new_month_diff_max = test.new_month_diff_max.fillna(24)\ntest.new_month_diff_lag_mean = test.new_month_diff_lag_mean.fillna(24)\ntest.new_month_diff_lag_min = test.new_month_diff_lag_min.fillna(24)\ntest.new_month_diff_lag_max = test.new_month_diff_lag_max.fillna(24)",
"_____no_output_____"
],
[
"for col in ['new_purchase_date_min','new_purchase_date_max']:\n train[col] = train[col].fillna(pd.to_datetime(1/9/2017))\n test[col] = test[col].fillna(pd.to_datetime(1/9/2017))",
"_____no_output_____"
],
[
"#Feature Engineering - Adding new features inspired by Chau's first kernel\ntrain['total_count_merid'] = train['count_newcount'] + train['count_histcount']\ntrain['total_count'] = train['new_countcount'] + train['hist_countcount']\ntrain['new_purchase_date_max'] = pd.to_datetime(train['new_purchase_date_max'])\ntrain['new_purchase_date_min'] = pd.to_datetime(train['new_purchase_date_min'])\ntrain['new_purchase_date_diff'] = (train['new_purchase_date_max'] - train['new_purchase_date_min']).dt.days\ntrain['new_purchase_date_average'] = train['new_purchase_date_diff']/train['new_card_id_size']\ntrain['new_purchase_date_uptonow'] = (pd.to_datetime('01/03/2018') - train['new_purchase_date_max']).dt.days\ntrain['new_purchase_date_uptomin'] = (pd.to_datetime('01/03/2018') - train['new_purchase_date_min']).dt.days\ntrain['new_first_buy'] = (train['new_purchase_date_min'] - train['first_active_month']).dt.days\nfor feature in ['new_purchase_date_max','new_purchase_date_min']:\n train[feature] = train[feature].astype(np.int64) * 1e-9\n\n#Feature Engineering - Adding new features inspired by Chau's first kernel\ntest['total_count_merid'] = test['count_newcount'] + test['count_histcount']\ntest['total_count'] = test['new_countcount'] + test['hist_countcount']\ntest['new_purchase_date_max'] = pd.to_datetime(test['new_purchase_date_max'])\ntest['new_purchase_date_min'] = pd.to_datetime(test['new_purchase_date_min'])\ntest['new_purchase_date_diff'] = (test['new_purchase_date_max'] - test['new_purchase_date_min']).dt.days\ntest['new_purchase_date_average'] = test['new_purchase_date_diff']/test['new_card_id_size']\ntest['new_purchase_date_uptonow'] = (pd.to_datetime('01/03/2018') - test['new_purchase_date_max']).dt.days\ntest['new_purchase_date_uptomin'] = (pd.to_datetime('01/03/2018') - test['new_purchase_date_min']).dt.days\ntest['new_first_buy'] = (test['new_purchase_date_min'] - test['first_active_month']).dt.days\nfor feature in ['new_purchase_date_max','new_purchase_date_min']:\n test[feature] = test[feature].astype(np.int64) * 1e-9\n \n#added new feature - Interactive\ntrain['card_id_total'] = train['new_card_id_size'] + train['hist_card_id_size']\ntrain['purchase_amount_total'] = train['new_purchase_amount_sum'] + train['hist_purchase_amount_sum']\n\ntest['card_id_total'] = test['new_card_id_size'] + test['hist_card_id_size']\ntest['purchase_amount_total'] = test['new_purchase_amount_sum'] + test['hist_purchase_amount_sum']\n\ngc.collect()",
"_____no_output_____"
],
[
"cols = ['new_freq_amount',]\nfor col in cols:\n train[col] = train[col].fillna(0)\n train[col] = pd.qcut(train[col], 5)\n label_enc.fit(list(train[col].values)) \n train[col] = label_enc.transform(list(train[col].values))\n \n test[col] = test[col].fillna(0)\n test[col] = pd.qcut(test[col], 5)\n label_enc.fit(list(test[col].values)) \n test[col] = label_enc.transform(list(test[col].values))\n\ntrain = train.drop(['new_freq_install'], axis = 1) \ntest = test.drop(['new_freq_install'], axis = 1) ",
"_____no_output_____"
],
[
"train.new_purchase_date_average = train.new_purchase_date_average.fillna(-1.0)\ntest.new_purchase_date_average = test.new_purchase_date_average.fillna(-1.0)",
"_____no_output_____"
],
[
"# last month of new over hist \ntrain['amountmean_ratiolastnew'] = train.new_purchase_amount_mean_last/train.hist_purchase_amount_mean\ntrain['amountsum_ratiolastnew'] = train.new_purchase_amount_sum_last/(train.hist_purchase_amount_sum/(train.hist_purchase_date_diff//30))\ntrain['transcount_ratiolastnew'] = train.new_card_id_size_last/(train.hist_transactions_count/(train.hist_purchase_date_diff//30))\n\ntest['amountmean_ratiolastnew'] = test.new_purchase_amount_mean_last/test.hist_purchase_amount_mean\ntest['amountsum_ratiolastnew'] = test.new_purchase_amount_sum_last/(test.hist_purchase_amount_sum/(test.hist_purchase_date_diff//30))\ntest['transcount_ratiolastnew'] = test.new_card_id_size_last/(test.hist_transactions_count/(test.hist_purchase_date_diff//30))",
"_____no_output_____"
],
[
"# last month of hist over hist \ntrain['amountmean_ratiolast'] = train.hist_purchase_amount_mean_last/train.hist_purchase_amount_mean\ntrain['amountsum_ratiolast'] = train.hist_purchase_amount_sum_last/(train.hist_purchase_amount_sum/(train.hist_purchase_date_diff//30))\ntrain['transcount_ratiolast'] = train.hist_card_id_size_last/(train.hist_transactions_count/(train.hist_purchase_date_diff//30))\n\ntest['amountmean_ratiolast'] = test.hist_purchase_amount_mean_last/test.hist_purchase_amount_mean\ntest['amountsum_ratiolast'] = test.hist_purchase_amount_sum_last/(test.hist_purchase_amount_sum/(test.hist_purchase_date_diff//30))\ntest['transcount_ratiolast'] = test.hist_card_id_size_last/(test.hist_transactions_count/(test.hist_purchase_date_diff//30))",
"_____no_output_____"
],
[
"# last 2 month of hist ratio\ntrain['amountmean_lastlast2'] = train.hist_purchase_amount_mean_last/train.hist_purchase_amount_mean_last2\ntrain['amountsum_lastlast2'] = train.hist_purchase_amount_sum_last/train.hist_purchase_amount_sum_last2\ntrain['transcount_lastlast2'] = train.hist_card_id_size_last/train.hist_card_id_size_last2\n\ntest['amountmean_lastlast2'] = test.hist_purchase_amount_mean_last/test.hist_purchase_amount_mean_last2\ntest['amountsum_lastlast2'] = test.hist_purchase_amount_sum_last/test.hist_purchase_amount_sum_last2\ntest['transcount_lastlast2'] = test.hist_card_id_size_last/test.hist_card_id_size_last2",
"_____no_output_____"
],
[
"# train['amountmean_ratiofirst'] = train.hist_purchase_amount_mean_first/train.hist_purchase_amount_mean\n# train['amountsum_ratiofirst'] = train.hist_purchase_amount_sum_first/train.hist_purchase_amount_sum\n# train['transcount_ratiofirst'] = train.hist_card_id_size_first/(train.hist_transactions_count/(train.hist_purchase_date_diff//30))\n\n# test['amountmean_ratiofirst'] = test.hist_purchase_amount_mean_first/test.hist_purchase_amount_mean\n# test['amountsum_ratiofirst'] = test.hist_purchase_amount_sum_first/test.hist_purchase_amount_sum\n# test['transcount_ratiofirst'] = test.hist_card_id_size_first/(test.hist_transactions_count/(test.hist_purchase_date_diff//30))",
"_____no_output_____"
],
[
"# train['amountmean_lastfirst'] = train.hist_purchase_amount_mean_last/train.hist_purchase_amount_mean_first\n# train['amountsum_lastfirst'] = train.hist_purchase_amount_sum_last/train.hist_purchase_amount_sum_first\n# train['transcount_lastfirst'] = train.hist_card_id_size_last/train.hist_card_id_size_first\n\n# test['amountmean_lastfirst'] = test.hist_purchase_amount_mean_last/test.hist_purchase_amount_mean_first\n# test['amountsum_lastfirst'] = test.hist_purchase_amount_sum_last/test.hist_purchase_amount_sum_first\n# test['transcount_lastfirst'] = test.hist_card_id_size_last/test.hist_card_id_size_first\n\ntrain = train.drop(['hist_purchase_amount_mean_last2','hist_purchase_amount_sum_last2','hist_card_id_size_last2'], axis = 1)\ntest = test.drop(['hist_purchase_amount_mean_last2','hist_purchase_amount_sum_last2','hist_card_id_size_last2'], axis = 1)",
"_____no_output_____"
],
[
"train = train.drop(['hist_card_id_size','new_card_id_size','card_id', 'first_active_month'], axis = 1)\ntest = test.drop(['hist_card_id_size','new_card_id_size','card_id', 'first_active_month'], axis = 1)",
"_____no_output_____"
],
[
"train.shape",
"_____no_output_____"
],
[
"# Remove the Outliers if any \ntrain['outliers'] = 0\ntrain.loc[train['target'] < -30, 'outliers'] = 1\ntrain['outliers'].value_counts()",
"_____no_output_____"
],
[
"for features in ['feature_1','feature_2','feature_3']:\n order_label = train.groupby([features])['outliers'].mean()\n train[features] = train[features].map(order_label)\n test[features] = test[features].map(order_label)",
"_____no_output_____"
],
[
"# Get the X and Y\ndf_train_columns = [c for c in train.columns if c not in ['target','outliers']] \ncat_features = [c for c in df_train_columns if 'feature_' in c] \n#df_train_columns",
"_____no_output_____"
],
[
"target = train['target']\ndel train['target']",
"_____no_output_____"
],
[
"import lightgbm as lgb\n\nparam = {'num_leaves': 31,\n 'min_data_in_leaf': 30, \n 'objective':'regression',\n 'max_depth': -1,\n 'learning_rate': 0.01,\n \"min_child_samples\": 20,\n \"boosting\": \"gbdt\",\n \"feature_fraction\": 0.9,\n \"bagging_freq\": 1,\n \"bagging_fraction\": 0.9 ,\n \"bagging_seed\": 11,\n \"metric\": 'rmse',\n \"lambda_l1\": 0.1,\n \"verbosity\": -1,\n \"nthread\": 4,\n \"random_state\": 4590}\n\nfolds = StratifiedKFold(n_splits=6, shuffle=True, random_state=4590)\noof = np.zeros(len(train))\npredictions = np.zeros(len(test))\nfeature_importance_df = pd.DataFrame()\n\nfor fold_, (trn_idx, val_idx) in enumerate(folds.split(train,train['outliers'].values)):\n print(\"fold {}\".format(fold_))\n trn_data = lgb.Dataset(train.iloc[trn_idx][df_train_columns], label=target.iloc[trn_idx])\n val_data = lgb.Dataset(train.iloc[val_idx][df_train_columns], label=target.iloc[val_idx])\n\n num_round = 10000\n clf = lgb.train(param, trn_data, num_round, valid_sets = [trn_data, val_data], verbose_eval=-1, early_stopping_rounds = 200)\n oof[val_idx] = clf.predict(train.iloc[val_idx][df_train_columns], num_iteration=clf.best_iteration)\n \n fold_importance_df = pd.DataFrame()\n fold_importance_df[\"Feature\"] = df_train_columns\n fold_importance_df[\"importance\"] = clf.feature_importance()\n fold_importance_df[\"fold\"] = fold_ + 1\n feature_importance_df = pd.concat([feature_importance_df, fold_importance_df], axis=0)\n \n predictions += clf.predict(test[df_train_columns], num_iteration=clf.best_iteration) / folds.n_splits\n\nnp.sqrt(mean_squared_error(oof, target))",
"fold 0\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1505]\ttraining's rmse: 3.26557\tvalid_1's rmse: 3.6424\nfold 1\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1455]\ttraining's rmse: 3.27453\tvalid_1's rmse: 3.63875\nfold 2\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1068]\ttraining's rmse: 3.33668\tvalid_1's rmse: 3.65562\nfold 3\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1330]\ttraining's rmse: 3.28814\tvalid_1's rmse: 3.65566\nfold 4\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1179]\ttraining's rmse: 3.30669\tvalid_1's rmse: 3.67016\nfold 5\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1115]\ttraining's rmse: 3.31959\tvalid_1's rmse: 3.64391\n"
],
[
"cols = (feature_importance_df[[\"Feature\", \"importance\"]]\n .groupby(\"Feature\")\n .mean()\n .sort_values(by=\"importance\", ascending=False)[:1000].index)\n\nbest_features = feature_importance_df.loc[feature_importance_df.Feature.isin(cols)]\n\nplt.figure(figsize=(14,25))\nsns.barplot(x=\"importance\",\n y=\"Feature\",\n data=best_features.sort_values(by=\"importance\",\n ascending=False))\nplt.title('LightGBM Features (avg over folds)')\nplt.tight_layout()\nplt.savefig('lgbm_importances.png')",
"_____no_output_____"
],
[
"features = [c for c in train.columns if c not in ['card_id', 'first_active_month','target','outliers']]\ncat_features = [c for c in features if 'feature_' in c]",
"_____no_output_____"
],
[
"param = {'num_leaves': 31,\n 'min_data_in_leaf': 30, \n 'objective':'regression',\n 'max_depth': -1,\n 'learning_rate': 0.01,\n \"min_child_samples\": 20,\n \"boosting\": \"gbdt\",\n \"feature_fraction\": 0.9,\n \"bagging_freq\": 1,\n \"bagging_fraction\": 0.9 ,\n \"bagging_seed\": 11,\n \"metric\": 'rmse',\n \"lambda_l1\": 0.1,\n \"verbosity\": -1,\n \"nthread\": 4,\n \"random_state\": 4590}\n\nfolds = RepeatedKFold(n_splits=6, n_repeats=2, random_state=4590)\noof_2 = np.zeros(len(train))\npredictions_2 = np.zeros(len(test))\nfeature_importance_df_2 = pd.DataFrame()\n\nfor fold_, (trn_idx, val_idx) in enumerate(folds.split(train.values, target.values)):\n print(\"fold {}\".format(fold_))\n trn_data = lgb.Dataset(train.iloc[trn_idx][features], label=target.iloc[trn_idx], categorical_feature=cat_features)\n val_data = lgb.Dataset(train.iloc[val_idx][features], label=target.iloc[val_idx], categorical_feature=cat_features)\n\n num_round = 10000\n clf_r = lgb.train(param, trn_data, num_round, valid_sets = [trn_data, val_data], verbose_eval=-1, early_stopping_rounds = 200)\n oof_2[val_idx] = clf_r.predict(train.iloc[val_idx][features], num_iteration=clf_r.best_iteration)\n \n fold_importance_df = pd.DataFrame()\n fold_importance_df[\"Feature\"] = features\n fold_importance_df[\"importance\"] = clf_r.feature_importance()\n fold_importance_df[\"fold\"] = fold_ + 1\n feature_importance_df_2 = pd.concat([feature_importance_df_2, fold_importance_df], axis=0)\n \n predictions_2 += clf_r.predict(test[features], num_iteration=clf_r.best_iteration) / (5 * 2)\n\nprint(\"CV score: {:<8.5f}\".format(mean_squared_error(oof_2, target)**0.5))",
"fold 0\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1104]\ttraining's rmse: 3.30605\tvalid_1's rmse: 3.76077\nfold 1\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[932]\ttraining's rmse: 3.36225\tvalid_1's rmse: 3.6531\nfold 2\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1615]\ttraining's rmse: 3.26659\tvalid_1's rmse: 3.56672\nfold 3\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1116]\ttraining's rmse: 3.32268\tvalid_1's rmse: 3.65988\nfold 4\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1036]\ttraining's rmse: 3.34059\tvalid_1's rmse: 3.64775\nfold 5\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1596]\ttraining's rmse: 3.25505\tvalid_1's rmse: 3.63986\nfold 6\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1327]\ttraining's rmse: 3.27605\tvalid_1's rmse: 3.75108\nfold 7\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1813]\ttraining's rmse: 3.22164\tvalid_1's rmse: 3.64546\nfold 8\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1493]\ttraining's rmse: 3.24582\tvalid_1's rmse: 3.74937\nfold 9\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1250]\ttraining's rmse: 3.32085\tvalid_1's rmse: 3.55607\nfold 10\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1143]\ttraining's rmse: 3.32387\tvalid_1's rmse: 3.63679\nfold 11\nTraining until validation scores don't improve for 200 rounds.\nEarly stopping, best iteration is:\n[1858]\ttraining's rmse: 3.22733\tvalid_1's rmse: 3.55876\nCV score: 3.65044 \n"
],
[
"cols = (feature_importance_df_2[[\"Feature\", \"importance\"]]\n .groupby(\"Feature\")\n .mean()\n .sort_values(by=\"importance\", ascending=False)[:1000].index)\n\nbest_features = feature_importance_df_2.loc[feature_importance_df_2.Feature.isin(cols)]\n\nplt.figure(figsize=(14,25))\nsns.barplot(x=\"importance\",\n y=\"Feature\",\n data=best_features.sort_values(by=\"importance\",\n ascending=False))\nplt.title('LightGBM Features (avg over folds)')\nplt.tight_layout()\nplt.savefig('lgbm_importances.png')",
"_____no_output_____"
],
[
"from sklearn.linear_model import BayesianRidge\n\ntrain_stack = np.vstack([oof,oof_2]).transpose()\ntest_stack = np.vstack([predictions, predictions_2]).transpose()\n\nfolds_stack = RepeatedKFold(n_splits=6, n_repeats=1, random_state=4590)\noof_stack = np.zeros(train_stack.shape[0])\npredictions_3 = np.zeros(test_stack.shape[0])\n\nfor fold_, (trn_idx, val_idx) in enumerate(folds_stack.split(train_stack,target)):\n print(\"fold {}\".format(fold_))\n trn_data, trn_y = train_stack[trn_idx], target.iloc[trn_idx].values\n val_data, val_y = train_stack[val_idx], target.iloc[val_idx].values\n \n clf_3 = BayesianRidge()\n clf_3.fit(trn_data, trn_y)\n \n oof_stack[val_idx] = clf_3.predict(val_data)\n predictions_3 += clf_3.predict(test_stack) / 6\n \nnp.sqrt(mean_squared_error(target.values, oof_stack))",
"fold 0\nfold 1\nfold 2\nfold 3\nfold 4\nfold 5\n"
],
[
"sample_submission = pd.read_csv('sample_submission.csv')\nsample_submission['target'] = predictions_3\n# combine = pd.read_csv('combining_submission.csv')\n# sample_submission['target'] = predictions_3*0.7 + combine['target']*0.3\nq = sample_submission['target'].quantile(0.002)\n# #sample_submission['target'] = sample_submission['target'].apply(lambda x: x if x > q else x*1.04)\n# sample_submission.loc[sample_submission.target < -19.3, 'target'] = -33.218750\n# for i in [2726,17430,28039,42686]:\n# sample_submission['target'][i] = -33.21875\nsample_submission.to_csv('submission.csv', index=False)",
"_____no_output_____"
],
[
"((sample_submission.target <= -30) & (sample_submission.target > -35)).sum()",
"_____no_output_____"
],
[
"sample_submission.iloc[108111]",
"_____no_output_____"
],
[
"q",
"_____no_output_____"
],
[
"sample_submission.loc[sample_submission.target < -19.5]",
"_____no_output_____"
],
[
"sample_submission.head(5)",
"_____no_output_____"
],
[
"my = pd.read_csv('submission (1).csv')\nmy['target'][96354] = -33.218750\nmy.to_csv('submission96354.csv', index=False)",
"_____no_output_____"
]
],
[
[
"## Classification",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, ExtraTreesClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.base import BaseEstimator, TransformerMixin, RegressorMixin, clone\nfrom sklearn.model_selection import KFold, cross_val_score, train_test_split, GridSearchCV, StratifiedKFold\nfrom sklearn.metrics import mean_squared_error, accuracy_score\nfrom sklearn.preprocessing import LabelEncoder",
"_____no_output_____"
],
[
"y_train = train['outliers']\ndel train['outliers']",
"_____no_output_____"
],
[
"train['target'] = target\ntest['target'] = predictions_3",
"_____no_output_____"
],
[
"models = [RandomForestClassifier(),ExtraTreesClassifier()]\nnames = [\"RF\", \"Xtree\"]\ndict_score = {}\nfor name, model in zip(names, models):\n model.fit(train, y_train)\n model_train_pred = model.predict(train)\n accy = round(accuracy_score(y_train, model_train_pred), 6)\n dict_score[name] = accy\n\nimport operator\ndict_score = sorted(dict_score.items(), key = operator.itemgetter(1), reverse = True)\ndict_score",
"_____no_output_____"
],
[
"Xtree = ExtraTreesClassifier()\nXtreeMd = Xtree.fit(train, y_train)\ny_pred = XtreeMd.predict(test)",
"_____no_output_____"
