hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
cb4b1719b72fe076819b5bdba34047ca29b5b7d1 | 63,279 | ipynb | Jupyter Notebook | kubeflow/notebooks/doppelganger/Doppelganger_A.ipynb | winnerineast/pipeline | ca8094d6f5a3d0bd2c80d18bb5080b8e7bc02182 | [
"Apache-2.0"
] | 1 | 2020-01-01T16:39:48.000Z | 2020-01-01T16:39:48.000Z | kubeflow/notebooks/doppelganger/Doppelganger_A.ipynb | winnerineast/pipeline | ca8094d6f5a3d0bd2c80d18bb5080b8e7bc02182 | [
"Apache-2.0"
] | null | null | null | kubeflow/notebooks/doppelganger/Doppelganger_A.ipynb | winnerineast/pipeline | ca8094d6f5a3d0bd2c80d18bb5080b8e7bc02182 | [
"Apache-2.0"
] | null | null | null | 62.28248 | 32,828 | 0.759035 | [
[
[
"# DOPPELGANGER #",
"_____no_output_____"
],
[
"## Ever wondered how your \"doppelganger\" dog would look like? ",
"_____no_output_____"
],
[
"# EXPERIMENT LOCALLY",
"_____no_output_____"
],
[
"### Prepare Environment\nInstall and import needed modules.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport os\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\nfrom tensorflow.keras.applications.xception import Xception\nfrom tensorflow.keras.applications.xception import preprocess_input\nfrom sklearn.decomposition import PCA\n\nimport matplotlib.pyplot as plt\n%matplotlib inline\n%config InlineBackend.figure_format = 'retina'",
"_____no_output_____"
]
],
[
[
"Set image path and explore enivornment.",
"_____no_output_____"
]
],
[
[
"images_path = 'code/training/Images'\nlen(os.listdir(os.path.join(images_path)))",
"_____no_output_____"
]
],
[
[
"Set parameters.",
"_____no_output_____"
]
],
[
[
"batch_size = 200\nimg_w_size = 299\nimg_h_size = 299",
"_____no_output_____"
]
],
[
[
"Build Data Generator",
"_____no_output_____"
]
],
[
[
"datagen = ImageDataGenerator(preprocessing_function=preprocess_input)\n\nimage_generator = datagen.flow_from_directory(\n images_path,\n target_size=(img_w_size, img_h_size),\n batch_size=batch_size,\n class_mode=None,\n shuffle=False)",
"_____no_output_____"
],
[
"images = image_generator.next()\nimages.shape",
"_____no_output_____"
]
],
[
[
"### Show a sample picture!",
"_____no_output_____"
]
],
[
[
"sample_image_idx = 1\nplt.imshow((images[sample_image_idx] + 1) / 2)",
"_____no_output_____"
]
],
[
[
"## Transform Images to Lower Feature Space (Bottleneck) ##",
"_____no_output_____"
]
],
[
[
"base_model = Xception(include_top=False,\n weights='imagenet',\n input_shape=(img_w_size, img_h_size, 3),\n pooling='avg')",
"_____no_output_____"
],
[
"bottlenecks = base_model.predict(images)\nbottlenecks.shape",
"_____no_output_____"
]
],
[
[
"### Show Bottleneck",
"_____no_output_____"
]
],
[
[
"plt.plot(bottlenecks[0])\nplt.show()",
"_____no_output_____"
],
[
"from sklearn.neighbors import DistanceMetric\ndist = DistanceMetric.get_metric('euclidean')",
"_____no_output_____"
]
],
[
[
"### Calculate pairwise distances",
"_____no_output_____"
]
],
[
[
"bn_dist = dist.pairwise(bottlenecks)\nbn_dist.shape",
"_____no_output_____"
]
],
[
[
"## Pre-Process Image Similarities ##",
"_____no_output_____"
]
],
[
[
"plt.imshow(bn_dist, cmap='gray')",
"_____no_output_____"
]
],
[
[
"Set visualization parameters.",
"_____no_output_____"
]
],
[
[
"n_rows = 5\nn_cols = 5\nn_result_images = n_rows * n_cols",
"_____no_output_____"
]
],
[
[
"# Find Similar Images #",
"_____no_output_____"
],
[
"## Define `image_search()`",
"_____no_output_____"
]
],
[
[
"def image_search(img_index, n_rows=n_rows, n_columns=n_cols):\n n_images = n_rows * n_cols\n\n # create Pandas Series with distances from image\n dist_from_sel = pd.Series(bn_dist[img_index])\n \n # sort Series and get top n_images\n retrieved_indexes = dist_from_sel.sort_values().head(n_images)\n retrieved_images = []\n \n # create figure, loop over closest images indices \n # and display them\n plt.figure(figsize=(10, 10))\n i = 1\n for idx in retrieved_indexes.index:\n plt.subplot(n_rows, n_cols, i)\n plt.imshow((images[idx] + 1) / 2)\n if i == 1:\n plt.title('Selected image')\n else:\n plt.title(\"Dist: {:0.4f}\".format(retrieved_indexes[idx]))\n i += 1\n retrieved_images += [images[idx]]\n \n plt.tight_layout()\n \n return np.array(retrieved_images)",
"_____no_output_____"
]
],
[
[
"## Perform Image Search",
"_____no_output_____"
]
],
[
[
"similar_to_idx = 0\nplt.imshow((images[similar_to_idx] + 1) / 2)\n\nsimilar_images_sorted = image_search(similar_to_idx)\nsimilar_images_sorted.shape",
"_____no_output_____"
]
],
[
[
"## Convert images to gray-scale ##",
"_____no_output_____"
]
],
[
[
"grayscaled_similar_images_sorted = similar_images_sorted.mean(axis=3)\nflattened_grayscale_images = grayscaled_similar_images_sorted.reshape(n_result_images, -1)\nflattened_grayscale_images.shape",
"_____no_output_____"
],
[
"_, h, w = grayscaled_similar_images_sorted.shape\n\n# Compute a PCA \nn_components = 10\n\npca = PCA(n_components=n_components, whiten=True).fit(flattened_grayscale_images)\n\n# apply PCA transformation to training data\npca_transformed = pca.transform(flattened_grayscale_images)",
"_____no_output_____"
]
],
[
[
"## Visualize Eigenfaces",
"_____no_output_____"
]
],
[
[
"def plot_gallery(images, titles, h, w, rows=n_rows, cols=n_cols):\n plt.figure()\n for i in range(rows * cols):\n plt.subplot(rows, cols, i + 1)\n plt.imshow(images[i].reshape(h, w), cmap=plt.cm.gray)\n plt.title(titles[i])\n plt.xticks(())\n plt.yticks(())\n\neigenfaces = pca.components_.reshape((n_components, h, w))\neigenface_titles = [\"eigenface {0}\".format(i) for i in range(eigenfaces.shape[0])]\nplot_gallery(eigenfaces, eigenface_titles, h, w, 3, 3)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Show Average Face",
"_____no_output_____"
]
],
[
[
"average_face = eigenfaces[9]\nplt.imshow((average_face + 1) / 2)",
"_____no_output_____"
]
],
[
[
"# BUILD CONTAINER",
"_____no_output_____"
]
],
[
[
"!cat code/training/doppelganger-train.py",
"# References:\r\n# https://towardsdatascience.com/an-intuitive-guide-to-deep-network-architectures-65fdc477db41\r\n# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.pairwise_distances.html\r\n\r\nimport numpy as np\r\nimport pandas as pd\r\nimport os\r\nimport time\r\nimport random\r\nimport json\r\n\r\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\r\nfrom tensorflow.keras.applications.xception import Xception\r\nfrom tensorflow.keras.applications.xception import preprocess_input\r\n\r\nfrom sklearn.neighbors import DistanceMetric\r\n\r\nimages_path = 'images/'\r\nbatch_size = 200\r\nimg_w_size = 299\r\nimg_h_size = 299\r\n\r\nprint('** LOADING IMAGES **')\r\ndatagen = ImageDataGenerator(preprocessing_function=preprocess_input)\r\nimage_generator = datagen.flow_from_directory(\r\n images_path,\r\n target_size=(img_w_size, img_h_size),\r\n batch_size=batch_size,\r\n class_mode=None,\r\n shuffle=False)\r\n# Note: This needs to stay up here because we use it later to resolve the image name\r\nimages = image_generator.next()\r\nprint('** LOADED IMAGES **')\r\n\r\n# TODO: Convert this to a dict instead of an array\r\n# See TODO's below for supporting tasks\r\npairwise_top_25 = {} \r\n\r\n\r\ndef train(num_result_images=25):\r\n # Convert 2D image matrix => 1D bottleneck vector\r\n print('\\n** GENERATING BOTTLENECKS bottlenecks.csv **')\r\n\r\n # Setup model to convert 2D image matrix => 1D bottleneck vector\r\n base_model = Xception(include_top=False,\r\n weights='imagenet',\r\n input_shape=(img_w_size, img_h_size, 3),\r\n pooling='avg')\r\n\r\n bottlenecks = base_model.predict(images)\r\n\r\n # TODO: Change this to json\r\n np.savetxt(\"bottleneck.csv\", bottlenecks, delimiter=\",\") \r\n print('\\n** GENERATED BOTTLENECKS to bottleneck.csv **')\r\n\r\n bottlenecks = np.loadtxt(\"bottleneck.csv\", delimiter=\",\")\r\n\r\n print('\\n** GENERATING PAIRWISE pairwise_top_25.json **')\r\n dist = DistanceMetric.get_metric('euclidean')\r\n\r\n # Calculate pairwise distance -- O(n^2)\r\n bottleneck_pairwise_dist = dist.pairwise(bottlenecks)\r\n\r\n # Find the top 100 similar images per image\r\n retrieved_images = []\r\n for image_idx in range(0, len(bottleneck_pairwise_dist)):\r\n retrieved_indexes = pd.Series(bottleneck_pairwise_dist[image_idx]).sort_values().head(num_result_images).index.tolist()\r\n retrieved_indexes_int = list(map(lambda index: int(index), retrieved_indexes))\r\n\r\n pairwise_top_25[image_idx] = retrieved_indexes_int\r\n\r\n with open('pairwise_top_25.json', 'w') as fp:\r\n json.dump(pairwise_top_25, fp)\r\n\r\n print('\\n** GENERATED PAIRWISE to pairwise_top_25.json **')\r\n\r\n\r\nif __name__== \"__main__\":\r\n start = time.time()\r\n print(\"Start time: \", start)\r\n try:\r\n train()\r\n finally:\r\n end = time.time()\r\n print(\"End time: \", end)\r\n print(\"Total time taken: \", (end - start))\r\n # test\r\n image_idx = 0\r\n top_25_images = pairwise_top_25[image_idx]\r\n print('\\n** MOST SIMILAR IMAGES **')\r\n print(top_25_images)\r\n"
],
[
"!cat code/training/Dockerfile",
"FROM python:3.6-slim\r\n\r\nCOPY images/ images/\r\n\r\nCOPY requirements.txt requirements.txt\r\nRUN pip install -r requirements.txt\r\n\r\nCOPY doppelganger-train.py doppelganger-train.py\r\nCMD [ \"python\", \"doppelganger-train.py\" ]\r\n\r\n"
],
[
"!cat code/training/doppelganger-train-deploy.yaml",
"apiVersion: v1\r\nkind: Pod\r\nmetadata:\r\n name: doppelganger-train\r\n namespace: deployment\r\n labels:\r\n app: doppelganger-train\r\n type: training\r\n framework: tensorflow\r\nspec:\r\n restartPolicy: OnFailure\r\n containers:\r\n - name: doppelganger-train\r\n image: antjebarth/doppelganger-train:1.0.0 \r\n imagePullPolicy: Always\r\n command:\r\n - \"python\"\r\n - \"doppelganger-train.py\"\r\n# env:\r\n# - name: AWS_REGION\r\n# value: eu-central-1\r\n"
]
],
[
[
"# RUN TRAINING POD\nDeploy the training job to Kubernetes",
"_____no_output_____"
]
],
[
[
"!kubectl create -f code/training/doppelganger-train-deploy.yaml",
"pod/doppelganger-train created\r\n"
],
[
"!kubectl logs doppelganger-train -c doppelganger-train --namespace deployment",
"/usr/local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:526: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\n/usr/local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:527: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\n/usr/local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:528: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\n/usr/local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:529: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\n/usr/local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:530: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\n/usr/local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:535: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\nWARNING:tensorflow:From /usr/local/lib/python3.6/site-packages/tensorflow/python/ops/resource_variable_ops.py:435: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\n** LOADING IMAGES **\nFound 20583 images belonging to 122 classes.\n** LOADED IMAGES **\nStart time: 1571675411.2719145\n\n** GENERATING BOTTLENECKS bottlenecks.csv **\nDownloading data from https://github.com/fchollet/deep-learning-models/releases/download/v0.4/xception_weights_tf_dim_ordering_tf_kernels_notop.h5\n83689472/83683744 [==============================] - 1s 0us/step\n2019-10-21 16:30:17.038400: I tensorflow/core/platform/cpu_feature_guard.cc:141] Your CPU supports instructions that this TensorFlow binary was not compiled to use: AVX2 AVX512F FMA\n2019-10-21 16:30:17.045826: I tensorflow/core/platform/profile_utils/cpu_utils.cc:94] CPU Frequency: 2000175000 Hz\n2019-10-21 16:30:17.047524: I tensorflow/compiler/xla/service/service.cc:150] XLA service 0x55659af62b80 executing computations on platform Host. Devices:\n2019-10-21 16:30:17.047571: I tensorflow/compiler/xla/service/service.cc:158] StreamExecutor device (0): <undefined>, <undefined>\n\n** GENERATED BOTTLENECKS to bottleneck.csv **\n\n** GENERATING PAIRWISE pairwise_top_25.json **\n\n** GENERATED PAIRWISE to pairwise_top_25.json **\nEnd time: 1571675450.5442653\nTotal time taken: 39.272350788116455\n\n** MOST SIMILAR IMAGES **\n[0, 53, 146, 180, 198, 65, 58, 169, 132, 197, 189, 185, 175, 187, 196, 85, 162, 104, 172, 117, 182, 159, 160, 193, 119]\n"
],
[
"!kubectl delete -f code/training/doppelganger-train-deploy.yaml",
"pod \"doppelganger-train\" deleted\r\n"
]
],
[
[
"# RUN INFERENCE POD\nUse the previously trained model and run an inference service on Kubernetes",
"_____no_output_____"
]
],
[
[
"!cat code/inference/DoppelgangerModel.py",
"import keras\r\nimport sys\r\nimport numpy as np\r\nimport json\r\n\r\nclass DoppelgangerModel(object):\r\n def __init__(self):\r\n print(\"\\n** LOADING MODEL from pairwise_top_25.json **\")\r\n with open('pairwise_top_25.json') as fp:\r\n self.model = json.load(fp) \r\n print(\"\\n** LOADED MODEL from pairwise_top_25.json **\")\r\n\r\n def predict(self, X, feature_names):\r\n similar_image_arr = self.model[str(int(X[0]))]\r\n return similar_image_arr\r\n\r\nif __name__== \"__main__\":\r\n model = DoppelgangerModel()\r\n print(model.predict([0], ['image_id']))\r\n"
],
[
"!cat code/inference/Dockerfile-v1",
"FROM python:3.6-slim\r\n\r\nCOPY ./requirements.txt /app/requirements.txt\r\nWORKDIR /app\r\nRUN pip install -r requirements.txt\r\nCOPY DoppelgangerModel.py /app\r\nCOPY pairwise_top_25_v1.json pairwise_top_25.json\r\n\r\nENV MODEL_NAME DoppelgangerModel \r\nENV API_TYPE REST\r\nENV SERVICE_TYPE MODEL\r\nENV PERSISTENCE 0\r\n\r\nEXPOSE 5000 \r\n\r\nCMD exec seldon-core-microservice $MODEL_NAME $API_TYPE --service-type $SERVICE_TYPE --persistence $PERSISTENCE\r\n"
],
[
"!cat code/inference/doppelganger-predict-deploy.yaml",
"apiVersion: machinelearning.seldon.io/v1alpha2\r\nkind: SeldonDeployment\r\nmetadata:\r\n generation: 1\r\n labels:\r\n app: seldon\r\n name: doppelganger-model\r\n namespace: deployment\r\nspec:\r\n annotations:\r\n seldon.io/engine-log-message-type: seldon.message.pair\r\n seldon.io/engine-log-messages-externally: \"false\"\r\n seldon.io/engine-log-requests: \"false\"\r\n seldon.io/engine-log-responses: \"false\"\r\n seldon.io/headless-svc: \"false\"\r\n name: doppelganger-model\r\n predictors:\r\n - componentSpecs:\r\n - spec:\r\n containers:\r\n - env:\r\n - name: LOG_LEVEL\r\n value: INFO\r\n - name: PREDICTIVE_UNIT_SERVICE_PORT\r\n value: \"5000\"\r\n - name: PREDICTIVE_UNIT_ID\r\n value: recommender\r\n - name: PREDICTOR_ID\r\n value: doppelganger-model \r\n - name: SELDON_DEPLOYMENT_ID\r\n value: doppelganger-model\r\n image: antjebarth/doppelganger-predict:1.0.0 \r\n imagePullPolicy: Always \r\n lifecycle:\r\n preStop:\r\n exec:\r\n command:\r\n - /bin/sh\r\n - -c\r\n - /bin/sleep 10\r\n livenessProbe:\r\n failureThreshold: 3\r\n initialDelaySeconds: 60\r\n periodSeconds: 5\r\n successThreshold: 1\r\n tcpSocket:\r\n port: http\r\n timeoutSeconds: 1\r\n name: recommender \r\n ports:\r\n - containerPort: 5000\r\n name: http\r\n protocol: TCP\r\n readinessProbe:\r\n failureThreshold: 3\r\n initialDelaySeconds: 20\r\n periodSeconds: 5\r\n successThreshold: 1\r\n tcpSocket:\r\n port: http\r\n timeoutSeconds: 1\r\n resources:\r\n requests:\r\n memory: 1Mi\r\n terminationMessagePath: /dev/termination-log\r\n terminationMessagePolicy: File\r\n volumeMounts:\r\n - mountPath: /etc/podinfo\r\n name: podinfo\r\n dnsPolicy: ClusterFirst\r\n restartPolicy: Always\r\n schedulerName: default-scheduler\r\n securityContext: {}\r\n terminationGracePeriodSeconds: 20\r\n volumes:\r\n - downwardAPI:\r\n defaultMode: 420\r\n items:\r\n - fieldRef:\r\n apiVersion: v1\r\n fieldPath: metadata.annotations\r\n path: annotations\r\n name: podinfo\r\n graph:\r\n children: []\r\n endpoint:\r\n service_host: localhost\r\n service_port: 5000\r\n type: REST\r\n implementation: UNKNOWN_IMPLEMENTATION\r\n name: recommender\r\n type: MODEL\r\n labels:\r\n fluentd: \"true\"\r\n version: v1\r\n name: doppelganger-model\r\n replicas: 1\r\n svcOrchSpec:\r\n env: []\r\n resources:\r\n requests:\r\n cpu: \"0.1\"\r\n"
]
],
[
[
"### Deploy the service",
"_____no_output_____"
]
],
[
[
"!kubectl create -f code/inference/doppelganger-predict-deploy.yaml",
"seldondeployment.machinelearning.seldon.io/doppelganger-model created\r\n"
]
],
[
[
"### Make a prediction",
"_____no_output_____"
]
],
[
[
"plt.imshow((images[0] + 1) / 2)",
"_____no_output_____"
]
],
[
[
"### Run a curl command to get a prediction from the REST API",
"_____no_output_____"
]
],
[
[
"!curl https://community.cloud.pipeline.ai/seldon/deployment/doppelganger-model/api/v0.1/predictions -d '{\"data\":{\"ndarray\":[[0]]}}' -H \"Content-Type: application/json\"",
"{\r\n \"meta\": {\r\n \"puid\": \"ulq75rkrs2ur6kbsm9g4t8lp2s\",\r\n \"tags\": {\r\n },\r\n \"routing\": {\r\n },\r\n \"requestPath\": {\r\n \"recommender\": \"antjebarth/doppelganger-predict:1.0.0\"\r\n },\r\n \"metrics\": []\r\n },\r\n \"data\": {\r\n \"names\": [],\r\n \"ndarray\": [\"http://c0198e9d-istiosystem-istio-2af2-1928351968.eu-central-1.elb.amazonaws.com/notebook/doppelganger/doppelganger/view/doppelganger/code/training/images/n02085620-Chihuahua/n02085620_10976.jpg\", \"http://c0198e9d-istiosystem-istio-2af2-1928351968.eu-central-1.elb.amazonaws.com/notebook/doppelganger/doppelganger/view/doppelganger/code/training/images/n02087046-toy_terrier/n02087046_3211.jpg\", \"http://c0198e9d-istiosystem-istio-2af2-1928351968.eu-central-1.elb.amazonaws.com/notebook/doppelganger/doppelganger/view/doppelganger/code/training/images/n02089973-English_foxhound/n02089973_2484.jpg\", \"http://c0198e9d-istiosystem-istio-2af2-1928351968.eu-central-1.elb.amazonaws.com/notebook/doppelganger/doppelganger/view/doppelganger/code/training/images/n02109047-Great_Dane/n02109047_9604.jpg\", \"http://c0198e9d-istiosystem-istio-2af2-1928351968.eu-central-1.elb.amazonaws.com/notebook/doppelganger/doppelganger/view/doppelganger/code/training/images/n02110185-Siberian_husky/n02110185_3406.jpg\"]\r\n }\r\n}"
]
],
[
[
"### Clean up",
"_____no_output_____"
]
],
[
[
"!kubectl delete -f code/inference/doppelganger-predict-deploy.yaml",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4b1d14324b82937603ca40e6a26bfa79827d77 | 4,286 | ipynb | Jupyter Notebook | murphy-book/chapter03/q10.ipynb | yusueliu/murphy-book | 71d62cc083a683fb861be1e5acb8eeb948b00c54 | [
"Apache-2.0"
] | 2 | 2019-03-25T22:22:23.000Z | 2019-09-29T20:46:58.000Z | murphy-book/chapter03/q10.ipynb | yusueliu/murphy-book | 71d62cc083a683fb861be1e5acb8eeb948b00c54 | [
"Apache-2.0"
] | null | null | null | murphy-book/chapter03/q10.ipynb | yusueliu/murphy-book | 71d62cc083a683fb861be1e5acb8eeb948b00c54 | [
"Apache-2.0"
] | 1 | 2021-12-24T01:14:12.000Z | 2021-12-24T01:14:12.000Z | 42.435644 | 490 | 0.567662 | [
[
[
"## Exercise 3.10 Taxicab (tramcar) problem\nSuppose you arrive in a new city and see a taxi numbered 100. How many taxis are there in this city? Let us assume taxis are numbered sequentially as integers starting from 0, up to some unknown upper bound $\\theta$. (We number taxis from 0 for simplicity; we can also count from 1 without changing the analysis.) Hence the likelihood function is $p(x) = U(0,\\theta)$, the uniform distribution. The goal is to estimate $\\theta$. We will use the Bayesian analysis from Exercise 3.9.",
"_____no_output_____"
],
[
"a) Suppose we see one taxi numbered 100, so $D = \\{100\\}, m = 100, N = 1$. Using an (improper) non-informative prior on θ of the form $p(\\theta) = Pa(\\theta|0, \\theta) \\propto 1/\\theta$, what is the posterior $p(\\theta|D)$?\n\n**Solution**: Using that of 3.9, the posterior $p(\\theta|D) = Pa(\\theta|1, 100)$.",
"_____no_output_____"
],
[
"b) Compute the posterior mean, mode and median number of taxis in the city, if such quantities exist.\n\n**Solution**:\nThe Pareto distribution $Pa(\\theta|1, 100)$ does not have a mean defined (since $\\mathbb{E}(\\theta|a, b) = \\frac{ab}{a-1}$). The mode of the distribution is at 100. \n\nThe median of the distribution is given by\n\n$$\n\\int_{\\mathrm{median}}^\\infty 100\\theta^{-2} = 0.5\n$$\nwhich gives median = 200.",
"_____no_output_____"
],
[
"(c) Rather than trying to compute a point estimate of the number of taxis, we can compute the predictive density over the next taxicab number using\n\n$$\np(D'|D, \\alpha) = \\int p(D'|\\theta)p(\\theta|D, \\alpha)d\\theta = p(D'|\\beta) \n$$\n\nwhere $\\alpha = (b, K)$ are the hyper-parameters, $\\beta = (c, N + K )$ are the updated hyper-parameters. Now\nconsider the case $D = \\{m\\}$, and $D' = \\{x\\}$. Using Equation 3.95, write down an expression for $p(x|D, \\alpha)$.\nAs above, use a non-informative prior $b = K = 0$.\n\n**Solution**:\n\nLet's compute the predictive density over the next taxi number: First, we need to compute the posterior $p(\\theta|D)$:\n\n$$\np(\\theta|D) = \\mathrm{Pareto}(\\theta|N + K, \\max(m, b)) = \\mathrm{Pareto}(\\theta|1 + 0, \\max(m, 0) = \\mathrm{Pareto}(\\theta|1, m)\n$$\n\nSince the posterior is a Pareto distribution like the prior, we can use it as a 'prior' for inference on $D'$ and use the expression of $p(D)$ (evidence) and the joint distribution $p(D, \\theta)$. So our new 'prior' has the following distribution $p(\\theta|D) = \\mathrm{Pareto}(\\theta, K'=1, b'=m)$. The number o samples is $N'=1$ and $m'=\\max(D') = x$. Now we can calculate the predictive distribution:\n\n\\begin{aligned}\np(x|D, \\alpha) & = \\frac{K'}{(N'+K')b'^{N'}}\\mathbb{I}(x\\le m) + \\frac{K'b'^{K'}}{(N'+K')m'^{N'+K'}}\\mathbb{I}(x > m) \\\\\n& = \\frac{1}{2m}\\mathbb{I}(x\\le m) + \\frac{m}{2x^2}\\mathbb{I}(x > m)\n\\end{aligned}",
"_____no_output_____"
],
[
"(d) Use the predictive density formula to compute the probability that the next taxi you will see (say, the next day) has number 100, 50 or 150, i.e., compute $p(x = 100|D,\\alpha)$, $p(x = 50|D,\\alpha)$, $p(x = 150|D, \\alpha)$.\n\n**Solution**:\n\nIf we suppose $m = 100$, $p(100|D, \\alpha) = 0.005$, $p(50|D, \\alpha) = 0.01$, $p(150 |D, \\alpha) = 0.002$",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb4b204441814c66eed021df16dca12d07a80669 | 2,233 | ipynb | Jupyter Notebook | sklearn/sklearn learning/demonstration/auto_examples_jupyter/manifold/plot_swissroll.ipynb | wangyendt/deeplearning_models | 47883b6c65b8d05a0d1c5737f1552df6476ded34 | [
"MIT"
] | 1 | 2020-06-04T11:10:27.000Z | 2020-06-04T11:10:27.000Z | sklearn/sklearn learning/demonstration/auto_examples_jupyter/manifold/plot_swissroll.ipynb | wangyendt/deeplearning_models | 47883b6c65b8d05a0d1c5737f1552df6476ded34 | [
"MIT"
] | null | null | null | sklearn/sklearn learning/demonstration/auto_examples_jupyter/manifold/plot_swissroll.ipynb | wangyendt/deeplearning_models | 47883b6c65b8d05a0d1c5737f1552df6476ded34 | [
"MIT"
] | null | null | null | 41.351852 | 1,146 | 0.540528 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n# Swiss Roll reduction with LLE\n\n\nAn illustration of Swiss Roll reduction\nwith locally linear embedding\n",
"_____no_output_____"
]
],
[
[
"# Author: Fabian Pedregosa -- <[email protected]>\n# License: BSD 3 clause (C) INRIA 2011\n\nprint(__doc__)\n\nimport matplotlib.pyplot as plt\n\n# This import is needed to modify the way figure behaves\nfrom mpl_toolkits.mplot3d import Axes3D\nAxes3D\n\n#----------------------------------------------------------------------\n# Locally linear embedding of the swiss roll\n\nfrom sklearn import manifold, datasets\nX, color = datasets.make_swiss_roll(n_samples=1500)\n\nprint(\"Computing LLE embedding\")\nX_r, err = manifold.locally_linear_embedding(X, n_neighbors=12,\n n_components=2)\nprint(\"Done. Reconstruction error: %g\" % err)\n\n#----------------------------------------------------------------------\n# Plot result\n\nfig = plt.figure()\n\nax = fig.add_subplot(211, projection='3d')\nax.scatter(X[:, 0], X[:, 1], X[:, 2], c=color, cmap=plt.cm.Spectral)\n\nax.set_title(\"Original data\")\nax = fig.add_subplot(212)\nax.scatter(X_r[:, 0], X_r[:, 1], c=color, cmap=plt.cm.Spectral)\nplt.axis('tight')\nplt.xticks([]), plt.yticks([])\nplt.title('Projected data')\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4b229d33cf2587c26dd2e1b5c009ea0fa66718 | 28,936 | ipynb | Jupyter Notebook | Week11_Padding_stride_pooling.ipynb | yahyanh21/Machine-Learning-Homework | f9c00f93096a748d1c74b106ca1a7a575a5bcf07 | [
"Apache-2.0"
] | null | null | null | Week11_Padding_stride_pooling.ipynb | yahyanh21/Machine-Learning-Homework | f9c00f93096a748d1c74b106ca1a7a575a5bcf07 | [
"Apache-2.0"
] | null | null | null | Week11_Padding_stride_pooling.ipynb | yahyanh21/Machine-Learning-Homework | f9c00f93096a748d1c74b106ca1a7a575a5bcf07 | [
"Apache-2.0"
] | null | null | null | 39.856749 | 261 | 0.498065 | [
[
[
"<a href=\"https://colab.research.google.com/github/yahyanh21/Machine-Learning-Homework/blob/main/Week11_Padding_stride_pooling.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!pip install d2l",
"Collecting d2l\n Downloading d2l-0.17.3-py3-none-any.whl (82 kB)\n\u001b[K |████████████████████████████████| 82 kB 411 kB/s \n\u001b[?25hCollecting requests==2.25.1\n Downloading requests-2.25.1-py2.py3-none-any.whl (61 kB)\n\u001b[K |████████████████████████████████| 61 kB 5.6 MB/s \n\u001b[?25hRequirement already satisfied: jupyter==1.0.0 in /usr/local/lib/python3.7/dist-packages (from d2l) (1.0.0)\nCollecting pandas==1.2.2\n Downloading pandas-1.2.2-cp37-cp37m-manylinux1_x86_64.whl (9.9 MB)\n\u001b[K |████████████████████████████████| 9.9 MB 32.6 MB/s \n\u001b[?25hCollecting matplotlib==3.3.3\n Downloading matplotlib-3.3.3-cp37-cp37m-manylinux1_x86_64.whl (11.6 MB)\n\u001b[K |████████████████████████████████| 11.6 MB 45.5 MB/s \n\u001b[?25hCollecting numpy==1.18.5\n Downloading numpy-1.18.5-cp37-cp37m-manylinux1_x86_64.whl (20.1 MB)\n\u001b[K |████████████████████████████████| 20.1 MB 1.2 MB/s \n\u001b[?25hRequirement already satisfied: jupyter-console in /usr/local/lib/python3.7/dist-packages (from jupyter==1.0.0->d2l) (5.2.0)\nRequirement already satisfied: notebook in /usr/local/lib/python3.7/dist-packages (from jupyter==1.0.0->d2l) (5.3.1)\nRequirement already satisfied: ipywidgets in /usr/local/lib/python3.7/dist-packages (from jupyter==1.0.0->d2l) (7.6.5)\nRequirement already satisfied: qtconsole in /usr/local/lib/python3.7/dist-packages (from jupyter==1.0.0->d2l) (5.2.2)\nRequirement already satisfied: ipykernel in /usr/local/lib/python3.7/dist-packages (from jupyter==1.0.0->d2l) (4.10.1)\nRequirement already satisfied: nbconvert in /usr/local/lib/python3.7/dist-packages (from jupyter==1.0.0->d2l) (5.6.1)\nRequirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib==3.3.3->d2l) (2.8.2)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib==3.3.3->d2l) (1.3.2)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.3 in /usr/local/lib/python3.7/dist-packages (from matplotlib==3.3.3->d2l) (3.0.6)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/dist-packages (from matplotlib==3.3.3->d2l) (0.11.0)\nRequirement already satisfied: pillow>=6.2.0 in /usr/local/lib/python3.7/dist-packages (from matplotlib==3.3.3->d2l) (7.1.2)\nRequirement already satisfied: pytz>=2017.3 in /usr/local/lib/python3.7/dist-packages (from pandas==1.2.2->d2l) (2018.9)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests==2.25.1->d2l) (2.10)\nRequirement already satisfied: urllib3<1.27,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests==2.25.1->d2l) (1.24.3)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests==2.25.1->d2l) (2021.10.8)\nRequirement already satisfied: chardet<5,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests==2.25.1->d2l) (3.0.4)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.7/dist-packages (from python-dateutil>=2.1->matplotlib==3.3.3->d2l) (1.15.0)\nRequirement already satisfied: traitlets>=4.1.0 in /usr/local/lib/python3.7/dist-packages (from ipykernel->jupyter==1.0.0->d2l) (5.1.1)\nRequirement already satisfied: tornado>=4.0 in /usr/local/lib/python3.7/dist-packages (from ipykernel->jupyter==1.0.0->d2l) (5.1.1)\nRequirement already satisfied: jupyter-client in /usr/local/lib/python3.7/dist-packages (from ipykernel->jupyter==1.0.0->d2l) (5.3.5)\nRequirement already satisfied: ipython>=4.0.0 in /usr/local/lib/python3.7/dist-packages (from ipykernel->jupyter==1.0.0->d2l) (5.5.0)\nRequirement already satisfied: decorator in /usr/local/lib/python3.7/dist-packages (from ipython>=4.0.0->ipykernel->jupyter==1.0.0->d2l) (4.4.2)\nRequirement already satisfied: pygments in /usr/local/lib/python3.7/dist-packages (from ipython>=4.0.0->ipykernel->jupyter==1.0.0->d2l) (2.6.1)\nRequirement already satisfied: simplegeneric>0.8 in /usr/local/lib/python3.7/dist-packages (from ipython>=4.0.0->ipykernel->jupyter==1.0.0->d2l) (0.8.1)\nRequirement already satisfied: prompt-toolkit<2.0.0,>=1.0.4 in /usr/local/lib/python3.7/dist-packages (from ipython>=4.0.0->ipykernel->jupyter==1.0.0->d2l) (1.0.18)\nRequirement already satisfied: setuptools>=18.5 in /usr/local/lib/python3.7/dist-packages (from ipython>=4.0.0->ipykernel->jupyter==1.0.0->d2l) (57.4.0)\nRequirement already satisfied: pexpect in /usr/local/lib/python3.7/dist-packages (from ipython>=4.0.0->ipykernel->jupyter==1.0.0->d2l) (4.8.0)\nRequirement already satisfied: pickleshare in /usr/local/lib/python3.7/dist-packages (from ipython>=4.0.0->ipykernel->jupyter==1.0.0->d2l) (0.7.5)\nRequirement already satisfied: wcwidth in /usr/local/lib/python3.7/dist-packages (from prompt-toolkit<2.0.0,>=1.0.4->ipython>=4.0.0->ipykernel->jupyter==1.0.0->d2l) (0.2.5)\nRequirement already satisfied: jupyterlab-widgets>=1.0.0 in /usr/local/lib/python3.7/dist-packages (from ipywidgets->jupyter==1.0.0->d2l) (1.0.2)\nRequirement already satisfied: widgetsnbextension~=3.5.0 in /usr/local/lib/python3.7/dist-packages (from ipywidgets->jupyter==1.0.0->d2l) (3.5.2)\nRequirement already satisfied: nbformat>=4.2.0 in /usr/local/lib/python3.7/dist-packages (from ipywidgets->jupyter==1.0.0->d2l) (5.1.3)\nRequirement already satisfied: ipython-genutils~=0.2.0 in /usr/local/lib/python3.7/dist-packages (from ipywidgets->jupyter==1.0.0->d2l) (0.2.0)\nRequirement already satisfied: jupyter-core in /usr/local/lib/python3.7/dist-packages (from nbformat>=4.2.0->ipywidgets->jupyter==1.0.0->d2l) (4.9.1)\nRequirement already satisfied: jsonschema!=2.5.0,>=2.4 in /usr/local/lib/python3.7/dist-packages (from nbformat>=4.2.0->ipywidgets->jupyter==1.0.0->d2l) (4.3.3)\nRequirement already satisfied: importlib-resources>=1.4.0 in /usr/local/lib/python3.7/dist-packages (from jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets->jupyter==1.0.0->d2l) (5.4.0)\nRequirement already satisfied: typing-extensions in /usr/local/lib/python3.7/dist-packages (from jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets->jupyter==1.0.0->d2l) (3.10.0.2)\nRequirement already satisfied: attrs>=17.4.0 in /usr/local/lib/python3.7/dist-packages (from jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets->jupyter==1.0.0->d2l) (21.4.0)\nRequirement already satisfied: importlib-metadata in /usr/local/lib/python3.7/dist-packages (from jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets->jupyter==1.0.0->d2l) (4.10.0)\nRequirement already satisfied: pyrsistent!=0.17.0,!=0.17.1,!=0.17.2,>=0.14.0 in /usr/local/lib/python3.7/dist-packages (from jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets->jupyter==1.0.0->d2l) (0.18.0)\nRequirement already satisfied: zipp>=3.1.0 in /usr/local/lib/python3.7/dist-packages (from importlib-resources>=1.4.0->jsonschema!=2.5.0,>=2.4->nbformat>=4.2.0->ipywidgets->jupyter==1.0.0->d2l) (3.7.0)\nRequirement already satisfied: Send2Trash in /usr/local/lib/python3.7/dist-packages (from notebook->jupyter==1.0.0->d2l) (1.8.0)\nRequirement already satisfied: terminado>=0.8.1 in /usr/local/lib/python3.7/dist-packages (from notebook->jupyter==1.0.0->d2l) (0.12.1)\nRequirement already satisfied: jinja2 in /usr/local/lib/python3.7/dist-packages (from notebook->jupyter==1.0.0->d2l) (2.11.3)\nRequirement already satisfied: pyzmq>=13 in /usr/local/lib/python3.7/dist-packages (from jupyter-client->ipykernel->jupyter==1.0.0->d2l) (22.3.0)\nRequirement already satisfied: ptyprocess in /usr/local/lib/python3.7/dist-packages (from terminado>=0.8.1->notebook->jupyter==1.0.0->d2l) (0.7.0)\nRequirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.7/dist-packages (from jinja2->notebook->jupyter==1.0.0->d2l) (2.0.1)\nRequirement already satisfied: testpath in /usr/local/lib/python3.7/dist-packages (from nbconvert->jupyter==1.0.0->d2l) (0.5.0)\nRequirement already satisfied: bleach in /usr/local/lib/python3.7/dist-packages (from nbconvert->jupyter==1.0.0->d2l) (4.1.0)\nRequirement already satisfied: mistune<2,>=0.8.1 in /usr/local/lib/python3.7/dist-packages (from nbconvert->jupyter==1.0.0->d2l) (0.8.4)\nRequirement already satisfied: pandocfilters>=1.4.1 in /usr/local/lib/python3.7/dist-packages (from nbconvert->jupyter==1.0.0->d2l) (1.5.0)\nRequirement already satisfied: entrypoints>=0.2.2 in /usr/local/lib/python3.7/dist-packages (from nbconvert->jupyter==1.0.0->d2l) (0.3)\nRequirement already satisfied: defusedxml in /usr/local/lib/python3.7/dist-packages (from nbconvert->jupyter==1.0.0->d2l) (0.7.1)\nRequirement already satisfied: webencodings in /usr/local/lib/python3.7/dist-packages (from bleach->nbconvert->jupyter==1.0.0->d2l) (0.5.1)\nRequirement already satisfied: packaging in /usr/local/lib/python3.7/dist-packages (from bleach->nbconvert->jupyter==1.0.0->d2l) (21.3)\nRequirement already satisfied: qtpy in /usr/local/lib/python3.7/dist-packages (from qtconsole->jupyter==1.0.0->d2l) (2.0.0)\nInstalling collected packages: numpy, requests, pandas, matplotlib, d2l\n Attempting uninstall: numpy\n Found existing installation: numpy 1.19.5\n Uninstalling numpy-1.19.5:\n Successfully uninstalled numpy-1.19.5\n Attempting uninstall: requests\n Found existing installation: requests 2.23.0\n Uninstalling requests-2.23.0:\n Successfully uninstalled requests-2.23.0\n Attempting uninstall: pandas\n Found existing installation: pandas 1.1.5\n Uninstalling pandas-1.1.5:\n Successfully uninstalled pandas-1.1.5\n Attempting uninstall: matplotlib\n Found existing installation: matplotlib 3.2.2\n Uninstalling matplotlib-3.2.2:\n Successfully uninstalled matplotlib-3.2.2\n\u001b[31mERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.\ngoogle-colab 1.0.0 requires pandas~=1.1.0; python_version >= \"3.0\", but you have pandas 1.2.2 which is incompatible.\ngoogle-colab 1.0.0 requires requests~=2.23.0, but you have requests 2.25.1 which is incompatible.\ndatascience 0.10.6 requires folium==0.2.1, but you have folium 0.8.3 which is incompatible.\nalbumentations 0.1.12 requires imgaug<0.2.7,>=0.2.5, but you have imgaug 0.2.9 which is incompatible.\u001b[0m\nSuccessfully installed d2l-0.17.3 matplotlib-3.3.3 numpy-1.18.5 pandas-1.2.2 requests-2.25.1\n"
],
[
"import torch\nfrom torch import nn\n\n\n# We define a convenience function to calculate the convolutional layer. This\n# function initializes the convolutional layer weights and performs\n# corresponding dimensionality elevations and reductions on the input and\n# output\ndef comp_conv2d(conv2d, X):\n # Here (1, 1) indicates that the batch size and the number of channels\n # are both 1\n X = X.reshape((1, 1) + X.shape)\n Y = conv2d(X)\n # Exclude the first two dimensions that do not interest us: examples and\n # channels\n return Y.reshape(Y.shape[2:])\n# Note that here 1 row or column is padded on either side, so a total of 2\n# rows or columns are added\nconv2d = nn.Conv2d(1, 1, kernel_size=3, padding=1)\nX = torch.rand(size=(8, 8))\ncomp_conv2d(conv2d, X).shape",
"_____no_output_____"
],
[
"# Here, we use a convolution kernel with a height of 5 and a width of 3. The\n# padding numbers on either side of the height and width are 2 and 1,\n# respectively\nconv2d = nn.Conv2d(1, 1, kernel_size=(5, 3), padding=(2, 1))\ncomp_conv2d(conv2d, X).shape",
"_____no_output_____"
],
[
"conv2d = nn.Conv2d(1, 1, kernel_size=3, padding=1, stride=2)\ncomp_conv2d(conv2d, X).shape",
"_____no_output_____"
],
[
"#Next, we will look at a slightly more complicated example.\nconv2d = nn.Conv2d(1, 1, kernel_size=(3, 5), padding=(0, 1), stride=(3, 4))\ncomp_conv2d(conv2d, X).shape",
"_____no_output_____"
],
[
"import torch\nfrom d2l import torch as d2l\n\ndef corr2d_multi_in(X, K):\n # First, iterate through the 0th dimension (channel dimension) of `X` and\n # `K`. Then, add them together\n return sum(d2l.corr2d(x, k) for x, k in zip(X, K))",
"_____no_output_____"
],
[
"X = torch.tensor([[[0.0, 1.0, 2.0], [3.0, 4.0, 5.0], [6.0, 7.0, 8.0]],\n [[1.0, 2.0, 3.0], [4.0, 5.0, 6.0], [7.0, 8.0, 9.0]]])\nK = torch.tensor([[[0.0, 1.0], [2.0, 3.0]], [[1.0, 2.0], [3.0, 4.0]]])\n\ncorr2d_multi_in(X, K)",
"_____no_output_____"
],
[
"def corr2d_multi_in_out(X, K):\n # Iterate through the 0th dimension of `K`, and each time, perform\n # cross-correlation operations with input `X`. All of the results are\n # stacked together\n return torch.stack([corr2d_multi_in(X, k) for k in K], 0)",
"_____no_output_____"
],
[
"K = torch.stack((K, K + 1, K + 2), 0)\nK.shape",
"_____no_output_____"
],
[
"corr2d_multi_in_out(X, K)",
"_____no_output_____"
],
[
"def corr2d_multi_in_out_1x1(X, K):\n c_i, h, w = X.shape\n c_o = K.shape[0]\n X = X.reshape((c_i, h * w))\n K = K.reshape((c_o, c_i))\n # Matrix multiplication in the fully-connected layer\n Y = torch.matmul(K, X)\n return Y.reshape((c_o, h, w))",
"_____no_output_____"
],
[
"X = torch.normal(0, 1, (3, 3, 3))\nK = torch.normal(0, 1, (2, 3, 1, 1))\n\nY1 = corr2d_multi_in_out_1x1(X, K)\nY2 = corr2d_multi_in_out(X, K)\nassert float(torch.abs(Y1 - Y2).sum()) < 1e-6",
"_____no_output_____"
],
[
"import torch\nfrom torch import nn\nfrom d2l import torch as d2l\n\ndef pool2d(X, pool_size, mode='max'):\n p_h, p_w = pool_size\n Y = torch.zeros((X.shape[0] - p_h + 1, X.shape[1] - p_w + 1))\n for i in range(Y.shape[0]):\n for j in range(Y.shape[1]):\n if mode == 'max':\n Y[i, j] = X[i: i + p_h, j: j + p_w].max()\n elif mode == 'avg':\n Y[i, j] = X[i: i + p_h, j: j + p_w].mean()\n return Y",
"_____no_output_____"
],
[
"X = torch.tensor([[0.0, 1.0, 2.0], [3.0, 4.0, 5.0], [6.0, 7.0, 8.0]])\npool2d(X, (2, 2))",
"_____no_output_____"
],
[
"pool2d(X, (2, 2), 'avg')",
"_____no_output_____"
],
[
"X = torch.arange(16, dtype=torch.float32).reshape((1, 1, 4, 4))\nX",
"_____no_output_____"
],
[
"pool2d = nn.MaxPool2d(3)\npool2d(X)",
"_____no_output_____"
],
[
"#The stride and padding can be manually specified.\n\npool2d = nn.MaxPool2d(3, padding=1, stride=2)\npool2d(X)",
"_____no_output_____"
],
[
"#Of course, we can specify an arbitrary rectangular pooling window and specify the padding and stride for height and width, respectively.\n\npool2d = nn.MaxPool2d((2, 3), stride=(2, 3), padding=(0, 1))\npool2d(X)",
"_____no_output_____"
],
[
"X = torch.cat((X, X + 1), 1)\nX",
"_____no_output_____"
],
[
"pool2d = nn.MaxPool2d(3, padding=1, stride=2)\npool2d(X)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4b767e82e39d8a80ff242ac98947bb8874bd9b | 11,750 | ipynb | Jupyter Notebook | notebooks/elasticsearch/tmdb/tale-of-two-queries (ES).ipynb | DmitryKey/hello-ltr | a5c07c666e7c5358c26a5ca4be486b0ba8588eaa | [
"Apache-2.0"
] | null | null | null | notebooks/elasticsearch/tmdb/tale-of-two-queries (ES).ipynb | DmitryKey/hello-ltr | a5c07c666e7c5358c26a5ca4be486b0ba8588eaa | [
"Apache-2.0"
] | null | null | null | notebooks/elasticsearch/tmdb/tale-of-two-queries (ES).ipynb | DmitryKey/hello-ltr | a5c07c666e7c5358c26a5ca4be486b0ba8588eaa | [
"Apache-2.0"
] | null | null | null | 31.417112 | 348 | 0.497617 | [
[
[
"# Basics & Prereqs (run once)\n\nIf you don't already have the downloaded dependencies; if you don't have TheMovieDB data indexed run this",
"_____no_output_____"
]
],
[
[
"from ltr.client.elastic_client import ElasticClient\nclient = ElasticClient()\n\nfrom ltr import download, index\nfrom ltr.index import rebuild\nfrom ltr.helpers.movies import indexable_movies\nfrom ltr import download\n\ncorpus='http://es-learn-to-rank.labs.o19s.com/tmdb.json'\ndownload([corpus], dest='data/');\n\nmovies=indexable_movies(movies='data/tmdb.json')\nrebuild(client, index='tmdb', doc_src=movies)",
"_____no_output_____"
]
],
[
[
"## Create Elastic Client",
"_____no_output_____"
]
],
[
[
"from ltr.client.elastic_client import ElasticClient\nclient = ElasticClient()",
"_____no_output_____"
]
],
[
[
"# Our Task: Optimizing \"Drama\" and \"Science Fiction\" queries\n\nIn this example we have two user queries\n\n- Drama\n- Science Fiction\n\nAnd we want to train a model to return the best movies for these movies when a user types them into our search bar.\n\nWe learn through analysis that searchers prefer newer science fiction, but older drama. Like a lot of search relevance problems, two queries need to be optimized in *different* directions",
"_____no_output_____"
],
[
"### Synthetic Judgment List Generation\n\nTo setup this example, we'll generate a judgment list that rewards new science fiction movies as more relevant; and old drama movies as relevant.",
"_____no_output_____"
]
],
[
[
"from ltr.date_genre_judgments import synthesize\njudgments = synthesize(client, judgmentsOutFile='data/genre_by_date_judgments.txt')",
"_____no_output_____"
]
],
[
[
"### Feature selection should be *easy!*\n\nNotice we have 4 proposed features, that seem like they should work! This should be a piece of cake...\n\n1. Release Year of a movie `release_year` - feature ID 1\n2. Is the movie Science Fiction `is_scifi` - feature ID 2\n3. Is the movie Drama `is_drama` - feature ID 3\n4. Does the search term match the genre field `is_genre_match` - feature ID 4\n",
"_____no_output_____"
]
],
[
[
"client.reset_ltr(index='tmdb')\n\nconfig = {\n \"featureset\": {\n \"features\": [\n {\n \"name\": \"release_year\",\n \"params\": [],\n \"template\": {\n \"function_score\": {\n \"field_value_factor\": {\n \"field\": \"release_year\",\n \"missing\": 2000\n },\n \"query\": { \"match_all\": {} }\n }\n }\n },\n {\n \"name\": \"is_sci_fi\",\n \"params\": [],\n \"template\": {\n \"constant_score\": {\n \"filter\": {\n \"match_phrase\": {\"genres\": \"Science Fiction\"}\n },\n \"boost\": 1.0 }\n }\n },\n {\n \"name\": \"is_drama\",\n \"params\": [],\n \"template\": {\n \"constant_score\": {\n \"filter\": {\n \"match_phrase\": {\"genres\": \"Drama\"}\n },\n \"boost\": 1.0 }\n }\n },\n {\n \"name\": \"is_genre_match\",\n \"params\": [\"keywords\"],\n \"template\": {\n \"constant_score\": {\n \"filter\": {\n \"match_phrase\": {\"genres\": \"{{keywords}}\"}\n },\n \"boost\": 1.0\n }\n }\n }\n ]\n },\n \"validation\": {\n \"params\": {\n \"keywords\": \"Science Fiction\"\n },\n \"index\": \"tmdb\"\n }\n}\n\nclient.create_featureset(index='tmdb', name='genre', ftr_config=config)",
"_____no_output_____"
]
],
[
[
"### Log from search engine -> to training set\n\nEach feature is a query to be scored against the judgment list",
"_____no_output_____"
]
],
[
[
"from ltr.judgments import judgments_open\nfrom ltr.log import FeatureLogger\nfrom itertools import groupby\n\nfrom ltr.log import FeatureLogger\nfrom ltr.judgments import judgments_open\nfrom itertools import groupby\n\nftr_logger=FeatureLogger(client, index='tmdb', feature_set='genre')\nwith judgments_open('data/genre_by_date_judgments.txt') as judgment_list:\n for qid, query_judgments in groupby(judgment_list, key=lambda j: j.qid):\n ftr_logger.log_for_qid(judgments=query_judgments, \n qid=qid,\n keywords=judgment_list.keywords(qid))\n",
"_____no_output_____"
]
],
[
[
"### Training - Guaraneed Perfect Search Results!\n\nWe'll train a LambdaMART model against this training data.",
"_____no_output_____"
]
],
[
[
"from ltr.ranklib import train\ntrainLog = train(client,\n training_set=ftr_logger.logged,\n metric2t='NDCG@10',\n index='tmdb',\n featureSet='genre',\n modelName='genre')\n\nprint()\nprint(\"Impact of each feature on the model\")\nfor ftrId, impact in trainLog.impacts.items():\n print(\"{} - {}\".format(ftrId, impact))\n \nprint(\"Perfect NDCG! {}\".format(trainLog.rounds[-1]))",
"_____no_output_____"
]
],
[
[
"### But this search sucks!\nTry searches for \"Science Fiction\" and \"Drama\"",
"_____no_output_____"
]
],
[
[
"from ltr.search import search\nsearch(client, keywords=\"science fiction\", modelName=\"genre\")",
"_____no_output_____"
]
],
[
[
"### Why didn't it work!?!? Training data\n\n1. Examine the training data, do we cover every example of a BAD result\n2. Examine the feature impacts, do any of the features the model uses even USE the keywords?\n\n### Ranklib only sees the data you give it, we don't have good enough coverage\n\nYou need to have feature coverage, especially over negative examples. Most documents in the index are negative! \n\nOne trick commonly used is to treat other queries positive results as this queries negative results. Indeed what we're missing here are negative examples for \"Science Fiction\" that are not science fiction movies. A glaring omission, we'll handle now... With the `autoNegate` flag, we'll add additional negative examples to the judgment list",
"_____no_output_____"
]
],
[
[
"from ltr import date_genre_judgments\ndate_genre_judgments.synthesize(client,\n judgmentsOutFile='data/genre_by_date_judgments.txt',\n autoNegate=True)\n\nfrom ltr.log import FeatureLogger\nfrom ltr.judgments import judgments_open\nfrom itertools import groupby\n\nftr_logger=FeatureLogger(client, index='tmdb', feature_set='genre')\nwith judgments_open('data/genre_by_date_judgments.txt') as judgment_list:\n for qid, query_judgments in groupby(judgment_list, key=lambda j: j.qid):\n ftr_logger.log_for_qid(judgments=query_judgments, \n qid=qid,\n keywords=judgment_list.keywords(qid))\n \n \nfrom ltr.ranklib import train\ntrainLog = train(client,\n training_set=ftr_logger.logged,\n metric2t='NDCG@10',\n index='tmdb',\n featureSet='genre',\n modelName='genre')\n\nprint()\nprint(\"Impact of each feature on the model\")\nfor ftrId, impact in trainLog.impacts.items():\n print(\"{} - {}\".format(ftrId, impact))\n \nprint(\"NDCG {}\".format(trainLog.rounds[-1]))",
"_____no_output_____"
]
],
[
[
"### Now try those queries...\n\nReplace keywords below with 'science fiction' or 'drama' and see how it works",
"_____no_output_____"
]
],
[
[
"from ltr.search import search\nsearch(client, keywords=\"drama\", modelName=\"genre\")",
"_____no_output_____"
],
[
"from ltr.search import search\nsearch(client, keywords=\"science fiction\", modelName=\"genre\")",
"_____no_output_____"
]
],
[
[
"### The next problem\n\n- Overfit to these two examples\n- We need many more queries, covering more use cases",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cb4b7f6015e3a05653be61f963060480abf1f110 | 988 | ipynb | Jupyter Notebook | shape pattern/sqaure pattern with stars.ipynb | mujahid2580/python-pattern-program | 8fc230f871b747abd70f819b2e6cfff72d93c2a7 | [
"MIT"
] | 1 | 2020-02-19T19:32:31.000Z | 2020-02-19T19:32:31.000Z | shape pattern/sqaure pattern with stars.ipynb | mujahid2580/pyhton-pattern-programming | 8fc230f871b747abd70f819b2e6cfff72d93c2a7 | [
"MIT"
] | null | null | null | shape pattern/sqaure pattern with stars.ipynb | mujahid2580/pyhton-pattern-programming | 8fc230f871b747abd70f819b2e6cfff72d93c2a7 | [
"MIT"
] | 2 | 2020-09-30T15:26:27.000Z | 2021-03-05T07:09:42.000Z | 17.333333 | 39 | 0.423077 | [
[
[
"for i in range(5):\n for j in range(5):\n print(\"*\",end=\" \")\n print()",
"* * * * * \n* * * * * \n* * * * * \n* * * * * \n* * * * * \n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
cb4b7f67a2e591a27f9e93cfff1a61d97bd8a0f5 | 315,699 | ipynb | Jupyter Notebook | KNNRecommendation.ipynb | shubhamchouksey/Movie-Recommendation | b682792410bfb34e80c0d642e38d9cac0e75af15 | [
"MIT"
] | 1 | 2020-04-05T09:10:53.000Z | 2020-04-05T09:10:53.000Z | KNNRecommendation.ipynb | shubhamchouksey/Movie-Recommendation | b682792410bfb34e80c0d642e38d9cac0e75af15 | [
"MIT"
] | null | null | null | KNNRecommendation.ipynb | shubhamchouksey/Movie-Recommendation | b682792410bfb34e80c0d642e38d9cac0e75af15 | [
"MIT"
] | 2 | 2020-05-07T06:30:50.000Z | 2021-11-23T15:37:54.000Z | 218.174845 | 162,724 | 0.882245 | [
[
[
"## Nearest Neighbor item based Collaborative Filtering\n\n\n\nSource: https://towardsdatascience.com",
"_____no_output_____"
]
],
[
[
"##Dataset url: https://grouplens.org/datasets/movielens/latest/\n\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"r_cols = ['user_id','movie_id','rating']\nmovies_df = pd.read_csv('u.item.csv', names=['movieId','title'],sep='|',usecols=range(2))\nm_cols = ['movie_id','title']\nrating_df=pd.read_csv('u.data.csv', names=['userId', 'movieId', 'rating'],usecols=range(3))",
"_____no_output_____"
],
[
"movies_df.head()",
"_____no_output_____"
],
[
"rating_df.head()",
"_____no_output_____"
],
[
"df = pd.merge(rating_df,movies_df,on='movieId')\ndf.head()",
"_____no_output_____"
],
[
"combine_movie_rating = df.dropna(axis = 0, subset = ['title'])\n# combine_movie_rating.shape\nmovie_ratingCount = (combine_movie_rating.\n groupby(by = ['title'])['rating'].\n count().\n reset_index().\n rename(columns = {'rating': 'totalRatingCount'})\n [['title', 'totalRatingCount']]\n )\nmovie_ratingCount.head()\n",
"_____no_output_____"
],
[
"rating_with_totalRatingCount = combine_movie_rating.merge(movie_ratingCount, left_on = 'title', right_on = 'title', how = 'left')\nrating_with_totalRatingCount.head()",
"_____no_output_____"
],
[
"pd.set_option('display.float_format', lambda x: '%.3f' % x)\nprint(movie_ratingCount['totalRatingCount'].describe())",
"count 1664.000\nmean 60.098\nstd 80.963\nmin 1.000\n25% 7.000\n50% 27.000\n75% 80.250\nmax 584.000\nName: totalRatingCount, dtype: float64\n"
],
[
"popularity_threshold = 50\nrating_popular_movie= rating_with_totalRatingCount.query('totalRatingCount >= @popularity_threshold')\nrating_popular_movie.head()",
"_____no_output_____"
],
[
"rating_popular_movie.shape",
"_____no_output_____"
],
[
"## First lets create a Pivot matrix\n\nmovie_features_df=rating_popular_movie.pivot_table(index='title',columns='userId',values='rating').fillna(0)\nmovie_features_df.head()",
"_____no_output_____"
],
[
"from scipy.sparse import csr_matrix\n\nmovie_features_df_matrix = csr_matrix(movie_features_df.values)\n\n# print(movie_features_df_matrix)\nfrom sklearn.neighbors import NearestNeighbors\n\n\nmodel_knn = NearestNeighbors(metric = 'cosine', algorithm = 'brute')\nmodel_knn.fit(movie_features_df_matrix)",
"_____no_output_____"
],
[
"movie_features_df.shape",
"_____no_output_____"
],
[
"# query_index = np.random.choice(movie_features_df.shape[0])\n# print(query_index)\nquery_index = movie_features_df.index.get_loc('Star Wars (1977)')\ndistances, indices = model_knn.kneighbors(movie_features_df.iloc[query_index,:].values.reshape(1, -1), n_neighbors = 6)\n",
"_____no_output_____"
],
[
"movie_features_df.head()",
"_____no_output_____"
],
[
"distances",
"_____no_output_____"
],
[
"indices",
"_____no_output_____"
],
[
"for i in range(0, len(distances.flatten())):\n if i == 0:\n print('Recommendations for {0}:\\n'.format(movie_features_df.index[query_index]))\n else:\n print('{0}: {1}, with distance of {2}:'.format(i, movie_features_df.index[indices.flatten()[i]], distances.flatten()[i]))",
"Recommendations for Star Wars (1977):\n\n1: Return of the Jedi (1983), with distance of 0.11648183086402542:\n2: Raiders of the Lost Ark (1981), with distance of 0.2359429772070084:\n3: Empire Strikes Back, The (1980), with distance of 0.24955008270687218:\n4: Toy Story (1995), with distance of 0.26622322826178724:\n5: Godfather, The (1972), with distance of 0.3034231233589749:\n"
]
],
[
[
"## Cosine Similarity\n\n\n\n",
"_____no_output_____"
]
],
[
[
"my_ratings = movie_features_df[0]\nmy_ratings = my_ratings.loc[my_ratings!=0]\nmy_ratings",
"_____no_output_____"
],
[
"simCandidates = pd.Series()\nfor i in range(0,len(my_ratings.index)):\n print(\"Adding sims for \",my_ratings.index[i],\"...\")\n query_index = movie_features_df.index.get_loc(my_ratings.index[i])\n# print(query_index)\n \n distances, indices = model_knn.kneighbors(movie_features_df.iloc[query_index,:].values.reshape(1, -1), n_neighbors = 6)\n distances = (1/(1+distances)) * my_ratings[i] \n# print(distances)\n \n sims = pd.Series(distances.flatten(),\n name=\"ratings\", index=movie_features_df.index[indices.flatten()])\n# sims = distances.map(lambda x: (1/x)*myRatings[i])\n print(sims)\n simCandidates = simCandidates.append(sims)\n\nprint('\\nsorting..\\n')\nsimCandidates.sort_values(inplace=True,ascending=False)\nprint(simCandidates.head(20))",
"Adding sims for Empire Strikes Back, The (1980) ...\ntitle\nEmpire Strikes Back, The (1980) 5.000\nRaiders of the Lost Ark (1981) 4.247\nIndiana Jones and the Last Crusade (1989) 4.090\nBack to the Future (1985) 4.011\nStar Wars (1977) 4.001\nTerminator, The (1984) 3.976\nName: ratings, dtype: float64\nAdding sims for Gone with the Wind (1939) ...\ntitle\nGone with the Wind (1939) 1.000\nWizard of Oz, The (1939) 0.746\nSound of Music, The (1965) 0.704\nCasablanca (1942) 0.704\nIt's a Wonderful Life (1946) 0.702\nBack to the Future (1985) 0.693\nName: ratings, dtype: float64\nAdding sims for Star Wars (1977) ...\ntitle\nStar Wars (1977) 5.000\nReturn of the Jedi (1983) 4.478\nRaiders of the Lost Ark (1981) 4.045\nEmpire Strikes Back, The (1980) 4.001\nToy Story (1995) 3.949\nGodfather, The (1972) 3.836\nName: ratings, dtype: float64\n\nsorting..\n\nEmpire Strikes Back, The (1980) 5.000\nStar Wars (1977) 5.000\nReturn of the Jedi (1983) 4.478\nRaiders of the Lost Ark (1981) 4.247\nIndiana Jones and the Last Crusade (1989) 4.090\nRaiders of the Lost Ark (1981) 4.045\nBack to the Future (1985) 4.011\nEmpire Strikes Back, The (1980) 4.001\nStar Wars (1977) 4.001\nTerminator, The (1984) 3.976\nToy Story (1995) 3.949\nGodfather, The (1972) 3.836\nGone with the Wind (1939) 1.000\nWizard of Oz, The (1939) 0.746\nSound of Music, The (1965) 0.704\nCasablanca (1942) 0.704\nIt's a Wonderful Life (1946) 0.702\nBack to the Future (1985) 0.693\ndtype: float64\n"
],
[
"simCandidates = simCandidates.groupby(simCandidates.index).sum()\nsimCandidates.sort_values(inplace=True,ascending=False)\nsimCandidates.head(10)",
"_____no_output_____"
],
[
"filteredSims = simCandidates.drop(my_ratings.index)\nfilteredSims.head(10)",
"_____no_output_____"
]
],
[
[
"This is the final Recommendation of movies of similar that i was like earlier such as `Empire Strikes Back, The (1980)`, `Gone with the Wind (1939)`, `Star Wars (1977)` ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb4b90606b6c150bc92a4033045aba4ff4980098 | 1,423 | ipynb | Jupyter Notebook | assignments/9_Estimating Errors/Mat 395-495 - Topic 9 - Estimating Errors - Assignment.ipynb | jagar2/Summer_2020_MAT-395-495_Scientific-Data-Analysis-and-Computing | e4b831460bddd34e7ad1d8888327c8d85b80e35e | [
"BSD-3-Clause"
] | 1 | 2021-11-10T15:34:37.000Z | 2021-11-10T15:34:37.000Z | assignments/9_Estimating Errors/Mat 395-495 - Topic 9 - Estimating Errors - Assignment.ipynb | jagar2/Summer_2020_MAT-395-495_Scientific-Data-Analysis-and-Computing | e4b831460bddd34e7ad1d8888327c8d85b80e35e | [
"BSD-3-Clause"
] | null | null | null | assignments/9_Estimating Errors/Mat 395-495 - Topic 9 - Estimating Errors - Assignment.ipynb | jagar2/Summer_2020_MAT-395-495_Scientific-Data-Analysis-and-Computing | e4b831460bddd34e7ad1d8888327c8d85b80e35e | [
"BSD-3-Clause"
] | 3 | 2020-08-06T15:11:50.000Z | 2022-01-05T20:21:09.000Z | 1,423 | 1,423 | 0.718201 | [
[
[
"**Assignment**: Topic 9, Estimating Errors\n\n**Due**: 21 hours after slide deck completed during class time\n\n**Submission**: Submit as Juptyer notebook (ipynb) which runs in Colab (https://colab.research.google.com/) via your shared google drive folder.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport scipy.stats as stats",
"_____no_output_____"
]
],
[
[
"# Problem 1",
"_____no_output_____"
],
[
"1) Calculate the chi-squared value (manually and, separately, using a python function) for the following observed data from an experiment. The expected value is 15 for each measurement.\n\n16, 18, 20, 14, 11, 12 ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb4b971eef4dad260314819f2cdb34e4b0f7d4bc | 24,551 | ipynb | Jupyter Notebook | 06-recursive-data.ipynb | heyrutvik/scala-notes | 82c1399980f49891e96fb0bfcc743b5372f03496 | [
"CC0-1.0"
] | 2 | 2021-04-29T08:59:12.000Z | 2021-06-22T02:40:42.000Z | 06-recursive-data.ipynb | heyrutvik/scala-notes | 82c1399980f49891e96fb0bfcc743b5372f03496 | [
"CC0-1.0"
] | null | null | null | 06-recursive-data.ipynb | heyrutvik/scala-notes | 82c1399980f49891e96fb0bfcc743b5372f03496 | [
"CC0-1.0"
] | 1 | 2021-04-27T20:06:42.000Z | 2021-04-27T20:06:42.000Z | 38.846519 | 487 | 0.576962 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
cb4b9e2d5120411e9092084d5f7e707e58365dbb | 35,528 | ipynb | Jupyter Notebook | notebook/2018-08-14_get_zscores_for_sharvani.ipynb | jfear/larval_gonad_ovary | b0941dbdd450aae5efd6ff60632e6eec7574ab69 | [
"MIT"
] | null | null | null | notebook/2018-08-14_get_zscores_for_sharvani.ipynb | jfear/larval_gonad_ovary | b0941dbdd450aae5efd6ff60632e6eec7574ab69 | [
"MIT"
] | null | null | null | notebook/2018-08-14_get_zscores_for_sharvani.ipynb | jfear/larval_gonad_ovary | b0941dbdd450aae5efd6ff60632e6eec7574ab69 | [
"MIT"
] | null | null | null | 31.920934 | 217 | 0.280652 | [
[
[
"# Zscores for Sharvani",
"_____no_output_____"
],
[
"Sharvani was looking at the initial run that I did, but she could not see some common genes in the biomarkers list. I want to put together the zscores show she can easily look there and see how the gene behaves.",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nfrom pathlib import Path\n\nfrom IPython.display import display, HTML, Markdown\nimport numpy as np\nimport pandas as pd\n\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\n# Project level imports\nfrom larval_gonad_ovary.notebook import Nb\nfrom larval_gonad_ovary.plotting import make_figs\nfrom larval_gonad_ovary.config import memory",
"_____no_output_____"
],
[
"# Setup notebook\nnbconfig = Nb.setup_notebook()",
"last updated: 2018-08-14 \nGit hash: eb7e3486aa1ed6cc3c23658afd54dacdb200f517\n"
],
[
"genes = pd.Series(nbconfig.fbgn2symbol, name='gene_symbol')",
"_____no_output_____"
],
[
"zscores = pd.read_parquet('../output/scrnaseq-wf/zscore_tpm.res.0.4.parquet')\nzscores.index.name = 'FBgn'\ndat = zscores.join(genes).set_index('gene_symbol', append=True)\ndat.sort_index(level='gene_symbol', inplace=True, )\ndat.to_csv('../output/2018-08-14_zscores.tsv', sep='\\t')",
"_____no_output_____"
],
[
"raw = pd.read_parquet('../output/scrnaseq-wf/raw.res.0.4.parquet')\ndat = raw.join(genes).set_index('gene_symbol', append=True)\ndat.sort_index(level='gene_symbol', inplace=True, )\ndat.to_csv('../output/2018-08-14_raw.tsv', sep='\\t')",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb4ba17e15068ebca12341d87126d10a36f5364b | 77,402 | ipynb | Jupyter Notebook | asc_2_preprocess.ipynb | bencxs/allstate-claims | 609263e149c39c8a37a3fd939f595cb6fa454226 | [
"Apache-2.0"
] | null | null | null | asc_2_preprocess.ipynb | bencxs/allstate-claims | 609263e149c39c8a37a3fd939f595cb6fa454226 | [
"Apache-2.0"
] | null | null | null | asc_2_preprocess.ipynb | bencxs/allstate-claims | 609263e149c39c8a37a3fd939f595cb6fa454226 | [
"Apache-2.0"
] | null | null | null | 37.410343 | 599 | 0.372419 | [
[
[
"# Data Preprocessing",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom IPython.display import display # Allows the use of display() for DataFrames\n# Show matplotlib plots inline (nicely formatted in the notebook)\n%matplotlib inline",
"_____no_output_____"
],
[
"# Load train and test datasets\ndf_train = pd.read_csv('train.csv')\ndf_test = pd.read_csv('test.csv')\n\nprint (\"Training dataset has {} rows and {} columns.\").format(*df_train.shape)\nprint (\"Test dataset has {} rows and {} columns.\").format(*df_test.shape)",
"Training dataset has 188318 rows and 132 columns.\nTest dataset has 125546 rows and 131 columns.\n"
],
[
"# Inspect the datasets\ndisplay(df_train.head())\ndisplay(df_train.describe())\ndisplay(df_test.head())\ndisplay(df_test.describe())",
"_____no_output_____"
]
],
[
[
"Since some of the categorical variables can have up to 120+ unique values, creating dummy variables would considerably increase our feature space, requiring lots of memory. To save space, we label encode them instead.",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import LabelEncoder\n\ndef label_encode(df):\n '''Label encode categorical features and normalize continuous features'''\n le = LabelEncoder()\n \n for col, col_data in df.iteritems():\n if col_data.dtype == 'object':\n le.fit(col_data)\n df[col] = le.transform(col_data) \n return df\n\ndf_train = label_encode(df_train)\ndf_test = label_encode(df_test)\ndisplay(df_train.head())\ndisplay(df_test.head())",
"_____no_output_____"
],
[
"# Transform\nfrom scipy.stats import boxcox\nfrom scipy.stats import skew\n\n# Get the list of columns to be normalized, ignoring the target variable\ndef col_to_norm(df, target):\n col_to_norm = []\n for col, col_data in df.iteritems():\n if col == target:\n continue\n elif col_data.dtype == 'float64':\n col_to_norm.append(col)\n return col_to_norm\n\n# First, we need to convert all negative values to positive\ndef force_positive(df, col_to_norm):\n for col in col_to_norm:\n if np.min(df[col]) < 1:\n df[col] = df[col] + np.abs(np.min(df[col])) + 2\n return df\n\n# Then, we perform the Box-Cox transformation\ndef transform_box_cox(df, col_to_norm):\n '''Perform Box-Cox transformation for easier model learning'''\n lambda_vals = []\n for col in col_to_norm:\n tformed = boxcox(df[col].values)\n df[col] = tformed[0] #By default, Box-Cox outputs array and lambda. We only want the array output.\n print \"Box-Cox lambda: \" + col + \" \" + str(tformed[1])\n lambda_vals.append(tformed[1])\n return df, lambda_vals\n\ndef normalize_data(df, col_to_norm):\n '''Normalize data with the standard score for normal distributions'''\n mean_std = []\n for col in col_to_norm:\n mean = np.mean(df[col])\n std = np.std(df[col])\n df[col] = df[col].apply(lambda x: (x - mean) / std)\n print col + \": \" + \"mean =\" + str(mean) + \", std =\" + str(std)\n mean_std.append((mean, std))\n return df, mean_std\n\ncol_to_norm = col_to_norm(df_train, 'loss')\ndf_train = force_positive(df_train, col_to_norm)\ndf_train, lambda_vals = transform_box_cox(df_train, col_to_norm)\ndf_train, mean_std = normalize_data(df_train, col_to_norm)\n\ndisplay(df_train.describe())\n\n# Calculate skewness of transformed data\nfor col in col_to_norm:\n skness = skew(df_train[col])\n print col + \" skewnesss: {0:.2f}\".format(skness)",
"Box-Cox lambda: cont1 0.978804078009\nBox-Cox lambda: cont2 0.985394106026\nBox-Cox lambda: cont3 0.927989179108\nBox-Cox lambda: cont4 0.63809540093\nBox-Cox lambda: cont5 0.00875379455225\nBox-Cox lambda: cont6 0.733624296659\nBox-Cox lambda: cont7 0.813827890905\nBox-Cox lambda: cont8 0.391199987333\nBox-Cox lambda: cont9 1.07531679336\nBox-Cox lambda: cont10 0.817228947796\nBox-Cox lambda: cont11 0.662348896623\nBox-Cox lambda: cont12 0.663914494335\nBox-Cox lambda: cont13 0.42148626027\nBox-Cox lambda: cont14 0.348057576963\ncont1: mean =4.40564090179, std =0.96481834825\ncont2: mean =3.08520889164, std =0.979958114703\ncont3: mean =3.2339260311, std =0.899464879319\ncont4: mean =2.04572849446, std =0.625465861536\ncont5: mean =1.09809662146, std =0.328271226806\ncont6: mean =3.20892175954, std =0.645886083975\ncont7: mean =4.03343732263, std =0.718009806485\ncont8: mean =1.59919963672, std =0.468664811541\ncont9: mean =7.48483417658, std =1.16585074136\ncont10: mean =3.72764435643, std =0.732902500411\ncont11: mean =2.6590288061, std =0.597672934139\ncont12: mean =2.66961326658, std =0.598595418791\ncont13: mean =2.48503209081, std =0.373551746176\ncont14: mean =1.57853396095, std =0.439618821592\n"
],
[
"# Transform test set with parameters done for training set\ndef transform_test_set(df, col_to_norm, lambda_vals, mean_std):\n for col in col_to_norm:\n if np.min(df[col]) < 1:\n df[col] = df[col] + np.abs(np.min(df[col])) + 2\n \n for col, val in zip(col_to_norm, lambda_vals):\n tformed = boxcox(df[col].values, lmbda=val)\n df[col] = tformed[0] #By default, Box-Cox outputs array and lambda. We only want the array output.\n \n for col, (mean, std) in zip(col_to_norm, mean_std):\n df[col] = df[col].apply(lambda x: (x - mean) / std)\n\n return df\n\ndf_test = transform_test_set(df_test, col_to_norm, lambda_vals, mean_std)\n\ndisplay(df_test.describe())",
"_____no_output_____"
],
[
"sns_plot = sns.pairplot(df_train)\nsns_plot.savefig(\"output_asc_after_norm.png\")",
"_____no_output_____"
],
[
"# Save preprocessed datasets to pickle\ndf_train.to_pickle('train_2.pickle')\ndf_test.to_pickle('test_2.pickle')\nprint \"Files saved.\"",
"Files saved.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb4ba67c52c49077d66f527b9c9f47a9a6a7fb6b | 11,118 | ipynb | Jupyter Notebook | jupyter/paddlepaddle/paddle_ocr_java.ipynb | docrozza/djl | 11d9974da5e08664061069ab40ac69b6dd2180b2 | [
"Apache-2.0"
] | 622 | 2021-04-21T06:23:58.000Z | 2022-03-31T04:35:49.000Z | jupyter/paddlepaddle/paddle_ocr_java.ipynb | lanking520/djl | 734683bf98a301121814ec1f369d2d4abbe9955c | [
"Apache-2.0"
] | 468 | 2021-04-20T17:40:38.000Z | 2022-03-31T21:58:17.000Z | jupyter/paddlepaddle/paddle_ocr_java.ipynb | lanking520/djl | 734683bf98a301121814ec1f369d2d4abbe9955c | [
"Apache-2.0"
] | 122 | 2021-04-22T08:28:48.000Z | 2022-03-28T09:24:40.000Z | 35.295238 | 319 | 0.600378 | [
[
[
"# PaddleOCR DJL example\n\nIn this tutorial, we will be using pretrained PaddlePaddle model from [PaddleOCR](https://github.com/PaddlePaddle/PaddleOCR) to do Optical character recognition (OCR) from the given image. There are three models involved in this tutorial:\n\n- Word detection model: used to detect the word block from the image\n- Word direction model: used to find if the text needs to rotate\n- Word recognition model: Used to recognize test from the word block\n\n## Import dependencies and classes\n\nPaddlePaddle is one of the Deep Engines that requires DJL hybrid mode to run inference. Itself does not contains NDArray operations and needs a supplemental DL framework to help with that. So we import Pytorch DL engine as well in here to do the processing works.",
"_____no_output_____"
]
],
[
[
"// %mavenRepo snapshots https://oss.sonatype.org/content/repositories/snapshots/\n\n%maven ai.djl:api:0.14.0\n%maven ai.djl.paddlepaddle:paddlepaddle-model-zoo:0.14.0\n%maven org.slf4j:slf4j-api:1.7.32\n%maven org.slf4j:slf4j-simple:1.7.32\n\n// second engine to do preprocessing and postprocessing\n%maven ai.djl.pytorch:pytorch-engine:0.14.0",
"_____no_output_____"
],
[
"import ai.djl.*;\nimport ai.djl.inference.Predictor;\nimport ai.djl.modality.Classifications;\nimport ai.djl.modality.cv.Image;\nimport ai.djl.modality.cv.ImageFactory;\nimport ai.djl.modality.cv.output.*;\nimport ai.djl.modality.cv.util.NDImageUtils;\nimport ai.djl.ndarray.*;\nimport ai.djl.ndarray.types.DataType;\nimport ai.djl.ndarray.types.Shape;\nimport ai.djl.repository.zoo.*;\nimport ai.djl.paddlepaddle.zoo.cv.objectdetection.PpWordDetectionTranslator;\nimport ai.djl.paddlepaddle.zoo.cv.imageclassification.PpWordRotateTranslator;\nimport ai.djl.paddlepaddle.zoo.cv.wordrecognition.PpWordRecognitionTranslator;\nimport ai.djl.translate.*;\nimport java.util.concurrent.ConcurrentHashMap;",
"_____no_output_____"
]
],
[
[
"## the Image\nFirstly, let's take a look at our sample image, a flight ticket:",
"_____no_output_____"
]
],
[
[
"String url = \"https://resources.djl.ai/images/flight_ticket.jpg\";\nImage img = ImageFactory.getInstance().fromUrl(url);\nimg.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"## Word detection model\n\nIn our word detection model, we load the model exported from [PaddleOCR](https://github.com/PaddlePaddle/PaddleOCR/blob/release/2.0/doc/doc_en/inference_en.md#convert-detection-model-to-inference-model). After that, we can spawn a DJL Predictor from it called detector.",
"_____no_output_____"
]
],
[
[
"var criteria1 = Criteria.builder()\n .optEngine(\"PaddlePaddle\")\n .setTypes(Image.class, DetectedObjects.class)\n .optModelUrls(\"https://resources.djl.ai/test-models/paddleOCR/mobile/det_db.zip\")\n .optTranslator(new PpWordDetectionTranslator(new ConcurrentHashMap<String, String>()))\n .build();\nvar detectionModel = criteria1.loadModel();\nvar detector = detectionModel.newPredictor();",
"_____no_output_____"
]
],
[
[
"Then, we can detect the word block from it. The original output from the model is a bitmap that marked all word regions. The `PpWordDetectionTranslator` convert the output bitmap into a rectangle bounded box for us to crop the image.",
"_____no_output_____"
]
],
[
[
"var detectedObj = detector.predict(img);\nImage newImage = img.duplicate();\nnewImage.drawBoundingBoxes(detectedObj);\nnewImage.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"As you can see above, the word block are very narrow and does not include the whole body of all words. Let's try to extend it a bit for a better result. `extendRect` extend the box height and width to a certain scale. `getSubImage` will crop the image and extract the word block.",
"_____no_output_____"
]
],
[
[
"Image getSubImage(Image img, BoundingBox box) {\n Rectangle rect = box.getBounds();\n double[] extended = extendRect(rect.getX(), rect.getY(), rect.getWidth(), rect.getHeight());\n int width = img.getWidth();\n int height = img.getHeight();\n int[] recovered = {\n (int) (extended[0] * width),\n (int) (extended[1] * height),\n (int) (extended[2] * width),\n (int) (extended[3] * height)\n };\n return img.getSubImage(recovered[0], recovered[1], recovered[2], recovered[3]);\n}\n\ndouble[] extendRect(double xmin, double ymin, double width, double height) {\n double centerx = xmin + width / 2;\n double centery = ymin + height / 2;\n if (width > height) {\n width += height * 2.0;\n height *= 3.0;\n } else {\n height += width * 2.0;\n width *= 3.0;\n }\n double newX = centerx - width / 2 < 0 ? 0 : centerx - width / 2;\n double newY = centery - height / 2 < 0 ? 0 : centery - height / 2;\n double newWidth = newX + width > 1 ? 1 - newX : width;\n double newHeight = newY + height > 1 ? 1 - newY : height;\n return new double[] {newX, newY, newWidth, newHeight};\n}",
"_____no_output_____"
]
],
[
[
"Let's try to extract one block out:",
"_____no_output_____"
]
],
[
[
"List<DetectedObjects.DetectedObject> boxes = detectedObj.items();\nvar sample = getSubImage(img, boxes.get(5).getBoundingBox());\nsample.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"## Word Direction model\n\nThis model is exported from [PaddleOCR](https://github.com/PaddlePaddle/PaddleOCR/blob/release/2.0/doc/doc_en/inference_en.md#convert-angle-classification-model-to-inference-model) that can help to identify if the image is required to rotate. The following code will load this model and create a rotateClassifier.",
"_____no_output_____"
]
],
[
[
"var criteria2 = Criteria.builder()\n .optEngine(\"PaddlePaddle\")\n .setTypes(Image.class, Classifications.class)\n .optModelUrls(\"https://resources.djl.ai/test-models/paddleOCR/mobile/cls.zip\")\n .optTranslator(new PpWordRotateTranslator())\n .build();\nvar rotateModel = criteria2.loadModel();\nvar rotateClassifier = rotateModel.newPredictor();",
"_____no_output_____"
]
],
[
[
"## Word Recgonition model\n\nThe word recognition model is exported from [PaddleOCR](https://github.com/PaddlePaddle/PaddleOCR/blob/release/2.0/doc/doc_en/inference_en.md#convert-recognition-model-to-inference-model) that can recognize the text on the image. Let's load this model as well.\n",
"_____no_output_____"
]
],
[
[
"var criteria3 = Criteria.builder()\n .optEngine(\"PaddlePaddle\")\n .setTypes(Image.class, String.class)\n .optModelUrls(\"https://resources.djl.ai/test-models/paddleOCR/mobile/rec_crnn.zip\")\n .optTranslator(new PpWordRecognitionTranslator())\n .build();\nvar recognitionModel = criteria3.loadModel();\nvar recognizer = recognitionModel.newPredictor();",
"_____no_output_____"
]
],
[
[
"Then we can try to play with these two models on the previous cropped image:",
"_____no_output_____"
]
],
[
[
"System.out.println(rotateClassifier.predict(sample));\nrecognizer.predict(sample);",
"_____no_output_____"
]
],
[
[
"Finally, let's run these models on the whole image and see the outcome. DJL offers a rich image toolkit that allows you to draw the text on image and display them.",
"_____no_output_____"
]
],
[
[
"Image rotateImg(Image image) {\n try (NDManager manager = NDManager.newBaseManager()) {\n NDArray rotated = NDImageUtils.rotate90(image.toNDArray(manager), 1);\n return ImageFactory.getInstance().fromNDArray(rotated);\n }\n}\n\nList<String> names = new ArrayList<>();\nList<Double> prob = new ArrayList<>();\nList<BoundingBox> rect = new ArrayList<>();\n\nfor (int i = 0; i < boxes.size(); i++) {\n Image subImg = getSubImage(img, boxes.get(i).getBoundingBox());\n if (subImg.getHeight() * 1.0 / subImg.getWidth() > 1.5) {\n subImg = rotateImg(subImg);\n }\n Classifications.Classification result = rotateClassifier.predict(subImg).best();\n if (\"Rotate\".equals(result.getClassName()) && result.getProbability() > 0.8) {\n subImg = rotateImg(subImg);\n }\n String name = recognizer.predict(subImg);\n names.add(name);\n prob.add(-1.0);\n rect.add(boxes.get(i).getBoundingBox());\n}\nnewImage.drawBoundingBoxes(new DetectedObjects(names, prob, rect));\nnewImage.getWrappedImage();",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4bb43fcdd155980aadf43a4a6b013863046745 | 87,634 | ipynb | Jupyter Notebook | kenya-financial-census-2009/financialDataSet.ipynb | achingachris/datasciencelearninghub | a6dfb66b2f449170e8b30696bce0c1760cb325d5 | [
"MIT"
] | 2 | 2019-09-26T05:48:08.000Z | 2019-09-26T08:19:09.000Z | kenya-financial-census-2009/financialDataSet.ipynb | ChrisAchinga/Introduction-to-data-science | a6dfb66b2f449170e8b30696bce0c1760cb325d5 | [
"MIT"
] | null | null | null | kenya-financial-census-2009/financialDataSet.ipynb | ChrisAchinga/Introduction-to-data-science | a6dfb66b2f449170e8b30696bce0c1760cb325d5 | [
"MIT"
] | 1 | 2020-02-19T16:08:40.000Z | 2020-02-19T16:08:40.000Z | 76.005204 | 24,428 | 0.709165 | [
[
[
"<a href=\"https://colab.research.google.com/github/ChrisAchinga/Introduction-to-data-science/blob/master/financialDataSet.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"# Import Libraries we use for analysis\nimport pandas as pd\n#add a document\nmyDataSet = pd.read_csv('https://bit.ly/FinancialDataset')\n\n#view observations\nmyDataSet.head()",
"_____no_output_____"
],
[
"#check more info\nmyDataSet.tail(10)",
"_____no_output_____"
],
[
"myDataSet.info()\n#information on datamy",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 23524 entries, 0 to 23523\nData columns (total 13 columns):\ncountry 23510 non-null object\nyear 23524 non-null int64\nuniqueid 23524 non-null object\nHas a Bank account 23488 non-null object\nType of Location 23509 non-null object\nCell Phone Access 23513 non-null object\nhousehold_size 23496 non-null float64\nRespondent Age 23490 non-null float64\ngender_of_respondent 23490 non-null object\nThe relathip with head 23520 non-null object\nmarital_status 23492 non-null object\nLevel of Educuation 23495 non-null object\nType of Job 23494 non-null object\ndtypes: float64(2), int64(1), object(10)\nmemory usage: 2.3+ MB\n"
],
[
"myDataSet.describe()\n#summary of statistics",
"_____no_output_____"
],
[
"myDataSet.shape\n",
"_____no_output_____"
],
[
"myDataSet.columns\n",
"_____no_output_____"
],
[
"myDataSet.year.unique()",
"_____no_output_____"
],
[
"myDataSet.marital_status.unique()",
"_____no_output_____"
],
[
"myDataSet[\"Cell Phone Access\"].unique()",
"_____no_output_____"
],
[
"myDataSet[\"Type of Job\"].unique()",
"_____no_output_____"
],
[
"myDataSet.gender_of_respondent.unique()",
"_____no_output_____"
],
[
"myDataSet.columns",
"_____no_output_____"
],
[
"#viewing unique values\nmyDataSet.uniqueid.unique()",
"_____no_output_____"
],
[
"myDataSet.dtypes\n",
"_____no_output_____"
],
[
"myDataSet[\"Level of Educuation\"].unique()",
"_____no_output_____"
],
[
"#import libraries for graphs\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"myDataSet.dropna(inplace = True)",
"_____no_output_____"
],
[
"#plotting a histogram using matplotlib and seaborn\n#create a figure and give dimensions\nplt.figure(figsize = (8, 5))\nsns.distplot(myDataSet['Respondent Age'], color='green')\n#title to the graph\nplt.title('Distribution for Respondent Age')\nplt.xlabel('Respondent')\nplt.ylabel('Frequency')\nplt.show()\n\n",
"_____no_output_____"
],
[
"myDataSet['Respondent Age'].mean()",
"_____no_output_____"
],
[
"#another one lol\n#create afigure and give dimensions\nplt.figure(figsize = (12, 8))\nsns.countplot(myDataSet['country'])\nplt.title('Country Chart', color = 'green', )\nplt.xlabel('Country')\nplt.ylabel('Frequency')\nplt.show()",
"_____no_output_____"
],
[
"#another one\nplt.figure(figsize = (12, 8))\nsns.countplot(myDataSet['marital_status'])",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4bc3560b7be07f4fd5ba5f92c2206015595e11 | 42,424 | ipynb | Jupyter Notebook | PythonJupyterNotebooks/Week11-Day1-lecture-p1.ipynb | nomadic-me/CodingCheatSheets | a4109cce611dcfc81668db3a18c2c50d75a66ba4 | [
"MIT"
] | null | null | null | PythonJupyterNotebooks/Week11-Day1-lecture-p1.ipynb | nomadic-me/CodingCheatSheets | a4109cce611dcfc81668db3a18c2c50d75a66ba4 | [
"MIT"
] | null | null | null | PythonJupyterNotebooks/Week11-Day1-lecture-p1.ipynb | nomadic-me/CodingCheatSheets | a4109cce611dcfc81668db3a18c2c50d75a66ba4 | [
"MIT"
] | 1 | 2022-03-10T03:11:03.000Z | 2022-03-10T03:11:03.000Z | 27.229782 | 149 | 0.312135 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv('./diabetes.csv')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"X = df.drop(columns='Outcome')\ny = df['Outcome']",
"_____no_output_____"
],
[
"X.head()",
"_____no_output_____"
],
[
"y.head()",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"split = train_test_split(X, y)",
"_____no_output_____"
],
[
"split",
"_____no_output_____"
],
[
"len(split)",
"_____no_output_____"
],
[
"split[0]",
"_____no_output_____"
],
[
"X.shape",
"_____no_output_____"
],
[
"split[1]",
"_____no_output_____"
],
[
"split[2]",
"_____no_output_____"
],
[
"split[3]",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression",
"_____no_output_____"
],
[
"classifier = LogisticRegression(random_state=0)",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = split\n\nclassifier.fit(X_train, y_train)",
"/opt/anaconda3/lib/python3.9/site-packages/sklearn/linear_model/_logistic.py:814: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n n_iter_i = _check_optimize_result(\n"
],
[
"classifier.coef_",
"_____no_output_____"
],
[
"X.columns",
"_____no_output_____"
],
[
"coefficients = pd.DataFrame({\n 'Feature': X.columns,\n 'Coefficient': classifier.coef_[0]\n})\ncoefficients",
"_____no_output_____"
],
[
"y_preds = classifier.predict(X_test)\ny_preds",
"_____no_output_____"
],
[
"results = pd.DataFrame({\"Prediction\": y_preds, \"Actual\": y_test}).reset_index(drop=True)",
"_____no_output_____"
],
[
"results",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix",
"_____no_output_____"
],
[
"tn, fp, fn, tp = confusion_matrix(results['Actual'], results['Prediction']).ravel()",
"_____no_output_____"
],
[
"tn",
"_____no_output_____"
],
[
"fp",
"_____no_output_____"
],
[
"fn",
"_____no_output_____"
],
[
"tp",
"_____no_output_____"
],
[
"len(y_test)",
"_____no_output_____"
],
[
"sum(y_test)",
"_____no_output_____"
],
[
"59/192",
"_____no_output_____"
],
[
"accuracy = (tp + tn) / (tp + fn + fp + tn)\naccuracy",
"_____no_output_____"
],
[
"precision = tp / (tp + fp)\nprecision",
"_____no_output_____"
],
[
"recall = tp / (tp + fn)\nrecall",
"_____no_output_____"
],
[
"from sklearn.metrics import classification_report\nprint(classification_report(results['Actual'], results['Prediction']))\n",
" precision recall f1-score support\n\n 0 0.78 0.89 0.84 133\n 1 0.65 0.44 0.53 59\n\n accuracy 0.76 192\n macro avg 0.72 0.67 0.68 192\nweighted avg 0.74 0.76 0.74 192\n\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4bdc6e3e42e7fd092be4982df0b48dff17d6cf | 165,859 | ipynb | Jupyter Notebook | notebooks/EMNIST_Notebook.ipynb | dhsong95/dacon-emnist-competition | 736d869edeea052b92b1667e05872d3adda79711 | [
"MIT"
] | null | null | null | notebooks/EMNIST_Notebook.ipynb | dhsong95/dacon-emnist-competition | 736d869edeea052b92b1667e05872d3adda79711 | [
"MIT"
] | null | null | null | notebooks/EMNIST_Notebook.ipynb | dhsong95/dacon-emnist-competition | 736d869edeea052b92b1667e05872d3adda79711 | [
"MIT"
] | null | null | null | 64.038224 | 32,418 | 0.556919 | [
[
[
"<a href=\"https://colab.research.google.com/github/dhsong95/dacon-emnist-competition/blob/master/notebooks/EMNIST_Notebook.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/gdrive')",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly&response_type=code\n\nEnter your authorization code:\n··········\nMounted at /content/gdrive\n"
],
[
"%ls /content/gdrive/'My Drive'/'Google Colaboratory'/dacon-emnist-competition/",
"\u001b[0m\u001b[01;34mdata\u001b[0m/ LICENSE \u001b[01;34mnotebooks\u001b[0m/ README.md\n"
],
[
"%cd /content/gdrive/'My Drive'/'Google Colaboratory'/dacon-emnist-competition/",
"/content/gdrive/My Drive/Google Colaboratory/dacon-emnist-competition\n"
],
[
"import os\nimport pandas as pd",
"_____no_output_____"
],
[
"datadir = 'data/'\n\ntrain = pd.read_csv(os.path.join(datadir, 'train.csv'))\ntest = pd.read_csv(os.path.join(datadir, 'test.csv'))\nsubmission = pd.read_csv(os.path.join(datadir, 'submission.csv'))",
"_____no_output_____"
],
[
"train.sample(10)",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"digit_columns = train.columns[3:]\ndigits = np.array(train.loc[:, digit_columns])\ndigits.shape",
"_____no_output_____"
],
[
"index = np.random.randint(digits.shape[0])\nplt.figure(figsize=(4, 4))\nplt.imshow(digits[index, :].reshape(28, 28))\nplt.title(f'digit {train.loc[index, \"digit\"]} / letter {train.loc[index, \"letter\"]}')\nplt.xticks([])\nplt.yticks([])\nplt.axis('off')\nplt.show()",
"_____no_output_____"
],
[
"x_train = train.drop(columns=['id', 'digit', 'letter']).values.reshape(-1, 28, 28, 1)\nx_train = x_train / 255.0\nx_train.shape",
"_____no_output_____"
],
[
"N = len(train['digit'])\nC = len(pd.unique(train['digit']))\n\ny_train = np.zeros(shape=(N, C))\nfor idx, digit in enumerate(train['digit']):\n y_train[idx, digit] = 1\ny_train.shape",
"_____no_output_____"
],
[
"from tensorflow import keras\nfrom tensorflow.keras.layers import Activation, BatchNormalization, Conv2D,\\\n Dense, Dropout, Flatten, MaxPool2D, LeakyReLU\nfrom tensorflow.keras.optimizers import Adam\nimport tensorflow as tf",
"_____no_output_____"
],
[
"class BasicCNN:\n def __init__(self, image_shape):\n self.model = self.build_model(image_shape)\n adam = Adam(learning_rate=1e-3)\n self.model.compile(\n loss='categorical_crossentropy',\n optimizer=adam,\n metrics=['accuracy']\n )\n\n def build_model(self, image_shape):\n model = keras.models.Sequential()\n\n model.add(Conv2D(128, 3, padding='same'))\n model.add(BatchNormalization())\n model.add(LeakyReLU(0.01))\n model.add(MaxPool2D())\n model.add(Dropout(rate=0.5))\n\n model.add(Conv2D(256, 3, padding='same'))\n model.add(BatchNormalization())\n model.add(LeakyReLU(0.01))\n model.add(MaxPool2D())\n model.add(Dropout(rate=0.5))\n\n model.add(Conv2D(512, 3, padding='same'))\n model.add(BatchNormalization())\n model.add(LeakyReLU(0.01))\n model.add(MaxPool2D())\n model.add(Dropout(rate=0.5))\n\n model.add(Flatten())\n\n model.add(Dense(512))\n model.add(BatchNormalization())\n model.add(Activation('relu'))\n\n model.add(Dense(10, activation='softmax'))\n\n\n inputs = keras.Input(image_shape[1:])\n outputs = model(inputs)\n\n print(model.summary())\n\n return keras.Model(inputs, outputs)\n\n def train(self, x_train, y_train, epochs):\n history = self.model.fit(\n x_train, y_train,\n epochs=epochs,\n validation_split=0.2\n )\n return history\n\n def predict(self, x_test):\n return self.model.predict(x_test)",
"_____no_output_____"
],
[
"basic_cnn = BasicCNN(x_train.shape)\nhistory = basic_cnn.train(x_train, y_train, 100)",
"Model: \"sequential_9\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d_18 (Conv2D) (None, 28, 28, 128) 1280 \n_________________________________________________________________\nbatch_normalization_36 (Batc (None, 28, 28, 128) 512 \n_________________________________________________________________\nleaky_re_lu_18 (LeakyReLU) (None, 28, 28, 128) 0 \n_________________________________________________________________\nmax_pooling2d_18 (MaxPooling (None, 14, 14, 128) 0 \n_________________________________________________________________\ndropout_36 (Dropout) (None, 14, 14, 128) 0 \n_________________________________________________________________\nconv2d_19 (Conv2D) (None, 14, 14, 256) 295168 \n_________________________________________________________________\nbatch_normalization_37 (Batc (None, 14, 14, 256) 1024 \n_________________________________________________________________\nleaky_re_lu_19 (LeakyReLU) (None, 14, 14, 256) 0 \n_________________________________________________________________\nmax_pooling2d_19 (MaxPooling (None, 7, 7, 256) 0 \n_________________________________________________________________\ndropout_37 (Dropout) (None, 7, 7, 256) 0 \n_________________________________________________________________\nconv2d_20 (Conv2D) (None, 7, 7, 512) 1180160 \n_________________________________________________________________\nbatch_normalization_38 (Batc (None, 7, 7, 512) 2048 \n_________________________________________________________________\nleaky_re_lu_20 (LeakyReLU) (None, 7, 7, 512) 0 \n_________________________________________________________________\nmax_pooling2d_20 (MaxPooling (None, 3, 3, 512) 0 \n_________________________________________________________________\ndropout_38 (Dropout) (None, 3, 3, 512) 0 \n_________________________________________________________________\nflatten_9 (Flatten) (None, 4608) 0 \n_________________________________________________________________\ndense_27 (Dense) (None, 512) 2359808 \n_________________________________________________________________\nbatch_normalization_39 (Batc (None, 512) 2048 \n_________________________________________________________________\ndense_28 (Dense) (None, 10) 5130 \n=================================================================\nTotal params: 3,847,178\nTrainable params: 3,844,362\nNon-trainable params: 2,816\n_________________________________________________________________\nNone\nEpoch 1/100\n52/52 [==============================] - 2s 32ms/step - loss: 2.4918 - accuracy: 0.2411 - val_loss: 2.7183 - val_accuracy: 0.1073\nEpoch 2/100\n52/52 [==============================] - 1s 24ms/step - loss: 1.8836 - accuracy: 0.3694 - val_loss: 3.4789 - val_accuracy: 0.1073\nEpoch 3/100\n52/52 [==============================] - 1s 24ms/step - loss: 1.5213 - accuracy: 0.4817 - val_loss: 3.8515 - val_accuracy: 0.1073\nEpoch 4/100\n52/52 [==============================] - 1s 24ms/step - loss: 1.3460 - accuracy: 0.5470 - val_loss: 4.4321 - val_accuracy: 0.1073\nEpoch 5/100\n52/52 [==============================] - 1s 24ms/step - loss: 1.2250 - accuracy: 0.5812 - val_loss: 5.7952 - val_accuracy: 0.1073\nEpoch 6/100\n52/52 [==============================] - 1s 24ms/step - loss: 1.1683 - accuracy: 0.6099 - val_loss: 5.1939 - val_accuracy: 0.1073\nEpoch 7/100\n52/52 [==============================] - 1s 24ms/step - loss: 1.0871 - accuracy: 0.6258 - val_loss: 5.7725 - val_accuracy: 0.1073\nEpoch 8/100\n52/52 [==============================] - 1s 24ms/step - loss: 1.0680 - accuracy: 0.6398 - val_loss: 4.9267 - val_accuracy: 0.1073\nEpoch 9/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.9909 - accuracy: 0.6740 - val_loss: 5.4179 - val_accuracy: 0.1073\nEpoch 10/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.9688 - accuracy: 0.6764 - val_loss: 6.6550 - val_accuracy: 0.1073\nEpoch 11/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.9054 - accuracy: 0.6868 - val_loss: 6.0247 - val_accuracy: 0.1073\nEpoch 12/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.8172 - accuracy: 0.7265 - val_loss: 4.2501 - val_accuracy: 0.1073\nEpoch 13/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.7672 - accuracy: 0.7350 - val_loss: 4.6209 - val_accuracy: 0.1073\nEpoch 14/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.7609 - accuracy: 0.7552 - val_loss: 3.6325 - val_accuracy: 0.1463\nEpoch 15/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.7343 - accuracy: 0.7491 - val_loss: 1.1989 - val_accuracy: 0.6244\nEpoch 16/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.7026 - accuracy: 0.7589 - val_loss: 1.1823 - val_accuracy: 0.6317\nEpoch 17/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.7118 - accuracy: 0.7558 - val_loss: 1.1149 - val_accuracy: 0.6146\nEpoch 18/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.6161 - accuracy: 0.7851 - val_loss: 1.4898 - val_accuracy: 0.4756\nEpoch 19/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.6516 - accuracy: 0.7790 - val_loss: 0.8211 - val_accuracy: 0.7317\nEpoch 20/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.5793 - accuracy: 0.7973 - val_loss: 0.9951 - val_accuracy: 0.6634\nEpoch 21/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.5737 - accuracy: 0.8022 - val_loss: 0.8202 - val_accuracy: 0.7561\nEpoch 22/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.5453 - accuracy: 0.8150 - val_loss: 0.7559 - val_accuracy: 0.7512\nEpoch 23/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.5138 - accuracy: 0.8217 - val_loss: 0.8438 - val_accuracy: 0.7366\nEpoch 24/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.5271 - accuracy: 0.8144 - val_loss: 1.0731 - val_accuracy: 0.6341\nEpoch 25/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.4906 - accuracy: 0.8339 - val_loss: 0.7774 - val_accuracy: 0.7390\nEpoch 26/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.4848 - accuracy: 0.8272 - val_loss: 0.6538 - val_accuracy: 0.7976\nEpoch 27/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.4713 - accuracy: 0.8321 - val_loss: 1.1042 - val_accuracy: 0.7024\nEpoch 28/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.4423 - accuracy: 0.8437 - val_loss: 0.7030 - val_accuracy: 0.7854\nEpoch 29/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.4459 - accuracy: 0.8443 - val_loss: 0.7196 - val_accuracy: 0.7902\nEpoch 30/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3910 - accuracy: 0.8620 - val_loss: 0.6347 - val_accuracy: 0.7976\nEpoch 31/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3845 - accuracy: 0.8584 - val_loss: 0.6546 - val_accuracy: 0.8049\nEpoch 32/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3738 - accuracy: 0.8694 - val_loss: 0.6061 - val_accuracy: 0.8073\nEpoch 33/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3900 - accuracy: 0.8657 - val_loss: 0.6076 - val_accuracy: 0.8049\nEpoch 34/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3852 - accuracy: 0.8700 - val_loss: 0.5923 - val_accuracy: 0.8024\nEpoch 35/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3281 - accuracy: 0.8895 - val_loss: 0.6483 - val_accuracy: 0.8049\nEpoch 36/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3032 - accuracy: 0.8962 - val_loss: 0.6629 - val_accuracy: 0.7829\nEpoch 37/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3019 - accuracy: 0.8944 - val_loss: 0.6510 - val_accuracy: 0.8049\nEpoch 38/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3021 - accuracy: 0.8919 - val_loss: 0.6518 - val_accuracy: 0.8024\nEpoch 39/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3594 - accuracy: 0.8736 - val_loss: 0.8004 - val_accuracy: 0.7683\nEpoch 40/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3683 - accuracy: 0.8663 - val_loss: 0.6333 - val_accuracy: 0.8024\nEpoch 41/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.3072 - accuracy: 0.8938 - val_loss: 0.5611 - val_accuracy: 0.8268\nEpoch 42/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.2580 - accuracy: 0.9109 - val_loss: 0.8642 - val_accuracy: 0.7488\nEpoch 43/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.2694 - accuracy: 0.9072 - val_loss: 0.5980 - val_accuracy: 0.8220\nEpoch 44/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.2702 - accuracy: 0.9017 - val_loss: 0.6952 - val_accuracy: 0.7976\nEpoch 45/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.2563 - accuracy: 0.8968 - val_loss: 0.6512 - val_accuracy: 0.8024\nEpoch 46/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.2240 - accuracy: 0.9249 - val_loss: 0.7794 - val_accuracy: 0.7707\nEpoch 47/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.2489 - accuracy: 0.9151 - val_loss: 0.5464 - val_accuracy: 0.8463\nEpoch 48/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1911 - accuracy: 0.9383 - val_loss: 0.5586 - val_accuracy: 0.8415\nEpoch 49/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1871 - accuracy: 0.9353 - val_loss: 0.6449 - val_accuracy: 0.8024\nEpoch 50/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.2035 - accuracy: 0.9304 - val_loss: 0.5563 - val_accuracy: 0.8195\nEpoch 51/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1812 - accuracy: 0.9353 - val_loss: 0.6363 - val_accuracy: 0.8220\nEpoch 52/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1978 - accuracy: 0.9243 - val_loss: 0.6822 - val_accuracy: 0.8049\nEpoch 53/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.2004 - accuracy: 0.9280 - val_loss: 0.6442 - val_accuracy: 0.7951\nEpoch 54/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1935 - accuracy: 0.9255 - val_loss: 0.5399 - val_accuracy: 0.8390\nEpoch 55/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1813 - accuracy: 0.9359 - val_loss: 0.5491 - val_accuracy: 0.8415\nEpoch 56/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1813 - accuracy: 0.9280 - val_loss: 0.5485 - val_accuracy: 0.8439\nEpoch 57/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1596 - accuracy: 0.9457 - val_loss: 0.6941 - val_accuracy: 0.8146\nEpoch 58/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.2006 - accuracy: 0.9353 - val_loss: 0.6389 - val_accuracy: 0.8341\nEpoch 59/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.2207 - accuracy: 0.9255 - val_loss: 0.6709 - val_accuracy: 0.8073\nEpoch 60/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1864 - accuracy: 0.9377 - val_loss: 0.5814 - val_accuracy: 0.8293\nEpoch 61/100\n52/52 [==============================] - 1s 23ms/step - loss: 0.1791 - accuracy: 0.9408 - val_loss: 0.5915 - val_accuracy: 0.8244\nEpoch 62/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1600 - accuracy: 0.9396 - val_loss: 0.6416 - val_accuracy: 0.8220\nEpoch 63/100\n52/52 [==============================] - 1s 23ms/step - loss: 0.1803 - accuracy: 0.9328 - val_loss: 0.5522 - val_accuracy: 0.8463\nEpoch 64/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1553 - accuracy: 0.9457 - val_loss: 0.6168 - val_accuracy: 0.8293\nEpoch 65/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1620 - accuracy: 0.9463 - val_loss: 0.6540 - val_accuracy: 0.8220\nEpoch 66/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1520 - accuracy: 0.9487 - val_loss: 0.5631 - val_accuracy: 0.8341\nEpoch 67/100\n52/52 [==============================] - 1s 25ms/step - loss: 0.1552 - accuracy: 0.9487 - val_loss: 0.5701 - val_accuracy: 0.8317\nEpoch 68/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1283 - accuracy: 0.9573 - val_loss: 0.6353 - val_accuracy: 0.8073\nEpoch 69/100\n52/52 [==============================] - 1s 25ms/step - loss: 0.1230 - accuracy: 0.9548 - val_loss: 0.6568 - val_accuracy: 0.8293\nEpoch 70/100\n52/52 [==============================] - 1s 25ms/step - loss: 0.1627 - accuracy: 0.9444 - val_loss: 0.5897 - val_accuracy: 0.8293\nEpoch 71/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1246 - accuracy: 0.9603 - val_loss: 0.5881 - val_accuracy: 0.8439\nEpoch 72/100\n52/52 [==============================] - 1s 25ms/step - loss: 0.1279 - accuracy: 0.9591 - val_loss: 0.5506 - val_accuracy: 0.8463\nEpoch 73/100\n52/52 [==============================] - 1s 25ms/step - loss: 0.1105 - accuracy: 0.9597 - val_loss: 0.5731 - val_accuracy: 0.8390\nEpoch 74/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1061 - accuracy: 0.9664 - val_loss: 0.5838 - val_accuracy: 0.8293\nEpoch 75/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0808 - accuracy: 0.9750 - val_loss: 0.5591 - val_accuracy: 0.8488\nEpoch 76/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1003 - accuracy: 0.9658 - val_loss: 0.5160 - val_accuracy: 0.8561\nEpoch 77/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1286 - accuracy: 0.9536 - val_loss: 0.7109 - val_accuracy: 0.8146\nEpoch 78/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1146 - accuracy: 0.9597 - val_loss: 0.5938 - val_accuracy: 0.8463\nEpoch 79/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1312 - accuracy: 0.9615 - val_loss: 0.7140 - val_accuracy: 0.8049\nEpoch 80/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0961 - accuracy: 0.9676 - val_loss: 0.6126 - val_accuracy: 0.8366\nEpoch 81/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1176 - accuracy: 0.9573 - val_loss: 0.5698 - val_accuracy: 0.8561\nEpoch 82/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1288 - accuracy: 0.9585 - val_loss: 0.6122 - val_accuracy: 0.8512\nEpoch 83/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1139 - accuracy: 0.9676 - val_loss: 0.6620 - val_accuracy: 0.8317\nEpoch 84/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1197 - accuracy: 0.9597 - val_loss: 0.5871 - val_accuracy: 0.8366\nEpoch 85/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0705 - accuracy: 0.9756 - val_loss: 0.6085 - val_accuracy: 0.8512\nEpoch 86/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0935 - accuracy: 0.9646 - val_loss: 0.6145 - val_accuracy: 0.8439\nEpoch 87/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0881 - accuracy: 0.9634 - val_loss: 0.6241 - val_accuracy: 0.8195\nEpoch 88/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0907 - accuracy: 0.9664 - val_loss: 0.5276 - val_accuracy: 0.8634\nEpoch 89/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.1156 - accuracy: 0.9621 - val_loss: 0.6162 - val_accuracy: 0.8463\nEpoch 90/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0750 - accuracy: 0.9768 - val_loss: 0.5875 - val_accuracy: 0.8512\nEpoch 91/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0902 - accuracy: 0.9695 - val_loss: 0.6031 - val_accuracy: 0.8610\nEpoch 92/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0749 - accuracy: 0.9744 - val_loss: 0.6459 - val_accuracy: 0.8488\nEpoch 93/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0888 - accuracy: 0.9670 - val_loss: 0.6803 - val_accuracy: 0.8268\nEpoch 94/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0747 - accuracy: 0.9756 - val_loss: 0.6533 - val_accuracy: 0.8512\nEpoch 95/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0643 - accuracy: 0.9792 - val_loss: 0.5826 - val_accuracy: 0.8537\nEpoch 96/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0711 - accuracy: 0.9762 - val_loss: 0.7371 - val_accuracy: 0.8122\nEpoch 97/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0671 - accuracy: 0.9750 - val_loss: 0.6156 - val_accuracy: 0.8439\nEpoch 98/100\n52/52 [==============================] - 1s 23ms/step - loss: 0.0674 - accuracy: 0.9762 - val_loss: 0.6226 - val_accuracy: 0.8512\nEpoch 99/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0644 - accuracy: 0.9762 - val_loss: 0.6747 - val_accuracy: 0.8561\nEpoch 100/100\n52/52 [==============================] - 1s 24ms/step - loss: 0.0561 - accuracy: 0.9835 - val_loss: 0.5960 - val_accuracy: 0.8585\n"
],
[
"train_loss = history.history['loss']\nval_loss = history.history['val_loss']\ntrain_acc = history.history['accuracy']\nval_acc = history.history['val_accuracy']",
"_____no_output_____"
],
[
"plt.figure(figsize=(8, 6))\nplt.plot(train_loss, label='train')\nplt.plot(val_loss, label='validation')\nplt.legend()\nplt.xlabel('epochs')\nplt.ylabel('loss')\nplt.title('Loss per Epochs')\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(8, 6))\nplt.plot(train_acc, label='train')\nplt.plot(val_acc, label='validation')\nplt.legend()\nplt.xlabel('epochs')\nplt.ylabel('accuracy')\nplt.title('Accuracy per Epochs')\nplt.show()",
"_____no_output_____"
],
[
"x_test = np.array(test.loc[:, digit_columns].values)\nx_test = x_test.reshape(-1, 28, 28, 1)\nx_test = x_test / 255.0\nx_test.shape",
"_____no_output_____"
],
[
"prediction = basic_cnn.predict(x_test)",
"_____no_output_____"
],
[
"prediction = np.argmax(prediction, axis=-1)",
"_____no_output_____"
],
[
"submission.head()",
"_____no_output_____"
],
[
"test.head()",
"_____no_output_____"
],
[
"submission['digit'] = prediction\nsubmission.head(10)",
"_____no_output_____"
],
[
"# submission.to_csv('data/submission_1.csv', index=False) # 0.83\nsubmission.to_csv('data/submissions/submission_2.csv', index=False)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4be394a12cf8a667b6a0e4c8431b70d8eb7909 | 310,409 | ipynb | Jupyter Notebook | MeetupOutubro2019/CMS-Open-Data-Tutorial-R-tidyverse.ipynb | clemencia/RJupyterCMS | 7f0063bdb66a6e7a1e5cc9aec76c9d2e7166c33c | [
"MIT"
] | 1 | 2019-10-25T20:20:39.000Z | 2019-10-25T20:20:39.000Z | MeetupOutubro2019/CMS-Open-Data-Tutorial-R-tidyverse.ipynb | clemencia/RStuff | 7f0063bdb66a6e7a1e5cc9aec76c9d2e7166c33c | [
"MIT"
] | null | null | null | MeetupOutubro2019/CMS-Open-Data-Tutorial-R-tidyverse.ipynb | clemencia/RStuff | 7f0063bdb66a6e7a1e5cc9aec76c9d2e7166c33c | [
"MIT"
] | 1 | 2020-02-18T01:18:10.000Z | 2020-02-18T01:18:10.000Z | 136.084612 | 49,646 | 0.844512 | [
[
[
"# Física de partículas ... com R e tidyverse\n\nEsse tutorial utiliza os dados abertos do experimento CMS do LHC [CMS Open Data](http://opendata.cern.ch/about/cms) Disponíveis no site [CERN Open Data portal](http://opendata.cern.ch).\n\nPara rodar esse tutorial offline, vide o arquivo [README](https://github.com/cms-opendata-education/cms-rmaterial-multiple-languages/blob/master/README.md), con instruções em inglés. Eu estou rodando o notebook na minha instalação local de R. \n\nTambém é possível copiar as linhas de código e colar na consola de comandos do RStudio, ou num script e logo rodá-lo.\n\n\n**Créditos:**\n * Adaptado do original de [Edith Villegas Garcia](https://github.com/edithvillegas), [Andrew John Lowe](https://github.com/andrewjohnlowe) e [Achintya Rao](https://github.com/RaoOfPhysics). \n\n * Traduzido ao português e adicionado o ajuste por [Clemencia Mora Herrera](https://github.com/clemencia).\n\n---\n",
"_____no_output_____"
],
[
"\n## Os dados:\n\nEste tutorial, introduce análise de dados com R usando dados divulgados ao público no portal **CMS Open Data**. \n\nA origem desses dados colisões de prótons do LHC no ano 2011 (Energia do centro-de-massa de 7 TeV). \nEstes dados contém medições de partículas do estado final, dois ***múons*** (uma versão um pouco mais pesada do elétron, comúm em raios cósmicos). \n\nA primeira imagem mostra um desenho esquemático do LHC e seus 4 principais experimentos.",
"_____no_output_____"
],
[
"<figure>\n <img src=\"https://github.com/cms-opendata-education/zboson-exercise/blob/master/images/LHC.png?raw=true\" alt=\"image missing\" style=\"height: 350px\" />\n <figcaption> Imagem 1: O LHC e seus 4 principais experimentos. © \n <a href=\"https://cds.cern.ch/record/1708847\">CERN</a> \n </figcaption>\n</figure>\n",
"_____no_output_____"
],
[
"No LHC prótons são acelerados a altíssimas velocidades e feitos colidir em pontos determinados (os 4 da figura acima), onde cada experimento com seus detectores registra e salva a informação dos produtos da colisão. A energia da colisão pode ser convertida em massa de novas partículas ($E=mc^2$) que podem decair em outras mais leves, deixando sinais nos instrumentos de medição em cada detector. Os sinais são traduzidos em momentum ($p=mv$), carga da partícula, energia e a sua direção de saida do ponto de interação.\n\n\n\nO seguinte é um vídeo que mostra como acontecem as colisões e medições no acelerador LHC.",
"_____no_output_____"
]
],
[
[
"library(IRdisplay)\ndisplay_html('<iframe width=\"560\" height=\"315\" src=\"https://www.youtube.com/embed/pQhbhpU9Wrg\" frameborder=\"0\" allow=\"accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture\" allowfullscreen></iframe>')",
"_____no_output_____"
]
],
[
[
"Se combinarmos a informação da energia e momentum dos dois múons para cada observação (_evento_), podemos observar que em certos valores de ***massa invariante*** (a energia de uma partícula de massa $m$ em reposo é $E=m c^2$ --> relatividade restrita isto é sempre válido no sistema de referência dela própia, então essa massa própria é constante para sistemas de referência diferentes) a frequência de observações é maior: isto quer dizer que existe uma partícula subatómica que decaiu em um par de múons e chamamos isso de uma \"ressonância\". Podemos inferir a presença dessas partículas indiretamente observando seus produtos de decaimento, os múons, e a sua frequência.\n",
"_____no_output_____"
],
[
"<figure>\n <img src=\"http://github.com/cms-opendata-education/zboson-exercise/blob/master/images/eventdisplay.png?raw=true\" alt=\"image missing\" style=\"height: 350px\" />\n <figcaption> Imagem 2: Visualização da detecção de dois múons em uma colisão no CMS. </figcaption>\n</figure>",
"_____no_output_____"
],
[
"<figure>\n <img src=\"http://github.com/cms-opendata-education/zboson-exercise/blob/master/images/CMS.jpg?raw=true\" alt=\"image missing\" style=\"height: 350px\" />\n <figcaption> Imagem 3: Estrutura do experimento CMS, aberto. © \n <a href=\"https://cds.cern.ch/record/1433717\">CERN</a> \n </figcaption>\n</figure>",
"_____no_output_____"
],
[
"<figure>\n <img src=\"http://github.com/cms-opendata-education/zboson-exercise/blob/master/images/CMS2.gif?raw=true\" alt=\"image missing\" style=\"height: 350px\" /> \n <figcaption>Imagem 4: Seção transversal do CMS, e como as partículas são detectadas nele. © \n <a href=\\\"https://cms-docdb.cern.ch/cgi-bin/PublicDocDB/ShowDocument?docid=4172\\\">CERN</a> \n </figcaption>\n</figure>",
"_____no_output_____"
],
[
"Nesse tutorial podemos criar um gráfico de frequências onde é possível perceber os picos correspondentes a algumas dessas partículas que tem preferência pelo decaimento em dois múons.",
"_____no_output_____"
],
[
"## Breve introdução a R\n\nR é uma linguagem de programação usada amplamente em estatística e ciência de dados. \n\n\n_\"R é a língua franca da estatística\"_ (W. Zeviani, UFPR)\n\n[ http://leg.ufpr.br/~walmes/cursoR/data-vis/slides/01-tidyverse.pdf ] \n\n\n\n \n### Os tipos de dados em R\n\nOs tipos básicos de dados em R são:\n- Logical -- lógicos ou booleanos: TRUE ou FALSE)\n- Numeric -- números em geral, números reais\n- Integer -- inteiros\n- Complex -- complexos\n- Character -- carateres ou sequências deles: letras, números como carater, símbolos e frases\n\nEm geral, sem ter que especificar, R assigna automátiamente um tipo às variáveis declaradas.\nQualquer número é tipo ```numeric```, mas para especificar ```integer``` temos que adicionar um aletra \"L\" no final do valor.\n\nA linha abaixo declara a variável ```a``` com valor $5$ de tipo inteiro.",
"_____no_output_____"
]
],
[
[
"a <- 5L",
"_____no_output_____"
]
],
[
[
"Para declarar variáveis complexas a sintaxe é a seguinte:",
"_____no_output_____"
]
],
[
[
"b <- 5 + 3i\nd <- 8 + 0i",
"_____no_output_____"
]
],
[
[
"As variáveis lógicas podem tomar o valor ```TRUE``` ou ```FALSE```, mas també pode ser assignado o valor resultante de uma expressão condicional, p.ex.:",
"_____no_output_____"
]
],
[
[
"c <- 3 > 5",
"_____no_output_____"
]
],
[
[
"As variáveis de carateres podem ser letras, frases ou outros carateres incluindo números entre aspas.",
"_____no_output_____"
]
],
[
[
"cr <- \"3!\"",
"_____no_output_____"
]
],
[
[
"Para saber o valor de cada variável eu simplesmente chamo o nome:",
"_____no_output_____"
]
],
[
[
"a\nb\nc\nd\ncr",
"_____no_output_____"
]
],
[
[
"#### Vetores\n\nÉ possível agrupar valores em variáveis vetoriais dessa forma:",
"_____no_output_____"
]
],
[
[
"a <- c(2, 3, 5)",
"_____no_output_____"
]
],
[
[
"Os vetores podem ser de qualquer tipo. Também podemos aplicar condições aos vetores para criar um vetor lógico:",
"_____no_output_____"
]
],
[
[
"a <- c(2, 5, 8, 3, 9)\nb <- a > 3",
"_____no_output_____"
]
],
[
[
"O vetor ```b``` é o resultado da avaliação da condição ```x>3``` para cada elemento ```x``` do vetor ```a```.",
"_____no_output_____"
]
],
[
[
"b",
"_____no_output_____"
]
],
[
[
"Para acessar algum elemento do vetor, podemos chamar o nome da variável vetor com o índice do elemento desejado. O contador começa de 1 (outras linguagens utilizam 0).\n\nEntáo o primeiro elemento de ```a``` será acessado assim:",
"_____no_output_____"
]
],
[
[
"a[1]",
"_____no_output_____"
]
],
[
[
"Também é possível acessar os elementos que satisfazem uma condição. A linha seguinte entrega o subconjunto (sub-vetor) dos elementos de ```a``` que tem valor maior que $3$.",
"_____no_output_____"
]
],
[
[
"c<-a[a>3]\nc",
"_____no_output_____"
]
],
[
[
"#### Matrices\n\nEm R podemos criar uma matriz a partir de vetores. As matrices são estruturas de dados em 2 dimensões. \n\nPodemos criar a matriz especificando os valores nela, o numero de linhas e colunas e se o preenchimento sera por filas ou por colunas.\n\nNeste exemplo começamos por um vetor do 1 ao 9:",
"_____no_output_____"
]
],
[
[
"a <- c(1:9)\na",
"_____no_output_____"
]
],
[
[
"Logo declaramos ```A``` uma matriz de 3x3 componentes, preenchidas por linha, com os 9 elementos de ```a```.",
"_____no_output_____"
]
],
[
[
"A <- matrix(a, nrow=3, ncol=3, byrow=TRUE)\n\nA",
"_____no_output_____"
]
],
[
[
"Para accessar os elementos da matriz, usamos colchetes com o numero de linha e coluna. Por exemplo para acessar o elemento na segunda linha, terceira coluna de ```A``` fazemos:",
"_____no_output_____"
]
],
[
[
"A[2,3]",
"_____no_output_____"
]
],
[
[
"Podemos acessar uma linha completa se especificamos só o primeiro número e deixamos em branco o índice das colunas, e viceversa. Por exemplo a chamada ```A[2,]``` retorna os valores da segunda linha de ```A```. ",
"_____no_output_____"
]
],
[
[
"A[2,]",
"_____no_output_____"
]
],
[
[
"As matrices podem ser acessadas com condições, como foi no caso dos vetores. ",
"_____no_output_____"
]
],
[
[
"# Criar um vetor de valores 1 a 25\na <- c(1:25)\n\n# Criar a matriz a partir desse vetor com 5 linhas e 5 colunas, preenchendo linha por linha.\nA <- matrix(a, nrow=5, ncol=5, byrow=TRUE)\n\n\n\n# Acessar os elementos de A que sejam maiores que 12\n# ao colocar a condição \"A>12\" nos colchetes\n# a variável nova é um vetor.\nC<-A[A>12]\n\nprint(C)",
" [1] 16 21 17 22 13 18 23 14 19 24 15 20 25\n"
],
[
"length(C)\n",
"_____no_output_____"
]
],
[
[
"#### Arrays (Arranjos)\n\nArrays são similares às matrices, mas podem ter mais de duas dimensões. \n\nPodem ser criadas, como as matrices, a partir de um vetor e especificando as dimensões escolhidas.",
"_____no_output_____"
]
],
[
[
"# Criar um vetor com valores 1 a 27\na <- c(1:27)\n\n# Criar um array a partir do vetor a\n# que contem 3 matrices de 3 linhas e 3 colunas .\nA <- array(a, dim=c(3,3,3))\n\n# Imprimir o array.\nprint(A)",
", , 1\n\n [,1] [,2] [,3]\n[1,] 1 4 7\n[2,] 2 5 8\n[3,] 3 6 9\n\n, , 2\n\n [,1] [,2] [,3]\n[1,] 10 13 16\n[2,] 11 14 17\n[3,] 12 15 18\n\n, , 3\n\n [,1] [,2] [,3]\n[1,] 19 22 25\n[2,] 20 23 26\n[3,] 21 24 27\n\n"
]
],
[
[
"#### Listas\n\nAs listas são como vetores, mas podem conter diferentes tipos de dados concomitantemente, e também vetores, entre seus elementos. ",
"_____no_output_____"
]
],
[
[
"l <- list(c(1,2,3),'a', 1, 1+5i)\n\nl",
"_____no_output_____"
]
],
[
[
"#### Data Frames\n\n\nData frames são como listas de vetores com o mesmo comprimento. São usados para armazenar dados em forma de tabela. \n\nPara criar um data frame podemos fazer, por exemplo:\n",
"_____no_output_____"
]
],
[
[
"data <- data.frame(\n Nome = c(\"Thereza\", \"Diana\"),\n Genero = c('F','F'),\n Idade = c(20, 23)\n)\n\ndata",
"_____no_output_____"
]
],
[
[
"(Para mim, aqui reside a beleza do R ... essa simplicidade!)\n\nPara acessar uma coluna em particular, é simplesmente usar o ```$``` e o nome da coluna. \nPor exemplo para ver os nomes:",
"_____no_output_____"
]
],
[
[
"data$Nome",
"_____no_output_____"
]
],
[
[
"Se queremos ver só uma linha (instância ou observação do seu experimento, medição) chamamos o número da linha",
"_____no_output_____"
]
],
[
[
"data[1,]",
"_____no_output_____"
]
],
[
[
"E R tem várias funções para importar arquivos (em formato texto, csv, até xls!) direto para data frames. \n\nComo não amar?",
"_____no_output_____"
],
[
"## Explorando o CMS Open Data\n\n\nAgora vamos à tarefa em mãos: analisar dados do CMS.\n\n---\n\n### Importar dados dos arquivos CSV \n\nNo portal do [CERN Open Data](http://opendata.cern.ch) tem vários conjuntos de dados disponíveis. Nós vamos usar dados que já foram reduzidos ao formato CSV (comma-separated values), importá-los em R e analizar seu conteúdo. \n\nOs dados desse tutorial vêm do seguinte registro: [http://opendata.cern.ch/record/545](http://opendata.cern.ch/record/545)\n\nPara importar usamos o seguinte comando:",
"_____no_output_____"
]
],
[
[
"mumu <- read.csv(\"http://opendata.cern.ch/record/545/files/Dimuon_DoubleMu.csv\")",
"_____no_output_____"
]
],
[
[
"O comando anterior carregou os dados do arquivo `Dimuon_DoubleMu.csv` numa variável chamada `mumu`, que é um data frame. \n\nPara olhar o conteúdo das primeiras 6 linhas podemos chamar a função `head` e para saber o número de observações usamos a função `nrow`",
"_____no_output_____"
]
],
[
[
"nrow(mumu)\nhead(mumu)",
"_____no_output_____"
]
],
[
[
"O nosso conjunto de dados tem 100 mil linhas (cada linha é um evento de colisão) e 21 colunas (cada coluna é uma variável da descrição ou das medições dos produtos finais do evento).\n\n\nNesse ponto já podemos chamar o *tidyverse*. Para ter uma visualização mais \"agradável\" dos dados podemos mudar de data frame para *tibble*.",
"_____no_output_____"
]
],
[
[
"require(tidyverse)\ntbmumu<- mumu %>% as_tibble()",
"Loading required package: tidyverse\nWarning message in as.POSIXlt.POSIXct(Sys.time()):\n\"unknown timezone 'zone/tz/2019b.1.0/zoneinfo/America/Sao_Paulo'\"-- \u001b[1mAttaching packages\u001b[22m --------------------------------------- tidyverse 1.2.1 --\n\u001b[32mv\u001b[39m \u001b[34mggplot2\u001b[39m 3.2.1 \u001b[32mv\u001b[39m \u001b[34mpurrr \u001b[39m 0.3.3\n\u001b[32mv\u001b[39m \u001b[34mtibble \u001b[39m 2.1.3 \u001b[32mv\u001b[39m \u001b[34mdplyr \u001b[39m 0.8.3\n\u001b[32mv\u001b[39m \u001b[34mtidyr \u001b[39m 1.0.0 \u001b[32mv\u001b[39m \u001b[34mstringr\u001b[39m 1.4.0\n\u001b[32mv\u001b[39m \u001b[34mreadr \u001b[39m 1.1.1 \u001b[32mv\u001b[39m \u001b[34mforcats\u001b[39m 0.4.0\n-- \u001b[1mConflicts\u001b[22m ------------------------------------------ tidyverse_conflicts() --\n\u001b[31mx\u001b[39m \u001b[34mdplyr\u001b[39m::\u001b[32mfilter()\u001b[39m masks \u001b[34mstats\u001b[39m::filter()\n\u001b[31mx\u001b[39m \u001b[34mdplyr\u001b[39m::\u001b[32mlag()\u001b[39m masks \u001b[34mstats\u001b[39m::lag()\n"
]
],
[
[
"O tidyverse inclui o pacote `magrittr`, que introduz o operador *pipe* (como tubulação, mas também \"ce ci n'est pas une pipe\") com símbolo `%>%` o que entrega o objeto da esquerda como argumento à função da direita. E com esses encanamentos é possível fazer várias operações sucessivas de forma concisa.\n\nEntão o código acima aplica a função `as_tibble` ao data frame `mumu` e o resultado (que é um *tibble*) é armazenado na variável `tbmumu`.\n\nLogo ao imprimir as primeiras 6 linhas da nossa tabela tipo *tibble* temos uma visualização que: \n\n * cabe na tela\n * dá informações sobre aquilo que não coube\n ",
"_____no_output_____"
]
],
[
[
"print(head(tbmumu))",
"\u001b[38;5;246m# A tibble: 6 x 21\u001b[39m\n Run Event type1 E1 px1 py1 pz1 pt1 eta1 phi1 Q1 type2\n \u001b[3m\u001b[38;5;246m<int>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<int>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<fct>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<int>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<fct>\u001b[39m\u001b[23m\n\u001b[38;5;250m1\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.46\u001b[38;5;246me\u001b[39m7 G 9.70 -\u001b[31m9\u001b[39m\u001b[31m.\u001b[39m\u001b[31m51\u001b[39m 0.366 1.86 9.52 0.194 3.10 -\u001b[31m1\u001b[39m G \n\u001b[38;5;250m2\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.51\u001b[38;5;246me\u001b[39m7 G 6.20 -\u001b[31m4\u001b[39m\u001b[31m.\u001b[39m\u001b[31m27\u001b[39m 0.456 -\u001b[31m4\u001b[39m\u001b[31m.\u001b[39m\u001b[31m48\u001b[39m 4.29 -\u001b[31m0\u001b[39m\u001b[31m.\u001b[39m\u001b[31m912\u001b[39m 3.04 -\u001b[31m1\u001b[39m G \n\u001b[38;5;250m3\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.56\u001b[38;5;246me\u001b[39m7 G 19.3 -\u001b[31m4\u001b[39m\u001b[31m.\u001b[39m\u001b[31m21\u001b[39m -\u001b[31m0\u001b[39m\u001b[31m.\u001b[39m\u001b[31m652\u001b[39m 18.8 4.26 2.19 -\u001b[31m2\u001b[39m\u001b[31m.\u001b[39m\u001b[31m99\u001b[39m -\u001b[31m1\u001b[39m G \n\u001b[38;5;250m4\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.57\u001b[38;5;246me\u001b[39m7 G 7.04 -\u001b[31m6\u001b[39m\u001b[31m.\u001b[39m\u001b[31m33\u001b[39m -\u001b[31m0\u001b[39m\u001b[31m.\u001b[39m\u001b[31m268\u001b[39m 3.08 6.33 0.469 -\u001b[31m3\u001b[39m\u001b[31m.\u001b[39m\u001b[31m10\u001b[39m -\u001b[31m1\u001b[39m G \n\u001b[38;5;250m5\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.59\u001b[38;5;246me\u001b[39m7 G 7.28 0.103 -\u001b[31m5\u001b[39m\u001b[31m.\u001b[39m\u001b[31m53\u001b[39m -\u001b[31m4\u001b[39m\u001b[31m.\u001b[39m\u001b[31m72\u001b[39m 5.53 -\u001b[31m0\u001b[39m\u001b[31m.\u001b[39m\u001b[31m774\u001b[39m -\u001b[31m1\u001b[39m\u001b[31m.\u001b[39m\u001b[31m55\u001b[39m -\u001b[31m1\u001b[39m G \n\u001b[38;5;250m6\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.46\u001b[38;5;246me\u001b[39m7 G 14.9 -\u001b[31m9\u001b[39m\u001b[31m.\u001b[39m\u001b[31m17\u001b[39m -\u001b[31m10\u001b[39m\u001b[31m.\u001b[39m\u001b[31m3\u001b[39m -\u001b[31m5\u001b[39m\u001b[31m.\u001b[39m\u001b[31m71\u001b[39m 13.8 -\u001b[31m0\u001b[39m\u001b[31m.\u001b[39m\u001b[31m402\u001b[39m -\u001b[31m2\u001b[39m\u001b[31m.\u001b[39m\u001b[31m30\u001b[39m -\u001b[31m1\u001b[39m T \n\u001b[38;5;246m# ... with 9 more variables: E2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, px2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, py2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, pz2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m,\n# pt2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, eta2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, phi2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, Q2 \u001b[3m\u001b[38;5;246m<int>\u001b[38;5;246m\u001b[23m, M \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m\u001b[39m\n"
]
],
[
[
"Do output acima podemos ver as primeiras 12 colunas, com seus tipos, com cores para valores negativos, e temos a informação adicional de 9 variáveis não mostradas. Podemos acessar as colunas e linhas da mesma forma do data frame.",
"_____no_output_____"
]
],
[
[
"# imprime os primeiros 6 elementos da coluna chamada E1\n# o que é retornado com esse operador é um vetor\nprint(head(tbmumu$E1))",
"[1] 9.6987 6.2039 19.2892 7.0427 7.2751 14.9422\n"
],
[
"# imprime a primeira linha de dados\n# retora um novo tibble que é sub-conjunto do original\nprint(tbmumu[1,])",
"\u001b[38;5;246m# A tibble: 1 x 21\u001b[39m\n Run Event type1 E1 px1 py1 pz1 pt1 eta1 phi1 Q1 type2\n \u001b[3m\u001b[38;5;246m<int>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<int>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<fct>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<int>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<fct>\u001b[39m\u001b[23m\n\u001b[38;5;250m1\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.46\u001b[38;5;246me\u001b[39m7 G 9.70 -\u001b[31m9\u001b[39m\u001b[31m.\u001b[39m\u001b[31m51\u001b[39m 0.366 1.86 9.52 0.194 3.10 -\u001b[31m1\u001b[39m G \n\u001b[38;5;246m# ... with 9 more variables: E2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, px2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, py2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, pz2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m,\n# pt2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, eta2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, phi2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, Q2 \u001b[3m\u001b[38;5;246m<int>\u001b[38;5;246m\u001b[23m, M \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m\u001b[39m\n"
],
[
"# Este outro exemplo retorna o subconjunto das 10 primeiras linhas\nprint(tbmumu[1:10,])",
"\u001b[38;5;246m# A tibble: 10 x 21\u001b[39m\n Run Event type1 E1 px1 py1 pz1 pt1 eta1 phi1 Q1\n \u001b[3m\u001b[38;5;246m<int>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<int>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<fct>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<int>\u001b[39m\u001b[23m\n\u001b[38;5;250m 1\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.46\u001b[38;5;246me\u001b[39m7 G 9.70 -\u001b[31m9\u001b[39m\u001b[31m.\u001b[39m\u001b[31m51\u001b[39m 0.366 1.86 9.52 0.194 3.10 -\u001b[31m1\u001b[39m\n\u001b[38;5;250m 2\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.51\u001b[38;5;246me\u001b[39m7 G 6.20 -\u001b[31m4\u001b[39m\u001b[31m.\u001b[39m\u001b[31m27\u001b[39m 0.456 -\u001b[31m4\u001b[39m\u001b[31m.\u001b[39m\u001b[31m48\u001b[39m 4.29 -\u001b[31m0\u001b[39m\u001b[31m.\u001b[39m\u001b[31m912\u001b[39m 3.04 -\u001b[31m1\u001b[39m\n\u001b[38;5;250m 3\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.56\u001b[38;5;246me\u001b[39m7 G 19.3 -\u001b[31m4\u001b[39m\u001b[31m.\u001b[39m\u001b[31m21\u001b[39m -\u001b[31m0\u001b[39m\u001b[31m.\u001b[39m\u001b[31m652\u001b[39m 18.8 4.26 2.19 -\u001b[31m2\u001b[39m\u001b[31m.\u001b[39m\u001b[31m99\u001b[39m -\u001b[31m1\u001b[39m\n\u001b[38;5;250m 4\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.57\u001b[38;5;246me\u001b[39m7 G 7.04 -\u001b[31m6\u001b[39m\u001b[31m.\u001b[39m\u001b[31m33\u001b[39m -\u001b[31m0\u001b[39m\u001b[31m.\u001b[39m\u001b[31m268\u001b[39m 3.08 6.33 0.469 -\u001b[31m3\u001b[39m\u001b[31m.\u001b[39m\u001b[31m10\u001b[39m -\u001b[31m1\u001b[39m\n\u001b[38;5;250m 5\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.59\u001b[38;5;246me\u001b[39m7 G 7.28 0.103 -\u001b[31m5\u001b[39m\u001b[31m.\u001b[39m\u001b[31m53\u001b[39m -\u001b[31m4\u001b[39m\u001b[31m.\u001b[39m\u001b[31m72\u001b[39m 5.53 -\u001b[31m0\u001b[39m\u001b[31m.\u001b[39m\u001b[31m774\u001b[39m -\u001b[31m1\u001b[39m\u001b[31m.\u001b[39m\u001b[31m55\u001b[39m -\u001b[31m1\u001b[39m\n\u001b[38;5;250m 6\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.46\u001b[38;5;246me\u001b[39m7 G 14.9 -\u001b[31m9\u001b[39m\u001b[31m.\u001b[39m\u001b[31m17\u001b[39m -\u001b[31m10\u001b[39m\u001b[31m.\u001b[39m\u001b[31m3\u001b[39m -\u001b[31m5\u001b[39m\u001b[31m.\u001b[39m\u001b[31m71\u001b[39m 13.8 -\u001b[31m0\u001b[39m\u001b[31m.\u001b[39m\u001b[31m402\u001b[39m -\u001b[31m2\u001b[39m\u001b[31m.\u001b[39m\u001b[31m30\u001b[39m -\u001b[31m1\u001b[39m\n\u001b[38;5;250m 7\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.47\u001b[38;5;246me\u001b[39m7 G 42.0 -\u001b[31m1\u001b[39m\u001b[31m.\u001b[39m\u001b[31m48\u001b[39m -\u001b[31m12\u001b[39m\u001b[31m.\u001b[39m\u001b[31m5\u001b[39m 40.0 12.6 1.88 -\u001b[31m1\u001b[39m\u001b[31m.\u001b[39m\u001b[31m69\u001b[39m -\u001b[31m1\u001b[39m\n\u001b[38;5;250m 8\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.47\u001b[38;5;246me\u001b[39m7 G 14.4 -\u001b[31m4\u001b[39m\u001b[31m.\u001b[39m\u001b[31m34\u001b[39m -\u001b[31m6\u001b[39m\u001b[31m.\u001b[39m\u001b[31m0\u001b[39m\u001b[31m7\u001b[39m -\u001b[31m12\u001b[39m\u001b[31m.\u001b[39m\u001b[31m3\u001b[39m 7.46 -\u001b[31m1\u001b[39m\u001b[31m.\u001b[39m\u001b[31m28\u001b[39m -\u001b[31m2\u001b[39m\u001b[31m.\u001b[39m\u001b[31m19\u001b[39m 1\n\u001b[38;5;250m 9\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.52\u001b[38;5;246me\u001b[39m7 G 9.76 2.39 -\u001b[31m3\u001b[39m\u001b[31m.\u001b[39m\u001b[31m78\u001b[39m 8.67 4.47 1.42 -\u001b[31m1\u001b[39m\u001b[31m.\u001b[39m\u001b[31m0\u001b[39m\u001b[31m1\u001b[39m -\u001b[31m1\u001b[39m\n\u001b[38;5;250m10\u001b[39m \u001b[4m1\u001b[24m\u001b[4m6\u001b[24m\u001b[4m5\u001b[24m617 7.52\u001b[38;5;246me\u001b[39m7 G 10.2 0.476 -\u001b[31m8\u001b[39m\u001b[31m.\u001b[39m\u001b[31m52\u001b[39m 5.52 8.53 0.609 -\u001b[31m1\u001b[39m\u001b[31m.\u001b[39m\u001b[31m51\u001b[39m -\u001b[31m1\u001b[39m\n\u001b[38;5;246m# ... with 10 more variables: type2 \u001b[3m\u001b[38;5;246m<fct>\u001b[38;5;246m\u001b[23m, E2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, px2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, py2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m,\n# pz2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, pt2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, eta2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, phi2 \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m, Q2 \u001b[3m\u001b[38;5;246m<int>\u001b[38;5;246m\u001b[23m, M \u001b[3m\u001b[38;5;246m<dbl>\u001b[38;5;246m\u001b[23m\u001b[39m\n"
]
],
[
[
"### Calcular a Massa invariante\n\nNossa tabela tem observações de colisões com 2 *múons* no estado final.\n\nComo vimos na tabela, temos valores para energia (E), o momentum linear (px, py, pz), a *pseudo-rapidez* (eta ou η, que tem relação com o ángulo polar) e o ángulo azimutal (phi ou φ). \n\nPodemos calcular a massa invariante, ou seja a energia equivalente em repouso que produziu esses múons, com a seguinte equação: \n\n$M = \\sqrt{(\\sum{E})^2 - ||\\sum{p}||^2}$\n\nonde $M$ é a massa invariante, $\\sum{E}$ é o total da soma das energias (cinética relativística) das partículas finais, e $\\sum{p}$ é o total da soma dos momentos lineares. \n\nNo nosso código, vamos calcular a massa invariante usando os valores de `px`, `py` e `pz` e a energia dos dois múons. Primeiramente precisamos calcular a soma vetorial do momentum. \n\nA função `mutate` do **tidyverse** faz o cálculo especificado para cada observação e _adiciona novas variáveis_, nesse casso `ptotal`, `E` e `mass`",
"_____no_output_____"
]
],
[
[
"tbmumu<-tbmumu%>%mutate(ptotal = sqrt((px1+px2)^2 + (py1+py2)^2 + (pz1+pz2)^2), \n E = E1+E2, \n mass = sqrt(E^2 - ptotal^2))\ntbmumu%>% select(Run, Event, ptotal,E, mass)%>%head()",
"Warning message in sqrt(E^2 - ptotal^2):\n\"NaNs produced\""
]
],
[
[
"É possível também definir uma função para fazer nosso cálculo:\n\n```\nmyfunctionname = function(arg1, arg2...)\n{\nstatements\nreturn(a) \n}\n```\n\nPor exemplo podemos definir uma função para a magnitude soma vetorial de dois vetores de 3 componentes, e outra função que entrega o resultado para a massa invariante a partir de `ptotal` e `E`",
"_____no_output_____"
]
],
[
[
"sumvecmag = function(x1,x2,y1,y2,z1,z2){\n x = x1+x2\n y = y1+y2\n z = z1+z2\n tot = sqrt(x^2+y^2+z^2)\n return(tot)\n}\n\ninvmass = function(ptot, E) {\n m = sqrt(E^2 - ptot^2)\n return(m)\n}",
"_____no_output_____"
]
],
[
[
"Agora podemos adicionar uma nova coluna calculada chamando as funções definidas :",
"_____no_output_____"
]
],
[
[
"tbmumu<- tbmumu %>% mutate( \n ptotal_f = sumvecmag( px1, px2, py1, py2 , pz1, pz2),\n E = E1 + E2,\n mass_f=invmass(ptotal_f,E)) \n\n# Visualizar as primeiras 6 linhas do tibble, selecionando só as colunas do meu interesse\nprint(head(tbmumu%>% select(ptotal,ptotal_f, E, mass, mass_f)))",
"Warning message in sqrt(E^2 - ptot^2):\n\"NaNs produced\""
]
],
[
[
"### Fazer um Histograma\n\nEm física de partículas trabalhamos com distribuições de frequências, quer dizer histogramas.\n\nNesse caso, quero olhar só uma porção dos dados, onde a variável massa esta entre 1.1 e 5 (GeV). Para isto posso utilizar a função `filter` do tidyverse, com operadores `%>%`\n",
"_____no_output_____"
]
],
[
[
"tbsel <- tbmumu%>% filter(mass>1.1 & mass < 5) ",
"_____no_output_____"
]
],
[
[
"A visualização do gráfico do histograma pode ser feita com a função própria básica do R",
"_____no_output_____"
]
],
[
[
"Sys.setlocale(locale = \"en_US.UTF-8\") #Para ter carateres de acentos\nlibrary(repr)\noptions(repr.plot.width=6,repr.plot.height=4 ) #Para ter gráficos de tamanho que caiba na tela",
"_____no_output_____"
],
[
"hist(tbsel$mass, breaks = 200, xlim=c(1,5),\n main=\"Histograma da Massa Invariante\",\n xlab = \"Massa (GeV)\",ylab=\"Frequência \", \n lty=\"blank\", \n col=\"purple\")",
"_____no_output_____"
]
],
[
[
"Observamos um pico maior perto do valor de $3.1$ GeV e outro pequeno perto de $3.7$ GeV.\nEsses valores correspondem às massas de duas partículas que decaem em dois múons ou mais específicamente, um múon e um anti-múon (múon carregado positivamente). \n\nOlhando na base de dados do [Particle Data Group](http://pdg.lbl.gov/), podemos ver que essas partículas são os **mésons** (partículas **hadrônicas** compostas de um quark e um anti-quark) ***J/ψ(1S)*** e ***ψ(2S)***, respectivamente. \n",
"_____no_output_____"
],
[
"### Graficando com o tidyverse\n\nPodemos condensar o processo de importar, manipular as variáveis e graficar o histograma com um código muito enxuto:\n\n",
"_____no_output_____"
]
],
[
[
"read_csv(\"http://opendata.cern.ch/record/545/files/Dimuon_DoubleMu.csv\", \n col_types = cols()) %>% \n mutate(ptotal = sqrt((px1+px2)^2 + (py1+py2)^2 + (pz1+pz2)^2), \n E = E1+E2, \n mass = sqrt(E^2 - ptotal^2)) %>% \n filter(mass >0.1 & mass<120) %>% \n ggplot(aes(mass)) + \n geom_histogram(bins = 250, fill = \"purple\", alpha = 0.5) + \n xlab(\"Massa (GeV)\") + \n ylab(\"Frequência\") + \n scale_x_continuous(trans = 'log10') +\n scale_y_continuous(trans = 'log10') +\n ggtitle(\"Espectro de di-múons no CMS\") + \n theme_bw() + \n theme(plot.title = element_text(hjust = 0.5))",
"Warning message in sqrt(E^2 - ptotal^2):\n“NaNs produced”Warning message:\n“Transformation introduced infinite values in continuous y-axis”Warning message:\n“Removed 3 rows containing missing values (geom_bar).”"