],
[
"sample_submission['outliers'] = y_pred\nsample_submission.loc[sample_submission['outliers'] == 1, 'target'] = -33.218750\nsample_submission = sample_submission.drop(['outliers'], axis = 1)\nsample_submission.to_csv('submission.csv', index=False)",
"_____no_output_____"
],
[
"sample_submission.loc[sample_submission['target'] == -33.21875][:40]",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05faaa0e3ce67562965fb945710ebf25c0baf51 | 92,742 | ipynb | Jupyter Notebook | Enem Sample.ipynb | otaciliojpereira/data-science | c98c7c1eb83ba4673a99b6d576830bbd958f10a3 | [
"BSD-4-Clause-UC"
] | null | null | null | Enem Sample.ipynb | otaciliojpereira/data-science | c98c7c1eb83ba4673a99b6d576830bbd958f10a3 | [
"BSD-4-Clause-UC"
] | null | null | null | Enem Sample.ipynb | otaciliojpereira/data-science | c98c7c1eb83ba4673a99b6d576830bbd958f10a3 | [
"BSD-4-Clause-UC"
] | null | null | null | 72.795918 | 1,352 | 0.680921 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d05fab4b7584e7de366df3b4a643095fd720a484 | 118,381 | ipynb | Jupyter Notebook | code_listings/04.05-Histograms-and-Binnings.ipynb | cesar-rocha/PythonDataScienceHandbook | 96c7f75d49b26a35bf76307bca86533061731859 | [
"MIT"
] | 1 | 2021-06-02T19:42:47.000Z | 2021-06-02T19:42:47.000Z | code_listings/04.05-Histograms-and-Binnings.ipynb | matt-staton/PythonDataScienceHandbook | 96c7f75d49b26a35bf76307bca86533061731859 | [
"MIT"
] | null | null | null | code_listings/04.05-Histograms-and-Binnings.ipynb | matt-staton/PythonDataScienceHandbook | 96c7f75d49b26a35bf76307bca86533061731859 | [
"MIT"
] | 1 | 2019-06-14T13:38:46.000Z | 2019-06-14T13:38:46.000Z | 462.425781 | 37,706 | 0.935775 | [
[
[
"# Histograms, Binnings, and Density",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt\nplt.style.use('seaborn-white')\n\ndata = np.random.randn(1000)",
"_____no_output_____"
],
[
"plt.hist(data);",
"_____no_output_____"
],
[
"plt.hist(data, bins=30, normed=True, alpha=0.5,\n histtype='stepfilled', color='steelblue',\n edgecolor='none');",
"_____no_output_____"
],
[
"x1 = np.random.normal(0, 0.8, 1000)\nx2 = np.random.normal(-2, 1, 1000)\nx3 = np.random.normal(3, 2, 1000)\n\nkwargs = dict(histtype='stepfilled', alpha=0.3, normed=True, bins=40)\n\nplt.hist(x1, **kwargs)\nplt.hist(x2, **kwargs)\nplt.hist(x3, **kwargs);",
"_____no_output_____"
],
[
"counts, bin_edges = np.histogram(data, bins=5)\nprint(counts)",
"[ 12 190 468 301 29]\n"
],
[
"mean = [0, 0]\ncov = [[1, 1], [1, 2]]\nx, y = np.random.multivariate_normal(mean, cov, 10000).T",
"_____no_output_____"
],
[
"plt.hist2d(x, y, bins=30, cmap='Blues')\ncb = plt.colorbar()\ncb.set_label('counts in bin')",
"_____no_output_____"
],
[
"counts, xedges, yedges = np.histogram2d(x, y, bins=30)",
"_____no_output_____"
],
[
"plt.hexbin(x, y, gridsize=30, cmap='Blues')\ncb = plt.colorbar(label='count in bin')",
"_____no_output_____"
],
[
"from scipy.stats import gaussian_kde\n\n# fit an array of size [Ndim, Nsamples]\ndata = np.vstack([x, y])\nkde = gaussian_kde(data)\n\n# evaluate on a regular grid\nxgrid = np.linspace(-3.5, 3.5, 40)\nygrid = np.linspace(-6, 6, 40)\nXgrid, Ygrid = np.meshgrid(xgrid, ygrid)\nZ = kde.evaluate(np.vstack([Xgrid.ravel(), Ygrid.ravel()]))\n\n# Plot the result as an image\nplt.imshow(Z.reshape(Xgrid.shape),\n origin='lower', aspect='auto',\n extent=[-3.5, 3.5, -6, 6],\n cmap='Blues')\ncb = plt.colorbar()\ncb.set_label(\"density\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d05fb7468a0b0c1523fbffcc5703b10c4f40de90 | 26,971 | ipynb | Jupyter Notebook | src_nlp/tensorflow/depreciated/word2vec_skipgram_test.ipynb | ashishpatel26/finch | bf2958c0f268575e5d51ad08fbc08b151cbea962 | [
"MIT"
] | 1 | 2021-02-23T03:25:43.000Z | 2021-02-23T03:25:43.000Z | nlp-models/tensorflow/word2vec_skipgram_test.ipynb | Jie-Yuan/finch | ed799c099daec4d57b88539c2382a62eba73d375 | [
"MIT"
] | null | null | null | nlp-models/tensorflow/word2vec_skipgram_test.ipynb | Jie-Yuan/finch | ed799c099daec4d57b88539c2382a62eba73d375 | [
"MIT"
] | 1 | 2020-10-15T21:34:17.000Z | 2020-10-15T21:34:17.000Z | 55.956432 | 237 | 0.59868 | [
[
[
"import string\nfrom word2vec_skipgram import SkipGram\n\n\nif __name__ == '__main__':\n with open('temp/ptb_train.txt') as f:\n text = f.read()\n sample_words = ['six', 'gold', 'japan', 'college']\n\n model = SkipGram(text, sample_words, useless_words=string.punctuation)\n model.fit()",
"Total words: 888723\nVocabulary size: 9251\nWord preprocessing completed ...\nWARNING:tensorflow:From /usr/local/lib/python3.6/site-packages/tensorflow/python/ops/nn_impl.py:1346: softmax_cross_entropy_with_logits (from tensorflow.python.ops.nn_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\n\nFuture major versions of TensorFlow will allow gradients to flow\ninto the labels input on backprop by default.\n\nSee tf.nn.softmax_cross_entropy_with_logits_v2.\n\nData Shuffled\nEpoch 1/10 | Batch 0/1545 | train loss: 3.6946\nNearest to [six]: enact, runkel, meantime, inch, was,\nNearest to [gold]: depress, genentech, unfortunate, pledge, arguing,\nNearest to [japan]: bernard, spouse, mailing, halt, sang,\nNearest to [college]: discounting, complete, private, freddie, defendant,\nEpoch 1/10 | Batch 50/1545 | train loss: 3.5260\nEpoch 1/10 | Batch 100/1545 | train loss: 3.2283\nEpoch 1/10 | Batch 150/1545 | train loss: 3.6859\nEpoch 1/10 | Batch 200/1545 | train loss: 3.5730\nEpoch 1/10 | Batch 250/1545 | train loss: 3.3671\nEpoch 1/10 | Batch 300/1545 | train loss: 3.8272\nEpoch 1/10 | Batch 350/1545 | train loss: 3.5215\nEpoch 1/10 | Batch 400/1545 | train loss: 3.8107\nEpoch 1/10 | Batch 450/1545 | train loss: 3.5948\nEpoch 1/10 | Batch 500/1545 | train loss: 3.8243\nEpoch 1/10 | Batch 550/1545 | train loss: 3.1904\nEpoch 1/10 | Batch 600/1545 | train loss: 3.4282\nEpoch 1/10 | Batch 650/1545 | train loss: 3.1884\nEpoch 1/10 | Batch 700/1545 | train loss: 3.8623\nEpoch 1/10 | Batch 750/1545 | train loss: 3.4765\nEpoch 1/10 | Batch 800/1545 | train loss: 3.0110\nEpoch 1/10 | Batch 850/1545 | train loss: 3.2705\nEpoch 1/10 | Batch 900/1545 | train loss: 3.4115\nEpoch 1/10 | Batch 950/1545 | train loss: 3.0006\nEpoch 1/10 | Batch 1000/1545 | train loss: 2.6920\nNearest to [six]: was, carnival, problem, troop, bank,\nNearest to [gold]: ounce, moderate, silver, december, ounces,\nNearest to [japan]: uncovered, emissions, workout, accompanied, greenspan,\nNearest to [college]: having, let, never, ask, man,\nEpoch 1/10 | Batch 1050/1545 | train loss: 3.7049\nEpoch 1/10 | Batch 1100/1545 | train loss: 3.4003\nEpoch 1/10 | Batch 1150/1545 | train loss: 4.2520\nEpoch 1/10 | Batch 1200/1545 | train loss: 2.9778\nEpoch 1/10 | Batch 1250/1545 | train loss: 3.2441\nEpoch 1/10 | Batch 1300/1545 | train loss: 2.9554\nEpoch 1/10 | Batch 1350/1545 | train loss: 3.0307\nEpoch 1/10 | Batch 1400/1545 | train loss: 2.9421\nEpoch 1/10 | Batch 1450/1545 | train loss: 3.1836\nEpoch 1/10 | Batch 1500/1545 | train loss: 2.6754\nData Shuffled\nEpoch 2/10 | Batch 0/1545 | train loss: 3.2930\nNearest to [six]: was, carnival, ships, donuts, attempts,\nNearest to [gold]: ounce, moderate, silver, commodity, platinum,\nNearest to [japan]: revamping, flexibility, greenspan, halt, urban,\nNearest to [college]: basketball, contemporary, mcduffie, things, characters,\nEpoch 2/10 | Batch 50/1545 | train loss: 3.6149\nEpoch 2/10 | Batch 100/1545 | train loss: 3.2603\nEpoch 2/10 | Batch 150/1545 | train loss: 2.7252\nEpoch 2/10 | Batch 200/1545 | train loss: 3.2109\nEpoch 2/10 | Batch 250/1545 | train loss: 3.8905\nEpoch 2/10 | Batch 300/1545 | train loss: 3.4359\nEpoch 2/10 | Batch 350/1545 | train loss: 2.9856\nEpoch 2/10 | Batch 400/1545 | train loss: 3.7584\nEpoch 2/10 | Batch 450/1545 | train loss: 2.8445\nEpoch 2/10 | Batch 500/1545 | train loss: 2.7886\nEpoch 2/10 | Batch 550/1545 | train loss: 2.7081\nEpoch 2/10 | Batch 600/1545 | train loss: 3.9816\nEpoch 2/10 | Batch 650/1545 | train loss: 3.2674\nEpoch 2/10 | Batch 700/1545 | train loss: 3.7068\nEpoch 2/10 | Batch 750/1545 | train loss: 2.9442\nEpoch 2/10 | Batch 800/1545 | train loss: 2.8798\nEpoch 2/10 | Batch 850/1545 | train loss: 2.9409\nEpoch 2/10 | Batch 900/1545 | train loss: 3.0363\nEpoch 2/10 | Batch 950/1545 | train loss: 3.4921\nEpoch 2/10 | Batch 1000/1545 | train loss: 3.0225\nNearest to [six]: ships, was, carnival, finland, emerge,\nNearest to [gold]: ounce, ounces, silver, platinum, commodity,\nNearest to [japan]: halt, revamping, greenspan, flexibility, exchanges,\nNearest to [college]: kids, basketball, parents, child, children,\nEpoch 2/10 | Batch 1050/1545 | train loss: 3.3520\nEpoch 2/10 | Batch 1100/1545 | train loss: 3.1030\nEpoch 2/10 | Batch 1150/1545 | train loss: 2.6124\nEpoch 2/10 | Batch 1200/1545 | train loss: 3.1420\nEpoch 2/10 | Batch 1250/1545 | train loss: 3.6545\nEpoch 2/10 | Batch 1300/1545 | train loss: 2.8047\nEpoch 2/10 | Batch 1350/1545 | train loss: 2.6423\nEpoch 2/10 | Batch 1400/1545 | train loss: 2.9904\nEpoch 2/10 | Batch 1450/1545 | train loss: 3.3502\nEpoch 2/10 | Batch 1500/1545 | train loss: 3.0796\nData Shuffled\nEpoch 3/10 | Batch 0/1545 | train loss: 2.6935\nNearest to [six]: ships, emerge, was, carnival, finland,\nNearest to [gold]: ounce, ounces, silver, platinum, commodity,\nNearest to [japan]: halt, greenspan, thereby, revamping, flexibility,\nNearest to [college]: basketball, football, kids, audiences, children,\nEpoch 3/10 | Batch 50/1545 | train loss: 2.9403\nEpoch 3/10 | Batch 100/1545 | train loss: 2.6900\nEpoch 3/10 | Batch 150/1545 | train loss: 3.1415\nEpoch 3/10 | Batch 200/1545 | train loss: 2.7785\nEpoch 3/10 | Batch 250/1545 | train loss: 3.2002\nEpoch 3/10 | Batch 300/1545 | train loss: 2.5229\nEpoch 3/10 | Batch 350/1545 | train loss: 2.8759\nEpoch 3/10 | Batch 400/1545 | train loss: 2.5635\nEpoch 3/10 | Batch 450/1545 | train loss: 2.8483\nEpoch 3/10 | Batch 500/1545 | train loss: 2.9317\nEpoch 3/10 | Batch 550/1545 | train loss: 2.4299\nEpoch 3/10 | Batch 600/1545 | train loss: 3.0184\nEpoch 3/10 | Batch 650/1545 | train loss: 2.8305\nEpoch 3/10 | Batch 700/1545 | train loss: 2.7454\nEpoch 3/10 | Batch 750/1545 | train loss: 3.0075\nEpoch 3/10 | Batch 800/1545 | train loss: 2.7318\nEpoch 3/10 | Batch 850/1545 | train loss: 2.6585\nEpoch 3/10 | Batch 900/1545 | train loss: 2.5398\nEpoch 3/10 | Batch 950/1545 | train loss: 2.8238\nEpoch 3/10 | Batch 1000/1545 | train loss: 2.9874\nNearest to [six]: ships, schedule, cruise, emerge, was,\nNearest to [gold]: ounce, ounces, platinum, bullion, silver,\nNearest to [japan]: thereby, halt, flexibility, greenspan, revamping,\nNearest to [college]: basketball, sports, football, audiences, colleges,\nEpoch 3/10 | Batch 1050/1545 | train loss: 2.8645\nEpoch 3/10 | Batch 1100/1545 | train loss: 2.8435\nEpoch 3/10 | Batch 1150/1545 | train loss: 2.8114\nEpoch 3/10 | Batch 1200/1545 | train loss: 3.1793\nEpoch 3/10 | Batch 1250/1545 | train loss: 2.7298\nEpoch 3/10 | Batch 1300/1545 | train loss: 3.3626\nEpoch 3/10 | Batch 1350/1545 | train loss: 2.9679\nEpoch 3/10 | Batch 1400/1545 | train loss: 2.8729\nEpoch 3/10 | Batch 1450/1545 | train loss: 2.7758\nEpoch 3/10 | Batch 1500/1545 | train loss: 2.4018\nData Shuffled\nEpoch 4/10 | Batch 0/1545 | train loss: 2.9856\nNearest to [six]: schedule, emerge, ships, finland, merged,\nNearest to [gold]: ounce, bullion, ounces, platinum, silver,\nNearest to [japan]: halt, exchanges, flexibility, thereby, greenspan,\nNearest to [college]: basketball, sports, parents, football, child,\nEpoch 4/10 | Batch 50/1545 | train loss: 2.4984\nEpoch 4/10 | Batch 100/1545 | train loss: 2.5274\nEpoch 4/10 | Batch 150/1545 | train loss: 2.6891\nEpoch 4/10 | Batch 200/1545 | train loss: 2.9773\nEpoch 4/10 | Batch 250/1545 | train loss: 2.8103\nEpoch 4/10 | Batch 300/1545 | train loss: 2.9578\nEpoch 4/10 | Batch 350/1545 | train loss: 2.8713\nEpoch 4/10 | Batch 400/1545 | train loss: 2.4680\nEpoch 4/10 | Batch 450/1545 | train loss: 2.7729\nEpoch 4/10 | Batch 500/1545 | train loss: 2.7352\nEpoch 4/10 | Batch 550/1545 | train loss: 2.6108\nEpoch 4/10 | Batch 600/1545 | train loss: 2.4470\nEpoch 4/10 | Batch 650/1545 | train loss: 2.9130\nEpoch 4/10 | Batch 700/1545 | train loss: 2.3081\nEpoch 4/10 | Batch 750/1545 | train loss: 2.4282\nEpoch 4/10 | Batch 800/1545 | train loss: 3.4740\nEpoch 4/10 | Batch 850/1545 | train loss: 2.8102\nEpoch 4/10 | Batch 900/1545 | train loss: 2.4646\nEpoch 4/10 | Batch 950/1545 | train loss: 2.4189\nEpoch 4/10 | Batch 1000/1545 | train loss: 2.6538\nNearest to [six]: schedule, emerge, merged, finland, ships,\nNearest to [gold]: ounce, bullion, platinum, ounces, silver,\nNearest to [japan]: halt, flexibility, revamping, exchanges, deeper,\nNearest to [college]: basketball, sports, football, child, junior,\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d05fc7e5730c597ecf8f0556d0439adcbf023349 | 3,426 | ipynb | Jupyter Notebook | Chapter01/Putting it All Together.ipynb | kraussc/scikit-learn-Cookbook-Second-Edition | 49a59e5478f6e478fbea5e85d3a35d35607b9b70 | [
"MIT"
] | 74 | 2017-11-28T08:50:53.000Z | 2022-01-25T05:52:34.000Z | Chapter01/Putting it All Together.ipynb | kraussc/scikit-learn-Cookbook-Second-Edition | 49a59e5478f6e478fbea5e85d3a35d35607b9b70 | [
"MIT"
] | 2 | 2019-02-25T05:57:58.000Z | 2020-09-23T12:11:28.000Z | Chapter01/Putting it All Together.ipynb | kraussc/scikit-learn-Cookbook-Second-Edition | 49a59e5478f6e478fbea5e85d3a35d35607b9b70 | [
"MIT"
] | 66 | 2017-12-07T00:18:07.000Z | 2022-03-13T11:18:21.000Z | 25.007299 | 103 | 0.571804 | [
[
[
"Putting it All Together",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn import datasets\n\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import accuracy_score\n\n#load the classifying models\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.svm import SVC\n\niris = datasets.load_iris()\nX = iris.data[:, :2] #load the first two features of the iris data \ny = iris.target #load the target of the iris data",
"_____no_output_____"
],
[
"from sklearn.neighbors import KNeighborsClassifier\nX_train, X_test, y_train, y_test = train_test_split(X, y, stratify=y, random_state = 0)",
"_____no_output_____"
],
[
"from sklearn.model_selection import cross_val_score\nknn_3_clf = KNeighborsClassifier(n_neighbors = 3)\nknn_5_clf = KNeighborsClassifier(n_neighbors = 5)\n\nknn_3_scores = cross_val_score(knn_3_clf, X_train, y_train, cv=10)\nknn_5_scores = cross_val_score(knn_5_clf, X_train, y_train, cv=10)",
"_____no_output_____"
],
[
"print \"knn_3 mean scores: \", knn_3_scores.mean(), \"knn_3 std: \",knn_3_scores.std()\nprint \"knn_5 mean scores: \", knn_5_scores.mean(), \" knn_5 std: \",knn_5_scores.std()",
"knn_3 mean scores: 0.798333333333 knn_3 std: 0.0908142181722\nknn_5 mean scores: 0.806666666667 knn_5 std: 0.0559320575496\n"
],
[
"all_scores = []\nfor n_neighbors in range(3,9,1):\n knn_clf = KNeighborsClassifier(n_neighbors = n_neighbors)\n all_scores.append((n_neighbors, cross_val_score(knn_clf, X_train, y_train, cv=10).mean()))\nsorted(all_scores, key = lambda x:x[1], reverse = True) ",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d05fcaf05f832cffd3364cb8b8363c7f5cca2625 | 80,239 | ipynb | Jupyter Notebook | assignment2/PyTorch.ipynb | Lion-HuangGz/CS231nAssignment | 9adfdeda1c892c468c7bab9661fe8eb4d1063510 | [
"MIT"
] | 419 | 2019-09-02T08:25:31.000Z | 2022-03-28T06:09:47.000Z | assignment2/PyTorch.ipynb | QNLM2015/CS231nAssignment | 9adfdeda1c892c468c7bab9661fe8eb4d1063510 | [
"MIT"
] | 9 | 2019-09-28T09:03:40.000Z | 2020-11-11T08:21:13.000Z | assignment2/PyTorch.ipynb | QNLM2015/CS231nAssignment | 9adfdeda1c892c468c7bab9661fe8eb4d1063510 | [
"MIT"
] | 164 | 2019-09-20T07:58:04.000Z | 2022-03-15T11:31:25.000Z | 40.917389 | 890 | 0.562395 | [
[
[
"# What's this PyTorch business?\n\nYou've written a lot of code in this assignment to provide a whole host of neural network functionality. Dropout, Batch Norm, and 2D convolutions are some of the workhorses of deep learning in computer vision. You've also worked hard to make your code efficient and vectorized.\n\nFor the last part of this assignment, though, we're going to leave behind your beautiful codebase and instead migrate to one of two popular deep learning frameworks: in this instance, PyTorch (or TensorFlow, if you choose to use that notebook).",
"_____no_output_____"
],
[
"### What is PyTorch?\n\nPyTorch is a system for executing dynamic computational graphs over Tensor objects that behave similarly as numpy ndarray. It comes with a powerful automatic differentiation engine that removes the need for manual back-propagation. \n\n### Why?\n\n* Our code will now run on GPUs! Much faster training. When using a framework like PyTorch or TensorFlow you can harness the power of the GPU for your own custom neural network architectures without having to write CUDA code directly (which is beyond the scope of this class).\n* We want you to be ready to use one of these frameworks for your project so you can experiment more efficiently than if you were writing every feature you want to use by hand. \n* We want you to stand on the shoulders of giants! TensorFlow and PyTorch are both excellent frameworks that will make your lives a lot easier, and now that you understand their guts, you are free to use them :) \n* We want you to be exposed to the sort of deep learning code you might run into in academia or industry.\n\n### PyTorch versions\nThis notebook assumes that you are using **PyTorch version 1.0**. In some of the previous versions (e.g. before 0.4), Tensors had to be wrapped in Variable objects to be used in autograd; however Variables have now been deprecated. In addition 1.0 also separates a Tensor's datatype from its device, and uses numpy-style factories for constructing Tensors rather than directly invoking Tensor constructors.",
"_____no_output_____"
],
[
"## How will I learn PyTorch?\n\nJustin Johnson has made an excellent [tutorial](https://github.com/jcjohnson/pytorch-examples) for PyTorch. \n\nYou can also find the detailed [API doc](http://pytorch.org/docs/stable/index.html) here. If you have other questions that are not addressed by the API docs, the [PyTorch forum](https://discuss.pytorch.org/) is a much better place to ask than StackOverflow.\n\n\n# Table of Contents\n\nThis assignment has 5 parts. You will learn PyTorch on **three different levels of abstraction**, which will help you understand it better and prepare you for the final project. \n\n1. Part I, Preparation: we will use CIFAR-10 dataset.\n2. Part II, Barebones PyTorch: **Abstraction level 1**, we will work directly with the lowest-level PyTorch Tensors. \n3. Part III, PyTorch Module API: **Abstraction level 2**, we will use `nn.Module` to define arbitrary neural network architecture. \n4. Part IV, PyTorch Sequential API: **Abstraction level 3**, we will use `nn.Sequential` to define a linear feed-forward network very conveniently. \n5. Part V, CIFAR-10 open-ended challenge: please implement your own network to get as high accuracy as possible on CIFAR-10. You can experiment with any layer, optimizer, hyperparameters or other advanced features. \n\nHere is a table of comparison:\n\n| API | Flexibility | Convenience |\n|---------------|-------------|-------------|\n| Barebone | High | Low |\n| `nn.Module` | High | Medium |\n| `nn.Sequential` | Low | High |",
"_____no_output_____"
],
[
"# Part I. Preparation\n\nFirst, we load the CIFAR-10 dataset. This might take a couple minutes the first time you do it, but the files should stay cached after that.\n\nIn previous parts of the assignment we had to write our own code to download the CIFAR-10 dataset, preprocess it, and iterate through it in minibatches; PyTorch provides convenient tools to automate this process for us.",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.optim as optim\nfrom torch.utils.data import DataLoader\nfrom torch.utils.data import sampler\n\nimport torchvision.datasets as dset\nimport torchvision.transforms as T\n\nimport numpy as np",
"_____no_output_____"
],
[
"NUM_TRAIN = 49000\n\n# The torchvision.transforms package provides tools for preprocessing data\n# and for performing data augmentation; here we set up a transform to\n# preprocess the data by subtracting the mean RGB value and dividing by the\n# standard deviation of each RGB value; we've hardcoded the mean and std.\ntransform = T.Compose([\n T.ToTensor(),\n T.Normalize((0.4914, 0.4822, 0.4465), (0.2023, 0.1994, 0.2010))\n ])\n\n# We set up a Dataset object for each split (train / val / test); Datasets load\n# training examples one at a time, so we wrap each Dataset in a DataLoader which\n# iterates through the Dataset and forms minibatches. We divide the CIFAR-10\n# training set into train and val sets by passing a Sampler object to the\n# DataLoader telling how it should sample from the underlying Dataset.\ncifar10_train = dset.CIFAR10('./cs231n/datasets', train=True, download=True,\n transform=transform)\nloader_train = DataLoader(cifar10_train, batch_size=64, \n sampler=sampler.SubsetRandomSampler(range(NUM_TRAIN)))\n\ncifar10_val = dset.CIFAR10('./cs231n/datasets', train=True, download=True,\n transform=transform)\nloader_val = DataLoader(cifar10_val, batch_size=64, \n sampler=sampler.SubsetRandomSampler(range(NUM_TRAIN, 50000)))\n\ncifar10_test = dset.CIFAR10('./cs231n/datasets', train=False, download=True, \n transform=transform)\nloader_test = DataLoader(cifar10_test, batch_size=64)",
"Files already downloaded and verified\nFiles already downloaded and verified\nFiles already downloaded and verified\n"
]
],
[
[
"You have an option to **use GPU by setting the flag to True below**. It is not necessary to use GPU for this assignment. Note that if your computer does not have CUDA enabled, `torch.cuda.is_available()` will return False and this notebook will fallback to CPU mode.\n\nThe global variables `dtype` and `device` will control the data types throughout this assignment. ",
"_____no_output_____"
]
],
[
[
"USE_GPU = True\n\ndtype = torch.float32 # we will be using float throughout this tutorial\n\nif USE_GPU and torch.cuda.is_available():\n device = torch.device('cuda')\nelse:\n device = torch.device('cpu')\n\n# Constant to control how frequently we print train loss\nprint_every = 100\n\nprint('using device:', device)",
"using device: cuda\n"
]
],
[
[
"# Part II. Barebones PyTorch\n\nPyTorch ships with high-level APIs to help us define model architectures conveniently, which we will cover in Part II of this tutorial. In this section, we will start with the barebone PyTorch elements to understand the autograd engine better. After this exercise, you will come to appreciate the high-level model API more.\n\nWe will start with a simple fully-connected ReLU network with two hidden layers and no biases for CIFAR classification. \nThis implementation computes the forward pass using operations on PyTorch Tensors, and uses PyTorch autograd to compute gradients. It is important that you understand every line, because you will write a harder version after the example.\n\nWhen we create a PyTorch Tensor with `requires_grad=True`, then operations involving that Tensor will not just compute values; they will also build up a computational graph in the background, allowing us to easily backpropagate through the graph to compute gradients of some Tensors with respect to a downstream loss. Concretely if x is a Tensor with `x.requires_grad == True` then after backpropagation `x.grad` will be another Tensor holding the gradient of x with respect to the scalar loss at the end.",
"_____no_output_____"
],
[
"### PyTorch Tensors: Flatten Function\nA PyTorch Tensor is conceptionally similar to a numpy array: it is an n-dimensional grid of numbers, and like numpy PyTorch provides many functions to efficiently operate on Tensors. As a simple example, we provide a `flatten` function below which reshapes image data for use in a fully-connected neural network.\n\nRecall that image data is typically stored in a Tensor of shape N x C x H x W, where:\n\n* N is the number of datapoints\n* C is the number of channels\n* H is the height of the intermediate feature map in pixels\n* W is the height of the intermediate feature map in pixels\n\nThis is the right way to represent the data when we are doing something like a 2D convolution, that needs spatial understanding of where the intermediate features are relative to each other. When we use fully connected affine layers to process the image, however, we want each datapoint to be represented by a single vector -- it's no longer useful to segregate the different channels, rows, and columns of the data. So, we use a \"flatten\" operation to collapse the `C x H x W` values per representation into a single long vector. The flatten function below first reads in the N, C, H, and W values from a given batch of data, and then returns a \"view\" of that data. \"View\" is analogous to numpy's \"reshape\" method: it reshapes x's dimensions to be N x ??, where ?? is allowed to be anything (in this case, it will be C x H x W, but we don't need to specify that explicitly). ",
"_____no_output_____"
]
],
[
[
"def flatten(x):\n N = x.shape[0] # read in N, C, H, W\n return x.view(N, -1) # \"flatten\" the C * H * W values into a single vector per image\n\ndef test_flatten():\n x = torch.arange(12).view(2, 1, 3, 2)\n print('Before flattening: ', x)\n print('After flattening: ', flatten(x))\n\ntest_flatten()",
"Before flattening: tensor([[[[ 0, 1],\n [ 2, 3],\n [ 4, 5]]],\n\n\n [[[ 6, 7],\n [ 8, 9],\n [10, 11]]]])\nAfter flattening: tensor([[ 0, 1, 2, 3, 4, 5],\n [ 6, 7, 8, 9, 10, 11]])\n"
]
],
[
[
"### Barebones PyTorch: Two-Layer Network\n\nHere we define a function `two_layer_fc` which performs the forward pass of a two-layer fully-connected ReLU network on a batch of image data. After defining the forward pass we check that it doesn't crash and that it produces outputs of the right shape by running zeros through the network.\n\nYou don't have to write any code here, but it's important that you read and understand the implementation.",
"_____no_output_____"
]
],
[
[
"import torch.nn.functional as F # useful stateless functions\n\ndef two_layer_fc(x, params):\n \"\"\"\n A fully-connected neural networks; the architecture is:\n NN is fully connected -> ReLU -> fully connected layer.\n Note that this function only defines the forward pass; \n PyTorch will take care of the backward pass for us.\n \n The input to the network will be a minibatch of data, of shape\n (N, d1, ..., dM) where d1 * ... * dM = D. The hidden layer will have H units,\n and the output layer will produce scores for C classes.\n \n Inputs:\n - x: A PyTorch Tensor of shape (N, d1, ..., dM) giving a minibatch of\n input data.\n - params: A list [w1, w2] of PyTorch Tensors giving weights for the network;\n w1 has shape (D, H) and w2 has shape (H, C).\n \n Returns:\n - scores: A PyTorch Tensor of shape (N, C) giving classification scores for\n the input data x.\n \"\"\"\n # first we flatten the image\n x = flatten(x) # shape: [batch_size, C x H x W]\n \n w1, w2 = params\n \n # Forward pass: compute predicted y using operations on Tensors. Since w1 and\n # w2 have requires_grad=True, operations involving these Tensors will cause\n # PyTorch to build a computational graph, allowing automatic computation of\n # gradients. Since we are no longer implementing the backward pass by hand we\n # don't need to keep references to intermediate values.\n # you can also use `.clamp(min=0)`, equivalent to F.relu()\n x = F.relu(x.mm(w1))\n x = x.mm(w2)\n return x\n \n\ndef two_layer_fc_test():\n hidden_layer_size = 42\n x = torch.zeros((64, 50), dtype=dtype) # minibatch size 64, feature dimension 50\n w1 = torch.zeros((50, hidden_layer_size), dtype=dtype)\n w2 = torch.zeros((hidden_layer_size, 10), dtype=dtype)\n scores = two_layer_fc(x, [w1, w2])\n print(scores.size()) # you should see [64, 10]\n\ntwo_layer_fc_test()",
"torch.Size([64, 10])\n"
]
],
[
[
"### Barebones PyTorch: Three-Layer ConvNet\n\nHere you will complete the implementation of the function `three_layer_convnet`, which will perform the forward pass of a three-layer convolutional network. Like above, we can immediately test our implementation by passing zeros through the network. The network should have the following architecture:\n\n1. A convolutional layer (with bias) with `channel_1` filters, each with shape `KW1 x KH1`, and zero-padding of two\n2. ReLU nonlinearity\n3. A convolutional layer (with bias) with `channel_2` filters, each with shape `KW2 x KH2`, and zero-padding of one\n4. ReLU nonlinearity\n5. Fully-connected layer with bias, producing scores for C classes.\n\nNote that we have **no softmax activation** here after our fully-connected layer: this is because PyTorch's cross entropy loss performs a softmax activation for you, and by bundling that step in makes computation more efficient.\n\n**HINT**: For convolutions: http://pytorch.org/docs/stable/nn.html#torch.nn.functional.conv2d; pay attention to the shapes of convolutional filters!",
"_____no_output_____"
]
],
[
[
"def three_layer_convnet(x, params):\n \"\"\"\n Performs the forward pass of a three-layer convolutional network with the\n architecture defined above.\n\n Inputs:\n - x: A PyTorch Tensor of shape (N, 3, H, W) giving a minibatch of images\n - params: A list of PyTorch Tensors giving the weights and biases for the\n network; should contain the following:\n - conv_w1: PyTorch Tensor of shape (channel_1, 3, KH1, KW1) giving weights\n for the first convolutional layer\n - conv_b1: PyTorch Tensor of shape (channel_1,) giving biases for the first\n convolutional layer\n - conv_w2: PyTorch Tensor of shape (channel_2, channel_1, KH2, KW2) giving\n weights for the second convolutional layer\n - conv_b2: PyTorch Tensor of shape (channel_2,) giving biases for the second\n convolutional layer\n - fc_w: PyTorch Tensor giving weights for the fully-connected layer. Can you\n figure out what the shape should be?\n - fc_b: PyTorch Tensor giving biases for the fully-connected layer. Can you\n figure out what the shape should be?\n \n Returns:\n - scores: PyTorch Tensor of shape (N, C) giving classification scores for x\n \"\"\"\n conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b = params\n scores = None\n ################################################################################\n # TODO: Implement the forward pass for the three-layer ConvNet. #\n ################################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n scores=F.relu_(F.conv2d(x,conv_w1,conv_b1,padding=2))\n scores=F.relu_(F.conv2d(scores,conv_w2,conv_b2,padding=1))\n scores=F.linear(flatten(scores),fc_w.T,fc_b)\n \n\n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ################################################################################\n # END OF YOUR CODE #\n ################################################################################\n return scores",
"_____no_output_____"
]
],
[
[
"After defining the forward pass of the ConvNet above, run the following cell to test your implementation.\n\nWhen you run this function, scores should have shape (64, 10).",
"_____no_output_____"
]
],
[
[
"def three_layer_convnet_test():\n x = torch.zeros((64, 3, 32, 32), dtype=dtype) # minibatch size 64, image size [3, 32, 32]\n\n conv_w1 = torch.zeros((6, 3, 5, 5), dtype=dtype) # [out_channel, in_channel, kernel_H, kernel_W]\n conv_b1 = torch.zeros((6,)) # out_channel\n conv_w2 = torch.zeros((9, 6, 3, 3), dtype=dtype) # [out_channel, in_channel, kernel_H, kernel_W]\n conv_b2 = torch.zeros((9,)) # out_channel\n\n # you must calculate the shape of the tensor after two conv layers, before the fully-connected layer\n fc_w = torch.zeros((9 * 32 * 32, 10))\n fc_b = torch.zeros(10)\n\n scores = three_layer_convnet(x, [conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b])\n print(scores.size()) # you should see [64, 10]\nthree_layer_convnet_test()",
"torch.Size([64, 10])\n"
]
],
[
[
"### Barebones PyTorch: Initialization\nLet's write a couple utility methods to initialize the weight matrices for our models.\n\n- `random_weight(shape)` initializes a weight tensor with the Kaiming normalization method.\n- `zero_weight(shape)` initializes a weight tensor with all zeros. Useful for instantiating bias parameters.\n\nThe `random_weight` function uses the Kaiming normal initialization method, described in:\n\nHe et al, *Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification*, ICCV 2015, https://arxiv.org/abs/1502.01852",
"_____no_output_____"
]
],
[
[
"def random_weight(shape):\n \"\"\"\n Create random Tensors for weights; setting requires_grad=True means that we\n want to compute gradients for these Tensors during the backward pass.\n We use Kaiming normalization: sqrt(2 / fan_in)\n \"\"\"\n if len(shape) == 2: # FC weight\n fan_in = shape[0]\n else:\n fan_in = np.prod(shape[1:]) # conv weight [out_channel, in_channel, kH, kW]\n # randn is standard normal distribution generator. \n w = torch.randn(shape, device=device, dtype=dtype) * np.sqrt(2. / fan_in)\n w.requires_grad = True\n return w\n\ndef zero_weight(shape):\n return torch.zeros(shape, device=device, dtype=dtype, requires_grad=True)\n\n# create a weight of shape [3 x 5]\n# you should see the type `torch.cuda.FloatTensor` if you use GPU. \n# Otherwise it should be `torch.FloatTensor`\nrandom_weight((3, 5))",
"_____no_output_____"
]
],
[
[
"### Barebones PyTorch: Check Accuracy\nWhen training the model we will use the following function to check the accuracy of our model on the training or validation sets.\n\nWhen checking accuracy we don't need to compute any gradients; as a result we don't need PyTorch to build a computational graph for us when we compute scores. To prevent a graph from being built we scope our computation under a `torch.no_grad()` context manager.",
"_____no_output_____"
]
],
[
[
"def check_accuracy_part2(loader, model_fn, params):\n \"\"\"\n Check the accuracy of a classification model.\n \n Inputs:\n - loader: A DataLoader for the data split we want to check\n - model_fn: A function that performs the forward pass of the model,\n with the signature scores = model_fn(x, params)\n - params: List of PyTorch Tensors giving parameters of the model\n \n Returns: Nothing, but prints the accuracy of the model\n \"\"\"\n split = 'val' if loader.dataset.train else 'test'\n print('Checking accuracy on the %s set' % split)\n num_correct, num_samples = 0, 0\n with torch.no_grad():\n for x, y in loader:\n x = x.to(device=device, dtype=dtype) # move to device, e.g. GPU\n y = y.to(device=device, dtype=torch.int64)\n scores = model_fn(x, params)\n _, preds = scores.max(1)\n num_correct += (preds == y).sum()\n num_samples += preds.size(0)\n acc = float(num_correct) / num_samples\n print('Got %d / %d correct (%.2f%%)' % (num_correct, num_samples, 100 * acc))",
"_____no_output_____"
]
],
[
[
"### BareBones PyTorch: Training Loop\nWe can now set up a basic training loop to train our network. We will train the model using stochastic gradient descent without momentum. We will use `torch.functional.cross_entropy` to compute the loss; you can [read about it here](http://pytorch.org/docs/stable/nn.html#cross-entropy).\n\nThe training loop takes as input the neural network function, a list of initialized parameters (`[w1, w2]` in our example), and learning rate.",
"_____no_output_____"
]
],
[
[
"def train_part2(model_fn, params, learning_rate):\n \"\"\"\n Train a model on CIFAR-10.\n \n Inputs:\n - model_fn: A Python function that performs the forward pass of the model.\n It should have the signature scores = model_fn(x, params) where x is a\n PyTorch Tensor of image data, params is a list of PyTorch Tensors giving\n model weights, and scores is a PyTorch Tensor of shape (N, C) giving\n scores for the elements in x.\n - params: List of PyTorch Tensors giving weights for the model\n - learning_rate: Python scalar giving the learning rate to use for SGD\n \n Returns: Nothing\n \"\"\"\n for t, (x, y) in enumerate(loader_train):\n # Move the data to the proper device (GPU or CPU)\n x = x.to(device=device, dtype=dtype)\n y = y.to(device=device, dtype=torch.long)\n\n # Forward pass: compute scores and loss\n scores = model_fn(x, params)\n loss = F.cross_entropy(scores, y)\n\n # Backward pass: PyTorch figures out which Tensors in the computational\n # graph has requires_grad=True and uses backpropagation to compute the\n # gradient of the loss with respect to these Tensors, and stores the\n # gradients in the .grad attribute of each Tensor.\n loss.backward()\n\n # Update parameters. We don't want to backpropagate through the\n # parameter updates, so we scope the updates under a torch.no_grad()\n # context manager to prevent a computational graph from being built.\n with torch.no_grad():\n for w in params:\n w -= learning_rate * w.grad\n\n # Manually zero the gradients after running the backward pass\n w.grad.zero_()\n\n if t % print_every == 0:\n print('Iteration %d, loss = %.4f' % (t, loss.item()))\n check_accuracy_part2(loader_val, model_fn, params)\n print()",