]
],
[
[
"Agora está ficando bom!\n\nA cadeia de comandos pode ser lida como uma frase, com uma sucessão de **ações** sobre os dados:\n\n\"**Leia** o arquivo, \n logo **mude** o conteúdo criando as novas variáveis `ptotal`,`E` e `mass`, \n logo **filtre** para ver só as observações no intervalo desejado, \n logo **grafique** com os parâmetros apropriados\"\n\nO pacote de gráficos do tidyverse é ``ggplot2``, onde as diferentes opções do gráfico estão sequenciadas com símbolo `+`. Neste caso eu escolhi usar uma escala `log-log` que permite ter uma visualização abrangente de várias ordens de grandeza, e assim observar vários picos de ressonância.",
"_____no_output_____"
],
[
"As opções do gráfico são:\n\n- `ggplot()` a função central do pacote `ggplot2`, que trabalha com o princípio de *camadas*:\n 1. `aes(mass)` quer dizer que vamos usar a variável `mass` \n 1. `geom_histogram()` tomar a variável e fazer o histograma\n 1. `xlab()` e `ylab()` os nomes dos eixos\n 1. `ggtitle()` título do gráfico\n 1. `theme_bw()` tema preto-e-branco, \n 1. `theme()` permite manipular alguns elementos específicos do gráfico",
"_____no_output_____"
],
[
"## Ajustando uma função ao pico do $J/\\psi$\n\nVoltando ao *tibble* onde já habiamos selecionado o intervalo que apresenta o pico do méson $J/\\psi$, podemos chamar a função hist sem graficar para ter só os resultados das frequências nos intervalos.",
"_____no_output_____"
]
],
[
[
"a<-hist(tbsel$mass,breaks=200,plot=FALSE)",
"_____no_output_____"
],
[
"mydf<- data.frame(x=a$mids, nobs=a$counts) \nprint(head(mydf))",
" x nobs\n1 1.11 123\n2 1.13 128\n3 1.15 112\n4 1.17 116\n5 1.19 142\n6 1.21 113\n"
],
[
"library(latex2exp)\nmydf %>%\n ggplot(aes(x,nobs, ymin=nobs-sqrt(nobs),ymax=nobs+sqrt(nobs))) + \n geom_point() + \n geom_errorbar() + \n xlab(\"Massa (GeV)\")+\n ylab(\"Frequência\")+\n ggtitle(TeX(\"Histograma do pico dos mésons J/$\\\\psi$ e $\\\\psi$\"))+\n theme_bw() + theme(plot.title = element_text(hjust = 0.5))",
"_____no_output_____"
]
],
[
[
"### A função que descreve os dados\n\nUma possível função para descrever esses dois picos seria a soma de uma gaussiana com média perto de $3.1$, outra com média perto de $3.7$ e uma reta decrescente como \"base\" (nosso *background*).",
"_____no_output_____"
]
],
[
[
"my2gausspluslin <- function(x, mean1, sigma1, norm1,mean2,sigma2,norm2,a,b) {\n \n f <- norm1 * exp(-1*((x-mean1)/sigma1)^2)+ norm2 * exp(-1*((x-mean2)/sigma2)^2) + a*x +b\n return(f)\n}\n",
"_____no_output_____"
]
],
[
[
"Vou chamar a função `nls` que faz a optimização dos parámetros da função com mínimos quadrados não lineares [documentação](https://stat.ethz.ch/R-manual/R-devel/library/stats/html/nls.html).",
"_____no_output_____"
]
],
[
[
"res <- nls( nobs ~ my2gausspluslin(x,mean1,sigma1,norm1,mean2,sigma2,norm2,a,b), \n data = mydf, \n start=list(mean1=3.1, sigma1=0.1, norm1=3000,mean2=3.7,sigma2=0.05,norm2=30,a=-10,b=100))\nsummary(res)",
"_____no_output_____"
]
],
[
[
"Aqui o resultado foi salvo na variável `res`, e posso aplicar esse resultado com a função `predict`. Vamos adicionar uma coluna do número calculado a partir da função ao data frame e salvar com um novo nome de `nexp` por *expected*, número esperado segundo um modelo de 2 gaussianas mais um fundo linear.",
"_____no_output_____"
]
],
[
[
"newdf<- mydf%>% mutate(nexp = predict(res,mydf))\nprint(head(newdf))",
" x nobs nexp\n1 1.11 123 132.9737\n2 1.13 128 132.3149\n3 1.15 112 131.6560\n4 1.17 116 130.9972\n5 1.19 142 130.3383\n6 1.21 113 129.6794\n"
]
],
[
[
"Graficamos a predição",
"_____no_output_____"
]
],
[
[
"newdf%>%\nggplot(aes(x,nexp))+\n geom_path(color=\"purple\")+\n xlab(\"Massa (GeV)\")+\n ylab(\"frequência\")+ \n ggtitle(\"Predição do ajuste da função gaussiana + reta decrescente\")+\n theme_bw() + theme(plot.title = element_text(hjust = 0.5))",
"_____no_output_____"
]
],
[
[
"### Resultado \n\nFinalmente graficamos os pontos das frequências observadas (com erros de distribuição de *Poisson*, $\\sigma_n =\\sqrt{n}$ ) junto com a linha da predição.",
"_____no_output_____"
]
],
[
[
"ggplot(newdf) +\n geom_path(aes(x,nexp),color=\"purple\")+\n geom_point(aes(x,nobs))+\n geom_errorbar(aes(x,nobs, ymin=nobs-sqrt(nobs),ymax=nobs+sqrt(nobs)))+\n xlab(\"Massa (GeV)\")+\n ylab(\"frequência\")+ \n ggtitle(\"Resultado dos dados com a função do ajuste\")+\n theme_bw() +theme(plot.title = element_text(hjust = 0.5))",
"Warning message:\n“Ignoring unknown aesthetics: y”"
]
],
[
[
"## Motivação!\n\nAgora estou entusiasmada, vamos dar uma olhada no pico de maior massa nesse espectro?\nÉ o pico do bóson Z, que é análogo a um fóton, só que com massa alta (para uma partícula subatómica).",
"_____no_output_____"
]
],
[
[
"tbZboson<-tbmumu %>% filter(mass>70 & mass <110)\n\n\ntbZboson %>% \nggplot(aes(mass)) + \n geom_histogram(bins = 80, fill = \"purple\", alpha = 0.5) + \n xlab(\"Massa (GeV)\") + \n ylab(\"Frequência\") + \n ggtitle(\"Pico do bóson Z\") + \n theme_bw() + \n theme(plot.title = element_text(hjust = 0.5))",
"_____no_output_____"
],
[
"tbZboson %>% filter(abs(eta1)<2.4 & abs(eta2)<2.4, pt1>20 & pt2>20, type1==\"G\" & type2==\"G\") %>%\nggplot(aes(mass)) + \n geom_histogram(bins = 80, fill = \"purple\", alpha = 0.5) + \n xlab(\"Massa (GeV)\") + \n ylab(\"Frequência\") + \n ggtitle(\"Pico do bóson Z\") + \n theme_bw() + \n theme(plot.title = element_text(hjust = 0.5))",
"_____no_output_____"
],
[
"zfilt<- tbZboson %>% filter(abs(eta1)<2.4 & abs(eta2)<2.4, pt1>20 & pt2>20, type1==\"G\" & type2==\"G\") \n\nzh<- hist(zfilt$mass,breaks=80,plot=FALSE)\n\nzdf<-data.frame(x=zh$mids,n=zh$counts)\nprint(head(zdf))",
" x n\n1 70.25 10\n2 70.75 8\n3 71.25 8\n4 71.75 10\n5 72.25 19\n6 72.75 9\n"
],
[
"breitwpluslin <- function(x,M,gamma,N,a,b){\n \n b<- a*x +b \n s<- N*( (2*sqrt(2)*M*gamma*sqrt(M**2*(M**2+gamma**2)))/(pi*sqrt(M**2+sqrt(M**2*(M**2+gamma**2)))) )/((x**2-M**2)**2+M**2*gamma**2)\n return(b+s)\n}",
"_____no_output_____"
],
[
"library(minpack.lm)\nresz <-nlsLM( n~ breitwpluslin(x,m,g,norm,a,b),\n data = zdf, \n start=list(m=90, g=3, norm =100,a=-10,b=100))\nsummary(resz)",
"_____no_output_____"
],
[
"newz<- zdf %>% mutate(nexp=predict(resz,zdf))\nprint(as_tibble(newz))",
"\u001b[38;5;246m# A tibble: 80 x 3\u001b[39m\n x n nexp\n \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<int>\u001b[39m\u001b[23m \u001b[3m\u001b[38;5;246m<dbl>\u001b[39m\u001b[23m\n\u001b[38;5;250m 1\u001b[39m 70.2 10 9.82\n\u001b[38;5;250m 2\u001b[39m 70.8 8 9.91\n\u001b[38;5;250m 3\u001b[39m 71.2 8 10.0 \n\u001b[38;5;250m 4\u001b[39m 71.8 10 10.2 \n\u001b[38;5;250m 5\u001b[39m 72.2 19 10.3 \n\u001b[38;5;250m 6\u001b[39m 72.8 9 10.5 \n\u001b[38;5;250m 7\u001b[39m 73.2 10 10.7 \n\u001b[38;5;250m 8\u001b[39m 73.8 10 10.9 \n\u001b[38;5;250m 9\u001b[39m 74.2 13 11.2 \n\u001b[38;5;250m10\u001b[39m 74.8 14 11.5 \n\u001b[38;5;246m# … with 70 more rows\u001b[39m\n"
],
[
"newz%>%\nggplot(aes(x,nexp))+\n geom_path(color=\"purple\")+\ngeom_point(aes(x,n))+\n geom_errorbar(aes(x,n, ymin=n-sqrt(n),ymax=n+sqrt(n)))+\n xlab(\"Massa (GeV)\")+\n ylab(\"frequência\")+ \n ggtitle(\"Predição do ajuste da função Breit-Wigner + reta decrescente\")+\n theme_bw() + theme(plot.title = element_text(hjust = 0.5))",
"Warning message:\n“Ignoring unknown aesthetics: y”"
]
],
[
[
"Ahhh amei! \nCorações roxinhos 4 ever 💜💜💜",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb4c0a533ecc7342ea3e2a60dc9fd63c59ebf444 | 41,387 | ipynb | Jupyter Notebook | ReembolsoSenadores.ipynb | robsonavr/python | 31c3986689eec9348795ca1d7e456853cfa89f12 | [
"MIT"
] | null | null | null | ReembolsoSenadores.ipynb | robsonavr/python | 31c3986689eec9348795ca1d7e456853cfa89f12 | [
"MIT"
] | null | null | null | ReembolsoSenadores.ipynb | robsonavr/python | 31c3986689eec9348795ca1d7e456853cfa89f12 | [
"MIT"
] | null | null | null | 41.222112 | 217 | 0.375335 | [
[
[
"#Análise de dados reembolso senadores 2018",
"_____no_output_____"
]
],
[
[
"#import bibliotecas\nimport pandas as pd",
"_____no_output_____"
],
[
"df_original = pd.read_csv(r'despesa_ceaps_2018.csv', \n encoding='latin_1', sep=';', decimal=',', thousands='.',\n skiprows=1)\n\ndisplay(df_original.head())",
"_____no_output_____"
],
[
"display(df_original.tail())",
"_____no_output_____"
],
[
"print(df_original.dtypes)",
"ANO int64\nMES int64\nSENADOR object\nTIPO_DESPESA object\nCNPJ_CPF object\nFORNECEDOR object\nDOCUMENTO object\nDATA object\nDETALHAMENTO object\nVALOR_REEMBOLSADO float64\nCOD_DOCUMENTO int64\ndtype: object\n"
],
[
"print(df_original.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 24492 entries, 0 to 24491\nData columns (total 11 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ANO 24492 non-null int64 \n 1 MES 24492 non-null int64 \n 2 SENADOR 24492 non-null object \n 3 TIPO_DESPESA 24492 non-null object \n 4 CNPJ_CPF 24492 non-null object \n 5 FORNECEDOR 24492 non-null object \n 6 DOCUMENTO 23476 non-null object \n 7 DATA 24492 non-null object \n 8 DETALHAMENTO 18882 non-null object \n 9 VALOR_REEMBOLSADO 24492 non-null float64\n 10 COD_DOCUMENTO 24492 non-null int64 \ndtypes: float64(1), int64(3), object(7)\nmemory usage: 2.1+ MB\nNone\n"
],
[
"print(f'A base tem um total de {df_original.shape[0]} linhas e {df_original.shape[1]} colunas')",
"A base tem um total de 24492 linhas e 11 colunas\n"
],
[
"print(df_original.describe())",
" ANO MES VALOR_REEMBOLSADO COD_DOCUMENTO\ncount 24492.0 24492.000000 24492.000000 2.449200e+04\nmean 2018.0 6.192226 1045.431215 2.100346e+06\nstd 0.0 3.305847 2718.840577 7.881637e+03\nmin 2018.0 1.000000 0.010000 2.085892e+06\n25% 2018.0 3.000000 125.000000 2.093630e+06\n50% 2018.0 6.000000 287.200000 2.100348e+06\n75% 2018.0 9.000000 1032.005000 2.107072e+06\nmax 2018.0 12.000000 103900.000000 2.118038e+06\n"
],
[
"#Total de reembolso\nprint(f\"Valor total reembolsado é de R$ {df_original['VALOR_REEMBOLSADO'].sum():,.2f}\")",
"Valor total reembolsado é de R$ 25,604,701.33\n"
],
[
"#Total de recibos por Senador\ndisplay(df_original['SENADOR'].value_counts())",
"_____no_output_____"
],
[
"#Valor total do reembolso por senador\ndisplay(df_original.groupby('SENADOR')['VALOR_REEMBOLSADO'].sum().sort_values(ascending=False))",
"_____no_output_____"
],
[
"#Os 5 maiores valores de reembolso\ndisplay(df_original.nlargest(5, 'VALOR_REEMBOLSADO').T)",
"_____no_output_____"
],
[
"#Os 5 menores valores de reembolso\ndisplay(df_original.nsmallest(5, 'VALOR_REEMBOLSADO').T)",
"_____no_output_____"
],
[
"#Quantidade de reembolso por tipo de despesa\ndf_original['TIPO_DESPESA'].value_counts()",
"_____no_output_____"
],
[
"#Valores totais em relação ao tipo de despesas\ndisplay(df_original.groupby('TIPO_DESPESA')['VALOR_REEMBOLSADO'].sum().sort_values(ascending=False))",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4c0ea897d0c0d03f6b0451e430434c90f9c36a | 531,914 | ipynb | Jupyter Notebook | _notebooks/2020-05-24-01-Relationships.ipynb | AntonovMikhail/chans_jupyter | c2cd1675408238ad5be81ba98994611d8c4e48ae | [
"Apache-2.0"
] | null | null | null | _notebooks/2020-05-24-01-Relationships.ipynb | AntonovMikhail/chans_jupyter | c2cd1675408238ad5be81ba98994611d8c4e48ae | [
"Apache-2.0"
] | null | null | null | _notebooks/2020-05-24-01-Relationships.ipynb | AntonovMikhail/chans_jupyter | c2cd1675408238ad5be81ba98994611d8c4e48ae | [
"Apache-2.0"
] | 1 | 2022-01-26T12:51:05.000Z | 2022-01-26T12:51:05.000Z | 1,065.95992 | 390,184 | 0.958505 | [
[
[
"# Relationships\n> A Summary of lecture \"Exploratory Data Analysis in Python\", via datacamp\n\n- toc: true \n- badges: true\n- comments: true\n- author: Chanseok Kang\n- categories: [Python, Datacamp]\n- image: images/brfss-boxplot.png",
"_____no_output_____"
],
[
"## Exploring relationships\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom empiricaldist import Pmf, Cdf",
"_____no_output_____"
],
[
"brfss_original = pd.read_hdf('./dataset/brfss.hdf5', 'brfss')",
"_____no_output_____"
]
],
[
[
"### PMF of age",
"_____no_output_____"
]
],
[
[
"# Extract age\nage = Pmf.from_seq(brfss_original['AGE'])\n\n# Plot the PMF\nage.bar()\n\n# Label the axes\nplt.xlabel('Age in years')\nplt.ylabel('PMF')",
"_____no_output_____"
]
],
[
[
"### Scatter plot",
"_____no_output_____"
]
],
[
[
"# Select the first 1000 respondents\nbrfss = brfss_original[:1000]\n\n# Extract age and weight\nage = brfss['AGE']\nweight = brfss['WTKG3']\n\n# Make a scatter plot\nplt.plot(age, weight, 'o', alpha=0.1)\n\nplt.xlabel('Age in years')\nplt.ylabel('Weight in kg')\n",
"_____no_output_____"
]
],
[
[
"### Jittering",
"_____no_output_____"
]
],
[
[
"# Select the first 1000 respondents\nbrfss = brfss_original[:1000]\n\n# Add jittering to age\nage = brfss['AGE'] + np.random.normal(0, 2.5, size=len(brfss))\n# Extract weight\nweight = brfss['WTKG3']\n\n# Make a scatter plot\nplt.plot(age, weight, 'o', markersize=4, alpha=0.2)\n\nplt.xlabel('Age in years')\nplt.ylabel('Weight in kg')",
"_____no_output_____"
]
],
[
[
"## Visualizing relationships\n",
"_____no_output_____"
],
[
"### Height and weight\n",
"_____no_output_____"
]
],
[
[
"# Drop rows with missing data\ndata = brfss_original.dropna(subset=['_HTMG10', 'WTKG3'])\n\n# Make a box plot\nsns.boxplot(x='_HTMG10', y='WTKG3', data=data, whis=10)\n\n# Plot the y-axis on a log scale\nplt.yscale('log')\n\n# Remove unneeded lines and label axes\nsns.despine(left=True, bottom=True)\nplt.xlabel('Height in cm')\nplt.ylabel('Weight in kg')\nplt.savefig('../images/brfss-boxplot.png')",
"_____no_output_____"
]
],
[
[
"### Distribution of income",
"_____no_output_____"
]
],
[
[
"# Extract income\nincome = brfss_original['INCOME2']\n\n# Plot the PMF\nPmf.from_seq(income).bar()\n\n# Label the axes\nplt.xlabel('Income level')\nplt.ylabel('PMF')",
"_____no_output_____"
]
],
[
[
"### Income and height",
"_____no_output_____"
]
],
[
[
"# Drop rows with missing data\ndata = brfss_original.dropna(subset=['INCOME2', 'HTM4'])\n\n# Make a violin plot\nsns.violinplot(x = 'INCOME2', y='HTM4', data=data, inner=None)\n\n# Remove unneeded lines and label axes\nsns.despine(left=True, bottom=True)\nplt.xlabel('Income level')\nplt.ylabel('Height in cm')",
"_____no_output_____"
]
],
[
[
"## Correlation",
"_____no_output_____"
],
[
"### Computing correlations\n",
"_____no_output_____"
]
],
[
[
"# Select columns\ncolumns = ['AGE', 'INCOME2', '_VEGESU1']\nsubset = brfss_original[columns]\n\n# Compute the correlation matrix\nprint(subset.corr())",
" AGE INCOME2 _VEGESU1\nAGE 1.000000 -0.015158 -0.009834\nINCOME2 -0.015158 1.000000 0.119670\n_VEGESU1 -0.009834 0.119670 1.000000\n"
]
],
[
[
"## Simple regression",
"_____no_output_____"
],
[
"### Income and vegetables",
"_____no_output_____"
]
],
[
[
"from scipy.stats import linregress",
"_____no_output_____"
],
[
"# Extract the variables\nsubset = brfss_original.dropna(subset=['INCOME2', '_VEGESU1'])\nxs = subset['INCOME2']\nys = subset['_VEGESU1']\n\n# Compute the linear regression\nres = linregress(xs, ys)\nprint(res)",
"LinregressResult(slope=0.06988048092105248, intercept=1.5287786243362973, rvalue=0.11967005884864361, pvalue=1.3785039162157718e-238, stderr=0.002110976356332355)\n"
]
],
[
[
"### Fit a line",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(10, 10))\n# Plot the scatter plot\nx_jitter = xs + np.random.normal(0, 0.15, len(xs))\nplt.plot(x_jitter, ys, 'o', alpha=0.2)\n\n# Plot the line of best fit\nfx = np.array([xs.min(), xs.max()])\nfy = res.intercept + res.slope * fx\nplt.plot(fx, fy, '-', alpha=0.7)\n\nplt.xlabel('Income code')\nplt.ylabel('Vegetable servings per day')\nplt.ylim([0, 6])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4c205856edfeb8254a1a410892695f151f0835 | 19,455 | ipynb | Jupyter Notebook | notebooks/user/pjayasundara/AEFI_with_uncertainty.ipynb | jtrauer/AuTuMN | 2e1defd0104bbecfe667b8ea5ecaf4bc6741905c | [
"BSD-2-Clause-FreeBSD"
] | null | null | null | notebooks/user/pjayasundara/AEFI_with_uncertainty.ipynb | jtrauer/AuTuMN | 2e1defd0104bbecfe667b8ea5ecaf4bc6741905c | [
"BSD-2-Clause-FreeBSD"
] | null | null | null | notebooks/user/pjayasundara/AEFI_with_uncertainty.ipynb | jtrauer/AuTuMN | 2e1defd0104bbecfe667b8ea5ecaf4bc6741905c | [
"BSD-2-Clause-FreeBSD"
] | 1 | 2019-10-22T04:47:34.000Z | 2019-10-22T04:47:34.000Z | 40.030864 | 199 | 0.539913 | [
[
[
"# Import packages\nimport os\nfrom matplotlib import pyplot as plt\nimport pandas as pd\nimport math \n\nimport numpy as np\n\n# Import AuTuMN modules\nfrom autumn.settings import Models, Region\nfrom autumn.settings.folders import OUTPUT_DATA_PATH\nfrom autumn.tools.project import get_project\nfrom autumn.tools import db\nfrom autumn.tools.plots.calibration.plots import calculate_r_hats, get_output_from_run_id\nfrom autumn.tools.plots.uncertainty.plots import _plot_uncertainty, _get_target_values\nfrom autumn.tools.plots.plotter.base_plotter import COLOR_THEME\nfrom autumn.tools.plots.utils import get_plot_text_dict, change_xaxis_to_date, REF_DATE, ALPHAS, COLORS, _apply_transparency, _plot_targets_to_axis\nfrom autumn.models.covid_19.stratifications.agegroup import AGEGROUP_STRATA\n\nimport matplotlib.patches as mpatches\n\nfrom autumn.tools.calibration.utils import get_uncertainty_df",
"_____no_output_____"
],
[
"# Specify model details\nmodel = Models.COVID_19\nregion = Region.MALAYSIA # http://www.autumn-data.com/app/covid_19/region/malaysia/run/1636441221-1a276cd.html\ndirname = \"2021-11-09\"",
"_____no_output_____"
],
[
"# get the relevant project and output data\nproject = get_project(model, region)\nproject_calib_dir = os.path.join(\n OUTPUT_DATA_PATH, \"calibrate\", project.model_name, project.region_name\n)\ncalib_path = os.path.join(project_calib_dir, dirname)\n# Load tables\nmcmc_tables = db.load.load_mcmc_tables(calib_path)\nmcmc_params = db.load.load_mcmc_params_tables(calib_path)\n\nuncertainty_df = get_uncertainty_df(calib_path, mcmc_tables, project.plots)\nscenario_list = uncertainty_df['scenario'].unique()\n\n# make output directories\noutput_dir = f\"{model}_{region}_{dirname}\"\nbase_dir = os.path.join(\"outputs\", output_dir)\nos.makedirs(base_dir, exist_ok=True)\ndirs_to_make = [\"MLE\", \"median\",\"csv_files\"]\nfor dir_to_make in dirs_to_make:\n os.makedirs(os.path.join(base_dir, dir_to_make), exist_ok=True)",
"_____no_output_____"
],
[
"titles = {\n \"notifications\": \"Daily number of notified Covid-19 cases\",\n \"infection_deaths\": \"Daily number of Covid-19 deaths\",\n \"accum_deaths\": \"Cumulative number of Covid-19 deaths\",\n \"incidence\": \"Daily incidence (incl. asymptomatics and undetected)\",\n \"hospital_occupancy\": \"Hospital beds occupied by Covid-19 patients\",\n \"icu_occupancy\": \"ICU beds occupied by Covid-19 patients\",\n \"new_hospital_admissions\": \"New hospital admissions\",\n \"cdr\": \"Proportion detected among symptomatics\",\n \"proportion_vaccinated\": \"Proportion vaccinated\",\n \"prop_incidence_strain_delta\": \"Proportion of Delta variant in new cases\",\n \"accum_notifications\": \"Cumulative Covid-19 notifications\"\n}\n\ndef plot_outputs(output_type, output_name, scenario_list, sc_linestyles, sc_colors, show_v_lines=False, x_min=590, x_max=775):\n\n # plot options\n title = titles[output_name]\n title_fontsize = 18\n label_font_size = 15\n linewidth = 3\n n_xticks = 10\n\n # initialise figure\n fig = plt.figure(figsize=(12, 8))\n plt.style.use(\"ggplot\")\n axis = fig.add_subplot()\n\n # prepare colors for ucnertainty\n n_scenarios_to_plot = len(scenario_list)\n uncertainty_colors = _apply_transparency(COLORS[:n_scenarios_to_plot], ALPHAS[:n_scenarios_to_plot])\n\n if output_type == \"MLE\":\n derived_output_tables = db.load.load_derived_output_tables(calib_path, column=output_name)\n for i, scenario in enumerate(scenario_list): \n linestyle = sc_linestyles[scenario]\n color = sc_colors[scenario]\n\n if output_type == \"MLE\":\n times, values = get_output_from_run_id(output_name, mcmc_tables, derived_output_tables, \"MLE\", scenario)\n axis.plot(times, values, color=color, linestyle=linestyle, linewidth=linewidth)\n elif output_type == \"median\":\n _plot_uncertainty(\n axis,\n uncertainty_df,\n output_name,\n scenario,\n x_max,\n x_min,\n [_, _, _, color],\n overlay_uncertainty=False,\n start_quantile=0,\n zorder=scenario + 1,\n linestyle=linestyle,\n linewidth=linewidth,\n )\n elif output_type == \"uncertainty\":\n scenario_colors = uncertainty_colors[i] \n _plot_uncertainty(\n axis,\n uncertainty_df,\n output_name,\n scenario,\n x_max,\n x_min,\n scenario_colors,\n overlay_uncertainty=True,\n start_quantile=0,\n zorder=scenario + 1,\n )\n else:\n print(\"Please use supported output_type option\")\n\n\n axis.set_xlim((x_min, x_max))\n axis.set_title(title, fontsize=title_fontsize)\n plt.setp(axis.get_yticklabels(), fontsize=label_font_size)\n plt.setp(axis.get_xticklabels(), fontsize=label_font_size)\n change_xaxis_to_date(axis, REF_DATE)\n plt.locator_params(axis=\"x\", nbins=n_xticks)\n \n if output_name == \"accum_notifications\":\n axis.set_ylabel(\"million\")\n \n\n if show_v_lines:\n release_dates = {}\n y_max = plt.gca().get_ylim()[1]\n linestyles = [\"dashdot\", \"solid\"]\n i = 0\n for time, date in release_dates.items():\n plt.vlines(time, ymin=0, ymax=y_max, linestyle=linestyles[i])\n text = f\"Lockdown relaxed on {date}\"\n plt.text(time - 5, .5*y_max, text, rotation=90, fontsize=11)\n i += 1\n \n return axis\n",
"_____no_output_____"
]
],
[
[
"# Scenario plots with single lines",
"_____no_output_____"
]
],
[
[
"output_names = [\"notifications\", \"icu_occupancy\",\"accum_deaths\",\"accum_notifications\"]\nscenario_x_min, scenario_x_max = 610, 913\n\nsc_to_plot = [0, 2]\nlegend = [\"With vaccine\", \"Without vaccine\"]\nlift_time = 731\nvaccine_time = 481 # 25 April, 2021\ntext_font = 14\n\nsc_colors = [COLOR_THEME[i] for i in scenario_list]\nsc_linestyles = [\"solid\"] * (len(scenario_list))\nfor output_type in [\"median\"]:\n for output_name in output_names:\n plot_outputs(output_type, output_name, sc_to_plot, sc_linestyles, sc_colors, False, x_min=scenario_x_min, x_max=scenario_x_max)\n path = os.path.join(base_dir, output_type, f\"{output_name}.png\")\n plt.legend(labels=legend, fontsize=text_font, facecolor=\"white\",loc = \"lower right\")\n \n ymax = plt.gca().get_ylim()[1]\n \n# if \"accum\" in output_name:\n# plt.vlines(x=vaccine_time,ymin=0,ymax=1.05*ymax, linestyle=\"dashed\") # 31 Dec 2021\n# plt.text(x=vaccine_time + 3, y=ymax, s=\"Vaccination starts\", fontsize = text_font, rotation=90, va=\"top\")\n \n# else:\n plt.vlines(x=lift_time,ymin=0,ymax=1.05*ymax, linestyle=\"dashed\") # 31 Dec 2021\n plt.text(x=(scenario_x_min + lift_time) / 2., y=1.* ymax, s=\"Vaccination phase\", ha=\"center\", fontsize = text_font)\n plt.text(x=lift_time + 3, y=ymax/2, s=\"Restrictions lifted\", fontsize = text_font, rotation=90, va=\"top\")\n \n \n \n plt.savefig(path)\n ",
"_____no_output_____"
]
],
[
[
"# Make Adverse Effects figures",
"_____no_output_____"
]
],
[
[
"params = project.param_set.baseline.to_dict()\nae_risk = {\n \"AstraZeneca\": params[\"vaccination_risk\"][\"tts_rate\"],\n \"mRNA\": params[\"vaccination_risk\"][\"myocarditis_rate\"]\n}",
"_____no_output_____"
],
[
"agg_agegroups = [\"10_14\",\"15_19\", \"20_29\", \"30_39\", \"40_49\", \"50_59\", \"60_69\", \"70_plus\"]\ntext_font = 12\n \nvacc_scenarios = {\n \"mRNA\": 2,\n \"AstraZeneca\": 2,\n}\n\nadverse_effects = {\n \"mRNA\": \"myocarditis\",\n \"AstraZeneca\": \"thrombosis with thrombocytopenia syndrome\",\n}\n\nadverse_effects_short= {\n \"mRNA\": \"myocarditis\",\n \"AstraZeneca\": \"tts\",\n}\n\nleft_title = \"COVID-19-associated hospitalisations prevented\"\n\ndef format_age_label(age_bracket):\n if age_bracket.startswith(\"70\"):\n return \"70+\"\n else:\n return age_bracket.replace(\"_\", \"-\")\n \n\n\ndef make_ae_figure(vacc_scenario, log_scale=False):\n trimmed_df = uncertainty_df[\n (uncertainty_df[\"scenario\"]==vacc_scenarios[vacc_scenario]) & (uncertainty_df[\"time\"]==913)\n ] \n \n right_title = f\"Cases of {adverse_effects[vacc_scenario]}\"\n \n fig = plt.figure(figsize=(10, 4))\n plt.style.use(\"default\")\n axis = fig.add_subplot() \n \n h_max = 0\n delta_agegroup = 1.2 if log_scale else 4000 \n barwidth = .7\n text_offset = 0.5 if log_scale else 20\n unc_color = \"black\"\n unc_lw = 1.\n \n for i, age_bracket in enumerate(agg_agegroups):\n y = len(agg_agegroups) - i - .5\n plt.text(x=delta_agegroup / 2, y=y, s=format_age_label(age_bracket), ha=\"center\", va=\"center\", fontsize=text_font)\n \n # get outputs\n hosp_output_name = f\"abs_diff_cumulative_hospital_admissionsXagg_age_{age_bracket}\"\n ae_output_name = f\"abs_diff_cumulative_{adverse_effects_short[vacc_scenario]}_casesXagg_age_{age_bracket}\"\n \n prev_hosp_df = trimmed_df[trimmed_df[\"type\"] == hosp_output_name]\n prev_hosp_values = [ # median, lower, upper\n float(prev_hosp_df['value'][prev_hosp_df[\"quantile\"] == q]) for q in [0.5, 0.025, 0.975]\n ]\n log_prev_hosp_values = [math.log10(v) for v in prev_hosp_values]\n \n ae_df = trimmed_df[trimmed_df[\"type\"] == ae_output_name]\n ae_values = [ # median, lower, upper\n - float(ae_df['value'][ae_df[\"quantile\"] == q]) for q in [0.5, 0.975, 0.025]\n ] \n log_ae_values = [max(math.log10(v), 0) for v in ae_values]\n \n if log_scale:\n plot_h_values = log_prev_hosp_values\n plot_ae_values = log_ae_values\n else:\n plot_h_values = prev_hosp_values\n plot_ae_values = ae_values\n \n h_max = max(plot_h_values[2], h_max) \n \n origin = 0\n # hospital\n rect = mpatches.Rectangle((origin, y - barwidth/2), width=-plot_h_values[0], height=barwidth, facecolor=\"cornflowerblue\")\n axis.add_patch(rect) \n plt.hlines(y=y, xmin=-plot_h_values[1], xmax=-plot_h_values[2], color=unc_color, linewidth=unc_lw)\n \n disp_val = int(prev_hosp_values[0])\n plt.text(x= -plot_h_values[0] - text_offset, y=y + barwidth/2, s=int(disp_val), ha=\"right\", va=\"center\", fontsize=text_font*.7) \n \n min_bar_length = 0\n if not log_scale:\n min_bar_length = 0 if vacc_scenario == \"Astrazeneca\" else 0\n \n rect = mpatches.Rectangle((delta_agegroup + origin, y - barwidth/2), width=max(plot_ae_values[0], min_bar_length), height=barwidth, facecolor=\"tab:red\")\n axis.add_patch(rect)\n plt.hlines(y=y, xmin=delta_agegroup + origin + plot_ae_values[1], xmax=delta_agegroup + origin + plot_ae_values[2], color=unc_color, linewidth=unc_lw)\n \n disp_val = int(ae_values[0])\n plt.text(x=delta_agegroup + origin + max(plot_ae_values[0], min_bar_length) + text_offset, y=y + barwidth/2, s=int(disp_val), ha=\"left\", va=\"center\", fontsize=text_font*.7) \n\n # main title\n axis.set_title(f\"Benefit/Risk analysis with {vacc_scenario} vaccine\", fontsize = text_font + 2)\n \n # x axis ticks\n if log_scale:\n max_val_display = math.ceil(h_max)\n else:\n magnitude = 500\n max_val_display = math.ceil(h_max / magnitude) * magnitude \n \n # sub-titles \n plt.text(x= - max_val_display / 2, y=len(agg_agegroups) + .3, s=left_title, ha=\"center\", fontsize=text_font)\n plt.text(x= max_val_display / 2 + delta_agegroup, y=len(agg_agegroups) + .3, s=right_title, ha=\"center\", fontsize=text_font)\n \n if log_scale:\n ticks = range(max_val_display + 1)\n rev_ticks = [-t for t in ticks]\n rev_ticks.reverse() \n x_ticks = rev_ticks + [delta_agegroup + t for t in ticks]\n \n labels = [10**(p) for p in range(max_val_display + 1)]\n rev_labels = [l for l in labels]\n rev_labels.reverse()\n x_labels = rev_labels + labels \n x_labels[max_val_display] = x_labels[max_val_display + 1] = 0\n else:\n n_ticks = 6\n x_ticks = [-max_val_display + j * (max_val_display/(n_ticks - 1)) for j in range(n_ticks)] + [delta_agegroup + j * (max_val_display/(n_ticks - 1)) for j in range(n_ticks)]\n rev_n_ticks = x_ticks[:n_ticks]\n rev_n_ticks.reverse()\n x_labels = [int(-v) for v in x_ticks[:n_ticks]] + [int(-v) for v in rev_n_ticks]\n \n plt.xticks(ticks=x_ticks, labels=x_labels)\n \n # x, y lims\n axis.set_xlim((-max_val_display, max_val_display + delta_agegroup))\n axis.set_ylim((0, len(agg_agegroups) + 1)) \n \n # remove axes\n axis.set_frame_on(False)\n axis.axes.get_yaxis().set_visible(False)\n\n log_ext = \"_log_scale\" if log_scale else \"\" \n path = os.path.join(base_dir, f\"{vacc_scenario}_adverse_effects{log_ext}.png\") \n plt.tight_layout()\n plt.savefig(path, dpi=600)\n\nfor vacc_scenario in [\"mRNA\", \"AstraZeneca\"]:\n for log_scale in [False,True]:\n make_ae_figure(vacc_scenario, log_scale) \n ",
"_____no_output_____"
]
],
[
[
"# Counterfactual no vaccine scenario",
"_____no_output_____"
]
],
[
[
"output_type = \"uncertainty\"\noutput_names = [\"notifications\", \"icu_occupancy\", \"accum_deaths\"]\nsc_to_plot = [0, 1]\nx_min, x_max = 400, 670\nvacc_start = 426\nfor output_name in output_names:\n axis = plot_outputs(output_type, output_name, sc_to_plot, sc_linestyles, sc_colors, False, x_min=400, x_max=670)\n y_max = plt.gca().get_ylim()[1]\n plt.vlines(x=vacc_start, ymin=0, ymax=y_max, linestyle=\"dashdot\")\n plt.text(x=vacc_start - 5, y=.6 * y_max, s=\"Vaccination starts\", rotation=90, fontsize=12)\n \n path = os.path.join(base_dir, f\"{output_name}_counterfactual.png\") \n plt.tight_layout()\n plt.savefig(path, dpi=600)\n",
"_____no_output_____"
]
],
[
[
"# number of lives saved",
"_____no_output_____"
]
],
[
[
"today = 660 # 21 Oct\ndf = uncertainty_df[(uncertainty_df[\"type\"] == \"accum_deaths\") & (uncertainty_df[\"quantile\"] == 0.5) & (uncertainty_df[\"time\"] == today)]\n\nbaseline = float(df[df[\"scenario\"] == 0][\"value\"])\ncounterfact = float(df[df[\"scenario\"] == 1][\"value\"])\n\nprint(counterfact - baseline)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4c27ab177e0f71ec5ad3319f9bf07d6129d771 | 26,986 | ipynb | Jupyter Notebook | Tutorial-BSSN_in_terms_of_ADM.ipynb | rhaas80/nrpytutorial | 4398cd6b5a071c8fb8b2b584a01f07a4591dd5f4 | [
"BSD-2-Clause"
] | null | null | null | Tutorial-BSSN_in_terms_of_ADM.ipynb | rhaas80/nrpytutorial | 4398cd6b5a071c8fb8b2b584a01f07a4591dd5f4 | [
"BSD-2-Clause"
] | null | null | null | Tutorial-BSSN_in_terms_of_ADM.ipynb | rhaas80/nrpytutorial | 4398cd6b5a071c8fb8b2b584a01f07a4591dd5f4 | [
"BSD-2-Clause"
] | null | null | null | 45.894558 | 528 | 0.559624 | [
[
[
"<script async src=\"https://www.googletagmanager.com/gtag/js?id=UA-59152712-8\"></script>\n<script>\n window.dataLayer = window.dataLayer || [];\n function gtag(){dataLayer.push(arguments);}\n gtag('js', new Date());\n\n gtag('config', 'UA-59152712-8');\n</script>\n\n# ADM Quantities in terms of BSSN Quantities\n## Author: Zach Etienne\n\n[comment]: <> (Abstract: TODO)\n\n**Notebook Status:** <font color='orange'><b> Self-Validated </b></font>\n\n**Validation Notes:** This tutorial notebook has been confirmed to be self-consistent with its corresponding NRPy+ module, as documented [below](#code_validation). **Additional validation tests may have been performed, but are as yet, undocumented. (TODO)**\n\n### NRPy+ Source Code for this module: [ADM_in_terms_of_BSSN.py](../edit/BSSN/BSSN_in_terms_of_ADM.py)\n\n## Introduction:\nThis module documents the conversion of ADM variables:\n\n$$\\left\\{\\gamma_{ij}, K_{ij}, \\alpha, \\beta^i\\right\\}$$\n\ninto BSSN variables\n\n$$\\left\\{\\bar{\\gamma}_{i j},\\bar{A}_{i j},\\phi, K, \\bar{\\Lambda}^{i}, \\alpha, \\beta^i, B^i\\right\\},$$ \n\nin the desired curvilinear basis (given by `reference_metric::CoordSystem`). Then it rescales the resulting BSSNCurvilinear variables (as defined in [the covariant BSSN formulation tutorial](Tutorial-BSSN_formulation.ipynb)) into the form needed for solving Einstein's equations with the BSSN formulation:\n\n$$\\left\\{h_{i j},a_{i j},\\phi, K, \\lambda^{i}, \\alpha, \\mathcal{V}^i, \\mathcal{B}^i\\right\\}.$$",
"_____no_output_____"
],
[
"# Table of Contents\n$$\\label{toc}$$ \n\nThis notebook is organized as follows\n\n1. [Step 1](#initializenrpy): Initialize core Python/NRPy+ modules; set desired output BSSN Curvilinear coordinate system set to Spherical\n1. [Step 2](#adm2bssn): Perform the ADM-to-BSSN conversion for 3-metric, extrinsic curvature, and gauge quantities\n 1. [Step 2.a](#adm2bssn_gamma): Convert ADM $\\gamma_{ij}$ to BSSN $\\bar{\\gamma}_{ij}$; rescale to get $h_{ij}$\n 1. [Step 2.b](#admexcurv_convert): Convert the ADM extrinsic curvature $K_{ij}$ to BSSN $\\bar{A}_{ij}$ and $K$; rescale to get $a_{ij}$, $K$.\n 1. [Step 2.c](#lambda): Define $\\bar{\\Lambda}^i$\n 1. [Step 2.d](#conformal): Define the conformal factor variable `cf`\n1. [Step 3](#code_validation): Code Validation against `BSSN.BSSN_in_terms_of_ADM` NRPy+ module\n1. [Step 4](#latex_pdf_output): Output this notebook to $\\LaTeX$-formatted PDF file",
"_____no_output_____"
],
[
"<a id='initializenrpy'></a>\n\n# Step 1: Initialize core Python/NRPy+ modules \\[Back to [top](#toc)\\]\n$$\\label{initializenrpy}$$\n",
"_____no_output_____"
]
],
[
[
"# Step 1: Import needed core NRPy+ modules\nimport sympy as sp # SymPy: The Python computer algebra package upon which NRPy+ depends\nimport NRPy_param_funcs as par # NRPy+: Parameter interface\nimport indexedexp as ixp # NRPy+: Symbolic indexed expression (e.g., tensors, vectors, etc.) support\nimport reference_metric as rfm # NRPy+: Reference metric support\nimport sys # Standard Python modules for multiplatform OS-level functions\nimport BSSN.BSSN_quantities as Bq # NRPy+: This module depends on the parameter EvolvedConformalFactor_cf,\n # which is defined in BSSN.BSSN_quantities\n\n# Step 1.a: Set DIM=3, as we're using a 3+1 decomposition of Einstein's equations\nDIM=3",
"_____no_output_____"
]
],
[
[
"<a id='adm2bssn'></a>\n\n# Step 2: Perform the ADM-to-BSSN conversion for 3-metric, extrinsic curvature, and gauge quantities \\[Back to [top](#toc)\\]\n$$\\label{adm2bssn}$$\n\nHere we convert ADM quantities to their BSSN Curvilinear counterparts.",
"_____no_output_____"
],
[
"<a id='adm2bssn_gamma'></a>\n\n## Step 2.a: Convert ADM $\\gamma_{ij}$ to BSSN $\\bar{\\gamma}_{ij}$; rescale to get $h_{ij}$ \\[Back to [top](#toc)\\]\n$$\\label{adm2bssn_gamma}$$\n\nWe have (Eqs. 2 and 3 of [Ruchlin *et al.*](https://arxiv.org/pdf/1712.07658.pdf)):\n$$\n\\bar{\\gamma}_{i j} = \\left(\\frac{\\bar{\\gamma}}{\\gamma}\\right)^{1/3} \\gamma_{ij},\n$$\nwhere we always make the choice $\\bar{\\gamma} = \\hat{\\gamma}$.\n\nAfter constructing $\\bar{\\gamma}_{ij}$, we rescale to get $h_{ij}$ according to the prescription described in the [the covariant BSSN formulation tutorial](Tutorial-BSSN_formulation.ipynb) (also [Ruchlin *et al.*](https://arxiv.org/pdf/1712.07658.pdf)):\n\n$$\nh_{ij} = (\\bar{\\gamma}_{ij} - \\hat{\\gamma}_{ij})/\\text{ReDD[i][j]}.\n$$",
"_____no_output_____"
]
],
[
[
"# Step 2: All ADM quantities were input into this function in the Spherical or Cartesian\n# basis, as functions of r,th,ph or x,y,z, respectively. In Steps 1 and 2 above,\n# we converted them to the xx0,xx1,xx2 basis, and as functions of xx0,xx1,xx2.\n# Here we convert ADM quantities to their BSSN Curvilinear counterparts:\n\n# Step 2.a: Convert ADM $\\gamma_{ij}$ to BSSN $\\bar{gamma}_{ij}$:\n# We have (Eqs. 2 and 3 of [Ruchlin *et al.*](https://arxiv.org/pdf/1712.07658.pdf)):\ndef gammabarDD_hDD(gammaDD):\n global gammabarDD,hDD\n if rfm.have_already_called_reference_metric_function == False:\n print(\"BSSN.BSSN_in_terms_of_ADM.hDD_given_ADM(): Must call reference_metric() first!\")\n sys.exit(1)\n # \\bar{gamma}_{ij} = (\\frac{\\bar{gamma}}{gamma})^{1/3}*gamma_{ij}.\n gammaUU, gammaDET = ixp.symm_matrix_inverter3x3(gammaDD)\n gammabarDD = ixp.zerorank2()\n hDD = ixp.zerorank2()\n for i in range(DIM):\n for j in range(DIM):\n gammabarDD[i][j] = (rfm.detgammahat/gammaDET)**(sp.Rational(1,3))*gammaDD[i][j]\n hDD[i][j] = (gammabarDD[i][j] - rfm.ghatDD[i][j]) / rfm.ReDD[i][j]",
"_____no_output_____"
]
],
[
[
"<a id='admexcurv_convert'></a>\n\n## Step 2.b: Convert the ADM extrinsic curvature $K_{ij}$ to BSSN quantities $\\bar{A}_{ij}$ and $K={\\rm tr}(K_{ij})$; rescale $\\bar{A}_{ij}$ to get $a_{ij}$ \\[Back to [top](#toc)\\]\n$$\\label{admexcurv_convert}$$\n\nConvert the ADM extrinsic curvature $K_{ij}$ to the trace-free extrinsic curvature $\\bar{A}_{ij}$, plus the trace of the extrinsic curvature $K$, where (Eq. 3 of [Baumgarte *et al.*](https://arxiv.org/pdf/1211.6632.pdf)):\n\\begin{align}\nK &= \\gamma^{ij} K_{ij} \\\\\n\\bar{A}_{ij} &= \\left(\\frac{\\bar{\\gamma}}{\\gamma}\\right)^{1/3} \\left(K_{ij} - \\frac{1}{3} \\gamma_{ij} K \\right)\n\\end{align}\n\nAfter constructing $\\bar{A}_{ij}$, we rescale to get $a_{ij}$ according to the prescription described in the [the covariant BSSN formulation tutorial](Tutorial-BSSN_formulation.ipynb) (also [Ruchlin *et al.*](https://arxiv.org/pdf/1712.07658.pdf)):\n\n$$\na_{ij} = \\bar{A}_{ij}/\\text{ReDD[i][j]}.\n$$",
"_____no_output_____"
]
],
[
[
"# Step 2.b: Convert the extrinsic curvature K_{ij} to the trace-free extrinsic\n# curvature \\bar{A}_{ij}, plus the trace of the extrinsic curvature K,\n# where (Eq. 3 of [Baumgarte *et al.*](https://arxiv.org/pdf/1211.6632.pdf)):\ndef trK_AbarDD_aDD(gammaDD,KDD):\n global trK,AbarDD,aDD\n if rfm.have_already_called_reference_metric_function == False:\n print(\"BSSN.BSSN_in_terms_of_ADM.trK_AbarDD(): Must call reference_metric() first!\")\n sys.exit(1)\n # \\bar{gamma}_{ij} = (\\frac{\\bar{gamma}}{gamma})^{1/3}*gamma_{ij}.\n gammaUU, gammaDET = ixp.symm_matrix_inverter3x3(gammaDD)\n # K = gamma^{ij} K_{ij}, and\n # \\bar{A}_{ij} &= (\\frac{\\bar{gamma}}{gamma})^{1/3}*(K_{ij} - \\frac{1}{3}*gamma_{ij}*K)\n trK = sp.sympify(0)\n for i in range(DIM):\n for j in range(DIM):\n trK += gammaUU[i][j]*KDD[i][j]\n\n AbarDD = ixp.zerorank2()\n aDD = ixp.zerorank2()\n for i in range(DIM):\n for j in range(DIM):\n AbarDD[i][j] = (rfm.detgammahat/gammaDET)**(sp.Rational(1,3))*(KDD[i][j] - sp.Rational(1,3)*gammaDD[i][j]*trK)\n aDD[i][j] = AbarDD[i][j] / rfm.ReDD[i][j]",
"_____no_output_____"
]
],
[
[
"<a id='lambda'></a>\n\n## Step 2.c: Assuming the ADM 3-metric $\\gamma_{ij}$ is given as an explicit function of `(xx0,xx1,xx2)`, convert to BSSN $\\bar{\\Lambda}^i$; rescale to compute $\\lambda^i$ \\[Back to [top](#toc)\\]\n$$\\label{lambda}$$\n\nTo define $\\bar{\\Lambda}^i$ we implement Eqs. 4 and 5 of [Baumgarte *et al.*](https://arxiv.org/pdf/1211.6632.pdf):\n$$\n\\bar{\\Lambda}^i = \\bar{\\gamma}^{jk}\\left(\\bar{\\Gamma}^i_{jk} - \\hat{\\Gamma}^i_{jk}\\right).\n$$\n\nThe [reference_metric.py](../edit/reference_metric.py) module provides us with exact, closed-form expressions for $\\hat{\\Gamma}^i_{jk}$, so here we need only compute exact expressions for $\\bar{\\Gamma}^i_{jk}$, based on $\\gamma_{ij}$ given as an explicit function of `(xx0,xx1,xx2)`. This is particularly useful when setting up initial data.\n\nAfter constructing $\\bar{\\Lambda}^i$, we rescale to get $\\lambda^i$ according to the prescription described in the [the covariant BSSN formulation tutorial](Tutorial-BSSN_formulation.ipynb) (also [Ruchlin *et al.*](https://arxiv.org/pdf/1712.07658.pdf)):\n\n$$\n\\lambda^i = \\bar{\\Lambda}^i/\\text{ReU[i]}.\n$$",
"_____no_output_____"
]
],
[
[
"# Step 2.c: Define \\bar{Lambda}^i (Eqs. 4 and 5 of [Baumgarte *et al.*](https://arxiv.org/pdf/1211.6632.pdf)):\ndef LambdabarU_lambdaU__exact_gammaDD(gammaDD):\n global LambdabarU,lambdaU\n\n # \\bar{Lambda}^i = \\bar{gamma}^{jk}(\\bar{Gamma}^i_{jk} - \\hat{Gamma}^i_{jk}).\n gammabarDD_hDD(gammaDD)\n gammabarUU, gammabarDET = ixp.symm_matrix_inverter3x3(gammabarDD)\n\n # First compute Christoffel symbols \\bar{Gamma}^i_{jk}, with respect to barred metric:\n GammabarUDD = ixp.zerorank3()\n for i in range(DIM):\n for j in range(DIM):\n for k in range(DIM):\n for l in range(DIM):\n GammabarUDD[i][j][k] += sp.Rational(1,2)*gammabarUU[i][l]*( sp.diff(gammabarDD[l][j],rfm.xx[k]) +\n sp.diff(gammabarDD[l][k],rfm.xx[j]) -\n sp.diff(gammabarDD[j][k],rfm.xx[l]) )\n # Next evaluate \\bar{Lambda}^i, based on GammabarUDD above and GammahatUDD\n # (from the reference metric):\n LambdabarU = ixp.zerorank1()\n for i in range(DIM):\n for j in range(DIM):\n for k in range(DIM):\n LambdabarU[i] += gammabarUU[j][k] * (GammabarUDD[i][j][k] - rfm.GammahatUDD[i][j][k])\n for i in range(DIM):\n # We evaluate LambdabarU[i] here to ensure proper cancellations. If these cancellations\n # are not applied, certain expressions (e.g., lambdaU[0] in StaticTrumpet) will\n # cause SymPy's (v1.5+) CSE algorithm to hang\n LambdabarU[i] = LambdabarU[i].doit()\n lambdaU = ixp.zerorank1()\n for i in range(DIM):\n lambdaU[i] = LambdabarU[i] / rfm.ReU[i]",
"_____no_output_____"
]
],
[
[
"<a id='conformal'></a>\n\n## Step 2.d: Define the conformal factor variable `cf` \\[Back to [top](#toc)\\]\n$$\\label{conformal}$$\n\nWe define the conformal factor variable `cf` based on the setting of the `\"BSSN_quantities::EvolvedConformalFactor_cf\"` parameter.\n\nFor example if `\"BSSN_quantities::EvolvedConformalFactor_cf\"` is set to `\"phi\"`, we can use Eq. 3 of [Ruchlin *et al.*](https://arxiv.org/pdf/1712.07658.pdf), which in arbitrary coordinates is written:\n\n$$\n\\phi = \\frac{1}{12} \\log\\left(\\frac{\\gamma}{\\bar{\\gamma}}\\right).\n$$\n\nAlternatively if `\"BSSN_quantities::EvolvedConformalFactor_cf\"` is set to `\"chi\"`, then\n$$\n\\chi = e^{-4 \\phi} = \\exp\\left(-4 \\frac{1}{12} \\left(\\frac{\\gamma}{\\bar{\\gamma}}\\right)\\right) \n= \\exp\\left(-\\frac{1}{3} \\log\\left(\\frac{\\gamma}{\\bar{\\gamma}}\\right)\\right) = \\left(\\frac{\\gamma}{\\bar{\\gamma}}\\right)^{-1/3}.\n$$\n\nFinally if `\"BSSN_quantities::EvolvedConformalFactor_cf\"` is set to `\"W\"`, then\n$$\nW = e^{-2 \\phi} = \\exp\\left(-2 \\frac{1}{12} \\log\\left(\\frac{\\gamma}{\\bar{\\gamma}}\\right)\\right) = \n\\exp\\left(-\\frac{1}{6} \\log\\left(\\frac{\\gamma}{\\bar{\\gamma}}\\right)\\right) = \n\\left(\\frac{\\gamma}{\\bar{\\gamma}}\\right)^{-1/6}.\n$$",
"_____no_output_____"
]
],
[
[
"# Step 2.d: Set the conformal factor variable cf, which is set\n# by the \"BSSN_quantities::EvolvedConformalFactor_cf\" parameter. For example if\n# \"EvolvedConformalFactor_cf\" is set to \"phi\", we can use Eq. 3 of\n# [Ruchlin *et al.*](https://arxiv.org/pdf/1712.07658.pdf),\n# which in arbitrary coordinates is written:\ndef cf_from_gammaDD(gammaDD):\n global cf\n\n # \\bar{Lambda}^i = \\bar{gamma}^{jk}(\\bar{Gamma}^i_{jk} - \\hat{Gamma}^i_{jk}).\n gammabarDD_hDD(gammaDD)\n gammabarUU, gammabarDET = ixp.symm_matrix_inverter3x3(gammabarDD)\n gammaUU, gammaDET = ixp.symm_matrix_inverter3x3(gammaDD)\n\n cf = sp.sympify(0)\n\n if par.parval_from_str(\"EvolvedConformalFactor_cf\") == \"phi\":\n # phi = \\frac{1}{12} log(\\frac{gamma}{\\bar{gamma}}).\n cf = sp.Rational(1,12)*sp.log(gammaDET/gammabarDET)\n elif par.parval_from_str(\"EvolvedConformalFactor_cf\") == \"chi\":\n # chi = exp(-4*phi) = exp(-4*\\frac{1}{12}*(\\frac{gamma}{\\bar{gamma}}))\n # = exp(-\\frac{1}{3}*log(\\frac{gamma}{\\bar{gamma}})) = (\\frac{gamma}{\\bar{gamma}})^{-1/3}.\n #\n cf = (gammaDET/gammabarDET)**(-sp.Rational(1,3))\n elif par.parval_from_str(\"EvolvedConformalFactor_cf\") == \"W\":\n # W = exp(-2*phi) = exp(-2*\\frac{1}{12}*log(\\frac{gamma}{\\bar{gamma}}))\n # = exp(-\\frac{1}{6}*log(\\frac{gamma}{\\bar{gamma}})) = (\\frac{gamma}{bar{gamma}})^{-1/6}.\n cf = (gammaDET/gammabarDET)**(-sp.Rational(1,6))\n else:\n print(\"Error EvolvedConformalFactor_cf type = \\\"\"+par.parval_from_str(\"EvolvedConformalFactor_cf\")+\"\\\" unknown.\")\n sys.exit(1)",
"_____no_output_____"
]
],
[
[
"<a id='betvet'></a>\n\n## Step 2.e: Rescale $\\beta^i$ and $B^i$ to compute $\\mathcal{V}^i={\\rm vet}^i$ and $\\mathcal{B}^i={\\rm bet}^i$, respectively \\[Back to [top](#toc)\\]\n$$\\label{betvet}$$\n\nWe rescale $\\beta^i$ and $B^i$ according to the prescription described in the [the covariant BSSN formulation tutorial](Tutorial-BSSN_formulation.ipynb) (also [Ruchlin *et al.*](https://arxiv.org/pdf/1712.07658.pdf)):\n\\begin{align}\n\\mathcal{V}^i &= \\beta^i/\\text{ReU[i]}\\\\\n\\mathcal{B}^i &= B^i/\\text{ReU[i]}.\n\\end{align}",
"_____no_output_____"
]
],
[
[
"# Step 2.e: Rescale beta^i and B^i according to the prescription described in\n# the [BSSN in curvilinear coordinates tutorial notebook](Tutorial-BSSNCurvilinear.ipynb)\n# (also [Ruchlin *et al.*](https://arxiv.org/pdf/1712.07658.pdf)):\n#\n# \\mathcal{V}^i &= beta^i/(ReU[i])\n# \\mathcal{B}^i &= B^i/(ReU[i])\ndef betU_vetU(betaU,BU):\n global vetU,betU\n\n if rfm.have_already_called_reference_metric_function == False:\n print(\"BSSN.BSSN_in_terms_of_ADM.bet_vet(): Must call reference_metric() first!\")\n sys.exit(1)\n vetU = ixp.zerorank1()\n betU = ixp.zerorank1()\n for i in range(DIM):\n vetU[i] = betaU[i] / rfm.ReU[i]\n betU[i] = BU[i] / rfm.ReU[i]",
"_____no_output_____"
]
],
[
[
"<a id='code_validation'></a>\n\n# Step 3: Code Validation against `BSSN.BSSN_in_terms_of_ADM` module \\[Back to [top](#toc)\\] \n$$\\label{code_validation}$$\n\nHere, as a code validation check, we verify agreement in the SymPy expressions for [UIUC initial data](Tutorial-ADM_Initial_Data-UIUC_BlackHole.ipynb) between\n1. this tutorial and \n2. the NRPy+ [BSSN.BSSN_in_terms_of_ADM](../edit/BSSN/BSSN_in_terms_of_ADM.py) module.\n\nAs no basis transformation is performed, we analyze these expressions in their native, Spherical coordinates.",
"_____no_output_____"
]
],
[
[
"# Step 3.a: Set the desired *output* coordinate system to Spherical:\npar.set_parval_from_str(\"reference_metric::CoordSystem\",\"Spherical\")\nrfm.reference_metric()\n\n# Step 3.b: Set up initial data; assume UIUC spinning black hole initial data\nimport BSSN.UIUCBlackHole as uibh\nuibh.UIUCBlackHole(ComputeADMGlobalsOnly=True)\n\n# Step 3.c: Call above functions to convert ADM to BSSN curvilinear\ngammabarDD_hDD( uibh.gammaSphDD)\ntrK_AbarDD_aDD( uibh.gammaSphDD,uibh.KSphDD)\nLambdabarU_lambdaU__exact_gammaDD(uibh.gammaSphDD)\ncf_from_gammaDD( uibh.gammaSphDD)\nbetU_vetU( uibh.betaSphU,uibh.BSphU)\n\n# Step 3.d: Now load the BSSN_in_terms_of_ADM module and perform the same conversion\nimport BSSN.BSSN_in_terms_of_ADM as BitoA\nBitoA.gammabarDD_hDD( uibh.gammaSphDD)\nBitoA.trK_AbarDD_aDD( uibh.gammaSphDD,uibh.KSphDD)\nBitoA.LambdabarU_lambdaU__exact_gammaDD(uibh.gammaSphDD)\nBitoA.cf_from_gammaDD( uibh.gammaSphDD)\nBitoA.betU_vetU( uibh.betaSphU,uibh.BSphU)\n\n# Step 3.e: Perform the consistency check\nprint(\"Consistency check between this tutorial notebook and BSSN.BSSN_in_terms_of_ADM NRPy+ module: ALL SHOULD BE ZERO.\")\n\nprint(\"cf - BitoA.cf = \" + str(cf - BitoA.cf))\nprint(\"trK - BitoA.trK = \" + str(trK - BitoA.trK))\n# alpha is the only variable that remains unchanged:\n# print(\"alpha - BitoA.alpha = \" + str(alpha - BitoA.alpha))\n\nfor i in range(DIM):\n print(\"vetU[\"+str(i)+\"] - BitoA.vetU[\"+str(i)+\"] = \" + str(vetU[i] - BitoA.vetU[i]))\n print(\"betU[\"+str(i)+\"] - BitoA.betU[\"+str(i)+\"] = \" + str(betU[i] - BitoA.betU[i]))\n print(\"lambdaU[\"+str(i)+\"] - BitoA.lambdaU[\"+str(i)+\"] = \" + str(lambdaU[i] - BitoA.lambdaU[i]))\n for j in range(DIM):\n print(\"hDD[\"+str(i)+\"][\"+str(j)+\"] - BitoA.hDD[\"+str(i)+\"][\"+str(j)+\"] = \"\n + str(hDD[i][j] - BitoA.hDD[i][j]))\n print(\"aDD[\"+str(i)+\"][\"+str(j)+\"] - BitoA.aDD[\"+str(i)+\"][\"+str(j)+\"] = \"\n + str(aDD[i][j] - BitoA.aDD[i][j]))",
"Consistency check between this tutorial notebook and BSSN.BSSN_in_terms_of_ADM NRPy+ module: ALL SHOULD BE ZERO.\ncf - BitoA.cf = 0\ntrK - BitoA.trK = 0\nvetU[0] - BitoA.vetU[0] = 0\nbetU[0] - BitoA.betU[0] = 0\nlambdaU[0] - BitoA.lambdaU[0] = 0\nhDD[0][0] - BitoA.hDD[0][0] = 0\naDD[0][0] - BitoA.aDD[0][0] = 0\nhDD[0][1] - BitoA.hDD[0][1] = 0\naDD[0][1] - BitoA.aDD[0][1] = 0\nhDD[0][2] - BitoA.hDD[0][2] = 0\naDD[0][2] - BitoA.aDD[0][2] = 0\nvetU[1] - BitoA.vetU[1] = 0\nbetU[1] - BitoA.betU[1] = 0\nlambdaU[1] - BitoA.lambdaU[1] = 0\nhDD[1][0] - BitoA.hDD[1][0] = 0\naDD[1][0] - BitoA.aDD[1][0] = 0\nhDD[1][1] - BitoA.hDD[1][1] = 0\naDD[1][1] - BitoA.aDD[1][1] = 0\nhDD[1][2] - BitoA.hDD[1][2] = 0\naDD[1][2] - BitoA.aDD[1][2] = 0\nvetU[2] - BitoA.vetU[2] = 0\nbetU[2] - BitoA.betU[2] = 0\nlambdaU[2] - BitoA.lambdaU[2] = 0\nhDD[2][0] - BitoA.hDD[2][0] = 0\naDD[2][0] - BitoA.aDD[2][0] = 0\nhDD[2][1] - BitoA.hDD[2][1] = 0\naDD[2][1] - BitoA.aDD[2][1] = 0\nhDD[2][2] - BitoA.hDD[2][2] = 0\naDD[2][2] - BitoA.aDD[2][2] = 0\n"
]
],
[
[
"<a id='latex_pdf_output'></a>\n\n# Step 5: Output this notebook to $\\LaTeX$-formatted PDF file \\[Back to [top](#toc)\\]\n$$\\label{latex_pdf_output}$$\n\nThe following code cell converts this Jupyter notebook into a proper, clickable $\\LaTeX$-formatted PDF file. After the cell is successfully run, the generated PDF may be found in the root NRPy+ tutorial directory, with filename [Tutorial-ADM_Initial_Data-Converting_Exact_ADM_Spherical_or_Cartesian_to_BSSNCurvilinear.pdf](Tutorial-ADM_Initial_Data-Converting_Exact_ADM_Spherical_or_Cartesian_to_BSSNCurvilinear.pdf) (Note that clicking on this link may not work; you may need to open the PDF file through another means.)",
"_____no_output_____"
]
],
[
[
"import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface\ncmd.output_Jupyter_notebook_to_LaTeXed_PDF(\"Tutorial-BSSN_in_terms_of_ADM\")",
"Created Tutorial-BSSN_in_terms_of_ADM.tex, and compiled LaTeX file to PDF\n file Tutorial-BSSN_in_terms_of_ADM.pdf\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4c30f190258ad5707c67e4c28ecb5885418cf4 | 11,310 | ipynb | Jupyter Notebook | .ipynb_checkpoints/tester-checkpoint.ipynb | prockwood/lambdata_prockwood | d721ee9a5e95d53e80673ff32f18d59ab6b3b7fb | [
"MIT"
] | null | null | null | .ipynb_checkpoints/tester-checkpoint.ipynb | prockwood/lambdata_prockwood | d721ee9a5e95d53e80673ff32f18d59ab6b3b7fb | [
"MIT"
] | null | null | null | .ipynb_checkpoints/tester-checkpoint.ipynb | prockwood/lambdata_prockwood | d721ee9a5e95d53e80673ff32f18d59ab6b3b7fb | [
"MIT"
] | null | null | null | 36.019108 | 139 | 0.233952 | [
[
[
"import pandas as pd\nimport numpy as np\nfrom lambdata_prockwood import helper_function as hp",
"_____no_output_____"
],
[
"df = pd.DataFrame({'a': np.array([1, 2, np.nan]), 'b': np.array(['x', None, 'z']), 'c': np.array([np.nan, np.nan, 999])})",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.isna().sum().sum()",
"_____no_output_____"
],
[
"hp.null_count(df)",
"_____no_output_____"
],
[
"for i in range(len(df)):\n print(df.iloc[i, :])",
"a 1.0\nb x\nc NaN\nName: 0, dtype: object\na 2.0\nb None\nc NaN\nName: 1, dtype: object\na NaN\nb z\nc 999.0\nName: 2, dtype: object\n"
],
[
"# def train_test_split(df, frac):\n# choice_len = round(len(df) * frac)\n# train_index = np.random.choice(len(df), choice_len, replace=False) \n# test_index = list(set(np.arange(len(df))) - set(train_index))\n\n# return (df.iloc[train_index], df.iloc[test_index])",
"_____no_output_____"
],
[
"train, test = hp.train_test_split(df, 0.66)\n\nprint(train, '\\n')\nprint(test)",
" a b c\n2 NaN z 999.0\n1 2.0 None NaN \n\n a b c\n0 1.0 x NaN\n"
],
[
"df_nums = pd.DataFrame({'a': np.array([1, 2, 3]), 'b': np.array([4,5,6]), 'c': np.array([7,8,9])})",
"_____no_output_____"
],
[
"df_nums.head()",
"_____no_output_____"
],
[
"hp.randomize(df_nums, 42)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4c3af53f889c4d1a39dd1bd64f1e18bb94c69c | 14,157 | ipynb | Jupyter Notebook | examples/tutorials/Part 4 - Federated Learning via Trusted Aggregator.ipynb | alexis-thual/PySyft | f34aba95776d57b9bf30252061a84b64fc23018b | [
"Apache-2.0"
] | null | null | null | examples/tutorials/Part 4 - Federated Learning via Trusted Aggregator.ipynb | alexis-thual/PySyft | f34aba95776d57b9bf30252061a84b64fc23018b | [
"Apache-2.0"
] | null | null | null | examples/tutorials/Part 4 - Federated Learning via Trusted Aggregator.ipynb | alexis-thual/PySyft | f34aba95776d57b9bf30252061a84b64fc23018b | [
"Apache-2.0"
] | null | null | null | 33.154567 | 453 | 0.586989 | [
[
[
"# Part 4: Federated Learning with Model Averaging\n\n**Recap**: In Part 2 of this tutorial, we trained a model using a very simple version of Federated Learning. This required each data owner to trust the model owner to be able to see their gradients.\n\n**Description:**In this tutorial, we'll show how to use the advanced aggregation tools from Part 3 to allow the weights to be aggregated by a trusted \"secure worker\" before the final resulting model is sent back to the model owner (us). \n\nIn this way, only the secure worker can see whose weights came from whom. We might be able to tell which parts of the model changed, but we do NOT know which worker (bob or alice) made which change, which creates a layer of privacy.\n\nAuthors:\n- Andrew Trask - Twitter: [@iamtrask](https://twitter.com/iamtrask)\n- Jason Mancuso - Twitter: [@jvmancuso](https://twitter.com/jvmancuso)",
"_____no_output_____"
]
],
[
[
"import torch\nimport syft as sy\nimport copy\nhook = sy.TorchHook(torch)\nfrom torch import nn\nfrom syft import optim",
"_____no_output_____"
]
],
[
[
"# Step 1: Create Data Owners\n\nFirst, we're going to create two data owners (Bob and Alice) each with a small amount of data. We're also going to initialize a secure machine called \"secure_worker\". In practice this could be secure hardware (such as Intel's SGX) or simply a trusted intermediary. ",
"_____no_output_____"
]
],
[
[
"# create a couple workers\n\nbob = sy.VirtualWorker(hook, id=\"bob\")\nalice = sy.VirtualWorker(hook, id=\"alice\")\nsecure_worker = sy.VirtualWorker(hook, id=\"secure_worker\")\n\nbob.add_workers([alice, secure_worker])\nalice.add_workers([bob, secure_worker])\nsecure_worker.add_workers([alice, bob])\n\n# A Toy Dataset\ndata = torch.tensor([[0,0],[0,1],[1,0],[1,1.]], requires_grad=True)\ntarget = torch.tensor([[0],[0],[1],[1.]], requires_grad=True)\n\n# get pointers to training data on each worker by\n# sending some training data to bob and alice\nbobs_data = data[0:2].send(bob)\nbobs_target = target[0:2].send(bob)\n\nalices_data = data[2:].send(alice)\nalices_target = target[2:].send(alice)",
"_____no_output_____"
]
],
[
[
"# Step 2: Create Our Model\n\nFor this example, we're going to train with a simple Linear model. We can initialize it normally using PyTorch's nn.Linear constructor.",
"_____no_output_____"
]
],
[
[
"# Iniitalize A Toy Model\nmodel = nn.Linear(2,1)",
"_____no_output_____"
]
],
[
[
"# Step 3: Send a Copy of the Model to Alice and Bob\n\nNext, we need to send a copy of the current model to Alice and Bob so that they can perform steps of learning on their own datasets.",
"_____no_output_____"
]
],
[
[
"bobs_model = model.copy().send(bob)\nalices_model = model.copy().send(alice)\n\nbobs_opt = optim.SGD(params=bobs_model.parameters(),lr=0.1)\nalices_opt = optim.SGD(params=alices_model.parameters(),lr=0.1)",
"_____no_output_____"
]
],
[
[
"# Step 4: Train Bob's and Alice's Models (in parallel)\n\nAs is conventional with Federated Learning via Secure Averaging, each data owner first trains their model for several iterations locally before the models are averaged together.",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n\n # Train Bob's Model\n bobs_opt.zero_grad()\n bobs_pred = bobs_model(bobs_data)\n bobs_loss = ((bobs_pred - bobs_target)**2).sum()\n bobs_loss.backward()\n\n bobs_opt.step(bobs_data.shape[0])\n bobs_loss = bobs_loss.get().data\n\n # Train Alice's Model\n alices_opt.zero_grad()\n alices_pred = alices_model(alices_data)\n alices_loss = ((alices_pred - alices_target)**2).sum()\n alices_loss.backward()\n\n alices_opt.step(alices_data.shape[0])\n alices_loss = alices_loss.get().data\n \n print(\"Bob:\" + str(bobs_loss) + \" Alice:\" + str(alices_loss))",
"Bob:tensor(0.4355) Alice:tensor(1.9072)\nBob:tensor(0.2525) Alice:tensor(0.5729)\nBob:tensor(0.1516) Alice:tensor(0.1775)\nBob:tensor(0.0956) Alice:tensor(0.0598)\nBob:tensor(0.0641) Alice:tensor(0.0244)\nBob:tensor(0.0460) Alice:tensor(0.0133)\nBob:tensor(0.0354) Alice:tensor(0.0096)\nBob:tensor(0.0288) Alice:tensor(0.0079)\nBob:tensor(0.0245) Alice:tensor(0.0070)\nBob:tensor(0.0215) Alice:tensor(0.0064)\n"
]
],
[
[
"# Step 5: Send Both Updated Models to a Secure Worker\n\nNow that each data owner has a partially trained model, it's time to average them together in a secure way. We achieve this by instructing Alice and Bob to send their model to the secure (trusted) server. \n\nNote that this use of our API means that each model is sent DIRECTLY to the secure_worker. We never see it.",
"_____no_output_____"
]
],
[
[
"alices_model.move(secure_worker)",
"_____no_output_____"
],
[
"bobs_model.move(secure_worker)",
"_____no_output_____"
]
],
[
[
"# Step 6: Average the Models",
"_____no_output_____"
],
[
"Finally, the last step for this training epoch is to average Bob and Alice's trained models together and then use this to set the values for our global \"model\". ",
"_____no_output_____"
]
],
[
[
"model.weight.data.set_(((alices_model.weight.data + bobs_model.weight.data) / 2).get())\nmodel.bias.data.set_(((alices_model.bias.data + bobs_model.bias.data) / 2).get())\n\"\"",
"_____no_output_____"
]
],
[
[
"# Rinse and Repeat\n\nAnd now we just need to iterate this multiple times!",
"_____no_output_____"
]
],
[
[
"iterations = 10\nworker_iters = 5\n\nfor a_iter in range(iterations):\n \n bobs_model = model.copy().send(bob)\n alices_model = model.copy().send(alice)\n\n bobs_opt = optim.SGD(params=bobs_model.parameters(),lr=0.1)\n alices_opt = optim.SGD(params=alices_model.parameters(),lr=0.1)\n\n for wi in range(worker_iters):\n\n # Train Bob's Model\n bobs_opt.zero_grad()\n bobs_pred = bobs_model(bobs_data)\n bobs_loss = ((bobs_pred - bobs_target)**2).sum()\n bobs_loss.backward()\n\n bobs_opt.step(bobs_data.shape[0])\n bobs_loss = bobs_loss.get().data\n\n # Train Alice's Model\n alices_opt.zero_grad()\n alices_pred = alices_model(alices_data)\n alices_loss = ((alices_pred - alices_target)**2).sum()\n alices_loss.backward()\n\n alices_opt.step(alices_data.shape[0])\n alices_loss = alices_loss.get().data\n \n alices_model.move(secure_worker)\n bobs_model.move(secure_worker)\n \n model.weight.data.set_(((alices_model.weight.data + bobs_model.weight.data) / 2).get())\n model.bias.data.set_(((alices_model.bias.data + bobs_model.bias.data) / 2).get())\n \n print(\"Bob:\" + str(bobs_loss) + \" Alice:\" + str(alices_loss))",
"Bob:tensor(0.0712) Alice:tensor(0.0141)\nBob:tensor(0.0684) Alice:tensor(0.0087)\nBob:tensor(0.0596) Alice:tensor(0.0056)\nBob:tensor(0.0506) Alice:tensor(0.0038)\nBob:tensor(0.0425) Alice:tensor(0.0027)\nBob:tensor(0.0356) Alice:tensor(0.0019)\nBob:tensor(0.0298) Alice:tensor(0.0015)\nBob:tensor(0.0248) Alice:tensor(0.0011)\nBob:tensor(0.0206) Alice:tensor(0.0009)\nBob:tensor(0.0171) Alice:tensor(0.0008)\n"
]
],
[
[
"Lastly, we want to make sure that our resulting model learned correctly, so we'll evaluate it on a test dataset. In this toy problem, we'll use the original data, but in practice we'll want to use new data to understand how well the model generalizes to unseen examples.",
"_____no_output_____"
]
],
[
[
"preds = model(data)\nloss = ((preds - target) ** 2).sum()",
"_____no_output_____"
],
[
"print(preds)\nprint(target)\nprint(loss.data)",
"tensor([[0.2274],\n [0.1693],\n [0.8352],\n [0.7771]], grad_fn=<AddmmBackward>)\ntensor([[0.],\n [0.],\n [1.],\n [1.]], requires_grad=True)\ntensor(0.1572)\n"
]
],
[
[
"In this toy example, the averaged model is underfitting relative to a plaintext model trained locally would behave, however we were able to train it without exposing each worker's training data. We were also able to aggregate the updated models from each worker on a trusted aggregator to prevent data leakage to the model owner.\n\nIn a future tutorial, we'll aim to do our trusted aggregation directly with the gradients, so that we can update the model with better gradient estimates and arrive at a stronger model.",
"_____no_output_____"
],
[
"# Congratulations!!! - Time to Join the Community!\n\nCongratulations on completing this notebook tutorial! If you enjoyed this and would like to join the movement toward privacy preserving, decentralized ownership of AI and the AI supply chain (data), you can do so in the following ways!\n\n### Star PySyft on GitHub\n\nThe easiest way to help our community is just by starring the Repos! This helps raise awareness of the cool tools we're building.\n\n- [Star PySyft](https://github.com/OpenMined/PySyft)\n\n### Join our Slack!\n\nThe best way to keep up to date on the latest advancements is to join our community! You can do so by filling out the form at [http://slack.openmined.org](http://slack.openmined.org)\n\n### Join a Code Project!\n\nThe best way to contribute to our community is to become a code contributor! At any time you can go to PySyft GitHub Issues page and filter for \"Projects\". This will show you all the top level Tickets giving an overview of what projects you can join! If you don't want to join a project, but you would like to do a bit of coding, you can also look for more \"one off\" mini-projects by searching for GitHub issues marked \"good first issue\".\n\n- [PySyft Projects](https://github.com/OpenMined/PySyft/issues?q=is%3Aopen+is%3Aissue+label%3AProject)\n- [Good First Issue Tickets](https://github.com/OpenMined/PySyft/issues?q=is%3Aopen+is%3Aissue+label%3A%22good+first+issue%22)\n\n### Donate\n\nIf you don't have time to contribute to our codebase, but would still like to lend support, you can also become a Backer on our Open Collective. All donations go toward our web hosting and other community expenses such as hackathons and meetups!\n\n[OpenMined's Open Collective Page](https://opencollective.com/openmined)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cb4c44732bc67da6f86e4760ff3b466d33d099e9 | 22,007 | ipynb | Jupyter Notebook | docs/params.ipynb | Nexuscompute/Cirq | 640ef8f82d6a56ec95361388ce7976e096cca906 | [
"Apache-2.0"
] | null | null | null | docs/params.ipynb | Nexuscompute/Cirq | 640ef8f82d6a56ec95361388ce7976e096cca906 | [
"Apache-2.0"
] | 4 | 2022-01-16T14:12:15.000Z | 2022-02-24T03:58:46.000Z | docs/params.ipynb | Nexuscompute/Cirq | 640ef8f82d6a56ec95361388ce7976e096cca906 | [
"Apache-2.0"
] | null | null | null | 33.343939 | 821 | 0.622257 | [
[
[
"##### Copyright 2022 The Cirq Developers",
"_____no_output_____"
]
],
[
[
"# @title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Parameter Sweeps",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://quantumai.google/cirq/params\"><img src=\"https://quantumai.google/site-assets/images/buttons/quantumai_logo_1x.png\" />View on QuantumAI</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/quantumlib/Cirq/blob/master/docs/params.ipynb\"><img src=\"https://quantumai.google/site-assets/images/buttons/colab_logo_1x.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/quantumlib/Cirq/blob/master/docs/params.ipynb\"><img src=\"https://quantumai.google/site-assets/images/buttons/github_logo_1x.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/Cirq/docs/params.ipynb\"><img src=\"https://quantumai.google/site-assets/images/buttons/download_icon_1x.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
]
],
[
[
"try:\n import cirq\nexcept ImportError:\n print(\"installing cirq...\")\n !pip install --quiet cirq\n print(\"installed cirq.\")\n import cirq",
"_____no_output_____"
]
],
[
[
"## Concept of Circuit Parameterization and Sweeps\n\nSuppose you have a quantum circuit and in this circuit there is a gate with some parameter. You might wish to run this circuit for different values of this parameter. An example of this type of circuit is a Rabi flop experiment. This experiment runs a set of quantum computations which 1) starts in $|0\\rangle$ state, 2) rotates the state by $\\theta$ about the $x$ axis, i.e. applies the gate $\\exp(i \\theta X)$, and 3) measures the state in the computational basis. Running this experiment for multiple values of $\\theta$, and plotting the probability of observing a $|1\\rangle$ outcome yields the quintessential $\\cos^2$ probability distribution as a function of the parameter $\\theta$. To support this type of experiment, Cirq provides the concept of parameterized circuits and parameter sweeps. \n\nThe next cell illustrates parameter sweeps with a simple example. Suppose you want to compare two quantum circuits that are identical except for a single exponentiated `cirq.Z` gate.",
"_____no_output_____"
]
],
[
[
"q0 = cirq.LineQubit(0)\n\ncircuit1 = cirq.Circuit([cirq.H(q0), cirq.Z(q0)**0.5, cirq.H(q0), cirq.measure(q0)])\nprint(f\"circuit1:\\n{circuit1}\")\n\ncircuit2 = cirq.Circuit([cirq.H(q0), cirq.Z(q0)**0.25, cirq.H(q0), cirq.measure(q0)])\nprint(f\"circuit2:\\n{circuit2}\")",
"_____no_output_____"
]
],
[
[
"You could run these circuits separately (either on hardware or in simulation), and collect statistics on the results of these circuits. However parameter sweeps can do this in a cleaner and more perfomant manner. \n\nFirst define a parameter, and construct a circuit that depends on this parameter. Cirq uses [SymPy](https://www.sympy.org/en/index.html){:external}, a symbolic mathematics package, to define parameters. In this example the Sympy parameter is `theta`, which is used to construct a parameterized circuit.",
"_____no_output_____"
]
],
[
[
"import sympy\n\ntheta = sympy.Symbol(\"theta\")\n\ncircuit = cirq.Circuit([cirq.H(q0), cirq.Z(q0)**theta, cirq.H(q0), cirq.measure(q0)])\nprint(f\"circuit:\\n{circuit}\")",
"_____no_output_____"
]
],
[
[
"Notice now that the circuit contains a `cirq.Z` gate that is raised to a power, but this power is the parameter `theta`. This is a \"parameterized circuit\". An equivalent way to construct this circuit, where the parameter is actually a parameter in the gate constructor's arguments, is:",
"_____no_output_____"
]
],
[
[
"circuit = cirq.Circuit(\n cirq.H(q0), cirq.ZPowGate(exponent=theta)(q0), cirq.H(q0), cirq.measure(q0)\n)\nprint(f\"circuit:\\n{circuit}\")",
"_____no_output_____"
]
],
[
[
"Note: You can check whether an object in Cirq is parameterized using `cirq.is_parameterized`:",
"_____no_output_____"
]
],
[
[
"cirq.is_parameterized(circuit)",
"_____no_output_____"
]
],
[
[
"Parameterized circuits are just like normal circuits; they just aren't defined in terms of gates that you can actually run on a quantum computer without the additional information about the values of the parameters. Following the example above, you can generate the two circuits (`circuit1` and `circuit2`) by using `cirq.resolve_parameter` and supplying the values that you want the parameter(s) to take:",
"_____no_output_____"
]
],
[
[
"# circuit1 has theta = 0.5\ncirq.resolve_parameters(circuit, {\"theta\": 0.5})\n# circuit2 has theta = 0.25\ncirq.resolve_parameters(circuit, {\"theta\": 0.25})",
"_____no_output_____"
]
],
[
[
"More interestingly, you can combine parameterized circuits with a list of parameter assignments when doing things like running circuits or simulating them. These lists of parameter assignements are called \"sweeps\". For example you can use a simulator's `run_sweep` method to run simulations for the parameters corresponding to the two circuits defined above. ",
"_____no_output_____"
]
],
[
[
"sim = cirq.Simulator()\nresults = sim.run_sweep(circuit, repetitions=25, params=[{\"theta\": 0.5}, {\"theta\": 0.25}])\nfor result in results:\n print(f\"param: {result.params}, result: {result}\")",
"_____no_output_____"
]
],
[
[
"To recap, you can construct parameterized circuits that depend on parameters that have not yet been assigned a value. These parameterized circuits can then be resolved to circuits with actual values via a dictionary that maps the sympy variable name to the value that parameter should take. You can also construct lists of dictionaries of parameter assignments, called sweeps, and pass this to many functions in Cirq that use circuits to do an action (such as `simulate` or `run`). For each of the elements in the sweep, the function will execute using the parameters as described by the element.",
"_____no_output_____"
],
[
"## Constructing Sweeps\n\nThe previous example constructed a sweep by simply constructing a list of parameter assignments, `[{\"theta\": 0.5}, {\"theta\": 0.25}]`. Cirq also provides other ways to construct sweeps. \n\nOne useful method for constructing parameter sweeps is `cirq.Linspace` which creates a sweep over a list of equally spaced elements. ",
"_____no_output_____"
]
],
[
[
"# Create a sweep over 5 equally spaced values from 0 to 2.5.\nparams = cirq.Linspace(key=\"theta\", start=0, stop=2.5, length=5)\nfor param in params:\n print(param)",
"_____no_output_____"
]
],
[
[
"Note: The `Linspace` sweep is composed of `cirq.ParamResolver` instances instead of simple dictionaries. However, you can think of them as effectively the same for most use cases. \n\nIf you need to explicitly and individually specify each parameter resolution, you can do it by constructing a list of dictionaries as before. However, you can also use `cirq.Points` to do this more succinctly.",
"_____no_output_____"
]
],
[
[
"params = cirq.Points(key=\"theta\", points=[0, 1, 3])\nfor param in params:\n print(param)",
"_____no_output_____"
]
],
[
[
"If you're working with parameterized circuits, it is very likely you'll need to keep track of multiple parameters. Two common use cases necessitate building a sweep from two constituent sweeps, where the new sweep includes: \n- Every possible combination of the elements of each sweep: A cartesian product. \n- A element-wise pairing of the two sweeps: A zip.\n\nThe following are examples of using the `*` and `+` operators to combine sweeps by cartesian product and zipping, respectively. ",
"_____no_output_____"
]
],
[
[
"sweep1 = cirq.Linspace(\"theta\", 0, 1, 5)\nsweep2 = cirq.Points(\"gamma\", [0, 3])\n# By taking the product of these two sweeps, you can sweep over all possible\n# combinations of the parameters.\nfor param in sweep1 * sweep2:\n print(param)",
"_____no_output_____"
],
[
"sweep1 = cirq.Points(\"theta\", [1, 2, 3])\nsweep2 = cirq.Points(\"gamma\", [0, 3, 4])\n# By taking the sum of these two sweeps, you can combine the sweeps\n# elementwise (similar to python's zip function):\nfor param in sweep1 + sweep2:\n print(param)",
"_____no_output_____"
]
],
[
[
"`cirq.Linspace` and `cirq.Points` are instances of the `cirq.Sweep` class, which explicitly supports cartesian product with the `*` operation, and zipping with the `+` operation. The `*` operation produces a `cirq.Product` object, and `+` produces a `cirq.Zip` object, both of which are also `Sweep`s. Other mathematical operations will not work in general *between sweeps*.",
"_____no_output_____"
],
[
"## Symbols and Expressions",
"_____no_output_____"
],
[
"[SymPy](https://www.sympy.org/en/index.html){:external} is a general symbolic mathematics toolset, and you can leverage this in Cirq to define more complex parameters than have been shown so far. For example, you can define an expression in Sympy and use it to construct circuits that depend on this expression:",
"_____no_output_____"
]
],
[
[
"# Construct an expression for 0.5 * a + 0.25:\nexpr = 0.5 * sympy.Symbol(\"a\") + 0.25\nprint(expr)",
"_____no_output_____"
],
[
"# Use the expression in the circuit:\ncircuit = cirq.Circuit(cirq.X(q0)**expr, cirq.measure(q0))\nprint(f\"circuit:\\n{circuit}\")",
"_____no_output_____"
]
],
[
[
"Both the exponents and parameter arguments of circuit operations can in fact be any general Sympy expression: The previous examples just used single-variable expressions. When you resolve parameters for this circuit, the expressions are evaluated under the given assignments to the variables in the expression. ",
"_____no_output_____"
]
],
[
[
"print(cirq.resolve_parameters(circuit, {\"a\": 0}))",
"_____no_output_____"
]
],
[
[
"Just as before, you can pass a sweep over variable values to `run` or `simulate`, and Cirq will evaluate the expression for each possible value. ",
"_____no_output_____"
]
],
[
[
"sim = cirq.Simulator()\nresults = sim.run_sweep(circuit, repetitions=25, params=cirq.Points('a', [0, 1]))\nfor result in results:\n print(f\"param: {result.params}, result: {result}\")",
"_____no_output_____"
]
],
[
[
"Sympy supports a large number of numeric functions and methods, which can be used to create fairly sophisticated expressions, like cosine, exponentiation, and more:",
"_____no_output_____"
]
],
[
[
"print(sympy.cos(sympy.Symbol(\"a\"))**sympy.Symbol(\"b\"))",
"_____no_output_____"
]
],
[
[
"Cirq can numerically evaluate all of the expressions Sympy can evalute. However, if you are running a parameterized circuit on a service (such as on a hardware backed quantum computing service) that service may not support evaluating all expressions. See documentation for the particular service you're using for details. \n\nAs a general workaround, you can instead use Cirq's flattening ability to evaluate the parameters before sending them off to the service.",
"_____no_output_____"
],
[
"### Flattening Expressions\n\nSuppose you build a circuit that includes multiple different expressions:",
"_____no_output_____"
]
],
[
[
"a = sympy.Symbol('a')\ncircuit = cirq.Circuit(cirq.X(q0)**(a / 4), cirq.Y(q0)**(1 - a / 2), cirq.measure(q0))\nprint(circuit)",
"_____no_output_____"
]
],
[
[
"Flattening replaces every expression in the circuit with a new symbol that is representative of the value of that expression. Additionally, it keeps track of the new symbols and provices a `cirq.ExpressionMap` object to map the old sympy expression objects to the new symbols that replaced them. ",
"_____no_output_____"
]
],
[
[
"# Flatten returns two objects, the circuit with new symbols, and the mapping from old to new values.\nc_flat, expr_map = cirq.flatten(circuit)\nprint(c_flat)\nprint(expr_map)",
"_____no_output_____"
]
],
[
[
"Notice that the new circuit has new symbols, `<a/2>` and `<1-a/2>`, which are explicitly not expressions. You can see this by looking at the value of the exponent in the first gate:",
"_____no_output_____"
]
],
[
[
"first_gate = c_flat[0][q0].gate\nprint(first_gate.exponent)\n# Note this is a symbol, not an expression\nprint(type(first_gate.exponent))",
"_____no_output_____"
]
],
[
[
"The second object returned by `cirq.flatten` is an object that can be used to map sweeps over the previous symbols to new sweeps over the new expression-symbols. The values assigned to the new expression symbols in the resulting sweep are the old expressions kept track of in the `ExpressionMap`, but resolved with the values provided by the original input sweep.",
"_____no_output_____"
]
],
[
[
"sweep = cirq.Linspace(a, start=0, stop=3, length=4)\nprint(f\"Old {sweep}\")\n\nnew_sweep = expr_map.transform_sweep(sweep)\nprint(f\"New {new_sweep}\")",
"_____no_output_____"
]
],
[
[
"To reinforce: The new sweep is over two new symbols, which each represent the values of the expressions in the original circuit. The values assigned to these new expression symbols is acquired by evaluating the expressions with `a` resolved to a value in `[0, 4]`, according to the old sweep. \n\nYou can use these new sweep elements to resolve the parameters of the flattened circuit:",
"_____no_output_____"
]
],
[
[
"for params in new_sweep:\n print(c_flat, '=>', end=' ')\n print(cirq.resolve_parameters(c_flat, params))",
"_____no_output_____"
]
],
[
[
"Using `cirq.flatten`, you can always take a parameterized circuit with any complicated expressions, plus a sweep, and produce an equivalent circuit with no expressions, only symbols, and a sweep for these new symbols. Because this is a common flow, Cirq provides `cirq.flatten_sweep` to do this in one step:",
"_____no_output_____"
]
],
[
[
"c_flat, new_sweep = cirq.flatten_with_sweep(circuit, sweep)\nprint(c_flat)\nprint(new_sweep)",
"_____no_output_____"
]
],
[
[
"You can then directly use these objects to run the sweeps. For example, you can use them to perform a simulation:",
"_____no_output_____"
]
],
[
[
"sim = cirq.Simulator()\nresults = sim.run_sweep(c_flat, repetitions=20, params=new_sweep)\nfor result in results:\n print(result.params, result)",
"_____no_output_____"
]
],
[
[
"You can see that the different flattened parameters have corresponding different results for their simulation.",
"_____no_output_____"
],
[
"# Summary\n\n- Cirq circuits can handle arbitrary Sympy expressions in place of exponents and parameter arguments in operations.\n- By providing one or a sequence of `ParamResolver`s or dictionaries that resolve the Sympy variables to values, `run`, `simulate`, and other functions can iterate efficiently over different parameter assignments for otherwise identical circuits. \n- Sweeps can be created succinctly with `cirq.Points` and `cirq.Linspace`, and composed with each other with `*` and `+`, to create `cirq.Product` and `cirq.Zip` sweeps. \n- When the service you're using does not support arbitrary expressions, you can flatten a circuit and sweep into a new circuit that doesn't have complex expressions, and a corresponding new sweep. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb4c4f29dababe31fe67a976468118a8a1cb09b2 | 7,335 | ipynb | Jupyter Notebook | scratch.ipynb | quantology/dicebox | 8bb04d372a400144a96692107242d770e3aa6a66 | [
"BSD-3-Clause"
] | null | null | null | scratch.ipynb | quantology/dicebox | 8bb04d372a400144a96692107242d770e3aa6a66 | [
"BSD-3-Clause"
] | null | null | null | scratch.ipynb | quantology/dicebox | 8bb04d372a400144a96692107242d770e3aa6a66 | [
"BSD-3-Clause"
] | null | null | null | 18.022113 | 92 | 0.443217 | [
[
[
"import re",
"_____no_output_____"
],
[
"re.match(r\"(adv)|(disadv)\\([^)]+\\)\", \"adv(asdf)\")",
"_____no_output_____"
],
[
"fnmatch = re.compile(r\"^(?P<fn>(adv)|(disadv))\\((?P<args>[^)]+)\\)$\")",
"_____no_output_____"
],
[
"from dicebox.core import parse",
"_____no_output_____"
],
[
"parse(\"5d8 + 10\")",
"_____no_output_____"
],
[
"import asteval",
"_____no_output_____"
],
[
"def parse_dice_term(s):\n n, sides = s.split(\"d\")\n return Dice({int(sides): int(n)})\n\ndef with_adv(d):\n return d.adv\ndef with_disadv(d):\n return d.disadv\n\n_symtable = {\"dice\": parse_dice_term, \"adv\": with_adv, \"disadv\": with_disadv}\n_dice_match = re.compile(r\"\\b([0-9]+d[0-9]+)\\b\")\n\ndef parse(s):\n eval_friendly = _dice_match.sub(r\"dice('\\1')\", s)\n return asteval.Interpreter(\n symtable=_symtable, minimal=True, use_numpy=False, builtins_readonly=True\n ).eval(eval_friendly)",
"_____no_output_____"
],
[
"parse(\"(5d6 - 3) // 5\")",
"_____no_output_____"
],
[
"\naeval.eval(\"dice('2d10')\")",
"_____no_output_____"
],
[
"dir(asteval)",
"_____no_output_____"
],
[
"asteval.asteval.a\n",
"_____no_output_____"
],
[
"\nasteval.a",
"_____no_output_____"
],
[
"parse_basic(\"10d6\")",
"_____no_output_____"
],
[
"dice_match.findall(\"adv(1d6+2d6)\")",
"_____no_output_____"
],
[
"m = fnmatch.match(\"adv(asdf)\")",
"_____no_output_____"
],
[
"m.group(\"fn\"), m.group(\"args\")",
"_____no_output_____"
],
[
"fn = m.group(0)",
"_____no_output_____"
],
[
"fn",
"_____no_output_____"
],
[
"m.groups()",
"_____no_output_____"
],
[
"r\"(adv)|(disadv)\\([^)]+\\)\"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4c5eedb56cb024276e087bdb570e5d0e22274b | 120,297 | ipynb | Jupyter Notebook | sphinx/tutorials/Completeness_Contours.ipynb | spencerhurt/rvsearch | 3ea322b54dcc2e87c57d3bc3af9ba5518ce40a18 | [
"MIT"
] | null | null | null | sphinx/tutorials/Completeness_Contours.ipynb | spencerhurt/rvsearch | 3ea322b54dcc2e87c57d3bc3af9ba5518ce40a18 | [
"MIT"
] | 11 | 2020-10-29T23:39:53.000Z | 2021-11-18T19:47:38.000Z | sphinx/tutorials/Completeness_Contours.ipynb | spencerhurt/rvsearch | 3ea322b54dcc2e87c57d3bc3af9ba5518ce40a18 | [
"MIT"
] | 1 | 2021-01-21T18:17:11.000Z | 2021-01-21T18:17:11.000Z | 847.161972 | 117,096 | 0.958328 | [
[
[
"# Completeness Contours\n\nIn this example we generate completeness contours from a suite of injection-recovery tests run using the `rvsearch inject` command",
"_____no_output_____"
]
],
[
[
"import os\n\nimport numpy as np\nimport pylab as pl\n\nimport rvsearch\nfrom rvsearch.inject import Completeness\nfrom rvsearch.plots import CompletenessPlots",
"_____no_output_____"
]
],
[
[
"Construct a `Completeness` object from the output of an injection-recovery run (recoveries.csv). If we want to convert to semi-major axis and msini on construction we need to pass a stellar mass.",
"_____no_output_____"
]
],
[
[
"recfile = os.path.join(rvsearch.DATADIR, 'recoveries.csv')\n\ncomp = Completeness.from_csv(recfile, 'inj_au', 'inj_msini', mstar=1.1)",
"_____no_output_____"
]
],
[
[
"Next we calculate completeness on a grid with the specified boundaries using a 2D moving average. See Howard & Fulton (2016) for a description of the moving average method.",
"_____no_output_____"
]
],
[
[
"xi, yi, zi = comp.completeness_grid(xlim=(0.05, 100), ylim=(1.0, 3e4), resolution=25)",
"_____no_output_____"
]
],
[
[
"Now contruct the `CompletenessPlots` object and plot the contours and individual injections",
"_____no_output_____"
]
],
[
[
"cp = CompletenessPlots(comp)\nfig = cp.completeness_plot(xlabel='$a$ [AU]', ylabel=r'M$\\sin{i}$ [M$_{\\oplus}$]')",
"_____no_output_____"
]
],
[
[
"We can also use the Completeness object to interpolate the completeness at any point in the grid.",
"_____no_output_____"
]
],
[
[
"comp.interpolate(1.0, 100, refresh=True)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4c67b3f719c93f9e0c9edcfef05f6c6e1fe153 | 14,491 | ipynb | Jupyter Notebook | aws_marketplace/using_algorithms/autogluon/autogluon_tabular_marketplace.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
] | null | null | null | aws_marketplace/using_algorithms/autogluon/autogluon_tabular_marketplace.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
] | null | null | null | aws_marketplace/using_algorithms/autogluon/autogluon_tabular_marketplace.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
] | 1 | 2021-05-04T22:09:07.000Z | 2021-05-04T22:09:07.000Z | 32.785068 | 356 | 0.59844 | [
[
[
"# AutoGluon-Tabular in AWS Marketplace\n\n[AutoGluon](https://github.com/awslabs/autogluon) automates machine learning tasks enabling you to easily achieve strong predictive performance in your applications. With just a few lines of code, you can train and deploy high-accuracy deep learning models on tabular, image, and text data.\nThis notebook shows how to use AutoGluon-Tabular in AWS Marketplace.\n\n### Contents:\n* [Step 1: Subscribe to AutoML algorithm from AWS Marketplace](#Step-1:-Subscribe-to-AutoML-algorithm-from-AWS-Marketplace)\n* [Step 2: Set up environment](#Step-2-:-Set-up-environment)\n* [Step 3: Prepare and upload data](#Step-3:-Prepare-and-upload-data)\n* [Step 4: Train a model](#Step-4:-Train-a-model)\n* [Step 5: Deploy the model and perform a real-time inference](#Step-5:-Deploy-the-model-and-perform-a-real-time-inference)\n* [Step 6: Use Batch Transform](#Step-6:-Use-Batch-Transform)\n* [Step 7: Clean-up](#Step-7:-Clean-up)",
"_____no_output_____"
],
[
"### Step 1: Subscribe to AutoML algorithm from AWS Marketplace\n\n1. Open [AutoGluon-Tabular listing from AWS Marketplace](https://aws.amazon.com/marketplace/pp/prodview-n4zf5pmjt7ism)\n2. Read the **Highlights** section and then **product overview** section of the listing.\n3. View **usage information** and then **additional resources**.\n4. Note the supported instance types and specify the same in the following cell.\n5. Next, click on **Continue to subscribe**.\n6. Review **End user license agreement**, **support terms**, as well as **pricing information**.\n7. Next, \"Accept Offer\" button needs to be clicked only if your organization agrees with EULA, pricing information as well as support terms. Once **Accept offer** button has been clicked, specify compatible training and inference types you wish to use. \n\n**Notes**: \n1. If **Continue to configuration** button is active, it means your account already has a subscription to this listing.\n2. Once you click on **Continue to configuration** button and then choose region, you will see that a product ARN will appear. This is the algorithm ARN that you need to specify in your training job. However, for this notebook, the algorithm ARN has been specified in **src/algorithm_arns.py** file and you do not need to specify the same explicitly.",
"_____no_output_____"
],
[
"### Step 2 : Set up environment",
"_____no_output_____"
]
],
[
[
"#Import necessary libraries.\nimport os\nimport boto3\nimport sagemaker\nfrom time import sleep\nfrom collections import Counter\nimport numpy as np\nimport pandas as pd\nfrom sagemaker import get_execution_role, local, Model, utils, fw_utils, s3\nfrom sagemaker.algorithm import AlgorithmEstimator\nfrom sagemaker.predictor import Predictor\nfrom sagemaker.serializers import CSVSerializer\nfrom sagemaker.deserializers import StringDeserializer\nfrom sklearn.metrics import accuracy_score, classification_report\nfrom IPython.core.display import display, HTML\nfrom IPython.core.interactiveshell import InteractiveShell\n\n# Print settings\nInteractiveShell.ast_node_interactivity = \"all\"\npd.set_option('display.max_columns', 500)\npd.set_option('display.max_rows', 10)\n\n# Account/s3 setup\nsession = sagemaker.Session()\nbucket = session.default_bucket()\nprefix = 'sagemaker/autogluon-tabular'\nregion = session.boto_region_name\nrole = get_execution_role()\n",
"_____no_output_____"
],
[
"compatible_training_instance_type='ml.m5.4xlarge' \ncompatible_inference_instance_type='ml.m5.4xlarge' ",
"_____no_output_____"
],
[
"#Specify algorithm ARN for AutoGluon-Tabular from AWS Marketplace. However, for this notebook, the algorithm ARN \n#has been specified in src/algorithm_arns.py file and you do not need to specify the same explicitly.\n\nfrom src.algorithm_arns import AlgorithmArnProvider\n\nalgorithm_arn = AlgorithmArnProvider.get_algorithm_arn(region)",
"_____no_output_____"
],
[
"import subprocess\nsubprocess.run(\"apt-get update -y\", shell=True)\nsubprocess.run(\"apt install unzip\", shell=True)",
"_____no_output_____"
]
],
[
[
"### Step 3: Get the data\n\nIn this example we'll use the direct-marketing dataset to build a binary classification model that predicts whether customers will accept or decline a marketing offer. \nFirst we'll download the data and split it into train and test sets. AutoGluon does not require a separate validation set (it uses bagged k-fold cross-validation).",
"_____no_output_____"
]
],
[
[
"# Download and unzip the data\nsubprocess.run(f\"aws s3 cp --region {region} s3://sagemaker-sample-data-{region}/autopilot/direct_marketing/bank-additional.zip .\", shell=True)\nsubprocess.run(\"unzip -qq -o bank-additional.zip\", shell=True)\nsubprocess.run(\"rm bank-additional.zip\", shell=True)\n\nlocal_data_path = './bank-additional/bank-additional-full.csv'\ndata = pd.read_csv(local_data_path)\n\n# Split train/test data\ntrain = data.sample(frac=0.7, random_state=42)\ntest = data.drop(train.index)\n\n# Split test X/y\nlabel = 'y'\ny_test = test[label]\nX_test = test.drop(columns=[label])",
"_____no_output_____"
]
],
[
[
"##### Check the data",
"_____no_output_____"
]
],
[
[
"train.head(3)\ntrain.shape\n\ntest.head(3)\ntest.shape\n\nX_test.head(3)\nX_test.shape",
"_____no_output_____"
]
],
[
[
"Upload the data to s3",
"_____no_output_____"
]
],
[
[
"train_file = 'train.csv'\ntrain.to_csv(train_file,index=False)\ntrain_s3_path = session.upload_data(train_file, key_prefix='{}/data'.format(prefix))\n\ntest_file = 'test.csv'\ntest.to_csv(test_file,index=False)\ntest_s3_path = session.upload_data(test_file, key_prefix='{}/data'.format(prefix))\n\nX_test_file = 'X_test.csv'\nX_test.to_csv(X_test_file,index=False)\nX_test_s3_path = session.upload_data(X_test_file, key_prefix='{}/data'.format(prefix))",
"_____no_output_____"
]
],
[
[
"### Step 4: Train a model\nNext, let us train a model.\n\n**Note:** Depending on how many underlying models are trained, `train_volume_size` may need to be increased so that they all fit on disk.",
"_____no_output_____"
]
],
[
[
"# Define required label and optional additional parameters\ninit_args = {\n 'label': 'y'\n}\n\n# Define additional parameters\nfit_args = {\n # Adding 'best_quality' to presets list will result in better performance (but longer runtime)\n 'presets': ['optimize_for_deployment'],\n}\n\n# Pass fit_args to SageMaker estimator hyperparameters\nhyperparameters = {\n 'init_args': init_args, \n 'fit_args': fit_args,\n 'feature_importance': True\n}\n\ntags = [{\n 'Key' : 'AlgorithmName',\n 'Value' : 'AutoGluon-Tabular'\n}]",
"_____no_output_____"
],
[
"algo = AlgorithmEstimator(algorithm_arn=algorithm_arn, \n role=role, \n instance_count=1, \n instance_type=compatible_training_instance_type, \n sagemaker_session=session, \n base_job_name='autogluon',\n hyperparameters=hyperparameters,\n train_volume_size=100) \n\ninputs = {'training': train_s3_path}\n\nalgo.fit(inputs)",
"_____no_output_____"
]
],
[
[
"### Step 5: Deploy the model and perform a real-time inference",
"_____no_output_____"
],
[
"##### Deploy a remote endpoint",
"_____no_output_____"
]
],
[
[
"%%time\n\npredictor = algo.deploy(initial_instance_count=1, \n instance_type=compatible_inference_instance_type, \n serializer=CSVSerializer(), \n deserializer=StringDeserializer())",
"_____no_output_____"
]
],
[
[
"##### Predict on unlabeled test data",
"_____no_output_____"
]
],
[
[
"results = predictor.predict(X_test.to_csv(index=False)).splitlines()\n\n# Check output\ny_results = np.array([i.split(\",\")[0] for i in results])\nprint(Counter(y_results))",
"_____no_output_____"
]
],
[
[
"##### Predict on data that includes label column\nPrediction performance metrics will be printed to endpoint logs.\n",
"_____no_output_____"
]
],
[
[
"results = predictor.predict(test.to_csv(index=False)).splitlines()\n\n# Check output\ny_results = np.array([i.split(\",\")[0] for i in results])\nprint(Counter(y_results))",
"_____no_output_____"
]
],
[
[
"##### Check that classification performance metrics match evaluation printed to endpoint logs as expected",
"_____no_output_____"
]
],
[
[
"y_results = np.array([i.split(\",\")[0] for i in results])\n\nprint(\"accuracy: {}\".format(accuracy_score(y_true=y_test, y_pred=y_results)))\nprint(classification_report(y_true=y_test, y_pred=y_results, digits=6))",
"_____no_output_____"
]
],
[
[
"### Step 6: Use Batch Transform",
"_____no_output_____"
],
[
"By including the label column in the test data, you can also evaluate prediction performance (In this case, passing `test_s3_path` instead of `X_test_s3_path`).",
"_____no_output_____"
]
],
[
[
"output_path = f's3://{bucket}/{prefix}/output/'\n\ntransformer = algo.transformer(instance_count=1, \n instance_type=compatible_inference_instance_type,\n strategy='MultiRecord',\n max_payload=6,\n max_concurrent_transforms=1, \n output_path=output_path)\n\ntransformer.transform(test_s3_path, content_type='text/csv', split_type='Line')\ntransformer.wait()",
"_____no_output_____"
]
],
[
[
"### Step 7: Clean-up",
"_____no_output_____"
],
[
"Once you have finished performing predictions, you can delete the endpoint to avoid getting charged for the same.",
"_____no_output_____"
]
],
[
[
"predictor.delete_endpoint()",
"_____no_output_____"
]
],
[
[
"Finally, if the AWS Marketplace subscription was created just for the experiment and you would like to unsubscribe to the product, here are the steps that can be followed.\nBefore you cancel the subscription, ensure that you do not have any [deployable model](https://console.aws.amazon.com/sagemaker/home#/models) created from the model-package or using the algorithm. Note - You can find this by looking at container associated with the model. \n\nSteps to un-subscribe to product from AWS Marketplace:\n1. Navigate to __Machine Learning__ tab on [__Your Software subscriptions page__](https://aws.amazon.com/marketplace/ai/library?productType=ml&ref_=lbr_tab_ml)\n2. Locate the listing that you would need to cancel subscription for, and then __Cancel Subscription__ can be clicked to cancel the subscription.\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb4c6cc21f9d0b1c29f052c3fb9cba738f51f4e1 | 57,312 | ipynb | Jupyter Notebook | notebook/covid19india.ipynb | Nikhil0504/CCT-App | edef16a6736d96147e83ad45c82f66206a25a1d1 | [
"MIT"
] | 1 | 2021-06-06T01:12:35.000Z | 2021-06-06T01:12:35.000Z | notebook/covid19india.ipynb | Nikhil0504/CCT-App | edef16a6736d96147e83ad45c82f66206a25a1d1 | [
"MIT"
] | 1 | 2021-06-20T18:38:55.000Z | 2021-06-21T07:35:07.000Z | notebook/covid19india.ipynb | Nikhil0504/CCT-App | edef16a6736d96147e83ad45c82f66206a25a1d1 | [
"MIT"
] | 1 | 2021-06-18T14:28:35.000Z | 2021-06-18T14:28:35.000Z | 37.754941 | 91 | 0.300949 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"url = \"https://api.covid19india.org/csv/latest/case_time_series.csv\"\r\nurl2 = \"https://api.covid19india.org/csv/latest/states.csv\"",
"_____no_output_____"
],
[
"c[\"Date\"] = pd.to_datetime(c[\"Date\"])\r\nstart_date = '2021-06-30'\r\nend_date = '2021-07-01'\r\nmask = (c['Date'] > start_date) & (c['Date'] <= end_date)\r\nc=pd.read_csv(url2)\r\nc.loc[mask]",
"_____no_output_____"
],
[
"c.head(-1)",
"_____no_output_____"
],
[
"pd.read_csv(\"https://api.covid19india.org/csv/latest/state_wise.csv\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb4c9337a821766bedabf5df60a538a21c9dbae6 | 2,248 | ipynb | Jupyter Notebook | ISBN978-4-7981-6720-6/list5_20.ipynb | zfukuoka/Copying_a_sutra | 2d0b1f781fc029ae0108b639e893708a8c45cee2 | [
"BSD-2-Clause"
] | null | null | null | ISBN978-4-7981-6720-6/list5_20.ipynb | zfukuoka/Copying_a_sutra | 2d0b1f781fc029ae0108b639e893708a8c45cee2 | [
"BSD-2-Clause"
] | null | null | null | ISBN978-4-7981-6720-6/list5_20.ipynb | zfukuoka/Copying_a_sutra | 2d0b1f781fc029ae0108b639e893708a8c45cee2 | [
"BSD-2-Clause"
] | null | null | null | 22.707071 | 253 | 0.446619 | [
[
[
"<a href=\"https://colab.research.google.com/github/zfukuoka/Copying_a_sutra/blob/master/ISBN978-4-7981-6720-6/list5_20.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Chapter 5 ディープラーニングの理論\n\n* 5.8 損失関数",
"_____no_output_____"
],
[
"### リスト 5.20 交差エントロピー誤差の計算をする関数",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndef cross_entropy(y, t):\n return - np.sum(t * np.log(y + 1e-7))",
"_____no_output_____"
]
],
[
[
"### リスト 5.21 交差エントロピー誤差を計算する",
"_____no_output_____"
]
],
[
[
"y = np.array([0.05, 0.9, 0.02, 0.02, 0.01])\nt = np.array([0, 1, 0, 0, 0])\nprint(cross_entropy(y, t))",
"0.1053604045467214\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4c969779462ffd852c52ed419d3b18a4368b02 | 12,766 | ipynb | Jupyter Notebook | 6-PCA/note/LSJU----NOTE-PCA.ipynb | PeterChenYijie/MachineLearningZeroToALL | b14005c3e0b5a39a0ba82db5c9791f682b5effd5 | [
"MIT"
] | 8 | 2018-04-20T09:10:20.000Z | 2019-02-16T07:50:32.000Z | 6-PCA/note/LSJU----NOTE-PCA.ipynb | DeepInDeeper/MachineLearningZeroToALL | b14005c3e0b5a39a0ba82db5c9791f682b5effd5 | [
"MIT"
] | null | null | null | 6-PCA/note/LSJU----NOTE-PCA.ipynb | DeepInDeeper/MachineLearningZeroToALL | b14005c3e0b5a39a0ba82db5c9791f682b5effd5 | [
"MIT"
] | 4 | 2020-01-27T00:55:59.000Z | 2021-03-25T00:07:56.000Z | 73.791908 | 1,097 | 0.650321 | [
[
[
"文章来自 作者:子实 更多机器学习笔记访问[这里](https://github.com/zlotus/notes-LSJU-machine-learning)",
"_____no_output_____"
],
[
"# 第十五讲:PCA的奇异值分解、独立成分分析\n\n回顾一下上一讲的内容——PCA算法,主要有三个步骤:\n1. 将数据正规化为零期望以及单位化方差;\n2. 计算协方差矩阵$\\displaystyle\\varSigma=\\frac{1}{m}x^{(i)}\\left(x^{(i)}\\right)^T$;\n3. 找到$\\varSigma$的前$k$个特征向量。\n\n在上一讲的最后,我们还介绍了PCA在面部识别中的应用。试想一下,在面部识别中$x^{(i)}\\in\\mathbb R^{10000}$,那么$\\varSigma\\in\\mathbb R^{100000000}$,而要求这种亿级别的矩阵的特征向量并不容易,这个问题我们先放一放。\n\n来看另一个例子,在前面([第五讲](https://github.com/zlotus/notes-LSJU-machine-learning/blob/master/chapter05.ipynb))我们讨论过关于垃圾邮件分类的问题,我们会构造一个词汇表向量,其每个分量所对应的单词如果在邮件中出现则置为$1$,反正则置为$0$。当我们对这种数据应用PCA时,产生的算法换了个名字,叫做**潜在语义索引(LSI: latent semantic analysis)**。不过在LSI中,我们通常跳过预处理阶段,因为正规化数据使它们具有相同的方差可能会大幅增加不参加单词的权重。比如说我们拿到了一个文档的内容$x^{(i)}$,现在希望知道这份文档与我们已有文档中的哪个相似度最高,也就是衡量两个高维输入向量所表示的文档的相似性$\\mathrm{sim}\\left(x^{(i)},x^{(j)}\\right), x\\in\\mathbb R^{50000}$。我们通常的做法是衡量这两个向量直接的夹角,如果夹角越小则认为他们越相似,于是$\\displaystyle\\mathrm{sim}\\left(x^{(i)},x^{(j)}\\right)=\\cos\\theta=\\frac{\\left(x^{(i)}\\right)^Tx^{(j)}}{\\left\\lVert x^{(i)}\\right\\rVert\\left\\lVert x^{(j)}\\right\\rVert}$。再来看上面这个分子,$x^{(i)}\\left(x^{(j)}\\right)^T=\\sum_kx_k^{(i)}x_k^{(j)}=\\sum_k1\\{\\textrm{文档i和j都包含词语k}\\}$,如果文档中没有词语重复,则该式结果为$0$。但是,如果文档$j$包含单词“study”而文档$j$包含单词“learn”,那么如果一篇介绍“study strategy”的文章和一篇介绍“method of learning”的文章在这种算法下就是无关的,我们现在想要使它们相关。于是一开始,“learn”向量与“study”向量是相互正交的,它们的内积为零,我们在这两个向量之间再找一个向量$u$,然后将“learn”与“study”投影在$u$上,此时两个向量的投影点在$u$上将相距很近,那么它们做内积时将会得到一个正数,表示它们相关。于是,如果算法再遇到一篇关于“政治”的文章和另一篇包含很多政治家名字的文章时,它会判断这两篇文章是相关的。\n\n## 奇异值分解(SVD: Singular Value Decomposition)\n\n我们引入奇异值分解(可以参考线性代数笔记中的[奇异值分解](http://nbviewer.jupyter.org/github/zlotus/notes-linear-algebra/blob/master/chapter30.ipynb))来解决一开始遇到的大矩阵求特征向量的问题。比如在潜在语义索引中的输入,是一个$50000$维的向量,那么其对应的$\\varSigma\\in\\mathbb R^{50000\\times50000}$,这种规模的矩阵太大了,我们需要使用另一种方法实现PCA。\n\n对矩阵$A\\in\\mathbb R^{m\\times n}$,总有$A=UDV^T,U\\in\\mathbb R^{m\\times m},D\\in\\mathbb R^{m\\times n},V^T\\in\\mathbb R^{n\\times n}$,其中$D=\\begin{bmatrix}\\sigma_1&&&\\\\&\\sigma_2&&\\\\&&\\ddots&\\\\&&&\\sigma_n\\end{bmatrix}$是一个对角矩阵,而$\\sigma_i$称为矩阵的奇异值。分解之后的矩阵为$\\begin{bmatrix}A\\\\m\\times n\\end{bmatrix}=\\begin{bmatrix}U\\\\m\\times m\\end{bmatrix}\\begin{bmatrix}D\\\\m\\times n\\end{bmatrix}\\begin{bmatrix}V^T\\\\n\\times n\\end{bmatrix}$。\n\n现在来观察协方差矩阵的定义式$\\displaystyle\\varSigma=\\sum_{i=1}^mx^{(i)}\\left(x^{(i)}\\right)^T$,在前面的章节中([第二讲](chapter02.ipynb))我们介绍过一种叫做“设计矩阵”的构造方式,也就是将每一个样本向量作为矩阵$X$的一行拼凑一个矩阵:$X=\\begin{bmatrix}—\\left(x^{(1)}\\right)^T—\\\\—\\left(x^{(2)}\\right)^T—\\\\\\vdots\\\\—\\left(x^{(m)}\\right)^T—\\end{bmatrix}$,则我们可以将协方差矩阵用设计矩阵来表示:$\\varSigma=\\begin{bmatrix}\\mid&\\mid&&\\mid\\\\x^{(1)}&x^{(2)}&\\cdots&x^{(m)}\\\\\\mid&\\mid&&\\mid\\end{bmatrix}\\begin{bmatrix}—\\left(x^{(1)}\\right)^T—\\\\—\\left(x^{(2)}\\right)^T—\\\\\\vdots\\\\—\\left(x^{(m)}\\right)^T—\\end{bmatrix}$。\n\n最后就是计算$\\varSigma$的前$k$个特征向量了,我们选择对$X$做奇异值分解$X=UDV^T$,而$X^TX=VDU^TUD^TV^T=VD^2V^T=\\varSigma$,于是在$D$中从大到小排列奇异值并在$V$中取前$k$个奇异值对应的特征向量即可。\n\n容易看出$X\\in\\mathbb R^{m\\times50000}$,做这种规模矩阵的奇异值分解会比直接计算$\\varSigma\\in\\mathbb R^{50000\\times50000}$的特征向量快很多。这就是使用SVD实现PCA算法的计算过程。\n\n(不过,值得注意的是,在不同的计算软件,甚至是在同一种软件的不同版本中,对SVD的计算可能遵循某种默认的维数约定,因为SVD经常会得到带有很多零元素的$U$和$D$,而软件可能会按照某种约定舍弃这些零元素。所以,在使用SVD时,需要注意这样的维数约定。)\n\n## 无监督学习各算法的对比\n\n我们在前面介绍因子分析模型时指出,它是对每个因子$z^{(i)}$进行高斯建模,是一种对概率密度进行估计的算法,它试图对训练样本$X$的概率密度进行建模;而在其后介绍的PCA则有所不同,它并不是一个概率算法,因为它并没有使用任何概率分布拟合训练集,而是直接去寻找子空间。从这里我们可以大致的看到,如何在解决问题时在因子分析与PCA间做取舍:如果目标就是降低数据维数、寻找数据所在的子空间,我们就更倾向于使用PCA;而因子分析会假设数据本来就在某子空间内,如果有维度很高的数据$X$,而我又想对$X$建模,那么就应该使用因子分析算法(比如做异常检测,我们可以建立关于$P(X)$的模型,如果有一个低概率事件,就可以将这个事件分解在各因子分布中,进而估计其异常情况)。这两种算法的共同点就是,它们都会假设数据位于或靠近某个低维的子空间。\n\n再来看回顾一开始介绍的两种无监督学习方法:混合高斯模型以及$k$-means算法。这两种算法的共同点是——它们都会假设数据聚集在位于某几个簇内。不同点是混合高斯模型是一种对概率密度进行估计的算法,而$k$-means则不是。所以,如果我们需要将数据分成簇并对每一个簇建模,那么我们就倾向于使用高斯混合模型;而如我我们只想将数据分簇,并不要求确定每个簇的概率系统,则就更倾向于使用$k$-means算法。\n\n综合上面的观点可以得到表格,便于记忆:\n\n$$\n\\begin{array}\n{c|c|c}\n&\\textbf{Model }P(x)&\\textbf{Not probabilistic}\\\\\\hline\n\\textbf{Subspace}&\\textrm{Factor Analysis}&\\textrm{PCA}\\\\\\hline\n\\textbf{Cluster}&\\textrm{Mixtures of Gaussians}&k\\textrm{-means}\n\\end{array}\n$$\n\n# 第十二部分:独立成分分析(Independent components analysis)\n\n接下来,我们将介绍独立成分分析(ICA: Independent components analysis)。类似于PCA,独立成分分析也会寻找一组新的基,用来重新表示训练样本,然而这两个算法的目标截然不同。\n\n举一个实际问题作为例子,在第一讲,我们介绍了一个在鸡尾酒会上从嘈杂的背景音中分离发言者音频的应用。假设有$n$个发言者在一个舞会上同时说话,而在屋子里的各话筒仅捕捉到这种$n$个发言者的声音叠加在一起的音频。但是,假设我们有$n$个不同的话筒,由于每个话筒到个发言者的距离都不相同,则这些话筒记录下来的是不同形式的发言者声音叠加。那么,通过使用这些话筒的音频记录,我们能否分离出这$n$个发言者各自的音频信号?\n\n为了正式的描述这个问题,我们假设数据$x\\in\\mathbb R^n$,这些数据由$n$个相互独立的来源生成,而我们能够观测到的数据为:\n\n$$\nx=As\n$$\n\n这里的$A$是一个未知的方阵,通常被称为**混合矩阵(mixing matrix)**。通过重复观测,我们得到一个数据集$\\left\\{x^{(i)};i=1,\\cdots,m\\right\\}$,而我们的目标是还原那一组生成“观测到的数据集($x^{(i)}=As^{(i)}$)”的声音源$s^{(i)}$。\n\n在鸡尾酒舞会问题中,$s^{(i)}$是一个$n$维向量,$s_j^{(i)}$表示发言者$j$在第$i$次音频采集时发出的声音。而$x^{(i)}$也是$n$维向量,$x_j^{(i)}$表示话筒$j$在第$i$次音频采样时记录下来的音频。\n\n令$W=A^{-1}$作为**分离矩阵(unmixing matrix)**。我们的目标就是求出$W$,进而使用$s^{(i)}=Wx^{(i)}$从话筒收集的音频中还原出各独立的音源。按照一贯的标记法,我们通常使用$w_i^T$表示$W$矩阵的第$i$行,则有$W=\\begin{bmatrix}—w_1^T—\\\\\\vdots\\\\—w_n^T—\\end{bmatrix}$。那么,对$w_i\\in\\mathbb R^n$,有第$j$个发言者音源可以使用$s_j^{(i)}=w_j^Tx^{(i)}$表示。\n\n## 1. ICA二义性\n\n$W=A^{-1}$能够做出什么程度的还原?如果没有关于音源和混合矩阵的先验经验,不难看出,矩阵$A$存在固有二义性使得它不可能只通过$x^{(i)}$就还原并对应出每一个$s^{(i)}$。\n\n* 令$P$为$n$阶置换矩阵,也就是$P$的每行每列都只有一个$1$,其余元素均为零。举个例子:$P=\\begin{bmatrix}0&1&0\\\\1&0&0\\\\0&0&1\\end{bmatrix},\\ P=\\begin{bmatrix}0&1\\\\1&0\\end{bmatrix},\\ P=\\begin{bmatrix}1&0\\\\0&1\\end{bmatrix}$。其作用是,对向量$z$,$Pz$会产生一个将$z$的各分量重新排列得到的的置换后的向量$z'$。对于给定的$x^{(i)}$,我们无法分辨出$W$和$PW$(也就是无法确定分离矩阵中的每一行向量对应哪一位发言者)。不难预料,音源也存在这种置换二义性,不过这种二义性对于大多数应用而言并不是重要问题。\n\n* 此外,我们也无法确定$w_i$的大小。比如当$A$变为$2A$,而每个$s^{(i)}$变为$0.5s^{(i)}$时,对于我们的观测值$x^{(i)}=2A\\cdot(0.5)s^{(i)}$没有任何影响。更广泛的,如果$A$的某列被加上了缩放因子$\\alpha$,而相应的音源又被缩放因子$\\frac{1}{\\alpha}$调整了大小,那么我们将无法从$x^{(i)}$这一单一条件中还原这次缩放调整。因此,我们无法还原音源的原有大小。不过,对于我们关心的问题(包括鸡尾酒舞会问题),这个二义性也并不重要。特别是在本例中,使用正缩放因子$\\alpha$调整发言者音源$s_j^{(i)}$的大小只会影响到发言者音量的大小。而且即使改变音源的符号也没关系,$s_j^{(i)}$和$-s_j^{(i)}$在扩音器上听起来是完全一样的。综上,如果算法得出的$w_i$被某非零缩放因子影响,那么使用$s_i=w_i^Tx$得到的相应音源也会受到这个缩放因子的影响,然而这种二义性也并不重要。(这个ICA二义性同样适用于后面介绍的脑MEG中。)\n\n我们不禁要问,上面提到的这两种情况是ICA涉及的所有可能的二义性吗?只要源$s_i$不服从高斯分布,那么答案就是肯定的。\n\n* 我们通过一个例子来看高斯分布下的数据会产生什么麻烦,考虑当$n=2$时的$s\\sim\\mathcal N(0,I)$,此处的$I$是一个二阶单位矩阵。注意到标准正态分布$\\mathcal N(0,I)$概率密度的等高线图是一组圆心在原点的正圆,其概率密度具有旋转对称性。假设我们观测到某些$x=As$,$A$是混合矩阵,因为源源服从$\\mathcal N(0,I)$,则混合后的$x$同样服从一个以$0$为期望、以$\\mathrm E\\left[xx^T\\right]=\\mathrm E\\left[Ass^TA^T\\right]=AA^T$为协方差的高斯分布。现在令$R$为任意正交矩阵(非正式的也叫作旋转矩阵或反射矩阵),则有$RR^T=R^TR=I$,并令$A'=AR$。那么如果源被$A'$混合(而不是被$A$混合),则我们会观测到$x'=A's$。而$x'$也服从高斯分布,同样以$0$围棋网、以$\\mathrm E\\left[x'\\left(x'\\right)^T\\right]=\\mathrm E\\left[A'ss^T\\left(A'\\right)^T\\right]=\\mathrm E\\left[ARss^T\\left(AR\\right)^T\\right]=ARR^TA^T=AA^T$。可以看到,不论混合矩阵是$A$还是$A'$,我们观测到的值都会服从同一个高斯分布$\\mathcal N\\left(0,AA^T\\right)$,于是,我们无法辨别观测到的随机变量是来自混合矩阵$A$还是$A'$。因此,混合矩阵中可以包含任意的旋转矩阵,而我们是无法从观测数据中看到这个旋转矩阵的痕迹的,所以我们也就无法完全还原出源了。\n\n上面的这段论述是基于“多元标准正态分布是旋转对称的”这一性质的。尽管ICA对于服从高斯分布的数据存在这样的缺陷,但只要数据不服从高斯分布,在数据充足的情况下,我们还是可以分离出$n$个独立的源的。\n\n## 2. 概率密度与线性变换\n\n在展开ICA算法的推导之前,我们先来简要介绍一下线性变换对概率密度的影响。\n\n假设有一个从$p_s(s)$中抽取的随机变量$s$,为了简约表达,令$s\\in\\mathbb R$为实数。现在按照$x=As$定义随机变量$x$($x\\in\\mathbb R,A\\in\\mathbb R$),令$p_x$为$x$的概率密度。那么,什么是$p_x$?\n\n令$W=A^{-1}$,要计算一个特定$x$的概率,也就是尝试计算$s=Wx$,之后再使用$p_s$估计再点$s$处的概率,从而得出$p_x(x)=p_s(Wx)$的结论——*然而这是一个错误的结论*。比如令$s\\sim\\mathrm{Uniform}[0,1]$服从均匀分布,则$s$的概率密度为$p_s(s)=1\\{0\\leq s\\leq1\\}$;再令$A=2$,则$x=2s$。显然$x$是一个在区间$[0,2]$上的均匀分布,则其概率密度函数为$p_x(x)=(0.5)1\\{0\\leq x\\leq2\\}$。而此时的$W=A^{-1}=0.5$,很明显,$p_s(Wx)$并不等于$p_x(x)$。此时,真正成立的是式子$p_x(x)=p_s(Wx)\\lvert W\\rvert$。\n\n一般的,若$s$是一个向量值,来自以$p_s$为概率密度的某分布,且对于可逆矩阵$A$有$x=As$,$A$的逆矩阵为$W=A^{-1}$,那么$x$的概率密度为:\n\n$$\np_x(x)=p_s(Wx)\\cdot\\lvert W\\rvert\n$$\n\n**注意:**如果曾经见过$A$将$[0,1]^n$映射到一组体积$\\lvert A\\rvert$的集合上(可以参考[克拉默法则、逆矩阵、体积](http://nbviewer.jupyter.org/github/zlotus/notes-linear-algebra/blob/master/chapter20.ipynb)),则就有另一种记忆上面这个关于$p_x$的公式的方法,这个方法同样可以一般化前面$1$维的例子。令$A\\in\\mathbb R^{n\\times n}$,令$W=A^{-1}$,再令$C_1=[0,1]^n$为$n$维超立方体,并定义$C_2=\\{As:\\ s\\in C_1\\}\\subseteq\\mathbb R^n$(即$C_2$是原像$C_1$在映射$A$作用下得到的像)。按照上面这些条件,在线性代数中有一个现成的标准公式可以使用(同时这也是行列式的一种定义方式):$C_2$的体积为$\\lvert A\\rvert$。假设$s$服从在$[0,1]^n$上的均匀分布,则其概率密度函数为$p_s(s)=1\\{s\\in C_1\\}$。很明显$x$为在$C_2$上的均匀分布,其概率密度为$\\displaystyle p_x(x)=\\frac{1\\{x\\in C_2\\}}{\\mathrm{Vol}(C_2)}$(因为它必须做从$C_2$到$1$的积分)。再利用“逆矩阵的行列式是原矩阵行列式的倒数”这一性质,则有$\\displaystyle\\frac{1}{\\mathrm{Vol}(C_2)}=\\frac{1}{\\lvert A\\rvert}=\\left\\lvert A^{-1}\\right\\rvert=\\lvert W\\rvert$。最终我们又得到$p_x(x)=1\\{x\\in C_2\\}\\lvert W\\rvert=1\\{Wx\\in C_1\\}\\lvert W\\rvert=p_s(Wx)\\lvert W\\rvert$。\n\n## 3. ICA算法\n\n现在我们可以开始推导ICA算法了。这个算法来自Bell和Sejnowski,我们这里给出的关于ICA的理解,会将其看做是一个用来最大化似然估计的算法(这与该算法原本的演绎不同,该演绎涉及到一个称为infomax原则的复杂的概念,不过在现代关于ICA的推导中已经没有必要提及了)。\n\n我们假设每个源$s_i$来自一个由概率密度函数$p_s$定义的分布,而且$s$的联合分布为:\n\n$$\np(s)=\\prod_{i=1}^np_s(s_i)\n$$\n\n注意到我们使用各源的边缘分布的乘积来对联合分布建模,也就是默认假设每一个源是独立的。使用上一节得到的公式能够得到关于$x=As=W^{-1}s$的概率密度:\n\n$$\np(x)=\\prod_{i=1}^np_s\\left(w_i^Tx\\right)\\cdot\\lvert W\\rvert\n$$\n\n接下来就剩为每一个独立的源$p_s$估计概率密度了。\n\n以前在概率论中我们学过,对于给定的实随机变量$z$,其累积分布函数(CDF: Cumulative Distribution Function)$F$可以使用概率密度函数(PDF: Probability Density Function)来计算:$F(z_0)=P(z\\leq z_0)=\\int_{-\\infty}^{z_0}p_z(z)\\mathrm dz$。当然,也可以通过对CDF求导得到$z$的概率密度函数$p_z(z)=F'(z)$。\n\n因此,要求出每个$s_i$的PDF,只需要求出它们对应的CDF的即可,而CDF是一个函数值从$0$到$1$的单调增函数。根据上一节的推导,我们知道ICA对服从高斯分布的数据无效,所以不能选择高斯分布的累积分布函数作为CDF。我们现在要选择一个合理的“默认”函数,其函数值也是从$0$缓慢的单调递增至$1$——这就是前面([第三讲](chapter03.ipynb))介绍的逻辑函数(即S型函数)$\\displaystyle g(s)=\\frac{1}{1+e^{-s}}$。于是有$p_s(s)=g'(s)$。\n\n(如果我们有关于源数据集的先验知识,已经知道源的PDF的形式,那么就可以用该PDF对应的CDF来代替上面的逻辑函数。如果不知道PDF的形式,那么逻辑函数就是一个很合理的默认函数,因为在处理很多问题时,逻辑函数都有具有良好的表现。并且,在本例中我们使用的输入观测数据集$x^{(i)}$要么已经被预处理为期望为$0$的数据,要么$x^{(i)}$在自然状态下就是期望为$0$的数据集,如音频信号。而零期望是必须的,因为我们假设了$p_s(s)=g'(s)$,表明$\\mathrm E[s]=0$——这里说明一下,对逻辑函数求导会得到一个对称函数,这也就是PDF,所以这个对称的PDF对应的随机变量必须保证期望为$0$——则有$\\mathrm E[x]=\\mathrm E[As]=0$。顺便再提一点,除了逻辑函数,PDF也经常选用[拉普拉斯分布](https://zh.wikipedia.org/wiki/%E6%8B%89%E6%99%AE%E6%8B%89%E6%96%AF%E5%88%86%E5%B8%83)/[Laplace distribution](https://en.wikipedia.org/wiki/Laplace_distribution) $\\displaystyle p(s)=\\frac{1}{2}e^{-\\lvert s\\rvert}$。)\n\n方阵$W$是模型中的参数,对已给定训练集$\\left\\{x^{(i)};i=1,\\cdots,m\\right\\}$,有对数似然函数为:\n\n$$\n\\mathscr l(W)=\\sum_{i=1}^m\\left(\\sum_{j=1}^n\\log g'\\left(w_j^Tx^{(i)}\\right)+\\log\\lvert W\\rvert\\right)\n$$\n\n我们的目标是用$W$最大化上面的函数,使用性质$\\nabla_W\\lvert W\\rvert=\\lvert W\\rvert\\left(W^{-1}\\right)^T$(参见[第二讲](chapter02.ipynb))对似然函数求导,便可以得到一个以$\\alpha$为学习速率的随机梯度上升的更新规则。对于训练集$x^{(i)}$的更新规则是:\n\n$$\nW:=W+\\alpha\\left(\\begin{bmatrix}\n1-2g\\left(w_1^Tx^{(i)}\\right)\\\\\n1-2g\\left(w_2^Tx^{(i)}\\right)\\\\\n\\vdots\\\\\n1-2g\\left(w_n^Tx^{(i)}\\right)\n\\end{bmatrix}\\left(x^{(i)}\\right)^T+\\left(W^T\\right)^{-1}\\right)\n$$\n\n在算法迭代收敛之后,通过$s^{(i)}=Wx^{(i)}$还原源即可。\n\n**注意:**在构造似然函数时,我们默认假设了各样本$x^{(i)}$是相互独立的(要注意,这不是指样本$x^{(i)}$的各分量间是相互独立的)。于是能够得到训练集的似然函数为$\\prod_ip\\left(x^{(i)};W\\right)$。这个假设在$x^{(i)}$表示演讲音频或其他基于时间序列的数据时是错误的,因为这种样本数据之间并不是相互独立的,不过这一点也可以证明在训练样本充足的前提下,存在相关关系的各样本并不会影响ICA的表现。不过,对于相关的训练样本,当我们使用随机梯度上升时,如果随机改变训练样本载入算法的次序,有时可能会加速算法的收敛过程。(建议准备多组经过乱序的训练集,然后用不同的顺序为模型加载样本,有时有利于快速收敛。)",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown"
]
] |
cb4c97993665fe48acc1967cbb1c3c110347c8a9 | 24,735 | ipynb | Jupyter Notebook | monthly/snapshots/template/2020-07-16 16.10.47.189610/template.ipynb | KyleOS/NotebookScheduler | 6a129eebd7b18830ec1fb560d350634e899db5e9 | [
"MIT"
] | null | null | null | monthly/snapshots/template/2020-07-16 16.10.47.189610/template.ipynb | KyleOS/NotebookScheduler | 6a129eebd7b18830ec1fb560d350634e899db5e9 | [
"MIT"
] | null | null | null | monthly/snapshots/template/2020-07-16 16.10.47.189610/template.ipynb | KyleOS/NotebookScheduler | 6a129eebd7b18830ec1fb560d350634e899db5e9 | [
"MIT"
] | null | null | null | 67.581967 | 15,520 | 0.78775 | [
[
[
"# Sales Report",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"filename = \"../data/sales_january.xlsx\"",
"_____no_output_____"
],
[
"# Parameters\nsnapshotDir = \"monthly/snapshots/template/2020-07-16 16.10.47.189610/\"\nfilename = \"data/sales_february.xlsx\"\n",
"_____no_output_____"
],
[
"data = pd.read_excel(filename, index_col=0)",
"_____no_output_____"
],
[
"filename.rsplit('/', 1)[1]",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"data.plot(kind=\"bar\", title=f\"Sales report from {filename.rsplit('/', 1)[1]}\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4c9d6a6dfb9f35243439dda47b87a13eff448d | 28,401 | ipynb | Jupyter Notebook | python/01_spatial_transformations.ipynb | zivy/ISBI2018_TUTORIAL | 4fa3d695982785f858fc35ac3ff02822bf5a1cdd | [
"Apache-2.0"
] | 26 | 2018-03-15T19:46:16.000Z | 2022-01-11T11:26:28.000Z | python/01_spatial_transformations.ipynb | zivy/ISBI2018_TUTORIAL | 4fa3d695982785f858fc35ac3ff02822bf5a1cdd | [
"Apache-2.0"
] | 1 | 2018-04-02T15:27:13.000Z | 2018-04-02T16:12:04.000Z | python/01_spatial_transformations.ipynb | zivy/ISBI2018_TUTORIAL | 4fa3d695982785f858fc35ac3ff02822bf5a1cdd | [
"Apache-2.0"
] | 16 | 2018-03-16T13:50:03.000Z | 2021-09-11T08:11:46.000Z | 41.461314 | 335 | 0.630506 | [
[
[
"<h1 align=\"center\">SimpleITK Spatial Transformations</h1>\n\n\n**Summary:**\n\n1. Points are represented by vector-like data types: Tuple, Numpy array, List.\n2. Matrices are represented by vector-like data types in row major order.\n3. Default transformation initialization as the identity transform.\n4. Angles specified in radians, distances specified in unknown but consistent units (nm,mm,m,km...).\n5. All global transformations **except translation** are of the form:\n$$T(\\mathbf{x}) = A(\\mathbf{x}-\\mathbf{c}) + \\mathbf{t} + \\mathbf{c}$$\n\n Nomenclature (when printing your transformation):\n\n * Matrix: the matrix $A$\n * Center: the point $\\mathbf{c}$\n * Translation: the vector $\\mathbf{t}$\n * Offset: $\\mathbf{t} + \\mathbf{c} - A\\mathbf{c}$\n6. Bounded transformations, BSplineTransform and DisplacementFieldTransform, behave as the identity transform outside the defined bounds.\n7. DisplacementFieldTransform:\n * Initializing the DisplacementFieldTransform using an image requires that the image's pixel type be sitk.sitkVectorFloat64.\n * Initializing the DisplacementFieldTransform using an image will \"clear out\" your image (your alias to the image will point to an empty, zero sized, image).\n8. Composite transformations are applied in stack order (first added, last applied).",
"_____no_output_____"
],
[
"## Transformation Types\n\nSimpleITK supports the following transformation types.\n\n<table width=\"100%\">\n<tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1TranslationTransform.html\">TranslationTransform</a></td><td>2D or 3D, translation</td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1VersorTransform.html\">VersorTransform</a></td><td>3D, rotation represented by a versor</td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1VersorRigid3DTransform.html\">VersorRigid3DTransform</a></td><td>3D, rigid transformation with rotation represented by a versor</td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1Euler2DTransform.html\">Euler2DTransform</a></td><td>2D, rigid transformation with rotation represented by a Euler angle</td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1Euler3DTransform.html\">Euler3DTransform</a></td><td>3D, rigid transformation with rotation represented by Euler angles</td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1Similarity2DTransform.html\">Similarity2DTransform</a></td><td>2D, composition of isotropic scaling and rigid transformation with rotation represented by a Euler angle</td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1Similarity3DTransform.html\">Similarity3DTransform</a></td><td>3D, composition of isotropic scaling and rigid transformation with rotation represented by a versor</td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1ScaleTransform.html\">ScaleTransform</a></td><td>2D or 3D, anisotropic scaling</td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1ScaleVersor3DTransform.html\">ScaleVersor3DTransform</a></td><td>3D, rigid transformation and anisotropic scale is <bf>added</bf> to the rotation matrix part (not composed as one would expect)</td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1ScaleSkewVersor3DTransform.html\">ScaleSkewVersor3DTransform</a></td><td>3D, rigid transformation with anisotropic scale and skew matrices <bf>added</bf> to the rotation matrix part (not composed as one would expect)</td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1AffineTransform.html\">AffineTransform</a></td><td>2D or 3D, affine transformation.</td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1BSplineTransform.html\">BSplineTransform</a></td><td>2D or 3D, deformable transformation represented by a sparse regular grid of control points. </td></tr>\n <tr><td><a href=\"http://www.itk.org/Doxygen/html/classitk_1_1DisplacementFieldTransform.html\">DisplacementFieldTransform</a></td><td>2D or 3D, deformable transformation represented as a dense regular grid of vectors.</td></tr>\n <tr><td><a href=\"http://www.itk.org/SimpleITKDoxygen/html/classitk_1_1simple_1_1Transform.html\">Transform</a></td>\n <td>A generic transformation. Can represent any of the SimpleITK transformations, and a <b>composite transformation</b> (stack of transformations concatenated via composition, last added, first applied). </td></tr>\n </table>",
"_____no_output_____"
]
],
[
[
"import SimpleITK as sitk\nimport utilities as util\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline \nfrom ipywidgets import interact, fixed\n\nOUTPUT_DIR = \"output\"",
"_____no_output_____"
]
],
[
[
"We will introduce the transformation types, starting with translation and illustrating how to move from a lower to higher parameter space (e.g. translation to rigid). \n\nWe start with the global transformations. All of them <b>except translation</b> are of the form:\n$$T(\\mathbf{x}) = A(\\mathbf{x}-\\mathbf{c}) + \\mathbf{t} + \\mathbf{c}$$\n\nIn ITK speak (when printing your transformation):\n<ul>\n<li>Matrix: the matrix $A$</li>\n<li>Center: the point $\\mathbf{c}$</li>\n<li>Translation: the vector $\\mathbf{t}$</li>\n<li>Offset: $\\mathbf{t} + \\mathbf{c} - A\\mathbf{c}$</li>\n</ul>",
"_____no_output_____"
],
[
"## TranslationTransform\n\nCreate a translation and then transform a point and use the inverse transformation to get the original back.",
"_____no_output_____"
]
],
[
[
"dimension = 2 \noffset = [2]*dimension # use a Python trick to create the offset list based on the dimension\ntranslation = sitk.TranslationTransform(dimension, offset)\nprint(translation)",
"_____no_output_____"
],
[
"point = [10, 11] if dimension==2 else [10, 11, 12] # set point to match dimension\ntransformed_point = translation.TransformPoint(point)\ntranslation_inverse = translation.GetInverse()\nprint('original point: ' + util.point2str(point) + '\\n'\n 'transformed point: ' + util.point2str(transformed_point) + '\\n'\n 'back to original: ' + util.point2str(translation_inverse.TransformPoint(transformed_point)))",
"_____no_output_____"
]
],
[
[
"## Euler2DTransform\n\nRigidly transform a 2D point using a Euler angle parameter specification.\n\nNotice that the dimensionality of the Euler angle based rigid transformation is associated with the class, unlike the translation which is set at construction.\n",
"_____no_output_____"
]
],
[
[
"point = [10, 11]\nrotation2D = sitk.Euler2DTransform()\nrotation2D.SetTranslation((7.2, 8.4))\nrotation2D.SetAngle(np.pi/2)\nprint('original point: ' + util.point2str(point) + '\\n'\n 'transformed point: ' + util.point2str(rotation2D.TransformPoint(point)))",
"_____no_output_____"
]
],
[
[
"## VersorTransform (rotation in 3D)\n\nRotation using a versor, vector part of unit quaternion, parameterization. Quaternion defined by rotation of $\\theta$ radians around axis $n$, is $q = [n*\\sin(\\frac{\\theta}{2}), \\cos(\\frac{\\theta}{2})]$.",
"_____no_output_____"
]
],
[
[
"# Use a versor:\nrotation1 = sitk.VersorTransform([0,0,1,0])\n\n# Use axis-angle:\nrotation2 = sitk.VersorTransform((0,0,1), np.pi)\n\n# Use a matrix:\nrotation3 = sitk.VersorTransform()\nrotation3.SetMatrix([-1, 0, 0, 0, -1, 0, 0, 0, 1]);\n\npoint = (10, 100, 1000)\n\np1 = rotation1.TransformPoint(point)\np2 = rotation2.TransformPoint(point)\np3 = rotation3.TransformPoint(point)\n\nprint('Points after transformation:\\np1=' + str(p1) + \n '\\np2='+ str(p2) + '\\np3='+ str(p3))",
"_____no_output_____"
]
],
[
[
"## Translation to Rigid [3D]\n\nWe only need to copy the translational component.",
"_____no_output_____"
]
],
[
[
"dimension = 3 \nt =(1,2,3) \ntranslation = sitk.TranslationTransform(dimension, t)\n\n# Copy the translational component.\nrigid_euler = sitk.Euler3DTransform()\nrigid_euler.SetTranslation(translation.GetOffset())\n\n# Apply the transformations to the same set of random points and compare the results.\nutil.print_transformation_differences(translation, rigid_euler)",
"_____no_output_____"
]
],
[
[
"## Rotation to Rigid [3D]\nCopy the matrix or versor and <b>center of rotation</b>.",
"_____no_output_____"
]
],
[
[
"rotation_center = (10, 10, 10)\nrotation = sitk.VersorTransform([0,0,1,0], rotation_center)\n\nrigid_versor = sitk.VersorRigid3DTransform()\nrigid_versor.SetRotation(rotation.GetVersor())\n#rigid_versor.SetCenter(rotation.GetCenter()) #intentional error, not copying center of rotation\n\n# Apply the transformations to the same set of random points and compare the results.\nutil.print_transformation_differences(rotation, rigid_versor)",
"_____no_output_____"
]
],
[
[
"In the cell above, when we don't copy the center of rotation we have a constant error vector, $\\mathbf{c}$ - A$\\mathbf{c}$.",
"_____no_output_____"
],
[
"## Similarity [2D]\n\nWhen the center of the similarity transformation is not at the origin the effect of the transformation is not what most of us expect. This is readily visible if we limit the transformation to scaling: $T(\\mathbf{x}) = s\\mathbf{x}-s\\mathbf{c} + \\mathbf{c}$. Changing the transformation's center results in scale + translation.",
"_____no_output_____"
]
],
[
[
"def display_center_effect(x, y, tx, point_list, xlim, ylim):\n tx.SetCenter((x,y))\n transformed_point_list = [ tx.TransformPoint(p) for p in point_list]\n\n plt.scatter(list(np.array(transformed_point_list).T)[0],\n list(np.array(transformed_point_list).T)[1],\n marker='^', \n color='red', label='transformed points')\n plt.scatter(list(np.array(point_list).T)[0],\n list(np.array(point_list).T)[1],\n marker='o', \n color='blue', label='original points')\n plt.xlim(xlim)\n plt.ylim(ylim)\n plt.legend(loc=(0.25,1.01))\n\n# 2D square centered on (0,0)\npoints = [np.array((-1.0,-1.0)), np.array((-1.0,1.0)), np.array((1.0,1.0)), np.array((1.0,-1.0))]\n\n# Scale by 2 \nsimilarity = sitk.Similarity2DTransform();\nsimilarity.SetScale(2)\n\ninteract(display_center_effect, x=(-10,10), y=(-10,10),tx = fixed(similarity), point_list = fixed(points), \n xlim = fixed((-10,10)),ylim = fixed((-10,10)));",
"_____no_output_____"
]
],
[
[
"## Rigid to Similarity [3D]\nCopy the translation, center, and matrix or versor.",
"_____no_output_____"
]
],
[
[
"rotation_center = (100, 100, 100)\ntheta_x = 0.0\ntheta_y = 0.0\ntheta_z = np.pi/2.0\ntranslation = (1,2,3)\n\nrigid_euler = sitk.Euler3DTransform(rotation_center, theta_x, theta_y, theta_z, translation)\n\nsimilarity = sitk.Similarity3DTransform()\nsimilarity.SetMatrix(rigid_euler.GetMatrix())\nsimilarity.SetTranslation(rigid_euler.GetTranslation())\nsimilarity.SetCenter(rigid_euler.GetCenter())\n\n# Apply the transformations to the same set of random points and compare the results.\nutil.print_transformation_differences(rigid_euler, similarity)",
"_____no_output_____"
]
],
[
[
"## Similarity to Affine [3D]\nCopy the translation, center and matrix.",
"_____no_output_____"
]
],
[
[
"rotation_center = (100, 100, 100)\naxis = (0,0,1)\nangle = np.pi/2.0\ntranslation = (1,2,3)\nscale_factor = 2.0\nsimilarity = sitk.Similarity3DTransform(scale_factor, axis, angle, translation, rotation_center)\n\naffine = sitk.AffineTransform(3)\naffine.SetMatrix(similarity.GetMatrix())\naffine.SetTranslation(similarity.GetTranslation())\naffine.SetCenter(similarity.GetCenter())\n\n# Apply the transformations to the same set of random points and compare the results.\nutil.print_transformation_differences(similarity, affine)",
"_____no_output_____"
]
],
[
[
"## Scale Transform\n\nJust as the case was for the similarity transformation above, when the transformations center is not at the origin, instead of a pure anisotropic scaling we also have translation ($T(\\mathbf{x}) = \\mathbf{s}^T\\mathbf{x}-\\mathbf{s}^T\\mathbf{c} + \\mathbf{c}$).",
"_____no_output_____"
]
],
[
[
"# 2D square centered on (0,0).\npoints = [np.array((-1.0,-1.0)), np.array((-1.0,1.0)), np.array((1.0,1.0)), np.array((1.0,-1.0))]\n\n# Scale by half in x and 2 in y.\nscale = sitk.ScaleTransform(2, (0.5,2));\n\n# Interactively change the location of the center.\ninteract(display_center_effect, x=(-10,10), y=(-10,10),tx = fixed(scale), point_list = fixed(points), \n xlim = fixed((-10,10)),ylim = fixed((-10,10)));",
"_____no_output_____"
]
],
[
[
"## Unintentional Misnomers (originally from ITK)\n\nTwo transformation types whose names may mislead you are ScaleVersor and ScaleSkewVersor. Basing your choices on expectations without reading the documentation will surprise you.\n\nScaleVersor - based on name expected a composition of transformations, in practice it is:\n$$T(x) = (R+S)(\\mathbf{x}-\\mathbf{c}) + \\mathbf{t} + \\mathbf{c},\\;\\; \\textrm{where } S= \\left[\\begin{array}{ccc} s_0-1 & 0 & 0 \\\\ 0 & s_1-1 & 0 \\\\ 0 & 0 & s_2-1 \\end{array}\\right]$$ \n\nScaleSkewVersor - based on name expected a composition of transformations, in practice it is:\n$$T(x) = (R+S+K)(\\mathbf{x}-\\mathbf{c}) + \\mathbf{t} + \\mathbf{c},\\;\\; \\textrm{where } S = \\left[\\begin{array}{ccc} s_0-1 & 0 & 0 \\\\ 0 & s_1-1 & 0 \\\\ 0 & 0 & s_2-1 \\end{array}\\right]\\;\\; \\textrm{and } K = \\left[\\begin{array}{ccc} 0 & k_0 & k_1 \\\\ k_2 & 0 & k_3 \\\\ k_4 & k_5 & 0 \\end{array}\\right]$$ \n\nNote that ScaleSkewVersor is is an over-parametrized version of the affine transform, 15 parameters (scale, skew, versor, translation) vs. 12 parameters (matrix, translation).",
"_____no_output_____"
],
[
"## Bounded Transformations\n\nSimpleITK supports two types of bounded non-rigid transformations, BSplineTransform (sparse representation) and \tDisplacementFieldTransform (dense representation).\n\nTransforming a point that is outside the bounds will return the original point - identity transform.",
"_____no_output_____"
],
[
"## BSpline\nUsing a sparse set of control points to control a free form deformation. Using the cell below it is clear that the BSplineTransform allows for folding and tearing.",
"_____no_output_____"
]
],
[
[
"# Create the transformation (when working with images it is easier to use the BSplineTransformInitializer function\n# or its object oriented counterpart BSplineTransformInitializerFilter).\ndimension = 2\nspline_order = 3\ndirection_matrix_row_major = [1.0,0.0,0.0,1.0] # identity, mesh is axis aligned\norigin = [-1.0,-1.0] \ndomain_physical_dimensions = [2,2]\n\nbspline = sitk.BSplineTransform(dimension, spline_order)\nbspline.SetTransformDomainOrigin(origin)\nbspline.SetTransformDomainDirection(direction_matrix_row_major)\nbspline.SetTransformDomainPhysicalDimensions(domain_physical_dimensions)\nbspline.SetTransformDomainMeshSize((4,3))\n\n# Random displacement of the control points.\noriginalControlPointDisplacements = np.random.random(len(bspline.GetParameters()))\nbspline.SetParameters(originalControlPointDisplacements)\n\n# Apply the BSpline transformation to a grid of points \n# starting the point set exactly at the origin of the BSpline mesh is problematic as\n# these points are considered outside the transformation's domain,\n# remove epsilon below and see what happens.\nnumSamplesX = 10\nnumSamplesY = 20\n \ncoordsX = np.linspace(origin[0]+np.finfo(float).eps, origin[0] + domain_physical_dimensions[0], numSamplesX)\ncoordsY = np.linspace(origin[1]+np.finfo(float).eps, origin[1] + domain_physical_dimensions[1], numSamplesY)\nXX, YY = np.meshgrid(coordsX, coordsY)\n\ninteract(util.display_displacement_scaling_effect, s= (-1.5,1.5), original_x_mat = fixed(XX), original_y_mat = fixed(YY),\n tx = fixed(bspline), original_control_point_displacements = fixed(originalControlPointDisplacements)); ",
"_____no_output_____"
]
],
[
[
"## DisplacementField\n\nA dense set of vectors representing the displacement inside the given domain. The most generic representation of a transformation.",
"_____no_output_____"
]
],
[
[
"# Create the displacement field. \n \n# When working with images the safer thing to do is use the image based constructor,\n# sitk.DisplacementFieldTransform(my_image), all the fixed parameters will be set correctly and the displacement\n# field is initialized using the vectors stored in the image. SimpleITK requires that the image's pixel type be \n# sitk.sitkVectorFloat64.\ndisplacement = sitk.DisplacementFieldTransform(2)\nfield_size = [10,20]\nfield_origin = [-1.0,-1.0] \nfield_spacing = [2.0/9.0,2.0/19.0] \nfield_direction = [1,0,0,1] # direction cosine matrix (row major order) \n\n# Concatenate all the information into a single list\ndisplacement.SetFixedParameters(field_size+field_origin+field_spacing+field_direction)\n# Set the interpolator, either sitkLinear which is default or nearest neighbor\ndisplacement.SetInterpolator(sitk.sitkNearestNeighbor)\n\noriginalDisplacements = np.random.random(len(displacement.GetParameters()))\ndisplacement.SetParameters(originalDisplacements)\n\ncoordsX = np.linspace(field_origin[0], field_origin[0]+(field_size[0]-1)*field_spacing[0], field_size[0])\ncoordsY = np.linspace(field_origin[1], field_origin[1]+(field_size[1]-1)*field_spacing[1], field_size[1])\nXX, YY = np.meshgrid(coordsX, coordsY)\n\ninteract(util.display_displacement_scaling_effect, s= (-1.5,1.5), original_x_mat = fixed(XX), original_y_mat = fixed(YY),\n tx = fixed(displacement), original_control_point_displacements = fixed(originalDisplacements)); ",
"_____no_output_____"
]
],
[
[
"## Composite transform (Transform)\n\nThe generic SimpleITK transform class. This class can represent both a single transformation (global, local), or a composite transformation (multiple transformations applied one after the other). This is the output typed returned by the SimpleITK registration framework. \n\nThe choice of whether to use a composite transformation or compose transformations on your own has subtle differences in the registration framework.\n\nComposite transforms enable a combination of a global transformation with multiple local/bounded transformations. This is useful if we want to apply deformations only in regions that deform while other regions are only effected by the global transformation.\n\nThe following code illustrates this, where the whole region is translated and subregions have different deformations.",
"_____no_output_____"
]
],
[
[
"# Global transformation.\ntranslation = sitk.TranslationTransform(2,(1.0,0.0))\n\n# Displacement in region 1.\ndisplacement1 = sitk.DisplacementFieldTransform(2)\nfield_size = [10,20]\nfield_origin = [-1.0,-1.0] \nfield_spacing = [2.0/9.0,2.0/19.0] \nfield_direction = [1,0,0,1] # direction cosine matrix (row major order) \n\n# Concatenate all the information into a single list.\ndisplacement1.SetFixedParameters(field_size+field_origin+field_spacing+field_direction)\ndisplacement1.SetParameters(np.ones(len(displacement1.GetParameters())))\n\n# Displacement in region 2.\ndisplacement2 = sitk.DisplacementFieldTransform(2)\nfield_size = [10,20]\nfield_origin = [1.0,-3] \nfield_spacing = [2.0/9.0,2.0/19.0] \nfield_direction = [1,0,0,1] #direction cosine matrix (row major order) \n\n# Concatenate all the information into a single list.\ndisplacement2.SetFixedParameters(field_size+field_origin+field_spacing+field_direction)\ndisplacement2.SetParameters(-1.0*np.ones(len(displacement2.GetParameters())))\n\n# Composite transform which applies the global and local transformations.\ncomposite = sitk.Transform(translation)\ncomposite.AddTransform(displacement1)\ncomposite.AddTransform(displacement2)\n\n# Apply the composite transformation to points in ([-1,-3],[3,1]) and \n# display the deformation using a quiver plot.\n \n# Generate points.\nnumSamplesX = 10\nnumSamplesY = 10 \ncoordsX = np.linspace(-1.0, 3.0, numSamplesX)\ncoordsY = np.linspace(-3.0, 1.0, numSamplesY)\nXX, YY = np.meshgrid(coordsX, coordsY)\n\n# Transform points and compute deformation vectors.\npointsX = np.zeros(XX.shape)\npointsY = np.zeros(XX.shape)\nfor index, value in np.ndenumerate(XX):\n px,py = composite.TransformPoint((value, YY[index]))\n pointsX[index]=px - value \n pointsY[index]=py - YY[index]\n \nplt.quiver(XX, YY, pointsX, pointsY); ",
"_____no_output_____"
]
],
[
[
"## Writing and Reading\n\nThe SimpleITK.ReadTransform() returns a SimpleITK.Transform . The content of the file can be any of the SimpleITK transformations or a composite (set of transformations). ",
"_____no_output_____"
]
],
[
[
"import os\n\n# Create a 2D rigid transformation, write it to disk and read it back.\nbasic_transform = sitk.Euler2DTransform()\nbasic_transform.SetTranslation((1.0,2.0))\nbasic_transform.SetAngle(np.pi/2)\n\nfull_file_name = os.path.join(OUTPUT_DIR, 'euler2D.tfm')\n\nsitk.WriteTransform(basic_transform, full_file_name)\n\n# The ReadTransform function returns an sitk.Transform no matter the type of the transform \n# found in the file (global, bounded, composite).\nread_result = sitk.ReadTransform(full_file_name)\n\nprint('Different types: '+ str(type(read_result) != type(basic_transform)))\nutil.print_transformation_differences(basic_transform, read_result)\n\n\n# Create a composite transform then write and read.\ndisplacement = sitk.DisplacementFieldTransform(2)\nfield_size = [10,20]\nfield_origin = [-10.0,-100.0] \nfield_spacing = [20.0/(field_size[0]-1),200.0/(field_size[1]-1)] \nfield_direction = [1,0,0,1] #direction cosine matrix (row major order)\n\n# Concatenate all the information into a single list.\ndisplacement.SetFixedParameters(field_size+field_origin+field_spacing+field_direction)\ndisplacement.SetParameters(np.random.random(len(displacement.GetParameters())))\n\ncomposite_transform = sitk.Transform(basic_transform)\ncomposite_transform.AddTransform(displacement)\n\nfull_file_name = os.path.join(OUTPUT_DIR, 'composite.tfm')\n\nsitk.WriteTransform(composite_transform, full_file_name)\nread_result = sitk.ReadTransform(full_file_name)\n\nutil.print_transformation_differences(composite_transform, read_result) ",
"_____no_output_____"
]
],
[
[
"<a href=\"02_images_and_resampling.ipynb\"><h2 align=right>Next »</h2></a>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb4ca12fb7e8ed4ff7ddbf87e94093baee9b2e0d | 588,560 | ipynb | Jupyter Notebook | Data analysis/Ign_dataset_Analysis.ipynb | acehanks/projects | d877b6124fe1d96d4c3f638ad780040064b86461 | [
"MIT"
] | null | null | null | Data analysis/Ign_dataset_Analysis.ipynb | acehanks/projects | d877b6124fe1d96d4c3f638ad780040064b86461 | [
"MIT"
] | null | null | null | Data analysis/Ign_dataset_Analysis.ipynb | acehanks/projects | d877b6124fe1d96d4c3f638ad780040064b86461 | [
"MIT"
] | null | null | null | 501.756181 | 111,469 | 0.923257 | [
[
[
"In this dataset, were are going to be exploring sales information provided.",
"_____no_output_____"
]
],
[
[
"# This Python 3 environment comes with many helpful analytics libraries installed\n# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python\n# For example, here's several helpful packages to load in \n\nimport numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\n\n# Input data files are available in the \"../input/\" directory.\n# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory\n\nfrom subprocess import check_output\nprint(check_output([\"ls\", \"../input\"]).decode(\"utf8\"))\n\n# Any results you write to the current directory are saved as output.",
"Video_Games_Sales_as_at_22_Dec_2016.csv\n\n"
]
],
[
[
"Let us start by importing some helper libraries and the dataset as well",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nfrom matplotlib import pyplot as plt\n%matplotlib inline\nfrom brewer2mpl import qualitative",
"_____no_output_____"
],
[
"df= pd.read_csv(\"../input/Video_Games_Sales_as_at_22_Dec_2016.csv\")\ndf.shape",
"_____no_output_____"
]
],
[
[
"Now that we know the shape of the dataset, let's have a peek at the data and try to find if they are any missing values.",
"_____no_output_____"
]
],
[
[
"df.head()",
"_____no_output_____"
]
],
[
[
"The cell below shows the data type of the columns",
"_____no_output_____"
]
],
[
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 16719 entries, 0 to 16718\nData columns (total 16 columns):\nName 16717 non-null object\nPlatform 16719 non-null object\nYear_of_Release 16450 non-null float64\nGenre 16717 non-null object\nPublisher 16665 non-null object\nNA_Sales 16719 non-null float64\nEU_Sales 16719 non-null float64\nJP_Sales 16719 non-null float64\nOther_Sales 16719 non-null float64\nGlobal_Sales 16719 non-null float64\nCritic_Score 8137 non-null float64\nCritic_Count 8137 non-null float64\nUser_Score 10015 non-null object\nUser_Count 7590 non-null float64\nDeveloper 10096 non-null object\nRating 9950 non-null object\ndtypes: float64(9), object(7)\nmemory usage: 2.0+ MB\n"
]
],
[
[
"The cell below shows the column name and the number of empty rows.",
"_____no_output_____"
]
],
[
[
"df.isnull().sum()",
"_____no_output_____"
]
],
[
[
"A list of all the columns in the dataset",
"_____no_output_____"
]
],
[
[
"df.columns.tolist()",
"_____no_output_____"
]
],
[
[
"Calculating the % of missing values ",
"_____no_output_____"
]
],
[
[
"df_na= ( df.isnull().sum() / len(df) ) * 100\ndf_na= df_na.drop(df_na[df_na == 0].index).sort_values(ascending= False)",
"_____no_output_____"
],
[
"f, ax= plt.subplots(figsize=(12, 8))\nplt.xticks(rotation='90')\nsns.barplot(x=df_na.index, y=df_na.values)\nax.set(title='Missing Values Plot', ylabel='% Missing')\n",
"_____no_output_____"
]
],
[
[
"Unique Gaming platforms",
"_____no_output_____"
]
],
[
[
"df.Platform.unique()",
"_____no_output_____"
],
[
"#df.Platform.value_counts()\nssc = df.Platform.value_counts()\nf, ax= plt.subplots(figsize=(12, 8))\nplt.xticks(rotation='90')\nsns.barplot(x=ssc.values, y=ssc.index, orient='h')\nax.set(title='Consoles by count', ylabel='Count')\nf.tight_layout()",
"_____no_output_____"
]
],
[
[
"dropping all NA values",
"_____no_output_____"
]
],
[
[
"\ndf_clean= df.dropna(axis=0)\ndf_clean.shape",
"_____no_output_____"
],
[
"ssc = df_clean.Platform.value_counts()\nf, ax= plt.subplots(figsize=(12, 8))\nplt.xticks(rotation='90')\nsns.barplot(x=ssc.values, y=ssc.index, orient='h')\nax.set(title='Consoles by count after dropping NAs', ylabel='Count')\nf.tight_layout()",
"_____no_output_____"
],
[
"#df['User_Score']= df['User_Score'].convert_objects(convert_numeric=True)\ndf_clean.User_Score= df_clean.User_Score.astype('float')\n#df.User_Score.dtype\n#df['User_Score'] = video['User_Score'].convert_objects(convert_numeric= True)",
"/opt/conda/lib/python3.6/site-packages/pandas/core/generic.py:3110: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n self[name] = value\n"
]
],
[
[
"Plot of the user score v the critic score of games. It appears the users and the critics agree on \ngames with score greater than 8.",
"_____no_output_____"
]
],
[
[
"sns.jointplot(x='User_Score', y='Critic_Score', data=df_clean, kind='hex', cmap='coolwarm', size=7)\n",
"_____no_output_____"
]
],
[
[
"Critic score v critic count. From this plot, we observe that few critics give scores above 80.",
"_____no_output_____"
]
],
[
[
"sns.jointplot(x='Critic_Score', y='Critic_Count', data=df_clean, kind='hex', cmap='plasma', size=7)",
"_____no_output_____"
]
],
[
[
"CORRELATION BETWEEN COLUMNS",
"_____no_output_____"
]
],
[
[
"stats=['Year_of_Release','NA_Sales', 'EU_Sales', 'JP_Sales', 'Other_Sales', \n 'Global_Sales', 'Critic_Score', 'Critic_Count', 'User_Score', 'User_Count', \n 'Rating']\ncorrmat = df_clean[stats].corr()\n\nf, ax = plt.subplots(figsize=(10, 7))\nplt.xticks(rotation='90')\nplt.title('correlation between columns')\nsns.heatmap(corrmat, square=True, linewidths=.5, annot=True)",
"_____no_output_____"
]
],
[
[
"Taking a look at Playstation",
"_____no_output_____"
]
],
[
[
"play= df_clean[(df_clean['Platform']== 'PS2') | (df_clean['Platform']== 'PS3')\n | (df_clean['Platform']== 'PS')| (df_clean['Platform']== 'PS4')]\nplay.shape",
"_____no_output_____"
]
],
[
[
"Playststation Global 1994-2016",
"_____no_output_____"
]
],
[
[
"sales_Play= play.groupby(['Year_of_Release', 'Platform'])['Global_Sales'].sum()\nsales_Play.unstack().plot(kind='bar',stacked=True, colormap= 'Oranges', grid=False)\nax.set(title='Playststation Global over the year', ylabel='Cumulative count')",
"_____no_output_____"
]
],
[
[
"Top selling genre for Playstation",
"_____no_output_____"
]
],
[
[
"sales_Play= play.groupby(['Genre', 'Platform'])['Global_Sales'].sum()\nsales_Play.unstack().plot(kind='bar',stacked=True, colormap= 'Oranges', grid=False)",
"_____no_output_____"
]
],
[
[
"Rating of the games made",
"_____no_output_____"
]
],
[
[
"sales_Play= play.groupby(['Rating', 'Platform'])['Global_Sales'].sum()\nsales_Play.unstack().plot(kind='bar',stacked=True, colormap= 'Oranges', grid=False)",
"_____no_output_____"
]
],
[
[
"Taking a closer look at Xbox ",
"_____no_output_____"
]
],
[
[
"xb= df_clean[(df_clean['Platform']== 'X360') | (df_clean['Platform']== 'XOne')\n | (df_clean['Platform']== 'XB')]\nxb.shape",
"_____no_output_____"
]
],
[
[
"Global sales of the Xbox consoles globally 1994-2016",
"_____no_output_____"
]
],
[
[
"sales_xb= xb.groupby(['Year_of_Release', 'Platform'])['Global_Sales'].sum()\nsales_xb.unstack().plot(kind='bar',stacked=True, colormap= 'Vega20', grid=False)",
"/opt/conda/lib/python3.6/site-packages/matplotlib/cbook/deprecation.py:106: MatplotlibDeprecationWarning: The Vega20 colormap was deprecated in version 2.0. Use tab20 instead.\n warnings.warn(message, mplDeprecation, stacklevel=1)\n"
]
],
[
[
"Top selling genre per Xbox console. The top selling genre is the shooter, which makes sense because of the \nhalo franchise.",
"_____no_output_____"
]
],
[
[
"sales_xb= xb.groupby(['Genre', 'Platform'])['Global_Sales'].sum()\nsales_xb.unstack().plot(kind='bar',stacked=True, colormap= 'Vega20', grid=False)",
"/opt/conda/lib/python3.6/site-packages/matplotlib/cbook/deprecation.py:106: MatplotlibDeprecationWarning: The Vega20 colormap was deprecated in version 2.0. Use tab20 instead.\n warnings.warn(message, mplDeprecation, stacklevel=1)\n"
]
],
[
[
"Rating and global sales",
"_____no_output_____"
]
],
[
[
"sales_xb= xb.groupby(['Rating', 'Platform'])['Global_Sales'].sum()\nsales_xb.unstack().plot(kind='bar',stacked=True, colormap= 'Vega20', grid=False)",
"/opt/conda/lib/python3.6/site-packages/matplotlib/cbook/deprecation.py:106: MatplotlibDeprecationWarning: The Vega20 colormap was deprecated in version 2.0. Use tab20 instead.\n warnings.warn(message, mplDeprecation, stacklevel=1)\n"
]
],
[
[
"Taking a closer look at nintendo ",
"_____no_output_____"
]
],
[
[
"nintendo= df_clean[(df_clean['Platform']== 'DS') | (df_clean['Platform']== 'Wii')\n | (df_clean['Platform']== 'GC')| (df_clean['Platform']== 'GBA')\n |(df_clean['Platform']== '3DS') | (df_clean['Platform']== 'WiiU')]\nnintendo.shape",
"_____no_output_____"
]
],
[
[
"Platform and total global sales from 1994-2016",
"_____no_output_____"
]
],
[
[
"nintendo_sales= nintendo.groupby(['Year_of_Release', 'Platform'])['Global_Sales'].sum()\nnintendo_sales.unstack().plot(kind='bar',stacked=True, colormap= 'Set1', grid=False)",
"_____no_output_____"
]
],
[
[
"Genre and total sales on platform. Nintendo looks to be selling alot of sports oriented games, especially on the \nWii. However the Wii U is struggling in sales.",
"_____no_output_____"
]
],
[
[
"nintendo_sales= nintendo.groupby(['Genre', 'Platform'])['Global_Sales'].sum()\nnintendo_sales.unstack().plot(kind='bar',stacked=True, colormap= 'Set1', grid=False)",
"_____no_output_____"
]
],
[
[
"Rating and total global sales. Nintendo sold majorly in the category of E (everyone)",
"_____no_output_____"
]
],
[
[
"nintendo_sales= nintendo.groupby(['Rating', 'Platform'])['Global_Sales'].sum()\nnintendo_sales.unstack().plot(kind='bar',stacked=True, colormap= 'Set1', grid=False)",
"_____no_output_____"
],
[
"current_gen= df_clean[(df_clean['Platform']== 'Wii') | (df_clean['Platform']== 'X360') | \n (df_clean['Platform']== 'PS3')]\ncurrent_gen.shape",
"_____no_output_____"
]
],
[
[
"Comparing the top selling platforms, last generation",
"_____no_output_____"
]
],
[
[
"current_gen_sales= current_gen.groupby(['Year_of_Release', 'Platform'])['Global_Sales'].sum()\ncurrent_gen_sales.unstack().plot(kind='bar',stacked=True, colormap= 'Set2', grid=False)",
"_____no_output_____"
],
[
"current_gen_sales= current_gen.groupby(['Genre', 'Platform'])['Global_Sales'].sum()\ncurrent_gen_sales.unstack().plot(kind='bar',stacked=True, colormap= 'Set2', grid=False)",
"_____no_output_____"
],
[
"current_gen_sales= current_gen.groupby(['Rating', 'Platform'])['Global_Sales'].sum()\ncurrent_gen_sales.unstack().plot(kind='bar',stacked=True, colormap= 'Set2', grid=False)",
"_____no_output_____"
]
],
[
[
"last generation sales from North America",
"_____no_output_____"
]
],
[
[
"current_gen_sales= current_gen.groupby(['Year_of_Release', 'Platform'])['NA_Sales'].sum()\ncurrent_gen_sales.unstack().plot(kind='bar',stacked=True, colormap= 'Blues', grid=False)",
"_____no_output_____"
]
],
[
[
"Last generation sales from Japan",
"_____no_output_____"
]
],
[
[
"current_gen_sales= current_gen.groupby(['Year_of_Release', 'Platform'])['JP_Sales'].sum()\ncurrent_gen_sales.unstack().plot(kind='bar',stacked=True, colormap= 'Blues', grid=False)",
"_____no_output_____"
]
],
[
[
"Last generation sales from EU",
"_____no_output_____"
]
],
[
[
"current_gen_sales= current_gen.groupby(['Year_of_Release', 'Platform'])['EU_Sales'].sum()\ncurrent_gen_sales.unstack().plot(kind='bar',stacked=True, colormap= 'Blues', grid=False)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4ca7fc44eb83a9292775ddddd833140e8e2db8 | 189,708 | ipynb | Jupyter Notebook | Python-Data-Science-Handbook/notebooks/04.02-Simple-Scatter-Plots.ipynb | Little-Potato-1990/learn_python | 9e54d150ef73e4bf53f8cd9b28a2a8bc65593fe1 | [
"Apache-2.0"
] | null | null | null | Python-Data-Science-Handbook/notebooks/04.02-Simple-Scatter-Plots.ipynb | Little-Potato-1990/learn_python | 9e54d150ef73e4bf53f8cd9b28a2a8bc65593fe1 | [
"Apache-2.0"
] | null | null | null | Python-Data-Science-Handbook/notebooks/04.02-Simple-Scatter-Plots.ipynb | Little-Potato-1990/learn_python | 9e54d150ef73e4bf53f8cd9b28a2a8bc65593fe1 | [
"Apache-2.0"
] | 1 | 2022-01-14T13:18:51.000Z | 2022-01-14T13:18:51.000Z | 500.548813 | 62,568 | 0.944942 | [
[
[
"<!--NAVIGATION-->\n< [简单的折线图](04.01-Simple-Line-Plots.ipynb) | [目录](Index.ipynb) | [误差可视化](04.03-Errorbars.ipynb) >\n\n<a href=\"https://colab.research.google.com/github/wangyingsm/Python-Data-Science-Handbook/blob/master/notebooks/04.02-Simple-Scatter-Plots.ipynb\"><img align=\"left\" src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open in Colab\" title=\"Open and Execute in Google Colaboratory\"></a>\n",
"_____no_output_____"
],
[
"# Simple Scatter Plots\n\n# 简单散点图",
"_____no_output_____"
],
[
"> Another commonly used plot type is the simple scatter plot, a close cousin of the line plot.\nInstead of points being joined by line segments, here the points are represented individually with a dot, circle, or other shape.\nWe’ll start by setting up the notebook for plotting and importing the functions we will use:\n\n另一种常用的图表类型是简单散点图,它是折线图的近亲。不像折线图,图中的点连接起来组成连线,散点图中的点都是独立分布的点状、圆圈或其他形状。本节开始我们也是首先将需要用到的图表工具和函数导入到notebook中:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nplt.style.use('seaborn-whitegrid')\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"## Scatter Plots with ``plt.plot``\n\n## 使用 `plt.plot` 绘制散点图\n\n> In the previous section we looked at ``plt.plot``/``ax.plot`` to produce line plots.\nIt turns out that this same function can produce scatter plots as well:\n\n在上一节中,我们介绍了`plt.plot`/`ax.plot`方法绘制折线图。这两个方法也可以同样用来绘制散点图:",
"_____no_output_____"
]
],
[
[
"x = np.linspace(0, 10, 30)\ny = np.sin(x)\n\nplt.plot(x, y, 'o', color='black');",
"_____no_output_____"
]
],
[
[
"> The third argument in the function call is a character that represents the type of symbol used for the plotting. Just as you can specify options such as ``'-'``, ``'--'`` to control the line style, the marker style has its own set of short string codes. The full list of available symbols can be seen in the documentation of ``plt.plot``, or in Matplotlib's online documentation. Most of the possibilities are fairly intuitive, and we'll show a number of the more common ones here:\n\n传递给函数的第三个参数是使用一个字符代表的图表绘制点的类型。就像你可以使用`'-'`或`'--'`来控制线条的风格那样,点的类型风格也可以使用短字符串代码来表示。所有可用的符号可以通过`plt.plot`文档或Matplotlib在线文档进行查阅。大多数的代码都是非常直观的,我们使用下面的例子可以展示那些最通用的符号:",
"_____no_output_____"
]
],
[
[
"rng = np.random.RandomState(0)\nfor marker in ['o', '.', ',', 'x', '+', 'v', '^', '<', '>', 's', 'd']:\n plt.plot(rng.rand(5), rng.rand(5), marker,\n label=\"marker='{0}'\".format(marker))\nplt.legend(numpoints=1)\nplt.xlim(0, 1.8);",
"_____no_output_____"
]
],
[
[
"> For even more possibilities, these character codes can be used together with line and color codes to plot points along with a line connecting them:\n\n而且这些符号代码可以和线条、颜色代码一起使用,这会在折线图的基础上绘制出散点:",
"_____no_output_____"
]
],
[
[
"plt.plot(x, y, '-ok');",
"_____no_output_____"
]
],
[
[
"> Additional keyword arguments to ``plt.plot`` specify a wide range of properties of the lines and markers:\n\n`plt.plot`还有很多额外的关键字参数用来指定广泛的线条和点的属性:",
"_____no_output_____"
]
],
[
[
"plt.plot(x, y, '-p', color='gray',\n markersize=15, linewidth=4,\n markerfacecolor='white',\n markeredgecolor='gray',\n markeredgewidth=2)\nplt.ylim(-1.2, 1.2);",
"_____no_output_____"
]
],
[
[
"> This type of flexibility in the ``plt.plot`` function allows for a wide variety of possible visualization options.\nFor a full description of the options available, refer to the ``plt.plot`` documentation.\n\n`plt.plot`函数的这种灵活性提供了很多的可视化选择。查阅`plt.plot`帮助文档获得完整的选项说明。",
"_____no_output_____"
],
[
"## Scatter Plots with ``plt.scatter``\n\n## 使用`plt.scatter`绘制散点图\n\n> A second, more powerful method of creating scatter plots is the ``plt.scatter`` function, which can be used very similarly to the ``plt.plot`` function:\n\n第二种更强大的绘制散点图的方法是使用`plt.scatter`函数,它的使用方法和`plt.plot`类似:",
"_____no_output_____"
]
],
[
[
"plt.scatter(x, y, marker='o');",
"_____no_output_____"
]
],
[
[
"> The primary difference of ``plt.scatter`` from ``plt.plot`` is that it can be used to create scatter plots where the properties of each individual point (size, face color, edge color, etc.) can be individually controlled or mapped to data.\n\n`plt.scatter`和`plt.plot`的主要区别在于,`plt.scatter`可以针对每个点设置不同属性(大小、填充颜色、边缘颜色等),还可以通过数据集合对这些属性进行设置。\n\n> Let's show this by creating a random scatter plot with points of many colors and sizes.\nIn order to better see the overlapping results, we'll also use the ``alpha`` keyword to adjust the transparency level:\n\n让我们通过一个随机值数据集绘制不同颜色和大小的散点图来说明。为了更好的查看重叠的结果,我们还使用了`alpha`关键字参数对点的透明度进行了调整:",
"_____no_output_____"
]
],
[
[
"rng = np.random.RandomState(0)\nx = rng.randn(100)\ny = rng.randn(100)\ncolors = rng.rand(100)\nsizes = 1000 * rng.rand(100)\n\nplt.scatter(x, y, c=colors, s=sizes, alpha=0.3,\n cmap='viridis')\nplt.colorbar(); # 显示颜色对比条",
"_____no_output_____"
]
],
[
[
"> Notice that the color argument is automatically mapped to a color scale (shown here by the ``colorbar()`` command), and that the size argument is given in pixels.\nIn this way, the color and size of points can be used to convey information in the visualization, in order to visualize multidimensional data.\n\n注意图表右边有一个颜色对比条(这里通过`colormap()`函数输出),图表中的点大小的单位是像素。使用这种方法,散点的颜色和大小都能用来展示数据信息,在希望展示多个维度数据集合的情况下很直观。\n\n> For example, we might use the Iris data from Scikit-Learn, where each sample is one of three types of flowers that has had the size of its petals and sepals carefully measured:\n\n例如,当我们使用Scikit-learn中的鸢尾花数据集,里面的每个样本都是三种鸢尾花中的其中一种,并带有仔细测量的花瓣和花萼的尺寸数据:",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import load_iris\niris = load_iris()\nfeatures = iris.data.T\n\nplt.scatter(features[0], features[1], alpha=0.2,\n s=100*features[3], c=iris.target, cmap='viridis')\nplt.xlabel(iris.feature_names[0])\nplt.ylabel(iris.feature_names[1]);",
"_____no_output_____"
]
],
[
[
"> We can see that this scatter plot has given us the ability to simultaneously explore four different dimensions of the data:\nthe (x, y) location of each point corresponds to the sepal length and width, the size of the point is related to the petal width, and the color is related to the particular species of flower.\nMulticolor and multifeature scatter plots like this can be useful for both exploration and presentation of data.\n\n我们可以从上图中看出,可以通过散点图同时展示该数据集的四个不同维度:图中的(x, y)位置代表每个样本的花萼的长度和宽度,散点的大小代表每个样本的花瓣的宽度,而散点的颜色代表一种特定的鸢尾花类型。如上图的多种颜色和多种属性的散点图对于我们分析和展示数据集时都非常有帮助。",
"_____no_output_____"
],
[
"## ``plot`` Versus ``scatter``: A Note on Efficiency\n\n## `plot` 和 `scatter` 对比:性能提醒\n\n> Aside from the different features available in ``plt.plot`` and ``plt.scatter``, why might you choose to use one over the other? While it doesn't matter as much for small amounts of data, as datasets get larger than a few thousand points, ``plt.plot`` can be noticeably more efficient than ``plt.scatter``.\nThe reason is that ``plt.scatter`` has the capability to render a different size and/or color for each point, so the renderer must do the extra work of constructing each point individually.\nIn ``plt.plot``, on the other hand, the points are always essentially clones of each other, so the work of determining the appearance of the points is done only once for the entire set of data.\nFor large datasets, the difference between these two can lead to vastly different performance, and for this reason, ``plt.plot`` should be preferred over ``plt.scatter`` for large datasets.\n\n除了上面说的`plt.plot`和`plt.scatter`对于每个散点不同属性的支持不同之外,还有别的因素影响对这两个函数的选择吗?对于小的数据集来说,两者并无差别,当数据集增长到几千个点时,`plt.plot`会明显比`plt.scatter`的性能要高。造成这个差异的原因是`plt.scatter`支持每个点使用不同的大小和颜色,因此渲染每个点时需要完成更多额外的工作。而`plt.plot`来说,每个点都是简单的复制另一个点产生,因此对于整个数据集来说,确定每个点的展示属性的工作仅需要进行一次即可。对于很大的数据集来说,这个差异会导致两者性能的巨大区别,因此,对于大数据集应该优先使用`plt.plot`函数。",
"_____no_output_____"
],
[
"<!--NAVIGATION-->\n< [简单的折线图](04.01-Simple-Line-Plots.ipynb) | [目录](Index.ipynb) | [误差可视化](04.03-Errorbars.ipynb) >\n\n<a href=\"https://colab.research.google.com/github/wangyingsm/Python-Data-Science-Handbook/blob/master/notebooks/04.02-Simple-Scatter-Plots.ipynb\"><img align=\"left\" src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open in Colab\" title=\"Open and Execute in Google Colaboratory\"></a>\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cb4cae63b85ad688ddc13760abc7702c90029f77 | 16,503 | ipynb | Jupyter Notebook | Thinkful Program/Model Preparation/Thinkful 15.8.ipynb | rleary90/sturdy-octo-happiness | 0b11c8575fe984f632f52f4e326defab5a28fd2e | [
"MIT"
] | null | null | null | Thinkful Program/Model Preparation/Thinkful 15.8.ipynb | rleary90/sturdy-octo-happiness | 0b11c8575fe984f632f52f4e326defab5a28fd2e | [
"MIT"
] | null | null | null | Thinkful Program/Model Preparation/Thinkful 15.8.ipynb | rleary90/sturdy-octo-happiness | 0b11c8575fe984f632f52f4e326defab5a28fd2e | [
"MIT"
] | null | null | null | 37.937931 | 406 | 0.438708 | [
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom sqlalchemy import create_engine\nimport warnings\n\nwarnings.filterwarnings('ignore')\nsns.set(style=\"whitegrid\")",
"_____no_output_____"
],
[
"postgres_user = 'dsbc_student'\npostgres_pw = '7*.8G9QH21'\npostgres_host = '142.93.121.174'\npostgres_port = '5432'\npostgres_db = 'useducation'\n\nengine = create_engine('postgresql://{}:{}@{}:{}/{}'.format(\n postgres_user, postgres_pw, postgres_host, postgres_port, postgres_db))\n\neducation_df = pd.read_sql_query('select * from useducation',con=engine)\n\nengine.dispose()",
"_____no_output_____"
],
[
"\nfill_list = [\"ENROLL\", \"TOTAL_REVENUE\", \"FEDERAL_REVENUE\",\n \"STATE_REVENUE\", \"LOCAL_REVENUE\", \"TOTAL_EXPENDITURE\",\n \"INSTRUCTION_EXPENDITURE\", \"SUPPORT_SERVICES_EXPENDITURE\",\n \"OTHER_EXPENDITURE\", \"CAPITAL_OUTLAY_EXPENDITURE\", \"GRADES_PK_G\",\n \"GRADES_KG_G\", \"GRADES_4_G\", \"GRADES_8_G\", \"GRADES_12_G\", \"GRADES_1_8_G\",\n \"GRADES_9_12_G\", \"GRADES_ALL_G\"]\n\nstates = education_df[\"STATE\"].unique()\n\nfor state in states:\n education_df.loc[education_df[\"STATE\"] == state, fill_list] = education_df.loc[education_df[\"STATE\"] == state, fill_list].interpolate()\n\neducation_df.dropna(inplace=True)",
"_____no_output_____"
],
[
"education_df[\"overall_score\"] = (education_df[\"GRADES_4_G\"]*((education_df[\"AVG_MATH_4_SCORE\"] + education_df[\"AVG_READING_4_SCORE\"])*0.5) + education_df[\"GRADES_8_G\"]\n * ((education_df[\"AVG_MATH_8_SCORE\"] + education_df[\"AVG_READING_8_SCORE\"])*0.5))/(education_df[\"GRADES_4_G\"] + education_df[\"GRADES_8_G\"])",
"_____no_output_____"
],
[
"education_df[[\"overall_score\", \"TOTAL_EXPENDITURE\", \"INSTRUCTION_EXPENDITURE\",\n \"SUPPORT_SERVICES_EXPENDITURE\", \"OTHER_EXPENDITURE\", \"CAPITAL_OUTLAY_EXPENDITURE\"]].corr()\n",
"_____no_output_____"
],
[
"from sklearn.decomposition import PCA\nfrom sklearn.preprocessing import StandardScaler\n\nX = education_df[[\"INSTRUCTION_EXPENDITURE\", \"SUPPORT_SERVICES_EXPENDITURE\",\n \"OTHER_EXPENDITURE\", \"CAPITAL_OUTLAY_EXPENDITURE\"]]\n\nX = StandardScaler().fit_transform(X)\n\nsklearn_pca = PCA(n_components=1)\neducation_df[\"pca_1\"] = sklearn_pca.fit_transform(X)\n\nprint(\n 'The percentage of total variance in the dataset explained by each',\n 'component from Sklearn PCA.\\n',\n sklearn_pca.explained_variance_ratio_)",
"The percentage of total variance in the dataset explained by each component from Sklearn PCA.\n [0.9430175]\n"
],
[
"education_df[[\"overall_score\", \"pca_1\", \"TOTAL_EXPENDITURE\", \"INSTRUCTION_EXPENDITURE\",\n \"SUPPORT_SERVICES_EXPENDITURE\", \"OTHER_EXPENDITURE\", \"CAPITAL_OUTLAY_EXPENDITURE\"]].corr()",
"_____no_output_____"
]
],
[
[
"Instruction expenditure variable is more correlated with the overall score than the first principal component. Hence using instruction expenditure makes more sense. PCA works best when the correlation between the variables are less than and equal to 0.8. In our case, all of the expenditure variables are highly correlated with each other. This may result in instable principal component estimations.",
"_____no_output_____"
]
]
] | [
"code",
"markdown"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb4caf4c5c8eff44f973a3f9409ea940bc677a0f | 92,046 | ipynb | Jupyter Notebook | models/aes256.ipynb | asinghani/crypto-accelerator | e1271ae213f6982d016b8795f85e9c5ecf2826e6 | [
"Apache-2.0"
] | 5 | 2021-01-13T07:37:41.000Z | 2021-12-13T01:55:50.000Z | models/aes256.ipynb | asinghani/crypto-accelerator | e1271ae213f6982d016b8795f85e9c5ecf2826e6 | [
"Apache-2.0"
] | null | null | null | models/aes256.ipynb | asinghani/crypto-accelerator | e1271ae213f6982d016b8795f85e9c5ecf2826e6 | [
"Apache-2.0"
] | null | null | null | 83.983577 | 193 | 0.72353 | [
[
[
"##### derived from https://github.com/bozhu/AES-Python\nimport copy\n\nSbox = (\n 0x63, 0x7C, 0x77, 0x7B, 0xF2, 0x6B, 0x6F, 0xC5, 0x30, 0x01, 0x67, 0x2B, 0xFE, 0xD7, 0xAB, 0x76,\n 0xCA, 0x82, 0xC9, 0x7D, 0xFA, 0x59, 0x47, 0xF0, 0xAD, 0xD4, 0xA2, 0xAF, 0x9C, 0xA4, 0x72, 0xC0,\n 0xB7, 0xFD, 0x93, 0x26, 0x36, 0x3F, 0xF7, 0xCC, 0x34, 0xA5, 0xE5, 0xF1, 0x71, 0xD8, 0x31, 0x15,\n 0x04, 0xC7, 0x23, 0xC3, 0x18, 0x96, 0x05, 0x9A, 0x07, 0x12, 0x80, 0xE2, 0xEB, 0x27, 0xB2, 0x75,\n 0x09, 0x83, 0x2C, 0x1A, 0x1B, 0x6E, 0x5A, 0xA0, 0x52, 0x3B, 0xD6, 0xB3, 0x29, 0xE3, 0x2F, 0x84,\n 0x53, 0xD1, 0x00, 0xED, 0x20, 0xFC, 0xB1, 0x5B, 0x6A, 0xCB, 0xBE, 0x39, 0x4A, 0x4C, 0x58, 0xCF,\n 0xD0, 0xEF, 0xAA, 0xFB, 0x43, 0x4D, 0x33, 0x85, 0x45, 0xF9, 0x02, 0x7F, 0x50, 0x3C, 0x9F, 0xA8,\n 0x51, 0xA3, 0x40, 0x8F, 0x92, 0x9D, 0x38, 0xF5, 0xBC, 0xB6, 0xDA, 0x21, 0x10, 0xFF, 0xF3, 0xD2,\n 0xCD, 0x0C, 0x13, 0xEC, 0x5F, 0x97, 0x44, 0x17, 0xC4, 0xA7, 0x7E, 0x3D, 0x64, 0x5D, 0x19, 0x73,\n 0x60, 0x81, 0x4F, 0xDC, 0x22, 0x2A, 0x90, 0x88, 0x46, 0xEE, 0xB8, 0x14, 0xDE, 0x5E, 0x0B, 0xDB,\n 0xE0, 0x32, 0x3A, 0x0A, 0x49, 0x06, 0x24, 0x5C, 0xC2, 0xD3, 0xAC, 0x62, 0x91, 0x95, 0xE4, 0x79,\n 0xE7, 0xC8, 0x37, 0x6D, 0x8D, 0xD5, 0x4E, 0xA9, 0x6C, 0x56, 0xF4, 0xEA, 0x65, 0x7A, 0xAE, 0x08,\n 0xBA, 0x78, 0x25, 0x2E, 0x1C, 0xA6, 0xB4, 0xC6, 0xE8, 0xDD, 0x74, 0x1F, 0x4B, 0xBD, 0x8B, 0x8A,\n 0x70, 0x3E, 0xB5, 0x66, 0x48, 0x03, 0xF6, 0x0E, 0x61, 0x35, 0x57, 0xB9, 0x86, 0xC1, 0x1D, 0x9E,\n 0xE1, 0xF8, 0x98, 0x11, 0x69, 0xD9, 0x8E, 0x94, 0x9B, 0x1E, 0x87, 0xE9, 0xCE, 0x55, 0x28, 0xDF,\n 0x8C, 0xA1, 0x89, 0x0D, 0xBF, 0xE6, 0x42, 0x68, 0x41, 0x99, 0x2D, 0x0F, 0xB0, 0x54, 0xBB, 0x16,\n)\n\nInvSbox = (\n 0x52, 0x09, 0x6A, 0xD5, 0x30, 0x36, 0xA5, 0x38, 0xBF, 0x40, 0xA3, 0x9E, 0x81, 0xF3, 0xD7, 0xFB,\n 0x7C, 0xE3, 0x39, 0x82, 0x9B, 0x2F, 0xFF, 0x87, 0x34, 0x8E, 0x43, 0x44, 0xC4, 0xDE, 0xE9, 0xCB,\n 0x54, 0x7B, 0x94, 0x32, 0xA6, 0xC2, 0x23, 0x3D, 0xEE, 0x4C, 0x95, 0x0B, 0x42, 0xFA, 0xC3, 0x4E,\n 0x08, 0x2E, 0xA1, 0x66, 0x28, 0xD9, 0x24, 0xB2, 0x76, 0x5B, 0xA2, 0x49, 0x6D, 0x8B, 0xD1, 0x25,\n 0x72, 0xF8, 0xF6, 0x64, 0x86, 0x68, 0x98, 0x16, 0xD4, 0xA4, 0x5C, 0xCC, 0x5D, 0x65, 0xB6, 0x92,\n 0x6C, 0x70, 0x48, 0x50, 0xFD, 0xED, 0xB9, 0xDA, 0x5E, 0x15, 0x46, 0x57, 0xA7, 0x8D, 0x9D, 0x84,\n 0x90, 0xD8, 0xAB, 0x00, 0x8C, 0xBC, 0xD3, 0x0A, 0xF7, 0xE4, 0x58, 0x05, 0xB8, 0xB3, 0x45, 0x06,\n 0xD0, 0x2C, 0x1E, 0x8F, 0xCA, 0x3F, 0x0F, 0x02, 0xC1, 0xAF, 0xBD, 0x03, 0x01, 0x13, 0x8A, 0x6B,\n 0x3A, 0x91, 0x11, 0x41, 0x4F, 0x67, 0xDC, 0xEA, 0x97, 0xF2, 0xCF, 0xCE, 0xF0, 0xB4, 0xE6, 0x73,\n 0x96, 0xAC, 0x74, 0x22, 0xE7, 0xAD, 0x35, 0x85, 0xE2, 0xF9, 0x37, 0xE8, 0x1C, 0x75, 0xDF, 0x6E,\n 0x47, 0xF1, 0x1A, 0x71, 0x1D, 0x29, 0xC5, 0x89, 0x6F, 0xB7, 0x62, 0x0E, 0xAA, 0x18, 0xBE, 0x1B,\n 0xFC, 0x56, 0x3E, 0x4B, 0xC6, 0xD2, 0x79, 0x20, 0x9A, 0xDB, 0xC0, 0xFE, 0x78, 0xCD, 0x5A, 0xF4,\n 0x1F, 0xDD, 0xA8, 0x33, 0x88, 0x07, 0xC7, 0x31, 0xB1, 0x12, 0x10, 0x59, 0x27, 0x80, 0xEC, 0x5F,\n 0x60, 0x51, 0x7F, 0xA9, 0x19, 0xB5, 0x4A, 0x0D, 0x2D, 0xE5, 0x7A, 0x9F, 0x93, 0xC9, 0x9C, 0xEF,\n 0xA0, 0xE0, 0x3B, 0x4D, 0xAE, 0x2A, 0xF5, 0xB0, 0xC8, 0xEB, 0xBB, 0x3C, 0x83, 0x53, 0x99, 0x61,\n 0x17, 0x2B, 0x04, 0x7E, 0xBA, 0x77, 0xD6, 0x26, 0xE1, 0x69, 0x14, 0x63, 0x55, 0x21, 0x0C, 0x7D,\n)\n\nRcon = (\n 0x00, 0x01, 0x02, 0x04, 0x08, 0x10, 0x20, 0x40,\n 0x80, 0x1B, 0x36, 0x6C, 0xD8, 0xAB, 0x4D, 0x9A,\n 0x2F, 0x5E, 0xBC, 0x63, 0xC6, 0x97, 0x35, 0x6A,\n 0xD4, 0xB3, 0x7D, 0xFA, 0xEF, 0xC5, 0x91, 0x39,\n)\n\n\ndef text2matrix(text): ##\n matrix = []\n for i in range(16):\n byte = (text >> (8 * (15 - i))) & 0xFF\n if i % 4 == 0:\n matrix.append([byte])\n else:\n matrix[i // 4].append(byte)\n \n #print(\"{:32x}\".format(text))\n #print([[hex(a) for a in m] for m in matrix])\n \n \"\"\"\n A B C D E F G H I J K L M N O P\n\n A B C D\n E F G H\n I J K L\n M N O P\n \"\"\"\n \n return matrix\n\n\ndef matrix2text(matrix): ##\n text = 0\n for i in range(4):\n for j in range(4):\n text |= (matrix[i][j] << (120 - 8 * (4 * i + j)))\n return text\n\n\nclass AES:\n def __init__(self, master_key, iv=None, aes256=True):\n self.num_rounds = 14 if aes256 else 10\n self.change_key(master_key)\n self.iv = iv\n \n def matrix_xor_elementwise(self, s, k): ##\n for i in range(4):\n for j in range(4):\n s[i][j] ^= k[i][j]\n \n \n # shifts / movements only\n def matrix_shift_rows(self, s): ##\n s[0][1], s[1][1], s[2][1], s[3][1] = s[1][1], s[2][1], s[3][1], s[0][1]\n s[0][2], s[1][2], s[2][2], s[3][2] = s[2][2], s[3][2], s[0][2], s[1][2]\n s[0][3], s[1][3], s[2][3], s[3][3] = s[3][3], s[0][3], s[1][3], s[2][3]\n\n def matrix_unshift_rows(self, s): ##\n s[0][1], s[1][1], s[2][1], s[3][1] = s[3][1], s[0][1], s[1][1], s[2][1]\n s[0][2], s[1][2], s[2][2], s[3][2] = s[2][2], s[3][2], s[0][2], s[1][2]\n s[0][3], s[1][3], s[2][3], s[3][3] = s[1][3], s[2][3], s[3][3], s[0][3]\n \n def matrix_sbox_lookup(self, s): ##\n for i in range(4):\n for j in range(4):\n s[i][j] = Sbox[s[i][j]]\n \n def matrix_invsbox_lookup(self, s): ##\n for i in range(4):\n for j in range(4):\n s[i][j] = InvSbox[s[i][j]]\n \n \n def mix_columns(self, s):\n xtime = lambda a: (((a << 1) ^ 0x1B) & 0xFF) if (a & 0x80) else (a << 1)\n for i in range(4):\n t = s[i][0] ^ s[i][1] ^ s[i][2] ^ s[i][3]\n u = s[i][0]\n s[i][0] ^= t ^ xtime(s[i][0] ^ s[i][1])\n s[i][1] ^= t ^ xtime(s[i][1] ^ s[i][2])\n s[i][2] ^= t ^ xtime(s[i][2] ^ s[i][3])\n s[i][3] ^= t ^ xtime(s[i][3] ^ u)\n\n\n def unmix_columns(self, s):\n xtime = lambda a: (((a << 1) ^ 0x1B) & 0xFF) if (a & 0x80) else (a << 1)\n\n for i in range(4):\n u = xtime(xtime(s[i][0] ^ s[i][2]))\n v = xtime(xtime(s[i][1] ^ s[i][3]))\n s[i][0] ^= u\n s[i][1] ^= v\n s[i][2] ^= u\n s[i][3] ^= v\n\n self.mix_columns(s)\n \n def encrypt(self, plaintext):\n if self.iv is not None:\n self.plain_state = text2matrix(plaintext ^ self.iv)\n else:\n self.plain_state = text2matrix(plaintext)\n\n self.matrix_xor_elementwise(self.plain_state, self.round_keys[0])\n \n #print([hex(x) for x in self.plain_state[0]])\n\n for i in range(1, self.num_rounds+1):\n ## CYCLE 1\n self.matrix_sbox_lookup(self.plain_state)\n self.matrix_shift_rows(self.plain_state)\n \n #print([hex(x) for x in self.plain_state[0]])\n \n ## CYCLE 2\n if i != self.num_rounds: self.mix_columns(self.plain_state)\n self.matrix_xor_elementwise(self.plain_state, self.round_keys[i])\n \n #print([hex(x) for x in self.plain_state[0]])\n \n if self.iv is not None:\n self.iv = matrix2text(self.plain_state)\n \n return matrix2text(self.plain_state)\n\n def decrypt(self, ciphertext):\n self.cipher_state = text2matrix(ciphertext)\n\n #print(hex(self.cipher_state[3][3]))\n \n for i in range(self.num_rounds, 0, -1):\n ## CYCLE 1\n self.matrix_xor_elementwise(self.cipher_state, self.round_keys[i])\n if i != self.num_rounds: self.unmix_columns(self.cipher_state)\n \n #print(hex(self.cipher_state[3][3]))\n \n ## CYCLE 2\n self.matrix_unshift_rows(self.cipher_state)\n self.matrix_invsbox_lookup(self.cipher_state)\n \n #print(hex(self.cipher_state[0][3]))\n \n self.matrix_xor_elementwise(self.cipher_state, self.round_keys[0])\n \n out = matrix2text(self.cipher_state)\n \n if self.iv is not None:\n out = out ^ self.iv\n self.iv = ciphertext\n\n return out\n\n\n def change_key(self, master_key):\n \n if (self.num_rounds == 14):\n self.round_keys = [text2matrix(master_key >> 128), text2matrix(master_key & ((1 << 128) - 1))]\n else:\n self.round_keys = [text2matrix(master_key)]\n \n last_key2 = self.round_keys[0]\n last_key = self.round_keys[1] if (self.num_rounds == 14) else self.round_keys[0]\n\n #print([hex(x) for x in last_key[0]])\n \n for i in range(1, self.num_rounds - len(self.round_keys) + 2):\n key = []\n\n aes256_alt = (i % 2 == 0) and (self.num_rounds == 14)\n \n # row 0\n s0 = Sbox[last_key[3][0]]\n s1 = Sbox[last_key[3][1]]\n s2 = Sbox[last_key[3][2]]\n s3 = Sbox[last_key[3][3]]\n \n last2 = last_key2 if (self.num_rounds == 14) else last_key\n \n round_const = Rcon[i // 2 + 1] if (self.num_rounds == 14) else Rcon[i]\n\n r0b0 = last2[0][0] ^ (s1 if not aes256_alt else s0) ^ (round_const if not aes256_alt else 0)\n r0b1 = last2[0][1] ^ (s2 if not aes256_alt else s1)\n r0b2 = last2[0][2] ^ (s3 if not aes256_alt else s2)\n r0b3 = last2[0][3] ^ (s0 if not aes256_alt else s3)\n key.append([r0b0, r0b1, r0b2, r0b3])\n\n # row 1\n r1b0 = last2[1][0] ^ r0b0\n r1b1 = last2[1][1] ^ r0b1\n r1b2 = last2[1][2] ^ r0b2\n r1b3 = last2[1][3] ^ r0b3\n key.append([r1b0, r1b1, r1b2, r1b3])\n\n # row 2\n r2b0 = last2[2][0] ^ r1b0\n r2b1 = last2[2][1] ^ r1b1\n r2b2 = last2[2][2] ^ r1b2\n r2b3 = last2[2][3] ^ r1b3\n key.append([r2b0, r2b1, r2b2, r2b3])\n\n # row 3\n r3b0 = last2[3][0] ^ r2b0\n r3b1 = last2[3][1] ^ r2b1\n r3b2 = last2[3][2] ^ r2b2\n r3b3 = last2[3][3] ^ r2b3\n key.append([r3b0, r3b1, r3b2, r3b3])\n \n \n self.round_keys.append(key)\n last_key2 = last_key\n last_key = key\n\n #print([hex(x) for x in key[0]])\n \n# 2d1541c695f88a16f8bfb5dbe3a95022\n\ndef packtext(s):\n s = s\n o = 0\n while len(s) > 0:\n o = (o << 8) | ord(s[0])\n s = s[1:]\n return o\n\ndef unpacktext(s):\n o = \"\"\n while s > 0:\n o = chr(s & 0xFF) + o\n s = s >> 8\n return o\n\n\nkey = \"abcd1234ABCD!@#$zyxwZYXW*1*2*3*4\"\nassert len(key) == 32\niv = \"54123892jsdkjsdj\"\nassert len(iv) == 16\n",
"_____no_output_____"
],
[
"string1 = \"helloworld123456\"\nstring2 = \"test string 1234\"\n\nprint(key)",
"abcd1234ABCD!@#$zyxwZYXW*1*2*3*4\n"
],
[
"#print(ek(key.encode(\"ascii\")))\n\na = AES(packtext(key), aes256=True)\n\nenc = a.encrypt(packtext(string1))\ndec = unpacktext(a.decrypt(enc))\n\nprint(packtext(key) >> 128, packtext(key) & ((1 << 128) - 1))\nprint(packtext(string1))\nprint(enc)\nprint(a.decrypt(enc))\nprint(unpacktext(a.decrypt(enc)))\n\nprint(hex(enc), dec)\n\n\nassert enc == 0x2d1541c695f88a16f8bfb5dbe3a95022\n\n##\n\n#print(ek(key.encode(\"ascii\")))\n\na = AES(packtext(key[:16]), aes256=False)\n\nenc = a.encrypt(packtext(string1))\ndec = unpacktext(a.decrypt(enc))\n\nprint(hex(enc), dec)\n\nassert enc == 0x1708271a0a18bb2e15bd658805297b8d",
"129445976579865719297921356551604413220 162796526750907890493247941453607545396\n138766332635719238849554048983485396278\n59925632134564593020758952134513872930\n138766332635719238849554048983485396278\nhelloworld123456\n0x2d1541c695f88a16f8bfb5dbe3a95022 helloworld123456\n0x1708271a0a18bb2e15bd658805297b8d helloworld123456\n"
],
[
"(packtext(string1) >> 128),(packtext(string1) & ((1 << 128) - 1))",
"_____no_output_____"
],
[
"\n\na = AES(packtext(key))\n\ne1 = hex(a.encrypt(packtext(string1)))\n#assert e1 == \"0x1708271a0a18bb2e15bd658805297b8d\"\ne2 = hex(a.encrypt(packtext(string2)))\n#assert e2 == \"0x482ac205196a804865262a0044915738\"\nprint(e1)\nprint(e2)\n\nprint(packtext(key), packtext(string1), int(e1, 0))\n\na = AES(packtext(key))\nprint(unpacktext(a.decrypt(int(e1, 0))))\n#assert(unpacktext(a.decrypt(int(e1, 0))) == string1)\nprint(unpacktext(a.decrypt(int(e2, 0))))\n#assert(unpacktext(a.decrypt(int(e2, 0))) == string2)\n\n",
"0x2d1541c695f88a16f8bfb5dbe3a95022\n0xaad2f0d199eba53714f739b449c90ca6\n44048183298989073697966048947243823604726133612511607082607857744689369393716 138766332635719238849554048983485396278 59925632134564593020758952134513872930\nhelloworld123456\ntest string 1234\n"
],
[
"hex(30614575354952859734368363414031006605)",
"_____no_output_____"
],
[
"a = AES(packtext(key), packtext(iv))\n\ne1 = hex(a.encrypt(packtext(string1)))\n#assert e1 == \"0x6cbaa5d41d87fc1cb2cde5f49c592554\"\ne2 = hex(a.encrypt(packtext(string2)))\n#assert e2 == \"0xb2b95376972f97140a84deda840144a2\"\nprint(e1)\nprint(e2)\n\na = AES(packtext(key), packtext(iv))\ndec1 = (unpacktext(a.decrypt(int(e1, 0))))\n#assert(dec1 == string1)\nprint(dec1)\ndec2 = (unpacktext(a.decrypt(int(e2, 0))))\n#assert(dec2 == string2)\nprint(dec2)\n\n",
"0x8ea7ae770057e1522436c05b3d7448b2\n0x683ec16c5385a750b7ecf9297f7b2e7e\nhelloworld123456\ntest string 1234\n"
],
[
"from Crypto.Cipher import AES as AE",
"_____no_output_____"
],
[
"print(key, len(key.encode()))\ncipher = AE.new(key.encode(), AE.MODE_ECB)\nciphertext = cipher.encrypt(string1 + string2)\nprint(ciphertext.hex()[:32])\nprint(ciphertext.hex()[32:])\nplaintext = cipher.decrypt(ciphertext)\nprint(plaintext)",
"abcd1234ABCD!@#$zyxwZYXW*1*2*3*4 32\n2d1541c695f88a16f8bfb5dbe3a95022\naad2f0d199eba53714f739b449c90ca6\nb'helloworld123456test string 1234'\n"
],
[
"cipher = AE.new(key.encode(), AE.MODE_CBC, iv)\nciphertext = cipher.encrypt(string1+string2)\nprint(ciphertext.hex()[:32])\nprint(ciphertext.hex()[32:])\n\ncipher = AE.new(key.encode(), AE.MODE_CBC, iv)\nplaintext = cipher.decrypt(ciphertext)\nprint(plaintext)",
"8ea7ae770057e1522436c05b3d7448b2\n683ec16c5385a750b7ecf9297f7b2e7e\nb'helloworld123456test string 1234'\n"
],
[
"import random\n# Generate 256-bit encrypt test-cases\n\nfor _1 in range(10):\n key = \"\".join([chr(random.randint(0x20, 0x7E)) for _ in range(32)]) # AES256 key\n print(\"setTopKey(BigInt(\\\"{}\\\"))\".format(packtext(key) >> 128))\n print(\"setKey(BigInt(\\\"{}\\\"))\".format(packtext(key) & ((1 << 128) - 1)))\n \n for _2 in range(10):\n plaintext = \"\".join([chr(random.randint(0x20, 0x7E)) for _ in range(16)])\n iv = \"\".join([chr(random.randint(0x20, 0x7E)) for _ in range(16)])\n \n c1 = AES(packtext(key))\n ct1 = c1.encrypt(packtext(plaintext))\n print(\"runSingleEncryptTest(BigInt(\\\"{}\\\"), BigInt(\\\"{}\\\"))\"\n .format(packtext(plaintext), ct1))\n \n c2 = AES(packtext(key), iv=packtext(iv))\n ct2 = c2.encrypt(packtext(plaintext))\n print(\"runSingleEncryptTest(BigInt(\\\"{}\\\"), BigInt(\\\"{}\\\"), iv=BigInt(\\\"{}\\\"))\"\n .format(packtext(plaintext), ct2, packtext(iv)))\n",
"setTopKey(BigInt(\"165406623308029099331681619155339851573\"))\nsetKey(BigInt(\"105658700701783706739513152549528565804\"))\nrunSingleEncryptTest(BigInt(\"67096222749627335679586640564596122443\"), BigInt(\"278013084954040053769286071749939562985\"))\nrunSingleEncryptTest(BigInt(\"67096222749627335679586640564596122443\"), BigInt(\"124392828678387920155199777039481918860\"), iv=BigInt(\"98873892024534219918523500662347411235\"))\nrunSingleEncryptTest(BigInt(\"157026096426878869629548132200093679224\"), BigInt(\"163243484665603839724123199786131420873\"))\nrunSingleEncryptTest(BigInt(\"157026096426878869629548132200093679224\"), BigInt(\"215429033216886894208538662194755851767\"), iv=BigInt(\"96349157545928519709772551141028812120\"))\nrunSingleEncryptTest(BigInt(\"166373065151925244287094997978767311159\"), BigInt(\"100156579591289555453087086047683728244\"))\nrunSingleEncryptTest(BigInt(\"166373065151925244287094997978767311159\"), BigInt(\"194120200536477666012091080169261089392\"), iv=BigInt(\"108157333658380454054498318112208077355\"))\nrunSingleEncryptTest(BigInt(\"110499670954322386381601909441930797110\"), BigInt(\"102222240809045086993654441052512555677\"))\nrunSingleEncryptTest(BigInt(\"110499670954322386381601909441930797110\"), BigInt(\"231084947047845461300851928589565444596\"), iv=BigInt(\"125374624503942989439423400246791253594\"))\nrunSingleEncryptTest(BigInt(\"65607026989468314640644831820621828456\"), BigInt(\"337014856448410650265565704669619312900\"))\nrunSingleEncryptTest(BigInt(\"65607026989468314640644831820621828456\"), BigInt(\"70713088363017165637531080709500480449\"), iv=BigInt(\"150646511700157512681192109204602907999\"))\nrunSingleEncryptTest(BigInt(\"137155563698573051078400931908527403111\"), BigInt(\"78162023916401621856550871408472858773\"))\nrunSingleEncryptTest(BigInt(\"137155563698573051078400931908527403111\"), BigInt(\"251638820343391361550120434757158528264\"), iv=BigInt(\"90799057287164454902192933441421650039\"))\nrunSingleEncryptTest(BigInt(\"133511723136542192548857540017589869381\"), BigInt(\"39457441307977837159360626063861568573\"))\nrunSingleEncryptTest(BigInt(\"133511723136542192548857540017589869381\"), BigInt(\"204363367207291739235609000488839690215\"), iv=BigInt(\"154441512936815436192291473001850094390\"))\nrunSingleEncryptTest(BigInt(\"96063665377643684811760407844284950094\"), BigInt(\"181984608953035131931515831717906854857\"))\nrunSingleEncryptTest(BigInt(\"96063665377643684811760407844284950094\"), BigInt(\"216942519331598054614872834162220287889\"), iv=BigInt(\"130899535989459668705237881758104167217\"))\nrunSingleEncryptTest(BigInt(\"109330993373986747972098701586578632572\"), BigInt(\"296445102448309294849700139138731719922\"))\nrunSingleEncryptTest(BigInt(\"109330993373986747972098701586578632572\"), BigInt(\"46448321193649798192293957285670425214\"), iv=BigInt(\"126796791335562948270921570881504041017\"))\nrunSingleEncryptTest(BigInt(\"49713183915796934637535093078866868567\"), BigInt(\"111114897401472490889755394727367630892\"))\nrunSingleEncryptTest(BigInt(\"49713183915796934637535093078866868567\"), BigInt(\"140211801387957503693780017173324423366\"), iv=BigInt(\"76125143772596278185459669916054943033\"))\nsetTopKey(BigInt(\"122778051174006138183611439216423819563\"))\nsetKey(BigInt(\"132217706756174712369348596715868276568\"))\nrunSingleEncryptTest(BigInt(\"78970336429486460126731434595304168486\"), BigInt(\"326503586831630566268512635441221688890\"))\nrunSingleEncryptTest(BigInt(\"78970336429486460126731434595304168486\"), BigInt(\"230050687648094419515121283581429151453\"), iv=BigInt(\"48341322053581213045475331077222853246\"))\nrunSingleEncryptTest(BigInt(\"75030237993503199601210132820597041275\"), BigInt(\"106892210648473842066282772805791715504\"))\nrunSingleEncryptTest(BigInt(\"75030237993503199601210132820597041275\"), BigInt(\"93975581168865498514909136739730283539\"), iv=BigInt(\"109454635006692842588267631611065624635\"))\nrunSingleEncryptTest(BigInt(\"100151078843611678114996437782905909617\"), BigInt(\"174753908814070885719198822956605417285\"))\nrunSingleEncryptTest(BigInt(\"100151078843611678114996437782905909617\"), BigInt(\"250233869718157262377863370266937489213\"), iv=BigInt(\"98748139721006564209796094724309667623\"))\nrunSingleEncryptTest(BigInt(\"105285064400588032305150819192230655545\"), BigInt(\"110130820901572993561352395204919213568\"))\nrunSingleEncryptTest(BigInt(\"105285064400588032305150819192230655545\"), BigInt(\"210518212846605243458833468803162870089\"), iv=BigInt(\"82901943608598843112151167623055882575\"))\nrunSingleEncryptTest(BigInt(\"127825002144326618660648983999584218737\"), BigInt(\"184387692927707904273417899599510642632\"))\nrunSingleEncryptTest(BigInt(\"127825002144326618660648983999584218737\"), BigInt(\"271280401692585408003905205545897148759\"), iv=BigInt(\"60429660458134526138895583236503448432\"))\nrunSingleEncryptTest(BigInt(\"96203916496365792578008528363995869564\"), BigInt(\"158834073141848908494392434369651524228\"))\nrunSingleEncryptTest(BigInt(\"96203916496365792578008528363995869564\"), BigInt(\"233217632812017140282922104455732146240\"), iv=BigInt(\"156159365843893576146976552556039450955\"))\nrunSingleEncryptTest(BigInt(\"97331619096166913493400647072254075750\"), BigInt(\"333316792481373814686691627696888945205\"))\nrunSingleEncryptTest(BigInt(\"97331619096166913493400647072254075750\"), BigInt(\"71200085093253058380738907101130517690\"), iv=BigInt(\"58689654860412084419911899163732502332\"))\nrunSingleEncryptTest(BigInt(\"84272912247171824117685484333891347240\"), BigInt(\"216708029396685165603776138941123487045\"))\nrunSingleEncryptTest(BigInt(\"84272912247171824117685484333891347240\"), BigInt(\"335308421966864773557468717813908454622\"), iv=BigInt(\"145329068009090883868684200440505186148\"))\nrunSingleEncryptTest(BigInt(\"144123966466981988907881762086638989404\"), BigInt(\"298204450278505092852585264598698224212\"))\nrunSingleEncryptTest(BigInt(\"144123966466981988907881762086638989404\"), BigInt(\"49234444439584230157807925442894647079\"), iv=BigInt(\"135967401536507141771863987370398203516\"))\nrunSingleEncryptTest(BigInt(\"47132410269165841034046254236622681140\"), BigInt(\"187311198518470079845025263797707919815\"))\nrunSingleEncryptTest(BigInt(\"47132410269165841034046254236622681140\"), BigInt(\"166959430304987997717166451823318976911\"), iv=BigInt(\"125172916725256928593240314851077668934\"))\nsetTopKey(BigInt(\"92247812687804810445351420209687066409\"))\nsetKey(BigInt(\"53456707853363221959168275173779522106\"))\nrunSingleEncryptTest(BigInt(\"96302223030712675621529609237165131632\"), BigInt(\"321318227697494351116496106091342520786\"))\nrunSingleEncryptTest(BigInt(\"96302223030712675621529609237165131632\"), BigInt(\"59414863249635110645235591302877369587\"), iv=BigInt(\"131933265036291212314430869052753472573\"))\nrunSingleEncryptTest(BigInt(\"83025782672586190724347227634570715227\"), BigInt(\"207516678719674375805977268087573992510\"))\nrunSingleEncryptTest(BigInt(\"83025782672586190724347227634570715227\"), BigInt(\"126490635052431371154182143178688420280\"), iv=BigInt(\"133439274772919260274212656736712667740\"))\nrunSingleEncryptTest(BigInt(\"154794037394970736262935712722354982177\"), BigInt(\"45065250452292359390457929304055739624\"))\nrunSingleEncryptTest(BigInt(\"154794037394970736262935712722354982177\"), BigInt(\"282541792795778101318340973334441293849\"), iv=BigInt(\"113338838852296199292414045531015620386\"))\nrunSingleEncryptTest(BigInt(\"161263414040015400546061821511741814309\"), BigInt(\"36407018247861487306439864606141826501\"))\nrunSingleEncryptTest(BigInt(\"161263414040015400546061821511741814309\"), BigInt(\"43940578567092878802161838871650839607\"), iv=BigInt(\"154596808529190741280701407878889962813\"))\nrunSingleEncryptTest(BigInt(\"134829573416288746467924114788056985651\"), BigInt(\"57355046475476387461359540920087758113\"))\nrunSingleEncryptTest(BigInt(\"134829573416288746467924114788056985651\"), BigInt(\"77437634203318601488147936257636418495\"), iv=BigInt(\"54936696978922616057984091387240607303\"))\nrunSingleEncryptTest(BigInt(\"158647029777377412064749202036938186852\"), BigInt(\"18515069827014187451606237225489730875\"))\nrunSingleEncryptTest(BigInt(\"158647029777377412064749202036938186852\"), BigInt(\"262545296785527298476511904278890384069\"), iv=BigInt(\"133366566852026215762339772402045107510\"))\nrunSingleEncryptTest(BigInt(\"100150428443545534336227365376514148412\"), BigInt(\"320164395144212553019456286970355638483\"))\nrunSingleEncryptTest(BigInt(\"100150428443545534336227365376514148412\"), BigInt(\"233063685282694200383867659658527707790\"), iv=BigInt(\"81592470417555605226606983753953657652\"))\nrunSingleEncryptTest(BigInt(\"167921134944282472817766804830552405060\"), BigInt(\"4784015971963504528811150619750622856\"))\nrunSingleEncryptTest(BigInt(\"167921134944282472817766804830552405060\"), BigInt(\"141508577953498742144921168308552800011\"), iv=BigInt(\"110935862395258655786913258597212175450\"))\nrunSingleEncryptTest(BigInt(\"109481798933316508018809221690635996005\"), BigInt(\"235486519028953880090254645239567755761\"))\nrunSingleEncryptTest(BigInt(\"109481798933316508018809221690635996005\"), BigInt(\"123103941655662217408332982071618447657\"), iv=BigInt(\"155864785010294375335897162317207730529\"))\nrunSingleEncryptTest(BigInt(\"91016811698542748019040400808872782137\"), BigInt(\"15488881413432671518949701320976961629\"))\nrunSingleEncryptTest(BigInt(\"91016811698542748019040400808872782137\"), BigInt(\"305314842882733500108802036216325076757\"), iv=BigInt(\"48211337328458829245686747285779858012\"))\nsetTopKey(BigInt(\"144118758467500981563581627004053704309\"))\nsetKey(BigInt(\"166502769971944992070326401537469674279\"))\nrunSingleEncryptTest(BigInt(\"92050502948036606825375445719570526576\"), BigInt(\"158557570141037954107141867703248059042\"))\nrunSingleEncryptTest(BigInt(\"92050502948036606825375445719570526576\"), BigInt(\"70356514895962446400520287116355281715\"), iv=BigInt(\"153091326020459869666733845833606855772\"))\nrunSingleEncryptTest(BigInt(\"145328220662700726972316821600917734769\"), BigInt(\"15680066339061957904205773048148689755\"))\nrunSingleEncryptTest(BigInt(\"145328220662700726972316821600917734769\"), BigInt(\"154694001003335741284640841311994855231\"), iv=BigInt(\"132228982031736961292323136812577078599\"))\nrunSingleEncryptTest(BigInt(\"115841145419462460901515935169093319207\"), BigInt(\"306227111158628649211183832250142463631\"))\nrunSingleEncryptTest(BigInt(\"115841145419462460901515935169093319207\"), BigInt(\"66897777979428918335800857398021349475\"), iv=BigInt(\"64235896970493973650956258977067260243\"))\nrunSingleEncryptTest(BigInt(\"125503848716026586757717826472512203624\"), BigInt(\"110707150385398737562173882911234230956\"))\nrunSingleEncryptTest(BigInt(\"125503848716026586757717826472512203624\"), BigInt(\"115798154822240013534860561012189712729\"), iv=BigInt(\"49680812233134513460530019047257498989\"))\nrunSingleEncryptTest(BigInt(\"160152547252691914155480216365105575477\"), BigInt(\"2597726447105103324743085741032785016\"))\nrunSingleEncryptTest(BigInt(\"160152547252691914155480216365105575477\"), BigInt(\"91296732392055152214359706551854573093\"), iv=BigInt(\"94770521353865477436960304566303803459\"))\nrunSingleEncryptTest(BigInt(\"135811749572751755746631189514791319878\"), BigInt(\"69279429350293873361060069171530705868\"))\nrunSingleEncryptTest(BigInt(\"135811749572751755746631189514791319878\"), BigInt(\"127076561984345966959163203812820168467\"), iv=BigInt(\"76105303301342195783408233248350432618\"))\nrunSingleEncryptTest(BigInt(\"102793709864788834824766533466816405886\"), BigInt(\"114835354912224935231399823738175454471\"))\nrunSingleEncryptTest(BigInt(\"102793709864788834824766533466816405886\"), BigInt(\"274993568576954929150925955624754965338\"), iv=BigInt(\"140090062122070889120573747219964048941\"))\nrunSingleEncryptTest(BigInt(\"85471725144511919501905120946471065124\"), BigInt(\"190336959163586597496291333252521520890\"))\nrunSingleEncryptTest(BigInt(\"85471725144511919501905120946471065124\"), BigInt(\"79100441389938778348785804812318499442\"), iv=BigInt(\"96101311341884548537920394130554780503\"))\nrunSingleEncryptTest(BigInt(\"125566433507886373734446940095875130710\"), BigInt(\"211677512440142857321283110820066665183\"))\nrunSingleEncryptTest(BigInt(\"125566433507886373734446940095875130710\"), BigInt(\"316881097353569713968363516730062445378\"), iv=BigInt(\"57433648346268316621009633776234689064\"))\nrunSingleEncryptTest(BigInt(\"100172291372980193328957768750888661862\"), BigInt(\"291894817882522504084695457838812725091\"))\nrunSingleEncryptTest(BigInt(\"100172291372980193328957768750888661862\"), BigInt(\"238262568718067245305651741169087619828\"), iv=BigInt(\"110546398128202584226497186020931893042\"))\nsetTopKey(BigInt(\"105300702783024393001970808958471469671\"))\nsetKey(BigInt(\"86655727264851750477146221934196115827\"))\nrunSingleEncryptTest(BigInt(\"147774883131807295560667226161132434553\"), BigInt(\"321914260466794533467751085977366295157\"))\nrunSingleEncryptTest(BigInt(\"147774883131807295560667226161132434553\"), BigInt(\"220731594492166217764285690628046710317\"), iv=BigInt(\"58665009528602297506299609771629113438\"))\nrunSingleEncryptTest(BigInt(\"143833157441559358923659591442635762297\"), BigInt(\"9756347448944277813663438886997593170\"))\nrunSingleEncryptTest(BigInt(\"143833157441559358923659591442635762297\"), BigInt(\"217193992289154557815962227837182721204\"), iv=BigInt(\"60409540617284294282934374931171868256\"))\nrunSingleEncryptTest(BigInt(\"116023118254215444282780126532834698606\"), BigInt(\"73367196278642926014107543850256698738\"))\nrunSingleEncryptTest(BigInt(\"116023118254215444282780126532834698606\"), BigInt(\"217590180426449043766762645610232643620\"), iv=BigInt(\"137545673908163024642440008343870792753\"))\nrunSingleEncryptTest(BigInt(\"122581211236767338022756408178980105767\"), BigInt(\"136164598132220643908380309457463934459\"))\nrunSingleEncryptTest(BigInt(\"122581211236767338022756408178980105767\"), BigInt(\"314990436844669193104952256999711388184\"), iv=BigInt(\"51103950646847686959579138811481908786\"))\nrunSingleEncryptTest(BigInt(\"56358028348622500149577952846906013742\"), BigInt(\"313525030324981085594014311956095662587\"))\nrunSingleEncryptTest(BigInt(\"56358028348622500149577952846906013742\"), BigInt(\"91460801006899787714186489830629826308\"), iv=BigInt(\"123868169984386422928201408955783926057\"))\nrunSingleEncryptTest(BigInt(\"128032512886312407427528295507939501635\"), BigInt(\"40929970675473756223130239321156612082\"))\nrunSingleEncryptTest(BigInt(\"128032512886312407427528295507939501635\"), BigInt(\"28396764880784692811999955945694669190\"), iv=BigInt(\"43056637467491370419948158900944714850\"))\nrunSingleEncryptTest(BigInt(\"143813261652958848032262010736504353846\"), BigInt(\"288643827536554114810405252943687876263\"))\nrunSingleEncryptTest(BigInt(\"143813261652958848032262010736504353846\"), BigInt(\"55001014962934086909653636445194322474\"), iv=BigInt(\"159904132166915511879988425328589093502\"))\nrunSingleEncryptTest(BigInt(\"158455804766988371773927432829872842564\"), BigInt(\"195547489053460679528331207974709666812\"))\nrunSingleEncryptTest(BigInt(\"158455804766988371773927432829872842564\"), BigInt(\"260889734786007591194028426549552906362\"), iv=BigInt(\"65331941433222028538643220932952072499\"))\nrunSingleEncryptTest(BigInt(\"65555027403514186255566590717533044567\"), BigInt(\"285722615659418279594375703976582059332\"))\nrunSingleEncryptTest(BigInt(\"65555027403514186255566590717533044567\"), BigInt(\"250915361643402476651120621931636298868\"), iv=BigInt(\"72351617708445218851738330040945954154\"))\nrunSingleEncryptTest(BigInt(\"43128988079304335116363921089480959021\"), BigInt(\"281601240773516449253909551648134721791\"))\nrunSingleEncryptTest(BigInt(\"43128988079304335116363921089480959021\"), BigInt(\"328589261371850031017858435095905676147\"), iv=BigInt(\"98951394596975363889206910803369289790\"))\nsetTopKey(BigInt(\"139898310075363065254141880687492082043\"))\nsetKey(BigInt(\"77356590837944944451891937368397404745\"))\nrunSingleEncryptTest(BigInt(\"165034503738745183658675874242194788650\"), BigInt(\"234059826897310701918743105428256372903\"))\nrunSingleEncryptTest(BigInt(\"165034503738745183658675874242194788650\"), BigInt(\"107290698524214986330682887201699672173\"), iv=BigInt(\"141205270985304504847081771868498504533\"))\nrunSingleEncryptTest(BigInt(\"72185447166563471536475176391609300042\"), BigInt(\"76182467917833890311166610488488153108\"))\nrunSingleEncryptTest(BigInt(\"72185447166563471536475176391609300042\"), BigInt(\"80953533920827713544427736006767880995\"), iv=BigInt(\"99942571298339826103522784465826695719\"))\nrunSingleEncryptTest(BigInt(\"154773493530304405098886859839061434696\"), BigInt(\"335528025606508861372090307743848708933\"))\nrunSingleEncryptTest(BigInt(\"154773493530304405098886859839061434696\"), BigInt(\"177909018401826144738683171190335613869\"), iv=BigInt(\"105425944660601973334312496746706251390\"))\nrunSingleEncryptTest(BigInt(\"85341690404780459810488495489231105064\"), BigInt(\"187724431734124348944445717836266975631\"))\nrunSingleEncryptTest(BigInt(\"85341690404780459810488495489231105064\"), BigInt(\"121341213365059002184248269169361664904\"), iv=BigInt(\"154752401893175440545140918768645195046\"))\nrunSingleEncryptTest(BigInt(\"102824196617529807237405400270203137326\"), BigInt(\"253437191447906218200203157075457959082\"))\nrunSingleEncryptTest(BigInt(\"102824196617529807237405400270203137326\"), BigInt(\"98223790639847963161474628239924387806\"), iv=BigInt(\"155921534160935251577872440779120257390\"))\nrunSingleEncryptTest(BigInt(\"56378895633503311049326323927139048762\"), BigInt(\"125442084943409434706100096019912886348\"))\nrunSingleEncryptTest(BigInt(\"56378895633503311049326323927139048762\"), BigInt(\"24370782635514691153400291356164125221\"), iv=BigInt(\"69371405386025988002792025487322207072\"))\nrunSingleEncryptTest(BigInt(\"166373546621428859745213258027579105056\"), BigInt(\"181764545435026669832560397294820599340\"))\nrunSingleEncryptTest(BigInt(\"166373546621428859745213258027579105056\"), BigInt(\"258795845866883904673066778070006020141\"), iv=BigInt(\"149451350735869947540213045283640001080\"))\nrunSingleEncryptTest(BigInt(\"104050047625182564961749806474139022419\"), BigInt(\"228919597648939913523242994500976903592\"))\nrunSingleEncryptTest(BigInt(\"104050047625182564961749806474139022419\"), BigInt(\"292761057334502842603685747044536189658\"), iv=BigInt(\"52386995763413729295686997042257880154\"))\nrunSingleEncryptTest(BigInt(\"103971392309493842654505563724505755187\"), BigInt(\"68460683669947061822832299341615707059\"))\nrunSingleEncryptTest(BigInt(\"103971392309493842654505563724505755187\"), BigInt(\"72419321800378853212069751693657486593\"), iv=BigInt(\"73430499209460787323845960412889361991\"))\nrunSingleEncryptTest(BigInt(\"68400137902462103858815641895395094063\"), BigInt(\"224060926362445377545161333666083813807\"))\nrunSingleEncryptTest(BigInt(\"68400137902462103858815641895395094063\"), BigInt(\"163991319550884138535006071676246109896\"), iv=BigInt(\"72154071568854749467338637830305233708\"))\nsetTopKey(BigInt(\"104247349376587814741480410013260592229\"))\nsetKey(BigInt(\"85331917377251684827634817512499265317\"))\nrunSingleEncryptTest(BigInt(\"46690964497811475382425372552086766371\"), BigInt(\"277639299278218823725198501946689568691\"))\nrunSingleEncryptTest(BigInt(\"46690964497811475382425372552086766371\"), BigInt(\"186220136498687452484901327486700017699\"), iv=BigInt(\"97347522406127781522233878992565594933\"))\nrunSingleEncryptTest(BigInt(\"52007310083760575088596027335259661144\"), BigInt(\"46396751440061351032855547555656154877\"))\nrunSingleEncryptTest(BigInt(\"52007310083760575088596027335259661144\"), BigInt(\"38252923443180308151957022237617150710\"), iv=BigInt(\"117601409069121589508366527458120714562\"))\nrunSingleEncryptTest(BigInt(\"90934407818756092258533231712795637553\"), BigInt(\"327998908434733462877758723868853064930\"))\nrunSingleEncryptTest(BigInt(\"90934407818756092258533231712795637553\"), BigInt(\"236603434241523520503963838240129371377\"), iv=BigInt(\"145075195224187493124581842870724742510\"))\nrunSingleEncryptTest(BigInt(\"56436700656582140253718595855045321250\"), BigInt(\"82911357177944411937014578870550983360\"))\nrunSingleEncryptTest(BigInt(\"56436700656582140253718595855045321250\"), BigInt(\"9420747822183795645120416560284346966\"), iv=BigInt(\"115908970826571219474485404424829482798\"))\nrunSingleEncryptTest(BigInt(\"112112766736868231073870461874201188653\"), BigInt(\"308679327824840528280100445031297016724\"))\nrunSingleEncryptTest(BigInt(\"112112766736868231073870461874201188653\"), BigInt(\"114551205058926394173053794453527228325\"), iv=BigInt(\"73493521647790233676373285370650503243\"))\nrunSingleEncryptTest(BigInt(\"109429876757069671493587997121055380572\"), BigInt(\"292383957741155182286062000758844614761\"))\nrunSingleEncryptTest(BigInt(\"109429876757069671493587997121055380572\"), BigInt(\"310410109046265094823133447546022872159\"), iv=BigInt(\"47100631128894681194470789172567941228\"))\nrunSingleEncryptTest(BigInt(\"114876656945771668991986888015865597288\"), BigInt(\"275464355846721860252861206767680523973\"))\nrunSingleEncryptTest(BigInt(\"114876656945771668991986888015865597288\"), BigInt(\"331145240149519496498771524048402947615\"), iv=BigInt(\"53601891797980248306660139617541960020\"))\nrunSingleEncryptTest(BigInt(\"119927259383002039236304450418170416978\"), BigInt(\"173921954505828005627332265854256976570\"))\nrunSingleEncryptTest(BigInt(\"119927259383002039236304450418170416978\"), BigInt(\"298292360361758347022674869792380628232\"), iv=BigInt(\"130550982929590240213831959083957252679\"))\nrunSingleEncryptTest(BigInt(\"96100860935322417641207248024005933685\"), BigInt(\"77944317088053364420652489699888541861\"))\nrunSingleEncryptTest(BigInt(\"96100860935322417641207248024005933685\"), BigInt(\"232469394746408162092020023261367010773\"), iv=BigInt(\"162483970292720195893271376670904171578\"))\nrunSingleEncryptTest(BigInt(\"162661643488633717100661128508688387932\"), BigInt(\"277585476573694295161359811330155741006\"))\nrunSingleEncryptTest(BigInt(\"162661643488633717100661128508688387932\"), BigInt(\"338167919000412163490406684729434972124\"), iv=BigInt(\"139856512216906073586590370399291992164\"))\nsetTopKey(BigInt(\"145158067007217270915560734093038730589\"))\nsetKey(BigInt(\"112165950040744394243239682832673897512\"))\nrunSingleEncryptTest(BigInt(\"69615439134636168826759173682134930521\"), BigInt(\"53796264360961930892199216825690069601\"))\nrunSingleEncryptTest(BigInt(\"69615439134636168826759173682134930521\"), BigInt(\"70455938478327941520885239864461516409\"), iv=BigInt(\"44369620297425237014444040569210493030\"))\nrunSingleEncryptTest(BigInt(\"158429189721203835052769617237146494504\"), BigInt(\"140577552565323124594289261790573026517\"))\nrunSingleEncryptTest(BigInt(\"158429189721203835052769617237146494504\"), BigInt(\"141111988498322318204186442154278694305\"), iv=BigInt(\"54744868177786698075084510609648736867\"))\nrunSingleEncryptTest(BigInt(\"131890654467718899036321392938664544365\"), BigInt(\"14072627741019424964012949120214778826\"))\nrunSingleEncryptTest(BigInt(\"131890654467718899036321392938664544365\"), BigInt(\"160665624392197162493967982194002683448\"), iv=BigInt(\"80211747691352905465350531716124522052\"))\nrunSingleEncryptTest(BigInt(\"128198608074272741371411740786204833071\"), BigInt(\"242894851216135252070013632358190421155\"))\nrunSingleEncryptTest(BigInt(\"128198608074272741371411740786204833071\"), BigInt(\"301664603141756414215206718009175800312\"), iv=BigInt(\"134549109845559205767702088700809519938\"))\nrunSingleEncryptTest(BigInt(\"77667659780060210916690952757717906813\"), BigInt(\"72777653910728127285336038575024197633\"))\nrunSingleEncryptTest(BigInt(\"77667659780060210916690952757717906813\"), BigInt(\"259854576201000205983900189967048632179\"), iv=BigInt(\"126636291938425900408394952048284482637\"))\nrunSingleEncryptTest(BigInt(\"167945884006820474559071776885122229558\"), BigInt(\"209132524653997492528513171459326618428\"))\nrunSingleEncryptTest(BigInt(\"167945884006820474559071776885122229558\"), BigInt(\"271069999151995277839021775476510416303\"), iv=BigInt(\"121459855537089886229292240917059289385\"))\nrunSingleEncryptTest(BigInt(\"61701773659864745786597916273386617382\"), BigInt(\"95453059838982436515689456771063126141\"))\nrunSingleEncryptTest(BigInt(\"61701773659864745786597916273386617382\"), BigInt(\"220695389291812775274607900793937819131\"), iv=BigInt(\"149466437369341063086336669039180803371\"))\nrunSingleEncryptTest(BigInt(\"131948051312143832739462049528293908781\"), BigInt(\"152367075685631214019656005759972421813\"))\nrunSingleEncryptTest(BigInt(\"131948051312143832739462049528293908781\"), BigInt(\"183670388828150343549849737803546643688\"), iv=BigInt(\"74900389197599132263062894247053312035\"))\nrunSingleEncryptTest(BigInt(\"105182185675207275405098740457032265781\"), BigInt(\"213587503791380765814217288913670594807\"))\nrunSingleEncryptTest(BigInt(\"105182185675207275405098740457032265781\"), BigInt(\"143176505085864141161300183769057215353\"), iv=BigInt(\"49437896243141068721293144368394955836\"))\nrunSingleEncryptTest(BigInt(\"112311885232103230813508850937277731945\"), BigInt(\"290467297379129703865528919910354983787\"))\nrunSingleEncryptTest(BigInt(\"112311885232103230813508850937277731945\"), BigInt(\"318250078659360019391381221033865583784\"), iv=BigInt(\"127788594959436361635148481019524702281\"))\nsetTopKey(BigInt(\"163974442724960837765969619176189796936\"))\nsetKey(BigInt(\"146684220244806559876708177990955195450\"))\nrunSingleEncryptTest(BigInt(\"129413525767193749178625174846628636781\"), BigInt(\"163327152124999420759968829649895435393\"))\nrunSingleEncryptTest(BigInt(\"129413525767193749178625174846628636781\"), BigInt(\"322117214364301306667746014641424691179\"), iv=BigInt(\"141175680459343522559016511353830589271\"))\nrunSingleEncryptTest(BigInt(\"69381582954002163299698793295313851988\"), BigInt(\"86315462702546425635885310782211585682\"))\nrunSingleEncryptTest(BigInt(\"69381582954002163299698793295313851988\"), BigInt(\"269000445162669049159247651045258638894\"), iv=BigInt(\"68347024221233067622301716318704460652\"))\nrunSingleEncryptTest(BigInt(\"75045878100669126511471474737864523330\"), BigInt(\"37300525774426581971343340512446920019\"))\nrunSingleEncryptTest(BigInt(\"75045878100669126511471474737864523330\"), BigInt(\"250134638824947254419102449817578112565\"), iv=BigInt(\"49567007721077322446499089046798758216\"))\nrunSingleEncryptTest(BigInt(\"116081188593574068323090741417420093490\"), BigInt(\"159645496789893426555950762147720292248\"))\nrunSingleEncryptTest(BigInt(\"116081188593574068323090741417420093490\"), BigInt(\"16919135083398345214865477624990086064\"), iv=BigInt(\"90784394131262621910598179015394156091\"))\nrunSingleEncryptTest(BigInt(\"155833066485944381451991727780070175077\"), BigInt(\"17484535405314064553123720223790054824\"))\nrunSingleEncryptTest(BigInt(\"155833066485944381451991727780070175077\"), BigInt(\"100719841578643816825056530027110205657\"), iv=BigInt(\"142669860548333121282233507031877240867\"))\nrunSingleEncryptTest(BigInt(\"73742854650410141125565636427220022899\"), BigInt(\"135680698802808417662405828772184497597\"))\nrunSingleEncryptTest(BigInt(\"73742854650410141125565636427220022899\"), BigInt(\"175739982145556329156613590980474232929\"), iv=BigInt(\"46987496798267429285567640327704226640\"))\nrunSingleEncryptTest(BigInt(\"123909952243944284767143670087817582698\"), BigInt(\"97115938898469588179942343645730493788\"))\nrunSingleEncryptTest(BigInt(\"123909952243944284767143670087817582698\"), BigInt(\"19211977660167816309411513536048483897\"), iv=BigInt(\"118650155260419033520368870073166941507\"))\nrunSingleEncryptTest(BigInt(\"47153384033232335454549472455073427032\"), BigInt(\"40728345119274514544591467553515812675\"))\nrunSingleEncryptTest(BigInt(\"47153384033232335454549472455073427032\"), BigInt(\"51619804595412432582108419391587526868\"), iv=BigInt(\"114533131247029577106426968317792311121\"))\nrunSingleEncryptTest(BigInt(\"104059537444847010377297945509558303020\"), BigInt(\"216971153444093099781709595572844155556\"))\nrunSingleEncryptTest(BigInt(\"104059537444847010377297945509558303020\"), BigInt(\"271481339979267997757723509031295699718\"), iv=BigInt(\"120068851995494730381781166260977476664\"))\nrunSingleEncryptTest(BigInt(\"76282090815363732879743121141401922341\"), BigInt(\"88429058959827480515333563729456876830\"))\nrunSingleEncryptTest(BigInt(\"76282090815363732879743121141401922341\"), BigInt(\"207073000568840245834622691551068012895\"), iv=BigInt(\"162489202141425824065606673908235648365\"))\nsetTopKey(BigInt(\"112259414818193981568056362832023864892\"))\nsetKey(BigInt(\"62859699897158771129210437379442308934\"))\nrunSingleEncryptTest(BigInt(\"58908175323807598008458580217284353626\"), BigInt(\"284763619537174650347558387084040849867\"))\nrunSingleEncryptTest(BigInt(\"58908175323807598008458580217284353626\"), BigInt(\"170871558668676746446943942632736612674\"), iv=BigInt(\"99897098132203959041632702303539642963\"))\nrunSingleEncryptTest(BigInt(\"105571243494121782413678411943880570445\"), BigInt(\"21959935367452524731706305099780543058\"))\nrunSingleEncryptTest(BigInt(\"105571243494121782413678411943880570445\"), BigInt(\"96877857061631265780991835304157047820\"), iv=BigInt(\"64449287013221498203685593539779440171\"))\nrunSingleEncryptTest(BigInt(\"146694479590301700520694956335296955946\"), BigInt(\"94949105892373633138439520091643283414\"))\nrunSingleEncryptTest(BigInt(\"146694479590301700520694956335296955946\"), BigInt(\"4975795596793341373519150408903930884\"), iv=BigInt(\"167660368907556566407894213424634670956\"))\nrunSingleEncryptTest(BigInt(\"145433382895315776931654860617672584239\"), BigInt(\"181148642007180847033588264907892925415\"))\nrunSingleEncryptTest(BigInt(\"145433382895315776931654860617672584239\"), BigInt(\"128398388520145577881057666459823891337\"), iv=BigInt(\"112222175273678293622564832392092148605\"))\nrunSingleEncryptTest(BigInt(\"117565982848034812278468239366114473842\"), BigInt(\"275777855901498633957025380354217543883\"))\nrunSingleEncryptTest(BigInt(\"117565982848034812278468239366114473842\"), BigInt(\"158305098936553270622129591755001099474\"), iv=BigInt(\"62832966829209041674762438646445857148\"))\nrunSingleEncryptTest(BigInt(\"85726432170429196814763396561970749240\"), BigInt(\"305617748993259395800991042877330126093\"))\nrunSingleEncryptTest(BigInt(\"85726432170429196814763396561970749240\"), BigInt(\"271453379884546126258513849116395818454\"), iv=BigInt(\"113401961353700986154221284904903583065\"))\nrunSingleEncryptTest(BigInt(\"96007645677366908440181914868440652885\"), BigInt(\"38976824621935894519807906441327087748\"))\nrunSingleEncryptTest(BigInt(\"96007645677366908440181914868440652885\"), BigInt(\"273401778943569273031446920318081631349\"), iv=BigInt(\"130547285826068319987528377227832614508\"))\nrunSingleEncryptTest(BigInt(\"142508861133615448118618431383194393123\"), BigInt(\"19346302594384151124530450971364789382\"))\nrunSingleEncryptTest(BigInt(\"142508861133615448118618431383194393123\"), BigInt(\"53781890268339887643383494841106730285\"), iv=BigInt(\"45538217319368713348311232180589900328\"))\nrunSingleEncryptTest(BigInt(\"42806521924956716715839884979539963505\"), BigInt(\"324633939673321927730998224373551550226\"))\nrunSingleEncryptTest(BigInt(\"42806521924956716715839884979539963505\"), BigInt(\"67507533347956637790351154905867128679\"), iv=BigInt(\"76187852392162154155651510271517864050\"))\nrunSingleEncryptTest(BigInt(\"166621525680933153608548403987783833695\"), BigInt(\"271110064386304907504650146373608500858\"))\nrunSingleEncryptTest(BigInt(\"166621525680933153608548403987783833695\"), BigInt(\"200673200101946374778194164281057220246\"), iv=BigInt(\"110680768615900245901540285371259304745\"))\n"