"_____no_output_____"
]
],
[
[
"### BareBones PyTorch: Train a Two-Layer Network\nNow we are ready to run the training loop. We need to explicitly allocate tensors for the fully connected weights, `w1` and `w2`. \n\nEach minibatch of CIFAR has 64 examples, so the tensor shape is `[64, 3, 32, 32]`. \n\nAfter flattening, `x` shape should be `[64, 3 * 32 * 32]`. This will be the size of the first dimension of `w1`. \nThe second dimension of `w1` is the hidden layer size, which will also be the first dimension of `w2`. \n\nFinally, the output of the network is a 10-dimensional vector that represents the probability distribution over 10 classes. \n\nYou don't need to tune any hyperparameters but you should see accuracies above 40% after training for one epoch.",
"_____no_output_____"
]
],
[
[
"hidden_layer_size = 4000\nlearning_rate = 1e-2\n\nw1 = random_weight((3 * 32 * 32, hidden_layer_size))\nw2 = random_weight((hidden_layer_size, 10))\n\ntrain_part2(two_layer_fc, [w1, w2], learning_rate)",
"Iteration 0, loss = 3.4106\nChecking accuracy on the val set\nGot 125 / 1000 correct (12.50%)\n\nIteration 100, loss = 2.0570\nChecking accuracy on the val set\nGot 382 / 1000 correct (38.20%)\n\nIteration 200, loss = 2.2069\nChecking accuracy on the val set\nGot 347 / 1000 correct (34.70%)\n\nIteration 300, loss = 1.6645\nChecking accuracy on the val set\nGot 405 / 1000 correct (40.50%)\n\nIteration 400, loss = 1.6819\nChecking accuracy on the val set\nGot 364 / 1000 correct (36.40%)\n\nIteration 500, loss = 1.7364\nChecking accuracy on the val set\nGot 443 / 1000 correct (44.30%)\n\nIteration 600, loss = 2.1000\nChecking accuracy on the val set\nGot 413 / 1000 correct (41.30%)\n\nIteration 700, loss = 1.6887\nChecking accuracy on the val set\nGot 451 / 1000 correct (45.10%)\n\n"
]
],
[
[
"### BareBones PyTorch: Training a ConvNet\n\nIn the below you should use the functions defined above to train a three-layer convolutional network on CIFAR. The network should have the following architecture:\n\n1. Convolutional layer (with bias) with 32 5x5 filters, with zero-padding of 2\n2. ReLU\n3. Convolutional layer (with bias) with 16 3x3 filters, with zero-padding of 1\n4. ReLU\n5. Fully-connected layer (with bias) to compute scores for 10 classes\n\nYou should initialize your weight matrices using the `random_weight` function defined above, and you should initialize your bias vectors using the `zero_weight` function above.\n\nYou don't need to tune any hyperparameters, but if everything works correctly you should achieve an accuracy above 42% after one epoch.",
"_____no_output_____"
]
],
[
[
"learning_rate = 3e-3\n\nchannel_1 = 32\nchannel_2 = 16\n\nconv_w1 = None\nconv_b1 = None\nconv_w2 = None\nconv_b2 = None\nfc_w = None\nfc_b = None\n\n################################################################################\n# TODO: Initialize the parameters of a three-layer ConvNet. #\n################################################################################\n# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\nconv_w1=random_weight((channel_1,3,5,5))\nconv_b1=zero_weight(channel_1)\nconv_w2=random_weight((channel_2,channel_1,3,3))\nconv_b2=zero_weight(channel_2)\nfc_w=random_weight((16*32*32,10))\nfc_b=zero_weight(10)\n\n# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n################################################################################\n# END OF YOUR CODE #\n################################################################################\n\nparams = [conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b]\ntrain_part2(three_layer_convnet, params, learning_rate)",
"Iteration 0, loss = 2.7253\nChecking accuracy on the val set\nGot 98 / 1000 correct (9.80%)\n\nIteration 100, loss = 1.9666\nChecking accuracy on the val set\nGot 358 / 1000 correct (35.80%)\n\nIteration 200, loss = 1.8562\nChecking accuracy on the val set\nGot 408 / 1000 correct (40.80%)\n\nIteration 300, loss = 1.6312\nChecking accuracy on the val set\nGot 422 / 1000 correct (42.20%)\n\nIteration 400, loss = 1.4729\nChecking accuracy on the val set\nGot 465 / 1000 correct (46.50%)\n\nIteration 500, loss = 1.3871\nChecking accuracy on the val set\nGot 470 / 1000 correct (47.00%)\n\nIteration 600, loss = 1.5025\nChecking accuracy on the val set\nGot 491 / 1000 correct (49.10%)\n\nIteration 700, loss = 1.5634\nChecking accuracy on the val set\nGot 496 / 1000 correct (49.60%)\n\n"
]
],
[
[
"# Part III. PyTorch Module API\n\nBarebone PyTorch requires that we track all the parameter tensors by hand. This is fine for small networks with a few tensors, but it would be extremely inconvenient and error-prone to track tens or hundreds of tensors in larger networks.\n\nPyTorch provides the `nn.Module` API for you to define arbitrary network architectures, while tracking every learnable parameters for you. In Part II, we implemented SGD ourselves. PyTorch also provides the `torch.optim` package that implements all the common optimizers, such as RMSProp, Adagrad, and Adam. It even supports approximate second-order methods like L-BFGS! You can refer to the [doc](http://pytorch.org/docs/master/optim.html) for the exact specifications of each optimizer.\n\nTo use the Module API, follow the steps below:\n\n1. Subclass `nn.Module`. Give your network class an intuitive name like `TwoLayerFC`. \n\n2. In the constructor `__init__()`, define all the layers you need as class attributes. Layer objects like `nn.Linear` and `nn.Conv2d` are themselves `nn.Module` subclasses and contain learnable parameters, so that you don't have to instantiate the raw tensors yourself. `nn.Module` will track these internal parameters for you. Refer to the [doc](http://pytorch.org/docs/master/nn.html) to learn more about the dozens of builtin layers. **Warning**: don't forget to call the `super().__init__()` first!\n\n3. In the `forward()` method, define the *connectivity* of your network. You should use the attributes defined in `__init__` as function calls that take tensor as input and output the \"transformed\" tensor. Do *not* create any new layers with learnable parameters in `forward()`! All of them must be declared upfront in `__init__`. \n\nAfter you define your Module subclass, you can instantiate it as an object and call it just like the NN forward function in part II.\n\n### Module API: Two-Layer Network\nHere is a concrete example of a 2-layer fully connected network:",
"_____no_output_____"
]
],
[
[
"class TwoLayerFC(nn.Module):\n def __init__(self, input_size, hidden_size, num_classes):\n super().__init__()\n # assign layer objects to class attributes\n self.fc1 = nn.Linear(input_size, hidden_size)\n # nn.init package contains convenient initialization methods\n # http://pytorch.org/docs/master/nn.html#torch-nn-init \n nn.init.kaiming_normal_(self.fc1.weight)\n self.fc2 = nn.Linear(hidden_size, num_classes)\n nn.init.kaiming_normal_(self.fc2.weight)\n \n def forward(self, x):\n # forward always defines connectivity\n x = flatten(x)\n scores = self.fc2(F.relu(self.fc1(x)))\n return scores\n\ndef test_TwoLayerFC():\n input_size = 50\n x = torch.zeros((64, input_size), dtype=dtype) # minibatch size 64, feature dimension 50\n model = TwoLayerFC(input_size, 42, 10)\n scores = model(x)\n print(scores.size()) # you should see [64, 10]\ntest_TwoLayerFC()",
"torch.Size([64, 10])\n"
]
],
[
[
"### Module API: Three-Layer ConvNet\nIt's your turn to implement a 3-layer ConvNet followed by a fully connected layer. The network architecture should be the same as in Part II:\n\n1. Convolutional layer with `channel_1` 5x5 filters with zero-padding of 2\n2. ReLU\n3. Convolutional layer with `channel_2` 3x3 filters with zero-padding of 1\n4. ReLU\n5. Fully-connected layer to `num_classes` classes\n\nYou should initialize the weight matrices of the model using the Kaiming normal initialization method.\n\n**HINT**: http://pytorch.org/docs/stable/nn.html#conv2d\n\nAfter you implement the three-layer ConvNet, the `test_ThreeLayerConvNet` function will run your implementation; it should print `(64, 10)` for the shape of the output scores.",
"_____no_output_____"
]
],
[
[
"class ThreeLayerConvNet(nn.Module):\n def __init__(self, in_channel, channel_1, channel_2, num_classes):\n super().__init__()\n ########################################################################\n # TODO: Set up the layers you need for a three-layer ConvNet with the #\n # architecture defined above. #\n ########################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n self.conv1=nn.Conv2d(in_channel,channel_1,5,padding=2)\n nn.init.kaiming_normal_(self.conv1.weight)\n self.conv2=nn.Conv2d(channel_1,channel_2,3,padding=1)\n nn.init.kaiming_normal_(self.conv2.weight)\n self.fc=nn.Linear(channel_2*32*32,num_classes)\n nn.init.kaiming_normal_(self.fc.weight)\n self.relu=nn.ReLU(inplace=True)\n\n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ########################################################################\n # END OF YOUR CODE # \n ########################################################################\n\n def forward(self, x):\n scores = None\n ########################################################################\n # TODO: Implement the forward function for a 3-layer ConvNet. you #\n # should use the layers you defined in __init__ and specify the #\n # connectivity of those layers in forward() #\n ########################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n scores=self.relu(self.conv1(x))\n scores=self.relu(self.conv2(scores))\n scores=self.fc(flatten(scores))\n\n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ########################################################################\n # END OF YOUR CODE #\n ########################################################################\n return scores\n\n\ndef test_ThreeLayerConvNet():\n x = torch.zeros((64, 3, 32, 32), dtype=dtype) # minibatch size 64, image size [3, 32, 32]\n model = ThreeLayerConvNet(in_channel=3, channel_1=12, channel_2=8, num_classes=10)\n scores = model(x)\n print(scores.size()) # you should see [64, 10]\ntest_ThreeLayerConvNet()",
"torch.Size([64, 10])\n"
]
],
[
[
"### Module API: Check Accuracy\nGiven the validation or test set, we can check the classification accuracy of a neural network. \n\nThis version is slightly different from the one in part II. You don't manually pass in the parameters anymore.",
"_____no_output_____"
]
],
[
[
"def check_accuracy_part34(loader, model):\n if loader.dataset.train:\n print('Checking accuracy on validation set')\n else:\n print('Checking accuracy on test set') \n num_correct = 0\n num_samples = 0\n model.eval() # set model to evaluation mode\n with torch.no_grad():\n for x, y in loader:\n x = x.to(device=device, dtype=dtype) # move to device, e.g. GPU\n y = y.to(device=device, dtype=torch.long)\n scores = model(x)\n _, preds = scores.max(1)\n num_correct += (preds == y).sum()\n num_samples += preds.size(0)\n acc = float(num_correct) / num_samples\n print('Got %d / %d correct (%.2f)' % (num_correct, num_samples, 100 * acc))",
"_____no_output_____"
]
],
[
[
"### Module API: Training Loop\nWe also use a slightly different training loop. Rather than updating the values of the weights ourselves, we use an Optimizer object from the `torch.optim` package, which abstract the notion of an optimization algorithm and provides implementations of most of the algorithms commonly used to optimize neural networks.",
"_____no_output_____"
]
],
[
[
"def train_part34(model, optimizer, epochs=1):\n \"\"\"\n Train a model on CIFAR-10 using the PyTorch Module API.\n \n Inputs:\n - model: A PyTorch Module giving the model to train.\n - optimizer: An Optimizer object we will use to train the model\n - epochs: (Optional) A Python integer giving the number of epochs to train for\n \n Returns: Nothing, but prints model accuracies during training.\n \"\"\"\n model = model.to(device=device) # move the model parameters to CPU/GPU\n for e in range(epochs):\n for t, (x, y) in enumerate(loader_train):\n model.train() # put model to training mode\n x = x.to(device=device, dtype=dtype) # move to device, e.g. GPU\n y = y.to(device=device, dtype=torch.long)\n\n scores = model(x)\n loss = F.cross_entropy(scores, y)\n\n # Zero out all of the gradients for the variables which the optimizer\n # will update.\n optimizer.zero_grad()\n\n # This is the backwards pass: compute the gradient of the loss with\n # respect to each parameter of the model.\n loss.backward()\n\n # Actually update the parameters of the model using the gradients\n # computed by the backwards pass.\n optimizer.step()\n\n if t % print_every == 0:\n print('Iteration %d, loss = %.4f' % (t, loss.item()))\n check_accuracy_part34(loader_val, model)\n print()",
"_____no_output_____"
]
],
[
[
"### Module API: Train a Two-Layer Network\nNow we are ready to run the training loop. In contrast to part II, we don't explicitly allocate parameter tensors anymore.\n\nSimply pass the input size, hidden layer size, and number of classes (i.e. output size) to the constructor of `TwoLayerFC`. \n\nYou also need to define an optimizer that tracks all the learnable parameters inside `TwoLayerFC`.\n\nYou don't need to tune any hyperparameters, but you should see model accuracies above 40% after training for one epoch.",
"_____no_output_____"
]
],
[
[
"hidden_layer_size = 4000\nlearning_rate = 1e-2\nmodel = TwoLayerFC(3 * 32 * 32, hidden_layer_size, 10)\noptimizer = optim.SGD(model.parameters(), lr=learning_rate)\n\ntrain_part34(model, optimizer)",
"Iteration 0, loss = 3.3422\nChecking accuracy on validation set\nGot 145 / 1000 correct (14.50)\n\nIteration 100, loss = 2.5751\nChecking accuracy on validation set\nGot 361 / 1000 correct (36.10)\n\nIteration 200, loss = 2.1785\nChecking accuracy on validation set\nGot 332 / 1000 correct (33.20)\n\nIteration 300, loss = 2.4100\nChecking accuracy on validation set\nGot 369 / 1000 correct (36.90)\n\nIteration 400, loss = 1.8031\nChecking accuracy on validation set\nGot 408 / 1000 correct (40.80)\n\nIteration 500, loss = 1.6054\nChecking accuracy on validation set\nGot 418 / 1000 correct (41.80)\n\nIteration 600, loss = 1.5121\nChecking accuracy on validation set\nGot 407 / 1000 correct (40.70)\n\nIteration 700, loss = 1.5409\nChecking accuracy on validation set\nGot 448 / 1000 correct (44.80)\n\n"
]
],
[
[
"### Module API: Train a Three-Layer ConvNet\nYou should now use the Module API to train a three-layer ConvNet on CIFAR. This should look very similar to training the two-layer network! You don't need to tune any hyperparameters, but you should achieve above above 45% after training for one epoch.\n\nYou should train the model using stochastic gradient descent without momentum.",
"_____no_output_____"
]
],
[
[
"learning_rate = 3e-3\nchannel_1 = 32\nchannel_2 = 16\n\nmodel = None\noptimizer = None\n################################################################################\n# TODO: Instantiate your ThreeLayerConvNet model and a corresponding optimizer #\n################################################################################\n# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\nmodel=ThreeLayerConvNet(3,channel_1,channel_2,10)\noptimizer=optim.SGD(model.parameters(),lr=learning_rate)\n\n# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n################################################################################\n# END OF YOUR CODE \n################################################################################\n\ntrain_part34(model, optimizer)",
"Iteration 0, loss = 4.2933\nChecking accuracy on validation set\nGot 152 / 1000 correct (15.20)\n\nIteration 100, loss = 1.8243\nChecking accuracy on validation set\nGot 359 / 1000 correct (35.90)\n\nIteration 200, loss = 1.9024\nChecking accuracy on validation set\nGot 406 / 1000 correct (40.60)\n\nIteration 300, loss = 1.7868\nChecking accuracy on validation set\nGot 433 / 1000 correct (43.30)\n\nIteration 400, loss = 1.5867\nChecking accuracy on validation set\nGot 454 / 1000 correct (45.40)\n\nIteration 500, loss = 1.4033\nChecking accuracy on validation set\nGot 462 / 1000 correct (46.20)\n\nIteration 600, loss = 1.6708\nChecking accuracy on validation set\nGot 470 / 1000 correct (47.00)\n\nIteration 700, loss = 1.3941\nChecking accuracy on validation set\nGot 483 / 1000 correct (48.30)\n\n"
]
],
[
[
"# Part IV. PyTorch Sequential API\n\nPart III introduced the PyTorch Module API, which allows you to define arbitrary learnable layers and their connectivity. \n\nFor simple models like a stack of feed forward layers, you still need to go through 3 steps: subclass `nn.Module`, assign layers to class attributes in `__init__`, and call each layer one by one in `forward()`. Is there a more convenient way? \n\nFortunately, PyTorch provides a container Module called `nn.Sequential`, which merges the above steps into one. It is not as flexible as `nn.Module`, because you cannot specify more complex topology than a feed-forward stack, but it's good enough for many use cases.\n\n### Sequential API: Two-Layer Network\nLet's see how to rewrite our two-layer fully connected network example with `nn.Sequential`, and train it using the training loop defined above.\n\nAgain, you don't need to tune any hyperparameters here, but you shoud achieve above 40% accuracy after one epoch of training.",
"_____no_output_____"
]
],
[
[
"# We need to wrap `flatten` function in a module in order to stack it\n# in nn.Sequential\nclass Flatten(nn.Module):\n def forward(self, x):\n return flatten(x)\n\nhidden_layer_size = 4000\nlearning_rate = 1e-2\n\nmodel = nn.Sequential(\n Flatten(),\n nn.Linear(3 * 32 * 32, hidden_layer_size),\n nn.ReLU(),\n nn.Linear(hidden_layer_size, 10),\n)\n\n# you can use Nesterov momentum in optim.SGD\noptimizer = optim.SGD(model.parameters(), lr=learning_rate,\n momentum=0.9, nesterov=True)\n\ntrain_part34(model, optimizer)",
"Iteration 0, loss = 2.3157\nChecking accuracy on validation set\nGot 149 / 1000 correct (14.90)\n\nIteration 100, loss = 1.7171\nChecking accuracy on validation set\nGot 381 / 1000 correct (38.10)\n\nIteration 200, loss = 1.8746\nChecking accuracy on validation set\nGot 407 / 1000 correct (40.70)\n\nIteration 300, loss = 2.0099\nChecking accuracy on validation set\nGot 393 / 1000 correct (39.30)\n\nIteration 400, loss = 1.5636\nChecking accuracy on validation set\nGot 461 / 1000 correct (46.10)\n\nIteration 500, loss = 1.8556\nChecking accuracy on validation set\nGot 436 / 1000 correct (43.60)\n\nIteration 600, loss = 1.8104\nChecking accuracy on validation set\nGot 457 / 1000 correct (45.70)\n\nIteration 700, loss = 1.8647\nChecking accuracy on validation set\nGot 430 / 1000 correct (43.00)\n\n"