],
[
"import random\n# Generate 256-bit decrypt test-cases\n\nfor _1 in range(10):\n key = \"\".join([chr(random.randint(0x20, 0x7E)) for _ in range(32)]) # AES256 key\n print(\"setTopKey(BigInt(\\\"{}\\\"))\".format(packtext(key) >> 128))\n print(\"setKey(BigInt(\\\"{}\\\"))\".format(packtext(key) & ((1 << 128) - 1)))\n \n for _2 in range(10):\n plaintext = \"\".join([chr(random.randint(0x20, 0x7E)) for _ in range(16)])\n iv = \"\".join([chr(random.randint(0x20, 0x7E)) for _ in range(16)])\n \n c1 = AES(packtext(key))\n ct1 = c1.encrypt(packtext(plaintext))\n print(\"runSingleDecryptTest(BigInt(\\\"{}\\\"), BigInt(\\\"{}\\\"))\"\n .format(ct1, packtext(plaintext)))\n \n c2 = AES(packtext(key), iv=packtext(iv))\n ct2 = c2.encrypt(packtext(plaintext))\n print(\"runSingleDecryptTest(BigInt(\\\"{}\\\"), BigInt(\\\"{}\\\"), iv=BigInt(\\\"{}\\\"))\"\n .format(ct2, packtext(plaintext), packtext(iv)))\n",
"setTopKey(BigInt(\"153423720557180764576530833117252168527\"))\nsetKey(BigInt(\"56218445522235001715694128441772617779\"))\nrunSingleDecryptTest(BigInt(\"218549502634221611235968548577572577803\"), BigInt(\"166611971413254361133535418193110267737\"))\nrunSingleDecryptTest(BigInt(\"235730131888855548816664693052185057147\"), BigInt(\"166611971413254361133535418193110267737\"), iv=BigInt(\"94640834602863508551148299614032913509\"))\nrunSingleDecryptTest(BigInt(\"27502652893792207678373774154216790380\"), BigInt(\"143755177493324503670103715785530488146\"))\nrunSingleDecryptTest(BigInt(\"162929894617595328179352778199365747800\"), BigInt(\"143755177493324503670103715785530488146\"), iv=BigInt(\"110945540262939000639196814610351356780\"))\nrunSingleDecryptTest(BigInt(\"236986021498394127807822385897946275530\"), BigInt(\"145240533470560153135250944230727237452\"))\nrunSingleDecryptTest(BigInt(\"188549392098996541011074000171597571433\"), BigInt(\"145240533470560153135250944230727237452\"), iv=BigInt(\"112051557271043465121891348645454309988\"))\nrunSingleDecryptTest(BigInt(\"265387625977907200902515798838182266039\"), BigInt(\"58934400645269100363997233936379296059\"))\nrunSingleDecryptTest(BigInt(\"222114255847365170030037140654272614990\"), BigInt(\"58934400645269100363997233936379296059\"), iv=BigInt(\"105198414541161314084714644634116694880\"))\nrunSingleDecryptTest(BigInt(\"275942770389822573089174624130587438677\"), BigInt(\"122612078291505405649135147562893062716\"))\nrunSingleDecryptTest(BigInt(\"641572733961256258987598946651427310\"), BigInt(\"122612078291505405649135147562893062716\"), iv=BigInt(\"96297664626890462383375710948320892510\"))\nrunSingleDecryptTest(BigInt(\"67284536501648177370117597804578138604\"), BigInt(\"131797436510946240948058796128411792704\"))\nrunSingleDecryptTest(BigInt(\"323900185345245704400868212054686220066\"), BigInt(\"131797436510946240948058796128411792704\"), iv=BigInt(\"149477833462439804258481655621072935716\"))\nrunSingleDecryptTest(BigInt(\"165826899790534397846357885068955552795\"), BigInt(\"72164679572343760766349977538711480403\"))\nrunSingleDecryptTest(BigInt(\"135947512898969047704031478400729788358\"), BigInt(\"72164679572343760766349977538711480403\"), iv=BigInt(\"80166125263802982357436095715243807047\"))\nrunSingleDecryptTest(BigInt(\"252090747071166803186910925319489242933\"), BigInt(\"45590585794126904825399653413954808692\"))\nrunSingleDecryptTest(BigInt(\"59453420073899438172119246565792877501\"), BigInt(\"45590585794126904825399653413954808692\"), iv=BigInt(\"116080374761863668652021206964705964128\"))\nrunSingleDecryptTest(BigInt(\"92279696658918275288031528979037854500\"), BigInt(\"158521479211790777067378978074067492397\"))\nrunSingleDecryptTest(BigInt(\"305689571781390855147021477320764053694\"), BigInt(\"158521479211790777067378978074067492397\"), iv=BigInt(\"129284750912696831536997105886238879067\"))\nrunSingleDecryptTest(BigInt(\"84458118486330259188139623961914048627\"), BigInt(\"74780356648721597611130635105769507691\"))\nrunSingleDecryptTest(BigInt(\"319753810934401583655436947249203808639\"), BigInt(\"74780356648721597611130635105769507691\"), iv=BigInt(\"150572641718498663527148527308644696128\"))\nsetTopKey(BigInt(\"123868271807515172555058581189087680114\"))\nsetKey(BigInt(\"102955219635434641563066841423867107954\"))\nrunSingleDecryptTest(BigInt(\"281795785750577999647377530112288647838\"), BigInt(\"113328434138355619485252337914748346475\"))\nrunSingleDecryptTest(BigInt(\"308002681203943089319079438841053013225\"), BigInt(\"113328434138355619485252337914748346475\"), iv=BigInt(\"149088128825229806572610123527944103243\"))\nrunSingleDecryptTest(BigInt(\"217252899750133859636070966489287512599\"), BigInt(\"72184532262705169610825046525347787566\"))\nrunSingleDecryptTest(BigInt(\"224696677253831228805325399056065841566\"), BigInt(\"72184532262705169610825046525347787566\"), iv=BigInt(\"145162044730835326216613344114642532911\"))\nrunSingleDecryptTest(BigInt(\"248666732591299517933258048481314734589\"), BigInt(\"60106971202901455297099851636532194364\"))\nrunSingleDecryptTest(BigInt(\"169344274617143608110841007155270950519\"), BigInt(\"60106971202901455297099851636532194364\"), iv=BigInt(\"119818276673654164464299410073743348293\"))\nrunSingleDecryptTest(BigInt(\"246099842808376845239658330648641840603\"), BigInt(\"61582151562690082480906343931617692485\"))\nrunSingleDecryptTest(BigInt(\"42445823227924301222619807094675217557\"), BigInt(\"61582151562690082480906343931617692485\"), iv=BigInt(\"166393344930285953707008236849985961520\"))\nrunSingleDecryptTest(BigInt(\"76856784693189720000872480006294616531\"), BigInt(\"162488696587148106113846488611869046309\"))\nrunSingleDecryptTest(BigInt(\"83593571632352393858839486930682533300\"), BigInt(\"162488696587148106113846488611869046309\"), iv=BigInt(\"100047029904361030145234109837035204734\"))\nrunSingleDecryptTest(BigInt(\"297898686194861965328226974201197933472\"), BigInt(\"114802561259731512192315954281336106621\"))\nrunSingleDecryptTest(BigInt(\"93207290257010406161232460363954348580\"), BigInt(\"114802561259731512192315954281336106621\"), iv=BigInt(\"58949149196106669511600812314369481065\"))\nrunSingleDecryptTest(BigInt(\"2512518542691890276155459484654672301\"), BigInt(\"86577420695092992870114604150701252216\"))\nrunSingleDecryptTest(BigInt(\"43130915696664580686681599170098516708\"), BigInt(\"86577420695092992870114604150701252216\"), iv=BigInt(\"87003591287270046127430421644195754863\"))\nrunSingleDecryptTest(BigInt(\"337636664611420278115554134168326779864\"), BigInt(\"92153886041987535827923328189963051608\"))\nrunSingleDecryptTest(BigInt(\"160989033093557566912706321987693736292\"), BigInt(\"92153886041987535827923328189963051608\"), iv=BigInt(\"102767993529467346163036418314068897391\"))\nrunSingleDecryptTest(BigInt(\"19220744988865170098447146597923921647\"), BigInt(\"110571590957633594191471220991723053620\"))\nrunSingleDecryptTest(BigInt(\"34661964237012947379594521372532995860\"), BigInt(\"110571590957633594191471220991723053620\"), iv=BigInt(\"165054262693186670979459270060833974895\"))\nrunSingleDecryptTest(BigInt(\"206391567113742882275988437748230259954\"), BigInt(\"80201158531750911014668651905531272507\"))\nrunSingleDecryptTest(BigInt(\"14379115669713424800533544277610251761\"), BigInt(\"80201158531750911014668651905531272507\"), iv=BigInt(\"167997747709005672596186123394131589171\"))\nsetTopKey(BigInt(\"141170488709997669980883493314906760790\"))\nsetKey(BigInt(\"117331233042432651200922368705131016481\"))\nrunSingleDecryptTest(BigInt(\"122386427588931712183942511911597349996\"), BigInt(\"142395728550364895458328671329824622638\"))\nrunSingleDecryptTest(BigInt(\"51122381198275130212389702570393678224\"), BigInt(\"142395728550364895458328671329824622638\"), iv=BigInt(\"110721903016845681388658550374558998827\"))\nrunSingleDecryptTest(BigInt(\"164938150602095199096055603830560292054\"), BigInt(\"92170356451076518321264370558915849034\"))\nrunSingleDecryptTest(BigInt(\"96760253566514847894048353647262541424\"), BigInt(\"92170356451076518321264370558915849034\"), iv=BigInt(\"149228243670362858031623964475134848556\"))\nrunSingleDecryptTest(BigInt(\"167267955175864463127707790457886226836\"), BigInt(\"105653569626599290251703698679877490274\"))\nrunSingleDecryptTest(BigInt(\"34616218110794349140981951644907259926\"), BigInt(\"105653569626599290251703698679877490274\"), iv=BigInt(\"127965240807250537437458373435069576287\"))\nrunSingleDecryptTest(BigInt(\"45278108943853410317319404236333264107\"), BigInt(\"74956915636512836230169706423485472888\"))\nrunSingleDecryptTest(BigInt(\"48677270029877316130997814340245536433\"), BigInt(\"74956915636512836230169706423485472888\"), iv=BigInt(\"60164553122237252311775468246101404004\"))\nrunSingleDecryptTest(BigInt(\"17948175539382043201242561793825634314\"), BigInt(\"123884027595266183753670117026636508496\"))\nrunSingleDecryptTest(BigInt(\"47959795078160563028823083331526135242\"), BigInt(\"123884027595266183753670117026636508496\"), iv=BigInt(\"139845069132391876768637380300511734575\"))\nrunSingleDecryptTest(BigInt(\"314608360262498036020137005323384336096\"), BigInt(\"77443803683478650211617309445789264468\"))\nrunSingleDecryptTest(BigInt(\"114365921248036210433389458233560217008\"), BigInt(\"77443803683478650211617309445789264468\"), iv=BigInt(\"80112950007752053898945071794801945142\"))\nrunSingleDecryptTest(BigInt(\"42088214585773760718491769947341896032\"), BigInt(\"157297213217992484017654351703680044383\"))\nrunSingleDecryptTest(BigInt(\"256793745649472096535406912280614758605\"), BigInt(\"157297213217992484017654351703680044383\"), iv=BigInt(\"53570335059364719520239312527650799480\"))\nrunSingleDecryptTest(BigInt(\"138013223102181546982853992504869569500\"), BigInt(\"144202404878531556716373068560410639176\"))\nrunSingleDecryptTest(BigInt(\"282245304923559219279405406649341175168\"), BigInt(\"144202404878531556716373068560410639176\"), iv=BigInt(\"46919936276051719367830937026757273442\"))\nrunSingleDecryptTest(BigInt(\"214528455627167744024491830659213431726\"), BigInt(\"154514081578017732659859073526811733349\"))\nrunSingleDecryptTest(BigInt(\"255364873013511385488230297089121954021\"), BigInt(\"154514081578017732659859073526811733349\"), iv=BigInt(\"63996606294192967683773470091311140401\"))\nrunSingleDecryptTest(BigInt(\"280474268500635035444428268078668485307\"), BigInt(\"142826238384702956212769987796070005615\"))\nrunSingleDecryptTest(BigInt(\"190411545180430503889116371575338854630\"), BigInt(\"142826238384702956212769987796070005615\"), iv=BigInt(\"49832691446804034490088408457020322920\"))\nsetTopKey(BigInt(\"62725181578095911680762976877684420217\"))\nsetKey(BigInt(\"163688100677607949424019329666349805142\"))\nrunSingleDecryptTest(BigInt(\"232080317522888178156787518268380447865\"), BigInt(\"128225180321623804463271030383636531044\"))\nrunSingleDecryptTest(BigInt(\"183703963784931197131953399464579462446\"), BigInt(\"128225180321623804463271030383636531044\"), iv=BigInt(\"56041704441530929243210640595845736264\"))\nrunSingleDecryptTest(BigInt(\"276205594545021372976777755871519377150\"), BigInt(\"102803769167805289604459493276624055384\"))\nrunSingleDecryptTest(BigInt(\"229557307734804068271183849234899460992\"), BigInt(\"102803769167805289604459493276624055384\"), iv=BigInt(\"94704333840277687074700399149299297077\"))\nrunSingleDecryptTest(BigInt(\"235281911570447845147312367700588203803\"), BigInt(\"149487346628120342312485655788229047598\"))\nrunSingleDecryptTest(BigInt(\"211721960873922386366881541031866949598\"), BigInt(\"149487346628120342312485655788229047598\"), iv=BigInt(\"82601008410276613364152046386351317563\"))\nrunSingleDecryptTest(BigInt(\"290417154849758027312137520532541459941\"), BigInt(\"133251526508447314904389833029781634928\"))\nrunSingleDecryptTest(BigInt(\"173130811234283549089088430546561802697\"), BigInt(\"133251526508447314904389833029781634928\"), iv=BigInt(\"65527505778115026156618496185188963378\"))\nrunSingleDecryptTest(BigInt(\"38693312486255006679919080098597625416\"), BigInt(\"159950134438401673316097216935510307173\"))\nrunSingleDecryptTest(BigInt(\"273530538854998285063493708237205723957\"), BigInt(\"159950134438401673316097216935510307173\"), iv=BigInt(\"121511860740724237242718690123796591179\"))\nrunSingleDecryptTest(BigInt(\"53016641836150165360084004145281102520\"), BigInt(\"113308112759372764020790569983827013419\"))\nrunSingleDecryptTest(BigInt(\"119982694485508847090134087027941887711\"), BigInt(\"113308112759372764020790569983827013419\"), iv=BigInt(\"105442230174382916513237043641929705324\"))\nrunSingleDecryptTest(BigInt(\"329927056234239594332351033663048790965\"), BigInt(\"60418239073838872902177903718209898846\"))\nrunSingleDecryptTest(BigInt(\"89468297827689021716711613854224317274\"), BigInt(\"60418239073838872902177903718209898846\"), iv=BigInt(\"132119216489614214498344302635881537651\"))\nrunSingleDecryptTest(BigInt(\"83396581569743402504928451418796493638\"), BigInt(\"51109774385660036879854446535271078737\"))\nrunSingleDecryptTest(BigInt(\"38898078084984214599307753841141699706\"), BigInt(\"51109774385660036879854446535271078737\"), iv=BigInt(\"69500924556855081440997562193849375045\"))\nrunSingleDecryptTest(BigInt(\"234605179313002016939009524629737926546\"), BigInt(\"147790199774269011913966003533090399581\"))\nrunSingleDecryptTest(BigInt(\"174495334081154160051969688234848845507\"), BigInt(\"147790199774269011913966003533090399581\"), iv=BigInt(\"166752361661866203030448627115744504150\"))\nrunSingleDecryptTest(BigInt(\"297149078752910844457527560506442578419\"), BigInt(\"113639405943559115429684375002312355681\"))\nrunSingleDecryptTest(BigInt(\"4032465419954272220491240392972748881\"), BigInt(\"113639405943559115429684375002312355681\"), iv=BigInt(\"140094422827738045265834870396947090282\"))\nsetTopKey(BigInt(\"118952080342839905069357327309775006060\"))\nsetKey(BigInt(\"96282309570230118939903290799173490772\"))\nrunSingleDecryptTest(BigInt(\"1484282520783611701304482040321108498\"), BigInt(\"74797597495106648476206203763909743484\"))\nrunSingleDecryptTest(BigInt(\"302499526744792374819540834462525971129\"), BigInt(\"74797597495106648476206203763909743484\"), iv=BigInt(\"52007347144317316092445557050776844126\"))\nrunSingleDecryptTest(BigInt(\"188489463515769540556506967537167967139\"), BigInt(\"118936724892449379493533609493264613197\"))\nrunSingleDecryptTest(BigInt(\"331538021628254565058475043361179520761\"), BigInt(\"118936724892449379493533609493264613197\"), iv=BigInt(\"118484835890859740037967884474184199019\"))\nrunSingleDecryptTest(BigInt(\"336434826785840365914789018766393259266\"), BigInt(\"108282639895829720466483521011237151797\"))\nrunSingleDecryptTest(BigInt(\"52903626931928728094674660876581145593\"), BigInt(\"108282639895829720466483521011237151797\"), iv=BigInt(\"149332474432541767020956366582382231139\"))\nrunSingleDecryptTest(BigInt(\"83599220187973177412281837960685496590\"), BigInt(\"142394754838043652243522727964244534566\"))\nrunSingleDecryptTest(BigInt(\"139965309715957403138228143330799109824\"), BigInt(\"142394754838043652243522727964244534566\"), iv=BigInt(\"54764706351920308741774511868722239098\"))\nrunSingleDecryptTest(BigInt(\"178324661467814642492371489042303255802\"), BigInt(\"49562346481301422008991513147775738959\"))\nrunSingleDecryptTest(BigInt(\"321719254768634911207401475310674301332\"), BigInt(\"49562346481301422008991513147775738959\"), iv=BigInt(\"168051147958836053494788574985712857189\"))\nrunSingleDecryptTest(BigInt(\"227699784991025695230469700174019288150\"), BigInt(\"47116163347832127239842385358787409771\"))\nrunSingleDecryptTest(BigInt(\"172081290614474022493441004631553386908\"), BigInt(\"47116163347832127239842385358787409771\"), iv=BigInt(\"154566021202760025906897223361505273724\"))\nrunSingleDecryptTest(BigInt(\"66894235942907365452698105529640647773\"), BigInt(\"102622854011702746738064287912698408250\"))\nrunSingleDecryptTest(BigInt(\"282962284283425782172053370179872279577\"), BigInt(\"102622854011702746738064287912698408250\"), iv=BigInt(\"45387249169225990046665353022719880766\"))\nrunSingleDecryptTest(BigInt(\"322220784450995683203775922160036092569\"), BigInt(\"56483330705799305279883302771069570900\"))\nrunSingleDecryptTest(BigInt(\"18994553654780697827241474827805943065\"), BigInt(\"56483330705799305279883302771069570900\"), iv=BigInt(\"134835075108500987287105389879108125995\"))\nrunSingleDecryptTest(BigInt(\"338045421514304143030485430796036044100\"), BigInt(\"47059415636419654187957854074921972274\"))\nrunSingleDecryptTest(BigInt(\"114003087337580882960856334439084042367\"), BigInt(\"47059415636419654187957854074921972274\"), iv=BigInt(\"57708614759715589996657923255338944347\"))\nrunSingleDecryptTest(BigInt(\"148214612050627014119958809622185822102\"), BigInt(\"121293134832359719338104704897215782246\"))\nrunSingleDecryptTest(BigInt(\"296267352598035952254676796838467811498\"), BigInt(\"121293134832359719338104704897215782246\"), iv=BigInt(\"104283211375133105298663890411682935342\"))\nsetTopKey(BigInt(\"92035048621904254527143255660444460122\"))\nsetKey(BigInt(\"88001200140511987241313678767516966177\"))\nrunSingleDecryptTest(BigInt(\"318871509022681004435400220129027018872\"), BigInt(\"98800267583566257543362696831952187711\"))\nrunSingleDecryptTest(BigInt(\"151996141220827732923297596026935982075\"), BigInt(\"98800267583566257543362696831952187711\"), iv=BigInt(\"134871841483513431479494729452175698733\"))\nrunSingleDecryptTest(BigInt(\"193443211952150335452884594144532526035\"), BigInt(\"132166675743905526506991908355024176507\"))\nrunSingleDecryptTest(BigInt(\"288306285176198770448133769064667151709\"), BigInt(\"132166675743905526506991908355024176507\"), iv=BigInt(\"53809971582352566258375389916792117586\"))\nrunSingleDecryptTest(BigInt(\"58159248240249357716395311058304529948\"), BigInt(\"153157877575782374302226042403644783156\"))\nrunSingleDecryptTest(BigInt(\"295876241009100907981696329086855924803\"), BigInt(\"153157877575782374302226042403644783156\"), iv=BigInt(\"105560980279826795792406803757952759148\"))\nrunSingleDecryptTest(BigInt(\"309847686522312833315543747350738296488\"), BigInt(\"125411195906500844898274555582687440689\"))\nrunSingleDecryptTest(BigInt(\"130232884163170298348810137479681945151\"), BigInt(\"125411195906500844898274555582687440689\"), iv=BigInt(\"82854705875659995679075017674629212493\"))\nrunSingleDecryptTest(BigInt(\"206326596710437918871475908320476413987\"), BigInt(\"83935448974186951806271132501068360043\"))\nrunSingleDecryptTest(BigInt(\"233702001480793409219116788906189364066\"), BigInt(\"83935448974186951806271132501068360043\"), iv=BigInt(\"82601252366754822522065879847075801906\"))\nrunSingleDecryptTest(BigInt(\"250953685564300709699698209001462696379\"), BigInt(\"70969046740193070630293641704492662061\"))\nrunSingleDecryptTest(BigInt(\"248942692356664295843664971534233304682\"), BigInt(\"70969046740193070630293641704492662061\"), iv=BigInt(\"120047675810775342544539102808465749112\"))\nrunSingleDecryptTest(BigInt(\"337921080826004622818448336521218489999\"), BigInt(\"121226652901858111851829224938856917848\"))\nrunSingleDecryptTest(BigInt(\"202653791792165728531464457578630373624\"), BigInt(\"121226652901858111851829224938856917848\"), iv=BigInt(\"168133998361695803330683792034969578018\"))\nrunSingleDecryptTest(BigInt(\"74047509106345038864351786681304422008\"), BigInt(\"134767733445623749493502519875986748449\"))\nrunSingleDecryptTest(BigInt(\"60398731637469221389656941053611067157\"), BigInt(\"134767733445623749493502519875986748449\"), iv=BigInt(\"78618055937784052693561000158459746601\"))\nrunSingleDecryptTest(BigInt(\"267172824540213750031883208545820586128\"), BigInt(\"80200970280333296091020332609568187766\"))\nrunSingleDecryptTest(BigInt(\"241827346595642935085930670833076254994\"), BigInt(\"80200970280333296091020332609568187766\"), iv=BigInt(\"68170588180714337745974435123931071053\"))\nrunSingleDecryptTest(BigInt(\"54170281921167225286481845336095388244\"), BigInt(\"97465503432015050445132956403784961343\"))\nrunSingleDecryptTest(BigInt(\"154808493016659862003138664475138308554\"), BigInt(\"97465503432015050445132956403784961343\"), iv=BigInt(\"69750116935583550005669014534751409229\"))\nsetTopKey(BigInt(\"128002757007842422470894842215213529961\"))\nsetKey(BigInt(\"69548045118977226954191747433298361213\"))\nrunSingleDecryptTest(BigInt(\"209903547188263354345984685530950755051\"), BigInt(\"73691029132779279909296784922745000830\"))\nrunSingleDecryptTest(BigInt(\"295260139529826266802409012339842211969\"), BigInt(\"73691029132779279909296784922745000830\"), iv=BigInt(\"130509033922303956185232881779061834847\"))\nrunSingleDecryptTest(BigInt(\"329340425489572172887597107841430873851\"), BigInt(\"44515473399730729602997967368677120867\"))\nrunSingleDecryptTest(BigInt(\"14710606167519072779316563629523596175\"), BigInt(\"44515473399730729602997967368677120867\"), iv=BigInt(\"97444772977935312466858030165420173646\"))\nrunSingleDecryptTest(BigInt(\"114159453566062220472049370453608121751\"), BigInt(\"58928481267787898399845851091992541273\"))\nrunSingleDecryptTest(BigInt(\"284031232158198592270853948312501630003\"), BigInt(\"58928481267787898399845851091992541273\"), iv=BigInt(\"78721491142168078743602182188694722681\"))\nrunSingleDecryptTest(BigInt(\"144300893987733331275200385098084371808\"), BigInt(\"131947730432506448087711597640827036245\"))\nrunSingleDecryptTest(BigInt(\"249346211773004259080788963519348681265\"), BigInt(\"131947730432506448087711597640827036245\"), iv=BigInt(\"165377035139205248304283492612184570656\"))\nrunSingleDecryptTest(BigInt(\"139913327972448915946213688082156891068\"), BigInt(\"150511268511849013652802664012572079219\"))\nrunSingleDecryptTest(BigInt(\"11350002121205355057032131273866056878\"), BigInt(\"150511268511849013652802664012572079219\"), iv=BigInt(\"153122480738357170403982401006420250660\"))\nrunSingleDecryptTest(BigInt(\"80450048282172535923437085100497235875\"), BigInt(\"125406492286212955387351731353073303339\"))\nrunSingleDecryptTest(BigInt(\"271171724154742926052138612820618518466\"), BigInt(\"125406492286212955387351731353073303339\"), iv=BigInt(\"82850426599513633912494280137468683352\"))\nrunSingleDecryptTest(BigInt(\"61165763769963880045118168436614308919\"), BigInt(\"127805736395737970331267593968991030649\"))\nrunSingleDecryptTest(BigInt(\"35757510848843344491734234865635839137\"), BigInt(\"127805736395737970331267593968991030649\"), iv=BigInt(\"146606963509630434044752908109306819693\"))\nrunSingleDecryptTest(BigInt(\"253113303712820421826710718167036395948\"), BigInt(\"165023190748515561656179914596299920222\"))\nrunSingleDecryptTest(BigInt(\"158262788832920256705284532733551046255\"), BigInt(\"165023190748515561656179914596299920222\"), iv=BigInt(\"129477007414161770121038780094731013742\"))\nrunSingleDecryptTest(BigInt(\"296798384579210841314505580733411534451\"), BigInt(\"132239595971098700244678640441697793868\"))\nrunSingleDecryptTest(BigInt(\"286471431097540587497990925977703193829\"), BigInt(\"132239595971098700244678640441697793868\"), iv=BigInt(\"115998436444977807692463514954612557887\"))\nrunSingleDecryptTest(BigInt(\"159138354310645719062911636298899672883\"), BigInt(\"141512589266156290882694369376424125252\"))\nrunSingleDecryptTest(BigInt(\"242811702333520393654742242268037420629\"), BigInt(\"141512589266156290882694369376424125252\"), iv=BigInt(\"144161062339646893135508241438696165410\"))\nsetTopKey(BigInt(\"50958361381197230942682606655286566750\"))\nsetKey(BigInt(\"98759641293778731167128858228168276809\"))\nrunSingleDecryptTest(BigInt(\"71353153941674797743141099985148812611\"), BigInt(\"49552205203474488379999033473254900057\"))\nrunSingleDecryptTest(BigInt(\"111864949012977898812827057263518478441\"), BigInt(\"49552205203474488379999033473254900057\"), iv=BigInt(\"46826408620850326257391872421777206564\"))\nrunSingleDecryptTest(BigInt(\"241457694887288713948163780701753671452\"), BigInt(\"141408396939971702974761488047773994342\"))\nrunSingleDecryptTest(BigInt(\"243761040529852074926633037538959507965\"), BigInt(\"141408396939971702974761488047773994342\"), iv=BigInt(\"61556469997387921410650439897473634168\"))\nrunSingleDecryptTest(BigInt(\"150355120039623514944232049444791734583\"), BigInt(\"83037526113724200294954523994414212167\"))\nrunSingleDecryptTest(BigInt(\"192341588198217448896681901179942504243\"), BigInt(\"83037526113724200294954523994414212167\"), iv=BigInt(\"141444014209029638507843270153794560067\"))\nrunSingleDecryptTest(BigInt(\"285002535655531243083898744918698100452\"), BigInt(\"101204706359119131025965438847705430361\"))\nrunSingleDecryptTest(BigInt(\"78089955612959110606960639100054560484\"), BigInt(\"101204706359119131025965438847705430361\"), iv=BigInt(\"118748527430914838332764886183330594126\"))\nrunSingleDecryptTest(BigInt(\"295119092125017147279810095637931881162\"), BigInt(\"86838110882384140300484213397304207941\"))\nrunSingleDecryptTest(BigInt(\"94952638223683536812625602439072154695\"), BigInt(\"86838110882384140300484213397304207941\"), iv=BigInt(\"97451202916176433937284739549804458287\"))\nrunSingleDecryptTest(BigInt(\"261609521377174502987762673905120218223\"), BigInt(\"94636534087358081484983187445330637616\"))\nrunSingleDecryptTest(BigInt(\"87301213465058958408472095445036834381\"), BigInt(\"94636534087358081484983187445330637616\"), iv=BigInt(\"118935997407486696387207507132750838898\"))\nrunSingleDecryptTest(BigInt(\"84433761358589692395606347585418266610\"), BigInt(\"163855286361633064062119068807991740708\"))\nrunSingleDecryptTest(BigInt(\"234307964554126407246036012622243025174\"), BigInt(\"163855286361633064062119068807991740708\"), iv=BigInt(\"104107279365891476095346872855878594892\"))\nrunSingleDecryptTest(BigInt(\"8546536104393451684491085527854519322\"), BigInt(\"81458096449433803431943122337948717131\"))\nrunSingleDecryptTest(BigInt(\"142470802489263648790775350544995714562\"), BigInt(\"81458096449433803431943122337948717131\"), iv=BigInt(\"67080972750139552238677299745654002299\"))\nrunSingleDecryptTest(BigInt(\"6606957205501255601958351588583973069\"), BigInt(\"80026160788189247428158007004320775217\"))\nrunSingleDecryptTest(BigInt(\"309407297516753129986324912042816233615\"), BigInt(\"80026160788189247428158007004320775217\"), iv=BigInt(\"150380036559108875176158623183315696420\"))\nrunSingleDecryptTest(BigInt(\"97208703913378913608649645431715500488\"), BigInt(\"122472798043044984926561702450766760525\"))\nrunSingleDecryptTest(BigInt(\"4088860886176830968185634923717426761\"), BigInt(\"122472798043044984926561702450766760525\"), iv=BigInt(\"57381660668077679741910809776506035492\"))\nsetTopKey(BigInt(\"137098592094472550963512223827006862397\"))\nsetKey(BigInt(\"66909872736550699471593395940214065211\"))\nrunSingleDecryptTest(BigInt(\"234956997023950404426676836204331065867\"), BigInt(\"89630672581739782334564775346268624246\"))\nrunSingleDecryptTest(BigInt(\"100147959450896392794196454554738191287\"), BigInt(\"89630672581739782334564775346268624246\"), iv=BigInt(\"49759531274784983218698603139769774163\"))\nrunSingleDecryptTest(BigInt(\"94316380160199662147726034481421477765\"), BigInt(\"146751329837191345835130429431074092363\"))\nrunSingleDecryptTest(BigInt(\"41688918216954383716479085052294433291\"), BigInt(\"146751329837191345835130429431074092363\"), iv=BigInt(\"138666338828976163272158392891960014661\"))\nrunSingleDecryptTest(BigInt(\"335061657502257153091715103941911320437\"), BigInt(\"158568982109217231076806994637928484457\"))\nrunSingleDecryptTest(BigInt(\"226595723248604112844824983343720546110\"), BigInt(\"158568982109217231076806994637928484457\"), iv=BigInt(\"138506895647877048846267194677343379539\"))\nrunSingleDecryptTest(BigInt(\"326911794820412877310928286070405973006\"), BigInt(\"74989574541708517035407161103061706076\"))\nrunSingleDecryptTest(BigInt(\"156195756850458702629955383454452144950\"), BigInt(\"74989574541708517035407161103061706076\"), iv=BigInt(\"128163965009946515929928225895500116516\"))\nrunSingleDecryptTest(BigInt(\"277337889856150875561241711576016129437\"), BigInt(\"142783873670352492073901532927474032995\"))\nrunSingleDecryptTest(BigInt(\"126566565993842578569143610361108835249\"), BigInt(\"142783873670352492073901532927474032995\"), iv=BigInt(\"49826361126526314785447260163997719337\"))\nrunSingleDecryptTest(BigInt(\"316955344939110174246215388887725793932\"), BigInt(\"136038953211723534738485012622276128613\"))\nrunSingleDecryptTest(BigInt(\"90292686255506238727333498990143031633\"), BigInt(\"136038953211723534738485012622276128613\"), iv=BigInt(\"129518545154515879282689118169271989556\"))\nrunSingleDecryptTest(BigInt(\"199197011527423793069787083185071771205\"), BigInt(\"66749291332815045920026924447511302215\"))\nrunSingleDecryptTest(BigInt(\"192260930637634278335881061008817798816\"), BigInt(\"66749291332815045920026924447511302215\"), iv=BigInt(\"145499339626913560334073984859762343979\"))\nrunSingleDecryptTest(BigInt(\"102924836233278343819359728676182210991\"), BigInt(\"120193767384556030590016434010248148828\"))\nrunSingleDecryptTest(BigInt(\"94769481360691161417305879876032192167\"), BigInt(\"120193767384556030590016434010248148828\"), iv=BigInt(\"82605837834741086258919862494898502455\"))\nrunSingleDecryptTest(BigInt(\"182994328471511870618616249224176934578\"), BigInt(\"122528923122056287596214476642286991992\"))\nrunSingleDecryptTest(BigInt(\"328468008110183793675372993675732948377\"), BigInt(\"122528923122056287596214476642286991992\"), iv=BigInt(\"99917122175052058070648109243688963196\"))\nrunSingleDecryptTest(BigInt(\"334285163740277474493270392924554553851\"), BigInt(\"148054616243632202761784660388735626049\"))\nrunSingleDecryptTest(BigInt(\"12629230989957337439148192221418531775\"), BigInt(\"148054616243632202761784660388735626049\"), iv=BigInt(\"162375985240771184817648067311760796216\"))\nsetTopKey(BigInt(\"110654790257517785980741224366444264527\"))\nsetKey(BigInt(\"58845459196370855455927954128718559289\"))\nrunSingleDecryptTest(BigInt(\"229765248428931195428545980852405935487\"), BigInt(\"94609819798992858568973191721193073261\"))\nrunSingleDecryptTest(BigInt(\"138216609730222498171086132417499016473\"), BigInt(\"94609819798992858568973191721193073261\"), iv=BigInt(\"114491649581627723044101808456928619069\"))\nrunSingleDecryptTest(BigInt(\"195828564130066576580235200932252445059\"), BigInt(\"109632069801380687798724386031388412740\"))\nrunSingleDecryptTest(BigInt(\"262843854248281442024676258661790553355\"), BigInt(\"109632069801380687798724386031388412740\"), iv=BigInt(\"94589054247708606340134179972062149215\"))\nrunSingleDecryptTest(BigInt(\"160629480918727366943957045226962115561\"), BigInt(\"115815749290314698202120313092763973432\"))\nrunSingleDecryptTest(BigInt(\"279194389319983380871529280053171651158\"), BigInt(\"115815749290314698202120313092763973432\"), iv=BigInt(\"45662340443608813551327124204655498326\"))\nrunSingleDecryptTest(BigInt(\"329602848185688921709277679530761651095\"), BigInt(\"121563152111150555032367672972495437634\"))\nrunSingleDecryptTest(BigInt(\"49516646351175962151549077671044562504\"), BigInt(\"121563152111150555032367672972495437634\"), iv=BigInt(\"150438736154781114212789084125585568604\"))\nrunSingleDecryptTest(BigInt(\"45004651977938973088812042460332091637\"), BigInt(\"151798307104471316342827140021386962029\"))\nrunSingleDecryptTest(BigInt(\"241047916121245304713995281738394962524\"), BigInt(\"151798307104471316342827140021386962029\"), iv=BigInt(\"161357469822204007325429960526271036505\"))\nrunSingleDecryptTest(BigInt(\"318425988133882029761974061256324676739\"), BigInt(\"158521539902723968128441071558877526064\"))\nrunSingleDecryptTest(BigInt(\"171949653302368177273160297301943029916\"), BigInt(\"158521539902723968128441071558877526064\"), iv=BigInt(\"152041977273773255643014041147597794128\"))\nrunSingleDecryptTest(BigInt(\"51493042985106360807155876797513842933\"), BigInt(\"139976357582311647208887731959447513919\"))\nrunSingleDecryptTest(BigInt(\"113590557373404902130046802768099165395\"), BigInt(\"139976357582311647208887731959447513919\"), iv=BigInt(\"78909063588819035320490519066101296928\"))\nrunSingleDecryptTest(BigInt(\"32438069645758105610870424781680708074\"), BigInt(\"129165102463029675449250506744536319050\"))\nrunSingleDecryptTest(BigInt(\"18332565019819378879030510509227134836\"), BigInt(\"129165102463029675449250506744536319050\"), iv=BigInt(\"143990063491777893639409194738625112902\"))\nrunSingleDecryptTest(BigInt(\"319139763863801670919740394802793479802\"), BigInt(\"133443456189340131109552709054619936565\"))\nrunSingleDecryptTest(BigInt(\"323829391730560382262843712587715523483\"), BigInt(\"133443456189340131109552709054619936565\"), iv=BigInt(\"58835178462470729826779068729057891108\"))\nrunSingleDecryptTest(BigInt(\"58483402385110061132228270281813236634\"), BigInt(\"121283605921318636740498325888855726420\"))\nrunSingleDecryptTest(BigInt(\"334275922866520223284449514336457504687\"), BigInt(\"121283605921318636740498325888855726420\"), iv=BigInt(\"48124425864484805353416525767816130886\"))\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4cc45c92301478942f4047d73288ae0d65381f | 47,396 | ipynb | Jupyter Notebook | nb/2019_winter/Lecture8.ipynb | samuelcheang0419/cme193 | 609e4655544292a28dbb9ca0301637b006970af2 | [
"MIT"
] | 15 | 2016-02-17T06:03:51.000Z | 2021-11-30T03:47:27.000Z | nb/2019_winter/Lecture8.ipynb | samuelcheang0419/cme193 | 609e4655544292a28dbb9ca0301637b006970af2 | [
"MIT"
] | null | null | null | nb/2019_winter/Lecture8.ipynb | samuelcheang0419/cme193 | 609e4655544292a28dbb9ca0301637b006970af2 | [
"MIT"
] | 33 | 2016-01-19T18:23:46.000Z | 2020-12-23T03:08:23.000Z | 33.097765 | 502 | 0.5603 | [
[
[
"# CME 193 - Lecture 8\n\nHere's what you've seen over the past 7 lectures:\n* Python Language Basics\n* NumPy - Arrays/Linear Algebra\n* SciPy - Sparse Linear Algebra/Optimization\n* DataFrames - Reading & Maniputlating tabular data\n* Scikit learn - Machine Learning Models & use with data\n* Ortools - More Optimization\n\nYou've now seen some tools for scientific computing in Python. How you add to them and what you do with them is up to you!\n\n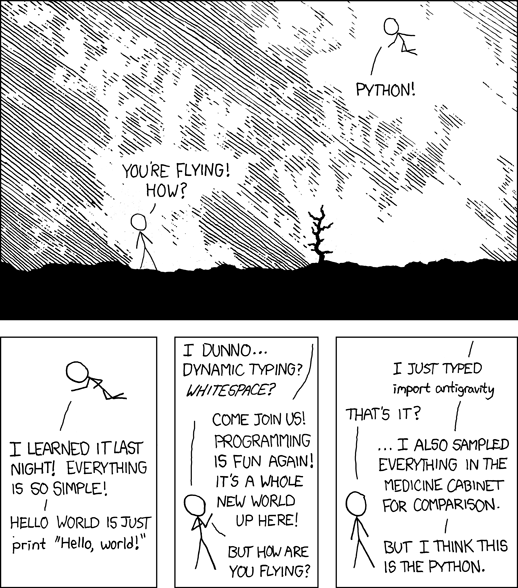\n\n(Maybe you've also had a bit of [this](https://xkcd.com/1987/) experience)\n\n## Today\n\n1. We'll revisit object oriented programming in Python\n2. We'll look at PyTorch (deep learning package)",
"_____no_output_____"
],
[
"# Object Oriented Programming - II\n\nRecall some of the basic terminology of [object oriented programming](https://en.wikipedia.org/wiki/Object-oriented_programming)\n* **Classes** are templates for objects (e.g., \"the Integers\" is a class)\n* **Objects** are specific instances of a class (e.g., \"2 is an integer\")\n* **Methods** are fuctions associated to objects of a class\n * the \"the square of 2\" may be expressed as `2.square()` (returns 4)\n * the \"addition of 1 to 2\" may be expressed as `2.add(1)` (returns 3)\n * the \"name of 2\" may be expressed as `2.name()` (returns \"two\")\n\nToday we'll use an extended example of univariate functions\n$$f:\\mathbb{R} \\to \\mathbb{R}$$\nto see how you might use object oriented programming for something like automatic differentiation, classical machine learning, or deep learning. Yes - you can maybe use a library like [Tensorflow](https://www.tensorflow.org/), [Keras](https://keras.io/), or [PyTorch](https://pytorch.org/), but it's more fun to understand how to do it yourself (and then maybe use someone else's fancy/high quality implementation).\n\nFirst thing to remember is that everything in Python is an object, even functions.",
"_____no_output_____"
]
],
[
[
"def f(x):\n return x\n\nisinstance(f, object)",
"_____no_output_____"
],
[
"isinstance(isinstance, object)",
"_____no_output_____"
],
[
"isinstance(object, object)",
"_____no_output_____"
]
],
[
[
"Once you create an object, it lives somewhere on your computer:",
"_____no_output_____"
]
],
[
[
"id(f) # memory address on your computer",
"_____no_output_____"
],
[
"x = 1000\nid(x)",
"_____no_output_____"
]
],
[
[
"You can check if two variables are referring to the same address using `is`",
"_____no_output_____"
]
],
[
[
"z = x\nprint(\"equality: {}\".format(z == x))\nprint(\"same address: {}\".format(z is x))",
"_____no_output_____"
],
[
"y = 1000\nprint(\"equality: {}\".format(y == x))\nprint(\"same address: {}\".format(y is x))",
"_____no_output_____"
]
],
[
[
"## Univariate functions\n\nLet's consider functions that send a real number to a real number\n$$f:\\mathbb{R} \\to \\mathbb{R}$$\nPerhaps these functions have some parameters $\\theta$, such as\n$$f(x; \\theta) = \\theta x$$\n(a linear function with slope $\\theta$), or\n$$g(x;\\theta) = \\theta_1 x + \\theta_0$$\n(linear function with slope $\\theta_1$ and intercept $\\theta_0$), or\n$$h(x;\\theta) = \\theta_0 \\exp(-\\theta_1 x^2)$$\nand so on. The point is that we can parameterize functions that have a similar form, and that there may be different numbers of parameters depending on the function.\n\nWhat might we want to be able to do with a function?\n1. Evaluate it (`y = f(x)`)\n2. Print it as a string `f(x) = \"3x + 2\"`\n3. Calculate a gradient\n4. add/multiply/exponentiate...\n\nWe could think of doint the above with methods like `f.evaluate(x)`, and `f.name()`, but we'll use the special methods `__call__` and `__str__` to be able to do things like call `f(x)` and `format(f)` just as we might do so with built-in objects. You can see the different special methods available to overload [here](https://docs.python.org/3/reference/datamodel.html)\n\nWe're going to create an abstract function class that all the other classes we create will inherit from. If you haven't seen object oriented programming before, think of this as a way to promise all our functions will be able to do certain things (or throw an error). We'll provide default implementations for some methods (these will get filled in later), and have some methods that will need to be implemented differently for each sub-class.\n\nFor more on classes and inheritance, see [here](https://thepythonguru.com/python-inheritance-and-polymorphism/). The idea of giving objects methods with the same name is one form of [polymorphism](https://stackoverflow.com/questions/1031273/what-is-polymorphism-what-is-it-for-and-how-is-it-used) - we'll see how this is actually quite useful and allows you to do things that would be difficult without object-oriented programming.",
"_____no_output_____"
]
],
[
[
"class AbstractUnivariate:\n def __init__(self):\n raise NotImplementedError\n \n def __call__(self, x):\n raise NotImplementedError\n \n def fmtstr(self, x=\"x\"):\n raise NotImplementedError\n \n def __str__(self):\n return self.fmtstr(\"x\")\n \n def gradient(self):\n raise NotImplementedError\n \n # the rest of these methods will be implemented when we write the appropriate functions\n def __add__(self, other):\n return SumFunction(self, other)\n \n def __mul__(self, other):\n return ProdFunction(self, other)\n \n def __rmul__(self, other):\n return ScaleFunction(other, self)\n \n def __pow__(self, n):\n return ComposeFunction(PowerFunction(1, n), self)",
"_____no_output_____"
]
],
[
[
"Now, to create a class that inherits from our abstract class, we just use the following syntax:",
"_____no_output_____"
]
],
[
[
"class ConstantFunction(AbstractUnivariate): # AbstractUnivariate indicates class to use for inheritance\n def __init__(self, c):\n self.c = c",
"_____no_output_____"
],
[
"f = ConstantFunction(3)",
"_____no_output_____"
]
],
[
[
"We can see there's a class hierarchy now:",
"_____no_output_____"
]
],
[
[
"print(isinstance(f, ConstantFunction))\nprint(isinstance(f, AbstractUnivariate))\nprint(isinstance(f, object))",
"_____no_output_____"
]
],
[
[
"If we haven't implemented the methods we promised we would, we'll get errors",
"_____no_output_____"
]
],
[
[
"f(1)",
"_____no_output_____"
]
],
[
[
"Let's go ahead an implement the promised methods",
"_____no_output_____"
]
],
[
[
"class ConstantFunction(AbstractUnivariate):\n def __init__(self, c):\n self.c = c\n \n def __call__(self, x):\n return self.c\n \n def fmtstr(self, x=\"x\"):\n return \"{}\".format(self.c)\n \n # __str__(self) uses default from abstract class\n \n def gradient(self):\n return ConstantFunction(0)\n \n # we inherit the other functions from the AbstractUnivariate class",
"_____no_output_____"
],
[
"f = ConstantFunction(3)\nprint(f)\nprint(f(1))\nprint(f(2))\nprint(f.gradient())",
"_____no_output_____"
]
],
[
[
"What is it this object does? It represents the constant function\n$$f: x \\mapsto c$$",
"_____no_output_____"
],
[
"Let's do something a little less trivial. Now we'll implement\n$$f: x \\mapsto ax + b$$",
"_____no_output_____"
]
],
[
[
"class AffineFunction(AbstractUnivariate):\n def __init__(self, a, b):\n self.a = a\n self.b = b\n \n def __call__(self, x):\n return self.a * x + self.b\n \n def fmtstr(self, x=\"x\"):\n s = \"{}\".format(x)\n if self.a != 1:\n s = \"{}*\".format(self.a) + s\n if self.b != 0:\n s = s + \" + {}\".format(self.b)\n return s\n \n def gradient(self):\n return ConstantFunction(self.a)",
"_____no_output_____"
],
[
"f = AffineFunction(1, 1)\nprint(f)\nprint(f(2))\nprint(f.gradient())\nprint(isinstance(f, AbstractUnivariate))",
"_____no_output_____"
]
],
[
[
"## Discussion\n\nLet's take ourselves back to calculus. At some point you learned that you can take any function\n$$y = ax + b$$\nand if you know the values of $a$ and $b$, and someone gives you a value for $x$, you can calculate the value of $y$. At some later point you learned the rule\n$$ \\frac{d}{dx}(ax + b) = a$$\nregardless of what values $a$ and $b$ take. The class `AffineFunction` defines the rules that you learned in math class. \n\nWhen you write something like\n```python\nf = AffineFunction(1,1)\n```\nYou are just choosing the values of $a$ and $b$. Now just like you would be able to use the rules of arithmetic and calculus to compute $y$ given $x$ or the gradient of the function, your computer can as well.\n\n**Summary**\n* Class definition gives mathematical rules for an equation of a certain form\n* Instance of class is choice of constants for a function of that type",
"_____no_output_____"
],
[
"# Exercise 1\n\nImplement classes for the following univariate function templates:\n1. `QuadraticFunction` -- $f: x \\mapsto a x^2 + bx + c$\n2. `ExponentialFunction` -- $f: x \\mapsto a e^{bx}$\n3. `PowerFunction` -- $f: x \\mapsto ax^n$\n\nMake sure to return derivatives that are also `AbstractUnivariate` sub-classes. Which class can I use to represent $f: x \\mapsto x^{-1}$?",
"_____no_output_____"
]
],
[
[
"# your code here\nfrom math import * # for math.exp",
"_____no_output_____"
]
],
[
[
"# More functions\n\nWe can do more than just encode standard functions - we can scale, add, multiply, and compose functions.\n\nScaling a function:\n$$ g(x)= a *f(x)$$",
"_____no_output_____"
]
],
[
[
"class ScaleFunction(AbstractUnivariate):\n def __init__(self, a, f):\n self.a = a\n if isinstance(f, AbstractUnivariate):\n self.f = f\n else:\n raise AssertionError(\"must input an AbstractUnivariate function\")\n \n def __call__(self, x):\n return self.a * self.f(x)\n \n def fmtstr(self, x=\"x\"):\n if self.a == 1:\n return self.f.fmtstr(x)\n else:\n return \"{}*({})\".format(self.a, self.f.fmtstr(x))\n \n def gradient(self):\n return ScaleFunction(self.a, self.f.gradient())",
"_____no_output_____"
],
[
"f = ExponentialFunction(1, 2)\nprint(f)\ng = ScaleFunction(2, f)\nprint(g)\nprint(g.gradient())\nprint(g(1))",
"_____no_output_____"
]
],
[
[
"Sum and product of two functions\n$$ h(x) = f(x) + g(x)$$\n$$ h(x) = f(x) * g(x)$$",
"_____no_output_____"
]
],
[
[
"class SumFunction(AbstractUnivariate):\n def __init__(self, f, g):\n if isinstance(f, AbstractUnivariate) and isinstance(g, AbstractUnivariate):\n self.f = f\n self.g = g\n else:\n raise AssertionError(\"must input AbstractUnivariate functions\")\n \n def __call__(self, x):\n return self.f(x) + self.g(x)\n \n def fmtstr(self, x=\"x\"):\n return \"{} + {}\".format(self.f.fmtstr(x), self.g.fmtstr(x))\n \n def gradient(self):\n return SumFunction(self.f.gradient(), self.g.gradient())",
"_____no_output_____"
],
[
"f = ExponentialFunction(1, 2)\ng = AffineFunction(2, 1)\nh = SumFunction(f, g)\nprint(h.fmtstr(x=\"y\"))\nprint(h(-1))",
"_____no_output_____"
],
[
"print(h.gradient())",
"_____no_output_____"
],
[
"class ProdFunction(AbstractUnivariate):\n def __init__(self, f, g):\n if isinstance(f, AbstractUnivariate) and isinstance(g, AbstractUnivariate):\n self.f = f\n self.g = g\n else:\n raise AssertionError(\"must input AbstractUnivariate functions\")\n \n def __call__(self, x):\n return self.f(x) * self.g(x)\n \n def fmtstr(self, x=\"x\"):\n return \"({}) * ({})\".format(self.f.fmtstr(x=x), self.g.fmtstr(x=x))\n \n # product rule (f*g)' = f'*g + f*g'\n def gradient(self):\n return SumFunction(ProdFunction(self.f.gradient(),self.g), ProdFunction(self.f, self.g.gradient()))",
"_____no_output_____"
],
[
"f = ExponentialFunction(1, 2)\ng = AffineFunction(2, 1)\nh = ProdFunction(f, g)\nprint(h)\nprint(h(-1))",
"_____no_output_____"
],
[
"print(h.gradient())",
"_____no_output_____"
]
],
[
[
"Compose Functions:\n$$h(x) = (g \\circ f)(x) = g(f(x))$$",
"_____no_output_____"
]
],
[
[
"class ComposeFunction(AbstractUnivariate):\n def __init__(self, g, f):\n if isinstance(f, AbstractUnivariate) and isinstance(g, AbstractUnivariate):\n self.f = f\n self.g = g\n else:\n raise AssertionError(\"must input AbstractUnivariate functions\")\n \n def __call__(self, x):\n return self.g(self.f(x))\n \n def fmtstr(self, x=\"x\"):\n return self.g.fmtstr(x=\"({})\".format(self.f.fmtstr(x)))\n \n # chain rule : g(f(x))' = g'(f(x))*f'(x)\n def gradient(self):\n return ProdFunction(ComposeFunction(self.g.gradient(), self.f), self.f.gradient())",
"_____no_output_____"
],
[
"f = PowerFunction(1,2)\nprint(f.fmtstr(\"x\"))\ng = ComposeFunction(f,f)\nprint(g)\nh = ComposeFunction(g, f)\nprint(h)\nprint(h(2)) # 2^(2*2*2) = 2^8 = 256",
"_____no_output_____"
],
[
"f = PowerFunction(1,2)\ng = ExponentialFunction(0.5, -1)\nh = ComposeFunction(g, f)\nprint(h)\nprint(h.gradient())",
"_____no_output_____"
]
],
[
[
"## Operator overloading makes everything better\n\nRecall how when we wrote the AbstractUnivariate class, we included some default methods\n```python\nclass AbstractUnivariate:\n # ...\n \n # the rest of these methods will be implemented when we write the appropriate functions\n def __add__(self, other):\n return SumFunction(self, other)\n \n def __mul__(self, other):\n return ProdFunction(self, other)\n \n def __rmul__(self, other):\n return ScaleFunction(other, self)\n \n def __pow__(self, n):\n return ComposeFunction(PowerFunction(1, n), self)\n```\n\nIf you think it is clunky to keep writing `SumFunction` or `ProdFunction` everywhere, you're not alone. Again, you can use the special methods above to [overload operators](https://docs.python.org/3/reference/datamodel.html#emulating-numeric-types)",
"_____no_output_____"
]
],
[
[
"f = ExponentialFunction(1, 2)\ng = AffineFunction(2, 1)\nprint(\"f = {}\".format(f))\nprint(\"g = {}\".format(g))\nprint(\"f + g = {}\".format(f+g))\nprint(\"f * g = {}\".format(f*g))\nprint(\"f^2 = {}\".format(f**2))\nprint(\"2*g = {}\".format(2*g))",
"_____no_output_____"
],
[
"f = ExponentialFunction(1, 2)\ng = AffineFunction(2, 1)\nh = f*g\nprint(h.gradient())",
"_____no_output_____"
]
],
[
[
"## What's going on?\n\nBecause we thought ahead to define addition, multiplication, scaling, and powers in our `AbstractUnivariate` class, every sub-class will implement those methods by default **without needing to write any extra code**.\n\nIf we hadn't done this, we would have had to copy and paste the same thing into every class definition to get the same behavior, **but we don't need to**. In fact, if we write a new basic univariate function class, e.g. `LogFunction`, we get addition, multiplication, etc., for free!",
"_____no_output_____"
],
[
"## Symbolic Functions\n\nJust for fun, let's create an `AbstractUnivariate` sub-class, which just holds a placeholder symbolic function",
"_____no_output_____"
]
],
[
[
"class SymbolicFunction(AbstractUnivariate):\n def __init__(self, name):\n if isinstance(name, str):\n self.name=name\n else:\n raise AssertionError(\"name must be string\")\n \n def __call__(self, x):\n return \"{}({})\".format(self.name, x)\n \n def fmtstr(self, x=\"x\"):\n return self.name + \"({})\".format(x)\n \n # product rule (f*g)' = f'*g + f*g'\n def gradient(self):\n return SymbolicFunction(self.name + \"'\")\n ",
"_____no_output_____"
],
[
"f = SymbolicFunction(\"f\")\nprint(f)\nprint(f.gradient())\ng = SymbolicFunction(\"g\")\nprint(g + f)",
"_____no_output_____"
]
],
[
[
"Now we can remind ourselves of product rule, and chain rule (which we encoded in `ProductFunction` and `ComposeFunction` classes)",
"_____no_output_____"
]
],
[
[
"f = SymbolicFunction(\"f\")\ng = SymbolicFunction(\"g\")\nprint((f*g).gradient())\nh = ComposeFunction(g, f)\nprint(h.gradient())",
"_____no_output_____"
]
],
[
[
"And we can derive quotient rule",
"_____no_output_____"
]
],
[
[
"f = SymbolicFunction(\"f\")\ng = SymbolicFunction(\"g\")\nh = f * g**-1\nprint(h)\nprint(h.gradient())",
"_____no_output_____"
]
],
[
[
"You can also add symbolic functions to non-symbolic ones:",
"_____no_output_____"
]
],
[
[
"f = SymbolicFunction(\"f\")\ng = AffineFunction(1, 2)\nh = f + g\nprint(h)\nprint(h.gradient())",
"_____no_output_____"
]
],
[
[
"## Summary\n\nYou're now on your way to having your own automatic differentiation library! Or your own symbolic computation library! You can probably see lots of ways to extend and improve what you've seen here:\n* Support Multivariate Functions\n* Add more \"basic functions\" such as trig functions, etc.\n* Reduce expressions when you are able to\n* ...\n\nYes, there are many libraries that do this very thing. Keywords are \"autodifferentiation\", \"symbolic math\". This sort of thing is used extensively in deep learning libraries, as well as optimization libraries.\n\n* [Sympy](https://www.sympy.org/en/index.html) for symbolic computation\n* [SciPy linear operators](https://docs.scipy.org/doc/scipy/reference/generated/scipy.sparse.linalg.LinearOperator.html) do something similar to HW1\n* [Sage](https://www.sagemath.org/) does a lot of symbolic math using Python\n* [Autodiff tools for Python](http://www.autodiff.org/?module=Tools&language=python)\n* [Autograd](https://github.com/HIPS/autograd) package\n* Most Deep learning libraries (see below) do some form of automatic differentiation\n\n\n### How was Object Oriented Programming Useful?\n\n**Class Inhertiance** allowed you to get functions like addition and multiplication for free once you defined the class everything inherited from\n\n**Polymorphism** enabled you to use any combination of `AbstractUnivariate` functions and still evaluate them, calculate derivatives, and format equations. Everyone played by the same rules.\n\n**Encapsulation** let you interact with functions without worrying about how they are implemented under the hood.\n\nIf you think back to HW1, we implicitly used polymorphism in the power method function (e.g., matrix-vector multiply always uses `dot()` no matter which class we're using)",
"_____no_output_____"
],
[
"# Exercise 2\n\nIgnoring our `SymbolicFunction` class, any sub-class of `AbstractUnivariate` is a real function $f:\\mathbb{R} \\to \\mathbb{R}$ that we can evaluate using `f(x)` syntax. One thing that you may wish to do is find roots of your function: $\\{x \\mid f(x) = 0\\}$.\n\nOne very classical algorithm for doing this is called [Newton's Method](https://en.wikipedia.org/wiki/Newton%27s_method), and has the basic pseudocode:\n```\ninitialize x_0\nwhile not converged:\n x_{k+1} = x_k - f(x_k)/f'(x_k)\n```\n\nWrite a function that implements Newton's method on any `AbstractUnivariate` function\n\nHint: use the `gradient()` method to get a function for derivatives",
"_____no_output_____"
]
],
[
[
"def find_root(f, x0=0.0, tol=1e-8):\n if isinstance(f, SymbolicFunction):\n raise AssertionError(\"can't handle symbolic input\")\n elif not isinstance(f, AbstractUnivariate):\n raise AssertionError(\"Input must be AbstractUnivariate\")\n x = x0\n # your code here\n \n return x",
"_____no_output_____"
]
],
[
[
"# Deep Learning\n\nAfter the first part of this lecture, you now have a pretty good idea of how to get started implementing a deep learning library. Recall that above we considered functions of the form\n$$f(x; \\theta): \\mathbb{R} \\to \\mathbb{R}$$\n\nTo get to machine learning, you need to handle multivariate input and output\n$$f(x; \\theta):\\mathbb{R}^p \\to \\mathbb{R}^k$$\nYou also need to be able to take the gradient of $f$ with respect to the parameters $\\theta$ (which we didn't do in our `AbstractUnivariate` class, but is straightforward), and then you can do things like optimize a loss function using your favorite optimization algorithm.\n\nIn deep learning, we have the exact same setup\n$$f(x; \\theta):\\mathbb{R}^p \\to \\mathbb{R}^k$$\nWhat makes deep learning a \"special case\" of machine learning is that the function $f$ is the composition of several/many functions\n$$f = f_n \\circ f_{n-1} \\circ \\dots \\circ f_1$$\nThis is what we mean by \"layers\", and you use chain rule to \"backpropagate\" gradients with respect to the parameters.\n\n**Disclaimer** If you really want to learn to use a deep learning library, you really should go through several tutorials and learn about the different functions that are used (and *why* they are used). This is beyond the scope of this course, but there are several courses at Stanford that are devoted to this.\n\n## Deep Learning Libraries\n\nSome popular libraries for deep learning are [Tensorflow](https://www.tensorflow.org/), [Keras](https://keras.io/), and [PyTorch](https://pytorch.org/). Each has their strengths and weaknesses. All of them do essientially the same thing: you define a function through composition using objects that are in many ways similar to what you just implemented. Then you choose a loss function and start optimizing the parameters in these functions using something like stochastic gradient descent.\n\nWe'll do an example in PyTorch, since it is higher-level than Tensorflow, and perhaps the most \"Pythonic\" of the libraries.\n\n```bash\nconda install pytorch pillow\n```\n\n## PyTorch\n\nWhat's a tensor? Conceptually identical to numpy array.\n\nWe'll consider the following network\n$$ x \\xrightarrow{w_1} h \\to ReLU(h) \\xrightarrow{w_2} y$$\nwhere $x$ is a 500-dimensional vector, $h$ is a 100-dimensional \"hidden layer\", and $y$ is a 10-dimensional vector. $w_1$ and $w_2$ are linear transformations (matrices), and ReLU refers to the function\n$$ReLU(x) = \\begin{cases}\nx & x > 0\\\\\n0 & x \\le 0\n\\end{cases}$$",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torch.autograd import Variable\n\ndtype = torch.FloatTensor\n# N - batch size\n# D_in - x dimension\n# H - h dimension\"\n# D_out - y dimension\nN, D_in, H, D_out = 64, 500, 100, 10\n\n# Setting requires_grad=False indicates that we do not need to compute gradients w.r.t var\n# during the backward pass.\nx = Variable(torch.randn(N, D_in).type(dtype), requires_grad = False)\ny = Variable(torch.randn(N, D_out).type(dtype), requires_grad = False)\n\n# Setting requires_grad=True indicates that we want to compute gradients with\n# respect to these Variables during the backward pass.\nw1 = Variable(torch.randn(D_in, H).type(dtype), requires_grad=True)\nw2 = Variable(torch.randn(H, D_out).type(dtype), requires_grad=True)",
"_____no_output_____"
],
[
"learning_rate = 1e-6\nfor t in range(10000):\n # Forward pass: compute predicted y using operations on Variables;\n y_pred = x.mm(w1).clamp(min=0).mm(w2) # clamp=ReLU\n \n # Compute and print loss using operations on Variables.\n # Now loss is a Variable of shape (1,) and loss.data is a Tensor of shape\n loss = (y_pred - y).pow(2).sum()\n\n\n # Use autograd to compute the backward pass. This call will compute the\n # gradient of loss with respect to all Variables with requires_grad=True.\n loss.backward()\n\n # Update weights using gradient descent; w1.data and w2.data are Tensors,\n # w1.grad and w2.grad are Variables and w1.grad.data and w2.grad.data are\n # Tensors.\n w1.data -= learning_rate * w1.grad.data\n w2.data -= learning_rate * w2.grad.data\n\n # Manually zero the gradients after running the backward pass\n w1.grad.data.zero_()\n w2.grad.data.zero_()\n print(\"Loss is: {}\".format(loss.data.numpy()), end='\\r')\n\nprint()\nprint(\"Final loss is {}\".format(loss.data[0]))",
"_____no_output_____"
]
],
[
[
"## That's still fairly cumbersome\n\n- When building neural networks, arrange the computation into layers, some of which have learnable parameters which will be optimized during learning.\n- Use the ``` torch.nn ``` package to define your layers\n- Create custom networks by subclassing the nn.Module\n- Really clean code!\n- Just create a class subclassing the nn.Module\n - specify layers in the ```__init__``` \n - define a forward pass by ```forward(self,x)``` method\n \nThis is analgous to how we created specific sub-classes of `AbstractUnivariate`, and got a lot for free through class inheritance, polymorphism, abstraction, etc.",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nfrom torchvision import datasets, transforms\nfrom torch.autograd import Variable\n\nclass TwoLayerNet(nn.Module):\n \n def __init__(self, D_in, H, D_out): # this defines the parameters, and stores them\n super(TwoLayerNet, self).__init__() # overrides class inheritance\n self.layer1 = nn.Linear(D_in, H) # initializes weights\n self.layer2 = nn.Linear(H, D_out)\n \n def forward(self, x): # this defines the composition of functions\n out = F.relu(self.layer1(x)) \n out = self.layer2(out)\n return out",
"_____no_output_____"
],
[
"# N is batch size; D_in is input dimension; H is hidden dimension; D_out is output dimension.\nN, D_in, H, D_out = 64, 1000, 100, 10\n\n# Create random Tensors to hold inputs and outputs, and wrap them in Variables\nx = Variable(torch.randn(N, D_in))\ny = Variable(torch.randn(N, D_out), requires_grad=False)\n\n# Construct our model by instantiating the class defined above\nmodel = TwoLayerNet(D_in, H, D_out) # we create our function f:x \\to y\n\n# Construct our loss function and an Optimizer. \nloss_fn = torch.nn.MSELoss(size_average=False)\noptimizer = torch.optim.SGD(model.parameters(), lr=1e-4)\nfor t in range(1000):\n # Forward pass: Compute predicted y by passing x to the model\n y_pred = model(x) # evaluate the f(x)\n\n # Compute and print loss\n loss = loss_fn(y_pred, y) # evaluate the loss\n\n # Zero gradients, perform a backward pass, and update the weights.\n optimizer.zero_grad()\n loss.backward()\n optimizer.step()\n \nprint(\"Final Loss is {}\".format(loss.data[0]))",
"_____no_output_____"
]
],
[
[
"## Training a CNN for Image Classification\n\nThe following example is ported from [PyTorch's Documentation](https://pytorch.org/tutorials/beginner/blitz/cifar10_tutorial.html#sphx-glr-beginner-blitz-cifar10-tutorial-py)\n\nThe basic task of the network is to classify images in the [CIFAR10 dataset](https://www.cs.toronto.edu/~kriz/cifar.html), which has 10 classes:\n\n```'plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck'```\n\n",
"_____no_output_____"
]
],
[
[
"import torch\nimport torchvision\nimport torchvision.transforms as transforms",
"_____no_output_____"
],
[
"# normalizes images to have pixel values between [-1,1]\n# turns image into \"tensor\" to be fed to network\ntransform = transforms.Compose(\n [transforms.ToTensor(),\n transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])",
"_____no_output_____"
],
[
"# get data\ntrainset = torchvision.datasets.CIFAR10(root='./data', train=True,\n download=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=4,\n shuffle=True, num_workers=2)\n\ntestset = torchvision.datasets.CIFAR10(root='./data', train=False,\n download=True, transform=transform)\ntestloader = torch.utils.data.DataLoader(testset, batch_size=4,\n shuffle=False, num_workers=2)",
"_____no_output_____"
],
[
"# Classes in the CIFAR10 dataset\nclasses = ('plane', 'car', 'bird', 'cat',\n 'deer', 'dog', 'frog', 'horse', 'ship', 'truck')",
"_____no_output_____"
]
],
[
[
"To visualize images:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"def imshow(img):\n img = img / 2 + 0.5 # unnormalize\n npimg = img.numpy()\n plt.imshow(np.transpose(npimg, (1, 2, 0)))",
"_____no_output_____"
],
[
"# get some random training images\ndataiter = iter(trainloader)\nimages, labels = dataiter.next()\n\n# show images\nimshow(torchvision.utils.make_grid(images))\n# print labels\nprint(' '.join('%5s' % classes[labels[j]] for j in range(4)))",
"_____no_output_____"
]
],
[
[
"$$ x \\xrightarrow{p_1 \\circ r_1 \\circ c_1} h_1 \\xrightarrow{p_2 \\circ r_2 \\circ c_2} h_2 \\xrightarrow{r_3 \\circ f_1} h_3 \\xrightarrow{r_4 \\circ f_2} h_4 \\xrightarrow{f_3} y$$\nwhere $c$ refers to a convolution (a type of linear transormation), $r$ a ReLU, $p$ a pool, and $f$ a (fully connected) linear transformation. $x$ is an input image, and $y$ is a vector of length 10 which you can think of as \"class probabilities\".\n\nYou might also write the above expression as the following composition of functions:\n$$y = f_3(r_4(f_2(r_3(f_1(p_2(r_2(c_2(p_1(r_1(c_1(x)))))))))))$$\nHow would you like to write out that chain rule by hand?",
"_____no_output_____"
]
],
[
[
"import torch.nn as nn\nimport torch.nn.functional as F",
"_____no_output_____"
],
[
"class Net(nn.Module):\n def __init__(self):\n super(Net, self).__init__()\n self.conv1 = nn.Conv2d(3, 6, 5)\n self.pool = nn.MaxPool2d(2, 2)\n self.conv2 = nn.Conv2d(6, 16, 5)\n self.fc1 = nn.Linear(16 * 5 * 5, 120)\n self.fc2 = nn.Linear(120, 84)\n self.fc3 = nn.Linear(84, 10)\n\n # composition of functions\n def forward(self, x):\n x = self.pool(F.relu(self.conv1(x)))\n x = self.pool(F.relu(self.conv2(x)))\n x = x.view(-1, 16 * 5 * 5) # flattens tensor\n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n x = self.fc3(x)\n return x",
"_____no_output_____"
],
[
"net = Net()",
"_____no_output_____"
]
],
[
[
"Now, we define a loss function and choose an optimimization algorithm",
"_____no_output_____"
]
],
[
[
"import torch.optim as optim\n\ncriterion = nn.CrossEntropyLoss()\noptimizer = optim.SGD(net.parameters(), lr=0.001, momentum=0.9)",
"_____no_output_____"
]
],
[
[
"Now, we can train the network",
"_____no_output_____"
]
],
[
[
"for epoch in range(2): # loop over the dataset multiple times\n\n running_loss = 0.0\n for i, data in enumerate(trainloader, 0):\n # get the inputs\n inputs, labels = data\n\n # zero the parameter gradients\n optimizer.zero_grad()\n\n # forward + backward + optimize\n outputs = net(inputs)\n loss = criterion(outputs, labels)\n loss.backward() # calculate gradient w.r.t. parameters\n optimizer.step() # update parameters\n\n # print statistics\n running_loss += loss.item()\n if i % 2000 == 1999: # print every 2000 mini-batches\n print('[%d, %5d] loss: %.3f' %\n (epoch + 1, i + 1, running_loss / 2000))\n running_loss = 0.0\n\nprint('Finished Training')",
"_____no_output_____"
]
],
[
[
"To test the classifier, we'll load a few images from our test set",
"_____no_output_____"
]
],
[
[
"dataiter = iter(testloader)\nimages, labels = dataiter.next()\n\n# print images\nimshow(torchvision.utils.make_grid(images))\nprint('GroundTruth: ', ' '.join('%5s' % classes[labels[j]] for j in range(4)))",
"_____no_output_____"
]
],
[
[
"Now we'll make predictions",
"_____no_output_____"
]
],
[
[
"outputs = net(images)\n\n_, predicted = torch.max(outputs, 1)\n\nprint('Predicted: ', ' '.join('%5s' % classes[predicted[j]]\n for j in range(4)))",
"_____no_output_____"
]
],
[
[
"To get accuracy over the whole test set (keep in mind, we expect 10% accuracy if we randomly guess a class):",
"_____no_output_____"
]
],
[
[
"correct = 0\ntotal = 0\nwith torch.no_grad():\n for data in testloader:\n images, labels = data\n outputs = net(images)\n _, predicted = torch.max(outputs.data, 1)\n total += labels.size(0)\n correct += (predicted == labels).sum().item()\n\nprint('Accuracy of the network on the 10000 test images: %d %%' % (\n 100 * correct / total))",
"_____no_output_____"
]
],
[
[
"## For more examples... \ncheck out [Pytorch Docs](http://pytorch.org/tutorials/beginner/deep_learning_60min_blitz.html)\n\n## To add your own function to PyTorch's autograd library\nIf you want to add your own functions to PyTorch's autograd library, see [here](https://pytorch.org/tutorials/beginner/examples_autograd/two_layer_net_custom_function.html).\nYou would write a class that inherits from `torch.autograd.Function`, and just need to implement `forward` and `backward` methods (conceptually similar to `eval` and `gradient`).",
"_____no_output_____"
],
[
"# Reminders\n\n* This is the last class\n* HW 2 is due - this is the last homework\n* After today, office hours will be by appointment\n\n# Course Conclusion\n\nYou've now seen the basics of Python, and have now seen some of the standard libraries for scientific computing and data science. Hopefully you may now have some ideas of how you can use Python for whatever problems interest you, and have some templates to get you started.\n\nTo continue on your Python journey, the best way to improve your skills and knowledge is to just try using it for whatever it is you're doing.\n\nIf you'd like to use Python for a specific task, and don't know how to get started, feel free to send me an email and I'll try to point you in a reasonable direction.",
"_____no_output_____"
],
[
"# Additional Resources\n\n## Object Oriented Programming\n\n* Beginner's guide to Object Oriented Programming in Python [here](https://stackabuse.com/object-oriented-programming-in-python/)\n\n## Image Processing\n\nIn this class, we've worked a lot with tabular data. Another important type of data to be able to work with is image data.\n\nSome options are\n* [scikit-image](https://scikit-image.org/)\n* [scipy](http://www.scipy-lectures.org/advanced/image_processing/index.html)\n* [Pillow](https://pillow.readthedocs.io)\n* [OpenCV](https://opencv-python-tutroals.readthedocs.io/en/latest/py_tutorials/py_tutorials.html)\n\nFor many examples, see the [Scikit-image gallery](http://scikit-image.org/docs/stable/auto_examples/). Other libraries also have examples.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cb4cc53a17a6150ff3760834578b46328c02598a | 1,832 | ipynb | Jupyter Notebook | chapter1/homework/computer/3-15/201611680862.ipynb | hpishacker/python_tutorial | 9005f0db9dae10bdc1d1c3e9e5cf2268036cd5bd | [
"MIT"
] | 76 | 2017-09-26T01:07:26.000Z | 2021-02-23T03:06:25.000Z | chapter1/homework/computer/3-15/201611680862.ipynb | hpishacker/python_tutorial | 9005f0db9dae10bdc1d1c3e9e5cf2268036cd5bd | [
"MIT"
] | 5 | 2017-12-10T08:40:11.000Z | 2020-01-10T03:39:21.000Z | chapter1/homework/computer/3-15/201611680862.ipynb | hacker-14/python_tutorial | 4a110b12aaab1313ded253f5207ff263d85e1b56 | [
"MIT"
] | 112 | 2017-09-26T01:07:30.000Z | 2021-11-25T19:46:51.000Z | 18.32 | 73 | 0.474345 | [
[
[
"n=int(input('please enter a positive ingeter'))\ni=1\ntotal=0\nwhile i<=n:\n a=int(input())\n total=total+a\n i=i+1\nprint(total)",
"please enter a positive ingeter5\n1\n2\n3\n4\n5\n15\n"
],
[
"input('if you don not want to enter anymore,please click Enter')\ni=input()\nwhile i!='':\n i=input()",
"if you don not want to enter anymore,please click Enter1\n2\n3\n\n"
],
[
"i=int(input())\nadd_total=i\nmulti_total=i\nwhile add_total>=i or multi_total>=(i*i):\n i=int(input())\n add_total=add_total+i\n multi_total=multi_total*i",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
cb4cccc28b567997f8146d67af0135caa7eb45d6 | 1,052 | ipynb | Jupyter Notebook | 8-Labs/Lab23/Untitled.ipynb | dustykat/engr-1330-psuedo-course | 3e7e31a32a1896fcb1fd82b573daa5248e465a36 | [
"CC0-1.0"
] | null | null | null | 8-Labs/Lab23/Untitled.ipynb | dustykat/engr-1330-psuedo-course | 3e7e31a32a1896fcb1fd82b573daa5248e465a36 | [
"CC0-1.0"
] | null | null | null | 8-Labs/Lab23/Untitled.ipynb | dustykat/engr-1330-psuedo-course | 3e7e31a32a1896fcb1fd82b573daa5248e465a36 | [
"CC0-1.0"
] | null | null | null | 19.481481 | 67 | 0.545627 | [
[
[
"import pandas \nimport numpy\nimport matplotlib.pyplot as plt\nmydogs=numpy.random.normal(70,20,2500000)\ndogweight = pandas.DataFrame({'Weight_lbs':mydogs})\ndogweight.to_csv('DogWeights.csv') # write to local directory",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
cb4cdad33e997b4d606290b442689b19e353340e | 23,119 | ipynb | Jupyter Notebook | CHSHGame/Workbook_CHSHGame.ipynb | vxfield/QuantumKatas | 4fd06ce5776164504725e564044241f155c4d9a1 | [
"MIT"
] | 1 | 2020-09-26T22:29:24.000Z | 2020-09-26T22:29:24.000Z | CHSHGame/Workbook_CHSHGame.ipynb | FingerLeakers/QuantumKatas | 4fd06ce5776164504725e564044241f155c4d9a1 | [
"MIT"
] | null | null | null | CHSHGame/Workbook_CHSHGame.ipynb | FingerLeakers/QuantumKatas | 4fd06ce5776164504725e564044241f155c4d9a1 | [
"MIT"
] | null | null | null | 36.580696 | 489 | 0.582681 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
cb4ce17bf7c58f04ffc03b56fd501f75d3fc3384 | 872,362 | ipynb | Jupyter Notebook | python/Post-processing the final result for the presentation..ipynb | ekvall93/kthLife | a8219ca6738a2a2130079b0d815d5e04ac45b3cd | [
"Apache-2.0"
] | null | null | null | python/Post-processing the final result for the presentation..ipynb | ekvall93/kthLife | a8219ca6738a2a2130079b0d815d5e04ac45b3cd | [
"Apache-2.0"
] | 8 | 2020-09-07T02:42:46.000Z | 2022-02-26T17:12:23.000Z | python/Post-processing the final result for the presentation..ipynb | ekvall93/kthLife | a8219ca6738a2a2130079b0d815d5e04ac45b3cd | [
"Apache-2.0"
] | null | null | null | 1,504.072414 | 836,160 | 0.954985 | [
[
[
"from utils import *\nimport gensim\nfrom sklearn.mixture import BayesianGaussianMixture\nimport json",
"_____no_output_____"
],
[
"df = pd.read_csv(\"assets/finalproduct/finalproductDf\")\ndf.drop([\"Unnamed: 0\"],axis=1, inplace=True)\nid_to_auth = pickle_o.load(\"assets/dictionaries/id_to_all_auths_2004\")\nauth_to_id = pickle_o.load(\"assets/dictionaries/auths_to_all_id_2004\")",
"_____no_output_____"
],
[
"Name = list(df.Author.values)\nkth_id = [auth_to_id[a] for a in Name]\ndf_only_auth = pd.DataFrame(data={\"Name\":Name, \"ID\":kth_id})\ndf_only_auth.to_csv(\"assets/finalproduct/onlyAuthors.csv\")",
"_____no_output_____"
],
[
"df_abs = pd.read_csv(\"assets/dataframes/all_authors_df_2004\")\ndf_abs.drop([\"Unnamed: 0\"],axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_abs.head()",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"list_of_dict = list()\nfor a, d in zip(df.Author.values, df.Doc_id.values):\n new_d = dict()\n new_d[\"name\"] = str(a)\n \n abstracts = list()\n all_d = d.split(\":\")\n new_d[\"docid\"] = all_d\n for ad in all_d:\n abst = df_abs[df_abs.Doc_id == int(ad)].Abstracts.values[0]\n abstracts.append(abst)\n new_d[\"abstracts\"] = abstracts\n list_of_dict.append(new_d)\n ",
"_____no_output_____"
],
[
"list_of_dict[0]",
"_____no_output_____"
],
[
"with open('assets/finalproduct/auth_to_abs.json', 'w') as fp:\n json.dump(list_of_dict, fp)",
"_____no_output_____"
],
[
"y = json.dumps(auth_to_abs)\n\n# the result is a Python dictionary:\nprint(y)",
"{\"Lundqvist, Mikael\": [\"Spontaneous oscillations measured by local field potentials, electroencephalograms and magnetoencephalograms exhibit a pronounced peak in the alpha band (8-12 Hz) in humans and primates. Both instantaneous power and phase of these ongoing oscillations have commonly been observed to correlate with psychophysical performance in stimulus detection tasks. We use a novel model-based approach to study the effect of prestimulus oscillations on detection rate. A previously developed biophysically detailed attractor network exhibits spontaneous oscillations in the alpha range before a stimulus is presented and transiently switches to gamma-like oscillations on successful detection. We demonstrate that both phase and power of the ongoing alpha oscillations modulate the probability of such state transitions. The power can either positively or negatively correlate with the detection rate, in agreement with experimental findings, depending on the underlying neural mechanism modulating the oscillatory power. Furthermore, the spatially distributed alpha oscillators of the network can be synchronized by global nonspecific weak excitatory signals. These synchronization events lead to transient increases in alpha-band power and render the network sensitive to the exact timing of target stimuli, making the alpha cycle function as a temporal mask in line with recent experimental observations. Our results are relevant to several studies that attribute a modulatory role to prestimulus alpha dynamics.\", \"Attractor neural networks are thought to underlie working memory functions in the cerebral cortex. Several such models have been proposed that successfully reproduce firing properties of neurons recorded from monkeys performing working memory tasks. However, the regular temporal structure of spike trains in these models is often incompatible with experimental data. Here, we show that the in vivo observations of bistable activity with irregular firing at the single cell level can be achieved in a large-scale network model with a modular structure in terms of several connected hypercolumns. Despite high irregularity of individual spike trains, the model shows population oscillations in the beta and gamma band in ground and active states, respectively. Irregular firing typically emerges in a high-conductance regime of balanced excitation and inhibition. Population oscillations can produce such a regime, but in previous models only a non-coding ground state was oscillatory. Due to the modular structure of our network, the oscillatory and irregular firing was maintained also in the active state without fine-tuning. Our model provides a novel mechanistic view of how irregular firing emerges in cortical populations as they go from beta to gamma oscillations during memory retrieval.\", \"Changes in oscillatory brain activity are strongly correlated with performance in cognitive tasks and modulations in specific frequency bands are associated with working memory tasks. Mesoscale network models allow the study of oscillations as an emergent feature of neuronal activity. Here we extend a previously developed attractor network model, shown to faithfully reproduce single-cell activity during retention and memory recall, with synaptic augmentation. This enables the network to function as a multi-item working memory by cyclic reactivation of up to six items. The reactivation happens at theta frequency, consistently with recent experimental findings, with increasing theta power for each additional item loaded in the network's memory. Furthermore, each memory reactivation is associated with gamma oscillations. Thus, single-cell spike trains as well as gamma oscillations in local groups are nested in the theta cycle. The network also exhibits an idling rhythm in the alpha/beta band associated with a noncoding global attractor. Put together, the resulting effect is increasing theta and gamma power and decreasing alpha/beta power with growing working memory load, rendering the network mechanisms involved a plausible explanation for this often reported behavior.\"], \"Herman, Pawel Andrzej\": [\"Spontaneous oscillations measured by local field potentials, electroencephalograms and magnetoencephalograms exhibit a pronounced peak in the alpha band (8-12 Hz) in humans and primates. Both instantaneous power and phase of these ongoing oscillations have commonly been observed to correlate with psychophysical performance in stimulus detection tasks. We use a novel model-based approach to study the effect of prestimulus oscillations on detection rate. A previously developed biophysically detailed attractor network exhibits spontaneous oscillations in the alpha range before a stimulus is presented and transiently switches to gamma-like oscillations on successful detection. We demonstrate that both phase and power of the ongoing alpha oscillations modulate the probability of such state transitions. The power can either positively or negatively correlate with the detection rate, in agreement with experimental findings, depending on the underlying neural mechanism modulating the oscillatory power. Furthermore, the spatially distributed alpha oscillators of the network can be synchronized by global nonspecific weak excitatory signals. These synchronization events lead to transient increases in alpha-band power and render the network sensitive to the exact timing of target stimuli, making the alpha cycle function as a temporal mask in line with recent experimental observations. Our results are relevant to several studies that attribute a modulatory role to prestimulus alpha dynamics.\", \"Quantifying neural and non-neural contributions to increased joint resistance in spasticity is essential for a better understanding of its pathophysiological mechanisms and evaluating different intervention strategies. However, direct measurement of spasticity-related manifestations, e.g., motoneuron and biophysical properties in humans, is extremely challenging. In this vein, we developed a forward neuromusculoskeletal model that accounts for dynamics of muscle spindles, motoneuron pools, muscle activation and musculotendon of wrist flexors and relies on the joint angle and resistant torque as the only input measurement variables. By modeling the stretch reflex pathway, neural and non-neural related properties of the spastic wrist flexors were estimated during the wrist extension test. Joint angle and resistant torque were collected from 17 persons with chronic stroke and healthy controls using NeuroFlexor, a motorized force measurement device during the passive wrist extension test. The model was optimized by tuning the passive and stretch reflex-related parameters to fit the measured torque in each participant. We found that persons with moderate and severe spasticity had significantly higher stiffness than controls. Among subgroups of stroke survivors, the increased neural component was mainly due to a lower muscle spindle rate at 50% of the motoneuron recruitment. The motoneuron pool threshold was highly correlated to the motoneuron pool gain in all subgroups. The model can describe the overall resistant behavior of the wrist joint during the test. Compared to controls, increased resistance was predominantly due to higher elasticity and neural components. We concluded that in combination with the NeuroFlexor measurement, the proposed neuromusculoskeletal model and optimization scheme served as suitable tools for investigating potential parameter changes along the stretch-reflex pathway in persons with spasticity.\", \"Quantifying neural and non-neural contributions to the joint resistance in spasticity is essential for a better evaluation of different intervention strategies such as botulinum toxin A (BoTN-A). However, direct measurement of muscle mechanical properties and spasticity-related parameters in humans is extremely challenging. The aim of this study was to use a previously developed musculoskeletal model and optimization scheme to evaluate the changes of neural and non-neural related properties of the spastic wrist flexors during passive wrist extension after BoTN-A injection. Data of joint angle and resistant torque were collected from 21 chronic stroke patients before, and 4 and 12 weeks post BoTN-A injection using NeuroFlexor, which is a motorized force measurement device to passively stretch wrist flexors. The model was optimized by tuning the passive and stretch-related parameters to fit the measured torque in each participant. It was found that stroke survivors exhibited decreased neural components at 4 weeks post BoNT-A injection, which returned to baseline levels after 12 weeks. The decreased neural component was mainly due to the increased motoneuron pool threshold, which is interpreted as a net excitatory and inhibitory inputs to the motoneuron pool. Though the linear stiffness and viscosity properties of wrist flexors were similar before and after treatment, increased exponential stiffness was observed over time which may indicate a decreased range of motion of the wrist joint. Using a combination of modeling and experimental measurement, valuable insights into the treatment responses, i.e., transmission of motoneurons, are provided by investigating potential parameter changes along the stretch reflex pathway in persons with chronic stroke.\", \"The olfactory sense is a particularly challenging domain for cognitive science investigations of perception, memory, and language. Although many studies show that odors often are difficult to describe verbally, little is known about the associations between olfactory percepts and the words that describe them. Quantitative models of how odor experiences are described in natural language are therefore needed to understand how odors are perceived and communicated. In this study, we develop a computational method to characterize the olfaction-related semantic content of words in a large text corpus of internet sites in English. We introduce two new metrics: olfactory association index (OAI, how strongly a word is associated with olfaction) and olfactory specificity index (OSI, how specific a word is in its description of odors). We validate the OAI and OSI metrics using psychophysical datasets by showing that terms with high OAI have high ratings of perceived olfactory association and are used to describe highly familiar odors. In contrast, terms with high OSI have high inter-individual consistency in how they are applied to odors. Finally, we analyze Dravnieks's (1985) dataset of odor ratings in terms of OAI and OSI. This analysis reveals that terms that are used broadly (applied often but with moderate ratings) tend to be olfaction-unrelated and abstract (e.g., \\u201cheavy-\\u009d or \\u201clight-\\u009d; low OAI and low OSI) while descriptors that are used selectively (applied seldom but with high ratings) tend to be olfaction-related (e.g., \\u201cvanilla-\\u009d or \\u201clicorice-\\u009d; high OAI). Thus, OAI and OSI provide behaviorally meaningful information about olfactory language. These statistical tools are useful for future studies of olfactory perception and cognition, and might help integrate research on odor perception, neuroimaging, and corpus-based linguistic models of semantic organization.\", \"Working memory is thought to result from sustained neuron spiking. However, computational models suggest complex dynamics with discrete oscillatory bursts. We analyzed local field potential (LFP) and spiking from the prefrontal cortex (PFC) of monkeys performing a working memory task. There were brief bursts of narrow-band gamma oscillations (45-100 Hz), varied in time and frequency, accompanying encoding and re-activation of sensory information. They appeared at a minority of recording sites associated with spiking reflecting the to-be-remembered items. Beta oscillations (20-35 Hz) also occurred in brief, variable bursts but reflected a default state interrupted by encoding and decoding. Only activity of neurons reflecting encoding/decoding correlated with changes in gamma burst rate. Thus, gamma bursts could gate access to, and prevent sensory interference with, working memory. This supports the hypothesis that working memory is manifested by discrete oscillatory dynamics and spiking, not sustained activity.\", \"One of the urgent challenges in the automated analysis and interpretation of electrical brain activity is the effective handling of uncertainties associated with the complexity and variability of brain dynamics, reflected in the nonstationary nature of brain signals such as electroencephalogram (EEG). This poses a severe problem for existing approaches to the classification task within brain-computer interface (BCI) systems. Recently emerged type-2 fuzzy logic (T2FL) methodology has shown a remarkable potential in dealing with uncertain information given limited insight into the nature of the data-generating mechanism. The objective of this work is, thus, to examine the applicability of the T2FL approach to the problem of EEG pattern recognition. In particular, the focus is two-fold: 1) the design methodology for the interval T2FL system (IT2FLS) that can robustly deal with inter-session as well as within-session manifestations of nonstationary spectral EEG correlates of motor imagery, and 2) the comprehensive examination of the proposed fuzzy classifier in both off-line and on-line EEG classification case studies. The on-line evaluation of the IT2FLS-controlled real-time neurofeedback over multiple recording sessions holds special importance for EEG-based BCI technology. In addition, a retrospective comparative analysis accounting for other popular BCI classifiers such as linear discriminant analysis, kernel Fisher discriminant, and support vector machines as well as a conventional type-1 FLS, simulated off-line on the recorded EEGs, has demonstrated the enhanced potential of the proposed IT2FLS approach to robustly handle uncertainty effects in BCI classification.\", \"Working memory (WM) activity is not as stationary or sustained as previously thought. There are brief bursts of gamma (similar to 50-120 Hz) and beta (similar to 20-35 Hz) oscillations, the former linked to stimulus information in spiking. We examined these dynamics in relation to readout and control mechanisms of WM. Monkeys held sequences of two objects in WM to match to subsequent sequences. Changes in beta and gamma bursting suggested their distinct roles. In anticipation of having to use an object for the match decision, there was an increase in gamma and spiking information about that object and reduced beta bursting. This readout signal was only seen before relevant test objects, and was related to premotor activity. When the objects were no longer needed, beta increased and gamma decreased together with object spiking information. Deviations from these dynamics predicted behavioral errors. Thus, beta could regulate gamma and the information in WM.\", \"Persistent spiking has been thought to underlie working memory (WM). However, virtually all of the evidence for this comes from studies that averaged spiking across time and across trials, which masks the details. On single trials, activity often occurs in sparse transient bursts. This has important computational and functional advantages. In addition, examination of more complex tasks reveals neural coding in WM is dynamic over the course of a trial. All this suggests that spiking is important for WM, but that its role is more complex than simply persistent spiking.\"]}\n"
],
[
"author = df.Author.values\nlist_of_author= list()\nfor i, a in enumerate(author):\n a_dict = dict()\n a_dict[\"id\"]= i\n a_dict[\"name\"]= a\n list_of_author.append(a_dict)\n",
"_____no_output_____"
],
[
"with open('assets/finalproduct/list_of_author.json', 'w') as fp:\n json.dump(list_of_author, fp)",
"_____no_output_____"
],
[
"nan_ix = [isinstance(i,float) for i in df.Department.values]\ndf.Department[nan_ix] = \"NaN\"\ndepartment = list(set(df.Department.values))",
"_____no_output_____"
],
[
"department = [make_name_noAscii(d) for d in department]",
"_____no_output_____"
],
[
"department_to_auth= list()\nfor i, d in enumerate(department):\n author = list(df[df.Department == d].Author.values)\n a_dict = dict()\n a_dict[\"department\"]= d\n a_dict[\"name\"]= author\n department_to_auth.append(a_dict)",
"_____no_output_____"
],
[
"with open('assets/finalproduct/department_to_auth.json', 'w') as fp:\n json.dump(department_to_auth, fp)",
"_____no_output_____"
],
[
"dep_list = list()\nfor i, d in enumerate(department):\n a_dict = dict()\n a_dict[\"id\"]= i\n a_dict[\"department\"]= d\n dep_list.append(a_dict)",
"_____no_output_____"
],
[
"with open('assets/finalproduct/departments.json', 'w') as fp:\n json.dump(dep_list, fp)",
"_____no_output_____"
],
[
"kth_school_s = pd.Series(np.array(df.Department)).value_counts().sort_values(ascending=False)\nplt.figure(figsize=(35,23))\nax = sns.barplot(kth_school_s.index,kth_school_s.values)\n\nax.set_xticklabels(ax.get_xticklabels(), rotation=50, ha=\"right\",fontsize=30)\nax.set_title(\"KTH authors distribution(departments)\",fontsize=50)\n\nax.set_ylabel(\"Counts\",fontsize=30)\nsns.set(font_scale=3)\nplt.gcf().subplots_adjust(bottom=0.40)\n#plt.show()\nplt.savefig(\"assets/figures/articleDepartmentFinal\")",
"_____no_output_____"
],
[
"len(kth_school_s.index)",
"_____no_output_____"
],
[
"39 - 5",
"_____no_output_____"
],
[
"kth_school_s.values.sum()",
"_____no_output_____"
],
[
"1744 - 884",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4cfa202058b0250c5ae6d5f4bc87052a1e4e3b | 873,544 | ipynb | Jupyter Notebook | arctic_cruise/Arctic_data_diurnal_plotting.ipynb | riddhimap/Saildrone | c6ecbdb1ad8ba50a59b72357be01422b8f0c0d73 | [
"Apache-2.0"
] | 3 | 2019-07-08T11:55:44.000Z | 2021-10-06T15:11:18.000Z | arctic_cruise/Arctic_data_diurnal_plotting.ipynb | riddhimap/Saildrone | c6ecbdb1ad8ba50a59b72357be01422b8f0c0d73 | [
"Apache-2.0"
] | null | null | null | arctic_cruise/Arctic_data_diurnal_plotting.ipynb | riddhimap/Saildrone | c6ecbdb1ad8ba50a59b72357be01422b8f0c0d73 | [
"Apache-2.0"
] | 3 | 2020-06-08T06:29:22.000Z | 2020-06-16T15:43:46.000Z | 646.590674 | 102,224 | 0.942534 | [
[
[
"import xarray as xr\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seawater as sw\nimport cartopy.crs as ccrs # import projections\nimport cartopy.feature as cf # import features\n\nfig_dir='C:/Users/gentemann/Google Drive/f_drive/docs/projects/misst-arctic/Saildrone/'\n\nicefile='C:/Users/gentemann/Google Drive/f_drive/docs/projects/misst-arctic/Ice Present.xlsx'\n\ndata_dir = 'F:/data/cruise_data/saildrone/2019_arctic/post_mission/'\n\nadir_sbe='F:/data/cruise_data/saildrone/2019_arctic/sbe56/sd-'\n\ndata_dir_sbe_combined = 'F:/data/cruise_data/saildrone/2019_arctic/post_mission_combined_fluxes/'",
"_____no_output_____"
],
[
"ds = xr.open_mfdataset(data_dir_sbe_combined+'*.nc',combine='nested',concat_dim='trajectory').load()\nds",
"_____no_output_____"
],
[
"# calculate density at different depth\n#import seawater as sw\n# tem=sw.dens0(ds.SAL_SBE37_MEAN,ds.TEMP_SBE37_MEAN)\n# ds['density_MEAN']=xr.DataArray(tem,dims=('time'),coords={'time':ds.time})\n",
"_____no_output_____"
],
[
"#make diruanl plot\nds2=ds#.isel(trajectory=0)\nxlon=ds2.lon\ntdif=ds2.TEMP_CTD_RBR_MEAN-ds2.TEMP_SBE37_MEAN\ntime_offset_to_lmt=(xlon/360.)*24.*60\nds2['tlmt']=ds2.lon\nfor i in range(2):\n ds2['tlmt'][i,:]= ds2.time.data+time_offset_to_lmt[i,:]*np.timedelta64(1,'m')# dt.timedelta(seconds=1)",
"_____no_output_____"
],
[
"tdif=ds2.TEMP_CTD_RBR_MEAN-ds2.TEMP_SBE37_MEAN\nfig,(ax1,ax2) =plt.subplots(1,2)\nfor i in range(2):\n cs=ax1.scatter(ds2.wspd_MEAN[i,:],tdif[i,:],c=ds2.time.dt.hour,s=.5)\nax1.set(xlabel='Wind Speed (ms$^{-1}$)', ylabel='RBR - SBE4 SST (K)')\nax1.set_xlim(0,15)\ncbar = fig.colorbar(cs,orientation='horizontal',ax=ax1)\ncbar.set_label('GMT Time (hrs)')\nfor i in range(2):\n cs2=ax2.scatter(ds2.time.dt.hour,tdif[i,:],c=ds2.wspd_MEAN[i,:],s=.5)\nax2.set(xlabel='GMT (hr)')\ncbar = fig.colorbar(cs2,orientation='horizontal',ax=ax2)\ncbar.set_label('Wind Speed (ms$^{-1}$)')\nfig.savefig(fig_dir+'figs/temp_buld_dw_data.png')\n\ntdif=ds2.TEMP_CTD_RBR_MEAN-ds2.TEMP_SBE37_MEAN\nfig,(ax1,ax2) =plt.subplots(1,2)\ncs=ax1.scatter(ds2.wspd_MEAN[0,:],tdif[i,:],c=ds2.time.dt.hour,s=.5)\nax1.set(xlabel='Wind Speed (ms$^{-1}$)', ylabel='RBR - SBE4 SST (K)')\nax1.set_xlim(0,15)\ncbar = fig.colorbar(cs,orientation='horizontal',ax=ax1)\ncbar.set_label('GMT Time (hrs)')\ncs2=ax2.scatter(ds2.time.dt.hour,tdif[0,:],c=ds2.wspd_MEAN[i,:],s=.5)\nax2.set(xlabel='GMT (hr)')\ncbar = fig.colorbar(cs2,orientation='horizontal',ax=ax2)\ncbar.set_label('Wind Speed (ms$^{-1}$)')\nfig.savefig(fig_dir+'figs/temp_buld_dw_data36.png')\n\ntdif=ds2.TEMP_CTD_RBR_MEAN-ds2.TEMP_SBE37_MEAN\nfig,(ax1,ax2) =plt.subplots(1,2)\ncs=ax1.scatter(ds2.wspd_MEAN[1,:],tdif[i,:],c=ds2.time.dt.hour,s=.5)\nax1.set(xlabel='Wind Speed (ms$^{-1}$)', ylabel='RBR - SBE4 SST (K)')\nax1.set_xlim(0,15)\ncbar = fig.colorbar(cs,orientation='horizontal',ax=ax1)\ncbar.set_label('GMT Time (hrs)')\ncs2=ax2.scatter(ds2.time.dt.hour,tdif[1,:],c=ds2.wspd_MEAN[i,:],s=.5)\nax2.set(xlabel='GMT (hr)')\ncbar = fig.colorbar(cs2,orientation='horizontal',ax=ax2)\ncbar.set_label('Wind Speed (ms$^{-1}$)')\nfig.savefig(fig_dir+'figs/temp_buld_dw_data37.png')",
"_____no_output_____"
],
[
"tdif=ds2.TEMP_CTD_RBR_MEAN-ds2.sea_water_temperature_01_mean\nfig,(ax1,ax2) =plt.subplots(1,2)\nfor i in range(2):\n cs=ax1.scatter(ds2.wspd_MEAN[i,:],tdif[i,:],c=ds2.time.dt.hour,s=.5)\nax1.set(xlabel='Wind Speed (ms$^{-1}$)', ylabel='RBR - SBE4 SST (K)')\nax1.set_xlim(0,15)\ncbar = fig.colorbar(cs,orientation='horizontal',ax=ax1)\ncbar.set_label('GMT Time (hrs)')\nfor i in range(2):\n cs2=ax2.scatter(ds2.time.dt.hour,tdif[i,:],c=ds2.wspd_MEAN[i,:],s=.5)\nax2.set(xlabel='GMT (hr)')\ncbar = fig.colorbar(cs2,orientation='horizontal',ax=ax2)\ncbar.set_label('Wind Speed (ms$^{-1}$)')\nfig.savefig(fig_dir+'figs/temp_rbr-sbe-buld_dw_data36.png')\n",
"_____no_output_____"
],
[
"tdif=ds2.TEMP_SBE37_MEAN-ds2.sea_water_temperature_01_mean\nfig,(ax1,ax2) =plt.subplots(1,2)\nfor i in range(2):\n cs=ax1.scatter(ds2.wspd_MEAN[i,:],tdif[i,:],c=ds2.time.dt.hour,s=.5)\nax1.set(xlabel='Wind Speed (ms$^{-1}$)', ylabel='SBE37 - SBE1 SST (K)')\nax1.set_xlim(0,15)\ncbar = fig.colorbar(cs,orientation='horizontal',ax=ax1)\ncbar.set_label('GMT Time (hrs)')\nfor i in range(2):\n cs2=ax2.scatter(ds2.time.dt.hour,tdif[i,:],c=ds2.wspd_MEAN[i,:],s=.5)\nax2.set(xlabel='GMT (hr)')\ncbar = fig.colorbar(cs2,orientation='horizontal',ax=ax2)\ncbar.set_label('Wind Speed (ms$^{-1}$)')\nfig.savefig(fig_dir+'figs/temp_sbe-sbe-buld_dw_data.png')",
"_____no_output_____"
],
[
"plt.scatter(ds2.wspd_MEAN,ds2.sea_water_temperature_01_std)",
"_____no_output_____"
],
[
"plt.scatter(ds2.wspd_MEAN,ds2.TEMP_SBE37_STDDEV)",
"_____no_output_____"
],
[
"#ICE VERIFIED FROM CAMERA\nt1='2019-06-22T14'\nt2='2019-06-23T00'\n#(ds2.sea_water_temperature_00_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s00')\n#(ds2.sea_water_temperature_01_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s01')\n(ds2.TEMP_SBE37_MEAN[0,:]-ds2.sea_water_temperature_01_mean[0,:]).sel(time=slice(t1,t2)).plot(label='sbe')\n#(ds2.wspd_MEAN[0,:]).sel(time=slice(t1,t2)).plot(label='sbe')\nplt.legend()",
"_____no_output_____"
],
[
"#Differences due to strong gradients in area and maybe shallow fresh layer\n#surface is COOLER than at depth\n#salinity drops significantly\n#deeper temperatures warmer from sbe56 05 as compared to sbe01\nt1='2019-07-17T00'\nt2='2019-07-18T00'\n#(ds2.sea_water_temperature_00_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s00')\n#(ds2.sea_water_temperature_01_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s01')\n(ds2.TEMP_SBE37_MEAN[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='sbe')\n(ds2.sea_water_temperature_00_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='sbe00')\n(ds2.sea_water_temperature_05_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='sbe05')\n(ds2.SAL_SBE37_MEAN[0,:]-28).sel(time=slice(t1,t2)).plot(label='salinity')\n#(ds2.wspd_MEAN[0,:]).sel(time=slice(t1,t2)).plot(label='sbe')\nplt.legend()",
"_____no_output_____"
],
[
"#Differences due to strong gradients in area and maybe shallow fresh layer\n#surface is COOLER than at depth\n#salinity drops significantly\n#deeper temperatures warmer from sbe56 05 as compared to sbe01\nimport seawater as sw\ntem=sw.dens0(ds.SAL_SBE37_MEAN,ds.sea_water_temperature_00_mean)\nds['density_MEAN_00']=xr.DataArray(tem,dims=('trajectory','time'),coords={'trajetcory':ds.trajectory,'time':ds.time})\ntem=sw.dens0(ds.SAL_SBE37_MEAN,ds.sea_water_temperature_01_mean)\nds['density_MEAN_01']=xr.DataArray(tem,dims=('trajectory','time'),coords={'trajetcory':ds.trajectory,'time':ds.time})\ntem=sw.dens0(ds.SAL_SBE37_MEAN,ds.sea_water_temperature_02_mean)\nds['density_MEAN_02']=xr.DataArray(tem,dims=('trajectory','time'),coords={'trajetcory':ds.trajectory,'time':ds.time})\ntem=sw.dens0(ds.SAL_SBE37_MEAN,ds.sea_water_temperature_04_mean)\nds['density_MEAN_04']=xr.DataArray(tem,dims=('trajectory','time'),coords={'trajetcory':ds.trajectory,'time':ds.time})\ntem=sw.dens0(ds.SAL_SBE37_MEAN,ds.sea_water_temperature_05_mean)\nds['density_MEAN_05']=xr.DataArray(tem,dims=('trajectory','time'),coords={'trajetcory':ds.trajectory,'time':ds.time})\ntem=sw.dens0(ds.SAL_SBE37_MEAN,ds.sea_water_temperature_06_mean)\nds['density_MEAN_06']=xr.DataArray(tem,dims=('trajectory','time'),coords={'trajetcory':ds.trajectory,'time':ds.time})\n\nt1='2019-07-17T00'\nt2='2019-07-18T00'\n#(ds2.sea_water_temperature_00_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s00')\n#(ds2.sea_water_temperature_01_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s01')\n(ds.density_MEAN[0,:]-ds.density_MEAN_06[0,:]).sel(time=slice(t1,t2)).plot(label='den')\n(ds.density_MEAN_00[0,:]-ds.density_MEAN_06[0,:]).sel(time=slice(t1,t2)).plot(label='den')\n(ds.density_MEAN_02[0,:]-ds.density_MEAN_06[0,:]).sel(time=slice(t1,t2)).plot(label='den')\n(ds.density_MEAN_04[0,:]-ds.density_MEAN_06[0,:]).sel(time=slice(t1,t2)).plot(label='den')\n(ds.density_MEAN_05[0,:]-ds.density_MEAN_06[0,:]).sel(time=slice(t1,t2)).plot(label='den')\n#(ds2.wspd_MEAN[0,:]).sel(time=slice(t1,t2)).plot(label='sbe')\nplt.legend()",
"_____no_output_____"
],
[
"t1='2019-10-01'\nt2='2019-10-11'\n(ds.density_MEAN[0,:]-ds.density_MEAN_06[0,:]).sel(time=slice(t1,t2)).plot(label='den')\n",
"_____no_output_____"
],
[
"t1='2019-07-04T18'\nt2='2019-07-05'\n(ds.sea_water_temperature_00_mean[0,:]-ds.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='den')\n#(ds.sea_water_temperature_05_mean[0,:]-ds.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='den')\n(ds.SAL_SBE37_MEAN[0,:]).sel(time=slice(t1,t2)).plot(label='den')\n#(ds.sea_water_temperature_05_mean[0,:]-ds.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='den')\n",
"_____no_output_____"
],
[
"#tdif=ds2.TEMP_SBE37_MEAN-ds2.sea_water_temperature_01_mean\nt1='2019-07-10T00'\nt2='2019-07-12T00'\n#(ds2.TEMP_AIR_MEAN[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='air')\n(ds2.sea_water_temperature_00_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s00')\n(ds2.sea_water_temperature_01_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s01')\n(ds2.TEMP_SBE37_MEAN[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='sbe')\n(ds2.sea_water_temperature_02_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s02')\n(ds2.sea_water_temperature_04_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s04')\nplt.legend()",
"_____no_output_____"
],
[
"(ds2.sea_water_temperature_04_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s04')\nplt.legend()",
"_____no_output_____"
],
[
"#tdif=ds2.TEMP_SBE37_MEAN-ds2.sea_water_temperature_01_mean\nt1='2019-07-08T18'\nt2='2019-07-10T00'\n(ds2.TEMP_AIR_MEAN[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='air')\n(ds2.sea_water_temperature_00_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s00')\n(ds2.sea_water_temperature_01_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s01')\n(ds2.TEMP_SBE37_MEAN[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='sbe')\n(ds2.sea_water_temperature_02_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s02')\n(ds2.sea_water_temperature_04_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s04')\nplt.legend()",
"_____no_output_____"
],
[
"#tdif=ds2.TEMP_SBE37_MEAN-ds2.sea_water_temperature_01_mean\nt1='2019-06-28T12'\nt2='2019-06-29T12'\n(ds2.TEMP_AIR_MEAN[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='air')\n(ds2.sea_water_temperature_00_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s00')\n(ds2.sea_water_temperature_01_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s01')\n(ds2.TEMP_SBE37_MEAN[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='sbe')\n(ds2.sea_water_temperature_02_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s02')\n(ds2.sea_water_temperature_04_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s04')\nplt.legend()",
"_____no_output_____"
],
[
"#tdif=ds2.TEMP_SBE37_MEAN-ds2.sea_water_temperature_01_mean\nt1='2019-06-05T18'\nt2='2019-06-06T05'\n(ds2.TEMP_AIR_MEAN[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='air')\n(ds2.sea_water_temperature_00_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s00')\n(ds2.sea_water_temperature_01_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s01')\n(ds2.TEMP_SBE37_MEAN[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='sbe')\n(ds2.sea_water_temperature_02_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s02')\n(ds2.sea_water_temperature_04_mean[0,:]-ds2.sea_water_temperature_06_mean[0,:]).sel(time=slice(t1,t2)).plot(label='s04')\nplt.legend()",
"_____no_output_____"
],
[
"tdif=ds.TEMP_SBE37_MEAN-ds.sea_water_temperature_00_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_CTD_RBR_MEAN-ds.sea_water_temperature_00_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_O2_RBR_MEAN-ds.sea_water_temperature_00_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)",
"[-0.01366711 -0.01452271] [0.12878561 0.1653436 ] [185060 198527]\n[-0.01363566 -0.01360196] [0.1452122 0.1905742] [187927 209898]\n[-0.00636237 0.00449801] [0.14480192 0.20057166] [187927 209898]\n"
],
[
"tdif=ds.TEMP_SBE37_MEAN-ds.sea_water_temperature_01_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_CTD_RBR_MEAN-ds.sea_water_temperature_01_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_O2_RBR_MEAN-ds.sea_water_temperature_01_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)",
"[-0.00592364 -0.00498099] [0.06595808 0.08145148] [185060 195642]\n[-0.00566546 -0.00381481] [0.07661844 0.10478591] [187927 209511]\n[0.00160783 0.01431352] [0.07614338 0.13102051] [187927 209511]\n"
],
[
"tdif=ds.TEMP_SBE37_MEAN-ds.sea_water_temperature_02_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_CTD_RBR_MEAN-ds.sea_water_temperature_02_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_O2_RBR_MEAN-ds.sea_water_temperature_02_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)",
"[0.01263407 0.00796006] [0.11068902 0.12335199] [185060 198527]\n[0.01304667 0.00900744] [0.09861127 0.10762502] [187927 209898]\n[0.02031996 0.02710741] [0.1043882 0.13512464] [187927 209898]\n"
],
[
"tdif=ds.TEMP_SBE37_MEAN-ds.sea_water_temperature_04_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_CTD_RBR_MEAN-ds.sea_water_temperature_04_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_O2_RBR_MEAN-ds.sea_water_temperature_04_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)",
"[0.0322524 0.00376437] [0.21888528 0.9619031 ] [185060 198527]\n[0.03276717 0.01047033] [0.2085751 0.96804988] [187927 209898]\n[0.04004046 0.0285703 ] [0.213239 0.96661893] [187927 209898]\n"
],
[
"tdif=ds.TEMP_SBE37_MEAN-ds.sea_water_temperature_05_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_CTD_RBR_MEAN-ds.sea_water_temperature_05_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_O2_RBR_MEAN-ds.sea_water_temperature_05_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)",
"[0.04187074 0.01061783] [0.25977723 0.96468803] [185060 198527]\n[0.04247323 0.01743482] [0.25020223 0.9689725 ] [187927 209898]\n[0.04974652 0.03553479] [0.25466871 0.96649315] [187927 209898]\n"
],
[
"tdif=ds.TEMP_SBE37_MEAN-ds.sea_water_temperature_06_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_CTD_RBR_MEAN-ds.sea_water_temperature_06_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)\ntdif=ds.TEMP_O2_RBR_MEAN-ds.sea_water_temperature_06_mean\nprint(tdif.mean('time').data,tdif.std('time').data,(np.isfinite(tdif)).sum('time').data)",
"[0.0526104 0.01798243] [0.29708388 0.96916961] [185060 198527]\n[0.05338097 0.02471025] [0.28785749 0.97156485] [187927 209898]\n[0.06065426 0.04281021] [0.29206886 0.96802942] [187927 209898]\n"
]
],
[
[
"# PLOT DIURANL WARMING",
"_____no_output_____"
]
],
[
[
"ds10=ds.isel(trajectory=0).resample(time='10min').mean()",
"_____no_output_____"
],
[
"plt.figure(figsize=(12,6))\nsubset=ds10.sel(time=slice('2019-06-15T08','2019-06-16'))\nfor i in range(2):\n var='sea_water_temperature_'+str(i).zfill(2)+'_mean'\n lvar=str(i).zfill(2)\n plt.plot(subset.time,subset[var]-subset.sea_water_temperature_06_mean,label=lvar,lw=3)\nvar='TEMP_SBE37_MEAN'\nlvar='SBE37'\nplt.plot(subset.time,subset[var]-subset.sea_water_temperature_06_mean,label=lvar,lw=3)\nfor i in range(2,7):\n var='sea_water_temperature_'+str(i).zfill(2)+'_mean'\n lvar=str(i).zfill(2)\n if i==3:\n continue\n plt.plot(subset.time,subset[var]-subset.sea_water_temperature_06_mean,label=lvar,lw=3)\nplt.legend()\nplt.ylabel('$\\Delta$ T (K)')\nplt.xlabel('Time (GMT)')\nplt.savefig(fig_dir+'figs/diurnal36_06-15.png')\n",
"_____no_output_____"
],
[
"plt.figure(figsize=(12,6))\nplt.plot(subset.time,subset.TEMP_AIR_MEAN-subset.sea_water_temperature_00_mean,label=lvar)\n",
"_____no_output_____"
],
[
"plt.figure(figsize=(12,6))\nsubset=ds10.sel(time=slice('2019-07-08T12','2019-07-10T12'))\nfor i in range(2):\n var='sea_water_temperature_'+str(i).zfill(2)+'_mean'\n lvar=str(i).zfill(2)\n plt.plot(subset.time,subset[var]-subset.sea_water_temperature_06_mean,label=lvar,lw=3)\nvar='TEMP_SBE37_MEAN'\nlvar='SBE37'\nplt.plot(subset.time,subset[var]-subset.sea_water_temperature_06_mean,label=lvar,lw=3)\nfor i in range(2,7):\n var='sea_water_temperature_'+str(i).zfill(2)+'_mean'\n lvar=str(i).zfill(2)\n if i==3:\n continue\n plt.plot(subset.time,subset[var]-subset.sea_water_temperature_06_mean,label=lvar,lw=3)\nplt.legend()\nplt.ylabel('$\\Delta$ T (K)')\nplt.xlabel('Time (GMT)')\nplt.savefig(fig_dir+'figs/diurnal36_07-08.png')\n",
"_____no_output_____"
],
[
"plt.figure(figsize=(12,6))\nplt.plot(subset.time,subset.TEMP_AIR_MEAN-subset.sea_water_temperature_00_mean,label=lvar)\n",
"_____no_output_____"
],
[
"plt.figure(figsize=(12,6))\nsubset=ds10.sel(time=slice('2019-05-15T12','2019-09-10T12'))\nplt.plot(subset.time,subset.TEMP_AIR_MEAN-subset.sea_water_temperature_00_mean,label='$\\Delta$T$_{air-sea}$')\nplt.plot(subset.time,subset.sea_water_temperature_00_mean-subset.sea_water_temperature_06_mean,label='$\\Delta$T$_{dw}$')\nplt.legend()\nplt.ylabel('$\\Delta$ T (K)')\nplt.xlabel('Time (GMT)')\nplt.savefig(fig_dir+'figs/diurnal36_airseatemp.png')\n",
"_____no_output_____"
],
[
"subset=ds.sel(time=slice('2019-07-07','2019-07-11'))\ntdif=subset.sea_water_temperature_00_mean-subset.sea_water_temperature_06_mean\ntdif[0,:].plot()",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(8,15))\nax = plt.axes(projection = ccrs.NorthPolarStereo(central_longitude=180.0)) # create a set of axes with Mercator projection\nfor i in range(1):\n ds2 = ds.isel(trajectory=i).sel(time=slice('2019-05-01','2019-09-15'))\n im2=ax.quiver(ds2.lon[::200].data,\n ds2.lat[::200].data,\n ds2.UWND_MEAN[::200].data,\n ds2.VWND_MEAN[::200].data,\n scale=140,transform=ccrs.PlateCarree())\n im=ax.scatter(ds2.lon,ds2.lat,\n c=ds2.TEMP_AIR_MEAN-ds2.sea_water_temperature_00_mean,\n s=.15,transform=ccrs.PlateCarree(),label=ds.trajectory[i].data,\n cmap='seismic',vmin=-2,vmax=2)\n ax.coastlines(resolution='10m') \n ax.set_extent([-180,-158,68,77])\n ax.legend()\n cax = fig.add_axes([0.45, 0.17, 0.3, 0.02])\n cbar = fig.colorbar(im,cax=cax, orientation='horizontal')\n cbar.set_label('SST ($^\\deg$C)')\nfig.savefig(fig_dir+'figs/map_nasa_data_air-sbe5600.png')",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(8,15))\nax = plt.axes(projection = ccrs.NorthPolarStereo(central_longitude=180.0)) # create a set of axes with Mercator projection\nfor i in range(1):\n ds2 = ds.isel(trajectory=i).sel(time=slice('2019-06-15','2019-06-16'))\n im2=ax.quiver(ds2.lon[::100].data,\n ds2.lat[::100].data,\n ds2.UWND_MEAN[::100].data,\n ds2.VWND_MEAN[::100].data,\n scale=20,transform=ccrs.PlateCarree())\n im=ax.scatter(ds2.lon,ds2.lat,\n c=ds2.TEMP_AIR_MEAN-ds2.sea_water_temperature_00_mean,\n s=.15,transform=ccrs.PlateCarree(),label=ds.trajectory[i].data,\n cmap='seismic',vmin=-2,vmax=2)\n ax.coastlines(resolution='10m') \n ax.set_extent([-175,-158,68,72])\n ax.legend()\n cax = fig.add_axes([0.45, 0.17, 0.3, 0.02])\n cbar = fig.colorbar(im,cax=cax, orientation='horizontal')\n cbar.set_label('SST ($^\\deg$C)')\nfig.savefig(fig_dir+'figs/map_nasa_data_air-sbe5600-06-15.png')",
"_____no_output_____"
],
[
"ig = plt.figure(figsize=(8,15))\nax = plt.axes(projection = ccrs.NorthPolarStereo(central_longitude=180.0)) # create a set of axes with Mercator projection\nfor i in range(1):\n ds2 = ds.isel(trajectory=i).sel(time=slice('2019-07-08','2019-07-10'))\n im2=ax.quiver(ds2.lon[::100].data,\n ds2.lat[::100].data,\n ds2.UWND_MEAN[::100].data,\n ds2.VWND_MEAN[::100].data,\n scale=100,transform=ccrs.PlateCarree())\n im=ax.scatter(ds2.lon,ds2.lat,\n c=ds2.sea_water_temperature_00_mean-ds2.sea_water_temperature_06_mean,\n s=.15,transform=ccrs.PlateCarree(),label=ds.trajectory[i].data,\n cmap='seismic',vmin=-2,vmax=2)\n ax.coastlines(resolution='10m') \n ax.set_extent([-173,-160,70,71])\n ax.legend()\n cax = fig.add_axes([0.45, 0.17, 0.3, 0.02])\n cbar = fig.colorbar(im,cax=cax, orientation='horizontal')\n cbar.set_label('SST ($^\\deg$C)')\nfig.savefig(fig_dir+'figs/map_nasa_data_air-sbe5600-07-10.png')",
"_____no_output_____"
],
[
"plt.quiver(ds2.lon[::100].data,\n ds2.lat[::100].data,\n ds2.UWND_MEAN[::100].data,\n ds2.VWND_MEAN[::100].data,\n scale=50)\nplt.scatter(ds2.lon,ds2.lat,\n c=ds2.sea_water_temperature_00_mean-ds2.sea_water_temperature_06_mean,\n s=.15,\n cmap='seismic',vmin=-2,vmax=2)\n",
"_____no_output_____"
],
[
"%matplotlib inline\nimport sys\nsys.path.append('./../../flux/')\nfrom coare3 import coare3\n",
"_____no_output_____"
],
[
"coare3",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4d21793336c6600f0ea79f71b3f638598b8bb4 | 220,957 | ipynb | Jupyter Notebook | src/puzzle/examples/msph/2018/the major.ipynb | PhilHarnish/forge | 663f19d759b94d84935c14915922070635a4af65 | [
"MIT"
] | 2 | 2020-08-18T18:43:09.000Z | 2020-08-18T20:05:59.000Z | src/puzzle/examples/msph/2018/the major.ipynb | PhilHarnish/forge | 663f19d759b94d84935c14915922070635a4af65 | [
"MIT"
] | null | null | null | src/puzzle/examples/msph/2018/the major.ipynb | PhilHarnish/forge | 663f19d759b94d84935c14915922070635a4af65 | [
"MIT"
] | null | null | null | 22.985228 | 91 | 0.479709 | [
[
[
"tiles = \"\"\"\nALS\nANK\nAPP\nATS\nBUR\nCAR\nCDR\nCIE\nDEV\nDIN\nEES\nELS\nERS\nFIE\nFLY\nFMA\nGHI\nGHM\nHLD\nHON\nHOU\nILS\nING\nING\nING\nIRR\nKIY\nLAN\nLAS\nLEM\nLEY\nLLC\nLYA\nMID\nNCH\nNDS\nOCK\nOND\nPUD\nRED\nRIC\nSBA\nSCR\nSOX\nSPR\nSQU\nTRI\nTYD\nUST\nVAL\n\"\"\".lower().split()",
"_____no_output_____"
],
[
"import forge\nfrom data import warehouse\nfrom puzzle.puzzlepedia import prod_config\nprod_config.init()\n\ntrie = warehouse.get('/words/unigram/trie')",
"_____no_output_____"
],
[
"import re\nfrom data.seek_sets import chain_seek_set",
"_____no_output_____"
],
[
"def walk(seek_set, acc, targets, pos=0):\n if pos >= len(targets):\n yield ' '.join(acc)\n return\n if targets:\n target = targets[pos]\n seek_set.set_length(target)\n for result, weight in trie.walk(seek_set, exact_match=False):\n if weight < 5e4:\n break\n acc.append(result)\n yield from walk(seek_set[result:], acc, targets, pos+1)\n acc.pop()\n\ndef process(tiles, targets):\n seek_set = chain_seek_set.ChainSeekSet(tiles, sum(targets))\n for result in walk(seek_set, [], targets):\n print(result)\n\ndef parse(s):\n parts = s.split(' ')\n result = []\n for p in parts:\n p = p.strip('’,;.‘^!-*')\n if p:\n result.append(int(p))\n return result",
"_____no_output_____"
],
[
"digits = parse(\"9\")\nprint(digits)\nprocess(tiles, digits)",
"[9]\ncardinals\nsquirrels\nspringing\nscrapping\nscrappers\nspringers\n"
],
[
"given = \"\"\"\nAK\tAKE\tARH\tAYI\tBE\tDA\tDO\tEA\t\tEI\tES\tETA\tETH\t\tFUS\t\tGR\tHEW\nHME\t\tIN\t\t\tLES\tLI\tMOL\tNB\tNEO\tNGO\tNIN\tOLE\t\tPA\tPAN\t\t\nPRA\t\tRC\tRIN\tRMY\tRNO\tSED\t\tSTA\tTAR\t\tTYP\tUSO\tUYT\tWIM\tWIT\t\t\n\"\"\".lower().split()",
"_____no_output_____"
],
[
"process(given, [11, 14])",
"a king\na kind\na kinda\na rhino\na klingon\na keane\na rhesus\na rhine\na kline\na keine\na khmer\na kelis\na kedar\na rhein\na kling\na kepada\na kernow\nin a\nin with\nin be\nin us\nin do\nin he\nin list\nin data\nin best\nin great\nin line\nin type\nin less\nin east\nin star\nin past\nin dog\nin bed\nin ring\nin bear\nin eat\nin nine\nin dot\nin pan\nin gray\nin earn\nin types\nin dose\nin bet\nin dad\nin pat\nin ear\nin ease\nin lie\nin pad\nin dam\nin bee\nin lip\nin leslie\nin pasta\nin lid\nin pale\nin lining\nin pant\nin grease\nin espanol\nin beg\nin staring\nin daring\nin starch\nin stalin\nin benin\nin espana\nin ealing\nin panty\nin panda\nin prada\nin bering\nin lesben\nin rinse\nin greased\nin mollie\nin witty\nin beaker\nin rinsed\nin pandas\nin bestality\nin panning\nin lipase\nin pastas\nin earch\nin dorint\nin paring\nin liberi\nin lista\nin typeset\nin doesn\nin stang\nin stale\nin lingo\nin esearch\nin fuses\nin beeing\nin libel\nin bening\nin darin\nin belies\nin greta\nin eased\nin dales\nin seddon\nin prado\nin molar\nin moles\nin does\nin eidos\nin sedaka\nin beaked\nin molest\nin praline\nin stata\nin estar\nin darcs\nin graying\nin bestar\nin panes\nin dahmer\nin listas\nin lidar\nin liber\nin dopant\nin witless\nin pango\nin padang\nin earing\nin espada\nin lesbe\nin dangos\nin listado\nin mollis\nin paling\nin bedale\nin pandan\nin fussed\nin listar\nin lipari\nin doling\nin palin\nin parco\nin typeid\nin parche\nin nines\nin done\nwith me\nwith meet\nwith medal\nwith metart\nwith merino\nwith medalist\nwith ewing\nwith melia\nwith meakin\nwith metar\nwith merch\nwith merci\nwith meine\nwith menino\nwith menina\nwith melita\nwith meines\ni no\ni net\ni near\ni naked\ni nest\ni neat\ni nearing\ni neale\ni nline\nbe a\nbe in\nbe with\nbe i\nbe us\nbe do\nbe he\nbe list\nbe data\nbe great\nbe line\nbe type\nbe less\nbe east\nbe star\nbe past\nbe dog\nbe ring\nbe pain\nbe eat\nbe nine\nbe dot\nbe paint\nbe pan\nbe gray\nbe earn\nbe doing\nbe types\nbe stainless\nbe dose\nbe dad\nbe pat\nbe ear\nbe ease\nbe lie\nbe pad\nbe dam\nbe inline\nbe lip\nbe typing\nbe leslie\nbe pasta\nbe lid\nbe pale\nbe lining\nbe staind\nbe pant\nbe grease\nbe espanol\nbe staring\nbe inning\nbe grin\nbe stains\nbe daring\nbe starch\nbe stalin\nbe espana\nbe ealing\nbe painless\nbe panty\nbe stain\nbe panda\nbe molina\nbe moline\nbe pains\nbe grind\nbe ingress\nbe prada\nbe rinse\nbe greased\nbe mollie\nbe dainty\nbe witty\nbe fusing\nbe rinsed\nbe pandas\nbe panning\nbe lipase\nbe staines\nbe pastas\nbe earch\nbe dorint\nbe paring\nbe panini\nbe ingres\nbe infuse\nbe lista\nbe typeset\nbe doesn\nbe paine\nbe stang\nbe stale\nbe lingo\nbe esearch\nbe fuses\nbe instal\nbe inlining\nbe darin\nbe greta\nbe inest\nbe eased\nbe indole\nbe molino\nbe dales\nbe seddon\nbe prado\nbe molar\nbe moles\nbe does\nbe instar\nbe eidos\nbe sedaka\nbe molest\nbe praline\nbe dainese\nbe stata\nbe estar\nbe darcs\nbe graying\nbe panes\nbe dahmer\nbe grins\nbe infuses\nbe listas\nbe lidar\nbe dopant\nbe witless\nbe pango\nbe padang\nbe earing\nbe espada\nbe insta\nbe dangos\nbe listado\nbe mollis\nbe paling\nbe pandan\nbe indoles\nbe fussed\nbe tarina\nbe listar\nbe lipari\nbe doling\nbe palin\nbe parco\nbe typeid\nbe daines\nbe parche\nbe nines\nbe done\nus of\nus on\nus or\nus one\nus oh\nus oak\nus ongoing\nus oakdale\nus oprah\nus obese\nus orinda\nus oakes\nus oline\ndo a\ndo in\ndo with\ndo i\ndo be\ndo us\ndo he\ndo list\ndo data\ndo best\ndo great\ndo line\ndo type\ndo being\ndo less\ndo east\ndo star\ndo past\ndo bed\ndo ring\ndo pain\ndo bear\ndo eat\ndo nine\ndo paint\ndo pan\ndo gray\ndo earn\ndo types\ndo stainless\ndo bet\ndo pat\ndo ear\ndo ease\ndo lie\ndo pad\ndo dam\ndo bee\ndo inline\ndo lip\ndo typing\ndo leslie\ndo pasta\ndo lid\ndo pale\ndo lining\ndo staind\ndo pant\ndo grease\ndo espanol\ndo beg\ndo staring\ndo inning\ndo grin\ndo stains\ndo daring\ndo starch\ndo stalin\ndo benin\ndo espana\ndo ealing\ndo painless\ndo panty\ndo stain\ndo panda\ndo molina\ndo moline\ndo pains\ndo grind\ndo ingress\ndo prada\ndo bering\ndo lesben\ndo rinse\ndo greased\ndo mollie\ndo dainty\ndo witty\ndo fusing\ndo beaker\ndo rinsed\ndo pandas\ndo bestality\ndo panning\ndo lipase\ndo staines\ndo pastas\ndo earch\ndo paring\ndo panini\ndo liberi\ndo ingres\ndo infuse\ndo lista\ndo typeset\ndo paine\ndo stang\ndo stale\ndo lingo\ndo esearch\ndo fuses\ndo beeing\ndo instal\ndo libel\ndo bening\ndo inlining\ndo darin\ndo belies\ndo greta\ndo inest\ndo eased\ndo molino\ndo dales\ndo molar\ndo moles\ndo instar\ndo sedaka\ndo beaked\ndo molest\ndo praline\ndo dainese\ndo stata\ndo estar\ndo darcs\ndo graying\ndo bestar\ndo panes\ndo dahmer\ndo grins\ndo infuses\ndo listas\ndo lidar\ndo liber\ndo witless\ndo pango\ndo padang\ndo earing\ndo espada\ndo lesbe\ndo insta\ndo dangos\ndo mollis\ndo paling\ndo bedale\ndo pandan\ndo fussed\ndo tarina\ndo listar\ndo lipari\ndo palin\ndo parco\ndo typeid\ndo daines\ndo parche\ndo nines\nhe we\nhe way\nhe west\nhe war\nhe window\nhe win\nhe wine\nhe wind\nhe winning\nhe wear\nhe wet\nhe wing\nhe wake\nhe wearing\nhe waking\nhe wines\nhe warhol\nhe weaning\nhe warhead\nhe weariness\nhe wakes\nhe winstar\nhe windom\nhe winless\nhe weiber\nhe wearch\nhe winline\nhe westar\nhe wearin\nhe weibel\nhe wakeling\nhe wakelin\nhe waker\nlist a\nlist at\nlist as\nlist am\nlist ad\nlist army\nlist arm\nlist ah\nlist adobe\nlist atari\nlist ahmed\nlist aarhus\nlist adoring\nlist alesse\nlist adorno\nlist arcing\nlist adorn\nlist aearch\nlist adamo\nlist agreing\nlist ahmet\nlist arche\nlist alesina\ndata re\ndata read\ndata research\ndata real\ndata role\ndata ring\ndata rest\ndata reality\ndata rear\ndata retain\ndata realise\ndata roles\ndata realised\ndata reset\ndata retains\ndata resins\ndata resin\ndata rearing\ndata realist\ndata rinse\ndata reseal\ndata rinsed\ndata raking\ndata reining\ndata rakesh\ndata reales\ndata reise\ndata rakes\ndata reseau\ndata resear\ndata researc\ndata reine\ndata raked\ndata reale\nbest a\nbest at\nbest as\nbest am\nbest ad\nbest army\nbest arm\nbest ah\nbest atari\nbest ahmed\nbest aarhus\nbest adoring\nbest alesse\nbest adorno\nbest adaline\nbest arcing\nbest adorn\nbest aearch\nbest aline\nbest adamo\nbest agreing\nbest ahmet\nbest arche\nbest alesina\ngreat a\ngreat art\ngreat arm\ngreat aretha\ngreat arpanet\ngreat arles\ngreat arline\nline of\nline on\nline or\nline oh\nline oak\nline ongoing\nline oakdale\nline oprah\nline obese\nline orinda\nline orcinus\nline oakes\ntype the\ntype a\ntype in\ntype i\ntype it\ntype at\ntype as\ntype if\ntype search\ntype their\ntype so\ntype am\ntype these\ntype she\ntype set\ntype say\ntype star\ntype thing\ntype ad\ntype sea\ntype thus\ntype ie\ntype army\ntype saying\ntype tag\ntype seat\ntype arm\ntype sin\ntype seal\ntype thin\ntype stars\ntype ah\ntype sole\ntype spain\ntype slip\ntype sing\ntype starring\ntype tale\ntype tap\ntype swim\ntype adobe\ntype sake\ntype tales\ntype staring\ntype spanning\ntype sealing\ntype stare\ntype sparc\ntype theta\ntype atari\ntype thinning\ntype starr\ntype sprang\ntype tango\ntype ahmed\ntype aarhus\ntype stardom\ntype starling\ntype sinus\ntype sling\ntype spans\ntype sparing\ntype soledad\ntype searing\ntype sparco\ntype seabed\ntype thine\ntype sinead\ntype sprains\ntype seine\ntype seibel\ntype sprain\ntype adoring\ntype soleus\ntype slingo\ntype alesse\ntype soles\ntype adorno\ntype irina\ntype sinning\ntype tapas\ntype adaline\ntype seadoo\ntype arcing\ntype tatarstan\ntype taliesin\ntype sayin\ntype spandau\ntype sinless\ntype adorn\ntype sparcs\ntype theist\ntype sealine\ntype searcg\ntype swims\ntype shewing\ntype searcn\ntype searcb\ntype sdarch\ntype searcu\ntype taint\ntype searct\ntype searcm\ntype starline\ntype taber\ntype talib\ntype tatars\ntype tatar\ntype idabel\ntype tarina\ntype spaeth\ntype tangos\ntype alist\ntype aline\ntype swiming\ntype seale\ntype searc\ntype seastar\ntype adamo\ntype astar\ntype ingot\ntype talese\ntype agreing\ntype sakes\ntype theanine\ntype talia\ntype ahmet\ntype thinline\ntype arche\ntype spanne\ntype sakar\ntype alesina\nbeing re\nbeing read\nbeing research\nbeing real\nbeing role\nbeing rest\nbeing reality\nbeing rear\nbeing restart\nbeing realise\nbeing roles\nbeing realised\nbeing reset\nbeing realist\nbeing reseal\nbeing rakesh\nbeing reales\nbeing reise\nbeing rakes\nbeing realidad\nbeing reseau\nbeing resear\nbeing researc\nbeing roleta\nbeing reine\nbeing raked\nbeing reale\nless tag\nless tap\nless tango\nless tapas\nless edina\nless taliesin\nless taint\nless taber\nless talib\nless tatars\nless tatar\nless tarina\nless tangos\nless talia\nless tainty\neast a\neast at\neast as\neast am\neast ad\neast army\neast arm\neast ah\neast adobe\neast atari\neast ahmed\neast aarhus\neast adoring\neast alesse\neast adorno\neast adaline\neast arcing\neast adorn\neast aline\neast adamo\neast agreing\neast ahmet\neast arche\neast alesina\nstar in\nstar i\nstar not\nstar my\nstar no\nstar now\nstar car\nstar none\nstar nor\nstar clip\nstar cake\nstar nose\nstar ceiling\nstar inline\nstar norm\nstar cease\nstar ceased\nstar nod\nstar inning\nstar chew\nstar chewing\nstar cakes\nstar cling\nstar nobel\nstar ingress\nstar ingres\nstar infuse\nstar norco\nstar cesar\nstar cline\nstar coles\nstar inlining\nstar chews\nstar cdata\nstar inest\nstar indole\nstar myles\nstar nosed\nstar infusing\nstar nolita\nstar nodal\nstar coleus\nstar myrinet\nstar infuses\nstar noline\nstar indain\nstar noakes\nstar mydata\nstar ctype\nstar indoles\nstar cinese\npast a\npast at\npast as\npast am\npast ad\npast army\npast arm\npast ah\npast adobe\npast atari\npast ahmed\npast aarhus\npast adoring\npast alesse\npast adorno\npast adaline\npast arcing\npast adorn\npast aearch\npast aline\npast adamo\npast agreing\npast ahmet\npast arche\npast alesina\ndog re\ndog read\ndog research\ndog real\ndog role\ndog rest\ndog reality\ndog rear\ndog retain\ndog restart\ndog realise\ndog roles\ndog realised\ndog reset\ndog retains\ndog resins\ndog resin\ndog realist\ndog rinse\ndog reseal\ndog rinsed\ndog rakesh\ndog reales\ndog reise\ndog rakes\ndog reseau\ndog resear\ndog researc\ndog roleta\ndog reine\ndog raked\ndog reale\nbed of\nbed a\nbed on\nbed or\nbed at\nbed as\nbed one\nbed am\nbed oh\nbed ad\nbed army\nbed arm\nbed oak\nbed ongoing\nbed ah\nbed atari\nbed oakdale\nbed ahmed\nbed oprah\nbed aarhus\nbed orinda\nbed orcinus\nbed adoring\nbed alesse\nbed adorno\nbed arcing\nbed adorn\nbed oakes\nbed aearch\nbed alist\nbed aline\nbed astar\nbed agreing\nbed ahmet\nbed arche\nbed oline\nbed alesina\nring re\nring read\nring research\nring real\nring role\nring rest\nring reality\nring rear\nring retain\nring restart\nring realise\nring roles\nring realised\nring reset\nring retains\nring resins\nring resin\nring realist\nring rinse\nring reseal\nring rinsed\nring rakesh\nring reales\nring reise\nring rakes\nring realidad\nring reseau\nring resear\nring researc\nring roleta\nring reine\nring raked\nring reale\npain a\npain with\npain be\npain us\npain do\npain he\npain list\npain data\npain best\npain great\npain line\npain type\npain less\npain east\npain star\npain dog\npain bed\npain ring\npain bear\npain eat\npain nine\npain dot\npain pan\npain gray\npain earn\npain types\npain dose\npain bet\npain dad\npain ear\npain ease\npain lie\npain dam\npain bee\npain lip\npain leslie\npain lid\npain lining\npain pant\npain grease\npain espanol\npain beg\npain staring\npain daring\npain starch\npain stalin\npain benin\npain espana\npain ealing\npain panty\npain panda\npain prada\npain bering\npain lesben\npain rinse\npain greased\npain mollie\npain witty\npain beaker\npain rinsed\npain pandas\npain bestality\npain panning\npain earch\npain dorint\npain liberi\npain lista\npain typeset\npain doesn\npain stang\npain stale\npain lingo\npain esearch\npain fuses\npain beeing\npain libel\npain bening\npain darin\npain belies\npain greta\npain eased\npain dales\npain seddon\npain prado\npain molar\npain moles\npain does\npain eidos\npain sedaka\npain beaked\npain molest\npain praline\npain stata\npain estar\npain darcs\npain graying\npain bestar\npain panes\npain dahmer\npain listas\npain lidar\npain liber\npain dopant\npain witless\npain earing\npain lesbe\npain dangos\npain listado\npain mollis\npain bedale\npain pandan\npain fussed\npain listar\npain doling\npain typeid\npain nines\npain done\nbear he\nbear head\nbear hi\nbear hear\nbear heat\nbear hearing\nbear hole\nbear ha\nbear healing\nbear hint\nbear hay\nbear heal\nbear holes\nbear hines\nbear hesse\nbear hearn\nbear heist\nbear heshe\nbear heine\nbear heilig\nbear holed\nbear heise\neat a\neat art\neat arm\neat aretha\neat arpanet\neat arles\neat arline\nnine the\nnine a\nnine in\nnine i\nnine it\nnine at\nnine as\nnine if\nnine search\nnine their\nnine so\nnine am\nnine these\nnine she\nnine set\nnine start\nnine say\nnine star\nnine thing\nnine ad\nnine sea\nnine thus\nnine ie\nnine army\nnine saying\nnine tag\nnine seat\nnine arm\nnine sin\nnine seal\nnine thin\nnine stars\nnine ah\nnine sole\nnine spain\nnine slip\nnine sing\nnine starring\nnine tale\nnine tap\nnine swim\nnine adobe\nnine sake\nnine tales\nnine staring\nnine sealing\nnine stare\nnine sparc\nnine theta\nnine atari\nnine starr\nnine sprang\nnine tango\nnine ahmed\nnine aarhus\nnine stardom\nnine starling\nnine sinus\nnine sling\nnine spans\nnine sparing\nnine soledad\nnine searing\nnine sparco\nnine seabed\nnine thine\nnine sinead\nnine sprains\nnine seine\nnine seibel\nnine sprain\nnine adoring\nnine soleus\nnine slingo\nnine alesse\nnine soles\nnine adorno\nnine irina\nnine tapas\nnine adaline\nnine seadoo\nnine arcing\nnine tatarstan\nnine taliesin\nnine sayin\nnine spandau\nnine sinless\nnine adorn\nnine sparcs\nnine theist\nnine sealine\nnine searcg\nnine swims\nnine shewing\nnine searcn\nnine searcb\nnine sdarch\nnine searcu\nnine taint\nnine searct\nnine searcm\nnine starline\nnine taber\nnine talib\nnine tatars\nnine tatar\nnine idabel\nnine tarina\nnine spaeth\nnine tangos\nnine alist\nnine aline\nnine swiming\nnine seale\nnine searc\nnine seastar\nnine adamo\nnine astar\nnine ingot\nnine talese\nnine starlit\nnine agreing\nnine sakes\nnine talia\nnine tainty\nnine ahmet\nnine thinline\nnine arche\nnine spanne\nnine sakar\nnine alesina\ndot a\ndot area\ndot art\ndot areas\ndot arm\ndot aretha\ndot arpanet\ndot arles\ndot arline\ndot areal\npaint a\npaint area\npaint art\npaint areas\npaint arm\npaint aretha\npaint arpanet\npaint arles\npaint arline\npaint areal\npan a\npan in\npan with\npan i\npan be\npan us\npan do\npan he\npan list\npan go\npan buy\npan data\npan best\npan great\npan line\npan type\npan being\npan less\npan east\npan star\npan god\npan past\npan bay\npan bar\npan dog\npan bed\npan bring\npan ring\npan pain\npan bus\npan going\npan bear\npan eat\npan beat\npan nine\npan dot\npan paint\npan pan\npan gray\npan earn\npan doing\npan types\npan stainless\npan dose\npan bet\npan dad\npan pat\npan ear\npan beast\npan ease\npan lie\npan pad\npan beam\npan bearing\npan dam\npan bean\npan bee\npan inline\npan lip\npan bind\npan typing\npan leslie\npan blessed\npan pasta\npan lid\npan baking\npan pale\npan bless\npan beastality\npan lining\npan staind\npan bake\npan baker\npan grease\npan beg\npan staring\npan bethesda\npan inning\npan grin\npan stains\npan daring\npan starch\npan stalin\npan benin\npan baked\npan ealing\npan painless\npan stain\npan goethe\npan molina\npan moline\npan pains\npan grind\npan ingress\npan prada\npan bering\npan lesben\npan rinse\npan greased\npan mollie\npan dainty\npan witty\npan fusing\npan beaker\npan rinsed\npan bling\npan bestality\npan lipase\npan staines\npan pastas\npan earch\npan dorint\npan paring\npan panini\npan liberi\npan ingres\npan infuse\npan lista\npan typeset\npan doesn\npan paine\npan stang\npan binning\npan stale\npan lingo\npan esearch\npan fuses\npan beeing\npan instal\npan libel\npan bening\npan brine\npan inlining\npan darin\npan belies\npan greta\npan inest\npan eased\npan indole\npan goring\npan molino\npan dales\npan seddon\npan betaine\npan prado\npan molar\npan moles\npan does\npan infusing\npan beale\npan instar\npan eidos\npan sedaka\npan beaked\npan molest\npan praline\npan dainese\npan stata\npan estar\npan darcs\npan graying\npan bestar\npan dahmer\npan grins\npan infuses\npan listas\npan lidar\npan liber\npan witless\npan brining\npan pango\npan padang\npan beset\npan earing\npan espada\npan indain\npan lesbe\npan insta\npan dangos\npan listado\npan mollis\npan paling\npan betas\npan brinda\npan bedale\npan indoles\npan fussed\npan tarina\npan listar\npan lipari\npan bethea\npan boles\npan doling\npan goole\npan palin\npan beane\npan parco\npan brines\npan typeid\npan daines\npan baying\npan beilin\npan blingo\npan parche\npan beaty\npan nines\npan busoni\npan done\ngray in\ngray i\ngray it\ngray if\ngray ie\ngray irina\ngray idabel\ngray ingot\nearn of\nearn on\nearn or\nearn one\nearn oh\nearn oak\nearn ongoing\nearn oakdale\nearn oprah\nearn obese\nearn orinda\nearn orcinus\nearn oakes\nearn oline\ndoing re\ndoing read\ndoing research\ndoing real\ndoing role\ndoing rest\ndoing reality\ndoing rear\ndoing restart\ndoing realise\ndoing roles\ndoing realised\ndoing reset\ndoing realist\ndoing reseal\ndoing rakesh\ndoing reales\ndoing reise\ndoing rakes\ndoing reseau\ndoing resear\ndoing researc\ndoing roleta\ndoing reine\ndoing raked\ndoing reale\ntypes a\ntypes in\ntypes with\ntypes i\ntypes be\ntypes us\ntypes do\ntypes he\ntypes list\ntypes data\ntypes best\ntypes great\ntypes line\ntypes being\ntypes less\ntypes east\ntypes star\ntypes past\ntypes dog\ntypes bed\ntypes ring\ntypes pain\ntypes bear\ntypes eat\ntypes nine\ntypes dot\ntypes paint\ntypes pan\ntypes gray\ntypes earn\ntypes doing\ntypes stainless\ntypes dose\ntypes bet\ntypes dad\ntypes pat\ntypes ear\ntypes ease\ntypes lie\ntypes pad\ntypes dam\ntypes bee\ntypes inline\ntypes lip\ntypes leslie\ntypes pasta\ntypes lid\ntypes pale\ntypes lining\ntypes staind\ntypes pant\ntypes grease\ntypes beg\ntypes staring\ntypes inning\ntypes grin\ntypes stains\ntypes daring\ntypes starch\ntypes stalin\ntypes benin\ntypes ealing\ntypes painless\ntypes stain\ntypes panda\ntypes molina\ntypes moline\ntypes pains\ntypes grind\ntypes prada\ntypes bering\ntypes lesben\ntypes rinse\ntypes greased\ntypes mollie\ntypes fusing\ntypes beaker\ntypes rinsed\ntypes pandas\ntypes panning\ntypes lipase\ntypes pastas\ntypes earch\ntypes dorint\ntypes paring\ntypes panini\ntypes liberi\ntypes infuse\ntypes lista\ntypes paine\ntypes stang\ntypes stale\ntypes lingo\ntypes beeing\ntypes instal\ntypes libel\ntypes bening\ntypes inlining\ntypes darin\ntypes greta\ntypes eased\ntypes indole\ntypes molino\ntypes dales\ntypes seddon\ntypes prado\ntypes molar\ntypes instar\ntypes eidos\ntypes sedaka\ntypes beaked\ntypes praline\ntypes stata\ntypes darcs\ntypes graying\ntypes bestar\ntypes dahmer\ntypes grins\ntypes listas\ntypes lidar\ntypes liber\ntypes dopant\ntypes witless\ntypes pango\ntypes padang\ntypes earing\ntypes lesbe\ntypes insta\ntypes dangos\ntypes listado\ntypes mollis\ntypes paling\ntypes bedale\ntypes pandan\ntypes indoles\ntypes fussed\ntypes tarina\ntypes listar\ntypes lipari\ntypes doling\ntypes palin\ntypes parco\ntypes parche\ntypes done\ndose do\ndose day\ndose deal\ndose dead\ndose dear\ndose dealing\ndose dinar\ndose dakine\ndose detain\ndose dinning\ndose dinesh\ndose deity\ndose dakar\ndose dearing\ndose deane\ndose dearch\ndose desing\ndose detalii\ndose detains\ndose deine\ndose dlese\nbet a\nbet area\nbet art\nbet areas\nbet arm\nbet aretha\nbet arpanet\nbet arles\nbet arline\nbet areal\ndad of\ndad on\ndad or\ndad one\ndad oh\ndad oak\ndad ongoing\ndad oprah\ndad obese\ndad orcinus\ndad oakes\ndad oline\npat a\npat area\npat art\npat areas\npat arm\npat aretha\npat arpanet\npat arles\npat arline\npat areal\near in\near i\near not\near my\near no\near now\near car\near none\near nor\near clip\near cake\near nose\near ceiling\near inline\near norm\near nod\near inning\near chew\near chewing\near cakes\near cling\near nobel\near ingress\near mylist\near ingres\near infuse\near norco\near cesar\near cline\near coles\near instal\near inlining\near chews\near cdata\near inest\near indole\near myles\near nosed\near infusing\near instar\near nolita\near nodal\near coleus\near myrinet\near infuses\near noline\near indain\near insta\near noakes\near mydata\near ctype\near indoles\near cinese\nease do\nease day\nease dinar\nease dakine\nease detain\nease dinning\nease dinesh\nease deity\nease dakar\nease desing\nease detalii\nease dlidos\nease detains\nease deine\nease dlese\nlie the\nlie a\nlie in\nlie i\nlie it\nlie at\nlie as\nlie if\nlie search\nlie their\nlie so\nlie am\nlie these\nlie she\nlie set\nlie start\nlie say\nlie star\nlie thing\nlie ad\nlie sea\nlie thus\nlie ie\nlie army\nlie saying\nlie tag\nlie seat\nlie arm\nlie sin\nlie seal\nlie thin\nlie stars\nlie ah\nlie sole\nlie spain\nlie sing\nlie starring\nlie tale\nlie tap\nlie swim\nlie adobe\nlie sake\nlie tales\nlie staring\nlie spanning\nlie stare\nlie sparc\nlie theta\nlie atari\nlie thinning\nlie starr\nlie sprang\nlie tango\nlie ahmed\nlie aarhus\nlie stardom\nlie sinus\nlie spans\nlie sparing\nlie soledad\nlie searing\nlie sparco\nlie seabed\nlie thine\nlie sinead\nlie sprains\nlie seine\nlie seibel\nlie sprain\nlie adoring\nlie soleus\nlie alesse\nlie soles\nlie adorno\nlie irina\nlie sinning\nlie tapas\nlie seadoo\nlie arcing\nlie tatarstan\nlie sayin\nlie spandau\nlie sinless\nlie adorn\nlie sparcs\nlie theist\nlie searcg\nlie swims\nlie shewing\nlie searcn\nlie searcb\nlie sdarch\nlie searcu\nlie taint\nlie searct\nlie searcm\nlie taber\nlie tatars\nlie tatar\nlie idabel\nlie tarina\nlie spaeth\nlie tangos\nlie swiming\nlie seale\nlie searc\nlie seastar\nlie adamo\nlie astar\nlie ingot\nlie talese\nlie agreing\nlie sakes\nlie theanine\nlie tainty\nlie ahmet\nlie arche\nlie spanne\nlie sakar\nlie alesina\npad of\npad a\npad on\npad or\npad at\npad as\npad one\npad am\npad oh\npad ad\npad army\npad arm\npad oak\npad ongoing\npad ah\npad adobe\npad atari\npad oakdale\npad ahmed\npad oprah\npad aarhus\npad obese\npad orinda\npad orcinus\npad adoring\npad alesse\npad adorno\npad arcing\npad adorn\npad oakes\npad aearch\npad alist\npad aline\npad astar\npad agreing\npad ahmet\npad arche\npad oline\npad alesina\ndam old\ndam ollie\ndam olnine\ndam olean\ndam oleari\ndam oline\nbee the\nbee a\nbee in\nbee i\nbee it\nbee at\nbee as\nbee if\nbee search\nbee their\nbee so\nbee am\nbee these\nbee she\nbee set\nbee start\nbee say\nbee star\nbee thing\nbee ad\nbee sea\nbee thus\nbee ie\nbee army\nbee saying\nbee tag\nbee seat\nbee arm\nbee sin\nbee seal\nbee thin\nbee stars\nbee ah\nbee sole\nbee spain\nbee slip\nbee sing\nbee starring\nbee tale\nbee tap\nbee swim\nbee sake\nbee tales\nbee staring\nbee spanning\nbee sealing\nbee stare\nbee sparc\nbee theta\nbee atari\nbee thinning\nbee starr\nbee sprang\nbee tango\nbee ahmed\nbee aarhus\nbee stardom\nbee starling\nbee sinus\nbee sling\nbee spans\nbee sparing\nbee soledad\nbee searing\nbee sparco\nbee thine\nbee sinead\nbee sprains\nbee seine\nbee sprain\nbee adoring\nbee soleus\nbee slingo\nbee alesse\nbee soles\nbee adorno\nbee irina\nbee sinning\nbee tapas\nbee adaline\nbee seadoo\nbee arcing\nbee tatarstan\nbee taliesin\nbee sayin\nbee spandau\nbee sinless\nbee adorn\nbee sparcs\nbee theist\nbee sealine\nbee searcg\nbee swims\nbee shewing\nbee searcn\nbee sdarch\nbee searcu\nbee taint\nbee searct\nbee searcm\nbee starline\nbee tatars\nbee tatar\nbee tarina\nbee spaeth\nbee tangos\nbee alist\nbee aline\nbee swiming\nbee seale\nbee searc\nbee seastar\nbee adamo\nbee astar\nbee ingot\nbee talese\nbee starlit\nbee agreing\nbee sakes\nbee theanine\nbee talia\nbee tainty\nbee ahmet\nbee thinline\nbee arche\nbee spanne\nbee sakar\nbee alesina\ninline of\ninline on\ninline or\ninline oh\ninline oak\ninline oakdale\ninline oprah\ninline obese\ninline orinda\ninline oakes\nlip and\nlip a\nlip at\nlip as\nlip am\nlip ad\nlip army\nlip rain\nlip raw\nlip arm\nlip radar\nlip ah\nlip rat\nlip andale\nlip adobe\nlip atari\nlip anakin\nlip ahmed\nlip aarhus\nlip rains\nlip raines\nlip antares\nlip radon\nlip adoring\nlip alesse\nlip adorno\nlip raine\nlip arcing\nlip rasta\nlip adorn\nlip antara\nlip aearch\nlip antari\nlip rafuse\nlip rahmen\nlip raring\nlip adamo\nlip astar\nlip agreing\nlip ahmet\nlip arche\nlip alesina\ntyping re\ntyping read\ntyping research\ntyping real\ntyping role\ntyping rest\ntyping rear\ntyping realise\ntyping roles\ntyping realised\ntyping reset\ntyping realist\ntyping reseal\ntyping rakesh\ntyping reales\ntyping reise\ntyping rakes\ntyping realidad\ntyping reseau\ntyping resear\ntyping researc\ntyping roleta\ntyping reine\ntyping raked\ntyping reale\nleslie the\nleslie a\nleslie in\nleslie i\nleslie it\nleslie at\nleslie as\nleslie if\nleslie search\nleslie their\nleslie so\nleslie am\nleslie these\nleslie she\nleslie set\nleslie start\nleslie say\nleslie star\nleslie thing\nleslie ad\nleslie sea\nleslie thus\nleslie ie\nleslie army\nleslie saying\nleslie tag\nleslie seat\nleslie arm\nleslie sin\nleslie thin\nleslie stars\nleslie ah\nleslie sole\nleslie spain\nleslie sing\nleslie starring\nleslie tap\nleslie swim\nleslie adobe\nleslie sake\nleslie staring\nleslie spanning\nleslie stare\nleslie sparc\nleslie theta\nleslie atari\nleslie thinning\nleslie starr\nleslie sprang\nleslie tango\nleslie ahmed\nleslie aarhus\nleslie stardom\nleslie sinus\nleslie spans\nleslie sparing\nleslie soledad\nleslie searing\nleslie sparco\nleslie seabed\nleslie thine\nleslie sinead\nleslie sprains\nleslie seine\nleslie sprain\nleslie adoring\nleslie soleus\nleslie soles\nleslie adorno\nleslie irina\nleslie sinning\nleslie tapas\nleslie seadoo\nleslie arcing\nleslie tatarstan\nleslie sayin\nleslie spandau\nleslie adorn\nleslie sparcs\nleslie theist\nleslie searcg\nleslie swims\nleslie shewing\nleslie searcn\nleslie searcb\nleslie sdarch\nleslie searcu\nleslie taint\nleslie searct\nleslie searcm\nleslie taber\nleslie tatars\nleslie tatar\nleslie tarina\nleslie spaeth\nleslie tangos\nleslie swiming\nleslie searc\nleslie seastar\nleslie adamo\nleslie astar\nleslie ingot\nleslie agreing\nleslie sakes\nleslie theanine\nleslie tainty\nleslie ahmet\nleslie arche\nleslie spanne\nleslie sakar\npasta a\npasta in\npasta with\npasta i\npasta be\npasta us\npasta do\npasta he\npasta data\npasta great\npasta line\npasta type\npasta being\npasta less\npasta dog\npasta bed\npasta ring\npasta bear\npasta eat\npasta nine\npasta dot\npasta pan\npasta gray\npasta earn\npasta doing\npasta types\npasta dose\npasta bet\npasta dad\npasta ear\npasta ease\npasta lie\npasta dam\npasta bee\npasta inline\npasta lip\npasta typing\npasta leslie\npasta lid\npasta lining\npasta pant\npasta grease\npasta espanol\npasta beg\npasta inning\npasta grin\npasta daring\npasta benin\npasta espana\npasta ealing\npasta panty\npasta panda\npasta molina\npasta moline\npasta grind\npasta ingress\npasta prada\npasta bering\npasta lesben\npasta rinse\npasta greased\npasta mollie\npasta dainty\npasta witty\npasta fusing\npasta beaker\npasta rinsed\npasta pandas\npasta panning\npasta earch\npasta dorint\npasta liberi\npasta ingres\npasta infuse\npasta typeset\npasta doesn\npasta lingo\npasta esearch\npasta fuses\npasta beeing\npasta libel\npasta bening\npasta inlining\npasta darin\npasta belies\npasta greta\npasta inest\npasta eased\npasta indole\npasta molino\npasta dales\npasta seddon\npasta prado\npasta molar\npasta moles\npasta does\npasta eidos\npasta sedaka\npasta beaked\npasta molest\npasta praline\npasta dainese\npasta estar\npasta darcs\npasta graying\npasta panes\npasta dahmer\npasta grins\npasta infuses\npasta lidar\npasta liber\npasta dopant\npasta witless\npasta earing\npasta lesbe\npasta dangos\npasta mollis\npasta bedale\npasta pandan\npasta indoles\npasta fussed\npasta tarina\npasta doling\npasta typeid\npasta daines\npasta nines\npasta done\nlid of\nlid a\nlid on\nlid or\nlid at\nlid as\nlid one\nlid am\nlid oh\nlid ad\nlid army\nlid arm\nlid oak\nlid ongoing\nlid ah\nlid adobe\nlid atari\nlid oakdale\nlid ahmed\nlid oprah\nlid aarhus\nlid obese\nlid orinda\nlid orcinus\nlid adoring\nlid alesse\nlid adorno\nlid arcing\nlid adorn\nlid oakes\nlid aearch\nlid astar\nlid agreing\nlid ahmet\nlid arche\nlid alesina\npale search\npale so\npale she\npale set\npale start\npale say\npale star\npale sea\npale saying\npale seat\npale sin\npale seal\npale stars\npale sole\npale slip\npale sing\npale starring\npale swim\npale sake\npale staring\npale spanning\npale sealing\npale stare\npale starr\npale sprang\npale stares\npale stardom\npale starling\npale sinus\npale sling\npale spans\npale soledad\npale searing\npale seabed\npale sinead\npale sprains\npale seine\npale seibel\npale sprain\npale soleus\npale slingo\npale soles\npale sinning\npale seadoo\npale sayin\npale spandau\npale sealine\npale searcg\npale swims\npale sesrch\npale shewing\npale searcn\npale searcb\npale sdarch\npale searcu\npale searct\npale searcm\npale starline\npale swiming\npale searc\npale seastar\npale starlit\npale sakes\npale spanne\npale sakar\nlining re\nlining read\nlining research\nlining real\nlining role\nlining rest\nlining rear\nlining retain\nlining restart\nlining roles\nlining reset\nlining retains\nlining resins\nlining resin\nlining rinse\nlining reseal\nlining rinsed\nlining rakesh\nlining reales\nlining reise\nlining rakes\nlining reseau\nlining resear\nlining researc\nlining roleta\nlining reine\nlining raked\nlining reale\nstaind of\nstaind a\nstaind on\nstaind or\nstaind at\nstaind as\nstaind one\nstaind am\nstaind oh\nstaind ad\nstaind army\nstaind arm\nstaind oak\nstaind ah\nstaind adobe\nstaind oakdale\nstaind ahmed\nstaind oprah\nstaind aarhus\nstaind obese\nstaind orinda\nstaind adoring\nstaind alesse\nstaind adorno\nstaind adorn\nstaind oakes\nstaind aearch\nstaind aline\nstaind agreing\nstaind ahmet\nstaind arche\nstaind oline\npant a\npant area\npant art\npant areas\npant arm\npant aretha\npant arles\npant arline\npant areal\ngrease do\ngrease day\ngrease dinar\ngrease dakine\ngrease detain\ngrease dinesh\ngrease deity\ngrease dakar\ngrease detalii\ngrease dlidos\ngrease detains\ngrease deine\ngrease dlese\nespanol ear\nespanol estado\nespanol estados\nespanol elise\nespanol estab\nespanol elist\nespanol estas\nespanol estar\nespanol eakins\nespanol egret\nespanol eearch\nespanol ebeling\nespanol erindale\nbeg re\nbeg read\nbeg research\nbeg real\nbeg role\nbeg rest\nbeg reality\nbeg rear\nbeg retain\nbeg restart\nbeg realise\nbeg roles\nbeg realised\nbeg reset\nbeg retains\nbeg resins\nbeg resin\nbeg realist\nbeg rinse\nbeg reseal\nbeg rinsed\nbeg rakesh\nbeg reales\nbeg reise\nbeg rakes\nbeg realidad\nbeg reseau\nbeg resear\nbeg researc\nbeg roleta\nbeg reine\nbeg raked\nbeg reale\nstaring re\nstaring read\nstaring research\nstaring real\nstaring role\nstaring rest\nstaring reality\nstaring rear\nstaring retain\nstaring restart\nstaring realise\nstaring roles\nstaring realised\nstaring reset\nstaring retains\nstaring resins\nstaring resin\nstaring rinse\nstaring reseal\nstaring rinsed\nstaring rakesh\nstaring reales\nstaring reise\nstaring rakes\nstaring realidad\nstaring reseau\nstaring resear\nstaring researc\nstaring roleta\nstaring reine\nstaring raked\nstaring reale\ninning re\ninning read\ninning research\ninning real\ninning role\ninning rest\ninning reality\ninning rear\ninning restart\ninning realise\ninning roles\ninning realised\ninning reset\ninning realist\ninning reseal\ninning rakesh\ninning reales\ninning reise\ninning rakes\ninning realidad\ninning reseau\ninning resear\ninning researc\ninning roleta\ninning reine\ninning raked\ninning reale\ngrin a\ngrin with\ngrin be\ngrin us\ngrin do\ngrin he\ngrin list\ngrin data\ngrin best\ngrin line\ngrin type\ngrin less\ngrin east\ngrin star\ngrin past\ngrin bed\ngrin bear\ngrin eat\ngrin nine\ngrin dot\ngrin pan\ngrin earn\ngrin types\ngrin dose\ngrin bet\ngrin dad\ngrin pat\ngrin ear\ngrin ease\ngrin lie\ngrin pad\ngrin dam\ngrin bee\ngrin lip\ngrin leslie\ngrin pasta\ngrin lid\ngrin pale\ngrin pant\ngrin espanol\ngrin starch\ngrin stalin\ngrin benin\ngrin espana\ngrin ealing\ngrin panty\ngrin panda\ngrin prada\ngrin lesben\ngrin rinse\ngrin mollie\ngrin witty\ngrin beaker\ngrin rinsed\ngrin pandas\ngrin bestality\ngrin lipase\ngrin pastas\ngrin earch\ngrin dorint\ngrin liberi\ngrin lista\ngrin typeset\ngrin doesn\ngrin stang\ngrin stale\ngrin lingo\ngrin esearch\ngrin fuses\ngrin beeing\ngrin libel\ngrin darin\ngrin belies\ngrin eased\ngrin dales\ngrin seddon\ngrin prado\ngrin molar\ngrin moles\ngrin does\ngrin eidos\ngrin sedaka\ngrin beaked\ngrin molest\ngrin praline\ngrin stata\ngrin estar\ngrin darcs\ngrin bestar\ngrin panes\ngrin dahmer\ngrin listas\ngrin lidar\ngrin liber\ngrin dopant\ngrin witless\ngrin pango\ngrin padang\ngrin espada\ngrin lesbe\ngrin dangos\ngrin listado\ngrin mollis\ngrin paling\ngrin bedale\ngrin pandan\ngrin fussed\ngrin listar\ngrin lipari\ngrin doling\ngrin palin\ngrin parco\ngrin typeid\ngrin parche\ngrin nines\ngrin done\ndaring re\ndaring read\ndaring research\ndaring real\ndaring role\ndaring rest\ndaring reality\ndaring rear\ndaring retain\ndaring restart\ndaring realise\ndaring roles\ndaring realised\ndaring reset\ndaring retains\ndaring resins\ndaring resin\ndaring realist\ndaring rinse\ndaring reseal\ndaring rinsed\ndaring rakesh\ndaring reales\ndaring reise\ndaring rakes\ndaring reseau\ndaring resear\ndaring researc\ndaring roleta\ndaring reine\ndaring raked\ndaring reale\nstarch me\nstarch meet\nstarch medal\nstarch metart\nstarch merino\nstarch ewing\nstarch melia\nstarch meakin\nstarch metar\nstarch meine\nstarch menino\nstarch menina\nstarch melita\nstarch meines\nstalin in\nstalin i\nstalin be\nstalin go\nstalin buy\nstalin best\nstalin being\nstalin god\nstalin bay\nstalin bar\nstalin bring\nstalin bus\nstalin going\nstalin bear\nstalin beat\nstalin bet\nstalin beam\nstalin bearing\nstalin bean\nstalin bind\nstalin blessed\nstalin baking\nstalin bless\nstalin bake\nstalin baker\nstalin bethesda\nstalin baked\nstalin goethe\nstalin ingress\nstalin ingres\nstalin infuse\nstalin binning\nstalin brine\nstalin inest\nstalin indole\nstalin goring\nstalin betaine\nstalin infusing\nstalin beale\nstalin bestar\nstalin infuses\nstalin brining\nstalin beset\nstalin indain\nstalin gopal\nstalin betas\nstalin brinda\nstalin indoles\nstalin bethea\nstalin boles\nstalin goole\nstalin beane\nstalin brines\nstalin baying\nstalin beaty\nstalin busoni\nbenin a\nbenin in\nbenin with\nbenin i\nbenin us\nbenin do\nbenin he\nbenin list\nbenin data\nbenin great\nbenin line\nbenin type\nbenin less\nbenin east\nbenin star\nbenin past\nbenin dog\nbenin ring\nbenin pain\nbenin eat\nbenin dot\nbenin paint\nbenin pan\nbenin gray\nbenin earn\nbenin doing\nbenin types\nbenin stainless\nbenin dose\nbenin dad\nbenin pat\nbenin ear\nbenin ease\nbenin lie\nbenin pad\nbenin dam\nbenin inline\nbenin lip\nbenin typing\nbenin leslie\nbenin pasta\nbenin lid\nbenin pale\nbenin staind\nbenin pant\nbenin grease\nbenin espanol\nbenin staring\nbenin grin\nbenin stains\nbenin daring\nbenin starch\nbenin stalin\nbenin espana\nbenin ealing\nbenin painless\nbenin panty\nbenin stain\nbenin panda\nbenin molina\nbenin moline\nbenin pains\nbenin grind\nbenin ingress\nbenin prada\nbenin rinse\nbenin greased\nbenin mollie\nbenin dainty\nbenin witty\nbenin fusing\nbenin rinsed\nbenin pandas\nbenin lipase\nbenin staines\nbenin pastas\nbenin earch\nbenin dorint\nbenin paring\nbenin ingres\nbenin infuse\nbenin lista\nbenin typeset\nbenin doesn\nbenin paine\nbenin stang\nbenin stale\nbenin lingo\nbenin esearch\nbenin fuses\nbenin instal\nbenin darin\nbenin greta\nbenin inest\nbenin eased\nbenin indole\nbenin molino\nbenin dales\nbenin seddon\nbenin prado\nbenin molar\nbenin moles\nbenin does\nbenin instar\nbenin eidos\nbenin sedaka\nbenin molest\nbenin praline\nbenin dainese\nbenin stata\nbenin estar\nbenin darcs\nbenin graying\nbenin panes\nbenin dahmer\nbenin grins\nbenin infuses\nbenin listas\nbenin lidar\nbenin dopant\nbenin witless\nbenin pango\nbenin padang\nbenin earing\nbenin espada\nbenin insta\nbenin dangos\nbenin listado\nbenin mollis\nbenin paling\nbenin pandan\nbenin indoles\nbenin fussed\nbenin tarina\nbenin listar\nbenin lipari\nbenin doling\nbenin palin\nbenin parco\nbenin typeid\nbenin daines\nbenin parche\nbenin done\nespana king\nespana kind\nespana kinda\nespana rhino\nespana klingon\nespana keane\nespana rhine\nespana kline\nespana keine\nespana khmer\nespana kelis\nespana kedar\nespana rhein\nespana kling\nespana kepada\nespana kernow\nealing of\nealing on\nealing or\nealing one\nealing oh\nealing oak\nealing oakdale\nealing oprah\nealing obese\nealing orinda\nealing orcinus\nealing oakes\npainless tag\npainless tap\npainless tango\npainless taber\npainless talib\npainless tatars\npainless tatar\npainless tarina\npainless tangos\npainless talia\npanty pm\npanty print\npanty pay\npanty pet\npanty pin\npanty paying\npanty pine\npanty pole\npanty pearce\npanty poles\npanty pines\npanty pinning\npanty peirce\npanty peseta\npanty pineal\npanty petal\npanty petaling\npanty pinole\npanty pease\npanty plist\npanty pinus\npanty pesetas\npanty polen\npanty pinless\npanty pindar\npanty prine\npanty polestar\npanty prins\npanty pineau\npanty plies\nstain a\nstain with\nstain be\nstain us\nstain do\nstain he\nstain data\nstain great\nstain line\nstain type\nstain less\nstain dog\nstain bed\nstain ring\nstain bear\nstain eat\nstain nine\nstain dot\nstain pan\nstain gray\nstain earn\nstain types\nstain dose\nstain bet\nstain dad\nstain pat\nstain ear\nstain ease\nstain lie\nstain pad\nstain dam\nstain bee\nstain lip\nstain leslie\nstain lid\nstain pale\nstain lining\nstain pant\nstain grease\nstain espanol\nstain beg\nstain daring\nstain benin\nstain espana\nstain ealing\nstain panty\nstain panda\nstain prada\nstain bering\nstain lesben\nstain rinse\nstain greased\nstain mollie\nstain witty\nstain beaker\nstain rinsed\nstain pandas\nstain panning\nstain lipase\nstain earch\nstain dorint\nstain paring\nstain liberi\nstain typeset\nstain doesn\nstain lingo\nstain esearch\nstain fuses\nstain beeing\nstain libel\nstain bening\nstain darin\nstain belies\nstain greta\nstain eased\nstain dales\nstain seddon\nstain prado\nstain molar\nstain moles\nstain does\nstain eidos\nstain sedaka\nstain beaked\nstain molest\nstain praline\nstain estar\nstain darcs\nstain graying\nstain panes\nstain dahmer\nstain lidar\nstain liber\nstain dopant\nstain witless\nstain pango\nstain padang\nstain earing\nstain espada\nstain lesbe\nstain dangos\nstain mollis\nstain paling\nstain bedale\nstain pandan\nstain fussed\nstain lipari\nstain doling\nstain palin\nstain parco\nstain typeid\nstain parche\nstain nines\nstain done\npanda a\npanda in\npanda with\npanda i\npanda be\npanda us\npanda do\npanda he\npanda list\npanda best\npanda great\npanda line\npanda type\npanda being\npanda less\npanda east\npanda star\npanda past\npanda dog\npanda bed\npanda ring\npanda pain\npanda bear\npanda eat\npanda nine\npanda dot\npanda paint\npanda pan\npanda gray\npanda earn\npanda doing\npanda types\npanda stainless\npanda dose\npanda bet\npanda pat\npanda ear\npanda ease\npanda lie\npanda pad\npanda bee\npanda inline\npanda lip\npanda typing\npanda leslie\npanda pasta\npanda lid\npanda pale\npanda lining\npanda staind\npanda grease\npanda beg\npanda staring\npanda inning\npanda grin\npanda stains\npanda starch\npanda stalin\npanda benin\npanda ealing\npanda painless\npanda stain\npanda molina\npanda moline\npanda pains\npanda grind\npanda ingress\npanda bering\npanda lesben\npanda rinse\npanda greased\npanda mollie\npanda witty\npanda fusing\npanda beaker\npanda rinsed\npanda bestality\npanda lipase\npanda staines\npanda pastas\npanda earch\npanda dorint\npanda paring\npanda panini\npanda liberi\npanda ingres\npanda infuse\npanda lista\npanda typeset\npanda doesn\npanda paine\npanda stang\npanda stale\npanda lingo\npanda esearch\npanda fuses\npanda beeing\npanda instal\npanda libel\npanda bening\npanda inlining\npanda belies\npanda greta\npanda inest\npanda eased\npanda indole\npanda molino\npanda seddon\npanda prado\npanda molar\npanda moles\npanda does\npanda instar\npanda eidos\npanda sedaka\npanda beaked\npanda molest\npanda praline\npanda stata\npanda estar\npanda graying\npanda bestar\npanda grins\npanda infuses\npanda listas\npanda liber\npanda witless\npanda pango\npanda earing\npanda lesbe\npanda insta\npanda listado\npanda mollis\npanda paling\npanda indoles\npanda fussed\npanda tarina\npanda listar\npanda lipari\npanda doling\npanda palin\npanda parco\npanda typeid\npanda parche\npanda nines\npanda done\nmolina klingon\nmolina keane\nmolina rhesus\nmolina kline\nmolina keine\nmolina khmer\nmolina kelis\nmolina kedar\nmolina rhein\nmolina kling\nmolina kepada\nmolina kernow\nmoline the\nmoline a\nmoline in\nmoline i\nmoline it\nmoline at\nmoline as\nmoline if\nmoline search\nmoline their\nmoline so\nmoline these\nmoline she\nmoline set\nmoline start\nmoline say\nmoline star\nmoline ad\nmoline sea\nmoline thus\nmoline ie\nmoline army\nmoline saying\nmoline tag\nmoline seat\nmoline arm\nmoline seal\nmoline stars\nmoline ah\nmoline sole\nmoline slip\nmoline starring\nmoline tale\nmoline tap\nmoline swim\nmoline adobe\nmoline sake\nmoline tales\nmoline spanning\nmoline sealing\nmoline stare\nmoline sparc\nmoline theta\nmoline starr\nmoline sprang\nmoline tango\nmoline ahmed\nmoline aarhus\nmoline starling\nmoline sling\nmoline spans\nmoline sparing\nmoline soledad\nmoline searing\nmoline sparco\nmoline seabed\nmoline seine\nmoline seibel\nmoline adoring\nmoline soleus\nmoline slingo\nmoline alesse\nmoline soles\nmoline adorno\nmoline irina\nmoline tapas\nmoline adaline\nmoline seadoo\nmoline tatarstan\nmoline sayin\nmoline spandau\nmoline adorn\nmoline sparcs\nmoline theist\nmoline sealine\nmoline searcg\nmoline swims\nmoline searcn\nmoline searcb\nmoline sdarch\nmoline searcu\nmoline searct\nmoline starline\nmoline taber\nmoline talib\nmoline tatars\nmoline tatar\nmoline idabel\nmoline tarina\nmoline spaeth\nmoline tangos\nmoline alist\nmoline aline\nmoline seale\nmoline searc\nmoline seastar\nmoline astar\nmoline ingot\nmoline talese\nmoline starlit\nmoline agreing\nmoline sakes\nmoline theanine\nmoline talia\nmoline ahmet\nmoline arche\nmoline spanne\nmoline sakar\npains tag\npains tale\npains tap\npains tales\npains tango\npains taber\npains talib\npains tatars\npains tatar\npains tarina\npains tangos\npains talese\npains talia\ngrind of\ngrind a\ngrind on\ngrind or\ngrind at\ngrind as\ngrind one\ngrind am\ngrind oh\ngrind ad\ngrind army\ngrind arm\ngrind oak\ngrind ah\ngrind adobe\ngrind oakdale\ngrind ahmed\ngrind oprah\ngrind aarhus\ngrind obese\ngrind orinda\ngrind alesse\ngrind adorno\ngrind adorn\ngrind oakes\ngrind aearch\ngrind alist\ngrind aline\ngrind astar\ngrind ahmet\ngrind arche\ngrind oline\ningress tale\ningress tap\ningress tales\ningress tango\ningress tapas\ningress taber\ningress talib\ningress tatars\ningress tatar\ningress tarina\ningress tangos\ningress talese\ningress talia\nprada a\nprada in\nprada with\nprada i\nprada be\nprada us\nprada do\nprada he\nprada list\nprada best\nprada great\nprada line\nprada type\nprada being\nprada less\nprada east\nprada star\nprada past\nprada dog\nprada bed\nprada ring\nprada pain\nprada bear\nprada eat\nprada nine\nprada dot\nprada paint\nprada pan\nprada gray\nprada earn\nprada doing\nprada types\nprada stainless\nprada dose\nprada bet\nprada pat\nprada ear\nprada ease\nprada lie\nprada pad\nprada bee\nprada inline\nprada lip\nprada typing\nprada leslie\nprada pasta\nprada lid\nprada pale\nprada lining\nprada staind\nprada pant\nprada grease\nprada espanol\nprada beg\nprada staring\nprada inning\nprada grin\nprada stains\nprada starch\nprada stalin\nprada benin\nprada espana\nprada ealing\nprada painless\nprada panty\nprada stain\nprada molina\nprada moline\nprada pains\nprada grind\nprada ingress\nprada bering\nprada lesben\nprada rinse\nprada greased\nprada mollie\nprada witty\nprada fusing\nprada beaker\nprada rinsed\nprada bestality\nprada panning\nprada lipase\nprada staines\nprada pastas\nprada earch\nprada dorint\nprada paring\nprada panini\nprada liberi\nprada ingres\nprada infuse\nprada lista\nprada typeset\nprada doesn\nprada paine\nprada stang\nprada stale\nprada lingo\nprada esearch\nprada fuses\nprada beeing\nprada instal\nprada libel\nprada bening\nprada inlining\nprada belies\nprada greta\nprada inest\nprada eased\nprada indole\nprada molino\nprada seddon\nprada molar\nprada moles\nprada does\nprada instar\nprada eidos\nprada sedaka\nprada beaked\nprada molest\nprada stata\nprada estar\nprada graying\nprada bestar\nprada panes\nprada grins\nprada infuses\nprada listas\nprada liber\nprada dopant\nprada witless\nprada pango\nprada earing\nprada lesbe\nprada insta\nprada listado\nprada mollis\nprada paling\nprada indoles\nprada fussed\nprada tarina\nprada listar\nprada lipari\nprada doling\nprada palin\nprada parco\nprada typeid\nprada parche\nprada nines\nprada done\nbering re\nbering read\nbering research\nbering real\nbering role\nbering rest\nbering reality\nbering rear\nbering retain\nbering restart\nbering realise\nbering roles\nbering realised\nbering reset\nbering retains\nbering resins\nbering resin\nbering realist\nbering rinse\nbering reseal\nbering rinsed\nbering rakesh\nbering reales\nbering reise\nbering rakes\nbering realidad\nbering reseau\nbering resear\nbering researc\nbering roleta\nbering reine\nbering raked\nbering reale\nlesben in\nlesben i\nlesben be\nlesben go\nlesben buy\nlesben best\nlesben being\nlesben god\nlesben bay\nlesben bar\nlesben bring\nlesben bus\nlesben going\nlesben bear\nlesben beat\nlesben bet\nlesben beast\nlesben beam\nlesben bearing\nlesben bean\nlesben inline\nlesben bind\nlesben baking\nlesben beastality\nlesben bake\nlesben baker\nlesben bethesda\nlesben baked\nlesben goethe\nlesben ingress\nlesben bling\nlesben ingres\nlesben infuse\nlesben binning\nlesben instal\nlesben brine\nlesben inest\nlesben goring\nlesben betaine\nlesben infusing\nlesben instar\nlesben bestar\nlesben infuses\nlesben brining\nlesben beset\nlesben indain\nlesben insta\nlesben gopal\nlesben betas\nlesben brinda\nlesben bethea\nlesben boles\nlesben goole\nlesben beane\nlesben brines\nlesben baying\nlesben beilin\nlesben blingo\nlesben beaty\nlesben busoni\nrinse do\nrinse day\nrinse deal\nrinse dead\nrinse dear\nrinse dealing\nrinse dinar\nrinse dakine\nrinse detain\nrinse dinning\nrinse dinesh\nrinse deity\nrinse dakar\nrinse deane\nrinse dearch\nrinse desing\nrinse detalii\nrinse dlidos\nrinse detains\nrinse deine\nrinse dlese\ngreased a\ngreased in\ngreased with\ngreased i\ngreased be\ngreased us\ngreased do\ngreased he\ngreased list\ngreased data\ngreased best\ngreased line\ngreased type\ngreased less\ngreased star\ngreased past\ngreased bed\ngreased pain\ngreased bear\ngreased nine\ngreased dot\ngreased paint\ngreased pan\ngreased types\ngreased bet\ngreased dad\ngreased pat\ngreased lie\ngreased pad\ngreased dam\ngreased bee\ngreased inline\ngreased lip\ngreased leslie\ngreased pasta\ngreased lid\ngreased pale\ngreased staind\ngreased pant\ngreased espanol\ngreased starch\ngreased stalin\ngreased benin\ngreased espana\ngreased painless\ngreased panty\ngreased stain\ngreased panda\ngreased molina\ngreased moline\ngreased pains\ngreased prada\ngreased lesben\ngreased mollie\ngreased dainty\ngreased witty\ngreased beaker\ngreased pandas\ngreased bestality\ngreased staines\ngreased dorint\ngreased panini\ngreased liberi\ngreased infuse\ngreased lista\ngreased typeset\ngreased doesn\ngreased paine\ngreased stang\ngreased stale\ngreased lingo\ngreased fuses\ngreased beeing\ngreased instal\ngreased libel\ngreased darin\ngreased belies\ngreased inest\ngreased indole\ngreased molino\ngreased dales\ngreased prado\ngreased molar\ngreased moles\ngreased does\ngreased instar\ngreased eidos\ngreased beaked\ngreased molest\ngreased praline\ngreased dainese\ngreased stata\ngreased estar\ngreased darcs\ngreased bestar\ngreased panes\ngreased dahmer\ngreased infuses\ngreased lidar\ngreased liber\ngreased dopant\ngreased witless\ngreased pango\ngreased padang\ngreased espada\ngreased lesbe\ngreased insta\ngreased dangos\ngreased listado\ngreased mollis\ngreased paling\ngreased bedale\ngreased pandan\ngreased indoles\ngreased tarina\ngreased listar\ngreased lipari\ngreased doling\ngreased palin\ngreased parco\ngreased typeid\ngreased daines\ngreased parche\ngreased nines\ngreased done\nmollie the\nmollie a\nmollie in\nmollie i\nmollie it\nmollie at\nmollie as\nmollie if\nmollie search\nmollie their\nmollie so\nmollie these\nmollie she\nmollie set\nmollie start\nmollie say\nmollie star\nmollie thing\nmollie ad\nmollie sea\nmollie thus\nmollie ie\nmollie army\nmollie saying\nmollie tag\nmollie seat\nmollie arm\nmollie sin\nmollie seal\nmollie thin\nmollie stars\nmollie ah\nmollie sole\nmollie spain\nmollie sing\nmollie starring\nmollie tale\nmollie tap\nmollie swim\nmollie adobe\nmollie sake\nmollie tales\nmollie staring\nmollie spanning\nmollie stare\nmollie sparc\nmollie theta\nmollie atari\nmollie thinning\nmollie starr\nmollie sprang\nmollie tango\nmollie ahmed\nmollie aarhus\nmollie sinus\nmollie spans\nmollie sparing\nmollie soledad\nmollie searing\nmollie sparco\nmollie seabed\nmollie thine\nmollie sinead\nmollie sprains\nmollie seine\nmollie seibel\nmollie sprain\nmollie adoring\nmollie soleus\nmollie alesse\nmollie soles\nmollie adorno\nmollie irina\nmollie sinning\nmollie tapas\nmollie seadoo\nmollie arcing\nmollie tatarstan\nmollie sayin\nmollie spandau\nmollie sinless\nmollie adorn\nmollie sparcs\nmollie theist\nmollie searcg\nmollie swims\nmollie shewing\nmollie searcn\nmollie searcb\nmollie sdarch\nmollie searcu\nmollie taint\nmollie searct\nmollie taber\nmollie tatars\nmollie tatar\nmollie idabel\nmollie tarina\nmollie spaeth\nmollie tangos\nmollie swiming\nmollie seale\nmollie searc\nmollie seastar\nmollie astar\nmollie ingot\nmollie talese\nmollie agreing\nmollie sakes\nmollie theanine\nmollie tainty\nmollie ahmet\nmollie arche\nmollie spanne\nmollie sakar\nmollie alesina\ndainty pm\ndainty print\ndainty pay\ndainty pet\ndainty paying\ndainty pole\ndainty pearce\ndainty poles\ndainty peirce\ndainty peseta\ndainty petal\ndainty petaling\ndainty pease\ndainty plist\ndainty pesetas\ndainty polen\ndainty prine\ndainty polestar\ndainty prins\ndainty plies\nwitty pm\nwitty print\nwitty pay\nwitty pet\nwitty pin\nwitty paying\nwitty pine\nwitty pole\nwitty pearce\nwitty poles\nwitty pines\nwitty pinning\nwitty peirce\nwitty peseta\nwitty pineal\nwitty petal\nwitty petaling\nwitty pinole\nwitty pease\nwitty plist\nwitty pinus\nwitty pesetas\nwitty polen\nwitty pinless\nwitty pindar\nwitty prine\nwitty polestar\nwitty prins\nwitty pineau\nwitty plies\nfusing re\nfusing read\nfusing research\nfusing real\nfusing role\nfusing rest\nfusing reality\nfusing rear\nfusing restart\nfusing realise\nfusing roles\nfusing realised\nfusing reset\nfusing realist\nfusing reseal\nfusing rakesh\nfusing reales\nfusing reise\nfusing rakes\nfusing realidad\nfusing reseau\nfusing resear\nfusing researc\nfusing roleta\nfusing reine\nfusing raked\nfusing reale\nbeaker in\nbeaker i\nbeaker not\nbeaker my\nbeaker no\nbeaker now\nbeaker car\nbeaker none\nbeaker nor\nbeaker clip\nbeaker cake\nbeaker nose\nbeaker ceiling\nbeaker inline\nbeaker norm\nbeaker cease\nbeaker ceased\nbeaker nod\nbeaker inning\nbeaker chew\nbeaker chewing\nbeaker cakes\nbeaker cling\nbeaker ingress\nbeaker mylist\nbeaker ingres\nbeaker infuse\nbeaker norco\nbeaker cesar\nbeaker cline\nbeaker coles\nbeaker instal\nbeaker inlining\nbeaker chews\nbeaker cdata\nbeaker inest\nbeaker indole\nbeaker myles\nbeaker nosed\nbeaker infusing\nbeaker instar\nbeaker nolita\nbeaker nodal\nbeaker coleus\nbeaker myrinet\nbeaker infuses\nbeaker noline\nbeaker indain\nbeaker insta\nbeaker noakes\nbeaker mydata\nbeaker ctype\nbeaker indoles\nbeaker cinese\nrinsed a\nrinsed in\nrinsed with\nrinsed i\nrinsed be\nrinsed us\nrinsed do\nrinsed he\nrinsed list\nrinsed data\nrinsed best\nrinsed great\nrinsed line\nrinsed type\nrinsed being\nrinsed less\nrinsed east\nrinsed star\nrinsed past\nrinsed dog\nrinsed bed\nrinsed pain\nrinsed bear\nrinsed eat\nrinsed nine\nrinsed dot\nrinsed paint\nrinsed pan\nrinsed gray\nrinsed earn\nrinsed doing\nrinsed types\nrinsed bet\nrinsed dad\nrinsed pat\nrinsed ear\nrinsed lie\nrinsed pad\nrinsed dam\nrinsed bee\nrinsed inline\nrinsed lip\nrinsed typing\nrinsed leslie\nrinsed pasta\nrinsed lid\nrinsed pale\nrinsed lining\nrinsed staind\nrinsed pant\nrinsed espanol\nrinsed beg\nrinsed inning\nrinsed grin\nrinsed starch\nrinsed stalin\nrinsed benin\nrinsed espana\nrinsed ealing\nrinsed painless\nrinsed panty\nrinsed stain\nrinsed panda\nrinsed molina\nrinsed moline\nrinsed pains\nrinsed grind\nrinsed ingress\nrinsed prada\nrinsed lesben\nrinsed mollie\nrinsed dainty\nrinsed witty\nrinsed fusing\nrinsed beaker\nrinsed pandas\nrinsed bestality\nrinsed panning\nrinsed staines\nrinsed earch\nrinsed panini\nrinsed ingres\nrinsed infuse\nrinsed lista\nrinsed typeset\nrinsed doesn\nrinsed paine\nrinsed stang\nrinsed stale\nrinsed lingo\nrinsed esearch\nrinsed fuses\nrinsed beeing\nrinsed instal\nrinsed libel\nrinsed bening\nrinsed inlining\nrinsed belies\nrinsed greta\nrinsed inest\nrinsed indole\nrinsed molino\nrinsed dales\nrinsed prado\nrinsed molar\nrinsed moles\nrinsed does\nrinsed instar\nrinsed eidos\nrinsed beaked\nrinsed molest\nrinsed praline\nrinsed dainese\nrinsed stata\nrinsed estar\nrinsed darcs\nrinsed graying\nrinsed bestar\nrinsed panes\nrinsed dahmer\nrinsed grins\nrinsed infuses\nrinsed lidar\nrinsed liber\nrinsed dopant\nrinsed witless\nrinsed pango\nrinsed padang\nrinsed espada\nrinsed lesbe\nrinsed insta\nrinsed dangos\nrinsed listado\nrinsed mollis\nrinsed paling\nrinsed bedale\nrinsed pandan\nrinsed indoles\nrinsed tarina\nrinsed listar\nrinsed doling\nrinsed palin\nrinsed parco\nrinsed typeid\nrinsed daines\nrinsed parche\nrinsed nines\nrinsed done\npandas tag\npandas tale\npandas tap\npandas tales\npandas tango\npandas tapas\npandas edina\npandas taliesin\npandas taint\npandas taber\npandas talib\npandas tatars\npandas tatar\npandas tarina\npandas tangos\npandas talese\npandas talia\npandas tainty\nbestality pm\nbestality print\nbestality pay\nbestality pet\nbestality pin\nbestality paying\nbestality pine\nbestality pole\nbestality pearce\nbestality poles\nbestality pines\nbestality pinning\nbestality peirce\nbestality peseta\nbestality pineal\nbestality petal\nbestality pinole\nbestality pease\nbestality pinus\nbestality pesetas\nbestality polen\nbestality pinless\nbestality pindar\nbestality prine\nbestality prins\nbestality pineau\npanning re\npanning read\npanning research\npanning real\npanning role\npanning rest\npanning reality\npanning rear\npanning retain\npanning restart\npanning realise\npanning roles\npanning realised\npanning reset\npanning retains\npanning resins\npanning resin\npanning realist\npanning rinse\npanning reseal\npanning rinsed\npanning rakesh\npanning reales\npanning reise\npanning rakes\npanning realidad\npanning reseau\npanning resear\npanning researc\npanning roleta\npanning reine\npanning raked\npanning reale\nlipase do\nlipase day\nlipase deal\nlipase dead\nlipase dear\nlipase dinar\nlipase dakine\nlipase detain\nlipase dinning\nlipase dinesh\nlipase deity\nlipase dakar\nlipase dearing\nlipase deane\nlipase dearch\nlipase desing\nlipase detains\nlipase deine\nlipase dlese\nstaines a\nstaines with\nstaines be\nstaines us\nstaines do\nstaines he\nstaines data\nstaines great\nstaines line\nstaines type\nstaines less\nstaines dog\nstaines bed\nstaines ring\nstaines bear\nstaines eat\nstaines nine\nstaines dot\nstaines pan\nstaines gray\nstaines earn\nstaines dose\nstaines bet\nstaines dad\nstaines pat\nstaines ear\nstaines ease\nstaines lie\nstaines pad\nstaines dam\nstaines bee\nstaines lip\nstaines leslie\nstaines lid\nstaines pale\nstaines lining\nstaines pant\nstaines grease\nstaines beg\nstaines daring\nstaines benin\nstaines ealing\nstaines panty\nstaines panda\nstaines prada\nstaines bering\nstaines lesben\nstaines rinse\nstaines greased\nstaines mollie\nstaines witty\nstaines beaker\nstaines rinsed\nstaines pandas\nstaines panning\nstaines lipase\nstaines earch\nstaines dorint\nstaines paring\nstaines liberi\nstaines lingo\nstaines beeing\nstaines libel\nstaines bening\nstaines darin\nstaines greta\nstaines eased\nstaines dales\nstaines seddon\nstaines prado\nstaines molar\nstaines eidos\nstaines sedaka\nstaines beaked\nstaines praline\nstaines darcs\nstaines graying\nstaines dahmer\nstaines lidar\nstaines liber\nstaines dopant\nstaines witless\nstaines pango\nstaines padang\nstaines earing\nstaines lesbe\nstaines dangos\nstaines mollis\nstaines paling\nstaines bedale\nstaines pandan\nstaines fussed\nstaines lipari\nstaines doling\nstaines palin\nstaines parco\nstaines typeid\nstaines parche\nstaines done\npastas edina\nearch me\nearch meet\nearch medal\nearch metart\nearch merino\nearch medalist\nearch ewing\nearch melia\nearch meakin\nearch metar\nearch meine\nearch menino\nearch menina\nearch melita\nearch meines\ndorint a\ndorint area\ndorint art\ndorint areas\ndorint arm\ndorint aretha\ndorint arpanet\ndorint arles\ndorint arline\ndorint areal\nparing re\nparing read\nparing research\nparing real\nparing role\nparing rest\nparing reality\nparing rear\nparing retain\nparing restart\nparing realise\nparing roles\nparing realised\nparing reset\nparing retains\nparing resins\nparing resin\nparing realist\nparing rinse\nparing reseal\nparing rinsed\nparing rakesh\nparing reales\nparing reise\nparing rakes\nparing realidad\nparing reseau\nparing resear\nparing researc\nparing roleta\nparing reine\nparing raked\nparing reale\npanini no\npanini net\npanini near\npanini naked\npanini nest\npanini neat\npanini nearing\npanini neale\npanini nline\nliberi no\nliberi net\nliberi near\nliberi naked\nliberi nine\nliberi nest\nliberi neat\nliberi neale\nliberi naking\nliberi nines\ningres a\ningres with\ningres be\ningres us\ningres do\ningres he\ningres list\ningres data\ningres best\ningres line\ningres type\ningres less\ningres east\ningres star\ningres past\ningres bed\ningres bear\ningres eat\ningres nine\ningres dot\ningres pan\ningres earn\ningres dose\ningres bet\ningres dad\ningres pat\ningres ear\ningres ease\ningres lie\ningres pad\ningres dam\ningres bee\ningres lip\ningres leslie\ningres pasta\ningres lid\ningres pale\ningres pant\ningres starch\ningres stalin\ningres benin\ningres ealing\ningres panty\ningres panda\ningres prada\ningres lesben\ningres rinse\ningres mollie\ningres witty\ningres beaker\ningres rinsed\ningres pandas\ningres bestality\ningres lipase\ningres pastas\ningres earch\ningres dorint\ningres liberi\ningres lista\ningres stang\ningres stale\ningres lingo\ningres beeing\ningres libel\ningres darin\ningres eased\ningres dales\ningres seddon\ningres prado\ningres molar\ningres eidos\ningres sedaka\ningres beaked\ningres praline\ningres stata\ningres darcs\ningres bestar\ningres dahmer\ningres listas\ningres lidar\ningres liber\ningres dopant\ningres witless\ningres pango\ningres padang\ningres lesbe\ningres dangos\ningres listado\ningres mollis\ningres paling\ningres bedale\ningres pandan\ningres fussed\ningres listar\ningres lipari\ningres doling\ningres palin\ningres parco\ningres typeid\ningres parche\ningres done\ninfuse the\ninfuse a\ninfuse in\ninfuse i\ninfuse it\ninfuse at\ninfuse as\ninfuse search\ninfuse their\ninfuse so\ninfuse am\ninfuse these\ninfuse she\ninfuse set\ninfuse start\ninfuse say\ninfuse star\ninfuse ad\ninfuse sea\ninfuse thus\ninfuse ie\ninfuse army\ninfuse saying\ninfuse tag\ninfuse seat\ninfuse arm\ninfuse seal\ninfuse stars\ninfuse ah\ninfuse sole\ninfuse slip\ninfuse starring\ninfuse tale\ninfuse tap\ninfuse swim\ninfuse adobe\ninfuse sake\ninfuse tales\ninfuse spanning\ninfuse sealing\ninfuse stare\ninfuse sparc\ninfuse theta\ninfuse starr\ninfuse sprang\ninfuse tango\ninfuse ahmed\ninfuse aarhus\ninfuse stardom\ninfuse starling\ninfuse sling\ninfuse spans\ninfuse sparing\ninfuse soledad\ninfuse searing\ninfuse sparco\ninfuse seabed\ninfuse seine\ninfuse seibel\ninfuse adoring\ninfuse soleus\ninfuse slingo\ninfuse alesse\ninfuse soles\ninfuse adorno\ninfuse irina\ninfuse tapas\ninfuse adaline\ninfuse seadoo\ninfuse tatarstan\ninfuse sayin\ninfuse spandau\ninfuse adorn\ninfuse sparcs\ninfuse theist\ninfuse sealine\ninfuse searcg\ninfuse swims\ninfuse searcn\ninfuse searcb\ninfuse sdarch\ninfuse searcu\ninfuse searct\ninfuse searcm\ninfuse starline\ninfuse taber\ninfuse talib\ninfuse tatars\ninfuse tatar\ninfuse idabel\ninfuse tarina\ninfuse spaeth\ninfuse tangos\ninfuse alist\ninfuse aline\ninfuse seale\ninfuse searc\ninfuse seastar\ninfuse adamo\ninfuse astar\ninfuse ingot\ninfuse talese\ninfuse starlit\ninfuse agreing\ninfuse sakes\ninfuse theanine\ninfuse talia\ninfuse ahmet\ninfuse arche\ninfuse spanne\ninfuse sakar\nlista a\nlista in\nlista with\nlista i\nlista be\nlista us\nlista do\nlista he\nlista data\nlista great\nlista type\nlista being\nlista less\nlista dog\nlista bed\nlista ring\nlista pain\nlista bear\nlista eat\nlista nine\nlista dot\nlista paint\nlista pan\nlista gray\nlista earn\nlista doing\nlista types\nlista dose\nlista bet\nlista dad\nlista pat\nlista ear\nlista ease\nlista pad\nlista dam\nlista bee\nlista typing\nlista pale\nlista pant\nlista grease\nlista espanol\nlista beg\nlista inning\nlista grin\nlista daring\nlista benin\nlista espana\nlista painless\nlista panty\nlista panda\nlista molina\nlista moline\nlista pains\nlista grind\nlista ingress\nlista prada\nlista bering\nlista lesben\nlista rinse\nlista greased\nlista dainty\nlista witty\nlista fusing\nlista beaker\nlista rinsed\nlista pandas\nlista panning\nlista earch\nlista dorint\nlista paring\nlista panini\nlista ingres\nlista infuse\nlista typeset\nlista doesn\nlista paine\nlista esearch\nlista fuses\nlista beeing\nlista bening\nlista darin\nlista greta\nlista inest\nlista eased\nlista indole\nlista molino\nlista dales\nlista seddon\nlista prado\nlista molar\nlista moles\nlista does\nlista eidos\nlista sedaka\nlista beaked\nlista molest\nlista dainese\nlista estar\nlista darcs\nlista graying\nlista panes\nlista dahmer\nlista grins\nlista infuses\nlista dopant\nlista witless\nlista pango\nlista padang\nlista earing\nlista espada\nlista lesbe\nlista dangos\nlista bedale\nlista pandan\nlista indoles\nlista fussed\nlista tarina\nlista parco\nlista typeid\nlista daines\nlista parche\nlista nines\nlista done\ntypeset a\ntypeset at\ntypeset as\ntypeset he\ntypeset am\ntypeset head\ntypeset ad\ntypeset hi\ntypeset hear\ntypeset army\ntypeset heat\ntypeset hearing\ntypeset hole\ntypeset arm\ntypeset ha\ntypeset healing\ntypeset ah\ntypeset adobe\ntypeset hint\ntypeset hay\ntypeset heal\ntypeset holes\ntypeset atari\ntypeset ahmed\ntypeset aarhus\ntypeset adoring\ntypeset alesse\ntypeset adorno\ntypeset adaline\ntypeset hearn\ntypeset arcing\ntypeset adorn\ntypeset heist\ntypeset aearch\ntypeset heine\ntypeset heilig\ntypeset alist\ntypeset aline\ntypeset holed\ntypeset adamo\ntypeset astar\ntypeset agreing\ntypeset ahmet\ntypeset arche\ntypeset alesina\ntypeset heise\ndoesn in\ndoesn i\ndoesn be\ndoesn go\ndoesn buy\ndoesn being\ndoesn god\ndoesn bay\ndoesn bar\ndoesn bring\ndoesn bus\ndoesn going\ndoesn bear\ndoesn beat\ndoesn bet\ndoesn beast\ndoesn beam\ndoesn bearing\ndoesn bean\ndoesn inline\ndoesn bind\ndoesn blessed\ndoesn baking\ndoesn bless\ndoesn beastality\ndoesn bake\ndoesn baker\ndoesn baked\ndoesn goethe\ndoesn bling\ndoesn infuse\ndoesn binning\ndoesn instal\ndoesn brine\ndoesn goring\ndoesn betaine\ndoesn infusing\ndoesn beale\ndoesn instar\ndoesn brining\ndoesn indain\ndoesn insta\ndoesn gopal\ndoesn betas\ndoesn brinda\ndoesn bethea\ndoesn boles\ndoesn goole\ndoesn beane\ndoesn baying\ndoesn beilin\ndoesn blingo\ndoesn beaty\ndoesn busoni\npaine the\npaine a\npaine in\npaine i\npaine it\npaine at\npaine as\npaine if\npaine search\npaine their\npaine so\npaine am\npaine these\npaine she\npaine set\npaine start\npaine say\npaine star\npaine ad\npaine sea\npaine thus\npaine ie\npaine army\npaine saying\npaine tag\npaine seat\npaine arm\npaine seal\npaine stars\npaine ah\npaine sole\npaine slip\npaine starring\npaine tale\npaine tap\npaine swim\npaine adobe\npaine sake\npaine tales\npaine spanning\npaine sealing\npaine stare\npaine theta\npaine starr\npaine sprang\npaine tango\npaine ahmed\npaine aarhus\npaine stardom\npaine starling\npaine sling\npaine spans\npaine soledad\npaine searing\npaine seabed\npaine seine\npaine seibel\npaine adoring\npaine soleus\npaine slingo\npaine alesse\npaine soles\npaine adorno\npaine irina\npaine adaline\npaine seadoo\npaine tatarstan\npaine sayin\npaine spandau\npaine adorn\npaine theist\npaine sealine\npaine searcg\npaine swims\npaine searcn\npaine searcb\npaine sdarch\npaine searcu\npaine searct\npaine searcm\npaine starline\npaine taber\npaine talib\npaine tatars\npaine tatar\npaine idabel\npaine tarina\npaine tangos\npaine alist\npaine aline\npaine seale\npaine searc\npaine seastar\npaine adamo\npaine astar\npaine ingot\npaine talese\npaine starlit\npaine agreing\npaine sakes\npaine theanine\npaine talia\npaine ahmet\npaine arche\npaine spanne\npaine sakar\nstang of\nstang on\nstang or\nstang one\nstang oh\nstang oak\nstang oakdale\nstang oprah\nstang obese\nstang orinda\nstang orcinus\nstang oakes\nstang oline\nstale search\nstale so\nstale she\nstale set\nstale start\nstale say\nstale star\nstale sea\nstale saying\nstale seat\nstale sin\nstale seal\nstale stars\nstale sole\nstale spain\nstale slip\nstale sing\nstale starring\nstale swim\nstale sake\nstale staring\nstale spanning\nstale sealing\nstale stare\nstale sparc\nstale starr\nstale sprang\nstale stares\nstale stardom\nstale starling\nstale sinus\nstale sling\nstale spans\nstale sparing\nstale soledad\nstale searing\nstale sparco\nstale seabed\nstale sinead\nstale sprains\nstale seine\nstale seibel\nstale sprain\nstale soleus\nstale slingo\nstale soles\nstale sinning\nstale seadoo\nstale sayin\nstale spandau\nstale sparcs\nstale sealine\nstale searcg\nstale swims\nstale sesrch\nstale shewing\nstale searcn\nstale searcb\nstale sdarch\nstale searcu\nstale searct\nstale searcm\nstale starline\nstale spaeth\nstale swiming\nstale searc\nstale starlit\nstale sakes\nstale spanne\nstale sakar\nlingo a\nlingo in\nlingo with\nlingo i\nlingo be\nlingo us\nlingo do\nlingo he\nlingo data\nlingo best\nlingo great\nlingo type\nlingo being\nlingo less\nlingo east\nlingo