]
],
[
[
"### Sequential API: Three-Layer ConvNet\nHere you should use `nn.Sequential` to define and train a three-layer ConvNet with the same architecture we used in Part III:\n\n1. Convolutional layer (with bias) with 32 5x5 filters, with zero-padding of 2\n2. ReLU\n3. Convolutional layer (with bias) with 16 3x3 filters, with zero-padding of 1\n4. ReLU\n5. Fully-connected layer (with bias) to compute scores for 10 classes\n\nYou should initialize your weight matrices using the `random_weight` function defined above, and you should initialize your bias vectors using the `zero_weight` function above.\n\nYou should optimize your model using stochastic gradient descent with Nesterov momentum 0.9.\n\nAgain, you don't need to tune any hyperparameters but you should see accuracy above 55% after one epoch of training.",
"_____no_output_____"
]
],
[
[
"channel_1 = 32\nchannel_2 = 16\nlearning_rate = 1e-2\n\nmodel = None\noptimizer = None\n\n################################################################################\n# TODO: Rewrite the 2-layer ConvNet with bias from Part III with the #\n# Sequential API. #\n################################################################################\n# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\nmodel=nn.Sequential(\n nn.Conv2d(3,channel_1,5,padding=2),\n nn.ReLU(inplace=True),\n nn.Conv2d(channel_1,channel_2,3,padding=1),\n nn.ReLU(inplace=True),\n Flatten(),\n nn.Linear(channel_2*32*32,10)\n)\n# for i in (0,2,5):\n# w_shape=model[i].weight.data.shape\n# b_shape=model[i].bias.data.shape\n# model[i].weight.data=random_weight(w_shape)\n# model[i].bias.data=zero_weight(b_shape)\n\noptimizer=optim.SGD(model.parameters(),nesterov=True,lr=learning_rate, momentum=0.9)\n\n# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n################################################################################\n# END OF YOUR CODE \n################################################################################\n\ntrain_part34(model, optimizer)",
"Iteration 0, loss = 2.3090\nChecking accuracy on validation set\nGot 150 / 1000 correct (15.00)\n\nIteration 100, loss = 1.4854\nChecking accuracy on validation set\nGot 416 / 1000 correct (41.60)\n\nIteration 200, loss = 1.3982\nChecking accuracy on validation set\nGot 476 / 1000 correct (47.60)\n\nIteration 300, loss = 1.1755\nChecking accuracy on validation set\nGot 486 / 1000 correct (48.60)\n\nIteration 400, loss = 1.5517\nChecking accuracy on validation set\nGot 531 / 1000 correct (53.10)\n\nIteration 500, loss = 1.3707\nChecking accuracy on validation set\nGot 554 / 1000 correct (55.40)\n\nIteration 600, loss = 1.5679\nChecking accuracy on validation set\nGot 579 / 1000 correct (57.90)\n\nIteration 700, loss = 1.4356\nChecking accuracy on validation set\nGot 555 / 1000 correct (55.50)\n\n"
]
],
[
[
"# Part V. CIFAR-10 open-ended challenge\n\nIn this section, you can experiment with whatever ConvNet architecture you'd like on CIFAR-10. \n\nNow it's your job to experiment with architectures, hyperparameters, loss functions, and optimizers to train a model that achieves **at least 70%** accuracy on the CIFAR-10 **validation** set within 10 epochs. You can use the check_accuracy and train functions from above. You can use either `nn.Module` or `nn.Sequential` API. \n\nDescribe what you did at the end of this notebook.\n\nHere are the official API documentation for each component. One note: what we call in the class \"spatial batch norm\" is called \"BatchNorm2D\" in PyTorch.\n\n* Layers in torch.nn package: http://pytorch.org/docs/stable/nn.html\n* Activations: http://pytorch.org/docs/stable/nn.html#non-linear-activations\n* Loss functions: http://pytorch.org/docs/stable/nn.html#loss-functions\n* Optimizers: http://pytorch.org/docs/stable/optim.html\n\n\n### Things you might try:\n- **Filter size**: Above we used 5x5; would smaller filters be more efficient?\n- **Number of filters**: Above we used 32 filters. Do more or fewer do better?\n- **Pooling vs Strided Convolution**: Do you use max pooling or just stride convolutions?\n- **Batch normalization**: Try adding spatial batch normalization after convolution layers and vanilla batch normalization after affine layers. Do your networks train faster?\n- **Network architecture**: The network above has two layers of trainable parameters. Can you do better with a deep network? Good architectures to try include:\n - [conv-relu-pool]xN -> [affine]xM -> [softmax or SVM]\n - [conv-relu-conv-relu-pool]xN -> [affine]xM -> [softmax or SVM]\n - [batchnorm-relu-conv]xN -> [affine]xM -> [softmax or SVM]\n- **Global Average Pooling**: Instead of flattening and then having multiple affine layers, perform convolutions until your image gets small (7x7 or so) and then perform an average pooling operation to get to a 1x1 image picture (1, 1 , Filter#), which is then reshaped into a (Filter#) vector. This is used in [Google's Inception Network](https://arxiv.org/abs/1512.00567) (See Table 1 for their architecture).\n- **Regularization**: Add l2 weight regularization, or perhaps use Dropout.\n\n### Tips for training\nFor each network architecture that you try, you should tune the learning rate and other hyperparameters. When doing this there are a couple important things to keep in mind:\n\n- If the parameters are working well, you should see improvement within a few hundred iterations\n- Remember the coarse-to-fine approach for hyperparameter tuning: start by testing a large range of hyperparameters for just a few training iterations to find the combinations of parameters that are working at all.\n- Once you have found some sets of parameters that seem to work, search more finely around these parameters. You may need to train for more epochs.\n- You should use the validation set for hyperparameter search, and save your test set for evaluating your architecture on the best parameters as selected by the validation set.\n\n### Going above and beyond\nIf you are feeling adventurous there are many other features you can implement to try and improve your performance. You are **not required** to implement any of these, but don't miss the fun if you have time!\n\n- Alternative optimizers: you can try Adam, Adagrad, RMSprop, etc.\n- Alternative activation functions such as leaky ReLU, parametric ReLU, ELU, or MaxOut.\n- Model ensembles\n- Data augmentation\n- New Architectures\n - [ResNets](https://arxiv.org/abs/1512.03385) where the input from the previous layer is added to the output.\n - [DenseNets](https://arxiv.org/abs/1608.06993) where inputs into previous layers are concatenated together.\n - [This blog has an in-depth overview](https://chatbotslife.com/resnets-highwaynets-and-densenets-oh-my-9bb15918ee32)\n\n### Have fun and happy training! ",
"_____no_output_____"
]
],
[
[
"################################################################################\n# TODO: # \n# Experiment with any architectures, optimizers, and hyperparameters. #\n# Achieve AT LEAST 70% accuracy on the *validation set* within 10 epochs. #\n# #\n# Note that you can use the check_accuracy function to evaluate on either #\n# the test set or the validation set, by passing either loader_test or #\n# loader_val as the second argument to check_accuracy. You should not touch #\n# the test set until you have finished your architecture and hyperparameter #\n# tuning, and only run the test set once at the end to report a final value. #\n################################################################################\nmodel = None\noptimizer = None\n\n# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\nclass AlexNet(nn.Module):\n def __init__(self, num_classes=10):\n super(AlexNet, self).__init__()\n self.relu=nn.ReLU(inplace=True)\n self.features = nn.Sequential(\n nn.Conv2d(3, 64, kernel_size=3, padding=1),\n self.relu,\n nn.MaxPool2d(kernel_size=2),\n\n nn.Conv2d(64, 192, kernel_size=3, padding=1),\n self.relu,\n nn.MaxPool2d(kernel_size=2),\n\n nn.Conv2d(192, 384, kernel_size=3, padding=1),\n self.relu,\n\n nn.Conv2d(384, 256, kernel_size=3, padding=1),\n self.relu,\n\n # nn.Conv2d(256, 256, kernel_size=3, padding=1),\n # nn.ReLU(inplace=True),\n # nn.MaxPool2d(kernel_size=2),\n )\n self.avgpool = nn.AdaptiveAvgPool2d((7, 7))\n self.classifier = nn.Sequential(\n nn.Dropout(),\n nn.Linear(256 * 7 * 7, 4096),\n nn.ReLU(inplace=True),\n nn.Dropout(),\n nn.Linear(4096, 4096),\n nn.ReLU(inplace=True),\n nn.Linear(4096, num_classes)\n )\n\n def forward(self, x):\n x = self.features(x)\n x: Tensor = self.avgpool(x)\n x = x.view(-1, 7 * 7 * 256)\n x = self.classifier(x)\n return x\n\nmodel=AlexNet()\noptimizer=optim.Adam(model.parameters())\n\n# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n################################################################################\n# END OF YOUR CODE \n################################################################################\n\n# You should get at least 70% accuracy\ntrain_part34(model, optimizer, epochs=10)",
"Iteration 0, loss = 2.2991\nChecking accuracy on validation set\nGot 87 / 1000 correct (8.70)\n\nIteration 100, loss = 1.7552\nChecking accuracy on validation set\nGot 297 / 1000 correct (29.70)\n\nIteration 200, loss = 1.5173\nChecking accuracy on validation set\nGot 422 / 1000 correct (42.20)\n\nIteration 300, loss = 1.8898\nChecking accuracy on validation set\nGot 462 / 1000 correct (46.20)\n\nIteration 400, loss = 1.2726\nChecking accuracy on validation set\nGot 454 / 1000 correct (45.40)\n\nIteration 500, loss = 1.3788\nChecking accuracy on validation set\nGot 517 / 1000 correct (51.70)\n\nIteration 600, loss = 1.4038\nChecking accuracy on validation set\nGot 549 / 1000 correct (54.90)\n\nIteration 700, loss = 1.3585\nChecking accuracy on validation set\nGot 585 / 1000 correct (58.50)\n\nIteration 0, loss = 1.2974\nChecking accuracy on validation set\nGot 521 / 1000 correct (52.10)\n\nIteration 100, loss = 1.1283\nChecking accuracy on validation set\nGot 595 / 1000 correct (59.50)\n\nIteration 200, loss = 1.0768\nChecking accuracy on validation set\nGot 561 / 1000 correct (56.10)\n\nIteration 300, loss = 1.3051\nChecking accuracy on validation set\nGot 578 / 1000 correct (57.80)\n\nIteration 400, loss = 1.1316\nChecking accuracy on validation set\nGot 614 / 1000 correct (61.40)\n\nIteration 500, loss = 1.3126\nChecking accuracy on validation set\nGot 610 / 1000 correct (61.00)\n\nIteration 600, loss = 0.9826\nChecking accuracy on validation set\nGot 615 / 1000 correct (61.50)\n\nIteration 700, loss = 1.0879\nChecking accuracy on validation set\nGot 642 / 1000 correct (64.20)\n\nIteration 0, loss = 0.8668\nChecking accuracy on validation set\nGot 649 / 1000 correct (64.90)\n\nIteration 100, loss = 0.6691\nChecking accuracy on validation set\nGot 640 / 1000 correct (64.00)\n\nIteration 200, loss = 1.0020\nChecking accuracy on validation set\nGot 651 / 1000 correct (65.10)\n\nIteration 300, loss = 1.0449\nChecking accuracy on validation set\nGot 658 / 1000 correct (65.80)\n\nIteration 400, loss = 1.0965\nChecking accuracy on validation set\nGot 689 / 1000 correct (68.90)\n\nIteration 500, loss = 0.8982\nChecking accuracy on validation set\nGot 690 / 1000 correct (69.00)\n\nIteration 600, loss = 1.0119\nChecking accuracy on validation set\nGot 683 / 1000 correct (68.30)\n\nIteration 700, loss = 1.0148\nChecking accuracy on validation set\nGot 714 / 1000 correct (71.40)\n\nIteration 0, loss = 0.9760\nChecking accuracy on validation set\nGot 701 / 1000 correct (70.10)\n\nIteration 100, loss = 1.0729\nChecking accuracy on validation set\nGot 723 / 1000 correct (72.30)\n\nIteration 200, loss = 0.9246\nChecking accuracy on validation set\nGot 723 / 1000 correct (72.30)\n\nIteration 300, loss = 1.0231\nChecking accuracy on validation set\nGot 688 / 1000 correct (68.80)\n\nIteration 400, loss = 0.7594\nChecking accuracy on validation set\nGot 702 / 1000 correct (70.20)\n\nIteration 500, loss = 0.9763\nChecking accuracy on validation set\nGot 720 / 1000 correct (72.00)\n\nIteration 600, loss = 0.8896\nChecking accuracy on validation set\nGot 723 / 1000 correct (72.30)\n\nIteration 700, loss = 0.8060\nChecking accuracy on validation set\nGot 722 / 1000 correct (72.20)\n\nIteration 0, loss = 0.7393\nChecking accuracy on validation set\nGot 701 / 1000 correct (70.10)\n\nIteration 100, loss = 0.7035\nChecking accuracy on validation set\nGot 735 / 1000 correct (73.50)\n\nIteration 200, loss = 0.6795\nChecking accuracy on validation set\nGot 729 / 1000 correct (72.90)\n\nIteration 300, loss = 0.6972\nChecking accuracy on validation set\nGot 727 / 1000 correct (72.70)\n\nIteration 400, loss = 1.0203\nChecking accuracy on validation set\nGot 740 / 1000 correct (74.00)\n\nIteration 500, loss = 0.5909\nChecking accuracy on validation set\nGot 747 / 1000 correct (74.70)\n\nIteration 600, loss = 0.7872\nChecking accuracy on validation set\nGot 708 / 1000 correct (70.80)\n\nIteration 700, loss = 0.9953\nChecking accuracy on validation set\nGot 726 / 1000 correct (72.60)\n\nIteration 0, loss = 0.6975\nChecking accuracy on validation set\nGot 729 / 1000 correct (72.90)\n\nIteration 100, loss = 0.9017\nChecking accuracy on validation set\nGot 758 / 1000 correct (75.80)\n\nIteration 200, loss = 0.8283\nChecking accuracy on validation set\nGot 742 / 1000 correct (74.20)\n\nIteration 300, loss = 0.5002\nChecking accuracy on validation set\nGot 745 / 1000 correct (74.50)\n\nIteration 400, loss = 0.6421\nChecking accuracy on validation set\nGot 756 / 1000 correct (75.60)\n\nIteration 500, loss = 0.5054\nChecking accuracy on validation set\nGot 764 / 1000 correct (76.40)\n\nIteration 600, loss = 0.6842\nChecking accuracy on validation set\nGot 724 / 1000 correct (72.40)\n\nIteration 700, loss = 0.6907\nChecking accuracy on validation set\nGot 762 / 1000 correct (76.20)\n\nIteration 0, loss = 0.5623\nChecking accuracy on validation set\nGot 733 / 1000 correct (73.30)\n\nIteration 100, loss = 0.5849\nChecking accuracy on validation set\nGot 763 / 1000 correct (76.30)\n\nIteration 200, loss = 0.9528\nChecking accuracy on validation set\nGot 759 / 1000 correct (75.90)\n\nIteration 300, loss = 0.5026\nChecking accuracy on validation set\nGot 755 / 1000 correct (75.50)\n\nIteration 400, loss = 0.6748\nChecking accuracy on validation set\nGot 758 / 1000 correct (75.80)\n\nIteration 500, loss = 0.8019\nChecking accuracy on validation set\nGot 775 / 1000 correct (77.50)\n\nIteration 600, loss = 0.8776\nChecking accuracy on validation set\nGot 771 / 1000 correct (77.10)\n\nIteration 700, loss = 0.3650\nChecking accuracy on validation set\nGot 748 / 1000 correct (74.80)\n\nIteration 0, loss = 0.7855\nChecking accuracy on validation set\nGot 771 / 1000 correct (77.10)\n\nIteration 100, loss = 0.6223\nChecking accuracy on validation set\nGot 770 / 1000 correct (77.00)\n\nIteration 200, loss = 0.5580\nChecking accuracy on validation set\nGot 767 / 1000 correct (76.70)\n\nIteration 300, loss = 0.4888\nChecking accuracy on validation set\nGot 750 / 1000 correct (75.00)\n\nIteration 400, loss = 0.9556\nChecking accuracy on validation set\nGot 764 / 1000 correct (76.40)\n\nIteration 500, loss = 0.5919\nChecking accuracy on validation set\nGot 762 / 1000 correct (76.20)\n\nIteration 600, loss = 0.6052\nChecking accuracy on validation set\nGot 766 / 1000 correct (76.60)\n\nIteration 700, loss = 0.5001\nChecking accuracy on validation set\nGot 762 / 1000 correct (76.20)\n\nIteration 0, loss = 0.5619\nChecking accuracy on validation set\nGot 768 / 1000 correct (76.80)\n\nIteration 100, loss = 0.5155\nChecking accuracy on validation set\nGot 758 / 1000 correct (75.80)\n\nIteration 200, loss = 0.5664\nChecking accuracy on validation set\nGot 773 / 1000 correct (77.30)\n\nIteration 300, loss = 0.5228\nChecking accuracy on validation set\nGot 749 / 1000 correct (74.90)\n\nIteration 400, loss = 0.5239\nChecking accuracy on validation set\nGot 759 / 1000 correct (75.90)\n\nIteration 500, loss = 0.5380\nChecking accuracy on validation set\nGot 769 / 1000 correct (76.90)\n\nIteration 600, loss = 0.4890\nChecking accuracy on validation set\nGot 785 / 1000 correct (78.50)\n\nIteration 700, loss = 0.5316\nChecking accuracy on validation set\nGot 766 / 1000 correct (76.60)\n\nIteration 0, loss = 0.6571\nChecking accuracy on validation set\nGot 768 / 1000 correct (76.80)\n\nIteration 100, loss = 0.6541\nChecking accuracy on validation set\nGot 766 / 1000 correct (76.60)\n\nIteration 200, loss = 0.4546\nChecking accuracy on validation set\nGot 766 / 1000 correct (76.60)\n\nIteration 300, loss = 0.4215\nChecking accuracy on validation set\nGot 783 / 1000 correct (78.30)\n\nIteration 400, loss = 0.3967\nChecking accuracy on validation set\nGot 778 / 1000 correct (77.80)\n\nIteration 500, loss = 0.6639\nChecking accuracy on validation set\nGot 778 / 1000 correct (77.80)\n\nIteration 600, loss = 0.9450\nChecking accuracy on validation set\nGot 775 / 1000 correct (77.50)\n\nIteration 700, loss = 0.5720\nChecking accuracy on validation set\nGot 789 / 1000 correct (78.90)\n\n"
]
],
[
[
"## Describe what you did \n\nIn the cell below you should write an explanation of what you did, any additional features that you implemented, and/or any graphs that you made in the process of training and evaluating your network.",
"_____no_output_____"
],
[
"TODO: Describe what you did",
"_____no_output_____"
],
[
"## Test set -- run this only once\n\nNow that we've gotten a result we're happy with, we test our final model on the test set (which you should store in best_model). Think about how this compares to your validation set accuracy.",
"_____no_output_____"
]
],
[
[
"best_model = model\ncheck_accuracy_part34(loader_test, best_model)",
"Checking accuracy on test set\nGot 7581 / 10000 correct (75.81)\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
d05fcf18cba3eac2b361e7ae66152980eaec08fa | 5,325 | ipynb | Jupyter Notebook | homework01/homework_differentiation.ipynb | DmitryUlyanov/Practical_DL | a707238a714ec240c742335be3ebe61bb2b85e9d | [
"MIT"
] | 4 | 2018-08-17T02:51:17.000Z | 2020-05-11T02:05:41.000Z | homework01/homework_differentiation.ipynb | victorlempitsky/Practical_DL | a707238a714ec240c742335be3ebe61bb2b85e9d | [
"MIT"
] | null | null | null | homework01/homework_differentiation.ipynb | victorlempitsky/Practical_DL | a707238a714ec240c742335be3ebe61bb2b85e9d | [
"MIT"
] | 2 | 2018-04-01T08:54:33.000Z | 2021-09-28T17:56:37.000Z | 21.385542 | 329 | 0.49784 | [
[
[
"# Homework 1: Differentiation",
"_____no_output_____"
],
[
"Since it easy to google every task please please please try to undestand what's going on. The \"just answer\" thing will be not counted, make sure to present derivation of your solution. It is absolutely OK if you found an answer on web then just exercise in $\\LaTeX$ copying it into here.",
"_____no_output_____"
],
[
"Useful links: \n[1](http://www.machinelearning.ru/wiki/images/2/2a/Matrix-Gauss.pdf)\n[2](http://www.atmos.washington.edu/~dennis/MatrixCalculus.pdf)\n[3](http://cal.cs.illinois.edu/~johannes/research/matrix%20calculus.pdf)\n[4](http://research.microsoft.com/en-us/um/people/cmbishop/prml/index.htm)",
"_____no_output_____"
],
[
"## ex. 1",
"_____no_output_____"
],
[
"$$ \ny = x^Tx, \\quad x \\in \\mathbb{R}^N \n$$",
"_____no_output_____"
],
[
"$$\n\\frac{dy}{dx} = \n$$ ",
"_____no_output_____"
],
[
"## ex. 2",
"_____no_output_____"
],
[
"$$ y = tr(AB) \\quad A,B \\in \\mathbb{R}^{N \\times N} $$ ",
"_____no_output_____"
],
[
"$$\n\\frac{dy}{dA} =\n$$",
"_____no_output_____"
],
[
"## ex. 3",
"_____no_output_____"
],
[
"$$ \ny = x^TAc , \\quad A\\in \\mathbb{R}^{N \\times N}, x\\in \\mathbb{R}^{N}, c\\in \\mathbb{R}^{N} \n$$",
"_____no_output_____"
],
[
"$$\n\\frac{dy}{dx} =\n$$",
"_____no_output_____"
],
[
"$$\n\\frac{dy}{dA} =\n$$ ",
"_____no_output_____"
],
[
"Hint for the latter (one of the ways): use *ex. 2* result and the fact \n$$\ntr(ABC) = tr (CAB)\n$$",
"_____no_output_____"
],
[
"## ex. 4",
"_____no_output_____"
],
[
"Classic matrix factorization example. Given matrix $X$ you need to find $A$, $S$ to approximate $X$. This can be done by simple gradient descent iteratively alternating $A$ and $S$ updates.\n$$\nJ = || X - AS ||_2^2 , \\quad A\\in \\mathbb{R}^{N \\times R} , \\quad S\\in \\mathbb{R}^{R \\times M}\n$$\n$$\n\\frac{dJ}{dS} = ? \n$$ ",
"_____no_output_____"
],
[
"### First approach\nUsing ex.2 and the fact:\n$$\n|| X ||_2^2 = tr(XX^T) \n$$ \nit is easy to derive gradients (you can find it in one of the refs). ",
"_____no_output_____"
],
[
"### Second approach\nYou can use *slightly different techniques* if they suits you. Take a look at this derivation:\n<img src=\"grad.png\">\n(excerpt from [Handbook of blind source separation, Jutten, page 517](https://books.google.ru/books?id=PTbj03bYH6kC&printsec=frontcover&dq=Handbook+of+Blind+Source+Separation&hl=en&sa=X&ved=0ahUKEwi-q_apiJDLAhULvXIKHVXJDWcQ6AEIHDAA#v=onepage&q=Handbook%20of%20Blind%20Source%20Separation&f=false), open for better picture).",
"_____no_output_____"
],
[
"### Third approach\nAnd finally we can use chain rule! **YOUR TURN** to do it.\nlet $ F = AS $ \n\n**Find**\n$$\n\\frac{dJ}{dF} = \n$$ \nand \n$$\n\\frac{dF}{dS} = \n$$ \n(the shape should be $ NM \\times RM )$.\n\nNow it is easy do get desired gradients:\n$$\n\\frac{dJ}{dS} = \n$$ ",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d05fd507986dff6a93945f092dacd2ae5074f76e | 23,977 | ipynb | Jupyter Notebook | ANALYSIS_engineered.ipynb | WhoIsJack/data-driven-analysis-lateralline | 0111c9e06e0cac57e4707b3fcb1553ca5ba4f236 | [
"MIT"
] | 3 | 2020-06-09T13:48:14.000Z | 2021-01-29T13:50:19.000Z | ANALYSIS_engineered.ipynb | WhoIsJack/data-driven-analysis-lateralline | 0111c9e06e0cac57e4707b3fcb1553ca5ba4f236 | [
"MIT"
] | null | null | null | ANALYSIS_engineered.ipynb | WhoIsJack/data-driven-analysis-lateralline | 0111c9e06e0cac57e4707b3fcb1553ca5ba4f236 | [
"MIT"
] | 2 | 2020-05-29T12:52:24.000Z | 2021-09-04T12:20:46.000Z | 31.799735 | 123 | 0.516578 | [
[
[
"<a id=top></a>\n\n# Analysis of Engineered Features",
"_____no_output_____"
],
[
"## Table of Contents\n\n**Note:** In this notebook, the engineered features are referred to as \"covariates\".\n\n----\n\n1. [Preparations](#prep)\n2. [Analysis of Covariates](#covar_analysis)\n 1. [Boxplots](#covar_analysis_boxplots)\n 2. [Forward Mapping (onto Shape Space)](#covar_analysis_fwdmap)\n 3. [Back Mapping (Tissue Consensus Map)](#covar_analysis_backmap)\n 4. [Covariate Correlations](#covar_analysis_correlations)\n3. [Covariate-Shape Relationships](#covar_fspace)\n 1. [Covariate-Shape Correlations](#covar_fspace_correlations)\n 2. [Covariate Relation Graph](#covar_fspace_graph)",
"_____no_output_____"
],
[
"<a id=prep></a>\n\n## 1. Preparations\n\n----",
"_____no_output_____"
]
],
[
[
"### Import modules\n\n# External, general\nfrom __future__ import division\nimport os, sys\nimport numpy as np\nnp.random.seed(42)\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n# External, specific\nimport pandas as pd\nimport ipywidgets as widgets\nfrom IPython.display import display, HTML\nfrom scipy.stats import linregress, pearsonr, gaussian_kde\nfrom scipy.spatial import cKDTree\nimport seaborn as sns\nsns.set_style('white')\nimport networkx as nx\n\n# Internal\nimport katachi.utilities.loading as ld\nimport katachi.utilities.plotting as kp",
"_____no_output_____"
],
[
"### Load data\n\n# Prep loader\nloader = ld.DataLoaderIDR()\nloader.find_imports(r\"data/experimentA/extracted_measurements/\", recurse=True, verbose=True)\n\n# Import embedded feature space\ndataset_suffix = \"shape_TFOR_pca_measured.tsv\"\n#dataset_suffix = \"shape_CFOR_pca_measured.tsv\"\n#dataset_suffix = \"tagRFPtUtrCH_TFOR_pca_measured.tsv\"\n#dataset_suffix = \"mKate2GM130_TFOR_pca_measured.tsv\"\nfspace_pca, prim_IDs, fspace_idx = loader.load_dataset(dataset_suffix)\nprint \"Imported feature space of shape:\", fspace_pca.shape\n\n# Import TFOR centroid locations\ncentroids = loader.load_dataset(\"_other_measurements.tsv\", IDs=prim_IDs)[0][:,3:6][:,::-1]\nprint \"Imported TFOR centroids of shape:\", centroids.shape\n \n# Import engineered features\ncovar_df, _, _ = loader.load_dataset(\"_other_measurements.tsv\", IDs=prim_IDs, force_df=True)\ndel covar_df['Centroids RAW X']; del covar_df['Centroids RAW Y']; del covar_df['Centroids RAW Z']\ncovar_names = list(covar_df.columns)\nprint \"Imported covariates of shape:\", covar_df.shape",
"_____no_output_____"
],
[
"### Report\nprint \"\\ncovar_df.head()\"\ndisplay(covar_df.head())\nprint \"\\ncovar_df.describe()\"\ndisplay(covar_df.describe())",
"_____no_output_____"
],
[
"### Z-standardize the covariates\n\ncovar_df_z = (covar_df - covar_df.mean()) / covar_df.std()",
"_____no_output_____"
]
],
[
[
"<a id=covar_analysis></a>\n\n## 2. Analysis of Covariates\n\n----",
"_____no_output_____"
],
[
"### Boxplots <a id=covar_analysis_boxplots></a>",
"_____no_output_____"
]
],
[
[
"### General boxplot of Covariates\n\n# Interactive selection of covariates\nwid = widgets.SelectMultiple(\n options=covar_names,\n value=covar_names,\n description='Covars',\n)\n\n# Interactive plot\[email protected](selected=wid, standardized=True)\ndef covariate_boxplot(selected=covar_names,\n standardized=True):\n\n # Select data\n if standardized:\n covar_df_plot = covar_df_z[list(selected)]\n else:\n covar_df_plot = covar_df[list(selected)]\n \n # Plot\n fig = plt.figure(figsize=(12,3))\n covar_df_plot.boxplot(grid=False)\n plt.tick_params(axis='both', which='major', labelsize=6)\n fig.autofmt_xdate()\n if standardized: plt.title(\"Boxplot of Covariates [standardized]\")\n if not standardized: plt.title(\"Boxplot of Covariates [raw]\")\n plt.show()",
"_____no_output_____"
]
],
[
[
"### Forward Mapping (onto Shape Space) <a id=covar_analysis_fwdmap></a>",
"_____no_output_____"
]
],
[
[
"### Interactive mapping of covariates onto PCA-transformed shape space\n\n# Set interactions\[email protected](covariate=covar_names,\n prim_ID=prim_IDs,\n PCx=(1, fspace_pca.shape[1], 1),\n PCy=(1, fspace_pca.shape[1], 1),\n standardized=False,\n show_all_prims=True)\n\n# Show \ndef show_PCs(covariate=covar_names[0], prim_ID=prim_IDs[0], \n PCx=1, PCy=2, standardized=False, show_all_prims=True): \n \n # Select covariate data\n if standardized:\n covar_df_plot = covar_df_z[covariate]\n else:\n covar_df_plot = covar_df[covariate]\n \n # Prep\n plt.figure(figsize=(9,7))\n \n # If all should be shown...\n if show_all_prims:\n \n # Plot\n plt.scatter(fspace_pca[:,PCx-1], fspace_pca[:,PCy-1],\n c=covar_df_plot, cmap=plt.cm.plasma,\n s=10, edgecolor='', alpha=0.75)\n \n # Cosmetics \n cbar = plt.colorbar()\n if standardized:\n cbar.set_label(covariate+\" [standardized]\", rotation=270, labelpad=15)\n else:\n cbar.set_label(covariate+\" [raw]\", rotation=270, labelpad=15)\n plt.xlabel(\"PC \"+str(PCx))\n plt.ylabel(\"PC \"+str(PCy))\n plt.title(\"PCA-Transformed Shape Space [All Prims]\")\n plt.show()\n \n # If individual prims should be shown...\n else:\n \n # Plot\n plt.scatter(fspace_pca[fspace_idx==prim_IDs.index(prim_ID), PCx-1], \n fspace_pca[fspace_idx==prim_IDs.index(prim_ID), PCy-1],\n c=covar_df_plot[fspace_idx==prim_IDs.index(prim_ID)], \n cmap=plt.cm.plasma, s=10, edgecolor='',\n vmin=covar_df_plot.min(), vmax=covar_df_plot.max())\n \n # Cosmetics\n cbar = plt.colorbar()\n if standardized:\n cbar.set_label(covariate+\" [standardized]\", rotation=270, labelpad=15)\n else:\n cbar.set_label(covariate+\" [raw]\", rotation=270, labelpad=15)\n plt.xlabel(\"PC \"+str(PCx))\n plt.ylabel(\"PC \"+str(PCy))\n plt.title(\"PCA-Transformed Shape Space [prim \"+prim_ID+\"]\")\n plt.show()",
"_____no_output_____"
]
],
[
[
"### Back Mapping (Tissue Consensus Map) <a id=covar_analysis_backmap></a>",
"_____no_output_____"
]
],
[
[
"### Interactive mapping of covariates onto centroids in TFOR\n\n# Axis range\nxlim = (-175, 15)\nylim = (- 25, 25)\n\n# Set interactions\[email protected](covariate=covar_names,\n standardized=['no','z'])\n\n# Plot\ndef centroid_backmap(covariate=covar_names[0],\n standardized='no'): \n\n # Select covariate data\n if standardized=='no':\n covar_df_plot = covar_df[covariate]\n elif standardized=='z':\n covar_df_plot = covar_df_z[covariate]\n \n # Init\n fig,ax = plt.subplots(1, figsize=(12,5))\n \n # Back-mapping plot\n #zord = np.argsort(covar_df_plot)\n zord = np.arange(len(covar_df_plot)); np.random.shuffle(zord) # Random is better!\n scat = ax.scatter(centroids[zord,2], centroids[zord,1],\n color=covar_df_plot[zord], cmap=plt.cm.plasma,\n edgecolor='', s=15, alpha=0.75)\n\n # Cosmetics\n ax.set_xlim(xlim)\n ax.set_ylim(ylim)\n ax.invert_yaxis() # To match images\n ax.set_xlabel('TFOR x')\n ax.set_ylabel('TFOR y')\n cbar = plt.colorbar(scat,ax=ax)\n if standardized:\n ax.set_title('Centroid Back-Mapping of '+covariate+' [standardized]')\n cbar.set_label(covariate+' [standardized]', rotation=270, labelpad=10)\n else:\n ax.set_title('Centroid Back-Mapping of '+covariate+' [raw]')\n cbar.set_label(covariate+' [raw]', rotation=270, labelpad=20)\n \n # Done\n plt.tight_layout()\n plt.show()",
"_____no_output_____"
],
[
"### Contour plot backmapping plot for publication\n\n# Set interactions\[email protected](covariate=covar_names,\n standardized=['no','z'])\n\n# Plot\ndef contour_backmap(covariate=covar_names[0],\n standardized='no'): \n\n # Settings\n xlim = (-130, 8)\n ylim = ( -19, 19)\n\n # Select covariate data\n if standardized=='no':\n covar_df_plot = covar_df[covariate]\n elif standardized=='z':\n covar_df_plot = covar_df_z[covariate]\n\n # Tools for smoothing on scatter\n from katachi.utilities.pcl_helpers import pcl_gaussian_smooth\n from scipy.spatial.distance import pdist, squareform\n\n # Cut off at prim contour outline\n kernel_prim = gaussian_kde(centroids[:,1:].T)\n f_prim = kernel_prim(centroids[:,1:].T)\n f_prim_mask = f_prim > f_prim.min() + (f_prim.max()-f_prim.min())*0.1\n plot_values = covar_df_plot[f_prim_mask]\n plot_centroids = centroids[f_prim_mask]\n\n # Smoothen\n pdists = squareform(pdist(plot_centroids[:,1:]))\n plot_values = pcl_gaussian_smooth(pdists, plot_values[:,np.newaxis], sg_percentile=0.5)[:,0]\n\n # Initialize figure\n fig, ax = plt.subplots(1, figsize=(8, 3.25))\n\n # Contourf plot\n cfset = ax.tricontourf(plot_centroids[:,2], plot_centroids[:,1], plot_values, 20, \n cmap='plasma')\n\n # Illustrative centroids from a single prim\n plt.scatter(centroids[fspace_idx==prim_IDs.index(prim_IDs[0]), 2], \n centroids[fspace_idx==prim_IDs.index(prim_IDs[0]), 1],\n c='', alpha=0.5)\n\n # Cosmetics\n ax.set_xlabel('TFOR x', fontsize=16)\n ax.set_ylabel('TFOR y', fontsize=16)\n plt.tick_params(axis='both', which='major', labelsize=13)\n plt.xlim(xlim); plt.ylim(ylim)\n ax.invert_yaxis() # To match images\n\n # Colorbar\n cbar = plt.colorbar(cfset, ax=ax, pad=0.01)\n cbar.set_label(covariate, rotation=270, labelpad=10, fontsize=16)\n cbar.ax.tick_params(labelsize=13)\n\n # Done\n plt.tight_layout()\n plt.show()",
"_____no_output_____"
]
],
[
[
"### Covariate Correlations <a id=covar_analysis_correlations></a>",
"_____no_output_____"
]
],
[
[
"### Interactive linear fitting plot\n\n# Set interaction\[email protected](covar_x=covar_names, \n covar_y=covar_names)\n\n# Plotting function\ndef corr_plot_covar(covar_x=covar_names[0], \n covar_y=covar_names[1]):\n\n # Prep\n plt.figure(figsize=(5,3))\n \n # Scatterplot\n plt.scatter(covar_df[covar_x], covar_df[covar_y],\n facecolor='darkblue', edgecolor='',\n s=5, alpha=0.5)\n plt.xlabel(covar_x)\n plt.ylabel(covar_y)\n \n # Linear regression and pearson\n fitted = linregress(covar_df[covar_x], covar_df[covar_y])\n pearson = pearsonr(covar_df[covar_x], covar_df[covar_y])\n \n # Report\n print \"Linear regression:\"\n for param,value in zip([\"slope\",\"intercept\",\"rvalue\",\"pvalue\",\"stderr\"], fitted):\n print \" {}:\\t{:.2e}\".format(param,value)\n print \"Pearson:\"\n print \" r:\\t{:.2e}\".format(pearson[0])\n print \" p:\\t{:.2e}\".format(pearson[1])\n \n # Add fit to plot\n xmin,xmax = (covar_df[covar_x].min(), covar_df[covar_x].max())\n ymin,ymax = (covar_df[covar_y].min(), covar_df[covar_y].max())\n ybot,ytop = (xmin*fitted[0]+fitted[1], xmax*fitted[0]+fitted[1])\n plt.plot([xmin,xmax], [ybot,ytop], c='blue', lw=2, alpha=0.5)\n \n # Cosmetics and show\n plt.xlim([xmin,xmax])\n plt.ylim([ymin,ymax])\n plt.show()",
"_____no_output_____"
],
[
"### Full pairwise correlation plot\n\n# Create the plot\nmclust = sns.clustermap(covar_df_z.corr(method='pearson'),\n figsize=(10, 10),\n cmap='RdBu')\n\n# Fix the y axis orientation\nmclust.ax_heatmap.set_yticklabels(mclust.ax_heatmap.get_yticklabels(),\n rotation=0)\n\n# Other cosmetics\nmclust.ax_heatmap.set_title(\"Pairwise Correlations Cluster Plot\", y=1.275)\nplt.ylabel(\"Pearson\\nCorr. Coef.\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"<a id=covar_fspace></a>\n\n## 3. Covariate-Shape Relationships\n\n----",
"_____no_output_____"
],
[
"### Covariate-Shape Correlations <a id=covar_fspace_correlations></a>",
"_____no_output_____"
]
],
[
[
"### Interactive linear fitting plot\n\n# Set interaction\[email protected](covar_x=covar_names, \n PC_y=range(1,fspace_pca.shape[1]+1))\n\n# Plotting function\ndef corr_plot_covar(covar_x=covar_names[0], \n PC_y=1):\n \n # Prep\n PC_y = int(PC_y)\n plt.figure(figsize=(5,3))\n \n # Scatterplot\n plt.scatter(covar_df[covar_x], fspace_pca[:, PC_y-1],\n facecolor='darkred', edgecolor='',\n s=5, alpha=0.5)\n plt.xlabel(covar_x)\n plt.ylabel(\"PC \"+str(PC_y))\n \n # Linear regression and pearson\n fitted = linregress(covar_df[covar_x], fspace_pca[:, PC_y-1])\n pearson = pearsonr(covar_df[covar_x], fspace_pca[:, PC_y-1])\n \n # Report\n print \"Linear regression:\"\n for param,value in zip([\"slope\",\"intercept\",\"rvalue\",\"pvalue\",\"stderr\"], fitted):\n print \" {}:\\t{:.2e}\".format(param,value)\n print \"Pearson:\"\n print \" r:\\t{:.2e}\".format(pearson[0])\n print \" p:\\t{:.2e}\".format(pearson[1])\n \n # Add fit to plot\n xmin,xmax = (covar_df[covar_x].min(), covar_df[covar_x].max())\n ymin,ymax = (fspace_pca[:, PC_y-1].min(), fspace_pca[:, PC_y-1].max())\n ybot,ytop = (xmin*fitted[0]+fitted[1], xmax*fitted[0]+fitted[1])\n plt.plot([xmin,xmax], [ybot,ytop], c='red', lw=2, alpha=0.5)\n \n # Cosmetics and show\n plt.xlim([xmin,xmax])\n plt.ylim([ymin,ymax])\n plt.show()",
"_____no_output_____"
],
[
"### Selected linear fits\n\n# Settings for TFOR PC 3\nif 'TFOR' in dataset_suffix:\n covar_x = 'Z Axis Length'\n PC_y = 3\n x_reduc = 0\n lbl_x = 'TFOR PC 3'\n lbl_y = 'Z Axis Length\\n(Cell Height)'\n\n# Settings for CFOR PC 1\nif 'CFOR' in dataset_suffix:\n covar_x = 'Sphericity'\n PC_y = 1\n x_reduc = 2\n lbl_x = 'CFOR PC 1'\n lbl_y = 'Sphericity'\n\n# Prep\nplt.figure(figsize=(6,4))\n\n# Scatterplot\nplt.scatter(fspace_pca[:, PC_y-1], covar_df[covar_x],\n facecolor='darkblue', edgecolor='',\n s=5, alpha=0.25)\nplt.xlabel(covar_x)\nplt.ylabel(\"PC \"+str(PC_y))\n\n# Linear regression and pearson\nfitted = linregress(fspace_pca[:, PC_y-1], covar_df[covar_x])\npearson = pearsonr(fspace_pca[:, PC_y-1], covar_df[covar_x])\n\n# Report\nprint \"Linear regression:\"\nfor param,value in zip([\"slope\",\"intercept\",\"rvalue\",\"pvalue\",\"stderr\"], fitted):\n print \" {}:\\t{:.2e}\".format(param,value)\nprint \"Pearson:\"\nprint \" r:\\t{:.2e}\".format(pearson[0])\nprint \" p:\\t{:.2e}\".format(pearson[1])\n\n# Add fit to plot\nymin,ymax = (covar_df[covar_x].min(), covar_df[covar_x].max())\nxmin,xmax = (fspace_pca[:, PC_y-1].min()-x_reduc, fspace_pca[:, PC_y-1].max())\nybot,ytop = (xmin*fitted[0]+fitted[1], xmax*fitted[0]+fitted[1])\nplt.plot([xmin,xmax], [ybot,ytop], c='black', lw=1, alpha=0.5)\n\n# Cosmetics\nplt.tick_params(axis='both', which='major', labelsize=16)\nplt.xlabel(lbl_x, fontsize=18)\nplt.ylabel(lbl_y, fontsize=18)\nplt.xlim([xmin,xmax])\nplt.ylim([ymin,ymax+0.05])\nplt.tight_layout()\n\n# Done\nplt.show()",
"_____no_output_____"
],
[
"### Full pairwise correlation plot \n\n# Prepare the pairwise correlation\nfspace_pca_z = (fspace_pca - fspace_pca.mean(axis=0)) / fspace_pca.std(axis=0)\nfspace_pca_z_df = pd.DataFrame(fspace_pca_z[:,:25])\npairwise_corr = covar_df_z.expanding(axis=1).corr(fspace_pca_z_df, pairwise=True).iloc[-1, :, :] # Ouf, pandas...\n\n# Create the plot\nmclust = sns.clustermap(pairwise_corr,\n figsize=(10, 10),\n col_cluster=False,\n cmap='RdBu')\n\n# Fix the y axis orientation\nmclust.ax_heatmap.set_yticklabels(mclust.ax_heatmap.get_yticklabels(),\n rotation=0)\n\n# Other cosmetics\nmclust.ax_heatmap.set_title(\"Pairwise Correlations Cluster Plot\", y=1.275)\nmclust.ax_heatmap.set_xticklabels(range(1,fspace_pca_z_df.shape[1]+1))\nplt.ylabel(\"Pearson\\nCorr. Coef.\")\n\n# Done\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Covariate Relation Graph <a id=covar_fspace_graph></a>",