star\nlingo past\nlingo dog\nlingo bed\nlingo ring\nlingo pain\nlingo bear\nlingo eat\nlingo nine\nlingo dot\nlingo paint\nlingo pan\nlingo gray\nlingo earn\nlingo doing\nlingo types\nlingo stainless\nlingo dose\nlingo bet\nlingo dad\nlingo pat\nlingo ear\nlingo ease\nlingo pad\nlingo dam\nlingo bee\nlingo typing\nlingo pasta\nlingo pale\nlingo staind\nlingo pant\nlingo grease\nlingo espanol\nlingo beg\nlingo staring\nlingo inning\nlingo grin\nlingo stains\nlingo daring\nlingo starch\nlingo benin\nlingo espana\nlingo painless\nlingo panty\nlingo stain\nlingo panda\nlingo molina\nlingo moline\nlingo pains\nlingo grind\nlingo ingress\nlingo prada\nlingo bering\nlingo lesben\nlingo rinse\nlingo greased\nlingo dainty\nlingo witty\nlingo fusing\nlingo beaker\nlingo rinsed\nlingo pandas\nlingo panning\nlingo staines\nlingo pastas\nlingo earch\nlingo dorint\nlingo paring\nlingo panini\nlingo ingres\nlingo infuse\nlingo typeset\nlingo doesn\nlingo paine\nlingo stale\nlingo esearch\nlingo fuses\nlingo instal\nlingo bening\nlingo darin\nlingo greta\nlingo inest\nlingo eased\nlingo indole\nlingo molino\nlingo dales\nlingo seddon\nlingo prado\nlingo molar\nlingo moles\nlingo does\nlingo instar\nlingo eidos\nlingo sedaka\nlingo beaked\nlingo molest\nlingo dainese\nlingo stata\nlingo estar\nlingo darcs\nlingo bestar\nlingo panes\nlingo dahmer\nlingo grins\nlingo infuses\nlingo dopant\nlingo witless\nlingo earing\nlingo espada\nlingo lesbe\nlingo insta\nlingo bedale\nlingo pandan\nlingo indoles\nlingo fussed\nlingo tarina\nlingo parco\nlingo typeid\nlingo daines\nlingo parche\nlingo nines\nlingo done\nesearch me\nesearch meet\nesearch medal\nesearch metart\nesearch merino\nesearch medalist\nesearch ewing\nesearch melia\nesearch meakin\nesearch metar\nesearch meine\nesearch menino\nesearch menina\nesearch melita\nfuses a\nfuses in\nfuses with\nfuses i\nfuses be\nfuses us\nfuses do\nfuses he\nfuses list\nfuses data\nfuses best\nfuses great\nfuses line\nfuses type\nfuses being\nfuses less\nfuses east\nfuses star\nfuses past\nfuses dog\nfuses bed\nfuses ring\nfuses pain\nfuses bear\nfuses eat\nfuses nine\nfuses dot\nfuses paint\nfuses pan\nfuses gray\nfuses earn\nfuses doing\nfuses stainless\nfuses dose\nfuses bet\nfuses dad\nfuses pat\nfuses ear\nfuses ease\nfuses lie\nfuses pad\nfuses dam\nfuses bee\nfuses inline\nfuses lip\nfuses typing\nfuses leslie\nfuses pasta\nfuses lid\nfuses pale\nfuses lining\nfuses staind\nfuses pant\nfuses grease\nfuses beg\nfuses staring\nfuses inning\nfuses grin\nfuses stains\nfuses daring\nfuses starch\nfuses stalin\nfuses benin\nfuses ealing\nfuses painless\nfuses panty\nfuses stain\nfuses panda\nfuses molina\nfuses moline\nfuses pains\nfuses grind\nfuses prada\nfuses bering\nfuses lesben\nfuses rinse\nfuses greased\nfuses mollie\nfuses dainty\nfuses witty\nfuses beaker\nfuses rinsed\nfuses pandas\nfuses bestality\nfuses panning\nfuses lipase\nfuses pastas\nfuses earch\nfuses dorint\nfuses paring\nfuses panini\nfuses liberi\nfuses lista\nfuses paine\nfuses stang\nfuses stale\nfuses lingo\nfuses beeing\nfuses instal\nfuses libel\nfuses bening\nfuses inlining\nfuses darin\nfuses greta\nfuses eased\nfuses indole\nfuses molino\nfuses dales\nfuses seddon\nfuses prado\nfuses molar\nfuses instar\nfuses eidos\nfuses sedaka\nfuses beaked\nfuses praline\nfuses stata\nfuses darcs\nfuses graying\nfuses bestar\nfuses dahmer\nfuses grins\nfuses listas\nfuses lidar\nfuses liber\nfuses dopant\nfuses witless\nfuses pango\nfuses padang\nfuses earing\nfuses lesbe\nfuses insta\nfuses dangos\nfuses listado\nfuses mollis\nfuses paling\nfuses bedale\nfuses pandan\nfuses indoles\nfuses tarina\nfuses listar\nfuses lipari\nfuses doling\nfuses palin\nfuses parco\nfuses typeid\nfuses parche\nfuses done\nbeeing of\nbeeing on\nbeeing or\nbeeing one\nbeeing oh\nbeeing oak\nbeeing oakdale\nbeeing oprah\nbeeing orinda\nbeeing orcinus\nbeeing oakes\nbeeing oline\ninstal in\ninstal i\ninstal it\ninstal if\ninstal ie\ninstal espanol\ninstal espana\ninstal esearch\ninstal irina\ninstal estar\ninstal espada\ninstal idabel\ninstal ingot\nlibel espanol\nlibel espana\nlibel esearch\nlibel estar\nlibel espada\nbening re\nbening read\nbening research\nbening real\nbening role\nbening rest\nbening reality\nbening rear\nbening retain\nbening restart\nbening realise\nbening roles\nbening realised\nbening reset\nbening retains\nbening resins\nbening resin\nbening realist\nbening rinse\nbening reseal\nbening rinsed\nbening rakesh\nbening reales\nbening reise\nbening rakes\nbening realidad\nbening reseau\nbening resear\nbening researc\nbening roleta\nbening reine\nbening raked\nbening reale\ninlining re\ninlining read\ninlining research\ninlining real\ninlining role\ninlining rest\ninlining rear\ninlining restart\ninlining roles\ninlining reset\ninlining reseal\ninlining rakesh\ninlining reales\ninlining reise\ninlining rakes\ninlining reseau\ninlining resear\ninlining researc\ninlining roleta\ninlining reine\ninlining raked\ninlining reale\ndarin a\ndarin in\ndarin with\ndarin i\ndarin be\ndarin us\ndarin do\ndarin he\ndarin list\ndarin best\ndarin great\ndarin line\ndarin type\ndarin being\ndarin less\ndarin east\ndarin star\ndarin past\ndarin dog\ndarin bed\ndarin pain\ndarin bear\ndarin eat\ndarin nine\ndarin dot\ndarin paint\ndarin pan\ndarin gray\ndarin earn\ndarin doing\ndarin types\ndarin stainless\ndarin dose\ndarin bet\ndarin pat\ndarin ear\ndarin ease\ndarin lie\ndarin pad\ndarin bee\ndarin inline\ndarin lip\ndarin typing\ndarin leslie\ndarin pasta\ndarin lid\ndarin pale\ndarin lining\ndarin staind\ndarin pant\ndarin grease\ndarin espanol\ndarin beg\ndarin inning\ndarin grin\ndarin stains\ndarin starch\ndarin stalin\ndarin benin\ndarin espana\ndarin ealing\ndarin painless\ndarin panty\ndarin stain\ndarin molina\ndarin moline\ndarin pains\ndarin grind\ndarin ingress\ndarin lesben\ndarin greased\ndarin mollie\ndarin witty\ndarin fusing\ndarin beaker\ndarin bestality\ndarin panning\ndarin lipase\ndarin staines\ndarin pastas\ndarin earch\ndarin panini\ndarin ingres\ndarin infuse\ndarin lista\ndarin typeset\ndarin doesn\ndarin paine\ndarin stang\ndarin stale\ndarin lingo\ndarin esearch\ndarin fuses\ndarin beeing\ndarin instal\ndarin libel\ndarin bening\ndarin inlining\ndarin belies\ndarin greta\ndarin inest\ndarin eased\ndarin indole\ndarin molino\ndarin seddon\ndarin prado\ndarin molar\ndarin moles\ndarin does\ndarin instar\ndarin eidos\ndarin sedaka\ndarin beaked\ndarin molest\ndarin praline\ndarin stata\ndarin estar\ndarin graying\ndarin bestar\ndarin panes\ndarin grins\ndarin infuses\ndarin listas\ndarin liber\ndarin dopant\ndarin witless\ndarin pango\ndarin lesbe\ndarin insta\ndarin listado\ndarin mollis\ndarin paling\ndarin indoles\ndarin fussed\ndarin tarina\ndarin listar\ndarin doling\ndarin palin\ndarin parco\ndarin typeid\ndarin parche\ndarin nines\ndarin done\nbelies a\nbelies in\nbelies with\nbelies i\nbelies us\nbelies do\nbelies he\nbelies data\nbelies great\nbelies type\nbelies less\nbelies east\nbelies star\nbelies past\nbelies dog\nbelies ring\nbelies pain\nbelies eat\nbelies nine\nbelies dot\nbelies paint\nbelies pan\nbelies gray\nbelies earn\nbelies doing\nbelies stainless\nbelies dose\nbelies dad\nbelies pat\nbelies ear\nbelies ease\nbelies pad\nbelies dam\nbelies typing\nbelies pasta\nbelies pale\nbelies staind\nbelies pant\nbelies grease\nbelies staring\nbelies inning\nbelies grin\nbelies stains\nbelies daring\nbelies starch\nbelies painless\nbelies panty\nbelies stain\nbelies panda\nbelies molina\nbelies moline\nbelies pains\nbelies grind\nbelies prada\nbelies rinse\nbelies greased\nbelies dainty\nbelies witty\nbelies fusing\nbelies rinsed\nbelies pandas\nbelies panning\nbelies pastas\nbelies earch\nbelies dorint\nbelies paring\nbelies panini\nbelies infuse\nbelies paine\nbelies stang\nbelies stale\nbelies instal\nbelies darin\nbelies greta\nbelies eased\nbelies indole\nbelies molino\nbelies dales\nbelies seddon\nbelies prado\nbelies molar\nbelies instar\nbelies eidos\nbelies sedaka\nbelies stata\nbelies darcs\nbelies graying\nbelies dahmer\nbelies grins\nbelies dopant\nbelies witless\nbelies pango\nbelies padang\nbelies earing\nbelies insta\nbelies dangos\nbelies pandan\nbelies indoles\nbelies fussed\nbelies tarina\nbelies parco\nbelies typeid\nbelies parche\nbelies done\ngreta a\ngreta in\ngreta with\ngreta i\ngreta be\ngreta us\ngreta do\ngreta he\ngreta list\ngreta data\ngreta best\ngreta line\ngreta type\ngreta less\ngreta east\ngreta star\ngreta past\ngreta bed\ngreta pain\ngreta bear\ngreta eat\ngreta nine\ngreta dot\ngreta paint\ngreta pan\ngreta earn\ngreta types\ngreta stainless\ngreta dose\ngreta bet\ngreta dad\ngreta pat\ngreta ear\ngreta ease\ngreta lie\ngreta pad\ngreta dam\ngreta bee\ngreta inline\ngreta lip\ngreta leslie\ngreta pasta\ngreta lid\ngreta pale\ngreta staind\ngreta pant\ngreta espanol\ngreta stains\ngreta starch\ngreta stalin\ngreta benin\ngreta espana\ngreta ealing\ngreta painless\ngreta panty\ngreta stain\ngreta panda\ngreta molina\ngreta moline\ngreta pains\ngreta prada\ngreta lesben\ngreta rinse\ngreta mollie\ngreta dainty\ngreta witty\ngreta beaker\ngreta rinsed\ngreta pandas\ngreta bestality\ngreta lipase\ngreta staines\ngreta pastas\ngreta earch\ngreta dorint\ngreta panini\ngreta liberi\ngreta infuse\ngreta lista\ngreta typeset\ngreta doesn\ngreta paine\ngreta stang\ngreta stale\ngreta lingo\ngreta esearch\ngreta fuses\ngreta beeing\ngreta instal\ngreta libel\ngreta darin\ngreta belies\ngreta inest\ngreta eased\ngreta indole\ngreta molino\ngreta dales\ngreta seddon\ngreta prado\ngreta molar\ngreta moles\ngreta does\ngreta instar\ngreta eidos\ngreta sedaka\ngreta beaked\ngreta molest\ngreta praline\ngreta dainese\ngreta stata\ngreta estar\ngreta darcs\ngreta bestar\ngreta panes\ngreta dahmer\ngreta infuses\ngreta listas\ngreta lidar\ngreta liber\ngreta dopant\ngreta witless\ngreta pango\ngreta padang\ngreta espada\ngreta lesbe\ngreta insta\ngreta dangos\ngreta listado\ngreta mollis\ngreta paling\ngreta bedale\ngreta pandan\ngreta indoles\ngreta fussed\ngreta tarina\ngreta listar\ngreta lipari\ngreta doling\ngreta palin\ngreta parco\ngreta typeid\ngreta daines\ngreta parche\ngreta nines\ngreta done\ninest a\ninest area\ninest art\ninest areas\ninest arm\ninest aretha\ninest arpanet\ninest arles\ninest arline\ninest areal\neased a\neased in\neased with\neased i\neased be\neased us\neased do\neased he\neased list\neased data\neased best\neased line\neased type\neased being\neased less\neased star\neased past\neased dog\neased bed\neased ring\neased pain\neased bear\neased nine\neased dot\neased paint\neased pan\neased gray\neased doing\neased types\neased bet\neased dad\neased pat\neased lie\neased pad\neased dam\neased bee\neased inline\neased lip\neased typing\neased leslie\neased pasta\neased lid\neased pale\neased lining\neased staind\neased pant\neased espanol\neased beg\neased staring\neased inning\neased grin\neased daring\neased starch\neased stalin\neased benin\neased espana\neased painless\neased panty\neased stain\neased panda\neased molina\neased moline\neased pains\neased grind\neased ingress\neased prada\neased bering\neased lesben\neased mollie\neased dainty\neased witty\neased fusing\neased beaker\neased pandas\neased bestality\neased panning\neased staines\neased dorint\neased paring\neased panini\neased liberi\neased ingres\neased infuse\neased lista\neased typeset\neased doesn\neased paine\neased stang\neased stale\neased lingo\neased fuses\neased beeing\neased instal\neased libel\neased bening\neased inlining\neased darin\neased belies\neased greta\neased inest\neased indole\neased molino\neased dales\neased prado\neased molar\neased moles\neased does\neased instar\neased eidos\neased beaked\neased molest\neased praline\neased dainese\neased stata\neased estar\neased darcs\neased graying\neased bestar\neased panes\neased dahmer\neased grins\neased infuses\neased lidar\neased liber\neased dopant\neased witless\neased pango\neased padang\neased espada\neased lesbe\neased insta\neased dangos\neased listado\neased mollis\neased paling\neased bedale\neased pandan\neased indoles\neased tarina\neased listar\neased lipari\neased doling\neased palin\neased parco\neased typeid\neased daines\neased parche\neased nines\neased done\nindole search\nindole so\nindole she\nindole set\nindole start\nindole say\nindole star\nindole sea\nindole saying\nindole seat\nindole seal\nindole stars\nindole sole\nindole slip\nindole starring\nindole swim\nindole sake\nindole spanning\nindole sealing\nindole stare\nindole sparc\nindole starr\nindole sprang\nindole stares\nindole starling\nindole sling\nindole spans\nindole sparing\nindole searing\nindole sparco\nindole seabed\nindole seine\nindole seibel\nindole soleus\nindole slingo\nindole soles\nindole sayin\nindole spandau\nindole sparcs\nindole sealine\nindole searcg\nindole swims\nindole sesrch\nindole searcn\nindole searcb\nindole sdarch\nindole searcu\nindole searct\nindole searcm\nindole starline\nindole spaeth\nindole searc\nindole seastar\nindole starlit\nindole sakes\nindole spanne\nindole sakar\nmolino let\nmolino leg\nmolino leaked\nmolino leaks\nmolino lenin\nmolino lestat\nmolino leben\nmolino lehmer\nmolino lerche\nmolino leprae\nmolino lening\nmolino leake\nmolino lewitt\ndales a\ndales in\ndales with\ndales i\ndales be\ndales us\ndales do\ndales he\ndales list\ndales best\ndales great\ndales line\ndales type\ndales being\ndales east\ndales star\ndales past\ndales dog\ndales bed\ndales ring\ndales pain\ndales bear\ndales eat\ndales nine\ndales dot\ndales paint\ndales pan\ndales gray\ndales earn\ndales doing\ndales types\ndales dose\ndales bet\ndales pat\ndales ear\ndales ease\ndales lie\ndales pad\ndales bee\ndales inline\ndales lip\ndales typing\ndales pasta\ndales lid\ndales lining\ndales staind\ndales pant\ndales grease\ndales espanol\ndales beg\ndales staring\ndales inning\ndales grin\ndales stains\ndales starch\ndales stalin\ndales benin\ndales espana\ndales ealing\ndales panty\ndales stain\ndales molina\ndales moline\ndales pains\ndales grind\ndales ingress\ndales bering\ndales rinse\ndales greased\ndales mollie\ndales witty\ndales fusing\ndales beaker\ndales rinsed\ndales bestality\ndales panning\ndales lipase\ndales staines\ndales pastas\ndales earch\ndales dorint\ndales paring\ndales panini\ndales liberi\ndales ingres\ndales infuse\ndales lista\ndales typeset\ndales doesn\ndales paine\ndales stang\ndales lingo\ndales esearch\ndales fuses\ndales beeing\ndales instal\ndales bening\ndales inlining\ndales belies\ndales greta\ndales inest\ndales eased\ndales molino\ndales seddon\ndales prado\ndales molar\ndales moles\ndales does\ndales instar\ndales eidos\ndales sedaka\ndales beaked\ndales molest\ndales praline\ndales stata\ndales estar\ndales graying\ndales bestar\ndales panes\ndales grins\ndales infuses\ndales listas\ndales liber\ndales dopant\ndales pango\ndales earing\ndales insta\ndales listado\ndales mollis\ndales paling\ndales fussed\ndales tarina\ndales listar\ndales lipari\ndales doling\ndales palin\ndales parco\ndales typeid\ndales parche\ndales nines\ndales done\nseddon in\nseddon i\nseddon be\nseddon go\nseddon buy\nseddon best\nseddon being\nseddon god\nseddon bay\nseddon bar\nseddon bring\nseddon bus\nseddon going\nseddon bear\nseddon beat\nseddon bet\nseddon beast\nseddon beam\nseddon bearing\nseddon bean\nseddon inline\nseddon bind\nseddon baking\nseddon bless\nseddon beastality\nseddon bake\nseddon baker\nseddon bethesda\nseddon baked\nseddon goethe\nseddon ingress\nseddon bling\nseddon ingres\nseddon infuse\nseddon binning\nseddon instal\nseddon brine\nseddon inest\nseddon goring\nseddon betaine\nseddon infusing\nseddon beale\nseddon instar\nseddon bestar\nseddon infuses\nseddon brining\nseddon beset\nseddon indain\nseddon insta\nseddon gopal\nseddon betas\nseddon brinda\nseddon bethea\nseddon boles\nseddon goole\nseddon beane\nseddon brines\nseddon baying\nseddon beilin\nseddon blingo\nseddon beaty\nseddon busoni\nprado a\nprado in\nprado with\nprado i\nprado be\nprado us\nprado he\nprado list\nprado data\nprado best\nprado great\nprado line\nprado type\nprado being\nprado less\nprado east\nprado star\nprado past\nprado bed\nprado ring\nprado pain\nprado bear\nprado eat\nprado nine\nprado paint\nprado pan\nprado gray\nprado earn\nprado types\nprado stainless\nprado bet\nprado pat\nprado ear\nprado ease\nprado lie\nprado pad\nprado dam\nprado bee\nprado inline\nprado lip\nprado typing\nprado leslie\nprado pasta\nprado lid\nprado pale\nprado lining\nprado staind\nprado pant\nprado grease\nprado espanol\nprado beg\nprado staring\nprado inning\nprado grin\nprado stains\nprado daring\nprado starch\nprado stalin\nprado benin\nprado espana\nprado ealing\nprado painless\nprado panty\nprado stain\nprado panda\nprado molina\nprado moline\nprado pains\nprado grind\nprado ingress\nprado bering\nprado lesben\nprado rinse\nprado greased\nprado mollie\nprado dainty\nprado witty\nprado fusing\nprado beaker\nprado rinsed\nprado pandas\nprado bestality\nprado panning\nprado lipase\nprado staines\nprado pastas\nprado earch\nprado paring\nprado panini\nprado liberi\nprado ingres\nprado infuse\nprado lista\nprado typeset\nprado paine\nprado stang\nprado stale\nprado lingo\nprado esearch\nprado fuses\nprado beeing\nprado instal\nprado libel\nprado bening\nprado inlining\nprado darin\nprado belies\nprado greta\nprado inest\nprado eased\nprado molino\nprado dales\nprado molar\nprado moles\nprado instar\nprado sedaka\nprado beaked\nprado molest\nprado dainese\nprado stata\nprado estar\nprado darcs\nprado graying\nprado bestar\nprado panes\nprado dahmer\nprado grins\nprado infuses\nprado listas\nprado lidar\nprado liber\nprado witless\nprado pango\nprado padang\nprado earing\nprado espada\nprado lesbe\nprado insta\nprado dangos\nprado mollis\nprado paling\nprado bedale\nprado pandan\nprado fussed\nprado tarina\nprado listar\nprado lipari\nprado palin\nprado parco\nprado typeid\nprado daines\nprado parche\nprado nines\nmolar he\nmolar head\nmolar hi\nmolar hear\nmolar heat\nmolar hearing\nmolar hole\nmolar ha\nmolar healing\nmolar hint\nmolar hay\nmolar heal\nmolar holes\nmolar hines\nmolar hesse\nmolar hearn\nmolar heist\nmolar heshe\nmolar heine\nmolar heilig\nmolar holed\nmolar heise\nmoles a\nmoles in\nmoles with\nmoles i\nmoles be\nmoles us\nmoles do\nmoles he\nmoles list\nmoles data\nmoles best\nmoles great\nmoles line\nmoles type\nmoles being\nmoles less\nmoles east\nmoles star\nmoles past\nmoles dog\nmoles bed\nmoles ring\nmoles pain\nmoles bear\nmoles eat\nmoles nine\nmoles dot\nmoles paint\nmoles pan\nmoles gray\nmoles earn\nmoles doing\nmoles stainless\nmoles dose\nmoles bet\nmoles dad\nmoles pat\nmoles ear\nmoles ease\nmoles lie\nmoles pad\nmoles bee\nmoles inline\nmoles lip\nmoles typing\nmoles leslie\nmoles pasta\nmoles lid\nmoles pale\nmoles lining\nmoles staind\nmoles pant\nmoles grease\nmoles beg\nmoles staring\nmoles inning\nmoles grin\nmoles stains\nmoles daring\nmoles starch\nmoles stalin\nmoles benin\nmoles ealing\nmoles painless\nmoles panty\nmoles stain\nmoles panda\nmoles pains\nmoles grind\nmoles prada\nmoles bering\nmoles lesben\nmoles rinse\nmoles greased\nmoles dainty\nmoles witty\nmoles fusing\nmoles beaker\nmoles rinsed\nmoles pandas\nmoles bestality\nmoles panning\nmoles lipase\nmoles pastas\nmoles earch\nmoles dorint\nmoles paring\nmoles panini\nmoles liberi\nmoles infuse\nmoles lista\nmoles paine\nmoles stang\nmoles stale\nmoles lingo\nmoles beeing\nmoles instal\nmoles libel\nmoles bening\nmoles inlining\nmoles darin\nmoles greta\nmoles eased\nmoles indole\nmoles dales\nmoles seddon\nmoles prado\nmoles instar\nmoles eidos\nmoles sedaka\nmoles beaked\nmoles praline\nmoles stata\nmoles darcs\nmoles graying\nmoles bestar\nmoles dahmer\nmoles grins\nmoles listas\nmoles lidar\nmoles liber\nmoles dopant\nmoles witless\nmoles pango\nmoles padang\nmoles earing\nmoles lesbe\nmoles insta\nmoles dangos\nmoles listado\nmoles paling\nmoles bedale\nmoles pandan\nmoles indoles\nmoles fussed\nmoles tarina\nmoles listar\nmoles lipari\nmoles doling\nmoles palin\nmoles parco\nmoles typeid\nmoles parche\nmoles done\ndoes a\ndoes in\ndoes with\ndoes i\ndoes be\ndoes us\ndoes he\ndoes list\ndoes data\ndoes best\ndoes great\ndoes line\ndoes type\ndoes being\ndoes less\ndoes east\ndoes star\ndoes past\ndoes bed\ndoes ring\ndoes pain\ndoes bear\ndoes eat\ndoes nine\ndoes paint\ndoes pan\ndoes gray\ndoes earn\ndoes stainless\ndoes bet\ndoes pat\ndoes ear\ndoes ease\ndoes lie\ndoes pad\ndoes dam\ndoes bee\ndoes inline\ndoes lip\ndoes typing\ndoes leslie\ndoes pasta\ndoes lid\ndoes pale\ndoes lining\ndoes staind\ndoes pant\ndoes grease\ndoes beg\ndoes staring\ndoes inning\ndoes grin\ndoes stains\ndoes daring\ndoes starch\ndoes stalin\ndoes benin\ndoes ealing\ndoes painless\ndoes panty\ndoes stain\ndoes panda\ndoes molina\ndoes moline\ndoes pains\ndoes grind\ndoes prada\ndoes bering\ndoes lesben\ndoes rinse\ndoes greased\ndoes mollie\ndoes dainty\ndoes witty\ndoes fusing\ndoes beaker\ndoes rinsed\ndoes pandas\ndoes bestality\ndoes panning\ndoes lipase\ndoes pastas\ndoes earch\ndoes paring\ndoes panini\ndoes liberi\ndoes infuse\ndoes lista\ndoes paine\ndoes stang\ndoes stale\ndoes lingo\ndoes beeing\ndoes instal\ndoes libel\ndoes bening\ndoes inlining\ndoes darin\ndoes greta\ndoes eased\ndoes molino\ndoes dales\ndoes molar\ndoes instar\ndoes sedaka\ndoes beaked\ndoes praline\ndoes stata\ndoes darcs\ndoes graying\ndoes bestar\ndoes dahmer\ndoes grins\ndoes listas\ndoes lidar\ndoes liber\ndoes witless\ndoes pango\ndoes padang\ndoes earing\ndoes lesbe\ndoes insta\ndoes dangos\ndoes mollis\ndoes paling\ndoes bedale\ndoes pandan\ndoes fussed\ndoes tarina\ndoes listar\ndoes lipari\ndoes palin\ndoes parco\ndoes typeid\ndoes parche\ninstar in\ninstar i\ninstar not\ninstar my\ninstar no\ninstar now\ninstar car\ninstar none\ninstar nor\ninstar clip\ninstar cake\ninstar nose\ninstar ceiling\ninstar inline\ninstar norm\ninstar cease\ninstar ceased\ninstar nod\ninstar inning\ninstar chew\ninstar cakes\ninstar cling\ninstar nobel\ninstar ingress\ninstar ingres\ninstar infuse\ninstar norco\ninstar cesar\ninstar cline\ninstar coles\ninstar inlining\ninstar chews\ninstar cdata\ninstar inest\ninstar indole\ninstar myles\ninstar nosed\ninstar nolita\ninstar nodal\ninstar coleus\ninstar myrinet\ninstar infuses\ninstar noline\ninstar noakes\ninstar mydata\ninstar ctype\ninstar indoles\neidos tag\neidos tale\neidos tap\neidos tales\neidos tango\neidos tapas\neidos edina\neidos taliesin\neidos taint\neidos taber\neidos talib\neidos tatars\neidos tatar\neidos tarina\neidos tangos\neidos talese\neidos talia\neidos tainty\nsedaka rhino\nsedaka rhesus\nsedaka rhine\nsedaka keine\nsedaka kelis\nsedaka kedar\nsedaka rhein\nsedaka kepada\nsedaka kernow\nbeaked of\nbeaked a\nbeaked on\nbeaked or\nbeaked at\nbeaked as\nbeaked one\nbeaked am\nbeaked oh\nbeaked ad\nbeaked army\nbeaked arm\nbeaked oak\nbeaked ongoing\nbeaked ah\nbeaked atari\nbeaked oakdale\nbeaked ahmed\nbeaked oprah\nbeaked aarhus\nbeaked orinda\nbeaked orcinus\nbeaked adoring\nbeaked alesse\nbeaked adorno\nbeaked arcing\nbeaked adorn\nbeaked oakes\nbeaked aearch\nbeaked alist\nbeaked aline\nbeaked astar\nbeaked agreing\nbeaked ahmet\nbeaked arche\nbeaked oline\nbeaked alesina\nmolest a\nmolest area\nmolest art\nmolest areas\nmolest aretha\nmolest arpanet\nmolest arles\nmolest arline\nmolest areal\npraline of\npraline on\npraline or\npraline oh\npraline oak\npraline ongoing\npraline oakdale\npraline obese\npraline orinda\npraline orcinus\npraline oakes\ndainese the\ndainese a\ndainese in\ndainese i\ndainese it\ndainese at\ndainese as\ndainese if\ndainese their\ndainese am\ndainese ad\ndainese thus\ndainese ie\ndainese army\ndainese tag\ndainese arm\ndainese ah\ndainese tale\ndainese tap\ndainese adobe\ndainese tales\ndainese theta\ndainese tango\ndainese ahmed\ndainese aarhus\ndainese adoring\ndainese alesse\ndainese adorno\ndainese irina\ndainese tapas\ndainese tatarstan\ndainese adorn\ndainese theist\ndainese taber\ndainese talib\ndainese tatars\ndainese tatar\ndainese tarina\ndainese tangos\ndainese alist\ndainese aline\ndainese astar\ndainese ingot\ndainese talese\ndainese agreing\ndainese theanine\ndainese talia\ndainese ahmet\ndainese arche\nstata re\nstata read\nstata research\nstata real\nstata role\nstata ring\nstata rest\nstata reality\nstata rear\nstata retain\nstata realise\nstata roles\nstata realised\nstata reset\nstata retains\nstata resins\nstata resin\nstata rearing\nstata rinse\nstata reseal\nstata rinsed\nstata raking\nstata reining\nstata rakesh\nstata reales\nstata reise\nstata rakes\nstata realidad\nstata reseau\nstata resear\nstata researc\nstata reine\nstata raked\nstata reale\nestar a\nestar in\nestar with\nestar i\nestar be\nestar us\nestar do\nestar he\nestar list\nestar best\nestar great\nestar line\nestar type\nestar being\nestar less\nestar east\nestar star\nestar past\nestar dog\nestar bed\nestar ring\nestar pain\nestar bear\nestar eat\nestar nine\nestar dot\nestar paint\nestar pan\nestar gray\nestar earn\nestar doing\nestar stainless\nestar dose\nestar bet\nestar dad\nestar pat\nestar ear\nestar ease\nestar lie\nestar pad\nestar dam\nestar bee\nestar inline\nestar lip\nestar typing\nestar leslie\nestar pasta\nestar lid\nestar pale\nestar lining\nestar staind\nestar pant\nestar grease\nestar beg\nestar staring\nestar inning\nestar grin\nestar stains\nestar daring\nestar starch\nestar stalin\nestar benin\nestar ealing\nestar painless\nestar panty\nestar stain\nestar panda\nestar molina\nestar moline\nestar pains\nestar grind\nestar prada\nestar bering\nestar lesben\nestar rinse\nestar greased\nestar mollie\nestar dainty\nestar witty\nestar fusing\nestar beaker\nestar rinsed\nestar pandas\nestar bestality\nestar panning\nestar lipase\nestar pastas\nestar earch\nestar dorint\nestar paring\nestar panini\nestar liberi\nestar infuse\nestar lista\nestar paine\nestar stang\nestar stale\nestar lingo\nestar beeing\nestar instal\nestar libel\nestar bening\nestar inlining\nestar darin\nestar greta\nestar eased\nestar indole\nestar molino\nestar dales\nestar seddon\nestar prado\nestar molar\nestar instar\nestar eidos\nestar sedaka\nestar beaked\nestar praline\nestar darcs\nestar graying\nestar bestar\nestar dahmer\nestar grins\nestar listas\nestar lidar\nestar liber\nestar dopant\nestar witless\nestar pango\nestar padang\nestar earing\nestar lesbe\nestar insta\nestar dangos\nestar listado\nestar mollis\nestar paling\nestar bedale\nestar pandan\nestar indoles\nestar fussed\nestar listar\nestar lipari\nestar doling\nestar palin\nestar parco\nestar typeid\nestar parche\nestar done\ndarcs tag\ndarcs tale\ndarcs tap\ndarcs tales\ndarcs tango\ndarcs tapas\ndarcs edina\ndarcs taliesin\ndarcs taint\ndarcs taber\ndarcs talib\ndarcs tatars\ndarcs tatar\ndarcs tarina\ndarcs tangos\ndarcs talese\ndarcs talia\ndarcs tainty\ngraying of\ngraying on\ngraying or\ngraying one\ngraying oh\ngraying oak\ngraying oakdale\ngraying oprah\ngraying obese\ngraying orinda\ngraying orcinus\ngraying oakes\ngraying oline\nbestar in\nbestar i\nbestar not\nbestar my\nbestar no\nbestar now\nbestar car\nbestar none\nbestar nor\nbestar clip\nbestar cake\nbestar nose\nbestar ceiling\nbestar inline\nbestar norm\nbestar cease\nbestar ceased\nbestar nod\nbestar inning\nbestar chew\nbestar chewing\nbestar cakes\nbestar cling\nbestar ingress\nbestar ingres\nbestar infuse\nbestar norco\nbestar cesar\nbestar cline\nbestar coles\nbestar inlining\nbestar chews\nbestar cdata\nbestar inest\nbestar indole\nbestar myles\nbestar nosed\nbestar infusing\nbestar nolita\nbestar nodal\nbestar coleus\nbestar myrinet\nbestar infuses\nbestar noline\nbestar indain\nbestar noakes\nbestar mydata\nbestar ctype\nbestar indoles\nbestar cinese\npanes a\npanes in\npanes with\npanes i\npanes be\npanes us\npanes do\npanes he\npanes list\npanes data\npanes best\npanes great\npanes line\npanes type\npanes being\npanes less\npanes east\npanes star\npanes past\npanes dog\npanes bed\npanes ring\npanes pain\npanes bear\npanes eat\npanes nine\npanes dot\npanes paint\npanes pan\npanes gray\npanes earn\npanes doing\npanes stainless\npanes dose\npanes bet\npanes dad\npanes pat\npanes ear\npanes ease\npanes lie\npanes pad\npanes dam\npanes bee\npanes inline\npanes lip\npanes typing\npanes leslie\npanes pasta\npanes lid\npanes pale\npanes lining\npanes staind\npanes grease\npanes beg\npanes staring\npanes inning\npanes grin\npanes stains\npanes daring\npanes starch\npanes stalin\npanes benin\npanes ealing\npanes painless\npanes stain\npanes molina\npanes moline\npanes pains\npanes grind\npanes prada\npanes bering\npanes lesben\npanes rinse\npanes greased\npanes mollie\npanes dainty\npanes witty\npanes fusing\npanes beaker\npanes rinsed\npanes bestality\npanes lipase\npanes pastas\npanes earch\npanes dorint\npanes paring\npanes panini\npanes liberi\npanes infuse\npanes lista\npanes paine\npanes stang\npanes stale\npanes lingo\npanes beeing\npanes instal\npanes libel\npanes bening\npanes inlining\npanes darin\npanes greta\npanes eased\npanes indole\npanes molino\npanes dales\npanes seddon\npanes prado\npanes molar\npanes instar\npanes eidos\npanes sedaka\npanes beaked\npanes praline\npanes stata\npanes darcs\npanes graying\npanes bestar\npanes dahmer\npanes grins\npanes listas\npanes lidar\npanes liber\npanes witless\npanes pango\npanes padang\npanes earing\npanes lesbe\npanes insta\npanes dangos\npanes listado\npanes mollis\npanes paling\npanes bedale\npanes indoles\npanes fussed\npanes tarina\npanes listar\npanes lipari\npanes doling\npanes palin\npanes parco\npanes typeid\npanes parche\npanes done\ndahmer in\ndahmer i\ndahmer not\ndahmer my\ndahmer no\ndahmer now\ndahmer car\ndahmer none\ndahmer nor\ndahmer clip\ndahmer cake\ndahmer nose\ndahmer ceiling\ndahmer inline\ndahmer norm\ndahmer cease\ndahmer ceased\ndahmer nod\ndahmer inning\ndahmer chew\ndahmer chewing\ndahmer cakes\ndahmer cling\ndahmer nobel\ndahmer ingress\ndahmer mylist\ndahmer ingres\ndahmer infuse\ndahmer norco\ndahmer cesar\ndahmer cline\ndahmer coles\ndahmer instal\ndahmer inlining\ndahmer chews\ndahmer inest\ndahmer indole\ndahmer myles\ndahmer nosed\ndahmer infusing\ndahmer instar\ndahmer nolita\ndahmer coleus\ndahmer myrinet\ndahmer infuses\ndahmer noline\ndahmer insta\ndahmer noakes\ndahmer ctype\ndahmer indoles\ndahmer cinese\ngrins tale\ngrins tap\ngrins tales\ngrins tango\ngrins tapas\ngrins taber\ngrins talib\ngrins tatars\ngrins tatar\ngrins tarina\ngrins tangos\ngrins talese\ngrins talia\ninfuses a\ninfuses with\ninfuses be\ninfuses us\ninfuses do\ninfuses he\ninfuses list\ninfuses data\ninfuses best\ninfuses great\ninfuses line\ninfuses type\ninfuses less\ninfuses east\ninfuses star\ninfuses past\ninfuses dog\ninfuses bed\ninfuses ring\ninfuses bear\ninfuses eat\ninfuses nine\ninfuses dot\ninfuses pan\ninfuses gray\ninfuses earn\ninfuses dose\ninfuses bet\ninfuses dad\ninfuses pat\ninfuses ear\ninfuses ease\ninfuses lie\ninfuses pad\ninfuses dam\ninfuses bee\ninfuses lip\ninfuses leslie\ninfuses pasta\ninfuses lid\ninfuses pale\ninfuses lining\ninfuses pant\ninfuses grease\ninfuses beg\ninfuses staring\ninfuses daring\ninfuses starch\ninfuses stalin\ninfuses benin\ninfuses ealing\ninfuses panty\ninfuses panda\ninfuses prada\ninfuses bering\ninfuses lesben\ninfuses rinse\ninfuses greased\ninfuses mollie\ninfuses witty\ninfuses beaker\ninfuses rinsed\ninfuses pandas\ninfuses bestality\ninfuses panning\ninfuses lipase\ninfuses pastas\ninfuses earch\ninfuses dorint\ninfuses paring\ninfuses liberi\ninfuses lista\ninfuses stang\ninfuses stale\ninfuses lingo\ninfuses beeing\ninfuses libel\ninfuses bening\ninfuses darin\ninfuses greta\ninfuses eased\ninfuses dales\ninfuses seddon\ninfuses prado\ninfuses molar\ninfuses eidos\ninfuses sedaka\ninfuses beaked\ninfuses praline\ninfuses stata\ninfuses darcs\ninfuses graying\ninfuses bestar\ninfuses dahmer\ninfuses listas\ninfuses lidar\ninfuses liber\ninfuses dopant\ninfuses witless\ninfuses pango\ninfuses padang\ninfuses earing\ninfuses lesbe\ninfuses dangos\ninfuses listado\ninfuses mollis\ninfuses paling\ninfuses bedale\ninfuses pandan\ninfuses listar\ninfuses lipari\ninfuses doling\ninfuses palin\ninfuses parco\ninfuses typeid\ninfuses parche\ninfuses done\nlistas edina\nlidar in\nlidar i\nlidar not\nlidar my\nlidar no\nlidar now\nlidar car\nlidar none\nlidar nor\nlidar cake\nlidar nose\nlidar norm\nlidar cease\nlidar ceased\nlidar nod\nlidar inning\nlidar chew\nlidar chewing\nlidar cakes\nlidar nobel\nlidar ingress\nlidar ingres\nlidar infuse\nlidar norco\nlidar cesar\nlidar coles\nlidar instal\nlidar chews\nlidar inest\nlidar indole\nlidar myles\nlidar nosed\nlidar infusing\nlidar instar\nlidar coleus\nlidar myrinet\nlidar infuses\nlidar insta\nlidar noakes\nlidar ctype\nlidar indoles\nlidar cinese\nliber in\nliber i\nliber not\nliber my\nliber no\nliber now\nliber car\nliber none\nliber nor\nliber cake\nliber nose\nliber norm\nliber cease\nliber ceased\nliber nod\nliber inning\nliber chew\nliber chewing\nliber cakes\nliber ingress\nliber ingres\nliber infuse\nliber norco\nliber cesar\nliber coles\nliber instal\nliber chews\nliber cdata\nliber inest\nliber indole\nliber myles\nliber nosed\nliber infusing\nliber instar\nliber nodal\nliber coleus\nliber myrinet\nliber infuses\nliber indain\nliber insta\nliber noakes\nliber mydata\nliber ctype\nliber indoles\nliber cinese\ndopant a\ndopant area\ndopant art\ndopant areas\ndopant arm\ndopant aretha\ndopant arles\ndopant arline\ndopant areal\nwitless tag\nwitless tap\nwitless tango\nwitless tapas\nwitless edina\nwitless taliesin\nwitless taint\nwitless taber\nwitless talib\nwitless tatars\nwitless tatar\nwitless tarina\nwitless tangos\nwitless talia\nwitless tainty\npango a\npango in\npango with\npango i\npango be\npango us\npango do\npango he\npango list\npango data\npango best\npango great\npango line\npango type\npango being\npango less\npango east\npango star\npango dog\npango bed\npango ring\npango bear\npango eat\npango nine\npango dot\npango pan\npango gray\npango earn\npango doing\npango types\npango stainless\npango dose\npango bet\npango dad\npango ear\npango ease\npango lie\npango dam\npango bee\npango inline\npango lip\npango typing\npango leslie\npango lid\npango lining\npango staind\npango pant\npango grease\npango espanol\npango beg\npango staring\npango inning\npango grin\npango stains\npango daring\npango starch\npango stalin\npango benin\npango espana\npango panty\npango stain\npango panda\npango molina\npango moline\npango grind\npango ingress\npango prada\npango bering\npango lesben\npango rinse\npango greased\npango mollie\npango dainty\npango witty\npango fusing\npango beaker\npango rinsed\npango pandas\npango bestality\npango panning\npango staines\npango earch\npango dorint\npango liberi\npango ingres\npango infuse\npango lista\npango typeset\npango doesn\npango stale\npango esearch\npango fuses\npango instal\npango libel\npango bening\npango inlining\npango darin\npango belies\npango greta\npango inest\npango eased\npango indole\npango molino\npango dales\npango seddon\npango prado\npango molar\npango moles\npango does\npango instar\npango eidos\npango sedaka\npango beaked\npango molest\npango praline\npango dainese\npango stata\npango estar\npango darcs\npango bestar\npango panes\npango dahmer\npango grins\npango infuses\npango listas\npango lidar\npango liber\npango dopant\npango witless\npango earing\npango lesbe\npango insta\npango listado\npango mollis\npango bedale\npango pandan\npango indoles\npango fussed\npango tarina\npango listar\npango typeid\npango daines\npango nines\npango done\npadang of\npadang on\npadang or\npadang one\npadang oh\npadang oak\npadang oprah\npadang obese\npadang orcinus\npadang oakes\npadang oline\nearing re\nearing role\nearing rest\nearing retain\nearing restart\nearing roles\nearing reset\nearing retains\nearing resins\nearing resin\nearing rinse\nearing rinsed\nearing rakesh\nearing reise\nearing rakes\nearing roleta\nearing reine\nearing raked\nespada a\nespada in\nespada with\nespada i\nespada be\nespada us\nespada do\nespada he\nespada list\nespada best\nespada great\nespada line\nespada type\nespada being\nespada less\nespada east\nespada star\nespada dog\nespada bed\nespada ring\nespada bear\nespada eat\nespada nine\nespada dot\nespada pan\nespada gray\nespada earn\nespada doing\nespada stainless\nespada dose\nespada bet\nespada ear\nespada ease\nespada lie\nespada bee\nespada inline\nespada lip\nespada typing\nespada leslie\nespada lid\nespada lining\nespada staind\nespada pant\nespada grease\nespada beg\nespada staring\nespada inning\nespada grin\nespada stains\nespada starch\nespada stalin\nespada benin\nespada ealing\nespada panty\nespada stain\nespada molina\nespada moline\nespada grind\nespada bering\nespada lesben\nespada rinse\nespada greased\nespada mollie\nespada witty\nespada fusing\nespada beaker\nespada rinsed\nespada bestality\nespada panning\nespada earch\nespada dorint\nespada liberi\nespada infuse\nespada lista\nespada stang\nespada stale\nespada lingo\nespada beeing\nespada instal\nespada libel\nespada bening\nespada inlining\nespada greta\nespada eased\nespada indole\nespada molino\nespada seddon\nespada prado\nespada molar\nespada instar\nespada eidos\nespada sedaka\nespada beaked\nespada praline\nespada stata\nespada graying\nespada bestar\nespada grins\nespada listas\nespada liber\nespada dopant\nespada witless\nespada earing\nespada lesbe\nespada insta\nespada listado\nespada mollis\nespada indoles\nespada fussed\nespada tarina\nespada listar\nespada doling\nespada typeid\nespada done\nlesbe a\nlesbe in\nlesbe with\nlesbe i\nlesbe us\nlesbe do\nlesbe he\nlesbe list\nlesbe data\nlesbe great\nlesbe line\nlesbe type\nlesbe east\nlesbe star\nlesbe past\nlesbe dog\nlesbe ring\nlesbe pain\nlesbe eat\nlesbe nine\nlesbe dot\nlesbe paint\nlesbe pan\nlesbe gray\nlesbe earn\nlesbe doing\nlesbe types\nlesbe dose\nlesbe dad\nlesbe pat\nlesbe ear\nlesbe ease\nlesbe lie\nlesbe pad\nlesbe dam\nlesbe inline\nlesbe lip\nlesbe typing\nlesbe pasta\nlesbe lid\nlesbe lining\nlesbe staind\nlesbe pant\nlesbe grease\nlesbe espanol\nlesbe staring\nlesbe inning\nlesbe grin\nlesbe stains\nlesbe daring\nlesbe starch\nlesbe stalin\nlesbe espana\nlesbe ealing\nlesbe panty\nlesbe stain\nlesbe panda\nlesbe molina\nlesbe moline\nlesbe pains\nlesbe grind\nlesbe ingress\nlesbe prada\nlesbe rinse\nlesbe greased\nlesbe mollie\nlesbe dainty\nlesbe witty\nlesbe fusing\nlesbe rinsed\nlesbe pandas\nlesbe panning\nlesbe lipase\nlesbe staines\nlesbe pastas\nlesbe earch\nlesbe dorint\nlesbe paring\nlesbe panini\nlesbe ingres\nlesbe infuse\nlesbe lista\nlesbe typeset\nlesbe doesn\nlesbe paine\nlesbe stang\nlesbe lingo\nlesbe esearch\nlesbe fuses\nlesbe instal\nlesbe inlining\nlesbe darin\nlesbe greta\nlesbe inest\nlesbe eased\nlesbe molino\nlesbe seddon\nlesbe prado\nlesbe molar\nlesbe moles\nlesbe does\nlesbe instar\nlesbe eidos\nlesbe sedaka\nlesbe molest\nlesbe praline\nlesbe dainese\nlesbe stata\nlesbe estar\nlesbe darcs\nlesbe graying\nlesbe panes\nlesbe dahmer\nlesbe grins\nlesbe infuses\nlesbe listas\nlesbe lidar\nlesbe dopant\nlesbe pango\nlesbe padang\nlesbe earing\nlesbe espada\nlesbe insta\nlesbe dangos\nlesbe listado\nlesbe mollis\nlesbe paling\nlesbe pandan\nlesbe fussed\nlesbe tarina\nlesbe listar\nlesbe lipari\nlesbe doling\nlesbe palin\nlesbe parco\nlesbe typeid\nlesbe daines\nlesbe parche\nlesbe nines\nlesbe done\ninsta a\ninsta with\ninsta be\ninsta us\ninsta do\ninsta he\ninsta data\ninsta great\ninsta line\ninsta type\ninsta less\ninsta dog\ninsta bed\ninsta ring\ninsta bear\ninsta eat\ninsta nine\ninsta dot\ninsta pan\ninsta gray\ninsta earn\ninsta types\ninsta dose\ninsta bet\ninsta dad\ninsta pat\ninsta ear\ninsta ease\ninsta lie\ninsta pad\ninsta dam\ninsta bee\ninsta lip\ninsta leslie\ninsta lid\ninsta pale\ninsta lining\ninsta pant\ninsta grease\ninsta espanol\ninsta beg\ninsta daring\ninsta benin\ninsta espana\ninsta ealing\ninsta panty\ninsta panda\ninsta prada\ninsta bering\ninsta lesben\ninsta rinse\ninsta greased\ninsta mollie\ninsta witty\ninsta beaker\ninsta rinsed\ninsta pandas\ninsta panning\ninsta lipase\ninsta earch\ninsta dorint\ninsta paring\ninsta liberi\ninsta typeset\ninsta doesn\ninsta lingo\ninsta esearch\ninsta fuses\ninsta beeing\ninsta libel\ninsta bening\ninsta darin\ninsta belies\ninsta greta\ninsta eased\ninsta dales\ninsta seddon\ninsta prado\ninsta molar\ninsta moles\ninsta does\ninsta eidos\ninsta sedaka\ninsta beaked\ninsta molest\ninsta praline\ninsta estar\ninsta darcs\ninsta graying\ninsta panes\ninsta dahmer\ninsta lidar\ninsta liber\ninsta dopant\ninsta witless\ninsta pango\ninsta padang\ninsta earing\ninsta espada\ninsta lesbe\ninsta dangos\ninsta mollis\ninsta paling\ninsta bedale\ninsta pandan\ninsta fussed\ninsta lipari\ninsta doling\ninsta palin\ninsta parco\ninsta typeid\ninsta parche\ninsta nines\ninsta done\ndangos tag\ndangos tale\ndangos tap\ndangos tales\ndangos tapas\ndangos edina\ndangos taliesin\ndangos taint\ndangos taber\ndangos talib\ndangos tatars\ndangos tatar\ndangos tarina\ndangos talese\ndangos talia\ndangos tainty\nlistado a\nlistado in\nlistado with\nlistado i\nlistado be\nlistado us\nlistado he\nlistado data\nlistado great\nlistado type\nlistado being\nlistado less\nlistado bed\nlistado ring\nlistado pain\nlistado bear\nlistado eat\nlistado nine\nlistado paint\nlistado pan\nlistado gray\nlistado earn\nlistado types\nlistado bet\nlistado pat\nlistado ear\nlistado ease\nlistado pad\nlistado dam\nlistado bee\nlistado typing\nlistado pale\nlistado pant\nlistado grease\nlistado espanol\nlistado beg\nlistado inning\nlistado grin\nlistado daring\nlistado benin\nlistado espana\nlistado painless\nlistado panty\nlistado panda\nlistado molina\nlistado moline\nlistado pains\nlistado grind\nlistado ingress\nlistado prada\nlistado bering\nlistado lesben\nlistado rinse\nlistado greased\nlistado dainty\nlistado witty\nlistado fusing\nlistado beaker\nlistado rinsed\nlistado pandas\nlistado panning\nlistado earch\nlistado paring\nlistado panini\nlistado ingres\nlistado infuse\nlistado typeset\nlistado paine\nlistado esearch\nlistado fuses\nlistado beeing\nlistado bening\nlistado darin\nlistado greta\nlistado inest\nlistado eased\nlistado molino\nlistado dales\nlistado molar\nlistado moles\nlistado sedaka\nlistado beaked\nlistado molest\nlistado dainese\nlistado estar\nlistado darcs\nlistado graying\nlistado panes\nlistado dahmer\nlistado grins\nlistado infuses\nlistado witless\nlistado pango\nlistado padang\nlistado earing\nlistado espada\nlistado lesbe\nlistado dangos\nlistado bedale\nlistado pandan\nlistado fussed\nlistado tarina\nlistado parco\nlistado typeid\nlistado daines\nlistado parche\nlistado nines\nmollis tag\nmollis tale\nmollis tap\nmollis tales\nmollis tango\nmollis tapas\nmollis edina\nmollis taint\nmollis taber\nmollis tatars\nmollis tatar\nmollis tarina\nmollis tangos\nmollis talese\nmollis tainty\npaling of\npaling on\npaling or\npaling one\npaling oh\npaling oak\npaling oakdale\npaling oprah\npaling obese\npaling orinda\npaling orcinus\npaling oakes\nbedale search\nbedale so\nbedale she\nbedale set\nbedale start\nbedale say\nbedale star\nbedale sea\nbedale saying\nbedale seat\nbedale sin\nbedale seal\nbedale stars\nbedale sole\nbedale spain\nbedale slip\nbedale sing\nbedale starring\nbedale swim\nbedale sake\nbedale staring\nbedale spanning\nbedale sealing\nbedale stare\nbedale sparc\nbedale starr\nbedale sprang\nbedale stares\nbedale stardom\nbedale starling\nbedale sinus\nbedale sling\nbedale spans\nbedale sparing\nbedale searing\nbedale sparco\nbedale sinead\nbedale sprains\nbedale seine\nbedale sprain\nbedale soleus\nbedale slingo\nbedale soles\nbedale sinning\nbedale seadoo\nbedale sayin\nbedale sparcs\nbedale sealine\nbedale searcg\nbedale swims\nbedale sesrch\nbedale shewing\nbedale searcn\nbedale searcu\nbedale searct\nbedale searcm\nbedale starline\nbedale spaeth\nbedale swiming\nbedale searc\nbedale seastar\nbedale starlit\nbedale sakes\nbedale spanne\nbedale sakar\npandan in\npandan i\npandan be\npandan go\npandan buy\npandan best\npandan being\npandan god\npandan bay\npandan bar\npandan bring\npandan bus\npandan going\npandan bear\npandan beat\npandan bet\npandan beast\npandan beam\npandan bearing\npandan bean\npandan inline\npandan bind\npandan blessed\npandan baking\npandan bless\npandan beastality\npandan bake\npandan baker\npandan baked\npandan goethe\npandan ingress\npandan bling\npandan ingres\npandan infuse\npandan binning\npandan instal\npandan brine\npandan inest\npandan indole\npandan goring\npandan betaine\npandan infusing\npandan beale\npandan instar\npandan bestar\npandan infuses\npandan brining\npandan beset\npandan insta\npandan gopal\npandan betas\npandan indoles\npandan bethea\npandan boles\npandan goole\npandan beane\npandan brines\npandan baying\npandan beilin\npandan blingo\npandan beaty\npandan busoni\nindoles a\nindoles with\nindoles be\nindoles us\nindoles he\nindoles list\nindoles data\nindoles best\nindoles great\nindoles line\nindoles type\nindoles east\nindoles star\nindoles past\nindoles bed\nindoles ring\nindoles bear\nindoles eat\nindoles nine\nindoles pan\nindoles gray\nindoles earn\nindoles types\nindoles bet\nindoles pat\nindoles ear\nindoles ease\nindoles lie\nindoles pad\nindoles dam\nindoles bee\nindoles lip\nindoles pasta\nindoles lid\nindoles lining\nindoles pant\nindoles grease\nindoles espanol\nindoles beg\nindoles staring\nindoles daring\nindoles starch\nindoles stalin\nindoles benin\nindoles espana\nindoles ealing\nindoles panty\nindoles panda\nindoles prada\nindoles bering\nindoles rinse\nindoles greased\nindoles mollie\nindoles witty\nindoles beaker\nindoles rinsed\nindoles pandas\nindoles bestality\nindoles panning\nindoles lipase\nindoles pastas\nindoles earch\nindoles paring\nindoles liberi\nindoles lista\nindoles typeset\nindoles stang\nindoles lingo\nindoles esearch\nindoles fuses\nindoles beeing\nindoles bening\nindoles darin\nindoles belies\nindoles greta\nindoles eased\nindoles molar\nindoles moles\nindoles sedaka\nindoles beaked\nindoles molest\nindoles praline\nindoles stata\nindoles estar\nindoles darcs\nindoles graying\nindoles bestar\nindoles panes\nindoles dahmer\nindoles listas\nindoles lidar\nindoles liber\nindoles pango\nindoles padang\nindoles earing\nindoles espada\nindoles dangos\nindoles mollis\nindoles paling\nindoles pandan\nindoles fussed\nindoles listar\nindoles lipari\nindoles palin\nindoles parco\nindoles typeid\nindoles parche\nindoles nines\nfussed a\nfussed in\nfussed with\nfussed i\nfussed be\nfussed us\nfussed do\nfussed he\nfussed list\nfussed data\nfussed best\nfussed great\nfussed line\nfussed type\nfussed being\nfussed less\nfussed east\nfussed star\nfussed past\nfussed dog\nfussed bed\nfussed ring\nfussed pain\nfussed bear\nfussed eat\nfussed nine\nfussed dot\nfussed paint\nfussed pan\nfussed gray\nfussed earn\nfussed doing\nfussed types\nfussed bet\nfussed dad\nfussed pat\nfussed ear\nfussed lie\nfussed pad\nfussed dam\nfussed bee\nfussed inline\nfussed lip\nfussed typing\nfussed leslie\nfussed pasta\nfussed lid\nfussed pale\nfussed lining\nfussed staind\nfussed pant\nfussed espanol\nfussed beg\nfussed staring\nfussed inning\nfussed grin\nfussed daring\nfussed starch\nfussed stalin\nfussed benin\nfussed espana\nfussed ealing\nfussed painless\nfussed panty\nfussed stain\nfussed panda\nfussed molina\nfussed moline\nfussed pains\nfussed grind\nfussed ingress\nfussed prada\nfussed bering\nfussed lesben\nfussed mollie\nfussed dainty\nfussed witty\nfussed beaker\nfussed pandas\nfussed bestality\nfussed panning\nfussed staines\nfussed earch\nfussed dorint\nfussed paring\nfussed panini\nfussed liberi\nfussed ingres\nfussed lista\nfussed typeset\nfussed doesn\nfussed paine\nfussed stang\nfussed stale\nfussed lingo\nfussed esearch\nfussed beeing\nfussed instal\nfussed libel\nfussed bening\nfussed inlining\nfussed darin\nfussed belies\nfussed greta\nfussed inest\nfussed indole\nfussed molino\nfussed dales\nfussed prado\nfussed molar\nfussed moles\nfussed does\nfussed instar\nfussed eidos\nfussed beaked\nfussed molest\nfussed praline\nfussed dainese\nfussed stata\nfussed estar\nfussed darcs\nfussed graying\nfussed bestar\nfussed panes\nfussed dahmer\nfussed grins\nfussed lidar\nfussed liber\nfussed dopant\nfussed witless\nfussed pango\nfussed padang\nfussed earing\nfussed espada\nfussed lesbe\nfussed insta\nfussed dangos\nfussed listado\nfussed mollis\nfussed paling\nfussed bedale\nfussed pandan\nfussed indoles\nfussed tarina\nfussed listar\nfussed lipari\nfussed doling\nfussed palin\nfussed parco\nfussed typeid\nfussed daines\nfussed parche\nfussed nines\nfussed done\ntarina klingon\ntarina keane\ntarina rhesus\ntarina kline\ntarina keine\ntarina khmer\ntarina kelis\ntarina kedar\ntarina rhein\ntarina kling\ntarina kepada\ntarina kernow\nlistar in\nlistar i\nlistar not\nlistar my\nlistar no\nlistar now\nlistar car\nlistar none\nlistar nor\nlistar cake\nlistar nose\nlistar norm\nlistar cease\nlistar ceased\nlistar nod\nlistar inning\nlistar chew\nlistar chewing\nlistar cakes\nlistar nobel\nlistar ingress\nlistar ingres\nlistar infuse\nlistar norco\nlistar cesar\nlistar coles\nlistar chews\nlistar cdata\nlistar inest\nlistar indole\nlistar myles\nlistar nosed\nlistar infusing\nlistar nodal\nlistar coleus\nlistar myrinet\nlistar infuses\nlistar indain\nlistar noakes\nlistar mydata\nlistar ctype\nlistar indoles\nlistar cinese\nlipari no\nlipari net\nlipari near\nlipari naked\nlipari nine\nlipari nest\nlipari neat\nlipari neale\nlipari naking\nlipari nines\ndoling of\ndoling on\ndoling or\ndoling one\ndoling oh\ndoling oak\ndoling oakdale\ndoling oprah\ndoling obese\ndoling orinda\ndoling orcinus\ndoling oakes\npalin in\npalin i\npalin be\npalin go\npalin buy\npalin best\npalin being\npalin god\npalin bay\npalin bar\npalin bring\npalin bus\npalin going\npalin bear\npalin beat\npalin bet\npalin beast\npalin beam\npalin bearing\npalin bean\npalin bind\npalin blessed\npalin baking\npalin bless\npalin bake\npalin baker\npalin bethesda\npalin baked\npalin goethe\npalin ingress\npalin ingres\npalin infuse\npalin binning\npalin instal\npalin brine\npalin inest\npalin indole\npalin goring\npalin betaine\npalin infusing\npalin beale\npalin instar\npalin bestar\npalin infuses\npalin brining\npalin beset\npalin indain\npalin insta\npalin betas\npalin brinda\npalin indoles\npalin bethea\npalin boles\npalin goole\npalin beane\npalin brines\npalin baying\npalin beaty\npalin busoni\nparco let\nparco leg\nparco leaking\nparco leaked\nparco leaks\nparco lenin\nparco lestat\nparco leben\nparco lehmer\nparco leprae\nparco lening\nparco lesedi\nparco leake\nparco lewitt\ntypeid of\ntypeid a\ntypeid on\ntypeid or\ntypeid at\ntypeid as\ntypeid one\ntypeid am\ntypeid oh\ntypeid ad\ntypeid army\ntypeid arm\ntypeid oak\ntypeid ongoing\ntypeid ah\ntypeid adobe\ntypeid atari\ntypeid oakdale\ntypeid ahmed\ntypeid oprah\ntypeid aarhus\ntypeid obese\ntypeid orinda\ntypeid orcinus\ntypeid adoring\ntypeid alesse\ntypeid adorno\ntypeid arcing\ntypeid adorn\ntypeid oakes\ntypeid aearch\ntypeid alist\ntypeid aline\ntypeid astar\ntypeid ahmet\ntypeid arche\ntypeid oline\ntypeid alesina\ndaines a\ndaines with\ndaines be\ndaines us\ndaines do\ndaines he\ndaines list\ndaines best\ndaines great\ndaines line\ndaines type\ndaines less\ndaines east\ndaines star\ndaines past\ndaines dog\ndaines bed\ndaines ring\ndaines bear\ndaines eat\ndaines nine\ndaines dot\ndaines pan\ndaines gray\ndaines earn\ndaines dose\ndaines bet\ndaines pat\ndaines ear\ndaines ease\ndaines lie\ndaines pad\ndaines bee\ndaines lip\ndaines leslie\ndaines pasta\ndaines lid\ndaines pale\ndaines lining\ndaines pant\ndaines grease\ndaines beg\ndaines staring\ndaines starch\ndaines stalin\ndaines benin\ndaines ealing\ndaines panty\ndaines bering\ndaines lesben\ndaines rinse\ndaines greased\ndaines mollie\ndaines witty\ndaines beaker\ndaines rinsed\ndaines bestality\ndaines panning\ndaines lipase\ndaines pastas\ndaines earch\ndaines dorint\ndaines paring\ndaines liberi\ndaines lista\ndaines stang\ndaines stale\ndaines lingo\ndaines beeing\ndaines libel\ndaines bening\ndaines greta\ndaines eased\ndaines seddon\ndaines prado\ndaines molar\ndaines eidos\ndaines sedaka\ndaines beaked\ndaines praline\ndaines stata\ndaines graying\ndaines bestar\ndaines listas\ndaines liber\ndaines dopant\ndaines witless\ndaines pango\ndaines earing\ndaines lesbe\ndaines listado\ndaines mollis\ndaines paling\ndaines fussed\ndaines listar\ndaines lipari\ndaines doling\ndaines palin\ndaines parco\ndaines typeid\ndaines parche\ndaines done\nparche we\nparche way\nparche west\nparche war\nparche window\nparche win\nparche wine\nparche wind\nparche winning\nparche wear\nparche wet\nparche wing\nparche wake\nparche wearing\nparche waking\nparche wines\nparche warhol\nparche weaning\nparche warhead\nparche weariness\nparche wakes\nparche winstar\nparche windom\nparche winless\nparche weiber\nparche winline\nparche westar\nparche wearin\nparche weibel\nparche wakeling\nparche wakelin\nparche waker\nnines a\nnines in\nnines with\nnines i\nnines be\nnines us\nnines do\nnines he\nnines list\nnines data\nnines best\nnines great\nnines line\nnines type\nnines being\nnines less\nnines east\nnines star\nnines past\nnines dog\nnines bed\nnines ring\nnines pain\nnines bear\nnines eat\nnines dot\nnines paint\nnines pan\nnines gray\nnines earn\nnines doing\nnines stainless\nnines dose\nnines bet\nnines dad\nnines pat\nnines ear\nnines ease\nnines lie\nnines pad\nnines dam\nnines bee\nnines inline\nnines lip\nnines typing\nnines leslie\nnines pasta\nnines lid\nnines pale\nnines staind\nnines pant\nnines grease\nnines beg\nnines staring\nnines grin\nnines stains\nnines daring\nnines starch\nnines stalin\nnines ealing\nnines painless\nnines panty\nnines stain\nnines panda\nnines molina\nnines moline\nnines pains\nnines grind\nnines prada\nnines bering\nnines lesben\nnines rinse\nnines greased\nnines mollie\nnines dainty\nnines witty\nnines fusing\nnines beaker\nnines rinsed\nnines pandas\nnines bestality\nnines lipase\nnines pastas\nnines earch\nnines dorint\nnines paring\nnines liberi\nnines infuse\nnines lista\nnines paine\nnines stang\nnines stale\nnines lingo\nnines beeing\nnines instal\nnines libel\nnines darin\nnines greta\nnines eased\nnines indole\nnines molino\nnines dales\nnines seddon\nnines prado\nnines molar\nnines instar\nnines eidos\nnines sedaka\nnines beaked\nnines praline\nnines stata\nnines darcs\nnines graying\nnines bestar\nnines dahmer\nnines grins\nnines listas\nnines lidar\nnines liber\nnines dopant\nnines witless\nnines pango\nnines padang\nnines earing\nnines lesbe\nnines insta\nnines dangos\nnines listado\nnines mollis\nnines paling\nnines bedale\nnines pandan\nnines indoles\nnines fussed\nnines tarina\nnines listar\nnines lipari\nnines doling\nnines palin\nnines parco\nnines typeid\nnines parche\nnines done\ndone of\ndone on\ndone or\ndone oh\ndone oak\ndone ongoing\ndone oakdale\ndone oprah\ndone obese\ndone orinda\ndone orcinus\ndone oakes\n"
],
[
"print(\"\"\"\nAK AKE ARH AYI BE DA DO EA EES EI ES ETA ETH EYB FUS GAR GR HEW\nHME HON IN KAN KEB LES LI MOL NB NEO NGO NIN OLE OOS PA PAN PLA\nPRA RAT RC RIN RMY RNO SED SNA STA TAR TLE TYP USO UYT WIM WIT\nYER\n\"\"\".strip().replace(' ', '\\t'))",
"AK\tAKE\tARH\tAYI\tBE\tDA\tDO\tEA\tEES\tEI\tES\tETA\tETH\tEYB\tFUS\tGAR\tGR\tHEW\nHME\tHON\tIN\tKAN\tKEB\tLES\tLI\tMOL\tNB\tNEO\tNGO\tNIN\tOLE\tOOS\tPA\tPAN\tPLA\nPRA\tRAT\tRC\tRIN\tRMY\tRNO\tSED\tSNA\tSTA\tTAR\tTLE\tTYP\tUSO\tUYT\tWIM\tWIT\nYER\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4d2d935434cafc450fde0784c3ecb9a64a1b35 | 27,614 | ipynb | Jupyter Notebook | Matrix Factorization/.ipynb_checkpoints/MatrixFactorization-checkpoint.ipynb | sakshisuman12/deeplearing | efa2769ee6f115fb138321e6a5f3ac633a567926 | [
"MIT"
] | null | null | null | Matrix Factorization/.ipynb_checkpoints/MatrixFactorization-checkpoint.ipynb | sakshisuman12/deeplearing | efa2769ee6f115fb138321e6a5f3ac633a567926 | [
"MIT"
] | null | null | null | Matrix Factorization/.ipynb_checkpoints/MatrixFactorization-checkpoint.ipynb | sakshisuman12/deeplearing | efa2769ee6f115fb138321e6a5f3ac633a567926 | [
"MIT"
] | null | null | null | 35.630968 | 141 | 0.533172 | [
[
[
"import numpy as np\nimport pandas as pd\nnp.random.seed(42)\nfrom sklearn.metrics import mean_squared_error\nimport time\n",
"_____no_output_____"
],
[
"names = ['user_id', 'movie_id', 'rating', 'timestamp']\ndf = pd.read_csv('./ml-100k/u.data', sep='\\t', names=names)\nprint(df.head())\nprint(df.shape)\n",
" user_id movie_id rating timestamp\n0 196 242 3 881250949\n1 186 302 3 891717742\n2 22 377 1 878887116\n3 244 51 2 880606923\n4 166 346 1 886397596\n(100000, 4)\n"
],
[
"n_users = df[\"user_id\"].unique().shape[0]\nn_movies = df[\"movie_id\"].unique().shape[0]\nratings = np.zeros((n_users, n_movies))\nfor row in df.itertuples():\n ratings[row[1] - 1, row[2] - 1] = row[3]\nprint(f\"ratings = {ratings}\")\nW = ratings.copy()\nW[W > 0] = 1\nprint(f\"W = {W}\")\nprint(W.shape)\n",
"ratings = [[5. 3. 4. ... 0. 0. 0.]\n [4. 0. 0. ... 0. 0. 0.]\n [0. 0. 0. ... 0. 0. 0.]\n ...\n [5. 0. 0. ... 0. 0. 0.]\n [0. 0. 0. ... 0. 0. 0.]\n [0. 5. 0. ... 0. 0. 0.]]\nW = [[1. 1. 1. ... 0. 0. 0.]\n [1. 0. 0. ... 0. 0. 0.]\n [0. 0. 0. ... 0. 0. 0.]\n ...\n [1. 0. 0. ... 0. 0. 0.]\n [0. 0. 0. ... 0. 0. 0.]\n [0. 1. 0. ... 0. 0. 0.]]\n(943, 1682)\n"
],
[
"def train_test_split(ratings, s=100, r=1000):\n test = np.zeros(ratings.shape)\n train = ratings.copy()\n found = False\n\n l = []\n for movie in range(ratings.shape[1]):\n l.append(np.sum(ratings[:, movie] != 0))\n l = np.array(l)\n top_r = np.argsort(-l)[:r]\n while not found:\n for user in range(ratings.shape[0]):\n test_ratings = np.random.choice(top_r.nonzero()[0], size=s, replace=False)\n train[user, test_ratings] = 0.\n test[user, test_ratings] = ratings[user, test_ratings]\n found = True\n for movie in range(ratings.shape[1]):\n if np.all(train[:, movie] == 0):\n found = False\n break\n\n mean_imputed_ratings = train.copy()\n \n for col in range(train.shape[1]):\n non_zero_cols = train[train[:, col] != 0, col]\n mean_imputed_ratings[:, col][test[:, col] != 0] = non_zero_cols.mean()\n\n # Check if train and test sets are disjoint\n assert(np.all((train * test) == 0)) \n return train, test, mean_imputed_ratings\n",
"_____no_output_____"
],
[
"def als_step(ratings, W, latent, fixed, lmd, cat=\"user\", basic=False):\n \n n, k, d = latent.shape[0], latent.shape[1], fixed.shape[0]\n lambdaI = lmd * np.eye(k)\n if not basic:\n for i in range(latent.shape[0]):\n if cat == \"user\":\n W_i, x_i = W[i, :].reshape(1, W[i, :].shape[0]), ratings[i, :] \n elif cat == \"movie\":\n W_i, x_i = W[:, i].reshape(1, W[:, i].shape[0]), ratings[:, i]\n latent[i, :] = np.linalg.solve(((fixed.T * W_i).dot(fixed) + lambdaI), (fixed.T * W_i).dot(x_i))\n else:\n fTf = fixed.T.dot(fixed)\n for i in range(latent.shape[0]):\n if cat == \"user\":\n x_i = ratings[i, :] \n elif cat == \"movie\":\n x_i = ratings[:, i]\n latent[i, :] = np.linalg.solve((fTf + lambdaI), fixed.T.dot(x_i))\n",
"_____no_output_____"
],
[
"def update_u_v(ratings, W, users, movies, epochs=10, n_factors=5, lmd=10, basic=False, debug=False):\n epoch = 0\n print(f\"Incremental epochs = {epochs}\")\n while epoch < epochs:\n als_step(ratings, W, users, movies, lmd, \"user\", basic)\n als_step(ratings, W, movies, users, lmd, \"movie\", basic)\n epoch += 1\n",
"_____no_output_____"
],
[
"def get_predictions(users, movies):\n predictions = np.zeros((users.shape[0], movies.shape[0]))\n for i in range(users.shape[0]):\n for j in range(movies.shape[0]):\n predictions[i, j] = users[i, :].dot(movies[j, :])\n return predictions\n",
"_____no_output_____"
],
[
"def get_mse(predictions, truths, tests=None):\n if tests is not None:\n non_zero_predictions = predictions[tests.nonzero()].flatten()\n non_zero_truths = truths[tests.nonzero()].flatten()\n else:\n non_zero_predictions = predictions[truths.nonzero()].flatten()\n non_zero_truths = truths[truths.nonzero()].flatten()\n return mean_squared_error(non_zero_predictions, non_zero_truths)\n",
"_____no_output_____"
],
[
"def get_best_hyperparameters(train, test, mean_imputed_ratings, epochs_list, k_list=[40], lmd_list=[0.1], basic=False, debug=False):\n start_time = time.time()\n \n epochs_list.sort()\n\n n, d = train.shape\n\n best_hyper_and_error = {}\n best_hyper_and_error[\"k\"] = k_list[0]\n best_hyper_and_error[\"lambda\"] = lmd_list[0]\n best_hyper_and_error[\"epochs\"] = 0\n best_hyper_and_error[\"train_error\"] = np.inf\n best_hyper_and_error[\"test_error\"] = np.inf\n best_hyper_and_error[\"mean_error\"] = np.inf\n \n W = train.copy()\n W[W > 0] = 1\n \n for k in k_list:\n for lmd in lmd_list:\n print(f\"k = {k} lambda = {lmd}\")\n train_error = []\n test_error = []\n mean_error = []\n\n users = np.random.random((n, k))\n movies = np.random.random((d, k))\n prev = 0\n for (i, epochs) in enumerate(epochs_list):\n if debug:\n print(f\"Total epochs = {epochs}\")\n\n update_u_v(train, W, users, movies, epochs - prev, k, lmd, basic, debug)\n\n predictions = get_predictions(users, movies)\n\n train_error.append(get_mse(predictions, train))\n test_error.append(get_mse(predictions, test))\n mean_error.append(get_mse(predictions, mean_imputed_ratings, test))\n if debug:\n print(f\"Train error = {train_error[-1]}\")\n print(f\"Test error = {test_error[-1]}\")\n print(f\"Mean error = {mean_error[-1]}\")\n prev = epochs\n min_test_error_index = np.argmin(test_error)\n if test_error[min_test_error_index] < best_hyper_and_error[\"test_error\"]:\n best_hyper_and_error[\"k\"] = k\n best_hyper_and_error[\"lambda\"] = lmd\n best_hyper_and_error[\"epochs\"] = epochs_list[min_test_error_index]\n best_hyper_and_error[\"train_error\"] = train_error[min_test_error_index]\n best_hyper_and_error[\"test_error\"] = test_error[min_test_error_index]\n best_hyper_and_error[\"mean_error\"] = mean_error[min_test_error_index]\n if debug:\n print(\"Current optimal hyperparameters are\")\n print(pd.Series(best_hyper_and_error))\n if debug:\n print(f\"Time elapsed = {time.strftime('%Hh %Mm %Ss', time.gmtime(time.time() - start_time))}\")\n print()\n return best_hyper_and_error\n",
"_____no_output_____"
],
[
"epochs_list = [1, 2, 5, 10]\nk_list = [5, 10, 20, 40, 80]\nlmd_list = [0.1, 2, 5, 10, 25, 50, 100]\ntrain, test, mean_imputed_ratings = train_test_split(ratings, 100, 1000)\n\nbest_hyper_and_error = get_best_hyperparameters(train, test, mean_imputed_ratings, epochs_list, k_list, lmd_list, False, True)\nprint(best_hyper_and_error)\n",
"k = 5 lambda = 0.1\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 0.7866717134258924\nTest error = 0.9777389549871205\nMean error = 0.29828981025650775\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 0.694584944798435\nTest error = 0.961878999205918\nMean error = 0.3291409454061807\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 0.6292339358546601\nTest error = 0.9137805501109458\nMean error = 0.39100401025976644\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 0.6113806263230467\nTest error = 0.9026566105892969\nMean error = 0.42920858386947197\nCurrent optimal hyperparameters are\nk 5.000000\nlambda 0.100000\nepochs 10.000000\ntrain_error 0.611381\ntest_error 0.902657\nmean_error 0.429209\ndtype: float64\nTime elapsed = 00h 00m 03s\n\nk = 5 lambda = 2\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 0.8085990706358384\nTest error = 0.9364964370884464\nMean error = 0.2458962175755709\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 0.7142316593397136\nTest error = 0.8952836217029992\nMean error = 0.258823539794594\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 0.6442373553474039\nTest error = 0.8498810194229777\nMean error = 0.3258175229611442\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 0.6281684727505704\nTest error = 0.8382719403297402\nMean error = 0.34497921615108446\nCurrent optimal hyperparameters are\nk 5.000000\nlambda 2.000000\nepochs 10.000000\ntrain_error 0.628168\ntest_error 0.838272\nmean_error 0.344979\ndtype: float64\nTime elapsed = 00h 00m 07s\n\nk = 5 lambda = 5\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 0.8343333093073558\nTest error = 0.9171247458565186\nMean error = 0.22106937481928657\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 0.7320226439612029\nTest error = 0.8694554344915387\nMean error = 0.24586656262029344\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 0.6810777897709868\nTest error = 0.8369783374984656\nMean error = 0.28761456851753514\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 0.6672904167634549\nTest error = 0.8236433053485409\nMean error = 0.30253079581882\nCurrent optimal hyperparameters are\nk 5.000000\nlambda 5.000000\nepochs 10.000000\ntrain_error 0.667290\ntest_error 0.823643\nmean_error 0.302531\ndtype: float64\nTime elapsed = 00h 00m 11s\n\nk = 5 lambda = 10\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 0.8950297594035256\nTest error = 0.9378779063950586\nMean error = 0.23070539536382811\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 0.8083422441392712\nTest error = 0.8878281969266383\nMean error = 0.22493365267780838\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 0.7397895360310702\nTest error = 0.83886922272727\nMean error = 0.26578496645891375\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 0.7350022647130001\nTest error = 0.8269582363236178\nMean error = 0.27513661463006883\nTime elapsed = 00h 00m 15s\n\nk = 5 lambda = 25\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 1.1174051286036577\nTest error = 1.1101402831423641\nMean error = 0.37213772656942007\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 1.0624287688703846\nTest error = 1.0437238350601181\nMean error = 0.3014931159334076\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 1.00469874796182\nTest error = 0.9873421413817645\nMean error = 0.30094379577983305\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 0.994163004329729\nTest error = 0.9765889805771533\nMean error = 0.3138885249789621\nTime elapsed = 00h 00m 19s\n\nk = 5 lambda = 50\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 1.5108519421702418\nTest error = 1.4591328254788256\nMean error = 0.6931225869819011\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 1.4525562511379855\nTest error = 1.3787982791470388\nMean error = 0.6075614083186599\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 1.453567209824128\nTest error = 1.356910209409129\nMean error = 0.5939531174990426\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 1.4612082460577471\nTest error = 1.359516679749117\nMean error = 0.603177220413788\nTime elapsed = 00h 00m 23s\n\nk = 5 lambda = 100\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 2.447859002080979\nTest error = 2.340323367504638\nMean error = 1.536564393450932\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 2.3658607245159797\nTest error = 2.2302049769559953\nMean error = 1.4221444588719403\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 2.4123431568677844\nTest error = 2.261099552546226\nMean error = 1.4548711350266323\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 2.418345238540401\nTest error = 2.265797248648805\nMean error = 1.4596533386561092\nTime elapsed = 00h 00m 27s\n\nk = 10 lambda = 0.1\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 0.6935320755442189\nTest error = 1.1221888446911301\nMean error = 0.44942044688312116\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 0.5800133341114407\nTest error = 1.134941864739319\nMean error = 0.5159351054022127\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 0.49376824306003186\nTest error = 1.1376586084087634\nMean error = 0.6394388148831088\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 0.4646196743338371\nTest error = 1.1565453130172092\nMean error = 0.7031740960062939\nTime elapsed = 00h 00m 32s\n\nk = 10 lambda = 2\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 0.7218156765771211\nTest error = 0.9668553198018257\nMean error = 0.29589989512930326\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 0.5819340677660454\nTest error = 0.9390692189560853\nMean error = 0.35739003373432127\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 0.5104694479665627\nTest error = 0.9225409128450367\nMean error = 0.45500436947731643\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 0.4900899706947794\nTest error = 0.9297882135877126\nMean error = 0.49112995113942964\nTime elapsed = 00h 00m 37s\n\nk = 10 lambda = 5\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 0.7765650685956231\nTest error = 0.9106318117322593\nMean error = 0.22834815326326485\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 0.6307210238885189\nTest error = 0.8710537642927173\nMean error = 0.2877141420468753\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 0.5568847514263804\nTest error = 0.8474575068281193\nMean error = 0.35820727968020105\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 0.5443243568234143\nTest error = 0.8484013848167224\nMean error = 0.3800143075192654\nTime elapsed = 00h 00m 42s\n\nk = 10 lambda = 10\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 0.8645566592136177\nTest error = 0.9184730971450342\nMean error = 0.21289753058731226\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 0.74411486992536\nTest error = 0.8763592622353835\nMean error = 0.22590617664747434\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 0.6636009163676926\nTest error = 0.8417732394607113\nMean error = 0.29401127247515924\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 0.647251306757177\nTest error = 0.8318302585728957\nMean error = 0.30874934660812847\nTime elapsed = 00h 00m 47s\n\nk = 10 lambda = 25\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 1.0705083313644062\nTest error = 1.0323640348722811\nMean error = 0.30533417058647716\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 1.0512634375071936\nTest error = 1.0144871742272552\nMean error = 0.2908023275637223\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 0.9904131031633877\nTest error = 0.9740561362871303\nMean error = 0.31171150420051913\nTotal epochs = 10\nIncremental epochs = 5\nTrain error = 0.9757938376909039\nTest error = 0.9690477425970436\nMean error = 0.3188765646912621\nTime elapsed = 00h 00m 51s\n\nk = 10 lambda = 50\nTotal epochs = 1\nIncremental epochs = 1\nTrain error = 1.4657820291193062\nTest error = 1.364846866679716\nMean error = 0.6120068221920445\nTotal epochs = 2\nIncremental epochs = 1\nTrain error = 1.474194125558719\nTest error = 1.3728700994139371\nMean error = 0.6050094556013683\nTotal epochs = 5\nIncremental epochs = 3\nTrain error = 1.4764150353901504\nTest error = 1.3724037281134283\nMean error = 0.6074659232374836\nTotal epochs = 10\nIncremental epochs = 5\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4d5dc3219ccb671a4172b6e37ee3979677f7e0 | 17,693 | ipynb | Jupyter Notebook | 1. question .ipynb | ramyuva/WEB2 | 17d7e513038199de282f37a074e234f05814faef | [
"Apache-2.0"
] | null | null | null | 1. question .ipynb | ramyuva/WEB2 | 17d7e513038199de282f37a074e234f05814faef | [
"Apache-2.0"
] | null | null | null | 1. question .ipynb | ramyuva/WEB2 | 17d7e513038199de282f37a074e234f05814faef | [
"Apache-2.0"
] | null | null | null | 34.28876 | 145 | 0.534957 | [
[
[
"import selenium\nimport pandas as pd\nfrom selenium import webdriver",
"_____no_output_____"
],
[
"driver=webdriver.Chrome(\"chromedriver.exe\")",
"_____no_output_____"
],
[
"driver.get(\"https://www.naukri.com/\")",
"_____no_output_____"
],
[
"search_job=driver.find_element_by_id('qsb-keyword-sugg')\nsearch_job.send_keys(\"Data Analyst\")\nsearch_loc=driver.find_element_by_xpath(\"//input[@id='qsb-location-sugg']\")\nsearch_loc.send_keys(\"Bangalore\")",
"_____no_output_____"
],
[
"search_btn=driver.find_element_by_xpath(\"//div[@class='search-btn']/button\")\nsearch_btn.click()",
"_____no_output_____"
],
[
"url=\"https://www.naukri.com/data-analyst-jobs-in-bangalore?k=data%20analyst&l=bangalore\"",
"_____no_output_____"
],
[
"driver.get(url)",
"_____no_output_____"
],
[
"job_title=[]\ncompany_names=[]\nlocations_list=[]\nexperience_list=[]",
"_____no_output_____"
],
[
"titles_tag=driver.find_elements_by_xpath(\"//a[@class='title fw500 ellipsis']\")\ntitles_tag[0:9]",
"_____no_output_____"
],
[
"for i in titles_tag:\n job_title.append(i.text)\njob_title[0:9]",
"_____no_output_____"
],
[
"company_tag=driver.find_elements_by_xpath(\"//a[@class='subTitle ellipsis fleft']\")\ncompany_tag[0:9]",
"_____no_output_____"
],
[
"for i in company_tag:\n company_names.append(i.text)\ncompany_names[0:9]",
"_____no_output_____"
],
[
"loc_tag=driver.find_elements_by_xpath(\"//li[@class='fleft grey-text br2 placeHolderLi location']/span[1]\")\nloc_tag[0:9]",
"_____no_output_____"
],
[
"for i in loc_tag:\n locations_list.append(i.text)\nlocations_list[0:9]",
"_____no_output_____"
],
[
"exp_tag=driver.find_elements_by_xpath(\"//li[@class='fleft grey-text br2 placeHolderLi experience']/span[1]\")\nexp_tag[0:9]",
"_____no_output_____"
],
[
"for i in exp_tag:\n experience_list.append(i.text)\nexperience_list[0:9]",
"_____no_output_____"
],
[
"import pandas as pd\nDataAnalyst=pd.DataFrame()\nDataAnalyst['JOB_NAME']=job_title[0:9]\nDataAnalyst['COMPANY']=company_names[0:9]\nDataAnalyst['JOB_LOCATION']=locations_list[0:9]\nDataAnalyst['EXPERIENCE']=experience_list[0:9]",
"_____no_output_____"
],
[
"DataAnalyst",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4d6c3214f078c378c8c54c13072e0d44c194cb | 126,593 | ipynb | Jupyter Notebook | mini_projects/presidential_election_Poland_2015.ipynb | maskubica/python | c37ff3ea8d5ff6fcd110a7d47de8cae16f5621b6 | [
"MIT"
] | null | null | null | mini_projects/presidential_election_Poland_2015.ipynb | maskubica/python | c37ff3ea8d5ff6fcd110a7d47de8cae16f5621b6 | [
"MIT"
] | null | null | null | mini_projects/presidential_election_Poland_2015.ipynb | maskubica/python | c37ff3ea8d5ff6fcd110a7d47de8cae16f5621b6 | [
"MIT"
] | null | null | null | 91.667632 | 70,370 | 0.690994 | [
[
[
"<a href=\"https://colab.research.google.com/github/maskubica/python/blob/master/mini_projects/presidential_election_Poland_2015.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\n\nimport matplotlib.pyplot as plt\n\n!pip install --upgrade geopandas\nimport geopandas as gpd",
"Collecting geopandas\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/f8/dd/c0a6429cc7692efd5c99420c9df525c40f472b50705871a770449027e244/geopandas-0.8.0-py2.py3-none-any.whl (962kB)\n\u001b[K |████████████████████████████████| 962kB 2.8MB/s \n\u001b[?25hRequirement already satisfied, skipping upgrade: shapely in /usr/local/lib/python3.6/dist-packages (from geopandas) (1.7.0)\nCollecting fiona\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/ec/20/4e63bc5c6e62df889297b382c3ccd4a7a488b00946aaaf81a118158c6f09/Fiona-1.8.13.post1-cp36-cp36m-manylinux1_x86_64.whl (14.7MB)\n\u001b[K |████████████████████████████████| 14.7MB 306kB/s \n\u001b[?25hCollecting pyproj>=2.2.0\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/e5/c3/071e080230ac4b6c64f1a2e2f9161c9737a2bc7b683d2c90b024825000c0/pyproj-2.6.1.post1-cp36-cp36m-manylinux2010_x86_64.whl (10.9MB)\n\u001b[K |████████████████████████████████| 10.9MB 47.7MB/s \n\u001b[?25hRequirement already satisfied, skipping upgrade: pandas>=0.23.0 in /usr/local/lib/python3.6/dist-packages (from geopandas) (1.0.5)\nRequirement already satisfied, skipping upgrade: attrs>=17 in /usr/local/lib/python3.6/dist-packages (from fiona->geopandas) (19.3.0)\nRequirement already satisfied, skipping upgrade: click<8,>=4.0 in /usr/local/lib/python3.6/dist-packages (from fiona->geopandas) (7.1.2)\nCollecting click-plugins>=1.0\n Downloading https://files.pythonhosted.org/packages/e9/da/824b92d9942f4e472702488857914bdd50f73021efea15b4cad9aca8ecef/click_plugins-1.1.1-py2.py3-none-any.whl\nCollecting munch\n Downloading https://files.pythonhosted.org/packages/cc/ab/85d8da5c9a45e072301beb37ad7f833cd344e04c817d97e0cc75681d248f/munch-2.5.0-py2.py3-none-any.whl\nRequirement already satisfied, skipping upgrade: six>=1.7 in /usr/local/lib/python3.6/dist-packages (from fiona->geopandas) (1.12.0)\nCollecting cligj>=0.5\n Downloading https://files.pythonhosted.org/packages/e4/be/30a58b4b0733850280d01f8bd132591b4668ed5c7046761098d665ac2174/cligj-0.5.0-py3-none-any.whl\nRequirement already satisfied, skipping upgrade: python-dateutil>=2.6.1 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.23.0->geopandas) (2.8.1)\nRequirement already satisfied, skipping upgrade: numpy>=1.13.3 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.23.0->geopandas) (1.18.5)\nRequirement already satisfied, skipping upgrade: pytz>=2017.2 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.23.0->geopandas) (2018.9)\nInstalling collected packages: click-plugins, munch, cligj, fiona, pyproj, geopandas\nSuccessfully installed click-plugins-1.1.1 cligj-0.5.0 fiona-1.8.13.post1 geopandas-0.8.0 munch-2.5.0 pyproj-2.6.1.post1\n"
],
[
"!wget \"https://prezydent2015.pkw.gov.pl/prezydent_2015_tura1.zip\"",
"--2020-06-30 09:29:11-- https://prezydent2015.pkw.gov.pl/prezydent_2015_tura1.zip\nResolving prezydent2015.pkw.gov.pl (prezydent2015.pkw.gov.pl)... 193.219.114.9\nConnecting to prezydent2015.pkw.gov.pl (prezydent2015.pkw.gov.pl)|193.219.114.9|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 3664617 (3.5M) [application/zip]\nSaving to: ‘prezydent_2015_tura1.zip’\n\nprezydent_2015_tura 100%[===================>] 3.49M 554KB/s in 7.4s \n\n2020-06-30 09:29:21 (483 KB/s) - ‘prezydent_2015_tura1.zip’ saved [3664617/3664617]\n\n"
],
[
"!unzip prezydent_2015_tura1.zip\n!ls",
"Archive: prezydent_2015_tura1.zip\n inflating: prezydent_2015_tura1.csv \n creating: __MACOSX/\n inflating: __MACOSX/._prezydent_2015_tura1.csv \n inflating: wyniki_tura1-1.xls \n inflating: __MACOSX/._wyniki_tura1-1.xls \n__MACOSX\t\t prezydent_2015_tura1.zip wyniki_tura1-1.xls\nprezydent_2015_tura1.csv sample_data\n"
],
[
"!wget ftp://91.223.135.109/prg/jednostki_administracyjne.zip",
"--2020-06-30 10:51:58-- ftp://91.223.135.109/prg/jednostki_administracyjne.zip\n => ‘jednostki_administracyjne.zip’\nConnecting to 91.223.135.109:21... connected.\nLogging in as anonymous ... Logged in!\n==> SYST ... done. ==> PWD ... done.\n==> TYPE I ... done. ==> CWD (1) /prg ... done.\n==> SIZE jednostki_administracyjne.zip ... 393290487\n==> PASV ... done. ==> RETR jednostki_administracyjne.zip ... done.\nLength: 393290487 (375M) (unauthoritative)\n\njednostki_administr 100%[===================>] 375.07M 975KB/s in 6m 33s \n\n2020-06-30 10:58:35 (977 KB/s) - ‘jednostki_administracyjne.zip’ saved [393290487]\n\n"
],
[
"!unzip jednostki_administracyjne.zip",
"Archive: jednostki_administracyjne.zip\n creating: PRG_jednostki_administracyjne_v40_SZPRG/\n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Gminy.dbf \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Gminy.prj \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Gminy.shp \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Gminy.shx \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Jednostki_ewidencyjne.dbf \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Jednostki_ewidencyjne.prj \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Jednostki_ewidencyjne.shp \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Jednostki_ewidencyjne.shx \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Obręby.dbf \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Obręby.prj \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Obręby.shp \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Obręby.shx \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Państwo.dbf \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Państwo.prj \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Państwo.shp \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Państwo.shx \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Powiaty.dbf \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Powiaty.prj \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Powiaty.shp \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Powiaty.shx \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Województwa.dbf \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Województwa.prj \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Województwa.shp \n inflating: PRG_jednostki_administracyjne_v40_SZPRG/Województwa.shx \n"
],
[
"mapa_woj = gpd.read_file('PRG_jednostki_administracyjne_v40_SZPRG/Województwa.shp')\nmapa_gmn = gpd.read_file('PRG_jednostki_administracyjne_v40_SZPRG/Gminy.shp')\n\nmapa_woj = mapa_woj[['JPT_KOD_JE', \"geometry\"]]\nmapa_gmn = mapa_gmn[['JPT_KOD_JE', \"geometry\"]]\n\nmapa_woj.head(20)\n\n",
"_____no_output_____"
],
[
"#df = pd.read_excel('wyniki_tura1-1.xls', dtype={'TERYT gminy': float})\n\ndf = pd.read_csv('prezydent_2015_tura1.csv', sep=\";\", encoding = \"windows-1250\", dtype={'TERYT gminy' : str})\n\ndf['TERYT gminy'] = df['TERYT gminy'].apply(lambda x: '0'+str(x) if len(str(x)) < 6 else str(x))\ndf['TERYT województwo'] = df['TERYT gminy'].str.slice(stop=2)\ndf = df.groupby('TERYT województwo').sum()\ndf['%Duda'] = df['Paweł Piotr Kukiz']/df['RAZEM']\ndf",
"_____no_output_____"
],
[
"dane_mapa_woj = pd.merge(mapa_woj, df, how='left', left_on='JPT_KOD_JE', right_on='TERYT województwo')\n\n\nfig, ax = plt.subplots(1, figsize = (8,8))\n \n# rysowanie mapy\ndane_mapa_woj.plot(column='%Duda', ax=ax, cmap='YlOrRd', linewidth=0.6, edgecolor='gray', alpha=0.5)\n \n# usuwamy osie\nax.axis('off')\n \n# pokazujemy obrazek\nplt.show()",
"_____no_output_____"
],
[
"df.loc[(df['Numer obwodu'] == 69) & (df['Województwo'] == 'dolnośląskie')]",
"_____no_output_____"
],
[
"np.sum(df['TERYT gminy'].values)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4d7100dfd2d807f9f20595c5068515db7c6321 | 33,071 | ipynb | Jupyter Notebook | temperature/temperature.ipynb | joangali1997/bcn-feb-2019-prework | 43886452cf015772eeb669c2a5634af01ca5d520 | [
"Unlicense"
] | null | null | null | temperature/temperature.ipynb | joangali1997/bcn-feb-2019-prework | 43886452cf015772eeb669c2a5634af01ca5d520 | [
"Unlicense"
] | null | null | null | temperature/temperature.ipynb | joangali1997/bcn-feb-2019-prework | 43886452cf015772eeb669c2a5634af01ca5d520 | [
"Unlicense"
] | null | null | null | 64.465887 | 19,192 | 0.791842 | [
[
[
"# Processor temperature\n\nWe have a temperature sensor in the processor of our company's server. We want to analyze the data provided to determinate whether we should change the cooling system for a better one. It is expensive and as a data analyst we cannot make decisions without a basis.\n\nWe provide the temperatures measured throughout the 24 hours of a day in a list-type data structure composed of 24 integers:\n```\ntemperatures_C = [33,66,65,0,59,60,62,64,70,76,80,69,80,83,68,79,61,53,50,49,53,48,45,39]\n```\n\n## Goals\n\n1. Treatment of lists\n2. Use of loop or list comprenhention\n3. Calculation of the mean, minimum and maximum.\n4. Filtering of lists.\n5. Interpolate an outlier.\n6. Logical operators.\n7. Print",
"_____no_output_____"
],
[
"## Temperature graph\nTo facilitate understanding, the temperature graph is shown below. You do not have to do anything in this section. The test starts in **Problem**.",
"_____no_output_____"
]
],
[
[
"# import\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n# axis x, axis y\ny = [33,66,65,0,59,60,62,64,70,76,80,81,80,83,90,79,61,53,50,49,53,48,45,39]\nx = list(range(len(y)))\n\n# plot\nplt.plot(x, y)\nplt.axhline(y=70, linewidth=1, color='r')\nplt.xlabel('hours')\nplt.ylabel('Temperature ºC')\nplt.title('Temperatures of our server throughout the day')",
"_____no_output_____"
]
],
[
[
"## Problem\n\nIf the sensor detects more than 4 hours with temperatures greater than or equal to 70ºC or any temperature above 80ºC or the average exceeds 65ºC throughout the day, we must give the order to change the cooling system to avoid damaging the processor.\n\nWe will guide you step by step so you can make the decision by calculating some intermediate steps:\n\n1. Minimum temperature\n2. Maximum temperature\n3. Temperatures equal to or greater than 70ºC\n4. Average temperatures throughout the day.\n5. If there was a sensor failure at 03:00 and we did not capture the data, how would you estimate the value that we lack? Correct that value in the list of temperatures.\n6. Bonus: Our maintenance staff is from the United States and does not understand the international metric system. Pass temperatures to Degrees Fahrenheit.\n\nFormula: F = 1.8 * C + 32\n\nweb: https://en.wikipedia.org/wiki/Conversion_of_units_of_temperature\n",
"_____no_output_____"
]
],
[
[
"# assign a variable to the list of temperatures\n\n# 1. Calculate the minimum of the list and print the value using print()\n\n\n# 2. Calculate the maximum of the list and print the value using print()\n\n\n# 3. Items in the list that are greater than 70ºC and print the result\n\n\n# 4. Calculate the mean temperature throughout the day and print the result\n\n\n# 5.1 Solve the fault in the sensor by estimating a value\n\n\n# 5.2 Update of the estimated value at 03:00 on the list\n\n\n\n# Bonus: convert the list of ºC to ºFarenheit\n\n",
"_____no_output_____"
],
[
"temperatures_list = [33,66,65,0,59,60,62,64,70,76,80,81,80,83,90,79,61,53,50,49,53,48,45,39]\nmin_temperature = 0\nmax_temperature = 0\n\nmax_temperature = max(temperatures_list)\nmin_temperature = min(temperatures_list)\n\nprint (\"The minimum temperature of the list is\", min_temperature)\nprint (\"The maximum temperature of the list is\", max_temperature)",
"The minimum temperature of the list is 0\nThe maximum temperature of the list is 90\n"
],
[
"greater_than_seventy = []\n\nfor i in temperatures_list:\n if i > 70:\n greater_than_seventy.append(i)\n\nprint (\"The items in the list that are greater than 70ºC are\", greater_than_seventy)",
"The items in the list that are greater than 70ºC are [76, 80, 81, 80, 83, 90, 79]\n"
],
[
"mean_temperature_24 = 0\n\nmean_temperature_24 = sum (temperatures_list) / len (temperatures_list)\n\nprint(\"The mean temperature is\", mean_temperature_24)",
"The mean temperature is 60.25\n"
],
[
"y = [33,66,65,0,59,60,62,64,70,76,80,81,80,83,90,79,61,53,50,49,53,48,45,39]\nx = ()\n\n#Struggled with the Value estimation\n\ntemperatures_list = y\nnew_temperatures_list = list(y)\nmean_temperature_24 = 60.25\n \n\nfor i in temperatures_list:\n mean_temperature_24 = ((sum (temperatures_list) -65) + int(x) / len (temperatures_list)\n int(x) = (60.25*24) - (sum(temperatures_list) -65)\n\n print(x) ",
"_____no_output_____"
],
[
"Farenheit_list = []\n\n#for loop to iterate through the temperatures_list/y list.\n#for loop inside the for loop to do the calculations through all the temperatures_list.\n#round to round the decimals\n\nfor i in y:\n Farenheit_list = [(round(i * 1.8)+32.1) for i in temperatures_list]\n \nprint(\"The temperatures list in Farenheit is the following:\", Farenheit_list)",
"The temperatures list in Farenheit is the following: [91.1, 151.1, 149.1, 32.1, 138.1, 140.1, 144.1, 147.1, 158.1, 169.1, 176.1, 178.1, 176.1, 181.1, 194.1, 174.1, 142.1, 127.1, 122.1, 120.1, 127.1, 118.1, 113.1, 102.1]\n"
]
],
[
[
"## Take the decision\nRemember that if the sensor detects more than 4 hours with temperatures greater than or equal to 70ºC or any temperature higher than 80ºC or the average was higher than 65ºC throughout the day, we must give the order to change the cooling system to avoid the danger of damaging the equipment:\n* more than 4 hours with temperatures greater than or equal to 70ºC\n* some temperature higher than 80ºC\n* average was higher than 65ºC throughout the day\nIf any of these three is met, the cooling system must be changed.\n",
"_____no_output_____"
]
],
[
[
"# Print True or False depending on whether you would change the cooling system or not\n\ntemperatures_list = [33,66,65,0,59,60,62,64,70,76,80,81,80,83,90,79,61,53,50,49,53,48,45,39]\navg_list = sum(temperatures_list) / len(temperatures_list)\nhours = 0\n\nfor i in temperatures_list:\n if i >= 70:\n hours += 1\n print(\"True\")\n elif i > 80:\n higher_80 += 1\n print (\"True\")\n elif avg_list>65:\n print(\"True\")\n else:\n print(\"False\")",
"False\nFalse\nFalse\nFalse\nFalse\nFalse\nFalse\nFalse\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nFalse\nFalse\nFalse\nFalse\nFalse\nFalse\nFalse\nFalse\n"
]
],
[
[
"## Future improvements\n1. We want the hours (not the temperatures) whose temperature exceeds 70ºC\n2. Condition that those hours are more than 4 consecutive and consecutive, not simply the sum of the whole set. Is this condition met?\n3. Average of each of the lists (ºC and ºF). How they relate?\n4. Standard deviation of each of the lists. How they relate?\n",
"_____no_output_____"
]
],
[
[
"# 1. We want the hours (not the temperatures) whose temperature exceeds 70ºC\n",
"_____no_output_____"
],
[
"hours_list = []\n\nfor i in temperatures_list:\n if i>70:\n print(i)\n \nprint (hours_list)",
"[]\n"
],
[
"hours = range(1,25)\n\nfor i in hours:\n hours = temperatures_list\n\nfor i in hours:\n print(i)\n ",
"1\n2\n3\n4\n5\n6\n7\n8\n9\n10\n11\n12\n13\n14\n15\n16\n17\n18\n19\n20\n21\n22\n23\n24\n"
],
[
"# 2. Condition that those hours are more than 4 consecutive and consecutive, not simply the sum of the whole set. Is this condition met?\n\n\n",
"_____no_output_____"
],
[
"# 3. Average of each of the lists (ºC and ºF). How they relate?\nFarenheit_list = [91.1, 151.1, 149.1, 32.1, 138.1, 140.1, 144.1, 147.1, 158.1, 169.1, 176.1, 178.1, 176.1, 181.1, 194.1, 174.1, 142.1, 127.1, 122.1, 120.1, 127.1, 118.1, 113.1, 102.1]\nmean_celsius = 0\nmean_farenheit = 0\n\nmean_celsius = sum (temperatures_list) / len (temperatures_list)\nmean_farenheit = sum (Farenheit_list) / len (Farenheit_list)\n\nprint(mean_celsius)\nprint(mean_farenheit)\n\nmeans_ratio = mean_celsius / mean_farenheit\nprint (\"1 farenheit degree equals to\",means_ratio, \"celsius degrees\")\n",
"12.5\n140.47499999999994\n1 farenheit degree equals to 0.08898380494749959 celsius degrees\n"
],
[
"# 4. Standard deviation of each of the lists. How they relate?\n\nsd_celsius = 0\nsd_farenheit = 0",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4d86c097431e0136579502b920bbfa66eeb818 | 12,034 | ipynb | Jupyter Notebook | DownloadSoundFiles/DownloadSoundFiles.ipynb | ptizei/iNaturalistDatasetGenerator | 908037382b902d417926764b2b4bae9ee21be185 | [
"BSD-3-Clause"
] | null | null | null | DownloadSoundFiles/DownloadSoundFiles.ipynb | ptizei/iNaturalistDatasetGenerator | 908037382b902d417926764b2b4bae9ee21be185 | [
"BSD-3-Clause"
] | null | null | null | DownloadSoundFiles/DownloadSoundFiles.ipynb | ptizei/iNaturalistDatasetGenerator | 908037382b902d417926764b2b4bae9ee21be185 | [
"BSD-3-Clause"
] | null | null | null | 21.722022 | 495 | 0.502825 | [
[
[
"import pandas as pd\nfrom pathlib import Path\nimport requests\nimport re\n\n",
"_____no_output_____"
],
[
"folderToOpen = Path(\"../Data/\")\nfileName = \"BemTeVi.csv\"\nfilePath = folderToOpen / fileName",
"_____no_output_____"
],
[
"folderForData = folderToOpen / filePath.stem\n#for child in folderForData.iterdir(): print(child)\nfname = 'bob.mp3'\nfname = str(folderForData.resolve()) + '\\\\' + fname\nfname\nfilePath.name",
"_____no_output_____"
],
[
"df_Pardal = pd.read_csv(filePath)",
"_____no_output_____"
],
[
"testPath = folderToOpen\nfor item in testPath.iterdir():\n if item.suffix:\n print(item.stem)\n pathToTest = testPath / item.stem\n if pathToTest.exists():\n print('Folder', item.stem, 'already exists')",
"\nFolder BemTeVi already exists\n\nFolder Pardal already exists\n\n\n"
],
[
"urlToDownload = df_Pardal.iloc[1,13]\nr = requests.get(urlToDownload, allow_redirects=True)",
"_____no_output_____"
],
[
"open('birb.3gp', 'wb').write(r.content)",
"_____no_output_____"
],
[
"df_Pardal.sound_url[1]",
"_____no_output_____"
],
[
"\nfor url in df_Pardal.sound_url:\n if isinstance(url,str):\n urlToDownload = url.split('?')[0]\n try:\n with requests.get(urlToDownload) as r:\n \n fname = ''\n if \"Content-Disposition\" in r.headers.keys():\n fname = re.findall(\"filename=(.+)\", r.headers[\"Content-Disposition\"])[0]\n else:\n fname = urlToDownload.split(\"/\")[-1]\n \n #print(fname)\n except RequestException as e:\n print(e)\n \n open(fname, 'wb').write(r.content)\n",
"_____no_output_____"
],
[
"r",
"_____no_output_____"
],
[
"r.headers",
"_____no_output_____"
],
[
"urlToDownload",
"_____no_output_____"
],
[
"url = urlToDownload.split('?')[0]\nurl",
"_____no_output_____"
],
[
"try:\n with requests.get(url) as r:\n\n fname = ''\n if \"Content-Disposition\" in r.headers.keys():\n fname = re.findall(\"filename=(.+)\", r.headers[\"Content-Disposition\"])[0]\n else:\n fname = url.split(\"/\")[-1]\n\n print(fname)\nexcept RequestException as e:\n print(e)",
"1798.3gp\n"
],
[
"urlToDownload.split('/')[-1]",
"_____no_output_____"
],
[
"i = 0",
"_____no_output_____"
],
[
"i++",
"_____no_output_____"
],
[
"i +=1",
"_____no_output_____"
],
[
"i",
"_____no_output_____"
],
[
"isinstance(url,str)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4d9517cd011f9029dfae3f044028ac863a76fd | 16,999 | ipynb | Jupyter Notebook | notebooks/Differential Equations and Markov Models.ipynb | rnowling/notebooks | cea286eae2379ec83ed0de08d8fa88cfd93e790a | [
"Apache-2.0"
] | 1 | 2015-07-20T18:21:38.000Z | 2015-07-20T18:21:38.000Z | notebooks/Differential Equations and Markov Models.ipynb | rnowling/notebooks | cea286eae2379ec83ed0de08d8fa88cfd93e790a | [
"Apache-2.0"
] | null | null | null | notebooks/Differential Equations and Markov Models.ipynb | rnowling/notebooks | cea286eae2379ec83ed0de08d8fa88cfd93e790a | [
"Apache-2.0"
] | null | null | null | 101.184524 | 12,419 | 0.830755 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
cb4da75d31ac9bce2ed1b639edafd104e2fe724e | 15,019 | ipynb | Jupyter Notebook | ml_source/src/blocktorch/docs/source/user_guide/data_actions.ipynb | blocktorch/blocktorch | 044aa269813ab22c5fd27f84272e5fb540fc522b | [
"MIT"
] | 1 | 2021-09-23T12:23:02.000Z | 2021-09-23T12:23:02.000Z | ml_source/src/blocktorch/docs/source/user_guide/data_actions.ipynb | blocktorch/blocktorch | 044aa269813ab22c5fd27f84272e5fb540fc522b | [
"MIT"
] | null | null | null | ml_source/src/blocktorch/docs/source/user_guide/data_actions.ipynb | blocktorch/blocktorch | 044aa269813ab22c5fd27f84272e5fb540fc522b | [
"MIT"
] | null | null | null | 37.083951 | 708 | 0.643984 | [
[
[
"# Understanding Data Actions\n\nblocktorch streamlines the creation and implementation of machine learning models for tabular data. One of the many features it offers is [data checks](https://blocktorch.alteryx.com/en/stable/user_guide/data_checks.html), which are geared towards determining the health of the data before we train a model on it. These data checks have associated actions with them and will be shown in this notebook. In our default data checks, we have the following checks:\n\n- `HighlyNullDataCheck`: Checks whether the rows or columns are highly null\n\n- `IDColumnsDataCheck`: Checks for columns that could be ID columns\n\n- `TargetLeakageDataCheck`: Checks if any of the input features have high association with the targets\n\n- `InvalidTargetDataCheck`: Checks if there are null or other invalid values in the target\n\n- `NoVarianceDataCheck`: Checks if either the target or any features have no variance\n\n- `NaturalLanguageNaNDataCheck`: Checks if any natural language columns have missing data\n\n- `DateTimeNaNDataCheck`: Checks if any datetime columns have missing data\n\n\nblocktorch has additional data checks that can be seen [here](https://blocktorch.alteryx.com/en/stable/api_index.html#data-checks), with usage examples [here](https://blocktorch.alteryx.com/en/stable/user_guide/data_checks.html). Below, we will walk through usage of blocktorch's default data checks and actions.\n\n\nFirst, we import the necessary requirements to demonstrate these checks.",
"_____no_output_____"
]
],
[
[
"import woodwork as ww\nimport pandas as pd\nfrom blocktorch import AutoMLSearch\nfrom blocktorch.demos import load_fraud\nfrom blocktorch.preprocessing import split_data",
"_____no_output_____"
]
],
[
[
"Let's look at the input feature data. blocktorch uses the [Woodwork](https://woodwork.alteryx.com/en/stable/) library to represent this data. The demo data that blocktorch returns is a Woodwork DataTable and DataColumn.",
"_____no_output_____"
]
],
[
[
"X, y = load_fraud(n_rows=1500)\nX",
"_____no_output_____"
]
],
[
[
"## Adding noise and unclean data\n\nThis data is already clean and compatible with blocktorch's ``AutoMLSearch``. In order to demonstrate blocktorch default data checks, we will add the following:\n\n- A column of mostly null values (<0.5% non-null)\n\n- A column with low/no variance\n\n- A row of null values\n\n- A missing target value\n\n\nWe will add the first two columns to the whole dataset and we will only add the last two to the training data. Note: these only represent some of the scenarios that blocktorch default data checks can catch.",
"_____no_output_____"
]
],
[
[
"# add a column with no variance in the data\nX['no_variance'] = [1 for _ in range(X.shape[0])]\n\n# add a column with >99.5% null values\nX['mostly_nulls'] = [None] * (X.shape[0] - 5) + [i for i in range(5)]\n\n# since we changed the data, let's reinitialize the woodwork datatable\nX.ww.init()\n# let's split some training and validation data\nX_train, X_valid, y_train, y_valid = split_data(X, y, problem_type='binary')",
"_____no_output_____"
],
[
"# let's copy the datetime at row 1 for future use\ndate = X_train.iloc[1]['datetime']\n\n# make row 1 all nan values\nX_train.iloc[1] = [None] * X_train.shape[1]\n\n# make one of the target values null\ny_train[990] = None\n\nX_train.ww.init()\ny_train = ww.init_series(y_train)\n# Let's take another look at the new X_train data\nX_train",
"_____no_output_____"
]
],
[
[
"If we call `AutoMLSearch.search()` on this data, the search will fail due to the columns and issues we've added above. Note: we use a try/except here to catch the resulting ValueError that AutoMLSearch raises.",
"_____no_output_____"
]
],
[
[
"automl = AutoMLSearch(X_train=X_train, y_train=y_train, problem_type='binary')\ntry:\n automl.search()\nexcept ValueError as e:\n # to make the error message more distinct\n print(\"=\" * 80, \"\\n\")\n print(\"Search errored out! Message received is: {}\".format(e))\n print(\"=\" * 80, \"\\n\")",
"_____no_output_____"
]
],
[
[
"We can use the `search_iterative()` function provided in blocktorch to determine what potential health issues our data has. We can see that this [search_iterative](https://blocktorch.alteryx.com/en/latest/autoapi/blocktorch/automl/index.html#blocktorch.automl.search_iterative) function is a public method available through `blocktorch.automl` and is different from the [search](https://blocktorch.alteryx.com/en/stable/autoapi/blocktorch/automl/index.html#blocktorch.automl.AutoMLSearch) function of the `AutoMLSearch` class in blocktorch. This `search_iterative()` function allows us to run the default data checks on the data, and, if there are no errors, automatically runs `AutoMLSearch.search()`.",
"_____no_output_____"
]
],
[
[
"from blocktorch.automl import search_iterative\nresults = search_iterative(X_train, y_train, problem_type='binary')\nresults",
"_____no_output_____"
]
],
[
[
"The return value of the `search_iterative` function above is a tuple. The first element is the `AutoMLSearch` object if it runs (and `None` otherwise), and the second element is a dictionary of potential warnings and errors that the default data checks find on the passed-in `X` and `y` data. In this dictionary, warnings are suggestions that the datachecks give that can useful to address to make the search better but will not break AutoMLSearch. On the flip side, errors will break AutoMLSearch and need to be addressed by the user.",
"_____no_output_____"
],
[
"## Addressing DataCheck errors\nWe will show that we can address errors to allow AutoMLSearch to run. However, ignoring warnings will come at the expense of performance.\n\nWe can print out the errors first to make it easier to read, and then we'll create new features and targets from the original training data.",
"_____no_output_____"
]
],
[
[
"results[1]['errors']",
"_____no_output_____"
],
[
"# copy the DataTables to new variables\nX_train_no_errors = X_train.copy()\ny_train_no_errors = y_train.copy()\n\n# We address the errors by looking at the resulting dictionary errors listed\n\n# first, let's address the `TARGET_HAS_NULL` error\ny_train_no_errors.fillna(False, inplace=True)\n\n# here, we address the `NO_VARIANCE` error \nX_train_no_errors.drop(\"no_variance\", axis=1, inplace=True)\n\n# lastly, we address the `DATETIME_HAS_NAN` error with the date we had saved earlier\nX_train_no_errors.iloc[1, 2] = date\n\n# let's reinitialize the Woodwork DataTable\nX_train_no_errors.ww.init()\nX_train_no_errors.head()",
"_____no_output_____"
]
],
[
[
"We can now run search on `X_train_no_errors` and `y_train_no_errors`. Note that the search here doesn't fail since we addressed the errors, but there will still exist warnings in the returned tuple. This search allows the `mostly_nulls` column to remain in the features during search.",
"_____no_output_____"
]
],
[
[
"results_no_errors = search_iterative(X_train_no_errors, y_train_no_errors, problem_type='binary')\nresults_no_errors",
"_____no_output_____"
]
],
[
[
"## Addressing all warnings and errors\nWe can look at the `actions` key of the dictionary in order to see how we can fix and clean all of the data. This will help us clean both the warnings and errors from the data and provide us with a better model.",
"_____no_output_____"
]
],
[
[
"results[1]['actions']",
"_____no_output_____"
]
],
[
[
"We note that there are four action tasks that we can take to clean the data. Three of the tasks ask us to drop a row or column in the features, while one task asks us to impute the target value. ",
"_____no_output_____"
]
],
[
[
"# The first action states to drop the row given by the action code\nX_train.drop(1477, axis=0, inplace=True)\n# we must also drop this for y since we are removing its associated feature input\ny_train.drop(index=1477, inplace=True)\n\nprint(\"The new length of X_train is {} and y_train is {}\".format(len(X_train),len(y_train)))",
"_____no_output_____"
],
[
"# Remove the 'mostly_nulls' column from X_train, which is the second action item\nX_train.drop('mostly_nulls', axis=1, inplace=True)\nX_train.head()",
"_____no_output_____"
],
[
"# Address the null in targets, which is the third action item\ny_train.fillna(False, inplace=True)\ny_train.isna().any()",
"_____no_output_____"
],
[
"# Finally, we can drop the 'no_variance' column, which is the final action item\nX_train.drop('no_variance', axis=1, inplace=True)\nX_train.head()",
"_____no_output_____"
],
[
"# let's reinitialize the dataframe using Woodwork and try the search again\nX_train.ww.init()\nresults_cleaned = search_iterative(X_train, y_train, problem_type='binary')",
"_____no_output_____"
]
],
[
[
"Note that this time, we do get an `AutoMLSearch` object returned to us, as well as an empty dictionary of warnings and errors. We can use the `AutoMLSearch` object as needed, and we can see that the resulting warning dictionary is empty.",
"_____no_output_____"
]
],
[
[
"aml = results_cleaned[0]\naml.rankings",
"_____no_output_____"
],
[
"data_check_results = results_cleaned[1]\ndata_check_results",
"_____no_output_____"
]
],
[
[
"## Comparing removing only errors versus removing both warnings and errors\nLet's see the differences in model performance when we remove only errors versus remove both warnings and errors. To do this, we compare the performance of the best pipelines on the validation data. Remember that in the search where we only address errors, we still have the `mostly_nulls` column present in the data, so we leave that column in the validation data for its respective search. We drop the other `no_variance` column from both searches.\n\nAdditionally, we do some logical type setting since we had added additional noise to just the training data. This allows the data to be of the same types in both training and validation.",
"_____no_output_____"
]
],
[
[
"# drop the no_variance column\nX_valid.drop(\"no_variance\", axis=1, inplace=True)\n\n# logical type management\nX_valid.ww.init(logical_types={\"customer_present\": \"Categorical\"})\ny_valid = ww.init_series(y_valid, logical_type=\"Categorical\")\n\nbest_pipeline_no_errors = results_no_errors[0].best_pipeline\nprint(\"Only dropping errors:\", best_pipeline_no_errors.score(X_valid, y_valid, [\"Log Loss Binary\"]), \"\\n\")\n\n# drop the mostly_nulls column and reinitialize the DataTable\nX_valid.drop(\"mostly_nulls\", axis=1, inplace=True)\nX_valid.ww.init()\n\nbest_pipeline_clean = results_cleaned[0].best_pipeline\nprint(\"Addressing all actions:\", best_pipeline_clean.score(X_valid, y_valid, [\"Log Loss Binary\"]), \"\\n\")",
"_____no_output_____"
]
],
[
[
"We can compare the differences in model performance when we address all action items (warnings and errors) in comparison to when we only address errors. While it isn't guaranteed that addressing all actions will always have better performance, we do recommend doing so since we only raise these issues when we believe the features have problems that could negatively impact or not benefit the search.\n\nIn the future, we aim to provide a helper function to allow users to quickly clean the data by taking in the list of actions and creating an appropriate pipeline of transformers to alter the data.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb4daad00aa61ceab9fbe0b523d43325cc2b2524 | 9,693 | ipynb | Jupyter Notebook | examples/plotting/notebook/glyphs.ipynb | bruns01/bokeh | 3e7c23ccd434043de2d4f37c258d97ec4e461168 | [
"BSD-3-Clause"
] | 17 | 2020-06-14T03:47:35.000Z | 2022-03-07T00:25:23.000Z | examples/plotting/notebook/glyphs.ipynb | bruns01/bokeh | 3e7c23ccd434043de2d4f37c258d97ec4e461168 | [
"BSD-3-Clause"
] | 12 | 2020-07-22T22:40:09.000Z | 2021-03-17T14:10:27.000Z | examples/plotting/notebook/glyphs.ipynb | bruns01/bokeh | 3e7c23ccd434043de2d4f37c258d97ec4e461168 | [
"BSD-3-Clause"
] | 8 | 2020-06-14T03:47:23.000Z | 2021-11-20T15:14:04.000Z | 23.757353 | 127 | 0.513566 | [
[
[
"This IPython Notebook contains simple examples of many of the basic vectorizable glyph functions that Bokeh provides. \n\nTo clear all previously rendered cell outputs, select from the menu:\n\n Cell -> All Output -> Clear",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom bokeh.plotting import figure, show, output_notebook\nfrom bokeh.layouts import gridplot\noutput_notebook()",
"_____no_output_____"
],
[
"N = 9\nx = np.linspace(-2, 2, N)\ny = x**2\nsizes = np.linspace(10, 20, N)\nxpts = np.array([-.09, -.12, .0, .12, .09])\nypts = np.array([-.1, .02, .1, .02, -.1])\nfigures = []",
"_____no_output_____"
],
[
"p = figure(title=\"annular_wedge\")\np.annular_wedge(x, y, 10, 20, 0.6, 4.1, color=\"#8888ee\",\n inner_radius_units=\"screen\", outer_radius_units=\"screen\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"annulus\")\np.annulus(x, y, 10, 20, color=\"#7FC97F\",\n inner_radius_units=\"screen\", outer_radius_units = \"screen\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"arc\")\np.arc(x, y, 20, 0.6, 4.1,\n radius_units=\"screen\", color=\"#BEAED4\", line_width=3)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"bezier\")\np.bezier(x, y, x+0.2, y, x+0.1, y+0.1, x-0.1, y-0.1,\n color=\"#D95F02\", line_width=2)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"circle\")\np.circle(x, y, radius=0.1, color=\"#3288BD\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"ellipse\")\np.ellipse(x, y, 15, 25, angle=-0.7, color=\"#1D91C0\",\n width_units=\"screen\", height_units=\"screen\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"line\")\np.line(x, y, color=\"#F46D43\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"multi_line\")\np.multi_line([xpts+xx for xx in x], [ypts+yy for yy in y], color=\"#8073AC\", line_width=2)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"multi_polygons\")\np.multi_polygons([[[xpts*2+xx, xpts+xx]] for xx in x], [[[ypts*3+yy, ypts+yy]] for yy in y], color=\"#FB9A99\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"oval\")\np.oval(x, y, 15, 25, angle=-0.7, color=\"#1D91C0\", \n width_units=\"screen\", height_units=\"screen\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"patch\")\np.patch(x, y, color=\"#A6CEE3\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"patches\")\np.patches([xpts+xx for xx in x], [ypts+yy for yy in y], color=\"#FB9A99\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"quad\")\np.quad(x, x-0.1, y, y-0.1, color=\"#B3DE69\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"quadratic\")\np.quadratic(x, y, x+0.2, y, x+0.1, y+0.1, color=\"#4DAF4A\", line_width=3)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"ray\")\np.ray(x, y, 45, -0.7, color=\"#FB8072\", line_width=2)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"rect\")\np.rect(x, y, 10, 20, color=\"#CAB2D6\", width_units=\"screen\", height_units=\"screen\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"segment\")\np.segment(x, y, x-0.1, y-0.1, color=\"#F4A582\", line_width=3)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"square\")\np.square(x, y, size=sizes, color=\"#74ADD1\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"wedge\")\np.wedge(x, y, 15, 0.6, 4.1, radius_units=\"screen\", color=\"#B3DE69\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"circle_x\")\np.scatter(x, y, marker=\"circle_x\", size=sizes, color=\"#DD1C77\", fill_color=None)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"triangle\")\np.scatter(x, y, marker=\"triangle\", size=sizes, color=\"#99D594\", line_width=2)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"circle\")\np.scatter(x, y, marker=\"o\", size=sizes, color=\"#80B1D3\", line_width=3)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"cross\")\np.scatter(x, y, marker=\"cross\", size=sizes, color=\"#E6550D\", line_width=2)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"diamond\")\np.scatter(x, y, marker=\"diamond\", size=sizes, color=\"#1C9099\", line_width=2)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"inverted_triangle\")\np.scatter(x, y, marker=\"inverted_triangle\", size=sizes, color=\"#DE2D26\")\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"square_x\")\np.scatter(x, y, marker=\"square_x\", size=sizes, color=\"#FDAE6B\",\n fill_color=None, line_width=2)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"asterisk\")\np.scatter(x, y, marker=\"asterisk\", size=sizes, color=\"#F0027F\", \n line_width=2)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"square_cross\")\np.scatter(x, y, marker=\"square_cross\", size=sizes, color=\"#7FC97F\",\n fill_color=None, line_width=2)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"diamond_cross\")\np.scatter(x, y, marker=\"diamond_cross\", size=sizes, color=\"#386CB0\",\n fill_color=None, line_width=2)\nfigures.append(p)",
"_____no_output_____"
],
[
"p = figure(title=\"circle_cross\")\np.scatter(x, y, marker=\"circle_cross\", size=sizes, color=\"#FB8072\",\n fill_color=None, line_width=2)\nfigures.append(p)",
"_____no_output_____"
],
[
"show(gridplot(figures, ncols=3, plot_width=200, plot_height=200))",
"_____no_output_____"
]
]
] | [
"raw",
"code"
] | [
[
"raw"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4dac7f79dd43cbf1eda3e874110f375c4a1c69 | 32,403 | ipynb | Jupyter Notebook | site/en-snapshot/guide/estimator.ipynb | NarimaneHennouni/docs-l10n | 39a48e0d5aa34950e29efd5c1f111c120185e9d9 | [
"Apache-2.0"
] | 2 | 2020-10-28T09:16:46.000Z | 2020-10-28T18:33:52.000Z | site/en-snapshot/guide/estimator.ipynb | NarimaneHennouni/docs-l10n | 39a48e0d5aa34950e29efd5c1f111c120185e9d9 | [
"Apache-2.0"
] | 1 | 2021-02-23T20:17:39.000Z | 2021-02-23T20:17:39.000Z | site/en-snapshot/guide/estimator.ipynb | NarimaneHennouni/docs-l10n | 39a48e0d5aa34950e29efd5c1f111c120185e9d9 | [
"Apache-2.0"
] | null | null | null | 37.330645 | 550 | 0.568805 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Estimators",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/guide/estimator\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/guide/estimator.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/guide/estimator.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs/site/en/guide/estimator.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"This document introduces `tf.estimator`—a high-level TensorFlow\nAPI. Estimators encapsulate the following actions:\n\n* training\n* evaluation\n* prediction\n* export for serving\n\nTensorFlow implements several pre-made Estimators. Custom estimators are still suported, but mainly as a backwards compatibility measure. **Custom estimators should not be used for new code**. All Estimators--whether pre-made or custom--are classes based on the `tf.estimator.Estimator` class.\n\nFor a quick example try [Estimator tutorials](../tutorials/estimator/linear.ipynb). For an overview of the API design, see the [white paper](https://arxiv.org/abs/1708.02637).",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"! pip install -U tensorflow_datasets",
"_____no_output_____"
],
[
"import tempfile\nimport os\n\nimport tensorflow as tf\nimport tensorflow_datasets as tfds",
"_____no_output_____"
]
],
[
[
"## Advantages\n\nSimilar to a `tf.keras.Model`, an `estimator` is a model-level abstraction. The `tf.estimator` provides some capabilities currently still under development for `tf.keras`. These are:\n\n * Parameter server based training\n * Full [TFX](http://tensorflow.org/tfx) integration.",
"_____no_output_____"
],
[
"## Estimators Capabilities\nEstimators provide the following benefits:\n\n* You can run Estimator-based models on a local host or on a distributed multi-server environment without changing your model. Furthermore, you can run Estimator-based models on CPUs, GPUs, or TPUs without recoding your model.\n* Estimators provide a safe distributed training loop that controls how and when to: \n * load data\n * handle exceptions\n * create checkpoint files and recover from failures\n * save summaries for TensorBoard\n\nWhen writing an application with Estimators, you must separate the data input\npipeline from the model. This separation simplifies experiments with\ndifferent data sets.",
"_____no_output_____"
],
[
"## Using pre-made Estimators\n\nPre-made Estimators enable you to work at a much higher conceptual level than the base TensorFlow APIs. You no longer have to worry about creating the computational graph or sessions since Estimators handle all the \"plumbing\" for you. Furthermore, pre-made Estimators let you experiment with different model architectures by making only minimal code changes. `tf.estimator.DNNClassifier`, for example, is a pre-made Estimator class that trains classification models based on dense, feed-forward neural networks.\n\nA TensorFlow program relying on a pre-made Estimator typically consists of the following four steps:",
"_____no_output_____"
],
[
"### 1. Write an input functions\n\nFor example, you might create one function to import the training set and another function to import the test set. Estimators expect their inputs to be formatted as a pair of objects:\n\n* A dictionary in which the keys are feature names and the values are Tensors (or SparseTensors) containing the corresponding feature data\n* A Tensor containing one or more labels\n\nThe `input_fn` should return a `tf.data.Dataset` that yields pairs in that format. \n\nFor example, the following code builds a `tf.data.Dataset` from the Titanic dataset's `train.csv` file:",
"_____no_output_____"
]
],
[
[
"def train_input_fn():\n titanic_file = tf.keras.utils.get_file(\"train.csv\", \"https://storage.googleapis.com/tf-datasets/titanic/train.csv\")\n titanic = tf.data.experimental.make_csv_dataset(\n titanic_file, batch_size=32,\n label_name=\"survived\")\n titanic_batches = (\n titanic.cache().repeat().shuffle(500)\n .prefetch(tf.data.experimental.AUTOTUNE))\n return titanic_batches",
"_____no_output_____"
]
],
[
[
"The `input_fn` is executed in a `tf.Graph` and can also directly return a `(features_dics, labels)` pair containing graph tensors, but this is error prone outside of simple cases like returning constants.",
"_____no_output_____"
],
[
"### 2. Define the feature columns.\n\nEach `tf.feature_column` identifies a feature name, its type, and any input pre-processing. \n\nFor example, the following snippet creates three feature columns.\n\n- The first uses the `age` feature directly as a floating-point input. \n- The second uses the `class` feature as a categorical input.\n- The third uses the `embark_town` as a categorical input, but uses the `hashing trick` to avoid the need to enumerate the options, and to set the number of options.\n\nFor further information, see the [feature columns tutorial](https://www.tensorflow.org/tutorials/keras/feature_columns).",
"_____no_output_____"
]
],
[
[
"age = tf.feature_column.numeric_column('age')\ncls = tf.feature_column.categorical_column_with_vocabulary_list('class', ['First', 'Second', 'Third']) \nembark = tf.feature_column.categorical_column_with_hash_bucket('embark_town', 32)",
"_____no_output_____"
]
],
[
[
"### 3. Instantiate the relevant pre-made Estimator.\n\nFor example, here's a sample instantiation of a pre-made Estimator named `LinearClassifier`:",
"_____no_output_____"
]
],
[
[
"model_dir = tempfile.mkdtemp()\nmodel = tf.estimator.LinearClassifier(\n model_dir=model_dir,\n feature_columns=[embark, cls, age],\n n_classes=2\n)",
"_____no_output_____"
]
],
[
[
"For further information, see the [linear classifier tutorial](https://www.tensorflow.org/tutorials/estimator/linear).",
"_____no_output_____"
],
[
"### 4. Call a training, evaluation, or inference method.\n\nAll Estimators provide `train`, `evaluate`, and `predict` methods.\n",
"_____no_output_____"
]
],
[
[
"model = model.train(input_fn=train_input_fn, steps=100)",
"_____no_output_____"
],
[
"result = model.evaluate(train_input_fn, steps=10)\n\nfor key, value in result.items():\n print(key, \":\", value)",
"_____no_output_____"
],
[
"for pred in model.predict(train_input_fn):\n for key, value in pred.items():\n print(key, \":\", value)\n break",
"_____no_output_____"
]
],
[
[
"### Benefits of pre-made Estimators\n\nPre-made Estimators encode best practices, providing the following benefits:\n\n* Best practices for determining where different parts of the computational graph should run, implementing strategies on a single machine or on a\n cluster.\n* Best practices for event (summary) writing and universally useful\n summaries.\n\nIf you don't use pre-made Estimators, you must implement the preceding features yourself.",
"_____no_output_____"
],
[
"## Custom Estimators\n\nThe heart of every Estimator—whether pre-made or custom—is its *model function*, `model_fn`, which is a method that builds graphs for training, evaluation, and prediction. When you are using a pre-made Estimator, someone else has already implemented the model function. When relying on a custom Estimator, you must write the model function yourself.\n\n> Note: A custom `model_fn` will still run in 1.x-style graph mode. This means there is no eager execution and no automatic control dependencies. You should plan to migrate away from `tf.estimator` with custom `model_fn`. The alternative APIs are `tf.keras` and `tf.distribute`. If you still need an `Estimator` for some part of your training you can use the `tf.keras.estimator.model_to_estimator` converter to create an `Estimator` from a `keras.Model`.",
"_____no_output_____"
],
[
"## Create an Estimator from a Keras model\n\nYou can convert existing Keras models to Estimators with `tf.keras.estimator.model_to_estimator`. This is helpful if you want to modernize your model code, but your training pipeline still requires Estimators. \n\nInstantiate a Keras MobileNet V2 model and compile the model with the optimizer, loss, and metrics to train with:",
"_____no_output_____"
],
[
"import tensorflow as tf\nimport tensorflow_datasets as tfds",
"_____no_output_____"
]
],
[
[
"keras_mobilenet_v2 = tf.keras.applications.MobileNetV2(\n input_shape=(160, 160, 3), include_top=False)\nkeras_mobilenet_v2.trainable = False\n\nestimator_model = tf.keras.Sequential([\n keras_mobilenet_v2,\n tf.keras.layers.GlobalAveragePooling2D(),\n tf.keras.layers.Dense(1)\n])\n\n# Compile the model\nestimator_model.compile(\n optimizer='adam',\n loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"Create an `Estimator` from the compiled Keras model. The initial model state of the Keras model is preserved in the created `Estimator`:",
"_____no_output_____"
]
],
[
[
"est_mobilenet_v2 = tf.keras.estimator.model_to_estimator(keras_model=estimator_model)",
"_____no_output_____"
]
],
[
[
"Treat the derived `Estimator` as you would with any other `Estimator`.",
"_____no_output_____"
]
],
[
[
"IMG_SIZE = 160 # All images will be resized to 160x160\n\ndef preprocess(image, label):\n image = tf.cast(image, tf.float32)\n image = (image/127.5) - 1\n image = tf.image.resize(image, (IMG_SIZE, IMG_SIZE))\n return image, label",
"_____no_output_____"
],
[
"def train_input_fn(batch_size):\n data = tfds.load('cats_vs_dogs', as_supervised=True)\n train_data = data['train']\n train_data = train_data.map(preprocess).shuffle(500).batch(batch_size)\n return train_data",
"_____no_output_____"
]
],
[
[
"To train, call Estimator's train function:",
"_____no_output_____"
]
],
[
[
"est_mobilenet_v2.train(input_fn=lambda: train_input_fn(32), steps=50)",
"_____no_output_____"
]
],
[
[
"Similarly, to evaluate, call the Estimator's evaluate function:",
"_____no_output_____"
]
],
[
[
"est_mobilenet_v2.evaluate(input_fn=lambda: train_input_fn(32), steps=10)",
"_____no_output_____"
]
],
[
[
"For more details, please refer to the documentation for `tf.keras.estimator.model_to_estimator`.",
"_____no_output_____"
],
[
"## Saving object-based checkpoints with Estimator\n\nEstimators by default save checkpoints with variable names rather than the object graph described in the [Checkpoint guide](checkpoint.ipynb). `tf.train.Checkpoint` will read name-based checkpoints, but variable names may change when moving parts of a model outside of the Estimator's `model_fn`. For forwards compatibility saving object-based checkpoints makes it easier to train a model inside an Estimator and then use it outside of one.",
"_____no_output_____"
]
],
[
[
"import tensorflow.compat.v1 as tf_compat",
"_____no_output_____"
],
[
"def toy_dataset():\n inputs = tf.range(10.)[:, None]\n labels = inputs * 5. + tf.range(5.)[None, :]\n return tf.data.Dataset.from_tensor_slices(\n dict(x=inputs, y=labels)).repeat().batch(2)",
"_____no_output_____"
],
[
"class Net(tf.keras.Model):\n \"\"\"A simple linear model.\"\"\"\n\n def __init__(self):\n super(Net, self).__init__()\n self.l1 = tf.keras.layers.Dense(5)\n\n def call(self, x):\n return self.l1(x)",
"_____no_output_____"
],
[
"def model_fn(features, labels, mode):\n net = Net()\n opt = tf.keras.optimizers.Adam(0.1)\n ckpt = tf.train.Checkpoint(step=tf_compat.train.get_global_step(),\n optimizer=opt, net=net)\n with tf.GradientTape() as tape:\n output = net(features['x'])\n loss = tf.reduce_mean(tf.abs(output - features['y']))\n variables = net.trainable_variables\n gradients = tape.gradient(loss, variables)\n return tf.estimator.EstimatorSpec(\n mode,\n loss=loss,\n train_op=tf.group(opt.apply_gradients(zip(gradients, variables)),\n ckpt.step.assign_add(1)),\n # Tell the Estimator to save \"ckpt\" in an object-based format.\n scaffold=tf_compat.train.Scaffold(saver=ckpt))\n\ntf.keras.backend.clear_session()\nest = tf.estimator.Estimator(model_fn, './tf_estimator_example/')\nest.train(toy_dataset, steps=10)",
"_____no_output_____"
]
],
[
[
"`tf.train.Checkpoint` can then load the Estimator's checkpoints from its `model_dir`.",
"_____no_output_____"
]
],
[
[
"opt = tf.keras.optimizers.Adam(0.1)\nnet = Net()\nckpt = tf.train.Checkpoint(\n step=tf.Variable(1, dtype=tf.int64), optimizer=opt, net=net)\nckpt.restore(tf.train.latest_checkpoint('./tf_estimator_example/'))\nckpt.step.numpy() # From est.train(..., steps=10)",
"_____no_output_____"
]
],
[
[
"## SavedModels from Estimators\n\nEstimators export SavedModels through [`tf.Estimator.export_saved_model`](https://www.tensorflow.org/api_docs/python/tf/estimator/Estimator#export_saved_model).",
"_____no_output_____"
]
],
[
[
"input_column = tf.feature_column.numeric_column(\"x\")\n\nestimator = tf.estimator.LinearClassifier(feature_columns=[input_column])\n\ndef input_fn():\n return tf.data.Dataset.from_tensor_slices(\n ({\"x\": [1., 2., 3., 4.]}, [1, 1, 0, 0])).repeat(200).shuffle(64).batch(16)\nestimator.train(input_fn)",
"_____no_output_____"
]
],
[
[
"To save an `Estimator` you need to create a `serving_input_receiver`. This function builds a part of a `tf.Graph` that parses the raw data received by the SavedModel. \n\nThe `tf.estimator.export` module contains functions to help build these `receivers`.\n",
"_____no_output_____"
],
[
"The following code builds a receiver, based on the `feature_columns`, that accepts serialized `tf.Example` protocol buffers, which are often used with [tf-serving](https://tensorflow.org/serving).",
"_____no_output_____"
]
],
[
[
"tmpdir = tempfile.mkdtemp()\n\nserving_input_fn = tf.estimator.export.build_parsing_serving_input_receiver_fn(\n tf.feature_column.make_parse_example_spec([input_column]))\n\nestimator_base_path = os.path.join(tmpdir, 'from_estimator')\nestimator_path = estimator.export_saved_model(estimator_base_path, serving_input_fn)",
"_____no_output_____"
]
],
[
[
"You can also load and run that model, from python:",
"_____no_output_____"
]
],
[
[
"imported = tf.saved_model.load(estimator_path)\n\ndef predict(x):\n example = tf.train.Example()\n example.features.feature[\"x\"].float_list.value.extend([x])\n return imported.signatures[\"predict\"](\n examples=tf.constant([example.SerializeToString()]))",
"_____no_output_____"
],
[
"print(predict(1.5))\nprint(predict(3.5))",
"_____no_output_____"
]
],
[
[
"`tf.estimator.export.build_raw_serving_input_receiver_fn` allows you to create input functions which take raw tensors rather than `tf.train.Example`s.",
"_____no_output_____"
],
[
"## Using `tf.distribute.Strategy` with Estimator (Limited support)\n\nSee the [Distributed training guide](guide/distributed_training.ipynb) for more info.\n\n`tf.estimator` is a distributed training TensorFlow API that originally supported the async parameter server approach. `tf.estimator` now supports `tf.distribute.Strategy`. If you're using `tf.estimator`, you can change to distributed training with very few changes to your code. With this, Estimator users can now do synchronous distributed training on multiple GPUs and multiple workers, as well as use TPUs. This support in Estimator is, however, limited. See [What's supported now](#estimator_support) section below for more details.\n\nThe usage of `tf.distribute.Strategy` with Estimator is slightly different than the Keras case. Instead of using `strategy.scope`, now we pass the strategy object into the [`RunConfig`](https://www.tensorflow.org/api_docs/python/tf/estimator/RunConfig) for the Estimator.\n\nHere is a snippet of code that shows this with a premade Estimator `LinearRegressor` and `MirroredStrategy`:\n",
"_____no_output_____"
]
],
[
[
"mirrored_strategy = tf.distribute.MirroredStrategy()\nconfig = tf.estimator.RunConfig(\n train_distribute=mirrored_strategy, eval_distribute=mirrored_strategy)\nregressor = tf.estimator.LinearRegressor(\n feature_columns=[tf.feature_column.numeric_column('feats')],\n optimizer='SGD',\n config=config)",
"_____no_output_____"
]
],
[
[
"We use a premade Estimator here, but the same code works with a custom Estimator as well. `train_distribute` determines how training will be distributed, and `eval_distribute` determines how evaluation will be distributed. This is another difference from Keras where we use the same strategy for both training and eval.\n\nNow we can train and evaluate this Estimator with an input function:\n",
"_____no_output_____"
]
],
[
[
"def input_fn():\n dataset = tf.data.Dataset.from_tensors(({\"feats\":[1.]}, [1.]))\n return dataset.repeat(1000).batch(10)\nregressor.train(input_fn=input_fn, steps=10)\nregressor.evaluate(input_fn=input_fn, steps=10)",
"_____no_output_____"
]
],
[
[
"Another difference to highlight here between Estimator and Keras is the input handling. In Keras, we mentioned that each batch of the dataset is split automatically across the multiple replicas. In Estimator, however, we do not do automatic splitting of batch, nor automatically shard the data across different workers. You have full control over how you want your data to be distributed across workers and devices, and you must provide an `input_fn` to specify how to distribute your data.\n\nYour `input_fn` is called once per worker, thus giving one dataset per worker. Then one batch from that dataset is fed to one replica on that worker, thereby consuming N batches for N replicas on 1 worker. In other words, the dataset returned by the `input_fn` should provide batches of size `PER_REPLICA_BATCH_SIZE`. And the global batch size for a step can be obtained as `PER_REPLICA_BATCH_SIZE * strategy.num_replicas_in_sync`.\n\nWhen doing multi worker training, you should either split your data across the workers, or shuffle with a random seed on each. You can see an example of how to do this in the [Multi-worker Training with Estimator](../tutorials/distribute/multi_worker_with_estimator.ipynb).",
"_____no_output_____"
],
[
"And similarly, you can use multi worker and parameter server strategies as well. The code remains the same, but you need to use `tf.estimator.train_and_evaluate`, and set `TF_CONFIG` environment variables for each binary running in your cluster.",
"_____no_output_____"
],
[
"<a name=\"estimator_support\"></a>\n### What's supported now?\n\nThere is limited support for training with Estimator using all strategies except `TPUStrategy`. Basic training and evaluation should work, but a number of advanced features such as `v1.train.Scaffold` do not. There may also be a number of bugs in this integration. At this time, we do not plan to actively improve this support, and instead are focused on Keras and custom training loop support. If at all possible, you should prefer to use `tf.distribute` with those APIs instead.\n\n| Training API \t| MirroredStrategy \t| TPUStrategy \t| MultiWorkerMirroredStrategy \t| CentralStorageStrategy \t| ParameterServerStrategy \t|\n|:---------------\t|:------------------\t|:-------------\t|:-----------------------------\t|:------------------------\t|:-------------------------\t|\n| Estimator API \t| Limited Support \t| Not supported \t| Limited Support \t| Limited Support \t| Limited Support \t|\n\n### Examples and Tutorials\nHere are some examples that show end to end usage of various strategies with Estimator:\n\n1. [Multi-worker Training with Estimator](../tutorials/distribute/multi_worker_with_estimator.ipynb) to train MNIST with multiple workers using `MultiWorkerMirroredStrategy`.\n2. [End to end example](https://github.com/tensorflow/ecosystem/tree/master/distribution_strategy) for multi worker training in tensorflow/ecosystem using Kubernetes templates. This example starts with a Keras model and converts it to an Estimator using the `tf.keras.estimator.model_to_estimator` API.\n3. Official [ResNet50](https://github.com/tensorflow/models/blob/master/official/vision/image_classification/resnet_imagenet_main.py) model, which can be trained using either `MirroredStrategy` or `MultiWorkerMirroredStrategy`.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cb4dc1c7561a789ee8e1a0e990d138eb14c97790 | 5,181 | ipynb | Jupyter Notebook | one_million/one_word_data_maker-test.ipynb | Sshanu/WSD | 74905c60e48ab1884bf5c8f208e21015f7f7fd21 | [
"MIT"
] | 29 | 2017-12-11T09:26:05.000Z | 2019-06-11T07:59:24.000Z | one_million/one_word_data_maker-test.ipynb | Sshanu/WSD | 74905c60e48ab1884bf5c8f208e21015f7f7fd21 | [
"MIT"
] | 1 | 2019-04-18T06:37:41.000Z | 2019-04-18T06:37:41.000Z | one_million/one_word_data_maker-test.ipynb | Sshanu/WSD | 74905c60e48ab1884bf5c8f208e21015f7f7fd21 | [
"MIT"
] | 7 | 2017-12-08T10:55:21.000Z | 2018-10-31T01:47:27.000Z | 20.724 | 95 | 0.476163 | [
[
[
"import pickle\nfrom nltk.corpus import wordnet as wn",
"_____no_output_____"
],
[
"with open('/data/aviraj/dataset/raw_preprocess_test','rb') as f:\n global_data=pickle.load(f)",
"_____no_output_____"
],
[
"with open('/data/aviraj/dataset/ALL.gold.key.txt','r') as f:\n data_key=f.readlines()",
"_____no_output_____"
],
[
"global_data[0][3]",
"_____no_output_____"
],
[
"def make_word_data(checkword):\n \n dataset_line=[]\n for i,list_ in enumerate(global_data): \n ind=[idx for idx,it in enumerate(list_[3]) if it==checkword]\n for ii in ind:\n if list_[2][ii] is not None:\n dataset_line.append([list_[2][ii],list_[1],list_[4]])\n \n print(len(dataset_line))\n with open('/data/aviraj/dataset/checkwords/'+checkword + '_data_test', 'wb') as f:\n pickle.dump(dataset_line, f)\n with open('/data/aviraj/dataset/checkwords/'+checkword + '_data_test', 'rb') as f:\n data_ = pickle.load(f)",
"_____no_output_____"
],
[
"test_words = ['force', 'make', 'open', 'place', 'point', 'serve', 'support']",
"_____no_output_____"
],
[
"for word in test_words:\n make_word_data(word)",
"1\n31\n4\n5\n11\n2\n12\n"
],
[
"with open('../Glove/vocab_glove', 'rb') as f:\n vocab = pickle.load(f)\n",
"_____no_output_____"
],
[
"train_words = []\nfor sent in global_data:\n train_words.extend(sent[1])",
"_____no_output_____"
],
[
"len(train_words), len(set(train_words)), len(vocab)",
"_____no_output_____"
],
[
"import collections\nunknown_words = []\nfor word in set(train_words):\n if word not in vocab:\n unknown_words.append(word)\n \nun_counter = collections.Counter(unknown_words)\nun_counter = dict(un_counter)\n\nsorted_un_counter = sorted(un_counter.items(), key=lambda x:x[1], reverse=True)\nsorted_un_counter",
"_____no_output_____"
],
[
"with open('million_unknown_words.pickle', 'wb') as f:\n pickle.dump(unknown_words, f)",
"_____no_output_____"
],
[
"len(sorted(global_data, key=lambda x:len(x[1]), reverse=True)[0][1])",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4dcac981e869ca3b0d91b00a8a085697b79b20 | 133,355 | ipynb | Jupyter Notebook | lgbm/lgbm_test.ipynb | nkelley-uga/misc_projects | ca9a07665e1e4c4dd6c8fb3f3223f9fbb9ee8dc4 | [
"MIT"
] | null | null | null | lgbm/lgbm_test.ipynb | nkelley-uga/misc_projects | ca9a07665e1e4c4dd6c8fb3f3223f9fbb9ee8dc4 | [
"MIT"
] | null | null | null | lgbm/lgbm_test.ipynb | nkelley-uga/misc_projects | ca9a07665e1e4c4dd6c8fb3f3223f9fbb9ee8dc4 | [
"MIT"
] | null | null | null | 99.966267 | 22,732 | 0.792861 | [
[
[
"# LightGBM Test Run\n\n### summary\n\n- 'gender' and 'device' are discrete strings -> dummies (keep both M and F to capture nan info)\n- 'drivers', 'vehicles', 'age', 'launch', and 'tenure' are all discrete numeric -> lgbm can handle\n- target variable 'outcome' is imbalanced at approximately 1:9 -> use lgbm imbalanced setting\n- multicolinearity with 'age', 'income', and 'tenure' -> lgbm (trees) is robust to colinearity",
"_____no_output_____"
],
[
"---\n\n# EDA",
"_____no_output_____"
]
],
[
[
"# mac install for lightgbm\n\n# !brew install lightgbm",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom sklearn.ensemble import RandomForestClassifier\nimport lightgbm\nfrom sklearn.model_selection import train_test_split, StratifiedKFold\nfrom sklearn import metrics\n\npd.set_option('display.max_rows', 50)\npd.set_option('display.max_columns', 50)\n\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"data = pd.read_csv('train.csv')\nprint(data.shape)\nlist(data.columns)",
"(10000, 10)\n"
],
[
"data.head()",
"_____no_output_____"
],
[
"def summary(df):\n '''exploratory data analysis of a dataframe'''\n \n import pandas as pd\n \n summary = {}\n summary['count'] = df.count()\n summary['dtypes'] = df.dtypes\n summary['null_sum'] = df.isnull().sum()\n summary['null_pct'] = df.isnull().mean()\n summary['mean'] = df.mean()\n summary['median'] = df.median()\n summary['min'] = df.min()\n summary['max'] = df.max()\n \n return pd.DataFrame(summary)",
"_____no_output_____"
],
[
"summary(data)",
"_____no_output_____"
],
[
"def numeric_hists(df):\n '''quick histograms for numeric features'''\n \n import pandas as pd \n import matplotlib.pyplot as plt\n \n for col in df.columns:\n if df[col].dtype in ['int64', 'float64']:\n print(f'{col}:')\n plt.hist(df[col])\n plt.show()",
"_____no_output_____"
],
[
"numeric_hists(data)",
"age:\n"
],
[
"data.corr()",
"_____no_output_____"
],
[
"for idx, i in enumerate(data.columns):\n if data[i].dtype in ['int64', 'float64']:\n for j in data.columns[idx+1:]:\n if data[j].dtype in ['int64', 'float64']:\n r = data[[i, j]].corr().values[0][1]\n if abs(r) > 0.2:\n print((i, j, r))",
"('age', 'income', 0.7472864357955471)\n('age', 'prior_ins_tenure', 0.6447341586065465)\n('income', 'prior_ins_tenure', 0.48653245183601684)\n"
],
[
"# missing gender has ~1/3 frequency of positive outcomes as overall data\n\nround(data[data['gender'].isna()]['outcome'].mean(), 2), round(data['outcome'].mean(), 2)",
"_____no_output_____"
]
],
[
[
"---\n\n# Transformations",
"_____no_output_____"
]
],
[
[
"# z_scores\n\ndef z_score(col):\n mu = col.mean()\n sig = col.std()\n return [(i-mu)/sig for i in col]\n\nfor col in ['cost_of_ad', 'income']:\n data[col] = z_score(data[col])",
"_____no_output_____"
],
[
"# dummies\n\ndevice = pd.get_dummies(data['device_type'], drop_first=True)\ndevice.columns = [f'device_{i}' for i in device.columns]\n\ngender = pd.get_dummies(data['gender'], drop_first=False)\n\ndata = data.drop(labels=['gender', 'device_type'], axis=1)\ndata = pd.concat([data, device, gender], axis=1)\ndata.head()",
"_____no_output_____"
],
[
"# feature correlations with outcome\n\ncorrelations = []\n\nfor col in data.columns:\n if col == 'outcome':\n pass\n else:\n if data[col].dtype in ['int64', 'float64', 'uint8']:\n correlations.append((col, np.corrcoef(data[col], data['outcome'])[0][1])) # extract r\n else:\n dummies = pd.get_dummies(data[col])\n for col in dummies.columns:\n correlations.append((col, np.corrcoef(dummies[col], data['outcome'])[0][1]))\n \ncorrelations = sorted(correlations, key = lambda x: x[1])\n\nplt.figure(figsize=(10, 10))\nplt.barh([i[0] for i in correlations],\n [i[1] for i in correlations])\nplt.title('Feature Correlations with Outcome', fontsize=18);",
"_____no_output_____"
],
[
"rfc = RandomForestClassifier(random_state=42)\n\nX_train = data.drop(labels=['outcome'], axis=1)\ny_train = data['outcome']\n\nrfc = rfc.fit(X_train, y_train)\n\nfeats = sorted(zip(rfc.feature_importances_, X_train.columns), key=lambda x: x[0])\n\nplt.figure(figsize=(10, 10))\nplt.barh([i[1] for i in feats],\n [i[0] for i in feats])\nplt.title('Feature Importances via Random Forest', fontsize=18);",
"_____no_output_____"
]
],
[
[
"---\n\n# Modeling",
"_____no_output_____"
]
],
[
[
"def run_kfold(X_train, y_train, model, kfold):\n idx = 1\n in_ = []\n out_ = []\n for train_index, test_index in kfold.split(X_train, y_train):\n try:\n model.fit(X_train.iloc[train_index], y_train.iloc[train_index])\n except:\n y_train = pd.Series(y_train)\n model.fit(X_train.iloc[train_index], y_train.iloc[train_index])\n train_fp = model.predict(X_train.iloc[train_index])\n test_fp = model.predict(X_train.iloc[test_index])\n in_.append(metrics.roc_auc_score(y_train.iloc[train_index], train_fp))\n out_.append(metrics.roc_auc_score(y_train.iloc[test_index], test_fp))\n idx += 1\n return np.mean(in_), np.mean(out_)\n\nskf = StratifiedKFold(n_splits=10, shuffle=True, random_state=42)",
"_____no_output_____"
],
[
"# default lgbm\n\nlgb = lightgbm.LGBMClassifier(random_state=42,\n is_unbalance=True)\n\nin_, out_ = run_kfold(X_train, y_train, model=lgb, kfold=skf)\nprint(f'Average in auc: {in_}')\nprint(f'Average oos auc: {out_}')",
"Average in auc: 0.9268453521764481\nAverage oos auc: 0.7424621492044641\n"
],
[
"# some manual tuning for faster, smaller trees\n\nlgb1 = lightgbm.LGBMClassifier(random_state=42,\n is_unbalance=True,\n learning_rate = 0.5,\n num_leaves = 2**3-1,\n num_trees = 50,\n min_data_in_leaf = 200,\n max_bin=2**3-1)\n\nin_, out_ = run_kfold(X_train, y_train, model=lgb1, kfold=skf)\nprint(f'Average in auc: {in_}')\nprint(f'Average oos auc: {out_}')",
"Average in auc: 0.8098501444330222\nAverage oos auc: 0.7636953916961224\n"
],
[
"# model for slower, smaller trees\n\nlgb2 = lightgbm.LGBMClassifier(random_state=42,\n is_unbalance=True,\n learning_rate = 0.1,\n num_leaves = 2**3-1,\n num_trees = 50,\n min_data_in_leaf = 10,\n max_bin=2**3-1)\n\nin_, out_ = run_kfold(X_train, y_train, model=lgb2, kfold=skf)\nprint(f'Average in auc: {in_}')\nprint(f'Average oos auc: {out_}')",
"Average in auc: 0.780120472401631\nAverage oos auc: 0.7643729753402707\n"
],
[
"# model for slower, bigger trees\n\nlgb3 = lightgbm.LGBMClassifier(random_state=42,\n is_unbalance=True,\n learning_rate = 0.005,\n num_leaves = 2**5-1,\n num_trees = 500,\n min_data_in_leaf = 100,\n max_bin=2**5-1)\n\nin_, out_ = run_kfold(X_train, y_train, model=lgb3, kfold=skf)\nprint(f'Average in auc: {in_}')\nprint(f'Average oos auc: {out_}')",
"Average in auc: 0.8169493667952074\nAverage oos auc: 0.7635953421935509\n"
],
[
"# ensembling\n\nin_all = []\nout_all = []\n\nfor train_index, test_index in skf.split(X_train, y_train):\n \n in_ = []\n out_ = []\n for model in [lgb1, lgb2, lgb3]:\n model.fit(X_train.iloc[train_index], y_train.iloc[train_index])\n in_.append(model.predict_proba(X_train.iloc[train_index])[:,1])\n out_.append(model.predict_proba(X_train.iloc[test_index])[:,1])\n \n in_pred = [1 if (i * in_[1][idx] * in_[2][idx])**(1/3) > 0.5\n else 0 for idx, i in enumerate(in_[0])]\n out_pred = [1 if (i * out_[1][idx] * out_[2][idx])**(1/3) > 0.5\n else 0 for idx, i in enumerate(out_[0])]\n\n in_all.append(metrics.roc_auc_score(y_train.iloc[train_index], in_pred))\n out_all.append(metrics.roc_auc_score(y_train.iloc[test_index], out_pred))\n \nprint(f'Average in auc: {np.mean(in_all)}')\nprint(f'Average oos auc: {np.mean(out_all)}')",
"Average in auc: 0.806352829704306\nAverage oos auc: 0.7721049801337093\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4dcaceb1aa8d997222811f85edbabacbb6e05f | 47,691 | ipynb | Jupyter Notebook | Notebook/Section 5 - RNN and NLP v2-experiment2.ipynb | PacktPublishing/Practical-Deep-Learning-with-PyTorch | 5507decaf76a73b3e2465f0fb0818d0b9c8dd83b | [
"MIT"
] | 17 | 2019-04-11T14:22:57.000Z | 2021-07-24T13:32:00.000Z | Notebook/Section 5 - RNN and NLP v2-experiment2.ipynb | PacktPublishing/Practical-Deep-Learning-with-PyTorch | 5507decaf76a73b3e2465f0fb0818d0b9c8dd83b | [
"MIT"
] | null | null | null | Notebook/Section 5 - RNN and NLP v2-experiment2.ipynb | PacktPublishing/Practical-Deep-Learning-with-PyTorch | 5507decaf76a73b3e2465f0fb0818d0b9c8dd83b | [
"MIT"
] | 14 | 2019-04-11T15:31:29.000Z | 2021-04-18T21:19:50.000Z | 88.810056 | 17,100 | 0.821035 | [
[
[
"<div class=\"alert alert-block alert-info\">\n<font size=\"5\"><b><center> Section 5</font></center>\n<br>\n<font size=\"5\"><b><center>Recurrent Neural Network in PyTorch with an Introduction to Natural Language Processing</font></center>\n</div>",
"_____no_output_____"
],
[
"Credit: This example is obtained from the following book:\n\nSubramanian, Vishnu. 2018. \"*Deep Learning with PyTorch: A Practical Approach to Building Neural Network Models Using PyTorch.*\" Birmingham, U.K., Packt Publishing.",
"_____no_output_____"
],
[
"# Simple Text Processing",
"_____no_output_____"
],
[
"## Typically Data Preprocessing Steps before Modeling Training for NLP Applications\n\n* Read the data from disk\n* Tokenize the text\n* Create a mapping from word to a unique integer\n* Convert the text into lists of integers\n* Load the data in whatever format your deep learning framework requires\n* Pad the text so that all the sequences are the same length, so you can process them in batch",
"_____no_output_____"
],
[
"## Word Embedding",
"_____no_output_____"
],
[
"Word embedding is a very popular way of representing text data in problems that are solved by deep learning algorithms\n\nWord embedding provides a dense representation of a word filled with floating numbers. \n\nIt drastically reduces the dimension of the dictionary\n\n",
"_____no_output_____"
],
[
"### `Torchtext` and Training word embedding by building a sentiment classifier",
"_____no_output_____"
],
[
"Torchtext takes a declarative approach to loading its data: \n\n * you tell torchtext how you want the data to look like, and torchtext handles it for you\n \n * Declaring a Field: The Field specifies how you want a certain field to be processed\n \nThe `Field` class is a fundamental component of torchtext and is what makes preprocessing very easy\n\n",
"_____no_output_____"
],
[
"### Load `torchtext.datasets`",
"_____no_output_____"
],
[
"# Use LSTM for Sentiment Classification",
"_____no_output_____"
],
[
"1. Preparing the data\n2. Creating the batches \n3. Creating the network \n4. Training the model",
"_____no_output_____"
]
],
[
[
"from torchtext import data, datasets\nfrom torchtext.vocab import GloVe,FastText,CharNGram\n\nTEXT = data.Field(lower=True, fix_length=100,batch_first=False)\nLABEL = data.Field(sequential=False,)\n\ntrain, test = datasets.imdb.IMDB.splits(TEXT, LABEL)\n\nTEXT.build_vocab(train, vectors=GloVe(name='6B', dim=300),max_size=10000,min_freq=10)\nLABEL.build_vocab(train,)",
"_____no_output_____"
],
[
"len(TEXT.vocab.vectors)",
"_____no_output_____"
],
[
"train_iter, test_iter = data.BucketIterator.splits((train, test), batch_size=32, device=-1)\ntrain_iter.repeat = False\ntest_iter.repeat = False",
"The `device` argument should be set by using `torch.device` or passing a string as an argument. This behavior will be deprecated soon and currently defaults to cpu.\nThe `device` argument should be set by using `torch.device` or passing a string as an argument. This behavior will be deprecated soon and currently defaults to cpu.\n"
],
[
"import torch.nn as nn\nimport torch.optim as optim\nimport torch.nn.functional as F\nfrom torch.autograd import Variable",
"_____no_output_____"
],
[
"class IMDBRnn(nn.Module):\n \n def __init__(self,vocab,hidden_size,n_cat,bs=1,nl=2):\n super().__init__()\n self.hidden_size = hidden_size\n self.bs = bs\n self.nl = nl\n self.e = nn.Embedding(n_vocab,hidden_size)\n self.rnn = nn.LSTM(hidden_size,hidden_size,nl)\n self.fc2 = nn.Linear(hidden_size,n_cat)\n self.softmax = nn.LogSoftmax(dim=-1)\n\n def forward(self,inp):\n bs = inp.size()[1]\n if bs != self.bs:\n self.bs = bs\n e_out = self.e(inp)\n h0 = c0 = Variable(e_out.data.new(*(self.nl,self.bs,self.hidden_size)).zero_())\n rnn_o,_ = self.rnn(e_out,(h0,c0))\n rnn_o = rnn_o[-1]\n fc = F.dropout(self.fc2(rnn_o),p=0.8)\n return self.softmax(fc)",
"_____no_output_____"
],
[
"n_vocab = len(TEXT.vocab)\nn_hidden = 100",
"_____no_output_____"
],
[
"model = IMDBRnn(n_vocab,n_hidden,n_cat=3,bs=32)\n#model = model.cuda()\n\noptimizer = optim.Adam(model.parameters(),lr=1e-3)\n\ndef fit(epoch,model,data_loader,phase='training',volatile=False):\n if phase == 'training':\n model.train()\n if phase == 'validation':\n model.eval()\n volatile=True\n running_loss = 0.0\n running_correct = 0\n for batch_idx , batch in enumerate(data_loader):\n text , target = batch.text , batch.label\n# if is_cuda:\n# text,target = text.cuda(),target.cuda()\n \n if phase == 'training':\n optimizer.zero_grad()\n output = model(text)\n loss = F.nll_loss(output,target)\n \n #running_loss += F.nll_loss(output,target,size_average=False).data[0]\n running_loss += F.nll_loss(output,target,size_average=False).data\n preds = output.data.max(dim=1,keepdim=True)[1]\n running_correct += preds.eq(target.data.view_as(preds)).cpu().sum()\n if phase == 'training':\n loss.backward()\n optimizer.step()\n \n loss = running_loss/len(data_loader.dataset)\n accuracy = 100. * running_correct/len(data_loader.dataset)\n print(\"epoch: \", epoch, \"loss: \", loss, \"accuracy: \", accuracy)\n #print(f'{phase} loss is {loss:{5}.{2}} and {phase} accuracy is {running_correct}/{len(data_loader.dataset)}{accuracy:{10}.{4}}')\n return loss,accuracy",
"_____no_output_____"
],
[
"import time\nstart = time.time()\n\ntrain_losses , train_accuracy = [],[]\nval_losses , val_accuracy = [],[]\n\nfor epoch in range(1,20):\n\n epoch_loss, epoch_accuracy = fit(epoch,model,train_iter,phase='training')\n val_epoch_loss , val_epoch_accuracy = fit(epoch,model,test_iter,phase='validation')\n train_losses.append(epoch_loss)\n train_accuracy.append(epoch_accuracy)\n val_losses.append(val_epoch_loss)\n val_accuracy.append(val_epoch_accuracy)\n \nend = time.time()\nprint((end-start)/60)\nprint(\"Execution Time: \", round(((end-start)/60),1), \"minutes\")",
"/Users/jeffrey/anaconda2/envs/dl/lib/python3.6/site-packages/torch/nn/_reduction.py:49: UserWarning: size_average and reduce args will be deprecated, please use reduction='sum' instead.\n warnings.warn(warning.format(ret))\n"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n\nplt.plot(range(1,len(train_losses)+1),train_losses,'bo',label = 'training loss')\nplt.plot(range(1,len(val_losses)+1),val_losses,'r',label = 'validation loss')\nplt.legend()",
"_____no_output_____"
],
[
"plt.plot(range(1,len(train_accuracy)+1),train_accuracy,'bo',label = 'train accuracy')\nplt.plot(range(1,len(val_accuracy)+1),val_accuracy,'r',label = 'val accuracy')\nplt.legend()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4dcc896f02d6dbd0f740641614c5ad92c2d8e2 | 2,251 | ipynb | Jupyter Notebook | config.ipynb | SkyMornKey/nightShot | c4d91f0af403c8ff4684fcb80a37247c120b59ba | [
"MIT"
] | 2 | 2019-02-23T04:25:27.000Z | 2019-06-07T15:34:24.000Z | config.ipynb | YCL92/nightShot | c4d91f0af403c8ff4684fcb80a37247c120b59ba | [
"MIT"
] | null | null | null | config.ipynb | YCL92/nightShot | c4d91f0af403c8ff4684fcb80a37247c120b59ba | [
"MIT"
] | null | null | null | 22.287129 | 62 | 0.534429 | [
[
[
"# Network Configration",
"_____no_output_____"
]
],
[
[
"class Config():\n # data path\n data_root = '/trainSets/Sony'\n save_root = None # intermediate saves\n\n # network parameters\n img_size = 512 # iuput image size\n batch_size = 1 # batch size\n lr = 1e-4 # learning rate\n lr_decay = 0.1 # learning rate decay ratio\n upd_freq = 2000 # learning rate update frequency\n max_epoch = 4000 # max epoch\n\n # other parameters\n num_workers = 8 # number of threads\n save_freq = 100 # save frequency\n val_freq = 50 # validation frequency",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
cb4dd525a857fcf543622568257c9c6d60eb992b | 414,916 | ipynb | Jupyter Notebook | ml4trading-2ed/12_gradient_boosting_machines/11_intraday_model.ipynb | JennEYoon/SEC-data | a532364f0d1314a29e701acbcb904ba01e19acb3 | [
"BSD-3-Clause"
] | null | null | null | ml4trading-2ed/12_gradient_boosting_machines/11_intraday_model.ipynb | JennEYoon/SEC-data | a532364f0d1314a29e701acbcb904ba01e19acb3 | [
"BSD-3-Clause"
] | 2 | 2020-10-27T19:44:15.000Z | 2020-11-03T23:55:36.000Z | ml4trading-2ed/12_gradient_boosting_machines/11_intraday_model.ipynb | JennEYoon/SEC-data | a532364f0d1314a29e701acbcb904ba01e19acb3 | [
"BSD-3-Clause"
] | null | null | null | 160.447022 | 70,476 | 0.858448 | [
[
[
"# Intraday Strategy, Part 2: Model Training & Signal Evaluation",
"_____no_output_____"
],
[
"In this notebook, we load the high-quality NASDAQ100 minute-bar trade-and-quote data generously provided by [Algoseek](https://www.algoseek.com/) (available [here](https://www.algoseek.com/ml4t-book-data.html)) and use the features engineered in the last notebook to train gradient boosting model that predicts the returns for the NASDAQ100 stocks over the next 1-minute bar. \n\n> Note that we will assume throughout that we can always buy (sell) at the first (last) trade price for a given bar at no cost and without market impact. This does certainly not reflect market reality, and is rather due to the challenges of simulating a trading strategy at this much higher intraday frequency in a realistic manner using open-source tools.\n\nNote also that this section has slightly changed from the version published in the book to permit replication using the Algoseek data sample.",
"_____no_output_____"
],
[
"## Imports & Settings",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"%matplotlib inline\n\nimport sys, os\nfrom pathlib import Path\nfrom time import time\nfrom tqdm import tqdm\n\nimport numpy as np\nimport pandas as pd\n\nfrom scipy.stats import spearmanr\nimport lightgbm as lgb\n\nimport matplotlib.pyplot as plt\nfrom matplotlib.ticker import FuncFormatter\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"Ensuring we can import `utils.py` in the repo's root directory:",
"_____no_output_____"
]
],
[
[
"sys.path.insert(1, os.path.join(sys.path[0], '..'))\nfrom utils import format_time",
"_____no_output_____"
],
[
"sns.set_style('whitegrid')\nidx = pd.IndexSlice\ndeciles = np.arange(.1, 1, .1)",
"_____no_output_____"
],
[
"# where we stored the features engineered in the previous notebook\ndata_store = 'data/algoseek.h5'",
"_____no_output_____"
],
[
"# where we'll store the model results\nresult_store = 'data/intra_day.h5'",
"_____no_output_____"
],
[
"# here we save the trained models\nmodel_path = Path('models/intraday')\nif not model_path.exists():\n model_path.mkdir(parents=True)",
"_____no_output_____"
]
],
[
[
"## Load Model Data",
"_____no_output_____"
]
],
[
[
"data = pd.read_hdf(data_store, 'model_data2')",
"_____no_output_____"
],
[
"data.info(null_counts=True)",
"<class 'pandas.core.frame.DataFrame'>\nMultiIndex: 30875649 entries, ('AAL', Timestamp('2015-01-02 09:30:00')) to ('YHOO', Timestamp('2017-06-16 15:59:00'))\nData columns (total 22 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 minute 30875649 non-null int64 \n 1 ret1min 30612848 non-null float64\n 2 ret2min 30302846 non-null float64\n 3 ret3min 30220887 non-null float64\n 4 ret4min 30141503 non-null float64\n 5 ret5min 30063236 non-null float64\n 6 ret6min 29983969 non-null float64\n 7 ret7min 29903822 non-null float64\n 8 ret8min 29824607 non-null float64\n 9 ret9min 29745431 non-null float64\n 10 ret10min 29666821 non-null float64\n 11 fwd1min 30875649 non-null float64\n 12 rup 30083777 non-null float64\n 13 rdown 30083777 non-null float64\n 14 BOP 30612848 non-null float64\n 15 CCI 28517773 non-null float64\n 16 MFI 30873719 non-null float64\n 17 STOCHRSI 30871639 non-null float64\n 18 slowd 30873302 non-null float64\n 19 slowk 30873302 non-null float64\n 20 NATR 30873719 non-null float64\n 21 trades_bid_ask 30083777 non-null float64\ndtypes: float64(21), int64(1)\nmemory usage: 5.2+ GB\n"
],
[
"data.sample(frac=.1).describe(percentiles=np.arange(.1, 1, .1))",
"_____no_output_____"
]
],
[
[
"## Model Training",
"_____no_output_____"
],
[
"### Helper functions",
"_____no_output_____"
]
],
[
[
"class MultipleTimeSeriesCV:\n \"\"\"Generates tuples of train_idx, test_idx pairs\n Assumes the MultiIndex contains levels 'symbol' and 'date'\n purges overlapping outcomes\"\"\"\n\n def __init__(self,\n n_splits=3,\n train_period_length=126,\n test_period_length=21,\n lookahead=None,\n date_idx='date',\n shuffle=False):\n self.n_splits = n_splits\n self.lookahead = lookahead\n self.test_length = test_period_length\n self.train_length = train_period_length\n self.shuffle = shuffle\n self.date_idx = date_idx\n\n def split(self, X, y=None, groups=None):\n unique_dates = X.index.get_level_values(self.date_idx).unique()\n days = sorted(unique_dates, reverse=True)\n split_idx = []\n for i in range(self.n_splits):\n test_end_idx = i * self.test_length\n test_start_idx = test_end_idx + self.test_length\n train_end_idx = test_start_idx + self.lookahead - 1\n train_start_idx = train_end_idx + self.train_length + self.lookahead - 1\n split_idx.append([train_start_idx, train_end_idx,\n test_start_idx, test_end_idx])\n\n dates = X.reset_index()[[self.date_idx]]\n for train_start, train_end, test_start, test_end in split_idx:\n\n train_idx = dates[(dates[self.date_idx] > days[train_start])\n & (dates[self.date_idx] <= days[train_end])].index\n test_idx = dates[(dates[self.date_idx] > days[test_start])\n & (dates[self.date_idx] <= days[test_end])].index\n if self.shuffle:\n np.random.shuffle(list(train_idx))\n yield train_idx.to_numpy(), test_idx.to_numpy()\n\n def get_n_splits(self, X, y, groups=None):\n return self.n_splits\n",
"_____no_output_____"
],
[
"def get_fi(model):\n fi = model.feature_importance(importance_type='gain')\n return (pd.Series(fi / fi.sum(),\n index=model.feature_name()))",
"_____no_output_____"
]
],
[
[
"### Categorical Variables",
"_____no_output_____"
]
],
[
[
"data['stock_id'] = pd.factorize(data.index.get_level_values('ticker'), sort=True)[0]",
"_____no_output_____"
],
[
"categoricals = ['stock_id']",
"_____no_output_____"
]
],
[
[
"### Custom Metric",
"_____no_output_____"
]
],
[
[
"def ic_lgbm(preds, train_data):\n \"\"\"Custom IC eval metric for lightgbm\"\"\"\n is_higher_better = True\n return 'ic', spearmanr(preds, train_data.get_label())[0], is_higher_better",
"_____no_output_____"
]
],
[
[
"### Cross-validation setup",
"_____no_output_____"
]
],
[
[
"DAY = 390 # number of minute bars in a trading day of 6.5 hrs (9:30 - 15:59)\nMONTH = 21 # trading days",
"_____no_output_____"
],
[
"def get_cv(n_splits=23):\n return MultipleTimeSeriesCV(n_splits=n_splits,\n lookahead=1,\n test_period_length=MONTH * DAY, # test for 1 month\n train_period_length=12 * MONTH * DAY, # train for 1 year\n date_idx='date_time')",
"_____no_output_____"
]
],
[
[
"Show train/validation periods:",
"_____no_output_____"
]
],
[
[
"for i, (train_idx, test_idx) in enumerate(get_cv().split(X=data)):\n train_dates = data.iloc[train_idx].index.unique('date_time')\n test_dates = data.iloc[test_idx].index.unique('date_time')\n print(train_dates.min(), train_dates.max(), test_dates.min(), test_dates.max())",
"2016-11-29 15:59:00 2017-11-29 15:59:00 2017-11-30 09:30:00 2017-12-29 15:59:00\n2016-10-28 15:47:00 2017-10-30 15:58:00 2017-10-30 15:59:00 2017-11-29 15:59:00\n2016-09-29 15:47:00 2017-09-29 15:58:00 2017-09-29 15:59:00 2017-10-30 15:58:00\n2016-08-30 15:47:00 2017-08-30 15:58:00 2017-08-30 15:59:00 2017-09-29 15:58:00\n2016-08-01 15:47:00 2017-08-01 15:58:00 2017-08-01 15:59:00 2017-08-30 15:58:00\n2016-06-30 15:47:00 2017-06-30 15:58:00 2017-06-30 15:59:00 2017-08-01 15:58:00\n2016-06-01 15:47:00 2017-06-01 15:58:00 2017-06-01 15:59:00 2017-06-30 15:58:00\n2016-05-02 15:47:00 2017-05-02 15:58:00 2017-05-02 15:59:00 2017-06-01 15:58:00\n2016-04-01 15:47:00 2017-03-31 15:58:00 2017-03-31 15:59:00 2017-05-02 15:58:00\n2016-03-02 15:47:00 2017-03-02 15:58:00 2017-03-02 15:59:00 2017-03-31 15:58:00\n2016-02-01 15:47:00 2017-01-31 15:58:00 2017-01-31 15:59:00 2017-03-02 15:58:00\n2015-12-30 15:47:00 2016-12-29 15:58:00 2016-12-29 15:59:00 2017-01-31 15:58:00\n2015-11-30 15:23:00 2016-11-29 15:58:00 2016-11-29 15:59:00 2016-12-29 15:58:00\n2015-10-29 15:09:00 2016-10-28 15:46:00 2016-10-28 15:47:00 2016-11-29 15:58:00\n2015-09-30 15:09:00 2016-09-29 15:46:00 2016-09-29 15:47:00 2016-10-28 15:46:00\n2015-08-31 15:09:00 2016-08-30 15:46:00 2016-08-30 15:47:00 2016-09-29 15:46:00\n2015-07-31 15:09:00 2016-08-01 15:46:00 2016-08-01 15:47:00 2016-08-30 15:46:00\n2015-07-01 15:09:00 2016-06-30 15:46:00 2016-06-30 15:47:00 2016-08-01 15:46:00\n2015-06-02 15:09:00 2016-06-01 15:46:00 2016-06-01 15:47:00 2016-06-30 15:46:00\n2015-05-01 15:09:00 2016-05-02 15:46:00 2016-05-02 15:47:00 2016-06-01 15:46:00\n2015-04-01 15:09:00 2016-04-01 15:46:00 2016-04-01 15:47:00 2016-05-02 15:46:00\n2015-03-03 15:09:00 2016-03-02 15:46:00 2016-03-02 15:47:00 2016-04-01 15:46:00\n2015-01-30 15:09:00 2016-02-01 15:46:00 2016-02-01 15:47:00 2016-03-02 15:46:00\n"
]
],
[
[
"### Train model",
"_____no_output_____"
]
],
[
[
"label = sorted(data.filter(like='fwd').columns)\nfeatures = data.columns.difference(label).tolist()\nlabel = label[0]",
"_____no_output_____"
],
[
"params = dict(objective='regression',\n metric=['rmse'],\n device='gpu',\n max_bin=63,\n gpu_use_dp=False,\n num_leaves=16,\n min_data_in_leaf=500,\n feature_fraction=.8,\n verbose=-1)",
"_____no_output_____"
],
[
"num_boost_round = 250",
"_____no_output_____"
],
[
"cv = get_cv(n_splits=23) # we have enough data for 23 different test periods",
"_____no_output_____"
],
[
"def get_scores(result):\n return pd.DataFrame({'train': result['training']['ic'],\n 'valid': result['valid_1']['ic']})",
"_____no_output_____"
]
],
[
[
"The following model-training loop will take more than 10 hours to run and also consumes substantial memory. If you run into resource constraints, you can modify the code, e.g., by:\n1. Only loading data required for one iteration.\n2. Shortening the training period to require less than one year.\n\nYou can also speed up the process by using fewer `n_splits`, which implies longer test periods.",
"_____no_output_____"
]
],
[
[
"start = time()\nfor fold, (train_idx, test_idx) in enumerate(cv.split(X=data), 1):\n # create lgb train set\n train_set = data.iloc[train_idx, :]\n lgb_train = lgb.Dataset(data=train_set.drop(label, axis=1),\n label=train_set[label],\n categorical_feature=categoricals)\n \n # create lgb test set\n test_set = data.iloc[test_idx, :]\n lgb_test = lgb.Dataset(data=test_set.drop(label, axis=1),\n label=test_set[label],\n categorical_feature=categoricals, \n reference=lgb_train)\n\n # train model\n evals_result = {}\n model = lgb.train(params=params,\n train_set=lgb_train,\n valid_sets=[lgb_train, lgb_test],\n feval=ic_lgbm,\n num_boost_round=num_boost_round,\n evals_result=evals_result,\n verbose_eval=50)\n model.save_model((model_path / f'{fold:02}.txt').as_posix())\n \n # get train/valid ic scores\n scores = get_scores(evals_result)\n scores.to_hdf(result_store, f'ic/{fold:02}')\n \n # get feature importance\n fi = get_fi(model)\n fi.to_hdf(result_store, f'fi/{fold:02}')\n \n # generate validation predictions\n X_test = test_set.loc[:, model.feature_name()]\n y_test = test_set.loc[:, [label]]\n y_test['pred'] = model.predict(X_test)\n y_test.to_hdf(result_store, f'predictions/{fold:02}')\n \n # compute average IC per minute\n by_minute = y_test.groupby(test_set.index.get_level_values('date_time'))\n daily_ic = by_minute.apply(lambda x: spearmanr(x[label], x.pred)[0]).mean()\n print(f'\\nFold: {fold:02} | {format_time(time()-start)} | IC per minute: {daily_ic:.2%}\\n')",
"[50]\ttraining's rmse: 0.0006962\ttraining's ic: 0.038731\tvalid_1's rmse: 0.000816226\tvalid_1's ic: 0.0543727\n[100]\ttraining's rmse: 0.000695586\ttraining's ic: 0.04416\tvalid_1's rmse: 0.000815993\tvalid_1's ic: 0.0552591\n[150]\ttraining's rmse: 0.000695027\ttraining's ic: 0.046986\tvalid_1's rmse: 0.000815898\tvalid_1's ic: 0.0557145\n[200]\ttraining's rmse: 0.000694592\ttraining's ic: 0.04948\tvalid_1's rmse: 0.000815859\tvalid_1's ic: 0.0561737\n[250]\ttraining's rmse: 0.000694165\ttraining's ic: 0.0517389\tvalid_1's rmse: 0.000815865\tvalid_1's ic: 0.0558025\n\nFold: 01 | 00:17:46 | IC per minute: 5.59%\n\n[50]\ttraining's rmse: 0.000699973\ttraining's ic: 0.0376039\tvalid_1's rmse: 0.000847957\tvalid_1's ic: 0.0416495\n[100]\ttraining's rmse: 0.000699303\ttraining's ic: 0.0426195\tvalid_1's rmse: 0.000847627\tvalid_1's ic: 0.043379\n[150]\ttraining's rmse: 0.000698748\ttraining's ic: 0.0457404\tvalid_1's rmse: 0.000847548\tvalid_1's ic: 0.043617\n[200]\ttraining's rmse: 0.000698298\ttraining's ic: 0.0482473\tvalid_1's rmse: 0.000847537\tvalid_1's ic: 0.0440953\n[250]\ttraining's rmse: 0.000697857\ttraining's ic: 0.0506102\tvalid_1's rmse: 0.000847582\tvalid_1's ic: 0.0439462\n\nFold: 02 | 00:35:29 | IC per minute: 4.45%\n\n[50]\ttraining's rmse: 0.000698592\ttraining's ic: 0.0370533\tvalid_1's rmse: 0.000706335\tvalid_1's ic: 0.0404773\n[100]\ttraining's rmse: 0.000697869\ttraining's ic: 0.0418831\tvalid_1's rmse: 0.000706128\tvalid_1's ic: 0.0413394\n[150]\ttraining's rmse: 0.000697354\ttraining's ic: 0.0452553\tvalid_1's rmse: 0.000706085\tvalid_1's ic: 0.0411713\n[200]\ttraining's rmse: 0.000696885\ttraining's ic: 0.0479669\tvalid_1's rmse: 0.000706038\tvalid_1's ic: 0.0413983\n[250]\ttraining's rmse: 0.000696456\ttraining's ic: 0.0503778\tvalid_1's rmse: 0.000706054\tvalid_1's ic: 0.0412612\n\nFold: 03 | 00:57:48 | IC per minute: 4.45%\n\n[50]\ttraining's rmse: 0.000701553\ttraining's ic: 0.0363031\tvalid_1's rmse: 0.000669637\tvalid_1's ic: 0.0326244\n[100]\ttraining's rmse: 0.000700849\ttraining's ic: 0.0413249\tvalid_1's rmse: 0.000669565\tvalid_1's ic: 0.0339486\n[150]\ttraining's rmse: 0.000700357\ttraining's ic: 0.0447981\tvalid_1's rmse: 0.000669562\tvalid_1's ic: 0.0343703\n[200]\ttraining's rmse: 0.000699884\ttraining's ic: 0.0476104\tvalid_1's rmse: 0.000669583\tvalid_1's ic: 0.0349983\n[250]\ttraining's rmse: 0.000699484\ttraining's ic: 0.0501712\tvalid_1's rmse: 0.000669543\tvalid_1's ic: 0.0355025\n\nFold: 04 | 01:24:05 | IC per minute: 3.83%\n\n[50]\ttraining's rmse: 0.000697019\ttraining's ic: 0.0354982\tvalid_1's rmse: 0.000697012\tvalid_1's ic: 0.0247309\n[100]\ttraining's rmse: 0.000696274\ttraining's ic: 0.0410205\tvalid_1's rmse: 0.000696904\tvalid_1's ic: 0.0271854\n[150]\ttraining's rmse: 0.000695755\ttraining's ic: 0.044584\tvalid_1's rmse: 0.000696912\tvalid_1's ic: 0.0276005\n[200]\ttraining's rmse: 0.000695313\ttraining's ic: 0.0474853\tvalid_1's rmse: 0.000696927\tvalid_1's ic: 0.0285591\n[250]\ttraining's rmse: 0.000694863\ttraining's ic: 0.0498696\tvalid_1's rmse: 0.000696917\tvalid_1's ic: 0.0285991\n\nFold: 05 | 01:50:23 | IC per minute: 3.13%\n\n[50]\ttraining's rmse: 0.00069678\ttraining's ic: 0.0350113\tvalid_1's rmse: 0.000701348\tvalid_1's ic: 0.0275999\n[100]\ttraining's rmse: 0.00069605\ttraining's ic: 0.0406079\tvalid_1's rmse: 0.000701289\tvalid_1's ic: 0.0297336\n[150]\ttraining's rmse: 0.000695473\ttraining's ic: 0.0441527\tvalid_1's rmse: 0.000701216\tvalid_1's ic: 0.0307175\n[200]\ttraining's rmse: 0.000694997\ttraining's ic: 0.0471703\tvalid_1's rmse: 0.000701244\tvalid_1's ic: 0.0314352\n[250]\ttraining's rmse: 0.000694559\ttraining's ic: 0.0492445\tvalid_1's rmse: 0.000701273\tvalid_1's ic: 0.0314369\n\nFold: 06 | 02:16:28 | IC per minute: 3.34%\n\n[50]\ttraining's rmse: 0.000702829\ttraining's ic: 0.0337797\tvalid_1's rmse: 0.000744246\tvalid_1's ic: 0.0246692\n[100]\ttraining's rmse: 0.00070212\ttraining's ic: 0.0385954\tvalid_1's rmse: 0.000744224\tvalid_1's ic: 0.0264151\n[150]\ttraining's rmse: 0.000701593\ttraining's ic: 0.0430637\tvalid_1's rmse: 0.000744229\tvalid_1's ic: 0.0275546\n[200]\ttraining's rmse: 0.000701114\ttraining's ic: 0.0458159\tvalid_1's rmse: 0.000744281\tvalid_1's ic: 0.0282104\n[250]\ttraining's rmse: 0.000700721\ttraining's ic: 0.0482636\tvalid_1's rmse: 0.000744313\tvalid_1's ic: 0.0283922\n\nFold: 07 | 02:42:44 | IC per minute: 3.28%\n\n[50]\ttraining's rmse: 0.000722509\ttraining's ic: 0.0334184\tvalid_1's rmse: 0.00062052\tvalid_1's ic: 0.032487\n[100]\ttraining's rmse: 0.000721876\ttraining's ic: 0.038585\tvalid_1's rmse: 0.000620422\tvalid_1's ic: 0.0333264\n[150]\ttraining's rmse: 0.000721342\ttraining's ic: 0.0423346\tvalid_1's rmse: 0.000620373\tvalid_1's ic: 0.0332792\n[200]\ttraining's rmse: 0.000720854\ttraining's ic: 0.0453648\tvalid_1's rmse: 0.000620391\tvalid_1's ic: 0.0344978\n[250]\ttraining's rmse: 0.00072039\ttraining's ic: 0.0475421\tvalid_1's rmse: 0.000620433\tvalid_1's ic: 0.0349232\n\nFold: 08 | 03:08:25 | IC per minute: 3.70%\n\n[50]\ttraining's rmse: 0.000752768\ttraining's ic: 0.0325142\tvalid_1's rmse: 0.0005842\tvalid_1's ic: 0.0271741\n[100]\ttraining's rmse: 0.000751985\ttraining's ic: 0.0374633\tvalid_1's rmse: 0.000584136\tvalid_1's ic: 0.0283447\n[150]\ttraining's rmse: 0.000751343\ttraining's ic: 0.0407396\tvalid_1's rmse: 0.000584099\tvalid_1's ic: 0.0289354\n[200]\ttraining's rmse: 0.000750835\ttraining's ic: 0.0439565\tvalid_1's rmse: 0.000584126\tvalid_1's ic: 0.0294128\n[250]\ttraining's rmse: 0.00075033\ttraining's ic: 0.0460732\tvalid_1's rmse: 0.000584183\tvalid_1's ic: 0.0293556\n\nFold: 09 | 03:34:14 | IC per minute: 3.21%\n\n[50]\ttraining's rmse: 0.000772983\ttraining's ic: 0.0315982\tvalid_1's rmse: 0.00063351\tvalid_1's ic: 0.0269043\n[100]\ttraining's rmse: 0.000772305\ttraining's ic: 0.0370821\tvalid_1's rmse: 0.000633424\tvalid_1's ic: 0.0295316\n[150]\ttraining's rmse: 0.000771751\ttraining's ic: 0.0402892\tvalid_1's rmse: 0.000633369\tvalid_1's ic: 0.0301651\n[200]\ttraining's rmse: 0.000771242\ttraining's ic: 0.0432137\tvalid_1's rmse: 0.000633349\tvalid_1's ic: 0.0312183\n[250]\ttraining's rmse: 0.000770771\ttraining's ic: 0.0455847\tvalid_1's rmse: 0.000633325\tvalid_1's ic: 0.0315627\n\nFold: 10 | 04:00:30 | IC per minute: 2.98%\n\n[50]\ttraining's rmse: 0.000832092\ttraining's ic: 0.0325253\tvalid_1's rmse: 0.000653653\tvalid_1's ic: 0.026781\n[100]\ttraining's rmse: 0.000831323\ttraining's ic: 0.0377314\tvalid_1's rmse: 0.000653568\tvalid_1's ic: 0.0289015\n[150]\ttraining's rmse: 0.000830753\ttraining's ic: 0.0411433\tvalid_1's rmse: 0.000653586\tvalid_1's ic: 0.0291601\n[200]\ttraining's rmse: 0.000830191\ttraining's ic: 0.043913\tvalid_1's rmse: 0.000653599\tvalid_1's ic: 0.0301002\n[250]\ttraining's rmse: 0.000829674\ttraining's ic: 0.0465464\tvalid_1's rmse: 0.000653658\tvalid_1's ic: 0.0303744\n\nFold: 11 | 04:26:17 | IC per minute: 2.94%\n\n[50]\ttraining's rmse: 0.000877395\ttraining's ic: 0.0320049\tvalid_1's rmse: 0.000721517\tvalid_1's ic: 0.0240198\n[100]\ttraining's rmse: 0.000876658\ttraining's ic: 0.0374841\tvalid_1's rmse: 0.00072146\tvalid_1's ic: 0.026157\n[150]\ttraining's rmse: 0.000876046\ttraining's ic: 0.0408182\tvalid_1's rmse: 0.000721393\tvalid_1's ic: 0.0272646\n[200]\ttraining's rmse: 0.000875495\ttraining's ic: 0.0441758\tvalid_1's rmse: 0.000721363\tvalid_1's ic: 0.0281185\n[250]\ttraining's rmse: 0.000875026\ttraining's ic: 0.0467237\tvalid_1's rmse: 0.00072137\tvalid_1's ic: 0.028905\n\nFold: 12 | 04:52:49 | IC per minute: 3.04%\n\n[50]\ttraining's rmse: 0.000886972\ttraining's ic: 0.0326955\tvalid_1's rmse: 0.000749551\tvalid_1's ic: 0.0260998\n[100]\ttraining's rmse: 0.000886233\ttraining's ic: 0.0374855\tvalid_1's rmse: 0.00074944\tvalid_1's ic: 0.0283205\n[150]\ttraining's rmse: 0.000885641\ttraining's ic: 0.0409926\tvalid_1's rmse: 0.000749411\tvalid_1's ic: 0.029227\n[200]\ttraining's rmse: 0.000885103\ttraining's ic: 0.0439042\tvalid_1's rmse: 0.000749372\tvalid_1's ic: 0.0297628\n[250]\ttraining's rmse: 0.000884651\ttraining's ic: 0.0465908\tvalid_1's rmse: 0.000749306\tvalid_1's ic: 0.0307105\n\nFold: 13 | 05:18:51 | IC per minute: 3.01%\n\n[50]\ttraining's rmse: 0.000892264\ttraining's ic: 0.0326621\tvalid_1's rmse: 0.00088496\tvalid_1's ic: 0.0215666\n[100]\ttraining's rmse: 0.000891562\ttraining's ic: 0.0366921\tvalid_1's rmse: 0.000884886\tvalid_1's ic: 0.0220376\n[150]\ttraining's rmse: 0.000890964\ttraining's ic: 0.0397876\tvalid_1's rmse: 0.000884839\tvalid_1's ic: 0.0227016\n[200]\ttraining's rmse: 0.000890451\ttraining's ic: 0.0430167\tvalid_1's rmse: 0.000884803\tvalid_1's ic: 0.0235889\n"
]
],
[
[
"## Signal Evaluation",
"_____no_output_____"
]
],
[
[
"with pd.HDFStore(result_store) as store:\n pred_keys = [k[1:] for k in store.keys() if k[1:].startswith('pred')]\n cv_predictions = pd.concat([store[k] for k in pred_keys]).sort_index()",
"_____no_output_____"
],
[
"cv_predictions.info(null_counts=True)",
"<class 'pandas.core.frame.DataFrame'>\nMultiIndex: 19648064 entries, ('AAL', Timestamp('2016-02-01 15:47:00')) to ('YHOO', Timestamp('2017-06-16 15:59:00'))\nData columns (total 2 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 fwd1min 19648064 non-null float64\n 1 pred 19648064 non-null float64\ndtypes: float64(2)\nmemory usage: 399.0+ MB\n"
],
[
"time_stamp = cv_predictions.index.get_level_values('date_time')\ndates = sorted(np.unique(time_stamp.date))",
"_____no_output_____"
]
],
[
[
"We have out-of-sample predictions for 484 days from February 2016 through December 2017:",
"_____no_output_____"
]
],
[
[
"print(f'# Days: {len(dates)} | First: {dates[0]} | Last: {dates[-1]}')",
"# Days: 484 | First: 2016-02-01 | Last: 2017-12-29\n"
]
],
[
[
"We only use minutes with at least 100 predictions:",
"_____no_output_____"
]
],
[
[
"n = cv_predictions.groupby('date_time').size()",
"_____no_output_____"
]
],
[
[
"There are ~700 periods, equivalent to a bit over a single trading day (0.67% of all periods in the sample), with fewer than 100 predictions over the 23 test months:",
"_____no_output_____"
]
],
[
[
"incomplete_minutes = n[n<100].index",
"_____no_output_____"
],
[
"print(f'{len(incomplete_minutes)} ({len(incomplete_minutes)/len(n):.2%})')",
"1255 (0.67%)\n"
],
[
"cv_predictions = cv_predictions[~time_stamp.isin(incomplete_minutes)]",
"_____no_output_____"
],
[
"cv_predictions.info(null_counts=True)",
"<class 'pandas.core.frame.DataFrame'>\nMultiIndex: 19571774 entries, ('AAL', Timestamp('2016-02-01 15:47:00')) to ('YHOO', Timestamp('2017-06-16 15:59:00'))\nData columns (total 2 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 fwd1min 19571774 non-null float64\n 1 pred 19571774 non-null float64\ndtypes: float64(2)\nmemory usage: 397.4+ MB\n"
]
],
[
[
"### Information Coefficient",
"_____no_output_____"
],
[
"#### Across all periods",
"_____no_output_____"
]
],
[
[
"ic = spearmanr(cv_predictions.fwd1min, cv_predictions.pred)[0]",
"_____no_output_____"
]
],
[
[
"#### By minute",
"_____no_output_____"
],
[
"We are making new predictions every minute, so it makes sense to look at the average performance across all short-term forecasts:",
"_____no_output_____"
]
],
[
[
"minutes = cv_predictions.index.get_level_values('date_time')\nby_minute = cv_predictions.groupby(minutes)",
"_____no_output_____"
],
[
"ic_by_minute = by_minute.apply(lambda x: spearmanr(x.fwd1min, x.pred)[0])\n\nminute_ic_mean = ic_by_minute.mean()\nminute_ic_median = ic_by_minute.median()\n\nprint(f'\\nAll periods: {ic:6.2%} | By Minute: {minute_ic_mean: 6.2%} (Median: {minute_ic_median: 6.2%})')",
"\nAll periods: 2.96% | By Minute: 3.21% (Median: 3.23%)\n"
]
],
[
[
"Plotted as a five-day rolling average, we see that the IC was mostly below the out-of-sample period mean, and increased during the last quarter of 2017 (as reflected in the validation results we observed while training the model).",
"_____no_output_____"
]
],
[
[
"ax = ic_by_minute.rolling(5*650).mean().plot(figsize=(14, 5), title='IC (5-day MA)', rot=0)\nax.axhline(minute_ic_mean, ls='--', lw=1, c='k')\nax.yaxis.set_major_formatter(FuncFormatter(lambda y, _: '{:.0%}'.format(y)))\nax.set_ylabel('Information Coefficient')\nax.set_xlabel('')\nsns.despine()\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"### Vectorized backtest of a naive strategey: financial performance by signal quantile",
"_____no_output_____"
],
[
"Alphalens does not work with minute-data, so we need to compute our own signal performance measures.",
"_____no_output_____"
],
[
"Unfortunately, Zipline's Pipeline also doesn't work for minute-data and Backtrader takes a very long time with such a large dataset. Hence, instead of an event-driven backtest of entry/exit rules as in previous examples, we can only create a rough sketch of the financial performance of a naive trading strategy driven by the model's predictions using vectorized backtesting (see Chapter 8 on the [ML4T workflow](../08_ml4t_workflow'). As we will see below, this does not produce particularly helpful results.",
"_____no_output_____"
],
[
"This naive strategy invests in equal-weighted portfolios of the stocks in each decile under the following assumptions (mentioned at the beginning of this notebook: \n1. Based on the predictions using inputs from the current and previous bars, we can enter positions at the first trade price in the following minute bar\n2. We exit all positions at the last price in that following minute bar\n3. There are no trading cost or market impact (slippage) of our trades (but we can check how sensitive the results would be).",
"_____no_output_____"
],
[
"#### Average returns by minute bar and signal quantile ",
"_____no_output_____"
],
[
"To this end, we compute the quintiles and deciles of the model's `fwd1min` predictions for each minute:",
"_____no_output_____"
]
],
[
[
"by_minute = cv_predictions.groupby(minutes, group_keys=False)",
"_____no_output_____"
],
[
"labels = list(range(1, 6))\ncv_predictions['quintile'] = by_minute.apply(lambda x: pd.qcut(x.pred, q=5, labels=labels).astype(int))",
"_____no_output_____"
],
[
"labels = list(range(1, 11))\ncv_predictions['decile'] = by_minute.apply(lambda x: pd.qcut(x.pred, q=10, labels=labels).astype(int))",
"_____no_output_____"
],
[
"cv_predictions.info(show_counts=True)",
"<class 'pandas.core.frame.DataFrame'>\nMultiIndex: 19571774 entries, ('AAL', Timestamp('2016-02-01 15:47:00')) to ('YHOO', Timestamp('2017-06-16 15:59:00'))\nData columns (total 4 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 fwd1min 19571774 non-null float64\n 1 pred 19571774 non-null float64\n 2 quintile 19571774 non-null int64 \n 3 decile 19571774 non-null int64 \ndtypes: float64(2), int64(2)\nmemory usage: 696.1+ MB\n"
]
],
[
[
"#### Descriptive statistics of intraday returns by quintile and decile of model predictions",
"_____no_output_____"
],
[
"Next, we compute the average one-minute returns for each quintile / decile and minute.",
"_____no_output_____"
]
],
[
[
"def compute_intraday_returns_by_quantile(predictions, quantile='quintile'):\n by_quantile = cv_predictions.reset_index().groupby(['date_time', quantile])\n return by_quantile.fwd1min.mean().unstack(quantile).sort_index()",
"_____no_output_____"
],
[
"intraday_returns = {'quintile': compute_intraday_returns_by_quantile(cv_predictions),\n 'decile': compute_intraday_returns_by_quantile(cv_predictions, quantile='decile')}",
"_____no_output_____"
],
[
"def summarize_intraday_returns(returns):\n summary = returns.describe(deciles)\n return pd.concat([summary.iloc[:1].applymap(lambda x: f'{x:,.0f}'),\n summary.iloc[1:].applymap(lambda x: f'{x:.4%}')])",
"_____no_output_____"
]
],
[
[
"The returns per minute, averaged over the 23-months period, increase by quintile/decile and range from -.3 (-.4) to .27 (.37) basis points for the bottom and top quintile (decile), respectively. While this aligns with the finding of a weakly positive rank correlation coefficient, it also suggests that such small gains are unlikely to survive the impact of trading costs.",
"_____no_output_____"
]
],
[
[
"summary = summarize_intraday_returns(intraday_returns['quintile'])\nsummary",
"_____no_output_____"
],
[
"summary = summarize_intraday_returns(intraday_returns['decile'])\nsummary",
"_____no_output_____"
]
],
[
[
"#### Cumulative Performance by Quantile",
"_____no_output_____"
],
[
"To simulate the performance of our naive strategy that trades all available stocks every minute, we simply assume that we can reinvest (including potential gains/losses) every minute. To check for the sensitivity with respect for trading cost, we can assume they are a constant number (fraction) of basis points, and subtract this number from the minute-bar returns.",
"_____no_output_____"
]
],
[
[
"def plot_cumulative_performance(returns, quantile='quintile', trading_costs_bp=0):\n \"\"\"Plot average return by quantile (in bp) as well as cumulative return, \n both net of trading costs (provided as basis points; 1bp = 0.01%) \n \"\"\"\n\n fig, axes = plt.subplots(figsize=(14, 4), ncols=2)\n\n sns.barplot(y='fwd1min', x=quantile,\n data=returns[quantile].mul(10000).sub(trading_costs_bp).stack().to_frame(\n 'fwd1min').reset_index(),\n ax=axes[0])\n axes[0].set_title(f'Avg. 1-min Return by Signal {quantile.capitalize()}')\n axes[0].set_ylabel('Return (bps)')\n axes[0].set_xlabel(quantile.capitalize())\n\n title = f'Cumulative Return by Signal {quantile.capitalize()}'\n (returns[quantile].sort_index().add(1).sub(trading_costs_bp/10000).cumprod().sub(1)\n .plot(ax=axes[1], title=title))\n\n axes[1].yaxis.set_major_formatter(\n FuncFormatter(lambda y, _: '{:.0%}'.format(y)))\n axes[1].set_xlabel('')\n axes[1].set_ylabel('Return')\n fig.suptitle(f'Average and Cumulative Performance (Net of Trading Cost: {trading_costs_bp:.2f}bp)')\n\n sns.despine()\n fig.tight_layout()",
"_____no_output_____"
]
],
[
[
"Without trading costs, the compounding of even fairly small gains leads to extremely large cumulative profits for the top quantile. However, these disappear as soon as we allow for minuscule trading costs that reduce the average quantile return close to zero.",
"_____no_output_____"
],
[
"##### Without trading costs",
"_____no_output_____"
]
],
[
[
"plot_cumulative_performance(intraday_returns, 'quintile', trading_costs_bp=0)",
"_____no_output_____"
],
[
"plot_cumulative_performance(intraday_returns, 'decile', trading_costs_bp=0)",
"_____no_output_____"
]
],
[
[
"##### With extremely low trading costs",
"_____no_output_____"
]
],
[
[
"# assuming costs of a fraction of a basis point, close to the average return of the top quantile\nplot_cumulative_performance(intraday_returns, 'quintile', trading_costs_bp=.2)",
"_____no_output_____"
],
[
"plot_cumulative_performance(intraday_returns, 'decile', trading_costs_bp=.3)",
"_____no_output_____"
]
],
[
[
"### Feature Importance",
"_____no_output_____"
],
[
"We'll take a quick look at the features that most contributed to improving the IC across the 23 folds:",
"_____no_output_____"
]
],
[
[
"with pd.HDFStore(result_store) as store:\n fi_keys = [k[1:] for k in store.keys() if k[1:].startswith('fi')]\n fi = pd.concat([store[k].to_frame(i) for i, k in enumerate(fi_keys, 1)], axis=1)",
"_____no_output_____"
]
],
[
[
"The top features from a conventional feature importance perspective are the ticker, followed by NATR, minute of the day, latest 1m return and the CCI:",
"_____no_output_____"
]
],
[
[
"fi.mean(1).nsmallest(25).plot.barh(figsize=(12, 8), title='LightGBM Feature Importance (gain)')\nsns.despine()\nplt.tight_layout();",
"_____no_output_____"
]
],
[
[
"Explore with greater accuracy and in more detail how feature values affect predictions using SHAP values as demonstrated in various other notebooks in this Chapter and the appendix!",
"_____no_output_____"
],
[
"## Conclusion\n\nWe have seen that a relatively simple gradient boosting model is able to achieve fairly consistent predictive performance that is significantly better than a random guess even on a very short horizon. \n\nHowever, the resulting economic gains of our naive strategy of frequently buying/(short-)selling the top/bottome quantiles are too small to overcome the inevitable transaction costs. On the one hand, this demonstrates the challenges of extracting value from a predictive signal. On the other hand, it shows that we need a more sophisticated backtesting platform so that we can even begin to design and evaluate a more sophisticated strategy that requires far fewer trades to exploit the signal in our ML predictions. \n\nIn addition, we would also want to work on improving the model by adding more informative feature, e.g. based on the quote/trade info contained in the Algoseek data, or by fine-tuning our model architecture and hyperparameter settings.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb4ddd35c99ca168bdea0b0a1ba4585d6f102c98 | 15,518 | ipynb | Jupyter Notebook | examples/user_guide/Network_Graphs.ipynb | ppwadhwa/holoviews | e8e2ec08c669295479f98bb2f46bbd59782786bf | [
"BSD-3-Clause"
] | 864 | 2019-11-13T08:18:27.000Z | 2022-03-31T13:36:13.000Z | examples/user_guide/Network_Graphs.ipynb | ppwadhwa/holoviews | e8e2ec08c669295479f98bb2f46bbd59782786bf | [
"BSD-3-Clause"
] | 1,117 | 2019-11-12T16:15:59.000Z | 2022-03-30T22:57:59.000Z | examples/user_guide/Network_Graphs.ipynb | ppwadhwa/holoviews | e8e2ec08c669295479f98bb2f46bbd59782786bf | [
"BSD-3-Clause"
] | 180 | 2019-11-19T16:44:44.000Z | 2022-03-28T22:49:18.000Z | 32.669474 | 645 | 0.61245 | [
[
[
"import numpy as np\nimport pandas as pd\nimport holoviews as hv\nimport networkx as nx\nfrom holoviews import opts\n\nhv.extension('bokeh')\n\ndefaults = dict(width=400, height=400)\nhv.opts.defaults(\n opts.EdgePaths(**defaults), opts.Graph(**defaults), opts.Nodes(**defaults))",
"_____no_output_____"
]
],
[
[
"Visualizing and working with network graphs is a common problem in many different disciplines. HoloViews provides the ability to represent and visualize graphs very simply and easily with facilities for interactively exploring the nodes and edges of the graph, especially using the bokeh plotting interface.\n\nThe ``Graph`` ``Element`` differs from other elements in HoloViews in that it consists of multiple sub-elements. The data of the ``Graph`` element itself are the abstract edges between the nodes. By default the element will automatically compute concrete ``x`` and ``y`` positions for the nodes and represent them using a ``Nodes`` element, which is stored on the Graph. The abstract edges and concrete node positions are sufficient to render the ``Graph`` by drawing straight-line edges between the nodes. In order to supply explicit edge paths we can also declare ``EdgePaths``, providing explicit coordinates for each edge to follow.\n\nTo summarize a ``Graph`` consists of three different components:\n\n* The ``Graph`` itself holds the abstract edges stored as a table of node indices.\n* The ``Nodes`` hold the concrete ``x`` and ``y`` positions of each node along with a node ``index``. The ``Nodes`` may also define any number of value dimensions, which can be revealed when hovering over the nodes or to color the nodes by.\n* The ``EdgePaths`` can optionally be supplied to declare explicit node paths.\n\n#### A simple Graph\n\nLet's start by declaring a very simple graph connecting one node to all others. If we simply supply the abstract connectivity of the ``Graph``, it will automatically compute a layout for the nodes using the ``layout_nodes`` operation, which defaults to a circular layout:",
"_____no_output_____"
]
],
[
[
"# Declare abstract edges\nN = 8\nnode_indices = np.arange(N, dtype=np.int32)\nsource = np.zeros(N, dtype=np.int32)\ntarget = node_indices\n\n\nsimple_graph = hv.Graph(((source, target),))\nsimple_graph",
"_____no_output_____"
]
],
[
[
"#### Accessing the nodes and edges\n\nWe can easily access the ``Nodes`` and ``EdgePaths`` on the ``Graph`` element using the corresponding properties:",
"_____no_output_____"
]
],
[
[
"simple_graph.nodes + simple_graph.edgepaths",
"_____no_output_____"
]
],
[
[
"#### Displaying directed graphs\n\nWhen specifying the graph edges the source and target node are listed in order, if the graph is actually a directed graph this may used to indicate the directionality of the graph. By setting ``directed=True`` as a plot option it is possible to indicate the directionality of each edge using an arrow:",
"_____no_output_____"
]
],
[
[
"simple_graph.relabel('Directed Graph').opts(directed=True, node_size=5, arrowhead_length=0.05)",
"_____no_output_____"
]
],
[
[
"The length of the arrows can be set as an fraction of the overall graph extent using the ``arrowhead_length`` option.",
"_____no_output_____"
],
[
"#### Supplying explicit paths\n\nNext we will extend this example by supplying explicit edges:",
"_____no_output_____"
]
],
[
[
"def bezier(start, end, control, steps=np.linspace(0, 1, 100)):\n return (1-steps)**2*start + 2*(1-steps)*steps*control+steps**2*end\n\nx, y = simple_graph.nodes.array([0, 1]).T\n\npaths = []\nfor node_index in node_indices:\n ex, ey = x[node_index], y[node_index]\n paths.append(np.column_stack([bezier(x[0], ex, 0), bezier(y[0], ey, 0)]))\n \nbezier_graph = hv.Graph(((source, target), (x, y, node_indices), paths))\nbezier_graph",
"_____no_output_____"
]
],
[
[
"## Interactive features",
"_____no_output_____"
],
[
"#### Hover and selection policies\n\nThanks to Bokeh we can reveal more about the graph by hovering over the nodes and edges. The ``Graph`` element provides an ``inspection_policy`` and a ``selection_policy``, which define whether hovering and selection highlight edges associated with the selected node or nodes associated with the selected edge, these policies can be toggled by setting the policy to ``'nodes'`` (the default) and ``'edges'``.",
"_____no_output_____"
]
],
[
[
"bezier_graph.relabel('Edge Inspection').opts(inspection_policy='edges')",
"_____no_output_____"
]
],
[
[
"In addition to changing the policy we can also change the colors used when hovering and selecting nodes:",
"_____no_output_____"
]
],
[
[
"bezier_graph.opts(\n opts.Graph(inspection_policy='nodes', tools=['hover', 'box_select'],\n edge_hover_line_color='green', node_hover_fill_color='red'))",
"_____no_output_____"
]
],
[
[
"#### Additional information\n\nWe can also associate additional information with the nodes and edges of a graph. By constructing the ``Nodes`` explicitly we can declare additional value dimensions, which are revealed when hovering and/or can be mapped to the color by setting the ``color`` to the dimension name ('Weight'). We can also associate additional information with each edge by supplying a value dimension to the ``Graph`` itself, which we can map to various style options, e.g. by setting the ``edge_color`` and ``edge_line_width``.",
"_____no_output_____"
]
],
[
[