"_____no_output_____"
]
],
[
[
"# Parameters\nnum_PCs = 8 # Number of PCs to include\ncorr_measure = 'pearsonr' # Correlation measure to use\nthreshold = 0.30 # Threshold to include a correlation as relevant\n\n# Get relevant data\nif corr_measure == 'pearsonr':\n covar_fspace_dists = pairwise_corr.get_values()[:, :num_PCs] # Retrieved from above!\nelse:\n raise NotImplementedError()\n\n# Generate the plot\nkp.covar_pc_bigraph(covar_fspace_dists, threshold, covar_names,\n height=0.6, verbose=True, show=False)\n\n# Done\nplt.show()",
"_____no_output_____"
]
],
[
[
"----\n[back to top](#top)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d05fda91dccb5e2e7bdf410b06bffd9a37303dbd | 70,545 | ipynb | Jupyter Notebook | 5-pandas-reshape.ipynb | terapyon/python-datahandling-tutorial | 1e8c0c2c7c5faafa28dd6f9d23e4deaf2caa1288 | [
"MIT"
] | 7 | 2017-12-30T18:28:02.000Z | 2021-05-28T12:03:53.000Z | 5-pandas-reshape.ipynb | terapyon/python-datahandling-tutorial | 1e8c0c2c7c5faafa28dd6f9d23e4deaf2caa1288 | [
"MIT"
] | 1 | 2018-01-08T09:41:30.000Z | 2018-01-14T07:48:01.000Z | 5-pandas-reshape.ipynb | terapyon/python-datahandling-tutorial | 1e8c0c2c7c5faafa28dd6f9d23e4deaf2caa1288 | [
"MIT"
] | null | null | null | 28.11678 | 86 | 0.326458 | [
[
[
"# pandasでデータの整形\n\nデータの整形、条件抽出、並べ替えを行います。",
"_____no_output_____"
],
[
"## 内容\n\n- データ抽出\n- データ型変換\n- 並べ替え\n- 不要なカラム削除\n- 組合せデータの挿入",
"_____no_output_____"
],
[
"## データ抽出",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"df = pd.read_excel(\"data/201704health.xlsx\")",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
],
[
[
"- 歩数が10000歩以上の日のみを抽出",
"_____no_output_____"
]
],
[
[
"df.loc[:, \"歩数\"] >= 10000",
"_____no_output_____"
],
[
"df_fill = df[df.loc[:, \"歩数\"] >= 10000]",
"_____no_output_____"
],
[
"df_fill",
"_____no_output_____"
],
[
"df_fill.shape",
"_____no_output_____"
],
[
"df.query('歩数 >= 10000 and 摂取カロリー <= 1800')",
"_____no_output_____"
]
],
[
[
"## データ型変換",
"_____no_output_____"
]
],
[
[
"df.loc[:, \"日付\"]",
"_____no_output_____"
],
[
"df.loc[:, \"date\"] = df.loc[:, \"日付\"].apply(lambda x: pd.to_datetime(x))",
"_____no_output_____"
]
],
[
[
"カラム 日付 に対して、applyメソッドを使うことで、データ変換し \"date\"カラムに挿入\napply は、データ一つづつに順次関数を適用するものです。\nlambda は Pythonの無名関数です。ここでは、引数を x とし、先程実行した日付型を返すpandasの関数 to_datetime を実行しています。",
"_____no_output_____"
]
],
[
[
"df.loc[:, \"date\"]",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.loc[:, \"摂取カロリー\"] = df.loc[:, \"摂取カロリー\"].astype(np.float32)",
"_____no_output_____"
],
[
"df = df.set_index(\"date\")",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"インデックをdateで置き換えました。",
"_____no_output_____"
],
[
"## 並べ替え",
"_____no_output_____"
]
],
[
[
"df.sort_values(by=\"歩数\")",
"_____no_output_____"
],
[
"df.sort_values(by=\"歩数\", ascending=False).head()",
"_____no_output_____"
]
],
[
[
"## 不要なカラム削除",
"_____no_output_____"
]
],
[
[
"df = df.drop(\"日付\", axis=1)",
"_____no_output_____"
],
[
"df.tail()",
"_____no_output_____"
]
],
[
[
"## 組合せデータの挿入",
"_____no_output_____"
],
[
"歩数 / 摂取カロリー という新たなカラムを追加します。",
"_____no_output_____"
]
],
[
[
"df.loc[:, \"歩数/カロリー\"] = df.loc[:, \"歩数\"] / df.loc[:, \"摂取カロリー\"]",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
],
[
[
"ここで計算した、歩数/カロリーを元に、\n新たに、運動指数カラムを作ります。\n3以下をLow, 3を超え6以下をMid、6を超えるのをHighとします。",
"_____no_output_____"
]
],
[
[
"def exercise_judge(ex):\n if ex <= 3.0:\n return \"Low\"\n elif 3 < ex <= 6.0:\n return \"Mid\"\n else:\n return \"High\"",
"_____no_output_____"
],
[
"df.loc[:, \"運動指数\"] = df.loc[:, \"歩数/カロリー\"].apply(exercise_judge)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
],
[
[
"### 別の章で使うのでデータをPickle形式で保存しておきます",
"_____no_output_____"
]
],
[
[
"df.to_pickle(\"data/df_201704health.pickle\")",
"_____no_output_____"
]
],
[
[
"One-hot Encording\n(データフレームの結合は次の次の章で)",
"_____no_output_____"
]
],
[
[
"df_moved = pd.get_dummies(df.loc[:, \"運動指数\"], prefix=\"運動\")",
"_____no_output_____"
],
[
"df_moved",
"_____no_output_____"
]
],
[
[
"### このデータもPickle形式で保存しておきます",
"_____no_output_____"
]
],
[
[
"df_moved.to_pickle(\"data/df_201704moved.pickle\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d05ffcfa9df119493c89aefd321dc74dd7e53a75 | 10,095 | ipynb | Jupyter Notebook | colab/lab_1_statistical_inference_cmunoz.ipynb | cmunozcortes/ds-fundamentals | 1648339d10238c6a9baa261ee7a367607e6385a2 | [
"MIT"
] | null | null | null | colab/lab_1_statistical_inference_cmunoz.ipynb | cmunozcortes/ds-fundamentals | 1648339d10238c6a9baa261ee7a367607e6385a2 | [
"MIT"
] | null | null | null | colab/lab_1_statistical_inference_cmunoz.ipynb | cmunozcortes/ds-fundamentals | 1648339d10238c6a9baa261ee7a367607e6385a2 | [
"MIT"
] | null | null | null | 23.15367 | 113 | 0.517781 | [
[
[
"**Due Date: Monday, October 19th, 11:59pm**\n\n- Fill out the missing parts.\n- Answer the questions (if any) in a separate document or by adding a new `Text` block inside the Colab.\n- Save the notebook by going to the menu and clicking `File` > `Download .ipynb`.\n- Make sure the saved version is showing your solutions.\n- Send the saved notebook by email to your TA.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nnp.random.seed(0)",
"_____no_output_____"
]
],
[
[
"Simulate a dataset of 100 coin flips for a coin with $p$ = P(head) = 0.6.",
"_____no_output_____"
]
],
[
[
"n = 100 # number of coin flips\np = 0.6 # probability of getting a head (not a fair coin)\n\n# A coin toss experiment can be modeled with a binomial distribution\n# if we set n=1 (one trial), which is equivalent to a Bernoulli distribution\ny = np.random.binomial(n=1, p=p, size=n)\ny",
"_____no_output_____"
]
],
[
[
"## Point Estimation",
"_____no_output_____"
],
[
"Estimate the value of $p$ using the data.",
"_____no_output_____"
]
],
[
[
"def estimator(y):\n return np.mean(y)\n\np_hat = estimator(y)\np_hat",
"_____no_output_____"
]
],
[
[
"## Bootstrap",
"_____no_output_____"
],
[
"Estimate the standard error of $\\hat{p}$ using bootstrap.",
"_____no_output_____"
]
],
[
[
"def bootstrap_se_est(y, stat_function, B=1000):\n # 1. Generate bootstrap samples from the observed/simulated data (i.e. y)\n # 2. Compute the statistic (using stat_function passed) on the bootstrap \n # samples\n # 3. Compute the standard error -> std dev\n t_boot_list = [stat_function(np.random.choice(y, len(y), replace=True)) \n for _ in range(B)]\n return np.std(t_boot_list)\n\nprint(\"Standard error of p_hat computed by bootstrap:\")\nprint(bootstrap_se_est(y, estimator))",
"Standard error of p_hat computed by bootstrap:\n0.04889048066853097\n"
]
],
[
[
"Validate the estimated standard error by computing it analytically.",
"_____no_output_____"
]
],
[
[
"def estimator_analytical_se(p, n):\n return np.sqrt(p * (1-p) / n)\n\nprint(\"Analytical standard error for the estimator: \", estimator_analytical_se(p, n))",
"Analytical standard error for the estimator: 0.04898979485566356\n"
]
],
[
[
"Estimate the 95% confidence interval for $p$.",
"_____no_output_____"
]
],
[
[
"def confidence_interval_95_for_p(y):\n ci_lower = estimator(y) - 1.96*bootstrap_se_est(y, estimator)\n ci_higher = estimator(y) + 1.96*bootstrap_se_est(y, estimator)\n return (ci_lower, ci_higher)\n\nlower, higher = confidence_interval_95_for_p(y)\nprint(\"95% confidence interval for p: ({},{})\".format(lower, higher))",
"95% confidence interval for p: (0.5254445619916019,0.717033857596202)\n"
]
],
[
[
"Validate the 95% confidence interval for $p$.",
"_____no_output_____"
]
],
[
[
"ci_contains_p_flags = []\nfor sim in range(1000):\n y = np.random.binomial(n=1, p=p, size=n)\n ci_lower, ci_higher = confidence_interval_95_for_p(y)\n if ci_lower < p and p < ci_higher:\n ci_contains_p_flags.append(1)\n else: \n ci_contains_p_flags.append(0)\n\ncoverage = np.mean(ci_contains_p_flags)\nprint(\"Coverage of 95% confidence interval for p: \", coverage)",
"Coverage of 95% confidence interval for p: 0.93\n"
]
],
[
[
"## Bayesian Inference",
"_____no_output_____"
],
[
"**[Optional]**\n\nEstimate $p$ using Bayesian inference. As the prior for $p$ use Normal(0.5, 0.1).",
"_____no_output_____"
]
],
[
[
"!pip install pystan",
"_____no_output_____"
],
[
"import pystan",
"_____no_output_____"
],
[
"model_code = \"\"\"\ndata {\n int<lower=0> n;\n int<lower=0,upper=1> y[n];\n}\nparameters {\n real<lower=0,upper=1> p;\n}\nmodel {\n p ~ normal(0.5, 0.1);\n for (i in 1:n)\n y[i] ~ bernoulli(p);\n}\n\"\"\"\n\nmodel = pystan.StanModel(model_code=model_code)\nfit = model.sampling(data={\"n\": n, \"y\": y}, iter=2000, chains=4, n_jobs=4)\nprint(fit.stansummary())",
"_____no_output_____"
]
],
[
[
"Compute the Bayesian inference results if our data contains 20 coin tosses instead.",
"_____no_output_____"
]
],
[
[
"n = 20\np = 0.6\ny = np.random.binomial(1, p, n)\n\nmodel = pystan.StanModel(model_code=model_code)\nfit = model.sampling(data={\"n\": n, \"y\": y}, iter=2000, chains=4, n_jobs=4)\nprint(fit.stansummary())",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d060164f3f2609a1bda996faf32f4b6284576b8e | 35,997 | ipynb | Jupyter Notebook | Imagenette Simple Self Attention.ipynb | RubensZimbres/SimpleSelfAttention | 0c301e2fbf0b49935bdf62f99f640eef1832032b | [
"Apache-2.0"
] | 218 | 2019-05-15T22:41:11.000Z | 2022-02-17T06:08:20.000Z | Imagenette Simple Self Attention.ipynb | RubensZimbres/SimpleSelfAttention | 0c301e2fbf0b49935bdf62f99f640eef1832032b | [
"Apache-2.0"
] | 3 | 2020-07-23T09:20:08.000Z | 2021-12-03T05:09:04.000Z | v0.1/Imagenette Simple Self Attention.ipynb | sdoria/SimpleSelfAttention | b725fd097cdbe0a5c0483536addb32e8cd2e2f9f | [
"Apache-2.0"
] | 25 | 2019-05-26T10:17:28.000Z | 2022-03-04T10:26:39.000Z | 27.775463 | 142 | 0.412562 | [
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F",
"_____no_output_____"
],
[
"import numpy",
"_____no_output_____"
],
[
"from fastai.script import *\nfrom fastai.vision import *\nfrom fastai.callbacks import *\nfrom fastai.distributed import *\nfrom fastprogress import fastprogress\nfrom torchvision.models import *\nfrom fastai.vision.models.xresnet import *\nfrom fastai.vision.models.xresnet2 import *\nfrom fastai.vision.models.presnet import *",
"_____no_output_____"
],
[
"torch.backends.cudnn.benchmark = True",
"_____no_output_____"
]
],
[
[
"# XResNet baseline",
"_____no_output_____"
]
],
[
[
"#https://github.com/fastai/fastai_docs/blob/master/dev_course/dl2/11_train_imagenette.ipynb",
"_____no_output_____"
],
[
"def noop(x): return x\n\nclass Flatten(nn.Module):\n def forward(self, x): return x.view(x.size(0), -1)\n\ndef conv(ni, nf, ks=3, stride=1, bias=False):\n return nn.Conv2d(ni, nf, kernel_size=ks, stride=stride, padding=ks//2, bias=bias)",
"_____no_output_____"
],
[
"act_fn = nn.ReLU(inplace=True)\n\ndef init_cnn(m):\n if getattr(m, 'bias', None) is not None: nn.init.constant_(m.bias, 0)\n if isinstance(m, (nn.Conv2d,nn.Linear)): nn.init.kaiming_normal_(m.weight)\n for l in m.children(): init_cnn(l)\n\ndef conv_layer(ni, nf, ks=3, stride=1, zero_bn=False, act=True):\n bn = nn.BatchNorm2d(nf)\n nn.init.constant_(bn.weight, 0. if zero_bn else 1.)\n layers = [conv(ni, nf, ks, stride=stride), bn]\n if act: layers.append(act_fn)\n return nn.Sequential(*layers)",
"_____no_output_____"
],
[
"class ResBlock(nn.Module):\n def __init__(self, expansion, ni, nh, stride=1):\n super().__init__()\n nf,ni = nh*expansion,ni*expansion\n layers = [conv_layer(ni, nh, 3, stride=stride),\n conv_layer(nh, nf, 3, zero_bn=True, act=False)\n ] if expansion == 1 else [\n conv_layer(ni, nh, 1),\n conv_layer(nh, nh, 3, stride=stride),\n conv_layer(nh, nf, 1, zero_bn=True, act=False)\n ]\n self.convs = nn.Sequential(*layers)\n self.idconv = noop if ni==nf else conv_layer(ni, nf, 1, act=False)\n self.pool = noop if stride==1 else nn.AvgPool2d(2, ceil_mode=True)\n\n def forward(self, x): return act_fn(self.convs(x) + self.idconv(self.pool(x)))",
"_____no_output_____"
],
[
"class XResNet(nn.Sequential):\n @classmethod\n def create(cls, expansion, layers, c_in=3, c_out=1000):\n nfs = [c_in, (c_in+1)*8, 64, 64]\n stem = [conv_layer(nfs[i], nfs[i+1], stride=2 if i==0 else 1)\n for i in range(3)]\n\n nfs = [64//expansion,64,128,256,512]\n res_layers = [cls._make_layer(expansion, nfs[i], nfs[i+1],\n n_blocks=l, stride=1 if i==0 else 2)\n for i,l in enumerate(layers)]\n res = cls(\n *stem,\n nn.MaxPool2d(kernel_size=3, stride=2, padding=1),\n *res_layers,\n nn.AdaptiveAvgPool2d(1), Flatten(),\n nn.Linear(nfs[-1]*expansion, c_out),\n )\n init_cnn(res)\n return res\n\n @staticmethod\n def _make_layer(expansion, ni, nf, n_blocks, stride):\n return nn.Sequential(\n *[ResBlock(expansion, ni if i==0 else nf, nf, stride if i==0 else 1)\n for i in range(n_blocks)])",
"_____no_output_____"
],
[
"def xresnet18 (**kwargs): return XResNet.create(1, [2, 2, 2, 2], **kwargs)\ndef xresnet34 (**kwargs): return XResNet.create(1, [3, 4, 6, 3], **kwargs)\ndef xresnet50 (**kwargs): return XResNet.create(4, [3, 4, 6, 3], **kwargs)\ndef xresnet101(**kwargs): return XResNet.create(4, [3, 4, 23, 3], **kwargs)\ndef xresnet152(**kwargs): return XResNet.create(4, [3, 8, 36, 3], **kwargs)",
"_____no_output_____"
]
],
[
[
"# XResNet with Self Attention",
"_____no_output_____"
]
],
[
[
"#Unmodified from https://github.com/fastai/fastai/blob/5c51f9eabf76853a89a9bc5741804d2ed4407e49/fastai/layers.py\ndef conv1d(ni:int, no:int, ks:int=1, stride:int=1, padding:int=0, bias:bool=False):\n \"Create and initialize a `nn.Conv1d` layer with spectral normalization.\"\n conv = nn.Conv1d(ni, no, ks, stride=stride, padding=padding, bias=bias)\n nn.init.kaiming_normal_(conv.weight)\n if bias: conv.bias.data.zero_()\n return spectral_norm(conv)\n\n\n\n# Adapted from SelfAttention layer at https://github.com/fastai/fastai/blob/5c51f9eabf76853a89a9bc5741804d2ed4407e49/fastai/layers.py\n# Inspired by https://arxiv.org/pdf/1805.08318.pdf\nclass SimpleSelfAttention(nn.Module):\n \n def __init__(self, n_in:int, ks=1):#, n_out:int):\n super().__init__()\n \n \n \n self.conv = conv1d(n_in, n_in, ks, padding=ks//2, bias=False)\n \n \n self.gamma = nn.Parameter(tensor([0.]))\n \n \n\n def forward(self,x):\n \n \n size = x.size()\n x = x.view(*size[:2],-1)\n o = torch.bmm(x.permute(0,2,1).contiguous(),self.conv(x))\n \n \n o = self.gamma * torch.bmm(x,o) + x\n \n \n return o.view(*size).contiguous() \n ",
"_____no_output_____"
],
[
"#unmodified from https://github.com/fastai/fastai/blob/9b9014b8967186dc70c65ca7dcddca1a1232d99d/fastai/vision/models/xresnet.py\n\ndef conv(ni, nf, ks=3, stride=1, bias=False):\n return nn.Conv2d(ni, nf, kernel_size=ks, stride=stride, padding=ks//2, bias=bias)\n\ndef noop(x): return x\n\ndef conv_layer(ni, nf, ks=3, stride=1, zero_bn=False, act=True):\n bn = nn.BatchNorm2d(nf)\n nn.init.constant_(bn.weight, 0. if zero_bn else 1.)\n layers = [conv(ni, nf, ks, stride=stride), bn]\n if act: layers.append(act_fn)\n return nn.Sequential(*layers)",
"_____no_output_____"
],
[
"# Modified from https://github.com/fastai/fastai/blob/9b9014b8967186dc70c65ca7dcddca1a1232d99d/fastai/vision/models/xresnet.py\n# Added self attention\nclass ResBlock(nn.Module):\n def __init__(self, expansion, ni, nh, stride=1,sa=False):\n super().__init__()\n \n \n nf,ni = nh*expansion,ni*expansion\n layers = [conv_layer(ni, nh, 3, stride=stride),\n conv_layer(nh, nf, 3, zero_bn=True, act=False)\n ] if expansion == 1 else [\n conv_layer(ni, nh, 1),\n conv_layer(nh, nh, 3, stride=stride),\n \n conv_layer(nh, nf, 1, zero_bn=True, act=False)\n \n ]\n \n self.sa = SimpleSelfAttention(nf,ks=1) if sa else noop\n \n self.convs = nn.Sequential(*layers)\n self.idconv = noop if ni==nf else conv_layer(ni, nf, 1, act=False)\n self.pool = noop if stride==1 else nn.AvgPool2d(2, ceil_mode=True)\n\n def forward(self, x): \n \n \n return act_fn(self.sa(self.convs(x)) + self.idconv(self.pool(x)))\n ",
"_____no_output_____"
],
[
"# Modified from https://github.com/fastai/fastai/blob/9b9014b8967186dc70c65ca7dcddca1a1232d99d/fastai/vision/models/xresnet.py\n# Added self attention\n\nclass XResNet_sa(nn.Sequential):\n @classmethod\n def create(cls, expansion, layers, c_in=3, c_out=1000):\n nfs = [c_in, (c_in+1)*8, 64, 64]\n stem = [conv_layer(nfs[i], nfs[i+1], stride=2 if i==0 else 1)\n for i in range(3)]\n\n nfs = [64//expansion,64,128,256,512]\n res_layers = [cls._make_layer(expansion, nfs[i], nfs[i+1],\n n_blocks=l, stride=1 if i==0 else 2, sa = True if i in[len(layers)-4] else False)\n for i,l in enumerate(layers)]\n res = cls(\n *stem,\n nn.MaxPool2d(kernel_size=3, stride=2, padding=1),\n *res_layers,\n \n nn.AdaptiveAvgPool2d(1), Flatten(),\n nn.Linear(nfs[-1]*expansion, c_out),\n )\n init_cnn(res)\n return res\n\n @staticmethod\n def _make_layer(expansion, ni, nf, n_blocks, stride, sa = False):\n return nn.Sequential(\n *[ResBlock(expansion, ni if i==0 else nf, nf, stride if i==0 else 1, sa if i in [n_blocks -1] else False)\n for i in range(n_blocks)])",
"_____no_output_____"
],
[
"def xresnet50_sa (**kwargs): return XResNet_sa.create(4, [3, 4, 6, 3], **kwargs)",
"_____no_output_____"
]
],
[
[
"# Data loading",
"_____no_output_____"
]
],
[
[
"#https://github.com/fastai/fastai/blob/master/examples/train_imagenette.py\n\ndef get_data(size, woof, bs, workers=None):\n if size<=128: path = URLs.IMAGEWOOF_160 if woof else URLs.IMAGENETTE_160\n elif size<=224: path = URLs.IMAGEWOOF_320 if woof else URLs.IMAGENETTE_320\n else : path = URLs.IMAGEWOOF if woof else URLs.IMAGENETTE\n path = untar_data(path)\n\n n_gpus = num_distrib() or 1\n if workers is None: workers = min(8, num_cpus()//n_gpus)\n\n return (ImageList.from_folder(path).split_by_folder(valid='val')\n .label_from_folder().transform(([flip_lr(p=0.5)], []), size=size)\n .databunch(bs=bs, num_workers=workers)\n .presize(size, scale=(0.35,1))\n .normalize(imagenet_stats))",
"_____no_output_____"
]
],
[
[
"# Train",
"_____no_output_____"
]
],
[
[
"opt_func = partial(optim.Adam, betas=(0.9,0.99), eps=1e-6)",
"_____no_output_____"
]
],
[
[
"## Imagewoof",
"_____no_output_____"
],
[
"### Image size = 256",
"_____no_output_____"
]
],
[
[
"image_size = 256\ndata = get_data(image_size,woof =True,bs=64)",
"_____no_output_____"
]
],
[
[
"#### Epochs = 5",
"_____no_output_____"
]
],
[
[
"# we use the same parameters for baseline and new model\nepochs = 5\nlr = 3e-3\nbs = 64\nmixup = 0",
"_____no_output_____"
]
],
[
[