"node_labels = ['Output']+['Input']*(N-1)\nnp.random.seed(7)\nedge_labels = np.random.rand(8)\n\nnodes = hv.Nodes((x, y, node_indices, node_labels), vdims='Type')\ngraph = hv.Graph(((source, target, edge_labels), nodes, paths), vdims='Weight')\n\n(graph + graph.opts(inspection_policy='edges', clone=True)).opts(\n opts.Graph(node_color='Type', edge_color='Weight', cmap='Set1',\n edge_cmap='viridis', edge_line_width=hv.dim('Weight')*10))",
"_____no_output_____"
]
],
[
[
"If you want to supply additional node information without speciying explicit node positions you may pass in a ``Dataset`` object consisting of various value dimensions.",
"_____no_output_____"
]
],
[
[
"node_info = hv.Dataset(node_labels, vdims='Label')\nhv.Graph(((source, target), node_info)).opts(node_color='Label', cmap='Set1')",
"_____no_output_____"
]
],
[
[
"## Working with NetworkX",
"_____no_output_____"
],
[
"NetworkX is a very useful library when working with network graphs and the Graph Element provides ways of importing a NetworkX Graph directly. Here we will load the Karate Club graph and use the ``circular_layout`` function provided by NetworkX to lay it out:",
"_____no_output_____"
]
],
[
[
"G = nx.karate_club_graph()\nhv.Graph.from_networkx(G, nx.layout.circular_layout).opts(tools=['hover'])",
"_____no_output_____"
]
],
[
[
"It is also possible to pass arguments to the NetworkX layout function as keywords to ``hv.Graph.from_networkx``, e.g. we can override the k-value of the Fruchteran Reingold layout",
"_____no_output_____"
]
],
[
[
"hv.Graph.from_networkx(G, nx.layout.fruchterman_reingold_layout, k=1)",
"_____no_output_____"
]
],
[
[
"Finally if we want to layout a Graph after it has already been constructed, the ``layout_nodes`` operation may be used, which also allows applying the ``weight`` argument to graphs which have not been constructed with networkx:",
"_____no_output_____"
]
],
[
[
"from holoviews.element.graphs import layout_nodes\n\ngraph = hv.Graph([\n ('a', 'b', 3),\n ('a', 'c', 0.2),\n ('c', 'd', 0.1),\n ('c', 'e', 0.7),\n ('c', 'f', 5),\n ('a', 'd', 0.3)\n], vdims='weight')\n\nlayout_nodes(graph, layout=nx.layout.fruchterman_reingold_layout, kwargs={'weight': 'weight'})",
"_____no_output_____"
]
],
[
[
"## Adding labels",
"_____no_output_____"
],
[
"If the ``Graph`` we have constructed has additional metadata we can easily use those as labels, we simply get a handle on the nodes, cast them to hv.Labels and then overlay them:",
"_____no_output_____"
]
],
[
[
"graph = hv.Graph.from_networkx(G, nx.layout.fruchterman_reingold_layout)\nlabels = hv.Labels(graph.nodes, ['x', 'y'], 'club')\n\n(graph * labels.opts(text_font_size='8pt', text_color='white', bgcolor='gray'))",
"_____no_output_____"
]
],
[
[
"## Animating graphs",
"_____no_output_____"
],
[
"Like all other elements ``Graph`` can be updated in a ``HoloMap`` or ``DynamicMap``. Here we animate how the Fruchterman-Reingold force-directed algorithm lays out the nodes in real time.",
"_____no_output_____"
]
],
[
[
"hv.HoloMap({i: hv.Graph.from_networkx(G, nx.spring_layout, iterations=i, seed=10) for i in range(5, 30, 5)},\n kdims='Iterations')",
"_____no_output_____"
]
],
[
[
"## Real world graphs",
"_____no_output_____"
],
[
"As a final example let's look at a slightly larger graph. We will load a dataset of a Facebook network consisting a number of friendship groups identified by their ``'circle'``. We will load the edge and node data using pandas and then color each node by their friendship group using many of the things we learned above.",
"_____no_output_____"
]
],
[
[
"kwargs = dict(width=800, height=800, xaxis=None, yaxis=None)\nopts.defaults(opts.Nodes(**kwargs), opts.Graph(**kwargs))\n\ncolors = ['#000000']+hv.Cycle('Category20').values\nedges_df = pd.read_csv('../assets/fb_edges.csv')\nfb_nodes = hv.Nodes(pd.read_csv('../assets/fb_nodes.csv')).sort()\nfb_graph = hv.Graph((edges_df, fb_nodes), label='Facebook Circles')\n\nfb_graph.opts(cmap=colors, node_size=10, edge_line_width=1,\n node_line_color='gray', node_color='circle')",
"_____no_output_____"
]
],
[
[
"## Bundling graphs",
"_____no_output_____"
],
[
"The datashader library provides algorithms for bundling the edges of a graph and HoloViews provides convenient wrappers around the libraries. Note that these operations need ``scikit-image`` which you can install using:\n\n```\nconda install scikit-image\n```\n\nor\n\n```\npip install scikit-image\n```",
"_____no_output_____"
]
],
[
[
"from holoviews.operation.datashader import datashade, bundle_graph\nbundled = bundle_graph(fb_graph)\nbundled",
"_____no_output_____"
]
],
[
[
"## Datashading graphs",
"_____no_output_____"
],
[
"For graphs with a large number of edges we can datashade the paths and display the nodes separately. This loses some of the interactive features but will let you visualize quite large graphs:",
"_____no_output_____"
]
],
[
[
"(datashade(bundled, normalization='linear', width=800, height=800) * bundled.nodes).opts(\n opts.Nodes(color='circle', size=10, width=1000, cmap=colors, legend_position='right'))",
"_____no_output_____"
]
],
[
[
"### Applying selections",
"_____no_output_____"
],
[
"Alternatively we can select the nodes and edges by an attribute that resides on either. In this case we will select the nodes and edges for a particular circle and then overlay just the selected part of the graph on the datashaded plot. Note that selections on the ``Graph`` itself will select all nodes that connect to one of the selected nodes. In this way a smaller subgraph can be highlighted and the larger graph can be datashaded.",
"_____no_output_____"
]
],
[
[
"datashade(bundle_graph(fb_graph), normalization='linear', width=800, height=800) *\\\nbundled.select(circle='circle15').opts(node_fill_color='white')",
"_____no_output_____"
]
],
[
[
"To select just nodes that are in 'circle15' set the ``selection_mode='nodes'`` overriding the default of 'edges':",
"_____no_output_____"
]
],
[
[
"bundled.select(circle='circle15', selection_mode='nodes')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4df054f94eed9414db0fdaa935fe9d57723b50 | 444,448 | ipynb | Jupyter Notebook | BitcoinDataAnalysis/AnalysisJunpyter.ipynb | KevinTieu9/Python-Experience | 026f7cebf57f1b1384a034123deeaacb5cb691df | [
"Apache-2.0"
] | 1 | 2021-06-24T20:25:31.000Z | 2021-06-24T20:25:31.000Z | BitcoinDataAnalysis/AnalysisJunpyter.ipynb | KevinTieu9/Python-Experience | 026f7cebf57f1b1384a034123deeaacb5cb691df | [
"Apache-2.0"
] | null | null | null | BitcoinDataAnalysis/AnalysisJunpyter.ipynb | KevinTieu9/Python-Experience | 026f7cebf57f1b1384a034123deeaacb5cb691df | [
"Apache-2.0"
] | null | null | null | 1,084.019512 | 162,884 | 0.953266 | [
[
[
"# Data Analysis of Bitcoin and Where it is Heading",
"_____no_output_____"
],
[
"# Graphing the whole Graph",
"_____no_output_____"
]
],
[
[
"#### Importing Pandas and others and Reading csv file\nimport os\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport seaborn as sns\nimport pandas as pd\nimport plotly.express as px\n\n##Remodified .CSV data to make managing data easier.\n##Some data cleaning.\nBitcoin = pd.read_csv('HistoricalData4.csv')\n\n##Created A daily Average for each day to work around with the data.\nBitcoin['Daily Average'] = Bitcoin.iloc[:, 2:4].sum(axis=1)/2\n\n##Just to let the viewer see the data that is coming out, truancated\n##because there is 2843 rows to show.... too much data, lol\nprint(Bitcoin[['Date', 'Low', 'High', 'Daily Average']])\nprint(Bitcoin[['Date', 'Open', 'Close']])\nprint(Bitcoin[['Date', 'Volume', 'Market Cap']])\n\n\n#Line graph plot to show, low, high, and Average price\nBitcoin.plot(x=\"Date\", y=[\"Low\", \"High\", \"Daily Average\"], figsize=(15, 20), title =\"Bitcoin Low, High, and Daily Average Prices.\", ylabel=\"Price in $\")\nplt.show()\n\n#Line graph to show traditonal Open and Close (although the selling and buying never sleeps for Cryptocurrency)\nBitcoin.plot(x=\"Date\", y=[\"Open\", \"Close\"], figsize=(15, 30), title =\"Open and Close Prices.\", ylabel=\"Price in $\")\nplt.show()\n\n#Line graph to Show Volume and Market Cap\n#These two indicators are important to understand how healthy or not healthy a particular stock or cryptocurrency is.\n#High Volume but decrease in price could mean people are because they see a decrease in price or cashing out\n#High Volume and higher price means that people are still buying the currency because there is value.\nBitcoin.plot(x=\"Date\", y=[\"Volume\", \"Market Cap\"], figsize=(15, 30), title =\"Volume and Market Cap.\", ylabel=\"Price in $\")\nplt.show()",
" Date Low High Daily Average\n0 2013-04-30 134.05 146.93 140.490\n1 2013-05-01 107.72 139.89 123.805\n2 2013-05-02 92.28 125.60 108.940\n3 2013-05-03 79.10 108.13 93.615\n4 2013-05-04 92.50 115.00 103.750\n... ... ... ... ...\n2838 2021-02-05 36658.76 38225.91 37442.335\n2839 2021-02-06 38138.39 40846.55 39492.470\n2840 2021-02-07 37446.15 39621.84 38533.995\n2841 2021-02-08 38076.32 46203.93 42140.125\n2842 2021-02-09 45166.96 48003.72 46585.340\n\n[2843 rows x 4 columns]\n Date Open Close\n0 2013-04-30 144.00 139.00\n1 2013-05-01 139.00 116.99\n2 2013-05-02 116.38 105.21\n3 2013-05-03 106.25 97.75\n4 2013-05-04 98.10 112.50\n... ... ... ...\n2838 2021-02-05 36931.55 38144.31\n2839 2021-02-06 38138.39 39266.01\n2840 2021-02-07 39250.19 38903.44\n2841 2021-02-08 38886.83 46196.46\n2842 2021-02-09 46184.99 46481.10\n\n[2843 rows x 3 columns]\n Date Volume Market Cap\n0 2013-04-30 0 1542813125\n1 2013-05-01 0 1298954594\n2 2013-05-02 0 1168517495\n3 2013-05-03 0 1085995169\n4 2013-05-04 0 1250316563\n... ... ... ...\n2838 2021-02-05 58598066402 710266752534\n2839 2021-02-06 71326033653 731192490093\n2840 2021-02-07 65500641143 724478833211\n2841 2021-02-08 101467222687 860342706260\n2842 2021-02-09 91809846886 865682956619\n\n[2843 rows x 3 columns]\n"
]
],
[
[
"## Thoughts on the Line Graph",
"_____no_output_____"
],
[
"This graph shows the whole entire graph since Bitcoin started being sold and used and the day I stopped collecting Data. As you can see, it isn't really useful for trying to extract specific Data because there is just so much Data and the data extremes like 0, and recently....bitcoin going past 50k makes it hard to view specific data unless this graphical picture is at least maybe 10 times bigger (if I were to guess.) You can't even see the minute difference between the 3 data sets.\n\n## Cryptocurrencies are said to be volatile, but are they though?\nOr is it only considered volatile if\n\n1: You invest in the wrong ones \n\n2: You continue to invest on a project that isn't ongoing (like Dogecoin), has an extremely high supply cap, or known to have problems (the people and company aren't trustworthy or the company or currency was hacked)\n\n3: You invest on unknown cryotocurrencies except that those currencies are just a deritivative of Bitcoin\n\n4: There is very few popular news around the cryptocurrency.",
"_____no_output_____"
],
[
"## There are 3 points in the graph that prove to be interesting because those are 3 points where Bitcoin skyrocketed\n\nThe last one needs no introduction. Elon Musk decided to invest more than $1 Billion dollars into Bitcoin. That alone has caused the cryotcurrency itself to skyrocket. [CNBC Link to Article About it!](https://www.cnbc.com/2021/02/08/tesla-buys-1point5-billion-in-bitcoin.html) While this graph doesn't refect the current day (the price is at 50k right now) it's still fairly accurate.\n\nOther than that, let's see if we can grab some data between the two other points, what caused it to go up close to 20k and then start dropping again around ~16k in the line graph? Lets create a closer line graph for that",
"_____no_output_____"
],
[
"# Bulls and Bear #1",
"_____no_output_____"
],
[
"The first point of the graph is interesting, it woentt close to about 1000 and quickly went close to 20000 in half a year. While I sold my bitcoin for a measely 700 back in the day, I stopped reading news about bitcoin, so I am not too sure what went on that cause it to signficantly jump, other than I know that bitcoin gets increasing worse/harder to grab as bitcoin solves ever harder math questions that take up a lot of energy. A perception of miners and non-miners are that these cryptocurrency mining is... energy intensive... and it is , if you are a first/second generation cryptocurrency that uses mining as currency \n\n[Bitcoin uses as much energy as Argentina](https://www.iflscience.com/technology/bitcoin-mining-now-uses-more-electricity-than-argentina/)",
"_____no_output_____"
]
],
[
[
"#### Importing Pandas and others and Reading csv file\nimport os\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport math\nimport seaborn as sns\nimport pandas as pd\nimport plotly.express as px\n\n\nBitcoin = pd.read_csv('HistoricalData4.csv')\n\n##Created A daily Average for each day to work around with the data.\nBitcoin['Daily Average'] = Bitcoin.iloc[:, 2:4].sum(axis=1)/2\n\n#Condensed line graph plot to show, low, high, and Average price\nBitcoin.plot(x=\"Date\", y=[\"Low\", \"High\", \"Daily Average\"], figsize=(15, 20), title =\"Bitcoin Low, High, and Daily Average Prices.\", ylabel=\"Price in $\")\n\nplt.ylim([1000, 20000])\nplt.xlim([1500, 2000])\nplt.xticks(visible = True)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Sorting around June 16, 2017 and August 21, 2018, [we get some various articles while searching through Google](https://www.google.com/search?q=bitcoin&client=firefox-b-1-d&tbs=cdr:1,cd_min:6/8/2017,cd_max:10/21/2018,sbd:1&tbm=nws&ei=ys0tYMT8EIXU-gSIh7WICA&start=0&sa=N&ved=0ahUKEwjEie_KrvLuAhUFqp4KHYhDDYE4ChDy0wMIhQE&biw=1280&bih=818&dpr=1) \n\n[Bitcoin Hits a New Record High, But Stops Short of $20,000](https://fortune.com/2017/12/17/bitcoin-record-high-short-of-20000/)\n\n[Why is bitcoin’s price so high?](https://techcrunch.com/2017/12/08/why-is-bitcoins-price-so-high/)\n\n[Bitcoin tops $16,000, and it's $271B market value passes Home Depot's](https://www.usatoday.com/story/money/2017/12/07/bitcoin-tops-15-000-and-its-259-b-market-value-tops-home-depot/929962001/)",
"_____no_output_____"
],
[
"## Bear Market of Bitcoin, and the Rise of Bitcoin Cash Derivative\n\nLater articles in 2018 point to [Bitcoin Falling Off its all time high of getting close to 20k and dropping down close to 8k](https://www.reuters.com/article/us-global-markets-bitcoin-idUSKBN1FM11M) due to possible regulatory clampdown similar to what is happening to another cryptocurrency called [Ripple](https://www.sec.gov/news/press-release/2020-338).\n\nIn 2017, it was also the year that [Bitcoin Cash](https://www.marketwatch.com/story/meet-bitcoin-cashthe-new-digital-currency-that-surged-122-in-less-than-a-day-2017-08-02) made a debut. In technical terms, is known as a fork/derivative. It follow the example of the original Bitcoin and made changes to it. Even so, [Bitcoin Cash as of this article is 700+](https://www.coindesk.com/price/bitcoin-cash) ",
"_____no_output_____"
],
[
"# What is the future of of Cryptocurrency?",
"_____no_output_____"
],
[
"With the Stockmarket and Cryptocurrencies, we get a time of low and highs. For most Cryptocurrencies, in typical stockmartket terms, it is in quite a bullish territory right now.\n\nBut the old addage applies here: Buy low, sell high. Right now might not be a good time to buy BTC. If news is an indicator, it can go any way right now. A few things to look at.\n\nElon Musk Spent 1B+ of Bitcoin\n\n[SEC is looking into regulating cryptocurrency](https://www.financemagnates.com/cryptocurrency/regulation/sec-commissioner-demands-clear-cryptocurrency-regulations/)\n\n[Visa is planning to include Crytocurrency in it's list of currencies allowed to be transacted.](https://www.forbes.com/sites/billybambrough/2021/02/03/visa-reveals-bitcoin-and-crypto-banking-roadmap-amid-race-to-reach-network-of-70-million/?sh=39b269b401cd)\n\nThe original type of crytocurrency are energy intensive, Bitcoin and Etherium are the worse offenders when it comes to energy consumption. New Crypto, like Cardano, choose to do away with the mining and choose to reward people who invest with more shares rather than having to have thousands of computers taking up the worlds energy.\n\n**A Linear Regression** points that the data has overexceed it's prediction expectations. From day 1 since the Data collected to recently, the current price of Bitcoin is an extreme outlier when it comes to it's price. The Linear Regression expects Bitcoin price to be ~10k, but it has pushed up to 50k recently.\n\nIf the pattern continues, the new linear regression will eventually move up closer to 50k to reflect what could be it's potential price, as of right now, the prediction isn't accurate.",
"_____no_output_____"
]
],
[
[
"import os\nimport matplotlib.pyplot as plt\nimport math\nimport numpy as np\nimport seaborn as sns\nimport pandas as pd\nimport plotly.express as px\nfrom sklearn.linear_model import LinearRegression\n\n##Remodified .CSV data to make managing data easier.\n##Some data cleaning.\nBitcoin = pd.read_csv('HistoricalData4.csv')\n\n##Created A daily Average for each day to work around with the data.\nBitcoin['Daily Average'] = Bitcoin.iloc[:, 2:4].sum(axis=1)/2\n\n\n\nBitcoin['Date'] = pd.to_datetime(Bitcoin['Date']).apply(lambda date: date.toordinal())\n\nX = Bitcoin[[\"Date\"]]\ny = Bitcoin[[\"Daily Average\"]]\n\nregressor = LinearRegression()\nregressor.fit(X, y)\n\ny_pred = regressor.predict(X)\n\nplt.scatter(X, y, color = 'red')\nplt.plot(X, regressor.predict(X), color='blue')\nplt.title('Simple Bitcoin Regression')\nplt.xlabel('Ordinal Date')\nplt.ylabel('Daily Average Price in $')\nplt.figsize=(15, 30)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## In the next installment, I'll be doing a few different cryptocurrencies to see if it follows a similar pattern.",
"_____no_output_____"
],
[
"My thought is to do one on Cardano/ADA as that is where I see the future of Crytocurrency that is outside of just currency itself (like Bitcoin.)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb4df080b47a0e193ea68821b0e1d49c44d77b4f | 18,652 | ipynb | Jupyter Notebook | FirstSteps.ipynb | okkhoy/JlML | bb70726034a2f654141a1391cff66694acac9006 | [
"MIT"
] | null | null | null | FirstSteps.ipynb | okkhoy/JlML | bb70726034a2f654141a1391cff66694acac9006 | [
"MIT"
] | null | null | null | FirstSteps.ipynb | okkhoy/JlML | bb70726034a2f654141a1391cff66694acac9006 | [
"MIT"
] | null | null | null | 26.722063 | 809 | 0.525091 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
cb4dfb93b251befc2887e813dd2d093a61aed089 | 35,191 | ipynb | Jupyter Notebook | notebooks/athermal_linear_elasticity.ipynb | ernoc/jax-md | e812a0662fe57b7a657d6729214202db1b05d4c4 | [
"ECL-2.0",
"Apache-2.0"
] | 2 | 2021-09-12T16:18:40.000Z | 2021-09-29T03:59:26.000Z | notebooks/athermal_linear_elasticity.ipynb | ernoc/jax-md | e812a0662fe57b7a657d6729214202db1b05d4c4 | [
"ECL-2.0",
"Apache-2.0"
] | null | null | null | notebooks/athermal_linear_elasticity.ipynb | ernoc/jax-md | e812a0662fe57b7a657d6729214202db1b05d4c4 | [
"ECL-2.0",
"Apache-2.0"
] | null | null | null | 40.035267 | 714 | 0.539001 | [
[
[
"<a href=\"https://colab.research.google.com/github/google/jax-md/blob/main/notebooks/athermal_linear_elasticity.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"#@title Imports and utility code\n!pip install jax-md\n\nimport numpy as onp\n\nimport jax.numpy as jnp\nfrom jax.config import config\nconfig.update('jax_enable_x64', True)\n\nfrom jax import random\nfrom jax import jit, lax, grad, vmap\nimport jax.scipy as jsp\n\nfrom jax_md import space, energy, smap, minimize, util, elasticity, quantity\nfrom jax_md.colab_tools import renderer\n\nf32 = jnp.float32\nf64 = jnp.float64\n\nfrom functools import partial\n\nimport matplotlib\nimport matplotlib.pyplot as plt\nplt.rcParams.update({'font.size': 16})\n\ndef format_plot(x, y): \n plt.grid(True)\n plt.xlabel(x, fontsize=20)\n plt.ylabel(y, fontsize=20)\n \ndef finalize_plot(shape=(1, 0.7)):\n plt.gcf().set_size_inches(\n shape[0] * 1.5 * plt.gcf().get_size_inches()[1], \n shape[1] * 1.5 * plt.gcf().get_size_inches()[1])\n \n\ndef run_minimization_while(energy_fn, R_init, shift, max_grad_thresh = 1e-12, max_num_steps=1000000, **kwargs):\n init,apply=minimize.fire_descent(jit(energy_fn), shift, **kwargs)\n apply = jit(apply)\n\n @jit\n def get_maxgrad(state):\n return jnp.amax(jnp.abs(state.force))\n\n @jit\n def cond_fn(val):\n state, i = val\n return jnp.logical_and(get_maxgrad(state) > max_grad_thresh, i<max_num_steps)\n\n @jit\n def body_fn(val):\n state, i = val\n return apply(state), i+1\n\n state = init(R_init)\n state, num_iterations = lax.while_loop(cond_fn, body_fn, (state, 0))\n\n return state.position, get_maxgrad(state), num_iterations\n\ndef run_minimization_while_neighbor_list(energy_fn, neighbor_fn, R_init, shift, \n max_grad_thresh = 1e-12, max_num_steps = 1000000, \n step_inc = 1000, verbose = False, **kwargs):\n nbrs = neighbor_fn.allocate(R_init)\n\n init,apply=minimize.fire_descent(jit(energy_fn), shift, **kwargs)\n apply = jit(apply)\n\n @jit\n def get_maxgrad(state):\n return jnp.amax(jnp.abs(state.force))\n\n @jit\n def body_fn(state_nbrs, t):\n state, nbrs = state_nbrs\n nbrs = neighbor_fn.update(state.position, nbrs)\n state = apply(state, neighbor=nbrs)\n return (state, nbrs), 0\n \n state = init(R_init, neighbor=nbrs)\n \n step = 0\n while step < max_num_steps:\n if verbose:\n print('minimization step {}'.format(step))\n rtn_state, _ = lax.scan(body_fn, (state, nbrs), step + jnp.arange(step_inc))\n new_state, nbrs = rtn_state\n # If the neighbor list overflowed, rebuild it and repeat part of \n # the simulation.\n if nbrs.did_buffer_overflow:\n print('Buffer overflow.')\n nbrs = neighbor_fn.allocate(state.position)\n else:\n state = new_state\n step += step_inc\n if get_maxgrad(state) <= max_grad_thresh:\n break\n\n if verbose:\n print('successfully finished {} steps.'.format(step*step_inc))\n\n return state.position, get_maxgrad(state), nbrs, step\n\ndef run_minimization_scan(energy_fn, R_init, shift, num_steps=5000, **kwargs):\n init,apply=minimize.fire_descent(jit(energy_fn), shift, **kwargs)\n apply = jit(apply)\n\n @jit\n def scan_fn(state, i):\n return apply(state), 0.\n\n state = init(R_init)\n state, _ = lax.scan(scan_fn,state,jnp.arange(num_steps))\n\n return state.position, jnp.amax(jnp.abs(state.force))\n\nkey = random.PRNGKey(0)\n",
"_____no_output_____"
]
],
[
[
"#Linear elasticity in athermal systems\n\n## The elastic modulus tensor\n\nAn global affine deformation is given to lowest order by a symmetric strain tensor $\\epsilon$, which transforms any vector $r$ according to\n\\begin{equation}\nr \\rightarrow (1 + \\epsilon) \\cdot r. \n\\end{equation}\nNote that in $d$ dimensions, the strain tensor has $d(d + 1)/2$ independent elements. Now, when a mechanically stable system (i.e. a system at a local energy minimum where there is zero net force on every particle) is subject to an affine deformation, it usually does not remain in mechanical equilibrium. Therefore, there is a secondary, nonaffine response that returns the system to mechanical equilibrium, though usually at a different energy than the undeformed state. \n\nThe change of energy can be written to quadratic order as\n\\begin{equation}\n\\frac{ \\Delta U}{V^0} = \\sigma^0_{ij}\\epsilon_{ji} + \\frac 12 C_{ijkl} \\epsilon_{ij} \\epsilon_{kl} + O\\left( \\epsilon^3 \\right)\n\\end{equation}\nwhere $C_{ijkl}$ is the $d × d × d × d$ elastic modulus tensor, $\\sigma^0$ is the $d × d$ symmetric stress tensor describing residual stresses in the initial state, and $V^0$ is the volume of the initial state. The symmetries of \u0005$\\epsilon_{ij}$ imply the following: \n\\begin{equation}\nC_{ijkl} = C_{jikl} = C_{ijlk} = C_{klij}\n\\end{equation}\nWhen no further symmetries are assumed, the number of independent elastic constants becomes $\\frac 18 d(d + 1)(d^2 + d + 2)$, which is 6 in two dimensions and 21 in three dimensions.\n\n\n##Linear response to an external force\n\nConsider a set of $N$ particles in $d$ dimensions with positions $R_0$. Using $u \\equiv R - R_0$ and assuming fixed boundary conditions, we can expand the energy about $R_0$:\n\\begin{equation}\nU = U^0 - F^0 u + \\frac 12 u H^0 u + O(u^3),\n\\end{equation}\nwhere $U^0$ is the energy at $R_0$, $F^0$ is the force, $F^0_\\mu \\equiv \\left. \\frac {\\partial U}{\\partial u_\\mu} \\right |_{u=0}$, and $H^0$ is the Hessian, $H^0 \\equiv \\left. \\frac{ \\partial^2 U}{\\partial u_\\mu \\partial u_\\nu}\\right|_{u=0}$. \nNote that here we are expanding in terms of the particle positions, where as above we were expanding in the global strain degrees of freedom.\nIf we assume that $R_0$ corresponds to a local energy minimum, then $F^0=0$. Dropping higher order terms, we have a system of coupled harmonic oscillators given by\n\\begin{equation}\n\\Delta U \\equiv U - U^0 = \\frac 12 u H^0 u.\n\\end{equation}\nThis is independent of the form or details of $U$. \n\nHooke's law for this system gives the net force $f$ as a result of displacing the particles by $u$: \n\\begin{equation}\nf = -H^0 u.\n\\end{equation}\nThus, if an *external* force $f_\\mathrm{ext}$ is applied, the particles will respond so that the total force is zero, i.e. $f = -f_\\mathrm{ext}$. This response is obtained by solving for $u$:\n\\begin{equation}\nu = (H^0)^{-1} f_\\mathrm{ext}.\n\\end{equation}\n\n\n## Response to an affine strain\n\nNow consider a strain tensor $\\epsilon = \\tilde \\epsilon \\gamma$, where $\\gamma$ is a scalar and will be used to explicitly take the limit of small strain for fixed $\\tilde \\epsilon$. Importantly, the strain tensor represents a deformation of the underlying space that the particles live in and thus is a degree of freedom that is independent of the $Nd$ particle degrees of freedom. Therefore, knowing the particle positions $R$ is not sufficient to describe the energy, we also need to know $\\gamma$ to specify the correct boundary conditions:\n\\begin{equation}\t\nU = U(R, \\gamma).\n\\end{equation}\n\nWe now have a system with $Nd+1$ variables $\\{R, \\gamma\\}$ that, like before, form a set of coupled harmonic oscillators. We can describe this using the so-called \"generalized Hessian\" matrix of second derivatives of the energy with respect to both $R$ and $\\gamma$. Specifically, Hooke's law reads \n\\begin{equation}\n \\left( \\begin{array}{ ccccc|c}\n\t&&&&&\\\\\n\t&&H^0 &&& -\\Xi \\\\\n\t&&&&& \\\\ \\hline\n\t&&-\\Xi^T &&&\\frac{\\partial ^2U}{\\partial \\gamma^2}\n\t\\end{array}\\right) \n \\left( \\begin{array}{ c}\n\t\\\\\n\tu \\\\\n\t\\\\ \\hline\n\t\\gamma\n\t\\end{array}\\right) \n =\n \\left( \\begin{array}{ c}\n\t\\\\\n\t0 \\\\\n\t\\\\ \\hline\n\t\\tilde \\sigma\n\t\\end{array}\\right),\n\\end{equation}\n\nwhere $u = R - R_0$ is the displacement of every particle, $\\Xi = -\\frac{ \\partial^2 U}{\\partial R \\partial \\gamma}$, and $\\tilde \\sigma$ is the induced stress caused by the deformation. (If there is prestress in the system, i.e. $\\sigma^0 = \\frac{\\partial U}{\\partial \\gamma} \\neq 0$, the total stress is $\\sigma = \\sigma^0 + \\tilde \\sigma$.) In this equation, $\\gamma$ is held fixed and the zero in the top of the right-hand-side imposes force balance after the deformation and resulting non-affine displacement of every particle. The non-affine displacement itself, $u$, and the induced stress $\\sigma$, are both unknown but can be solved for. First, the non-affine response is \n\\begin{equation}\nu = (H^0)^{-1} \\Xi \\; \\gamma,\n\\end{equation}\nwhere we note that in the limit of small $\\gamma$, the force induced on every particle due to the affine deformation is $\\Xi \\; \\gamma$. Second, the induced stress is\n\\begin{equation}\n\\tilde \\sigma = \\frac{\\partial ^2U}{\\partial \\gamma^2} \\gamma - \\Xi^T u = \\left(\\frac{\\partial ^2U}{\\partial \\gamma^2} - \\Xi^T (H^0)^{-1} \\Xi \\right) \\gamma.\n\\end{equation}\n\nSimilarly, the change in energy is \n\\begin{equation}\n\\frac{\\Delta U}{V^0} = \\sigma^0 \\gamma + \\frac 1{2V^0} \\left(\\frac{\\partial ^2U}{\\partial \\gamma^2} - \\Xi^T (H^0)^{-1} \\Xi \\right) \\gamma^2,\n\\end{equation}\nwhere $\\sigma^0$ is the prestress in the system per unit volume. Comparing this to the above definition of the the elastic modulus tensor, we see that the elastic constant associated with the deformation $\\tilde \\epsilon$ is \n\\begin{equation}\nC(\\tilde \\epsilon) = \\frac 1{V^0} \\left( \\frac{\\partial^2 U}{\\partial \\gamma^2} - \\Xi^T (H^0)^{-1} \\Xi \\right).\n\\end{equation}\n\n$C(\\tilde \\epsilon)$ is related to $C_{ijkl}$ by summing $C(\\tilde \\epsilon) = C_{ijkl}\\tilde \\epsilon_{ij} \\tilde \\epsilon_{kl}$. So, if $\\tilde \\epsilon_{ij} = \\delta_{0i}\\delta_{0j}$, then $C_{0000} = C(\\tilde \\epsilon)$.\n\nThe internal code in `jax_md.elasticity` repeats this calculation for different $\\tilde \\epsilon$ to back out the different independent elastic constants. ",
"_____no_output_____"
],
[
"#First example\n",
"_____no_output_____"
],
[
"As a first example, let's consider a 3d system of 128 soft spheres. The elastic modulus tensor is only defined for systems that are at a local energy minimum, so we start by minimizing the energy.",
"_____no_output_____"
]
],
[
[
"N = 128\ndimension = 3\n\nbox_size = quantity.box_size_at_number_density(N, 1.4, dimension)\ndisplacement, shift = space.periodic(box_size)\n\nenergy_fn = energy.soft_sphere_pair(displacement)\n\nkey, split = random.split(key)\nR_init = random.uniform(split, (N,dimension), minval=0.0, maxval=box_size, dtype=f64)\n\nR, max_grad, niters = run_minimization_while(energy_fn, R_init, shift)\nprint('Minimized the energy in {} minimization steps and reached a final \\\nmaximum gradient of {}'.format(niters, max_grad))",
"_____no_output_____"
]
],
[
[
"We can now calculate the elastic modulus tensor",
"_____no_output_____"
]
],
[
[
"emt_fn = jit(elasticity.athermal_moduli(energy_fn, check_convergence=True))\nC, converged = emt_fn(R,box_size)\nprint(converged)",
"_____no_output_____"
]
],
[
[
"The elastic modulus tensor gives a quantitative prediction for how the energy should change if we deform the system according to a strain tensor\n\\begin{equation}\n\\frac{ \\Delta U}{V^0} = \\sigma^0\\epsilon + \\frac 12 \\epsilon C \\epsilon + O\\left(\\epsilon^3\\right)\n\\end{equation}\nTo test this, we define $\\epsilon = \\tilde \\epsilon \\gamma$ for a randomly chosen strain tensor $\\tilde \\epsilon$ and for $\\gamma << 1$. Ignoring terms of order $\\gamma^3$ and higher, we have\n\\begin{equation}\n\\frac{ \\Delta U}{V^0} - \\sigma^0\\epsilon = \\left[\\frac 12 \\tilde \\epsilon C \\tilde \\epsilon \\right] \\gamma^2\n\\end{equation}\nThus, we can test our calculation of $C$ by plotting $\\frac{ \\Delta U}{V^0} - \\sigma^0\\epsilon$ as a function of $\\gamma$ for our randomly chosen $\\tilde \\epsilon$ and comparing it to the line $\\left[\\frac 12 \\tilde \\epsilon C \\tilde \\epsilon \\right] \\gamma^2$. \n\nFirst, generate a random $\\tilde \\epsilon$ and calculate $U$ for different $\\gamma$. ",
"_____no_output_____"
]
],
[
[
"key, split = random.split(key)\n#Pick a random (symmetric) strain tensor\nstrain_tensor = random.uniform(split, (dimension,dimension), minval=-1, maxval=1, dtype=f64)\nstrain_tensor = (strain_tensor + strain_tensor.T) / 2.0\n\n#Define a function to calculate the energy at a given strain\ndef get_energy_at_strain(gamma, strain_tensor, R_init, box):\n R_init = space.transform(space.inverse(box),R_init)\n new_box = jnp.matmul(jnp.eye(strain_tensor.shape[0]) + gamma * strain_tensor, box)\n displacement, shift = space.periodic_general(new_box, fractional_coordinates=True)\n energy_fn = energy.soft_sphere_pair(displacement, sigma=1.0)\n R_final, _, _ = run_minimization_while(energy_fn, R_init, shift)\n return energy_fn(R_final)\n\ngammas = jnp.logspace(-7,-4,50)\nUs = vmap(get_energy_at_strain, in_axes=(0,None,None,None))(gammas, strain_tensor, R, box_size * jnp.eye(dimension))",
"_____no_output_____"
]
],
[
[
"Plot $\\frac{ \\Delta U}{V^0} - \\sigma^0\\epsilon$ and $\\left[\\frac 12 \\tilde \\epsilon C \\tilde \\epsilon \\right] \\gamma^2$ as functinos of $\\gamma$. While there may be disagreements for very small $\\gamma$ due to numerical precision or at large $\\gamma$ due to higher-order terms becoming relevant, there should be a region of quantitative agreement.",
"_____no_output_____"
]
],
[
[
"U_0 = energy_fn(R)\nstress_0 = -quantity.stress(energy_fn, R, box_size)\nV_0 = quantity.volume(dimension, box_size)\n\n#Plot \\Delta E/V - sigma*epsilon\ny1 = (Us - U_0)/V_0 - gammas * jnp.einsum('ij,ji->',stress_0,strain_tensor)\nplt.plot(jnp.abs(gammas), y1, lw=3, label=r'$\\Delta U/V^0 - \\sigma^0 \\epsilon$')\n\n#Plot 0.5 * epsilon*C*epsilon\ny2 = 0.5 * jnp.einsum('ij,ijkl,kl->',strain_tensor, C, strain_tensor) * gammas**2\nplt.plot(jnp.abs(gammas), y2, ls='--', lw=3, label=r'$(1/2) \\epsilon C \\epsilon$')\n\nplt.xscale('log')\nplt.yscale('log')\nplt.legend()\nformat_plot('$\\gamma$','')\nfinalize_plot()",
"_____no_output_____"
]
],
[
[
"To test the accuracy of this agreement, we first define:\n\\begin{equation}\nT(\\gamma) = \\frac{ \\Delta U}{V^0} - \\sigma^0\\epsilon - \\frac 12 \\epsilon C \\epsilon \\sim O\\left(\\gamma^3\\right)\n\\end{equation}\nwhich should be proportional to $\\gamma^3$ for small $\\gamma$ (note that this expected scaling should break down when the y-axis approaches machine precision). This is a prediction of scaling only, so we plot a line proportional to $\\gamma^3$ to compare the slopes.",
"_____no_output_____"
]
],
[
[
"#Plot the difference, which should scales as gamma**3\nplt.plot(jnp.abs(gammas), jnp.abs(y1-y2), label=r'$T(\\gamma)$')\n#Plot gamma**3 for reference\nplt.plot(jnp.abs(gammas), jnp.abs(gammas**3), 'black', label=r'slope = $\\gamma^3$ (for reference)')\n\nplt.xscale('log')\nplt.yscale('log')\nplt.legend()\nformat_plot('$\\gamma$','')\nfinalize_plot()",
"_____no_output_____"
]
],
[
[
"Save `C` for later testing.",
"_____no_output_____"
]
],
[
[
"C_3d = C",
"_____no_output_____"
]
],
[
[
"#Example with neighbor lists",
"_____no_output_____"
],
[
"As a second example, consider a much larger systems that is implemented using neighbor lists.",
"_____no_output_____"
]
],
[
[
"N = 5000\ndimension = 2\n\nbox_size = quantity.box_size_at_number_density(N, 1.3, dimension)\nbox = box_size * jnp.eye(dimension)\ndisplacement, shift = space.periodic_general(box, fractional_coordinates=True)\n\nsigma = jnp.array([[1.0, 1.2], [1.2, 1.4]])\nN_2 = int(N / 2)\nspecies = jnp.where(jnp.arange(N) < N_2, 0, 1)\n\nneighbor_fn, energy_fn = energy.soft_sphere_neighbor_list(\n displacement, box_size, species=species, sigma=sigma, dr_threshold = 0.1, \n fractional_coordinates = True)\n\nkey, split = random.split(key)\nR_init = random.uniform(split, (N,dimension), minval=0.0, maxval=1.0, dtype=f64) \n\nR, max_grad, nbrs, niters = run_minimization_while_neighbor_list(energy_fn, neighbor_fn, R_init, shift)\nprint('Minimized the energy in {} minimization steps and reached a final \\\nmaximum gradient of {}'.format(niters, max_grad))",
"_____no_output_____"
]
],
[
[
"We have to pass the neighbor list to `emt_fn`.",
"_____no_output_____"
]
],
[
[
"emt_fn = jit(elasticity.athermal_moduli(energy_fn, check_convergence=True))\nC, converged = emt_fn(R,box,neighbor=nbrs)\nprint(converged)",
"_____no_output_____"
]
],
[
[
"We can time the calculation of the compiled function.",
"_____no_output_____"
]
],
[
[
"%timeit emt_fn(R,box,neighbor=nbrs)",
"_____no_output_____"
]
],
[
[
"Repeat the same tests as above. NOTE: this may take a few minutes. ",
"_____no_output_____"
]
],
[
[
"key, split = random.split(key)\n#Pick a random (symmetric) strain tensor\nstrain_tensor = random.uniform(split, (dimension,dimension), minval=-1, maxval=1, dtype=f64)\nstrain_tensor = (strain_tensor + strain_tensor.T) / 2.0\n\ndef get_energy_at_strain(gamma, strain_tensor, R_init, box):\n new_box = jnp.matmul(jnp.eye(strain_tensor.shape[0]) + gamma * strain_tensor, box)\n displacement, shift = space.periodic_general(new_box, fractional_coordinates=True)\n\n neighbor_fn, energy_fn = energy.soft_sphere_neighbor_list(\n displacement, box_size, species=species, sigma=sigma, dr_threshold = 0.1, \n fractional_coordinates = True, capacity_multiplier = 1.5)\n \n R_final, _, nbrs, _ = run_minimization_while_neighbor_list(energy_fn, neighbor_fn, R_init, shift)\n return energy_fn(R_final, neighbor=nbrs)\n\ngammas = jnp.logspace(-7,-3,20)\nUs = jnp.array([ get_energy_at_strain(gamma, strain_tensor, R, box) for gamma in gammas])",
"_____no_output_____"
],
[
"U_0 = energy_fn(R, neighbor=nbrs)\nstress_0 = -quantity.stress(energy_fn, R, box, neighbor=nbrs)\nV_0 = quantity.volume(dimension, box)\n\n#Plot \\Delta E/V - sigma*epsilon\ny1 = (Us - U_0)/V_0 - gammas * jnp.einsum('ij,ji->',stress_0,strain_tensor)\nplt.plot(jnp.abs(gammas), y1, lw=3, label=r'$\\Delta U/V^0 - \\sigma^0 \\epsilon$')\n\n#Plot 0.5 * epsilon*C*epsilon\ny2 = 0.5 * jnp.einsum('ij,ijkl,kl->',strain_tensor, C, strain_tensor) * gammas**2\nplt.plot(jnp.abs(gammas), y2, ls='--', lw=3, label=r'$(1/2) \\epsilon C \\epsilon$')\n\nplt.xscale('log')\nplt.yscale('log')\nplt.legend()\nformat_plot('$\\gamma$','')\nfinalize_plot()",
"_____no_output_____"
],
[
"#Plot the difference, which should scales as gamma**3\nplt.plot(jnp.abs(gammas), jnp.abs(y1-y2), label=r'$T(\\gamma)$')\n#Plot gamma**3 for reference\nplt.plot(jnp.abs(gammas), jnp.abs(gammas**3), 'black', label=r'slope = $\\gamma^3$ (for reference)')\n\nplt.xscale('log')\nplt.yscale('log')\nplt.legend()\nformat_plot('$\\gamma$','')\nfinalize_plot()",
"_____no_output_____"
]
],
[
[
"Save `C` for later testing.",
"_____no_output_____"
]
],
[
[
"C_2d = C",
"_____no_output_____"
]
],
[
[
"#Mandel notation",
"_____no_output_____"
],
[
"Mandel notation is a way to represent symmetric second-rank tensors and fourth-rank tensors with so-called \"minor symmetries\", i.e. $T_{ijkl} = T_{ijlk} = T_{jilk}$. The idea is to map pairs of indices so that $(i,i) \\rightarrow i$ and $(i,j) \\rightarrow K - i - j$ for $i\\neq j$, where $K = d(d+1)/2$ is the number of independent pairs $(i,j)$ for tensors with $d$ elements along each axis. Thus, second-rank tensors become first-rank tensors, and fourth-rank tensors become second-rank tensors, according to:\n\n\\begin{align}\nM_{m(i,j)} &= T_{ij} w(i,j) \\\\\nM_{m(i,j),m(k,l)} &= T_{ijkl} w(i,j) w(k,l).\n\\end{align}\n\nHere, $m(i,j)$ is the mapping function described above, and w(i,j) is a weight that preserves summation rules and is given by \n\\begin{align}\nw(i,j) = \\delta_{ij} + \\sqrt{2} (\\delta_{ij}-1).\n\\end{align}\n\nWe can convert strain tensors, stress tensors, and elastic modulus tensors to and from Mandel notation using the functions `elasticity.tensor_to_mandel` and `elasticity.mandel_to_tensor`. ",
"_____no_output_____"
],
[
"First, lets copy one of the previously calculated elastic modulus tensors and define a random strain tensor.",
"_____no_output_____"
]
],
[
[
"#This can be 2 or 3 depending on which of the above solutions has been calculated\ndimension = 3 \nif dimension == 2:\n C = C_2d\nelse:\n C = C_3d\nkey, split = random.split(key)\ne = random.uniform(key, (dimension,dimension), minval=-1, maxval=1, dtype=f64)\ne = (e + e.T)/2.",
"_____no_output_____"
]
],
[
[
"Convert `e` and `C` to Mental notation",
"_____no_output_____"
]
],
[
[
"e_m = jit(elasticity.tensor_to_mandel)(e)\nC_m = jit(elasticity.tensor_to_mandel)(C)\nprint(e_m)\nprint(C_m)",
"_____no_output_____"
]
],
[
[
"Using \"bar\" notation to represent Mandel vectors and matrices, we have\n\\begin{equation}\n\\frac{ \\Delta U}{V^0} = \\bar \\sigma_i^0 \\bar\\epsilon_i + \\frac 12 \\bar \\epsilon_i \\bar C_{ij} \\bar\\epsilon_j + O\\left(\\bar \\epsilon^3\\right)\n\\end{equation}\nWe can explicity test that the sums are equivalent to the sums involving the original tensors",
"_____no_output_____"
]
],
[
[
"sum_m = jnp.einsum('i,ij,j->',e_m, C_m, e_m)\nsum_t = jnp.einsum('ij,ijkl,kl->',e, C, e)\nprint('Relative error is {}, which should be very close to 0'.format((sum_t-sum_m)/sum_t))",
"_____no_output_____"
]
],
[
[
"Finally, we can convert back to the full tensors and check that they are unchanged.",
"_____no_output_____"
]
],
[
[
"C_new = jit(elasticity.mandel_to_tensor)(C_m)\nprint('Max error in C is {}, which should be very close to 0.'.format(jnp.max(jnp.abs(C-C_new))))\n\ne_new = jit(elasticity.mandel_to_tensor)(e_m)\nprint('Max error in e is {}, which should be very close to 0.'.format(jnp.max(jnp.abs(e-e_new))))",
"_____no_output_____"
]
],
[
[
"# Isotropic elastic constants\n",
"_____no_output_____"
],
[
"The calculation of the elastic modulus tensor does not make any assumptions about the underlying symmetries in the material. However, for isotropic systems, only two constants are needed to completely describe the elastic behavior. These are often taken to be the bulk modulus, $B$, and the shear modulus, $G$, or the Young's modulus, $E$, and the Poisson's ratio, $\\nu$. The function `elasticity.extract_isotropic_moduli` extracts these values, as well as the longitudinal modulus, $M$, from an elastic modulus tensor. \n\nImportantly, since there is not guarantee that `C` is calculated from a truely isotropic systems, these are \"orientation-averaged\" values. For example, there are many directions in which you can shear a system, and the shear modulus that is returned represents and average over all these orientations. This can be an effective way to average over small fluctuations in an \"almost isotropic\" system, but the values lose their typical meaning when the systems is highly anisotropic.",
"_____no_output_____"
]
],
[
[
"elasticity.extract_isotropic_moduli(C)",
"_____no_output_____"
]
],
[
[
"# Gradients",
"_____no_output_____"
],
[
"The calculation of the elastic modulus tensor is fully differentiable:",
"_____no_output_____"
]
],
[
[
"def setup(N,dimension,key):\n box_size = quantity.box_size_at_number_density(N, 1.4, dimension)\n box = box_size * jnp.eye(dimension)\n displacement, shift = space.periodic_general(box, fractional_coordinates=True)\n R_init = random.uniform(key, (N,dimension), minval=0.0, maxval=1.0, dtype=f64)\n\n def run(sigma):\n energy_fn = energy.soft_sphere_pair(displacement, sigma=sigma)\n R, max_grad = run_minimization_scan(energy_fn, R_init, shift, num_steps=1000)\n emt_fn = jit(elasticity.athermal_moduli(energy_fn))\n C = emt_fn(R,box)\n return elasticity.extract_isotropic_moduli(C)['G']\n return run\n\nkey, split = random.split(key)\nN = 50\ndimension = 2\nrun = setup(N, dimension, split)",
"_____no_output_____"
],
[
"sigma = jnp.linspace(1.0,1.4,N)\nprint(run(sigma))\nprint(grad(run)(sigma))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cb4dfd7eabae04a58fd1ececb2b010c93b02fc3b | 133,959 | ipynb | Jupyter Notebook | EMSEDataAnalytics-master/EMSE6992_Assignments/HW1.ipynb | dadebruce/dadebruce.github.io | 12a92a83e1681b712dc914e021c71ea40c6ca078 | [
"CC-BY-3.0"
] | null | null | null | EMSEDataAnalytics-master/EMSE6992_Assignments/HW1.ipynb | dadebruce/dadebruce.github.io | 12a92a83e1681b712dc914e021c71ea40c6ca078 | [
"CC-BY-3.0"
] | null | null | null | EMSEDataAnalytics-master/EMSE6992_Assignments/HW1.ipynb | dadebruce/dadebruce.github.io | 12a92a83e1681b712dc914e021c71ea40c6ca078 | [
"CC-BY-3.0"
] | null | null | null | 66.946027 | 26,424 | 0.790428 | [
[
[
"## This assignment\n\nIn this assignment, you'll learn (or review):\n\n* How to set up Jupyter on your own computer.\n* Python basics, like defining functions.\n* How to use the `numpy` library to compute with arrays of numbers.",
"_____no_output_____"
],
[
"# 2. Python\n\nPython is the main programming language we'll use in this course. We assume you have some experience with Python or can learn it yourself, but here is a brief review.\n\nBelow are some simple Python code fragments.\n\nYou should feel confident explaining what each fragment is doing. If not,\nplease brush up on your Python. There a number of tutorials online (search\nfor \"Python tutorial\"). https://docs.python.org/3/tutorial/ is a good place to\nstart.",
"_____no_output_____"
]
],
[
[
"2 + 2",
"_____no_output_____"
],
[
"# This is a comment.\n# In Python, the ** operator performs exponentiation.\nimport math\nmath.e**(-2)",
"_____no_output_____"
],
[
"print(\"Hello\" + \",\", \"world!\")\n\"Hello, cell output!\"",
"Hello, world!\n"
],
[
"def add2(x):\n \"\"\"This docstring explains what this function does: it adds 2 to a number.\"\"\"\n return x + 2",
"_____no_output_____"
],
[
"def makeAdder(amount):\n \"\"\"Make a function that adds the given amount to a number.\"\"\"\n def addAmount(x):\n return x + amount\n return addAmount\n\nadd3 = makeAdder(3)\nadd3(4)",
"_____no_output_____"
],
[
"# add4 is very similar to add2, but it's been created using a lambda expression.\nadd4 = lambda x: x + 4\nadd4(5)",
"_____no_output_____"
],
[
"sameAsMakeAdder = lambda amount: lambda x: x + amount\nadd5 = sameAsMakeAdder(5)\nadd5(6)",
"_____no_output_____"
],
[
"def fib(n):\n if n <= 1:\n return 1\n # Functions can call themselves recursively.\n return fib(n-1) + fib(n-2)\n\nfib(4)",
"_____no_output_____"
],
[
"# A for loop repeats a block of code once for each\n# element in a given collection.\nfor i in range(5):\n if i % 2 == 0:\n print(2**i)\n else:\n print(\"Odd power of 2\")",
"1\nOdd power of 2\n4\nOdd power of 2\n16\n"
],
[
"# A list comprehension is a convenient way to apply a function\n# to each element in a given collection.\n# The String method join appends together all its arguments\n# separated by the given string. So we append each element produced\n# by the list comprehension, each separated by a newline (\"\\n\").\nprint(\"\\n\".join([str(2**i) if i % 2 == 0 else \"Odd power of 2\" for i in range(5)]))",
"1\nOdd power of 2\n4\nOdd power of 2\n16\n"
]
],
[
[
"#### Question 1\n\n##### Question 1a\nWrite a function nums_reversed that takes in an integer `n` and returns a string\ncontaining the numbers 1 through `n` including `n` in reverse order, separated\nby spaces. For example:\n\n >>> nums_reversed(5)\n '5 4 3 2 1'\n\n***Note:*** The ellipsis (`...`) indicates something you should fill in. It *doesn't* necessarily imply you should replace it with only one line of code.",
"_____no_output_____"
]
],
[
[
"def nums_reversed(n):\n lst1 = list(range(n + 1))\n lst2 = lst1[1:]\n lst3 = lst2[::-1]\n print(lst3)\n\nnums_reversed(4)",
"[4, 3, 2, 1]\n"
],
[
"_ = ok.grade('q01a')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"##### Question 1b\n\nWrite a function `string_splosion` that takes in a non-empty string like\n`\"Code\"` and returns a long string containing every prefix of the input.\nFor example:\n\n >>> string_splosion('Code')\n 'CCoCodCode'\n >>> string_splosion('data!')\n 'ddadatdatadata!'\n >>> string_splosion('hi')\n 'hhi'\n",
"_____no_output_____"
]
],
[
[
"def string_splosion(string):\n \n ans = ''\n \n for i in range(len(string)):\n ans = ans + string[:(i+1)]\n \n return ans\n\nstring_splosion('data!')",
"_____no_output_____"
],
[
"_ = ok.grade('q01b')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"##### Question 1c\n\nWrite a function `double100` that takes in a list of integers\nand returns `True` only if the list has two `100`s next to each other.\n\n >>> double100([100, 2, 3, 100])\n False\n >>> double100([2, 3, 100, 100, 5])\n True\n",
"_____no_output_____"
]
],
[
[
"def double100(nums):\n if 100 in nums:\n first = nums.index(100)\n if nums[first + 1] == 100:\n return True\n else:\n return False\n\ndouble100([2, 3, 100, 100, 5])",
"_____no_output_____"
],
[
"_ = ok.grade('q01c')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"##### Question 1d\n\nWrite a function `median` that takes in a list of numbers\nand returns the median element of the list. If the list has even\nlength, it returns the mean of the two elements in the middle.\n\n >>> median([5, 4, 3, 2, 1])\n 3\n >>> median([ 40, 30, 10, 20 ])\n 25",
"_____no_output_____"
]
],
[
[
"def median(number_list):\n half = len(number_list) // 2\n \n if len(number_list) % 2 == 0:\n right = number_list[half]\n left = number_list[half - 1]\n return (right + left) / 2\n else:\n return number_list[half]\n\nmedian([ 40, 30, 10, 20 ])",
"_____no_output_____"
],
[
"_ = ok.grade('q01d')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"# 3. `NumPy`\n\nThe `NumPy` library lets us do fast, simple computing with numbers in Python.",
"_____no_output_____"
],
[
"## 3.1. Arrays\n\nThe basic `NumPy` data type is the array, a homogeneously-typed sequential collection (a list of things that all have the same type). Arrays will most often contain strings, numbers, or other arrays.",
"_____no_output_____"
],
[
"Let's create some arrays:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\narray1 = np.array([2, 3, 4, 5])\narray2 = np.arange(4)\narray1, array2",
"_____no_output_____"
]
],
[
[
"Math operations on arrays happen *element-wise*. Here's what we mean:",
"_____no_output_____"
]
],
[
[
"array1 * 2",
"_____no_output_____"
],
[
"array1 * array2",
"_____no_output_____"
],
[
"array1 ** array2",
"_____no_output_____"
]
],
[
[
"This is not only very convenient (fewer `for` loops!) but also fast. `NumPy` is designed to run operations on arrays much faster than equivalent Python code on lists. Data science sometimes involves working with large datasets where speed is important - even the constant factors!",
"_____no_output_____"
],
[
"**Jupyter pro-tip**: Pull up the docs for any function in Jupyter by running a cell with\nthe function name and a `?` at the end:",
"_____no_output_____"
]
],
[
[
"np.arange?",
"_____no_output_____"
]
],
[
[
"**Another Jupyter pro-tip**: Pull up the docs for any function in Jupyter by typing the function\nname, then `<Shift>-<Tab>` on your keyboard. Super convenient when you forget the order\nof the arguments to a function. You can press `<Tab>` multiple tabs to expand the docs.\n\nTry it on the function below:",
"_____no_output_____"
]
],
[
[
"np.linspace",
"_____no_output_____"
]
],
[
[
"#### Question 2\nUsing the `np.linspace` function, create an array called `xs` that contains\n100 evenly spaced points between `0` and `2 * np.pi`. Then, create an array called `ys` that\ncontains the value of $ \\sin{x} $ at each of those 100 points.\n\n*Hint:* Use the `np.sin` function. You should be able to define each variable with one line of code.)",
"_____no_output_____"
]
],
[
[
"xs = np.linspace(0, 2 * np.pi)\nys = np.sin(np.linspace(0, 2 * np.pi))",
"_____no_output_____"
],
[
"_ = ok.grade('q02')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"The `plt.plot` function from another library called `matplotlib` lets us make plots. It takes in\nan array of x-values and a corresponding array of y-values. It makes a scatter plot of the (x, y) pairs and connects points with line segments. If you give it enough points, it will appear to create a smooth curve.\n\nLet's plot the points you calculated in the previous question:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nplt.plot(xs, ys);",
"_____no_output_____"
]
],
[
[
"This is a useful recipe for plotting any function:\n1. Use `linspace` or `arange` to make a range of x-values.\n2. Apply the function to each point to produce y-values.\n3. Plot the points.",
"_____no_output_____"
],
[
"You might remember from calculus that the derivative of the `sin` function is the `cos` function. That means that the slope of the curve you plotted above at any point `xs[i]` is given by `cos(xs[i])`. You can try verifying this by plotting `cos` in the next cell.",
"_____no_output_____"
]
],
[
[
"yc = np.cos(np.linspace(0, 2 * np.pi))\n\nplt.plot(xs, yc);",
"_____no_output_____"
]
],
[
[
"Calculating derivatives is an important operation in data science, but it can be difficult. We can have computers do it for us using a simple idea called *numerical differentiation*.\n\nConsider the `i`th point `(xs[i], ys[i])`. The slope of `sin` at `xs[i]` is roughly the slope of the line connecting `(xs[i], ys[i])` to the nearby point `(xs[i+1], ys[i+1])`. That slope is:\n\n (ys[i+1] - ys[i]) / (xs[i+1] - xs[i])\n\nIf the difference between `xs[i+1]` and `xs[i]` were infinitessimal, we'd have exactly the derivative. In numerical differentiation we take advantage of the fact that it's often good enough to use \"really small\" differences instead.",
"_____no_output_____"
],
[
"#### Question 3\n\nDefine a function called `derivative` that takes in an array of x-values and their\ncorresponding y-values and computes the slope of the line connecting each point to the next point.\n\n >>> derivative(np.array([0, 1, 2]), np.array([2, 4, 6]))\n np.array([2., 2.])\n >>> derivative(np.arange(5), np.arange(5) ** 2)\n np.array([0., 2., 4., 6.])\n\nNotice that the output array has one less element than the inputs since we can't\nfind the slope for the last point.\n\nIt's possible to do this in one short line using [slicing](http://pythoncentral.io/how-to-slice-listsarrays-and-tuples-in-python/), but feel free to use whatever method you know.\n\n**Then**, use your `derivative` function to compute the slopes for each point in `xs`, `ys`.\nStore the slopes in an array called `slopes`.",
"_____no_output_____"
]
],
[
[
"def derivative(xvals, yvals):\n y_dim = np.diff(yvals)\n x_dim = np.diff(xvals)\n slopes = y_dim / x_dim\n return slopes\n\nslopes = derivative(xs, ys)\n\nderivative(np.array([0, 1, 2]), np.array([2, 4, 6]))",
"_____no_output_____"
],
[
"_ = ok.grade('q03')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"#### Question 4\nPlot the slopes you computed. Then plot `cos` on top of your plot, calling `plt.plot` again in the same cell. Did numerical differentiation work?\n\n*Note:* Since we have only 99 slopes, you'll need to take off the last x-value before plotting to avoid an error.",
"_____no_output_____"
]
],
[
[
"plt.plot(xs[:-1], slopes);\nplt.plot(xs, yc);",
"_____no_output_____"
]
],
[
[
"In the plot above, it's probably not clear which curve is which. Examine the cell below to see how to plot your results with a legend.",
"_____no_output_____"
]
],
[
[
"plt.plot(xs[:-1], slopes, label=\"Numerical derivative\")\nplt.plot(xs[:-1], np.cos(xs[:-1]), label=\"True derivative\")\n# You can just call plt.legend(), but the legend will cover up\n# some of the graph. Use bbox_to_anchor=(x,y) to set the x-\n# and y-coordinates of the center-left point of the legend,\n# where, for example, (0, 0) is the bottom-left of the graph\n# and (1, .5) is all the way to the right and halfway up.\nplt.legend(bbox_to_anchor=(1, .5), loc=\"center left\");",
"_____no_output_____"
]
],
[
[
"## 3.2. Multidimensional Arrays\nA multidimensional array is a primitive version of a table, containing only one kind of data and having no column labels. A 2-dimensional array is useful for working with *matrices* of numbers.",
"_____no_output_____"
]
],
[
[
"# The zeros function creates an array with the given shape.\n# For a 2-dimensional array like this one, the first\n# coordinate says how far the array goes *down*, and the\n# second says how far it goes *right*.\narray3 = np.zeros((4, 5))\narray3",
"_____no_output_____"
],
[
"# The shape attribute returns the dimensions of the array.\narray3.shape",
"_____no_output_____"
],
[
"# You can think of array3 as an array containing 4 arrays, each\n# containing 5 zeros. Accordingly, we can set or get the third\n# element of the second array in array 3 using standard Python\n# array indexing syntax twice:\narray3[1][2] = 7\narray3",
"_____no_output_____"
],
[
"# This comes up so often that there is special syntax provided\n# for it. The comma syntax is equivalent to using multiple\n# brackets:\narray3[1, 2] = 8\narray3",
"_____no_output_____"
]
],
[
[
"Arrays allow you to assign to multiple places at once. The special character `:` means \"everything.\"",
"_____no_output_____"
]
],
[
[
"array4 = np.zeros((3, 5))\narray4[:, 2] = 5\narray4",
"_____no_output_____"
]
],
[
[
"In fact, you can use arrays of indices to assign to multiple places. Study the next example and make sure you understand how it works.",
"_____no_output_____"
]
],
[
[
"array5 = np.zeros((3, 5))\nrows = np.array([1, 0, 2])\ncols = np.array([3, 1, 4])\n\n# Indices (1,3), (0,1), and (2,4) will be set.\narray5[rows, cols] = 3\narray5",
"_____no_output_____"
]
],
[
[
"#### Question 5\nCreate a 50x50 array called `twice_identity` that contains all zeros except on the\ndiagonal, where it contains the value `2`.\n\nStart by making a 50x50 array of all zeros, then set the values. Use indexing, not a `for` loop! (Don't use `np.eye` either, though you might find that function useful later.)",
"_____no_output_____"
]
],
[
[
"twice_identity = np.identity(50) * 2\ntwice_identity",
"_____no_output_____"
],
[
"_ = ok.grade('q05')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"# 4. A Picture Puzzle",
"_____no_output_____"
],
[
"Your boss has given you some strange text files. He says they're images,\nsome of which depict a summer scene and the rest a winter scene.\n\nHe demands that you figure out how to determine whether a given\ntext file represents a summer scene or a winter scene.\n\nYou receive 10 files, `1.txt` through `10.txt`. Peek at the files in a text\neditor of your choice.",
"_____no_output_____"
],
[
"#### Question 6\nHow do you think the contents of the file are structured? Take your best guess.",
"_____no_output_____"
],
[
"**Much like the MNIST dataset, these files are probably structured as pixels where the data represents a greyscale value or the level to which each pixel is dark or has color.**",
"_____no_output_____"
],
[
"#### Question 7\nCreate a function called `read_file_lines` that takes in a filename as its argument.\nThis function should return a Python list containing the lines of the\nfile as strings. That is, if `1.txt` contains:\n\n```\n1 2 3\n3 4 5\n7 8 9\n```\n\nthe return value should be: `['1 2 3\\n', '3 4 5\\n', '7 8 9\\n']`.\n\n**Then**, use the `read_file_lines` function on the file `1.txt`, reading the contents\ninto a variable called `file1`.\n\n*Hint:* Check out [this Stack Overflow page](http://stackoverflow.com/questions/3277503/how-to-read-a-file-line-by-line-into-a-list-with-python) on reading lines of files.",
"_____no_output_____"
]
],
[
[
"def read_file_lines(filename):\n with open(filename) as f:\n lines = f.readlines()\n file1 = [x.strip() for x in lines] \n return file1\n\nfile1 = read_file_lines('C:\\\\Users\\\\davei\\\\Documents\\\\EMSEDataAnalytics\\\\EMSE6992_Assignments\\\\data\\\\HW1\\\\1.txt')",
"_____no_output_____"
],
[
"_ = ok.grade('q07')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"Each file begins with a line containing two numbers. After checking the length of\na file, you could notice that the product of these two numbers equals the number of\nlines in each file (other than the first one).\n\nThis suggests the rows represent elements in a 2-dimensional grid. In fact, each\ndataset represents an image!\n\nOn the first line, the first of the two numbers is\nthe height of the image (in pixels) and the second is the width (again in pixels).\n\nEach line in the rest of the file contains the pixels of the image.\nEach pixel is a triplet of numbers denoting how much red, green, and blue\nthe pixel contains, respectively.\n\nIn image processing, each column in one of these image files is called a *channel*\n(disregarding line 1). So there are 3 channels: red, green, and blue.\n\n#### Question 8\nDefine a function called `lines_to_image` that takes in the contents of a\nfile as a list (such as `file1`). It should return an array containing integers of\nshape `(n_rows, n_cols, 3)`. That is, it contains the pixel triplets organized in the\ncorrect number of rows and columns.\n\nFor example, if the file originally contained:\n\n```\n4 2\n0 0 0\n10 10 10\n2 2 2\n3 3 3\n4 4 4\n5 5 5\n6 6 6\n7 7 7\n```\n\nThe resulting array should be a *3-dimensional* array that looks like this:\n\n```\narray([\n [ [0,0,0], [10,10,10] ],\n [ [2,2,2], [3,3,3] ],\n [ [4,4,4], [5,5,5] ],\n [ [6,6,6], [7,7,7] ]\n])\n```\n\nThe string method `split` and the function `np.reshape` might be useful.\n\n**Important note:** You must call `.astype(np.uint8)` on the final array before\nreturning so that `numpy` will recognize the array represents an image.\n\nOnce you've defined the function, set `image1` to the result of calling\n`lines_to_image` on `file1`.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndef lines_to_image(lines_to_image):\n file = lines_to_image[1:]\n \n lst = []\n \n for line in file:\n new_file1 = line.split()\n new_file2 = list(new_file1)\n new_file2 = [int(i) for i in new_file2]\n lst.append(new_file2)\n \n array = np.array([lst])\n new_array = array.reshape((array.shape[0], array.shape[1], 3))\n \n return new_array\n\nimage1 = lines_to_image(file1)\nimage1.shape",
"_____no_output_____"
],
[
"_ = ok.grade('q08')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"#### Question 9\n\nImages in `numpy` are simply arrays, but we can also display them them as\nactual images in this notebook.\n\nUse the provided `show_images` function to display `image1`. You may call it\nlike `show_images(image1)`. If you later have multiple images to display, you\ncan call `show_images([image1, image2])` to display them all at once.\n\nThe resulting image should look almost completely black. Why do you suppose\nthat is?",
"_____no_output_____"
]
],
[
[
"def show_images(images, ncols=2, figsize=(10, 7), **kwargs):\n \"\"\"\n Shows one or more color images.\n \n images: Image or list of images. Each image is a 3-dimensional\n array, where dimension 1 indexes height and dimension 2\n the width. Dimension 3 indexes the 3 color values red,\n blue, and green (so it always has length 3).\n \"\"\"\n def show_image(image, axis=plt):\n plt.imshow(image, **kwargs)\n \n if not (isinstance(images, list) or isinstance(images, tuple)):\n images = [images]\n images = [image.astype(np.uint8) for image in images]\n \n nrows = math.ceil(len(images) / ncols)\n ncols = min(len(images), ncols)\n \n plt.figure(figsize=figsize)\n for i, image in enumerate(images):\n axis = plt.subplot2grid(\n (nrows, ncols),\n (i // ncols, i % ncols),\n )\n axis.tick_params(bottom='off', left='off', top='off', right='off',\n labelleft='off', labelbottom='off')\n axis.grid(False)\n show_image(image, axis)",
"_____no_output_____"
],
[
"show_images(image1)",
"C:\\Users\\davei\\Anaconda3\\lib\\site-packages\\matplotlib\\cbook\\__init__.py:424: MatplotlibDeprecationWarning: \nPassing one of 'on', 'true', 'off', 'false' as a boolean is deprecated; use an actual boolean (True/False) instead.\n warn_deprecated(\"2.2\", \"Passing one of 'on', 'true', 'off', 'false' as a \"\n"
]
],
[
[
"#### Question 10\n\nIf you look at the data, you'll notice all the numbers lie between 0 and 10.\nIn `NumPy`, a color intensity is an integer ranging from 0 to 255, where 0 is\nno color (black). That's why the image is almost black. To see the image,\nwe'll need to rescale the numbers in the data to have a larger range.\n\nDefine a function `expand_image_range` that takes in an image. It returns a\n**new copy** of the image with the following transformation:\n \n old value | new value\n ========= | =========\n 0 | 12\n 1 | 37\n 2 | 65\n 3 | 89\n 4 | 114\n 5 | 137\n 6 | 162\n 7 | 187\n 8 | 214\n 9 | 240\n 10 | 250\n\nThis expands the color range of the image. For example, a pixel that previously\nhad the value `[5 5 5]` (almost-black) will now have the value `[137 137 137]`\n(gray).\n\nSet `expanded1` to the expanded `image1`, then display it with `show_images`.\n\n[This page](https://docs.scipy.org/doc/numpy/reference/arrays.indexing.html#boolean-array-indexing)\nfrom the numpy docs has some useful information that will allow you\nto use indexing instead of `for` loops.\n\nHowever, the slickest implementation uses one very short line of code.\n*Hint:* If you index an array with another array or list as in question 5, your\narray (or list) of indices can contain repeats, as in `array1[[0, 1, 0]]`.\nInvestigate what happens in that case.",
"_____no_output_____"
]
],
[
[
"# This array is provided for your convenience.\ntransformed = np.array([12, 37, 65, 89, 114, 137, 162, 187, 214, 240, 250])\n\ndef expand_image_range(image):\n ...\n\nexpanded1 = ...\nshow_images(expanded1)",
"_____no_output_____"
],
[
"_ = ok.grade('q10')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"#### Question 11\n\nEureka! You've managed to reveal the image that the text file represents.\n\nNow, define a function called `reveal_file` that takes in a filename\nand returns an expanded image. This should be relatively easy since you've\ndefined functions for each step in the process.\n\nThen, set `expanded_images` to a list of all the revealed images. There are\n10 images to reveal (including the one you just revealed).\n\nFinally, use `show_images` to display the `expanded_images`.",
"_____no_output_____"
]
],
[
[
"def reveal_file(filename):\n ...\n\nfilenames = ['1.txt', '2.txt', '3.txt', '4.txt', '5.txt',\n '6.txt', '7.txt', '8.txt', '9.txt', '10.txt']\nexpanded_images = ...\n\nshow_images(expanded_images, ncols=5)",
"_____no_output_____"
]
],
[
[
"Notice that 5 of the above images are of summer scenes; the other 5\nare of winter.\n\nThink about how you'd distinguish between pictures of summer and winter. What\nqualities of the image seem to signal to your brain that the image is one of\nsummer? Of winter?\n\nOne trait that seems specific to summer pictures is that the colors are warmer.\nLet's see if the proportion of pixels of each color in the image can let us\ndistinguish between summer and winter pictures.",
"_____no_output_____"
],
[
"#### Question 12\nTo simplify things, we can categorize each pixel according to its most intense\n(highest-value) channel. (Remember, red, green, and blue are the 3 channels.)\nFor example, we could just call a `[2 4 0]` pixel \"green.\" If a pixel has a\ntie between several channels, let's count it as none of them.\n\nWrite a function `proportion_by_channel`. It takes in an image. It assigns\neach pixel to its greatest-intensity channel: red, green, or blue. Then\nthe function returns an array of length three containing the proportion of\npixels categorized as red, the proportion categorized as green, and the\nproportion categorized as blue (respectively). (Again, don't count pixels\nthat are tied between 2 or 3 colors as any category, but do count them\nin the denominator when you're computing proportions.)\n\nFor example:\n\n```\n>>> test_im = np.array([\n [ [5, 2, 2], [2, 5, 10] ] \n])\n>>> proportion_by_channel(test_im)\narray([ 0.5, 0, 0.5 ])\n\n# If tied, count neither as the highest\n>>> test_im = np.array([\n [ [5, 2, 5], [2, 50, 50] ] \n])\n>>> proportion_by_channel(test_im)\narray([ 0, 0, 0 ])\n```\n\nThen, set `image_proportions` to the result of `proportion_by_channel` called\non each image in `expanded_images` as a 2d array.\n\n*Hint:* It's fine to use a `for` loop, but for a difficult challenge, try\navoiding it. (As a side benefit, your code will be much faster.) Our solution\nuses the `NumPy` functions `np.reshape`, `np.sort`, `np.argmax`, and `np.bincount`.",
"_____no_output_____"
]
],
[
[
"def proportion_by_channel(image):\n ...\n\nimage_proportions = ...\nimage_proportions",
"_____no_output_____"
],
[
"_ = ok.grade('q12')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"Let's plot the proportions you computed above on a bar chart:",
"_____no_output_____"
]
],
[
[
"# You'll learn about Pandas and DataFrames soon.\nimport pandas as pd\npd.DataFrame({\n 'red': image_proportions[:, 0],\n 'green': image_proportions[:, 1],\n 'blue': image_proportions[:, 2]\n }, index=pd.Series(['Image {}'.format(n) for n in range(1, 11)], name='image'))\\\n .iloc[::-1]\\\n .plot.barh();",
"_____no_output_____"
]
],
[
[
"#### Question 13\n\nWhat do you notice about the colors present in the summer images compared to\nthe winter ones?\n\nUse this info to write a function `summer_or_winter`. It takes in an image and\nreturns `True` if the image is a summer image and `False` if the image is a\nwinter image.\n\n**Do not hard-code the function to the 10 images you currently have (eg.\n`if image1, return False`).** We will run your function on other images\nthat we've reserved for testing.\n\nYou must classify all of the 10 provided images correctly to pass the test\nfor this function.\n",
"_____no_output_____"
]
],
[
[
"def summer_or_winter(image):\n ...",
"_____no_output_____"
],
[
"_ = ok.grade('q13')\n_ = ok.backup()",
"_____no_output_____"
]
],
[
[
"Congrats! You've created your very first classifier for this class.",
"_____no_output_____"
],
[
"#### Question 14\n\n1. How do you think your classification function will perform\n in general?\n2. Why do you think it will perform that way?\n3. What do you think would most likely give you false positives?\n4. False negatives?",
"_____no_output_____"
],
[
"*Write your answer here, replacing this text.*",
"_____no_output_____"
],
[
"**Final note:** While our approach here is simplistic, skin color segmentation\n-- figuring out which parts of the image belong to a human body -- is a\nkey step in many algorithms such as face detection.",
"_____no_output_____"
],
[
"# Optional: Our code to encode images",
"_____no_output_____"
],
[
"Here are the functions we used to generate the text files for this assignment.\n\nFeel free to send not-so-secret messages to your friends if you'd like.",
"_____no_output_____"
]
],
[
[
"import skimage as sk\nimport skimage.io as skio",
"_____no_output_____"
],
[
"def read_image(filename):\n '''Reads in an image from a filename'''\n return skio.imread(filename)",
"_____no_output_____"
],
[
"def compress_image(im):\n '''Takes an image as an array and compresses it to look black.'''\n res = im / 25\n return res.astype(np.uint8)",
"_____no_output_____"
],
[
"def to_text_file(im, filename):\n '''\n Takes in an image array and a filename for the resulting text file.\n \n Creates the encoded text file for later decoding.\n '''\n h, w, c = im.shape\n to_rgb = ' '.join\n to_row = '\\n'.join\n to_lines = '\\n'.join\n \n rgb = [[to_rgb(triplet) for triplet in row] for row in im.astype(str)]\n lines = to_lines([to_row(row) for row in rgb])\n\n with open(filename, 'w') as f:\n f.write('{} {}\\n'.format(h, w))\n f.write(lines)\n f.write('\\n')",
"_____no_output_____"
],
[
"summers = skio.imread_collection('orig/summer/*.jpg')\nwinters = skio.imread_collection('orig/winter/*.jpg')\nlen(summers)",
"_____no_output_____"
],
[
"sum_nums = np.array([ 5, 6, 9, 3, 2, 11, 12])\nwin_nums = np.array([ 10, 7, 8, 1, 4, 13, 14])\n\nfor im, n in zip(summers, sum_nums):\n to_text_file(compress_image(im), '{}.txt'.format(n))\nfor im, n in zip(winters, win_nums):\n to_text_file(compress_image(im), '{}.txt'.format(n))",
"_____no_output_____"
]
],
[
[
"# 5. Submitting this assignment\n",
"_____no_output_____"
],
[
"First, run this cell to run all the autograder tests at once so you can double-\ncheck your work.",
"_____no_output_____"
]
],
[
[
"_ = ok.grade_all()",
"_____no_output_____"
]
],
[
[
"Now, run this code in your terminal to make a\n[git commit](https://www.atlassian.com/git/tutorials/saving-changes/git-commit)\nthat saves a snapshot of your changes in `git`. The last line of the cell\nruns [git push](http://stackoverflow.com/questions/2745076/what-are-the-differences-between-git-commit-and-git-push), which will send your work to your personal Github repo.",
"_____no_output_____"
],
[
"```\n# Tell git to commit all the changes so far\ngit add -A\n\n# Tell git to make the commit\ngit commit -m \"hw1 finished\"\n\n# Send your updates to your personal private repo\ngit push origin master\n```",
"_____no_output_____"
],
[
"Finally, we'll submit the assignment to OkPy so that the staff will know to\ngrade it. You can submit as many times as you want and you can choose which\nsubmission you want us to grade by going to https://okpy.org/cal/data100/sp17/.",
"_____no_output_____"
]
],
[
[
"# Now, we'll submit to okpy\n_ = ok.submit()",
"_____no_output_____"
]
],
[
[
"Congrats! You are done with homework 1.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb4e02f01984ba0a5de97f603c5bc060d9dae1d4 | 18,510 | ipynb | Jupyter Notebook | COVID-19/COVID19 Analysis across countries and weeks.ipynb | Thejineaswar/Exploratory_Data_Analysis | 13a8a7128db510c282f7f51f4db9ff400445eef1 | [
"MIT"
] | null | null | null | COVID-19/COVID19 Analysis across countries and weeks.ipynb | Thejineaswar/Exploratory_Data_Analysis | 13a8a7128db510c282f7f51f4db9ff400445eef1 | [
"MIT"
] | null | null | null | COVID-19/COVID19 Analysis across countries and weeks.ipynb | Thejineaswar/Exploratory_Data_Analysis | 13a8a7128db510c282f7f51f4db9ff400445eef1 | [
"MIT"
] | null | null | null | 30.85 | 323 | 0.559427 | [
[
[
"# COVID-19 Analysis across countries and weeks",
"_____no_output_____"
],
[
"In this study, the focus is on the country cases. This analysis examines at the case growth, case proportion and weekly growth.",
"_____no_output_____"
],
[
"# This kerenel will be updated frequently to keep it up to date",
"_____no_output_____"
],
[
"# Library and dataset imports",
"_____no_output_____"
]
],
[
[
"#Importing the libraries\nimport pandas as pd\nimport numpy as np\n\nimport matplotlib.pyplot as plt\nimport plotly.express as px\nfrom plotly.subplots import make_subplots\nimport plotly.graph_objects as go",
"_____no_output_____"
]
],
[
[
"The data used for this study is taken from the Johns Hopkins CSSE data repository. The following files were used for the analysis:\n1. time_series_covid19_confirmed_global.csv-> https://rb.gy/uktxf3\n2. time_series_covid19_deaths_global.csv -> https://rb.gy/qnjgsj\n3. time_series_covid19_recovered_global.csv-> https://rb.gy/dxfjfl",
"_____no_output_____"
]
],
[
[
"#Importing the datasets\nurl='https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv'\nconfirmed=pd.read_csv(url,error_bad_lines=False)\ndeath=pd.read_csv('https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv',\n error_bad_lines=False)\nrecovered=pd.read_csv('https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_recovered_global.csv',\n error_bad_lines=False)\n",
"_____no_output_____"
]
],
[
[
"# Case proportions of countries",
"_____no_output_____"
],
[
"The function country_with_cases will return a dataframe with just the country name and the number of cases. This function also adds up all the province value into one to make the overall data just in terms of the Country/Region. The main reason this is done is because the Province/State contains a lot of null values",
"_____no_output_____"
]
],
[
[
"def country_with_cases(dataset):\n is_province=dataset.loc[dataset['Province/State'].isna()==False]\n is_province=is_province['Country/Region'].unique()\n df=dataset.copy()\n Country=df['Country/Region'].values\n temp=df.drop(columns=['Province/State','Country/Region','Lat','Long'])\n values=temp.values\n cases=[]\n for i in range(0,len(values)):\n cases.append(values[i][values.shape[1]-1])\n new_df=pd.concat([pd.DataFrame(Country),pd.DataFrame(cases)],axis=1)\n new_df.columns=['Country','Cases']\n index=[]\n is_province_sums=[]\n for i in is_province:\n temp=new_df.loc[new_df['Country']==i]\n index.append(temp.index)\n s=np.sum(temp['Cases'])\n is_province_sums.append(s)\n for i in index:\n new_df.drop(i,axis=0,inplace=True)\n countries_with_province=pd.concat([pd.DataFrame(is_province),pd.DataFrame(is_province_sums)],axis=1)\n countries_with_province.columns=['Country','Cases']\n All_country_cases=pd.concat([new_df,countries_with_province],axis=0)\n All_country_cases.reset_index(inplace=True)\n return All_country_cases",
"_____no_output_____"
],
[
"confirmed_tree=country_with_cases(confirmed)",
"_____no_output_____"
],
[
"def tree_map(df,color_scale,title):\n #The df is the one which only has country and cases\n fig = px.treemap(df,path=['Country'], values='Cases',color='Cases',\n color_continuous_scale=color_scale,\n title=title)\n fig.show()",
"_____no_output_____"
],
[
"tree_map(confirmed_tree,'amp','Confirmed cases across countries')",
"_____no_output_____"
],
[
"recovered_tree=country_with_cases(recovered)\ntree_map(recovered_tree,'Greens','Recovery across countries')",
"_____no_output_____"
],
[
"death_tree=country_with_cases(death)\ntree_map(death_tree,'Reds','Deaths across countries')",
"_____no_output_____"
]
],
[
[
"# Visualising the Country growth ",
"_____no_output_____"
]
],
[
[
"def top_10(df):# This function will only work \n df_descending=df.sort_values(by='Cases', ascending=False)\n df_descending=df_descending.reset_index()\n top=df_descending.iloc[:10 :]\n return top['Country'].values",
"_____no_output_____"
]
],
[
[
"The rate function converts the structire of the dataframe in general. In the dataset the dates are the column making it not so convenient for visualising the data. This function transforms all the dates into one column and this makes plotting very much easier",
"_____no_output_____"
]
],
[
[
"def rate(df): \n is_province=df.loc[confirmed['Province/State'].isna()==False]\n is_province=is_province['Country/Region'].unique()\n copy=df.copy()\n final=[]\n index=[]\n for i in is_province: \n temp=copy.loc[copy['Country/Region']==i]\n index=copy.loc[copy['Country/Region']==i].index\n temp=temp.sum(axis=0)\n final.append(temp)\n copy.drop(index,inplace=True)\n new_df=pd.DataFrame(final)\n new_df['Country/Region']=is_province\n total=pd.concat([copy,new_df],axis=0)\n total.reset_index(inplace=True)\n total.drop(columns=['Province/State'],inplace=True)\n t=pd.melt(total,id_vars=['Country/Region','index','Lat','Long'],var_name=\"Date\", value_name=\"Value\")\n return t",
"_____no_output_____"
],
[
"c=rate(confirmed)\nfig = px.line(c, x=\"Date\", y=\"Value\", color=\"Country/Region\",\n title='Confirmed cases across countries')\nfig.update_layout(showlegend=False)\nfig.show()",
"_____no_output_____"
],
[
"r=rate(recovered)\nfig = px.line(r, x=\"Date\", y=\"Value\", color=\"Country/Region\",\n title='Confirmed cases across countries')\nfig.update_layout(showlegend=False)\n\nfig.show()",
"_____no_output_____"
],
[
"d=rate(death)\nfig = px.line(d, x=\"Date\", y=\"Value\", color=\"Country/Region\",\n title='Deaths across countries')\nfig.update_layout(showlegend=False)\n\nfig.show()",
"_____no_output_____"
]
],
[
[
"# Analysing the top 10 countries ",
"_____no_output_____"
],
[
"We will analyse 10 countries with the highest confirmed case",
"_____no_output_____"
]
],
[
[
"top_ten=top_10(confirmed_tree)",
"_____no_output_____"
],
[
"def stacked_line_subplots(confirmed,death,recovered,countries):\n subplot_title=[]\n for i in countries:\n subplot_title.append('Cases in {}'.format(i))\n subplot_title=tuple(subplot_title)\n fig = make_subplots(rows=len(countries), cols=1,subplot_titles=subplot_title)\n #countries=['India','US','Yemen','Angola']\n dates=confirmed.columns\n dates=np.delete(dates,[0,2,3])\n dfs=[confirmed,death,recovered]\n row=1\n for i in range(len(countries)):\n value=[]\n for j in dfs:\n temp=j.loc[j['Country/Region']==countries[i]].values\n temp=np.delete(temp,[0,1,2,3])\n value.append(temp)\n if(i==0):\n fig.append_trace(go.Scatter(x=dates,y=value[1],mode='lines',name='Death',\n line_color='red',stackgroup='covid',legendgroup=\"group1\"),row=row,col=1)\n fig.append_trace(go.Scatter(x=dates,y=value[2],mode='lines',name='Recovered',\n line_color='green',stackgroup='covid',legendgroup=\"group2\"),row=row,col=1)\n fig.append_trace(go.Scatter(x=dates,y=value[0],mode='lines',name='Confirmed',\n line_color='blue',stackgroup='covid',legendgroup=\"group3\"),row=row,col=1)\n else:\n fig.append_trace(go.Scatter(x=dates,y=value[1],mode='lines',name='Death',\n line_color='red',stackgroup='covid', showlegend=False,\n legendgroup=\"group1\"),row=row,col=1)\n fig.append_trace(go.Scatter(x=dates,y=value[2],mode='lines',name='Recovered',\n line_color='green',stackgroup='covid', showlegend=False,\n legendgroup=\"group2\"),row=row,col=1)\n fig.append_trace(go.Scatter(x=dates,y=value[0],mode='lines',name='Confirmed',\n line_color='blue',stackgroup='covid', showlegend=False,\n legendgroup=\"group3\"),row=row,col=1)\n \n\n row+=1\n \n fig.update_layout(height=2000, width=800,\n title_text=\"Cases across the top 10 countries\")\n \n fig.show()\n ",
"_____no_output_____"
]
],
[
[
"This stacked line graph in a way shows the ratio of the cases in a country. The blue area in way indicates the active cases(when both red and green areas are present), green indicates recovered and red indicates the deaths",
"_____no_output_____"
]
],
[
[
"stacked_line_subplots(confirmed,death,recovered,top_ten)",
"_____no_output_____"
]
],
[
[
"# Visuals on the World Map",
"_____no_output_____"
]
],
[
[
"def world_map(df,title,color):\n dates=df.columns\n dates=np.delete(dates,[0,2,3])\n country=df['Country/Region']\n lat=df['Lat']\n long=df['Long']\n transformed=pd.melt(df,id_vars=['Province/State','Country/Region','Lat','Long'],\n var_name=\"Date\", value_name=\"Value\")\n fig = px.scatter_geo(transformed, lat=\"Lat\",lon=\"Long\", color=\"Value\",\n hover_name=\"Country/Region\", size=transformed[\"Value\"],\n animation_frame=\"Date\",\n projection=\"natural earth\",title=title,\n color_continuous_scale=color)\n fig.show()",
"_____no_output_____"
],
[
"world_map(confirmed,'Confirmed cases with time series','Jet')",
"_____no_output_____"
],
[
"world_map(recovered,'Recovered cases with time series','YlGn')",
"_____no_output_____"
],
[
"world_map(death,'Death Cases with time series','Burg')",
"_____no_output_____"
]
],
[
[
"# Weekly analysis on case growth",
"_____no_output_____"
],
[
"The function weekly_trend() mainly creates a new column named week. This column indicates the week \nnumber of a particular day. For this, the dates have been converted to days since the first reported day. The function will return the a dataframe of one particular week",
"_____no_output_____"
]
],
[
[
"from datetime import date\ndef weekly_trend(df,number):\n c=rate(df)\n date_values=c['Date'].values\n first_date=date_values[0].split('/')\n first=list(map(int, first_date))\n for i in range(len(date_values)):\n random=date_values[i].split('/')\n random=list(map(int, random))\n delta=date(2020,random[0],random[1])-date(2020,first[0],first[1])\n date_values[i]=delta.days\n c['Date']=date_values\n c['Weeks']=c['Date']//7\n confirmed_week=c.loc[c['Weeks']==number]\n confirmed_week=confirmed_week.sort_values(by='Value',ascending=False) \n return confirmed_week\n \n",
"_____no_output_____"
]
],
[
[
"The function top_3_trends returns the top 3 countries. These are the countries which have the highest growth in the week",
"_____no_output_____"
]
],
[
[
"def top_3_trends(week,week_number):\n last_day=week.loc[week['Date']==((week_number*7)+6)]\n first_day=week.loc[week['Date']==(week_number*7)]\n first_day_values=first_day['Value'].values\n last_day['Value']=last_day['Value']-first_day_values\n last_day=last_day.sort_values(by='Value',ascending=False)\n countries=last_day['Country/Region'].unique()[:3]\n return countries\n ",
"_____no_output_____"
]
],
[
[
"This function plots the top 3 countries in a subplot",
"_____no_output_____"
]
],
[
[
"def top_3(week,week_number,subject):\n top_3=top_3_trends(week,week_number)\n week=week.sort_values(by='Date',ascending=True)\n \n subplot_title=[]\n for i in top_3:\n subplot_title.append( '{}'.format(i))\n subplot_title=tuple(subplot_title)\n fig = make_subplots(rows=1, cols=3,subplot_titles=subplot_title)\n \n col=1\n for i in range(0,3):\n temp=week.loc[week['Country/Region']==top_3[i]]\n temp['Value']-=temp.iloc[0,5]\n fig.append_trace(go.Scatter(x=temp['Date'],y=temp['Value'],mode='lines',showlegend=False\n ),col=col,row=1)\n \n col+=1\n \n fig.update_layout(title_text=\"Week {} trends of {} in top 3 countries\".format(week_number,subject))\n \n fig.show()\n \n ",
"_____no_output_____"
],
[
"for i in range(1,22):\n week=weekly_trend(confirmed,i)\n top_3(week,i,'Confirmed Cases')\n#The same can be done for Recovered and death cases, but the the kernel becomes to long and redundant",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb4e04c600978e1b9a14b3c6014dfcb68e897434 | 56,928 | ipynb | Jupyter Notebook | docs/notebooks/Common_Gotchas_in_JAX.ipynb | GregCT/jax | 9580fd1cb2d2850081b99797982ebc0f007461ad | [
"ECL-2.0",
"Apache-2.0"
] | null | null | null | docs/notebooks/Common_Gotchas_in_JAX.ipynb | GregCT/jax | 9580fd1cb2d2850081b99797982ebc0f007461ad | [
"ECL-2.0",
"Apache-2.0"
] | null | null | null | docs/notebooks/Common_Gotchas_in_JAX.ipynb | GregCT/jax | 9580fd1cb2d2850081b99797982ebc0f007461ad | [
"ECL-2.0",
"Apache-2.0"
] | null | null | null | 29.089423 | 670 | 0.554402 | [
[
[
"# 🔪 JAX - The Sharp Bits 🔪\n\n[](https://colab.research.google.com/github/google/jax/blob/master/docs/notebooks/Common_Gotchas_in_JAX.ipynb)",
"_____no_output_____"
],
[
"*levskaya@ mattjj@*\n\nWhen walking about the countryside of [Italy](https://iaml.it/blog/jax-intro), the people will not hesitate to tell you that __JAX__ has _\"una anima di pura programmazione funzionale\"_.\n\n__JAX__ is a language for __expressing__ and __composing__ __transformations__ of numerical programs. __JAX__ is also able to __compile__ numerical programs for CPU or accelerators (GPU/TPU). \nJAX works great for many numerical and scientific programs, but __only if they are written with certain constraints__ that we describe below.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom jax import grad, jit\nfrom jax import lax\nfrom jax import random\nimport jax\nimport jax.numpy as jnp\nimport matplotlib as mpl\nfrom matplotlib import pyplot as plt\nfrom matplotlib import rcParams\nrcParams['image.interpolation'] = 'nearest'\nrcParams['image.cmap'] = 'viridis'\nrcParams['axes.grid'] = False",
"_____no_output_____"
]
],
[
[
"## 🔪 Pure functions",
"_____no_output_____"
],
[
"JAX transformation and compilation are designed to work only on Python functions that are functionally pure: all the input data is passed through the function parameters, all the results are output through the function results. A pure function will always return the same result if invoked with the same inputs. \n\nHere are some examples of functions that are not functionally pure for which JAX behaves differently than the Python interpreter. Note that these behaviors are not guaranteed by the JAX system; the proper way to use JAX is to use it only on functionally pure Python functions.",
"_____no_output_____"
]
],
[
[
"def impure_print_side_effect(x):\n print(\"Executing function\") # This is a side-effect \n return x\n\n# The side-effects appear during the first run \nprint (\"First call: \", jit(impure_print_side_effect)(4.))\n\n# Subsequent runs with parameters of same type and shape may not show the side-effect\n# This is because JAX now invokes a cached compilation of the function\nprint (\"Second call: \", jit(impure_print_side_effect)(5.))\n\n# JAX re-runs the Python function when the type or shape of the argument changes\nprint (\"Third call, different type: \", jit(impure_print_side_effect)(jnp.array([5.])))",
"Executing function\nFirst call: 4.0\nSecond call: 5.0\nExecuting function\nThird call, different type: [5.]\n"
],
[
"g = 0.\ndef impure_uses_globals(x):\n return x + g\n\n# JAX captures the value of the global during the first run\nprint (\"First call: \", jit(impure_uses_globals)(4.))\ng = 10. # Update the global\n\n# Subsequent runs may silently use the cached value of the globals\nprint (\"Second call: \", jit(impure_uses_globals)(5.))\n\n# JAX re-runs the Python function when the type or shape of the argument changes\n# This will end up reading the latest value of the global\nprint (\"Third call, different type: \", jit(impure_uses_globals)(jnp.array([4.])))",
"First call: 4.0\nSecond call: 5.0\nThird call, different type: [14.]\n"
],
[
"g = 0.\ndef impure_saves_global(x):\n global g\n g = x\n return x\n\n# JAX runs once the transformed function with special Traced values for arguments\nprint (\"First call: \", jit(impure_saves_global)(4.))\nprint (\"Saved global: \", g) # Saved global has an internal JAX value",
"First call: 4.0\nSaved global: Traced<ShapedArray(float32[], weak_type=True):JaxprTrace(level=-1/1)>\n"
]
],
[
[
"A Python function can be functionally pure even if it actually uses stateful objects internally, as long as it does not read or write external state:",
"_____no_output_____"
]
],
[
[
"def pure_uses_internal_state(x):\n state = dict(even=0, odd=0)\n for i in range(10):\n state['even' if i % 2 == 0 else 'odd'] += x\n return state['even'] + state['odd']\n\nprint(jit(pure_uses_internal_state)(5.))",
"_____no_output_____"
]
],
[
[
"It is not recommended to use iterators in any JAX function you want to `jit` or in any control-flow primitive. The reason is that an iterator is a python object which introduces state to retrieve the next element. Therefore, it is incompatible with JAX functional programming model. In the code below, there are some examples of incorrect attempts to use iterators with JAX. Most of them return an error, but some give unexpected results.",
"_____no_output_____"
]
],
[
[
"import jax.numpy as jnp\nimport jax.lax as lax\nfrom jax import make_jaxpr\n\n# lax.fori_loop\narray = jnp.arange(10)\nprint(lax.fori_loop(0, 10, lambda i,x: x+array[i], 0)) # expected result 45\niterator = iter(range(10))\nprint(lax.fori_loop(0, 10, lambda i,x: x+next(iterator), 0)) # unexpected result 0\n\n# lax.scan\ndef func11(arr, extra):\n ones = jnp.ones(arr.shape) \n def body(carry, aelems):\n ae1, ae2 = aelems\n return (carry + ae1 * ae2 + extra, carry)\n return lax.scan(body, 0., (arr, ones)) \nmake_jaxpr(func11)(jnp.arange(16), 5.)\n# make_jaxpr(func11)(iter(range(16)), 5.) # throws error\n\n# lax.cond\narray_operand = jnp.array([0.])\nlax.cond(True, lambda x: x+1, lambda x: x-1, array_operand)\niter_operand = iter(range(10))\n# lax.cond(True, lambda x: next(x)+1, lambda x: next(x)-1, iter_operand) # throws error",
"45\n0\n"
]
],
[
[
"## 🔪 In-Place Updates",
"_____no_output_____"
],
[
"In Numpy you're used to doing this:",
"_____no_output_____"
]
],
[
[
"numpy_array = np.zeros((3,3), dtype=np.float32)\nprint(\"original array:\")\nprint(numpy_array)\n\n# In place, mutating update\nnumpy_array[1, :] = 1.0\nprint(\"updated array:\")\nprint(numpy_array)",
"original array:\n[[0. 0. 0.]\n [0. 0. 0.]\n [0. 0. 0.]]\nupdated array:\n[[0. 0. 0.]\n [1. 1. 1.]\n [0. 0. 0.]]\n"
]
],
[
[
"If we try to update a JAX device array in-place, however, we get an __error__! (☉_☉)",
"_____no_output_____"
]
],
[
[
"jax_array = jnp.zeros((3,3), dtype=jnp.float32)\n\n# In place update of JAX's array will yield an error!\ntry:\n jax_array[1, :] = 1.0\nexcept Exception as e:\n print(\"Exception {}\".format(e))",
"Exception '<class 'jax.interpreters.xla.DeviceArray'>' object does not support item assignment. JAX arrays are immutable; perhaps you want jax.ops.index_update or jax.ops.index_add instead?\n"
]
],
[
[
"__What gives?!__ \n\nAllowing mutation of variables in-place makes program analysis and transformation very difficult. JAX requires a pure functional expression of a numerical program. \n\nInstead, JAX offers the _functional_ update functions: [__index_update__](https://jax.readthedocs.io/en/latest/_autosummary/jax.ops.index_update.html#jax.ops.index_update), [__index_add__](https://jax.readthedocs.io/en/latest/_autosummary/jax.ops.index_add.html#jax.ops.index_add), [__index_min__](https://jax.readthedocs.io/en/latest/_autosummary/jax.ops.index_min.html#jax.ops.index_min), [__index_max__](https://jax.readthedocs.io/en/latest/_autosummary/jax.ops.index_max.html#jax.ops.index_max), and the [__index__](https://jax.readthedocs.io/en/latest/_autosummary/jax.ops.index.html#jax.ops.index) helper.\n\n️⚠️ inside `jit`'d code and `lax.while_loop` or `lax.fori_loop` the __size__ of slices can't be functions of argument _values_ but only functions of argument _shapes_ -- the slice start indices have no such restriction. See the below __Control Flow__ Section for more information on this limitation.",
"_____no_output_____"
]
],
[
[
"from jax.ops import index, index_add, index_update",
"_____no_output_____"
]
],
[
[
"### index_update",
"_____no_output_____"
],
[
"If the __input values__ of __index_update__ aren't reused, __jit__-compiled code will perform these operations _in-place_.",
"_____no_output_____"
]
],
[
[
"jax_array = jnp.zeros((3, 3))\nprint(\"original array:\")\nprint(jax_array)\n\nnew_jax_array = index_update(jax_array, index[1, :], 1.)\n\nprint(\"old array unchanged:\")\nprint(jax_array)\n\nprint(\"new array:\")\nprint(new_jax_array)",
"original array:\n[[0. 0. 0.]\n [0. 0. 0.]\n [0. 0. 0.]]\nold array unchanged:\n[[0. 0. 0.]\n [0. 0. 0.]\n [0. 0. 0.]]\nnew array:\n[[0. 0. 0.]\n [1. 1. 1.]\n [0. 0. 0.]]\n"
]
],
[
[
"### index_add",
"_____no_output_____"
],
[
"If the __input values__ of __index_update__ aren't reused, __jit__-compiled code will perform these operations _in-place_.",
"_____no_output_____"
]
],
[
[
"print(\"original array:\")\njax_array = jnp.ones((5, 6))\nprint(jax_array)\n\nnew_jax_array = index_add(jax_array, index[::2, 3:], 7.)\nprint(\"new array post-addition:\")\nprint(new_jax_array)",
"original array:\n[[1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1.]]\nnew array post-addition:\n[[1. 1. 1. 8. 8. 8.]\n [1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 8. 8. 8.]\n [1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 8. 8. 8.]]\n"
]
],
[
[
"## 🔪 Out-of-Bounds Indexing",
"_____no_output_____"
],
[
"In Numpy, you are used to errors being thrown when you index an array outside of its bounds, like this:",
"_____no_output_____"
]
],
[
[
"try:\n np.arange(10)[11]\nexcept Exception as e:\n print(\"Exception {}\".format(e))",
"Exception index 11 is out of bounds for axis 0 with size 10\n"
]
],
[
[
"However, raising an error from code running on an accelerator can be difficult or impossible. Therefore, JAX must choose some non-error behavior for out of bounds indexing (akin to how invalid floating point arithmetic results in `NaN`). When the indexing operation is an array index update (e.g. `index_add` or `scatter`-like primitives), updates at out-of-bounds indices will be skipped; when the operation is an array index retrieval (e.g. NumPy indexing or `gather`-like primitives) the index is clamped to the bounds of the array since __something__ must be returned. For example, the last value of the array will be returned from this indexing operation:",
"_____no_output_____"
]
],
[
[
"jnp.arange(10)[11]",
"_____no_output_____"
]
],
[
[
"Note that due to this behavior for index retrieval, functions like `jnp.nanargmin` and `jnp.nanargmax` return -1 for slices consisting of NaNs whereas Numpy would throw an error.\n\nNote also that, as the two behaviors described above are not inverses of each other, reverse-mode automatic differentiation (which turns index updates into index retrievals and vice versa) [will not preserve the semantics of out of bounds indexing](https://github.com/google/jax/issues/5760). Thus it may be a good idea to think of out-of-bounds indexing in JAX as a case of [undefined behavior](https://en.wikipedia.org/wiki/Undefined_behavior).",
"_____no_output_____"
],
[
"## 🔪 Non-array inputs: NumPy vs. JAX\n\nNumPy is generally happy accepting Python lists or tuples as inputs to its API functions:",
"_____no_output_____"
]
],
[
[
"np.sum([1, 2, 3])",
"_____no_output_____"
]
],
[
[
"JAX departs from this, generally returning a helpful error:",
"_____no_output_____"
]
],
[
[
"try:\n jnp.sum([1, 2, 3])\nexcept TypeError as e:\n print(f\"TypeError: {e}\")",
"TypeError: sum requires ndarray or scalar arguments, got <class 'list'> at position 0.\n"
]
],
[
[
"This is a deliberate design choice, because passing lists or tuples to traced functions can lead to silent performance degradation that might otherwise be difficult to detect.\n\nFor example, consider the following permissive version of `jnp.sum` that allows list inputs:",
"_____no_output_____"
]
],
[
[
"def permissive_sum(x):\n return jnp.sum(jnp.array(x))\n\nx = list(range(10))\npermissive_sum(x)",
"_____no_output_____"
]
],
[
[
"The output is what we would expect, but this hides potential performance issues under the hood. In JAX's tracing and JIT compilation model, each element in a Python list or tuple is treated as a separate JAX variable, and individually processed and pushed to device. This can be seen in the jaxpr for the ``permissive_sum`` function above:",
"_____no_output_____"
]
],
[
[
"make_jaxpr(permissive_sum)(x)",
"_____no_output_____"
]
],
[
[
"Each entry of the list is handled as a separate input, resulting in a tracing & compilation overhead that grows linearly with the size of the list. To prevent surprises like this, JAX avoids implicit conversions of lists and tuples to arrays.\n\nIf you would like to pass a tuple or list to a JAX function, you can do so by first explicitly converting it to an array:",
"_____no_output_____"
]
],
[
[
"jnp.sum(jnp.array(x))",
"_____no_output_____"
]
],
[
[
"## 🔪 Random Numbers",
"_____no_output_____"
],
[
"> _If all scientific papers whose results are in doubt because of bad \n> `rand()`s were to disappear from library shelves, there would be a \n> gap on each shelf about as big as your fist._ - Numerical Recipes",
"_____no_output_____"
],
[
"### RNGs and State\nYou're used to _stateful_ pseudorandom number generators (PRNGs) from numpy and other libraries, which helpfully hide a lot of details under the hood to give you a ready fountain of pseudorandomness:",
"_____no_output_____"
]
],
[
[
"print(np.random.random())\nprint(np.random.random())\nprint(np.random.random())",
"0.08960303423860538\n0.6720478073539145\n0.24536720985284477\n"
]
],
[
[
"Underneath the hood, numpy uses the [Mersenne Twister](https://en.wikipedia.org/wiki/Mersenne_Twister) PRNG to power its pseudorandom functions. The PRNG has a period of $2^{19937}-1$ and at any point can be described by __624 32bit unsigned ints__ and a __position__ indicating how much of this \"entropy\" has been used up.",
"_____no_output_____"
]
],
[
[
"np.random.seed(0)\nrng_state = np.random.get_state()\n#print(rng_state)\n# --> ('MT19937', array([0, 1, 1812433255, 1900727105, 1208447044,\n# 2481403966, 4042607538, 337614300, ... 614 more numbers..., \n# 3048484911, 1796872496], dtype=uint32), 624, 0, 0.0)",
"_____no_output_____"
]
],
[
[
"This pseudorandom state vector is automagically updated behind the scenes every time a random number is needed, \"consuming\" 2 of the uint32s in the Mersenne twister state vector:",
"_____no_output_____"
]
],
[
[
"_ = np.random.uniform()\nrng_state = np.random.get_state()\n#print(rng_state) \n# --> ('MT19937', array([2443250962, 1093594115, 1878467924,\n# ..., 2648828502, 1678096082], dtype=uint32), 2, 0, 0.0)\n\n# Let's exhaust the entropy in this PRNG statevector\nfor i in range(311):\n _ = np.random.uniform()\nrng_state = np.random.get_state()\n#print(rng_state) \n# --> ('MT19937', array([2443250962, 1093594115, 1878467924,\n# ..., 2648828502, 1678096082], dtype=uint32), 624, 0, 0.0)\n\n# Next call iterates the RNG state for a new batch of fake \"entropy\".\n_ = np.random.uniform()\nrng_state = np.random.get_state()\n# print(rng_state) \n# --> ('MT19937', array([1499117434, 2949980591, 2242547484, \n# 4162027047, 3277342478], dtype=uint32), 2, 0, 0.0)",
"_____no_output_____"
]
],
[
[
"The problem with magic PRNG state is that it's hard to reason about how it's being used and updated across different threads, processes, and devices, and it's _very easy_ to screw up when the details of entropy production and consumption are hidden from the end user.\n\nThe Mersenne Twister PRNG is also known to have a [number](https://cs.stackexchange.com/a/53475) of problems, it has a large 2.5Kb state size, which leads to problematic [initialization issues](https://dl.acm.org/citation.cfm?id=1276928). It [fails](http://www.pcg-random.org/pdf/toms-oneill-pcg-family-v1.02.pdf) modern BigCrush tests, and is generally slow.",
"_____no_output_____"
],
[
"### JAX PRNG",
"_____no_output_____"
],
[
"JAX instead implements an _explicit_ PRNG where entropy production and consumption are handled by explicitly passing and iterating PRNG state. JAX uses a modern [Threefry counter-based PRNG](https://github.com/google/jax/blob/master/design_notes/prng.md) that's __splittable__. That is, its design allows us to __fork__ the PRNG state into new PRNGs for use with parallel stochastic generation.\n\nThe random state is described by two unsigned-int32s that we call a __key__:",
"_____no_output_____"
]
],
[
[
"from jax import random\nkey = random.PRNGKey(0)\nkey",
"_____no_output_____"
]
],
[
[
"JAX's random functions produce pseudorandom numbers from the PRNG state, but __do not__ change the state! \n\nReusing the same state will cause __sadness__ and __monotony__, depriving the enduser of __lifegiving chaos__:",
"_____no_output_____"
]
],
[
[
"print(random.normal(key, shape=(1,)))\nprint(key)\n# No no no!\nprint(random.normal(key, shape=(1,)))\nprint(key)",
"[-0.20584226]\n[0 0]\n[-0.20584226]\n[0 0]\n"
]
],
[
[
"Instead, we __split__ the PRNG to get usable __subkeys__ every time we need a new pseudorandom number:",
"_____no_output_____"
]
],
[
[
"print(\"old key\", key)\nkey, subkey = random.split(key)\nnormal_pseudorandom = random.normal(subkey, shape=(1,))\nprint(\" \\---SPLIT --> new key \", key)\nprint(\" \\--> new subkey\", subkey, \"--> normal\", normal_pseudorandom)",
"old key [0 0]\n \\---SPLIT --> new key [4146024105 967050713]\n \\--> new subkey [2718843009 1272950319] --> normal [-1.2515389]\n"
]
],
[
[
"We propagate the __key__ and make new __subkeys__ whenever we need a new random number:",
"_____no_output_____"
]
],
[
[
"print(\"old key\", key)\nkey, subkey = random.split(key)\nnormal_pseudorandom = random.normal(subkey, shape=(1,))\nprint(\" \\---SPLIT --> new key \", key)\nprint(\" \\--> new subkey\", subkey, \"--> normal\", normal_pseudorandom)",
"old key [4146024105 967050713]\n \\---SPLIT --> new key [2384771982 3928867769]\n \\--> new subkey [1278412471 2182328957] --> normal [-0.58665055]\n"
]
],
[
[
"We can generate more than one __subkey__ at a time:",
"_____no_output_____"
]
],
[
[
"key, *subkeys = random.split(key, 4)\nfor subkey in subkeys:\n print(random.normal(subkey, shape=(1,)))",
"[-0.37533438]\n[0.98645043]\n[0.14553197]\n"
]
],
[
[
"## 🔪 Control Flow",
"_____no_output_____"
],
[
"### ✔ python control_flow + autodiff ✔\n\nIf you just want to apply `grad` to your python functions, you can use regular python control-flow constructs with no problems, as if you were using [Autograd](https://github.com/hips/autograd) (or Pytorch or TF Eager).",
"_____no_output_____"
]
],
[
[
"def f(x):\n if x < 3:\n return 3. * x ** 2\n else:\n return -4 * x\n\nprint(grad(f)(2.)) # ok!\nprint(grad(f)(4.)) # ok!",
"12.0\n-4.0\n"
]
],
[
[
"### python control flow + JIT\n\nUsing control flow with `jit` is more complicated, and by default it has more constraints.\n\nThis works:",
"_____no_output_____"
]
],
[
[
"@jit\ndef f(x):\n for i in range(3):\n x = 2 * x\n return x\n\nprint(f(3))",
"24\n"
]
],
[
[
"So does this:",
"_____no_output_____"
]
],
[
[
"@jit\ndef g(x):\n y = 0.\n for i in range(x.shape[0]):\n y = y + x[i]\n return y\n\nprint(g(jnp.array([1., 2., 3.])))",
"6.0\n"
]
],
[
[
"But this doesn't, at least by default:",
"_____no_output_____"
]
],
[
[
"@jit\ndef f(x):\n if x < 3:\n return 3. * x ** 2\n else:\n return -4 * x\n\n# This will fail!\ntry:\n f(2)\nexcept Exception as e:\n print(\"Exception {}\".format(e))",
"Exception Abstract value passed to `bool`, which requires a concrete value. The function to be transformed can't be traced at the required level of abstraction. If using `jit`, try using `static_argnums` or applying `jit` to smaller subfunctions instead.\n"
]
],
[
[
"__What gives!?__\n\nWhen we `jit`-compile a function, we usually want to compile a version of the function that works for many different argument values, so that we can cache and reuse the compiled code. That way we don't have to re-compile on each function evaluation.\n\nFor example, if we evaluate an `@jit` function on the array `jnp.array([1., 2., 3.], jnp.float32)`, we might want to compile code that we can reuse to evaluate the function on `jnp.array([4., 5., 6.], jnp.float32)` to save on compile time.\n\nTo get a view of your Python code that is valid for many different argument values, JAX traces it on _abstract values_ that represent sets of possible inputs. There are [multiple different levels of abstraction](https://github.com/google/jax/blob/master/jax/abstract_arrays.py), and different transformations use different abstraction levels.\n\nBy default, `jit` traces your code on the `ShapedArray` abstraction level, where each abstract value represents the set of all array values with a fixed shape and dtype. For example, if we trace using the abstract value `ShapedArray((3,), jnp.float32)`, we get a view of the function that can be reused for any concrete value in the corresponding set of arrays. That means we can save on compile time.\n\nBut there's a tradeoff here: if we trace a Python function on a `ShapedArray((), jnp.float32)` that isn't committed to a specific concrete value, when we hit a line like `if x < 3`, the expression `x < 3` evaluates to an abstract `ShapedArray((), jnp.bool_)` that represents the set `{True, False}`. When Python attempts to coerce that to a concrete `True` or `False`, we get an error: we don't know which branch to take, and can't continue tracing! The tradeoff is that with higher levels of abstraction we gain a more general view of the Python code (and thus save on re-compilations), but we require more constraints on the Python code to complete the trace.\n\nThe good news is that you can control this tradeoff yourself. By having `jit` trace on more refined abstract values, you can relax the traceability constraints. For example, using the `static_argnums` argument to `jit`, we can specify to trace on concrete values of some arguments. Here's that example function again:",
"_____no_output_____"
]
],
[
[
"def f(x):\n if x < 3:\n return 3. * x ** 2\n else:\n return -4 * x\n\nf = jit(f, static_argnums=(0,))\n\nprint(f(2.))",
"12.0\n"
]
],
[
[
"Here's another example, this time involving a loop:",
"_____no_output_____"
]
],
[
[
"def f(x, n):\n y = 0.\n for i in range(n):\n y = y + x[i]\n return y\n\nf = jit(f, static_argnums=(1,))\n\nf(jnp.array([2., 3., 4.]), 2)",
"_____no_output_____"
]
],
[
[
"In effect, the loop gets statically unrolled. JAX can also trace at _higher_ levels of abstraction, like `Unshaped`, but that's not currently the default for any transformation",
"_____no_output_____"
],
[
"️⚠️ **functions with argument-__value__ dependent shapes**\n\nThese control-flow issues also come up in a more subtle way: numerical functions we want to __jit__ can't specialize the shapes of internal arrays on argument _values_ (specializing on argument __shapes__ is ok). As a trivial example, let's make a function whose output happens to depend on the input variable `length`.",
"_____no_output_____"
]
],
[
[
"def example_fun(length, val):\n return jnp.ones((length,)) * val\n# un-jit'd works fine\nprint(example_fun(5, 4))\n\nbad_example_jit = jit(example_fun)\n# this will fail:\ntry:\n print(bad_example_jit(10, 4))\nexcept Exception as e:\n print(\"Exception {}\".format(e))\n# static_argnums tells JAX to recompile on changes at these argument positions:\ngood_example_jit = jit(example_fun, static_argnums=(0,))\n# first compile\nprint(good_example_jit(10, 4))\n# recompiles\nprint(good_example_jit(5, 4))",
"[4. 4. 4. 4. 4.]\nException Shapes must be 1D sequences of concrete values of integer type, got (Traced<ShapedArray(int32[], weak_type=True):JaxprTrace(level=-1/1)>,).\nIf using `jit`, try using `static_argnums` or applying `jit` to smaller subfunctions.\n[4. 4. 4. 4. 4. 4. 4. 4. 4. 4.]\n[4. 4. 4. 4. 4.]\n"
]
],
[
[
"`static_argnums` can be handy if `length` in our example rarely changes, but it would be disastrous if it changed a lot! \n\nLastly, if your function has global side-effects, JAX's tracer can cause weird things to happen. A common gotcha is trying to print arrays inside __jit__'d functions:",
"_____no_output_____"
]
],
[
[
"@jit\ndef f(x):\n print(x)\n y = 2 * x\n print(y)\n return y\nf(2)",
"Traced<ShapedArray(int32[], weak_type=True):JaxprTrace(level=-1/1)>\nTraced<ShapedArray(int32[]):JaxprTrace(level=-1/1)>\n"
]
],
[
[
"### Structured control flow primitives\n\nThere are more options for control flow in JAX. Say you want to avoid re-compilations but still want to use control flow that's traceable, and that avoids un-rolling large loops. Then you can use these 4 structured control flow primitives:\n\n - `lax.cond` _differentiable_\n - `lax.while_loop` __fwd-mode-differentiable__\n - `lax.fori_loop` __fwd-mode-differentiable__\n - `lax.scan` _differentiable_",
"_____no_output_____"
],
[
"#### cond\npython equivalent:\n\n```python\ndef cond(pred, true_fun, false_fun, operand):\n if pred:\n return true_fun(operand)\n else:\n return false_fun(operand)\n```",
"_____no_output_____"
]
],
[
[
"from jax import lax\n\noperand = jnp.array([0.])\nlax.cond(True, lambda x: x+1, lambda x: x-1, operand)\n# --> array([1.], dtype=float32)\nlax.cond(False, lambda x: x+1, lambda x: x-1, operand)\n# --> array([-1.], dtype=float32)",
"_____no_output_____"
]
],
[
[
"#### while_loop\n\npython equivalent:\n```\ndef while_loop(cond_fun, body_fun, init_val):\n val = init_val\n while cond_fun(val):\n val = body_fun(val)\n return val\n```",
"_____no_output_____"
]
],
[
[
"init_val = 0\ncond_fun = lambda x: x<10\nbody_fun = lambda x: x+1\nlax.while_loop(cond_fun, body_fun, init_val)\n# --> array(10, dtype=int32)",
"_____no_output_____"
]
],
[
[
"#### fori_loop\npython equivalent:\n```\ndef fori_loop(start, stop, body_fun, init_val):\n val = init_val\n for i in range(start, stop):\n val = body_fun(i, val)\n return val\n```",
"_____no_output_____"
]
],
[
[
"init_val = 0\nstart = 0\nstop = 10\nbody_fun = lambda i,x: x+i\nlax.fori_loop(start, stop, body_fun, init_val)\n# --> array(45, dtype=int32)",
"_____no_output_____"
]
],
[
[
"#### Summary\n\n$$\n\\begin{array} {r|rr} \n\\hline \\\n\\textrm{construct} \n& \\textrm{jit} \n& \\textrm{grad} \\\\\n\\hline \\\n\\textrm{if} & ❌ & ✔ \\\\\n\\textrm{for} & ✔* & ✔\\\\\n\\textrm{while} & ✔* & ✔\\\\\n\\textrm{lax.cond} & ✔ & ✔\\\\\n\\textrm{lax.while_loop} & ✔ & \\textrm{fwd}\\\\\n\\textrm{lax.fori_loop} & ✔ & \\textrm{fwd}\\\\\n\\textrm{lax.scan} & ✔ & ✔\\\\\n\\hline\n\\end{array}\n$$\n<center>$\\ast$ = argument-__value__-independent loop condition - unrolls the loop </center>",
"_____no_output_____"
],
[
"## 🔪 NaNs",
"_____no_output_____"
],
[
"### Debugging NaNs\n\nIf you want to trace where NaNs are occurring in your functions or gradients, you can turn on the NaN-checker by:\n\n* setting the `JAX_DEBUG_NANS=True` environment variable;\n\n* adding `from jax.config import config` and `config.update(\"jax_debug_nans\", True)` near the top of your main file;\n\n* adding `from jax.config import config` and `config.parse_flags_with_absl()` to your main file, then set the option using a command-line flag like `--jax_debug_nans=True`;\n\nThis will cause computations to error-out immediately on production of a NaN. Switching this option on adds a nan check to every floating point type value produced by XLA. That means values are pulled back to the host and checked as ndarrays for every primitive operation not under an `@jit`. For code under an `@jit`, the output of every `@jit` function is checked and if a nan is present it will re-run the function in de-optimized op-by-op mode, effectively removing one level of `@jit` at a time.\n\nThere could be tricky situations that arise, like nans that only occur under a `@jit` but don't get produced in de-optimized mode. In that case you'll see a warning message print out but your code will continue to execute.\n\nIf the nans are being produced in the backward pass of a gradient evaluation, when an exception is raised several frames up in the stack trace you will be in the backward_pass function, which is essentially a simple jaxpr interpreter that walks the sequence of primitive operations in reverse. In the example below, we started an ipython repl with the command line `env JAX_DEBUG_NANS=True ipython`, then ran this:",
"_____no_output_____"
],
[
"```\nIn [1]: import jax.numpy as jnp\n\nIn [2]: jnp.divide(0., 0.)\n---------------------------------------------------------------------------\nFloatingPointError Traceback (most recent call last)\n<ipython-input-2-f2e2c413b437> in <module>()\n----> 1 jnp.divide(0., 0.)\n\n.../jax/jax/numpy/lax_numpy.pyc in divide(x1, x2)\n 343 return floor_divide(x1, x2)\n 344 else:\n--> 345 return true_divide(x1, x2)\n 346\n 347\n\n.../jax/jax/numpy/lax_numpy.pyc in true_divide(x1, x2)\n 332 x1, x2 = _promote_shapes(x1, x2)\n 333 return lax.div(lax.convert_element_type(x1, result_dtype),\n--> 334 lax.convert_element_type(x2, result_dtype))\n 335\n 336\n\n.../jax/jax/lax.pyc in div(x, y)\n 244 def div(x, y):\n 245 r\"\"\"Elementwise division: :math:`x \\over y`.\"\"\"\n--> 246 return div_p.bind(x, y)\n 247\n 248 def rem(x, y):\n\n... stack trace ...\n\n.../jax/jax/interpreters/xla.pyc in handle_result(device_buffer)\n 103 py_val = device_buffer.to_py()\n 104 if np.any(np.isnan(py_val)):\n--> 105 raise FloatingPointError(\"invalid value\")\n 106 else:\n 107 return DeviceArray(device_buffer, *result_shape)\n\nFloatingPointError: invalid value\n```",
"_____no_output_____"
],
[
"The nan generated was caught. By running `%debug`, we can get a post-mortem debugger. This also works with functions under `@jit`, as the example below shows.",
"_____no_output_____"
],
[
"```\nIn [4]: from jax import jit\n\nIn [5]: @jit\n ...: def f(x, y):\n ...: a = x * y\n ...: b = (x + y) / (x - y)\n ...: c = a + 2\n ...: return a + b * c\n ...:\n\nIn [6]: x = jnp.array([2., 0.])\n\nIn [7]: y = jnp.array([3., 0.])\n\nIn [8]: f(x, y)\nInvalid value encountered in the output of a jit function. Calling the de-optimized version.\n---------------------------------------------------------------------------\nFloatingPointError Traceback (most recent call last)\n<ipython-input-8-811b7ddb3300> in <module>()\n----> 1 f(x, y)\n\n ... stack trace ...\n\n<ipython-input-5-619b39acbaac> in f(x, y)\n 2 def f(x, y):\n 3 a = x * y\n----> 4 b = (x + y) / (x - y)\n 5 c = a + 2\n 6 return a + b * c\n\n.../jax/jax/numpy/lax_numpy.pyc in divide(x1, x2)\n 343 return floor_divide(x1, x2)\n 344 else:\n--> 345 return true_divide(x1, x2)\n 346\n 347\n\n.../jax/jax/numpy/lax_numpy.pyc in true_divide(x1, x2)\n 332 x1, x2 = _promote_shapes(x1, x2)\n 333 return lax.div(lax.convert_element_type(x1, result_dtype),\n--> 334 lax.convert_element_type(x2, result_dtype))\n 335\n 336\n\n.../jax/jax/lax.pyc in div(x, y)\n 244 def div(x, y):\n 245 r\"\"\"Elementwise division: :math:`x \\over y`.\"\"\"\n--> 246 return div_p.bind(x, y)\n 247\n 248 def rem(x, y):\n\n ... stack trace ...\n```",
"_____no_output_____"
],
[
"When this code sees a nan in the output of an `@jit` function, it calls into the de-optimized code, so we still get a clear stack trace. And we can run a post-mortem debugger with `%debug` to inspect all the values to figure out the error.\n\n⚠️ You shouldn't have the NaN-checker on if you're not debugging, as it can introduce lots of device-host round-trips and performance regressions!",
"_____no_output_____"
],
[
"## Double (64bit) precision\n\nAt the moment, JAX by default enforces single-precision numbers to mitigate the Numpy API's tendency to aggressively promote operands to `double`. This is the desired behavior for many machine-learning applications, but it may catch you by surprise!",
"_____no_output_____"
]
],
[
[
"x = random.uniform(random.PRNGKey(0), (1000,), dtype=jnp.float64)\nx.dtype",
"_____no_output_____"
]
],
[
[
"To use double-precision numbers, you need to set the `jax_enable_x64` configuration variable __at startup__. \n\nThere are a few ways to do this:\n\n1. You can enable 64bit mode by setting the environment variable `JAX_ENABLE_X64=True`.\n\n2. You can manually set the `jax_enable_x64` configuration flag at startup:\n\n ```python\n # again, this only works on startup!\n from jax.config import config\n config.update(\"jax_enable_x64\", True)\n ```\n\n3. You can parse command-line flags with `absl.app.run(main)`\n\n ```python\n from jax.config import config\n config.config_with_absl()\n ```\n\n4. If you want JAX to run absl parsing for you, i.e. you don't want to do `absl.app.run(main)`, you can instead use\n\n ```python\n from jax.config import config\n if __name__ == '__main__':\n # calls config.config_with_absl() *and* runs absl parsing\n config.parse_flags_with_absl()\n ```\n\nNote that #2-#4 work for _any_ of JAX's configuration options.\n\nWe can then confirm that `x64` mode is enabled:",
"_____no_output_____"
]
],
[
[
"import jax.numpy as jnp\nfrom jax import random\nx = random.uniform(random.PRNGKey(0), (1000,), dtype=jnp.float64)\nx.dtype # --> dtype('float64')",
"_____no_output_____"
]
],
[
[
"### Caveats\n⚠️ XLA doesn't support 64-bit convolutions on all backends!",
"_____no_output_____"
],
[
"## Fin.\n\nIf something's not covered here that has caused you weeping and gnashing of teeth, please let us know and we'll extend these introductory _advisos_!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb4e11c3b68c922cf66e4eea6dca77add53aa15b | 22,327 | ipynb | Jupyter Notebook | identify_domain_training_data.ipynb | gkovacs/tmi-browsing-behavior-prediction | 0e7d44574003272ce457bf221bfa3e92002ad217 | [
"MIT"
] | 1 | 2019-12-16T11:44:18.000Z | 2019-12-16T11:44:18.000Z | identify_domain_training_data.ipynb | gkovacs/browsing-behavior-reconstuction-analysis | 0e7d44574003272ce457bf221bfa3e92002ad217 | [
"MIT"
] | null | null | null | identify_domain_training_data.ipynb | gkovacs/browsing-behavior-reconstuction-analysis | 0e7d44574003272ce457bf221bfa3e92002ad217 | [
"MIT"
] | null | null | null | 46.417879 | 548 | 0.656156 | [
[
[
"# noexport\n\nimport os\nos.system('export_notebook identify_domain_training_data.ipynb')",
"_____no_output_____"
],
[
"from tmilib import *\nimport csv",
"_____no_output_____"
],
[
"import sys\nnum_prev_enabled = int(sys.argv[1])\nnum_labels_enabled = 2 + num_prev_enabled\ndata_version = 4 + num_prev_enabled\nprint 'num_prev_enabled', num_prev_enabled\nprint 'data_version', data_version",
"_____no_output_____"
],
[
"twenty_letters = [\"a\",\"b\",\"c\",\"d\",\"e\",\"f\",\"g\",\"h\",\"i\",\"j\",\"k\",\"l\",\"m\",\"n\",\"o\",\"p\",\"q\",\"r\",\"s\",\"t\"]\n#domain_to_letter = {x:twenty_letters[i] for i,x in enumerate(top_domains)}\ndomain_id_to_letter = {domain_to_id(x):twenty_letters[i] for i,x in enumerate(top_n_domains_by_visits(20))}\n#print domain_id_to_letter\n#print domain_to_letter",
"_____no_output_____"
],
[
"productivity_letters = {-2: 'v', -1: 'w', 0: 'x', 1: 'y', 2: 'z'}\ndomain_id_to_productivity_letter = [productivity_letters[x] for x in get_domain_id_to_productivity()]\n#print domain_id_to_productivity[:10]\n#print domain_id_to_productivity_letter[:10]",
"_____no_output_____"
],
[
"def get_row_names():\n output_row_names = [\n 'label',\n 'spanlen',\n 'since_cur',\n 'cur_domain_letter',\n 'cur_domain_productivity',\n 'to_next',\n 'next_domain_letter',\n 'next_domain_productivity',\n 'n_eq_c',\n ]\n for idx_p_zeroidx in range(num_prev_enabled):\n sp = str(idx_p_zeroidx + 1)\n new_feature_names_for_p = [\n 'since_prev' + sp,\n 'prev' + sp +'_domain_letter',\n 'prev' + sp + '_domain_productivity',\n 'n_eq_p' + sp,\n ]\n output_row_names.extend(new_feature_names_for_p)\n return tuple(output_row_names)\n\nrow_names = get_row_names()\nprint row_names",
"('label', 'spanlen', 'since_cur', 'cur_domain_letter', 'cur_domain_productivity', 'to_next', 'next_domain_letter', 'next_domain_productivity', 'n_eq_c', 'since_prev1', 'prev1_domain_letter', 'prev1_productivity', 'n_eq_p1', 'since_prev2', 'prev2_domain_letter', 'prev2_productivity', 'n_eq_p2')\n"
],
[
"def get_rows_for_user(user):\n output = []\n #ordered_visits = get_history_ordered_visits_corrected_for_user(user)\n ordered_visits = get_history_ordered_visits_corrected_for_user(user)\n ordered_visits = exclude_bad_visits(ordered_visits)\n #active_domain_at_time = get_active_domain_at_time_for_user(user)\n active_seconds_set = set(get_active_insession_seconds_for_user(user))\n active_second_to_domain_id = {int(k):v for k,v in get_active_second_to_domain_id_for_user(user).viewitems()}\n prev_domain_ids = [-1]*8\n domain_id_to_most_recent_visit = {}\n total_items = 0\n skipped_items = 0\n for idx,visit in enumerate(ordered_visits):\n if idx+1 >= len(ordered_visits):\n break\n \n next_visit = ordered_visits[idx+1]\n cur_domain = url_to_domain(visit['url'])\n cur_domain_id = domain_to_id(cur_domain)\n next_domain = url_to_domain(next_visit['url'])\n next_domain_id = domain_to_id(next_domain)\n\n cur_time_sec = int(round(visit['visitTime'] / 1000.0))\n next_time_sec = int(round(next_visit['visitTime'] / 1000.0))\n \n domain_id_to_most_recent_visit[cur_domain_id] = cur_time_sec\n if prev_domain_ids[0] != cur_domain_id:\n #prev_domain_ids = ([cur_domain_id] + [x for x in prev_domain_ids if x != cur_domain_id])[:4]\n if cur_domain_id in prev_domain_ids:\n prev_domain_ids.remove(cur_domain_id)\n prev_domain_ids.insert(0, cur_domain_id)\n while len(prev_domain_ids) > 8:\n prev_domain_ids.pop()\n # prev_domain_ids includes the current one\n\n if cur_time_sec > next_time_sec:\n continue\n\n prev1_domain_id = prev_domain_ids[1]\n prev2_domain_id = prev_domain_ids[2]\n prev3_domain_id = prev_domain_ids[3]\n prev4_domain_id = prev_domain_ids[4]\n prev5_domain_id = prev_domain_ids[5]\n prev6_domain_id = prev_domain_ids[6]\n prev7_domain_id = prev_domain_ids[7]\n n_eq_c = 'T' if (next_domain_id == cur_domain_id) else 'F'\n n_eq_p1 = 'T' if (next_domain_id == prev1_domain_id) else 'F'\n n_eq_p2 = 'T' if (next_domain_id == prev2_domain_id) else 'F'\n n_eq_p3 = 'T' if (next_domain_id == prev3_domain_id) else 'F'\n n_eq_p4 = 'T' if (next_domain_id == prev4_domain_id) else 'F'\n n_eq_p5 = 'T' if (next_domain_id == prev5_domain_id) else 'F'\n n_eq_p6 = 'T' if (next_domain_id == prev6_domain_id) else 'F'\n n_eq_p7 = 'T' if (next_domain_id == prev7_domain_id) else 'F'\n \n for time_sec in xrange(cur_time_sec+1, next_time_sec):\n if time_sec not in active_seconds_set:\n continue\n ref_domain_id = active_second_to_domain_id[time_sec]\n total_items += 1\n label = None\n available_labels = (\n (cur_domain_id, 'c'),\n (next_domain_id, 'n'),\n (prev1_domain_id, 'p1'),\n (prev2_domain_id, 'p2'),\n (prev3_domain_id, 'p3'),\n (prev4_domain_id, 'p4'),\n (prev5_domain_id, 'p5'),\n (prev6_domain_id, 'p6'),\n (prev7_domain_id, 'p7'),\n )[:num_labels_enabled]\n # c p n p q r s t\n for label_value,label_name in available_labels:\n if ref_domain_id == label_value:\n label = label_name\n break\n if label == None:\n skipped_items += 1\n continue\n \n next_domain_letter = domain_id_to_letter.get(next_domain_id, 'u')\n cur_domain_letter = domain_id_to_letter.get(cur_domain_id, 'u')\n prev1_domain_letter = domain_id_to_letter.get(prev1_domain_id, 'u')\n prev2_domain_letter = domain_id_to_letter.get(prev2_domain_id, 'u')\n prev3_domain_letter = domain_id_to_letter.get(prev3_domain_id, 'u')\n prev4_domain_letter = domain_id_to_letter.get(prev4_domain_id, 'u')\n prev5_domain_letter = domain_id_to_letter.get(prev5_domain_id, 'u')\n prev6_domain_letter = domain_id_to_letter.get(prev6_domain_id, 'u')\n prev7_domain_letter = domain_id_to_letter.get(prev7_domain_id, 'u')\n \n next_domain_productivity = domain_id_to_productivity_letter[next_domain_id]\n cur_domain_productivity = domain_id_to_productivity_letter[cur_domain_id]\n prev1_domain_productivity = domain_id_to_productivity_letter[prev1_domain_id]\n prev2_domain_productivity = domain_id_to_productivity_letter[prev2_domain_id]\n prev3_domain_productivity = domain_id_to_productivity_letter[prev3_domain_id]\n prev4_domain_productivity = domain_id_to_productivity_letter[prev4_domain_id]\n prev5_domain_productivity = domain_id_to_productivity_letter[prev5_domain_id]\n prev6_domain_productivity = domain_id_to_productivity_letter[prev6_domain_id]\n prev7_domain_productivity = domain_id_to_productivity_letter[prev7_domain_id]\n \n since_cur = time_sec - cur_time_sec\n to_next = next_time_sec - time_sec\n spanlen = since_cur + to_next\n prev1_domain_last_visit = domain_id_to_most_recent_visit.get(prev1_domain_id, 0)\n prev2_domain_last_visit = domain_id_to_most_recent_visit.get(prev2_domain_id, 0)\n prev3_domain_last_visit = domain_id_to_most_recent_visit.get(prev3_domain_id, 0)\n prev3_domain_last_visit = domain_id_to_most_recent_visit.get(prev3_domain_id, 0)\n prev4_domain_last_visit = domain_id_to_most_recent_visit.get(prev4_domain_id, 0)\n prev5_domain_last_visit = domain_id_to_most_recent_visit.get(prev5_domain_id, 0)\n prev6_domain_last_visit = domain_id_to_most_recent_visit.get(prev6_domain_id, 0)\n prev7_domain_last_visit = domain_id_to_most_recent_visit.get(prev7_domain_id, 0)\n \n since_prev1 = time_sec - prev1_domain_last_visit\n since_prev2 = time_sec - prev2_domain_last_visit\n since_prev3 = time_sec - prev3_domain_last_visit\n since_prev4 = time_sec - prev4_domain_last_visit\n since_prev5 = time_sec - prev5_domain_last_visit\n since_prev6 = time_sec - prev6_domain_last_visit\n since_prev7 = time_sec - prev7_domain_last_visit\n \n since_cur = log(since_cur)\n to_next = log(to_next)\n spanlen = log(spanlen)\n since_prev1 = log(since_prev1)\n since_prev2 = log(since_prev2)\n since_prev3 = log(since_prev3)\n since_prev4 = log(since_prev4)\n since_prev5 = log(since_prev5)\n since_prev6 = log(since_prev6)\n since_prev7 = log(since_prev7)\n \n cached_locals = locals()\n output.append([cached_locals[row_name] for row_name in row_names])\n #print 'user', user, 'guaranteed error', float(skipped_items)/total_items, 'skipped', skipped_items, 'total', total_items\n return {\n 'rows': output,\n 'skipped_items': skipped_items,\n 'total_items': total_items,\n }\n",
"_____no_output_____"
],
[
"def create_domainclass_data_for_users(users, filename):\n if sdir_exists(filename):\n print 'already exists', filename\n return\n outfile = csv.writer(open(sdir_path(filename), 'w'))\n outfile.writerow(row_names)\n total_items = 0\n skipped_items = 0\n for user in users:\n data = get_rows_for_user(user)\n total_items += data['total_items']\n if total_items == 0:\n print user, 'no items'\n continue\n skipped_items += data['skipped_items']\n print user, 'skipped', float(data['skipped_items'])/data['total_items'], 'skipped', data['skipped_items'], 'total', data['total_items']\n outfile.writerows(data['rows'])\n print 'guaranteed error', float(skipped_items) / total_items, 'skipped', skipped_items, 'total', total_items\n\n",
"_____no_output_____"
],
[
"create_domainclass_data_for_users(get_training_users(), 'domainclass_cpn_train_v' + str(data_version) +'.csv')\ncreate_domainclass_data_for_users(get_test_users(), 'domainclass_cpn_test_v' + str(data_version) + '.csv')",
"g34wuNJgSm skipped 0.0511520305124 skipped 8181 total 159935\nqM6L9Z5Ss9 skipped 0.0544641624085 skipped 9465 total 173784\n8QkC8G5H2H skipped 0.0111096881603 skipped 347 total 31234\nqsSxPV131T skipped 0.0307532826538 skipped 4183 total 136018\n3aWG01QnPf skipped 0.0146279003596 skipped 1131 total 77318\nxtOJXxtOqe skipped 0.100018194856 skipped 9345 total 93433\napytwOyBle skipped 0.00880281690141 skipped 5 total 568\nfx8NZArV8y skipped 0.0126476809575 skipped 4903 total 387660\nsEFFqggDAp skipped 0.00886187385163 skipped 2426 total 273757\nq589WvazlK skipped 0.013971354998 skipped 1313 total 93978\nnYeXJLKoUm skipped 0.0123773463841 skipped 8179 total 660804\n89uAYhqXqL skipped 0.0117496151873 skipped 1603 total 136430\nb3NWn44W69 skipped 0.0287169806442 skipped 2221 total 77341\nV8VCDqL7Sm skipped 0.0711222133195 skipped 1509 total 21217\nQM7GFqLIYB skipped 0.0229500153017 skipped 9074 total 395381\nOQZ2XlXq2F skipped 0.10353935552 skipped 588 total 5679\nRe25RopoXx skipped 0.00181233601771 skipped 239 total 131874\nXqdAvyooV5 skipped 0.0457314055313 skipped 6930 total 151537\nNPrpJqWMkb skipped 0.025837916364 skipped 498 total 19274\nXHyA0IcHIX skipped 0.0195386360771 skipped 1240 total 63464\nfblHu0OZkZ skipped 0.00180940688657 skipped 535 total 295677\no30E6A3d3o skipped 0.0517527251543 skipped 9239 total 178522\nOc1iqDqhTN skipped 0.0325385055983 skipped 4147 total 127449\nsXuYR7xPL2 skipped 0.00313366276968 skipped 52 total 16594\npOYN6mJH0T skipped 0.0129254657635 skipped 2141 total 165642\nqVpbz10wnQ skipped 0.0120328747219 skipped 265 total 22023\nPwBfIrHd8Z skipped 0.0235296342882 skipped 4976 total 211478\nq9TQALCrVo skipped 0.0198114344238 skipped 6577 total 331980\n0sDxoGSuf6 skipped 0.0567935093132 skipped 6461 total 113763\nI7pqdKa4NJ skipped 0.00479097232285 skipped 2116 total 441664\nqYSuZxSVNC skipped 0.0931694332548 skipped 17046 total 182957\nfmFkk3ur1N skipped 0.00632200291785 skipped 377 total 59633\n8c3d0IF3zD skipped 0.00172370344237 skipped 171 total 99205\nE53DGPZ188 skipped 0.0303886149332 skipped 4702 total 154729\n8ngKlSW0Wv skipped 0.0397522956715 skipped 5052 total 127087\nqVSOPmVR59 skipped 0.00624332623168 skipped 1222 total 195729\nNHtxolRPTS skipped 0.0186803492379 skipped 4722 total 252779\n4546ZN0A5f skipped 0.027020152612 skipped 2762 total 102220\nrlWaaPKvSX skipped 0.00891347076839 skipped 780 total 87508\nLvC0lAqsGl skipped 0.0564422620484 skipped 7253 total 128503\nVYfLv7b8hg skipped 0.0174244172629 skipped 906 total 51996\ndwCCD1uofp skipped 0.00207739018241 skipped 351 total 168962\nehkM3inRFq skipped 0.027554304103 skipped 1370 total 49720\noHo0KJayR3 skipped 0.0344631615761 skipped 9460 total 274496\nTfAZTnnzST skipped 0.00162582043717 skipped 81 total 49821\nxDhU4lHDlP skipped 0.0100642580535 skipped 473 total 46998\nKS6zl4omuv skipped 0.0534786095978 skipped 10268 total 192002\naMoAdTi22n skipped 0.0552200447678 skipped 17318 total 313618\npEUnqUDtBT skipped 0.0370817003316 skipped 2337 total 63023\nTHH9Ar2ypm skipped 0.0162536959867 skipped 2820 total 173499\nNf91QP2i7x skipped 0.0287330184829 skipped 2464 total 85755\n4ZbDxUSz5z skipped 0.0161026685223 skipped 3448 total 214126\nb8DDWibd85 skipped 0.0693695842277 skipped 8726 total 125790\nPfcYy82N2O skipped 0.00354855461312 skipped 82 total 23108\nEU6AdYpVvP skipped 0.0786152046163 skipped 35545 total 452139\nO3Zx6P958u skipped 0.0192293380302 skipped 5426 total 282173\nyrH3HQNrho skipped 0.0546242520239 skipped 2483 total 45456\nhGCJGBh8Ah skipped 0.0128289908742 skipped 679 total 52927\nKTJOgTS6K4 skipped 0.00358822673714 skipped 148 total 41246\nMLancLTOqE skipped 0.0190618090129 skipped 6819 total 357731\nY0KTMpMY6x skipped 0.00498222772475 skipped 998 total 200312\npNiXtUQkEz skipped 0.0259660067816 skipped 4878 total 187861\n9NMeyGG4Dc skipped 0.0101700060716 skipped 134 total 13176\nDGhdRc6hcE skipped 0.00659352042199 skipped 1655 total 251004\n2bK78oKaW6 skipped 0.0457673559791 skipped 10807 total 236129\n6lkHBDiou2 skipped 0.0457910611461 skipped 1336 total 29176\nZqYweFWTDB skipped 0.00709458815153 skipped 1116 total 157303\nXyJ4fbLuxp skipped 0.0520835527471 skipped 7418 total 142425\n7fg6XHTfAp skipped 0.00526290107837 skipped 755 total 143457\nHfLuMki1JN skipped 0.0126601586333 skipped 249 total 19668\n4qWTb2hyaL skipped 0.0356548800773 skipped 3987 total 111822\nRHPAsraTxD skipped 0.0585658920313 skipped 8130 total 138818\nXasdQ0RMDo skipped 0.00340672960124 skipped 1105 total 324358\nLkAJvBy8fA skipped 0.016869232958 skipped 830 total 49202\nCpkJ8L4Xwg skipped 0.0118650137271 skipped 3479 total 293215\nXFH3VkbR3W skipped 0.00999211433139 skipped 1850 total 185146\ni1VbZ3etn4 skipped 0.0368324817153 skipped 8823 total 239544\nyvAg28ridg skipped 0.0153559901025 skipped 4729 total 307958\nQUgoBB7Rp4 skipped 0.0242856209333 skipped 10406 total 428484\nHUduBl984Q skipped 0.0133478997717 skipped 1906 total 142794\nOtHgO83x97 skipped 0.0474692021808 skipped 12581 total 265035\nVXKLcUCkFv skipped 0.00517640648413 skipped 836 total 161502\n7JOMMv7Kqf skipped 0.0110955673098 skipped 3006 total 270919\nXepiGGFf3W skipped 0.0750511655768 skipped 15035 total 200330\nhDDqPeyL7R skipped 0.0674321999679 skipped 12191 total 180789\n2JilVuENVz skipped 0.00107981478431 skipped 118 total 109278\nkhps2gAfwV skipped 0.0682947914415 skipped 6320 total 92540\ny9QE4DHHJN skipped 0.0209022523153 skipped 2837 total 135727\nnRansgMcBn skipped 0.00860549932385 skipped 210 total 24403\nGXaxkQQNgE skipped 0.0504289302811 skipped 3192 total 63297\nkT7S3wmYZO skipped 0.00113018949202 skipped 244 total 215893\ncLsK8oQYmZ skipped 0.0273514498172 skipped 2012 total 73561\n00LnQeq1SQ skipped 0.178916377324 skipped 28805 total 160997\nguaranteed error 0.0284647950423 skipped 427158 total 15006537\nUrZvGUsI8N skipped 0.0185366595969 skipped 1949 total 105143\n3a3FX1s9S6 skipped 0.0237283459136 skipped 1204 total 50741\nTfAZTnnzST skipped 0.00162582043717 skipped 81 total 49821\nN3buswFgSO skipped 0.008070196623 skipped 401 total 49689\nsEFFqggDAp skipped 0.00886187385163 skipped 2426 total 273757\nfx8NZArV8y skipped 0.0126476809575 skipped 4903 total 387660\nsXuYR7xPL2 skipped 0.00313366276968 skipped 52 total 16594\nb3NWn44W69 skipped 0.0287169806442 skipped 2221 total 77341\nIAm4ofgNKb skipped 0.00257761636919 skipped 291 total 112895\n8ngKlSW0Wv skipped 0.0397522956715 skipped 5052 total 127087\nMLancLTOqE skipped 0.0190618090129 skipped 6819 total 357731\nr6qiC7PoI1 skipped 0.0139485028114 skipped 3019 total 216439\nnYeXJLKoUm skipped 0.0123773463841 skipped 8179 total 660804\ndwBGfodTyh skipped 0.0172710642419 skipped 3210 total 185860\n2e7ex5t0MT skipped 0.0787736630538 skipped 9034 total 114683\nMUHeUeRBTJ skipped 0.0251972493127 skipped 6764 total 268442\nkhps2gAfwV skipped 0.0682947914415 skipped 6320 total 92540\nqgV6lFlkSa skipped 0.103702013996 skipped 14093 total 135899\nZiXJx6z1Rl skipped 0.0334348584922 skipped 977 total 29221\n60qS4pDWkC skipped 0.022043866186 skipped 2189 total 99302\nS6XIUa9DJ8 skipped 0.0329564378999 skipped 7144 total 216771\nZXWhHg9RZG skipped 0.00494588644315 skipped 1121 total 226653\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4e19855f295b8e117201e48db4334e198ae349 | 164,614 | ipynb | Jupyter Notebook | source_code/Titanic-Data-Visualization.ipynb | akhilayaragoppa/akhilayaragoppa | d60d09316abaa5b2f8538176d9830af394e80d25 | [
"MIT"
] | null | null | null | source_code/Titanic-Data-Visualization.ipynb | akhilayaragoppa/akhilayaragoppa | d60d09316abaa5b2f8538176d9830af394e80d25 | [
"MIT"
] | 5 | 2020-07-31T19:57:42.000Z | 2022-03-25T19:37:47.000Z | source_code/Titanic-Data-Visualization.ipynb | akhilayaragoppa/akhilayaragoppa | d60d09316abaa5b2f8538176d9830af394e80d25 | [
"MIT"
] | null | null | null | 99.705633 | 14,686 | 0.795607 | [
[
[
"from matplotlib import pyplot as plt\nimport pandas as pd",
"_____no_output_____"
],
[
"titanic_df = pd.read_csv('train.csv')",
"_____no_output_____"
],
[
"titanic_df.head()",
"_____no_output_____"
],
[
"titanic_df.columns",
"_____no_output_____"
],
[
"male_df = titanic_df[titanic_df['Sex'] == 'male']",
"_____no_output_____"
],
[
"female_df = titanic_df[titanic_df['Sex'] == 'female']",
"_____no_output_____"
],
[
"male_df.head()",
"_____no_output_____"
],
[
"female_df.head()",
"_____no_output_____"
],
[
"plt.subplot(1,2,1)\nplt.bar([0,1],[sum(male_df['Survived'] == 0), sum(male_df['Survived'] == 1)])\nplt.ylim(ymax = 500)\nplt.xticks([0,1],['Not-survived','Survived'])\nplt.legend(['Male'])\nplt.subplot(1,2,2)\nplt.bar([0,1],[sum(female_df['Survived'] == 0), sum(female_df['Survived'] == 1)],color = 'orange')\nplt.ylim(ymax = 500)\nplt.xticks([0,1],['Not-survived','Survived'])\nplt.legend(['Female'])\nplt.show()",
"_____no_output_____"
],
[
"print(len(male_df))\nprint(len(female_df))",
"577\n314\n"
],
[
"survivor_df = titanic_df[titanic_df['Survived'] == 1]\nnon_survivor_df = titanic_df[titanic_df['Survived'] == 0]",
"_____no_output_____"
],
[
"plt.subplot(1,2,1)\nplt.bar([0,1],[sum(survivor_df['Sex'] == 'male'), sum(survivor_df['Sex'] == 'female')])\nplt.ylim(ymax = 500)\nplt.xticks([0,1],['Male','Female'])\nplt.legend(['Survived'])\nplt.subplot(1,2,2)\nplt.bar([0,1],[sum(non_survivor_df['Sex'] == 'male'), sum(non_survivor_df['Sex'] == 'female')],color = 'orange')\nplt.ylim(ymax = 500)\nplt.xticks([0,1],['Male','Female'])\nplt.legend(['Not-survived'])\nplt.show()",
"_____no_output_____"
],
[
"plt.subplot(1,2,1)\nplt.bar([0,1,2],[sum(survivor_df['Pclass'] == 1), sum(survivor_df['Pclass'] == 2), sum(survivor_df['Pclass'] == 3)])\nplt.ylim(ymax = 400)\nplt.xticks([0,1,2],['Class 1','Class 2', 'Class 3'])\nplt.legend(['Survived'])\nplt.subplot(1,2,2)\nplt.bar([0,1,2],[sum(non_survivor_df['Pclass'] == 1), sum(non_survivor_df['Pclass'] == 2), sum(non_survivor_df['Pclass'] == 3)], color = 'orange')\nplt.ylim(ymax = 400)\nplt.xticks([0,1,2],['Class 1','Class 2', 'Class 3'])\nplt.legend(['Not-survived'])\nplt.show()",
"_____no_output_____"
],
[
"plt.subplot(2,1,1)\nplt.hist(survivor_df[survivor_df.Age > 1].Age,40,edgecolor = 'black')\nplt.xlabel('Age')\nplt.ylim(ymax = 50)\nplt.xlim(xmax = 85)\nplt.legend(['Survivors'])\nplt.subplot(2,1,2)\nplt.hist(non_survivor_df[non_survivor_df.Age > 1].Age,40,edgecolor = 'black', color = 'orange')\nplt.xlabel('Age')\nplt.ylim(ymax = 50)\nplt.xlim(xmax = 85)\nplt.legend(['Non-Survivors'])\nplt.show()",
"_____no_output_____"
],
[
"plt.subplot(2,1,1)\nplt.hist([len(name) for name in survivor_df['Name']], 20,edgecolor='black')\nplt.ylim(ymax = 85)\nplt.xlim(xmax = 85)\nplt.legend(['Survivors'])\nplt.subplot(2,1,2)\nplt.hist([len(name) for name in non_survivor_df['Name']], 20,edgecolor='black',color = 'orange')\nplt.ylim(ymax = 85)\nplt.xlim(xmax = 85)\nplt.xlabel('Name Length')\nplt.legend(['Non-survivors'])\nplt.show()",
"_____no_output_____"
],
[
"plt.subplot(2,1,1)\nplt.hist([len(name) for name in female_df['Name']], 20,edgecolor='black')\nplt.ylim(ymax = 90)\nplt.xlim(xmax = 85)\nplt.legend(['Female'])\nplt.subplot(2,1,2)\nplt.hist([len(name) for name in male_df['Name']], 20,edgecolor='black',color = 'orange')\nplt.ylim(ymax = 90)\nplt.xlim(xmax = 85)\nplt.xlabel('Name Length')\nplt.legend(['Male'])\nplt.show()",
"_____no_output_____"
],
[
"# Embarked\nset(titanic_df.Embarked)",
"_____no_output_____"
],
[
"plt.subplot(1,2,1)\nplt.bar([0,1,2],[sum(survivor_df['Embarked'] == 'Q'), sum(survivor_df['Embarked'] == 'C'), sum(survivor_df['Embarked'] == 'S')])\nplt.ylim(ymax = 450)\nplt.xticks([0,1,2],['Queenstown','Cherbourg', 'Southampton'], rotation = 'vertical')\nplt.legend(['Survived'])\nplt.subplot(1,2,2)\nplt.bar([0,1,2],[sum(non_survivor_df['Embarked'] == 'Q'), sum(non_survivor_df['Embarked'] == 'C'), sum(non_survivor_df['Embarked'] == 'S')], color = 'orange')\nplt.ylim(ymax = 450)\nplt.xticks([0,1,2],['Queenstown','Cherbourg', 'Southampton'], rotation='vertical')\nplt.legend(['Not-survived'])\nplt.show()",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"x = np.arange(3)\nfig, ax1 = plt.subplots()\n\nwidth = 0.3\nplt.xticks(x + width/2, ['Queenstown','Cherbourg', 'Southampton'])\nsurvivors = ax1.bar(x, [sum(survivor_df['Embarked'] == 'Q'), sum(survivor_df['Embarked'] == 'C'), sum(survivor_df['Embarked'] == 'S')], width)\nplt.ylabel('No. of Survivors')\nplt.ylim(ymax = 450)\n\nax2 = ax1.twinx()\nnon_survivors = ax2.bar(x + width, [sum(non_survivor_df['Embarked'] == 'Q'), sum(non_survivor_df['Embarked'] == 'C'), sum(non_survivor_df['Embarked'] == 'S')], width, color='orange')\nplt.ylabel('No. of non-survivors')\nplt.ylim(ymax = 450)\n\nplt.legend([survivors, non_survivors],['Survivors','Non-survivors'])\nfigure = plt.gcf()\nplt.show()",
"_____no_output_____"
],
[
"x = np.arange(2)\nfig, ax1 = plt.subplots()\n\nwidth = 0.2\nplt.xticks(x + width/2, ['Male','Female'])\nsurvivors = ax1.bar(x, [sum(survivor_df['Sex'] == 'male'), sum(survivor_df['Sex'] == 'female')], width)\nplt.ylabel('No. of People')\nplt.ylim(ymax = 480)\n\nax2 = ax1.twinx()\nnon_survivors = ax2.bar(x + width, [sum(non_survivor_df['Sex'] == 'male'), sum(non_survivor_df['Sex'] == 'female')], width, color='orange')\nplt.ylim(ymax = 480)\n\nplt.legend([survivors, non_survivors],['Survivors','Non-survivors'],loc=9)\nfigure = plt.gcf()\nplt.show()",
"_____no_output_____"
],
[
"sum(survivor_df['Age'] > 65)",
"_____no_output_____"
],
[
"titanic_df.describe()",
"_____no_output_____"
],
[
"survivor_df.describe()",
"_____no_output_____"
],
[
"non_survivor_df.describe()",
"_____no_output_____"
],
[
"sum(survivor_df.SibSp > 1)",
"_____no_output_____"
],
[
"sum(non_survivor_df.SibSp > 1)",
"_____no_output_____"
],
[
"sum(survivor_df.SibSp == 0)",
"_____no_output_____"
],
[
"sum(non_survivor_df.SibSp == 0)",
"_____no_output_____"
],
[
"sum(survivor_df.SibSp == 1)",
"_____no_output_____"
],
[
"sum(non_survivor_df.SibSp == 1)",
"_____no_output_____"
],
[
"sum(titanic_df.Age > 65)",
"_____no_output_____"
],
[
"np.arange(3)",
"_____no_output_____"
],
[
"class1 = titanic_df.loc[titanic_df.Pclass == 1]",
"_____no_output_____"
],
[
"plt.bar([1,2,3], [sum(class1.Embarked == 'Q'), sum(class1.Embarked == 'C'), sum(class1.Embarked == 'S')])\nplt.xticks([1,2,3],['Queenstown','Cherbourg','Southampton'])\n\nplt.show()",
"_____no_output_____"
],
[
"class2 = titanic_df.loc[titanic_df.Pclass == 2]",
"_____no_output_____"
],
[
"plt.bar([1,2,3], [sum(class2.Embarked == 'Q'), sum(class2.Embarked == 'C'), sum(class2.Embarked == 'S')])\nplt.xticks([1,2,3],['Queenstown','Cherbourg','Southampton'])\n\nplt.show()",
"_____no_output_____"
],
[
"plt.subplot(2,1,1)\nplt.hist(survivor_df.Fare, 35,edgecolor='black')\nplt.ylim(ymax = 340)\nplt.xlim(xmax = 300)\nplt.legend(['Survivors'])\nplt.subplot(2,1,2)\nplt.hist(non_survivor_df.Fare, 20,edgecolor='black',color = 'orange')\nplt.ylim(ymax = 340)\nplt.xlim(xmax = 300)\nplt.xlabel('Ticket Fare')\nplt.legend(['Non-survivors'])\nplt.show()",
"_____no_output_____"
],
[
"sum(titanic_df.Fare > 300)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4e1b7e048a8f874c5fdc69f982435845247363 | 4,478 | ipynb | Jupyter Notebook | Python Fundamentals/Module_3_Required_Code_Python_Fundamentals.ipynb | gracieforthman/pythonteachingcode | 34fdbcd58209566fd40f9fdabdcd00d18625bbce | [
"MIT"
] | 1 | 2021-01-19T17:16:57.000Z | 2021-01-19T17:16:57.000Z | Python Fundamentals/Module_3_Required_Code_Python_Fundamentals.ipynb | gracieforthman/pythonteachingcode | 34fdbcd58209566fd40f9fdabdcd00d18625bbce | [
"MIT"
] | null | null | null | Python Fundamentals/Module_3_Required_Code_Python_Fundamentals.ipynb | gracieforthman/pythonteachingcode | 34fdbcd58209566fd40f9fdabdcd00d18625bbce | [
"MIT"
] | 1 | 2021-01-19T17:20:40.000Z | 2021-01-19T17:20:40.000Z | 38.603448 | 329 | 0.587092 | [
[
[
"\n# Module 3 Required Coding Activity \nIntroduction to Python (Unit 2) Fundamentals \n\nAll course .ipynb Jupyter Notebooks are available from the project files download topic in Module 1, Section 1.\n\nThis is an activity from the Jupyter Notebook **`Practice_MOD03_IntroPy.ipynb`** which you may have already completed.\n\n| Assignment Requirements | \n|:-------------------------------| \n| **NOTE:** This program requires **`print`** output and using code syntax used in module 3: **`if`**, **`input`**, **`def`**, **`return`**, **`for`**/**`in`** keywords, **`.lower()`** and **`.upper()`** method, **`.append`**, **`.pop`**, **`.split`** methods, **`range`** and **`len`** functions | \n\n## Program: poem mixer \nThis program takes string input and then prints out a mixed order version of the string \n\n\n**Program Parts** \n- **program flow** gathers the word list, modifies the case and order, and prints \n - get string input, input like a poem, verse or saying \n - split the string into a list of individual words \n - determine the length of the list\n - Loop the length of the list by index number and for each list index: \n - if a word is short (3 letters or less) make the word in the list lowercase \n - if a word is long (7 letters or more) make the word in the list uppercase \n - **call the word_mixer** function with the modified list \n - print the return value from the word_mixer function \n\n- **word_mixer** Function has 1 argument: an original list of string words, containing greater than 5 words and the function returns a new list. \n - sort the original list \n - create a new list \n - Loop while the list is longer than 5 words: \n - *in each loop pop a word from the sorted original list and append to the new list* \n - pop the word 5th from the end of the list and append to the new list \n - pop the first word in the list and append to the new list \n - pop the last word in the list and append to the new list \n - **return** the new list on exiting the loop\n\n\n\n\n\n\n **input example** *(beginning of William Blake poem, \"The Fly\")*\n\n >enter a saying or poem: `Little fly, Thy summer’s play My thoughtless hand Has brushed away. Am not I A fly like thee? Or art not thou A man like me?` \n\n\n**output example** \n>`or BRUSHED thy not Little thou me? SUMMER’S thee? like THOUGHTLESS play i a not hand a my fly am man`\n\n\n**alternative output** in each loop in the function that creates the new list add a \"\\\\n\" to the list \n```\n or BRUSHED thy \n not Little thou \n me? SUMMER’S thee? \n like THOUGHTLESS play \n i a not \n hand a my \n fly am man\n```\n\n",
"_____no_output_____"
]
],
[
[
"# [] create poem mixer\n# [] copy and paste in edX assignment page\n\n\n\n",
"_____no_output_____"
]
],
[
[
"Submit this by creating a python file (.py) and submitting it in D2L. Be sure to test that it works. \n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb4e225c08b47959bbe6f5fdc01e1b273a68ac28 | 51,768 | ipynb | Jupyter Notebook | study_roadmaps/1_getting_started_roadmap/2_elemental_features_of_monk/5) Feature - Switch modes without reloading experiment - train, eval, infer.ipynb | shubham7169/monk_v1 | 2d63ba9665160cc7758ba0541baddf87c1cfa578 | [
"Apache-2.0"
] | 7 | 2020-07-26T08:37:29.000Z | 2020-10-30T10:23:11.000Z | study_roadmaps/1_getting_started_roadmap/2_elemental_features_of_monk/5) Feature - Switch modes without reloading experiment - train, eval, infer.ipynb | aayush-fadia/monk_v1 | 4234eecede3427efc952461408e2d14ef5fa0e57 | [
"Apache-2.0"
] | null | null | null | study_roadmaps/1_getting_started_roadmap/2_elemental_features_of_monk/5) Feature - Switch modes without reloading experiment - train, eval, infer.ipynb | aayush-fadia/monk_v1 | 4234eecede3427efc952461408e2d14ef5fa0e57 | [
"Apache-2.0"
] | null | null | null | 24.804983 | 410 | 0.500019 | [
[
[
"<a href=\"https://colab.research.google.com/github/Tessellate-Imaging/monk_v1/blob/master/study_roadmaps/1_getting_started_roadmap/2_elemental_features_of_monk/5)%20Feature%20-%20Switch%20modes%20without%20reloading%20experiment%20-%20train%2C%20eval%2C%20infer.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Goals\n\n### 1. Understand how continuously and manually switching between training and externally validating help improve training\n\n### 2. Experiment and learning how switching between train and val modes can help choose right hyper-parameters\n\n\n### 2. Steps\n - You will use mxnet gluon backend for this example\n - You will first train a classifier using default params\n \n - You will switch mode from train to val for the first time here\n - You will then validate to check accuracy\n \n - You will switch mode from val to train here\n - You will reduce the learning rate (Need notfocus on how it is done for now)\n - You will retrain again using this new lr\n \n - You will switch mode from train to val for the second time here\n - You will then validate to check accuracy\n \n - You will again switch mode from val to train here\n - You will further change the learning rate (Need notfocus on how it is done for now)\n - You will retrain again using this newest lr\n \n - You will switch mode from train to val for the final time here\n - You will then validate to check accuracy",
"_____no_output_____"
],
[
"# Table of Contents\n\n\n## [0. Install](#0)\n\n\n## [1. Train a classifier using default settings](#1)\n\n\n## [2. Switch mode from train to eval and validate](#2)\n\n\n## [3. Switch back mode, reduce lr, retrain](#3)\n\n\n## [4. Switch mode from train to eval and re-validate](#4)\n\n\n## [5. Switch back mode, change lr further, retrain](#5)\n\n\n## [6. Switch mode from train to eval and re-validate](#6)",
"_____no_output_____"
],
[
"<a id='0'></a>\n# Install Monk\n \n - git clone https://github.com/Tessellate-Imaging/monk_v1.git\n \n - cd monk_v1/installation/Linux && pip install -r requirements_cu9.txt\n - (Select the requirements file as per OS and CUDA version)",
"_____no_output_____"
]
],
[
[
"!git clone https://github.com/Tessellate-Imaging/monk_v1.git",
"_____no_output_____"
],
[
"# If using Colab install using the commands below\n!cd monk_v1/installation/Misc && pip install -r requirements_colab.txt\n\n# If using Kaggle uncomment the following command\n#!cd monk_v1/installation/Misc && pip install -r requirements_kaggle.txt\n\n# Select the requirements file as per OS and CUDA version when using a local system or cloud\n#!cd monk_v1/installation/Linux && pip install -r requirements_cu9.txt",
"_____no_output_____"
]
],
[
[
"## Dataset - Malarial cell images\n - Credits: https://www.kaggle.com/iarunava/cell-images-for-detecting-malaria",
"_____no_output_____"
]
],
[
[
"! wget --load-cookies /tmp/cookies.txt \"https://docs.google.com/uc?export=download&confirm=$(wget --save-cookies /tmp/cookies.txt --keep-session-cookies --no-check-certificate 'https://docs.google.com/uc?export=download&id=1mMEtGIK8UZNCrErXRJR-kutNTaN1zxjC' -O- | sed -rn 's/.*confirm=([0-9A-Za-z_]+).*/\\1\\n/p')&id=1mMEtGIK8UZNCrErXRJR-kutNTaN1zxjC\" -O malaria_cell.zip && rm -rf /tmp/cookies.txt",
"_____no_output_____"
],
[
"! unzip -qq malaria_cell.zip",
"_____no_output_____"
],
[
"! wget --load-cookies /tmp/cookies.txt \"https://docs.google.com/uc?export=download&confirm=$(wget --save-cookies /tmp/cookies.txt --keep-session-cookies --no-check-certificate 'https://docs.google.com/uc?export=download&id=1WHpd7M-E_EiXmdjOr48BfvlUtMRPV6PM' -O- | sed -rn 's/.*confirm=([0-9A-Za-z_]+).*/\\1\\n/p')&id=1WHpd7M-E_EiXmdjOr48BfvlUtMRPV6PM\" -O malaria_cell_val.zip && rm -rf /tmp/cookies.txt",
"_____no_output_____"
],
[
"! unzip -qq malaria_cell_val.zip",
"_____no_output_____"
]
],
[
[
"# Imports \n\n - Using single mxnet-gluoncv backend for this tutorial",
"_____no_output_____"
]
],
[
[
"# Monk\nimport os\nimport sys\nsys.path.append(\"monk_v1/monk/\");",
"_____no_output_____"
],
[
"#Using mxnet-gluon backend \nfrom gluon_prototype import prototype",
"_____no_output_____"
]
],
[
[
"<a id='1'></a>\n# Train a classifier using default settings",
"_____no_output_____"
],
[
"### Creating and managing experiments\n - Provide project name\n - Provide experiment name",
"_____no_output_____"
]
],
[
[
"gtf = prototype(verbose=1);\ngtf.Prototype(\"Malaria-Cell\", \"exp-switch-modes\");",
"Mxnet Version: 1.5.0\n\nExperiment Details\n Project: Malaria-Cell\n Experiment: exp-switch-modes\n Dir: /home/abhi/Desktop/Work/tess_tool/gui/v0.3/finetune_models/Organization/development/v5.0_blocks/study_roadmap/change_post_num_layers/2_elemental_features_of_monk/workspace/Malaria-Cell/exp-switch-modes/\n\n"
]
],
[
[
"### This creates files and directories as per the following structure\n \n \n workspace\n |\n |--------Malaria-Cell\n |\n |\n |-----exp-switch-modes\n |\n |-----experiment-state.json\n |\n |-----output\n |\n |------logs (All training logs and graphs saved here)\n |\n |------models (all trained models saved here)",
"_____no_output_____"
],
[
"### Load Dataset",
"_____no_output_____"
]
],
[
[
"gtf.Default(dataset_path=\"malaria_cell\", \n model_name=\"resnet18_v1\", \n num_epochs=5);\n\n#Read the summary generated once you run this cell. ",
"Dataset Details\n Train path: malaria_cell\n Val path: None\n CSV train path: None\n CSV val path: None\n\nDataset Params\n Input Size: 224\n Batch Size: 4\n Data Shuffle: True\n Processors: 4\n Train-val split: 0.7\n\nPre-Composed Train Transforms\n[{'RandomHorizontalFlip': {'p': 0.8}}, {'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]\n\nPre-Composed Val Transforms\n[{'RandomHorizontalFlip': {'p': 0.8}}, {'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]\n\nDataset Numbers\n Num train images: 1411\n Num val images: 605\n Num classes: 2\n\nModel Params\n Model name: resnet18_v1\n Use Gpu: True\n Use pretrained: True\n Freeze base network: True\n\nModel Details\n Loading pretrained model\n Model Loaded on device\n Model name: resnet18_v1\n Num of potentially trainable layers: 41\n Num of actual trainable layers: 1\n\nOptimizer\n Name: sgd\n Learning rate: 0.01\n Params: {'lr': 0.01, 'momentum': 0, 'weight_decay': 0, 'momentum_dampening_rate': 0, 'clipnorm': 0.0, 'clipvalue': 0.0}\n\n\n\nLearning rate scheduler\n Name: steplr\n Params: {'step_size': 1, 'gamma': 0.98, 'last_epoch': -1}\n\nLoss\n Name: softmaxcrossentropy\n Params: {'weight': None, 'batch_axis': 0, 'axis_to_sum_over': -1, 'label_as_categories': True, 'label_smoothing': False}\n\nTraining params\n Num Epochs: 5\n\nDisplay params\n Display progress: True\n Display progress realtime: True\n Save Training logs: True\n Save Intermediate models: True\n Intermediate model prefix: intermediate_model_\n\n"
]
],
[
[
"### From summary current Learning rate: 0.01",
"_____no_output_____"
]
],
[
[
"#Start Training\ngtf.Train();\n\n#Read the training summary generated once you run the cell and training is completed",
"Training Start\n Epoch 1/5\n ----------\n"
]
],
[
[
"<a id='2'></a>\n# Switch mode from train to eval and validate",
"_____no_output_____"
]
],
[
[
"gtf.Switch_Mode(eval_infer=True)",
"_____no_output_____"
]
],
[
[
"### Load the validation dataset",
"_____no_output_____"
]
],
[
[
"gtf.Dataset_Params(dataset_path=\"malaria_cell_val\");\ngtf.Dataset();",
"Dataset Details\n Test path: malaria_cell_val\n CSV test path: None\n\nDataset Params\n Input Size: 224\n Processors: 4\n\nPre-Composed Test Transforms\n[{'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]\n\nDataset Numbers\n Num test images: 1011\n Num classes: 2\n\n"
]
],
[
[
"### Run validation",
"_____no_output_____"
]
],
[
[
"accuracy, class_based_accuracy = gtf.Evaluate();",
"Testing\n"
]
],
[
[
"### Accuracy now is - 65.08% when learning rate is 0.01\n(Can change when you run the exp)",
"_____no_output_____"
],
[
"<a id='3'></a>\n# Switch back mode, reduce lr, retrain",
"_____no_output_____"
]
],
[
[
"gtf.Switch_Mode(train=True)",
"_____no_output_____"
]
],
[
[
"## Reduce learning rate from 0.01 to 0.001",
"_____no_output_____"
]
],
[
[
"# This part of code will be taken up again in upcoming sections\ngtf.update_learning_rate(0.001);\ngtf.Reload();",
"Update: Learning Rate - 0.001\n\nPre-Composed Train Transforms\n[{'RandomHorizontalFlip': {'p': 0.8}}, {'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]\n\nPre-Composed Val Transforms\n[{'RandomHorizontalFlip': {'p': 0.8}}, {'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]\n\nDataset Numbers\n Num train images: 1411\n Num val images: 605\n Num classes: 2\n\nModel Details\n Loading pretrained model\n Model Loaded on device\n Model name: resnet18_v1\n Num of potentially trainable layers: 61\n Num of actual trainable layers: 1\n\n"
],
[
"#Start Training\ngtf.Train();\n\n#Read the training summary generated once you run the cell and training is completed",
"Training Start\n Epoch 1/5\n ----------\n"
]
],
[
[
"<a id='4'></a>\n# Switch mode from train to eval and re-validate",
"_____no_output_____"
]
],
[
[
"gtf.Switch_Mode(eval_infer=True)",
"_____no_output_____"
]
],
[
[
"### Load the validation dataset",
"_____no_output_____"
]
],
[
[
"gtf.Dataset_Params(dataset_path=\"malaria_cell_val\");\ngtf.Dataset();",
"Dataset Details\n Test path: malaria_cell_val\n CSV test path: None\n\nDataset Params\n Input Size: 224\n Processors: 4\n\nPre-Composed Test Transforms\n[{'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]\n\nDataset Numbers\n Num test images: 1011\n Num classes: 2\n\n"
]
],
[
[
"### Run validation",
"_____no_output_____"
]
],
[
[
"accuracy, class_based_accuracy = gtf.Evaluate();",
"Testing\n"
]
],
[
[
"### Accuracy now is - 58.85% when learning rate is 0.001 \n(Can change when you run the exp)\n - Thus reducing learning rate didn't help our case",
"_____no_output_____"
],
[
"<a id='5'></a>\n# Switch back mode, change lr, retrain",
"_____no_output_____"
]
],
[
[
"gtf.Switch_Mode(train=True)",
"_____no_output_____"
]
],
[
[
"## Update the learning rate again",
"_____no_output_____"
]
],
[
[
"# This part of code will be taken up again in upcoming sections\ngtf.update_learning_rate(0.1);\ngtf.Reload();",
"Update: Learning Rate - 0.1\n\nPre-Composed Train Transforms\n[{'RandomHorizontalFlip': {'p': 0.8}}, {'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]\n\nPre-Composed Val Transforms\n[{'RandomHorizontalFlip': {'p': 0.8}}, {'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]\n\nDataset Numbers\n Num train images: 1411\n Num val images: 605\n Num classes: 2\n\nModel Details\n Loading pretrained model\n Model Loaded on device\n Model name: resnet18_v1\n Num of potentially trainable layers: 61\n Num of actual trainable layers: 1\n\n"
],
[
"#Start Training\ngtf.Train();\n\n#Read the training summary generated once you run the cell and training is completed",
"Training Start\n Epoch 1/5\n ----------\n"
]
],
[
[
"<a id='6'></a>\n# Switch mode from train to eval and re-validate",
"_____no_output_____"
]
],
[
[
"gtf.Switch_Mode(eval_infer=True)",
"_____no_output_____"
]
],
[
[
"### Load the validation dataset",
"_____no_output_____"
]
],
[
[
"gtf.Dataset_Params(dataset_path=\"malaria_cell_val\");\ngtf.Dataset();",
"Dataset Details\n Test path: malaria_cell_val\n CSV test path: None\n\nDataset Params\n Input Size: 224\n Processors: 4\n\nPre-Composed Test Transforms\n[{'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]\n\nDataset Numbers\n Num test images: 1011\n Num classes: 2\n\n"
]
],
[
[
"### Run validation",
"_____no_output_____"
]
],
[
[
"accuracy, class_based_accuracy = gtf.Evaluate();",
"Testing\n"
]
],
[
[
"### Accuracy now is - 49.85% when learning rate is 0.1, even lower\n(Can change when you run the exp)\n - Thus increasing learning rate didn't help our case",
"_____no_output_____"
],
[
"### LR 0.01 worked best for us\n - That's how manual hyper-parameter tuning can be done using switch modes",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb4e259c05258cf986a1940057d70f9bc6d2abbe | 1,747 | ipynb | Jupyter Notebook | day1-Mon/seminar-04-word2vec/arxiv_parser.ipynb | thaarres/mlhep2018 | 2538e63c6d181caddad52b50e53914488a1134bb | [
"Apache-2.0"
] | null | null | null | day1-Mon/seminar-04-word2vec/arxiv_parser.ipynb | thaarres/mlhep2018 | 2538e63c6d181caddad52b50e53914488a1134bb | [
"Apache-2.0"
] | null | null | null | day1-Mon/seminar-04-word2vec/arxiv_parser.ipynb | thaarres/mlhep2018 | 2538e63c6d181caddad52b50e53914488a1134bb | [
"Apache-2.0"
] | null | null | null | 27.296875 | 118 | 0.497424 | [
[
[
"import os\ncategories = ['hep-th']\n\nfor cat in categories:\n if not os.path.exists('data/' + cat):\n os.makedirs('data/' + cat)",
"_____no_output_____"
],
[
"import arxivpy\nfrom tqdm import tqdm_notebook as tqdm\nfor start_index in tqdm(range(0, 10**10, 200)):\n articles = arxivpy.query(search_query=['hep-ex', 'hep-lat', 'hep-ph', 'hep-th'],\n start_index=start_index, max_index=start_index + 200, results_per_iteration=100,\n wait_time=5.0, sort_by='lastUpdatedDate') \n for article in articles:\n path = 'abstracts/' + article['term'] + '/'\n try:\n filename = article['id'].split('/')[1]\n except:\n filename = article['id']\n if not os.path.exists(path):\n os.makedirs(path)\n with open(path + filename + '.txt','w+') as outfile:\n outfile.write(article['abstract'])",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
cb4e26b1f1138036b61ee8c70b830dfaf125b771 | 26,138 | ipynb | Jupyter Notebook | random-number-generator/random_number_generator_tutorial.ipynb | fimoziq/tutorials | f47f1b59bf3c9e9f79d530c6fc8ca36c0d9ea93b | [
"MIT"
] | 670 | 2020-07-23T11:33:36.000Z | 2022-03-31T16:38:11.000Z | random-number-generator/random_number_generator_tutorial.ipynb | terragord7/tutorials | a5c3f1fed6c5c4d23f59a41c024f7499055c8d81 | [
"MIT"
] | 3 | 2021-01-03T16:36:39.000Z | 2022-02-17T06:05:43.000Z | random-number-generator/random_number_generator_tutorial.ipynb | terragord7/tutorials | a5c3f1fed6c5c4d23f59a41c024f7499055c8d81 | [
"MIT"
] | 281 | 2020-07-23T06:37:28.000Z | 2022-03-30T07:33:48.000Z | 25.575342 | 309 | 0.431326 | [
[
[
"<a href=\"https://colab.research.google.com/github/towardsai/tutorials/blob/master/random-number-generator/random_number_generator_tutorial.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Random Number Generator Tutorial with Python\r\n\r\n* Tutorial: https://towardsai.net/p/data-science/random-number-generator-tutorial-with-python-3b35986132c7\r\n\r\n* Github: https://github.com/towardsai/tutorials/tree/master/random-number-generator",
"_____no_output_____"
],
[
"## Generating pseudorandom numbers with Python's standard library\r\n\r\nPython has a built-in module called random to generate a variety of pseudorandom numbers. Although it is recommended that this module should not be used for security purposes like cryptographic uses this will do for machine learning and data science. This module uses a PRNG called Mersenne Twister.",
"_____no_output_____"
],
[
"### Importing module: random",
"_____no_output_____"
]
],
[
[
"import random",
"_____no_output_____"
]
],
[
[
"### Random numbers within a range",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nrandom.seed(25)\r\n\r\n#generating random number between 10 and 20(both excluded)\r\nprint(random.randrange(10, 20))\r\n\r\n#generating random number between 10 and 20(both included)\r\nprint(random.randint(10, 20))",
"16\n10\n"
]
],
[
[
"### Random element from a sequence",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 2\r\nrandom.seed(2)\r\n\r\n#setting up the sequence\r\nmyseq = ['Towards', 'AI', 'is', 1]\r\n\r\n#randomly choosing an element from the sequence\r\nrandom.choice(myseq)",
"_____no_output_____"
]
],
[
[
"### Multiple random selections with different possibilities",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nrandom.seed(25)\r\n\r\n#setting up the sequence\r\nmyseq = ['Towards', 'AI', 'is', 1]\r\n\r\n#random selection of length 15\r\n#10 time higher possibility of selecting 'Towards'\r\n#5 time higher possibility of selecting 'AI'\r\n#2 time higher possibility of selecting 'is'\r\n#2 time higher possibility of selecting 1\r\nrandom.choices(myseq, weights=[10, 5, 2, 2], k = 15)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"### Random element from a sequence without replacement",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nrandom.seed(25)\r\n\r\n#setting up the sequence\r\nmyseq = ['Towards', 'AI', 'is', 1]\r\n\r\n#randomly choosing an element from the sequence\r\nrandom.sample(myseq, 2)",
"_____no_output_____"
],
[
"#initialize the seed to 25\r\nrandom.seed(25)\r\n\r\n#setting up the sequence\r\nmyseq = ['Towards', 'AI', 'is', 1]\r\n\r\n#randomly choosing an element from the sequence\r\n#you are trying to choose 5 random elements from a sequence of lenth 4\r\n#since the selection is without replacement it is not possible and hence the error\r\nrandom.sample(myseq, 35)",
"_____no_output_____"
]
],
[
[
"### Rearrange the sequence",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nrandom.seed(25)\r\n\r\n#setting up the sequence\r\nmyseq = ['Towards', 'AI', 'is', 1]\r\n\r\n#rearranging the order of elements of the list\r\nrandom.shuffle(myseq)\r\nmyseq",
"_____no_output_____"
]
],
[
[
"### Floating-point random number",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nrandom.seed(25)\r\n\r\n#random float number between 0 and 1 \r\nrandom.random()",
"_____no_output_____"
]
],
[
[
"### Real-valued distributions",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nrandom.seed(25)\r\n\r\n#random float number between 10 and 20 (both included)\r\nprint(random.uniform(10, 20))\r\n\r\n#random float number mean 10 standard deviation 4\r\nprint(random.gauss(10, 4))",
"13.76962302390386\n16.90247841037158\n"
]
],
[
[
"## Generating pseudorandom numbers with Numpy",
"_____no_output_____"
]
],
[
[
"#importing random module from numpy\r\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"### Uniform distributed floating values",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#single uniformly distributed random number\r\nnp.random.rand()",
"_____no_output_____"
],
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#uniformly distributed random numbers of length 10: 1-D array\r\nnp.random.rand(10)",
"_____no_output_____"
],
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#uniformly distributed random numbers of 2 rows and 3 columns: 2-D array\r\nnp.random.rand(2, 3)",
"_____no_output_____"
]
],
[
[
"### Normal distributed floating values",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#single normally distributed random number\r\nnp.random.randn()",
"_____no_output_____"
],
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#normally distributed random numbers of length 10: 1-D array\r\nnp.random.randn(10)",
"_____no_output_____"
],
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#normally distributed random numbers of 2 rows and 3 columns: 2-D array\r\nnp.random.randn(2, 3)",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"### Uniformly distributed integers in a given range",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#single uniformly distributed random integer between 10 and 20\r\nnp.random.randint(10, 20)",
"_____no_output_____"
],
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#uniformly distributed random integer between 0 to 100 of length 10: 1-D array\r\nnp.random.randint(100, size=(10))",
"_____no_output_____"
],
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#uniformly distributed random integer between 0 to 100 of 2 rows and 3 columns: 2-D array\r\nnp.random.randint(100, size=(2, 3))",
"_____no_output_____"
]
],
[
[
"### Random elements from a defined list",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nrandom.seed(25)\r\n\r\n#setting up the sequence\r\nmyseq = ['Towards', 'AI', 'is', 1]\r\n\r\n#randomly choosing an element from the sequence\r\nnp.random.choice(myseq)",
"_____no_output_____"
],
[
"#initialize the seed to 25\r\nrandom.seed(25)\r\n\r\n#setting up the sequence\r\nmyseq = ['Towards', 'AI', 'is', 1]\r\n\r\n#randomly choosing elements from the sequence: 2-D array\r\nnp.random.choice(myseq, size=(2, 3))",
"_____no_output_____"
],
[
"#initialize the seed to 25\r\nrandom.seed(25)\r\n\r\n#setting up the sequence\r\nmyseq = ['Towards', 'AI', 'is', 1]\r\n\r\n#randomly choosing elements from the sequence with defined probabilities\r\n#The probability for the value to be 'Towards' is set to be 0.1\r\n#The probability for the value to be 'AI' is set to be 0.6\r\n#The probability for the value to be 'is' is set to be 0.05\r\n#The probability for the value to be 1 is set to be 0.25\r\n#0.1 + 0.6 + 0.05 + 0.25 = 1\r\nnp.random.choice(myseq, p=[0.1, 0.6, 0.05, 0.25], size=(2, 3))",
"_____no_output_____"
]
],
[
[
"### Binomial distributed values",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#10 number of trials with probability of 0.5 each\r\nnp.random.binomial(n=10, p=0.5, size=10)",
"_____no_output_____"
]
],
[
[
"### Poisson Distribution values",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#rate 2 and size 10\r\nnp.random.poisson(lam=2, size=10)",
"_____no_output_____"
]
],
[
[
"### Chi Square distribution",
"_____no_output_____"
]
],
[
[
"#initialize the seed to 25\r\nnp.random.seed(25)\r\n\r\n#degree of freedom 2 and size (2, 3)\r\nnp.random.chisquare(df=2, size=(2, 3))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4e2a8bfa527cc86df08a022d4384894a57742f | 13,474 | ipynb | Jupyter Notebook | docs/tutorials/ctapipe_handson.ipynb | pgrespan/ctapipe | 9eeaaebc741c48bba9e5a457587c4d5138f79320 | [
"BSD-3-Clause"
] | null | null | null | docs/tutorials/ctapipe_handson.ipynb | pgrespan/ctapipe | 9eeaaebc741c48bba9e5a457587c4d5138f79320 | [
"BSD-3-Clause"
] | null | null | null | docs/tutorials/ctapipe_handson.ipynb | pgrespan/ctapipe | 9eeaaebc741c48bba9e5a457587c4d5138f79320 | [
"BSD-3-Clause"
] | null | null | null | 20.477204 | 141 | 0.533101 | [
[
[
"# Getting Started with ctapipe\n\nThis hands-on was presented at the Paris CTA Consoritum meeting (K. Kosack)",
"_____no_output_____"
],
[
"## Part 1: load and loop over data",
"_____no_output_____"
]
],
[
[
"from ctapipe.io import event_source\nfrom ctapipe import utils\nfrom matplotlib import pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"path = utils.get_dataset_path(\"gamma_test_large.simtel.gz\")",
"_____no_output_____"
],
[
"for event in event_source(path, max_events=4):\n print(event.count, event.r0.event_id, event.mc.energy)",
"_____no_output_____"
],
[
"event",
"_____no_output_____"
],
[
"event.r0",
"_____no_output_____"
],
[
"for event in event_source(path, max_events=4):\n print(event.count, event.r0.tels_with_data)",
"_____no_output_____"
],
[
"event.r0.tel[2]",
"_____no_output_____"
],
[
"r0tel = event.r0.tel[2]",
"_____no_output_____"
],
[
"r0tel.waveform",
"_____no_output_____"
],
[
"r0tel.waveform.shape",
"_____no_output_____"
]
],
[
[
"note that this is ($N_{channels}$, $N_{pixels}$, $N_{samples}$)",
"_____no_output_____"
]
],
[
[
"plt.pcolormesh(r0tel.waveform[0])",
"_____no_output_____"
],
[
"plt.plot(r0tel.waveform[0,10])",
"_____no_output_____"
],
[
"from ipywidgets import interact\n\n@interact\ndef view_waveform(chan=0, pix_id=200):\n plt.plot(r0tel.waveform[chan, pix_id])",
"_____no_output_____"
]
],
[
[
"try making this compare 2 waveforms",
"_____no_output_____"
],
[
"## Part 2: Explore the instrument description\nThis is all well and good, but we don't really know what camera or telescope this is... how do we get instrumental description info?\n\nCurrently this is returned *inside* the event (it will soon change to be separate in next version or so)",
"_____no_output_____"
]
],
[
[
"subarray = event.inst.subarray # soon EventSource will give you event, subarray separate",
"_____no_output_____"
],
[
"subarray",
"_____no_output_____"
],
[
"subarray.peek()",
"_____no_output_____"
],
[
"subarray.to_table()",
"_____no_output_____"
],
[
"subarray.tel[2]",
"_____no_output_____"
],
[
"subarray.tel[2].camera",
"_____no_output_____"
],
[
"subarray.tel[2].optics",
"_____no_output_____"
],
[
"tel = subarray.tel[2]",
"_____no_output_____"
],
[
"tel.camera",
"_____no_output_____"
],
[
"tel.optics",
"_____no_output_____"
],
[
"tel.camera.geometry.pix_x",
"_____no_output_____"
],
[
"tel.camera.geometry.to_table()",
"_____no_output_____"
],
[
"tel.optics.mirror_area",
"_____no_output_____"
],
[
"from ctapipe.visualization import CameraDisplay",
"_____no_output_____"
],
[
"disp = CameraDisplay(tel.camera.geometry)",
"_____no_output_____"
],
[
"disp = CameraDisplay(tel.camera.geometry)\ndisp.image = r0tel.waveform[0,:,10] # display channel 0, sample 0 (try others like 10)",
"_____no_output_____"
]
],
[
[
" ** aside: ** show demo using a CameraDisplay in interactive mode in ipython rather than notebook",
"_____no_output_____"
],
[
"## Part 3: Apply some calibration and trace integration",
"_____no_output_____"
]
],
[
[
"from ctapipe.calib import CameraCalibrator",
"_____no_output_____"
],
[
"calib = CameraCalibrator(subarray=subarray)",
"_____no_output_____"
],
[
"for event in event_source(path, max_events=4):\n calib(event) # fills in r1, dl0, and dl1\n print(event.dl1.tel.keys())",
"_____no_output_____"
],
[
"event.dl1.tel[2]",
"_____no_output_____"
],
[
"dl1tel = event.dl1.tel[2]",
"_____no_output_____"
],
[
"dl1tel.image.shape # note this will be gain-selected in next version, so will be just 1D array of 1855",
"_____no_output_____"
],
[
"dl1tel.pulse_time",
"_____no_output_____"
],
[
"CameraDisplay(tel.camera.geometry, image=dl1tel.image)",
"_____no_output_____"
],
[
"CameraDisplay(tel.camera.geometry, image=dl1tel.pulse_time)",
"_____no_output_____"
]
],
[
[
"Now for Hillas Parameters",
"_____no_output_____"
]
],
[
[
"from ctapipe.image import hillas_parameters, tailcuts_clean",
"_____no_output_____"
],
[
"image = dl1tel.image\nmask = tailcuts_clean(tel.camera.geometry, image, picture_thresh=10, boundary_thresh=5)\nmask",
"_____no_output_____"
],
[
"CameraDisplay(tel.camera.geometry, image=mask)",
"_____no_output_____"
],
[
"cleaned = image.copy()\ncleaned[~mask] = 0 ",
"_____no_output_____"
],
[
"disp = CameraDisplay(tel.camera.geometry, image=cleaned)\ndisp.cmap = plt.cm.coolwarm\ndisp.add_colorbar()\nplt.xlim(-1.0,0)\nplt.ylim(0,1.0)",
"_____no_output_____"
],
[
"params = hillas_parameters(tel.camera.geometry, cleaned)\nprint(params)",
"_____no_output_____"
],
[
"disp = CameraDisplay(tel.camera.geometry, image=cleaned)\ndisp.cmap = plt.cm.coolwarm\ndisp.add_colorbar()\nplt.xlim(-1.0,0)\nplt.ylim(0,1.0)\ndisp.overlay_moments(params, color='white', lw=2)",
"_____no_output_____"
]
],
[
[
"## Part 4: Let's put it all together: \n- loop over events, selecting only telescopes of the same type (e.g. LST:LSTCam)\n- for each event, apply calibration/trace integration\n- calculate Hillas parameters \n- write out all hillas paremeters to a file that can be loaded with Pandas",
"_____no_output_____"
],
[
"first let's select only those telescopes with LST:LSTCam",
"_____no_output_____"
]
],
[
[
"subarray.telescope_types",
"_____no_output_____"
],
[
"subarray.get_tel_ids_for_type(\"LST_LST_LSTCam\")",
"_____no_output_____"
]
],
[
[
"Now let's write out program",
"_____no_output_____"
]
],
[
[
"data = utils.get_dataset_path(\"gamma_test_large.simtel.gz\") \nsource = event_source(data, allowed_tels=[1,2,3,4], max_events=10) # remove the max_events limit to get more stats",
"_____no_output_____"
],
[
"for event in source:\n calib(event)\n \n for tel_id, tel_data in event.dl1.tel.items():\n tel = event.inst.subarray.tel[tel_id]\n mask = tailcuts_clean(tel.camera.geometry, tel_data.image)\n params = hillas_parameters(tel.camera.geometry[mask], tel_data.image[mask])",
"_____no_output_____"
],
[
"from ctapipe.io import HDF5TableWriter\n",
"_____no_output_____"
],
[
"with HDF5TableWriter(filename='hillas.h5', group_name='dl1', overwrite=True) as writer:\n \n for event in event_source(data, allowed_tels=[1,2,3,4], max_events=10):\n calib(event)\n \n for tel_id, tel_data in event.dl1.tel.items():\n tel = event.inst.subarray.tel[tel_id]\n mask = tailcuts_clean(tel.camera.geometry, tel_data.image)\n params = hillas_parameters(tel.camera.geometry[mask], tel_data.image[mask])\n writer.write(\"hillas\", params)",
"_____no_output_____"
]
],
[
[
"### We can now load in the file we created and plot it",
"_____no_output_____"
]
],
[
[
"!ls *.h5",
"_____no_output_____"
],
[
"import pandas as pd\n\nhillas = pd.read_hdf(\"hillas.h5\", key='/dl1/hillas')\nhillas",
"_____no_output_____"
],
[
"_ = hillas.hist(figsize=(8,8))",
"_____no_output_____"
]
],
[
[
"If you do this yourself, loop over more events to get better statistics",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb4e3562970d9dd149ed14216fdbaffeaafcfd12 | 7,846 | ipynb | Jupyter Notebook | QuestionDetector.ipynb | daveshap/DialogActDetector | 76cde6ccb019223295fb75decdb8fbacd22909ad | [
"MIT"
] | 2 | 2021-08-09T20:03:13.000Z | 2021-09-20T14:54:58.000Z | QuestionDetector.ipynb | daveshap/DialogActDetector | 76cde6ccb019223295fb75decdb8fbacd22909ad | [
"MIT"
] | null | null | null | QuestionDetector.ipynb | daveshap/DialogActDetector | 76cde6ccb019223295fb75decdb8fbacd22909ad | [
"MIT"
] | 1 | 2021-12-06T21:50:07.000Z | 2021-12-06T21:50:07.000Z | 34.716814 | 238 | 0.463548 | [
[
[
"<a href=\"https://colab.research.google.com/github/daveshap/QuestionDetector/blob/main/QuestionDetector.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Compile Training Data\nNote: Generate the raw data with [this notebook](https://github.com/daveshap/QuestionDetector/blob/main/DownloadGutenbergTop100.ipynb)",
"_____no_output_____"
]
],
[
[
"import re\nimport random\n\ndatafile = '/content/drive/My Drive/Gutenberg/sentence_data.txt'\ncorpusfile = '/content/drive/My Drive/Gutenberg/corpus_data.txt'\ntestfile = '/content/drive/My Drive/Gutenberg/test_data.txt'\nsample_cnt = 3000\ntest_cnt = 30\n\nquestions = list()\nexclamations = list()\nother = list()\n\nwith open(datafile, 'r', encoding='utf-8') as infile:\n body = infile.read()\nsentences = re.split('\\n\\n', body)\n\nfor i in sentences:\n if 'í' in i or 'á' in i:\n continue \n if '?' in i:\n questions.append(i)\n elif '!' in i:\n exclamations.append(i)\n else:\n other.append(i)\n\ndef flatten_sentence(text):\n text = text.lower()\n fa = re.findall('[\\w\\s]',text)\n return ''.join(fa)\n\n\ndef compose_corpus(data, count, label):\n result = ''\n random.seed()\n subset = random.sample(data, count)\n for i in subset:\n result += '<|SENTENCE|> %s <|LABEL|> %s <|END|>\\n\\n' % (flatten_sentence(i), label)\n return result\n\ncorpus = compose_corpus(questions, sample_cnt, 'question')\ncorpus += compose_corpus(exclamations, sample_cnt, 'other')\ncorpus += compose_corpus(other, sample_cnt, 'other')\n\nwith open(corpusfile, 'w', encoding='utf-8') as outfile:\n outfile.write(corpus)\nprint('Done!', corpusfile)\n\ncorpus = compose_corpus(questions, test_cnt, 'question')\ncorpus += compose_corpus(exclamations, test_cnt, 'other')\ncorpus += compose_corpus(other, test_cnt, 'other')\n\nwith open(testfile, 'w', encoding='utf-8') as outfile:\n outfile.write(corpus)\nprint('Done!', testfile)",
"_____no_output_____"
]
],
[
[
"# Finetune Model\nFinetune GPT-2",
"_____no_output_____"
]
],
[
[
"!pip install tensorflow-gpu==1.15.0 --quiet\n!pip install gpt-2-simple --quiet\n\nimport gpt_2_simple as gpt2\n\n# note: manually mount your google drive in the file explorer to the left\n\nmodel_dir = '/content/drive/My Drive/GPT2/models'\ncheckpoint_dir = '/content/drive/My Drive/GPT2/checkpoint'\n#model_name = '124M'\nmodel_name = '355M'\n#model_name = '774M'\n\n\ngpt2.download_gpt2(model_name=model_name, model_dir=model_dir)\nprint('\\n\\nModel is ready!')\n\nrun_name = 'QuestionDetector'\nstep_cnt = 4000\n\nsess = gpt2.start_tf_sess()\n\ngpt2.finetune(sess,\n dataset=corpusfile,\n model_name=model_name,\n model_dir=model_dir,\n checkpoint_dir=checkpoint_dir,\n steps=step_cnt,\n restore_from='fresh', # start from scratch\n #restore_from='latest', # continue from last work\n run_name=run_name,\n print_every=50,\n sample_every=1000,\n save_every=1000\n )",
"_____no_output_____"
]
],
[
[
"# Test Results\n\n| Run | Model | Steps | Samples | Last Loss | Avg Loss | Accuracy |\n|---|---|---|---|---|---|---|\n| 01 | 124M | 2000 | 9000 | 0.07 | 0.69 | 71.4% |\n| 02 | 355M | 2000 | 9000 | 0.24 | 1.63 | 66% |\n| 03 | 355M | 4000 | 9000 | 0.06 | 0.83 | 58% |\n| 04 | 355M | 4000 | 9000 | 0.11 | 0.68 | 74.4% |\n\nLarger models seem to need more steps and/or data. Seems to perform very high on questions and less good on others. Test 04 was reduced to 2 classes. \n\n",
"_____no_output_____"
]
],
[
[
"right = 0\nwrong = 0\n\nprint('Loading test set...')\nwith open(testfile, 'r', encoding='utf-8') as file:\n test_set = file.readlines()\n\nfor t in test_set:\n t = t.strip()\n if t == '':\n continue\n prompt = t.split('<|LABEL|>')[0] + '<|LABEL|>'\n expect = t.split('<|LABEL|>')[1].replace('<|END|>', '').strip()\n #print('\\nPROMPT:', prompt)\n response = gpt2.generate(sess, \n return_as_list=True,\n length=30, # prevent it from going too crazy\n prefix=prompt,\n model_name=model_name,\n model_dir=model_dir,\n truncate='\\n', # stop inferring here\n include_prefix=False,\n checkpoint_dir=checkpoint_dir,)[0]\n response = response.strip()\n if expect in response:\n right += 1\n else:\n wrong += 1\n print('right:', right, '\\twrong:', wrong, '\\taccuracy:', right / (right+wrong))\n #print('RESPONSE:', response)\n\nprint('\\n\\nModel:', model_name)\nprint('Samples:', max_samples)\nprint('Steps:', step_cnt)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4e570f0b0feacc4e0f5a9e4f8488e4b734c633 | 824,676 | ipynb | Jupyter Notebook | practicals/week_3.ipynb | max-de-rooij/8dm50-machine-learning | ae4832a70dcb36da2ae69fcfd9c212d1b46996b8 | [
"MIT"
] | null | null | null | practicals/week_3.ipynb | max-de-rooij/8dm50-machine-learning | ae4832a70dcb36da2ae69fcfd9c212d1b46996b8 | [
"MIT"
] | null | null | null | practicals/week_3.ipynb | max-de-rooij/8dm50-machine-learning | ae4832a70dcb36da2ae69fcfd9c212d1b46996b8 | [
"MIT"
] | null | null | null | 1,120.483696 | 729,161 | 0.736564 | [
[
[
"# Preliminaries\n\nThe `pandas` library allows the user several data structures for different data manipulation tasks:\n1. Data storage through its `Series` and `DataFrame` data structures.\n2. Data filtering using multiple methods from the package.\n3. Reading data from many different file formats such as `csv`, `txt`, `xlsx`, ...\n\nBelow we provide a brief overview of the `pandas` functionalities needed for these exercises. The complete documentation can be found on the [`pandas` website](https://pandas.pydata.org/).\n\n## Pandas data structures\n\n### Series\nThe Pandas Series data structure is similar to a one-dimensional array. It can store any type of data. The values are mutable but the size not.\n\nTo create `Series`, we call the `pd.Series()` method and pass an array. A `Series` may also be created from a numpy array.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nfirst_series = pd.Series([1,10,100,1000])\n\nprint(first_series)\n\nteams = np.array(['PSV','Ajax','Feyenoord','Twente'])\nsecond_series = pd.Series(teams)\n\nprint('\\n')\nprint(second_series)",
"0 1\n1 10\n2 100\n3 1000\ndtype: int64\n\n\n0 PSV\n1 Ajax\n2 Feyenoord\n3 Twente\ndtype: object\n"
]
],
[
[
"### DataFrame\nOne can think of a `DataFrame` as a table with rows and columns (2D structure). The columns can be of a different type (as opposed to `numpy` arrays) and the size of the `DataFrame` is mutable.\n\nTo create `DataFrame`, we call the `pd.DataFrame()` method and we can create it from scratch or we can convert a numpy array or a list into a `DataFrame`.",
"_____no_output_____"
]
],
[
[
"# DataFrame from scratch\nfirst_dataframe = pd.DataFrame({\n \"Position\": [1, 2, 3, 4],\n \"Team\": ['PSV','Ajax','Feyenoord','Twente'],\n \"GF\": [80, 75, 75, 70],\n \"GA\": [30, 25, 40, 60],\n \"Points\": [79, 78, 70, 66]\n})\n\nprint(\"From scratch: \\n {} \\n\".format(first_dataframe))\n\n# DataFrme from a list\ndata = [[1, 2, 3, 4], ['PSV','Ajax','Feyenoord','Twente'], \n [80, 75, 75, 70], [30, 25, 40, 60], [79, 78, 70, 66]]\ncolumns = [\"Position\", \"Team\", \"GF\", \"GA\", \"Points\"]\n\nsecond_dataframe = pd.DataFrame(data, index=columns)\n\nprint(\"From list: \\n {} \\n\".format(second_dataframe.T)) # the '.T' operator is explained later on\n\n# DataFrame from numpy array\ndata = np.array([[1, 2, 3, 4], ['PSV','Ajax','Feyenoord','Twente'], \n [80, 75, 75, 70], [30, 25, 40, 60], [79, 78, 70, 66]])\ncolumns = [\"Position\", \"Team\", \"GF\", \"GA\", \"Points\"]\n\nthird_dataframe = pd.DataFrame(data.T, columns=columns)\n\nprint(\"From numpy array: \\n {} \\n\".format(third_dataframe))",
"From scratch: \n Position Team GF GA Points\n0 1 PSV 80 30 79\n1 2 Ajax 75 25 78\n2 3 Feyenoord 75 40 70\n3 4 Twente 70 60 66 \n\nFrom list: \n Position Team GF GA Points\n0 1 PSV 80 30 79\n1 2 Ajax 75 25 78\n2 3 Feyenoord 75 40 70\n3 4 Twente 70 60 66 \n\nFrom numpy array: \n Position Team GF GA Points\n0 1 PSV 80 30 79\n1 2 Ajax 75 25 78\n2 3 Feyenoord 75 40 70\n3 4 Twente 70 60 66 \n\n"
]
],
[
[
"### DataFrame attributes\nThis section gives a quick overview of some of the `pandas.DataFrame` attributes such as `T`, `index`, `columns`, `iloc`, `loc`, `shape` and `values`.",
"_____no_output_____"
]
],
[
[
"# transpose the index and columns\nprint(third_dataframe.T)",
" 0 1 2 3\nPosition 1 2 3 4\nTeam PSV Ajax Feyenoord Twente\nGF 80 75 75 70\nGA 30 25 40 60\nPoints 79 78 70 66\n"
],
[
"# index makes reference to the row labels\nprint(third_dataframe.index)",
"RangeIndex(start=0, stop=4, step=1)\n"
],
[
"# columns makes reference to the column labels\nprint(third_dataframe.columns)",
"Index(['Position', 'Team', 'GF', 'GA', 'Points'], dtype='object')\n"
],
[
"# iloc allows to access the index by integer-location (e.g. all team names, which are in the second columm)\nprint(third_dataframe.iloc[:,1])",
"0 PSV\n1 Ajax\n2 Feyenoord\n3 Twente\nName: Team, dtype: object\n"
],
[
"# loc allows to access the index by label(s)-location (e.g. all team names, which are in the \"Team\" columm)\nprint(third_dataframe.loc[0, 'Team'])",
"PSV\n"
],
[
"# shape returns a tuple with the DataFrame dimension, similar to numpy\nprint(third_dataframe.shape)",
"(4, 5)\n"
],
[
"# values return a Numpy representation of the DataFrame data\nprint(third_dataframe.values)",
"[['1' 'PSV' '80' '30' '79']\n ['2' 'Ajax' '75' '25' '78']\n ['3' 'Feyenoord' '75' '40' '70']\n ['4' 'Twente' '70' '60' '66']]\n"
]
],
[
[
"### DataFrame methods\nThis section gives a quick overview of some of the `pandas.DataFrame` methods such as `head`, `describe`, `concat`, `groupby`,`rename`, `filter`, `drop` and `isna`. To import data from CSV or MS Excel files, we can make use of `read_csv` and `read_excel`, respectively.",
"_____no_output_____"
]
],
[
[
"# print the first few rows in your dataset with head()\nprint(third_dataframe.head()) # In this case, it is not very useful because we don't have thousands of rows",
" Position Team GF GA Points\n0 1 PSV 80 30 79\n1 2 Ajax 75 25 78\n2 3 Feyenoord 75 40 70\n3 4 Twente 70 60 66\n"
],
[
"# get the summary statistics of the DataFrame with describe()\nprint(third_dataframe.describe())",
" Position Team GF GA Points\ncount 4 4 4 4 4\nunique 4 4 3 4 4\ntop 2 Ajax 75 40 70\nfreq 1 1 2 1 1\n"
],
[
"# concatenate (join) DataFrame objects using concat()\n\n# first, we will split the above DataFrame in two different ones\ndf_a = third_dataframe.loc[[0,1],:]\ndf_b = third_dataframe.loc[[2,3],:]\n\nprint(df_a)\nprint('\\n')\n\nprint(df_b)\nprint('\\n')\n\n# now, we concatenate both datasets\ndf = pd.concat([df_a, df_b])\n\nprint(df)",
" Position Team GF GA Points\n0 1 PSV 80 30 79\n1 2 Ajax 75 25 78\n\n\n Position Team GF GA Points\n2 3 Feyenoord 75 40 70\n3 4 Twente 70 60 66\n\n\n Position Team GF GA Points\n0 1 PSV 80 30 79\n1 2 Ajax 75 25 78\n2 3 Feyenoord 75 40 70\n3 4 Twente 70 60 66\n"
],
[
"# group the data by certain variable via groupby()\n# here, we have grouped the data by goals for, which in this case is 75\n\ngroup = df.groupby('GF')\n\nprint(group.get_group('75'))",
" Position Team GF GA Points\n1 2 Ajax 75 25 78\n2 3 Feyenoord 75 40 70\n"
],
[
"# rename() helps you change the column or index names\nprint(df.rename(columns={'Position':'Pos','Team':'Club'}))",
" Pos Club GF GA Points\n0 1 PSV 80 30 79\n1 2 Ajax 75 25 78\n2 3 Feyenoord 75 40 70\n3 4 Twente 70 60 66\n"
],
[
"# build a subset of rows or columns of your dataset according to labels via filter()\n# here, items refer to the variable names: 'Team' and 'Points'; to select columns, we specify axis=1\nprint(df.filter(items=['Team', 'Points'], axis=1))",
" Team Points\n0 PSV 79\n1 Ajax 78\n2 Feyenoord 70\n3 Twente 66\n"
],
[
"# dropping some labels\nprint(df.drop(columns=['GF', 'GA']))",
" Position Team Points\n0 1 PSV 79\n1 2 Ajax 78\n2 3 Feyenoord 70\n3 4 Twente 66\n"
],
[
"# search for NA (not available) entries in the DataFrame\nprint(df.isna()) # No NA values\nprint('\\n')\n\n# create a pandas Series with a NA value\n# the Series as W (winnin matches)\ntmp = pd.Series([np.NaN, 25, 24, 19], name=\"W\")\n\n# concatenate the Series with the DataFrame\ndf = pd.concat([df,tmp], axis = 1)\nprint(df)\nprint('\\n')\n\n# again, check for NA entries\nprint(df.isna())",
" Position Team GF GA Points\n0 False False False False False\n1 False False False False False\n2 False False False False False\n3 False False False False False\n\n\n Position Team GF GA Points W\n0 1 PSV 80 30 79 NaN\n1 2 Ajax 75 25 78 25.0\n2 3 Feyenoord 75 40 70 24.0\n3 4 Twente 70 60 66 19.0\n\n\n Position Team GF GA Points W\n0 False False False False False True\n1 False False False False False False\n2 False False False False False False\n3 False False False False False False\n"
]
],
[
[
"## Dataset\n\nFor this week exercises we will use a dataset from the Genomics of Drug Sensitivity in Cancer (GDSC) project (https://www.cancerrxgene.org/). In this study (['Iorio et al., Cell, 2016']()), 265 compounds were tested on 1001 cancer cell lines for which different types of -omics data (RNA expression, DNA methylation, Copy Number Alteration, DNA sequencing) are available. This is a valuable resource to look for biomarkers of drugs sensitivity in order to try to understand why cancer patients responds very differently to cancer drugs and find ways to assign the optimal treatment to each patient.\n\nFor this exercise we will use a subset of the data, focusing the response to the drug YM155 (Sepantronium bromide) on four cancer types, for a total of 148 cancer cell lines.\n\n| ID | Cancer type |\n|-------------|----------------------------------|\n| COAD/READ | Colorectal adenocarcinoma |\n| NB | Neuroblastoma |\n| KIRC | Kidney renal clear cell carcinoma|\n| BRCA | Breast carcinoma |\n\nWe will use the RNA expression data (RMA normalised). Only genes with high variability across cell lines (variance > 5, resulting in 238 genes) have been kept.\n\nDrugs have been tested at different concentration, measuring each time the viability of the cells. Drug sensitivity is measured using the natural log of the fitted IC50 metric, which is defined as the half maximal inhibitory concentration. A lower IC50 corresponds to a more sensitive cell line because a lower amount of drug is sufficient to have a strong response, while a higher IC50 corresponds to a more resistant cell line because more drug is needed for killing the cells.\n\nBased on the IC50 metric, cells can be classified as sensitive or resistant. The classification is done by computing the $z$-score across all cell lines in the GDSC for each drug, and considering as sensitive the ones with $z$-score < 0 and resistant the ones with $z$-score > 0.\n\nThe dataset is originally provided as 3 files ([original source](https://www.sciencedirect.com/science/article/pii/S0092867416307462?via%3Dihub)) :\n\n`GDSC_RNA_expression.csv`: gene expression matrix with the cell lines in the rows (148) and the genes in the columns (238).\n\n`GDSC_drug_response.csv`: vector with the cell lines response to the drug YM155 in terms of log(IC50) and as classification in sensitive or resistant.\n\n`GDSC_metadata.csv`: metadata for the 148 cell lines including name, COSMIC ID and tumor type (using the classification from ['The Cancer Genome Atlas TCGA'](https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga))\n\nFor convenience, we provide the data already curated.\n\n`RNA_expression_curated.csv`: [148 cell lines , 238 genes]\n\n`drug_response_curated.csv`: [148 cell lines , YM155 drug]\n\nThe curated data cam be read as `pandas` `DataFrame`s in the following way:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ngene_expression = pd.read_csv(\"./data/RNA_expression_curated.csv\", sep=',', header=0, index_col=0)\ndrug_response = pd.read_csv(\"./data/drug_response_curated.csv\", sep=',', header=0, index_col=0)",
"_____no_output_____"
]
],
[
[
"You can use the `DataFrame`s directly as inputs to the the `sklearn` models. The advantage over using `numpy` arrays is that the variable are annotated, i.e. each input and output has a name.",
"_____no_output_____"
],
[
"## Tools\nThe `scikit-learn` library provides the required tools for linear regression/classification and shrinkage, as well as for logistic regression.",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LinearRegression\nfrom sklearn.linear_model import Ridge\nfrom sklearn.linear_model import Lasso\nfrom sklearn.linear_model import LogisticRegression",
"_____no_output_____"
]
],
[
[
"Note that the notation used for the hyperparameters in the `scikit-learn` library is different from the one used in the lecture. More specifically, in the lecture $\\alpha$ is the tunable parameter to select the compromise between Ridge and Lasso. Whereas, `scikit-learn` library refers to `alpha` as the tunable parameter $\\lambda$. Please check the documentation for more details.",
"_____no_output_____"
],
[
"# Exercises\n\n## Selection of the hyperparameter\n\nImplement cross-validation (using `sklearn.grid_search.GridSearchCV`) to select the `alpha` hyperparameter of `sklearn.linear_model.Lasso`. \n\n\n## Feature selection\n\nLook at the features selected using the hyperparameter which corresponds to the minimum cross-validation error.\n\n<p><font color='#770a0a'>Is the partition in training and validation sets playing a role in the selection of the hyperparameter? How will this affect the selection of the relevant features?</font></p>\n\n**Answer**: The partition in itself has no direct relation to the selection of the hyperparameter (see the graph with selection frequency), as these partitions are averaged in the hyperparameter selection. Nevertheless, the selected features may be sensitive to this partition. Therefore, it is useful to repeat cross-validation multiple times (using bootstrap).\n\n\n<p><font color='#770a0a'>Should the value of the intercept also be shrunk to zero with Lasso and Ridge regression? Motivate your answer.</font></p>\n\n**Answer**: No, this should not be done, because then the optimization procedure would become dependent on the origin chosen for the output variable $\\mathbf{y}$. For example, adding a constant value to your training $\\mathbf{y}$, would not result in an addition of this constant value for the predictions. This would be the case for a non-penalized intercept.\n\n## Bias-variance \n\nShow the effect of the regularization on the parameter estimates in terms of bias and variance. For this you can repeat the optimization 100 times using bootstrap and visualise the profile of the Lasso regression coefficient over a grid of the hyperparameter, optionally including the variability as error bars.\n\n<p><font color='#770a0a'>Based on the visual analysis of the plot, what are your observation on bias and variance in relation to model complexity? Motivate your answer.</font></p>\n\n**Answer**: For a low $\\alpha$, many parameters are included, leading to a complex model with high variance. As $\\alpha$ increases, the amount and values of the parameters decrease, leading to a less complex model. A less and less complex model increases the bias, but decreases the variance. \n\n\n## Logistic regression\n\n<p><font color='#770a0a'>Write the expression of the objective function for the penalized logistic regression with $L_1$ and $L_2$ regularisation (as in Elastic net).</font></p>\n\n**Logistic Regression with Elastic net**\n\n$$\\max_{\\beta_0, \\beta} \\left\\{ \\sum^{N}_{i=1} \\left[y_i\\left(\\beta_0 + \\beta^T x_i\\right) - \\log{\\left(1+e^{\\beta_0 + \\beta^T x_i}\\right)}\\right]-\\left[\\lambda_1 \\sum_{j=1}^{p} |\\beta_j | + \\lambda_2 \\sum_{j=1}^{p} \\beta_j^2 \\right] \\right\\}$$\n",
"_____no_output_____"
],
[
"**Selection of the Hyperparameter $\\alpha$**",
"_____no_output_____"
]
],
[
[
"import sys\nsys.path.append('code/')\nfrom week_3_utils import *\nfrom sklearn.model_selection import train_test_split\nimport matplotlib.pyplot as plt\n\nX_train, X_test, y_train, y_test = train_test_split(gene_expression, drug_response, test_size=0.2, random_state=40)\n\nalpha_range = np.linspace(10e-4,1,num=100)\nmodel = cv_lasso(alpha_range,folds=5)\nmodel.fit(X_train, y_train)\nprint(model.best_estimator_)",
"Pipeline(steps=[('normalize', StandardScaler()),\n ('lasso', Lasso(alpha=0.4752727272727273))])\n"
]
],
[
[
"**Feature Selection**",
"_____no_output_____"
]
],
[
[
"features = gene_expression.columns\ncounter = np.zeros((1,len(features)))\namt_of_rep = 5\nfor ix in range(amt_of_rep):\n alpha_range = np.linspace(10e-4,1,num=50)\n model = cv_lasso(alpha_range,folds=5)\n model.fit(X_train, y_train)\n\n coefficients = model.best_estimator_.named_steps['lasso'].coef_\n nonzero_coef = np.array((coefficients != 0.)).astype(int)\n counter = counter+nonzero_coef\n print(f'{ix} of {amt_of_rep-1}')\n\n \n",
"0 of 4\n1 of 4\n2 of 4\n3 of 4\n4 of 4\n"
],
[
"counter = counter.ravel()\nfeatures_in_plot = features[counter != 0]\ncounters_in_plot = counter[counter != 0]\n#print(features_in_plot)\nplt.bar(list(range(0,4*len(features_in_plot),4)), counters_in_plot/amt_of_rep, tick_label=features_in_plot)\nplt.xticks(rotation=30)\nplt.ylabel('Fraction of Selection')\nplt.show()\n",
"Index(['PRSS3', 'GAL', 'CDH17', 'ABCB1', 'CYR61', 'FABP1'], dtype='object')\n"
]
],
[
[
"**Bias-Variance**",
"_____no_output_____"
]
],
[
[
"from sklearn.utils import resample\nfrom week_3_utils import lasso_estimator\nn_bootstrap = 100\nsamplesize = 80\nalpha_range = np.linspace(0,3,num=100)\ncoef = np.zeros((len(alpha_range),n_bootstrap,len(gene_expression.columns)))\n\nfor j in range(n_bootstrap):\n x_bs, y_bs = resample(X_train, y_train, replace=True, n_samples=samplesize)\n for i,alpha in enumerate(alpha_range):\n model_bs = lasso_estimator(alpha=alpha)\n model_bs.fit(x_bs, y_bs)\n coef[i,j,:] = model_bs.named_steps['lasso'].coef_\n\naverage_coef = np.mean(coef, axis=1)\nstd_coef = np.std(coef, axis=1)\nfor k in range(len(gene_expression.columns)):\n plt.plot(alpha_range, average_coef[:,k],linewidth=0.5)\nplt.xlabel('alpha')\nplt.ylabel('Coefficients')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4e61604e7d44c15a90dadc245da0ad8d7d0a55 | 1,015,701 | ipynb | Jupyter Notebook | .ipynb_checkpoints/demo-checkpoint.ipynb | laxmaniron/aiimautomaticoneshotdetection | 144927cca4605e25579f07c90db93e52b4c21edc | [
"MIT"
] | null | null | null | .ipynb_checkpoints/demo-checkpoint.ipynb | laxmaniron/aiimautomaticoneshotdetection | 144927cca4605e25579f07c90db93e52b4c21edc | [
"MIT"
] | null | null | null | .ipynb_checkpoints/demo-checkpoint.ipynb | laxmaniron/aiimautomaticoneshotdetection | 144927cca4605e25579f07c90db93e52b4c21edc | [
"MIT"
] | null | null | null | 943.960037 | 411,536 | 0.946617 | [
[
[
"This is a demo illustrating an application of the OS2D method on one image.\nDemo assumes the OS2D code is [installed](./INSTALL.md).",
"_____no_output_____"
]
],
[
[
"import os\nimport argparse\nimport matplotlib.pyplot as plt\n\nimport torch\nimport torchvision.transforms as transforms\n\nfrom os2d.modeling.model import build_os2d_from_config\nfrom os2d.config import cfg\nimport os2d.utils.visualization as visualizer\nfrom os2d.structures.feature_map import FeatureMapSize\nfrom os2d.utils import setup_logger, read_image, get_image_size_after_resize_preserving_aspect_ratio\n\nlogger = setup_logger(\"OS2D\")",
"_____no_output_____"
],
[
"# use GPU if have available\ncfg.is_cuda = torch.cuda.is_available()",
"_____no_output_____"
]
],
[
[
"Download the trained model (is the script does not work download from [Google Drive](https://drive.google.com/open?id=1l_aanrxHj14d_QkCpein8wFmainNAzo8) and put to models/os2d_v2-train.pth). See [README](./README.md) to get links for other released models.",
"_____no_output_____"
]
],
[
[
"!./os2d/utils/wget_gdrive.sh models/os2d_v2-train.pth 1l_aanrxHj14d_QkCpein8wFmainNAzo8",
"--2020-05-02 17:51:17-- https://docs.google.com/uc?export=download&confirm=&id=1l_aanrxHj14d_QkCpein8wFmainNAzo8\nResolving docs.google.com (docs.google.com)... 209.85.233.194, 2a00:1450:4010:c01::c2\nConnecting to docs.google.com (docs.google.com)|209.85.233.194|:443...connected.\nHTTP request sent, awaiting response...302 Moved Temporarily\nLocation: https://doc-04-60-docs.googleusercontent.com/docs/securesc/klc24bgsvh27l2s553lm5mif6bdal7bk/7u9jiamn68gk60nq9463uivp5449u3nr/1588431075000/02695332612409210478/02864383199933671631Z/1l_aanrxHj14d_QkCpein8wFmainNAzo8?e=download [following]\n--2020-05-02 17:51:18-- https://doc-04-60-docs.googleusercontent.com/docs/securesc/klc24bgsvh27l2s553lm5mif6bdal7bk/7u9jiamn68gk60nq9463uivp5449u3nr/1588431075000/02695332612409210478/02864383199933671631Z/1l_aanrxHj14d_QkCpein8wFmainNAzo8?e=download\nResolving doc-04-60-docs.googleusercontent.com (doc-04-60-docs.googleusercontent.com)...173.194.221.132, 2a00:1450:4010:c05::84\nConnecting to doc-04-60-docs.googleusercontent.com (doc-04-60-docs.googleusercontent.com)|173.194.221.132|:443...connected.\nHTTP request sent, awaiting response...302 Found\nLocation: https://docs.google.com/nonceSigner?nonce=pvee233ijmg3u&continue=https://doc-04-60-docs.googleusercontent.com/docs/securesc/klc24bgsvh27l2s553lm5mif6bdal7bk/7u9jiamn68gk60nq9463uivp5449u3nr/1588431075000/02695332612409210478/02864383199933671631Z/1l_aanrxHj14d_QkCpein8wFmainNAzo8?e%3Ddownload&hash=vcmeprk2g2dpia9mbhtbvtegknmmo1eh [following]\n--2020-05-02 17:51:18-- https://docs.google.com/nonceSigner?nonce=pvee233ijmg3u&continue=https://doc-04-60-docs.googleusercontent.com/docs/securesc/klc24bgsvh27l2s553lm5mif6bdal7bk/7u9jiamn68gk60nq9463uivp5449u3nr/1588431075000/02695332612409210478/02864383199933671631Z/1l_aanrxHj14d_QkCpein8wFmainNAzo8?e%3Ddownload&hash=vcmeprk2g2dpia9mbhtbvtegknmmo1eh\nConnecting to docs.google.com (docs.google.com)|209.85.233.194|:443...connected.\nHTTP request sent, awaiting response...302 Found\nLocation: https://doc-04-60-docs.googleusercontent.com/docs/securesc/klc24bgsvh27l2s553lm5mif6bdal7bk/7u9jiamn68gk60nq9463uivp5449u3nr/1588431075000/02695332612409210478/02864383199933671631Z/1l_aanrxHj14d_QkCpein8wFmainNAzo8?e=download&nonce=pvee233ijmg3u&user=02864383199933671631Z&hash=78345uus286hhvnat5cg2pqdpkqkvqiv [following]\n--2020-05-02 17:51:19-- https://doc-04-60-docs.googleusercontent.com/docs/securesc/klc24bgsvh27l2s553lm5mif6bdal7bk/7u9jiamn68gk60nq9463uivp5449u3nr/1588431075000/02695332612409210478/02864383199933671631Z/1l_aanrxHj14d_QkCpein8wFmainNAzo8?e=download&nonce=pvee233ijmg3u&user=02864383199933671631Z&hash=78345uus286hhvnat5cg2pqdpkqkvqiv\nConnecting to doc-04-60-docs.googleusercontent.com (doc-04-60-docs.googleusercontent.com)|173.194.221.132|:443...connected.\nHTTP request sent, awaiting response...200 OK\nLength: unspecified [application/octet-stream]\nSaving to: ‘models/os2d_v2-train.pth’\n\nmodels/os2d_v2-trai [ <=> ] 39,01M 31,7MB/s in 1,2s \n\n2020-05-02 17:51:20 (31,7 MB/s) - ‘models/os2d_v2-train.pth’ saved [40904550]\n\n"
],
[
"cfg.init.model = \"models/os2d_v2-train.pth\"\nnet, box_coder, criterion, img_normalization, optimizer_state = build_os2d_from_config(cfg)",
"2020-05-02 17:51:32,089 OS2D INFO: Building the OS2D model\n2020-05-02 17:51:34,424 OS2D INFO: Creating model on one GPU\n2020-05-02 17:51:34,453 OS2D INFO: Reading model file models/os2d_v2-train.pth\n2020-05-02 17:51:34,543 OS2D INFO: Loaded complete model from checkpoint\n2020-05-02 17:51:34,546 OS2D INFO: Cannot find 'optimizer' in the checkpoint file. Initializing optimizer from scratch.\n2020-05-02 17:51:34,549 OS2D INFO: OS2D has 139 blocks of 10169478 parameters (before freezing)\n2020-05-02 17:51:34,551 OS2D INFO: OS2D has 139 blocks of 10169478 trainable parameters\n"
]
],
[
[
"Get the image where to detect and two class images.",
"_____no_output_____"
]
],
[
[
"input_image = read_image(\"data/demo/input_image.jpg\")\nclass_images = [read_image(\"data/demo/class_image_0.jpg\"),\n read_image(\"data/demo/class_image_1.jpg\")]\nclass_ids = [0, 1]",
"_____no_output_____"
]
],
[
[
"Use torchvision to convert images to torch.Tensor and to apply normalization.",
"_____no_output_____"
]
],
[
[
"transform_image = transforms.Compose([\n transforms.ToTensor(),\n transforms.Normalize(img_normalization[\"mean\"], img_normalization[\"std\"])\n ])",
"_____no_output_____"
]
],
[
[
"Prepare the input image",
"_____no_output_____"
]
],
[
[
"h, w = get_image_size_after_resize_preserving_aspect_ratio(h=input_image.size[1],\n w=input_image.size[0],\n target_size=1500)\ninput_image = input_image.resize((w, h))\n\ninput_image_th = transform_image(input_image)\ninput_image_th = input_image_th.unsqueeze(0)\nif cfg.is_cuda:\n input_image_th = input_image_th.cuda()\n",
"_____no_output_____"
]
],
[
[
"Prepare the class images",
"_____no_output_____"
]
],
[
[
"class_images_th = []\nfor class_image in class_images:\n h, w = get_image_size_after_resize_preserving_aspect_ratio(h=class_image.size[1],\n w=class_image.size[0],\n target_size=cfg.model.class_image_size)\n class_image = class_image.resize((w, h))\n\n class_image_th = transform_image(class_image)\n if cfg.is_cuda:\n class_image_th = class_image_th.cuda()\n\n class_images_th.append(class_image_th)",
"_____no_output_____"
]
],
[
[
"Run the network with one command",
"_____no_output_____"
]
],
[
[
"with torch.no_grad():\n loc_prediction_batch, class_prediction_batch, _, fm_size, transform_corners_batch = net(images=input_image_th, class_images=class_images_th)",
"_____no_output_____"
]
],
[
[
"Alternatively one can run the stages of the model separatly, which is convenient, e.g., for sharing class feature extraction between many input images.",
"_____no_output_____"
]
],
[
[
"# with torch.no_grad():\n# feature_map = net.net_feature_maps(input_image_th)\n\n# class_feature_maps = net.net_label_features(class_images_th)\n# class_head = net.os2d_head_creator.create_os2d_head(class_feature_maps)\n\n# loc_prediction_batch, class_prediction_batch, _, fm_size, transform_corners_batch = net(class_head=class_head,\n# feature_maps=feature_map)\n",
"_____no_output_____"
]
],
[
[
"Convert image organized in batches into images organized in pyramid levels. Not needed in the demo, but essential for multiple images in a batch and multiple pyramid levels.",
"_____no_output_____"
]
],
[
[
"\nimage_loc_scores_pyramid = [loc_prediction_batch[0]]\nimage_class_scores_pyramid = [class_prediction_batch[0]]\nimg_size_pyramid = [FeatureMapSize(img=input_image_th)]\ntransform_corners_pyramid = [transform_corners_batch[0]]\n",
"_____no_output_____"
]
],
[
[
"Decode network outputs into detection boxes",
"_____no_output_____"
]
],
[
[
"boxes = box_coder.decode_pyramid(image_loc_scores_pyramid, image_class_scores_pyramid,\n img_size_pyramid, class_ids,\n nms_iou_threshold=cfg.eval.nms_iou_threshold,\n nms_score_threshold=cfg.eval.nms_score_threshold,\n transform_corners_pyramid=transform_corners_pyramid)\n\n# remove some fields to lighten visualization \nboxes.remove_field(\"default_boxes\")\n",
"_____no_output_____"
],
[
"# Note that the system outputs the correaltions that lie in the [-1, 1] segment as the detection scores (the higher the better the detection).\nscores = boxes.get_field(\"scores\")",
"_____no_output_____"
]
],
[
[
"Show class images",
"_____no_output_____"
]
],
[
[
"figsize = (8, 8)\nfig=plt.figure(figsize=figsize)\ncolumns = len(class_images)\nfor i, class_image in enumerate(class_images):\n fig.add_subplot(1, columns, i + 1)\n plt.imshow(class_image)\n plt.axis('off')\n",
"_____no_output_____"
]
],
[
[
"Show fixed number of detections that are above a certain threshold. Yellow rectangles show detection boxes. Each box has a class label and the detection scores (the higher the better the detection). Red parallelograms illustrate the affine transformations that align class images to the input image at the location of detection.",
"_____no_output_____"
]
],
[
[
"plt.rcParams[\"figure.figsize\"] = figsize\n\ncfg.visualization.eval.max_detections = 8\ncfg.visualization.eval.score_threshold = float(\"-inf\")\nvisualizer.show_detections(boxes, input_image,\n cfg.visualization.eval)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4e651e6a43a495f4cacfd6dcbc5233a71b3341 | 84,984 | ipynb | Jupyter Notebook | Classification/Radius Neighbors/RadiusNeighborsClassifier_MinMaxScaler.ipynb | shreepad-nade/ds-seed | 93ddd3b73541f436b6832b94ca09f50872dfaf10 | [
"Apache-2.0"
] | 53 | 2021-08-28T07:41:49.000Z | 2022-03-09T02:20:17.000Z | Classification/Radius Neighbors/RadiusNeighborsClassifier_MinMaxScaler.ipynb | shreepad-nade/ds-seed | 93ddd3b73541f436b6832b94ca09f50872dfaf10 | [
"Apache-2.0"
] | 142 | 2021-07-27T07:23:10.000Z | 2021-08-25T14:57:24.000Z | Classification/Radius Neighbors/RadiusNeighborsClassifier_MinMaxScaler.ipynb | shreepad-nade/ds-seed | 93ddd3b73541f436b6832b94ca09f50872dfaf10 | [
"Apache-2.0"
] | 38 | 2021-07-27T04:54:08.000Z | 2021-08-23T02:27:20.000Z | 116.736264 | 47,454 | 0.846795 | [
[
[
"# RadiusNeighborsClassifier with MinMaxScaler ",
"_____no_output_____"
],
[
"This Code template is for the Classification task using a simple Radius Neighbor Classifier, with data being scaled by MinMaxScaler. It implements learning based on the number of neighbors within a fixed radius r of each training point, where r is a floating-point value specified by the user.",
"_____no_output_____"
],
[
"### Required Packages",
"_____no_output_____"
]
],
[
[
"!pip install imblearn",
"_____no_output_____"
],
[
"import warnings \r\nimport numpy as np \r\nimport pandas as pd \r\nimport matplotlib.pyplot as plt \r\nimport seaborn as se \r\nfrom imblearn.over_sampling import RandomOverSampler\r\nfrom sklearn.preprocessing import LabelEncoder, MinMaxScaler \r\nfrom sklearn.model_selection import train_test_split \r\nfrom sklearn.neighbors import RadiusNeighborsClassifier\r\nfrom sklearn.pipeline import make_pipeline\r\nfrom sklearn.metrics import classification_report,plot_confusion_matrix\r\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"### Initialization\n\nFilepath of CSV file",
"_____no_output_____"
]
],
[
[
"#filepath\r\nfile_path= \"\"",
"_____no_output_____"
]
],
[
[
"List of features which are required for model training .",
"_____no_output_____"
]
],
[
[
"#x_values\nfeatures=[]",
"_____no_output_____"
]
],
[
[
"Target feature for prediction.",
"_____no_output_____"
]
],
[
[
"#y_value\ntarget=''",
"_____no_output_____"
]
],
[
[
"### Data Fetching\n\nPandas is an open-source, BSD-licensed library providing high-performance, easy-to-use data manipulation and data analysis tools.\n\nWe will use panda's library to read the CSV file using its storage path.And we use the head function to display the initial row or entry.",
"_____no_output_____"
]
],
[
[
"df=pd.read_csv(file_path)\ndf.head()",
"_____no_output_____"
]
],
[
[
"### Feature Selections\n\nIt is the process of reducing the number of input variables when developing a predictive model. Used to reduce the number of input variables to both reduce the computational cost of modelling and, in some cases, to improve the performance of the model.\n\nWe will assign all the required input features to X and target/outcome to Y.",
"_____no_output_____"
]
],
[
[
"X = df[features]\nY = df[target]",
"_____no_output_____"
]
],
[
[
"### Data Preprocessing\n\nSince the majority of the machine learning models in the Sklearn library doesn't handle string category data and Null value, we have to explicitly remove or replace null values. The below snippet have functions, which removes the null value if any exists. And convert the string classes data in the datasets by encoding them to integer classes.\n",
"_____no_output_____"
]
],
[
[
"def NullClearner(df):\n if(isinstance(df, pd.Series) and (df.dtype in [\"float64\",\"int64\"])):\n df.fillna(df.mean(),inplace=True)\n return df\n elif(isinstance(df, pd.Series)):\n df.fillna(df.mode()[0],inplace=True)\n return df\n else:return df\ndef EncodeX(df):\n return pd.get_dummies(df)\ndef EncodeY(df):\n if len(df.unique())<=2:\n return df\n else:\n un_EncodedT=np.sort(pd.unique(df), axis=-1, kind='mergesort')\n df=LabelEncoder().fit_transform(df)\n EncodedT=[xi for xi in range(len(un_EncodedT))]\n print(\"Encoded Target: {} to {}\".format(un_EncodedT,EncodedT))\n return df",
"_____no_output_____"
],
[
"x=X.columns.to_list()\nfor i in x:\n X[i]=NullClearner(X[i]) \nX=EncodeX(X)\nY=EncodeY(NullClearner(Y))\nX.head()",
"Encoded Target: ['DrugY' 'drugA' 'drugB' 'drugC' 'drugX'] to [0, 1, 2, 3, 4]\n"
]
],
[
[
"#### Correlation Map\n\nIn order to check the correlation between the features, we will plot a correlation matrix. It is effective in summarizing a large amount of data where the goal is to see patterns.",
"_____no_output_____"
]
],
[
[
"f,ax = plt.subplots(figsize=(18, 18))\nmatrix = np.triu(X.corr())\nse.heatmap(X.corr(), annot=True, linewidths=.5, fmt= '.1f',ax=ax, mask=matrix)\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Distribution Of Target Variable",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize = (10,6))\nse.countplot(Y)",
"_____no_output_____"
]
],
[
[
"### Data Splitting\n\nThe train-test split is a procedure for evaluating the performance of an algorithm. The procedure involves taking a dataset and dividing it into two subsets. The first subset is utilized to fit/train the model. The second subset is used for prediction. The main motive is to estimate the performance of the model on new data.",
"_____no_output_____"
]
],
[
[
"x_train,x_test,y_train,y_test=train_test_split(X,Y,test_size=0.2,random_state=123)",
"_____no_output_____"
]
],
[
[
"#### Handling Target Imbalance\n\nThe challenge of working with imbalanced datasets is that most machine learning techniques will ignore, and in turn have poor performance on, the minority class, although typically it is performance on the minority class that is most important.\n\nOne approach to addressing imbalanced datasets is to oversample the minority class. The simplest approach involves duplicating examples in the minority class.We will perform overspampling using imblearn library. ",
"_____no_output_____"
]
],
[
[
"x_train,y_train = RandomOverSampler(random_state=123).fit_resample(x_train, y_train)",
"_____no_output_____"
]
],
[
[
"### Model\n\nRadiusNeighborsClassifier implements learning based on the number of neighbors within a fixed radius of each training point, where is a floating-point value specified by the user.\nIn cases where the data is not uniformly sampled, radius-based neighbors classification can be a better choice.\n\n#### Tuning parameters\n\n> **radius**: Range of parameter space to use by default for radius_neighbors queries.\n\n> **algorithm**: Algorithm used to compute the nearest neighbors:\n\n> **leaf_size**: Leaf size passed to BallTree or KDTree. \n\n> **p**: Power parameter for the Minkowski metric.\n\n> **metric**: the distance metric to use for the tree. \n\n> **outlier_label**: label for outlier samples \n\n> **weights**: weight function used in prediction.\n\nFor more information refer: [API](https://scikit-learn.org/stable/modules/generated/sklearn.neighbors.RadiusNeighborsClassifier.html)\n\n#### Data Rescaling\n\nMinMaxScaler subtracts the minimum value in the feature and then divides by the range, where range is the difference between the original maximum and original minimum.",
"_____no_output_____"
]
],
[
[
"# Build Model here\nmodel = make_pipeline(MinMaxScaler(),RadiusNeighborsClassifier(n_jobs=-1))\nmodel.fit(x_train, y_train)",
"_____no_output_____"
]
],
[
[
"#### Model Accuracy\n\nscore() method return the mean accuracy on the given test data and labels.\n\nIn multi-label classification, this is the subset accuracy which is a harsh metric since you require for each sample that each label set be correctly predicted.",
"_____no_output_____"
]
],
[
[
"print(\"Accuracy score {:.2f} %\\n\".format(model.score(x_test,y_test)*100))",
"Accuracy score 37.50 %\n\n"
]
],
[
[
"#### Confusion Matrix\n\nA confusion matrix is utilized to understand the performance of the classification model or algorithm in machine learning for a given test set where results are known.",
"_____no_output_____"
]
],
[
[
"plot_confusion_matrix(model,x_test,y_test,cmap=plt.cm.Blues)",
"_____no_output_____"
]
],
[
[
"#### Classification Report\nA Classification report is used to measure the quality of predictions from a classification algorithm. How many predictions are True, how many are False.\n\n* **where**:\n - Precision:- Accuracy of positive predictions.\n - Recall:- Fraction of positives that were correctly identified.\n - f1-score:- percent of positive predictions were correct\n - support:- Support is the number of actual occurrences of the class in the specified dataset.",
"_____no_output_____"
]
],
[
[
"print(classification_report(y_test,model.predict(x_test)))",
" precision recall f1-score support\n\n 0 0.00 0.00 0.00 23\n 1 0.12 0.50 0.20 2\n 2 0.22 0.67 0.33 3\n 3 0.60 1.00 0.75 3\n 4 0.50 1.00 0.67 9\n\n accuracy 0.38 40\n macro avg 0.29 0.63 0.39 40\nweighted avg 0.18 0.38 0.24 40\n\n"
]
],
[
[
" #### Creator: Viraj Jayant, Github: [Profile](https://github.com/Viraj-Jayant/)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb4e6f7c1cf8c76ecaea3b77e93101842d414c14 | 73,699 | ipynb | Jupyter Notebook | Simulation.ipynb | PuneethKouloorkar/MD_openmm | 0dec746d251469e9b3bb68bb8d17394806472af6 | [
"MIT"
] | null | null | null | Simulation.ipynb | PuneethKouloorkar/MD_openmm | 0dec746d251469e9b3bb68bb8d17394806472af6 | [
"MIT"
] | null | null | null | Simulation.ipynb | PuneethKouloorkar/MD_openmm | 0dec746d251469e9b3bb68bb8d17394806472af6 | [
"MIT"
] | null | null | null | 187.053299 | 31,956 | 0.9043 | [
[
[
"# SIMULATE THE SYSTEM",
"_____no_output_____"
]
],
[
[
"import simtk.openmm as mm # Main OpenMM functionality\nimport simtk.openmm.app as app # Application layer (handy interface)\nimport simtk.unit as unit # Unit/quantity handling\nimport mdtraj\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport os",
"_____no_output_____"
],
[
"cwd = cwd = os.getcwd() \nsolvent_solute_system = os.path.join(cwd, 'files/6a5j_protein/solv.pdb') \n\nmolecule = app.PDBFile(solvent_solute_system) # Load the solvated peptide",
"_____no_output_____"
]
],
[
[
"### (a) System object from topology of solvated peptide",
"_____no_output_____"
]
],
[
[
"forcefield = app.ForceField(\"amber14/protein.ff14SB.xml\", \"amber14/tip3p.xml\")\n\nsystem = forcefield.createSystem(\n molecule.topology,\n nonbondedMethod=app.PME, # Non-bonded interactions\n nonbondedCutoff=1 * unit.nanometer, # Cut-off of non-bonded interactions \n constraints=app.HBonds\n )",
"_____no_output_____"
]
],
[
[
"### (b) Simulation using Langevin integrator in timesteps of 2 fs at 300k",
"_____no_output_____"
]
],
[
[
"integrator = mm.LangevinIntegrator(300.*unit.kelvin, 1./unit.picosecond, 2.*unit.femtoseconds)\n\nsimulation = app.Simulation(\n molecule.topology, # Topology\n system, # System\n integrator, # Integrator\n mm.Platform.getPlatformByName('CPU') # Platform = 'CPU' or 'CUDA'\n)",
"_____no_output_____"
],
[
"simulation.context.setPositions(molecule.positions) # Add the current atomic positions of the solvated peptide \n # to the context of the simulation.",
"_____no_output_____"
]
],
[
[
"### (c) Energy minimization of the system",
"_____no_output_____"
]
],
[
[
"simulation.minimizeEnergy()",
"_____no_output_____"
],
[
"state = simulation.context.getState(getPositions=True) # New co-ordinates\nmolecule.positions = state.getPositions() ",
"_____no_output_____"
],
[
"minimized_system = os.path.join(cwd, 'files/6a5j_protein/min.pdb')\n\nwith open(minimized_system, \"w\") as file_:\n molecule.writeFile(\n molecule.topology, molecule.positions,\n file=file_\n )\n \n# One can visualize this in VMD and see the difference between original and the minimised atoms",
"_____no_output_____"
]
],
[
[
"### (c) Equilibration of system in the NVT ensemble for 100 ps",
"_____no_output_____"
]
],
[
[
"molecule = app.PDBFile(minimized_system)\nsimulation.context.setPositions(molecule.positions)",
"_____no_output_____"
],
[
"run_length = 50000 # 50000 * 2 fs = 100 ps\n\nequilibration_log = os.path.join(cwd, 'files/6a5j_protein/equilibration.log')\n\nsimulation.reporters.append(\n app.StateDataReporter(\n equilibration_log, 500, step=True, # 500 = Write every 500th step\n potentialEnergy=True, totalEnergy=True,\n temperature=True, progress=True,\n remainingTime=True, speed=True,\n totalSteps=run_length,\n separator='\\t')\n )",
"_____no_output_____"
],
[
"simulation.step(run_length) # Run the simulation",
"_____no_output_____"
],
[
"save_state = os.path.join(cwd, 'files/6a5j_protein/eq.xml')\nsimulation.saveState(save_state)",
"_____no_output_____"
],
[
"pot_e = []\ntot_e = []\ntemperature = []\n\nwith open(equilibration_log) as file_:\n for line in file_:\n if line.startswith(\"#\"):\n continue\n pot_e_, tot_e_, temperature_ = line.split()[2:5]\n pot_e.append(float(pot_e_))\n tot_e.append(float(tot_e_))\n temperature.append(float(temperature_))",
"_____no_output_____"
],
[
"plt.rcParams[\"figure.figsize\"] = (10,7)\nt = range(1, 101)\n\nfig, ax = plt.subplots()\nax.plot(t, [x / 1000 for x in pot_e], label=\"potential\")\nax.plot(t, [x / 1000 for x in tot_e], label=\"total\")\n\nax.set(**{\n \"xlabel\": \"time / ps\",\n \"xlim\": (0, 100),\n \"ylabel\": \"energy / 10$^{3}$ kJ mol$^{-1}$\"\n })\n\nax.legend(\n framealpha=1,\n edgecolor=\"k\",\n fancybox=False\n)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nax.plot(t, temperature)\n\nax.set(**{\n \"xlabel\": \"time / ps\",\n \"xlim\": (0, 100),\n \"ylabel\": \"temperature / K\"\n })",
"_____no_output_____"
]
],
[
[
"### (d) Production run (in CUDA)",
"_____no_output_____"
]
],
[
[
"simulation.loadState(save_state)\n\nsimulation = os.path.join(cwd, 'files/6a5j_protein/simulation.log')\n\nsimulation.reporters = [] # Reset the simulation reporters\n\nrun_length = 375000000 # 375000000 * 2 fs = 750 ns\nsimulation.reporters.append( \n app.StateDataReporter( # State reporter that appends potential energy\n simulation, 5000, step=True,\n potentialEnergy=True,\n temperature=True, progress=True,\n remainingTime=True, speed=True,\n totalSteps=run_length,\n separator='\\t')\n )\n\nproduction_dcd = os.path.join(cwd, 'files/6a5j_protein/prod_run.dcd')\n\nsimulation.reporters.append(mdtraj.reporters.DCDReporter(\n production_dcd, 5000, # Structure reporter that appends positions\n atomSubset=range(260)) # atomSubset = Save just the peptide atoms\n ) ",
"_____no_output_____"
],
[
"simulation.step(run_length)\n# One can visualize the saved .dcd file in VMD and compare with th original",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb4e71efff440725ba72bec3c5d88c7a103ad09e | 356,812 | ipynb | Jupyter Notebook | causal-inference-for-the-brave-and-true/03-Stats-Review-The-Most-Dangerous-Equation.ipynb | qiringji/python-causality-handbook | add5ab57a8e755242bdbc3d4d0ee00867f6a1e55 | [
"MIT"
] | 1 | 2021-07-07T03:57:54.000Z | 2021-07-07T03:57:54.000Z | causal-inference-for-the-brave-and-true/03-Stats-Review-The-Most-Dangerous-Equation.ipynb | adebayoj/python-causality-handbook | bae5790bba173c89dedacbe6bcd3d65c1dc20a07 | [
"MIT"
] | null | null | null | causal-inference-for-the-brave-and-true/03-Stats-Review-The-Most-Dangerous-Equation.ipynb | adebayoj/python-causality-handbook | bae5790bba173c89dedacbe6bcd3d65c1dc20a07 | [
"MIT"
] | null | null | null | 388.684096 | 153,780 | 0.926659 | [
[
[
"# 03 - Stats Review: The Most Dangerous Equation\n\nIn his famous article of 2007, Howard Wainer writes about very dangerous equations:\n\n\"Some equations are dangerous if you know them, and others are dangerous if you do not. The first category may pose danger because the secrets within its bounds open doors behind which lies terrible peril. The obvious winner in this is Einstein’s ionic equation \\\\(E = MC^2\\\\), for it provides a measure of the enormous energy hidden within ordinary matter. \\[...\\] Instead I am interested in equations that unleash their danger not when we know about them, but rather when we do not. Kept close at hand, these equations allow us to understand things clearly, but their absence leaves us dangerously ignorant.\"\n\nThe equation he talks about is Moivre’s equation:\n\n$\nSE = \\dfrac{\\sigma}{\\sqrt{n}} \n$\n\nwhere \\\\(SE\\\\) is the standard error of the mean, \\\\(\\sigma\\\\) is the standard deviation and \\\\(n\\\\) is the sample size. Sounds like a piece of math the brave and true should master, so let's get to it.\n\nTo see why not knowing this equation is very dangerous, let's take a look at some education data. I've compiled data on ENEM scores (Brazilian standardised high school scores, similar to SAT) from different schools for a period of 3 years. I also did some cleaning on the data to keep only the information relevant to us. The original data can be downloaded in the [Inep website](http://portal.inep.gov.br/web/guest/microdados#).\n\nIf we look at the top performing school, something catches the eye: those schools have a fairly small number of students. ",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')\n\nimport pandas as pd\nimport numpy as np\nfrom scipy import stats\nimport seaborn as sns\nfrom matplotlib import pyplot as plt\nfrom matplotlib import style\nstyle.use(\"fivethirtyeight\")\n\ndf = pd.read_csv(\"./data/enem_scores.csv\")\ndf.sort_values(by=\"avg_score\", ascending=False).head(10)",
"_____no_output_____"
]
],
[
[
"Looking at it from another angle, we can separate only the 1% top schools and study them. What are they like? Perhaps we can learn something from the best and replicate it elsewhere. And sure enough, if we look at the top 1% schools, we figure out they have, on average, fewer students.",
"_____no_output_____"
]
],
[
[
"plot_data = (df\n .assign(top_school = df[\"avg_score\"] >= np.quantile(df[\"avg_score\"], .99))\n [[\"top_school\", \"number_of_students\"]]\n .query(f\"number_of_students<{np.quantile(df['number_of_students'], .98)}\")) # remove outliers\n\nplt.figure(figsize=(6,6))\nsns.boxplot(x=\"top_school\", y=\"number_of_students\", data=plot_data)\nplt.title(\"Number of Students of 1% Top Schools (Right)\");",
"_____no_output_____"
]
],
[
[
"One natural conclusion that follows is that small schools lead to higher academic performance. This makes intuitive sense, since we believe that less students per teacher allows the teacher to give focused attention to each student. But what does this have to do with Moivre’s equation? And why is it dangerous? \n\nWell, it becomes dangerous once people start to make important and expensive decisions based on this information. In his article, Howard continues:\n\n\"In the 1990s, it became popular to champion reductions in the size of schools. Numerous philanthropic organisations and government agencies funded the division of larger schools based on the fact that students at small schools are over represented in groups with high test scores.\"\n\nWhat people forgot to do was to look also at the bottom 1% of schools. If we do that, lo and behold! They also have very few students!",
"_____no_output_____"
]
],
[
[
"q_99 = np.quantile(df[\"avg_score\"], .99)\nq_01 = np.quantile(df[\"avg_score\"], .01)\n\nplot_data = (df\n .sample(10000)\n .assign(Group = lambda d: np.select([d[\"avg_score\"] > q_99, d[\"avg_score\"] < q_01],\n [\"Top\", \"Bottom\"], \"Middle\")))\nplt.figure(figsize=(10,5))\nsns.scatterplot(y=\"avg_score\", x=\"number_of_students\", hue=\"Group\", data=plot_data)\nplt.title(\"ENEM Score by Number of Students in the School\");",
"_____no_output_____"
]
],
[
[
"What we are seeing above is exactly what is expected according to the Moivre’s equation. As the number of students grows, the average score becomes more and more precise. Schools with very few samples can have very high and very low scores simply due to chance. This is less likley to occur with large schools. Moivre’s equation talks about a fundamental fact about the reality of information and records in the form of data: it is always imprecise. The question then becomes how imprecise.\n\nStatistics is the science that deals with these imprecisions so they don't catch us off-guard. As Taleb puts it in his book, Fooled by Randomness:\n\n> Probability is not a mere computation of odds on the dice or more complicated variants; it is the acceptance of the lack of certainty in our knowledge and the development of methods for dealing with our ignorance.\n\nOne way to quantify our uncertainty is the **variance of our estimates**. Variance tells us how much observation deviates from their central and most probably value. As indicated by Moivre’s equation, this uncertainty shrinks as the amount of data we observe increases. This makes sense, right? If we see lots and lots of students performing excellently at a school, we can be more confident that this is indeed a good school. However, if we see a school with only 10 students and 8 of them perform well, we need to be more suspicious. It could be that, by chance, that school got some above average students.\n\nThe beautiful triangular plot we see above tells exactly this story. It shows us how our estimates of the school performance has a huge variance when the sample sizes are small. It also shows that variance shrinks as the sample size increases. This is true for the average score in a school, but it is also true about any summary statistics that we have, including the ATE we so often want to estimate.\n\n## The Standard Error of Our Estimates\n\nSince this is just a review on statistics, I'll take the liberty to go a bit faster now. If you are not familiar with distributions, variance and standard errors, please, do read on, but keep in mind that you might need some additional resources. I suggest you google any MIT course on introduction to statistics. They are usually quite good.\n\nIn the previous section, we estimated the average treatment effect \\\\(E[Y_1-Y_0]\\\\) as the difference in the means between the treated and the untreated \\\\(E[Y|T=1]-E[Y|T=0]\\\\). As our motivating example, we figured out the \\\\(ATE\\\\) for online classes. We also saw that it was a negative impact, that is, online classes made students perform about 5 points worse than the students with face to face classes. Now, we get to see if this impact is statistically significant.\n\nTo do so, we need to estimate the \\\\(SE\\\\). We already have \\\\(n\\\\), our sample size. To get the estimate for the standard deviation we can do the following\n\n$\n\\hat{\\sigma}=\\frac{1}{N-1}\\sum_{i=0}^N (x-\\bar{x})^2\n$\n\nwhere \\\\(\\bar{x}\\\\) is the mean of \\\\(x\\\\). Fortunately for us, most programming software already implements this. In Pandas, we can use the method [std](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.std.html).",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv(\"./data/online_classroom.csv\")\nonline = data.query(\"format_ol==1\")[\"falsexam\"]\nface_to_face = data.query(\"format_ol==0 & format_blended==0\")[\"falsexam\"]\n\ndef se(y: pd.Series):\n return y.std() / np.sqrt(len(y))\n\nprint(\"SE for Online:\", se(online))\nprint(\"SE for Face to Face:\", se(face_to_face))",
"SE for Online: 1.5371593973041635\nSE for Face to Face: 0.8723511456319106\n"
]
],
[
[
"## Confidence Intervals\n\nThe standard error of our estimate is a measure of confidence. To understand exactly what it means, we need to go into turbulent and polemic statistical waters. For one view of statistics, the frequentist view, we would say that the data we have is nothing more than a manifestation of a true data generating process. This process is abstract and ideal. It is governed by true parameters that are unchanging but also unknown to us. In the context of the students test, if we could run multiple experiments and collect multiple datasets, all would resemble the true underlying data generating process, but wouldn't be exactly like it. This is very much like Plato's writing on the Forms:\n\n> Each [of the essential forms] manifests itself in a great variety of combinations, with actions, with material things, and with one another, and each seems to be many\n\nTo better grasp this, let's suppose we have a true abstract distribution of students' test score. This is a normal distribution with true mean of 74 and true standard deviation of 2. From this distribution, we can run 10000 experiments. On each one, we collect 500 samples. Some experiment data will have a mean lower than the true one, some will be higher. If we plot them in a histogram, we can see that means of the experiments are distributed around the true mean.",
"_____no_output_____"
]
],
[
[
"true_std = 2\ntrue_mean = 74\n\nn = 500\ndef run_experiment(): \n return np.random.normal(true_mean,true_std, 500)\n\nnp.random.seed(42)\n\nplt.figure(figsize=(8,5))\nfreq, bins, img = plt.hist([run_experiment().mean() for _ in range(10000)], bins=40, label=\"Experiment Means\")\nplt.vlines(true_mean, ymin=0, ymax=freq.max(), linestyles=\"dashed\", label=\"True Mean\", color=\"orange\")\nplt.legend();\n",
"_____no_output_____"
]
],
[
[
"Notice that we are talking about the mean of means here. So, by chance, we could have an experiment where the mean is somewhat below or above the true mean. This is to say that we can never be sure that the mean of our experiment matches the true platonic and ideal mean. However, **with the standard error, we can create an interval that will contain the true mean 95% of the time**.\n\nIn real life, we don't have the luxury of simulating the same experiment with multiple datasets. We often only have one. But we can draw on the intuition above to construct what we call **confidence intervals**. Confidence intervals come with a probability attached to them. The most common one is 95%. This probability tells us how many of the hypothetical confidence intervals we would build from different studies contain the true mean. For example, the 95% confidence intervals computed from many similar studies would contain the true mean 95% of the time. \n\nTo calculate the confidence interval, we use what is called the **central limit theorem**. This theorem states that **means of experiments are normally distributed**. From statistical theory, we know that 95% of the mass of a normal distribution is between 2 standard deviations above and below the mean. Technically, 1.96, but 2 is close enough. \n\n\n\nThe Standard Error of the mean serves as our estimate of the distribution of the experiment means. So, if we multiply it by 2 and add and subtract it from the mean of one of our experiments, we will construct a 95% confidence interval for the true mean.",
"_____no_output_____"
]
],
[
[
"np.random.seed(321)\nexp_data = run_experiment()\nexp_se = exp_data.std() / np.sqrt(len(exp_data))\nexp_mu = exp_data.mean()\nci = (exp_mu - 2 * exp_se, exp_mu + 2 * exp_se)\nprint(ci)",
"(73.82718114045632, 74.17341543460314)\n"
],
[
"x = np.linspace(exp_mu - 4*exp_se, exp_mu + 4*exp_se, 100)\ny = stats.norm.pdf(x, exp_mu, exp_se)\nplt.plot(x, y)\nplt.vlines(ci[1], ymin=0, ymax=1)\nplt.vlines(ci[0], ymin=0, ymax=1, label=\"95% CI\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"Of course, we don't need to restrict ourselves to the 95% confidence interval. We could generate the 99% interval by finding what we need to multiply the standard deviation by so the interval contains 99% of the mass of a normal distribution. \n\nThe function `ppf` in python gives us the inverse of the CDF. So, `ppf(0.5)` will return 0.0, saying that 50% of the mass of the standard normal distribution is below 0.0. By the same token, if we plug 99.5%, we will have the value `z`, such that 99.5% of the distribution mass falls below this value. In other words, 0.05% of the mass falls above this value. Instead of multiplying the standard error by 2 like we did to find the 95% CI, we will multiply it by `z`, which will result in the 99% CI.",
"_____no_output_____"
]
],
[
[
"from scipy import stats\nz = stats.norm.ppf(.995)\nprint(z)\nci = (exp_mu - z * exp_se, exp_mu + z * exp_se)\nci",
"2.5758293035489004\n"
],
[
"x = np.linspace(exp_mu - 4*exp_se, exp_mu + 4*exp_se, 100)\ny = stats.norm.pdf(x, exp_mu, exp_se)\nplt.plot(x, y)\nplt.vlines(ci[1], ymin=0, ymax=1)\nplt.vlines(ci[0], ymin=0, ymax=1, label=\"99% CI\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"Back to our classroom experiment, we can construct the confidence interval for the mean exam score for both the online and face to face students' group",
"_____no_output_____"
]
],
[
[
"def ci(y: pd.Series):\n return (y.mean() - 2 * se(y), y.mean() + 2 * se(y))\n\nprint(\"95% CI for Online:\", ci(online))\nprint(\"95% for Face to Face:\", ci(face_to_face))",
"95% CI for Online: (70.56094429049804, 76.7095818797147)\n95% for Face to Face: (76.80278229206951, 80.29218687459715)\n"
]
],
[
[
"What we can see is that the 95% CI of the groups don't overlap. The lower end of the CI for Face to Face class is above the upper end of the CI for online classes. This is evidence that our result is not by chance, and that the true mean for students in face to face clases is higher than the true mean for students in online classes. In other words, there is a significant causal decrease in academic performance when switching from face to face to online classes.\n\nAs a recap, confidence intervals are a way to place uncertainty around our estimates. The smaller the sample size, the larger the standard error and the wider the confidence interval. Finally, you should always be suspicious of measurements without any uncertainty metric attached to it. Since they are super easy to compute, lack of confidence intervals signals either some bad intentions or simply lack of knowledge, which is equally concerning. \n\n\n\nOne final word of caution here. Confidence intervals are trickier to interpret than at first glance. For instance, I **shouldn't** say that this particular 95% confidence interval contains the true population mean with 95% chance. That's because in frequentist statistics, the one that uses confidence intervals, the population mean is regarded as a true population constant. So it either is or isn't in our particular confidence interval. In other words, our particular confidence interval either contains or doesn't contain the true mean. If it does, the chance of containing it would be 100%, not 95%. If it doesn't, the chance would be 0%. Rather, in confidence intervals, the 95% refers to the frequency that such confidence intervals, computed in many many studies, contain the true mean. 95% is our confidence in the algorithm used to compute the 95% CI, not on the particular interval itself.\n\nNow, having said that, as an Economist (statisticians, please look away now), I think this purism is not very useful. In practice, you will see people saying that the particular confidence interval contains the true mean 95% of the time. Although wrong, this is not very harmful, as it still places a precise degree of uncertainty in our estimates. Moreover, if we switch to Bayesian statistics and use probable intervals instead of confidence intervals, we would be able to say that the interval contains the distribution mean 95% of the time. Also, from what I've seen in practice, with decent sample sizes, bayesian probability intervals are more similar to confidence intervals than both bayesian and frequentists would like to admit. So, if my word counts for anything, feel free to say whatever you want about your confidence interval. I don't care if you say they contain the true mean 95% of the time. Just, please, never forget to place them around your estimates, otherwise you will look silly. \n\n\n## Hypothesis Testing\n\nAnother way to incorporate uncertainty is to state a hypothesis test: is the difference in means statistically different from zero (or any other value)? To do so, we will recall that the sum or difference of 2 normal distributions is also a normal distribution. The resulting mean will be the sum or difference between the two distributions, while the variance will always be the sum of the variance:\n\n$\nN(\\mu_1, \\sigma_1^2) - N(\\mu_2, \\sigma_2^2) = N(\\mu_1 - \\mu_2, \\sigma_1^2 + \\sigma_2^2)\n$\n\n$\nN(\\mu_1, \\sigma_1^2) + N(\\mu_2, \\sigma_2^2) = N(\\mu_1 + \\mu_2, \\sigma_1^2 + \\sigma_2^2)\n$\n\nIf you don't recall, its OK. We can always use code and simulated data to check:",
"_____no_output_____"
]
],
[
[
"np.random.seed(123)\nn1 = np.random.normal(4, 3, 30000)\nn2 = np.random.normal(1, 4, 30000)\nn_diff = n2 - n1\nsns.distplot(n1, hist=False, label=\"N(4,3)\")\nsns.distplot(n2, hist=False, label=\"N(1,4)\")\nsns.distplot(n_diff, hist=False, label=f\"N(4,3) - N(1,4) = N(-1, 5)\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"If we take the distribution of the means of our 2 groups and subtract one from the other, we will have a third distribution. The mean of this final distribution will be the difference in the means and the standard deviation of this distribution will be the square root of the sum of the standard deviations.\n\n$\n\\mu_{diff} = \\mu_1 - \\mu_2\n$\n\n$\nSE_{diff} = \\sqrt{SE_1 + SE_2} = \\sqrt{\\sigma_1^2/n_1 + \\sigma_2^2/n_2}\n$\n\nLet's return to our classroom example. We will construct this distribution of the difference. Of course, once we have it, building the 95% CI is very easy.",
"_____no_output_____"
]
],
[
[
"diff_mu = online.mean() - face_to_face.mean()\ndiff_se = np.sqrt(face_to_face.var()/len(face_to_face) + online.var()/len(online))\nci = (diff_mu - 1.96*diff_se, diff_mu + 1.96*diff_se)\nprint(ci)",
"(-8.376410208363385, -1.4480327880905248)\n"
],
[
"x = np.linspace(diff_mu - 4*diff_se, diff_mu + 4*diff_se, 100)\ny = stats.norm.pdf(x, diff_mu, diff_se)\nplt.plot(x, y)\nplt.vlines(ci[1], ymin=0, ymax=.05)\nplt.vlines(ci[0], ymin=0, ymax=.05, label=\"95% CI\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"With this at hand, we can say that we are 95% confident that the true difference between the online and face to face group falls between -8.37 and -1.44. We can also construct a **z statistic** by dividing the difference in mean by the \\\\\\(SE\\\\\\\\) of the differences.\n\n$\nz = \\dfrac{\\mu_{diff} - H_{0}}{SE_{diff}} = \\dfrac{(\\mu_1 - \\mu_2) - H_{0}}{\\sqrt{\\sigma_1^2/n_1 + \\sigma_2^2/n_2}}\n$\n\nWhere \\\\(H_0\\\\) is the value which we want to test our difference against.\n\nThe z statistic is a measure of how extreme the observed difference is. To test our hypothesis that the difference in the means is statistically different from zero, we will use contradiction. We will assume that the opposite is true, that is, we will assume that the difference is zero. This is called a null hypothesis, or \\\\(H_0\\\\). Then, we will ask ourselves \"is it likely that we would observe such a difference if the true difference were indeed zero?\" In statistical math terms, we can translate this question to checking how far from zero is our z statistic. \n\nUnder \\\\(H_0\\\\), the z statistic follows a standard normal distribution. So, if the difference is indeed zero, we would see the z statistic within 2 standard deviations of the mean 95% of the time. The direct consequence of this is that if z falls above or below 2 standard deviations, we can reject the null hypothesis with 95% confidence.\n\nLet's see how this looks like in our classroom example.",
"_____no_output_____"
]
],
[
[
"z = diff_mu / diff_se\nprint(z)",
"-2.7792810791031224\n"
],
[
"x = np.linspace(-4,4,100)\ny = stats.norm.pdf(x, 0, 1)\nplt.plot(x, y, label=\"Standard Normal\")\nplt.vlines(z, ymin=0, ymax=.05, label=\"Z statistic\", color=\"C1\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"This looks like a pretty extreme value. Indeed, it is above 2, which means there is less than a 5% chance that we would see such an extreme value if there were no difference in the groups. This again leads us to conclude that switching from face to face to online classes causes a statistically significant drop in academic performance.\n\nOne final interesting thing about hypothesis tests is that it is less conservative than checking if the 95% CI from the treated and untreated group overlaps. In other words, if the confidence intervals in the two groups overlap, it can still be the case that the result is statistically significant. For example, let's pretend that the face-to-face group has an average score of 74 and standard error of 7 and the online group has an average score of 71 with a standard error of 1. ",
"_____no_output_____"
]
],
[
[
"cont_mu, cont_se = (71, 1)\ntest_mu, test_se = (74, 7)\n\ndiff_mu = test_mu - cont_mu\ndiff_se = np.sqrt(cont_se + cont_se)\n\nprint(\"Control 95% CI:\", (cont_mu-1.96*cont_se, cont_mu+1.96*cont_se))\nprint(\"Test 95% CI:\", (test_mu-1.96*test_se, test_mu+1.96*test_se))\nprint(\"Diff 95% CI:\", (diff_mu-1.96*diff_se, diff_mu+1.96*diff_se))",
"Control 95% CI: (69.04, 72.96)\nTest 95% CI: (60.28, 87.72)\nDiff 95% CI: (0.22814141774873375, 5.771858582251266)\n"
]
],
[
[
"If we construct the confidence intervals for these groups, they overlap. The upper bound for the 95% CI of the online group is 72.96 and the lower bound for the face-to-face group is 60.28. However, once we compute the 95% confidence interval for the difference between the groups, we can see that it does not contain zero. In summary, even though the individual confidence intervals overlap, the difference can still be statistically different from zero.\n\n## P-values\n\nI've said previously that there is less than 5% chance that we would observe such an extreme value if the difference between online and face to face groups were actually zero. But can we estimate exactly what is that chance? How likely are we to observe such an extreme value? Enters p-values!\n\nJust like with confidence intervals (and most frequentist statistics, as a matter of fact) the true definition of p-values can be very confusing. So, to not take any risks, I'll copy the definition from Wikipedia: \"the p-value is the probability of obtaining test results at least as extreme as the results actually observed during the test, assuming that the null hypothesis is correct\". \n\nTo put it more succinctly, the p-value is the probability of seeing such data, given that the null-hypothesis is true. It measures how unlikely it is that you are seeing a measurement if the null-hypothesis is true. Naturally, this often gets confused with the probability of the null-hypothesis being true. Note the difference here. The p-value is NOT \\\\(P(H_0|data)\\\\), but rather \\\\(P(data|H_0)\\\\).\n\nBut don't let this complexity fool you. In practical terms, they are pretty straightforward to use.\n\n\n\nTo get the p-value, we need to compute the area under the standard normal distribution before or after the z statistic. Fortunately, we have a computer to do this calculation for us. We can simply plug the z statistic in the CDF of the standard normal distribution.",
"_____no_output_____"
]
],
[
[
"print(\"P-value:\", stats.norm.cdf(z))",
"P-value: 0.0027239680835563383\n"
]
],
[
[
"This means that there is only a 0.2% chance of observing this extreme z statistic if the difference was zero. Notice how the p-value is interesting because it avoids us having to specify a confidence level, like 95% or 99%. But, if we wish to report one, from the p-value, we know exactly at which confidence our test will pass or fail. For instance, with a p-value of 0.0027, we know that we have significance up to the 0.2% level. So, while the 95% CI and the 99% CI for the difference will neither contain zero, the 99.9% CI will.",
"_____no_output_____"
]
],
[
[
"diff_mu = online.mean() - face_to_face.mean()\ndiff_se = np.sqrt(face_to_face.var()/len(face_to_face) + online.var()/len(online))\nprint(\"95% CI:\", (diff_mu - stats.norm.ppf(.975)*diff_se, diff_mu + stats.norm.ppf(.975)*diff_se))\nprint(\"99% CI:\", (diff_mu - stats.norm.ppf(.995)*diff_se, diff_mu + stats.norm.ppf(.995)*diff_se))\nprint(\"99.9% CI:\", (diff_mu - stats.norm.ppf(.9995)*diff_se, diff_mu + stats.norm.ppf(.9995)*diff_se))",
"95% CI: (-8.376346553082909, -1.4480964433710017)\n99% CI: (-9.46485353526404, -0.3595894611898709)\n99.9% CI: (-10.728040658245558, 0.9035976617916459)\n"
]
],
[
[
"## Keys Ideas\n\nWe've seen how important it is to know Moivre’s equation and we used it to place a degree of certainty around our estimates. Namely, we figured out that the online classes cause a decrease in academic performance compared to face to face classes. We also saw that this was a statistically significant result. We did it by comparing the Confidence Intervals of the means for the 2 groups, by looking at the confidence interval for the difference, by doing a hypothesis test and by looking at the p-value. Let's wrap everything up in a single function that does A/B testing comparison like the one we did above",
"_____no_output_____"
]
],
[
[
"def AB_test(test: pd.Series, control: pd.Series, confidence=0.95, h0=0):\n mu1, mu2 = test.mean(), control.mean()\n se1, se2 = test.std() / np.sqrt(len(test)), control.std() / np.sqrt(len(control))\n \n diff = mu1 - mu2\n se_diff = np.sqrt(test.var()/len(test) + control.var()/len(control))\n \n z_stats = (diff-h0)/se_diff\n p_value = stats.norm.cdf(z_stats)\n \n def critial(se): return -se*stats.norm.ppf((1 - confidence)/2)\n \n print(f\"Test {confidence*100}% CI: {mu1} +- {critial(se1)}\")\n print(f\"Control {confidence*100}% CI: {mu2} +- {critial(se2)}\")\n print(f\"Test-Control {confidence*100}% CI: {diff} +- {critial(se_diff)}\")\n print(f\"Z Statistic {z_stats}\")\n print(f\"P-Value {p_value}\")\n \nAB_test(online, face_to_face)",
"Test 95.0% CI: 73.63526308510637 +- 3.0127770572134565\nControl 95.0% CI: 78.54748458333333 +- 1.7097768273108005\nTest-Control 95.0% CI: -4.912221498226955 +- 3.4641250548559537\nZ Statistic -2.7792810791031224\nP-Value 0.0027239680835563383\n"
]
],
[
[
"Since our function is generic enough, we can test other null hypotheses. For instance, can we try to reject that the difference between online and face to face class performance is -1. With the results we get, we can say with 95% confidence that the difference is greater than -1. But we can't say it with 99% confidence:",
"_____no_output_____"
]
],
[
[
"AB_test(online, face_to_face, h0=-1)",
"Test 95.0% CI: 73.63526308510637 +- 3.0127770572134565\nControl 95.0% CI: 78.54748458333333 +- 1.7097768273108005\nTest-Control 95.0% CI: -4.912221498226955 +- 3.4641250548559537\nZ Statistic -2.2134920404560883\nP-Value 0.013431870694630114\n"
]
],
[
[
"## References\n\nI like to think of this entire book as a tribute to Joshua Angrist, Alberto Abadie and Christopher Walters for their amazing Econometrics class. Most of the ideas here are taken from their classes at the American Economic Association. Watching them is what is keeping me sane during this tough year of 2020.\n* [Cross-Section Econometrics](https://www.aeaweb.org/conference/cont-ed/2017-webcasts)\n* [Mastering Mostly Harmless Econometrics](https://www.aeaweb.org/conference/cont-ed/2020-webcasts)\n\nI'll also like to reference the amazing books from Angrist. They have shown me that Econometrics, or 'Metrics as they call it, is not only extremely useful but also profoundly fun.\n\n* [Mostly Harmless Econometrics](https://www.mostlyharmlesseconometrics.com/)\n* [Mastering 'Metrics](https://www.masteringmetrics.com/)\n\nMy final reference is Miguel Hernan and Jamie Robins' book. It has been my trustworthy companion in the most thorny causal questions I had to answer.\n\n* [Causal Inference Book](https://www.hsph.harvard.edu/miguel-hernan/causal-inference-book/)\n\nIn this particular section, I've also referenced The [Most Dangerous Equation](https://www.researchgate.net/publication/255612702_The_Most_Dangerous_Equation), by Howard Wainer.\n\nFinally, if you are curious about the correct interpretation of the statistical concepts we've discussed here, I recommend reading the paper by Greenland et al, 2016: [Statistical tests, P values, confidence intervals, and power: a guide to misinterpretations](https://link.springer.com/content/pdf/10.1007/s10654-016-0149-3.pdf).\n\n\n\n## Contribute\n\nCausal Inference for the Brave and True is an open-source material on causal inference, the statistics of science. It uses only free software, based in Python. Its goal is to be accessible monetarily and intellectually.\nIf you found this book valuable and you want to support it, please go to [Patreon](https://www.patreon.com/causal_inference_for_the_brave_and_true). If you are not ready to contribute financially, you can also help by fixing typos, suggesting edits or giving feedback on passages you didn't understand. Just go to the book's repository and [open an issue](https://github.com/matheusfacure/python-causality-handbook/issues). Finally, if you liked this content, please share it with others who might find it useful and give it a [star on GitHub](https://github.com/matheusfacure/python-causality-handbook/stargazers).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb4e97e87aa6b84bc3955e8790517e1b80ab1ef2 | 156,427 | ipynb | Jupyter Notebook | 3_GenderClassification/notebooks/dockerizer-model-import-gender-classifier.ipynb | Shuu-Ri/aws-nlp-workshop | 888171694c5623a7fd5e19baa3e8cbc22c09bc69 | [
"MIT-0"
] | 54 | 2018-07-18T09:30:45.000Z | 2021-11-29T08:48:54.000Z | 3_GenderClassification/notebooks/dockerizer-model-import-gender-classifier.ipynb | Shuu-Ri/aws-nlp-workshop | 888171694c5623a7fd5e19baa3e8cbc22c09bc69 | [
"MIT-0"
] | null | null | null | 3_GenderClassification/notebooks/dockerizer-model-import-gender-classifier.ipynb | Shuu-Ri/aws-nlp-workshop | 888171694c5623a7fd5e19baa3e8cbc22c09bc69 | [
"MIT-0"
] | 29 | 2018-07-19T21:21:33.000Z | 2021-11-29T08:48:56.000Z | 65.205085 | 12,333 | 0.655264 | [
[
[
"# Gender Prediction, using Pre-trained Keras Model\n\nDeep Neural Networks can be used to extract features in the input and derive higher level abstractions. This technique is used regularly in vision, speech and text analysis. In this exercise, we use a pre-trained model deep learning model that would identify low level features in texts containing people's names, and would be able to classify them in one of two categories - Male or Female.\n",
"_____no_output_____"
],
[
"## Network Architecture\nThe problem we are trying to solve is to predict whether a given name belongs to a male or female. We will use supervised learning, where the character sequence making up the names would be `X` variable, and the flag indicating **Male(M)** or **Female(F)** would be `Y` variable.\n\nWe use a stacked 2-Layer LSTM model and a final dense layer with softmax activation as our network architecture. We use categorical cross-entropy as loss function, with an Adam optimizer. We also add a 20% dropout layer is added for regularization to avoid over-fitting. ",
"_____no_output_____"
],
[
"## Dependencies\n* The model was built using Keras, therefore we need to include Keras deep learning library to build the network locally, in order to be able to test, prior to hosting the model. \n* While running on SageMaker Notebook Instance, we choose conda_tensorflow kernel, so that Keras code is compiled to use tensorflow in the backend. \n* If you choose P2 and P3 class of instances for your Notebook, using Tensorflow ensures the low level code takes advantage of all available GPUs. So further dependencies needs to be installed.\n",
"_____no_output_____"
]
],
[
[
"import os\nimport time\nimport numpy as np\nimport keras\nfrom keras.models import load_model\nimport boto3",
"Using TensorFlow backend.\n"
]
],
[
[
"## Model testing\nTo test the validity of the model, we do some local testing.<p>\nThe model was built to be able to process one-hot encoded data representing names, therefore we need to do same pre-processing on our test data (one-hot encoding using the same character indices)<p>\nWe feed this one-hot encoded test data to the model, and the `predict` generates a vector, similar to the training labels vector we used before. Except in this case, it contains what model thinks the gender represented by each of the test records.<p>\nTo present data intutitively, we simply map it back to `Male` / `Female`, from the `0` / `1` flag. ",
"_____no_output_____"
]
],
[
[
"!tar -zxvf ../pretrained-model/model.tar.gz -C ../pretrained-model/ ",
"lstm-gender-classifier-model.h5\nlstm-gender-classifier-indices.npy\n"
],
[
"model = load_model('../pretrained-model/lstm-gender-classifier-model.h5')\nchar_indices = np.load('../pretrained-model/lstm-gender-classifier-indices.npy').item()\nmax_name_length = char_indices['max_name_length']\nchar_indices.pop('max_name_length', None)\nalphabet_size = len(char_indices)\nprint(char_indices)\nprint(max_name_length)\nprint(alphabet_size)",
"{'p': 15, 'v': 21, 'd': 3, 'f': 5, 'm': 12, 's': 18, 'l': 11, 'j': 9, 'g': 6, 'w': 22, 'x': 23, 'q': 16, 'n': 13, 'k': 10, 'i': 8, 'r': 17, 'e': 4, 'z': 25, 'u': 20, 'h': 7, 'b': 1, 'y': 24, 'a': 0, 'c': 2, 't': 19, 'o': 14}\n15\n26\n"
],
[
"names_test = [\"Tom\",\"Allie\",\"Jim\",\"Sophie\",\"John\",\"Kayla\",\"Mike\",\"Amanda\",\"Andrew\"]\nnum_test = len(names_test)\n\nX_test = np.zeros((num_test, max_name_length, alphabet_size))\n\nfor i,name in enumerate(names_test):\n name = name.lower()\n for t, char in enumerate(name):\n X_test[i, t,char_indices[char]] = 1\n\npredictions = model.predict(X_test)\n\nfor i,name in enumerate(names_test):\n print(\"{} ({})\".format(names_test[i],\"M\" if predictions[i][0]>predictions[i][1] else \"F\"))",
"Tom (M)\nAllie (F)\nJim (M)\nSophie (F)\nJohn (M)\nKayla (F)\nMike (M)\nAmanda (F)\nAndrew (M)\n"
]
],
[
[
"## Model saving\nIn order to deploy the model behind an hosted endpoint, we need to save the model fileto an S3 location.<p>\n \nWe can obtain the name of the S3 bucket from the execution role we attached to this Notebook instance. This should work if the policies granting read permission to IAM policies was granted, as per the documentation.\n\nIf for some reason, it fails to fetch the associated bucket name, it asks the user to enter the name of the bucket. If asked, use the bucket that you created in Module-3, such as 'smworkshop-firstname-lastname'.<p>\n \nIt is important to ensure that this is the same S3 bucket, to which you provided access in the Execution role used while creating this Notebook instance.",
"_____no_output_____"
]
],
[
[
"sts = boto3.client('sts')\niam = boto3.client('iam')\n\n\ncaller = sts.get_caller_identity()\naccount = caller['Account']\narn = caller['Arn']\nrole = arn[arn.find(\"/AmazonSageMaker\")+1:arn.find(\"/SageMaker\")]\ntimestamp = role[role.find(\"Role-\")+5:]\npolicyarn = \"arn:aws:iam::{}:policy/service-role/AmazonSageMaker-ExecutionPolicy-{}\".format(account, timestamp)\n\ns3bucketname = \"\"\npolicystatements = []\n\ntry:\n policy = iam.get_policy(\n PolicyArn=policyarn\n )['Policy']\n policyversion = policy['DefaultVersionId']\n policystatements = iam.get_policy_version(\n PolicyArn = policyarn, \n VersionId = policyversion\n )['PolicyVersion']['Document']['Statement']\nexcept Exception as e:\n s3bucketname=input(\"Which S3 bucket do you want to use to host training data and model? \")\n \nfor stmt in policystatements:\n action = \"\"\n actions = stmt['Action']\n for act in actions:\n if act == \"s3:ListBucket\":\n action = act\n break\n if action == \"s3:ListBucket\":\n resource = stmt['Resource'][0]\n s3bucketname = resource[resource.find(\":::\")+3:]\n\nprint(s3bucketname)",
"smworkshop-john-doe\n"
],
[
"s3 = boto3.resource('s3')\ns3.meta.client.upload_file('../pretrained-model/model.tar.gz', s3bucketname, 'model/model.tar.gz')",
"_____no_output_____"
]
],
[
[
"# Model hosting\n\nAmazon SageMaker provides a powerful orchestration framework that you can use to productionize any of your own machine learning algorithm, using any machine learning framework and programming languages.<p>\nThis is possible because SageMaker, as a manager of containers, have standarized ways of interacting with your code running inside a Docker container. Since you are free to build a docker container using whatever code and depndency you like, this gives you freedom to bring your own machinery.<p>\nIn the following steps, we'll containerize the prediction code and host the model behind an API endpoint.<p>\nThis would allow us to use the model from web-application, and put it into real use.<p>\nThe boilerplate code, which we affectionately call the `Dockerizer` framework, was made available on this Notebook instance by the Lifecycle Configuration that you used. Just look into the folder and ensure the necessary files are available as shown.<p>\n \n <home> \n |\n ├── container\n │\n ├── byoa\n | |\n │ ├── train\n | |\n │ ├── predictor.py\n | |\n │ ├── serve\n | |\n │ ├── nginx.conf\n | |\n │ └── wsgi.py\n |\n ├── build_and_push.sh\n │ \n ├── Dockerfile.cpu\n │ \n └── Dockerfile.gpu",
"_____no_output_____"
]
],
[
[
"os.chdir('../container')\nos.getcwd()\n!ls -Rl ",
".:\r\ntotal 16\r\n-rwxrwxrwx 1 root root 1382 Aug 16 07:39 build_and_push.sh\r\ndrwxrwxrwx 2 root root 4096 Aug 16 07:39 byoa\r\n-rw-rw-rw- 1 root root 1872 Aug 16 07:39 Dockerfile.cpu\r\n-rw-rw-rw- 1 root root 1938 Aug 16 07:39 Dockerfile.gpu\r\n\r\n./byoa:\r\ntotal 20\r\n-rwxrwxrwx 1 root root 687 Aug 16 07:39 nginx.conf\r\n-rwxrwxrwx 1 root root 2887 Aug 16 07:39 predictor.py\r\n-rwxrwxrwx 1 root root 2429 Aug 16 07:39 serve\r\n-rwxrwxrwx 1 root root 2336 Aug 16 07:39 train\r\n-rwxrwxrwx 1 root root 202 Aug 16 07:39 wsgi.py\r\n"
]
],
[
[
"* `Dockerfile` describes the container image and the accompanying script `build_and_push.sh` does the heavy lifting of building the container, and uploading it into an Amazon ECR repository\n* Sagemaker containers that we'll be building serves prediction request using a Flask based application. `wsgi.py` is a wrapper to invoke the Flask application, while `nginx.conf` is the configuration for the nginx front end and `serve` is the program that launches the gunicorn server. These files can be used as-is, and are required to build the webserver stack serving prediction requests, following the architecture as shown:\n\n<details>\n<summary><strong>Request serving stack (expand to view diagram)</strong></summary><p>\n \n</p></details>\n\n* The file named `predictor.py` is where we need to package the code for generating inference using the trained model that was saved into an S3 bucket location by the training code during the training job run.<p>\n* We'll write code into this file using Jupyter magic command - `writefile`.<p><br>\nFirst part of the file would contain the necessary imports, as ususal. ",
"_____no_output_____"
]
],
[
[
"%%writefile byoa/predictor.py\n# This is the file that implements a flask server to do inferences. It's the file that you will modify to\n# implement the scoring for your own algorithm.\n\nfrom __future__ import print_function\n\nimport os\nimport json\nimport pickle\nfrom io import StringIO\nimport sys\nimport signal\nimport traceback\n\nimport numpy as np\n\nimport keras\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout\nfrom keras.layers import Embedding\nfrom keras.layers import LSTM\nfrom keras.models import load_model\nimport flask\n\nimport tensorflow as tf\n\nimport pandas as pd\n\nfrom os import listdir, sep\nfrom os.path import abspath, basename, isdir\nfrom sys import argv",
"Overwriting byoa/predictor.py\n"
]
],
[
[
"When run within an instantiated container, SageMaker makes the trained model available locally at `/opt/ml`",
"_____no_output_____"
]
],
[
[
"%%writefile -a byoa/predictor.py\n\nprefix = '/opt/ml/'\nmodel_path = os.path.join(prefix, 'model')",
"Appending to byoa/predictor.py\n"
]
],
[
[
"The machinery to produce inference is wrapped around in a Pythonic class structure, within a `Singleton` class, aptly named - `ScoringService`.<p>\nWe create `Class` variables in this class to hold loaded model, character indices, tensor-flow graph, and anything else that needs to be referenced while generating prediction. ",
"_____no_output_____"
]
],
[
[
"%%writefile -a byoa/predictor.py\n\n# A singleton for holding the model. This simply loads the model and holds it.\n# It has a predict function that does a prediction based on the model and the input data.\n\nclass ScoringService(object):\n model_type = None # Where we keep the model type, qualified by hyperparameters used during training\n model = None # Where we keep the model when it's loaded\n graph = None\n indices = None # Where we keep the indices of Alphabet when it's loaded",
"Appending to byoa/predictor.py\n"
]
],
[
[
"Generally, we have to provide class methods to load the model and related artefacts from the model path as assigned by SageMaker within the running container.<p>\nNotice here that SageMaker copies the artefacts from the S3 location (as defined during model creation) into the container local file system.",
"_____no_output_____"
]
],
[
[
"%%writefile -a byoa/predictor.py\n\n @classmethod\n def get_indices(cls):\n #Get the indices for Alphabet for this instance, loading it if it's not already loaded\n if cls.indices == None:\n model_type='lstm-gender-classifier'\n index_path = os.path.join(model_path, '{}-indices.npy'.format(model_type))\n if os.path.exists(index_path):\n cls.indices = np.load(index_path).item()\n else:\n print(\"Character Indices not found.\")\n return cls.indices\n\n @classmethod\n def get_model(cls):\n #Get the model object for this instance, loading it if it's not already loaded\n if cls.model == None:\n model_type='lstm-gender-classifier'\n mod_path = os.path.join(model_path, '{}-model.h5'.format(model_type))\n if os.path.exists(mod_path):\n cls.model = load_model(mod_path)\n cls.model._make_predict_function()\n cls.graph = tf.get_default_graph()\n else:\n print(\"LSTM Model not found.\")\n return cls.model",
"Appending to byoa/predictor.py\n"
]
],
[
[
"Finally, inside another clas method, named `predict`, we provide the code that we used earlier to generate prediction.<p>\nOnly difference with our previous test prediciton (in development notebook) is that in this case, the predictor will grab the data from the `input` variable, which in turn is obtained from the HTTP request payload.",
"_____no_output_____"
]
],
[
[
"%%writefile -a byoa/predictor.py\n\n @classmethod\n def predict(cls, input):\n\n mod = cls.get_model()\n ind = cls.get_indices()\n\n result = {}\n\n if mod == None:\n print(\"Model not loaded.\")\n else:\n if 'max_name_length' not in ind:\n max_name_length = 15\n alphabet_size = 26\n else:\n max_name_length = ind['max_name_length']\n ind.pop('max_name_length', None)\n alphabet_size = len(ind)\n\n inputs_list = input.strip('\\n').split(\",\")\n num_inputs = len(inputs_list)\n\n X_test = np.zeros((num_inputs, max_name_length, alphabet_size))\n\n for i,name in enumerate(inputs_list):\n name = name.lower().strip('\\n')\n for t, char in enumerate(name):\n if char in ind:\n X_test[i, t,ind[char]] = 1\n\n with cls.graph.as_default():\n predictions = mod.predict(X_test)\n\n for i,name in enumerate(inputs_list):\n result[name] = 'M' if predictions[i][0]>predictions[i][1] else 'F'\n print(\"{} ({})\".format(inputs_list[i],\"M\" if predictions[i][0]>predictions[i][1] else \"F\"))\n\n return json.dumps(result)",
"Appending to byoa/predictor.py\n"
]
],
[
[
"With the prediction code captured, we move on to define the flask app, and provide a `ping`, which SageMaker uses to conduct health check on container instances that are responsible behind the hosted prediction endpoint.<p>\nHere we can have the container return healthy response, with status code `200` when everythings goes well.<p>\nFor simplicity, we are only validating whether model has been loaded in this case. In practice, this provides opportunity extensive health check (including any external dependency check), as required.",
"_____no_output_____"
]
],
[
[
"%%writefile -a byoa/predictor.py\n\n# The flask app for serving predictions\napp = flask.Flask(__name__)\n\[email protected]('/ping', methods=['GET'])\ndef ping():\n #Determine if the container is working and healthy.\n # Declare it healthy if we can load the model successfully.\n health = ScoringService.get_model() is not None and ScoringService.get_indices() is not None\n status = 200 if health else 404\n return flask.Response(response='\\n', status=status, mimetype='application/json')\n",
"Appending to byoa/predictor.py\n"
]
],
[
[
"Last but not the least, we define a `transformation` method that would intercept the HTTP request coming through to the SageMaker hosted endpoint.<p>\nHere we have the opportunity to decide what type of data we accept with the request. In this particular example, we are accepting only `CSV` formatted data, decoding the data, and invoking prediction.<p>\nThe response is similarly funneled backed to the caller with MIME type of `CSV`.<p>\nYou are free to choose any or multiple MIME types for your requests and response. However if you choose to do so, it is within this method that we have to transform the back to and from the format that is suitable to passed for prediction.",
"_____no_output_____"
]
],
[
[
"%%writefile -a byoa/predictor.py\n\n\[email protected]('/invocations', methods=['POST'])\ndef transformation():\n #Do an inference on a single batch of data\n data = None\n\n # Convert from CSV to pandas\n if flask.request.content_type == 'text/csv':\n data = flask.request.data.decode('utf-8')\n else:\n return flask.Response(response='This predictor only supports CSV data', status=415, mimetype='text/plain')\n\n print('Invoked with {} records'.format(data.count(\",\")+1))\n\n # Do the prediction\n predictions = ScoringService.predict(data)\n\n result = \"\"\n for prediction in predictions:\n result = result + prediction\n\n return flask.Response(response=result, status=200, mimetype='text/csv')",
"Appending to byoa/predictor.py\n"
]
],
[
[
"Note that in containerizing our custom LSTM Algorithm, where we used `Keras` as our framework of our choice, we did not have to interact directly with the SageMaker API, even though SageMaker API doesn't support `Keras`.<p>\nThis serves to show the power and flexibility offered by containerized machine learning pipeline on SageMaker.",
"_____no_output_____"
],
[
"## Container publishing\n\nIn order to host and deploy the trained model using SageMaker, we need to build the `Docker` containers, publish it to `Amazon ECR` repository, and then either use SageMaker console or API to created the endpoint configuration and deploy the stages.<p>\n\nConceptually, the steps required for publishing are:<p>\n1. Make the`predictor.py` files executable\n2. Create an ECR repository within your default region\n3. Build a docker container with an identifieable name\n4. Tage the image and publish to the ECR repository\n<p><br>\nAll of these are conveniently encapsulated inside `build_and_push` script. We simply run it with the unique name of our production run.",
"_____no_output_____"
]
],
[
[
"run_type='cpu'\ninstance_class = \"p3\" if run_type.lower()=='gpu' else \"c4\"\ninstance_type = \"ml.{}.8xlarge\".format(instance_class)\n\npipeline_name = 'gender-classifier'\nrun=input(\"Enter run version: \")\n\nrun_name = pipeline_name+\"-\"+run\nif run_type == \"cpu\":\n !cp \"Dockerfile.cpu\" \"Dockerfile\"\n\nif run_type == \"gpu\":\n !cp \"Dockerfile.gpu\" \"Dockerfile\"\n \n!sh build_and_push.sh $run_name",
"Enter run version: 1\nWARNING! Using --password via the CLI is insecure. Use --password-stdin.\nLogin Succeeded\nSending build context to Docker daemon 25.6kB\nStep 1/13 : FROM ubuntu:16.04\n16.04: Pulling from library/ubuntu\n\n\u001b[1B9e426c26: Pulling fs layer \n\u001b[1Bb260b73b: Pulling fs layer \n\u001b[1B65fd1143: Pulling fs layer \n\u001b[1Ba07f8222: Pulling fs layer \n\u001b[1BDigest: sha256:3097ac92b852f878f802c22a38f97b097b4084dbef82893ba453ba0297d76a6a\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[5A\u001b[1K\u001b[K\u001b[4A\u001b[1K\u001b[K\u001b[3A\u001b[1K\u001b[K\u001b[2A\u001b[1K\u001b[K\u001b[1A\u001b[1K\u001b[K\nStatus: Downloaded newer image for ubuntu:16.04\n ---> 7aa3602ab41e\nStep 2/13 : MAINTAINER Binoy Das <[email protected]>\n ---> Running in 74a633b2ea23\nRemoving intermediate container 74a633b2ea23\n ---> c4265b4a021a\nStep 3/13 : RUN apt-get update && apt-get install -y --no-install-recommends apt-utils build-essential curl libfreetype6-dev libpng12-dev libzmq3-dev libhdf5-dev libcurl3-dev libgtk2.0-0 pkg-config python3-dev python3-pip rsync software-properties-common unzip gzip wget vim git nginx ca-certificates && apt-get clean && rm -rf /var/lib/apt/lists/*\n ---> Running in e04af221dd9e\nGet:1 http://archive.ubuntu.com/ubuntu xenial InRelease [247 kB]\nGet:2 http://security.ubuntu.com/ubuntu xenial-security InRelease [107 kB]\nGet:3 http://security.ubuntu.com/ubuntu xenial-security/universe Sources [88.1 kB]\nGet:4 http://archive.ubuntu.com/ubuntu xenial-updates InRelease [109 kB]\nGet:5 http://security.ubuntu.com/ubuntu xenial-security/main amd64 Packages [688 kB]\nGet:6 http://archive.ubuntu.com/ubuntu xenial-backports InRelease [107 kB]\nGet:7 http://archive.ubuntu.com/ubuntu xenial/universe Sources [9802 kB]\nGet:8 http://security.ubuntu.com/ubuntu xenial-security/restricted amd64 Packages [12.7 kB]\nGet:9 http://security.ubuntu.com/ubuntu xenial-security/universe amd64 Packages [465 kB]\nGet:10 http://security.ubuntu.com/ubuntu xenial-security/multiverse amd64 Packages [3746 B]\nGet:11 http://archive.ubuntu.com/ubuntu xenial/main amd64 Packages [1558 kB]\nGet:12 http://archive.ubuntu.com/ubuntu xenial/restricted amd64 Packages [14.1 kB]\nGet:13 http://archive.ubuntu.com/ubuntu xenial/universe amd64 Packages [9827 kB]\nGet:14 http://archive.ubuntu.com/ubuntu xenial/multiverse amd64 Packages [176 kB]\nGet:15 http://archive.ubuntu.com/ubuntu xenial-updates/universe Sources [275 kB]\nGet:16 http://archive.ubuntu.com/ubuntu xenial-updates/main amd64 Packages [1073 kB]\nGet:17 http://archive.ubuntu.com/ubuntu xenial-updates/restricted amd64 Packages [13.1 kB]\nGet:18 http://archive.ubuntu.com/ubuntu xenial-updates/universe amd64 Packages [876 kB]\nGet:19 http://archive.ubuntu.com/ubuntu xenial-updates/multiverse amd64 Packages [18.8 kB]\nGet:20 http://archive.ubuntu.com/ubuntu xenial-backports/main amd64 Packages [7343 B]\nGet:21 http://archive.ubuntu.com/ubuntu xenial-backports/universe amd64 Packages [8086 B]\nFetched 25.5 MB in 3s (8459 kB/s)\nReading package lists...\nReading package lists...\nBuilding dependency tree...\nReading state information...\ngzip is already the newest version (1.6-4ubuntu1).\nThe following additional packages will be installed:\n binutils bzip2 cpp cpp-5 dh-python distro-info-data dpkg-dev fontconfig\n fontconfig-config fonts-dejavu-core g++ g++-5 gcc gcc-5 gir1.2-glib-2.0\n git-man hdf5-helpers iso-codes libaec-dev libaec0 libapt-inst2.0 libasan2\n libasn1-8-heimdal libatk1.0-0 libatk1.0-data libatomic1 libavahi-client3\n libavahi-common-data libavahi-common3 libc-dev-bin libc6-dev libcairo2\n libcc1-0 libcilkrts5 libcups2 libcurl3 libcurl3-gnutls libdatrie1\n libdbus-1-3 libdbus-glib-1-2 libdpkg-perl liberror-perl libexpat1\n libexpat1-dev libffi6 libfontconfig1 libfreetype6 libgcc-5-dev libgd3\n libgdbm3 libgdk-pixbuf2.0-0 libgdk-pixbuf2.0-common libgeoip1 libgfortran3\n libgirepository-1.0-1 libglib2.0-0 libgmp10 libgnutls30 libgomp1 libgpm2\n libgraphite2-3 libgssapi-krb5-2 libgssapi3-heimdal libgtk2.0-common\n libharfbuzz0b libhcrypto4-heimdal libhdf5-10 libhdf5-cpp-11\n libheimbase1-heimdal libheimntlm0-heimdal libhogweed4 libhx509-5-heimdal\n libicu55 libidn11 libisl15 libitm1 libjbig0 libjpeg-dev libjpeg-turbo8\n libjpeg-turbo8-dev libjpeg8 libjpeg8-dev libk5crypto3 libkeyutils1\n libkrb5-26-heimdal libkrb5-3 libkrb5support0 libldap-2.4-2 liblsan0 libmpc3\n libmpdec2 libmpfr4 libmpx0 libnettle6 libp11-kit0 libpango-1.0-0\n libpangocairo-1.0-0 libpangoft2-1.0-0 libperl5.22 libpixman-1-0 libpng12-0\n libpopt0 libpython3-dev libpython3-stdlib libpython3.5 libpython3.5-dev\n libpython3.5-minimal libpython3.5-stdlib libquadmath0 libroken18-heimdal\n librtmp1 libsasl2-2 libsasl2-modules-db libsodium18 libsqlite3-0 libssl1.0.0\n libstdc++-5-dev libsz2 libtasn1-6 libthai-data libthai0 libtiff5 libtsan0\n libubsan0 libvpx3 libwind0-heimdal libx11-6 libx11-data libxau6\n libxcb-render0 libxcb-shm0 libxcb1 libxcomposite1 libxcursor1 libxdamage1\n libxdmcp6 libxext6 libxfixes3 libxi6 libxinerama1 libxml2 libxpm4 libxrandr2\n libxrender1 libxslt1.1 libzmq5 linux-libc-dev lsb-release make mime-support\n nginx-common nginx-core openssl patch perl perl-modules-5.22\n python-apt-common python-pip-whl python3 python3-apt python3-dbus python3-gi\n python3-minimal python3-pycurl python3-software-properties python3.5\n python3.5-dev python3.5-minimal shared-mime-info ucf vim-common vim-runtime\n xz-utils zlib1g-dev\nSuggested packages:\n binutils-doc bzip2-doc cpp-doc gcc-5-locales debian-keyring g++-multilib\n g++-5-multilib gcc-5-doc libstdc++6-5-dbg gcc-multilib manpages-dev autoconf\n automake libtool flex bison gdb gcc-doc gcc-5-multilib libgcc1-dbg\n libgomp1-dbg libitm1-dbg libatomic1-dbg libasan2-dbg liblsan0-dbg\n libtsan0-dbg libubsan0-dbg libcilkrts5-dbg libmpx0-dbg libquadmath0-dbg\n gettext-base git-daemon-run | git-daemon-sysvinit git-doc git-el git-email\n git-gui gitk gitweb git-arch git-cvs git-mediawiki git-svn isoquery\n glibc-doc cups-common libcurl4-doc libcurl3-dbg libidn11-dev libkrb5-dev\n libldap2-dev librtmp-dev libssl-dev libgd-tools geoip-bin gnutls-bin gpm\n krb5-doc krb5-user librsvg2-common gvfs libhdf5-doc libstdc++-5-doc lsb\n make-doc fcgiwrap nginx-doc ssl-cert ed diffutils-doc perl-doc\n libterm-readline-gnu-perl | libterm-readline-perl-perl python3-doc\n python3-tk python3-venv python3-apt-dbg python-apt-doc python-dbus-doc\n python3-dbus-dbg libcurl4-gnutls-dev python-pycurl-doc python3-pycurl-dbg\n python3.5-venv python3.5-doc binfmt-support openssh-client openssh-server\n zip ctags vim-doc vim-scripts vim-gnome-py2 | vim-gtk-py2 | vim-gtk3-py2\n | vim-athena-py2 | vim-nox-py2\nRecommended packages:\n fakeroot libalgorithm-merge-perl less ssh-client manpages manpages-dev dbus\n libfile-fcntllock-perl geoip-database libglib2.0-data xdg-user-dirs\n hicolor-icon-theme libgtk2.0-bin krb5-locales libsasl2-modules xml-core file\n netbase rename python3-setuptools python3-wheel unattended-upgrades\nThe following NEW packages will be installed:\n apt-utils binutils build-essential bzip2 ca-certificates cpp cpp-5 curl\n dh-python distro-info-data dpkg-dev fontconfig fontconfig-config\n fonts-dejavu-core g++ g++-5 gcc gcc-5 gir1.2-glib-2.0 git git-man\n hdf5-helpers iso-codes libaec-dev libaec0 libapt-inst2.0 libasan2\n libasn1-8-heimdal libatk1.0-0 libatk1.0-data libatomic1 libavahi-client3\n libavahi-common-data libavahi-common3 libc-dev-bin libc6-dev libcairo2\n libcc1-0 libcilkrts5 libcups2 libcurl3 libcurl3-gnutls libcurl4-openssl-dev\n libdatrie1 libdbus-1-3 libdbus-glib-1-2 libdpkg-perl liberror-perl libexpat1\n libexpat1-dev libffi6 libfontconfig1 libfreetype6 libfreetype6-dev\n libgcc-5-dev libgd3 libgdbm3 libgdk-pixbuf2.0-0 libgdk-pixbuf2.0-common\n libgeoip1 libgfortran3 libgirepository-1.0-1 libglib2.0-0 libgmp10\n libgnutls30 libgomp1 libgpm2 libgraphite2-3 libgssapi-krb5-2\n libgssapi3-heimdal libgtk2.0-0 libgtk2.0-common libharfbuzz0b\n libhcrypto4-heimdal libhdf5-10 libhdf5-cpp-11 libhdf5-dev\n libheimbase1-heimdal libheimntlm0-heimdal libhogweed4 libhx509-5-heimdal\n libicu55 libidn11 libisl15 libitm1 libjbig0 libjpeg-dev libjpeg-turbo8\n libjpeg-turbo8-dev libjpeg8 libjpeg8-dev libk5crypto3 libkeyutils1\n libkrb5-26-heimdal libkrb5-3 libkrb5support0 libldap-2.4-2 liblsan0 libmpc3\n libmpdec2 libmpfr4 libmpx0 libnettle6 libp11-kit0 libpango-1.0-0\n libpangocairo-1.0-0 libpangoft2-1.0-0 libperl5.22 libpixman-1-0 libpng12-0\n libpng12-dev libpopt0 libpython3-dev libpython3-stdlib libpython3.5\n libpython3.5-dev libpython3.5-minimal libpython3.5-stdlib libquadmath0\n libroken18-heimdal librtmp1 libsasl2-2 libsasl2-modules-db libsodium18\n libsqlite3-0 libssl1.0.0 libstdc++-5-dev libsz2 libtasn1-6 libthai-data\n libthai0 libtiff5 libtsan0 libubsan0 libvpx3 libwind0-heimdal libx11-6\n libx11-data libxau6 libxcb-render0 libxcb-shm0 libxcb1 libxcomposite1\n libxcursor1 libxdamage1 libxdmcp6 libxext6 libxfixes3 libxi6 libxinerama1\n libxml2 libxpm4 libxrandr2 libxrender1 libxslt1.1 libzmq3-dev libzmq5\n linux-libc-dev lsb-release make mime-support nginx nginx-common nginx-core\n openssl patch perl perl-modules-5.22 pkg-config python-apt-common\n python-pip-whl python3 python3-apt python3-dbus python3-dev python3-gi\n python3-minimal python3-pip python3-pycurl python3-software-properties\n python3.5 python3.5-dev python3.5-minimal rsync shared-mime-info\n software-properties-common ucf unzip vim vim-common vim-runtime wget\n xz-utils zlib1g-dev\n"
]
],
[
[
"## Orchestration\n\nAt this point, we can head to ECS console, grab the ARN for the repository where we published the docker image, and use SageMaker console to create hosted model, and endpoint.<p>\nHowever, it is often more convenient to automate these steps. In this notebook we do exactly that using `boto3 SageMaker` API.<p>\nFollowing are the steps:<p>\n \n* First we create a model hosting definition, by providing the S3 location to the model artifact, and ARN to the ECR image of the container.\n* Using the model hosting definition, our next step is to create configuration of a hosted endpoint that will be used to serve prediciton generation requests. \n* Creating the endpoint is the last step in the ML cycle, that prepares your model to serve client reqests from applications.\n* We wait until the provision is completed and the endpoint in service. At this point we can send request to this endpoint and obtain gender predictions.\n",
"_____no_output_____"
]
],
[
[
"import sagemaker\nsm_role = sagemaker.get_execution_role()\nprint(\"Using Role {}\".format(sm_role))\nacc = boto3.client('sts').get_caller_identity().get('Account')\nreg = boto3.session.Session().region_name\nsagemaker = boto3.client('sagemaker')\n\n#Check if model already exists\nmodel_name = \"{}-model\".format(run_name)\nmodels = sagemaker.list_models(NameContains=model_name)['Models']\nmodel_exists = False\nif len(models) > 0:\n for model in models:\n if model['ModelName'] == model_name:\n model_exists = True\n break\n#Delete model, if chosen\nif model_exists == True: \n choice = input(\"Model already exists, do you want to delete and create a fresh one (Y/N) ? \")\n if choice.upper()[0:1] == \"Y\":\n sagemaker.delete_model(ModelName = model_name)\n model_exists = False\n else:\n print(\"Model - {} already exists\".format(model_name))\n\nif model_exists == False: \n model_response = sagemaker.create_model(\n ModelName=model_name,\n PrimaryContainer={\n 'Image': '{}.dkr.ecr.{}.amazonaws.com/{}:latest'.format(acc, reg, run_name),\n 'ModelDataUrl': 's3://{}/model/model.tar.gz'.format(s3bucketname)\n },\n ExecutionRoleArn=sm_role,\n Tags=[\n {\n 'Key': 'Name',\n 'Value': model_name\n }\n ]\n )\n print(\"{} Created at {}\".format(model_response['ModelArn'], \n model_response['ResponseMetadata']['HTTPHeaders']['date']))",
"Using Role arn:aws:iam::741855114961:role/service-role/AmazonSageMaker-ExecutionRole-20180815T114786\nModel already exists, do you want to delete and create a fresh one (Y/N) ? Y\narn:aws:sagemaker:us-east-1:741855114961:model/gender-classifier-1-model Created at Thu, 16 Aug 2018 07:52:24 GMT\n"
],
[
"#Check if endpoint configuration already exists\nendpoint_config_name = \"{}-endpoint-config\".format(run_name)\nendpoint_configs = sagemaker.list_endpoint_configs(NameContains=endpoint_config_name)['EndpointConfigs']\nendpoint_config_exists = False\nif len(endpoint_configs) > 0:\n for endpoint_config in endpoint_configs:\n if endpoint_config['EndpointConfigName'] == endpoint_config_name:\n endpoint_config_exists = True\n break\n \n#Delete endpoint configuration, if chosen\nif endpoint_config_exists == True: \n choice = input(\"Endpoint Configuration already exists, do you want to delete and create a fresh one (Y/N) ? \")\n if choice.upper()[0:1] == \"Y\":\n sagemaker.delete_endpoint_config(EndpointConfigName = endpoint_config_name)\n endpoint_config_exists = False\n else:\n print(\"Endpoint Configuration - {} already exists\".format(endpoint_config_name))\n \nif endpoint_config_exists == False: \n endpoint_config_response = sagemaker.create_endpoint_config(\n EndpointConfigName=endpoint_config_name,\n ProductionVariants=[\n {\n 'VariantName': 'default',\n 'ModelName': model_name,\n 'InitialInstanceCount': 1,\n 'InstanceType': instance_type,\n 'InitialVariantWeight': 1\n },\n ],\n Tags=[\n {\n 'Key': 'Name',\n 'Value': endpoint_config_name\n }\n ]\n )\n print(\"{} Created at {}\".format(endpoint_config_response['EndpointConfigArn'], \n endpoint_config_response['ResponseMetadata']['HTTPHeaders']['date']))",
"Endpoint Configuration already exists, do you want to delete and create a fresh one (Y/N) ? Y\narn:aws:sagemaker:us-east-1:741855114961:endpoint-config/gender-classifier-1-endpoint-config Created at Thu, 16 Aug 2018 07:52:27 GMT\n"
],
[
"from ipywidgets import widgets\nfrom IPython.display import display\n\n#Check if endpoint already exists\nendpoint_name = \"{}-endpoint\".format(run_name)\nendpoints = sagemaker.list_endpoints(NameContains=endpoint_name)['Endpoints']\nendpoint_exists = False\nif len(endpoints) > 0:\n for endpoint in endpoints:\n if endpoint['EndpointName'] == endpoint_name:\n endpoint_exists = True\n break\n \n#Delete endpoint, if chosen\nif endpoint_exists == True: \n choice = input(\"Endpoint already exists, do you want to delete and create a fresh one (Y/N) ? \")\n if choice.upper()[0:1] == \"Y\":\n sagemaker.delete_endpoint(EndpointName = endpoint_name)\n print(\"Deleting Endpoint - {} ...\".format(endpoint_name))\n waiter = sagemaker.get_waiter('endpoint_deleted')\n waiter.wait(EndpointName=endpoint_name,\n WaiterConfig = {'Delay':1,'MaxAttempts':100})\n endpoint_exists = False\n print(\"Endpoint - {} deleted\".format(endpoint_name))\n \n else:\n print(\"Endpoint - {} already exists\".format(endpoint_name))\n \nif endpoint_exists == False: \n\n endpoint_response = sagemaker.create_endpoint(\n EndpointName=endpoint_name,\n EndpointConfigName=endpoint_config_name,\n Tags=[\n {\n 'Key': 'string',\n 'Value': endpoint_name\n }\n ]\n )\n status='Creating'\n sleep = 3\n\n print(\"{} Endpoint : {}\".format(status,endpoint_name))\n bar = widgets.FloatProgress(min=0, description=\"Progress\") # instantiate the bar\n display(bar) # display the bar\n\n while status != 'InService' and status != 'Failed' and status != 'OutOfService': \n endpoint_response = sagemaker.describe_endpoint(\n EndpointName=endpoint_name\n )\n status = endpoint_response['EndpointStatus']\n time.sleep(sleep)\n bar.value = bar.value + 1 \n if bar.value >= bar.max-1:\n bar.max = int(bar.max*1.05)\n if status != 'InService' and status != 'Failed' and status != 'OutOfService': \n print(\".\", end='')\n\n bar.max = bar.value \n html = widgets.HTML(\n value=\"<H2>Endpoint <b><u>{}</b></u> - {}</H2>\".format(endpoint_response['EndpointName'], status)\n )\n display(html)",
"Endpoint already exists, do you want to delete and create a fresh one (Y/N) ? Y\nDeleting Endpoint - gender-classifier-1-endpoint ...\nEndpoint - gender-classifier-1-endpoint deleted\nCreating Endpoint : gender-classifier-1-endpoint\n"
]
],
[
[
"At the end we run a quick test to validate we are able to generate meaningful predicitions using the hosted endpoint, as we did locally using the model on the Notebbok instance.",
"_____no_output_____"
]
],
[
[
"!aws sagemaker-runtime invoke-endpoint --endpoint-name \"$run_name-endpoint\" --body 'Tom,Allie,Jim,Sophie,John,Kayla,Mike,Amanda,Andrew' --content-type text/csv outfile\n!cat outfile",
"{\n \"ContentType\": \"text/csv; charset=utf-8\",\n \"InvokedProductionVariant\": \"default\"\n}\n{\"Sophie\": \"F\", \"Mike\": \"M\", \"Tom\": \"M\", \"Andrew\": \"M\", \"John\": \"M\", \"Amanda\": \"F\", \"Kayla\": \"F\", \"Allie\": \"F\", \"Jim\": \"M\"}"
]
],
[
[
"Head back to Module-3 of the workshop now, to the section titled - `Integration`, and follow the steps described.<p>\nYou'll need to copy the endpoint name from the output of the cell below, to use in the Lambda function that will send request to this hosted endpoint.",
"_____no_output_____"
]
],
[
[
"print(endpoint_response\n ['EndpointName'])",
"gender-classifier-1-endpoint\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4e9df4eb92cfb9a1e066ecdb2e7452fc970715 | 136,911 | ipynb | Jupyter Notebook | docs/notebooks/Carver.ipynb | IMULMUL/fuzzingbook | b16f5bc5ff8b72486dd5b6236f3a9fbd5c0b4334 | [
"MIT"
] | 728 | 2018-09-21T03:51:04.000Z | 2022-03-28T09:35:04.000Z | docs/notebooks/Carver.ipynb | IMULMUL/fuzzingbook | b16f5bc5ff8b72486dd5b6236f3a9fbd5c0b4334 | [
"MIT"
] | 103 | 2018-09-02T12:26:32.000Z | 2022-02-09T07:19:08.000Z | docs/notebooks/Carver.ipynb | IMULMUL/fuzzingbook | b16f5bc5ff8b72486dd5b6236f3a9fbd5c0b4334 | [
"MIT"
] | 157 | 2018-09-02T08:00:50.000Z | 2022-03-27T22:04:50.000Z | 31.517265 | 1,433 | 0.579413 | [
[
[
"# Carving Unit Tests\n\nSo far, we have always generated _system input_, i.e. data that the program as a whole obtains via its input channels. If we are interested in testing only a small set of functions, having to go through the system can be very inefficient. This chapter introduces a technique known as _carving_, which, given a system test, automatically extracts a set of _unit tests_ that replicate the calls seen during the unit test. The key idea is to _record_ such calls such that we can _replay_ them later – as a whole or selectively. On top, we also explore how to synthesize API grammars from carved unit tests; this means that we can _synthesize API tests without having to write a grammar at all._",
"_____no_output_____"
],
[
"**Prerequisites**\n\n* Carving makes use of dynamic traces of function calls and variables, as introduced in the [chapter on configuration fuzzing](ConfigurationFuzzer.ipynb).\n* Using grammars to test units was introduced in the [chapter on API fuzzing](APIFuzzer.ipynb).",
"_____no_output_____"
]
],
[
[
"import bookutils",
"_____no_output_____"
],
[
"import APIFuzzer",
"_____no_output_____"
]
],
[
[
"## Synopsis\n<!-- Automatically generated. Do not edit. -->\n\nTo [use the code provided in this chapter](Importing.ipynb), write\n\n```python\n>>> from fuzzingbook.Carver import <identifier>\n```\n\nand then make use of the following features.\n\n\nThis chapter provides means to _record and replay function calls_ during a system test. Since individual function calls are much faster than a whole system run, such \"carving\" mechanisms have the potential to run tests much faster.\n\n### Recording Calls\n\nThe `CallCarver` class records all calls occurring while it is active. It is used in conjunction with a `with` clause:\n\n```python\n>>> with CallCarver() as carver:\n>>> y = my_sqrt(2)\n>>> y = my_sqrt(4)\n```\nAfter execution, `called_functions()` lists the names of functions encountered:\n\n```python\n>>> carver.called_functions()\n['my_sqrt', '__exit__']\n```\nThe `arguments()` method lists the arguments recorded for a function. This is a mapping of the function name to a list of lists of arguments; each argument is a pair (parameter name, value).\n\n```python\n>>> carver.arguments('my_sqrt')\n[[('x', 2)], [('x', 4)]]\n```\nComplex arguments are properly serialized, such that they can be easily restored.\n\n### Synthesizing Calls\n\nWhile such recorded arguments already could be turned into arguments and calls, a much nicer alternative is to create a _grammar_ for recorded calls. This allows to synthesize arbitrary _combinations_ of arguments, and also offers a base for further customization of calls.\n\nThe `CallGrammarMiner` class turns a list of carved executions into a grammar.\n\n```python\n>>> my_sqrt_miner = CallGrammarMiner(carver)\n>>> my_sqrt_grammar = my_sqrt_miner.mine_call_grammar()\n>>> my_sqrt_grammar\n{'<start>': ['<call>'],\n '<call>': ['<my_sqrt>'],\n '<my_sqrt-x>': ['2', '4'],\n '<my_sqrt>': ['my_sqrt(<my_sqrt-x>)']}\n```\nThis grammar can be used to synthesize calls.\n\n```python\n>>> fuzzer = GrammarCoverageFuzzer(my_sqrt_grammar)\n>>> fuzzer.fuzz()\n'my_sqrt(4)'\n```\nThese calls can be executed in isolation, effectively extracting unit tests from system tests:\n\n```python\n>>> eval(fuzzer.fuzz())\n1.414213562373095\n```\n",
"_____no_output_____"
],
[
"## System Tests vs Unit Tests\n\nRemember the URL grammar introduced for [grammar fuzzing](Grammars.ipynb)? With such a grammar, we can happily test a Web browser again and again, checking how it reacts to arbitrary page requests.\n\nLet us define a very simple \"web browser\" that goes and downloads the content given by the URL.",
"_____no_output_____"
]
],
[
[
"import urllib.parse",
"_____no_output_____"
],
[
"def webbrowser(url):\n \"\"\"Download the http/https resource given by the URL\"\"\"\n import requests # Only import if needed\n\n r = requests.get(url)\n return r.text",
"_____no_output_____"
]
],
[
[
"Let us apply this on [fuzzingbook.org](https://www.fuzzingbook.org/) and measure the time, using the [Timer class](Timer.ipynb):",
"_____no_output_____"
]
],
[
[
"from Timer import Timer",
"_____no_output_____"
],
[
"with Timer() as webbrowser_timer:\n fuzzingbook_contents = webbrowser(\n \"http://www.fuzzingbook.org/html/Fuzzer.html\")\n\nprint(\"Downloaded %d bytes in %.2f seconds\" %\n (len(fuzzingbook_contents), webbrowser_timer.elapsed_time()))",
"Downloaded 421620 bytes in 0.34 seconds\n"
],
[
"fuzzingbook_contents[:100]",
"_____no_output_____"
]
],
[
[
"A full webbrowser, of course, would also render the HTML content. We can achieve this using these commands (but we don't, as we do not want to replicate the entire Web page here):\n\n\n```python\nfrom IPython.display import HTML, display\nHTML(fuzzingbook_contents)\n```",
"_____no_output_____"
],
[
"Having to start a whole browser (or having it render a Web page) again and again means lots of overhead, though – in particular if we want to test only a subset of its functionality. In particular, after a change in the code, we would prefer to test only the subset of functions that is affected by the change, rather than running the well-tested functions again and again.",
"_____no_output_____"
],
[
"Let us assume we change the function that takes care of parsing the given URL and decomposing it into the individual elements – the scheme (\"http\"), the network location (`\"www.fuzzingbook.com\"`), or the path (`\"/html/Fuzzer.html\"`). This function is named `urlparse()`:",
"_____no_output_____"
]
],
[
[
"from urllib.parse import urlparse",
"_____no_output_____"
],
[
"urlparse('https://www.fuzzingbook.com/html/Carver.html')",
"_____no_output_____"
]
],
[
[
"You see how the individual elements of the URL – the _scheme_ (`\"http\"`), the _network location_ (`\"www.fuzzingbook.com\"`), or the path (`\"//html/Carver.html\"`) are all properly identified. Other elements (like `params`, `query`, or `fragment`) are empty, because they were not part of our input.",
"_____no_output_____"
],
[
"The interesting thing is that executing only `urlparse()` is orders of magnitude faster than running all of `webbrowser()`. Let us measure the factor:",
"_____no_output_____"
]
],
[
[
"runs = 1000\nwith Timer() as urlparse_timer:\n for i in range(runs):\n urlparse('https://www.fuzzingbook.com/html/Carver.html')\n\navg_urlparse_time = urlparse_timer.elapsed_time() / 1000\navg_urlparse_time",
"_____no_output_____"
]
],
[
[
"Compare this to the time required by the webbrowser",
"_____no_output_____"
]
],
[
[
"webbrowser_timer.elapsed_time()",
"_____no_output_____"
]
],
[
[
"The difference in time is huge:",
"_____no_output_____"
]
],
[
[
"webbrowser_timer.elapsed_time() / avg_urlparse_time",
"_____no_output_____"
]
],
[
[
"Hence, in the time it takes to run `webbrowser()` once, we can have _tens of thousands_ of executions of `urlparse()` – and this does not even take into account the time it takes the browser to render the downloaded HTML, to run the included scripts, and whatever else happens when a Web page is loaded. Hence, strategies that allow us to test at the _unit_ level are very promising as they can save lots of overhead.",
"_____no_output_____"
],
[
"## Carving Unit Tests\n\nTesting methods and functions at the unit level requires a very good understanding of the individual units to be tested as well as their interplay with other units. Setting up an appropriate infrastructure and writing unit tests by hand thus is demanding, yet rewarding. There is, however, an interesting alternative to writing unit tests by hand. The technique of _carving_ automatically _converts system tests into unit tests_ by means of recording and replaying function calls:\n\n1. During a system test (given or generated), we _record_ all calls into a function, including all arguments and other variables the function reads.\n2. From these, we synthesize a self-contained _unit test_ that reconstructs the function call with all arguments.\n3. This unit test can be executed (replayed) at any time with high efficiency.\n\nIn the remainder of this chapter, let us explore these steps.",
"_____no_output_____"
],
[
"## Recording Calls\n\nOur first challenge is to record function calls together with their arguments. (In the interest of simplicity, we restrict ourself to arguments, ignoring any global variables or other non-arguments that are read by the function.) To record calls and arguments, we use the mechanism [we introduced for coverage](Coverage.ipynb): By setting up a tracer function, we track all calls into individual functions, also saving their arguments. Just like `Coverage` objects, we want to use `Carver` objects to be able to be used in conjunction with the `with` statement, such that we can trace a particular code block:\n\n```python\nwith Carver() as carver:\n function_to_be_traced()\nc = carver.calls()\n```\n\nThe initial definition supports this construct:",
"_____no_output_____"
],
[
"\\todo{Get tracker from [dynamic invariants](DynamicInvariants.ipynb)}",
"_____no_output_____"
]
],
[
[
"import sys",
"_____no_output_____"
],
[
"class Carver(object):\n def __init__(self, log=False):\n self._log = log\n self.reset()\n\n def reset(self):\n self._calls = {}\n\n # Start of `with` block\n def __enter__(self):\n self.original_trace_function = sys.gettrace()\n sys.settrace(self.traceit)\n return self\n\n # End of `with` block\n def __exit__(self, exc_type, exc_value, tb):\n sys.settrace(self.original_trace_function)",
"_____no_output_____"
]
],
[
[
"The actual work takes place in the `traceit()` method, which records all calls in the `_calls` attribute. First, we define two helper functions:",
"_____no_output_____"
]
],
[
[
"import inspect",
"_____no_output_____"
],
[
"def get_qualified_name(code):\n \"\"\"Return the fully qualified name of the current function\"\"\"\n name = code.co_name\n module = inspect.getmodule(code)\n if module is not None:\n name = module.__name__ + \".\" + name\n return name",
"_____no_output_____"
],
[
"def get_arguments(frame):\n \"\"\"Return call arguments in the given frame\"\"\"\n # When called, all arguments are local variables\n arguments = [(var, frame.f_locals[var]) for var in frame.f_locals]\n arguments.reverse() # Want same order as call\n return arguments",
"_____no_output_____"
],
[
"class CallCarver(Carver):\n def add_call(self, function_name, arguments):\n \"\"\"Add given call to list of calls\"\"\"\n if function_name not in self._calls:\n self._calls[function_name] = []\n self._calls[function_name].append(arguments)\n\n # Tracking function: Record all calls and all args\n def traceit(self, frame, event, arg):\n if event != \"call\":\n return None\n\n code = frame.f_code\n function_name = code.co_name\n qualified_name = get_qualified_name(code)\n arguments = get_arguments(frame)\n\n self.add_call(function_name, arguments)\n if qualified_name != function_name:\n self.add_call(qualified_name, arguments)\n\n if self._log:\n print(simple_call_string(function_name, arguments))\n\n return None",
"_____no_output_____"
]
],
[
[
"Finally, we need some convenience functions to access the calls:",
"_____no_output_____"
]
],
[
[
"class CallCarver(CallCarver):\n def calls(self):\n \"\"\"Return a dictionary of all calls traced.\"\"\"\n return self._calls\n\n def arguments(self, function_name):\n \"\"\"Return a list of all arguments of the given function\n as (VAR, VALUE) pairs.\n Raises an exception if the function was not traced.\"\"\"\n return self._calls[function_name]\n\n def called_functions(self, qualified=False):\n \"\"\"Return all functions called.\"\"\"\n if qualified:\n return [function_name for function_name in self._calls.keys()\n if function_name.find('.') >= 0]\n else:\n return [function_name for function_name in self._calls.keys()\n if function_name.find('.') < 0]",
"_____no_output_____"
]
],
[
[
"### Recording my_sqrt()",
"_____no_output_____"
],
[
"Let's try out our new `Carver` class – first on a very simple function:",
"_____no_output_____"
]
],
[
[
"from Intro_Testing import my_sqrt",
"_____no_output_____"
],
[
"with CallCarver() as sqrt_carver:\n my_sqrt(2)\n my_sqrt(4)",
"_____no_output_____"
]
],
[
[
"We can retrieve all calls seen...",
"_____no_output_____"
]
],
[
[
"sqrt_carver.calls()",
"_____no_output_____"
],
[
"sqrt_carver.called_functions()",
"_____no_output_____"
]
],
[
[
"... as well as the arguments of a particular function:",
"_____no_output_____"
]
],
[
[
"sqrt_carver.arguments(\"my_sqrt\")",
"_____no_output_____"
]
],
[
[
"We define a convenience function for nicer printing of these lists:",
"_____no_output_____"
]
],
[
[
"def simple_call_string(function_name, argument_list):\n \"\"\"Return function_name(arg[0], arg[1], ...) as a string\"\"\"\n return function_name + \"(\" + \\\n \", \".join([var + \"=\" + repr(value)\n for (var, value) in argument_list]) + \")\"",
"_____no_output_____"
],
[
"for function_name in sqrt_carver.called_functions():\n for argument_list in sqrt_carver.arguments(function_name):\n print(simple_call_string(function_name, argument_list))",
"my_sqrt(x=2)\nmy_sqrt(x=4)\n__exit__(self=<__main__.CallCarver object at 0x7f836f7a4518>, exc_type=None, exc_value=None, tb=None)\n"
]
],
[
[
"This is a syntax we can directly use to invoke `my_sqrt()` again:",
"_____no_output_____"
]
],
[
[
"eval(\"my_sqrt(x=2)\")",
"_____no_output_____"
]
],
[
[
"### Carving urlparse()",
"_____no_output_____"
],
[
"What happens if we apply this to `webbrowser()`?",
"_____no_output_____"
]
],
[
[
"with CallCarver() as webbrowser_carver:\n webbrowser(\"http://www.example.com\")",
"_____no_output_____"
]
],
[
[
"We see that retrieving a URL from the Web requires quite some functionality:",
"_____no_output_____"
]
],
[
[
"function_list = webbrowser_carver.called_functions(qualified=True)\nlen(function_list)",
"_____no_output_____"
],
[
"print(function_list[:50])",
"['requests.api.get', 'requests.api.request', 'requests.sessions.__init__', 'requests.utils.default_headers', 'requests.utils.default_user_agent', 'requests.structures.__init__', 'collections.abc.update', 'abc.__instancecheck__', '_weakrefset.__contains__', 'requests.structures.__setitem__', 'requests.hooks.default_hooks', 'requests.hooks.<dictcomp>', 'requests.cookies.cookiejar_from_dict', 'http.cookiejar.__init__', 'threading.RLock', 'http.cookiejar.__iter__', 'requests.cookies.<listcomp>', 'http.cookiejar.deepvalues', 'http.cookiejar.vals_sorted_by_key', 'requests.adapters.__init__', 'urllib3.util.retry.__init__', 'urllib3.util.retry.<listcomp>', 'requests.adapters.init_poolmanager', 'urllib3.poolmanager.__init__', 'urllib3.request.__init__', 'urllib3._collections.__init__', 'requests.sessions.mount', 'requests.sessions.<listcomp>', 'requests.sessions.__enter__', 'requests.sessions.request', 'requests.models.__init__', 'requests.sessions.prepare_request', 'requests.cookies.merge_cookies', 'requests.cookies.update', 'requests.utils.get_netrc_auth', 'collections.abc.get', 'os.__getitem__', 'os.encode', 'requests.utils.<genexpr>', 'posixpath.expanduser', 'posixpath._get_sep', 'collections.abc.__contains__', 'os.decode', 'genericpath.exists', 'urllib.parse.urlparse', 'urllib.parse._coerce_args', 'urllib.parse.urlsplit', 'urllib.parse._splitnetloc', 'urllib.parse._checknetloc', 'urllib.parse.<genexpr>']\n"
]
],
[
[
"Among several other functions, we also have a call to `urlparse()`:",
"_____no_output_____"
]
],
[
[
"urlparse_argument_list = webbrowser_carver.arguments(\"urllib.parse.urlparse\")\nurlparse_argument_list",
"_____no_output_____"
]
],
[
[
"Again, we can convert this into a well-formatted call:",
"_____no_output_____"
]
],
[
[
"urlparse_call = simple_call_string(\"urlparse\", urlparse_argument_list[0])\nurlparse_call",
"_____no_output_____"
]
],
[
[
"Again, we can re-execute this call:",
"_____no_output_____"
]
],
[
[
"eval(urlparse_call)",
"_____no_output_____"
]
],
[
[
"We now have successfully carved the call to `urlparse()` out of the `webbrowser()` execution.",
"_____no_output_____"
],
[
"## Replaying Calls",
"_____no_output_____"
],
[
"Replaying calls in their entirety and in all generality is tricky, as there are several challenges to be addressed. These include:\n\n1. We need to be able to _access_ individual functions. If we access a function by name, the name must be in scope. If the name is not visible (for instance, because it is a name internal to the module), we must make it visible.\n\n2. Any _resources_ accessed outside of arguments must be recorded and reconstructed for replay as well. This can be difficult if variables refer to external resources such as files or network resources.\n\n3. _Complex objects_ must be reconstructed as well.\n\nThese constraints make carving hard or even impossible if the function to be tested interacts heavily with its environment. To illustrate these issues, consider the `email.parser.parse()` method that is invoked in `webbrowser()`:",
"_____no_output_____"
]
],
[
[
"email_parse_argument_list = webbrowser_carver.arguments(\"email.parser.parse\")",
"_____no_output_____"
]
],
[
[
"Calls to this method look like this:",
"_____no_output_____"
]
],
[
[
"email_parse_call = simple_call_string(\n \"email.parser.parse\",\n email_parse_argument_list[0])\nemail_parse_call",
"_____no_output_____"
]
],
[
[
"We see that `email.parser.parse()` is part of a `email.parser.Parser` object and it gets a `StringIO` object. Both are non-primitive values. How could we possibly reconstruct them?",
"_____no_output_____"
],
[
"### Serializing Objects\n\nThe answer to the problem of complex objects lies in creating a _persistent_ representation that can be _reconstructed_ at later points in time. This process is known as _serialization_; in Python, it is also known as _pickling_. The `pickle` module provides means to create a serialized representation of an object. Let us apply this on the `email.parser.Parser` object we just found:",
"_____no_output_____"
]
],
[
[
"import pickle ",
"_____no_output_____"
],
[
"parser_object = email_parse_argument_list[0][0][1]\nparser_object",
"_____no_output_____"
],
[
"pickled = pickle.dumps(parser_object)\npickled",
"_____no_output_____"
]
],
[
[
"From this string representing the serialized `email.parser.Parser` object, we can recreate the Parser object at any time:",
"_____no_output_____"
]
],
[
[
"unpickled_parser_object = pickle.loads(pickled)\nunpickled_parser_object",
"_____no_output_____"
]
],
[
[
"The serialization mechanism allows us to produce a representation for all objects passed as parameters (assuming they can be pickled, that is). We can now extend the `simple_call_string()` function such that it automatically pickles objects. Additionally, we set it up such that if the first parameter is named `self` (i.e., it is a class method), we make it a method of the `self` object.",
"_____no_output_____"
]
],
[
[
"def call_value(value):\n value_as_string = repr(value)\n if value_as_string.find('<') >= 0:\n # Complex object\n value_as_string = \"pickle.loads(\" + repr(pickle.dumps(value)) + \")\"\n return value_as_string",
"_____no_output_____"
],
[
"def call_string(function_name, argument_list):\n \"\"\"Return function_name(arg[0], arg[1], ...) as a string, pickling complex objects\"\"\"\n if len(argument_list) > 0:\n (first_var, first_value) = argument_list[0]\n if first_var == \"self\":\n # Make this a method call\n method_name = function_name.split(\".\")[-1]\n function_name = call_value(first_value) + \".\" + method_name\n argument_list = argument_list[1:]\n\n return function_name + \"(\" + \\\n \", \".join([var + \"=\" + call_value(value)\n for (var, value) in argument_list]) + \")\"",
"_____no_output_____"
]
],
[
[
"Let us apply the extended `call_string()` method to create a call for `email.parser.parse()`, including pickled objects:",
"_____no_output_____"
]
],
[
[
"call = call_string(\"email.parser.parse\", email_parse_argument_list[0])\nprint(call)",
"pickle.loads(b'\\x80\\x03cemail.parser\\nParser\\nq\\x00)\\x81q\\x01}q\\x02(X\\x06\\x00\\x00\\x00_classq\\x03chttp.client\\nHTTPMessage\\nq\\x04X\\x06\\x00\\x00\\x00policyq\\x05cemail._policybase\\nCompat32\\nq\\x06)\\x81q\\x07ub.').parse(fp=pickle.loads(b'\\x80\\x03c_io\\nStringIO\\nq\\x00)\\x81q\\x01(X\\xb2\\x00\\x00\\x00Connection: close\\r\\nContent-Length: 245\\r\\nContent-Type: text/html\\r\\nLocation: http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9\\r\\n\\r\\nq\\x02X\\x01\\x00\\x00\\x00\\nq\\x03K\\xb2Ntq\\x04b.'), headersonly=False)\n"
]
],
[
[
"With this call involvimng the pickled object, we can now re-run the original call and obtain a valid result:",
"_____no_output_____"
]
],
[
[
"eval(call)",
"_____no_output_____"
]
],
[
[
"### All Calls\n\nSo far, we have seen only one call of `webbrowser()`. How many of the calls within `webbrowser()` can we actually carve and replay? Let us try this out and compute the numbers.",
"_____no_output_____"
]
],
[
[
"import traceback",
"_____no_output_____"
],
[
"import enum\nimport socket",
"_____no_output_____"
],
[
"all_functions = set(webbrowser_carver.called_functions(qualified=True))\ncall_success = set()\nrun_success = set()",
"_____no_output_____"
],
[
"exceptions_seen = set()\n\nfor function_name in webbrowser_carver.called_functions(qualified=True):\n for argument_list in webbrowser_carver.arguments(function_name):\n try:\n call = call_string(function_name, argument_list)\n call_success.add(function_name)\n\n result = eval(call)\n run_success.add(function_name)\n\n except Exception as exc:\n exceptions_seen.add(repr(exc))\n # print(\"->\", call, file=sys.stderr)\n # traceback.print_exc()\n # print(\"\", file=sys.stderr)\n continue",
"_____no_output_____"
],
[
"print(\"%d/%d calls (%.2f%%) successfully created and %d/%d calls (%.2f%%) successfully ran\" % (\n len(call_success), len(all_functions), len(\n call_success) * 100 / len(all_functions),\n len(run_success), len(all_functions), len(run_success) * 100 / len(all_functions)))",
"287/338 calls (84.91%) successfully created and 118/338 calls (34.91%) successfully ran\n"
]
],
[
[
"About half of the calls succeed. Let us take a look into some of the error messages we get:",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print(list(exceptions_seen)[i])",
"SyntaxError('invalid syntax', ('<string>', 1, 16, \"requests.utils.<lambda>(k='no_proxy')\"))\nCannotSendHeader()\nSyntaxError('invalid syntax', ('<string>', 1, 18, \"urllib3.util.url.<listcomp>(.0=pickle.loads(b'\\\\x80\\\\x03cbuiltins\\\\niter\\\\nq\\\\x00X\\\\x00\\\\x00\\\\x00\\\\x00q\\\\x01\\\\x85q\\\\x02Rq\\\\x03.'))\"))\nPicklingError(\"Can't pickle <class 'method_descriptor'>: attribute lookup method_descriptor on builtins failed\",)\nTypeError(\"can't pickle _thread.RLock objects\",)\nTypeError(\"wrapper __setitem__ doesn't take keyword arguments\",)\nAttributeError(\"module 'urllib.request' has no attribute 'ip2num'\",)\nValueError('Timeout value connect was <object object at 0x7f836f0d1660>, but it must be an int, float or None.',)\nSyntaxError('invalid syntax', ('<string>', 1, 18, \"urllib3.response.<genexpr>(.0=pickle.loads(b'\\\\x80\\\\x03cbuiltins\\\\niter\\\\nq\\\\x00]q\\\\x01X\\\\x07\\\\x00\\\\x00\\\\x00chunkedq\\\\x02a\\\\x85q\\\\x03Rq\\\\x04K\\\\x01b.'))\"))\nTypeError('__contains__() takes no keyword arguments',)\n"
]
],
[
[
"We see that:\n\n* **A large majority of calls could be converted into call strings.** If this is not the case, this is mostly due to having unserialized objects being passed.\n* **About half of the calls could be executed.** The error messages for the failing runs are varied; the most frequent being that some internal name is invoked that is not in scope.",
"_____no_output_____"
],
[
"Our carving mechanism should be taken with a grain of salt: We still do not cover the situation where external variables and values (such as global variables) are being accessed, and the serialization mechanism cannot recreate external resources. Still, if the function of interest falls among those that _can_ be carved and replayed, we can very effectively re-run its calls with their original arguments.",
"_____no_output_____"
],
[
"## Mining API Grammars from Carved Calls\n\nSo far, we have used carved calls to replay exactly the same invocations as originally encountered. However, we can also _mutate_ carved calls to effectively fuzz APIs with previously recorded arguments.\n\nThe general idea is as follows:\n\n1. First, we record all calls of a specific function from a given execution of the program.\n2. Second, we create a grammar that incorporates all these calls, with separate rules for each argument and alternatives for each value found; this allows us to produce calls that arbitrarily _recombine_ these arguments.\n\nLet us explore these steps in the following sections.",
"_____no_output_____"
],
[
"### From Calls to Grammars\n\nLet us start with an example. The `power(x, y)` function returns $x^y$; it is but a wrapper around the equivalent `math.pow()` function. (Since `power()` is defined in Python, we can trace it – in contrast to `math.pow()`, which is implemented in C.)",
"_____no_output_____"
]
],
[
[
"import math",
"_____no_output_____"
],
[
"def power(x, y):\n return math.pow(x, y)",
"_____no_output_____"
]
],
[
[
"Let us invoke `power()` while recording its arguments:",
"_____no_output_____"
]
],
[
[
"with CallCarver() as power_carver:\n z = power(1, 2)\n z = power(3, 4)",
"_____no_output_____"
],
[
"power_carver.arguments(\"power\")",
"_____no_output_____"
]
],
[
[
"From this list of recorded arguments, we could now create a grammar for the `power()` call, with `x` and `y` expanding into the values seen:",
"_____no_output_____"
]
],
[
[
"from Grammars import START_SYMBOL, is_valid_grammar, new_symbol, extend_grammar",
"_____no_output_____"
],
[
"POWER_GRAMMAR = {\n \"<start>\": [\"power(<x>, <y>)\"],\n \"<x>\": [\"1\", \"3\"],\n \"<y>\": [\"2\", \"4\"]\n}\n\nassert is_valid_grammar(POWER_GRAMMAR)",
"_____no_output_____"
]
],
[
[
"When fuzzing with this grammar, we then get arbitrary combinations of `x` and `y`; aiming for coverage will ensure that all values are actually tested at least once:",
"_____no_output_____"
]
],
[
[
"from GrammarCoverageFuzzer import GrammarCoverageFuzzer",
"_____no_output_____"
],
[
"power_fuzzer = GrammarCoverageFuzzer(POWER_GRAMMAR)\n[power_fuzzer.fuzz() for i in range(5)]",
"_____no_output_____"
]
],
[
[
"What we need is a method to automatically convert the arguments as seen in `power_carver` to the grammar as seen in `POWER_GRAMMAR`. This is what we define in the next section.",
"_____no_output_____"
],
[
"### A Grammar Miner for Calls\n\nWe introduce a class `CallGrammarMiner`, which, given a `Carver`, automatically produces a grammar from the calls seen. To initialize, we pass the carver object:",
"_____no_output_____"
]
],
[
[
"class CallGrammarMiner(object):\n def __init__(self, carver, log=False):\n self.carver = carver\n self.log = log",
"_____no_output_____"
]
],
[
[
"#### Initial Grammar\n\nThe initial grammar produces a single call. The possible `<call>` expansions are to be constructed later:",
"_____no_output_____"
]
],
[
[
"import copy ",
"_____no_output_____"
],
[
"class CallGrammarMiner(CallGrammarMiner):\n CALL_SYMBOL = \"<call>\"\n\n def initial_grammar(self):\n return extend_grammar(\n {START_SYMBOL: [self.CALL_SYMBOL],\n self.CALL_SYMBOL: []\n })",
"_____no_output_____"
],
[
"m = CallGrammarMiner(power_carver)\ninitial_grammar = m.initial_grammar()\ninitial_grammar",
"_____no_output_____"
]
],
[
[
"#### A Grammar from Arguments\n\nLet us start by creating a grammar from a list of arguments. The method `mine_arguments_grammar()` creates a grammar for the arguments seen during carving, such as these:",
"_____no_output_____"
]
],
[
[
"arguments = power_carver.arguments(\"power\")\narguments",
"_____no_output_____"
]
],
[
[
"The `mine_arguments_grammar()` method iterates through the variables seen and creates a mapping `variables` of variable names to a set of values seen (as strings, going through `call_value()`). In a second step, it then creates a grammar with a rule for each variable name, expanding into the values seen.",
"_____no_output_____"
]
],
[
[
"class CallGrammarMiner(CallGrammarMiner):\n def var_symbol(self, function_name, var, grammar):\n return new_symbol(grammar, \"<\" + function_name + \"-\" + var + \">\")\n\n def mine_arguments_grammar(self, function_name, arguments, grammar):\n var_grammar = {}\n\n variables = {}\n for argument_list in arguments:\n for (var, value) in argument_list:\n value_string = call_value(value)\n if self.log:\n print(var, \"=\", value_string)\n\n if value_string.find(\"<\") >= 0:\n var_grammar[\"<langle>\"] = [\"<\"]\n value_string = value_string.replace(\"<\", \"<langle>\")\n\n if var not in variables:\n variables[var] = set()\n variables[var].add(value_string)\n\n var_symbols = []\n for var in variables:\n var_symbol = self.var_symbol(function_name, var, grammar)\n var_symbols.append(var_symbol)\n var_grammar[var_symbol] = list(variables[var])\n\n return var_grammar, var_symbols",
"_____no_output_____"
],
[
"m = CallGrammarMiner(power_carver)\nvar_grammar, var_symbols = m.mine_arguments_grammar(\n \"power\", arguments, initial_grammar)",
"_____no_output_____"
],
[
"var_grammar",
"_____no_output_____"
]
],
[
[
"The additional return value `var_symbols` is a list of argument symbols in the call:",
"_____no_output_____"
]
],
[
[
"var_symbols",
"_____no_output_____"
]
],
[
[
"#### A Grammar from Calls\n\nTo get the grammar for a single function (`mine_function_grammar()`), we add a call to the function:",
"_____no_output_____"
]
],
[
[
"class CallGrammarMiner(CallGrammarMiner):\n def function_symbol(self, function_name, grammar):\n return new_symbol(grammar, \"<\" + function_name + \">\")\n\n def mine_function_grammar(self, function_name, grammar):\n arguments = self.carver.arguments(function_name)\n\n if self.log:\n print(function_name, arguments)\n\n var_grammar, var_symbols = self.mine_arguments_grammar(\n function_name, arguments, grammar)\n\n function_grammar = var_grammar\n function_symbol = self.function_symbol(function_name, grammar)\n\n if len(var_symbols) > 0 and var_symbols[0].find(\"-self\") >= 0:\n # Method call\n function_grammar[function_symbol] = [\n var_symbols[0] + \".\" + function_name + \"(\" + \", \".join(var_symbols[1:]) + \")\"]\n else:\n function_grammar[function_symbol] = [\n function_name + \"(\" + \", \".join(var_symbols) + \")\"]\n\n if self.log:\n print(function_symbol, \"::=\", function_grammar[function_symbol])\n\n return function_grammar, function_symbol",
"_____no_output_____"
],
[
"m = CallGrammarMiner(power_carver)\nfunction_grammar, function_symbol = m.mine_function_grammar(\n \"power\", initial_grammar)\nfunction_grammar",
"_____no_output_____"
]
],
[
[
"The additionally returned `function_symbol` holds the name of the function call just added:",
"_____no_output_____"
]
],
[
[
"function_symbol",
"_____no_output_____"
]
],
[
[
"#### A Grammar from all Calls\n\nLet us now repeat the above for all function calls seen during carving. To this end, we simply iterate over all function calls seen:",
"_____no_output_____"
]
],
[
[
"power_carver.called_functions()",
"_____no_output_____"
],
[
"class CallGrammarMiner(CallGrammarMiner):\n def mine_call_grammar(self, function_list=None, qualified=False):\n grammar = self.initial_grammar()\n fn_list = function_list\n if function_list is None:\n fn_list = self.carver.called_functions(qualified=qualified)\n\n for function_name in fn_list:\n if function_list is None and (function_name.startswith(\"_\") or function_name.startswith(\"<\")):\n continue # Internal function\n\n # Ignore errors with mined functions\n try:\n function_grammar, function_symbol = self.mine_function_grammar(\n function_name, grammar)\n except:\n if function_list is not None:\n raise\n\n if function_symbol not in grammar[self.CALL_SYMBOL]:\n grammar[self.CALL_SYMBOL].append(function_symbol)\n grammar.update(function_grammar)\n\n assert is_valid_grammar(grammar)\n return grammar",
"_____no_output_____"
]
],
[
[
"The method `mine_call_grammar()` is the one that clients can and should use – first for mining...",
"_____no_output_____"
]
],
[
[
"m = CallGrammarMiner(power_carver)\npower_grammar = m.mine_call_grammar()\npower_grammar",
"_____no_output_____"
]
],
[
[
"...and then for fuzzing:",
"_____no_output_____"
]
],
[
[
"power_fuzzer = GrammarCoverageFuzzer(power_grammar)\n[power_fuzzer.fuzz() for i in range(5)]",
"_____no_output_____"
]
],
[
[
"With this, we have successfully extracted a grammar from a recorded execution; in contrast to \"simple\" carving, our grammar allows us to _recombine_ arguments and thus to fuzz at the API level.",
"_____no_output_____"
],
[
"## Fuzzing Web Functions\n\nLet us now apply our grammar miner on a larger API – the `urlparse()` function we already encountered during carving.",
"_____no_output_____"
]
],
[
[
"with CallCarver() as webbrowser_carver:\n webbrowser(\"https://www.fuzzingbook.org\")\n webbrowser(\"http://www.example.com\")",
"_____no_output_____"
]
],
[
[
"We can mine a grammar from the calls encountered:",
"_____no_output_____"
]
],
[
[
"m = CallGrammarMiner(webbrowser_carver)\nwebbrowser_grammar = m.mine_call_grammar()",
"_____no_output_____"
]
],
[
[
"This is a rather large grammar:",
"_____no_output_____"
]
],
[
[
"call_list = webbrowser_grammar['<call>']\nlen(call_list)",
"_____no_output_____"
],
[
"print(call_list[:20])",
"['<webbrowser>', '<default_headers>', '<default_user_agent>', '<update>', '<default_hooks>', '<cookiejar_from_dict>', '<RLock>', '<deepvalues>', '<vals_sorted_by_key>', '<init_poolmanager>', '<mount>', '<prepare_request>', '<merge_cookies>', '<get_netrc_auth>', '<encode>', '<expanduser>', '<exists>', '<urlparse>', '<urlsplit>', '<getpreferredencoding>']\n"
]
],
[
[
"Here's the rule for the `urlsplit()` function:",
"_____no_output_____"
]
],
[
[
"webbrowser_grammar[\"<urlsplit>\"]",
"_____no_output_____"
]
],
[
[
"Here are the arguments. Note that although we only passed `http://www.fuzzingbook.org` as a parameter, we also see the `https:` variant. That is because opening the `http:` URL automatically redirects to the `https:` URL, which is then also processed by `urlsplit()`.",
"_____no_output_____"
]
],
[
[
"webbrowser_grammar[\"<urlsplit-url>\"]",
"_____no_output_____"
]
],
[
[
"There also is some variation in the `scheme` argument:",
"_____no_output_____"
]
],
[
[
"webbrowser_grammar[\"<urlsplit-scheme>\"]",
"_____no_output_____"
]
],
[
[
"If we now apply a fuzzer on these rules, we systematically cover all variations of arguments seen, including, of course, combinations not seen during carving. Again, we are fuzzing at the API level here.",
"_____no_output_____"
]
],
[
[
"urlsplit_fuzzer = GrammarCoverageFuzzer(\n webbrowser_grammar, start_symbol=\"<urlsplit>\")\nfor i in range(5):\n print(urlsplit_fuzzer.fuzz())",
"urlsplit('http://www.example.com', '', True)\nurlsplit('https://www.fuzzingbook.org', '', True)\nurlsplit('https://www.fuzzingbook.org/', '', True)\nurlsplit('http://www.example.com/', '', True)\nurlsplit('http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', '', True)\n"
]
],
[
[
"Just as seen with carving, running tests at the API level is orders of magnitude faster than executing system tests. Hence, this calls for means to fuzz at the method level:",
"_____no_output_____"
]
],
[
[
"from urllib.parse import urlsplit",
"_____no_output_____"
],
[
"from Timer import Timer",
"_____no_output_____"
],
[
"with Timer() as urlsplit_timer:\n urlsplit('http://www.fuzzingbook.org/', 'http', True)\nurlsplit_timer.elapsed_time()",
"_____no_output_____"
],
[
"with Timer() as webbrowser_timer:\n webbrowser(\"http://www.fuzzingbook.org\")\nwebbrowser_timer.elapsed_time()",
"_____no_output_____"
],
[
"webbrowser_timer.elapsed_time() / urlsplit_timer.elapsed_time()",
"_____no_output_____"
]
],
[
[
"But then again, the caveats encountered during carving apply, notably the requirement to recreate the original function environment. If we also alter or recombine arguments, we get the additional risk of _violating an implicit precondition_ – that is, invoking a function with arguments the function was never designed for. Such _false alarms_, resulting from incorrect invocations rather than incorrect implementations, must then be identified (typically manually) and wed out (for instance, by altering or constraining the grammar). The huge speed gains at the API level, however, may well justify this additional investment.",
"_____no_output_____"
],
[
"## Synopsis\n\nThis chapter provides means to _record and replay function calls_ during a system test. Since individual function calls are much faster than a whole system run, such \"carving\" mechanisms have the potential to run tests much faster.",
"_____no_output_____"
],
[
"### Recording Calls\n\nThe `CallCarver` class records all calls occurring while it is active. It is used in conjunction with a `with` clause:",
"_____no_output_____"
]
],
[
[
"with CallCarver() as carver:\n y = my_sqrt(2)\n y = my_sqrt(4)",
"_____no_output_____"
]
],
[
[
"After execution, `called_functions()` lists the names of functions encountered:",
"_____no_output_____"
]
],
[
[
"carver.called_functions()",
"_____no_output_____"
]
],
[
[
"The `arguments()` method lists the arguments recorded for a function. This is a mapping of the function name to a list of lists of arguments; each argument is a pair (parameter name, value).",
"_____no_output_____"
]
],
[
[
"carver.arguments('my_sqrt')",
"_____no_output_____"
]
],
[
[
"Complex arguments are properly serialized, such that they can be easily restored.",
"_____no_output_____"
],
[
"### Synthesizing Calls\n\nWhile such recorded arguments already could be turned into arguments and calls, a much nicer alternative is to create a _grammar_ for recorded calls. This allows to synthesize arbitrary _combinations_ of arguments, and also offers a base for further customization of calls.",
"_____no_output_____"
],
[
"The `CallGrammarMiner` class turns a list of carved executions into a grammar.",
"_____no_output_____"
]
],
[
[
"my_sqrt_miner = CallGrammarMiner(carver)\nmy_sqrt_grammar = my_sqrt_miner.mine_call_grammar()\nmy_sqrt_grammar",
"_____no_output_____"
]
],
[
[
"This grammar can be used to synthesize calls.",
"_____no_output_____"
]
],
[
[
"fuzzer = GrammarCoverageFuzzer(my_sqrt_grammar)\nfuzzer.fuzz()",
"_____no_output_____"
]
],
[
[
"These calls can be executed in isolation, effectively extracting unit tests from system tests:",
"_____no_output_____"
]
],
[
[
"eval(fuzzer.fuzz())",
"_____no_output_____"
]
],
[
[
"## Lessons Learned\n\n* _Carving_ allows for effective replay of function calls recorded during a system test.\n* A function call can be _orders of magnitude faster_ than a system invocation.\n* _Serialization_ allows to create persistent representations of complex objects.\n* Functions that heavily interact with their environment and/or access external resources are difficult to carve.\n* From carved calls, one can produce API grammars that arbitrarily combine carved arguments.",
"_____no_output_____"
],
[
"## Next Steps\n\nIn the next chapter, we will discuss [how to reduce failure-inducing inputs](Reducer.ipynb).",
"_____no_output_____"
],
[
"## Background\n\nCarving was invented by Elbaum et al. \\cite{Elbaum2006} and originally implemented for Java. In this chapter, we follow several of their design choices (including recording and serializing method arguments only).\n\nThe combination of carving and fuzzing at the API level is described in \\cite{Kampmann2018}.",
"_____no_output_____"
],
[
"## Exercises\n\n### Exercise 1: Carving for Regression Testing\n\nSo far, during carving, we only have looked into reproducing _calls_, but not into actually checking the _results_ of these calls. This is important for _regression testing_ – i.e. checking whether a change to code does not impede existing functionality. We can build this by recording not only _calls_, but also _return values_ – and then later compare whether the same calls result in the same values. This may not work on all occasions; values that depend on time, randomness, or other external factors may be different. Still, for functionality that abstracts from these details, checking that nothing has changed is an important part of testing.",
"_____no_output_____"
],
[
"Our aim is to design a class `ResultCarver` that extends `CallCarver` by recording both calls and return values.\n\nIn a first step, create a `traceit()` method that also tracks return values by extending the `traceit()` method. The `traceit()` event type is `\"return\"` and the `arg` parameter is the returned value. Here is a prototype that only prints out the returned values:",
"_____no_output_____"
]
],
[
[
"class ResultCarver(CallCarver):\n def traceit(self, frame, event, arg):\n if event == \"return\":\n if self._log:\n print(\"Result:\", arg)\n\n super().traceit(frame, event, arg)\n # Need to return traceit function such that it is invoked for return\n # events\n return self.traceit",
"_____no_output_____"
],
[
"with ResultCarver(log=True) as result_carver:\n my_sqrt(2)",
"my_sqrt(x=2)\nResult: 1.414213562373095\n__exit__(self=<__main__.ResultCarver object at 0x7f836c9ddcc0>, exc_type=None, exc_value=None, tb=None)\n"
]
],
[
[
"#### Part 1: Store function results\n\nExtend the above code such that results are _stored_ in a way that associates them with the currently returning function (or method). To this end, you need to keep track of the _current stack of called functions_.",
"_____no_output_____"
],
[
"**Solution.** Here's a solution, building on the above:",
"_____no_output_____"
]
],
[
[
"class ResultCarver(CallCarver):\n def reset(self):\n super().reset()\n self._call_stack = []\n self._results = {}\n\n def add_result(self, function_name, arguments, result):\n key = simple_call_string(function_name, arguments)\n self._results[key] = result\n\n def traceit(self, frame, event, arg):\n if event == \"call\":\n code = frame.f_code\n function_name = code.co_name\n qualified_name = get_qualified_name(code)\n self._call_stack.append(\n (function_name, qualified_name, get_arguments(frame)))\n\n if event == \"return\":\n result = arg\n (function_name, qualified_name, arguments) = self._call_stack.pop()\n self.add_result(function_name, arguments, result)\n if function_name != qualified_name:\n self.add_result(qualified_name, arguments, result)\n if self._log:\n print(\n simple_call_string(\n function_name,\n arguments),\n \"=\",\n result)\n\n # Keep on processing current calls\n super().traceit(frame, event, arg)\n\n # Need to return traceit function such that it is invoked for return\n # events\n return self.traceit",
"_____no_output_____"
],
[
"with ResultCarver(log=True) as result_carver:\n my_sqrt(2)\nresult_carver._results",
"my_sqrt(x=2)\nmy_sqrt(x=2) = 1.414213562373095\n__exit__(self=<__main__.ResultCarver object at 0x7f83707afba8>, exc_type=None, exc_value=None, tb=None)\n"
]
],
[
[
"#### Part 2: Access results\n\n Give it a method `result()` that returns the value recorded for that particular function name and result:\n\n```python\nclass ResultCarver(CallCarver):\n def result(self, function_name, argument):\n \"\"\"Returns the result recorded for function_name(argument\"\"\"\n```",
"_____no_output_____"
],
[
"**Solution.** This is mostly done in the code for part 1:",
"_____no_output_____"
]
],
[
[
"class ResultCarver(ResultCarver):\n def result(self, function_name, argument):\n key = simple_call_string(function_name, arguments)\n return self._results[key]",
"_____no_output_____"
]
],
[
[
"#### Part 3: Produce assertions\n\nFor the functions called during `webbrowser()` execution, create a set of _assertions_ that check whether the result returned is still the same. Test this for `urllib.parse.urlparse()` and `urllib.parse.urlsplit()`.",
"_____no_output_____"
],
[
"**Solution.** Not too hard now:",
"_____no_output_____"
]
],
[
[
"with ResultCarver() as webbrowser_result_carver:\n webbrowser(\"http://www.example.com\")",
"_____no_output_____"
],
[
"for function_name in [\"urllib.parse.urlparse\", \"urllib.parse.urlsplit\"]:\n for arguments in webbrowser_result_carver.arguments(function_name):\n try:\n call = call_string(function_name, arguments)\n result = webbrowser_result_carver.result(function_name, arguments)\n print(\"assert\", call, \"==\", call_value(result))\n except Exception:\n continue",
"assert urllib.parse.urlparse(url='http://www.example.com', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://www.example.com/', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://www.example.com/', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://www.example.com/', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://www.example.com/', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://www.example.com/', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://www.example.com/', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://www.example.com/', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://www.example.com/', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://www.example.com/', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://www.example.com/', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://www.example.com/', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlparse(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == ParseResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', params='', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://www.example.com/', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\nassert urllib.parse.urlsplit(url='http://fritz.box/tools/kids_not_allowed.lua?account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', scheme='', allow_fragments=True) == SplitResult(scheme='http', netloc='fritz.box', path='/tools/kids_not_allowed.lua', query='account=default-2a02:810c:c240:15dc:a924:3ce5:9b4f:f8b9', fragment='')\n"
]
],
[
[
"We can run these assertions:",
"_____no_output_____"
]
],
[
[
"from urllib.parse import SplitResult, ParseResult, urlparse, urlsplit",
"_____no_output_____"
],
[
"assert urlparse(\n url='http://www.example.com',\n scheme='',\n allow_fragments=True) == ParseResult(\n scheme='http',\n netloc='www.example.com',\n path='',\n params='',\n query='',\n fragment='')\nassert urlsplit(\n url='http://www.example.com',\n scheme='',\n allow_fragments=True) == SplitResult(\n scheme='http',\n netloc='www.example.com',\n path='',\n query='',\n fragment='')",
"_____no_output_____"
]
],
[
[
"We can now add these carved tests to a _regression test suite_ which would be run after every change to ensure that the functionality of `urlparse()` and `urlsplit()` is not changed.",
"_____no_output_____"
],
[
"### Exercise 2: Abstracting Arguments\n\nWhen mining an API grammar from executions, set up an abstraction scheme to widen the range of arguments to be used during testing. If the values for an argument, all conform to some type `T`. abstract it into `<T>`. For instance, if calls to `foo(1)`, `foo(2)`, `foo(3)` have been seen, the grammar should abstract its calls into `foo(<int>)`, with `<int>` being appropriately defined.\n\nDo this for a number of common types: integers, positive numbers, floating-point numbers, host names, URLs, mail addresses, and more.",
"_____no_output_____"
],
[
"**Solution.** Left to the reader.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cb4ea1656e99433a2e514d733bc1375961356aec | 9,793 | ipynb | Jupyter Notebook | Python_IE534/hw2/.ipynb_checkpoints/Custom_OneLayer_CNN_SGD-checkpoint.ipynb | Rothdyt/codes-for-courses | a2dfea516ebc7cabef31a5169533b6da352e7ccb | [
"MIT"
] | 4 | 2018-09-23T00:00:13.000Z | 2018-11-02T22:56:35.000Z | Python_IE534/hw2/.ipynb_checkpoints/Custom_OneLayer_CNN_SGD-checkpoint.ipynb | Rothdyt/codes-for-courses | a2dfea516ebc7cabef31a5169533b6da352e7ccb | [
"MIT"
] | null | null | null | Python_IE534/hw2/.ipynb_checkpoints/Custom_OneLayer_CNN_SGD-checkpoint.ipynb | Rothdyt/codes-for-courses | a2dfea516ebc7cabef31a5169533b6da352e7ccb | [
"MIT"
] | null | null | null | 27.585915 | 258 | 0.498315 | [
[
[
"# Import Module\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport h5py",
"C:\\Users\\LEN\\Anaconda3\\lib\\site-packages\\h5py\\__init__.py:34: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
],
[
"# Read data, which has a size of N * 784 and N * 1\nMNIST = h5py.File(\"..\\MNISTdata.hdf5\",'r')\nx_train = np.float32(MNIST['x_train'][:])\nx_test = np.float32(MNIST['x_test'][:])\ny_train = np.int32(MNIST['y_train'][:,0])\ny_test = np.int32(MNIST['y_test'][:,0])",
"_____no_output_____"
],
[
"# Reshape samples as 28 * 28 images\nx_trainnew = np.reshape(x_train, (len(x_train),28,28))\nx_testnew = np.reshape(x_test, (len(x_test),28,28))",
"_____no_output_____"
],
[
"# Build activate functions\nrelu = lambda x: x*(x>0)\n\n# Input a m * n matrix, output a m * n matrix whose rows are transformed and normalized\ndef softmax(X):\n Xexp = np.exp(X)\n return Xexp / np.sum(Xexp,axis=1,keepdims=True)",
"_____no_output_____"
],
[
"# Initialize the parameters\ndef param_init(input_size, kernel_size, output_size):\n lx = input_size # 2-dim\n lk = kernel_size # 2-dim\n lh = (lx[0]-lk[0]+1, lx[1]-lk[1]+1) # Hidden layer size, 2-dim\n ly = output_size # 1-dim\n K = np.random.randn(lk[0],lk[1]) / max(lx)\n W = np.random.randn(ly,lh[0],lh[1]) / max(lx)\n b = np.zeros(ly)\n \n return K,W,b",
"_____no_output_____"
],
[
"K,W,b = param_init((28,28),(3,3),10)",
"_____no_output_____"
],
[
"# Build the forward step\n# Model: Z = X * K → H = relu(Z) → U = WH + b → Yhat = softmax(U)\ndef Convolution(image, kernel):\n d1,d2 = image.shape\n k1,k2 = kernel.shape\n output_a = d1 - k1 + 1\n output_b = d2 - k2 + 1\n conv = np.zeros((output_a,output_b))\n for a in range(output_a):\n for b in range(output_b):\n conv[a,b] = np.sum(np.multiply(image[a:a+k1,b:b+k2], kernel))\n return conv\n\ndef forward_prop(X,K,W,b):\n # Input to Hidden layer\n Z = Convolution(X,K) # Shape: (lx[0]-lk[0]+1, lx[1]-lk[1]+1)\n H = relu(Z) # Shape: (lx[0]-lk[0]+1, lx[1]-lk[1]+1)\n \n # Hidden layer to Output\n U = np.sum(np.multiply(W,H), axis=(1,2)) + b # Shape: (1 * ly)\n U.shape = (1,W.shape[0])\n Yhat = softmax(U) # Shape: (1 * ly)\n \n return Z, H, Yhat",
"_____no_output_____"
],
[
"N = x_trainnew.shape[0]\nr = np.random.randint(N)\nx_samp = x_trainnew[r,:,:]\n\nY_oh = np.array(pd.get_dummies(np.squeeze(y_train)))\ny_samp = Y_oh[[r]]",
"_____no_output_____"
],
[
"Z, H, Yhat = forward_prop(x_samp,K,W,b)",
"_____no_output_____"
],
[
"# Build the back-propagation step\ndef back_prop(K,W,b,Z,H,Yhat,X,Y,alpha):\n bDel = Y - Yhat # Length ly\n bDel = np.squeeze(bDel)\n WDel = np.tensordot(bDel, H, axes=0) # Shape (ly, lx[0]-lk[0]+1, lx[1]-lk[1]+1)\n HDel = np.tensordot(bDel, W, axes=1) # Shape (lx[0]-lk[0]+1, lx[1]-lk[1]+1)\n ZDel = np.multiply(HDel,(lambda x:(x>0))(Z)) # Shape (lx[0]-lk[0]+1, lx[1]-lk[1]+1)\n KDel = Convolution(X,ZDel) # Shape: (lk[0], lk[1])\n #KDel = np.zeros(KDel.shape)\n #WDel = np.zeros(WDel.shape)\n #bDel = np.zeros(bDel.shape)\n \n bn = b + alpha * bDel # Length ly\n Wn = W + alpha * WDel # Shape (ly, lx[0]-lk[0]+1, lx[1]-lk[1]+1)\n Kn = K + alpha * KDel # Shape (1k[0], lk[1])\n \n return Kn,Wn,bn",
"_____no_output_____"
],
[
"alpha = 0.01\nbDel,WDel,KDel = back_prop(K,W,b,Z,H,Yhat,x_samp,y_samp,alpha)",
"_____no_output_____"
],
[
"# Build the complete Neural Network\ndef TwoLayer_CNN_train(X, Y, ChannelSize = (3,3), NumChannel = 1, OrigAlpha = 0.01, num_epochs = 10): \n # Recode Y as One-Hot\n Y_oh = np.array(pd.get_dummies(np.squeeze(Y)))\n \n # Indicate number of units per layer\n N = X.shape[0] # Number of samples\n xsize = X.shape[1:] # Size of every sample\n ksize = ChannelSize # Size of the channel\n ysize = Y_oh.shape[1] # Number of classes\n \n # Initialized the parameters\n K,W,b = param_init(xsize,ksize,ysize)\n \n # Run 20 train iterations, record the error every time\n for epoch in range(num_epochs):\n if epoch <= 5:\n alpha = OrigAlpha\n elif epoch <= 10: \n alpha = OrigAlpha * 1e-1\n elif epoch <= 15:\n alpha = OrigAlpha * 1e-2\n else:\n alpha = OrigAlpha * 1e-3\n total_cor = 0\n for n in range(int(N/6)):\n r = np.random.randint(N)\n x_samp = X[r,:,:]\n y_samp = Y_oh[[r]]\n # Forward\n Z, H, Yhat = forward_prop(x_samp,K,W,b)\n pred = np.argmax(Yhat)\n if pred==Y[r]:\n total_cor += 1\n # Backward\n K,W,b = back_prop(K,W,b,Z,H,Yhat,x_samp,y_samp,alpha)\n print(\"Training Accuracy: \",total_cor / np.float(N/6))\n return K,W,b",
"_____no_output_____"
],
[
"K,W,b = TwoLayer_CNN_train(x_trainnew, y_train, OrigAlpha=0.01, num_epochs=10)",
"Training Accuracy: 0.8487\nTraining Accuracy: 0.8944\nTraining Accuracy: 0.9117\nTraining Accuracy: 0.9087\nTraining Accuracy: 0.9111\nTraining Accuracy: 0.9134\nTraining Accuracy: 0.9269\nTraining Accuracy: 0.9222\nTraining Accuracy: 0.9241\nTraining Accuracy: 0.9285\n"
],
[
"# For a given neural network, predict an input X\ndef predict_NN(X,K,W,b):\n X_predprob = forward_prop(X,K,W,b)[2]\n X_pred = X_predprob.argmax(axis=1) # Take the biggest probability as its choice\n return X_pred",
"_____no_output_____"
],
[
"# Predict on test set\n# Still has problems!\ny_predtest = predict_NN(x_testnew,K,W,b)\nnp.sum(y_predtest == y_test) / x_testnew.shape[0]",
"_____no_output_____"
],
[
"Ut = np.array([1,2,3])\nUt.shape = (1,3)\nWt = np.array([[[1,1],[2,2]],[[3,3],[4,4]],[[5,5],[6,6]]])\nHt = np.array([[0.3,0.3],[0.4,0.4]])\nkt = np.sum(np.multiply(Wt,Ht),axis=(1,2))",
"_____no_output_____"
],
[
"np.tensordot(Ut,Wt,axes=1)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4eb6fea22b8fb2e58a59b9ba7e90b5395f20f5 | 720,093 | ipynb | Jupyter Notebook | lectures/ng/Lecture-07-Support-Vector-Machines.ipynb | tgrasty/CSCI574-Machine-Learning | bcf797262852c4b46a6702c69f69724b0b9e93f6 | [
"CC-BY-3.0"
] | null | null | null | lectures/ng/Lecture-07-Support-Vector-Machines.ipynb | tgrasty/CSCI574-Machine-Learning | bcf797262852c4b46a6702c69f69724b0b9e93f6 | [
"CC-BY-3.0"
] | null | null | null | lectures/ng/Lecture-07-Support-Vector-Machines.ipynb | tgrasty/CSCI574-Machine-Learning | bcf797262852c4b46a6702c69f69724b0b9e93f6 | [
"CC-BY-3.0"
] | 1 | 2021-09-17T17:03:58.000Z | 2021-09-17T17:03:58.000Z | 449.215845 | 245,088 | 0.939306 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport matplotlib\n%matplotlib inline\n",
"_____no_output_____"
],
[
"matplotlib.rcParams['figure.figsize'] = (12, 8) # set default figure size, 8in by 6in",
"_____no_output_____"
]
],
[
[
"This week, you will be learning about the support vector machine (SVM) algorithm. SVMs are considered by many to be the most powerful 'black box' learning algorithm, and by posing a cleverly-chosen optimization objective, one of the most widely used learning algorithms today.",
"_____no_output_____"
],
[
"# Video W7 01: Optimziation Objective\n\n[YouTube Video Link](https://www.youtube.com/watch?v=r3uBEDCqIN0&list=PLZ9qNFMHZ-A4rycgrgOYma6zxF4BZGGPW&index=71)\n\n\nOne way of understanding SVM is that it is a simple modification of logistic regression (just as the logistic regression is\na simple extension of linear regression, and neural networks are a way of extending the concepts of logistic regression, etc.)\n\nRecall that the basic modification we made for logistic regression was to pass our hypothesis through a logistic (or sigmoid)\nfunction. This caused the output from our hypothesis to always be bound to a value in the range from 0.0 to 1.0\n\n$$\nh_{\\theta}(x) = \\frac{1}{1 + e^{-\\theta^T x}}\n$$",
"_____no_output_____"
]
],
[
[
"def sigmoid(z):\n return 1.0 / (1.0 + np.e**-z)\n\nplt.figure()\nx = np.linspace(-5.0, 5.0)\nz = sigmoid(x)\nplt.plot(x, z)\nplt.axis([-5, 5, 0, 1])\nplt.grid()\nplt.xlabel('$z = \\\\theta^Tx$', fontsize=20)\nplt.text(-1, 0.85, '$h_{\\\\theta}(x) = g(z)$', fontsize=20);",
"_____no_output_____"
]
],
[
[
"Recall that for a single input/output pair $(x, y)$, the objective or cost function for logistic regression has the following form:\n\n$$\n-y \\;\\; \\textrm{log} \\frac{1}{1 + e^{-\\theta^T x}} - (1 - y) \\;\\; \\textrm{log} \\; \\big(1 - \\frac{1}{1 + e^{-\\theta^T x}}\\big)\n$$\n\nThis expression will only involve either the left or right side, depending on whether $y = 1$ or $y = 0$ (recall that in logistic\nregression, we are performming a classification, where each training example is either in the class, or it is not in the class).\nSo for example, if we want $y = 1$, then we want $\\theta^Tx \\gg 0$. The curve for the function when $y = 1$ looks like the\nfollowing:",
"_____no_output_____"
]
],
[
[
"z = np.linspace(-3.0, 3.0)\ny = -np.log( 1.0 / (1.0 + np.exp(-z)) )\n\nplt.figure()\nplt.plot(z, y)\nplt.xlabel('$z$', fontsize=20)\nplt.grid()\nplt.text(1, 3, '$-log \\\\; \\\\frac{1}{1 + e^{-z}}$', fontsize=20);",
"_____no_output_____"
]
],
[
[
"Likewise, for the case where $y = 0$ then we want $\\theta^T x \\ll 0$. The curve for the objective function when $y = 0$ similarly\nlooks like the following:",
"_____no_output_____"
]
],
[
[
"z = np.linspace(-3.0, 3.0)\ny = -np.log( 1.0 - (1.0 / (1.0 + np.exp(-z))) )\n\nplt.figure()\nplt.plot(z, y)\nplt.xlabel('$z$', fontsize=20)\nplt.grid()\nplt.text(-3, 3, '$-log \\\\; ( 1 - \\\\frac{1}{1 + e^{-z}} )$', fontsize=20);",
"_____no_output_____"
]
],
[
[
"The full cost function we were trying to minimize, then, for logistic regression was:\n\n$$\n\\frac{1}{m} \\big[ \\sum_{i=1}^{m} y^{(i)} \\big( - \\textrm{log} \\; h_{\\theta}(x^{(i)} \\big) + \n(1 - y^{(i)}) \\big( - \\textrm{log} \\; (1 - h_{\\theta}(x^{(i)})) \\big) \\big] + \\frac{\\lambda}{2m} \\sum_{j=1}^{n} \\theta_j^2\n$$\n\nFor the support vector machine, we change the terms relating to the hypothesis to functions $\\textrm{cost}_1$ and\n$\\textrm{cost}_0$:\n\n$$\n\\frac{1}{m} \\big[ \\sum_{i=1}^{m} y^{(i)} \\; \\textrm{cost}_1( \\theta^{T} x^{(i)} )\n+ (1 - y^{(i)}) \\; \\textrm{cost}_0 ( \\theta^{T} x^{(i)} ) \\big] \n+ \\frac{\\lambda}{2m} \\sum_{j=1}^{n} \\theta_j^2\n$$\n\nas described in the video, by convention in SVM we remove the division by $m$ and we parameterize the regularization and cost\nterms a bit differently. Usually you will see the objective function for SVM specified in this slightly different but equivalent\nform:\n\n$$\n\\underset{\\theta}{\\textrm{min}} \\; C\n\\sum_{i=1}^{m} \\big[ y^{(i)} \\; \\textrm{cost}_1( \\theta^{T} x^{(i)} )\n+ (1 - y^{(i)}) \\; \\textrm{cost}_0 ( \\theta^{T} x^{(i)} ) \\big]\n+ \\frac{1}{2} \\sum_{j=1}^{n} \\theta_j^2\n$$\n\n\nIn this formulation of the objective function, the term C is being used as the regularization parameter. But now, the larger\nthe value of C, the more emphasis that is placed on the cost terms (and the less that is placed on the regularization terms.",
"_____no_output_____"
],
[
"# Video W7 02: Large Margin Intuition\n\n[YouTube Video Link](https://www.youtube.com/watch?v=yjH3ZSPqLhU&list=PLZ9qNFMHZ-A4rycgrgOYma6zxF4BZGGPW&index=72)\n\nBecause of the form of the $\\textrm{cost}_0$ and $\\textrm{cost}_1$ functions (which we haven't specified yet), these\nnaturally favor cost functions that give wide margins between the hypothesis outputs for $y=0$ and $y=1$.\n\nIntiutively, as shown in the video, the objective function that we have defined will find decision boundaries that maximize\nthe margin between the negative and positive examples. This is where the name large margin classifier comes from. The\nterm `support vector` from the name for SVM also refers to the mathematical properties of these objective functions that\ntry and maximize this margin between positive and negative examples. \n\nThe $cost_0$ and $cost_1$ functions described in the video are basically the same idea of using\nwhat are known as rectified linear units (ReLU) in neural network. Here we give a linear\nactivation response when the value is above (or below) some threshold, and 0 otherwise.\n\nThe following figures compare a possible implementation of the discussed $cost_0$ and $cost_1$ functions\nto this type of threshold RELU activation function.",
"_____no_output_____"
]
],
[
[
"def cost_0(z):\n return np.where(z > -1, z+1, 0)\n \ndef cost_1(z):\n return np.where(z < 1, -z+1, 0)",
"_____no_output_____"
],
[
"z = np.linspace(-3.0, 3.0)\ny = -np.log( 1.0 / (1.0 + np.exp(-z)) )\n\n# logistic cost function, for y=1\nplt.figure()\nplt.plot(z, y, label='logistic cost function $y=1$')\nplt.xlabel('$z$', fontsize=20)\nplt.grid()\n\n# cost_1 function, RELU like\ny = cost_1(z)\nplt.plot(z, y, label='$cost_1$ RELU approximation for SVM')\nplt.legend();",
"_____no_output_____"
],
[
"z = np.linspace(-3.0, 3.0)\ny = -np.log( 1.0 - (1.0 / (1.0 + np.exp(-z))) )\n\n# logistic cost function, for y=0\nplt.figure()\nplt.plot(z, y, label='logistic cost function $y=0$')\nplt.xlabel('$z$', fontsize=20)\nplt.grid()\n\n# cost_1 function, RELU like\ny = cost_0(z)\nplt.plot(z, y, label='$cost_0$ RELU approximation for SVM')\nplt.legend();",
"_____no_output_____"
]
],
[
[
"# Video W7 03: Mathematics Behind Large Margin Intuition (Optional)\n\n[YouTube Video Link](https://www.youtube.com/watch?v=Jm49m7ey34o&list=PLZ9qNFMHZ-A4rycgrgOYma6zxF4BZGGPW&index=73)\n\nThe nitty gritty of the mathematics behind how the SVM optimization finds large margin decision boundaries is not necessary\nfor using SVM well. But at least watch the video to get a bit of a feel for what happens behind the scenes when creating an SVM\nand how it finds such decision boundaries given our definition of the cost function.",
"_____no_output_____"
],
[
"# Video W7 04: Kernels I\n\n[YouTube Video Link](https://www.youtube.com/watch?v=0Fg2U6LN3pg&list=PLZ9qNFMHZ-A4rycgrgOYma6zxF4BZGGPW&index=74)\n\nThis video starts by giving a good explanation of what are known as gaussian feature kernels. When we looked at logistic\nregression, we did examine using nonlinear features to produce more complex decision boundaries. Kernel methods, used\nmost commonly with SVM systems, allow us to create sets of nonlinear features, but in a more directed and less random way\nthan simply using polynomial combinations of basic features.\n\nThe [gaussian function](https://en.wikipedia.org/wiki/Gaussian_function) discussed in the video is related to gaussian\nor normal distributions that you may be familiar with (e.g. the standard 'bell curve' distribution). For a single\nfeature, the gaussian function is usually specified in terms of $\\mu$, the mean or location of the feature, and \n$\\sigma^2$ the square of the deviation of the feature. So for example, as given in the video, we can think of\nthe similarity measure for a system that has a single feature, with a landmar at the point $\\mu = 0$ as follows:\n\n$$\nf(x) = exp \\Big(- \\frac{(x - \\mu)^2}{2\\sigma^2} \\Big)\n$$\n\nThe expression $(x - \\mu)^2$ is really just an expression of the distance from some input $x$ to the landmark. So when\nwhen we only have a single feature, and our landmark is at the origin point 0 (e.g. $\\mu = 0$) then we have:",
"_____no_output_____"
]
],
[
[
"def gauss(x, mu, sigma):\n return np.exp(- (x - mu)**2.0 / (2 * sigma**2.0))\n\nx = np.linspace(-3.0, 3.0)\nplt.figure()\nplt.plot(x, gauss(x, 0.0, 1.0), 'k-')\nplt.xlabel('$x_1$', fontsize=20)\nplt.ylabel('gaussian similarity function');",
"_____no_output_____"
]
],
[
[
"This is the basic gaussian distribution, with a mean of 0 and a standard deviation of 1. In the context of a gaussian kernel \nfunction, we will return a similarity of 1.0 for any feature that is exactly the same as our landmark ($\\mu$ or 0 in this case),\nand we will return lesser values, eventually approaching 0, as we get a further distance from the landmark $\\mu$ location of 0.\n\nIn the videos, the linear algebra norm simply calculates the distance from an input $x$ when we have more than 1 feature. So for\nexample, when we have 2 features, or 2 dimensional space, we need to visualize the gaussian function using a 3 dimensional plot, \nwhere we plot our two features $x_1$ and $x_2$ on two orthogonal axis, and plot the gaussian function on the 3rd orthogonal\nz axis.\n\nSo for example, as shown in the video, if we have 2 features, and our landmark is located at the position where $x_1 = 3$\nand $x_2 = 5$, e.g. \n$$\nl^{(1)} =\n\\begin{bmatrix}\n3 \\\\\n5 \\\\\n\\end{bmatrix}\n$$\nWe will simply have a gaussian function in 2 dimensions (features) that has a value of 1.0 exactly at that $\\mu$ location (in\n2 dimensions), and falls away in the bell curve shape in both dimensions as a function of the $\\sigma$ (the deviation value).\nSo the gaussian similiarity function written in the video is:\n\n$$\nf_1 = \\mathrm{exp} \\Big(- \\frac{ \\|x - l^{(1)}\\|^2 }{2 \\sigma^2} \\Big)\n$$\n\nThe top part of the fraction is simply calculating the distance between some point $x$ and the landmark location in a 2 or higher\ndimensional space (e.g. the sum of all of the differences for each individual dimension, then squaring this sum, in \nlinear algebra this is simply the square of the norm of the difference of these two vectors). So for a two dimensional feature, the gaussian function can\nbe plotted on a 3D plot in python as follows:",
"_____no_output_____"
]
],
[
[
"# first plot as a contour map\n\ndef gauss(x, mu, sigma):\n \"\"\"A multi-dimensional version of the gaussian function. x and mu are n dimensional feature vectors, so\n we take the linear algebra norm of the difference and square this).\"\"\"\n from numpy.linalg import norm\n return np.exp(- norm(x - mu, axis=1)**2.0 / (2 * sigma**2.0))\n\n# the landmark, I have been calling it mu\nmu = np.array([3, 5])\nsigma = 1.0\n\n# we create a mesh so we can plot our gaussin function in 3d\nx1_min, x1_max = -2.0, 8.0\nx2_min, x2_max = 0.0, 10.0\nh = 0.02 # step size in the mesh\nx1, x2 = np.meshgrid(np.arange(x1_min, x1_max, h),\n np.arange(x2_min, x2_max, h))\n\nx = np.c_[x1.ravel(), x2.ravel()]\nZ = gauss(x, mu, sigma)\nZ = Z.reshape(x1.shape)",
"_____no_output_____"
],
[
"# plot the 2 feature dimensional gaussian as a contour map\nplt.contourf(x1, x2, Z, cmap=plt.cm.jet, alpha=0.8)\nplt.colorbar()\nplt.xlabel('$x_1$', fontsize=20)\nplt.ylabel('$x_2$', fontsize=20);",
"_____no_output_____"
],
[
"# now plot as as a 3D surface plot\nfrom mpl_toolkits.mplot3d import Axes3D\n\nZ = Z.reshape(x1.shape)\nfig = plt.figure(figsize=(12,12))\nax = fig.gca(projection='3d')\nsurf = ax.plot_surface(x1, x2, Z, cmap=plt.cm.jet)\nax.set_xlabel('$x_1$', fontsize=20)\nax.set_ylabel('$x_2$', fontsize=20);",
"_____no_output_____"
]
],
[
[
"You should try changing the landmark location $\\mu$ and the sigma value (which controls how fast the change in distance affects\nthe value of the gaussian function) in the previous cell.\n\nAs shown in the video (closer to the end), gaussian kernels allow for nonlinear decision boundaries. But unlike creating\nan (exponential) combination of polynomial fetures, we can simply pick an appropriate number of gausian kernels that will\nlikely produce a good enough decision boundary for our given set of data. As discussed in this video, a good way \nof thinking of the gaussian kernels is as landmarks that are chosen (we discuss how to choose the landmarks in the next video)\nand features are then simply similarity measures to the chosen set of landmarks.",
"_____no_output_____"
],
[
"# Video W7 05: Kernels II\n\n[YouTube Video Link](https://www.youtube.com/watch?v=P9Xjvr2JfOk&index=75&list=PLZ9qNFMHZ-A4rycgrgOYma6zxF4BZGGPW)\n\nIn practice, landmarks for gaussian kernels are chosen by putting landmarks at each of the training example locations.\nThus the number of landmarks will grow linearly with the size of our training set data (instead of being a combinatorial\nexplosion in terms of the number of input features, as creating polynomial terms from the features tends to do).",
"_____no_output_____"
],
[
"# Video W7 06: Using an SVM\n\n[YouTube Video Link](https://www.youtube.com/watch?v=wtno4WSDTlY&list=PLZ9qNFMHZ-A4rycgrgOYma6zxF4BZGGPW&index=76)\n\n\nThis video shows using SVM packages for octave/matlab. In this class we have been using Python, of course. There are many good\nimplementations of SVM in Python. For example, the libsvm mentioned in the video is actually a language neutral implementation\nof svm, and there are extensions available to use libsvm in python:\n[libsvm](https://www.csie.ntu.edu.tw/~cjlin/libsvm/)\n\nAs of the creation of this notebook, however (Fall 2015), I do recommend using the svm implementation\nin the scikit learn library. It is the most mature and has the best (most consistent) user interface. You will have to install\nscikit learn in your environment in order to use it. If you are using the enthought Python environemnt, it should have been\ninstalled for you. The documentation for the scikit learn svm library is [here](http://scikit-learn.org/stable/modules/svm.html).\n\nAs discussed in this video, sometimes we want to do an SVM classification, but not ues any complex kernels, e.g. a\nstraightforward linear SVM classifier. If you want to do this, you should use the `SVC` in scikit learn with a linear kernel to\ndo a linear SVM classifier. For example, recall that in assignment 03 we used logistic regression to classify\nexam score data (with a single class, admit or not admit) using a linear decision boundary. The data looked like this:",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv('../../data/assg-03-data.csv', names=['exam1', 'exam2', 'admitted'])\nx = data[['exam1', 'exam2']].values\ny = data.admitted.values\nm = y.size\n\nprint((x.shape))\nprint((y.shape))\nprint(m)\nX = np.ones( (3, m) )\nX[1:,:] = x.T # the second column contains the raw inputs\nX = X.T\n\nneg_indexes = np.where(y==0)[0]\npos_indexes = np.where(y==1)[0]\nplt.plot(x[neg_indexes, 0], x[neg_indexes, 1], 'yo', label='Not admitted')\nplt.plot(x[pos_indexes, 0], x[pos_indexes, 1], 'r^', label='Admitted')\nplt.title('Admit/No Admit as a function of Exam Scores')\nplt.xlabel('Exam 1 score')\nplt.ylabel('Exam 2 score')\nplt.legend();",
"(100, 2)\n(100,)\n100\n"
]
],
[
[
"Before we do a linear SVM, lets use the logistic regression functions from scikit learn to perform a logistic regression. The\nscikit learn uses C rather than the $\\lambda$ to specify the amount of regularization. In our assignment we didn't use any regularization. We can get similar theta parameters by using a large C, which will do the optimization using only the cost, without\nmuch weight for the regularization. But try it with more regularization (e.g. smaller C values), and you will see that the \ndecision boundary is still basically the same.",
"_____no_output_____"
]
],
[
[
"from sklearn import linear_model\nlogreg = linear_model.LogisticRegression(C=1e6, solver='lbfgs')\nlogreg.fit(x, y)\nprint((logreg.coef_)) # show the coefficients that were fitted to the data by logistic regression\nprint((logreg.intercept_))",
"[[0.20623222 0.2014719 ]]\n[-25.16138556]\n"
]
],
[
[
"Notice that for scikit learn we don't have to add in the column of 1's to the input data. By default, most scikit learn functions\nwill assume they need to add in such an intercept parameter. So there will only be two theta parameters in this case, but the \nparameter corresponding to the intercept value is in a separate constant after we fit our model to the training data.\n\nHere is a plot of the decision boundary that was found.",
"_____no_output_____"
]
],
[
[
"# display the decision boundary for the coeficients\nneg_indexes = np.where(y==0)[0]\npos_indexes = np.where(y==1)[0]\n\n# visualize the data points of the two categories\nplt.plot(x[neg_indexes, 0], x[neg_indexes, 1], 'yo', label='Not admitted')\nplt.plot(x[pos_indexes, 0], x[pos_indexes, 1], 'r^', label='Admitted')\nplt.title('Admit/No Admit as a function of Exam Scores')\nplt.xlabel('Exam 1 score')\nplt.ylabel('Exam 2 score')\nplt.legend()\n\n# add the decision boundary line\ndec_xpts = np.arange(30, 93)\ntheta = logreg.coef_[0]\ndec_ypts = - (logreg.intercept_ + theta[0] * dec_xpts) / theta[1]\nplt.plot(dec_xpts, dec_ypts, 'b-');",
"_____no_output_____"
]
],
[
[
"Now lets use the linear SVM classifier from scikit learn to perform the same classification.",
"_____no_output_____"
]
],
[
[
"from sklearn import svm\nlinclf = svm.SVC(kernel='linear', C=1e6)\nlinclf.fit(x, y)\nprint((linclf.coef_)) # show the coefficients that were fitted to the data by logistic regression\nprint((linclf.intercept_))",
"[[64.17436109 67.1465802 ]]\n[-8028.53017612]\n"
]
],
[
[
"Notice that the parameters found for the model andthe intercept are a bit different, but these do actually correspond to basically\nabout the same decision boundary as before. If we plot it you can see this is the case:",
"_____no_output_____"
]
],
[
[
"# display the decision boundary for the coeficients\nneg_indexes = np.where(y==0)[0]\npos_indexes = np.where(y==1)[0]\n\n# visualize the data points of the two categories\nplt.plot(x[neg_indexes, 0], x[neg_indexes, 1], 'yo', label='Not admitted')\nplt.plot(x[pos_indexes, 0], x[pos_indexes, 1], 'r^', label='Admitted')\nplt.title('Admit/No Admit as a function of Exam Scores')\nplt.xlabel('Exam 1 score')\nplt.ylabel('Exam 2 score')\nplt.legend()\n\n# add the decision boundary line\ndec_xpts = np.arange(30, 93)\ntheta = linclf.coef_[0]\ndec_ypts = - (linclf.intercept_ + theta[0] * dec_xpts) / theta[1]\nplt.plot(dec_xpts, dec_ypts, 'b-');",
"_____no_output_____"
]
],
[
[
"And finally, lets use an SVM with a gaussian kernel. It is not so interesting to use a nonlinear classifier with the\nprevious data, so lets make up some data, similar to the data shown in our companion video\nof a class surrounded by another class. Here we use a function from the scikit learn library that can beused to creat data\nat random. The data has 2 features, and only 2 classes (either positivie or negative, e.g. admitted or not\nadmitted). The random data generated from this function is centered at the origin (0, 0). The further away the data is from\nthe center, the more probable it is in another class (using a gaussian probability function). Thus with two classes we tend to get\na class inside surrounded by another class, with a basically circular decision boundary.",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import make_gaussian_quantiles\nX, Y = make_gaussian_quantiles(n_features=2, n_classes=2)\n\nneg_indexes = np.where(Y==0)[0]\npos_indexes = np.where(Y==1)[0]\nplt.plot(X[neg_indexes, 0], X[neg_indexes, 1], 'yo', label='negative examples')\nplt.plot(X[pos_indexes, 0], X[pos_indexes, 1], 'r^', label='positive examples');",
"_____no_output_____"
]
],
[
[
"Here then we will use a SVM with gaussian kernels to create a classifier for the data. Note that we specify 'rbf' for the\nkernel, these are radial basis functions kernels. Radial basis function kernels include \ngaussian functions, as well as the polynomial functions\ndiscussed in our companion videos. You specify the gamma, degree and coef0 parameters to get the different types of kernel \nfunctions that were discussed. I believe that by specifying a gamma of 1.0 we will be using simple gaussian kernel functions as\nwere shown in our videos.",
"_____no_output_____"
]
],
[
[
"from sklearn import svm\nrbfclf = svm.SVC(kernel='rbf', gamma=1.0)\nrbfclf.fit(X, Y)",
"_____no_output_____"
],
[
"# Now display the results. We don't really have simple theta parameters anymore, the parameters are specifying\n# relative values of the gaussian kernels now. In fact, rbclf.coef_ will not be defined for non linear kernels.\n# Here we use an alternative method to visualize the decision boundary that was discovered.\n\n# create a mesh to plot in\nx_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1\ny_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1\nh = .02 # step size in the mesh\nxx, yy = np.meshgrid(np.arange(x_min, x_max, h),\n np.arange(y_min, y_max, h))\n\nZ = rbfclf.predict(np.c_[xx.ravel(), yy.ravel()])\n\n# Put the result into a color plot\nZ = Z.reshape(xx.shape)\nplt.contourf(xx, yy, Z, cmap=plt.cm.Paired, alpha=0.8)\n\n# plot the original data\nneg_indexes = np.where(Y==0)[0]\npos_indexes = np.where(Y==1)[0]\nplt.plot(X[neg_indexes, 0], X[neg_indexes, 1], 'yo', label='negative examples')\nplt.plot(X[pos_indexes, 0], X[pos_indexes, 1], 'r^', label='positive examples')\nplt.legend();",
"_____no_output_____"
]
],
[
[
"# More SciKit-Learn Examples\n\n## Linear SVC on iris dataset\n\nUse a LinearSVC (non-kernel) based SVM. LinearSVC will be much faster than using SVC and specifying a 'linear'\nkernel.",
"_____no_output_____"
]
],
[
[
"from sklearn import datasets\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.svm import LinearSVC",
"_____no_output_____"
],
[
"iris = datasets.load_iris()\nX = iris['data'][:, (2, 3)] # petal length, petal width\ny = (iris['target'] == 2).astype(np.float64) # Iris-virginica",
"_____no_output_____"
],
[
"C = 1.0\nsvmclf = Pipeline((\n (\"scaler\", StandardScaler()),\n (\"linear_svc\", LinearSVC(C=C, loss='hinge')),\n))\n\nsvmclf.fit(X, y)",
"_____no_output_____"
],
[
"svmclf.predict([[5.5, 1.7]])",
"_____no_output_____"
],
[
"# visualize resulting decision boundary\nx_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1\ny_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1\nh = .02 # step size in the mesh\nxx, yy = np.meshgrid(np.arange(x_min, x_max, h),\n np.arange(y_min, y_max, h))\n\nZ = svmclf.predict(np.c_[xx.ravel(), yy.ravel()])\n\n# Put the result into a color plot\nZ = Z.reshape(xx.shape)\nplt.contourf(xx, yy, Z, cmap=plt.cm.Paired, alpha=0.8)\n\n\n# plot the original data\n# plot the original data\nneg_indexes = np.where(y==0)[0]\npos_indexes = np.where(y==1)[0]\nplt.plot(X[neg_indexes, 0], X[neg_indexes, 1], 'yo', label='other')\nplt.plot(X[pos_indexes, 0], X[pos_indexes, 1], 'r^', label='Iris-virginica')\nplt.legend();\nplt.title('C = %f' % C)",
"_____no_output_____"
]
],
[
[
"## Moon data using Generated Polynomial Features\n\nExample using the PolynomialFeatures class to create all feature combinations up to degree 3 polynomials here.",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import make_moons\nfrom sklearn.preprocessing import PolynomialFeatures",
"_____no_output_____"
],
[
"X, y = make_moons(noise=0.1)",
"_____no_output_____"
],
[
"d = 3 # polynomial degree\nC = 10\n\npolynomial_svm_clf = Pipeline((\n ('poly_features', PolynomialFeatures(degree=d)),\n ('scaler', StandardScaler()),\n ('svm_clf', LinearSVC(C=C, loss='hinge')),\n))\n\npolynomial_svm_clf.fit(X, y)",
"_____no_output_____"
],
[
"# visualize resulting decision boundary\nx_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1\ny_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1\nh = .02 # step size in the mesh\nxx, yy = np.meshgrid(np.arange(x_min, x_max, h),\n np.arange(y_min, y_max, h))\n\nZ = polynomial_svm_clf.predict(np.c_[xx.ravel(), yy.ravel()])\n\n# Put the result into a color plot\nZ = Z.reshape(xx.shape)\nplt.contourf(xx, yy, Z, cmap=plt.cm.Paired, alpha=0.8)\n\n\n# plot the original data\n# plot the original data\nneg_indexes = np.where(y==0)[0]\npos_indexes = np.where(y==1)[0]\nplt.plot(X[neg_indexes, 0], X[neg_indexes, 1], 'yo', label='negative class')\nplt.plot(X[pos_indexes, 0], X[pos_indexes, 1], 'r^', label='positive class')\nplt.legend();\nplt.title('C = %f, degree=%d' % (C, d))",
"_____no_output_____"
]
],
[
[
"## Polynimial Kernel",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import SVC",
"_____no_output_____"
],
[
"d = 10\nc0 = 100\nC = 5\npoly_kernel_svm_clf = Pipeline((\n ('scalar', StandardScaler()),\n ('svm_clf', SVC(kernel='poly', degree=d, coef0=c0, C=C)),\n))\n\npoly_kernel_svm_clf.fit(X, y)",
"_____no_output_____"
],
[
"# visualize resulting decision boundary\nx_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1\ny_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1\nh = .02 # step size in the mesh\nxx, yy = np.meshgrid(np.arange(x_min, x_max, h),\n np.arange(y_min, y_max, h))\n\nZ = poly_kernel_svm_clf.predict(np.c_[xx.ravel(), yy.ravel()])\n\n# Put the result into a color plot\nZ = Z.reshape(xx.shape)\nplt.contourf(xx, yy, Z, cmap=plt.cm.Paired, alpha=0.8)\n\n\n# plot the original data\n# plot the original data\nneg_indexes = np.where(y==0)[0]\npos_indexes = np.where(y==1)[0]\nplt.plot(X[neg_indexes, 0], X[neg_indexes, 1], 'yo', label='negative class')\nplt.plot(X[pos_indexes, 0], X[pos_indexes, 1], 'r^', label='positive class')\nplt.legend();\nplt.title('C = %f, degree=%d, c0=%f' % (C, d, c0))",
"_____no_output_____"
]
],
[
[
"## Gaussian RBF Kernel",
"_____no_output_____"
]
],
[
[
"g = 5\nC = 0.001\nrbf_kernel_svm_clf = Pipeline((\n ('scaler', StandardScaler()),\n ('svm_clf', SVC(kernel='rbf', gamma=g, C=C)),\n))\n\nrbf_kernel_svm_clf.fit(X, y)",
"_____no_output_____"
],
[
"# visualize resulting decision boundary\nx_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1\ny_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1\nh = .02 # step size in the mesh\nxx, yy = np.meshgrid(np.arange(x_min, x_max, h),\n np.arange(y_min, y_max, h))\n\nZ = rbf_kernel_svm_clf.predict(np.c_[xx.ravel(), yy.ravel()])\n\n# Put the result into a color plot\nZ = Z.reshape(xx.shape)\nplt.contourf(xx, yy, Z, cmap=plt.cm.Paired, alpha=0.8)\n\n\n# plot the original data\n# plot the original data\nneg_indexes = np.where(y==0)[0]\npos_indexes = np.where(y==1)[0]\nplt.plot(X[neg_indexes, 0], X[neg_indexes, 1], 'yo', label='negative class')\nplt.plot(X[pos_indexes, 0], X[pos_indexes, 1], 'r^', label='positive class')\nplt.legend();\nplt.title('C = %f, degree=%d, c0=%f' % (C, d, c0))",
"_____no_output_____"
]
],
[
[
"# Versions and Acknowledgements",
"_____no_output_____"
]
],
[
[
"import sys\nsys.path.append(\"../../src\") # add our class modules to the system PYTHON_PATH\n\nfrom ml_python_class.custom_funcs import version_information\nversion_information()",
" Module Versions\n-------------------- ------------------------------------------------------------\n matplotlib: ['3.3.0']\n numpy: ['1.18.5']\n pandas: ['1.0.5']\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4ece9e3687e1681f9f48c58bc96555dd631cde | 15,857 | ipynb | Jupyter Notebook | notebooks/foretold-submission.ipynb | bmillwood/ergo | 34be736f1979ad7f1f130bb90728270cb58dbfe8 | [
"MIT"
] | 93 | 2020-04-16T03:49:55.000Z | 2022-03-26T14:56:29.000Z | notebooks/foretold-submission.ipynb | seanjtaylor/ergo | 56835cdf9140cbb3fd0e804fd14c5209bcb6489b | [
"MIT"
] | 326 | 2020-03-25T17:49:11.000Z | 2021-03-25T03:19:51.000Z | notebooks/foretold-submission.ipynb | seanjtaylor/ergo | 56835cdf9140cbb3fd0e804fd14c5209bcb6489b | [
"MIT"
] | 26 | 2020-03-25T03:18:58.000Z | 2022-03-18T21:19:11.000Z | 116.595588 | 12,632 | 0.884278 | [
[
[
"import numpy as np\nimport scipy.stats\nimport seaborn\nimport ergo",
"_____no_output_____"
]
],
[
[
"## Testing the internal method used to build a CDF from a bag of samples:\n\nGenerate some samples, and convert to Foretold CDF:",
"_____no_output_____"
]
],
[
[
"loc = 50\nscale = 5\nsamples = np.random.normal(loc, scale, size=2000)\ncdf = ergo.foretold.ForetoldCdf.from_samples(samples, length=10)\ncdf",
"_____no_output_____"
]
],
[
[
"Compare the CDF derived from samples with the true CDF:",
"_____no_output_____"
]
],
[
[
"xs = np.linspace(loc - scale * 4, loc + scale * 4, 100)\nys = scipy.stats.norm.cdf(xs, loc=loc, scale=scale)\nseaborn.lineplot(xs, ys);\nseaborn.lineplot(cdf.xs, cdf.ys);",
"_____no_output_____"
]
],
[
[
"## Testing submission:\n\nSubmit samples as a prediction (measurement) of a [question](https://www.foretold.io/c/f45577e4-f1b0-4bba-8cf6-63944e63d70c/m/cf86da3f-c257-4787-b526-3ef3cb670cb4) outcome (measureable):",
"_____no_output_____"
]
],
[
[
"token = \"YOUR-TOKEN\"\nforetold = ergo.Foretold(token)\nquestion = foretold.get_question(\"cf86da3f-c257-4787-b526-3ef3cb670cb4\")\nresponse = question.submit_from_samples(samples, length=20)\nresponse",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4ecec5e413599348f1ffee30101d13af4b4245 | 15,291 | ipynb | Jupyter Notebook | lectures/lec-07-vectors-distances-regression.ipynb | gaby-chu/info3350-s22 | b5912187a00a5910f54f2461320438fc09b508d5 | [
"CC0-1.0"
] | null | null | null | lectures/lec-07-vectors-distances-regression.ipynb | gaby-chu/info3350-s22 | b5912187a00a5910f54f2461320438fc09b508d5 | [
"CC0-1.0"
] | null | null | null | lectures/lec-07-vectors-distances-regression.ipynb | gaby-chu/info3350-s22 | b5912187a00a5910f54f2461320438fc09b508d5 | [
"CC0-1.0"
] | null | null | null | 45.106195 | 379 | 0.623242 | [
[
[
"# INFO 3350/6350\n\n## Lecture 07: Vectorization, distance metrics, and regression\n\n## To do\n\n* Read HDA ch. 5 and Grimmer and Stewart for Monday (a lot of reading)\n* HW3 (gender and sentiment; dictionary methods) due by Thursday night at 11:59.\n* Extra credit for good, consistent answers on Ed\n* Study groups are great for homeworks.\n* Questions?\n\n## Definitions\n\n* What is a **vector**?\n * An ordered collection of numbers that locate a point in space relative to a shared reference point (called the *origin*).\n * We can also think of vectors as representing the quantified *features* of an object.\n * Vectors are usually written as *row matrices*, or just as lists: $vec = [1.0, 0.5, 3.0, 1.2]$\n * Vectors have as many *dimensions* as there are features of the object to represent.\n * The number of features to represent is a choice of the experiment. There is no correct choice, though some choices are better than others for a given purpose.\n* What is **vectorization**?\n * The process of transforming an object into its vector representation, typically by measuring some of the object's properties.\n \n## Why would we want to do this?\n\nOne goal of humanistic inquiry and of scientific research is to compare objects, so that we can gather them into types and compare any one object to others that we observe. Think of biological species or literary genres or historical eras. But how can we measure the difference or similarity between objects that are, after all, always necessarily individual and unique?\n\n* Measuring the *properties* of objects lets us compare those objects to one another.\n * But ... *which* properties?\n * Example: We counted words by type to compare gender and sentiment in novels.\n* Establishing a vector representation allows us to define a **distance metric** between objects that aren't straightforwardly spatial.\n * \"Distance\" is a metaphor. Ditto \"similarity.\"\n * Nothing is, in itself, like or unlike anything else. \n * We sometimes seek to assert that objects are similar by erasing aspects of their particularity.\n * Measuring similarity and difference are (always and only) interpretive interventions.\n \n## A spatial example\n\nConsider this map of central campus:\n\n\n\n**How far apart are Gates Hall (purple star) and the clock tower (orange star)?**\n\nWhat do we need to know or define in order to answer this question?\n\n* Where is each building in physical space.\n * Latitude/longitude; meters north/south and east/west of the book store; etc.\n* How do we want to measure the distance between them (walking, driving, flying, tunneling, ...). Minutes or miles?\n\nNormal, boring answer: about 0.4 miles on foot via Campus Rd and Ho Plaza, or a bit less if you cut some corners, or less than 0.3 miles if you can fly.\n\n| Clock tower | Gates Hall |\n| --- | --- | \n|  |  |\n\nMore interesting version: How far apart are these buildings conceptually? Architecturally? Historically? \n\n* What are the features and metrics you would use to answer this question?\n* This is a lot more like the problem of comparing texts.\n\n## A textual example",
"_____no_output_____"
]
],
[
[
"text = '''\\\nMy cat likes water.\nThe dog eats food.\nThe dog and the cat play together.\nA dog and a cat meet another dog and cat.\nThe end.'''\n\n# Print with sentence numbers\nfor line in enumerate(text.split('\\n')):\n print(line)",
"(0, 'My cat likes water.')\n(1, 'The dog eats food.')\n(2, 'The dog and the cat play together.')\n(3, 'A dog and a cat meet another dog and cat.')\n(4, 'The end.')\n"
]
],
[
[