"##### Baseline",
"_____no_output_____"
]
],
[
[
"m = xresnet50(c_out=10)",
"_____no_output_____"
],
[
"learn = (Learner(data, m, wd=1e-2, opt_func=opt_func,\n metrics=[accuracy,top_k_accuracy],\n bn_wd=False, true_wd=True,\n loss_func = LabelSmoothingCrossEntropy())\n )",
"_____no_output_____"
],
[
"if mixup: learn = learn.mixup(alpha=mixup)",
"_____no_output_____"
],
[
"learn = learn.to_fp16(dynamic=True)",
"_____no_output_____"
],
[
"learn.fit_one_cycle(epochs, lr, div_factor=10, pct_start=0.3)",
"_____no_output_____"
],
[
"learn.fit_one_cycle(epochs, lr, div_factor=10, pct_start=0.3)",
"_____no_output_____"
],
[
"learn.fit_one_cycle(epochs, lr, div_factor=10, pct_start=0.3)",
"_____no_output_____"
],
[
"learn.fit_one_cycle(epochs, lr, div_factor=10, pct_start=0.3)",
"_____no_output_____"
],
[
"learn.fit_one_cycle(epochs, lr, div_factor=10, pct_start=0.3)",
"_____no_output_____"
],
[
"results = [61.8,64.8,57.4,62.4,63,61.8, 57.6,63,62.6, 64.8] #included some from previous notebook iteration",
"_____no_output_____"
],
[
"np.mean(results), np.std(results), np.min(results), np.max(results)",
"_____no_output_____"
]
],
[
[
"##### New model",
"_____no_output_____"
]
],
[
[
"m = xresnet50_sa(c_out=10)",
"_____no_output_____"
],
[
"learn = None\ngc.collect()",
"_____no_output_____"
],
[
"learn = (Learner(data, m, wd=1e-2, opt_func=opt_func,\n metrics=[accuracy,top_k_accuracy],\n bn_wd=False, true_wd=True,\n loss_func = LabelSmoothingCrossEntropy())\n )",
"_____no_output_____"
],
[
"\nif mixup: learn = learn.mixup(alpha=mixup)",
"_____no_output_____"
],
[
"learn = learn.to_fp16(dynamic=True)",
"_____no_output_____"
],
[
"learn.fit_one_cycle(5, lr, div_factor=10, pct_start=0.3)",
"_____no_output_____"
],
[
"learn.fit_one_cycle(5, lr, div_factor=10, pct_start=0.3)",
"_____no_output_____"
],
[
"learn.fit_one_cycle(5, lr, div_factor=10, pct_start=0.3)",
"_____no_output_____"
],
[
"learn.fit_one_cycle(5, lr, div_factor=10, pct_start=0.3)",
"_____no_output_____"
],
[
"results = [67.4,65.8,70.6,65.8,67.8,69,65.6,66.4, 67.8,70.2]",
"_____no_output_____"
],
[
"np.mean(results), np.std(results), np.min(results), np.max(results)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d06022f113817d7ca2cb6118177b30ebf3a72c67 | 14,484 | ipynb | Jupyter Notebook | BFS_DFS_simple_example/BFS_DFS_simple_example.ipynb | amirhpd/Python_Basics | 8f613330623d7b83199ead19a3a225978f88d839 | [
"MIT"
] | null | null | null | BFS_DFS_simple_example/BFS_DFS_simple_example.ipynb | amirhpd/Python_Basics | 8f613330623d7b83199ead19a3a225978f88d839 | [
"MIT"
] | null | null | null | BFS_DFS_simple_example/BFS_DFS_simple_example.ipynb | amirhpd/Python_Basics | 8f613330623d7b83199ead19a3a225978f88d839 | [
"MIT"
] | null | null | null | 75.832461 | 10,368 | 0.828708 | [
[
[
"Software License Agreement (MIT License) <br />\nCopyright (c) 2020, Amirhossein Pakdaman.",
"_____no_output_____"
],
[
"## Simple DFS, BFS \n**Problem**: Implement a search tree with the following characteristics:\n1. The initial state contains value 10.\n2. At each step two successors are created, the value one of them is one unit smaller than its parent and the other is one unit larger.\n3. Search tree continues up to 3 levels of depth.",
"_____no_output_____"
]
],
[
[
"import IPython\nIPython.core.display.Image(\"tree.png\", embed=True)",
"_____no_output_____"
]
],
[
[
"### BFS",
"_____no_output_____"
]
],
[
[
"import queue\n\nclass Node:\n def __init__(self,value,parent,depth):\n self.value = value\n self.parent = parent\n self.depth = depth\n \nparent = Node(10,None,0)\nfrontier = queue.Queue()\nfrontier.put(parent)\n\nwhile frontier:\n current_node = frontier.get()\n \n if current_node.depth > 3:\n break\n \n frontier.put(Node(current_node.value-1, current_node, current_node.depth+1))\n frontier.put(Node(current_node.value+1, current_node, current_node.depth+1))\n print(current_node.value)",
"10\n9\n11\n8\n10\n10\n12\n7\n9\n9\n11\n9\n11\n11\n13\n"
]
],
[
[
"### DFS",
"_____no_output_____"
]
],
[
[
"class Node:\n def __init__(self,value,parent,depth):\n self.value = value\n self.parent = parent\n self.depth = depth\n \nparent = Node(10,None,0)\nfrontier = []\nfrontier.append(parent)\n\nwhile frontier:\n current_node = frontier.pop()\n \n if current_node.depth > 3:\n current_node = frontier.pop()\n if len(frontier) == 0:\n break\n current_node = frontier.pop()\n \n frontier.append(Node(current_node.value+1, current_node, current_node.depth+1))\n frontier.append(Node(current_node.value-1, current_node, current_node.depth+1))\n print(current_node.value)\n",
"10\n9\n8\n7\n9\n10\n9\n11\n11\n10\n9\n11\n12\n11\n13\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0603251d20bbbd5a88f63abd184430a7286e83d | 28,953 | ipynb | Jupyter Notebook | Python Workshop/Functions.ipynb | CalPolyPat/Python-Workshop | b240a8223a2970fbf807f0c2f5ff109d72a4870a | [
"MIT"
] | null | null | null | Python Workshop/Functions.ipynb | CalPolyPat/Python-Workshop | b240a8223a2970fbf807f0c2f5ff109d72a4870a | [
"MIT"
] | null | null | null | Python Workshop/Functions.ipynb | CalPolyPat/Python-Workshop | b240a8223a2970fbf807f0c2f5ff109d72a4870a | [
"MIT"
] | null | null | null | 73.113636 | 15,020 | 0.778848 | [
[
[
"##Functions\nLet's say that we have some code that does some task, but the code is 25 lines long, we need to run it over 1000 items and it doesn't work in a loop. How in the world will we handle this situation? That is where functions come in really handy. Functions are a generalized block of code that allow you to run code over and over while changing its parameters if you so choose. Functions may take **(arguments)** that you are allowed to change when you call the function. It may also **return** a value.\n\nA function must be defined before you can call it. To define a function, we use the following syntax:\n\n def <function name>(arg0, arg1, arg3,...):\n #code here must be indented.\n #you can use arg0,...,argn within the function\n #you can also return things\n return 1\n #This code returns 1 no matter what you tell the function\n \nFunctions can take as many arguments as you wish, but they may only return 1 thing. A simple example of a familiar function is any mathematical function. Take sin(x), it is a function that takes one argument x and returns one value based on the input. Let's get familiar with functions.",
"_____no_output_____"
]
],
[
[
"def add1(x):\n return x+1\n\nprint(add1(1))\n\ndef xsq(x):\n return x**2\n\nprint(xsq(5))\n\nfor i in range(0,10):\n print(xsq(i))",
"2\n25\n0\n1\n4\n9\n16\n25\n36\n49\n64\n81\n"
]
],
[
[
"The true power of functions is being able to call it as many times as we would like. In the previous example, we called the square function, xsq in a loop 10 times. Let's check out some more complicated examples.",
"_____no_output_____"
]
],
[
[
"def removefs(data):\n newdata=''\n for d in data:\n if(d==\"f\" or d==\"F\"):\n pass\n else:\n newdata+=(d)\n return newdata",
"_____no_output_____"
],
[
"print(removefs('ffffffFFFFFg'))",
"g\n"
],
[
"intro='''##Functions\nLet's say that we have some code that does some task, but the code is 25 lines long, we need to run it over 1000 items and it doesn't work in a loop. How in the world will we handle this situation? That is where functions come in really handy. Functions are a generalized block of code that allow you to run code over and over while changing its parameters if you so choose. Functions may take **(arguments)** that you are allowed to change when you call the function. It may also **return** a value.\n\nA function must be defined before you can call it. To define a function, we use the following syntax:\n\n def <function name>(arg0, arg1, arg3,...):\n #code here must be indented.\n #you can use arg0,...,argn within the function\n #you can also return things\n return 1\n #This code returns 1 no matter what you tell the function\n \nFunctions can take as many arguments as you wish, but they may only return 1 thing. A simple example of a familiar function is any mathematical function. Take sin(x), it is a function that takes one argument x and returns one value based on the input. Let's get familiar with functions.\"'''\nprint(removefs(intro))",
"##unctions\nLet's say that we have some code that does some task, but the code is 25 lines long, we need to run it over 1000 items and it doesn't work in a loop. How in the world will we handle this situation? That is where unctions come in really handy. unctions are a generalized block o code that allow you to run code over and over while changing its parameters i you so choose. unctions may take **(arguments)** that you are allowed to change when you call the unction. It may also **return** a value.\n\nA unction must be deined beore you can call it. To deine a unction, we use the ollowing syntax:\n\n de <unction name>(arg0, arg1, arg3,...):\n #code here must be indented.\n #you can use arg0,...,argn within the unction\n #you can also return things\n return 1\n #This code returns 1 no matter what you tell the unction\n \nunctions can take as many arguments as you wish, but they may only return 1 thing. A simple example o a amiliar unction is any mathematical unction. Take sin(x), it is a unction that takes one argument x and returns one value based on the input. Let's get amiliar with unctions.\"\n"
],
[
"def removevowels(data):\n newdata = ''\n for d in data:\n if(d=='a' or d=='e' or d=='i' or d=='o' or d=='u' or d=='y'):\n pass\n else:\n newdata+=d\n return newdata",
"_____no_output_____"
],
[
"print(removevowels(intro))",
"##Fnctns\nLt's s tht w hv sm cd tht ds sm tsk, bt th cd s 25 lns lng, w nd t rn t vr 1000 tms nd t dsn't wrk n lp. Hw n th wrld wll w hndl ths sttn? Tht s whr fnctns cm n rll hnd. Fnctns r gnrlzd blck f cd tht llw t rn cd vr nd vr whl chngng ts prmtrs f s chs. Fnctns m tk **(rgmnts)** tht r llwd t chng whn cll th fnctn. It m ls **rtrn** vl.\n\nA fnctn mst b dfnd bfr cn cll t. T dfn fnctn, w s th fllwng sntx:\n\n df <fnctn nm>(rg0, rg1, rg3,...):\n #cd hr mst b ndntd.\n # cn s rg0,...,rgn wthn th fnctn\n # cn ls rtrn thngs\n rtrn 1\n #Ths cd rtrns 1 n mttr wht tll th fnctn\n \nFnctns cn tk s mn rgmnts s wsh, bt th m nl rtrn 1 thng. A smpl xmpl f fmlr fnctn s n mthmtcl fnctn. Tk sn(x), t s fnctn tht tks n rgmnt x nd rtrns n vl bsd n th npt. Lt's gt fmlr wth fnctns.\"\n"
]
],
[
[
"So clearly we can do some powerful things. Now let's see why these functions have significant power over loops.",
"_____no_output_____"
]
],
[
[
"def fib(n):\n a,b = 1,1\n for i in range(n-1):\n a,b = b,a+b\n return a\n\ndef printfib(n):\n for i in range(0,n):\n print(fib(i))\n\nprintfib(15)",
"1\n1\n1\n2\n3\n5\n8\n13\n21\n34\n55\n89\n144\n233\n377\n"
]
],
[
[
"Here, using loops within functions allows to generate the fibonacci sequence. We then write a function to print out the first n numbers.",
"_____no_output_____"
],
[
"##Exercises\n1. Write a function that takes two arguments and returns a value that uses the arguments.\n2. Write a power function. It should take two arguments and returns the first argument to the power of the second argument.\n\n3. is a semi-guided exercise. If you are stumped ask for help.\n\n3a. Write a function that takes the cost of a dinner as an argument and returns the cost after a .075% sales tax is added.\n\n3b. Write a function that takes the cost of a dinner and tax and adds a 20% tip to the total, then returns the total.\n\n3c. Write a function that takes a list of food names(choose them yourself) as an argument and returns the cost of purchasing all those items.\n\n3d. Write a function that takes a list of food names as an argument and returns the total cost of having a meal including tax and tip.\n\n4 . In the next cell is a 1000-digit number, write a function to solve Project Euler #8 https://projecteuler.net/problem=8",
"_____no_output_____"
]
],
[
[
"thoudigits = 7316717653133062491922511967442657474235534919493496983520312774506326239578318016984801869478851843858615607891129494954595017379583319528532088055111254069874715852386305071569329096329522744304355766896648950445244523161731856403098711121722383113622298934233803081353362766142828064444866452387493035890729629049156044077239071381051585930796086670172427121883998797908792274921901699720888093776657273330010533678812202354218097512545405947522435258490771167055601360483958644670632441572215539753697817977846174064955149290862569321978468622482839722413756570560574902614079729686524145351004748216637048440319989000889524345065854122758866688116427171479924442928230863465674813919123162824586178664583591245665294765456828489128831426076900422421902267105562632111110937054421750694165896040807198403850962455444362981230987879927244284909188845801561660979191338754992005240636899125607176060588611646710940507754100225698315520005593572972571636269561882670428252483600823257530420752963450\n",
"_____no_output_____"
]
],
[
[
"##Lambda\nNext we will look at a special type of function called a lambda. A lambda is a single line, single expression function. It is perfect for evaluating mathematical expressions like x^2 and e^sin(x^cos(x)). To write a lambda function, we use the following syntax:\n\n func = lambda <args>:<expression>\n \nfor example:\n\n xsq = lambda x:x**2\n xsq(4) #returns 16\n\nLambdas will return the result of the expression. Let's check it out.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n#^^^Some junk we will learn later^^^\n\nfunc = lambda x:np.exp(np.sin(x**np.cos(x)))\n#^^^The important part^^^\n\nplt.plot(np.linspace(0,10,1000), func(np.linspace(0,10,1000)))\n#^^^We will learn this next^^^",
"_____no_output_____"
]
],
[
[
"##Exercises\n\n1. Write a lambda for x^n where x and n are arguments.\n\n2. Write a function that removes all instances of the letters \"p\", \"h\", \"y\", \"s\", \"i\", \"c\", \"s\" from any string. Then prints the new string out.\n\n3. Write a function that does the same thing as **in**, that is, write a function that takes two arguments, a variable and a list and check if the variable is in the list. If it is, return True, otherwise, return False.\n\n4. The factorial function takes a number n and returns the product n\\*(n-1)\\*(n-2)... Write this function.\n\n5. If you want to retrieve the 4th digit of a number, first convert it to a string using the str() command, then take the value at index [3]. Using this information and your factorial function from 4. solve Project Euler #20 https://projecteuler.net/problem=20.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0603d44026c8be1a7321f5630647a6afaf3bdf2 | 6,530 | ipynb | Jupyter Notebook | notebooks/session2_inclass_rdkm.ipynb | Rysias/cds-visual | 3a513aa039909b4169706bcdfb10fbf7b2f0ee2a | [
"MIT"
] | null | null | null | notebooks/session2_inclass_rdkm.ipynb | Rysias/cds-visual | 3a513aa039909b4169706bcdfb10fbf7b2f0ee2a | [
"MIT"
] | null | null | null | notebooks/session2_inclass_rdkm.ipynb | Rysias/cds-visual | 3a513aa039909b4169706bcdfb10fbf7b2f0ee2a | [
"MIT"
] | null | null | null | 20.40625 | 123 | 0.502603 | [
[
[
"# Exploring colour channels",
"_____no_output_____"
],
[
"In this session, we'll be looking at how to explore the different colour channels that compris an image.",
"_____no_output_____"
]
],
[
[
"# We need to include the home directory in our path, so we can read in our own module.\nimport os\n\n# image processing tools\nimport cv2\nimport numpy as np\n\n# utility functions for this course\nimport sys\nsys.path.append(os.path.join(\"..\", \"..\", \"CDS-VIS\"))\nfrom utils.imutils import jimshow\nfrom utils.imutils import jimshow_channel\n\n# plotting tool\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Rotation",
"_____no_output_____"
]
],
[
[
"filename = os.path.join(\"..\", \"..\", \"CDS-VIS\", \"img\", \"terasse.jpeg\")",
"_____no_output_____"
],
[
"image = cv2.imread(filename)",
"_____no_output_____"
],
[
"image.shape",
"_____no_output_____"
],
[
"jimshow(image)",
"_____no_output_____"
]
],
[
[
"## Splitting channels",
"_____no_output_____"
]
],
[
[
"(B, G, R) = cv2.split(image)",
"_____no_output_____"
],
[
"jimshow_channel(R, \"Red\")",
"_____no_output_____"
]
],
[
[
"__Empty numpy array__",
"_____no_output_____"
]
],
[
[
"zeros = np.zeros(image.shape[:2], dtype = \"uint8\")",
"_____no_output_____"
],
[
"jimshow(cv2.merge([zeros, zeros, R]))",
"_____no_output_____"
],
[
"jimshow(cv2.merge([zeros, G, zeros]))",
"_____no_output_____"
],
[
"jimshow(cv2.merge([B, zeros, zeros]))",
"_____no_output_____"
]
],
[
[
"## Histograms",
"_____no_output_____"
]
],
[
[
"jimshow_channel(cv2.cvtColor(image, cv2.COLOR_BGR2GRAY), \"Greyscale\")",
"_____no_output_____"
]
],
[
[
"__A note on ```COLOR_BRG2GRAY```__",
"_____no_output_____"
]
],
[
[
"greyed_image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)",
"_____no_output_____"
]
],
[
[
"```greyed_image.flatten() != image.flatten()```",
"_____no_output_____"
],
[
"## A quick greyscale histogram using matplotlib",
"_____no_output_____"
]
],
[
[
"# Create figure\nplt.figure()\n# Add histogram\nplt.hist(image.flatten(), 256, [0,256])\n# Plot title\nplt.title(\"Greyscale histogram\")\nplt.xlabel(\"Bins\")\nplt.ylabel(\"# of Pixels\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Plotting color histograms",
"_____no_output_____"
],
[
"```cv2.calcHist(images, channels, mask, histSize, ranges[, hist[, accumulate]])```\n\n- images : it is the source image of type uint8 or float32 represented as “[img]”.\n- channels : it is the index of channel for which we calculate histogram. \n - For grayscale image, its value is [0] and\n - color image, you can pass [0], [1] or [2] to calculate histogram of blue, green or red channel respectively.\n- mask : mask image. To find histogram of full image, it is given as “None”.\n- histSize : this represents our BIN count. For full scale, we pass [256].\n- ranges : this is our RANGE. Normally, it is [0,256].",
"_____no_output_____"
]
],
[
[
"# split channels\nchannels = cv2.split(image)\n# names of colours\ncolors = (\"b\", \"g\", \"r\")\n# create plot\nplt.figure()\n# add title\nplt.title(\"Histogram\")\n# Add xlabel\nplt.xlabel(\"Bins\")\n# Add ylabel\nplt.ylabel(\"# of Pixels\")\n\n# for every tuple of channel, colour\nfor (channel, color) in zip(channels, colors):\n # Create a histogram\n hist = cv2.calcHist([channel], [0], None, [256], [0, 256])\n # Plot histogram\n plt.plot(hist, color=color)\n # Set limits of x-axis\n plt.xlim([0, 256])\n# Show plot\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.