"Let us stipulate that we want to compare these five sentences according to their \"*dogness*\" and \"*catness*.\" We care about those two aspects alone, nothing else.\n\nLet's develop some intuitions here:\n\n* Sentences 0 and 1 are as far apart as can be: 0 is about cats, 1 is about dogs.\n* Sentence 2 lies between 0 and 1. It contains a mix of dogness and catness.\n* Sentence 3 is kind of like sentence 2, but it has twice as much of both dogness and catness.\n * How different are sentences 2 and 3? (There's no objectively correct answer.)\n* Sentence 4 is a zero point. It has no dogness or catness.\n\n### Count relevant words\n\n||**cat**|**dog**|\n|---|---|---|\n|**sent**| | |\n|0|1|0|\n|1|0|1|\n|2|1|1|\n|3|2|2|\n|4|0|0|\n\nThe **vector representation** of sentence 0 is `[1, 0]`. The vector representation of sentence 3 is `[2, 2]`. And so on ...\n### Visualize (scatter plot)\n\nSketch this by hand ...\n\n### Distance measures\n\nHow far apart are sentences 0 and 1 (and all the rest)?\n\n#### Manhattan distance\n\n* Also called \"city block\" distance. \n* Not much used, but easy to understand and to compute (which matters for very large data sets). \n* Sum of the absolute difference in each dimension.\n\nFor **sentences 0 and 1**, the Manhattan distance = |1| + |-1| = 2.\n\n#### Euclidean distance\n\n* Straight-line or \"as the crow flies\" distance. \n* Widely used in data science, but not always the best choice for textual data.\n\nRecall the Pythagorean theorem for the hypotenuse of a triangle: $a^2 = b^2 + c^2$ or $a = \\sqrt{b^2 +c^2}$.\n\nFor **sentences 0 and 1**, the Euclidean distance = $\\sqrt{1^2 + 1^2} = \\sqrt{2} = 1.414$.\n\nOK, but what about the Euclidean distance between **sentence 0 and sentence 3**? Well, that distance = $\\sqrt{1^2 + 2^2} = \\sqrt{5} = 2.24$.\n\nAnd between **sentences 2 and 3** (both balanced 50:50 between dogs and cats)? That's 1.4 again, the same as the distance between sentences 0 and 1 (which, recall, are totally divergent in dog/cat content).\n\nAn obvious improvement in this case would be to **normalize word counts by document length**.\n\n#### Cosine distance\n\nMaybe instead of distance, we could measure the difference in **direction** from the origin between points.\n\n* **Sentences 0 and 1** are 90 degrees apart.\n* **Sentences 2 and 3** are 0 degrees apart.\n* **Sentences 0 and 1** are each 45 degrees away from **sentences 2 and 3**.\n\nNow, recall the values of the **cosine** of an angle between 0 and 90 degrees. (Sketch by hand)\n\nSo, the cosines of the angles between sentences are:\n\nsentences|angle|cosine\n---|---|---\n0 and 1|90|0\n2 and 3|0|1\n0 and 2|45|0.707\n0 and 3|45|0.707\n1 and 2|45|0.707\n\nWe could then transform these cosine **similarities** into **distances** by subtracting them from 1, so that the most *dissimilar* sentences (like 0 and 1) have the greatest distance between them.\n\nThe big advantage here is that we don't need to worry about getting length normalization right. Cosine distance is often a good choice for text similarity tasks.\n\n#### Higher dimensions\n\nAll of these metrics can be calculated in arbitrarily many dimensions. Which is good, because textual data is often very high-dimensional. Imagine counting the occurrences of each word type in a large corpus of novels or historical documents. Can easily be tens of thousands of dimensions.\n\n## In the real world\n\n* There's nothing wrong with any of these vectorizations and distance metrics, exactly, but they're not state of the art.\n* If you've done some recent NLP work, you'll know that, at the very least, you'd want to use static word embeddings in place of raw tokens.\n * This allows you to capture the similarity of meaning between, e.g., \"cat\" and \"kitten.\"\n* If you were especially ambitious, you'd be looking at something like BERT or ELMo or GPT-2/3, etc.\n * These transformer-based methods allow for *contextual* embeddings, that is, they represent a word token differently depending on the context in which it appears, so that the representation of \"bank\" in \"my money is in the bank\" is different from the the representation of \"bank\" in \"we walked along the bank of the river.\"\n * We'll touch on contextual embeddings near the end of the semester.\n* And then you might want features that correspond to aspects of a text other than the specific words it contains.\n * When was it written?\n * By *whom* was it written?\n * How long is it?\n * In what style is it written?\n * Who read it?\n * How much did it cost?\n * How many people read or reviewed it?\n * What else did its readers also read?\n * And so on ...\n\nHere, though, we're trying to grasp the *idea* behind document similarity, on which all of these methods depend: transform text into a numeric representation of its features (often, a representation of its content or meaning), then quantify the difference or similarity between those numeric representations.\n\n## In the problem set world\n\nWe'll dig into how, as a practical matter, we can vectorize texts and calclulate distance metrics in this week's problem set.\n\nWe'll use `scikit-learn` to implement vectorization and distance metrics. The `scikit-learn` API almost always involves *three* steps:\n\n1. Instantiate a learning object (such as a vectorizer, regressor, classifier, etc.). This is the object that will hold the parameters of your fitted model.\n1. Call the instantiated learning object's `.fit()` method, passing in your data. This allows the model to learn the optimal parameters from your data.\n1. Call the fitted model's `.transform()` or `.predict()` method, passing in either the same data from the `fit` step or new data. This step uses the fitted model to generate outputs given the input data you supply.\n\nFor example:",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import CountVectorizer\n\n# get example text as one doc per line\ndocs = [sent for sent in text.split('\\n')]\n\n# instantiate vectorizer object\n# note setup options\nvectorizer = CountVectorizer(\n vocabulary=['cat', 'dog']\n)\n\n# fit to data\nvectorizer.fit(docs)\n\n# transform docs to features\nfeatures = vectorizer.transform(docs)\n\n# print output feature matrix\nprint(vectorizer.get_feature_names_out())\nprint(features.toarray())",
"_____no_output_____"
],
[
"# calculate distances\nfrom sklearn.metrics.pairwise import euclidean_distances, cosine_distances, cosine_similarity\nimport numpy as np\n\nprint(\"Euclidean distances\")\nprint(np.round(euclidean_distances(features),2))\n\nprint(\"\\nCosine distances\")\nprint(np.round(cosine_distances(features),2))\n\nprint(\"\\nCosine **similarities**\")\nprint(np.round(cosine_similarity(features),2))",
"_____no_output_____"
],
[
"# FYI, a heatmap vis\nimport seaborn as sns\n\nprint(\"Euclidean distances\")\nsns.heatmap(\n euclidean_distances(features),\n annot=True,\n square=True\n);",
"_____no_output_____"
]
],
[
[
"## Regression\n\nWe are often interested in the relationships between measured properties of texts, or between a textual property and some other variable (year of publication, number of sales, and so on).\n\nMaybe the most basic way to measure the relationship between two variables is to use **linear regression**. The idea is to calculate a straight line through your data such that the average distance between the observed data points and the line is as small as possible. \n\n(Sketch what this looks like)\n\nYou can then calculate the **coefficient of determination**, written $r^2$ (\"r squared\"), which measures the fraction of the variation in the dependent (y) variable that is predictable from the independent (x) variable.\n\n$r^2$ = 1 - (sum of squared residuals)/(sum of squared values)\n\nAn $r^2$ value of 1 indicates perfect correlation between the variables; zero means no correlation. \n\n* There's a *lot* more to this. We'll spend some time on it later in the semester.\n* For now, focus on the fact that regression is a way to calculate a line of best fit through a data set.\n* Notice that we could also try to find something like a \"line of *worst* fit,\" which we could think of as the dividing line between two regions of feature space. This would be something like the line on which we are least likely to encounter any actual data points. \n * Think about what use-value such a dividing line might have ...",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb4ed4b7904be1ae6e09b1047a1cded302dffb3f | 154,567 | ipynb | Jupyter Notebook | Python/2. Python Basics (cont.)/4. Visualizations/Networks Science and Advanced Visualizations/networkx_holoviews_pyvis_bokeh_advanced/5b_networkx_Network-Algorithms-II.ipynb | okara83/Becoming-a-Data-Scientist | f09a15f7f239b96b77a2f080c403b2f3e95c9650 | [
"MIT"
] | null | null | null | Python/2. Python Basics (cont.)/4. Visualizations/Networks Science and Advanced Visualizations/networkx_holoviews_pyvis_bokeh_advanced/5b_networkx_Network-Algorithms-II.ipynb | okara83/Becoming-a-Data-Scientist | f09a15f7f239b96b77a2f080c403b2f3e95c9650 | [
"MIT"
] | null | null | null | Python/2. Python Basics (cont.)/4. Visualizations/Networks Science and Advanced Visualizations/networkx_holoviews_pyvis_bokeh_advanced/5b_networkx_Network-Algorithms-II.ipynb | okara83/Becoming-a-Data-Scientist | f09a15f7f239b96b77a2f080c403b2f3e95c9650 | [
"MIT"
] | 2 | 2022-02-09T15:41:33.000Z | 2022-02-11T07:47:40.000Z | 230.353204 | 51,480 | 0.907154 | [
[
[
"## Algorithm - II",
"_____no_output_____"
],
[
"### Clustering, Link Analysis, Node Classification, Link Prediction",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport networkx as nx\nimport seaborn as sns\nsns.set()\n%matplotlib inline",
"_____no_output_____"
],
[
"import warnings\nimport matplotlib.cbook\nwarnings.filterwarnings(\"ignore\",category=matplotlib.cbook.mplDeprecation)",
"_____no_output_____"
],
[
"G = nx.karate_club_graph()\nnx.draw(G, node_size = 500, node_color = \"lightblue\", with_labels = True)",
"_____no_output_____"
]
],
[
[
"### Clustering\n\nAlgorithms to characterize the number of triangles in a graph.\n\n- ```triangles(G[, nodes])``` \tCompute the number of triangles.\n- ```transitivity(G)``` \tCompute graph transitivity, the fraction of all possible triangles present in G.\n- ```clustering(G[, nodes, weight])``` \tCompute the clustering coefficient for nodes.\n- ```average_clustering(G[, nodes, weight, …])``` \tCompute the average clustering coefficient for the graph G.\n- ```square_clustering(G[, nodes])``` \tCompute the squares clustering coefficient for nodes.\n- ```generalized_degree(G[, nodes])``` \tCompute the generalized degree for nodes.",
"_____no_output_____"
]
],
[
[
"nx.triangles(G)",
"_____no_output_____"
],
[
"nx.transitivity(G)",
"_____no_output_____"
],
[
"nx.clustering(G)",
"_____no_output_____"
]
],
[
[
"--------------",
"_____no_output_____"
],
[
"### Link Analysis\n\n#### PageRank\n\nPageRank analysis of graph structure.\n\n- ```pagerank(G[, alpha, personalization, …])``` \tReturns the PageRank of the nodes in the graph.\n- ```pagerank_numpy(G[, alpha, personalization, …])``` \tReturns the PageRank of the nodes in the graph.\n- ```pagerank_scipy(G[, alpha, personalization, …])``` \tReturns the PageRank of the nodes in the graph.\n- ```google_matrix(G[, alpha, personalization, …])``` \tReturns the Google matrix of the graph.",
"_____no_output_____"
]
],
[
[
"nx.pagerank(G)",
"_____no_output_____"
],
[
"nx.google_matrix(G)",
"_____no_output_____"
]
],
[
[
"------------",
"_____no_output_____"
],
[
"#### Hits\n\nHubs and authorities analysis of graph structure.\n\n- ```hits(G[, max_iter, tol, nstart, normalized])``` \tReturns HITS hubs and authorities values for nodes.\n- ```hits_numpy(G[, normalized])``` \tReturns HITS hubs and authorities values for nodes.\n- ```hits_scipy(G[, max_iter, tol, normalized])``` \tReturns HITS hubs and authorities values for nodes.\n- ```hub_matrix(G[, nodelist])``` \tReturns the HITS hub matrix.\n- ```authority_matrix(G[, nodelist])``` \tReturns the HITS authority matrix.\n",
"_____no_output_____"
]
],
[
[
"nx.hits(G)",
"_____no_output_____"
],
[
"nx.hub_matrix(G)",
"_____no_output_____"
],
[
"nx.authority_matrix(G)",
"_____no_output_____"
]
],
[
[
"----------------",
"_____no_output_____"
],
[
"### Node Classification\n\nThis module provides the functions for node classification problem.\n\nThe functions in this module are not imported into the top level networkx namespace. You can access these functions by importing the ```networkx.algorithms.node_classification``` modules, then accessing the functions as attributes of node_classification. For example:",
"_____no_output_____"
]
],
[
[
"import networkx as nx\nfrom networkx.algorithms import node_classification\nG = nx.balanced_tree(3,3)\nnx.draw(G, node_size = 500, node_color = \"lightgreen\", with_labels = True)",
"_____no_output_____"
],
[
"G.node[1]['label'] = 'A'\nG.node[2]['label'] = 'B'\nG.node[3]['label'] = 'C'\nL = node_classification.harmonic_function(G)\nprint(L)",
"_____no_output_____"
],
[
"LL = {}\nfor n,l in zip(G.nodes(),L):\n LL.update({n:l})",
"_____no_output_____"
],
[
"nx.draw(G, node_size = 500, labels = LL, node_color = \"lightgreen\", with_labels = True)",
"_____no_output_____"
]
],
[
[
"--------------",
"_____no_output_____"
],
[
"### Link Prediction\n\nLink prediction algorithms.\n\n- ```resource_allocation_index(G[, ebunch])``` \tCompute the resource allocation index of all node pairs in ebunch.\n- ```jaccard_coefficient(G[, ebunch])``` \tCompute the Jaccard coefficient of all node pairs in ebunch.\n- ```adamic_adar_index(G[, ebunch])``` \tCompute the Adamic-Adar index of all node pairs in ebunch.\n- ```preferential_attachment(G[, ebunch])``` \tCompute the preferential attachment score of all node pairs in ebunch.\n- ```cn_soundarajan_hopcroft(G[, ebunch, community])``` \tCount the number of common neighbors of all node pairs in ebunch\n- ```ra_index_soundarajan_hopcroft(G[, ebunch, …])``` \tCompute the resource allocation index of all node pairs in ebunch using community information.\n- ```within_inter_cluster(G[, ebunch, delta, …])``` \tCompute the ratio of within- and inter-cluster common neighb",
"_____no_output_____"
]
],
[
[
"G = nx.karate_club_graph()\nnx.draw(G, node_size = 500, node_color = \"lightblue\", with_labels = True)",
"_____no_output_____"
],
[
"preds = nx.resource_allocation_index(G, [(0,10),(9, 18), (11, 12),(30,27),(16,26)])\nfor u, v, p in preds:\n print('(%d, %d) -> %.8f' % (u, v, p))\n",
"(0, 10) -> 0.58333333\n(9, 18) -> 0.05882353\n(11, 12) -> 0.06250000\n(30, 27) -> 0.05882353\n(16, 26) -> 0.00000000\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cb4edf93e2b1fcfe5b90327129ae9d887b9d6dda | 56,249 | ipynb | Jupyter Notebook | 05_Deep_Learning/sol/[HW2]TrainingNN.ipynb | wjh1065/goormNLP | ed6aeef6f76507f3e1a2abb15abdad33074bdaaa | [
"MIT"
] | null | null | null | 05_Deep_Learning/sol/[HW2]TrainingNN.ipynb | wjh1065/goormNLP | ed6aeef6f76507f3e1a2abb15abdad33074bdaaa | [
"MIT"
] | null | null | null | 05_Deep_Learning/sol/[HW2]TrainingNN.ipynb | wjh1065/goormNLP | ed6aeef6f76507f3e1a2abb15abdad33074bdaaa | [
"MIT"
] | null | null | null | 28.581809 | 184 | 0.416221 | [
[
[
"# **[HW2] Training Neural Network**\n1. Prerequisite\n2. Activation\n3. Optimizer\n4. Regularization\n5. FC vs Conv\n6. Do it by yourself\n\n이번 실습에서는 지난 시간에 배웠던 MLP-layer의 component들을 하나씩 바꿔가며 activation, optimizer, regularization, convolution layer등의 중요성을 하나씩 익혀가는 시간을 갖도록 하겠습니다. ",
"_____no_output_____"
],
[
"# 1. Prerequisite\n\n본격적인 실습을 진행하기 이전, 지난 [HW1.2 Logistic Regression vs MLP]에서 진행했던것과 동일하게 \\\\\nMnist dataset에 대해서 DataLoader와 Trainer class를 생성해두겠습니다.",
"_____no_output_____"
],
[
"\n\n## Import packages",
"_____no_output_____"
],
[
"런타임의 유형을 변경해줍니다.\n\n상단 메뉴에서 [런타임]->[런타임유형변경]->[하드웨어가속기]->[GPU]\n\n변경 이후 아래의 cell을 실행 시켰을 때, torch.cuda.is_avialable()이 True가 나와야 합니다.\n\n",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torchvision\nimport torch.optim as optim\nfrom torch.utils import data\nprint(torch.__version__)\nprint(torch.cuda.is_available())",
"1.10.0+cu111\nTrue\n"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport scipy as sp\nfrom sklearn.datasets import fetch_openml\nfrom sklearn.model_selection import train_test_split\n\nnp.set_printoptions(precision=3)\nnp.set_printoptions(suppress=True)",
"_____no_output_____"
]
],
[
[
"## Load Dataset",
"_____no_output_____"
]
],
[
[
"mnist = fetch_openml('mnist_784', cache=False)\nX = mnist.data.astype('float32').values\ny = mnist.target.astype('int64').values\nX /= 255.0\nprint(X.shape)\nprint(y.shape)",
"(70000, 784)\n(70000,)\n"
]
],
[
[
"## Split Dataset\n\n학습과 평가를 위한 dataset으로 나눕니다.",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)\nprint(X_train.shape)\nprint(y_train.shape)\nprint(X_test.shape)\nprint(y_test.shape)",
"(56000, 784)\n(56000,)\n(14000, 784)\n(14000,)\n"
]
],
[
[
"## Pytorch Dataset ",
"_____no_output_____"
]
],
[
[
"class CustomDataset(torch.utils.data.Dataset):\n def __init__(self, X, y):\n super(CustomDataset, self).__init__()\n self.X = X\n self.y = y\n \n def __getitem__(self, index):\n x = self.X[index]\n y = self.y[index]\n x = torch.from_numpy(x).float()\n y = torch.from_numpy(np.array(y)).long()\n return x, y\n\n def __len__(self):\n return len(self.X)",
"_____no_output_____"
],
[
"train_dataset = CustomDataset(X_train, y_train)\ntest_dataset = CustomDataset(X_test, y_test)\n\nprint(len(train_dataset))\nprint(train_dataset.X.shape)\nprint(len(test_dataset))\nprint(test_dataset.X.shape)",
"56000\n(56000, 784)\n14000\n(14000, 784)\n"
]
],
[
[
"## DataLoader\n",
"_____no_output_____"
]
],
[
[
"batch_size = 64\n\n# shuffle the train data\ntrain_dataloader = torch.utils.data.DataLoader(train_dataset, batch_size=batch_size, shuffle=True)\n\n# do not shuffle the val & test data\ntest_dataloader = torch.utils.data.DataLoader(test_dataset, batch_size=batch_size, shuffle=False)\n\n# dataset size // batch_size\nprint(len(train_dataloader))\nprint(len(test_dataloader))",
"875\n219\n"
]
],
[
[
"## Trainer\n",
"_____no_output_____"
]
],
[
[
"class Trainer():\n def __init__(self, trainloader, testloader, model, optimizer, criterion, device):\n \"\"\"\n trainloader: train data's loader\n testloader: test data's loader\n model: model to train\n optimizer: optimizer to update your model\n criterion: loss function\n \"\"\"\n self.trainloader = trainloader\n self.testloader = testloader\n self.model = model\n self.optimizer = optimizer\n self.criterion = criterion\n self.device = device\n \n def train(self, epoch = 1):\n self.model.train()\n for e in range(epoch):\n running_loss = 0.0 \n for i, data in enumerate(self.trainloader, 0): \n inputs, labels = data \n # model에 input으로 tensor를 gpu-device로 보낸다\n inputs = inputs.to(self.device) \n labels = labels.to(self.device)\n # zero the parameter gradients\n self.optimizer.zero_grad() \n # forward + backward + optimize\n outputs = self.model(inputs) \n loss = self.criterion(outputs, labels) \n loss.backward() \n self.optimizer.step() \n running_loss += loss.item()\n \n print('epoch: %d loss: %.3f' % (e + 1, running_loss / len(self.trainloader)))\n running_loss = 0.0\n \n def test(self):\n self.model.eval() \n correct = 0\n for inputs, labels in self.testloader:\n inputs = inputs.to(self.device)\n labels = labels.to(self.device)\n output = self.model(inputs) \n pred = output.max(1, keepdim=True)[1] # get the index of the max \n correct += pred.eq(labels.view_as(pred)).sum().item()\n test_acc = correct / len(self.testloader.dataset)\n print('test_acc: %.3f' %(test_acc))",
"_____no_output_____"
]
],
[
[
"# 2. Activation Function\n\n이번 section에서는 가장 대표적으로 사용되는 sigmoid function과 relu function을 사용해보고 비교해보도록 하겠습니다.\n\n\n",
"_____no_output_____"
],
[
"- input: 784\n- hidden: 32 or (32, 32)\n- output: 10\n- **activation: sigmoid or relu**\n- optimizer: sgd\n- loss: cross-entropy",
"_____no_output_____"
],
[
"## 2-layer Network + Sigmoid",
"_____no_output_____"
]
],
[
[
"class MLP(nn.Module):\n def __init__(self, \n input_dim=784, \n hidden_dim=32, \n output_dim=10):\n super(MLP, self).__init__()\n self.fc1 = nn.Linear(input_dim, hidden_dim)\n self.fc2 = nn.Linear(hidden_dim, output_dim)\n\n def forward(self, x):\n x = self.fc1(x)\n x = F.sigmoid(x)\n x = self.fc2(x)\n return x\n\nmodel = MLP()\ncriterion = nn.CrossEntropyLoss()\noptimizer = optim.SGD(model.parameters(), lr=0.01)\ndevice = torch.device('cuda')\nmodel.to(device)",
"_____no_output_____"
],
[
"trainer = Trainer(trainloader = train_dataloader,\n testloader = test_dataloader,\n model = model,\n criterion = criterion,\n optimizer = optimizer,\n device = device)\n\ntrainer.train(epoch = 10)",
"/usr/local/lib/python3.7/dist-packages/torch/nn/functional.py:1806: UserWarning: nn.functional.sigmoid is deprecated. Use torch.sigmoid instead.\n warnings.warn(\"nn.functional.sigmoid is deprecated. Use torch.sigmoid instead.\")\n"
],
[
"trainer.test()",
"test_acc: 0.881\n"
]
],
[
[
"## 2-layer Network + ReLU",
"_____no_output_____"
]
],
[
[
"class MLP(nn.Module):\n def __init__(self, \n input_dim=784, \n hidden_dim=32, \n output_dim=10):\n super(MLP, self).__init__()\n self.fc1 = nn.Linear(input_dim, hidden_dim)\n self.fc2 = nn.Linear(hidden_dim, output_dim)\n\n def forward(self, x):\n x = self.fc1(x)\n x = F.relu(x)\n x = self.fc2(x)\n return x\n\nmodel = MLP()\ncriterion = nn.CrossEntropyLoss()\noptimizer = optim.SGD(model.parameters(), lr=0.01)\ndevice = torch.device('cuda')\nmodel.to(device)",
"_____no_output_____"
],
[
"trainer = Trainer(trainloader = train_dataloader,\n testloader = test_dataloader,\n model = model,\n criterion = criterion,\n optimizer = optimizer,\n device = device)\n\ntrainer.train(epoch = 10)",
"epoch: 1 loss: 1.406\nepoch: 2 loss: 0.543\nepoch: 3 loss: 0.413\nepoch: 4 loss: 0.365\nepoch: 5 loss: 0.338\nepoch: 6 loss: 0.320\nepoch: 7 loss: 0.306\nepoch: 8 loss: 0.295\nepoch: 9 loss: 0.285\nepoch: 10 loss: 0.276\n"
],
[
"trainer.test()",
"test_acc: 0.924\n"
]
],
[
[
"#### Q1. Activation Function에 따라 성능의 차이가 있나요? 있다면, 왜 차이가 발생했을까요?\n\n\n",
"_____no_output_____"
],
[
"## 3-layer Network + Sigmoid",
"_____no_output_____"
]
],
[
[
"class MLP(nn.Module):\n def __init__(self, \n input_dim=784, \n hidden_dim=(32,32), \n output_dim=10):\n super(MLP, self).__init__()\n self.fc1 = nn.Linear(input_dim, hidden_dim[0])\n self.fc2 = nn.Linear(hidden_dim[0], hidden_dim[1])\n self.fc3 = nn.Linear(hidden_dim[1], output_dim)\n\n def forward(self, x):\n x = self.fc1(x)\n x = F.sigmoid(x)\n x = self.fc2(x)\n x = F.sigmoid(x)\n x = self.fc3(x)\n return x\n\nmodel = MLP()\ncriterion = nn.CrossEntropyLoss()\noptimizer = optim.SGD(model.parameters(), lr=0.01)\ndevice = torch.device('cuda')\nmodel.to(device)",
"_____no_output_____"
],
[
"trainer = Trainer(trainloader = train_dataloader,\n testloader = test_dataloader,\n model = model,\n criterion = criterion,\n optimizer = optimizer,\n device = device)\n\ntrainer.train(epoch = 10)",
"/usr/local/lib/python3.7/dist-packages/torch/nn/functional.py:1806: UserWarning: nn.functional.sigmoid is deprecated. Use torch.sigmoid instead.\n warnings.warn(\"nn.functional.sigmoid is deprecated. Use torch.sigmoid instead.\")\n"
],
[
"trainer.test()",
"test_acc: 0.601\n"
]
],
[
[
"## 3-layer Network + ReLU",
"_____no_output_____"
]
],
[
[
"class MLP(nn.Module):\n def __init__(self, \n input_dim=784, \n hidden_dim=(32,32), \n output_dim=10):\n super(MLP, self).__init__()\n self.fc1 = nn.Linear(input_dim, hidden_dim[0])\n self.fc2 = nn.Linear(hidden_dim[0], hidden_dim[1])\n self.fc3 = nn.Linear(hidden_dim[1], output_dim)\n\n def forward(self, x):\n x = self.fc1(x)\n x = F.relu(x)\n x = self.fc2(x)\n x = F.relu(x)\n x = self.fc3(x)\n return x\n\nmodel = MLP()\ncriterion = nn.CrossEntropyLoss()\noptimizer = optim.SGD(model.parameters(), lr=0.01)\ndevice = torch.device('cuda')\nmodel.to(device)",
"_____no_output_____"
],
[
"trainer = Trainer(trainloader = train_dataloader,\n testloader = test_dataloader,\n model = model,\n criterion = criterion,\n optimizer = optimizer,\n device = device)\n\ntrainer.train(epoch = 10)",
"epoch: 1 loss: 1.685\nepoch: 2 loss: 0.581\nepoch: 3 loss: 0.422\nepoch: 4 loss: 0.373\nepoch: 5 loss: 0.345\nepoch: 6 loss: 0.324\nepoch: 7 loss: 0.305\nepoch: 8 loss: 0.289\nepoch: 9 loss: 0.274\nepoch: 10 loss: 0.260\n"
],
[
"trainer.test()",
"test_acc: 0.928\n"
]
],
[
[
"#### Q2. Activation function 별로 Layer 수를 늘리는 것이 성능이 어떻게 변하나요? 양상이 다르게 나타난다면 왜 그럴까요?",
"_____no_output_____"
],
[
"\n#### Q3. Activation function이 존재하지 않는다면 어떤 일이 일어날까요?",
"_____no_output_____"
],
[
"# 3. Optimization\n\n이번 section에서는 sgd, momentum, Adam등의 optimizer를 사용해보고 성능을 비교해보도록 하겠습니다.\n\n\n",
"_____no_output_____"
],
[
"- input: 784\n- hidden: (32, 32)\n- output: 10\n- activation: relu\n- **optimizer: sgd or momentum or adam**\n- loss: cross-entropy",
"_____no_output_____"
]
],
[
[
"class MLP(nn.Module):\n def __init__(self, \n input_dim=784, \n hidden_dim=(32,32), \n output_dim=10):\n super(MLP, self).__init__()\n self.fc1 = nn.Linear(input_dim, hidden_dim[0])\n self.fc2 = nn.Linear(hidden_dim[0], hidden_dim[1])\n self.fc3 = nn.Linear(hidden_dim[1], output_dim)\n\n def forward(self, x):\n x = self.fc1(x)\n x = F.relu(x)\n x = self.fc2(x)\n x = F.relu(x)\n x = self.fc3(x)\n return x",
"_____no_output_____"
]
],
[
[
"## 3-layer Network + ReLU + SGD",
"_____no_output_____"
]
],
[
[
"model = MLP()\noptimizer = optim.SGD(model.parameters(), lr=0.01)\ncriterion = nn.CrossEntropyLoss()\ndevice = torch.device('cuda')\nmodel.to(device)",
"_____no_output_____"
],
[
"trainer = Trainer(trainloader = train_dataloader,\n testloader = test_dataloader,\n model = model,\n criterion = criterion,\n optimizer = optimizer,\n device = device)\n\ntrainer.train(epoch = 10)",
"epoch: 1 loss: 1.927\nepoch: 2 loss: 0.727\nepoch: 3 loss: 0.467\nepoch: 4 loss: 0.381\nepoch: 5 loss: 0.342\nepoch: 6 loss: 0.315\nepoch: 7 loss: 0.297\nepoch: 8 loss: 0.282\nepoch: 9 loss: 0.268\nepoch: 10 loss: 0.258\n"
],
[
"trainer.test()",
"test_acc: 0.928\n"
]
],
[
[
"## 3-layer Network + ReLU + Momentum\n\n\n\n",
"_____no_output_____"
]
],
[
[
"model = MLP()\noptimizer = optim.SGD(model.parameters(), lr=0.01, momentum=0.99)\ncriterion = nn.CrossEntropyLoss()\ndevice = torch.device('cuda')\nmodel.to(device)",
"_____no_output_____"
],
[
"trainer = Trainer(trainloader = train_dataloader,\n testloader = test_dataloader,\n model = model,\n criterion = criterion,\n optimizer = optimizer,\n device = device)\n\ntrainer.train(epoch = 10)",
"epoch: 1 loss: 0.561\nepoch: 2 loss: 0.239\nepoch: 3 loss: 0.188\nepoch: 4 loss: 0.182\nepoch: 5 loss: 0.160\nepoch: 6 loss: 0.145\nepoch: 7 loss: 0.142\nepoch: 8 loss: 0.150\nepoch: 9 loss: 0.127\nepoch: 10 loss: 0.124\n"
],
[
"trainer.test()",
"test_acc: 0.956\n"
]
],
[
[
"## 3-layer Network + ReLU + Adam\n\n",
"_____no_output_____"
]
],
[
[
"model = MLP()\noptimizer = optim.Adam(model.parameters(), lr=0.01)\ncriterion = nn.CrossEntropyLoss()\ndevice = torch.device('cuda')\nmodel.to(device)",
"_____no_output_____"
],
[
"trainer = Trainer(trainloader = train_dataloader,\n testloader = test_dataloader,\n model = model,\n criterion = criterion,\n optimizer = optimizer,\n device = device)\n\ntrainer.train(epoch = 10)",
"epoch: 1 loss: 0.300\nepoch: 2 loss: 0.178\nepoch: 3 loss: 0.150\nepoch: 4 loss: 0.143\nepoch: 5 loss: 0.131\nepoch: 6 loss: 0.127\nepoch: 7 loss: 0.126\nepoch: 8 loss: 0.116\nepoch: 9 loss: 0.119\nepoch: 10 loss: 0.111\n"
],
[
"trainer.test()",
"test_acc: 0.959\n"
]
],
[
[
"",
"_____no_output_____"
],
[
"#### Q4. Optimizer 별로 수렴 속도가 어떻게 다른가요? \n##### Q4.1 수렴 속도가 다르다면 sgd와 momentum의 차이는 왜 발생할까요? \n##### Q4.2 수렴 속도가 다르다면 momentum과 Adam의 차이는 왜 발생할까요?",
"_____no_output_____"
],
[
"## 4. Regularization\n\n이번 section에서는 image data에서 주로 사용되는 batch-normalization을 어떻게 사용하는지를 확인해보겠습니다.\n\n",
"_____no_output_____"
],
[
"- input: 784\n- hidden: 32 or (32, 32)\n- output: 10\n- activation: relu\n- optimizer: adam\n- **regularizer: batch_norm**\n- loss: cross-entropy",
"_____no_output_____"
],
[
"## 3-layer Network + ReLU + Adam + batch_norm",
"_____no_output_____"
]
],
[
[
"class MLP(nn.Module):\n def __init__(self, \n input_dim=784, \n hidden_dim=(32,32), \n output_dim=10):\n super(MLP, self).__init__()\n self.fc1 = nn.Linear(input_dim, hidden_dim[0])\n self.bn1 = nn.BatchNorm1d(hidden_dim[0])\n self.fc2 = nn.Linear(hidden_dim[0], hidden_dim[1])\n self.bn2 = nn.BatchNorm1d(hidden_dim[1])\n self.fc3 = nn.Linear(hidden_dim[1], output_dim)\n\n def forward(self, x):\n x = self.fc1(x)\n x = self.bn1(x)\n x = F.relu(x)\n x = self.fc2(x)\n x = self.bn2(x)\n x = F.relu(x)\n x = self.fc3(x)\n return x",
"_____no_output_____"
],
[
"model = MLP()\noptimizer = optim.Adam(model.parameters(), lr=0.01)\ncriterion = nn.CrossEntropyLoss()\ndevice = torch.device('cuda')\nmodel.to(device)",
"_____no_output_____"
],
[
"trainer = Trainer(trainloader = train_dataloader,\n testloader = test_dataloader,\n model = model,\n criterion = criterion,\n optimizer = optimizer,\n device = device)\n\ntrainer.train(epoch = 10)",
"epoch: 1 loss: 0.263\nepoch: 2 loss: 0.150\nepoch: 3 loss: 0.122\nepoch: 4 loss: 0.106\nepoch: 5 loss: 0.099\nepoch: 6 loss: 0.088\nepoch: 7 loss: 0.086\nepoch: 8 loss: 0.077\nepoch: 9 loss: 0.073\nepoch: 10 loss: 0.073\n"
],
[
"trainer.test()",
"test_acc: 0.972\n"
],
[
"def count_parameters(model):\n print(sum(p.numel() for p in model.parameters() if p.requires_grad))\n return sum(p.numel() for p in model.parameters() if p.requires_grad)\n\ncount_parameters(model)",
"26634\n"
]
],
[
[
"#### Q5. Batch-normalization을 사용하기 전 후로 성능이 어떻게 변화했나요? 왜 이러한 변화가 일어났을까요?\n",
"_____no_output_____"
],
[
"# 5. Fully-Connected Layer vs Convolution Layer\n\n지금까지 model의 다양한 node를 바꿔가며 mnist의 성능 변화를 확인해보는 실습을 진행해 보았습니다. \\\\\n비록, fully-connected network가 mnist 데이터에서 높은 성능을 내는데는 문제가 없었지만, 모든 layer를 fully-connected layer로 만드는 것은 엄청난 파라미터와 연산량을 필요로 하기 때문에 더욱 큰 고화질의 이미지 데이터를 처리하는데는 적합하지 않습니다. \\\\ \n\n따라서, 이번 section에서는 이미지 데이터 처리에 주로 사용되는 convolution layer를 사용해보고 파라미터 수와 성능이 어떻게 변화하는지 확인해보도록 하겠습니다. ",
"_____no_output_____"
],
[
"## Convolution Operation\n\n\n",
"_____no_output_____"
],
[
"### Q6. Input이 (H, W, C) 일 때, stride S의 2개의 (F * F) convolutional filter를 적용하면 output이 어떻게 되나요?",
"_____no_output_____"
]
],
[
[
"class Conv(nn.Module):\n def __init__(self, \n input_dim=784, \n output_dim=10):\n super(Conv, self).__init__()\n self.conv1 = nn.Conv2d(in_channels=1,\n out_channels=8,\n kernel_size=7,\n stride=2)\n self.conv2 = nn.Conv2d(in_channels=8,\n out_channels=8,\n kernel_size=7,\n stride=2)\n self.fc = nn.Linear(3*3*8, output_dim)\n\n def forward(self, x):\n # should reshape data into image\n x = x.reshape(-1, 1, 28, 28)\n x = self.conv1(x)\n x = F.relu(x)\n x = self.conv2(x)\n x = F.relu(x)\n x = x.reshape(-1, 3*3*8)\n x = self.fc(x)\n return x",
"_____no_output_____"
],
[
"model = Conv()\noptimizer = optim.Adam(model.parameters(), lr=0.01)\ncriterion = nn.CrossEntropyLoss()\ndevice = torch.device('cuda')\nmodel.to(device)",
"_____no_output_____"
],
[
"trainer = Trainer(trainloader = train_dataloader,\n testloader = test_dataloader,\n model = model,\n criterion = criterion,\n optimizer = optimizer,\n device = device)\n\ntrainer.train(epoch = 10)",
"epoch: 1 loss: 0.226\nepoch: 2 loss: 0.103\nepoch: 3 loss: 0.090\nepoch: 4 loss: 0.085\nepoch: 5 loss: 0.081\nepoch: 6 loss: 0.081\nepoch: 7 loss: 0.082\nepoch: 8 loss: 0.077\nepoch: 9 loss: 0.077\nepoch: 10 loss: 0.071\n"
],
[
"trainer.test()",
"test_acc: 0.973\n"
],
[
"count_parameters(model)",
"4274\n"
]
],
[
[
"##### Q7. covolution operation은 image데이터를 다루는데 있어서 fully-connected layer에 비해 어떤 점에서 효과적일까요?\n",
"_____no_output_____"
],
[
"## 6. Do It By Yourself\n\n위에서 했던 실습들과 수업에 배웠던 다양한 network component들을 참조해서 20,000개 이하의 파라미터로 98%의 accuracy를 달성해보세요!",
"_____no_output_____"
]
],
[
[
"class CustomModel(nn.Module):\n def __init__(self, \n input_dim=784, \n output_dim=10):\n super(CustomModel, self).__init__()\n\n # [64, 1, 28, 28] => [64, 3, 22, 22]\n self.conv1 = nn.Conv2d(in_channels=1,\n out_channels=3,\n kernel_size=7,\n stride=1)\n \n self.bn1 = nn.BatchNorm2d(3)\n\n # [64, 3, 22, 22] => [64, 8, 16, 16]\n self.conv2 = nn.Conv2d(in_channels=3,\n out_channels=7,\n kernel_size=7,\n stride=1)\n \n self.bn2 = nn.BatchNorm2d(7)\n\n\n self.fc = nn.Linear(16*16*7, output_dim)\n\n def forward(self, x):\n # should reshape data into image\n x = x.reshape(-1, 1, 28, 28)\n \n x = self.conv1(x)\n x = self.bn1(x)\n x = F.relu(x)\n\n x = self.conv2(x)\n x = self.bn2(x)\n x = F.relu(x)\n\n\n x = x.reshape(-1, 16*16*7)\n x = self.fc(x)\n return x",
"_____no_output_____"
],
[
"model = CustomModel()\ncount_parameters(model)",
"19136\n"
],
[
"optimizer = optim.Adam(model.parameters(), lr=0.01)\ncriterion = nn.CrossEntropyLoss()\ndevice = torch.device('cuda')\nmodel.to(device)",
"_____no_output_____"
],
[
"trainer = Trainer(trainloader = train_dataloader,\n testloader = test_dataloader,\n model = model,\n criterion = criterion,\n optimizer = optimizer,\n device = device)\n\ntrainer.train(epoch = 10)",
"epoch: 1 loss: 0.162\nepoch: 2 loss: 0.070\nepoch: 3 loss: 0.058\nepoch: 4 loss: 0.053\nepoch: 5 loss: 0.048\nepoch: 6 loss: 0.041\nepoch: 7 loss: 0.041\nepoch: 8 loss: 0.036\nepoch: 9 loss: 0.030\nepoch: 10 loss: 0.027\n"
],
[
"trainer.test()",
"test_acc: 0.981\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4ee18b05fc399125eea1c8be6596b2f9c46255 | 506,384 | ipynb | Jupyter Notebook | 12-Scraping-APIs/lecture-12-exercise-solutions.ipynb | macrodatascience/2019-datascience-lectures | 6e49d0d0e52247cf748966274cc8679d3409b60d | [
"MIT"
] | 36 | 2018-01-09T18:25:45.000Z | 2021-08-08T02:15:43.000Z | 12-Scraping-APIs/lecture-12-exercise-solutions.ipynb | macrodatascience/2019-datascience-lectures | 6e49d0d0e52247cf748966274cc8679d3409b60d | [
"MIT"
] | 1 | 2018-11-16T10:49:32.000Z | 2018-11-17T12:35:17.000Z | 12-Scraping-APIs/lecture-12-exercise-solutions.ipynb | macrodatascience/2019-datascience-lectures | 6e49d0d0e52247cf748966274cc8679d3409b60d | [
"MIT"
] | 41 | 2018-01-10T20:40:25.000Z | 2020-08-20T03:25:15.000Z | 205.679935 | 319,246 | 0.669496 | [
[
[
"## Exercise 1: Exceptional Olympians\n\nScrape data from [this wikipedia site](https://en.wikipedia.org/wiki/List_of_multiple_Olympic_medalists) about exceptional Olympic medalists. \n\n1. Download the html using urllib. \n2. Parse this html with BeautifulSoup.\n3. Extract the html that corresponds to the big table from the soup.\n4. Parse the table into a pandas dataframe. Hint: both the \"No.\" and the \"Total.\" column use row-spans which are tricky to parse, both with a pandas reader and manually. For the purpose of this exercise, exclude all rows that are not easy to parse (the first one is Bjørn Dæhlie).\n5. Create a table that shows for each country how many gold, silver, bronze, and total medals it won in that list.",
"_____no_output_____"
]
],
[
[
"from bs4 import BeautifulSoup\nimport urllib.request\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"Download the html using urllib. \nParse this html with BeautifulSoup.",
"_____no_output_____"
]
],
[
[
"url = \"https://en.wikipedia.org/wiki/List_of_multiple_Olympic_medalists\"\n\nreq = urllib.request.Request(url)\nwith urllib.request.urlopen(req) as response:\n html = response.read()\n\nprint(html)\n\nclass_soup = BeautifulSoup(html, 'html.parser')",
"b'<!DOCTYPE html>\\n<html class=\"client-nojs\" lang=\"en\" dir=\"ltr\">\\n<head>\\n<meta charset=\"UTF-8\"/>\\n<title>List of multiple Olympic medalists - Wikipedia</title>\\n<script>document.documentElement.className = document.documentElement.className.replace( /(^|\\\\s)client-nojs(\\\\s|$)/, \"$1client-js$2\" );</script>\\n<script>(window.RLQ=window.RLQ||[]).push(function(){mw.config.set({\"wgCanonicalNamespace\":\"\",\"wgCanonicalSpecialPageName\":false,\"wgNamespaceNumber\":0,\"wgPageName\":\"List_of_multiple_Olympic_medalists\",\"wgTitle\":\"List of multiple Olympic medalists\",\"wgCurRevisionId\":825763339,\"wgRevisionId\":825763339,\"wgArticleId\":18855244,\"wgIsArticle\":true,\"wgIsRedirect\":false,\"wgAction\":\"view\",\"wgUserName\":null,\"wgUserGroups\":[\"*\"],\"wgCategories\":[\"Pages using citations with format and no URL\",\"Articles with hCards\",\"Incomplete lists from May 2012\",\"Lists of Olympic medalists\",\"Olympic Games medal tables\"],\"wgBreakFrames\":false,\"wgPageContentLanguage\":\"en\",\"wgPageContentModel\":\"wikitext\",\"wgSeparatorTransformTable\":[\"\",\"\"],\"wgDigitTransformTable\":[\"\",\"\"],\"wgDefaultDateFormat\":\"dmy\",\"wgMonthNames\":[\"\",\"January\",\"February\",\"March\",\"April\",\"May\",\"June\",\"July\",\"August\",\"September\",\"October\",\"November\",\"December\"],\"wgMonthNamesShort\":[\"\",\"Jan\",\"Feb\",\"Mar\",\"Apr\",\"May\",\"Jun\",\"Jul\",\"Aug\",\"Sep\",\"Oct\",\"Nov\",\"Dec\"],\"wgRelevantPageName\":\"List_of_multiple_Olympic_medalists\",\"wgRelevantArticleId\":18855244,\"wgRequestId\":\"WoVmuApAMFoAAGYM798AAABA\",\"wgIsProbablyEditable\":true,\"wgRelevantPageIsProbablyEditable\":true,\"wgRestrictionEdit\":[],\"wgRestrictionMove\":[],\"wgFlaggedRevsParams\":{\"tags\":{}},\"wgStableRevisionId\":null,\"wgWikiEditorEnabledModules\":[],\"wgBetaFeaturesFeatures\":[],\"wgMediaViewerOnClick\":true,\"wgMediaViewerEnabledByDefault\":true,\"wgPopupsShouldSendModuleToUser\":true,\"wgPopupsConflictsWithNavPopupGadget\":false,\"wgVisualEditor\":{\"pageLanguageCode\":\"en\",\"pageLanguageDir\":\"ltr\",\"pageVariantFallbacks\":\"en\",\"usePageImages\":true,\"usePageDescriptions\":true},\"wgPreferredVariant\":\"en\",\"wgMFExpandAllSectionsUserOption\":false,\"wgMFDisplayWikibaseDescriptions\":{\"search\":true,\"nearby\":true,\"watchlist\":true,\"tagline\":false},\"wgRelatedArticles\":null,\"wgRelatedArticlesUseCirrusSearch\":true,\"wgRelatedArticlesOnlyUseCirrusSearch\":false,\"wgULSCurrentAutonym\":\"English\",\"wgNoticeProject\":\"wikipedia\",\"wgCentralNoticeCookiesToDelete\":[],\"wgCentralNoticeCategoriesUsingLegacy\":[\"Fundraising\",\"fundraising\"],\"wgCategoryTreePageCategoryOptions\":\"{\\\\\"mode\\\\\":0,\\\\\"hideprefix\\\\\":20,\\\\\"showcount\\\\\":true,\\\\\"namespaces\\\\\":false}\",\"wgWikibaseItemId\":\"Q6629540\",\"wgScoreNoteLanguages\":{\"arabic\":\"\\xd8\\xa7\\xd9\\x84\\xd8\\xb9\\xd8\\xb1\\xd8\\xa8\\xd9\\x8a\\xd8\\xa9\",\"catalan\":\"catal\\xc3\\xa0\",\"deutsch\":\"Deutsch\",\"english\":\"English\",\"espanol\":\"espa\\xc3\\xb1ol\",\"italiano\":\"italiano\",\"nederlands\":\"Nederlands\",\"norsk\":\"norsk\",\"portugues\":\"portugu\\xc3\\xaas\",\"suomi\":\"suomi\",\"svenska\":\"svenska\",\"vlaams\":\"West-Vlams\"},\"wgScoreDefaultNoteLanguage\":\"nederlands\",\"wgCentralAuthMobileDomain\":false,\"wgCodeMirrorEnabled\":false,\"wgVisualEditorToolbarScrollOffset\":0,\"wgVisualEditorUnsupportedEditParams\":[\"undo\",\"undoafter\",\"veswitched\"],\"wgEditSubmitButtonLabelPublish\":true});mw.loader.state({\"ext.gadget.charinsert-styles\":\"ready\",\"ext.globalCssJs.user.styles\":\"ready\",\"ext.globalCssJs.site.styles\":\"ready\",\"site.styles\":\"ready\",\"noscript\":\"ready\",\"user.styles\":\"ready\",\"user\":\"ready\",\"user.options\":\"ready\",\"user.tokens\":\"loading\",\"ext.cite.styles\":\"ready\",\"wikibase.client.init\":\"ready\",\"ext.visualEditor.desktopArticleTarget.noscript\":\"ready\",\"ext.uls.interlanguage\":\"ready\",\"ext.wikimediaBadges\":\"ready\",\"mediawiki.legacy.shared\":\"ready\",\"mediawiki.legacy.commonPrint\":\"ready\",\"mediawiki.sectionAnchor\":\"ready\",\"mediawiki.skinning.interface\":\"ready\",\"skins.vector.styles\":\"ready\",\"ext.globalCssJs.user\":\"ready\",\"ext.globalCssJs.site\":\"ready\"});mw.loader.implement(\"user.tokens@1dqfd7l\",function($,jQuery,require,module){/*@nomin*/mw.user.tokens.set({\"editToken\":\"+\\\\\\\\\",\"patrolToken\":\"+\\\\\\\\\",\"watchToken\":\"+\\\\\\\\\",\"csrfToken\":\"+\\\\\\\\\"});\\n});mw.loader.load([\"ext.cite.a11y\",\"site\",\"mediawiki.page.startup\",\"mediawiki.user\",\"mediawiki.hidpi\",\"mediawiki.page.ready\",\"jquery.tablesorter\",\"mediawiki.toc\",\"mediawiki.searchSuggest\",\"ext.gadget.teahouse\",\"ext.gadget.ReferenceTooltips\",\"ext.gadget.watchlist-notice\",\"ext.gadget.DRN-wizard\",\"ext.gadget.charinsert\",\"ext.gadget.refToolbar\",\"ext.gadget.extra-toolbar-buttons\",\"ext.gadget.switcher\",\"ext.centralauth.centralautologin\",\"mmv.head\",\"mmv.bootstrap.autostart\",\"ext.popups\",\"ext.visualEditor.desktopArticleTarget.init\",\"ext.visualEditor.targetLoader\",\"ext.eventLogging.subscriber\",\"ext.wikimediaEvents\",\"ext.navigationTiming\",\"ext.uls.eventlogger\",\"ext.uls.init\",\"ext.uls.interface\",\"ext.centralNotice.geoIP\",\"ext.centralNotice.startUp\",\"skins.vector.js\"]);});</script>\\n<link rel=\"stylesheet\" href=\"/w/load.php?debug=false&lang=en&modules=ext.cite.styles%7Cext.uls.interlanguage%7Cext.visualEditor.desktopArticleTarget.noscript%7Cext.wikimediaBadges%7Cmediawiki.legacy.commonPrint%2Cshared%7Cmediawiki.sectionAnchor%7Cmediawiki.skinning.interface%7Cskins.vector.styles%7Cwikibase.client.init&only=styles&skin=vector\"/>\\n<script async=\"\" src=\"/w/load.php?debug=false&lang=en&modules=startup&only=scripts&skin=vector\"></script>\\n<meta name=\"ResourceLoaderDynamicStyles\" content=\"\"/>\\n<link rel=\"stylesheet\" href=\"/w/load.php?debug=false&lang=en&modules=ext.gadget.charinsert-styles&only=styles&skin=vector\"/>\\n<link rel=\"stylesheet\" href=\"/w/load.php?debug=false&lang=en&modules=site.styles&only=styles&skin=vector\"/>\\n<meta name=\"generator\" content=\"MediaWiki 1.31.0-wmf.20\"/>\\n<meta name=\"referrer\" content=\"origin\"/>\\n<meta name=\"referrer\" content=\"origin-when-crossorigin\"/>\\n<meta name=\"referrer\" content=\"origin-when-cross-origin\"/>\\n<link rel=\"alternate\" href=\"android-app://org.wikipedia/http/en.m.wikipedia.org/wiki/List_of_multiple_Olympic_medalists\"/>\\n<link rel=\"alternate\" type=\"application/x-wiki\" title=\"Edit this page\" href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit\"/>\\n<link rel=\"edit\" title=\"Edit this page\" href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit\"/>\\n<link rel=\"apple-touch-icon\" href=\"/static/apple-touch/wikipedia.png\"/>\\n<link rel=\"shortcut icon\" href=\"/static/favicon/wikipedia.ico\"/>\\n<link rel=\"search\" type=\"application/opensearchdescription+xml\" href=\"/w/opensearch_desc.php\" title=\"Wikipedia (en)\"/>\\n<link rel=\"EditURI\" type=\"application/rsd+xml\" href=\"//en.wikipedia.org/w/api.php?action=rsd\"/>\\n<link rel=\"license\" href=\"//creativecommons.org/licenses/by-sa/3.0/\"/>\\n<link rel=\"canonical\" href=\"https://en.wikipedia.org/wiki/List_of_multiple_Olympic_medalists\"/>\\n<link rel=\"dns-prefetch\" href=\"//login.wikimedia.org\"/>\\n<link rel=\"dns-prefetch\" href=\"//meta.wikimedia.org\" />\\n<!--[if lt IE 9]><script src=\"/w/load.php?debug=false&lang=en&modules=html5shiv&only=scripts&skin=vector&sync=1\"></script><![endif]-->\\n</head>\\n<body class=\"mediawiki ltr sitedir-ltr mw-hide-empty-elt ns-0 ns-subject page-List_of_multiple_Olympic_medalists rootpage-List_of_multiple_Olympic_medalists skin-vector action-view\">\\t\\t<div id=\"mw-page-base\" class=\"noprint\"></div>\\n\\t\\t<div id=\"mw-head-base\" class=\"noprint\"></div>\\n\\t\\t<div id=\"content\" class=\"mw-body\" role=\"main\">\\n\\t\\t\\t<a id=\"top\"></a>\\n\\t\\t\\t<div id=\"siteNotice\" class=\"mw-body-content\"><!-- CentralNotice --></div><div class=\"mw-indicators mw-body-content\">\\n</div>\\n<h1 id=\"firstHeading\" class=\"firstHeading\" lang=\"en\">List of multiple Olympic medalists</h1>\\t\\t\\t<div id=\"bodyContent\" class=\"mw-body-content\">\\n\\t\\t\\t\\t<div id=\"siteSub\" class=\"noprint\">From Wikipedia, the free encyclopedia</div>\\t\\t\\t\\t<div id=\"contentSub\"></div>\\n\\t\\t\\t\\t\\t\\t\\t\\t<div id=\"jump-to-nav\" class=\"mw-jump\">\\n\\t\\t\\t\\t\\tJump to:\\t\\t\\t\\t\\t<a href=\"#mw-head\">navigation</a>, \\t\\t\\t\\t\\t<a href=\"#p-search\">search</a>\\n\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t<div id=\"mw-content-text\" lang=\"en\" dir=\"ltr\" class=\"mw-content-ltr\"><div class=\"mw-parser-output\"><p>The page lists various individuals who have won multiple <a href=\"/wiki/Olympic_medal\" title=\"Olympic medal\">Olympic medals</a> at either the <a href=\"/wiki/Summer_Olympic_Games\" title=\"Summer Olympic Games\">Summer Olympic Games</a> or the <a href=\"/wiki/Winter_Olympic_Games\" title=\"Winter Olympic Games\">Winter Olympic Games</a>.</p>\\n<p>As of August 13, 2016, American swimmer <a href=\"/wiki/Michael_Phelps\" title=\"Michael Phelps\">Michael Phelps</a> has won the most Olympic medals with 28 medals (23 gold, 3 silver, 2 bronze). He is also the most decorated Olympian in individual events, with 16 medals (13 gold, 2 silver, 1 bronze). Norwegian biathlete <a href=\"/wiki/Ole_Einar_Bj%C3%B8rndalen\" title=\"Ole Einar Bj\\xc3\\xb8rndalen\">Ole Einar Bj\\xc3\\xb8rndalen</a> is the most decorated Winter Olympian, with 13 medals (8 gold, 4 silver, and 1 bronze).</p>\\n<p></p>\\n<div id=\"toc\" class=\"toc\">\\n<div class=\"toctitle\" lang=\"en\" dir=\"ltr\" xml:lang=\"en\">\\n<h2>Contents</h2>\\n</div>\\n<ul>\\n<li class=\"toclevel-1 tocsection-1\"><a href=\"#List_of_Olympic_medals_over_career\"><span class=\"tocnumber\">1</span> <span class=\"toctext\">List of Olympic medals over career</span></a>\\n<ul>\\n<li class=\"toclevel-2 tocsection-2\"><a href=\"#Timeline\"><span class=\"tocnumber\">1.1</span> <span class=\"toctext\">Timeline</span></a></li>\\n</ul>\\n</li>\\n<li class=\"toclevel-1 tocsection-3\"><a href=\"#List_of_most_career_medals_in_individual_events\"><span class=\"tocnumber\">2</span> <span class=\"toctext\">List of most career medals in individual events</span></a></li>\\n<li class=\"toclevel-1 tocsection-4\"><a href=\"#Athletes_with_medals_in_different_disciplines\"><span class=\"tocnumber\">3</span> <span class=\"toctext\">Athletes with medals in different disciplines</span></a>\\n<ul>\\n<li class=\"toclevel-2 tocsection-5\"><a href=\"#In_the_Summer_and_Winter_Games\"><span class=\"tocnumber\">3.1</span> <span class=\"toctext\">In the Summer and Winter Games</span></a></li>\\n<li class=\"toclevel-2 tocsection-6\"><a href=\"#In_the_Summer_Games\"><span class=\"tocnumber\">3.2</span> <span class=\"toctext\">In the Summer Games</span></a>\\n<ul>\\n<li class=\"toclevel-3 tocsection-7\"><a href=\"#Swimming_and_water_polo\"><span class=\"tocnumber\">3.2.1</span> <span class=\"toctext\">Swimming and water polo</span></a></li>\\n<li class=\"toclevel-3 tocsection-8\"><a href=\"#Others_in_Summer_Games\"><span class=\"tocnumber\">3.2.2</span> <span class=\"toctext\">Others in Summer Games</span></a></li>\\n</ul>\\n</li>\\n<li class=\"toclevel-2 tocsection-9\"><a href=\"#In_the_Winter_Games\"><span class=\"tocnumber\">3.3</span> <span class=\"toctext\">In the Winter Games</span></a>\\n<ul>\\n<li class=\"toclevel-3 tocsection-10\"><a href=\"#Cross-country_skiing_and_Nordic_combined\"><span class=\"tocnumber\">3.3.1</span> <span class=\"toctext\">Cross-country skiing and Nordic combined</span></a></li>\\n<li class=\"toclevel-3 tocsection-11\"><a href=\"#Others_in_Winter_Games\"><span class=\"tocnumber\">3.3.2</span> <span class=\"toctext\">Others in Winter Games</span></a></li>\\n</ul>\\n</li>\\n</ul>\\n</li>\\n<li class=\"toclevel-1 tocsection-12\"><a href=\"#See_also\"><span class=\"tocnumber\">4</span> <span class=\"toctext\">See also</span></a></li>\\n<li class=\"toclevel-1 tocsection-13\"><a href=\"#References\"><span class=\"tocnumber\">5</span> <span class=\"toctext\">References</span></a></li>\\n</ul>\\n</div>\\n<p></p>\\n<h2><span class=\"mw-headline\" id=\"List_of_Olympic_medals_over_career\">List of Olympic medals over career</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=1\" title=\"Edit section: List of Olympic medals over career\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h2>\\n<p>This is a <b>list of multiple Olympic medalists</b>, listing people who have won <b>seven or more</b> <a href=\"/wiki/Olympic_Games\" title=\"Olympic Games\">Olympic</a> medals. Medals won in the <a href=\"/wiki/1906_Intercalated_Games\" title=\"1906 Intercalated Games\">1906 Intercalated Games</a> are not included. It includes top-three placings in 1896 and 1900, before medals were awarded for top-three placings. When an athlete has won medals for more than one nation, for simplicity only the last nation he or she represented is mentioned here. The Olympics listed for each athlete only include games when they won medals. See the particular article on the athlete for more details on when and for what nation an athlete competed.</p>\\n<p>In those instances where more than one athlete has the same number of total medals, the first tiebreaker is the number of gold medals, then the number of silver medals. Where two or more athletes have exactly the same number of gold, silver and bronze medals, the ranking is shown as a tie and the athletes are shown in order by career years and name.</p>\\n<table class=\"wikitable sortable\">\\n<tr>\\n<th>No.</th>\\n<th style=\"width:7.8em;\">Athlete</th>\\n<th style=\"width:8.2em;\">Nation</th>\\n<th style=\"width:5.6em;\">Sport</th>\\n<th>Years</th>\\n<th>Games</th>\\n<th>Gender</th>\\n<th style=\"background-color:gold; width:3.5em; font-weight:bold;\">Gold</th>\\n<th style=\"background-color:silver; width:3.5em; font-weight:bold;\">Silver</th>\\n<th style=\"background-color:#cc9966; width:3.5em; font-weight:bold;\">Bronze</th>\\n<th style=\"width:3.5em;\">Total</th>\\n</tr>\\n<tr>\\n<td>1</td>\\n<td align=\"left\"><span class=\"sortkey\">Phelps, Michael</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Michael_Phelps\" title=\"Michael Phelps\">Michael Phelps</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2004\\xe2\\x80\\x932016</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>23</td>\\n<td>3</td>\\n<td>2</td>\\n<td>28</td>\\n</tr>\\n<tr>\\n<td>2</td>\\n<td align=\"left\"><span class=\"sortkey\">Latynina, Larisa</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Larisa_Latynina\" title=\"Larisa Latynina\">Larisa Latynina</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1956\\xe2\\x80\\x931964</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>9</td>\\n<td>5</td>\\n<td>4</td>\\n<td>18</td>\\n</tr>\\n<tr>\\n<td>3</td>\\n<td align=\"left\"><span class=\"sortkey\">Andrianov, Nikolai</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Nikolai_Andrianov\" title=\"Nikolai Andrianov\">Nikolai Andrianov</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1972\\xe2\\x80\\x931980</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>7</td>\\n<td>5</td>\\n<td>3</td>\\n<td>15</td>\\n</tr>\\n<tr>\\n<td>4</td>\\n<td align=\"left\"><span class=\"sortkey\">Bjorndalen, Ole Einar</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ole_Einar_Bj%C3%B8rndalen\" title=\"Ole Einar Bj\\xc3\\xb8rndalen\">Ole Einar Bj\\xc3\\xb8rndalen</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">Norway</a></td>\\n<td><a href=\"/wiki/Biathlon_at_the_Winter_Olympics\" title=\"Biathlon at the Winter Olympics\">Biathlon</a></td>\\n<td>1998\\xe2\\x80\\x932014</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>8</td>\\n<td>4</td>\\n<td>1</td>\\n<td rowspan=\"4\">13</td>\\n</tr>\\n<tr>\\n<td>5</td>\\n<td align=\"left\"><span class=\"sortkey\">Shakhlin, Boris</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Boris_Shakhlin\" title=\"Boris Shakhlin\">Boris Shakhlin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1956\\xe2\\x80\\x931964</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>7</td>\\n<td>4</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>6</td>\\n<td align=\"left\"><span class=\"sortkey\">Mangiarotti, Edoardo</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Edoardo_Mangiarotti\" title=\"Edoardo Mangiarotti\">Edoardo Mangiarotti</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/22px-Flag_of_Italy.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/33px-Flag_of_Italy.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/44px-Flag_of_Italy.svg.png 2x\" data-file-width=\"1500\" data-file-height=\"1000\" /> <a href=\"/wiki/Italy_at_the_Olympics\" title=\"Italy at the Olympics\">Italy</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_Summer_Olympics\" title=\"Fencing at the Summer Olympics\">Fencing</a></td>\\n<td>1936\\xe2\\x80\\x931960</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>5</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>7</td>\\n<td align=\"left\"><span class=\"sortkey\">Ono, Takashi</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Takashi_Ono\" title=\"Takashi Ono\">Takashi Ono</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/22px-Flag_of_Japan.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/33px-Flag_of_Japan.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/44px-Flag_of_Japan.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Japan_at_the_Olympics\" title=\"Japan at the Olympics\">Japan</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1952\\xe2\\x80\\x931964</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>4</td>\\n<td>4</td>\\n</tr>\\n<tr>\\n<td>8</td>\\n<td align=\"left\"><span class=\"sortkey\">Nurmi, Paavo</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Paavo_Nurmi\" title=\"Paavo Nurmi\">Paavo Nurmi</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/22px-Flag_of_Finland.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/33px-Flag_of_Finland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/44px-Flag_of_Finland.svg.png 2x\" data-file-width=\"1800\" data-file-height=\"1100\" /> <a href=\"/wiki/Finland_at_the_Olympics\" title=\"Finland at the Olympics\">Finland</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>1920\\xe2\\x80\\x931928</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>9</td>\\n<td>3</td>\\n<td>0</td>\\n<td rowspan=\"10\">12</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">9</td>\\n<td align=\"left\"><span class=\"sortkey\">Fischer, Birgit</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Birgit_Fischer\" title=\"Birgit Fischer\">Birgit Fischer</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/22px-Flag_of_East_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/33px-Flag_of_East_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/44px-Flag_of_East_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/East_Germany_at_the_Olympics\" title=\"East Germany at the Olympics\">East Germany</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">Germany</a></td>\\n<td><a href=\"/wiki/Canoeing_at_the_Summer_Olympics\" title=\"Canoeing at the Summer Olympics\">Canoeing</a></td>\\n<td>1980\\xe2\\x80\\x932004</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>8</td>\\n<td>4</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Dahlie, Bjorn</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Bj%C3%B8rn_D%C3%A6hlie\" title=\"Bj\\xc3\\xb8rn D\\xc3\\xa6hlie\">Bj\\xc3\\xb8rn D\\xc3\\xa6hlie</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">Norway</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1992\\xe2\\x80\\x931998</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>8</td>\\n<td>4</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">11</td>\\n<td align=\"left\"><span class=\"sortkey\">Kato, Sawao</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Sawao_Kato\" title=\"Sawao Kato\">Sawao Kato</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/22px-Flag_of_Japan.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/33px-Flag_of_Japan.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/44px-Flag_of_Japan.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Japan_at_the_Olympics\" title=\"Japan at the Olympics\">Japan</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1968\\xe2\\x80\\x931976</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>8</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Thompson, Jenny</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Jenny_Thompson\" title=\"Jenny Thompson\">Jenny Thompson</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1992\\xe2\\x80\\x932004</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>8</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>13</td>\\n<td align=\"left\"><span class=\"sortkey\">Bjorgen, Marit</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Marit_Bj%C3%B8rgen\" title=\"Marit Bj\\xc3\\xb8rgen\">Marit Bj\\xc3\\xb8rgen</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">Norway</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>2002\\xe2\\x80\\x932018</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>6</td>\\n<td>4</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>14</td>\\n<td align=\"left\"><span class=\"sortkey\">Lochte, Ryan</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ryan_Lochte\" title=\"Ryan Lochte\">Ryan Lochte</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2004\\xe2\\x80\\x932016</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>3</td>\\n<td>3</td>\\n</tr>\\n<tr>\\n<td>15</td>\\n<td align=\"left\"><span class=\"sortkey\">Torres, Dara</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Dara_Torres\" title=\"Dara Torres\">Dara Torres</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1984\\xe2\\x80\\x932008</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>4</td>\\n<td>4</td>\\n<td>4</td>\\n</tr>\\n<tr>\\n<td>16</td>\\n<td align=\"left\"><span class=\"sortkey\">Nemov, Alexei</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Alexei_Nemov\" title=\"Alexei Nemov\">Alexei Nemov</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/22px-Flag_of_Russia.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/33px-Flag_of_Russia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/44px-Flag_of_Russia.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Russia_at_the_Olympics\" title=\"Russia at the Olympics\">Russia</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1996\\xe2\\x80\\x932000</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>2</td>\\n<td>6</td>\\n</tr>\\n<tr>\\n<td>17</td>\\n<td align=\"left\"><span class=\"sortkey\">Coughlin, Natalie</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Natalie_Coughlin\" title=\"Natalie Coughlin\">Natalie Coughlin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2004\\xe2\\x80\\x932012</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>3</td>\\n<td>4</td>\\n<td>5</td>\\n</tr>\\n<tr>\\n<td>18</td>\\n<td align=\"left\"><span class=\"sortkey\">Spitz, Mark</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Mark_Spitz\" title=\"Mark Spitz\">Mark Spitz</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1968\\xe2\\x80\\x931972</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>9</td>\\n<td>1</td>\\n<td>1</td>\\n<td rowspan=\"5\">11</td>\\n</tr>\\n<tr>\\n<td>19</td>\\n<td align=\"left\"><span class=\"sortkey\">Biondi, Matt</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Matt_Biondi\" title=\"Matt Biondi\">Matt Biondi</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1984\\xe2\\x80\\x931992</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>8</td>\\n<td>2</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>20</td>\\n<td align=\"left\"><span class=\"sortkey\">Caslavska, Vera</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/V%C4%9Bra_%C4%8C%C3%A1slavsk%C3%A1\" title=\"V\\xc4\\x9bra \\xc4\\x8c\\xc3\\xa1slavsk\\xc3\\xa1\">V\\xc4\\x9bra \\xc4\\x8c\\xc3\\xa1slavsk\\xc3\\xa1</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/cb/Flag_of_the_Czech_Republic.svg/22px-Flag_of_the_Czech_Republic.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/cb/Flag_of_the_Czech_Republic.svg/33px-Flag_of_the_Czech_Republic.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/cb/Flag_of_the_Czech_Republic.svg/44px-Flag_of_the_Czech_Republic.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Czechoslovakia_at_the_Olympics\" title=\"Czechoslovakia at the Olympics\">Czechoslovakia</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1960\\xe2\\x80\\x931968</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>7</td>\\n<td>4</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td>21</td>\\n<td align=\"left\"><span class=\"sortkey\">Chukarin, Viktor</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Viktor_Chukarin\" title=\"Viktor Chukarin\">Viktor Chukarin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1952\\xe2\\x80\\x931956</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>7</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>22</td>\\n<td align=\"left\"><span class=\"sortkey\">Osburn, Carl</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Carl_Osburn\" title=\"Carl Osburn\">Carl Osburn</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/22px-Flag_of_the_United_States_%281912-1959%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/33px-Flag_of_the_United_States_%281912-1959%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/44px-Flag_of_the_United_States_%281912-1959%29.svg.png 2x\" data-file-width=\"1900\" data-file-height=\"1000\" /> <a href=\"/wiki/United_States_at_the_1924_Summer_Olympics\" title=\"United States at the 1924 Summer Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Shooting_at_the_Summer_Olympics\" title=\"Shooting at the Summer Olympics\">Shooting</a></td>\\n<td>1912\\xe2\\x80\\x931924</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>4</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>23</td>\\n<td align=\"left\"><span class=\"sortkey\">Lewis, Carl</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Carl_Lewis\" title=\"Carl Lewis\">Carl Lewis</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>1984\\xe2\\x80\\x931996</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>9</td>\\n<td>1</td>\\n<td>0</td>\\n<td rowspan=\"13\">10</td>\\n</tr>\\n<tr>\\n<td>24</td>\\n<td align=\"left\"><span class=\"sortkey\">Gerevich, Aladar</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Alad%C3%A1r_Gerevich\" title=\"Alad\\xc3\\xa1r Gerevich\">Alad\\xc3\\xa1r Gerevich</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/22px-Flag_of_Hungary.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/33px-Flag_of_Hungary.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/44px-Flag_of_Hungary.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Hungary_at_the_Olympics\" title=\"Hungary at the Olympics\">Hungary</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_Summer_Olympics\" title=\"Fencing at the Summer Olympics\">Fencing</a></td>\\n<td>1932\\xe2\\x80\\x931960</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>7</td>\\n<td>1</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>25</td>\\n<td align=\"left\"><span class=\"sortkey\">Werth, Isabell</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Isabell_Werth\" title=\"Isabell Werth\">Isabell Werth</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">Germany</a></td>\\n<td><a href=\"/wiki/Equestrian_at_the_Summer_Olympics\" title=\"Equestrian at the Summer Olympics\">Equestrian</a></td>\\n<td>1992\\xe2\\x80\\x932016</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>6</td>\\n<td>4</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td>26</td>\\n<td align=\"left\"><span class=\"sortkey\">Nakayama, Akinori</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Akinori_Nakayama\" title=\"Akinori Nakayama\">Akinori Nakayama</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/22px-Flag_of_Japan.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/33px-Flag_of_Japan.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/44px-Flag_of_Japan.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Japan_at_the_Olympics\" title=\"Japan at the Olympics\">Japan</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1968\\xe2\\x80\\x931972</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>27</td>\\n<td align=\"left\"><span class=\"sortkey\">Scherbo, Vitaly</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Vitaly_Scherbo\" title=\"Vitaly Scherbo\">Vitaly Scherbo</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/22px-Olympic_flag.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/33px-Olympic_flag.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/44px-Olympic_flag.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Unified_Team_at_the_Olympics\" title=\"Unified Team at the Olympics\">Unified Team</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/85/Flag_of_Belarus.svg/22px-Flag_of_Belarus.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/85/Flag_of_Belarus.svg/33px-Flag_of_Belarus.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/8/85/Flag_of_Belarus.svg/44px-Flag_of_Belarus.svg.png 2x\" data-file-width=\"900\" data-file-height=\"450\" /> <a href=\"/wiki/Belarus_at_the_Olympics\" title=\"Belarus at the Olympics\">Belarus</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1992\\xe2\\x80\\x931996</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>0</td>\\n<td>4</td>\\n</tr>\\n<tr>\\n<td>28</td>\\n<td align=\"left\"><span class=\"sortkey\">W\\xc3\\xbcst, Ireen</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ireen_W%C3%BCst\" title=\"Ireen W\\xc3\\xbcst\">Ireen W\\xc3\\xbcst</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/22px-Flag_of_the_Netherlands.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/33px-Flag_of_the_Netherlands.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/44px-Flag_of_the_Netherlands.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Netherlands_at_the_Olympics\" title=\"Netherlands at the Olympics\">Netherlands</a></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_Winter_Olympics\" title=\"Speed skating at the Winter Olympics\">Speed skating</a></td>\\n<td>2006\\xe2\\x80\\x932018</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>5</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">29</td>\\n<td align=\"left\"><span class=\"sortkey\">Keleti, Agnes</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/%C3%81gnes_Keleti\" title=\"\\xc3\\x81gnes Keleti\">\\xc3\\x81gnes Keleti</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/64/Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg/22px-Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/64/Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg/33px-Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/6/64/Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg/44px-Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Hungary_at_the_1956_Summer_Olympics\" title=\"Hungary at the 1956 Summer Olympics\">Hungary</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1952\\xe2\\x80\\x931956</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>5</td>\\n<td>3</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Hall Jr., Gary</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Gary_Hall_Jr.\" title=\"Gary Hall Jr.\">Gary Hall Jr.</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1996\\xe2\\x80\\x932004</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>3</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>31</td>\\n<td align=\"left\"><span class=\"sortkey\">Astakhova, Polina</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Polina_Astakhova\" title=\"Polina Astakhova\">Polina Astakhova</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1956\\xe2\\x80\\x931964</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>5</td>\\n<td>2</td>\\n<td>3</td>\\n</tr>\\n<tr>\\n<td>32</td>\\n<td align=\"left\"><span class=\"sortkey\">Smetanina, Raisa</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Raisa_Smetanina\" title=\"Raisa Smetanina\">Raisa Smetanina</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/22px-Olympic_flag.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/33px-Olympic_flag.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/44px-Olympic_flag.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Unified_Team_at_the_Olympics\" title=\"Unified Team at the Olympics\">Unified Team</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1976\\xe2\\x80\\x931992</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>4</td>\\n<td>5</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>33</td>\\n<td align=\"left\"><span class=\"sortkey\">Dityatin, Alexander</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Alexander_Dityatin\" title=\"Alexander Dityatin\">Alexander Dityatin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1976\\xe2\\x80\\x931980</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>3</td>\\n<td>6</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>34</td>\\n<td align=\"left\"><span class=\"sortkey\">Belmondo, Stefania</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Stefania_Belmondo\" title=\"Stefania Belmondo\">Stefania Belmondo</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/22px-Flag_of_Italy.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/33px-Flag_of_Italy.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/44px-Flag_of_Italy.svg.png 2x\" data-file-width=\"1500\" data-file-height=\"1000\" /> <a href=\"/wiki/Italy_at_the_Olympics\" title=\"Italy at the Olympics\">Italy</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1992\\xe2\\x80\\x932002</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>2</td>\\n<td>3</td>\\n<td>5</td>\\n</tr>\\n<tr>\\n<td>35</td>\\n<td align=\"left\"><span class=\"sortkey\">Almsick, Franziska</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Franziska_van_Almsick\" title=\"Franziska van Almsick\">Franziska van Almsick</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">Germany</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1992\\xe2\\x80\\x932004</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>0</td>\\n<td>4</td>\\n<td>6</td>\\n</tr>\\n<tr>\\n<td rowspan=\"3\">36</td>\\n<td align=\"left\"><span class=\"sortkey\">Van Innis, Hubert</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Hubert_Van_Innis\" title=\"Hubert Van Innis\">Hubert Van Innis</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/9/92/Flag_of_Belgium_%28civil%29.svg/22px-Flag_of_Belgium_%28civil%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/9/92/Flag_of_Belgium_%28civil%29.svg/33px-Flag_of_Belgium_%28civil%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/9/92/Flag_of_Belgium_%28civil%29.svg/44px-Flag_of_Belgium_%28civil%29.svg.png 2x\" data-file-width=\"450\" data-file-height=\"300\" /> <a href=\"/wiki/Belgium_at_the_Olympics\" title=\"Belgium at the Olympics\">Belgium</a></td>\\n<td><a href=\"/wiki/Archery_at_the_Summer_Olympics\" title=\"Archery at the Summer Olympics\">Archery</a></td>\\n<td>1900\\xe2\\x80\\x931920</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>3</td>\\n<td>0</td>\\n<td rowspan=\"19\">9</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Yegorova, Lyubov</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Lyubov_Yegorova_(cross-country_skier)\" title=\"Lyubov Yegorova (cross-country skier)\">Lyubov Yegorova</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/22px-Olympic_flag.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/33px-Olympic_flag.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/44px-Olympic_flag.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Unified_Team_at_the_Olympics\" title=\"Unified Team at the Olympics\">Unified Team</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/22px-Flag_of_Russia.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/33px-Flag_of_Russia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/44px-Flag_of_Russia.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Russia_at_the_Olympics\" title=\"Russia at the Olympics\">Russia</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1992\\xe2\\x80\\x931994</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>6</td>\\n<td>3</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Felix, Allyson</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Allyson_Felix\" title=\"Allyson Felix\">Allyson Felix</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>2004\\xe2\\x80\\x932016</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>6</td>\\n<td>3</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td>39</td>\\n<td align=\"left\"><span class=\"sortkey\">Vezzali, Valentina</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Valentina_Vezzali\" title=\"Valentina Vezzali\">Valentina Vezzali</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/22px-Flag_of_Italy.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/33px-Flag_of_Italy.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/44px-Flag_of_Italy.svg.png 2x\" data-file-width=\"1500\" data-file-height=\"1000\" /> <a href=\"/wiki/Italy_at_the_Olympics\" title=\"Italy at the Olympics\">Italy</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_Summer_Olympics\" title=\"Fencing at the Summer Olympics\">Fencing</a></td>\\n<td>1996\\xe2\\x80\\x932012</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>6</td>\\n<td>1</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">40</td>\\n<td align=\"left\"><span class=\"sortkey\">Comaneci, Nadia</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Nadia_Com%C4%83neci\" title=\"Nadia Com\\xc4\\x83neci\">Nadia Com\\xc4\\x83neci</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/7f/Flag_of_Romania_%281965-1989%29.svg/22px-Flag_of_Romania_%281965-1989%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/7f/Flag_of_Romania_%281965-1989%29.svg/33px-Flag_of_Romania_%281965-1989%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/7/7f/Flag_of_Romania_%281965-1989%29.svg/44px-Flag_of_Romania_%281965-1989%29.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Romania_at_the_1980_Summer_Olympics\" title=\"Romania at the 1980 Summer Olympics\">Romania</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1976\\xe2\\x80\\x931980</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>5</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Thorpe, Ian</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ian_Thorpe\" title=\"Ian Thorpe\">Ian Thorpe</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/22px-Flag_of_Australia.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/33px-Flag_of_Australia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/44px-Flag_of_Australia.svg.png 2x\" data-file-width=\"1280\" data-file-height=\"640\" /> <a href=\"/wiki/Australia_at_the_Olympics\" title=\"Australia at the Olympics\">Australia</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2000\\xe2\\x80\\x932004</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>42</td>\\n<td align=\"left\"><span class=\"sortkey\">Pechstein, Claudia</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Claudia_Pechstein\" title=\"Claudia Pechstein\">Claudia Pechstein</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">Germany</a></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_Winter_Olympics\" title=\"Speed skating at the Winter Olympics\">Speed skating</a></td>\\n<td>1992\\xe2\\x80\\x932006</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>5</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>43</td>\\n<td align=\"left\"><span class=\"sortkey\">Tsukahara, Mitsuo</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Mitsuo_Tsukahara\" title=\"Mitsuo Tsukahara\">Mitsuo Tsukahara</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/22px-Flag_of_Japan.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/33px-Flag_of_Japan.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/44px-Flag_of_Japan.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Japan_at_the_Olympics\" title=\"Japan at the Olympics\">Japan</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1968\\xe2\\x80\\x931976</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>1</td>\\n<td>3</td>\\n</tr>\\n<tr>\\n<td>44</td>\\n<td align=\"left\"><span class=\"sortkey\">Popov, Alexander</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Alexander_Popov_(swimmer)\" title=\"Alexander Popov (swimmer)\">Alexander Popov</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/22px-Olympic_flag.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/33px-Olympic_flag.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/44px-Olympic_flag.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Unified_Team_at_the_Olympics\" title=\"Unified Team at the Olympics\">Unified Team</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/22px-Flag_of_Russia.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/33px-Flag_of_Russia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/44px-Flag_of_Russia.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Russia_at_the_Olympics\" title=\"Russia at the Olympics\">Russia</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1992\\xe2\\x80\\x932000</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>5</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">45</td>\\n<td align=\"left\"><span class=\"sortkey\">Jernberg, Sixten</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Sixten_Jernberg\" title=\"Sixten Jernberg\">Sixten Jernberg</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/22px-Flag_of_Sweden.svg.png\" width=\"22\" height=\"14\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/33px-Flag_of_Sweden.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/44px-Flag_of_Sweden.svg.png 2x\" data-file-width=\"1600\" data-file-height=\"1000\" /> <a href=\"/wiki/Sweden_at_the_Olympics\" title=\"Sweden at the Olympics\">Sweden</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1956\\xe2\\x80\\x931964</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>4</td>\\n<td>3</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Tourischeva, Ludmilla</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ludmilla_Tourischeva\" title=\"Ludmilla Tourischeva\">Ludmilla Tourischeva</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1968\\xe2\\x80\\x931976</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>4</td>\\n<td>3</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">47</td>\\n<td align=\"left\"><span class=\"sortkey\">Grunsven, Anky</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Anky_van_Grunsven\" title=\"Anky van Grunsven\">Anky van Grunsven</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/22px-Flag_of_the_Netherlands.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/33px-Flag_of_the_Netherlands.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/44px-Flag_of_the_Netherlands.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Netherlands_at_the_Olympics\" title=\"Netherlands at the Olympics\">Netherlands</a></td>\\n<td><a href=\"/wiki/Equestrian_at_the_Summer_Olympics\" title=\"Equestrian at the Summer Olympics\">Equestrian</a></td>\\n<td>1992\\xe2\\x80\\x932012</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>3</td>\\n<td>5</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Jones, Leisel</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Leisel_Jones\" title=\"Leisel Jones\">Leisel Jones</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/22px-Flag_of_Australia.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/33px-Flag_of_Australia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/44px-Flag_of_Australia.svg.png 2x\" data-file-width=\"1280\" data-file-height=\"640\" /> <a href=\"/wiki/Australia_at_the_Olympics\" title=\"Australia at the Olympics\">Australia</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2000\\xe2\\x80\\x932012</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>3</td>\\n<td>5</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>49</td>\\n<td align=\"left\"><span class=\"sortkey\">Gaudini, Giulio</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Giulio_Gaudini\" title=\"Giulio Gaudini\">Giulio Gaudini</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/ad/Flag_of_Italy_%281861-1946%29.svg/22px-Flag_of_Italy_%281861-1946%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/ad/Flag_of_Italy_%281861-1946%29.svg/33px-Flag_of_Italy_%281861-1946%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/ad/Flag_of_Italy_%281861-1946%29.svg/44px-Flag_of_Italy_%281861-1946%29.svg.png 2x\" data-file-width=\"1500\" data-file-height=\"1000\" /> <a href=\"/wiki/Italy_at_the_1936_Summer_Olympics\" title=\"Italy at the 1936 Summer Olympics\">Italy</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_Summer_Olympics\" title=\"Fencing at the Summer Olympics\">Fencing</a></td>\\n<td>1928\\xe2\\x80\\x931936</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>3</td>\\n<td>4</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>50</td>\\n<td align=\"left\"><span class=\"sortkey\">Swahn, Alfred</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Alfred_Swahn\" title=\"Alfred Swahn\">Alfred Swahn</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/22px-Flag_of_Sweden.svg.png\" width=\"22\" height=\"14\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/33px-Flag_of_Sweden.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/44px-Flag_of_Sweden.svg.png 2x\" data-file-width=\"1600\" data-file-height=\"1000\" /> <a href=\"/wiki/Sweden_at_the_Olympics\" title=\"Sweden at the Olympics\">Sweden</a></td>\\n<td><a href=\"/wiki/Shooting_at_the_Summer_Olympics\" title=\"Shooting at the Summer Olympics\">Shooting</a></td>\\n<td>1908\\xe2\\x80\\x931924</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>3</td>\\n<td>3</td>\\n<td>3</td>\\n</tr>\\n<tr>\\n<td>51</td>\\n<td align=\"left\"><span class=\"sortkey\">Voronin, Mikhail</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Mikhail_Voronin\" title=\"Mikhail Voronin\">Mikhail Voronin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1968\\xe2\\x80\\x931972</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>2</td>\\n<td>6</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>52</td>\\n<td align=\"left\"><span class=\"sortkey\">Savolainen, Heikki</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Heikki_Savolainen_(gymnast)\" title=\"Heikki Savolainen (gymnast)\">Heikki Savolainen</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/22px-Flag_of_Finland.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/33px-Flag_of_Finland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/44px-Flag_of_Finland.svg.png 2x\" data-file-width=\"1800\" data-file-height=\"1100\" /> <a href=\"/wiki/Finland_at_the_Olympics\" title=\"Finland at the Olympics\">Finland</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1928\\xe2\\x80\\x931952</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>2</td>\\n<td>1</td>\\n<td>6</td>\\n</tr>\\n<tr>\\n<td>53</td>\\n<td align=\"left\"><span class=\"sortkey\">Titov, Yuri</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Yuri_Titov\" title=\"Yuri Titov\">Yuri Titov</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1956\\xe2\\x80\\x931964</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>1</td>\\n<td>5</td>\\n<td>3</td>\\n</tr>\\n<tr>\\n<td>54</td>\\n<td align=\"left\"><span class=\"sortkey\">Ottey, Merlene</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Merlene_Ottey\" title=\"Merlene Ottey\">Merlene Ottey</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/22px-Flag_of_Jamaica.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/33px-Flag_of_Jamaica.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/44px-Flag_of_Jamaica.svg.png 2x\" data-file-width=\"600\" data-file-height=\"300\" /> <a href=\"/wiki/Jamaica_at_the_Olympics\" title=\"Jamaica at the Olympics\">Jamaica</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>1980\\xe2\\x80\\x932000</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>0</td>\\n<td>3</td>\\n<td>6</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">55</td>\\n<td align=\"left\"><span class=\"sortkey\">Ewry, Ray</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ray_Ewry\" title=\"Ray Ewry\">Ray Ewry</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/22px-Flag_of_the_United_States_%281896-1908%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/33px-Flag_of_the_United_States_%281896-1908%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/44px-Flag_of_the_United_States_%281896-1908%29.svg.png 2x\" data-file-width=\"1024\" data-file-height=\"539\" /> <a href=\"/wiki/United_States_at_the_1908_Summer_Olympics\" title=\"United States at the 1908 Summer Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>1900\\xe2\\x80\\x931908</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>8</td>\\n<td>0</td>\\n<td>0</td>\\n<td rowspan=\"37\">8</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Bolt, Usain</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Usain_Bolt\" title=\"Usain Bolt\">Usain Bolt</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/22px-Flag_of_Jamaica.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/33px-Flag_of_Jamaica.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/44px-Flag_of_Jamaica.svg.png 2x\" data-file-width=\"600\" data-file-height=\"300\" /> <a href=\"/wiki/Jamaica_at_the_Olympics\" title=\"Jamaica at the Olympics\">Jamaica</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>2008\\xe2\\x80\\x932016</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>8</td>\\n<td>0</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td>57</td>\\n<td align=\"left\"><span class=\"sortkey\">Fredriksson, Gert</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Gert_Fredriksson\" title=\"Gert Fredriksson\">Gert Fredriksson</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/22px-Flag_of_Sweden.svg.png\" width=\"22\" height=\"14\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/33px-Flag_of_Sweden.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/44px-Flag_of_Sweden.svg.png 2x\" data-file-width=\"1600\" data-file-height=\"1000\" /> <a href=\"/wiki/Sweden_at_the_Olympics\" title=\"Sweden at the Olympics\">Sweden</a></td>\\n<td><a href=\"/wiki/Canoeing_at_the_Summer_Olympics\" title=\"Canoeing at the Summer Olympics\">Canoeing</a></td>\\n<td>1948\\xe2\\x80\\x931960</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>1</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">58</td>\\n<td align=\"left\"><span class=\"sortkey\">Klimke, Reiner</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Reiner_Klimke\" title=\"Reiner Klimke\">Reiner Klimke</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/9/9a/German_Olympic_flag_%281959-1968%29.svg/22px-German_Olympic_flag_%281959-1968%29.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/9/9a/German_Olympic_flag_%281959-1968%29.svg/33px-German_Olympic_flag_%281959-1968%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/9/9a/German_Olympic_flag_%281959-1968%29.svg/44px-German_Olympic_flag_%281959-1968%29.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/United_Team_of_Germany_at_the_Olympics\" class=\"mw-redirect\" title=\"United Team of Germany at the Olympics\">United Team of Germany</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/West_Germany_at_the_Olympics\" title=\"West Germany at the Olympics\">West Germany</a></td>\\n<td><a href=\"/wiki/Equestrian_at_the_Summer_Olympics\" title=\"Equestrian at the Summer Olympics\">Equestrian</a></td>\\n<td>1964\\xe2\\x80\\x931988</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>0</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><a href=\"/wiki/Viktor_Ahn\" class=\"mw-redirect\" title=\"Viktor Ahn\">Ahn Hyun-Soo</a><br />\\n<a href=\"/wiki/Viktor_Ahn\" class=\"mw-redirect\" title=\"Viktor Ahn\">Viktor Ahn</a></td>\\n<td><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/0/09/Flag_of_South_Korea.svg/22px-Flag_of_South_Korea.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/0/09/Flag_of_South_Korea.svg/33px-Flag_of_South_Korea.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/0/09/Flag_of_South_Korea.svg/44px-Flag_of_South_Korea.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/South_Korea_at_the_Olympics\" title=\"South Korea at the Olympics\">South Korea</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/22px-Flag_of_Russia.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/33px-Flag_of_Russia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/44px-Flag_of_Russia.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Russia_at_the_Olympics\" title=\"Russia at the Olympics\">Russia</a></td>\\n<td><a href=\"/wiki/Short_track_speed_skating_at_the_Winter_Olympics\" title=\"Short track speed skating at the Winter Olympics\">Short track speed skating</a></td>\\n<td>2006-2014</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>6</td>\\n<td>0</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>60</td>\\n<td align=\"left\"><span class=\"sortkey\">Ritola, Ville</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ville_Ritola\" title=\"Ville Ritola\">Ville Ritola</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/22px-Flag_of_Finland.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/33px-Flag_of_Finland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/44px-Flag_of_Finland.svg.png 2x\" data-file-width=\"1800\" data-file-height=\"1100\" /> <a href=\"/wiki/Finland_at_the_Olympics\" title=\"Finland at the Olympics\">Finland</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>1924\\xe2\\x80\\x931928</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>3</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td>61</td>\\n<td align=\"left\"><span class=\"sortkey\">Lipa, Elisabeta</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Elisabeta_Lip%C4%83\" title=\"Elisabeta Lip\\xc4\\x83\">Elisabeta Lip\\xc4\\x83</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/73/Flag_of_Romania.svg/22px-Flag_of_Romania.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/73/Flag_of_Romania.svg/33px-Flag_of_Romania.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/7/73/Flag_of_Romania.svg/44px-Flag_of_Romania.svg.png 2x\" data-file-width=\"600\" data-file-height=\"400\" /> <a href=\"/wiki/Romania_at_the_Olympics\" title=\"Romania at the Olympics\">Romania</a></td>\\n<td><a href=\"/wiki/Rowing_at_the_Summer_Olympics\" title=\"Rowing at the Summer Olympics\">Rowing</a></td>\\n<td>1984\\xe2\\x80\\x932000</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>5</td>\\n<td>2</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">62</td>\\n<td align=\"left\"><span class=\"sortkey\">Wiggins, Bradley</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Bradley_Wiggins\" title=\"Bradley Wiggins\">Bradley Wiggins</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/22px-Flag_of_the_United_Kingdom.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/33px-Flag_of_the_United_Kingdom.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/44px-Flag_of_the_United_Kingdom.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Great_Britain_at_the_Olympics\" title=\"Great Britain at the Olympics\">Great Britain</a></td>\\n<td><a href=\"/wiki/Cycling_at_the_Summer_Olympics\" title=\"Cycling at the Summer Olympics\">Cycling</a></td>\\n<td>2000\\xe2\\x80\\x932016</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>1</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Adrian, Nathan</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Nathan_Adrian\" title=\"Nathan Adrian\">Nathan Adrian</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2008\\xe2\\x80\\x932016</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>1</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td rowspan=\"3\">64</td>\\n<td align=\"left\"><span class=\"sortkey\">Carlberg, Vilhelm</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Vilhelm_Carlberg\" title=\"Vilhelm Carlberg\">Vilhelm Carlberg</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/22px-Flag_of_Sweden.svg.png\" width=\"22\" height=\"14\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/33px-Flag_of_Sweden.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/44px-Flag_of_Sweden.svg.png 2x\" data-file-width=\"1600\" data-file-height=\"1000\" /> <a href=\"/wiki/Sweden_at_the_Olympics\" title=\"Sweden at the Olympics\">Sweden</a></td>\\n<td><a href=\"/wiki/Shooting_at_the_Summer_Olympics\" title=\"Shooting at the Summer Olympics\">Shooting</a></td>\\n<td>1908\\xe2\\x80\\x931924</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>4</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Fraser, Dawn</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Dawn_Fraser\" title=\"Dawn Fraser\">Dawn Fraser</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/22px-Flag_of_Australia.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/33px-Flag_of_Australia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/44px-Flag_of_Australia.svg.png 2x\" data-file-width=\"1280\" data-file-height=\"640\" /> <a href=\"/wiki/Australia_at_the_Olympics\" title=\"Australia at the Olympics\">Australia</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1956\\xe2\\x80\\x931964</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>4</td>\\n<td>4</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Ender, Kornelia</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Kornelia_Ender\" title=\"Kornelia Ender\">Kornelia Ender</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/22px-Flag_of_East_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/33px-Flag_of_East_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/44px-Flag_of_East_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/East_Germany_at_the_Olympics\" title=\"East Germany at the Olympics\">East Germany</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1972\\xe2\\x80\\x931976</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>4</td>\\n<td>4</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td rowspan=\"3\">67</td>\\n<td align=\"left\"><span class=\"sortkey\">Olsen, Otto</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Otto_Olsen_(sport_shooter)\" title=\"Otto Olsen (sport shooter)\">Otto Olsen</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">Norway</a></td>\\n<td><a href=\"/wiki/Shooting_at_the_Summer_Olympics\" title=\"Shooting at the Summer Olympics\">Shooting</a></td>\\n<td>1920\\xe2\\x80\\x931924</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Miez, Georges</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Georges_Miez\" title=\"Georges Miez\">Georges Miez</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/f3/Flag_of_Switzerland.svg/20px-Flag_of_Switzerland.svg.png\" width=\"20\" height=\"20\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/f3/Flag_of_Switzerland.svg/30px-Flag_of_Switzerland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/f/f3/Flag_of_Switzerland.svg/40px-Flag_of_Switzerland.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"1000\" /> <a href=\"/wiki/Switzerland_at_the_Olympics\" title=\"Switzerland at the Olympics\">Switzerland</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1924\\xe2\\x80\\x931936</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Gross, Rico</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ricco_Gro%C3%9F\" title=\"Ricco Gro\\xc3\\x9f\">Ricco Gro\\xc3\\x9f</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">Germany</a></td>\\n<td><a href=\"/wiki/Biathlon_at_the_Winter_Olympics\" title=\"Biathlon at the Winter Olympics\">Biathlon</a></td>\\n<td>1992\\xe2\\x80\\x932006</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>4</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"8\">70</td>\\n<td align=\"left\"><span class=\"sortkey\">Matthes, Roland</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Roland_Matthes\" title=\"Roland Matthes\">Roland Matthes</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/22px-Flag_of_East_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/33px-Flag_of_East_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/44px-Flag_of_East_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/East_Germany_at_the_Olympics\" title=\"East Germany at the Olympics\">East Germany</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1968\\xe2\\x80\\x931976</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Schmitt, Allison</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Allison_Schmitt\" title=\"Allison Schmitt\">Allison Schmitt</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2008-2016</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>4</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Kulakova, Galina</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Galina_Kulakova\" title=\"Galina Kulakova\">Galina Kulakova</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1972\\xe2\\x80\\x931980</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>4</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Aamodt, Kjetil Andre</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Kjetil_Andr%C3%A9_Aamodt\" title=\"Kjetil Andr\\xc3\\xa9 Aamodt\">Kjetil Andr\\xc3\\xa9 Aamodt</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">Norway</a></td>\\n<td><a href=\"/wiki/Alpine_skiing_at_the_Winter_Olympics\" title=\"Alpine skiing at the Winter Olympics\">Alpine skiing</a></td>\\n<td>1992\\xe2\\x80\\x932006</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>4</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Fischer, Sven</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Sven_Fischer\" title=\"Sven Fischer\">Sven Fischer</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">Germany</a></td>\\n<td><a href=\"/wiki/Biathlon_at_the_Winter_Olympics\" title=\"Biathlon at the Winter Olympics\">Biathlon</a></td>\\n<td>1994\\xe2\\x80\\x932006</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>4</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Bruijn, Inge</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Inge_de_Bruijn\" title=\"Inge de Bruijn\">Inge de Bruijn</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/22px-Flag_of_the_Netherlands.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/33px-Flag_of_the_Netherlands.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/44px-Flag_of_the_Netherlands.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Netherlands_at_the_Olympics\" title=\"Netherlands at the Olympics\">Netherlands</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2000\\xe2\\x80\\x932004</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>4</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Lezak, Jason</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Jason_Lezak\" title=\"Jason Lezak\">Jason Lezak</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2000\\xe2\\x80\\x932012</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Kramer, Sven</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Sven_Kramer\" title=\"Sven Kramer\">Sven Kramer</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/22px-Flag_of_the_Netherlands.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/33px-Flag_of_the_Netherlands.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/44px-Flag_of_the_Netherlands.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Netherlands_at_the_Olympics\" title=\"Netherlands at the Olympics\">Netherlands</a></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_Winter_Olympics\" title=\"Speed skating at the Winter Olympics\">Speed skating</a></td>\\n<td>2006\\xe2\\x80\\x932018</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>4</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>77</td>\\n<td align=\"left\"><span class=\"sortkey\">Trillini, Giovanna</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Giovanna_Trillini\" title=\"Giovanna Trillini\">Giovanna Trillini</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/22px-Flag_of_Italy.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/33px-Flag_of_Italy.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/44px-Flag_of_Italy.svg.png 2x\" data-file-width=\"1500\" data-file-height=\"1000\" /> <a href=\"/wiki/Italy_at_the_Olympics\" title=\"Italy at the Olympics\">Italy</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_Summer_Olympics\" title=\"Fencing at the Summer Olympics\">Fencing</a></td>\\n<td>1992\\xe2\\x80\\x932008</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>4</td>\\n<td>1</td>\\n<td>3</td>\\n</tr>\\n<tr>\\n<td>78</td>\\n<td align=\"left\"><span class=\"sortkey\">Kovacs, Katalin</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Katalin_Kov%C3%A1cs\" title=\"Katalin Kov\\xc3\\xa1cs\">Katalin Kov\\xc3\\xa1cs</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/22px-Flag_of_Hungary.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/33px-Flag_of_Hungary.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/44px-Flag_of_Hungary.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Hungary_at_the_Olympics\" title=\"Hungary at the Olympics\">Hungary</a></td>\\n<td><a href=\"/wiki/Canoeing_at_the_Summer_Olympics\" title=\"Canoeing at the Summer Olympics\">Canoeing</a></td>\\n<td>2000\\xe2\\x80\\x932012</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>3</td>\\n<td>5</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td rowspan=\"5\">79</td>\\n<td align=\"left\"><span class=\"sortkey\">Ducret, Roger</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Roger_Ducret\" title=\"Roger Ducret\">Roger Ducret</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/22px-Flag_of_France.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/33px-Flag_of_France.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/44px-Flag_of_France.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/France_at_the_Olympics\" title=\"France at the Olympics\">France</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_Summer_Olympics\" title=\"Fencing at the Summer Olympics\">Fencing</a></td>\\n<td>1920\\xe2\\x80\\x931928</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>3</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Cattiau, Philippe</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Philippe_Cattiau\" title=\"Philippe Cattiau\">Philippe Cattiau</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/22px-Flag_of_France.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/33px-Flag_of_France.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/44px-Flag_of_France.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/France_at_the_Olympics\" title=\"France at the Olympics\">France</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_Summer_Olympics\" title=\"Fencing at the Summer Olympics\">Fencing</a></td>\\n<td>1920\\xe2\\x80\\x931936</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>3</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Enke, Karin</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Karin_Enke\" title=\"Karin Enke\">Karin Enke</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/22px-Flag_of_East_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/33px-Flag_of_East_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/44px-Flag_of_East_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/East_Germany_at_the_Olympics\" title=\"East Germany at the Olympics\">East Germany</a></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_Winter_Olympics\" title=\"Speed skating at the Winter Olympics\">Speed skating</a></td>\\n<td>1980\\xe2\\x80\\x931988</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>3</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Niemann-Stirnemann, Gunda</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Gunda_Niemann-Stirnemann\" title=\"Gunda Niemann-Stirnemann\">Gunda Niemann-Stirnemann</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">Germany</a></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_Winter_Olympics\" title=\"Speed skating at the Winter Olympics\">Speed skating</a></td>\\n<td>1992\\xe2\\x80\\x931998</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>3</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Thomas, Petria</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Petria_Thomas\" title=\"Petria Thomas\">Petria Thomas</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/22px-Flag_of_Australia.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/33px-Flag_of_Australia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/44px-Flag_of_Australia.svg.png 2x\" data-file-width=\"1280\" data-file-height=\"640\" /> <a href=\"/wiki/Australia_at_the_Olympics\" title=\"Australia at the Olympics\">Australia</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1996\\xe2\\x80\\x932004</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>3</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>84</td>\\n<td align=\"left\"><span class=\"sortkey\">Campbell-Brown, Veronica</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Veronica_Campbell-Brown\" title=\"Veronica Campbell-Brown\">Veronica Campbell-Brown</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/22px-Flag_of_Jamaica.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/33px-Flag_of_Jamaica.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/44px-Flag_of_Jamaica.svg.png 2x\" data-file-width=\"600\" data-file-height=\"300\" /> <a href=\"/wiki/Jamaica_at_the_Olympics\" title=\"Jamaica at the Olympics\">Jamaica</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>2000-2016</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>3</td>\\n<td>3</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>85</td>\\n<td align=\"left\"><span class=\"sortkey\">Babashoff, Shirley</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Shirley_Babashoff\" title=\"Shirley Babashoff\">Shirley Babashoff</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1972\\xe2\\x80\\x931976</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>6</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td>86</td>\\n<td align=\"left\"><span class=\"sortkey\">Oneill, Susie</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Susie_O%27Neill\" title=\"Susie O\\'Neill\">Susie O\\'Neill</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/22px-Flag_of_Australia.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/33px-Flag_of_Australia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/44px-Flag_of_Australia.svg.png 2x\" data-file-width=\"1280\" data-file-height=\"640\" /> <a href=\"/wiki/Australia_at_the_Olympics\" title=\"Australia at the Olympics\">Australia</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1992\\xe2\\x80\\x932000</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>4</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td rowspan=\"4\">87</td>\\n<td align=\"left\"><span class=\"sortkey\">Korondi, Margit</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Margit_Korondi\" title=\"Margit Korondi\">Margit Korondi</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/64/Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg/22px-Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/64/Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg/33px-Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/6/64/Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg/44px-Flag_of_Hungary_%281946-1949%2C_1956-1957%29.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Hungary_at_the_1956_Summer_Olympics\" title=\"Hungary at the 1956 Summer Olympics\">Hungary</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1952\\xe2\\x80\\x931956</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>2</td>\\n<td>4</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Muratova, Sofia</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Sofia_Muratova\" title=\"Sofia Muratova\">Sofia Muratova</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1956\\xe2\\x80\\x931960</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>2</td>\\n<td>4</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Sautin, Dmitri</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Dmitri_Sautin\" title=\"Dmitri Sautin\">Dmitri Sautin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/22px-Olympic_flag.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/33px-Olympic_flag.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/44px-Olympic_flag.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Unified_Team_at_the_Olympics\" title=\"Unified Team at the Olympics\">Unified Team</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/22px-Flag_of_Russia.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/33px-Flag_of_Russia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/44px-Flag_of_Russia.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Russia_at_the_Olympics\" title=\"Russia at the Olympics\">Russia</a></td>\\n<td><a href=\"/wiki/Diving_at_the_Summer_Olympics\" title=\"Diving at the Summer Olympics\">Diving</a></td>\\n<td>1992\\xe2\\x80\\x932008</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>2</td>\\n<td>2</td>\\n<td>4</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Ohno, Apolo</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Apolo_Ohno\" title=\"Apolo Ohno\">Apolo Ohno</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Short_track_speed_skating_at_the_Winter_Olympics\" title=\"Short track speed skating at the Winter Olympics\">Short track speed skating</a></td>\\n<td>2002\\xe2\\x80\\x932010</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>2</td>\\n<td>2</td>\\n<td>4</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">91</td>\\n<td align=\"left\"><span class=\"sortkey\">Hoy, Chris</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Chris_Hoy\" title=\"Chris Hoy\">Chris Hoy</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/22px-Flag_of_the_United_Kingdom.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/33px-Flag_of_the_United_Kingdom.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/44px-Flag_of_the_United_Kingdom.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Great_Britain_at_the_Olympics\" title=\"Great Britain at the Olympics\">Great Britain</a></td>\\n<td><a href=\"/wiki/Cycling_at_the_Summer_Olympics\" title=\"Cycling at the Summer Olympics\">Cycling</a></td>\\n<td>2000\\xe2\\x80\\x932012</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>1</td>\\n<td>0</td>\\n<td rowspan=\"40\">7</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Kenny, Jason</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Jason_Kenny\" title=\"Jason Kenny\">Jason Kenny</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/22px-Flag_of_the_United_Kingdom.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/33px-Flag_of_the_United_Kingdom.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/44px-Flag_of_the_United_Kingdom.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Great_Britain_at_the_Olympics\" title=\"Great Britain at the Olympics\">Great Britain</a></td>\\n<td><a href=\"/wiki/Cycling_at_the_Summer_Olympics\" title=\"Cycling at the Summer Olympics\">Cycling</a></td>\\n<td>2008\\xe2\\x80\\x932016</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>1</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td>93</td>\\n<td align=\"left\"><span class=\"sortkey\">Kovacs, Pal</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/P%C3%A1l_Kov%C3%A1cs\" title=\"P\\xc3\\xa1l Kov\\xc3\\xa1cs\">P\\xc3\\xa1l Kov\\xc3\\xa1cs</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/22px-Flag_of_Hungary.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/33px-Flag_of_Hungary.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/44px-Flag_of_Hungary.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Hungary_at_the_Olympics\" title=\"Hungary at the Olympics\">Hungary</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_Summer_Olympics\" title=\"Fencing at the Summer Olympics\">Fencing</a></td>\\n<td>1936\\xe2\\x80\\x931960</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>0</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">94</td>\\n<td align=\"left\"><span class=\"sortkey\">Endo, Yukio</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Yukio_Endo\" title=\"Yukio Endo\">Yukio Endo</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/22px-Flag_of_Japan.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/33px-Flag_of_Japan.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/44px-Flag_of_Japan.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Japan_at_the_Olympics\" title=\"Japan at the Olympics\">Japan</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1960\\xe2\\x80\\x931968</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>2</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Peirsol, Aaron</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Aaron_Peirsol\" title=\"Aaron Peirsol\">Aaron Peirsol</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2000\\xe2\\x80\\x932008</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>2</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td rowspan=\"7\">96</td>\\n<td align=\"left\"><span class=\"sortkey\">Lee, Willis Augustus</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Willis_Augustus_Lee\" title=\"Willis Augustus Lee\">Willis Augustus Lee</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/22px-Flag_of_the_United_States_%281912-1959%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/33px-Flag_of_the_United_States_%281912-1959%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/44px-Flag_of_the_United_States_%281912-1959%29.svg.png 2x\" data-file-width=\"1900\" data-file-height=\"1000\" /> <a href=\"/wiki/United_States_at_the_1920_Summer_Olympics\" title=\"United States at the 1920 Summer Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Shooting_at_the_Summer_Olympics\" title=\"Shooting at the Summer Olympics\">Shooting</a></td>\\n<td>1920</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>1</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Thunberg, Clas</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Clas_Thunberg\" title=\"Clas Thunberg\">Clas Thunberg</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/22px-Flag_of_Finland.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/33px-Flag_of_Finland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/44px-Flag_of_Finland.svg.png 2x\" data-file-width=\"1800\" data-file-height=\"1100\" /> <a href=\"/wiki/Finland_at_the_Olympics\" title=\"Finland at the Olympics\">Finland</a></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_Winter_Olympics\" title=\"Speed skating at the Winter Olympics\">Speed skating</a></td>\\n<td>1924\\xe2\\x80\\x931928</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>5</td>\\n<td>1</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Winkler, Hans Gunter</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Hans_G%C3%BCnter_Winkler\" title=\"Hans G\\xc3\\xbcnter Winkler\">Hans G\\xc3\\xbcnter Winkler</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/9/9a/German_Olympic_flag_%281959-1968%29.svg/22px-German_Olympic_flag_%281959-1968%29.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/9/9a/German_Olympic_flag_%281959-1968%29.svg/33px-German_Olympic_flag_%281959-1968%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/9/9a/German_Olympic_flag_%281959-1968%29.svg/44px-German_Olympic_flag_%281959-1968%29.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/United_Team_of_Germany_at_the_Olympics\" class=\"mw-redirect\" title=\"United Team of Germany at the Olympics\">United Team of Germany</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/West_Germany_at_the_Olympics\" title=\"West Germany at the Olympics\">West Germany</a></td>\\n<td><a href=\"/wiki/Equestrian_at_the_Summer_Olympics\" title=\"Equestrian at the Summer Olympics\">Equestrian</a></td>\\n<td>1956\\xe2\\x80\\x931976</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>1</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Jager, Tom</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Tom_Jager\" title=\"Tom Jager\">Tom Jager</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1984\\xe2\\x80\\x931992</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>1</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Egerszegi, Krisztina</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Krisztina_Egerszegi\" title=\"Krisztina Egerszegi\">Krisztina Egerszegi</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/22px-Flag_of_Hungary.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/33px-Flag_of_Hungary.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/44px-Flag_of_Hungary.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Hungary_at_the_Olympics\" title=\"Hungary at the Olympics\">Hungary</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1988\\xe2\\x80\\x931996</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>5</td>\\n<td>1</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Lazutina, Larisa</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Larisa_Lazutina\" title=\"Larisa Lazutina\">Larisa Lazutina</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/22px-Olympic_flag.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/33px-Olympic_flag.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/44px-Olympic_flag.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Unified_Team_at_the_Olympics\" title=\"Unified Team at the Olympics\">Unified Team</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/22px-Flag_of_Russia.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/33px-Flag_of_Russia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/44px-Flag_of_Russia.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Russia_at_the_Olympics\" title=\"Russia at the Olympics\">Russia</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1992\\xe2\\x80\\x931998</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>5</td>\\n<td>1</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Wu, Minxia</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Minxia_Wu\" class=\"mw-redirect\" title=\"Minxia Wu\">Minxia Wu</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fa/Flag_of_the_People%27s_Republic_of_China.svg/22px-Flag_of_the_People%27s_Republic_of_China.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fa/Flag_of_the_People%27s_Republic_of_China.svg/33px-Flag_of_the_People%27s_Republic_of_China.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/f/fa/Flag_of_the_People%27s_Republic_of_China.svg/44px-Flag_of_the_People%27s_Republic_of_China.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/China_at_the_Olympics\" title=\"China at the Olympics\">China</a></td>\\n<td><a href=\"/wiki/Diving_at_the_Summer_Olympics\" title=\"Diving at the Summer Olympics\">Diving</a></td>\\n<td>2004-2016</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>5</td>\\n<td>1</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">103</td>\\n<td align=\"left\"><span class=\"sortkey\">Gaudin, Lucien</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Lucien_Gaudin\" title=\"Lucien Gaudin\">Lucien Gaudin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/22px-Flag_of_France.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/33px-Flag_of_France.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/44px-Flag_of_France.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/France_at_the_Olympics\" title=\"France at the Olympics\">France</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_Summer_Olympics\" title=\"Fencing at the Summer Olympics\">Fencing</a></td>\\n<td>1920\\xe2\\x80\\x931928</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>3</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Patzaichin, Ivan</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ivan_Patzaichin\" title=\"Ivan Patzaichin\">Ivan Patzaichin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/7f/Flag_of_Romania_%281965-1989%29.svg/22px-Flag_of_Romania_%281965-1989%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/7f/Flag_of_Romania_%281965-1989%29.svg/33px-Flag_of_Romania_%281965-1989%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/7/7f/Flag_of_Romania_%281965-1989%29.svg/44px-Flag_of_Romania_%281965-1989%29.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Romania_at_the_1984_Summer_Olympics\" title=\"Romania at the 1984 Summer Olympics\">Romania</a></td>\\n<td><a href=\"/wiki/Canoeing_at_the_Summer_Olympics\" title=\"Canoeing at the Summer Olympics\">Canoeing</a></td>\\n<td>1968\\xe2\\x80\\x931984</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>3</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">105</td>\\n<td align=\"left\"><span class=\"sortkey\">Liberg, Einar</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Einar_Liberg\" title=\"Einar Liberg\">Einar Liberg</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">Norway</a></td>\\n<td><a href=\"/wiki/Shooting_at_the_Summer_Olympics\" title=\"Shooting at the Summer Olympics\">Shooting</a></td>\\n<td>1908\\xe2\\x80\\x931924</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>2</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Ballangrud, Ivar</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ivar_Ballangrud\" title=\"Ivar Ballangrud\">Ivar Ballangrud</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">Norway</a></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_Winter_Olympics\" title=\"Speed skating at the Winter Olympics\">Speed skating</a></td>\\n<td>1928\\xe2\\x80\\x931936</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>4</td>\\n<td>2</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"4\">107</td>\\n<td align=\"left\"><span class=\"sortkey\">Daniels, Charles</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Charles_Daniels_(swimmer)\" title=\"Charles Daniels (swimmer)\">Charles Daniels</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/22px-Flag_of_the_United_States_%281896-1908%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/33px-Flag_of_the_United_States_%281896-1908%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/44px-Flag_of_the_United_States_%281896-1908%29.svg.png 2x\" data-file-width=\"1024\" data-file-height=\"539\" /> <a href=\"/wiki/United_States_at_the_1908_Summer_Olympics\" title=\"United States at the 1908 Summer Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1904\\xe2\\x80\\x931908</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>1</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Spooner, Lloyd</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Lloyd_Spooner\" title=\"Lloyd Spooner\">Lloyd Spooner</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/22px-Flag_of_the_United_States_%281912-1959%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/33px-Flag_of_the_United_States_%281912-1959%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/44px-Flag_of_the_United_States_%281912-1959%29.svg.png 2x\" data-file-width=\"1900\" data-file-height=\"1000\" /> <a href=\"/wiki/United_States_at_the_1920_Summer_Olympics\" title=\"United States at the 1920 Summer Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Shooting_at_the_Summer_Olympics\" title=\"Shooting at the Summer Olympics\">Shooting</a></td>\\n<td>1920</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>1</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Kitajima, Kosuke</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Kosuke_Kitajima\" title=\"Kosuke Kitajima\">Kosuke Kitajima</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/22px-Flag_of_Japan.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/33px-Flag_of_Japan.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/44px-Flag_of_Japan.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Japan_at_the_Olympics\" title=\"Japan at the Olympics\">Japan</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2004\\xe2\\x80\\x932012</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>1</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Trickett, Libby</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Libby_Trickett\" title=\"Libby Trickett\">Libby Trickett</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/22px-Flag_of_Australia.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/33px-Flag_of_Australia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/44px-Flag_of_Australia.svg.png 2x\" data-file-width=\"1280\" data-file-height=\"640\" /> <a href=\"/wiki/Australia_at_the_Olympics\" title=\"Australia at the Olympics\">Australia</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2004\\xe2\\x80\\x932012</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>4</td>\\n<td>1</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>111</td>\\n<td><a href=\"/wiki/Charlotte_Kalla\" title=\"Charlotte Kalla\">Charlotte Kalla</a></td>\\n<td><a href=\"/wiki/Sweden\" title=\"Sweden\">Sweden</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing\" title=\"Cross-country skiing\">Cross-country skiing</a></td>\\n<td>2010-2018</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>3</td>\\n<td>4</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td rowspan=\"1\">112</td>\\n<td align=\"left\"><span class=\"sortkey\">Hakulinen, Veikko</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Veikko_Hakulinen\" title=\"Veikko Hakulinen\">Veikko Hakulinen</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/22px-Flag_of_Finland.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/33px-Flag_of_Finland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/44px-Flag_of_Finland.svg.png 2x\" data-file-width=\"1800\" data-file-height=\"1100\" /> <a href=\"/wiki/Finland_at_the_Olympics\" title=\"Finland at the Olympics\">Finland</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1952\\xe2\\x80\\x931960</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>3</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"4\">113</td>\\n<td align=\"left\"><span class=\"sortkey\">Mantyranta, Eero</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Eero_M%C3%A4ntyranta\" title=\"Eero M\\xc3\\xa4ntyranta\">Eero M\\xc3\\xa4ntyranta</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/22px-Flag_of_Finland.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/33px-Flag_of_Finland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/44px-Flag_of_Finland.svg.png 2x\" data-file-width=\"1800\" data-file-height=\"1100\" /> <a href=\"/wiki/Finland_at_the_Olympics\" title=\"Finland at the Olympics\">Finland</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1960\\xe2\\x80\\x931968</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>3</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Szewinska, Irena</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Irena_Szewi%C5%84ska\" title=\"Irena Szewi\\xc5\\x84ska\">Irena Szewi\\xc5\\x84ska</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/1/12/Flag_of_Poland.svg/22px-Flag_of_Poland.svg.png\" width=\"22\" height=\"14\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/1/12/Flag_of_Poland.svg/33px-Flag_of_Poland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/1/12/Flag_of_Poland.svg/44px-Flag_of_Poland.svg.png 2x\" data-file-width=\"1280\" data-file-height=\"800\" /> <a href=\"/wiki/Poland_at_the_Olympics\" title=\"Poland at the Olympics\">Poland</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>1964\\xe2\\x80\\x931976</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>3</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Andersson, Agneta</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Agneta_Andersson\" title=\"Agneta Andersson\">Agneta Andersson</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/22px-Flag_of_Sweden.svg.png\" width=\"22\" height=\"14\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/33px-Flag_of_Sweden.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/44px-Flag_of_Sweden.svg.png 2x\" data-file-width=\"1600\" data-file-height=\"1000\" /> <a href=\"/wiki/Sweden_at_the_Olympics\" title=\"Sweden at the Olympics\">Sweden</a></td>\\n<td><a href=\"/wiki/Canoeing_at_the_Summer_Olympics\" title=\"Canoeing at the Summer Olympics\">Canoeing</a></td>\\n<td>1984\\xe2\\x80\\x931996</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>3</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Hoogenband, Pieter</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Pieter_van_den_Hoogenband\" title=\"Pieter van den Hoogenband\">Pieter van den Hoogenband</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/22px-Flag_of_the_Netherlands.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/33px-Flag_of_the_Netherlands.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/44px-Flag_of_the_Netherlands.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Netherlands_at_the_Olympics\" title=\"Netherlands at the Olympics\">Netherlands</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2000\\xe2\\x80\\x932004</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>3</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td rowspan=\"3\">118</td>\\n<td align=\"left\"><span class=\"sortkey\">Strickland, Shirley</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Shirley_Strickland\" title=\"Shirley Strickland\">Shirley Strickland</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/22px-Flag_of_Australia.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/33px-Flag_of_Australia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/44px-Flag_of_Australia.svg.png 2x\" data-file-width=\"1280\" data-file-height=\"640\" /> <a href=\"/wiki/Australia_at_the_Olympics\" title=\"Australia at the Olympics\">Australia</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>1948\\xe2\\x80\\x931956</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>3</td>\\n<td>1</td>\\n<td>3</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Amanar, Simona</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Simona_Am%C3%A2nar\" title=\"Simona Am\\xc3\\xa2nar\">Simona Am\\xc3\\xa2nar</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/73/Flag_of_Romania.svg/22px-Flag_of_Romania.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/73/Flag_of_Romania.svg/33px-Flag_of_Romania.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/7/73/Flag_of_Romania.svg/44px-Flag_of_Romania.svg.png 2x\" data-file-width=\"600\" data-file-height=\"400\" /> <a href=\"/wiki/Romania_at_the_Olympics\" title=\"Romania at the Olympics\">Romania</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1996-2000</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>3</td>\\n<td>1</td>\\n<td>3</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Gottwald, Felix</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Felix_Gottwald\" title=\"Felix Gottwald\">Felix Gottwald</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/41/Flag_of_Austria.svg/22px-Flag_of_Austria.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/41/Flag_of_Austria.svg/33px-Flag_of_Austria.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/4/41/Flag_of_Austria.svg/44px-Flag_of_Austria.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Austria_at_the_Olympics\" title=\"Austria at the Olympics\">Austria</a></td>\\n<td><a href=\"/wiki/Nordic_combined_at_the_Winter_Olympics\" title=\"Nordic combined at the Winter Olympics\">Nordic combined</a></td>\\n<td>2002\\xe2\\x80\\x932010</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>3</td>\\n<td>1</td>\\n<td>3</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">120</td>\\n<td align=\"left\"><span class=\"sortkey\">Kirvesniemi, Marja-Liisa</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Marja-Liisa_Kirvesniemi\" title=\"Marja-Liisa Kirvesniemi\">Marja-Liisa Kirvesniemi</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/22px-Flag_of_Finland.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/33px-Flag_of_Finland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/44px-Flag_of_Finland.svg.png 2x\" data-file-width=\"1800\" data-file-height=\"1100\" /> <a href=\"/wiki/Finland_at_the_Olympics\" title=\"Finland at the Olympics\">Finland</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1984\\xe2\\x80\\x931994</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>3</td>\\n<td>0</td>\\n<td>4</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Valbe, Yelena</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Yelena_V%C3%A4lbe\" title=\"Yelena V\\xc3\\xa4lbe\">Yelena V\\xc3\\xa4lbe</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/22px-Olympic_flag.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/33px-Olympic_flag.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/44px-Olympic_flag.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Unified_Team_at_the_Olympics\" title=\"Unified Team at the Olympics\">Unified Team</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/22px-Flag_of_Russia.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/33px-Flag_of_Russia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/44px-Flag_of_Russia.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Russia_at_the_Olympics\" title=\"Russia at the Olympics\">Russia</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1992\\xe2\\x80\\x931998</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>3</td>\\n<td>0</td>\\n<td>4</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">122</td>\\n<td align=\"left\"><span class=\"sortkey\">Marzi, Gustavo</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Gustavo_Marzi\" title=\"Gustavo Marzi\">Gustavo Marzi</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/ad/Flag_of_Italy_%281861-1946%29.svg/22px-Flag_of_Italy_%281861-1946%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/ad/Flag_of_Italy_%281861-1946%29.svg/33px-Flag_of_Italy_%281861-1946%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/ad/Flag_of_Italy_%281861-1946%29.svg/44px-Flag_of_Italy_%281861-1946%29.svg.png 2x\" data-file-width=\"1500\" data-file-height=\"1000\" /> <a href=\"/wiki/Italy_at_the_1936_Summer_Olympics\" title=\"Italy at the 1936 Summer Olympics\">Italy</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_Summer_Olympics\" title=\"Fencing at the Summer Olympics\">Fencing</a></td>\\n<td>1928\\xe2\\x80\\x931936</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>2</td>\\n<td>5</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Gorokhovskaya, Maria</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Maria_Gorokhovskaya\" title=\"Maria Gorokhovskaya\">Maria Gorokhovskaya</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d3/Flag_of_the_Soviet_Union_%281936-1955%29.svg/22px-Flag_of_the_Soviet_Union_%281936-1955%29.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d3/Flag_of_the_Soviet_Union_%281936-1955%29.svg/33px-Flag_of_the_Soviet_Union_%281936-1955%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d3/Flag_of_the_Soviet_Union_%281936-1955%29.svg/44px-Flag_of_the_Soviet_Union_%281936-1955%29.svg.png 2x\" data-file-width=\"600\" data-file-height=\"300\" /> <a href=\"/wiki/Soviet_Union_at_the_1952_Summer_Olympics\" title=\"Soviet Union at the 1952 Summer Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1952</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>5</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td rowspan=\"3\">124</td>\\n<td align=\"left\"><span class=\"sortkey\">Beard, Amanda</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Amanda_Beard\" title=\"Amanda Beard\">Amanda Beard</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1996\\xe2\\x80\\x932004</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Khorkina, Svetlana</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Svetlana_Khorkina\" title=\"Svetlana Khorkina\">Svetlana Khorkina</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/22px-Flag_of_Russia.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/33px-Flag_of_Russia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/44px-Flag_of_Russia.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Russia_at_the_Olympics\" title=\"Russia at the Olympics\">Russia</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1996\\xe2\\x80\\x932004</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Coventry, Kirsty</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Kirsty_Coventry\" title=\"Kirsty Coventry\">Kirsty Coventry</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/6a/Flag_of_Zimbabwe.svg/22px-Flag_of_Zimbabwe.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/6a/Flag_of_Zimbabwe.svg/33px-Flag_of_Zimbabwe.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/6/6a/Flag_of_Zimbabwe.svg/44px-Flag_of_Zimbabwe.svg.png 2x\" data-file-width=\"840\" data-file-height=\"420\" /> <a href=\"/wiki/Zimbabwe_at_the_Olympics\" title=\"Zimbabwe at the Olympics\">Zimbabwe</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2004\\xe2\\x80\\x932008</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">127</td>\\n<td align=\"left\"><span class=\"sortkey\">Ujlaki-Rejto, Ildiko</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ildik%C3%B3_%C3%9Ajlaky-Rejt%C5%91\" title=\"Ildik\\xc3\\xb3 \\xc3\\x9ajlaky-Rejt\\xc5\\x91\">Ildik\\xc3\\xb3 \\xc3\\x9ajlaky-Rejt\\xc5\\x91</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/22px-Flag_of_Hungary.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/33px-Flag_of_Hungary.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/44px-Flag_of_Hungary.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Hungary_at_the_Olympics\" title=\"Hungary at the Olympics\">Hungary</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_Summer_Olympics\" title=\"Fencing at the Summer Olympics\">Fencing</a></td>\\n<td>1960\\xe2\\x80\\x931976</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>3</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Janz, Karin</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Karin_B%C3%BCttner-Janz\" title=\"Karin B\\xc3\\xbcttner-Janz\">Karin Janz</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/22px-Flag_of_East_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/33px-Flag_of_East_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/44px-Flag_of_East_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/East_Germany_at_the_Olympics\" title=\"East Germany at the Olympics\">East Germany</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1968\\xe2\\x80\\x931972</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>3</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td rowspan=\"3\">129</td>\\n<td align=\"left\"><span class=\"sortkey\">Miller, Shannon</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Shannon_Miller\" title=\"Shannon Miller\">Shannon Miller</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1992\\xe2\\x80\\x931996</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>2</td>\\n<td>3</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Di Centa, Manuela</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Manuela_Di_Centa\" title=\"Manuela Di Centa\">Manuela Di Centa</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/22px-Flag_of_Italy.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/33px-Flag_of_Italy.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/44px-Flag_of_Italy.svg.png 2x\" data-file-width=\"1500\" data-file-height=\"1000\" /> <a href=\"/wiki/Italy_at_the_Olympics\" title=\"Italy at the Olympics\">Italy</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1992\\xe2\\x80\\x931998</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>2</td>\\n<td>2</td>\\n<td>3</td>\\n</tr>\\n</table>\\n<h3><span class=\"mw-headline\" id=\"Timeline\">Timeline</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=2\" title=\"Edit section: Timeline\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h3>\\n<p>This is a progressive list of Olympians that have held the record for most medals won. Medals won in the <a href=\"/wiki/1906_Intercalated_Games\" title=\"1906 Intercalated Games\">1906 Intercalated Games</a> are not included. It includes top-three placings in 1896 and 1900, before medals were awarded for top-three placings. All record-holders have competed at Summer Games rather than Winter Games.</p>\\n<table class=\"wikitable\">\\n<tr>\\n<th>Medal count</th>\\n<th>Date</th>\\n<th>Athlete</th>\\n<th>Nation</th>\\n<th>Sport</th>\\n<th>Record medal event</th>\\n<th>Earlier medal events</th>\\n</tr>\\n<tr>\\n<th rowspan=\"6\">1</th>\\n<td rowspan=\"6\">6 April 1896<sup id=\"cite_ref-ioc1896_1-0\" class=\"reference\"><a href=\"#cite_note-ioc1896-1\">[1]</a></sup></td>\\n<td align=\"left\"><a href=\"/wiki/James_Brendan_Connolly\" title=\"James Brendan Connolly\">James Connolly</a></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/22px-Flag_of_the_United_States_%281891-1896%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/33px-Flag_of_the_United_States_%281891-1896%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/44px-Flag_of_the_United_States_%281891-1896%29.svg.png 2x\" data-file-width=\"1024\" data-file-height=\"539\" /> <a href=\"/wiki/United_States_at_the_1896_Summer_Olympics\" title=\"United States at the 1896 Summer Olympics\">United States</a></td>\\n<td rowspan=\"3\"><a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics\" title=\"Athletics at the 1896 Summer Olympics\">Athletics</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_triple_jump\" title=\"Athletics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s triple jump\">Triple jump G</a></td>\\n<td rowspan=\"6\">\\xe2\\x80\\x93</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><a href=\"/wiki/Alexandre_Tuff%C3%A8re\" title=\"Alexandre Tuff\\xc3\\xa8re\">Alexandre Tuff\\xc3\\xa8re</a></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/22px-Flag_of_France.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/33px-Flag_of_France.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/c/c3/Flag_of_France.svg/44px-Flag_of_France.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/France_at_the_1896_Summer_Olympics\" title=\"France at the 1896 Summer Olympics\">France</a></td>\\n<td>Triple jump S</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><a href=\"/wiki/Ioannis_Persakis\" title=\"Ioannis Persakis\">Ioannis Persakis</a></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/6e/Flag_of_Greece_%281822-1978%29.svg/22px-Flag_of_Greece_%281822-1978%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/6e/Flag_of_Greece_%281822-1978%29.svg/33px-Flag_of_Greece_%281822-1978%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/6/6e/Flag_of_Greece_%281822-1978%29.svg/44px-Flag_of_Greece_%281822-1978%29.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"800\" /> <a href=\"/wiki/Greece_at_the_1896_Summer_Olympics\" title=\"Greece at the 1896 Summer Olympics\">Greece</a></td>\\n<td>Triple jump B</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><a href=\"/wiki/Robert_Garrett\" title=\"Robert Garrett\">Robert Garrett</a></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/22px-Flag_of_the_United_States_%281891-1896%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/33px-Flag_of_the_United_States_%281891-1896%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/44px-Flag_of_the_United_States_%281891-1896%29.svg.png 2x\" data-file-width=\"1024\" data-file-height=\"539\" /> <a href=\"/wiki/United_States_at_the_1896_Summer_Olympics\" title=\"United States at the 1896 Summer Olympics\">United States</a></td>\\n<td rowspan=\"3\"><a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics\" title=\"Athletics at the 1896 Summer Olympics\">Athletics</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_discus_throw\" title=\"Athletics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s discus throw\">Discus G</a></td>\\n</tr>\\n<tr>\\n<td align=\"left\"><a href=\"/wiki/Panagiotis_Paraskevopoulos\" title=\"Panagiotis Paraskevopoulos\">Panagiotis Paraskevopoulos</a></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/6e/Flag_of_Greece_%281822-1978%29.svg/22px-Flag_of_Greece_%281822-1978%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/6e/Flag_of_Greece_%281822-1978%29.svg/33px-Flag_of_Greece_%281822-1978%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/6/6e/Flag_of_Greece_%281822-1978%29.svg/44px-Flag_of_Greece_%281822-1978%29.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"800\" /> <a href=\"/wiki/Greece_at_the_1896_Summer_Olympics\" title=\"Greece at the 1896 Summer Olympics\">Greece</a></td>\\n<td>Discus S</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><a href=\"/wiki/Sotirios_Versis\" title=\"Sotirios Versis\">Sotirios Versis</a></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/6e/Flag_of_Greece_%281822-1978%29.svg/22px-Flag_of_Greece_%281822-1978%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/6e/Flag_of_Greece_%281822-1978%29.svg/33px-Flag_of_Greece_%281822-1978%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/6/6e/Flag_of_Greece_%281822-1978%29.svg/44px-Flag_of_Greece_%281822-1978%29.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"800\" /> <a href=\"/wiki/Greece_at_the_1896_Summer_Olympics\" title=\"Greece at the 1896 Summer Olympics\">Greece</a></td>\\n<td>Discus B</td>\\n</tr>\\n<tr>\\n<th rowspan=\"2\">2</th>\\n<td rowspan=\"3\">7 April 1896<sup id=\"cite_ref-ioc1896_1-1\" class=\"reference\"><a href=\"#cite_note-ioc1896-1\">[1]</a></sup></td>\\n<td align=\"left\">Robert Garrett</td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/22px-Flag_of_the_United_States_%281891-1896%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/33px-Flag_of_the_United_States_%281891-1896%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/44px-Flag_of_the_United_States_%281891-1896%29.svg.png 2x\" data-file-width=\"1024\" data-file-height=\"539\" /> <a href=\"/wiki/United_States_at_the_1896_Summer_Olympics\" title=\"United States at the 1896 Summer Olympics\">United States</a></td>\\n<td rowspan=\"2\"><a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics\" title=\"Athletics at the 1896 Summer Olympics\">Athletics</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_long_jump\" title=\"Athletics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s long jump\">Long jump S</a></td>\\n<td rowspan=\"2\" align=\"left\"><i>See above</i></td>\\n</tr>\\n<tr>\\n<td align=\"left\">James Connolly</td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/22px-Flag_of_the_United_States_%281891-1896%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/33px-Flag_of_the_United_States_%281891-1896%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/44px-Flag_of_the_United_States_%281891-1896%29.svg.png 2x\" data-file-width=\"1024\" data-file-height=\"539\" /> <a href=\"/wiki/United_States_at_the_1896_Summer_Olympics\" title=\"United States at the 1896 Summer Olympics\">United States</a></td>\\n<td>Long jump B</td>\\n</tr>\\n<tr>\\n<th rowspan=\"3\">3</th>\\n<td align=\"left\">Robert Garrett</td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/22px-Flag_of_the_United_States_%281891-1896%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/33px-Flag_of_the_United_States_%281891-1896%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c8/Flag_of_the_United_States_%281891-1896%29.svg/44px-Flag_of_the_United_States_%281891-1896%29.svg.png 2x\" data-file-width=\"1024\" data-file-height=\"539\" /> <a href=\"/wiki/United_States_at_the_1896_Summer_Olympics\" title=\"United States at the 1896 Summer Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics\" title=\"Athletics at the 1896 Summer Olympics\">Athletics</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_shot_put\" title=\"Athletics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s shot put\">Shot put G</a></td>\\n<td align=\"left\"><a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics\" title=\"Athletics at the 1896 Summer Olympics\">1896</a> \\xe2\\x80\\x93 <a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_discus_throw\" title=\"Athletics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s discus throw\">1 G</a>, <a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_long_jump\" title=\"Athletics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s long jump\">2 S</a></td>\\n</tr>\\n<tr>\\n<td rowspan=\"5\">9 April 1896<sup id=\"cite_ref-ioc1896_1-2\" class=\"reference\"><a href=\"#cite_note-ioc1896-1\">[1]</a></sup></td>\\n<td align=\"left\"><a href=\"/wiki/Carl_Schuhmann\" title=\"Carl Schuhmann\">Carl Schuhmann</a></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/e/ec/Flag_of_the_German_Empire.svg/22px-Flag_of_the_German_Empire.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/e/ec/Flag_of_the_German_Empire.svg/33px-Flag_of_the_German_Empire.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/e/ec/Flag_of_the_German_Empire.svg/44px-Flag_of_the_German_Empire.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"800\" /> <a href=\"/wiki/Germany_at_the_1896_Summer_Olympics\" title=\"Germany at the 1896 Summer Olympics\">Germany</a></td>\\n<td rowspan=\"2\"><a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics\" title=\"Gymnastics at the 1896 Summer Olympics\">Gymnastics</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_vault\" title=\"Gymnastics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s vault\">Vault G</a></td>\\n<td align=\"left\" valign=\"top\"><a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics\" title=\"Gymnastics at the 1896 Summer Olympics\">1896</a> \\xe2\\x80\\x93 <a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_team_parallel_bars\" title=\"Gymnastics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s team parallel bars\">1 G</a>, <a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_team_horizontal_bar\" title=\"Gymnastics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s team horizontal bar\">2 G</a></td>\\n</tr>\\n<tr>\\n<td rowspan=\"4\" align=\"left\"><a href=\"/wiki/Hermann_Weing%C3%A4rtner\" title=\"Hermann Weing\\xc3\\xa4rtner\">Hermann Weing\\xc3\\xa4rtner</a></td>\\n<td rowspan=\"4\" align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/e/ec/Flag_of_the_German_Empire.svg/22px-Flag_of_the_German_Empire.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/e/ec/Flag_of_the_German_Empire.svg/33px-Flag_of_the_German_Empire.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/e/ec/Flag_of_the_German_Empire.svg/44px-Flag_of_the_German_Empire.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"800\" /> <a href=\"/wiki/Germany_at_the_1896_Summer_Olympics\" title=\"Germany at the 1896 Summer Olympics\">Germany</a></td>\\n<td>Vault B</td>\\n<td rowspan=\"4\" align=\"left\" valign=\"top\"><a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics\" title=\"Gymnastics at the 1896 Summer Olympics\">1896</a> \\xe2\\x80\\x93 <a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_team_parallel_bars\" title=\"Gymnastics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s team parallel bars\">1 G</a>, <a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_team_horizontal_bar\" title=\"Gymnastics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s team horizontal bar\">2 G</a></td>\\n</tr>\\n<tr>\\n<th>4</th>\\n<td rowspan=\"3\"><a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics\" title=\"Gymnastics at the 1896 Summer Olympics\">Gymnastics</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_pommel_horse\" title=\"Gymnastics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s pommel horse\">Pommel horse S</a></td>\\n</tr>\\n<tr>\\n<th>5</th>\\n<td><a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_rings\" title=\"Gymnastics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s rings\">Rings S</a></td>\\n</tr>\\n<tr>\\n<th rowspan=\"3\">6</th>\\n<td><a href=\"/wiki/Gymnastics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_horizontal_bar\" title=\"Gymnastics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s horizontal bar\">Horizontal bar G</a></td>\\n</tr>\\n<tr>\\n<td>16 July 1900</td>\\n<td align=\"left\">Robert Garrett</td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/22px-Flag_of_the_United_States_%281896-1908%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/33px-Flag_of_the_United_States_%281896-1908%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/44px-Flag_of_the_United_States_%281896-1908%29.svg.png 2x\" data-file-width=\"1024\" data-file-height=\"539\" /> <a href=\"/wiki/United_States_at_the_1900_Summer_Olympics\" title=\"United States at the 1900 Summer Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_1900_Summer_Olympics\" title=\"Athletics at the 1900 Summer Olympics\">Athletics</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_1900_Summer_Olympics_%E2%80%93_Men%27s_standing_triple_jump\" title=\"Athletics at the 1900 Summer Olympics \\xe2\\x80\\x93 Men\\'s standing triple jump\">Standing triple jump B</a></td>\\n<td align=\"left\" valign=\"top\"><a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics\" title=\"Athletics at the 1896 Summer Olympics\">1896</a> \\xe2\\x80\\x93 <a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_discus_throw\" title=\"Athletics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s discus throw\">1 G</a>, <a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_long_jump\" title=\"Athletics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s long jump\">2 S</a>, <a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_shot_put\" title=\"Athletics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s shot put\">3 G</a>, <a href=\"/wiki/Athletics_at_the_1896_Summer_Olympics_%E2%80%93_Men%27s_high_jump\" title=\"Athletics at the 1896 Summer Olympics \\xe2\\x80\\x93 Men\\'s high jump\">4 S</a><br />\\n<a href=\"/wiki/Athletics_at_the_1900_Summer_Olympics\" title=\"Athletics at the 1900 Summer Olympics\">1900</a> \\xe2\\x80\\x93 <a href=\"/wiki/Athletics_at_the_1900_Summer_Olympics_%E2%80%93_Men%27s_shot_put\" title=\"Athletics at the 1900 Summer Olympics \\xe2\\x80\\x93 Men\\'s shot put\">5 B</a></td>\\n</tr>\\n<tr>\\n<td>3 September 1904</td>\\n<td rowspan=\"3\" align=\"left\"><a href=\"/wiki/Ray_Ewry\" title=\"Ray Ewry\">Ray Ewry</a></td>\\n<td rowspan=\"3\" align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/22px-Flag_of_the_United_States_%281896-1908%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/33px-Flag_of_the_United_States_%281896-1908%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/44px-Flag_of_the_United_States_%281896-1908%29.svg.png 2x\" data-file-width=\"1024\" data-file-height=\"539\" /> <a href=\"/wiki/United_States_at_the_1904_Summer_Olympics\" title=\"United States at the 1904 Summer Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_1904_Summer_Olympics\" title=\"Athletics at the 1904 Summer Olympics\">Athletics</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_1904_Summer_Olympics_%E2%80%93_Men%27s_standing_triple_jump\" title=\"Athletics at the 1904 Summer Olympics \\xe2\\x80\\x93 Men\\'s standing triple jump\">Standing triple jump G</a></td>\\n<td rowspan=\"3\" align=\"left\" valign=\"top\"><a href=\"/wiki/Athletics_at_the_1900_Summer_Olympics\" title=\"Athletics at the 1900 Summer Olympics\">1900</a> \\xe2\\x80\\x93 <a href=\"/wiki/Athletics_at_the_1900_Summer_Olympics_%E2%80%93_Men%27s_standing_high_jump\" title=\"Athletics at the 1900 Summer Olympics \\xe2\\x80\\x93 Men\\'s standing high jump\">1 G</a>, <a href=\"/wiki/Athletics_at_the_1900_Summer_Olympics_%E2%80%93_Men%27s_standing_long_jump\" title=\"Athletics at the 1900 Summer Olympics \\xe2\\x80\\x93 Men\\'s standing long jump\">2 G</a>, <a href=\"/wiki/Athletics_at_the_1900_Summer_Olympics_%E2%80%93_Men%27s_standing_triple_jump\" title=\"Athletics at the 1900 Summer Olympics \\xe2\\x80\\x93 Men\\'s standing triple jump\">3 G</a><br />\\n<a href=\"/wiki/Athletics_at_the_1904_Summer_Olympics\" title=\"Athletics at the 1904 Summer Olympics\">1904</a> \\xe2\\x80\\x93 <a href=\"/wiki/Athletics_at_the_1904_Summer_Olympics_%E2%80%93_Men%27s_standing_high_jump\" title=\"Athletics at the 1904 Summer Olympics \\xe2\\x80\\x93 Men\\'s standing high jump\">4 G</a>, <a href=\"/wiki/Athletics_at_the_1904_Summer_Olympics_%E2%80%93_Men%27s_standing_long_jump\" title=\"Athletics at the 1904 Summer Olympics \\xe2\\x80\\x93 Men\\'s standing long jump\">5 G</a></td>\\n</tr>\\n<tr>\\n<th>7</th>\\n<td>20 July 1908</td>\\n<td rowspan=\"2\"><a href=\"/wiki/Athletics_at_the_1908_Summer_Olympics\" title=\"Athletics at the 1908 Summer Olympics\">Athletics</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_1908_Summer_Olympics_%E2%80%93_Men%27s_standing_long_jump\" title=\"Athletics at the 1908 Summer Olympics \\xe2\\x80\\x93 Men\\'s standing long jump\">Standing long jump G</a></td>\\n</tr>\\n<tr>\\n<th rowspan=\"2\">8</th>\\n<td>23 July 1908</td>\\n<td><a href=\"/wiki/Athletics_at_the_1908_Summer_Olympics_%E2%80%93_Men%27s_standing_high_jump\" title=\"Athletics at the 1908 Summer Olympics \\xe2\\x80\\x93 Men\\'s standing high jump\">Standing high jump G</a></td>\\n</tr>\\n<tr>\\n<td>29 July 1920</td>\\n<td rowspan=\"4\" align=\"left\"><a href=\"/wiki/Carl_Osburn\" title=\"Carl Osburn\">Carl Osburn</a></td>\\n<td rowspan=\"4\" align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/22px-Flag_of_the_United_States_%281912-1959%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/33px-Flag_of_the_United_States_%281912-1959%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/f/f5/Flag_of_the_United_States_%281912-1959%29.svg/44px-Flag_of_the_United_States_%281912-1959%29.svg.png 2x\" data-file-width=\"1900\" data-file-height=\"1000\" /> <a href=\"/wiki/United_States_at_the_1920_Summer_Olympics\" title=\"United States at the 1920 Summer Olympics\">United States</a></td>\\n<td rowspan=\"3\"><a href=\"/wiki/Shooting_at_the_1920_Summer_Olympics\" title=\"Shooting at the 1920 Summer Olympics\">Shooting</a></td>\\n<td><a href=\"/wiki/Shooting_at_the_1920_Summer_Olympics_%E2%80%93_Men%27s_300_and_600_metre_team_military_rifle,_prone\" title=\"Shooting at the 1920 Summer Olympics \\xe2\\x80\\x93 Men\\'s 300 and 600 metre team military rifle, prone\">Team 300 m / 600 m military rifle, prone G</a></td>\\n<td rowspan=\"4\" align=\"left\" valign=\"top\"><a href=\"/wiki/Shooting_at_the_1912_Summer_Olympics\" title=\"Shooting at the 1912 Summer Olympics\">1912</a> \\xe2\\x80\\x93 <a href=\"/wiki/Shooting_at_the_1912_Summer_Olympics_%E2%80%93_Men%27s_50_metre_team_small-bore_rifle\" title=\"Shooting at the 1912 Summer Olympics \\xe2\\x80\\x93 Men\\'s 50 metre team small-bore rifle\">1 B</a>, <a href=\"/wiki/Shooting_at_the_1912_Summer_Olympics_%E2%80%93_Men%27s_300_metre_military_rifle,_three_positions\" title=\"Shooting at the 1912 Summer Olympics \\xe2\\x80\\x93 Men\\'s 300 metre military rifle, three positions\">2 S</a>, <a href=\"/wiki/Shooting_at_the_1912_Summer_Olympics_%E2%80%93_Men%27s_600_metre_free_rifle\" title=\"Shooting at the 1912 Summer Olympics \\xe2\\x80\\x93 Men\\'s 600 metre free rifle\">3 S</a>, <a href=\"/wiki/Shooting_at_the_1912_Summer_Olympics_%E2%80%93_Men%27s_team_rifle\" title=\"Shooting at the 1912 Summer Olympics \\xe2\\x80\\x93 Men\\'s team rifle\">4 G</a><br />\\n<a href=\"/wiki/Archery_at_the_1920_Summer_Olympics\" title=\"Archery at the 1920 Summer Olympics\">1920</a> \\xe2\\x80\\x93 <a href=\"/wiki/Shooting_at_the_1920_Summer_Olympics_%E2%80%93_Men%27s_100_metre_team_running_deer,_single_shots\" title=\"Shooting at the 1920 Summer Olympics \\xe2\\x80\\x93 Men\\'s 100 metre team running deer, single shots\">5 B</a>, <a href=\"/wiki/Shooting_at_the_1920_Summer_Olympics_%E2%80%93_Men%27s_300_metre_team_military_rifle,_standing\" title=\"Shooting at the 1920 Summer Olympics \\xe2\\x80\\x93 Men\\'s 300 metre team military rifle, standing\">6 S</a>, <a href=\"/wiki/Shooting_at_the_1920_Summer_Olympics_%E2%80%93_Men%27s_300_metre_team_military_rifle,_prone\" title=\"Shooting at the 1920 Summer Olympics \\xe2\\x80\\x93 Men\\'s 300 metre team military rifle, prone\">7 G</a></td>\\n</tr>\\n<tr>\\n<th>9</th>\\n<td>30 July 1920</td>\\n<td><a href=\"/wiki/Shooting_at_the_1920_Summer_Olympics_%E2%80%93_Men%27s_300_metre_military_rifle,_standing\" title=\"Shooting at the 1920 Summer Olympics \\xe2\\x80\\x93 Men\\'s 300 metre military rifle, standing\">300 m military rifle, standing G</a></td>\\n</tr>\\n<tr>\\n<th>10</th>\\n<td>31 July 1920</td>\\n<td><a href=\"/wiki/Shooting_at_the_1920_Summer_Olympics_%E2%80%93_Men%27s_team_free_rifle\" title=\"Shooting at the 1920 Summer Olympics \\xe2\\x80\\x93 Men\\'s team free rifle\">Team free rifle G</a></td>\\n</tr>\\n<tr>\\n<th rowspan=\"2\">11</th>\\n<td>27 June 1924</td>\\n<td><a href=\"/wiki/Shooting_at_the_1924_Summer_Olympics\" title=\"Shooting at the 1924 Summer Olympics\">Shooting</a></td>\\n<td><a href=\"/wiki/Shooting_at_the_1924_Summer_Olympics_%E2%80%93_Men%27s_600_metre_free_rifle\" title=\"Shooting at the 1924 Summer Olympics \\xe2\\x80\\x93 Men\\'s 600 metre free rifle\">600 m free rifle S</a></td>\\n</tr>\\n<tr>\\n<td>3 August 1928</td>\\n<td rowspan=\"2\" align=\"left\"><a href=\"/wiki/Paavo_Nurmi\" title=\"Paavo Nurmi\">Paavo Nurmi</a></td>\\n<td rowspan=\"2\" align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/22px-Flag_of_Finland.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/33px-Flag_of_Finland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/44px-Flag_of_Finland.svg.png 2x\" data-file-width=\"1800\" data-file-height=\"1100\" /> <a href=\"/wiki/Finland_at_the_1928_Summer_Olympics\" title=\"Finland at the 1928 Summer Olympics\">Finland</a></td>\\n<td rowspan=\"2\"><a href=\"/wiki/Athletics_at_the_1928_Summer_Olympics\" title=\"Athletics at the 1928 Summer Olympics\">Athletics</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_1928_Summer_Olympics_%E2%80%93_Men%27s_5000_metres\" title=\"Athletics at the 1928 Summer Olympics \\xe2\\x80\\x93 Men\\'s 5000 metres\">5000 m S</a></td>\\n<td rowspan=\"2\" align=\"left\" valign=\"top\"><a href=\"/wiki/Athletics_at_the_1920_Summer_Olympics\" title=\"Athletics at the 1920 Summer Olympics\">1920</a> \\xe2\\x80\\x93 <a href=\"/wiki/Athletics_at_the_1920_Summer_Olympics_%E2%80%93_Men%27s_5000_metres\" title=\"Athletics at the 1920 Summer Olympics \\xe2\\x80\\x93 Men\\'s 5000 metres\">1 S</a>, <a href=\"/wiki/Athletics_at_the_1920_Summer_Olympics_%E2%80%93_Men%27s_10000_metres\" class=\"mw-redirect\" title=\"Athletics at the 1920 Summer Olympics \\xe2\\x80\\x93 Men\\'s 10000 metres\">2 G</a>, <a href=\"/wiki/Athletics_at_the_1920_Summer_Olympics_%E2%80%93_Men%27s_individual_cross_country\" title=\"Athletics at the 1920 Summer Olympics \\xe2\\x80\\x93 Men\\'s individual cross country\">3 G</a>, <a href=\"/wiki/Athletics_at_the_1920_Summer_Olympics_%E2%80%93_Men%27s_team_cross_country\" title=\"Athletics at the 1920 Summer Olympics \\xe2\\x80\\x93 Men\\'s team cross country\">4 G</a><br />\\n<a href=\"/wiki/Athletics_at_the_1924_Summer_Olympics\" title=\"Athletics at the 1924 Summer Olympics\">1924</a> \\xe2\\x80\\x93 <a href=\"/wiki/Athletics_at_the_1924_Summer_Olympics_%E2%80%93_Men%27s_1500_metres\" title=\"Athletics at the 1924 Summer Olympics \\xe2\\x80\\x93 Men\\'s 1500 metres\">5 G</a>, <a href=\"/wiki/Athletics_at_the_1924_Summer_Olympics_%E2%80%93_Men%27s_5000_metres\" title=\"Athletics at the 1924 Summer Olympics \\xe2\\x80\\x93 Men\\'s 5000 metres\">6 G</a>, <a href=\"/wiki/Athletics_at_the_1924_Summer_Olympics_%E2%80%93_Men%27s_individual_cross_country\" title=\"Athletics at the 1924 Summer Olympics \\xe2\\x80\\x93 Men\\'s individual cross country\">7 G</a>, <a href=\"/wiki/Athletics_at_the_1924_Summer_Olympics_%E2%80%93_Men%27s_team_cross_country\" title=\"Athletics at the 1924 Summer Olympics \\xe2\\x80\\x93 Men\\'s team cross country\">8 G</a>, <a href=\"/wiki/Athletics_at_the_1924_Summer_Olympics_%E2%80%93_Men%27s_3000_metre_team_race\" class=\"mw-redirect\" title=\"Athletics at the 1924 Summer Olympics \\xe2\\x80\\x93 Men\\'s 3000 metre team race\">9 G</a><br />\\n<a href=\"/wiki/Athletics_at_the_1928_Summer_Olympics\" title=\"Athletics at the 1928 Summer Olympics\">1928</a> \\xe2\\x80\\x93 <a href=\"/wiki/Athletics_at_the_1928_Summer_Olympics_%E2%80%93_Men%27s_10000_metres\" class=\"mw-redirect\" title=\"Athletics at the 1928 Summer Olympics \\xe2\\x80\\x93 Men\\'s 10000 metres\">10 G</a></td>\\n</tr>\\n<tr>\\n<th rowspan=\"2\">12</th>\\n<td>4 August 1928</td>\\n<td><a href=\"/wiki/Athletics_at_the_1928_Summer_Olympics_%E2%80%93_Men%27s_3000_metre_steeplechase\" class=\"mw-redirect\" title=\"Athletics at the 1928 Summer Olympics \\xe2\\x80\\x93 Men\\'s 3000 metre steeplechase\">3000 m steeplechase S</a></td>\\n</tr>\\n<tr>\\n<td>2 September 1960</td>\\n<td rowspan=\"2\" align=\"left\"><a href=\"/wiki/Edoardo_Mangiarotti\" title=\"Edoardo Mangiarotti\">Edoardo Mangiarotti</a></td>\\n<td rowspan=\"2\" align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/22px-Flag_of_Italy.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/33px-Flag_of_Italy.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/44px-Flag_of_Italy.svg.png 2x\" data-file-width=\"1500\" data-file-height=\"1000\" /> <a href=\"/wiki/Italy_at_the_1960_Summer_Olympics\" title=\"Italy at the 1960 Summer Olympics\">Italy</a></td>\\n<td rowspan=\"2\"><a href=\"/wiki/Fencing_at_the_1960_Summer_Olympics\" title=\"Fencing at the 1960 Summer Olympics\">Fencing</a></td>\\n<td><a href=\"/wiki/Fencing_at_the_1960_Summer_Olympics_%E2%80%93_Men%27s_team_foil\" title=\"Fencing at the 1960 Summer Olympics \\xe2\\x80\\x93 Men\\'s team foil\">Team foil S</a></td>\\n<td rowspan=\"2\" align=\"left\" valign=\"top\"><a href=\"/wiki/Fencing_at_the_1936_Summer_Olympics\" title=\"Fencing at the 1936 Summer Olympics\">1936</a> \\xe2\\x80\\x93 <a href=\"/wiki/Fencing_at_the_1936_Summer_Olympics_%E2%80%93_Men%27s_team_%C3%A9p%C3%A9e\" title=\"Fencing at the 1936 Summer Olympics \\xe2\\x80\\x93 Men\\'s team \\xc3\\xa9p\\xc3\\xa9e\">1 G</a><br />\\n<a href=\"/wiki/Fencing_at_the_1948_Summer_Olympics\" title=\"Fencing at the 1948 Summer Olympics\">1948</a> \\xe2\\x80\\x93 <a href=\"/wiki/Fencing_at_the_1948_Summer_Olympics_%E2%80%93_Men%27s_team_foil\" title=\"Fencing at the 1948 Summer Olympics \\xe2\\x80\\x93 Men\\'s team foil\">2 S</a>, <a href=\"/wiki/Fencing_at_the_1948_Summer_Olympics_%E2%80%93_Men%27s_team_%C3%A9p%C3%A9e\" title=\"Fencing at the 1948 Summer Olympics \\xe2\\x80\\x93 Men\\'s team \\xc3\\xa9p\\xc3\\xa9e\">3 S</a>, <a href=\"/wiki/Fencing_at_the_1948_Summer_Olympics_%E2%80%93_Men%27s_%C3%A9p%C3%A9e\" title=\"Fencing at the 1948 Summer Olympics \\xe2\\x80\\x93 Men\\'s \\xc3\\xa9p\\xc3\\xa9e\">4 B</a><br />\\n<a href=\"/wiki/Fencing_at_the_1952_Summer_Olympics\" title=\"Fencing at the 1952 Summer Olympics\">1952</a> \\xe2\\x80\\x93 <a href=\"/wiki/Fencing_at_the_1952_Summer_Olympics_%E2%80%93_Men%27s_team_foil\" title=\"Fencing at the 1952 Summer Olympics \\xe2\\x80\\x93 Men\\'s team foil\">5 S</a>, <a href=\"/wiki/Fencing_at_the_1952_Summer_Olympics_%E2%80%93_Men%27s_foil\" title=\"Fencing at the 1952 Summer Olympics \\xe2\\x80\\x93 Men\\'s foil\">6 S</a>, <a href=\"/wiki/Fencing_at_the_1952_Summer_Olympics_%E2%80%93_Men%27s_team_%C3%A9p%C3%A9e\" title=\"Fencing at the 1952 Summer Olympics \\xe2\\x80\\x93 Men\\'s team \\xc3\\xa9p\\xc3\\xa9e\">7 G</a>, <a href=\"/wiki/Fencing_at_the_1952_Summer_Olympics_%E2%80%93_Men%27s_%C3%A9p%C3%A9e\" title=\"Fencing at the 1952 Summer Olympics \\xe2\\x80\\x93 Men\\'s \\xc3\\xa9p\\xc3\\xa9e\">8 G</a><br />\\n<a href=\"/wiki/Fencing_at_the_1956_Summer_Olympics\" title=\"Fencing at the 1956 Summer Olympics\">1956</a> \\xe2\\x80\\x93 <a href=\"/wiki/Fencing_at_the_1956_Summer_Olympics_%E2%80%93_Men%27s_team_foil\" title=\"Fencing at the 1956 Summer Olympics \\xe2\\x80\\x93 Men\\'s team foil\">9 G</a>, <a href=\"/wiki/Fencing_at_the_1956_Summer_Olympics_%E2%80%93_Men%27s_team_%C3%A9p%C3%A9e\" title=\"Fencing at the 1956 Summer Olympics \\xe2\\x80\\x93 Men\\'s team \\xc3\\xa9p\\xc3\\xa9e\">10 G</a>, <a href=\"/wiki/Fencing_at_the_1956_Summer_Olympics_%E2%80%93_Men%27s_%C3%A9p%C3%A9e\" title=\"Fencing at the 1956 Summer Olympics \\xe2\\x80\\x93 Men\\'s \\xc3\\xa9p\\xc3\\xa9e\">11 B</a><br /></td>\\n</tr>\\n<tr>\\n<th rowspan=\"2\">13</th>\\n<td>9 September 1960</td>\\n<td><a href=\"/wiki/Fencing_at_the_1960_Summer_Olympics_%E2%80%93_Men%27s_team_%C3%A9p%C3%A9e\" title=\"Fencing at the 1960 Summer Olympics \\xe2\\x80\\x93 Men\\'s team \\xc3\\xa9p\\xc3\\xa9e\">Team \\xc3\\xa9p\\xc3\\xa9e G</a></td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">21 October 1964</td>\\n<td rowspan=\"6\" align=\"left\"><a href=\"/wiki/Larisa_Latynina\" title=\"Larisa Latynina\">Larisa Latynina</a></td>\\n<td rowspan=\"6\" align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/72/Flag_of_the_Soviet_Union_%281955-1980%29.svg/22px-Flag_of_the_Soviet_Union_%281955-1980%29.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/72/Flag_of_the_Soviet_Union_%281955-1980%29.svg/33px-Flag_of_the_Soviet_Union_%281955-1980%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/7/72/Flag_of_the_Soviet_Union_%281955-1980%29.svg/44px-Flag_of_the_Soviet_Union_%281955-1980%29.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_1964_Summer_Olympics\" title=\"Soviet Union at the 1964 Summer Olympics\">Soviet Union</a></td>\\n<td rowspan=\"6\"><a href=\"/wiki/Gymnastics_at_the_1964_Summer_Olympics\" title=\"Gymnastics at the 1964 Summer Olympics\">Gymnastics</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_1964_Summer_Olympics_%E2%80%93_Women%27s_team_competition\" class=\"mw-redirect\" title=\"Gymnastics at the 1964 Summer Olympics \\xe2\\x80\\x93 Women\\'s team competition\">Team G</a></td>\\n<td rowspan=\"6\" align=\"left\" valign=\"top\"><a href=\"/wiki/Gymnastics_at_the_1956_Summer_Olympics\" title=\"Gymnastics at the 1956 Summer Olympics\">1956</a> \\xe2\\x80\\x93 1 G, <a href=\"/wiki/Gymnastics_at_the_1956_Summer_Olympics_%E2%80%93_Women%27s_team_portable_apparatus\" title=\"Gymnastics at the 1956 Summer Olympics \\xe2\\x80\\x93 Women\\'s team portable apparatus\">2 B</a>, 3 G, 4 G, 5 G, <a href=\"/wiki/Gymnastics_at_the_1956_Summer_Olympics_%E2%80%93_Women%27s_uneven_bars\" title=\"Gymnastics at the 1956 Summer Olympics \\xe2\\x80\\x93 Women\\'s uneven bars\">6 S</a><br />\\n<a href=\"/wiki/Gymnastics_at_the_1960_Summer_Olympics\" title=\"Gymnastics at the 1960 Summer Olympics\">1960</a> \\xe2\\x80\\x93 7 G, 8 G, 9 S, 10 S, <a href=\"/wiki/Gymnastics_at_the_1960_Summer_Olympics_%E2%80%93_Women%27s_vault\" title=\"Gymnastics at the 1960 Summer Olympics \\xe2\\x80\\x93 Women\\'s vault\">11 B</a>, 12 G</td>\\n</tr>\\n<tr>\\n<th>14</th>\\n<td><a href=\"/wiki/Gymnastics_at_the_1964_Summer_Olympics_%E2%80%93_Women%27s_individual_all-around\" title=\"Gymnastics at the 1964 Summer Olympics \\xe2\\x80\\x93 Women\\'s individual all-around\">All-around S</a></td>\\n</tr>\\n<tr>\\n<th>15</th>\\n<td rowspan=\"2\">22 October 1964</td>\\n<td><a href=\"/wiki/Gymnastics_at_the_1964_Summer_Olympics_%E2%80%93_Women%27s_vault\" title=\"Gymnastics at the 1964 Summer Olympics \\xe2\\x80\\x93 Women\\'s vault\">Vault S</a></td>\\n</tr>\\n<tr>\\n<th>16</th>\\n<td><a href=\"/wiki/Gymnastics_at_the_1964_Summer_Olympics_%E2%80%93_Women%27s_uneven_bars\" title=\"Gymnastics at the 1964 Summer Olympics \\xe2\\x80\\x93 Women\\'s uneven bars\">Uneven bars B</a></td>\\n</tr>\\n<tr>\\n<th>17</th>\\n<td rowspan=\"2\">23 October 1964</td>\\n<td><a href=\"/wiki/Gymnastics_at_the_1964_Summer_Olympics_%E2%80%93_Women%27s_balance_beam\" title=\"Gymnastics at the 1964 Summer Olympics \\xe2\\x80\\x93 Women\\'s balance beam\">Balance beam B</a></td>\\n</tr>\\n<tr>\\n<th rowspan=\"2\">18</th>\\n<td><a href=\"/wiki/Gymnastics_at_the_1964_Summer_Olympics_%E2%80%93_Women%27s_floor_exercise\" title=\"Gymnastics at the 1964 Summer Olympics \\xe2\\x80\\x93 Women\\'s floor exercise\">Floor exercise G</a></td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">31 July 2012</td>\\n<td rowspan=\"11\" align=\"left\"><a href=\"/wiki/Michael_Phelps\" title=\"Michael Phelps\">Michael Phelps</a></td>\\n<td rowspan=\"11\" align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_2012_Summer_Olympics\" title=\"United States at the 2012 Summer Olympics\">United States</a></td>\\n<td rowspan=\"11\"><a href=\"/wiki/Swimming_at_the_2012_Summer_Olympics\" title=\"Swimming at the 2012 Summer Olympics\">Swimming</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_2012_Summer_Olympics_%E2%80%93_Men%27s_200_metre_butterfly\" title=\"Swimming at the 2012 Summer Olympics \\xe2\\x80\\x93 Men\\'s 200 metre butterfly\">200 m butterfly S</a></td>\\n<td rowspan=\"11\" align=\"left\" valign=\"top\"><a href=\"/wiki/Swimming_at_the_2004_Summer_Olympics\" title=\"Swimming at the 2004 Summer Olympics\">2004</a> \\xe2\\x80\\x93 <a href=\"/wiki/Swimming_at_the_2004_Summer_Olympics_%E2%80%93_Men%27s_400_metre_individual_medley\" title=\"Swimming at the 2004 Summer Olympics \\xe2\\x80\\x93 Men\\'s 400 metre individual medley\">1 G</a>, <a href=\"/wiki/Swimming_at_the_2004_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_100_metre_freestyle_relay\" title=\"Swimming at the 2004 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 100 metre freestyle relay\">2 B</a>, <a href=\"/wiki/Swimming_at_the_2004_Summer_Olympics_%E2%80%93_Men%27s_200_metre_freestyle\" title=\"Swimming at the 2004 Summer Olympics \\xe2\\x80\\x93 Men\\'s 200 metre freestyle\">3 B</a>, <a href=\"/wiki/Swimming_at_the_2004_Summer_Olympics_%E2%80%93_Men%27s_200_metre_butterfly\" title=\"Swimming at the 2004 Summer Olympics \\xe2\\x80\\x93 Men\\'s 200 metre butterfly\">4 G</a>, <a href=\"/wiki/Swimming_at_the_2004_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_200_metre_freestyle_relay\" title=\"Swimming at the 2004 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 200 metre freestyle relay\">5 G</a>, <a href=\"/wiki/Swimming_at_the_2004_Summer_Olympics_%E2%80%93_Men%27s_200_metre_individual_medley\" title=\"Swimming at the 2004 Summer Olympics \\xe2\\x80\\x93 Men\\'s 200 metre individual medley\">6 G</a>, <a href=\"/wiki/Swimming_at_the_2004_Summer_Olympics_%E2%80%93_Men%27s_100_metre_butterfly\" title=\"Swimming at the 2004 Summer Olympics \\xe2\\x80\\x93 Men\\'s 100 metre butterfly\">7 G</a>, <a href=\"/wiki/Swimming_at_the_2004_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_100_metre_medley_relay\" title=\"Swimming at the 2004 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 100 metre medley relay\">8 G</a><br />\\n<a href=\"/wiki/Swimming_at_the_2008_Summer_Olympics\" title=\"Swimming at the 2008 Summer Olympics\">2008</a> \\xe2\\x80\\x93 <a href=\"/wiki/Swimming_at_the_2008_Summer_Olympics_%E2%80%93_Men%27s_400_metre_individual_medley\" title=\"Swimming at the 2008 Summer Olympics \\xe2\\x80\\x93 Men\\'s 400 metre individual medley\">9 G</a>, <a href=\"/wiki/Swimming_at_the_2008_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_100_metre_freestyle_relay\" title=\"Swimming at the 2008 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 100 metre freestyle relay\">10 G</a>, <a href=\"/wiki/Swimming_at_the_2008_Summer_Olympics_%E2%80%93_Men%27s_200_metre_freestyle\" title=\"Swimming at the 2008 Summer Olympics \\xe2\\x80\\x93 Men\\'s 200 metre freestyle\">11 G</a>, <a href=\"/wiki/Swimming_at_the_2008_Summer_Olympics_%E2%80%93_Men%27s_200_metre_butterfly\" title=\"Swimming at the 2008 Summer Olympics \\xe2\\x80\\x93 Men\\'s 200 metre butterfly\">12 G</a>, <a href=\"/wiki/Swimming_at_the_2008_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_200_metre_freestyle_relay\" title=\"Swimming at the 2008 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 200 metre freestyle relay\">13 G</a>, <a href=\"/wiki/Swimming_at_the_2008_Summer_Olympics_%E2%80%93_Men%27s_200_metre_individual_medley\" title=\"Swimming at the 2008 Summer Olympics \\xe2\\x80\\x93 Men\\'s 200 metre individual medley\">14 G</a>, <a href=\"/wiki/Swimming_at_the_2008_Summer_Olympics_%E2%80%93_Men%27s_100_metre_butterfly\" title=\"Swimming at the 2008 Summer Olympics \\xe2\\x80\\x93 Men\\'s 100 metre butterfly\">15 G</a>, <a href=\"/wiki/Swimming_at_the_2008_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_100_metre_medley_relay\" title=\"Swimming at the 2008 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 100 metre medley relay\">16 G</a><br />\\n<a href=\"/wiki/Swimming_at_the_2012_Summer_Olympics\" title=\"Swimming at the 2012 Summer Olympics\">2012</a> \\xe2\\x80\\x93 <a href=\"/wiki/Swimming_at_the_2012_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_100_metre_freestyle_relay\" title=\"Swimming at the 2012 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 100 metre freestyle relay\">17 S</a></td>\\n</tr>\\n<tr>\\n<th>19</th>\\n<td><a href=\"/wiki/Swimming_at_the_2012_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_200_metre_freestyle_relay\" title=\"Swimming at the 2012 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 200 metre freestyle relay\">4 \\xc3\\x97 200 m freestyle G</a></td>\\n</tr>\\n<tr>\\n<th>20</th>\\n<td>2 August 2012</td>\\n<td><a href=\"/wiki/Swimming_at_the_2012_Summer_Olympics_%E2%80%93_Men%27s_200_metre_individual_medley\" title=\"Swimming at the 2012 Summer Olympics \\xe2\\x80\\x93 Men\\'s 200 metre individual medley\">200 m individual medley G</a></td>\\n</tr>\\n<tr>\\n<th>21</th>\\n<td>3 August 2012</td>\\n<td><a href=\"/wiki/Swimming_at_the_2012_Summer_Olympics_%E2%80%93_Men%27s_100_metre_butterfly\" title=\"Swimming at the 2012 Summer Olympics \\xe2\\x80\\x93 Men\\'s 100 metre butterfly\">100 m butterfly G</a></td>\\n</tr>\\n<tr>\\n<th>22</th>\\n<td>4 August 2012</td>\\n<td><a href=\"/wiki/Swimming_at_the_2012_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_100_metre_medley_relay\" title=\"Swimming at the 2012 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 100 metre medley relay\">4 \\xc3\\x97 100 m medley relay G</a></td>\\n</tr>\\n<tr>\\n<th>23</th>\\n<td>7 August 2016</td>\\n<td><a href=\"/wiki/Swimming_at_the_2016_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_100_metre_freestyle_relay\" title=\"Swimming at the 2016 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 100 metre freestyle relay\">4 \\xc3\\x97 100 m freestyle relay G</a></td>\\n</tr>\\n<tr>\\n<th>24</th>\\n<td rowspan=\"2\">9 August 2016</td>\\n<td><a href=\"/wiki/Swimming_at_the_2016_Summer_Olympics_%E2%80%93_Men%27s_200_metre_butterfly\" title=\"Swimming at the 2016 Summer Olympics \\xe2\\x80\\x93 Men\\'s 200 metre butterfly\">200 m butterfly G</a></td>\\n</tr>\\n<tr>\\n<th>25</th>\\n<td><a href=\"/wiki/Swimming_at_the_2016_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_200_metre_freestyle_relay\" title=\"Swimming at the 2016 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 200 metre freestyle relay\">4 \\xc3\\x97 200 m freestyle relay G</a></td>\\n</tr>\\n<tr>\\n<th>26</th>\\n<td>11 August 2016</td>\\n<td><a href=\"/wiki/Swimming_at_the_2016_Summer_Olympics_%E2%80%93_Men%27s_200_metre_individual_medley\" title=\"Swimming at the 2016 Summer Olympics \\xe2\\x80\\x93 Men\\'s 200 metre individual medley\">200 m individual medley G</a></td>\\n</tr>\\n<tr>\\n<th>27</th>\\n<td>12 August 2016</td>\\n<td><a href=\"/wiki/Swimming_at_the_2016_Summer_Olympics_%E2%80%93_Men%27s_100_metre_butterfly\" title=\"Swimming at the 2016 Summer Olympics \\xe2\\x80\\x93 Men\\'s 100 metre butterfly\">100 m butterfly S</a></td>\\n</tr>\\n<tr>\\n<th>28</th>\\n<td>13 August 2016</td>\\n<td><a href=\"/wiki/Swimming_at_the_2016_Summer_Olympics_%E2%80%93_Men%27s_4_%C3%97_100_metre_medley_relay\" title=\"Swimming at the 2016 Summer Olympics \\xe2\\x80\\x93 Men\\'s 4 \\xc3\\x97 100 metre medley relay\">4 \\xc3\\x97 100 m medley relay G</a></td>\\n</tr>\\n</table>\\n<p>Legend: G = Gold, S = Silver, B = Bronze.</p>\\n<h2><span class=\"mw-headline\" id=\"List_of_most_career_medals_in_individual_events\">List of most career medals in individual events</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=3\" title=\"Edit section: List of most career medals in individual events\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h2>\\n<p>This list contains only medals won in individual events, no relays or team events count for this section.</p>\\n<table class=\"wikitable sortable\">\\n<tr>\\n<th>No.</th>\\n<th>Athlete</th>\\n<th>Nation</th>\\n<th>Sport</th>\\n<th>Years</th>\\n<th>Games</th>\\n<th>Sex</th>\\n<th style=\"background-color:gold; width:4.0em; font-weight:bold;\">Gold</th>\\n<th style=\"background-color:silver; width:4.0em; font-weight:bold;\">Silver</th>\\n<th style=\"background-color:#cc9966; width:4.0em; font-weight:bold;\">Bronze</th>\\n<th style=\"width:4.0em;\">Total</th>\\n</tr>\\n<tr>\\n<td>1</td>\\n<td align=\"left\"><span class=\"sortkey\">Phelps, Michael</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Michael_Phelps\" title=\"Michael Phelps\">Michael Phelps</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2004\\xe2\\x80\\x932016</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>13</td>\\n<td>2</td>\\n<td>1</td>\\n<td>16</td>\\n</tr>\\n<tr>\\n<td>2</td>\\n<td align=\"left\"><span class=\"sortkey\">Latynina, Larisa</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Larisa_Latynina\" title=\"Larisa Latynina\">Larisa Latynina</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1956\\xe2\\x80\\x931964</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>6</td>\\n<td>5</td>\\n<td>3</td>\\n<td>14</td>\\n</tr>\\n<tr>\\n<td>3</td>\\n<td align=\"left\"><span class=\"sortkey\">Andrianov, Nikolai</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Nikolai_Andrianov\" title=\"Nikolai Andrianov\">Nikolai Andrianov</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1972\\xe2\\x80\\x931980</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>3</td>\\n<td>3</td>\\n<td>12</td>\\n</tr>\\n<tr>\\n<td>4</td>\\n<td align=\"left\"><span class=\"sortkey\">Shakhlin, Boris</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Boris_Shakhlin\" title=\"Boris Shakhlin\">Boris Shakhlin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1956\\xe2\\x80\\x931964</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>2</td>\\n<td>2</td>\\n<td rowspan=\"3\">10</td>\\n</tr>\\n<tr>\\n<td>5</td>\\n<td align=\"left\"><span class=\"sortkey\">Ono, Takashi</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Takashi_Ono\" title=\"Takashi Ono\">Takashi Ono</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/22px-Flag_of_Japan.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/33px-Flag_of_Japan.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/44px-Flag_of_Japan.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Japan_at_the_Olympics\" title=\"Japan at the Olympics\">Japan</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1952\\xe2\\x80\\x931964</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>3</td>\\n<td>3</td>\\n<td>4</td>\\n</tr>\\n<tr>\\n<td>6</td>\\n<td align=\"left\"><span class=\"sortkey\">Nemov, Alexei</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Alexei_Nemov\" title=\"Alexei Nemov\">Alexei Nemov</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/22px-Flag_of_Russia.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/33px-Flag_of_Russia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/f/f3/Flag_of_Russia.svg/44px-Flag_of_Russia.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Russia_at_the_Olympics\" title=\"Russia at the Olympics\">Russia</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1996\\xe2\\x80\\x932000</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>3</td>\\n<td>2</td>\\n<td>5</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">7</td>\\n<td align=\"left\"><span class=\"sortkey\">Nurmi, Paavo</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Paavo_Nurmi\" title=\"Paavo Nurmi\">Paavo Nurmi</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/22px-Flag_of_Finland.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/33px-Flag_of_Finland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/44px-Flag_of_Finland.svg.png 2x\" data-file-width=\"1800\" data-file-height=\"1100\" /> <a href=\"/wiki/Finland_at_the_Olympics\" title=\"Finland at the Olympics\">Finland</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>1920\\xe2\\x80\\x931928</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>6</td>\\n<td>3</td>\\n<td>0</td>\\n<td rowspan=\"7\">9</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Dahlie, Bjorn</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Bj%C3%B8rn_D%C3%A6hlie\" title=\"Bj\\xc3\\xb8rn D\\xc3\\xa6hlie\">Bj\\xc3\\xb8rn D\\xc3\\xa6hlie</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">Norway</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1992\\xe2\\x80\\x931998</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>6</td>\\n<td>3</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td rowspan=\"3\">9</td>\\n<td align=\"left\"><span class=\"sortkey\">Chukarin, Viktor</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Viktor_Chukarin\" title=\"Viktor Chukarin\">Viktor Chukarin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1952\\xe2\\x80\\x931956</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Kato, Sawao</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Sawao_Kato\" title=\"Sawao Kato\">Sawao Kato</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/22px-Flag_of_Japan.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/33px-Flag_of_Japan.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/44px-Flag_of_Japan.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Japan_at_the_Olympics\" title=\"Japan at the Olympics\">Japan</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1968\\xe2\\x80\\x931976</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Bjorndalen, Ole Einar</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ole_Einar_Bj%C3%B8rndalen\" title=\"Ole Einar Bj\\xc3\\xb8rndalen\">Ole Einar Bj\\xc3\\xb8rndalen</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">Norway</a></td>\\n<td><a href=\"/wiki/Biathlon_at_the_Winter_Olympics\" title=\"Biathlon at the Winter Olympics\">Biathlon</a></td>\\n<td>1998\\xe2\\x80\\x932014</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>5</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>12</td>\\n<td align=\"left\"><span class=\"sortkey\">Scherbo, Vitaly</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Vitaly_Scherbo\" title=\"Vitaly Scherbo\">Vitaly Scherbo</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/22px-Olympic_flag.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/33px-Olympic_flag.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/44px-Olympic_flag.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Unified_Team_at_the_Olympics\" title=\"Unified Team at the Olympics\">Unified Team</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/85/Flag_of_Belarus.svg/22px-Flag_of_Belarus.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/85/Flag_of_Belarus.svg/33px-Flag_of_Belarus.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/8/85/Flag_of_Belarus.svg/44px-Flag_of_Belarus.svg.png 2x\" data-file-width=\"900\" data-file-height=\"450\" /> <a href=\"/wiki/Belarus_at_the_Olympics\" title=\"Belarus at the Olympics\">Belarus</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1992\\xe2\\x80\\x931996</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>5</td>\\n<td>0</td>\\n<td>4</td>\\n</tr>\\n<tr>\\n<td>13</td>\\n<td align=\"left\"><span class=\"sortkey\">W\\xc3\\xbcst, Ireen</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ireen_W%C3%BCst\" title=\"Ireen W\\xc3\\xbcst\">Ireen W\\xc3\\xbcst</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/22px-Flag_of_the_Netherlands.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/33px-Flag_of_the_Netherlands.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/20/Flag_of_the_Netherlands.svg/44px-Flag_of_the_Netherlands.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Netherlands_at_the_Olympics\" title=\"Netherlands at the Olympics\">Netherlands</a></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_Winter_Olympics\" title=\"Speed skating at the Winter Olympics\">Speed skating</a></td>\\n<td>2006\\xe2\\x80\\x932018</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>4</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>14</td>\\n<td align=\"left\"><span class=\"sortkey\">Ewry, Ray</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ray_Ewry\" title=\"Ray Ewry\">Ray Ewry</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/22px-Flag_of_the_United_States_%281896-1908%29.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/33px-Flag_of_the_United_States_%281896-1908%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/f/fc/Flag_of_the_United_States_%281896-1908%29.svg/44px-Flag_of_the_United_States_%281896-1908%29.svg.png 2x\" data-file-width=\"1024\" data-file-height=\"539\" /> <a href=\"/wiki/United_States_at_the_1908_Summer_Olympics\" title=\"United States at the 1908 Summer Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>1900\\xe2\\x80\\x931908</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>8</td>\\n<td>0</td>\\n<td>0</td>\\n<td rowspan=\"7\">8</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">15</td>\\n<td align=\"left\"><span class=\"sortkey\">Caslavska, Vera</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/V%C4%9Bra_%C4%8C%C3%A1slavsk%C3%A1\" title=\"V\\xc4\\x9bra \\xc4\\x8c\\xc3\\xa1slavsk\\xc3\\xa1\">V\\xc4\\x9bra \\xc4\\x8c\\xc3\\xa1slavsk\\xc3\\xa1</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/cb/Flag_of_the_Czech_Republic.svg/22px-Flag_of_the_Czech_Republic.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/cb/Flag_of_the_Czech_Republic.svg/33px-Flag_of_the_Czech_Republic.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/cb/Flag_of_the_Czech_Republic.svg/44px-Flag_of_the_Czech_Republic.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Czechoslovakia_at_the_Olympics\" title=\"Czechoslovakia at the Olympics\">Czechoslovakia</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1960\\xe2\\x80\\x931968</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>7</td>\\n<td>1</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Lewis, Carl</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Carl_Lewis\" title=\"Carl Lewis\">Carl Lewis</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>1984\\xe2\\x80\\x931996</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>7</td>\\n<td>1</td>\\n<td>0</td>\\n</tr>\\n<tr>\\n<td>17</td>\\n<td align=\"left\"><span class=\"sortkey\">Bj\\xc3\\xb8rgen, Marit</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Marit_Bj%C3%B8rgen\" title=\"Marit Bj\\xc3\\xb8rgen\">Marit Bj\\xc3\\xb8rgen</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">Norway</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>2002\\xe2\\x80\\x932018</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>4</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\">18</td>\\n<td align=\"left\"><span class=\"sortkey\">Nakayama, Akinori</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Akinori_Nakayama\" title=\"Akinori Nakayama\">Akinori Nakayama</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/22px-Flag_of_Japan.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/33px-Flag_of_Japan.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/9/9e/Flag_of_Japan.svg/44px-Flag_of_Japan.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Japan_at_the_Olympics\" title=\"Japan at the Olympics\">Japan</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1968\\xe2\\x80\\x931972</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>4</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Pechstein, Claudia</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Claudia_Pechstein\" title=\"Claudia Pechstein\">Claudia Pechstein</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">Germany</a></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_Winter_Olympics\" title=\"Speed skating at the Winter Olympics\">Speed skating</a></td>\\n<td>1992\\xe2\\x80\\x932006</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>4</td>\\n<td>2</td>\\n<td>2</td>\\n</tr>\\n<tr>\\n<td>20</td>\\n<td align=\"left\"><span class=\"sortkey\">Dityatin, Alexander</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Alexander_Dityatin\" title=\"Alexander Dityatin\">Alexander Dityatin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1976\\xe2\\x80\\x931980</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>2</td>\\n<td>5</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"3\">21</td>\\n<td align=\"left\"><span class=\"sortkey\">Thunberg, Clas</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Clas_Thunberg\" title=\"Clas Thunberg\">Clas Thunberg</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/22px-Flag_of_Finland.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/33px-Flag_of_Finland.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Flag_of_Finland.svg/44px-Flag_of_Finland.svg.png 2x\" data-file-width=\"1800\" data-file-height=\"1100\" /> <a href=\"/wiki/Finland_at_the_Olympics\" title=\"Finland at the Olympics\">Finland</a></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_Winter_Olympics\" title=\"Speed skating at the Winter Olympics\">Speed skating</a></td>\\n<td>1924-1928</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>5</td>\\n<td>1</td>\\n<td>1</td>\\n<td rowspan=\"10\">7</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Comaneci, Nadia</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Nadia_Com%C4%83neci\" title=\"Nadia Com\\xc4\\x83neci\">Nadia Com\\xc4\\x83neci</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/7f/Flag_of_Romania_%281965-1989%29.svg/22px-Flag_of_Romania_%281965-1989%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/7/7f/Flag_of_Romania_%281965-1989%29.svg/33px-Flag_of_Romania_%281965-1989%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/7/7f/Flag_of_Romania_%281965-1989%29.svg/44px-Flag_of_Romania_%281965-1989%29.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Romania_at_the_1980_Summer_Olympics\" title=\"Romania at the 1980 Summer Olympics\">Romania</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1976\\xe2\\x80\\x931980</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>5</td>\\n<td>1</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Egerszegi, Krisztina</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Krisztina_Egerszegi\" title=\"Krisztina Egerszegi\">Krisztina Egerszegi</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/22px-Flag_of_Hungary.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/33px-Flag_of_Hungary.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/44px-Flag_of_Hungary.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Hungary_at_the_Olympics\" title=\"Hungary at the Olympics\">Hungary</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>1988\\xe2\\x80\\x931996</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>5</td>\\n<td>1</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>24</td>\\n<td align=\"left\"><span class=\"sortkey\">Jernberg, Sixten</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Sixten_Jernberg\" title=\"Sixten Jernberg\">Sixten Jernberg</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/22px-Flag_of_Sweden.svg.png\" width=\"22\" height=\"14\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/33px-Flag_of_Sweden.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/44px-Flag_of_Sweden.svg.png 2x\" data-file-width=\"1600\" data-file-height=\"1000\" /> <a href=\"/wiki/Sweden_at_the_Olympics\" title=\"Sweden at the Olympics\">Sweden</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1956\\xe2\\x80\\x931964</td>\\n<td>Winter</td>\\n<td>M</td>\\n<td>3</td>\\n<td>3</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td rowspan=\"3\">25</td>\\n<td align=\"left\"><span class=\"sortkey\">Voronin, Mikhail</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Mikhail_Voronin\" title=\"Mikhail Voronin\">Mikhail Voronin</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a></td>\\n<td><a href=\"/wiki/Gymnastics_at_the_Summer_Olympics\" title=\"Gymnastics at the Summer Olympics\">Gymnastics</a></td>\\n<td>1968\\xe2\\x80\\x931972</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>2</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Smetanina, Raisa</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Raisa_Smetanina\" title=\"Raisa Smetanina\">Raisa Smetanina</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">Soviet Union</a><br />\\n<img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/22px-Olympic_flag.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/33px-Olympic_flag.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a7/Olympic_flag.svg/44px-Olympic_flag.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Unified_Team_at_the_Olympics\" title=\"Unified Team at the Olympics\">Unified Team</a></td>\\n<td><a href=\"/wiki/Cross-country_skiing_at_the_Winter_Olympics\" title=\"Cross-country skiing at the Winter Olympics\">Cross-country skiing</a></td>\\n<td>1976\\xe2\\x80\\x931992</td>\\n<td>Winter</td>\\n<td>F</td>\\n<td>2</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td align=\"left\"><span class=\"sortkey\">Coventry, Kirsty</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Kirsty_Coventry\" title=\"Kirsty Coventry\">Kirsty Coventry</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/6a/Flag_of_Zimbabwe.svg/22px-Flag_of_Zimbabwe.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/6/6a/Flag_of_Zimbabwe.svg/33px-Flag_of_Zimbabwe.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/6/6a/Flag_of_Zimbabwe.svg/44px-Flag_of_Zimbabwe.svg.png 2x\" data-file-width=\"840\" data-file-height=\"420\" /> <a href=\"/wiki/Zimbabwe_at_the_Olympics\" title=\"Zimbabwe at the Olympics\">Zimbabwe</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2004\\xe2\\x80\\x932008</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>2</td>\\n<td>4</td>\\n<td>1</td>\\n</tr>\\n<tr>\\n<td>27</td>\\n<td align=\"left\"><span class=\"sortkey\">Lochte, Ryan</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Ryan_Lochte\" title=\"Ryan Lochte\">Ryan Lochte</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">United States</a></td>\\n<td><a href=\"/wiki/Swimming_at_the_Summer_Olympics\" title=\"Swimming at the Summer Olympics\">Swimming</a></td>\\n<td>2004\\xe2\\x80\\x932012</td>\\n<td>Summer</td>\\n<td>M</td>\\n<td>2</td>\\n<td>2</td>\\n<td>3</td>\\n</tr>\\n<tr>\\n<td>28</td>\\n<td align=\"left\"><span class=\"sortkey\">Ottey, Merlene</span><span class=\"vcard\"><span class=\"fn\"><a href=\"/wiki/Merlene_Ottey\" title=\"Merlene Ottey\">Merlene Ottey</a></span></span></td>\\n<td align=\"left\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/22px-Flag_of_Jamaica.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/33px-Flag_of_Jamaica.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/0/0a/Flag_of_Jamaica.svg/44px-Flag_of_Jamaica.svg.png 2x\" data-file-width=\"600\" data-file-height=\"300\" /> <a href=\"/wiki/Jamaica_at_the_Olympics\" title=\"Jamaica at the Olympics\">Jamaica</a></td>\\n<td><a href=\"/wiki/Athletics_at_the_Summer_Olympics\" title=\"Athletics at the Summer Olympics\">Athletics</a></td>\\n<td>1980\\xe2\\x80\\x932000</td>\\n<td>Summer</td>\\n<td>F</td>\\n<td>0</td>\\n<td>2</td>\\n<td>5</td>\\n</tr>\\n</table>\\n<h2><span class=\"mw-headline\" id=\"Athletes_with_medals_in_different_disciplines\">Athletes with medals in different disciplines</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=4\" title=\"Edit section: Athletes with medals in different disciplines\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h2>\\n<div role=\"note\" class=\"hatnote navigation-not-searchable plainlinks\">This list is <a href=\"/wiki/Wikipedia:WikiProject_Lists#Incomplete_lists\" title=\"Wikipedia:WikiProject Lists\">incomplete</a>; you can help by <a class=\"external text\" href=\"//en.wikipedia.org/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit\">expanding it</a>.</div>\\n<h3><span class=\"mw-headline\" id=\"In_the_Summer_and_Winter_Games\">In the Summer and Winter Games</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=5\" title=\"Edit section: In the Summer and Winter Games\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h3>\\n<table class=\"wikitable\">\\n<tr>\\n<th colspan=\"2\" rowspan=\"2\">Athlete (Nation)</th>\\n<th colspan=\"3\">Summer Games</th>\\n<th colspan=\"3\">Winter Games</th>\\n<th rowspan=\"2\">Ref</th>\\n</tr>\\n<tr>\\n<th>Year</th>\\n<th>Medal</th>\\n<th>Event</th>\\n<th>Year</th>\\n<th>Medal</th>\\n<th>Event</th>\\n</tr>\\n<tr>\\n<td><a href=\"/wiki/File:Eddie_eagan.jpg\" class=\"image\"><img alt=\"Eddie eagan.jpg\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/b2/Eddie_eagan.jpg/75px-Eddie_eagan.jpg\" width=\"75\" height=\"105\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/b/b2/Eddie_eagan.jpg/113px-Eddie_eagan.jpg 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/b/b2/Eddie_eagan.jpg/150px-Eddie_eagan.jpg 2x\" data-file-width=\"215\" data-file-height=\"300\" /></a></td>\\n<td><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/Eddie_Eagan\" title=\"Eddie Eagan\">Eddie Eagan</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">USA</a>)</span></td>\\n<td><a href=\"/wiki/1920_Summer_Olympics\" title=\"1920 Summer Olympics\">1920 Antwerp</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">01 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/16px-Gold_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/24px-Gold_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/32px-Gold_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Gold</span></td>\\n<td><a href=\"/wiki/Boxing_at_the_1920_Summer_Olympics_%E2%80%93_Men%27s_light_heavyweight\" title=\"Boxing at the 1920 Summer Olympics \\xe2\\x80\\x93 Men\\'s light heavyweight\">Boxing (light heavyweight)</a></td>\\n<td><a href=\"/wiki/1932_Winter_Olympics\" title=\"1932 Winter Olympics\">1932 Lake Placid</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">01 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/16px-Gold_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/24px-Gold_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/32px-Gold_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Gold</span></td>\\n<td><a href=\"/wiki/Bobsleigh_at_the_1932_Winter_Olympics_%E2%80%93_Four-man\" title=\"Bobsleigh at the 1932 Winter Olympics \\xe2\\x80\\x93 Four-man\">Bobsleigh (four-man)</a></td>\\n<td align=\"center\"><sup id=\"cite_ref-2\" class=\"reference\"><a href=\"#cite_note-2\">[2]</a></sup></td>\\n</tr>\\n<tr>\\n<td><a href=\"/wiki/File:Jacob_Tullin_Thams.jpeg\" class=\"image\"><img alt=\"Jacob Tullin Thams.jpeg\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/25/Jacob_Tullin_Thams.jpeg/75px-Jacob_Tullin_Thams.jpeg\" width=\"75\" height=\"98\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/25/Jacob_Tullin_Thams.jpeg/113px-Jacob_Tullin_Thams.jpeg 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/25/Jacob_Tullin_Thams.jpeg/150px-Jacob_Tullin_Thams.jpeg 2x\" data-file-width=\"480\" data-file-height=\"626\" /></a></td>\\n<td><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Jacob_Tullin_Thams\" title=\"Jacob Tullin Thams\">Jacob Tullin Thams</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">NOR</a>)</span></td>\\n<td><a href=\"/wiki/1936_Summer_Olympics\" title=\"1936 Summer Olympics\">1936 Berlin</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">02 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/16px-Silver_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/24px-Silver_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/32px-Silver_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Silver</span></td>\\n<td><a href=\"/wiki/Sailing_at_the_1936_Summer_Olympics\" title=\"Sailing at the 1936 Summer Olympics\">Sailing (8-metre)</a></td>\\n<td><a href=\"/wiki/1924_Winter_Olympics\" title=\"1924 Winter Olympics\">1924 Chamonix</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">01 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/16px-Gold_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/24px-Gold_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/32px-Gold_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Gold</span></td>\\n<td><a href=\"/wiki/Ski_jumping_at_the_1924_Winter_Olympics\" title=\"Ski jumping at the 1924 Winter Olympics\">Ski jumping (individual large hill)</a></td>\\n<td align=\"center\"><sup id=\"cite_ref-3\" class=\"reference\"><a href=\"#cite_note-3\">[3]</a></sup></td>\\n</tr>\\n<tr>\\n<td rowspan=\"4\"><a href=\"/wiki/File:Bundesarchiv_Bild_183-Z0301-022,_Christa_Rothenburger.jpg\" class=\"image\"><img alt=\"Bundesarchiv Bild 183-Z0301-022, Christa Rothenburger.jpg\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/cd/Bundesarchiv_Bild_183-Z0301-022%2C_Christa_Rothenburger.jpg/75px-Bundesarchiv_Bild_183-Z0301-022%2C_Christa_Rothenburger.jpg\" width=\"75\" height=\"107\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/cd/Bundesarchiv_Bild_183-Z0301-022%2C_Christa_Rothenburger.jpg/113px-Bundesarchiv_Bild_183-Z0301-022%2C_Christa_Rothenburger.jpg 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/cd/Bundesarchiv_Bild_183-Z0301-022%2C_Christa_Rothenburger.jpg/150px-Bundesarchiv_Bild_183-Z0301-022%2C_Christa_Rothenburger.jpg 2x\" data-file-width=\"562\" data-file-height=\"800\" /></a></td>\\n<td rowspan=\"4\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/22px-Flag_of_East_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/33px-Flag_of_East_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Flag_of_East_Germany.svg/44px-Flag_of_East_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Christa_Luding-Rothenburger\" title=\"Christa Luding-Rothenburger\">Christa Luding-Rothenburger</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/East_Germany_at_the_Olympics\" title=\"East Germany at the Olympics\">GDR</a>)</span></td>\\n<td rowspan=\"4\"><a href=\"/wiki/1988_Summer_Olympics\" title=\"1988 Summer Olympics\">1988 Seoul</a></td>\\n<td rowspan=\"4\"><span style=\"display:none;\" class=\"sortkey\">02 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/16px-Silver_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/24px-Silver_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/32px-Silver_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Silver</span></td>\\n<td rowspan=\"4\"><a href=\"/wiki/Cycling_at_the_1988_Summer_Olympics#Women\\'s_events\" title=\"Cycling at the 1988 Summer Olympics\">Cycling (sprint)</a></td>\\n<td><a href=\"/wiki/1984_Winter_Olympics\" title=\"1984 Winter Olympics\">1984 Sarajevo</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">01 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/16px-Gold_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/24px-Gold_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/32px-Gold_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Gold</span></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_1984_Winter_Olympics#500_m_2\" title=\"Speed skating at the 1984 Winter Olympics\">Speed skating (500 m)</a></td>\\n<td rowspan=\"4\" align=\"center\"><sup id=\"cite_ref-4\" class=\"reference\"><a href=\"#cite_note-4\">[4]</a></sup></td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\"><a href=\"/wiki/1988_Winter_Olympics\" title=\"1988 Winter Olympics\">1988 Calgary</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">01 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/16px-Gold_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/24px-Gold_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/32px-Gold_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Gold</span></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_1988_Winter_Olympics#1,000_m_2\" title=\"Speed skating at the 1988 Winter Olympics\">Speed skating (1000 m)</a></td>\\n</tr>\\n<tr>\\n<td><span style=\"display:none;\" class=\"sortkey\">02 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/16px-Silver_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/24px-Silver_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/32px-Silver_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Silver</span></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_1988_Winter_Olympics#500_m_2\" title=\"Speed skating at the 1988 Winter Olympics\">Speed skating (500 m)</a></td>\\n</tr>\\n<tr>\\n<td><a href=\"/wiki/1992_Winter_Olympics\" title=\"1992 Winter Olympics\">1992 Albertville</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">03 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/16px-Bronze_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/24px-Bronze_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/32px-Bronze_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Bronze</span></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_1992_Winter_Olympics#500_m_2\" title=\"Speed skating at the 1992 Winter Olympics\">Speed skating (500 m)</a></td>\\n</tr>\\n<tr>\\n<td rowspan=\"4\"><a href=\"/wiki/File:Hughes0004_024.jpg\" class=\"image\"><img alt=\"Hughes0004 024.jpg\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/5/51/Hughes0004_024.jpg/75px-Hughes0004_024.jpg\" width=\"75\" height=\"95\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/5/51/Hughes0004_024.jpg/113px-Hughes0004_024.jpg 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/5/51/Hughes0004_024.jpg/150px-Hughes0004_024.jpg 2x\" data-file-width=\"1308\" data-file-height=\"1654\" /></a></td>\\n<td rowspan=\"4\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/c/cf/Flag_of_Canada.svg/22px-Flag_of_Canada.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/c/cf/Flag_of_Canada.svg/33px-Flag_of_Canada.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/c/cf/Flag_of_Canada.svg/44px-Flag_of_Canada.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"500\" /> <a href=\"/wiki/Clara_Hughes\" title=\"Clara Hughes\">Clara Hughes</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Canada_at_the_Olympics\" title=\"Canada at the Olympics\">CAN</a>)</span></td>\\n<td rowspan=\"4\"><a href=\"/wiki/1996_Summer_Olympics\" title=\"1996 Summer Olympics\">1996 Atlanta</a></td>\\n<td rowspan=\"2\"><span style=\"display:none;\" class=\"sortkey\">03 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/16px-Bronze_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/24px-Bronze_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/32px-Bronze_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Bronze</span></td>\\n<td rowspan=\"2\"><a href=\"/wiki/Cycling_at_the_1996_Summer_Olympics_%E2%80%93_Women%27s_road_race\" class=\"mw-redirect\" title=\"Cycling at the 1996 Summer Olympics \\xe2\\x80\\x93 Women\\'s road race\">Cycling (road race)</a></td>\\n<td><a href=\"/wiki/2002_Winter_Olympics\" title=\"2002 Winter Olympics\">2002 Salt Lake City</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">03 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/16px-Bronze_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/24px-Bronze_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/32px-Bronze_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Bronze</span></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_2002_Winter_Olympics_%E2%80%93_Women%27s_5000_metres\" title=\"Speed skating at the 2002 Winter Olympics \\xe2\\x80\\x93 Women\\'s 5000 metres\">Speed skating (5000 m)</a></td>\\n<td rowspan=\"4\" align=\"center\"><sup id=\"cite_ref-5\" class=\"reference\"><a href=\"#cite_note-5\">[5]</a></sup></td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\"><a href=\"/wiki/2006_Winter_Olympics\" title=\"2006 Winter Olympics\">2006 Turin</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">02 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/16px-Silver_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/24px-Silver_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/32px-Silver_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Silver</span></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_2006_Winter_Olympics_%E2%80%93_Women%27s_team_pursuit\" title=\"Speed skating at the 2006 Winter Olympics \\xe2\\x80\\x93 Women\\'s team pursuit\">Speed skating (team pursuit)</a></td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\"><span style=\"display:none;\" class=\"sortkey\">03 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/16px-Bronze_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/24px-Bronze_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/32px-Bronze_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Bronze</span></td>\\n<td rowspan=\"2\"><a href=\"/wiki/Cycling_at_the_1996_Summer_Olympics_%E2%80%93_Women%27s_time_trial\" title=\"Cycling at the 1996 Summer Olympics \\xe2\\x80\\x93 Women\\'s time trial\">Cycling (time trial)</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">01 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/16px-Gold_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/24px-Gold_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/32px-Gold_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Gold</span></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_2006_Winter_Olympics_%E2%80%93_Women%27s_5000_metres\" title=\"Speed skating at the 2006 Winter Olympics \\xe2\\x80\\x93 Women\\'s 5000 metres\">Speed skating (5000 m)</a></td>\\n</tr>\\n<tr>\\n<td><a href=\"/wiki/2010_Winter_Olympics\" title=\"2010 Winter Olympics\">2010 Vancouver</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">03 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/16px-Bronze_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/24px-Bronze_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/8/89/Bronze_medal_icon.svg/32px-Bronze_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Bronze</span></td>\\n<td><a href=\"/wiki/Speed_skating_at_the_2010_Winter_Olympics_%E2%80%93_Women%27s_5000_metres\" title=\"Speed skating at the 2010 Winter Olympics \\xe2\\x80\\x93 Women\\'s 5000 metres\">Speed skating (5000 m)</a></td>\\n</tr>\\n<tr>\\n<td rowspan=\"2\"><a href=\"/wiki/File:Lauryn_Williams_Osaka07.jpg\" class=\"image\"><img alt=\"Lauryn Williams Osaka07.jpg\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c9/Lauryn_Williams_Osaka07.jpg/75px-Lauryn_Williams_Osaka07.jpg\" width=\"75\" height=\"75\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c9/Lauryn_Williams_Osaka07.jpg/113px-Lauryn_Williams_Osaka07.jpg 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c9/Lauryn_Williams_Osaka07.jpg/150px-Lauryn_Williams_Osaka07.jpg 2x\" data-file-width=\"250\" data-file-height=\"250\" /></a></td>\\n<td rowspan=\"2\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/Lauryn_Williams\" title=\"Lauryn Williams\">Lauryn Williams</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">USA</a>)</span></td>\\n<td><a href=\"/wiki/2004_Summer_Olympics\" title=\"2004 Summer Olympics\">2004 Athens</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">02 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/16px-Silver_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/24px-Silver_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/32px-Silver_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Silver</span></td>\\n<td><a href=\"/wiki/Athletics_at_the_2004_Summer_Olympics_%E2%80%93_Women%27s_100_metres\" title=\"Athletics at the 2004 Summer Olympics \\xe2\\x80\\x93 Women\\'s 100 metres\">Athletics (100 m)</a></td>\\n<td rowspan=\"2\"><a href=\"/wiki/2014_Winter_Olympics\" title=\"2014 Winter Olympics\">2014 Sochi</a></td>\\n<td rowspan=\"2\"><span style=\"display:none;\" class=\"sortkey\">02 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/16px-Silver_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/24px-Silver_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/2/2e/Silver_medal_icon.svg/32px-Silver_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Silver</span></td>\\n<td rowspan=\"2\"><a href=\"/wiki/Bobsleigh_at_the_2014_Winter_Olympics_%E2%80%93_Two-woman\" title=\"Bobsleigh at the 2014 Winter Olympics \\xe2\\x80\\x93 Two-woman\">Bobsleigh (two-woman)</a></td>\\n<td rowspan=\"2\" align=\"center\"><sup id=\"cite_ref-6\" class=\"reference\"><a href=\"#cite_note-6\">[6]</a></sup></td>\\n</tr>\\n<tr>\\n<td><a href=\"/wiki/2012_Summer_Olympics\" title=\"2012 Summer Olympics\">2012 London</a></td>\\n<td><span style=\"display:none;\" class=\"sortkey\">01 !</span><span class=\"sorttext\"><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/16px-Gold_medal_icon.svg.png\" width=\"16\" height=\"16\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/24px-Gold_medal_icon.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/4/47/Gold_medal_icon.svg/32px-Gold_medal_icon.svg.png 2x\" data-file-width=\"16\" data-file-height=\"16\" /> Gold</span></td>\\n<td><a href=\"/wiki/Athletics_at_the_2012_Summer_Olympics_%E2%80%93_Women%27s_4_%C3%97_100_metres_relay\" title=\"Athletics at the 2012 Summer Olympics \\xe2\\x80\\x93 Women\\'s 4 \\xc3\\x97 100 metres relay\">Athletics (4 \\xc3\\x97 100 m relay)</a></td>\\n</tr>\\n</table>\\n<ul>\\n<li><a href=\"/wiki/Gillis_Grafstr%C3%B6m\" title=\"Gillis Grafstr\\xc3\\xb6m\">Gillis Grafstr\\xc3\\xb6m</a> became the first person to win a medal in the <i>same event</i> in Summer and Winter Olympics, winning <a href=\"/wiki/Figure_skating\" title=\"Figure skating\">figure skating</a> golds at the 1920 Olympics and at the first Winter Olympics in 1924.</li>\\n<li><a href=\"/wiki/Eddie_Eagan\" title=\"Eddie Eagan\">Eddie Eagan</a> became the first person to win a medal in the <a href=\"/wiki/Winter_Olympics\" class=\"mw-redirect\" title=\"Winter Olympics\">Winter Olympics</a> and in the <a href=\"/wiki/Summer_Olympics\" class=\"mw-redirect\" title=\"Summer Olympics\">Summer Olympics</a> in <i>different events</i>. He is the only Summer and Winter medalist to win <i>gold medals in different events</i>.</li>\\n<li><a href=\"/wiki/Christa_Luding-Rothenburger\" title=\"Christa Luding-Rothenburger\">Christa Luding-Rothenburger</a> is the only person to win medals at the Winter and Summer Games in the <i>same year.</i> (This feat is no longer possible due to the staggering of the Winter and Summer Olympic years). She is also the first person to win medals in successive Winter and Summer Games or vice versa.</li>\\n<li><a href=\"/wiki/Clara_Hughes\" title=\"Clara Hughes\">Clara Hughes</a> is the first person to win <i>multiple medals in both</i> Summer and Winter Games and holds the highest number of medals of any Olympian to win medals in both the Summer and Winter Games.</li>\\n</ul>\\n<h3><span class=\"mw-headline\" id=\"In_the_Summer_Games\">In the Summer Games</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=6\" title=\"Edit section: In the Summer Games\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h3>\\n<h4><span class=\"mw-headline\" id=\"Swimming_and_water_polo\">Swimming and water polo</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=7\" title=\"Edit section: Swimming and water polo\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h4>\\n<ul>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/22px-Flag_of_the_United_Kingdom.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/33px-Flag_of_the_United_Kingdom.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/44px-Flag_of_the_United_Kingdom.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Paul_Radmilovic\" title=\"Paul Radmilovic\">Paul Radmilovic</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Great_Britain_at_the_Olympics\" title=\"Great Britain at the Olympics\">GBR</a>)</span></li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/Johnny_Weissmuller\" title=\"Johnny Weissmuller\">Johnny Weissmuller</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">USA</a>)</span></li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/9/92/Flag_of_Belgium_%28civil%29.svg/22px-Flag_of_Belgium_%28civil%29.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/9/92/Flag_of_Belgium_%28civil%29.svg/33px-Flag_of_Belgium_%28civil%29.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/9/92/Flag_of_Belgium_%28civil%29.svg/44px-Flag_of_Belgium_%28civil%29.svg.png 2x\" data-file-width=\"450\" data-file-height=\"300\" /> <a href=\"/wiki/G%C3%A9rard_Blitz_(swimmer)\" title=\"G\\xc3\\xa9rard Blitz (swimmer)\">G\\xc3\\xa9rard Blitz</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Belgium_at_the_Olympics\" title=\"Belgium at the Olympics\">BEL</a>)</span></li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/Tim_Shaw_(swimmer)\" title=\"Tim Shaw (swimmer)\">Tim Shaw</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">USA</a>)</span></li>\\n</ul>\\n<h4><span class=\"mw-headline\" id=\"Others_in_Summer_Games\">Others in Summer Games</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=8\" title=\"Edit section: Others in Summer Games\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h4>\\n<ul>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/Morris_Kirksey\" title=\"Morris Kirksey\">Morris Kirksey</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">USA</a>)</span> (athletics and rugby)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/22px-Flag_of_Australia.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/33px-Flag_of_Australia.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/44px-Flag_of_Australia.svg.png 2x\" data-file-width=\"1280\" data-file-height=\"640\" /> <a href=\"/wiki/Edwin_Flack\" title=\"Edwin Flack\">Edwin Flack</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Australia_at_the_Olympics\" title=\"Australia at the Olympics\">AUS</a>)</span> (athletics and tennis)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/Karch_Kiraly\" title=\"Karch Kiraly\">Karch Kiraly</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">USA</a>)</span> (indoor volleyball and beach volleyball)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/41/Flag_of_Austria.svg/22px-Flag_of_Austria.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/4/41/Flag_of_Austria.svg/33px-Flag_of_Austria.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/4/41/Flag_of_Austria.svg/44px-Flag_of_Austria.svg.png 2x\" data-file-width=\"900\" data-file-height=\"600\" /> <a href=\"/wiki/Otto_Herschmann\" title=\"Otto Herschmann\">Otto Herschmann</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Austria_at_the_Olympics\" title=\"Austria at the Olympics\">AUT</a>)</span> (swimming and fencing)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Carl_Schuhmann\" title=\"Carl Schuhmann\">Carl Schuhmann</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">GER</a>)</span> (gymnastics and wrestling)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/22px-Flag_of_the_United_Kingdom.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/33px-Flag_of_the_United_Kingdom.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/ae/Flag_of_the_United_Kingdom.svg/44px-Flag_of_the_United_Kingdom.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Rebecca_Romero\" title=\"Rebecca Romero\">Rebecca Romero</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Great_Britain_at_the_Olympics\" title=\"Great Britain at the Olympics\">GBR</a>)</span> (cycling and rowing)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Roswitha_Krause\" title=\"Roswitha Krause\">Roswitha Krause</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">GER</a>)</span> (swimming and handball)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/Walter_W._Winans\" title=\"Walter W. Winans\">Walter W. Winans</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">USA</a>)</span> (shooting and sculpture)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/22px-Flag_of_Hungary.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/33px-Flag_of_Hungary.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/c/c1/Flag_of_Hungary.svg/44px-Flag_of_Hungary.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Alfr%C3%A9d_Haj%C3%B3s\" title=\"Alfr\\xc3\\xa9d Haj\\xc3\\xb3s\">Alfr\\xc3\\xa9d Haj\\xc3\\xb3s</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Hungary_at_the_Olympics\" title=\"Hungary at the Olympics\">HUN</a>)</span> (swimming and architecture)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/Conn_Findlay\" title=\"Conn Findlay\">Conn Findlay</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">USA</a>)</span> (rowing and sailing)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/22px-Flag_of_Sweden.svg.png\" width=\"22\" height=\"14\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/33px-Flag_of_Sweden.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/44px-Flag_of_Sweden.svg.png 2x\" data-file-width=\"1600\" data-file-height=\"1000\" /> <a href=\"/wiki/Daniel_Norling\" title=\"Daniel Norling\">Daniel Norling</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Sweden_at_the_Olympics\" title=\"Sweden at the Olympics\">SWE</a>)</span> (gymnastics and equestrian)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Fritz_Hofmann_(athlete)\" title=\"Fritz Hofmann (athlete)\">Fritz Hofmann</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">GER</a>)</span> (gymnastics and athletics)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/22px-Flag_of_Sweden.svg.png\" width=\"22\" height=\"14\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/33px-Flag_of_Sweden.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/44px-Flag_of_Sweden.svg.png 2x\" data-file-width=\"1600\" data-file-height=\"1000\" /> <a href=\"/wiki/Oswald_Holmberg\" title=\"Oswald Holmberg\">Oswald Holmberg</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Sweden_at_the_Olympics\" title=\"Sweden at the Olympics\">SWE</a>)</span> (gymnastics and tug of war)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/22px-Flag_of_Sweden.svg.png\" width=\"22\" height=\"14\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/33px-Flag_of_Sweden.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/4/4c/Flag_of_Sweden.svg/44px-Flag_of_Sweden.svg.png 2x\" data-file-width=\"1600\" data-file-height=\"1000\" /> <a href=\"/wiki/Gustaf_Dyrssen\" title=\"Gustaf Dyrssen\">Gustaf Dyrssen</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Sweden_at_the_Olympics\" title=\"Sweden at the Olympics\">SWE</a>)</span> (modern pentathlon and fencing)</li>\\n</ul>\\n<h3><span class=\"mw-headline\" id=\"In_the_Winter_Games\">In the Winter Games</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=9\" title=\"Edit section: In the Winter Games\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h3>\\n<h4><span class=\"mw-headline\" id=\"Cross-country_skiing_and_Nordic_combined\">Cross-country skiing and Nordic combined</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=10\" title=\"Edit section: Cross-country skiing and Nordic combined\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h4>\\n<ul>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Thorleif_Haug\" title=\"Thorleif Haug\">Thorleif Haug</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">NOR</a>)</span></li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Johan_Gr%C3%B8ttumsbr%C3%A5ten\" title=\"Johan Gr\\xc3\\xb8ttumsbr\\xc3\\xa5ten\">Johan Gr\\xc3\\xb8ttumsbr\\xc3\\xa5ten</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">NOR</a>)</span></li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Thoralf_Str%C3%B8mstad\" title=\"Thoralf Str\\xc3\\xb8mstad\">Thoralf Str\\xc3\\xb8mstad</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">NOR</a>)</span></li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/22px-Flag_of_Norway.svg.png\" width=\"22\" height=\"16\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/33px-Flag_of_Norway.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Flag_of_Norway.svg/44px-Flag_of_Norway.svg.png 2x\" data-file-width=\"1100\" data-file-height=\"800\" /> <a href=\"/wiki/Oddbj%C3%B8rn_Hagen\" title=\"Oddbj\\xc3\\xb8rn Hagen\">Oddbj\\xc3\\xb8rn Hagen</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Norway_at_the_Olympics\" title=\"Norway at the Olympics\">NOR</a>)</span></li>\\n</ul>\\n<h4><span class=\"mw-headline\" id=\"Others_in_Winter_Games\">Others in Winter Games</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=11\" title=\"Edit section: Others in Winter Games\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h4>\\n<ul>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/22px-Flag_of_the_Soviet_Union.svg.png\" width=\"22\" height=\"11\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/33px-Flag_of_the_Soviet_Union.svg.png 1.5x, //upload.wikimedia.org/wikipedia/commons/thumb/a/a9/Flag_of_the_Soviet_Union.svg/44px-Flag_of_the_Soviet_Union.svg.png 2x\" data-file-width=\"1200\" data-file-height=\"600\" /> <a href=\"/wiki/Anfisa_Reztsova\" title=\"Anfisa Reztsova\">Anfisa Reztsova</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Soviet_Union_at_the_Olympics\" title=\"Soviet Union at the Olympics\">URS</a>)</span> (biathlon and cross-country skiing)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/22px-Flag_of_Germany.svg.png\" width=\"22\" height=\"13\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/33px-Flag_of_Germany.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/b/ba/Flag_of_Germany.svg/44px-Flag_of_Germany.svg.png 2x\" data-file-width=\"1000\" data-file-height=\"600\" /> <a href=\"/wiki/Susi_Erdmann\" title=\"Susi Erdmann\">Susi Erdmann</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Germany_at_the_Olympics\" title=\"Germany at the Olympics\">GER</a>)</span> (luge and bobsleigh)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/22px-Flag_of_Italy.svg.png\" width=\"22\" height=\"15\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/33px-Flag_of_Italy.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/0/03/Flag_of_Italy.svg/44px-Flag_of_Italy.svg.png 2x\" data-file-width=\"1500\" data-file-height=\"1000\" /> <a href=\"/wiki/Gerda_Weissensteiner\" title=\"Gerda Weissensteiner\">Gerda Weissensteiner</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/Italy_at_the_Olympics\" title=\"Italy at the Olympics\">ITA</a>)</span> (luge and bobsleigh)</li>\\n<li><img alt=\"\" src=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/22px-Flag_of_the_United_States.svg.png\" width=\"22\" height=\"12\" class=\"thumbborder\" srcset=\"//upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/33px-Flag_of_the_United_States.svg.png 1.5x, //upload.wikimedia.org/wikipedia/en/thumb/a/a4/Flag_of_the_United_States.svg/44px-Flag_of_the_United_States.svg.png 2x\" data-file-width=\"1235\" data-file-height=\"650\" /> <a href=\"/wiki/Eric_Flaim\" title=\"Eric Flaim\">Eric Flaim</a> <span style=\"font-size:90%;\">(<a href=\"/wiki/United_States_at_the_Olympics\" title=\"United States at the Olympics\">USA</a>)</span> (long track speedskating and short track speedskating)</li>\\n</ul>\\n<h2><span class=\"mw-headline\" id=\"See_also\">See also</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=12\" title=\"Edit section: See also\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h2>\\n<ul>\\n<li><a href=\"/wiki/List_of_multiple_Olympic_medalists_at_a_single_Games\" title=\"List of multiple Olympic medalists at a single Games\">List of multiple Olympic medalists at a single Games</a></li>\\n<li><a href=\"/wiki/List_of_multiple_Olympic_medalists_in_one_event\" title=\"List of multiple Olympic medalists in one event\">List of multiple Olympic medalists in one event</a></li>\\n<li><a href=\"/wiki/List_of_multiple_Olympic_gold_medalists\" title=\"List of multiple Olympic gold medalists\">List of multiple Olympic gold medalists</a></li>\\n<li><a href=\"/wiki/List_of_multiple_Olympic_gold_medalists_at_a_single_Games\" title=\"List of multiple Olympic gold medalists at a single Games\">List of multiple Olympic gold medalists at a single Games</a></li>\\n<li><a href=\"/wiki/List_of_multiple_Olympic_gold_medalists_in_one_event\" title=\"List of multiple Olympic gold medalists in one event\">List of multiple Olympic gold medalists in one event</a></li>\\n<li><a href=\"/wiki/List_of_multiple_Winter_Olympic_medallists\" title=\"List of multiple Winter Olympic medallists\">List of multiple Winter Olympic medallists</a></li>\\n<li><a href=\"/wiki/List_of_athletes_with_the_most_appearances_at_Olympic_Games\" title=\"List of athletes with the most appearances at Olympic Games\">List of athletes with the most appearances at Olympic Games</a></li>\\n<li><a href=\"/wiki/All-time_Olympic_Games_medal_table\" title=\"All-time Olympic Games medal table\">All-time Olympic Games medal table</a></li>\\n<li><a href=\"/wiki/Leonidas_of_Rhodes\" title=\"Leonidas of Rhodes\">Leonidas of Rhodes</a></li>\\n</ul>\\n<h2><span class=\"mw-headline\" id=\"References\">References</span><span class=\"mw-editsection\"><span class=\"mw-editsection-bracket\">[</span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit&section=13\" title=\"Edit section: References\">edit</a><span class=\"mw-editsection-bracket\">]</span></span></h2>\\n<div class=\"reflist\" style=\"list-style-type: decimal;\">\\n<div class=\"mw-references-wrap\">\\n<ol class=\"references\">\\n<li id=\"cite_note-ioc1896-1\"><span class=\"mw-cite-backlink\">^ <a href=\"#cite_ref-ioc1896_1-0\"><sup><i><b>a</b></i></sup></a> <a href=\"#cite_ref-ioc1896_1-1\"><sup><i><b>b</b></i></sup></a> <a href=\"#cite_ref-ioc1896_1-2\"><sup><i><b>c</b></i></sup></a></span> <span class=\"reference-text\"><cite class=\"citation book\"><a href=\"/wiki/Pierre_de_Coubertin\" title=\"Pierre de Coubertin\">Coubertin, Pierre de</a>; Timoleon J. Philimon; N. G. Politis; Ch. Anninos (1897). <a rel=\"nofollow\" class=\"external text\" href=\"http://www.la84foundation.org/6oic/OfficialReports/1896/1896part2.pdf\">\"Second Part: The Olympic Games in 1896\"</a> <span style=\"font-size:85%;\">(PDF)</span>. <i>The Olympic Games, B.C. 776 \\xe2\\x80\\x93 A.D. 1896</i> <span style=\"font-size:85%;\">(<a href=\"/wiki/PDF\" class=\"mw-redirect\" title=\"PDF\">PDF</a>)</span><span style=\"display:none;font-size:100%\" class=\"error citation-comment\"><code style=\"color:inherit; border:inherit; padding:inherit;\">|format=</code> requires <code style=\"color:inherit; border:inherit; padding:inherit;\">|url=</code> (<a href=\"/wiki/Help:CS1_errors#format_missing_url\" title=\"Help:CS1 errors\">help</a>)</span>. IOC official Olympic reports. translated by A. v. K. London: Grevel. pp. 55\\xe2\\x80\\x9381 (passim)<span class=\"reference-accessdate\">. Retrieved <span class=\"nowrap\">13 August</span> 2008</span>.</cite><span title=\"ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Abook&rft.genre=bookitem&rft.atitle=Second+Part%3A+The+Olympic+Games+in+1896&rft.btitle=The+Olympic+Games%2C+B.C.+776+%E2%80%93+A.D.+1896&rft.place=London&rft.series=IOC+official+Olympic+reports&rft.pages=55-81+%28passim%29&rft.pub=Grevel&rft.date=1897&rft.aulast=Coubertin&rft.aufirst=Pierre+de&rft.au=Timoleon+J.+Philimon&rft.au=N.+G.+Politis&rft.au=Ch.+Anninos&rft_id=http%3A%2F%2Fwww.la84foundation.org%2F6oic%2FOfficialReports%2F1896%2F1896part2.pdf&rfr_id=info%3Asid%2Fen.wikipedia.org%3AList+of+multiple+Olympic+medalists\" class=\"Z3988\"><span style=\"display:none;\"> </span></span> (note: the source gives dates in the <a href=\"/wiki/Julian_calendar\" title=\"Julian calendar\">Julian calendar</a>; this table uses the <a href=\"/wiki/Gregorian_calendar\" title=\"Gregorian calendar\">Gregorian calendar</a>)</span></li>\\n<li id=\"cite_note-2\"><span class=\"mw-cite-backlink\"><b><a href=\"#cite_ref-2\">^</a></b></span> <span class=\"reference-text\"><cite class=\"citation web\"><a rel=\"nofollow\" class=\"external text\" href=\"http://www.sports-reference.com/olympics/athletes/ea/eddie-eagan-1.html\">\"Eddie Eagan\"</a>. sports-reference.com<span class=\"reference-accessdate\">. Retrieved <span class=\"nowrap\">9 May</span> 2012</span>.</cite><span title=\"ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Abook&rft.genre=unknown&rft.btitle=Eddie+Eagan&rft.pub=sports-reference.com&rft_id=http%3A%2F%2Fwww.sports-reference.com%2Folympics%2Fathletes%2Fea%2Feddie-eagan-1.html&rfr_id=info%3Asid%2Fen.wikipedia.org%3AList+of+multiple+Olympic+medalists\" class=\"Z3988\"><span style=\"display:none;\"> </span></span></span></li>\\n<li id=\"cite_note-3\"><span class=\"mw-cite-backlink\"><b><a href=\"#cite_ref-3\">^</a></b></span> <span class=\"reference-text\"><cite class=\"citation web\"><a rel=\"nofollow\" class=\"external text\" href=\"http://www.sports-reference.com/olympics/athletes/th/jacob-tullin-thams-1.html\">\"Jacob Tullin Thams\"</a>. sports-reference.com<span class=\"reference-accessdate\">. Retrieved <span class=\"nowrap\">9 May</span> 2012</span>.</cite><span title=\"ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Abook&rft.genre=unknown&rft.btitle=Jacob+Tullin+Thams&rft.pub=sports-reference.com&rft_id=http%3A%2F%2Fwww.sports-reference.com%2Folympics%2Fathletes%2Fth%2Fjacob-tullin-thams-1.html&rfr_id=info%3Asid%2Fen.wikipedia.org%3AList+of+multiple+Olympic+medalists\" class=\"Z3988\"><span style=\"display:none;\"> </span></span></span></li>\\n<li id=\"cite_note-4\"><span class=\"mw-cite-backlink\"><b><a href=\"#cite_ref-4\">^</a></b></span> <span class=\"reference-text\"><cite class=\"citation web\"><a rel=\"nofollow\" class=\"external text\" href=\"http://www.sports-reference.com/olympics/athletes/ro/christa-rothenburger-luding-1.html\">\"Christa Rothenburger-Luding\"</a>. sports-reference.com<span class=\"reference-accessdate\">. Retrieved <span class=\"nowrap\">9 May</span> 2012</span>.</cite><span title=\"ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Abook&rft.genre=unknown&rft.btitle=Christa+Rothenburger-Luding&rft.pub=sports-reference.com&rft_id=http%3A%2F%2Fwww.sports-reference.com%2Folympics%2Fathletes%2Fro%2Fchrista-rothenburger-luding-1.html&rfr_id=info%3Asid%2Fen.wikipedia.org%3AList+of+multiple+Olympic+medalists\" class=\"Z3988\"><span style=\"display:none;\"> </span></span></span></li>\\n<li id=\"cite_note-5\"><span class=\"mw-cite-backlink\"><b><a href=\"#cite_ref-5\">^</a></b></span> <span class=\"reference-text\"><cite class=\"citation web\"><a rel=\"nofollow\" class=\"external text\" href=\"http://www.sports-reference.com/olympics/athletes/hu/clara-hughes-1.html\">\"Clara Hughes\"</a>. sports-reference.com<span class=\"reference-accessdate\">. Retrieved <span class=\"nowrap\">9 May</span> 2012</span>.</cite><span title=\"ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Abook&rft.genre=unknown&rft.btitle=Clara+Hughes&rft.pub=sports-reference.com&rft_id=http%3A%2F%2Fwww.sports-reference.com%2Folympics%2Fathletes%2Fhu%2Fclara-hughes-1.html&rfr_id=info%3Asid%2Fen.wikipedia.org%3AList+of+multiple+Olympic+medalists\" class=\"Z3988\"><span style=\"display:none;\"> </span></span></span></li>\\n<li id=\"cite_note-6\"><span class=\"mw-cite-backlink\"><b><a href=\"#cite_ref-6\">^</a></b></span> <span class=\"reference-text\"><cite class=\"citation news\">Borden, Sam (19 February 2014). <a rel=\"nofollow\" class=\"external text\" href=\"https://www.nytimes.com/2014/02/20/sports/olympics/canada-catches-us-in-final-bobsled-run.html?ref=sports&_r=0\">\"Canada Catches U.S. in Final Bobsled Run\"</a>. <i>New York Times</i><span class=\"reference-accessdate\">. Retrieved <span class=\"nowrap\">19 February</span> 2014</span>.</cite><span title=\"ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.jtitle=New+York+Times&rft.atitle=Canada+Catches+U.S.+in+Final+Bobsled+Run&rft.date=2014-02-19&rft.aulast=Borden&rft.aufirst=Sam&rft_id=https%3A%2F%2Fwww.nytimes.com%2F2014%2F02%2F20%2Fsports%2Folympics%2Fcanada-catches-us-in-final-bobsled-run.html%3Fref%3Dsports%26_r%3D0&rfr_id=info%3Asid%2Fen.wikipedia.org%3AList+of+multiple+Olympic+medalists\" class=\"Z3988\"><span style=\"display:none;\"> </span></span></span></li>\\n</ol>\\n</div>\\n</div>\\n<ul>\\n<li><cite class=\"citation web\"><a rel=\"nofollow\" class=\"external text\" href=\"http://www.olympic.org/uk/athletes/results/search_r_uk.asp\">\"Olympic Medal Winners\"</a>. <a href=\"/wiki/International_Olympic_Committee\" title=\"International Olympic Committee\">International Olympic Committee</a><span class=\"reference-accessdate\">. Retrieved <span class=\"nowrap\">3 April</span> 2007</span>.</cite><span title=\"ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Abook&rft.genre=unknown&rft.btitle=Olympic+Medal+Winners&rft.pub=International+Olympic+Committee&rft_id=http%3A%2F%2Fwww.olympic.org%2Fuk%2Fathletes%2Fresults%2Fsearch_r_uk.asp&rfr_id=info%3Asid%2Fen.wikipedia.org%3AList+of+multiple+Olympic+medalists\" class=\"Z3988\"><span style=\"display:none;\"> </span></span></li>\\n<li><cite class=\"citation web\"><a rel=\"nofollow\" class=\"external text\" href=\"https://web.archive.org/web/20070318010246/http://www.databaseolympics.com/index.htm\">\"databaseOlympics.com\"</a>. databaseSports.com. Archived from <a rel=\"nofollow\" class=\"external text\" href=\"http://www.databaseolympics.com/index.htm\">the original</a> on 18 March 2007<span class=\"reference-accessdate\">. Retrieved <span class=\"nowrap\">20 March</span> 2007</span>.</cite><span title=\"ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Abook&rft.genre=unknown&rft.btitle=databaseOlympics.com&rft.pub=databaseSports.com&rft_id=http%3A%2F%2Fwww.databaseolympics.com%2Findex.htm&rfr_id=info%3Asid%2Fen.wikipedia.org%3AList+of+multiple+Olympic+medalists\" class=\"Z3988\"><span style=\"display:none;\"> </span></span></li>\\n<li>See also references in the articles on each athlete.</li>\\n</ul>\\n<div role=\"navigation\" class=\"navbox\" aria-labelledby=\"Olympics_and_Paralympics_statistics\" style=\"padding:3px\">\\n<table class=\"nowraplinks collapsible autocollapse navbox-inner\" style=\"border-spacing:0;background:transparent;color:inherit\">\\n<tr>\\n<th scope=\"col\" class=\"navbox-title\" colspan=\"2\" style=\"background: #BFD7FF\">\\n<div class=\"plainlinks hlist navbar mini\">\\n<ul>\\n<li class=\"nv-view\"><a href=\"/wiki/Template:Olympics_statistics\" title=\"Template:Olympics statistics\"><abbr title=\"View this template\" style=\";background: #BFD7FF;background:none transparent;border:none;-moz-box-shadow:none;-webkit-box-shadow:none;box-shadow:none;\">v</abbr></a></li>\\n<li class=\"nv-talk\"><a href=\"/wiki/Template_talk:Olympics_statistics\" title=\"Template talk:Olympics statistics\"><abbr title=\"Discuss this template\" style=\";background: #BFD7FF;background:none transparent;border:none;-moz-box-shadow:none;-webkit-box-shadow:none;box-shadow:none;\">t</abbr></a></li>\\n<li class=\"nv-edit\"><a class=\"external text\" href=\"//en.wikipedia.org/w/index.php?title=Template:Olympics_statistics&action=edit\"><abbr title=\"Edit this template\" style=\";background: #BFD7FF;background:none transparent;border:none;-moz-box-shadow:none;-webkit-box-shadow:none;box-shadow:none;\">e</abbr></a></li>\\n</ul>\\n</div>\\n<div id=\"Olympics_and_Paralympics_statistics\" style=\"font-size:114%;margin:0 4em\"><a href=\"/wiki/Olympic_Games\" title=\"Olympic Games\">Olympics</a> and <a href=\"/wiki/Paralympic_Games\" title=\"Paralympic Games\">Paralympics</a> statistics</div>\\n</th>\\n</tr>\\n<tr>\\n<th scope=\"row\" class=\"navbox-group\" style=\"width:1%;text-align:center; background: #DDEBFF\">Medalists and medal tables</th>\\n<td class=\"navbox-list navbox-odd hlist\" style=\"text-align:left;border-left-width:2px;border-left-style:solid;width:100%;padding:0px\">\\n<div style=\"padding:0em 0.25em\">\\n<ul>\\n<li><a href=\"/wiki/Lists_of_Olympic_medalists\" title=\"Lists of Olympic medalists\">Lists of Olympic medalists</a></li>\\n<li><a href=\"/wiki/Lists_of_Paralympic_medalists\" title=\"Lists of Paralympic medalists\">Lists of Paralympic medalists</a></li>\\n<li><a href=\"/wiki/All-time_Olympic_Games_medal_table\" title=\"All-time Olympic Games medal table\">All-time Olympic Games medal table</a></li>\\n<li><a href=\"/wiki/All-time_Paralympic_Games_medal_table\" title=\"All-time Paralympic Games medal table\">All-time Paralympic Games medal table</a></li>\\n</ul>\\n</div>\\n</td>\\n</tr>\\n<tr>\\n<th scope=\"row\" class=\"navbox-group\" style=\"width:1%;text-align:center; background: #DDEBFF\">Multiple medalists</th>\\n<td class=\"navbox-list navbox-even hlist\" style=\"text-align:left;border-left-width:2px;border-left-style:solid;width:100%;padding:0px\">\\n<div style=\"padding:0em 0.25em\">\\n<ul>\\n<li><a class=\"mw-selflink selflink\">Multiple Olympic medalists</a>\\n<ul>\\n<li><a href=\"/wiki/List_of_multiple_Olympic_medalists_at_a_single_Games\" title=\"List of multiple Olympic medalists at a single Games\">at a single Games</a></li>\\n<li><a href=\"/wiki/List_of_multiple_Olympic_medalists_in_one_event\" title=\"List of multiple Olympic medalists in one event\">in one event</a></li>\\n<li><a href=\"/wiki/List_of_multiple_Winter_Olympic_medallists\" title=\"List of multiple Winter Olympic medallists\">at Winter Olympics</a></li>\\n</ul>\\n</li>\\n</ul>\\n</div>\\n</td>\\n</tr>\\n<tr>\\n<th scope=\"row\" class=\"navbox-group\" style=\"width:1%;text-align:center; background: #DDEBFF\">Multiple gold medalists</th>\\n<td class=\"navbox-list navbox-odd hlist\" style=\"text-align:left;border-left-width:2px;border-left-style:solid;width:100%;padding:0px\">\\n<div style=\"padding:0em 0.25em\">\\n<ul>\\n<li><a href=\"/wiki/List_of_multiple_Olympic_gold_medalists\" title=\"List of multiple Olympic gold medalists\">Multiple Olympic gold medalists</a>\\n<ul>\\n<li><a href=\"/wiki/List_of_multiple_Olympic_gold_medalists_at_a_single_Games\" title=\"List of multiple Olympic gold medalists at a single Games\">at a single Games</a></li>\\n<li><a href=\"/wiki/List_of_multiple_Olympic_gold_medalists_in_one_event\" title=\"List of multiple Olympic gold medalists in one event\">in one event</a></li>\\n</ul>\\n</li>\\n<li><a href=\"/wiki/List_of_multiple_Paralympic_gold_medalists\" title=\"List of multiple Paralympic gold medalists\">Multiple Paralympic gold medalists</a>\\n<ul>\\n<li><a href=\"/wiki/List_of_multiple_Paralympic_gold_medalists_at_a_single_Games\" title=\"List of multiple Paralympic gold medalists at a single Games\">at a single Games</a></li>\\n</ul>\\n</li>\\n</ul>\\n</div>\\n</td>\\n</tr>\\n<tr>\\n<th scope=\"row\" class=\"navbox-group\" style=\"width:1%;text-align:center; background: #DDEBFF\">Other statistics</th>\\n<td class=\"navbox-list navbox-even hlist\" style=\"text-align:left;border-left-width:2px;border-left-style:solid;width:100%;padding:0px\">\\n<div style=\"padding:0em 0.25em\">\\n<ul>\\n<li><a href=\"/wiki/List_of_athletes_with_Olympic_medals_in_different_disciplines\" title=\"List of athletes with Olympic medals in different disciplines\">Olympic medals in different sports</a>\\n<ul>\\n<li><a href=\"/wiki/List_of_multiple_Olympic_medalists#Athletes_with_medals_in_different_disciplines\" title=\"List of multiple Olympic medalists\">at Summer and Winter Olympics</a></li>\\n</ul>\\n</li>\\n<li><a href=\"/wiki/List_of_athletes_with_the_most_appearances_at_Olympic_Games\" title=\"List of athletes with the most appearances at Olympic Games\">Most appearances at Olympic Games</a></li>\\n<li><a href=\"/wiki/List_of_medal_sweeps_in_Olympic_athletics\" title=\"List of medal sweeps in Olympic athletics\">Medal sweeps in Olympic athletics</a></li>\\n<li><a href=\"/wiki/List_of_athletes_who_competed_in_both_the_Summer_and_Winter_Olympic_games\" title=\"List of athletes who competed in both the Summer and Winter Olympic games\">Athletes at both the Summer and Winter Olympics</a></li>\\n<li><a href=\"/wiki/List_of_ties_for_medals_at_the_Olympics\" title=\"List of ties for medals at the Olympics\">Ties for medals at the Olympics</a></li>\\n<li><a href=\"/wiki/List_of_stripped_Olympic_medals\" title=\"List of stripped Olympic medals\">Stripped Olympic medals</a></li>\\n<li><a href=\"/wiki/List_of_Olympic_medals_by_host_nation\" title=\"List of Olympic medals by host nation\">Olympic medals by host nation</a></li>\\n<li><a href=\"/wiki/List_of_Olympic_medalist_families\" title=\"List of Olympic medalist families\">Olympic medalist families</a></li>\\n</ul>\\n</div>\\n</td>\\n</tr>\\n</table>\\n</div>\\n\\n\\n<!-- \\nNewPP limit report\\nParsed by mw1325\\nCached time: 20180215103610\\nCache expiry: 1900800\\nDynamic content: false\\nCPU time usage: 1.672 seconds\\nReal time usage: 1.777 seconds\\nPreprocessor visited node count: 34561/1000000\\nPreprocessor generated node count: 0/1500000\\nPost\\xe2\\x80\\x90expand include size: 159304/2097152 bytes\\nTemplate argument size: 32222/2097152 bytes\\nHighest expansion depth: 15/40\\nExpensive parser function count: 1/500\\nLua time usage: 0.877/10.000 seconds\\nLua memory usage: 3.68 MB/50 MB\\n-->\\n<!--\\nTransclusion expansion time report (%,ms,calls,template)\\n100.00% 1402.375 1 -total\\n 81.09% 1137.131 632 Template:Country_alias\\n 74.72% 1047.815 190 Template:FlagIOC\\n 73.84% 1035.512 190 Template:FlagIOC2\\n 12.03% 168.721 31 Template:FlagIOCathlete\\n 11.78% 165.221 31 Template:FlagIOC2athlete\\n 4.68% 65.630 1 Template:Reflist\\n 3.42% 47.955 158 Template:Sortname\\n 2.42% 33.881 1 Template:Cite_book\\n 1.64% 22.981 1 Template:Expand_list\\n-->\\n</div>\\n<!-- Saved in parser cache with key enwiki:pcache:idhash:18855244-0!canonical and timestamp 20180215103608 and revision id 825763339\\n -->\\n<noscript><img src=\"//en.wikipedia.org/wiki/Special:CentralAutoLogin/start?type=1x1\" alt=\"\" title=\"\" width=\"1\" height=\"1\" style=\"border: none; position: absolute;\" /></noscript></div>\\t\\t\\t\\t\\t<div class=\"printfooter\">\\n\\t\\t\\t\\t\\t\\tRetrieved from \"<a dir=\"ltr\" href=\"https://en.wikipedia.org/w/index.php?title=List_of_multiple_Olympic_medalists&oldid=825763339\">https://en.wikipedia.org/w/index.php?title=List_of_multiple_Olympic_medalists&oldid=825763339</a>\"\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t<div id=\"catlinks\" class=\"catlinks\" data-mw=\"interface\"><div id=\"mw-normal-catlinks\" class=\"mw-normal-catlinks\"><a href=\"/wiki/Help:Category\" title=\"Help:Category\">Categories</a>: <ul><li><a href=\"/wiki/Category:Lists_of_Olympic_medalists\" title=\"Category:Lists of Olympic medalists\">Lists of Olympic medalists</a></li><li><a href=\"/wiki/Category:Olympic_Games_medal_tables\" title=\"Category:Olympic Games medal tables\">Olympic Games medal tables</a></li></ul></div><div id=\"mw-hidden-catlinks\" class=\"mw-hidden-catlinks mw-hidden-cats-hidden\">Hidden categories: <ul><li><a href=\"/wiki/Category:Pages_using_citations_with_format_and_no_URL\" title=\"Category:Pages using citations with format and no URL\">Pages using citations with format and no URL</a></li><li><a href=\"/wiki/Category:Articles_with_hCards\" title=\"Category:Articles with hCards\">Articles with hCards</a></li><li><a href=\"/wiki/Category:Incomplete_lists_from_May_2012\" title=\"Category:Incomplete lists from May 2012\">Incomplete lists from May 2012</a></li></ul></div></div>\\t\\t\\t\\t<div class=\"visualClear\"></div>\\n\\t\\t\\t\\t\\t\\t\\t</div>\\n\\t\\t</div>\\n\\t\\t<div id=\"mw-navigation\">\\n\\t\\t\\t<h2>Navigation menu</h2>\\n\\t\\t\\t<div id=\"mw-head\">\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t<div id=\"p-personal\" role=\"navigation\" class=\"\" aria-labelledby=\"p-personal-label\">\\n\\t\\t\\t\\t\\t\\t<h3 id=\"p-personal-label\">Personal tools</h3>\\n\\t\\t\\t\\t\\t\\t<ul>\\n\\t\\t\\t\\t\\t\\t\\t<li id=\"pt-anonuserpage\">Not logged in</li><li id=\"pt-anontalk\"><a href=\"/wiki/Special:MyTalk\" title=\"Discussion about edits from this IP address [n]\" accesskey=\"n\">Talk</a></li><li id=\"pt-anoncontribs\"><a href=\"/wiki/Special:MyContributions\" title=\"A list of edits made from this IP address [y]\" accesskey=\"y\">Contributions</a></li><li id=\"pt-createaccount\"><a href=\"/w/index.php?title=Special:CreateAccount&returnto=List+of+multiple+Olympic+medalists\" title=\"You are encouraged to create an account and log in; however, it is not mandatory\">Create account</a></li><li id=\"pt-login\"><a href=\"/w/index.php?title=Special:UserLogin&returnto=List+of+multiple+Olympic+medalists\" title=\"You're encouraged to log in; however, it's not mandatory. [o]\" accesskey=\"o\">Log in</a></li>\\t\\t\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t<div id=\"left-navigation\">\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t<div id=\"p-namespaces\" role=\"navigation\" class=\"vectorTabs\" aria-labelledby=\"p-namespaces-label\">\\n\\t\\t\\t\\t\\t\\t<h3 id=\"p-namespaces-label\">Namespaces</h3>\\n\\t\\t\\t\\t\\t\\t<ul>\\n\\t\\t\\t\\t\\t\\t\\t<li id=\"ca-nstab-main\" class=\"selected\"><span><a href=\"/wiki/List_of_multiple_Olympic_medalists\" title=\"View the content page [c]\" accesskey=\"c\">Article</a></span></li><li id=\"ca-talk\"><span><a href=\"/wiki/Talk:List_of_multiple_Olympic_medalists\" rel=\"discussion\" title=\"Discussion about the content page [t]\" accesskey=\"t\">Talk</a></span></li>\\t\\t\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t<div id=\"p-variants\" role=\"navigation\" class=\"vectorMenu emptyPortlet\" aria-labelledby=\"p-variants-label\">\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t<h3 id=\"p-variants-label\">\\n\\t\\t\\t\\t\\t\\t\\t<span>Variants</span>\\n\\t\\t\\t\\t\\t\\t</h3>\\n\\t\\t\\t\\t\\t\\t<div class=\"menu\">\\n\\t\\t\\t\\t\\t\\t\\t<ul>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t<div id=\"right-navigation\">\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t<div id=\"p-views\" role=\"navigation\" class=\"vectorTabs\" aria-labelledby=\"p-views-label\">\\n\\t\\t\\t\\t\\t\\t<h3 id=\"p-views-label\">Views</h3>\\n\\t\\t\\t\\t\\t\\t<ul>\\n\\t\\t\\t\\t\\t\\t\\t<li id=\"ca-view\" class=\"collapsible selected\"><span><a href=\"/wiki/List_of_multiple_Olympic_medalists\">Read</a></span></li><li id=\"ca-edit\" class=\"collapsible\"><span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=edit\" title=\"Edit this page [e]\" accesskey=\"e\">Edit</a></span></li><li id=\"ca-history\" class=\"collapsible\"><span><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=history\" title=\"Past revisions of this page [h]\" accesskey=\"h\">View history</a></span></li>\\t\\t\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t<div id=\"p-cactions\" role=\"navigation\" class=\"vectorMenu emptyPortlet\" aria-labelledby=\"p-cactions-label\">\\n\\t\\t\\t\\t\\t\\t<h3 id=\"p-cactions-label\"><span>More</span></h3>\\n\\t\\t\\t\\t\\t\\t<div class=\"menu\">\\n\\t\\t\\t\\t\\t\\t\\t<ul>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t<div id=\"p-search\" role=\"search\">\\n\\t\\t\\t\\t\\t\\t<h3>\\n\\t\\t\\t\\t\\t\\t\\t<label for=\"searchInput\">Search</label>\\n\\t\\t\\t\\t\\t\\t</h3>\\n\\t\\t\\t\\t\\t\\t<form action=\"/w/index.php\" id=\"searchform\">\\n\\t\\t\\t\\t\\t\\t\\t<div id=\"simpleSearch\">\\n\\t\\t\\t\\t\\t\\t\\t\\t<input type=\"search\" name=\"search\" placeholder=\"Search Wikipedia\" title=\"Search Wikipedia [f]\" accesskey=\"f\" id=\"searchInput\"/><input type=\"hidden\" value=\"Special:Search\" name=\"title\"/><input type=\"submit\" name=\"fulltext\" value=\"Search\" title=\"Search Wikipedia for this text\" id=\"mw-searchButton\" class=\"searchButton mw-fallbackSearchButton\"/><input type=\"submit\" name=\"go\" value=\"Go\" title=\"Go to a page with this exact name if it exists\" id=\"searchButton\" class=\"searchButton\"/>\\t\\t\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t\\t\\t</form>\\n\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t</div>\\n\\t\\t\\t</div>\\n\\t\\t\\t<div id=\"mw-panel\">\\n\\t\\t\\t\\t<div id=\"p-logo\" role=\"banner\"><a class=\"mw-wiki-logo\" href=\"/wiki/Main_Page\" title=\"Visit the main page\"></a></div>\\n\\t\\t\\t\\t\\t\\t<div class=\"portal\" role=\"navigation\" id=\\'p-navigation\\' aria-labelledby=\\'p-navigation-label\\'>\\n\\t\\t\\t<h3 id=\\'p-navigation-label\\'>Navigation</h3>\\n\\t\\t\\t<div class=\"body\">\\n\\t\\t\\t\\t\\t\\t\\t\\t<ul>\\n\\t\\t\\t\\t\\t<li id=\"n-mainpage-description\"><a href=\"/wiki/Main_Page\" title=\"Visit the main page [z]\" accesskey=\"z\">Main page</a></li><li id=\"n-contents\"><a href=\"/wiki/Portal:Contents\" title=\"Guides to browsing Wikipedia\">Contents</a></li><li id=\"n-featuredcontent\"><a href=\"/wiki/Portal:Featured_content\" title=\"Featured content \\xe2\\x80\\x93 the best of Wikipedia\">Featured content</a></li><li id=\"n-currentevents\"><a href=\"/wiki/Portal:Current_events\" title=\"Find background information on current events\">Current events</a></li><li id=\"n-randompage\"><a href=\"/wiki/Special:Random\" title=\"Load a random article [x]\" accesskey=\"x\">Random article</a></li><li id=\"n-sitesupport\"><a href=\"https://donate.wikimedia.org/wiki/Special:FundraiserRedirector?utm_source=donate&utm_medium=sidebar&utm_campaign=C13_en.wikipedia.org&uselang=en\" title=\"Support us\">Donate to Wikipedia</a></li><li id=\"n-shoplink\"><a href=\"//shop.wikimedia.org\" title=\"Visit the Wikipedia store\">Wikipedia store</a></li>\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t\\t\\t</div>\\n\\t\\t</div>\\n\\t\\t\\t<div class=\"portal\" role=\"navigation\" id=\\'p-interaction\\' aria-labelledby=\\'p-interaction-label\\'>\\n\\t\\t\\t<h3 id=\\'p-interaction-label\\'>Interaction</h3>\\n\\t\\t\\t<div class=\"body\">\\n\\t\\t\\t\\t\\t\\t\\t\\t<ul>\\n\\t\\t\\t\\t\\t<li id=\"n-help\"><a href=\"/wiki/Help:Contents\" title=\"Guidance on how to use and edit Wikipedia\">Help</a></li><li id=\"n-aboutsite\"><a href=\"/wiki/Wikipedia:About\" title=\"Find out about Wikipedia\">About Wikipedia</a></li><li id=\"n-portal\"><a href=\"/wiki/Wikipedia:Community_portal\" title=\"About the project, what you can do, where to find things\">Community portal</a></li><li id=\"n-recentchanges\"><a href=\"/wiki/Special:RecentChanges\" title=\"A list of recent changes in the wiki [r]\" accesskey=\"r\">Recent changes</a></li><li id=\"n-contactpage\"><a href=\"//en.wikipedia.org/wiki/Wikipedia:Contact_us\" title=\"How to contact Wikipedia\">Contact page</a></li>\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t\\t\\t</div>\\n\\t\\t</div>\\n\\t\\t\\t<div class=\"portal\" role=\"navigation\" id=\\'p-tb\\' aria-labelledby=\\'p-tb-label\\'>\\n\\t\\t\\t<h3 id=\\'p-tb-label\\'>Tools</h3>\\n\\t\\t\\t<div class=\"body\">\\n\\t\\t\\t\\t\\t\\t\\t\\t<ul>\\n\\t\\t\\t\\t\\t<li id=\"t-whatlinkshere\"><a href=\"/wiki/Special:WhatLinksHere/List_of_multiple_Olympic_medalists\" title=\"List of all English Wikipedia pages containing links to this page [j]\" accesskey=\"j\">What links here</a></li><li id=\"t-recentchangeslinked\"><a href=\"/wiki/Special:RecentChangesLinked/List_of_multiple_Olympic_medalists\" rel=\"nofollow\" title=\"Recent changes in pages linked from this page [k]\" accesskey=\"k\">Related changes</a></li><li id=\"t-upload\"><a href=\"/wiki/Wikipedia:File_Upload_Wizard\" title=\"Upload files [u]\" accesskey=\"u\">Upload file</a></li><li id=\"t-specialpages\"><a href=\"/wiki/Special:SpecialPages\" title=\"A list of all special pages [q]\" accesskey=\"q\">Special pages</a></li><li id=\"t-permalink\"><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&oldid=825763339\" title=\"Permanent link to this revision of the page\">Permanent link</a></li><li id=\"t-info\"><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&action=info\" title=\"More information about this page\">Page information</a></li><li id=\"t-wikibase\"><a href=\"https://www.wikidata.org/wiki/Special:EntityPage/Q6629540\" title=\"Link to connected data repository item [g]\" accesskey=\"g\">Wikidata item</a></li><li id=\"t-cite\"><a href=\"/w/index.php?title=Special:CiteThisPage&page=List_of_multiple_Olympic_medalists&id=825763339\" title=\"Information on how to cite this page\">Cite this page</a></li>\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t\\t\\t</div>\\n\\t\\t</div>\\n\\t\\t\\t<div class=\"portal\" role=\"navigation\" id=\\'p-coll-print_export\\' aria-labelledby=\\'p-coll-print_export-label\\'>\\n\\t\\t\\t<h3 id=\\'p-coll-print_export-label\\'>Print/export</h3>\\n\\t\\t\\t<div class=\"body\">\\n\\t\\t\\t\\t\\t\\t\\t\\t<ul>\\n\\t\\t\\t\\t\\t<li id=\"coll-create_a_book\"><a href=\"/w/index.php?title=Special:Book&bookcmd=book_creator&referer=List+of+multiple+Olympic+medalists\">Create a book</a></li><li id=\"coll-download-as-rdf2latex\"><a href=\"/w/index.php?title=Special:ElectronPdf&page=List+of+multiple+Olympic+medalists&action=show-download-screen\">Download as PDF</a></li><li id=\"t-print\"><a href=\"/w/index.php?title=List_of_multiple_Olympic_medalists&printable=yes\" title=\"Printable version of this page [p]\" accesskey=\"p\">Printable version</a></li>\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t\\t\\t</div>\\n\\t\\t</div>\\n\\t\\t\\t<div class=\"portal\" role=\"navigation\" id=\\'p-lang\\' aria-labelledby=\\'p-lang-label\\'>\\n\\t\\t\\t<h3 id=\\'p-lang-label\\'>Languages</h3>\\n\\t\\t\\t<div class=\"body\">\\n\\t\\t\\t\\t\\t\\t\\t\\t<ul>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t<div class=\"after-portlet after-portlet-lang\"><span class=\"wb-langlinks-add wb-langlinks-link\"><a href=\"https://www.wikidata.org/wiki/Special:EntityPage/Q6629540#sitelinks-wikipedia\" title=\"Add interlanguage links\" class=\"wbc-editpage\">Add links</a></span></div>\\t\\t\\t</div>\\n\\t\\t</div>\\n\\t\\t\\t\\t</div>\\n\\t\\t</div>\\n\\t\\t\\t\\t<div id=\"footer\" role=\"contentinfo\">\\n\\t\\t\\t\\t\\t\\t<ul id=\"footer-info\">\\n\\t\\t\\t\\t\\t\\t\\t\\t<li id=\"footer-info-lastmod\"> This page was last edited on 15 February 2018, at 07:33.</li>\\n\\t\\t\\t\\t\\t\\t\\t\\t<li id=\"footer-info-copyright\">Text is available under the <a rel=\"license\" href=\"//en.wikipedia.org/wiki/Wikipedia:Text_of_Creative_Commons_Attribution-ShareAlike_3.0_Unported_License\">Creative Commons Attribution-ShareAlike License</a><a rel=\"license\" href=\"//creativecommons.org/licenses/by-sa/3.0/\" style=\"display:none;\"></a>;\\nadditional terms may apply. By using this site, you agree to the <a href=\"//wikimediafoundation.org/wiki/Terms_of_Use\">Terms of Use</a> and <a href=\"//wikimediafoundation.org/wiki/Privacy_policy\">Privacy Policy</a>. Wikipedia\\xc2\\xae is a registered trademark of the <a href=\"//www.wikimediafoundation.org/\">Wikimedia Foundation, Inc.</a>, a non-profit organization.</li>\\n\\t\\t\\t\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t\\t<ul id=\"footer-places\">\\n\\t\\t\\t\\t\\t\\t\\t\\t<li id=\"footer-places-privacy\"><a href=\"https://wikimediafoundation.org/wiki/Privacy_policy\" class=\"extiw\" title=\"wmf:Privacy policy\">Privacy policy</a></li>\\n\\t\\t\\t\\t\\t\\t\\t\\t<li id=\"footer-places-about\"><a href=\"/wiki/Wikipedia:About\" title=\"Wikipedia:About\">About Wikipedia</a></li>\\n\\t\\t\\t\\t\\t\\t\\t\\t<li id=\"footer-places-disclaimer\"><a href=\"/wiki/Wikipedia:General_disclaimer\" title=\"Wikipedia:General disclaimer\">Disclaimers</a></li>\\n\\t\\t\\t\\t\\t\\t\\t\\t<li id=\"footer-places-contact\"><a href=\"//en.wikipedia.org/wiki/Wikipedia:Contact_us\">Contact Wikipedia</a></li>\\n\\t\\t\\t\\t\\t\\t\\t\\t<li id=\"footer-places-developers\"><a href=\"https://www.mediawiki.org/wiki/Special:MyLanguage/How_to_contribute\">Developers</a></li>\\n\\t\\t\\t\\t\\t\\t\\t\\t<li id=\"footer-places-cookiestatement\"><a href=\"https://wikimediafoundation.org/wiki/Cookie_statement\">Cookie statement</a></li>\\n\\t\\t\\t\\t\\t\\t\\t\\t<li id=\"footer-places-mobileview\"><a href=\"//en.m.wikipedia.org/w/index.php?title=List_of_multiple_Olympic_medalists&mobileaction=toggle_view_mobile\" class=\"noprint stopMobileRedirectToggle\">Mobile view</a></li>\\n\\t\\t\\t\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t<ul id=\"footer-icons\" class=\"noprint\">\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t<li id=\"footer-copyrightico\">\\n\\t\\t\\t\\t\\t\\t<a href=\"https://wikimediafoundation.org/\"><img src=\"/static/images/wikimedia-button.png\" srcset=\"/static/images/wikimedia-button-1.5x.png 1.5x, /static/images/wikimedia-button-2x.png 2x\" width=\"88\" height=\"31\" alt=\"Wikimedia Foundation\"/></a>\\t\\t\\t\\t\\t</li>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t<li id=\"footer-poweredbyico\">\\n\\t\\t\\t\\t\\t\\t<a href=\"//www.mediawiki.org/\"><img src=\"/static/images/poweredby_mediawiki_88x31.png\" alt=\"Powered by MediaWiki\" srcset=\"/static/images/poweredby_mediawiki_132x47.png 1.5x, /static/images/poweredby_mediawiki_176x62.png 2x\" width=\"88\" height=\"31\"/></a>\\t\\t\\t\\t\\t</li>\\n\\t\\t\\t\\t\\t\\t\\t\\t\\t</ul>\\n\\t\\t\\t\\t\\t\\t<div style=\"clear: both;\"></div>\\n\\t\\t</div>\\n\\t\\t<script>(window.RLQ=window.RLQ||[]).push(function(){mw.config.set({\"wgPageParseReport\":{\"limitreport\":{\"cputime\":\"1.672\",\"walltime\":\"1.777\",\"ppvisitednodes\":{\"value\":34561,\"limit\":1000000},\"ppgeneratednodes\":{\"value\":0,\"limit\":1500000},\"postexpandincludesize\":{\"value\":159304,\"limit\":2097152},\"templateargumentsize\":{\"value\":32222,\"limit\":2097152},\"expansiondepth\":{\"value\":15,\"limit\":40},\"expensivefunctioncount\":{\"value\":1,\"limit\":500},\"entityaccesscount\":{\"value\":0,\"limit\":400},\"timingprofile\":[\"100.00% 1402.375 1 -total\",\" 81.09% 1137.131 632 Template:Country_alias\",\" 74.72% 1047.815 190 Template:FlagIOC\",\" 73.84% 1035.512 190 Template:FlagIOC2\",\" 12.03% 168.721 31 Template:FlagIOCathlete\",\" 11.78% 165.221 31 Template:FlagIOC2athlete\",\" 4.68% 65.630 1 Template:Reflist\",\" 3.42% 47.955 158 Template:Sortname\",\" 2.42% 33.881 1 Template:Cite_book\",\" 1.64% 22.981 1 Template:Expand_list\"]},\"scribunto\":{\"limitreport-timeusage\":{\"value\":\"0.877\",\"limit\":\"10.000\"},\"limitreport-memusage\":{\"value\":3862169,\"limit\":52428800}},\"cachereport\":{\"origin\":\"mw1325\",\"timestamp\":\"20180215103610\",\"ttl\":1900800,\"transientcontent\":false}}});});</script><script>(window.RLQ=window.RLQ||[]).push(function(){mw.config.set({\"wgBackendResponseTime\":87,\"wgHostname\":\"mw1255\"});});</script>\\n\\t</body>\\n</html>\\n'\n"
]
],
[
[
"Extract the html that corresponds to the big table from the soup.",
"_____no_output_____"
]
],
[
[
"# we can retreive all tables, our desired table is the first one:\ntable_html = class_soup(\"table\")[0]\ntable_html",
"_____no_output_____"
]
],
[
[
"Parse the table into a pandas dataframe. ",
"_____no_output_____"
]
],
[
[
"athletes = pd.read_html(str(table_html), header=0)[0]\nathletes.head(15)",
"_____no_output_____"
]
],
[
[
"For cases where the row is screwed up, the bronze column is NaN, which we can use to filter:",
"_____no_output_____"
]
],
[
[
"athletes = athletes[pd.notnull(athletes[\"Bronze\"])]",
"_____no_output_____"
]
],
[
[
"Subset to the relevant columns: ",
"_____no_output_____"
]
],
[
[
"athletes = athletes[[\"Nation\", \"Gold\", \"Silver\", \"Bronze\"]]\nathletes.head(10)",
"_____no_output_____"
]
],
[
[
"Grouping, summing, calculating the total, and sorting: ",
"_____no_output_____"
]
],
[
[
"countries = athletes.groupby(\"Nation\").sum()",
"_____no_output_____"
],
[
"countries[\"Total\"] = countries[\"Gold\"] + countries[\"Silver\"] + countries[\"Bronze\"]\ncountries.sort_values(\"Total\", ascending=False)",
"_____no_output_____"
]
],
[
[
"## Exercise 2 – APIs\n\nUse the [Open Notify API](http://open-notify.org/Open-Notify-API/People-In-Space/) to find out how many people are in space right now.",
"_____no_output_____"
]
],
[
[
"import requests \n\nurl = \"http://api.open-notify.org/astros.json\"\n\nr = requests.get(url)\ndata = r.json()\ndata",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4ef5ae5369c3cf5a0885adc152c3ed5d86d2b8 | 9,193 | ipynb | Jupyter Notebook | attention/Attention_Basics.ipynb | subham73/deep-learning-v2-pytorch | a200ee8d644fa176fc6f2b663cac6cc0207c1b40 | [
"MIT"
] | 4,850 | 2018-09-04T19:40:22.000Z | 2022-03-31T10:21:49.000Z | attention/Attention_Basics.ipynb | subham73/deep-learning-v2-pytorch | a200ee8d644fa176fc6f2b663cac6cc0207c1b40 | [
"MIT"
] | 220 | 2018-09-15T20:30:55.000Z | 2022-03-30T04:45:30.000Z | attention/Attention_Basics.ipynb | subham73/deep-learning-v2-pytorch | a200ee8d644fa176fc6f2b663cac6cc0207c1b40 | [
"MIT"
] | 5,729 | 2018-09-04T22:07:30.000Z | 2022-03-31T11:52:07.000Z | 31.268707 | 335 | 0.624171 | [
[
[
"# Attention Basics\nIn this notebook, we look at how attention is implemented. We will focus on implementing attention in isolation from a larger model. That's because when implementing attention in a real-world model, a lot of the focus goes into piping the data and juggling the various vectors rather than the concepts of attention themselves.\n\nWe will implement attention scoring as well as calculating an attention context vector.\n\n## Attention Scoring\n### Inputs to the scoring function\nLet's start by looking at the inputs we'll give to the scoring function. We will assume we're in the first step in the decoding phase. The first input to the scoring function is the hidden state of decoder (assuming a toy RNN with three hidden nodes -- not usable in real life, but easier to illustrate):",
"_____no_output_____"
]
],
[
[
"dec_hidden_state = [5,1,20]",
"_____no_output_____"
]
],
[
[
"Let's visualize this vector:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\n# Let's visualize our decoder hidden state\nplt.figure(figsize=(1.5, 4.5))\nsns.heatmap(np.transpose(np.matrix(dec_hidden_state)), annot=True, cmap=sns.light_palette(\"purple\", as_cmap=True), linewidths=1)",
"_____no_output_____"
]
],
[
[
"Our first scoring function will score a single annotation (encoder hidden state), which looks like this:",
"_____no_output_____"
]
],
[
[
"annotation = [3,12,45] #e.g. Encoder hidden state",
"_____no_output_____"
],
[
"# Let's visualize the single annotation\nplt.figure(figsize=(1.5, 4.5))\nsns.heatmap(np.transpose(np.matrix(annotation)), annot=True, cmap=sns.light_palette(\"orange\", as_cmap=True), linewidths=1)",
"_____no_output_____"
]
],
[
[
"### IMPLEMENT: Scoring a Single Annotation\nLet's calculate the dot product of a single annotation. NumPy's [dot()](https://docs.scipy.org/doc/numpy/reference/generated/numpy.dot.html) is a good candidate for this operation",
"_____no_output_____"
]
],
[
[
"def single_dot_attention_score(dec_hidden_state, enc_hidden_state):\n # TODO: return the dot product of the two vectors\n return \n \nsingle_dot_attention_score(dec_hidden_state, annotation)",
"_____no_output_____"
]
],
[
[
"\n### Annotations Matrix\nLet's now look at scoring all the annotations at once. To do that, here's our annotation matrix:",
"_____no_output_____"
]
],
[
[
"annotations = np.transpose([[3,12,45], [59,2,5], [1,43,5], [4,3,45.3]])",
"_____no_output_____"
]
],
[
[
"And it can be visualized like this (each column is a hidden state of an encoder time step):",
"_____no_output_____"
]
],
[
[
"# Let's visualize our annotation (each column is an annotation)\nax = sns.heatmap(annotations, annot=True, cmap=sns.light_palette(\"orange\", as_cmap=True), linewidths=1)",
"_____no_output_____"
]
],
[
[
"### IMPLEMENT: Scoring All Annotations at Once\nLet's calculate the scores of all the annotations in one step using matrix multiplication. Let's continue to use the dot scoring method\n\n<img src=\"images/scoring_functions.png\" />\n\nTo do that, we'll have to transpose `dec_hidden_state` and [matrix multiply](https://docs.scipy.org/doc/numpy/reference/generated/numpy.matmul.html) it with `annotations`.",
"_____no_output_____"
]
],
[
[
"def dot_attention_score(dec_hidden_state, annotations):\n # TODO: return the product of dec_hidden_state transpose and annotations\n return \n \nattention_weights_raw = dot_attention_score(dec_hidden_state, annotations)\nattention_weights_raw",
"_____no_output_____"
]
],
[
[
"Looking at these scores, can you guess which of the four vectors will get the most attention from the decoder at this time step?\n\n## Softmax\nNow that we have our scores, let's apply softmax:\n<img src=\"images/softmax.png\" />",
"_____no_output_____"
]
],
[
[
"def softmax(x):\n x = np.array(x, dtype=np.float128)\n e_x = np.exp(x)\n return e_x / e_x.sum(axis=0) \n\nattention_weights = softmax(attention_weights_raw)\nattention_weights",
"_____no_output_____"
]
],
[
[
"Even when knowing which annotation will get the most focus, it's interesting to see how drastic softmax makes the end score become. The first and last annotation had the respective scores of 927 and 929. But after softmax, the attention they'll get is 0.12 and 0.88 respectively.\n\n# Applying the scores back on the annotations\nNow that we have our scores, let's multiply each annotation by its score to proceed closer to the attention context vector. This is the multiplication part of this formula (we'll tackle the summation part in the latter cells)\n\n<img src=\"images/Context_vector.png\" />",
"_____no_output_____"
]
],
[
[
"def apply_attention_scores(attention_weights, annotations):\n # TODO: Multiple the annotations by their weights\n return\n\napplied_attention = apply_attention_scores(attention_weights, annotations)\napplied_attention",
"_____no_output_____"
]
],
[
[
"Let's visualize how the context vector looks now that we've applied the attention scores back on it:",
"_____no_output_____"
]
],
[
[
"# Let's visualize our annotations after applying attention to them\nax = sns.heatmap(applied_attention, annot=True, cmap=sns.light_palette(\"orange\", as_cmap=True), linewidths=1)",
"_____no_output_____"
]
],
[
[
"Contrast this with the raw annotations visualized earlier in the notebook, and we can see that the second and third annotations (columns) have been nearly wiped out. The first annotation maintains some of its value, and the fourth annotation is the most pronounced.\n\n# Calculating the Attention Context Vector\nAll that remains to produce our attention context vector now is to sum up the four columns to produce a single attention context vector\n",
"_____no_output_____"
]
],
[
[
"def calculate_attention_vector(applied_attention):\n return np.sum(applied_attention, axis=1)\n\nattention_vector = calculate_attention_vector(applied_attention)\nattention_vector",
"_____no_output_____"
],
[
"# Let's visualize the attention context vector\nplt.figure(figsize=(1.5, 4.5))\nsns.heatmap(np.transpose(np.matrix(attention_vector)), annot=True, cmap=sns.light_palette(\"Blue\", as_cmap=True), linewidths=1)",
"_____no_output_____"
]
],
[
[
"Now that we have the context vector, we can concatenate it with the hidden state and pass it through a hidden layer to produce the the result of this decoding time step.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cb4eff10d814862ee0c7235f2312a787661bf8e1 | 11,021 | ipynb | Jupyter Notebook | module-1/Intro-Pandas/your-code/main.ipynb | ruesaintdenis/daft-miami-0120-labs | a9f91b0f60d4ed172c3e2d1966628ed8e5e0df78 | [
"MIT"
] | null | null | null | module-1/Intro-Pandas/your-code/main.ipynb | ruesaintdenis/daft-miami-0120-labs | a9f91b0f60d4ed172c3e2d1966628ed8e5e0df78 | [
"MIT"
] | null | null | null | module-1/Intro-Pandas/your-code/main.ipynb | ruesaintdenis/daft-miami-0120-labs | a9f91b0f60d4ed172c3e2d1966628ed8e5e0df78 | [
"MIT"
] | 1 | 2020-02-12T20:06:11.000Z | 2020-02-12T20:06:11.000Z | 23.498934 | 114 | 0.451048 | [
[
[
"# Introduction to Pandas\n\nComplete the following set of exercises to solidify your knowledge of Pandas fundamentals.",
"_____no_output_____"
],
[
"#### 1. Import Numpy and Pandas and alias them to `np` and `pd` respectively.",
"_____no_output_____"
]
],
[
[
"# your code here\n\nimport numpy as np\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"#### 2. Create a Pandas Series containing the elements of the list below.",
"_____no_output_____"
]
],
[
[
"lst = [5.7, 75.2, 74.4, 84.0, 66.5, 66.3, 55.8, 75.7, 29.1, 43.7]",
"_____no_output_____"
],
[
"# your code here\nlstseries = pd.Series(lst)\nprint(lstseries)",
"0 5.7\n1 75.2\n2 74.4\n3 84.0\n4 66.5\n5 66.3\n6 55.8\n7 75.7\n8 29.1\n9 43.7\ndtype: float64\n"
]
],
[
[
"#### 3. Use indexing to return the third value in the Series above.\n\n*Hint: Remember that indexing begins at 0.*",
"_____no_output_____"
]
],
[
[
"# your code here\nprint(lstseries[2])",
"74.4\n"
]
],
[
[
"#### 4. Create a Pandas DataFrame from the list of lists below. Each sublist should be represented as a row.",
"_____no_output_____"
]
],
[
[
"b = [[53.1, 95.0, 67.5, 35.0, 78.4],\n [61.3, 40.8, 30.8, 37.8, 87.6],\n [20.6, 73.2, 44.2, 14.6, 91.8],\n [57.4, 0.1, 96.1, 4.2, 69.5],\n [83.6, 20.5, 85.4, 22.8, 35.9],\n [49.0, 69.0, 0.1, 31.8, 89.1],\n [23.3, 40.7, 95.0, 83.8, 26.9],\n [27.6, 26.4, 53.8, 88.8, 68.5],\n [96.6, 96.4, 53.4, 72.4, 50.1],\n [73.7, 39.0, 43.2, 81.6, 34.7]]",
"_____no_output_____"
],
[
"# your code here\n\nbdf = pd.DataFrame(b)\nprint(bdf)",
" 0 1 2 3 4\n0 53.1 95.0 67.5 35.0 78.4\n1 61.3 40.8 30.8 37.8 87.6\n2 20.6 73.2 44.2 14.6 91.8\n3 57.4 0.1 96.1 4.2 69.5\n4 83.6 20.5 85.4 22.8 35.9\n5 49.0 69.0 0.1 31.8 89.1\n6 23.3 40.7 95.0 83.8 26.9\n7 27.6 26.4 53.8 88.8 68.5\n8 96.6 96.4 53.4 72.4 50.1\n9 73.7 39.0 43.2 81.6 34.7\n"
]
],
[
[
"#### 5. Rename the data frame columns based on the names in the list below.",
"_____no_output_____"
]
],
[
[
"colnames = ['Score_1', 'Score_2', 'Score_3', 'Score_4', 'Score_5']",
"_____no_output_____"
],
[
"# your code here\nbdf.columns = colnames\nprint(bdf)",
" Score_1 Score_2 Score_3 Score_4 Score_5\n0 53.1 95.0 67.5 35.0 78.4\n1 61.3 40.8 30.8 37.8 87.6\n2 20.6 73.2 44.2 14.6 91.8\n3 57.4 0.1 96.1 4.2 69.5\n4 83.6 20.5 85.4 22.8 35.9\n5 49.0 69.0 0.1 31.8 89.1\n6 23.3 40.7 95.0 83.8 26.9\n7 27.6 26.4 53.8 88.8 68.5\n8 96.6 96.4 53.4 72.4 50.1\n9 73.7 39.0 43.2 81.6 34.7\n"
]
],
[
[
"#### 6. Create a subset of this data frame that contains only the Score 1, 3, and 5 columns.",
"_____no_output_____"
]
],
[
[
"# your code here\nnewbdf = bdf[['Score_1','Score_3','Score_5']]\nprint(newbdf)\n\n",
" Score_1 Score_3 Score_5\n0 53.1 67.5 78.4\n1 61.3 30.8 87.6\n2 20.6 44.2 91.8\n3 57.4 96.1 69.5\n4 83.6 85.4 35.9\n5 49.0 0.1 89.1\n6 23.3 95.0 26.9\n7 27.6 53.8 68.5\n8 96.6 53.4 50.1\n9 73.7 43.2 34.7\n"
]
],
[
[
"#### 7. From the original data frame, calculate the average Score_3 value.",
"_____no_output_____"
]
],
[
[
"# your code here\n\nbdf['Score_3'].mean()",
"_____no_output_____"
]
],
[
[
"#### 8. From the original data frame, calculate the maximum Score_4 value.",
"_____no_output_____"
]
],
[
[
"# your code here\nbdf['Score_4'].max()",
"_____no_output_____"
]
],
[
[
"#### 9. From the original data frame, calculate the median Score 2 value.",
"_____no_output_____"
]
],
[
[
"# your code here\nbdf['Score_2'].median()",
"_____no_output_____"
]
],
[
[
"#### 10. Create a Pandas DataFrame from the dictionary of product orders below.",
"_____no_output_____"
]
],
[
[
"orders = {'Description': ['LUNCH BAG APPLE DESIGN',\n 'SET OF 60 VINTAGE LEAF CAKE CASES ',\n 'RIBBON REEL STRIPES DESIGN ',\n 'WORLD WAR 2 GLIDERS ASSTD DESIGNS',\n 'PLAYING CARDS JUBILEE UNION JACK',\n 'POPCORN HOLDER',\n 'BOX OF VINTAGE ALPHABET BLOCKS',\n 'PARTY BUNTING',\n 'JAZZ HEARTS ADDRESS BOOK',\n 'SET OF 4 SANTA PLACE SETTINGS'],\n 'Quantity': [1, 24, 1, 2880, 2, 7, 1, 4, 10, 48],\n 'UnitPrice': [1.65, 0.55, 1.65, 0.18, 1.25, 0.85, 11.95, 4.95, 0.19, 1.25],\n 'Revenue': [1.65, 13.2, 1.65, 518.4, 2.5, 5.95, 11.95, 19.8, 1.9, 60.0]}",
"_____no_output_____"
],
[
"# your code here\nordersdf = pd.DataFrame(orders)\n# x.head()\nprint(ordersdf)",
" Description Quantity UnitPrice Revenue\n0 LUNCH BAG APPLE DESIGN 1 1.65 1.65\n1 SET OF 60 VINTAGE LEAF CAKE CASES 24 0.55 13.20\n2 RIBBON REEL STRIPES DESIGN 1 1.65 1.65\n3 WORLD WAR 2 GLIDERS ASSTD DESIGNS 2880 0.18 518.40\n4 PLAYING CARDS JUBILEE UNION JACK 2 1.25 2.50\n5 POPCORN HOLDER 7 0.85 5.95\n6 BOX OF VINTAGE ALPHABET BLOCKS 1 11.95 11.95\n7 PARTY BUNTING 4 4.95 19.80\n8 JAZZ HEARTS ADDRESS BOOK 10 0.19 1.90\n9 SET OF 4 SANTA PLACE SETTINGS 48 1.25 60.00\n"
]
],
[
[
"#### 11. Calculate the total quantity ordered and revenue generated from these orders.",
"_____no_output_____"
]
],
[
[
"# your code here\ntotal_qty = ordersdf['Quantity'].sum()\nprint(total_qty)\n\ntotal_revenue = ordersdf['Revenue'].sum()\nprint(total_revenue)",
"2978\n637.0\n"
]
],
[
[
"#### 12. Obtain the prices of the most expensive and least expensive items ordered and print the difference.",
"_____no_output_____"
]
],
[
[
"# your code here\nexpensive = ordersdf['UnitPrice'].max()\nprint(expensive)\naffordable = ordersdf['UnitPrice'].min()\nprint(affordable)\n\ndiff = expensive - affordable \nprint(diff)",
"11.95\n0.18\n11.77\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4eff9fe68dd6c421b38ec49c9b0c5c98dcb236 | 13,799 | ipynb | Jupyter Notebook | Notebooks/04_Accuracy_Assessment_Seasonal.ipynb | eloise-b/wofs-validation | 9010c82b66a5df04305a09092f6d47be7db714a6 | [
"Apache-2.0"
] | 1 | 2021-03-15T03:25:23.000Z | 2021-03-15T03:25:23.000Z | Notebooks/04_Accuracy_Assessment_Seasonal.ipynb | eloise-b/wofs-validation | 9010c82b66a5df04305a09092f6d47be7db714a6 | [
"Apache-2.0"
] | null | null | null | Notebooks/04_Accuracy_Assessment_Seasonal.ipynb | eloise-b/wofs-validation | 9010c82b66a5df04305a09092f6d47be7db714a6 | [
"Apache-2.0"
] | null | null | null | 30.327473 | 382 | 0.61287 | [
[
[
"# Seasonal Accuracy Assessment of Water Observations from Space (WOfS) Product in Africa<img align=\"right\" src=\"../Supplementary_data/DE_Africa_Logo_Stacked_RGB_small.jpg\">",
"_____no_output_____"
],
[
"## Description\nNow that we have run WOfS classification for each AEZs in Africa, its time to conduct seasonal accuracy assessment for each AEZ in Africa which is already compiled and stored in the following folder:`Results/WOfS_Assessment/Point_Based/ValidPoints_Per_AEZ`.\n\nAccuracy assessment for WOfS product in Africa includes generating a confusion error matrix for a WOFL binary classification.\nThe inputs for the estimating the accuracy of WOfS derived product are a binary classification WOFL layer showing water/non-water and a shapefile containing validation points collected by [Collect Earth Online](https://collect.earth/) tool. Validation points are the ground truth or actual data while the extracted value for each location from WOFL is the predicted value. \n\nThis notebook will explain how you can perform seasonal accuracy assessment for WOfS starting with `Western` AEZ using collected ground truth dataset. It will output a confusion error matrix containing overall, producer's and user's accuracy, along with the F1 score for each class.",
"_____no_output_____"
],
[
"## Getting started\n\nTo run this analysis, run all the cells in the notebook, starting with the \"Load packages\" cell.",
"_____no_output_____"
],
[
"### Load packages\nImport Python packages that are used for the analysis.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport sys\nimport os\nimport rasterio\nimport xarray\nimport glob\nimport numpy as np\nimport pandas as pd\nimport seaborn as sn\nimport geopandas as gpd\nimport matplotlib.pyplot as plt\nimport scipy, scipy.ndimage\nimport warnings\nwarnings.filterwarnings(\"ignore\") #this will suppress the warnings for multiple UTM zones in your AOI \n\nsys.path.append(\"../Scripts\")\nfrom geopandas import GeoSeries, GeoDataFrame\nfrom shapely.geometry import Point\nfrom sklearn.metrics import confusion_matrix, accuracy_score \nfrom sklearn.metrics import plot_confusion_matrix, f1_score \nfrom deafrica_plotting import map_shapefile,display_map, rgb\nfrom deafrica_spatialtools import xr_rasterize\nfrom deafrica_datahandling import wofs_fuser, mostcommon_crs,load_ard,deepcopy\nfrom deafrica_dask import create_local_dask_cluster",
"_____no_output_____"
]
],
[
[
"### Analysis Parameters ",
"_____no_output_____"
],
[
"- CEO : groundtruth points containing valid points in each AEZ containing WOfS assigned classes, WOfS clear observations and the labels identified by analyst in each calendar month \n- input_data : dataframe for further analysis and accuracy assessment ",
"_____no_output_____"
],
[
"### Load the Dataset",
"_____no_output_____"
],
[
"Validation points that are valid for each AEZ ",
"_____no_output_____"
]
],
[
[
"#Read the valid ground truth data \nCEO = 'Results/WOfS_Assessment/Point_Based/ValidPoints_Per_AEZ/ValidationPoints_Western.csv'\n\ndf = pd.read_csv(CEO,delimiter=\",\")",
"_____no_output_____"
],
[
"#explore the dataframe\ndf.columns",
"_____no_output_____"
],
[
"#rename a column in dataframe \ninput_data = df.drop(['Unnamed: 0'], axis=1)\ninput_data = input_data.rename(columns={'WATERFLAG':'ACTUAL'})",
"_____no_output_____"
],
[
"#The table contains each calendar month as well as CEO and WOfS lables for each validation points \ninput_data",
"_____no_output_____"
],
[
"#Counting the number of rows in valid points dataframe \ncount = input_data.groupby('PLOT_ID',as_index=False,sort=False).last()",
"_____no_output_____"
],
[
"count",
"_____no_output_____"
]
],
[
[
"From the table, choose those rows that are in Wet season and also choose those in Dry season, then save them in separate tables. ",
"_____no_output_____"
]
],
[
[
"#setting the months that are identified as wet in the AEZ using Climatology dataset \nWetMonth = [5,6,7,8,9,10]",
"_____no_output_____"
],
[
"#identifying the points that are in wet season and counting their numbers \nWet_Season = input_data[input_data['MONTH'].isin(WetMonth)]\ncount_Wet_Season = Wet_Season.groupby('PLOT_ID',as_index=False,sort=False).last()\ncount_Wet_Season",
"_____no_output_____"
],
[
"#setting the months that are identified as dry in the AEZ using Climatology dataset then counting the points that are in dry season \nDry_Season = input_data[~input_data['MONTH'].isin(WetMonth)]\ncount_Dry_Season = Dry_Season.groupby('PLOT_ID',as_index=False,sort=False).last()\ncount_Dry_Season",
"_____no_output_____"
]
],
[
[
"Some points are in both dry and wet seasons as the number of points show.",
"_____no_output_____"
],
[
"### Create a Confusion Matrix ",
"_____no_output_____"
]
],
[
[
"confusion_matrix = pd.crosstab(Wet_Season['ACTUAL'],Wet_Season['PREDICTION'],rownames=['ACTUAL'],colnames=['PREDICTION'],margins=True)\nconfusion_matrix",
"_____no_output_____"
]
],
[
[
"`Producer's Accuracy` is the map-maker accuracy showing the probability that a certain class on the ground is classified. Producer's accuracy complements error of omission. ",
"_____no_output_____"
]
],
[
[
"confusion_matrix[\"Producer's\"] = [confusion_matrix.loc[0][0] / confusion_matrix.loc[0]['All'] * 100, confusion_matrix.loc[1][1] / confusion_matrix.loc[1]['All'] *100, np.nan]\nconfusion_matrix",
"_____no_output_____"
]
],
[
[
"`User's Accuracy` is the map-user accuracy showing how often the class on the map will actually be present on the ground. `User's accuracy` shows the reliability. It is calculated based on the total number of correct classification for a particular class over the total number of classified sites.",
"_____no_output_____"
]
],
[
[
"users_accuracy = pd.Series([confusion_matrix[0][0] / confusion_matrix[0]['All'] * 100,\n confusion_matrix[1][1] / confusion_matrix[1]['All'] * 100]).rename(\"User's\")\n\nconfusion_matrix = confusion_matrix.append(users_accuracy)\nconfusion_matrix ",
"_____no_output_____"
]
],
[
[
"`Overal Accuracy` shows what proportion of reference(actual) sites mapped correctly.",
"_____no_output_____"
]
],
[
[
"confusion_matrix.loc[\"User's\", \"Producer's\"] = (confusion_matrix[0][0] + confusion_matrix[1][1]) / confusion_matrix['All']['All'] * 100\nconfusion_matrix",
"_____no_output_____"
],
[
"input_data['PREDICTION'] = input_data['PREDICTION'] .astype(str).astype(int)",
"_____no_output_____"
]
],
[
[
"The F1 score is the harmonic mean of the precision and recall, where an F1 score reaches its best value at 1(perfect precision and recall), and is calculated as:",
"_____no_output_____"
]
],
[
[
"fscore = pd.Series([(2*(confusion_matrix.loc[\"User's\"][0]*confusion_matrix.loc[0][\"Producer's\"]) / (confusion_matrix.loc[\"User's\"][0] + confusion_matrix.loc[0][\"Producer's\"])) / 100,\n f1_score(input_data['ACTUAL'],input_data['PREDICTION'])]).rename(\"F-score\")\nconfusion_matrix = confusion_matrix.append(fscore)",
"_____no_output_____"
],
[
"confusion_matrix",
"_____no_output_____"
],
[
"confusion_matrix = confusion_matrix.round(decimals=2)",
"_____no_output_____"
],
[
"confusion_matrix = confusion_matrix.rename(columns={'0':'NoWater','1':'Water', 0:'NoWater',1:'Water','All':'Total'},index={'0':'NoWater','1':'Water',0:'NoWater',1:'Water','All':'Total'})",
"_____no_output_____"
],
[
"confusion_matrix",
"_____no_output_____"
],
[
"confusion_matrix.to_csv('../Results/WOfS_Assessment/Point_Based/ConfusionMatrix/Western_WetSeason_confusion_matrix.csv')",
"_____no_output_____"
]
],
[
[
"***\n\n## Additional information\n\n**License:** The code in this notebook is licensed under the [Apache License, Version 2.0](https://www.apache.org/licenses/LICENSE-2.0). \nDigital Earth Africa data is licensed under the [Creative Commons by Attribution 4.0](https://creativecommons.org/licenses/by/4.0/) license.\n\n**Contact:** If you need assistance, please post a question on the [Open Data Cube Slack channel](http://slack.opendatacube.org/) or on the [GIS Stack Exchange](https://gis.stackexchange.com/questions/ask?tags=open-data-cube) using the `open-data-cube` tag (you can view previously asked questions [here](https://gis.stackexchange.com/questions/tagged/open-data-cube)).\nIf you would like to report an issue with this notebook, you can file one on [Github](https://github.com/digitalearthafrica/deafrica-sandbox-notebooks).\n\n**Last modified:** January 2020\n\n**Compatible datacube version:** ",
"_____no_output_____"
],
[
"## Tags\nBrowse all available tags on the DE Africa User Guide's [Tags Index](https://) (placeholder as this does not exist yet)",
"_____no_output_____"
]
],
[
[
"**Tags**: :index:`WOfS`, :index:`fractional cover`, :index:`deafrica_plotting`, :index:`deafrica_datahandling`, :index:`display_map`, :index:`wofs_fuser`, :index:`WOFL`, :index:`masking`",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"raw"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"raw"
]
] |
cb4f00aadab276f7da16308304e2b79d4be641a8 | 2,859 | ipynb | Jupyter Notebook | split.ipynb | elek/asf-project-stat | afa8567e8e404b429c4aee11ae34ec359458f8fc | [
"Apache-2.0"
] | null | null | null | split.ipynb | elek/asf-project-stat | afa8567e8e404b429c4aee11ae34ec359458f8fc | [
"Apache-2.0"
] | null | null | null | split.ipynb | elek/asf-project-stat | afa8567e8e404b429c4aee11ae34ec359458f8fc | [
"Apache-2.0"
] | null | null | null | 23.825 | 171 | 0.541448 | [
[
[
"import pandas as pd\nimport os",
"_____no_output_____"
],
[
"contribution = pd.read_csv(\"github-contribution.csv.gz\")",
"_____no_output_____"
],
[
"asfgithub = pd.read_csv(\"asf-github-to-apache.csv.gz\")\nmembership = pd.read_csv(\"asf-membership.csv.gz\")",
"_____no_output_____"
],
[
"commits = pd.read_csv(\"github-commit.csv.gz\",encoding = \"iso-8859-1\")\n",
"_____no_output_____"
],
[
"asfgithubmembership = pd.merge(asfgithub, membership, how=\"left\")",
"_____no_output_____"
],
[
"def asfproject(name):\n parts = name.split(\"-\")\n if parts[0] == \"incubator\" and len(parts) > 1:\n return parts[1]\n else:\n return parts[0]\n \ncontribution[\"project\"] = contribution.repo.apply(asfproject)",
"_____no_output_____"
],
[
"projects = pd.read_csv(\"asf-project.csv.gz\")\nfor index,row in projects.iterrows():\n project = row[\"name\"]\n if not os.path.exists(project):\n os.makedirs(project)\n \n# project_contribution.to_csv(os.path.join(project_contribution = contribution[contribution.project == project], \"github-contribution.csv.gz\"), index=False)\n# asfgithubmembership[asfgithubmembership.project == project].to_csv(os.path.join(project, \"asf-github-membership.csv.gz\"), index=False)\n commits[commits.repo.apply(asfproject) == project].to_csv(os.path.join(project, \"github-commit.csv.gz\"), index=False)\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4f04bdd4f30e506f55a245a36aa788fb1429ab | 4,494 | ipynb | Jupyter Notebook | docs/source/examples/plotting/REE_v_radii.ipynb | bomtuckle/pyrolite | c0af0ade14ff26b4e9fdd5a033b27e73df085c55 | [
"BSD-3-Clause"
] | 69 | 2019-02-25T00:17:53.000Z | 2022-03-31T17:26:48.000Z | docs/source/examples/plotting/REE_v_radii.ipynb | bomtuckle/pyrolite | c0af0ade14ff26b4e9fdd5a033b27e73df085c55 | [
"BSD-3-Clause"
] | 68 | 2018-07-20T09:01:01.000Z | 2022-03-31T16:28:36.000Z | docs/source/examples/plotting/REE_v_radii.ipynb | bomtuckle/pyrolite | c0af0ade14ff26b4e9fdd5a033b27e73df085c55 | [
"BSD-3-Clause"
] | 24 | 2018-10-02T04:32:10.000Z | 2021-11-10T08:24:17.000Z | 29.761589 | 387 | 0.508011 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\nREE Radii Plots\n============================\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom pyrolite.plot import pyroplot",
"_____no_output_____"
]
],
[
[
"Here we generate some example data, using the\n:func:`~pyrolite.util.synthetic.example_spider_data` function (based on EMORB,\nhere normalised to Primitive Mantle);\n\n\n",
"_____no_output_____"
]
],
[
[
"from pyrolite.util.synthetic import example_spider_data\n\ndf = example_spider_data(noise_level=0.1, nobs=20)",
"_____no_output_____"
]
],
[
[
"Where data is specified, the default plot is a line-based spiderplot:\n\n",
"_____no_output_____"
]
],
[
[
"ax = df.pyroplot.REE(color=\"0.5\", figsize=(8, 4))\nplt.show()",
"_____no_output_____"
]
],
[
[
"This behaviour can be modified (see spiderplot docs) to provide e.g. filled ranges:\n\n\n",
"_____no_output_____"
]
],
[
[
"df.pyroplot.REE(mode=\"fill\", color=\"0.5\", alpha=0.5, figsize=(8, 4))\nplt.show()",
"_____no_output_____"
]
],
[
[
"The plotting axis can be specified to use exisiting axes:\n\n\n",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(1, 2, sharey=True, figsize=(12, 4))\n\ndf.pyroplot.REE(ax=ax[0])\n# we can also change the index of the second axes\nanother_df = example_spider_data(noise_level=0.2, nobs=20) # some 'nosier' data\nanother_df.pyroplot.REE(ax=ax[1], color=\"k\", index=\"radii\")\n\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
],
[
[
"If you're just after a plotting template, you can use\n:func:`~pyrolite.plot.spider.REE_v_radii` to get a formatted axis which can be used\nfor subsequent plotting:\n\n\n",
"_____no_output_____"
]
],
[
[
"from pyrolite.plot.spider import REE_v_radii\n\nax = REE_v_radii(index=\"radii\") # radii mode will put ionic radii on the x axis\nplt.show()",
"_____no_output_____"
]
],
[
[
".. seealso::\n\n Examples:\n `Ionic Radii <ionic_radii.html>`__,\n `Spider Diagrams <spider.html>`__,\n `lambdas: Parameterising REE Profiles <lambdas.html>`__\n\n Functions:\n :func:`~pyrolite.geochem.ind.get_ionic_radii`,\n :func:`pyrolite.plot.pyroplot.REE`,\n :func:`pyrolite.plot.pyroplot.spider`,\n :func:`~pyrolite.geochem.pyrochem.lambda_lnREE`\n\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb4f077c10a19dcf2b5813d92828bd31e91b741d | 115,338 | ipynb | Jupyter Notebook | Chapter11-PCA/Ch-11.ipynb | cameronmcewan/Python-Data-Analysis-Third-Edition | d465a0d60cde07630dd8c69d7f1b5e944b587350 | [
"MIT"
] | null | null | null | Chapter11-PCA/Ch-11.ipynb | cameronmcewan/Python-Data-Analysis-Third-Edition | d465a0d60cde07630dd8c69d7f1b5e944b587350 | [
"MIT"
] | null | null | null | Chapter11-PCA/Ch-11.ipynb | cameronmcewan/Python-Data-Analysis-Third-Edition | d465a0d60cde07630dd8c69d7f1b5e944b587350 | [
"MIT"
] | null | null | null | 141.345588 | 20,970 | 0.87156 | [
[
[
"## Principal Component Analysis ",
"_____no_output_____"
]
],
[
[
"# Import numpy \nimport numpy as np\n\n# Import linear algebra module\nfrom scipy import linalg as la\n \n# Create dataset\ndata=np.array([[7., 4., 3.],\n [4., 1., 8.],\n [6., 3., 5.],\n [8., 6., 1.],\n [8., 5., 7.],\n [7., 2., 9.],\n [5., 3., 3.],\n [9., 5., 8.],\n [7., 4., 5.],\n [8., 2., 2.]])\n \n# 1. Calculate the covariance matrix\n# Center your data\ndata -= data.mean(axis=0)\ncov = np.cov(data, rowvar=False)\n \n# 2. Calculate eigenvalues and eigenvector of the covariance matrix\nevals, evecs = la.eig(cov)\nprint(\"Eigenvalues:\", evals)\nprint(\"Eigenvector:\", evecs)\n \n# 3. Multiply the original data matrix with Eigenvector matrix. \n# Sort the Eigen values and vector and select components\nnum_components=2\nsorted_key = np.argsort(evals)[::-1][:num_components]\nevals, evecs = evals[sorted_key], evecs[:, sorted_key]\nprint(\"Sorted and Selected Eigen Values:\", evals)\nprint(\"Sorted and Selected Eigen Vector:\", evecs)\n \n# Multiply original data and Eigen vector\nprincipal_components=np.dot(data,evecs)\nprint(\"Principal Components:\", principal_components)",
"Eigenvalues: [0.74992815+0.j 3.67612927+0.j 8.27394258+0.j]\nEigenvector: [[-0.70172743 0.69903712 -0.1375708 ]\n [ 0.70745703 0.66088917 -0.25045969]\n [ 0.08416157 0.27307986 0.95830278]]\nSorted and Selected Eigen Values: [8.27394258+0.j 3.67612927+0.j]\nSorted and Selected Eigen Vector: [[-0.1375708 0.69903712]\n [-0.25045969 0.66088917]\n [ 0.95830278 0.27307986]]\nPrincipal Components: [[-2.15142276 -0.17311941]\n [ 3.80418259 -2.88749898]\n [ 0.15321328 -0.98688598]\n [-4.7065185 1.30153634]\n [ 1.29375788 2.27912632]\n [ 4.0993133 0.1435814 ]\n [-1.62582148 -2.23208282]\n [ 2.11448986 3.2512433 ]\n [-0.2348172 0.37304031]\n [-2.74637697 -1.06894049]]\n"
],
[
"# Import pandas and PCA\nimport pandas as pd\n\n# Import principal component analysis\nfrom sklearn.decomposition import PCA\n \n# Create dataset\ndata=np.array([[7., 4., 3.],\n [4., 1., 8.],\n [6., 3., 5.],\n [8., 6., 1.],\n [8., 5., 7.],\n [7., 2., 9.],\n [5., 3., 3.],\n [9., 5., 8.],\n [7., 4., 5.],\n [8., 2., 2.]])\n \n# Create and fit PCA Model\npca_model = PCA(n_components=2)\ncomponents = pca_model.fit_transform(data)\n \ncomponents_df = pd.DataFrame(data = components, \n columns = ['principal_component_1', 'principal_component_2'])\nprint(components_df)",
" principal_component_1 principal_component_2\n0 2.151423 -0.173119\n1 -3.804183 -2.887499\n2 -0.153213 -0.986886\n3 4.706518 1.301536\n4 -1.293758 2.279126\n5 -4.099313 0.143581\n6 1.625821 -2.232083\n7 -2.114490 3.251243\n8 0.234817 0.373040\n9 2.746377 -1.068940\n"
]
],
[
[
"## Finding Number of Clusters",
"_____no_output_____"
],
[
"### The Elbow Method",
"_____no_output_____"
]
],
[
[
"# import pandas\nimport pandas as pd\n\n# import matplotlib\nimport matplotlib.pyplot as plt\n\n# import K-means \nfrom sklearn.cluster import KMeans\n \n# Create a DataFrame\ndata=pd.DataFrame({\"X\":[12,15,18,10,8,9,12,20],\n \"Y\":[6,16,17,8,7,6,9,18]})\n \nwcss_list = []\n# Run a loop for different value of number of cluster\nfor i in range(1, 6):\n # Create and fit the KMeans model\n kmeans_model = KMeans(n_clusters = i, random_state = 123)\n kmeans_model.fit(data)\n # Add the WCSS or inertia of the clusters to the score_list\n wcss_list.append(kmeans_model.inertia_)\n\n# Plot the inertia(WCSS) and number of clusters \nplt.plot(range(1, 6), wcss_list, marker='*')\n# set title of the plot\nplt.title('Selecting Optimum Number of Clusters using Elbow Method')\n# Set x-axis label\nplt.xlabel('Number of Clusters K')\n# Set y-axis label\nplt.ylabel('Within-Cluster Sum of the Squares(Inertia)')\n# Display plot\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Silhouette Method",
"_____no_output_____"
]
],
[
[
"# import pandas \nimport pandas as pd\n\n# import matplotlib for data visualization\nimport matplotlib.pyplot as plt\n\n# import k-means for performing clustering\nfrom sklearn.cluster import KMeans\n\n# import silhouette score\nfrom sklearn.metrics import silhouette_score\n \n# Create a DataFrame\ndata=pd.DataFrame({\"X\":[12,15,18,10,8,9,12,20],\n \"Y\":[6,16,17,8,7,6,9,18]})\n \nscore_list = []\n\n# Run a loop for different value of number of cluster\nfor i in range(2, 6):\n # Create and fit the KMeans model\n kmeans_model = KMeans(n_clusters = i, random_state = 123)\n kmeans_model.fit(data)\n \n # Make predictions\n pred=kmeans_model.predict(data)\n \n # Calculate the Silhouette Score\n score = silhouette_score (data, pred, metric='euclidean')\n \n # Add the Silhouette score of the clusters to the score_list\n score_list.append(score)\n \n# Plot the Silhouette score and number of cluster \nplt.bar(range(2, 6), score_list)\n# Set title of the plot \nplt.title('Silhouette Score Plot')\n# Set x-axis label\nplt.xlabel('Number of Clusters K')\n# Set y-axis label\nplt.ylabel('Silhouette Scores')\n# Display plot\nplt.show()",
"_____no_output_____"
]
],
[
[
"## K-Means Clustering",
"_____no_output_____"
]
],
[
[
"# import pandas \nimport pandas as pd\n\n# import matplotlib for data visualization\nimport matplotlib.pyplot as plt\n\n# Import K-means\nfrom sklearn.cluster import KMeans\n\n# Create a DataFrame\ndata=pd.DataFrame({\"X\":[12,15,18,10,8,9,12,20],\n \"Y\":[6,16,17,8,7,6,9,18]})\n \n# Define number of clusters\nnum_clusters = 2\n \n# Create and fit the KMeans model\nkm = KMeans(n_clusters=num_clusters)\nkm.fit(data)\n \n# Predict the target variable\npred=km.predict(data)\n \n# Plot the Clusters\nplt.scatter(data.X,data.Y,c=pred, marker=\"o\", cmap=\"bwr_r\")\n# Set title of the plot \nplt.title('K-Means Clustering')\n# Set x-axis label\nplt.xlabel('X-Axis Values')\n# Set y-axis label\nplt.ylabel('Y-Axis Values')\n# Display the plot\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Hierarchical Clustering",
"_____no_output_____"
]
],
[
[
"# import pandas \nimport pandas as pd\n\n# import matplotlib for data visualization\nimport matplotlib.pyplot as plt\n\n# Import dendrogram\nfrom scipy.cluster.hierarchy import dendrogram\nfrom scipy.cluster.hierarchy import linkage\n \n# Create a DataFrame\ndata=pd.DataFrame({\"X\":[12,15,18,10,8,9,12,20],\n \"Y\":[6,16,17,8,7,6,9,18]})\n\n# create dendrogram using ward linkage\ndendrogram_plot = dendrogram(linkage(data, method = 'ward'))\n\n# Set title of the plot\nplt.title('Hierarchical Clustering: Dendrogram')\n\n# Set x-axis label\nplt.xlabel('Data Items')\n\n# Set y-axis label\nplt.ylabel('Distance')\n\n# Display the plot\nplt.show()",
"_____no_output_____"
],
[
"# import pandas \nimport pandas as pd\n\n# import matplotlib for data visualization\nimport matplotlib.pyplot as plt\n\n# Import Agglomerative Clustering\nfrom sklearn.cluster import AgglomerativeClustering\n \n# Create a DataFrame\ndata=pd.DataFrame({\"X\":[12,15,18,10,8,9,12,20],\n \"Y\":[6,16,17,8,7,6,9,18]})\n \n# Specify number of clusters\nnum_clusters = 2\n \n# Create agglomerative clustering model\nac = AgglomerativeClustering(n_clusters = num_clusters, linkage='ward')\n \n# Fit the Agglomerative Clustering model\nac.fit(data)\n \n# Predict the target variable\npred=ac.labels_\n \n# Plot the Clusters\nplt.scatter(data.X,data.Y,c=pred, marker=\"o\")\n\n# Set title of the plot \nplt.title('Agglomerative Clustering')\n# Set x-axis label\nplt.xlabel('X-Axis Values')\n# Set y-axis label\nplt.ylabel('Y-Axis Values')\n# Display the plot\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"## DBSCAN Clustering",
"_____no_output_____"
]
],
[
[
"# import pandas \nimport pandas as pd\n\n# import matplotlib for data visualization\nimport matplotlib.pyplot as plt\n\n# Import DBSCAN clustering model\nfrom sklearn.cluster import DBSCAN\n\n# import make_moons dataset\nfrom sklearn.datasets import make_moons\n \n# Generate some random moon data\nfeatures, label = make_moons(n_samples = 2000)\n \n# Create DBSCAN clustering model\ndb = DBSCAN()\n \n# Fit the Spectral Clustering model\ndb.fit(features)\n \n# Predict the target variable\npred_label=db.labels_\n \n# Plot the Clusters\nplt.scatter(features[:, 0], features[:, 1], c=pred_label, marker=\"o\",cmap=\"bwr_r\")\n# Set title of the plot\nplt.title('DBSCAN Clustering')\n# Set x-axis label\nplt.xlabel('X-Axis Values')\n# Set y-axis label\nplt.ylabel('Y-Axis Values')\n# Display the plot\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Spectral Clustering",
"_____no_output_____"
]
],
[
[
"# import pandas \nimport pandas as pd\n\n# import matplotlib for data visualization\nimport matplotlib.pyplot as plt\n\n# Import Spectral Clustering\nfrom sklearn.cluster import SpectralClustering\n \n# Create a DataFrame\ndata=pd.DataFrame({\"X\":[12,15,18,10,8,9,12,20],\n \"Y\":[6,16,17,8,7,6,9,18]})\n \n# Specify number of clusters\nnum_clusters = 2\n \n# Create Spectral Clustering model\nsc=SpectralClustering(num_clusters, affinity='rbf', n_init=100, assign_labels='discretize')\n \n# Fit the Spectral Clustering model\nsc.fit(data)\n \n# Predict the target variable\npred=sc.labels_\n \n# Plot the Clusters\nplt.scatter(data.X,data.Y,c=pred, marker=\"o\")\n# Set title of the plot\nplt.title('Spectral Clustering')\n# Set x-axis label\nplt.xlabel('X-Axis Values')\n# Set y-axis label\nplt.ylabel('Y-Axis Values')\n# Display the plot\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Cluster Performance Evaluation",
"_____no_output_____"
]
],
[
[
"# Import libraries\nimport pandas as pd\n \n# read the dataset\ndiabetes = pd.read_csv(\"diabetes.csv\")\n \n \n# Show top 5-records\ndiabetes.head()",
"_____no_output_____"
],
[
"# split dataset in two parts: feature set and target label \nfeature_set = ['pregnant', 'insulin', 'bmi', 'age','glucose','bp','pedigree']\nfeatures = diabetes[feature_set] \ntarget = diabetes.label\n \n# partition data into training and testing set \nfrom sklearn.model_selection import train_test_split\nfeature_train, feature_test, target_train, target_test = train_test_split(features, target, test_size=0.3, random_state=1)",
"_____no_output_____"
],
[
"# Import K-means Clustering\nfrom sklearn.cluster import KMeans\n\n# Import metrics module for performance evaluation\nfrom sklearn.metrics import davies_bouldin_score\nfrom sklearn.metrics import silhouette_score\nfrom sklearn.metrics import adjusted_rand_score\nfrom sklearn.metrics import jaccard_score\nfrom sklearn.metrics import f1_score\nfrom sklearn.metrics import fowlkes_mallows_score\n \n# Specify the number of clusters\nnum_clusters = 2\n \n# Create and fit the KMeans model\nkm = KMeans(n_clusters=num_clusters)\nkm.fit(feature_train)\n \n# Predict the target variable\npredictions=km.predict(feature_test)\n \n# Calculate internal performance evaluation measures\nprint(\"Davies-Bouldin Index:\", davies_bouldin_score(feature_test, predictions))\nprint(\"Silhouette Coefficient:\", silhouette_score(feature_test, predictions))\n\n# Calculate External performance evaluation measures\nprint(\"Adjusted Rand Score:\", adjusted_rand_score(target_test, predictions))\nprint(\"Jaccard Score:\", jaccard_score(target_test, predictions))\nprint(\"F-Measure(F1-Score):\", f1_score(target_test, predictions))\nprint(\"Fowlkes Mallows Score:\", fowlkes_mallows_score(target_test, predictions))",
"Davies-Bouldin Index: 0.7916877512521092\nSilhouette Coefficient: 0.5365443098840619\nAdjusted Rand Score: 0.03789319261940484\nJaccard Score: 0.29411764705882354\nF-Measure(F1-Score): 0.4545454545454546\nFowlkes Mallows Score: 0.6041244457314743\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb4f0b0e03655a5bccd8e8fa71d8889cc1efb609 | 3,186 | ipynb | Jupyter Notebook | examples/shortest.ipynb | carolina-mesquita-hp/trimesh | 90af69803958bdffee55a7fa3f43bdadfac95415 | [
"MIT"
] | 1,882 | 2015-04-21T06:51:06.000Z | 2022-03-31T03:16:12.000Z | examples/shortest.ipynb | carolina-mesquita-hp/trimesh | 90af69803958bdffee55a7fa3f43bdadfac95415 | [
"MIT"
] | 1,400 | 2016-01-22T14:05:04.000Z | 2022-03-31T19:13:55.000Z | examples/shortest.ipynb | carolina-mesquita-hp/trimesh | 90af69803958bdffee55a7fa3f43bdadfac95415 | [
"MIT"
] | 465 | 2015-05-29T21:27:38.000Z | 2022-03-31T17:45:46.000Z | 21.099338 | 67 | 0.515694 | [
[
[
"\"\"\"\nshortest.ipynb\n----------------\n\nGiven a mesh and two vertex indices find the shortest path\nbetween the two vertices while only traveling along edges\nof the mesh.\n\"\"\"\n\nimport trimesh\nimport numpy as np\nimport networkx as nx",
"_____no_output_____"
],
[
"# test on a sphere mesh\nmesh = trimesh.primitives.Sphere()",
"_____no_output_____"
],
[
"# edges without duplication\nedges = mesh.edges_unique",
"_____no_output_____"
],
[
"# the actual length of each unique edge\nlength = mesh.edges_unique_length",
"_____no_output_____"
],
[
"# create the graph with edge attributes for length\ng = nx.Graph()\nfor edge, L in zip(edges, length):\n g.add_edge(*edge, length=L)",
"_____no_output_____"
],
[
"# arbitrary indices of mesh.vertices to test with\nstart = 0\nend = int(len(mesh.vertices) / 2.0)",
"_____no_output_____"
],
[
"# run the shortest path query using length for edge weight\npath = nx.shortest_path(g,\n source=start,\n target=end,\n weight='length')",
"_____no_output_____"
],
[
"# VISUALIZE RESULT\n# make the sphere white\nmesh.visual.face_colors = [255,255,255,255]\n# Path3D with the path between the points\npath_visual = trimesh.load_path(mesh.vertices[path])",
"_____no_output_____"
],
[
"# create a scene with the mesh, path, and points\nscene = trimesh.Scene([path_visual, mesh ])",
"_____no_output_____"
],
[
"scene.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4f18e29fef054f0f2b247504b071d30f444945 | 12,681 | ipynb | Jupyter Notebook | resources/q-learning-one-dim-world.ipynb | subaochen/subaochen.github.io | 3857215d18b83cf12a9fa92b5b06bf1e817fa634 | [
"MIT"
] | null | null | null | resources/q-learning-one-dim-world.ipynb | subaochen/subaochen.github.io | 3857215d18b83cf12a9fa92b5b06bf1e817fa634 | [
"MIT"
] | 92 | 2019-07-06T13:29:19.000Z | 2022-03-04T15:15:30.000Z | resources/q-learning-one-dim-world.ipynb | subaochen/subaochen.github.io | 3857215d18b83cf12a9fa92b5b06bf1e817fa634 | [
"MIT"
] | 3 | 2019-09-05T07:48:08.000Z | 2021-10-05T02:47:16.000Z | 37.187683 | 1,010 | 0.533081 | [
[
[
"<h1>Table of Contents<span class=\"tocSkip\"></span></h1>\n<div class=\"toc\"><ul class=\"toc-item\"><li><span><a href=\"#定义系统参数\" data-toc-modified-id=\"定义系统参数-1\"><span class=\"toc-item-num\">1 </span>定义系统参数</a></span></li><li><span><a href=\"#Q表的创建函数,初始化为0\" data-toc-modified-id=\"Q表的创建函数,初始化为0-2\"><span class=\"toc-item-num\">2 </span>Q表的创建函数,初始化为0</a></span></li><li><span><a href=\"#策略\" data-toc-modified-id=\"策略-3\"><span class=\"toc-item-num\">3 </span>策略</a></span></li><li><span><a href=\"#和环境的交互\" data-toc-modified-id=\"和环境的交互-4\"><span class=\"toc-item-num\">4 </span>和环境的交互</a></span></li><li><span><a href=\"#更新环境\" data-toc-modified-id=\"更新环境-5\"><span class=\"toc-item-num\">5 </span>更新环境</a></span></li><li><span><a href=\"#游戏的实现\" data-toc-modified-id=\"游戏的实现-6\"><span class=\"toc-item-num\">6 </span>游戏的实现</a></span></li><li><span><a href=\"#执行强化学习训练\" data-toc-modified-id=\"执行强化学习训练-7\"><span class=\"toc-item-num\">7 </span>执行强化学习训练</a></span></li></ul></div>",
"_____no_output_____"
],
[
"Q-Learning是增强学习中model free的的重要算法,其基本思想是通过Q表记录并更新状态-行动的价值,使得最后获得一个“完美”的Q表:当agent处于任意状态时,查询该Q表即可获知如何行动。\n\n下面通过一个非常简单的小例子来说明Q Learning的思想(本案例主要参考了: https://morvanzhou.github.io/tutorials/ )。这是一个来自一维世界的agent,它只能在一个固定长度的线段上左右运动,每次只能运动一格,当运动到线段的最右边时才会获得奖励:+1的reward。初始时,agent位于线段的最左边,它并不知道在线段的最右边有个“宝物”可以获得reward。\n\n下面的一篇文章可以参考:https://blog.csdn.net/Young_Gy/article/details/73485518",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport time",
"_____no_output_____"
]
],
[
[
"## 定义系统参数",
"_____no_output_____"
]
],
[
[
"N_STATES = 6 # the length of the 1 dimensional world\nACTIONS = ['left', 'right'] # available actions\nEPSILON = 0.9 # greedy police,这里的意思是,即便在Q表中有对应的(最佳)Q价值,也有10%的概率随机选取action\nALPHA = 0.1 # learning rate\nGAMMA = 0.9 # discount factor\nMAX_EPISODES = 7 # maximum episodes\nFRESH_TIME = 0.01 # fresh time for one move\n\nTERMINAL='bang' # 终止状态,当agent遇到最右边的宝物时设置此状态\nDEBUG=True # 调试时设置为True则打印更多的信息",
"_____no_output_____"
]
],
[
[
"## Q表的创建函数,初始化为0\n本案例Q表的结构如下,其中最左边的一列是状态,本案例有6个状态,即agent可以在6个格子内左右移动:\n\n| |left|right|\n|---|---|---|\n|0|0|0|\n|1|0|0|\n|2|0|0|\n|3|0|0|\n|4|0|0|\n|5|0|0|\n",
"_____no_output_____"
]
],
[
[
"def build_q_table(n_states, actions):\n table = pd.DataFrame(\n np.zeros((n_states, len(actions))), # q_table initial values\n columns=actions, # actions's name\n )\n print(table) # show table\n return table",
"_____no_output_____"
]
],
[
[
"## 策略\n这是增强学习中的策略部分,这里的策略很简单:如果平均随机采样值大于设定的epsilon或者当前状态的所有动作价值为0则随机游走探索(随机选取动作),否则从Q表选取价值最大的动作。我们的目标是不断优化Q表中的动作价值。",
"_____no_output_____"
]
],
[
[
"def choose_action(state, q_table):\n # This is how to choose an action\n state_actions = q_table.iloc[state, :]\n # 如果当前状态的所有动作的价值为0,则随机选取动作\n # 如果平均随机采样值 > EPSILON,则随机选取动作\n if (np.random.uniform() > EPSILON) or ((state_actions == 0).all()): \n action_name = np.random.choice(ACTIONS)\n else: # act greedy\n action_name = state_actions.idxmax()\n return action_name",
"_____no_output_____"
]
],
[
[
"## 和环境的交互\n环境接受agent的action并执行之,然后给出下一个状态和相应的reward。只有agent走到了最右边,环境才给予+1的reward,其他情况下reward=0。",
"_____no_output_____"
]
],
[
[
"def get_env_feedback(S, A):\n # This is how agent will interact with the environment\n # S_: next status\n # R: reward to action A\n if A == 'right': # move right\n if S == N_STATES - 2: # terminate\n S_ = TERMINAL\n R = 1\n else:\n S_ = S + 1\n R = 0\n else: # move left\n R = 0\n if S == 0:\n S_ = S # reach the wall\n else:\n S_ = S - 1\n return S_, R",
"_____no_output_____"
]
],
[
[
"## 更新环境\n这是agent和环境交互的一部分,绘制环境。",
"_____no_output_____"
]
],
[
[
"def update_env(S, episode, step_counter):\n # This is how environment be updated\n env_list = ['-']*(N_STATES-1) + ['T'] # '---------T' our environment\n if S == TERMINAL:\n interaction = 'Episode %s: total_steps = %s' % (episode+1, step_counter)\n print('\\r{}'.format(interaction), end='')\n time.sleep(2)\n print('\\r ', end='')\n else:\n env_list[S] = 'o'\n interaction = ''.join(env_list)\n print('\\r{}'.format(interaction), end='')\n time.sleep(FRESH_TIME)",
"_____no_output_____"
]
],
[
[
"## 游戏的实现\nrl = reinforcement learning\n\n这里重点区分两个概念:\n\n* q_predict,Q预测,即当前(S,A)在Q表中的值(简称Q价值),表达了在S状态下如果采取A动作的价值多少。这是在环境还没有接收并执行A动作时的Q价值,即此时A动作还没有真正执行,因此是一个预测值,或者说是上一轮(S,A)后的Q真实,如果存在上一轮的话。\n* q_target,Q真实,即(S,A)执行后的Q价值:环境接收并执行了A动作,给出了S_(下一个动作)和R(reward),则根据Q Learning算法的更新公式可计算q_target。之所以叫做Q真实,是因为这个时候A动作已经被环境执行了,这是确凿发生的事实产生的Q价值。\n\n画个图来进一步理解:\n\n\n\n下图说明了Q Learning的算法(see: https://www.cse.unsw.edu.au/~cs9417ml/RL1/algorithms.html ):\n",
"_____no_output_____"
]
],
[
[
"def rl():\n # main part of RL loop\n q_table = build_q_table(N_STATES, ACTIONS)\n for episode in range(MAX_EPISODES):\n step_counter = 0\n S = 0\n is_terminated = False\n update_env(S, episode, step_counter)\n while not is_terminated:\n A = choose_action(S, q_table)\n # Q表中当前(S,A)对应的值称为Q预测,即当前的(S,A)组合的价值。\n q_predict = q_table.loc[S, A]\n S_, R = get_env_feedback(S, A) # take action & get next state and reward\n if S_ != TERMINAL:\n q_target = R + GAMMA * q_table.iloc[S_, :].max() # next state is not terminal\n else:\n q_target = R # next state is terminal\n is_terminated = True # terminate this episode\n\n q_table.loc[S, A] = q_predict + ALPHA * (q_target - q_predict) # update\n if DEBUG == True and q_target != q_predict:\n print(' %s episode,S(%s),A(%s),R(%.6f),S_(%s),q_p(%.6f),q_t(%.6f),q_tab[S,A](%.6f)' % (episode,S,A,R,S_,q_predict,q_target,q_table.loc[S,A]))\n #print(q_table)\n S = S_ # move to next state\n\n update_env(S, episode, step_counter+1)\n step_counter += 1\n return q_table",
"_____no_output_____"
]
],
[
[
"## 执行强化学习训练\n遗憾的是,还不知道在jupyter中如何不换行持续显示训练的过程,请高手指点。目前可以通过打开DEBUG开关观察agent的训练过程。",
"_____no_output_____"
]
],
[
[
"q_table = rl()\nprint('\\r\\nQ-table after training:\\n')\nprint(q_table)",
" left right\n0 0.0 0.0\n1 0.0 0.0\n2 0.0 0.0\n3 0.0 0.0\n4 0.0 0.0\n5 0.0 0.0\n----oT 0 episode,S(4),A(right),R(1.000000),S_(bang),q_p(0.000000),q_t(1.000000),q_tab[S,A](0.100000)\n---o-T 1 episode,S(3),A(right),R(0.000000),S_(4),q_p(0.000000),q_t(0.090000),q_tab[S,A](0.009000)\n----oT 1 episode,S(4),A(right),R(1.000000),S_(bang),q_p(0.100000),q_t(1.000000),q_tab[S,A](0.190000)\n--o--T 2 episode,S(2),A(right),R(0.000000),S_(3),q_p(0.000000),q_t(0.008100),q_tab[S,A](0.000810)\n---o-T 2 episode,S(3),A(right),R(0.000000),S_(4),q_p(0.009000),q_t(0.171000),q_tab[S,A](0.025200)\n----oT 2 episode,S(4),A(right),R(1.000000),S_(bang),q_p(0.190000),q_t(1.000000),q_tab[S,A](0.271000)\n-o---T 3 episode,S(1),A(right),R(0.000000),S_(2),q_p(0.000000),q_t(0.000729),q_tab[S,A](0.000073)\n--o--T 3 episode,S(2),A(right),R(0.000000),S_(3),q_p(0.000810),q_t(0.022680),q_tab[S,A](0.002997)\n---o-T 3 episode,S(3),A(right),R(0.000000),S_(4),q_p(0.025200),q_t(0.243900),q_tab[S,A](0.047070)\n----oT 3 episode,S(4),A(right),R(1.000000),S_(bang),q_p(0.271000),q_t(1.000000),q_tab[S,A](0.343900)\no----T 4 episode,S(0),A(right),R(0.000000),S_(1),q_p(0.000000),q_t(0.000066),q_tab[S,A](0.000007)\n-o---T 4 episode,S(1),A(right),R(0.000000),S_(2),q_p(0.000073),q_t(0.002697),q_tab[S,A](0.000335)\n--o--T 4 episode,S(2),A(right),R(0.000000),S_(3),q_p(0.002997),q_t(0.042363),q_tab[S,A](0.006934)\n---o-T 4 episode,S(3),A(right),R(0.000000),S_(4),q_p(0.047070),q_t(0.309510),q_tab[S,A](0.073314)\n----oT 4 episode,S(4),A(right),R(1.000000),S_(bang),q_p(0.343900),q_t(1.000000),q_tab[S,A](0.409510)\no----T 5 episode,S(0),A(right),R(0.000000),S_(1),q_p(0.000007),q_t(0.000302),q_tab[S,A](0.000036)\n-o---T 5 episode,S(1),A(right),R(0.000000),S_(2),q_p(0.000335),q_t(0.006240),q_tab[S,A](0.000926)\n--o--T 5 episode,S(2),A(right),R(0.000000),S_(3),q_p(0.006934),q_t(0.065983),q_tab[S,A](0.012839)\n---o-T 5 episode,S(3),A(right),R(0.000000),S_(4),q_p(0.073314),q_t(0.368559),q_tab[S,A](0.102839)\n----oT 5 episode,S(4),A(right),R(1.000000),S_(bang),q_p(0.409510),q_t(1.000000),q_tab[S,A](0.468559)\no----T 6 episode,S(0),A(right),R(0.000000),S_(1),q_p(0.000036),q_t(0.000833),q_tab[S,A](0.000116)\n-o---T 6 episode,S(1),A(right),R(0.000000),S_(2),q_p(0.000926),q_t(0.011555),q_tab[S,A](0.001989)\n--o--T 6 episode,S(2),A(right),R(0.000000),S_(3),q_p(0.012839),q_t(0.092555),q_tab[S,A](0.020810)\n---o-T 6 episode,S(3),A(right),R(0.000000),S_(4),q_p(0.102839),q_t(0.421703),q_tab[S,A](0.134725)\n----oT 6 episode,S(4),A(right),R(1.000000),S_(bang),q_p(0.468559),q_t(1.000000),q_tab[S,A](0.521703)\n \nQ-table after training:\n\n left right\n0 0.0 0.000116\n1 0.0 0.001989\n2 0.0 0.020810\n3 0.0 0.134725\n4 0.0 0.521703\n5 0.0 0.000000\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4f2152910112edde853d43a601d781b942a519 | 10,875 | ipynb | Jupyter Notebook | site/en-snapshot/hub/tutorials/senteval_for_universal_sentence_encoder_cmlm.ipynb | serbible/docs-l10n | 60c70a3167b2f751c0ef29b9a9fb8b5ad5bb7d02 | [
"Apache-2.0"
] | null | null | null | site/en-snapshot/hub/tutorials/senteval_for_universal_sentence_encoder_cmlm.ipynb | serbible/docs-l10n | 60c70a3167b2f751c0ef29b9a9fb8b5ad5bb7d02 | [
"Apache-2.0"
] | null | null | null | site/en-snapshot/hub/tutorials/senteval_for_universal_sentence_encoder_cmlm.ipynb | serbible/docs-l10n | 60c70a3167b2f751c0ef29b9a9fb8b5ad5bb7d02 | [
"Apache-2.0"
] | null | null | null | 43.674699 | 480 | 0.557241 | [
[
[
"**Copyright 2021 The TensorFlow Hub Authors.**\n\nLicensed under the Apache License, Version 2.0 (the \"License\");",
"_____no_output_____"
]
],
[
[
"# Copyright 2021 The TensorFlow Hub Authors. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n# ==============================================================================",
"_____no_output_____"
]
],
[
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://tfhub.dev/google/universal-sentence-encoder-cmlm/en-base/1\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/hub/blob/master/examples/colab/senteval_for_universal_sentence_encoder_cmlm.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/hub/blob/master/examples/colab/senteval_for_universal_sentence_encoder_cmlm.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/hub/examples/colab/senteval_for_universal_sentence_encoder_cmlm.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n <td>\n <a href=\"https://tfhub.dev/google/universal-sentence-encoder-cmlm/en-base/1\"><img src=\"https://www.tensorflow.org/images/hub_logo_32px.png\" />See TF Hub model</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"#Universal Sentence Encoder SentEval demo\nThis colab demostrates the [Universal Sentence Encoder CMLM model](https://tfhub.dev/google/universal-sentence-encoder-cmlm/en-base/1) using the [SentEval](https://github.com/facebookresearch/SentEval) toolkit, which is a library for measuring the quality of sentence embeddings. The SentEval toolkit includes a diverse set of downstream tasks that are able to evaluate the generalization power of an embedding model and to evaluate the linguistic properties encoded.\n\nRun the first two code blocks to setup the environment, in the third code block you can pick a SentEval task to evaluate the model. A GPU runtime is recommended to run this Colab.\n\nTo learn more about the Universal Sentence Encoder CMLM model, see https://openreview.net/forum?id=WDVD4lUCTzU.",
"_____no_output_____"
]
],
[
[
"#@title Install dependencies\n!pip install --quiet tensorflow_text==2.7.3\n!pip install --quiet torch==1.8.1",
"_____no_output_____"
]
],
[
[
"## Download SentEval and task data\nThis step download SentEval from github and execute the data script to download the task data. It may take up to 5 minutes to complete.",
"_____no_output_____"
]
],
[
[
"#@title Install SentEval and download task data\n!rm -rf ./SentEval\n!git clone https://github.com/facebookresearch/SentEval.git\n!cd $PWD/SentEval/data/downstream && bash get_transfer_data.bash > /dev/null 2>&1",
"_____no_output_____"
]
],
[
[
"#Execute a SentEval evaulation task\nThe following code block executes a SentEval task and output the results, choose one of the following tasks to evaluate the USE CMLM model:\n\n```\nMR\tCR\tSUBJ\tMPQA\tSST\tTREC\tMRPC\tSICK-E\n```\n\nSelect a model, params and task to run. The rapid prototyping params can be used for reducing computation time for faster result.\n\nIt typically takes 5-15 mins to complete a task with the **'rapid prototyping'** params and up to an hour with the **'slower, best performance'** params.\n\n```\nparams = {'task_path': PATH_TO_DATA, 'usepytorch': True, 'kfold': 5}\nparams['classifier'] = {'nhid': 0, 'optim': 'rmsprop', 'batch_size': 128,\n 'tenacity': 3, 'epoch_size': 2}\n```\n\nFor better result, use the slower **'slower, best performance'** params, computation may take up to 1 hour:\n\n```\nparams = {'task_path': PATH_TO_DATA, 'usepytorch': True, 'kfold': 10}\nparams['classifier'] = {'nhid': 0, 'optim': 'adam', 'batch_size': 16,\n 'tenacity': 5, 'epoch_size': 6}\n```\n",
"_____no_output_____"
]
],
[
[
"import os\nos.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'\n\nimport sys\nsys.path.append(f'{os.getcwd()}/SentEval')\n\nimport tensorflow as tf\n\n# Prevent TF from claiming all GPU memory so there is some left for pytorch.\ngpus = tf.config.list_physical_devices('GPU')\nif gpus:\n # Memory growth needs to be the same across GPUs.\n for gpu in gpus:\n tf.config.experimental.set_memory_growth(gpu, True)\n\nimport tensorflow_hub as hub\nimport tensorflow_text\nimport senteval\nimport time\n\nPATH_TO_DATA = f'{os.getcwd()}/SentEval/data'\nMODEL = 'https://tfhub.dev/google/universal-sentence-encoder-cmlm/en-base/1' #@param ['https://tfhub.dev/google/universal-sentence-encoder-cmlm/en-base/1', 'https://tfhub.dev/google/universal-sentence-encoder-cmlm/en-large/1']\nPARAMS = 'rapid prototyping' #@param ['slower, best performance', 'rapid prototyping']\nTASK = 'CR' #@param ['CR','MR', 'MPQA', 'MRPC', 'SICKEntailment', 'SNLI', 'SST2', 'SUBJ', 'TREC']\n\nparams_prototyping = {'task_path': PATH_TO_DATA, 'usepytorch': True, 'kfold': 5}\nparams_prototyping['classifier'] = {'nhid': 0, 'optim': 'rmsprop', 'batch_size': 128,\n 'tenacity': 3, 'epoch_size': 2}\n\nparams_best = {'task_path': PATH_TO_DATA, 'usepytorch': True, 'kfold': 10}\nparams_best['classifier'] = {'nhid': 0, 'optim': 'adam', 'batch_size': 16,\n 'tenacity': 5, 'epoch_size': 6}\n\nparams = params_best if PARAMS == 'slower, best performance' else params_prototyping\n\npreprocessor = hub.KerasLayer(\n \"https://tfhub.dev/tensorflow/bert_en_uncased_preprocess/3\")\nencoder = hub.KerasLayer(\n \"https://tfhub.dev/google/universal-sentence-encoder-cmlm/en-base/1\")\n\ninputs = tf.keras.Input(shape=tf.shape(''), dtype=tf.string)\noutputs = encoder(preprocessor(inputs))\n\nmodel = tf.keras.Model(inputs=inputs, outputs=outputs)\n\ndef prepare(params, samples):\n return\n\ndef batcher(_, batch):\n batch = [' '.join(sent) if sent else '.' for sent in batch]\n return model.predict(tf.constant(batch))[\"default\"]\n\n\nse = senteval.engine.SE(params, batcher, prepare)\nprint(\"Evaluating task %s with %s parameters\" % (TASK, PARAMS))\nstart = time.time()\nresults = se.eval(TASK)\nend = time.time()\nprint('Time took on task %s : %.1f. seconds' % (TASK, end - start))\nprint(results)\n",
"_____no_output_____"
]
],
[
[
"#Learn More\n\n* Find more text embedding models on [TensorFlow Hub](https://tfhub.dev)\n* See also the [Multilingual Universal Sentence Encoder CMLM model](https://tfhub.dev/google/universal-sentence-encoder-cmlm/multilingual-base-br/1)\n* Check out other [Universal Sentence Encoder models](https://tfhub.dev/google/collections/universal-sentence-encoder/1)\n\n## Reference\n\n* Ziyi Yang, Yinfei Yang, Daniel Cer, Jax Law, Eric Darve. [Universal Sentence Representations Learning with Conditional Masked Language Model. November 2020](https://openreview.net/forum?id=WDVD4lUCTzU)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb4f35e8166e51f5854dd05b281861f002ef1005 | 65,840 | ipynb | Jupyter Notebook | HW2/ex3.ipynb | hansenbeast/2019-SYSU-DM | 39250fe21681ea445d38d521679fb4b9d6b0d345 | [
"MIT"
] | 2 | 2021-06-21T02:54:49.000Z | 2021-07-15T07:26:41.000Z | HW2/ex3.ipynb | hansenbeast/2019-SYSU-DM | 39250fe21681ea445d38d521679fb4b9d6b0d345 | [
"MIT"
] | null | null | null | HW2/ex3.ipynb | hansenbeast/2019-SYSU-DM | 39250fe21681ea445d38d521679fb4b9d6b0d345 | [
"MIT"
] | 1 | 2019-06-04T06:20:59.000Z | 2019-06-04T06:20:59.000Z | 160.194647 | 13,340 | 0.86657 | [
[
[
"import tensorflow as tf\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn import preprocessing \n\n# Try to find value for W and b to compute y_data = x_data * W + b \n\n# Define dimensions\nd = 2 # Size of the parameter space\nN = 50 # Number of data sample\n\n# Model parameters\nW = tf.Variable(tf.zeros([d, 1], tf.float32), name=\"weights\")\nb = tf.Variable(tf.zeros([1], tf.float32), name=\"biases\")\n\n# Model input and output\nx = tf.placeholder(tf.float32, shape=[None, d])\ny = tf.placeholder(tf.float32, shape=[None, 1])\n\n# hypothesis\nlinear_regression_model = tf.add(tf.matmul(x, W), b)\n# cost/loss function\nloss = tf.reduce_mean(tf.square(linear_regression_model - y)) / 2\n\n# optimizer\noptimizer = tf.train.GradientDescentOptimizer(learning_rate=0.00015)\ntrain = optimizer.minimize(loss)\n\n# 导入训练集和测试集\ntraining_filename = \"dataForTraining.txt\"\ntesting_filename = \"dataForTesting.txt\"\ntraining_dataset = np.loadtxt(training_filename)\ntesting_dataset = np.loadtxt(testing_filename)\ndataset = np.vstack((training_dataset,testing_dataset))\n\n# 保存训练集中的参数(均值、方差)直接使用其对象转换测试集数据\n# 特征缩放\nmin_max_scaler = preprocessing.MinMaxScaler() \n# 标准化\nnormal_scaler = preprocessing.StandardScaler().fit(training_dataset)\n# 归一化\ndataset = min_max_scaler.fit_transform(dataset)\n# 标准化\ntraining_dataset = normal_scaler.transform(training_dataset)\ntesting_dataset = normal_scaler.transform(testing_dataset)\n\nprint(np.mean(training_dataset,axis=0))\nprint(np.std(training_dataset,axis=0))\nprint(np.mean(testing_dataset,axis=0))\nprint(np.std(testing_dataset,axis=0))\n\nx_train = np.array(training_dataset[:,:2])\ny_train = np.array(training_dataset[:,2:3])\nx_test = np.array(testing_dataset[:,:2])\ny_test = np.array(testing_dataset[:,2:3])\nprint(\"Training data shape:\")\nprint(x_train.shape)\nprint(\"Testing data shape:\")\nprint(x_test.shape)\nprint('')\nprint(\"normalized training data:\")\nprint(x_train)\nprint('')\nprint(\"normalized testing data:\")\nprint(x_test)\nprint('')\n\nmini_batch_size = 1\nn_batch = N // mini_batch_size + (N % mini_batch_size != 0)\nprint(n_batch)\n\nsave_step_loss = {\"step\":[],\"train_loss\":[],\"test_loss\":[]}# 保存step和loss用于可视化操作\ninit = tf.global_variables_initializer()\nwith tf.Session() as sess:\n sess.run(init) # reset values to wrong\n steps = 1500001\n for i in range(steps):\n i_batch = (i % n_batch)*mini_batch_size\n batch = x_train[i_batch:i_batch+mini_batch_size], y_train[i_batch:i_batch+mini_batch_size]\n sess.run(train, {x: batch[0], y:batch[1]})\n# random_index = np.random.choice(N)\n# sess.run(train, {x: [x_train[random_index]], y:[y_train[random_index]]})\n if i % 100000 == 0:\n # evaluate training accuracy\n print(\"iteration times: %s\" % i)\n curr_W, curr_b, curr_train_loss = sess.run([W, b, loss], {x: x_train, y: y_train})\n print(\"W: %s \\nb: %s \\nTrain Loss: %s\" % (curr_W, curr_b, curr_train_loss))\n # Accuracy computation\n curr_test_loss = sess.run(loss,{x:x_test,y:y_test})\n print(\"Test Loss: %s\\n\" % curr_test_loss)\n save_step_loss[\"step\"].append(i)\n save_step_loss[\"train_loss\"].append(curr_train_loss)\n save_step_loss[\"test_loss\"].append(curr_test_loss)\n\n#画图损失函数变化曲线\nplt.plot(save_step_loss[\"step\"],save_step_loss[\"train_loss\"],label='Training Loss')\nplt.plot(save_step_loss[\"step\"],save_step_loss[\"test_loss\"],label='Testing Loss')\nplt.xlabel('Iteration times')\nplt.ylabel('Loss')\nplt.legend()\nplt.show()\n#画图损失函数变化曲线\nplt.plot(save_step_loss[\"step\"][1:],save_step_loss[\"train_loss\"][1:],label='Training Loss')\nplt.plot(save_step_loss[\"step\"][1:],save_step_loss[\"test_loss\"][1:],label='Testing Loss')\nplt.xlabel('Iteration times')\nplt.ylabel('Loss')\nplt.legend()\nplt.show()\n#画图损失函数变化曲线\nplt.plot(save_step_loss[\"step\"][3:],save_step_loss[\"train_loss\"][3:],label='Training Loss')\nplt.plot(save_step_loss[\"step\"][3:],save_step_loss[\"test_loss\"][3:],label='Testing Loss')\nplt.xlabel('Iteration times')\nplt.ylabel('Loss')\nplt.legend()\nplt.show()\n#画图损失函数变化曲线\nplt.plot(save_step_loss[\"step\"][5:],save_step_loss[\"train_loss\"][5:],label='Training Loss')\nplt.plot(save_step_loss[\"step\"][5:],save_step_loss[\"test_loss\"][5:],label='Testing Loss')\nplt.xlabel('Iteration times')\nplt.ylabel('Loss')\nplt.legend()\nplt.show()\n\nprint('Train Loss:\\n',save_step_loss[\"train_loss\"])\nprint('')\nprint('Test Loss:\\n',save_step_loss[\"test_loss\"])",
"[1.42108547e-16 4.44089210e-17 4.56856775e-16]\n[1. 1. 1.]\n[-0.28034986 -0.65473668 0.42167664]\n[0.69835876 0.59887862 0.68026069]\nTraining data shape:\n(50, 2)\nTesting data shape:\n(10, 2)\n\nnormalized training data:\n[[-0.00635354 -1.13312693]\n [ 0.7481291 -0.92751508]\n [ 0.54958103 -0.34494816]\n [ 0.86725793 -0.75617187]\n [-0.12548237 -1.30447015]\n [ 0.39074258 -1.61288793]\n [-0.64170733 -1.40042235]\n [-0.99909384 -0.57797492]\n [-0.91967462 -0.07079902]\n [-0.36374005 -0.79044051]\n [-0.68141695 0.3781202 ]\n [ 0.86725793 1.09433483]\n [ 1.62174057 2.53361782]\n [-0.16519199 0.82018569]\n [-1.03880346 0.1862158 ]\n [-0.48286889 0.44323062]\n [ 2.29680397 1.50555854]\n [-0.04606315 -0.85897779]\n [ 0.03335607 0.47749926]\n [ 0.39074258 0.10739793]\n [ 1.30406367 1.16287212]\n [-0.99909384 -0.76302559]\n [-0.5225785 -0.53685255]\n [-0.48286889 -0.54370628]\n [-1.31677074 -0.79386737]\n [-1.43589958 -1.44154472]\n [ 1.22464444 1.29994669]\n [ 1.70115979 2.53361782]\n [ 0.58929065 0.58030519]\n [ 0.27161375 -0.39292426]\n [-1.55502842 -0.27641087]\n [-0.08577276 0.78591705]\n [ 0.66870987 0.13481284]\n [-1.59473803 -0.57454806]\n [ 1.93941746 0.85445433]\n [ 1.22464444 0.48435299]\n [ 0.82754832 1.49870481]\n [-1.03880346 -0.16332435]\n [-1.19764191 -0.42376603]\n [-0.60199772 0.58715892]\n [-0.95938423 -0.12905571]\n [-0.32403044 -0.15647062]\n [-0.40344966 -0.44775408]\n [-1.31677074 -0.2352885 ]\n [ 0.27161375 1.20056762]\n [ 0.4304522 1.84824497]\n [ 0.78783871 -0.50258391]\n [ 0.23190414 -0.76302559]\n [ 1.66145018 -1.15368812]\n [-1.59473803 -1.44154472]]\n\nnormalized testing data:\n[[-0.32403044 -1.44839844]\n [ 0.1127753 -0.40663171]\n [ 0.35103297 -0.25242282]\n [-1.27706113 0.06627555]\n [-0.8402554 -1.29761642]\n [-0.87996501 -1.41755667]\n [ 1.06580599 0.18278894]\n [ 0.23190414 -0.17017808]\n [-1.03880346 -0.6190973 ]\n [-0.2049016 -1.1845299 ]]\n\n50\niteration times: 0\nW: [[-1.2158955e-06]\n [-2.1684990e-04]] \nb: [0.00019137] \nTrain Loss: 0.49986112\nTest Loss: 0.3200914\n\niteration times: 100000\nW: [[ 0.9029868]\n [-1.1202316]] \nb: [1.5226635e-07] \nTrain Loss: 5.4115448e-05\nTest Loss: 0.00185396\n\niteration times: 200000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.555532e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 300000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.555591e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 400000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.5555615e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 500000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.555532e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 600000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.555591e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 700000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.5555615e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 800000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.555532e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 900000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.555591e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 1000000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.5555615e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 1100000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.555532e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 1200000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.555591e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 1300000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.5555615e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 1400000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.555532e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\niteration times: 1500000\nW: [[ 0.90379936]\n [-1.1210359 ]] \nb: [3.555591e-07] \nTrain Loss: 5.3752112e-05\nTest Loss: 0.0018809683\n\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
cb4f38c1a22859fe2f22adb8c19d1a5d4d654cfa | 5,140 | ipynb | Jupyter Notebook | test/jupyter_book.ipynb | michelmetran/jupyter_book | a2e99a6646914f5545bf9e3c9550ef1a93efdee2 | [
"MIT"
] | null | null | null | test/jupyter_book.ipynb | michelmetran/jupyter_book | a2e99a6646914f5545bf9e3c9550ef1a93efdee2 | [
"MIT"
] | null | null | null | test/jupyter_book.ipynb | michelmetran/jupyter_book | a2e99a6646914f5545bf9e3c9550ef1a93efdee2 | [
"MIT"
] | null | null | null | 20.725806 | 101 | 0.521984 | [
[
[
"<br>\n\n# Introdução",
"_____no_output_____"
]
],
[
[
"#!pip3 install jupyter-book --upgrade",
"_____no_output_____"
],
[
"import os\nimport shutil",
"_____no_output_____"
],
[
"os.getcwd()",
"_____no_output_____"
],
[
"shutil.rmtree(\n path=os.path.join('..', 'my_test_book'),\n ignore_errors=True,\n onerror=print('Deu ruin!'),\n)",
"_____no_output_____"
],
[
"# Create schema\n!jupyter-book create ../my_test_book/",
"_____no_output_____"
],
[
"# Build\n!jupyter-book build ../my_test_book/",
"_____no_output_____"
],
[
"a = 'file:///home/michel/Codes/case_office/jupyter_book/my_test_book/_build/html/index.html'\na",
"_____no_output_____"
]
],
[
[
"<br>\n\n# Publicação",
"_____no_output_____"
],
[
"<br>\n\n## Alternativa 1\n\nPublicar em uma nova *branch*.\n\nPor meio do *package* **ghp-import** é possível deixar o livro na *branch* **gh-pages**.",
"_____no_output_____"
]
],
[
[
"#!pip3 install ghp-import --upgrade",
"_____no_output_____"
],
[
"from ghp_import import ghp_import",
"_____no_output_____"
]
],
[
[
"<br>\n\nSendo possível fazer isso por meio do terminal.",
"_____no_output_____"
]
],
[
[
"#!ghp-import -n -p -f ../docs/_build/html",
"_____no_output_____"
]
],
[
[
"<br>\n\nOu por meio do python.",
"_____no_output_____"
]
],
[
[
"ghp_import(\n #srcdir='docs',\n #srcdir='../docs/my_test_book/_build/html',\n srcdir='../docs/my_test_book/_build/html',\n push=True,\n #cname='example.com'\n)",
"_____no_output_____"
]
],
[
[
"<br>\n\n## Alternativa 2: docs\n\nCopiando o conteúdo do *build* para a pasta *docs* e definindo ela no gitpages.",
"_____no_output_____"
]
],
[
[
"src = os.path.join('..', 'my_test_book', '_build', 'html')\ndocs_path = os.path.join('..', 'docs')\n\nshutil.copytree(src, docs_path) ",
"_____no_output_____"
],
[
"# Create file .nojerkyll\nopen(os.path.join(docs_path, '.nojekyll'), mode='w').close()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb4f4fe4738c02369877099d533a04b9f4346e4e | 4,046 | ipynb | Jupyter Notebook | ipynb/Qatar.ipynb | RobertRosca/oscovida.github.io | d609949076e3f881e38ec674ecbf0887e9a2ec25 | [
"CC-BY-4.0"
] | null | null | null | ipynb/Qatar.ipynb | RobertRosca/oscovida.github.io | d609949076e3f881e38ec674ecbf0887e9a2ec25 | [
"CC-BY-4.0"
] | null | null | null | ipynb/Qatar.ipynb | RobertRosca/oscovida.github.io | d609949076e3f881e38ec674ecbf0887e9a2ec25 | [
"CC-BY-4.0"
] | null | null | null | 28.293706 | 159 | 0.50692 | [
[
[
"# Qatar\n\n* Homepage of project: https://oscovida.github.io\n* [Execute this Jupyter Notebook using myBinder](https://mybinder.org/v2/gh/oscovida/binder/master?filepath=ipynb/Qatar.ipynb)",
"_____no_output_____"
]
],
[
[
"import datetime\nimport time\n\nstart = datetime.datetime.now()\nprint(f\"Notebook executed on: {start.strftime('%d/%m/%Y %H:%M:%S%Z')} {time.tzname[time.daylight]}\")",
"_____no_output_____"
],
[
"%config InlineBackend.figure_formats = ['svg']\nfrom oscovida import *",
"_____no_output_____"
],
[
"overview(\"Qatar\");",
"_____no_output_____"
],
[
"# load the data\ncases, deaths, region_label = get_country_data(\"Qatar\")\n\n# compose into one table\ntable = compose_dataframe_summary(cases, deaths)\n\n# show tables with up to 500 rows\npd.set_option(\"max_rows\", 500)\n\n# display the table\ntable",
"_____no_output_____"
]
],
[
[
"# Explore the data in your web browser\n\n- If you want to execute this notebook, [click here to use myBinder](https://mybinder.org/v2/gh/oscovida/binder/master?filepath=ipynb/Qatar.ipynb)\n- and wait (~1 to 2 minutes)\n- Then press SHIFT+RETURN to advance code cell to code cell\n- See http://jupyter.org for more details on how to use Jupyter Notebook",
"_____no_output_____"
],
[
"# Acknowledgements:\n\n- Johns Hopkins University provides data for countries\n- Robert Koch Institute provides data for within Germany\n- Open source and scientific computing community for the data tools\n- Github for hosting repository and html files\n- Project Jupyter for the Notebook and binder service\n- The H2020 project Photon and Neutron Open Science Cloud ([PaNOSC](https://www.panosc.eu/))\n\n--------------------",
"_____no_output_____"
]
],
[
[
"print(f\"Download of data from Johns Hopkins university: cases at {fetch_cases_last_execution()} and \"\n f\"deaths at {fetch_deaths_last_execution()}.\")",
"_____no_output_____"
],
[
"# to force a fresh download of data, run \"clear_cache()\"",
"_____no_output_____"
],
[
"print(f\"Notebook execution took: {datetime.datetime.now()-start}\")\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb4f57bd13880cbd69f67227a27529dbee19f0d8 | 40,984 | ipynb | Jupyter Notebook | Week 2/Residual_Networks_v2a.ipynb | Suparno1998/convolutional-nn | 569b443b35e3547067b9a5f6cc5b6c48914051f5 | [
"MIT"
] | 1 | 2020-04-02T08:06:28.000Z | 2020-04-02T08:06:28.000Z | Week 2/Residual_Networks_v2a.ipynb | Suparno1998/convolutional-nn | 569b443b35e3547067b9a5f6cc5b6c48914051f5 | [
"MIT"
] | null | null | null | Week 2/Residual_Networks_v2a.ipynb | Suparno1998/convolutional-nn | 569b443b35e3547067b9a5f6cc5b6c48914051f5 | [
"MIT"
] | null | null | null | 43.880086 | 428 | 0.593134 | [
[
[
"# Residual Networks\n\nWelcome to the second assignment of this week! You will learn how to build very deep convolutional networks, using Residual Networks (ResNets). In theory, very deep networks can represent very complex functions; but in practice, they are hard to train. Residual Networks, introduced by [He et al.](https://arxiv.org/pdf/1512.03385.pdf), allow you to train much deeper networks than were previously practically feasible.\n\n**In this assignment, you will:**\n- Implement the basic building blocks of ResNets. \n- Put together these building blocks to implement and train a state-of-the-art neural network for image classification. ",
"_____no_output_____"
],
[
"## <font color='darkblue'>Updates</font>\n\n#### If you were working on the notebook before this update...\n* The current notebook is version \"2a\".\n* You can find your original work saved in the notebook with the previous version name (\"v2\") \n* To view the file directory, go to the menu \"File->Open\", and this will open a new tab that shows the file directory.\n\n#### List of updates\n* For testing on an image, replaced `preprocess_input(x)` with `x=x/255.0` to normalize the input image in the same way that the model's training data was normalized.\n* Refers to \"shallower\" layers as those layers closer to the input, and \"deeper\" layers as those closer to the output (Using \"shallower\" layers instead of \"lower\" or \"earlier\").\n* Added/updated instructions.\n",
"_____no_output_____"
],
[
"This assignment will be done in Keras. \n\nBefore jumping into the problem, let's run the cell below to load the required packages.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom keras import layers\nfrom keras.layers import Input, Add, Dense, Activation, ZeroPadding2D, BatchNormalization, Flatten, Conv2D, AveragePooling2D, MaxPooling2D, GlobalMaxPooling2D\nfrom keras.models import Model, load_model\nfrom keras.preprocessing import image\nfrom keras.utils import layer_utils\nfrom keras.utils.data_utils import get_file\nfrom keras.applications.imagenet_utils import preprocess_input\nimport pydot\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\nfrom keras.utils import plot_model\nfrom resnets_utils import *\nfrom keras.initializers import glorot_uniform\nimport scipy.misc\nfrom matplotlib.pyplot import imshow\n%matplotlib inline\n\nimport keras.backend as K\nK.set_image_data_format('channels_last')\nK.set_learning_phase(1)",
"Using TensorFlow backend.\n"
]
],
[
[
"## 1 - The problem of very deep neural networks\n\nLast week, you built your first convolutional neural network. In recent years, neural networks have become deeper, with state-of-the-art networks going from just a few layers (e.g., AlexNet) to over a hundred layers.\n\n* The main benefit of a very deep network is that it can represent very complex functions. It can also learn features at many different levels of abstraction, from edges (at the shallower layers, closer to the input) to very complex features (at the deeper layers, closer to the output). \n* However, using a deeper network doesn't always help. A huge barrier to training them is vanishing gradients: very deep networks often have a gradient signal that goes to zero quickly, thus making gradient descent prohibitively slow. \n* More specifically, during gradient descent, as you backprop from the final layer back to the first layer, you are multiplying by the weight matrix on each step, and thus the gradient can decrease exponentially quickly to zero (or, in rare cases, grow exponentially quickly and \"explode\" to take very large values). \n* During training, you might therefore see the magnitude (or norm) of the gradient for the shallower layers decrease to zero very rapidly as training proceeds: ",
"_____no_output_____"
],
[
"<img src=\"images/vanishing_grad_kiank.png\" style=\"width:450px;height:220px;\">\n<caption><center> <u> <font color='purple'> **Figure 1** </u><font color='purple'> : **Vanishing gradient** <br> The speed of learning decreases very rapidly for the shallower layers as the network trains </center></caption>\n\nYou are now going to solve this problem by building a Residual Network!",
"_____no_output_____"
],
[
"## 2 - Building a Residual Network\n\nIn ResNets, a \"shortcut\" or a \"skip connection\" allows the model to skip layers: \n\n<img src=\"images/skip_connection_kiank.png\" style=\"width:650px;height:200px;\">\n<caption><center> <u> <font color='purple'> **Figure 2** </u><font color='purple'> : A ResNet block showing a **skip-connection** <br> </center></caption>\n\nThe image on the left shows the \"main path\" through the network. The image on the right adds a shortcut to the main path. By stacking these ResNet blocks on top of each other, you can form a very deep network. \n\nWe also saw in lecture that having ResNet blocks with the shortcut also makes it very easy for one of the blocks to learn an identity function. This means that you can stack on additional ResNet blocks with little risk of harming training set performance. \n \n(There is also some evidence that the ease of learning an identity function accounts for ResNets' remarkable performance even more so than skip connections helping with vanishing gradients).\n\nTwo main types of blocks are used in a ResNet, depending mainly on whether the input/output dimensions are same or different. You are going to implement both of them: the \"identity block\" and the \"convolutional block.\"",
"_____no_output_____"
],
[
"### 2.1 - The identity block\n\nThe identity block is the standard block used in ResNets, and corresponds to the case where the input activation (say $a^{[l]}$) has the same dimension as the output activation (say $a^{[l+2]}$). To flesh out the different steps of what happens in a ResNet's identity block, here is an alternative diagram showing the individual steps:\n\n<img src=\"images/idblock2_kiank.png\" style=\"width:650px;height:150px;\">\n<caption><center> <u> <font color='purple'> **Figure 3** </u><font color='purple'> : **Identity block.** Skip connection \"skips over\" 2 layers. </center></caption>\n\nThe upper path is the \"shortcut path.\" The lower path is the \"main path.\" In this diagram, we have also made explicit the CONV2D and ReLU steps in each layer. To speed up training we have also added a BatchNorm step. Don't worry about this being complicated to implement--you'll see that BatchNorm is just one line of code in Keras! \n\nIn this exercise, you'll actually implement a slightly more powerful version of this identity block, in which the skip connection \"skips over\" 3 hidden layers rather than 2 layers. It looks like this: \n\n<img src=\"images/idblock3_kiank.png\" style=\"width:650px;height:150px;\">\n<caption><center> <u> <font color='purple'> **Figure 4** </u><font color='purple'> : **Identity block.** Skip connection \"skips over\" 3 layers.</center></caption>",
"_____no_output_____"
],
[
"Here are the individual steps.\n\nFirst component of main path: \n- The first CONV2D has $F_1$ filters of shape (1,1) and a stride of (1,1). Its padding is \"valid\" and its name should be `conv_name_base + '2a'`. Use 0 as the seed for the random initialization. \n- The first BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2a'`.\n- Then apply the ReLU activation function. This has no name and no hyperparameters. \n\nSecond component of main path:\n- The second CONV2D has $F_2$ filters of shape $(f,f)$ and a stride of (1,1). Its padding is \"same\" and its name should be `conv_name_base + '2b'`. Use 0 as the seed for the random initialization. \n- The second BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2b'`.\n- Then apply the ReLU activation function. This has no name and no hyperparameters. \n\nThird component of main path:\n- The third CONV2D has $F_3$ filters of shape (1,1) and a stride of (1,1). Its padding is \"valid\" and its name should be `conv_name_base + '2c'`. Use 0 as the seed for the random initialization. \n- The third BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2c'`. \n- Note that there is **no** ReLU activation function in this component. \n\nFinal step: \n- The `X_shortcut` and the output from the 3rd layer `X` are added together.\n- **Hint**: The syntax will look something like `Add()([var1,var2])`\n- Then apply the ReLU activation function. This has no name and no hyperparameters. \n\n**Exercise**: Implement the ResNet identity block. We have implemented the first component of the main path. Please read this carefully to make sure you understand what it is doing. You should implement the rest. \n- To implement the Conv2D step: [Conv2D](https://keras.io/layers/convolutional/#conv2d)\n- To implement BatchNorm: [BatchNormalization](https://faroit.github.io/keras-docs/1.2.2/layers/normalization/) (axis: Integer, the axis that should be normalized (typically the 'channels' axis))\n- For the activation, use: `Activation('relu')(X)`\n- To add the value passed forward by the shortcut: [Add](https://keras.io/layers/merge/#add)",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: identity_block\n\ndef identity_block(X, f, filters, stage, block):\n \"\"\"\n Implementation of the identity block as defined in Figure 4\n \n Arguments:\n X -- input tensor of shape (m, n_H_prev, n_W_prev, n_C_prev)\n f -- integer, specifying the shape of the middle CONV's window for the main path\n filters -- python list of integers, defining the number of filters in the CONV layers of the main path\n stage -- integer, used to name the layers, depending on their position in the network\n block -- string/character, used to name the layers, depending on their position in the network\n \n Returns:\n X -- output of the identity block, tensor of shape (n_H, n_W, n_C)\n \"\"\"\n \n # defining name basis\n conv_name_base = 'res' + str(stage) + block + '_branch'\n bn_name_base = 'bn' + str(stage) + block + '_branch'\n \n # Retrieve Filters\n F1, F2, F3 = filters\n \n # Save the input value. You'll need this later to add back to the main path. \n X_shortcut = X\n \n # First component of main path\n X = Conv2D(filters = F1, kernel_size = (1, 1), strides = (1,1), padding = 'valid', name = conv_name_base + '2a', kernel_initializer = glorot_uniform(seed=0))(X)\n X = BatchNormalization(axis = 3, name = bn_name_base + '2a')(X)\n X = Activation('relu')(X)\n \n ### START CODE HERE ###\n \n # Second component of main path (≈3 lines)\n X = Conv2D(filters = F2, kernel_size = (f, f), strides = (1,1), padding = 'same', name = conv_name_base + '2b', kernel_initializer = glorot_uniform(seed=0))(X)\n X = BatchNormalization(axis = 3, name = bn_name_base + '2b')(X)\n X = Activation('relu')(X)\n\n # Third component of main path (≈2 lines)\n X = Conv2D(filters = F3, kernel_size = (1, 1), strides = (1,1), padding = 'valid', name = conv_name_base + '2c', kernel_initializer = glorot_uniform(seed=0))(X)\n X = BatchNormalization(axis = 3, name = bn_name_base + '2c')(X)\n\n # Final step: Add shortcut value to main path, and pass it through a RELU activation (≈2 lines)\n R = Add()([X_shortcut,X])\n X = Activation('relu')(R)\n \n ### END CODE HERE ###\n \n return X",
"_____no_output_____"
],
[
"tf.reset_default_graph()\n\nwith tf.Session() as test:\n np.random.seed(1)\n A_prev = tf.placeholder(\"float\", [3, 4, 4, 6])\n X = np.random.randn(3, 4, 4, 6)\n A = identity_block(A_prev, f = 2, filters = [2, 4, 6], stage = 1, block = 'a')\n test.run(tf.global_variables_initializer())\n out = test.run([A], feed_dict={A_prev: X, K.learning_phase(): 0})\n print(\"out = \" + str(out[0][1][1][0]))",
"out = [ 0.94822985 0. 1.16101444 2.747859 0. 1.36677003]\n"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **out**\n </td>\n <td>\n [ 0.94822985 0. 1.16101444 2.747859 0. 1.36677003]\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"## 2.2 - The convolutional block\n\nThe ResNet \"convolutional block\" is the second block type. You can use this type of block when the input and output dimensions don't match up. The difference with the identity block is that there is a CONV2D layer in the shortcut path: \n\n<img src=\"images/convblock_kiank.png\" style=\"width:650px;height:150px;\">\n<caption><center> <u> <font color='purple'> **Figure 4** </u><font color='purple'> : **Convolutional block** </center></caption>\n\n* The CONV2D layer in the shortcut path is used to resize the input $x$ to a different dimension, so that the dimensions match up in the final addition needed to add the shortcut value back to the main path. (This plays a similar role as the matrix $W_s$ discussed in lecture.) \n* For example, to reduce the activation dimensions's height and width by a factor of 2, you can use a 1x1 convolution with a stride of 2. \n* The CONV2D layer on the shortcut path does not use any non-linear activation function. Its main role is to just apply a (learned) linear function that reduces the dimension of the input, so that the dimensions match up for the later addition step. \n\nThe details of the convolutional block are as follows. \n\nFirst component of main path:\n- The first CONV2D has $F_1$ filters of shape (1,1) and a stride of (s,s). Its padding is \"valid\" and its name should be `conv_name_base + '2a'`. Use 0 as the `glorot_uniform` seed.\n- The first BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2a'`.\n- Then apply the ReLU activation function. This has no name and no hyperparameters. \n\nSecond component of main path:\n- The second CONV2D has $F_2$ filters of shape (f,f) and a stride of (1,1). Its padding is \"same\" and it's name should be `conv_name_base + '2b'`. Use 0 as the `glorot_uniform` seed.\n- The second BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2b'`.\n- Then apply the ReLU activation function. This has no name and no hyperparameters. \n\nThird component of main path:\n- The third CONV2D has $F_3$ filters of shape (1,1) and a stride of (1,1). Its padding is \"valid\" and it's name should be `conv_name_base + '2c'`. Use 0 as the `glorot_uniform` seed.\n- The third BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2c'`. Note that there is no ReLU activation function in this component. \n\nShortcut path:\n- The CONV2D has $F_3$ filters of shape (1,1) and a stride of (s,s). Its padding is \"valid\" and its name should be `conv_name_base + '1'`. Use 0 as the `glorot_uniform` seed.\n- The BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '1'`. \n\nFinal step: \n- The shortcut and the main path values are added together.\n- Then apply the ReLU activation function. This has no name and no hyperparameters. \n \n**Exercise**: Implement the convolutional block. We have implemented the first component of the main path; you should implement the rest. As before, always use 0 as the seed for the random initialization, to ensure consistency with our grader.\n- [Conv2D](https://keras.io/layers/convolutional/#conv2d)\n- [BatchNormalization](https://keras.io/layers/normalization/#batchnormalization) (axis: Integer, the axis that should be normalized (typically the features axis))\n- For the activation, use: `Activation('relu')(X)`\n- [Add](https://keras.io/layers/merge/#add)",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: convolutional_block\n\ndef convolutional_block(X, f, filters, stage, block, s = 2):\n \"\"\"\n Implementation of the convolutional block as defined in Figure 4\n \n Arguments:\n X -- input tensor of shape (m, n_H_prev, n_W_prev, n_C_prev)\n f -- integer, specifying the shape of the middle CONV's window for the main path\n filters -- python list of integers, defining the number of filters in the CONV layers of the main path\n stage -- integer, used to name the layers, depending on their position in the network\n block -- string/character, used to name the layers, depending on their position in the network\n s -- Integer, specifying the stride to be used\n \n Returns:\n X -- output of the convolutional block, tensor of shape (n_H, n_W, n_C)\n \"\"\"\n \n # defining name basis\n conv_name_base = 'res' + str(stage) + block + '_branch'\n bn_name_base = 'bn' + str(stage) + block + '_branch'\n \n # Retrieve Filters\n F1, F2, F3 = filters\n \n # Save the input value\n X_shortcut = X\n\n\n ##### MAIN PATH #####\n # First component of main path \n X = Conv2D(F1,kernel_size= (1, 1), strides = (s,s),padding = 'valid', name = conv_name_base + '2a', kernel_initializer = glorot_uniform(seed=0))(X)\n X = BatchNormalization(axis = 3, name = bn_name_base + '2a')(X)\n X = Activation('relu')(X)\n \n ### START CODE HERE ###\n\n # Second component of main path (≈3 lines)\n X = Conv2D(F2,kernel_size= (f, f), strides = (1,1), name = conv_name_base + '2b',padding = 'same', kernel_initializer = glorot_uniform(seed=0))(X)\n X = BatchNormalization(axis = 3, name = bn_name_base + '2b')(X)\n X = Activation('relu')(X)\n\n # Third component of main path (≈2 lines)\n X = Conv2D(F3,kernel_size= (1, 1), strides = (1,1),padding = 'valid', name = conv_name_base + '2c', kernel_initializer = glorot_uniform(seed=0))(X)\n X = BatchNormalization(axis = 3, name = bn_name_base + '2c')(X)\n\n ##### SHORTCUT PATH #### (≈2 lines)\n X_shortcut = Conv2D(F3,kernel_size= (1, 1), strides = (s,s),padding = 'valid', name = conv_name_base + '1', kernel_initializer = glorot_uniform(seed=0))(X_shortcut)\n X_shortcut = BatchNormalization(axis = 3, name = bn_name_base + '1')(X_shortcut)\n\n # Final step: Add shortcut value to main path, and pass it through a RELU activation (≈2 lines)\n X = Add()([X,X_shortcut])\n X = Activation('relu')(X)\n \n ### END CODE HERE ###\n \n return X",
"_____no_output_____"
],
[
"tf.reset_default_graph()\n\nwith tf.Session() as test:\n np.random.seed(1)\n A_prev = tf.placeholder(\"float\", [3, 4, 4, 6])\n X = np.random.randn(3, 4, 4, 6)\n A = convolutional_block(A_prev, f = 2, filters = [2, 4, 6], stage = 1, block = 'a')\n test.run(tf.global_variables_initializer())\n out = test.run([A], feed_dict={A_prev: X, K.learning_phase(): 0})\n print(\"out = \" + str(out[0][1][1][0]))",
"out = [ 0.09018463 1.23489773 0.46822017 0.0367176 0. 0.65516603]\n"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **out**\n </td>\n <td>\n [ 0.09018463 1.23489773 0.46822017 0.0367176 0. 0.65516603]\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"## 3 - Building your first ResNet model (50 layers)\n\nYou now have the necessary blocks to build a very deep ResNet. The following figure describes in detail the architecture of this neural network. \"ID BLOCK\" in the diagram stands for \"Identity block,\" and \"ID BLOCK x3\" means you should stack 3 identity blocks together.\n\n<img src=\"images/resnet_kiank.png\" style=\"width:850px;height:150px;\">\n<caption><center> <u> <font color='purple'> **Figure 5** </u><font color='purple'> : **ResNet-50 model** </center></caption>\n\nThe details of this ResNet-50 model are:\n- Zero-padding pads the input with a pad of (3,3)\n- Stage 1:\n - The 2D Convolution has 64 filters of shape (7,7) and uses a stride of (2,2). Its name is \"conv1\".\n - BatchNorm is applied to the 'channels' axis of the input.\n - MaxPooling uses a (3,3) window and a (2,2) stride.\n- Stage 2:\n - The convolutional block uses three sets of filters of size [64,64,256], \"f\" is 3, \"s\" is 1 and the block is \"a\".\n - The 2 identity blocks use three sets of filters of size [64,64,256], \"f\" is 3 and the blocks are \"b\" and \"c\".\n- Stage 3:\n - The convolutional block uses three sets of filters of size [128,128,512], \"f\" is 3, \"s\" is 2 and the block is \"a\".\n - The 3 identity blocks use three sets of filters of size [128,128,512], \"f\" is 3 and the blocks are \"b\", \"c\" and \"d\".\n- Stage 4:\n - The convolutional block uses three sets of filters of size [256, 256, 1024], \"f\" is 3, \"s\" is 2 and the block is \"a\".\n - The 5 identity blocks use three sets of filters of size [256, 256, 1024], \"f\" is 3 and the blocks are \"b\", \"c\", \"d\", \"e\" and \"f\".\n- Stage 5:\n - The convolutional block uses three sets of filters of size [512, 512, 2048], \"f\" is 3, \"s\" is 2 and the block is \"a\".\n - The 2 identity blocks use three sets of filters of size [512, 512, 2048], \"f\" is 3 and the blocks are \"b\" and \"c\".\n- The 2D Average Pooling uses a window of shape (2,2) and its name is \"avg_pool\".\n- The 'flatten' layer doesn't have any hyperparameters or name.\n- The Fully Connected (Dense) layer reduces its input to the number of classes using a softmax activation. Its name should be `'fc' + str(classes)`.\n\n**Exercise**: Implement the ResNet with 50 layers described in the figure above. We have implemented Stages 1 and 2. Please implement the rest. (The syntax for implementing Stages 3-5 should be quite similar to that of Stage 2.) Make sure you follow the naming convention in the text above. \n\nYou'll need to use this function: \n- Average pooling [see reference](https://keras.io/layers/pooling/#averagepooling2d)\n\nHere are some other functions we used in the code below:\n- Conv2D: [See reference](https://keras.io/layers/convolutional/#conv2d)\n- BatchNorm: [See reference](https://keras.io/layers/normalization/#batchnormalization) (axis: Integer, the axis that should be normalized (typically the features axis))\n- Zero padding: [See reference](https://keras.io/layers/convolutional/#zeropadding2d)\n- Max pooling: [See reference](https://keras.io/layers/pooling/#maxpooling2d)\n- Fully connected layer: [See reference](https://keras.io/layers/core/#dense)\n- Addition: [See reference](https://keras.io/layers/merge/#add)",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: ResNet50\n\ndef ResNet50(input_shape = (64, 64, 3), classes = 6):\n \"\"\"\n Implementation of the popular ResNet50 the following architecture:\n CONV2D -> BATCHNORM -> RELU -> MAXPOOL -> CONVBLOCK -> IDBLOCK*2 -> CONVBLOCK -> IDBLOCK*3\n -> CONVBLOCK -> IDBLOCK*5 -> CONVBLOCK -> IDBLOCK*2 -> AVGPOOL -> TOPLAYER\n\n Arguments:\n input_shape -- shape of the images of the dataset\n classes -- integer, number of classes\n\n Returns:\n model -- a Model() instance in Keras\n \"\"\"\n \n # Define the input as a tensor with shape input_shape\n X_input = Input(input_shape)\n\n \n # Zero-Padding\n X = ZeroPadding2D((3, 3))(X_input)\n \n # Stage 1\n X = Conv2D(64, (7, 7), strides = (2, 2), name = 'conv1', kernel_initializer = glorot_uniform(seed=0))(X)\n X = BatchNormalization(axis = 3, name = 'bn_conv1')(X)\n X = Activation('relu')(X)\n X = MaxPooling2D((3, 3), strides=(2, 2))(X)\n\n # Stage 2\n X = convolutional_block(X, f = 3, filters = [64, 64, 256], stage = 2, block='a', s = 1)\n X = identity_block(X, 3, [64, 64, 256], stage=2, block='b')\n X = identity_block(X, 3, [64, 64, 256], stage=2, block='c')\n\n ### START CODE HERE ###\n #[128,128,512], \"f\" is 3, \"s\" is 2 and the block is \"a\".\n #The 3 identity blocks use three sets of filters of size [128,128,512], \"f\" is 3 and the blocks are \"b\", \"c\" and \"d\".\n # Stage 3 (≈4 lines)\n X = convolutional_block(X,f=3,s=2,filters = [128,128,512],stage = 3,block = 'a')\n X = identity_block(X,3,[128,128,512],stage = 3,block = 'b')\n X = identity_block(X,3,[128,128,512],stage = 3,block = 'c')\n X = identity_block(X,3,[128,128,512],stage = 3,block = 'd')\n #The convolutional block uses three sets of filters of size [256, 256, 1024], \"f\" is 3, \"s\" is 2 and the block is \"a\".\n #The 5 identity blocks use three sets of filters of size [256, 256, 1024], \"f\" is 3 and the blocks are \"b\", \"c\", \"d\", \"e\" and \"f\".\n # Stage 4 (≈6 lines)\n X = convolutional_block(X, f=3, filters=[256, 256, 1024], stage=4, block='a', s=2)\n X = identity_block(X, 3, [256, 256, 1024], stage=4, block='b')\n X = identity_block(X, 3, [256, 256, 1024], stage=4, block='c')\n X = identity_block(X, 3, [256, 256, 1024], stage=4, block='d')\n X = identity_block(X, 3, [256, 256, 1024], stage=4, block='e')\n X = identity_block(X, 3, [256, 256, 1024], stage=4, block='f')\n \n # Stage 5 (≈3 lines)\n X = X = convolutional_block(X, f=3, filters=[512, 512, 2048], stage=5, block='a', s=2)\n X = identity_block(X, 3, [512, 512, 2048], stage=5, block='b')\n X = identity_block(X, 3, [512, 512, 2048], stage=5, block='c')\n\n # AVGPOOL (≈1 line). Use \"X = AveragePooling2D(...)(X)\"\n X = AveragePooling2D(pool_size=(2, 2), padding='same')(X)\n \n ### END CODE HERE ###\n\n # output layer\n X = Flatten()(X)\n X = Dense(classes, activation='softmax', name='fc' + str(classes), kernel_initializer = glorot_uniform(seed=0))(X)\n \n \n # Create model\n model = Model(inputs = X_input, outputs = X, name='ResNet50')\n\n return model",
"_____no_output_____"
]
],
[
[
"Run the following code to build the model's graph. If your implementation is not correct you will know it by checking your accuracy when running `model.fit(...)` below.",
"_____no_output_____"
]
],
[
[
"model = ResNet50(input_shape = (64, 64, 3), classes = 6)",
"_____no_output_____"
]
],
[
[
"As seen in the Keras Tutorial Notebook, prior training a model, you need to configure the learning process by compiling the model.",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"The model is now ready to be trained. The only thing you need is a dataset.",
"_____no_output_____"
],
[
"Let's load the SIGNS Dataset.\n\n<img src=\"images/signs_data_kiank.png\" style=\"width:450px;height:250px;\">\n<caption><center> <u> <font color='purple'> **Figure 6** </u><font color='purple'> : **SIGNS dataset** </center></caption>\n",
"_____no_output_____"
]
],
[
[
"X_train_orig, Y_train_orig, X_test_orig, Y_test_orig, classes = load_dataset()\n\n# Normalize image vectors\nX_train = X_train_orig/255.\nX_test = X_test_orig/255.\n\n# Convert training and test labels to one hot matrices\nY_train = convert_to_one_hot(Y_train_orig, 6).T\nY_test = convert_to_one_hot(Y_test_orig, 6).T\n\nprint (\"number of training examples = \" + str(X_train.shape[0]))\nprint (\"number of test examples = \" + str(X_test.shape[0]))\nprint (\"X_train shape: \" + str(X_train.shape))\nprint (\"Y_train shape: \" + str(Y_train.shape))\nprint (\"X_test shape: \" + str(X_test.shape))\nprint (\"Y_test shape: \" + str(Y_test.shape))",
"number of training examples = 1080\nnumber of test examples = 120\nX_train shape: (1080, 64, 64, 3)\nY_train shape: (1080, 6)\nX_test shape: (120, 64, 64, 3)\nY_test shape: (120, 6)\n"
]
],
[
[
"Run the following cell to train your model on 2 epochs with a batch size of 32. On a CPU it should take you around 5min per epoch. ",
"_____no_output_____"
]
],
[
[
"model.fit(X_train, Y_train, epochs = 2, batch_size = 32)",
"Epoch 1/2\n 256/1080 [======>.......................] - ETA: 183s - loss: 3.5132 - acc: 0.2070"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n ** Epoch 1/2**\n </td>\n <td>\n loss: between 1 and 5, acc: between 0.2 and 0.5, although your results can be different from ours.\n </td>\n </tr>\n <tr>\n <td>\n ** Epoch 2/2**\n </td>\n <td>\n loss: between 1 and 5, acc: between 0.2 and 0.5, you should see your loss decreasing and the accuracy increasing.\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"Let's see how this model (trained on only two epochs) performs on the test set.",
"_____no_output_____"
]
],
[
[
"preds = model.evaluate(X_test, Y_test)\nprint (\"Loss = \" + str(preds[0]))\nprint (\"Test Accuracy = \" + str(preds[1]))",
"_____no_output_____"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **Test Accuracy**\n </td>\n <td>\n between 0.16 and 0.25\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"For the purpose of this assignment, we've asked you to train the model for just two epochs. You can see that it achieves poor performances. Please go ahead and submit your assignment; to check correctness, the online grader will run your code only for a small number of epochs as well.",
"_____no_output_____"
],
[
"After you have finished this official (graded) part of this assignment, you can also optionally train the ResNet for more iterations, if you want. We get a lot better performance when we train for ~20 epochs, but this will take more than an hour when training on a CPU. \n\nUsing a GPU, we've trained our own ResNet50 model's weights on the SIGNS dataset. You can load and run our trained model on the test set in the cells below. It may take ≈1min to load the model.",
"_____no_output_____"
]
],
[
[
"model = load_model('ResNet50.h5') ",
"_____no_output_____"
],
[
"preds = model.evaluate(X_test, Y_test)\nprint (\"Loss = \" + str(preds[0]))\nprint (\"Test Accuracy = \" + str(preds[1]))",
"_____no_output_____"
]
],
[
[
"ResNet50 is a powerful model for image classification when it is trained for an adequate number of iterations. We hope you can use what you've learnt and apply it to your own classification problem to perform state-of-the-art accuracy.\n\nCongratulations on finishing this assignment! You've now implemented a state-of-the-art image classification system! ",
"_____no_output_____"
],
[
"## 4 - Test on your own image (Optional/Ungraded)",
"_____no_output_____"
],
[
"If you wish, you can also take a picture of your own hand and see the output of the model. To do this:\n 1. Click on \"File\" in the upper bar of this notebook, then click \"Open\" to go on your Coursera Hub.\n 2. Add your image to this Jupyter Notebook's directory, in the \"images\" folder\n 3. Write your image's name in the following code\n 4. Run the code and check if the algorithm is right! ",
"_____no_output_____"
]
],
[
[
"img_path = 'images/my_image.jpg'\nimg = image.load_img(img_path, target_size=(64, 64))\nx = image.img_to_array(img)\nx = np.expand_dims(x, axis=0)\nx = x/255.0\nprint('Input image shape:', x.shape)\nmy_image = scipy.misc.imread(img_path)\nimshow(my_image)\nprint(\"class prediction vector [p(0), p(1), p(2), p(3), p(4), p(5)] = \")\nprint(model.predict(x))",
"_____no_output_____"
]
],
[
[
"You can also print a summary of your model by running the following code.",
"_____no_output_____"
]
],
[
[
"model.summary()",
"_____no_output_____"
]
],
[
[
"Finally, run the code below to visualize your ResNet50. You can also download a .png picture of your model by going to \"File -> Open...-> model.png\".",
"_____no_output_____"
]
],
[
[
"plot_model(model, to_file='model.png')\nSVG(model_to_dot(model).create(prog='dot', format='svg'))",
"_____no_output_____"
]
],
[
[
"## What you should remember\n- Very deep \"plain\" networks don't work in practice because they are hard to train due to vanishing gradients. \n- The skip-connections help to address the Vanishing Gradient problem. They also make it easy for a ResNet block to learn an identity function. \n- There are two main types of blocks: The identity block and the convolutional block. \n- Very deep Residual Networks are built by stacking these blocks together.",
"_____no_output_____"
],
[
"### References \n\nThis notebook presents the ResNet algorithm due to He et al. (2015). The implementation here also took significant inspiration and follows the structure given in the GitHub repository of Francois Chollet: \n\n- Kaiming He, Xiangyu Zhang, Shaoqing Ren, Jian Sun - [Deep Residual Learning for Image Recognition (2015)](https://arxiv.org/abs/1512.03385)\n- Francois Chollet's GitHub repository: https://github.com/fchollet/deep-learning-models/blob/master/resnet50.py\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb4f5e373942b4dd98120c06b1e11e89383276bb | 208,781 | ipynb | Jupyter Notebook | Regression/Support Vector Machine/NuSVR_StandardScaler_QuantileTransformer.ipynb | mohityogesh44/ds-seed | e124f0078faf97568951e19e4302451ad0c7cf6c | [
"Apache-2.0"
] | null | null | null | Regression/Support Vector Machine/NuSVR_StandardScaler_QuantileTransformer.ipynb | mohityogesh44/ds-seed | e124f0078faf97568951e19e4302451ad0c7cf6c | [
"Apache-2.0"
] | null | null | null | Regression/Support Vector Machine/NuSVR_StandardScaler_QuantileTransformer.ipynb | mohityogesh44/ds-seed | e124f0078faf97568951e19e4302451ad0c7cf6c | [
"Apache-2.0"
] | null | null | null | 83.345709 | 84,342 | 0.727911 | [
[
[
"# Nu-Support Vector Regression with StandardScaler & Quantile Transformer\n",
"_____no_output_____"
],
[
"This Code template is for regression analysis using a Nu-Support Vector Regressor(NuSVR) based on the Support Vector Machine algorithm with Quantile Transformer as Feature Transformation Technique and StandardScaler for Feature Scaling in a pipeline.",
"_____no_output_____"
],
[
"### Required Packages",
"_____no_output_____"
]
],
[
[
"import warnings \nimport numpy as np \nimport pandas as pd \nimport matplotlib.pyplot as plt\nimport seaborn as se \nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.preprocessing import StandardScaler, QuantileTransformer\nfrom sklearn.model_selection import train_test_split \nfrom sklearn.svm import NuSVR \nfrom sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"### Initialization\n\nFilepath of CSV file",
"_____no_output_____"
]
],
[
[
"file_path= \"\"",
"_____no_output_____"
]
],
[
[
"List of features which are required for model training .",
"_____no_output_____"
]
],
[
[
"features =[]",
"_____no_output_____"
]
],
[
[
"Target feature for prediction.",
"_____no_output_____"
]
],
[
[
"target=''",
"_____no_output_____"
]
],
[
[
"### Data Fetching\nPandas is an open-source, BSD-licensed library providing high-performance, easy-to-use data manipulation and data analysis tools.\n\nWe will use panda's library to read the CSV file using its storage path.And we use the head function to display the initial row or entry.",
"_____no_output_____"
]
],
[
[
"df=pd.read_csv(file_path)\ndf.head()",
"_____no_output_____"
]
],
[
[
"### Feature Selections\n\nIt is the process of reducing the number of input variables when developing a predictive model. Used to reduce the number of input variables to both reduce the computational cost of modelling and, in some cases, to improve the performance of the model.\n\nWe will assign all the required input features to X and target/outcome to Y.",
"_____no_output_____"
]
],
[
[
"X=df[features]\nY=df[target]",
"_____no_output_____"
]
],
[
[
"### Data Preprocessing\n\nSince the majority of the machine learning models in the Sklearn library doesn't handle string category data and Null value, we have to explicitly remove or replace null values. The below snippet have functions, which removes the null value if any exists. And convert the string classes data in the datasets by encoding them to integer classes.\n",
"_____no_output_____"
]
],
[
[
"def NullClearner(df):\n if(isinstance(df, pd.Series) and (df.dtype in [\"float64\",\"int64\"])):\n df.fillna(df.mean(),inplace=True)\n return df\n elif(isinstance(df, pd.Series)):\n df.fillna(df.mode()[0],inplace=True)\n return df\n else:return df\ndef EncodeX(df):\n return pd.get_dummies(df)",
"_____no_output_____"
]
],
[
[
"Calling preprocessing functions on the feature and target set.\n",
"_____no_output_____"
]
],
[
[
"x=X.columns.to_list()\nfor i in x:\n X[i]=NullClearner(X[i])\nY=NullClearner(Y)\nX=EncodeX(X)\nX.head()",
"_____no_output_____"
]
],
[
[
"#### Correlation Map\n\nIn order to check the correlation between the features, we will plot a correlation matrix. It is effective in summarizing a large amount of data where the goal is to see patterns.",
"_____no_output_____"
]
],
[
[
"f,ax = plt.subplots(figsize=(18, 18))\nmatrix = np.triu(X.corr())\nse.heatmap(X.corr(), annot=True, linewidths=.5, fmt= '.1f',ax=ax, mask=matrix)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Data Splitting\n\nThe train-test split is a procedure for evaluating the performance of an algorithm. The procedure involves taking a dataset and dividing it into two subsets. The first subset is utilized to fit/train the model. The second subset is used for prediction. The main motive is to estimate the performance of the model on new data.",
"_____no_output_____"
]
],
[
[
"X_train,X_test,y_train,y_test=train_test_split(X,Y,test_size=0.2,random_state=123)",
"_____no_output_____"
]
],
[
[
"### Model\nSupport vector machines (SVMs) are a set of supervised learning methods used for classification, regression and outliers detection.\n\nA Support Vector Machine is a discriminative classifier formally defined by a separating hyperplane. In other terms, for a given known/labelled data points, the SVM outputs an appropriate hyperplane that classifies the inputted new cases based on the hyperplane. In 2-Dimensional space, this hyperplane is a line separating a plane into two segments where each class or group occupied on either side.\n\nHere we will use NuSVR, the NuSVR implementation is based on libsvm. Similar to NuSVC, for regression, uses a parameter nu to control the number of support vectors. However, unlike NuSVC, where nu replaces C, here nu replaces the parameter epsilon of epsilon-SVR. \n#### Model Tuning Parameters\n\n 1. nu : float, default=0.5\n> An upper bound on the fraction of training errors and a lower bound of the fraction of support vectors. Should be in the interval (0, 1]. By default 0.5 will be taken.\n\n 2. C : float, default=1.0\n> Regularization parameter. The strength of the regularization is inversely proportional to C. Must be strictly positive. The penalty is a squared l2 penalty.\n\n 3. kernel : {‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’, ‘precomputed’}, default=’rbf’\n> Specifies the kernel type to be used in the algorithm. It must be one of ‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’, ‘precomputed’ or a callable. If none is given, ‘rbf’ will be used. If a callable is given it is used to pre-compute the kernel matrix from data matrices; that matrix should be an array of shape (n_samples, n_samples).\n\n 4. gamma : {‘scale’, ‘auto’} or float, default=’scale’\n> Gamma is a hyperparameter that we have to set before the training model. Gamma decides how much curvature we want in a decision boundary.\n\n 5. degree : int, default=3\n> Degree of the polynomial kernel function (‘poly’). Ignored by all other kernels.Using degree 1 is similar to using a linear kernel. Also, increasing degree parameter leads to higher training times.\n\n#### Rescaling technique\nStandardize features by removing the mean and scaling to unit variance\n\nThe standard score of a sample x is calculated as:\n\n z = (x - u) / s\n\nwhere u is the mean of the training samples or zero if with_mean=False, and s is the standard deviation of the training samples or one if with_std=False.\n\n\n#### Feature Transformation\n\nTransform features using quantiles information.\n\nThis method transforms the features to follow a uniform or a normal distribution. Therefore, for a given feature, this transformation tends to spread out the most frequent values. It also reduces the impact of (marginal) outliers: this is therefore a robust preprocessing scheme.The transformation is applied on each feature independently.",
"_____no_output_____"
]
],
[
[
"model=make_pipeline(StandardScaler(),QuantileTransformer(),NuSVR())\nmodel.fit(X_train,y_train)",
"_____no_output_____"
]
],
[
[
"#### Model Accuracy\n\nWe will use the trained model to make a prediction on the test set.Then use the predicted value for measuring the accuracy of our model.\n\n> **score**: The **score** function returns the coefficient of determination <code>R<sup>2</sup></code> of the prediction.",
"_____no_output_____"
]
],
[
[
"print(\"Accuracy score {:.2f} %\\n\".format(model.score(X_test,y_test)*100))",
"Accuracy score 93.55 %\n\n"
]
],
[
[
"> **r2_score**: The **r2_score** function computes the percentage variablility explained by our model, either the fraction or the count of correct predictions. \n\n> **mae**: The **mean abosolute error** function calculates the amount of total error(absolute average distance between the real data and the predicted data) by our model. \n\n> **mse**: The **mean squared error** function squares the error(penalizes the model for large errors) by our model. ",
"_____no_output_____"
]
],
[
[
"y_pred=model.predict(X_test)\nprint(\"R2 Score: {:.2f} %\".format(r2_score(y_test,y_pred)*100))\nprint(\"Mean Absolute Error {:.2f}\".format(mean_absolute_error(y_test,y_pred)))\nprint(\"Mean Squared Error {:.2f}\".format(mean_squared_error(y_test,y_pred)))",
"R2 Score: 93.55 %\nMean Absolute Error 3.29\nMean Squared Error 18.66\n"
]
],
[
[
"#### Prediction Plot\n\nFirst, we make use of a scatter plot to plot the actual observations, with x_train on the x-axis and y_train on the y-axis.\nFor the regression line, we will use x_train on the x-axis and then the predictions of the x_train observations on the y-axis.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(14,10))\nplt.plot(range(20),y_test[0:20], color = \"green\")\nplt.plot(range(20),model.predict(X_test[0:20]), color = \"red\")\nplt.legend([\"Actual\", \"prediction\"]) \nplt.title(\"Predicted vs True Value\")\nplt.xlabel(\"Record number\")\nplt.ylabel(target)\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Creator: Vamsi Mukkamala , Github: [Profile](https://github.com/vmc99)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb4f65a10fe82604916fd6470ad7cffe874ba9bd | 2,250 | ipynb | Jupyter Notebook | Image_PreProcessing.ipynb | Harsh120/Sort-Contour-By-Distance | 2b80102da2330a5fb01826c9e6af3ea9e4433694 | [
"MIT"
] | null | null | null | Image_PreProcessing.ipynb | Harsh120/Sort-Contour-By-Distance | 2b80102da2330a5fb01826c9e6af3ea9e4433694 | [
"MIT"
] | null | null | null | Image_PreProcessing.ipynb | Harsh120/Sort-Contour-By-Distance | 2b80102da2330a5fb01826c9e6af3ea9e4433694 | [
"MIT"
] | null | null | null | 22.727273 | 133 | 0.544 | [
[
[
"import numpy as np\nimport cv2\n",
"_____no_output_____"
],
[
"image = cv2.imread('alphabet.jpg')",
"_____no_output_____"
],
[
"gray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)\ngray = 255*(gray < 128).astype(np.uint8) # To invert the text to white\ncoords = cv2.findNonZero(gray) # Find all non-zero points (text)\nx, y, w, h = cv2.boundingRect(coords) # Find minimum spanning bounding box\nrect = image[y:y+h, x:x+w] # Crop the image - note we do this on the original image",
"_____no_output_____"
],
[
"img = cv2.resize(rect, None, fx=1.2, fy=1.2, interpolation=cv2.INTER_CUBIC)\nimg = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\nkernel = np.ones((1, 1), np.uint8)\nimg = cv2.dilate(img, kernel, iterations=1)\nimg = cv2.erode(img, kernel, iterations=1)\nimg = cv2.adaptiveThreshold(cv2.bilateralFilter(img, 9, 75, 75), 255, cv2.ADAPTIVE_THRESH_GAUSSIAN_C, cv2.THRESH_BINARY, 31, 2)",
"_____no_output_____"
],
[
"cv2.imwrite(\"rect.png\", img) # Save the image",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.